hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
d0f213ab9c594050596b4c9ebe35e43c36c066ba | 7,582 | ipynb | Jupyter Notebook | notebook/notes_ICsubbox_profiling.ipynb | changhoonhahn/mNuCosmoMap | 7cd2a8640022c582637d59c891e1787a95b78e32 | [
"MIT"
] | null | null | null | notebook/notes_ICsubbox_profiling.ipynb | changhoonhahn/mNuCosmoMap | 7cd2a8640022c582637d59c891e1787a95b78e32 | [
"MIT"
] | null | null | null | notebook/notes_ICsubbox_profiling.ipynb | changhoonhahn/mNuCosmoMap | 7cd2a8640022c582637d59c891e1787a95b78e32 | [
"MIT"
] | null | null | null | 34.153153 | 128 | 0.502374 | [
[
[
"import os\nimport numpy as np\n\nfrom mnucosmomap import util as UT\nfrom mnucosmomap import catalogs as mNuCats",
"_____no_output_____"
],
[
"%load_ext line_profiler",
"_____no_output_____"
],
[
"fullbox = mNuCats.mNuICs(1, sim='paco')\nx, y, z = fullbox['Position'].T\nvx, vy, vz = fullbox['Velocity'].T\n\nnside = 8\nL_subbox = 1000./float(nside) # L_subbox\nL_res = 1000./512.\nL_halfres = 0.5 * L_res\nN_partside = 512/nside\nN_subbox = (N_partside)**3",
"_____no_output_____"
],
[
"def method1(isubbox): \n i_x = ((isubbox % nside**2) % nside) \n i_y = ((isubbox % nside**2) // nside) \n i_z = (isubbox // nside**2) \n\n xmin = L_subbox * float(i_x) + L_halfres\n xmax = (L_subbox * float(i_x+1) + L_halfres) % 1000.\n ymin = L_subbox * float(i_y) + L_halfres\n ymax = (L_subbox * float(i_y+1) + L_halfres) % 1000.\n zmin = L_subbox * float(i_z) + L_halfres\n zmax = (L_subbox * float(i_z+1) + L_halfres) % 1000.\n if xmin <= xmax: xlim = ((x >= xmin) & (x < xmax))\n else: xlim = ((x >= xmin) | (x < xmax))\n if ymin <= ymax: ylim = ((y >= ymin) & (y < ymax))\n else: ylim = ((y >= ymin) | (y < ymax))\n if zmin <= zmax: zlim = ((z >= zmin) & (z < zmax))\n else: zlim = ((z >= zmin) | (z < zmax))\n in_subbox = (xlim & ylim & zlim)\n assert np.sum(in_subbox) == N_subbox\n\n ID_sub = fullbox['ID'][in_subbox]\n x_subbox = x[in_subbox]\n y_subbox = y[in_subbox]\n z_subbox = z[in_subbox]\n x_sub = (x_subbox - i_x * L_subbox) % 1000.\n y_sub = (y_subbox - i_y * L_subbox) % 1000.\n z_sub = (z_subbox - i_z * L_subbox) % 1000.\n\n vx_subbox = vx[in_subbox]\n vy_subbox = vy[in_subbox]\n vz_subbox = vz[in_subbox]\n subbox_ID = np.zeros((N_partside, N_partside, N_partside))\n subbox_pos = np.zeros((3, N_partside, N_partside, N_partside))\n subbox_vel = np.zeros((3, N_partside, N_partside, N_partside))\n for j_z in range(N_partside): \n #print('j_z = %i , %f < z < %f' % (j_z, L_res* float(j_z) + L_halfres, L_res * float(j_z + 1) + L_halfres))\n zlim_sub = ((z_sub > L_res* float(j_z) + L_halfres) & \n (z_sub < L_res * float(j_z + 1) + L_halfres))\n for j_y in range(N_partside): \n #print('j_y = %i , %f < y < %f' % (j_y, L_res* float(j_y) + L_halfres, L_res * float(j_y + 1) + L_halfres))\n ylim_sub = ((y_sub > L_res * float(j_y) + L_halfres) & \n (y_sub < L_res * float(j_y + 1) + L_halfres))\n #for j_x in range(N_partside): \n j_x_sorted = np.argsort(x_sub[ylim_sub & zlim_sub])\n subbox_ID[:,j_y,j_z] = ID_sub[ylim_sub & zlim_sub][j_x_sorted]\n subbox_pos[0,:,j_y,j_z] = x_subbox[ylim_sub & zlim_sub][j_x_sorted]\n subbox_pos[1,:,j_y,j_z] = y_subbox[ylim_sub & zlim_sub][j_x_sorted]\n subbox_pos[2,:,j_y,j_z] = z_subbox[ylim_sub & zlim_sub][j_x_sorted]\n subbox_vel[0,:,j_y,j_z] = vx_subbox[ylim_sub & zlim_sub][j_x_sorted]\n subbox_vel[1,:,j_y,j_z] = vy_subbox[ylim_sub & zlim_sub][j_x_sorted]\n subbox_vel[2,:,j_y,j_z] = vz_subbox[ylim_sub & zlim_sub][j_x_sorted]\n return None",
"_____no_output_____"
],
[
"def method2(isubbox): \n i_x = ((isubbox % nside**2) % nside) \n i_y = ((isubbox % nside**2) // nside) \n i_z = (isubbox // nside**2) \n\n xmin = L_subbox * float(i_x) + L_halfres\n xmax = (L_subbox * float(i_x+1) + L_halfres) % 1000.\n ymin = L_subbox * float(i_y) + L_halfres\n ymax = (L_subbox * float(i_y+1) + L_halfres) % 1000.\n zmin = L_subbox * float(i_z) + L_halfres\n zmax = (L_subbox * float(i_z+1) + L_halfres) % 1000.\n if xmin <= xmax: xlim = ((x >= xmin) & (x < xmax))\n else: xlim = ((x >= xmin) | (x < xmax))\n if ymin <= ymax: ylim = ((y >= ymin) & (y < ymax))\n else: ylim = ((y >= ymin) | (y < ymax))\n if zmin <= zmax: zlim = ((z >= zmin) & (z < zmax))\n else: zlim = ((z >= zmin) | (z < zmax))\n in_subbox = (xlim & ylim & zlim)\n assert np.sum(in_subbox) == N_subbox\n\n ID_sub = fullbox['ID'][in_subbox]\n x_subbox = x[in_subbox]\n y_subbox = y[in_subbox]\n z_subbox = z[in_subbox]\n x_sub = (x_subbox - i_x * L_subbox) % 1000.\n y_sub = (y_subbox - i_y * L_subbox) % 1000.\n z_sub = (z_subbox - i_z * L_subbox) % 1000.\n\n vx_subbox = vx[in_subbox]\n vy_subbox = vy[in_subbox]\n vz_subbox = vz[in_subbox]\n \n subbox_ID = np.zeros((N_partside, N_partside, N_partside))\n subbox_pos = np.zeros((3, N_partside, N_partside, N_partside))\n subbox_vel = np.zeros((3, N_partside, N_partside, N_partside))\n\n j_x = ((x_sub - L_halfres) // L_res).astype(int) \n j_y = ((y_sub - L_halfres) // L_res).astype(int) \n j_z = ((z_sub - L_halfres) // L_res).astype(int) \n subbox_ID[j_x,j_y,j_z] = ID_sub\n subbox_pos[0,j_x,j_y,j_z] = x_subbox\n subbox_pos[1,j_x,j_y,j_z] = y_subbox\n subbox_pos[2,j_x,j_y,j_z] = z_subbox\n subbox_vel[0,j_x,j_y,j_z] = vx_subbox\n subbox_vel[1,j_x,j_y,j_z] = vy_subbox\n subbox_vel[2,j_x,j_y,j_z] = vz_subbox\n return None",
"_____no_output_____"
],
[
"%timeit -n 3 method1(0)",
"3 loops, best of 3: 6.43 s per loop\n"
],
[
"%timeit -n 3 method2(0)",
"3 loops, best of 3: 2.21 s per loop\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f2141899d0ce537bbe2af18d4535ea703c8228 | 35,061 | ipynb | Jupyter Notebook | Day-2/Task-2/PY0101EN-2-2-Lists.ipynb | reddyprasade/2020-PY-101- | bde564f8a5ef6d2c19bb6322385ef21eb1e5cc75 | [
"CC0-1.0"
] | 1 | 2020-09-16T06:22:06.000Z | 2020-09-16T06:22:06.000Z | Day-2/Task-2/PY0101EN-2-2-Lists.ipynb | reddyprasade/2020-PY-101- | bde564f8a5ef6d2c19bb6322385ef21eb1e5cc75 | [
"CC0-1.0"
] | null | null | null | Day-2/Task-2/PY0101EN-2-2-Lists.ipynb | reddyprasade/2020-PY-101- | bde564f8a5ef6d2c19bb6322385ef21eb1e5cc75 | [
"CC0-1.0"
] | 1 | 2020-10-10T18:40:56.000Z | 2020-10-10T18:40:56.000Z | 24.656118 | 375 | 0.428596 | [
[
[
"<h1>Lists in Python</h1>",
"_____no_output_____"
],
[
"<p><strong>Welcome!</strong> This notebook will teach you about the lists in the Python Programming Language. By the end of this lab, you'll know the basics list operations in Python, including indexing, list operations and copy/clone list.</p> ",
"_____no_output_____"
],
[
"<h2>Table of Contents</h2>\n<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <ul>\n <li>\n <a href=\"#dataset\">About the Dataset</a>\n </li>\n <li>\n <a href=\"#list\">Lists</a>\n <ul>\n <li><a href=\"index\">Indexing</a></li>\n <li><a href=\"content\">List Content</a></li>\n <li><a href=\"op\">List Operations</a></li>\n <li><a href=\"co\">Copy and Clone List</a></li>\n </ul>\n </li>\n <li>\n <a href=\"#quiz\">Quiz on Lists</a>\n </li>\n </ul>\n <p>\n Estimated time needed: <strong>15 min</strong>\n </p>\n</div>\n\n<hr>",
"_____no_output_____"
],
[
"<h2 id=\"#dataset\">About the Dataset</h2>",
"_____no_output_____"
],
[
"Imagine you received album recommendations from your friends and compiled all of the recommandations into a table, with specific information about each album.\n\nThe table has one row for each movie and several columns:\n\n- **artist** - Name of the artist\n- **album** - Name of the album\n- **released_year** - Year the album was released\n- **length_min_sec** - Length of the album (hours,minutes,seconds)\n- **genre** - Genre of the album\n- **music_recording_sales_millions** - Music recording sales (millions in USD) on [SONG://DATABASE](http://www.song-database.com/)\n- **claimed_sales_millions** - Album's claimed sales (millions in USD) on [SONG://DATABASE](http://www.song-database.com/)\n- **date_released** - Date on which the album was released\n- **soundtrack** - Indicates if the album is the movie soundtrack (Y) or (N)\n- **rating_of_friends** - Indicates the rating from your friends from 1 to 10\n<br>\n<br>\n\nThe dataset can be seen below:\n\n<font size=\"1\">\n<table font-size:xx-small style=\"width:70%\">\n <tr>\n <th>Artist</th>\n <th>Album</th> \n <th>Released</th>\n <th>Length</th>\n <th>Genre</th> \n <th>Music recording sales (millions)</th>\n <th>Claimed sales (millions)</th>\n <th>Released</th>\n <th>Soundtrack</th>\n <th>Rating (friends)</th>\n </tr>\n <tr>\n <td>Michael Jackson</td>\n <td>Thriller</td> \n <td>1982</td>\n <td>00:42:19</td>\n <td>Pop, rock, R&B</td>\n <td>46</td>\n <td>65</td>\n <td>30-Nov-82</td>\n <td></td>\n <td>10.0</td>\n </tr>\n <tr>\n <td>AC/DC</td>\n <td>Back in Black</td> \n <td>1980</td>\n <td>00:42:11</td>\n <td>Hard rock</td>\n <td>26.1</td>\n <td>50</td>\n <td>25-Jul-80</td>\n <td></td>\n <td>8.5</td>\n </tr>\n <tr>\n <td>Pink Floyd</td>\n <td>The Dark Side of the Moon</td> \n <td>1973</td>\n <td>00:42:49</td>\n <td>Progressive rock</td>\n <td>24.2</td>\n <td>45</td>\n <td>01-Mar-73</td>\n <td></td>\n <td>9.5</td>\n </tr>\n <tr>\n <td>Whitney Houston</td>\n <td>The Bodyguard</td> \n <td>1992</td>\n <td>00:57:44</td>\n <td>Soundtrack/R&B, soul, pop</td>\n <td>26.1</td>\n <td>50</td>\n <td>25-Jul-80</td>\n <td>Y</td>\n <td>7.0</td>\n </tr>\n <tr>\n <td>Meat Loaf</td>\n <td>Bat Out of Hell</td> \n <td>1977</td>\n <td>00:46:33</td>\n <td>Hard rock, progressive rock</td>\n <td>20.6</td>\n <td>43</td>\n <td>21-Oct-77</td>\n <td></td>\n <td>7.0</td>\n </tr>\n <tr>\n <td>Eagles</td>\n <td>Their Greatest Hits (1971-1975)</td> \n <td>1976</td>\n <td>00:43:08</td>\n <td>Rock, soft rock, folk rock</td>\n <td>32.2</td>\n <td>42</td>\n <td>17-Feb-76</td>\n <td></td>\n <td>9.5</td>\n </tr>\n <tr>\n <td>Bee Gees</td>\n <td>Saturday Night Fever</td> \n <td>1977</td>\n <td>1:15:54</td>\n <td>Disco</td>\n <td>20.6</td>\n <td>40</td>\n <td>15-Nov-77</td>\n <td>Y</td>\n <td>9.0</td>\n </tr>\n <tr>\n <td>Fleetwood Mac</td>\n <td>Rumours</td> \n <td>1977</td>\n <td>00:40:01</td>\n <td>Soft rock</td>\n <td>27.9</td>\n <td>40</td>\n <td>04-Feb-77</td>\n <td></td>\n <td>9.5</td>\n </tr>\n</table></font>",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"<h2 id=\"list\">Lists</h2>",
"_____no_output_____"
],
[
"We are going to take a look at lists in Python. \n* A list is a sequenced collection of different objects such as integers, strings, Bool, Float,complex and other lists as well. \n* The address of each element within a list is called an <b>index</b>.\n* An index is used to access and refer to Element/items within a list.\n* List will allow us to Perfrom `index`,`Slice`,`Extended Slice` and we asign a Element to it as well.\n* List is Mutable(Which we can Change at any time), We Can Add, Delete, modify the Element.\n* List is having different Methods ",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/ListsIndex.png\" width=\"1000\" />",
"_____no_output_____"
],
[
" To create a list, type the list within square brackets <b>[ ]</b>, with your content inside the parenthesis and separated by commas. Let’s try it!",
"_____no_output_____"
]
],
[
[
"# Create a list\n\nL = [\"Michael Jackson\", 10.1, 1982]\nL",
"_____no_output_____"
]
],
[
[
"We can use negative and regular indexing with a list :",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/ListsNeg.png\" width=\"1000\" />",
"_____no_output_____"
]
],
[
[
"L[0],L[-3]",
"_____no_output_____"
],
[
"# Print the elements on each index\n\nprint('the same element using negative and positive indexing:\\n Postive:',L[0],\n'\\n Negative:' , L[-3] )\nprint('the same element using negative and positive indexing:\\n Postive:',L[1],\n'\\n Negative:' , L[-2] )\nprint('the same element using negative and positive indexing:\\n Postive:',L[2],\n'\\n Negative:' , L[-1] )",
"_____no_output_____"
],
[
"# Print the elements on each slice\nL[0:2] # Start and End-1-->Slice",
"_____no_output_____"
],
[
"L[1:]",
"_____no_output_____"
]
],
[
[
"<h3 id=\"content\">List Content</h3>",
"_____no_output_____"
],
[
"Lists can contain strings, floats, and integers. We can nest other lists, and we can also nest tuples and other data structures. The same indexing conventions apply for nesting: \n",
"_____no_output_____"
]
],
[
[
"# Sample List\n\nSample_list = [\"Michael Jackson\", 10.1, 1982,2j+3,True ,[1, 2], (\"A\", 1)]\nSample_list",
"_____no_output_____"
],
[
"Sample_list[1],Sample_list[-6]",
"_____no_output_____"
],
[
"Sample_list[2]",
"_____no_output_____"
],
[
"Sample_list[0:5]",
"_____no_output_____"
],
[
"Sample_list[-5:-1]",
"_____no_output_____"
]
],
[
[
"<h3 id=\"op\">List Operations</h3>",
"_____no_output_____"
],
[
" We can also perform slicing in lists. For example, if we want the last two elements, we use the following command:",
"_____no_output_____"
]
],
[
[
"# Sample List\n\nL = [\"Michael Jackson\", 10.1,1982,\"MJ\",1]\nL",
"_____no_output_____"
]
],
[
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/ListsSlice.png\" width=\"1000\">",
"_____no_output_____"
]
],
[
[
"# List slicing\n\nL[3:5]",
"_____no_output_____"
]
],
[
[
"We can use the method <code>extend</code> to add new elements to the list will be add at last:",
"_____no_output_____"
]
],
[
[
"# Use extend to add elements to list\n\nL = [ \"Michael Jackson\", 10.2]\nL.extend(['pop', 10])\nL",
"_____no_output_____"
]
],
[
[
"Another similar method is <code>append</code>. If we apply <code>append</code> instead of <code>extend</code>, we add one element to the list:",
"_____no_output_____"
]
],
[
[
"# Use append to add elements to list\n\nL = [ \"Michael Jackson\", 10.2]\nL.append(['pop', 10])\nL",
"_____no_output_____"
]
],
[
[
" Each time we apply a method, the list changes. If we apply <code>extend</code> we add two new elements to the list. The list <code>L</code> is then modified by adding two new elements:",
"_____no_output_____"
]
],
[
[
"# Use extend to add elements to list\n\nL = [ \"Michael Jackson\", 10.2]\nL.extend(['pop', 10])\nL",
"_____no_output_____"
]
],
[
[
"If we append the list <code>['a','b']</code> we have one new element consisting of a nested list:",
"_____no_output_____"
]
],
[
[
"# Use append to add elements to list\n\nL.append(['a','b'])\nL",
"_____no_output_____"
]
],
[
[
"As lists are mutable, we can change them. For example, we can change the first element as follows:",
"_____no_output_____"
]
],
[
[
"# Change the element based on the index\n\nA = [\"disco\", 10, 1.2]\nprint('Before change:', A)",
"_____no_output_____"
],
[
"A[0]",
"_____no_output_____"
],
[
"A[0] = 'hard rock' # Mutable \nprint('After change:', A)",
"_____no_output_____"
]
],
[
[
" We can also delete an element of a list using the <code>del</code> command:",
"_____no_output_____"
]
],
[
[
"# Delete the element based on the index\n\nprint('Before change:', A)\ndel(A[0])",
"_____no_output_____"
],
[
"print('After change:', A)",
"_____no_output_____"
]
],
[
[
"We can convert a string to a list using <code>split</code>. For example, the method <code>split</code> translates every group of characters separated by a space into an element in a list:",
"_____no_output_____"
]
],
[
[
"# Split the string, default is by space\n\n'hard rock'.split()",
"_____no_output_____"
]
],
[
[
"We can use the split function to separate strings on a specific character. We pass the character we would like to split on into the argument, which in this case is a comma. The result is a list, and each element corresponds to a set of characters that have been separated by a comma: ",
"_____no_output_____"
]
],
[
[
"# Split the string by comma\n\n'A,B,C,D'.split(',')",
"_____no_output_____"
]
],
[
[
"<h3 id=\"co\">Copy and Clone List</h3>",
"_____no_output_____"
],
[
"When we set one variable <b>B</b> equal to <b>A</b>; both <b>A</b> and <b>B</b> are referencing the same list in memory:",
"_____no_output_____"
]
],
[
[
"# Copy (copy by reference) the list A\n\nA = [\"hard rock\", 10, 1.2]\nB = A # copy by reference\nprint('A:', A)\nprint('B:', B)",
"_____no_output_____"
]
],
[
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/ListsRef.png\" width=\"1000\" align=\"center\">",
"_____no_output_____"
]
],
[
[
"id(A)",
"_____no_output_____"
],
[
"id(B)",
"_____no_output_____"
]
],
[
[
"Initially, the value of the first element in <b>B</b> is set as hard rock. If we change the first element in <b>A</b> to <b>banana</b>, we get an unexpected side effect. As <b>A</b> and <b>B</b> are referencing the same list, if we change list <b>A</b>, then list <b>B</b> also changes. If we check the first element of <b>B</b> we get banana instead of hard rock:",
"_____no_output_____"
]
],
[
[
"# Examine the copy by reference\n\nprint('B[0]:', B[0])",
"_____no_output_____"
],
[
"A[0] = \"banana\"",
"_____no_output_____"
],
[
"A",
"_____no_output_____"
],
[
"print('B[0]:', B[0])",
"_____no_output_____"
],
[
"B",
"_____no_output_____"
]
],
[
[
"This is demonstrated in the following figure: ",
"_____no_output_____"
],
[
"<img src = \"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/ListsRefGif.gif\" width=\"1000\" />",
"_____no_output_____"
],
[
"You can clone list **A** by using the following syntax:",
"_____no_output_____"
]
],
[
[
"# Clone (clone by value) the list A\n\nB = A[:]\nB",
"_____no_output_____"
]
],
[
[
" Variable **B** references a new copy or clone of the original list; this is demonstrated in the following figure:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/ListsVal.gif\" width=\"1000\" />",
"_____no_output_____"
],
[
"Now if you change <b>A</b>, <b>B</b> will not change: ",
"_____no_output_____"
]
],
[
[
"print('B[0]:', B[0])\nA[0] = \"hard rock\"\nprint('B[0]:', B[0])",
"_____no_output_____"
],
[
"A",
"_____no_output_____"
],
[
"B",
"_____no_output_____"
],
[
"li = list(range(25,40)) # hear 25 is Starting Element and 39 Is Ending Element\nli",
"_____no_output_____"
],
[
"li.append(10.25)",
"_____no_output_____"
],
[
"li",
"_____no_output_____"
],
[
"li.clear()",
"_____no_output_____"
],
[
"li",
"_____no_output_____"
],
[
"li_1 = [10,20,30,'hi','hello',True,2.5]\nli_1",
"_____no_output_____"
],
[
"li_2 = li_1.copy()\nli_2",
"_____no_output_____"
],
[
"li_1",
"_____no_output_____"
],
[
"li_1.append(10)\nli_1",
"_____no_output_____"
],
[
"li_1.count(10)",
"_____no_output_____"
],
[
"li",
"_____no_output_____"
],
[
"li.extend(li_1)",
"_____no_output_____"
],
[
"li",
"_____no_output_____"
],
[
"li_1",
"_____no_output_____"
],
[
"li_2",
"_____no_output_____"
],
[
"co = [10,20,30,40,50] \nco",
"_____no_output_____"
],
[
"co.index(30)",
"_____no_output_____"
],
[
"co[1]",
"_____no_output_____"
],
[
"co.insert(1,\"Hello\")",
"_____no_output_____"
],
[
"co",
"_____no_output_____"
],
[
"co.pop() # it will remove last element",
"_____no_output_____"
],
[
"co.pop(1) # This Is Used Remove 1 position Element",
"_____no_output_____"
],
[
"co",
"_____no_output_____"
],
[
"co.remove('hi')",
"_____no_output_____"
],
[
"co.remove('hello')",
"_____no_output_____"
],
[
"co",
"_____no_output_____"
],
[
"co.reverse()\nco",
"_____no_output_____"
],
[
"li",
"_____no_output_____"
],
[
"li.remove(2.5)",
"_____no_output_____"
],
[
"li.sort()\nli",
"_____no_output_____"
]
],
[
[
"|Methods|Description|\n|--------|----------|\n|**Append()**|it is used to add the element at last in a list|\n|**clear()**| it is used to Clear the all the elemnts in a list|\n|**copy()**| it is used to copy all the elements in to list|\n|**count()**|We are counting perticular element is Reparting in list|\n|**extend()**|Add Multiple Values to the Existing list|\n|**index()**| which used for find the first occurance of element in a list|\n|**pop()**| it used for remove the last element|\n|**pop(postion)**|it is used for remove perticular element|\n|**remove(Element)**|it remove perticular Remove|\n|**reverse()**|it is used for reverse order element|\n|**sort()**|it i will work for the only perticular data type only|",
"_____no_output_____"
],
[
"### Nested List",
"_____no_output_____"
]
],
[
[
"a = [[10,20,30],\n [2.5,3.5,4.5],\n [True,False,True]]",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"a[0]",
"_____no_output_____"
],
[
"a[0][1]",
"_____no_output_____"
],
[
"a[1]",
"_____no_output_____"
],
[
"a[2]=10",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
]
],
[
[
"<h2 id=\"quiz\">Quiz on List</h2>",
"_____no_output_____"
],
[
"Create a list <code>a_list</code>, with the following elements <code>1</code>, <code>hello</code>, <code>[1,2,3]</code> and <code>True</code>. ",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n\n",
"_____no_output_____"
]
],
[
[
"Double-click <b>here</b> for the solution.\n\n<!-- Your answer is below:\na_list = [1, 'hello', [1, 2, 3] , True]\na_list\n-->",
"_____no_output_____"
],
[
"Find the value stored at index 1 of <code>a_list</code>.",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n",
"_____no_output_____"
]
],
[
[
"Double-click <b>here</b> for the solution.\n\n<!-- Your answer is below:\na_list[1]\n-->",
"_____no_output_____"
],
[
"Retrieve the elements stored at index 1, 2 and 3 of <code>a_list</code>.",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n",
"_____no_output_____"
]
],
[
[
"Double-click <b>here</b> for the solution.\n\n<!-- Your answer is below:\na_list[1:4]\n-->",
"_____no_output_____"
],
[
"Concatenate the following lists <code>A = [1, 'a']</code> and <code>B = [2, 1, 'd']</code>:",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n",
"_____no_output_____"
]
],
[
[
"Double-click <b>here</b> for the solution.\n\n<!-- Your answer is below:\nA = [1, 'a'] \nB = [2, 1, 'd']\nA + B\n-->",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0f2153b70593d613a93953da2a3cc7bff189695 | 65,499 | ipynb | Jupyter Notebook | notebooks/03_scrape/06_Goofing.ipynb | jfilter/ptf | 94e76a6f26344ba37c41793523e70ba75863c5c0 | [
"MIT"
] | 10 | 2020-05-20T07:25:21.000Z | 2021-05-20T15:14:55.000Z | notebooks/03_scrape/06_Goofing.ipynb | jfilter/ptf | 94e76a6f26344ba37c41793523e70ba75863c5c0 | [
"MIT"
] | null | null | null | notebooks/03_scrape/06_Goofing.ipynb | jfilter/ptf | 94e76a6f26344ba37c41793523e70ba75863c5c0 | [
"MIT"
] | 2 | 2020-04-28T15:07:41.000Z | 2021-05-06T14:30:01.000Z | 28.293305 | 569 | 0.53381 | [
[
[
"! wget http://corpora.linguistik.uni-erlangen.de/someweta/german_web_social_media_2018-12-21.model -P /mnt/data2/ptf",
"--2019-05-28 11:39:33-- http://corpora.linguistik.uni-erlangen.de/someweta/german_web_social_media_2018-12-21.model\nResolving corpora.linguistik.uni-erlangen.de (corpora.linguistik.uni-erlangen.de)... 131.188.76.25\nConnecting to corpora.linguistik.uni-erlangen.de (corpora.linguistik.uni-erlangen.de)|131.188.76.25|:80... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 114351304 (109M)\nSaving to: ‘/mnt/data2/ptf/german_web_social_media_2018-12-21.model’\n\ngerman_web_social_m 100%[===================>] 109.05M 3.76MB/s in 29s \n\n2019-05-28 11:40:02 (3.75 MB/s) - ‘/mnt/data2/ptf/german_web_social_media_2018-12-21.model’ saved [114351304/114351304]\n\n"
],
[
"from someweta import ASPTagger\n\nmodel = \"/mnt/data2/ptf/german_web_social_media_2018-12-21.model\"\n\n\n# future versions will have sensible default values\nasptagger = ASPTagger(beam_size=5, iterations=10)\nasptagger.load(model)\n\n",
"Ein\tART\nSatz\tNN\nist\tVAFIN\neine\tART\nListe\tNN\nvon\tAPPR\nTokens\tNN\n.\t$.\n\nZeitfliegen\tNN\nmögen\tVMFIN\neinen\tART\nPfeil\tNN\n.\t$.\n\n"
],
[
"sentences = ['Wer dürfen Atommacht sein, wer nicht. Da treffen sich Regierung, Atommacht und Anwärter auf die Bombe.',\n 'Über was werden da verhandeln?',\n 'Die Bombe selbst stehen nicht zur Disposition, für die Atommacht, sondern der Verfügungsanspruch eines Anwärter.',\n 'Der Besitz dieser Bombe verändern die politisch Option eines Staat, und damit auch die militärisch , in der Folge die politisch Option der existierend Atommacht.',\n 'Bereits der Wille zur Bombe werden deshalb von den real Atommacht beaufsichtigen. Diese Macht verhalten sich zum Wille eines ausländisch Souverän wie Polizei. Wer nicht gehorchen werden bestrafen.',\n 'Das können diese Macht, weil diese in der Lage sein ihre Anspruch an das Wohlverhalten anderer Regierung wirtschaftlich und militärisch zu erzwingen.',\n 'Von wegen hier gehen es um den Schutz vor einer militärisch Bedrohung.',\n 'Die Fähigkeit zu atomar Eskalation stehen doch nur für den Angeklagte zur Disposition.',\n 'Was bleiben? Die auch atomar Überlegenheit der selbsternannt Weltpolizist die sich als Helfer der Menschheit feiern lassen.',\n 'Und die Öffentlichkeit? Die finden wie immer alles toll, was die eigen Regierung machen. Auch kritisch; Da haben man sich über den Tisch zeihen lassen. Beweis: Die Aufhebung der Sanktion. Sein das nicht bereits ein einknick der eigen Herr?',\n 'So konstruktiv sein national Opportunismus,',\n 'Die Bombe in \"unseren\" Hand? Aber sicher, wir sein doch die Guter!',\n 'Alle anderen, wenn es so sagen werden im politisch Rundfunk, sein die Böses.',\n '(i.) Sein \"Satoshi Nakamoto\" nicht der Name einer real Person, die den Bitcoin erfinden haben, sondern ein virtuell Nickname. Ob sich dahint eine real Person, eine real Organisation oder ein Computerprogramm verbergen, weiss kein Schwein.',\n '(ii.) Sein Bitcoins nicht \"mathematisch selten\", sondern mit der gegenwärtig verfügbar Computer-Rechenleistung allenfalls mit einig, energetisch sauteuer Registerschiebe-Aufwand in Mikroprozessor auffindbar.',\n 'Ob es Bitcoins im Überfluss geben, sofern das gegenwärtig weltweit Forscher ernährend, physikalisch Konstrukt von Quantencomputer Realität werden, können \"mathematisch\" bis heute weder beweisen, noch widerlegen werden.',\n '(iiien.) Erzeugen Bitcoins realweltlich nichts, sondern reduzieren erwas zuvor sauteuer Erzeugtes.',\n 'Bitcoins sein die elektrisch Heizlüfter unter den Währung.',\n 'Die reduzieren, Sommer wie Winter, aufwendig-geordnet erschaffen, elektrisch Energie zu popelig-ungeordnet Wärmeenergie.',\n 'Bitcoins machen das, was mittels Klimakonferenz reduzieren werden sollen.',\n '(iv.) Eine einzig, mittels Bitcoin-Heizlüfter vorgenommen Transaktion benötigen zur Zeit 215 kWh elektrisch Energie.https://motherboard.vice....',\n 'Ein deutsch Haushalt verbraten ohne Bitcoin im Durchschnitt 3107 kWh, also schlapp 14 Bitcoin-Transaktion, elektrisch Energie pro Jahr.https://www.musterhaushal...',\n 'P.S.:',\n 'Wer wissen mögen, wie die virtuell \"begehrenswert\" Bitcoins \"gebären\" werden, der können sich sehr einfach ein realweltlich Bild davon machen.\"Photo: Life inside of China’s massiv and remote bitcoinen min\"https://qz.com/1026605/ph...',\n 'Die Idee von bitcoin sein doch die Abschaffung gewöhnlich Währung. Das einzig, was man also tun muss, sein den investitionshyp aussitzen, bis cryptowährung zum Standard werden, international, und dann sein es auch egal, ob ein Bitcoin 500.000 Dollar wert sein, oder?^^',\n 'Und wenn der Bitcoin zwingen sein, so teuer zu bleiben, weil eben so viele Depp so viel investieren, wirdsen halt eine anderer Global Währung. Was ich damit sagen wollen: die cryptowährung Bitcoin an sich sein, glauben ich, zum scheit verurteilen, beziehungsweise besitzen nur ein sehr kurz Zeitfenster, in dem sie einem was nützen. Sein halt so‘n spannend Übergangsprodukt',\n 'Bitcoins werden auf Null oder nahe Null fallen.Das werden passieren.',\n 'Schon zweihundern Kommentar. Das zeigen tatsächlich die Expertise der Deutsch. Toll!Dies sein ein Fachgebiet in das man sich mindestens ein Jahr einarbeiten müssen und das drei Stunde täglich. Alles Andere sein Mumpitz. Gelten für den gesamt Kryptomarkt.Viele Akademiker. Nur mal so am Rand.',\n 'Wer damit real Geld machen, haben es verdienen. Wer seins verlieren auch.',\n '\"Derzeit vergehen kein Tag ohne Facebook-Schlagzeile.\".',\n 'Dann lassen es doch einfach!',\n 'Wer entscheiden, was Fake News sein? Herr Kleber? Fake News sein von der Meinungsfreiheit decken.',\n 'Für anonym Account geben es keine Meinungsfreiheit.',\n 'Es sein ein leidig Thema mit diesem Facebook. Das einzig, was man als Einzelner dagegen tun können, sein der Boykott des Netzwerk.',\n 'Ich halten ja Twitter für eine groß Haß- und Fakenewsschleuder als Facebook. Allerdings sein auf Twitter hauptsächlich Politiker, Journalist und \"Aktivist\" unterwegs, während Facebook mehr so etwas für das gemein Volk sein.',\n 'Deshalb werden wohl auch mehr auf Facebook herumhacken, als auf Twitter. Der Pöbel haben ruhig zu sein.',\n 'Die Regierung mögen so gern handlungsfähig erscheinen, die Mitglied und die angeschlossen Medium beeilen sich, täglich neu \"Grausamkeit\" gegen Flüchtling zu verkünden ohne dabei die Kanzlerin und ihr \"Schaff\" weiter zu beschädigen.',\n 'Dabei sein offensichtlich: eine EU-Normalverteilung sein genauso wenig in Sicht wie eine Einigung mit Griechenland oder gar der Türkei.',\n 'In den Syriengespräch haben man sich nicht nur ins moralisch sondern auch ins diplomatisch Abseits manövrieren.',\n 'Die fortgesetzt Unterstützung für das Regime in Kiew und die beständig Wiederholung der dort verkünden Dogma engen die Handlungsoption für eine Einigung mit Russland entscheidend ein.',\n 'Amerika werden nicht helfen sondern erst mal wählen.',\n 'Nein, die Regierung sein nicht handlungsfähig.',\n 'Und so greifen man zu den verblieben Mittel:',\n 'Diffamierung der AfD wie zuvor schon der Pirat.',\n 'Angriff der auf Aussöhnung mit Russland bedachen Kraft.',\n 'Beide haben zuletzt etwas ungeschickt agieren bzw. nicht mit der an Verzweiflung grenzend Aggressivität der Medium hier rechnen.',\n 'Ein Witz- werden so niemals funktionieren, und das wissen die Beteilgten genau! Verzweiflungsreflex der CDU angesichts befürchtet massiv Stimmeneinbruch bei den Wahl im März.',\n 'Ein Witz?',\n 'Oder eher eine wirkungslos \"Beruhigungspille\" für den Wahlpöbel...',\n 'Erst gar nicht reinlassen sein die gut Lösung.',\n 'Das bedeuten 50-70 Milliarde pro Jahr an Beamten- und Materialaufwand, aber vor allem ein Anstieg der Stückgutkosten, da die lean production, Basis des Erfolg der Deutsch Industrie im Wettbewerb mit den Billiglohnland, nicht mit unkalkulierbar Transportzeit klar kommen.',\n 'Im Klartext Wirtschaftskrise. Nun mögen dem Beschäftigungslosen diese weniger schlimm erscheinen als eine Flüchtlingskrise, wir Arbeitenden werden aber ganz gerne unsere Job behalten.',\n 'Ich denken, man sollen es so machen, wie etwa die Israeli oder die Australier.',\n 'Wenn die Heimatstaat ihre Bürger nicht mehr zurück haben wollen, oder der Herkunftstaat unbekannt sein, sollen man in Drittstaat abschieben, mit denen man zu diesem Zweck entsprechend Vertrag machen.',\n 'Vielleicht fallen dem Migrant dann ja noch rechtzeitig sein Heimatland ein oder wo er seine Papier hintun haben, wenn er etwa nach Uganda abschieben werden sollen.',\n 'ich fragen mich, auf welcher Basis werden denn das alles prüfen.',\n 'Wenn einer erkären er sein Syrer, leider ohne Papier, muss das doch irgendwie prüfen werden, ihm stringent Frage stellen werden, zur Mitarbeit veranlassen werden.',\n 'Wenn sich dann rausstellen, er sein kein Syrer, er wollen sich nicht äussern, wo er eigentlich',\n 'herkommen, dann muss man doch den Antrag negativ bescheiden. Wer sein Herkunftsland nicht preisgeben, sich verweigern, wieso haben derjenige überhaupt ein Anrecht auf Asyl ? Wer wollen denn was von wem ?',\n 'Es gehen nicht um \"links\", \"Linkskurs\" oder das Gegenteil.',\n 'Es gehen um Politik für die eigen Bevölkerung.',\n 'Es gehen um Politik für die Deutsch von deutsch Politiker oder um',\n 'keine Politik für die Deutsch von deutsch Politiker.',\n 'Das sein die Alternative.',\n 'Und die SPD haben sich entscheiden.',\n 'Wahlergebnis von Parteivorsitzender im Bereich von 90% oder gar mehr',\n 'sein ein Indiz für stalinistisch Struktur innerhalb einer Partei.',\n 'https://www.youtube.com/w...',\n 'Unser Gottesgeschenk?!?',\n 'Mit Nahles und der jetzig Parteispitze werden die SPD leider den Weg der französisch, niederländisch, österreichisch und italienisch Sozialdemokrat gehen. Alles andere sein eine Überraschung. Die Delegierte können aber zeigen, dass die SPD DIE Demokratiepartei sein und Simone Lange ihre Stimme geben. Nur Mut: Ein Personeller Neuanfang sein alternativlos.',\n 'Ich stimmen Ihnen zu. Aber ich glauben nicht, dass das, was Sie aufzeigen, an einer Persönlichkeit festzumachen sein.',\n 'Insgesamt meinen ich, dass unsere Gesellschaft in einem anderer Fahrwasser denken und fühlen muss. Wir dürfen nicht die Verhältnis aus der Zeit des tief Menschenelends mit heute bei uns vergleichen und deshalb zeitgerecht Lösung finden. Auf dem Weg der Suche müssen gerecht Kompromiss finden werden.',\n 'Der feudalistisch Überfluss und die Zügellosigkeit der Gewinn- und Luxussucht sein die drastisch Gegenwart der Vergangenheit mit allem menschlich Elend weltweit.',\n 'Sein Las Vegas ein Vorbild, in dem Armut und Elend im Dunkele liegen?',\n 'Na bitten, und Söder gehen dann nach Berlin und werden Innenminister in der GroKo und können so sein Talent beim Management von Migration und Terrorbekämpfung mal richtig unter Beweis stellen....',\n 'Das Bild sagen mehr als tausend Wort. Go, Jo!',\n \"Sein sowieso flabbergasted in Anbetracht der Vorstellung, dieser blass Franke sollen ausgerechnet MP in Bayern werden. Dageg sein ja Stephan Weil ne Partymaus. Passt auch überhaupt nicht in die Reihe irgendwie. Bei Söder weißen du immer schon vorher, was er sagen werden und zwar genau wie er's sagen werden. Ein Politroboter vor dem Herr und genauso gucken er da ja auch drein. Also wie immer eigentlich.\",\n 'Herrmann werden doch bei der Bundestagswahl komplett verbrennen. Söder sein kein Thema, wenn dem nicht so sein.',\n 'Mich werden eher interessieren, ob und welche politisch-inhaltlich Differenz es zwischen den Personalie geben.',\n 'Gegenfrage, gehen es in Bayern und seiner Führungskamarilla jemals um Politisch-Inhaltliches?',\n 'Eine sachlich Diskussion sein doch gar nicht erwünscht.Was haben ich denn jetzt schon wieder bös schreiben?',\n 'Dass sein Faschos hassen? Egal wie sie sich verkleiden und unter welchem Banner sie Meinung löschen?',\n 'Meinungsfreiheit nur noch für Journalist, die dann auch mal Falschzitat kommentieren dürfen?',\n 'Gabriel und Merkel schaden dem Ansehen DeutschlandsEntfernt. Bitte äußern Sie sich zum Thema des Artikel. Die Redaktion/cs',\n '`Das Deutschen-Gen...Das Deutschen-Gen scheinen das Gen der Intoleranz zu sein, mit der ein Deutsche seine Meinung gegenüber Anderen in Forum verteidigen.',\n 'Können man tagtäglich bei der ZEIT beobachten.',\n 'Kürzen. Wir bitten Sie, sich in den Kommentar wieder dem Thema des Artikel zuwenden und weit Anmerkung zur Moderation direkt an [email protected] zu richten, damit im Forum eine sachlich Diskussion ermöglichen werden. Die Redaktion/cs',\n 'Liebe - Sarrazin - MitläuferWenn Herr Sarrazin sich zu Haus oder in seiner Kneipe mit seinen \"dämlich Ansicht“ privat äußern - sein das \"unter Meinungsfreiheit\" noch hinnehmen - kein Hahn werden nach ihm krähen. Aber er nutzen seine exponieren Stellung zum Provozieren, um sein Buch möglichst oft zu verkaufen. Das sein nicht o.k. Für diese Provokation muss er entsprechend Kritik aushalten - die er doch so selbstverständlich an anderen üben. Die zahllos Mitläufer hier auf den Kommentarseite sollen nicht \"stellvertretend für ihn\" so beleidigt tun.',\n 'Vergessen Sie nicht, vor ca. 40 Jahr haben wir Deutsch herablassend die Einwanderung von \"dumm Türke\" wünschen, damit die Drecksarbeit machen werden.',\n 'Da finden wir die Niedrigstlohn für Türke o.k. – die kommen ja aus den doof Ecke der Türkei. Wo sein Herr Sarrazin damals, als es besorgt Stimme zu dieser arrogant Einwanderungspolitik geben.',\n 'Dass heute viele Mensch in diesem \"tollen Deutschland\" für Niedrigstlohn arbeiten, auf dem Lohnniveau damalig Einwanderer und noch darunt, sein das eigentlich Problem - und daran sein die \"deutsch Rassegene, wir sein ja was Gute\" ganz erheblich Schuld. Diese doof deutsch Niedriglöhner sein nämlich auch bald die Moor …wie heute die Türke. Das sein die Angst.',\n 'Übrigens: Als „reinrassig Deutsch“ kennen und mögen ich eine ganz Menge (hoch)intelligent, erfolgreich und obendrein auch noch sehr sympathisch Türke aus Region am Marmarameer bis nach Anatolien (wo ja die Doofen wohnen).',\n 'warum?Warum haben sich chinesen, russen, thaisen, italien integrieren?',\n 'Das sein die Frage, die zu diskutieren sein. Doch das wollen die Medium doch gar nicht, wie das wiederholen Löschen dieser Frage bei der ZEIT zeigen.',\n 'MP3 sein doch total Schrot. selbst im Auto. Zum Glück können meine neu Karre jetzt FLAC abspielen, vorher gehen zwar WAV, aber ich müssen extra konvertieren.',\n 'Selb schuld, wer seinen Ohr MP3 antun. FLAC bieten alle Vorteil: Tagging, Komprimierung, keinen Qualitätsverlust.',\n 'MP3´s haben bei gut Quellqualität kaum Qualitätsverlust. Um das dann noch überhaupt zu merken, brauchen man erstens ein sehr gut Gehör und zweitens mindestens ein gut Abspielgerät. Aber das Sie gleich sich ne neu Karre anschaffen, um FlAC zu hören... xD',\n 'Irgendwo gaanz tief unten in den Katakombe der Zeit.de-Redaktion haben jemand jetzt sehr glücklich da er/sie sehr lange darauf warten, dieses Wortspiel im Titel erscheinen...',\n 'Ich haben mir mal die Mühe machen und bei Spotify nach den von ihnen erwähnen Künstler machen.',\n 'Hugo Alfven, Thomas Arne, Carles Baguer, Mily Balakirev, Jiri Antonin Benda, William Sterndal Bennett finden sich alle bei Spotify, was ja klar sagen das solche Dienst nicht nur den Mainstream bedienen mögen.']",
"_____no_output_____"
],
[
"sentences = [s.split() for s in sentences]",
"_____no_output_____"
],
[
"for sentence in sentences:\n tagged_sentence = asptagger.tag_sentence(sentence)\n print(\"\\n\".join([\"\\t\".join(t) for t in tagged_sentence]), \"\\n\", sep=\"\")",
"Wer\tPWS\ndürfen\tVMFIN\nAtommacht\tNN\nsein,\tNE\nwer\tPWS\nnicht.\tPTKNEG\nDa\tADV\ntreffen\tVVFIN\nsich\tPRF\nRegierung,\tADJA\nAtommacht\tNN\nund\tKON\nAnwärter\tNN\nauf\tAPPR\ndie\tART\nBombe.\tNN\n\nÜber\tAPPR\nwas\tPWS\nwerden\tVAFIN\nda\tADV\nverhandeln?\tNN\n\nDie\tART\nBombe\tNN\nselbst\tADV\nstehen\tVVFIN\nnicht\tPTKNEG\nzur\tAPPRART\nDisposition,\tNN\nfür\tAPPR\ndie\tART\nAtommacht,\tNN\nsondern\tKON\nder\tART\nVerfügungsanspruch\tNN\neines\tART\nAnwärter.\tNN\n\nDer\tART\nBesitz\tNN\ndieser\tPDAT\nBombe\tNN\nverändern\tVVFIN\ndie\tART\npolitisch\tADJD\nOption\tNN\neines\tART\nStaat,\tTRUNC\nund\tKON\ndamit\tPAV\nauch\tADV\ndie\tART\nmilitärisch\tADJD\n,\t$,\nin\tAPPR\nder\tPRELS\nFolge\tNN\ndie\tART\npolitisch\tADJD\nOption\tNN\nder\tART\nexistierend\tADJD\nAtommacht.\tNN\n\nBereits\tADV\nder\tART\nWille\tNN\nzur\tAPPRART\nBombe\tNN\nwerden\tVAFIN\ndeshalb\tPAV\nvon\tAPPR\nden\tART\nreal\tADJA\nAtommacht\tNN\nbeaufsichtigen.\tEMOIMG\nDiese\tPDS\nMacht\tNN\nverhalten\tVVFIN\nsich\tPRF\nzum\tAPPRART\nWille\tNN\neines\tART\nausländisch\tADJD\nSouverän\tADJD\nwie\tKOKOM\nPolizei.\tNE\nWer\tPWS\nnicht\tPTKNEG\ngehorchen\tVVINF\nwerden\tVAFIN\nbestrafen.\tNN\n\nDas\tPDS\nkönnen\tVMFIN\ndiese\tPDAT\nMacht,\tNN\nweil\tKOUS\ndiese\tPDS\nin\tAPPR\nder\tART\nLage\tNN\nsein\tVAINF\nihre\tPPOSAT\nAnspruch\tNN\nan\tAPPR\ndas\tART\nWohlverhalten\tNN\nanderer\tADJA\nRegierung\tNN\nwirtschaftlich\tADJD\nund\tKON\nmilitärisch\tADJD\nzu\tAPPR\nerzwingen.\tNE\n\nVon\tAPPR\nwegen\tNN\nhier\tADV\ngehen\tVVFIN\nes\tPPER\num\tAPPR\nden\tART\nSchutz\tNN\nvor\tAPPR\neiner\tART\nmilitärisch\tADJD\nBedrohung.\tNN\n\nDie\tART\nFähigkeit\tNN\nzu\tAPPR\natomar\tADJA\nEskalation\tNN\nstehen\tVVFIN\ndoch\tPTKMA\nnur\tPTKIFG\nfür\tAPPR\nden\tART\nAngeklagte\tNN\nzur\tAPPRART\nDisposition.\tNN\n\nWas\tPWS\nbleiben?\tNE\nDie\tART\nauch\tADV\natomar\tADJA\nÜberlegenheit\tNN\nder\tART\nselbsternannt\tADJA\nWeltpolizist\tNN\ndie\tPRELS\nsich\tPRF\nals\tAPPR\nHelfer\tNN\nder\tART\nMenschheit\tNN\nfeiern\tVVFIN\nlassen.\tNE\n\nUnd\tKON\ndie\tART\nÖffentlichkeit?\tNN\nDie\tART\nfinden\tNN\nwie\tKOKOM\nimmer\tADV\nalles\tPIS\ntoll,\tNN\nwas\tPRELS\ndie\tART\neigen\tADJA\nRegierung\tNN\nmachen.\tADR\nAuch\tADV\nkritisch;\tADR\nDa\tADV\nhaben\tVAFIN\nman\tPIS\nsich\tPRF\nüber\tAPPR\nden\tART\nTisch\tNN\nzeihen\tADJA\nlassen.\tNN\nBeweis:\tKON\nDie\tART\nAufhebung\tNN\nder\tART\nSanktion.\tNN\nSein\tPPOSAT\ndas\tPDS\nnicht\tPTKNEG\nbereits\tADV\nein\tART\neinknick\tNN\nder\tART\neigen\tADJA\nHerr?\tNN\n\nSo\tADV\nkonstruktiv\tADJD\nsein\tVAINF\nnational\tADJD\nOpportunismus,\tNN\n\nDie\tART\nBombe\tNN\nin\tAPPR\n\"unseren\"\tART\nHand?\tNN\nAber\tADV\nsicher,\tADR\nwir\tPPER\nsein\tVAINF\ndoch\tPTKMA\ndie\tART\nGuter!\tNN\n\nAlle\tPIAT\nanderen,\tPIS\nwenn\tKOUS\nes\tPPER\nso\tPTKMA\nsagen\tVVINF\nwerden\tVAFIN\nim\tAPPRART\npolitisch\tADJD\nRundfunk,\tNN\nsein\tPPOSAT\ndie\tART\nBöses.\tNN\n\n(i.)\tVAFIN\nSein\tPPOSAT\n\"Satoshi\tNN\nNakamoto\"\tKON\nnicht\tPTKNEG\nder\tART\nName\tNN\neiner\tART\nreal\tADJA\nPerson,\tNN\ndie\tART\nden\tART\nBitcoin\tNN\nerfinden\tVVINF\nhaben,\tNE\nsondern\tKON\nein\tART\nvirtuell\tADJD\nNickname.\tNN\nOb\tKOUS\nsich\tPRF\ndahint\tVVFIN\neine\tART\nreal\tADJD\nPerson,\tNN\neine\tART\nreal\tADJA\nOrganisation\tNN\noder\tKON\nein\tART\nComputerprogramm\tNN\nverbergen,\tEMOIMG\nweiss\tADV\nkein\tPIAT\nSchwein.\tNN\n\n(ii.)\tHST\nSein\tPPOSAT\nBitcoins\tNN\nnicht\tPTKNEG\n\"mathematisch\tNN\nselten\",\tADJD\nsondern\tKON\nmit\tAPPR\nder\tART\ngegenwärtig\tADJD\nverfügbar\tADJD\nComputer-Rechenleistung\tNN\nallenfalls\tADV\nmit\tAPPR\neinig,\tART\nenergetisch\tADJD\nsauteuer\tADJA\nRegisterschiebe-Aufwand\tNN\nin\tAPPR\nMikroprozessor\tNN\nauffindbar.\tVVPP\n\nOb\tKOUS\nes\tPPER\nBitcoins\tNN\nim\tAPPRART\nÜberfluss\tNN\ngeben,\tADR\nsofern\tKOUS\ndas\tART\ngegenwärtig\tADJD\nweltweit\tADJD\nForscher\tNN\nernährend,\tNE\nphysikalisch\tADJD\nKonstrukt\tNN\nvon\tAPPR\nQuantencomputer\tNN\nRealität\tNN\nwerden,\tTRUNC\nkönnen\tVMFIN\n\"mathematisch\"\tNN\nbis\tAPPR\nheute\tADV\nweder\tKON\nbeweisen,\tTRUNC\nnoch\tKON\nwiderlegen\tVVINF\nwerden.\tNN\n\n(iiien.)\tHST\nErzeugen\tNN\nBitcoins\tNN\nrealweltlich\tADJD\nnichts,\tNN\nsondern\tKON\nreduzieren\tVVINF\nerwas\tVAFIN\nzuvor\tADV\nsauteuer\tADJD\nErzeugtes.\tNE\n\nBitcoins\tNN\nsein\tPPOSAT\ndie\tART\nelektrisch\tADJD\nHeizlüfter\tNN\nunter\tAPPR\nden\tART\nWährung.\tNN\n\nDie\tART\nreduzieren,\tNN\nSommer\tNN\nwie\tKOKOM\nWinter,\tTRUNC\naufwendig-geordnet\tADJA\nerschaffen,\tHST\nelektrisch\tADJD\nEnergie\tNN\nzu\tAPPR\npopelig-ungeordnet\tADJA\nWärmeenergie.\tNN\n\nBitcoins\tNN\nmachen\tVVFIN\ndas,\tNN\nwas\tPWS\nmittels\tAPPR\nKlimakonferenz\tNN\nreduzieren\tVVINF\nwerden\tVAFIN\nsollen.\tPIS\n\n(iv.)\tNE\nEine\tART\neinzig,\tNN\nmittels\tAPPR\nBitcoin-Heizlüfter\tNN\nvorgenommen\tVVPP\nTransaktion\tNN\nbenötigen\tVVFIN\nzur\tAPPRART\nZeit\tNN\n215\tCARD\nkWh\tNN\nelektrisch\tADJD\nEnergie.https://motherboard.vice....\tEMOASC\n\nEin\tART\ndeutsch\tADJA\nHaushalt\tNN\nverbraten\tVVFIN\nohne\tAPPR\nBitcoin\tNE\nim\tAPPRART\nDurchschnitt\tNN\n3107\tCARD\nkWh,\tNN\nalso\tADV\nschlapp\tADJD\n14\tCARD\nBitcoin-Transaktion,\tNN\nelektrisch\tADJD\nEnergie\tNN\npro\tAPPR\nJahr.https://www.musterhaushal...\tNN\n\nP.S.:\tHST\n\nWer\tPWS\nwissen\tVVFIN\nmögen,\tADV\nwie\tKOKOM\ndie\tART\nvirtuell\tADJD\n\"begehrenswert\"\tADJA\nBitcoins\tNN\n\"gebären\"\tART\nwerden,\tTRUNC\nder\tPDS\nkönnen\tVMFIN\nsich\tPRF\nsehr\tPTKIFG\neinfach\tADJD\nein\tART\nrealweltlich\tADJD\nBild\tNN\ndavon\tPAV\nmachen.\"Photo:\tNE\nLife\tFM\ninside\tFM\nof\tFM\nChina’s\tFM\nmassiv\tFM\nand\tFM\nremote\tFM\nbitcoinen\tFM\nmin\"https://qz.com/1026605/ph...\tNN\n\nDie\tART\nIdee\tNN\nvon\tAPPR\nbitcoin\tNE\nsein\tVAINF\ndoch\tPTKMA\ndie\tART\nAbschaffung\tNN\ngewöhnlich\tADJD\nWährung.\tTRUNC\nDas\tART\neinzig,\tNN\nwas\tPRELS\nman\tPIS\nalso\tADV\ntun\tVVINF\nmuss,\tNN\nsein\tVAINF\nden\tART\ninvestitionshyp\tADJA\naussitzen,\tTRUNC\nbis\tAPPR\ncryptowährung\tNN\nzum\tAPPRART\nStandard\tNN\nwerden,\tART\ninternational,\tTRUNC\nund\tKON\ndann\tADV\nsein\tVAINF\nes\tPPER\nauch\tADV\negal,\tADV\nob\tKOUS\nein\tART\nBitcoin\tNE\n500.000\tCARD\nDollar\tNN\nwert\tADJD\nsein,\tNE\noder?^^\tNE\n\nUnd\tKON\nwenn\tKOUS\nder\tART\nBitcoin\tNN\nzwingen\tVVINF\nsein,\tNE\nso\tADV\nteuer\tADJD\nzu\tAPPR\nbleiben,\tNE\nweil\tKOUS\neben\tADV\nso\tADV\nviele\tPIAT\nDepp\tNN\nso\tADV\nviel\tPIAT\ninvestieren,\tNN\nwirdsen\tVVFIN\nhalt\tPTKMA\neine\tART\nanderer\tADJA\nGlobal\tNN\nWährung.\tADR\nWas\tPWS\nich\tPPER\ndamit\tPAV\nsagen\tVVFIN\nwollen:\tNE\ndie\tART\ncryptowährung\tNN\nBitcoin\tNE\nan\tAPPR\nsich\tPRF\nsein,\tNE\nglauben\tVVFIN\nich,\tNN\nzum\tAPPRART\nscheit\tADJA\nverurteilen,\tNN\nbeziehungsweise\tKON\nbesitzen\tVVFIN\nnur\tPTKIFG\nein\tART\nsehr\tPTKIFG\nkurz\tADJD\nZeitfenster,\tNN\nin\tAPPR\ndem\tART\nsie\tPPER\neinem\tART\nwas\tPIAT\nnützen.\tNN\nSein\tPPOSAT\nhalt\tNN\nso‘n\tEMOIMG\nspannend\tADJD\nÜbergangsprodukt\tNN\n\nBitcoins\tNN\nwerden\tVAFIN\nauf\tAPPR\nNull\tCARD\noder\tKON\nnahe\tAPPR\nNull\tCARD\nfallen.Das\tNN\nwerden\tVAFIN\npassieren.\tVVPP\n\nSchon\tADV\nzweihundern\tADR\nKommentar.\tADR\nDas\tART\nzeigen\tVVFIN\ntatsächlich\tADJD\ndie\tART\nExpertise\tNN\nder\tART\nDeutsch.\tNN\nToll!Dies\tNE\nsein\tVAINF\nein\tART\nFachgebiet\tNN\nin\tAPPR\ndas\tPRELS\nman\tPIS\nsich\tPRF\nmindestens\tADV\nein\tART\nJahr\tNN\neinarbeiten\tVVINF\nmüssen\tVMFIN\nund\tKON\ndas\tART\ndrei\tCARD\nStunde\tNN\ntäglich.\tADR\nAlles\tPIAT\nAndere\tPIS\nsein\tPPOSAT\nMumpitz.\tNN\nGelten\tNN\nfür\tAPPR\nden\tART\ngesamt\tADJA\nKryptomarkt.Viele\tNN\nAkademiker.\tEMOIMG\nNur\tPTKIFG\nmal\tPTKMA\nso\tADV\nam\tAPPRART\nRand.\tNN\n\nWer\tPWS\ndamit\tPAV\nreal\tADJD\nGeld\tNN\nmachen,\tNE\nhaben\tVAFIN\nes\tPPER\nverdienen.\tADV\nWer\tPWS\nseins\tNN\nverlieren\tVVFIN\nauch.\tNE\n\n\"Derzeit\tNN\nvergehen\tVVFIN\nkein\tPIAT\nTag\tNN\nohne\tAPPR\nFacebook-Schlagzeile.\".\tNN\n\nDann\tADV\nlassen\tVVFIN\nes\tPPER\ndoch\tPTKMA\neinfach!\tPIS\n\nWer\tPWS\nentscheiden,\tNN\nwas\tPWS\nFake\tNN\nNews\tNN\nsein?\tPPOSAT\nHerr\tNN\nKleber?\tNE\nFake\tNN\nNews\tNN\nsein\tVAINF\nvon\tAPPR\nder\tART\nMeinungsfreiheit\tNN\ndecken.\t$.\n\nFür\tAPPR\nanonym\tADJD\nAccount\tNN\ngeben\tVVFIN\nes\tPPER\nkeine\tPIAT\nMeinungsfreiheit.\tNN\n\nEs\tPPER\nsein\tVAINF\nein\tART\nleidig\tADJA\nThema\tNN\nmit\tAPPR\ndiesem\tPDAT\nFacebook.\tNN\nDas\tART\neinzig,\tNN\nwas\tPRELS\nman\tPIS\nals\tAPPR\nEinzelner\tNN\ndagegen\tPAV\ntun\tVVFIN\nkönnen,\tNE\nsein\tPPOSAT\nder\tART\nBoykott\tNN\ndes\tART\nNetzwerk.\tNN\n\nIch\tPPER\nhalten\tVVFIN\nja\tPTKMA\nTwitter\tNE\nfür\tAPPR\neine\tART\ngroß\tADJD\nHaß-\tTRUNC\nund\tKON\nFakenewsschleuder\tNN\nals\tAPPR\nFacebook.\tTRUNC\nAllerdings\tADV\nsein\tVAINF\nauf\tAPPR\nTwitter\tNE\nhauptsächlich\tADJD\nPolitiker,\tNE\nJournalist\tNN\nund\tKON\n\"Aktivist\"\tTRUNC\nunterwegs,\tHST\nwährend\tAPPR\nFacebook\tNN\nmehr\tADV\nso\tPTKMA\netwas\tPIS\nfür\tAPPR\ndas\tART\ngemein\tADJD\nVolk\tNN\nsein.\tNE\n\nDeshalb\tPAV\nwerden\tVAFIN\nwohl\tPTKMA\nauch\tADV\nmehr\tPIS\nauf\tAPPR\nFacebook\tNE\nherumhacken,\tNE\nals\tKOKOM\nauf\tAPPR\nTwitter.\tNN\nDer\tART\nPöbel\tNN\nhaben\tVAFIN\nruhig\tADJD\nzu\tAPPR\nsein.\tVAINF\n\nDie\tART\nRegierung\tNN\nmögen\tVMFIN\nso\tADV\ngern\tADV\nhandlungsfähig\tADJD\nerscheinen,\tNE\ndie\tART\nMitglied\tNN\nund\tKON\ndie\tART\nangeschlossen\tADJA\nMedium\tNN\nbeeilen\tVVFIN\nsich,\tADR\ntäglich\tADJD\nneu\tADJD\n\"Grausamkeit\"\tNN\ngegen\tAPPR\nFlüchtling\tNN\nzu\tPTKZU\nverkünden\tVVINF\nohne\tAPPR\ndabei\tPAV\ndie\tART\nKanzlerin\tNN\nund\tKON\nihr\tPPOSAT\n\"Schaff\"\tNN\nweiter\tADV\nzu\tAPPR\nbeschädigen.\tNN\n\nDabei\tPAV\nsein\tPPOSAT\noffensichtlich:\tNN\neine\tART\nEU-Normalverteilung\tNN\nsein\tVAINF\ngenauso\tADV\nwenig\tPIS\nin\tAPPR\nSicht\tNN\nwie\tKOKOM\neine\tART\nEinigung\tNN\nmit\tAPPR\nGriechenland\tNE\noder\tKON\ngar\tPTKIFG\nder\tART\nTürkei.\tNN\n\nIn\tAPPR\nden\tART\nSyriengespräch\tNN\nhaben\tVAFIN\nman\tPIS\nsich\tPRF\nnicht\tPTKNEG\nnur\tADV\nins\tAPPRART\nmoralisch\tADJD\nsondern\tKON\nauch\tADV\nins\tAPPRART\ndiplomatisch\tADJD\nAbseits\tAPPR\nmanövrieren.\tNE\n\nDie\tART\nfortgesetzt\tADJA\nUnterstützung\tNN\nfür\tAPPR\ndas\tART\nRegime\tNN\nin\tAPPR\nKiew\tNE\nund\tKON\ndie\tART\nbeständig\tADJD\nWiederholung\tNN\nder\tART\ndort\tADV\nverkünden\tADJA\nDogma\tNN\nengen\tVVFIN\ndie\tART\nHandlungsoption\tNN\nfür\tAPPR\neine\tART\nEinigung\tNN\nmit\tAPPR\nRussland\tNE\nentscheidend\tADJD\nein.\tEMOASC\n\nAmerika\tNE\nwerden\tVAFIN\nnicht\tPTKNEG\nhelfen\tVVINF\nsondern\tKON\nerst\tADV\nmal\tPTKMA\nwählen.\tPIS\n\nNein,\tVVIMP\ndie\tART\nRegierung\tNN\nsein\tVAINF\nnicht\tPTKNEG\nhandlungsfähig.\t$.\n\nUnd\tKON\nso\tADV\ngreifen\tVVFIN\nman\tPIS\nzu\tAPPR\nden\tART\nverblieben\tADJA\nMittel:\tNN\n\nDiffamierung\tNN\nder\tART\nAfD\tNN\nwie\tKOKOM\nzuvor\tADV\nschon\tADV\nder\tART\nPirat.\tNN\n\nAngriff\tNN\nder\tART\nauf\tAPPR\nAussöhnung\tNN\nmit\tAPPR\nRussland\tNE\nbedachen\tADJA\nKraft.\tNN\n\nBeide\tPIS\nhaben\tVAFIN\nzuletzt\tADV\netwas\tPIS\nungeschickt\tADJD\nagieren\tVVFIN\nbzw.\tKON\nnicht\tPTKNEG\nmit\tAPPR\nder\tART\nan\tAPPR\nVerzweiflung\tNN\ngrenzend\tADJD\nAggressivität\tNN\nder\tART\nMedium\tNN\nhier\tADV\nrechnen.\tPIS\n\nEin\tART\nWitz-\tTRUNC\nwerden\tVAFIN\nso\tADV\nniemals\tADV\nfunktionieren,\tNN\nund\tKON\ndas\tART\nwissen\tVVFIN\ndie\tART\nBeteilgten\tNN\ngenau!\tART\nVerzweiflungsreflex\tNN\nder\tART\nCDU\tNE\nangesichts\tAPPR\nbefürchtet\tVVFIN\nmassiv\tADJD\nStimmeneinbruch\tNN\nbei\tAPPR\nden\tART\nWahl\tNN\nim\tAPPRART\nMärz.\tNE\n\nEin\tART\nWitz?\tNN\n\nOder\tKON\neher\tADV\neine\tART\nwirkungslos\tADJD\n\"Beruhigungspille\"\tNN\nfür\tAPPR\nden\tART\nWahlpöbel...\tNN\n\nErst\tADV\ngar\tPTKIFG\nnicht\tPTKNEG\nreinlassen\tVVFIN\nsein\tPPOSAT\ndie\tART\ngut\tADJD\nLösung.\tNN\n\nDas\tPDS\nbedeuten\tVVFIN\n50-70\tCARD\nMilliarde\tNN\npro\tAPPR\nJahr\tNN\nan\tAPPR\nBeamten-\tTRUNC\nund\tKON\nMaterialaufwand,\tNN\naber\tADV\nvor\tAPPR\nallem\tPIS\nein\tART\nAnstieg\tNN\nder\tART\nStückgutkosten,\tNN\nda\tADV\ndie\tART\nlean\tADJA\nproduction,\tNE\nBasis\tNN\ndes\tART\nErfolg\tNN\nder\tART\nDeutsch\tNN\nIndustrie\tNN\nim\tAPPRART\nWettbewerb\tNN\nmit\tAPPR\nden\tART\nBilliglohnland,\tNN\nnicht\tPTKNEG\nmit\tAPPR\nunkalkulierbar\tADJD\nTransportzeit\tNN\nklar\tADJD\nkommen.\tTRUNC\n\nIm\tAPPRART\nKlartext\tNN\nWirtschaftskrise.\tNN\nNun\tADV\nmögen\tVMFIN\ndem\tART\nBeschäftigungslosen\tNN\ndiese\tPDS\nweniger\tPIAT\nschlimm\tADJD\nerscheinen\tVVFIN\nals\tKOKOM\neine\tART\nFlüchtlingskrise,\tNN\nwir\tPPER\nArbeitenden\tNN\nwerden\tVAFIN\naber\tADV\nganz\tPTKIFG\ngerne\tADV\nunsere\tPPOSAT\nJob\tNN\nbehalten.\tNE\n\nIch\tPPER\ndenken,\tVVFIN\nman\tPIS\nsollen\tVMFIN\nes\tPPER\nso\tADV\nmachen,\tTRUNC\nwie\tKOKOM\netwa\tADV\ndie\tART\nIsraeli\tNN\noder\tKON\ndie\tART\nAustralier.\tNN\n\nWenn\tKOUS\ndie\tART\nHeimatstaat\tNN\nihre\tPPOSAT\nBürger\tNN\nnicht\tPTKNEG\nmehr\tPTKMWL\nzurück\tPTKVZ\nhaben\tVAFIN\nwollen,\tTRUNC\noder\tKON\nder\tART\nHerkunftstaat\tNN\nunbekannt\tADJD\nsein,\tADV\nsollen\tVMFIN\nman\tPIS\nin\tAPPR\nDrittstaat\tNN\nabschieben,\tNN\nmit\tAPPR\ndenen\tPDS\nman\tPIS\nzu\tAPPR\ndiesem\tPDAT\nZweck\tNN\nentsprechend\tADJD\nVertrag\tNN\nmachen.\tNE\n\nVielleicht\tADV\nfallen\tVVFIN\ndem\tART\nMigrant\tNN\ndann\tADV\nja\tPTKMA\nnoch\tPTKMWL\nrechtzeitig\tADJD\nsein\tPPOSAT\nHeimatland\tNN\nein\tPTKVZ\noder\tKON\nwo\tPWAV\ner\tPPER\nseine\tPPOSAT\nPapier\tNN\nhintun\tVVFIN\nhaben,\tKOKOM\nwenn\tKOUS\ner\tPPER\netwa\tADV\nnach\tAPPR\nUganda\tNE\nabschieben\tVVINF\nwerden\tVAFIN\nsollen.\tPIS\n\nich\tPPER\nfragen\tVVFIN\nmich,\tPRF\nauf\tAPPR\nwelcher\tPWAT\nBasis\tNN\nwerden\tVAFIN\ndenn\tPTKMA\ndas\tPDS\nalles\tPIS\nprüfen.\tTRUNC\n\nWenn\tKOUS\neiner\tPIS\nerkären\tVVFIN\ner\tPPER\nsein\tPPOSAT\nSyrer,\tNN\nleider\tADV\nohne\tAPPR\nPapier,\tNN\nmuss\tVMFIN\ndas\tPDS\ndoch\tPTKMA\nirgendwie\tADV\nprüfen\tVVINF\nwerden,\tTRUNC\nihm\tPPER\nstringent\tADJD\nFrage\tNN\nstellen\tVVFIN\nwerden,\tNN\nzur\tAPPRART\nMitarbeit\tNN\nveranlassen\tVVFIN\nwerden.\tNE\n\nWenn\tKOUS\nsich\tPRF\ndann\tADV\nrausstellen,\tVVFIN\ner\tPPER\nsein\tPPOSAT\nkein\tPIAT\nSyrer,\tNN\ner\tPPER\nwollen\tVMFIN\nsich\tPRF\nnicht\tPTKNEG\näussern,\tTRUNC\nwo\tPWAV\ner\tPPER\neigentlich\tADJD\n\nherkommen,\tHST\ndann\tADV\nmuss\tVMFIN\nman\tPIS\ndoch\tPTKMA\nden\tART\nAntrag\tNN\nnegativ\tADJD\nbescheiden.\tTRUNC\nWer\tPWS\nsein\tPPOSAT\nHerkunftsland\tNN\nnicht\tPTKNEG\npreisgeben,\tTRUNC\nsich\tPRF\nverweigern,\tADR\nwieso\tPWAV\nhaben\tVAFIN\nderjenige\tPDS\nüberhaupt\tADV\nein\tART\nAnrecht\tNN\nauf\tAPPR\nAsyl\tNN\n?\t$.\nWer\tPWS\nwollen\tVMFIN\ndenn\tPTKMA\nwas\tPIS\nvon\tAPPR\nwem\tPIS\n?\t$.\n\nEs\tPPER\ngehen\tVVFIN\nnicht\tPTKNEG\num\tAPPR\n\"links\",\tART\n\"Linkskurs\"\tNN\noder\tKON\ndas\tART\nGegenteil.\tNN\n\nEs\tPPER\ngehen\tVVFIN\num\tAPPR\nPolitik\tNN\nfür\tAPPR\ndie\tART\neigen\tADJA\nBevölkerung.\tNN\n\nEs\tPPER\ngehen\tVVFIN\num\tAPPR\nPolitik\tNN\nfür\tAPPR\ndie\tART\nDeutsch\tNN\nvon\tAPPR\ndeutsch\tNN\nPolitiker\tNN\noder\tKON\num\tPTKVZ\n\nkeine\tPIAT\nPolitik\tNN\nfür\tAPPR\ndie\tART\nDeutsch\tNN\nvon\tAPPR\ndeutsch\tADJD\nPolitiker.\tNE\n\nDas\tPDS\nsein\tVAFIN\ndie\tART\nAlternative.\tNN\n\nUnd\tKON\ndie\tART\nSPD\tNE\nhaben\tVAFIN\nsich\tPRF\nentscheiden.\tVVPP\n\nWahlergebnis\tNN\nvon\tAPPR\nParteivorsitzender\tNN\nim\tAPPRART\nBereich\tNN\nvon\tAPPR\n90%\tTRUNC\noder\tKON\ngar\tPTKIFG\nmehr\tPIS\n\nsein\tPPOSAT\nein\tART\nIndiz\tNN\nfür\tAPPR\nstalinistisch\tADJA\nStruktur\tNN\ninnerhalb\tAPPR\neiner\tART\nPartei.\tNN\n\nhttps://www.youtube.com/w...\tURL\n\nUnser\tPPOSAT\nGottesgeschenk?!?\tNN\n\nMit\tAPPR\nNahles\tNE\nund\tKON\nder\tART\njetzig\tADJD\nParteispitze\tNN\nwerden\tVAFIN\ndie\tART\nSPD\tNE\nleider\tADV\nden\tART\nWeg\tNN\nder\tART\nfranzösisch,\tNN\nniederländisch,\tEMOIMG\nösterreichisch\tADJD\nund\tKON\nitalienisch\tADJD\nSozialdemokrat\tNN\ngehen.\tNE\nAlles\tPIAT\nandere\tPIS\nsein\tVAINF\neine\tART\nÜberraschung.\tNN\nDie\tART\nDelegierte\tNN\nkönnen\tVMFIN\naber\tADV\nzeigen,\tNN\ndass\tKOUS\ndie\tART\nSPD\tNE\nDIE\tART\nDemokratiepartei\tNN\nsein\tVAINF\nund\tKON\nSimone\tNE\nLange\tNE\nihre\tPPOSAT\nStimme\tNN\ngeben.\tADJD\nNur\tPTKIFG\nMut:\tADR\nEin\tART\nPersoneller\tADJA\nNeuanfang\tNN\nsein\tVAINF\nalternativlos.\tNN\n\nIch\tPPER\nstimmen\tVVFIN\nIhnen\tPPER\nzu.\tPRF\nAber\tADV\nich\tPPER\nglauben\tVVFIN\nnicht,\tPIS\ndass\tKOUS\ndas,\tPDS\nwas\tPWS\nSie\tPPER\naufzeigen,\tADR\nan\tAPPR\neiner\tART\nPersönlichkeit\tNN\nfestzumachen\tVVIZU\nsein.\tTRUNC\n\nInsgesamt\tADV\nmeinen\tPPOSAT\nich,\tNN\ndass\tKOUS\nunsere\tPPOSAT\nGesellschaft\tNN\nin\tAPPR\neinem\tART\nanderer\tADJA\nFahrwasser\tNN\ndenken\tVVFIN\nund\tKON\nfühlen\tVVFIN\nmuss.\tADR\nWir\tPPER\ndürfen\tVMFIN\nnicht\tPTKNEG\ndie\tART\nVerhältnis\tNN\naus\tAPPR\nder\tART\nZeit\tNN\ndes\tART\ntief\tADJA\nMenschenelends\tNN\nmit\tAPPR\nheute\tADV\nbei\tAPPR\nuns\tPPER\nvergleichen\tVVINF\nund\tKON\ndeshalb\tPAV\nzeitgerecht\tVVPPER\nLösung\tNN\nfinden.\tNN\nAuf\tAPPR\ndem\tART\nWeg\tNN\nder\tART\nSuche\tNN\nmüssen\tVMFIN\ngerecht\tADJD\nKompromiss\tNN\nfinden\tVVFIN\nwerden.\tNN\n\nDer\tART\nfeudalistisch\tADJD\nÜberfluss\tNN\nund\tKON\ndie\tART\nZügellosigkeit\tNN\nder\tART\nGewinn-\tTRUNC\nund\tKON\nLuxussucht\tNN\nsein\tVAINF\ndie\tART\ndrastisch\tADJA\nGegenwart\tNN\nder\tART\nVergangenheit\tNN\nmit\tAPPR\nallem\tPIS\nmenschlich\tADJD\nElend\tNN\nweltweit.\tNE\n\nSein\tPPOSAT\nLas\tNN\nVegas\tNE\nein\tART\nVorbild,\tNN\nin\tAPPR\ndem\tART\nArmut\tNN\nund\tKON\nElend\tNN\nim\tAPPRART\nDunkele\tADJA\nliegen?\tNN\n\nNa\tITJ\nbitten,\tNE\nund\tKON\nSöder\tNE\ngehen\tVVFIN\ndann\tADV\nnach\tAPPR\nBerlin\tNE\nund\tKON\nwerden\tVAFIN\nInnenminister\tNN\nin\tAPPR\nder\tART\nGroKo\tNN\nund\tKON\nkönnen\tVMFIN\nso\tADV\nsein\tVAINF\nTalent\tNN\nbeim\tAPPRART\nManagement\tNN\nvon\tAPPR\nMigration\tNN\nund\tKON\nTerrorbekämpfung\tNN\nmal\tPTKMA\nrichtig\tADJD\nunter\tAPPR\nBeweis\tNN\nstellen....\tNE\n\nDas\tART\nBild\tNN\nsagen\tVVFIN\nmehr\tPIAT\nals\tKOKOM\ntausend\tCARD\nWort.\tNN\nGo,\tXY\nJo!\tXY\n\nSein\tPPOSAT\nsowieso\tADV\nflabbergasted\tADJD\nin\tAPPR\nAnbetracht\tNN\nder\tART\nVorstellung,\tNN\ndieser\tPDAT\nblass\tADJA\nFranke\tNN\nsollen\tVMFIN\nausgerechnet\tADV\nMP\tNN\nin\tAPPR\nBayern\tNE\nwerden.\tNN\nDageg\tNE\nsein\tVAINF\nja\tPTKMA\nStephan\tNE\nWeil\tNE\nne\tART\nPartymaus.\tNN\nPasst\tVVFIN\nauch\tPTKMA\nüberhaupt\tADV\nnicht\tPTKNEG\nin\tAPPR\ndie\tART\nReihe\tNN\nirgendwie.\tPIS\nBei\tAPPR\nSöder\tNE\nweißen\tADJA\ndu\tPPER\nimmer\tADV\nschon\tPTKMA\nvorher,\tNN\nwas\tPWS\ner\tPPER\nsagen\tVVINF\nwerden\tVAFIN\nund\tKON\nzwar\tADV\ngenau\tADJD\nwie\tKOKOM\ner's\tNE\nsagen\tVVFIN\nwerden.\tNN\nEin\tART\nPolitroboter\tNN\nvor\tAPPR\ndem\tART\nHerr\tNN\nund\tKON\ngenauso\tADV\ngucken\tVVFIN\ner\tPPER\nda\tADV\nja\tPTKMA\nauch\tADV\ndrein.\tADR\nAlso\tADV\nwie\tKOKOM\nimmer\tADV\neigentlich.\tEMOASC\n\nHerrmann\tNE\nwerden\tVAFIN\ndoch\tADV\nbei\tAPPR\nder\tART\nBundestagswahl\tNN\nkomplett\tADJD\nverbrennen.\tTRUNC\nSöder\tNE\nsein\tVAINF\nkein\tPIAT\nThema,\tKOKOM\nwenn\tKOUS\ndem\tPDS\nnicht\tPTKNEG\nso\tADV\nsein.\tADJD\n\nMich\tPPER\nwerden\tVAFIN\neher\tADV\ninteressieren,\tNN\nob\tKOUS\nund\tKON\nwelche\tPWAT\npolitisch-inhaltlich\tADJD\nDifferenz\tNN\nes\tPPER\nzwischen\tAPPR\nden\tART\nPersonalie\tNN\ngeben.\tNE\n\nGegenfrage,\tNN\ngehen\tVVFIN\nes\tPPER\nin\tAPPR\nBayern\tNE\nund\tKON\nseiner\tPPOSAT\nFührungskamarilla\tNN\njemals\tADV\num\tAPPR\nPolitisch-Inhaltliches?\tNN\n\nEine\tART\nsachlich\tADJD\nDiskussion\tNN\nsein\tVAINF\ndoch\tPTKMA\ngar\tPTKIFG\nnicht\tPTKNEG\nerwünscht.Was\tVVPP\nhaben\tVAFIN\nich\tPPER\ndenn\tPTKMA\njetzt\tADV\nschon\tPTKMA\nwieder\tADV\nbös\tAKW\nschreiben?\tNE\n\nDass\tKOUS\nsein\tPPOSAT\nFaschos\tNN\nhassen?\tNE\nEgal\tADV\nwie\tKOKOM\nsie\tPPER\nsich\tPRF\nverkleiden\tVVINF\nund\tKON\nunter\tAPPR\nwelchem\tPWAT\nBanner\tNN\nsie\tPPER\nMeinung\tNN\nlöschen?\tNE\n\nMeinungsfreiheit\tNN\nnur\tPTKIFG\nnoch\tADV\nfür\tAPPR\nJournalist,\tPIS\ndie\tPDS\ndann\tADV\nauch\tADV\nmal\tPTKMA\nFalschzitat\tNN\nkommentieren\tVVINF\ndürfen?\tNN\n\nGabriel\tNE\nund\tKON\nMerkel\tNE\nschaden\tVVFIN\ndem\tART\nAnsehen\tNN\nDeutschlandsEntfernt.\tNN\nBitte\tADV\näußern\tVVFIN\nSie\tPPER\nsich\tPRF\nzum\tAPPRART\nThema\tNN\ndes\tART\nArtikel.\tPIS\nDie\tART\nRedaktion/cs\tNN\n\n`Das\tHST\nDeutschen-Gen...Das\tNN\nDeutschen-Gen\tNN\nscheinen\tVVFIN\ndas\tART\nGen\tNN\nder\tART\nIntoleranz\tNN\nzu\tAPPR\nsein,\tNN\nmit\tAPPR\nder\tART\nein\tART\nDeutsche\tADJA\nseine\tPPOSAT\nMeinung\tNN\ngegenüber\tAPPO\nAnderen\tNN\nin\tAPPR\nForum\tNN\nverteidigen.\tVVPP\n\nKönnen\tVMFIN\nman\tPIS\ntagtäglich\tADJD\nbei\tAPPR\nder\tART\nZEIT\tNN\nbeobachten.\t$.\n\nKürzen.\tHST\nWir\tPPER\nbitten\tVVFIN\nSie,\tNE\nsich\tPRF\nin\tAPPR\nden\tART\nKommentar\tNN\nwieder\tADV\ndem\tART\nThema\tNN\ndes\tART\nArtikel\tNN\nzuwenden\tVVFIN\nund\tKON\nweit\tPTKIFG\nAnmerkung\tNN\nzur\tAPPRART\nModeration\tNN\ndirekt\tADJD\nan\tAPPR\[email protected]\tNN\nzu\tAPPR\nrichten,\tTRUNC\ndamit\tPAV\nim\tAPPRART\nForum\tNN\neine\tART\nsachlich\tADJD\nDiskussion\tNN\nermöglichen\tVVFIN\nwerden.\tADJD\nDie\tART\nRedaktion/cs\tNN\n\nLiebe\tNN\n-\t$(\nSarrazin\tNE\n-\t$(\nMitläuferWenn\tNE\nHerr\tNN\nSarrazin\tNE\nsich\tPRF\nzu\tAPPR\nHaus\tNN\noder\tKON\nin\tAPPR\nseiner\tPPOSAT\nKneipe\tNN\nmit\tAPPR\nseinen\tPPOSAT\n\"dämlich\tNN\nAnsicht“\tADV\nprivat\tADJD\näußern\tVVINF\n-\t$(\nsein\tPPOSAT\ndas\tART\n\"unter\tNN\nMeinungsfreiheit\"\tEMOIMG\nnoch\tADV\nhinnehmen\tVVINF\n-\t$(\nkein\tPIAT\nHahn\tNN\nwerden\tVAFIN\nnach\tAPPR\nihm\tPPER\nkrähen.\tPIS\nAber\tADV\ner\tPPER\nnutzen\tVVFIN\nseine\tPPOSAT\nexponieren\tVVINF\nStellung\tNN\nzum\tAPPRART\nProvozieren,\tNN\num\tAPPR\nsein\tPPOSAT\nBuch\tNN\nmöglichst\tADV\noft\tADV\nzu\tAPPR\nverkaufen.\tXY\nDas\tPDS\nsein\tVAINF\nnicht\tPTKNEG\no.k.\tADJD\nFür\tAPPR\ndiese\tPDAT\nProvokation\tNN\nmuss\tVMFIN\ner\tPPER\nentsprechend\tAPPR\nKritik\tNN\naushalten\tVVFIN\n-\t$(\ndie\tPRELS\ner\tPPER\ndoch\tPTKMA\nso\tADV\nselbstverständlich\tADJD\nan\tAPPR\nanderen\tPIAT\nüben.\tNN\nDie\tART\nzahllos\tADJA\nMitläufer\tNN\nhier\tADV\nauf\tAPPR\nden\tART\nKommentarseite\tNN\nsollen\tVMFIN\nnicht\tPTKNEG\n\"stellvertretend\tADJD\nfür\tAPPR\nihn\"\tPPER\nso\tADV\nbeleidigt\tADJD\ntun.\tEMOASC\n\nVergessen\tVVIMP\nSie\tPPER\nnicht,\tADR\nvor\tAPPR\nca.\tADV\n40\tCARD\nJahr\tNN\nhaben\tVAFIN\nwir\tPPER\nDeutsch\tNN\nherablassend\tADJD\ndie\tART\nEinwanderung\tNN\nvon\tAPPR\n\"dumm\tTRUNC\nTürke\"\tHST\nwünschen,\tHST\ndamit\tKOUS\ndie\tART\nDrecksarbeit\tNN\nmachen\tVVFIN\nwerden.\tNN\n\nDa\tADV\nfinden\tVVFIN\nwir\tPPER\ndie\tART\nNiedrigstlohn\tNN\nfür\tAPPR\nTürke\tNN\no.k.\tEMOASC\n–\t$(\ndie\tPDS\nkommen\tVVFIN\nja\tPTKMA\naus\tAPPR\nden\tART\ndoof\tADJA\nEcke\tNN\nder\tART\nTürkei.\tNN\nWo\tPWAV\nsein\tPPOSAT\nHerr\tNN\nSarrazin\tNE\ndamals,\tNE\nals\tKOUS\nes\tPPER\nbesorgt\tADJD\nStimme\tNN\nzu\tAPPR\ndieser\tPDAT\narrogant\tADJD\nEinwanderungspolitik\tNN\ngeben.\tADJD\n\nDass\tKOUS\nheute\tADV\nviele\tPIAT\nMensch\tNN\nin\tAPPR\ndiesem\tPDAT\n\"tollen\tNN\nDeutschland\"\tHST\nfür\tAPPR\nNiedrigstlohn\tNN\narbeiten,\tNN\nauf\tAPPR\ndem\tART\nLohnniveau\tNN\ndamalig\tADJD\nEinwanderer\tNN\nund\tKON\nnoch\tADV\ndarunt,\tPAV\nsein\tVAINF\ndas\tART\neigentlich\tPTKIFG\nProblem\tNN\n-\t$(\nund\tKON\ndaran\tPAV\nsein\tPPOSAT\ndie\tART\n\"deutsch\tNN\nRassegene,\tVVFIN\nwir\tPPER\nsein\tPPOSAT\nja\tPTKMA\nwas\tPWS\nGute\"\tNE\nganz\tPTKIFG\nerheblich\tADJD\nSchuld.\tNE\nDiese\tPDAT\ndoof\tADJD\ndeutsch\tNN\nNiedriglöhner\tNN\nsein\tPPOSAT\nnämlich\tADV\nauch\tADV\nbald\tADV\ndie\tART\nMoor\tNN\n…wie\tVAFIN\nheute\tADV\ndie\tART\nTürke.\tNN\nDas\tPDS\nsein\tPPOSAT\ndie\tART\nAngst.\tNN\n\nÜbrigens:\tADR\nAls\tAPPR\n„reinrassig\tART\nDeutsch“\tNN\nkennen\tVVINF\nund\tKON\nmögen\tVMFIN\nich\tPPER\neine\tART\nganz\tPTKIFG\nMenge\tNN\n(hoch)intelligent,\tEMOIMG\nerfolgreich\tADJD\nund\tKON\nobendrein\tADV\nauch\tADV\nnoch\tADV\nsehr\tPTKIFG\nsympathisch\tADJD\nTürke\tNN\naus\tAPPR\nRegion\tNN\nam\tAPPRART\nMarmarameer\tNE\nbis\tAPPR\nnach\tAPPR\nAnatolien\tNE\n(wo\tXY\nja\tPTKMA\ndie\tART\nDoofen\tADJA\nwohnen).\tNN\n\nwarum?Warum\tPIS\nhaben\tVAFIN\nsich\tPRF\nchinesen,\tADR\nrussen,\tHST\nthaisen,\tHST\nitalien\tNE\nintegrieren?\tNE\n\nDas\tPDS\nsein\tVAFIN\ndie\tART\nFrage,\tNN\ndie\tART\nzu\tPTKZU\ndiskutieren\tVVINF\nsein.\tNE\nDoch\tPTKMA\ndas\tPDS\nwollen\tVMFIN\ndie\tART\nMedium\tNN\ndoch\tADV\ngar\tPTKIFG\nnicht,\tNN\nwie\tKOKOM\ndas\tPDS\nwiederholen\tVVFIN\nLöschen\tNN\ndieser\tPDAT\nFrage\tNN\nbei\tAPPR\nder\tART\nZEIT\tNN\nzeigen.\t$.\n\nMP3\tNN\nsein\tPPOSAT\ndoch\tPTKMA\ntotal\tADJD\nSchrot.\tTRUNC\nselbst\tADV\nim\tAPPRART\nAuto.\tNN\nZum\tAPPRART\nGlück\tNN\nkönnen\tVMFIN\nmeine\tPPOSAT\nneu\tADJD\nKarre\tNN\njetzt\tADV\nFLAC\tART\nabspielen,\tNN\nvorher\tADV\ngehen\tVVFIN\nzwar\tADV\nWAV,\tADR\naber\tADV\nich\tPPER\nmüssen\tVMFIN\nextra\tADV\nkonvertieren.\tNN\n\nSelb\tNE\nschuld,\tNE\nwer\tPWS\nseinen\tPPOSAT\nOhr\tNN\nMP3\tTRUNC\nantun.\tHST\nFLAC\tNN\nbieten\tVVFIN\nalle\tPIAT\nVorteil:\tNN\nTagging,\tHST\nKomprimierung,\tADV\nkeinen\tPIAT\nQualitätsverlust.\tNN\n\nMP3´s\tHST\nhaben\tVAFIN\nbei\tAPPR\ngut\tADJA\nQuellqualität\tNN\nkaum\tPTKIFG\nQualitätsverlust.\tNN\nUm\tKOUI\ndas\tPDS\ndann\tADV\nnoch\tADV\nüberhaupt\tADV\nzu\tAPPR\nmerken,\tNE\nbrauchen\tVVFIN\nman\tPIS\nerstens\tADV\nein\tART\nsehr\tPTKIFG\ngut\tADJD\nGehör\tNN\nund\tKON\nzweitens\tADV\nmindestens\tADV\nein\tART\ngut\tADJA\nAbspielgerät.\tNN\nAber\tADV\ndas\tPRELS\nSie\tPPER\ngleich\tADJD\nsich\tPRF\nne\tART\nneu\tADJD\nKarre\tNN\nanschaffen,\tNN\num\tAPPR\nFlAC\tNE\nzu\tAPPR\nhören...\tNE\nxD\tEMOASC\n\nIrgendwo\tADV\ngaanz\tVVPPER\ntief\tADJD\nunten\tADV\nin\tAPPR\nden\tART\nKatakombe\tNN\nder\tART\nZeit.de-Redaktion\tNN\nhaben\tVAFIN\njemand\tPIS\njetzt\tADV\nsehr\tPTKIFG\nglücklich\tADJD\nda\tADV\ner/sie\tVVPPER\nsehr\tPTKIFG\nlange\tADJD\ndarauf\tPAV\nwarten,\tNN\ndieses\tPDAT\nWortspiel\tNN\nim\tAPPRART\nTitel\tNN\nerscheinen...\tVVPP\n\nIch\tPPER\nhaben\tVAFIN\nmir\tPRF\nmal\tPTKMA\ndie\tART\nMühe\tNN\nmachen\tVVINF\nund\tKON\nbei\tAPPR\nSpotify\tNE\nnach\tAPPR\nden\tART\nvon\tAPPR\nihnen\tPPER\nerwähnen\tVVFIN\nKünstler\tNN\nmachen.\tNE\n\nHugo\tNE\nAlfven,\tNE\nThomas\tNE\nArne,\tNE\nCarles\tNE\nBaguer,\tNE\nMily\tNE\nBalakirev,\tNE\nJiri\tNE\nAntonin\tNE\nBenda,\tNE\nWilliam\tNE\nSterndal\tNE\nBennett\tNE\nfinden\tVVFIN\nsich\tPRF\nalle\tPIS\nbei\tAPPR\nSpotify,\tNE\nwas\tPWS\nja\tPTKMA\nklar\tADJD\nsagen\tVVFIN\ndas\tART\nsolche\tPIAT\nDienst\tNN\nnicht\tPTKNEG\nnur\tPTKIFG\nden\tART\nMainstream\tNN\nbedienen\tVVINF\nmögen.\tTRUNC\n\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0f218a5df3fbaa5cb698b99d184e02714f2d87e | 10,551 | ipynb | Jupyter Notebook | tf_practice.ipynb | quinn-dougherty/DS-Unit-4-Sprint-4-Deep-Learning | 887123e5e72112681d5b4f8d9c62cf783eea6015 | [
"MIT"
] | null | null | null | tf_practice.ipynb | quinn-dougherty/DS-Unit-4-Sprint-4-Deep-Learning | 887123e5e72112681d5b4f8d9c62cf783eea6015 | [
"MIT"
] | null | null | null | tf_practice.ipynb | quinn-dougherty/DS-Unit-4-Sprint-4-Deep-Learning | 887123e5e72112681d5b4f8d9c62cf783eea6015 | [
"MIT"
] | null | null | null | 30.232092 | 105 | 0.370486 | [
[
[
"import sys\nimport tensorflow as tf\nfrom sklearn.datasets import load_boston\nimport pandas as pd\nimport matplotlib.pyplot as plt\nplt.style.use('dark_background')\nboston = load_boston()\n''' THIS is how you print the name of a function from within the function\n print(sys._getframe().f_code.co_name) \n\n'''#print([x for x in dir(boston) if '_' not in x])\n\n#print(boston.DESCR)\n\ndef load_dat(dat: pd.DataFrame) -> pd.DataFrame: \n return (dat.rename(columns={k: name \n for k,name \n in enumerate(['CRIM', 'ZN', 'INDUS', 'CHAS', \n 'NOX', 'RM', 'AGE', 'DIS', 'RAD', \n 'TAX', 'PTRATIO', 'B', 'LSTAT'])}\n )).assign(ones=pd.np.ones(dat.shape[0]))\n\ny_ = pd.DataFrame(boston.target, columns=['MEDV'])\n\nX_ = load_dat(pd.DataFrame(boston.data))\n\nX = tf.constant(X_.values, tf.float32, name='X') \ny = tf.constant(y_.values, tf.float32, name='y')\nprint(X, y)\nprint([x for x in dir(X) if '_' not in x])\n\nX_.head()\n\n",
"Tensor(\"X:0\", shape=(506, 14), dtype=float32) Tensor(\"y:0\", shape=(506, 1), dtype=float32)\n['consumers', 'device', 'dtype', 'eval', 'graph', 'name', 'op', 'shape']\n"
],
[
"# def predict(X: tf.Tensor, y: tf.Tensor) -> tf.Tensor: \n# ''' will return yhat as a tensor '''\n# beta = tf.ones((X.shape[1],1))\n# #print(beta)\n# o = tf.matmul(a=X, b=beta)\n\n# with tf.Session() as sess: \n# x = sess.run(o)\n# return tf.constant(x, name='yhat')\n\n# predict(X, y)\n\n",
"_____no_output_____"
],
[
"gam = tf.pow(2., -10, name='gamma')\n\nbeta = tf.ones((X.shape[1],1))\n\n'''\nloss = tf.reduce_sum(tf.pow(tf.subtract(y, \n o), \n tf.constant(2.)))\n# we want grad_loss to be a 14 x 1 vector \ngradloss_ = tf.multiply(beta, \n tf.reduce_sum(tf.subtract(y, o), axis=1))\n\ngradloss = tf.multiply(tf.constant(-2, tf.float32), \n gradloss_r)\n#gradloss_r = tf.multiply(beta, X)'''\n\n\n'''from wikip\n\ngrad(loss(beta)) == 2 * X^T * (Xbeta - y)\n'''\ngradloss = tf.multiply(tf.constant(2, tf.float32), \n tf.matmul(a=X, \n b=tf.subtract(o, \n y), \n transpose_a=True))\n\n\n\ngam = tf.pow(2., -10, name='gamma')\n\nbeta = tf.ones((X.shape[1],1))\n\ndef gradloss(beta: tf.Tensor) -> tf.Tensor: \n '''from wikip\n\n grad(loss(beta)) == 2 * X^T * (Xbeta - y)\n '''\n o = tf.matmul(a=X, b=beta)\n return tf.multiply(tf.constant(2, tf.float32), \n tf.matmul(a=X, \n b=tf.subtract(o, \n y), \n transpose_a=True))\n\ndef beta_next(beta_prev: tf.Tensor) -> tf.Tensor: \n return tf.subtract(beta_prev, tf.multiply(gam, gradloss(beta_prev)))\n\nwith tf.Session() as sess: \n x = sess.run(gradloss(beta_next(beta_next(beta_next(beta)))))\n print(x)",
"[[-1.19036155e+23]\n [-2.64888255e+23]\n [-3.21022706e+23]\n [-1.81797777e+21]\n [-1.51157275e+22]\n [-1.65864962e+23]\n [-1.89500159e+24]\n [-9.51447751e+22]\n [-2.93692774e+23]\n [-1.17169084e+25]\n [-4.95573293e+23]\n [-9.46718438e+24]\n [-3.54167722e+23]\n [-2.65633497e+22]]\n"
],
[
"with tf.Session() as sess:\n assert \n print(sess.run(tf.multiply(tf.constant([9,8,7,6,5]), tf.constant([1,2,3,4,5]))))",
"[ 9 16 21 24 25]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d0f22e509741ddc3a15aa05575f60147c312667b | 12,401 | ipynb | Jupyter Notebook | Dzien01/01-jupyter-intro.ipynb | gitgeoman/PAD2022 | 49266a5f130971a8b9b7fda890f0f43e6d6d64fe | [
"MIT"
] | null | null | null | Dzien01/01-jupyter-intro.ipynb | gitgeoman/PAD2022 | 49266a5f130971a8b9b7fda890f0f43e6d6d64fe | [
"MIT"
] | null | null | null | Dzien01/01-jupyter-intro.ipynb | gitgeoman/PAD2022 | 49266a5f130971a8b9b7fda890f0f43e6d6d64fe | [
"MIT"
] | null | null | null | 25.308163 | 819 | 0.510765 | [
[
[
"import numpy as np\nimport time # biblioteka do obsługi czasu",
"_____no_output_____"
],
[
"#sprawdzanie wersji biblioteki numpy\nprint(np.__version__) ",
"1.20.3\n"
],
[
"#deklaracja zmiennych w python\nx= 10\ny=-2.5\nprint(time.time())",
"1639990590.4254217\n"
],
[
"#deklaracja pustej klasy\nclass TestClass:\n pass\n",
"_____no_output_____"
]
],
[
[
"#Przykład formatowania tekstu w notebooku\n<a href='https://alx.pl'> Nagłówek 1 </a>\n## Nagłówek 2\n### Nagłówek 3\nGdy piszemy jakiś tekst w snippet to może on być **pogrubiony**, *kursywa* albo ***jedno i drugie***\nMożna również tworzyć w snippetach listy:\n- li1\n- li2\n\nMoga być one numerowane:\n1. li1\n2. li3",
"_____no_output_____"
],
[
"# Maginczne komendy\nMagiczne komendy to dodataki/usprawnienia do pythona w postaci funkcji które umożliwiają realizację okreslonych operacji. ",
"_____no_output_____"
]
],
[
[
"#lista magicznych komend wywoujemy przy użyciu poniższej komendy\n%lsmagic",
"_____no_output_____"
],
[
"#gdy trzeba doinstalowaać jakieś biblioteki zewnętrzene to można to zrobić przy użyciu takiej metody\n\n####Wykonywanie polceń systemu operacyjnego\n",
"_____no_output_____"
],
[
"# instalacja dodatkowej biblioteki\n!pip install pydotplus ",
"Requirement already satisfied: pydotplus in c:\\users\\kurs\\anaconda3\\lib\\site-packages (2.0.2)\nRequirement already satisfied: pyparsing>=2.0.1 in c:\\users\\kurs\\anaconda3\\lib\\site-packages (from pydotplus) (3.0.4)\n"
],
[
"x = 1.2342532345\nx",
"_____no_output_____"
],
[
"# zmieni liczbę cyfr po przecinku domyślnie wyświetlanych\n%precision 3",
"_____no_output_____"
],
[
"#pomiar szybkości kodu\na=time.time()\nsum(range (1,1_000_001))\nb=time.time()\nprint(a-b)",
"-0.047652482986450195\n"
],
[
"%time sum(range (1,1_000_001))",
"Wall time: 47.7 ms\n"
],
[
"%%time\nsum(range (1,1_000_001))\nsum(range (1,1_000_001))",
"_____no_output_____"
],
[
"%%timeit -r 10 -n 10\nresult=[]\nfor x in range(1,1_000_001):\n if x%2==0:\n result.append(x)",
"129 ms ± 5.8 ms per loop (mean ± std. dev. of 10 runs, 10 loops each)\n"
],
[
"%%timeit -r 10 -n 10\n[x for x in range(1,1_000_001) if x%2==0]",
"104 ms ± 5.48 ms per loop (mean ± std. dev. of 10 runs, 10 loops each)\n"
],
[
"#obliczenie zysku czasu (wyszło ok 18%)\n104/129",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f23f9aeec7ae6df47d29d9e6ba9389677a227c | 31,886 | ipynb | Jupyter Notebook | Post_Group/com_matrix_and_network_creation.ipynb | Lally09-fx/net2020 | 042bdd384cd9f8a5978c1281545349c9884897a3 | [
"CC0-1.0"
] | null | null | null | Post_Group/com_matrix_and_network_creation.ipynb | Lally09-fx/net2020 | 042bdd384cd9f8a5978c1281545349c9884897a3 | [
"CC0-1.0"
] | null | null | null | Post_Group/com_matrix_and_network_creation.ipynb | Lally09-fx/net2020 | 042bdd384cd9f8a5978c1281545349c9884897a3 | [
"CC0-1.0"
] | 13 | 2020-12-05T18:43:10.000Z | 2020-12-30T22:37:43.000Z | 69.017316 | 6,800 | 0.454494 | [
[
[
"# Import the data",
"_____no_output_____"
]
],
[
[
"import pandas as pd \nimport numpy as np \nimport networkx as nx\n\nimport statsmodels\nimport statsmodels.api as sm\nimport scipy.stats as stats\n\nimport matplotlib.pyplot as plt\n\n# import the csv file with JUST the politicians post\ncomDB = pd.read_csv(r\"/Users/tassan-mazzoccoarthur/Desktop/NETWORK SCIENCE (MOD. B)/1_Project/database/com_liwc.csv\", sep='\\t', engine='python')\n\ndf = pd.DataFrame(data=comDB)\ndf",
"_____no_output_____"
],
[
"df_trunc = df[df['c_rating']=='positivo']\ndf_trunc",
"_____no_output_____"
]
],
[
[
"# Matrix creation",
"_____no_output_____"
]
],
[
[
"## Matrix creation with parties, politicians, posts",
"_____no_output_____"
],
[
"#Setting variables\nsize_df= len(df)\nparties=df['p_PARTITO'].unique().tolist()\npol=df['p_politician'].unique().tolist()\npost=df['p_id'].unique().tolist()\n\n#Matrix shape= root/party/party.....pol/pol/pol/...../post/post/.../com/com\ntotal_size=1+len(parties)+len(pol)+len(post)+size_df\nshift_size=1+len(parties)+len(pol)+len(post)\nmatrix = np.identity(total_size,dtype=int)\n\nfor i in range(size_df):\n #for each post with have to add 1 to the link party-politician, 1 to the link politician-post, 1 to the link post-comment\n index_party=parties.index(df['p_PARTITO'][i])+1 #add 1 for the root element\n index_pol=pol.index(df['p_politician'][i])+1+len(parties)\n index_post=post.index(df['p_id'][i])+1+len(parties)+len(pol)\n\n #We fill the 1rst half of the matrix\n matrix[0][index_party]+=1 #add 1 to link root-party\n matrix[index_party][index_pol]+=1 #add 1 to the link party-politician\n matrix[index_pol][i+1+len(parties)+len(pol)]+=1 #1 to the link politician-post\n matrix[index_post][i+shift_size]+=1 #1 to the link post-comment\n #now we fill the other half (lower-left one)\n matrix[index_party][0]+=1\n matrix[index_pol][index_party]+=1\n matrix[i+1+len(parties)+len(pol)][index_pol]+=1 \n matrix[i+shift_size][index_post]+=1\n\nm=np.asmatrix(matrix)\nprint(m.shape)",
"(88393, 88393)\n"
]
],
[
[
"## Topic matrix:",
"_____no_output_____"
]
],
[
[
"#connect comments together on same topic -> only post and com matrix\ntopic=df['c_topic'].unique().tolist()\n\n#Matrix creation\n#topic/topic/...com/com\ntotal_size=len(topic)+size_df\nshift_size=len(topic)\ntopic_matrix = np.identity(total_size,dtype=int)\nfor i in range(size_df):\n index_topic=topic.index(df['c_topic'][i])+shift_size\n topic_matrix[index_topic][i+shift_size]+=1 #1 to the link comment-topic\n topic_matrix[i+shift_size][index_topic]+=1\n \nm_topic=np.asmatrix(topic_matrix)\nprint(m_topic.shape)\n\n#NEED TO ADD LINK BETWEEN TOPICS???? \n#CREATE A ROOT????\n#ADD C_RATING",
"(78450, 78450)\n"
],
[
"m_topic",
"_____no_output_____"
],
[
"topic=df['c_topic'].unique().tolist()\nlen(topic)",
"_____no_output_____"
],
[
"G_topic = nx.from_numpy_matrix(m_topic)",
"_____no_output_____"
],
[
"nx.write_graphml(G_topic, \"topic_com_attributes_networkx_export.graphml\") ",
"_____no_output_____"
]
],
[
[
"# Adding attributes",
"_____no_output_____"
]
],
[
[
"import math\n#create a dictionnary of attributes to update a networkx graph \n# you should change the elements of the \"attributes\" array to select the attributes you want\ndef create_attributes_dict_com(pandas_df, index_shift): #index_shift -> value to shift the index of the nodes we focus our attention on\n#for example: if the matrix is topic/topic/topic.....comments/comments/comments the shift size will be the number of topic\n#note: the shape of the dictionnary to update the node with index 0 is :\n# attrs = {0: {\"p_PARTITO\": df_post['p_PARTITO'][0], \"p_politician\": df_post['p_politician'][0]}}\n attributes=[\"p_PARTITO\"]\n #attributes=[\"p_PARTITO\",\"p_politician\",\"p_favoriteCount\",\"p_shareCount\",\"p_replyCount\",\"p_numComments\",\"p_rating\",\"p_topic\",\"p_campagna\",\"c_rating\"]\n att_dict = {} #final dict to be returned\n for index in pandas_df.index:\n temp_dict={}\n for att in attributes:\n #we need to delete the \"nan\" values\n if(isinstance(pandas_df[att][index], float)):\n if(not math.isnan(pandas_df[att][index])):\n temp_dict[att]=pandas_df[att][index]\n else:\n temp_dict[att]=pandas_df[att][index]\n temp_dict[\"label\"]=index+shift_size #we set the node label to its index in the dataframe\n temp_dict[\"type\"]=\"com\" #we set the node type\n att_dict[index+index_shift]=temp_dict.copy() #don't fprget that the good index in the dictionnary is + shift_size\n return att_dict",
"_____no_output_____"
],
[
"#create a dictionnary of attributes for the topics\ndef add_com_topic_att(pandas_df, topic): #param = lists\n att_dict = {}\n for i in range(len(topic)):\n #att_dict[i+1]={\"label\":topic[i], \"type\":\"topic\"} #this line if we have a root element\n att_dict[i]={\"label\":topic[i], \"type\":\"topic\"}\n return att_dict",
"_____no_output_____"
],
[
"attributes_dict_com=create_attributes_dict_com(df,shift_size) #we create the dict for the comments\nmissing_attributes_dict_com=add_com_topic_att(df,topic) #we create the dict for the topic...\nattributes_dict_com.update(missing_attributes_dict_com) #and add it to the one for the comments\nnx.set_node_attributes(G, attributes_dict_com) #finally we set the nodes' attributes in the graph",
"_____no_output_____"
],
[
"nx.write_graphml(G_topic, \"topic_com_attributes_networkx_export.graphml\") #export the graph as a GraphML XML file",
"_____no_output_____"
]
],
[
[
"# Some cheks",
"_____no_output_____"
]
],
[
[
"import networkx as nx\n#First let's try with a small matrix\nm_trunc=m[:40000,:40000]\nnp.set_printoptions(linewidth=200)\nprint(m_trunc)\nG_trunc = nx.from_numpy_matrix(m_trunc)",
"[[ 1 10245 18760 ... 0 0 0]\n [10245 1 0 ... 0 0 0]\n [18760 0 1 ... 0 0 0]\n ...\n [ 0 0 0 ... 1 0 0]\n [ 0 0 0 ... 0 1 0]\n [ 0 0 0 ... 0 0 1]]\n"
],
[
"nx.write_graphml(G_trunc, \"com_attributes_networkx_export.graphml\") ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0f242d1cf13dc113299902b68130d3a27fb87c5 | 12,605 | ipynb | Jupyter Notebook | titanic/titanic_kaggle.ipynb | iriyagupta/DA_repo | e9b47e49c36b99c36cb2340a98e2d67b3bd581dd | [
"MIT"
] | null | null | null | titanic/titanic_kaggle.ipynb | iriyagupta/DA_repo | e9b47e49c36b99c36cb2340a98e2d67b3bd581dd | [
"MIT"
] | null | null | null | titanic/titanic_kaggle.ipynb | iriyagupta/DA_repo | e9b47e49c36b99c36cb2340a98e2d67b3bd581dd | [
"MIT"
] | null | null | null | 30.373494 | 92 | 0.353114 | [
[
[
"import pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"train_data = pd.read_csv(\"data/train.csv\", index_col=\"PassengerId\")\ntest_data = pd.read_csv(\"data/test.csv\",index_col=\"PassengerId\")",
"_____no_output_____"
],
[
"train_data.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 891 entries, 1 to 891\nData columns (total 11 columns):\nSurvived 891 non-null int64\nPclass 891 non-null int64\nName 891 non-null object\nSex 891 non-null object\nAge 714 non-null float64\nSibSp 891 non-null int64\nParch 891 non-null int64\nTicket 891 non-null object\nFare 891 non-null float64\nCabin 204 non-null object\nEmbarked 889 non-null object\ndtypes: float64(2), int64(4), object(5)\nmemory usage: 83.5+ KB\n"
],
[
"train_data.head()",
"_____no_output_____"
],
[
"train_data.isnull().sum()",
"_____no_output_____"
],
[
"train_data.describe()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f246067b5e756dbde7641557285143869c852d | 84,799 | ipynb | Jupyter Notebook | New Thing - Same Shit/15_Linear_Discriminant_Analysis.ipynb | subhadeep-123/Machine-Learning-Algorithms | 3db4f0c9703a58c56a8cf560dcb41dc5654e36ff | [
"MIT"
] | 7 | 2019-06-21T12:00:42.000Z | 2021-04-19T21:14:34.000Z | New Thing - Same Shit/15_Linear_Discriminant_Analysis.ipynb | subhadeep-123/Machine-Learning-Algorithms | 3db4f0c9703a58c56a8cf560dcb41dc5654e36ff | [
"MIT"
] | null | null | null | New Thing - Same Shit/15_Linear_Discriminant_Analysis.ipynb | subhadeep-123/Machine-Learning-Algorithms | 3db4f0c9703a58c56a8cf560dcb41dc5654e36ff | [
"MIT"
] | 1 | 2021-06-17T14:48:08.000Z | 2021-06-17T14:48:08.000Z | 67.676776 | 21,540 | 0.669194 | [
[
[
"import glob\nimport warnings\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom matplotlib.colors import ListedColormap\nfrom sklearn.metrics import classification_report\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.linear_model import LogisticRegression\n\n%matplotlib inline\n\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"file = glob.iglob('*.csv')\ndf = pd.read_csv(*file)\n\nprint(f\"The Dimension of the data is - {df.shape}\")",
"The Dimension of the data is - (178, 14)\n"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.tail()",
"_____no_output_____"
],
[
"X = df.iloc[:, :-1].values\nY = df.iloc[:, -1].values",
"_____no_output_____"
],
[
"X",
"_____no_output_____"
],
[
"Y",
"_____no_output_____"
],
[
"print(\"Size of X: {}\".format(X.shape))\nprint(\"Size of Y: {}\".format(Y.shape))",
"Size of X: (178, 13)\nSize of Y: (178,)\n"
],
[
"X_train, X_test, Y_train, Y_test = train_test_split(X,\n Y,\n test_size = 0.2,\n random_state = 0)",
"_____no_output_____"
],
[
"print(\"Size of X_train: {}\".format(X_train.shape))\nprint(\"Size of X_test: {}\".format(X_test.shape))\nprint(\"Size of Y_train: {}\".format(Y_train.shape))\nprint(\"Size of Y_test: {}\".format(Y_test.shape))",
"Size of X_train: (142, 13)\nSize of X_test: (36, 13)\nSize of Y_train: (142,)\nSize of Y_test: (36,)\n"
],
[
"sc = StandardScaler()\nX_train = sc.fit_transform(X_train)\nX_test = sc.transform(X_test)",
"_____no_output_____"
],
[
"X_train",
"_____no_output_____"
],
[
"X_test",
"_____no_output_____"
],
[
"lda = LDA(solver = 'eigen',\n n_components = 2)\nX_train = lda.fit_transform(X_train, Y_train)\nX_test = lda.transform(X_test)",
"_____no_output_____"
],
[
"X_train",
"_____no_output_____"
],
[
"X_test",
"_____no_output_____"
],
[
"classifier = LogisticRegression(verbose = 1,\n random_state = 42,\n n_jobs = -1)\nclassifier.fit(X_train, Y_train)",
"[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers.\n[Parallel(n_jobs=-1)]: Done 1 out of 1 | elapsed: 1.8s finished\n"
],
[
"y_pred = classifier.predict(X_test)\ny_pred",
"_____no_output_____"
],
[
"cm = confusion_matrix(Y_test, y_pred)\ncm",
"_____no_output_____"
],
[
"acc = accuracy_score(Y_test, y_pred)\nprint(f\"The accuracy of the model is - {acc*100:.3f}%\")",
"The accuracy of the model is - 100.000%\n"
],
[
"report = classification_report(Y_test, y_pred)\nprint(report)",
" precision recall f1-score support\n\n 1 1.00 1.00 1.00 14\n 2 1.00 1.00 1.00 16\n 3 1.00 1.00 1.00 6\n\n accuracy 1.00 36\n macro avg 1.00 1.00 1.00 36\nweighted avg 1.00 1.00 1.00 36\n\n"
],
[
"# Visualizing the Training Set Results\n\nfigure = plt.figure(figsize = (10,10))\n\nx_set, y_set = X_train, Y_train\n\nX1, X2 = np.meshgrid(np.arange(start = x_set[:, 0].min() - 1,\n stop = x_set[:, 0].max() + 1,\n step = 0.01),\n np.arange(start = x_set[:, 1].min() - 1,\n stop = x_set[:, 1].max() + 1,\n step = 0.01))\n\nplt.contourf(X1,\n X2,\n classifier.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),\n camp = ListedColormap(('red', 'green', 'blue')),\n alpha = 0.4\n )\n\nfor i, j in enumerate(np.unique(y_set)):\n plt.scatter(x_set[y_set == j, 0],\n x_set[y_set == j, 1],\n color = ListedColormap(('red', 'green', 'blue'))(i),\n label = j,\n s = 15,\n marker = '*'\n )\n \nplt.xlim(X1.min(), X1.max())\nplt.xlim(X2.min(), X2.max())\nplt.title('Linear Discriminant analysis (PCA) - Train')\nplt.xlabel('PC1')\nplt.ylabel('PC2')\nplt.legend()",
"_____no_output_____"
],
[
"# Visualizing the Test Set Results\n\nfigure = plt.figure(figsize = (10,10))\n\nx_set, y_set = X_test, Y_test\n\nX1, X2 = np.meshgrid(np.arange(start = x_set[:, 0].min() - 1,\n stop = x_set[:, 0].max() + 1,\n step = 0.01),\n np.arange(start = x_set[:, 1].min() - 1,\n stop = x_set[:, 1].max() + 1,\n step = 0.01))\n\nplt.contourf(X1,\n X2,\n classifier.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),\n camp = ListedColormap(('red', 'green', 'blue')),\n alpha = 0.4\n )\n\nfor i, j in enumerate(np.unique(y_set)):\n plt.scatter(x_set[y_set == j, 0],\n x_set[y_set == j, 1],\n color = ListedColormap(('red', 'green', 'blue'))(i),\n label = j,\n s = 15,\n marker = '*'\n )\n \nplt.xlim(X1.min(), X1.max())\nplt.xlim(X2.min(), X2.max())\nplt.title('Linear Discriminant analysis (PCA) - Test')\nplt.xlabel('PC1')\nplt.ylabel('PC2')\nplt.legend()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f24c7134aa9d667f2e2b67365647df68dc9a54 | 9,004 | ipynb | Jupyter Notebook | examples/make_your_own.ipynb | rafaelschlatter/strava-heatmap | 1462dfa263cf8f58232f8846ec5ed517151d9798 | [
"MIT"
] | 2 | 2020-06-06T13:08:04.000Z | 2020-06-15T19:41:01.000Z | examples/make_your_own.ipynb | rafaelschlatter/strava-heatmap | 1462dfa263cf8f58232f8846ec5ed517151d9798 | [
"MIT"
] | 1 | 2020-08-06T06:13:48.000Z | 2020-08-06T06:13:48.000Z | examples/make_your_own.ipynb | rafaelschlatter/strava-heatmap | 1462dfa263cf8f58232f8846ec5ed517151d9798 | [
"MIT"
] | null | null | null | 32.505415 | 229 | 0.545869 | [
[
[
"## Make your own heatmap based on Strava activities\nThis notebook shows you how to create your own heatmap based on your Strava activities.\n\nYou need to create a Strava API application in order to use their API. Follow the instructions on this page to create your app: <https://medium.com/@annthurium/getting-started-with-the-strava-api-a-tutorial-f3909496cd2d>\n\nAfter setting up the app, note down the following information (you will need it to run this notebook):\n- Client id\n- Client secret\n\n**Note:** Strava imposes some request limits (30'000/day, and 600/every 15min). ",
"_____no_output_____"
]
],
[
[
"!pip install stravaio folium",
"_____no_output_____"
],
[
"import os\nimport logging\nimport json\nimport urllib\nimport requests\nimport folium\nfrom stravaio import StravaIO",
"_____no_output_____"
],
[
"# Paste your client id and client secret here.\nSTRAVA_CLIENT_ID = \"ENTER-YOUR-CLIENT-ID\"\nSTRAVA_CLIENT_SECRET = \"ENTER-YOUR-CLIENT-SECRET\"",
"_____no_output_____"
]
],
[
[
"### Authorization with Strava\nThe cell below creates the proper authorization link using the Stravaio Python library, which is used later to retrieve activities.\nIt is important to run this cell, just pasting the access_token from your Strava settings will not work, because Stravaio needs to be authorized.\n\n- Run the cell below and click the link that is printed, when prompted click \"Authorize\" on the website that opens\n- After you click \"Authorize\" you see something like, \"This site can't be reached\"\n- Stay on that page and look at the URL\n- The URL will show the authorization code (the bit after \"code=\" in the URL) and scope you accepted\n- Copy the code and paste it below and continue the notebook execution\n\nMore detailed info can be found here:\n- <https://developers.strava.com/docs/getting-started/>\n- <https://developers.strava.com/docs/authentication/>",
"_____no_output_____"
]
],
[
[
"params_oauth = {\n \"client_id\": STRAVA_CLIENT_ID,\n \"response_type\": \"code\",\n \"redirect_uri\": f\"http://localhost:8000/authorization_successful\",\n \"scope\": \"read,profile:read_all,activity:read\",\n \"state\": 'https://github.com/sladkovm/strava-http', # Sladkovm is the author of the Stravaio library\n \"approval_prompt\": \"force\"\n}\nvalues_url = urllib.parse.urlencode(params_oauth)\nbase_url = 'https://www.strava.com/oauth/authorize'\nauthorize_url = base_url + '?' + values_url\nprint(authorize_url)",
"_____no_output_____"
],
[
"# Paste the code from the URL here. Afterwards there are no manual steps anymore.\nAUTHORIZATION_CODE = \"ENTER-YOUR-AUTHORIZATION-CODE\"",
"_____no_output_____"
]
],
[
[
"The following cell retrieves an access token using the authorization code. That access token can then be used to retrieve Strava data.",
"_____no_output_____"
]
],
[
[
"payload = {\n \"client_id\": STRAVA_CLIENT_ID,\n \"client_secret\": STRAVA_CLIENT_SECRET,\n \"grant_type\": \"authorization_code\",\n \"code\": AUTHORIZATION_CODE,\n}\n\nresponse = requests.request(\n \"POST\", \"https://www.strava.com/api/v3/oauth/token\", data=payload\n)\n\nresponse = json.loads(response.text)\nTOKEN = response[\"access_token\"]",
"_____no_output_____"
],
[
"!pip install stravaio folium",
"_____no_output_____"
],
[
"client = StravaIO(access_token=TOKEN)\nathlete = client.get_logged_in_athlete()\nactivities = client.get_logged_in_athlete_activities(after=20170101)",
"_____no_output_____"
],
[
"m = folium.Map(\n tiles=\"cartodbpositron\",\n location=[59.925, 10.728123],\n zoom_start=11.5,\n control_scale=True\n)\nfolium.TileLayer(\"cartodbpositron\").add_to(m)\nfolium.TileLayer(\"cartodbdark_matter\").add_to(m)\nfolium.LayerControl().add_to(m)",
"_____no_output_____"
],
[
"def downsample(l, n):\n \"\"\"Returns every nth element from list l. Returns the\n original list if n is set to 1.\n Used to reduce the number of GPS points per activity,\n to improve performance of the website.\n \"\"\"\n \n return l[0::n]\n\ndef map_activities(activities, folium_map, opacity=0.5, weight=1):\n if len(activities) == 0:\n logging.info(\"No activities found, returning empty folium map.\")\n return folium_map\n\n counter = 0\n for a in activities:\n if a.type == \"Workout\":\n continue\n streams = client.get_activity_streams(a.id, athlete.id)\n try:\n points = list(zip(streams.lat, streams.lng))\n points = downsample(l=points, n=2)\n if a.type == \"Run\":\n folium.PolyLine(\n locations=points, color=\"#ff9933\", opacity=opacity, weight=weight\n ).add_to(folium_map)\n elif a.type == \"Ride\":\n folium.PolyLine(\n locations=points, color=\"#0066ff\", opacity=opacity, weight=weight\n ).add_to(folium_map)\n elif a.type == \"NordicSki\":\n folium.PolyLine(\n locations=points, color=\"#00ffff\", opacity=opacity, weight=weight\n ).add_to(folium_map)\n elif a.type == \"AlpineSki\":\n folium.PolyLine(\n locations=points, color=\"#00ccff\", opacity=opacity, weight=weight\n ).add_to(folium_map)\n elif a.type == \"Canoeing\":\n folium.PolyLine(\n locations=points, color=\"#00ff55\", opacity=opacity, weight=weight\n ).add_to(folium_map)\n elif a.type == \"IceSkate\":\n folium.PolyLine(\n locations=points, color=\"#f6ff00\", opacity=opacity, weight=weight\n ).add_to(folium_map)\n else:\n folium.PolyLine(\n locations=points, color=\"#cc00ff\", opacity=opacity, weight=weight\n ).add_to(folium_map)\n logging.critical(\"Mapped activity with id: {}\".format(a.id))\n except Exception:\n logging.error(\"Could not map activity with id: {}\".format(a.id))\n \n return folium_map",
"_____no_output_____"
],
[
"m = map_activities(\n activities=activities,\n folium_map=m,\n opacity=0.5,\n weight=2\n)",
"_____no_output_____"
],
[
"m",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f283def778f15725c68cbd6bc07d3407aedf0c | 608,325 | ipynb | Jupyter Notebook | Data Science/Data_Preprocessing/Notebooks/INFO7390_Assignment_3_Mini_Project_One_Part_Two.ipynb | RushabhNisher/Data | a7479ec1cba6846d491ee9ec11ae5db1a7abff32 | [
"Unlicense"
] | 1 | 2021-04-12T00:32:40.000Z | 2021-04-12T00:32:40.000Z | Data Science/Data_Preprocessing/Notebooks/INFO7390_Assignment_3_Mini_Project_One_Part_Two.ipynb | RushabhNisher/Data | a7479ec1cba6846d491ee9ec11ae5db1a7abff32 | [
"Unlicense"
] | null | null | null | Data Science/Data_Preprocessing/Notebooks/INFO7390_Assignment_3_Mini_Project_One_Part_Two.ipynb | RushabhNisher/Data | a7479ec1cba6846d491ee9ec11ae5db1a7abff32 | [
"Unlicense"
] | null | null | null | 320.846519 | 133,452 | 0.917367 | [
[
[
"# <p style=\"text-align: center;\"> Part Two: Scaling & Normalization </p>",
"_____no_output_____"
]
],
[
[
"from IPython.display import HTML\nfrom IPython.display import Image\nImage(url= \"https://miro.medium.com/max/3316/1*yR54MSI1jjnf2QeGtt57PA.png\")",
"_____no_output_____"
],
[
"HTML('''<script>\ncode_show=true; \nfunction code_toggle() {\n if (code_show){\n $('div.input').hide();\n } else {\n $('div.input').show();\n }\n code_show = !code_show\n} \n$( document ).ready(code_toggle);\n</script>\nThe raw code for this IPython notebook is by default hidden for easier reading.\nTo toggle on/off the raw code, click <a href=\"javascript:code_toggle()\">here</a>.''')",
"_____no_output_____"
]
],
[
[
"# <p style=\"text-align: center;\"> Table of Contents </p>\n- ## 1. [Introduction](#Introduction)\n - ### 1.1 [Abstract](#abstract)\n - ### 1.2 [Importing Libraries](#importing_libraries)\n- ## 2. [Data Scaling](#data_scaling)\n - ### 2.1 [Standardization](#standardization)\n - ### 2.2 [Normalization](#normalization)\n - ### 2.3 [The Big Question – Normalize or Standardize?](#the_big_question)\n - ### 2.4 [Implementation](#implementation)\n - #### 2.4.1 [Original Distributions](#original_distributions)\n - #### 2.4.2 [Adding a Feature with Much Larger Values](#larger_values)\n - #### 2.4.3 [MinMaxScaler](#min_max_scaler)\n - #### 2.4.4 [StandardScaler](#standard_scaler)\n - #### 2.4.5 [RobustScaler](#robust_scaler)\n - #### 2.4.6 [Normalizer](#normalizer)\n - #### 2.4.7 [Combined Plot](#combined_plot)\n- ## 3. [Conclusion](#Conclusion)\n- ## 4. [Contribution](#Contribution)\n- ## 5. [Citation](#Citation)\n- ## 6. [License](#License)",
"_____no_output_____"
],
[
"# <p style=\"text-align: center;\"> 1.0 Introduction </p> <a id='Introduction'></a>\n\n# 1.1 Abstract <a id='abstract'></a>\n\nWelcome to the Data Cleaning\n\n[Back to top](#Introduction)",
"_____no_output_____"
],
[
"# 1.2 Importing Libraries <a id='importing_libraries'></a>\n\nThis is the official start to any Data Science or Machine Learning Project. A Python library is a reusable chunk of code that you may want to include in your programs/ projects. \nIn this step we import a few libraries that are required in our program. Some major libraries that are used are Numpy, Pandas, MatplotLib, Seaborn, Sklearn etc.\n\n[Back to top](#Introduction)",
"_____no_output_____"
]
],
[
[
"# modules we'll use\nimport pandas as pd\nimport numpy as np\n\n# for Box-Cox Transformation\nfrom scipy import stats\n\n# for min_max scaling\nfrom sklearn import preprocessing\nfrom mlxtend.preprocessing import minmax_scaling\n\n# plotting modules\nimport seaborn as sns\nimport matplotlib\nimport matplotlib.pyplot as plt\n\nfrom astropy.table import Table, Column\n\nimport warnings\nwarnings.filterwarnings('ignore')\n\n%matplotlib inline\nmatplotlib.style.use('ggplot')\n\nnp.random.seed(34)",
"_____no_output_____"
]
],
[
[
"# 2.0 Data Scaling <a id='data_scaling'></a>\n\n## Why Should we Use Feature Scaling?\nThe first question we need to address – why do we need to scale the variables in our dataset? Some machine learning algorithms are sensitive to feature scaling while others are virtually invariant to it. \nMachine learning models learn a mapping from input variables to an output variable. As such, the scale and distribution of the data drawn from the domain may be different for each variable. Input variables may have different units (e.g. feet, kilometers, and hours) that, in turn, may mean the variables have different scales.\n\n\n### Gradient Descent Based Algorithms\nMachine learning algorithms like linear regression, logistic regression, neural network, etc. that use gradient descent as an optimization technique require data to be scaled. Take a look at the formula for gradient descent below:\n\n\n\nThe presence of feature value X in the formula will affect the step size of the gradient descent. The difference in ranges of features will cause different step sizes for each feature. To ensure that the gradient descent moves smoothly towards the minima and that the steps for gradient descent are updated at the same rate for all the features, we scale the data before feeding it to the model.\n\n> Having features on a similar scale can help the gradient descent converge more quickly towards the minima.\n\n### Distance-Based Algorithms\nDistance algorithms like KNN, K-means, and SVM are most affected by the range of features. This is because behind the scenes they are using distances between data points to determine their similarity.\n\nFor example, let’s say we have data containing high school CGPA scores of students (ranging from 0 to 5) and their future incomes (in thousands Dollars):\n\n\n\nSince both the features have different scales, there is a chance that higher weightage is given to features with higher magnitude. This will impact the performance of the machine learning algorithm and obviously, we do not want our algorithm to be biassed towards one feature.\n\n> Therefore, we scale our data before employing a distance based algorithm so that all the features contribute equally to the result.\n\n\n\nThe effect of scaling is conspicuous when we compare the Euclidean distance between data points for students A and B, and between B and C, before and after scaling as shown below:\n\n\n\nScaling has brought both the features into the picture and the distances are now more comparable than they were before we applied scaling.\n\n### Tree-Based Algorithms\n\nTree-based algorithms, on the other hand, are fairly insensitive to the scale of the features. Think about it, a decision tree is only splitting a node based on a single feature. The decision tree splits a node on a feature that increases the homogeneity of the node. This split on a feature is not influenced by other features.\n\nSo, there is virtually no effect of the remaining features on the split. This is what makes them invariant to the scale of the features!\n\nOne of the reasons that it's easy to get confused between scaling and normalization is because the terms are sometimes used interchangeably and, to make it even more confusing, they are very similar! In both cases, you're transforming the values of numeric variables so that the transformed data points have specific helpful properties. \n\n\n[Back to top](#Introduction)",
"_____no_output_____"
],
[
"## 2.1 Standardization <a id='standardization'></a>\n\n**Scaling (Standardization):** Change in the range of your data.\n\nDifferences in the scales across input variables may increase the difficulty of the problem being modeled. A model with large weight values is often unstable, meaning that it may suffer from poor performance during learning and sensitivity to input values resulting in higher generalization error.\n\nThis means that you're transforming your data so that it fits within a specific scale, like 0-100 or 0-1. You want to scale data when you're using methods based on measures of how far apart data points are, like support vector machines (SVM) or k-nearest neighbors (KNN). With these algorithms, a change of \"1\" in any numeric feature is given the same importance.\n\nFor example, you might be looking at the prices of some products in both Yen and US Dollars. One US Dollar is worth about 100 Yen, but if you don't scale your prices, methods like SVM or KNN will consider a difference in price of 1 Yen as important as a difference of 1 US Dollar! This clearly doesn't fit with our intuitions of the world. With currency, you can convert between currencies. But what about if you're looking at something like height and weight? It's not entirely clear how many pounds should equal one inch (or how many kilograms should equal one meter).\n\nBy scaling your variables, you can help compare different variables on equal footing\n\nStandardization is scaling a technique where the values are centered around the mean with a unit standard deviation. This means that the mean of the attribute becomes zero and the resultant distribution has a unit standard deviation.\n\nHere’s the formula for standardization:\n\n\n\n- Mu is the mean of the feature values and \n- Sigma is the standard deviation of the feature values. Note that in this case, the values are not restricted to a particular range.\n\n[Back to top](#Introduction)",
"_____no_output_____"
]
],
[
[
"# generate 1000 data points randomly drawn from an exponential distribution\noriginal_data = np.random.exponential(size=1000)\n\n# mix-max scale the data between 0 and 1\nscaled_data = minmax_scaling(original_data, columns=[0])\n\n# plot both together to compare\nfig, ax = plt.subplots(1,2)\nsns.distplot(original_data, ax=ax[0])\nax[0].set_title(\"Original Data\")\nsns.distplot(scaled_data, ax=ax[1])\nax[1].set_title(\"Scaled data\")",
"_____no_output_____"
]
],
[
[
"## 2.2 Normalization <a id='normalization'></a>\n\n**Normalization:** Change in the shape of the distribution of data.\n\nNormalization scales each input variable separately to the range 0-1, which is the range for floating-point values where we have the most precision. Normalization requires that you know or are able to accurately estimate the minimum and maximum observable values. You may be able to estimate these values from your available data.\n\nScaling just changes the range of your data. Normalization is a more radical transformation. The point of normalization is to change your observations so that they can be described as a normal distribution.\n\nNormal distribution: Also known as the \"bell curve\", this is a specific statistical distribution where a roughly equal observations fall above and below the mean, the mean and the median are the same, and there are more observations closer to the mean. The normal distribution is also known as the Gaussian distribution.\n\nIn general, you'll normalize your data if you're going to be using a machine learning or statistics technique that assumes your data is normally distributed. Some examples of these include linear discriminant analysis (LDA) and Gaussian naive Bayes. (Pro tip: any method with \"Gaussian\" in the name probably assumes normality.)\n\nNormalization is a scaling technique in which values are shifted and rescaled so that they end up ranging between 0 and 1. It is also known as Min-Max scaling.\n\nHere’s the formula for normalization:\n\n\n\nHere, Xmax and Xmin are the maximum and the minimum values of the feature respectively.\n\n- When the value of X is the minimum value in the column, the numerator will be 0, and hence X’ is 0\n- On the other hand, when the value of X is the maximum value in the column, the numerator is equal to the denominator and thus the value of X’ is 1\n- If the value of X is between the minimum and the maximum value, then the value of X’ is between 0 and 1\n\n**PS:-** The method we're using to normalize here is called the Box-Cox Transformation. \n\n\nNow, the big question in your mind must be when should we use normalization and when should we use standardization? Let’s find out!\n\n[Back to top](#Introduction)",
"_____no_output_____"
]
],
[
[
"# normalize the exponential data with boxcox\nnormalized_data = stats.boxcox(original_data)\n\n# plot both together to compare\nfig, ax=plt.subplots(1,2)\nsns.distplot(original_data, ax=ax[0])\nax[0].set_title(\"Original Data\")\nsns.distplot(normalized_data[0], ax=ax[1])\nax[1].set_title(\"Normalized data\")",
"_____no_output_____"
]
],
[
[
"## 2.3 The Big Question – Normalize or Standardize? <a id='the_big_question'></a>\n\nNormalization vs. standardization is an eternal question among machine learning newcomers. Let me elaborate on the answer in this section.\n\n- Normalization is good to use when you know that the distribution of your data does not follow a Gaussian distribution. This can be useful in algorithms that do not assume any distribution of the data like K-Nearest Neighbors and Neural Networks.\n\n- Standardization, on the other hand, can be helpful in cases where the data follows a Gaussian distribution. However, this does not have to be necessarily true. Also, unlike normalization, standardization does not have a bounding range. So, even if you have outliers in your data, they will not be affected by standardization.\n\nHowever, at the end of the day, the choice of using normalization or standardization will depend on your problem and the machine learning algorithm you are using. There is no hard and fast rule to tell you when to normalize or standardize your data. You can always start by fitting your model to raw, normalized and standardized data and compare the performance for best results.\n\nIt is a good practice to fit the scaler on the training data and then use it to transform the testing data. This would avoid any data leakage during the model testing process. Also, the scaling of target values is generally not required.\n\n[Back to top](#Introduction)",
"_____no_output_____"
],
[
"## 2.4 Implementation <a id='implementation'></a>\n\nThis is all good in theory, but how do we implement it in real life. The sklearn library has various modules in the preprocessing section which implement these in different ways. The 4, that are most widely used and that we're going to implement here are:- \n\n- **MinMaxScalar:** The MinMaxScaler transforms features by scaling each feature to a given range. This range can be set by specifying the feature_range parameter (default at (0,1)). This scaler works better for cases where the distribution is not Gaussian or the standard deviation is very small. However, it is sensitive to outliers, so if there are outliers in the data, you might want to consider another scaler. \n> x_scaled = (x-min(x)) / (max(x)–min(x))\n- **StandardScaler:** Sklearn its main scaler, the StandardScaler, uses a strict definition of standardization to standardize data. It purely centers the data by using the following formula, where u is the mean and s is the standard deviation. \n> x_scaled = (x — u) / s\n\n- **RobustScalar:** If your data contains many outliers, scaling using the mean and standard deviation of the data is likely to not work very well. In these cases, you can use the RobustScaler. It removes the median and scales the data according to the quantile range. The exact formula of the RobustScaler is not specified by the documentation. By default, the scaler uses the Inter Quartile Range (IQR), which is the range between the 1st quartile and the 3rd quartile. The quantile range can be manually set by specifying the quantile_range parameter when initiating a new instance of the RobustScaler. \n\n\n- **Normalizer:** \n - **‘l1’:** The l1 norm uses the sum of all the values as and thus gives equal penalty to all parameters, enforcing sparsity.\n > x_normalized = x / sum(X)\n - **‘l2’:** The l2 norm uses the square root of the sum of all the squared values. This creates smoothness and rotational invariance. Some models, like PCA, assume rotational invariance, and so l2 will perform better.\n > x_normalized = x / sqrt(sum((i\\**2) for i in X))\n \n \n**`TLDR`**\n- Use MinMaxScaler as your default\n- Use RobustScaler if you have outliers and can handle a larger range\n- Use StandardScaler if you need normalized features\n- Use Normalizer sparingly - it normalizes rows, not columns\n\n[Back to top](#Introduction)",
"_____no_output_____"
],
[
"### 2.4.1 Original Distributions <a id='original_distributions'></a>\n\nLet's make several types of random distributions. We're doing this because when we deal with real world data, the data is not necessarily in a normal (Gaussian) distribution. Each type of scaling may have a different effect depending on the type of the distribution, thus we take examples of 5 different type of distributions here.\n\n- **Beta:** The Beta distribution is a probability distribution on probabilities.\n- **Exponential:** The exponential distribution is a probability distribution which represents the time between events in a Poisson process.\n- **Normal (Platykurtic):** The term \"platykurtic\" refers to a statistical distribution in which the excess kurtosis value is negative. For this reason, a platykurtic distribution will have thinner tails than a normal distribution, resulting in fewer extreme positive or negative events.\n- **Normal (Leptokurtic):** Leptokurtic distributions are statistical distributions with kurtosis over three. It is one of three major categories found in kurtosis analysis.\n- **Bimodal:** The bimodal distribution has two peaks. \n\n[Back to top](#Introduction)",
"_____no_output_____"
]
],
[
[
"#create columns of various distributions\ndf = pd.DataFrame({ \n 'beta': np.random.beta(5, 1, 1000) * 60, # beta\n 'exponential': np.random.exponential(10, 1000), # exponential\n 'normal_p': np.random.normal(10, 2, 1000), # normal platykurtic\n 'normal_l': np.random.normal(10, 10, 1000), # normal leptokurtic\n})\n\n# make bimodal distribution\nfirst_half = np.random.normal(20, 3, 500) \nsecond_half = np.random.normal(-20, 3, 500) \nbimodal = np.concatenate([first_half, second_half])\n\ndf['bimodal'] = bimodal\n\n# create list of column names to use later\ncol_names = list(df.columns)",
"_____no_output_____"
]
],
[
[
"After defining the distributions, lets visualize them",
"_____no_output_____"
]
],
[
[
"# plot original distribution plot\nfig, (ax1) = plt.subplots(ncols=1, figsize=(10, 8))\nax1.set_title('Original Distributions')\n\nsns.kdeplot(df['beta'], ax=ax1)\nsns.kdeplot(df['exponential'], ax=ax1)\nsns.kdeplot(df['normal_p'], ax=ax1)\nsns.kdeplot(df['normal_l'], ax=ax1)\nsns.kdeplot(df['bimodal'], ax=ax1);",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
],
[
"df.plot()",
"_____no_output_____"
]
],
[
[
"As we can clearly see from the statistics and the plots, all values are in the same ball park. But what happens if we disturb this by adding a feature with much larger values.",
"_____no_output_____"
],
[
"### 2.4.2 Adding a Feature with Much Larger Values <a id='larger_values'></a>\n\nThis feature could be home prices, for example.\n\n[Back to Top](#Introduction)",
"_____no_output_____"
]
],
[
[
"normal_big = np.random.normal(1000000, 10000, (1000,1)) # normal distribution of large values\ndf['normal_big'] = normal_big\ncol_names.append('normal_big')\ndf['normal_big'].plot(kind='kde')",
"_____no_output_____"
],
[
"df.normal_big.mean()",
"_____no_output_____"
]
],
[
[
"We've got a normalish distribution with a mean near 1,000,0000. But if we put this on the same plot as the original distributions, you can't even see the earlier columns.",
"_____no_output_____"
]
],
[
[
"# plot original distribution plot with larger value feature\nfig, (ax1) = plt.subplots(ncols=1, figsize=(10, 8))\nax1.set_title('Original Distributions')\n\nsns.kdeplot(df['beta'], ax=ax1)\nsns.kdeplot(df['exponential'], ax=ax1)\nsns.kdeplot(df['normal_p'], ax=ax1)\nsns.kdeplot(df['normal_l'], ax=ax1)\nsns.kdeplot(df['bimodal'], ax=ax1);\nsns.kdeplot(df['normal_big'], ax=ax1);",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
]
],
[
[
"The new, high-value distribution is way to the right. And here's a plot of the values.",
"_____no_output_____"
]
],
[
[
"df.plot()",
"_____no_output_____"
]
],
[
[
"### 2.4.3 MinMaxScaler <a id='min_max_scaler'></a>\n\nMinMaxScaler subtracts the column mean from each value and then divides by the range.\n\n[Back to Top](#Introduction)",
"_____no_output_____"
]
],
[
[
"mm_scaler = preprocessing.MinMaxScaler()\ndf_mm = mm_scaler.fit_transform(df)\n\ndf_mm = pd.DataFrame(df_mm, columns=col_names)\n\nfig, (ax1) = plt.subplots(ncols=1, figsize=(10, 8))\nax1.set_title('After MinMaxScaler')\n\nsns.kdeplot(df_mm['beta'], ax=ax1)\nsns.kdeplot(df_mm['exponential'], ax=ax1)\nsns.kdeplot(df_mm['normal_p'], ax=ax1)\nsns.kdeplot(df_mm['normal_l'], ax=ax1)\nsns.kdeplot(df_mm['bimodal'], ax=ax1)\nsns.kdeplot(df_mm['normal_big'], ax=ax1);",
"_____no_output_____"
],
[
"df_mm.describe()",
"_____no_output_____"
]
],
[
[
"Notice how the shape of each distribution remains the same, but now the values are between 0 and 1. Our feature with much larger values was brought into scale with our other features.",
"_____no_output_____"
],
[
"### 2.4.4 StandardScaler <a id='standard_scaler'></a>\n\nStandardScaler is scales each column to have 0 mean and unit variance.\n\n[Back to Top](#Introduction)",
"_____no_output_____"
]
],
[
[
"s_scaler = preprocessing.StandardScaler()\ndf_s = s_scaler.fit_transform(df)\n\ndf_s = pd.DataFrame(df_s, columns=col_names)\n\nfig, (ax1) = plt.subplots(ncols=1, figsize=(10, 8))\nax1.set_title('After StandardScaler')\n\nsns.kdeplot(df_s['beta'], ax=ax1)\nsns.kdeplot(df_s['exponential'], ax=ax1)\nsns.kdeplot(df_s['normal_p'], ax=ax1)\nsns.kdeplot(df_s['normal_l'], ax=ax1)\nsns.kdeplot(df_s['bimodal'], ax=ax1)\nsns.kdeplot(df_s['normal_big'], ax=ax1);",
"_____no_output_____"
]
],
[
[
"You can see that all features now have 0 mean.",
"_____no_output_____"
]
],
[
[
"df_s.describe()",
"_____no_output_____"
]
],
[
[
"### 2.4.5 RobustScaler <a id='robust_scaler'></a>\n\nRobustScaler subtracts the column median and divides by the interquartile range.\n\n[Back to Top](#Introduction)",
"_____no_output_____"
]
],
[
[
"r_scaler = preprocessing.RobustScaler()\ndf_r = r_scaler.fit_transform(df)\n\ndf_r = pd.DataFrame(df_r, columns=col_names)\n\nfig, (ax1) = plt.subplots(ncols=1, figsize=(10, 8))\nax1.set_title('After RobustScaler')\n\nsns.kdeplot(df_r['beta'], ax=ax1)\nsns.kdeplot(df_r['exponential'], ax=ax1)\nsns.kdeplot(df_r['normal_p'], ax=ax1)\nsns.kdeplot(df_r['normal_l'], ax=ax1)\nsns.kdeplot(df_r['bimodal'], ax=ax1)\nsns.kdeplot(df_r['normal_big'], ax=ax1);",
"_____no_output_____"
],
[
"df_r.describe()",
"_____no_output_____"
]
],
[
[
"Although the range of values for each feature is much smaller than for the original features, it's larger and varies more than for MinMaxScaler. The bimodal distribution values are now compressed into two small groups. Standard and RobustScalers have pretty much the same ranges.",
"_____no_output_____"
],
[
"### 2.4.6 Normalizer <a id='normalizer'></a>\n\nNote that normalizer operates on the rows, not the columns. It applies l2 normalization by default.\n\n[Back to Top](#Introduction)",
"_____no_output_____"
]
],
[
[
"n_scaler = preprocessing.Normalizer()\ndf_n = n_scaler.fit_transform(df)\n\ndf_n = pd.DataFrame(df_n, columns=col_names)\n\nfig, (ax1) = plt.subplots(ncols=1, figsize=(10, 8))\nax1.set_title('After Normalizer')\n\nsns.kdeplot(df_n['beta'], ax=ax1)\nsns.kdeplot(df_n['exponential'], ax=ax1)\nsns.kdeplot(df_n['normal_p'], ax=ax1)\nsns.kdeplot(df_n['normal_l'], ax=ax1)\nsns.kdeplot(df_n['bimodal'], ax=ax1)\nsns.kdeplot(df_n['normal_big'], ax=ax1);",
"_____no_output_____"
],
[
"df_n.describe()",
"_____no_output_____"
]
],
[
[
"Normalizer also moved the features to similar scales. Notice that the range for our much larger feature's values is now extremely small and clustered around .9999999999.",
"_____no_output_____"
],
[
"### 2.4.7 Combined Plot <a id='combined_plot'></a>\n\nLet's look at our original and transformed distributions together. We'll exclude Normalizer because you generally want to tranform your features, not your samples.\n\n[Back to Top](#Introduction)",
"_____no_output_____"
]
],
[
[
"# Combined plot.\n\nfig, (ax0, ax1, ax2, ax3) = plt.subplots(ncols=4, figsize=(20, 8))\n\n\nax0.set_title('Original Distributions')\n\nsns.kdeplot(df['beta'], ax=ax0)\nsns.kdeplot(df['exponential'], ax=ax0)\nsns.kdeplot(df['normal_p'], ax=ax0)\nsns.kdeplot(df['normal_l'], ax=ax0)\nsns.kdeplot(df['bimodal'], ax=ax0)\nsns.kdeplot(df['normal_big'], ax=ax0);\n\n\nax1.set_title('After MinMaxScaler')\n\nsns.kdeplot(df_mm['beta'], ax=ax1)\nsns.kdeplot(df_mm['exponential'], ax=ax1)\nsns.kdeplot(df_mm['normal_p'], ax=ax1)\nsns.kdeplot(df_mm['normal_l'], ax=ax1)\nsns.kdeplot(df_mm['bimodal'], ax=ax1)\nsns.kdeplot(df_mm['normal_big'], ax=ax1);\n\n\nax2.set_title('After RobustScaler')\n\nsns.kdeplot(df_r['beta'], ax=ax2)\nsns.kdeplot(df_r['exponential'], ax=ax2)\nsns.kdeplot(df_r['normal_p'], ax=ax2)\nsns.kdeplot(df_r['normal_l'], ax=ax2)\nsns.kdeplot(df_r['bimodal'], ax=ax2)\nsns.kdeplot(df_r['normal_big'], ax=ax2);\n\n\nax3.set_title('After StandardScaler')\n\nsns.kdeplot(df_s['beta'], ax=ax3)\nsns.kdeplot(df_s['exponential'], ax=ax3)\nsns.kdeplot(df_s['normal_p'], ax=ax3)\nsns.kdeplot(df_s['normal_l'], ax=ax3)\nsns.kdeplot(df_s['bimodal'], ax=ax3)\nsns.kdeplot(df_s['normal_big'], ax=ax3);",
"_____no_output_____"
]
],
[
[
"You can see that after any transformation the distributions are on a similar scale. Also notice that MinMaxScaler doesn't distort the distances between the values in each feature.",
"_____no_output_____"
],
[
"# <p style=\"text-align: center;\">Conclusion<p><a id='Conclusion'></a>\n \nWe have used various data Scaling and preprocessing techniques in this notebook. As listed below\n\n- Use MinMaxScaler as your default\n- Use RobustScaler if you have outliers and can handle a larger range\n- Use StandardScaler if you need normalized features\n- Use Normalizer sparingly - it normalizes rows, not columns\n\n[Back to top](#Introduction)",
"_____no_output_____"
],
[
"# <p style=\"text-align: center;\">Contribution<p><a id='Contribution'></a>\n\nThis was a fun project in which we explore the idea of Data cleaning and Data Preprocessing. We take inspiration from kaggle learning course and create our own notebook enhancing the same idea and supplementing it with our own contributions from our experiences and past projects.\n \n- Code by self : 65%\n- Code from external Sources : 35%\n\n[Back to top](#Introduction)",
"_____no_output_____"
],
[
"# <p style=\"text-align: center;\">Citation<p><a id='Citation'></a>\n\n- https://www.kaggle.com/alexisbcook/scaling-and-normalization\n- https://scikit-learn.org/stable/modules/preprocessing.html\n- https://www.analyticsvidhya.com/blog/2020/04/feature-scaling-machine-learning-normalization-standardization/\n- https://kharshit.github.io/blog/2018/03/23/scaling-vs-normalization\n- https://www.kaggle.com/discdiver/guide-to-scaling-and-standardizing\n- https://docs.google.com/spreadsheets/d/1woVi7wq13628HJ-tN6ApaRGVZ85OdmHsDBKLAf5ylaQ/edit#gid=0\n- https://towardsdatascience.com/preprocessing-with-sklearn-a-complete-and-comprehensive-guide-670cb98fcfb9\n- https://www.kaggle.com/rpsuraj/outlier-detection-techniques-simplified?select=insurance.csv\n- https://statisticsbyjim.com/basics/remove-outliers/\n- https://statisticsbyjim.com/basics/outliers/",
"_____no_output_____"
],
[
"# <p style=\"text-align: center;\">License<p><a id='License'></a>\nCopyright (c) 2020 Manali Sharma, Rushabh Nisher\n\nPermission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the \"Software\"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:\n\nThe above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.\n\nTHE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.\n\n[Back to top](#Introduction)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0f28429055f3f2e1455541e3cca716d7cf8eb2a | 3,748 | ipynb | Jupyter Notebook | tests/notebooks/mirror/script_to_ipynb/knitr-spin.ipynb | kiendang/jupytext | b36e07504db398cb922525a8514d863c563d8679 | [
"MIT"
] | null | null | null | tests/notebooks/mirror/script_to_ipynb/knitr-spin.ipynb | kiendang/jupytext | b36e07504db398cb922525a8514d863c563d8679 | [
"MIT"
] | null | null | null | tests/notebooks/mirror/script_to_ipynb/knitr-spin.ipynb | kiendang/jupytext | b36e07504db398cb922525a8514d863c563d8679 | [
"MIT"
] | null | null | null | 20.593407 | 88 | 0.522946 | [
[
[
"The below derives from\nhttps://github.com/yihui/knitr/blob/master/inst/examples/knitr-spin.R\n\nThis is a special R script which can be used to generate a report. You can\nwrite normal text in roxygen comments.\n\nFirst we set up some options (you do not have to do this):",
"_____no_output_____"
]
],
[
[
"library(knitr)\nopts_chunk$set(fig.path = 'figure/silk-')",
"_____no_output_____"
]
],
[
[
"The report begins here.",
"_____no_output_____"
]
],
[
[
"# boring examples as usual\nset.seed(123)\nx = rnorm(5)\nmean(x)",
"_____no_output_____"
]
],
[
[
"You can not use here the special syntax {{code}} to embed inline expressions, e.g.",
"_____no_output_____"
]
],
[
[
"{{mean(x) + 2}}",
"_____no_output_____"
]
],
[
[
"is the mean of x plus 2.\nThe code itself may contain braces, but these are not checked. Thus,\nperfectly valid (though very strange) R code such as `{{2 + 3}} - {{4 - 5}}`\ncan lead to errors because `2 + 3}} - {{4 - 5` will be treated as inline code.\n\nNow we continue writing the report. We can draw plots as well.",
"_____no_output_____"
]
],
[
[
"par(mar = c(4, 4, .1, .1)); plot(x)",
"_____no_output_____"
]
],
[
[
"Actually you do not have to write chunk options, in which case knitr will use\ndefault options. For example, the code below has no options attached:",
"_____no_output_____"
]
],
[
[
"var(x)\nquantile(x)",
"_____no_output_____"
]
],
[
[
"And you can also write two chunks successively like this:",
"_____no_output_____"
]
],
[
[
"sum(x^2) # chi-square distribution with df 5",
"_____no_output_____"
],
[
"sum((x - mean(x))^2) # df is 4 now",
"_____no_output_____"
]
],
[
[
"Done. Call spin('knitr-spin.R') to make silk from sow's ear now and knit a\nlovely purse.",
"_____no_output_____"
]
],
[
[
"# /* you can write comments between /* and */ like C comments (the preceding #\n# is optional)\nSys.sleep(60)\n# */",
"_____no_output_____"
],
[
"# /* there is no inline comment; you have to write block comments */",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0f28d700af9215093f2fc85ab6d93b59630f12d | 146,168 | ipynb | Jupyter Notebook | project_notebook.ipynb | classofai/Route-Planner | d831aeabe9b38f8102910a5e485aa5947c528e63 | [
"MIT"
] | null | null | null | project_notebook.ipynb | classofai/Route-Planner | d831aeabe9b38f8102910a5e485aa5947c528e63 | [
"MIT"
] | null | null | null | project_notebook.ipynb | classofai/Route-Planner | d831aeabe9b38f8102910a5e485aa5947c528e63 | [
"MIT"
] | null | null | null | 50.057534 | 15,208 | 0.634462 | [
[
[
"# Implementing a Route Planner\nIn this project you will use A\\* search to implement a \"Google-maps\" style route planning algorithm.",
"_____no_output_____"
],
[
"## The Map",
"_____no_output_____"
]
],
[
[
"# Run this cell first!\n\nfrom helpers import Map, load_map_10, load_map_40, show_map\nimport math\n\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"### Map Basics",
"_____no_output_____"
]
],
[
[
"map_10 = load_map_10()\nshow_map(map_10)",
"_____no_output_____"
]
],
[
[
"The map above (run the code cell if you don't see it) shows a disconnected network of 10 intersections. The two intersections on the left are connected to each other but they are not connected to the rest of the road network. This map is quite literal in its expression of distance and connectivity. On the graph above, the edge between 2 nodes(intersections) represents a literal straight road not just an abstract connection of 2 cities.\n\nThese `Map` objects have two properties you will want to use to implement A\\* search: `intersections` and `roads`\n\n**Intersections**\n\nThe `intersections` are represented as a dictionary. \n\nIn this example, there are 10 intersections, each identified by an x,y coordinate. The coordinates are listed below. You can hover over each dot in the map above to see the intersection number.",
"_____no_output_____"
],
[
"map_10.intersections",
"_____no_output_____"
],
[
"**Roads**\n\nThe `roads` property is a list where `roads[i]` contains a list of the intersections that intersection `i` connects to.",
"_____no_output_____"
]
],
[
[
"# this shows that intersection 0 connects to intersections 7, 6, and 5\nmap_10.roads[0] ",
"_____no_output_____"
],
[
"# This shows the full connectivity of the map\nmap_10.roads",
"_____no_output_____"
],
[
"# map_40 is a bigger map than map_10\nmap_40 = load_map_40()\nshow_map(map_40)",
"_____no_output_____"
]
],
[
[
"### Advanced Visualizations\n\nThe map above shows a network of roads which spans 40 different intersections (labeled 0 through 39). \n\nThe `show_map` function which generated this map also takes a few optional parameters which might be useful for visualizaing the output of the search algorithm you will write.\n\n* `start` - The \"start\" node for the search algorithm.\n* `goal` - The \"goal\" node.\n* `path` - An array of integers which corresponds to a valid sequence of intersection visits on the map.",
"_____no_output_____"
]
],
[
[
"# run this code, note the effect of including the optional\n# parameters in the function call.\nshow_map(map_40, start=5, goal=34, path=[5,16,37,12,34])",
"_____no_output_____"
]
],
[
[
"## The Algorithm\n### Writing your algorithm\nThe algorithm written will be responsible for generating a `path` like the one passed into `show_map` above. In fact, when called with the same map, start and goal, as above you algorithm should produce the path `[5, 16, 37, 12, 34]`. However you must complete several methods before it will work.\n\n```bash\n> PathPlanner(map_40, 5, 34).path\n[5, 16, 37, 12, 34]\n```",
"_____no_output_____"
]
],
[
[
"# Do not change this cell\n# When you write your methods correctly this cell will execute\n# without problems\nclass PathPlanner():\n \"\"\"Construct a PathPlanner Object\"\"\"\n def __init__(self, M, start=None, goal=None):\n \"\"\" \"\"\"\n self.map = M\n self.start= start\n self.goal = goal\n self.closedSet = self.create_closedSet() if goal != None and start != None else None\n self.openSet = self.create_openSet() if goal != None and start != None else None\n self.cameFrom = self.create_cameFrom() if goal != None and start != None else None\n self.gScore = self.create_gScore() if goal != None and start != None else None\n self.fScore = self.create_fScore() if goal != None and start != None else None\n self.path = self.run_search() if self.map and self.start != None and self.goal != None else None\n \n def get_path(self):\n \"\"\" Reconstructs path after search \"\"\"\n if self.path:\n return self.path \n else :\n self.run_search()\n return self.path\n \n def reconstruct_path(self, current):\n \"\"\" Reconstructs path after search \"\"\"\n total_path = [current]\n while current in self.cameFrom.keys():\n current = self.cameFrom[current]\n total_path.append(current)\n return total_path\n \n def _reset(self):\n \"\"\"Private method used to reset the closedSet, openSet, cameFrom, gScore, fScore, and path attributes\"\"\"\n self.closedSet = None\n self.openSet = None\n self.cameFrom = None\n self.gScore = None\n self.fScore = None\n self.path = self.run_search() if self.map and self.start and self.goal else None\n\n def run_search(self):\n \"\"\" \"\"\"\n if self.map == None:\n raise(ValueError, \"Must create map before running search. Try running PathPlanner.set_map(start_node)\")\n if self.goal == None:\n raise(ValueError, \"Must create goal node before running search. Try running PathPlanner.set_goal(start_node)\")\n if self.start == None:\n raise(ValueError, \"Must create start node before running search. Try running PathPlanner.set_start(start_node)\")\n\n self.closedSet = self.closedSet if self.closedSet != None else self.create_closedSet()\n self.openSet = self.openSet if self.openSet != None else self.create_openSet()\n self.cameFrom = self.cameFrom if self.cameFrom != None else self.create_cameFrom()\n self.gScore = self.gScore if self.gScore != None else self.create_gScore()\n self.fScore = self.fScore if self.fScore != None else self.create_fScore()\n\n while not self.is_open_empty():\n current = self.get_current_node()\n\n if current == self.goal:\n self.path = [x for x in reversed(self.reconstruct_path(current))]\n return self.path\n else:\n self.openSet.remove(current)\n self.closedSet.add(current)\n\n for neighbor in self.get_neighbors(current):\n if neighbor in self.closedSet:\n continue # Ignore the neighbor which is already evaluated.\n\n if not neighbor in self.openSet: # Discover a new node\n self.openSet.add(neighbor)\n \n # The distance from start to a neighbor\n #the \"dist_between\" function may vary as per the solution requirements.\n if self.get_tenative_gScore(current, neighbor) >= self.get_gScore(neighbor):\n continue # This is not a better path.\n\n # This path is the best until now. Record it!\n self.record_best_path_to(current, neighbor)\n print(\"No Path Found\")\n self.path = None\n return False",
"_____no_output_____"
]
],
[
[
"Create the following methods:",
"_____no_output_____"
]
],
[
[
"def create_closedSet(self):\n \"\"\" Creates and returns a data structure suitable to hold the set of nodes already evaluated\"\"\"\n # TODO: return a data structure suitable to hold the set of nodes already evaluated\n return set()",
"_____no_output_____"
],
[
"def create_openSet(self):\n \"\"\" Creates and returns a data structure suitable to hold the set of currently discovered nodes \n that are not evaluated yet. Initially, only the start node is known.\"\"\"\n if self.start != None:\n # TODO: return a data structure suitable to hold the set of currently discovered nodes \n # that are not evaluated yet. Make sure to include the start node.\n self.open_set = set()\n self.open_set.add(self.start)\n return self.open_set\n \n raise(ValueError, \"Must create start node before creating an open set. Try running PathPlanner.set_start(start_node)\")",
"_____no_output_____"
],
[
"def create_cameFrom(self):\n \"\"\"Creates and returns a data structure that shows which node can most efficiently be reached from another,\n for each node.\"\"\"\n # TODO: return a data structure that shows which node can most efficiently be reached from another,\n # for each node. \n self.come_from = {}\n return self.come_from",
"_____no_output_____"
],
[
"def create_gScore(self):\n \"\"\"Creates and returns a data structure that holds the cost of getting from the start node to that node, for each node.\n The cost of going from start to start is zero.\"\"\"\n # TODO: a data structure that holds the cost of getting from the start node to that node, for each node.\n # for each node. The cost of going from start to start is zero. The rest of the node's values should be set to infinity.\n self.gScore = {}\n nodes_index = len(self.map.roads)\n for node in range(nodes_index):\n if node == self.start:\n self.gScore[node] = 0\n continue\n self.gScore[node] = math.inf \n return self.gScore ",
"_____no_output_____"
],
[
"def create_fScore(self):\n \"\"\"Creates and returns a data structure that holds the total cost of getting from the start node to the goal\n by passing by that node, for each node. That value is partly known, partly heuristic.\n For the first node, that value is completely heuristic.\"\"\"\n # TODO: a data structure that holds the total cost of getting from the start node to the goal\n # by passing by that node, for each node. That value is partly known, partly heuristic.\n # For the first node, that value is completely heuristic. The rest of the node's value should be \n # set to infinity.\n self.fScore = {}\n nodes_index = len(self.map.roads)\n for node in range(nodes_index):\n if node == self.start:\n self.fScore[node] = heuristic_cost_estimate(self, self.start)\n continue\n self.fScore[node] = math.inf \n return self.fScore \n",
"_____no_output_____"
],
[
"def set_map(self, M):\n \"\"\"Method used to set map attribute \"\"\"\n self._reset(self)\n self.start = None\n self.goal = None\n # TODO: Set map to new value. \n self.map = M",
"_____no_output_____"
],
[
"def set_start(self, start):\n \"\"\"Method used to set start attribute \"\"\"\n self._reset(self)\n # TODO: Set start value. Remember to remove goal, closedSet, openSet, cameFrom, gScore, fScore, \n # and path attributes' values.\n self.start = start",
"_____no_output_____"
],
[
"def set_goal(self, goal):\n \"\"\"Method used to set goal attribute \"\"\"\n self._reset(self)\n # TODO: Set goal value. \n self.goal = goal",
"_____no_output_____"
],
[
"def get_current_node(self):\n \"\"\" Returns the node in the open set with the lowest value of f(node).\"\"\"\n # TODO: Return the node in the open set with the lowest value of f(node).\n current_node = {}\n for node in self.open_set:\n if node in self.fScore.keys():\n calculate_fscore(self, node)\n current_node[node] = self.fScore[node]\n current = min(current_node, key=current_node.get)\n return current ",
"_____no_output_____"
],
[
"def get_neighbors(self, node):\n \"\"\"Returns the neighbors of a node\"\"\"\n # TODO: Return the neighbors of a node\n return self.map.roads[node]",
"_____no_output_____"
],
[
"def get_gScore(self, node):\n \"\"\"Returns the g Score of a node\"\"\"\n # TODO: Return the g Score of a node\n return self.gScore[node]",
"_____no_output_____"
],
[
"def get_tenative_gScore(self, current, neighbor):\n \"\"\"Returns the tenative g Score of a node\"\"\"\n # TODO: Return the g Score of the current node \n # plus distance from the current node to it's neighbors\n tenative_gScore = self.gScore[current] + distance(self, current, neighbor)\n return tenative_gScore\n",
"_____no_output_____"
],
[
"def is_open_empty(self):\n \"\"\"returns True if the open set is empty. False otherwise. \"\"\"\n # TODO: Return True if the open set is empty. False otherwise.\n return len(self.open_set) == 0",
"_____no_output_____"
],
[
"def distance(self, node_1, node_2):\n \"\"\" Computes the Euclidean L2 Distance\"\"\"\n # TODO: Compute and return the Euclidean L2 Distance\n x1, y1 = self.map.intersections[node_1]\n x2, y2 = self.map.intersections[node_2]\n euclidian_dist = math.sqrt( pow((x2-x1),2) + pow((y2-y1),2))\n return euclidian_dist",
"_____no_output_____"
],
[
"def heuristic_cost_estimate(self, node):\n \"\"\" Returns the heuristic cost estimate of a node \"\"\"\n # TODO: Return the heuristic cost estimate of a node\n x1, y1 = self.map.intersections[node]\n x2, y2 = self.map.intersections[self.goal]\n heuristic_cost_node = math.sqrt( pow((x2-x1),2) + pow((y2-y1),2))\n return heuristic_cost_node",
"_____no_output_____"
],
[
"def calculate_fscore(self, node):\n \"\"\"Calculate the f score of a node. \"\"\"\n # TODO: Calculate and returns the f score of a node. \n # REMEMBER F = G + H\n self.gScore[node] = get_gScore(self, node)\n self.fScore[node] = self.gScore[node] + heuristic_cost_estimate(self, node)\n return self.fScore\n ",
"_____no_output_____"
],
[
"def record_best_path_to(self, current, neighbor):\n \"\"\"Record the best path to a node \"\"\"\n # TODO: Record the best path to a node, by updating cameFrom, gScore, and fScore\n self.come_from[neighbor] = current\n self.gScore[neighbor] = get_tenative_gScore(self, current, neighbor)\n self.fScore[neighbor] = self.gScore[neighbor] + heuristic_cost_estimate(self, neighbor)\n ",
"_____no_output_____"
],
[
"PathPlanner.create_closedSet = create_closedSet\nPathPlanner.create_openSet = create_openSet\nPathPlanner.create_cameFrom = create_cameFrom\nPathPlanner.create_gScore = create_gScore\nPathPlanner.create_fScore = create_fScore\n#PathPlanner._reset = _reset\nPathPlanner.set_map = set_map\nPathPlanner.set_start = set_start\nPathPlanner.set_goal = set_goal\nPathPlanner.get_current_node = get_current_node\nPathPlanner.get_neighbors = get_neighbors\nPathPlanner.get_gScore = get_gScore\nPathPlanner.get_tenative_gScore = get_tenative_gScore\nPathPlanner.is_open_empty = is_open_empty\nPathPlanner.distance = distance\nPathPlanner.heuristic_cost_estimate = heuristic_cost_estimate\nPathPlanner.calculate_fscore = calculate_fscore\nPathPlanner.record_best_path_to = record_best_path_to",
"_____no_output_____"
],
[
"planner = PathPlanner(map_40, 5, 34)\npath = planner.path\nif path == [5, 16, 37, 12, 34]:\n print(\"great! Your code works for these inputs!\")\nelse:\n print(\"something is off, your code produced the following:\")\n print(path)",
"great! Your code works for these inputs!\n"
]
],
[
[
"### Testing your Code\nIf the code below produces no errors, your algorithm is behaving correctly. You are almost ready to submit! Before you submit, go through the following submission checklist:\n\n**Submission Checklist**\n\n1. Does my code pass all tests?\n2. Does my code implement `A*` search and not some other search algorithm?\n3. Do I use an **admissible heuristic** to direct search efforts towards the goal?\n4. Do I use data structures which avoid unnecessarily slow lookups?\n\nWhen you can answer \"yes\" to all of these questions, submit by pressing the Submit button in the lower right!",
"_____no_output_____"
]
],
[
[
"from test import test\n\ntest(PathPlanner)",
"All tests pass! Congratulations!\n"
]
],
[
[
"## Questions\n\n**Instructions** Answer the following questions in your own words. We do not you expect you to know all of this knowledge on the top of your head. We expect you to do research and ask question. However do not merely copy and paste the answer from a google or stackoverflow. Read the information and understand it first. Then use your own words to explain the answer.",
"_____no_output_____"
],
[
"- How would you explain A-Star to a family member(layman)?\n\n** ANSWER **: \n\n A-star algorithm has a brain/extra knowledge which helps in making smart choice at each step and thereby leading to destination without exploring much unwanted paths",
"_____no_output_____"
],
[
"- How does A-Star search algorithm differ from Uniform cost search? What about Best First search?\n\n** ANSWER **: \n \n A-star algorithm has uses f which is sum of (each step cost(g) + estimated goal cost(h)), A-star has extra knowledge/information about goal. \n \n Uniform cost search it keep on exploring nodes in uniform way in each direction, which slows down search.\n \n Best First Search is like A-star without extra knowledge/brain, it keeps on exploring neighboring nodes with lowest cost until it leads to destination/goal.",
"_____no_output_____"
],
[
"- What is a heuristic?\n\n** ANSWER **:\n\n A heuristic is a estimated movement cost from given node to the goal, it's usually a smart guess which is always less which is always less than actual cost from given node to the goal node.",
"_____no_output_____"
],
[
"- What is a consistent heuristic?\n\n** ANSWER **:\n\n A heuristic is consistent if estimated cost from the current node to the goal is less than or equal to the the cost from the current node to a successor node, plus the estimated cost from the successor node to the goal",
"_____no_output_____"
],
[
"- What is a admissible heuristic? \n\n** ANSWER **:\n\n A heuristic is admissible if the estimated cost is never more than the actual cost from the current node to the goal node.\n i.e. A heuristic function is admissible if it never overestimates the distance to the goal.",
"_____no_output_____"
],
[
"- ___ admissible heuristic are consistent.\n*CHOOSE ONE*\n - All\n - Some\n - None\n \n** ANSWER **:\n \n Some",
"_____no_output_____"
],
[
"- ___ Consistent heuristic are admissible.\n*CHOOSE ONE*\n - All\n - Some\n - None\n \n** ANSWER **:\n\n All",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0f290aeb333407ccab31e75b4969878b4438b4d | 138,453 | ipynb | Jupyter Notebook | courses/dl1/lesson3-rossman.ipynb | jaskinn/fastai | c49bfa30468e8e5e10e19dd8b0363aee1c5b8744 | [
"Apache-2.0"
] | null | null | null | courses/dl1/lesson3-rossman.ipynb | jaskinn/fastai | c49bfa30468e8e5e10e19dd8b0363aee1c5b8744 | [
"Apache-2.0"
] | null | null | null | courses/dl1/lesson3-rossman.ipynb | jaskinn/fastai | c49bfa30468e8e5e10e19dd8b0363aee1c5b8744 | [
"Apache-2.0"
] | null | null | null | 34.423918 | 12,640 | 0.459318 | [
[
[
"# Structured and time series data",
"_____no_output_____"
],
[
"This notebook contains an implementation of the third place result in the Rossman Kaggle competition as detailed in Guo/Berkhahn's [Entity Embeddings of Categorical Variables](https://arxiv.org/abs/1604.06737).\n\nThe motivation behind exploring this architecture is it's relevance to real-world application. Most data used for decision making day-to-day in industry is structured and/or time-series data. Here we explore the end-to-end process of using neural networks with practical structured data problems.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n%reload_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"from fastai.structured import *\nfrom fastai.column_data import *\nnp.set_printoptions(threshold=50, edgeitems=20)\n\nPATH='data/rossmann/'",
"_____no_output_____"
]
],
[
[
"## Create datasets",
"_____no_output_____"
],
[
"In addition to the provided data, we will be using external datasets put together by participants in the Kaggle competition. You can download all of them [here](http://files.fast.ai/part2/lesson14/rossmann.tgz).\n\nFor completeness, the implementation used to put them together is included below.",
"_____no_output_____"
]
],
[
[
"def concat_csvs(dirname):\n path = f'{PATH}{dirname}'\n filenames=glob(f\"{PATH}/*.csv\")\n\n wrote_header = False\n with open(f\"{path}.csv\",\"w\") as outputfile:\n for filename in filenames:\n name = filename.split(\".\")[0]\n with open(filename) as f:\n line = f.readline()\n if not wrote_header:\n wrote_header = True\n outputfile.write(\"file,\"+line)\n for line in f:\n outputfile.write(name + \",\" + line)\n outputfile.write(\"\\n\")",
"_____no_output_____"
],
[
"# concat_csvs('googletrend')\n# concat_csvs('weather')",
"_____no_output_____"
]
],
[
[
"Feature Space:\n* train: Training set provided by competition\n* store: List of stores\n* store_states: mapping of store to the German state they are in\n* List of German state names\n* googletrend: trend of certain google keywords over time, found by users to correlate well w/ given data\n* weather: weather\n* test: testing set",
"_____no_output_____"
]
],
[
[
"table_names = ['train', 'store', 'store_states', 'state_names', \n 'googletrend', 'weather', 'test']",
"_____no_output_____"
]
],
[
[
"We'll be using the popular data manipulation framework `pandas`. Among other things, pandas allows you to manipulate tables/data frames in python as one would in a database.\n\nWe're going to go ahead and load all of our csv's as dataframes into the list `tables`.",
"_____no_output_____"
]
],
[
[
"tables = [pd.read_csv(f'{PATH}{fname}.csv', low_memory=False) for fname in table_names]",
"_____no_output_____"
],
[
"from IPython.display import HTML",
"_____no_output_____"
]
],
[
[
"We can use `head()` to get a quick look at the contents of each table:\n* train: Contains store information on a daily basis, tracks things like sales, customers, whether that day was a holdiay, etc.\n* store: general info about the store including competition, etc.\n* store_states: maps store to state it is in\n* state_names: Maps state abbreviations to names\n* googletrend: trend data for particular week/state\n* weather: weather conditions for each state\n* test: Same as training table, w/o sales and customers\n",
"_____no_output_____"
]
],
[
[
"for t in tables: display(t.head())",
"_____no_output_____"
]
],
[
[
"This is very representative of a typical industry dataset.\n\nThe following returns summarized aggregate information to each table accross each field.",
"_____no_output_____"
]
],
[
[
"for t in tables: display(DataFrameSummary(t).summary())",
"_____no_output_____"
]
],
[
[
"## Data Cleaning / Feature Engineering",
"_____no_output_____"
],
[
"As a structured data problem, we necessarily have to go through all the cleaning and feature engineering, even though we're using a neural network.",
"_____no_output_____"
]
],
[
[
"train, store, store_states, state_names, googletrend, weather, test = tables",
"_____no_output_____"
],
[
"len(train),len(test)",
"_____no_output_____"
]
],
[
[
"We turn state Holidays to booleans, to make them more convenient for modeling. We can do calculations on pandas fields using notation very similar (often identical) to numpy.",
"_____no_output_____"
]
],
[
[
"train.StateHoliday = train.StateHoliday!='0'\ntest.StateHoliday = test.StateHoliday!='0'",
"_____no_output_____"
]
],
[
[
"`join_df` is a function for joining tables on specific fields. By default, we'll be doing a left outer join of `right` on the `left` argument using the given fields for each table.\n\nPandas does joins using the `merge` method. The `suffixes` argument describes the naming convention for duplicate fields. We've elected to leave the duplicate field names on the left untouched, and append a \"\\_y\" to those on the right.",
"_____no_output_____"
]
],
[
[
"def join_df(left, right, left_on, right_on=None, suffix='_y'):\n if right_on is None: right_on = left_on\n return left.merge(right, how='left', left_on=left_on, right_on=right_on, \n suffixes=(\"\", suffix))",
"_____no_output_____"
]
],
[
[
"Join weather/state names.",
"_____no_output_____"
]
],
[
[
"weather = join_df(weather, state_names, \"file\", \"StateName\")",
"_____no_output_____"
]
],
[
[
"In pandas you can add new columns to a dataframe by simply defining it. We'll do this for googletrends by extracting dates and state names from the given data and adding those columns.\n\nWe're also going to replace all instances of state name 'NI' to match the usage in the rest of the data: 'HB,NI'. This is a good opportunity to highlight pandas indexing. We can use `.loc[rows, cols]` to select a list of rows and a list of columns from the dataframe. In this case, we're selecting rows w/ statename 'NI' by using a boolean list `googletrend.State=='NI'` and selecting \"State\".",
"_____no_output_____"
]
],
[
[
"googletrend['Date'] = googletrend.week.str.split(' - ', expand=True)[0]\ngoogletrend['State'] = googletrend.file.str.split('_', expand=True)[2]\ngoogletrend.loc[googletrend.State=='NI', \"State\"] = 'HB,NI'",
"_____no_output_____"
]
],
[
[
"The following extracts particular date fields from a complete datetime for the purpose of constructing categoricals.\n\nYou should *always* consider this feature extraction step when working with date-time. Without expanding your date-time into these additional fields, you can't capture any trend/cyclical behavior as a function of time at any of these granularities. We'll add to every table with a date field.",
"_____no_output_____"
]
],
[
[
"add_datepart(weather, \"Date\", drop=False)\nadd_datepart(googletrend, \"Date\", drop=False)\nadd_datepart(train, \"Date\", drop=False)\nadd_datepart(test, \"Date\", drop=False)",
"_____no_output_____"
]
],
[
[
"The Google trends data has a special category for the whole of the Germany - we'll pull that out so we can use it explicitly.",
"_____no_output_____"
]
],
[
[
"trend_de = googletrend[googletrend.file == 'Rossmann_DE']",
"_____no_output_____"
]
],
[
[
"Now we can outer join all of our data into a single dataframe. Recall that in outer joins everytime a value in the joining field on the left table does not have a corresponding value on the right table, the corresponding row in the new table has Null values for all right table fields. One way to check that all records are consistent and complete is to check for Null values post-join, as we do here.\n\n*Aside*: Why note just do an inner join?\nIf you are assuming that all records are complete and match on the field you desire, an inner join will do the same thing as an outer join. However, in the event you are wrong or a mistake is made, an outer join followed by a null-check will catch it. (Comparing before/after # of rows for inner join is equivalent, but requires keeping track of before/after row #'s. Outer join is easier.)",
"_____no_output_____"
]
],
[
[
"store = join_df(store, store_states, \"Store\")\nlen(store[store.State.isnull()])",
"_____no_output_____"
],
[
"joined = join_df(train, store, \"Store\")\njoined_test = join_df(test, store, \"Store\")\nlen(joined[joined.StoreType.isnull()]),len(joined_test[joined_test.StoreType.isnull()])",
"_____no_output_____"
],
[
"joined = join_df(joined, googletrend, [\"State\",\"Year\", \"Week\"])\njoined_test = join_df(joined_test, googletrend, [\"State\",\"Year\", \"Week\"])\nlen(joined[joined.trend.isnull()]),len(joined_test[joined_test.trend.isnull()])",
"_____no_output_____"
],
[
"joined = joined.merge(trend_de, 'left', [\"Year\", \"Week\"], suffixes=('', '_DE'))\njoined_test = joined_test.merge(trend_de, 'left', [\"Year\", \"Week\"], suffixes=('', '_DE'))\nlen(joined[joined.trend_DE.isnull()]),len(joined_test[joined_test.trend_DE.isnull()])",
"_____no_output_____"
],
[
"joined = join_df(joined, weather, [\"State\",\"Date\"])\njoined_test = join_df(joined_test, weather, [\"State\",\"Date\"])\nlen(joined[joined.Mean_TemperatureC.isnull()]),len(joined_test[joined_test.Mean_TemperatureC.isnull()])",
"_____no_output_____"
],
[
"for df in (joined, joined_test):\n for c in df.columns:\n if c.endswith('_y'):\n if c in df.columns: df.drop(c, inplace=True, axis=1)",
"_____no_output_____"
]
],
[
[
"Next we'll fill in missing values to avoid complications with `NA`'s. `NA` (not available) is how Pandas indicates missing values; many models have problems when missing values are present, so it's always important to think about how to deal with them. In these cases, we are picking an arbitrary *signal value* that doesn't otherwise appear in the data.",
"_____no_output_____"
]
],
[
[
"for df in (joined,joined_test):\n df['CompetitionOpenSinceYear'] = df.CompetitionOpenSinceYear.fillna(1900).astype(np.int32)\n df['CompetitionOpenSinceMonth'] = df.CompetitionOpenSinceMonth.fillna(1).astype(np.int32)\n df['Promo2SinceYear'] = df.Promo2SinceYear.fillna(1900).astype(np.int32)\n df['Promo2SinceWeek'] = df.Promo2SinceWeek.fillna(1).astype(np.int32)",
"_____no_output_____"
]
],
[
[
"Next we'll extract features \"CompetitionOpenSince\" and \"CompetitionDaysOpen\". Note the use of `apply()` in mapping a function across dataframe values.",
"_____no_output_____"
]
],
[
[
"for df in (joined,joined_test):\n df[\"CompetitionOpenSince\"] = pd.to_datetime(dict(year=df.CompetitionOpenSinceYear, \n month=df.CompetitionOpenSinceMonth, day=15))\n df[\"CompetitionDaysOpen\"] = df.Date.subtract(df.CompetitionOpenSince).dt.days",
"_____no_output_____"
]
],
[
[
"We'll replace some erroneous / outlying data.",
"_____no_output_____"
]
],
[
[
"for df in (joined,joined_test):\n df.loc[df.CompetitionDaysOpen<0, \"CompetitionDaysOpen\"] = 0\n df.loc[df.CompetitionOpenSinceYear<1990, \"CompetitionDaysOpen\"] = 0",
"_____no_output_____"
]
],
[
[
"We add \"CompetitionMonthsOpen\" field, limiting the maximum to 2 years to limit number of unique categories.",
"_____no_output_____"
]
],
[
[
"for df in (joined,joined_test):\n df[\"CompetitionMonthsOpen\"] = df[\"CompetitionDaysOpen\"]//30\n df.loc[df.CompetitionMonthsOpen>24, \"CompetitionMonthsOpen\"] = 24\njoined.CompetitionMonthsOpen.unique()",
"_____no_output_____"
]
],
[
[
"Same process for Promo dates.",
"_____no_output_____"
]
],
[
[
"for df in (joined,joined_test):\n df[\"Promo2Since\"] = pd.to_datetime(df.apply(lambda x: Week(\n x.Promo2SinceYear, x.Promo2SinceWeek).monday(), axis=1).astype(pd.datetime))\n df[\"Promo2Days\"] = df.Date.subtract(df[\"Promo2Since\"]).dt.days",
"_____no_output_____"
],
[
"for df in (joined,joined_test):\n df.loc[df.Promo2Days<0, \"Promo2Days\"] = 0\n df.loc[df.Promo2SinceYear<1990, \"Promo2Days\"] = 0\n df[\"Promo2Weeks\"] = df[\"Promo2Days\"]//7\n df.loc[df.Promo2Weeks<0, \"Promo2Weeks\"] = 0\n df.loc[df.Promo2Weeks>25, \"Promo2Weeks\"] = 25\n df.Promo2Weeks.unique()",
"_____no_output_____"
],
[
"joined.to_feather(f'{PATH}joined')\njoined_test.to_feather(f'{PATH}joined_test')",
"_____no_output_____"
]
],
[
[
"## Durations",
"_____no_output_____"
],
[
"It is common when working with time series data to extract data that explains relationships across rows as opposed to columns, e.g.:\n* Running averages\n* Time until next event\n* Time since last event\n\nThis is often difficult to do with most table manipulation frameworks, since they are designed to work with relationships across columns. As such, we've created a class to handle this type of data.\n\nWe'll define a function `get_elapsed` for cumulative counting across a sorted dataframe. Given a particular field `fld` to monitor, this function will start tracking time since the last occurrence of that field. When the field is seen again, the counter is set to zero.\n\nUpon initialization, this will result in datetime na's until the field is encountered. This is reset every time a new store is seen. We'll see how to use this shortly.",
"_____no_output_____"
]
],
[
[
"def get_elapsed(fld, pre):\n day1 = np.timedelta64(1, 'D')\n last_date = np.datetime64()\n last_store = 0\n res = []\n\n for s,v,d in zip(df.Store.values,df[fld].values, df.Date.values):\n if s != last_store:\n last_date = np.datetime64()\n last_store = s\n if v: last_date = d\n res.append(((d-last_date).astype('timedelta64[D]') / day1))\n df[pre+fld] = res",
"_____no_output_____"
]
],
[
[
"We'll be applying this to a subset of columns:",
"_____no_output_____"
]
],
[
[
"columns = [\"Date\", \"Store\", \"Promo\", \"StateHoliday\", \"SchoolHoliday\"]",
"_____no_output_____"
],
[
"df = train[columns]",
"_____no_output_____"
],
[
"df = test[columns]",
"_____no_output_____"
]
],
[
[
"Let's walk through an example.\n\nSay we're looking at School Holiday. We'll first sort by Store, then Date, and then call `add_elapsed('SchoolHoliday', 'After')`:\nThis will apply to each row with School Holiday:\n* A applied to every row of the dataframe in order of store and date\n* Will add to the dataframe the days since seeing a School Holiday\n* If we sort in the other direction, this will count the days until another holiday.",
"_____no_output_____"
]
],
[
[
"fld = 'SchoolHoliday'\ndf = df.sort_values(['Store', 'Date'])\nget_elapsed(fld, 'After')\ndf = df.sort_values(['Store', 'Date'], ascending=[True, False])\nget_elapsed(fld, 'Before')",
"_____no_output_____"
]
],
[
[
"We'll do this for two more fields.",
"_____no_output_____"
]
],
[
[
"fld = 'StateHoliday'\ndf = df.sort_values(['Store', 'Date'])\nget_elapsed(fld, 'After')\ndf = df.sort_values(['Store', 'Date'], ascending=[True, False])\nget_elapsed(fld, 'Before')",
"_____no_output_____"
],
[
"fld = 'Promo'\ndf = df.sort_values(['Store', 'Date'])\nget_elapsed(fld, 'After')\ndf = df.sort_values(['Store', 'Date'], ascending=[True, False])\nget_elapsed(fld, 'Before')",
"_____no_output_____"
]
],
[
[
"We're going to set the active index to Date.",
"_____no_output_____"
]
],
[
[
"df = df.set_index(\"Date\")",
"_____no_output_____"
]
],
[
[
"Then set null values from elapsed field calculations to 0.",
"_____no_output_____"
]
],
[
[
"columns = ['SchoolHoliday', 'StateHoliday', 'Promo']",
"_____no_output_____"
],
[
"for o in ['Before', 'After']:\n for p in columns:\n a = o+p\n df[a] = df[a].fillna(0).astype(int)",
"_____no_output_____"
]
],
[
[
"Next we'll demonstrate window functions in pandas to calculate rolling quantities.\n\nHere we're sorting by date (`sort_index()`) and counting the number of events of interest (`sum()`) defined in `columns` in the following week (`rolling()`), grouped by Store (`groupby()`). We do the same in the opposite direction.",
"_____no_output_____"
]
],
[
[
"bwd = df[['Store']+columns].sort_index().groupby(\"Store\").rolling(7, min_periods=1).sum()",
"_____no_output_____"
],
[
"fwd = df[['Store']+columns].sort_index(ascending=False\n ).groupby(\"Store\").rolling(7, min_periods=1).sum()",
"_____no_output_____"
]
],
[
[
"Next we want to drop the Store indices grouped together in the window function.\n\nOften in pandas, there is an option to do this in place. This is time and memory efficient when working with large datasets.",
"_____no_output_____"
]
],
[
[
"bwd.drop('Store',1,inplace=True)\nbwd.reset_index(inplace=True)",
"_____no_output_____"
],
[
"fwd.drop('Store',1,inplace=True)\nfwd.reset_index(inplace=True)",
"_____no_output_____"
],
[
"df.reset_index(inplace=True)",
"_____no_output_____"
]
],
[
[
"Now we'll merge these values onto the df.",
"_____no_output_____"
]
],
[
[
"df = df.merge(bwd, 'left', ['Date', 'Store'], suffixes=['', '_bw'])\ndf = df.merge(fwd, 'left', ['Date', 'Store'], suffixes=['', '_fw'])",
"_____no_output_____"
],
[
"df.drop(columns,1,inplace=True)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"It's usually a good idea to back up large tables of extracted / wrangled features before you join them onto another one, that way you can go back to it easily if you need to make changes to it.",
"_____no_output_____"
]
],
[
[
"df.to_feather(f'{PATH}df')",
"_____no_output_____"
],
[
"df = pd.read_feather(f'{PATH}df')",
"_____no_output_____"
],
[
"df[\"Date\"] = pd.to_datetime(df.Date)",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"joined = join_df(joined, df, ['Store', 'Date'])",
"_____no_output_____"
],
[
"joined_test = join_df(joined_test, df, ['Store', 'Date'])",
"_____no_output_____"
]
],
[
[
"The authors also removed all instances where the store had zero sale / was closed. We speculate that this may have cost them a higher standing in the competition. One reason this may be the case is that a little exploratory data analysis reveals that there are often periods where stores are closed, typically for refurbishment. Before and after these periods, there are naturally spikes in sales that one might expect. By ommitting this data from their training, the authors gave up the ability to leverage information about these periods to predict this otherwise volatile behavior.",
"_____no_output_____"
]
],
[
[
"joined = joined[joined.Sales!=0]",
"_____no_output_____"
]
],
[
[
"We'll back this up as well.",
"_____no_output_____"
]
],
[
[
"joined.reset_index(inplace=True)\njoined_test.reset_index(inplace=True)",
"_____no_output_____"
],
[
"joined.to_feather(f'{PATH}joined')\njoined_test.to_feather(f'{PATH}joined_test')",
"_____no_output_____"
]
],
[
[
"We now have our final set of engineered features.\n\nWhile these steps were explicitly outlined in the paper, these are all fairly typical feature engineering steps for dealing with time series data and are practical in any similar setting.",
"_____no_output_____"
],
[
"## Create features",
"_____no_output_____"
]
],
[
[
"joined = pd.read_feather(f'{PATH}joined')\njoined_test = pd.read_feather(f'{PATH}joined_test')",
"_____no_output_____"
],
[
"joined.head().T.head(40)",
"_____no_output_____"
]
],
[
[
"Now that we've engineered all our features, we need to convert to input compatible with a neural network.\n\nThis includes converting categorical variables into contiguous integers or one-hot encodings, normalizing continuous features to standard normal, etc...",
"_____no_output_____"
]
],
[
[
"cat_vars = ['Store', 'DayOfWeek', 'Year', 'Month', 'Day', 'StateHoliday', 'CompetitionMonthsOpen',\n 'Promo2Weeks', 'StoreType', 'Assortment', 'PromoInterval', 'CompetitionOpenSinceYear', 'Promo2SinceYear',\n 'State', 'Week', 'Events', 'Promo_fw', 'Promo_bw', 'StateHoliday_fw', 'StateHoliday_bw',\n 'SchoolHoliday_fw', 'SchoolHoliday_bw']\n\ncontin_vars = ['CompetitionDistance', 'Max_TemperatureC', 'Mean_TemperatureC', 'Min_TemperatureC',\n 'Max_Humidity', 'Mean_Humidity', 'Min_Humidity', 'Max_Wind_SpeedKm_h', \n 'Mean_Wind_SpeedKm_h', 'CloudCover', 'trend', 'trend_DE',\n 'AfterStateHoliday', 'BeforeStateHoliday', 'Promo', 'SchoolHoliday']\n\nn = len(joined); n",
"_____no_output_____"
],
[
"dep = 'Sales'\njoined = joined[cat_vars+contin_vars+[dep, 'Date']].copy()",
"_____no_output_____"
],
[
"joined_test[dep] = 0\njoined_test = joined_test[cat_vars+contin_vars+[dep, 'Date', 'Id']].copy()",
"_____no_output_____"
],
[
"for v in cat_vars: joined[v] = joined[v].astype('category').cat.as_ordered()",
"_____no_output_____"
],
[
"apply_cats(joined_test, joined)",
"_____no_output_____"
],
[
"for v in contin_vars:\n joined[v] = joined[v].fillna(0).astype('float32')\n joined_test[v] = joined_test[v].fillna(0).astype('float32')",
"_____no_output_____"
]
],
[
[
"We're going to run on a sample.",
"_____no_output_____"
]
],
[
[
"idxs = get_cv_idxs(n, val_pct=150000/n)\njoined_samp = joined.iloc[idxs].set_index(\"Date\")\nsamp_size = len(joined_samp); samp_size",
"_____no_output_____"
]
],
[
[
"To run on the full dataset, use this instead:",
"_____no_output_____"
]
],
[
[
"samp_size = n\njoined_samp = joined.set_index(\"Date\")",
"_____no_output_____"
]
],
[
[
"We can now process our data...",
"_____no_output_____"
]
],
[
[
"joined_samp.head(2)",
"_____no_output_____"
],
[
"df, y, nas, mapper = proc_df(joined_samp, 'Sales', do_scale=True)\nyl = np.log(y)",
"_____no_output_____"
],
[
"joined_test = joined_test.set_index(\"Date\")",
"_____no_output_____"
],
[
"df_test, _, nas, mapper = proc_df(joined_test, 'Sales', do_scale=True, skip_flds=['Id'],\n mapper=mapper, na_dict=nas)",
"_____no_output_____"
],
[
"df.head(2)",
"_____no_output_____"
]
],
[
[
"In time series data, cross-validation is not random. Instead, our holdout data is generally the most recent data, as it would be in real application. This issue is discussed in detail in [this post](http://www.fast.ai/2017/11/13/validation-sets/) on our web site.\n\nOne approach is to take the last 25% of rows (sorted by date) as our validation set.",
"_____no_output_____"
]
],
[
[
"train_ratio = 0.75\n# train_ratio = 0.9\ntrain_size = int(samp_size * train_ratio); train_size\nval_idx = list(range(train_size, len(df)))",
"_____no_output_____"
]
],
[
[
"An even better option for picking a validation set is using the exact same length of time period as the test set uses - this is implemented here:",
"_____no_output_____"
]
],
[
[
"val_idx = np.flatnonzero(\n (df.index<=datetime.datetime(2014,9,17)) & (df.index>=datetime.datetime(2014,8,1)))",
"_____no_output_____"
],
[
"val_idx=[0]",
"_____no_output_____"
]
],
[
[
"## DL",
"_____no_output_____"
],
[
"We're ready to put together our models.\n\nRoot-mean-squared percent error is the metric Kaggle used for this competition.",
"_____no_output_____"
]
],
[
[
"def inv_y(a): return np.exp(a)\n\ndef exp_rmspe(y_pred, targ):\n targ = inv_y(targ)\n pct_var = (targ - inv_y(y_pred))/targ\n return math.sqrt((pct_var**2).mean())\n\nmax_log_y = np.max(yl)\ny_range = (0, max_log_y*1.2)",
"_____no_output_____"
]
],
[
[
"We can create a ModelData object directly from out data frame.",
"_____no_output_____"
]
],
[
[
"md = ColumnarModelData.from_data_frame(PATH, val_idx, df, yl.astype(np.float32), cat_flds=cat_vars, bs=128,\n test_df=df_test)",
"_____no_output_____"
]
],
[
[
"Some categorical variables have a lot more levels than others. Store, in particular, has over a thousand!",
"_____no_output_____"
]
],
[
[
"cat_sz = [(c, len(joined_samp[c].cat.categories)+1) for c in cat_vars]",
"_____no_output_____"
],
[
"cat_sz",
"_____no_output_____"
]
],
[
[
"We use the *cardinality* of each variable (that is, its number of unique values) to decide how large to make its *embeddings*. Each level will be associated with a vector with length defined as below.",
"_____no_output_____"
]
],
[
[
"emb_szs = [(c, min(50, (c+1)//2)) for _,c in cat_sz]",
"_____no_output_____"
],
[
"emb_szs",
"_____no_output_____"
],
[
"m = md.get_learner(emb_szs, len(df.columns)-len(cat_vars),\n 0.04, 1, [1000,500], [0.001,0.01], y_range=y_range)\nlr = 1e-3",
"_____no_output_____"
],
[
"m.lr_find()",
"_____no_output_____"
],
[
"m.sched.plot(100)",
"_____no_output_____"
]
],
[
[
"### Sample",
"_____no_output_____"
]
],
[
[
"m = md.get_learner(emb_szs, len(df.columns)-len(cat_vars),\n 0.04, 1, [1000,500], [0.001,0.01], y_range=y_range)\nlr = 1e-3",
"_____no_output_____"
],
[
"m.fit(lr, 3, metrics=[exp_rmspe])",
"_____no_output_____"
],
[
"m.fit(lr, 5, metrics=[exp_rmspe], cycle_len=1)",
"_____no_output_____"
],
[
"m.fit(lr, 2, metrics=[exp_rmspe], cycle_len=4)",
"_____no_output_____"
]
],
[
[
"### All",
"_____no_output_____"
]
],
[
[
"m = md.get_learner(emb_szs, len(df.columns)-len(cat_vars),\n 0.04, 1, [1000,500], [0.001,0.01], y_range=y_range)\nlr = 1e-3",
"_____no_output_____"
],
[
"m.fit(lr, 1, metrics=[exp_rmspe])",
"_____no_output_____"
],
[
"m.fit(lr, 3, metrics=[exp_rmspe])",
"_____no_output_____"
],
[
"m.fit(lr, 3, metrics=[exp_rmspe], cycle_len=1)",
"_____no_output_____"
]
],
[
[
"### Test",
"_____no_output_____"
]
],
[
[
"m = md.get_learner(emb_szs, len(df.columns)-len(cat_vars),\n 0.04, 1, [1000,500], [0.001,0.01], y_range=y_range)\nlr = 1e-3",
"_____no_output_____"
],
[
"m.fit(lr, 3, metrics=[exp_rmspe])",
"_____no_output_____"
],
[
"m.fit(lr, 3, metrics=[exp_rmspe], cycle_len=1)",
"_____no_output_____"
],
[
"m.save('val0')",
"_____no_output_____"
],
[
"m.load('val0')",
"_____no_output_____"
],
[
"x,y=m.predict_with_targs()",
"_____no_output_____"
],
[
"exp_rmspe(x,y)",
"_____no_output_____"
],
[
"pred_test=m.predict(True)",
"_____no_output_____"
],
[
"pred_test = np.exp(pred_test)",
"_____no_output_____"
],
[
"joined_test['Sales']=pred_test",
"_____no_output_____"
],
[
"csv_fn=f'{PATH}tmp/sub.csv'",
"_____no_output_____"
],
[
"joined_test[['Id','Sales']].to_csv(csv_fn, index=False)",
"_____no_output_____"
],
[
"FileLink(csv_fn)",
"_____no_output_____"
]
],
[
[
"## RF",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestRegressor",
"_____no_output_____"
],
[
"((val,trn), (y_val,y_trn)) = split_by_idx(val_idx, df.values, yl)",
"_____no_output_____"
],
[
"m = RandomForestRegressor(n_estimators=40, max_features=0.99, min_samples_leaf=2,\n n_jobs=-1, oob_score=True)\nm.fit(trn, y_trn);",
"_____no_output_____"
],
[
"preds = m.predict(val)\nm.score(trn, y_trn), m.score(val, y_val), m.oob_score_, exp_rmspe(preds, y_val)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0f292efd22d1a3661afe38c88ad90fd0268f9e8 | 2,877 | ipynb | Jupyter Notebook | test/ipynb/python/Pyspark_yarn_example.ipynb | binder-oilgains/beakerx | 794e627ecdfaf5e956a5f13f65047dd4fbfd8887 | [
"Apache-2.0"
] | 1,491 | 2017-03-30T03:05:05.000Z | 2022-03-27T04:26:02.000Z | test/ipynb/python/Pyspark_yarn_example.ipynb | binder-oilgains/beakerx | 794e627ecdfaf5e956a5f13f65047dd4fbfd8887 | [
"Apache-2.0"
] | 3,268 | 2015-01-01T00:10:26.000Z | 2017-05-05T18:59:41.000Z | test/ipynb/python/Pyspark_yarn_example.ipynb | binder-oilgains/beakerx | 794e627ecdfaf5e956a5f13f65047dd4fbfd8887 | [
"Apache-2.0"
] | 287 | 2017-04-03T01:30:06.000Z | 2022-03-17T06:09:15.000Z | 21.631579 | 140 | 0.529718 | [
[
[
"### Run spark in yarn client mode",
"_____no_output_____"
],
[
"We run yarn by the following instruction https://hadoop.apache.org/docs/r3.1.2/hadoop-project-dist/hadoop-common/SingleCluster.html\n\nAdditionally we have to add the following property to etc/hadoop/yarn-site.xml\n\n```\n<property>\n <name>yarn.nodemanager.vmem-pmem-ratio</name>\n <value>5</value>\n</property>\n```\n\nor\n\n```\n<property>\n <name>yarn.nodemanager.pmem-check-enabled</name>\n <value>false</value>\n</property>\n\n<property>\n <name>yarn.nodemanager.vmem-check-enabled</name>\n <value>false</value>\n</property>\n```",
"_____no_output_____"
]
],
[
[
"import os\nos.environ[\"HADOOP_CONF_DIR\"] = PATH_TO_HADOOP_CONF_DIR",
"_____no_output_____"
],
[
"%%spark --yarn\nfrom pyspark.sql import SparkSession\nSparkSession.builder \\\n.appName(\"SparkYarnBeakerxSupport3\")",
"_____no_output_____"
],
[
"import random\ndef inside(p):\n x, y = random.random(), random.random()\n return x*x + y*y < 1\n\nNUM_SAMPLES =100000000\n\ncount = sc.parallelize(range(0, NUM_SAMPLES)).filter(inside).count()\nprint(\"Pi is roughly %f\" % (4.0 * count / NUM_SAMPLES))",
"_____no_output_____"
],
[
"spark.stop()",
"_____no_output_____"
],
[
"sc.stop()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0f29b6909719cf2691d6b1a32c4bac00c14f843 | 208,012 | ipynb | Jupyter Notebook | courses/machine_learning/deepdive2/computer_vision_fun/solutions/classifying_images_with_a_nn_and_dnn_model.ipynb | ssensalo/training-data-analyst | ddd0c422880634df43c3fab5dae1d2e3edc9d3d2 | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive2/computer_vision_fun/solutions/classifying_images_with_a_nn_and_dnn_model.ipynb | ssensalo/training-data-analyst | ddd0c422880634df43c3fab5dae1d2e3edc9d3d2 | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive2/computer_vision_fun/solutions/classifying_images_with_a_nn_and_dnn_model.ipynb | ssensalo/training-data-analyst | ddd0c422880634df43c3fab5dae1d2e3edc9d3d2 | [
"Apache-2.0"
] | null | null | null | 270.848958 | 35,180 | 0.90602 | [
[
[
"# Classifying Images with a NN and DNN Model\n\n## Introduction\n\nIn this notebook, you learn how to build a neural network to classify the tf-flowers dataset using a Deep Neural Network Model.\n\n## Learning Objectives\n\n* Define Helper Functions.\n* Train and evaluate a Neural Network (NN) model.\n* Train and evaluate a Deep Neural Network model.\n\n\nEach learning objective will correspond to a __#TODO__ in the [student lab notebook](../labs/classifying_images_with_a_nn_and_dnn_model.ipynb) -- try to complete that notebook first before reviewing this solution notebook.\n",
"_____no_output_____"
]
],
[
[
"# Import and print the installed version of TensorFlow\nimport tensorflow as tf\nprint(tf.version.VERSION)",
"2.6.3\n"
]
],
[
[
"## Defining Helper Functions\n#### Reading and Preprocessing image data",
"_____no_output_____"
]
],
[
[
"# Helper functions\ndef training_plot(metrics, history):\n f, ax = plt.subplots(1, len(metrics), figsize=(5*len(metrics), 5))\n for idx, metric in enumerate(metrics):\n ax[idx].plot(history.history[metric], ls='dashed')\n ax[idx].set_xlabel(\"Epochs\")\n ax[idx].set_ylabel(metric)\n ax[idx].plot(history.history['val_' + metric]);\n ax[idx].legend([metric, 'val_' + metric])\n\n# Call model.predict() on a few images in the evaluation dataset\ndef plot_predictions(filename):\n f, ax = plt.subplots(3, 5, figsize=(25,15))\n dataset = (tf.data.TextLineDataset(filename).\n map(decode_csv))\n for idx, (img, label) in enumerate(dataset.take(15)):\n ax[idx//5, idx%5].imshow((img.numpy()));\n batch_image = tf.reshape(img, [1, IMG_HEIGHT, IMG_WIDTH, IMG_CHANNELS])\n batch_pred = model.predict(batch_image)\n pred = batch_pred[0]\n label = CLASS_NAMES[label.numpy()]\n pred_label_index = tf.math.argmax(pred).numpy()\n pred_label = CLASS_NAMES[pred_label_index]\n prob = pred[pred_label_index]\n ax[idx//5, idx%5].set_title('{}: {} ({:.4f})'.format(label, pred_label, prob))\n\ndef show_trained_weights(model):\n # CLASS_NAMES is ['daisy', 'dandelion', 'roses', 'sunflowers', 'tulips']\n LAYER = 1 # Layer 0 flattens the image, layer=1 is the first dense layer\n WEIGHT_TYPE = 0 # 0 for weight, 1 for bias\n\n f, ax = plt.subplots(1, 5, figsize=(15,15))\n for flower in range(len(CLASS_NAMES)):\n weights = model.layers[LAYER].get_weights()[WEIGHT_TYPE][:, flower]\n min_wt = tf.math.reduce_min(weights).numpy()\n max_wt = tf.math.reduce_max(weights).numpy()\n flower_name = CLASS_NAMES[flower]\n print(\"Scaling weights for {} in {} to {}\".format(\n flower_name, min_wt, max_wt))\n weights = (weights - min_wt)/(max_wt - min_wt)\n ax[flower].imshow(weights.reshape(IMG_HEIGHT, IMG_WIDTH, 3));\n ax[flower].set_title(flower_name);",
"_____no_output_____"
],
[
"# The import statement combines two operations; it searches for the named module, then it binds the results of that search\nimport matplotlib.pylab as plt\nimport numpy as np\nimport tensorflow as tf\nIMG_HEIGHT = 224\nIMG_WIDTH = 224\nIMG_CHANNELS = 3\n\ndef read_and_decode(filename, reshape_dims):\n # TODO 1: Read the file\n img = tf.io.read_file(filename)\n # Convert the compressed string to a 3D uint8 tensor.\n img = tf.image.decode_jpeg(img, channels=IMG_CHANNELS)\n # Use `convert_image_dtype` to convert to floats in the [0,1] range.\n img = tf.image.convert_image_dtype(img, tf.float32)\n # Resize the image to the desired size.\n return tf.image.resize(img, reshape_dims)\n\nCLASS_NAMES = [item.numpy().decode(\"utf-8\") for item in \n tf.strings.regex_replace(\n tf.io.gfile.glob(\"gs://practical-ml-vision-book/flowers_5_jpeg/flower_photos/*\"),\n \"gs://practical-ml-vision-book/flowers_5_jpeg/flower_photos/\", \"\")]\nCLASS_NAMES = [item for item in CLASS_NAMES if item.find(\".\") == -1]\nprint(\"These are the available classes:\", CLASS_NAMES)\n\n# the label is the index into CLASS_NAMES array\ndef decode_csv(csv_row):\n record_defaults = [\"path\", \"flower\"]\n filename, label_string = tf.io.decode_csv(csv_row, record_defaults)\n img = read_and_decode(filename, [IMG_HEIGHT, IMG_WIDTH])\n label = tf.argmax(tf.math.equal(CLASS_NAMES, label_string))\n return img, label",
"These are the available classes: ['daisy', 'dandelion', 'roses', 'sunflowers', 'tulips']\n"
]
],
[
[
"## Train and evaluate a Neural Network (NN) model\n\nOne way to get a more complex method is to interpose one or more Dense layers in between the input and output. The model now has three layers. A layer with trainable weights such as the one recently added, that is neither the input nor the output, is called a hidden layer.",
"_____no_output_____"
],
[
"In Keras, you introduce the activation function with tf.keras.activations.\n\nThe Rectified Linear Unit (ReLU) is the most commonly used activation function for hidden layers – other commonly used activation functions include sigmoid, tanh, and elu.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport numpy as np\nimport matplotlib.pylab as plt\n\nfig, ax = plt.subplots(1, 3, figsize=(10,5))\nx = np.arange(-10.0, 10.0, 0.1)\ny = tf.keras.activations.sigmoid(x)\nax[0].plot(x, y);\nax[0].set_title(\"sigmoid\")\ny = tf.keras.activations.relu(x)\nax[1].plot(x, y);\nax[1].set_title(\"relu\")\ny = tf.keras.activations.elu(x)\nax[2].plot(x, y);\nax[2].set_title(\"elu\");",
"_____no_output_____"
],
[
"model = tf.keras.Sequential([\n tf.keras.layers.Flatten(input_shape=(IMG_WIDTH, IMG_HEIGHT, 3)),\n tf.keras.layers.Dense(128, activation='relu'),\n tf.keras.layers.Dense(len(CLASS_NAMES), activation='softmax')\n])\nmodel.summary()\ntf.keras.utils.plot_model(model, show_shapes=True, show_layer_names=False)",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nflatten (Flatten) (None, 150528) 0 \n_________________________________________________________________\ndense (Dense) (None, 128) 19267712 \n_________________________________________________________________\ndense_1 (Dense) (None, 5) 645 \n=================================================================\nTotal params: 19,268,357\nTrainable params: 19,268,357\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"model = tf.keras.Sequential([\n tf.keras.layers.Flatten(input_shape=(IMG_HEIGHT, IMG_WIDTH, 3)),\n tf.keras.layers.Dense(len(CLASS_NAMES), activation='softmax')\n])\nmodel.summary()",
"Model: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nflatten_1 (Flatten) (None, 150528) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 5) 752645 \n=================================================================\nTotal params: 752,645\nTrainable params: 752,645\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"BATCH_SIZE = 32\n\ntrain_dataset = (tf.data.TextLineDataset(\n \"gs://practical-ml-vision-book/flowers_5_jpeg/flower_photos/train_set.csv\").\n map(decode_csv)).batch(BATCH_SIZE)\n\neval_dataset = (tf.data.TextLineDataset(\n \"gs://practical-ml-vision-book/flowers_5_jpeg/flower_photos/eval_set.csv\").\n map(decode_csv)).batch(BATCH_SIZE)\n\n# NN with one hidden layer\nmodel = tf.keras.Sequential([\n tf.keras.layers.Flatten(input_shape=(IMG_HEIGHT, IMG_WIDTH, IMG_CHANNELS)),\n tf.keras.layers.Dense(128, activation=tf.keras.activations.relu),\n tf.keras.layers.Dense(len(CLASS_NAMES), activation='softmax')\n])\nmodel.compile(optimizer='adam',\n loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),\n metrics=['accuracy'])\n# TODO 2: Train a Neural Network model\nhistory = model.fit(train_dataset, validation_data=eval_dataset, epochs=10)",
"2022-02-17 14:48:37.005204: I tensorflow/compiler/mlir/mlir_graph_optimization_pass.cc:185] None of the MLIR Optimization Passes are enabled (registered 2)\n"
],
[
"training_plot(['loss', 'accuracy'], history)",
"_____no_output_____"
]
],
[
[
"## Training the neural network\n\nTraining the neural network is similar to training the linear model. Compile the model passing in the optimizer, the loss, and the metrics. Then, call model.fit() passing in the datasets.",
"_____no_output_____"
]
],
[
[
"# parameterize to the values in the previous cell\ndef train_and_evaluate(batch_size = 32,\n lrate = 0.001, # default in Adam constructor\n l1 = 0,\n l2 = 0,\n num_hidden = 128):\n regularizer = tf.keras.regularizers.l1_l2(l1, l2)\n\n train_dataset = (tf.data.TextLineDataset(\n \"gs://practical-ml-vision-book/flowers_5_jpeg/flower_photos/train_set.csv\").\n map(decode_csv)).batch(batch_size)\n\n eval_dataset = (tf.data.TextLineDataset(\n \"gs://practical-ml-vision-book/flowers_5_jpeg/flower_photos/eval_set.csv\").\n map(decode_csv)).batch(32) # this doesn't matter\n\n # NN with one hidden layers\n model = tf.keras.Sequential([\n tf.keras.layers.Flatten(input_shape=(IMG_HEIGHT, IMG_WIDTH, IMG_CHANNELS)),\n tf.keras.layers.Dense(num_hidden,\n kernel_regularizer=regularizer, \n activation=tf.keras.activations.relu),\n tf.keras.layers.Dense(len(CLASS_NAMES), \n kernel_regularizer=regularizer,\n activation='softmax')\n ])\n model.compile(optimizer=tf.keras.optimizers.Adam(learning_rate=lrate),\n loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),\n metrics=['accuracy'])\n \n history = model.fit(train_dataset, validation_data=eval_dataset, epochs=10)\n training_plot(['loss', 'accuracy'], history)\n return model",
"_____no_output_____"
]
],
[
[
"First, train your model by using 128 hidden layers.",
"_____no_output_____"
]
],
[
[
"model = train_and_evaluate(batch_size=32, lrate=0.0001, l1=0, l2=0, num_hidden=128)",
"Epoch 1/10\n104/104 [==============================] - 157s 2s/step - loss: 1.8989 - accuracy: 0.3512 - val_loss: 1.3971 - val_accuracy: 0.4378\nEpoch 2/10\n104/104 [==============================] - 122s 1s/step - loss: 1.3643 - accuracy: 0.4464 - val_loss: 1.2772 - val_accuracy: 0.4486\nEpoch 3/10\n104/104 [==============================] - 110s 1s/step - loss: 1.2519 - accuracy: 0.5006 - val_loss: 1.2926 - val_accuracy: 0.4703\nEpoch 4/10\n104/104 [==============================] - 114s 1s/step - loss: 1.1849 - accuracy: 0.5315 - val_loss: 1.3158 - val_accuracy: 0.4676\nEpoch 5/10\n104/104 [==============================] - 122s 1s/step - loss: 1.1232 - accuracy: 0.5636 - val_loss: 1.3247 - val_accuracy: 0.4649\nEpoch 6/10\n104/104 [==============================] - 122s 1s/step - loss: 1.0699 - accuracy: 0.5918 - val_loss: 1.3222 - val_accuracy: 0.4757\nEpoch 7/10\n104/104 [==============================] - 115s 1s/step - loss: 1.0254 - accuracy: 0.6100 - val_loss: 1.3207 - val_accuracy: 0.4811\nEpoch 8/10\n104/104 [==============================] - 112s 1s/step - loss: 0.9800 - accuracy: 0.6291 - val_loss: 1.3283 - val_accuracy: 0.4676\nEpoch 9/10\n104/104 [==============================] - 108s 1s/step - loss: 0.9390 - accuracy: 0.6461 - val_loss: 1.3506 - val_accuracy: 0.4649\nEpoch 10/10\n104/104 [==============================] - 109s 1s/step - loss: 0.9068 - accuracy: 0.6658 - val_loss: 1.3453 - val_accuracy: 0.4784\n"
]
],
[
[
"You would normally expect that adding layers to a model will improve the ability of the model to fit the training data, and thus lower the loss. Notice that it is not always the case though.",
"_____no_output_____"
]
],
[
[
"model = train_and_evaluate(batch_size=32, lrate=0.0001, l1=0, l2=0, num_hidden=256)",
"Epoch 1/10\n104/104 [==============================] - 114s 1s/step - loss: 2.4669 - accuracy: 0.3509 - val_loss: 1.4723 - val_accuracy: 0.4324\nEpoch 2/10\n104/104 [==============================] - 106s 1s/step - loss: 1.3723 - accuracy: 0.4409 - val_loss: 1.6079 - val_accuracy: 0.4514\nEpoch 3/10\n104/104 [==============================] - 118s 1s/step - loss: 1.3005 - accuracy: 0.4673 - val_loss: 1.5676 - val_accuracy: 0.4568\nEpoch 4/10\n104/104 [==============================] - 107s 1s/step - loss: 1.2473 - accuracy: 0.4976 - val_loss: 1.5239 - val_accuracy: 0.4405\nEpoch 5/10\n104/104 [==============================] - 111s 1s/step - loss: 1.1703 - accuracy: 0.5294 - val_loss: 1.5198 - val_accuracy: 0.4432\nEpoch 6/10\n104/104 [==============================] - 109s 1s/step - loss: 1.1074 - accuracy: 0.5600 - val_loss: 1.5674 - val_accuracy: 0.4486\nEpoch 7/10\n104/104 [==============================] - 106s 1s/step - loss: 1.0581 - accuracy: 0.5758 - val_loss: 1.5704 - val_accuracy: 0.4459\nEpoch 8/10\n104/104 [==============================] - 109s 1s/step - loss: 1.0213 - accuracy: 0.5994 - val_loss: 1.5222 - val_accuracy: 0.4351\nEpoch 9/10\n104/104 [==============================] - 110s 1s/step - loss: 1.0000 - accuracy: 0.6173 - val_loss: 1.4565 - val_accuracy: 0.4595\nEpoch 10/10\n104/104 [==============================] - 110s 1s/step - loss: 0.9855 - accuracy: 0.6276 - val_loss: 1.3454 - val_accuracy: 0.4973\n"
]
],
[
[
"## Train and evaluate a Deep Neural Network model\n\nNow train a DNN. You need to parameterize the number of layers, and the number of nodes in each layer.",
"_____no_output_____"
]
],
[
[
"# parameterize to the values in the previous cell\ndef train_and_evaluate(batch_size = 32,\n lrate = 0.0001,\n l1 = 0,\n l2 = 0.001,\n num_hidden = [64, 16]):\n regularizer = tf.keras.regularizers.l1_l2(l1, l2)\n\n train_dataset = (tf.data.TextLineDataset(\n \"gs://practical-ml-vision-book/flowers_5_jpeg/flower_photos/train_set.csv\").\n map(decode_csv)).batch(batch_size)\n\n eval_dataset = (tf.data.TextLineDataset(\n \"gs://practical-ml-vision-book/flowers_5_jpeg/flower_photos/eval_set.csv\").\n map(decode_csv)).batch(32) # this doesn't matter\n\n # NN with multiple hidden layers\n layers = [tf.keras.layers.Flatten(\n input_shape=(IMG_HEIGHT, IMG_WIDTH, IMG_CHANNELS),\n name='input_pixels')]\n layers = layers + [\n tf.keras.layers.Dense(nodes,\n kernel_regularizer=regularizer, \n activation=tf.keras.activations.relu,\n name='hidden_dense_{}'.format(hno))\n for hno, nodes in enumerate(num_hidden)\n ]\n layers = layers + [\n tf.keras.layers.Dense(len(CLASS_NAMES), \n kernel_regularizer=regularizer,\n activation='softmax',\n name='flower_prob')\n ]\n\n model = tf.keras.Sequential(layers, name='flower_classification')\n model.compile(optimizer=tf.keras.optimizers.Adam(learning_rate=lrate),\n loss=tf.keras.losses.SparseCategoricalCrossentropy(\n from_logits=False),\n metrics=['accuracy'])\n print(model.summary())\n history = model.fit(train_dataset, validation_data=eval_dataset, epochs=10)\n training_plot(['loss', 'accuracy'], history)\n return model",
"_____no_output_____"
],
[
"# TODO 3: Train and evaluate a DNN model\nmodel = train_and_evaluate(lrate=0.0001, l2=0.001, num_hidden = [64, 16])",
"Model: \"flower_classification\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_pixels (Flatten) (None, 150528) 0 \n_________________________________________________________________\nhidden_dense_0 (Dense) (None, 64) 9633856 \n_________________________________________________________________\nhidden_dense_1 (Dense) (None, 16) 1040 \n_________________________________________________________________\nflower_prob (Dense) (None, 5) 85 \n=================================================================\nTotal params: 9,634,981\nTrainable params: 9,634,981\nNon-trainable params: 0\n_________________________________________________________________\nNone\nEpoch 1/10\n104/104 [==============================] - 121s 1s/step - loss: 1.8870 - accuracy: 0.3139 - val_loss: 1.5853 - val_accuracy: 0.3676\nEpoch 2/10\n104/104 [==============================] - 109s 1s/step - loss: 1.6866 - accuracy: 0.3785 - val_loss: 1.5293 - val_accuracy: 0.4054\nEpoch 3/10\n104/104 [==============================] - 113s 1s/step - loss: 1.5494 - accuracy: 0.4415 - val_loss: 1.6159 - val_accuracy: 0.4027\nEpoch 4/10\n104/104 [==============================] - 111s 1s/step - loss: 1.4291 - accuracy: 0.4864 - val_loss: 1.4335 - val_accuracy: 0.4351\nEpoch 5/10\n104/104 [==============================] - 107s 1s/step - loss: 1.4071 - accuracy: 0.5018 - val_loss: 1.9287 - val_accuracy: 0.4054\nEpoch 6/10\n104/104 [==============================] - 104s 1s/step - loss: 1.4099 - accuracy: 0.5039 - val_loss: 2.3087 - val_accuracy: 0.3486\nEpoch 7/10\n104/104 [==============================] - 106s 1s/step - loss: 1.4112 - accuracy: 0.5088 - val_loss: 1.7884 - val_accuracy: 0.3838\nEpoch 8/10\n104/104 [==============================] - 104s 996ms/step - loss: 1.3400 - accuracy: 0.5291 - val_loss: 1.6256 - val_accuracy: 0.4216\nEpoch 9/10\n104/104 [==============================] - 105s 1s/step - loss: 1.2254 - accuracy: 0.5694 - val_loss: 1.6722 - val_accuracy: 0.4081\nEpoch 10/10\n104/104 [==============================] - 109s 1s/step - loss: 1.2065 - accuracy: 0.5806 - val_loss: 1.5633 - val_accuracy: 0.4270\n"
]
],
[
[
"Congrats! You've completed the lab!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d0f29ecd8021dc2755e121243164531efce7b578 | 4,081 | ipynb | Jupyter Notebook | notebooks/ui/eprint.ipynb | bworstell/gee_tools | 4123395cf5bad39f20613afb37d041186d9516fe | [
"MIT"
] | null | null | null | notebooks/ui/eprint.ipynb | bworstell/gee_tools | 4123395cf5bad39f20613afb37d041186d9516fe | [
"MIT"
] | null | null | null | notebooks/ui/eprint.ipynb | bworstell/gee_tools | 4123395cf5bad39f20613afb37d041186d9516fe | [
"MIT"
] | null | null | null | 19.81068 | 176 | 0.474148 | [
[
[
"import ee",
"_____no_output_____"
],
[
"from geetools import ui",
"_____no_output_____"
]
],
[
[
"## Test objects",
"_____no_output_____"
]
],
[
[
"point = ee.Geometry.Point([-72, -42])\nimage = ee.Image.constant(0)\ndate = ee.Date('2000-1-1')",
"_____no_output_____"
]
],
[
[
"## `getInfo`\nGet information asynchronously. It always returns a `dict` with one element: `info`. Inside this element will be the result of the object. The result is a modified `dict`",
"_____no_output_____"
]
],
[
[
"infop = ui.getInfo(point)",
"_____no_output_____"
],
[
"infop.get()",
"_____no_output_____"
],
[
"infop()",
"_____no_output_____"
],
[
"info_date = ui.getInfo(date)",
"_____no_output_____"
],
[
"info_date()",
"_____no_output_____"
]
],
[
[
"## `eprint`\nPrint Earth Engine object",
"_____no_output_____"
]
],
[
[
"ui.eprint(point, notebook=True, do_async=True)",
"_____no_output_____"
],
[
"ui.eprint(point, date, notebook=True, do_async=True)",
"_____no_output_____"
],
[
"ui.eprint(image, date, notebook=False, do_async=True)",
"Cannot make async printing outside a Jupyter environment\n{ 'bands': [ { 'crs': 'EPSG:4326',\n 'crs_transform': [1.0, 0.0, 0.0, 0.0, 1.0, 0.0],\n 'data_type': { 'max': 0,\n 'min': 0,\n 'precision': 'int',\n 'type': 'PixelType'},\n 'id': 'constant'}],\n 'type': 'Image'}\n\n'2000-01-01T00:00:00'\n\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0f29f66c57020df1d3cc18244d41793a8e2851c | 6,576 | ipynb | Jupyter Notebook | data-analysis/data-frames.ipynb | dsoto/computing-tutorial | de029ee92e6ad1f0705de27a1bfe61a1d16806ce | [
"CC-BY-4.0"
] | 1 | 2016-02-15T13:34:42.000Z | 2016-02-15T13:34:42.000Z | data-analysis/data-frames.ipynb | dsoto/computing-tutorial | de029ee92e6ad1f0705de27a1bfe61a1d16806ce | [
"CC-BY-4.0"
] | null | null | null | data-analysis/data-frames.ipynb | dsoto/computing-tutorial | de029ee92e6ad1f0705de27a1bfe61a1d16806ce | [
"CC-BY-4.0"
] | null | null | null | 24.355556 | 85 | 0.385797 | [
[
[
"Often the data we are using would fit well in a spreadsheet.\nData that is appropriate for a spreadsheet is called tabular data.\nPandas is a powerful python library for working with tabular data.",
"_____no_output_____"
]
],
[
[
"data = '''\nhousehold,dorm,phone_energy,laptop_energy\nA,tuscany,10,50\nB,sauv,30,60\nC,tuscany,12,45\nD,sauv,20,50\n'''\n\n%matplotlib inline\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\nfrom io import StringIO\nfrom tabulate import tabulate\n\ndf = pd.read_csv(StringIO(data))\ndf",
"_____no_output_____"
],
[
"# you can select an element from the table much like you do in a spreadsheet\n# by using the column name and the row number\ndf['household'][1]",
"_____no_output_____"
],
[
"# you can get a column\ndf['household']",
"_____no_output_____"
],
[
"# you can do math and store it in a new column\ndf['total_energy'] = df['phone_energy'] * df['laptop_energy']\ndf",
"_____no_output_____"
]
],
[
[
"You can learn about the many other abilities of this library at\nhttp://pandas.pydata.org/",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0f2a1f5194b13971eb2ed04d41342c4965dbb8c | 635,638 | ipynb | Jupyter Notebook | Notebooks/GD_placement.ipynb | AndOleAnd/Capstone_N_A_P | f619ed31171d83ebdc080776ce06055b580c6705 | [
"MIT"
] | null | null | null | Notebooks/GD_placement.ipynb | AndOleAnd/Capstone_N_A_P | f619ed31171d83ebdc080776ce06055b580c6705 | [
"MIT"
] | 38 | 2020-12-11T19:35:25.000Z | 2021-06-16T08:34:09.000Z | Notebooks/GD_placement.ipynb | AndOleAnd/Capstone_N_A_P | f619ed31171d83ebdc080776ce06055b580c6705 | [
"MIT"
] | null | null | null | 938.903988 | 78,268 | 0.950519 | [
[
[
"import sys\nsys.path.append('../Scripts')\n\nfrom capstone_functions import *",
"_____no_output_____"
]
],
[
[
"## Gradient Descent exploration\n### This notebook has many example of running gradient descent with different hyper parameters",
"_____no_output_____"
],
[
"### Epoch Choice\nThis calls our main pipeline function that loads raw data performs all adjustments and creates centroids via GD\nThe point of the next two is to see the difference extending the epochs makes to the final scores. \nThe Score on test set is most important method to validate performance",
"_____no_output_____"
]
],
[
[
"ambulance_placement_pipeline(input_path='../Inputs/', output_path='../Outputs/', crash_source_csv='Train',\n outlier_filter=0.005, \n holdout_strategy='random', holdout_test_size=0.2,\n test_period_date_start='2018-01-01', test_period_date_end='2019-12-31',\n tw_cluster_strategy='saturday_2', placement_method='gradient_descent', verbose=1,\n lr=3e-2, n_epochs=400)",
"4 clusters created\n4 clusters created\nusing gradient descent clustering\n4 placement sets created\nsubmission dataframe created\nTotal size of test set: 1264\nData points in test period: 1264\nTotal size of train set: 6318\nData points in test period: 4937\nScore on test set: 0.053058366125481406\nScore on train set: 0.03891002335438391 (avg distance per accident)\nsubmission dataframe created\n2020128_Train_0.005_saturday_2_gradient_descent.csv saved in ../Outputs/\n"
],
[
"\nambulance_placement_pipeline(input_path='../Inputs/', output_path='../Outputs/', crash_source_csv='Train',\n outlier_filter=0.005, \n holdout_strategy='random', holdout_test_size=0.2,\n test_period_date_start='2018-01-01', test_period_date_end='2019-12-31',\n tw_cluster_strategy='saturday_2', placement_method='gradient_descent', verbose=1,\n lr=3e-2, n_epochs=800)",
"4 clusters created\n4 clusters created\nusing gradient descent clustering\n4 placement sets created\nsubmission dataframe created\nTotal size of test set: 1264\nData points in test period: 1264\nTotal size of train set: 6318\nData points in test period: 4937\nScore on test set: 0.053653958785398\nScore on train set: 0.03904091820556965 (avg distance per accident)\nsubmission dataframe created\n2020128_Train_0.005_saturday_2_gradient_descent.csv saved in ../Outputs/\n"
]
],
[
[
"### This appears to show that increasing epochs improves final score. ",
"_____no_output_____"
],
[
"### Learning Rate\n## What if instead we change the learning rate to smaller steps",
"_____no_output_____"
]
],
[
[
"ambulance_placement_pipeline(input_path='../Inputs/', output_path='../Outputs/', crash_source_csv='Train',\n outlier_filter=0.005, \n holdout_strategy='random', holdout_test_size=0.2,\n test_period_date_start='2018-01-01', test_period_date_end='2019-12-31',\n tw_cluster_strategy='saturday_2', placement_method='gradient_descent', verbose=1,\n lr=3e-3, n_epochs=400)",
"4 clusters created\n4 clusters created\nusing gradient descent clustering\n4 placement sets created\nsubmission dataframe created\nTotal size of test set: 1264\nData points in test period: 1264\nTotal size of train set: 6318\nData points in test period: 4937\nScore on test set: 0.053616697671851066\nScore on train set: 0.03925874537430147 (avg distance per accident)\nsubmission dataframe created\n2020128_Train_0.005_saturday_2_gradient_descent.csv saved in ../Outputs/\n"
]
],
[
[
"### Compared to the first run, the score has not improved with smaller steps. Perhaps there are local minimal.",
"_____no_output_____"
],
[
"## Performance of GD based on holdout size\n\nThe first two example used 0.2 holdout but our model will perform better with more data so it will be good to see how much holdout data is required",
"_____no_output_____"
]
],
[
[
"ambulance_placement_pipeline(input_path='../Inputs/', output_path='../Outputs/', crash_source_csv='Train',\n outlier_filter=0.005, \n holdout_strategy='random', holdout_test_size=0.05,\n test_period_date_start='2018-01-01', test_period_date_end='2019-12-31',\n tw_cluster_strategy='saturday_2', placement_method='gradient_descent', verbose=1,\n lr=3e-2, n_epochs=800)",
"4 clusters created\n4 clusters created\nusing gradient descent clustering\n4 placement sets created\nsubmission dataframe created\nTotal size of test set: 316\nData points in test period: 316\nTotal size of train set: 6318\nData points in test period: 5866\nScore on test set: 0.06513533400595606\nScore on train set: 0.04002475085708951 (avg distance per accident)\nsubmission dataframe created\n2020128_Train_0.005_saturday_2_gradient_descent.csv saved in ../Outputs/\n"
]
],
[
[
"### Changing holdout size makes a big difference to test score (but this is not comparable across runs for different sizes)\n### Train score is then our indicateor if model is improving but we will not be able to understand overfitting. Submitting to zindi will be one solution to this. In this case train score also got worse. But it is not necessarily comparable across holdout sizes",
"_____no_output_____"
],
[
"### We can look deeper into how the model is handling each tw cluster and changing over time with additonal output.",
"_____no_output_____"
]
],
[
[
"ambulance_placement_pipeline(input_path='../Inputs/', output_path='../Outputs/', crash_source_csv='Train',\n outlier_filter=0.005, \n holdout_strategy='random', holdout_test_size=0.1,\n test_period_date_start='2018-01-01', test_period_date_end='2019-12-31',\n tw_cluster_strategy='saturday_2', placement_method='gradient_descent', verbose=1,\n lr=3e-2, n_epochs=800)",
"4 clusters created\n4 clusters created\nusing gradient descent clustering\nVal loss: 0.06301859766244888\nVal loss: 0.059005241841077805\nVal loss: 0.05927123501896858\nVal loss: 0.05977119877934456\nVal loss: 0.059426069259643555\nVal loss: 0.05930862948298454\nVal loss: 0.05885466933250427\nVal loss: 0.05877223610877991\n"
]
],
[
[
"### From the charts it appears that most improvement in train loss and validation loss occurs in the first epochs\n### perhaps we can reduce epochs to save time. Mini batch size optimization is also something to consider. Learning rate could also be further explored",
"_____no_output_____"
],
[
"## Finally What does changing the tw_cluster_strategy do. We can run again with a different set.\n### spoiler: it got worse. even though on other placement methods (k-means) off_peak_split outperformmed 'saturday_2'\nThis should be further investigated to find best strategy",
"_____no_output_____"
]
],
[
[
"ambulance_placement_pipeline(input_path='../Inputs/', output_path='../Outputs/', crash_source_csv='Train',\n outlier_filter=0.005, \n holdout_strategy='random', holdout_test_size=0.2,\n test_period_date_start='2018-01-01', test_period_date_end='2019-12-31',\n tw_cluster_strategy='off_peak_split', placement_method='k_means', verbose=10)",
"5 clusters created\n5 clusters created\nusing k-means clustering\n"
]
],
[
[
"### Rerunning model with small holdout set to have best out put for zindi.",
"_____no_output_____"
]
],
[
[
"#best\nambulance_placement_pipeline(input_path='../Inputs/', output_path='../Outputs/', crash_source_csv='Train',\n outlier_filter=0.005, \n holdout_strategy='random', holdout_test_size=0.005,\n test_period_date_start='2018-01-01', test_period_date_end='2019-12-31',\n tw_cluster_strategy='holiday_simple', placement_method='gradient_descent', verbose=0,\n lr=3e-3, n_epochs=400)",
"4 clusters created\n4 clusters created\nusing gradient descent clustering\n4 placement sets created\nsubmission dataframe created\nTotal size of test set: 32\nData points in test period: 32\nTotal size of train set: 6318\nData points in test period: 6142\nScore on test set: 0.045116526210462554\nScore on train set: 0.03930937487050926 (avg distance per accident)\nsubmission dataframe created\n2020128_Train_0.005_holiday_simple_gradient_descent.csv saved in ../Outputs/\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f2a8929e9f7b68915e5bdb36a3bedd3b7f687a | 8,102 | ipynb | Jupyter Notebook | 波士頓房價分析練習.ipynb | biolytic1996/HAHABANAN-PAGE | 3b2b973d1078bcd75d6faefa127be5c42e62a098 | [
"MIT"
] | null | null | null | 波士頓房價分析練習.ipynb | biolytic1996/HAHABANAN-PAGE | 3b2b973d1078bcd75d6faefa127be5c42e62a098 | [
"MIT"
] | null | null | null | 波士頓房價分析練習.ipynb | biolytic1996/HAHABANAN-PAGE | 3b2b973d1078bcd75d6faefa127be5c42e62a098 | [
"MIT"
] | null | null | null | 86.191489 | 5,738 | 0.821649 | [
[
[
"<a href=\"https://colab.research.google.com/github/biolytic1996/HAHABANAN-PAGE/blob/master/%E6%B3%A2%E5%A3%AB%E9%A0%93%E6%88%BF%E5%83%B9%E5%88%86%E6%9E%90%E7%B7%B4%E7%BF%92.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"**用sklearn做波士頓房價線性回歸**",
"_____no_output_____"
],
[
"1. 前置練習 <br>\n 先自己創建一組線性二維數據,且自己設定y=mx+b的m與b <br>\n 再用matplotlib做直線\n",
"_____no_output_____"
]
],
[
[
">>> import numpy as np //引入numpy資源庫\n>>> import matplotlib.pyplot as plt //引入matplotlib資源庫\n>>> x=np.linspace(0,5,50) // x軸在0-50之間產生50個點\n>>> y=1.5*x+0.8 // 斜率設1.5\n>>> plt.scatter(x,y)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
d0f2b6b35979fd52f701967880aeab7b5404f88b | 7,682 | ipynb | Jupyter Notebook | machine-learning/naive-bayes-from-scratch.ipynb | ParvathiRajan/learning | bb112015d4513414bf86c7392c12b13f8d0fdd21 | [
"MIT"
] | 54 | 2017-09-10T17:28:21.000Z | 2021-12-17T14:55:04.000Z | machine-learning/naive-bayes-from-scratch.ipynb | parvathirajan/learning | bb112015d4513414bf86c7392c12b13f8d0fdd21 | [
"MIT"
] | 1 | 2019-07-04T21:57:14.000Z | 2019-07-04T21:57:14.000Z | machine-learning/naive-bayes-from-scratch.ipynb | parvathirajan/learning | bb112015d4513414bf86c7392c12b13f8d0fdd21 | [
"MIT"
] | 36 | 2017-11-13T16:54:58.000Z | 2022-02-07T11:20:20.000Z | 28.557621 | 124 | 0.408748 | [
[
[
"# Naive Bayes from scratch",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"def get_accuracy(x: pd.DataFrame, y: pd.Series, y_hat: pd.Series):\n correct = y_hat == y\n acc = np.sum(correct) / len(y)\n cond = y == 1\n y1 = len(y[cond])\n y0 = len(y[~cond])\n\n print(f'Class 0: tested {y0}, correctly classified {correct[~cond].sum()}')\n print(f'Class 1: tested {y1}, correctly classified {correct[cond].sum()}')\n print(f'Overall: tested {len(y)}, correctly classified {correct.sum()}')\n print(f'Accuracy = {acc:.2f}')",
"_____no_output_____"
],
[
"class Classifier:\n def __init__(self, dataset: str = None, mle: bool=True):\n if dataset:\n x_train, y_train = reader(f'datasets/{dataset}-train.txt')\n x_test, y_test = reader(f'datasets/{dataset}-test.txt')\n self.train(x_train, y_train, mle)\n print('Training accuracy')\n print('=' * 10)\n self.accuracy(x_train, y_train)\n print('Test accuracy')\n print('=' * 10)\n self.accuracy(x_test, y_test)\n \n def accuracy(self, x: pd.DataFrame, y: pd.DataFrame) -> None:\n y_hat = self.predict(x)\n get_accuracy(x, y, y_hat)",
"_____no_output_____"
],
[
"class NB(Classifier):\n def __init__(self, dataset: str = None, mle: bool=True):\n self.prior = None\n self.p_xi_given_y = {0: {}, 1: {}}\n self.prior_x = {}\n self.cols = None\n super().__init__(dataset, mle)\n \n def train(self, x: pd.DataFrame, y: pd.Series, mle: bool=True):\n adj_den = 0 if mle else 2\n adj_num = 0 if mle else 1\n self.prior = y.value_counts().to_dict()\n for c in [0, 1]:\n self.prior[c] += adj_num\n self.prior[c] /= (len(y) + adj_den)\n \n self.cols = x.columns\n for col in x.columns:\n self.prior_x[col] = (x[col].value_counts() / len(y)).to_dict()\n \n cond = y == 1\n y1 = np.sum(cond)\n y0 = len(y) - y1\n y1 += adj_den\n y0 += adj_den\n x_pos = x[cond]\n x_neg = x[~cond]\n for cls in [0, 1]:\n for col in x.columns:\n x_cls = x_pos if cls == 1 else x_neg\n y_cls = y1 if cls == 1 else y0\n x1 = len(x_cls.query(f'{col} == 1'))\n x0 = len(x_cls.query(f'{col} == 0'))\n \n x1 += adj_num\n x0 += adj_num\n \n self.p_xi_given_y[cls][col] = {\n 0: x0 / y_cls,\n 1: x1 / y_cls\n }\n \n def predict(self, x: pd.DataFrame) -> pd.Series:\n out = []\n for _, row in x.iterrows():\n m = {}\n for cls in [0, 1]:\n m[cls] = np.log([self.prior[0]] + [\n self.p_xi_given_y[cls][col][row[col]]\n for col in x.columns\n ]).sum()\n out.append(1 if m[1] >= m[0] else 0)\n return pd.Series(out)\n \n def _get_ind(self, col):\n num = self.prior_x[col][0] * self.p_xi_given_y[1][col][1]\n den = self.prior_x[col][1] * self.p_xi_given_y[1][col][0]\n return num / den\n \n def most_indicative(self):\n return pd.Series({\n col: self._get_ind(col)\n for col in self.cols\n }).sort_values(ascending=False)",
"_____no_output_____"
],
[
"x = pd.DataFrame({'x1': [0, 0, 1, 1], 'x2': [0, 1, 0, 1]})\ny = pd.Series([0, 0, 1, 1])",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"nb = NB()\nnb.train(x, y)\nnb.accuracy(x, y)",
"Class 0: tested 2, correctly classified 2\nClass 1: tested 2, correctly classified 2\nOverall: tested 4, correctly classified 4\nAccuracy = 1.00\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f2bcff656d8f428302fbd3b8acad2345c2727e | 8,195 | ipynb | Jupyter Notebook | Simple CNN from CSV.ipynb | JanderHungrige/Workshop | eaf9b4f04ecd10f7125218f65f8202d7cb87c8b3 | [
"MIT"
] | null | null | null | Simple CNN from CSV.ipynb | JanderHungrige/Workshop | eaf9b4f04ecd10f7125218f65f8202d7cb87c8b3 | [
"MIT"
] | null | null | null | Simple CNN from CSV.ipynb | JanderHungrige/Workshop | eaf9b4f04ecd10f7125218f65f8202d7cb87c8b3 | [
"MIT"
] | null | null | null | 28.754386 | 309 | 0.505552 | [
[
[
"# Create simple CNN network",
"_____no_output_____"
],
[
"Import all important libraries",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom tensorflow import keras\nimport matplotlib as plt\nimport pandas as pd \n# Import stuff fof preprocessing\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator",
"_____no_output_____"
]
],
[
[
"Now load the IDmap ",
"_____no_output_____"
]
],
[
[
"IDmap = {} # id: hand gesture\\n\",\n# open the csv file\nf = open(\"./IDmapping.csv\", mode='r') \nfileContents = f.read() #reading the content\\n\",\nfileContents = fileContents.split('\\\\n') # splitting the content at the line break \\\\n and saving it into a list\\n\",\n \nfor i in range(len(fileContents)-1):\n fileContents[i] = fileContents[i].split(',') # splitting each list entry at the ,\\n\",\n IDmap[fileContents[i][0]] = fileContents[i][1] #create dictionary with map ID\"\nprint(fileContents[:][0])",
"Faust,1\nOffen,2\n\n"
],
[
" colnames=['file', 'label'] \n # Read data from file \n data = pd.read_csv('./Hand_Annotations_2.csv',dtype=str,names=colnames, header=None)\n # Preview the first 5 lines of the loaded data \n data.head()",
"_____no_output_____"
]
],
[
[
"Now create ImageDataGenerator",
"_____no_output_____"
]
],
[
[
"datagen=ImageDataGenerator(rescale=1./255)",
"_____no_output_____"
]
],
[
[
"# flow_from_dataframe\n\nnow we can use load the data from a CSV or Json file.\n\nParameters:\n - **dataframe** Pandas DataFrame which contains the filenames and classes or numeric data to be treated as target values.\n - **directory** Path to the folder which contains all the images,None if x_col contains absolute paths pointing to each image instead of just filenames.\n - **x_col** The column in the dataframe that has the filenames of the images\n - **y_col** The column/columns in the dataframe in the filename that will be treated as raw target values if class_mode=”raw” (useful for regression tasks) or they will be treated as name of the classes if class_mode is “binary”/”categorical” or they will be ignored if class_mode is “input”/None.\n - **class_mode** In addition to all the class_modes previously available in flow_from_directory, there is “raw”.\n - **drop_duplicates** Boolean, whether to drop duplicate rows based on filename,True by default.\n\n\nSo you can put either all the images in one folder and point to the folder with the `directory` parameter or you have them somewhere scattered and point to them with a full path (with extension like *.jpeg) in the CSV and parameter `directroy=None`",
"_____no_output_____"
]
],
[
[
"train_generator=datagen.flow_from_dataframe(dataframe=data,\n directory=None, \n x_col=colnames[0], \n y_col=colnames[1],\n class_indices=IDmap,\n class_mode=\"categorical\", target_size=(224,224), batch_size=32)",
"Found 0 images belonging to 0 classes.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f2c281079403a32f932b96ed92feb1e50b5140 | 62,537 | ipynb | Jupyter Notebook | repofordummies.ipynb | caseybecker/python_resources | c4c853214c613d8a7c1506b6be007e70d63aa8e9 | [
"MIT"
] | null | null | null | repofordummies.ipynb | caseybecker/python_resources | c4c853214c613d8a7c1506b6be007e70d63aa8e9 | [
"MIT"
] | 1 | 2018-11-30T04:32:59.000Z | 2018-11-30T04:32:59.000Z | repofordummies.ipynb | caseybecker/python_resources | c4c853214c613d8a7c1506b6be007e70d63aa8e9 | [
"MIT"
] | 23 | 2018-11-30T03:35:31.000Z | 2018-12-05T23:09:10.000Z | 856.671233 | 60,604 | 0.950717 | [
[
[
"https://www.codecademy.com/learn/technical-interview-practice-python\n\nWhy practice technical interview problems in Python?\nYou'll need to pass a technical interview if you want to be hired for a technical role. Don't worry — these interviews are pretty predictable, and the same kinds of problems appear again and again. Even if you don't have a technical interview scheduled just yet, practicing these common problems will help you grow as a programmer and problem solver, and will help you write cleaner and better code.\nTake-Away Skills:\nAfter completing this course, you'll be ready to ace coding interviews anywhere and you'll write more efficient code!\nNote on Prerequisites:\nA basic understanding of Python is required to start this course, and any more advanced prerequisites will be noted along the way.\n",
"_____no_output_____"
]
],
[
[
"pwd",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
d0f2cc38f00bfea2bd75a643940cdf1a6921d9fb | 11,940 | ipynb | Jupyter Notebook | notebooks/pandas/advanced-custom-lambda.ipynb | tactlabs/mlnotes | 2393443a749ef3e9161f93287ded85bac1e858ee | [
"CC0-1.0"
] | null | null | null | notebooks/pandas/advanced-custom-lambda.ipynb | tactlabs/mlnotes | 2393443a749ef3e9161f93287ded85bac1e858ee | [
"CC0-1.0"
] | null | null | null | notebooks/pandas/advanced-custom-lambda.ipynb | tactlabs/mlnotes | 2393443a749ef3e9161f93287ded85bac1e858ee | [
"CC0-1.0"
] | null | null | null | 27.575058 | 132 | 0.342127 | [
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"# Check the student passed either math or passed at least in 2 subjects. If no condidition matched, consider them as failed\ndef pass_math_or_two_subjects(row):\n if(row.maths > 34):\n return 'Pass'\n if(row.language > 34 and row.science > 34):\n return 'Pass'\n return 'Fail'",
"_____no_output_____"
],
[
"df = pd.read_csv('abc.csv')",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df['special_result'] = df.apply(pass_math_or_two_subjects, axis=1)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"# Filter those candidates whoever passed the given criteria\ndf_passed = df[df.apply(pass_math_or_two_subjects, axis=1) == 'Pass']",
"_____no_output_____"
],
[
"df_passed",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f300cab3843427cfb667f4efb5c17336542c5c | 686,642 | ipynb | Jupyter Notebook | notebooks/parameter-tuning/default-experiment-metrics.ipynb | AntoniaLbg/leach-et-al-2022 | 9c8c26b0c583f9e6fd0fe4c4dda5ec59057bb713 | [
"Apache-2.0"
] | 1 | 2021-07-07T10:04:11.000Z | 2021-07-07T10:04:11.000Z | notebooks/parameter-tuning/default-experiment-metrics.ipynb | AntoniaLbg/leach-et-al-2022 | 9c8c26b0c583f9e6fd0fe4c4dda5ec59057bb713 | [
"Apache-2.0"
] | null | null | null | notebooks/parameter-tuning/default-experiment-metrics.ipynb | AntoniaLbg/leach-et-al-2022 | 9c8c26b0c583f9e6fd0fe4c4dda5ec59057bb713 | [
"Apache-2.0"
] | 1 | 2021-07-27T15:23:56.000Z | 2021-07-27T15:23:56.000Z | 329.957713 | 291,660 | 0.902313 | [
[
[
"## Import dependencies",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport sys\nimport pandas as pd\nimport matplotlib\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.axes_grid1.inset_locator import inset_axes\nimport seaborn as sn\nimport scipy as sp\nfrom tqdm import tqdm\nimport glob\n\nfrom fair import *\nfrom fair.scripts.data_retrieval import *\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"definition used to round output tables to given sig figs.",
"_____no_output_____"
]
],
[
[
"def round_to_sf(x,sf):\n if x==0:\n return 0\n if np.isnan(x):\n return '-'\n else:\n num= round(x, sf - int(np.floor(np.log10(abs(x)))))\n if abs(num)>10**sf:\n return str(int(num))\n else:\n return str(num)",
"_____no_output_____"
]
],
[
[
"# I. Default parameter simulated concentrations\nHere we run historical emissions to test how the default parameter set simulates the historical evolution of concentrations.",
"_____no_output_____"
]
],
[
[
"## first we view & create a latex table for the default parameter set:\ndefault_params = get_gas_parameter_defaults()\nparams_table = default_params.default.T.sort_index().rename(dict(a1='$a_1$',a2='$a_2$',a3='$a_3$',a4='$a_4$',\n tau1='$\\tau_1$',tau2='$\\tau_2$',tau3='$\\tau_3$',tau4='$\\tau_4$',\n r0='$r_0$',rC='$r_u$',rT='$r_T$',rA='$r_a$',PI_conc='PI\\_conc',\n f1='$f_1$',f2='$f_2$',f3='$f_3$'),axis=1)\nparams_table.index.name='agent'\nparams_table.columns.name='parameter'\nparams_table.index = [x.replace('_','\\_') for x in params_table.index]\n\nparams_table.applymap(lambda x:round_to_sf(x,2)).replace(np.nan,'')#.to_latex('../../docs/manuscript/tables/TabS2',escape=False,bold_rows=True)",
"_____no_output_____"
]
],
[
[
"### data retrieval",
"_____no_output_____"
],
[
"#### concentrations\n\nWMGHG concentrations are from the CMIP6 concentration dataset, [Meinshausen et al., 2017](https://www.geosci-model-dev.net/10/2057/2017/). For some species, these are extended using data from NOAA.\n\nReference:\n\nMeinshausen, M., Vogel, E., Nauels, A., Lorbacher, K., Meinshausen, N., Etheridge, D. M., … Weiss, R. (2017). Historical greenhouse gas concentrations for climate modelling (CMIP6). Geoscientific Model Development, 10(5), 2057–2116. https://doi.org/10.5194/gmd-10-2057-2017",
"_____no_output_____"
]
],
[
[
"import ftplib\n\n## import concentrations from official CMIP6 timeseries:\nCMIP6_conc_ftp = ftplib.FTP('data.iac.ethz.ch','anonymous')\nCMIP6_conc_ftp.cwd('CMIP6/input4MIPs/UoM/GHGConc/CMIP/yr/atmos/UoM-CMIP-1-1-0/GHGConc/gr3-GMNHSH/v20160701')\nCMIP6_ftp_list = [x for x in CMIP6_conc_ftp.nlst() if x[-3:]=='csv']\nWMGHG_concs = pd.DataFrame(dict(zip(['_'.join(x.split('_')[3:-8]) for x in CMIP6_ftp_list],[pd.read_csv('ftp://data.iac.ethz.ch/CMIP6/input4MIPs/UoM/GHGConc/CMIP/yr/atmos/UoM-CMIP-1-1-0/GHGConc/gr3-GMNHSH/v20160701/'+x,usecols=[0,1],index_col=0).iloc[:,0] for x in CMIP6_ftp_list])))\nWMGHG_concs = WMGHG_concs[[x for x in WMGHG_concs.columns if x[-2:]!='eq']] # remove \"equivalent\" concentrations\n\nWMGHG_concs['halon1202'] = 0\nWMGHG_concs.loc[1765:2014,'halon1202'] = pd.read_csv('http://www.pik-potsdam.de/~mmalte/rcps/data/RCP45_MIDYEAR_CONCENTRATIONS.DAT',skiprows=38,delim_whitespace=True,index_col=0)['HALON1202'].loc[1765:2014].values",
"_____no_output_____"
],
[
"## we extend CO2, CH4 & N2O out to 2019 using the NOAA ESRL data\nNOAA_molefrac = pd.read_csv('https://www.esrl.noaa.gov/gmd/aggi/NOAA_MoleFractions_2020.csv',skiprows=2,index_col=0,skipfooter=5).iloc[1:].replace('nd',np.nan).apply(pd.to_numeric).rename(dict(CO2='carbon_dioxide',CH4='methane',N2O='nitrous_oxide'),axis=1)\n\nWMGHG_concs = WMGHG_concs.reindex(np.arange(2020))\n\nfor species in ['carbon_dioxide','methane','nitrous_oxide']:\n \n # scale the NOAA data to join seamlessly (scale factors are almost exactly 1)\n scale_factor = WMGHG_concs.loc[2010:2014,species].mean() / NOAA_molefrac.loc[2010:2015,species].mean()\n WMGHG_concs.loc[2015:2019,species] = NOAA_molefrac.loc[2015:2020,species].values * scale_factor",
"/nfs/a65/pmcjs/miniconda3/envs/leach2021/lib/python3.7/site-packages/ipykernel_launcher.py:2: ParserWarning: Falling back to the 'python' engine because the 'c' engine does not support skipfooter; you can avoid this warning by specifying engine='python'.\n \n"
],
[
"WMGHG_concs.drop(np.arange(1750),inplace=True)",
"_____no_output_____"
],
[
"# rescale all GHGs to be in ppb (bar CO2)\nWMGHG_concs[WMGHG_concs.columns.drop(['carbon_dioxide','methane','nitrous_oxide'])] *= 1/1000",
"_____no_output_____"
]
],
[
[
"#### emissions & forcing\n\nEmissions & external forcing are taken from the RCMIP protocol.\n\nReference:\n\nNicholls, Z. R. J., Meinshausen, M., Lewis, J., Gieseke, R., Dommenget, D., Dorheim, K., … Xie, Z. (2020). Reduced complexity model intercomparison project phase 1: Protocol, results and initial observations. Geoscientific Model Development Discussions, 1–33. https://doi.org/10.5194/gmd-2019-375",
"_____no_output_____"
]
],
[
[
"## emissions\ndef get_SSP_emms(ssp):\n emms = RCMIP_to_FaIR_input_emms(ssp).interpolate().loc[1750:2100]\n rebase_species = ['so2','nox','co','nmvoc','bc','nh3','oc','nox_avi','methyl_bromide','methyl_chloride','chcl3','ch2cl2']\n emms.loc[:,rebase_species] -= emms.loc[1750,rebase_species]\n return emms\n\nchoose_ssps=['ssp119','ssp126','ssp245','ssp370','ssp585']\nSSP_emms = pd.concat([get_SSP_emms(x) for x in choose_ssps],axis=1,keys=choose_ssps)",
"_____no_output_____"
],
[
"## forcing\nSSP_forc = pd.concat([get_RCMIP_forc(x) for x in choose_ssps],axis=1,keys=choose_ssps).loc[:2100]",
"_____no_output_____"
]
],
[
[
"## run the model!",
"_____no_output_____"
]
],
[
[
"default_SSP_run = run_FaIR(emissions_in=SSP_emms,forcing_in=SSP_forc)",
"Integrating 5 scenarios, 1 gas cycle parameter sets, 1 thermal response parameter sets, over ['bc', 'bc|aci', 'bc|bc_on_snow', 'c2f6', 'c3f8', 'c4f10', 'c5f12', 'c6f14', 'c7f16', 'c8f18', 'c_c4f8', 'carbon_dioxide', 'carbon_tetrachloride', 'carbon_tetrachloride|o3', 'cf4', 'cfc11', 'cfc113', 'cfc113|o3', 'cfc114', 'cfc114|o3', 'cfc115', 'cfc115|o3', 'cfc11|o3', 'cfc12', 'cfc12|o3', 'ch2cl2', 'ch2cl2|o3', 'ch3ccl3', 'ch3ccl3|o3', 'chcl3', 'chcl3|o3', 'co', 'co|o3', 'halon1202', 'halon1202|o3', 'halon1211', 'halon1211|o3', 'halon1301', 'halon1301|o3', 'halon2402', 'halon2402|o3', 'hcfc141b', 'hcfc141b|o3', 'hcfc142b', 'hcfc142b|o3', 'hcfc22', 'hcfc22|o3', 'hfc125', 'hfc134a', 'hfc143a', 'hfc152a', 'hfc227ea', 'hfc23', 'hfc236fa', 'hfc245fa', 'hfc32', 'hfc365mfc', 'hfc4310mee', 'methane', 'methane|strat_h2o', 'methane|o3', 'methyl_bromide', 'methyl_bromide|o3', 'methyl_chloride', 'methyl_chloride|o3', 'nf3', 'nh3', 'nitrous_oxide', 'nitrous_oxide|o3', 'nmvoc', 'nmvoc|o3', 'nox', 'nox_avi', 'nox_avi|contrails', 'nox|o3', 'oc', 'oc|aci', 'sf6', 'so2', 'so2f2', 'so2|aci'] forcing agents, between 1750 and 2100...\n"
]
],
[
[
"## plot the results",
"_____no_output_____"
]
],
[
[
"## get MAGICC7.1.0 data to benchmark\nMAGICC_defaults = pd.read_csv('../../aux/input-data/RCMIP/data_results_phase-1_magicc7_rcmip_phase-1_magicc7.1.0.beta_v1-0-0.csv').drop(['Model','Unit','Climatemodel','Region'],axis=1).set_index(['Scenario','Variable']).reindex(['esm-'+x+'-allGHG' for x in choose_ssps],level=0)\nRCMIP_outputmap = pd.read_csv('../../aux/FaIRv2.0.0-alpha_RCMIP_inputmap.csv',index_col=0)\n\nMAGICC_defaults = MAGICC_defaults.rename(RCMIP_outputmap.reset_index().set_index('RCMIP_concs_key')['index'].to_dict(),level=1).reindex(RCMIP_outputmap.index,level=1).T\nMAGICC_defaults.index = MAGICC_defaults.index.astype(int)\nMAGICC_defaults.rename(dict(zip(['esm-'+x+'-allGHG' for x in choose_ssps],choose_ssps)),axis=1,level=0,inplace=True)",
"_____no_output_____"
],
[
"## get FaIRv1.5 data to benchmark\nFaIR_defaults = pd.concat([pd.read_csv('../../aux/input-data/RCMIP/rcmip-master-data-results-phase-1-fair/data/results/phase-1/fair/rcmip_phase-1_fair-1.5-default-'+x+'_v1-0-1.csv') for x in ['esm-'+x+'-allGHG' for x in choose_ssps]]).drop(['Model','Unit','Climatemodel','Region'],axis=1).set_index(['Scenario','Variable'])\n\nFaIR_defaults = FaIR_defaults.rename(RCMIP_outputmap.reset_index().set_index('RCMIP_concs_key')['index'].to_dict(),level=1).reindex(RCMIP_outputmap.index,level=1).T\nFaIR_defaults.index = [int(x[:4]) for x in FaIR_defaults.index]\nFaIR_defaults.rename(dict(zip(['esm-'+x+'-allGHG' for x in choose_ssps],choose_ssps)),axis=1,level=0,inplace=True)",
"_____no_output_____"
],
[
"## set plot rcParams\nmatplotlib.rcParams['font.family']='Helvetica'\nmatplotlib.rcParams['font.size']=11\n\nmatplotlib.rcParams['axes.formatter.limits']=-3,3\n\nmatplotlib.rcParams['legend.frameon']=False\n\nplt.rcParams['pdf.fonttype'] = 42",
"_____no_output_____"
],
[
"## & plot!\ncolors= {'ssp245':'#7570b3','ssp370':'#d95f02','ssp585':'#e7298a','ssp119':'#66a61e','ssp126':'#1b9e77','history':'grey'}\nmap_conc_names = dict(zip(WMGHG_concs.columns,['C$_2$F$_6$','C$_3$F$_8$','C$_4$F$_{10}$','C$_5$F$_{12}$','C$_6$F$_{14}$','C$_7$F$_{16}$','C$_8$F$_{18}$','cC$_4$F$_{8}$','CO$_2$','CCl$_4$','CF$_4$','CFC113','CFC114','CFC115','CFC11','CFC12','CH$_2$Cl$_2$','CH$_3$CCl$_3$','CHCl$_3$','Halon1211','Halon1301','Halon2402','HCFC141b', 'HCFC142b', 'HCFC22', 'HFC125','HFC134a', 'HFC143a', 'HFC152a', 'HFC227ea', 'HFC236fa', 'HFC23','HFC245fa', 'HFC32', 'HFC365mfc', 'HFC4310mee','CH$_4$','CH$_3$Br','CH$_3$Cl','NF$_3$','N$_2$O','SF$_6$','SO$_2$F$_2$','Halon1202']))\n\nfig,ax = plt.subplots(8,6,figsize=(15,15))\n\nwith plt.rc_context({\"lines.linewidth\": 0.75,\"lines.markersize\":4,\"lines.markerfacecolor\":'none',\"lines.markeredgewidth\":0.5}):\n\n for i,gas in enumerate(WMGHG_concs.columns):\n \n ax.flatten()[i].plot(WMGHG_concs.loc[1850:,gas].iloc[::10],'o',color='k')\n \n for ssp in choose_ssps:\n ax.flatten()[i].plot(default_SSP_run['C'].loc[2014:2100,(ssp,'default',gas)],color=colors[ssp],label=ssp)\n\n ax.flatten()[i].plot(MAGICC_defaults.loc[2014:2100,(ssp,gas)]*RCMIP_outputmap.loc[gas,'RCMIP_concs_scaling'],color=colors[ssp],ls=':')\n \n try: # need exceptions for FaIR as not all gases were included as this point.\n ax.flatten()[i].plot(FaIR_defaults.loc[2014:2100,(ssp,gas)]*RCMIP_outputmap.loc[gas,'RCMIP_concs_scaling'],color=colors[ssp],ls='-.')\n except:\n pass\n\n ax.flatten()[i].plot(default_SSP_run['C'].loc[1850:2014,('ssp245','default',gas)],color=colors['history'],label='historical')\n \n ax.flatten()[i].plot(MAGICC_defaults.loc[1850:2014,(ssp,gas)]*RCMIP_outputmap.loc[gas,'RCMIP_concs_scaling'],color=colors['history'],ls=':')\n \n try:\n ax.flatten()[i].plot(FaIR_defaults.loc[1850:2014,(ssp,gas)]*RCMIP_outputmap.loc[gas,'RCMIP_concs_scaling'],color=colors['history'],ls='-.')\n except:\n pass\n \n ax.flatten()[i].text(0.5,0.98,map_conc_names[gas],transform=ax.flatten()[i].transAxes,va='bottom',ha='center',fontsize=12,fontweight='bold')\n \n if gas in ['carbon_dioxide','methane','nitrous_oxide']:\n ax1 = inset_axes(ax.flatten()[i],width=\"100%\",height=\"100%\",bbox_to_anchor=(0.05,0.43,0.5,0.6),bbox_transform=ax.flatten()[i].transAxes)\n ax1.plot(default_SSP_run['C'].loc[1850:2014,('ssp245','default',gas)],color=colors['history'])\n ax1.plot(MAGICC_defaults.loc[1850:2014,(ssp,gas)]*RCMIP_outputmap.loc[gas,'RCMIP_concs_scaling'],color=colors['history'],ls=':')\n ax1.plot(FaIR_defaults.loc[1850:2014,(ssp,gas)]*RCMIP_outputmap.loc[gas,'RCMIP_concs_scaling'],color=colors['history'],ls='-.')\n ax1.plot(WMGHG_concs.loc[1850:,gas].iloc[::10],'o',color='k')\n ax1.set_xticklabels([])\n ax1.tick_params(left=False,labelleft=False,right=True,labelright=True)\n ax1.ticklabel_format(axis='y',style=\"plain\")\n ax1.set_xlim(1850,2014)\n\n [a.tick_params(labelbottom=False) for a in ax.flatten()]\n [a.tick_params(labelbottom=True) for a in ax.flatten()[-11:]]\n [a.ticklabel_format(style=\"plain\") for a in ax.flatten()[-11:]]\n [a.set_xlabel('year') for a in ax.flatten()[-11:]]\n [a.set_xlim(1850,2100) for a in ax.flatten()]\n [a.spines[pos].set_visible(False) for pos in ['right','top'] for a in ax.flatten()]\n \n ax.flatten()[-6].plot([],[],'k',label='FaIRv2.0.0')\n ax.flatten()[-6].plot([],[],'k:',label='MAGICC7.1.0-beta')\n ax.flatten()[-6].plot([],[],'k-.',label='FaIRv1.5')\n\n# fig.subplots_adjust(hspace=0.1)\n \n plt.tight_layout(h_pad=0,w_pad=0)\n \n ax.flatten()[-6].legend(loc=(1.05,0),labelspacing=0.1,prop={'size':9})\n\n [a.set_visible(False) for a in ax.flatten()[-5:]]\n\n \n[fig.savefig('../../docs/manuscript/figures/Fig2.'+x,dpi=600,bbox_inches='tight') for x in ['png','pdf']]\n''",
"/nfs/a65/pmcjs/miniconda3/envs/leach2021/lib/python3.7/site-packages/ipykernel_launcher.py:58: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.\nfindfont: Font family ['Helvetica'] not found. Falling back to DejaVu Sans.\nfindfont: Font family ['Helvetica'] not found. Falling back to DejaVu Sans.\nfindfont: Font family ['Helvetica'] not found. Falling back to DejaVu Sans.\n"
]
],
[
[
"# I. Default parameter metrics\nHere we compute GWP values for each gas in the FaIRv2.0.0-alpha namelist; under a scenario of concentrations fixed at their present day (2014) levels. These include the impact due to all forcing (direct through radiative effects + indirect through any atmospheric chemistry).",
"_____no_output_____"
]
],
[
[
"historical_concrun = WMGHG_concs.dropna().copy()\n## add in aerosol emissions\naer_species = ['so2','nox','co','nmvoc','bc','nh3','oc','nox_avi']\nhistorical_concrun = pd.concat([historical_concrun,get_SSP_emms('ssp245').loc[:2014,aer_species]],axis=1)\nhistorical_concrun = pd.concat([historical_concrun],axis=1,keys=['historical'])\n\nhistorical_forc = pd.concat([get_RCMIP_forc('ssp245').loc[historical_concrun.index]],axis=1,keys=['historical'])\n\n## extend both series into the future, but fixed @ 2014 levels\nhistorical_concrun = historical_concrun.reindex(np.arange(1750,2516)).interpolate(limit=501,limit_direction='forward')\nhistorical_forc = historical_forc.reindex(np.arange(1750,2516)).interpolate(limit=501,limit_direction='forward')",
"_____no_output_____"
],
[
"## concentration-driven run over history\nhist_run = run_FaIR(concentrations_in=historical_concrun, forcing_in=historical_forc, aer_concs_in=aer_species)",
"Integrating 1 scenarios, 1 gas cycle parameter sets, 1 thermal response parameter sets, over ['bc', 'bc|aci', 'bc|bc_on_snow', 'c2f6', 'c3f8', 'c4f10', 'c5f12', 'c6f14', 'c7f16', 'c8f18', 'c_c4f8', 'carbon_dioxide', 'carbon_tetrachloride', 'carbon_tetrachloride|o3', 'cf4', 'cfc11', 'cfc113', 'cfc113|o3', 'cfc114', 'cfc114|o3', 'cfc115', 'cfc115|o3', 'cfc11|o3', 'cfc12', 'cfc12|o3', 'ch2cl2', 'ch2cl2|o3', 'ch3ccl3', 'ch3ccl3|o3', 'chcl3', 'chcl3|o3', 'co', 'co|o3', 'halon1202', 'halon1202|o3', 'halon1211', 'halon1211|o3', 'halon1301', 'halon1301|o3', 'halon2402', 'halon2402|o3', 'hcfc141b', 'hcfc141b|o3', 'hcfc142b', 'hcfc142b|o3', 'hcfc22', 'hcfc22|o3', 'hfc125', 'hfc134a', 'hfc143a', 'hfc152a', 'hfc227ea', 'hfc23', 'hfc236fa', 'hfc245fa', 'hfc32', 'hfc365mfc', 'hfc4310mee', 'methane', 'methane|strat_h2o', 'methane|o3', 'methyl_bromide', 'methyl_bromide|o3', 'methyl_chloride', 'methyl_chloride|o3', 'nf3', 'nh3', 'nitrous_oxide', 'nitrous_oxide|o3', 'nmvoc', 'nmvoc|o3', 'nox', 'nox_avi', 'nox_avi|contrails', 'nox|o3', 'oc', 'oc|aci', 'sf6', 'so2', 'so2f2', 'so2|aci'] forcing agents, between 1750 and 2515...\n"
],
[
"## obtain corresponding emissions & reset aerosol emissions\nhist_emms = hist_run['Emissions'].droplevel(axis=1,level=1)\nhist_emms.loc[:2014,('historical',aer_species)] = get_SSP_emms('ssp245').loc[:2014,aer_species].values\nhist_emms.loc[2015:,('historical',aer_species)] = hist_emms.loc[2014,('historical',aer_species)].values",
"_____no_output_____"
],
[
"## run emissions to check consistency\nhist_run_emms = run_FaIR(emissions_in=hist_emms, forcing_in=historical_forc)",
"Integrating 1 scenarios, 1 gas cycle parameter sets, 1 thermal response parameter sets, over ['bc', 'bc|aci', 'bc|bc_on_snow', 'c2f6', 'c3f8', 'c4f10', 'c5f12', 'c6f14', 'c7f16', 'c8f18', 'c_c4f8', 'carbon_dioxide', 'carbon_tetrachloride', 'carbon_tetrachloride|o3', 'cf4', 'cfc11', 'cfc113', 'cfc113|o3', 'cfc114', 'cfc114|o3', 'cfc115', 'cfc115|o3', 'cfc11|o3', 'cfc12', 'cfc12|o3', 'ch2cl2', 'ch2cl2|o3', 'ch3ccl3', 'ch3ccl3|o3', 'chcl3', 'chcl3|o3', 'co', 'co|o3', 'halon1202', 'halon1202|o3', 'halon1211', 'halon1211|o3', 'halon1301', 'halon1301|o3', 'halon2402', 'halon2402|o3', 'hcfc141b', 'hcfc141b|o3', 'hcfc142b', 'hcfc142b|o3', 'hcfc22', 'hcfc22|o3', 'hfc125', 'hfc134a', 'hfc143a', 'hfc152a', 'hfc227ea', 'hfc23', 'hfc236fa', 'hfc245fa', 'hfc32', 'hfc365mfc', 'hfc4310mee', 'methane', 'methane|strat_h2o', 'methane|o3', 'methyl_bromide', 'methyl_bromide|o3', 'methyl_chloride', 'methyl_chloride|o3', 'nf3', 'nh3', 'nitrous_oxide', 'nitrous_oxide|o3', 'nmvoc', 'nmvoc|o3', 'nox', 'nox_avi', 'nox_avi|contrails', 'nox|o3', 'oc', 'oc|aci', 'sf6', 'so2', 'so2f2', 'so2|aci'] forcing agents, between 1750 and 2515...\n"
],
[
"## run over each gas species, perturbing each by 1t in 2015\ngas_mass_conversion_factors = pd.Series(index=hist_emms.columns.levels[1],dtype=float)\ngas_mass_conversion_factors.loc[:] = 1\ngas_mass_conversion_factors.loc['carbon_dioxide'] = (1/1000)/(44.01/12.01)\ngas_mass_conversion_factors.loc['nitrous_oxide'] = 28/44\n\nrf_results = []\n\nfor gas_species in hist_emms.columns.levels[1]:\n \n pert_emms = hist_emms.copy()\n pert_emms.loc[2015,('historical',gas_species)] += gas_mass_conversion_factors.loc[gas_species]/1e6\n \n pert_result = run_FaIR(emissions_in=pert_emms, forcing_in=historical_forc, show_run_info=False)\n \n rf_results += [(pert_result['RF'].loc[:,('historical','default','Total')]-hist_run_emms['RF'].loc[:,('historical','default','Total')]).rename(gas_species)]\n \nrf_results = pd.concat(rf_results,axis=1)",
"100%|██████████| 765/765 [00:00<00:00, 2395.30 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2411.74 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2406.14 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2410.23 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2408.74 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2395.27 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2402.66 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2401.23 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2397.19 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2403.85 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2416.76 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2413.48 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2413.65 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2410.79 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2399.82 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2402.50 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2410.87 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2410.36 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2406.90 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2393.91 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2401.53 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2388.75 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2395.24 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2407.69 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2405.70 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2394.59 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2396.24 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2413.20 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2402.47 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2386.94 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2404.02 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2404.06 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2398.38 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2397.07 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2402.42 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2397.61 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2410.40 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2409.80 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2398.35 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2409.12 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2403.94 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2394.80 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2407.36 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2396.37 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2397.04 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2408.10 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2397.88 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2400.70 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2416.45 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2403.86 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2398.85 timestep/s]\n100%|██████████| 765/765 [00:00<00:00, 2407.93 timestep/s]\n"
],
[
"AGWP = rf_results.cumsum().loc[2015+np.array([5,10,20,50,100,500])]\nAGWP.index = np.array([5,10,20,50,100,500])\nGWP = AGWP.apply(lambda x: x/AGWP.carbon_dioxide)",
"_____no_output_____"
],
[
"print('GWP value over various timescales:')\nGWP.index.name = 'timescale / years'\nGWP.columns.name = 'agent'\n\nGWP.T.applymap(lambda x:round_to_sf(x,2))#.to_latex('../../docs/manuscript/tables/TabS3',escape=True,bold_rows=True)",
"GWP value over various timescales:\n"
]
],
[
[
"# Supplement I. Methane lifetime over history + RCP8.5 extension\nA demonstration of the state-dependent lifetime of methane over RCP history + extended to 2100 with RCP8.5. We use RCP8.5 since this is (at least, appears to be) the most commonly discussed scenario in methane sensitivity literature.",
"_____no_output_____"
]
],
[
[
"RCP85_emms = RCMIP_to_FaIR_input_emms('rcp85').dropna(how='all').dropna(axis=1,how='all')\nRCP85_emms = pd.concat([RCP85_emms],axis=1,keys=['RCP8.5'])\nrebase_species = ['so2','nox','co','nmvoc','bc','nh3','oc','nox_avi','methyl_bromide','methyl_chloride','chcl3','ch2cl2']\nrebase_species = list(set(rebase_species).intersection(RCP85_emms.columns.levels[1]))\nRCP85_emms.loc[:,('RCP8.5',rebase_species)] -= RCP85_emms.loc[1765,('RCP8.5',rebase_species)]\n\nRCP85_forc = pd.concat([get_RCMIP_forc('rcp85',['Radiative Forcing|Anthropogenic|Albedo Change','Radiative Forcing|Natural']).dropna()],axis=1,keys=['RCP8.5'])",
"_____no_output_____"
],
[
"RCP85_run = run_FaIR(emissions_in=RCP85_emms,\n forcing_in=RCP85_forc,\n gas_parameters=get_gas_parameter_defaults().reindex(RCP85_emms.columns.levels[1],axis=1,level=1))",
"Integrating 1 scenarios, 1 gas cycle parameter sets, 1 thermal response parameter sets, over ['bc', 'c2f6', 'c6f14', 'carbon_dioxide', 'carbon_tetrachloride', 'cf4', 'cfc11', 'cfc113', 'cfc114', 'cfc115', 'cfc12', 'ch3ccl3', 'co', 'halon1202', 'halon1211', 'halon1301', 'halon2402', 'hcfc141b', 'hcfc142b', 'hcfc22', 'hfc125', 'hfc134a', 'hfc143a', 'hfc227ea', 'hfc23', 'hfc245fa', 'hfc32', 'hfc4310mee', 'methane', 'methyl_bromide', 'methyl_chloride', 'nh3', 'nitrous_oxide', 'nmvoc', 'nox', 'oc', 'sf6', 'so2'] forcing agents, between 1765 and 2500...\n"
],
[
"CH4_lifetime = RCP85_run['alpha'].xs('methane',axis=1,level=2).droplevel(axis=1,level=1)*RCP85_run['gas_parameters'].loc['tau1',('default','methane')]",
"_____no_output_____"
],
[
"sn.lineplot(data=CH4_lifetime.loc[1850:2100],palette=['k'])\nsn.despine()\nplt.xlabel('year')\nplt.ylabel('CH$_4$ lifetime / yrs')\nplt.gca().ticklabel_format(style='plain')\nplt.xlim(1850,2100)\n\n[plt.savefig('../../docs/manuscript/figures/FigS2.'+x,dpi=600,bbox_inches='tight') for x in ['png','pdf']]\n''",
"_____no_output_____"
],
[
"# comparison with Holmes et al., 2013\n\n## 2010 values:\nprint('Holmes 2010:',1/(1/120+1/150+1/200+1/11.2))\nprint('FaIRv2.0.0-alpha 2010:',CH4_lifetime.loc[2010].values[0],end='\\n\\n')\n\nprint('Holmes 2010-2100 change:',(1/120+1/150+1/200+1/11.2)/(1/120+1/150+1/200+1/(11.2*1.129)))\nprint('FaIRv2.0.0-alpha 2010-2100 change:',(CH4_lifetime.loc[2100]/CH4_lifetime.loc[2010]).values[0])",
"Holmes 2010: 9.15032679738562\nFaIRv2.0.0-alpha 2010: 8.902796846929608\n\nHolmes 2010-2100 change: 1.102961458892039\nFaIRv2.0.0-alpha 2010-2100 change: 1.0991068995747364\n"
]
],
[
[
"# Supplement II. FaIRv2.0.0 additivity\nVery brief test of how linear FaIR actually is. Non-linearity in FaIR only arises from the CO2 & CH4 cycles. The climate response of FaIR is linear in forcing. Here we test the linearity over history by carrying out several CO2 / CH4 pulse response experiments.",
"_____no_output_____"
]
],
[
[
"# default_SSP_run = run_FaIR(emissions_in=SSP_emms,forcing_in=SSP_forc)\n\nbase_emms = RCMIP_to_FaIR_input_emms('ssp245').interpolate().loc[1750:2500]\nrebase_species = ['so2','nox','co','nmvoc','bc','nh3','oc','nox_avi','methyl_bromide','methyl_chloride','chcl3','ch2cl2']\nbase_emms.loc[:,rebase_species] -= base_emms.loc[1750,rebase_species]\nbase_emms = pd.concat([base_emms],axis=1,keys=['ssp245'])\n\nexperiments = []\n\n# scale methane by 28 (GWP100) for closer comparison\npulse_scaling = dict(carbon_dioxide=12/44,methane=1000/28)\n\nfor species in ['carbon_dioxide','methane']:\n for pulse_size in [0]+list(np.arange(0.01,0.1,0.01))+list(np.arange(0.1,1,0.1))+list(np.arange(1,10,1))+list(np.arange(10,100,10))+list(np.arange(100,1001,100)):\n experiment = base_emms.copy()\n experiment.loc[2019,('ssp245',species)] += pulse_size*pulse_scaling[species]\n experiments += [experiment.rename(dict(ssp245=species+'_'+str(pulse_size)),axis=1,level=0)]\n \nexperiments = pd.concat(experiments,axis=1)",
"_____no_output_____"
],
[
"pulse_runs = run_FaIR(emissions_in=experiments,\n forcing_in=pd.concat([get_RCMIP_forc('ssp245')]*experiments.columns.levels[0].size,axis=1,keys=experiments.columns.levels[0]))",
"Integrating 94 scenarios, 1 gas cycle parameter sets, 1 thermal response parameter sets, over ['bc', 'bc|aci', 'bc|bc_on_snow', 'c2f6', 'c3f8', 'c4f10', 'c5f12', 'c6f14', 'c7f16', 'c8f18', 'c_c4f8', 'carbon_dioxide', 'carbon_tetrachloride', 'carbon_tetrachloride|o3', 'cf4', 'cfc11', 'cfc113', 'cfc113|o3', 'cfc114', 'cfc114|o3', 'cfc115', 'cfc115|o3', 'cfc11|o3', 'cfc12', 'cfc12|o3', 'ch2cl2', 'ch2cl2|o3', 'ch3ccl3', 'ch3ccl3|o3', 'chcl3', 'chcl3|o3', 'co', 'co|o3', 'halon1202', 'halon1202|o3', 'halon1211', 'halon1211|o3', 'halon1301', 'halon1301|o3', 'halon2402', 'halon2402|o3', 'hcfc141b', 'hcfc141b|o3', 'hcfc142b', 'hcfc142b|o3', 'hcfc22', 'hcfc22|o3', 'hfc125', 'hfc134a', 'hfc143a', 'hfc152a', 'hfc227ea', 'hfc23', 'hfc236fa', 'hfc245fa', 'hfc32', 'hfc365mfc', 'hfc4310mee', 'methane', 'methane|strat_h2o', 'methane|o3', 'methyl_bromide', 'methyl_bromide|o3', 'methyl_chloride', 'methyl_chloride|o3', 'nf3', 'nh3', 'nitrous_oxide', 'nitrous_oxide|o3', 'nmvoc', 'nmvoc|o3', 'nox', 'nox_avi', 'nox_avi|contrails', 'nox|o3', 'oc', 'oc|aci', 'sf6', 'so2', 'so2f2', 'so2|aci'] forcing agents, between 1750 and 2500...\n"
]
],
[
[
"### nonlinearities in terms of scaled anomalies",
"_____no_output_____"
]
],
[
[
"## compute the pulse experiment anomalies relative to the baseline\npulse_temp_anomalies = (pulse_runs['T'] - pulse_runs['T'].carbon_dioxide_0.values)\npulse_temp_anomalies.columns = pd.MultiIndex.from_tuples([(x[:14],float(x[15:])) if x[0]=='c' else (x[:7],float(x[8:])) for x in pulse_temp_anomalies.columns.levels[0]])\npulse_temp_anomalies = pulse_temp_anomalies.drop(0,axis=1,level=1)\n\npulse_temp_anomalies_scaled = pulse_temp_anomalies.apply(lambda x: x*1000/x.name[1])\n\nCO2_RF_anomalies = (pulse_runs['RF'].xs('carbon_dioxide',axis=1,level=2) - pulse_runs['RF'].xs('carbon_dioxide',axis=1,level=2).carbon_dioxide_0.values)\nCO2_RF_anomalies.columns = pd.MultiIndex.from_tuples([(x[:14],float(x[15:])) if x[0]=='c' else (x[:7],float(x[8:])) for x in CO2_RF_anomalies.columns.levels[0]])\nCO2_RF_anomalies = CO2_RF_anomalies.drop(0,axis=1,level=1)\n\nCO2_RF_anomalies_scaled = CO2_RF_anomalies.apply(lambda x: x*1000/x.name[1])\n\nCH4_RF_anomalies = (pulse_runs['RF'].xs('methane',axis=1,level=2) - pulse_runs['RF'].xs('methane',axis=1,level=2).carbon_dioxide_0.values)\nCH4_RF_anomalies.columns = pd.MultiIndex.from_tuples([(x[:14],float(x[15:])) if x[0]=='c' else (x[:7],float(x[8:])) for x in CH4_RF_anomalies.columns.levels[0]])\nCH4_RF_anomalies = CH4_RF_anomalies.drop(0,axis=1,level=1)\n\nCH4_RF_anomalies_scaled = CH4_RF_anomalies.apply(lambda x: x*1000/x.name[1])\n\nCO2_C_anomalies = (pulse_runs['C'].xs('carbon_dioxide',axis=1,level=2) - pulse_runs['C'].xs('carbon_dioxide',axis=1,level=2).carbon_dioxide_0.values)\nCO2_C_anomalies.columns = pd.MultiIndex.from_tuples([(x[:14],float(x[15:])) if x[0]=='c' else (x[:7],float(x[8:])) for x in CO2_C_anomalies.columns.levels[0]])\nCO2_C_anomalies = CO2_C_anomalies.drop(0,axis=1,level=1)\n\nCO2_C_anomalies_scaled = CO2_C_anomalies.apply(lambda x: x*1000/x.name[1])\n\nCH4_C_anomalies = (pulse_runs['C'].xs('methane',axis=1,level=2) - pulse_runs['C'].xs('methane',axis=1,level=2).carbon_dioxide_0.values)\nCH4_C_anomalies.columns = pd.MultiIndex.from_tuples([(x[:14],float(x[15:])) if x[0]=='c' else (x[:7],float(x[8:])) for x in CH4_C_anomalies.columns.levels[0]])\nCH4_C_anomalies = CH4_C_anomalies.drop(0,axis=1,level=1)\n\nCH4_C_anomalies_scaled = CH4_C_anomalies.apply(lambda x: x*1000/x.name[1])\n\nCO2_alph_anomalies = pulse_runs['alpha'].xs('carbon_dioxide',axis=1,level=2).sub(pulse_runs['alpha'].xs('carbon_dioxide',axis=1,level=2).carbon_dioxide_0,axis=0)\nCO2_alph_anomalies.columns = pd.MultiIndex.from_tuples([(x[:14],float(x[15:])) if x[0]=='c' else (x[:7],float(x[8:])) for x in CO2_alph_anomalies.columns.levels[0]])\nCO2_alph_anomalies = CO2_alph_anomalies.drop(0,axis=1,level=1)\n\nCO2_alph_anomalies_scaled = CO2_alph_anomalies.apply(lambda x: x*1000/x.name[1])\n\nCH4_alph_anomalies = pulse_runs['alpha'].xs('methane',axis=1,level=2).sub(pulse_runs['alpha'].xs('methane',axis=1,level=2).carbon_dioxide_0,axis=0)\nCH4_alph_anomalies.columns = pd.MultiIndex.from_tuples([(x[:14],float(x[15:])) if x[0]=='c' else (x[:7],float(x[8:])) for x in CH4_alph_anomalies.columns.levels[0]])\nCH4_alph_anomalies = CH4_alph_anomalies.drop(0,axis=1,level=1)\n\nCH4_alph_anomalies_scaled = CH4_alph_anomalies.apply(lambda x: x*1000/x.name[1])\n\nanomalies = pd.concat([pulse_temp_anomalies_scaled,\n CO2_RF_anomalies_scaled,\n CH4_RF_anomalies_scaled,\n CO2_C_anomalies_scaled,\n CH4_C_anomalies_scaled,\n CO2_alph_anomalies_scaled,\n CH4_alph_anomalies_scaled],\n axis=1,\n keys=['T',r'RF$_{\\mathrm{CO}_2}$',r'RF$_{\\mathrm{CH}_4}$',r'C$_{\\mathrm{CO}_2}$',r'C$_{\\mathrm{CH}_4}$',r'$\\alpha_{\\mathrm{CO}_2}$',r'$\\alpha_{\\mathrm{CH}_4}$'],\n names=['variable']).rename(dict(carbon_dioxide='CO$_2$',methane='CH$_4$'),axis=1,level=1).loc[2020:].sort_index(axis=1).stack(level=[0,1,2]).reset_index().rename({'level_0':'time','level_2':'pulse_type','level_3':'pulse_size',0:'value'},axis=1)\n\nanomalies.time -= 2019\n\n# set relative to small pulse limit\n## comment out if absolute anomalies (ie. relative to reference) desired\npulse_temp_anomalies_scaled = pulse_temp_anomalies_scaled.apply(lambda x: x-pulse_temp_anomalies_scaled.loc[:,(x.name[0],0.01)])\nCO2_RF_anomalies_scaled = CO2_RF_anomalies_scaled.apply(lambda x: x-CO2_RF_anomalies_scaled.loc[:,(x.name[0],0.01)])\nCH4_RF_anomalies_scaled = CH4_RF_anomalies_scaled.apply(lambda x: x-CH4_RF_anomalies_scaled.loc[:,(x.name[0],0.01)])\nCO2_C_anomalies_scaled = CO2_C_anomalies_scaled.apply(lambda x: x-CO2_C_anomalies_scaled.loc[:,(x.name[0],0.01)])\nCH4_C_anomalies_scaled = CH4_C_anomalies_scaled.apply(lambda x: x-CH4_C_anomalies_scaled.loc[:,(x.name[0],0.01)])\nCO2_alph_anomalies_scaled = CO2_alph_anomalies_scaled.apply(lambda x: x-CO2_alph_anomalies_scaled.loc[:,(x.name[0],0.01)])\nCH4_alph_anomalies_scaled = CH4_alph_anomalies_scaled.apply(lambda x: x-CH4_alph_anomalies_scaled.loc[:,(x.name[0],0.01)])\n\nanomalies_rel = pd.concat([pulse_temp_anomalies_scaled,\n CO2_RF_anomalies_scaled,\n CH4_RF_anomalies_scaled,\n CO2_C_anomalies_scaled,\n CH4_C_anomalies_scaled,\n CO2_alph_anomalies_scaled,\n CH4_alph_anomalies_scaled],\n axis=1,\n keys=['T',r'RF$_{\\mathrm{CO}_2}$',r'RF$_{\\mathrm{CH}_4}$',r'C$_{\\mathrm{CO}_2}$',r'C$_{\\mathrm{CH}_4}$',r'$\\alpha_{\\mathrm{CO}_2}$',r'$\\alpha_{\\mathrm{CH}_4}$'],\n names=['variable']).rename(dict(carbon_dioxide='CO$_2$ - relative',methane='CH$_4$ - relative'),axis=1,level=1).loc[2020:].sort_index(axis=1).stack(level=[0,1,2]).reset_index().rename({'level_0':'time','level_2':'pulse_type','level_3':'pulse_size',0:'value'},axis=1)\n\nanomalies_rel.time -= 2019",
"_____no_output_____"
],
[
"plot_df = pd.concat([anomalies,anomalies_rel])\nplot_df.head()",
"_____no_output_____"
],
[
"g=sn.FacetGrid(plot_df.query('pulse_size in [1,10,100,200,500,1000]').sort_values(['pulse_type','variable']),col='variable',row='pulse_type',hue='pulse_size',palette=[(x,x,x) for x in np.arange(0,1,1/7)[::-1]],margin_titles=True,sharey=False)\ng.map(sn.lineplot,'time','value')\n\ng.set_titles(col_template=\"{col_name}\",row_template='pulse type = {row_name}',fontweight='bold').set(xlim=[0,480])\n[a.set_ylabel('anomaly / ppb') for a in g.axes[:,2]]\n[a.set_ylabel('anomaly / ppm') for a in g.axes[:,3]]\n[a.set_ylabel('anomaly / W m$^{-2}$') for a in g.axes[:,4]]\n[a.set_ylabel('anomaly / K') for a in g.axes[:,-1]]\n[a.set_ylabel('anomaly / -') for a in g.axes[:,0]]\n\ng.axes[0,0].legend(title='pulse size / GtCO$_2$-eq')\n\n[plt.savefig('../../docs/manuscript/figures/FigS3.'+x,dpi=600,bbox_inches='tight') for x in ['png','pdf']]\n''",
"findfont: Font family ['Helvetica'] not found. Falling back to DejaVu Sans.\n"
]
],
[
[
"### measuring nonlinearities in a relative sense:\nMarked out to prevent from running.",
"_____no_output_____"
],
[
"## measuring extent of nonlinearity as anomalies relative to 1000 GtC-eq pulse, normalised by 1000 GtC-eq pulse anomaly\nCO2_T_nonlin = pulse_temp_anomalies_scaled.loc[2020:,'carbon_dioxide'].sub(pulse_temp_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0).div(pulse_temp_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0)\nCH4_T_nonlin = pulse_temp_anomalies_scaled.loc[2020:,'methane'].sub(pulse_temp_anomalies_scaled.loc[2020:,('methane',1000)],axis=0).div(pulse_temp_anomalies_scaled.loc[2020:,('methane',1000)],axis=0)\n\nCO2_CO2_RF_nonlin = CO2_RF_anomalies_scaled.loc[2020:,'carbon_dioxide'].sub(CO2_RF_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0).div(CO2_RF_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0)\nCO2_CH4_RF_nonlin = CO2_RF_anomalies_scaled.loc[2020:,'methane'].sub(CO2_RF_anomalies_scaled.loc[2020:,('methane',1000)],axis=0).div(CO2_RF_anomalies_scaled.loc[2020:,('methane',1000)],axis=0)\n\nCH4_CO2_RF_nonlin = CH4_RF_anomalies_scaled.loc[2020:,'carbon_dioxide'].sub(CH4_RF_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0).div(CH4_RF_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0)\nCH4_CH4_RF_nonlin = CH4_RF_anomalies_scaled.loc[2020:,'methane'].sub(CH4_RF_anomalies_scaled.loc[2020:,('methane',1000)],axis=0).div(CH4_RF_anomalies_scaled.loc[2020:,('methane',1000)],axis=0)\n\nCO2_CO2_C_nonlin = CO2_C_anomalies_scaled.loc[2020:,'carbon_dioxide'].sub(CO2_C_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0).div(CO2_C_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0)\nCO2_CH4_C_nonlin = CO2_C_anomalies_scaled.loc[2020:,'methane'].sub(CO2_C_anomalies_scaled.loc[2020:,('methane',1000)],axis=0).div(CO2_C_anomalies_scaled.loc[2020:,('methane',1000)],axis=0)\n\nCH4_CO2_C_nonlin = CH4_C_anomalies_scaled.loc[2020:,'carbon_dioxide'].sub(CH4_C_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0).div(CH4_C_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0)\nCH4_CH4_C_nonlin = CH4_C_anomalies_scaled.loc[2020:,'methane'].sub(CH4_C_anomalies_scaled.loc[2020:,('methane',1000)],axis=0).div(CH4_C_anomalies_scaled.loc[2020:,('methane',1000)],axis=0)\n\nCO2_CO2_alph_nonlin = CO2_alph_anomalies_scaled.loc[2020:,'carbon_dioxide'].sub(CO2_alph_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0).div(CO2_alph_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0)\nCO2_CH4_alph_nonlin = CO2_alph_anomalies_scaled.loc[2020:,'methane'].sub(CO2_alph_anomalies_scaled.loc[2020:,('methane',1000)],axis=0).div(CO2_alph_anomalies_scaled.loc[2020:,('methane',1000)],axis=0)\n\nCH4_CO2_alph_nonlin = CH4_alph_anomalies_scaled.loc[2020:,'carbon_dioxide'].sub(CH4_alph_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0).div(CH4_alph_anomalies_scaled.loc[2020:,('carbon_dioxide',1000)],axis=0)\nCH4_CH4_alph_nonlin = CH4_alph_anomalies_scaled.loc[2020:,'methane'].sub(CH4_alph_anomalies_scaled.loc[2020:,('methane',1000)],axis=0).div(CH4_alph_anomalies_scaled.loc[2020:,('methane',1000)],axis=0)\n\nnonlinearities = pd.concat([pd.concat([CO2_T_nonlin,CH4_T_nonlin],axis=1,keys=['CO2','CH4'],names=['pulse_type']),\n pd.concat([CO2_CO2_RF_nonlin,CO2_CH4_RF_nonlin],axis=1,keys=['CO2','CH4'],names=['pulse_type']),\n pd.concat([CH4_CO2_RF_nonlin,CH4_CO2_RF_nonlin],axis=1,keys=['CO2','CH4'],names=['pulse_type']),\n pd.concat([CO2_CO2_C_nonlin,CO2_CH4_C_nonlin],axis=1,keys=['CO2','CH4'],names=['pulse_type']),\n pd.concat([CH4_CO2_C_nonlin,CH4_CH4_C_nonlin],axis=1,keys=['CO2','CH4'],names=['pulse_type']),\n pd.concat([CO2_CO2_alph_nonlin,CO2_CH4_alph_nonlin],axis=1,keys=['CO2','CH4'],names=['pulse_type']),\n pd.concat([CH4_CO2_alph_nonlin,CH4_CH4_alph_nonlin],axis=1,keys=['CO2','CH4'],names=['pulse_type'])],\n axis=1,\n keys=['T','RF$_{\\text{CO}_2}$','RF$_{\\text{CH}_4}$','C$_{\\text{CO}_2}$','C$_{\\text{CH}_4}$','$\\alpha_{\\text{CO}_2}$','$\\alpha_{\\text{CH}_4}$'],\n names=['variable']).sort_index(axis=1).stack(level=[0,1,2]).reset_index().rename({'level_0':'time','level_3':'pulse_size',0:'value'},axis=1)\n\nnonlinearities.time -= 2019",
"_____no_output_____"
],
[
"from mpl_toolkits.axes_grid1.inset_locator import inset_axes\n\nclass SymPowerNorm(matplotlib.colors.Normalize):\n def __init__(self, vmin=None, vmax=None, order=1, clip=False):\n self.order = order\n matplotlib.colors.Normalize.__init__(self, vmin, vmax, clip)\n\n def __call__(self, value, clip=None):\n # I'm ignoring masked values and all kinds of edge cases to make a\n # simple example...\n x, y = [abs(self.vmin) / self.vmin * abs(self.vmin)**self.order , abs(self.vmax) / self.vmax * abs(self.vmax)**self.order], [0,1]\n return np.ma.masked_array(np.interp(abs(value) / value * abs(value)**self.order, x, y))\n\ndef mapplot(x,y,z,**kwargs):\n data = pd.concat([x,y,z],axis=1).set_index(['time','pulse_size']).unstack().droplevel(0,axis=1)\n norm=matplotlib.colors.Normalize(vmin=-0.5,vmax=0.5)#SymPowerNorm(order=1,vmin=-0.5,vmax=0.5)\n plt.pcolormesh(data.index,data.columns,data.values.T,shading='auto',norm=norm,cmap='RdBu_r')\n\ng=sn.FacetGrid(nonlinearities,col='variable',row='pulse_type',margin_titles=True,despine=False,gridspec_kws=dict(hspace=0.1,wspace=0.1))\ng.map(mapplot,'time','pulse_size','value')\n\ng.set_titles(col_template=\"{col_name}\",row_template='pulse type = {row_name}',fontweight='bold')\ng.set(yscale='log')\n[a.set_ylabel('pulse size / GtC-eq') for a in g.axes[:,0]]\n[a.set_xlabel('year') for a in g.axes[-1,:]]\n\naxins = inset_axes(g.axes[-1,-1], width=\"5%\",height=\"100%\",loc='lower left',bbox_to_anchor=(1.2, 0.55, 1, 1),bbox_transform=g.axes[-1,-1].transAxes,borderpad=0)\nplt.colorbar(cax=axins,extend='both')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0f31d02a8207f92c84310db38ff3be3230fad6f | 161,577 | ipynb | Jupyter Notebook | doc/LectureNotes/chapter4.ipynb | Shield94/Physics321 | 9875a3bf840b0fa164b865a3cb13073aff9094ca | [
"CC0-1.0"
] | 20 | 2020-01-09T17:41:16.000Z | 2022-03-09T00:48:58.000Z | doc/LectureNotes/chapter4.ipynb | Shield94/Physics321 | 9875a3bf840b0fa164b865a3cb13073aff9094ca | [
"CC0-1.0"
] | 6 | 2020-01-08T03:47:53.000Z | 2020-12-15T15:02:57.000Z | doc/LectureNotes/chapter4.ipynb | Shield94/Physics321 | 9875a3bf840b0fa164b865a3cb13073aff9094ca | [
"CC0-1.0"
] | 33 | 2020-01-10T20:40:55.000Z | 2022-02-11T20:28:41.000Z | 26.947465 | 482 | 0.530725 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0f33c420779a3381aa44ef92b9b6f8a6ec8d9a0 | 284,385 | ipynb | Jupyter Notebook | notebooks/02. NSynth (Audio Classification by Generating Images).ipynb | vinhnemo/fastai_audio | 7873fe9909889ebf31a72637468c6bac83ac3880 | [
"Apache-2.0"
] | 168 | 2018-11-29T08:09:01.000Z | 2020-04-20T12:35:59.000Z | notebooks/02. NSynth (Audio Classification by Generating Images).ipynb | vinhnemo/fastai_audio | 7873fe9909889ebf31a72637468c6bac83ac3880 | [
"Apache-2.0"
] | 8 | 2019-01-31T19:19:26.000Z | 2020-03-05T06:34:43.000Z | notebooks/02. NSynth (Audio Classification by Generating Images).ipynb | vinhnemo/fastai_audio | 7873fe9909889ebf31a72637468c6bac83ac3880 | [
"Apache-2.0"
] | 48 | 2018-12-02T10:32:53.000Z | 2020-04-05T02:02:28.000Z | 587.572314 | 170,824 | 0.862422 | [
[
[
"%reload_ext autoreload\n%autoreload 2\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Image Generation from Audio",
"_____no_output_____"
]
],
[
[
"from pathlib import Path\nfrom IPython.display import Audio\nimport librosa\nimport librosa.display\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom utils import read_file, transform_path",
"_____no_output_____"
],
[
"DATA = Path('data')\n\n# these folders must be in place\nNSYNTH_AUDIO = DATA/'nsynth_audio'\nTRAIN_AUDIO_PATH = NSYNTH_AUDIO/'train'\nVALID_AUDIO_PATH = NSYNTH_AUDIO/'valid'\n\n# these folders will be created\nNSYNTH_IMAGES = DATA/'nsynth_images'\nTRAIN_IMAGE_PATH = NSYNTH_IMAGES/'train'\nVALID_IMAGE_PATH = NSYNTH_IMAGES/'valid'",
"_____no_output_____"
],
[
"train_acoustic_fnames = [f.name for f in TRAIN_AUDIO_PATH.iterdir()\n if 'acoustic' in f.name]\nvalid_acoustic_fnames = [f.name for f in VALID_AUDIO_PATH.iterdir()\n if 'acoustic' in f.name]\nlen(train_acoustic_fnames), len(valid_acoustic_fnames)",
"_____no_output_____"
],
[
"fn = train_acoustic_fnames[8]; fn",
"_____no_output_____"
],
[
"Audio(str(TRAIN_AUDIO_PATH/fn))",
"_____no_output_____"
],
[
"x, sr = read_file(fn, TRAIN_AUDIO_PATH)\nx.shape, sr, x.dtype",
"_____no_output_____"
],
[
"def log_mel_spec_tfm(fname, src_path, dst_path):\n x, sample_rate = read_file(fname, src_path)\n \n n_fft = 1024\n hop_length = 256\n n_mels = 40\n fmin = 20\n fmax = sample_rate / 2 \n \n mel_spec_power = librosa.feature.melspectrogram(x, sr=sample_rate, n_fft=n_fft, \n hop_length=hop_length, \n n_mels=n_mels, power=2.0, \n fmin=fmin, fmax=fmax)\n mel_spec_db = librosa.power_to_db(mel_spec_power, ref=np.max)\n dst_fname = dst_path / (fname[:-4] + '.png')\n plt.imsave(dst_fname, mel_spec_db)",
"_____no_output_____"
],
[
"log_mel_spec_tfm(fn, TRAIN_AUDIO_PATH, Path('.'))\nimg = plt.imread(fn[:-4] + '.png')\nplt.imshow(img, origin='lower');",
"_____no_output_____"
],
[
"# TRAIN files took 10m43s\n\n# transform_path(TRAIN_AUDIO_PATH, TRAIN_IMAGE_PATH, log_mel_spec_tfm, \n# fnames=train_acoustic_fnames, delete=True)",
"_____no_output_____"
],
[
"# VALID files took 0m31s\n# transform_path(VALID_AUDIO_PATH, VALID_IMAGE_PATH, log_mel_spec_tfm, \n# fnames=valid_acoustic_fnames, delete=True)",
"_____no_output_____"
]
],
[
[
"### Run Image Classifier",
"_____no_output_____"
]
],
[
[
"import fastai\nfastai.__version__",
"_____no_output_____"
],
[
"from fastai.vision import *",
"_____no_output_____"
],
[
"instrument_family_pattern = r'(\\w+)_\\w+_\\d+-\\d+-\\d+.png$'\n\ndata = (ImageItemList.from_folder(NSYNTH_IMAGES)\n .split_by_folder()\n .label_from_re(instrument_family_pattern)\n .databunch())\ndata.c, data.classes",
"_____no_output_____"
],
[
"xs, ys = data.one_batch()\nxs.shape, ys.shape",
"_____no_output_____"
],
[
"xs.min(), xs.max(), xs.mean(), xs.std()",
"_____no_output_____"
],
[
"data.show_batch(3, figsize=(8,4), hide_axis=False)",
"_____no_output_____"
],
[
"learn = create_cnn(data, models.resnet18, metrics=accuracy)\nlearn.fit_one_cycle(3)",
"_____no_output_____"
],
[
"interp = ClassificationInterpretation.from_learner(learn)",
"_____no_output_____"
],
[
"interp.plot_confusion_matrix(figsize=(10, 10), dpi=60)",
"_____no_output_____"
],
[
"interp.most_confused(min_val=20)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f357eb1584baa62a69f78328d8a9c119d7c635 | 92,970 | ipynb | Jupyter Notebook | jupyter notebook/Multiple Logistic Regression.ipynb | jooyeongkang/allocation-ventilator | 343c6287b5a816597a56d6282352fd44e1cd8394 | [
"MIT"
] | null | null | null | jupyter notebook/Multiple Logistic Regression.ipynb | jooyeongkang/allocation-ventilator | 343c6287b5a816597a56d6282352fd44e1cd8394 | [
"MIT"
] | null | null | null | jupyter notebook/Multiple Logistic Regression.ipynb | jooyeongkang/allocation-ventilator | 343c6287b5a816597a56d6282352fd44e1cd8394 | [
"MIT"
] | null | null | null | 39.662969 | 14,132 | 0.50085 | [
[
[
"import os \nimport pandas as pd\n\nfrom pandas_profiling import ProfileReport\nfrom pandas_profiling.utils.cache import cache_file\nfrom collections import Counter\nimport seaborn as sn\nimport random\nimport statistics \nimport statsmodels.api as sm\n\nimport numpy as np",
"/home/joel/anaconda3/lib/python3.7/site-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
],
[
"box_file_dir = os.path.join(os.getcwd(), \"..\", \"..\", \"Box\")\nfile_path_csv = os.path.join(box_file_dir, \"covid_pts_enc_level_labs_dx_2021-02-02_deid.csv\")",
"_____no_output_____"
],
[
"df = pd.read_csv(file_path_csv, index_col=False)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"def latinx(row):\n if row.ethnicity_display == 'Hispanic or Latino' and row.race_display == 'White':\n return \"Hispanic\"\n elif row.ethnicity_display == 'Not Hispanic or Latino' and row.race_display == 'White': \n return \"White\"\n else:\n return row.race_display\n ",
"_____no_output_____"
],
[
"df['race_display'] = df.apply(lambda row: latinx(row), axis=1)",
"_____no_output_____"
],
[
"vent_df = df[~df['vent_hours_summed'].isnull()]",
"_____no_output_____"
],
[
"len(vent_df)",
"_____no_output_____"
],
[
"Counter(vent_df['race_display'])",
"_____no_output_____"
],
[
"icu_df = df[~df['icu_hours_summed'].isnull()]",
"_____no_output_____"
],
[
"Counter(icu_df['race_display'])",
"_____no_output_____"
],
[
"working_df = icu_df[~icu_df['qSOFA_score'].isnull()] \nCounter(working_df['race_display'])",
"_____no_output_____"
],
[
"data = icu_df[['age_at_admit', 'pO2_Art', \n 'qSOFA_score','race_display',\n 'vent_hours_summed', 'zip_cust_table', 'heartfailure_com_flag',\n 'cancer_com_flag','gender','WBC','Mean_Arterial_Pressure',\n 'Bili_Total','CAD_com_flag','CKD_com_flag','COPD_com_flag',\n 'Creatinine', 'FiO2/Percent','Glasgow_Coma_Score','diabetes_com_flag',\n 'hypertension_com_flag','length_of_stay','discharge_disposition_display','Platelet', 'deid_empi_encounter']]",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"working_df[['race_display', 'age_at_admit']].groupby('race_display').agg(['mean', 'count'])",
"_____no_output_____"
],
[
"# only 236 patients with all tests\nallo_df = data[['pO2_Art', 'Creatinine', 'FiO2/Percent', \n 'Glasgow_Coma_Score', 'Platelet', 'Mean_Arterial_Pressure',\n 'Bili_Total', 'deid_empi_encounter']].dropna()",
"_____no_output_____"
],
[
"list_of_patients = list(allo_df['deid_empi_encounter'])",
"_____no_output_____"
],
[
"adjusted_patients = data[data['deid_empi_encounter'].isin(list_of_patients)]",
"_____no_output_____"
],
[
"def calculate_sofa(row):\n count = 0\n \n # need to implement Fi02/po2\n \n if row.Platelet >= 100 and row.Platelet <= 149:\n count += 1 \n \n elif row.Platelet >= 50 and row.Platelet <= 99:\n count += 2\n \n elif row.Platelet >= 20 and row.Platelet <= 49:\n count += 3\n \n elif row.Platelet < 20:\n count += 4\n \n # Glasgow\n if row.Glasgow_Coma_Score == 13 or row.Glasgow_Coma_Score == 14:\n count += 1 \n \n elif row.Glasgow_Coma_Score >= 10 and row.Glasgow_Coma_Score <= 12:\n count += 2\n \n elif row.Glasgow_Coma_Score >= 6 and row.Glasgow_Coma_Score <= 9:\n count += 3\n \n elif row.Glasgow_Coma_Score < 6:\n count += 4\n \n # Bilirubin \n \n if float(row.Bili_Total) >= 1.2 and float(row.Bili_Total) <= 1.9:\n count += 1 \n \n elif float(row.Bili_Total) >= 2.0 and float(row.Bili_Total) <= 5.9:\n count += 2\n \n elif float(row.Bili_Total) >= 6.0 and float(row.Bili_Total) <= 11.9:\n count += 3\n \n elif float(row.Bili_Total) >= 12.0:\n count += 4 \n \n # Need to implement Mean artieral pressure later \n \n # Creatinine \n \n if row.Creatinine >= 1.2 and row.Creatinine <= 1.9:\n count += 1 \n \n elif row.Creatinine >= 2.0 and row.Creatinine <= 3.4:\n count += 2\n \n elif row.Creatinine >= 3.5 and row.Creatinine <= 4.9:\n count += 3\n \n elif row.Creatinine >= 5.0:\n count += 4 \n \n return count ",
"_____no_output_____"
],
[
"allo_df['sofa'] = allo_df.apply(lambda row: calculate_sofa(row), axis = 1)",
"_____no_output_____"
],
[
"adjusted_patients['sofa'] = allo_df.apply(lambda row: calculate_sofa(row), axis = 1)",
"/home/joel/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"allo_df['sofa'].describe()",
"_____no_output_____"
],
[
"adjusted_patients['sofa'].describe()",
"_____no_output_____"
],
[
"#https://www.mdcalc.com/sequential-organ-failure-assessment-sofa-score#evidence\nsofa_mortality_calibration = {\n 0: 0,\n 1: 0 ,\n 2: 6.4,\n 3: 6.4,\n 4: 20.2,\n 5: 20.2,\n 6: 21.5,\n 7: 21.5,\n 8: 33.3,\n 9: 33.3 ,\n 10: 50.0,\n 11: 50.0 ,\n 12: 95.2,\n 13: 95.2 ,\n 14: 95.2 , \n \n}",
"_____no_output_____"
],
[
"# still need to check corrobate \n# digging onto various studies on measuring qSOFA for different comorbidities \n# Min linked a paper about influenza \n# can use these values \n\nqsofa_mortality_calibration = {\n 0: 0.6,\n 1: 5 ,\n 2: 10,\n 3: 24,\n \n}",
"_____no_output_____"
],
[
"working_df.dtypes",
"_____no_output_____"
],
[
"def comorbidity_count(row):\n count = 0 \n \n if row.COPD_com_flag == 1:\n count += 1 \n \n if row.asthma_com_flag == 1:\n count += 1 \n \n if row.diabetes_com_flag == 1:\n count += 1 \n \n if row.hypertension_com_flag == 1:\n count += 1 \n \n if row.CAD_com_flag == 1:\n count += 1 \n \n if row.heartfailure_com_flag == 1: \n count += 1 \n \n if row.CKD_com_flag == 1:\n count += 1 \n \n if row.cancer_com_flag == 1:\n count += 1 \n \n return count",
"_____no_output_____"
],
[
"working_df[['COPD_com_flag', 'asthma_com_flag', 'diabetes_com_flag',\n 'hypertension_com_flag', 'CAD_com_flag', 'heartfailure_com_flag',\n 'CKD_com_flag', 'cancer_com_flag']] = working_df[['COPD_com_flag', 'asthma_com_flag', 'diabetes_com_flag',\n 'hypertension_com_flag', 'CAD_com_flag', 'heartfailure_com_flag',\n 'CKD_com_flag', 'cancer_com_flag']].fillna(0)",
"/home/joel/anaconda3/lib/python3.7/site-packages/pandas/core/frame.py:3069: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n self[k1] = value[k2]\n"
],
[
"working_df[['COPD_com_flag', 'asthma_com_flag', 'diabetes_com_flag',\n 'hypertension_com_flag', 'CAD_com_flag', 'heartfailure_com_flag',\n 'CKD_com_flag', 'cancer_com_flag']] = working_df[['COPD_com_flag', 'asthma_com_flag', 'diabetes_com_flag',\n 'hypertension_com_flag', 'CAD_com_flag', 'heartfailure_com_flag',\n 'CKD_com_flag', 'cancer_com_flag']].astype(int)",
"_____no_output_____"
],
[
"working_df['total_comorbidities'] = working_df.apply(lambda row: comorbidity_count(row), axis=1)",
"/home/joel/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"working_df['cancer_com_flag'].dtype",
"_____no_output_____"
],
[
"working_df['has_comorbidity'] = working_df.total_comorbidities.apply(lambda x: 1 if x >= 1 else 0)",
"/home/joel/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"working_df['has_comorbidity2'] = working_df.total_comorbidities.apply(lambda x: 1 if x >= 2 else 0)",
"/home/joel/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"working_df['life_years'] = working_df.age_at_admit.apply(lambda x: 100 - x)",
"/home/joel/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"Counter(adjusted_patients['discharge_disposition_display'])",
"_____no_output_____"
],
[
"class Allocation(object):\n # Code will be adjusted for SOFA. Currently using qSOFA \n # Only looking at State Level CSC for vent allocation \n def __init__(self, patients, scarcity, sofa_calibration):\n self.patients = patients.copy() \n self.patients['death'] = [0 for _ in range(len(self.patients))]\n self.patients['allocated_vent'] = [\"no\" for _ in range(len(self.patients))]\n \n self.num_vents = int(len(patients) * scarcity) \n \n self.mortality_model = sofa_calibration\n \n def allocate(self, row):\n prob = self.mortality_model[row.qSOFA_score]\n \n \n death = np.random.binomial(size=1, n=1, p=prob*.01)[0]\n #print(death)\n \n if death == 1 or row.discharge_disposition_display == 'Expired':\n return death, 'yes'\n \n else:\n #print('yup yup')\n return death, 'yes'\n \n def check_expiration(self, df):\n\n temp_df = df.copy()\n for i, row in df.iterrows():\n row = row.copy()\n if (pd.isna(row.vent_hours_summed)) or row.discharge_disposition_display == 'Expired':\n \n temp_df.loc[i, 'death'] = 1\n\n else:\n \n temp_df.loc[i, 'death'] = 0\n \n \n return temp_df\n \n def __run_allocation(self, df2):\n \n for i, row in df2.iterrows():\n row = row.copy()\n if self.num_vents == 0:\n #print('out')\n break\n \n mortality, allocate_cond = self.allocate(row)\n \n df2.loc[i, 'death'] = mortality\n \n df2.loc[i, 'allocated_vent'] = allocate_cond\n \n self.num_vents -= 1 \n \n non_allocated = df2[df2['allocated_vent']=='no']\n allocated = df2[df2['allocated_vent']=='yes']\n \n adj_df = self.check_expiration(non_allocated)\n \n return pd.concat([allocated, adj_df])\n \n \n def lottery(self):\n temp_patients = self.patients.copy()\n \n temp_patients.sample(frac=1)\n \n out_df = self.__run_allocation(temp_patients)\n return out_df \n \n def youngest(self):\n temp_patients = self.patients.copy()\n \n temp_patients.sort_values(by=['age_at_admit'], ascending=True, inplace=True)\n \n out_df = self.__run_allocation(temp_patients)\n return out_df \n \n # pandas function\n def __age_categorization(self, row):\n if row.age_at_admit < 50:\n return 1 \n elif row.age_at_admit < 70: \n return 2 \n elif row.age_at_admit < 85:\n return 3 \n else:\n return 4\n \n \n \n def maryland(self):\n temp_patients = self.patients.copy()\n temp_patients['age_cat'] = temp_patients.apply(lambda row: self.__age_categorization(row)\n , axis=1)\n \n temp_patients.sort_values(by=['qSOFA_score', 'total_comorbidities', 'age_cat'], \n ascending=[True, True, True], inplace=True)\n \n out_df = self.__run_allocation(temp_patients)\n return out_df \n \n def new_york(self):\n temp_patients = self.patients.copy()\n\n groups = [df for _, df in temp_patients.groupby('qSOFA_score')]\n random.shuffle(groups)\n\n grouped = pd.concat(groups).reset_index(drop=True)\n grouped = grouped.sort_values('qSOFA_score', ascending=True)\n \n out_df = self.__run_allocation(grouped)\n return out_df \n \n \n def max_lives_saved(self):\n temp_patients = self.patients.copy()\n \n temp_patients.sort_values(by=['qSOFA_score'], ascending=True, inplace=True)\n \n out_df = self.__run_allocation(temp_patients)\n return out_df \n \n def max_life_years(self):\n temp_patients = self.patients.copy()\n\n temp_patients.sort_values(by=['qSOFA_score', 'life_years'], ascending=[True,False], inplace=True)\n\n out_df = self.__run_allocation(temp_patients)\n return out_df \n \n def sickest_first(self):\n temp_patients = self.patients.copy()\n \n temp_patients.sort_values(by=['qSOFA_score'], ascending=False, inplace=True)\n \n out_df = self.__run_allocation(temp_patients)\n return out_df \n ",
"_____no_output_____"
],
[
"zip_df = pd.read_csv('zip_code_data.csv', index_col=False)\nzip_df['zip_code'] = zip_df.zip_code.apply(lambda x: x.strip('ZCTA5 '))",
"_____no_output_____"
],
[
"working_df = pd.merge(working_df, zip_df, left_on='zip_cust_table', right_on='zip_code', how='inner')",
"_____no_output_____"
]
],
[
[
"### Baseline",
"_____no_output_____"
]
],
[
[
"Counter(working_df['discharge_disposition_display'])",
"_____no_output_____"
],
[
"def latinx(row):\n if row.ethnicity_display == 'Hispanic or Latino' and row.race_display == 'White':\n return \"Hispanic\"\n elif row.ethnicity_display == 'Not Hispanic or Latino' and row.race_display == 'White': \n return \"White\"\n else:\n return row.race_display",
"_____no_output_____"
],
[
"working_df['race_display'] = df.apply(lambda row: latinx(row), axis=1)",
"_____no_output_____"
],
[
"# later think about the mortality rate as well \n# summarize what I'm going to do and send to Victoria ",
"_____no_output_____"
],
[
"len(working_df)",
"_____no_output_____"
],
[
"# compute other descriptive stats for this groupby \n# final analysis \nworking_df[['race_display', 'age_at_admit']].groupby('race_display').agg(['mean', 'std', 'count']).round(2)",
"_____no_output_____"
],
[
"Counter(working_df['qSOFA_score'])",
"_____no_output_____"
],
[
"len(working_df['zip_cust_table'].unique())",
"_____no_output_____"
],
[
"# zip code demo eda \nc = Counter(working_df['zip_cust_table'])\nalist = c.most_common()\nsum_patient = list(filter(lambda x: x[0][2] == '7', alist))\n\nprint(len(sum_patient))\nnum_p = 0 \nfor x in sum_patient:\n num_p += x[1]\n \nnum_p",
"37\n"
],
[
"c = Counter(working_df['zip_cust_table'])\nalist = c.most_common()\nn_alist = list(filter(lambda x: x[1] > 1, alist))\nprint(len(n_alist))\n#n_alist",
"60\n"
],
[
"sn.distplot(working_df['qSOFA_score'])",
"/home/joel/anaconda3/lib/python3.7/site-packages/seaborn/distributions.py:2551: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n"
],
[
"race_count = Counter(working_df['race_display'])\nrace_count",
"_____no_output_____"
],
[
"working_df['poverty_rate'] = working_df['poverty_rate'].astype(float)\nworking_df['median_income'] = working_df['median_income'].astype(float)",
"_____no_output_____"
],
[
"bins = [0, 6, 12, 18,24,30,36,40]\nbin_conv = [i+1 for i in range(len(bins))]\nworking_df['zip_binned_by_poverty'] = np.searchsorted(bins, working_df['poverty_rate'].values)\n#temp_df['zip_binned_by_poverty'] = np.searchsorted(bins, temp_df['poverty_rate'].values)",
"_____no_output_____"
],
[
"bins = [20000, 40000, 60000, 80000,100000]\nbin_conv = [i+1 for i in range(len(bins))]\nworking_df['zip_binned_by_income'] = np.searchsorted(bins, working_df['median_income'].values)",
"_____no_output_____"
],
[
"working_df['death'] = working_df.discharge_disposition_display.apply(lambda row: 1 if row == 'Expired' else 0)",
"_____no_output_____"
],
[
"Counter(working_df['death'])",
"_____no_output_____"
],
[
"data_for_model_X = working_df[['zip_binned_by_income', \n 'zip_binned_by_poverty', \n 'race_display',\n 'total_comorbidities', \n 'has_comorbidity',\n 'has_comorbidity2',\n 'age_at_admit', \n 'qSOFA_score', \n ]]\nupdated_data_for_model_X = pd.get_dummies(data_for_model_X)\ndata_for_model_y = working_df['death']",
"_____no_output_____"
],
[
"data_for_model_X.dtypes",
"_____no_output_____"
],
[
"log_reg = sm.Logit(data_for_model_y, updated_data_for_model_X).fit()",
"Warning: Maximum number of iterations has been exceeded.\n Current function value: 0.456956\n Iterations: 35\n"
],
[
"print(log_reg.summary())",
" Logit Regression Results \n==============================================================================\nDep. Variable: death No. Observations: 652\nModel: Logit Df Residuals: 637\nMethod: MLE Df Model: 14\nDate: Tue, 04 May 2021 Pseudo R-squ.: 0.1469\nTime: 15:46:59 Log-Likelihood: -297.94\nconverged: False LL-Null: -349.22\nCovariance Type: nonrobust LLR p-value: 1.519e-15\n==============================================================================================================\n coef std err z P>|z| [0.025 0.975]\n--------------------------------------------------------------------------------------------------------------\nzip_binned_by_income -0.1587 0.164 -0.970 0.332 -0.479 0.162\nzip_binned_by_poverty -0.0456 0.134 -0.340 0.734 -0.308 0.217\ntotal_comorbidities 0.0175 0.151 0.116 0.908 -0.279 0.314\nhas_comorbidity 0.5008 0.404 1.241 0.215 -0.290 1.292\nhas_comorbidity2 0.2534 0.349 0.726 0.468 -0.430 0.937\nage_at_admit 0.0460 0.007 6.364 0.000 0.032 0.060\nqSOFA_score 0.5434 0.145 3.758 0.000 0.260 0.827\nrace_display_American Indian/Alaska Native -31.6672 7.87e+05 -4.03e-05 1.000 -1.54e+06 1.54e+06\nrace_display_Asian -3.6108 1.269 -2.846 0.004 -6.098 -1.124\nrace_display_Black or African American -4.6172 0.973 -4.746 0.000 -6.524 -2.711\nrace_display_Decline to Specify -23.7085 6784.832 -0.003 0.997 -1.33e+04 1.33e+04\nrace_display_Hispanic -4.6186 0.925 -4.993 0.000 -6.432 -2.806\nrace_display_Other Race -4.3989 1.011 -4.349 0.000 -6.381 -2.417\nrace_display_Unknown -5.5549 1.129 -4.920 0.000 -7.768 -3.342\nrace_display_White -4.6257 0.929 -4.982 0.000 -6.446 -2.806\n==============================================================================================================\n"
],
[
"for table in log_reg.summary().tables:\n print(table.as_latex_tabular())",
"\\begin{center}\n\\begin{tabular}{lclc}\n\\toprule\n\\textbf{Dep. Variable:} & death & \\textbf{ No. Observations: } & 652 \\\\\n\\textbf{Model:} & Logit & \\textbf{ Df Residuals: } & 637 \\\\\n\\textbf{Method:} & MLE & \\textbf{ Df Model: } & 14 \\\\\n\\textbf{Date:} & Tue, 04 May 2021 & \\textbf{ Pseudo R-squ.: } & 0.1469 \\\\\n\\textbf{Time:} & 15:46:59 & \\textbf{ Log-Likelihood: } & -297.94 \\\\\n\\textbf{converged:} & False & \\textbf{ LL-Null: } & -349.22 \\\\\n\\textbf{Covariance Type:} & nonrobust & \\textbf{ LLR p-value: } & 1.519e-15 \\\\\n\\bottomrule\n\\end{tabular}\n%\\caption{Logit Regression Results}\n\\end{center}\n\\begin{center}\n\\begin{tabular}{lcccccc}\n\\toprule\n & \\textbf{coef} & \\textbf{std err} & \\textbf{z} & \\textbf{P$> |$z$|$} & \\textbf{[0.025} & \\textbf{0.975]} \\\\\n\\midrule\n\\textbf{zip\\_binned\\_by\\_income} & -0.1587 & 0.164 & -0.970 & 0.332 & -0.479 & 0.162 \\\\\n\\textbf{zip\\_binned\\_by\\_poverty} & -0.0456 & 0.134 & -0.340 & 0.734 & -0.308 & 0.217 \\\\\n\\textbf{total\\_comorbidities} & 0.0175 & 0.151 & 0.116 & 0.908 & -0.279 & 0.314 \\\\\n\\textbf{has\\_comorbidity} & 0.5008 & 0.404 & 1.241 & 0.215 & -0.290 & 1.292 \\\\\n\\textbf{has\\_comorbidity2} & 0.2534 & 0.349 & 0.726 & 0.468 & -0.430 & 0.937 \\\\\n\\textbf{age\\_at\\_admit} & 0.0460 & 0.007 & 6.364 & 0.000 & 0.032 & 0.060 \\\\\n\\textbf{qSOFA\\_score} & 0.5434 & 0.145 & 3.758 & 0.000 & 0.260 & 0.827 \\\\\n\\textbf{race\\_display\\_American Indian/Alaska Native} & -31.6672 & 7.87e+05 & -4.03e-05 & 1.000 & -1.54e+06 & 1.54e+06 \\\\\n\\textbf{race\\_display\\_Asian} & -3.6108 & 1.269 & -2.846 & 0.004 & -6.098 & -1.124 \\\\\n\\textbf{race\\_display\\_Black or African American} & -4.6172 & 0.973 & -4.746 & 0.000 & -6.524 & -2.711 \\\\\n\\textbf{race\\_display\\_Decline to Specify} & -23.7085 & 6784.832 & -0.003 & 0.997 & -1.33e+04 & 1.33e+04 \\\\\n\\textbf{race\\_display\\_Hispanic} & -4.6186 & 0.925 & -4.993 & 0.000 & -6.432 & -2.806 \\\\\n\\textbf{race\\_display\\_Other Race} & -4.3989 & 1.011 & -4.349 & 0.000 & -6.381 & -2.417 \\\\\n\\textbf{race\\_display\\_Unknown} & -5.5549 & 1.129 & -4.920 & 0.000 & -7.768 & -3.342 \\\\\n\\textbf{race\\_display\\_White} & -4.6257 & 0.929 & -4.982 & 0.000 & -6.446 & -2.806 \\\\\n\\bottomrule\n\\end{tabular}\n\\end{center}\n"
],
[
"params = log_reg.params\nconf = log_reg.conf_int()\nconf['Odds Ratio'] = params\nconf.columns = ['5%', '95%', 'Odds Ratio']\nprint(np.exp(conf))",
" 5% 95% Odds Ratio\nzip_binned_by_income 0.619255 1.175702 8.532643e-01\nzip_binned_by_poverty 0.734656 1.242609 9.554528e-01\ntotal_comorbidities 0.756419 1.369123 1.017660e+00\nhas_comorbidity 0.748006 3.639985 1.650070e+00\nhas_comorbidity2 0.650263 2.552781 1.288402e+00\nage_at_admit 1.032313 1.061950 1.047026e+00\nqSOFA_score 1.296882 2.286150 1.721879e+00\nrace_display_American Indian/Alaska Native 0.000000 inf 1.766573e-14\nrace_display_Asian 0.002248 0.325027 2.703084e-02\nrace_display_Black or African American 0.001468 0.066494 9.879973e-03\nrace_display_Decline to Specify 0.000000 inf 5.052744e-11\nrace_display_Hispanic 0.001610 0.060471 9.866908e-03\nrace_display_Other Race 0.001693 0.089224 1.229125e-02\nrace_display_Unknown 0.000423 0.035365 3.868562e-03\nrace_display_White 0.001587 0.060463 9.796797e-03\n"
],
[
"conf[['5%', '95%', 'Odds Ratio' ]] = np.exp(conf[['5%', '95%', 'Odds Ratio']])\nconf ",
"/home/joel/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:1: RuntimeWarning: overflow encountered in exp\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"print(conf.round(2).to_latex(index=True))",
"\\begin{tabular}{lrrr}\n\\toprule\n{} & 5\\% & 95\\% & Odds Ratio \\\\\n\\midrule\nzip\\_binned\\_by\\_income & 0.62 & 1.18 & 0.85 \\\\\nzip\\_binned\\_by\\_poverty & 0.73 & 1.24 & 0.96 \\\\\ntotal\\_comorbidities & 0.76 & 1.37 & 1.02 \\\\\nhas\\_comorbidity & 0.75 & 3.64 & 1.65 \\\\\nhas\\_comorbidity2 & 0.65 & 2.55 & 1.29 \\\\\nage\\_at\\_admit & 1.03 & 1.06 & 1.05 \\\\\nqSOFA\\_score & 1.30 & 2.29 & 1.72 \\\\\nrace\\_display\\_American Indian/Alaska Native & 0.00 & inf & 0.00 \\\\\nrace\\_display\\_Asian & 0.00 & 0.33 & 0.03 \\\\\nrace\\_display\\_Black or African American & 0.00 & 0.07 & 0.01 \\\\\nrace\\_display\\_Decline to Specify & 0.00 & inf & 0.00 \\\\\nrace\\_display\\_Hispanic & 0.00 & 0.06 & 0.01 \\\\\nrace\\_display\\_Other Race & 0.00 & 0.09 & 0.01 \\\\\nrace\\_display\\_Unknown & 0.00 & 0.04 & 0.00 \\\\\nrace\\_display\\_White & 0.00 & 0.06 & 0.01 \\\\\n\\bottomrule\n\\end{tabular}\n\n"
],
[
"from sklearn.metrics import roc_auc_score\nroc_auc_score(data_for_model_y, log_reg.predict(updated_data_for_model_X))",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f379c1962486b69e63abc0280a087f8a89899f | 14,000 | ipynb | Jupyter Notebook | learning resources/4-labs/week6-Linear Regression/Week 7 Linear Regression.ipynb | linksdl/acs_project_maching-learning | 8cc034ec07432c3a79cefc2eacc8e9a77019748d | [
"Apache-2.0"
] | 1 | 2021-12-10T14:10:56.000Z | 2021-12-10T14:10:56.000Z | learning resources/4-labs/week6-Linear Regression/Week 7 Linear Regression.ipynb | linksdl/acs_project_maching-learning | 8cc034ec07432c3a79cefc2eacc8e9a77019748d | [
"Apache-2.0"
] | null | null | null | learning resources/4-labs/week6-Linear Regression/Week 7 Linear Regression.ipynb | linksdl/acs_project_maching-learning | 8cc034ec07432c3a79cefc2eacc8e9a77019748d | [
"Apache-2.0"
] | 1 | 2020-11-04T02:29:40.000Z | 2020-11-04T02:29:40.000Z | 28.629857 | 369 | 0.472143 | [
[
[
"### In this lab, we will implement Linear Regression using Least-square Solution. We will use the same example as we did in the class (Slide 18 from the linear regression slides). There are 5 steps. Let's implement them using only numpy step by step. ",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"import numpy as np ",
"_____no_output_____"
]
],
[
[
"We are given the dataset: {(0,0), (0,1), (1,0)} and asked to find the \\\nleast-squares solution for the parameters in the regression of \\\nthe function: y = w1 +w2^2",
"_____no_output_____"
]
],
[
[
"# creating the input and target in numpy arrays \ninputs = np.array([[0], [0], [1]])\ntargets = np.array([[0], [1], [0]])\nprint('inputs shape :',np.shape(inputs))\nprint('targets shape :',np.shape(targets))",
"inputs shape : (3, 1)\ntargets shape : (3, 1)\n"
],
[
"# now let's do the steps to find the solution \n# Step 1: evaluate the basis on the points\ninputs = np.concatenate((np.ones((np.shape(inputs)[0],1)),inputs),axis=1)\nprint('inputs shape :',np.shape(inputs))\nprint(inputs)",
"inputs shape : (3, 2)\n[[1. 0.]\n [1. 0.]\n [1. 1.]]\n"
],
[
"# step 2: compute -> transpose(inputs) * inputs \nq_matrix = np.dot(np.transpose(inputs),inputs)\nprint('q_matrix shape :',np.shape(q_matrix))\nprint(q_matrix)",
"q_matrix shape : (2, 2)\n[[3. 1.]\n [1. 1.]]\n"
],
[
"# step 3: invert q_matrix\nq_inverse = np.linalg.inv(q_matrix)\nprint('q_inverse shape :',np.shape(q_inverse))\nprint(q_inverse)",
"q_inverse shape : (2, 2)\n[[ 0.5 -0.5]\n [-0.5 1.5]]\n"
],
[
"# step 4: Compute the pseudo-inverse -> q_inverse * transpose(inputs)\nq_pseudo = np.dot(q_inverse,np.transpose(inputs))\nprint('q_pseudo shape :',np.shape(q_pseudo))\nprint(q_pseudo.astype(np.float16))",
"q_pseudo shape : (2, 3)\n[[ 0.5 0.5 0. ]\n [-0.5 -0.5 1. ]]\n"
],
[
"# step 5: compute w = q_pseudo * targets\nweights = np.dot(q_pseudo,targets)\nprint('w shape :',np.shape(weights))\nprint(weights)",
"w shape : (2, 1)\n[[ 0.5]\n [-0.5]]\n"
]
],
[
[
"#### Now, let's implement the steps but on a real dataset. we will work on the auto-mpg dataset. This consists of a collection of a number of datapoints about certain cars (weight, horsepower, etc.), with the aim being to predict the fuel efficiency in miles per gallon (mpg) in for each car.",
"_____no_output_____"
]
],
[
[
"\"\"\"\nYou are asked to\n - load the dataset text file (auto-mpg.txt) as numpy array \n - prerocess the dataset (normalise, split it into train and test sets)\n - find the least-squares solution for the parameters (weights vector)\n - test the found parameters on the test set and calculate the error\n\nThe following comments and codes are meant to guide you. \n\"\"\"",
"_____no_output_____"
],
[
"\"\"\"\nPlease note: This dataset has one problem. There are missing values \nin it (labelled with question marks ‘?’). The np.loadtxt() method doesn’t\nlike these, and we don’t know what to do with them, anyway,manually edit \nthe file and delete all lines where there is a ? in that line. The linear\nregressor can’t do much with the names of the cars either, but since they \nappear in quotes(\") we will tell np.loadtxt that they are comments\n\n\nBelow are the attribute Information for the dataset:\n\n 1. mpg: continuous \n 2. cylinders: multi-valued discrete\n 3. displacement: continuous\n 4. horsepower: continuous\n 5. weight: continuous\n 6. acceleration: continuous\n 7. model year: multi-valued discrete\n 8. origin: multi-valued discrete\n 9. car name: string (unique for each instance)\n\nPlease note: the first column is our target (mpg)\n\"\"\"",
"_____no_output_____"
],
[
"# TODO: load the dataset file using np.loadtxt()\nimport pandas as pd\n\ndf = pd.read_csv(\"auto-mpg.txt\", delimiter=' ')\ndf.head()\n# data = np.loadtxt(\"auto-mpg.txt\", delimiter=' ', usecols=range(5))",
"/Users/shengdaolin_sh/dir_install/Aanconda3/anaconda3/envs/ml_env/lib/python3.6/site-packages/ipykernel_launcher.py:4: ParserWarning: Falling back to the 'python' engine because the 'c' engine does not support regex separators (separators > 1 char and different from '\\s+' are interpreted as regex); you can avoid this warning by specifying engine='python'.\n after removing the cwd from sys.path.\n"
],
[
"# TODO: Normalise the dataset. You can do this easily in numpy \n# by using np.mean and np.var. The only place where care is needed \n# is along which axis the mean and variance are computed: \n# axis=0 sums down the columns and axis=1 sums across the rows.\n\nnormalised_date = None",
"_____no_output_____"
],
[
"# TODO: Now separate the data into training and testing sets,\n\ntraining, testing = None, None \n\n# And split each set into inputs and targets hint: slicing the array\ntrainin, traintgt = None, None\ntestin, testtgt = None, None",
"_____no_output_____"
],
[
"# TODO: Use the training set to find the weights vector.\n# you need to implement the previous 5 steps on the training set \n# and find the weights vector (this is called training). \n# To make it simple we define a function that takes \n# two args: inputs and targets and return the weights vector\n\ndef linreg(inputs,targets):\n # you should implement the 5 steps here\n \n weights = None\n\n \n return weights\n\n",
"_____no_output_____"
],
[
"# test your implementation \nweights = linreg(trainin,traintgt) \nweights",
"_____no_output_____"
],
[
"# TODO: Testing the found weights on the testing set \n# you can do this by \n# - testout = (testin*weights)\n# - error = sum((testout - testtgt)**2)\n\ntestout = None\nerror = None \n",
"_____no_output_____"
],
[
"\"\"\"\n You can try to re-train the model without the normalising the data \n and see if this makes any different on the error value\n\"\"\"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f37a4e3ae9061a8ec943375a03b5bb794ec9ca | 411,354 | ipynb | Jupyter Notebook | module3-databackedassertions/GKwolek_3rd_assignment_LS_DS_113_Making_Data_backed_Assertions_Assignment.ipynb | grzegorzkwolek/DS-Unit-1-Sprint-1-Dealing-With-Data | c269e8c0c66bd58adced523656e7757c2eb6391f | [
"MIT"
] | null | null | null | module3-databackedassertions/GKwolek_3rd_assignment_LS_DS_113_Making_Data_backed_Assertions_Assignment.ipynb | grzegorzkwolek/DS-Unit-1-Sprint-1-Dealing-With-Data | c269e8c0c66bd58adced523656e7757c2eb6391f | [
"MIT"
] | null | null | null | module3-databackedassertions/GKwolek_3rd_assignment_LS_DS_113_Making_Data_backed_Assertions_Assignment.ipynb | grzegorzkwolek/DS-Unit-1-Sprint-1-Dealing-With-Data | c269e8c0c66bd58adced523656e7757c2eb6391f | [
"MIT"
] | null | null | null | 295.30079 | 240,708 | 0.878214 | [
[
[
"<a href=\"https://colab.research.google.com/github/grzegorzkwolek/DS-Unit-1-Sprint-1-Dealing-With-Data/blob/master/GKwolek_3rd_assignment_LS_DS_113_Making_Data_backed_Assertions_Assignment.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Lambda School Data Science - Making Data-backed Assertions\n\nThis is, for many, the main point of data science - to create and support reasoned arguments based on evidence. It's not a topic to master in a day, but it is worth some focused time thinking about and structuring your approach to it.",
"_____no_output_____"
],
[
"## Assignment - what's going on here?\n\nConsider the data in `persons.csv` (already prepared for you, in the repo for the week). It has four columns - a unique id, followed by age (in years), weight (in lbs), and exercise time (in minutes/week) of 1200 (hypothetical) people.\n\nTry to figure out which variables are possibly related to each other, and which may be confounding relationships.\n\nTry and isolate the main relationships and then communicate them using crosstabs and graphs. Share any cool graphs that you make with the rest of the class in Slack!",
"_____no_output_____"
]
],
[
[
"# TODO - your code here\n# Use what we did live in lecture as an example\n\n# HINT - you can find the raw URL on GitHub and potentially use that\n# to load the data with read_csv, or you can upload it yourself",
"_____no_output_____"
],
[
"import pandas as pd\ndf = pd.read_csv('https://raw.githubusercontent.com/LambdaSchool/DS-Unit-1-Sprint-1-Dealing-With-Data/0cb02007e8a7f3193cd33daa6cb2ed4158e73aed/module3-databackedassertions/persons.csv')",
"_____no_output_____"
],
[
"#it looks like \"id\" is missing as a column label\ndf = df.rename(columns = {'Unnamed: 0':'ID'})\nprint (df)",
" ID age weight exercise_time\n0 0 44 118 192\n1 1 41 161 35\n2 2 46 128 220\n3 3 39 216 57\n4 4 28 116 182\n5 5 58 103 165\n6 6 55 161 107\n7 7 21 188 37\n8 8 55 216 79\n9 9 50 127 267\n10 10 21 160 228\n11 11 43 102 78\n12 12 73 209 44\n13 13 27 165 48\n14 14 21 169 171\n15 15 36 131 194\n16 16 49 171 191\n17 17 69 172 147\n18 18 18 122 271\n19 19 55 157 111\n20 20 19 218 28\n21 21 34 143 24\n22 22 20 116 267\n23 23 20 159 241\n24 24 32 117 181\n25 25 71 103 21\n26 26 21 164 229\n27 27 79 189 38\n28 28 72 149 110\n29 29 26 117 279\n... ... ... ... ...\n1170 1170 22 199 102\n1171 1171 56 139 158\n1172 1172 27 111 252\n1173 1173 74 161 142\n1174 1174 30 137 244\n1175 1175 61 164 86\n1176 1176 57 213 103\n1177 1177 38 174 103\n1178 1178 67 150 118\n1179 1179 69 140 184\n1180 1180 34 124 147\n1181 1181 41 142 18\n1182 1182 49 160 256\n1183 1183 70 133 198\n1184 1184 57 210 27\n1185 1185 35 124 48\n1186 1186 61 170 30\n1187 1187 48 160 125\n1188 1188 71 129 21\n1189 1189 35 126 201\n1190 1190 70 156 104\n1191 1191 29 105 174\n1192 1192 51 122 194\n1193 1193 33 106 112\n1194 1194 64 122 184\n1195 1195 47 195 1\n1196 1196 29 210 22\n1197 1197 29 147 15\n1198 1198 25 149 185\n1199 1199 67 154 84\n\n[1200 rows x 4 columns]\n"
],
[
"#putting ID as an index (just removing duplication)\ndf = df.set_index(\"ID\")\ndf.head()",
"_____no_output_____"
],
[
"df.plot.scatter(\"exercise_time\", \"weight\")",
"_____no_output_____"
],
[
"df.plot.scatter(\"age\", \"exercise_time\")",
"_____no_output_____"
],
[
"!pip install pandas==0.23.4",
"Requirement already satisfied: pandas==0.23.4 in /usr/local/lib/python3.6/dist-packages (0.23.4)\nRequirement already satisfied: pytz>=2011k in /usr/local/lib/python3.6/dist-packages (from pandas==0.23.4) (2018.9)\nRequirement already satisfied: numpy>=1.9.0 in /usr/local/lib/python3.6/dist-packages (from pandas==0.23.4) (1.16.5)\nRequirement already satisfied: python-dateutil>=2.5.0 in /usr/local/lib/python3.6/dist-packages (from pandas==0.23.4) (2.5.3)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.6/dist-packages (from python-dateutil>=2.5.0->pandas==0.23.4) (1.12.0)\n"
],
[
"#(the charts above are to help me understanding a notch more of the data, and validate soem assumptions)\n# back to the exercise we did during the class\nage_bins = pd.cut(df['age'], 5, precision=0)\nweight_bins = pd.cut(df['weight'], 8, precision = 0)\npd.crosstab(weight_bins, age_bins, normalize = \"index\")",
"_____no_output_____"
],
[
"pd.crosstab(weight_bins, age_bins, normalize = \"columns\")",
"_____no_output_____"
],
[
"#possibly giving up too early, but crosstabs don't seem to be the right approach for this dataset. Back to charts.\n\nimport matplotlib.pyplot as plt\nplt.style.use('dark_background')",
"_____no_output_____"
],
[
"age_weight_col_CT = pd.crosstab(age_bins, weight_bins, normalize = \"columns\")\nage_weight_col_CT.plot(kind = \"bar\")",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"cols = ['age', 'weight', 'exercise_time']\ndf2 = df[cols]\n",
"_____no_output_____"
],
[
"df.plot(figsize = (15, 10))",
"_____no_output_____"
],
[
"df = df.set_index(\"age\")",
"_____no_output_____"
],
[
"df.sort_index?\n#(inplace=True)\ndf.head(200)",
"_____no_output_____"
]
],
[
[
"### Assignment questions\n\nAfter you've worked on some code, answer the following questions in this text block:\n\n1. What are the variable types in the data?\nAll continous variables, represented as discreet ones.\n\n2. What are the relationships between the variables? The higher exercise time the lower is the weight (and the lower maximum weight)\n\n\n3. Which relationships are \"real\", and which spurious? The relation between age and weight seems spurious. There is a relation between exercise time and weight as well as the age and exercise time (neagtive, beyond age 60).\n\n",
"_____no_output_____"
],
[
"## Stretch goals and resources\n\nFollowing are *optional* things for you to take a look at. Focus on the above assignment first, and make sure to commit and push your changes to GitHub.\n\n- [Spurious Correlations](http://tylervigen.com/spurious-correlations)\n- [NIH on controlling for confounding variables](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4017459/)\n\nStretch goals:\n\n- Produce your own plot inspired by the Spurious Correlation visualizations (and consider writing a blog post about it - both the content and how you made it)\n- Pick one of the techniques that NIH highlights for confounding variables - we'll be going into many of them later, but see if you can find which Python modules may help (hint - check scikit-learn)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d0f39265abb38edaf1bc6a2521becdeb6a0ec692 | 70,684 | ipynb | Jupyter Notebook | src/interfaces/python/examples/tutorial/Visualize Graphical Models.ipynb | stopfer/opengm | f3835a4d3048d0c4660c46c50dfd4f796b6da6c1 | [
"MIT"
] | 318 | 2015-01-07T15:22:02.000Z | 2022-01-22T10:10:29.000Z | src/interfaces/python/examples/tutorial/Visualize Graphical Models.ipynb | stopfer/opengm | f3835a4d3048d0c4660c46c50dfd4f796b6da6c1 | [
"MIT"
] | 89 | 2015-03-24T14:33:01.000Z | 2020-07-10T13:59:13.000Z | src/interfaces/python/examples/tutorial/Visualize Graphical Models.ipynb | stopfer/opengm | f3835a4d3048d0c4660c46c50dfd4f796b6da6c1 | [
"MIT"
] | 119 | 2015-01-13T08:35:03.000Z | 2022-03-01T01:49:08.000Z | 453.102564 | 30,766 | 0.935077 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0f39565cc23a33968a2546964c3379aeac297a8 | 81,709 | ipynb | Jupyter Notebook | Week 3/Day 4/Materi Hari 4 Pekan 3.ipynb | algonacci/SanbercodeDataScience | 4e4af11f38e6e559144af8d1fa096c775abb69df | [
"MIT"
] | null | null | null | Week 3/Day 4/Materi Hari 4 Pekan 3.ipynb | algonacci/SanbercodeDataScience | 4e4af11f38e6e559144af8d1fa096c775abb69df | [
"MIT"
] | null | null | null | Week 3/Day 4/Materi Hari 4 Pekan 3.ipynb | algonacci/SanbercodeDataScience | 4e4af11f38e6e559144af8d1fa096c775abb69df | [
"MIT"
] | null | null | null | 173.112288 | 18,940 | 0.874494 | [
[
[
"import pandas as pd\n\ndf = pd.read_csv('vgsales.csv')\ndf.head()",
"_____no_output_____"
],
[
"# extract data tentang banyaknya setiap jenis genre dalam data\ndata = df['Genre'].value_counts()\ncategory = data.index\nfrequency = data.values\n\n# plot data\nfig, ax = plt.subplots()\nax.bar(category, frequency)\nax.set_xticklabels(category, rotation=90)\nplt.show()",
"C:\\Users\\Vulcan\\AppData\\Local\\Temp/ipykernel_7796/1768862595.py:9: UserWarning: FixedFormatter should only be used together with FixedLocator\n ax.set_xticklabels(category, rotation=90)\n"
],
[
"fig, ax = plt.subplots()\n\n# pembuatan plot\nax.scatter(df['JP_Sales'], df['EU_Sales'], c='g')\n\n# axis labeling\nax.set_xlabel('Penjualan di Jepang')\nax.set_ylabel('Penjualan di Europa')\nax.set_title('Penjualan Semua Video Games')\n\nplt.show()",
"_____no_output_____"
],
[
"action = df[df['Genre']=='Action']['Global_Sales']\nsport = df[df['Genre']=='Sports']['Global_Sales']\n\nfig, ax = plt.subplots(ncols=2, figsize=(16, 8))\nax[0].hist(action, bins=50)\nax[0].set_xlabel('Penjualan Global Game Action')\nax[0].set_ylabel('Frekuensi')\nax[0].set_title('Distribusi Game Action Global')\n\nax[1].hist(sport, bins=50, color='g')\nax[1].set_xlabel('Penjualan Global Game Sports')\nax[1].set_ylabel('Frekuensi')\nax[1].set_title('Distribusi Penjualan Game Sports Global')\nplt.show()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport pandas as pd\ndf = pd.read_csv('Iris.csv', index_col=0)\ndf_group = df.groupby('Species').mean()\ndf_group.head()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(12,8))\nax.bar(df_group.index, df_group['SepalLengthCm'], label='Sepal Length')\nax.bar(df_group.index, df_group['SepalWidthCm'], bottom=df_group['SepalLengthCm'], label='Sepal Width')\nax.bar(df_group.index, df_group['PetalLengthCm'], bottom=df_group['SepalLengthCm']+df_group['SepalWidthCm'], label='Petal Length')\nax.bar(df_group.index, df_group['PetalWidthCm'], bottom=df_group['SepalLengthCm']+df_group['SepalWidthCm']+df_group['PetalLengthCm'], label='Petal Width')\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"df_mean = df.mean()\ndf_mean",
"C:\\Users\\Vulcan\\AppData\\Local\\Temp/ipykernel_7796/4125945695.py:1: FutureWarning: Dropping of nuisance columns in DataFrame reductions (with 'numeric_only=None') is deprecated; in a future version this will raise TypeError. Select only valid columns before calling the reduction.\n df_mean = df.mean()\n"
],
[
"fig, ax = plt.subplots(figsize=(12,8))\n\nfor i in range(len(df_mean)):\n ax.bar(df_mean.index[i], df_mean.values[i], yerr=df.std()[i])\n\nplt.show()",
"C:\\Users\\Vulcan\\AppData\\Local\\Temp/ipykernel_7796/2660526071.py:4: FutureWarning: Dropping of nuisance columns in DataFrame reductions (with 'numeric_only=None') is deprecated; in a future version this will raise TypeError. Select only valid columns before calling the reduction.\n ax.bar(df_mean.index[i], df_mean.values[i], yerr=df.std()[i])\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f3a817c9a1965f6a48b7f1d06361f11d6889df | 62,908 | ipynb | Jupyter Notebook | _posts/DS/Deep-Learning/Deep-Learning-Coursera/4. Convolutional Neural Networks/Convolution model - Step by Step - v2.ipynb | wansook0316/wansook.github.io | 9a5c2cb84d7a9d64f7dfc7e9d4110d4e5af67efb | [
"MIT"
] | 136 | 2018-04-02T11:08:06.000Z | 2022-02-27T21:31:17.000Z | _posts/DS/Deep-Learning/Deep-Learning-Coursera/4. Convolutional Neural Networks/Convolution model - Step by Step - v2.ipynb | wansook0316/wansook.github.io | 9a5c2cb84d7a9d64f7dfc7e9d4110d4e5af67efb | [
"MIT"
] | 1 | 2019-01-20T06:47:19.000Z | 2019-01-20T06:47:19.000Z | _posts/DS/Deep-Learning/Deep-Learning-Coursera/4. Convolutional Neural Networks/Convolution model - Step by Step - v2.ipynb | wansook0316/wansook.github.io | 9a5c2cb84d7a9d64f7dfc7e9d4110d4e5af67efb | [
"MIT"
] | 201 | 2018-04-19T22:06:50.000Z | 2022-03-13T16:21:58.000Z | 41.170157 | 5,306 | 0.557608 | [
[
[
"# Convolutional Neural Networks: Step by Step\n\nWelcome to Course 4's first assignment! In this assignment, you will implement convolutional (CONV) and pooling (POOL) layers in numpy, including both forward propagation and (optionally) backward propagation. \n\n**Notation**:\n- Superscript $[l]$ denotes an object of the $l^{th}$ layer. \n - Example: $a^{[4]}$ is the $4^{th}$ layer activation. $W^{[5]}$ and $b^{[5]}$ are the $5^{th}$ layer parameters.\n\n\n- Superscript $(i)$ denotes an object from the $i^{th}$ example. \n - Example: $x^{(i)}$ is the $i^{th}$ training example input.\n \n \n- Lowerscript $i$ denotes the $i^{th}$ entry of a vector.\n - Example: $a^{[l]}_i$ denotes the $i^{th}$ entry of the activations in layer $l$, assuming this is a fully connected (FC) layer.\n \n \n- $n_H$, $n_W$ and $n_C$ denote respectively the height, width and number of channels of a given layer. If you want to reference a specific layer $l$, you can also write $n_H^{[l]}$, $n_W^{[l]}$, $n_C^{[l]}$. \n- $n_{H_{prev}}$, $n_{W_{prev}}$ and $n_{C_{prev}}$ denote respectively the height, width and number of channels of the previous layer. If referencing a specific layer $l$, this could also be denoted $n_H^{[l-1]}$, $n_W^{[l-1]}$, $n_C^{[l-1]}$. \n\nWe assume that you are already familiar with `numpy` and/or have completed the previous courses of the specialization. Let's get started!",
"_____no_output_____"
],
[
"## 1 - Packages\n\nLet's first import all the packages that you will need during this assignment. \n- [numpy](www.numpy.org) is the fundamental package for scientific computing with Python.\n- [matplotlib](http://matplotlib.org) is a library to plot graphs in Python.\n- np.random.seed(1) is used to keep all the random function calls consistent. It will help us grade your work.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport h5py\nimport matplotlib.pyplot as plt\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (5.0, 4.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n%load_ext autoreload\n%autoreload 2\n\nnp.random.seed(1)",
"_____no_output_____"
]
],
[
[
"## 2 - Outline of the Assignment\n\nYou will be implementing the building blocks of a convolutional neural network! Each function you will implement will have detailed instructions that will walk you through the steps needed:\n\n- Convolution functions, including:\n - Zero Padding\n - Convolve window \n - Convolution forward\n - Convolution backward (optional)\n- Pooling functions, including:\n - Pooling forward\n - Create mask \n - Distribute value\n - Pooling backward (optional)\n \nThis notebook will ask you to implement these functions from scratch in `numpy`. In the next notebook, you will use the TensorFlow equivalents of these functions to build the following model:\n\n<img src=\"images/model.png\" style=\"width:800px;height:300px;\">\n\n**Note** that for every forward function, there is its corresponding backward equivalent. Hence, at every step of your forward module you will store some parameters in a cache. These parameters are used to compute gradients during backpropagation. ",
"_____no_output_____"
],
[
"## 3 - Convolutional Neural Networks\n\nAlthough programming frameworks make convolutions easy to use, they remain one of the hardest concepts to understand in Deep Learning. A convolution layer transforms an input volume into an output volume of different size, as shown below. \n\n<img src=\"images/conv_nn.png\" style=\"width:350px;height:200px;\">\n\nIn this part, you will build every step of the convolution layer. You will first implement two helper functions: one for zero padding and the other for computing the convolution function itself. ",
"_____no_output_____"
],
[
"### 3.1 - Zero-Padding\n\nZero-padding adds zeros around the border of an image:\n\n<img src=\"images/PAD.png\" style=\"width:600px;height:400px;\">\n<caption><center> <u> <font color='purple'> **Figure 1** </u><font color='purple'> : **Zero-Padding**<br> Image (3 channels, RGB) with a padding of 2. </center></caption>\n\nThe main benefits of padding are the following:\n\n- It allows you to use a CONV layer without necessarily shrinking the height and width of the volumes. This is important for building deeper networks, since otherwise the height/width would shrink as you go to deeper layers. An important special case is the \"same\" convolution, in which the height/width is exactly preserved after one layer. \n\n- It helps us keep more of the information at the border of an image. Without padding, very few values at the next layer would be affected by pixels as the edges of an image.\n\n**Exercise**: Implement the following function, which pads all the images of a batch of examples X with zeros. [Use np.pad](https://docs.scipy.org/doc/numpy/reference/generated/numpy.pad.html). Note if you want to pad the array \"a\" of shape $(5,5,5,5,5)$ with `pad = 1` for the 2nd dimension, `pad = 3` for the 4th dimension and `pad = 0` for the rest, you would do:\n```python\na = np.pad(a, ((0,0), (1,1), (0,0), (3,3), (0,0)), 'constant', constant_values = (..,..))\n```",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: zero_pad\n\ndef zero_pad(X, pad):\n \"\"\"\n Pad with zeros all images of the dataset X. The padding is applied to the height and width of an image, \n as illustrated in Figure 1.\n \n Argument:\n X -- python numpy array of shape (m, n_H, n_W, n_C) representing a batch of m images\n pad -- integer, amount of padding around each image on vertical and horizontal dimensions\n \n Returns:\n X_pad -- padded image of shape (m, n_H + 2*pad, n_W + 2*pad, n_C)\n \"\"\"\n \n ### START CODE HERE ### (≈ 1 line)\n npad = ((0, 0), (pad,pad), (pad,pad),(0,0))\n X_pad = np.pad(X,npad,'constant',constant_values = 0)\n ### END CODE HERE ###\n \n return X_pad",
"_____no_output_____"
],
[
"np.random.seed(1)\nx = np.random.randn(4, 3, 3, 2)\nx_pad = zero_pad(x, 2)\nprint (\"x.shape =\", x.shape)\nprint (\"x_pad.shape =\", x_pad.shape)\nprint (\"x[1,1] =\", x[1,1])\nprint (\"x_pad[1,1] =\", x_pad[1,1])\n#first image, first column, every row, every channel:\n#print (\"x_pad[1,1,:,:] =\", x_pad[1,1,:,:])\n#first image, first column, first row, every channel\n#print (\"x_pad[1,1,1,:] =\", x_pad[1,1,1,:])\n#first image, first column, every row, first channel\n#print (\"x_pad[1,1,1,1] =\", x_pad[1,1,1,1])\n\n\nfig, axarr = plt.subplots(1, 2)\naxarr[0].set_title('x')\naxarr[0].imshow(x[0,:,:,0])\naxarr[1].set_title('x_pad')\naxarr[1].imshow(x_pad[0,:,:,0])",
"x.shape = (4, 3, 3, 2)\nx_pad.shape = (4, 7, 7, 2)\nx[1,1] = [[ 0.90085595 -0.68372786]\n [-0.12289023 -0.93576943]\n [-0.26788808 0.53035547]]\nx_pad[1,1] = [[ 0. 0.]\n [ 0. 0.]\n [ 0. 0.]\n [ 0. 0.]\n [ 0. 0.]\n [ 0. 0.]\n [ 0. 0.]]\n"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **x.shape**:\n </td>\n <td>\n (4, 3, 3, 2)\n </td>\n </tr>\n <tr>\n <td>\n **x_pad.shape**:\n </td>\n <td>\n (4, 7, 7, 2)\n </td>\n </tr>\n <tr>\n <td>\n **x[1,1]**:\n </td>\n <td>\n [[ 0.90085595 -0.68372786]\n [-0.12289023 -0.93576943]\n [-0.26788808 0.53035547]]\n </td>\n </tr>\n <tr>\n <td>\n **x_pad[1,1]**:\n </td>\n <td>\n [[ 0. 0.]\n [ 0. 0.]\n [ 0. 0.]\n [ 0. 0.]\n [ 0. 0.]\n [ 0. 0.]\n [ 0. 0.]]\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"### 3.2 - Single step of convolution \n\nIn this part, implement a single step of convolution, in which you apply the filter to a single position of the input. This will be used to build a convolutional unit, which: \n\n- Takes an input volume \n- Applies a filter at every position of the input\n- Outputs another volume (usually of different size)\n\n<img src=\"images/Convolution_schematic.gif\" style=\"width:500px;height:300px;\">\n<caption><center> <u> <font color='purple'> **Figure 2** </u><font color='purple'> : **Convolution operation**<br> with a filter of 2x2 and a stride of 1 (stride = amount you move the window each time you slide) </center></caption>\n\nIn a computer vision application, each value in the matrix on the left corresponds to a single pixel value, and we convolve a 3x3 filter with the image by multiplying its values element-wise with the original matrix, then summing them up and adding a bias. In this first step of the exercise, you will implement a single step of convolution, corresponding to applying a filter to just one of the positions to get a single real-valued output. \n\nLater in this notebook, you'll apply this function to multiple positions of the input to implement the full convolutional operation. \n\n**Exercise**: Implement conv_single_step(). [Hint](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.sum.html).\n",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: conv_single_step\n\ndef conv_single_step(a_slice_prev, W, b):\n \"\"\"\n Apply one filter defined by parameters W on a single slice (a_slice_prev) of the output activation \n of the previous layer.\n \n Arguments:\n a_slice_prev -- slice of input data of shape (f, f, n_C_prev)\n W -- Weight parameters contained in a window - matrix of shape (f, f, n_C_prev)\n b -- Bias parameters contained in a window - matrix of shape (1, 1, 1)\n \n Returns:\n Z -- a scalar value, result of convolving the sliding window (W, b) on a slice x of the input data\n \"\"\"\n\n ### START CODE HERE ### (≈ 2 lines of code)\n # Element-wise product between a_slice and W. Do not add the bias yet.\n #GAC:(element-wise multiplication)\n s = np.multiply(a_slice_prev,W)\n # Sum over all entries of the volume s.\n Z = np.sum(s,axis=None)\n # Add bias b to Z. Cast b to a float() so that Z results in a scalar value.\n Z = Z+ np.float(b)\n ### END CODE HERE ###\n\n return Z",
"_____no_output_____"
],
[
"np.random.seed(1)\na_slice_prev = np.random.randn(4, 4, 3)\nW = np.random.randn(4, 4, 3)\nb = np.random.randn(1, 1, 1)\n\nZ = conv_single_step(a_slice_prev, W, b)\nprint(\"Z =\", Z)",
"Z = -6.99908945068\n"
]
],
[
[
"**Expected Output**:\n<table>\n <tr>\n <td>\n **Z**\n </td>\n <td>\n -6.99908945068\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"### 3.3 - Convolutional Neural Networks - Forward pass\n\nIn the forward pass, you will take many filters and convolve them on the input. Each 'convolution' gives you a 2D matrix output. You will then stack these outputs to get a 3D volume: \n\n<center>\n<video width=\"620\" height=\"440\" src=\"images/conv_kiank.mp4\" type=\"video/mp4\" controls>\n</video>\n</center>\n\n**Exercise**: Implement the function below to convolve the filters W on an input activation A_prev. This function takes as input A_prev, the activations output by the previous layer (for a batch of m inputs), F filters/weights denoted by W, and a bias vector denoted by b, where each filter has its own (single) bias. Finally you also have access to the hyperparameters dictionary which contains the stride and the padding. \n\n**Hint**: \n1. To select a 2x2 slice at the upper left corner of a matrix \"a_prev\" (shape (5,5,3)), you would do:\n```python\na_slice_prev = a_prev[0:2,0:2,:]\n```\nThis will be useful when you will define `a_slice_prev` below, using the `start/end` indexes you will define.\n2. To define a_slice you will need to first define its corners `vert_start`, `vert_end`, `horiz_start` and `horiz_end`. This figure may be helpful for you to find how each of the corner can be defined using h, w, f and s in the code below.\n\n<img src=\"images/vert_horiz_kiank.png\" style=\"width:400px;height:300px;\">\n<caption><center> <u> <font color='purple'> **Figure 3** </u><font color='purple'> : **Definition of a slice using vertical and horizontal start/end (with a 2x2 filter)** <br> This figure shows only a single channel. </center></caption>\n\n\n**Reminder**:\nThe formulas relating the output shape of the convolution to the input shape is:\n$$ n_H = \\lfloor \\frac{n_{H_{prev}} - f + 2 \\times pad}{stride} \\rfloor +1 $$\n$$ n_W = \\lfloor \\frac{n_{W_{prev}} - f + 2 \\times pad}{stride} \\rfloor +1 $$\n$$ n_C = \\text{number of filters used in the convolution}$$\n\nFor this exercise, we won't worry about vectorization, and will just implement everything with for-loops.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: conv_forward\n\ndef conv_forward(A_prev, W, b, hparameters):\n \"\"\"\n Implements the forward propagation for a convolution function\n \n Arguments:\n A_prev -- output activations of the previous layer, numpy array of shape (m, n_H_prev, n_W_prev, n_C_prev)\n W -- Weights, numpy array of shape (f, f, n_C_prev, n_C)\n b -- Biases, numpy array of shape (1, 1, 1, n_C)\n hparameters -- python dictionary containing \"stride\" and \"pad\"\n \n Returns:\n Z -- conv output, numpy array of shape (m, n_H, n_W, n_C)\n cache -- cache of values needed for the conv_backward() function\n \"\"\"\n \n ### START CODE HERE ###\n # Retrieve dimensions from A_prev's shape (≈1 line) \n (m, n_H_prev, n_W_prev, n_C_prev) = A_prev.shape\n #print(\"m =\", m)\n #print(\"n_H_prev =\", n_H_prev)\n #print(\"n_W_prev =\", n_W_prev)\n #print(\"n_C_prev =\", n_C_prev)\n \n # Retrieve dimensions from W's shape (≈1 line)\n (f, f, n_C_prev, n_C) = W.shape\n #print(\"f =\", f)\n #print(\"n_C =\", n_C)\n \n # Retrieve information from \"hparameters\" (≈2 lines)\n stride = hparameters['stride']\n pad = hparameters['pad']\n #print(\"stride =\", stride)\n #print(\"pad =\", pad)\n \n # Compute the dimensions of the CONV output volume using the formula given above. Hint: use int() to floor. (≈2 lines)\n n_H = np.int((n_H_prev-f+2*pad)/stride)+1\n n_W = np.int((n_W_prev-f+2*pad)/stride)+1\n #print(\"n_H =\", n_H)\n #print(\"n_W =\", n_W)\n \n # Initialize the output volume Z with zeros. (≈1 line)\n Z = np.zeros((m, n_H, n_W, n_C))\n #print (\"Z.shape =\", Z.shape)\n \n # Create A_prev_pad by padding A_prev\n A_prev_pad = zero_pad(A_prev, pad) #padded image of shape (m, n_H_prev + 2*pad, n_W_prev + 2*pad, n_C_prev)\n #print (\"A_prev_pad.shape =\", A_prev_pad.shape)\n \n for i in range(m): # loop over the batch of training examples\n a_prev_pad = A_prev_pad[i,:,:,:] # Select ith training example's padded activation\n #print (\"a_prev_pad.shape =\", a_prev_pad.shape)\n for h in range(n_H): # loop over vertical axis of the output volume\n for w in range(n_W): # loop over horizontal axis of the output volume\n for c in range(n_C): # loop over channels (= #filters) of the output volume\n \n # Find the corners of the current \"slice\" (≈4 lines)\n # (GAC) using h, w, f and s\n vert_start = h*stride\n vert_end = h*stride + f\n horiz_start = w*stride\n horiz_end = w*stride + f\n #print (\"vert_start =\", vert_start)\n #print (\"vert_end =\", vert_end)\n #print (\"horiz_start =\", horiz_start)\n #print (\"horiz_end =\", horiz_end)\n \n # Use the corners to define the (3D) slice of a_prev_pad (See Hint above the cell). (≈1 line)\n a_slice_prev = a_prev_pad[vert_start:vert_end,horiz_start:horiz_end,:]\n #print (\"a_slice_prev.shape =\", a_slice_prev.shape)\n \n # Convolve the (3D) slice with the correct filter W and bias b, to get back one output neuron. (≈1 line)\n Z[i, h, w, c] = conv_single_step(a_slice_prev, W[:,:,:,c], b[:,:,:,c])\n \n ### END CODE HERE ###\n \n # Making sure your output shape is correct\n assert(Z.shape == (m, n_H, n_W, n_C))\n \n # Save information in \"cache\" for the backprop\n cache = (A_prev, W, b, hparameters)\n \n return Z, cache",
"_____no_output_____"
],
[
"np.random.seed(1)\nA_prev = np.random.randn(10,4,4,3)\nW = np.random.randn(2,2,3,8)\nb = np.random.randn(1,1,1,8)\nhparameters = {\"pad\" : 2,\n \"stride\": 2}\n\nZ, cache_conv = conv_forward(A_prev, W, b, hparameters)\nprint(\"Z's mean =\", np.mean(Z))\nprint(\"Z[3,2,1] =\", Z[3,2,1])\nprint(\"cache_conv[0][1][2][3] =\", cache_conv[0][1][2][3])",
"Z's mean = 0.0489952035289\nZ[3,2,1] = [-0.61490741 -6.7439236 -2.55153897 1.75698377 3.56208902 0.53036437\n 5.18531798 8.75898442]\ncache_conv[0][1][2][3] = [-0.20075807 0.18656139 0.41005165]\n"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **Z's mean**\n </td>\n <td>\n 0.0489952035289\n </td>\n </tr>\n <tr>\n <td>\n **Z[3,2,1]**\n </td>\n <td>\n [-0.61490741 -6.7439236 -2.55153897 1.75698377 3.56208902 0.53036437\n 5.18531798 8.75898442]\n </td>\n </tr>\n <tr>\n <td>\n **cache_conv[0][1][2][3]**\n </td>\n <td>\n [-0.20075807 0.18656139 0.41005165]\n </td>\n </tr>\n\n</table>\n",
"_____no_output_____"
],
[
"Finally, CONV layer should also contain an activation, in which case we would add the following line of code:\n\n```python\n# Convolve the window to get back one output neuron\nZ[i, h, w, c] = ...\n# Apply activation\nA[i, h, w, c] = activation(Z[i, h, w, c])\n```\n\nYou don't need to do it here. \n",
"_____no_output_____"
],
[
"## 4 - Pooling layer \n\nThe pooling (POOL) layer reduces the height and width of the input. It helps reduce computation, as well as helps make feature detectors more invariant to its position in the input. The two types of pooling layers are: \n\n- Max-pooling layer: slides an ($f, f$) window over the input and stores the max value of the window in the output.\n\n- Average-pooling layer: slides an ($f, f$) window over the input and stores the average value of the window in the output.\n\n<table>\n<td>\n<img src=\"images/max_pool1.png\" style=\"width:500px;height:300px;\">\n<td>\n\n<td>\n<img src=\"images/a_pool.png\" style=\"width:500px;height:300px;\">\n<td>\n</table>\n\nThese pooling layers have no parameters for backpropagation to train. However, they have hyperparameters such as the window size $f$. This specifies the height and width of the fxf window you would compute a max or average over. \n\n### 4.1 - Forward Pooling\nNow, you are going to implement MAX-POOL and AVG-POOL, in the same function. \n\n**Exercise**: Implement the forward pass of the pooling layer. Follow the hints in the comments below.\n\n**Reminder**:\nAs there's no padding, the formulas binding the output shape of the pooling to the input shape is:\n$$ n_H = \\lfloor \\frac{n_{H_{prev}} - f}{stride} \\rfloor +1 $$\n$$ n_W = \\lfloor \\frac{n_{W_{prev}} - f}{stride} \\rfloor +1 $$\n$$ n_C = n_{C_{prev}}$$",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: pool_forward\n\ndef pool_forward(A_prev, hparameters, mode = \"max\"):\n \"\"\"\n Implements the forward pass of the pooling layer\n \n Arguments:\n A_prev -- Input data, numpy array of shape (m, n_H_prev, n_W_prev, n_C_prev)\n hparameters -- python dictionary containing \"f\" and \"stride\"\n mode -- the pooling mode you would like to use, defined as a string (\"max\" or \"average\")\n \n Returns:\n A -- output of the pool layer, a numpy array of shape (m, n_H, n_W, n_C)\n cache -- cache used in the backward pass of the pooling layer, contains the input and hparameters \n \"\"\"\n \n # Retrieve dimensions from the input shape\n (m, n_H_prev, n_W_prev, n_C_prev) = A_prev.shape\n \n # Retrieve hyperparameters from \"hparameters\"\n f = hparameters[\"f\"]\n stride = hparameters[\"stride\"]\n \n # Define the dimensions of the output\n n_H = int(1 + (n_H_prev - f) / stride)\n n_W = int(1 + (n_W_prev - f) / stride)\n n_C = n_C_prev\n \n # Initialize output matrix A\n A = np.zeros((m, n_H, n_W, n_C)) \n \n ### START CODE HERE ###\n for i in range(m): # loop over the training examples\n for h in range(n_H): # loop on the vertical axis of the output volume\n for w in range(n_W): # loop on the horizontal axis of the output volume\n for c in range (n_C): # loop over the channels of the output volume\n \n # Find the corners of the current \"slice\" (≈4 lines)\n vert_start = h * stride\n vert_end = vert_start + f\n horiz_start = w * stride\n horiz_end = horiz_start + f\n \n # Use the corners to define the current slice on the ith training example of A_prev, channel c. (≈1 line)\n #a_prev = A_prev[i,:,:,:]\n #a_slice_prev = a_prev[vert_start:vert_end,horiz_start:horiz_end,c]\n a_slice_prev = A_prev[i, vert_start:vert_end, horiz_start:horiz_end, c]\n \n # Compute the pooling operation on the slice. Use an if statment to differentiate the modes. Use np.max/np.mean.\n #Z[i, h, w, c] = conv_single_step(a_slice_prev, W[:,:,:,c], b[:,:,:,c])\n if mode == \"max\":\n A[i, h, w, c] = np.max(a_slice_prev)\n elif mode == \"average\":\n A[i, h, w, c] = np.mean(a_slice_prev)\n \n ### END CODE HERE ###\n \n # Store the input and hparameters in \"cache\" for pool_backward()\n cache = (A_prev, hparameters)\n \n # Making sure your output shape is correct\n assert(A.shape == (m, n_H, n_W, n_C))\n \n return A, cache",
"_____no_output_____"
],
[
"np.random.seed(1)\nA_prev = np.random.randn(2, 4, 4, 3)\nhparameters = {\"stride\" : 2, \"f\": 3}\n\nA, cache = pool_forward(A_prev, hparameters)\nprint(\"mode = max\")\nprint(\"A =\", A)\nprint()\nA, cache = pool_forward(A_prev, hparameters, mode = \"average\")\nprint(\"mode = average\")\nprint(\"A =\", A)",
"mode = max\nA = [[[[ 1.74481176 0.86540763 1.13376944]]]\n\n\n [[[ 1.13162939 1.51981682 2.18557541]]]]\n\nmode = average\nA = [[[[ 0.02105773 -0.20328806 -0.40389855]]]\n\n\n [[[-0.22154621 0.51716526 0.48155844]]]]\n"
]
],
[
[
"**Expected Output:**\n<table>\n\n <tr>\n <td>\n A =\n </td>\n <td>\n [[[[ 1.74481176 0.86540763 1.13376944]]]\n\n\n [[[ 1.13162939 1.51981682 2.18557541]]]]\n\n </td>\n </tr>\n <tr>\n <td>\n A =\n </td>\n <td>\n [[[[ 0.02105773 -0.20328806 -0.40389855]]]\n\n\n [[[-0.22154621 0.51716526 0.48155844]]]]\n\n </td>\n </tr>\n\n</table>\n",
"_____no_output_____"
],
[
"Congratulations! You have now implemented the forward passes of all the layers of a convolutional network. \n\nThe remainer of this notebook is optional, and will not be graded.\n",
"_____no_output_____"
],
[
"## 5 - Backpropagation in convolutional neural networks (OPTIONAL / UNGRADED)\n\nIn modern deep learning frameworks, you only have to implement the forward pass, and the framework takes care of the backward pass, so most deep learning engineers don't need to bother with the details of the backward pass. The backward pass for convolutional networks is complicated. If you wish however, you can work through this optional portion of the notebook to get a sense of what backprop in a convolutional network looks like. \n\nWhen in an earlier course you implemented a simple (fully connected) neural network, you used backpropagation to compute the derivatives with respect to the cost to update the parameters. Similarly, in convolutional neural networks you can to calculate the derivatives with respect to the cost in order to update the parameters. The backprop equations are not trivial and we did not derive them in lecture, but we briefly presented them below.\n\n### 5.1 - Convolutional layer backward pass \n\nLet's start by implementing the backward pass for a CONV layer. \n\n#### 5.1.1 - Computing dA:\nThis is the formula for computing $dA$ with respect to the cost for a certain filter $W_c$ and a given training example:\n\n$$ dA += \\sum _{h=0} ^{n_H} \\sum_{w=0} ^{n_W} W_c \\times dZ_{hw} \\tag{1}$$\n\nWhere $W_c$ is a filter and $dZ_{hw}$ is a scalar corresponding to the gradient of the cost with respect to the output of the conv layer Z at the hth row and wth column (corresponding to the dot product taken at the ith stride left and jth stride down). Note that at each time, we multiply the the same filter $W_c$ by a different dZ when updating dA. We do so mainly because when computing the forward propagation, each filter is dotted and summed by a different a_slice. Therefore when computing the backprop for dA, we are just adding the gradients of all the a_slices. \n\nIn code, inside the appropriate for-loops, this formula translates into:\n```python\nda_prev_pad[vert_start:vert_end, horiz_start:horiz_end, :] += W[:,:,:,c] * dZ[i, h, w, c]\n```\n\n#### 5.1.2 - Computing dW:\nThis is the formula for computing $dW_c$ ($dW_c$ is the derivative of one filter) with respect to the loss:\n\n$$ dW_c += \\sum _{h=0} ^{n_H} \\sum_{w=0} ^ {n_W} a_{slice} \\times dZ_{hw} \\tag{2}$$\n\nWhere $a_{slice}$ corresponds to the slice which was used to generate the acitivation $Z_{ij}$. Hence, this ends up giving us the gradient for $W$ with respect to that slice. Since it is the same $W$, we will just add up all such gradients to get $dW$. \n\nIn code, inside the appropriate for-loops, this formula translates into:\n```python\ndW[:,:,:,c] += a_slice * dZ[i, h, w, c]\n```\n\n#### 5.1.3 - Computing db:\n\nThis is the formula for computing $db$ with respect to the cost for a certain filter $W_c$:\n\n$$ db = \\sum_h \\sum_w dZ_{hw} \\tag{3}$$\n\nAs you have previously seen in basic neural networks, db is computed by summing $dZ$. In this case, you are just summing over all the gradients of the conv output (Z) with respect to the cost. \n\nIn code, inside the appropriate for-loops, this formula translates into:\n```python\ndb[:,:,:,c] += dZ[i, h, w, c]\n```\n\n**Exercise**: Implement the `conv_backward` function below. You should sum over all the training examples, filters, heights, and widths. You should then compute the derivatives using formulas 1, 2 and 3 above. ",
"_____no_output_____"
]
],
[
[
"def conv_backward(dZ, cache):\n \"\"\"\n Implement the backward propagation for a convolution function\n \n Arguments:\n dZ -- gradient of the cost with respect to the output of the conv layer (Z), numpy array of shape (m, n_H, n_W, n_C)\n cache -- cache of values needed for the conv_backward(), output of conv_forward()\n \n Returns:\n dA_prev -- gradient of the cost with respect to the input of the conv layer (A_prev),\n numpy array of shape (m, n_H_prev, n_W_prev, n_C_prev)\n dW -- gradient of the cost with respect to the weights of the conv layer (W)\n numpy array of shape (f, f, n_C_prev, n_C)\n db -- gradient of the cost with respect to the biases of the conv layer (b)\n numpy array of shape (1, 1, 1, n_C)\n \"\"\"\n \n ### START CODE HERE ###\n # Retrieve information from \"cache\"\n (A_prev, W, b, hparameters) = cache\n \n # Retrieve dimensions from A_prev's shape\n (m, n_H_prev, n_W_prev, n_C_prev) = A_prev.shape\n \n # Retrieve dimensions from W's shape\n (f, f, n_C_prev, n_C) = W.shape\n \n # Retrieve information from \"hparameters\"\n stride = hparameters['stride']\n pad = hparameters['pad']\n \n # Retrieve dimensions from dZ's shape\n (m, n_H, n_W, n_C) = dZ.shape\n \n # Initialize dA_prev, dW, db with the correct shapes\n dA_prev = np.zeros((m, n_H_prev, n_W_prev, n_C_prev)) \n dW = np.zeros((f, f, n_C_prev, n_C))\n db = np.zeros((1, 1, 1, n_C))\n\n # Pad A_prev and dA_prev\n A_prev_pad = zero_pad(A_prev, pad)\n dA_prev_pad = zero_pad(dA_prev, pad)\n \n for i in range(m): # loop over the training examples\n \n # select ith training example from A_prev_pad and dA_prev_pad\n a_prev_pad = A_prev_pad[i,:,:,:]\n da_prev_pad = dA_prev_pad[i,:,:,:]\n \n for h in range(n_H): # loop over vertical axis of the output volume\n for w in range(n_W): # loop over horizontal axis of the output volume\n for c in range(n_C): # loop over the channels of the output volume\n \n # Find the corners of the current \"slice\"\n vert_start = h*stride\n vert_end = vert_start+f\n horiz_start = w*stride\n horiz_end = horiz_start+f\n \n # Use the corners to define the slice from a_prev_pad\n a_slice = a_prev_pad[vert_start:vert_end,horiz_start:horiz_end,:]\n\n # Update gradients for the window and the filter's parameters using the code formulas given above\n da_prev_pad[vert_start:vert_end, horiz_start:horiz_end, :] += W[:,:,:,c] * dZ[i, h, w, c]\n dW[:,:,:,c] += a_slice * dZ[i, h, w, c]\n db[:,:,:,c] += dZ[i, h, w, c]\n \n # Set the ith training example's dA_prev to the unpaded da_prev_pad (Hint: use X[pad:-pad, pad:-pad, :])\n dA_prev[i, :, :, :] = da_prev_pad[pad:-pad, pad:-pad, :]\n ### END CODE HERE ###\n \n # Making sure your output shape is correct\n assert(dA_prev.shape == (m, n_H_prev, n_W_prev, n_C_prev))\n \n return dA_prev, dW, db",
"_____no_output_____"
],
[
"np.random.seed(1)\ndA, dW, db = conv_backward(Z, cache_conv)\nprint(\"dA_mean =\", np.mean(dA))\nprint(\"dW_mean =\", np.mean(dW))\nprint(\"db_mean =\", np.mean(db))",
"dA_mean = 1.45243777754\ndW_mean = 1.72699145831\ndb_mean = 7.83923256462\n"
]
],
[
[
"** Expected Output: **\n<table>\n <tr>\n <td>\n **dA_mean**\n </td>\n <td>\n 1.45243777754\n </td>\n </tr>\n <tr>\n <td>\n **dW_mean**\n </td>\n <td>\n 1.72699145831\n </td>\n </tr>\n <tr>\n <td>\n **db_mean**\n </td>\n <td>\n 7.83923256462\n </td>\n </tr>\n\n</table>\n",
"_____no_output_____"
],
[
"## 5.2 Pooling layer - backward pass\n\nNext, let's implement the backward pass for the pooling layer, starting with the MAX-POOL layer. Even though a pooling layer has no parameters for backprop to update, you still need to backpropagation the gradient through the pooling layer in order to compute gradients for layers that came before the pooling layer. \n\n### 5.2.1 Max pooling - backward pass \n\nBefore jumping into the backpropagation of the pooling layer, you are going to build a helper function called `create_mask_from_window()` which does the following: \n\n$$ X = \\begin{bmatrix}\n1 && 3 \\\\\n4 && 2\n\\end{bmatrix} \\quad \\rightarrow \\quad M =\\begin{bmatrix}\n0 && 0 \\\\\n1 && 0\n\\end{bmatrix}\\tag{4}$$\n\nAs you can see, this function creates a \"mask\" matrix which keeps track of where the maximum of the matrix is. True (1) indicates the position of the maximum in X, the other entries are False (0). You'll see later that the backward pass for average pooling will be similar to this but using a different mask. \n\n**Exercise**: Implement `create_mask_from_window()`. This function will be helpful for pooling backward. \nHints:\n- [np.max()]() may be helpful. It computes the maximum of an array.\n- If you have a matrix X and a scalar x: `A = (X == x)` will return a matrix A of the same size as X such that:\n```\nA[i,j] = True if X[i,j] = x\nA[i,j] = False if X[i,j] != x\n```\n- Here, you don't need to consider cases where there are several maxima in a matrix.",
"_____no_output_____"
]
],
[
[
"def create_mask_from_window(x):\n \"\"\"\n Creates a mask from an input matrix x, to identify the max entry of x.\n \n Arguments:\n x -- Array of shape (f, f)\n \n Returns:\n mask -- Array of the same shape as window, contains a True at the position corresponding to the max entry of x.\n \"\"\"\n \n ### START CODE HERE ### (≈1 line)\n mask = (x==np.max(x))\n ### END CODE HERE ###\n \n return mask",
"_____no_output_____"
],
[
"np.random.seed(1)\nx = np.random.randn(2,3)\nmask = create_mask_from_window(x)\nprint('x = ', x)\nprint(\"mask = \", mask)",
"x = [[ 1.62434536 -0.61175641 -0.52817175]\n [-1.07296862 0.86540763 -2.3015387 ]]\nmask = [[ True False False]\n [False False False]]\n"
]
],
[
[
"**Expected Output:** \n\n<table> \n<tr> \n<td>\n\n**x =**\n</td>\n\n<td>\n\n[[ 1.62434536 -0.61175641 -0.52817175] <br>\n [-1.07296862 0.86540763 -2.3015387 ]]\n\n </td>\n</tr>\n\n<tr> \n<td>\n**mask =**\n</td>\n<td>\n[[ True False False] <br>\n [False False False]]\n</td>\n</tr>\n\n\n</table>",
"_____no_output_____"
],
[
"Why do we keep track of the position of the max? It's because this is the input value that ultimately influenced the output, and therefore the cost. Backprop is computing gradients with respect to the cost, so anything that influences the ultimate cost should have a non-zero gradient. So, backprop will \"propagate\" the gradient back to this particular input value that had influenced the cost. ",
"_____no_output_____"
],
[
"### 5.2.2 - Average pooling - backward pass \n\nIn max pooling, for each input window, all the \"influence\" on the output came from a single input value--the max. In average pooling, every element of the input window has equal influence on the output. So to implement backprop, you will now implement a helper function that reflects this.\n\nFor example if we did average pooling in the forward pass using a 2x2 filter, then the mask you'll use for the backward pass will look like: \n$$ dZ = 1 \\quad \\rightarrow \\quad dZ =\\begin{bmatrix}\n1/4 && 1/4 \\\\\n1/4 && 1/4\n\\end{bmatrix}\\tag{5}$$\n\nThis implies that each position in the $dZ$ matrix contributes equally to output because in the forward pass, we took an average. \n\n**Exercise**: Implement the function below to equally distribute a value dz through a matrix of dimension shape. [Hint](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ones.html)",
"_____no_output_____"
]
],
[
[
"def distribute_value(dz, shape):\n \"\"\"\n Distributes the input value in the matrix of dimension shape\n \n Arguments:\n dz -- input scalar\n shape -- the shape (n_H, n_W) of the output matrix for which we want to distribute the value of dz\n \n Returns:\n a -- Array of size (n_H, n_W) for which we distributed the value of dz\n \"\"\"\n \n ### START CODE HERE ###\n # Retrieve dimensions from shape (≈1 line)\n (n_H, n_W) = shape\n \n # Compute the value to distribute on the matrix (≈1 line)\n average = dz / (n_H * n_W)\n \n # Create a matrix where every entry is the \"average\" value (≈1 line)\n a = np.ones((n_H, n_W))*average\n ### END CODE HERE ###\n \n return a",
"_____no_output_____"
],
[
"a = distribute_value(2, (2,2))\nprint('distributed value =', a)",
"distributed value = [[ 0.5 0.5]\n [ 0.5 0.5]]\n"
]
],
[
[
"**Expected Output**: \n\n<table> \n<tr> \n<td>\ndistributed_value =\n</td>\n<td>\n[[ 0.5 0.5]\n<br\\> \n[ 0.5 0.5]]\n</td>\n</tr>\n</table>",
"_____no_output_____"
],
[
"### 5.2.3 Putting it together: Pooling backward \n\nYou now have everything you need to compute backward propagation on a pooling layer.\n\n**Exercise**: Implement the `pool_backward` function in both modes (`\"max\"` and `\"average\"`). You will once again use 4 for-loops (iterating over training examples, height, width, and channels). You should use an `if/elif` statement to see if the mode is equal to `'max'` or `'average'`. If it is equal to 'average' you should use the `distribute_value()` function you implemented above to create a matrix of the same shape as `a_slice`. Otherwise, the mode is equal to '`max`', and you will create a mask with `create_mask_from_window()` and multiply it by the corresponding value of dZ.",
"_____no_output_____"
]
],
[
[
"def pool_backward(dA, cache, mode = \"max\"):\n \"\"\"\n Implements the backward pass of the pooling layer\n \n Arguments:\n dA -- gradient of cost with respect to the output of the pooling layer, same shape as A\n cache -- cache output from the forward pass of the pooling layer, contains the layer's input and hparameters \n mode -- the pooling mode you would like to use, defined as a string (\"max\" or \"average\")\n \n Returns:\n dA_prev -- gradient of cost with respect to the input of the pooling layer, same shape as A_prev\n \"\"\"\n \n ### START CODE HERE ###\n \n # Retrieve information from cache (≈1 line)\n (A_prev, hparameters) = cache\n \n # Retrieve hyperparameters from \"hparameters\" (≈2 lines)\n stride = hparameters['stride']\n f = hparameters['f']\n \n # Retrieve dimensions from A_prev's shape and dA's shape (≈2 lines)\n m, n_H_prev, n_W_prev, n_C_prev = A_prev.shape\n m, n_H, n_W, n_C = dA.shape\n \n # Initialize dA_prev with zeros (≈1 line)\n dA_prev = np.zeros((m, n_H_prev, n_W_prev, n_C_prev))\n \n for i in range(m): # loop over the training examples\n \n # select training example from A_prev (≈1 line)\n a_prev = A_prev[i,:,:,:]\n \n for h in range(n_H): # loop on the vertical axis\n for w in range(n_W): # loop on the horizontal axis\n for c in range(n_C): # loop over the channels (depth)\n \n # Find the corners of the current \"slice\" (≈4 lines)\n vert_start = h*stride\n vert_end = vert_start+f\n horiz_start = w*stride\n horiz_end = horiz_start+f\n \n # Compute the backward propagation in both modes.\n if mode == \"max\":\n \n # Use the corners and \"c\" to define the current slice from a_prev (≈1 line)\n a_prev_slice = a_prev[vert_start:vert_end,horiz_start:horiz_end,c]\n # Create the mask from a_prev_slice (≈1 line)\n mask = create_mask_from_window(a_prev_slice)\n # Set dA_prev to be dA_prev + (the mask multiplied by the correct entry of dA) (≈1 line)\n dA_prev[i, vert_start: vert_end, horiz_start: horiz_end, c] += mask * dA[i,h,w,c] \n \n elif mode == \"average\":\n \n # Get the value a from dA (≈1 line)\n da = dA[i, h, w, c]\n # Define the shape of the filter as fxf (≈1 line)\n shape = (f,f)\n # Distribute it to get the correct slice of dA_prev. i.e. Add the distributed value of da. (≈1 line)\n dA_prev[i, vert_start: vert_end, horiz_start: horiz_end, c] += distribute_value(da, shape)\n \n ### END CODE ###\n \n # Making sure your output shape is correct\n assert(dA_prev.shape == A_prev.shape)\n \n return dA_prev",
"_____no_output_____"
],
[
"np.random.seed(1)\nA_prev = np.random.randn(5, 5, 3, 2)\nhparameters = {\"stride\" : 1, \"f\": 2}\nA, cache = pool_forward(A_prev, hparameters)\ndA = np.random.randn(5, 4, 2, 2)\n\ndA_prev = pool_backward(dA, cache, mode = \"max\")\nprint(\"mode = max\")\nprint('mean of dA = ', np.mean(dA))\nprint('dA_prev[1,1] = ', dA_prev[1,1]) \nprint()\ndA_prev = pool_backward(dA, cache, mode = \"average\")\nprint(\"mode = average\")\nprint('mean of dA = ', np.mean(dA))\nprint('dA_prev[1,1] = ', dA_prev[1,1]) ",
"mode = max\nmean of dA = 0.145713902729\ndA_prev[1,1] = [[ 0. 0. ]\n [ 5.05844394 -1.68282702]\n [ 0. 0. ]]\n\nmode = average\nmean of dA = 0.145713902729\ndA_prev[1,1] = [[ 0.08485462 0.2787552 ]\n [ 1.26461098 -0.25749373]\n [ 1.17975636 -0.53624893]]\n"
]
],
[
[
"**Expected Output**: \n\nmode = max:\n<table> \n<tr> \n<td>\n\n**mean of dA =**\n</td>\n\n<td>\n\n0.145713902729\n\n </td>\n</tr>\n\n<tr> \n<td>\n**dA_prev[1,1] =** \n</td>\n<td>\n[[ 0. 0. ] <br>\n [ 5.05844394 -1.68282702] <br>\n [ 0. 0. ]]\n</td>\n</tr>\n</table>\n\nmode = average\n<table> \n<tr> \n<td>\n\n**mean of dA =**\n</td>\n\n<td>\n\n0.145713902729\n\n </td>\n</tr>\n\n<tr> \n<td>\n**dA_prev[1,1] =** \n</td>\n<td>\n[[ 0.08485462 0.2787552 ] <br>\n [ 1.26461098 -0.25749373] <br>\n [ 1.17975636 -0.53624893]]\n</td>\n</tr>\n</table>",
"_____no_output_____"
],
[
"### Congratulations !\n\nCongratulation on completing this assignment. You now understand how convolutional neural networks work. You have implemented all the building blocks of a neural network. In the next assignment you will implement a ConvNet using TensorFlow.",
"_____no_output_____"
]
],
[
[
"!tar cvfz notebook.tar.gz *",
"Convolution model - Application - v1-Copy1.ipynb\nConvolution model - Application - v1.ipynb\nConvolution model - Step by Step - v1.ipynb\nConvolution model - Step by Step - v2-Copy1.ipynb\nConvolution model - Step by Step - v2.ipynb\n__pycache__/\n__pycache__/cnn_utils.cpython-36.pyc\ncnn_utils.py\ndatasets/\ndatasets/train_signs.h5\ndatasets/test_signs.h5\nimages/\nimages/PAD.png\nimages/model.png\nimages/conv_nn.png\nimages/Convolution_schematic.gif\nimages/vert_horiz_kiank.png\nimages/conv_kiank.mp4\nimages/max_pool.png\nimages/conv1.png\nimages/ave-pool.png\nimages/thumbs_up.jpg\nimages/ave_pool1.png\nimages/conv.png\nimages/a_pool.png\nimages/max_pool1.png\nimages/SIGNS.png\nimages/average_pool.png\nnotebook.tar.gz\n"
],
[
"!tar cvfz notebook.tar.gz *",
"Convolution model - Application - v1-Copy1.ipynb\nConvolution model - Application - v1.ipynb\nConvolution model - Step by Step - v1.ipynb\nConvolution model - Step by Step - v2-Copy1.ipynb\nConvolution model - Step by Step - v2.ipynb\n__pycache__/\n__pycache__/cnn_utils.cpython-36.pyc\ncnn_utils.py\ndatasets/\ndatasets/train_signs.h5\ndatasets/test_signs.h5\nimages/\nimages/PAD.png\nimages/model.png\nimages/conv_nn.png\nimages/Convolution_schematic.gif\nimages/vert_horiz_kiank.png\nimages/conv_kiank.mp4\nimages/max_pool.png\nimages/conv1.png\nimages/ave-pool.png\nimages/thumbs_up.jpg\nimages/ave_pool1.png\nimages/conv.png\nimages/a_pool.png\nimages/max_pool1.png\nimages/SIGNS.png\nimages/average_pool.png\nnotebook.tar.gz\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
d0f3b494c794ae08bb901b85f1ff0d2897dbea5d | 570,756 | ipynb | Jupyter Notebook | empenhos_requests_ufrn_budget.ipynb | igorabrandao/UFRN-budget-application | 19067dbf7207b48c2ae62e39d83520854451da3b | [
"MIT"
] | 1 | 2018-12-01T19:15:02.000Z | 2018-12-01T19:15:02.000Z | empenhos_requests_ufrn_budget.ipynb | igorabrandao/UFRN-budget-application | 19067dbf7207b48c2ae62e39d83520854451da3b | [
"MIT"
] | null | null | null | empenhos_requests_ufrn_budget.ipynb | igorabrandao/UFRN-budget-application | 19067dbf7207b48c2ae62e39d83520854451da3b | [
"MIT"
] | null | null | null | 75.168708 | 40,048 | 0.614679 | [
[
[
"# Ciência de dados - Unidade 3\n\n*Por: Débora Azevedo, Eliseu Jayro, Francisco de Paiva e Igor Brandão*\n\n### Objetivos\n\nO objetivo desse projeto é explorar os [datasets da UFRN](http://dados.ufrn.br/group/despesas-e-orcamento) contendo informações sobre requisições de material, requisições de manutenção e empenhos sob o contexto da [diminuição de verba](https://g1.globo.com/educacao/noticia/rio-grande-do-norte-veja-a-evolucao-do-orcamento-repassado-pelo-mec-as-duas-universidades-federais-do-estado.ghtml) que a UFRN recentemente vem sofrendo devido a crise financeira.\n\nDe acordo com a pesquisa feita pelo nosso grupo, as fontes dizem que os cortes atingem [principalmente serviços terceirizados](https://g1.globo.com/educacao/noticia/90-das-universidades-federais-tiveram-perda-real-no-orcamento-em-cinco-anos-verba-nacional-encolheu-28.ghtml) como limpeza, manutenção e segurança, além de benefícios para estudantes de baixa renda, dado que estas [não são despesas obrigatórias] (https://g1.globo.com/educacao/noticia/salario-de-professores-das-universidades-federais-e-despesa-obrigatoria-mas-auxilio-estudantil-nao-entenda-a-diferenca.ghtml), ao contrário do pagamento de aposentadorias e pensões e o pagamento de pessoal ativo, no entanto, em [entrevista](http://www.tribunadonorte.com.br/noticia/na-s-vamos-receber-o-ma-nimo-diz-reitora-da-ufrn/399980), a atual reitora disse que o setor mais afetado seria o de obras e sua gestão, o que pode ser uma informação mais confiável, visto que até 2017 todo o orçamento era destinado diretamente as universidades federais, portanto eles decidiam como todos os gastos eram feitos. Isso mudou em 2018, já que o Ministério da Educação adotou uma nova metodologia que restringe ainda mais os gastos à \"matriz Andifes\" de forma que 50% do orçamento passou a ser gerenciado pelo próprio ministério da educação, logo a comparação do orçamento de 2018 com os anteriores deixa de ser possível.\n",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"# 0 - Importando as bibliotecas\n\nAqui utilizaremos o *pip* para instalar as bibliotecas necessárias para executar o notebook, sendo estas:\n\n- pandas\n- numpy\n- matplotlib\n- wordcloud",
"_____no_output_____"
]
],
[
[
"!pip install pandas\n!pip install numpy\n!pip install matplotlib\n!pip install wordcloud",
"Requirement already satisfied: pandas in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (0.23.4)\nRequirement already satisfied: python-dateutil>=2.5.0 in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from pandas) (2.7.5)\nRequirement already satisfied: pytz>=2011k in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from pandas) (2018.7)\nRequirement already satisfied: numpy>=1.9.0 in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from pandas) (1.15.4)\nRequirement already satisfied: six>=1.5 in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from python-dateutil>=2.5.0->pandas) (1.11.0)\nRequirement already satisfied: numpy in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (1.15.4)\nRequirement already satisfied: matplotlib in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (3.0.1)\nRequirement already satisfied: numpy>=1.10.0 in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from matplotlib) (1.15.4)\nRequirement already satisfied: cycler>=0.10 in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from matplotlib) (0.10.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from matplotlib) (1.0.1)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from matplotlib) (2.3.0)\nRequirement already satisfied: python-dateutil>=2.1 in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from matplotlib) (2.7.5)\nRequirement already satisfied: six in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from cycler>=0.10->matplotlib) (1.11.0)\nRequirement already satisfied: setuptools in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from kiwisolver>=1.0.1->matplotlib) (40.5.0)\nRequirement already satisfied: wordcloud in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (1.5.0)\nRequirement already satisfied: numpy>=1.6.1 in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from wordcloud) (1.15.4)\nRequirement already satisfied: pillow in c:\\users\\usuario\\anaconda3\\envs\\tfp36\\lib\\site-packages (from wordcloud) (5.3.0)\n"
]
],
[
[
"# 1 - Lendo os datasets\n\nNessa seção nós iremos importar os datasets contendo informações sobre requisiçoes de manutenção, requisições de material de serviço e empenhos, todos disponíveis no site de dados da UFRN.\n\nNa célula abaixo nós definimos uma lista com os arquivos que iremos precisar, lemos todos eles e os guardamos em um dicionário.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom os import path\n\n# Lista com o nome dos arquivos de todos os datasets que iremos utilizar\ndataset_names = ['requisicaomanutencao.csv', 'requisicaomaterialservico.csv', 'empenhos.csv']\n\n# Pasta em que os datasets se encontram\ndataset_path = 'datasets'\n\n# Dicionário onde eles serão armazenados\ndata = {}\n\n# Loop que itera sobre todos os nomes definidos e armazena os dados lidos no dicionário\nfor name in dataset_names:\n data[name[:-4]] = pd.read_csv(path.join(dataset_path, name), sep=';', low_memory=False)\n ",
"_____no_output_____"
],
[
"# Mostrando 'requisicaomanutencao.csv'\ndata['requisicaomanutencao']",
"_____no_output_____"
],
[
"# Mostrando 'requisicaomaterialservico.csv'\ndata['requisicaomaterialservico']",
"_____no_output_____"
],
[
"# Mostrando 'empenhos.csv'\ndata['empenhos']",
"_____no_output_____"
]
],
[
[
"# 2 - Explorando e limpando os datasets\n\nNessa seção é feita a análise das diferentes colunas dos datasets para identificar seus significados e suas utilidades para os problemas que iremos analisar. Sendo feita essa análise, nós então limpamos os datasets para que eles se tornem mais legíveis e mais fáceis de manusear.",
"_____no_output_____"
],
[
"## 2.1 - Requisição de manutenção\n\nTrata-se de um dataset listando todas as requisições de manutenções da UFRN desde 2005. Lembrando que iremos analisar apenas dados de 2008 a 2017, que são os anos em que temos o valor da verba total da UFRN.",
"_____no_output_____"
]
],
[
[
"maintenance_data = data['requisicaomanutencao']\nprint(maintenance_data.head())\nprint(maintenance_data.divisao.unique())",
" numero ano divisao \\\n0 7075 2018 Serviços Gerais \n1 7074 2018 Serviços Gerais \n2 7073 2018 Serviços Gerais \n3 7072 2018 Serviços Gerais \n4 7071 2018 Instalações Elétricas e Telecomunicações \n\n id_unidade_requisitante nome_unidade_requisitante \\\n0 1840 ASSESSORIA ADMINISTRATIVA DO CCHLA \n1 203 DEPARTAMENTO DE NUTRIÇÃO \n2 389 ADMINISTRAÇÃO DO CENTRO DE CIÊNCIAS DA SAÚDE \n3 184 DEPARTAMENTO DE BIOFÍSICA E FARMACOLOGIA \n4 389 ADMINISTRAÇÃO DO CENTRO DE CIÊNCIAS DA SAÚDE \n\n id_unidade_custo nome_unidade_custo data_cadastro \\\n0 141 DEPARTAMENTO DE HISTÓRIA 19/09/2018 \n1 441 CENTRO DE CIÊNCIAS DA SAÚDE 19/09/2018 \n2 441 CENTRO DE CIÊNCIAS DA SAÚDE 19/09/2018 \n3 184 DEPARTAMENTO DE BIOFÍSICA E FARMACOLOGIA 19/09/2018 \n4 441 CENTRO DE CIÊNCIAS DA SAÚDE 19/09/2018 \n\n descricao \\\n0 SOLICITO PEDREIRO PARA RECOMPOR 4 AZULEJOS NA ... \n1 É NECESSÁRIO PINTOR PARA COMPLEMENTAR SERVIÇO ... \n2 SOLICITAMOS CONSERTO DE VAZAMENTO DE INFILTRAÇ... \n3 É NECESSÁRIO PINTOR PARA COMPLEMENTAR SERVIÇO ... \n4 SOLICITAMOS INSTALAÇÃO DE REDE ELÉTRICA, COM P... \n\n local \\\n0 SALA 801 - HISTÓRIA / PRÉDIO DO CCHLA \n1 Lab de Educação Alimentar \n2 CCS/Sala de convivência dos alunos do Curso de... \n3 DEPARTAMENTO DE BIOFÍSICA E FARMACOLOGIA/ LABO... \n4 CCS/Sala de convivência dos alunos do Curso de... \n\n usuario status \n0 AMANDA SANTOS LIMA DA CUNHA PENDENTE AUTORIZAÇÃO DIREÇÃO CENTRO \n1 JONATAS HENRIQUE CAMARA DA SILVA ENVIADA \n2 WILSON SILVA DE SOUZA JÚNIOR EM ROTA VISITA \n3 JONATAS HENRIQUE CAMARA DA SILVA ENVIADA \n4 WILSON SILVA DE SOUZA JÚNIOR PEDIDO MATERIAL REALIZADO \n['Serviços Gerais' 'Instalações Elétricas e Telecomunicações'\n 'Instalações Hidráulicas e Sanitárias' 'Viário' 'Ar condicionado'\n 'Outros' nan]\n"
]
],
[
[
"### 2.11 - Descrevendo as colunas e valores\n\nObservando o resultado da célula acima, podemos fazer as seguintes conclusões sobre as colunas:\n\n- <span style=\"color:red\"><b>numero</b></span>: ID da requisição, não é relevante para o problema.\n\n- **ano**: Ano em que foi feita requisição de manutenção\n\n- **divisão**: Diz a divisão para qual a manutenção foi requisitada, assume os seguintes valores: 'Serviços Gerais', 'Instalações Elétricas e Telecomunicações', 'Instalações Hidráulicas e Sanitárias', 'Viário', 'Ar condicionado', 'Outros'.\n\n- **id_unidade_requisitante**: ID da unidade que fez a requisição.\n\n- **nome_unidade_requisitante**: Nome da unidade que fez a requisição.\n\n- **id_unidade_custo**: ID da unidade para qual o custo será destinado (pode ser igual a requisitante).\n\n- **nome_unidade_custo**: Nome da unidade para qual o custo será destinado (poder ser igual a requisitante).\n\n- **data_cadastro**: Data em que a requisição foi cadastrada.\n\n- **descricao**: Descrição da requisição, geralmente uma justificativa para aquela manutenção.\n\n- **local**: Local exato em que será feito a manutenção, pode ser uma sala, laboratório etc\n\n- <span style=\"color:red\"><b>usuario</b></span>: Usuário que solicitou a manutenção. Provavelmente não tem utilidade para nosso problema.\n\n- **status**: Diz o status atual da requisição. Pode ajudar na análise de custos, considerando apenas as que já foram aprovadas, comparando a proporção de aprovadas e reprovadas para cada setor.",
"_____no_output_____"
],
[
"### 2.12 - Removendo colunas desnecessárias\n\n- <span style=\"color:red\"><b>numero</b></span>: É apenas o ID da requisição\n- <span style=\"color:red\"><b>usuario</b></span>: Não precisamos saber o usuário para nossa análise",
"_____no_output_____"
]
],
[
[
"def remove_cols(df_input, dropped_columns):\n '''\n This functions receives a dataframe and a list of column names as input. It checks if each column exist,\n and if they do, they're removed.\n '''\n for dropped_column in dropped_columns:\n if dropped_column in df_input:\n df_input = df_input.drop([dropped_column], axis=1)\n return df_input\n\n\nmaintenance_dropped = ['numero', 'usuario']\nmaintenance_data = remove_cols(maintenance_data, maintenance_dropped)\nmaintenance_data.head()",
"_____no_output_____"
]
],
[
[
"### 2.13 - Removendo outliers e valores desnecessários\n\nAqui iremos analisar os valores do nosso dataset e determinar quais podemos remover ou modificar de modo a facilitar a nossa análise.",
"_____no_output_____"
]
],
[
[
"print(maintenance_data.status.value_counts())",
"FINALIZADA 67211\nAGUARDANDO_EXECUCAO 3649\nATENDIDA 1471\nNEGADA 1383\nAUTORIZADA 957\nCADASTRADA 794\nEM ROTA VISITA 759\nESTORNADA 743\nAGUARDANDO VISITA 685\nPEDIDO MATERIAL REALIZADO 679\nAGUARDANDO AVALIAÇÃO REQUISITANTE 610\nENVIADA 522\nSERVIÇO AVALIADO 199\nSERVIÇO NÃO EXECUTADO 143\nPENDENTE AUTORIZAÇÃO CHEFE UNIDADE 104\nARQUIVADA 84\nEM_EXECUCAO 67\nAGUARDANDO MATERIAL 54\nMATERIAL RECEBIDO 53\nATENDIDA DIMAN 48\nPENDENTE AUTORIZAÇÃO CHEFIA 41\nRETORNADA 39\nAGUARDANDO PEDIDO MATERIAL 21\nENVIADA_PROJETO 13\nPENDENTE AUTORIZAÇÃO DIREÇÃO CENTRO 10\nAUTORIZADA PARA INFRA 7\nPENDENTE_RECEBIMENTO_SIN 1\nRECEBIDA 1\nENVIADA_LICITACAO 1\nName: status, dtype: int64\n"
]
],
[
[
"**Observação:**\n\nChecando os status, podemos perceber que a maioria dos valores ocorrem um número muito pequeno de vezes e não precisamos deles para nossa análise, portanto iremos eliminar os valores com 800 ocorrências ou menos",
"_____no_output_____"
]
],
[
[
"maintenance_data = maintenance_data.groupby('status').filter(lambda x: len(x) > 800)\nmaintenance_data.status.value_counts()",
"_____no_output_____"
]
],
[
[
"**Observação:**\n\nSobram portanto 5 valores possíveis para a coluna **status**. Porém, para nossa análise de custos, precisamos saber apenas se a requisição foi negada ou autorizada. Analisando os status restantes, podemos considerar que toda requisição que tiver valor diferente de negada pode ser considerada como autorizada.",
"_____no_output_____"
]
],
[
[
"def convert_status(status_val):\n '''Converts the value of all strings in the status column to AUTORIZADA, unless their value is NEGADA.'''\n if status_val == 'NEGADA':\n return status_val\n else:\n return 'AUTORIZADA'\n\nmaintenance_data['status'] = maintenance_data['status'].apply(convert_status)\nmaintenance_data.status.value_counts()",
"_____no_output_____"
],
[
"maintenance_data.info()\nprint(maintenance_data.divisao.value_counts())\nprint(maintenance_data.nome_unidade_custo.value_counts())",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 74671 entries, 13 to 80339\nData columns (total 10 columns):\nano 74671 non-null int64\ndivisao 74652 non-null object\nid_unidade_requisitante 74671 non-null int64\nnome_unidade_requisitante 74671 non-null object\nid_unidade_custo 74671 non-null int64\nnome_unidade_custo 74671 non-null object\ndata_cadastro 74671 non-null object\ndescricao 74671 non-null object\nlocal 53327 non-null object\nstatus 74671 non-null object\ndtypes: int64(3), object(7)\nmemory usage: 6.3+ MB\nServiços Gerais 21978\nInstalações Elétricas e Telecomunicações 19376\nOutros 17501\nInstalações Hidráulicas e Sanitárias 15265\nViário 485\nAr condicionado 47\nName: divisao, dtype: int64\nCENTRO DE CIÊNCIAS DA SAÚDE 7179\nRESIDENCIAS UNIVERSITÁRIAS 7169\nUNIVERSIDADE FEDERAL DO RIO GRANDE DO NORTE 2314\nDEPARTAMENTO DE FÍSICA TEÓRICA E EXPERIMENTAL 1682\nPRÓ-REITORIA DE ADMINISTRAÇÃO (PROAD) 1655\nDIRETORIA DO RESTAURANTE UNIVERSITÁRIO 1569\nADMINISTRAÇÃO DO CCET 1306\nDEPARTAMENTO DE GEOLOGIA 1290\nDEPARTAMENTO DE BIOQUÍMICA 1181\nCENTRO DE TECNOLOGIA 1156\nCENTRO DE BIOCIÊNCIAS 1104\nADMINISTRAÇÃO DO CB 1081\nESCOLA DE MÚSICA 1026\nSUPERINTENDÊNCIA DE INFRAESTRUTURA 991\nDEPARTAMENTO DE FISIOLOGIA E COMPORTAMENTO 957\nSETOR DE COMPRAS E MANUTENÇÃO - CCSA 933\nNÚCLEO DE EDUCAÇÃO DA INFÂNCIA 932\nBIBLIOTECA CENTRAL ZILA MAMEDE 928\nPRÓ-REITORIA DE GESTÃO DE PESSOAS 926\nDEPARTAMENTO DE ENGENHARIA DE COMPUTACAO E AUTOMACAO 918\nDEPARTAMENTO DE MICROBIOLOGIA E PARASITOLOGIA 895\nMATERIAIS FORNECIDOS PELA SUPIN / INFRA-ESTRUTURA 839\nGASTOS COMUNS 838\nASSESSORIA ADMINISTRATIVA DO CCHLA 764\nGABINETE DO REITOR 744\nDEPARTAMENTO DE OCEANOGRAFIA E LIMNOLOGIA 719\nCENTRO DE CIÊNCIAS EXATAS E DA TERRA 712\nPRÓ-REITORIA DE ASSUNTOS ESTUDANTIS 703\nINSTITUTO DO CÉREBRO 690\nDAS/PROGESP - DIRETORIA DE ATENÇÃO À SAÚDE DO SERVIDOR 684\n ... \nCHEFIA DE GABINETE 1\nCOORDENADORIA DE CONCURSOS 1\nPROJETOS DE ENGENHARIA 1\nDPO - ELÉTRICA 1\nCCS - SETOR DO PATRIMÔNIO 1\nDAS - COORDENADORIA DE PROMOÇÃO DA SEGURANÇA DO TRABALHO E VIGILÂNCIA AMBIENTAL 1\nDPRF/UFRN - SINFOR 1\nCOMUNICA - TV UNIVERSITÁRIA 1\nPROGRAMA DE PÓS-GRADUAÇÃO EM SISTEMAS E COMPUTAÇÃO 1\nPROPESQ - ASSESSORIA TÉCNICA 1\nTVU/ARQUIVO 1\nDIRETORIA DE DESENVOLVIMENTO PEDAGÓGICO - SETOR DE ACOMPANHAMENTO DOS CURSOS DE GRADUAÇÃO 1\nCOORDENAÇÃO NPITI (NÚCLEO DE PESQUISAS E INOVAÇÃO EM TECNOLOGIA DA INFORMAÇÃO) - IMD 1\nDEPARTAMENTO DE CONTABILIDADE E FINANÇAS 1\nESCOLA DE GOVERNO 1\nLABORATORIO DE ARQUIOLOGIA 1\nCNPQ/IMPORTAÇÃO 1\nSUPERVISÃO ACADÊMICO-ADMINISTRATIVA DO SETOR I CCSA 1\nPROGRAMA DE PÓS-GRADUAÇÃO EM GEOGRAFIA 1\nAPOIO FINANCEIRO A GRUPOS DE PESQUISA, NOVOS PESQUISADORES E BIOTÉRIO 1\nLABORATÓRIO DE QUÍMICA I/DQ 1\nPROGRAMA DE PÓS-GRADUAÇÃO EM FILOSOFIA 1\nSECRETARIA ADMINISTRATIVA / ENGENHARIA DE MATERIAIS 1\nREDE GIGANATAL 1\nGESTÃO DE CONVÊNIOS 1\nADMINISTRAÇÃO DO CERES - CAICÓ 1\nSIM-FAMEB-UFBA SUBCONT. Nº 3P50AI030639-15S1/07 1\nDCF - DIREÇÃO 1\nPROJETOS ACADÊMICOS - PROAE 1\nMINC - 62ª REUNIÃO DA SBPC 1\nName: nome_unidade_custo, Length: 647, dtype: int64\n"
]
],
[
[
"### 2.14 - Lidando com valores nulos\n\nAqui nós utilizaremos o método [pandas.DataFrame.info](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.info.html) para verificar quais os valores nulos das colunas de nosso dataset. A partir disso, dependendo da quantidade de colunas com valores nulos e do tipo de dado, nós iremos decidir o que fazer com esses valores.",
"_____no_output_____"
]
],
[
[
"maintenance_data.info()\nmaintenance_data.divisao.value_counts()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 74671 entries, 13 to 80339\nData columns (total 10 columns):\nano 74671 non-null int64\ndivisao 74652 non-null object\nid_unidade_requisitante 74671 non-null int64\nnome_unidade_requisitante 74671 non-null object\nid_unidade_custo 74671 non-null int64\nnome_unidade_custo 74671 non-null object\ndata_cadastro 74671 non-null object\ndescricao 74671 non-null object\nlocal 53327 non-null object\nstatus 74671 non-null object\ndtypes: int64(3), object(7)\nmemory usage: 6.3+ MB\n"
]
],
[
[
"**Observação**\n\nUtilizando o método [pandas.DataFrame.info](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.info.html) percebemos que existem muitos valores **NULL** na coluna *local* e alguns poucos na coluna *divisao*. Para a coluna *local*, iremos preencher as linhas **nulas** com seus valores de *nome_unidade_custo*. Para a coluna *divisao*, iremos preencher com o valor 'Outros', que é um dos mais comuns.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nmaintenance_data['local'] = np.where(maintenance_data.local.isnull(), maintenance_data.nome_unidade_custo, maintenance_data.local)\nmaintenance_data['divisao'] = maintenance_data['divisao'].fillna('Outros')\n\nmaintenance_data.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 74671 entries, 13 to 80339\nData columns (total 10 columns):\nano 74671 non-null int64\ndivisao 74671 non-null object\nid_unidade_requisitante 74671 non-null int64\nnome_unidade_requisitante 74671 non-null object\nid_unidade_custo 74671 non-null int64\nnome_unidade_custo 74671 non-null object\ndata_cadastro 74671 non-null object\ndescricao 74671 non-null object\nlocal 74671 non-null object\nstatus 74671 non-null object\ndtypes: int64(3), object(7)\nmemory usage: 6.3+ MB\n"
],
[
"# Resultado final da limpeza\nmaintenance_data.head()",
"_____no_output_____"
]
],
[
[
"## 2.2 - Requisição de material de serviço\n\nEsse dataset lista todas as requisições de materiais e serviços contratados pela UFRN desde 2008.",
"_____no_output_____"
]
],
[
[
"material_request_data = data['requisicaomaterialservico']\nprint('===== Primeiras linhas =====')\nprint(material_request_data.head())\nprint('===== Contagem de valores de natureza_despesa =====')\nprint(material_request_data.natureza_despesa.value_counts())\nprint('===== Contagem de valores de status =====')\nprint(material_request_data.status.value_counts())",
"===== Primeiras linhas =====\n numero ano id_unidade_requisitante \\\n0 1786 2018 160 \n1 1785 2018 2640 \n2 1784 2018 1481 \n3 1783 2018 6799 \n4 1782 2018 4894 \n\n nome_unidade_requisitante id_unidade_custo \\\n0 DEPARTAMENTO DE ECONOMIA - DEPEC 434 \n1 SETOR DE COMPRAS E MANUTENÇÃO - CCSA 2640 \n2 PRÓ-REITORIA DE PESQUISA 1481 \n3 FNDE - PRONATEC - EAJ 6799 \n4 DEPARTAMENTO DE GEOFÍSICA 4894 \n\n nome_unidade_custo data_envio numero_contrato \\\n0 PROGRAMA DE PÓS-GRADUAÇÃO EM ECONOMIA 19/09/2018 60/2014 \n1 SETOR DE COMPRAS E MANUTENÇÃO - CCSA 18/09/2018 25/2018 \n2 PRÓ-REITORIA DE PESQUISA 18/09/2018 60/2014 \n3 FNDE - PRONATEC - EAJ 18/09/2018 18/2015 \n4 DEPARTAMENTO DE GEOFÍSICA 18/09/2018 25/2018 \n\n contratado natureza_despesa valor \\\n0 PRAIAMAR EMPREENDIMENTOS TURISTICOS LTDA SERV. PESSOA JURÍDICA 290.0 \n1 INSTRUCON COMÉRCIO E SERVIÇOS LTDA - ME SERV. PESSOA JURÍDICA 4433.0 \n2 PRAIAMAR EMPREENDIMENTOS TURISTICOS LTDA SERV. PESSOA JURÍDICA 290.0 \n3 PROTÁSIO LOCAÇÃO E TURISMO LTDA SERV. PESSOA JURÍDICA 7013.3 \n4 INSTRUCON COMÉRCIO E SERVIÇOS LTDA - ME SERV. PESSOA JURÍDICA 2316.0 \n\n observacoes status \n0 A professora ADRYANE GORAYEB (Requisição de Ho... ENVIADA \n1 SERVIÇOS DE MANUTENÇÃO PREVENTIVA E CORRETIVA,... ENVIADA \n2 GENAUTO CARVALHO DE FRANÇA FILHO\\r\\nChegada di... ENVIADA \n3 Condução de alunos para a Feira Internacional ... ENVIADA \n4 MANUTENÇÃO CORRETIVA (COM CARGA DE GÁS , SUBST... ENVIADA \n===== Contagem de valores de natureza_despesa =====\nSERV. PESSOA JURÍDICA 24188\nName: natureza_despesa, dtype: int64\n===== Contagem de valores de status =====\nLIQUIDADA 11645\nEM_LIQUIDACAO 5988\nENVIADA 5335\nESTORNADA 2856\nFINALIZADA 1543\nCADASTRADA 695\nA_EMPENHAR 144\nEMPENHO_ANULADO 139\nNEGADA 136\nPENDENTE ATENDIMENTO 45\nPENDENTE AUTORIZAÇÃO 37\nAUTORIZADA 36\nCANCELADA\\n 1\nName: status, dtype: int64\n"
]
],
[
[
"### 2.21 - Descrevendo as colunas e valores\n\nObservando o resultado da célula acima, podemos fazer as seguintes conclusões sobre as colunas:\n\n- <span style=\"color:red\"><b>numero</b></span>: ID da requisição, não é relevante.\n\n- **ano**: Ano em que foi feita a requisição.\n\n- **id_unidade_requisitante**: ID da unidade que fez a requisição, toda unidade tem um ID único.\n\n- **nome_unidade_requisitante**: Nome da unidade que fez a requisição.\n\n- **id_unidade_custo**: ID da unidade para qual os custos serão destinados, pode ser diferente da requisitante.\n\n- **nome_unidade_custo**: Nome da unidade para qual os custos serão destinados, pode ser diferente da requisitante.\n\n- **data_envio**: Data em que a requisição foi enviada.\n\n- <span style=\"color:red\"><b>numero_contrato</b></span>: Aparentemente as requisições são feitas por meio de contratos, esse é o número do contrato.\n\n- **contratado**: Empresa contratada para fornecer o material.\n\n- <span style=\"color:red\"><b>natureza_despesa</b></span>: Em todas as linhas analisadas, essa coluna tem o valor 'SERV. PESSOA JURÍDICA'.\n\n- **valor**: Valor pedido pela requisição.\n\n- **observacoes**: Comentário feito pela pessoa que fez a requisição, explicando o motivo desta\n\n- **status**: O status atual da requisição, está diretamente ligada ao empenho e pode assumir os seguintes valores: 'ENVIADA', 'PENDENTE ATENDIMENTO', 'CADASTRADA', 'ESTORNADA', 'LIQUIDADA', 'PENDENTE AUTORIZAÇÃO', 'FINALIZADA', 'EM_LIQUIDACAO', 'NEGADA', 'A_EMPENHAR', 'EMPENHO_ANULADO', 'AUTORIZADA', 'CANCELADA\\n'.",
"_____no_output_____"
],
[
"### 2.22 - Removendo colunas desnecessárias\n\nAs seguintes colunas serão dropadas\n\n- <span style=\"color:red\"><b>numero</b></span>: Trata-se apenas do ID da requisição, não é necessário\n\n- <span style=\"color:red\"><b>numero_contrato</b></span>: Informação desnecessária para a análise\n\n- <span style=\"color:red\"><b>natureza_despesa</b></span>: Possui o mesmo valor em todas as linhas",
"_____no_output_____"
]
],
[
[
"material_dropped = ['numero' ,'natureza_despesa', 'numero_contrato']\nmaterial_request_data = remove_cols(material_request_data, material_dropped)\n\nprint(material_request_data.head())",
" ano id_unidade_requisitante nome_unidade_requisitante \\\n0 2018 160 DEPARTAMENTO DE ECONOMIA - DEPEC \n1 2018 2640 SETOR DE COMPRAS E MANUTENÇÃO - CCSA \n2 2018 1481 PRÓ-REITORIA DE PESQUISA \n3 2018 6799 FNDE - PRONATEC - EAJ \n4 2018 4894 DEPARTAMENTO DE GEOFÍSICA \n\n id_unidade_custo nome_unidade_custo data_envio \\\n0 434 PROGRAMA DE PÓS-GRADUAÇÃO EM ECONOMIA 19/09/2018 \n1 2640 SETOR DE COMPRAS E MANUTENÇÃO - CCSA 18/09/2018 \n2 1481 PRÓ-REITORIA DE PESQUISA 18/09/2018 \n3 6799 FNDE - PRONATEC - EAJ 18/09/2018 \n4 4894 DEPARTAMENTO DE GEOFÍSICA 18/09/2018 \n\n contratado valor \\\n0 PRAIAMAR EMPREENDIMENTOS TURISTICOS LTDA 290.0 \n1 INSTRUCON COMÉRCIO E SERVIÇOS LTDA - ME 4433.0 \n2 PRAIAMAR EMPREENDIMENTOS TURISTICOS LTDA 290.0 \n3 PROTÁSIO LOCAÇÃO E TURISMO LTDA 7013.3 \n4 INSTRUCON COMÉRCIO E SERVIÇOS LTDA - ME 2316.0 \n\n observacoes status \n0 A professora ADRYANE GORAYEB (Requisição de Ho... ENVIADA \n1 SERVIÇOS DE MANUTENÇÃO PREVENTIVA E CORRETIVA,... ENVIADA \n2 GENAUTO CARVALHO DE FRANÇA FILHO\\r\\nChegada di... ENVIADA \n3 Condução de alunos para a Feira Internacional ... ENVIADA \n4 MANUTENÇÃO CORRETIVA (COM CARGA DE GÁS , SUBST... ENVIADA \n"
]
],
[
[
"### 2.23 - Removendo outliers e valores desnecessários\n\nAqui iremos analisar os dados do nosso dataset e determinar quais podemos remover ou modificar de modo a facilitar a nossa análise.",
"_____no_output_____"
]
],
[
[
"print(material_request_data.status.value_counts())",
"LIQUIDADA 11645\nEM_LIQUIDACAO 5988\nENVIADA 5335\nESTORNADA 2856\nFINALIZADA 1543\nCADASTRADA 695\nA_EMPENHAR 144\nEMPENHO_ANULADO 139\nNEGADA 136\nPENDENTE ATENDIMENTO 45\nPENDENTE AUTORIZAÇÃO 37\nAUTORIZADA 36\nCANCELADA\\n 1\nName: status, dtype: int64\n"
]
],
[
[
"**Observação:**\nVerificando a contagem de valores da coluna *status*, percebemos que grande parte dos valores possíveis tem um número muito pequeno de ocorrências no dataset. Esses valores com poucas ocorrências influenciam pouco na nossa análise, portanto iremos eliminá-los.",
"_____no_output_____"
]
],
[
[
"allowed_status = ['LIQUIDADA', 'EM_LIQUIDACAO', 'ENVIADA', 'ESTORNADA', 'FINALIZADA', 'CADASTRADA']\nmaterial_request_data = material_request_data[material_request_data.status.isin(allowed_status)]\n\nprint(material_request_data.status.value_counts())",
"LIQUIDADA 11645\nEM_LIQUIDACAO 5988\nENVIADA 5335\nESTORNADA 2856\nFINALIZADA 1543\nCADASTRADA 695\nName: status, dtype: int64\n"
]
],
[
[
"### 2.24 - Lidando com valores nulos\n\nAqui nós utilizaremos o método [pandas.DataFrame.info](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.info.html) para verificar quais os valores nulos das colunas de nosso dataset. A partir disso, dependendo da quantidade de colunas com valores nulos e do tipo de dado, nós iremos decidir o que fazer com esses valores.",
"_____no_output_____"
]
],
[
[
"material_request_data.info()\nmaterial_request_data[material_request_data.data_envio.isnull()].head(n=20)",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 28062 entries, 0 to 28599\nData columns (total 10 columns):\nano 28062 non-null int64\nid_unidade_requisitante 28062 non-null int64\nnome_unidade_requisitante 28062 non-null object\nid_unidade_custo 28062 non-null int64\nnome_unidade_custo 28062 non-null object\ndata_envio 20202 non-null object\ncontratado 28062 non-null object\nvalor 28062 non-null float64\nobservacoes 20161 non-null object\nstatus 28062 non-null object\ndtypes: float64(1), int64(3), object(6)\nmemory usage: 2.4+ MB\n"
]
],
[
[
"- **data_envio**: Possui vários valores nulos. Como a maioria deles está bem separado um do outro e o dataset está ordenado por data, podemos preenchê-los usando o valor dessa coluna em linhas anteriores.\n- **observacoes**: Algumas observações também tem valores nulos, iremos simplesmente settar esses para uma string vazia.",
"_____no_output_____"
]
],
[
[
"material_request_data.data_envio = material_request_data.data_envio.fillna(method='ffill')\nmaterial_request_data.observacoes = material_request_data.observacoes.fillna('')\n\nmaterial_request_data.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 28062 entries, 0 to 28599\nData columns (total 10 columns):\nano 28062 non-null int64\nid_unidade_requisitante 28062 non-null int64\nnome_unidade_requisitante 28062 non-null object\nid_unidade_custo 28062 non-null int64\nnome_unidade_custo 28062 non-null object\ndata_envio 28062 non-null object\ncontratado 28062 non-null object\nvalor 28062 non-null float64\nobservacoes 28062 non-null object\nstatus 28062 non-null object\ndtypes: float64(1), int64(3), object(6)\nmemory usage: 2.4+ MB\n"
]
],
[
[
"## 2.3 - Empenhos\n\nDataset contendo a relação de todos os empenhos efetuados pela UFRN desde 2001.\n\nO empenho da despesa importa em deduzir do saldo de determinada dotação orçamentária a parcela necessária à execução das atividades do órgão. É a forma de comprometimento de recursos orçamentários. Nenhuma despesa poderá ser realizada sem prévio empenho (art. 60 da Lei n° 4.320/64), sendo realizado após autorização do Ordenador de Despesa em cada Unidade Gestora Executora.",
"_____no_output_____"
]
],
[
[
"empenhos_data = data['empenhos']\n\nprint(empenhos_data.head())\nprint(empenhos_data.data.value_counts())",
" cod_empenho ano modalidade id_unidade_gestora \\\n0 8000116 2018 ordinário 440 \n1 803463 2018 estimativo 605 \n2 803457 2018 ordinário 605 \n3 803449 2018 ordinário 605 \n4 803448 2018 ordinário 605 \n\n nome_unidade_gestora data \\\n0 CENTRO DE BIOCIÊNCIAS 29/08/2018 \n1 UNIVERSIDADE FEDERAL DO RIO GRANDE DO NORTE 19/09/2018 \n2 UNIVERSIDADE FEDERAL DO RIO GRANDE DO NORTE 19/09/2018 \n3 UNIVERSIDADE FEDERAL DO RIO GRANDE DO NORTE 19/09/2018 \n4 UNIVERSIDADE FEDERAL DO RIO GRANDE DO NORTE 19/09/2018 \n\n programa_trabalho_resumido \\\n0 FUNCIONAMENTO DE INSTITUIÇÕES FEDERAIS DE ENSI... \n1 FUNCIONAMENTO DE INSTITUIÇÕES FEDERAIS DE EDUC... \n2 CAPES - APOIO A CAPACITAÇÃO E FORMAÇÃO INICIAL... \n3 CAPES - APOIO A CAPACITAÇÃO E FORMAÇÃO INICIAL... \n4 CAPES - APOIO A CAPACITAÇÃO E FORMAÇÃO INICIAL... \n\n fonte_recurso \\\n0 RECURSOS ORDINÁRIOS - MANUT. E DESENV. DO ENSINO \n1 RECURSOS ORDINÁRIOS - MANUT. E DESENV. DO ENSINO \n2 CAPES - PROGRAMAS EDUC. A DISTÂNCIA \n3 CAPES - PROGRAMAS EDUC. A DISTÂNCIA \n4 CAPES - PROGRAMAS EDUC. A DISTÂNCIA \n\n plano_interno esfera \\\n0 UFRN - GESTAO DA UNIDADE/ FUNCIONAMENTO DOS CU... FISCAL \n1 UFRN-GESTAO DA UNIDADE - ESCOLAS TECNICAS / FU... FISCAL \n2 CAPES - UAB TRADICIONAL FISCAL \n3 CAPES - UAB TRADICIONAL FISCAL \n4 CAPES - UAB TRADICIONAL FISCAL \n\n ... valor_empenho \\\n0 ... 140.0 \n1 ... 200.0 \n2 ... 5090.0 \n3 ... 29900.0 \n4 ... 1340.0 \n\n valor_reforcado valor_anulado valor_cancelado saldo_empenho \\\n0 NaN NaN NaN 140.0 \n1 NaN NaN NaN 170.7 \n2 NaN NaN NaN 5090.0 \n3 NaN NaN NaN 29900.0 \n4 NaN NaN NaN 1340.0 \n\n processo documento_associado licitacao convenio \\\n0 23077.52608/2017-88 NaN PR 92/2017 - UFRN NaN \n1 NaN NaN IN 8/2017 - UFRN NaN \n2 23077.56906/2016-66 NaN PR 72/2016 - UFRN NaN \n3 23077.56906/2016-66 NaN PR 72/2016 - UFRN NaN \n4 23077.56906/2016-66 NaN PR 72/2016 - UFRN NaN \n\n observacoes \n0 REQ.MAT.25342/2018\\r\\nPREGÃO 92/2017 - UFRN\\r\\n \n1 NaN \n2 MATERIAL DE EXPEDIENTE. \n3 MATERIAL DE EXPEDIENTE. \n4 MATERIAL DE EXPEDIENTE. \n\n[5 rows x 22 columns]\n"
]
],
[
[
"### 2.31 - Descrevendo as colunas e valores\n\n- <span style=\"color:red\"><b>cod_empenho</b></span>: ID do empenho;\n\n- **ano**: Ano em que foi solicitado o empenho;\n\n- **modalidade**: O empenho da despesa pode assumir três tipos diferentes: \n - a) Ordinário – a despesa com valor exato deve ser liquidada e paga de uma única vez;\n - b) Estimativo – O valor total da despesa é estimado, podendo ser liquidado e pago em parcelas mensais;\n - c) Global – a despesa total é conhecida e seu pagamento é parcelado, de acordo com cronograma de execução. \n\n- **id_unidade_getora**: ID da unidade orçamentária ou administrativa investida de poder para gerir créditos orçamentários e/ou recursos financeiros;\n\n- **nome_unidade_gestora**: Nome da unidade orçamentária ou administrativa investida de poder para gerir créditos orçamentários e/ou recursos financeiros;\n\n- **data**: Data em que foi feito o empenho;\n\n- **programa_trabalho_resumido**: Resumo do programa/trabalho para qual o empenho será destinado;\n\n- **fonte_recurso**: De onde vem os recursos usados no empenho;\n\n- **plano_interno**: Plano associado ao orçamento de um órgão;\n\n- **esfera**: Pode assumir os seguintes valores: 'FISCAL', 'SEGURIDADE', 'INVESTIMENTO', 'CUSTEIO';\n\n- **natureza_despesa**: Para que tipo de obra foi feito o empenho. Podemos verificar a despesa para desenvolvimento de software, entre os valores dessas colunas temos: 'MAT. CONSUMO', 'SERV. PESSOA JURÍDICA', 'EQUIP. MATERIAL PERMANENTE', 'OBRAS E INSTALAÇÕES', 'PASSAGENS', 'SERVIÇOS DE TECNOLOGIA DA INFORMAÇÃO E COMUNICAÇÃO', 'DESENVOLVIMENTO DE SOFTWARE', 'DIV.EXERCÍCIOS ANTERIORES', 'SERV. PESSOA FÍSICA' 'LOC. MÃO-DE-OBRA', 'SERVIÇOS / UG-GESTÃO' etc.\n\n- **creador**: O beneficiário do empenho;\n\n- **valor_empenho**: Valor total do empenho;\n\n- **valor_reforcado**: O Empenho poderá ser reforçado quando o valor empenhado for insuficiente para atender à despesa a ser realizada, e caso o valor do empenho exceda o montante da despesa realizada, o empenho deverá ser anulado parcialmente. Será anulado totalmente quando o objeto do contrato não tiver sido cumprido, ou ainda, no caso de ter sido emitido incorretamente. Portanto este se trata de um valor adicional ao valor inicial;\n\n- **valor_cancelado**: Valor do empenho que foi cancelado em relação ao total;\n\n- **valor_anulado**: Semelhante ao valor cancelado, porém deve anular a totalidade de valor_empenho ou valor_reforcado.\n\n- **saldo_empenho**: Valor final do empenho\n\n- <span style=\"color:red\"><b>processo</b></span>: Número do processo do empenho DROPAR\n\n- <span style=\"color:red\"><b>documento_associado</b></span>: Documento associado ao processo DROPAR\n\n- <span style=\"color:red\"><b>licitacao</b></span>: DROPAR\n\n- <span style=\"color:red\"><b>convenio</b></span>: DROPAR (?) talvez JOIN com outro dataset \n\n- <span style=\"color:red\"><b>observacoes</b></span>: DROPAR",
"_____no_output_____"
],
[
"### 2.32 - Removendo colunas desnecessárias\n\nIremos remover as seguintes colunas:\n\n- <span style=\"color:red\"><b>cod_empenho</b></span>: Trata-se apenas do ID do empenho, não é necessário\n- <span style=\"color:red\"><b>processo</b></span>: Não adiciona informação relevante ao estudo\n- <span style=\"color:red\"><b>documento_associado</b></span>: Não adiciona informação relevante ao estudo\n- <span style=\"color:red\"><b>licitacao</b></span>: Não adiciona informação relevante ao estudo\n- <span style=\"color:red\"><b>convenio</b></span>: Não adiciona informação relevante ao estudo\n- <span style=\"color:red\"><b>observacoes</b></span>: Não adiciona informação relevante ao estudo\n\n\nPodemos observar também diversas colunas com valores nulos ou repetidos, que serão investigadas mais a fundo em uma seção futura.",
"_____no_output_____"
]
],
[
[
"empenhos_dropped = ['cod_empenho', 'processo', 'documento_associado', 'licitacao', 'convenio', 'observacoes']\nempenhos_data = remove_cols(empenhos_data, empenhos_dropped)\n\nprint(empenhos_data.head())",
" ano modalidade id_unidade_gestora \\\n4809 2017 global 605 \n4810 2017 global 605 \n4811 2017 ordinário 605 \n4812 2017 ordinário 605 \n4813 2017 ordinário 605 \n\n nome_unidade_gestora data \\\n4809 UNIVERSIDADE FEDERAL DO RIO GRANDE DO NORTE 29/12/2017 \n4810 UNIVERSIDADE FEDERAL DO RIO GRANDE DO NORTE 29/12/2017 \n4811 UNIVERSIDADE FEDERAL DO RIO GRANDE DO NORTE 29/12/2017 \n4812 UNIVERSIDADE FEDERAL DO RIO GRANDE DO NORTE 29/12/2017 \n4813 UNIVERSIDADE FEDERAL DO RIO GRANDE DO NORTE 29/12/2017 \n\n programa_trabalho_resumido fonte_recurso \\\n4809 MTPS - FOMENTO E FORTALECIMENTO DA ECONOMIA NA... TESOURO - ORDINÁRIO \n4810 MTPS - FOMENTO E FORTALECIMENTO DA ECONOMIA NA... TESOURO - ORDINÁRIO \n4811 REESTRUTURA E EXPANSÃO DAS IFES - CONSOLIDAÇÃO TESOURO - EDUCAÇÃO \n4812 REESTRUTURA E EXPANSÃO DAS IFES - CONSOLIDAÇÃO TESOURO - EDUCAÇÃO \n4813 REESTRUTURA E EXPANSÃO DAS IFES - CONSOLIDAÇÃO TESOURO - EDUCAÇÃO \n\n plano_interno esfera \\\n4809 MT - TED SENAES FISCAL \n4810 MT - TED SENAES FISCAL \n4811 UFRN-GESTAO DA UNIDADE :PDE - RESTRUTURACAO E ... FISCAL \n4812 UFRN-GESTAO DA UNIDADE :PDE - RESTRUTURACAO E ... FISCAL \n4813 UFRN-GESTAO DA UNIDADE :PDE - RESTRUTURACAO E ... FISCAL \n\n natureza_despesa \\\n4809 SERV. PESSOA JURÍDICA \n4810 SERV. PESSOA JURÍDICA \n4811 EQUIP. MATERIAL PERMANENTE \n4812 EQUIP. MATERIAL PERMANENTE \n4813 EQUIP. MATERIAL PERMANENTE \n\n credor valor_empenho \\\n4809 FUNDAÇÃO NORTE-RIO-GRANDENSE DE PESQUISA E CUL... 22727.27 \n4810 FUNDAÇÃO NORTE-RIO-GRANDENSE DE PESQUISA E CUL... 177272.73 \n4811 NADJA MARINA PIRES - EPP 2699.00 \n4812 CCK COMERCIAL EIRELI 322.88 \n4813 DIGISERVI TRADING LTDA - ME 540.47 \n\n valor_reforcado valor_anulado valor_cancelado saldo_empenho \n4809 NaN NaN NaN 8264.48 \n4810 NaN NaN NaN 0.00 \n4811 NaN NaN NaN 2699.00 \n4812 NaN NaN NaN 322.88 \n4813 NaN NaN NaN 540.47 \n"
]
],
[
[
"### 2.33 - Removendo outliers e valores desnecessários\n\nO dataset de empenhos nos dá valores desde 2001 até 2018, porém estamos trabalhando com dados de 2008 a 2017, logo podemos remover todas as linhas cuja coluna **ano** tem valor menor que 2008 e maior que 2017.",
"_____no_output_____"
]
],
[
[
"# Defining a vector with the years we'll analyse\nyears = [2008, 2009, 2010, 2011, 2012, 2013, 2014, 2015, 2016, 2017]\n\nempenhos_data = empenhos_data[empenhos_data.ano.isin(years)]",
"_____no_output_____"
]
],
[
[
"### 2.34 - Lidando com valores nulos\n\nAqui nós utilizaremos o método [pandas.DataFrame.info](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.info.html) para verificar quais os valores nulos das colunas de nosso dataset. A partir disso, dependendo da quantidade de colunas com valores nulos e do tipo de dado, nós iremos decidir o que fazer com esses valores.",
"_____no_output_____"
]
],
[
[
"empenhos_data.info()\n\nempenhos_data[empenhos_data.valor_anulado.notnull()].head()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 158860 entries, 4809 to 163668\nData columns (total 16 columns):\nano 158860 non-null int64\nmodalidade 158860 non-null object\nid_unidade_gestora 158860 non-null int64\nnome_unidade_gestora 158860 non-null object\ndata 158860 non-null object\nprograma_trabalho_resumido 158860 non-null object\nfonte_recurso 158860 non-null object\nplano_interno 158860 non-null object\nesfera 158860 non-null object\nnatureza_despesa 158860 non-null object\ncredor 158860 non-null object\nvalor_empenho 158860 non-null float64\nvalor_reforcado 3321 non-null float64\nvalor_anulado 8851 non-null float64\nvalor_cancelado 4620 non-null float64\nsaldo_empenho 158860 non-null float64\ndtypes: float64(5), int64(2), object(9)\nmemory usage: 20.6+ MB\n"
]
],
[
[
"**Observação**:\n\nAs colunas **valor_anulado**, **valor_reforcado** e **valor_cancelado** todas possuem uma quantidade muito pequena de valores não-nulos. Como as colunas **valor_empenho** e **saldo_empenho** possuem todos os valores, nós não precisamos das outras para fazermos nossa análise, logo podemos dropá-las.",
"_____no_output_____"
]
],
[
[
"valores_drop = ['valor_reforcado', 'valor_anulado', 'valor_cancelado']\nempenhos_data = remove_cols(empenhos_data, valores_drop)\n\nempenhos_data.head()",
"_____no_output_____"
]
],
[
[
"# 3 - Visualizando os dados\n\nNessa seção iremos utilizar a biblioteca *matplotlib* para plottar gráficos a fim de visualizar nossos dados.",
"_____no_output_____"
],
[
"## 3.1 - Orçamento da UFRN\n\nEm nossa análise, iremos utilizar os dados do valor total de repasses do governo federal para a UFRN de 2006 a 2018 para comparar investimentos da universidade nesses anos. Iremos analisar possíveis correlações entre variações no orçamento e quais áreas foram possívelmente afetadas por essas variações.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n\nyears = [2008, 2009, 2010, 2011, 2012, 2013, 2014, 2015, 2016, 2017]\nbudget = [62010293, 136021308, 203664331, 172999177, 221801098, 246858171, 228864259, 207579799, 230855480, 186863902]",
"_____no_output_____"
],
[
"# Plottagem do orçamento da UFRN de 2008 a 2017, podemos perceber que caiu em todos os anos desde 2013, exceto por 2016.\nbudget_scaled = [value / 1000000 for value in budget]\n\nplt.rcParams['figure.figsize'] = (11, 7)\n\nplt.plot(years, budget_scaled, 'r')\nplt.scatter(years, budget_scaled, color='green')\nplt.xlabel(\"Ano\")\nplt.ylabel(\"Orçamento (em milhões de reais)\")\nplt.xticks(years)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 3.2 - Requisição de manutenção\n\nEsse dataset não possui valores de custo, portanto iremos analisar apenas a quantidade de requisições por ano, seus *status*, *divisao* e *descricao*.",
"_____no_output_____"
]
],
[
[
"autorized_count_year = []\ndenied_count_year = []\n\nfor year in years:\n status_count = maintenance_data[maintenance_data.ano == year].status.value_counts()\n autorized_count_year.append(status_count['AUTORIZADA'])\n denied_count_year.append(status_count['NEGADA'])",
"_____no_output_____"
],
[
"import datetime\nfrom matplotlib.dates import date2num\n\nbar_width = 0.2\n# Shifts each year by bar_width to make sure bars are drawn some space appart from each other\nyears_shifted_left = [year - bar_width for year in years]\nyears_shifted_right = [year + bar_width for year in years]\n\nax = plt.subplot(111)\nax.bar(years_shifted_left, autorized_count_year, width=bar_width, color='g', align='center')\nax.bar(years, denied_count_year, width=bar_width, color='r', align='center')\n\nlegends = ['Autorizadas', 'Negadas']\n\nplt.legend(legends)\nplt.ylabel(\"Quantidade\")\nplt.xlabel(\"Ano\")\nplt.xticks(years)\n\nplt.title(\"Manutenções autorizadas x negadas de 2008 a 2017\")\n\nplt.show()",
"_____no_output_____"
],
[
"divisao_year_count = []\n# Keeps all unique values for 'divisao' column.\ndivisao_values = maintenance_data.divisao.unique()\n\nfor year in years:\n maintenance_data_year = maintenance_data[maintenance_data.ano == year]\n divisao_year_count.append(maintenance_data_year.divisao.value_counts())\n\n# If a key doesn't exist in the count, we add it.\nfor possible_value in divisao_values:\n for year_count in divisao_year_count:\n if possible_value not in year_count.index:\n year_count[possible_value] = 0",
"_____no_output_____"
],
[
"bar_width = 0.15\n# Shifts each year by bar_width to make sure bars are drawn some space appart from each other\n\nax = plt.subplot(111)\ncolors = ['red', 'green', 'blue', 'orange', 'grey', 'black']\nshifts = [-3, -2, -1, 0, 1, 2]\n\nfor i, divisao in enumerate(divisao_values):\n total_divisao_count = []\n for year_count in divisao_year_count:\n total_divisao_count.append(year_count[divisao])\n \n years_shifted = [year - shifts[i] * bar_width for year in years]\n ax.bar(years_shifted, total_divisao_count, width=bar_width, color=colors[i], align='center')\n\nplt.legend(divisao_values)\nplt.ylabel(\"Quantidade\")\nplt.xlabel(\"Ano\")\nplt.xticks(years)\n\nplt.title(\"Proporção dos tipos de manutenção de 2008 a 2017.\")\n\nplt.show()",
"_____no_output_____"
],
[
"from wordcloud import WordCloud\n\ntext = ''\n\nremove_list = ['de', 'na', 'da', 'para', 'um', 'solicito', 'solicitamos', 'vossa', 'senhoria', 'que', 'encontra', 'se', 'dos',\n 'uma', 'ao', '-se', 'das', 'nos', 'nas', 'não', 'está', 'encontra-se', 'solicita-se', 'procurar', 'gilvan',\n 'em', 'frente']\n\nfor descricao in maintenance_data.descricao:\n word_list = descricao.split()\n descricao = ' '.join([i for i in word_list if i.lower() not in remove_list])\n text += descricao + '\\n'\n\nwordcloud = WordCloud().generate(text)\n\nplt.figure()\nplt.imshow(wordcloud, interpolation=\"bilinear\")\nplt.axis(\"off\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 3.3 - Requisição de material",
"_____no_output_____"
]
],
[
[
"# Considerando que o orçamento começou a diminuir em 2013, ainda tivemos picos de gasto em materiais em 2013 e 2016, porém\n# também tivemos grandes baixas em 2015 e 2017 que são justamente os dois anos que tiveram as maiores baixas de orçamento,\n# indicando que a UFRN pode ter sofrido pelo corte de gastos.\n\nmaterial_spending = []\nfor year in years:\n material_spending.append(material_request_data[material_request_data.ano == year].valor.sum() / 1000000)\n\nplt.plot(years, material_spending, 'r')\nplt.scatter(years, material_spending, color='green')\n\nplt.xlabel(\"Ano\")\nplt.ylabel(\"Gasto com material (em milhões de reais)\")\nplt.xticks(years)\n\nplt.title(\"Valor gasto com material na UFRN de 2008 a 2017.\")\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 3.4 - Empenhos",
"_____no_output_____"
]
],
[
[
"valor_year = []\nsaldo_year = []\n\nfor year in years:\n valor_year.append(empenhos_data[empenhos_data.ano == year].valor_empenho.sum() / 1000000)\n saldo_year.append(empenhos_data[empenhos_data.ano == year].saldo_empenho.sum() / 1000000)\n \nplt.plot(years, valor_year, 'r', label='Valor pedido')\nplt.scatter(years, valor_year, color='blue')\n\nplt.title(\"Valor total pedido pelos empenhos da UFRN de 2006 a 2017.\")\n\nplt.xlabel('Ano')\nplt.ylabel('Valor total (milhões)')\nplt.xticks(years)\n\nplt.show()",
"_____no_output_____"
],
[
"# A plotagem dos valores do saldo não nos dá uma boa visualização, pois o intervalo entre os valores é pequeno demais,\n# o que faz com que a variação em proporção seja grande, mas em valor não.\nplt.plot(years, saldo_year, 'g')\nplt.scatter(years, saldo_year, color='blue')\n\nplt.title(\"Valor total empenhado pela UFRN de 2006 a 2017.\")\n\nplt.xlabel('Ano')\nplt.ylabel('Saldo (milhões)')\nplt.xticks(years)\n\nplt.show()",
"_____no_output_____"
],
[
"# O gráfico de barras nos dá uma visualização melhor. Podemos observar que não há grande variação no valor total dos empenhos\n# anuais da UFRN, mas ainda assim, eles seguem tendência de variação semelhante ao valor dos orçamentos.\nplt.bar(years, saldo_year)\n\nplt.title(\"Saldo autorizado pelos empenhos da UFRN de 2006 a 2017.\")\n\nplt.xlabel(\"Ano\")\nplt.ylabel(\"Gastos (em milhões de reais)\")\nplt.xticks(years)\n\nplt.show()",
"_____no_output_____"
],
[
"bar_width = 0.2\n# Shifts each year by bar_width to make sure bars are drawn some space appart from each other\nyears_shifted_left = [year - bar_width for year in years]\nyears_shifted_right = [year + bar_width for year in years]\n\nax = plt.subplot(111)\nax.bar(years_shifted_left, valor_year, width=bar_width, color='g', align='center')\nax.bar(years_shifted_right, saldo_year, width=bar_width, color='b', align='center')\nax.bar(years, budget_scaled, width=bar_width, color='r', align='center')\n\nlegends = ['Valor solicitado', 'Valor empenhado', 'Orçamento total']\n\nplt.legend(legends)\nplt.ylabel(\"Valor (milhões)\")\nplt.xlabel(\"Ano\")\nplt.xticks(years)\n\nplt.title(\"Valor pedido vs. Valor empenhado vs. Orçamento\")\n\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0f3ba5b9ca8d6647c85722362a0a43c05fbb4ce | 19,298 | ipynb | Jupyter Notebook | docs/tutorials/deconvolution.ipynb | Majoburo/spaxlet | 9eaf52b996dd6f64401e95eedfa50785b6f8cc85 | [
"MIT"
] | null | null | null | docs/tutorials/deconvolution.ipynb | Majoburo/spaxlet | 9eaf52b996dd6f64401e95eedfa50785b6f8cc85 | [
"MIT"
] | null | null | null | docs/tutorials/deconvolution.ipynb | Majoburo/spaxlet | 9eaf52b996dd6f64401e95eedfa50785b6f8cc85 | [
"MIT"
] | null | null | null | 44.983683 | 896 | 0.620686 | [
[
[
"# Deconvolution Tutorial\n\n## Introduction\n\nThere are several problems with the standard initialization performed in the [Quickstart Guide](../0-quickstart.ipynb):\n\n1. The models exist in a frame with a narrow model PSF while the the observed scene will have a much wider PSF. So the initial models will be spread out over a larger region, which causes more blending and an increased number of iterations for convergence.\n\n1. The initial morphologies for `ExtendedSource`s and `MultibandSource`s are determined using a combined \"detection coadd,\" which weights each observed image with the SED at the center of each source. Due to different seeing in each band, this results in artificial color gradients in the detection coadd that produce a less accurate initial model.\n\nOne way to solve these problems is to deconvolve the observations into the model frame where the PSF is the same in each band, resulting in more accurate initial morphologies and colors. This is not a trivial task, as deconvolution of a noisy image is an ill-defined operation and numerical divergences dominate the matching kernel when matching a wider PSF to a narrower PSF in Fourier space.\n\nTo avoid the numerical instability of deconvolution kernels created in k-space we instead use scarlet itself to model the kernel and deconvolve the image. There is a computational cost to this procedure and creating the deconvolution kernel for use with a single blend is not advisable, as the cost to generate it is greater than the time saved. However, there are some situations where the following procedure is quite useful, including deblending a large number of blends from survey data where the PSF is well-behaved. For example, we have experimented with HSC data and found that if we calculate the deconvolution kernel at the center of a 4k$\\times$4k patch, we can use the result to deconvolve _all_ of the blends from the same coadd. This is possible because the deconvolution doesn't have to be exact, we just require it to be better for _initialization_ than the observed images.",
"_____no_output_____"
]
],
[
[
"# Import Packages and setup\nfrom functools import partial\n\nimport numpy as np\nimport scarlet\nimport scarlet.display as display\n\n%matplotlib inline\nimport matplotlib\nimport matplotlib.pyplot as plt\n# use a good colormap and don't interpolate the pixels\nmatplotlib.rc('image', cmap='inferno', interpolation='none', origin='lower')",
"_____no_output_____"
]
],
[
[
"## Load and Display Data\n\nWe load the same example data set used in the quickstart guide.",
"_____no_output_____"
]
],
[
[
"# Load the sample images\ndata = np.load(\"../../data/hsc_cosmos_35.npz\")\nimages = data[\"images\"]\nfilters = data[\"filters\"]\ncatalog = data[\"catalog\"]\nweights = 1/data[\"variance\"]\n# Note that unlike in the quickstart guide,\n# we set psfs the data[\"psfs\"] image\n# not a scarlet.PSF object.\npsfs = data[\"psfs\"]",
"_____no_output_____"
]
],
[
[
"## Generate the PSF models\n\nUnlike the [Quickstart Guide](../0-quickstart.ipynb), we cannot use the pixel integrated model PSF because the [error function](https://docs.scipy.org/doc/scipy-0.14.0/reference/generated/scipy.special.erf.html) in scipy used to integrate the gaussian goes to zero too quickly to match an observed PSF. So instead we use a gaussian with a similar $\\sigma=1/\\sqrt{2}$ for our model. We then make this the _observed_ PSF, since this is the seeing that we want to deconvolve our observed images into.",
"_____no_output_____"
]
],
[
[
"py, px = np.array(psfs.shape[1:])//2\nmodel_psf = scarlet.psf.gaussian(py, px, 1/np.sqrt(2), bbox=scarlet.Box(psfs.shape), integrate=False)[0]\nmodel_psf = model_psf/model_psf.sum()\nmodel_psf = np.array([model_psf]*psfs.shape[0])\nmodel_frame = scarlet.Frame(psfs.shape,channels=filters)\n\npsf_observation = scarlet.PsfObservation(model_psf, channels=filters).match(psfs)",
"_____no_output_____"
]
],
[
[
"## Matching the PSFs\n\n### Algorithm\n\nTo understand how the matching algorithm works it is useful to understand how convolutions are performed in scarlet. We can define the observed PSF $P$ by convolving the model PSF $M$ with the difference kernel $D$, giving us\n\n$P = M * D$,\n\nwhere `*` is the convolution operator. The difference kernel is calculated in k-space using the ratio $\\tilde{P}/\\tilde{D}$, which is well defined as long as $P$ is wider than $M$ in real space. Then the `Observation.render` method is used to convolve the model with $D$ to match it with the observed seeing.\n\nFor deconvolution we require the opposite, namely\n\n$M = P * D$\n\nAs mentioned in the [Introduction](#Introduction) this is numerically unstable because in k-space $\\tilde{D}/\\tilde{P}$ diverges in the wings as $\\tilde{P}$ is narrower than $\\tilde{D}$. Modeling the deconvolution kernel with scarlet is possible because of the commutivity of the convolution operation, where\n\n$M = D * P$.\n\nIn this case we can define $M$ as the observation we seek to match, make $D$ the model we want to fit, and then convolve the model ($D$) with $P$ in each iteration to match the \"data.\" In this way we can fit the deconvolution kernel needed to deconvolve from the observation seeing to the model frame.\n\n## An implementation\n\nChoosing the correct parameters for PSF matching is a bit of a black art in itself, another reason why deconvolution should only be done when deblending large datasets and the payoff is greater than the cost. For 41$\\times$41 pixel HSC PSFs we've found the following initialization script to work well, however the configuration for your observations may differ substantially.\n\nWe introduce the `PSFDiffKernel` class, which acts like a scarlet `Component` used to model the scene, however in this case there is a \"source\" for each band since we want out deconvolution kernels to be mono-chromatic.",
"_____no_output_____"
]
],
[
[
"# Parameters used to initial and configure the fit.\nmax_iter = 300\ne_rel = 1e-5\nmorph_step = 1e-2\n# We should be able to improve our initial guess if we model the\n# width of the observed PSF and calculate an analytic solution\n# for the deconvolution kernel, however for now just using the\n# observed PSF works well.\ninit_guess = psfs.copy()\n\n\npsf_kernels = [\n scarlet.PSFDiffKernel(model_frame, init_guess, band, morph_step)\n for band in range(len(filters))\n]\n\npsf_blend = scarlet.Blend(psf_kernels, psf_observation)\n%time psf_blend.fit(max_iter, e_rel=e_rel)\nplt.plot(psf_blend.loss, \".-\")\nplt.title(\"$\\Delta$loss: {:.3e}, e_rel:{:.3e}\".format(psf_blend.loss[-2]-psf_blend.loss[-1], (psf_blend.loss[-2]-psf_blend.loss[-1])/np.abs(psf_blend.loss[-1])))\nplt.show()\n\nfor band, src in enumerate(psf_blend.sources):\n residual = psfs[band]-psf_observation.render(psf_blend.get_model())[band]\n print(\"{}: chi^2={:.3f}, max(abs)={:.3f}\".format(filters[band], np.sum(residual**2), np.max(np.abs(residual))))\n fig, ax = plt.subplots(1, 2, figsize=(7, 3))\n ax[0].imshow(src.get_model()[band], cmap=\"Greys_r\")\n ax[0].set_title(\"{} band kernel\".format(filters[band]))\n vmax = np.max(np.abs(residual))\n im = ax[1].imshow(residual, vmin=-vmax, vmax=vmax, cmap=\"seismic\")\n ax[1].set_title(\"residual\")\n plt.colorbar(im, ax=ax[1])\n plt.show()",
"_____no_output_____"
]
],
[
[
"The residual is created by convolving the observed PSF with the deconvolution kernel and comparing it to the model PSF. We see that the kernel isn't perfect and that it tends to overshoot the center of the model PSF, but the result is good enough to improve our initialization. One thing that we've noticed is that if we set our relative error too low then the ringing in the wings of bright objects is too large while running for too long makes the images crisper at the cost of amplifying the noise to the point where it isn't useful for faint (and even moderately faint) sources.\n\nWe now create the frame for our model, using an analytic PSF, and an observation for the deconvolved image. This is a `DeconvolvedObservation` class, which sets the deconvolution kernel.",
"_____no_output_____"
]
],
[
[
"# This is the frame for our model\nmodel_psf = scarlet.PSF(partial(scarlet.psf.gaussian, sigma=1/np.sqrt(2)), shape=(None, 11, 11))\nmodel_frame = scarlet.Frame(\n images.shape,\n psfs=model_psf,\n channels=filters)\n\n# This object will perform the deconvolution\ndeconvolved = scarlet.DeconvolvedObservation(\n images,\n psfs=model_psf,\n weights=weights,\n channels=filters).match(model_frame, psf_blend.get_model())\n\n# These are the observations that we want to model\nobservation = scarlet.Observation(\n images,\n psfs=scarlet.PSF(psfs),\n weights=weights,\n channels=filters).match(model_frame)",
"_____no_output_____"
]
],
[
[
"Let's take a look at the result:",
"_____no_output_____"
]
],
[
[
"model = deconvolved.images\n\nfig, ax = plt.subplots(1, 2, figsize=(15,7))\n\nnorm = display.AsinhMapping(minimum=np.min(images), stretch=np.max(images)*0.055, Q=10)\nrgb = display.img_to_rgb(images, norm=norm)\nax[0].imshow(rgb)\nax[0].set_title(\"Observed\")\nfor center in catalog:\n ax[0].plot(center[1], center[0], \"wx\")\n\nnorm = display.AsinhMapping(minimum=np.min(model), stretch=np.max(model)*0.055, Q=10)\nrgb = display.img_to_rgb(model, norm=norm)\nax[1].imshow(rgb)\nax[1].set_title(\"Deconvolved\")\nfor center in catalog:\n ax[1].plot(center[1], center[0], \"wx\")\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"In the case the result isn't great due to the bright star at the center. We could try to fit the model a bit better to supress the ringing but it turns out this is usually unnecessary and not worth the extra computation time.\n\nTo see how this is useful lets take a look at the detection coadds for the brightest 3 sources with and without deconvolution. These detection coadds are built internally for all extended and multiband sources, but it's a useful exercise to build them separately just to take a look at them. The red x's in the plots below mark the location of the source whose SED was used to make that particular detection coadd:",
"_____no_output_____"
]
],
[
[
"# We just define a rough estimate of the background RMS needed\n# for `build_detection_coadd`.\nbg_rms=np.zeros((len(images),))\nbg_rms[:] = 1e-3\nfor center in catalog[:4]:\n center = (center[1], center[0])\n figure, ax = plt.subplots(1, 2, figsize=(10, 5))\n # Build the deconvolved coadd\n sed = scarlet.source.get_psf_sed(center, deconvolved, model_frame)\n detect, bg_cutoff = scarlet.source.build_detection_coadd(sed, bg_rms, deconvolved)\n # display\n ax[1].imshow(np.log10(detect), cmap=\"Greys_r\")\n ax[1].plot(center[1], center[0], \"rx\")\n ax[1].set_title(\"deconvolved detection coadd\")\n # Build the coadd without deconvolution\n sed = scarlet.source.get_psf_sed(center, observation, model_frame)\n detect, bg_cutoff = scarlet.source.build_detection_coadd(sed, bg_rms, observation)\n #display\n ax[0].imshow(np.log10(detect), cmap=\"Greys_r\")\n ax[0].plot(center[1], center[0], \"rx\")\n ax[0].set_title(\"detection coadd\")\n plt.show()",
"_____no_output_____"
]
],
[
[
"We see that the ringing in the PSF doesn't really matter, as it's at the same amplitude as the noise and our initial requirement of monotonicity will trim the model to the inner region that doesn't ring, achieving our goal of making the initial models compact and allowing them to grow if necessary. So next we'll initialize our sources using both the deconvolved and original observations and compare them:",
"_____no_output_____"
]
],
[
[
"# Build the sources without deconvolution\nsources = []\nfor k,src in enumerate(catalog):\n if k == 1:\n new_source = scarlet.MultiComponentSource(model_frame, (src['y'], src['x']), observation)\n else:\n new_source = scarlet.ExtendedSource(model_frame, (src['y'], src['x']), observation)\n sources.append(new_source)\n\n# Build the convolved sources\ndeconvolved_sources = []\nfor k,src in enumerate(catalog):\n if k == 1:\n new_source = scarlet.MultiComponentSource(model_frame, (src['y'], src['x']), deconvolved)\n else:\n new_source = scarlet.ExtendedSource(model_frame, (src['y'], src['x']), deconvolved)\n deconvolved_sources.append(new_source)",
"_____no_output_____"
],
[
"norm = display.AsinhMapping(minimum=np.min(images), stretch=np.max(images)*0.055, Q=10)\ndisplay.show_sources(sources[:4],\n norm=norm,\n observation=observation,\n show_rendered=True,\n show_observed=True)\nplt.show()\n\ndisplay.show_sources(deconvolved_sources[:3],\n norm=norm,\n observation=observation,\n show_rendered=True,\n show_observed=True)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Notice that the deconvovled initial models use much smaller boxes while still capturing all of the features in the true observations. The better initial guess and smaller boxes will make it much faster to deblend:",
"_____no_output_____"
]
],
[
[
"# Fit the non-deconvolved blend\nblend = scarlet.Blend(sources, observation)\n%time blend.fit(200)\nprint(\"scarlet ran for {0} iterations to logL = {1}\".format(len(blend.loss), -blend.loss[-1]))\nplt.plot(-np.array(blend.loss))\nplt.title(\"Regular initialization\")\nplt.xlabel('Iteration')\nplt.ylabel('log-Likelihood')\nplt.show()",
"_____no_output_____"
],
[
"# Fit the deconvolved blend\ndeconvolved_blend = scarlet.Blend(deconvolved_sources, observation)\n%time deconvolved_blend.fit(200)\nprint(\"scarlet ran for {0} iterations to logL = {1}\".format(len(deconvolved_blend.loss), -deconvolved_blend.loss[-1]))\nplt.plot(-np.array(deconvolved_blend.loss))\nplt.title(\"Deconvolved initialization\")\nplt.xlabel('Iteration')\nplt.ylabel('log-Likelihood')\nplt.show()",
"_____no_output_____"
]
],
[
[
"So we see that using the deconvolved images for initialization cut our runtime in half for this particular blend (this difference might not be as pronounced in the notebook environment because the default initialization is executed first, heating up the processors before the second blend is run). Looking at the residuals we see that the final models are comparable, so when the same kernel can be used on multiple blends this method proves to be quite useful.",
"_____no_output_____"
]
],
[
[
"norm = display.AsinhMapping(minimum=np.min(images), stretch=np.max(images)*0.055, Q=10)\n# Display the convolved model\nscarlet.display.show_scene(blend.sources,\n norm=norm,\n observation=observation,\n show_rendered=True,\n show_observed=True,\n show_residual=True)\nplt.show()\n# Display the deconvolved model\nscarlet.display.show_scene(deconvolved_blend.sources,\n norm=norm,\n observation=observation,\n show_rendered=True,\n show_observed=True,\n show_residual=True)\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f3cc32bd868dfbc40efaa1d6cccf757169e796 | 13,266 | ipynb | Jupyter Notebook | Sessions/Session13/Day2/02-Fast-GPs.ipynb | Messicka/LSSTC-DSFP-Sessions | f53c28e703a3cc3b07b3d1c5e3c07fc2cd577fd6 | [
"MIT"
] | null | null | null | Sessions/Session13/Day2/02-Fast-GPs.ipynb | Messicka/LSSTC-DSFP-Sessions | f53c28e703a3cc3b07b3d1c5e3c07fc2cd577fd6 | [
"MIT"
] | null | null | null | Sessions/Session13/Day2/02-Fast-GPs.ipynb | Messicka/LSSTC-DSFP-Sessions | f53c28e703a3cc3b07b3d1c5e3c07fc2cd577fd6 | [
"MIT"
] | null | null | null | 30.356979 | 758 | 0.56543 | [
[
[
"# Fast GP implementations",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"%config InlineBackend.figure_format = 'retina'",
"_____no_output_____"
],
[
"from matplotlib import rcParams\nrcParams[\"figure.dpi\"] = 100\nrcParams[\"figure.figsize\"] = 12, 4",
"_____no_output_____"
]
],
[
[
"## Benchmarking GP codes\nImplemented the right way, GPs can be super fast! Let's compare the time it takes to evaluate our GP likelihood and the time it takes to evaluate the likelihood computed with the snazzy ``george`` and ``celerite`` packages. We'll learn how to use both along the way. Let's create a large, fake dataset for these tests:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nnp.random.seed(0)\nt = np.linspace(0, 10, 10000)\ny = np.random.randn(10000)\nsigma = np.ones(10000)",
"_____no_output_____"
]
],
[
[
"### Our GP",
"_____no_output_____"
]
],
[
[
"def ExpSquaredCovariance(t, A=1.0, l=1.0, tprime=None):\n \"\"\"\n Return the ``N x M`` exponential squared\n covariance matrix.\n \n \"\"\"\n if tprime is None:\n tprime = t\n TPrime, T = np.meshgrid(tprime, t)\n return A ** 2 * np.exp(-0.5 * (T - TPrime) ** 2 / l ** 2)\n\n\ndef ln_gp_likelihood(t, y, sigma=0, A=1.0, l=1.0):\n \"\"\"\n Return the log of the GP likelihood for a datatset y(t)\n with uncertainties sigma, modeled with a Squared Exponential\n Kernel with amplitude A and lengthscale l.\n \n \"\"\"\n # The covariance and its determinant\n npts = len(t)\n K = ExpSquaredCovariance(t, A=A, l=l) + sigma ** 2 * np.eye(npts)\n \n # The log marginal likelihood\n log_like = -0.5 * np.dot(y.T, np.linalg.solve(K, y))\n log_like -= 0.5 * np.linalg.slogdet(K)[1]\n log_like -= 0.5 * npts * np.log(2 * np.pi)\n \n return log_like",
"_____no_output_____"
]
],
[
[
"Time to evaluate the GP likelihood:",
"_____no_output_____"
]
],
[
[
"%%time\nln_gp_likelihood(t, y, sigma)",
"CPU times: user 1min 41s, sys: 3.82 s, total: 1min 45s\nWall time: 18 s\n"
]
],
[
[
"### george",
"_____no_output_____"
],
[
"Let's time how long it takes to do the same operation using the ``george`` package (``pip install george``).\n\nThe kernel we'll use is\n\n```python\nkernel = amp ** 2 * george.kernels.ExpSquaredKernel(tau ** 2)\n```\n\nwhere ``amp = 1`` and ``tau = 1`` in this case.\n\nTo instantiate a GP using ``george``, simply run\n\n```python\ngp = george.GP(kernel)\n```\n\nThe ``george`` package pre-computes a lot of matrices that are re-used in different operations, so before anything else, we'll ask it to compute the GP model for our timeseries:\n\n```python\ngp.compute(t, sigma)\n```\n\nNote that we've only given it the time array and the uncertainties, so as long as those remain the same, you don't have to re-compute anything. This will save you a lot of time in the long run!\n\nFinally, the log likelihood is given by ``gp.log_likelihood(y)``.\n\nHow do the speeds compare? Did you get the same value of the likelihood?",
"_____no_output_____"
]
],
[
[
"import george",
"_____no_output_____"
],
[
"%%time\nkernel = george.kernels.ExpSquaredKernel(1.0)\ngp = george.GP(kernel)\ngp.compute(t, sigma)",
"CPU times: user 24.2 s, sys: 690 ms, total: 24.9 s\nWall time: 4.32 s\n"
],
[
"%%time\nprint(gp.log_likelihood(y))",
"-14095.32136897017\nCPU times: user 294 ms, sys: 50.5 ms, total: 345 ms\nWall time: 128 ms\n"
]
],
[
[
"``george`` also offers a fancy GP solver called the HODLR solver, which makes some approximations that dramatically speed up the matrix algebra. Let's instantiate the GP object again by passing the keyword ``solver=george.HODLRSolver`` and re-compute the log likelihood. How long did that take? Did we get the same value for the log likelihood?",
"_____no_output_____"
]
],
[
[
"%%time\ngp = george.GP(kernel, solver=george.HODLRSolver)\ngp.compute(t, sigma)",
"CPU times: user 42.8 ms, sys: 41.8 ms, total: 84.7 ms\nWall time: 84.2 ms\n"
],
[
"%%time\ngp.log_likelihood(y)",
"CPU times: user 6.74 ms, sys: 3.29 ms, total: 10 ms\nWall time: 8.94 ms\n"
]
],
[
[
"### celerite",
"_____no_output_____"
],
[
"The ``george`` package is super useful for GP modeling, and I recommend you read over the [docs and examples](https://george.readthedocs.io/en/latest/). It implements several different [kernels](https://george.readthedocs.io/en/latest/user/kernels/) that come in handy in different situations, and it has support for multi-dimensional GPs. But if all you care about are GPs in one dimension (in this case, we're only doing GPs in the time domain, so we're good), then ``celerite`` is what it's all about:\n\n```bash\npip install celerite\n```\n\nCheck out the [docs](https://celerite.readthedocs.io/en/stable/) here, as well as several tutorials. There is also a [paper](https://arxiv.org/abs/1703.09710) that discusses the math behind ``celerite``. The basic idea is that for certain families of kernels, there exist **extremely efficient** methods of factorizing the covariance matrices. Whereas GP fitting typically scales with the number of datapoints $N$ as $N^3$, ``celerite`` is able to do everything in order $N$ (!!!) This is a **huge** advantage, especially for datasets with tens or hundreds of thousands of data points. Using ``george`` or any homebuilt GP model for datasets larger than about ``10,000`` points is simply intractable, but with ``celerite`` you can do it in a breeze.\n\nNext we repeat the timing tests, but this time using ``celerite``. Note that the Exponential Squared Kernel is not available in ``celerite``, because it doesn't have the special form needed to make its factorization fast. Instead, we'll use the ``Matern 3/2`` kernel, which is qualitatively similar and can be approximated quite well in terms of the ``celerite`` basis functions:\n\n```python\nkernel = celerite.terms.Matern32Term(np.log(1), np.log(1))\n```\n\nNote that ``celerite`` accepts the **log** of the amplitude and the **log** of the timescale. Other than this, we can compute the likelihood using the same syntax as ``george``.\n\nHow much faster did it run? Is the value of the likelihood different from what you found above? Why?",
"_____no_output_____"
]
],
[
[
"import celerite\nfrom celerite import terms",
"_____no_output_____"
],
[
"%%time\nkernel = terms.Matern32Term(np.log(1), np.log(1))\ngp = celerite.GP(kernel)\ngp.compute(t, sigma)",
"_____no_output_____"
],
[
"%%time\ngp.log_likelihood(y)",
"_____no_output_____"
]
],
[
[
"<div style=\"background-color: #D6EAF8; border-left: 15px solid #2E86C1;\">\n <h1 style=\"line-height:2.5em; margin-left:1em;\">Exercise (the one and only)</h1>\n</div>\n\nLet's use what we've learned about GPs in a real application: fitting an exoplanet transit model in the presence of correlated noise.\n\nHere is a (fictitious) light curve for a star with a transiting planet: ",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nt, y, yerr = np.loadtxt(\"data/sample_transit.txt\", unpack=True)\nplt.errorbar(t, y, yerr=yerr, fmt=\".k\", capsize=0)\nplt.xlabel(\"time\")\nplt.ylabel(\"relative flux\");",
"_____no_output_____"
]
],
[
[
"There is a transit visible to the eye at $t = 0$, which (say) is when you'd expect the planet to transit if its orbit were perfectly periodic. However, a recent paper claims that the planet shows transit timing variations, which are indicative of a second, perturbing planet in the system, and that a transit at $t = 0$ can be ruled out at 3 $\\sigma$. **Your task is to verify this claim.**\n\nAssume you have no prior information on the planet other than the transit occurs in the observation window, the depth of the transit is somewhere in the range $(0, 1)$, and the transit duration is somewhere between $0.1$ and $1$ day. You don't know the exact process generating the noise, but you are certain that there's correlated noise in the dataset, so you'll have to pick a reasonable kernel and estimate its hyperparameters.\n\n\nFit the transit with a simple inverted Gaussian with three free parameters:\n\n```python\ndef transit_shape(depth, t0, dur):\n return -depth * np.exp(-0.5 * (t - t0) ** 2 / (0.2 * dur) ** 2)\n```\n\n*HINT: I borrowed heavily from [this tutorial](https://celerite.readthedocs.io/en/stable/tutorials/modeling/) in the celerite documentation, so you might want to take a look at it...*",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0f3f1fe9ef6c4465ce6c0389bbfa852f022141d | 136,420 | ipynb | Jupyter Notebook | bootcamps/automl/featuretools_predict-customer-churn/churn/2. Prediction Engineering.ipynb | yobibytes/quant_trading | 6db6815f671431612030b266205a588c135c0856 | [
"Apache-2.0"
] | 2 | 2020-02-11T12:03:24.000Z | 2020-02-11T12:04:02.000Z | bootcamps/automl/featuretools_predict-customer-churn/churn/2. Prediction Engineering.ipynb | yobibytes/quant_trading | 6db6815f671431612030b266205a588c135c0856 | [
"Apache-2.0"
] | 8 | 2020-11-13T18:54:26.000Z | 2022-02-10T02:17:31.000Z | bootcamps/automl/featuretools_predict-customer-churn/churn/2. Prediction Engineering.ipynb | yobibytes/quant_trading | 6db6815f671431612030b266205a588c135c0856 | [
"Apache-2.0"
] | 1 | 2020-02-11T12:04:04.000Z | 2020-02-11T12:04:04.000Z | 49.320318 | 20,572 | 0.580685 | [
[
[
"# Introduction: Prediction Engineering: Labeling Historical Examples\n\nIn this notebook, we will develop a method for labeling customer transactions data for a customer churn prediction problem. The objective of labeling is to create a set of historical examples of what we want to predict based on the business need: in this problem, our goal is to predict customer churn, so we want to create labeled examples of past churn from the data.\n\nThe end outcome of this notebook is a set of labels each with an associated cutoff time in a table called a label times table. These labels with cutoff times can later be used in Featuretools for automated feature engineering. These features in turn will be used to train a predictive model to forecast customer churn, a common need for subscription-based business models, and one for which machine learning is well-suited.\n\nThe process of prediction engineering is shown below:\n\n\n\n## Definition of Churn: Prediction Problems\n\nThe definition of churn is __a customer going without an active membership for a certain number of days.__ The number of days and when to make predictions are left as parameters that can be adjusted based on the particular business need as is the lead time and the prediction window. In this notebook, we'll make labels for two scenarios:\n\n1. Monthly churn\n * Prediction date = first of month\n * Number of days to churn = 31\n * Lead time = 1 month\n * Prediction window = 1 month\n2. Bimonthly churn\n * Prediction date = first and fifteenth of month\n * Number of days to churn = 14\n * Lead time = 2 weeks\n * Prediction window = 2 weeks\n \nThe problem parameters with details filled in for the first situation are shown below:\n\n\n\n### Dataset\n\nThe [data (publicly available)](https://www.kaggle.com/c/kkbox-churn-prediction-challenge/data) consists of customer transactions for [KKBOX](https://www.kkbox.com), the leading music subscription streaming service in Asia.\nFor each customer, we have background information (in `members`), logs of listening behavior (in `logs`), and transactions information (in `trans`). The only data we need for labeling is the _transactions information_.\n\nThe transactions data consists of a number of variables, the most important of which are customer id (`msno`), the date of transaction (`transaction_date`), and the expiration date of the membership (`membership_expire_date`). Using these columns, we can find each churn for each customer and the corresponding date on which it occurred. Let's look at a few typical examples of customer transaction data to illustrate how to find a churn example. For these examples, we will use the first prediction problem.\n\n## Churn Examples\n\n__Example 1:__\n\n```\n(transaction_date, membership_expire_date, is_cancel)\n\n(2017-01-01, 2017-02-28, false)\n\n(2017-02-25, 0217-03-15, false)\n\n(2017-04-31, 3117-05-20, false)\n```\nThis customer is a churn because they go without a membership for over 31 days, from 03-15 to 04-31. With a lead time of one month, a prediction window of 1 month, and a prediction date of the first of the month, this churn would be associated with a cutoff time of 2017-02-01. \n\n__Example 2:__\n```\n(transaction_date, membership_expire_date, is_cancel)\n\n(2017-01-01, 2017-02-28, false)\n\n(2017-02-25, 2017-04-03, false)\n\n(2017-03-15, 2017-03-16, true)\n\n(2017-04-01, 3117-06-31, false)\n```\n\nThis customer is not a churn. Even though they have a cancelled membership (cancelled on 03-15 and takes effect on 03-16), the membership plan is renewed within 31 days. \n\n__Example 3:__\n```\n(transaction_date, membership_expire_date, is_cancel)\n\n(2017-05-31, 2017-06-31, false)\n\n(2017-07-01, 2017-08-01, false)\n\n(2017-08-01, 2017-09-01, false)\n\n(2017-10-15, 2017-11-15, false)\n```\nThis customer is a churn because they go without a membership for over 31 days, from 09-01 to 10-15. The associated cutoff time of this churn in 2017-09-01. \n\nThese three examples illustrate different situations that occur in the data. Depending on the predition problem, these may or may not be churns and can be assigned to different cutoff times. \n\n# Approach\n\nGiven the data above, to find each example of churn, we need to find the difference between one `membership_expire_date` and the next `transaction_date`. If this period is greater than the days selected for a churn, then this is a positive example of churn. For each churn, we can find the exact date on which it occurred by adding the number of days for a churn to the `membership_expire_date` associated with the churn. We create a set of cutoff times using the prediction date parameter and then for each positive label, determine the cutoff time for the churn. As an example, if the churn occurs on 09-15 with a lead time of 1 month and a prediction window of 1 month, then this churn gets the cutoff time 08-01. Cutoff times where the customer was active 1-2 months out (for this problem) will receive a negative label, and, cutoff times where we cannot determine whether the customer was active or was a churn, will not be labeled. \n\nWe can very rapidly label customer transactions by shifting each `transaction_date` back by one and matching it to the previous `membership_expire_date`. We then find the difference in days between these two (`transaction` - `expire`) and if the difference is greater than the number of days established for churn, this is a positive label. Once we have these positive labels, associating them with a cutoff time is straightforward. \n\nIf this is not clear, we'll shortly see how to do it in code which should clear things up! \n\nThe general framework is implemented in two functions:\n\n1. `label_customer(customer_id, transactions, **params)`\n2. `make_label_times(transactions, **params)` \n\nThe first takes a single member and returns a table of cutoff times for the member along with the associated labels. The second goes through all of the customers and applies the `customer_to_label_times` function to each one. The end outcome is a single table consisting of the label times for each customer. Since we already partitioned the data, we can run this function over multiple partitions in parallel to rapidly label all the data.\n\n## Cutoff Times\n\nA critical part of the label times table is the cutoff time associated with each label. This time at which we make a prediction are referred to as _cutoff_ times and they represent when all our data for making features for that particular label must be before. For instance, if our cutoff time is July 1, and we want to make predictions of churn during the month of August, all of our features for this label must be made with data from before July 1. Cutoff times are a critical consideration when feature engineering for time-series problems to prevent data leakage. Later when we go to perform automated feature engineering, Featuretools will automatically filter data based on the cutoff times so we don't have to worry about invalid training data.\n\n### Outcome\n\nOur overall goal is to build two functions that will generate labels for customers. We can then run this function over our partitions in parallel (our data has been partitioned in 1000 segments, each containing a random subset of customers). Once the label dataframes with cutoff times have been created, we can use them for automated feature engineering using Featuretools.\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"### Data Storage \n\nAll of the data is stored and written to AWS S3. The work was completed on AWS EC2 instances which makes retrieving and writing data to S3 extremely fast. The data is publicly readable from the bucket but you'll have to configure AWS with your credentials. \n* For reading, run `aws configure` from the command line and fill in the details\n* For writing with the `s3fs` library, you'll need to provide your credentials as below\n\nThe benefits of using S3 are that if we shut off our machines, we don't have to worry about losing any of the data. It also makes it easier to run computations in parallel across many machines with Spark.\n",
"_____no_output_____"
]
],
[
[
"PARTITION = '100'\nBASE_DIR = 's3://customer-churn-spark/'\nPARTITION_DIR = BASE_DIR + 'p' + PARTITION\n\nmembers = pd.read_csv(f'{PARTITION_DIR}/members.csv', \n parse_dates=['registration_init_time'], infer_datetime_format = True)\ntrans = pd.read_csv(f'{PARTITION_DIR}/transactions.csv',\n parse_dates=['transaction_date', 'membership_expire_date'], infer_datetime_format = True)\nlogs = pd.read_csv(f'{PARTITION_DIR}/logs.csv', parse_dates = ['date'])\n\ntrans.head()",
"_____no_output_____"
]
],
[
[
"The transactions table is all we will need to make labels. \n\nThe next cell is needed for writing data back to S3.",
"_____no_output_____"
]
],
[
[
"import s3fs\n\n# Credentials\nwith open('/data/credentials.txt', 'r') as f:\n info = f.read().strip().split(',')\n key = info[0]\n secret = info[1]\n\nfs = s3fs.S3FileSystem(key=key, secret=secret)",
"_____no_output_____"
]
],
[
[
"# Churn for One Customer\n\nThe function below takes in a single customer's transactions along with a number of parameters that define the prediction problem. \n\n* `prediction_date`: when we want to make predictions\n* `churn_days`: the number of days without a membership required for a churn\n* `lead_time`: how long in advance to predict churn\n* `prediction_window`: the length of time we are considering for a churn . \n\nThe return from `label_customer` is a label_times dataframe for the customer which has cutoff times for the specified `prediction_date` and the label at each prediction time. Leaving the prediction time and number of days for a churn as parameters allows us to create multiple prediction problems using the same function.",
"_____no_output_____"
]
],
[
[
"def label_customer(customer_id, customer_transactions, prediction_date, churn_days, \n lead_time = 1, prediction_window = 1, return_trans = False):\n \"\"\"\n Make label times for a single customer. Returns a dataframe of labels with times, the binary label, \n and the number of days until the next churn.\n \n Params\n --------\n customer_id (str): unique id for the customer\n customer_transactions (dataframe): transactions dataframe for the customer\n prediction_date (str): time at which predictions are made. Either \"MS\" for the first of the month\n or \"SMS\" for the first and fifteenth of each month \n churn_days (int): integer number of days without an active membership required for a churn. A churn is\n defined by exceeding this number of days without an active membership.\n lead_time (int): number of periods in advance to make predictions for. Defaults to 1 (preditions for one offset)\n prediction_window(int): number of periods over which to consider churn. Defaults to 1.\n return_trans (boolean): whether or not to return the transactions for analysis. Defaults to False.\n \n Return\n --------\n label_times (dataframe): a table of customer id, the cutoff times at the specified frequency, the \n label for each cutoff time, the number of days until the next churn for each\n cutoff time, and the date on which the churn itself occurred.\n transactions (dataframe): [optional] dataframe of customer transactions if return_trans = True. Useful\n for making sure that the function performed as expected\n \n \"\"\"\n \n assert(prediction_date in ['MS', 'SMS']), \"Prediction day must be either 'MS' or 'SMS'\"\n assert(customer_transactions['msno'].unique() == [customer_id]), \"Transactions must be for only customer\"\n \n # Don't modify original\n transactions = customer_transactions.copy()\n \n # Make sure to sort chronalogically\n transactions.sort_values(['transaction_date', 'membership_expire_date'], inplace = True)\n \n # Create next transaction date by shifting back one transaction\n transactions['next_transaction_date'] = transactions['transaction_date'].shift(-1)\n \n # Find number of days between membership expiration and next transaction\n transactions['difference_days'] = (transactions['next_transaction_date'] - \n transactions['membership_expire_date']).\\\n dt.total_seconds() / (3600 * 24)\n \n # Determine which transactions are associated with a churn\n transactions['churn'] = transactions['difference_days'] > churn_days\n \n # Find date of each churn\n transactions.loc[transactions['churn'] == True, \n 'churn_date'] = transactions.loc[transactions['churn'] == True, \n 'membership_expire_date'] + pd.Timedelta(churn_days + 1, 'd')\n \n # Range for cutoff times is from first to (last + 1 month) transaction\n first_transaction = transactions['transaction_date'].min()\n last_transaction = transactions['transaction_date'].max()\n start_date = pd.datetime(first_transaction.year, first_transaction.month, 1)\n \n # Handle December\n if last_transaction.month == 12:\n end_date = pd.datetime(last_transaction.year + 1, 1, 1)\n else:\n end_date = pd.datetime(last_transaction.year, last_transaction.month + 1, 1)\n \n # Make label times dataframe with cutoff times corresponding to prediction date\n label_times = pd.DataFrame({'cutoff_time': pd.date_range(start_date, end_date, freq = prediction_date),\n 'msno': customer_id\n })\n \n # Use the lead time and prediction window parameters to establish the prediction window \n # Prediction window is for each cutoff time\n label_times['prediction_window_start'] = label_times['cutoff_time'].shift(-lead_time)\n label_times['prediction_window_end'] = label_times['cutoff_time'].shift(-(lead_time + prediction_window))\n \n previous_churn_date = None\n\n # Iterate through every cutoff time\n for i, row in label_times.iterrows():\n \n # Default values if unknown\n churn_date = pd.NaT\n label = np.nan\n # Find the window start and end\n window_start = row['prediction_window_start']\n window_end = row['prediction_window_end']\n # Determine if there were any churns during the prediction window\n churns = transactions.loc[(transactions['churn_date'] >= window_start) & \n (transactions['churn_date'] < window_end), 'churn_date']\n\n # Positive label if there was a churn during window\n if not churns.empty:\n label = 1\n churn_date = churns.values[0]\n\n # Find number of days until next churn by \n # subsetting to cutoff times before current churn and after previous churns\n if not previous_churn_date:\n before_idx = label_times.loc[(label_times['cutoff_time'] <= churn_date)].index\n else:\n before_idx = label_times.loc[(label_times['cutoff_time'] <= churn_date) & \n (label_times['cutoff_time'] > previous_churn_date)].index\n\n # Calculate days to next churn for cutoff times before current churn\n label_times.loc[before_idx, 'days_to_churn'] = (churn_date - label_times.loc[before_idx, \n 'cutoff_time']).\\\n dt.total_seconds() / (3600 * 24)\n previous_churn_date = churn_date\n # No churns, but need to determine if an active member\n else:\n # Find transactions before the end of the window that were not cancelled\n transactions_before = transactions.loc[(transactions['transaction_date'] < window_end) & \n (transactions['is_cancel'] == False)].copy()\n # If the membership expiration date for this membership is after the window start, the custom has not churned\n if np.any(transactions_before['membership_expire_date'] >= window_start):\n label = 0\n\n # Assign values\n label_times.loc[i, 'label'] = label\n label_times.loc[i, 'churn_date'] = churn_date\n \n # Handle case with no churns\n if not np.any(label_times['label'] == 1):\n label_times['days_to_churn'] = np.nan\n label_times['churn_date'] = pd.NaT\n \n if return_trans:\n return label_times.drop(columns = ['msno']), transactions\n \n return label_times[['msno', 'cutoff_time', 'label', 'days_to_churn', 'churn_date']].copy()",
"_____no_output_____"
]
],
[
[
"Let's take a look at the output of this function for a typical customer. We'll take the use case of making predictions on the first of each month with 31 days required for a churn, a lead time of 1 month, and a prediction window of 1 month.",
"_____no_output_____"
]
],
[
[
"CUSTOMER_ID = trans.iloc[8, 0]\ncustomer_transactions = trans.loc[trans['msno'] == CUSTOMER_ID].copy()\n\nlabel_times, cust_transactions = label_customer(CUSTOMER_ID, customer_transactions, \n prediction_date = 'MS', churn_days = 31, \n lead_time = 1, prediction_window = 1, return_trans = True)\nlabel_times.head(10)",
"_____no_output_____"
]
],
[
[
"To make sure the function worked, we'll want to take a look at the transactions.",
"_____no_output_____"
]
],
[
[
"cust_transactions.iloc[3:10, -7:]",
"_____no_output_____"
]
],
[
[
"We see that the churn occurred on 2016-03-16 as the customer went 98 days between an active membership from 2016-02-14 to 2016-05-22. The actual churn occurs 31 days from when the membership expires. The churn is only associated with one cutoff time, 2016-02-01. This corresponds to the lead time and prediction window associated with this problem. ",
"_____no_output_____"
],
[
"Let's see the function in use for the other prediction problem, making predictions on the first and fifteenth of each month with churn defined as more than 14 days without an active membership. The lead time is set to two weeks (one prediction period) and the prediction window is also set to two weeks. To change the prediction problem, all we need to do is alter the parameters.",
"_____no_output_____"
]
],
[
[
"CUSTOMER_ID = trans.iloc[100, 0]\ncustomer_transactions = trans.loc[trans['msno'] == CUSTOMER_ID].copy()\n\nlabel_times, cust_transactions = label_customer(CUSTOMER_ID, customer_transactions, \n prediction_date = 'SMS', churn_days = 14, \n lead_time = 1, prediction_window = 1, return_trans = True)\nlabel_times.head(12)",
"_____no_output_____"
]
],
[
[
"There are several times when we can't determine if the customer churned or not because of the way the problem has been set up. ",
"_____no_output_____"
]
],
[
[
"cust_transactions.iloc[:10, -7:]",
"_____no_output_____"
]
],
[
[
"Looking at the churn on 2016-03-15, it was assigned to the `cutoff_time` of 2016-03-01 as expected with a lead time of two weeks and a prediction window of two weeks. (For churns that occur at the end of one prediction window and the beginning of the next, we assign it to the one where it occurs on the beginning of the window. This can be quickly changed by altering the logic of the function.)\n\nThe function works as designed, we can pass in different parameters and rapidly make prediction problems. We also have the number of days to the churn which means we could formulate the problem as regression instead of classification. ",
"_____no_output_____"
],
[
"# Churn for All Customers\n\nNext, we take the function which works for one customer and apply it to all customers in a dataset. This requires a loop through the customers by grouping the customer transactions and applying `label_customer` to each customer's transactions. ",
"_____no_output_____"
]
],
[
[
"def make_label_times(transactions, prediction_date, churn_days, \n lead_time = 1, prediction_window = 1,):\n \"\"\"\n Make labels for an entire series of transactions. \n \n Params\n --------\n transactions (dataframe): table of customer transactions\n prediction_date (str): time at which predictions are made. Either \"MS\" for the first of the month\n or \"SMS\" for the first and fifteenth of each month \n churn_days (int): integer number of days without an active membership required for a churn. A churn is\n defined by exceeding this number of days without an active membership.\n lead_time (int): number of periods in advance to make predictions for. Defaults to 1 (preditions for one offset)\n prediction_window(int): number of periods over which to consider churn. Defaults to 1.\n Return\n --------\n label_times (dataframe): a table with customer ids, cutoff times, binary label, regression label, \n and date of churn. This table can then be used for feature engineering.\n \"\"\"\n \n label_times = []\n transactions = transactions.sort_values(['msno', 'transaction_date'])\n \n # Iterate through each customer and find labels\n for customer_id, customer_transactions in transactions.groupby('msno'):\n lt_cust = label_customer(customer_id, customer_transactions,\n prediction_date, churn_days, \n lead_time, prediction_window)\n \n label_times.append(lt_cust)\n \n # Concatenate into a single dataframe\n return pd.concat(label_times)",
"_____no_output_____"
]
],
[
[
"Let's look at examples of using this function for both prediction problems.\n\n## First Prediction Problem\n\nThe defintion of the first prediction problem is as follows:\n\n* Monthly churn\n * Prediction date = first of month\n * Number of days to churn = 31\n * Lead time = 1 month\n * Prediction window = 1 month",
"_____no_output_____"
]
],
[
[
"label_times = make_label_times(trans, prediction_date = 'MS', churn_days = 31,\n lead_time = 1, prediction_window = 1)",
"_____no_output_____"
],
[
"label_times.tail(10)",
"_____no_output_____"
],
[
"label_times.shape",
"_____no_output_____"
],
[
"label_times['label'].value_counts()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n\nplt.style.use('fivethirtyeight')\n\nlabel_times['label'].value_counts().plot.bar(color = 'r');\nplt.xlabel('Label'); plt.ylabel('Count'); plt.title('Label Distribution with Monthly Predictions');",
"_____no_output_____"
]
],
[
[
"This is an imbalanced classification problem. There are far more instances of customers not churning than of customers churning. This is not necessarily an issue as long as we are smart about the choices of metrics we use for modeling. \n\n\n## Second Prediction Problem\n\nTo demonstrate how to quickly change the problem parameters, we can use the labeling function for a different prediction problem. The parameters are defined below:\n\n* Bimonthly churn\n * Prediction date = first and fifteenth of month\n * Number of days to churn = 14\n * Lead time = 2 weeks\n * Prediction window = 2 weeks",
"_____no_output_____"
]
],
[
[
"label_times = make_label_times(trans, prediction_date = 'SMS', churn_days = 14,\n lead_time = 1, prediction_window = 1)\nlabel_times.tail(10)",
"_____no_output_____"
],
[
"label_times.shape",
"_____no_output_____"
],
[
"label_times['label'].value_counts().plot.bar(color = 'r');\nplt.xlabel('Label'); plt.ylabel('Count'); plt.title('Label Distribution with Bimonthly Predictions');",
"_____no_output_____"
],
[
"label_times['label'].isnull().sum()",
"_____no_output_____"
]
],
[
[
"There are quite a few missing labels, which occur when there is no next transaction for the customer (we don't know if the last entry for the customer is a churn or not). We won't be able to use these examples when training a model although we can make predictions for them.",
"_____no_output_____"
],
[
"# Parallelizing Labeling\n\nNow that we have a function that can make a label times table out of customer transactions, we need to label all of the customer transactions in our dataset. We already broke the data into 1000 partitions, so we can parallelize this operation using Spark with PySpark. The basic idea is to write a function that makes the label times for one partition, and then run this in parallel across all the partitions using either multiple cores on a single machine, or a cluster of machines. \n\nThe function below takes in a partition number, reads the transactions data from S3, creates the label times table for both prediction problems, and writes the label times back to S3. We can run this function in parallel over multiple partitions at once since the customers are independent of one another. That is, the labels for one customer do not depend on the data for any other customer. ",
"_____no_output_____"
]
],
[
[
"def partition_to_labels(partition_number, prediction_dates = ['MS', 'SMS'], churn_periods= [31, 14],\n lead_times = [1, 1], prediction_windows = [1, 1]):\n \"\"\"Make labels for all customers in one partition\n Either for one month or twice a month\n \n Params\n --------\n partition (int): number of partition\n label_type (list of str): either 'MS' for monthly labels or\n 'SMS' for bimonthly labels\n churn_periods(list of int): number of days with no active membership to be considered a churn\n lead_times (list of int): lead times in number of periods\n prediction_windows (list of int): prediction windows in number of periods\n \n Returns\n --------\n None: saves the label dataframes with the appropriate name to the partition directory\n \"\"\"\n partition_dir = BASE_DIR + 'p' + str(partition_number)\n \n # Read in data and filter anomalies\n trans = pd.read_csv(f'{partition_dir}/transactions.csv',\n parse_dates=['transaction_date', 'membership_expire_date'], \n infer_datetime_format = True)\n \n # Deal with data inconsistencies\n rev = trans[(trans['membership_expire_date'] < trans['transaction_date']) | \n ((trans['is_cancel'] == 0) & (trans['membership_expire_date'] == trans['transaction_date']))]\n rev_members = rev['msno'].unique()\n \n # Remove data errors\n trans = trans.loc[~trans['msno'].isin(rev_members)]\n\n # Create both sets of lables\n for prediction_date, churn_days, lead_time, prediction_window in zip(prediction_dates, churn_periods, lead_times, prediction_windows):\n \n cutoff_list = []\n \n # Make label times for all customers\n cutoff_list.append(make_label_times(trans, prediction_date = prediction_date, \n churn_days = churn_days, lead_time = lead_time,\n prediction_window = prediction_window))\n # Turn into a dataframe\n cutoff_times = pd.concat(cutoff_list)\n cutoff_times = cutoff_times.drop_duplicates(subset = ['msno', 'cutoff_time'])\n \n # Encode in order to write to s3\n bytes_to_write = cutoff_times.to_csv(None, index = False).encode()\n\n # Write cutoff times to S3\n with fs.open(f'{partition_dir}/{prediction_date}-{churn_days}_labels.csv', 'wb') as f:\n f.write(bytes_to_write)",
"_____no_output_____"
],
[
"partition_to_labels(1, prediction_dates = ['MS'], churn_periods = [31], \n lead_times = [1], prediction_windows = [1])",
"_____no_output_____"
],
[
"label_times = pd.read_csv('s3://customer-churn-spark/p1/MS-31_labels.csv')\nlabel_times.tail(10)",
"_____no_output_____"
],
[
"partition_to_labels(1, prediction_dates = ['SMS'], churn_periods = [14],\n lead_times = [1], prediction_windows = [1])\nlabel_times = pd.read_csv('s3://customer-churn-spark/p1/SMS-14_labels.csv')\nlabel_times.head(10)",
"_____no_output_____"
]
],
[
[
"## Spark for Parallelization\n\nThe below code uses Spark to parallelize the label making. This particular implementation uses a single machine although the same idea can be extended to a cluster of machines.",
"_____no_output_____"
]
],
[
[
"import findspark\nfindspark.init('/usr/local/spark/')\n\nimport pyspark\n\nconf = pyspark.SparkConf()\n\n# Enable logging\nconf.set('spark.eventLog.enabled', True);\nconf.set('spark.eventLog.dir', '/data/churn/tmp/');\n\n# Use all cores on a single machine\nconf.set('spark.num.executors', 1)\nconf.set('spark.executor.memory', '56g')\nconf.set('spark.executor.cores', 15)\n\n# Make sure to specify correct spark master ip\nsc = pyspark.SparkContext(master = 'spark://ip-172-31-23-133.ec2.internal:7077',\n appName = 'labeling', conf = conf)\n\nsc",
"_____no_output_____"
],
[
"from timeit import default_timer as timer\n\n# Parallelize making all labels in Spark\nstart = timer()\nsc.parallelize(list(range(1000)), numSlices=1000).\\\n map(partition_to_labels).collect()\nsc.stop()\nend = timer()",
"_____no_output_____"
]
],
[
[
"While Spark is running, you can navigate to localhost:4040 to see the details of the particular job, or to localhost:8080 to see the overview of the cluster. This is useful for diagnosing the state of a spark operation.",
"_____no_output_____"
]
],
[
[
"print(f'{round(end - start)} seconds elapsed.')",
"40354 seconds elapsed.\n"
],
[
"labels = pd.read_csv(f's3://customer-churn-spark/p980/MS-31_labels.csv')\nlabels.tail(10)",
"_____no_output_____"
],
[
"labels = pd.read_csv(f's3://customer-churn-spark/p980/SMS-14_labels.csv')\nlabels.tail(10)",
"_____no_output_____"
]
],
[
[
"# Conclusions\n\nIn this notebook, we implemented prediction engineering for the customer churn use case. After defining the business need, we translated it into a task that can be solved with machine learning and created a set of label times. We saw how to define functions with parameters so we could solve multiple prediction problems without needing to re-write the entire code. Although we only worked through two problems, there are numerous others that could be solved with the same data and approach.\n\n\nThe label times contain cutoff times for a specific prediction problem along with the associated label. The label times can now be used to make features for each label by filtering the data to before the cutoff time. This ensures that any features made are valid and will automatically be taken care of in Featuretools. \n\nThe general procedure for making labels is:\n\n1. Define the business requirement: predict customers who will churn during a specified period of time\n2. Translate the business requirement into a machine learning problem: given historical customer data, build a model to predict which customers will churn depending on several parameters\n3. Make labels along with cutoff times corresponding to the machine learning problem: develop functions that take in parameters so the same function can be used for multiple prediction problems.\n4. Label all past historical data: parallelize operations by partitioning data into independent subsets\n\nThis approach can be extended to other problems. Although the exact syntax is specific to this use case, the overall approach is designed to be general purpose.\n\n## Next Steps\n\nWith a complete set of label times, we can now make features for each label using the cutoff times to ensure our features are valid. However, instead of the painstaking and error-prone process of making features by hand, we can use automated feature engineering in [Featuretools](https://github.com/Featuretools/featuretools) to automated this process. Featuretools will build hundreds of relevant features using only a few lines of code and will automatically filter the data to ensure that all of our features are valid. The feature engineering pipeline is developed in the `Feature Engineering` notebook.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0f3f3cc62eec1058942867f94259d33a35092ee | 385,503 | ipynb | Jupyter Notebook | machineLearning/applications/anomaly_detection/2 - fraud_detection_with_classical_machine_learning/fraud detection.ipynb | ljyang100/dataScience | ad2b243673c570c18d83ab1a0cd1bb4694c17eac | [
"MIT"
] | 2 | 2020-12-10T02:05:29.000Z | 2021-05-30T15:23:56.000Z | machineLearning/applications/anomaly_detection/2 - fraud_detection_with_classical_machine_learning/fraud detection.ipynb | ljyang100/dataScience | ad2b243673c570c18d83ab1a0cd1bb4694c17eac | [
"MIT"
] | null | null | null | machineLearning/applications/anomaly_detection/2 - fraud_detection_with_classical_machine_learning/fraud detection.ipynb | ljyang100/dataScience | ad2b243673c570c18d83ab1a0cd1bb4694c17eac | [
"MIT"
] | 1 | 2020-04-21T11:18:18.000Z | 2020-04-21T11:18:18.000Z | 177.897093 | 138,648 | 0.883684 | [
[
[
"## Reference\nData Camp course",
"_____no_output_____"
],
[
"## Course Description\n* A typical organization loses an estimated 5% of its yearly revenue to fraud. \n* Apply supervised learning algorithms to detect fraudulent behavior similar to past ones,as well as unsupervised learning methods to discover new types of fraud activities. \n* Deal with highly imbalanced datasets. \n* The course provides a mix of technical and theoretical insights and shows you hands-on how to practically implement fraud detection models. \n* Tips and advise from real-life experience to help you prevent making common mistakes in fraud analytics.\n* Examples of fraud: insurance fraud, credit card fraud, identify theft, money laundering, tax evasion, product warranty, healthcare fraud. ",
"_____no_output_____"
],
[
"## Introduction and preparing your data\n\n* Typical challenges associated with fraud detection.\n* Resample your data in a smart way, to tackle problems with imbalanced data. ",
"_____no_output_____"
],
[
"### Checking the fraud to non-fraud ratio\n* Fraud occurrences are fortunately an extreme minority in these transactions.\n* However, Machine Learning algorithms usually work best when the different classes contained in the dataset are more or less equally present. If there are few cases of fraud, then there's little data to learn how to identify them. This is known as **class imbalance** (or skewed class), and it's one of the main challenges of fraud detection.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndf = pd.read_csv(\"creditcard_sampledata_3.csv\") \n#This is dieferent from the data in the course. But it will be corrected\n#in the following cells.\n\nocc = df['Class'].value_counts() #good for counting categorical data\nprint(occ)\nprint(occ / len(df.index))",
"0 5000\n1 50\nName: Class, dtype: int64\n0 0.990099\n1 0.009901\nName: Class, dtype: float64\n"
]
],
[
[
"### Plotting your data\nVisualize the fraud to non-fraud ratio. ",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport pandas as pd\ndf = pd.read_csv(\"creditcard_sampledata_3.csv\")\n#print(df.columns) #It is not df.colnames. \ndf = df.drop(['Unnamed: 0'],axis = 1)\n# print(df.head())\ny=df['Class'].values\nX=df.drop(['Class'],axis = 1).values\n\ndef plot_data(X, y):\n plt.scatter(X[y == 0, 0], X[y == 0, 1], label=\"Class #0\", alpha=0.5, linewidth=0.15)\n plt.scatter(X[y == 1, 0], X[y == 1, 1], label=\"Class #1\", alpha=0.5, linewidth=0.15, c='r')\n plt.legend()\n return plt.show()\n\n# X, y = prep_data(df) #original code \n\nplot_data(X, y)\nlen(X[y==0,0])\n",
"_____no_output_____"
]
],
[
[
"### Applying SMOTE\n* Re-balance the data using the Synthetic Minority Over-sampling Technique (SMOTE). \n* Unlike ROS, SMOTE does not create exact copies of observations, but creates new, synthetic, samples that are quite similar to the existing observations in the minority class. \n* Visualize the result and compare it to the original data, such that we can see the effect of applying SMOTE very clearly.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport pandas as pd\nfrom imblearn.over_sampling import SMOTE\ndf = pd.read_csv(\"creditcard_sampledata_3.csv\")\n#print(df.columns) #It is not df.colnames. \ndf = df.drop(['Unnamed: 0'],axis = 1)\n# print(df.head())\ny=df['Class'].values\nX=df.drop(['Class'],axis = 1).values\n#my code above\n\nmethod = SMOTE(kind='regular')\nX_resampled, y_resampled = method.fit_sample(X, y)\nplot_data(X_resampled, y_resampled)\nprint(X.shape)\nprint(y.shape)",
"_____no_output_____"
]
],
[
[
"### Compare SMOTE to original data\n* Compare those results of SMOTE to the original data, to get a good feeling for what has actually happened.\n* Have a look at the value counts again of our old and new data, and let's plot the two scatter plots of the data side by side. * Use the function compare_plot() (not defined here), which takes the following arguments: X, y, X_resampled, y_resampled, method=''. The function plots the original data in a scatter plot, along with the resampled side by side.",
"_____no_output_____"
]
],
[
[
"print(pd.value_counts(pd.Series(y)))\n\nprint(pd.value_counts(pd.Series(y_resampled)))\n\ncompare_plot(X, y, X_resampled, y_resampled, method='SMOTE')\n# This fundtion is not defined here. But the result picture is as below\n#The compare_plot should be implemented by the subplot defined on dataframe, or by the subplot way summarized elsewhere. ",
"0 5000\n1 50\ndtype: int64\n1 5000\n0 5000\ndtype: int64\n"
]
],
[
[
"",
"_____no_output_____"
],
[
"### Exploring the traditional way to catch fraud\n* Try finding fraud cases in our credit card dataset the \"old way\". First you'll define threshold values using common statistics, to split fraud and non-fraud. Then, use those thresholds on your features to detect fraud. This is common practice within fraud analytics teams.\n\n* Statistical thresholds are often determined by looking at the mean values of observations. \n* Check whether feature means differ between fraud and non-fraud cases. Then, use that information to create common sense thresholds.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nfrom imblearn.over_sampling import SMOTE\ndf = pd.read_csv(\"creditcard_sampledata_3.csv\")\n#print(df.columns) #It is not df.colnames. \ndf = df.drop(['Unnamed: 0'],axis = 1)\n#print(df.head())\ny=df['Class'].values\nX=df.drop(['Class'],axis = 1).values\n#my code above\n\n# Run a groupby command on our labels and obtain the mean for each feature\ndf.groupby('Class').mean()\n\n# Implement a rule for stating which cases are flagged as fraud\ndf['flag_as_fraud'] = np.where(np.logical_and(df['V1'] < -3, df['V3'] < -5), 1, 0)\n\n# Create a crosstab of flagged fraud cases versus the actual fraud cases\nprint(pd.crosstab(df.Class, df.flag_as_fraud, rownames=['Actual Fraud'], colnames=['Flagged Fraud']))",
"Flagged Fraud 0 1\nActual Fraud \n0 4984 16\n1 28 22\n"
]
],
[
[
"Not bad, with this rule, we detect 22 out of 50 fraud cases, but can't detect the other 28, and get 16 false positives. In the next exercise, we'll see how this measures up to a machine learning model. ",
"_____no_output_____"
],
[
"### Using ML classification to catch fraud\n* Use a simple machine learning model on our credit card data instead.\n* Implement a Logistic Regression model. ",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import classification_report\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)\n\n# Fit a logistic regression model to our data\nmodel = LogisticRegression()\nmodel.fit(X_train, y_train)\n\n# Obtain model predictions\npredicted = model.predict(X_test)\n\n# Print the classifcation report and confusion matrix\nprint('Classification report:\\n', classification_report(y_test, predicted))\nconf_mat = confusion_matrix(y_true=y_test, y_pred=predicted)\nprint('Confusion matrix:\\n', conf_mat)",
"C:\\Users\\ljyan\\Anaconda3\\lib\\site-packages\\sklearn\\linear_model\\logistic.py:433: FutureWarning: Default solver will be changed to 'lbfgs' in 0.22. Specify a solver to silence this warning.\n FutureWarning)\n"
]
],
[
[
"* We are getting much less false positives, so that's an improvement. \n* We're catching a higher percentage of fraud cases, so that is also better than before. ",
"_____no_output_____"
],
[
"### Logistic regression combined with SMOTE",
"_____no_output_____"
]
],
[
[
"# This is the pipeline module we need for this from imblearn\nfrom imblearn.pipeline import Pipeline \n\n# Define which resampling method and which ML model to use in the pipeline\nresampling = SMOTE(kind='borderline2')\nmodel = LogisticRegression()\n\n# Define the pipeline, tell it to combine SMOTE with the Logistic Regression model\npipeline = Pipeline([('SMOTE', resampling), ('Logistic Regression', model)])",
"_____no_output_____"
]
],
[
[
"### Using a pipeline\nTreat the pipeline as if it were a single machine learning model. Our data X and y are already defined, and the pipeline is defined in the previous exercise. ",
"_____no_output_____"
]
],
[
[
"# Split your data X and y, into a training and a test set and fit the pipeline onto the training data\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)\n\n# Fit your pipeline onto your training set and obtain predictions by fitting the model onto the test data \npipeline.fit(X_train, y_train) \npredicted = pipeline.predict(X_test)\n\n# Obtain the results from the classification report and confusion matrix \nprint('Classifcation report:\\n', classification_report(y_test, predicted))\nconf_mat = confusion_matrix(y_true=y_test, y_pred=predicted)\nprint('Confusion matrix:\\n', conf_mat)",
"Classifcation report:\n precision recall f1-score support\n\n 0 1.00 1.00 1.00 1505\n 1 0.67 1.00 0.80 10\n\n micro avg 1.00 1.00 1.00 1515\n macro avg 0.83 1.00 0.90 1515\nweighted avg 1.00 1.00 1.00 1515\n\nConfusion matrix:\n [[1500 5]\n [ 0 10]]\n"
]
],
[
[
"* The SMOTE slightly improves our results. We now manage to find all cases of fraud, but we have a slightly higher number of false positives, albeit only 7 cases.\n* Remember, not in all cases does resampling necessarily lead to better results. **When the fraud cases are very spread and scattered over the data, using SMOTE can introduce a bit of bias.** Nearest neighbors aren't necessarily also fraud cases, so the synthetic samples might 'confuse' the model slightly. \n* In the next chapters, we'll learn how to also adjust our machine learning models to better detect the minority fraud cases.",
"_____no_output_____"
],
[
"## Fraud detection using labelled data\n* Flag fraudulent transactions with supervised learning. \n* Use classifiers, adjust them and compare them to find the most efficient fraud detection model.",
"_____no_output_____"
],
[
"### Natural hit rate\n* Explore how prevalent fraud is in the dataset, to understand what the \"natural accuracy\" is, if we were to predict everything as non-fraud. \n* It's is important to understand which level of \"accuracy\" you need to \"beat\" in order to get a better prediction than by doing nothing. \n* Create a random forest classifier for fraud detection. That will serve as the \"baseline\" model that you're going to try to improve in the upcoming exercises.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport pandas as pd\nfrom imblearn.over_sampling import SMOTE\ndf = pd.read_csv(\"creditcard_sampledata_2.csv\")\n#print(df.columns) #It is not df.colnames. \ndf = df.drop(['Unnamed: 0'],axis = 1)\n# print(df.head())\ny=df['Class'].values\nX=df.drop(['Class'],axis = 1).values\n#extra code above\n\n# Count the total number of observations from the length of y\ntotal_obs = len(y)\n\n# Count the total number of non-fraudulent observations \nnon_fraud = [i for i in y if i == 0]\ncount_non_fraud = non_fraud.count(0)\n\n# Calculate the percentage of non fraud observations in the dataset\npercentage = (float(count_non_fraud)/float(total_obs)) * 100\n\n# Print the percentage: this is our \"natural accuracy\" by doing nothing\nprint(percentage)\n",
"95.8904109589041\n"
]
],
[
[
"This tells us that by doing nothing, we would be correct in 95.9% of the cases. So now you understand, that if we get an accuracy of less than this number, our model does not actually add any value in predicting how many cases are correct. ",
"_____no_output_____"
],
[
"### Random Forest Classifier - part 1",
"_____no_output_____"
]
],
[
[
"print(X.shape)\nprint(y.shape)",
"(7300, 29)\n(7300,)\n"
],
[
"from sklearn.ensemble import RandomForestClassifier\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)\nmodel = RandomForestClassifier(random_state=5) ",
"_____no_output_____"
]
],
[
[
"### Random Forest Classifier - part 2\nSee how our Random Forest model performs without doing anything special to it. ",
"_____no_output_____"
]
],
[
[
"model.fit(X_train, y_train)\npredicted = model.predict(X_test)",
"C:\\Users\\ljyan\\Anaconda3\\lib\\site-packages\\sklearn\\ensemble\\forest.py:246: FutureWarning: The default value of n_estimators will change from 10 in version 0.20 to 100 in 0.22.\n \"10 in version 0.20 to 100 in 0.22.\", FutureWarning)\n"
],
[
"from sklearn.metrics import accuracy_score\n\nmodel.fit(X_train, y_train)\npredicted = model.predict(X_test)\nprint(accuracy_score(y_test, predicted))",
"C:\\Users\\ljyan\\Anaconda3\\lib\\site-packages\\sklearn\\ensemble\\forest.py:246: FutureWarning: The default value of n_estimators will change from 10 in version 0.20 to 100 in 0.22.\n \"10 in version 0.20 to 100 in 0.22.\", FutureWarning)\n"
]
],
[
[
"### Performance metrics for the RF model\n* In the previous exercises you obtained an accuracy score for your random forest model. This time, we know accuracy can be misleading in the case of fraud detection. \n* With highly imbalanced fraud data, the AUROC curve is a more reliable performance metric, used to compare different classifiers. Moreover, the classification report tells you about the precision and recall of your model, whilst the confusion matrix actually shows how many fraud cases you can predict correctly. So let's get these performance metrics.\n* Continue working on the same random forest model from the previous exercise. The model, defined as model = RandomForestClassifier(random_state=5) has been fitted to the training data already, and X_train, y_train, X_test, y_test are available.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report, confusion_matrix, roc_auc_score\n\npredicted = model.predict(X_test)\n\nprobs = model.predict_proba(X_test)\n\n# Print the ROC curve, classification report and confusion matrix\nprint(roc_auc_score(y_test, probs[:,1]))\nprint(classification_report(y_test, predicted))\nprint(confusion_matrix(y_test, predicted))",
"0.9397777068096268\n precision recall f1-score support\n\n 0 0.99 1.00 1.00 2099\n 1 0.97 0.80 0.88 91\n\n micro avg 0.99 0.99 0.99 2190\n macro avg 0.98 0.90 0.94 2190\nweighted avg 0.99 0.99 0.99 2190\n\n[[2097 2]\n [ 18 73]]\n"
]
],
[
[
"You have now obtained more meaningful performance metrics that tell us how well the model performs, given the highly imbalanced data that you're working with. The model predicts 76 cases of fraud, out of which 73 are actual fraud. You have only 3 false positives. This is really good, and as a result you have a very high precision score. You do however, don't catch 18 cases of actual fraud. Recall is therefore not as good as precision. Let's try to improve that in the following exercises. ",
"_____no_output_____"
],
[
"### Plotting the Precision Recall Curve\n* Plot a Precision-Recall curve, to investigate the trade-off between the two in your model. In this curve Precision and Recall are inversely related; as Precision increases, Recall falls and vice-versa. A balance between these two needs to be achieved in your model, otherwise you might end up with many false positives, or not enough actual fraud cases caught. To achieve this and to compare performance, the precision-recall curves come in handy.\n\n* The Random Forest Classifier is available as model, and the predictions as predicted. You can simply obtain the average precision score and the PR curve from the sklearn package. T\n\n* The function plot_pr_curve() plots the results.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import average_precision_score\n# Calculate average precision and the PR curve\naverage_precision = average_precision_score(y_test, predicted) ",
"_____no_output_____"
],
[
"# Calculate average precision and the PR curve\naverage_precision = average_precision_score(y_test, predicted)\n\n# Obtain precision and recall\nprecision, recall, _ = precision_recall_curve(y_test, predicted)\n\n# Plot the recall precision tradeoff\nplot_pr_curve(recall, precision, average_precision)\n#This function is unavailable. ",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"### Model adjustments\n* A simple way to adjust the random forest model to deal with highly imbalanced fraud data, is to use the **class_weights option **when defining your sklearn model. However, as you will see, it is a bit of a blunt force mechanism and might not work for your very special case.\n\n* Explore the weight = \"balanced_subsample\" mode the Random Forest model from the earlier exercise. ",
"_____no_output_____"
]
],
[
[
"model = RandomForestClassifier(class_weight='balanced_subsample', random_state=5)\n\nmodel.fit(X_train, y_train)\n\n# Obtain the predicted values and probabilities from the model \npredicted = model.predict(X_test)\nprobs = model.predict_proba(X_test)\n\nprint(roc_auc_score(y_test, probs[:,1]))\nprint(classification_report(y_test, predicted))\nprint(confusion_matrix(y_test, predicted))",
"0.9463271364176556\n precision recall f1-score support\n\n 0 0.99 1.00 1.00 2099\n 1 0.99 0.80 0.88 91\n\n micro avg 0.99 0.99 0.99 2190\n macro avg 0.99 0.90 0.94 2190\nweighted avg 0.99 0.99 0.99 2190\n\n[[2098 1]\n [ 18 73]]\n"
]
],
[
[
"* The model results don't improve drastically. We now have 3 less false positives, but now 19 in stead of 18 false negatives, i.e. cases of fraud we are not catching. If we mostly care about catching fraud, and not so much about the false positives, this does actually not improve our model at all, albeit a simple option to try. \n* In the next exercises we will see how to more smartly tweak your model to focus on reducing false negatives and catch more fraud. \n",
"_____no_output_____"
],
[
"### Adjusting your Random Forest to fraud detection\n* Explore the options for the random forest classifier, as we'll assign weights and tweak the shape of the decision trees in the forest. \n* Define weights manually, to be able to off-set that imbalance slightly. In our case we have 300 fraud to 7000 non-fraud cases, so by setting the weight ratio to 1:12, we get to a 1/3 fraud to 2/3 non-fraud ratio, which is good enough for training the model on.\n",
"_____no_output_____"
]
],
[
[
"# Change the model options\nmodel = RandomForestClassifier(bootstrap=True, class_weight={0:1, 1:12}, criterion='entropy',\n max_depth=10, \n min_samples_leaf=10, \n \n # Change the number of trees to use\n n_estimators=20, n_jobs=-1, random_state=5)\n\n# Run the function get_model_results\n# get_model_results(X_train, y_train, X_test, y_test, model)\n#This function fits the model to your training data, predicts and obtains performance metrics \n#similar to the steps you did in the previous exercises.",
"_____no_output_____"
]
],
[
[
"* By smartly defining more options in the model, you can obtain better predictions. You have effectively reduced the number of false negatives, i.e. you are catching more cases of fraud, whilst keeping the number of false positives low. \n* In this exercise you've manually changed the options of the model. There is a smarter way of doing it, by using GridSearchCV, which you'll see in the next exercise! ",
"_____no_output_____"
],
[
"### GridSearchCV to find optimal parameters\nWith GridSearchCV you can define which performance metric to score the options on. Since for fraud detection we are mostly interested in catching as many fraud cases as possible, you can optimize your model settings to get the **best possible Recall score.** If you also cared about reducing the number of false positives, you could optimize on F1-score, this gives you that nice Precision-Recall trade-off. ",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import GridSearchCV\n# Define the parameter sets to test\nparam_grid = {'n_estimators': [1, 30], 'max_features': ['auto', 'log2'], 'max_depth': [4, 8], 'criterion': ['gini', 'entropy']\n}\n\nmodel = RandomForestClassifier(random_state=5)\n\nCV_model = GridSearchCV(estimator=model, param_grid=param_grid, cv=5, scoring='recall', n_jobs=-1)\n\nCV_model.fit(X_train, y_train)\nCV_model.best_params_",
"_____no_output_____"
]
],
[
[
"### Model results using GridSearchCV\n* You discovered that the best parameters for your model are that the split criterion should be set to 'gini', the number of estimators (trees) should be 30, the maximum depth of the model should be 8 and the maximum features should be set to \"log2\".\n\n* Let's give this a try and see how well our model performs. You can use the get_model_results() function again to save time.",
"_____no_output_____"
]
],
[
[
"# Input the optimal parameters in the model\nmodel = RandomForestClassifier(class_weight={0:1,1:12}, criterion='gini',\n max_depth=8, max_features='log2', min_samples_leaf=10, n_estimators=30, n_jobs=-1, random_state=5)\n\n# Get results from your model\n# get_model_results(X_train, y_train, X_test, y_test, model)",
"_____no_output_____"
]
],
[
[
"<script.py> output:\n precision recall f1-score support\n \n 0.0 0.99 1.00 1.00 2099\n 1.0 0.95 0.84 0.89 91\n \n micro avg 0.99 0.99 0.99 2190\n macro avg 0.97 0.92 0.94 2190\n weighted avg 0.99 0.99 0.99 2190\n \n [[2095 4]\n [ 15 76]]",
"_____no_output_____"
],
[
"* The number of false positives has now been slightly reduced even further, which means we are catching more cases of fraud. \n* However, you see that the number of false positives actually went up. That is that Precision-Recall trade-off in action. \n* To decide which final model is best, you need to take into account how bad it is not to catch fraudsters, versus how many false positives the fraud analytics team can deal with. Ultimately, this final decision should be made by you and the fraud team together. ",
"_____no_output_____"
],
[
"### Logistic Regression\n* Combine three algorithms into one model with the VotingClassifier. This allows us to benefit from the different aspects from all models, and hopefully improve overall performance and detect more fraud. The first model, the Logistic Regression, has a slightly higher recall score than our optimal Random Forest model, but gives a lot more false positives.\n* You'll also add a Decision Tree with balanced weights to it. The data is already split into a training and test set, i.e. X_train, y_train, X_test, y_test are available.\n* In order to understand how the Voting Classifier can potentially improve your original model, you should check the standalone results of the Logistic Regression model first.",
"_____no_output_____"
]
],
[
[
"# Define the Logistic Regression model with weights\nmodel = LogisticRegression(class_weight={0:1, 1:15}, random_state=5)\n\n# Get the model results\n# get_model_results(X_train, y_train, X_test, y_test, model)",
"_____no_output_____"
]
],
[
[
" precision recall f1-score support\n\n 0.0 0.99 0.98 0.99 2099\n 1.0 0.63 0.88 0.73 91\n\n` micro avg 0.97 0.97 0.97 2190\n macro avg 0.81 0.93 0.86 2190\nweighted avg 0.98 0.97 0.98 2190\n`\n\n`\n[[2052 47]\n [ 11 80]]\n`",
"_____no_output_____"
],
[
"The Logistic Regression has quite different performance from the Random Forest. More false positives, but also a better Recall. It will therefore will a useful addition to the Random Forest in an ensemble model. ",
"_____no_output_____"
],
[
"### Voting Classifier\n* Combine three machine learning models into one, to improve our Random Forest fraud detection model from before. You'll combine our usual Random Forest model, with the Logistic Regression from the previous exercise, with a simple Decision Tree. \n* Use the short cut get_model_results() to see the immediate result of the ensemble model. ",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import VotingClassifier\nfrom sklearn.tree import DecisionTreeClassifier\n\n# Define the three classifiers to use in the ensemble\nclf1 = LogisticRegression(class_weight={0:1, 1:15}, random_state=5)\nclf2 = RandomForestClassifier(class_weight={0:1, 1:12}, criterion='gini', max_depth=8, max_features='log2',\n min_samples_leaf=10, n_estimators=30, n_jobs=-1, random_state=5)\nclf3 = DecisionTreeClassifier(random_state=5, class_weight=\"balanced\")\n\n# Combine the classifiers in the ensemble model\nensemble_model = VotingClassifier(estimators=[('lr', clf1), ('rf', clf2), ('dt', clf3)], voting='hard')\n\n# Get the results \n# get_model_results(X_train, y_train, X_test, y_test, ensemble_model)",
"_____no_output_____"
]
],
[
[
"<script.py> output:\n precision recall f1-score support\n \n 0.0 0.99 1.00 0.99 2099\n 1.0 0.90 0.86 0.88 91\n \n micro avg 0.99 0.99 0.99 2190\n macro avg 0.95 0.93 0.94 2190\n weighted avg 0.99 0.99 0.99 2190\n \n [[2090 9]\n [ 13 78]]\n",
"_____no_output_____"
],
[
"* By combining the classifiers, you can take the best of multiple models. You've increased the cases of fraud you are catching from 76 to 78, and you only have 5 extra false positives in return.\n* If you do care about catching as many fraud cases as you can, whilst keeping the false positives low, this is a pretty good trade-off. \n* The Logistic Regression as a standalone was quite bad in terms of false positives, and the Random Forest was worse in terms of false negatives. By combining these together you indeed managed to improve performance. ",
"_____no_output_____"
],
[
"### Adjust weights within the Voting Classifier\n* The Voting Classifier allows you to improve your fraud detection performance, by combining good aspects from multiple models. Now let's try to adjust the weights we give to these models. By increasing or decreasing weights you can play with how much emphasis you give to a particular model relative to the rest. This comes in handy when a certain model has overall better performance than the rest, but you still want to combine aspects of the others to further improve your results.\n\n* The data is already split into a training and test set, and clf1, clf2 and clf3 are available and defined as before, i.e. they are the Logistic Regression, the Random Forest model and the Decision Tree respectively.",
"_____no_output_____"
]
],
[
[
"ensemble_model = VotingClassifier(estimators=[('lr', clf1), ('rf', clf2), ('gnb', clf3)], voting='soft', weights=[1, 4, 1], flatten_transform=True)\n\n# Get results \n# get_model_results(X_train, y_train, X_test, y_test, ensemble_model)",
"_____no_output_____"
]
],
[
[
"<script.py> output:\n precision recall f1-score support\n \n 0.0 0.99 1.00 1.00 2099\n 1.0 0.94 0.85 0.89 91\n \n micro avg 0.99 0.99 0.99 2190\n macro avg 0.97 0.92 0.94 2190\n weighted avg 0.99 0.99 0.99 2190\n \n [[2094 5]\n [ 14 77]]",
"_____no_output_____"
],
[
"The weight option allows you to play with the individual models to get the best final mix for your fraud detection model. Now that you have finalized fraud detection with supervised learning, let's have a look at how fraud detection can be done when you don't have any labels to train on. ",
"_____no_output_____"
],
[
"## Fraud detection using unlabelled data\n* Use unsupervised learning techniques to detect fraud. \n* Segment customers, use K-means clustering and other clustering algorithms to find suspicious occurrences in your data. ",
"_____no_output_____"
],
[
"### Exploring your data\n* Look at bank payment transaction data. \n* Distinguish normal from abnormal (thus potentially fraudulent) behavior. As a fraud analyst to understand what is \"normal\", you need to have a good understanding of the data and its characteristics. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndf = pd.read_csv('banksim.csv')\ndf = df.drop(['Unnamed: 0'],axis = 1)\nprint(df.head())\nprint(df.groupby('category').mean())",
" age gender category amount fraud\n0 3 F es_transportation 49.71 0\n1 4 F es_health 39.29 0\n2 3 F es_transportation 18.76 0\n3 4 M es_transportation 13.95 0\n4 2 M es_transportation 49.87 0\n amount fraud\ncategory \nes_barsandrestaurants 43.841793 0.022472\nes_contents 55.170000 0.000000\nes_fashion 59.780769 0.020619\nes_food 35.216050 0.000000\nes_health 126.604704 0.242798\nes_home 120.688317 0.208333\nes_hotelservices 172.756245 0.548387\nes_hyper 46.788180 0.125000\nes_leisure 229.757600 1.000000\nes_otherservices 149.648960 0.600000\nes_sportsandtoys 157.251737 0.657895\nes_tech 132.852862 0.179487\nes_transportation 27.422014 0.000000\nes_travel 231.818656 0.944444\nes_wellnessandbeauty 66.167078 0.060606\n"
]
],
[
[
"Even from simple group by, we can find that the majority of fraud is observed in travel, leisure and sports related transactions. ",
"_____no_output_____"
],
[
"### Customer segmentation\n* Check whether there are any obvious patterns for the clients in this data, thus whether you need to segment your data into groups, or whether the data is rather homogenous.\n\n* There is not a lot client information available; However, there is data on **age ** available, so let's see whether there is any significant difference between behavior of age groups.",
"_____no_output_____"
]
],
[
[
"# Group by age groups and get the mean\nprint(df.groupby('age').mean())",
" amount fraud\nage \n0 49.468935 0.050000\n1 35.622829 0.026648\n2 37.228665 0.028718\n3 37.279338 0.023283\n4 36.197985 0.035966\n5 37.547521 0.023990\n6 36.700852 0.022293\nU 39.117000 0.000000\n"
],
[
"# Group by age groups and get the mean\ndf.groupby('age').mean()\n\n# Count the values of the observations in each age group\nprint(df['age'].value_counts())",
"2 2333\n3 1718\n4 1279\n5 792\n1 713\n6 314\n0 40\nU 11\nName: age, dtype: int64\n"
]
],
[
[
"* Does it make sense to divide your data into age segments before running a fraud detection algorithm? \n* No, the age groups who are the largest are relatively similar. As you can see the average amount spent as well as fraud occurrence is rather similar across groups. Age group '0' stands out but since there are only 40 cases, it does not make sense to split these out in a separate group and run a separate model on them. ",
"_____no_output_____"
],
[
"### Using statistics to define normal behavior\n* In the previous exercises we saw that fraud is more prevalent in certain transaction categories, but that there is no obvious way to segment our data into for example age groups.\n* This time, let's investigate the average amounts spend in normal transactions versus fraud transactions. This gives you an idea of how fraudulent transactions differ structurally from normal transactions.",
"_____no_output_____"
]
],
[
[
"# Create two dataframes with fraud and non-fraud data \ndf_fraud = df.loc[df.fraud == 1] \ndf_non_fraud = df.loc[df.fraud == 0]\n\n# Plot histograms of the amounts in fraud and non-fraud data \nplt.hist(df_fraud.amount, alpha=0.5, label='fraud')\nplt.hist(df_non_fraud.amount, alpha=0.5, label='nonfraud')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"* As the number fraud observations is much smaller, it is difficult to see the full distribution. \n* Nonetheless, you can see that the fraudulent transactions tend to be on the larger side relative to normal observations.\n* This helps us later in detecting fraud from non-fraud. In the next chapter you're going to implement a clustering model to distinguish between normal and abnormal transactions, when the fraud labels are no longer available. ",
"_____no_output_____"
],
[
"### Scaling the data\nFor ML algorithms using distance based metrics, it is crucial to always scale your data, as features using different scales will distort your results. K-means uses the Euclidian distance to assess distance to cluster centroids, therefore you first need to scale your data before continuing to implement the algorithm. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndf = pd.read_csv('banksim_adj.csv')\nX = df.drop(['Unnamed: 0'],axis = 1).values\ny = df['fraud'].values\nprint(df.head())\n#extra code above. The data might not be same as the DataCamp\n\n\nfrom sklearn.preprocessing import MinMaxScaler\n\nX = np.array(df).astype(np.float)\n\nscaler = MinMaxScaler()\nX_scaled = scaler.fit_transform(X)",
" Unnamed: 0 age amount fraud M es_barsandrestaurants es_contents \\\n0 0 3 49.71 0 0 0 0 \n1 1 4 39.29 0 0 0 0 \n2 2 3 18.76 0 0 0 0 \n3 3 4 13.95 0 1 0 0 \n4 4 2 49.87 0 1 0 0 \n\n es_fashion es_food es_health es_home es_hotelservices es_hyper \\\n0 0 0 0 0 0 0 \n1 0 0 1 0 0 0 \n2 0 0 0 0 0 0 \n3 0 0 0 0 0 0 \n4 0 0 0 0 0 0 \n\n es_leisure es_otherservices es_sportsandtoys es_tech es_transportation \\\n0 0 0 0 0 1 \n1 0 0 0 0 0 \n2 0 0 0 0 1 \n3 0 0 0 0 1 \n4 0 0 0 0 1 \n\n es_travel \n0 0 \n1 0 \n2 0 \n3 0 \n4 0 \n"
]
],
[
[
"### K-means clustering\n* For fraud detection, K-means clustering is straightforward to implement and relatively powerful in predicting suspicious cases. It is a good algorithm to start with when working on fraud detection problems. \n* However, fraud data is oftentimes very large, especially when you are working with transaction data. MiniBatch K-means is an efficient way to implement K-means on a large dataset, which you will use in this exercise.",
"_____no_output_____"
]
],
[
[
"# Import MiniBatchKmeans \nfrom sklearn.cluster import MiniBatchKMeans\n\nkmeans = MiniBatchKMeans(n_clusters=8, random_state=0)\n\nkmeans.fit(X_scaled)",
"_____no_output_____"
]
],
[
[
"### Elbow method\n* It is important to get the number of clusters right, especially when you want to **use the outliers of those clusters as fraud predictions**. \n* Apply the Elbow method and see what the optimal number of clusters should be based on this method.",
"_____no_output_____"
]
],
[
[
"clustno = range(1, 10)\n\nkmeans = [MiniBatchKMeans(n_clusters=i) for i in clustno]\nscore = [kmeans[i].fit(X_scaled).score(X_scaled) for i in range(len(kmeans))]\n\nplt.plot(clustno, score)\nplt.xlabel('Number of Clusters')\nplt.ylabel('Score')\nplt.title('Elbow Curve')\nplt.show()",
"_____no_output_____"
]
],
[
[
"The optimal number of clusters should probably be at around 3 clusters, as that is where the elbow is in the curve. ",
"_____no_output_____"
],
[
"### Detecting outliers\n* Use the K-means algorithm to predict fraud, and compare those predictions to the actual labels that are saved, to sense check our results.\n\n* The fraudulent transactions are typically flagged as the observations that are furthest aways from the cluster centroid. \n* How to determine the cut-off in this exercise. \n",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(X_scaled, y, test_size=0.3, random_state=0)\n\nkmeans = MiniBatchKMeans(n_clusters=3, random_state=42).fit(X_train)\n\nX_test_clusters = kmeans.predict(X_test)\nX_test_clusters_centers = kmeans.cluster_centers_\ndist = [np.linalg.norm(x-y) for x, y in zip(X_test, X_test_clusters_centers[X_test_clusters])]\n# np.linagl.norm calculate the 'norm' of a vector or a matrix. \n\n\n# Create fraud predictions based on outliers on clusters \nkm_y_pred = np.array(dist)\nkm_y_pred[dist >= np.percentile(dist, 95)] = 1\nkm_y_pred[dist < np.percentile(dist, 95)] = 0\n\nprint(len(X_test))\nprint(len(X_test_clusters))\nprint(X_test_clusters)\nprint('--------------------')\nprint(X_test_clusters_centers)\nprint(len(dist))",
"2157\n2157\n[2 1 1 ... 2 2 2]\n--------------------\n[[0.46208979 0.50503256 0.11993546 0.0071048 1. 0.01243339\n 0.0017762 0.01420959 0.02664298 0. 0. 0.0017762\n 0.01243339 0. 0.0017762 0.0053286 0.0017762 0.89520426\n 0. ]\n [0.58159793 0.49215686 0.36850912 0.18823529 0.27058824 0.06470588\n 0.00588235 0.05294118 0.28823529 0.3 0.01764706 0.00588235\n 0.02941176 0. 0. 0.08235294 0.01764706 0.\n 0.01764706]\n [0.49623688 0.494003 0.11833627 0.005997 0. 0.\n 0. 0.0029985 0.00749625 0. 0. 0.00149925\n 0.0029985 0. 0. 0.00149925 0.00149925 0.97901049\n 0. ]]\n2157\n"
]
],
[
[
"### Checking model results\nIn the previous exercise you've flagged all observations to be fraud, if they are in the top 5th percentile in distance from the cluster centroid. I.e. these are the very outliers of the three clusters. For this exercise you have the scaled data and labels already split into training and test set, so y_test is available. The predictions from the previous exercise, km_y_pred, are also available. Let's create some performance metrics and see how well you did.",
"_____no_output_____"
]
],
[
[
"# Obtain the ROC score\nprint(roc_auc_score(y_test, km_y_pred))\n#output: 0.8197704982668266",
"0.934936176504411\n"
],
[
"# Obtain the ROC score\nprint(roc_auc_score(y_test, km_y_pred))\n\n# Create a confusion matrix\nkm_cm = confusion_matrix(y_test, km_y_pred)\n\n# Plot the confusion matrix in a figure to visualize results \n# plot_confusion_matrix(km_cm)",
"0.934936176504411\n"
]
],
[
[
"",
"_____no_output_____"
],
[
"Question \nIf you were to decrease the percentile used as a cutoff point in the previous exercise to 93% instead of 95%, what would that do to your prediction results? \nThe number of fraud cases caught increases, but false positives also increase. ",
"_____no_output_____"
],
[
"### DB scan\n* Explore using a density based clustering method (DBSCAN) to detect fraud. The advantage of DBSCAN is that you do not need to define the number of clusters beforehand. Also, DBSCAN can handle weirdly shaped data (i.e. non-convex) much better than K-means can. \n* This time, you are **not going to take the outliers of the clusters and use that for fraud, but take the smallest clusters in the data and label those as fraud**. You again have the scaled dataset, i.e. X_scaled available. ",
"_____no_output_____"
]
],
[
[
"from sklearn.cluster import DBSCAN\n\n# Initialize and fit the DBscan model\ndb = DBSCAN(eps=0.9, min_samples=10, n_jobs=-1).fit(X_scaled)\n\n# Obtain the predicted labels and calculate number of clusters\npred_labels = db.labels_\nn_clusters = len(set(pred_labels)) - (1 if -1 in labels else 0)\n\n# # Print performance metrics for DBscan\n# print('Estimated number of clusters: %d' % n_clusters)\n# print(\"Homogeneity: %0.3f\" % homogeneity_score(labels, pred_labels))\n# print(\"Silhouette Coefficient: %0.3f\" % silhouette_score(X_scaled, pred_labels))",
"_____no_output_____"
]
],
[
[
"output: \n`\nEstimated number of clusters: 18\nHomogeneity: 0.633\nSilhouette Coefficient: 0.707\n` \n\nThe number of clusters is much higher than with K-means. For fraud detection this is for now OK, as we are only interested in the smallest clusters, since those are considered as abnormal. Now let's have a look at those clusters and decide which one to flag as fraud. ",
"_____no_output_____"
],
[
"### Assessing smallest clusters\n* Check the clusters that came out of DBscan, and flag certain clusters as fraud:\n* Figure out how big the clusters are, and filter out the smallest. Then take the smallest ones and flag those as fraud. \n* Check with the original labels whether this does actually do a good job in detecting fraud. \n\nAvailable are the DBscan model predictions, so n_clusters is available as well as the cluster labels, which are saved under pred_labels. ",
"_____no_output_____"
]
],
[
[
"counts = np.bincount(pred_labels[pred_labels >= 0])\n\nprint(counts)\n",
"[3252 105 2714 46 174 119 112 79 52 74 43 24 45 42\n 11 19 16 13 10 15 35 10 40 19 11]\n"
]
],
[
[
"output:\n [3252 145 2714 55 174 119 122 98 54 15 76 15 43 25\n 51 47 42 15 25 20 19 10]\n",
"_____no_output_____"
]
],
[
[
"# Count observations in each cluster number\ncounts = np.bincount(pred_labels[pred_labels>=0])\n\n# Sort the sample counts of the clusters and take the top 3 smallest clusters\nsmallest_clusters = np.argsort(counts)[:3]\n\n# Print the results \nprint(\"The smallest clusters are clusters:\") \nprint(smallest_clusters)",
"_____no_output_____"
]
],
[
[
"output:\n The smallest clusters are clusters:\n [21 17 9] ",
"_____no_output_____"
]
],
[
[
"# Count observations in each cluster number\ncounts = np.bincount(pred_labels[pred_labels>=0])\n\n# Sort the sample counts of the clusters and take the top 3 smallest clusters\nsmallest_clusters = np.argsort(counts)[:3]\n\n# Print the counts of the smallest clusters only\nprint(\"Their counts are:\") \nprint(counts[smallest_clusters])",
"Their counts are:\n[10 10 11]\n"
]
],
[
[
"<script.py> output:\n Their counts are:\n [10 15 15] \n \n So now we know which smallest clusters you could flag as fraud. If you were to take more of the smallest clusters, you cast your net wider and catch more fraud, but most likely also more false positives. It is up to the fraud analyst to find the right amount of cases to flag and to investigate. In the next exercise you'll check the results with the actual labels. \n \n### Checking results\nIn this exercise you're going to check the results of your DBscan fraud detection model. In reality, you often don't have reliable labels and this where a fraud analyst can help you validate the results. He/She can check your results and see whether the cases you flagged are indeed suspicious. You can also check historically known cases of fraud and see whether your model flags them.\n\nIn this case, you'll use the fraud labels to check your model results. The predicted cluster numbers are available under pred_labels as well as the original fraud labels labels. ",
"_____no_output_____"
]
],
[
[
"# Create a dataframe of the predicted cluster numbers and fraud labels\ndf = pd.DataFrame({'clusternr':pred_labels,'fraud':labels})\n\n# Create a condition flagging fraud for the smallest clusters \ndf['predicted_fraud'] = np.where((df['clusternr']==21) | (df['clusternr']==17) | (df['clusternr']==9), 1, 0)\n\n# Run a crosstab on the results \nprint(pd.crosstab(df.fraud, df.predicted_fraud, rownames=['Actual Fraud'], colnames=['Flagged Fraud']))",
"_____no_output_____"
]
],
[
[
"output: \n` Flagged Fraud 0 1\n Actual Fraud \n 0 6973 16\n 1 176 24\n `",
"_____no_output_____"
],
[
"How does this compare to the K-means model?\n* The good thing is: our of all flagged cases, roughly 2/3 are actually fraud! Since you only take the three smallest clusters, by definition you flag less cases of fraud, so you catch less but also have less false positives. However, you are missing quite a lot of fraud cases. \n* Increasing the amount of smallest clusters you flag could improve that, at the cost of more false positives of course. ",
"_____no_output_____"
],
[
"## Fraud detection using text\nUse text data, text mining and topic modeling to detect fraudulent behavior. ",
"_____no_output_____"
],
[
"### Word search with dataframes\n* Work with text data, containing emails from Enron employees. \n* Using string operations on dataframes, you can easily sift through messy email data and create flags based on word-hits.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndf = pd.read_csv('enron_emails_clean.csv',index_col = 0)\n# Find all cleaned emails that contain 'sell enron stock'\nmask = df['clean_content'].str.contains('sell enron stock', na=False)\n# Select the data from df that contain the searched for words \nprint(df.loc[mask])",
" From \\\nMessage-ID \n<6336501.1075841154311.JavaMail.evans@thyme> ('[email protected]') \n\n To \\\nMessage-ID \n<6336501.1075841154311.JavaMail.evans@thyme> ('[email protected]') \n\n Date \\\nMessage-ID \n<6336501.1075841154311.JavaMail.evans@thyme> 2002-02-01 14:53:35 \n\n content \\\nMessage-ID \n<6336501.1075841154311.JavaMail.evans@thyme> \\nJoint Venture: A 1997 Enron Meeting Belies O... \n\n clean_content \nMessage-ID \n<6336501.1075841154311.JavaMail.evans@thyme> joint venture enron meeting belies officers cl... \n"
]
],
[
[
"### Using list of terms\n* Search on more than one term. \n* Create a full \"fraud dictionary\" of terms that could potentially flag fraudulent clients and/or transactions. Fraud analysts often will have an idea what should be in such a dictionary. In this exercise you're going to flag a multitude of terms, and in the next exercise you'll create a new flag variable out of it. The 'flag' can be used either directly in a machine learning model as a feature, or as an additional filter on top of your machine learning model results. ",
"_____no_output_____"
]
],
[
[
"# Create a list of terms to search for\nsearchfor = ['enron stock', 'sell stock', 'stock bonus', 'sell enron stock']\n\n# Filter cleaned emails on searchfor list and select from df \nfiltered_emails = df.loc[df['clean_content'].str.contains('|'.join(searchfor), na=False)]\n# print(filtered_emails)",
"_____no_output_____"
]
],
[
[
"### Creating a flag\nThis time you are going to create an actual flag variable that gives a 1 when the emails get a hit on the search terms of interest, and 0 otherwise. This is the last step you need to make in order to actually use the text data content as a feature in a machine learning model, or as an actual flag on top of model results. You can continue working with the dataframe df containing the emails, and the searchfor list is the one defined in the last exercise. \n",
"_____no_output_____"
]
],
[
[
"import numpy as np\n# Create flag variable where the emails match the searchfor terms\ndf['flag'] = np.where((df['clean_content'].str.contains('|'.join(searchfor)) == True), 1, 0)\n\n# Count the values of the flag variable\ncount = df['flag'].value_counts()\nprint(count)",
"0 1776\n1 314\nName: flag, dtype: int64\n"
]
],
[
[
"You have now managed to search for a list of strings in several lines of text data. These skills come in handy when you want to flag certain words based on what you discovered in your topic model, or when you know beforehand what you want to search for. In the next exercises you're going to learn how to clean text data and to create your own topic model to further look for indications of fraud in your text data. ",
"_____no_output_____"
],
[
"### Removing stopwords\nIn the following exercises you're going to clean the Enron emails, in order to be able to use the data in a topic model. Text cleaning can be challenging, so you'll learn some steps to do this well. The dataframe containing the emails df is available. In a first step you need to define the list of stopwords and punctuations that are to be removed in the next exercise from the text data. Let's give it a try. ",
"_____no_output_____"
]
],
[
[
"# Import nltk packages and string \nfrom nltk.corpus import stopwords \nimport string\n\n# Define stopwords to exclude\nstop = set(stopwords.words('english'))\n# stop.update((\"to\",\"cc\",\"subject\",\"http\",\"from\",\"sent\", \"ect\", \"u\", \"fwd\", \"www\", \"com\"))\n\n# Define punctuations to exclude and lemmatizer\nexclude = set(string.punctuation)",
"_____no_output_____"
]
],
[
[
"The following is the stop contents. However, stop = set(stopwords('english')) has problems to run. \n{'a',\n 'about',\n 'above',\n 'after',\n 'again',\n 'against',\n 'ain',\n 'all',\n 'am',\n .\n .\n .\n 'y',\n 'you',\n \"you'd\",\n \"you'll\",\n \"you're\",\n \"you've\",\n 'your',\n 'yours',\n 'yourself',\n 'yourselves'\n }",
"_____no_output_____"
],
[
"### Cleaning text data\nNow that you've defined the stopwords and punctuations, let's use these to clean our enron emails in the dataframe df further. The lists containing stopwords and punctuations are available under stop and exclude There are a few more steps to take before you have cleaned data, such as \"lemmatization\" of words, and stemming the verbs. The verbs in the email data are already stemmed, and the lemmatization is already done for you in this exercise.",
"_____no_output_____"
]
],
[
[
"# Import the lemmatizer from nltk\nfrom nltk.stem.wordnet import WordNetLemmatizer\nlemma = WordNetLemmatizer()\n\n# Define word cleaning function\ndef clean(text, stop):\n text = text.rstrip()\n # Remove stopwords\n stop_free = \" \".join([word for word in text.lower().split() if ((word not in stop) and (not word.isdigit()))])\n # Remove punctuations\n punc_free = ''.join(word for word in stop_free if word not in exclude)\n # Lemmatize all words\n normalized = \" \".join(lemma.lemmatize(word) for word in punc_free.split()) \n return normalized\n\n# Import the lemmatizer from nltk\nfrom nltk.stem.wordnet import WordNetLemmatizer\nlemma = WordNetLemmatizer()",
"_____no_output_____"
],
[
"# Import the lemmatizer from nltk\nfrom nltk.stem.wordnet import WordNetLemmatizer\nlemma = WordNetLemmatizer()\n\n# Define word cleaning function\ndef clean(text, stop):\n text = text.rstrip()\n stop_free = \" \".join([i for i in text.lower().split() if((i not in stop) and (not i.isdigit()))])\n punc_free = ''.join(i for i in stop_free if i not in exclude)\n normalized = \" \".join(lemma.lemmatize(i) for i in punc_free.split()) \n return normalized\n\n# Clean the emails in df and print results\ntext_clean=[]\nfor text in df['clean_content']:\n text_clean.append(clean(text, stop).split()) \nprint(text_clean)\n",
"_____no_output_____"
]
],
[
[
"Now that you have cleaned your data entirely with the necessary steps, including splitting the text into words, removing stopwords and punctuations, and lemmatizing your words. You are now ready to run a topic model on this data. In the following exercises you're going to explore how to do that. \n\n### Create dictionary and corpus\nIn order to run an LDA topic model, you first need to define your dictionary and corpus first, as those need to go into the model. You're going to continue working on the cleaned text data that you've done in the previous exercises. That means that text_clean is available for you already to continue working with, and you'll use that to create your dictionary and corpus.\n\nThis exercise will take a little longer to execute than usual.",
"_____no_output_____"
]
],
[
[
"# Import the packages\nimport gensim\nfrom gensim import corpora\n\n# Define the dictionary\ndictionary = corpora.Dictionary(text_clean)\n\n# Define the corpus\ncorpus = [dictionary.doc2bow(text) for text in text_clean]\n\n# Print corpus and dictionary\nprint(dictionary)\nprint(corpus) ",
"_____no_output_____"
]
],
[
[
"Dictionary(8948 unique tokens: ['conducted', 'read', 'wil', 'daniel', 'piazze']...) \n[[(0, 1), (1, 2), (2, 1), (3, 1), (4, 2), (5, 1), (6, 2), (7, 1), (8, 1), (9, 1), (10, 5), (11, 2), (12, 1), (13, 1), (14, 1), (15, 1), (16, 1), (17, 1), (18, 1), (19, 1), (20, 1), (21, 1), (22, 1), (23, 1), (24, 1),....] total length. \nNote doc2bow is doc to bag of words. ",
"_____no_output_____"
],
[
"### LDA model (It is is not linear discriminant analysis)\nNow it's time to build the LDA model. Using the dictionary and corpus, you are ready to discover which topics are present in the Enron emails. With a quick print of words assigned to the topics, you can do a first exploration about whether there are any obvious topics that jump out. Be mindful that the topic model is heavy to calculate so it will take a while to run. Let's give it a try!",
"_____no_output_____"
]
],
[
[
"ldamodel = gensim.models.ldamodel.LdaModel(corpus, num_topics=5, id2word=dictionary, passes=5)\n\n# Save the topics and top 5 words\ntopics = ldamodel.print_topics(num_words=5)\n\n# Print the results\nfor topic in topics:\n print(topic)",
"(0, '0.043*\"enron\" + 0.023*\"company\" + 0.022*\"employee\" + 0.019*\"million\" + 0.016*\"energy\"')\n(1, '0.019*\"enron\" + 0.013*\"company\" + 0.010*\"energy\" + 0.008*\"com\" + 0.007*\"power\"')\n(2, '0.040*\"enron\" + 0.017*\"stock\" + 0.011*\"option\" + 0.010*\"com\" + 0.010*\"dynegy\"')\n(3, '0.015*\"enron\" + 0.009*\"company\" + 0.009*\"com\" + 0.006*\"said\" + 0.006*\"http\"')\n(4, '0.023*\"enron\" + 0.011*\"company\" + 0.010*\"said\" + 0.007*\"mr\" + 0.005*\"stock\"')\n"
]
],
[
[
"`(0, '0.024*\"enron\" + 0.015*\"ect\" + 0.011*\"com\" + 0.007*\"hou\" + 0.005*\"company\"')\n (1, '0.032*\"enron\" + 0.011*\"com\" + 0.009*\"diabetes\" + 0.008*\"message\" + 0.006*\"please\"')\n (2, '0.031*\"enron\" + 0.011*\"company\" + 0.010*\"said\" + 0.007*\"mr\" + 0.005*\"partnership\"')\n (3, '0.021*\"enron\" + 0.012*\"employee\" + 0.010*\"company\" + 0.009*\"million\" + 0.009*\"com\"')\n (4, '0.040*\"error\" + 0.021*\"database\" + 0.018*\"borland\" + 0.018*\"engine\" + 0.018*\"initialize\"') \n` \nYou have now successfully created your first topic model on the Enron email data. However, the print of words doesn't really give you enough information to find a topic that might lead you to signs of fraud. You'll therefore need to closely inspect the model results in order to be able to detect anything that can be related to fraud in your data. ",
"_____no_output_____"
],
[
"Below are visualisation results from the pyLDAvis library available. Have a look at topic 1 and 3 from the LDA model on the Enron email data. Which one would you research further for fraud detection purposes and why? \n\nTopic 1 seems to discuss the employee share option program, and seems to point to internal conversation (with \"please, may, know\" etc), so this is more likely to be related to the internal accounting fraud and trading stock with insider knowledge. Topic 3 seems to be more related to general news around Enron.",
"_____no_output_____"
],
[
"### Finding fraudsters based on topic\nIn this exercise you're going to link the results from the topic model back to your original data. You now learned that you want to flag everything related to topic 3. As you will see, this is actually not that straightforward. You'll be given the function get_topic_details() which takes the arguments ldamodel and corpus. It retrieves the details of the topics for each line of text. With that function, you can append the results back to your original data. If you want to learn more detail on how to work with the model results, which is beyond the scope of this course, you're highly encouraged to read this article (https://www.machinelearningplus.com/nlp/topic-modeling-gensim-python/).\n\nAvailable for you are the dictionary and corpus, the text data text_clean as well as your model results ldamodel. Also defined is get_topic_details().",
"_____no_output_____"
]
],
[
[
"# Run get_topic_details function and check the results\nprint(get_topic_details(ldamodel, corpus))",
"_____no_output_____"
],
[
"# Add original text to topic details in a dataframe\ncontents = pd.DataFrame({'Original text': text_clean})\ntopic_details = pd.concat([get_topic_details(ldamodel, corpus), contents], axis=1)\ntopic_details.head()",
"_____no_output_____"
],
[
"# Add original text to topic details in a dataframe\ncontents = pd.DataFrame({'Original text':text_clean})\ntopic_details = pd.concat([get_topic_details(ldamodel, corpus), contents], axis=1)\n\n# Create flag for text highest associated with topic 3\ntopic_details['flag'] = np.where((topic_details['Dominant_Topic'] == 3.0), 1, 0)\nprint(topic_details.head())",
"_____no_output_____"
]
],
[
[
"You have now flagged all data that is highest associated with topic 3, that seems to cover internal conversation about enron stock options. You are a true detective. With these exercises you have demonstrated that text mining and topic modeling can be a powerful tool for fraud detection.",
"_____no_output_____"
],
[
"### Summary\n* We may apply all types of machine learning algorithms to handle anomaly and fraud detection. \n * Supervised learning such as classification algorithms, neural network, etc. \n * Unsupervised learning such as clustering algorithms. \n * All the linear or nonlinear dimension reduction techniques that can be used directly to handle anomaly detection, or can be combined with other supervised/unsupervised learning algorithm. \n * Natural language processing.\n \n* Directly constructing Gaussian distribution (or other contributions) and flag outliers. \n\n* Use network analysis for fraud or anomaly detection. \n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d0f428d267d633d717acaa8dabbfb3435bfd3f9b | 21,698 | ipynb | Jupyter Notebook | tools/colab/profiling_tpus_in_colab.ipynb | zhtmike/tpu | 72a501051c2f854566910f13028936ff7a90057b | [
"Apache-2.0"
] | 5,098 | 2018-02-09T16:56:49.000Z | 2022-03-31T13:50:40.000Z | tools/colab/profiling_tpus_in_colab.ipynb | zhtmike/tpu | 72a501051c2f854566910f13028936ff7a90057b | [
"Apache-2.0"
] | 550 | 2018-02-07T05:30:06.000Z | 2022-03-13T22:00:09.000Z | tools/colab/profiling_tpus_in_colab.ipynb | zhtmike/tpu | 72a501051c2f854566910f13028936ff7a90057b | [
"Apache-2.0"
] | 1,920 | 2018-02-07T23:44:49.000Z | 2022-03-29T03:11:08.000Z | 37.801394 | 347 | 0.568854 | [
[
[
"<a href=\"https://colab.research.google.com/github/tensorflow/tpu/blob/master/tools/colab/profiling_tpus_in_colab.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"##### Copyright 2018 The TensorFlow Hub Authors.",
"_____no_output_____"
],
[
"Copyright 2019-2020 Google LLC\n\nLicensed under the Apache License, Version 2.0 (the \"License\");\nyou may not use this file except in compliance with the License.\nYou may obtain a copy of the License at\n\n http://www.apache.org/licenses/LICENSE-2.0\n\nUnless required by applicable law or agreed to in writing, software\ndistributed under the License is distributed on an \"AS IS\" BASIS,\nWITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\nSee the License for the specific language governing permissions and\nlimitations under the License.\n\n\n",
"_____no_output_____"
],
[
"# Profiling TPUs in Colab <a href=\"https://cloud.google.com/tpu/\"><img valign=\"middle\" src=\"https://raw.githubusercontent.com/GoogleCloudPlatform/tensorflow-without-a-phd/master/tensorflow-rl-pong/images/tpu-hexagon.png\" width=\"50\"></a>\nAdapted from [TPU colab example](https://colab.sandbox.google.com/notebooks/tpu.ipynb).",
"_____no_output_____"
],
[
"## Overview\nThis example works through training a model to classify images of\nflowers on Google's lightning-fast Cloud TPUs. Our model takes as input a photo of a flower and returns whether it is a daisy, dandelion, rose, sunflower, or tulip. A key objective of this colab is to show you how to set up and run TensorBoard, the program used for visualizing and analyzing program performance on Cloud TPU.\n\nThis notebook is hosted on GitHub. To view it in its original repository, after opening the notebook, select **File > View on GitHub**.",
"_____no_output_____"
],
[
"## Instructions\n\n<h3><a href=\"https://cloud.google.com/tpu/\"><img valign=\"middle\" src=\"https://raw.githubusercontent.com/GoogleCloudPlatform/tensorflow-without-a-phd/master/tensorflow-rl-pong/images/tpu-hexagon.png\" width=\"50\"></a> Train on TPU </h3>\n\n* Create a Cloud Storage bucket for your TensorBoard logs at http://console.cloud.google.com/storage. Give yourself Storage Legacy Bucket Owner permission on the bucket.\nYou will need to provide the bucket name when launching TensorBoard in the **Training** section. \n\nNote: User input is required when launching and viewing TensorBoard, so do not use **Runtime > Run all** to run through the entire colab. \n",
"_____no_output_____"
],
[
"## Authentication for connecting to GCS bucket for logging.",
"_____no_output_____"
]
],
[
[
"import os\nIS_COLAB_BACKEND = 'COLAB_GPU' in os.environ # this is always set on Colab, the value is 0 or 1 depending on GPU presence\nif IS_COLAB_BACKEND:\n from google.colab import auth\n # Authenticates the Colab machine and also the TPU using your\n # credentials so that they can access your private GCS buckets.\n auth.authenticate_user()",
"_____no_output_____"
]
],
[
[
"## Updating tensorboard_plugin_profile",
"_____no_output_____"
]
],
[
[
"!pip install -U pip install -U tensorboard_plugin_profile==2.3.0",
"_____no_output_____"
]
],
[
[
"## Enabling and testing the TPU\n\nFirst, you'll need to enable TPUs for the notebook:\n\n- Navigate to Edit→Notebook Settings\n- select TPU from the Hardware Accelerator drop-down\n\nNext, we'll check that we can connect to the TPU:",
"_____no_output_____"
]
],
[
[
"%tensorflow_version 2.x\nimport tensorflow as tf\nprint(\"Tensorflow version \" + tf.__version__)\n\ntry:\n tpu = tf.distribute.cluster_resolver.TPUClusterResolver() # TPU detection\n print('Running on TPU ', tpu.cluster_spec().as_dict()['worker'])\nexcept ValueError:\n raise BaseException('ERROR: Not connected to a TPU runtime; please see the previous cell in this notebook for instructions!')\n\ntf.config.experimental_connect_to_cluster(tpu)\ntf.tpu.experimental.initialize_tpu_system(tpu)\ntpu_strategy = tf.distribute.experimental.TPUStrategy(tpu)",
"_____no_output_____"
],
[
"import re\nimport numpy as np\nfrom matplotlib import pyplot as plt",
"_____no_output_____"
]
],
[
[
"\n## Input data\n\nOur input data is stored on Google Cloud Storage. To more fully use the parallelism TPUs offer us, and to avoid bottlenecking on data transfer, we've stored our input data in TFRecord files, 230 images per file.\n\nBelow, we make heavy use of `tf.data.experimental.AUTOTUNE` to optimize different parts of input loading.\n\nAll of these techniques are a bit overkill for our (small) dataset, but demonstrate best practices for using TPUs.\n",
"_____no_output_____"
]
],
[
[
"AUTO = tf.data.experimental.AUTOTUNE\n\nIMAGE_SIZE = [331, 331]\n\nbatch_size = 16 * tpu_strategy.num_replicas_in_sync\n\ngcs_pattern = 'gs://flowers-public/tfrecords-jpeg-331x331/*.tfrec'\nvalidation_split = 0.19\nfilenames = tf.io.gfile.glob(gcs_pattern)\nsplit = len(filenames) - int(len(filenames) * validation_split)\ntrain_fns = filenames[:split]\nvalidation_fns = filenames[split:]\n \ndef parse_tfrecord(example):\n features = {\n \"image\": tf.io.FixedLenFeature([], tf.string), # tf.string means bytestring\n \"class\": tf.io.FixedLenFeature([], tf.int64), # shape [] means scalar\n \"one_hot_class\": tf.io.VarLenFeature(tf.float32),\n }\n example = tf.io.parse_single_example(example, features)\n decoded = tf.image.decode_jpeg(example['image'], channels=3)\n normalized = tf.cast(decoded, tf.float32) / 255.0 # convert each 0-255 value to floats in [0, 1] range\n image_tensor = tf.reshape(normalized, [*IMAGE_SIZE, 3])\n one_hot_class = tf.reshape(tf.sparse.to_dense(example['one_hot_class']), [5])\n return image_tensor, one_hot_class\n\ndef load_dataset(filenames):\n # Read from TFRecords. For optimal performance, we interleave reads from multiple files.\n records = tf.data.TFRecordDataset(filenames, num_parallel_reads=AUTO)\n return records.map(parse_tfrecord, num_parallel_calls=AUTO)\n\ndef get_training_dataset():\n dataset = load_dataset(train_fns)\n\n # Create some additional training images by randomly flipping and\n # increasing/decreasing the saturation of images in the training set. \n def data_augment(image, one_hot_class):\n modified = tf.image.random_flip_left_right(image)\n modified = tf.image.random_saturation(modified, 0, 2)\n return modified, one_hot_class\n augmented = dataset.map(data_augment, num_parallel_calls=AUTO)\n\n # Prefetch the next batch while training (autotune prefetch buffer size).\n return augmented.repeat().shuffle(2048).batch(batch_size).prefetch(AUTO) \n\ntraining_dataset = get_training_dataset()\nvalidation_dataset = load_dataset(validation_fns).batch(batch_size).prefetch(AUTO)",
"_____no_output_____"
]
],
[
[
"Let's take a peek at the training dataset we've created:",
"_____no_output_____"
]
],
[
[
"CLASSES = ['daisy', 'dandelion', 'roses', 'sunflowers', 'tulips']\n\ndef display_one_flower(image, title, subplot, color):\n plt.subplot(subplot)\n plt.axis('off')\n plt.imshow(image)\n plt.title(title, fontsize=16, color=color)\n \n# If model is provided, use it to generate predictions.\ndef display_nine_flowers(images, titles, title_colors=None):\n subplot = 331\n plt.figure(figsize=(13,13))\n for i in range(9):\n color = 'black' if title_colors is None else title_colors[i]\n display_one_flower(images[i], titles[i], 331+i, color)\n plt.tight_layout()\n plt.subplots_adjust(wspace=0.1, hspace=0.1)\n plt.show()\n\ndef get_dataset_iterator(dataset, n_examples):\n return dataset.unbatch().batch(n_examples).as_numpy_iterator()\n\ntraining_viz_iterator = get_dataset_iterator(training_dataset, 9)",
"_____no_output_____"
],
[
"# Re-run this cell to show a new batch of images\nimages, classes = next(training_viz_iterator)\nclass_idxs = np.argmax(classes, axis=-1) # transform from one-hot array to class number\nlabels = [CLASSES[idx] for idx in class_idxs]\ndisplay_nine_flowers(images, labels)",
"_____no_output_____"
]
],
[
[
"## Model\nTo get maxmimum accuracy, we leverage a pretrained image recognition model (here, [Xception](http://openaccess.thecvf.com/content_cvpr_2017/papers/Chollet_Xception_Deep_Learning_CVPR_2017_paper.pdf)). We drop the ImageNet-specific top layers (`include_top=false`), and add a max pooling and a softmax layer to predict our 5 classes.",
"_____no_output_____"
]
],
[
[
"def create_model():\n pretrained_model = tf.keras.applications.Xception(input_shape=[*IMAGE_SIZE, 3], include_top=False)\n pretrained_model.trainable = True\n model = tf.keras.Sequential([\n pretrained_model,\n tf.keras.layers.GlobalAveragePooling2D(),\n tf.keras.layers.Dense(5, activation='softmax')\n ])\n model.compile(\n optimizer='adam',\n loss = 'categorical_crossentropy',\n metrics=['accuracy']\n )\n return model\n\nwith tpu_strategy.scope(): # creating the model in the TPUStrategy scope means we will train the model on the TPU\n model = create_model()\nmodel.summary()",
"_____no_output_____"
]
],
[
[
"## Training",
"_____no_output_____"
],
[
"Calculate the number of images in each dataset. Rather than actually load the data to do so (expensive), we rely on hints in the filename. This is used to calculate the number of batches per epoch.\n",
"_____no_output_____"
]
],
[
[
"def count_data_items(filenames):\n # The number of data items is written in the name of the .tfrec files, i.e. flowers00-230.tfrec = 230 data items\n n = [int(re.compile(r\"-([0-9]*)\\.\").search(filename).group(1)) for filename in filenames]\n return np.sum(n)\n\nn_train = count_data_items(train_fns)\nn_valid = count_data_items(validation_fns)\ntrain_steps = count_data_items(train_fns) // batch_size\nprint(\"TRAINING IMAGES: \", n_train, \", STEPS PER EPOCH: \", train_steps)\nprint(\"VALIDATION IMAGES: \", n_valid)",
"_____no_output_____"
]
],
[
[
"Calculate and show a learning rate schedule. We start with a fairly low rate, as we're using a pre-trained model and don't want to undo all the fine work put into training it.",
"_____no_output_____"
]
],
[
[
"EPOCHS = 12\n\nstart_lr = 0.00001\nmin_lr = 0.00001\nmax_lr = 0.00005 * tpu_strategy.num_replicas_in_sync\nrampup_epochs = 5\nsustain_epochs = 0\nexp_decay = .8\n\ndef lrfn(epoch):\n if epoch < rampup_epochs:\n return (max_lr - start_lr)/rampup_epochs * epoch + start_lr\n elif epoch < rampup_epochs + sustain_epochs:\n return max_lr\n else:\n return (max_lr - min_lr) * exp_decay**(epoch-rampup_epochs-sustain_epochs) + min_lr\n \nlr_callback = tf.keras.callbacks.LearningRateScheduler(lambda epoch: lrfn(epoch), verbose=True)\n\nrang = np.arange(EPOCHS)\ny = [lrfn(x) for x in rang]\nplt.plot(rang, y)\nprint('Learning rate per epoch:')",
"_____no_output_____"
]
],
[
[
"Train the model. While the first epoch will be quite a bit slower as we must XLA-compile the execution graph and load the data, later epochs should complete in ~5s.",
"_____no_output_____"
]
],
[
[
"# Load the TensorBoard notebook extension.\n%load_ext tensorboard",
"_____no_output_____"
],
[
"# Get TPU profiling service address. This address will be needed for capturing\n# profile information with TensorBoard in the following steps.\nservice_addr = tpu.get_master().replace(':8470', ':8466')\nprint(service_addr)",
"_____no_output_____"
],
[
"# Launch TensorBoard.\n%tensorboard --logdir=gs://bucket-name # Replace the bucket-name variable with your own gcs bucket",
"_____no_output_____"
]
],
[
[
"The TensorBoard UI is displayed in a browser window. In this colab, perform the following steps to prepare to capture profile information.\n1. Click on the dropdown menu box on the top right side and scroll down and click PROFILE. A new window appears that shows: **No profile data was found** at the top.\n1. Click on the CAPTURE PROFILE button. A new dialog appears. The top input line shows: **Profile Service URL or TPU name**. Copy and paste the Profile Service URL (the service_addr value shown before launching TensorBoard) into the top input line. While still on the dialog box, start the training with the next step.\n1. Click on the next colab cell to start training the model.\n1. Watch the output from the training until several epochs have completed. This allows time for the profile data to start being collected. Return to the dialog box and click on the CAPTURE button. If the capture succeeds, the page will auto refresh and redirect you to the profiling results.",
"_____no_output_____"
]
],
[
[
"history = model.fit(training_dataset, validation_data=validation_dataset,\n steps_per_epoch=train_steps, epochs=EPOCHS, callbacks=[lr_callback])\n\nfinal_accuracy = history.history[\"val_accuracy\"][-5:]\nprint(\"FINAL ACCURACY MEAN-5: \", np.mean(final_accuracy))",
"_____no_output_____"
],
[
"def display_training_curves(training, validation, title, subplot):\n ax = plt.subplot(subplot)\n ax.plot(training)\n ax.plot(validation)\n ax.set_title('model '+ title)\n ax.set_ylabel(title)\n ax.set_xlabel('epoch')\n ax.legend(['training', 'validation'])\n\nplt.subplots(figsize=(10,10))\nplt.tight_layout()\ndisplay_training_curves(history.history['accuracy'], history.history['val_accuracy'], 'accuracy', 211)\ndisplay_training_curves(history.history['loss'], history.history['val_loss'], 'loss', 212)",
"_____no_output_____"
]
],
[
[
"Accuracy goes up and loss goes down. Looks good!",
"_____no_output_____"
],
[
"## Next steps\n\nMore TPU/Keras examples include:\n- [Shakespeare in 5 minutes with Cloud TPUs and Keras](https://colab.research.google.com/github/tensorflow/tpu/blob/master/tools/colab/shakespeare_with_tpu_and_keras.ipynb)\n- [Fashion MNIST with Keras and TPUs](https://colab.research.google.com/github/tensorflow/tpu/blob/master/tools/colab/fashion_mnist.ipynb)\n\nWe'll be sharing more examples of TPU use in Colab over time, so be sure to check back for additional example links, or [follow us on Twitter @GoogleColab](https://twitter.com/googlecolab).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d0f44642dddd28426d278a428efd4c1651322b01 | 664,390 | ipynb | Jupyter Notebook | tf2-handson.ipynb | msavardi/tf2.0-handson | 5dfe760875290e19a1e25617b92537339eacb115 | [
"MIT"
] | null | null | null | tf2-handson.ipynb | msavardi/tf2.0-handson | 5dfe760875290e19a1e25617b92537339eacb115 | [
"MIT"
] | null | null | null | tf2-handson.ipynb | msavardi/tf2.0-handson | 5dfe760875290e19a1e25617b92537339eacb115 | [
"MIT"
] | null | null | null | 208.338037 | 87,008 | 0.889568 | [
[
[
"# Deep learning hands-on\n\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"## What frameworks do\n\n- Tensor math\n- Common network operations/layers\n- Gradient of common operations\n- Backpropagation\n- Optimizer\n- GPU implementation of the above\n- usally: data loading, serialization (saving/loading) of models\n- sometime: distributed computing\n- why not: production deployment on clusters/end devices/low power nodes\n\n\n**TensorFlow** is an end-to-end open source platform for machine learning. It has a comprehensive, flexible ecosystem of tools, libraries and community resources that lets researchers push the state-of-the-art in ML and developers easily build and deploy ML powered applications.\n\n[ref](https://www.tensorflow.org/tutorials): tf documentation",
"_____no_output_____"
],
[
"# Tensors\n\nA Tensor is a multi-dimensional array. Similar to NumPy ndarray objects, Tensor objects have a data type and a shape. Additionally, Tensors can reside in accelerator (like GPU) memory. TensorFlow offers a rich library of operations (tf.add, tf.matmul, tf.linalg.inv etc.) that consume and produce Tensors. These operations automatically convert native Python types. \n\n\n",
"_____no_output_____"
]
],
[
[
"%pylab inline\nimport tensorflow as tf",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"print(tf.add(1, 2))\nprint(tf.add([1, 2], [3, 4]))\nprint(tf.square(5))\nprint(tf.reduce_sum([1, 2, 3]))\n\n# Operator overloading is also supported\nprint(tf.square(2) + tf.square(3))",
"tf.Tensor(3, shape=(), dtype=int32)\ntf.Tensor([4 6], shape=(2,), dtype=int32)\ntf.Tensor(25, shape=(), dtype=int32)\ntf.Tensor(6, shape=(), dtype=int32)\ntf.Tensor(13, shape=(), dtype=int32)\n"
]
],
[
[
"\n\nEach Tensor has a shape and a datatype\n",
"_____no_output_____"
]
],
[
[
"x = tf.matmul([[1]], [[2, 3]])\nprint(x.shape)\nprint(x.dtype)",
"(1, 2)\n<dtype: 'int32'>\n"
]
],
[
[
"The most obvious differences between NumPy arrays and TensorFlow Tensors are:\n\n - Tensors can be backed by accelerator memory (like GPU, TPU).\n - Tensors are immutable.\n\n",
"_____no_output_____"
],
[
"## GPU acceleration\n\nMany TensorFlow operations can be accelerated by using the GPU for computation. Without any annotations, TensorFlow automatically decides whether to use the GPU or CPU for an operation (and copies the tensor between CPU and GPU memory if necessary). Tensors produced by an operation are typically backed by the memory of the device on which the operation executed. For example:\n",
"_____no_output_____"
]
],
[
[
"x = tf.random.uniform([3, 3])\n\nprint(\"Is there any GPUs available?\"),\nprint(tf.config.list_physical_devices('GPU'))\n\nprint(\"Is the Tensor on GPU #0: \"),\nprint(x.device.endswith('GPU:0'))",
"Is there any GPUs available?\n[]\nIs the Tensor on GPU #0: \nFalse\n"
]
],
[
[
"## Explicit Device Placement\n\nThe term \"placement\" in TensorFlow refers to how individual operations are assigned (placed on) a device for execution. As mentioned above, when there is no explicit guidance provided, TensorFlow automatically decides which device to execute an operation, and copies Tensors to that device if needed. However, TensorFlow operations can be explicitly placed on specific devices using the tf.device context manager. For example:\n",
"_____no_output_____"
]
],
[
[
"def time_matmul(x):\n %timeit tf.matmul(x, x) \n \n# Force execution on CPU\nprint(\"On CPU:\")\nwith tf.device(\"CPU:0\"):\n x = tf.random.uniform([1000, 1000])\n assert x.device.endswith(\"CPU:0\")\n time_matmul(x)\n\n# Force execution on GPU #0 if available\nif tf.test.is_gpu_available():\n with tf.device(\"GPU:0\"): # Or GPU:1 for the 2nd GPU, GPU:2 for the 3rd etc.\n x = tf.random.uniform([1000, 1000])\n assert x.device.endswith(\"GPU:0\")\n time_matmul(x)",
"On CPU:\n7.62 ms ± 1.12 ms per loop (mean ± std. dev. of 7 runs, 100 loops each)\nWARNING:tensorflow:From <ipython-input-6-750e9424349b>:12: is_gpu_available (from tensorflow.python.framework.test_util) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.config.list_physical_devices('GPU')` instead.\n"
]
],
[
[
"# Derivatives of a function - Computing gradients\n\nTensorFlow provides APIs for automatic differentiation - computing the derivative of a function. \n\nTo differentiate automatically, TensorFlow needs to remember what operations happen in what order during the forward pass. Then, during the backward pass, TensorFlow traverses this list of operations in reverse order to compute gradients.\n\n## Gradient tapes\n\nTensorFlow provides `tf.GradientTape` API for automatic differentiation; that is, computing the gradient of a computation with respect to its input variables. TensorFlow \"records\" all operations executed inside the context of a `tf.GradientTape` onto a \"tape\". TensorFlow then uses that tape and the gradients associated with each recorded operation to compute the gradients of a \"recorded\" computation using reverse mode differentiation.\n\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"# With scalars:\nx = tf.constant(3.0)\n# y = x ^ 2\nwith tf.GradientTape() as t:\n t.watch(x)\n y = x * x\n# dy = 2x\ndy_dx = t.gradient(y, x)\ndy_dx.numpy() ",
"_____no_output_____"
],
[
"# Using matrices:\nx = tf.constant([3.0, 3.0])\n\nwith tf.GradientTape() as t:\n t.watch(x)\n z = tf.multiply(x, x)\n\nprint(z)\n\n# Find derivative of z with respect to the original input tensor x\nprint(t.gradient(z, x))",
"tf.Tensor([9. 9.], shape=(2,), dtype=float32)\ntf.Tensor([6. 6.], shape=(2,), dtype=float32)\n"
],
[
"x = tf.constant([3.0, 3.0])\n\nwith tf.GradientTape() as t:\n t.watch(x)\n y = tf.multiply(x, x)\n z = tf.multiply(y, y)\n\n# Use the tape to compute the derivative of z with respect to the\n# intermediate value y.\n# dz_dx = 2 * y, where y = x ^ 2\nprint(t.gradient(z, y))",
"tf.Tensor([18. 18.], shape=(2,), dtype=float32)\n"
]
],
[
[
"## Recording control flow\n\nBecause tapes record operations as they are executed, Python control flow (using ifs and whiles for example) is naturally handled:",
"_____no_output_____"
]
],
[
[
"def f(x, y):\n output = 1.0\n for i in range(y):\n if i > 1 and i < 5:\n output *= x \n return output\n\ndef grad(x, y):\n with tf.GradientTape() as t:\n t.watch(x)\n out = f(x, y)\n return t.gradient(out, x)\n\nx = tf.constant(2.0)\n\nprint(grad(x, 6).numpy()) # 12.0\nprint(grad(x, 5).numpy()) # 12.0\nprint(grad(x, 4).numpy()) # 4.0\n",
"12.0\n12.0\n4.0\n"
]
],
[
[
"## Higher-order gradients\n\nOperations inside of the `GradientTape` context manager are recorded for automatic differentiation. If gradients are computed in that context, then the gradient computation is recorded as well. As a result, the exact same API works for higher-order gradients as well. For example:",
"_____no_output_____"
]
],
[
[
"x = tf.Variable(1.0) # Create a Tensorflow variable initialized to 1.0\n\nwith tf.GradientTape() as t:\n with tf.GradientTape() as t2:\n y = x * x * x\n\n # Compute the gradient inside the 't' context manager\n # which means the gradient computation is differentiable as well.\n dy_dx = t2.gradient(y, x)\nd2y_dx2 = t.gradient(dy_dx, x)\n\nprint(\"dy_dx:\", dy_dx.numpy()) # 3.0\nprint(\"d2y_dx2:\", d2y_dx2.numpy()) # 6.0",
"dy_dx: 3.0\nd2y_dx2: 6.0\n"
],
[
"def f(x):\n return tf.square(tf.sin(x))\n\nx = tf.Variable(3.1415/2)\nwith tf.GradientTape() as t:\n y = f(x)\n \nt.gradient(y,x)",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\nx = tf.constant(np.linspace(0,2*np.pi,100))\nwith tf.GradientTape() as t:\n t.watch(x)\n y = f(x)\n \ng = t.gradient(y,x)\nplt.plot(x.numpy(), g.numpy(), x.numpy(), y.numpy())",
"_____no_output_____"
]
],
[
[
"# Canny edge\n\nCanny, John. \"A computational approach to edge detection.\" IEEE Transactions on pattern analysis and machine intelligence 6 (1986): 679-698. (27000+ citations)\nCanny:\n- assume noisy step edges\n- construct an edge detector using an optimal linear filter\n\nThis is actually a simple neural network...",
"_____no_output_____"
]
],
[
[
"# package loading, helper functions\n\nimport os\nimport pickle\nimport numpy as np\nimport scipy.ndimage as ndi\nimport sklearn\nimport tensorflow.keras \nimport tensorflow.keras.backend as K\nimport tensorflow as tf\nimport matplotlib.pyplot as plt\n\n#Source(\"digraph {X->Y; Y->Z;}\")\nfrom graphviz import Source\nfrom scipy.stats import kde\nfrom sklearn import decomposition\n\n\ndef tshow(v, ax=None, **keys):\n if isinstance(v, tf.Tensor):\n v = v.float_val\n if v.ndim == 1:\n v = v.reshape(28, 28)\n if v.ndim == 3 and v.shape[0] == 1:\n v = v[0]\n if v.ndim == 3 and v.shape[2] == 1:\n v = v[:, :, 0]\n if v.ndim == 4:\n v = v[0, 0]\n v = v - amin(v)\n v /= amax(v)\n if ax is not None:\n ax.imshow(v, **keys)\n else:\n imshow(v, **keys)\n\n \ndef showrow(*args, **kw):\n figsize(*kw.get(\"fs\", (15, 5)))\n if \"fs\" in kw:\n del kw[\"fs\"]\n for i, im in enumerate(args):\n subplot(1, len(args), i+1)\n tshow(im, **kw)\n\ndef showgrid(images, rows=4, cols=4, cmap=plt.cm.gray, size=(7, 7)):\n if size is not None:\n figsize(*size)\n for i in range(rows*cols):\n subplot(rows, cols, i+1)\n xticks([])\n yticks([])\n tshow(images[i], cmap=cmap)",
"_____no_output_____"
]
],
[
[
"The Process of Canny edge detection algorithm can be broken down to 5 different steps:\n\n- Apply Gaussian filter to smooth the image in order to remove the noise\n- Find the intensity gradients of the image\n- Apply non-maximum suppression to get rid of spurious response to edge detection\n- Apply double threshold to determine potential edges\n- Track edge by hysteresis: Finalize the detection of edges by suppressing all the other edges that are weak and not connected to strong edges.\n\n",
"_____no_output_____"
]
],
[
[
"length = 50\npos = np.random.randint(length, size=10000)\nsignal = np.random.rand(10000,length) / 5\ngt = np.zeros((10000,length))\n\nfor j,i in enumerate(pos):\n signal[j, i:] = signal[j, i:] + 0.5\n gt[j, i] = 1\n\nfigure(figsize=(14,4))\nplt.plot(signal[15])\nplt.plot(gt[15])",
"_____no_output_____"
],
[
"from tensorflow.keras.models import Sequential\nfrom tensorflow.keras.layers import Conv1D, Flatten\nfrom tensorflow.keras.optimizers import SGD",
"_____no_output_____"
],
[
"# Canny model\nmodel = Sequential()\nmodel.add(Conv1D(1, 33, input_shape=(length, 1), activation=\"sigmoid\", padding='same'))\nmodel.add(Flatten())\n\nmodel.compile(SGD(lr=.1, momentum=0.9, decay=1e-5), loss='categorical_crossentropy')\nmodel.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv1d (Conv1D) (None, 50, 1) 34 \n_________________________________________________________________\nflatten (Flatten) (None, 50) 0 \n=================================================================\nTotal params: 34\nTrainable params: 34\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"K.set_value(model.optimizer.lr, 1e-3)\nmodel.fit(x = np.expand_dims(signal, 3), y=gt, batch_size=100, epochs=100)",
"/usr/local/lib/python3.7/site-packages/ipykernel_launcher.py:2: DeprecationWarning: Both axis > a.ndim and axis < -a.ndim - 1 are deprecated and will raise an AxisError in the future.\n \n"
],
[
"figure(figsize=(14,4))\nylim(ymax=1)\n\na = 128\nplt.plot(signal[a,:].reshape((length)))\nplt.plot(gt[a,:], '-.')\nplt.plot(model.predict(signal[a,:].reshape((1,length,1))).reshape(length))",
"_____no_output_____"
]
],
[
[
"## Kanny kernel",
"_____no_output_____"
]
],
[
[
"plt.plot(model.get_layer(index=0).get_weights( )[0].flatten())",
"_____no_output_____"
]
],
[
[
"There is a large and rich theory of linear filters in signal processing and image processing:\n\n - mathematical properties: superposition, decomposition, impulse response\n - frequency domain analysis\n - optimal design for given tasks\n\nNB: \"optimal linear\" is not the same as \"optimal\"\n\n# No free lunch theorem!",
"_____no_output_____"
],
[
"A model, a family of function, a neural net architecture+regularizer, a likelihood model+prior, are all forms of inductive bias.\nAnd we know that there is no free lunch, meaning that:\n- There is no learning without inductive bias.\n- There is no neural net training without an architecture.\n- There is no statistical estimation without a likelihood model.\n- There is no non-parametric estimation without regularization.\n\nWithout some sort of inductive bias\n- There is no estimation of probability distribution.\n- There is no estimation of entropy.\n- There is no estimation of mutual information.\n- There is no estimation of conditional independence.\n- There is no measure of complexity\n- There is no estimation of minimum description length.\n\nWhich means that none of these things are well defined quantities (except perhaps in the asymptotic case of infinite data. But who cares about that). \nThe estimation of all of these quantities is subjectively dependent upon your choice of model.\nYou may say: \"the entropy of my data is well defined. It's `H = -SUM_x P(x) log P(x)`\nYes, but what is `P(x)`? \nYou only know `P(x)` through a bunch of samples. \n\nWhich mean you need to estimate a model of `P(x)` from your data.\nWhich means your model will necessarily have some sort of inductive bias, some sort of arbitrariness in it.\n\n\nUltimately, all measures of distributions, information, entropy, complexity and dependency are in the eye of the beholder.\n\nThe subjectivity of those quantities also exists when applied to physical systems. The entropy of a physical system is also in the eyes of the beholder.",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.layers import BatchNormalization, Activation, ZeroPadding1D ",
"_____no_output_____"
],
[
"# DL model\nmodel = Sequential()\nmodel.add(ZeroPadding1D(input_shape=(length, 1), padding=(8,1)))\nmodel.add(Conv1D(4, 7))\nmodel.add(BatchNormalization(momentum=0.1))\nmodel.add(Activation('relu'))\nmodel.add(ZeroPadding1D(padding=(2,1)))\nmodel.add(Conv1D(1, 7))\nmodel.add(Flatten())\nmodel.add(Activation('sigmoid'))\n\nmodel.compile(SGD(lr=.1, momentum=0.9, decay=1e-5), loss='categorical_crossentropy')\nmodel.summary()",
"Model: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nzero_padding1d (ZeroPadding1 (None, 59, 1) 0 \n_________________________________________________________________\nconv1d_1 (Conv1D) (None, 53, 4) 32 \n_________________________________________________________________\nbatch_normalization (BatchNo (None, 53, 4) 16 \n_________________________________________________________________\nactivation (Activation) (None, 53, 4) 0 \n_________________________________________________________________\nzero_padding1d_1 (ZeroPaddin (None, 56, 4) 0 \n_________________________________________________________________\nconv1d_2 (Conv1D) (None, 50, 1) 29 \n_________________________________________________________________\nflatten_1 (Flatten) (None, 50) 0 \n_________________________________________________________________\nactivation_1 (Activation) (None, 50) 0 \n=================================================================\nTotal params: 77\nTrainable params: 69\nNon-trainable params: 8\n_________________________________________________________________\n"
],
[
"K.set_value(model.optimizer.lr, 1e-3)\nmodel.fit(x = np.expand_dims(signal, 3), \n y=gt, \n batch_size=200, \n epochs=50)",
"/usr/local/lib/python3.7/site-packages/ipykernel_launcher.py:2: DeprecationWarning: Both axis > a.ndim and axis < -a.ndim - 1 are deprecated and will raise an AxisError in the future.\n \n"
],
[
"figure(figsize=(14,4))\nylim(ymax=1)\na = 10\nplt.plot(signal[a,:].reshape((length)))\nplt.plot(gt[a,:], '-.')\nplt.plot(model.predict(signal[a,:].reshape((1,length,1))).reshape(length))",
"_____no_output_____"
],
[
"for i, l in enumerate(model.layers):\n print (i, l.name)",
"0 zero_padding1d\n1 conv1d_1\n2 batch_normalization\n3 activation\n4 zero_padding1d_1\n5 conv1d_2\n6 flatten_1\n7 activation_1\n"
]
],
[
[
"## How does it works?",
"_____no_output_____"
]
],
[
[
"figure(figsize=(14,4))\n\nx = signal[a,:].reshape((1,length,1))\nlayer = model.layers[1]\nf = K.function([model.get_input_at(0)], [layer.get_output_at(0)])\nplt.plot(f([x])[0][0,:,:]) # your activation tensor\nplt.plot(gt[a,:], '-.')",
"_____no_output_____"
],
[
"figure(figsize=(14,6))\n\nlayer = model.layers[2]\nf = K.function([model.get_input_at(0)], [layer.get_output_at(0)])\nplt.plot(f([x])[0][0,:,:]) # your activation tensor\nplt.plot(gt[a,:], '-.')",
"_____no_output_____"
],
[
"figure(figsize=(14,4))\n\nlayer = model.layers[5]\nf = K.function([model.get_input_at(0)], [layer.get_output_at(0)])\nplt.plot(f([x])[0][0,:,:]) # your activation tensor\nplt.plot(gt[a,:], '-.')",
"_____no_output_____"
]
],
[
[
"- Zero padding on the boundary creates spurious edge responses.\n- DL can automatically suppress these for you (beware when benchmarking)\n- How would you pick weights to get rid of such spurious responses?",
"_____no_output_____"
]
],
[
[
"# DL model\nmodel = Sequential()\nmodel.add(ZeroPadding1D(input_shape=(length, 1), padding=(8,1)))\nmodel.add(Conv1D(16, 7))\nmodel.add(BatchNormalization(momentum=0.1))\nmodel.add(Activation('relu'))\nmodel.add(ZeroPadding1D(padding=(2,1)))\nmodel.add(Conv1D(1, 7))\nmodel.add(Flatten())\nmodel.add(Activation('sigmoid'))\n\nmodel.compile(SGD(lr=1e-2, momentum=0.9, decay=1e-5), loss='categorical_crossentropy')\n\nmodel.fit(x = np.expand_dims(signal, 3), y=gt, batch_size=200, epochs=50)",
"/usr/local/lib/python3.7/site-packages/ipykernel_launcher.py:14: DeprecationWarning: Both axis > a.ndim and axis < -a.ndim - 1 are deprecated and will raise an AxisError in the future.\n \n"
],
[
"figure(figsize=(18,6))\n\nx = np.zeros(length)\nx[int(length/2):] = 1\nx = x.reshape((1,length,1))\n\nlayer = model.layers[4]\nf = K.function([model.get_input_at(0)], [layer.get_output_at(0)])\nplt.plot(f([x])[0][0,6:,:]) # your activation tensor\nplt.plot(7*x.flatten(), '-.', linewidth=4)",
"_____no_output_____"
]
],
[
[
"- DL discovered another neat trick: Instead of a single edge localizer, train two localization filters in the first layer (plus multiple suppression filters again).\n- Each localization filter is offset from the desired peak by one pixel.\n- The padding on the second convolutional layer means that the spurious\nedges on the boundary are only surrounded by one peak.",
"_____no_output_____"
],
[
"# SVM \n\n Let's try to implement a standard L2-regularized support vector machine (SVM)\n \n \n \n where:\n \n ",
"_____no_output_____"
]
],
[
[
"from sklearn import svm\nfrom tensorflow.keras.layers import Input, Dense, Activation\nfrom tensorflow.keras.models import Model\nfrom tensorflow.keras.regularizers import L1L2\nimport tensorflow.keras.backend as K",
"_____no_output_____"
],
[
"def make_meshgrid(x, y, h=.02):\n \"\"\"Create a mesh of points to plot in\n\n Parameters\n ----------\n x: data to base x-axis meshgrid on\n y: data to base y-axis meshgrid on\n h: stepsize for meshgrid, optional\n\n Returns\n -------\n xx, yy : ndarray\n \"\"\"\n x_min, x_max = x.min() - 1, x.max() + 1\n y_min, y_max = y.min() - 1, y.max() + 1\n xx, yy = np.meshgrid(np.arange(x_min, x_max, h),\n np.arange(y_min, y_max, h))\n return xx, yy\n\n\ndef plot_contours(ax, clf, xx, yy, **params):\n \"\"\"Plot the decision boundaries for a classifier.\n\n Parameters\n ----------\n ax: matplotlib axes object\n clf: a classifier\n xx: meshgrid ndarray\n yy: meshgrid ndarray\n params: dictionary of params to pass to contourf, optional\n \"\"\"\n Z = clf.predict(np.c_[xx.ravel(), yy.ravel()]) > .5\n Z = Z.reshape(xx.shape)\n out = ax.contourf(xx, yy, Z, **params)\n return out\n",
"_____no_output_____"
],
[
"rng = np.random.RandomState(0)\nn_samples_1 = 1000\nn_samples_2 = 1000\nX = np.r_[1. * rng.randn(n_samples_1, 2),\n 1. * rng.randn(n_samples_2, 2) + [2, 2]]\ny = [0] * (n_samples_1) + [1] * (n_samples_2)\n\n# we create an instance of SVM and fit out data. We do not scale our\n# data since we want to plot the support vectors\nC = 1.0 # SVM regularization parameter\nmodel = svm.LinearSVC(C=C)\nclf = model.fit(X, y)\n\nX0, X1 = X[:, 0], X[:, 1]\nxx, yy = make_meshgrid(X0, X1)\n\nplot_contours(plt, clf, xx, yy,\n cmap=plt.cm.coolwarm, alpha=0.8)\nplt.scatter(X0, X1, c=y, cmap=plt.cm.coolwarm, s=20, edgecolors='k')\n\nplt.show()",
"_____no_output_____"
],
[
"def hinge(y_true, y_pred):\n return K.mean(K.maximum(1. - y_true * y_pred, 0.), axis=-1)\n\nl2 = 1/float(C)\ninputs = Input(shape=(2,))\nx = Dense(1, activation=None, kernel_regularizer=L1L2(l1=0,l2=l2))(inputs)\npredictions = Activation('linear')(x)\n\nmodel = Model(inputs=inputs, outputs=predictions)\nmodel.compile(optimizer='sgd',\n loss=hinge,\n metrics=['accuracy'])\n\nmodel.summary()\n\n# Wx + b",
"Model: \"model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 2)] 0 \n_________________________________________________________________\ndense (Dense) (None, 1) 3 \n_________________________________________________________________\nactivation_4 (Activation) (None, 1) 0 \n=================================================================\nTotal params: 3\nTrainable params: 3\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"K.set_value(model.optimizer.lr, 1e-3)\nhistory = model.fit(x=X, y=np.array(y), \n batch_size=100, \n epochs=100, \n shuffle=True, \n validation_split=.3)",
"Epoch 1/100\n14/14 [==============================] - 0s 14ms/step - loss: 3.8389 - accuracy: 0.4507 - val_loss: 6.2210 - val_accuracy: 0.0000e+00\nEpoch 2/100\n14/14 [==============================] - 0s 2ms/step - loss: 3.6745 - accuracy: 0.4536 - val_loss: 5.9683 - val_accuracy: 0.0000e+00\nEpoch 3/100\n14/14 [==============================] - 0s 3ms/step - loss: 3.5189 - accuracy: 0.4564 - val_loss: 5.7268 - val_accuracy: 0.0000e+00\nEpoch 4/100\n14/14 [==============================] - 0s 3ms/step - loss: 3.3718 - accuracy: 0.4579 - val_loss: 5.4958 - val_accuracy: 0.0000e+00\nEpoch 5/100\n14/14 [==============================] - 0s 3ms/step - loss: 3.2326 - accuracy: 0.4600 - val_loss: 5.2750 - val_accuracy: 0.0000e+00\nEpoch 6/100\n14/14 [==============================] - 0s 3ms/step - loss: 3.1010 - accuracy: 0.4607 - val_loss: 5.0638 - val_accuracy: 0.0000e+00\nEpoch 7/100\n14/14 [==============================] - 0s 3ms/step - loss: 2.9764 - accuracy: 0.4636 - val_loss: 4.8616 - val_accuracy: 0.0000e+00\nEpoch 8/100\n14/14 [==============================] - 0s 3ms/step - loss: 2.8586 - accuracy: 0.4657 - val_loss: 4.6682 - val_accuracy: 0.0000e+00\nEpoch 9/100\n14/14 [==============================] - 0s 3ms/step - loss: 2.7472 - accuracy: 0.4679 - val_loss: 4.4830 - val_accuracy: 0.0000e+00\nEpoch 10/100\n14/14 [==============================] - 0s 3ms/step - loss: 2.6417 - accuracy: 0.4693 - val_loss: 4.3057 - val_accuracy: 0.0000e+00\nEpoch 11/100\n14/14 [==============================] - 0s 3ms/step - loss: 2.5419 - accuracy: 0.4764 - val_loss: 4.1359 - val_accuracy: 0.0000e+00\nEpoch 12/100\n14/14 [==============================] - 0s 2ms/step - loss: 2.4476 - accuracy: 0.4814 - val_loss: 3.9733 - val_accuracy: 0.0000e+00\nEpoch 13/100\n14/14 [==============================] - 0s 3ms/step - loss: 2.3583 - accuracy: 0.4829 - val_loss: 3.8175 - val_accuracy: 0.0000e+00\nEpoch 14/100\n14/14 [==============================] - 0s 3ms/step - loss: 2.2738 - accuracy: 0.4900 - val_loss: 3.6682 - val_accuracy: 0.0000e+00\nEpoch 15/100\n14/14 [==============================] - 0s 3ms/step - loss: 2.1939 - accuracy: 0.4957 - val_loss: 3.5251 - val_accuracy: 0.0000e+00\nEpoch 16/100\n14/14 [==============================] - 0s 3ms/step - loss: 2.1182 - accuracy: 0.5000 - val_loss: 3.3878 - val_accuracy: 0.0000e+00\nEpoch 17/100\n14/14 [==============================] - 0s 3ms/step - loss: 2.0466 - accuracy: 0.5064 - val_loss: 3.2562 - val_accuracy: 0.0000e+00\nEpoch 18/100\n14/14 [==============================] - 0s 2ms/step - loss: 1.9789 - accuracy: 0.5114 - val_loss: 3.1300 - val_accuracy: 0.0000e+00\nEpoch 19/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.9148 - accuracy: 0.5150 - val_loss: 3.0089 - val_accuracy: 0.0000e+00\nEpoch 20/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.8541 - accuracy: 0.5229 - val_loss: 2.8926 - val_accuracy: 0.0000e+00\nEpoch 21/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.7966 - accuracy: 0.5279 - val_loss: 2.7810 - val_accuracy: 0.0000e+00\nEpoch 22/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.7422 - accuracy: 0.5329 - val_loss: 2.6739 - val_accuracy: 0.0000e+00\nEpoch 23/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.6908 - accuracy: 0.5393 - val_loss: 2.5710 - val_accuracy: 0.0000e+00\nEpoch 24/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.6421 - accuracy: 0.5500 - val_loss: 2.4721 - val_accuracy: 0.0000e+00\nEpoch 25/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.5959 - accuracy: 0.5543 - val_loss: 2.3771 - val_accuracy: 0.0000e+00\nEpoch 26/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.5522 - accuracy: 0.5636 - val_loss: 2.2858 - val_accuracy: 0.0000e+00\nEpoch 27/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.5109 - accuracy: 0.5700 - val_loss: 2.1980 - val_accuracy: 0.0000e+00\nEpoch 28/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.4717 - accuracy: 0.5750 - val_loss: 2.1136 - val_accuracy: 0.0017\nEpoch 29/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.4346 - accuracy: 0.5821 - val_loss: 2.0324 - val_accuracy: 0.0017\nEpoch 30/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.3995 - accuracy: 0.5893 - val_loss: 1.9543 - val_accuracy: 0.0017\nEpoch 31/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.3662 - accuracy: 0.5971 - val_loss: 1.8791 - val_accuracy: 0.0017\nEpoch 32/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.3347 - accuracy: 0.6043 - val_loss: 1.8067 - val_accuracy: 0.0017\nEpoch 33/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.3048 - accuracy: 0.6129 - val_loss: 1.7370 - val_accuracy: 0.0017\nEpoch 34/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.2765 - accuracy: 0.6271 - val_loss: 1.6698 - val_accuracy: 0.0017\nEpoch 35/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.2497 - accuracy: 0.6350 - val_loss: 1.6051 - val_accuracy: 0.0017\nEpoch 36/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.2243 - accuracy: 0.6457 - val_loss: 1.5428 - val_accuracy: 0.0017\nEpoch 37/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.2002 - accuracy: 0.6479 - val_loss: 1.4827 - val_accuracy: 0.0017\nEpoch 38/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.1773 - accuracy: 0.6543 - val_loss: 1.4248 - val_accuracy: 0.0017\nEpoch 39/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.1556 - accuracy: 0.6607 - val_loss: 1.3688 - val_accuracy: 0.0017\nEpoch 40/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.1351 - accuracy: 0.6664 - val_loss: 1.3149 - val_accuracy: 0.0017\nEpoch 41/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.1156 - accuracy: 0.6743 - val_loss: 1.2629 - val_accuracy: 0.0017\nEpoch 42/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.0971 - accuracy: 0.6829 - val_loss: 1.2126 - val_accuracy: 0.0017\nEpoch 43/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.0796 - accuracy: 0.6886 - val_loss: 1.1642 - val_accuracy: 0.0017\nEpoch 44/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.0629 - accuracy: 0.6943 - val_loss: 1.1173 - val_accuracy: 0.0017\nEpoch 45/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.0471 - accuracy: 0.6986 - val_loss: 1.0720 - val_accuracy: 0.0017\nEpoch 46/100\n14/14 [==============================] - 0s 2ms/step - loss: 1.0321 - accuracy: 0.7007 - val_loss: 1.0283 - val_accuracy: 0.0017\nEpoch 47/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.0178 - accuracy: 0.7029 - val_loss: 0.9860 - val_accuracy: 0.0017\nEpoch 48/100\n14/14 [==============================] - 0s 3ms/step - loss: 1.0043 - accuracy: 0.7050 - val_loss: 0.9451 - val_accuracy: 0.0017\nEpoch 49/100\n14/14 [==============================] - 0s 4ms/step - loss: 0.9915 - accuracy: 0.7057 - val_loss: 0.9056 - val_accuracy: 0.0017\nEpoch 50/100\n14/14 [==============================] - 0s 3ms/step - loss: 0.9792 - accuracy: 0.7064 - val_loss: 0.8674 - val_accuracy: 0.0017\nEpoch 51/100\n14/14 [==============================] - 0s 3ms/step - loss: 0.9676 - accuracy: 0.7100 - val_loss: 0.8304 - val_accuracy: 0.0017\nEpoch 52/100\n14/14 [==============================] - 0s 2ms/step - loss: 0.9566 - accuracy: 0.7114 - val_loss: 0.7946 - val_accuracy: 0.0050\nEpoch 53/100\n14/14 [==============================] - 0s 2ms/step - loss: 0.9461 - accuracy: 0.7121 - val_loss: 0.7599 - val_accuracy: 0.0117\nEpoch 54/100\n14/14 [==============================] - 0s 3ms/step - loss: 0.9361 - accuracy: 0.7136 - val_loss: 0.7263 - val_accuracy: 0.0233\nEpoch 55/100\n14/14 [==============================] - 0s 3ms/step - loss: 0.9266 - accuracy: 0.7157 - val_loss: 0.6938 - val_accuracy: 0.0550\nEpoch 56/100\n14/14 [==============================] - 0s 3ms/step - loss: 0.9175 - accuracy: 0.7221 - val_loss: 0.6622 - val_accuracy: 0.0967\nEpoch 57/100\n14/14 [==============================] - 0s 2ms/step - loss: 0.9089 - accuracy: 0.7393 - val_loss: 0.6317 - val_accuracy: 0.1533\n"
],
[
"plt.plot(history.history['loss'])\nplt.plot(history.history['val_loss'])\nplt.plot(history.history['accuracy'])\nplt.plot(history.history['val_accuracy'])\nplt.title('model loss')\nplt.ylabel('loss')\nplt.xlabel('epoch')\nplt.legend(['train', 'test'], loc='upper right');",
"_____no_output_____"
],
[
"plot_contours(plt, model, xx, yy,\n cmap=plt.cm.coolwarm, alpha=0.8)\nplt.scatter(X0, X1, c=y, cmap=plt.cm.coolwarm, s=20, edgecolors='k')",
"_____no_output_____"
]
],
[
[
"# IIR Filters and their DL Equivalents",
"_____no_output_____"
]
],
[
[
"# simple IIR filter\n\ndef simple_iir(xs):\n value = xs[0]\n output = zeros(len(xs))\n for i in range(len(xs)):\n output[i] = value\n value = 0.7 * value + 0.3 * xs[i]\n return output",
"_____no_output_____"
],
[
"xs = rand(100) * (rand(100) < 0.1)\n\nfigure(figsize=(14,5))\nplot(xs)\nplot(simple_iir(xs))",
"_____no_output_____"
]
],
[
[
"## IIR Filters\n\nIIR filters are simple linear filters.\n\nUnlike FIR filters, the output is a linear function of both inputs and past output values.\n\nIIR filters can approximate FIR filters well.\n\nIIR filters are the linear equivalent of *recurrent neural networks*.\n",
"_____no_output_____"
]
],
[
[
"inputs = array([(rand(200) < 0.1).astype('f') for i in range(10000)]).reshape(-1, 1, 200)\noutputs = array([4.0*ndi.gaussian_filter(s, 3.0) for s in inputs]).reshape(-1, 1, 200)\n\nfigure(figsize=(14,5))\nplot(inputs[0,0]); plot(outputs[0,0])",
"_____no_output_____"
]
],
[
[
"### Simple NN",
"_____no_output_____"
]
],
[
[
"length = 200\n\nmodel = Sequential()\nmodel.add(Conv1D(1, 33, input_shape=(length, 1), activation=\"sigmoid\", padding='same'))\nmodel.add(Flatten())\n\nmodel.compile(SGD(lr=.1, momentum=0.9, decay=1e-5), loss='mse')\nmodel.summary()",
"_____no_output_____"
],
[
"#K.set_value(model.optimizer.lr, 1e-2)\nmodel.fit(x=inputs.reshape((-1, 200,1)), y=outputs.reshape((-1, 200)), batch_size=100, epochs=100)",
"_____no_output_____"
],
[
"pred = model.predict(inputs.reshape((-1, 200,1)))\n\nfigure(figsize=(15,5))\nplot(inputs[0,0])\nplot(pred[0], linewidth=2, color=\"green\")\nplot(outputs[0,0], linewidth=8, alpha=0.3, color=\"red\")",
"_____no_output_____"
]
],
[
[
"### Recurrent neural network - LSTM",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.layers import LSTM, Bidirectional",
"_____no_output_____"
],
[
"length = 200\n\nmodel = Sequential()\nmodel.add(Bidirectional(LSTM(4, input_shape=(length, 1), activation=\"sigmoid\", return_sequences=True)))\nmodel.add(Conv1D(1, 8, padding='same'))\n\nmodel.compile(SGD(lr=.1, momentum=0.9, decay=1e-5), loss='mse')\nmodel.build(input_shape=(None, 200,1))",
"_____no_output_____"
],
[
"model.summary()",
"_____no_output_____"
],
[
"K.set_value(model.optimizer.lr, 1e-1)\nmodel.fit(x=inputs.reshape((-1, 200,1)), \n y=outputs.reshape((-1, 200,1)), \n batch_size=1000, \n epochs=15)",
"_____no_output_____"
],
[
"pred = model.predict(inputs.reshape((-1, 200,1)))\n\nfigure(figsize=(15,5))\nplot(inputs[0,0])\nplot(pred[0], linewidth=2, color=\"green\")\nplot(outputs[0,0], linewidth=8, alpha=0.3, color=\"red\")",
"_____no_output_____"
]
],
[
[
"# Handsome input pipelines with the `tf.data` API \n\nThe `tf.data` API offers functions for data pipelining and related operations. We can build pipelines, map preprocessing functions, shuffle or batch a dataset and much more.",
"_____no_output_____"
],
[
"## From tensors",
"_____no_output_____"
]
],
[
[
"dataset = tf.data.Dataset.from_tensor_slices([8, 3, 0, 8, 2, 1])\niter(dataset).next().numpy()",
"_____no_output_____"
]
],
[
[
"## Batch and Shuffle",
"_____no_output_____"
]
],
[
[
"# Shuffle\ndataset = tf.data.Dataset.from_tensor_slices([8, 3, 0, 8, 2, 1]).shuffle(6)\niter(dataset).next().numpy()",
"_____no_output_____"
],
[
"# Batch\ndataset = tf.data.Dataset.from_tensor_slices([8, 3, 0, 8, 2, 1]).batch(2)\niter(dataset).next().numpy()",
"_____no_output_____"
],
[
"# Shuffle and Batch\ndataset = tf.data.Dataset.from_tensor_slices([8, 3, 0, 8, 2, 1]).shuffle(6).batch(2)\niter(dataset).next().numpy()",
"_____no_output_____"
]
],
[
[
"## Zipping Two Datsets",
"_____no_output_____"
]
],
[
[
"dataset0 = tf.data.Dataset.from_tensor_slices([8, 3, 0, 8, 2, 1])\ndataset1 = tf.data.Dataset.from_tensor_slices([1, 2, 3, 4, 5, 6])\ndataset = tf.data.Dataset.zip((dataset0, dataset1))\niter(dataset).next()",
"_____no_output_____"
]
],
[
[
"## Mapping External Functions",
"_____no_output_____"
]
],
[
[
"def into_2(num):\n return num * 2\n\ndataset = tf.data.Dataset.from_tensor_slices([8, 3, 0, 8, 2, 1]).map(into_2)",
"_____no_output_____"
]
],
[
[
"## ImageDataGenerator\n\nThis is one of the best features of the `tensorflow.keras` API (in my opinion). The `ImageDataGenerator` is capable of generating dataset slices while batching and preprocessing along with data augmentation in real-time.\n\nThe Generator allows data flow directly from directories or from dataframes.\n\nA misconception about data augmentation in `ImageDataGenerator` is that, it adds more data to the existing dataset. Although that is the actual definition of data augmentation, in `ImageDataGenerator`, the images in the dataset are transformed dynamically at different steps in training so that the model can be trained on noisy data it hasn’t seen.",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.preprocessing.image import ImageDataGenerator\n\ntrain_datagen = ImageDataGenerator(\n rescale=1./255,\n shear_range=0.2,\n zoom_range=0.2,\n horizontal_flip=True\n)",
"_____no_output_____"
]
],
[
[
"Here, the rescaling is done on all the samples (for normalizing), while the other parameters are for augmentation.",
"_____no_output_____"
]
],
[
[
"train_generator = train_datagen.flow_from_directory(\n 'data/train',\n target_size=(150, 150),\n batch_size=32,\n class_mode='binary'\n)",
"_____no_output_____"
]
],
[
[
"# Custom Layers\n\nNeural Nets are known for many layer deep networks wherein the layers can be of different types. TensorFlow contains many predefined layers (like Dense, LSTM, etc.). But for more complex architectures, the logic of a layer is much more complex than a primary layer. For such instances, TensorFlow allows building custom layers. This can be done by subclassing the tf.keras.layers.Layer class.",
"_____no_output_____"
]
],
[
[
"class CustomDense(tf.keras.layers.Layer):\n def __init__(self, num_outputs):\n super(CustomDense, self).__init__()\n self.num_outputs = num_outputs\n\n def build(self, input_shape):\n self.kernel = self.add_weight(\n \"kernel\",\n shape=[int(input_shape[-1]),\n self.num_outputs]\n )\n\n def call(self, input):\n return tf.matmul(input, self.kernel)",
"_____no_output_____"
]
],
[
[
"As stated in the documentation, The best way to implement your own layer is extending the `tf.keras.Layer` class and implementing:\n\n- `__init__` , where you can do all input-independent initialization.\n- `build`, where you know the shapes of the input tensors and can do the rest of the initialization.\n- `call`, where you do the forward computation.\n\nAlthough the kernel initialization can be done in `__init__` itself, it is considered better to be initialized in build as otherwise, you would have to explicitly specify the input_shape on every instance of a new layer creation.",
"_____no_output_____"
],
[
"# Custom Training\n\nThe `tf.keras` Sequential and the Model API makes training models easier. However, most of the time while training complex models, custom loss functions are used. Moreover, the model training can also differ from the default (for eg. applying gradients separately to different model components).\n\nTensorFlow’s automatic differentiation helps calculating gradients in an efficient way. These primitives are used in defining custom training loops.\n\n",
"_____no_output_____"
]
],
[
[
"def train(model, inputs, outputs, learning_rate):\n with tf.GradientTape() as t:\n # Computing Losses from Model Prediction\n current_loss = loss(outputs, model(inputs)) # Gradients for Trainable Variables with Obtained Losses\n dW, db = t.gradient(current_loss, [model.W, model.b]) # Applying Gradients to Weights\n model.W.assign_sub(learning_rate * dW)\n model.b.assign_sub(learning_rate * db)",
"_____no_output_____"
]
],
[
[
"This loop can be repeated for multiple epochs and with a more customised setting as per the use case.",
"_____no_output_____"
],
[
"# Autoencoders\n\npredict the input, given that same input. \n\n\\begin{equation}\\label{eq:}\nF_{W,b}(x) \\approx x\n\\end{equation}\n\nThe goal is to learn a compressed representation of the data, thus find structure. This can be done by limiting the number of hidden units in the model. Those kind of autoencoders are called *undercomplete*.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0f44edf97caf7a957596718ac28abe258f370fd | 78,096 | ipynb | Jupyter Notebook | ml_algos/linear_reg_lasso.ipynb | igor-davidyuk/ds_interview | 832d1b742304e6f160fe9538e5966bd9864b3189 | [
"MIT"
] | null | null | null | ml_algos/linear_reg_lasso.ipynb | igor-davidyuk/ds_interview | 832d1b742304e6f160fe9538e5966bd9864b3189 | [
"MIT"
] | null | null | null | ml_algos/linear_reg_lasso.ipynb | igor-davidyuk/ds_interview | 832d1b742304e6f160fe9538e5966bd9864b3189 | [
"MIT"
] | null | null | null | 221.863636 | 39,576 | 0.904989 | [
[
[
"from typing import List, Union\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"# Lasso just adds L1 regularization\n",
"_____no_output_____"
]
],
[
[
"class LinRegLasso:\n def _init(self, n_feat: int) -> None:\n self.weights = np.ones((n_feat + 1)) # (n_feat + 1,) weights + bias\n \n def predict(self, feature_vector: Union[np.ndarray, List[int]]) -> float:\n '''\n feature_vector may be a list or have shape (n_feat,)\n or it may be a bunch of vectors (n_vec, nfeat)\n '''\n feature_vector = np.array(feature_vector)\n assert feature_vector.shape[-1] == self.weights.shape[0] - 1\n if len(feature_vector.shape) == 1:\n feature_vector = feature_vector[np.newaxis,]\n \n return self.weights @ np.concatenate((feature_vector.T, [[1]*feature_vector.shape[0]]))\n \n def mse(self, X, Y):\n Y_hat = self.predict(X)\n return np.sum((Y - Y_hat)**2) / Y.shape[0]\n\n def _update_weights(self, X, Y, lr, wd):\n '''\n X: (n_samples, n_features)\n Y: (n_samples,)\n self.weights: (n_features + 1)\n \n Cost function is MSE: (y - W*X - b)**2;\n its derivative with resp to any x is -2*X*(y - W*X - b),\n and with resp to b is -2*(y - W*X - b).\n \n Regularisation function is L1 |W|;\n its derivative is SIGN(w)\n '''\n predictions = self.predict(X)\n error = Y - predictions # (n_samples,)\n X_with_bias = np.concatenate((X.T, [[1]*X.shape[0]])).T\n updates = -2 * X_with_bias.T @ error / Y.shape[0]\n regression_term = np.sign(self.weights)\n \n self.weights -= lr * updates + wd * regression_term\n \n def fit(self, X, Y, n_epochs: int, lr: float, wd: float):\n self._init(X.shape[-1])\n for i in range(n_epochs):\n self._update_weights(X, Y, lr, wd)\n mse = self.mse(X, Y)\n print(f'epoch: {i}, \\t MSE: {mse}')\n ",
"_____no_output_____"
],
[
"import random\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom matplotlib.pylab import rcParams\nrcParams['figure.figsize'] = 12, 10\n\n#Define input array with angles from 60deg to 300deg converted to radians\nx = np.array([i*np.pi/180 for i in range(60,300,4)])\nnp.random.seed(10) #Setting seed for reproducibility\ny = np.sin(x) + np.random.normal(0,0.15,len(x))\nplt.plot(x,y,'.')",
"_____no_output_____"
],
[
"lrs = LinRegLasso()\nalpha = 0.0001\nlr = 0.08\nepochs = 100\n\nlrs.fit(x[:,np.newaxis],y, epochs, lr, alpha)\nlrs.weights",
"epoch: 0, \t MSE: 17.28968604018978\nepoch: 1, \t MSE: 14.626147202039622\nepoch: 2, \t MSE: 12.378658694531488\nepoch: 3, \t MSE: 10.479716191022815\nepoch: 4, \t MSE: 8.877331485218843\nepoch: 5, \t MSE: 7.523061332680313\nepoch: 6, \t MSE: 6.380218738551775\nepoch: 7, \t MSE: 5.413992777056065\nepoch: 8, \t MSE: 4.598539218214396\nepoch: 9, \t MSE: 3.9088044230822674\nepoch: 10, \t MSE: 3.3266204439073426\nepoch: 11, \t MSE: 2.833923743313259\nepoch: 12, \t MSE: 2.4179748665052587\nepoch: 13, \t MSE: 2.065723301420112\nepoch: 14, \t MSE: 1.7682633907098035\nepoch: 15, \t MSE: 1.5161442311623887\nepoch: 16, \t MSE: 1.3017589498895028\nepoch: 17, \t MSE: 1.1212745163895848\nepoch: 18, \t MSE: 0.9675475720623493\nepoch: 19, \t MSE: 0.8381392683863532\nepoch: 20, \t MSE: 0.7276930057626166\nepoch: 21, \t MSE: 0.6347172215712844\nepoch: 22, \t MSE: 0.555169674708247\nepoch: 23, \t MSE: 0.48819497063429\nepoch: 24, \t MSE: 0.4307222812313734\nepoch: 25, \t MSE: 0.3823168375814191\nepoch: 26, \t MSE: 0.3406294855489011\nepoch: 27, \t MSE: 0.3054976601025762\nepoch: 28, \t MSE: 0.2751113766668806\nepoch: 29, \t MSE: 0.24947885720760934\nepoch: 30, \t MSE: 0.22719538774215067\nepoch: 31, \t MSE: 0.2083713644082345\nepoch: 32, \t MSE: 0.1919086709679473\nepoch: 33, \t MSE: 0.1779740609937118\nepoch: 34, \t MSE: 0.1657029460117539\nepoch: 35, \t MSE: 0.15528837778235108\nepoch: 36, \t MSE: 0.1460448948904327\nepoch: 37, \t MSE: 0.13817260766072617\nepoch: 38, \t MSE: 0.1311244446383004\nepoch: 39, \t MSE: 0.1250957011628041\nepoch: 40, \t MSE: 0.11964700010480936\nepoch: 41, \t MSE: 0.11496190797845003\nepoch: 42, \t MSE: 0.1106854598635149\nepoch: 43, \t MSE: 0.10698587038632806\nepoch: 44, \t MSE: 0.10357480584693664\nepoch: 45, \t MSE: 0.10060364592069727\nepoch: 46, \t MSE: 0.09783701182454695\nepoch: 47, \t MSE: 0.09540931753098268\nepoch: 48, \t MSE: 0.09312754392558133\nepoch: 49, \t MSE: 0.09110982674125985\nepoch: 50, \t MSE: 0.08919723874805703\nepoch: 51, \t MSE: 0.08749278541472007\nepoch: 52, \t MSE: 0.08586513362848656\nepoch: 53, \t MSE: 0.08440353145280079\nepoch: 54, \t MSE: 0.08299909761681269\nepoch: 55, \t MSE: 0.0817287688386036\nepoch: 56, \t MSE: 0.08050201889712326\nepoch: 57, \t MSE: 0.07938489800498058\nepoch: 58, \t MSE: 0.0783019504125196\nepoch: 59, \t MSE: 0.0773096876776223\nepoch: 60, \t MSE: 0.07634507548628225\nepoch: 61, \t MSE: 0.07545632756630384\nepoch: 62, \t MSE: 0.0745906828265676\nepoch: 63, \t MSE: 0.0737891777382282\nepoch: 64, \t MSE: 0.07300757358764424\nepoch: 65, \t MSE: 0.07228072738227141\nepoch: 66, \t MSE: 0.07157148928886212\nepoch: 67, \t MSE: 0.0709094158743937\nepoch: 68, \t MSE: 0.07026326769775444\nepoch: 69, \t MSE: 0.06965806889846705\nepoch: 70, \t MSE: 0.06906751803399105\nepoch: 71, \t MSE: 0.06851277348182165\nepoch: 72, \t MSE: 0.06797166685053134\nepoch: 73, \t MSE: 0.06746206644448868\nepoch: 74, \t MSE: 0.06696526867880499\nepoch: 75, \t MSE: 0.066496346827004\nepoch: 76, \t MSE: 0.06603950595761333\nepoch: 77, \t MSE: 0.06560744854915922\nepoch: 78, \t MSE: 0.0651868244409165\nepoch: 79, \t MSE: 0.06478832784880968\nepoch: 80, \t MSE: 0.06440066571874699\nepoch: 81, \t MSE: 0.0640328330832463\nepoch: 82, \t MSE: 0.06367526948336952\nepoch: 83, \t MSE: 0.06333553376069448\nepoch: 84, \t MSE: 0.06300552600786866\nepoch: 85, \t MSE: 0.06269159228507007\nepoch: 86, \t MSE: 0.062386864886926394\nepoch: 87, \t MSE: 0.06209666661146153\nepoch: 88, \t MSE: 0.061815170067919256\nepoch: 89, \t MSE: 0.061546835370024686\nepoch: 90, \t MSE: 0.06128671403625909\nepoch: 91, \t MSE: 0.061038539411680824\nepoch: 92, \t MSE: 0.06079810604087325\nepoch: 93, \t MSE: 0.0605685354376613\nepoch: 94, \t MSE: 0.06034625068797903\nepoch: 95, \t MSE: 0.06013385859413892\nepoch: 96, \t MSE: 0.05992831426058195\nepoch: 97, \t MSE: 0.05973179178364991\nepoch: 98, \t MSE: 0.05954169685614971\nepoch: 99, \t MSE: 0.05935984006696908\n"
]
],
[
[
"# Just to check the result we can use scikit-learn\nThe difference may be explained by lr scheduler in scikit probably",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import Lasso\nlassoreg = Lasso(alpha=alpha, max_iter=epochs)\nlassoreg.fit(x[:,np.newaxis],y)\ny_pred = lassoreg.predict(x[:,np.newaxis])\n\ny_hat = lrs.predict(x[:,np.newaxis])\nplt.plot(x,y,'.')\nplt.plot(x,y_hat,'.')\nplt.plot(x,y_pred,'.')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f4555c5936858d2c3ad270b8e2af0f7c50d284 | 2,511 | ipynb | Jupyter Notebook | notebooks/Syft - Paillier Encrypted Linear Classification.ipynb | ivuckovic/PySyft | 17acc82fc8533188816bb0a5b82c1a529ceb6914 | [
"Apache-2.0"
] | null | null | null | notebooks/Syft - Paillier Encrypted Linear Classification.ipynb | ivuckovic/PySyft | 17acc82fc8533188816bb0a5b82c1a529ceb6914 | [
"Apache-2.0"
] | null | null | null | notebooks/Syft - Paillier Encrypted Linear Classification.ipynb | ivuckovic/PySyft | 17acc82fc8533188816bb0a5b82c1a529ceb6914 | [
"Apache-2.0"
] | null | null | null | 20.925 | 344 | 0.554361 | [
[
[
"# Paillier Encrypted Linear Classification Example\n\nDISCLAIMER: This is a proof-of-concept implementation. It does not represent a remotely product ready implementation or follow proper conventions for security, convenience, or scalability. It is part of a broader proof-of-concept demonstrating the vision of the OpenMined project, its major moving parts, and how they might work together.",
"_____no_output_____"
]
],
[
[
"from syft.he.paillier import KeyPair\nfrom syft.nn.linear import LinearClassifier\nfrom syft.he.keys import Paillier\nimport numpy as np",
"_____no_output_____"
],
[
"pk, sk = Paillier()\nmodel = LinearClassifier(n_inputs=4, n_labels=2)\nepochs = 3",
"_____no_output_____"
],
[
"model = model.encrypt(pk)",
"_____no_output_____"
],
[
"input = np.array([[0,0,1,1],[0,0,1,0]])\ntarget = np.array([[0,1],[0,0]])",
"_____no_output_____"
],
[
"for i in range(epochs):\n model.learn(input, target, alpha=0.5)",
"_____no_output_____"
],
[
"model = model.decrypt(sk)",
"_____no_output_____"
],
[
"for value in input:\n print(model.forward(value))",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f4630d6e650584e18a6ee6b880e6d55a30be5b | 27,520 | ipynb | Jupyter Notebook | Logistic_Regression/logistic_regression_newtons_method-master/NewtonCode.ipynb | DillipKS/The-Math-of-Intelligence-course | fc0f33e638fdbd05e93d54d38ed8493808f6ec74 | [
"Apache-2.0"
] | null | null | null | Logistic_Regression/logistic_regression_newtons_method-master/NewtonCode.ipynb | DillipKS/The-Math-of-Intelligence-course | fc0f33e638fdbd05e93d54d38ed8493808f6ec74 | [
"Apache-2.0"
] | null | null | null | Logistic_Regression/logistic_regression_newtons_method-master/NewtonCode.ipynb | DillipKS/The-Math-of-Intelligence-course | fc0f33e638fdbd05e93d54d38ed8493808f6ec74 | [
"Apache-2.0"
] | 1 | 2019-09-08T07:58:13.000Z | 2019-09-08T07:58:13.000Z | 40.77037 | 1,444 | 0.570313 | [
[
[
"# Newton's Method for Logistic Regression - The Math of Intelligence (Week 2)\n\n\n\n\n## Our Task\n\nWe're going to compute the probability that someone has Diabetes given their height, weight, and blood pressure. We'll generate this data ourselves (toy data), plot it, learn a logistic regression curve using Newton's Method for Optimization, then use that curve to predict the probability someone new with these 3 features has diabetes. We'll use Calculus, Probability Theory, Statistics, and Linear Algebra to do this. Get ready, ish is about to go down.\n\n## What is Logistic regression?\n\nLogistic regression is named for the function used at the core of the method, the logistic function. In linear regression, the outcome (dependent variable) is continuous. It can have any one of an infinite number of possible values. In logistic regression, the outcome (dependent variable) has only a limited number of possible values. Logistic Regression is used when response variable is categorical in nature.\n\nThe logistic function, also called the sigmoid function is an S-shaped curve that can take any real-valued number and map it into a value between 0 and 1, but never exactly at those limits.\n\n\n\nWhere e is the base of the natural logarithms (Euler’s number or the EXP() function in your spreadsheet) and value is the actual numerical value that you want to transform. E is a really convenient number for math, for example Whenever you take the derivative of e^x (that's e to the x), you get e^x back again. It's the only function on Earth that will do that.\n\nLogistic regression uses an equation as the representation, similar to linear regression. The central premise of Logistic Regression is the assumption that your input space can be separated into two nice ‘regions’, one for each class, by a linear(read: straight) boundary. Your data must be linearly seperable in n dimensions\n\n\n\nSo if we had the following function\n\n\n\nGiven some point (a,b), if we plugged it in, the equation could output a positive result (for one class), negative result (for the other class), or 0 (the point lies right on the decision boundary).\n\nSo we have a function that outputs a value in (-infinity, +infinity) given an input data point. But how do we map this to the probability P_+, that goes from [0, 1]? The answer, is in the odds function.\n\n\n\nLet $P(X)$ denote the probability of an event X occurring. In that case, the odds ratio (OR(X)) is defined by \n\n\n\nwhich is essentially the ratio of the probability of the event happening, vs. it not happening. It is clear that probability and odds convey the exact same information. But as $P(X)$ goes from 0 to 1, OR(X) goes from 0 to infinity\n\nInput values (x) are combined linearly using weights or coefficient values (referred to as the Greek capital letter Beta) to predict an output value (y). A key difference from linear regression is that the output value being modeled is a binary values (0 or 1) (discrete) rather than a numeric value (continuous)\n\n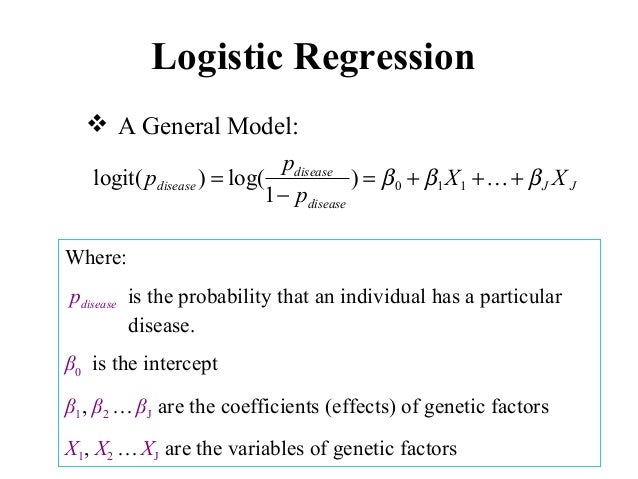\n\nHowever, we are still not quite there yet, since our boundary function gives a value from –infinity to infinity. So what we do, is take the logarithm of OR(X), called the log-odds function. Mathematically, as OR(X) goes from 0 to infinity, log(OR(X)) goes from –infinity to infinity\n\nWe are modeling the probability that an input (X) belongs to the default class (Y=1). The probability prediction must be transformed into a binary values (0 or 1) in order to actually make a probability prediction. Logistic regression is a linear method, but the predictions are transformed using the logistic function. The impact of this is that we can no longer understand the predictions as a linear combination of the inputs as we can with linear regression.\n\nWait, how is the boundary function computed? Well we want to maximize the likelihood that a random data point gets classified correctly. We call this Maximimum likelihood estimation.\n\nMaximum likelihood estimation is a general approach to estimating parameters in statistical models by maximizing the likelihood function. MLE applied to deep networks gets a special name “Backpropagation\". MLE is defined as\n\nL(θ|X)=f(X|θ)\n\nNewton's Method is an optimization algorithm. You can use this algorithm to find maximum (or minimum) of many different functions, including the likelihood function. You can obtain maximum likelihood estimates using different methods and using an optimization algorithm is one of them. \n\n## Why use Newton's Method for optimizing?\n\n- Newton’s method usually converges faster than gradient\ndescent when maximizing logistic regression log\nlikelihood.\n- Each iteration is more expensive than gradient descent\nbecause of calculating inverse of Hessian\n- As long as data points are not very large, Newton’s\nmethods are preferred\n\n## What are some other good examples of logistic regression + newton's method? \n\n1 https://github.com/hhl60492/newton_logistic/blob/master/main.py (Spam Classification)\n2 https://github.com/yangarbiter/logistic_regression_newton-cg (Click Through Rate Classification)",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\n#matrix math\nimport numpy as np\n#data manipulation\nimport pandas as pd\n#matrix data structure\nfrom patsy import dmatrices\n#for error logging\nimport warnings",
"_____no_output_____"
]
],
[
[
"<a name=\"data_setup\"></a>\n## Setup\n\n### Parameter / Data Setup\n\nIn the below cells, there are various parameters and options to play with involving data creation, algorithm settings, and what model you want to try and fit.",
"_____no_output_____"
]
],
[
[
"#outputs probability between 0 and 1, used to help define our logistic regression curve\ndef sigmoid(x):\n '''Sigmoid function of x.'''\n return 1/(1+np.exp(-x))",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"#makes the random numbers predictable\n#(pseudo-)random numbers work by starting with a number (the seed), \n#multiplying it by a large number, then taking modulo of that product. \n#The resulting number is then used as the seed to generate the next \"random\" number. \n#When you set the seed (every time), it does the same thing every time, giving you the same numbers.\n#good for reproducing results for debugging\n\n\nnp.random.seed(0) # set the seed\n\n##Step 1 - Define model parameters (hyperparameters)\n\n## algorithm settings\n#the minimum threshold for the difference between the predicted output and the actual output\n#this tells our model when to stop learning, when our prediction capability is good enough\ntol=1e-8 # convergence tolerance\n\nlam = None # l2-regularization\n#how long to train for?\nmax_iter = 20 # maximum allowed iterations\n\n## data creation settings\n#Covariance measures how two variables move together. \n#It measures whether the two move in the same direction (a positive covariance) \n#or in opposite directions (a negative covariance). \nr = 0.95 # covariance between x and z\nn = 1000 # number of observations (size of dataset to generate) \nsigma = 1 # variance of noise - how spread out is the data?\n\n## model settings\nbeta_x, beta_z, beta_v = -4, .9, 1 # true beta coefficients\nvar_x, var_z, var_v = 1, 1, 4 # variances of inputs\n\n## the model specification you want to fit\nformula = 'y ~ x + z + v + np.exp(x) + I(v**2 + z)'",
"_____no_output_____"
],
[
"## Step 2 - Generate and organize our data\n\n#The multivariate normal, multinormal or Gaussian distribution is a generalization of the one-dimensional normal \n#distribution to higher dimensions. Such a distribution is specified by its mean and covariance matrix.\n#so we generate input values - (x, v, z) using normal distributions\n\n#A probability distribution is a function that provides us the probabilities of all \n#possible outcomes of a stochastic process. \n\n#lets keep x and z closely related (height and weight)\nx, z = np.random.multivariate_normal([0,0], [[var_x,r],[r,var_z]], n).T\n#blood presure\nv = np.random.normal(0,var_v,n)**3\n\n#create a pandas dataframe (easily parseable object for manipulation)\nA = pd.DataFrame({'x' : x, 'z' : z, 'v' : v})\n#compute the log odds for our 3 independent variables\n#using the sigmoid function \nA['log_odds'] = sigmoid(A[['x','z','v']].dot([beta_x,beta_z,beta_v]) + sigma*np.random.normal(0,1,n))\n\n\n#compute the probability sample from binomial distribution\n#A binomial random variable is the number of successes x has in n repeated trials of a binomial experiment. \n#The probability distribution of a binomial random variable is called a binomial distribution. \nA['y'] = [np.random.binomial(1,p) for p in A.log_odds]\n\n#create a dataframe that encompasses our input data, model formula, and outputs\ny, X = dmatrices(formula, A, return_type='dataframe')\n\n#print it\nX.head(10)",
"/home/dks/.local/lib/python3.5/site-packages/ipykernel/__main__.py:4: RuntimeWarning: overflow encountered in exp\n"
]
],
[
[
"<a name=\"algorithms\"></a>\n<hr>\n### Algorithm Setup\n\nWe begin with a quick function for catching singular matrix errors that we will use to decorate our Newton steps.",
"_____no_output_____"
]
],
[
[
"#like dividing by zero (Wtff omgggggg universe collapses)\ndef catch_singularity(f):\n '''Silences LinAlg Errors and throws a warning instead.'''\n \n def silencer(*args, **kwargs):\n try:\n return f(*args, **kwargs)\n except np.linalg.LinAlgError:\n warnings.warn('Algorithm terminated - singular Hessian!')\n return args[0]\n return silencer\n",
"_____no_output_____"
]
],
[
[
"<a name=\"newton\"></a>\n<hr>\n### Explanation of a Single Newton Step\n\nRecall that Newton's method for maximizing / minimizing a given function $f(\\beta)$ iteratively computes the following estimate:\n\n$$\n\\beta^+ = \\beta - Hf(\\beta)^{-1}\\nabla f(\\beta)\n$$\n\nThe Hessian of the log-likelihood for logistic regression is given by:\n\nhessian of our function = negative tranpose of (N times (p+1) times (N x N diagional matrix of weights, each is p*(1-p) times X again\n\n\n$$\nHf(\\beta) = -X^TWX\n$$\nand the gradient is:\n\ngradient of our function = tranpose of X times (column vector - N vector of probabilities)\n\n$$\n\\nabla f(\\beta) = X^T(y-p)\n$$\nwhere\n$$\nW := \\text{diag}\\left(p(1-p)\\right)\n$$\nand $p$ are the predicted probabilites computed at the current value of $\\beta$.\n\n### Connection to Iteratively Reweighted Least Squares (IRLS)\n*For logistic regression, this step is actually equivalent to computing a weighted least squares estimator at each iteration!* \nThe method of least squares is about estimating\nparameters by minimizing the squared discrepancies\nbetween observed data, on the one hand, and their\nexpected values on the other\n\nI.e.,\n$$\n\\beta^+ = \\arg\\min_\\beta (z-X\\beta)^TW(z-X\\beta)\n$$\nwith $W$ as before and the *adjusted response* $z$ is given by\n$$\nz := X\\beta + W^{-1}(y-p)\n$$\n\n**Takeaway:** This is fun, but in fact it can be leveraged to derive asymptotics and inferential statistics about the computed MLE $\\beta^*$!\n\n### Our implementations\nBelow we implement a single step of Newton's method, and we compute $Hf(\\beta)^{-1}\\nabla f(\\beta)$ using `np.linalg.lstsq(A,b)` to solve the equation $Ax = b$. Note that this does not require us to compute the actual inverse of the Hessian.",
"_____no_output_____"
]
],
[
[
"@catch_singularity\ndef newton_step(curr, X, lam=None):\n '''One naive step of Newton's Method'''\n \n #how to compute inverse? http://www.mathwarehouse.com/algebra/matrix/images/square-matrix/inverse-matrix.gif\n \n ## compute necessary objects\n #create probability matrix, miniminum 2 dimensions, tranpose (flip it)\n p = np.array(sigmoid(X.dot(curr[:,0])), ndmin=2).T\n #create weight matrix from it\n W = np.diag((p*(1-p))[:,0])\n #derive the hessian \n hessian = X.T.dot(W).dot(X)\n #derive the gradient\n grad = X.T.dot(y-p)\n \n ## regularization step (avoiding overfitting)\n if lam:\n # Return the least-squares solution to a linear matrix equation\n step, *_ = np.linalg.lstsq(hessian + lam*np.eye(curr.shape[0]), grad)\n else:\n step, *_ = np.linalg.lstsq(hessian, grad)\n \n ## update our \n beta = curr + step\n \n return beta",
"_____no_output_____"
]
],
[
[
"Next, we implement Newton's method in a *slightly* different way; this time, at each iteration, we actually compute the full inverse of the Hessian using `np.linalg.inv()`.",
"_____no_output_____"
]
],
[
[
"@catch_singularity\ndef alt_newton_step(curr, X, lam=None):\n '''One naive step of Newton's Method'''\n \n ## compute necessary objects\n p = np.array(sigmoid(X.dot(curr[:,0])), ndmin=2).T\n W = np.diag((p*(1-p))[:,0])\n hessian = X.T.dot(W).dot(X)\n grad = X.T.dot(y-p)\n \n ## regularization\n if lam:\n #Compute the inverse of a matrix.\n step = np.dot(np.linalg.inv(hessian + lam*np.eye(curr.shape[0])), grad)\n else:\n step = np.dot(np.linalg.inv(hessian), grad)\n \n ## update our weights\n beta = curr + step\n \n return beta",
"_____no_output_____"
]
],
[
[
"<a name=\"conv\"></a>\n<hr>\n### Convergence Setup\n\nFirst we implement coefficient convergence; this stopping criterion results in convergence whenever\n$$\n\\|\\beta^+ - \\beta\\|_\\infty < \\epsilon\n$$\nwhere $\\epsilon$ is a given tolerance.",
"_____no_output_____"
]
],
[
[
"def check_coefs_convergence(beta_old, beta_new, tol, iters):\n '''Checks whether the coefficients have converged in the l-infinity norm.\n Returns True if they have converged, False otherwise.'''\n #calculate the change in the coefficients\n coef_change = np.abs(beta_old - beta_new)\n \n #if change hasn't reached the threshold and we have more iterations to go, keep training\n return not (np.any(coef_change>tol) & (iters < max_iter))",
"_____no_output_____"
]
],
[
[
"<a name=\"numerics\"></a>\n<hr>\n## Numerical Examples\n\n### Standard Newton with Coefficient Convergence",
"_____no_output_____"
]
],
[
[
"## initial conditions\n#initial coefficients (weight values), 2 copies, we'll update one\nbeta_old, beta = np.ones((len(X.columns),1)), np.zeros((len(X.columns),1))\n\n#num iterations we've done so far\niter_count = 0\n#have we reached convergence?\ncoefs_converged = False\n\n#if we haven't reached convergence... (training step)\nwhile not coefs_converged:\n \n #set the old coefficients to our current\n beta_old = beta\n #perform a single step of newton's optimization on our data, set our updated beta values\n beta = newton_step(beta, X, lam=lam)\n #increment the number of iterations\n iter_count += 1\n \n #check for convergence between our old and new beta values\n coefs_converged = check_coefs_convergence(beta_old, beta, tol, iter_count)\n \nprint('Iterations : {}'.format(iter_count))\nprint('Beta : {}'.format(beta))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f46c49845fbab26f266fd928e5cf013520687d | 1,577 | ipynb | Jupyter Notebook | unconditional/main_bs900.ipynb | minhtannguyen/ffjord | f3418249eaa4647f4339aea8d814cf2ce33be141 | [
"MIT"
] | null | null | null | unconditional/main_bs900.ipynb | minhtannguyen/ffjord | f3418249eaa4647f4339aea8d814cf2ce33be141 | [
"MIT"
] | null | null | null | unconditional/main_bs900.ipynb | minhtannguyen/ffjord | f3418249eaa4647f4339aea8d814cf2ce33be141 | [
"MIT"
] | null | null | null | 29.203704 | 390 | 0.621433 | [
[
[
"import os\nos.environ['CUDA_VISIBLE_DEVICES']='0,1,2,3,4,5'",
"_____no_output_____"
],
[
"%run -p ../train_cnf.py --data mnist --dims 64,64,64 --strides 1,1,1,1 --num_blocks 2 --layer_type concat --multiscale True --rademacher True --batch_size 900 --test_batch_size 500 --save ../experiments_published/cnf_published_baseline_bs900_1 --seed 1 --conditional False --controlled_tol False --log_freq 10",
"_____no_output_____"
],
[
"# %run -p ../train_cnf.py --data mnist --dims 64,64,64 --strides 1,1,1,1 --num_blocks 2 --layer_type concat --multiscale True --rademacher True --batch_size 900 --test_batch_size 900 --save ../experiments_published/cnf_published_baseline_bs900 --resume ../experiments_published/cnf_published_baseline_bs900/current_checkpt.pth --conditional False --controlled_tol False --log_freq 10",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d0f470dace2bf38b3ce50db81e5129e2b9ea22f5 | 41,570 | ipynb | Jupyter Notebook | dataset/turkish/TrainEn2TrTestTr/Untitled.ipynb | feyzaakyurek/newsframing | 29c916aacb0c361e8e3169f5bf8d76766289a417 | [
"Apache-2.0"
] | 9 | 2020-05-18T19:28:51.000Z | 2022-03-04T21:02:08.000Z | dataset/turkish/TrainEn2TrTestTr/Untitled.ipynb | feyzaakyurek/newsframing | 29c916aacb0c361e8e3169f5bf8d76766289a417 | [
"Apache-2.0"
] | null | null | null | dataset/turkish/TrainEn2TrTestTr/Untitled.ipynb | feyzaakyurek/newsframing | 29c916aacb0c361e8e3169f5bf8d76766289a417 | [
"Apache-2.0"
] | null | null | null | 35.408859 | 90 | 0.325427 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"train = pd.read_csv('train-Copy1.tsv', sep='\\t',header=None)\nturkish = pd.read_csv('train-en-to-tr.txt', sep='\\n',header=None)\n",
"_____no_output_____"
],
[
"train[1] = turkish",
"_____no_output_____"
],
[
"train",
"_____no_output_____"
],
[
"train.to_csv('train.tsv', header=False, index=False, sep='\\t')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0f4749b2a3aed7ecdd5cac243112700be9b4254 | 9,336 | ipynb | Jupyter Notebook | Trabalho.ipynb | Clayton100/Phyton | e4198eceb257dac569892756338d509e7995fff3 | [
"Unlicense"
] | null | null | null | Trabalho.ipynb | Clayton100/Phyton | e4198eceb257dac569892756338d509e7995fff3 | [
"Unlicense"
] | null | null | null | Trabalho.ipynb | Clayton100/Phyton | e4198eceb257dac569892756338d509e7995fff3 | [
"Unlicense"
] | null | null | null | 27.139535 | 222 | 0.441624 | [
[
[
"<a href=\"https://colab.research.google.com/github/Clayton100/Phyton/blob/main/Trabalho.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"A=int(input (\"Digite um número \"))\nB=int(input (\"Digite um número \"))\nC=A+B\nprint (C)",
"Digite um número 8\nDigite um número 6\n14\n"
],
[
"A=int(input (\"Digite um número \"))\nB=int(input (\"Digite um número \"))\nC=A+B\nD=A-B\nE=A/B\nF=A*B\nprint (C)\nprint (D)\nprint (E)\nprint (F)",
"Digite um número 5\nDigite um número 2\n7\n3\n2.5\n10\n"
],
[
"Dist=float(input (\"Digite a distância percorrida \"))\nComb= float(input (\"Digite o combustível gasto \"))\nConsumo=Dist/Comb\nprint (Consumo)",
"Digite a distância percorrida 10\nDigite o combustível gasto 5\n2.0\n"
],
[
"Nvend = input ( \"Digite o nome do vendedor\" ) \nSalF = float ( input ( \"Digite o salário fixo\" ))\nVendasmes = float ( input ( \"Digite o valor das suas vendas mensais\")) \nComissao = Vendasmes * 0.15 \nSalT = SalF + Comissao \nprint (Nvend) \nprint (\"O seu salário é de R$ %.2f\" %SalF) \nprint (\"O seu salário total é de R $%.2f\" %SalT)",
"Digite o nome do vendedorClayton\nDigite o salário fixo1700\nDigite o valor das suas vendas mensais1000\nClayton\nO seu salário é de R$ 1700.00\nO seu salário total é de R $1850.00\n"
],
[
"Aluno= str(input(\"Digite o nome do aluno\"))\nA1= float(input (\"Digite o valor da primeira prova\"))\nA2= float(input (\"Digite o valor da segunda prova\"))\nA3= float(input (\"Digite o valor da terceira prova\"))\nMedia=(A1+A2+A3)/3\nprint (Aluno)\nprint (Media)",
"Digite o nome do alunoCinthia\nDigite o valor da primeira prova8\nDigite o valor da segunda prova9\nDigite o valor da terceira prova10\nCinthia\n9.0\n"
],
[
"A = float ( input ( \"Digite um número inteiro \" )) \nB = float (input ( \"Digite um número inteiro \" ))\nC=A\nA=B\nB=C\nprint (\"O primeiro número trocado é: %.0f \" %A)\nprint (\"O segundo número trocado é:%.0f \"%B)",
"Digite um número inteiro 5\nDigite um número inteiro 6\nO primeiro número trocado é: 6 \nO segundo número trocado é:5 \n"
],
[
" \nTempC = float (input ( \"Digite o valor da temperatura em ° C\" )) \nTempF = 9 * TempC / 5 + 32 \nprint( \"A temperatura em fahrenheit é;% .0f\" % TempF)",
"Digite o valor da temperatura em ° C20\nA temperatura em fahrenheit é; 68\n"
],
[
"Dol= float ( input(\"Digite a quantidade de dólares que você possui \"))\nCotacao= float ( input(\"Digite a cotação do dólar atual \"))\nVreais = Dol*Cotacao\nprint (\"O valor que você possui em reais é: R$%.2f\" %Vreais)",
"Digite a quantidade de dólares que você possui 300\nDigite a cotação do dólar atual 5\nO valor que você possui em reais é: R$1500.00\n"
],
[
"Dep=float(input (\"Digite o valor depositado\"))\nJuro=0.7/100\nRend=Dep+Dep*Juro\nprint (\"O valor após 1 mês é: R$%.2f \" %Rend)",
"Digite o valor depositado1000\nO valor após 1 mês é: R$1007.00 \n"
],
[
"Compra= float(input (\"Digite o valor da compra\"))\nPrest=Compra/5\nprint (Compra)\nprint (Prest)",
"Digite o valor da compra500\n500.0\n100.0\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f48518190e03d7b43928405fd3562ed267e7b5 | 9,196 | ipynb | Jupyter Notebook | Content/1.python/1.python_fundamentals/01.Python-Basics/2.variables-and-data-types/variables_and_data_types.ipynb | becodeorg/ai_fundamentals_1.0 | 2b8f90511e1e0190e01916f411726013269e210c | [
"MIT"
] | null | null | null | Content/1.python/1.python_fundamentals/01.Python-Basics/2.variables-and-data-types/variables_and_data_types.ipynb | becodeorg/ai_fundamentals_1.0 | 2b8f90511e1e0190e01916f411726013269e210c | [
"MIT"
] | null | null | null | Content/1.python/1.python_fundamentals/01.Python-Basics/2.variables-and-data-types/variables_and_data_types.ipynb | becodeorg/ai_fundamentals_1.0 | 2b8f90511e1e0190e01916f411726013269e210c | [
"MIT"
] | null | null | null | 26.501441 | 574 | 0.521096 | [
[
[
"# Variables and data types\nVariables are used to temporarily store a value in the computer's memory. We can then use it later, when we need it in the program. We can compare that to a small box in which we could store information. \n\nSince Python is a dynamically typed language, Python values, not variables, carry type. This has implications for many aspects of the way the language functions.\n\nAll variables in Python hold references to objects, and these references are passed to functions; a function cannot change the value of variable references in its calling function (but see below for exceptions). Some people (including Guido van Rossum himself) have called this parameter-passing scheme \"Call by object reference.\" An object reference means a name, and the passed reference is an \"alias\", i.e. a copy of the reference to the same object, just as in C/C++. The object's value may be changed in the called function with the \"alias\", for example:\n\n\n\n## Naming and code writing conventions\nAs in many programming languages, to name a variable, some conventions must be respected.\n\n#### 1. The name of the variable must start with a **letter or an underscore**. The variable can not start with a number or a hyphen. \n\n❌ Bad example :\n```py\n# Do not do this\n2Name = \"James\" \n\n# Do not do this\n-name = \"James\"\n\n```\n\n✅ Good example :\n```py\n# Do this\nname = \"James\" \n\n# Do this\n_name = \"James\"\n\n```\n#### 2. **Never put space between words.** \n\n❌ Bad example :\n```py\n# Do not do this\nMy name = \"Odile\" \n```\n\n✅ Good example :\n```py\n# DO is\nmy_name = \"Odile\"\n\n```\n\n#### 3. No accents on the names of variables. **Use only English**\n\n❌ Bad example :\n```py\n# Do not do this\nprénom = \"Odile\" \n```\n\n✅ Good example :\n```py\n# Do this\nfirst_name = \"Odile\" \n```\n\n#### 4. **Always give an explicit name** to the variable.\n\n❌ Bad example :\n```py\n# Do not do this\na = \"Odile\" \n\n# Do not do this\nfstnme = \"Odile\"\n\n```\n\n✅ Good example :\n```py\n# Do this\nfirst_name = \"Odile\" \n\n# Do this\nmagic_potion = 42\n\n```",
"_____no_output_____"
],
[
"**Try it for yourself by clicking on the Run button**",
"_____no_output_____"
]
],
[
[
"# BAD \n2name = \"James\"",
"_____no_output_____"
],
[
"# BAD\n-name = \"Bond\"",
"_____no_output_____"
],
[
"# BAD\nMy name = \"bond\"",
"_____no_output_____"
]
],
[
[
"The print() function allows us to display the result. ",
"_____no_output_____"
]
],
[
[
"# GOOD\nname = \"Bond\"\nprint(\"Your name is\", name)",
"_____no_output_____"
]
],
[
[
"To format the text, you can use the format() method in the class string",
"_____no_output_____"
]
],
[
[
"last_name = \"Bond\"\nfirst_name = \"James\"\ntext = \"My name is {}, {} {}.\".format(last_name, first_name, last_name)\nprint(text)",
"My name is Bond, James Bond.\n"
]
],
[
[
"Another example. Replace the value of the variable ``age`` with your age and the variable ``firstname`` with your First name.",
"_____no_output_____"
]
],
[
[
"age = \"34\"\nfirst_name =\"Ludovic\"\ntext = \"Hello, my name is {} and i am {}\".format(first_name, age)\nprint(text)",
"Hello, my name is Ludovic and i am 34\n"
]
],
[
[
"## Data types\n\nSince Python is a high-level language, it has a dynamic variable typing. \nBy dynamics, understand that it is the computer that deals with defining what type of variable should be used. \nTo be perfectly accurate, **it is not the variable that is typed (unlike Java) but its content**\n\nIn java we declare a variable like this:\n```java\nString fisrtName = \"James\"\n```\nWe define the type of variable ourselves. \n\n With python we declare a variable like this:\n```py\nfirst_name = \"James\"\n```\nAnd so, it's python that will define what type will be used.",
"_____no_output_____"
]
],
[
[
"first_name = \"James\" # String\nlast_name = \"Bond\" # String\nage = 39 # Integer\nweigth = 81.56 # Float\ndouble_agent = True # Boolean\nlogin = \"007\" # String\nagent = [first_name, last_name, age, weigth, double_agent, login] # List\nprint(agent)",
"['James', 'Bond', 39, 81.56, True, '007']\n"
]
],
[
[
"1. Note that ``True`` and ``False`` are written with the first letter uppercase.\n2. ``login`` is a string.\n3. ``List`` is like an array, but you can store values of different types.",
"_____no_output_____"
],
[
"Here is a not limited list of the types we use most often. These are the most frequently used. \nFor tuples, dictionaries and sets we'll see them later\n\n|Name | Type | Description |\n|:---------|:---------:|------------:|\n|Integers | int | Whole numbers, such as : **1, 67, 5000** |\n|Floating point | float | Decimal point numbers, such as : **1.89, 0.67, 9.99999** |\n|Strings | str | Ordered sequence of characters : **\"Hello\", \"10\"** |\n|Lists | list | Ordered sequence of objects : **[\"hello\", 10, 56.89]** |\n|Dictionaries | dict | Unordered (Key : Value) pairs : **{\"Key1\": value, \"name\" : \"Peter}** |\n|Tuples | tuple | Ordered sequence of objects (immutable) : **(\"hello\", 10, 56.89)** |\n|Sets | set | Unordered collections of unique objects : **{1,2}** |\n|Booleans |bool| Logical value : **True or False** |\n",
"_____no_output_____"
],
[
"**There is a native pyhton function that allows you to know what type of data you have. This is the type() function** ",
"_____no_output_____"
]
],
[
[
"print(first_name, type(first_name))\nprint(last_name, type(last_name))\nprint(age, type(age))\nprint(weigth, type(weigth))\nprint(agent, type(agent))",
"James <class 'str'>\nBond <class 'str'>\n39 <class 'int'>\n81.56 <class 'float'>\n['James', 'Bond', 39, 81.56, True, '007'] <class 'list'>\n"
]
],
[
[
"## [Finished? Practice with these exercises](./drill-variables-data-types.ipynb).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0f4b4e39c47fabb954a1ea43e534dc781e22c07 | 77,911 | ipynb | Jupyter Notebook | 0909_3_rate_check.ipynb | root4kaido/Cornell-Birdcall-Identification | 186008965ad7e8797fc181a2836bb63aacb324e4 | [
"MIT"
] | 1 | 2020-11-21T12:03:07.000Z | 2020-11-21T12:03:07.000Z | 0909_3_rate_check.ipynb | root4kaido/Cornell-Birdcall-Identification | 186008965ad7e8797fc181a2836bb63aacb324e4 | [
"MIT"
] | null | null | null | 0909_3_rate_check.ipynb | root4kaido/Cornell-Birdcall-Identification | 186008965ad7e8797fc181a2836bb63aacb324e4 | [
"MIT"
] | null | null | null | 36.492272 | 362 | 0.373336 | [
[
[
"!pip install /home/knikaido/work/Cornell-Birdcall-Identification/data/resnest50-fast-package/resnest-0.0.6b20200701/resnest/\n!pip install torch==1.4.0\n!pip install opencv-python\n!pip install slackweb\n!pip install torchvision==0.2.2\n!pip install torch_summary",
"Defaulting to user installation because normal site-packages is not writeable\nProcessing /home/knikaido/work/Cornell-Birdcall-Identification/data/resnest50-fast-package/resnest-0.0.6b20200701/resnest\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from resnest==0.0.6b20200908) (1.17.4)\nRequirement already satisfied: tqdm in /home/user/.local/lib/python3.6/site-packages (from resnest==0.0.6b20200908) (4.19.9)\nRequirement already satisfied: nose in /home/user/.local/lib/python3.6/site-packages (from resnest==0.0.6b20200908) (1.3.7)\nRequirement already satisfied: torch>=1.0.0 in /home/user/.local/lib/python3.6/site-packages (from resnest==0.0.6b20200908) (1.4.0)\nRequirement already satisfied: Pillow in /usr/local/lib/python3.6/dist-packages (from resnest==0.0.6b20200908) (7.1.2)\nRequirement already satisfied: scipy in /usr/local/lib/python3.6/dist-packages (from resnest==0.0.6b20200908) (1.1.0)\nRequirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from resnest==0.0.6b20200908) (2.24.0)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests->resnest==0.0.6b20200908) (2020.4.5.2)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->resnest==0.0.6b20200908) (3.0.4)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->resnest==0.0.6b20200908) (2.9)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /home/user/.local/lib/python3.6/site-packages (from requests->resnest==0.0.6b20200908) (1.24.3)\nBuilding wheels for collected packages: resnest\n Building wheel for resnest (setup.py) ... \u001b[?25ldone\n\u001b[?25h Created wheel for resnest: filename=resnest-0.0.6b20200908-py3-none-any.whl size=30754 sha256=13a96a321fe3e9a358a1357f441e745428c6ccaf0734f94499d0ec7654deac22\n Stored in directory: /tmp/pip-ephem-wheel-cache-zyrrdww9/wheels/98/b8/20/14b175a058326076510265be935570257f33b40bafba7255a9\nSuccessfully built resnest\nInstalling collected packages: resnest\n Attempting uninstall: resnest\n Found existing installation: resnest 0.0.6b20200908\n Uninstalling resnest-0.0.6b20200908:\n Successfully uninstalled resnest-0.0.6b20200908\nSuccessfully installed resnest-0.0.6b20200908\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\nDefaulting to user installation because normal site-packages is not writeable\nRequirement already satisfied: torch==1.4.0 in /home/user/.local/lib/python3.6/site-packages (1.4.0)\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\nDefaulting to user installation because normal site-packages is not writeable\nRequirement already satisfied: opencv-python in /home/user/.local/lib/python3.6/site-packages (4.4.0.42)\nRequirement already satisfied: numpy>=1.13.3 in /usr/local/lib/python3.6/dist-packages (from opencv-python) (1.17.4)\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\nDefaulting to user installation because normal site-packages is not writeable\nRequirement already satisfied: slackweb in /home/user/.local/lib/python3.6/site-packages (1.0.5)\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\nDefaulting to user installation because normal site-packages is not writeable\nRequirement already satisfied: torchvision==0.2.2 in /home/user/.local/lib/python3.6/site-packages (0.2.2)\nRequirement already satisfied: torch in /home/user/.local/lib/python3.6/site-packages (from torchvision==0.2.2) (1.4.0)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from torchvision==0.2.2) (1.11.0)\nRequirement already satisfied: tqdm==4.19.9 in /home/user/.local/lib/python3.6/site-packages (from torchvision==0.2.2) (4.19.9)\nRequirement already satisfied: pillow>=4.1.1 in /usr/local/lib/python3.6/dist-packages (from torchvision==0.2.2) (7.1.2)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from torchvision==0.2.2) (1.17.4)\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\nDefaulting to user installation because normal site-packages is not writeable\nRequirement already satisfied: torch_summary in /home/user/.local/lib/python3.6/site-packages (1.4.2)\n\u001b[33mWARNING: You are using pip version 20.1.1; however, version 20.2.2 is available.\nYou should consider upgrading via the '/usr/bin/python3 -m pip install --upgrade pip' command.\u001b[0m\n"
],
[
"from pathlib import Path\nimport numpy as np\nimport pandas as pd\nimport typing as tp\nimport yaml\nimport random\nimport os\nimport sys\nimport soundfile as sf\nimport librosa\nimport cv2\nimport matplotlib.pyplot as plt\nimport time\nimport pickle\n\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.utils.data as data\nimport resnest.torch as resnest_torch\n\nfrom torchvision import models\n\nfrom sklearn.model_selection import StratifiedKFold\nfrom sklearn.metrics import f1_score\nfrom radam import RAdam\nfrom resnet import ResNet, Bottleneck\n\n\npd.options.display.max_rows = 500\npd.options.display.max_columns = 500",
"_____no_output_____"
],
[
"with open('0909_2_config.yml', 'r') as yml:\n settings = yaml.safe_load(yml)",
"_____no_output_____"
],
[
"def set_seed(seed: int = 42):\n random.seed(seed)\n np.random.seed(seed)\n os.environ[\"PYTHONHASHSEED\"] = str(seed)\n torch.manual_seed(seed)\n torch.cuda.manual_seed(seed) # type: ignore\n# torch.backends.cudnn.deterministic = True # type: ignore\n# torch.backends.cudnn.benchmark = True # type: ignore\n ",
"_____no_output_____"
],
[
"# def progress_bar(i):\n# pro_bar = ('=' * i) + (' ' * (pro_size - i))\n# print('\\r[{0}] {1}%'.format(pro_bar, i / pro_size * 100.), end='')",
"_____no_output_____"
],
[
"# ROOT = Path.cwd().parent\n# INPUT_ROOT = ROOT / \"input\"\nINPUT_ROOT = Path(\"/home/knikaido/work/Cornell-Birdcall-Identification/data\")\nRAW_DATA = INPUT_ROOT / \"birdsong_recognition\"\nTRAIN_AUDIO_DIR = RAW_DATA / \"train_audio\"\nTRAIN_RESAMPLED_AUDIO_DIRS = [\n INPUT_ROOT / \"birdsong-resampled-train-audio-{:0>2}\".format(i) for i in range(5)\n]\nTEST_AUDIO_DIR = RAW_DATA / \"test_audio\"",
"_____no_output_____"
],
[
"BIRD_CODE = {\n 'aldfly': 0, 'ameavo': 1, 'amebit': 2, 'amecro': 3, 'amegfi': 4,\n 'amekes': 5, 'amepip': 6, 'amered': 7, 'amerob': 8, 'amewig': 9,\n 'amewoo': 10, 'amtspa': 11, 'annhum': 12, 'astfly': 13, 'baisan': 14,\n 'baleag': 15, 'balori': 16, 'banswa': 17, 'barswa': 18, 'bawwar': 19,\n 'belkin1': 20, 'belspa2': 21, 'bewwre': 22, 'bkbcuc': 23, 'bkbmag1': 24,\n 'bkbwar': 25, 'bkcchi': 26, 'bkchum': 27, 'bkhgro': 28, 'bkpwar': 29,\n 'bktspa': 30, 'blkpho': 31, 'blugrb1': 32, 'blujay': 33, 'bnhcow': 34,\n 'boboli': 35, 'bongul': 36, 'brdowl': 37, 'brebla': 38, 'brespa': 39,\n 'brncre': 40, 'brnthr': 41, 'brthum': 42, 'brwhaw': 43, 'btbwar': 44,\n 'btnwar': 45, 'btywar': 46, 'buffle': 47, 'buggna': 48, 'buhvir': 49,\n 'bulori': 50, 'bushti': 51, 'buwtea': 52, 'buwwar': 53, 'cacwre': 54,\n 'calgul': 55, 'calqua': 56, 'camwar': 57, 'cangoo': 58, 'canwar': 59,\n 'canwre': 60, 'carwre': 61, 'casfin': 62, 'caster1': 63, 'casvir': 64,\n 'cedwax': 65, 'chispa': 66, 'chiswi': 67, 'chswar': 68, 'chukar': 69,\n 'clanut': 70, 'cliswa': 71, 'comgol': 72, 'comgra': 73, 'comloo': 74,\n 'commer': 75, 'comnig': 76, 'comrav': 77, 'comred': 78, 'comter': 79,\n 'comyel': 80, 'coohaw': 81, 'coshum': 82, 'cowscj1': 83, 'daejun': 84,\n 'doccor': 85, 'dowwoo': 86, 'dusfly': 87, 'eargre': 88, 'easblu': 89,\n 'easkin': 90, 'easmea': 91, 'easpho': 92, 'eastow': 93, 'eawpew': 94,\n 'eucdov': 95, 'eursta': 96, 'evegro': 97, 'fiespa': 98, 'fiscro': 99,\n 'foxspa': 100, 'gadwal': 101, 'gcrfin': 102, 'gnttow': 103, 'gnwtea': 104,\n 'gockin': 105, 'gocspa': 106, 'goleag': 107, 'grbher3': 108, 'grcfly': 109,\n 'greegr': 110, 'greroa': 111, 'greyel': 112, 'grhowl': 113, 'grnher': 114,\n 'grtgra': 115, 'grycat': 116, 'gryfly': 117, 'haiwoo': 118, 'hamfly': 119,\n 'hergul': 120, 'herthr': 121, 'hoomer': 122, 'hoowar': 123, 'horgre': 124,\n 'horlar': 125, 'houfin': 126, 'houspa': 127, 'houwre': 128, 'indbun': 129,\n 'juntit1': 130, 'killde': 131, 'labwoo': 132, 'larspa': 133, 'lazbun': 134,\n 'leabit': 135, 'leafly': 136, 'leasan': 137, 'lecthr': 138, 'lesgol': 139,\n 'lesnig': 140, 'lesyel': 141, 'lewwoo': 142, 'linspa': 143, 'lobcur': 144,\n 'lobdow': 145, 'logshr': 146, 'lotduc': 147, 'louwat': 148, 'macwar': 149,\n 'magwar': 150, 'mallar3': 151, 'marwre': 152, 'merlin': 153, 'moublu': 154,\n 'mouchi': 155, 'moudov': 156, 'norcar': 157, 'norfli': 158, 'norhar2': 159,\n 'normoc': 160, 'norpar': 161, 'norpin': 162, 'norsho': 163, 'norwat': 164,\n 'nrwswa': 165, 'nutwoo': 166, 'olsfly': 167, 'orcwar': 168, 'osprey': 169,\n 'ovenbi1': 170, 'palwar': 171, 'pasfly': 172, 'pecsan': 173, 'perfal': 174,\n 'phaino': 175, 'pibgre': 176, 'pilwoo': 177, 'pingro': 178, 'pinjay': 179,\n 'pinsis': 180, 'pinwar': 181, 'plsvir': 182, 'prawar': 183, 'purfin': 184,\n 'pygnut': 185, 'rebmer': 186, 'rebnut': 187, 'rebsap': 188, 'rebwoo': 189,\n 'redcro': 190, 'redhea': 191, 'reevir1': 192, 'renpha': 193, 'reshaw': 194,\n 'rethaw': 195, 'rewbla': 196, 'ribgul': 197, 'rinduc': 198, 'robgro': 199,\n 'rocpig': 200, 'rocwre': 201, 'rthhum': 202, 'ruckin': 203, 'rudduc': 204,\n 'rufgro': 205, 'rufhum': 206, 'rusbla': 207, 'sagspa1': 208, 'sagthr': 209,\n 'savspa': 210, 'saypho': 211, 'scatan': 212, 'scoori': 213, 'semplo': 214,\n 'semsan': 215, 'sheowl': 216, 'shshaw': 217, 'snobun': 218, 'snogoo': 219,\n 'solsan': 220, 'sonspa': 221, 'sora': 222, 'sposan': 223, 'spotow': 224,\n 'stejay': 225, 'swahaw': 226, 'swaspa': 227, 'swathr': 228, 'treswa': 229,\n 'truswa': 230, 'tuftit': 231, 'tunswa': 232, 'veery': 233, 'vesspa': 234,\n 'vigswa': 235, 'warvir': 236, 'wesblu': 237, 'wesgre': 238, 'weskin': 239,\n 'wesmea': 240, 'wessan': 241, 'westan': 242, 'wewpew': 243, 'whbnut': 244,\n 'whcspa': 245, 'whfibi': 246, 'whtspa': 247, 'whtswi': 248, 'wilfly': 249,\n 'wilsni1': 250, 'wiltur': 251, 'winwre3': 252, 'wlswar': 253, 'wooduc': 254,\n 'wooscj2': 255, 'woothr': 256, 'y00475': 257, 'yebfly': 258, 'yebsap': 259,\n 'yehbla': 260, 'yelwar': 261, 'yerwar': 262, 'yetvir': 263\n}\n\nINV_BIRD_CODE = {v: k for k, v in BIRD_CODE.items()}",
"_____no_output_____"
],
[
"train = pd.read_csv(RAW_DATA / \"train.csv\")\n# train = pd.read_csv(TRAIN_RESAMPLED_AUDIO_DIRS[0] / \"train_mod.csv\")\n\ntrain_rate = train[['ebird_code', 'filename', 'rating']].sort_values('rating')\ntrain_rate[train_rate['rating'] == 2.0]",
"_____no_output_____"
],
[
"train_rate['rating'].value_counts()",
"_____no_output_____"
],
[
"len(train_rate[train_rate['rating'] <= 1.5]) / len(train_rate)",
"_____no_output_____"
],
[
"train['secondary_labels'].value_counts()",
"_____no_output_____"
],
[
"train.columns",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f4b6794ef5e642f8d4a7a6d58e29e9447958ca | 285,321 | ipynb | Jupyter Notebook | mwdsbe/.ipynb_checkpoints/tf-idf-alg-test-nick-checkpoint.ipynb | BinnyDaBin/MWDSBE | aa0de50f2289e47f7c2e9134334b23c3b5594f0c | [
"MIT"
] | null | null | null | mwdsbe/.ipynb_checkpoints/tf-idf-alg-test-nick-checkpoint.ipynb | BinnyDaBin/MWDSBE | aa0de50f2289e47f7c2e9134334b23c3b5594f0c | [
"MIT"
] | 10 | 2021-03-10T01:06:45.000Z | 2022-02-26T21:02:40.000Z | mwdsbe/.ipynb_checkpoints/tf-idf-alg-test-nick-checkpoint.ipynb | BinnyDaBin/MWDSBE | aa0de50f2289e47f7c2e9134334b23c3b5594f0c | [
"MIT"
] | null | null | null | 41.380856 | 304 | 0.375405 | [
[
[
"# TF-IDF",
"_____no_output_____"
]
],
[
[
"import mwdsbe\nimport mwdsbe.datasets.licenses as licenses\nimport schuylkill as skool\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# import registry\nregistry = mwdsbe.load_registry() # geopandas df",
"_____no_output_____"
],
[
"registry.head()",
"_____no_output_____"
],
[
"# import license data\nlicense = licenses.CommercialActivityLicenses.download()",
"_____no_output_____"
],
[
"license.head()",
"_____no_output_____"
],
[
"mini_registry = registry[:5]",
"_____no_output_____"
],
[
"# clean company_name and dba_name of clean datasets\nignore = ['inc', 'group', 'llc', 'corp', 'pc', 'incorporated', 'ltd']\nregistry = skool.clean_strings(registry, ['company_name', 'dba_name'], True, ignore)\nlicense = skool.clean_strings(license, ['company_name'], True, ignore)",
"_____no_output_____"
],
[
"from ftfy import fix_text",
"_____no_output_____"
],
[
"import re\n\ndef ngrams(string, n=3):\n string = re.sub(r'[,-./]|\\sBD',r'', string)\n ngrams = zip(*[string[i:] for i in range(n)])\n return [''.join(ngram) for ngram in ngrams]",
"_____no_output_____"
],
[
"ngrams(registry['company_name'].iloc[0])",
"_____no_output_____"
],
[
"from scipy.sparse import csr_matrix\nimport sparse_dot_topn.sparse_dot_topn as ct",
"_____no_output_____"
],
[
"def awesome_cossim_top(A, B, ntop, lower_bound=0):\n # force A and B as a CSR matrix.\n # If they have already been CSR, there is no overhead\n A = A.tocsr()\n B = B.tocsr()\n M, _ = A.shape\n _, N = B.shape\n \n idx_dtype = np.int32\n \n nnz_max = M*ntop\n \n indptr = np.zeros(M+1, dtype=idx_dtype)\n indices = np.zeros(nnz_max, dtype=idx_dtype)\n data = np.zeros(nnz_max, dtype=A.dtype)\n\n ct.sparse_dot_topn(\n M, N, np.asarray(A.indptr, dtype=idx_dtype),\n np.asarray(A.indices, dtype=idx_dtype),\n A.data,\n np.asarray(B.indptr, dtype=idx_dtype),\n np.asarray(B.indices, dtype=idx_dtype),\n B.data,\n ntop,\n lower_bound,\n indptr, indices, data)\n\n return csr_matrix((data,indices,indptr),shape=(M,N))",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import TfidfVectorizer\n\ncompany_names = registry['company_name'].iloc[:10]\nvectorizer = TfidfVectorizer(min_df=1, analyzer=ngrams)\ntf_idf_matrix = vectorizer.fit_transform(company_names)",
"_____no_output_____"
],
[
"tf_idf_matrix",
"_____no_output_____"
],
[
"def get_matches_df(sparse_matrix, name_vector, top=100):\n non_zeros = sparse_matrix.nonzero()\n \n sparserows = non_zeros[0]\n sparsecols = non_zeros[1]\n \n if top:\n nr_matches = top\n else:\n nr_matches = sparsecols.size\n \n left_side = np.empty([nr_matches], dtype=object)\n right_side = np.empty([nr_matches], dtype=object)\n similairity = np.zeros(nr_matches)\n \n for index in range(0, nr_matches):\n left_side[index] = name_vector[sparserows[index]]\n right_side[index] = name_vector[sparsecols[index]]\n similairity[index] = sparse_matrix.data[index]\n \n return pd.DataFrame({'left_side': left_side,\n 'right_side': right_side,\n 'similairity': similairity})",
"_____no_output_____"
],
[
"all_company_names = pd.concat([registry['company_name'].dropna(), license['company_name'].dropna()]).unique()",
"_____no_output_____"
],
[
"len(all_company_names)",
"_____no_output_____"
],
[
"vectorizer = TfidfVectorizer(min_df=1, analyzer=ngrams)\ntf_idf_matrix = vectorizer.fit_transform(all_company_names)",
"_____no_output_____"
],
[
"import time\nt1 = time.time()\nmatches = awesome_cossim_top(tf_idf_matrix, tf_idf_matrix.transpose(), 10, 0.85)\nt = time.time()-t1\nprint(\"SELFTIMED:\", t)",
"SELFTIMED: 86.46712231636047\n"
],
[
"matches",
"_____no_output_____"
],
[
"def get_matches_df(sparse_matrix, name_vector, top=100):\n non_zeros = sparse_matrix.nonzero()\n \n sparserows = non_zeros[0]\n sparsecols = non_zeros[1]\n \n if top:\n nr_matches = top\n else:\n nr_matches = sparsecols.size\n \n left_side = np.empty([nr_matches], dtype=object)\n right_side = np.empty([nr_matches], dtype=object)\n similairity = np.zeros(nr_matches)\n \n for index in range(0, nr_matches):\n left_side[index] = name_vector[sparserows[index]]\n right_side[index] = name_vector[sparsecols[index]]\n similairity[index] = sparse_matrix.data[index]\n \n return pd.DataFrame({'left_side': left_side,\n 'right_side': right_side,\n 'similarity': similairity})",
"_____no_output_____"
],
[
"matches_df = get_matches_df(matches, all_company_names, top=100000)",
"_____no_output_____"
],
[
"pd.options.display.max_rows = 999",
"_____no_output_____"
],
[
"matches_df.loc[(matches_df['similairity'] < 0.99999) & (matches_df['similairity'] > 0.94)]",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f4bb5b4b8edb4e4b4194c67c4b81e346b448e0 | 12,458 | ipynb | Jupyter Notebook | tf_align_model_input_feature_percentiles.ipynb | aimalz/justice | 2edcb471cd01d6659a498bcd0209cb5dae83375a | [
"MIT"
] | 1 | 2018-09-17T01:30:34.000Z | 2018-09-17T01:30:34.000Z | tf_align_model_input_feature_percentiles.ipynb | aimalz/justice | 2edcb471cd01d6659a498bcd0209cb5dae83375a | [
"MIT"
] | 38 | 2018-06-12T18:55:25.000Z | 2019-01-13T21:13:09.000Z | tf_align_model_input_feature_percentiles.ipynb | aimalz/justice | 2edcb471cd01d6659a498bcd0209cb5dae83375a | [
"MIT"
] | 2 | 2018-08-19T21:38:30.000Z | 2018-09-09T19:00:03.000Z | 36.3207 | 444 | 0.577139 | [
[
[
"# Generating percentiles for TensorFlow model input features\n\nThe current TensorFlow model uses histogram-like percentile features, which are kind of a continuous version of one-hot features.\n\nFor example, if key cutoff points are `[-3, 1, 0, 2, 10]`, we might encode a value `x` as `sigma((x - cutoff) / scale)`. If `sigma` is the sigmoid function, `x = 0.1`, and `scale = 0.1`, then we'd get `[1, 1, 0.73, 0, 0]`, in other words `x` is definitely above the first 2 points, mostly above the third, and below the fourth and fifth. If we increase `scale` to `2.0`, then values are less discrete: `[0.82, 0.63, 0.51, 0.28, 0.01]`.\n\nThis notebook generates appropriate cutoff points for these, to reflect most data encountered.",
"_____no_output_____"
]
],
[
[
"# Different options for soft-onehot function.\n%matplotlib inline\nimport numpy as np\nfrom matplotlib import pyplot as plt\nx = np.linspace(-10, 10, 100)\ncutoff = 1.0\nsigmoid = lambda x: 1/(1+np.exp(-x))\nscale = 2.0\nlogit = (x - cutoff) / scale\nplt.plot(x, sigmoid(logit))\nplt.plot(x, np.exp(- logit * logit))",
"_____no_output_____"
],
[
"NUM_LCS = 10_000 # key parameter, turn it down if you want this notebook to finish faster.\n\n# Settings determining type of features extracted.\nwindow_size = 10\nband_time_diff = 4.0",
"_____no_output_____"
],
[
"from matplotlib import pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport tensorflow as tf\n\nfrom justice.datasets import plasticc_data\n\nsource = plasticc_data.PlasticcBcolzSource.get_default()\nbcolz_source = plasticc_data.PlasticcBcolzSource.get_default()\nmeta_table = bcolz_source.get_table('test_set_metadata')\n%time all_ids = meta_table['object_id'][:]",
"_____no_output_____"
],
[
"%%time\nimport random\nsample_ids = random.Random(828372).sample(list(all_ids), NUM_LCS)\n\nlcs = []\n_chunk_sz = 100\nfor start in range(0, len(sample_ids), _chunk_sz):\n lcs.extend(plasticc_data.PlasticcDatasetLC.bcolz_get_lcs_by_obj_ids(\n bcolz_source=source,\n dataset=\"test_set\",\n obj_ids=sample_ids[start:start + _chunk_sz]\n ))",
"_____no_output_____"
],
[
"%%time\n\nfrom justice.features import band_settings_params\nfrom justice.features import dense_extracted_features\nfrom justice.features import feature_combinators\nfrom justice.features import metadata_features\nfrom justice.features import per_point_dataset\nfrom justice.features import raw_value_features\n\nbatch_size = 32\nrve = raw_value_features.RawValueExtractor(\n window_size=window_size,\n band_settings=band_settings_params.BandSettings(lcs[0].expected_bands)\n)\nmve = metadata_features.MetadataValueExtractor()\ndata_gen = per_point_dataset.PerPointDatasetGenerator(\n extract_fcn=feature_combinators.combine([rve.extract, mve.extract]),\n batch_size=batch_size,\n)\n\ndef input_fn():\n return data_gen.make_dataset_lcs(lcs)\n\ndef per_band_model_fn(band_features, params):\n batch_size = params[\"batch_size\"]\n window_size = params[\"window_size\"]\n wf = dense_extracted_features.WindowFeatures(\n band_features, batch_size=batch_size, window_size=window_size, band_time_diff=band_time_diff)\n dflux_dt = wf.dflux_dt(clip_magnitude=None)\n init_layer = dense_extracted_features.initial_layer(wf, include_flux_and_time=True)\n init_layer_masked = wf.masked(init_layer, value_if_masked=0, expected_extra_dims=[3])\n return {\n \"initial_layer\": init_layer_masked,\n \"in_window\": wf.in_window,\n }\n\ndef model_fn(features, labels, mode, params):\n band_settings = band_settings_params.BandSettings.from_params(params)\n per_band_data = band_settings.per_band_sub_model_fn(\n per_band_model_fn, features, params=params\n )\n predictions = {\n 'band_{}.{}'.format(band, name): tensor\n for band, tensor_dict in zip(band_settings.bands, per_band_data)\n for name, tensor in tensor_dict.items()\n }\n predictions['time'] = features['time']\n predictions['object_id'] = features['object_id']\n return tf.estimator.EstimatorSpec(\n mode=mode, predictions=predictions, loss=tf.constant(0.0), train_op=tf.no_op()\n )\n\nparams = {\n 'batch_size': batch_size,\n 'window_size': window_size,\n 'flux_scale_epsilon': 0.5,\n 'lc_bands': lcs[0].expected_bands,\n}\nestimator = tf.estimator.Estimator(\n model_fn=model_fn,\n params=params\n)\npredictions = list(estimator.predict(input_fn=input_fn, yield_single_examples=True))\nprint(f\"Got {len(predictions)} predictions.\")",
"_____no_output_____"
],
[
"predictions[4]",
"_____no_output_____"
],
[
"def get_values_df(band):\n arrays = [x[f\"band_{band}.initial_layer\"] for x in predictions if x[f\"band_{band}.in_window\"]]\n return pd.DataFrame(np.concatenate(arrays, axis=0), columns=[\"dflux_dt\", \"dflux\", \"dtime\"])\ndf = get_values_df(lcs[0].expected_bands[0])\ndf.hist('dflux_dt', bins=32)\ndf.hist('dflux', bins=32)\ndf.hist('dtime', bins=32)",
"_____no_output_____"
]
],
[
[
"## Really messy code to get a histogram with mostly-unique bins.\n\nBecause we want fixed-size arrays for TensorFlow code, we want a set of e.g. 32 unique cutoff points that reflect a good distribution of cutoffs. However its is really messy, because there tend to be strong peaks in the histogram which are repeated frequently.",
"_____no_output_____"
]
],
[
[
"import collections\nimport scipy.optimize\n\ndef _some_duplicates(non_unique, unique, num_desired):\n to_duplicate_candidates = non_unique.tolist()\n for x in unique:\n to_duplicate_candidates.remove(x)\n unique = unique.tolist()\n while len(unique) < num_desired:\n assert len(unique) <= num_desired\n to_duplicate = random.choice(to_duplicate_candidates)\n unique.insert(unique.index(to_duplicate), to_duplicate)\n return unique\n\ndef unique_percentiles(array, num_desired):\n partition_size = 100.0 / num_desired\n epsilon = 0.05 * partition_size\n \n solution = None\n optimal_solution = None\n\n def _actual_unique(vals):\n nonlocal solution, optimal_solution\n if optimal_solution is not None:\n return 0 # stop optimization, or at least return quickly\n num_points_base, perturb = vals\n num_points = int(round(num_desired * num_points_base))\n perturb = abs(perturb)\n q = np.linspace(0, 100, int(num_points))\n rng = np.random.RandomState(int(1e6 * perturb))\n noise = rng.normal(loc=0, scale=min(1.0, 10 * perturb) * epsilon, size=q.shape)\n noise[0] = 0\n noise[-1] = 0\n q += noise\n non_unique = np.percentile(array, q=q, interpolation='linear')\n unique = np.unique(non_unique)\n result = abs(num_desired - len(unique))\n if num_desired == len(unique):\n optimal_solution = unique\n elif len(unique) <= num_desired <= len(unique) + 1:\n solution = _some_duplicates(non_unique, unique, num_desired)\n return (4 if len(unique) > num_desired else 1) * result + perturb\n \n res = scipy.optimize.minimize(\n _actual_unique,\n x0=[1.0, 0.1],\n options={'maxiter': 1000, 'rhobeg': 0.3},\n tol=1e-6,\n method='COBYLA')\n if optimal_solution is None and solution is None:\n raise ValueError(f\"Could not find deduplicated percentiles!\")\n return optimal_solution if optimal_solution is not None else solution\n\ndesired_num_cutoffs = 32\nall_solutions = []\nfor band in lcs[0].expected_bands:\n df = get_values_df(band)\n for i, column in enumerate(df.columns):\n print(band, column)\n percentiles = np.array(unique_percentiles(df[column], desired_num_cutoffs), dtype=np.float32)\n median_scale = np.median(percentiles[1:] - percentiles[:-1])\n all_solutions.append({\n 'band': band,\n 'column_index': i,\n 'column': column,\n 'median_scale': float(median_scale),\n 'cutoffs': percentiles,\n })\n\nwith_settings = {\n 'window_size': window_size,\n 'band_time_diff': band_time_diff,\n 'desired_num_cutoffs': desired_num_cutoffs,\n 'solutions': all_solutions\n}",
"_____no_output_____"
]
],
[
[
"## Save to nicely-formatted JSON\n\nWrites numpy arrays as strings, then rewrites those strings.",
"_____no_output_____"
]
],
[
[
"import datetime\nimport json\n\nfrom justice import path_util\n\nclass ArrayPreEncoder(json.JSONEncoder):\n def default(self, obj):\n if isinstance(obj, np.ndarray):\n return \"<<<<{}>>>>\".format(\", \".join(f\"{x:.8f}\" for x in obj.tolist()))\n else:\n print(obj)\n return json.JSONEncoder.default(self, obj)\n\ndef _encode(x):\n result = json.dumps(x, indent=2, cls=ArrayPreEncoder).replace('\"<<<<', '[').replace('>>>>\"', ']')\n json.loads(result) # error if not decodable\n return result\n\nnow = datetime.datetime.now()\npath = path_util.data_dir / 'tf_align_model' / 'feature_extraction' / (\n f\"cutoffs__window_sz-{window_size}__{now.year:04d}-{now.month:02d}-{now.day:02d}.json\")\npath.parent.mkdir(parents=True, exist_ok=True)\nwith open(str(path), 'w') as f:\n f.write(_encode(with_settings))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f4c13d9c72efec4c57e32d3bb48c173f1bca9f | 9,792 | ipynb | Jupyter Notebook | examples/1. Introductory tutorial.ipynb | KarrLab/bcforms | 053765e83b89430f288539e0a211012d23bf2e6f | [
"MIT"
] | null | null | null | examples/1. Introductory tutorial.ipynb | KarrLab/bcforms | 053765e83b89430f288539e0a211012d23bf2e6f | [
"MIT"
] | null | null | null | examples/1. Introductory tutorial.ipynb | KarrLab/bcforms | 053765e83b89430f288539e0a211012d23bf2e6f | [
"MIT"
] | null | null | null | 23.09434 | 723 | 0.529514 | [
[
[
"`BcForms` is a toolkit for concretely describing the primary structure of macromolecular complexes, including non-canonical monomeric forms and intra and inter-subunit crosslinks. `BcForms` includes a textual grammar for describing complexes and a Python library, a command line program, and a REST API for validating and manipulating complexes described in this grammar. `BcForms` represents complexes as sets of subunits, with their stoichiometries, and covalent crosslinks which link the subunits. DNA, RNA, and protein subunits can be represented using `BpForms`. Small molecule subunits can be represented using `openbabel.OBMol`, typically imported from SMILES or InChI.",
"_____no_output_____"
],
[
"This notebook illustrates how to use the `BcForms` Python library via some simple. Please see the second tutorial for more details and more examples. Please also see the [documentation](https://docs.karrlab.org/bcforms/) for more information about the `BcForms` grammar and more instructions for using the `BcForms` website, JSON REST API, and command line interface.",
"_____no_output_____"
],
[
"# Import BpForms and BcForms libraries",
"_____no_output_____"
]
],
[
[
"import bcforms\nimport bpforms",
"_____no_output_____"
]
],
[
[
"# Create complexes from their string representations",
"_____no_output_____"
]
],
[
[
"form_1 = bcforms.BcForm().from_str('2 * subunit_a + 3 * subunit_b')\nform_1.set_subunit_attribute('subunit_a', 'structure',\n bpforms.ProteinForm().from_str('CAAAAAAAA'))\nform_1.set_subunit_attribute('subunit_b', 'structure',\n bpforms.ProteinForm().from_str('AAAAAAAAC'))",
"_____no_output_____"
],
[
"form_2 = bcforms.BcForm().from_str(\n '2 * subunit_a'\n '| x-link: [type: disulfide | l: subunit_a(1)-1 | r: subunit_a(2)-1]')\nform_2.set_subunit_attribute('subunit_a', 'structure',\n bpforms.ProteinForm().from_str('CAAAAAAAA'))",
"_____no_output_____"
]
],
[
[
"# Create complexes programmatically",
"_____no_output_____"
]
],
[
[
"form_1_b = bcforms.BcForm()\nform_1_b.subunits.append(bcforms.core.Subunit('subunit_a', 2,\n bpforms.ProteinForm().from_str('CAAAAAAAA')))\nform_1_b.subunits.append(bcforms.core.Subunit('subunit_b', 3,\n bpforms.ProteinForm().from_str('AAAAAAAAC')))",
"_____no_output_____"
],
[
"form_2_b = bcforms.BcForm()\nsubunit = bcforms.core.Subunit('subunit_a', 2,\n bpforms.ProteinForm().from_str('CAAAAAAAA'))\nform_2_b.subunits.append(subunit)\nform_2_b.crosslinks.append(bcforms.core.OntologyCrosslink(\n 'disulfide', 'subunit_a', 1, 'subunit_a', 1, 1, 2))",
"_____no_output_____"
]
],
[
[
"# Get properties of polymers",
"_____no_output_____"
],
[
"## Subunits",
"_____no_output_____"
]
],
[
[
"form_1.subunits",
"_____no_output_____"
]
],
[
[
"## Crosslinks",
"_____no_output_____"
]
],
[
[
"form_2.crosslinks ",
"_____no_output_____"
]
],
[
[
"# Get the string representation of a complex",
"_____no_output_____"
]
],
[
[
"str(form_1_b) ",
"_____no_output_____"
]
],
[
[
"# Check equality of complexes",
"_____no_output_____"
]
],
[
[
"form_1_b.is_equal(form_1)",
"_____no_output_____"
]
],
[
[
"# Calculate properties of a complex",
"_____no_output_____"
],
[
"## Molecular structure",
"_____no_output_____"
]
],
[
[
"form_1.get_structure()[0]",
"_____no_output_____"
]
],
[
[
"## SMILES representation",
"_____no_output_____"
]
],
[
[
"form_1.export('smiles') ",
"_____no_output_____"
]
],
[
[
"## Formula",
"_____no_output_____"
]
],
[
[
"form_1.get_formula()",
"_____no_output_____"
]
],
[
[
"## Charge",
"_____no_output_____"
]
],
[
[
"form_1.get_charge()",
"_____no_output_____"
]
],
[
[
"## Molecular weight",
"_____no_output_____"
]
],
[
[
"form_1.get_mol_wt()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f4d198c8f185d94d7bd746a780fe1c0ecf5cbc | 26,032 | ipynb | Jupyter Notebook | adanet/examples/tutorials/adanet_tpu.ipynb | gitter-badger/adanet | 91fdf069ff370e34f55a57398745dd247dafc8a3 | [
"Apache-2.0"
] | 1 | 2020-08-14T02:22:14.000Z | 2020-08-14T02:22:14.000Z | adanet/examples/tutorials/adanet_tpu.ipynb | gitter-badger/adanet | 91fdf069ff370e34f55a57398745dd247dafc8a3 | [
"Apache-2.0"
] | 1 | 2019-03-04T16:57:55.000Z | 2019-03-04T16:57:55.000Z | adanet/examples/tutorials/adanet_tpu.ipynb | gitter-badger/adanet | 91fdf069ff370e34f55a57398745dd247dafc8a3 | [
"Apache-2.0"
] | null | null | null | 39.442424 | 283 | 0.507183 | [
[
[
"##### Copyright 2018 The AdaNet Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# AdaNet on TPU",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/adanet/blob/master/adanet/examples/tutorials/adanet_tpu.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/adanet/blob/master/adanet/examples/tutorials/adanet_tpu.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n</table>\n",
"_____no_output_____"
],
[
"AdaNet supports training on Google's custom machine learning accelerators known\nas Tensor Processing Units (TPU). Conveniently, we provide `adanet.TPUEstimator`\nwhich handles TPU support behind the scenes. There are only a few minor changes\nneeded to switch from `adanet.Estimator` to `adanet.TPUEstimator`. We highlight\nthe necessary changes in this tutorial.\n\nIf the reader is not familiar with AdaNet, it is reccommended they take a look\nat\n[The AdaNet Objective](https://colab.sandbox.google.com/github/tensorflow/adanet/blob/master/adanet/examples/tutorials/adanet_objective.ipynb)\nand in particular\n[Customizing AdaNet](https://colab.sandbox.google.com/github/tensorflow/adanet/blob/master/adanet/examples/tutorials/adanet_objective.ipynb)\nas this tutorial builds upon the latter.\n\n**NOTE: you must provide a valid GCS bucket to use TPUEstimator.**\n\nTo begin, we import the necessary packages, obtain the Colab's TPU master\naddress, and give the TPU permissions to write to our GCS Bucket. Follow the\ninstructions\n[here](https://colab.sandbox.google.com/notebooks/tpu.ipynb#scrollTo=_pQCOmISAQBu)\nto connect to a Colab TPU runtime.\n\n",
"_____no_output_____"
]
],
[
[
"#@test {\"skip\": true}\n# If you're running this in Colab, first install the adanet package:\n!pip install adanet",
"_____no_output_____"
],
[
"from __future__ import absolute_import\nfrom __future__ import division\nfrom __future__ import print_function\n\nimport functools\nimport json\nimport os\nimport six\nimport time\n\nimport adanet\nfrom google.colab import auth\nimport tensorflow as tf\n\nBUCKET = '' #@param {type: 'string'}\nMODEL_DIR = 'gs://{}/{}'.format(\n BUCKET, time.strftime('adanet-tpu-estimator/%Y-%m-%d-%H-%M-%S'))\n\nMASTER = ''\nif 'COLAB_TPU_ADDR' in os.environ:\n auth.authenticate_user()\n\n MASTER = 'grpc://' + os.environ['COLAB_TPU_ADDR']\n\n # Authenticate TPU to use GCS Bucket.\n with tf.Session(MASTER) as sess:\n with open('/content/adc.json', 'r') as file_:\n auth_info = json.load(file_)\n tf.contrib.cloud.configure_gcs(sess, credentials=auth_info)\n\n\n# The random seed to use.\nRANDOM_SEED = 42",
"_____no_output_____"
]
],
[
[
"## Fashion MNIST\n\nWe focus again on the Fashion MNIST dataset and download the data via Keras.",
"_____no_output_____"
]
],
[
[
"(x_train, y_train), (x_test, y_test) = (\n tf.keras.datasets.fashion_mnist.load_data())",
"Downloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/train-labels-idx1-ubyte.gz\n32768/29515 [=================================] - 0s 0us/step\n40960/29515 [=========================================] - 0s 0us/step\nDownloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/train-images-idx3-ubyte.gz\n26427392/26421880 [==============================] - 0s 0us/step\n26435584/26421880 [==============================] - 0s 0us/step\nDownloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/t10k-labels-idx1-ubyte.gz\n16384/5148 [===============================================================================================] - 0s 0us/step\nDownloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/t10k-images-idx3-ubyte.gz\n4423680/4422102 [==============================] - 0s 0us/step\n4431872/4422102 [==============================] - 0s 0us/step\n"
]
],
[
[
"## `input_fn` Changes\n\nThere are two minor changes we must make to `input_fn` to support running on\nTPU:\n\n1. TPUs dynamically shard the input data depending on the number of cores used.\n Because of this, we augment `input_fn` to take a dictionary `params`\n argument. When running on TPU, `params` contains a `batch_size` field with\n the appropriate batch size.\n\n1. Once the input is batched, we drop the last batch if it is smaller than\n `batch_size`. This can simply be done by specifying `drop_remainder=True` to\n the\n [`tf.data.DataSet.batch()`](https://www.tensorflow.org/api_docs/python/tf/data/Dataset#batch)\n function. It is important to specify this option since TPUs do not support\n dynamic shapes. Note that we only drop the remainder batch during training\n since evaluation is still done on the CPU.",
"_____no_output_____"
]
],
[
[
"FEATURES_KEY = \"images\"\n\n\ndef generator(images, labels):\n \"\"\"Returns a generator that returns image-label pairs.\"\"\"\n\n def _gen():\n for image, label in zip(images, labels):\n yield image, label\n\n return _gen\n\n\ndef preprocess_image(image, label):\n \"\"\"Preprocesses an image for an `Estimator`.\"\"\"\n image = image / 255.\n image = tf.reshape(image, [28, 28, 1])\n features = {FEATURES_KEY: image}\n return features, label\n\n\ndef input_fn(partition, training, batch_size):\n \"\"\"Generate an input_fn for the Estimator.\"\"\"\n\n def _input_fn(params): # TPU: specify `params` argument.\n\n # TPU: get the TPU set `batch_size`, if available.\n batch_size_ = params.get(\"batch_size\", batch_size)\n\n if partition == \"train\":\n dataset = tf.data.Dataset.from_generator(\n generator(x_train, y_train), (tf.float32, tf.int32), ((28, 28), ()))\n elif partition == \"predict\":\n dataset = tf.data.Dataset.from_generator(\n generator(x_test[:10], y_test[:10]), (tf.float32, tf.int32),\n ((28, 28), ()))\n else:\n dataset = tf.data.Dataset.from_generator(\n generator(x_test, y_test), (tf.float32, tf.int32), ((28, 28), ()))\n\n if training:\n dataset = dataset.shuffle(10 * batch_size_, seed=RANDOM_SEED).repeat()\n\n # TPU: drop the remainder batch when training on TPU.\n dataset = dataset.map(preprocess_image).batch(\n batch_size_, drop_remainder=training)\n iterator = dataset.make_one_shot_iterator()\n features, labels = iterator.get_next()\n return features, labels\n\n return _input_fn",
"_____no_output_____"
]
],
[
[
"## `model_fn` Changes\n\nWe use a similar CNN architecture as used in the\n[Customizing AdaNet](https://colab.sandbox.google.com/github/tensorflow/adanet/blob/master/adanet/examples/tutorials/customizing_adanet.ipynb)\ntutorial. The only TPU specific change we need to make is wrap the `optimizer`\nin a\n[`tf.contrib.tpu.CrossShardOptimizer`](https://www.google.com/search?q=cross+shard+optimizer&oq=cross+shard+optimizer&aqs=chrome.0.0j69i57.2391j0j7&sourceid=chrome&ie=UTF-8).",
"_____no_output_____"
]
],
[
[
"#@title Define the Builder and Generator\nclass SimpleCNNBuilder(adanet.subnetwork.Builder):\n \"\"\"Builds a CNN subnetwork for AdaNet.\"\"\"\n\n def __init__(self, learning_rate, max_iteration_steps, seed):\n \"\"\"Initializes a `SimpleCNNBuilder`.\n\n Args:\n learning_rate: The float learning rate to use.\n max_iteration_steps: The number of steps per iteration.\n seed: The random seed.\n\n Returns:\n An instance of `SimpleCNNBuilder`.\n \"\"\"\n self._learning_rate = learning_rate\n self._max_iteration_steps = max_iteration_steps\n self._seed = seed\n\n def build_subnetwork(self,\n features,\n logits_dimension,\n training,\n iteration_step,\n summary,\n previous_ensemble=None):\n \"\"\"See `adanet.subnetwork.Builder`.\"\"\"\n images = list(features.values())[0]\n kernel_initializer = tf.keras.initializers.he_normal(seed=self._seed)\n x = tf.keras.layers.Conv2D(\n filters=16,\n kernel_size=3,\n padding=\"same\",\n activation=\"relu\",\n kernel_initializer=kernel_initializer)(\n images)\n x = tf.keras.layers.MaxPool2D(pool_size=2, strides=2)(x)\n x = tf.keras.layers.Flatten()(x)\n x = tf.keras.layers.Dense(\n units=64, activation=\"relu\", kernel_initializer=kernel_initializer)(\n x)\n\n logits = tf.keras.layers.Dense(\n units=10, activation=None, kernel_initializer=kernel_initializer)(\n x)\n\n complexity = tf.constant(1)\n\n return adanet.Subnetwork(\n last_layer=x,\n logits=logits,\n complexity=complexity,\n persisted_tensors={})\n\n def build_subnetwork_train_op(self,\n subnetwork,\n loss,\n var_list,\n labels,\n iteration_step,\n summary,\n previous_ensemble=None):\n \"\"\"See `adanet.subnetwork.Builder`.\"\"\"\n\n learning_rate = tf.train.cosine_decay(\n learning_rate=self._learning_rate,\n global_step=iteration_step,\n decay_steps=self._max_iteration_steps)\n optimizer = tf.train.MomentumOptimizer(learning_rate, .9)\n # TPU: wrap the optimizer in a CrossShardOptimizer.\n optimizer = tf.contrib.tpu.CrossShardOptimizer(optimizer)\n return optimizer.minimize(loss=loss, var_list=var_list)\n\n def build_mixture_weights_train_op(self, loss, var_list, logits, labels,\n iteration_step, summary):\n \"\"\"See `adanet.subnetwork.Builder`.\"\"\"\n return tf.no_op(\"mixture_weights_train_op\")\n\n @property\n def name(self):\n \"\"\"See `adanet.subnetwork.Builder`.\"\"\"\n return \"simple_cnn\"\n\n\nclass SimpleCNNGenerator(adanet.subnetwork.Generator):\n \"\"\"Generates a `SimpleCNN` at each iteration.\"\"\"\n\n def __init__(self, learning_rate, max_iteration_steps, seed=None):\n \"\"\"Initializes a `Generator` that builds `SimpleCNNs`.\n\n Args:\n learning_rate: The float learning rate to use.\n max_iteration_steps: The number of steps per iteration.\n seed: The random seed.\n\n Returns:\n An instance of `Generator`.\n \"\"\"\n self._seed = seed\n self._dnn_builder_fn = functools.partial(\n SimpleCNNBuilder,\n learning_rate=learning_rate,\n max_iteration_steps=max_iteration_steps)\n\n def generate_candidates(self, previous_ensemble, iteration_number,\n previous_ensemble_reports, all_reports):\n \"\"\"See `adanet.subnetwork.Generator`.\"\"\"\n seed = self._seed\n # Change the seed according to the iteration so that each subnetwork\n # learns something different.\n if seed is not None:\n seed += iteration_number\n return [self._dnn_builder_fn(seed=seed)]",
"_____no_output_____"
]
],
[
[
"## Launch TensorBoard\n\nLet's run [TensorBoard](https://www.tensorflow.org/guide/summaries_and_tensorboard) to visualize model training over time. We'll use [ngrok](https://ngrok.com/) to tunnel traffic to localhost.\n\n*The instructions for setting up Tensorboard were obtained from https://www.dlology.com/blog/quick-guide-to-run-tensorboard-in-google-colab/*\n\nRun the next cells and follow the link to see the TensorBoard in a new tab.",
"_____no_output_____"
]
],
[
[
"#@test {\"skip\": true}\n\nget_ipython().system_raw(\n 'tensorboard --logdir {} --host 0.0.0.0 --port 6006 &'\n .format(MODEL_DIR)\n)\n\n# Install ngrok binary.\n! wget https://bin.equinox.io/c/4VmDzA7iaHb/ngrok-stable-linux-amd64.zip\n! unzip ngrok-stable-linux-amd64.zip\n\nprint(\"Follow this link to open TensorBoard in a new tab.\")\nget_ipython().system_raw('./ngrok http 6006 &')\n! curl -s http://localhost:4040/api/tunnels | python3 -c \\\n \"import sys, json; print(json.load(sys.stdin)['tunnels'][0]['public_url'])\"\n\n",
"_____no_output_____"
]
],
[
[
"## Using `adanet.TPUEstimator` to Train and Evaluate\n\nFinally, we switch from `adanet.Estimator` to `adanet.TPUEstimator`. There are\ntwo last changes needed:\n\n1. Update the `RunConfig` to be a\n [`tf.contrib.tpu.RunConfig`](https://www.tensorflow.org/api_docs/python/tf/contrib/tpu/RunConfig).\n We supply the TPU `master` address and set `iterations_per_loop=200`. This\n choice is fairly arbitrary in our case. A good practice is to set it to the\n number of steps in between summary writes and metric evals.\n1. Finally, we specify the `use_tpu` and `batch_size` parameters\n `adanet.TPUEstimator`.\n\nThere is an important thing to note about the `batch_size`: each TPU chip\nconsists of 2 cores with 4 shards each. In the\n[Customizing AdaNet](https://colab.sandbox.google.com/github/tensorflow/adanet/blob/master/adanet/examples/tutorials/customizing_adanet.ipynb)\ntutorial, a `batch_size` of 64 was used. To be consistent we use the same\n`batch_size` per shard and drop the number of training steps accordingly. In\nother words, since we're running on one TPU we set `batch_size=64*8=512` and\n`train_steps=1000`. In the ideal case, since we drop the `train_steps` by 5x,\nthis means we're **training 5x faster!**",
"_____no_output_____"
]
],
[
[
"#@title AdaNet Parameters\nLEARNING_RATE = 0.25 #@param {type:\"number\"}\nTRAIN_STEPS = 1000 #@param {type:\"integer\"}\nBATCH_SIZE = 512 #@param {type:\"integer\"}\nADANET_ITERATIONS = 2 #@param {type:\"integer\"}\n\n# TPU: switch `tf.estimator.RunConfig` to `tf.contrib.tpu.RunConfig`.\n# The main required changes are specifying `tpu_config` and `master`.\nconfig = tf.contrib.tpu.RunConfig(\n tpu_config=tf.contrib.tpu.TPUConfig(iterations_per_loop=200),\n master=MASTER,\n save_checkpoints_steps=200,\n save_summary_steps=200,\n tf_random_seed=RANDOM_SEED)\n\nhead = tf.contrib.estimator.multi_class_head(\n n_classes=10, loss_reduction=tf.losses.Reduction.SUM_OVER_BATCH_SIZE)\nmax_iteration_steps = TRAIN_STEPS // ADANET_ITERATIONS\n# TPU: switch `adanet.Estimator` to `adanet.TPUEstimator`.\ntry:\n estimator = adanet.TPUEstimator(\n head=head,\n subnetwork_generator=SimpleCNNGenerator(\n learning_rate=LEARNING_RATE,\n max_iteration_steps=max_iteration_steps,\n seed=RANDOM_SEED),\n max_iteration_steps=max_iteration_steps,\n evaluator=adanet.Evaluator(\n input_fn=input_fn(\"train\", training=False, batch_size=BATCH_SIZE),\n steps=None),\n adanet_loss_decay=.99,\n config=config,\n model_dir=MODEL_DIR,\n # TPU: specify `use_tpu` and the batch_size parameters.\n use_tpu=True,\n train_batch_size=BATCH_SIZE,\n eval_batch_size=32)\nexcept tf.errors.InvalidArgumentError as e:\n six.raise_from(\n Exception(\n \"Invalid GCS Bucket: you must provide a valid GCS bucket in the \"\n \"`BUCKET` form field of the first cell.\"), e)\n\nresults, _ = tf.estimator.train_and_evaluate(\n estimator,\n train_spec=tf.estimator.TrainSpec(\n input_fn=input_fn(\"train\", training=True, batch_size=BATCH_SIZE),\n max_steps=TRAIN_STEPS),\n eval_spec=tf.estimator.EvalSpec(\n input_fn=input_fn(\"test\", training=False, batch_size=BATCH_SIZE),\n steps=None,\n start_delay_secs=1,\n throttle_secs=1,\n ))\n\nprint(\"Accuracy:\", results[\"accuracy\"])\nprint(\"Loss:\", results[\"average_loss\"])",
"Accuracy: 0.8913\nLoss: 0.298405\n"
]
],
[
[
"## Conclusion\n\nThat was easy! With very few changes we were able to transform our original\nestimator into one which can harness the power of TPUs.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0f4ebe1cae2d669a9e5ed39bd8afbb41a81fb77 | 6,910 | ipynb | Jupyter Notebook | 4. Time Series/Udacity/03_time_windows.ipynb | AmirRazaMBA/TensorFlow-Certification | ec0990007cff6daf36beac6d00d95c81cdf80353 | [
"MIT"
] | 1 | 2020-11-20T14:46:45.000Z | 2020-11-20T14:46:45.000Z | 4. Time Series/Coursera/Exam Prep/03_time_windows.ipynb | AmirRazaMBA/TF_786 | ec0990007cff6daf36beac6d00d95c81cdf80353 | [
"MIT"
] | null | null | null | 4. Time Series/Coursera/Exam Prep/03_time_windows.ipynb | AmirRazaMBA/TF_786 | ec0990007cff6daf36beac6d00d95c81cdf80353 | [
"MIT"
] | 1 | 2021-11-17T02:40:23.000Z | 2021-11-17T02:40:23.000Z | 21.936508 | 161 | 0.473661 | [
[
[
"# Time windows",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf",
"_____no_output_____"
]
],
[
[
"## Time Windows\n\nFirst, we will train a model to forecast the next step given the previous 20 steps, therefore, we need to create a dataset of 20-step windows for training.",
"_____no_output_____"
]
],
[
[
"dataset = tf.data.Dataset.range(10)\nfor val in dataset:\n print(val.numpy())",
"0\n1\n2\n3\n4\n5\n6\n7\n8\n9\n"
],
[
"dataset = tf.data.Dataset.range(10)\ndataset = dataset.window(5, shift=1)\nfor window_dataset in dataset:\n for val in window_dataset:\n print(val.numpy(), end=\" \")\n print()",
"0 1 2 3 4 \n1 2 3 4 5 \n2 3 4 5 6 \n3 4 5 6 7 \n4 5 6 7 8 \n5 6 7 8 9 \n6 7 8 9 \n7 8 9 \n8 9 \n9 \n"
],
[
"dataset = tf.data.Dataset.range(10)\ndataset = dataset.window(5, shift=1, drop_remainder=True)\nfor window_dataset in dataset:\n for val in window_dataset:\n print(val.numpy(), end=\" \")\n print()",
"0 1 2 3 4 \n1 2 3 4 5 \n2 3 4 5 6 \n3 4 5 6 7 \n4 5 6 7 8 \n5 6 7 8 9 \n"
],
[
"dataset = tf.data.Dataset.range(10)\ndataset = dataset.window(5, shift=1, drop_remainder=True)\ndataset = dataset.flat_map(lambda window: window.batch(5))\nfor window in dataset:\n print(window.numpy())",
"[0 1 2 3 4]\n[1 2 3 4 5]\n[2 3 4 5 6]\n[3 4 5 6 7]\n[4 5 6 7 8]\n[5 6 7 8 9]\n"
],
[
"dataset = tf.data.Dataset.range(10)\ndataset = dataset.window(5, shift=1, drop_remainder=True)\ndataset = dataset.flat_map(lambda window: window.batch(5))\ndataset = dataset.map(lambda window: (window[:-1], window[-1:]))\nfor x, y in dataset:\n print(x.numpy(), y.numpy())",
"[0 1 2 3] [4]\n[1 2 3 4] [5]\n[2 3 4 5] [6]\n[3 4 5 6] [7]\n[4 5 6 7] [8]\n[5 6 7 8] [9]\n"
],
[
"dataset = tf.data.Dataset.range(10)\ndataset = dataset.window(5, shift=1, drop_remainder=True)\ndataset = dataset.flat_map(lambda window: window.batch(5))\ndataset = dataset.map(lambda window: (window[:-1], window[-1:]))\ndataset = dataset.shuffle(buffer_size=10)\nfor x, y in dataset:\n print(x.numpy(), y.numpy())",
"[2 3 4 5] [6]\n[1 2 3 4] [5]\n[0 1 2 3] [4]\n[5 6 7 8] [9]\n[4 5 6 7] [8]\n[3 4 5 6] [7]\n"
],
[
"dataset = tf.data.Dataset.range(10)\ndataset = dataset.window(5, shift=1, drop_remainder=True)\ndataset = dataset.flat_map(lambda window: window.batch(5))\ndataset = dataset.map(lambda window: (window[:-1], window[-1:]))\ndataset = dataset.shuffle(buffer_size=10)\ndataset = dataset.batch(2).prefetch(1)\nfor x, y in dataset:\n print(\"x =\", x.numpy())\n print(\"y =\", y.numpy())",
"x = [[1 2 3 4]\n [4 5 6 7]]\ny = [[5]\n [8]]\nx = [[0 1 2 3]\n [5 6 7 8]]\ny = [[4]\n [9]]\nx = [[3 4 5 6]\n [2 3 4 5]]\ny = [[7]\n [6]]\n"
],
[
"def window_dataset(series, window_size, batch_size=32,\n shuffle_buffer=1000):\n dataset = tf.data.Dataset.from_tensor_slices(series)\n dataset = dataset.window(window_size + 1, shift=1, drop_remainder=True)\n dataset = dataset.flat_map(lambda window: window.batch(window_size + 1))\n dataset = dataset.shuffle(shuffle_buffer)\n dataset = dataset.map(lambda window: (window[:-1], window[-1]))\n dataset = dataset.batch(batch_size).prefetch(1)\n return dataset",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f50a961252f8cfae25200cc31a9bb71f5ac15b | 1,739 | ipynb | Jupyter Notebook | Jour 1/hello.ipynb | bellash13/SmartAcademyPython | 44d0f6db0fcdcbbf1449a45b073a2b3182a19714 | [
"MIT"
] | null | null | null | Jour 1/hello.ipynb | bellash13/SmartAcademyPython | 44d0f6db0fcdcbbf1449a45b073a2b3182a19714 | [
"MIT"
] | null | null | null | Jour 1/hello.ipynb | bellash13/SmartAcademyPython | 44d0f6db0fcdcbbf1449a45b073a2b3182a19714 | [
"MIT"
] | null | null | null | 17.927835 | 48 | 0.493387 | [
[
[
"print(\"Hello world!\")",
"Hello world!\n"
],
[
"def direBonjour():\n print(\"Hello World!\")\n \ndireBonjour()",
"Hello World!\n"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
d0f52f26a0cff6c2aeb66e223a519a8cbfdc9c84 | 50,156 | ipynb | Jupyter Notebook | H20 ML Notebooks/H20 Finalized Chunking Procedure.ipynb | Pandula1234/Data-Analysis-Intern | 6c6a25e9b4587be1103e9c4b0c50fcb8c04d3062 | [
"Apache-2.0"
] | null | null | null | H20 ML Notebooks/H20 Finalized Chunking Procedure.ipynb | Pandula1234/Data-Analysis-Intern | 6c6a25e9b4587be1103e9c4b0c50fcb8c04d3062 | [
"Apache-2.0"
] | null | null | null | H20 ML Notebooks/H20 Finalized Chunking Procedure.ipynb | Pandula1234/Data-Analysis-Intern | 6c6a25e9b4587be1103e9c4b0c50fcb8c04d3062 | [
"Apache-2.0"
] | null | null | null | 35.954122 | 283 | 0.387192 | [
[
[
"## Data Procesing",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"import h2o\nfrom h2o.estimators.random_forest import H2ORandomForestEstimator\nh2o.init()",
"Checking whether there is an H2O instance running at http://localhost:54321 . connected.\n"
],
[
"%%time\ntrain_new = pd.read_csv(\"D:\\\\DC Universe\\\\Ucsc\\\\Third Year\\\\ENH 3201 Industrial Placements\\\\H20 Applications\\\\H20 ML Notebooks\\\\H20Csv\\\\Titanic\\\\titanic_train.csv\")\ntest = pd.read_csv(\"D:\\\\DC Universe\\\\Ucsc\\\\Third Year\\\\ENH 3201 Industrial Placements\\\\H20 Applications\\\\H20 ML Notebooks\\\\H20Csv\\\\Titanic\\\\titanic_test.csv\")\nsubs = pd.read_csv('D:\\\\DC Universe\\\\Ucsc\\\\Third Year\\\\ENH 3201 Industrial Placements\\\\H20 Applications\\\\H20 ML Notebooks\\\\H20Csv\\\\Titanic\\\\gender_submission.csv')\n\n#train_new.to_pickle(\"D:\\\\DC Universe\\\\Ucsc\\\\Third Year\\\\ENH 3201 Industrial Placements\\\\H20 Applications\\\\H20 ML Notebooks\\\\H20Csv\\\\Titanic\\\\train_set.pkl\")",
"Wall time: 31.5 ms\n"
],
[
"%%time\ntrain = pd.read_pickle(\"D:\\\\DC Universe\\\\Ucsc\\\\Third Year\\\\ENH 3201 Industrial Placements\\\\H20 Applications\\\\H20 ML Notebooks\\\\H20Csv\\\\Titanic\\\\train_set.pkl\")",
"Wall time: 4.06 ms\n"
],
[
"drop_elements = ['PassengerId', 'Name', 'Ticket', 'Cabin', 'SibSp','Parch']\ntrain = train.drop(drop_elements, axis = 1)\ntest = test.drop(drop_elements, axis = 1)\n\ndef checkNull_fillData(df):\n for col in df.columns:\n if len(df.loc[df[col].isnull() == True]) != 0:\n if df[col].dtype == \"float64\" or df[col].dtype == \"int64\":\n df.loc[df[col].isnull() == True,col] = df[col].mean()\n else:\n df.loc[df[col].isnull() == True,col] = df[col].mode()[0]\n \ncheckNull_fillData(train)\ncheckNull_fillData(test)\n\nstr_list = [] \nnum_list = []\nfor colname, colvalue in train.iteritems():\n if type(colvalue[1]) == str:\n str_list.append(colname)\n else:\n num_list.append(colname)\n \ntrain = pd.get_dummies(train, columns=str_list)\ntest = pd.get_dummies(test, columns=str_list)\n\ntrain = h2o.H2OFrame(train)\ntest = h2o.H2OFrame(test)",
"Parse progress: |████████████████████████████████████████████████████████████████| (done) 100%\nParse progress: |████████████████████████████████████████████████████████████████| (done) 100%\n"
],
[
"train.describe()",
"Rows:891\nCols:9\n\n\n"
],
[
"test.describe()",
"Rows:418\nCols:8\n\n\n"
],
[
"train1, valid1, new_data1 = train.split_frame(ratios = [.7, .15], seed = 1234)",
"_____no_output_____"
],
[
"#train.columns\ntest.columns",
"_____no_output_____"
],
[
"predictors = [\"Age\",\"Embarked_C\",\"Pclass\",\"Embarked_Q\",\"Sex_male\"]\nresponse = \"Fare\"",
"_____no_output_____"
],
[
"titanic = H2ORandomForestEstimator(model_id=\"titanic\", ntrees = 1, seed = 1234)\ntitanic.train(x = predictors, y = response, training_frame = train1, validation_frame = valid1)",
"drf Model Build progress: |██████████████████████████████████████████████████████| (done) 100%\nModel Details\n=============\nH2ORandomForestEstimator : Distributed Random Forest\nModel Key: titanic\n\n\nModel Summary: \n"
],
[
"model_path = h2o.save_model(model=titanic, path=\"D:\\\\DC Universe\\\\Ucsc\\\\Third Year\\\\ENH 3201 Industrial Placements\\\\H20 Applications\\\\H20 ML Notebooks\\\\H20Csv\\\\Titanic\\\\\", force=True)\nprint (model_path)",
"D:\\DC Universe\\Ucsc\\Third Year\\ENH 3201 Industrial Placements\\H20 Applications\\H20 ML Notebooks\\H20Csv\\Titanic\\titanic\n"
],
[
"titanic = h2o.load_model(\"D:\\\\DC Universe\\\\Ucsc\\\\Third Year\\\\ENH 3201 Industrial Placements\\\\H20 Applications\\\\H20 ML Notebooks\\\\H20Csv\\\\Titanic\\\\titanic\")",
"_____no_output_____"
],
[
"titanic",
"Model Details\n=============\nH2ORandomForestEstimator : Distributed Random Forest\nModel Key: titanic\n\n\nModel Summary: \n"
],
[
"train2, valid2, new_data2 = test.split_frame(ratios = [.7, .15], seed = 1234)",
"_____no_output_____"
],
[
"# Checkpoint on the same dataset. This shows how to train an additional\n# 9 trees on top of the first 1. To do this, set ntrees equal to 10.\ntitanic_continued = H2ORandomForestEstimator(model_id = 'titanic_new',\n checkpoint = titanic,\n ntrees = 4,\n seed = 1234)\ntitanic_continued.train(x = predictors, y = response, training_frame = train2, validation_frame = valid2)",
"drf Model Build progress: |██████████████████████████████████████████████████████| (done) 100%\nModel Details\n=============\nH2ORandomForestEstimator : Distributed Random Forest\nModel Key: titanic_new\n\n\nModel Summary: \n"
],
[
"col_count=len(train.columns)\ncol_count",
"_____no_output_____"
],
[
"row_count=len(train)\n# split_ratio = int(input(\"Enter Split Ratio >>>\"))",
"_____no_output_____"
],
[
"#split_ratio = int(input(\"Enter Split Ratio >>>\"))\n#import modin.pandas as mpd",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f53be8eea6ed99235c53553d1b9bb8c5ae745b | 28,862 | ipynb | Jupyter Notebook | database/notebooks/01-set-up-database.ipynb | BoxesFullOfPepe/serverless-full-stack-apps-azure-sql | d568744084d4a5ad99b69a34ae16aa2b03776eaa | [
"MIT"
] | null | null | null | database/notebooks/01-set-up-database.ipynb | BoxesFullOfPepe/serverless-full-stack-apps-azure-sql | d568744084d4a5ad99b69a34ae16aa2b03776eaa | [
"MIT"
] | null | null | null | database/notebooks/01-set-up-database.ipynb | BoxesFullOfPepe/serverless-full-stack-apps-azure-sql | d568744084d4a5ad99b69a34ae16aa2b03776eaa | [
"MIT"
] | null | null | null | 33.251152 | 547 | 0.419063 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0f541b5628ae44e8e0600957067e6832cc7f842 | 179,841 | ipynb | Jupyter Notebook | notebooks_tf2/3_ConvolutionalNetwork/2_CustomizedTrainingLoops.ipynb | kattoyoshi/Tensorflow-Examples | 6b0d4315d397917dc6e6e6f3448d3223fb0e1816 | [
"MIT"
] | null | null | null | notebooks_tf2/3_ConvolutionalNetwork/2_CustomizedTrainingLoops.ipynb | kattoyoshi/Tensorflow-Examples | 6b0d4315d397917dc6e6e6f3448d3223fb0e1816 | [
"MIT"
] | null | null | null | notebooks_tf2/3_ConvolutionalNetwork/2_CustomizedTrainingLoops.ipynb | kattoyoshi/Tensorflow-Examples | 6b0d4315d397917dc6e6e6f3448d3223fb0e1816 | [
"MIT"
] | null | null | null | 386.754839 | 84,504 | 0.923927 | [
[
[
"# MNIST Classification with CNN -- Customized training loops --\nIn this notebook, I describe how to implement CNN using tf.keras.\nTraining loop is defined by customized step-by-step training loops.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport time\nimport numpy as np\nimport sklearn\nfrom sklearn.model_selection import train_test_split\nimport matplotlib\nimport matplotlib.pyplot as plt\nimport os\n%matplotlib inline\n\nprint(\"tensorflow version: \", tf.__version__)\nprint(\"numpy version: \", np.__version__)\nprint(\"scikit learn version: \", sklearn.__version__)\nprint(\"matplotlib version: \", matplotlib.__version__)",
"tensorflow version: 2.3.0\nnumpy version: 1.19.1\nscikit learn version: 0.23.2\nmatplotlib version: 3.3.0\n"
]
],
[
[
"## 1. Load data & preprocessing\nIn this notebook, I use pre-defined mnist dataset.",
"_____no_output_____"
]
],
[
[
"(X_train, y_train), (X_test, y_test) = tf.keras.datasets.mnist.load_data()",
"_____no_output_____"
],
[
"# Split original training dataset into train/validation dataset\nX_train, X_val, y_train, y_val = train_test_split(X_train, y_train, test_size=0.2, shuffle=True)",
"_____no_output_____"
],
[
"# Normalize Intensity\nX_train = X_train / 255.\nX_val = X_val / 255.\nX_test = X_test / 255.",
"_____no_output_____"
],
[
"# Convert into 4d tensor shape\nX_train = X_train.reshape((*X_train.shape, 1))\nX_val = X_val.reshape((*X_val.shape, 1))\nX_test = X_test.reshape((*X_test.shape, 1))",
"_____no_output_____"
],
[
"# Convert into one-hot\ny_train = tf.keras.utils.to_categorical(y_train)\ny_val = tf.keras.utils.to_categorical(y_val)\ny_test = tf.keras.utils.to_categorical(y_test)",
"_____no_output_____"
]
],
[
[
"## 2. Create tf.data.Dataset",
"_____no_output_____"
]
],
[
[
"train_batch_size = 64\ntest_batch_size = 1\n\n# Build source dataset for training\nX_train_dataset = tf.data.Dataset.from_tensor_slices(X_train)\ny_train_dataset = tf.data.Dataset.from_tensor_slices(y_train)\ntrain_dataset = tf.data.Dataset.zip((X_train_dataset, y_train_dataset)).batch(train_batch_size)\n\n# Build source dataset for validation\nX_valid_dataset = tf.data.Dataset.from_tensor_slices(X_val)\ny_valid_dataset = tf.data.Dataset.from_tensor_slices(y_val)\nvalidation_dataset = tf.data.Dataset.zip((X_valid_dataset, y_valid_dataset)).batch(train_batch_size)\n\n# Build source dataset for test\nX_test_dataset = tf.data.Dataset.from_tensor_slices(X_test)\ny_test_dataset = tf.data.Dataset.from_tensor_slices(y_test)\ntest_dataset = tf.data.Dataset.zip((X_test_dataset, y_test_dataset)).batch(test_batch_size)",
"_____no_output_____"
]
],
[
[
"Debug dataset",
"_____no_output_____"
]
],
[
[
"def visualize_batch(X_batch, y_batch, y_pred=None):\n assert len(X_batch) == len(y_batch)\n \n n_col = 10\n if len(X_batch) % n_col ==0:\n n_row=len(X_batch)//n_col\n else:\n n_row=len(X_batch)//n_col + 1\n \n fig = plt.figure(figsize=(20,15))\n \n for idx in range(len(y_batch)):\n if y_pred is not None:\n ax = fig.add_subplot(n_row, n_col, idx+1, title=\"gt={}, pred={}\".format(np.argmax(y_batch[idx]), y_pred[idx]))\n else:\n ax = fig.add_subplot(n_row, n_col, idx+1, title=\"gt={}\".format(np.argmax(y_batch[idx])))\n ax.imshow(X_batch[idx].reshape(28,28), cmap='gray')\n ax.axes.xaxis.set_visible(False)\n ax.axes.yaxis.set_visible(False)\n plt.show()",
"_____no_output_____"
],
[
"for X_batch, y_batch in train_dataset:\n visualize_batch(X_batch.numpy(), y_batch.numpy())\n break",
"_____no_output_____"
]
],
[
[
"## 3. Create CNN model\nNetwork structure:\n[CONV(32) - BN - RELU] - MAXPOOL - [CONV(64) - BN - RELU] - MAXPOOL - [FC(1024) - BN - RELU] - DROPOUT - FC(10) - SOFTMAX\n\nThe weight initialization rule are following: \n- Layer with relu activation: He initialization \n- Others: Xavier initialization ",
"_____no_output_____"
],
[
"### Define network\nBy creating model as an inherited class of tf.keras.Model, we can easily train model by fit() method. ",
"_____no_output_____"
]
],
[
[
"class ConvNet(tf.keras.Model):\n def __init__(self, conv_filters=[32, 64], units=1024, num_class=10, dropout_rate=0.2):\n super(ConvNet, self).__init__(name=\"ConvNet\")\n self.conv1 = tf.keras.layers.Conv2D(conv_filters[0], 3, 1, 'same', \n kernel_initializer=tf.keras.initializers.he_normal())\n self.bn1 = tf.keras.layers.BatchNormalization()\n self.relu1 = tf.keras.layers.Activation(\"relu\")\n self.pool1 = tf.keras.layers.MaxPool2D(2, 2, padding=\"same\")\n \n self.conv2 = tf.keras.layers.Conv2D(conv_filters[1], 3, 1, 'same', \n kernel_initializer=tf.keras.initializers.he_normal())\n self.bn2 = tf.keras.layers.BatchNormalization()\n self.relu2 = tf.keras.layers.Activation(\"relu\")\n self.pool2 = tf.keras.layers.MaxPool2D(2, 2, padding=\"same\")\n \n self.flattern = tf.keras.layers.Flatten()\n\n self.dense3 = tf.keras.layers.Dense(units, kernel_initializer=tf.keras.initializers.he_normal())\n self.bn3 = tf.keras.layers.BatchNormalization()\n self.relu3 = tf.keras.layers.Activation(\"relu\")\n \n self.dropout = tf.keras.layers.Dropout(rate=dropout_rate)\n \n self.dense4 = tf.keras.layers.Dense(num_class, kernel_initializer=tf.keras.initializers.glorot_normal())\n \n def call(self, inputs, training=True):\n x = self.conv1(inputs)\n x = self.bn1(x, training)\n x = self.relu1(x)\n x = self.pool1(x)\n \n x = self.conv2(x)\n x = self.bn2(x, training)\n x = self.relu2(x)\n x = self.pool2(x)\n \n x = self.flattern(x)\n \n x = self.dense3(x)\n x = self.bn3(x, training)\n x = self.relu3(x)\n \n x = self.dropout(x, training)\n \n x = self.dense4(x)\n \n return x",
"_____no_output_____"
]
],
[
[
"## 4. Training",
"_____no_output_____"
]
],
[
[
"lr = 0.0001\nepochs = 10\n\n# -------------------------------------------------------------\n\nloss = tf.keras.losses.BinaryCrossentropy(from_logits=True)\naccuracy = tf.keras.metrics.CategoricalAccuracy()\noptimizer = tf.keras.optimizers.Adam(lr)\n\n# Define model\nmodel = ConvNet()\n\[email protected]\ndef train_on_batch(model, X, y, accuracy_i):\n # Open a GradientTape\n with tf.GradientTape() as tape:\n logits = model(X)\n loss_value = loss(y, logits)\n\n # Coumpute gradients\n gradients = tape.gradient(loss_value, model.trainable_weights)\n\n # Apply back propagation\n optimizer.apply_gradients(zip(gradients, model.trainable_weights))\n \n # Update the runnning accuracy\n accuracy_i.update_state(y, logits)\n return loss_value\n\[email protected]\ndef eval_on_batch(model, X, y, accuracy_i):\n logits = model(X, training=False)\n loss_value = loss(y, logits)\n accuracy_i.update_state(y, logits)\n return loss_value\n\n\nfor epoch in range(epochs):\n \n # Training\n train_losses = []\n accuracy.reset_states()\n \n for step, (X, y) in enumerate(train_dataset):\n train_loss = train_on_batch(model, X, y, accuracy)\n train_losses.append(train_loss)\n \n train_accuracy = accuracy.result()\n train_loss = np.average(train_losses)\n \n # Validation\n accuracy.reset_states()\n validation_losses = []\n \n for X, y in validation_dataset:\n val_loss = eval_on_batch(model, X, y, accuracy)\n validation_losses.append(val_loss)\n\n validation_accuracy = accuracy.result()\n validation_loss = np.average(validation_losses)\n \n print('Epoch: {}, loss: {:.5f}, accuracy: {:.5f}, val_loss: {:.5f}, val_accuracy: {:.5f}'.format(\n epoch, train_loss, train_accuracy, validation_loss, validation_accuracy\n ))\n \n model.save_weights('./checkpoints_2/model_{:04d}.ckpt'.format(epoch))",
"WARNING:tensorflow:Layer ConvNet is casting an input tensor from dtype float64 to the layer's dtype of float32, which is new behavior in TensorFlow 2. The layer has dtype float32 because its dtype defaults to floatx.\n\nIf you intended to run this layer in float32, you can safely ignore this warning. If in doubt, this warning is likely only an issue if you are porting a TensorFlow 1.X model to TensorFlow 2.\n\nTo change all layers to have dtype float64 by default, call `tf.keras.backend.set_floatx('float64')`. To change just this layer, pass dtype='float64' to the layer constructor. If you are the author of this layer, you can disable autocasting by passing autocast=False to the base Layer constructor.\n\nEpoch: 0, loss: 0.06656, accuracy: 0.94671, val_loss: 0.01883, val_accuracy: 0.97908\nEpoch: 1, loss: 0.01470, accuracy: 0.98481, val_loss: 0.01210, val_accuracy: 0.98567\nEpoch: 2, loss: 0.00918, accuracy: 0.99175, val_loss: 0.00973, val_accuracy: 0.98750\nEpoch: 3, loss: 0.00635, accuracy: 0.99469, val_loss: 0.00871, val_accuracy: 0.98875\nEpoch: 4, loss: 0.00443, accuracy: 0.99713, val_loss: 0.00777, val_accuracy: 0.98992\nEpoch: 5, loss: 0.00315, accuracy: 0.99848, val_loss: 0.00750, val_accuracy: 0.99067\nEpoch: 6, loss: 0.00219, accuracy: 0.99933, val_loss: 0.00705, val_accuracy: 0.99100\nEpoch: 7, loss: 0.00148, accuracy: 0.99969, val_loss: 0.00689, val_accuracy: 0.99142\nEpoch: 8, loss: 0.00099, accuracy: 0.99992, val_loss: 0.00756, val_accuracy: 0.99058\nEpoch: 9, loss: 0.00070, accuracy: 0.99998, val_loss: 0.00723, val_accuracy: 0.99108\n"
]
],
[
[
"## 5. Test",
"_____no_output_____"
]
],
[
[
"accuracy.reset_states()\ntest_losses = []\n\nfor X, y in test_dataset:\n test_loss = eval_on_batch(model, X, y, accuracy)\n test_losses.append(test_loss)\n\ntest_accuracy = accuracy.result()\ntest_loss = np.average(test_losses)\n\nprint('loss: {:.5f}, accuracy: {:.5f}'.format(test_loss, test_accuracy))",
"loss: 0.00718, accuracy: 0.99010\n"
],
[
"index = np.random.choice(np.arange(0, len(y_test)), size=60)\ntest_input = X_test[index]\ny_true = y_test[index]\n\npredicted = model.predict(test_input)\npredicted_label = np.argmax(predicted, axis=1)\n\nvisualize_batch(test_input, y_true, predicted_label)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0f552fe4f2eb79b50c2620b2838416a8fb8ca44 | 7,903 | ipynb | Jupyter Notebook | Clash Activity/last_played_wars.ipynb | BeyondBoy1/WookieForce | 9c47bc60d0672e929ce9bcbd47931e235ff81036 | [
"MIT"
] | 1 | 2021-05-31T11:45:48.000Z | 2021-05-31T11:45:48.000Z | Clash Activity/last_played_wars.ipynb | BeyondBoy1/WookieForce | 9c47bc60d0672e929ce9bcbd47931e235ff81036 | [
"MIT"
] | null | null | null | Clash Activity/last_played_wars.ipynb | BeyondBoy1/WookieForce | 9c47bc60d0672e929ce9bcbd47931e235ff81036 | [
"MIT"
] | null | null | null | 32.126016 | 120 | 0.486398 | [
[
[
"import pandas as pd \nimport numpy as np \nimport datetime as datetime",
"_____no_output_____"
],
[
"#read in last_played_wars csv\nlast_played_wars = pd.read_csv(\"updated_last_played_wars.csv\")\n\n# last_played_wars[\"Participation\"] = last_played_wars[\"Joined Wars\"] / last_played_wars[\"Total Wars\"]\nlast_played_wars = last_played_wars[[\"Name\", \"Tag\", \"Last War\", \"Town Hall\", \"Clan\", \"Total Wars\"]]\nlast_played_wars.head()\n",
"_____no_output_____"
],
[
"#Split Last Played Wars by clan\n#Sheer Force\n\n#Create a copy of Sheer Force only to manipulate\nsf_lpw = last_played_wars.loc[last_played_wars.Clan == \"Sheer Force\"].copy()\n#Joined Wars of clan member\nsf_lpw[\"Joined Wars\"] = sf_lpw[\"Total Wars\"]\n#get maximum number of wars this season\nsf_max = sf_lpw[\"Total Wars\"].max()\nsf_lpw[\"Total Wars\"] = sf_max\n#find participation of members\nsf_lpw[\"Participation\"] = sf_lpw[\"Joined Wars\"]/sf_max\n#get < 50% participation members and add to slackers\nsheer_force_slackers = sf_lpw.loc[sf_lpw.Participation < .5].copy()\nsheer_force_slackers.head()\nsf = sheer_force_slackers.to_csv(r\"sf.csv\", index = True, header = True)",
"_____no_output_____"
],
[
"#Dark Matter\ndm_lpw = last_played_wars.loc[last_played_wars.Clan == \"Dark Matter\"].copy()\ndm_lpw[\"Joined Wars\"] = dm_lpw[\"Total Wars\"]\ndm_max = dm_lpw[\"Total Wars\"].max()\ndm_lpw[\"Total Wars\"] = dm_max\ndm_lpw[\"Participation\"] = dm_lpw[\"Joined Wars\"]/dm_max\ndark_matter_slackers = dm_lpw.loc[dm_lpw.Participation < .5].copy()\ndark_matter_slackers.head()\ndm = dark_matter_slackers.to_csv(r\"dm.csv\", index = True, header = True)",
"_____no_output_____"
],
[
"#Mini Matter\nmm_lpw = last_played_wars.loc[last_played_wars.Clan == \"Mini Matter\"].copy()\nmm_lpw[\"Joined Wars\"] = mm_lpw[\"Total Wars\"]\nmm_max = mm_lpw[\"Total Wars\"].max()\nmm_lpw[\"Total Wars\"] = mm_max\nmm_lpw[\"Participation\"] = mm_lpw[\"Joined Wars\"]/mm_max\nmini_matter_slackers = mm_lpw.loc[mm_lpw.Participation < .5].copy()\nmini_matter_slackers.head()\nmm = mini_matter_slackers.to_csv(r\"mm.csv\", index = True, header = True)",
"_____no_output_____"
],
[
"#Legendary Monks\nlm_lpw = last_played_wars.loc[last_played_wars.Clan == \"Legendary Monks\"].copy()\nlm_lpw[\"Joined Wars\"] = lm_lpw[\"Total Wars\"]\nlm_max = lm_lpw[\"Total Wars\"].max()\nlm_lpw[\"Total Wars\"] = lm_max\nlm_lpw[\"Participation\"] = lm_lpw[\"Joined Wars\"]/lm_max\nlegendary_monks_slackers = lm_lpw.loc[lm_lpw.Participation < .5].copy()\nlegendary_monks_slackers.head()\nlm = legendary_monks_slackers.to_csv(r\"lm.csv\", index = True, header = True)",
"_____no_output_____"
],
[
"#Golden Clan\nkbwf_lpw = last_played_wars.loc[last_played_wars.Clan == \"Golden Clan\"].copy()\nkbwf_lpw[\"Joined Wars\"] = kbwf_lpw[\"Total Wars\"]\nkbwf_max = kbwf_lpw[\"Total Wars\"].max()\nkbwf_lpw[\"Total Wars\"] = kbwf_max\nkbwf_lpw[\"Participation\"] = kbwf_lpw[\"Joined Wars\"]/kbwf_max\nkiller_black_slackers = kbwf_lpw.loc[kbwf_lpw.Participation < .5].copy()\nkiller_black_slackers.head()\nkbwf = killer_black_slackers.to_csv(r\"gc.csv\", index = True, header = True)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f55418a81cae56d7d7f3602d5872a31431f45b | 27,634 | ipynb | Jupyter Notebook | task4(LSTM_names_generation).ipynb | Rojanson/stepik-dl-nlp | e32748fbccd0868a8e4a131e4749188935ed524f | [
"MIT"
] | null | null | null | task4(LSTM_names_generation).ipynb | Rojanson/stepik-dl-nlp | e32748fbccd0868a8e4a131e4749188935ed524f | [
"MIT"
] | null | null | null | task4(LSTM_names_generation).ipynb | Rojanson/stepik-dl-nlp | e32748fbccd0868a8e4a131e4749188935ed524f | [
"MIT"
] | null | null | null | 61.959641 | 14,424 | 0.728668 | [
[
[
"import os\nimport re\nimport collections\nimport copy\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nfrom IPython.display import clear_output\nfrom random import sample\n\nimport dlnlputils\nfrom dlnlputils.data import tokenize_corpus, build_vocabulary\nimport torch\n\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")",
"_____no_output_____"
],
[
"# Если Вы запускаете ноутбук на colab или kaggle, добавьте в начало пути ./stepik-dl-nlp\nwith open('datasets/russian_names.txt') as input_file:\n names = input_file.read()[:-1].split('\\n')\n names = [' ' + line for line in names]\nhandled_text = names",
"_____no_output_____"
],
[
"#all unique characters go here\ntokens = list(set(''.join(handled_text)))\n\nnum_tokens = len(tokens)\nprint ('num_tokens =', num_tokens)",
"num_tokens = 53\n"
],
[
"token_to_id = {token: idx for idx, token in enumerate(tokens)}",
"_____no_output_____"
],
[
"assert len(tokens) == len(token_to_id), \"dictionaries must have same size\"\n\nfor i in range(num_tokens):\n assert token_to_id[tokens[i]] == i, \"token identifier must be it's position in tokens list\"\n\nprint(\"Seems alright!\")",
"Seems alright!\n"
],
[
"def to_matrix(data, token_to_id, max_len=None, dtype='int32', batch_first = True):\n \"\"\"Casts a list of names into rnn-digestable matrix\"\"\"\n \n max_len = max_len or max(map(len, data))\n data_ix = np.zeros([len(data), max_len], dtype) + token_to_id[' ']\n\n for i in range(len(data)):\n line_ix = [token_to_id[c] for c in data[i]]\n data_ix[i, :len(line_ix)] = line_ix\n \n if not batch_first: # convert [batch, time] into [time, batch]\n data_ix = np.transpose(data_ix)\n\n return data_ix",
"_____no_output_____"
],
[
"print(handled_text[3])\nprint(to_matrix(handled_text[3:5], token_to_id)[0])\nprint(len(handled_text[3]))",
" Abaidulin\n[ 3 37 5 19 40 6 44 47 40 28 3]\n10\n"
],
[
"import torch, torch.nn as nn\nimport torch.nn.functional as F",
"_____no_output_____"
],
[
"from IPython.display import clear_output\nfrom random import sample\n\nclass CharLSTMLoop(nn.Module):\n def __init__(self, num_tokens=num_tokens, emb_size=32, hidden_size=128):\n super(self.__class__, self).__init__()\n self.emb = nn.Embedding(num_tokens, emb_size)\n self.LSTM = nn.LSTM(input_size=emb_size, hidden_size=hidden_size, num_layers=1, batch_first=True)\n self.hid_to_logits = nn.Linear(hidden_size, num_tokens)\n \n def forward(self, x, h=None, c=None):\n if h is not None and c is not None:\n out_put, (h_new, c_new) = self.LSTM(self.emb(x), (h, c))\n else:\n out_put, (h_new, c_new) = self.LSTM(self.emb(x))\n \n next_logits = self.hid_to_logits(out_put)\n next_logp = F.log_softmax(next_logits, dim=-1)\n \n return next_logp, h_new, c_new",
"_____no_output_____"
],
[
"MAX_LENGTH = max(map(len, handled_text))\n\nmodel = CharLSTMLoop()\nmodel = model.to(device)\nopt = torch.optim.Adam(model.parameters())\nhistory = []\n\nbest_loss = 6\n\nbest_model_wts = copy.deepcopy(model.state_dict())\n\nfor i in range(4000):\n batch_ix = to_matrix(sample(handled_text, 32), token_to_id, max_len=MAX_LENGTH)\n batch_ix = torch.tensor(batch_ix, dtype=torch.int64).to(device)\n\n logp_seq, _, _ = model(batch_ix)\n \n # compute loss\n predictions_logp = logp_seq[:, :-1]\n actual_next_tokens = batch_ix[:, 1:]\n\n loss = -torch.mean(torch.gather(predictions_logp, dim=2, index=actual_next_tokens[:,:,None]))\n\n if loss < best_loss:\n best_loss = loss\n best_model_wts = copy.deepcopy(model.state_dict())\n \n # train with backprop\n loss.backward()\n opt.step()\n opt.zero_grad()\n \n history.append(loss.cpu().data.numpy())\n if (i + 1) % 20 == 0:\n clear_output(True)\n plt.plot(history, label='loss')\n plt.legend()\n plt.show()\n\nassert np.mean(history[:25]) > np.mean(history[-25:]), \"RNN didn't converge.\"",
"_____no_output_____"
],
[
"model.load_state_dict(best_model_wts)",
"_____no_output_____"
],
[
"def generate_sample(char_rnn, seed_phrase=' ', max_length=MAX_LENGTH, temperature=1.0):\n '''\n The function generates text given a phrase of length at least SEQ_LENGTH.\n :param seed_phrase: prefix characters. The RNN is asked to continue the phrase\n :param max_length: maximum output length, including seed_phrase\n :param temperature: coefficient for sampling. higher temperature produces more chaotic outputs,\n smaller temperature converges to the single most likely output\n '''\n \n x_sequence = [[token_to_id[token] for token in seed_phrase]]\n x_sequence = torch.tensor(x_sequence, dtype=torch.int64)\n \n h_t = None\n c_t = None\n if len(seed_phrase) > 1:\n _, h_t, c_t = model.forward(x_sequence[:, :-1], h_t)\n \n for _ in range(max_length - len(seed_phrase)):\n logp_next, h_t, c_t = model.forward(x_sequence[:, -1].unsqueeze(-1), h_t, c_t)\n p_next = F.softmax(logp_next / temperature, dim=-1).data.numpy()[0]\n \n next_ix = np.random.choice(len(tokens), p=p_next[0])\n next_ix = torch.tensor([[next_ix]], dtype=torch.int64)\n x_sequence = torch.cat([x_sequence, next_ix], dim=1)\n \n return ''.join([tokens[ix] for ix in x_sequence[0].data.numpy()])\n\n\nmodel = model.to('cpu')\nfor _ in range(100):\n print(generate_sample(model, seed_phrase=' ', temperature=0.5))",
" Martyushkin \n Halushkin \n Baidanov \n Baidanoff \n Zimatov \n Vakachev \n Zhituhin \n Velikov \n Abdulazyan \n Makhorev \n Jablonov \n Atamahov \n Dubin \n Mashkov \n Bakalov \n Tovashenko \n Rozhankin \n Andrusichev \n Lapinsky \n Valennikov \n Mokhin \n Voloseev \n Beloishtein \n Zhitin \n Belovarov \n Babuhov \n Zherkov \n Guzin \n Zinov \n Martyshev \n Glushin \n Tsaramovsky \n Batsevich \n Shainer \n Abdulladzhanoff \n Babakov \n Babashov \n Awaloff \n Litin \n Babakhin \n Abitov \n Turkovich \n Grashin \n Anorin \n Bakhtin \n Maksyukin \n Haleikov \n Ponitsky \n Gulin \n Andrukhin \n Lihomenko \n Antoshkin \n Ryzhenko \n Shadrahmanoff \n Valtsev \n Badurin \n Poltushkin \n Awduroff \n Gudin \n Jarikhin \n Andrusikov \n Rahmanov \n Abdugoff \n Gulia \n Dizhanov \n Babarov \n Turkov \n Abdrazyzhin \n Pavlushkin \n Babashev \n Patsev \n Rahmanov \n Averkovich \n Zhidin \n Muzykin \n Awdulichin \n Rakhmanovich \n Guzikov \n Babashev \n Antoshin \n Marykov \n Rahmanov \n Peserev \n Zhivailin \n Maksievsky \n Bakhtin \n Elenko \n Minzhenko \n Averkov \n Badyanoff \n Valkov \n Tuma \n Tumin \n Baban \n Belomoshev \n Rakhmat \n Prigalin \n Makholin \n Vaisin \n Tikhonushin \n"
],
[
"torch.save(model, 'Names_generation.pt')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f556d3dbb3d8c6447b8c5e6c56f0eeff55c667 | 742,239 | ipynb | Jupyter Notebook | Lab_2_1_Thresholding.ipynb | power9939/CE6003 | f7e4c32195d8af8f2a0c5391d5a3ed057f5e5b79 | [
"MIT"
] | null | null | null | Lab_2_1_Thresholding.ipynb | power9939/CE6003 | f7e4c32195d8af8f2a0c5391d5a3ed057f5e5b79 | [
"MIT"
] | null | null | null | Lab_2_1_Thresholding.ipynb | power9939/CE6003 | f7e4c32195d8af8f2a0c5391d5a3ed057f5e5b79 | [
"MIT"
] | null | null | null | 912.9631 | 251,304 | 0.954674 | [
[
[
"# \"Thresholding\"\n\n# Recap\n\nThis is the Lab on Thresholding for Classical Image Segmention in CE6003. You should complete the tasks in this lab as part of the Thresholding section of the lesson.\n\nPlease remember this lab must be completed before taking the quiz at the end of this lesson.\n",
"_____no_output_____"
],
[
"First, if we haven't already done so, we need to clone the various images and resources needed to run these labs into our workspace.",
"_____no_output_____"
]
],
[
[
"#!git clone https://github.com/EmdaloTechnologies/CE6003.git",
"_____no_output_____"
]
],
[
[
"# Introduction\n\nIn this lab we introduce our first image segmentation project where we will use thresholding operations to segment a relatively simple image. We will work through this project using the types of image processing techniques such projects typically need and then segment an image. \n\nAt the end of the lab we'll review the work we've done and assess what types of images and projects this approach is effective for.\n",
"_____no_output_____"
],
[
"# Goal\n\nIn this tutorial we will learn about three key items:\n* The general image processing algorithms that are required for most image processing projects; e.g. denoising, \n* Using our first classical segmentation technique on images (thresholding);\n* How to use Otsu's Method to automatically find a suitable threshold level to segment an image.\n",
"_____no_output_____"
],
[
"# Background\n\nImage segmentation is the process of partitioning a digital image into multiple segments to make the image easier to analyze. Often we are looking to locate objects and boundaries in the original image. \n\nA more precise way of looking at it is to say that image segmentation's goal is to assign a label to every pixel in an image such that pixels with the same label share certain characteristics. \n\nFor example, these images show a typical road scene on the left and a segmented version of the image on the right where the cars have been separated from the road, the buildings, the people in the scene, etc.\n\n<p float=\"center\">\n <img src=\"https://github.com/EmdaloTechnologies/CE6003/blob/master/images/lab2/bg-road.png?raw=1\" width=\"450\" />\n</p>\n",
"_____no_output_____"
],
[
"# Our First Segmenter\n\nSo, now that we've seen what is possible, let's start by solving our first segmentation problem.\n\nLet's look at this image of a starfish. Let's examine it in its original color, in grayscale and in black and white.\n\nColour | Grayscale | Black & White\n:--------------------------------:|:------------------------------------:|:---------------------------:\n |  | \n\nWe are searching these images for clues as to how we might be able to segment them into two regions - the 'starfish' region and the 'not starfish' region.\n\nIt turns out we can segment this image into a region approximating the starfish and a background region (the 'not starfish; region) using thresholding, along with general purpose image processing techniques such as denoising, morphological operations, and some contour detection and drawing. Finally, once we've established a boundary for the starfish, we can fill our starfish shape. After that we'll use some bitwise operations to overlay our segmented image over the original image.",
"_____no_output_____"
],
[
"First, lets use OpenCV's fastN1MeansDenoisingColored routine to denoise the image. We're using a larger 'h' and 'hColor' value than typically used as the image is more noisy than images typically used with these technique. (This should make more sense as we go forward into the CNN segmentation examples).",
"_____no_output_____"
]
],
[
[
"# First import OpenCV, NumPY and MatPlotLib as we will use these libraries\nimport cv2\nimport numpy as np\nimport matplotlib.pyplot as plt\n\n%matplotlib inline\n\n# Load a color image\n#img = cv2.imread(\"/content/CE6003/images/lab2/starfish.png\")\nimg = cv2.imread(\"./images/lab2/starfish.png\")\n\n#plt.imshow(img, cmap='gray')\n#plt.imshow(img)\nplt.imshow(cv2.cvtColor(img, cv2.COLOR_BGR2RGB))",
"_____no_output_____"
],
[
"# Apply some blurring to reduce noise\n\n# h is the Parameter regulating filter strength for luminance component. \n# Bigger h value perfectly removes noise but also removes image details, \n# smaller h value preserves details but also preserves some noise\n\n# Hint: I recommend using larger h and hColor values than typical to remove noise at the\n# expense of losing image details\n\n# YOUR CODE HERE\n# Experiment with setting h and hColor to a suitable value.\n# Exercise: Insert code here to set values for h and hColor. \n# https://docs.opencv.org/2.4/modules/photo/doc/denoising.html\n#\n# h – Parameter regulating filter strength for luminance component. Bigger h value perfectly \n# removes noise but also removes image details, smaller h value preserves details but also \n# preserves some noise\n#\n# hColor – The same as h but for color components. For most images value equals 10 will be enough\n# to remove colored noise and do not distort colors\n#\nh = 20\nhColor = 10\n# END YOUR CODE HERE\n\n# Default values\ntemplateWindowSize = 7\nsearchWindowSize = 21\n \nblur = cv2.fastNlMeansDenoisingColored(img, None, h, hColor,templateWindowSize,searchWindowSize)\n \nplt.imshow(cv2.cvtColor(blur, cv2.COLOR_BGR2RGB))",
"_____no_output_____"
]
],
[
[
"After applying the fastN1MeansDenoisingColored routine above, you should end up with an image similar to the one on the right here. You may need to vary the h and hColor parameters to observe the effect of changing them on the blurred image.\n\nYour blurred image should look like this one.\n\n<img src=\"https://github.com/EmdaloTechnologies/CE6003/blob/master/images/lab2/starfish_blur.png?raw=1\" alt=\"Blurred Starfish\" align=\"left\" style=\"width: 300px;\"/>\n",
"_____no_output_____"
],
[
"Now, lets run a morphological operation on the blurred image. \n\nFor this example, we are going to generate a gradient. \n\nThis builds on dilation and erosion. You can read more about erosion and dilation \nin the 'Basics' section of Lesson 2.\n\nToday we are going to use them to generate an outline for our starfish.\n\n\n# Edge Detection\nInstead of using a gradient, you could use an edge detection such as Sobol, Laplacian and Canny here in combination with adjusting the image denoising step above. I'll leave those as an exercise for the reader for now!",
"_____no_output_____"
]
],
[
[
"# Apply a morphological gradient (dilate the image, erode the image, and take the difference\n\nelKernel = cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (13,13))\n\n# YOUR CODE HERE\n# Exercise: Use openCV's morphologyEx to generate a gradient using the kernel above\ngradient = cv2.morphologyEx(blur, cv2.MORPH_GRADIENT, elKernel)\n# END YOUR CODE HERE\n\nplt.imshow(cv2.cvtColor(gradient, cv2.COLOR_BGR2RGB))",
"_____no_output_____"
]
],
[
[
"After applying the gradient morphology routine above, you should end up with an image similar to the one shown here. The outline of the starfish should be starting to emerge at this point.\n \n<img src=\"https://github.com/EmdaloTechnologies/CE6003/blob/master/images/lab2/starfish_grad.png?raw=1\" alt=\"Gradient Starfish\" align=\"left\" style=\"width: 300px;\"/>\n\n",
"_____no_output_____"
],
[
"We now have completed the pre-processing of our image. \n\nFrom this point onwards, we are concerning ourselves with:\n\na) filling the region of interest, and\n\nb) removing artefacts from the image which we are not interested in.\n\nThere are quite a few approaches we can take to this (including not doing them at all), but today lets apply OTSU's threshold to convert the image to black and white, and perform a closing operation to 'fill in' the starfish and then perform some erosion to remove parts of the image that we consider noise.\n\n## OTSU Thresholding\nWhen converting from a grayscale image to a black and white image, selecting a good threshold value can be a time-consuming and manual process. There are a number of automatic thresholding techniques available - and Otsu's method thresholding is one of the better known techniques.\n\nConceptually simple, and relatively low cost computationally, Otsu's threshold iterate through all the possible threshold values to find the threshold value where the sum of foreground and background spreads is at its minimum. \n",
"_____no_output_____"
]
],
[
[
"# Apply Otsu's method - or you can adjust the level at which thresholding occurs\n# and see what the effect of this is\n\n# Convert gradient to grayscale\ngray_gradient = cv2.cvtColor(gradient, cv2.COLOR_BGR2GRAY)\n\n\n# YOUR CODE HERE\n# Exercise: Generate a matrix called otsu using OpenCV's threshold() function. Use\n# Otsu's method.\n# Otsu's thresholding\nret, otsu = cv2.threshold(gray_gradient,0,255,cv2.THRESH_BINARY+cv2.THRESH_OTSU)\nplt.imshow(otsu, cmap='gray')\n# END YOUR CODE HERE",
"_____no_output_____"
],
[
" \n# Apply a closing operation - we're using a large kernel here. By all means adjust the size of this kernel\n# and observe the effects\nclosingKernel = cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (33,33))\nclose = cv2.morphologyEx(otsu, cv2.MORPH_CLOSE, closingKernel)\n\nplt.imshow(close, cmap='gray')",
"_____no_output_____"
],
[
"# Erode smaller artefacts out of the image - play with iterations to see how it works\n \n# YOUR CODE HERE\n# Exercise: Generate a matrix called eroded using cv2.erode() function over the 'close' matrix.\n# Experiment until your output image is similar to the image below\n\n# Taking a matrix of size n as the kernel \nn = 5\nkernel = np.ones((n,n), np.uint8) \n\neroded = cv2.erode(close,kernel,iterations = 5)\n# END YOUR CODE HERE\n\nplt.imshow(eroded, cmap='gray')",
"_____no_output_____"
]
],
[
[
"After switching to black and white and applying our closing and erosion operations, our simplified starfish is starting to emerge\n \nOriginal Image | B&W Image | After Closing | After Erosion\n:------------------------:|:------------------------------:|:-------------------------------:|:--------------\n |  |  | \n",
"_____no_output_____"
],
[
"So, now we've effectively segmented our image. \n\nNow, let's post-process the image to find the contours that represent the edge of the starfish. We'll just use the intuition that the starfish is the largest object in the scene.\n\nThen we'lll do a little image manipulation to generate a colour representing the starfish, another colour representing the background (i.e. not the starfish) and then merge those colours with the original image.\n\nYou'll notice the closing and erosion steps are not perfect - they're not supposed to be. They are good enough to feed into the findContours routine. By all means, tune them further to get better quality input into findContours.\n\nIn the findContours routine we're using cv2.RETR_EXTERNAL. This is to reduce the complexity of post-processing by only reporting 'external' contours (i.e. we'll attempt to suppress contours that are inside other contours).",
"_____no_output_____"
]
],
[
[
"p = int(img.shape[1] * 0.05)\neroded[:, 0:p] = 0\neroded[:, img.shape[1] - p:] = 0\n\nplt.imshow(eroded, cmap='gray')\n",
"_____no_output_____"
],
[
"# from https://www.programcreek.com/python/example/70440/cv2.findContours\nmajor = cv2.__version__.split('.')[0]\nprint(major)",
"4\n"
],
[
"# YOUR CODE HERE\n# Exercise: Find the contours - just external contours to keep post-processing simple\ncontours, _ = cv2.findContours(eroded, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)\n# END YOUR CODE HERE",
"_____no_output_____"
],
[
"\n\n# Sort the candidates by size, and just keep the largest one\ncontours = sorted(contours, key=cv2.contourArea, reverse=True)[:1]\n\n# Lets create two images, initially all zeros (i.e. black)\n# One image will be filled with 'Blue' wherever we think there's some starfish\n# The other image will be filled with 'Green' whereever we think there's not some starfish\nh, w, num_c = img.shape\nsegmask = np.zeros((h, w, num_c), np.uint8)\nstencil = np.zeros((h, w, num_c), np.uint8)\n\n# I know we've only one contour, but - in general - we'd expect to have more contours to deal with\nfor c in contours:\n # Fill in the starfish shape into segmask\n cv2.drawContours(segmask, [c], 0, (255, 0, 0), -1)\n # Lets fill in the starfish shape into stencil as well\n # and then re-arrange the colors using numpy\n cv2.drawContours(stencil, [c], 0, (255, 0, 0), -1)\n stencil[np.where((stencil==[0,0,0]).all(axis=2))] = [0, 255, 0]\n stencil[np.where((stencil==[255,0,0]).all(axis=2))] = [0, 0, 0]\n\n# Now, lets create a mask image by bitwise ORring segmask and stencil together\nmask = cv2.bitwise_or(stencil, segmask)\n\nplt.imshow(cv2.cvtColor(mask, cv2.COLOR_BGR2RGB))",
"_____no_output_____"
]
],
[
[
"You should have generated a reasonable mask representing our image as having two parts - a 'starfish' and 'not a starfish'. It should look like the final mask in the image below.\n \nStarfish Mask | Not Starfish Mask | Final Mask\n:-------------------------------:|:--------------------------------:|:-------------------------------\n |  | \n",
"_____no_output_____"
]
],
[
[
"# Now, lets just blend our original image with our mask\n\n# YOUR CODE HERE\n# Exercise: Blend the original image 'img' and our mask 'mask'\n# in any way you see fit, and store it in a variable called output\n# Hint: You'll find answers at the bottom of the lab. \noutput = cv2.bitwise_or(mask, img)\n# END YOUR CODE HERE",
"_____no_output_____"
],
[
"plt.imshow(cv2.cvtColor(output, cv2.COLOR_BGR2RGB))",
"_____no_output_____"
]
],
[
[
"After you blend the original image with your mask you should see an image similar to the image shown here.\n\n<img src=\"https://github.com/EmdaloTechnologies/CE6003/blob/master/images/lab2/starfish_segmented.png?raw=1\" alt=\"Segmented Starfish\" align=\"left\" style=\"width: 300px;\"/>\n\n",
"_____no_output_____"
],
[
"# Conclusion\n\nSo, that completes the first of the four labs to this module. \n\nTo summarise , we''ve learned some basic image processing techniques, such as morphological operations like erosion and dilation, contour detection and we've used these techniques in combination with Otsu's thresholding method to segment an image.\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d0f55944b04950299a6cf73e65ad604a1b3ec53e | 112,079 | ipynb | Jupyter Notebook | Tensorflow/Courses/0_Intro_to_tf/2_Week1_Assignmen_normalized.ipynb | maxtcurie/Artificial_Intelligence | f49ccb28165736950a8c800be8e6d01dde1fd3e9 | [
"MIT"
] | null | null | null | Tensorflow/Courses/0_Intro_to_tf/2_Week1_Assignmen_normalized.ipynb | maxtcurie/Artificial_Intelligence | f49ccb28165736950a8c800be8e6d01dde1fd3e9 | [
"MIT"
] | null | null | null | Tensorflow/Courses/0_Intro_to_tf/2_Week1_Assignmen_normalized.ipynb | maxtcurie/Artificial_Intelligence | f49ccb28165736950a8c800be8e6d01dde1fd3e9 | [
"MIT"
] | null | null | null | 50.124776 | 432 | 0.35433 | [
[
[
"# Week 1 Assignment: Housing Prices\n\nIn this exercise you'll try to build a neural network that predicts the price of a house according to a simple formula.\n\nImagine that house pricing is as easy as:\n\nA house has a base cost of 50k, and every additional bedroom adds a cost of 50k. This will make a 1 bedroom house cost 100k, a 2 bedroom house cost 150k etc.\n\nHow would you create a neural network that learns this relationship so that it would predict a 7 bedroom house as costing close to 400k etc.\n\nHint: Your network might work better if you scale the house price down. You don't have to give the answer 400...it might be better to create something that predicts the number 4, and then your answer is in the 'hundreds of thousands' etc.",
"_____no_output_____"
]
],
[
[
"import keras\nimport numpy as np",
"_____no_output_____"
],
[
"# GRADED FUNCTION: house_model\ndef normalize(xs,ys):\n return xs,ys*0.01\ndef un_normalize(xs,ys):\n return xs,ys*100.\ndef house_model():\n ### START CODE HERE\n \n # Define input and output tensors with the values for houses with 1 up to 6 bedrooms\n # Hint: Remember to explictly set the dtype as float\n #xs:number of bedrooms\n xs = np.arange(6,dtype=float)\n #ys:cost of house\n ys = 50+50*xs\n\n print(xs)\n print(ys)\n xs,ys=normalize(xs,ys)\n print(xs)\n print(ys)\n # Define your model (should be a model with 1 dense layer and 1 unit)\n model = keras.Sequential([keras.layers.Dense(units=1,input_shape=[1])])\n \n # Compile your model\n # Set the optimizer to Stochastic Gradient Descent\n # and use Mean Squared Error as the loss function\n model.compile(optimizer='sgd',loss='mean_squared_error')\n \n # Train your model for 1000 epochs by feeding the i/o tensors\n model.fit(xs,ys,epochs=1000)\n \n ### END CODE HERE\n return model",
"_____no_output_____"
]
],
[
[
"Now that you have a function that returns a compiled and trained model when invoked, use it to get the model to predict the price of houses: ",
"_____no_output_____"
]
],
[
[
"# Get your trained model\nmodel = house_model()",
"[0. 1. 2. 3. 4. 5.]\n[ 50. 100. 150. 200. 250. 300.]\n[0. 1. 2. 3. 4. 5.]\n[0.5 1. 1.5 2. 2.5 3. ]\nEpoch 1/1000\n1/1 [==============================] - 0s 236ms/step - loss: 0.6199\nEpoch 2/1000\n1/1 [==============================] - 0s 997us/step - loss: 0.4340\nEpoch 3/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.3138\nEpoch 4/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.2360\nEpoch 5/1000\n1/1 [==============================] - 0s 1ms/step - loss: 0.1855\nEpoch 6/1000\n1/1 [==============================] - 0s 998us/step - loss: 0.1526\nEpoch 7/1000\n1/1 [==============================] - 0s 996us/step - loss: 0.1310\nEpoch 8/1000\n1/1 [==============================] - 0s 4ms/step - loss: 0.1167\nEpoch 9/1000\n1/1 [==============================] - 0s 5ms/step - loss: 0.1071\nEpoch 10/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.1005\nEpoch 11/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0959\nEpoch 12/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0926\nEpoch 13/1000\n1/1 [==============================] - 0s 3ms/step - loss: 0.0901\nEpoch 14/1000\n1/1 [==============================] - 0s 996us/step - loss: 0.0881\nEpoch 15/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0865\nEpoch 16/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0851\nEpoch 17/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0838\nEpoch 18/1000\n1/1 [==============================] - 0s 996us/step - loss: 0.0827\nEpoch 19/1000\n1/1 [==============================] - 0s 3ms/step - loss: 0.0816\nEpoch 20/1000\n1/1 [==============================] - 0s 3ms/step - loss: 0.0806\nEpoch 21/1000\n1/1 [==============================] - 0s 3ms/step - loss: 0.0796\nEpoch 22/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0786\nEpoch 23/1000\n1/1 [==============================] - 0s 998us/step - loss: 0.0777\nEpoch 24/1000\n1/1 [==============================] - 0s 998us/step - loss: 0.0768\nEpoch 25/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0759\nEpoch 26/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0750\nEpoch 27/1000\n1/1 [==============================] - 0s 996us/step - loss: 0.0741\nEpoch 28/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0732\nEpoch 29/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0723\nEpoch 30/1000\n1/1 [==============================] - 0s 3ms/step - loss: 0.0715\nEpoch 31/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0706\nEpoch 32/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0698\nEpoch 33/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0690\nEpoch 34/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0682\nEpoch 35/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0674\nEpoch 36/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0666\nEpoch 37/1000\n1/1 [==============================] - 0s 1ms/step - loss: 0.0658\nEpoch 38/1000\n1/1 [==============================] - 0s 997us/step - loss: 0.0650\nEpoch 39/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0642\nEpoch 40/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0635\nEpoch 41/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0627\nEpoch 42/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0620\nEpoch 43/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0613\nEpoch 44/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0606\nEpoch 45/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0598\nEpoch 46/1000\n1/1 [==============================] - 0s 996us/step - loss: 0.0591\nEpoch 47/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0584\nEpoch 48/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0577\nEpoch 49/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0571\nEpoch 50/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0564\nEpoch 51/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0557\nEpoch 52/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0551\nEpoch 53/1000\n1/1 [==============================] - 0s 3ms/step - loss: 0.0544\nEpoch 54/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0538\nEpoch 55/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0531\nEpoch 56/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0525\nEpoch 57/1000\n1/1 [==============================] - 0s 997us/step - loss: 0.0519\nEpoch 58/1000\n1/1 [==============================] - 0s 996us/step - loss: 0.0513\nEpoch 59/1000\n1/1 [==============================] - 0s 997us/step - loss: 0.0507\nEpoch 60/1000\n1/1 [==============================] - 0s 997us/step - loss: 0.0501\nEpoch 61/1000\n1/1 [==============================] - 0s 998us/step - loss: 0.0495\nEpoch 62/1000\n1/1 [==============================] - 0s 997us/step - loss: 0.0489\nEpoch 63/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0483\nEpoch 64/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0478\nEpoch 65/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0472\nEpoch 66/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0467\nEpoch 67/1000\n1/1 [==============================] - 0s 1ms/step - loss: 0.0461\nEpoch 68/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0456\nEpoch 69/1000\n1/1 [==============================] - 0s 1ms/step - loss: 0.0450\nEpoch 70/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0445\nEpoch 71/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0440\nEpoch 72/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0434\nEpoch 73/1000\n1/1 [==============================] - 0s 998us/step - loss: 0.0429\nEpoch 74/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0424\nEpoch 75/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0419\nEpoch 76/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0414\nEpoch 77/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0409\nEpoch 78/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0405\nEpoch 79/1000\n1/1 [==============================] - 0s 994us/step - loss: 0.0400\nEpoch 80/1000\n1/1 [==============================] - 0s 991us/step - loss: 0.0395\nEpoch 81/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0391\nEpoch 82/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0386\nEpoch 83/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0381\nEpoch 84/1000\n1/1 [==============================] - 0s 998us/step - loss: 0.0377\nEpoch 85/1000\n1/1 [==============================] - 0s 998us/step - loss: 0.0372\nEpoch 86/1000\n1/1 [==============================] - 0s 1ms/step - loss: 0.0368\nEpoch 87/1000\n1/1 [==============================] - 0s 1ms/step - loss: 0.0364\nEpoch 88/1000\n1/1 [==============================] - 0s 1ms/step - loss: 0.0359\nEpoch 89/1000\n1/1 [==============================] - 0s 996us/step - loss: 0.0355\nEpoch 90/1000\n1/1 [==============================] - 0s 507us/step - loss: 0.0351\nEpoch 91/1000\n1/1 [==============================] - 0s 1ms/step - loss: 0.0347\nEpoch 92/1000\n1/1 [==============================] - 0s 998us/step - loss: 0.0343\nEpoch 93/1000\n1/1 [==============================] - 0s 997us/step - loss: 0.0339\nEpoch 94/1000\n1/1 [==============================] - 0s 997us/step - loss: 0.0335\nEpoch 95/1000\n1/1 [==============================] - 0s 997us/step - loss: 0.0331\nEpoch 96/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0327\nEpoch 97/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0323\nEpoch 98/1000\n1/1 [==============================] - 0s 3ms/step - loss: 0.0319\nEpoch 99/1000\n1/1 [==============================] - 0s 3ms/step - loss: 0.0315\nEpoch 100/1000\n1/1 [==============================] - 0s 2ms/step - loss: 0.0312\nEpoch 101/1000\n"
]
],
[
[
"Now that your model has finished training it is time to test it out! You can do so by running the next cell.",
"_____no_output_____"
]
],
[
[
"new_x = 7.0\nprediction = model.predict([new_x])[0]*100.\nprint(50+7*50)\nprint(prediction)",
"400\n[400.1458]\n"
]
],
[
[
"If everything went as expected you should see a prediction value very close to 4. **If not, try adjusting your code before submitting the assignment.** Notice that you can play around with the value of `new_y` to get different predictions. In general you should see that the network was able to learn the linear relationship between `x` and `y`, so if you use a value of 8.0 you should get a prediction close to 4.5 and so on.",
"_____no_output_____"
],
[
"**Congratulations on finishing this week's assignment!**\n\nYou have successfully coded a neural network that learned the linear relationship between two variables. Nice job!\n\n**Keep it up!**",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0f5718d8ff4345c4468bdfccfe0516570c89348 | 18,050 | ipynb | Jupyter Notebook | intro-to-tflearn/TFLearn_Digit_Recognition.ipynb | tanmaysinha987/deep-Learning | 6905ccb8d5131e43fb252d717cdc1c4525e0041d | [
"MIT"
] | null | null | null | intro-to-tflearn/TFLearn_Digit_Recognition.ipynb | tanmaysinha987/deep-Learning | 6905ccb8d5131e43fb252d717cdc1c4525e0041d | [
"MIT"
] | null | null | null | intro-to-tflearn/TFLearn_Digit_Recognition.ipynb | tanmaysinha987/deep-Learning | 6905ccb8d5131e43fb252d717cdc1c4525e0041d | [
"MIT"
] | null | null | null | 64.928058 | 7,342 | 0.757784 | [
[
[
"# Handwritten Number Recognition with TFLearn and MNIST\n\nIn this notebook, we'll be building a neural network that recognizes handwritten numbers 0-9. \n\nThis kind of neural network is used in a variety of real-world applications including: recognizing phone numbers and sorting postal mail by address. To build the network, we'll be using the **MNIST** data set, which consists of images of handwritten numbers and their correct labels 0-9.\n\nWe'll be using [TFLearn](http://tflearn.org/), a high-level library built on top of TensorFlow to build the neural network. We'll start off by importing all the modules we'll need, then load the data, and finally build the network.",
"_____no_output_____"
]
],
[
[
"# Import Numpy, TensorFlow, TFLearn, and MNIST data\nimport numpy as np\nimport tensorflow as tf\nimport tflearn\nimport tflearn.datasets.mnist as mnist",
"_____no_output_____"
]
],
[
[
"## Retrieving training and test data\n\nThe MNIST data set already contains both training and test data. There are 55,000 data points of training data, and 10,000 points of test data.\n\nEach MNIST data point has:\n1. an image of a handwritten digit and \n2. a corresponding label (a number 0-9 that identifies the image)\n\nWe'll call the images, which will be the input to our neural network, **X** and their corresponding labels **Y**.\n\nWe're going to want our labels as *one-hot vectors*, which are vectors that holds mostly 0's and one 1. It's easiest to see this in a example. As a one-hot vector, the number 0 is represented as [1, 0, 0, 0, 0, 0, 0, 0, 0, 0], and 4 is represented as [0, 0, 0, 0, 1, 0, 0, 0, 0, 0].\n\n### Flattened data\n\nFor this example, we'll be using *flattened* data or a representation of MNIST images in one dimension rather than two. So, each handwritten number image, which is 28x28 pixels, will be represented as a one dimensional array of 784 pixel values. \n\nFlattening the data throws away information about the 2D structure of the image, but it simplifies our data so that all of the training data can be contained in one array whose shape is [55000, 784]; the first dimension is the number of training images and the second dimension is the number of pixels in each image. This is the kind of data that is easy to analyze using a simple neural network.",
"_____no_output_____"
]
],
[
[
"# Retrieve the training and test data\ntrainX, trainY, testX, testY = mnist.load_data(one_hot=True)",
"_____no_output_____"
]
],
[
[
"## Visualize the training data\n\nProvided below is a function that will help you visualize the MNIST data. By passing in the index of a training example, the function `show_digit` will display that training image along with it's corresponding label in the title.",
"_____no_output_____"
]
],
[
[
"# Visualizing the data\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n# Function for displaying a training image by it's index in the MNIST set\ndef show_digit(index):\n label = trainY[index].argmax(axis=0)\n # Reshape 784 array into 28x28 image\n image = trainX[index].reshape([28,28])\n plt.title('Training data, index: %d, Label: %d' % (index, label))\n plt.imshow(image, cmap='gray_r')\n plt.show()\n \n# Display the first (index 0) training image\nshow_digit(0)",
"_____no_output_____"
]
],
[
[
"## Building the network\n\nTFLearn lets you build the network by defining the layers in that network. \n\nFor this example, you'll define:\n\n1. The input layer, which tells the network the number of inputs it should expect for each piece of MNIST data. \n2. Hidden layers, which recognize patterns in data and connect the input to the output layer, and\n3. The output layer, which defines how the network learns and outputs a label for a given image.\n\nLet's start with the input layer; to define the input layer, you'll define the type of data that the network expects. For example,\n\n```\nnet = tflearn.input_data([None, 100])\n```\n\nwould create a network with 100 inputs. The number of inputs to your network needs to match the size of your data. For this example, we're using 784 element long vectors to encode our input data, so we need **784 input units**.\n\n\n### Adding layers\n\nTo add new hidden layers, you use \n\n```\nnet = tflearn.fully_connected(net, n_units, activation='ReLU')\n```\n\nThis adds a fully connected layer where every unit (or node) in the previous layer is connected to every unit in this layer. The first argument `net` is the network you created in the `tflearn.input_data` call, it designates the input to the hidden layer. You can set the number of units in the layer with `n_units`, and set the activation function with the `activation` keyword. You can keep adding layers to your network by repeated calling `tflearn.fully_connected(net, n_units)`. \n\nThen, to set how you train the network, use:\n\n```\nnet = tflearn.regression(net, optimizer='sgd', learning_rate=0.1, loss='categorical_crossentropy')\n```\n\nAgain, this is passing in the network you've been building. The keywords: \n\n* `optimizer` sets the training method, here stochastic gradient descent\n* `learning_rate` is the learning rate\n* `loss` determines how the network error is calculated. In this example, with categorical cross-entropy.\n\nFinally, you put all this together to create the model with `tflearn.DNN(net)`.",
"_____no_output_____"
],
[
"**Exercise:** Below in the `build_model()` function, you'll put together the network using TFLearn. You get to choose how many layers to use, how many hidden units, etc.\n\n**Hint:** The final output layer must have 10 output nodes (one for each digit 0-9). It's also recommended to use a `softmax` activation layer as your final output layer. ",
"_____no_output_____"
]
],
[
[
"# Define the neural network\ndef build_model():\n # This resets all parameters and variables, leave this here\n tf.reset_default_graph()\n \n #### Your code ####\n # Include the input layer, hidden layer(s), and set how you want to train the model\n net=tflearn.input_data(None,trainX.shape[1])\n net=tflearn.fully_connected(net,128,activation='ReLU')\n # This model assumes that your network is named \"net\" \n model = tflearn.DNN(net)\n return model",
"_____no_output_____"
],
[
"# Build the model\nmodel = build_model()",
"_____no_output_____"
]
],
[
[
"## Training the network\n\nNow that we've constructed the network, saved as the variable `model`, we can fit it to the data. Here we use the `model.fit` method. You pass in the training features `trainX` and the training targets `trainY`. Below I set `validation_set=0.1` which reserves 10% of the data set as the validation set. You can also set the batch size and number of epochs with the `batch_size` and `n_epoch` keywords, respectively. \n\nToo few epochs don't effectively train your network, and too many take a long time to execute. Choose wisely!",
"_____no_output_____"
]
],
[
[
"# Training\nmodel.fit(trainX, trainY, validation_set=0.1, show_metric=True, batch_size=100, n_epoch=20)",
"_____no_output_____"
]
],
[
[
"## Testing\nAfter you're satisified with the training output and accuracy, you can then run the network on the **test data set** to measure it's performance! Remember, only do this after you've done the training and are satisfied with the results.\n\nA good result will be **higher than 95% accuracy**. Some simple models have been known to get up to 99.7% accuracy!",
"_____no_output_____"
]
],
[
[
"# Compare the labels that our model predicts with the actual labels\n\n# Find the indices of the most confident prediction for each item. That tells us the predicted digit for that sample.\npredictions = np.array(model.predict(testX)).argmax(axis=1)\n\n# Calculate the accuracy, which is the percentage of times the predicated labels matched the actual labels\nactual = testY.argmax(axis=1)\ntest_accuracy = np.mean(predictions == actual, axis=0)\n\n# Print out the result\nprint(\"Test accuracy: \", test_accuracy)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f5757a2e58c29fe4fa133bab0d19ad604fa2ae | 33,833 | ipynb | Jupyter Notebook | 03-import.ipynb | WillRhB/PythonLesssons | e2f268145a715acd5d63668752a4d61617a4c562 | [
"MIT"
] | null | null | null | 03-import.ipynb | WillRhB/PythonLesssons | e2f268145a715acd5d63668752a4d61617a4c562 | [
"MIT"
] | null | null | null | 03-import.ipynb | WillRhB/PythonLesssons | e2f268145a715acd5d63668752a4d61617a4c562 | [
"MIT"
] | null | null | null | 359.925532 | 32,056 | 0.929359 | [
[
[
"import datalook",
"_____no_output_____"
],
[
"help(datalook.analyse)",
"Help on function analyse in module datalook:\n\nanalyse(filename, outfile=None)\n Displays the mean, maxima and minimum value for each weather station.\n Creates a figure of three subplots, showing values for mean, maxima and minimum value with axis = 0, \n y axis labels, and a tight layout.\n\n"
],
[
"datalook.analyse('data/weather-02.csv')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d0f57d3ca4d274bce0b52a7ab93366aa468a0f84 | 71,367 | ipynb | Jupyter Notebook | docs/Pandas.ipynb | hotbaby/mmn | 8dfefc2f41e3cee96efb9e81a3cba97ec3cb0f29 | [
"MIT"
] | 3 | 2020-12-09T02:52:02.000Z | 2021-01-12T15:07:17.000Z | docs/Pandas.ipynb | hotbaby/mmn | 8dfefc2f41e3cee96efb9e81a3cba97ec3cb0f29 | [
"MIT"
] | null | null | null | docs/Pandas.ipynb | hotbaby/mmn | 8dfefc2f41e3cee96efb9e81a3cba97ec3cb0f29 | [
"MIT"
] | 2 | 2020-11-18T02:31:18.000Z | 2021-01-09T01:37:52.000Z | 22.822833 | 98 | 0.31841 | [
[
[
"# Pandas",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"Pandas提供了3种数据类型,分别是`Series`、`DataFrame`和`Panel`。\n\n* `Series`用于保存一维数据\n* `DataFrame` 用于保存二维数据\n* `Panel`用于保存三维或者可变维数据",
"_____no_output_____"
],
[
"## Series数据结构",
"_____no_output_____"
],
[
"`Series`本质上是一个带索引的一维数组。",
"_____no_output_____"
],
[
"指定索引:",
"_____no_output_____"
]
],
[
[
"s = pd.Series([1,3,2,4], index=['a', 'b', 'c', 'd'])",
"_____no_output_____"
],
[
"s.index",
"_____no_output_____"
],
[
"s.values",
"_____no_output_____"
]
],
[
[
"默认索引:",
"_____no_output_____"
]
],
[
[
"s = pd.Series([1, 3, 2, 4])",
"_____no_output_____"
],
[
"s.index",
"_____no_output_____"
],
[
"s.values",
"_____no_output_____"
]
],
[
[
"## DataFrame数据结构",
"_____no_output_____"
],
[
"### 创建DataFrame",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({'x': ['a', 'b', 'c'],\n 'y': range(1, 4),\n 'z': [2, 5, 3]})",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df.values",
"_____no_output_____"
]
],
[
[
"### 查看数据内容",
"_____no_output_____"
],
[
"* `df.info()` 查看DataFrame属性信息\n* `df.head()` 查看DataFrame前五行数据信息\n* `df.tail()` 查看DataFrame后五行数据信息",
"_____no_output_____"
],
[
"### 选取多列",
"_____no_output_____"
],
[
"* df.loc\n* df.iloc",
"_____no_output_____"
]
],
[
[
"df[['x', 'y']]\ndf.loc[:, ['x', 'y']]\ndf.iloc[:, [0, 1]]",
"_____no_output_____"
]
],
[
[
"### 单行过滤",
"_____no_output_____"
]
],
[
[
"df[df.z>=3]",
"_____no_output_____"
]
],
[
[
"### 重新定义列名",
"_____no_output_____"
]
],
[
[
"df.rename(columns={'x': 'X'}, inplace=True)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.columns = ['X', 'Y', 'Z']",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
],
[
[
"### 数据的多重索引",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({\n 'X': list('ABCABC'),\n 'year': [2010] * 3 + [2011] * 3,\n 'Value': [1, 3, 4, 3, 5, 2]\n})",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.set_index(['X', 'year'])",
"_____no_output_____"
]
],
[
[
"## 表格的变换",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({\n 'X': list('ABC'),\n '2010': [1, 3, 4],\n '2011': [3, 5, 2]\n})",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df_melt = pd.melt(df, id_vars='X', var_name='year', value_name='value')\ndf_melt",
"_____no_output_____"
]
],
[
[
"* `id.vars('X')`表示由标识变量构成的向量,用于标识观测的变量\n* `variable_name('year')`表示用于保存原始变量名的变量名称\n* `value.name('value')`表示用于保存原始值的名称",
"_____no_output_____"
]
],
[
[
"df_pivot = df_melt.pivot_table(index='X', columns='year', values='value')\ndf_pivot.reset_index(inplace=True)\ndf_pivot",
"_____no_output_____"
]
],
[
[
"## 变量的变换",
"_____no_output_____"
],
[
"* `apply`的操作对象是`DataFrame`的某一列(`axis`=0)或者某一列(`axis`=1)\n* `applymap`的操作对象是元素级,作用于每个`DataFrame`的每个数据",
"_____no_output_____"
],
[
"## 表格的排序",
"_____no_output_____"
],
[
"`df.sort_values(by, ascending=True)`",
"_____no_output_____"
]
],
[
[
"df",
"_____no_output_____"
],
[
"df.sort_values('2010', ascending=False)",
"_____no_output_____"
],
[
"df.sort_values('2011', ascending=True)",
"_____no_output_____"
],
[
"df.sort_values(by=['X', '2010'], ascending=False)",
"_____no_output_____"
]
],
[
[
"## 表格拼接",
"_____no_output_____"
]
],
[
[
"df1 = pd.DataFrame({\n 'x': ['a', 'b', 'c'],\n 'y': range(1, 4),\n})",
"_____no_output_____"
],
[
"df2 = pd.DataFrame({\n 'z': ['B', 'D', 'H'],\n 'g': [2, 5, 3]\n})",
"_____no_output_____"
],
[
"df3 = pd.DataFrame({\n 'x': ['g', 'd'],\n 'y': [2, 5]\n})",
"_____no_output_____"
]
],
[
[
"横轴方向连接",
"_____no_output_____"
]
],
[
[
"pd.concat([df1, df2], axis=1)",
"_____no_output_____"
]
],
[
[
"纵轴方向连接",
"_____no_output_____"
]
],
[
[
"pd.concat([df1, df3], axis=0).reset_index()",
"_____no_output_____"
]
],
[
[
"## 表的融合",
"_____no_output_____"
]
],
[
[
"df1 = pd.DataFrame({\n 'x': list('abc'),\n 'y': range(1, 4)\n})",
"_____no_output_____"
],
[
"df2 = pd.DataFrame({\n 'x': list('abd'),\n 'z': [2, 5, 3]\n})",
"_____no_output_____"
],
[
"df3 = pd.DataFrame({\n 'g': list('abd'),\n 'z': [2, 5, 3]\n})",
"_____no_output_____"
],
[
"df1",
"_____no_output_____"
],
[
"df2",
"_____no_output_____"
],
[
"df3",
"_____no_output_____"
]
],
[
[
"只保留左表的所有数据",
"_____no_output_____"
]
],
[
[
"pd.merge(df1, df2, how='left', on='x')",
"_____no_output_____"
]
],
[
[
"只保留右表的数据",
"_____no_output_____"
]
],
[
[
"pd.merge(df1, df2, how='right', on='x')",
"_____no_output_____"
]
],
[
[
"保留两个表中公共的部分信息",
"_____no_output_____"
]
],
[
[
"pd.merge(df1, df2, how='inner', on='x')",
"_____no_output_____"
]
],
[
[
"保留两个表的所有信息",
"_____no_output_____"
]
],
[
[
"pd.merge(df1, df2, how='outer', on='x')",
"_____no_output_____"
]
],
[
[
"## 表格分组操作",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({\n 'X': list('ABC'),\n '2010': [1, 3, 4],\n '2011': [3, 5, 2]\n})",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
],
[
[
"按行或列操作",
"_____no_output_____"
],
[
"按行求和",
"_____no_output_____"
]
],
[
[
"df[['2010', '2011']].apply(lambda x: x.sum(), axis=1)",
"_____no_output_____"
]
],
[
[
"按列求和",
"_____no_output_____"
]
],
[
[
"df[['2010', '2011']].apply(lambda x: x.sum(), axis=0)",
"_____no_output_____"
]
],
[
[
"多列运算",
"_____no_output_____"
]
],
[
[
"df['2010_2011'] = df[['2010', '2011']].apply(lambda x: x['2010'] + 2 * x['2011'], axis=1)\ndf",
"_____no_output_____"
]
],
[
[
"分组操作",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({\n 'X': list('ABC'),\n '2010': [1, 3, 4],\n '2011': [3, 5, 2]\n})",
"_____no_output_____"
],
[
"df_melt = pd.melt(df, id_vars=['X'], var_name='year', value_name='value')",
"_____no_output_____"
],
[
"df_melt",
"_____no_output_____"
]
],
[
[
"按`year`分组求均值",
"_____no_output_____"
]
],
[
[
"df_melt.groupby('year').mean()",
"_____no_output_____"
]
],
[
[
"按`year`和`x`两列分组求均值",
"_____no_output_____"
]
],
[
[
"df_melt.groupby(['year', 'X']).mean()",
"_____no_output_____"
],
[
"df_melt.groupby(['year', 'X'], as_index=False).mean()",
"_____no_output_____"
]
],
[
[
"分组聚合",
"_____no_output_____"
]
],
[
[
"df_melt.groupby(['X', 'year']).aggregate([np.mean, np.median])",
"_____no_output_____"
]
],
[
[
"分组运算:`transform()`函数",
"_____no_output_____"
]
],
[
[
"df_melt['percentage'] = df_melt.groupby('X')['value'].transform(lambda x: x/s.sum())",
"_____no_output_____"
],
[
"df_melt",
"_____no_output_____"
]
],
[
[
"分组筛选:`filter()`函数",
"_____no_output_____"
]
],
[
[
"df_melt.groupby('X').filter(lambda x: x['value'].mean()>2)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f58113c19137414e35b3b452343126a7fc8518 | 205,011 | ipynb | Jupyter Notebook | cohortbalancer-function.ipynb | csrinesh/balance_cohort | ea96f81cbd163e9a5e31ad611e7d33f8fa2e9c74 | [
"MIT"
] | null | null | null | cohortbalancer-function.ipynb | csrinesh/balance_cohort | ea96f81cbd163e9a5e31ad611e7d33f8fa2e9c74 | [
"MIT"
] | null | null | null | cohortbalancer-function.ipynb | csrinesh/balance_cohort | ea96f81cbd163e9a5e31ad611e7d33f8fa2e9c74 | [
"MIT"
] | 1 | 2022-02-08T02:55:02.000Z | 2022-02-08T02:55:02.000Z | 406.767857 | 129,288 | 0.93173 | [
[
[
"from numpy.random import Generator\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport random\nfrom scipy.optimize import minimize \nfrom scipy.stats import gaussian_kde\nfrom scipy.stats import ks_2samp\nfrom scipy.special import rel_entr\nfrom scipy.stats import entropy\nimport scipy\nfrom optimparallel import minimize_parallel",
"_____no_output_____"
],
[
"config = {\n 'nbase': 500,\n 'neca': 2000,\n 'nwaypoints': 20,\n 'optimizeon': 'entropy'\n}",
"_____no_output_____"
]
],
[
[
"# Toy Datasets",
"_____no_output_____"
]
],
[
[
"# For Base cohort\nage = 45\nlab1 = 3.4\nlab2 = 5000\npcond1 = 0.3\npcat1 = 0.5\n\nmean = [age, lab1, lab2]\ncov = [[14, 3, 100], \n [3, 2, 50],\n [100, 50, 25000]]\n\n\nnp.random.seed(42)\nx = np.random.multivariate_normal(mean, cov, config['nbase'])\nx[x< 0] = 0\n\nlab3 = np.random.beta(1, 2, size=config['nbase'])\nx = np.concatenate((x, lab3.reshape(config['nbase'], -1)), axis=1)\n\ncond1 = np.random.binomial(1, pcond1, size=config['nbase']).reshape(config['nbase'], -1)\nx = np.concatenate((x, cond1), axis=1)\n\ncat1 = np.random.binomial(3, pcat1, size=config['nbase']).reshape(config['nbase'], -1)\nx = np.concatenate((x, cat1), axis=1)\n\ndata_base = pd.DataFrame(x, columns=['age', 'lab1', 'lab2', 'lab3', 'cond1', 'cat1'])",
"_____no_output_____"
],
[
"# For External cohort\nage = 50\nlab1 = 5.5\nlab2 = 4800\npcond1 = 0.5\npcat1 = 0.7\nfactor = 1.5\n\nmean = [age, lab1, lab2]\ncov = [[20, 5, 150], \n [5, 4, 100],\n [150, 100, 55000]]\n\n\nx = np.random.multivariate_normal(mean, cov, config['neca'])\nx[x< 0] = 0\n\nlab3 = factor*np.random.beta(1, 2, size=config['neca'])\nx = np.concatenate((x, lab3.reshape(config['neca'], -1)), axis=1)\n\ncond1 = np.random.binomial(1, pcond1, size=config['neca']).reshape(config['neca'], -1)\nx = np.concatenate((x, cond1), axis=1)\n\ncat1 = np.random.binomial(3, pcat1, size=config['neca']).reshape(config['neca'], -1)\nx = np.concatenate((x, cat1), axis=1)\n\ndata_eca = pd.DataFrame(x, columns=['age', 'lab1', 'lab2', 'lab3', 'cond1', 'cat1'])",
"_____no_output_____"
],
[
"# Define Cohen's d: Standardized Mean Difference\ndef cohensd(d1, d2):\n num = np.mean(d1) - np.mean(d2)\n den = np.sqrt((np.std(d1) ** 2 + np.std(d2) ** 2) / 2)\n cohens_d = num / den\n return cohens_d",
"_____no_output_____"
],
[
"# Plot data distribution\ndef plot_dist(data1, data2, col, ax=None):\n if not ax:\n _, ax = plt.subplots(1,1)\n \n ax.hist(data1[col], density=True, fill='black', histtype='stepfilled', \n edgecolor='black', bins= 20, linewidth=1.2, label=data1.name)\n ax.hist(data2[col], density=True, bins= 20, fill='blue', histtype='stepfilled',\n edgecolor='blue', linewidth=1.2, label=data2.name, alpha=0.7)\n ax.legend(loc='best')\n ax.set_title(col)\n return ",
"_____no_output_____"
],
[
"# View the Starting distribution\n\n_, ax = plt.subplots(3,2, figsize=(15, 10))\n\ndata_base.name = 'RCT Cohort'\ndata_eca.name = 'ECA Cohort'\nplot_dist(data_base, data_eca, 'age', ax[0][0])\nplot_dist(data_base, data_eca, 'lab1', ax[0][1])\nplot_dist(data_base, data_eca, 'lab2', ax[1][0])\nplot_dist(data_base, data_eca, 'lab3', ax[1][1])\nplot_dist(data_base, data_eca, 'cond1', ax[2][0])\nplot_dist(data_base, data_eca, 'cat1', ax[2][1])",
"_____no_output_____"
],
[
"# Define main function here\ndef genetic_matching(df_base, df_eca, optimize=True, pweights=None):\n # Set the plotting style\n try:\n plt.style.use('neuroblu')\n except:\n print('neuroblu style not found. Using default style!')\n \n # Define loss function\n def calc_loss(w): \n est_den = {}\n for c in data_base.columns:\n est_den[c] = gaussian_kde(data_eca[c], weights=w)\n\n\n if config['optimizeon'] == 'least_squares':\n loss = 0\n for c in data_base.columns:\n loss += (kde_base[c] - est_den[c](waypoints[c])*barwidth[c])**2 \n\n return sum(loss)\n\n elif config['optimizeon'] == 'rel_entr':\n loss = 0\n for c in data_base.columns:\n loss += rel_entr(kde_base[c], est_den[c](waypoints[c])*barwidth[c]) \n\n return sum(loss)\n\n elif config['optimizeon'] == 'entropy':\n loss = 0\n for c in data_base.columns:\n loss += entropy(kde_base[c], est_den[c](waypoints[c])*barwidth[c]) \n\n return loss\n else:\n raise NotImplemented\n \n if optimize:\n # Precompute parameters required for KDE estimation\n dist_density, waypoints, barwidth, kde_base = {}, {}, {}, {}\n\n for c in df_base.columns:\n dist_density[c] = gaussian_kde(df_base[c])\n\n b = max(df_base[c].max(), df_eca[c].max())\n a = min(df_base[c].min(), df_eca[c].min())\n waypoints[c] = np.linspace(a, b, config['nwaypoints'])\n\n barwidth[c] = np.diff(waypoints[c])[0]\n\n kde_base[c] = dist_density[c](waypoints[c])*barwidth[c]\n\n # Optimization\n bounds = list(zip(np.zeros(config['neca']), np.ones(config['neca'])))\n weights = np.ones(config['neca'])/config['neca']\n\n wopt = minimize(calc_loss, weights, bounds=bounds, method=\"L-BFGS-B\", \n options={'gtol':'1e-3', 'maxiter': 200, 'disp': True})\n p = wopt.x/sum(wopt.x)\n \n # Plot weights\n fig = plt.figure()\n ax = fig.add_subplot(1, 1, 1)\n ax.hist(p, bins=20)\n ax.set_title('Distribution of weights')\n ax.set_yscale('log')\n\n # Results of optimization\n print('Total Loss:', wopt.fun)\n print('Has optimization converged:', wopt.success)\n \n print('\\nSampling from distribution ..')\n df_eca_study = data_eca.sample(n=config['nbase'], replace=True, weights=p, \n random_state=42)\n df_eca_study.name = 'Optimized ECA Cohort'\n \n f, ax = plt.subplots(len(df_base.columns),2, figsize=(15, 20))\n\n for i, col in enumerate(df_base.columns):\n plot_dist(df_base, df_eca, col, ax[i][0])\n plot_dist(df_base, df_eca_study, col, ax[i][1])\n \n f.suptitle('Before (left) and after(right) Matching') \n \n return df_eca_study",
"_____no_output_____"
],
[
"data_eca_study = genetic_matching(data_base, data_eca)",
"RUNNING THE L-BFGS-B CODE\n\n * * *\n\nMachine precision = 2.220D-16\n N = 2000 M = 10\n\nAt X0 0 variables are exactly at the bounds\n\nAt iterate 0 f= 2.22630D+00 |proj g|= 9.99500D-01\n\nAt iterate 1 f= 6.33250D-01 |proj g|= 3.20153D-02\n\nAt iterate 2 f= 6.30402D-01 |proj g|= 2.14964D-02\n\nAt iterate 3 f= 6.28251D-01 |proj g|= 1.46053D-02\n\nAt iterate 4 f= 5.91055D-01 |proj g|= 1.10376D-02\n\nAt iterate 5 f= 3.37441D-01 |proj g|= 1.18596D-02\n\nAt iterate 6 f= 2.17999D-01 |proj g|= 1.41561D-02\n\n * * *\n\nTit = total number of iterations\nTnf = total number of function evaluations\nTnint = total number of segments explored during Cauchy searches\nSkip = number of BFGS updates skipped\nNact = number of active bounds at final generalized Cauchy point\nProjg = norm of the final projected gradient\nF = final function value\n\n * * *\n\n N Tit Tnf Tnint Skip Nact Projg F\n 2000 6 8 2647 0 1237 1.416D-02 2.180D-01\n F = 0.21799895405045799 \n\nSTOP: TOTAL NO. of f AND g EVALUATIONS EXCEEDS LIMIT \nTotal Loss: 0.217998954050458\nHas optimization converged: False\n\nSampling from distribution ..\n"
],
[
"for col in data_base.columns:\n print(f'Distribution of {col}')\n print('Before balancing', ks_2samp(data_base[col], data_eca[col]))\n print('After balancing', ks_2samp(data_base[col], data_eca_study[col]))\n print('\\n')",
"Distribution of age\nBefore balancing KstestResult(statistic=0.4615, pvalue=1.2212453270876722e-15)\nAfter balancing KstestResult(statistic=0.102, pvalue=0.010968966298223139)\n\n\nDistribution of lab1\nBefore balancing KstestResult(statistic=0.459, pvalue=1.2212453270876722e-15)\nAfter balancing KstestResult(statistic=0.11, pvalue=0.004686682589297701)\n\n\nDistribution of lab2\nBefore balancing KstestResult(statistic=0.406, pvalue=1.2212453270876722e-15)\nAfter balancing KstestResult(statistic=0.186, pvalue=5.6534932411175595e-08)\n\n\nDistribution of lab3\nBefore balancing KstestResult(statistic=0.259, pvalue=1.2212453270876722e-15)\nAfter balancing KstestResult(statistic=0.086, pvalue=0.04950261174890187)\n\n\nDistribution of cond1\nBefore balancing KstestResult(statistic=0.2135, pvalue=1.2212453270876722e-15)\nAfter balancing KstestResult(statistic=0.014, pvalue=0.9999999998936827)\n\n\nDistribution of cat1\nBefore balancing KstestResult(statistic=0.2625, pvalue=1.2212453270876722e-15)\nAfter balancing KstestResult(statistic=0.138, pvalue=0.0001434246303518553)\n\n\n"
],
[
"for col in data_base.columns:\n print(f'Distribution of {col}')\n print('Before balancing', cohensd(data_base[col], data_eca[col]))\n print('After balancing', cohensd(data_base[col], data_eca_study[col]))\n print('\\n')",
"Distribution of age\nBefore balancing -1.2072887363365403\nAfter balancing -0.18460449201331947\n\n\nDistribution of lab1\nBefore balancing -1.127079119575515\nAfter balancing -0.1807599351241777\n\n\nDistribution of lab2\nBefore balancing 0.8934273136316265\nAfter balancing 0.31464657671400276\n\n\nDistribution of lab3\nBefore balancing -0.6267566209648288\nAfter balancing -0.140457191185468\n\n\nDistribution of cond1\nBefore balancing -0.44759118866934916\nAfter balancing 0.031150362222411618\n\n\nDistribution of cat1\nBefore balancing -0.7026411298035246\nAfter balancing -0.3578078178825893\n\n\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f5817394cfa8c10f779aa073af8866ee7d82f0 | 158,877 | ipynb | Jupyter Notebook | SentimentAnalysisOnTwitterWithR.ipynb | JPedro-loureiro/Sentiment-Analysis-on-Twitter-R | de04d233c62ee5811044957b5e2058de187f627b | [
"MIT"
] | null | null | null | SentimentAnalysisOnTwitterWithR.ipynb | JPedro-loureiro/Sentiment-Analysis-on-Twitter-R | de04d233c62ee5811044957b5e2058de187f627b | [
"MIT"
] | null | null | null | SentimentAnalysisOnTwitterWithR.ipynb | JPedro-loureiro/Sentiment-Analysis-on-Twitter-R | de04d233c62ee5811044957b5e2058de187f627b | [
"MIT"
] | null | null | null | 148.206157 | 43,236 | 0.790945 | [
[
[
"## Índice\n\n### 1 - Análise de Sentimentos: uma introdução\n### 2 - Carregando os pacotes necessários\n### 3 - Conectando com a API do Twitter\n #### 3.1 - Autenticação\n #### 3.2 - Obtendo os tweets\n### 4 - Text Mining\n### 5 - Manipulação de dados\n#### 5.1 - Tokenização\n#### 5.2 - Núvem de palavras\n### 6 - Removendo as Stopwords\n### 7 - Dicionário Léxico\n### 8 - Resultados\n#### 8.1 - Top 10 tweets \"positivos\"\n#### 8.2 - Top 10 tweets \"negativos\"\n#### 8.3 - Overview da análise de sentimentos\n#### 8.4 - Análise de sentimentos em série temporal\n### 9 - Considerações finais",
"_____no_output_____"
],
[
"### 1 - Análise de Sentimentos: uma introdução\n\nA análise de sentimentos é a tarefa que tem por objetivo analisar e identificar de maneira quantitativa o estado emocional e afetivo do autor através de seu texto. Existem diversos tipos de textos (acadêmicos, literários, jornalísticos e etc.) e neste artigo iremos analisar o que as pessoas dizem em suas contas do Twitter sobre o time de futebol do Clube de Regatas do Flamengo.\n",
"_____no_output_____"
],
[
"### 2 - Carregando os pacotes necessários\nO primeiro passo a ser dado é carregar os pacotes que nos auxiliaram nesta jornada!",
"_____no_output_____"
]
],
[
[
"#Install\ninstall.packages(\"twitteR\")\ninstall.packages(\"rvest\")\ninstall.packages(\"tidyverse\")\ninstall.packages(\"tidytext\")\ninstall.packages(\"tm\") # for text mining\ninstall.packages(\"SnowballC\") # for text stemming\ninstall.packages(\"wordcloud\") # word-cloud generator \ninstall.packages(\"RColorBrewer\") # color palettes\ninstall.packages(\"stopwords\")\ninstall.packages(\"lexiconPT\")",
"_____no_output_____"
],
[
"library(twitteR)\nlibrary(rvest)\nlibrary(tidyverse)\nlibrary(tidytext)\nlibrary(tm)\nlibrary(SnowballC)\nlibrary(wordcloud)\nlibrary(RColorBrewer)\nlibrary(stopwords)\nlibrary(lexiconPT)",
"Warning message:\n\"package 'twitteR' was built under R version 3.6.1\"Warning message:\n\"package 'rvest' was built under R version 3.6.1\"Loading required package: xml2\nWarning message:\n\"package 'xml2' was built under R version 3.6.1\"Warning message:\n\"package 'tidyverse' was built under R version 3.6.1\"-- Attaching packages --------------------------------------- tidyverse 1.2.1 --\nv ggplot2 3.2.0 v purrr 0.3.2\nv tibble 2.1.3 v dplyr 0.8.3\nv tidyr 0.8.3 v stringr 1.4.0\nv readr 1.3.1 v forcats 0.4.0\nWarning message:\n\"package 'tibble' was built under R version 3.6.1\"Warning message:\n\"package 'tidyr' was built under R version 3.6.1\"Warning message:\n\"package 'readr' was built under R version 3.6.1\"Warning message:\n\"package 'purrr' was built under R version 3.6.1\"Warning message:\n\"package 'dplyr' was built under R version 3.6.1\"Warning message:\n\"package 'stringr' was built under R version 3.6.1\"Warning message:\n\"package 'forcats' was built under R version 3.6.1\"-- Conflicts ------------------------------------------ tidyverse_conflicts() --\nx dplyr::filter() masks stats::filter()\nx readr::guess_encoding() masks rvest::guess_encoding()\nx dplyr::id() masks twitteR::id()\nx dplyr::lag() masks stats::lag()\nx dplyr::location() masks twitteR::location()\nx purrr::pluck() masks rvest::pluck()\nWarning message:\n\"package 'tidytext' was built under R version 3.6.1\"Warning message:\n\"package 'tm' was built under R version 3.6.1\"Loading required package: NLP\n\nAttaching package: 'NLP'\n\nThe following object is masked from 'package:ggplot2':\n\n annotate\n\nWarning message:\n\"package 'wordcloud' was built under R version 3.6.1\"Loading required package: RColorBrewer\nWarning message:\n\"package 'stopwords' was built under R version 3.6.1\"\nAttaching package: 'stopwords'\n\nThe following object is masked from 'package:tm':\n\n stopwords\n\n"
]
],
[
[
"### 3 - Conectando com a API do Twitter\n#### 3.1 - Autenticação\nApós instalarmos e carregarmos todos os pacotes necessários, iremos efetuar a conexão com o Twitter através do pacote \"TwitteR\". Para isso precisamos das chaves de autenticação da API do twitter.",
"_____no_output_____"
]
],
[
[
"#API's and Token's keys\nAPI_key = \"hhrTn58RfcBoxjpGdvQbLTf6v\"\nAPI_secret_key = \"r6zON2bTvFv8qlaEDWWaIowRknxRxc97EGENdSBfdXZlFmkmrm\"\nAccess_token = \"4928132291-OFnrrwGy5FKHLPePkUsxLA5exJwrbJ7l51azMWV\"\nAccess_token_secret = \"q7inW05npl3S70yRihMkJdasF0roZyrlzZW46D1XTSbDd\"\n\n#OAuth\nsetup_twitter_oauth(API_key, API_secret_key, Access_token, Access_token_secret)",
"[1] \"Using direct authentication\"\n"
]
],
[
[
"#### 3.2 - Obtendo os Tweets\nEm seguida, iremos criar uma função que solicita ao usuário uma palavra chave ou hashtag a ser buscada, a quantidade de tweets por dia, o idioma, a localidade e as datas iniciais e finais nas quais a pesquisa será feita. De posse destas informações, a função irá buscar os tweets de acordo com os parâmetros estabelecidos e armazenas as informações numa tabela chamada \"tweets_df\"",
"_____no_output_____"
]
],
[
[
"#Search Twitters\nsearch_twiter <- function(){\n #This function search for a specific number of tweets per day in a range of dates\n #Return a Tibble whith 3 columns: 1- TweetID; 2- Date; 3- Tweet;\n \n #Search string\n search_string <- paste(readline(prompt = \"A wordkey or a hashtag: \"), \" -filter:retweets\") #An exemple: \"flamengo -filter:retweets\"\n \n #Number of tweets per day\n n_tweets <- as.integer(readline(prompt = \"The number of tweets per day: \"))\n \n #Language\n language <- readline(prompt = \"Language (ex.: pt): \")\n \n #Locale\n loc <- readline(prompt = \"Tweets locale (ex.: br): \")\n \n #Date range: yyyy-mm-dd\n ini_date <- as.Date(readline(prompt = \"The inicial date (yyyy-mm-dd): \"), format = \"%Y-%m-%d\") #Inicial date\n final_date <- as.Date(readline(prompt = \"The final date (yyyy-mm-dd): \"), format = \"%Y-%m-%d\") #Final date\n n_days <- as.integer(final_date - ini_date)\n \n #Searching\n tweets <- c()\n date <- c()\n for(day in 0:n_days){\n tweets <- c(tweets, searchTwitter(searchString = search_string, n = n_tweets,\n lang = language, locale = loc,\n since = as.character(ini_date + day),\n until = as.character(ini_date + day + 1)))\n \n date <- c(date, rep(ini_date + day, n_tweets))\n }\n \n tweets_df <- tibble(tweetID = 1:length(tweets),\n date = format(as.Date(date, origin = \"1970-01-01\"), \"%d-%m-%Y\"),\n tweets)\n \n return(tweets_df)\n}\n\ntweets_df <- search_twiter()",
"A wordkey or a hashtag: flamengo\nThe number of tweets per day: 500\nLanguage (ex.: pt): pt\nTweets locale (ex.: br): br\nThe inicial date (yyyy-mm-dd): 2019-07-16\nThe final date (yyyy-mm-dd): 2019-07-23\n"
]
],
[
[
"Vamos dar uma olhada nos tweets que coletamos!",
"_____no_output_____"
]
],
[
[
"head(tweets_df)",
"_____no_output_____"
]
],
[
[
"### 4 - Text Mining\nNote que temos 3500 observações e 3 colunas na nossa tabela. Porém, a terceira coluna é uma lista que contém todas as informações sobre os tweets coletados. É importante ter em mente que um tweet não é representado apenas pelo seu texto, mas sim por outras informações também, como por exemplo, o seu ID, o autor, a data de publicação e etc. Porém, nos interessa apenas o texto contido no tweet. Então, iremos criar outra função que irá extrair o texto e através de Text Mining, vamos \"limpar\" este texto. Ou seja, vamos remover hashtags, links, pontuações, dígitos e tudo aquilo que não interessa para a nossa análise.",
"_____no_output_____"
]
],
[
[
"#Cleaning tweets\nclean_tweets <- function(tweets){\n \n #Getting tweets texts\n tweets = sapply(tweets, function(x) x$text)\n \n #Remove retweet entities\n tweets = gsub('(RT|via)((?:\\\\b\\\\W*@\\\\w+)+)', \"\", tweets)\n \n #Remove Hashtags\n tweets = gsub('#\\\\w+', \"\", tweets)\n \n #Remove links\n tweets = gsub('http\\\\w+', \"\", tweets)\n \n #Remove punctuations\n tweets = gsub('[[:punct:]]', \"\", tweets)\n \n #Remove numbers\n tweets = gsub('[[:digit:]]', \"\", tweets)\n \n #Remove line break\n tweets = gsub('\\n', \"\", tweets)\n \n #lower case\n tweets = tolower(tweets)\n}\n\ntweets_df$tweets <- clean_tweets(tweets_df$tweets)",
"_____no_output_____"
]
],
[
[
"Embora os tweets ainda apresentem alguns caracteres estranhos, podemos dizer que temos nossos textos devidamente coletados. Estes caracteres estranhos serão tratados mais adiante.",
"_____no_output_____"
]
],
[
[
"head(tweets_df)",
"_____no_output_____"
]
],
[
[
"### 5 - Manipulação de dados\n#### 5.1 - Tokenização\nNeste tópico iremos fazer a manipulação dos dados. O objetivo aqui é ter uma tabela onde cada registro possui uma única palavra, mantendo sempre a referência a qual tweet esta palavra pertence. Isso será muito útil para as análises seguintes.",
"_____no_output_____"
]
],
[
[
"#Tokenization \ntweets_tokens <- tweets_df %>%\n unnest_tokens(words, tweets)\n\nhead(tweets_tokens, 25)",
"_____no_output_____"
]
],
[
[
"#### 5.2 - Núvem de palavras\nA fim de visualizar as palavras que são mais comuns dentre os tweets, vamos criar uma outra função que gera uma núvem de palavras.",
"_____no_output_____"
]
],
[
[
"wc <- function(tweets_tokens){\n plot <- tweets_tokens %>%\n group_by(words) %>%\n summarise(freq = n()) %>%\n filter(words != \"ifood\")\n \n wordcloud(words = plot$words, freq = plot$freq, min.freq = 1,\n max.words=200, random.order=FALSE, rot.per=0.35, \n colors=brewer.pal(8, \"Dark2\"))\n}\n\nwc(tweets_tokens)",
"_____no_output_____"
]
],
[
[
"### 6 - Stopwords\nNote que a palavra mais comum é \"flamengo\" pois é o tema da nossa pesquisa e é seguida por várias outras palavras que não tem muito peso semântico.Essas palavras são conhecidas como \"stopwords\". As stopwords são palavras que são comumente usadas e não possuem grande relevância para a semântica do texto. Como por exemplo, as palavras \"a\", \"o\", \"onde\", \"de\" e etc. Então, iremos remove-las.\nPara tal, iremos usar um conjunto de dados do pacote \"stopwords\" que contém alguns exemplos de stopwords em português. Após carregar esses dados, vamos usar a função \"anti_join\" para excluir todas as palavras em comum entre os tweets e o conjunto de stopwords.",
"_____no_output_____"
]
],
[
[
"#Stopwords\n\nstopwords <- stopwords(language = \"pt\", source = \"stopwords-iso\")\nstopwords <- tibble(stopwords)\n\nhead(stopwords)\n\ntweets_tokens <- tweets_tokens %>%\n anti_join(stopwords, by = c(\"words\" = \"stopwords\"))",
"_____no_output_____"
]
],
[
[
"### 7 - Dicionário Léxico\nAté aqui, o que fizemos foi apenas limpar e manipular os dados. Neste tópico daremos início à análise de sentimentos.\nO primeiro passo consiste em carregarmos o dicionário léxico. Este dicionário léxico contém uma coluna que indica qual a polaridade das palavras, ou seja, se uma palavra é tida como positiva (+1), neutra (0) ou negativa (-1). O objetivo é conseguir quantificar o valor semântico de cada palavra, e por conseguinte, de cada tweet.",
"_____no_output_____"
]
],
[
[
"#LexiconPT\nlex_pt <- oplexicon_v3.0\nhead(lex_pt)",
"_____no_output_____"
]
],
[
[
"Em seguida iremos cruzar as palavras dos tweets com as palavras do dicionário léxico, atribuindo uma polaridade para cada palavra dos tweets. Para isso, usaremos a função \"inner_join\".",
"_____no_output_____"
]
],
[
[
"tweets_tokens <- tweets_tokens %>%\n inner_join(lex_pt, by = c(\"words\" = \"term\")) %>%\n select(tweetID, date, words, polarity)\n\nhead(tweets_tokens)",
"_____no_output_____"
]
],
[
[
"Agora iremos visualizar novamente uma núvem de palavras para entender como a retirada das stopwords e cruzamento com o dicionário léxico afetaram nossos tweets.",
"_____no_output_____"
]
],
[
[
"wc(tweets_tokens)",
"_____no_output_____"
]
],
[
[
"### 8 - Resultados\nApós feita a polarização das palavras contidas nos tweets, vamos utilizar esses dados para extrair melhores informações dos textos.",
"_____no_output_____"
],
[
"#### 8.1 - Top 10 tweets \"positivos\"",
"_____no_output_____"
]
],
[
[
"#Most positive tweets\ntop_10_pos <- tweets_tokens %>%\n group_by(tweetID) %>%\n summarise(polarity = sum(polarity)) %>%\n arrange(desc(polarity)) %>%\n head(10)\n\n #Ordering\n top_10_pos$tweetID <- factor(top_10_pos$tweetID,\n levels = unique(top_10_pos$tweetID[order(top_10_pos$polarity,\n decreasing = FALSE)]))\n #Plot\n ggplot(top_10_pos, aes(tweetID, polarity)) +\n geom_col(fill = \"cornflowerblue\") +\n coord_flip() +\n xlab(\"Tweet ID\") + ylab(\"Polarity\") + ggtitle(\"Top 10 positive Polarity x Tweet ID\")",
"_____no_output_____"
]
],
[
[
"#### 8.2 - Top 10 tweets \"negativos\"",
"_____no_output_____"
]
],
[
[
"#Most negative tweets\ntop_10_neg <- tweets_tokens %>%\n group_by(tweetID) %>%\n summarise(polarity = sum(polarity)) %>%\n arrange(polarity) %>%\n head(10)\n\n #Ordering\n top_10_neg$tweetID <- factor(top_10_neg$tweetID,\n levels = unique(top_10_neg$tweetID)[order(top_10_neg$polarity,\n decreasing = TRUE)])\n #Plot\n ggplot(top_10_neg, aes(tweetID, polarity)) +\n geom_col(fill = \"lightcoral\") +\n coord_flip() +\n xlab(\"Tweet ID\") + ylab(\"Polarity\") + ggtitle(\"Top 10 negative Polarity x Tweet ID\")",
"_____no_output_____"
]
],
[
[
"#### 8.3 - Overview da análise de sentimentos\nO gráfico a seguir categoriza os tweets como \"positivos\", \"negativos\" e \"neutros\" e mostra qual a quantidade de tweets por categoria.",
"_____no_output_____"
]
],
[
[
"#Sentiment Analysis Overview\n\ntweets_tokens %>%\n group_by(tweetID) %>%\n summarise(polarity = sum(polarity)) %>%\n mutate(class = ifelse(polarity > 0, \"positive\",\n ifelse(polarity < 0, \"negative\", \"neutral\"))) %>%\n count(class) %>%\n ggplot(aes(factor(class), n, fill = class)) +\n geom_col() +\n xlab(\"Class\") + ylab(\"Tweet ID\") + ggtitle(\"Class x Tweet ID\")",
"_____no_output_____"
]
],
[
[
"#### 8.4 - Análise de sentimentos em série temporal\nEste gráfico mostra como foi a sentimento dos usuários em relação ao tema ao longo do período determinado na busca.",
"_____no_output_____"
]
],
[
[
"#Time series\ntweets_tokens %>%\n group_by(date) %>%\n summarise(polarity = sum(polarity)) %>%\n ggplot(aes(date, polarity, group = 1)) +\n geom_line() +\n ggtitle(\"Polarity x Date\")",
"_____no_output_____"
]
],
[
[
"### 9 - Considerações finais",
"_____no_output_____"
],
[
"Por fim, conseguimos extrair insights relevantes de como andou o ânimo dos rubro negros na semana do dia 16/07 a 23/07 de 2019.\nVamos analisar o gráfico do ítem 8.4. Entre os dias 16 e 17 temos uma leve crescida no índice de polaridade pois o torcedor vivia a expectativa do jogo decisivo entre Flamengo e Atlético Paranaense pela Copa do Brasil (https://globoesporte.globo.com/pr/futebol/copa-do-brasil/noticia/veja-os-melhores-momentos-gols-e-os-penaltis-de-flamengo-x-athletico-pela-copa-do-brasil.ghtml). Expectativa esta que não se confirmou e isso explica a forte insatisfação da torcida nos dias seguintes. Junto à derrota nas penalidades máximas, no dia 21 o time empatou com o Corinthians num jogo moroso o que deixou a exigente torcida mais insatisfeita ainda. Porém, no dia seguinte, a diretoria do clube anunciou uma contratação de peso! Felipe Luiz se torna então jogador do flamengo(https://esportes.r7.com/prisma/cosme-rimoli/por-copa-do-qatar-e-salario-europeu-filipe-luis-e-do-flamengo-22072019). O lateral veio pra ocupar um setor do campo que a torcida apontava carências já há um bom tempo o que explica a forte acensão do humor dos rubro negros.\nVale ressaltar que o método aqui adotado para a análise de sentimentos pode não ser o mais preciso pois depende muito da qualidade do dicionário léxico. Porém, podemos ver que os resultados coincidem com os acontecimentos da agitada semana no ninho do urubu.\n\nDeixo a cargo do leitor adivinhar qual o time que o autor torce. Espero que você não precise fazer uma análise de sentimentos para descobrir! Rs.\n\nObrigado!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0f59042474dbf63c97b77a6b16370a1658f37ec | 142,997 | ipynb | Jupyter Notebook | Dimensionality Reduction/LDA/LDA_Algo.ipynb | Abhi10398/Machine-Learning | 6ec5e522e9eca7eef76a4f7a95007b27bf5e0fb8 | [
"Apache-2.0"
] | null | null | null | Dimensionality Reduction/LDA/LDA_Algo.ipynb | Abhi10398/Machine-Learning | 6ec5e522e9eca7eef76a4f7a95007b27bf5e0fb8 | [
"Apache-2.0"
] | null | null | null | Dimensionality Reduction/LDA/LDA_Algo.ipynb | Abhi10398/Machine-Learning | 6ec5e522e9eca7eef76a4f7a95007b27bf5e0fb8 | [
"Apache-2.0"
] | null | null | null | 317.066519 | 42,534 | 0.919355 | [
[
[
"# Linear Discriminate Analysis (LDA)",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\n# data generation\n\nmean1=np.array([0,0])\nmean2=np.array([4,5])\nvar=np.array([[1,0.1],[0.1,1]])\nnp.random.seed(0)\ndata1=np.random.multivariate_normal(mean1,var,500)\ndata2=np.random.multivariate_normal(mean2,var,500)\ndata=np.concatenate((data1,data2))\nlabel=np.concatenate((np.zeros(data1.shape[0]),np.ones(data2.shape[0])))\n\nplt.figure()\nplt.scatter(data[:,0],data[:,1],c=label)\nplt.title('Data visualization')\nplt.figure()\nplt.scatter(data[:,0],np.zeros(data.shape[0]),c=label)\nplt.title('distribution in x direction')\nplt.figure()\nplt.scatter(data[:,1],np.zeros(data.shape[0]),c=label)\nplt.title('distribution in y direction')",
"_____no_output_____"
],
[
"# perform 2-class and m-class LDA\ndef LDA(data,label): \n id={}\n data_l={}\n mean_l={}\n cov_l={}\n S_w=np.zeros((data.shape[1],data.shape[1]))\n\n\n cls=np.unique(label)\n for i in cls:\n id[i]=np.where(label==i)[0]\n data_l[i]=data[id[i],:]\n mean_l[i]=np.mean(data_l[i],axis=0)\n cov_l[i]=(data_l[i]-mean_l[i]).T @ (data_l[i]-mean_l[i])\n S_w=S_w+cov_l[i]\n\n\n S_w=S_w/len(data_l)\n\n if len(data_l)==2:\n S_b=(mean_l[0]-mean_l[1]).T @ (mean_l[0]-mean_l[1])\n w=np.linalg.inv(S_w) @ (mean_l[0]-mean_l[1])\n #print(w.shape)\n\n else:\n S_t=np.cov(data,rowvar=False)\n S_b=np.zeros((data.shape[1],data.shape[1]))\n for i in cls:\n S_b += len(data_l[i])*(mean_l[i]-np.mean(data,axis=0)).T@(mean_l[i]-np.mean(data,axis=0))\n \n #S_b = S_t-S_w # insert your code here\n u,_,_= np.linalg.svd(np.linalg.inv(S_w)@S_b)\n w=u[:,:len(data_l)-1]\n\n return w",
"_____no_output_____"
],
[
"# after LDA projection\n\nw=LDA(data,label)\nplt.figure()\nplt.scatter(data @ w,np.zeros(data.shape[0]),c=label)\n",
"_____no_output_____"
],
[
"#classification using LDA\n#use k-nearest neighbour classifier after dimensionality reduction\n\n\nfrom sklearn.neighbors import KNeighborsClassifier\n\nLDA_data= data @ w[:,np.newaxis]\nk=5\nknn = KNeighborsClassifier(n_neighbors=k)\nknn.fit(LDA_data, label)\n\nprint('KNN Training accuracy =',knn.score(LDA_data,label)*100)\n\n# test data\nnp.random.seed(0)\ndata1=np.random.multivariate_normal(mean1,var,50)\ndata2=np.random.multivariate_normal(mean2,var,50)\ndata_tst=np.concatenate((data1,data2))\ntst_label=np.concatenate((np.zeros(data1.shape[0]),np.ones(data2.shape[0])))\n\nprint('KNN Testing accuracy =',knn.score(data_tst@ w[:,np.newaxis],tst_label)*100)\n\n",
"KNN Training accuracy = 100.0\nKNN Testing accuracy = 100.0\n"
]
],
[
[
"## LDA multiclass \n1. 3 class Sythetic data\n2. Homework: Mnist 3 class and 10 class\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\nmean1=np.array([0,0])\nmean2=np.array([4,5])\nmean3=np.array([-5,-4])\nvar=np.array([[1,0.1],[0.1,1]])\nnp.random.seed(0)\ndata1=np.random.multivariate_normal(mean1,var,500)\ndata2=np.random.multivariate_normal(mean2,var,500)\ndata3=np.random.multivariate_normal(mean3,var,500)\ndata=np.concatenate((data1,data2,data3))\nlabel=np.concatenate((np.zeros(data1.shape[0]),np.ones(data2.shape[0]),np.ones(data3.shape[0])+1))\n\nplt.figure()\nplt.scatter(data[:,0],data[:,1],c=label)\nplt.title('Data visualization')\nplt.figure()\nplt.scatter(data[:,0],np.zeros(data.shape[0]),c=label)\nplt.title('distribution in x direction')\nplt.figure()\nplt.scatter(data[:,1],np.zeros(data.shape[0]),c=label)\nplt.title('distribution in y direction')\n",
"_____no_output_____"
],
[
"# after projection\nw=LDA(data,label)\nprint(w.shape)\nplt.figure()\nplt.scatter(data @ w[:,0],np.zeros(data.shape[0]),c=label) # by performing 1D projection",
"(2, 2)\n"
],
[
"# testing (using KNN)\n\nfrom sklearn.neighbors import KNeighborsClassifier\n\nLDA_data= data @ w\nk=5\nknn = KNeighborsClassifier(n_neighbors=k)\nknn.fit(LDA_data, label)\n\nprint('KNN Training accuracy =',knn.score(LDA_data,label)*100)\n\n# test data\nnp.random.seed(0)\ndata1=np.random.multivariate_normal(mean1,var,50)\ndata2=np.random.multivariate_normal(mean2,var,50)\ndata3=np.random.multivariate_normal(mean3,var,50)\ndata_tst=np.concatenate((data1,data2,data3))\ntst_label=np.concatenate((np.zeros(data1.shape[0]),np.ones(data2.shape[0]),np.ones(data2.shape[0])+1))\n\nprint('KNN Testing accuracy =',knn.score(data_tst@ w,tst_label)*100)\n",
"KNN Training accuracy = 99.93333333333332\nKNN Testing accuracy = 100.0\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0f59bad238276b80fdc6ca23d601cdcf34a1758 | 5,983 | ipynb | Jupyter Notebook | compss/programming_model/bindings/python/src/pycompss/tests/resources/notebook/simple.ipynb | eflows4hpc/compss | c497f6d34722103c6c8f83ebc314b495573ce054 | [
"Apache-2.0"
] | null | null | null | compss/programming_model/bindings/python/src/pycompss/tests/resources/notebook/simple.ipynb | eflows4hpc/compss | c497f6d34722103c6c8f83ebc314b495573ce054 | [
"Apache-2.0"
] | null | null | null | compss/programming_model/bindings/python/src/pycompss/tests/resources/notebook/simple.ipynb | eflows4hpc/compss | c497f6d34722103c6c8f83ebc314b495573ce054 | [
"Apache-2.0"
] | null | null | null | 18.696875 | 91 | 0.489721 | [
[
[
"# Test suite for Jupyter-notebook",
"_____no_output_____"
],
[
"## First step\nImport ipycompss library",
"_____no_output_____"
]
],
[
[
"import pycompss.interactive as ipycompss",
"_____no_output_____"
]
],
[
[
"## Second step \nInitialize COMPSs runtime",
"_____no_output_____"
]
],
[
[
"ipycompss.start(graph=True, trace=True, debug=True)",
"_____no_output_____"
]
],
[
[
"## Third step\nImport task module before annotating functions or methods ",
"_____no_output_____"
]
],
[
[
"from pycompss.api.task import task",
"_____no_output_____"
]
],
[
[
"Other declarations",
"_____no_output_____"
]
],
[
[
"MY_GLOBAL = 2",
"_____no_output_____"
],
[
"class myClass(object):\n def __init__(self):\n self.value = 1",
"_____no_output_____"
],
[
"def divide(a, b):\n def is_zero(value):\n if value == 0:\n raise True\n else:\n return False\n\n if not is_zero(b):\n return a / b\n else:\n raise Exception(\"Can not divide by 0.\")",
"_____no_output_____"
],
[
"def multiply(a, b):\n return a * b",
"_____no_output_____"
]
],
[
[
"## Fourth step\nDeclare functions and decorate with @task those that should be tasks ",
"_____no_output_____"
]
],
[
[
"@task(returns=int)\ndef test(val1):\n return multiply(val1, val1)",
"_____no_output_____"
],
[
"@task(returns=int)\ndef test2(val2, val3):\n return val2 + val3",
"_____no_output_____"
],
[
"@task(returns=int)\ndef test3(val2, val3):\n return divide(val2, val3)",
"_____no_output_____"
]
],
[
[
"## Fifth step\nInvoke tasks ",
"_____no_output_____"
]
],
[
[
"a = test(2)",
"_____no_output_____"
],
[
"b = test2(a, 5)",
"_____no_output_____"
],
[
"c = test3(a, 2)",
"_____no_output_____"
]
],
[
[
"## Sixt step \nImport compss_wait_on module and synchronize tasks ",
"_____no_output_____"
]
],
[
[
"from pycompss.api.api import compss_wait_on",
"_____no_output_____"
],
[
"result = compss_wait_on(b)",
"_____no_output_____"
]
],
[
[
"### Only those results being sychronized with compss_wait_on will have a valid value ",
"_____no_output_____"
]
],
[
[
"print(\"Results: \")\nprint(\"a: \", a)\nprint(\"b: \", b)\nprint(\"c: \", c)\nprint(\"result: \", result)",
"_____no_output_____"
]
],
[
[
"### Stop COMPSs runtime. All data will be synchronized in the main program ",
"_____no_output_____"
]
],
[
[
"ipycompss.stop(sync=True)",
"_____no_output_____"
],
[
"print(\"Results after stopping PyCOMPSs: \")\nprint(\"a: \", a)\nprint(\"b: \", b)\nprint(\"c: \", c)\nprint(\"result: \", result)",
"_____no_output_____"
]
],
[
[
"### CHECK THE RESULTS FOR THE TEST ",
"_____no_output_____"
]
],
[
[
"from pycompss.runtime.management.classes import Future\n\nif a == 4 and isinstance(b, Future) and result == 9 and c == 2:\n print(\"RESULT=EXPECTED\")\nelse:\n raise Exception(\"RESULT=UNEXPECTED\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f5a08e5d5b3ba97a79dd8645de837579939a4a | 324,092 | ipynb | Jupyter Notebook | Project1.ipynb | Howie4PP/Airbnb_Data_Analyst | 513e01f50ed0f1f57a0216dd4344924a10d99574 | [
"CNRI-Python"
] | null | null | null | Project1.ipynb | Howie4PP/Airbnb_Data_Analyst | 513e01f50ed0f1f57a0216dd4344924a10d99574 | [
"CNRI-Python"
] | null | null | null | Project1.ipynb | Howie4PP/Airbnb_Data_Analyst | 513e01f50ed0f1f57a0216dd4344924a10d99574 | [
"CNRI-Python"
] | null | null | null | 106.996368 | 96,692 | 0.777909 | [
[
[
"## AirBnB Data anaylst",
"_____no_output_____"
],
[
"### 1. Business Understanding:",
"_____no_output_____"
],
[
"This project aims to follow the CRISP-DM to address the three questions which related to business or real-world applications. The dataset is picked up from Kaggle, contributed by AirBnB, which contains the rent data about Seattle. I would like to process whole data and try to find some valuable data to help owner understand which feature could improve the rate of rent, moreover, whether train a model to predict. Three questions are:",
"_____no_output_____"
],
[
"1. What features influence the rating of the house?\n2. When is the most popular time for this area?\n3. Could we create a model to predict the price?",
"_____no_output_____"
],
[
"### 2. Data Understanding",
"_____no_output_____"
],
[
"Dataset comes from Kaggle",
"_____no_output_____"
]
],
[
[
"# import libraries\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n% matplotlib inline\n\n#load data\nlisting = pd.read_csv('listings.csv')",
"_____no_output_____"
],
[
"## to check the feature of the dataset, then fliter\nlisting.columns.values ",
"_____no_output_____"
]
],
[
[
"### Q1. What features influence the rating of house?",
"_____no_output_____"
],
[
"### 2.1 Pre-processing data - the null value columns",
"_____no_output_____"
]
],
[
[
"## check the missing value\nlisting.isnull().mean().sort_values() ",
"_____no_output_____"
],
[
"## calculate how many column has missing value\nmiss_num = np.sum(listing.isnull().mean() != 0) \nmiss_num ",
"_____no_output_____"
],
[
"# visulize the missing data\nplt.figure(figsize=(20,8))\n(listing.isnull().sum(axis = 0).sort_values(ascending=False)[:miss_num]/len(listing)*100).plot(kind='bar', facecolor='b');\nplt.title('Proportion of missing values per feature')\nplt.ylabel('% of missing values')\nplt.xlabel(\"Features\");",
"_____no_output_____"
]
],
[
[
"As we can see above, the features of \"security_deposit\", \"monthly_price\", \"square_feet\" and \"license\" are missing more than 50% values, and these features are unnecessary for the project, thus, I will drop them. Technically, I will make a copy of original data.",
"_____no_output_____"
]
],
[
[
"#back up dataset\nlisting_backup = listing.copy()",
"_____no_output_____"
],
[
"# drop the columns which lose more than 50% missing value\nlisting = listing.drop(['security_deposit', 'monthly_price', 'square_feet', 'license'], axis = 1)",
"_____no_output_____"
],
[
"## check again, I have dropped them all\nlisting.isnull().mean().sort_values() ",
"_____no_output_____"
],
[
"# check and summary the column of missing data\nmiss_num = np.sum(listing.isnull().mean() != 0)\nmiss_num",
"_____no_output_____"
],
[
"# visulize the missing data to check\nplt.figure(figsize=(20,8))\n(listing.isnull().sum(axis = 0).sort_values(ascending=False)[:miss_num]/len(listing)*100).plot(kind='bar', facecolor='b');\nplt.title('Proportion of missing values per feature')\nplt.ylabel('% of missing values')\nplt.xlabel(\"Features\");",
"_____no_output_____"
],
[
"# check the total features\nlisting.shape",
"_____no_output_____"
]
],
[
[
"Currently, the dataset has 88 features. Although the column of 'Weekly_price' quite close 50%, it may will be used, just keep it first. However, I think the dataset still contain some outliers and useless data, I will continue to process.",
"_____no_output_____"
],
[
"### 2.2 Processing the outliers and useless data",
"_____no_output_____"
],
[
"#### 2.2.1, As the final target is prediction. The column of \"id\" and \"note\" are doesn't help to predict, so drop its",
"_____no_output_____"
]
],
[
[
"def drop_columns(m_columns):\n listing.drop(columns = m_columns, inplace = True)",
"_____no_output_____"
],
[
"drop_columns = ['id', 'notes']\ndrop_columns(drop_columns)",
"_____no_output_____"
]
],
[
[
"#### 2.2.2 When I check the features, I found the values of some columns are link, which also could be defined as useless data.",
"_____no_output_____"
]
],
[
[
"## the column contains key word of 'url'\nurl_col=[col for col in listing.columns.values if 'url' in col]\nurl_col\ndrop_columns(url_col)",
"_____no_output_____"
]
],
[
[
"#### 2.2.3 Furthermore, there are some wired values, which means whole column only one value. I think those data won't help me to predict, drop it!",
"_____no_output_____"
]
],
[
[
"unique_col=[col for col in listing.columns.values if listing[col].nunique()==1]\nunique_col\ndrop_columns(unique_col)",
"_____no_output_____"
]
],
[
[
"#### 2.2.4 Next, after checking again, I think some columns that contains 'host', which may not be relevant for the prediction. For instance, 'host_id', 'host_name', 'host_since', 'host_location', 'host_about', 'host_verifications', 'host_neighbourhood'. Same operate as before. Drop it.",
"_____no_output_____"
]
],
[
[
"drop_cols=['host_id', 'host_name', 'host_since', 'host_location', 'host_about', 'host_verifications', 'host_neighbourhood']\ndrop_columns(drop_cols)",
"_____no_output_____"
]
],
[
[
"#### 2.2.5 Lastly, the unstructured and redundant data. Some columns, such as City, State, Street, smart_location, latitude and longitude, could be represented by other value - zipcode. Therefore, they can be dropped",
"_____no_output_____"
]
],
[
[
"drop_cols=['city','street','state','smart_location','latitude','longitude', 'neighbourhood_cleansed']\ndrop_cols ## It shows what is the columns I will drop\ndrop_columns(drop_cols)",
"_____no_output_____"
],
[
"#drop columns containing unstructured data\ndrop_cols=['name','summary', 'space', 'description', 'neighborhood_overview','transit', 'first_review', 'last_review', 'calendar_updated']\ndrop_cols\ndrop_columns(drop_cols)",
"_____no_output_____"
],
[
"# review_scores_rating is likely a combination of the other review scores so no need to keep them all.\n# furthermore, I want to find which features has a high relationship with rating, I would like to assume another review scores \n# has an exterme influence \nlisting.drop(['review_scores_accuracy', 'review_scores_cleanliness', 'review_scores_checkin','reviews_per_month',\n 'review_scores_communication', 'review_scores_location', 'review_scores_value'\n ], axis=1, inplace=True)",
"_____no_output_____"
],
[
"## Now, we have 38 features after dropping columns, \n## then next step, I will transfer categorical data and fill up missing value of some columns\nlisting.shape ",
"_____no_output_____"
],
[
"# check the sample\nlisting.head()",
"_____no_output_____"
]
],
[
[
"### 2.3 Filling up missing value and transferring categorical data",
"_____no_output_____"
],
[
"##### 2.3.1 transferring categorical data",
"_____no_output_____"
]
],
[
[
"# Creating a table to visual the total value\npd.merge(pd.DataFrame(listing.dtypes,columns=['datatype']).reset_index(),\n pd.DataFrame(pd.DataFrame(listing.iloc[0]).reset_index()),on='index')",
"_____no_output_____"
],
[
"# get the name of columns which contain categorical data\nlisting.select_dtypes(include=['object']).columns",
"_____no_output_____"
],
[
"# to check unique value of the 'zip' column\nlisting.zipcode.unique()",
"_____no_output_____"
],
[
"# there is a outlier in zipcode, which need fix\nlisting.zipcode.replace('99\\n98122','98122',inplace=True)",
"_____no_output_____"
],
[
"# summary the categorical columns and check the values\ncategorical_col = ['host_response_time', 'host_response_rate', 'host_acceptance_rate',\n 'host_is_superhost', 'host_has_profile_pic', 'host_identity_verified',\n 'neighbourhood', \n 'neighbourhood_group_cleansed', 'zipcode', 'is_location_exact',\n 'property_type', 'room_type', 'bed_type', 'amenities', 'price',\n 'weekly_price', 'cleaning_fee', 'extra_people', \n 'instant_bookable',\n 'cancellation_policy', 'require_guest_profile_picture',\n 'require_guest_phone_verification']\n\nfor i in categorical_col:\n print(i,\":\", listing[i].unique(), \"\\n\")",
"host_response_time : ['within a few hours' 'within an hour' nan 'within a day'\n 'a few days or more'] \n\nhost_response_rate : ['96%' '98%' '67%' nan '100%' '71%' '97%' '60%' '50%' '31%' '90%' '70%'\n '88%' '80%' '63%' '33%' '99%' '75%' '83%' '94%' '58%' '43%' '93%' '92%'\n '40%' '57%' '89%' '95%' '78%' '81%' '91%' '38%' '86%' '30%' '56%' '76%'\n '64%' '82%' '17%' '87%' '25%' '69%' '53%' '65%' '68%' '55%'] \n\nhost_acceptance_rate : ['100%' nan '0%'] \n\nhost_is_superhost : ['f' 't' nan] \n\nhost_has_profile_pic : ['t' 'f' nan] \n\nhost_identity_verified : ['t' 'f' nan] \n\nneighbourhood : ['Queen Anne' nan 'Ballard' 'Phinney Ridge' 'Fremont' 'Lower Queen Anne'\n 'Westlake' 'Wallingford' 'Green Lake' 'Minor' 'Madrona'\n 'Harrison/Denny-Blaine' 'Leschi' 'University District' 'Roosevelt'\n 'Madison Park' 'Capitol Hill' 'Atlantic' 'North Beacon Hill'\n 'Central Business District' 'Pike Place Market' 'Pike Market' 'Eastlake'\n 'Portage Bay' 'South Lake Union' 'Magnolia' 'Belltown'\n 'International District' 'Yesler Terrace' 'First Hill' 'Pioneer Square'\n 'Gatewood' 'The Junction' 'Arbor Heights' 'Alki' 'North Admiral'\n 'Crown Hill' 'Genesee' 'Fairmount Park' 'Interbay' 'Industrial District'\n 'Holly Park' 'South Beacon Hill' 'Greenwood' 'Bitter Lake' 'Fauntleroy'\n 'Mount Baker' 'Columbia City' 'Seward Park' 'Brighton' 'South Delridge'\n 'Highland Park' 'High Point' 'View Ridge' 'Windermere' 'Dunlap'\n 'Rainier Beach' 'North Delridge' 'Maple Leaf' 'Bryant' 'Ravenna'\n 'Wedgewood' 'Riverview' 'Montlake' 'Broadway' 'Stevens' 'Victory Heights'\n 'Mathews Beach' 'Cedar Park' 'Meadowbrook' 'Olympic Hills'\n 'North Beach/Blue Ridge' 'Broadview' 'Haller Lake' 'Pinehurst'\n 'Licton Springs' 'North College Park' 'Laurelhurst' 'Seaview'\n 'Georgetown' 'South Park' 'Roxhill'] \n\nneighbourhood_group_cleansed : ['Queen Anne' 'Ballard' 'Other neighborhoods' 'Cascade' 'Central Area'\n 'University District' 'Downtown' 'Magnolia' 'West Seattle' 'Interbay'\n 'Beacon Hill' 'Rainier Valley' 'Delridge' 'Seward Park' 'Northgate'\n 'Capitol Hill' 'Lake City'] \n\nzipcode : ['98119' '98109' '98107' '98117' nan '98103' '98105' '98115' '98101'\n '98122' '98112' '98144' '98121' '98102' '98199' '98104' '98134' '98136'\n '98126' '98146' '98116' '98177' '98118' '98108' '98133' '98106' '98178'\n '98125'] \n\nis_location_exact : ['t' 'f'] \n\nproperty_type : ['Apartment' 'House' 'Cabin' 'Condominium' 'Camper/RV' 'Bungalow'\n 'Townhouse' 'Loft' 'Boat' 'Bed & Breakfast' 'Other' 'Dorm' 'Treehouse'\n 'Yurt' 'Chalet' 'Tent' nan] \n\nroom_type : ['Entire home/apt' 'Private room' 'Shared room'] \n\nbed_type : ['Real Bed' 'Futon' 'Pull-out Sofa' 'Airbed' 'Couch'] \n\namenities : ['{TV,\"Cable TV\",Internet,\"Wireless Internet\",\"Air Conditioning\",Kitchen,Heating,\"Family/Kid Friendly\",Washer,Dryer}'\n '{TV,Internet,\"Wireless Internet\",Kitchen,\"Free Parking on Premises\",\"Buzzer/Wireless Intercom\",Heating,\"Family/Kid Friendly\",Washer,Dryer,\"Smoke Detector\",\"Carbon Monoxide Detector\",\"First Aid Kit\",\"Safety Card\",\"Fire Extinguisher\",Essentials}'\n '{TV,\"Cable TV\",Internet,\"Wireless Internet\",\"Air Conditioning\",Kitchen,\"Free Parking on Premises\",\"Pets Allowed\",\"Pets live on this property\",Dog(s),Cat(s),\"Hot Tub\",\"Indoor Fireplace\",Heating,\"Family/Kid Friendly\",Washer,Dryer,\"Smoke Detector\",\"Carbon Monoxide Detector\",Essentials,Shampoo}'\n ...\n '{\"Cable TV\",\"Wireless Internet\",Kitchen,\"Free Parking on Premises\",Breakfast,\"Pets live on this property\",Dog(s),Cat(s),Heating,Washer,Dryer,\"Smoke Detector\",\"First Aid Kit\",\"Safety Card\",\"Fire Extinguisher\",Essentials,Hangers,\"Hair Dryer\",Iron,\"Laptop Friendly Workspace\"}'\n '{TV,\"Wireless Internet\",Kitchen,\"Free Parking on Premises\",\"Elevator in Building\",Heating,\"Smoke Detector\",Essentials,Hangers,\"Hair Dryer\",\"Laptop Friendly Workspace\"}'\n '{TV,\"Cable TV\",Internet,\"Wireless Internet\",Kitchen,\"Free Parking on Premises\",\"Pets live on this property\",Dog(s),\"Elevator in Building\",\"Hot Tub\",\"Indoor Fireplace\",\"Buzzer/Wireless Intercom\",Heating,\"Family/Kid Friendly\",Washer,Dryer,\"Smoke Detector\",Essentials,Shampoo,Hangers,Iron,\"Laptop Friendly Workspace\"}'] \n\nprice : ['$85.00' '$150.00' '$975.00' '$100.00' '$450.00' '$120.00' '$80.00'\n '$60.00' '$90.00' '$95.00' '$99.00' '$245.00' '$165.00' '$461.00'\n '$109.00' '$66.00' '$200.00' '$700.00' '$110.00' '$75.00' '$600.00'\n '$45.00' '$300.00' '$175.00' '$325.00' '$222.00' '$159.00' '$125.00'\n '$348.00' '$148.00' '$350.00' '$349.00' '$160.00' '$130.00' '$137.00'\n '$20.00' '$145.00' '$400.00' '$70.00' '$170.00' '$465.00' '$50.00'\n '$59.00' '$47.00' '$55.00' '$65.00' '$25.00' '$40.00' '$149.00' '$129.00'\n '$105.00' '$218.00' '$126.00' '$115.00' '$225.00' '$89.00' '$134.00'\n '$375.00' '$97.00' '$197.00' '$135.00' '$180.00' '$69.00' '$195.00'\n '$224.00' '$338.00' '$79.00' '$49.00' '$57.00' '$96.00' '$295.00'\n '$53.00' '$35.00' '$133.00' '$61.00' '$52.00' '$275.00' '$199.00'\n '$435.00' '$250.00' '$116.00' '$39.00' '$98.00' '$73.00' '$190.00'\n '$68.00' '$196.00' '$209.00' '$749.00' '$285.00' '$235.00' '$103.00'\n '$143.00' '$142.00' '$335.00' '$499.00' '$156.00' '$94.00' '$219.00'\n '$91.00' '$74.00' '$140.00' '$104.00' '$122.00' '$158.00' '$42.00'\n '$185.00' '$680.00' '$119.00' '$575.00' '$139.00' '$259.00' '$166.00'\n '$215.00' '$249.00' '$210.00' '$439.00' '$155.00' '$54.00' '$254.00'\n '$310.00' '$490.00' '$255.00' '$144.00' '$46.00' '$445.00' '$395.00'\n '$88.00' '$425.00' '$114.00' '$117.00' '$107.00' '$76.00' '$179.00'\n '$84.00' '$265.00' '$92.00' '$550.00' '$111.00' '$141.00' '$775.00'\n '$78.00' '$48.00' '$82.00' '$169.00' '$58.00' '$240.00' '$189.00'\n '$500.00' '$41.00' '$43.00' '$44.00' '$30.00' '$112.00' '$63.00' '$28.00'\n '$93.00' '$306.00' '$29.00' '$83.00' '$118.00' '$128.00' '$168.00'\n '$62.00' '$380.00' '$157.00' '$203.00' '$56.00' '$330.00' '$67.00'\n '$72.00' '$81.00' '$124.00' '$108.00' '$220.00' '$37.00' '$270.00'\n '$87.00' '$287.00' '$279.00' '$257.00' '$163.00' '$64.00' '$38.00'\n '$557.00' '$299.00' '$311.00' '$357.00' '$138.00' '$183.00' '$229.00'\n '$333.00' '$147.00' '$280.00' '$178.00' '$480.00' '$172.00' '$999.00'\n '$320.00' '$127.00' '$405.00' '$899.00' '$239.00' '$131.00' '$113.00'\n '$106.00' '$475.00' '$205.00' '$181.00' '$399.00' '$193.00' '$444.00'\n '$187.00' '$186.00' '$171.00' '$230.00' '$167.00' '$151.00' '$132.00'\n '$365.00' '$121.00' '$188.00' '$276.00' '$290.00' '$237.00' '$77.00'\n '$174.00' '$153.00' '$269.00' '$495.00' '$101.00' '$370.00' '$136.00'\n '$51.00' '$34.00' '$26.00' '$86.00' '$177.00' '$260.00' '$36.00'\n '$244.00' '$525.00' '$750.00' '$415.00' '$162.00' '$256.00' '$182.00'\n '$212.00' '$360.00' '$198.00' '$673.00' '$204.00' '$152.00' '$420.00'\n '$71.00' '$252.00' '$1,000.00' '$33.00' '$498.00' '$217.00' '$207.00'\n '$228.00' '$950.00' '$102.00' '$27.00' '$146.00' '$545.00' '$22.00'\n '$31.00' '$449.00' '$232.00' '$154.00' '$359.00'] \n\nweekly_price : [nan '$1,000.00' '$650.00' '$800.00' '$575.00' '$360.00' '$500.00'\n '$595.00' '$1,575.00' '$4,100.00' '$750.00' '$568.00' '$3,000.00'\n '$350.00' '$699.00' '$1,325.00' '$2,500.00' '$450.00' '$950.00'\n '$2,100.00' '$540.00' '$700.00' '$1,140.00' '$815.00' '$1,195.00'\n '$3,035.00' '$351.00' '$250.00' '$1,300.00' '$900.00' '$903.00' '$599.00'\n '$1,221.00' '$945.00' '$549.00' '$910.00' '$555.00' '$1,800.00' '$590.00'\n '$1,172.00' '$1,500.00' '$665.00' '$1,200.00' '$499.00' '$525.00'\n '$530.00' '$550.00' '$274.00' '$559.00' '$420.00' '$400.00' '$1,100.00'\n '$390.00' '$244.00' '$300.00' '$1,065.00' '$1,250.00' '$698.00' '$536.00'\n '$1,260.00' '$490.00' '$488.00' '$600.00' '$1,950.00' '$625.00'\n '$2,900.00' '$440.00' '$690.00' '$582.00' '$805.00' '$232.00' '$715.00'\n '$640.00' '$850.00' '$560.00' '$588.00' '$1,600.00' '$895.00' '$925.00'\n '$1,750.00' '$1,400.00' '$720.00' '$1,015.00' '$6,300.00' '$428.00'\n '$1,650.00' '$425.00' '$692.00' '$3,150.00' '$541.00' '$1,005.00'\n '$730.00' '$1,040.00' '$1,380.00' '$915.00' '$725.00' '$990.00'\n '$1,050.00' '$770.00' '$1,460.00' '$630.00' '$619.00' '$896.00' '$432.00'\n '$790.00' '$1,470.00' '$275.00' '$393.00' '$395.00' '$325.00' '$3,250.00'\n '$685.00' '$833.00' '$1,205.00' '$670.00' '$200.00' '$2,399.00'\n '$1,225.00' '$370.00' '$1,096.00' '$419.00' '$875.00' '$339.00' '$825.00'\n '$2,950.00' '$678.00' '$2,000.00' '$785.00' '$620.00' '$696.00' '$482.00'\n '$999.00' '$1,900.00' '$446.00' '$775.00' '$675.00' '$375.00' '$637.00'\n '$349.00' '$3,950.00' '$760.00' '$1,495.00' '$225.00' '$285.00' '$795.00'\n '$399.00' '$899.00' '$726.00' '$1,090.00' '$340.00' '$1,485.00' '$749.00'\n '$405.00' '$1,077.00' '$1,890.00' '$629.00' '$1,150.00' '$460.00'\n '$510.00' '$585.00' '$2,250.00' '$930.00' '$1,350.00' '$554.00' '$455.00'\n '$2,700.00' '$480.00' '$385.00' '$475.00' '$695.00' '$175.00' '$680.00'\n '$150.00' '$259.00' '$262.00' '$755.00' '$580.00' '$430.00' '$165.00'\n '$445.00' '$780.00' '$762.00' '$827.00' '$643.00' '$292.00' '$333.00'\n '$1,850.00' '$309.00' '$738.00' '$553.00' '$485.00' '$1,499.00' '$843.00'\n '$476.00' '$235.00' '$845.00' '$660.00' '$2,037.00' '$628.00' '$1,159.00'\n '$1,529.00' '$495.00' '$398.00' '$345.00' '$920.00' '$552.00' '$315.00'\n '$889.00' '$1,160.00' '$381.00' '$863.00' '$659.00' '$280.00' '$3,100.00'\n '$1,700.00' '$610.00' '$506.00' '$799.00' '$860.00' '$4,000.00' '$980.00'\n '$4,499.00' '$1,295.00' '$2,200.00' '$298.00' '$255.00' '$320.00'\n '$1,975.00' '$737.00' '$238.00' '$305.00' '$1,071.00' '$2,398.00'\n '$260.00' '$960.00' '$1,530.00' '$881.00' '$5,499.00' '$1,045.00'\n '$672.00' '$452.00' '$1,299.00' '$1,490.00' '$890.00' '$1,524.00'\n '$1,029.00' '$1,176.00' '$1,089.00' '$240.00' '$1,265.00' '$869.00'\n '$2,300.00' '$1,008.00' '$1,009.00' '$1,224.00' '$520.00' '$993.00'\n '$577.00' '$830.00' '$914.00' '$839.00' '$1,365.00' '$1,149.00' '$623.00'\n '$931.00' '$387.00' '$528.00' '$1,133.00' '$389.00' '$458.00' '$571.00'\n '$290.00' '$1,999.00' '$635.00' '$1,875.00' '$2,125.00' '$270.00'\n '$982.00' '$330.00' '$470.00' '$558.00' '$1,375.00' '$100.00' '$299.00'\n '$268.00' '$411.00' '$1,075.00' '$840.00' '$343.00' '$357.00' '$1,899.00'\n '$1,550.00' '$1,599.00' '$645.00' '$744.00' '$1,327.00' '$524.00'\n '$161.00' '$893.00' '$410.00' '$439.00' '$1,110.00' '$256.00' '$226.00'\n '$732.00' '$230.00' '$745.00' '$905.00' '$607.00' '$615.00' '$214.00'\n '$512.00' '$220.00' '$565.00' '$740.00' '$329.00' '$327.00' '$457.00'\n '$964.00' '$1,390.00' '$535.00' '$210.00' '$605.00' '$655.00' '$167.00'\n '$173.00' '$208.00' '$539.00' '$4,800.00' '$415.00' '$985.00' '$1,590.00'\n '$548.00' '$3,500.00' '$464.00' '$472.00' '$406.00' '$756.00' '$613.00'\n '$286.00' '$269.00' '$279.00' '$267.00' '$310.00' '$422.00' '$1,311.00'\n '$417.00' '$1,261.00' '$940.00' '$363.00' '$855.00' '$710.00' '$3,600.00'\n '$449.00' '$1,363.00' '$1,148.00' '$679.00' '$995.00' '$567.00'\n '$1,450.00' '$409.00' '$631.00' '$380.00' '$519.00' '$1,020.00' '$435.00'\n '$1,239.00' '$1,475.00' '$594.00' '$714.00' '$704.00' '$949.00' '$589.00'\n '$887.00' '$735.00' '$413.00' '$759.00' '$687.00' '$1,107.00' '$1,830.00'\n '$773.00' '$1,925.00' '$202.00' '$975.00' '$347.00' '$355.00' '$647.00'\n '$1,190.00' '$2,800.00' '$1,025.00' '$2,211.00' '$562.00' '$689.00'\n '$587.00' '$888.00' '$1,120.00'] \n\ncleaning_fee : [nan '$40.00' '$300.00' '$125.00' '$25.00' '$15.00' '$150.00' '$95.00'\n '$85.00' '$89.00' '$35.00' '$250.00' '$200.00' '$65.00' '$100.00'\n '$80.00' '$99.00' '$50.00' '$20.00' '$55.00' '$75.00' '$30.00' '$60.00'\n '$120.00' '$78.00' '$12.00' '$45.00' '$10.00' '$264.00' '$180.00'\n '$90.00' '$7.00' '$131.00' '$8.00' '$5.00' '$185.00' '$199.00' '$175.00'\n '$110.00' '$155.00' '$111.00' '$72.00' '$105.00' '$160.00' '$13.00'\n '$275.00' '$28.00' '$70.00' '$209.00' '$82.00' '$195.00' '$145.00'\n '$22.00' '$225.00' '$169.00' '$119.00' '$29.00' '$140.00' '$61.00'\n '$49.00' '$108.00' '$6.00' '$26.00' '$83.00' '$18.00' '$19.00' '$117.00'\n '$112.00' '$58.00' '$16.00' '$170.00' '$64.00' '$113.00' '$79.00'\n '$130.00' '$96.00' '$149.00' '$164.00' '$159.00' '$32.00' '$184.00'\n '$109.00' '$107.00' '$274.00' '$143.00' '$88.00' '$229.00' '$38.00'\n '$69.00' '$135.00' '$59.00' '$101.00' '$67.00' '$240.00' '$137.00'\n '$134.00' '$21.00' '$189.00' '$9.00' '$17.00' '$106.00' '$24.00'\n '$165.00' '$39.00' '$68.00' '$27.00' '$87.00' '$42.00' '$71.00' '$194.00'\n '$129.00' '$210.00' '$178.00' '$76.00' '$97.00' '$179.00' '$52.00'\n '$142.00' '$230.00'] \n\nextra_people : ['$5.00' '$0.00' '$25.00' '$15.00' '$30.00' '$10.00' '$20.00' '$50.00'\n '$60.00' '$75.00' '$100.00' '$35.00' '$40.00' '$45.00' '$7.00' '$14.00'\n '$55.00' '$18.00' '$29.00' '$12.00' '$19.00' '$8.00' '$21.00' '$26.00'\n '$17.00' '$44.00' '$9.00' '$80.00' '$200.00' '$28.00' '$85.00' '$250.00'\n '$13.00' '$16.00' '$300.00' '$33.00' '$49.00' '$22.00' '$27.00' '$68.00'\n '$71.00' '$48.00' '$6.00' '$36.00' '$175.00'] \n\ninstant_bookable : ['f' 't'] \n\ncancellation_policy : ['moderate' 'strict' 'flexible'] \n\nrequire_guest_profile_picture : ['f' 't'] \n\nrequire_guest_phone_verification : ['f' 't'] \n\n"
],
[
"## transfer the columns which contains 'f', 't'\nBinary_cols = ['host_is_superhost','host_has_profile_pic','host_identity_verified','is_location_exact','instant_bookable','require_guest_profile_picture','require_guest_phone_verification']\nfor i in Binary_cols:\n listing[i] = listing[i].map(lambda x: 1 if x == 't' else 0)",
"_____no_output_____"
],
[
"# to check whether the value is change\nlisting['host_is_superhost'].unique()",
"_____no_output_____"
],
[
"# same with last step, hot-coding to dummny variables\nencode_cols=['host_response_time','neighbourhood_group_cleansed','zipcode', 'neighbourhood'\n ,'property_type','room_type','bed_type','cancellation_policy']\nlisting=pd.get_dummies(data=listing, columns=encode_cols)",
"_____no_output_____"
],
[
"listing.shape # Cheking, after convert to dummy variables",
"_____no_output_____"
],
[
"# to check the sample\nlisting.head()",
"_____no_output_____"
]
],
[
[
"#### 2.3.2 Engineering for 'amenities' column",
"_____no_output_____"
],
[
"Next I will engineer 'amenities' column to extract categorical variables. Since the amenities column is in the form of list of amenities, I will extract each amenity and it would be its own categorical feature for each listing.",
"_____no_output_____"
]
],
[
[
"# process the data of amenities column, splite it first\nlisting.amenities = listing.amenities.str.replace(\"[{}]\", \"\") \namenities_col = listing.amenities.str.get_dummies(sep = \",\")\nlisting_cleaned = pd.merge(listing, amenities_col, left_index=True, right_index=True)",
"_____no_output_____"
],
[
"# drop the amenities column now that is has been onehot encoded\nlisting_cleaned.drop(['amenities'], axis=1, inplace = True)",
"_____no_output_____"
],
[
"# to check the amenities column already split\nlisting_cleaned.shape\nlisting_cleaned.head(3)",
"_____no_output_____"
]
],
[
[
"#### 2.3.3 Checking missing value and filling up",
"_____no_output_____"
]
],
[
[
"# check what column has missing value\nlisting_cleaned.isnull().mean().sort_values() ",
"_____no_output_____"
],
[
"# summary the missing value column and to find out what is value of them, ready to fill up\nmissing_cols = ['beds','host_listings_count','host_total_listings_count','bedrooms','bathrooms',\n 'host_response_rate','review_scores_rating', \n 'cleaning_fee']\n\nfor x in missing_cols:\n print(x,\":\", listing_cleaned[x].unique(), \"\\n\")",
"beds : [ 1. 7. 2. 3. 15. 5. 4. 6. 9. 8. nan 10.] \n\nhost_listings_count : [ 3. 6. 2. 1. 5. 9. 4. 36. 354. 8. 37. 7. 34. 10.\n 13. 21. 11. 169. 48. 502. 12. nan 18. 19. 15. 84. 163. 17.] \n\nhost_total_listings_count : [ 3. 6. 2. 1. 5. 9. 4. 36. 354. 8. 37. 7. 34. 10.\n 13. 21. 11. 169. 48. 502. 12. nan 18. 19. 15. 84. 163. 17.] \n\nbedrooms : [ 1. 5. 0. 3. 2. 6. 4. nan 7.] \n\nbathrooms : [1. 4.5 2. 3.5 1.5 2.5 3. 0.5 4. 0. 8. nan 5. ] \n\nhost_response_rate : ['96%' '98%' '67%' nan '100%' '71%' '97%' '60%' '50%' '31%' '90%' '70%'\n '88%' '80%' '63%' '33%' '99%' '75%' '83%' '94%' '58%' '43%' '93%' '92%'\n '40%' '57%' '89%' '95%' '78%' '81%' '91%' '38%' '86%' '30%' '56%' '76%'\n '64%' '82%' '17%' '87%' '25%' '69%' '53%' '65%' '68%' '55%'] \n\nreview_scores_rating : [ 95. 96. 97. nan 92. 99. 98. 100. 80. 85. 84. 88. 75. 90.\n 91. 94. 93. 77. 89. 71. 82. 87. 73. 83. 86. 76. 78. 66.\n 74. 60. 64. 70. 68. 72. 40. 79. 55. 20. 67. 57. 65. 53.\n 81.] \n\ncleaning_fee : [nan '$40.00' '$300.00' '$125.00' '$25.00' '$15.00' '$150.00' '$95.00'\n '$85.00' '$89.00' '$35.00' '$250.00' '$200.00' '$65.00' '$100.00'\n '$80.00' '$99.00' '$50.00' '$20.00' '$55.00' '$75.00' '$30.00' '$60.00'\n '$120.00' '$78.00' '$12.00' '$45.00' '$10.00' '$264.00' '$180.00'\n '$90.00' '$7.00' '$131.00' '$8.00' '$5.00' '$185.00' '$199.00' '$175.00'\n '$110.00' '$155.00' '$111.00' '$72.00' '$105.00' '$160.00' '$13.00'\n '$275.00' '$28.00' '$70.00' '$209.00' '$82.00' '$195.00' '$145.00'\n '$22.00' '$225.00' '$169.00' '$119.00' '$29.00' '$140.00' '$61.00'\n '$49.00' '$108.00' '$6.00' '$26.00' '$83.00' '$18.00' '$19.00' '$117.00'\n '$112.00' '$58.00' '$16.00' '$170.00' '$64.00' '$113.00' '$79.00'\n '$130.00' '$96.00' '$149.00' '$164.00' '$159.00' '$32.00' '$184.00'\n '$109.00' '$107.00' '$274.00' '$143.00' '$88.00' '$229.00' '$38.00'\n '$69.00' '$135.00' '$59.00' '$101.00' '$67.00' '$240.00' '$137.00'\n '$134.00' '$21.00' '$189.00' '$9.00' '$17.00' '$106.00' '$24.00'\n '$165.00' '$39.00' '$68.00' '$27.00' '$87.00' '$42.00' '$71.00' '$194.00'\n '$129.00' '$210.00' '$178.00' '$76.00' '$97.00' '$179.00' '$52.00'\n '$142.00' '$230.00'] \n\n"
]
],
[
[
"As we can see, for the host_acceptance_rate column, there are only 3 unique values, which including Nan. Hence, I will \ndrop this column as it does not offer an useful information.",
"_____no_output_____"
]
],
[
[
"#drop host_acceptance_rate column\nlisting_cleaned.drop(columns='host_acceptance_rate',inplace=True)",
"_____no_output_____"
]
],
[
[
"Weekly price be dropped. In my mind, firstly, it lost almost 50% data, it is not an accurate data. Next, it could be \nthe target value to be predicted, but not an independant value.",
"_____no_output_____"
]
],
[
[
"# drop the weekly_price column\nlisting_cleaned.drop(columns='weekly_price',inplace=True)",
"_____no_output_____"
],
[
"# Transfer String to float\ndollar_cols = ['cleaning_fee', 'extra_people', 'price']\nlisting_cleaned[dollar_cols]=listing_cleaned[dollar_cols].replace('[\\$,]', '', regex=True).astype(float)\npercent_cols = ['host_response_rate']\nlisting_cleaned[percent_cols]=listing_cleaned[percent_cols].replace('%', '', regex=True).astype(float)\nlisting_cleaned[percent_cols]=listing_cleaned[percent_cols]/100",
"_____no_output_____"
],
[
"# to check whether string already transfer to float\nlisting_cleaned.cleaning_fee.head()",
"_____no_output_____"
],
[
"# to check whether string already transfer to float\nlisting_cleaned.host_response_rate.head()",
"_____no_output_____"
]
],
[
[
"There are many missing values in review related features because these are new listings and don't get reviewed yet. In reality, travellers usually prefer listings with high number of reviews and high review scores. It might be possible to use a form of clustering to make a better estimate for this feature by looking at the other review scores for the same listing in the cases where these aren't missing. Considering this, I will replace missing values in review related features with mean value of each column.",
"_____no_output_____"
]
],
[
[
"# fill up missing value with mean of their column\nfor x in missing_cols:\n listing_cleaned[x] = listing_cleaned[x].fillna(listing_cleaned[x].mean())",
"_____no_output_____"
],
[
"# check whether the missing value is appear\nlisting_cleaned.isnull().mean().sort_values() ",
"_____no_output_____"
]
],
[
[
"### 3. Modelling",
"_____no_output_____"
],
[
"Base on the Q1 of business understanding, I will check which feature influnce the rating of the house. \nThus, 'review_scores_rating' would be the target(dependent) variable. ",
"_____no_output_____"
]
],
[
[
"# split the data into different column, prepare to create a model\ny = listing_cleaned['review_scores_rating']\nX = listing_cleaned.drop(['review_scores_rating'], axis = 1)",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.tree import DecisionTreeRegressor ### I use Decision Tree as there are a lot of features\nfrom sklearn.grid_search import GridSearchCV",
"_____no_output_____"
],
[
"# finding the best parameters\ndt = DecisionTreeRegressor(random_state=42)\nparameters = { 'max_depth': [20, 30, 40],\n 'min_samples_split': [2, 3],\n 'min_samples_leaf': [1, 2]}\ngridCV = GridSearchCV(estimator = dt, param_grid = parameters, cv = 5)",
"_____no_output_____"
],
[
"# training model with dataset\ngridCV.fit(X,y)",
"_____no_output_____"
],
[
"# to find out what is the best features\ngridCV.best_params_",
"_____no_output_____"
],
[
"# list the important features\nimportance_t = pd.DataFrame(data=[X.columns,gridCV.best_estimator_.feature_importances_]).T\nimportance_t.columns = ['feature','importance']\nimportance_t.sort_values(by='importance',ascending=False)[:5]",
"_____no_output_____"
]
],
[
[
"Apparently, many reiviews that has a extermely imporant coefficients with the rating! More reviews which means the house always be the best choice, and people would follow this trend when they are choosing. \nBesides, we can see that price is an important factor for rating (I think so, price always play a key role, which not a surprise). \nFor most of the properties, cleaning fee is included in the price, so the higher the cleaning fee, the higher the price. Most of people select house from AirBnB as they need a convenient and cleaning house. Therefore, it makes sense both the price and cleaning fee are on top 5 list. \n365 day-availability and host is superhost are the key role in rating, which Straightly reflect the conception of consume of current customer. They hope that they could rent the house at anytime if they need.",
"_____no_output_____"
],
[
"### Q2. When is the most popular time for this area?",
"_____no_output_____"
],
[
"#### 4.1 Loading data",
"_____no_output_____"
],
[
"We are mainly looking at the calendar dataset. As seen during the Data Exploration stage, the dataset contains the listings, dates, availability and the price.",
"_____no_output_____"
]
],
[
[
"# loading data\ncalendar = pd.read_csv('calendar.csv')\ncalendar.head()",
"_____no_output_____"
]
],
[
[
"#### 4.2 Pre-processing data",
"_____no_output_____"
]
],
[
[
"# a lot of missing value in 'Price', but we are not interest it. :)\ncalendar.isnull().sum() ",
"_____no_output_____"
],
[
"# drop the column that I dont use\nalendar.drop(columns = 'price').head(5)",
"_____no_output_____"
],
[
"# to find out when is busy days\npopular_time=calendar[calendar['available']=='f']",
"_____no_output_____"
],
[
"#group dataset by date\npopular_time=popular_time.groupby('date')['listing_id'].count()",
"_____no_output_____"
],
[
"# visualize Data\ndf=pd.DataFrame({'date': popular_time.index, 'count': popular_time.values })\ndf\n# plot trend of unavailable listings\nplt.figure(figsize=(30,20))\nplt.plot(df['count'], 'o-')\nplt.title('Nonavailable days')\nplt.show()",
"_____no_output_____"
]
],
[
[
"It could no be clearly see from this graph, we still need to sort it",
"_____no_output_____"
]
],
[
[
"# to list the data that what I need\npopular_time.sort_values(ascending=False).head(50)",
"_____no_output_____"
]
],
[
[
"As we can see, January and July are the popular time for this area. Then, combining the results of question 1 and question 2, whatever is a host or a customer, they could make a strategy that how to improve the rating or attract the customer.",
"_____no_output_____"
],
[
"### Q3.Could we create a model to predict the price?",
"_____no_output_____"
],
[
"Actually same with Q1, I would like to use DecisionTreeModel to train the dataset. The parameters will be used, which tested at Q1.",
"_____no_output_____"
]
],
[
[
"# We use the data after cleaning\ny_st = listing_cleaned.price\nX_st = listing_cleaned.drop(['price'], axis = 1)\n\nX_train_st, X_test_st, y_train_st, y_test_st = train_test_split(X_st, y_st, test_size = .20, random_state=42)\n\n# Check the datasets\nprint(X_train_st.shape, y_train_st.shape)\nprint( X_test_st.shape, y_test_st.shape)",
"(3054, 223) (3054,)\n(764, 223) (764,)\n"
],
[
"# import the libary\nfrom sklearn.metrics import r2_score",
"_____no_output_____"
],
[
"# initial model and train model\nregressor = DecisionTreeRegressor(max_depth=30, min_samples_leaf=2, min_samples_split= 2, random_state=42)\nregressor = regressor.fit(X_train_st, y_train_st)\npred = regressor.predict(X_test_st)",
"_____no_output_____"
],
[
"# Check how good the model is by calculating r-squared on the test set\nprint(\"The r-squared for the model is:\", r2_score(y_test_st, pred))",
"The r-squared for the model is: 0.412996290368991\n"
]
],
[
[
"It shows above, there is a model that could be created, which is decision tree. The reason is features is not too many or less, and avoid overfitting. We could see, the R2 score is 0.41, which means 41% of the variance in Y can be predicted from X. Futhermore, the dataset just normal processing and the model does not train quite good, if spending more time to adjust the model, I think the score will be increase.",
"_____no_output_____"
],
[
"### 5. Conclusion",
"_____no_output_____"
],
[
"In this project, I have analized the dataset about renting house from the Kaggle, which provided by AirBnb. The main motivation is to find out more insights for Airbnb business owners and customer in Seattle. \n\nThrough the analyst, we can see, a rating for a house is quite important. To improve the rating, number of reviews, cleaning fee, price, available days and house's owner is the Top 5 feature which has a big influence for attacting customer. The popular time for the customer is January and July.\n\nFuthuremore, we could use scientice tool, such as data analyst to mining more valuable information",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0f5a1815d92d611de808a65d3394bcb6b8432b1 | 23,622 | ipynb | Jupyter Notebook | Notebooks/Preview_Data_Visualization.ipynb | BrunoGeraldine/Python_Zero_To_DS | d487fcf16d0c35fca4bf124d29beca9e275f51e2 | [
"MIT"
] | null | null | null | Notebooks/Preview_Data_Visualization.ipynb | BrunoGeraldine/Python_Zero_To_DS | d487fcf16d0c35fca4bf124d29beca9e275f51e2 | [
"MIT"
] | null | null | null | Notebooks/Preview_Data_Visualization.ipynb | BrunoGeraldine/Python_Zero_To_DS | d487fcf16d0c35fca4bf124d29beca9e275f51e2 | [
"MIT"
] | null | null | null | 26.631342 | 128 | 0.46262 | [
[
[
"## Import LIbraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"## Loading Data",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv( 'datasets/kc_house_data.csv' )",
"_____no_output_____"
],
[
"# Ploting data\ndata.head()",
"_____no_output_____"
]
],
[
[
"# Business questions:",
"_____no_output_____"
],
[
"## 1. How many homes are available for purchase?",
"_____no_output_____"
]
],
[
[
"# Counting number of lines the same number of houses\n# len(data['id'].unique())\n# Or using .drop function (remove od duplicados e conta novamente)\n# Estrategy:\n# 1. Select \"id\" column;\n# 2. Remov repeated value;\n# 3. Count the number of unique values.\nhouses = len(data['id'].drop_duplicates())\nprint('The number of avaible houses are {}'.format(houses))",
"The number of avaible houses are 21436\n"
]
],
[
[
"## 2. How many attributes do houses have?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Count the number os columns;\n# 2. Remove values \"id\", \"date\" (they are not attributes):\nattributes = len(data.drop(['id', 'date'], axis = 1).columns)\nprint('The houses have {} attributes'.format(attributes))",
"The houses have 19 attributes\n"
]
],
[
[
"## 3. What are the attributes of houses?",
"_____no_output_____"
]
],
[
[
"## Estrategy:\n# 1. Plot the columns.\nattributes2 = data.drop(['id', 'date'], axis=1).columns\nprint('The attributes are: {}'.format(attributes2))",
"The attributes are: Index(['price', 'bedrooms', 'bathrooms', 'sqft_living', 'sqft_lot', 'floors',\n 'waterfront', 'view', 'condition', 'grade', 'sqft_above',\n 'sqft_basement', 'yr_built', 'yr_renovated', 'zipcode', 'lat', 'long',\n 'sqft_living15', 'sqft_lot15'],\n dtype='object')\n"
]
],
[
[
"## 4. What is the most expensive house (house with the highest sale price)?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Arrange the price column from highest to lowest \n# 2. Apply reset index to recount rows and ensure correct result;\n# 3. Collect the value and id from the first row.\n\ndata[['id','price']].sort_values('price', ascending=False).head() # 1\nexphouse = data[['id','price']].sort_values('price', ascending=False).reset_index(drop=True)['id'][0] # 2 and 3\nhighestprice = data[['id','price']].sort_values('price', ascending=False).reset_index(drop=True)['price'][0]\nprint('The most expensive House is: id {} price U${}'.format(exphouse, highestprice))",
"The most expensive House is: id 6762700020 price U$7700000.0\n"
]
],
[
[
"## 5. Which house has the most bedrooms?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Arrange the bedroom column from highest to lowest;\n# 2. Apply reset index to recount rows and ensure correct result;\n# 3. Collect the value and id from the first row.\ndata[[\"id\",\"bedrooms\"]].sort_values(\"bedrooms\", ascending=False).head() # 1\nbedroomsid = data[['id','bedrooms']].sort_values('bedrooms', ascending=False).reset_index(drop=True)['id'][0] # 2 and 3\nbedrooms1 = data[['id','bedrooms']].sort_values('bedrooms', ascending=False).reset_index(drop=True)['bedrooms'][0]\nprint('The house with the most bedrooms is {} and have {} bedroomns'.format(bedroomsid, bedrooms1))",
"The house with the most bedrooms is 2402100895 and have 33 bedroomns\n"
]
],
[
[
"# 6. What is the sum total of bedrooms in the dataset?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Filter the columns \"bedrooms\" and sum this value.\nbedroomsnumber = data['bedrooms'].sum()\nprint('There are a total of {} bedrooms'.format(bedroomsnumber))",
"There are a total of 72854 bedrooms\n"
]
],
[
[
"# 7. How many houses have 2 bathrooms?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Find the number of houses whit 2 bathrooms; \n# 2. Select columns \"id\" and \"bathroom\";\n# 3. Sum the number of houses.\ndata.loc[data['bathrooms'] == 2, ['id', 'bathrooms']] # 1\nbethroomsum = data.loc[data['bathrooms'] == 2, ['id', 'bathrooms']].shape # 2 and 3\nprint('There are a total of {} bethrooms'.format(bethroomsum))",
"There are a total of (1930, 23) bethrooms\n"
]
],
[
[
"# 8. What is the average price of all houses in the dataset?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Find the average price of houses.\n# Ps.: Use numpy's round() function to select only 2 number after point.\naverageprice = np.round(data['price'].mean(), 2)\nprint('The average price of the houses is: U${}'.format(averageprice ))",
"The average price of the houses is: U$540088.14\n"
],
[
"# the function data.dtypes show us what types variables we have.\ndata.dtypes",
"_____no_output_____"
]
],
[
[
"# 9. What is the average price only houses whit 2 bathroom?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Find only the average price of houses whit 2 bathroms.\navg_bath = np.round(data.loc[data['bathrooms'] == 2, 'price'].mean(), 2)\nprint('The average price for houses whit 2 bathrooms is U${}'.format(avg_bath))",
"The average price for houses whit 2 bathrooms is U$457889.72\n"
]
],
[
[
"# 10. What is the minimum price between 3 bedroom homes?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Select only 3 bedroom house and arrange ascending by price\nmin_price_bed = data.loc[data['bedrooms'] == 3, 'price'].min(),\nprint('The minimum price for houses whit 3 bedrooms is U${}'.format(min_pricebed))",
"The minimum price for houses whit 3 bedrooms is U$82000.0\n"
]
],
[
[
"# 11. How many homes have more than 300 square meters in the living room?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Select only houses whit mor than 300ft² of living room and read the number of lines.\ndata['m2'] = data['sqft_living'] * 0.093\nsqft_300 = len(data.loc[data['m2'] > 300, 'id'])\nprint('There are {} houses whit mor than 300ft² in the living room.'.format(sqft_300))",
"There are 2258 houses whit mor than 300ft² in the living room.\n"
]
],
[
[
"#### Se quiser fazer a converção de ft² para m² basta usar o seguinte raciocionio: (1 ft² = 0.093 m²)\n#### data['m²']=data['sqft_living'] * 0.093 - (aqui substituimos a variavel sqft_living pela m² ja convertendo o valor)\n#### len(data.loc[data['m²'] > 300, 'id'])",
"_____no_output_____"
],
[
"# 12. How many homes have more than 2 floors?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Select only houses whit mor than 300ft² of living room and read the number of lines.\nfloor_2 = data.loc[data['floors'] > 2, 'id'].size\nprint('There are {} houses whit mor than 2 floors.'.format(floor_2))",
"There are 782 houses whit mor than 2 floors.\n"
]
],
[
[
"# 13. How many houses have a waterfront view?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Select only houses whit waterfront view and read the number of lines.\nwaterfront_view = len(data.loc[data['waterfront'] != 0, 'id'])\nprint('There are {} houses whit waterfront view.'.format(waterfront_view))",
"There are 163 houses whit waterfront view.\n"
]
],
[
[
"# 14. Of the houses with a waterfront view, how many have 3 bedrooms?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Select only houses whit waterfront view and read how many have 3 bedrooms.\ndata.columns\nwaterfront_bed = data.loc[(data['waterfront'] != 0) & (data['bedrooms'] == 3), \"id\"].size\nprint('Of the houses whit waterfront, {} houses have 3 bedrooms.'.format(waterfront_bed))",
"Of the houses whit waterfront, 64 houses have 3 bedrooms.\n"
]
],
[
[
"# 15. Of the houses with more than 300 square meters of living room, how many have more than 2 bathrooms?",
"_____no_output_____"
]
],
[
[
"# Estrategy:\n# 1. Select only houses whit mor than 300m² of livingo room and mor than 2 bedrooms.\nhouse_300m_2bat = data[(data['m2']>300) & (data['bathrooms']>2)].shape[0]\nprint('Of the houses whit 300 square meters of living room, {} houses have 2 bathrooms.'.format(house_300m_2bat))",
"Of the houses whit 300 square meters of living room, 2201 houses have 2 bathrooms.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f5a574e5f82563f6ce5949e04664a334544ea2 | 118,888 | ipynb | Jupyter Notebook | src/notebooks/199-matplotlib-style-sheets.ipynb | s-lasch/The-Python-Graph-Gallery | 1df060780e5e9cf763815581aad15da20f5a4213 | [
"0BSD"
] | 1 | 2022-01-28T09:36:36.000Z | 2022-01-28T09:36:36.000Z | src/notebooks/199-matplotlib-style-sheets.ipynb | preguza/The-Python-Graph-Gallery | 4645ec59eaa6b8c8e2ff4eee86516ee3a7933b4d | [
"0BSD"
] | null | null | null | src/notebooks/199-matplotlib-style-sheets.ipynb | preguza/The-Python-Graph-Gallery | 4645ec59eaa6b8c8e2ff4eee86516ee3a7933b4d | [
"0BSD"
] | null | null | null | 550.407407 | 38,552 | 0.721309 | [
[
[
"## List of themes",
"_____no_output_____"
],
[
"The list of available `matplotlib` themes is stored in a list called `plt.style.available`. There are 26 of them.\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nplt.style.available",
"_____no_output_____"
]
],
[
[
"## Scatterplot",
"_____no_output_____"
],
[
"The [scatterplot section](https://python-graph-gallery.com/scatter-plot/) of the gallery explains in depth how to build a basic scatterplot with `matplotlib`. It is pretty straightforward thanks to the `plot()` function.",
"_____no_output_____"
]
],
[
[
"# Create a dataset:\nimport numpy as np\nimport pandas as pd\ndf=pd.DataFrame({'x': range(1,101), 'y': np.random.randn(100)*15+range(1,101) })\n \n# plot\nplt.plot( 'x', 'y', data=df, linestyle='none', marker='o')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"## Apply a theme",
"_____no_output_____"
],
[
"Now, let's make this chart a bit prettier thanks to the style called `fivethirtyheight`. In case you don't know it already, [FiveThirtyHeight](https://fivethirtyeight.com) is an online newspaper that often displays some very nice dataviz articles.",
"_____no_output_____"
]
],
[
[
"plt.style.use('fivethirtyeight')\nplt.plot( 'x', 'y', data=df, linestyle='none', marker='o')\nplt.title('Scatterplot with the five38 theme', fontsize=12)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Apply the style on a barchart",
"_____no_output_____"
],
[
"You can apply the same exact tip for any kind of chart to make it look better. Here is a barchart example coming from the barchart section of the gallery. It uses the `dark_background` theme to demo another type of customization.",
"_____no_output_____"
]
],
[
[
"# create dataset\nheight = [3, 12, 5, 18, 45]\nbars = ('A', 'B', 'C', 'D', 'E')\ny_pos = np.arange(len(bars))\n \n# Create horizontal bars\nplt.barh(y_pos, height)\n \n# Create names on the x-axis\nplt.yticks(y_pos, bars)\n \n# Show graphic\nplt.style.use('dark_background')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d0f5a9c4ca0bbcc7bf2cfc8b95bf5a9f97ea75d9 | 45,541 | ipynb | Jupyter Notebook | various/searchFromMQL.ipynb | dirkroorda/text-fabric-data | 877ce9b7bc3dc31ea5b04076e9675725b46ba383 | [
"MIT"
] | 4 | 2017-02-04T21:53:42.000Z | 2017-09-12T21:53:56.000Z | various/searchFromMQL.ipynb | Dans-labs/text-fabric-data | 877ce9b7bc3dc31ea5b04076e9675725b46ba383 | [
"MIT"
] | 4 | 2017-03-21T17:48:36.000Z | 2017-06-19T14:10:58.000Z | various/searchFromMQL.ipynb | Dans-labs/text-fabric-data | 877ce9b7bc3dc31ea5b04076e9675725b46ba383 | [
"MIT"
] | 5 | 2017-01-04T21:12:57.000Z | 2017-07-17T10:49:42.000Z | 35.859055 | 184 | 0.501197 | [
[
[
"<img align=\"right\" src=\"tf-small.png\"/>\n\n# Search from MQL\n\nThese are examples of\n[MQL](https://shebanq.ancient-data.org/static/docs/MQL-Query-Guide.pdf)\nqueries on\n[SHEBANQ](https://shebanq.ancient-data.org/hebrew/queries), \nnow expressed\nas Text-Fabric search templates.\n\nFor more basic examples, see\n[searchTutorial](https://github.com/etcbc/text-fabric/blob/master/docs/searchTutorial.ipynb).\n\n*Search* in Text-Fabric is a template based way of looking for structural patterns in your dataset.",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"from tf.fabric import Fabric",
"_____no_output_____"
],
[
"ETCBC = 'hebrew/etcbc4c'\nTF = Fabric( modules=ETCBC )",
"This is Text-Fabric 2.2.0\nApi reference : https://github.com/ETCBC/text-fabric/wiki/Api\nTutorial : https://github.com/ETCBC/text-fabric/blob/master/docs/tutorial.ipynb\nData sources : https://github.com/ETCBC/text-fabric-data\nData docs : https://etcbc.github.io/text-fabric-data\nShebanq docs : https://shebanq.ancient-data.org/text\nSlack team : https://shebanq.slack.com/signup\nQuestions? Ask [email protected] for an invite to Slack\n107 features found and 0 ignored\n"
],
[
"api = TF.load('''\n rela function pdp\n''')\napi.makeAvailableIn(globals())",
" 0.00s loading features ...\n | 0.21s B rela from /Users/dirk/github/text-fabric-data/hebrew/etcbc4c\n | 0.08s B function from /Users/dirk/github/text-fabric-data/hebrew/etcbc4c\n | 0.13s B pdp from /Users/dirk/github/text-fabric-data/hebrew/etcbc4c\n | 0.00s Feature overview: 102 nodes; 4 edges; 1 configs; 7 computeds\n 5.26s All features loaded/computed - for details use loadLog()\n"
]
],
[
[
"# By Oliver Glanz\n\n[Oliver Glanz: PP with adjective followed by noun](https://shebanq.ancient-data.org/hebrew/query?version=4b&id=547)\n```\nselect all objects where\n[phrase FOCUS typ = PP\n [word sp= prep]\n [word sp=adjv]\n [word sp=subs]\n]\n```\n64 results having 251 words.",
"_____no_output_____"
]
],
[
[
"query = '''\nphrase typ=PP\n word sp=prep\n <: word sp=adjv\n <: word sp=subs\n'''\nS.study(query)\nS.showPlan()\nS.count(progress=1000, limit=-1)\nfor r in S.fetch(amount=10):\n print(S.glean(r))",
" 0.00s Checking search template ...\n 0.00s loading features ...\n | 0.20s B sp from /Users/dirk/github/text-fabric-data/hebrew/etcbc4c\n | 0.22s B typ from /Users/dirk/github/text-fabric-data/hebrew/etcbc4c\n 0.43s All additional features loaded - for details use loadLog()\n 0.43s Setting up search space for 4 objects ...\n 2.56s Constraining search space with 5 relations ...\n 2.64s Setting up retrieval plan ...\n 2.64s Ready to deliver results from 256 nodes\nIterate over S.fetch() to get the results\nSee S.showPlan() to interpret the results\n 2.65s The results are connected to the original search template as follows:\n 0 \n 1 R0 phrase typ=PP\n 2 R1 word sp=prep\n 3 R2 <: word sp=adjv\n 4 R3 <: word sp=subs\n 5 \n 0.00s Counting results per 1000 up to the end of the results ...\n 0.00s Done: 64 results\nphrase[אֵ֖ת רַבַּ֣ת בְּנֵֽי־עַמֹּ֑ון וְ...] אֵ֖ת רַבַּ֣ת בְּנֵֽי־\nphrase[לָכֶ֜ם יִרְאֵ֤י שְׁמִי֙ ] לָכֶ֜ם יִרְאֵ֤י שְׁמִי֙ \nphrase[עַ֥ל רִגְעֵי־אֶ֑רֶץ ] עַ֥ל רִגְעֵי־ אֶ֑רֶץ \nphrase[אֶת־חַלְלֵי־חָ֑רֶב ] אֶת־ חַלְלֵי־ חָ֑רֶב \nphrase[לַחֲכַם־לֵ֭ב ] לַ חֲכַם־ לֵ֭ב \nphrase[לְמָ֣רֵי נָֽפֶשׁ׃ ] לְ מָ֣רֵי נָֽפֶשׁ׃ \nphrase[לְיִשְׁרֵי־לֵֽב׃ ] לְ יִשְׁרֵי־ לֵֽב׃ \nphrase[עִם־יְפֵ֥ה עֵינַ֖יִם וְטֹ֣וב ...] עִם־ יְפֵ֥ה עֵינַ֖יִם \nphrase[עִם־מְלֵ֥א יָמִֽים׃ ] עִם־ מְלֵ֥א יָמִֽים׃ \nphrase[אֶת־חַלְלֵי־חָֽרֶב׃ ] אֶת־ חַלְלֵי־ חָֽרֶב׃ \n"
]
],
[
[
"The number of results is right. The number of words that SHEBANQ reports\nis the number of words in the phrases of the result. Let us count them:",
"_____no_output_____"
]
],
[
[
"print(sum([len(L.d(r[0], otype='word')) for r in S.fetch()]))",
"251\n"
]
],
[
[
"# By Martijn Naaijer\n\n[Martijn Naaijer: Object clauses with >CR](https://shebanq.ancient-data.org/hebrew/query?version=4b&id=997)\n\n```\nSelect all objects where \n\n[clause rela = Objc\n [word focus first lex = '>CR']\n]\n```\n\n157 results",
"_____no_output_____"
]
],
[
[
"query = '''\nverse\n clause rela=Objc\n =: word lex=>CR\n'''\nS.study(query)\nS.showPlan()\nS.count(progress=1000, limit=-1)\nfor r in sorted(S.fetch(), key=lambda x: C.rank.data[x[0]-1])[0:10]:\n print(S.glean(r))",
" 0.00s Checking search template ...\n 0.00s loading features ...\n | 0.19s B lex from /Users/dirk/github/text-fabric-data/hebrew/etcbc4c\n 0.20s All additional features loaded - for details use loadLog()\n 0.20s Setting up search space for 3 objects ...\n 0.94s Constraining search space with 3 relations ...\n 0.96s Setting up retrieval plan ...\n 0.96s Ready to deliver results from 284 nodes\nIterate over S.fetch() to get the results\nSee S.showPlan() to interpret the results\n 0.96s The results are connected to the original search template as follows:\n 0 \n 1 R0 verse\n 2 R1 clause rela=Objc\n 3 R2 =: word lex=>CR\n 4 \n 0.00s Counting results per 1000 up to the end of the results ...\n 0.00s Done: 96 results\nGenesis 14:24 clause[אֲשֶׁ֣ר אָֽכְל֣וּ הַנְּעָרִ֔ים ] אֲשֶׁ֣ר \nGenesis 18:17 clause[אֲשֶׁ֖ר אֲנִ֥י עֹשֶֽׂה׃ ] אֲשֶׁ֖ר \nGenesis 24:3 clause[אֲשֶׁ֨ר לֹֽא־תִקַּ֤ח אִשָּׁה֙ לִ...] אֲשֶׁ֨ר \nGenesis 34:11 clause[אֲשֶׁ֥ר תֹּאמְר֛וּ אֵלַ֖י ] אֲשֶׁ֥ר \nGenesis 39:23 clause[אֲשֶׁר־ה֥וּא עֹשֶׂ֖ה ] אֲשֶׁר־\nGenesis 41:28 clause[אֲשֶׁ֧ר הָאֱלֹהִ֛ים עֹשֶׂ֖ה ] אֲשֶׁ֧ר \nGenesis 41:55 clause[אֲשֶׁר־יֹאמַ֥ר לָכֶ֖ם ] אֲשֶׁר־\nGenesis 44:5 clause[אֲשֶׁ֥ר עֲשִׂיתֶֽם׃ ] אֲשֶׁ֥ר \nExodus 4:12 clause[אֲשֶׁ֥ר תְּדַבֵּֽר׃ ] אֲשֶׁ֥ר \nExodus 5:21 clause[אֲשֶׁ֧ר הִבְאַשְׁתֶּ֣ם אֶת־רֵיחֵ֗נוּ בְּ...] אֲשֶׁ֧ר \n"
]
],
[
[
"We have fewer cases: 96 instead of 157.\nWe are working on the ETCBC version 4c, and the query has been executed against 4b.\nThere have been coding updates that are relevant to this query, e.g. in Genesis 43:27, which is in the results\non SHEBANQ, but not here. In 4c the `rela` is `Attr`, and not `Objc`.",
"_____no_output_____"
]
],
[
[
"query = '''\nverse book=Genesis chapter=43 verse=27\n clause\n =: word lex=>CR\n'''\nS.study(query)\nS.showPlan()\nS.count(progress=1000, limit=-1)\nresults = sorted(S.fetch(), key=lambda x: C.rank.data[x[0]-1])\nfor r in results:\n print(r[1], F.rela.v(r[1]), S.glean(r))",
" 0.00s Checking search template ...\n 0.00s loading features ...\n 0.00s All additional features loaded - for details use loadLog()\n 0.01s Setting up search space for 3 objects ...\n 0.75s Constraining search space with 3 relations ...\n 0.84s Setting up retrieval plan ...\n 0.84s Ready to deliver results from 3 nodes\nIterate over S.fetch() to get the results\nSee S.showPlan() to interpret the results\n 0.85s The results are connected to the original search template as follows:\n 0 \n 1 R0 verse book=Genesis chapter=43 verse=27\n 2 R1 clause\n 3 R2 =: word lex=>CR\n 4 \n 0.00s Counting results per 1000 up to the end of the results ...\n 0.00s Done: 1 results\n431688 Attr Genesis 43:27 clause[אֲשֶׁ֣ר אֲמַרְתֶּ֑ם ] אֲשֶׁ֣ר \n"
]
],
[
[
"# By Cody Kingham\n\n[Cody Kingham: MI Hierarchies. p.18n49. First Person Verbs in Narrative](https://shebanq.ancient-data.org/hebrew/query?version=4b&id=1050)\n\n```\nSELECT ALL OBJECTS WHERE\n\n[book\n [clause txt = 'N'\n [word FOCUS sp = verb\n [word ps = p1\n ]\n ]\n ]\n]\nOR\n[book\n [clause txt = '?N'\n [word FOCUS sp = verb\n [word ps = p1\n ]\n ]\n ]\n]\n```\n\n273 results.",
"_____no_output_____"
]
],
[
[
"query = '''\nbook\n clause txt=N|?N\n word sp=verb ps=p1\n'''\nS.study(query)\nS.showPlan()\nS.count(progress=1000, limit=-1)\nfor r in sorted(S.fetch(), key=lambda x: C.rank.data[x[0]-1])[0:10]:\n print(S.glean(r))",
" 0.00s Checking search template ...\n 0.00s loading features ...\n | 0.21s B ps from /Users/dirk/github/text-fabric-data/hebrew/etcbc4c\n | 0.04s B txt from /Users/dirk/github/text-fabric-data/hebrew/etcbc4c\n 0.26s All additional features loaded - for details use loadLog()\n 0.26s Setting up search space for 3 objects ...\n 0.99s Constraining search space with 2 relations ...\n 1.01s Setting up retrieval plan ...\n 1.04s Ready to deliver results from 557 nodes\nIterate over S.fetch() to get the results\nSee S.showPlan() to interpret the results\n 1.04s The results are connected to the original search template as follows:\n 0 \n 1 R0 book\n 2 R1 clause txt=N|?N\n 3 R2 word sp=verb ps=p1\n 4 \n 0.00s Counting results per 1000 up to the end of the results ...\n 0.05s Done: 273 results\n clause[וְאֶת־יְהֹושׁ֣וּעַ צִוֵּ֔יתִי בָּ...] צִוֵּ֔יתִי \n clause[וָאֶתְחַנַּ֖ן אֶל־יְהוָ֑ה בָּ...] אֶתְחַנַּ֖ן \n clause[וָאֶשְׁלַ֤ח מַלְאָכִים֙ מִמִּדְבַּ֣ר ...] אֶשְׁלַ֤ח \n clause[וַנֵּ֣שֶׁב בַּגָּ֔יְא ...] נֵּ֣שֶׁב \n clause[וַנֵּ֜פֶן ] נֵּ֜פֶן \n clause[וַנַּ֥ךְ אֹתֹ֛ו וְאֶת־...] נַּ֥ךְ \n clause[וַנִּלְכֹּ֤ד אֶת־כָּל־עָרָיו֙ ...] נִּלְכֹּ֤ד \n clause[וַֽנַּחֲרֵם֙ אֶת־כָּל־עִ֣יר ...] נַּחֲרֵם֙ \n clause[וַנָּ֥סָב אֶת־הַר־שֵׂעִ֖יר ...] נָּ֥סָב \n clause[לֹ֥א הִשְׁאַ֖רְנוּ שָׂרִֽיד׃ ] הִשְׁאַ֖רְנוּ \n"
]
],
[
[
"# By Reinoud Oosting\n\n[Reinoud Oosting: to go + object marker](https://shebanq.ancient-data.org/hebrew/query?version=4b&id=755)\n\n```\nSelect all objects\nwhere\n [clause\n [phrase function = Pred OR function = PreC\n [word FOCUS sp = verb AND vs = qal AND lex = \"HLK[\" ]\n ]\n ..\n [phrase FOCUS\n [word First lex = \">T\"]\n ]\n ]\nOR\n [clause\n [phrase FOCUS\n [word First lex = \">T\" ]\n ]\n..\n [phrase function = Pred OR function = PreC\n [word FOCUS sp = verb AND vs = qal AND lex = \"HLK[\"]\n ]\n ]\n ```\n \n 4 results.\n \n This is a case where we can simplify greatly because we are not hampered\n by automatic constraints on the order of the phrases.",
"_____no_output_____"
]
],
[
[
"query = '''\nclause\n p1:phrase function=Pred|PreC\n word sp=verb vs=qal lex=HLK[\n p2:phrase\n =: word lex=>T\n p1 # p2\n'''\n\nS.study(query)\nS.showPlan()\nS.count(progress=1000, limit=-1)\nfor r in sorted(S.fetch(), key=lambda x: C.rank.data[x[0]-1])[0:10]:\n print(S.glean(r))",
" 0.00s Checking search template ...\n 0.00s loading features ...\n | 0.21s B vs from /Users/dirk/github/text-fabric-data/hebrew/etcbc4c\n 0.21s All additional features loaded - for details use loadLog()\n 0.21s Setting up search space for 5 objects ...\n 1.84s Constraining search space with 6 relations ...\n 1.85s Setting up retrieval plan ...\n 1.89s Ready to deliver results from 430023 nodes\nIterate over S.fetch() to get the results\nSee S.showPlan() to interpret the results\n 1.89s The results are connected to the original search template as follows:\n 0 \n 1 R0 clause\n 2 R1 p1:phrase function=Pred|PreC\n 3 R2 word sp=verb vs=qal lex=HLK[\n 4 R3 p2:phrase\n 5 R4 =: word lex=>T\n 6 p1 # p2\n 7 \n 0.00s Counting results per 1000 up to the end of the results ...\n 0.02s Done: 4 results\nclause[וַנֵּ֡לֶךְ אֵ֣ת כָּל־הַ...] phrase[נֵּ֡לֶךְ ] נֵּ֡לֶךְ phrase[אֵ֣ת כָּל־הַמִּדְבָּ֣ר הַ...] אֵ֣ת \nclause[וְאֶת־בֵּ֤ית יְהוָה֙ אֲנִ֣י ...] phrase[הֹלֵ֔ךְ ] הֹלֵ֔ךְ phrase[אֶת־בֵּ֤ית יְהוָה֙ ] אֶת־\nclause[הַהֹלְכִ֖ים אֹותָֽךְ׃ ] phrase[הֹלְכִ֖ים ] הֹלְכִ֖ים phrase[אֹותָֽךְ׃ ] אֹותָֽךְ׃ \nclause[אֵ֤ת אֲשֶׁר־בְּחֻקַּי֙ תֵּלֵ֔כוּ ] phrase[תֵּלֵ֔כוּ ] תֵּלֵ֔כוּ phrase[אֵ֤ת אֲשֶׁר־] אֵ֤ת \n"
]
],
[
[
"# By Reinoud Oosting (ii)\n\n[Reinoud Oosting: To establish covenant](https://shebanq.ancient-data.org/hebrew/query?version=4b&id=1485)\n\n```\nselect all objects\nwhere\n\n [clause\n [phrase function = Pred OR function = PreC\n [word FOCUS sp = verb AND vs = hif AND lex = \"QWM[\" ]\n ]\n ..\n [phrase function = Objc\n [word FOCUS lex = \"BRJT/\" ]\n ]\n ]\nOR\n [clause\n [phrase function = Objc\n [word FOCUS lex = \"BRJT/\" ]\n ]\n..\n [phrase function = Pred OR function = PreC\n [word FOCUS sp = verb AND vs = hif AND lex = \"QWM[\"]\n ]\n \n]\n```\n\n13 results",
"_____no_output_____"
]
],
[
[
"query = '''\nclause\n phrase function=Pred|PreC\n word sp=verb vs=hif lex=QWM[\n phrase function=Objc\n word lex=BRJT/\n'''\n\nS.study(query)\nS.showPlan()\nS.count(progress=1000, limit=-1)",
" 0.00s Checking search template ...\n 0.00s Setting up search space for 5 objects ...\n 1.94s Constraining search space with 4 relations ...\n 1.95s Setting up retrieval plan ...\n 1.96s Ready to deliver results from 65 nodes\nIterate over S.fetch() to get the results\nSee S.showPlan() to interpret the results\n 1.96s The results are connected to the original search template as follows:\n 0 \n 1 R0 clause\n 2 R1 phrase function=Pred|PreC\n 3 R2 word sp=verb vs=hif lex=QWM[\n 4 R3 phrase function=Objc\n 5 R4 word lex=BRJT/\n 6 \n 0.00s Counting results per 1000 up to the end of the results ...\n 0.00s Done: 13 results\n"
],
[
"resultsx = sorted((L.u(r[0], otype='verse')+r for r in S.fetch()), key=lambda r: sortKey(r[0]))\nfor r in resultsx:\n print(S.glean(r))",
"Genesis 6:18 clause[וַהֲקִמֹתִ֥י אֶת־בְּרִיתִ֖י אִתָּ֑ךְ ] phrase[הֲקִמֹתִ֥י ] הֲקִמֹתִ֥י phrase[אֶת־בְּרִיתִ֖י ] בְּרִיתִ֖י \nGenesis 9:9 clause[הִנְנִ֥י מֵקִ֛ים אֶת־בְּרִיתִ֖י אִתְּכֶ֑ם ...] phrase[מֵקִ֛ים ] מֵקִ֛ים phrase[אֶת־בְּרִיתִ֖י ] בְּרִיתִ֖י \nGenesis 9:11 clause[וַהֲקִמֹתִ֤י אֶת־בְּרִיתִי֙ אִתְּכֶ֔ם ] phrase[הֲקִמֹתִ֤י ] הֲקִמֹתִ֤י phrase[אֶת־בְּרִיתִי֙ ] בְּרִיתִי֙ \nGenesis 17:7 clause[וַהֲקִמֹתִ֨י אֶת־בְּרִיתִ֜י בֵּינִ֣י ...] phrase[הֲקִמֹתִ֨י ] הֲקִמֹתִ֨י phrase[אֶת־בְּרִיתִ֜י ] בְּרִיתִ֜י \nGenesis 17:19 clause[וַהֲקִמֹתִ֨י אֶת־בְּרִיתִ֥י אִתֹּ֛ו ...] phrase[הֲקִמֹתִ֨י ] הֲקִמֹתִ֨י phrase[אֶת־בְּרִיתִ֥י ] בְּרִיתִ֥י \nGenesis 17:21 clause[וְאֶת־בְּרִיתִ֖י אָקִ֣ים אֶת־...] phrase[אָקִ֣ים ] אָקִ֣ים phrase[אֶת־בְּרִיתִ֖י ] בְּרִיתִ֖י \nExodus 6:4 clause[וְגַ֨ם הֲקִמֹ֤תִי אֶת־בְּרִיתִי֙ ...] phrase[הֲקִמֹ֤תִי ] הֲקִמֹ֤תִי phrase[אֶת־בְּרִיתִי֙ ] בְּרִיתִי֙ \nLeviticus 26:9 clause[וַהֲקִימֹתִ֥י אֶת־בְּרִיתִ֖י אִתְּכֶֽם׃ ] phrase[הֲקִימֹתִ֥י ] הֲקִימֹתִ֥י phrase[אֶת־בְּרִיתִ֖י ] בְּרִיתִ֖י \nDeuteronomy 8:18 clause[לְמַ֨עַן הָקִ֧ים אֶת־בְּרִיתֹ֛ו ] phrase[לְמַ֨עַן הָקִ֧ים ] הָקִ֧ים phrase[אֶת־בְּרִיתֹ֛ו ] בְּרִיתֹ֛ו \n2_Kings 23:3 clause[לְהָקִ֗ים אֶת־דִּבְרֵי֙ הַ...] phrase[לְהָקִ֗ים ] הָקִ֗ים phrase[אֶת־דִּבְרֵי֙ הַבְּרִ֣ית הַ...] בְּרִ֣ית \nJeremiah 34:18 clause[אֲשֶׁ֤ר לֹֽא־הֵקִ֨ימוּ֙ אֶת־דִּבְרֵ֣י ...] phrase[הֵקִ֨ימוּ֙ ] הֵקִ֨ימוּ֙ phrase[אֶת־דִּבְרֵ֣י הַבְּרִ֔ית ] בְּרִ֔ית \nEzekiel 16:60 clause[וַהֲקִמֹותִ֥י לָ֖ךְ בְּרִ֥ית עֹולָֽם׃ ] phrase[הֲקִמֹותִ֥י ] הֲקִמֹותִ֥י phrase[בְּרִ֥ית עֹולָֽם׃ ] בְּרִ֥ית \nEzekiel 16:62 clause[וַהֲקִימֹותִ֥י אֲנִ֛י אֶת־בְּרִיתִ֖י ...] phrase[הֲקִימֹותִ֥י ] הֲקִימֹותִ֥י phrase[אֶת־בְּרִיתִ֖י ] בְּרִיתִ֖י \n"
]
],
[
[
"# By Reinoud Oosting (iii)\n\n[Reinoud Oosting: To find grace in sight of](https://shebanq.ancient-data.org/hebrew/query?version=4b&id=1484)\n\n```\nselect all objects\nwhere\n\n [clause\n [phrase FOCUS function = Pred OR function = PreC\n [word sp = verb AND vs = qal AND lex = \"MY>[\" ]\n ]\n ..\n [phrase function = Objc\n [word FOCUS lex = \"XN/\" ]\n ]\n[phrase function = Cmpl\n[word FOCUS lex = \"B\"]\n[word FOCUS lex = \"<JN/\"]\n]\n ]\nOR\n [clause\n [phrase function = Objc\n [word FOCUS lex = \"XN/\" ]\n ]\n[phrase function = Cmpl\n[word FOCUS lex = \"B\"]\n[word FOCUS lex = \"<JN/\"]\n..\n [phrase function = Pred OR function = PreC\n [word FOCUS sp = verb AND vs = qal AND lex = \"MY>[\"]\n ]\n ]\n]\n\n```\n\n38 results",
"_____no_output_____"
]
],
[
[
"query = '''\nclause\n p1:phrase function=Pred|PreC\n word sp=verb vs=qal lex=MY>[\n p2:phrase function=Objc\n word lex=XN/\n p3:phrase function=Cmpl\n word lex=B\n <: word lex=<JN/\n p2 << p3\n'''\n\nS.study(query)\nS.showPlan(details=True)\nS.count(progress=1000, limit=-1)",
" 0.00s Checking search template ...\n 0.00s Setting up search space for 8 objects ...\n 3.44s Constraining search space with 9 relations ...\n 3.45s Setting up retrieval plan ...\n 3.46s Ready to deliver results from 16034 nodes\nIterate over S.fetch() to get the results\nSee S.showPlan() to interpret the results\nSearch with 8 objects and 9 relations\nResults are instantiations of the following objects:\nnode 0-clause ( 38 choices)\nnode 1-phrase ( 38 choices)\nnode 2-word ( 38 choices)\nnode 3-phrase ( 38 choices)\nnode 4-word ( 38 choices)\nnode 5-phrase ( 38 choices)\nnode 6-word ( 15768 choices)\nnode 7-word ( 38 choices)\nInstantiations are computed along the following relations:\nnode 0-clause ( 38 choices)\nedge 0-clause [[ 5-phrase ( 1.0 choices)\nedge 5-phrase [[ 6-word ( 1.0 choices)\nedge 6-word <: 7-word ( 0.0 choices)\nedge 7-word ]] 5-phrase ( 1.0 choices)\nedge 0-clause [[ 3-phrase ( 1.0 choices)\nedge 3-phrase << 5-phrase ( 18.6 choices)\nedge 3-phrase [[ 4-word ( 1.0 choices)\nedge 0-clause [[ 1-phrase ( 1.0 choices)\nedge 1-phrase [[ 2-word ( 1.0 choices)\n 3.50s The results are connected to the original search template as follows:\n 0 \n 1 R0 clause\n 2 R1 p1:phrase function=Pred|PreC\n 3 R2 word sp=verb vs=qal lex=MY>[\n 4 R3 p2:phrase function=Objc\n 5 R4 word lex=XN/\n 6 R5 p3:phrase function=Cmpl\n 7 R6 word lex=B\n 8 R7 <: word lex=<JN/\n 9 p2 << p3\n10 \n 0.00s Counting results per 1000 up to the end of the results ...\n 0.00s Done: 38 results\n"
]
],
[
[
"# By Stephen Ku\n\n[Stephen Ku: Verbless Clauses](https://shebanq.ancient-data.org/hebrew/query?version=4&id=1314)\n\n```\nSELECT ALL OBJECTS WHERE \n\n[clause \n [phrase function IN (Subj) \n [phrase_atom NOT rela IN (Appo,Para,Spec)\n [word FOCUS pdp IN (subs,nmpr,prps,prde,prin,adjv)\n ]\n ]\n ]\n NOTEXIST [phrase function IN (Pred)]\n ..\n NOTEXIST [phrase function IN (Pred)]\n [phrase function IN (PreC)\n NOTEXIST [word pdp IN (prep)]\n [word FOCUS pdp IN (subs,nmpr,prin,adjv) AND ls IN (card,ordn)]\n ]\n]\n```\n\n1441 results with 1244 words in those results.\n\nWe do not have the `NOTEXIST` operator, and we cannot say `NOT rela IN`,\nso we are at a disadvantage here.\nLet's see what we can do.\nWe can use additional processing to furnish the template and weed out results.\n\nThe first thing is: we have to fetch all possible values of the `rela` feature,\nin order to see what other values than `Appo`, `Para`, `Spec` it can take.\n\nThe function `freqList()` gives us a frequency list of values, we only need the values\nother than the indicated ones, separated by a `|`.\n\nWe also need to consult the relation legend to pick the proper ordering between the\ntwo phrases.",
"_____no_output_____"
]
],
[
[
"excludedRela = {'Appo', 'Para', 'Spec'}\n'|'.join(x[0] for x in F.rela.freqList() if x[0] not in excludedRela)",
"_____no_output_____"
],
[
"print(S.relationLegend)",
" = left equal to right (as node)\n # left unequal to right (as node)\n < left before right (in canonical node ordering)\n > left after right (in canonical node ordering)\n == left occupies same slots as right\n && left has overlapping slots with right\n ## left and right do not have the same slot set\n || left and right do not have common slots\n [[ left embeds right\n ]] left embedded in right\n << left completely before right\n >> left completely after right\n =: left and right start at the same slot\n := left and right end at the same slot\n :: left and right start and end at the same slot\n <: left immediately before right\n :> left immediately after right\n =k: left and right start at k-nearly the same slot\n :k= left and right end at k-nearly the same slot\n :k: left and right start and end at k-near slots\n <k: left k-nearly before right\n :k> left k-nearly after right\n-distributional_parent> edge feature \"distributional_parent\"\n<distributional_parent- edge feature \"distributional_parent\" (opposite direction)\n -functional_parent> edge feature \"functional_parent\"\n <functional_parent- edge feature \"functional_parent\" (opposite direction)\n -mother> edge feature \"mother\"\n <mother- edge feature \"mother\" (opposite direction)\nThe grid feature \"oslots\" cannot be used in searches.\nSurely, one of the above relations on nodes and/or slots will suit you better!\n"
],
[
"query = '''\nclause\n p1:phrase function=Subj\n phrase_atom rela=NA|rec|par|Adju|Attr|adj|Coor|atr|dem|Resu|Objc|Link|mod|Subj|RgRc|ReVo|Cmpl|PrAd|PreC|Sfxs\n word pdp=subs|nmpr|prps|prde|prin|adjv\n p2:phrase function=PreC\n word pdp=subs|nmpr|prin|adjv ls=card|ordn\n\n p1 << p2\n'''\n\nS.study(query)\nS.showPlan()\nS.count(progress=1000, limit=-1)\nfor r in sorted(S.fetch(), key=lambda x: C.rank.data[x[0]-1])[0:10]:\n print(S.glean(r))",
" 0.00s Checking search template ...\n 0.00s loading features ...\n | 0.14s B ls from /Users/dirk/github/text-fabric-data/hebrew/etcbc4c\n 0.15s All additional features loaded - for details use loadLog()\n 0.15s Setting up search space for 6 objects ...\n 2.55s Constraining search space with 6 relations ...\n 2.56s Setting up retrieval plan ...\n 2.58s Ready to deliver results from 573297 nodes\nIterate over S.fetch() to get the results\nSee S.showPlan() to interpret the results\n 2.58s The results are connected to the original search template as follows:\n 0 \n 1 R0 clause\n 2 R1 p1:phrase function=Subj\n 3 R2 phrase_atom rela=NA|rec|par|Adju|Attr|adj|Coor|atr|dem|Resu|Objc|Link|mod|Subj|RgRc|ReVo|Cmpl|PrAd|PreC|Sfxs\n 4 R3 word pdp=subs|nmpr|prps|prde|prin|adjv\n 5 R4 p2:phrase function=PreC\n 6 R5 word pdp=subs|nmpr|prin|adjv ls=card|ordn\n 7 \n 8 p1 << p2\n 9 \n 0.00s Counting results per 1000 up to the end of the results ...\n | 0.02s 1000\n | 0.04s 2000\n 0.05s Done: 2588 results\nclause[הֵ֤ן הָֽאָדָם֙ הָיָה֙ כְּ...] phrase[הָֽאָדָם֙ ] phrase_atom[הָֽאָדָם֙ ] אָדָם֙ phrase[כְּאַחַ֣ד מִמֶּ֔נּוּ ] אַחַ֣ד \nclause[וַיִּֽהְי֣וּ יְמֵי־אָדָ֗ם שְׁמֹנֶ֥ה ...] phrase[יְמֵי־אָדָ֗ם ] phrase_atom[יְמֵי־אָדָ֗ם ] יְמֵי־ phrase[שְׁמֹנֶ֥ה מֵאֹ֖ת שָׁנָ֑ה ] שְׁמֹנֶ֥ה \nclause[וַיִּֽהְי֣וּ יְמֵי־אָדָ֗ם שְׁמֹנֶ֥ה ...] phrase[יְמֵי־אָדָ֗ם ] phrase_atom[יְמֵי־אָדָ֗ם ] אָדָ֗ם phrase[שְׁמֹנֶ֥ה מֵאֹ֖ת שָׁנָ֑ה ] שְׁמֹנֶ֥ה \nclause[וַיִּֽהְי֣וּ יְמֵי־אָדָ֗ם שְׁמֹנֶ֥ה ...] phrase[יְמֵי־אָדָ֗ם ] phrase_atom[יְמֵי־אָדָ֗ם ] יְמֵי־ phrase[שְׁמֹנֶ֥ה מֵאֹ֖ת שָׁנָ֑ה ] מֵאֹ֖ת \nclause[וַיִּֽהְי֣וּ יְמֵי־אָדָ֗ם שְׁמֹנֶ֥ה ...] phrase[יְמֵי־אָדָ֗ם ] phrase_atom[יְמֵי־אָדָ֗ם ] אָדָ֗ם phrase[שְׁמֹנֶ֥ה מֵאֹ֖ת שָׁנָ֑ה ] מֵאֹ֖ת \nclause[וַיִּֽהְי֞וּ כָּל־יְמֵ֤י אָדָם֙ ...] phrase[כָּל־יְמֵ֤י אָדָם֙ ] phrase_atom[כָּל־יְמֵ֤י אָדָם֙ ] כָּל־ phrase[תְּשַׁ֤ע מֵאֹות֙ שָׁנָ֔ה וּשְׁלֹשִׁ֖ים ...] תְּשַׁ֤ע \nclause[וַיִּֽהְי֞וּ כָּל־יְמֵ֤י אָדָם֙ ...] phrase[כָּל־יְמֵ֤י אָדָם֙ ] phrase_atom[כָּל־יְמֵ֤י אָדָם֙ ] יְמֵ֤י phrase[תְּשַׁ֤ע מֵאֹות֙ שָׁנָ֔ה וּשְׁלֹשִׁ֖ים ...] תְּשַׁ֤ע \nclause[וַיִּֽהְי֞וּ כָּל־יְמֵ֤י אָדָם֙ ...] phrase[כָּל־יְמֵ֤י אָדָם֙ ] phrase_atom[כָּל־יְמֵ֤י אָדָם֙ ] אָדָם֙ phrase[תְּשַׁ֤ע מֵאֹות֙ שָׁנָ֔ה וּשְׁלֹשִׁ֖ים ...] תְּשַׁ֤ע \nclause[וַיִּֽהְי֞וּ כָּל־יְמֵ֤י אָדָם֙ ...] phrase[כָּל־יְמֵ֤י אָדָם֙ ] phrase_atom[כָּל־יְמֵ֤י אָדָם֙ ] כָּל־ phrase[תְּשַׁ֤ע מֵאֹות֙ שָׁנָ֔ה וּשְׁלֹשִׁ֖ים ...] מֵאֹות֙ \nclause[וַיִּֽהְי֞וּ כָּל־יְמֵ֤י אָדָם֙ ...] phrase[כָּל־יְמֵ֤י אָדָם֙ ] phrase_atom[כָּל־יְמֵ֤י אָדָם֙ ] יְמֵ֤י phrase[תְּשַׁ֤ע מֵאֹות֙ שָׁנָ֔ה וּשְׁלֹשִׁ֖ים ...] מֵאֹות֙ \n"
]
],
[
[
"We have too many results, because we have not posed the restrictions by the `NOTEXIST` operator.\nLet's weed out the results that do not satisfy those criteria.\nThat is, essentially, throwing away those clauses \n\n* that have a phrase with `function=Pred` after the phrase with `function=Pred`\n* where the second phrase has a preposition",
"_____no_output_____"
]
],
[
[
"indent(reset=True)\nproperResults = []\nresultWords = set()\n\nfor r in S.fetch():\n clause = r[0]\n phrase1 = r[1]\n phrase2 = r[4]\n word1 = r[3]\n word2 = r[5]\n phrases = [p for p in L.d(clause, otype='phrase') if sortKey(p) > sortKey(phrase1)]\n \n words2 = L.d(phrase2, otype='word')\n if any(F.function.v(phrase) == 'Pred' for phrase in phrases): continue\n if any(F.pdp.v(word) == 'prep' for word in words2): continue\n resultWords |= {word1, word2}\n properResults.append(r)\n\ninfo('Found {} proper results with {} words in it'.format(len(properResults), len(resultWords)))",
" 0.17s Found 2307 proper results with 2133 words in it\n"
]
],
[
[
"We have still many more results than the MQL query on SHEBANQ.\n\nLet us have a look at some results words and compare them with the result words on SHEBANQ.\nIt is handy to fetch from SHEBANQ the csv file with query results.",
"_____no_output_____"
]
],
[
[
"resultsx = sorted((L.u(r[0], otype='verse')+r for r in properResults), key=lambda r: sortKey(r[0]))\nresultWordsx = [(L.u(w, otype='verse')[0], w) for w in sortNodes(resultWords)]\nfor r in resultWordsx[0:30]:\n print(S.glean(r))",
"Genesis 5:4 יְמֵי־\nGenesis 5:4 אָדָ֗ם \nGenesis 5:4 שְׁמֹנֶ֥ה \nGenesis 5:4 מֵאֹ֖ת \nGenesis 5:5 כָּל־\nGenesis 5:5 יְמֵ֤י \nGenesis 5:5 אָדָם֙ \nGenesis 5:5 תְּשַׁ֤ע \nGenesis 5:5 מֵאֹות֙ \nGenesis 5:5 שְׁלֹשִׁ֖ים \nGenesis 5:8 כָּל־\nGenesis 5:8 יְמֵי־\nGenesis 5:8 שֵׁ֔ת \nGenesis 5:8 שְׁתֵּ֤ים \nGenesis 5:8 עֶשְׂרֵה֙ \nGenesis 5:8 תְשַׁ֥ע \nGenesis 5:8 מֵאֹ֖ות \nGenesis 5:11 כָּל־\nGenesis 5:11 יְמֵ֣י \nGenesis 5:11 אֱנֹ֔ושׁ \nGenesis 5:11 חָמֵ֣שׁ \nGenesis 5:11 תְשַׁ֥ע \nGenesis 5:11 מֵאֹ֖ות \nGenesis 5:14 כָּל־\nGenesis 5:14 יְמֵ֣י \nGenesis 5:14 קֵינָ֔ן \nGenesis 5:14 עֶ֣שֶׂר \nGenesis 5:14 תְשַׁ֥ע \nGenesis 5:14 מֵאֹ֖ות \nGenesis 5:17 כָּל־\n"
]
],
[
[
"In the list from SHEBANQ we see this:",
"_____no_output_____"
]
],
[
[
"Genesis,5,4,יְמֵי־,,JWM/,day,subs,2169,m,pl,NA,NA,451,1,326,9\nGenesis,5,4,אָדָ֗ם ,,>DM===/,Adam,nmpr,2170,m,sg,NA,NA,451,1,326,9\nGenesis,5,4,שְׁמֹנֶ֥ה ,,CMNH/,eight,subs,2175,unknown,sg,NA,NA,453,1,326,9\nGenesis,5,4,מֵאֹ֖ת ,,M>H/,hundred,subs,2176,f,pl,NA,NA,453,1,326,9\nGenesis,5,5,כָּל־,,KL/,whole,subs,2185,m,sg,NA,NA,455,1,328,11\nGenesis,5,5,יְמֵ֤י ,,JWM/,day,subs,2186,m,pl,NA,NA,455,1,328,11\nGenesis,5,5,אָדָם֙ ,,>DM===/,Adam,nmpr,2187,m,sg,NA,NA,455,1,328,11\nGenesis,5,5,תְּשַׁ֤ע ,,TC</,nine,subs,2190,unknown,sg,NA,NA,457,1,328,11\nGenesis,5,5,מֵאֹות֙ ,,M>H/,hundred,subs,2191,f,pl,NA,NA,457,1,328,11\nGenesis,5,5,שְׁלֹשִׁ֖ים ,,CLC/,three,subs,2194,m,pl,NA,NA,457,1,328,11\nGenesis,5,8,כָּל־,,KL/,whole,subs,2230,m,sg,NA,NA,465,1,334,17\nGenesis,5,8,יְמֵי־,,JWM/,day,subs,2231,m,pl,NA,NA,465,1,334,17\nGenesis,5,8,שֵׁ֔ת ,,CT==/,,nmpr,2232,m,sg,NA,NA,465,1,334,17\nGenesis,5,8,שְׁתֵּ֤ים ,,CNJM/,two,subs,2233,f,du,NA,NA,465,1,334,17\nGenesis,5,8,עֶשְׂרֵה֙ ,,<FRH/,ten,subs,2234,unknown,sg,NA,NA,465,1,334,17\nGenesis,5,8,תְשַׁ֥ע ,,TC</,nine,subs,2237,unknown,sg,NA,NA,465,1,334,17\nGenesis,5,8,מֵאֹ֖ות ,,M>H/,hundred,subs,2238,f,pl,NA,NA,465,1,334,17\nGenesis,5,11,כָּל־,,KL/,whole,subs,2272,m,sg,NA,NA,473,1,340,23\nGenesis,5,11,יְמֵ֣י ,,JWM/,day,subs,2273,m,pl,NA,NA,473,1,340,23\nGenesis,5,11,אֱנֹ֔ושׁ ,,>NWC==/,,nmpr,2274,m,sg,NA,NA,473,1,340,23\nGenesis,5,11,חָמֵ֣שׁ ,,XMC/,five,subs,2275,unknown,sg,NA,NA,473,1,340,23\nGenesis,5,11,תְשַׁ֥ע ,,TC</,nine,subs,2278,unknown,sg,NA,NA,473,1,340,23\nGenesis,5,11,מֵאֹ֖ות ,,M>H/,hundred,subs,2279,f,pl,NA,NA,473,1,340,23\nGenesis,5,14,כָּל־,,KL/,whole,subs,2312,m,sg,NA,NA,481,1,346,29\nGenesis,5,14,יְמֵ֣י ,,JWM/,day,subs,2313,m,pl,NA,NA,481,1,346,29\nGenesis,5,14,קֵינָ֔ן ,,QJNN/,,nmpr,2314,m,sg,NA,NA,481,1,346,29\nGenesis,5,14,תְשַׁ֥ע ,,TC</,nine,subs,2318,unknown,sg,NA,NA,481,1,346,29\nGenesis,5,14,מֵאֹ֖ות ,,M>H/,hundred,subs,2319,f,pl,NA,NA,481,1,346,29\nGenesis,5,17,כָּל־,,KL/,whole,subs,2355,m,sg,NA,NA,489,1,352,35",
"_____no_output_____"
]
],
[
[
"The first thing we miss in the SHEBANQ output is\n\n```\nGenesis 5:14 עֶ֣שֶׂר\n```\n\nand in SHEBANQ we see that this word has not been marked with `ls=card|ordn`,\nwhile in the newer ETCBC4c it is!\n\nI have conducted a SHEBANQ query for numerals here\n[Dirk Roorda: numerals](https://shebanq.ancient-data.org/hebrew/query?id=1487),\nin versions 4 and 4b, \nand quite something happened with the encoding of numerals between those versions.\n\nLet us also find the numerals in 4c:",
"_____no_output_____"
]
],
[
[
"S.study('''\nword ls=card|ordn\n''')",
" 0.00s Checking search template ...\n 0.00s Setting up search space for 1 objects ...\n 0.61s Constraining search space with 0 relations ...\n 0.61s Setting up retrieval plan ...\n 0.61s Ready to deliver results from 7013 nodes\nIterate over S.fetch() to get the results\nSee S.showPlan() to interpret the results\n"
]
],
[
[
"So we have for the amount of numerals in the ETCBC versions:\n\n4|4b|4c\n---|---|---\n6839|7014|7013\n\nOn the basis of these numbers, this cannot be the sole cause of the discrepancy.",
"_____no_output_____"
],
[
"# By Dirk Roorda\n\n[Dirk Roorda: Yesh](https://shebanq.ancient-data.org/hebrew/query?version=4b&id=556)\n\n```\nselect all objects where\n[book [chapter [verse\n[clause\n [clause_atom\n [phrase\n [phrase_atom\n [word focus lex=\"JC/\" OR lex=\">JN/\"]\n ]\n ]\n ]\n]\n]]]\n```\n\n926 results",
"_____no_output_____"
]
],
[
[
"query = '''\nverse\n clause\n clause_atom\n phrase\n phrase_atom\n word lex=JC/|>JN/\n'''\n\nS.study(query)\nS.showPlan()\nS.count(progress=1000, limit=-1)\nfor r in sorted(S.fetch(), key=lambda x: C.rank.data[x[0]-1])[0:10]:\n print(S.glean(r))",
" 0.00s Checking search template ...\n 0.00s Setting up search space for 6 objects ...\n 0.77s Constraining search space with 5 relations ...\n 0.81s Setting up retrieval plan ...\n 0.82s Ready to deliver results from 5415 nodes\nIterate over S.fetch() to get the results\nSee S.showPlan() to interpret the results\n 0.82s The results are connected to the original search template as follows:\n 0 \n 1 R0 verse\n 2 R1 clause\n 3 R2 clause_atom\n 4 R3 phrase\n 5 R4 phrase_atom\n 6 R5 word lex=JC/|>JN/\n 7 \n 0.00s Counting results per 1000 up to the end of the results ...\n 0.02s Done: 926 results\nGenesis 2:5 clause[וְאָדָ֣ם אַ֔יִן ] clause_atom[וְאָדָ֣ם אַ֔יִן ] phrase[אַ֔יִן ] phrase_atom[אַ֔יִן ] אַ֔יִן \nGenesis 5:24 clause[וְאֵינֶ֕נּוּ ] clause_atom[וְאֵינֶ֕נּוּ ] phrase[אֵינֶ֕נּוּ ] phrase_atom[אֵינֶ֕נּוּ ] אֵינֶ֕נּוּ \nGenesis 7:8 clause[אֲשֶׁ֥ר אֵינֶ֖נָּה טְהֹרָ֑ה ] clause_atom[אֲשֶׁ֥ר אֵינֶ֖נָּה טְהֹרָ֑ה ] phrase[אֵינֶ֖נָּה ] phrase_atom[אֵינֶ֖נָּה ] אֵינֶ֖נָּה \nGenesis 11:30 clause[אֵ֥ין לָ֖הּ וָלָֽד׃ ] clause_atom[אֵ֥ין לָ֖הּ וָלָֽד׃ ] phrase[אֵ֥ין ] phrase_atom[אֵ֥ין ] אֵ֥ין \nGenesis 18:24 clause[אוּלַ֥י יֵ֛שׁ חֲמִשִּׁ֥ים צַדִּיקִ֖ם בְּ...] clause_atom[אוּלַ֥י יֵ֛שׁ חֲמִשִּׁ֥ים צַדִּיקִ֖ם בְּ...] phrase[יֵ֛שׁ ] phrase_atom[יֵ֛שׁ ] יֵ֛שׁ \nGenesis 19:31 clause[וְאִ֨ישׁ אֵ֤ין בָּ...] clause_atom[וְאִ֨ישׁ אֵ֤ין בָּ...] phrase[אֵ֤ין ] phrase_atom[אֵ֤ין ] אֵ֤ין \nGenesis 20:7 clause[וְאִם־אֵֽינְךָ֣ מֵשִׁ֗יב ] clause_atom[וְאִם־אֵֽינְךָ֣ מֵשִׁ֗יב ] phrase[אֵֽינְךָ֣ ] phrase_atom[אֵֽינְךָ֣ ] אֵֽינְךָ֣ \nGenesis 20:11 clause[רַ֚ק אֵין־יִרְאַ֣ת אֱלֹהִ֔ים בַּ...] clause_atom[רַ֚ק אֵין־יִרְאַ֣ת אֱלֹהִ֔ים בַּ...] phrase[אֵין־] phrase_atom[אֵין־] אֵין־\nGenesis 23:8 clause[אִם־יֵ֣שׁ אֶֽת־נַפְשְׁכֶ֗ם ] clause_atom[אִם־יֵ֣שׁ אֶֽת־נַפְשְׁכֶ֗ם ] phrase[יֵ֣שׁ ] phrase_atom[יֵ֣שׁ ] יֵ֣שׁ \nGenesis 24:23 clause[הֲיֵ֧שׁ בֵּית־אָבִ֛יךְ מָקֹ֥ום ...] clause_atom[הֲיֵ֧שׁ בֵּית־אָבִ֛יךְ מָקֹ֥ום ...] phrase[יֵ֧שׁ ] phrase_atom[יֵ֧שׁ ] יֵ֧שׁ \n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d0f5b4c2633904053f94f9219a40dfdecb9723a8 | 79,743 | ipynb | Jupyter Notebook | Chat-Bots.ipynb | SUSHRUTM/NLP-CHATBOT | 3344c81a66d2847c337e5b6374540c8c61de6fd5 | [
"MIT"
] | null | null | null | Chat-Bots.ipynb | SUSHRUTM/NLP-CHATBOT | 3344c81a66d2847c337e5b6374540c8c61de6fd5 | [
"MIT"
] | null | null | null | Chat-Bots.ipynb | SUSHRUTM/NLP-CHATBOT | 3344c81a66d2847c337e5b6374540c8c61de6fd5 | [
"MIT"
] | null | null | null | 41.468019 | 22,014 | 0.602636 | [
[
[
"## Question and Answer Chatbot",
"_____no_output_____"
],
[
"----\n\n------",
"_____no_output_____"
],
[
"## Loading the Data\n\nUsing the Babi Data Set from Facebook Research.\nhttps://research.fb.com/downloads/babi/\n\n## Paper Reference\n\n- Jason Weston, Antoine Bordes, Sumit Chopra, Tomas Mikolov, Alexander M. Rush,\n \"Towards AI-Complete Question Answering: A Set of Prerequisite Toy Tasks\",\n http://arxiv.org/abs/1502.05698\n",
"_____no_output_____"
]
],
[
[
"import pickle\nimport numpy as np",
"_____no_output_____"
],
[
"with open(\"train_qa.txt\", \"rb\") as fp: # Unpickling\n train_data = pickle.load(fp)",
"_____no_output_____"
],
[
"with open(\"test_qa.txt\", \"rb\") as fp: # Unpickling\n test_data = pickle.load(fp)",
"_____no_output_____"
]
],
[
[
"----",
"_____no_output_____"
],
[
"## Exploring the Format of the Data",
"_____no_output_____"
]
],
[
[
"type(test_data)",
"_____no_output_____"
],
[
"type(train_data)",
"_____no_output_____"
],
[
"len(test_data)",
"_____no_output_____"
],
[
"len(train_data)",
"_____no_output_____"
],
[
"train_data[0]",
"_____no_output_____"
],
[
"' '.join(train_data[0][0])",
"_____no_output_____"
],
[
"' '.join(train_data[0][1])",
"_____no_output_____"
],
[
"train_data[0][2]",
"_____no_output_____"
]
],
[
[
"-----\n\n## Setting up Vocabulary of All Words",
"_____no_output_____"
]
],
[
[
"# Create a set that holds the vocab words\nvocab = set()",
"_____no_output_____"
],
[
"all_data = test_data + train_data",
"_____no_output_____"
],
[
"for story, question , answer in all_data:\n # In case you don't know what a union of sets is:\n # https://www.programiz.com/python-programming/methods/set/union\n vocab = vocab.union(set(story))\n vocab = vocab.union(set(question))",
"_____no_output_____"
],
[
"vocab.add('no')\nvocab.add('yes')",
"_____no_output_____"
],
[
"vocab",
"_____no_output_____"
],
[
"vocab_len = len(vocab) + 1 #we add an extra space to hold a 0 for Keras's pad_sequences",
"_____no_output_____"
],
[
"max_story_len = max([len(data[0]) for data in all_data])",
"_____no_output_____"
],
[
"max_story_len",
"_____no_output_____"
],
[
"max_question_len = max([len(data[1]) for data in all_data])",
"_____no_output_____"
],
[
"max_question_len",
"_____no_output_____"
]
],
[
[
"## Vectorizing the Data",
"_____no_output_____"
]
],
[
[
"vocab",
"_____no_output_____"
],
[
"# Reserve 0 for pad_sequences\nvocab_size = len(vocab) + 1",
"_____no_output_____"
]
],
[
[
"-----------",
"_____no_output_____"
]
],
[
[
"from keras.preprocessing.sequence import pad_sequences\nfrom keras.preprocessing.text import Tokenizer",
"Using TensorFlow backend.\n"
],
[
"# integer encode sequences of words\ntokenizer = Tokenizer(filters=[])\ntokenizer.fit_on_texts(vocab)",
"_____no_output_____"
],
[
"tokenizer.word_index",
"_____no_output_____"
],
[
"train_story_text = []\ntrain_question_text = []\ntrain_answers = []\n\nfor story,question,answer in train_data:\n train_story_text.append(story)\n train_question_text.append(question)",
"_____no_output_____"
],
[
"train_story_seq = tokenizer.texts_to_sequences(train_story_text)",
"_____no_output_____"
],
[
"len(train_story_text)",
"_____no_output_____"
],
[
"len(train_story_seq)",
"_____no_output_____"
],
[
"# word_index = tokenizer.word_index",
"_____no_output_____"
]
],
[
[
"### Functionalize Vectorization",
"_____no_output_____"
]
],
[
[
"def vectorize_stories(data, word_index=tokenizer.word_index, max_story_len=max_story_len,max_question_len=max_question_len):\n '''\n INPUT: \n \n data: consisting of Stories,Queries,and Answers\n word_index: word index dictionary from tokenizer\n max_story_len: the length of the longest story (used for pad_sequences function)\n max_question_len: length of the longest question (used for pad_sequences function)\n\n\n OUTPUT:\n \n Vectorizes the stories,questions, and answers into padded sequences. We first loop for every story, query , and\n answer in the data. Then we convert the raw words to an word index value. Then we append each set to their appropriate\n output list. Then once we have converted the words to numbers, we pad the sequences so they are all of equal length.\n \n Returns this in the form of a tuple (X,Xq,Y) (padded based on max lengths)\n '''\n \n \n # X = STORIES\n X = []\n # Xq = QUERY/QUESTION\n Xq = []\n # Y = CORRECT ANSWER\n Y = []\n \n \n for story, query, answer in data:\n \n # Grab the word index for every word in story\n x = [word_index[word.lower()] for word in story]\n # Grab the word index for every word in query\n xq = [word_index[word.lower()] for word in query]\n \n # Grab the Answers (either Yes/No so we don't need to use list comprehension here)\n # Index 0 is reserved so we're going to use + 1\n y = np.zeros(len(word_index) + 1)\n \n # Now that y is all zeros and we know its just Yes/No , we can use numpy logic to create this assignment\n #\n y[word_index[answer]] = 1\n \n # Append each set of story,query, and answer to their respective holding lists\n X.append(x)\n Xq.append(xq)\n Y.append(y)\n \n # Finally, pad the sequences based on their max length so the RNN can be trained on uniformly long sequences.\n \n # RETURN TUPLE FOR UNPACKING\n return (pad_sequences(X, maxlen=max_story_len),pad_sequences(Xq, maxlen=max_question_len), np.array(Y))",
"_____no_output_____"
],
[
"inputs_train, queries_train, answers_train = vectorize_stories(train_data)",
"_____no_output_____"
],
[
"inputs_test, queries_test, answers_test = vectorize_stories(test_data)",
"_____no_output_____"
],
[
"inputs_test",
"_____no_output_____"
],
[
"queries_test",
"_____no_output_____"
],
[
"answers_test",
"_____no_output_____"
],
[
"sum(answers_test)",
"_____no_output_____"
],
[
"tokenizer.word_index['yes']",
"_____no_output_____"
],
[
"tokenizer.word_index['no']",
"_____no_output_____"
]
],
[
[
"## Creating the Model",
"_____no_output_____"
]
],
[
[
"from keras.models import Sequential, Model\nfrom keras.layers.embeddings import Embedding\nfrom keras.layers import Input, Activation, Dense, Permute, Dropout\nfrom keras.layers import add, dot, concatenate\nfrom keras.layers import LSTM",
"_____no_output_____"
]
],
[
[
"### Placeholders for Inputs\n\nRecall we technically have two inputs, stories and questions. So we need to use placeholders. `Input()` is used to instantiate a Keras tensor.\n",
"_____no_output_____"
]
],
[
[
"input_sequence = Input((max_story_len,))\nquestion = Input((max_question_len,))",
"_____no_output_____"
]
],
[
[
"### Building the Networks\n\nTo understand why we chose this setup, make sure to read the paper we are using:\n\n* Sainbayar Sukhbaatar, Arthur Szlam, Jason Weston, Rob Fergus,\n \"End-To-End Memory Networks\",\n http://arxiv.org/abs/1503.08895",
"_____no_output_____"
],
[
"## Encoders\n\n### Input Encoder m",
"_____no_output_____"
]
],
[
[
"# Input gets embedded to a sequence of vectors\ninput_encoder_m = Sequential()\ninput_encoder_m.add(Embedding(input_dim=vocab_size,output_dim=64))\ninput_encoder_m.add(Dropout(0.3))\n\n# This encoder will output:\n# (samples, story_maxlen, embedding_dim)",
"_____no_output_____"
]
],
[
[
"### Input Encoder c",
"_____no_output_____"
]
],
[
[
"# embed the input into a sequence of vectors of size query_maxlen\ninput_encoder_c = Sequential()\ninput_encoder_c.add(Embedding(input_dim=vocab_size,output_dim=max_question_len))\ninput_encoder_c.add(Dropout(0.3))\n# output: (samples, story_maxlen, query_maxlen)",
"_____no_output_____"
]
],
[
[
"### Question Encoder",
"_____no_output_____"
]
],
[
[
"# embed the question into a sequence of vectors\nquestion_encoder = Sequential()\nquestion_encoder.add(Embedding(input_dim=vocab_size,\n output_dim=64,\n input_length=max_question_len))\nquestion_encoder.add(Dropout(0.3))\n# output: (samples, query_maxlen, embedding_dim)",
"_____no_output_____"
]
],
[
[
"### Encode the Sequences",
"_____no_output_____"
]
],
[
[
"# encode input sequence and questions (which are indices)\n# to sequences of dense vectors\ninput_encoded_m = input_encoder_m(input_sequence)\ninput_encoded_c = input_encoder_c(input_sequence)\nquestion_encoded = question_encoder(question)",
"_____no_output_____"
]
],
[
[
"##### Use dot product to compute the match between first input vector seq and the query",
"_____no_output_____"
]
],
[
[
"# shape: `(samples, story_maxlen, query_maxlen)`\nmatch = dot([input_encoded_m, question_encoded], axes=(2, 2))\nmatch = Activation('softmax')(match)",
"_____no_output_____"
]
],
[
[
"#### Add this match matrix with the second input vector sequence",
"_____no_output_____"
]
],
[
[
"# add the match matrix with the second input vector sequence\nresponse = add([match, input_encoded_c]) # (samples, story_maxlen, query_maxlen)\nresponse = Permute((2, 1))(response) # (samples, query_maxlen, story_maxlen)",
"_____no_output_____"
]
],
[
[
"#### Concatenate",
"_____no_output_____"
]
],
[
[
"# concatenate the match matrix with the question vector sequence\nanswer = concatenate([response, question_encoded])",
"_____no_output_____"
],
[
"answer",
"_____no_output_____"
],
[
"# Reduce with RNN (LSTM)\nanswer = LSTM(32)(answer) # (samples, 32)",
"_____no_output_____"
],
[
"# Regularization with Dropout\nanswer = Dropout(0.5)(answer)\nanswer = Dense(vocab_size)(answer) # (samples, vocab_size)",
"_____no_output_____"
],
[
"# we output a probability distribution over the vocabulary\nanswer = Activation('softmax')(answer)\n\n# build the final model\nmodel = Model([input_sequence, question], answer)\nmodel.compile(optimizer='rmsprop', loss='categorical_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"model.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) (None, 156) 0 \n__________________________________________________________________________________________________\ninput_2 (InputLayer) (None, 6) 0 \n__________________________________________________________________________________________________\nsequential_1 (Sequential) multiple 2432 input_1[0][0] \n__________________________________________________________________________________________________\nsequential_3 (Sequential) (None, 6, 64) 2432 input_2[0][0] \n__________________________________________________________________________________________________\ndot_1 (Dot) (None, 156, 6) 0 sequential_1[1][0] \n sequential_3[1][0] \n__________________________________________________________________________________________________\nactivation_1 (Activation) (None, 156, 6) 0 dot_1[0][0] \n__________________________________________________________________________________________________\nsequential_2 (Sequential) multiple 228 input_1[0][0] \n__________________________________________________________________________________________________\nadd_1 (Add) (None, 156, 6) 0 activation_1[0][0] \n sequential_2[1][0] \n__________________________________________________________________________________________________\npermute_1 (Permute) (None, 6, 156) 0 add_1[0][0] \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 6, 220) 0 permute_1[0][0] \n sequential_3[1][0] \n__________________________________________________________________________________________________\nlstm_1 (LSTM) (None, 32) 32384 concatenate_1[0][0] \n__________________________________________________________________________________________________\ndropout_4 (Dropout) (None, 32) 0 lstm_1[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 38) 1254 dropout_4[0][0] \n__________________________________________________________________________________________________\nactivation_2 (Activation) (None, 38) 0 dense_1[0][0] \n==================================================================================================\nTotal params: 38,730\nTrainable params: 38,730\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
],
[
"# train\nhistory = model.fit([inputs_train, queries_train], answers_train,batch_size=32,epochs=120,validation_data=([inputs_test, queries_test], answers_test))",
"Train on 10000 samples, validate on 1000 samples\nEpoch 1/120\n10000/10000 [==============================] - 7s 701us/step - loss: 0.8846 - acc: 0.4966 - val_loss: 0.6938 - val_acc: 0.4970\nEpoch 2/120\n10000/10000 [==============================] - 4s 364us/step - loss: 0.7022 - acc: 0.4987 - val_loss: 0.6935 - val_acc: 0.5030\nEpoch 3/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.6958 - acc: 0.5042 - val_loss: 0.6937 - val_acc: 0.4970\nEpoch 4/120\n10000/10000 [==============================] - 3s 348us/step - loss: 0.6946 - acc: 0.5097 - val_loss: 0.6977 - val_acc: 0.4970\nEpoch 5/120\n10000/10000 [==============================] - 3s 345us/step - loss: 0.6943 - acc: 0.5073 - val_loss: 0.6932 - val_acc: 0.5030\nEpoch 6/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.6954 - acc: 0.4873 - val_loss: 0.6938 - val_acc: 0.4970\nEpoch 7/120\n10000/10000 [==============================] - 3s 345us/step - loss: 0.6946 - acc: 0.4970 - val_loss: 0.6953 - val_acc: 0.4970\nEpoch 8/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.6948 - acc: 0.4955 - val_loss: 0.6939 - val_acc: 0.4970\nEpoch 9/120\n10000/10000 [==============================] - 3s 346us/step - loss: 0.6944 - acc: 0.4937 - val_loss: 0.6933 - val_acc: 0.5030\nEpoch 10/120\n10000/10000 [==============================] - 4s 360us/step - loss: 0.6939 - acc: 0.5011 - val_loss: 0.6937 - val_acc: 0.4970\nEpoch 11/120\n10000/10000 [==============================] - 4s 365us/step - loss: 0.6941 - acc: 0.5051 - val_loss: 0.6934 - val_acc: 0.5030\nEpoch 12/120\n10000/10000 [==============================] - 4s 352us/step - loss: 0.6941 - acc: 0.5014 - val_loss: 0.6955 - val_acc: 0.4970\nEpoch 13/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.6936 - acc: 0.5104 - val_loss: 0.6943 - val_acc: 0.4940\nEpoch 14/120\n10000/10000 [==============================] - 3s 350us/step - loss: 0.6938 - acc: 0.5045 - val_loss: 0.6938 - val_acc: 0.4950\nEpoch 15/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.6914 - acc: 0.5216 - val_loss: 0.6944 - val_acc: 0.5030\nEpoch 16/120\n10000/10000 [==============================] - 4s 351us/step - loss: 0.6830 - acc: 0.5467 - val_loss: 0.6825 - val_acc: 0.5250\nEpoch 17/120\n10000/10000 [==============================] - 4s 356us/step - loss: 0.6663 - acc: 0.5840 - val_loss: 0.6656 - val_acc: 0.6020\nEpoch 18/120\n10000/10000 [==============================] - 4s 368us/step - loss: 0.6404 - acc: 0.6339 - val_loss: 0.6247 - val_acc: 0.6690\nEpoch 19/120\n10000/10000 [==============================] - 4s 364us/step - loss: 0.6049 - acc: 0.6829 - val_loss: 0.5708 - val_acc: 0.7210\nEpoch 20/120\n10000/10000 [==============================] - 4s 356us/step - loss: 0.5569 - acc: 0.7290 - val_loss: 0.5159 - val_acc: 0.7460\nEpoch 21/120\n10000/10000 [==============================] - 4s 358us/step - loss: 0.5180 - acc: 0.7549 - val_loss: 0.4775 - val_acc: 0.7870\nEpoch 22/120\n10000/10000 [==============================] - 4s 354us/step - loss: 0.4891 - acc: 0.7774 - val_loss: 0.4449 - val_acc: 0.7970\nEpoch 23/120\n10000/10000 [==============================] - 4s 354us/step - loss: 0.4528 - acc: 0.8020 - val_loss: 0.4142 - val_acc: 0.8190\nEpoch 24/120\n10000/10000 [==============================] - 4s 354us/step - loss: 0.4253 - acc: 0.8161 - val_loss: 0.4205 - val_acc: 0.8280\nEpoch 25/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.4009 - acc: 0.8354 - val_loss: 0.4094 - val_acc: 0.8280\nEpoch 26/120\n10000/10000 [==============================] - 3s 348us/step - loss: 0.3815 - acc: 0.8432 - val_loss: 0.3919 - val_acc: 0.8240\nEpoch 27/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.3653 - acc: 0.8496 - val_loss: 0.3926 - val_acc: 0.8450\nEpoch 28/120\n10000/10000 [==============================] - 4s 351us/step - loss: 0.3535 - acc: 0.8549 - val_loss: 0.3939 - val_acc: 0.8430\nEpoch 29/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.3435 - acc: 0.8581 - val_loss: 0.3716 - val_acc: 0.8320\nEpoch 30/120\n10000/10000 [==============================] - 4s 350us/step - loss: 0.3403 - acc: 0.8603 - val_loss: 0.3677 - val_acc: 0.8340\nEpoch 31/120\n10000/10000 [==============================] - 4s 351us/step - loss: 0.3302 - acc: 0.8570 - val_loss: 0.3681 - val_acc: 0.8430\nEpoch 32/120\n10000/10000 [==============================] - 3s 348us/step - loss: 0.3295 - acc: 0.8593 - val_loss: 0.3476 - val_acc: 0.8380\nEpoch 33/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.3239 - acc: 0.8628 - val_loss: 0.3521 - val_acc: 0.8430\nEpoch 34/120\n10000/10000 [==============================] - 4s 350us/step - loss: 0.3171 - acc: 0.8677 - val_loss: 0.3443 - val_acc: 0.8390\nEpoch 35/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.3168 - acc: 0.8629 - val_loss: 0.3507 - val_acc: 0.8340\nEpoch 36/120\n10000/10000 [==============================] - 4s 351us/step - loss: 0.3121 - acc: 0.8664 - val_loss: 0.3558 - val_acc: 0.8310\nEpoch 37/120\n10000/10000 [==============================] - 4s 351us/step - loss: 0.3107 - acc: 0.8662 - val_loss: 0.3411 - val_acc: 0.8430\nEpoch 38/120\n10000/10000 [==============================] - 4s 355us/step - loss: 0.3061 - acc: 0.8698 - val_loss: 0.3460 - val_acc: 0.8400\nEpoch 39/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.3065 - acc: 0.8671 - val_loss: 0.3493 - val_acc: 0.8400\nEpoch 40/120\n10000/10000 [==============================] - 4s 352us/step - loss: 0.3060 - acc: 0.8688 - val_loss: 0.3446 - val_acc: 0.8410\nEpoch 41/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.3000 - acc: 0.8696 - val_loss: 0.3542 - val_acc: 0.8450\nEpoch 42/120\n10000/10000 [==============================] - 4s 354us/step - loss: 0.3039 - acc: 0.8665 - val_loss: 0.3692 - val_acc: 0.8350\nEpoch 43/120\n10000/10000 [==============================] - 3s 348us/step - loss: 0.3015 - acc: 0.8695 - val_loss: 0.3513 - val_acc: 0.8400\nEpoch 44/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.2986 - acc: 0.8694 - val_loss: 0.3577 - val_acc: 0.8320\nEpoch 45/120\n10000/10000 [==============================] - 3s 346us/step - loss: 0.2952 - acc: 0.8730 - val_loss: 0.3496 - val_acc: 0.8400\nEpoch 46/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.2969 - acc: 0.8681 - val_loss: 0.3424 - val_acc: 0.8450\nEpoch 47/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.2923 - acc: 0.8721 - val_loss: 0.3549 - val_acc: 0.8280\nEpoch 48/120\n10000/10000 [==============================] - 3s 346us/step - loss: 0.2911 - acc: 0.8732 - val_loss: 0.4681 - val_acc: 0.8140\nEpoch 49/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.2917 - acc: 0.8703 - val_loss: 0.3502 - val_acc: 0.8390\nEpoch 50/120\n10000/10000 [==============================] - 3s 344us/step - loss: 0.2900 - acc: 0.8746 - val_loss: 0.3515 - val_acc: 0.8400\nEpoch 51/120\n10000/10000 [==============================] - 4s 352us/step - loss: 0.2855 - acc: 0.8757 - val_loss: 0.3499 - val_acc: 0.8360\nEpoch 52/120\n10000/10000 [==============================] - 4s 354us/step - loss: 0.2864 - acc: 0.8735 - val_loss: 0.3531 - val_acc: 0.8410\nEpoch 53/120\n10000/10000 [==============================] - 4s 356us/step - loss: 0.2864 - acc: 0.8772 - val_loss: 0.3905 - val_acc: 0.8270\nEpoch 54/120\n10000/10000 [==============================] - 3s 346us/step - loss: 0.2857 - acc: 0.8752 - val_loss: 0.3618 - val_acc: 0.8390\nEpoch 55/120\n10000/10000 [==============================] - 3s 346us/step - loss: 0.2819 - acc: 0.8742 - val_loss: 0.3501 - val_acc: 0.8380\nEpoch 56/120\n10000/10000 [==============================] - 3s 348us/step - loss: 0.2853 - acc: 0.8775 - val_loss: 0.3484 - val_acc: 0.8400\nEpoch 57/120\n10000/10000 [==============================] - 4s 355us/step - loss: 0.2767 - acc: 0.8804 - val_loss: 0.3463 - val_acc: 0.8410\nEpoch 58/120\n10000/10000 [==============================] - 4s 355us/step - loss: 0.2802 - acc: 0.8780 - val_loss: 0.3763 - val_acc: 0.8350\nEpoch 59/120\n10000/10000 [==============================] - 3s 350us/step - loss: 0.2844 - acc: 0.8777 - val_loss: 0.3483 - val_acc: 0.8420\nEpoch 60/120\n10000/10000 [==============================] - 4s 350us/step - loss: 0.2759 - acc: 0.8828 - val_loss: 0.3819 - val_acc: 0.8340\nEpoch 61/120\n10000/10000 [==============================] - 4s 351us/step - loss: 0.2722 - acc: 0.8799 - val_loss: 0.3596 - val_acc: 0.8400\nEpoch 62/120\n10000/10000 [==============================] - 4s 366us/step - loss: 0.2704 - acc: 0.8845 - val_loss: 0.3751 - val_acc: 0.8400\nEpoch 63/120\n10000/10000 [==============================] - 4s 372us/step - loss: 0.2691 - acc: 0.8854 - val_loss: 0.3745 - val_acc: 0.8430\nEpoch 64/120\n10000/10000 [==============================] - 4s 356us/step - loss: 0.2698 - acc: 0.8865 - val_loss: 0.3562 - val_acc: 0.8400\nEpoch 65/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.2674 - acc: 0.8875 - val_loss: 0.3534 - val_acc: 0.8400\nEpoch 66/120\n10000/10000 [==============================] - 4s 356us/step - loss: 0.2622 - acc: 0.8888 - val_loss: 0.3763 - val_acc: 0.8390\nEpoch 67/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.2593 - acc: 0.8885 - val_loss: 0.3670 - val_acc: 0.8470\nEpoch 68/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.2567 - acc: 0.8937 - val_loss: 0.3699 - val_acc: 0.8560\nEpoch 69/120\n10000/10000 [==============================] - 4s 361us/step - loss: 0.2573 - acc: 0.8951 - val_loss: 0.3676 - val_acc: 0.8430\nEpoch 70/120\n10000/10000 [==============================] - 4s 364us/step - loss: 0.2489 - acc: 0.8962 - val_loss: 0.3564 - val_acc: 0.8510\nEpoch 71/120\n10000/10000 [==============================] - 4s 363us/step - loss: 0.2479 - acc: 0.8961 - val_loss: 0.3605 - val_acc: 0.8460\nEpoch 72/120\n10000/10000 [==============================] - 4s 353us/step - loss: 0.2406 - acc: 0.9026 - val_loss: 0.3605 - val_acc: 0.8560\nEpoch 73/120\n10000/10000 [==============================] - 4s 354us/step - loss: 0.2404 - acc: 0.9020 - val_loss: 0.3490 - val_acc: 0.8510\nEpoch 74/120\n10000/10000 [==============================] - 4s 358us/step - loss: 0.2374 - acc: 0.9045 - val_loss: 0.3400 - val_acc: 0.8470\nEpoch 75/120\n10000/10000 [==============================] - 4s 381us/step - loss: 0.2299 - acc: 0.9060 - val_loss: 0.3453 - val_acc: 0.8490\nEpoch 76/120\n10000/10000 [==============================] - 4s 352us/step - loss: 0.2301 - acc: 0.9046 - val_loss: 0.3372 - val_acc: 0.8490\nEpoch 77/120\n10000/10000 [==============================] - 4s 353us/step - loss: 0.2250 - acc: 0.9076 - val_loss: 0.3354 - val_acc: 0.8510\nEpoch 78/120\n10000/10000 [==============================] - 3s 346us/step - loss: 0.2147 - acc: 0.9087 - val_loss: 0.3416 - val_acc: 0.8490\nEpoch 79/120\n10000/10000 [==============================] - 4s 351us/step - loss: 0.2111 - acc: 0.9119 - val_loss: 0.3774 - val_acc: 0.8520\nEpoch 80/120\n10000/10000 [==============================] - 4s 351us/step - loss: 0.2148 - acc: 0.9139 - val_loss: 0.3209 - val_acc: 0.8650\nEpoch 81/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.2045 - acc: 0.9158 - val_loss: 0.3157 - val_acc: 0.8650\nEpoch 82/120\n10000/10000 [==============================] - 3s 345us/step - loss: 0.1916 - acc: 0.9194 - val_loss: 0.3012 - val_acc: 0.8700\nEpoch 83/120\n10000/10000 [==============================] - 3s 350us/step - loss: 0.1881 - acc: 0.9228 - val_loss: 0.2922 - val_acc: 0.8670\nEpoch 84/120\n10000/10000 [==============================] - 3s 348us/step - loss: 0.1783 - acc: 0.9266 - val_loss: 0.2849 - val_acc: 0.8770\nEpoch 85/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.1933 - acc: 0.9209 - val_loss: 0.3006 - val_acc: 0.8730\nEpoch 86/120\n10000/10000 [==============================] - 4s 352us/step - loss: 0.1824 - acc: 0.9279 - val_loss: 0.2729 - val_acc: 0.8810\nEpoch 87/120\n10000/10000 [==============================] - 3s 350us/step - loss: 0.1779 - acc: 0.9282 - val_loss: 0.2774 - val_acc: 0.8840\nEpoch 88/120\n10000/10000 [==============================] - 3s 345us/step - loss: 0.1710 - acc: 0.9303 - val_loss: 0.2758 - val_acc: 0.8810\nEpoch 89/120\n10000/10000 [==============================] - 3s 348us/step - loss: 0.1658 - acc: 0.9345 - val_loss: 0.2854 - val_acc: 0.8880\nEpoch 90/120\n10000/10000 [==============================] - 4s 358us/step - loss: 0.1637 - acc: 0.9347 - val_loss: 0.2634 - val_acc: 0.8930\nEpoch 91/120\n10000/10000 [==============================] - 4s 351us/step - loss: 0.1577 - acc: 0.9358 - val_loss: 0.2546 - val_acc: 0.8910\nEpoch 92/120\n10000/10000 [==============================] - 4s 354us/step - loss: 0.1611 - acc: 0.9387 - val_loss: 0.2445 - val_acc: 0.9080\nEpoch 93/120\n10000/10000 [==============================] - 4s 368us/step - loss: 0.1560 - acc: 0.9381 - val_loss: 0.2369 - val_acc: 0.9040\nEpoch 94/120\n10000/10000 [==============================] - 4s 352us/step - loss: 0.1470 - acc: 0.9409 - val_loss: 0.2764 - val_acc: 0.8950\nEpoch 95/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.1475 - acc: 0.9428 - val_loss: 0.2634 - val_acc: 0.8980\nEpoch 96/120\n10000/10000 [==============================] - 3s 350us/step - loss: 0.1418 - acc: 0.9454 - val_loss: 0.2367 - val_acc: 0.9070\nEpoch 97/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.1436 - acc: 0.9453 - val_loss: 0.2460 - val_acc: 0.9120\nEpoch 98/120\n10000/10000 [==============================] - 4s 350us/step - loss: 0.1434 - acc: 0.9430 - val_loss: 0.2593 - val_acc: 0.9130\nEpoch 99/120\n10000/10000 [==============================] - 3s 348us/step - loss: 0.1348 - acc: 0.9465 - val_loss: 0.2851 - val_acc: 0.9000\nEpoch 100/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.1406 - acc: 0.9431 - val_loss: 0.2609 - val_acc: 0.9040\nEpoch 101/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.1342 - acc: 0.9478 - val_loss: 0.2705 - val_acc: 0.9050\nEpoch 102/120\n10000/10000 [==============================] - 3s 346us/step - loss: 0.1352 - acc: 0.9475 - val_loss: 0.2505 - val_acc: 0.9010\nEpoch 103/120\n10000/10000 [==============================] - 3s 345us/step - loss: 0.1291 - acc: 0.9502 - val_loss: 0.2708 - val_acc: 0.9080\nEpoch 104/120\n10000/10000 [==============================] - 4s 351us/step - loss: 0.1250 - acc: 0.9523 - val_loss: 0.2634 - val_acc: 0.9120\nEpoch 105/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.1203 - acc: 0.9519 - val_loss: 0.2725 - val_acc: 0.9070\nEpoch 106/120\n10000/10000 [==============================] - 4s 350us/step - loss: 0.1187 - acc: 0.9540 - val_loss: 0.2557 - val_acc: 0.9170\nEpoch 107/120\n10000/10000 [==============================] - 3s 347us/step - loss: 0.1182 - acc: 0.9531 - val_loss: 0.2664 - val_acc: 0.9090\nEpoch 108/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.1181 - acc: 0.9530 - val_loss: 0.2334 - val_acc: 0.9130\nEpoch 109/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.1158 - acc: 0.9554 - val_loss: 0.2899 - val_acc: 0.9120\nEpoch 110/120\n10000/10000 [==============================] - 3s 350us/step - loss: 0.1167 - acc: 0.9567 - val_loss: 0.2754 - val_acc: 0.9090\nEpoch 111/120\n10000/10000 [==============================] - 4s 350us/step - loss: 0.1120 - acc: 0.9561 - val_loss: 0.2898 - val_acc: 0.9100\nEpoch 112/120\n10000/10000 [==============================] - 3s 349us/step - loss: 0.1118 - acc: 0.9588 - val_loss: 0.2541 - val_acc: 0.9140\nEpoch 113/120\n10000/10000 [==============================] - 3s 346us/step - loss: 0.1083 - acc: 0.9583 - val_loss: 0.2511 - val_acc: 0.9110\nEpoch 114/120\n10000/10000 [==============================] - 4s 359us/step - loss: 0.1131 - acc: 0.9560 - val_loss: 0.2496 - val_acc: 0.9180\nEpoch 115/120\n10000/10000 [==============================] - 4s 364us/step - loss: 0.1050 - acc: 0.9599 - val_loss: 0.3021 - val_acc: 0.9170\nEpoch 116/120\n10000/10000 [==============================] - 3s 342us/step - loss: 0.1038 - acc: 0.9619 - val_loss: 0.2673 - val_acc: 0.9160\nEpoch 117/120\n"
]
],
[
[
"### Saving the Model",
"_____no_output_____"
]
],
[
[
"filename = 'chatbot_120_epochs.h5'\nmodel.save(filename)",
"_____no_output_____"
]
],
[
[
"## Evaluating the Model\n\n### Plotting Out Training History",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline\nprint(history.history.keys())\n# summarize history for accuracy\nplt.plot(history.history['acc'])\nplt.plot(history.history['val_acc'])\nplt.title('model accuracy')\nplt.ylabel('accuracy')\nplt.xlabel('epoch')\nplt.legend(['train', 'test'], loc='upper left')\nplt.show()",
"dict_keys(['val_loss', 'val_acc', 'loss', 'acc'])\n"
]
],
[
[
"### Evaluating on Given Test Set",
"_____no_output_____"
]
],
[
[
"model.load_weights(filename)\npred_results = model.predict(([inputs_test, queries_test]))",
"_____no_output_____"
],
[
"test_data[0][0]",
"_____no_output_____"
],
[
"story =' '.join(word for word in test_data[0][0])\nprint(story)",
"Mary got the milk there . John moved to the bedroom .\n"
],
[
"query = ' '.join(word for word in test_data[0][1])\nprint(query)",
"Is John in the kitchen ?\n"
],
[
"print(\"True Test Answer from Data is:\",test_data[0][2])",
"True Test Answer from Data is: no\n"
],
[
"#Generate prediction from model\nval_max = np.argmax(pred_results[0])\n\nfor key, val in tokenizer.word_index.items():\n if val == val_max:\n k = key\n\nprint(\"Predicted answer is: \", k)\nprint(\"Probability of certainty was: \", pred_results[0][val_max])",
"Predicted answer is: no\nProbability of certainty was: 0.9999999\n"
]
],
[
[
"## Writing Own Stories and Questions\n\nRemember you can only use words from the existing vocab",
"_____no_output_____"
]
],
[
[
"vocab",
"_____no_output_____"
],
[
"# Note the whitespace of the periods\nmy_story = \"John left the kitchen . Sandra dropped the football in the garden .\"\nmy_story.split()",
"_____no_output_____"
],
[
"my_question = \"Is the football in the garden ?\"",
"_____no_output_____"
],
[
"my_question.split()",
"_____no_output_____"
],
[
"mydata = [(my_story.split(),my_question.split(),'yes')]",
"_____no_output_____"
],
[
"my_story,my_ques,my_ans = vectorize_stories(mydata)",
"_____no_output_____"
],
[
"pred_results = model.predict(([ my_story, my_ques]))",
"_____no_output_____"
],
[
"#Generate prediction from model\nval_max = np.argmax(pred_results[0])\n\nfor key, val in tokenizer.word_index.items():\n if val == val_max:\n k = key\n\nprint(\"Predicted answer is: \", k)\nprint(\"Probability of certainty was: \", pred_results[0][val_max])",
"Predicted answer is: yes\nProbability of certainty was: 0.97079676\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f5c3fce8497f8794c206be851854a8fb1b91da | 18,775 | ipynb | Jupyter Notebook | FantasyBaseball - Combine and Remove.ipynb | MBoerenko/Final-Project | 29c4f2fe3dd8b5b7d60071912456abe7d98184a9 | [
"MIT"
] | null | null | null | FantasyBaseball - Combine and Remove.ipynb | MBoerenko/Final-Project | 29c4f2fe3dd8b5b7d60071912456abe7d98184a9 | [
"MIT"
] | null | null | null | FantasyBaseball - Combine and Remove.ipynb | MBoerenko/Final-Project | 29c4f2fe3dd8b5b7d60071912456abe7d98184a9 | [
"MIT"
] | null | null | null | 29.018547 | 149 | 0.388229 | [
[
[
"# Dependencies and Setup\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# Load Batter Data\nbatters_data_to_load = \"All Batters Data/Batters_2010-2019.csv\"\nbatters_leaders_data_to_load = \"Data - Lead Batters/Batters_leaders__2010-2019.csv\"\nbatters2019_data_to_load = \"Data Sets/mlb-player-stats-Batters (2019).csv\"\nbatters_leaders_2019_data_to_load = \"Data Sets/Batter Leaders/Batter Leaders 2019.csv\"",
"_____no_output_____"
],
[
"# Read batters Data File and store into Pandas DataFrames\nbatters_data = pd.read_csv(batters_data_to_load)\nbatters_leaders_data = pd.read_csv(batters_leaders_data_to_load)\nbatters2019_data = pd.read_csv(batters2019_data_to_load)\nbatters_leaders_2019 = pd.read_csv(batters_leaders_2019_data_to_load)",
"_____no_output_____"
],
[
"batters2019_data['Year'] = 2019\nbatters_leaders_2019['Year'] = 2019",
"_____no_output_____"
],
[
"print(len(batters_data))\nprint(len(batters2019_data))",
"6850\n694\n"
],
[
"print(len(batters_leaders_data))\nprint(len(batters_leaders_2019))",
"1367\n135\n"
],
[
"batters_leaders_data.head()",
"_____no_output_____"
],
[
"batters_leaders_2019.head()",
"_____no_output_____"
],
[
"#combine all batters datasets into one\nbatters_without_leaders = pd.concat([batters_data, batters_leaders_data])\nbatters2019_without_leaders = pd.concat([batters2019_data, batters_leaders_2019])",
"_____no_output_____"
],
[
"print(len(batters_without_leaders))\nprint(len(batters2019_without_leaders))",
"8217\n829\n"
],
[
"batters_without_leaders_df = batters_without_leaders.drop_duplicates(subset=['Player','Year'], keep=False)",
"_____no_output_____"
],
[
"batters2019_without_leaders_df = batters2019_without_leaders.drop_duplicates(subset=['Player'], keep=False)\nprint(len(batters_without_leaders_df))\nprint(len(batters2019_without_leaders_df))",
"4386\n448\n"
],
[
"batters_without_leaders_df.rename({\"Unnamed: 0\":\"a\"}, axis=\"columns\", inplace=True)\n\n# Then, drop the column as usual.\nbatters_without_leaders_df.drop([\"a\"], axis=1, inplace=True)",
"C:\\Users\\ssbne\\anaconda3\\envs\\PythonData\\lib\\site-packages\\pandas\\core\\frame.py:4304: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n errors=errors,\nC:\\Users\\ssbne\\anaconda3\\envs\\PythonData\\lib\\site-packages\\pandas\\core\\frame.py:4170: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n errors=errors,\n"
],
[
"#Write larger data sets to a CSV File\nbatters_without_leaders_df.to_csv(\"Arun Data/batters_without_leaders 2010-2019.csv\")\nbatters2019_without_leaders_df.to_csv(\"Arun Data/batters_without_leaders 2019 Only.csv\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f5c71fdfd6e3d07430113c48f4e601b33db166 | 37,879 | ipynb | Jupyter Notebook | 17 - Monitor Data Drift.ipynb | ldtanh/MS-Azure-DP-100 | 099a474d00aec1d68d3e123ee827eb081fa0cfdd | [
"MIT"
] | null | null | null | 17 - Monitor Data Drift.ipynb | ldtanh/MS-Azure-DP-100 | 099a474d00aec1d68d3e123ee827eb081fa0cfdd | [
"MIT"
] | null | null | null | 17 - Monitor Data Drift.ipynb | ldtanh/MS-Azure-DP-100 | 099a474d00aec1d68d3e123ee827eb081fa0cfdd | [
"MIT"
] | null | null | null | 63.76936 | 8,266 | 0.653027 | [
[
[
"# Monitoring Data Drift\n\nOver time, models can become less effective at predicting accurately due to changing trends in feature data. This phenomenon is known as *data drift*, and it's important to monitor your machine learning solution to detect it so you can retrain your models if necessary.\n\nIn this lab, you'll configure data drift monitoring for datasets.",
"_____no_output_____"
],
[
"## Before you start\n\nIn addition to the latest version of the **azureml-sdk** and **azureml-widgets** packages, you'll need the **azureml-datadrift** package to run the code in this notebook. Run the cell below to verify that it is installed.",
"_____no_output_____"
]
],
[
[
"!pip show azureml-datadrift",
"Name: azureml-datadrift\r\nVersion: 1.34.0\r\nSummary: Contains functionality for data drift detection for various datasets used in machine learning.\r\nHome-page: https://docs.microsoft.com/python/api/overview/azure/ml/?view=azure-ml-py\r\nAuthor: Microsoft Corp\r\nAuthor-email: None\r\nLicense: https://aka.ms/azureml-sdk-license\r\nLocation: /anaconda/envs/azureml_py36/lib/python3.6/site-packages\r\nRequires: pandas, lightgbm, msrest, pyspark, scipy, azureml-telemetry, jsonpickle, numpy, matplotlib, azureml-dataset-runtime, scikit-learn, azureml-core, azureml-pipeline-core\r\nRequired-by: \r\n"
]
],
[
[
"## Connect to your workspace\n\nWith the required SDK packages installed, now you're ready to connect to your workspace.\n\n> **Note**: If you haven't already established an authenticated session with your Azure subscription, you'll be prompted to authenticate by clicking a link, entering an authentication code, and signing into Azure.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Workspace\n\n# Load the workspace from the saved config file\nws = Workspace.from_config()\nprint('Ready to work with', ws.name)",
"Ready to work with azml-ws\n"
]
],
[
[
"## Create a *baseline* dataset\n\nTo monitor a dataset for data drift, you must register a *baseline* dataset (usually the dataset used to train your model) to use as a point of comparison with data collected in the future. ",
"_____no_output_____"
]
],
[
[
"from azureml.core import Datastore, Dataset\n\n\n# Upload the baseline data\ndefault_ds = ws.get_default_datastore()\ndefault_ds.upload_files(files=['./data/diabetes.csv', './data/diabetes2.csv'],\n target_path='diabetes-baseline',\n overwrite=True, \n show_progress=True)\n\n# Create and register the baseline dataset\nprint('Registering baseline dataset...')\nbaseline_data_set = Dataset.Tabular.from_delimited_files(path=(default_ds, 'diabetes-baseline/*.csv'))\nbaseline_data_set = baseline_data_set.register(workspace=ws, \n name='diabetes baseline',\n description='diabetes baseline data',\n tags = {'format':'CSV'},\n create_new_version=True)\n\nprint('Baseline dataset registered!')",
"Uploading an estimated of 2 files\nUploading ./data/diabetes.csv\nUploaded ./data/diabetes.csv, 1 files out of an estimated total of 2\nUploading ./data/diabetes2.csv\nUploaded ./data/diabetes2.csv, 2 files out of an estimated total of 2\nUploaded 2 files\nRegistering baseline dataset...\nBaseline dataset registered!\n"
]
],
[
[
"## Create a *target* dataset\n\nOver time, you can collect new data with the same features as your baseline training data. To compare this new data to the baseline data, you must define a target dataset that includes the features you want to analyze for data drift as well as a timestamp field that indicates the point in time when the new data was current -this enables you to measure data drift over temporal intervals. The timestamp can either be a field in the dataset itself, or derived from the folder and filename pattern used to store the data. For example, you might store new data in a folder hierarchy that consists of a folder for the year, containing a folder for the month, which in turn contains a folder for the day; or you might just encode the year, month, and day in the file name like this: *data_2020-01-29.csv*; which is the approach taken in the following code:",
"_____no_output_____"
]
],
[
[
"import datetime as dt\nimport pandas as pd\n\nprint('Generating simulated data...')\n\n# Load the smaller of the two data files\ndata = pd.read_csv('data/diabetes2.csv')\n\n# We'll generate data for the past 6 weeks\nweeknos = reversed(range(6))\n\nfile_paths = []\nfor weekno in weeknos:\n \n # Get the date X weeks ago\n data_date = dt.date.today() - dt.timedelta(weeks=weekno)\n \n # Modify data to ceate some drift\n data['Pregnancies'] = data['Pregnancies'] + 1\n data['Age'] = round(data['Age'] * 1.2).astype(int)\n data['BMI'] = data['BMI'] * 1.1\n \n # Save the file with the date encoded in the filename\n file_path = 'data/diabetes_{}.csv'.format(data_date.strftime(\"%Y-%m-%d\"))\n data.to_csv(file_path)\n file_paths.append(file_path)\n\n# Upload the files\npath_on_datastore = 'diabetes-target'\ndefault_ds.upload_files(files=file_paths,\n target_path=path_on_datastore,\n overwrite=True,\n show_progress=True)\n\n# Use the folder partition format to define a dataset with a 'date' timestamp column\npartition_format = path_on_datastore + '/diabetes_{date:yyyy-MM-dd}.csv'\ntarget_data_set = Dataset.Tabular.from_delimited_files(path=(default_ds, path_on_datastore + '/*.csv'),\n partition_format=partition_format)\n\n# Register the target dataset\nprint('Registering target dataset...')\ntarget_data_set = target_data_set.with_timestamp_columns('date').register(workspace=ws,\n name='diabetes target',\n description='diabetes target data',\n tags = {'format':'CSV'},\n create_new_version=True)\n\nprint('Target dataset registered!')",
"Generating simulated data...\nUploading an estimated of 6 files\nUploading data/diabetes_2021-09-29.csv\nUploaded data/diabetes_2021-09-29.csv, 1 files out of an estimated total of 6\nUploading data/diabetes_2021-10-06.csv\nUploaded data/diabetes_2021-10-06.csv, 2 files out of an estimated total of 6\nUploading data/diabetes_2021-10-13.csv\nUploaded data/diabetes_2021-10-13.csv, 3 files out of an estimated total of 6\nUploading data/diabetes_2021-10-20.csv\nUploaded data/diabetes_2021-10-20.csv, 4 files out of an estimated total of 6\nUploading data/diabetes_2021-10-27.csv\nUploaded data/diabetes_2021-10-27.csv, 5 files out of an estimated total of 6\nUploading data/diabetes_2021-11-03.csv\nUploaded data/diabetes_2021-11-03.csv, 6 files out of an estimated total of 6\nUploaded 6 files\nRegistering target dataset...\nTarget dataset registered!\n"
]
],
[
[
"## Create a data drift monitor\n\nNow you're ready to create a data drift monitor for the diabetes data. The data drift monitor will run periodicaly or on-demand to compare the baseline dataset with the target dataset, to which new data will be added over time.\n\n### Create a compute target\n\nTo run the data drift monitor, you'll need a compute target. Run the following cell to specify a compute cluster (if it doesn't exist, it will be created).\n\n> **Important**: Change *your-compute-cluster* to the name of your compute cluster in the code below before running it! Cluster names must be globally unique names between 2 to 16 characters in length. Valid characters are letters, digits, and the - character.",
"_____no_output_____"
]
],
[
[
"from azureml.core.compute import ComputeTarget, AmlCompute\nfrom azureml.core.compute_target import ComputeTargetException\n\ncluster_name = \"anhldt-compute2\"\n\ntry:\n # Check for existing compute target\n training_cluster = ComputeTarget(workspace=ws, name=cluster_name)\n print('Found existing cluster, use it.')\nexcept ComputeTargetException:\n # If it doesn't already exist, create it\n try:\n compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_DS11_V2', max_nodes=2)\n training_cluster = ComputeTarget.create(ws, cluster_name, compute_config)\n training_cluster.wait_for_completion(show_output=True)\n except Exception as ex:\n print(ex)\n ",
"InProgress...\nSucceededProvisioning operation finished, operation \"Succeeded\"\nSucceeded\nAmlCompute wait for completion finished\n\nMinimum number of nodes requested have been provisioned\n"
]
],
[
[
"> **Note**: Compute instances and clusters are based on standard Azure virtual machine images. For this exercise, the *Standard_DS11_v2* image is recommended to achieve the optimal balance of cost and performance. If your subscription has a quota that does not include this image, choose an alternative image; but bear in mind that a larger image may incur higher cost and a smaller image may not be sufficient to complete the tasks. Alternatively, ask your Azure administrator to extend your quota.\n\n### Define the data drift monitor\n\nNow you're ready to use a **DataDriftDetector** class to define the data drift monitor for your data. You can specify the features you want to monitor for data drift, the name of the compute target to be used to run the monitoring process, the frequency at which the data should be compared, the data drift threshold above which an alert should be triggered, and the latency (in hours) to allow for data collection.",
"_____no_output_____"
]
],
[
[
"from azureml.datadrift import DataDriftDetector\n\n# set up feature list\nfeatures = ['Pregnancies', 'Age', 'BMI']\n\n# set up data drift detector\nmonitor = DataDriftDetector.create_from_datasets(ws, 'mslearn-diabates-drift', baseline_data_set, target_data_set,\n compute_target=cluster_name, \n frequency='Week', \n feature_list=features, \n drift_threshold=.3, \n latency=24)\nmonitor",
"_____no_output_____"
]
],
[
[
"## Backfill the data drift monitor\n\nYou have a baseline dataset and a target dataset that includes simulated weekly data collection for six weeks. You can use this to backfill the monitor so that it can analyze data drift between the original baseline and the target data.\n\n> **Note** This may take some time to run, as the compute target must be started to run the backfill analysis. The widget may not always update to show the status, so click the link to observe the experiment status in Azure Machine Learning studio!",
"_____no_output_____"
]
],
[
[
"from azureml.widgets import RunDetails\n\nbackfill = monitor.backfill(dt.datetime.now() - dt.timedelta(weeks=6), dt.datetime.now())\n\nRunDetails(backfill).show()\nbackfill.wait_for_completion()",
"_____no_output_____"
]
],
[
[
"## Analyze data drift\n\nYou can use the following code to examine data drift for the points in time collected in the backfill run.",
"_____no_output_____"
]
],
[
[
"drift_metrics = backfill.get_metrics()\nfor metric in drift_metrics:\n print(metric, drift_metrics[metric])",
"start_date 2021-09-19\nend_date 2021-11-07\nfrequency Week\nDatadrift percentage {'days_from_start': [7, 14, 21, 28, 35, 42], 'drift_percentage': [74.19152901127207, 87.23985219136877, 91.74192122865539, 94.96492628559955, 97.58354951107833, 99.23199438682525]}\n"
]
],
[
[
"You can also visualize the data drift metrics in [Azure Machine Learning studio](https://ml.azure.com) by following these steps:\n\n1. On the **Datasets** page, view the **Dataset monitors** tab.\n2. Click the data drift monitor you want to view.\n3. Select the date range over which you want to view data drift metrics (if the column chart does not show multiple weeks of data, wait a minute or so and click **Refresh**).\n4. Examine the charts in the **Drift overview** section at the top, which show overall drift magnitude and the drift contribution per feature.\n5. Explore the charts in the **Feature detail** section at the bottom, which enable you to see various measures of drift for individual features.\n\n> **Note**: For help understanding the data drift metrics, see the [How to monitor datasets](https://docs.microsoft.com/azure/machine-learning/how-to-monitor-datasets#understanding-data-drift-results) in the Azure Machine Learning documentation.\n\n## Explore further\n\nThis lab is designed to introduce you to the concepts and principles of data drift monitoring. To learn more about monitoring data drift using datasets, see the [Detect data drift on datasets](https://docs.microsoft.com/azure/machine-learning/how-to-monitor-datasets) in the Azure machine Learning documentation.\n\nYou can also collect data from published services and use it as a target dataset for datadrift monitoring. See [Collect data from models in production](https://docs.microsoft.com/azure/machine-learning/how-to-enable-data-collection) for details.\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0f5d0de4406eccfa13f0e068e2afa19e2ad5327 | 225,548 | ipynb | Jupyter Notebook | tools/100-gitcount.ipynb | AndrewAnnex/pysal | c7a3b84b1a277baf3a8fb109d2c0e8af6302863c | [
"BSD-3-Clause"
] | null | null | null | tools/100-gitcount.ipynb | AndrewAnnex/pysal | c7a3b84b1a277baf3a8fb109d2c0e8af6302863c | [
"BSD-3-Clause"
] | null | null | null | tools/100-gitcount.ipynb | AndrewAnnex/pysal | c7a3b84b1a277baf3a8fb109d2c0e8af6302863c | [
"BSD-3-Clause"
] | 1 | 2021-04-09T04:29:21.000Z | 2021-04-09T04:29:21.000Z | 64.608422 | 513 | 0.572375 | [
[
[
"# PySAL Change Log Statistics",
"_____no_output_____"
],
[
"## Approach\n- get date of last gh release of each package -> github_released\n- get date of last pypi release of each package -> pypi_released\n- get data of last meta-release -> start_date\n- for each package\n - get issues between start_date and package_released in master\n - get pulls between start_date and package_released in master",
"_____no_output_____"
]
],
[
[
"from release_info import (issues_closed_since, packages,\n is_pull_request,\n sorted_by_field,\n clone_masters\n )\nimport datetime",
"_____no_output_____"
],
[
"clone_masters()",
"libpysal 4.3.0\ngit clone --branch master https://github.com/pysal/libpysal.git tmp/libpysal\naccess 1.1.1\ngit clone --branch master https://github.com/pysal/access.git tmp/access\nesda 2.3.1\ngit clone --branch master https://github.com/pysal/esda.git tmp/esda\ngiddy 2.3.3\ngit clone --branch master https://github.com/pysal/giddy.git tmp/giddy\ninequality 1.0.0\ngit clone --branch master https://github.com/pysal/inequality.git tmp/inequality\npointpats 2.2.0\ngit clone --branch master https://github.com/pysal/pointpats.git tmp/pointpats\nsegregation 1.3.0\ngit clone --branch master https://github.com/pysal/segregation.git tmp/segregation\nspaghetti 1.5.0\ngit clone --branch master https://github.com/pysal/spaghetti.git tmp/spaghetti\nmgwr 2.1.1\ngit clone --branch master https://github.com/pysal/mgwr.git tmp/mgwr\nspglm 1.0.7\ngit clone --branch master https://github.com/pysal/spglm.git tmp/spglm\nspint 1.0.6\ngit clone --branch master https://github.com/pysal/spint.git tmp/spint\nspreg 1.1.1\ngit clone --branch master https://github.com/pysal/spreg.git tmp/spreg\nspvcm 0.3.0\ngit clone --branch master https://github.com/pysal/spvcm.git tmp/spvcm\ntobler 0.3.1\ngit clone --branch master https://github.com/pysal/tobler.git tmp/tobler\nmapclassify 2.3.0\ngit clone --branch master https://github.com/pysal/mapclassify.git tmp/mapclassify\nsplot 1.1.3\ngit clone --branch master https://github.com/pysal/splot.git tmp/splot\npysal 2.3.0\ngit clone --branch master https://github.com/pysal/pysal.git tmp/pysal\n"
],
[
"packages",
"_____no_output_____"
],
[
"#packages['pysal'] = '2.2.0'",
"_____no_output_____"
],
[
"release_date = '2020-07-27'\nstart_date = '2020-02-09'\nsince_date = '--since=\"{start}\"'.format(start=start_date)\nsince_date\nsince = datetime.datetime.strptime(start_date+\" 0:0:0\", \"%Y-%m-%d %H:%M:%S\")\nsince",
"_____no_output_____"
],
[
"issues = {}\nfor package in packages:\n issues[package] = issues_closed_since(since, project=f'pysal/{package}')",
"_____no_output_____"
],
[
"pulls = {}\nfor package in packages:\n pulls[package] = issues_closed_since(since, project=f'pysal/{package}',\n pulls=True)",
"_____no_output_____"
],
[
"len(issues)",
"_____no_output_____"
],
[
"len(pulls)",
"_____no_output_____"
],
[
"pulls['pysal']",
"_____no_output_____"
],
[
"import pickle \n\npickle.dump( issues, open( \"issues_closed.p\", \"wb\" ) )\n\npickle.dump( pulls, open(\"pulls_closed.p\", \"wb\"))",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f5d3ec8d30c9befe76cf0d02fa793bf8b6c9d5 | 16,930 | ipynb | Jupyter Notebook | courses/machine_learning/deepdive/03_tensorflow/e_ai_platform.ipynb | Glairly/introduction_to_tensorflow | aa0a44d9c428a6eb86d1f79d73f54c0861b6358d | [
"Apache-2.0"
] | 2 | 2022-01-06T11:52:57.000Z | 2022-01-09T01:53:56.000Z | courses/machine_learning/deepdive/03_tensorflow/e_ai_platform.ipynb | Glairly/introduction_to_tensorflow | aa0a44d9c428a6eb86d1f79d73f54c0861b6358d | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive/03_tensorflow/e_ai_platform.ipynb | Glairly/introduction_to_tensorflow | aa0a44d9c428a6eb86d1f79d73f54c0861b6358d | [
"Apache-2.0"
] | null | null | null | 34.410569 | 554 | 0.577377 | [
[
[
"# Scaling up ML using Cloud AI Platform\n\nIn this notebook, we take a previously developed TensorFlow model to predict taxifare rides and package it up so that it can be run in Cloud AI Platform. For now, we'll run this on a small dataset. The model that was developed is rather simplistic, and therefore, the accuracy of the model is not great either. However, this notebook illustrates *how* to package up a TensorFlow model to run it within Cloud AI Platform. \n\nLater in the course, we will look at ways to make a more effective machine learning model.",
"_____no_output_____"
],
[
"## Environment variables for project and bucket\n\nNote that:\n<ol>\n<li> Your project id is the *unique* string that identifies your project (not the project name). You can find this from the GCP Console dashboard's Home page. My dashboard reads: <b>Project ID:</b> cloud-training-demos </li>\n<li> Cloud training often involves saving and restoring model files. If you don't have a bucket already, I suggest that you create one from the GCP console (because it will dynamically check whether the bucket name you want is available). A common pattern is to prefix the bucket name by the project id, so that it is unique. Also, for cost reasons, you might want to use a single region bucket. </li>\n</ol>\n<b>Change the cell below</b> to reflect your Project ID and bucket name.\n",
"_____no_output_____"
]
],
[
[
"!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst",
"_____no_output_____"
],
[
"import os\nPROJECT = 'cloud-training-demos' # REPLACE WITH YOUR PROJECT ID\nBUCKET = 'cloud-training-demos-ml' # REPLACE WITH YOUR BUCKET NAME\nREGION = 'us-central1' # REPLACE WITH YOUR BUCKET REGION e.g. us-central1",
"_____no_output_____"
],
[
"# For Python Code\n# Model Info\nMODEL_NAME = 'taxifare'\n# Model Version\nMODEL_VERSION = 'v1'\n# Training Directory name\nTRAINING_DIR = 'taxi_trained'",
"_____no_output_____"
],
[
"# For Bash Code\nos.environ['PROJECT'] = PROJECT\nos.environ['BUCKET'] = BUCKET\nos.environ['REGION'] = REGION\nos.environ['MODEL_NAME'] = MODEL_NAME\nos.environ['MODEL_VERSION'] = MODEL_VERSION\nos.environ['TRAINING_DIR'] = TRAINING_DIR \nos.environ['TFVERSION'] = '2.5' # Tensorflow version",
"_____no_output_____"
],
[
"%%bash\ngcloud config set project $PROJECT\ngcloud config set compute/region $REGION",
"_____no_output_____"
]
],
[
[
"## Packaging up the code\n\nTake your code and put into a standard Python package structure. <a href=\"taxifare/trainer/model.py\">model.py</a> and <a href=\"taxifare/trainer/task.py\">task.py</a> containing the Tensorflow code from earlier (explore the <a href=\"taxifare/trainer/\">directory structure</a>).",
"_____no_output_____"
]
],
[
[
"%%bash\nfind ${MODEL_NAME}",
"_____no_output_____"
],
[
"%%bash\ncat ${MODEL_NAME}/trainer/model.py",
"_____no_output_____"
]
],
[
[
"## Find absolute paths to your data",
"_____no_output_____"
],
[
"Note the absolute paths below. ",
"_____no_output_____"
]
],
[
[
"%%bash\necho \"Working Directory: ${PWD}\"\necho \"Head of taxi-train.csv\"\nhead -1 $PWD/taxi-train.csv\necho \"Head of taxi-valid.csv\"\nhead -1 $PWD/taxi-valid.csv",
"_____no_output_____"
]
],
[
[
"## Running the Python module from the command-line",
"_____no_output_____"
],
[
"#### Clean model training dir/output dir",
"_____no_output_____"
]
],
[
[
"%%bash\n# This is so that the trained model is started fresh each time. However, this needs to be done before \nrm -rf $PWD/${TRAINING_DIR}",
"_____no_output_____"
],
[
"%%bash\n# Setup python so it sees the task module which controls the model.py\nexport PYTHONPATH=${PYTHONPATH}:${PWD}/${MODEL_NAME}\n# Currently set for python 2. To run with python 3 \n# 1. Replace 'python' with 'python3' in the following command\n# 2. Edit trainer/task.py to reflect proper module import method \npython -m trainer.task \\\n --train_data_paths=\"${PWD}/taxi-train*\" \\\n --eval_data_paths=${PWD}/taxi-valid.csv \\\n --output_dir=${PWD}/${TRAINING_DIR} \\\n --train_steps=1000 --job-dir=./tmp",
"_____no_output_____"
],
[
"%%bash\nls $PWD/${TRAINING_DIR}/export/exporter/",
"_____no_output_____"
],
[
"%%writefile ./test.json\n{\"pickuplon\": -73.885262,\"pickuplat\": 40.773008,\"dropofflon\": -73.987232,\"dropofflat\": 40.732403,\"passengers\": 2}",
"_____no_output_____"
],
[
"%%bash\nsudo find \"/usr/lib/google-cloud-sdk/lib/googlecloudsdk/command_lib/ml_engine\" -name '*.pyc' -delete",
"_____no_output_____"
],
[
"%%bash\n# This model dir is the model exported after training and is used for prediction\n#\nmodel_dir=$(ls ${PWD}/${TRAINING_DIR}/export/exporter | tail -1)\n# predict using the trained model\ngcloud ai-platform local predict \\\n --model-dir=${PWD}/${TRAINING_DIR}/export/exporter/${model_dir} \\\n --json-instances=./test.json",
"_____no_output_____"
]
],
[
[
"#### Clean model training dir/output dir",
"_____no_output_____"
]
],
[
[
"%%bash\n# This is so that the trained model is started fresh each time. However, this needs to be done before \nrm -rf $PWD/${TRAINING_DIR}",
"_____no_output_____"
]
],
[
[
"## Running locally using gcloud",
"_____no_output_____"
]
],
[
[
"%%bash\n# Use Cloud Machine Learning Engine to train the model in local file system\ngcloud ai-platform local train \\\n --module-name=trainer.task \\\n --package-path=${PWD}/${MODEL_NAME}/trainer \\\n -- \\\n --train_data_paths=${PWD}/taxi-train.csv \\\n --eval_data_paths=${PWD}/taxi-valid.csv \\\n --train_steps=1000 \\\n --output_dir=${PWD}/${TRAINING_DIR} ",
"_____no_output_____"
],
[
"%%bash\nls $PWD/${TRAINING_DIR}",
"_____no_output_____"
]
],
[
[
"## Submit training job using gcloud\n\nFirst copy the training data to the cloud. Then, launch a training job.\n\nAfter you submit the job, go to the cloud console (http://console.cloud.google.com) and select <b>AI Platform | Jobs</b> to monitor progress. \n\n<b>Note:</b> Don't be concerned if the notebook stalls (with a blue progress bar) or returns with an error about being unable to refresh auth tokens. This is a long-lived Cloud job and work is going on in the cloud. Use the Cloud Console link (above) to monitor the job.",
"_____no_output_____"
]
],
[
[
"%%bash\n# Clear Cloud Storage bucket and copy the CSV files to Cloud Storage bucket\necho $BUCKET\ngsutil -m rm -rf gs://${BUCKET}/${MODEL_NAME}/smallinput/\ngsutil -m cp ${PWD}/*.csv gs://${BUCKET}/${MODEL_NAME}/smallinput/",
"_____no_output_____"
],
[
"%%bash\nOUTDIR=gs://${BUCKET}/${MODEL_NAME}/smallinput/${TRAINING_DIR}\nJOBNAME=${MODEL_NAME}_$(date -u +%y%m%d_%H%M%S)\necho $OUTDIR $REGION $JOBNAME\n# Clear the Cloud Storage Bucket used for the training job\ngsutil -m rm -rf $OUTDIR\ngcloud ai-platform jobs submit training $JOBNAME \\\n --region=$REGION \\\n --module-name=trainer.task \\\n --package-path=${PWD}/${MODEL_NAME}/trainer \\\n --job-dir=$OUTDIR \\\n --staging-bucket=gs://$BUCKET \\\n --scale-tier=BASIC \\\n --runtime-version 2.3 \\\n --python-version 3.5 \\\n -- \\\n --train_data_paths=\"gs://${BUCKET}/${MODEL_NAME}/smallinput/taxi-train*\" \\\n --eval_data_paths=\"gs://${BUCKET}/${MODEL_NAME}/smallinput/taxi-valid*\" \\\n --output_dir=$OUTDIR \\\n --train_steps=10000",
"_____no_output_____"
]
],
[
[
"Don't be concerned if the notebook appears stalled (with a blue progress bar) or returns with an error about being unable to refresh auth tokens. This is a long-lived Cloud job and work is going on in the cloud. \n\n<b>Use the Cloud Console link to monitor the job and do NOT proceed until the job is done.</b>",
"_____no_output_____"
]
],
[
[
"%%bash\ngsutil ls gs://${BUCKET}/${MODEL_NAME}/smallinput",
"_____no_output_____"
]
],
[
[
"## Train on larger dataset\n\nI have already followed the steps below and the files are already available. <b> You don't need to do the steps in this comment. </b> In the next chapter (on feature engineering), we will avoid all this manual processing by using Cloud Dataflow.\n\nGo to http://bigquery.cloud.google.com/ and type the query:\n<pre>\nSELECT\n (tolls_amount + fare_amount) AS fare_amount,\n pickup_longitude AS pickuplon,\n pickup_latitude AS pickuplat,\n dropoff_longitude AS dropofflon,\n dropoff_latitude AS dropofflat,\n passenger_count*1.0 AS passengers,\n 'nokeyindata' AS key\nFROM\n [nyc-tlc:yellow.trips]\nWHERE\n trip_distance > 0\n AND fare_amount >= 2.5\n AND pickup_longitude > -78\n AND pickup_longitude < -70\n AND dropoff_longitude > -78\n AND dropoff_longitude < -70\n AND pickup_latitude > 37\n AND pickup_latitude < 45\n AND dropoff_latitude > 37\n AND dropoff_latitude < 45\n AND passenger_count > 0\n AND ABS(HASH(pickup_datetime)) % 1000 == 1\n</pre>\n\nNote that this is now 1,000,000 rows (i.e. 100x the original dataset). Export this to CSV using the following steps (Note that <b>I have already done this and made the resulting GCS data publicly available</b>, so you don't need to do it.):\n<ol>\n<li> Click on the \"Save As Table\" button and note down the name of the dataset and table.\n<li> On the BigQuery console, find the newly exported table in the left-hand-side menu, and click on the name.\n<li> Click on \"Export Table\"\n<li> Supply your bucket name and give it the name train.csv (for example: gs://cloud-training-demos-ml/taxifare/ch3/train.csv). Note down what this is. Wait for the job to finish (look at the \"Job History\" on the left-hand-side menu)\n<li> In the query above, change the final \"== 1\" to \"== 2\" and export this to Cloud Storage as valid.csv (e.g. gs://cloud-training-demos-ml/taxifare/ch3/valid.csv)\n<li> Download the two files, remove the header line and upload it back to GCS.\n</ol>\n\n<p/>\n<p/>\n\n## Run Cloud training on 1-million row dataset\n\nThis took 60 minutes and uses as input 1-million rows. The model is exactly the same as above. The only changes are to the input (to use the larger dataset) and to the Cloud MLE tier (to use STANDARD_1 instead of BASIC -- STANDARD_1 is approximately 10x more powerful than BASIC). At the end of the training the loss was 32, but the RMSE (calculated on the validation dataset) was stubbornly at 9.03. So, simply adding more data doesn't help.",
"_____no_output_____"
]
],
[
[
"%%bash\n\nOUTDIR=gs://${BUCKET}/${MODEL_NAME}/${TRAINING_DIR}\nJOBNAME=${MODEL_NAME}_$(date -u +%y%m%d_%H%M%S)\nCRS_BUCKET=cloud-training-demos # use the already exported data\necho $OUTDIR $REGION $JOBNAME\ngsutil -m rm -rf $OUTDIR\ngcloud ai-platform jobs submit training $JOBNAME \\\n --region=$REGION \\\n --module-name=trainer.task \\\n --package-path=${PWD}/${MODEL_NAME}/trainer \\\n --job-dir=$OUTDIR \\\n --staging-bucket=gs://$BUCKET \\\n --scale-tier=STANDARD_1 \\\n --runtime-version 2.3 \\\n --python-version 3.5 \\\n -- \\\n --train_data_paths=\"gs://${CRS_BUCKET}/${MODEL_NAME}/ch3/train.csv\" \\\n --eval_data_paths=\"gs://${CRS_BUCKET}/${MODEL_NAME}/ch3/valid.csv\" \\\n --output_dir=$OUTDIR \\\n --train_steps=100000",
"_____no_output_____"
]
],
[
[
"## Challenge Exercise\n\nModify your solution to the challenge exercise in d_trainandevaluate.ipynb appropriately. Make sure that you implement training and deployment. Increase the size of your dataset by 10x since you are running on the cloud. Does your accuracy improve?",
"_____no_output_____"
],
[
"Copyright 2021 Google Inc. Licensed under the Apache License, Version 2.0 (the \"License\"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0f614a1fe8a862a3a0a601aa9ad604ce3f6aa57 | 188,816 | ipynb | Jupyter Notebook | finding_donors_PT.ipynb | aronkst/finding_donors | 013dd6afb3e29d8cc742972c4a77606564499bdc | [
"MIT"
] | null | null | null | finding_donors_PT.ipynb | aronkst/finding_donors | 013dd6afb3e29d8cc742972c4a77606564499bdc | [
"MIT"
] | null | null | null | finding_donors_PT.ipynb | aronkst/finding_donors | 013dd6afb3e29d8cc742972c4a77606564499bdc | [
"MIT"
] | null | null | null | 146.142415 | 51,456 | 0.847132 | [
[
[
"# Nanodegree Engenheiro de Machine Learning\n## Aprendizado Supervisionado\n## Projeto: Encontrando doadores para a *CharityML*",
"_____no_output_____"
],
[
"Seja bem-vindo ao segundo projeto do Nanodegree Engenheiro de Machine Learning! Neste notebook, você receberá alguns códigos de exemplo e será seu trabalho implementar as funcionalidades adicionais necessárias para a conclusão do projeto. As seções cujo cabeçalho começa com **'Implementação'** indicam que o bloco de código posterior requer funcionalidades adicionais que você deve desenvolver. Para cada parte do projeto serão fornecidas instruções e as diretrizes da implementação estarão marcadas no bloco de código com uma expressão `'TODO'`. \nPor favor, leia cuidadosamente as instruções!\n\nAlém de implementações de código, você terá de responder questões relacionadas ao projeto e à sua implementação. Cada seção onde você responderá uma questão terá um cabeçalho com o termo **'Questão X'**. Leia com atenção as questões e forneça respostas completas nas caixas de texto que começam com o termo **'Resposta:'**. A submissão do seu projeto será avaliada baseada nas suas resostas para cada uma das questões além das implementações que você disponibilizar.\n\n>**Nota:** Por favor, especifique QUAL A VERSÃO DO PYTHON utilizada por você para a submissão deste notebook. As células \"Code\" e \"Markdown\" podem ser executadas utilizando o atalho do teclado **Shift + Enter**. Além disso, as células \"Markdown\" podem ser editadas clicando-se duas vezes na célula.\n\n>**Python 2.7**",
"_____no_output_____"
],
[
"## Iniciando\n\nNeste projeto, você utilizará diversos algoritmos de aprendizado supervisionado para modelar com precisão a remuneração de indivíduos utilizando dados coletados no censo americano de 1994. Você escolherá o algoritmo mais adequado através dos resultados preliminares e irá otimizá-lo para modelagem dos dados. O seu objetivo com esta implementação é construir um modelo que pode predizer com precisão se um indivíduo possui uma remuneração superior a $50,000. Este tipo de tarefa pode surgir em organizações sem fins lucrativos que sobrevivem de doações. Entender a remuneração de um indivíduo pode ajudar a organização o montante mais adequado para uma solicitação de doação, ou ainda se eles realmente deveriam entrar em contato com a pessoa. Enquanto pode ser uma tarefa difícil determinar a faixa de renda de uma pesssoa de maneira direta, nós podemos inferir estes valores através de outros recursos disponíveis publicamente. \n\nO conjunto de dados para este projeto se origina do [Repositório de Machine Learning UCI](https://archive.ics.uci.edu/ml/datasets/Census+Income) e foi cedido por Ron Kohavi e Barry Becker, após a sua publicação no artigo _\"Scaling Up the Accuracy of Naive-Bayes Classifiers: A Decision-Tree Hybrid\"_. Você pode encontrar o artigo de Ron Kohavi [online](https://www.aaai.org/Papers/KDD/1996/KDD96-033.pdf). Os dados que investigaremos aqui possuem algumas pequenas modificações se comparados com os dados originais, como por exemplo a remoção da funcionalidade `'fnlwgt'` e a remoção de registros inconsistentes.\n",
"_____no_output_____"
],
[
"----\n## Explorando os dados\nExecute a célula de código abaixo para carregas as bibliotecas Python necessárias e carregas os dados do censo. Perceba que a última coluna deste cojunto de dados, `'income'`, será o rótulo do nosso alvo (se um indivíduo possui remuneração igual ou maior do que $50,000 anualmente). Todas as outras colunas são dados de cada indívduo na base de dados do censo.",
"_____no_output_____"
]
],
[
[
"# Importe as bibliotecas necessárias para o projeto.\nimport numpy as np\nimport pandas as pd\nfrom time import time\nfrom IPython.display import display # Permite a utilização da função display() para DataFrames.\n\n# Importação da biblioteca de visualização visuals.py\nimport visuals as vs\n\n# Exibição amigável para notebooks\n%matplotlib inline\n\n# Carregando os dados do Censo\ndata = pd.read_csv(\"census.csv\")\n\n# Sucesso - Exibindo o primeiro registro\ndisplay(data.head(n=1))",
"_____no_output_____"
]
],
[
[
"### Implementação: Explorando os Dados\n\nUma investigação superficial da massa de dados determinará quantos indivíduos se enquadram em cada grupo e nos dirá sobre o percentual destes indivúdos com remuneração anual superior à \\$50,000. No código abaixo, você precisará calcular o seguinte:\n- O número total de registros, `'n_records'`\n- O número de indivíduos com remuneração anual superior à \\$50,000, `'n_greater_50k'`.\n- O número de indivíduos com remuneração anual até \\$50,000, `'n_at_most_50k'`.\n- O percentual de indivíduos com remuneração anual superior à \\$50,000, `'greater_percent'`.\n\n** DICA: ** Você pode precisar olhar a tabela acima para entender como os registros da coluna `'income'` estão formatados.",
"_____no_output_____"
]
],
[
[
"# Usado para a divisão retornar um float\nfrom __future__ import division\n\n# TODO: Número total de registros.\nn_records = data['age'].count()\n\n# TODO: Número de registros com remuneração anual superior à $50,000\nn_greater_50k = data[data['income'] == '>50K']['age'].count()\n\n# TODO: O número de registros com remuneração anual até $50,000\nn_at_most_50k = data[data['income'] == '<=50K']['age'].count()\n\n# TODO: O percentual de indivíduos com remuneração anual superior à $50,000\ngreater_percent = (data[data['income'] == '>50K']['age'].count() * 100) / data['age'].count()\n\n# Exibindo os resultados\nprint \"Total number of records: {}\".format(n_records)\nprint \"Individuals making more than $50,000: {}\".format(n_greater_50k)\nprint \"Individuals making at most $50,000: {}\".format(n_at_most_50k)\nprint \"Percentage of individuals making more than $50,000: {:.2f}%\".format(greater_percent)",
"Total number of records: 45222\nIndividuals making more than $50,000: 11208\nIndividuals making at most $50,000: 34014\nPercentage of individuals making more than $50,000: 24.78%\n"
]
],
[
[
"** Explorando as colunas **\n* **age**: contínuo. \n* **workclass**: Private, Self-emp-not-inc, Self-emp-inc, Federal-gov, Local-gov, State-gov, Without-pay, Never-worked. \n* **education**: Bachelors, Some-college, 11th, HS-grad, Prof-school, Assoc-acdm, Assoc-voc, 9th, 7th-8th, 12th, Masters, 1st-4th, 10th, Doctorate, 5th-6th, Preschool. \n* **education-num**: contínuo. \n* **marital-status**: Married-civ-spouse, Divorced, Never-married, Separated, Widowed, Married-spouse-absent, Married-AF-spouse. \n* **occupation**: Tech-support, Craft-repair, Other-service, Sales, Exec-managerial, Prof-specialty, Handlers-cleaners, Machine-op-inspct, Adm-clerical, Farming-fishing, Transport-moving, Priv-house-serv, Protective-serv, Armed-Forces. \n* **relationship**: Wife, Own-child, Husband, Not-in-family, Other-relative, Unmarried. \n* **race**: Black, White, Asian-Pac-Islander, Amer-Indian-Eskimo, Other. \n* **sex**: Female, Male. \n* **capital-gain**: contínuo. \n* **capital-loss**: contínuo. \n* **hours-per-week**: contínuo. \n* **native-country**: United-States, Cambodia, England, Puerto-Rico, Canada, Germany, Outlying-US(Guam-USVI-etc), India, Japan, Greece, South, China, Cuba, Iran, Honduras, Philippines, Italy, Poland, Jamaica, Vietnam, Mexico, Portugal, Ireland, France, Dominican-Republic, Laos, Ecuador, Taiwan, Haiti, Columbia, Hungary, Guatemala, Nicaragua, Scotland, Thailand, Yugoslavia, El-Salvador, Trinadad&Tobago, Peru, Hong, Holand-Netherlands.",
"_____no_output_____"
],
[
"----\n## Preparando os dados\nAntes de que os dados possam ser utilizados como input para algoritmos de machine learning, muitas vezes eles precisam ser tratados, formatados e reestruturados — este processo é conhecido como **pré-processamento**. Felizmente neste conjunto de dados não existem registros inconsistentes para tratamento, porém algumas colunas precisam ser ajustadas. Este pré-processamento pode ajudar muito com o resultado e poder de predição de quase todos os algoritmos de aprendizado.",
"_____no_output_____"
],
[
"### Transformando os principais desvios das colunas contínuas\nUm conjunto de dados pode conter ao menos uma coluna onde os valores tendem a se próximar para um único número, mas também podem conter registros com o mesmo atributo contendo um valor muito maior ou muito menor do que esta tendência. Algoritmos podem ser sensíveis para estes casos de distribuição de valores e este fator pode prejudicar sua performance se a distribuição não estiver normalizada de maneira adequada. Com o conjunto de dados do censo, dois atributos se encaixam nesta descrição: '`capital-gain'` e `'capital-loss'`.\n\nExecute o código da célula abaixo para plotar um histograma destes dois atributos. Repare na distribuição destes valores.",
"_____no_output_____"
]
],
[
[
"# Dividindo os dados entre features e coluna alvo\nincome_raw = data['income']\nfeatures_raw = data.drop('income', axis = 1)\n\n# Visualizando os principais desvios das colunas contínuas entre os dados\nvs.distribution(data)",
"_____no_output_____"
]
],
[
[
"Para atributos com distribuição muito distorcida, tais como `'capital-gain'` e `'capital-loss'`, é uma prática comum aplicar uma <a href=\"https://en.wikipedia.org/wiki/Data_transformation_(statistics)\">transformação logarítmica</a> nos dados para que os valores muito grandes e muito pequenos não afetem a performance do algoritmo de aprendizado. Usar a transformação logarítmica reduz significativamente os limites dos valores afetados pelos outliers (valores muito grandes ou muito pequenos). Deve-se tomar cuidado ao aplicar esta transformação, poir o logaritmo de `0` é indefinido, portanto temos que incrementar os valores em uma pequena quantia acima de `0` para aplicar o logaritmo adequadamente.\n\nExecute o código da célula abaixo para realizar a transformação nos dados e visualizar os resultados. De novo, note os valores limite e como os valores estão distribuídos.",
"_____no_output_____"
]
],
[
[
"# Aplicando a transformação de log nos registros distorcidos.\nskewed = ['capital-gain', 'capital-loss']\nfeatures_log_transformed = pd.DataFrame(data = features_raw)\nfeatures_log_transformed[skewed] = features_raw[skewed].apply(lambda x: np.log(x + 1))\n\n# Visualizando as novas distribuições após a transformação.\nvs.distribution(features_log_transformed, transformed = True)",
"_____no_output_____"
]
],
[
[
"### Normalizando atributos numéricos\nAlém das transformações em atributos distorcidos, é uma boa prática comum realizar algum tipo de adaptação de escala nos atributos numéricos. Ajustar a escala nos dados não modifica o formato da distribuição de cada coluna (tais como `'capital-gain'` ou `'capital-loss'` acima); no entanto, a normalização garante que cada atributo será tratado com o mesmo peso durante a aplicação de aprendizado supervisionado. Note que uma vez aplicada a escala, a observação dos dados não terá o significado original, como exemplificado abaixo.\n\nExecute o código da célula abaixo para normalizar cada atributo numérico, nós usaremos ara isso a [`sklearn.preprocessing.MinMaxScaler`](http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.MinMaxScaler.html).",
"_____no_output_____"
]
],
[
[
"# Importando sklearn.preprocessing.StandardScaler\nfrom sklearn.preprocessing import MinMaxScaler\n\n# Inicializando um aplicador de escala e aplicando em seguida aos atributos\nscaler = MinMaxScaler() # default=(0, 1)\nnumerical = ['age', 'education-num', 'capital-gain', 'capital-loss', 'hours-per-week']\n\nfeatures_log_minmax_transform = pd.DataFrame(data = features_log_transformed)\nfeatures_log_minmax_transform[numerical] = scaler.fit_transform(features_log_transformed[numerical])\n\n# Exibindo um exemplo de registro com a escala aplicada\ndisplay(features_log_minmax_transform.head(n=5))",
"_____no_output_____"
]
],
[
[
"### Implementação: Pré-processamento dos dados\n\nA partir da tabela em **Explorando os dados** acima, nós podemos observar que existem diversos atributos não-numéricos para cada registro. Usualmente, algoritmos de aprendizado esperam que os inputs sejam numéricos, o que requer que os atributos não numéricos (chamados de *variáveis de categoria*) sejam convertidos. Uma maneira popular de converter as variáveis de categoria é utilizar a estratégia **one-hot encoding**. Esta estratégia cria uma variável para cada categoria possível de cada atributo não numérico. Por exemplo, assuma que `algumAtributo` possuí três valores possíveis: `A`, `B`, ou `C`. Nós então transformamos este atributo em três novos atributos: `algumAtributo_A`, `algumAtributo_B` e `algumAtributo_C`.\n\n\n| | algumAtributo | | algumAtributo_A | algumAtributo_B | algumAtributo_C |\n| :-: | :-: | | :-: | :-: | :-: |\n| 0 | B | | 0 | 1 | 0 |\n| 1 | C | ----> one-hot encode ----> | 0 | 0 | 1 |\n| 2 | A | | 1 | 0 | 0 |\n\nAlém disso, assim como os atributos não-numéricos, precisaremos converter a coluna alvo não-numérica, `'income'`, para valores numéricos para que o algoritmo de aprendizado funcione. Uma vez que só existem duas categorias possíveis para esta coluna (\"<=50K\" e \">50K\"), nós podemos evitar a utilização do one-hot encoding e simplesmente transformar estas duas categorias para `0` e `1`, respectivamente. No trecho de código abaixo, você precisará implementar o seguinte:\n - Utilizar [`pandas.get_dummies()`](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.get_dummies.html?highlight=get_dummies#pandas.get_dummies) para realizar o one-hot encoding nos dados da `'features_log_minmax_transform'`.\n - Converter a coluna alvo `'income_raw'` para re.\n - Transforme os registros com \"<=50K\" para `0` e os registros com \">50K\" para `1`.",
"_____no_output_____"
]
],
[
[
"# TODO: Utilize o one-hot encoding nos dados em 'features_log_minmax_transform' utilizando pandas.get_dummies()\nfeatures_final = pd.get_dummies(features_log_minmax_transform)\n\n# TODO: Faça o encode da coluna 'income_raw' para valores numéricos\nincome = income_raw.apply(lambda d: 0 if d == '<=50K' else 1)\n\n# Exiba o número de colunas depois do one-hot encoding\nencoded = list(features_final.columns)\nprint \"{} total features after one-hot encoding.\".format(len(encoded))\n\n# Descomente a linha abaixo para ver as colunas após o encode\n# print encoded",
"103 total features after one-hot encoding.\n"
]
],
[
[
"### Embaralhar e dividir os dados\nAgora todas as _variáveis de categoria_ foram convertidas em atributos numéricos e todos os atributos numéricos foram normalizados. Como sempre, nós agora dividiremos os dados entre conjuntos de treinamento e de teste. 80% dos dados serão utilizados para treinamento e 20% para teste.\n\nExecute o código da célula abaixo para realizar divisão.",
"_____no_output_____"
]
],
[
[
"# Importar train_test_split\nfrom sklearn.cross_validation import train_test_split\n\n# Dividir os 'atributos' e 'income' entre conjuntos de treinamento e de testes.\nX_train, X_test, y_train, y_test = train_test_split(features_final, \n income, \n test_size = 0.2, \n random_state = 0)\n\n# Show the results of the split\nprint \"Training set has {} samples.\".format(X_train.shape[0])\nprint \"Testing set has {} samples.\".format(X_test.shape[0])",
"Training set has 36177 samples.\nTesting set has 9045 samples.\n"
]
],
[
[
"----\n## Avaliando a performance do modelo\nNesta seção nós investigaremos quatro algoritmos diferentes e determinaremos qual deles é melhor para a modelagem dos dados. Três destes algoritmos serão algoritmos de aprendizado supervisionado de sua escolha e o quarto algoritmo é conhecido como *naive predictor*.",
"_____no_output_____"
],
[
"### Métricas e o Naive predictor\n\n*CharityML*, equpada com sua pesquisa, sabe que os indivíduos que fazem mais do que \\$50,000 possuem maior probabilidade de doar para a sua campanha de caridade. Por conta disto, a *CharityML* está particularmente interessada em predizer com acurácia quais indivíduos possuem remuneração acima de \\$50,000. Parece que utilizar **acurácia (accuracy)** como uma métrica para avaliar a performance de um modelo é um parâmetro adequado. Além disso, identificar alguém que *não possui* remuneração acima de \\$50,000 como alguém que recebe acima deste valor seria ruim para a *CharityML*, uma vez que eles estão procurando por indivíduos que desejam doar. Com isso, a habilidade do modelo em predizer com preisão aqueles que possuem a remuneração acima dos \\$50,000 é *mais importante* do que a habilidade de realizar o **recall** destes indivíduos. Nós podemos utilizar a fórmula **F-beta score** como uma métrica que considera ambos: precision e recall.\n\n\n$$ F_{\\beta} = (1 + \\beta^2) \\cdot \\frac{precision \\cdot recall}{\\left( \\beta^2 \\cdot precision \\right) + recall} $$\n\nEm particular, quando $\\beta = 0.5$, maior ênfase é atribuída para a variável precision. Isso é chamado de **F$_{0.5}$ score** (ou F-score, simplificando).\n\nAnalisando a distribuição de classes (aqueles que possuem remuneração até \\$50,000 e aqueles que possuem remuneração superior), fica claro que a maioria dos indivíduos não possui remuneração acima de \\$50,000. Isto pode ter grande impacto na **acurácia (accuracy)**, uma vez que nós poderíamos simplesmente dizer *\"Esta pessoa não possui remuneração acima de \\$50,000\"* e estar certos em boa parte das vezes, sem ao menos olhar os dados! Fazer este tipo de afirmação seria chamado de **naive**, uma vez que não consideramos nenhuma informação para balisar este argumento. É sempre importante considerar a *naive prediction* para seu conjunto de dados, para ajudar a estabelecer um benchmark para análise da performance dos modelos. Com isso, sabemos que utilizar a naive prediction não traria resultado algum: Se a predição apontasse que todas as pessoas possuem remuneração inferior à \\$50,000, a *CharityML* não identificaria ninguém como potencial doador. \n\n\n\n#### Nota: Revisando: accuracy, precision e recall\n\n** Accuracy ** mede com que frequência o classificador faz a predição correta. É a proporção entre o número de predições corretas e o número total de predições (o número de registros testados).\n\n** Precision ** informa qual a proporção de mensagens classificamos como spam eram realmente spam. Ou seja, é a proporção de verdadeiros positivos (mensagens classificadas como spam que eram realmente spam) sobre todos os positivos (todas as palavras classificadas como spam, independente se a classificação estava correta), em outras palavras, é a proporção\n\n`[Verdadeiros positivos/(Verdadeiros positivos + Falso positivos)]`\n\n** Recall(sensibilidade)** nos informa qual a proporção das mensagens que eram spam que foram corretamente classificadas como spam. É a proporção entre os verdadeiros positivos (classificados como spam, que realmente eram spam) sobre todas as palavras que realmente eram spam. Em outras palavras, é a proporção entre\n\n`[Verdadeiros positivos/(Verdadeiros positivos + Falso negativos)]`\n\nPara problemas de classificação distorcidos em suas distribuições, como no nosso caso, por exemplo, se tivéssemos 100 mensagems de texto e apenas 2 fossem spam e todas as outras não fossem, a \"accuracy\" por si só não seria uma métrica tão boa. Nós poderiamos classificar 90 mensagems como \"não-spam\" (incluindo as 2 que eram spam mas que teriam sido classificadas como não-spam e, por tanto, seriam falso negativas.) e 10 mensagems como spam (todas as 10 falso positivas) e ainda assim teriamos uma boa pontuação de accuracy. Para estess casos, precision e recall são muito úteis. Estas duas métricas podem ser combinadas para resgatar o F1 score, que é calculado através da média(harmônica) dos valores de precision e de recall. Este score pode variar entre 0 e 1, sendo 1 o melhor resultado possível para o F1 score (consideramos a média harmônica pois estamos lidando com proporções).",
"_____no_output_____"
]
],
[
[
"TP = np.sum(income) # Contando pois este é o caso \"naive\". Note que 'income' são os dados 'income_raw' convertido para valores numéricos durante o passo de pré-processamento de dados.\nFP = income.count() - TP # Específico para o caso naive\n\nTN = 0 # Sem predições negativas para o caso naive\nFN = 0 # Sem predições negativas para o caso naive\n\n# TODO: Calcular accuracy, precision e recall\naccuracy = TP / income.count()\nrecall = TP / (TP + FN)\nprecision = TP / (TP + FP)\n\n# TODO: Calcular o F-score utilizando a fórmula acima para o beta = 0.5 e os valores corretos de precision e recall.\nfscore = (1 + (0.5 ** 2)) * ((precision * recall) / (((0.5 ** 2) * precision) + recall))\n\n# Exibir os resultados \nprint \"Naive Predictor: [Accuracy score: {:.4f}, F-score: {:.4f}]\".format(accuracy, fscore)",
"Naive Predictor: [Accuracy score: 0.2478, F-score: 0.2917]\n"
]
],
[
[
"### Questão 1 - Performance do Naive Predictor\n* Se escolhessemos um modelo que sempre prediz que um indivíduo possui remuneração acima de $50,000, qual seria a accuracy e o F-score considerando este conjunto de dados? Você deverá utilizar o código da célula abaixo e atribuir os seus resultados para as variáveis `'accuracy'` e `'fscore'` que serão usadas posteriormente.\n\n** Por favor, note ** que o propósito ao gerar um naive predictor é simplesmente exibir como um modelo sem nenhuma inteligência se comportaria. No mundo real, idealmente o seu modelo de base será o resultado de um modelo anterior ou poderia ser baseado em um paper no qual você se basearia para melhorar. Quando não houver qualquer benchmark de modelo, utilizar um naive predictor será melhor do que uma escolha aleatória.\n\n** DICA: ** \n\n* Quando temos um modelo que sempre prediz '1' (e.x o indivíduo possui remuneração superior à 50k) então nosso modelo não terá Verdadeiros Negativos ou Falso Negativos, pois nós não estaremos afirmando que qualquer dos valores é negativo (ou '0') durante a predição. Com isso, nossa accuracy neste caso se torna o mesmo valor da precision (Verdadeiros positivos/ (Verdadeiros positivos + Falso positivos)) pois cada predição que fizemos com o valor '1' que deveria ter o valor '0' se torna um falso positivo; nosso denominador neste caso é o número total de registros.\n* Nossa pontuação de Recall(Verdadeiros positivos/(Verdadeiros Positivos + Falsos negativos)) será 1 pois não teremos Falsos negativos.",
"_____no_output_____"
],
[
"### Modelos de Aprendizado Supervisionado\n**Estes são alguns dos modelos de aprendizado supervisionado disponíveis em** [`scikit-learn`](http://scikit-learn.org/stable/supervised_learning.html)\n- Gaussian Naive Bayes (GaussianNB)\n- Decision Trees (Árvores de decisão)\n- Ensemble Methods (Bagging, AdaBoost, Random Forest, Gradient Boosting)\n- K-Nearest Neighbors (KNeighbors)\n- Stochastic Gradient Descent Classifier (SGDC)\n- Support Vector Machines (SVM)\n- Logistic Regression",
"_____no_output_____"
],
[
"### Questão 2 - Aplicação do Modelo\nListe três dos modelos de aprendizado supervisionado acima que são apropriados para este problema que você irá testar nos dados do censo. Para cada modelo escolhido\n\n- Descreva uma situação do mundo real onde este modelo pode ser utilizado. \n- Quais são as vantagems da utilização deste modelo; quando ele performa bem?\n- Quais são as fraquesas do modelo; quando ele performa mal?\n- O que torna este modelo um bom candidato para o problema, considerando o que você sabe sobre o conjunto de dados?\n\n** DICA: **\n\nEstruture sua resposta no mesmo formato acima^, com 4 partes para cada um dos modelos que você escolher. Por favor, inclua referências em cada uma das respostas.",
"_____no_output_____"
],
[
"**Resposta: **\n\n**Gaussian Naive Bayes**\n\n- **Descreva uma situação do mundo real onde este modelo pode ser utilizado.** Detectar span.\n\n- **Quais são as vantagens da utilização deste modelo; quando ele performa bem?** Implementação fácil. Ele é rápido.\n\n- **Quais são as fraquezas do modelo; quando ele performa mal?** Dados com muitos atributos podem não funcionar tão bem. Atributos são independentes.\n\n- **O que torna este modelo um bom candidato para o problema, considerando o que você sabe sobre o conjunto de dados?** É um modelo bem conhecido e pode ser usado para muitos tipos de dados, e também é rápido.\n\n**Referências: **http://scikit-learn.org/stable/modules/svm.html , https://en.wikipedia.org/wiki/Naive_Bayes_classifier , https://pt.wikipedia.org/wiki/Teorema_de_Bayes\n\n**Decision Trees**\n\n- **Descreva uma situação do mundo real onde este modelo pode ser utilizado.** Decidir quando jogar futebol com base nos dados, quando está frio ou quente, está chovendo ou ensolarado, e mais tipos de dados disponíveis.\n\n- **Quais são as vantagens da utilização deste modelo; quando ele performa bem?** Simples de entender, interpretar e analisar. Árvores podem ser visualizadas em forma gráfica. Requer pouca (ou nenhuma) preparação e modificação de dados. Capaz de lidar com dados numéricos e texto.\n\n- **Quais são as fraquezas do modelo; quando ele performa mal?** A árvore de decisão podem ser complexas e não generalizar bem os dados. As árvores de decisão podem ficar instáveis com pequenas variações nos dados, e podem resultar na geração de uma árvore diferente.\n\n- **O que torna este modelo um bom candidato para o problema, considerando o que você sabe sobre o conjunto de dados?** Eu realmente gosto de árvores de decisão e acredito que com a quantidade de dados e seus tipos, pode ser uma excelente escolha para o projeto.\n\n**Referências: **http://scikit-learn.org/stable/modules/tree.html , https://en.wikipedia.org/wiki/Decision_tree\n\n**Logistic Regression**\n\n- **Descreva uma situação do mundo real onde este modelo pode ser utilizado.** Prever se você pode ganhar ou perder em ações.\n\n- **Quais são as vantagens da utilização deste modelo; quando ele performa bem?** É simples. É rápido. Funciona com grandes e pequenas quantidades de dados. É fácil de entender.\n\n- **Quais são as fraquezas do modelo; quando ele performa mal?** Não pode ser usado quando os dados não são binários.\n\n- **O que torna este modelo um bom candidato para o problema, considerando o que você sabe sobre o conjunto de dados?** É um modelo relativamente mais simples que os outros modelo e tem um excelente desempenho.\n\n**Referências: **http://scikit-learn.org/stable/modules/linear_model.html#logistic-regression , https://en.wikipedia.org/wiki/Logistic_regression , https://pt.wikipedia.org/wiki/Regress%C3%A3o_log%C3%ADstica",
"_____no_output_____"
],
[
"### Implementação - Criando um Pipeline de Treinamento e Predição\nPara avaliar adequadamente a performance de cada um dos modelos que você escolheu é importante que você crie um pipeline de treinamento e predição que te permite de maneira rápida e eficiente treinar os modelos utilizando vários tamanhos de conjuntos de dados para treinamento, além de performar predições nos dados de teste. Sua implementação aqui será utilizada na próxima seção. No bloco de código abaixo, você precisará implementar o seguinte:\n - Importar `fbeta_score` e `accuracy_score` de [`sklearn.metrics`](http://scikit-learn.org/stable/modules/classes.html#sklearn-metrics-metrics).\n - Adapte o algoritmo para os dados de treinamento e registre o tempo de treinamento. \n - Realize predições nos dados de teste `X_test`, e também nos 300 primeiros pontos de treinamento `X_train[:300]`.\n - Registre o tempo total de predição. \n - Calcule a acurácia tanto para o conjundo de dados de treino quanto para o conjunto de testes.\n - Calcule o F-score para os dois conjuntos de dados: treino e testes. \n - Garanta que você configurou o parâmetro `beta`! ",
"_____no_output_____"
]
],
[
[
"# TODO: Import two metrics from sklearn - fbeta_score and accuracy_score\nfrom sklearn.metrics import fbeta_score, accuracy_score\n\ndef train_predict(learner, sample_size, X_train, y_train, X_test, y_test): \n '''\n inputs:\n - learner: the learning algorithm to be trained and predicted on\n - sample_size: the size of samples (number) to be drawn from training set\n - X_train: features training set\n - y_train: income training set\n - X_test: features testing set\n - y_test: income testing set\n '''\n \n results = {}\n \n # TODO: Fit the learner to the training data using slicing with 'sample_size' using .fit(training_features[:], training_labels[:])\n start = time() # Get start time\n learner = learner.fit(X_train[:sample_size], y_train[:sample_size])\n end = time() # Get end time\n \n # TODO: Calculate the training time\n results['train_time'] = end - start\n \n # TODO: Get the predictions on the test set(X_test),\n # then get predictions on the first 300 training samples(X_train) using .predict()\n start = time() # Get start time\n predictions_test = learner.predict(X_test)\n predictions_train = learner.predict(X_train[:300])\n end = time() # Get end time\n \n # TODO: Calculate the total prediction time\n results['pred_time'] = end - start\n \n # TODO: Compute accuracy on the first 300 training samples which is y_train[:300]\n results['acc_train'] = accuracy_score(y_train[:300], predictions_train)\n \n # TODO: Compute accuracy on test set using accuracy_score()\n results['acc_test'] = accuracy_score(y_test, predictions_test)\n \n # TODO: Compute F-score on the the first 300 training samples using fbeta_score()\n results['f_train'] = fbeta_score(y_train[:300], predictions_train, 0.5)\n \n # TODO: Compute F-score on the test set which is y_test\n results['f_test'] = fbeta_score(y_test, predictions_test, 0.5)\n \n # Success\n print \"{} trained on {} samples.\".format(learner.__class__.__name__, sample_size)\n \n # Return the results\n return results",
"_____no_output_____"
]
],
[
[
"### Implementação: Validação inicial do modelo\nNo código da célular, você precisará implementar o seguinte:\n- Importar os três modelos de aprendizado supervisionado que você escolheu na seção anterior \n- Inicializar os três modelos e armazená-los em `'clf_A'`, `'clf_B'`, e `'clf_C'`. \n - Utilize um `'random_state'` para cada modelo que você utilizar, caso seja fornecido.\n - **Nota:** Utilize as configurações padrão para cada modelo - você otimizará um modelo específico em uma seção posterior\n- Calcule o número de registros equivalentes à 1%, 10%, e 100% dos dados de treinamento.\n - Armazene estes valores em `'samples_1'`, `'samples_10'`, e `'samples_100'` respectivamente.\n\n**Nota:** Dependendo do algoritmo de sua escolha, a implementação abaixo pode demorar algum tempo para executar!",
"_____no_output_____"
]
],
[
[
"# TODO: Importe os três modelos de aprendizado supervisionado da sklearn\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.linear_model import LogisticRegression\n\n# TODO: Inicialize os três modelos\nclf_A = GaussianNB()\nclf_B = DecisionTreeClassifier(random_state=8)\nclf_C = LogisticRegression(random_state=8)\n\n# TODO: Calcule o número de amostras para 1%, 10%, e 100% dos dados de treinamento\n# HINT: samples_100 é todo o conjunto de treinamento e.x.: len(y_train)\n# HINT: samples_10 é 10% de samples_100\n# HINT: samples_1 é 1% de samples_100\nsamples_100 = len(y_train)\nsamples_10 = int(samples_100 * 0.1)\nsamples_1 = int(samples_100 * 0.01)\n\n# Colete os resultados dos algoritmos de aprendizado\nresults = {}\nfor clf in [clf_A, clf_B, clf_C]:\n clf_name = clf.__class__.__name__\n results[clf_name] = {}\n for i, samples in enumerate([samples_1, samples_10, samples_100]):\n results[clf_name][i] = \\\n train_predict(clf, samples, X_train, y_train, X_test, y_test)\n\n# Run metrics visualization for the three supervised learning models chosen\nvs.evaluate(results, accuracy, fscore)",
"GaussianNB trained on 361 samples.\nGaussianNB trained on 3617 samples.\nGaussianNB trained on 36177 samples.\nDecisionTreeClassifier trained on 361 samples.\nDecisionTreeClassifier trained on 3617 samples.\nDecisionTreeClassifier trained on 36177 samples.\nLogisticRegression trained on 361 samples.\nLogisticRegression trained on 3617 samples.\nLogisticRegression trained on 36177 samples.\n"
]
],
[
[
"----\n## Melhorando os resultados\nNesta seção final, você irá escolher o melhor entre os três modelos de aprendizado supervisionado para utilizar nos dados dos estudantes. Você irá então realizar uma busca grid para otimização em todo o conjunto de dados de treino (`X_train` e `y_train`) fazendo o tuning de pelo menos um parâmetro para melhorar o F-score anterior do modelo.",
"_____no_output_____"
],
[
"### Questão 3 - Escolhendo o melhor modelo\n\n* Baseado na validação anterior, em um ou dois parágrafos explique para a *CharityML* qual dos três modelos você acredita ser o mais apropriado para a tarefa de identificar indivíduos com remuneração anual superior à \\$50,000. \n\n** DICA: ** \nAnalise o gráfico do canto inferior esquerdo da célula acima(a visualização criada através do comando `vs.evaluate(results, accuracy, fscore)`) e verifique o F score para o conjunto de testes quando 100% do conjunto de treino é utilizado. Qual modelo possui o maior score? Sua resposta deve abranger os seguintes pontos:\n* métricas - F score no conjunto de testes quando 100% dos dados de treino são utilizados, \n* tempo de predição/treinamento \n* a adequação do algoritmo para este cojunto de dados.",
"_____no_output_____"
],
[
"**Resposta: **\n\nO modelo GaussianNB teve um desempenho melhor e o modelo DecisionTreeClassifier teve um melhor precisão, os dois quando estavam utilizando o Subset. Mas o LogisticRegression, utilizando o Set, teve uma precisão maior e desempenho melhor que os outros dois.\n\nDos modelos escolhidos para realizar os testes, o que tem a melhor desempenho e precisão para a identificação de quem poderá ser doador é o modelo LogisticRegression, ele é o melhor utilizando o Set, tando no Accuracy e F-score.",
"_____no_output_____"
],
[
"### Questão 4 - Descrevendo o modelo nos termos de Layman\n \n* Em um ou dois parágrafos, explique para a *CharityML*, nos termos de layman, como o modelo final escolhido deveria funcionar. Garanta que você está descrevendo as principais vantagens do modelo, tais como o modo de treinar o modelo e como o modelo realiza a predição. Evite a utilização de jargões matemáticos avançados, como por exemplo a descrição de equações. \n\n** DICA: **\n\nQuando estiver explicando seu modelo, cite as fontes externas utilizadas, caso utilize alguma.",
"_____no_output_____"
],
[
"**Resposta: ** \n\nO modelo que escolhemos usar é o Regressão Logística, muito utilizado em Machine Learning, para classificação binaria e tem origem no campo da estatística. A Regressão Logística é um modelo estatístico de classificação binária, que retorna um resultado binário (0 e 1), medindo a relação entre variáveis dependentes e variáveis de predição, assim conseguindo estimar suas probabilidades.\n\nMais especificamente para essa analise, o modelo Regressão Logística ira pegar todos os atributos e seus valores de nossos dados, que seria a idade, educação, profissão, e os demais atributos, analisar, e tentar encontrar uma combinação entre eles, e categorizando essa analise com 0 para como um não possível doador e 1 para um possível doador, fazendo isso para todas as linhas de nossos dados, e retornando como resultado dessa essa analise, a quantidade e quais são os possíveis doadores que ganham mais de $50.000,00 por ano, ou não.\n\n**Referências: **https://en.wikipedia.org/wiki/Logistic_regression , https://pt.wikipedia.org/wiki/Regress%C3%A3o_log%C3%ADstica",
"_____no_output_____"
],
[
"### Implementação: Tuning do modelo\nRefine o modelo escolhido. Utilize uma busca grid (`GridSearchCV`) com pleo menos um parâmetro importante refinado com pelo menos 3 valores diferentes. Você precisará utilizar todo o conjunto de treinamento para isso. Na célula de código abaixo, você precisará implementar o seguinte:\n- Importar [`sklearn.grid_search.GridSearchCV`](http://scikit-learn.org/0.17/modules/generated/sklearn.grid_search.GridSearchCV.html) e [`sklearn.metrics.make_scorer`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.make_scorer.html).\n- Inicializar o classificador escolhido por você e armazená-lo em `clf`.\n - Configurar um `random_state` se houver um disponível para o mesmo estado que você configurou anteriormente.\n- Criar um dicionário dos parâmetros que você quer otimizar para o modelo escolhido.\n - Exemplo: `parâmetro = {'parâmetro' : [lista de valores]}`.\n - **Nota:** Evite otimizar o parâmetro `max_features` se este parâmetro estiver disponível! \n- Utilize `make_scorer` para criar um objeto de pontuação `fbeta_score` (com $\\beta = 0.5$).\n- Realize a busca gride no classificador `clf` utilizando o `'scorer'` e armazene-o na variável `grid_obj`. \n- Adeque o objeto da busca grid aos dados de treino (`X_train`, `y_train`) e armazene em `grid_fit`.\n\n**Nota:** Dependendo do algoritmo escolhido e da lista de parâmetros, a implementação a seguir pode levar algum tempo para executar! ",
"_____no_output_____"
]
],
[
[
"# TODO: Importar 'GridSearchCV', 'make_scorer', e qualquer biblioteca necessária\nfrom sklearn.grid_search import GridSearchCV\nfrom sklearn.metrics import fbeta_score, make_scorer\n\n# TODO: Inicializar o classificador\nclf = LogisticRegression(random_state=8)\n\n# TODO: Criar a lista de parâmetros que você quer otimizar, utilizando um dicionário, caso necessário.\n# HINT: parameters = {'parameter_1': [value1, value2], 'parameter_2': [value1, value2]}\nparameters = {'penalty': ['l2'], 'C': [0.01, 0.1, 1, 10, 100], 'max_iter': [500, 1000, 1500], 'verbose': [1, 5, 10]}\n\n# TODO: Criar um objeto fbeta_score utilizando make_scorer()\nscorer = make_scorer(fbeta_score, beta=0.5)\n\n# TODO: Realizar uma busca grid no classificador utilizando o 'scorer' como o método de score no GridSearchCV() \ngrid_obj = GridSearchCV(clf, parameters, scorer)\n\n# TODO: Adequar o objeto da busca grid como os dados para treinamento e encontrar os parâmetros ótimos utilizando fit() \ngrid_fit = grid_obj.fit(X_train, y_train)\n\n# Recuperar o estimador\nbest_clf = grid_fit.best_estimator_\n\n# Realizar predições utilizando o modelo não otimizado e modelar\npredictions = (clf.fit(X_train, y_train)).predict(X_test)\nbest_predictions = best_clf.predict(X_test)\n\n# Reportar os scores de antes e de depois\nprint \"Unoptimized model\\n------\"\nprint \"Accuracy score on testing data: {:.4f}\".format(accuracy_score(y_test, predictions))\nprint \"F-score on testing data: {:.4f}\".format(fbeta_score(y_test, predictions, beta = 0.5))\nprint \"\\nOptimized Model\\n------\"\nprint \"Final accuracy score on the testing data: {:.4f}\".format(accuracy_score(y_test, best_predictions))\nprint \"Final F-score on the testing data: {:.4f}\".format(fbeta_score(y_test, best_predictions, beta = 0.5))",
"/home/aron/.local/lib/python2.7/site-packages/sklearn/grid_search.py:42: DeprecationWarning: This module was deprecated in version 0.18 in favor of the model_selection module into which all the refactored classes and functions are moved. This module will be removed in 0.20.\n DeprecationWarning)\n"
]
],
[
[
"### Questão 5 - Validação final do modelo\n\n* Qual é a accuracy e o F-score do modelo otimizado utilizando os dados de testes?\n* Estes scores são melhores ou piores do que o modelo antes da otimização? \n* Como os resultados do modelo otimizado se comparam aos benchmarks do naive predictor que você encontrou na **Questão 1**?_\n\n**Nota:** Preencha a tabela abaixo com seus resultados e então responda as questões no campo **Resposta** ",
"_____no_output_____"
],
[
"#### Resultados:\n\n| Metric | Unoptimized Model | Optimized Model |\n| :------------: | :---------------: | :-------------: | \n| Accuracy Score | 0.8419 | 0.8420 |\n| F-score | 0.6832 | 0.6842 |\n",
"_____no_output_____"
],
[
"**Resposta: **\n\n- O Accuracy e F-score do modelo otimizado utilizando os dados de de teste são 0.8420 e 0.6842 respectivamente.\n- As precisões são melhores com o modelo otimizado. Teve uma pequeno aumento tanto no accuracy e F-score.\n- O Accuracy e F-score encontrados na Questão 1 é muito inferior aos do modelo otimizado, sendo que o modelo otimizado é mais que o dobro do que encontrado na Questão 1, uma melhoria muito significativa.",
"_____no_output_____"
],
[
"----\n## Importância dos atributos\n\nUma tarefa importante quando realizamos aprendizado supervisionado em um conjunto de dados como os dados do censo que estudamos aqui é determinar quais atributos fornecem maior poder de predição. Focando no relacionamento entre alguns poucos atributos mais importantes e na label alvo nós simplificamos muito o nosso entendimento do fenômeno, que é a coisa mais importante a se fazer. No caso deste projeto, isso significa que nós queremos identificar um pequeno número de atributos que possuem maior chance de predizer se um indivíduo possui renda anual superior à \\$50,000.\n\nEscolha um classificador da scikit-learn (e.x.: adaboost, random forests) que possua o atributo `feature_importance_`, que é uma função que calcula o ranking de importância dos atributos de acordo com o classificador escolhido. Na próxima célula python ajuste este classificador para o conjunto de treinamento e utilize este atributo para determinar os 5 atributos mais importantes do conjunto de dados do censo.",
"_____no_output_____"
],
[
"### Questão 6 - Observação da Relevância dos Atributos\nQuando **Exploramos os dados**, vimos que existem treze atributos disponíveis para cada registro nos dados do censo. Destes treze atributos, quais os 5 atributos que você acredita que são os mais importantes para predição e em que ordem você os ranquearia? Por quê?",
"_____no_output_____"
],
[
"**Resposta:**\n\n- 1) capital-gain - Para conseguir ter uma renda alta, é muito importante ter uma ganho de capital alto.\n- 2) capital-loss - Para conseguir ter uma renda alta, é necessário não perder dinheiro, uma perda de valor alto pode prejudicar muito sua renda.\n- 3) education-num - Representação numérica da educação, quanto maior for o numero, maior é seu grau de educação e seu salario, como por exemplo, Graduado e Doutor.\n- 4) occupation - Profissões variam muito, e seus salários também, existem profissões que tem um salario muito alto, por necessitar de um conhecimento maior, enquanto outras profissões que não necessitam muito conhecimento que recebem menos.\n- 5) age - Todas as pessoas que estão começando sua vida profissional tendem a ganhar menos, e ao passar dos anos, este começa a ganhar mais.",
"_____no_output_____"
],
[
"### Implementação - Extraindo a importância do atributo\nEscolha um algoritmo de aprendizado supervisionado da `sciki-learn` que possui o atributo `feature_importance_` disponível. Este atributo é uma função que ranqueia a importância de cada atributo dos registros do conjunto de dados quando realizamos predições baseadas no algoritmo escolhido.\n\nNa célula de código abaixo, você precisará implementar o seguinte:\n - Importar um modelo de aprendizado supervisionado da sklearn se este for diferente dos três usados anteriormente. \n - Treinar o modelo supervisionado com todo o conjunto de treinamento.\n - Extrair a importância dos atributos utilizando `'.feature_importances_'`.",
"_____no_output_____"
]
],
[
[
"# TODO: Importar um modelo de aprendizado supervisionado que tenha 'feature_importances_'\nfrom sklearn.tree import DecisionTreeClassifier\n\nmodel = DecisionTreeClassifier(random_state=8)\n\n# TODO: Treinar o modelo utilizando o conjunto de treinamento com .fit(X_train, y_train)\nmodel = model.fit(X_train, y_train)\n\n# TODO: Extrair a importância dos atributos utilizando .feature_importances_ \nimportances = model.feature_importances_\n\n# Plotar\nvs.feature_plot(importances, X_train, y_train)",
"_____no_output_____"
]
],
[
[
"### Questão 7 - Extraindo importância dos atributos\n\nObserve a visualização criada acima que exibe os cinco atributos mais relevantes para predizer se um indivíduo possui remuneração igual ou superior à \\$50,000 por ano.\n\n* Como estes cinco atributos se comparam com os 5 atributos que você discutiu na **Questão 6**? \n* Se você estivesse próximo da mesma resposta, como esta visualização confirma o seu raciocínio? \n* Se você não estava próximo, por que você acha que estes atributos são mais relevantes? ",
"_____no_output_____"
],
[
"**Resposta:**\n\nDos cinco que eu listei e dos cinco atributos mais relevantes, 3 foram iguais, são eles capital-gain, education-num e age, e os outros dois são marital-status e hours-per-week. A minha resposta esta próxima ao mostrado no gráfico acima, pois são atributos muitos importantes para conseguir uma renda acima de $50.000,00.\n\nO atributo relacionado a casamento foi uma surpresa, mas analisando, é um atributo importante, pois pessoas tente a casar quando já estão com um situação financeira maior, o que possibilita a uma renda anual alta.\n\nO atributo idade se reflete que no começo da vida profissional, se ganha menos, e são poucos os jovens que guardam dinheiro e/ou investem, isso tende a acontecer quando eles já estão com alguns anos de experiencia na vida profissional.\n\nO atributo da educação é um dos principais atributos, pois quanto mais se estuda, mais a chance de você ganhar mais, e conseguir uma renda alta.\n\nO atributo de Ganho de Capital, que podem ser investimentos, facilita muito ter uma renda anual alta, pois é um dinheiro extra, não relacionado ao trabalho do possível doador.\n\nO atributo Horas Trabalhadas por Semana demonstra que, quanto mais horas se trabalha, mais ganha, pois irá ter um salario maior, que resulta a uma ótima renda anual.",
"_____no_output_____"
],
[
"### Selecionando atributos\n\nComo um modelo performa se nós só utilizamos um subconjunto de todos os atributos disponíveis nos dados? Com menos atributos necessários para treinar, a expectativa é que o treinamento e a predição sejam executados em um tempo muito menor — com o custo da redução nas métricas de performance. A partir da visualização acima, nós vemos que os cinco atributos mais importantes contribuem para mais de 50% da importância de **todos** os atributos presentes nos dados. Isto indica que nós podemos tentar *reduzir os atributos* e simplificar a informação necessária para o modelo aprender. O código abaixo utilizará o mesmo modelo otimizado que você encontrou anteriormente e treinará o modelo com o mesmo conjunto de dados de treinamento, porém apenas com *os cinco atributos mais importantes*",
"_____no_output_____"
]
],
[
[
"# Importar a funcionalidade para clonar um modelo\nfrom sklearn.base import clone\n\n# Reduzir a quantidade de atributos\nX_train_reduced = X_train[X_train.columns.values[(np.argsort(importances)[::-1])[:5]]]\nX_test_reduced = X_test[X_test.columns.values[(np.argsort(importances)[::-1])[:5]]]\n\n# Treinar o melhor modelo encontrado com a busca grid anterior\nclf = (clone(best_clf)).fit(X_train_reduced, y_train)\n\n# Fazer novas predições\nreduced_predictions = clf.predict(X_test_reduced)\n\n# Reportar os scores do modelo final utilizando as duas versões dos dados.\nprint \"Final Model trained on full data\\n------\"\nprint \"Accuracy on testing data: {:.4f}\".format(accuracy_score(y_test, best_predictions))\nprint \"F-score on testing data: {:.4f}\".format(fbeta_score(y_test, best_predictions, beta = 0.5))\nprint \"\\nFinal Model trained on reduced data\\n------\"\nprint \"Accuracy on testing data: {:.4f}\".format(accuracy_score(y_test, reduced_predictions))\nprint \"F-score on testing data: {:.4f}\".format(fbeta_score(y_test, reduced_predictions, beta = 0.5))",
"[LibLinear]Final Model trained on full data\n------\nAccuracy on testing data: 0.8420\nF-score on testing data: 0.6842\n\nFinal Model trained on reduced data\n------\nAccuracy on testing data: 0.8271\nF-score on testing data: 0.6499\n"
]
],
[
[
"### Questão 8 - Efeitos da seleção de atributos\n\n* Como o F-score do modelo final e o accuracy score do conjunto de dados reduzido utilizando apenas cinco atributos se compara aos mesmos indicadores utilizando todos os atributos? \n* Se o tempo de treinamento é uma variável importante, você consideraria utilizar os dados enxutos como seu conjunto de treinamento? \n",
"_____no_output_____"
],
[
"**Resposta:**\n\nOs dois modelo são parecidos em comparação aos valores, pois a diferença é muito pequena, para Accuracy, é de menos de 0.02, enquanto F-score, é de menos de 0.04, uma redução aceitável se o tempo for muito importante. Mas se o tempo não é problema, ainda é melhor continuar usando todos os atributos, pois uma diferença pequena pode ser importante no resultado final.",
"_____no_output_____"
],
[
"> **Nota**: Uma vez que você tenha concluído toda a implementação de código e respondido cada uma das questões acima, você poderá finalizar o seu trabalho exportando o iPython Notebook como um documento HTML. Você pode fazer isso utilizando o menu acima navegando para \n**File -> Download as -> HTML (.html)**. Inclua este documento junto do seu notebook como sua submissão.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0f634fe82dae7a39171f076bbf03021846b035f | 24,824 | ipynb | Jupyter Notebook | week1_intro/primer/recap_tensorflow.ipynb | minori111/Practical_RL | e403a8c738574c1b52178b2fd6f196d19ffb4cea | [
"MIT"
] | 2 | 2019-02-13T15:47:11.000Z | 2019-02-26T19:50:11.000Z | week1_intro/primer/recap_tensorflow.ipynb | minori111/Practical_RL | e403a8c738574c1b52178b2fd6f196d19ffb4cea | [
"MIT"
] | 2 | 2019-02-18T19:33:36.000Z | 2019-02-19T15:03:35.000Z | week1_intro/primer/recap_tensorflow.ipynb | minori111/Practical_RL | e403a8c738574c1b52178b2fd6f196d19ffb4cea | [
"MIT"
] | 1 | 2019-02-14T21:12:04.000Z | 2019-02-14T21:12:04.000Z | 29.980676 | 347 | 0.581977 | [
[
[
"# Going deeper with Tensorflow\n\nIn this seminar, we're going to play with [Tensorflow](https://www.tensorflow.org/) and see how it helps us build deep learning models.\n\nIf you're running this notebook outside the course environment, you'll need to install tensorflow:\n* `pip install tensorflow` should install cpu-only TF on Linux & Mac OS\n* If you want GPU support from offset, see [TF install page](https://www.tensorflow.org/install/)",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\ngpu_options = tf.GPUOptions(allow_growth=True, per_process_gpu_memory_fraction=0.1)\ns = tf.InteractiveSession(config=tf.ConfigProto(gpu_options=gpu_options))",
"_____no_output_____"
]
],
[
[
"# Warming up\nFor starters, let's implement a python function that computes the sum of squares of numbers from 0 to N-1.\n* Use numpy or python\n* An array of numbers 0 to N - numpy.arange(N)",
"_____no_output_____"
]
],
[
[
"import numpy as np\ndef sum_squares(N):\n return <student.Implement_me()>",
"_____no_output_____"
],
[
"%%time\nsum_squares(10**8)",
"_____no_output_____"
]
],
[
[
"# Tensorflow teaser\n\nDoing the very same thing",
"_____no_output_____"
]
],
[
[
"#I gonna be your function parameter\nN = tf.placeholder('int64', name=\"input_to_your_function\")\n\n#i am a recipe on how to produce sum of squares of arange of N given N\nresult = tf.reduce_sum((tf.range(N)**2))",
"_____no_output_____"
],
[
"%%time\n#example of computing the same as sum_squares\nprint(result.eval({N:10**8}))",
"_____no_output_____"
]
],
[
[
"# How does it work?\n1. define placeholders where you'll send inputs;\n2. make symbolic graph: a recipe for mathematical transformation of those placeholders;\n3. compute outputs of your graph with particular values for each placeholder\n * output.eval({placeholder:value}) \n * s.run(output, {placeholder:value})\n\n* So far there are two main entities: \"placeholder\" and \"transformation\"\n* Both can be numbers, vectors, matrices, tensors, etc.\n* Both can be int32/64, floats of booleans (uint8) of various size.\n\n* You can define new transformations as an arbitrary operation on placeholders and other transformations\n * tf.reduce_sum(tf.arange(N)\\**2) are 3 sequential transformations of placeholder N\n * There's a tensorflow symbolic version for every numpy function\n * `a+b, a/b, a**b, ...` behave just like in numpy\n * np.mean -> tf.reduce_mean\n * np.arange -> tf.range\n * np.cumsum -> tf.cumsum\n * If if you can't find the op you need, see the [docs](https://www.tensorflow.org/api_docs/python).\n \n \nStill confused? We gonna fix that.",
"_____no_output_____"
]
],
[
[
"#Default placeholder that can be arbitrary float32 scalar, vector, matrix, etc.\narbitrary_input = tf.placeholder('float32')\n\n#Input vector of arbitrary length\ninput_vector = tf.placeholder('float32',shape=(None,))\n\n#Input vector that _must_ have 10 elements and integer type\nfixed_vector = tf.placeholder('int32',shape=(10,))\n\n#Matrix of arbitrary n_rows and 15 columns (e.g. a minibatch your data table)\ninput_matrix = tf.placeholder('float32',shape=(None,15))\n\n#You can generally use None whenever you don't need a specific shape\ninput1 = tf.placeholder('float64',shape=(None,100,None))\ninput2 = tf.placeholder('int32',shape=(None,None,3,224,224))",
"_____no_output_____"
],
[
"#elementwise multiplication\ndouble_the_vector = input_vector*2\n\n#elementwise cosine\nelementwise_cosine = tf.cos(input_vector)\n\n#difference between squared vector and vector itself\nvector_squares = input_vector**2 - input_vector\n",
"_____no_output_____"
],
[
"#Practice time: create two vectors of type float32\nmy_vector = <student.init_float32_vector()>\nmy_vector2 = <student.init_one_more_such_vector()>",
"_____no_output_____"
],
[
"#Write a transformation(recipe):\n#(vec1)*(vec2) / (sin(vec1) +1)\nmy_transformation = <student.implementwhatwaswrittenabove()>",
"_____no_output_____"
],
[
"print(my_transformation)\n#it's okay, it's a symbolic graph",
"_____no_output_____"
],
[
"#\ndummy = np.arange(5).astype('float32')\n\nmy_transformation.eval({my_vector:dummy,my_vector2:dummy[::-1]})",
"_____no_output_____"
]
],
[
[
"### Visualizing graphs\n\nIt's often useful to visualize the computation graph when debugging or optimizing. \nInteractive visualization is where tensorflow really shines as compared to other frameworks. \n\nThere's a special instrument for that, called Tensorboard. You can launch it from console:\n\n```tensorboard --logdir=/tmp/tboard --port=7007```\n\nIf you're pathologically afraid of consoles, try this:\n\n```os.system(\"tensorboard --logdir=/tmp/tboard --port=7007 &\"```\n\n_(but don't tell anyone we taught you that)_",
"_____no_output_____"
]
],
[
[
"# launch tensorflow the ugly way, uncomment if you need that\nimport os\nport = 6000 + os.getuid()\nprint(\"Port: %d\" % port)\n#!killall tensorboard\nos.system(\"tensorboard --logdir=./tboard --port=%d &\" % port)\n\n# show graph to tensorboard\nwriter = tf.summary.FileWriter(\"./tboard\", graph=tf.get_default_graph())\nwriter.close()",
"_____no_output_____"
]
],
[
[
"One basic functionality of tensorboard is drawing graphs. Once you've run the cell above, go to `localhost:7007` in your browser and switch to _graphs_ tab in the topbar. \n\nHere's what you should see:\n\n<img src=\"https://s12.postimg.org/a374bmffx/tensorboard.png\" width=480>\n\nTensorboard also allows you to draw graphs (e.g. learning curves), record images & audio ~~and play flash games~~. This is useful when monitoring learning progress and catching some training issues.\n\nOne researcher said:\n```\nIf you spent last four hours of your worktime watching as your algorithm prints numbers and draws figures, you're probably doing deep learning wrong.\n```",
"_____no_output_____"
],
[
"You can read more on tensorboard usage [here](https://www.tensorflow.org/get_started/graph_viz)",
"_____no_output_____"
],
[
"# Do It Yourself\n\n__[2 points max]__",
"_____no_output_____"
]
],
[
[
"# Quest #1 - implement a function that computes a mean squared error of two input vectors\n# Your function has to take 2 vectors and return a single number\n\n<student.define_inputs_and_transformations()>\n\nmse =<student.define_transformation()>\n\ncompute_mse = lambda vector1, vector2: <how to run you graph?>",
"_____no_output_____"
],
[
"# Tests\nfrom sklearn.metrics import mean_squared_error\n\nfor n in [1,5,10,10**3]:\n \n elems = [np.arange(n),np.arange(n,0,-1), np.zeros(n),\n np.ones(n),np.random.random(n),np.random.randint(100,size=n)]\n \n for el in elems:\n for el_2 in elems:\n true_mse = np.array(mean_squared_error(el,el_2))\n my_mse = compute_mse(el,el_2)\n if not np.allclose(true_mse,my_mse):\n print('Wrong result:')\n print('mse(%s,%s)' % (el,el_2))\n print(\"should be: %f, but your function returned %f\" % (true_mse,my_mse))\n raise ValueError(\"Что-то не так\")\n\nprint(\"All tests passed\") ",
"_____no_output_____"
]
],
[
[
"# variables\n\nThe inputs and transformations have no value outside function call. This isn't too comfortable if you want your model to have parameters (e.g. network weights) that are always present, but can change their value over time.\n\nTensorflow solves this with `tf.Variable` objects.\n* You can assign variable a value at any time in your graph\n* Unlike placeholders, there's no need to explicitly pass values to variables when `s.run(...)`-ing\n* You can use variables the same way you use transformations \n ",
"_____no_output_____"
]
],
[
[
"#creating shared variable\nshared_vector_1 = tf.Variable(initial_value=np.ones(5))",
"_____no_output_____"
],
[
"#initialize variable(s) with initial values\ns.run(tf.global_variables_initializer())\n\n#evaluating shared variable (outside symbolic graph)\nprint(\"initial value\", s.run(shared_vector_1))\n\n# within symbolic graph you use them just as any other inout or transformation, not \"get value\" needed",
"_____no_output_____"
],
[
"#setting new value\ns.run(shared_vector_1.assign(np.arange(5)))\n\n#getting that new value\nprint(\"new value\", s.run(shared_vector_1))\n",
"_____no_output_____"
]
],
[
[
"# tf.gradients - why graphs matter\n* Tensorflow can compute derivatives and gradients automatically using the computation graph\n* Gradients are computed as a product of elementary derivatives via chain rule:\n\n$$ {\\partial f(g(x)) \\over \\partial x} = {\\partial f(g(x)) \\over \\partial g(x)}\\cdot {\\partial g(x) \\over \\partial x} $$\n\nIt can get you the derivative of any graph as long as it knows how to differentiate elementary operations",
"_____no_output_____"
]
],
[
[
"my_scalar = tf.placeholder('float32')\n\nscalar_squared = my_scalar**2\n\n#a derivative of scalar_squared by my_scalar\nderivative = tf.gradients(scalar_squared, my_scalar)[0]",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n\nx = np.linspace(-3,3)\nx_squared, x_squared_der = s.run([scalar_squared,derivative],\n {my_scalar:x})\n\nplt.plot(x, x_squared,label=\"x^2\")\nplt.plot(x, x_squared_der, label=\"derivative\")\nplt.legend();",
"_____no_output_____"
]
],
[
[
"# Why that rocks",
"_____no_output_____"
]
],
[
[
"my_vector = tf.placeholder('float32',[None])\n\n#Compute the gradient of the next weird function over my_scalar and my_vector\n#warning! Trying to understand the meaning of that function may result in permanent brain damage\n\nweird_psychotic_function = tf.reduce_mean((my_vector+my_scalar)**(1+tf.nn.moments(my_vector,[0])[1]) + 1./ tf.atan(my_scalar))/(my_scalar**2 + 1) + 0.01*tf.sin(2*my_scalar**1.5)*(tf.reduce_sum(my_vector)* my_scalar**2)*tf.exp((my_scalar-4)**2)/(1+tf.exp((my_scalar-4)**2))*(1.-(tf.exp(-(my_scalar-4)**2))/(1+tf.exp(-(my_scalar-4)**2)))**2\n\nder_by_scalar = <student.compute_grad_over_scalar()>\nder_by_vector = <student.compute_grad_over_vector()>",
"_____no_output_____"
],
[
"#Plotting your derivative\nscalar_space = np.linspace(1, 7, 100)\n\ny = [s.run(weird_psychotic_function, {my_scalar:x, my_vector:[1, 2, 3]})\n for x in scalar_space]\n\nplt.plot(scalar_space, y, label='function')\n\ny_der_by_scalar = [s.run(der_by_scalar, {my_scalar:x, my_vector:[1, 2, 3]})\n for x in scalar_space]\n\nplt.plot(scalar_space, y_der_by_scalar, label='derivative')\nplt.grid()\nplt.legend();",
"_____no_output_____"
]
],
[
[
"# Almost done - optimizers\n\nWhile you can perform gradient descent by hand with automatic grads from above, tensorflow also has some optimization methods implemented for you. Recall momentum & rmsprop?",
"_____no_output_____"
]
],
[
[
"y_guess = tf.Variable(np.zeros(2,dtype='float32'))\ny_true = tf.range(1,3,dtype='float32')\n\nloss = tf.reduce_mean((y_guess - y_true + tf.random_normal([2]))**2) \n\noptimizer = tf.train.MomentumOptimizer(0.01,0.9).minimize(loss,var_list=y_guess)\n\n#same, but more detailed:\n#updates = [[tf.gradients(loss,y_guess)[0], y_guess]]\n#optimizer = tf.train.MomentumOptimizer(0.01,0.9).apply_gradients(updates)",
"_____no_output_____"
],
[
"from IPython.display import clear_output\n\ns.run(tf.global_variables_initializer())\n\nguesses = [s.run(y_guess)]\n\nfor _ in range(100):\n s.run(optimizer)\n guesses.append(s.run(y_guess))\n \n clear_output(True)\n plt.plot(*zip(*guesses),marker='.')\n plt.scatter(*s.run(y_true),c='red')\n plt.show()",
"_____no_output_____"
]
],
[
[
"# Logistic regression example\nImplement the regular logistic regression training algorithm\n\nTips:\n* Use a shared variable for weights\n* X and y are potential inputs\n* Compile 2 functions:\n * `train_function(X, y)` - returns error and computes weights' new values __(through updates)__\n * `predict_fun(X)` - just computes probabilities (\"y\") given data\n \n \nWe shall train on a two-class MNIST dataset\n* please note that target `y` are `{0,1}` and not `{-1,1}` as in some formulae",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import load_digits\nmnist = load_digits(2)\n\nX,y = mnist.data, mnist.target\n\nprint(\"y [shape - %s]:\" % (str(y.shape)), y[:10])\nprint(\"X [shape - %s]:\" % (str(X.shape)))",
"_____no_output_____"
],
[
"print('X:\\n',X[:3,:10])\nprint('y:\\n',y[:10])\nplt.imshow(X[0].reshape([8,8]))",
"_____no_output_____"
],
[
"# inputs and shareds\nweights = <student.code_variable()>\ninput_X = <student.code_placeholder()>\ninput_y = <student.code_placeholder()>",
"_____no_output_____"
],
[
"predicted_y = <predicted probabilities for input_X>\nloss = <logistic loss (scalar, mean over sample)>\n\noptimizer = <optimizer that minimizes loss>",
"_____no_output_____"
],
[
"train_function = <compile function that takes X and y, returns log loss and updates weights>\npredict_function = <compile function that takes X and computes probabilities of y>",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y)",
"_____no_output_____"
],
[
"from sklearn.metrics import roc_auc_score\n\nfor i in range(5):\n <run optimizer operation>\n loss_i = <compute loss at iteration i>\n \n print(\"loss at iter %i:%.4f\" % (i, loss_i))\n \n print(\"train auc:\",roc_auc_score(y_train, predict_function(X_train)))\n print(\"test auc:\",roc_auc_score(y_test, predict_function(X_test)))\n\n \nprint (\"resulting weights:\")\nplt.imshow(shared_weights.get_value().reshape(8, -1))\nplt.colorbar();",
"_____no_output_____"
]
],
[
[
"# Bonus: my1stNN\nYour ultimate task for this week is to build your first neural network [almost] from scratch and pure tensorflow.\n\nThis time you will same digit recognition problem, but at a larger scale\n* images are now 28x28\n* 10 different digits\n* 50k samples\n\nNote that you are not required to build 152-layer monsters here. A 2-layer (one hidden, one output) NN should already have ive you an edge over logistic regression.\n\n__[bonus score]__\nIf you've already beaten logistic regression with a two-layer net, but enthusiasm still ain't gone, you can try improving the test accuracy even further! The milestones would be 95%/97.5%/98.5% accuraсy on test set.\n\n__SPOILER!__\nAt the end of the notebook you will find a few tips and frequently made mistakes. If you feel enough might to shoot yourself in the foot without external assistance, we encourage you to do so, but if you encounter any unsurpassable issues, please do look there before mailing us.",
"_____no_output_____"
]
],
[
[
"from mnist import load_dataset\n\n#[down]loading the original MNIST dataset.\n#Please note that you should only train your NN on _train sample,\n# _val can be used to evaluate out-of-sample error, compare models or perform early-stopping\n# _test should be hidden under a rock untill final evaluation... But we both know it is near impossible to catch you evaluating on it.\nX_train,y_train,X_val,y_val,X_test,y_test = load_dataset()\n\nprint (X_train.shape,y_train.shape)",
"_____no_output_____"
],
[
"plt.imshow(X_train[0,0])",
"_____no_output_____"
],
[
"<here you could just as well create computation graph>",
"_____no_output_____"
],
[
"<this may or may not be a good place to evaluating loss and optimizer>",
"_____no_output_____"
],
[
"<this may be a perfect cell to write a training&evaluation loop in>",
"_____no_output_____"
],
[
"<predict & evaluate on test here, right? No cheating pls.>",
"_____no_output_____"
]
],
[
[
"```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n\n# SPOILERS!\n\nRecommended pipeline\n\n* Adapt logistic regression from previous assignment to classify some number against others (e.g. zero vs nonzero)\n* Generalize it to multiclass logistic regression.\n - Either try to remember lecture 0 or google it.\n - Instead of weight vector you'll have to use matrix (feature_id x class_id)\n - softmax (exp over sum of exps) can implemented manually or as T.nnet.softmax (stable)\n - probably better to use STOCHASTIC gradient descent (minibatch)\n - in which case sample should probably be shuffled (or use random subsamples on each iteration)\n* Add a hidden layer. Now your logistic regression uses hidden neurons instead of inputs.\n - Hidden layer uses the same math as output layer (ex-logistic regression), but uses some nonlinearity (sigmoid) instead of softmax\n - You need to train both layers, not just output layer :)\n - Do not initialize layers with zeros (due to symmetry effects). A gaussian noize with small sigma will do.\n - 50 hidden neurons and a sigmoid nonlinearity will do for a start. Many ways to improve. \n - In ideal casae this totals to 2 .dot's, 1 softmax and 1 sigmoid\n - __make sure this neural network works better than logistic regression__\n \n* Now's the time to try improving the network. Consider layers (size, neuron count), nonlinearities, optimization methods, initialization - whatever you want, but please avoid convolutions for now.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0f663d4f388f85f8febeefc6017442fb996fe3b | 2,833 | ipynb | Jupyter Notebook | nb/2. Perceptron (vector-input).ipynb | tivvit/nn-basics | fd7d2993c3d4632c580c37dce18f9e4994fa4258 | [
"MIT"
] | null | null | null | nb/2. Perceptron (vector-input).ipynb | tivvit/nn-basics | fd7d2993c3d4632c580c37dce18f9e4994fa4258 | [
"MIT"
] | null | null | null | nb/2. Perceptron (vector-input).ipynb | tivvit/nn-basics | fd7d2993c3d4632c580c37dce18f9e4994fa4258 | [
"MIT"
] | null | null | null | 16.964072 | 62 | 0.44264 | [
[
[
"# Multi input perceptron",
"_____no_output_____"
]
],
[
[
"w = [.5, .8, .75]\nb = 3\nx = [3, 4, 4]",
"_____no_output_____"
]
],
[
[
"## Python",
"_____no_output_____"
]
],
[
[
"%time\ns = 0\nfor i, xi in enumerate(x):\n s += (w[i] * xi)\nr = s + b\nr",
"CPU times: user 0 ns, sys: 0 ns, total: 0 ns\nWall time: 16 µs\n"
]
],
[
[
"## Numpy",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"%time\nnp.add(np.dot(w, x), b)",
"CPU times: user 0 ns, sys: 0 ns, total: 0 ns\nWall time: 14.3 µs\n"
]
],
[
[
"## Tensorflow",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"%time\nwith tf.Session() as sess:\n res = tf.add(tf.reduce_sum(tf.multiply(w, x)), b)\n print(sess.run(res))",
"CPU times: user 0 ns, sys: 0 ns, total: 0 ns\nWall time: 17.6 µs\n10.7\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0f67741d63c230c27dd222fe3080ec4d8e3e630 | 19,189 | ipynb | Jupyter Notebook | Notebooks/VAE_clustering_empty.ipynb | lasofivec/dataflowr | 68697f57968c4b8bc2669375f257a55057125f96 | [
"Apache-2.0"
] | null | null | null | Notebooks/VAE_clustering_empty.ipynb | lasofivec/dataflowr | 68697f57968c4b8bc2669375f257a55057125f96 | [
"Apache-2.0"
] | null | null | null | Notebooks/VAE_clustering_empty.ipynb | lasofivec/dataflowr | 68697f57968c4b8bc2669375f257a55057125f96 | [
"Apache-2.0"
] | null | null | null | 38.149105 | 524 | 0.562822 | [
[
[
"# VAE for MNIST clustering and generation\n\nThe goal of this notebook is to explore some recent works dealing with variational auto-encoder (VAE).\n\nWe will use MNIST dataset and a basic VAE architecture. ",
"_____no_output_____"
]
],
[
[
"import os\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torchvision\nfrom torchvision import transforms\nfrom torchvision.utils import save_image\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nfrom sklearn.metrics.cluster import normalized_mutual_info_score\n\ndef show(img):\n npimg = img.numpy()\n plt.imshow(np.transpose(npimg, (1,2,0)), interpolation='nearest')",
"_____no_output_____"
],
[
"# Device configuration\ndevice = torch.device('cuda' if torch.cuda.is_available() else 'cpu')\n\n# Create a directory if not exists\nsample_dir = 'samples'\nif not os.path.exists(sample_dir):\n os.makedirs(sample_dir)",
"_____no_output_____"
],
[
"batch_size = 128\n#to be modified\ndata_dir = '/home/mlelarge/data'\n# MNIST dataset\ndataset = torchvision.datasets.MNIST(root=data_dir,\n train=True,\n transform=transforms.ToTensor(),\n download=True)\n\n# Data loader\ndata_loader = torch.utils.data.DataLoader(dataset=dataset,\n batch_size=batch_size, \n shuffle=True)\n\ntest_loader = torch.utils.data.DataLoader(\n torchvision.datasets.MNIST(data_dir, train=False, download=True, transform=transforms.ToTensor()),\n batch_size=10, shuffle=False)",
"_____no_output_____"
]
],
[
[
"# Variational Autoencoders\n\nConsider a latent variable model with a data variable $x\\in \\mathcal{X}$ and a latent variable $z\\in \\mathcal{Z}$, $p(z,x) = p(z)p_\\theta(x|z)$. Given the data $x_1,\\dots, x_n$, we want to train the model by maximizing the marginal log-likelihood:\n\\begin{eqnarray*}\n\\mathcal{L} = \\mathbf{E}_{p_d(x)}\\left[\\log p_\\theta(x)\\right]=\\mathbf{E}_{p_d(x)}\\left[\\log \\int_{\\mathcal{Z}}p_{\\theta}(x|z)p(z)dz\\right],\n \\end{eqnarray*}\n where $p_d$ denotes the empirical distribution of $X$: $p_d(x) =\\frac{1}{n}\\sum_{i=1}^n \\delta_{x_i}(x)$.\n\n To avoid the (often) difficult computation of the integral above, the idea behind variational methods is to instea maximize a lower bound to the log-likelihood:\n \\begin{eqnarray*}\n\\mathcal{L} \\geq L(p_\\theta(x|z),q(z|x)) =\\mathbf{E}_{p_d(x)}\\left[\\mathbf{E}_{q(z|x)}\\left[\\log p_\\theta(x|z)\\right]-\\mathrm{KL}\\left( q(z|x)||p(z)\\right)\\right].\n \\end{eqnarray*}\n Any choice of $q(z|x)$ gives a valid lower bound. Variational autoencoders replace the variational posterior $q(z|x)$ by an inference network $q_{\\phi}(z|x)$ that is trained together with $p_{\\theta}(x|z)$ to jointly maximize $L(p_\\theta,q_\\phi)$. The variational posterior $q_{\\phi}(z|x)$ is also called the encoder and the generative model $p_{\\theta}(x|z)$, the decoder or generator.\n\nThe first term $\\mathbf{E}_{q(z|x)}\\left[\\log p_\\theta(x|z)\\right]$ is the negative reconstruction error. Indeed under a gaussian assumption i.e. $p_{\\theta}(x|z) = \\mathcal{N}(\\mu_{\\theta}(z), 1)$ the term $\\log p_\\theta(x|z)$ reduced to $\\propto \\|x-\\mu_\\theta(z)\\|^2$, which is often used in practice. The term $\\mathrm{KL}\\left( q(z|x)||p(z)\\right)$ can be seen as a regularization term, where the variational posterior $q_\\phi(z|x)$ should be matched to the prior $p(z)= \\mathcal{N}(0,1)$.\n\nVariational Autoencoders were introduced by [Kingma and Welling](https://arxiv.org/abs/1312.6114), see also [Doersch](https://arxiv.org/abs/1606.05908) for a tutorial.\n\nThere are vairous examples of VAE in pytorch available [here](https://github.com/pytorch/examples/tree/master/vae) or [here](https://github.com/yunjey/pytorch-tutorial/blob/master/tutorials/03-advanced/variational_autoencoder/main.py#L38-L65). The code below is taken from this last source.",
"_____no_output_____"
]
],
[
[
"# Hyper-parameters\nimage_size = 784\nh_dim = 400\nz_dim = 20\nnum_epochs = 15\nlearning_rate = 1e-3\n\n# VAE model\nclass VAE(nn.Module):\n def __init__(self, image_size=784, h_dim=400, z_dim=20):\n super(VAE, self).__init__()\n self.fc1 = nn.Linear(image_size, h_dim)\n self.fc2 = nn.Linear(h_dim, z_dim)\n self.fc3 = nn.Linear(h_dim, z_dim)\n self.fc4 = nn.Linear(z_dim, h_dim)\n self.fc5 = nn.Linear(h_dim, image_size)\n \n def encode(self, x):\n h = F.relu(self.fc1(x))\n return self.fc2(h), self.fc3(h)\n \n def reparameterize(self, mu, log_var):\n std = torch.exp(log_var/2)\n eps = torch.randn_like(std)\n return mu + eps * std\n\n def decode(self, z):\n h = F.relu(self.fc4(z))\n return torch.sigmoid(self.fc5(h))\n \n def forward(self, x):\n mu, log_var = self.encode(x)\n z = self.reparameterize(mu, log_var)\n x_reconst = self.decode(z)\n return x_reconst, mu, log_var\n\nmodel = VAE().to(device)\noptimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)",
"_____no_output_____"
]
],
[
[
"Here for the loss, instead of MSE for the reconstruction loss, we take BCE. The code below is still from the pytorch tutorial (with minor modifications to avoid warnings!).",
"_____no_output_____"
]
],
[
[
"# Start training\nfor epoch in range(num_epochs):\n for i, (x, _) in enumerate(data_loader):\n # Forward pass\n x = x.to(device).view(-1, image_size)\n x_reconst, mu, log_var = model(x)\n \n # Compute reconstruction loss and kl divergence\n # For KL divergence, see Appendix B in VAE paper\n reconst_loss = F.binary_cross_entropy(x_reconst, x, reduction='sum')\n kl_div = - 0.5 * torch.sum(1 + log_var - mu.pow(2) - log_var.exp())\n \n # Backprop and optimize\n loss = reconst_loss + kl_div\n optimizer.zero_grad()\n loss.backward()\n optimizer.step()\n \n if (i+1) % 10 == 0:\n print (\"Epoch[{}/{}], Step [{}/{}], Reconst Loss: {:.4f}, KL Div: {:.4f}\" \n .format(epoch+1, num_epochs, i+1, len(data_loader), reconst_loss.item()/batch_size, kl_div.item()/batch_size))",
"_____no_output_____"
]
],
[
[
"Let see how our network reconstructs our last batch. We display pairs of original digits and reconstructed version.",
"_____no_output_____"
]
],
[
[
"mu, _ = model.encode(x) \nout = model.decode(mu)\nx_concat = torch.cat([x.view(-1, 1, 28, 28), out.view(-1, 1, 28, 28)], dim=3)\nout_grid = torchvision.utils.make_grid(x_concat).cpu().data\nshow(out_grid)",
"_____no_output_____"
]
],
[
[
"Let see now, how our network generates new samples.",
"_____no_output_____"
]
],
[
[
"with torch.no_grad():\n z = torch.randn(16, z_dim).to(device)\n out = model.decode(z).view(-1, 1, 28, 28)\n\nout_grid = torchvision.utils.make_grid(out).cpu()\nshow(out_grid)",
"_____no_output_____"
]
],
[
[
"Not great, but we did not train our network for long... That being said, we have no control of the generated digits. In the rest of this jupyter, we explore ways to generates zeros, ones, twos and so on. As a by product, we show how our VAE will allow us to do clustering.\n\nThe main idea is to build what we call a Gumbel VAE as described below.",
"_____no_output_____"
],
[
"# Gumbel VAE\n\nImplement a VAE where you add a categorical variable $c\\in \\{0,\\dots 9\\}$ so that your latent variable model is $p(c,z,x) = p(c)p(z)p_{\\theta}(x|,c,z)$ and your variational posterior is $q_{\\phi}(c|x)q_{\\phi}(z|x)$ as described in this NIPS [paper](https://arxiv.org/abs/1804.00104). Make minimal modifications to previous architecture...\n\nThe idea is that you incorporates a categorical variable in your latent space. You hope that this categorical variable will encode the class of the digit, so that your network can use it for a better reconstruction. Moreover, if things work as planed, you will then be able to generate digits conditionally to the class, i.e. you can choose the class thanks to the latent categorical variable $c$ and then generate digits from this class.\n\nAs noticed above, in order to sample random variables while still being able to use backpropagation required us to use the reparameterization trick which is easy for Gaussian random variables. For categorical random variables, the reparameterization trick is explained in this [paper](https://arxiv.org/abs/1611.01144). This is implemented in pytorch thanks to [F.gumbel_softmax](https://pytorch.org/docs/stable/nn.html?highlight=gumbel_softmax#torch.nn.functional.gumbel_softmax)",
"_____no_output_____"
]
],
[
[
"n_classes = 10\n\nclass VAE_Gumbel(nn.Module):\n def __init__(self, image_size=784, h_dim=400, z_dim=20, n_classes = 10):\n super(VAE_Gumbel, self).__init__()\n #\n # your code here\n #\n \n \n def encode(self, x):\n #\n # your code here / use F.log_softmax\n #\n \n \n def reparameterize(self, mu, log_var):\n std = torch.exp(log_var/2)\n eps = torch.randn_like(std)\n return mu + eps * std\n\n def decode(self, z, y_onehot):\n #\n # your code here / use torch.cat \n #\n \n \n def forward(self, x):\n #\n # your code here / use F.gumbel_softmax\n #\n \n\nmodel_G = VAE_Gumbel().to(device)\noptimizer = torch.optim.Adam(model_G.parameters(), lr=learning_rate)",
"_____no_output_____"
]
],
[
[
"You need to modify the loss to take into account the categorical random variable with an uniform prior on $\\{0,\\dots 9\\}$, see Appendix A.2 in the NIPS [paper](https://arxiv.org/abs/1804.00104)",
"_____no_output_____"
]
],
[
[
"def train_G(model, data_loader=data_loader,num_epochs=num_epochs, beta = 1., verbose=True):\n nmi_scores = []\n model.train(True)\n for epoch in range(num_epochs):\n all_labels = []\n all_labels_est = []\n for i, (x, labels) in enumerate(data_loader):\n # Forward pass\n x = x.to(device).view(-1, image_size)\n #\n # your code here\n #\n \n reconst_loss = F.binary_cross_entropy(x_reconst, x, reduction='sum')\n #\n # your code here\n #\n\n # Backprop and optimize\n loss = # your code here\n optimizer.zero_grad()\n loss.backward()\n optimizer.step()\n \n if verbose:\n if (i+1) % 10 == 0:\n print (\"Epoch[{}/{}], Step [{}/{}], Reconst Loss: {:.4f}, KL Div: {:.4f}, Entropy: {:.4f}\" \n .format(epoch+1, num_epochs, i+1, len(data_loader), reconst_loss.item()/batch_size,\n kl_div.item()/batch_size, H_cat.item()/batch_size))",
"_____no_output_____"
],
[
"train_G(model_G,num_epochs=10,verbose=True)",
"_____no_output_____"
],
[
"x,_ = next(iter(data_loader))\nx = x[:24,:,:,:].to(device)\nout, _, _, log_p = model_G(x.view(-1, image_size)) \nx_concat = torch.cat([x.view(-1, 1, 28, 28), out.view(-1, 1, 28, 28)], dim=3)\nout_grid = torchvision.utils.make_grid(x_concat).cpu().data\nshow(out_grid)",
"_____no_output_____"
]
],
[
[
"This was for reconstruction, but we care more about generation. For each category, we are generating 8 samples thanks to the following matrix, so that in the end, we should have on each line only one digit represented.",
"_____no_output_____"
]
],
[
[
"matrix = np.zeros((8,n_classes))\nmatrix[:,0] = 1\nfinal = matrix[:]\nfor i in range(1,n_classes):\n final = np.vstack((final,np.roll(matrix,i)))",
"_____no_output_____"
],
[
"with torch.no_grad():\n z = torch.randn(8*n_classes, z_dim).to(device)\n y_onehot = torch.tensor(final).type(torch.FloatTensor).to(device)\n out = model_G.decode(z,y_onehot).view(-1, 1, 28, 28)\n\nout_grid = torchvision.utils.make_grid(out).cpu()\nshow(out_grid)",
"_____no_output_____"
]
],
[
[
"It does not look like our original idea is working...\n\nTo check that our network is not using the categorical variable, we can track the [normalized mutual information](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.normalized_mutual_info_score.html) between the true labels and the labels 'predicted' by our network (just by taking the category with maximal probability). Change your training loop to return the normalized mutual information (NMI) for each epoch. Plot the curve to check that the NMI is actually decreasing.",
"_____no_output_____"
],
[
"In order to force our network to use the categorical variable, we will change the loss following this ICLR [paper](https://openreview.net/forum?id=Sy2fzU9gl)\n\nImplement this change in the training loop and plot the new NMI curve after 10 epochs. For $\\beta = 20$, you should see that NMI increases. But reconstruction starts to be bad and generation is still poor.\n\nThis is explained in this [paper](https://arxiv.org/abs/1804.03599) and a solution is proposed see Section 5. Implement the solution described in Section 3 equation (7) if the NIPS [paper](https://arxiv.org/abs/1804.00104) ",
"_____no_output_____"
]
],
[
[
"model_G = VAE_Gumbel().to(device)\noptimizer = torch.optim.Adam(model_G.parameters(), lr=learning_rate)",
"_____no_output_____"
],
[
"def train_G_modified_loss(model, data_loader=data_loader,num_epochs=num_epochs, beta , C_z_fin, C_c_fin, verbose=True):\n #\n # your code here\n #",
"_____no_output_____"
],
[
"with torch.no_grad():\n z = torch.randn(8*n_classes, z_dim).to(device)\n y_onehot = torch.tensor(final).type(torch.FloatTensor).to(device)\n out = model_G.decode(z,y_onehot).view(-1, 1, 28, 28)\nout_grid = torchvision.utils.make_grid(out).cpu()\nshow(out_grid)",
"_____no_output_____"
],
[
"i = 1\nwith torch.no_grad():\n plt.plot()\n z = torch.randn(8, z_dim).to(device)\n y_onehot = torch.tensor(np.roll(matrix,i)).type(torch.FloatTensor).to(device)\n out = model_G.decode(z,y_onehot).view(-1, 1, 28, 28)\n out_grid = torchvision.utils.make_grid(out).cpu()\n show(out_grid)",
"_____no_output_____"
],
[
"x,_ = next(iter(data_loader))\nx = x[:24,:,:,:].to(device)\nout, _, _, log_p = model_G(x.view(-1, image_size)) \nx_concat = torch.cat([x.view(-1, 1, 28, 28), out.view(-1, 1, 28, 28)], dim=3)\nout_grid = torchvision.utils.make_grid(x_concat).cpu().data\nshow(out_grid)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0f6838063b7a01a5eb5908029de5838e0b10ca2 | 2,812 | ipynb | Jupyter Notebook | examples/EmbeddedFeatures.ipynb | igvteam/igv-notebook | 1d85c58b21dda0e34589939d5be2191333bf501f | [
"MIT"
] | null | null | null | examples/EmbeddedFeatures.ipynb | igvteam/igv-notebook | 1d85c58b21dda0e34589939d5be2191333bf501f | [
"MIT"
] | null | null | null | examples/EmbeddedFeatures.ipynb | igvteam/igv-notebook | 1d85c58b21dda0e34589939d5be2191333bf501f | [
"MIT"
] | null | null | null | 24.034188 | 127 | 0.382646 | [
[
[
"# Embedded features example\n\n**Example illustrates embedding features directly in an IGV track configuration, as opposed to loading data from a file**",
"_____no_output_____"
]
],
[
[
"!pip install igv-notebook",
"_____no_output_____"
],
[
"import igv_notebook\n\nigv_notebook.init()\n\nb = igv_notebook.Browser({\n \"genome\": \"hg19\",\n \"locus\": \"chr20:1,233,645-1,235,507\"\n})\n\nb.load_track({\n \"name\": \"Copy number\",\n \"type\": \"seg\",\n \"displayMode\": \"EXPANDED\",\n \"height\": 50,\n \"isLog\": True,\n \"features\": [\n {\n \"chr\": \"chr20\",\n \"start\": 1233820,\n \"end\": 1235000,\n \"value\": 0.8239,\n \"sample\": \"TCGA-OR-A5J2-01\"\n },\n {\n \"chr\": \"chr20\",\n \"start\": 1234500,\n \"end\": 1235180,\n \"value\": -0.8391,\n \"sample\": \"TCGA-OR-A5J3-01\"\n }\n ]\n})\n\nb.load_track({\n \"name\": \"Annotations\",\n \"type\": \"annotation\",\n \"displayMode\": \"EXPANDED\",\n \"features\": [\n {\n \"chr\": \"chr20\",\n \"start\": 1233820,\n \"end\": 1234000,\n \"name\": 'Feature 1',\n \"color\": 'rgb(100,100,0)'\n },\n {\n \"chr\": \"chr20\",\n \"start\": 1234500,\n \"end\": 1235180,\n \"name\": 'Feature 2',\n \"color\": \"red\"\n }\n ]\n})\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
]
] |
d0f683a27d864225a00cd2e921cef4fe20e012c8 | 7,945 | ipynb | Jupyter Notebook | SpringSemester2021/00_Preparation/Data Science Ex 00 - Preparation.ipynb | KretschiGL/DataScienceLecture | e6bbb3efd531b08aa4757fb6e89d12e959678a44 | [
"MIT"
] | 1 | 2021-05-09T11:02:35.000Z | 2021-05-09T11:02:35.000Z | SpringSemester2021/00_Preparation/Data Science Ex 00 - Preparation.ipynb | KretschiGL/DataScienceLecture | e6bbb3efd531b08aa4757fb6e89d12e959678a44 | [
"MIT"
] | null | null | null | SpringSemester2021/00_Preparation/Data Science Ex 00 - Preparation.ipynb | KretschiGL/DataScienceLecture | e6bbb3efd531b08aa4757fb6e89d12e959678a44 | [
"MIT"
] | 1 | 2020-05-26T15:35:40.000Z | 2020-05-26T15:35:40.000Z | 30.675676 | 207 | 0.606293 | [
[
[
"# Data Science Ex 00 - Preparation",
"_____no_output_____"
],
[
"23.02.2021, Lukas Kretschmar ([email protected]) ",
"_____no_output_____"
],
[
"## Let's have some Fun with Data Science!",
"_____no_output_____"
],
[
"Welcome to Data Science.\nWe will use an interactive environment where you can mix text and code, with the awesome feature that you can execute the code.",
"_____no_output_____"
],
[
"## Pre-Installation",
"_____no_output_____"
],
[
"We will work with Anaconda.\nYou can download the package at the following location: https://www.anaconda.com/products/individual\n\nInstall the distribution for your operating system that uses **Python 3.8**.\n\n**Note:** Anaconda needs up to **6 GB** of disk space. So, make sure you have this amount of storage available on your machine.",
"_____no_output_____"
],
[
"## Installation",
"_____no_output_____"
],
[
"Please follow the installation instructions provided under the following link: https://docs.anaconda.com/anaconda/install/\n\nIf you don't want to install Anaconda just for your user profile, you can use the instrations under https://docs.anaconda.com/anaconda/install/multi-user/",
"_____no_output_____"
],
[
"### Known Issues",
"_____no_output_____"
],
[
"#### Non-unicode characters in user name (e.g., ä, ö, ü, etc.)",
"_____no_output_____"
],
[
"We have encountered problems with students having non-unicode characters in their installation path (e.g., ä, ö, ü, é, etc.).\nThis might be a problem for you if you use the default location which points to your user profile (e.g., C:\\Users\\\\[your user name]).\nPlease choose a location that does only contain ASCII characters.",
"_____no_output_____"
],
[
"**Solution:**\n- Choose a location that contains only ASCII characters\n- Install Anaconda for multiple-users (https://docs.anaconda.com/anaconda/install/multi-user/)",
"_____no_output_____"
],
[
"If you've installed Anaconda nevertheless to a \"non-suitable\" location, there exists a simple workaround.\nIn this case you have to change the default security settings on your notebook server and open the website everytime by hand (or you try to find the url that your notebook server hosted).\nYou'll find the instructions at the end of this document.",
"_____no_output_____"
],
[
"## Post-Installation",
"_____no_output_____"
],
[
"### Update",
"_____no_output_____"
],
[
"After the installation is complete, you should also run an update to ensure that all packages are up-to-date.\nTo do so, open an **Anaconda Prompt with elevated privileges (administrator rights)** and enter the following\n```\nconda update --all\n```",
"_____no_output_____"
],
[
"### Configuration",
"_____no_output_____"
],
[
"Juypter Notebooks opens the file browser in a specific directory.\nPer default, it's your *My Documents* folder.\nYou can change the starting location to a different path by editing the configuration.\nSo, the [following](https://stackoverflow.com/questions/35254852/how-to-change-the-jupyter-start-up-folder) steps are only necessary, if you want Jupyter Notebooks to start from a specific location.\n\nOpen an **Anaconda Prompt** and enter the following\n```\njupyter notebook --generate-config\n```\nThis command will generate a configuration for your Jupyter installation at *C:\\Users\\yourusername\\\\.jupyter\\jupyter_notebook_config.py* (for the nerds of you - yeah, it's a python code file).\nThe location on a Mac is probably at a similar location.\nOpen the file with a text editor and search for the line\n``` python\n#c.NotebookApp.notebook_dir = ''\n```\nRemove the \\# at the beginning (this is the character for code comments) and enter the path you want Jupyter to start per default.\nAnd use / within your path and not \\\\ as it is common on windows systems.\nOtherwise the path might not work.\nYour entry should now look like\n``` python\nc.NotebookApp.notebook_dir = 'path/to/your/folder'\n```",
"_____no_output_____"
],
[
"### Change the security settings",
"_____no_output_____"
],
[
"**PLEASE NOTE: This step is only necessary, if your notebooks won't start property (e.g., installation at a location with unicode characters).**\nIf your Jupyter Lab or Jupyter Notebook starts, you must not change the security settings.",
"_____no_output_____"
],
[
"Within the configuration, you'll find the following line\n``` python\n# c.NotebookApp.token = '<generated>'\n```\nPer default, a new token is generated everytime you start a new server.\n\nNow, you can either set the token to a fixed value, like\n``` python\nc.NotebookApp.token = 'ffed3a68-f5b2-47a3-bb11-df8711c5aab3'\n```\n*Note: This is just an example. You can choose your own token value.*\n\nor to none (security is disabled)\n``` python\nc.NotebookApp.token = ''\n```\n\nIn the first case, your server will always run at\n- **JupyterLab:** http://localhost:8888/lab?token=ffed3a68-f5b2-47a3-bb11-df8711c5aab3\n- **Jupyter Notebook:** http://localhost:8888/tree?token=ffed3a68-f5b2-47a3-bb11-df8711c5aab3\n\nIn the second case, your server will always run at\n- **JupyterLab:** http://localhost:8888/lab\n- **Juypter Notebook:** http://localhost:8888/tree\n\nPlease note: The port (8888) might be incremented by 1 if 8888 is already blocked.\nThus, if http://localhost:8888/lab is already used, the next server will be hosted at http://localhost:8889/lab",
"_____no_output_____"
],
[
"### Run Anaconda",
"_____no_output_____"
],
[
"Check that your installation is running by starting **Anaconda**.\nYou should be able to get to the following screen.\n\n<img src=\"./AnacondaNavigator.png\" style=\"height:600px\" />\n\nAnd then try to start either **JupyterLab** or **Jupyter Notebook**.\nBoth tools will open a new browser tab.",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0f6852488cf1fde386b99fae4d2238bfe65b6f3 | 2,330 | ipynb | Jupyter Notebook | examples/Dataverse/UploadToDataverse.ipynb | haeussma/PyEnzyme | d3a9fcf144c0075d48458ffc63dd5d77b267ce68 | [
"BSD-2-Clause"
] | null | null | null | examples/Dataverse/UploadToDataverse.ipynb | haeussma/PyEnzyme | d3a9fcf144c0075d48458ffc63dd5d77b267ce68 | [
"BSD-2-Clause"
] | null | null | null | examples/Dataverse/UploadToDataverse.ipynb | haeussma/PyEnzyme | d3a9fcf144c0075d48458ffc63dd5d77b267ce68 | [
"BSD-2-Clause"
] | null | null | null | 26.781609 | 328 | 0.61588 | [
[
[
"# Upload to Dataverse\n\nPyEnzyme offers the upload to any Dataverse installation that supports the official [EnzymeML metadatablock](https://doi.org/10.18419/darus-2105) by utilizing the Dataverse API [PyDaRUS](https://github.com/JR-1991/pyDaRUS) to map all relevant fields and perform upload. The following steps will be done in this example:\n\n- Convert an EnzymeML spreadsheet to an `EnzymeMLDocument`\n- Upload the dataset to Dataverse",
"_____no_output_____"
]
],
[
[
"import pyenzyme as pe",
"_____no_output_____"
],
[
"# Load the EnzymeMLDocument\nenzmldoc = pe.EnzymeMLDocument.fromTemplate(\"EnzymeML_Template_Example.xlsm\")",
"_____no_output_____"
],
[
"# Upload it to Dataverse (Dataset is private)\nenzmldoc.uploadToDataverse(dataverse_name=\"playground\")",
"_____no_output_____"
]
],
[
[
"For reasons of data quality, the resulting dataset cant be viewed on the web. In order to visit examples that have utilized the method, see the [EnzymeML at Work](https://darus.uni-stuttgart.de/dataverse/enzymeml_at_work) collection.",
"_____no_output_____"
],
[
"------",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d0f688faac9f3afbe71c0b3a0aaa2e5105b10fd6 | 389,064 | ipynb | Jupyter Notebook | santana/EDA/new_featuresBruno.ipynb | joaopedromattos/DMC2020 | 85838edfc5c0a8d28e2d7863aaa8225d00173166 | [
"MIT"
] | null | null | null | santana/EDA/new_featuresBruno.ipynb | joaopedromattos/DMC2020 | 85838edfc5c0a8d28e2d7863aaa8225d00173166 | [
"MIT"
] | null | null | null | santana/EDA/new_featuresBruno.ipynb | joaopedromattos/DMC2020 | 85838edfc5c0a8d28e2d7863aaa8225d00173166 | [
"MIT"
] | 1 | 2020-08-24T22:47:36.000Z | 2020-08-24T22:47:36.000Z | 113.694915 | 23,932 | 0.778049 | [
[
[
"<h1>Table of Contents<span class=\"tocSkip\"></span></h1>\n<div class=\"toc\"><ul class=\"toc-item\"><li><span><a href=\"#My-ideia:\" data-toc-modified-id=\"My-ideia:-1\">My ideia:</a></span><ul class=\"toc-item\"><li><span><a href=\"#1\" data-toc-modified-id=\"1-1.1\">1</a></span></li><li><span><a href=\"#2\" data-toc-modified-id=\"2-1.2\">2</a></span></li><li><span><a href=\"#1\" data-toc-modified-id=\"1-1.3\">1</a></span></li></ul></li><li><span><a href=\"#Train-model\" data-toc-modified-id=\"Train-model-2\">Train model</a></span><ul class=\"toc-item\"><li><span><a href=\"#--First-normal-xgb\" data-toc-modified-id=\"--First-normal-xgb-2.1\">- First normal xgb</a></span></li></ul></li></ul></div>",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set()\nimport numpy as np\nimport pandas as pd\npd.set_option('display.max_columns', 50)\n\nimport sys \nfrom utils import read_data, process_time, merge_data ",
"_____no_output_____"
],
[
"infos, items, orders = read_data(data_dir='../../main/datasets/') ",
"_____no_output_____"
],
[
"infos.head(20)",
"_____no_output_____"
],
[
"items.head(20)",
"_____no_output_____"
],
[
"print(orders.shape)\norders.head(20)",
"(2181955, 5)\n"
],
[
"df = merge_data(orders, items, infos)\nprocess_time(df) # Modifies in place ",
"_____no_output_____"
],
[
"print(df.shape)\ndf.head(20) ",
"(2181955, 18)\n"
],
[
"# Since data is ordered by time, this works:\nfirst_pair_week_item = df.groupby([\"itemID\"])[\"group_backwards\"].first()\nfirst_pair_week_item.head(20)",
"_____no_output_____"
],
[
"for pair_week in range(13, 0, -1):\n group = df.query(\"group_backwards == @pair_week\")\n total = group[\"order\"].sum()\n total_revenue = (group[\"salesPrice\"]*group[\"order\"]).sum()\n\n new_items = first_pair_week_item[first_pair_week_item == pair_week].index\n new_items_total = group.loc[group[\"itemID\"].isin(new_items), \"order\"].sum()\n new_items_revenue = group.loc[group[\"itemID\"].isin(new_items), [\"order\", \"salesPrice\"]]\n new_items_revenue = (new_items_revenue[\"order\"]*new_items_revenue[\"salesPrice\"]).sum()\n\n print(\"group backwards:\", pair_week, \n \"\\tamount of new items\", len(new_items), \n # \"\\n\",\n \"\\t% of sales new items:\", f\"{100*new_items_total/total:.2f} %\",\n \"\\t% of revenue new items:\", f\"{100*new_items_revenue/total_revenue:.2f}, %\",\n #\"\\n\\n\"\n )\nprint(\"New items in DMC test\", len(items)-df[\"itemID\"].nunique())",
"group backwards: 13 \tamount of new items 1431 \t% of sales new items: 100.00 % \t% of revenue new items: 100.00, %\ngroup backwards: 12 \tamount of new items 729 \t% of sales new items: 42.80 % \t% of revenue new items: 56.79, %\ngroup backwards: 11 \tamount of new items 371 \t% of sales new items: 28.23 % \t% of revenue new items: 36.18, %\ngroup backwards: 10 \tamount of new items 533 \t% of sales new items: 36.82 % \t% of revenue new items: 41.80, %\ngroup backwards: 9 \tamount of new items 785 \t% of sales new items: 47.24 % \t% of revenue new items: 52.29, %\ngroup backwards: 8 \tamount of new items 909 \t% of sales new items: 52.43 % \t% of revenue new items: 48.72, %\ngroup backwards: 7 \tamount of new items 716 \t% of sales new items: 42.38 % \t% of revenue new items: 45.98, %\ngroup backwards: 6 \tamount of new items 661 \t% of sales new items: 37.07 % \t% of revenue new items: 40.05, %\ngroup backwards: 5 \tamount of new items 785 \t% of sales new items: 46.28 % \t% of revenue new items: 45.09, %\ngroup backwards: 4 \tamount of new items 671 \t% of sales new items: 39.19 % \t% of revenue new items: 39.51, %\ngroup backwards: 3 \tamount of new items 794 \t% of sales new items: 42.00 % \t% of revenue new items: 38.68, %\ngroup backwards: 2 \tamount of new items 727 \t% of sales new items: 35.75 % \t% of revenue new items: 32.34, %\ngroup backwards: 1 \tamount of new items 728 \t% of sales new items: 34.39 % \t% of revenue new items: 36.46, %\nNew items in DMC test 623\n"
],
[
"for pair_week in range(13, 0, -1):\n group = df.query(\"group_backwards == @pair_week\")\n total = group[\"order\"].sum()\n\n new_items = first_pair_week_item[first_pair_week_item == pair_week].index\n new_items_counts = group.loc[group[\"itemID\"].isin(new_items)]\n new_items_counts = new_items_counts.groupby(\"itemID\")[\"order\"].sum().values\n pl = sns.violinplot(new_items_counts)\n plt.title(f\"pair_week {pair_week}\")\n plt.savefig(f'pair_week_{pair_week}.png', bbox_inches='tight')\n plt.show()",
"_____no_output_____"
],
[
"from tabulate import tabulate",
"_____no_output_____"
],
[
"for pair_week in range(13, 0, -1):\n group = df.query(\"group_backwards == @pair_week\")\n total = group[\"order\"].sum()\n\n new_items = first_pair_week_item[first_pair_week_item == pair_week].index\n new_items_counts = group.loc[group[\"itemID\"].isin(new_items)]\n new_items_counts = new_items_counts.groupby(\"itemID\")[\"order\"].sum()\n print(f\"pair_week {pair_week}\")\n print(tabulate(pd.DataFrame(new_items_counts.describe())))\n print(\"\\n\\n\")",
"pair_week 13\n----- ---------\ncount 1431\nmean 60.4913\nstd 235.209\nmin 1\n25% 1\n50% 3\n75% 13\nmax 3022\n----- ---------\n\n\n\npair_week 12\n----- ---------\ncount 729\nmean 63.4678\nstd 148.621\nmin 1\n25% 1\n50% 4\n75% 64\nmax 2523\n----- ---------\n\n\n\npair_week 11\n----- ---------\ncount 371\nmean 80.5067\nstd 192.055\nmin 1\n25% 1\n50% 4\n75% 58\nmax 2049\n----- ---------\n\n\n\npair_week 10\n----- ---------\ncount 533\nmean 94.1351\nstd 153.654\nmin 1\n25% 1\n50% 20\n75% 149\nmax 1575\n----- ---------\n\n\n\npair_week 9\n----- --------\ncount 785\nmean 141.001\nstd 225.788\nmin 1\n25% 8\n50% 52\n75% 203\nmax 3290\n----- --------\n\n\n\npair_week 8\n----- --------\ncount 909\nmean 155.911\nstd 288.306\nmin 1\n25% 2\n50% 56\n75% 189\nmax 2269\n----- --------\n\n\n\npair_week 7\n----- --------\ncount 716\nmean 125.041\nstd 215.195\nmin 1\n25% 7\n50% 75.5\n75% 155\nmax 2550\n----- --------\n\n\n\npair_week 6\n----- --------\ncount 661\nmean 132.702\nstd 192.326\nmin 1\n25% 6\n50% 81\n75% 175\nmax 1874\n----- --------\n\n\n\npair_week 5\n----- --------\ncount 785\nmean 147.122\nstd 230.814\nmin 1\n25% 7\n50% 81\n75% 203\nmax 3020\n----- --------\n\n\n\npair_week 4\n----- --------\ncount 671\nmean 136.903\nstd 179.701\nmin 1\n25% 4\n50% 69\n75% 207\nmax 1528\n----- --------\n\n\n\npair_week 3\n----- --------\ncount 794\nmean 142.03\nstd 229.347\nmin 1\n25% 6\n50% 72.5\n75% 203\nmax 3728\n----- --------\n\n\n\npair_week 2\n----- --------\ncount 727\nmean 135.398\nstd 183.151\nmin 1\n25% 5.5\n50% 86\n75% 202\nmax 1650\n----- --------\n\n\n\npair_week 1\n----- --------\ncount 728\nmean 141.021\nstd 221.834\nmin 1\n25% 6\n50% 70\n75% 183\nmax 2401\n----- --------\n\n\n\n"
]
],
[
[
"Therefore, around 35% of a fortnight sales is NEW ITEMS.\n\nThat is, items that never appeared in the dataset until now. Aproximately the same thing should happen for DMC (maybe a bit less)\n\n---\n\n<br>\n<br>\n<br>\n<br>\n<br>\n<br>",
"_____no_output_____"
],
[
"# My ideia:\n\n- Divide into TWO seperate problems:\n - 1) Predict the amount of items already seen with tradional approach\n - 2) Predict new items with a model dedicated to new items ONLY for our test fortnight.\n- This is further broken down into:\n\n## 1\n- Use a binary classifier to first find out if a item sold or not (given that it's not completely new)\n- If it sold, use normal model with good features (Tobias, Sasaki, Dora)\n- Otherwise, predict 0.\n\nWe will start from group backwards 11 (first data will be 10, since for 11 all null)\n\n## 2\n- Make it work somehow: start off with easy stats.\n\nLet's go!",
"_____no_output_____"
],
[
"---\n\n<br>",
"_____no_output_____"
],
[
"## 1",
"_____no_output_____"
]
],
[
[
"def create_dataset(target_week, final_test=False):\n if final_test:\n train_items = df.query('(@target_week + 1) <= group_backwards <= 13')[\"itemID\"].unique()\n full = df.query('group_backwards >= (@target_week)').reset_index(drop = True)\n else:\n train_items = df.query('(@target_week + 2) <= group_backwards <= 13')[\"itemID\"].unique()\n full = df.query('group_backwards >= (@target_week)').reset_index(drop = True)\n # only return instances where that item has already appeared.\n full = full.query(\"itemID in @train_items\")\n\n return full\n\n#full_val = create_dataset(2)\nfull_sub = create_dataset(1, final_test=True)\n\n# ok, so the proper is the way above, but this is SHIT, but turns out the other version\n# is good *m\nfull_sub = create_dataset(0, final_test=True)",
"_____no_output_____"
],
[
"X_full_val = pd.DataFrame(\n index=pd.MultiIndex.from_product([\n full_sub[\"itemID\"].unique(), \n range(13, 0, -1)\n ], names=[\"itemID\", \"group_backwards\"]\n)).reset_index()",
"_____no_output_____"
],
[
"X_full_val = pd.merge(X_full_val, items, left_on=\"itemID\", right_on=\"itemID\", validate=\"m:1\")\nX_full_val .head()",
"_____no_output_____"
]
],
[
[
"**OBS**: From now on, salesPrice and order in X_train means the SUM\n\n**OBS**: We remove it when defining x/y later on.",
"_____no_output_____"
]
],
[
[
"cols = [\"order\", \"salesPrice\"]\nextra = full_sub.groupby([\"group_backwards\", \"itemID\"], \n as_index=False)[cols].sum()\n\n\nX_full_val = pd.merge(X_full_val, extra, on=[\"group_backwards\", \"itemID\"], how=\"left\",\n validate=\"1:1\")\n\n# 0 so the features generated will make sense\nX_full_val.fillna(0, inplace=True)\nX_full_val.tail()",
"_____no_output_____"
],
[
"# X_full_val.loc[X_full_val[\"group_backwards\"] == 1, \"order\"].sum()",
"_____no_output_____"
]
],
[
[
"---\n\n<br>\n\nBrute force some features",
"_____no_output_____"
]
],
[
[
"def feat_amount_sold(groupby):\n return groupby[\"order\"].sum()\n\ndef feat_value(groupby):\n return groupby[\"salesPrice\"].sum()\n\ndef amount_of_transactions():\n return groupby.size()\n\nfunc_feats_past_time = {\n \"quantidade_vend_sum_{}\": feat_amount_sold,\n \"valor_vend_sum_{}\": feat_value,\n}\n\n# These are features generated only from the last 14 days. Currently it's the same,\n# but who knows.\nfunc_feats_past_fortnight = {\n \"quantidade_vend_sum_{}\": feat_amount_sold,\n \"valor_vend_sum_{}\": feat_value,\n}\n\n# TODO\n#func_feats_other = {\n# \"quantidade_trans_sum\": amount_of_transactions,\n#}",
"_____no_output_____"
],
[
"def apply_all_item_feats(X, past_time, func_feats):\n \"\"\"Only works for data that hasnt been grouped! ie, \"Orders\" like.\n \n Calculates features that depend on the item ID and it's derivatives\n \"\"\"\n cols = [\"itemID\", \"brand\", \"manufacturer\", \"category1\", \"category2\", \"category3\"]\n # All theses columns above depend only on the item, so we can just get the first\n feats = X.groupby([\"itemID\"]).first()\n\n for col in cols:\n groupby = X.loc[past_time].groupby(col)\n for name, func in func_feats.items():\n feat = func(groupby)\n feats = pd.merge(feats, feat.rename(name.format(col)),\n left_on=col, right_index=True)\n\n return feats.drop(columns=X.loc[past_time].columns.drop(\"itemID\"),\n errors=\"ignore\")\n ",
"_____no_output_____"
],
[
"# Test the features:\ntemp = X_full_val.query(\"group_backwards >= 12\").index\ntemp_results = apply_all_item_feats(X_full_val, temp, func_feats_past_time)\n\n# temp_results.head()\ntemp_results.loc[[450, 108, 10224]]",
"_____no_output_____"
],
[
"def generate_all(X, stop_week, start_fortnight):\n new_X = X.copy()\n new_past_cols = None\n\n # Start off with 12 because we don't generate data for the first fortnight due to time lag.\n for fortnight in range(start_fortnight, stop_week-1, -1):\n # here is the line that COULD BE LEAK if we >=\n past_time = X[\"group_backwards\"] > fortnight\n \n this_fortnight = X[\"group_backwards\"] == fortnight\n \n feats = apply_all_item_feats(X, past_time, func_feats_past_time)\n # if fortnight == stop_week: return feats\n if new_past_cols is None:\n new_past_cols = feats.columns\n for col in new_past_cols:\n new_X[col] = np.nan\n # Kind of bad, I know. There is a way a better way, but I want to sleep.\n merged = pd.merge(X.loc[this_fortnight], feats, on=\"itemID\", validate=\"m:1\",\n how=\"left\")\n new_X.loc[this_fortnight, new_past_cols] = merged[new_past_cols].values\n\n return new_X",
"_____no_output_____"
],
[
"stop_week = 1\nstart_fortnight = 8\n#start_fortnight = 5\nX_full_val_feats = generate_all(X_full_val, stop_week, start_fortnight)\nX_full_val_feats = X_full_val_feats.query(\"group_backwards <= @start_fortnight\")",
"_____no_output_____"
],
[
"X_full_val_feats.head(6)",
"_____no_output_____"
],
[
"X_full_val_feats.tail()",
"_____no_output_____"
],
[
"# Since we filled missing orders/sales price with 0, impossible to have NaN\nX_full_val_feats.isna().sum().sum()",
"_____no_output_____"
]
],
[
[
"# Train model",
"_____no_output_____"
]
],
[
[
"full_train = X_full_val_feats.query(\"group_backwards > 1\")\ntrain = X_full_val_feats.query(\"group_backwards > 2\")\nval = X_full_val_feats.query(\"group_backwards == 2\")\nsub = X_full_val_feats.query(\"group_backwards == 1\")\n\ndrop_cols = [\"order\", \"salesPrice\"]\nx_full_train = full_train.drop(columns=drop_cols)\nx_train = train.drop(columns=drop_cols)\nx_val = val.drop(columns=drop_cols)\nx_sub = sub.drop(columns=drop_cols)\n\ny_full_train = full_train[\"order\"]\ny_train = train[\"order\"]\ny_val = val[\"order\"]\ny_sub = sub[\"order\"]",
"_____no_output_____"
],
[
"weights = infos.set_index('itemID')['simulationPrice'].to_dict()\nw_full_train = full_train['itemID'].map(weights)\nw_train = train['itemID'].map(weights)\nw_val = val['itemID'].map(weights)\nw_sub = sub['itemID'].map(weights)\n\n# Create binary targets\ny_full_train_bin = (y_full_train > 0).astype(int)\ny_train_bin = (y_train > 0).astype(int)\ny_val_bin = (y_val > 0).astype(int)\ny_sub_bin = (y_sub > 0).astype(int)",
"_____no_output_____"
],
[
"# Not that good, but doable... maybe\n# for idx, (y_temp, name) in enumerate(zip([y_train_bin, y_val_bin, y_sub_bin], \n# [\"train\", \"val\", \"sub\"]), 1):\n# plt.subplot(1, 3, idx)\n# sns.countplot(y_temp)\n# plt.title(name)",
"_____no_output_____"
]
],
[
[
"<hr>",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import mean_squared_error as mse\ndef evaluate(prediction, target, simulationPrice):\n return np.sum((prediction - np.maximum(prediction - target, 0) * 1.6) * simulationPrice)",
"_____no_output_____"
]
],
[
[
"## - First normal xgb",
"_____no_output_____"
]
],
[
[
"import xgboost as xgb",
"_____no_output_____"
],
[
"xgb.__version__",
"_____no_output_____"
],
[
"# custom objective\n\ndef gradient(prediction, dtrain):\n y = dtrain.get_label()\n# prediction.astype(int)\n# prediction = np.minimum(prediction.astype(int), 1)\n return -2 * (prediction - np.maximum(prediction - y, 0) * 1.6) * (1 - (prediction > y) * 1.6)\n\ndef hessian(prediction, dtrain):\n y = dtrain.get_label()\n# prediction.prediction(int)\n# prediction = np.minimum(prediction.astype(int), 1)\n return -2 * (1 - (prediction > y) * 1.6) ** 2\n\ndef objective(prediction, dtrain):\n w = dtrain.get_weight()\n grad = gradient(prediction, dtrain) * w\n hess = hessian(prediction, dtrain) * w\n return grad, hess",
"_____no_output_____"
],
[
"# custom feval\n\ndef feval(prediction, dtrain):\n prediction = prediction.astype(int)\n# predt = np.minimum(predt.astype(int), 1)\n target = dtrain.get_label()\n simulationPrice = dtrain.get_weight()\n return 'feval', np.sum((prediction - np.maximum(prediction - target, 0) * 1.6) * simulationPrice)",
"_____no_output_____"
],
[
"dtrain = xgb.DMatrix(x_train, y_train, w_train, missing=0)\ndval = xgb.DMatrix(x_val, y_val, w_val, missing=0)\ndsub = xgb.DMatrix(x_sub, y_sub, w_sub, missing=0)\ndfulltrain = xgb.DMatrix(x_full_train, y_full_train, w_full_train, missing=0)\n# specify parameters via map\nparam = {\n 'max_depth':10,\n 'eta':0.005,\n 'objective':'reg:squarederror',\n 'disable_default_eval_metric': 1,\n# 'tree_method' : 'gpu_hist',\n}\nnum_round = 100\nbst = xgb.train(param, dtrain,\n num_round,\n early_stopping_rounds = 30,\n evals = [(dtrain, 'train'), (dval, 'val')],\n# obj = objective,\n feval = feval,\n maximize = True,\n )",
"_____no_output_____"
],
[
"prediction = bst.predict(dsub, ntree_limit=bst.best_ntree_limit).astype(int)\nevaluate(prediction, y_sub, w_sub)",
"_____no_output_____"
],
[
"# retrain!",
"_____no_output_____"
],
[
"bst_sub = xgb.train(param, dfulltrain,\n num_boost_round = bst.best_ntree_limit, early_stopping_rounds=30,\n # obj = objective,\n feval = feval, maximize = True,\n evals = [(dfulltrain, 'ftrain')],\n verbose_eval = False,\n)",
"_____no_output_____"
],
[
"prediction = bst_sub.predict(dsub, ntree_limit=bst_sub.best_ntree_limit).astype(int)\nevaluate(prediction, y_sub, w_sub)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f699dbcc060eb6c1c083435cedd6fcc7835c4d | 39,281 | ipynb | Jupyter Notebook | notebooks/book1/binom_dist_plot.ipynb | peterchang0414/pyprobml | 4f5bb63e4423ecbfc2615b5aa794f529a0439bf8 | [
"MIT"
] | null | null | null | notebooks/book1/binom_dist_plot.ipynb | peterchang0414/pyprobml | 4f5bb63e4423ecbfc2615b5aa794f529a0439bf8 | [
"MIT"
] | null | null | null | notebooks/book1/binom_dist_plot.ipynb | peterchang0414/pyprobml | 4f5bb63e4423ecbfc2615b5aa794f529a0439bf8 | [
"MIT"
] | null | null | null | 173.809735 | 8,660 | 0.908913 | [
[
[
"# Plots the pmfs of binomial distributions with varying probability of success parameter",
"_____no_output_____"
]
],
[
[
"try:\n import jax\nexcept:\n %pip install jax jaxlib\n import jax\nimport jax.numpy as jnp\n\ntry:\n import matplotlib.pyplot as plt\nexcept:\n %pip install matplotlib\n import matplotlib.pyplot as plt\n\ntry:\n import seaborn as sns\nexcept:\n %pip install seaborn\n import seaborn as sns\n\ntry:\n from scipy.stats import binom\nexcept:\n %pip install scipy\n from scipy.stats import binom",
"_____no_output_____"
],
[
"import os\n\nLATEXIFY = \"LATEXIFY\" in os.environ\n\nif LATEXIFY:\n import sys\n\n sys.path.append(\"scripts\")\n from plot_utils import latexify, savefig\n\n latexify(width_scale_factor=2, fig_height=1.5)",
"_____no_output_____"
],
[
"N = 10\nthetas = [0.25, 0.5, 0.75, 0.9]\nx = jnp.arange(0, N + 1)\n\n\ndef make_graph(data):\n plt.figure()\n x = data[\"x\"]\n n = data[\"n\"]\n theta = data[\"theta\"]\n\n probs = binom.pmf(x, n, theta)\n title = r\"$\\theta=\" + str(theta) + \"$\"\n\n plt.bar(x, probs, align=\"center\")\n plt.xlim([min(x) - 0.5, max(x) + 0.5])\n plt.ylim([0, 0.4])\n plt.xticks(x)\n plt.xlabel(\"$x$\")\n plt.ylabel(\"$p(x)$\")\n plt.title(title)\n sns.despine()\n if LATEXIFY:\n savefig(\"binomDistTheta\" + str(int(theta * 100)) + \"_latexified.pdf\")\n\n\nfor theta in thetas:\n data = {\"x\": x, \"n\": N, \"theta\": theta}\n make_graph(data)",
"WARNING:absl:No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)\n"
]
],
[
[
"## Demo\nYou can see different examples of binomial distributions by changing the theta in the following demo.",
"_____no_output_____"
]
],
[
[
"from ipywidgets import interact\n\n\n@interact(theta=(0.1, 0.9))\ndef generate_random(theta):\n n = 10\n data = {\"x\": jnp.arange(0, n + 1), \"n\": n, \"theta\": theta}\n make_graph(data)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f6bb0f10564d85e0cef7b102b0d04045554269 | 225,749 | ipynb | Jupyter Notebook | wine-qualty.ipynb | FernandoNici/Write-a-data-science-blog-post | 0f2b8c284e80a3d370868dcc351f6ea4b97eef33 | [
"FTL",
"CNRI-Python"
] | null | null | null | wine-qualty.ipynb | FernandoNici/Write-a-data-science-blog-post | 0f2b8c284e80a3d370868dcc351f6ea4b97eef33 | [
"FTL",
"CNRI-Python"
] | null | null | null | wine-qualty.ipynb | FernandoNici/Write-a-data-science-blog-post | 0f2b8c284e80a3d370868dcc351f6ea4b97eef33 | [
"FTL",
"CNRI-Python"
] | null | null | null | 419.607807 | 129,656 | 0.928053 | [
[
[
"import sys\n!{sys.executable} -m pip install numpy pandas matplotlib sklearn seaborn",
"_____no_output_____"
]
],
[
[
"### 1 - Step\n\n**1.** Looking for missing values.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\n\n#%matplotlib inline\n%matplotlib notebook \n\ndf = pd.read_csv('./winequality-red.csv')\n\ndf_missig_values = df[df.isna() == True].value_counts()\n\nprint(\"Checking if contains missing values: {}\".format(len(df_missig_values)))\nprint(\"Size: {} rows and {} features\".format(df.shape[0], df.shape[1]))",
"Checking if contains missing values: 0\nSize: 1599 rows and 12 features\n"
],
[
"df.describe()\n",
"_____no_output_____"
]
],
[
[
"#### Dataframe info\n\n- Dataframe have 1599 records\n- Data columns: total 12 columns\n- There is no missing values\n- There is no categorical feature",
"_____no_output_____"
],
[
"### 2 - Step\n\n**2.** Correlation between the features",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\n%matplotlib inline\n\nplt.subplots(figsize=(10, 8))\n\nsns.heatmap(df.corr(), annot=True, fmt=\".3f\", linewidths=.5);",
"_____no_output_____"
],
[
"# Normalize the dataframe to return proportions rather than frequencies\ndef normalize_data(dataframe, column, ascending):\n '''\n Parameters\n ----------\n dataframe: DataFrame\n Data structure with labeled axes\n column: str\n Column label to use for normalization\n ascending: bool\n Sort ascending (True) or descending (False)\n \n Returns\n -------\n dataframe: DataFrame\n Dataframe normalized with proportions according to the column\n '''\n return dataframe[column].value_counts(normalize=True).sort_values(ascending=ascending).to_frame()",
"_____no_output_____"
],
[
"plt.figure(figsize=(15,6))\n \nquality_count = normalize_data(df, 'quality', False)\nquality_count = quality_count.rename(columns={'quality':'Percentage'})\n\nax = sns.barplot(x=quality_count.index, y='Percentage', data=quality_count, palette=\"pastel\")\n\n# Annotate the point xy with number formatted like percentage\n# For more details look on https://matplotlib.org/3.3.3/api/_as_gen/matplotlib.axes.Axes.annotate.html#matplotlib.axes.Axes.annotate \nfor p in ax.patches:\n ax.annotate('{:.1f} %'.format((p.get_height() * 100)),\n (p.get_x() + p.get_width() / 2., p.get_height()),\n ha='center', va='center',\n xytext = (0,9),\n textcoords='offset points')\n \nplt.xlabel(\"Quality\", fontsize=15)\nplt.ylabel(\"Percentage\", fontsize=15)\nplt.title(\"Chart of Quality\", fontsize=20)\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"### 3 - Step\n\n**3.** Transform into percentage values",
"_____no_output_____"
]
],
[
[
"def convert_value_into_percentage(fraction_number):\n '''\n Parameters\n ----------\n fraction_number: float\n Number in decimal form\n \n Returns\n -------\n float\n the percentage calculated\n '''\n return fraction_number * 100",
"_____no_output_____"
],
[
"quality_count = normalize_data(df, 'quality', True)\n\n# I sorted to look better on the pie chart\nquality_count = quality_count.rename(columns={'quality':'Percentage'}).sort_values(by='Percentage').reindex([3, 5, 4, 7, 8, 6])\n\n# Apply the function to transform\nquality_count['Percentage'] = quality_count['Percentage'].apply(convert_value_into_percentage)\n\n\n# Building a plot\nfig1, ax1 = plt.subplots(figsize=(6, 6))\n\nwedges, texts, autotexts = ax1.pie(quality_count['Percentage'], autopct='%1.1f%%', startangle=0, textprops=dict(color=\"w\"))\n\nax1.legend(wedges, quality_count.index,\n title=\"Quality level\",\n loc=\"center left\",\n bbox_to_anchor=(1, 0, 0, 1))\n\nplt.setp(autotexts, size=10, weight=\"bold\")\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 4 - Step\n\n**4.** Looking on alcohol features distribution using box plot",
"_____no_output_____"
]
],
[
[
"f, ax = plt.subplots(1, 1, figsize=(12, 6))\nsns.despine(left=True)\n\nsns.boxplot(x=df['alcohol'])\n#sns.boxplot(x=df['citric acid'], ax=ax[1])\n#sns.boxplot(x=df['sulphates'], ax=ax[2])\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 5 - Step\n\n**5.** Looking the alcohol features distribution bases on quality",
"_____no_output_____"
]
],
[
[
"sns.catplot(x=\"quality\", y=\"alcohol\", data=df)\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f6d5fbb5f1f23cb9d9106a3df10cf0ce5e8113 | 21,980 | ipynb | Jupyter Notebook | assignments/2020/assignment3_colab/assignment3/NetworkVisualization-PyTorch.ipynb | benkmoore/cs231n.github.io | 143e8864aed5ebe6b1ffaa407faf58d60de0adb9 | [
"MIT"
] | 2 | 2021-03-03T02:49:00.000Z | 2021-09-17T06:53:13.000Z | assignments/2020/assignment3_colab/assignment3/NetworkVisualization-PyTorch.ipynb | benkmoore/cs231n.github.io | 143e8864aed5ebe6b1ffaa407faf58d60de0adb9 | [
"MIT"
] | 32 | 2020-09-17T19:43:53.000Z | 2022-03-12T00:55:26.000Z | assignments/2020/assignment3_colab/assignment3/NetworkVisualization-PyTorch.ipynb | benkmoore/cs231n.github.io | 143e8864aed5ebe6b1ffaa407faf58d60de0adb9 | [
"MIT"
] | 1 | 2020-09-24T19:57:47.000Z | 2020-09-24T19:57:47.000Z | 40.478821 | 736 | 0.62343 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0f6d982245fd47b2bf40f94ace9b7b41c208339 | 152,353 | ipynb | Jupyter Notebook | Diabetes-Data-Analysis-main/Diabetes Data Analysis.ipynb | Lilasrinivasreddy/NLP_Speech_recog | bb7797cbacd9945ad120305a135fb8e5ed3bae99 | [
"MIT"
] | null | null | null | Diabetes-Data-Analysis-main/Diabetes Data Analysis.ipynb | Lilasrinivasreddy/NLP_Speech_recog | bb7797cbacd9945ad120305a135fb8e5ed3bae99 | [
"MIT"
] | null | null | null | Diabetes-Data-Analysis-main/Diabetes Data Analysis.ipynb | Lilasrinivasreddy/NLP_Speech_recog | bb7797cbacd9945ad120305a135fb8e5ed3bae99 | [
"MIT"
] | null | null | null | 119.21205 | 35,220 | 0.840279 | [
[
[
"<h1 align=\"center\">Diabetes Data Analysis</h1>\n\nAll information regarding the features and dataset can be found in this research arcticle:\nImpact of HbA1c Measurement on Hospital Readmission Rates: Analysis of 70,000 Clinical Database Patient Records\n\nIn this project we want to know how different features affect diabetes in general.\nFor this kernel, we will be using a diabetes readmission dataset to explore the different frameworks for model explainability\n\n\nMachine learning models that can be used in the medical field should be interpretable.\nHumans should know why these models decided on a conclusion.\nThe problem is the more complex an ML model gets the less interpretable it gets.\nIn this kernel we will examine techniques and frameworks in interpreting ML models.",
"_____no_output_____"
]
],
[
[
"#import libraries\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"# read the file and create a pandas dataframe\ndata = pd.read_csv('dataset/diabetic_data.csv')",
"_____no_output_____"
],
[
"# check the dimensions of the data\ndata.shape",
"_____no_output_____"
],
[
"# first 5 rows of data\ndata.head()",
"_____no_output_____"
]
],
[
[
"we get discriptive statistics of numerical variable",
"_____no_output_____"
]
],
[
[
"#discribtion of numerical data\ndata[['time_in_hospital','num_lab_procedures','num_procedures','num_medications',\n 'number_outpatient','number_emergency','number_inpatient',\n 'number_diagnoses']].describe()",
"_____no_output_____"
],
[
"#no of unique patient\nlen(np.unique(data['patient_nbr']))",
"_____no_output_____"
]
],
[
[
"+ Remove duplicate recod based on patient_nbr column",
"_____no_output_____"
]
],
[
[
"#remove duplicate patient recods\ndata = data.drop_duplicates(subset = 'patient_nbr', keep = 'first')",
"_____no_output_____"
]
],
[
[
"+ Plot some column data",
"_____no_output_____"
]
],
[
[
"# the response variable 'readmitted' in the original dataset contains three categories.\n# 11% of patients were readmitted within 30 days (<30)\n# 35% of patients were readmitted after 30 days (>30)\n# 54% of patients were never readmitted (NO)\ndata.groupby('readmitted').size().plot(kind='bar')\nplt.ylabel('Count')\nplt.title(\"Distribution of Readmitted Data\")\nplt.show()",
"_____no_output_____"
],
[
"data['readmitted'] = pd.Series([0 if val in ['NO', '>30'] else val \n for val in data['readmitted']], index=data.index)\ndata['readmitted'] = pd.Series([1 if val in ['<30'] else val \n for val in data['readmitted']], index=data.index)",
"_____no_output_____"
],
[
"#values counts of readmited column\ndata.readmitted.value_counts()",
"_____no_output_____"
],
[
"# the response variable 'readmitted' in the original dataset contains three categories.\n# 11% of patients were readmitted within 30 days (<30)\n# 35% of patients were readmitted after 30 days (>30)\n# 54% of patients were never readmitted (NO)\ndata.groupby('readmitted').size().plot(kind='bar')\nplt.ylabel('Count')\nplt.title(\"Distribution of Readmitted Data\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"+ Drop unnecessary column from data",
"_____no_output_____"
]
],
[
[
"# remove irrelevant features\ndata.drop(['encounter_id','patient_nbr', 'weight', 'payer_code','max_glu_serum','A1Cresult'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"# remove rows that have NA in 'race', 'diag_1', 'diag_2', or 'diag_3' and 'gender'\n# remove rows that have invalid values in 'gender'\ndata = data[data['race'] != '?']\ndata = data[data['diag_1'] != '?']\ndata = data[data['diag_2'] != '?']\ndata = data[data['diag_3'] != '?']\ndata = data[data['gender'] != 'Unknown/Invalid']",
"_____no_output_____"
],
[
"# check 'age' feature\ndata.groupby('age').size().plot(kind='bar')\nplt.ylabel('Count')\nplt.title(\"Distribution of Age\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"> from above graph we see that 60 to 100 age group data is large so we make single group for this",
"_____no_output_____"
]
],
[
[
"# Recategorize 'age' so that the population is more evenly distributed\ndata['age'] = pd.Series(['[20-60)' if val in ['[20-30)', '[30-40)', '[40-50)', '[50-60)'] else val \n for val in data['age']], index=data.index)\ndata['age'] = pd.Series(['[60-100)' if val in ['[60-70)','[70-80)','[80-90)', '[90-100)'] else val \n for val in data['age']], index=data.index)\ndata.groupby('age').size().plot(kind='bar')\nplt.ylabel('Count')\nplt.title(\"Distribution of Age\")\nplt.show()",
"_____no_output_____"
],
[
"data.groupby('number_outpatient').size().plot(kind='bar')\nplt.ylabel('Count')\nplt.title(\"number_outpatient v/s Count \")\nplt.show()",
"_____no_output_____"
],
[
"data.groupby('number_emergency').size().plot(kind='bar')\nplt.title(\"number_emergency v/s Count\")\nplt.ylabel('Count')\nplt.show()",
"_____no_output_____"
],
[
"data.groupby('number_inpatient').size().plot(kind='bar')\nplt.title(\"number_inpatient v/s Count\")\nplt.ylabel('Count')\nplt.show()",
"_____no_output_____"
],
[
"# remove the other medications\ndata.drop(['metformin', 'repaglinide', 'nateglinide', 'chlorpropamide', 'glimepiride', \n 'acetohexamide', 'glipizide', 'glyburide', 'tolbutamide', 'pioglitazone', \n 'rosiglitazone', 'acarbose', 'miglitol', 'troglitazone', 'tolazamide', 'examide', \n 'citoglipton', 'glyburide-metformin', 'glipizide-metformin', 'glimepiride-pioglitazone',\n 'metformin-rosiglitazone', 'metformin-pioglitazone','insulin'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"# Recategorize 'age' so that the population is more evenly distributed\ndata['discharge_disposition_id'] = pd.Series(['Home' if val in [1] else val \n for val in data['discharge_disposition_id']], index=data.index)\n\ndata['discharge_disposition_id'] = pd.Series(['Anather' if val in [2,3,4,5,6] else val \n for val in data['discharge_disposition_id']], index=data.index)\n\ndata['discharge_disposition_id'] = pd.Series(['Expired' if val in [11,19,20,21] else val \n for val in data['discharge_disposition_id']], index=data.index)\n\ndata['discharge_disposition_id'] = pd.Series(['NaN' if val in [18,25,26] else val\n for val in data['discharge_disposition_id']], index=data.index)\n \ndata['discharge_disposition_id'] = pd.Series(['other' if val in [7,8,9,10,12,13,14,15,16,17,22,23,24,27,28,29,30] else val \n for val in data['discharge_disposition_id']], index=data.index)",
"_____no_output_____"
],
[
"# original 'admission_source_id' contains 25 levels\n# reduce 'admission_source_id' into 3 categories\ndata['admission_source_id'] = pd.Series(['Emergency Room' if val == 7 else 'Referral' if val in [1,2,3] else \n 'NaN' if val in [15,17,20,21] else 'Other source' \n for val in data['admission_source_id']], index=data.index)",
"_____no_output_____"
],
[
"# original 'admission_type_id' contains 8 levels\n# reduce 'admission_type_id' into 2 categories\ndata['admission_type_id'] = pd.Series(['Emergency' if val == 1 else 'Other type' \n for val in data['admission_type_id']], index=data.index)",
"_____no_output_____"
],
[
"# Extract codes related to heart disease\ndata = data.loc[data['diag_1'].isin(['410','411','412','413','414','415','420','421','422','423','424','425','426','427','428','429','430']) | data['diag_2'].isin(['410','411','412','413','414','415','420','421','422','423','424','425','426','427','428','429','430']) | data['diag_3'].isin(['410','411','412','413','414','415','420','421','422','423','424','425','426','427','428','429','430'])]\n",
"_____no_output_____"
],
[
"data.shape",
"_____no_output_____"
],
[
"import random\n#create variable emergency visits\ndata['emergency_visits'] = [random.randint(0, 5) for _ in range(26703)]",
"_____no_output_____"
],
[
"#create variable emergency visits\ndata['acuity_of_admission'] = [random.randint(1, 5) for _ in range(26703)]",
"_____no_output_____"
],
[
"#create variable emergency visits\ndata['comorbidity'] = [random.randint(1, 15) for _ in range(26703)]",
"_____no_output_____"
],
[
"categarical_colmun=[\"age\",\"race\",\"gender\",\"medical_specialty\",\"change\",\"diabetesMed\",\"discharge_disposition_id\",\"admission_source_id\",\"admission_type_id\",\"diag_1\",\"diag_2\",\"diag_3\"]\ndtypes = {c: 'category' for c in categarical_colmun}\ndata=data.astype(dtypes)",
"_____no_output_____"
],
[
"# conver categarical variable into categary code\nfor i in categarical_colmun:\n data[i]=data[i].cat.codes",
"_____no_output_____"
],
[
"# apply square root transformation on right skewed count data to reduce the effects of extreme values.\n# here log transformation is not appropriate because the data is Poisson distributed and contains many zero values.\ndata['number_outpatient'] = data['number_outpatient'].apply(lambda x: np.sqrt(x + 0.5))\ndata['number_emergency'] = data['number_emergency'].apply(lambda x: np.sqrt(x + 0.5))\ndata['number_inpatient'] = data['number_inpatient'].apply(lambda x: np.sqrt(x + 0.5))",
"_____no_output_____"
],
[
"# feature scaling, features are standardized to have zero mean and unit variance\nfeature_scale_cols = ['time_in_hospital', 'num_lab_procedures', 'num_procedures', 'num_medications', \n 'number_diagnoses', 'number_inpatient', 'number_emergency', 'number_outpatient']\n\nfrom sklearn import preprocessing\nscaler = preprocessing.StandardScaler().fit(data[feature_scale_cols])\ndata_scaler = scaler.transform(data[feature_scale_cols])\n\ndata_scaler_df = pd.DataFrame(data=data_scaler, columns=feature_scale_cols, index=data.index)\ndata.drop(feature_scale_cols, axis=1, inplace=True)\ndata = pd.concat([data, data_scaler_df], axis=1)",
"_____no_output_____"
],
[
"# create X (features) and y (response)\nX = data.drop(['readmitted'], axis=1)\ny = data['readmitted']",
"_____no_output_____"
],
[
"y.value_counts()",
"_____no_output_____"
]
],
[
[
"#### Find Top Features in data",
"_____no_output_____"
]
],
[
[
"# split X and y into cross-validation (75%) and testing (25%) data sets\nfrom sklearn.model_selection import train_test_split\nX_cv, X_test, y_cv, y_test = train_test_split(X, y, test_size=0.25)",
"_____no_output_____"
],
[
"# fit Random Forest model to the cross-validation data\nfrom sklearn.ensemble import RandomForestClassifier\nforest = RandomForestClassifier()\nforest.fit(X_cv, y_cv)\nimportances = forest.feature_importances_\n\n# make importance relative to the max importance\nfeature_importance = 100.0 * (importances / importances.max())\nsorted_idx = np.argsort(feature_importance)\nfeature_names = list(X_cv.columns.values)\nfeature_names_sort = [feature_names[indice] for indice in sorted_idx]\npos = np.arange(sorted_idx.shape[0]) + .5\nprint('Top 10 features are: ')\nfor feature in feature_names_sort[::-1][:10]:\n print(feature)\n\n# plot the result\nplt.figure(figsize=(12, 10))\nplt.barh(pos, feature_importance[sorted_idx], align='center')\nplt.yticks(pos, feature_names_sort)\nplt.title('Relative Feature Importance', fontsize=20)\nplt.show()",
"Top 10 features are: \nnum_lab_procedures\ndiag_3\ndiag_2\nnum_medications\ndiag_1\ncomorbidity\ntime_in_hospital\nemergency_visits\nacuity_of_admission\nmedical_specialty\n"
]
],
[
[
"### Use over sampling method for handle imbalanace data",
"_____no_output_____"
]
],
[
[
"from imblearn.over_sampling import SMOTE\nfrom sklearn import metrics\nfrom collections import Counter",
"_____no_output_____"
],
[
"oversample = SMOTE()\nX, y = oversample.fit_resample(X, y)",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = .3)",
"_____no_output_____"
]
],
[
[
"### Logistic Regession",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nclf=LogisticRegression(solver='saga')\nclf.fit(X_train, y_train)\nprint(clf.score(X_test, y_test))",
"0.683537340118278\n"
],
[
"from sklearn.metrics import cohen_kappa_score\ny_pred=clf.predict(X_test)\ncohen_kappa_score(y_test, y_pred)",
"_____no_output_____"
],
[
"#Classification Score\nfrom sklearn.metrics import classification_report\nprint(classification_report(y_test, y_pred))",
" precision recall f1-score support\n\n 0 0.68 0.71 0.69 7304\n 1 0.69 0.66 0.67 7238\n\n accuracy 0.68 14542\n macro avg 0.68 0.68 0.68 14542\nweighted avg 0.68 0.68 0.68 14542\n\n"
]
],
[
[
"### Random Forest ",
"_____no_output_____"
]
],
[
[
"rf = RandomForestClassifier()\nrf.fit(X_train, y_train)\nprint(rf.score(X_test, y_test))",
"0.9360473112364186\n"
],
[
"# cohen kappa score\nfrom sklearn.metrics import cohen_kappa_score\ny_pred=rf.predict(X_test)\ncohen_kappa_score(y_test, y_pred)",
"_____no_output_____"
],
[
"#Classification Score\nfrom sklearn.metrics import classification_report\nprint(classification_report(y_test, y_pred))",
" precision recall f1-score support\n\n 0 0.90 0.98 0.94 7304\n 1 0.97 0.90 0.93 7238\n\n accuracy 0.94 14542\n macro avg 0.94 0.94 0.94 14542\nweighted avg 0.94 0.94 0.94 14542\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0f6d99e836f70b907c0b9521b4ac74149bfc49e | 13,664 | ipynb | Jupyter Notebook | tracking-tensorflow-ssd_mobilenet_v2_coco_2018_03_29.ipynb | chaitanya9807/GuidedVision | 89915c27a73fc841b367de232064032950ca5207 | [
"MIT"
] | 1 | 2019-12-07T18:27:51.000Z | 2019-12-07T18:27:51.000Z | tracking-tensorflow-ssd_mobilenet_v2_coco_2018_03_29.ipynb | chaitanya9807/GuidedVision | 89915c27a73fc841b367de232064032950ca5207 | [
"MIT"
] | null | null | null | tracking-tensorflow-ssd_mobilenet_v2_coco_2018_03_29.ipynb | chaitanya9807/GuidedVision | 89915c27a73fc841b367de232064032950ca5207 | [
"MIT"
] | null | null | null | 39.953216 | 221 | 0.484704 | [
[
[
"import cv2 as cv\nfrom scipy.spatial import distance\nimport numpy as np\nfrom collections import OrderedDict",
"_____no_output_____"
]
],
[
[
"##### Object Tracking Class",
"_____no_output_____"
]
],
[
[
"class Tracker:\n def __init__(self, maxLost = 30): # maxLost: maximum object lost counted when the object is being tracked\n self.nextObjectID = 0 # ID of next object\n self.objects = OrderedDict() # stores ID:Locations\n self.lost = OrderedDict() # stores ID:Lost_count\n \n self.maxLost = maxLost # maximum number of frames object was not detected.\n \n def addObject(self, new_object_location):\n self.objects[self.nextObjectID] = new_object_location # store new object location\n self.lost[self.nextObjectID] = 0 # initialize frame_counts for when new object is undetected\n \n self.nextObjectID += 1\n \n def removeObject(self, objectID): # remove tracker data after object is lost\n del self.objects[objectID]\n del self.lost[objectID]\n \n @staticmethod\n def getLocation(bounding_box):\n xlt, ylt, xrb, yrb = bounding_box\n return (int((xlt + xrb) / 2.0), int((ylt + yrb) / 2.0))\n \n def update(self, detections):\n \n if len(detections) == 0: # if no object detected in the frame\n lost_ids = list(self.lost.keys())\n for objectID in lost_ids:\n self.lost[objectID] +=1\n if self.lost[objectID] > self.maxLost: self.removeObject(objectID)\n \n return self.objects\n \n new_object_locations = np.zeros((len(detections), 2), dtype=\"int\") # current object locations\n \n for (i, detection) in enumerate(detections): new_object_locations[i] = self.getLocation(detection)\n \n if len(self.objects)==0:\n for i in range(0, len(detections)): self.addObject(new_object_locations[i])\n else:\n objectIDs = list(self.objects.keys())\n previous_object_locations = np.array(list(self.objects.values()))\n \n D = distance.cdist(previous_object_locations, new_object_locations) # pairwise distance between previous and current\n \n row_idx = D.min(axis=1).argsort() # (minimum distance of previous from current).sort_as_per_index\n \n cols_idx = D.argmin(axis=1)[row_idx] # index of minimum distance of previous from current\n \n assignedRows, assignedCols = set(), set()\n \n for (row, col) in zip(row_idx, cols_idx):\n \n if row in assignedRows or col in assignedCols:\n continue\n \n objectID = objectIDs[row]\n self.objects[objectID] = new_object_locations[col]\n self.lost[objectID] = 0\n \n assignedRows.add(row)\n assignedCols.add(col)\n \n unassignedRows = set(range(0, D.shape[0])).difference(assignedRows)\n unassignedCols = set(range(0, D.shape[1])).difference(assignedCols)\n \n \n if D.shape[0]>=D.shape[1]:\n for row in unassignedRows:\n objectID = objectIDs[row]\n self.lost[objectID] += 1\n \n if self.lost[objectID] > self.maxLost:\n self.removeObject(objectID)\n \n else:\n for col in unassignedCols:\n self.addObject(new_object_locations[col])\n \n return self.objects\n",
"_____no_output_____"
]
],
[
[
"#### Loading Object Detector Model",
"_____no_output_____"
],
[
"##### Tensorflow model for Object Detection and Tracking\n\nHere, the SSD Object Detection Model is used.\n\nFor more details about single shot detection (SSD), refer the following:\n - **Liu, W., Anguelov, D., Erhan, D., Szegedy, C., Reed, S., Fu, C. Y., & Berg, A. C. (2016, October). Ssd: Single shot multibox detector. In European conference on computer vision (pp. 21-37). Springer, Cham.**\n - Research paper link: https://arxiv.org/abs/1512.02325\n - The pretrained model: https://github.com/opencv/opencv/wiki/TensorFlow-Object-Detection-API#use-existing-config-file-for-your-model",
"_____no_output_____"
]
],
[
[
"model_info = {\"config_path\":\"./tensorflow_model_dir/ssd_mobilenet_v2_coco_2018_03_29.pbtxt\",\n \"model_weights_path\":\"./tensorflow_model_dir/ssd_mobilenet_v2_coco_2018_03_29/frozen_inference_graph.pb\",\n \"object_names\": {0: 'background',\n 1: 'person', 2: 'bicycle', 3: 'car', 4: 'motorcycle', 5: 'airplane', 6: 'bus',\n 7: 'train', 8: 'truck', 9: 'boat', 10: 'traffic light', 11: 'fire hydrant',\n 13: 'stop sign', 14: 'parking meter', 15: 'bench', 16: 'bird', 17: 'cat',\n 18: 'dog', 19: 'horse', 20: 'sheep', 21: 'cow', 22: 'elephant', 23: 'bear',\n 24: 'zebra', 25: 'giraffe', 27: 'backpack', 28: 'umbrella', 31: 'handbag',\n 32: 'tie', 33: 'suitcase', 34: 'frisbee', 35: 'skis', 36: 'snowboard',\n 37: 'sports ball', 38: 'kite', 39: 'baseball bat', 40: 'baseball glove',\n 41: 'skateboard', 42: 'surfboard', 43: 'tennis racket', 44: 'bottle',\n 46: 'wine glass', 47: 'cup', 48: 'fork', 49: 'knife', 50: 'spoon',\n 51: 'bowl', 52: 'banana', 53: 'apple', 54: 'sandwich', 55: 'orange',\n 56: 'broccoli', 57: 'carrot', 58: 'hot dog', 59: 'pizza', 60: 'donut',\n 61: 'cake', 62: 'chair', 63: 'couch', 64: 'potted plant', 65: 'bed',\n 67: 'dining table', 70: 'toilet', 72: 'tv', 73: 'laptop', 74: 'mouse',\n 75: 'remote', 76: 'keyboard', 77: 'cell phone', 78: 'microwave', 79: 'oven',\n 80: 'toaster', 81: 'sink', 82: 'refrigerator', 84: 'book', 85: 'clock',\n 86: 'vase', 87: 'scissors', 88: 'teddy bear', 89: 'hair drier', 90: 'toothbrush'},\n \"confidence_threshold\": 0.5,\n \"threshold\": 0.4\n }\n\nnet = cv.dnn.readNetFromTensorflow(model_info[\"model_weights_path\"], model_info[\"config_path\"])",
"_____no_output_____"
],
[
"np.random.seed(12345)\n\nbbox_colors = {key: np.random.randint(0, 255, size=(3,)).tolist() for key in model_info['object_names'].keys()}",
"_____no_output_____"
]
],
[
[
"##### Instantiate the Tracker Class",
"_____no_output_____"
]
],
[
[
"maxLost = 5 # maximum number of object losts counted when the object is being tracked\ntracker = Tracker(maxLost = maxLost)",
"_____no_output_____"
]
],
[
[
"##### Initiate opencv video capture object\n\nThe `video_src` can take two values:\n1. If `video_src=0`: OpenCV accesses the camera connected through USB\n2. If `video_src='video_file_path'`: OpenCV will access the video file at the given path (can be MP4, AVI, etc format)",
"_____no_output_____"
]
],
[
[
"video_src = \"./data/video_test5.mp4\"#0\ncap = cv.VideoCapture(video_src)",
"_____no_output_____"
]
],
[
[
"##### Start object detection and tracking",
"_____no_output_____"
]
],
[
[
"(H, W) = (None, None) # input image height and width for the network\nwriter = None\nwhile(True):\n \n ok, image = cap.read()\n \n if not ok:\n print(\"Cannot read the video feed.\")\n break\n \n if W is None or H is None: (H, W) = image.shape[:2]\n \n blob = cv.dnn.blobFromImage(image, size=(300, 300), swapRB=True, crop=False)\n net.setInput(blob)\n detections = net.forward()\n \n detections_bbox = [] # bounding box for detections\n \n boxes, confidences, classIDs = [], [], []\n \n for detection in detections[0, 0, :, :]:\n classID = detection[1]\n confidence = detection[2]\n\n if confidence > model_info['confidence_threshold']:\n box = detection[3:7] * np.array([W, H, W, H])\n \n (left, top, right, bottom) = box.astype(\"int\")\n width = right - left + 1\n height = bottom - top + 1\n\n boxes.append([int(left), int(top), int(width), int(height)])\n confidences.append(float(confidence))\n classIDs.append(int(classID))\n \n indices = cv.dnn.NMSBoxes(boxes, confidences, model_info[\"confidence_threshold\"], model_info[\"threshold\"])\n \n if len(indices)>0:\n for i in indices.flatten():\n x, y, w, h = boxes[i][0], boxes[i][1], boxes[i][2], boxes[i][3]\n \n detections_bbox.append((x, y, x+w, y+h))\n \n clr = [int(c) for c in bbox_colors[classIDs[i]]]\n cv.rectangle(image, (x, y), (x+w, y+h), clr, 2)\n \n label = \"{}:{:.4f}\".format(model_info[\"object_names\"][classIDs[i]], confidences[i])\n (label_width, label_height), baseLine = cv.getTextSize(label, cv.FONT_HERSHEY_SIMPLEX, 0.5, 2)\n y_label = max(y, label_height)\n cv.rectangle(image, (x, y_label-label_height),\n (x+label_width, y_label+baseLine), (255, 255, 255), cv.FILLED)\n cv.putText(image, label, (x, y_label), cv.FONT_HERSHEY_SIMPLEX, 0.5, clr, 2)\n \n objects = tracker.update(detections_bbox) # update tracker based on the newly detected objects\n \n for (objectID, centroid) in objects.items():\n text = \"ID {}\".format(objectID)\n cv.putText(image, text, (centroid[0] - 10, centroid[1] - 10), cv.FONT_HERSHEY_SIMPLEX,\n 0.5, (0, 255, 0), 2)\n cv.circle(image, (centroid[0], centroid[1]), 4, (0, 255, 0), -1)\n \n cv.imshow(\"image\", image)\n \n if cv.waitKey(1) & 0xFF == ord('q'):\n break\n \n if writer is None:\n fourcc = cv.VideoWriter_fourcc(*\"MJPG\")\n writer = cv.VideoWriter(\"output.avi\", fourcc, 30, (W, H), True)\n writer.write(image)\nwriter.release()\ncap.release()\ncv.destroyWindow(\"image\")",
"Cannot read the video feed.\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0f6df395af409e643ada7e8fd7dcf2aae8d773f | 374,674 | ipynb | Jupyter Notebook | notebooks/Back_up/SIR_modeling_Working_nested.ipynb | VikasRajashekar/DataScienceProject | e3130332b9eb5338524f637a30dd1801e96898cf | [
"MIT"
] | null | null | null | notebooks/Back_up/SIR_modeling_Working_nested.ipynb | VikasRajashekar/DataScienceProject | e3130332b9eb5338524f637a30dd1801e96898cf | [
"MIT"
] | null | null | null | notebooks/Back_up/SIR_modeling_Working_nested.ipynb | VikasRajashekar/DataScienceProject | e3130332b9eb5338524f637a30dd1801e96898cf | [
"MIT"
] | null | null | null | 665.49556 | 71,672 | 0.944589 | [
[
[
"import pandas as pd\nimport numpy as np\n\nfrom datetime import datetime\nimport pandas as pd \n\nfrom scipy import optimize\nfrom scipy import integrate\n\n%matplotlib inline\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\n\nimport seaborn as sns\n\n\nsns.set(style=\"darkgrid\")\n\nmpl.rcParams['figure.figsize'] = (16, 9)\npd.set_option('display.max_rows', 500)\n\n\n",
"_____no_output_____"
],
[
"# try to parse the dates right at the beginning \n# it works out of the box if the date was stored ISO YYYY-MM-DD format\n\ndf_analyse=pd.read_csv('../data/processed/COVID_small_flat_table.csv',sep=';') \ndf_analyse.sort_values('date',ascending=True).head()",
"_____no_output_____"
],
[
"def SIR_model(SIR,beta,gamma):\n ''' Simple SIR model\n S: susceptible population\n I: infected people\n R: recovered people\n beta: \n \n overall condition is that the sum of changes (differnces) sum up to 0\n dS+dI+dR=0\n S+I+R= N (constant size of population)\n \n '''\n \n S,I,R=SIR\n dS_dt=-beta*S*I/N0 #S*I is the \n dI_dt=beta*S*I/N0-gamma*I\n dR_dt=gamma*I\n return([dS_dt,dI_dt,dR_dt])\n",
"_____no_output_____"
]
],
[
[
"# Simulative approach to calculate SIR curves",
"_____no_output_____"
]
],
[
[
"# set some basic parameters\n# beta/gamma is denoted as 'basic reproduction number'\ndef simulate_SIR(country='Germany'):\n N0=1000000 #max susceptible population\n beta=0.4 # infection spread dynamics\n gamma=0.1 # recovery rate\n\n\n # condition I0+S0+R0=N0\n I0=df_analyse[country][35]\n S0=N0-I0\n R0=0\n SIR=np.array([S0,I0,R0])\n propagation_rates_sumulation=pd.DataFrame(columns={'susceptible':S0,\n 'infected':I0,\n 'recoverd':R0})\n\n\n\n for each_t in np.arange(100):\n\n new_delta_vec=SIR_model(SIR,beta,gamma)\n\n SIR=SIR+new_delta_vec\n\n propagation_rates_sumulation=propagation_rates_sumulation.append({'susceptible':SIR[0],\n 'infected':SIR[1],\n 'recovered':SIR[2]}, ignore_index=True)\n\n\n fig, ax1 = plt.subplots(1, 1)\n\n ax1.plot(propagation_rates_sumulation.index,propagation_rates_sumulation.infected,label='infected',color='k')\n ax1.plot(propagation_rates_sumulation.index,propagation_rates_sumulation.recovered,label='recovered')\n ax1.plot(propagation_rates_sumulation.index,propagation_rates_sumulation.susceptible,label='susceptible')\n\n ax1.set_ylim(10, 1000000)\n ax1.set_yscale('linear')\n ax1.set_title('Szenario SIR simulations (demonstration purposes only)',size=16)\n ax1.set_xlabel('time in days',size=16)\n ax1.legend(loc='best',\n prop={'size': 16});",
"_____no_output_____"
],
[
"simulate_SIR()",
"_____no_output_____"
],
[
"simulate_SIR('Italy')",
"_____no_output_____"
]
],
[
[
"# Fitting the parameters of SIR model",
"_____no_output_____"
]
],
[
[
"# ensure re-initialization \ndef dynamic_beda_SIR(country='Germany'):\n \n def SIR_model_t(SIR,t,beta,gamma):\n ''' Simple SIR model\n S: susceptible population\n t: time step, mandatory for integral.odeint\n I: infected people\n R: recovered people\n beta: \n\n overall condition is that the sum of changes (differnces) sum up to 0\n dS+dI+dR=0\n S+I+R= N (constant size of population)\n\n '''\n\n S,I,R=SIR\n dS_dt=-beta*S*I/N0 #S*I is the \n dI_dt=beta*S*I/N0-gamma*I\n dR_dt=gamma*I\n return dS_dt,dI_dt,dR_dt\n\n def fit_odeint(x, beta, gamma):\n '''\n helper function for the integration\n '''\n return integrate.odeint(SIR_model_t, (S0, I0, R0), t, args=(beta, gamma))[:,1] # we only would like to get dI\n\n ydata = np.array(df_analyse[country][35:])\n t=np.arange(len(ydata))\n I0=ydata[0]\n S0=N0-I0\n R0=0\n beta=0.4 \n\n popt, pcov = optimize.curve_fit(fit_odeint, t, ydata)\n perr = np.sqrt(np.diag(pcov))\n\n print('standard deviation errors : ',str(perr), ' start infect:',ydata[0])\n print(\"Optimal parameters: beta =\", popt[0], \" and gamma = \", popt[1])\n\n\n fitted=fit_odeint(t, *popt)\n\n plt.semilogy(t, ydata, 'o')\n plt.semilogy(t, fitted)\n plt.title(\"Fit of SIR model for Germany cases\")\n plt.ylabel(\"Population infected\")\n plt.xlabel(\"Days\")\n plt.show()\n print(\"Optimal parameters: beta =\", popt[0], \" and gamma = \", popt[1])\n print(\"Basic Reproduction Number R0 \" , popt[0]/ popt[1])\n print(\"This ratio is derived as the expected number of new infections (these new infections are sometimes called secondary infections from a single \\ infection in a population where all subjects are susceptible. @wiki\")",
"_____no_output_____"
],
[
"dynamic_beda_SIR('Italy')",
"/home/vikas/anaconda3/envs/ds/lib/python3.6/site-packages/ipykernel_launcher.py:19: RuntimeWarning: overflow encountered in double_scalars\n/home/vikas/anaconda3/envs/ds/lib/python3.6/site-packages/ipykernel_launcher.py:20: RuntimeWarning: overflow encountered in double_scalars\n/home/vikas/anaconda3/envs/ds/lib/python3.6/site-packages/ipykernel_launcher.py:21: RuntimeWarning: overflow encountered in double_scalars\n"
],
[
"dynamic_beda_SIR()",
"/home/vikas/anaconda3/envs/ds/lib/python3.6/site-packages/scipy/integrate/odepack.py:247: ODEintWarning: Excess work done on this call (perhaps wrong Dfun type). Run with full_output = 1 to get quantitative information.\n warnings.warn(warning_msg, ODEintWarning)\n"
]
],
[
[
"# Dynamic beta in SIR (infection rate)",
"_____no_output_____"
]
],
[
[
"def dynamic_beta_SIR(country='Germany'):\n t_initial=28\n t_intro_measures=14\n t_hold=21\n t_relax=21\n\n ydata = np.array(df_analyse[country][35:])\n I0=ydata[0]\n N0=1000000\n S0=N0-I0\n R0=0\n\n beta_max=0.4\n beta_min=0.11\n gamma=0.1\n pd_beta=np.concatenate((np.array(t_initial*[beta_max]),\n np.linspace(beta_max,beta_min,t_intro_measures),\n np.array(t_hold*[beta_min]),\n np.linspace(beta_min,beta_max,t_relax),\n ))\n\n SIR=np.array([S0,I0,R0])\n propagation_rates_dynamic=pd.DataFrame(columns={'susceptible':S0,\n 'infected':I0,\n 'recoverd':R0})\n\n\n\n for each_beta in pd_beta:\n\n new_delta_vec=SIR_model(SIR,each_beta,gamma)\n\n SIR=SIR+new_delta_vec\n\n propagation_rates_dynamic=propagation_rates_dynamic.append({'susceptible':SIR[0],\n 'infected':SIR[1],\n 'recovered':SIR[2]}, ignore_index=True)\n\n fig, ax1 = plt.subplots(1, 1)\n\n ax1.plot(propagation_rates_dynamic.index,propagation_rates_dynamic.infected,label='infected',linewidth=3)\n\n t_phases=np.array([t_initial,t_intro_measures,t_hold,t_relax]).cumsum()\n ax1.bar(np.arange(len(ydata)),ydata, width=0.8,label=' current infected Germany',color='r')\n ax1.axvspan(0,t_phases[0], facecolor='b', alpha=0.2,label='no measures')\n ax1.axvspan(t_phases[0],t_phases[1], facecolor='b', alpha=0.3,label='hard measures introduced')\n ax1.axvspan(t_phases[1],t_phases[2], facecolor='b', alpha=0.4,label='hold measures')\n ax1.axvspan(t_phases[2],t_phases[3], facecolor='b', alpha=0.5,label='relax measures')\n ax1.axvspan(t_phases[3],len(propagation_rates_dynamic.infected), facecolor='b', alpha=0.6,label='repead hard measures')\n\n ax1.set_ylim(10, 1.5*max(propagation_rates_dynamic.infected))\n ax1.set_yscale('log')\n ax1.set_title('Szenario SIR simulations (demonstration purposes only)',size=16)\n ax1.set_xlabel('time in days',size=16)\n ax1.legend(loc='best',\n prop={'size': 16});\n",
"_____no_output_____"
],
[
"dynamic_beta_SIR()",
"_____no_output_____"
],
[
"dynamic_beta_SIR('Italy')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0f6e27a23e6eda86872f48659ff2ed5f2c8a6f5 | 3,805 | ipynb | Jupyter Notebook | Create_Job_Submission_Script.ipynb | aboussetta/deploy-ai-edge-iot-smart-queuing-system-app | efeb7719dd7533fb39b2ac9c600ed045ec8eb725 | [
"MIT"
] | null | null | null | Create_Job_Submission_Script.ipynb | aboussetta/deploy-ai-edge-iot-smart-queuing-system-app | efeb7719dd7533fb39b2ac9c600ed045ec8eb725 | [
"MIT"
] | null | null | null | Create_Job_Submission_Script.ipynb | aboussetta/deploy-ai-edge-iot-smart-queuing-system-app | efeb7719dd7533fb39b2ac9c600ed045ec8eb725 | [
"MIT"
] | null | null | null | 36.586538 | 308 | 0.60184 | [
[
[
"# Step 2: Create Job Submission Script\n\nThe next step is to create our job submission script. In the cell below, you will need to complete the job submission script and run the cell to generate the file using the magic `%%writefile` command. Your main task is to complete the following items of the script:\n\n* Create a variable `MODEL` and assign it the value of the first argument passed to the job submission script.\n* Create a variable `DEVICE` and assign it the value of the second argument passed to the job submission script.\n* Create a variable `VIDEO` and assign it the value of the third argument passed to the job submission script.\n* Create a variable `PEOPLE` and assign it the value of the sixth argument passed to the job submission script.",
"_____no_output_____"
]
],
[
[
"%%writefile queue_job.sh\n#!/bin/bash\n\nexec 1>/output/stdout.log 2>/output/stderr.log\n\n model=args.model\n device=args.device\n video_file=args.video\n max_people=args.max_people\n threshold=args.threshold\n output_path=args.output_path\n\n# TODO: Create MODEL variable\nMODEL=$2\n# TODO: Create DEVICE variable\nDEVICE=$1\n# TODO: Create VIDEO variable\nVIDEO=$3\nQUEUE=$4\nOUTPUT=$5\n# TODO: Create PEOPLE variable\nPEOPLE=$6\n\nmkdir -p $5\n\nif echo \"$DEVICE\" | grep -q \"FPGA\"; then # if device passed in is FPGA, load bitstream to program FPGA\n #Environment variables and compilation for edge compute nodes with FPGAs\n export AOCL_BOARD_PACKAGE_ROOT=/opt/intel/openvino/bitstreams/a10_vision_design_sg2_bitstreams/BSP/a10_1150_sg2\n\n source /opt/altera/aocl-pro-rte/aclrte-linux64/init_opencl.sh\n aocl program acl0 /opt/intel/openvino/bitstreams/a10_vision_design_sg2_bitstreams/2020-2_PL2_FP16_MobileNet_Clamp.aocx\n\n export CL_CONTEXT_COMPILER_MODE_INTELFPGA=3\nfi\n\npython3 person_detect.py --model ${MODEL} \\\n --device ${DEVICE} \\\n --video ${VIDEO} \\\n --queue_param ${QUEUE} \\\n --output_path ${OUTPUT}\\\n --max_people ${PEOPLE} \\\n\ncd /output\n\ntar zcvf output.tgz *",
"_____no_output_____"
]
],
[
[
"# Next Step\n\nNow that you've run the above cell and created your job submission script, you will work through each scenarios notebook in the next three workspaces. In each of these notebooks, you will submit jobs to Intel's DevCloud to load and run inference on each type of hardware and then review the results.\n\n**Note**: As a reminder, if you need to make any changes to the job submission script, you can come back to this workspace to edit and run the above cell to overwrite the file with your changes.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0f6ea6bd91ae66bc7111c732d30b9a502f03ef5 | 124,615 | ipynb | Jupyter Notebook | examples/source-interdiction/results-notebooks/Learning-Curves.ipynb | arfc/annsa | e6791d1c72bbdd3a792080ff261568813375f3a3 | [
"BSD-3-Clause"
] | 7 | 2019-03-11T12:34:21.000Z | 2021-05-07T14:49:23.000Z | examples/source-interdiction/results-notebooks/Learning-Curves.ipynb | arfc/annsa | e6791d1c72bbdd3a792080ff261568813375f3a3 | [
"BSD-3-Clause"
] | 67 | 2018-08-20T21:49:09.000Z | 2019-12-04T17:52:03.000Z | examples/source-interdiction/results-notebooks/Learning-Curves.ipynb | arfc/annsa | e6791d1c72bbdd3a792080ff261568813375f3a3 | [
"BSD-3-Clause"
] | 6 | 2018-10-25T17:41:25.000Z | 2020-12-26T18:09:53.000Z | 276.922222 | 54,132 | 0.911816 | [
[
[
"import matplotlib.pyplot as plt\nimport os\n\nimport numpy as np\nimport itertools\nfrom glob import glob\nimport pandas as pd\nfrom itertools import product\nimport os\n\nfrom annsa.model_classes import f1\nfrom tensorflow.python.keras.models import load_model\nfrom pandas import read_csv\nfrom sklearn.metrics import auc\n\nfrom matplotlib.lines import Line2D",
"Using TensorFlow backend.\n"
]
],
[
[
"#### Import model, training function ",
"_____no_output_____"
]
],
[
[
"def plot_learning_curve_points(sizes, errors, label='None', linestyle='-', color='k', marker='.', linewidth=2):\n '''\n Plots the learning curve. \n \n Inputs:\n sizes : list, int\n List of traning dataset sizes\n errors : list, float \n List of final errors for some metric\n \n \n '''\n average = np.average(errors, axis=1)\n std = np.var(errors)\n plt.plot(sizes, average, label=label, linestyle=linestyle, color=color, linewidth=linewidth,)\n plt.scatter(np.array([[size]*5 for size in sizes]).flatten(),\n np.array(errors).flatten(),\n color=color,\n marker=marker)",
"_____no_output_____"
],
[
"import matplotlib.colors\n\ndef categorical_cmap(nc, nsc, cmap=\"tab10\", continuous=False):\n if nc > plt.get_cmap(cmap).N:\n raise ValueError(\"Too many categories for colormap.\")\n if continuous:\n ccolors = plt.get_cmap(cmap)(np.linspace(0,1,nc))\n else:\n ccolors = plt.get_cmap(cmap)(np.arange(nc, dtype=int))\n cols = np.zeros((nc*nsc, 3))\n for i, c in enumerate(ccolors):\n chsv = matplotlib.colors.rgb_to_hsv(c[:3])\n arhsv = np.tile(chsv,nsc).reshape(nsc,3)\n arhsv[:,1] = np.linspace(chsv[1],0.25,nsc)\n arhsv[:,2] = np.linspace(chsv[2],1,nsc)\n rgb = matplotlib.colors.hsv_to_rgb(arhsv)\n cols[i*nsc:(i+1)*nsc,:] = rgb \n cmap = matplotlib.colors.ListedColormap(cols)\n return cmap\n\nc1 = categorical_cmap(5,1, cmap=\"tab10\")\nplt.scatter(np.arange(5*1),np.ones(5*1)+1, c=np.arange(5*1), s=180, cmap=c1)\n",
"_____no_output_____"
],
[
"line_colors = {'caednn' : c1.colors[0],\n 'daednn' : c1.colors[1],\n 'dnn' : c1.colors[2],\n 'cnn' : c1.colors[3],\n }\n\nline_styles = {'test' : '-',\n 'train' : '--',}\n\nmarker_styles = {'test' : '',\n 'train' : '',}",
"_____no_output_____"
],
[
"dependencies = {'f1' : f1}",
"_____no_output_____"
]
],
[
[
"# All models Full",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(10,5))\nmatplotlib.rcParams.update({'font.size': 22})\ndataset_modes = ['test', 'train']\nmodels = [\n 'dnn',\n 'cnn',\n 'caednn',\n 'daednn',\n]\nmodel_modes = ['full']\ntrain_sizes = [\n '50',\n '100',\n '500',\n '1000',\n '5000',\n '10000',\n '15000',\n '20000',\n ]\n\nerrors_all = {}\n\nfor model, model_mode, dataset_mode in product (models, model_modes, dataset_modes):\n if dataset_mode == 'train':\n loss = 'f1'\n else:\n loss = 'val_f1'\n\n errors = []\n for train_size in train_sizes:\n tmp_error = []\n \n identifier = '-final_trainsize'\n if model == 'cnn':\n identifier = '-final-reluupdate_trainsize'\n \n \n file_path = os.path.join(\n '..',\n 'final_training_notebooks',\n 'final-models-keras',\n 'learningcurve-'+model+'-'+model_mode+identifier+train_size+'_'+'*.log',)\n for tmp_file_path in glob(file_path):\n history_temp = read_csv(tmp_file_path)\n tmp_error.append(history_temp.tail(1).iloc[0][loss])\n errors.append(np.array(tmp_error))\n errors = np.array(errors)\n errors_all[dataset_mode + '_' + model] = np.average(errors, axis=1)\n plot_learning_curve_points([int(train_size) for train_size in train_sizes],\n errors,\n label=model+' '+dataset_mode+'ing set',\n linestyle=line_styles[dataset_mode],\n color=line_colors[model],\n marker=marker_styles[dataset_mode],\n linewidth=2)\n\ncustom_lines = [Line2D([0], [0], color=c1.colors[3], lw=4),\n Line2D([0], [0], color=c1.colors[2], lw=4),\n Line2D([0], [0], color=c1.colors[0], lw=4),\n Line2D([0], [0], color=c1.colors[1], lw=4),\n Line2D([0], [0], color='k', linestyle=line_styles['test'], marker=marker_styles['test'], markersize=15, lw=2),\n Line2D([0], [0], color='k', linestyle=line_styles['train'], marker=marker_styles['train'], markersize=15, lw=2),\n ]\nplt.legend(custom_lines,\n ['CNN', 'DNN', 'CAE', 'DAE', 'Validation', 'Training'], \n prop={'size': 15})\n\nplt.ylim([0,1.1])\nplt.xlabel('Number of Examples')\nplt.ylabel('F1 Score')\nplt.xticks([0, 5000, 10000, 15000, 20000], [0, 5000, 10000, 15000, 20000])",
"_____no_output_____"
],
[
"for item in errors_all:\n print(item, round((auc([int(train_size) for train_size in train_sizes], errors_all[item]))/20000., 2))",
"test_dnn 0.74\ntrain_dnn 0.76\ntest_cnn 0.88\ntrain_cnn 0.93\ntest_caednn 0.89\ntrain_caednn 0.94\ntest_daednn 0.74\ntrain_daednn 0.76\n"
]
],
[
[
"# All models Easy",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(10,5))\n\ndataset_modes = ['test', 'train']\nmodels = [\n 'dnn',\n 'cnn',\n 'caednn',\n 'daednn',\n]\nmodel_modes = ['easy']\ntrain_sizes = [\n '50',\n '100',\n '500',\n '1000',\n '5000',\n '10000',\n '15000',\n '20000',\n ]\n\n\nfor model, model_mode, dataset_mode in product (models, model_modes, dataset_modes):\n if dataset_mode == 'train':\n loss = 'f1'\n else:\n loss = 'val_f1'\n\n errors = []\n for train_size in train_sizes:\n tmp_error = []\n identifier = '-final_trainsize'\n if model == 'cnn':\n identifier = '-final-reluupdate_trainsize'\n file_path = os.path.join(\n '..',\n 'final_training_notebooks',\n 'final-models-keras',\n 'learningcurve-'+model+'-'+model_mode+identifier+train_size+'_'+'*.log',)\n for tmp_file_path in glob(file_path):\n history_temp = read_csv(tmp_file_path)\n tmp_error.append(history_temp.tail(1).iloc[0][loss])\n errors.append(np.array(tmp_error))\n errors = np.array(errors)\n errors_all[dataset_mode + '_' + model] = np.average(errors, axis=1)\n \n plot_learning_curve_points([int(train_size) for train_size in train_sizes],\n errors,\n label=model+' '+dataset_mode+'ing set',\n linestyle=line_styles[dataset_mode],\n color=line_colors[model],\n marker=marker_styles[dataset_mode],\n linewidth=2)\n\ncustom_lines = [Line2D([0], [0], color=c1.colors[3], lw=4),\n Line2D([0], [0], color=c1.colors[2], lw=4),\n Line2D([0], [0], color=c1.colors[0], lw=4),\n Line2D([0], [0], color=c1.colors[1], lw=4),\n Line2D([0], [0], color='k', linestyle=line_styles['test'], marker=marker_styles['test'], markersize=15, lw=2),\n Line2D([0], [0], color='k', linestyle=line_styles['train'], marker=marker_styles['train'], markersize=15, lw=2),\n ]\nplt.legend(custom_lines,\n ['CNN', 'DNN', 'CAE', 'DAE', 'validation', 'Training'], \n prop={'size': 15})\nplt.ylim([0,1.1])\nplt.xlabel('Number of Examples')\nplt.ylabel('F1 Score')\nplt.xticks([0, 5000, 10000, 15000, 20000], [0, 5000, 10000, 15000, 20000])",
"_____no_output_____"
],
[
"for item in errors_all:\n print(item, round((auc([int(train_size) for train_size in train_sizes], errors_all[item]))/20000., 2))",
"test_dnn 0.88\ntrain_dnn 0.87\ntest_cnn 0.98\ntrain_cnn 0.98\ntest_caednn 0.97\ntrain_caednn 0.98\ntest_daednn 0.82\ntrain_daednn 0.81\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0f6f56cc473a479b2d1c32d6f8de1b9fbded0b5 | 15,579 | ipynb | Jupyter Notebook | course_2/course_material/Part_7_Deep_Learning/S54_L390/9. TensorFlow_MNIST_Learning_rate_Part_2_Solution.ipynb | Alexander-Meldrum/learning-data-science | a87cf6be80c67a8d1b57a96c042bdf423ba0a142 | [
"MIT"
] | null | null | null | course_2/course_material/Part_7_Deep_Learning/S54_L390/9. TensorFlow_MNIST_Learning_rate_Part_2_Solution.ipynb | Alexander-Meldrum/learning-data-science | a87cf6be80c67a8d1b57a96c042bdf423ba0a142 | [
"MIT"
] | null | null | null | course_2/course_material/Part_7_Deep_Learning/S54_L390/9. TensorFlow_MNIST_Learning_rate_Part_2_Solution.ipynb | Alexander-Meldrum/learning-data-science | a87cf6be80c67a8d1b57a96c042bdf423ba0a142 | [
"MIT"
] | null | null | null | 47.066465 | 911 | 0.64863 | [
[
[
"# Exercises\n\n### 9. Adjust the learning rate. Try a value of 0.02. Does it make a difference?\n\n** Solution **\n\nThis is the simplest exercise and you have do that before. \n\nFind the line: \n\n optimize = tf.train.AdamOptimizer(learning_rate=0.001).minimize(mean_loss)\n \nAnd change the learning_rate to 0.02.\n\nWhile Adam adapts to the problem, if the orders of magnitude are too different, it may not have time to adjust accordingly. We start overfitting before we can reach a neat solution.\n\nTherefore, for this problem, even 0.02 is a **HIGH** starting learning rate. \n\nIt's a good practice to try 0.001, 0.0001, and 0.00001. If it makes no difference, pick whatever, otherwise it makes sense to fiddle with the learning rate.",
"_____no_output_____"
],
[
"## Deep Neural Network for MNIST Classification\n\nWe'll apply all the knowledge from the lectures in this section to write a deep neural network. The problem we've chosen is referred to as the \"Hello World\" for machine learning because for most students it is their first example. The dataset is called MNIST and refers to handwritten digit recognition. You can find more about it on Yann LeCun's website (Director of AI Research, Facebook). He is one of the pioneers of what we've been talking about and of more complex approaches that are widely used today, such as covolutional networks. The dataset provides 28x28 images of handwritten digits (1 per image) and the goal is to write an algorithm that detects which digit is written. Since there are only 10 digits, this is a classification problem with 10 classes. In order to exemplify what we've talked about in this section, we will build a network with 2 hidden layers between inputs and outputs.",
"_____no_output_____"
],
[
"## Import the relevant packages",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport tensorflow as tf\nfrom tensorflow.examples.tutorials.mnist import input_data\n\n# TensorFLow includes a data provider for MNIST that we'll use.\n# This function automatically downloads the MNIST dataset to the chosen directory. \n# The dataset is already split into training, validation, and test subsets. \n# Furthermore, it preprocess it into a particularly simple and useful format.\n# Every 28x28 image is flattened into a vector of length 28x28=784, where every value\n# corresponds to the intensity of the color of the corresponding pixel.\n# The samples are grayscale (but standardized from 0 to 1), so a value close to 0 is almost white and a value close to\n# 1 is almost purely black. This representation (flattening the image row by row into\n# a vector) is slightly naive but as you'll see it works surprisingly well.\n# Since this is a classification problem, our targets are categorical.\n# Recall from the lecture on that topic that one way to deal with that is to use one-hot encoding.\n# With it, the target for each individual sample is a vector of length 10\n# which has nine 0s and a single 1 at the position which corresponds to the correct answer.\n# For instance, if the true answer is \"1\", the target will be [0,0,0,1,0,0,0,0,0,0] (counting from 0).\n# Have in mind that the very first time you execute this command it might take a little while to run\n# because it has to download the whole dataset. Following commands only extract it so they're faster.\nmnist = input_data.read_data_sets(\"MNIST_data/\", one_hot=True)",
"Extracting MNIST_data/train-images-idx3-ubyte.gz\nExtracting MNIST_data/train-labels-idx1-ubyte.gz\nExtracting MNIST_data/t10k-images-idx3-ubyte.gz\nExtracting MNIST_data/t10k-labels-idx1-ubyte.gz\n"
]
],
[
[
"## Outline the model\n\nThe whole code is in one cell, so you can simply rerun this cell (instead of the whole notebook) and train a new model.\nThe tf.reset_default_graph() function takes care of clearing the old parameters. From there on, a completely new training starts.",
"_____no_output_____"
]
],
[
[
"input_size = 784\noutput_size = 10\n# Use same hidden layer size for both hidden layers. Not a necessity.\nhidden_layer_size = 50\n\n# Reset any variables left in memory from previous runs.\ntf.reset_default_graph()\n\n# As in the previous example - declare placeholders where the data will be fed into.\ninputs = tf.placeholder(tf.float32, [None, input_size])\ntargets = tf.placeholder(tf.float32, [None, output_size])\n\n# Weights and biases for the first linear combination between the inputs and the first hidden layer.\n# Use get_variable in order to make use of the default TensorFlow initializer which is Xavier.\nweights_1 = tf.get_variable(\"weights_1\", [input_size, hidden_layer_size])\nbiases_1 = tf.get_variable(\"biases_1\", [hidden_layer_size])\n\n# Operation between the inputs and the first hidden layer.\n# We've chosen ReLu as our activation function. You can try playing with different non-linearities.\noutputs_1 = tf.nn.relu(tf.matmul(inputs, weights_1) + biases_1)\n\n# Weights and biases for the second linear combination.\n# This is between the first and second hidden layers.\nweights_2 = tf.get_variable(\"weights_2\", [hidden_layer_size, hidden_layer_size])\nbiases_2 = tf.get_variable(\"biases_2\", [hidden_layer_size])\n\n# Operation between the first and the second hidden layers. Again, we use ReLu.\noutputs_2 = tf.nn.relu(tf.matmul(outputs_1, weights_2) + biases_2)\n\n# Weights and biases for the final linear combination.\n# That's between the second hidden layer and the output layer.\nweights_3 = tf.get_variable(\"weights_3\", [hidden_layer_size, output_size])\nbiases_3 = tf.get_variable(\"biases_3\", [output_size])\n\n# Operation between the second hidden layer and the final output.\n# Notice we have not used an activation function because we'll use the trick to include it directly in \n# the loss function. This works for softmax and sigmoid with cross entropy.\noutputs = tf.matmul(outputs_2, weights_3) + biases_3\n\n# Calculate the loss function for every output/target pair.\n# The function used is the same as applying softmax to the last layer and then calculating cross entropy\n# with the function we've seen in the lectures. This function, however, combines them in a clever way, \n# which makes it both faster and more numerically stable (when dealing with very small numbers).\n# Logits here means: unscaled probabilities (so, the outputs, before they are scaled by the softmax)\n# Naturally, the labels are the targets.\nloss = tf.nn.softmax_cross_entropy_with_logits(logits=outputs, labels=targets)\n\n# Get the average loss\nmean_loss = tf.reduce_mean(loss)\n\n# Define the optimization step. Using adaptive optimizers such as Adam in TensorFlow\n# is as simple as that.\noptimize = tf.train.AdamOptimizer(learning_rate=0.02).minimize(mean_loss)\n\n# Get a 0 or 1 for every input in the batch indicating whether it output the correct answer out of the 10.\nout_equals_target = tf.equal(tf.argmax(outputs, 1), tf.argmax(targets, 1))\n\n# Get the average accuracy of the outputs.\naccuracy = tf.reduce_mean(tf.cast(out_equals_target, tf.float32))\n\n# Declare the session variable.\nsess = tf.InteractiveSession()\n\n# Initialize the variables. Default initializer is Xavier.\ninitializer = tf.global_variables_initializer()\nsess.run(initializer)\n\n# Batching\nbatch_size = 100\n\n# Calculate the number of batches per epoch for the training set.\nbatches_number = mnist.train._num_examples // batch_size\n\n# Basic early stopping. Set a miximum number of epochs.\nmax_epochs = 15\n\n# Keep track of the validation loss of the previous epoch.\n# If the validation loss becomes increasing, we want to trigger early stopping.\n# We initially set it at some arbitrarily high number to make sure we don't trigger it\n# at the first epoch\nprev_validation_loss = 9999999.\n\nimport time\nstart_time = time.time()\n\n# Create a loop for the epochs. Epoch_counter is a variable which automatically starts from 0.\nfor epoch_counter in range(max_epochs):\n \n # Keep track of the sum of batch losses in the epoch.\n curr_epoch_loss = 0.\n \n # Iterate over the batches in this epoch.\n for batch_counter in range(batches_number):\n \n # Input batch and target batch are assigned values from the train dataset, given a batch size\n input_batch, target_batch = mnist.train.next_batch(batch_size)\n \n # Run the optimization step and get the mean loss for this batch.\n # Feed it with the inputs and the targets we just got from the train dataset\n _, batch_loss = sess.run([optimize, mean_loss], \n feed_dict={inputs: input_batch, targets: target_batch})\n \n # Increment the sum of batch losses.\n curr_epoch_loss += batch_loss\n \n # So far curr_epoch_loss contained the sum of all batches inside the epoch\n # We want to find the average batch losses over the whole epoch\n # The average batch loss is a good proxy for the current epoch loss\n curr_epoch_loss /= batches_number\n \n # At the end of each epoch, get the validation loss and accuracy\n # Get the input batch and the target batch from the validation dataset\n input_batch, target_batch = mnist.validation.next_batch(mnist.validation._num_examples)\n \n # Run without the optimization step (simply forward propagate)\n validation_loss, validation_accuracy = sess.run([mean_loss, accuracy], \n feed_dict={inputs: input_batch, targets: target_batch})\n \n # Print statistics for the current epoch\n # Epoch counter + 1, because epoch_counter automatically starts from 0, instead of 1\n # We format the losses with 3 digits after the dot\n # We format the accuracy in percentages for easier interpretation\n print('Epoch '+str(epoch_counter+1)+\n '. Mean loss: '+'{0:.3f}'.format(curr_epoch_loss)+\n '. Validation loss: '+'{0:.3f}'.format(validation_loss)+\n '. Validation accuracy: '+'{0:.2f}'.format(validation_accuracy * 100.)+'%')\n \n # Trigger early stopping if validation loss begins increasing.\n if validation_loss > prev_validation_loss:\n break\n \n # Store this epoch's validation loss to be used as previous validation loss in the next iteration.\n prev_validation_loss = validation_loss\n\n# Not essential, but it is nice to know when the algorithm stopped working in the output section, rather than check the kernel\nprint('End of training.')\n\n#Add the time it took the algorithm to train\nprint(\"Training time: %s seconds\" % (time.time() - start_time))",
"Epoch 1. Mean loss: 0.284. Validation loss: 0.213. Validation accuracy: 94.28%\nEpoch 2. Mean loss: 0.195. Validation loss: 0.182. Validation accuracy: 95.14%\nEpoch 3. Mean loss: 0.169. Validation loss: 0.167. Validation accuracy: 95.64%\nEpoch 4. Mean loss: 0.161. Validation loss: 0.166. Validation accuracy: 95.88%\nEpoch 5. Mean loss: 0.150. Validation loss: 0.190. Validation accuracy: 95.68%\nEnd of training.\nTraining time: 6.1423585414886475 seconds\n"
]
],
[
[
"## Test the model\n\nAs we discussed in the lectures, after training on the training and validation sets, we test the final prediction power of our model by running it on the test dataset that the algorithm has not seen before.\n\nIt is very important to realize that fiddling with the hyperparameters overfits the validation dataset. The test is the absolute final instance. You should not test before you are completely done with adjusting your model.",
"_____no_output_____"
]
],
[
[
"input_batch, target_batch = mnist.test.next_batch(mnist.test._num_examples)\ntest_accuracy = sess.run([accuracy], \n feed_dict={inputs: input_batch, targets: target_batch})\n\n# Test accuracy is a list with 1 value, so we want to extract the value from it, using x[0]\n# Uncomment the print to see how it looks before the manipulation\n# print (test_accuracy)\ntest_accuracy_percent = test_accuracy[0] * 100.\n\n# Print the test accuracy formatted in percentages\nprint('Test accuracy: '+'{0:.2f}'.format(test_accuracy_percent)+'%')",
"Test accuracy: 95.68%\n"
]
],
[
[
"Using the initial model and hyperparameters given in this notebook, the final test accuracy should be roughly between 97% and 98%. Each time the code is rerunned, we get a different accuracy as the batches are shuffled, the weights are initialized in a different way, etc.\n\nFinally, we have intentionally reached a suboptimal solution, so you can have space to build on it.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0f6fb2a50eb10076b418957badc40e21a6cacb5 | 6,738 | ipynb | Jupyter Notebook | Class_exercises_and_projects/exercise1.ipynb | ayomideoj/ayomideCSC102 | 5b0e5523d529fab152302b345a75bad4a038cada | [
"MIT"
] | null | null | null | Class_exercises_and_projects/exercise1.ipynb | ayomideoj/ayomideCSC102 | 5b0e5523d529fab152302b345a75bad4a038cada | [
"MIT"
] | null | null | null | Class_exercises_and_projects/exercise1.ipynb | ayomideoj/ayomideCSC102 | 5b0e5523d529fab152302b345a75bad4a038cada | [
"MIT"
] | null | null | null | 20.732308 | 71 | 0.461264 | [
[
[
"\n# Integer number\nnum = 100\nprint(num)\nprint(\"Data type of variable num is\",type(num))\n\n#float number\nfnum = 34.45\nprint(fnum)\nprint(\"Data type variable fnum is\",type(fnum))\n\n#complex number\ncnum = 3 + 4j\nprint(cnum)\nprint(\"Data type variable cnum is\", type(cnum))",
"100\nData type of variable num is <class 'int'>\n34.45\nData type variable fnum is <class 'float'>\n(3+4j)\nData type variable cnum is <class 'complex'>\n"
],
[
"#python program to print strings and type\nstr1 = \"Hi my name is Matthew. I am String\"\nstr2 = \"Hi my name is OJ. I am also a String\"\n\nprint(str1)\nprint(type(str1))\n\nprint(str2)\nprint(type(str2))\n",
"Hi my name is Matthew. I am String\n<class 'str'>\nHi my name is OJ. I am also a String\n<class 'str'>\n"
],
[
"# tuple of integers\nt1 = (1,2,3,4,5)\nprint(t1)\n\n#tuple of strings\nt2 = (\"Nifemi\",\"Gina\",\"Marho\")\n\nfor s in t2:\n print(s)\n\n#tuple of mixed type elements\nt3 =(2,\"Ebube\",45,\"Jeffery\")\n\n'''\nPrint a specific element\nindexes start with zero\n'''\nprint(t3[2])\n",
"(1, 2, 3, 4, 5)\nNifemi\nGina\nMarho\n45\n"
],
[
"#list of integers\nlis1 =[1,2,3,4,5]\nprint(lis1)\n\n#list of strings\nlis2 =[\"Mouse\",\"Keyboard\",\"Monitor\"]\n\n#loop through list elements\nfor x in lis2:\n print(x)\n\n#list of mixed type elements\nlis3 =[20,\"CSC102\",39,\"Python Programming\"]\n'''Print a specific element in list indexes start with zero'''\n\nprint(\"Element at index 3 is:\",lis3[3])\n",
"[1, 2, 3, 4, 5]\nMouse\nKeyboard\nMonitor\nElement at index 3 is: Python Programming\n"
],
[
"#Dictionary example\ndict ={1:\"Maryam\",\"lastname\":\"Shefiu\",\"age\":25}\n\nprint(dict[1])\nprint(dict[\"lastname\"])\nprint(dict[\"age\"])\n",
"Maryam\nShefiu\n25\n"
],
[
"# Set Example\nmyset ={\"Joseph\", \"Adaobi\", \"Kamara\",\"Ugochi\"}\n#loop through set\nfor a in myset:\n print(a)\n\n#checking whether 2 exists in myset\nprint(2 in myset)\n\n#adding new element\nmyset.add(99)\n\nprint(myset)\n",
"Adaobi\nUgochi\nJoseph\nKamara\nFalse\n{99, 'Ugochi', 'Joseph', 'Kamara', 'Adaobi'}\n"
],
[
"#Simple Interest Problems\nP=1000\nR=1\nT=2\n\nA= (P*(1 + ((R / 100) * T)))\nprint(\"Amount is\", A)\nSI = A-P\nprint(\"Simple Interest is\", SI)\n",
"Amount is 1020.0\nSimple Interest is 20.0\n"
],
[
"#Solving Quadratic Equations\nimport cmath\n\na=1\nb=5\nc=6\n\n#calculate the discriminant\nd = (b**2)-(4*a*c)\n\nsol1=(-b-cmath.sqrt(d))/(2*a)\nsol2=(-b+cmath.sqrt(d))/(2*a)\n\nprint('The solutions are', sol1,sol2)\n",
"The solutions are (-3+0j) (-2+0j)\n"
],
[
"# Python program to find the area of a traingle\na = float(input('Enter first side: '))\nb = float(input('Enter second side: '))\nc = float(input('Enter third side: '))\n\n#calculate the semi-perimeter\ns = (a+b+c)/2\n\n#calculate the area\narea = (s*(s-a)*(s-b)*(s-c))**0.5\nprint('The area of the traingle is %0.2f' %area)\n",
"Enter first side: 5\nEnter second side: 6\nEnter third side: 7\nThe area of the traingle is 14.70\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f7142476c2fa39b326a2258fb7897c1a875447 | 2,417 | ipynb | Jupyter Notebook | data/geojson/get_counties_geojson_data.ipynb | geodesign/ubiquitous-octo-parakeet | 9c77b6564f74751112b126b913affb1312de30fe | [
"MIT"
] | null | null | null | data/geojson/get_counties_geojson_data.ipynb | geodesign/ubiquitous-octo-parakeet | 9c77b6564f74751112b126b913affb1312de30fe | [
"MIT"
] | null | null | null | data/geojson/get_counties_geojson_data.ipynb | geodesign/ubiquitous-octo-parakeet | 9c77b6564f74751112b126b913affb1312de30fe | [
"MIT"
] | null | null | null | 22.588785 | 111 | 0.539512 | [
[
[
"url = 'https://eric.clst.org/assets/wiki/uploads/Stuff/gz_2010_us_050_00_500k.json'",
"_____no_output_____"
],
[
"!wget {url}",
"--2019-06-10 22:44:31-- https://eric.clst.org/assets/wiki/uploads/Stuff/gz_2010_us_050_00_500k.json\nResolving eric.clst.org (eric.clst.org)... 162.144.177.108\nConnecting to eric.clst.org (eric.clst.org)|162.144.177.108|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 23543838 (22M) [application/json]\nSaving to: ‘gz_2010_us_050_00_500k.json’\n\ngz_2010_us_050_00_5 100%[===================>] 22.45M 31.2MB/s in 0.7s \n\n2019-06-10 22:44:32 (31.2 MB/s) - ‘gz_2010_us_050_00_500k.json’ saved [23543838/23543838]\n\n"
],
[
"!mv gz_2010_us_050_00_500k.json counties.json",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d0f717aa2e6fd5c49444769af244343c93c7cf53 | 54,833 | ipynb | Jupyter Notebook | notebooks/02.3-The-Core-Language-of-Python.ipynb | jpgill86/python-for-neuroscientists | bda03e9c93eeec507f03e410b35e2a6469c6aad4 | [
"CC-BY-4.0"
] | null | null | null | notebooks/02.3-The-Core-Language-of-Python.ipynb | jpgill86/python-for-neuroscientists | bda03e9c93eeec507f03e410b35e2a6469c6aad4 | [
"CC-BY-4.0"
] | null | null | null | notebooks/02.3-The-Core-Language-of-Python.ipynb | jpgill86/python-for-neuroscientists | bda03e9c93eeec507f03e410b35e2a6469c6aad4 | [
"CC-BY-4.0"
] | null | null | null | 30.412091 | 1,029 | 0.47548 | [
[
[
"<a href=\"https://colab.research.google.com/github/jpgill86/python-for-neuroscientists/blob/master/notebooks/02.3-The-Core-Language-of-Python.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# The Core Language of Python: Part 3",
"_____no_output_____"
],
[
"## For Loops and List Comprehensions",
"_____no_output_____"
],
[
"A `for` loop can **iterate** over a list or tuple, performing calculations for each item in the sequence. Like `if` statements, `for` loops require a **colon** to terminate the first line and consistent **indentation** (typically four spaces) below it for the block of code that will be executed for each item in the sequence. Each item in the sequence is assigned a temporary variable name that can be used within the block. In the example below, this temporary variable is called `i`:",
"_____no_output_____"
]
],
[
[
"my_list = [0, 1, 2, 3, 4, 5]\n\n# print the square of each item in my_list\n# - colon and indentation are important!\nfor i in my_list:\n print(i**2)",
"0\n1\n4\n9\n16\n25\n"
]
],
[
[
"Be careful what name you give the iterator variable, since its value will be overwritten again and again with the items in the sequence. If the variable had a value before the `for` loop, it will be lost, which may not be what you intended.",
"_____no_output_____"
]
],
[
[
"i = 'abc'\nprint(f'i at the start = {i}')\n\nfor i in my_list:\n print(i**2)\n\nprint(f'i at the end = {i}')",
"i at the start = abc\n0\n1\n4\n9\n16\n25\ni at the end = 5\n"
]
],
[
[
"If you wanted to store a result from each step of the `for` loop in another list, one way you could do it is\n\n1. Initialize another variable as an empty list (`another_list = []`), and then\n2. Append a result to the new list in each step (`another_list.append`).\n\nFor example:",
"_____no_output_____"
]
],
[
[
"another_list = []\nfor i in my_list:\n another_list.append(i**2)\n\nanother_list",
"_____no_output_____"
]
],
[
[
"If the calculation of the result (in this example, squaring `i`) is fairly simple, you can perform the same work using a more concise notation called **list comprehension**. The simplest version of list comprehension takes the form `[f(i) for i in my_list]`, where `f(i)` is some function or transformation of the list item `i`. Notice list comprehensions are enclosed in square brackets (`[]`) because they create lists. Here is a list comprehension equivalent to the example above:",
"_____no_output_____"
]
],
[
[
"# basic list comprehension\n# - this means \"square the item for each item in my_list\"\nanother_list = [i**2 for i in my_list]\nanother_list",
"_____no_output_____"
]
],
[
[
"All of the work is completed in a single line of code. Elegant!\n\nList comprehensions can be more complex than this. Suppose we modified the `for` loop to append a result only if the list item is an even number (`i % 2 == 0` means that `i` divided by 2 must have a remainder of 0):",
"_____no_output_____"
]
],
[
[
"another_list = []\nfor i in my_list:\n if i % 2 == 0:\n # append only if i is even\n another_list.append(i**2)\n\n# the squares of 0, 2, 4\nanother_list",
"_____no_output_____"
]
],
[
[
"To do this with list comprehension, just add `if i % 2 == 0` to the end:",
"_____no_output_____"
]
],
[
[
"# list comprehension with conditional\n# - this means \"square the item for each item in my_list if it is even (otherwise skip it)\"\nanother_list = [i**2 for i in my_list if i % 2 == 0]\nanother_list",
"_____no_output_____"
]
],
[
[
"Suppose we modify the `for` loop further to perform a different calculation if the list item is an odd number:",
"_____no_output_____"
]
],
[
[
"another_list = []\nfor i in my_list:\n if i % 2 == 0:\n # square if i is even\n another_list.append(i**2)\n else:\n # add 100 if i is odd\n another_list.append(i+100)\n\nanother_list",
"_____no_output_____"
]
],
[
[
"This can be done with list comprehension by moving the `if i % 2 == 0` to an earlier position, just after `i**2`, and adding `else i+100`:",
"_____no_output_____"
]
],
[
[
"# list comprehension with complex conditional\n# - this means \"square the item if it is even, otherwise add 100 to it, for each item in my_list\"\nanother_list = [i**2 if i%2==0 else i+100 for i in my_list]\nanother_list",
"_____no_output_____"
]
],
[
[
"The results stored in `another_list` could be something other than a calculation using `i`. For example, strings:",
"_____no_output_____"
]
],
[
[
"# this means \"store the string 'less than 2' if the item is less than 2, otherwise store '2 or greater', for each item in my_list\"\nanother_list = ['less than 2' if i < 2 else '2 or greater' for i in my_list]\nanother_list",
"_____no_output_____"
]
],
[
[
"## While Loops",
"_____no_output_____"
],
[
"A `while` loop is another way to repeatedly perform calculations. Whereas `for` loops execute code for each item in a sequence, `while` loops execute code for as long as a condition is true. For example:",
"_____no_output_____"
]
],
[
[
"x = 0\n\nwhile x < 5:\n print(x)\n x = x + 1\n\nprint(f'final value of x = {x}')",
"0\n1\n2\n3\n4\nfinal value of x = 5\n"
]
],
[
[
"Generally, this means that the code within the `while` loop should take steps toward making the condition no longer true, even if it is unknown ahead of time how many steps that may require. In the simple example above, `x` was incremented each step until `x` was no longer less than 5. A more practical example would be a piece of code that reads a text file of unknown length one line at a time using a `while` loop that continues until a request for the next line yields nothing.\n\nBe warned: **If the condition never ceases to be true, the `while` loop will never stop**, which is probably not what you want!\n\nTry executing the code cell below, which will start an infinite loop because `x` is never incremented. You will see the icon in the left margin of the code cell spin and spin endlessly as the computer keeps executing the code within the `while` loop again and again, never stopping because the condition `x < 5` never stops being true. For this to end, you must **manually interrupt the code execution**, which you can do two ways:\n\n1. Click the spinning stop icon in the left margin, or\n2. Use the \"Runtime\" menu at the top of the page and click \"Interrupt execution\".\n\nColab executes cells one at a time, so until you interrupt the execution of this cell, you will not be able to run any other code!",
"_____no_output_____"
]
],
[
[
"# this will run forever until interrupted!\nx = 0\nwhile x < 5:\n pass # do nothing",
"_____no_output_____"
]
],
[
[
"## Dictionaries",
"_____no_output_____"
],
[
"Dictionaries are another important data type in Python. Dictionaries store **key-value pairs**, where each piece of data (the **value**) is assigned a name (the **key**) for easy access.\n\nDictionaries are created using curly braces (`{}`). (This is different from the use of curly braces in f-strings!) Inside of the curly braces, key-value pairs are separated from one another by commas, and colons separate each key from its value. For example:",
"_____no_output_____"
]
],
[
[
"# create a new dictionary using curly braces {}\nmy_dict = {'genus': 'Aplysia', 'species': 'californica', 'mass': 150}\nmy_dict",
"_____no_output_____"
]
],
[
[
"The syntax for extracting a piece of data from a dictionary is similar to indexing into lists. It uses square brackets after the dictionary name (not curly braces like you might guess), but instead of a number indicating position, a key should be provided.",
"_____no_output_____"
]
],
[
[
"# select items by key\nmy_dict['species']",
"_____no_output_____"
]
],
[
[
"Like changing the value of an item in a list via its index, the value of an item in a dictionary can be changed via its key:",
"_____no_output_____"
]
],
[
[
"# change values by key\nmy_dict['mass'] = 300\nmy_dict",
"_____no_output_____"
]
],
[
[
"New key-value pairs can be added to a dictionary the same way. In fact, you can start with an empty dictionary and build it up one key-value pair at a time:",
"_____no_output_____"
]
],
[
[
"my_dict2 = {}\n\nmy_dict2['genus'] = 'Aplysia'\nmy_dict2['species'] = 'californica'\nmy_dict2['mass'] = 300\n\nmy_dict2",
"_____no_output_____"
]
],
[
[
"Values can have any data type. Most basic data types are valid for keys too, but an important exception is lists: **lists are not allowed to be dictionary keys**. Tuples, on the other hand, are allowed to be keys. This is because keys must be immutable (uneditable), which is a property that tuples have but lists do not.",
"_____no_output_____"
]
],
[
[
"# lists cannot be keys, so this is NOT allowed\n# - the error \"unhashable type\" is a consequence of the fact that lists are not immutable (they can be changed)\nmy_dict2[['x', 'y', 'z']] = [1, 2, 3]",
"_____no_output_____"
],
[
"# tuples can be keys, so this IS allowed\nmy_dict2[('x', 'y', 'z')] = [1, 2, 3]\n\nmy_dict2",
"_____no_output_____"
]
],
[
[
"You can delete a key-value pair from a dictionary using the `del` keyword:",
"_____no_output_____"
]
],
[
[
"del my_dict2['species']\n\nmy_dict2",
"_____no_output_____"
]
],
[
[
"As a matter of fact, **`del` is how you unset any variable**:",
"_____no_output_____"
]
],
[
[
"del my_dict2\n\n# now my_dict2 is not defined\nmy_dict2",
"_____no_output_____"
]
],
[
[
"Just like lists and tuples, `for` loops can iterate over a dictionary. In its simplest form, this actually iterates over the dictionary's keys. In the example below, we choose to use the name `k`, rather than `i`, for the temporary variable to reflect this. To access the value associated with key `k`, we must use `my_dict[k]`.",
"_____no_output_____"
]
],
[
[
"# iterate over keys\nfor k in my_dict:\n print(f'key: {k} --> value: {my_dict[k]}')",
"key: genus --> value: Aplysia\nkey: species --> value: californica\nkey: mass --> value: 300\n"
]
],
[
[
"The dictionary method `items()` returns a (special type of) list of tuples, where each tuple is a key-value pair:",
"_____no_output_____"
]
],
[
[
"# using list() here to simplify how the list of tuples is displayed\nlist(my_dict.items())",
"_____no_output_____"
]
],
[
[
"When using a `for` loop to iterate over any list of tuples (or a list of lists, or a tuple of lists, or a tuple of tuples...) such as this, you can assign a temporary variable name to each item in the inner tuple/list. This is an example of what is called **unpacking**. For example:",
"_____no_output_____"
]
],
[
[
"list_of_tuples = [\n ('a', 1),\n ('b', 2),\n ('c', 3),\n]\n\nfor (letter, number) in list_of_tuples:\n print(letter, number)",
"a 1\nb 2\nc 3\n"
]
],
[
[
"In the example above, the parentheses around the iterator variables `(letter, number)` are actually optional, so the loop could be written without them:",
"_____no_output_____"
]
],
[
[
"for letter, number in list_of_tuples:\n print(letter, number)",
"a 1\nb 2\nc 3\n"
]
],
[
[
"Using the list of tuples produced by `my_dict.items()` and unpacking each key-value tuple into `k,v`, we can write the `for` loop this way:",
"_____no_output_____"
]
],
[
[
"# iterate over key-value pairs\nfor k,v in my_dict.items():\n print(f'key: {k} --> value: {v}')",
"key: genus --> value: Aplysia\nkey: species --> value: californica\nkey: mass --> value: 300\n"
]
],
[
[
"Notice this does exactly the same thing as\n```python\nfor k in my_dict:\n print(f'key: {k} --> value: {my_dict[k]}')\n```\nseen earlier, except the version using `for k,v in my_dict.items()` conveniently assigns the name `v` to the value of each key-value pair, so that `my_dict[k]` does not need to be typed out.\n\nLike list comprehensions, there exists a concise way for constructing dictionaries from a sequence, called **dictionary comprehension**. The syntax is similar, but a key and a value must be computed for each iteration, separated by a colon.\n\nTo set up an example, here is a long way of constructing a dictionary of squares using a `for` loop, where the keys are string versions of the numbers:",
"_____no_output_____"
]
],
[
[
"my_list = [0, 1, 2, 3, 4, 5]\n\nsquares = {}\nfor i in my_list:\n # store keys as strings and values as integers\n squares[str(i)] = i**2\n\nsquares",
"_____no_output_____"
]
],
[
[
"Here is a dictionary comprehension that does the same thing. Note it is enclosed with curly braces because it produces a dictionary, and a colon separates a key and its value.",
"_____no_output_____"
]
],
[
[
"# basic dictionary comprehension\n# - this means \"pair a string version of the item with its square for each item in my_list\"\nsquares = {str(i): i**2 for i in my_list}\n\nsquares",
"_____no_output_____"
]
],
[
[
"Like list comprehension, conditionals are allowed for the values:",
"_____no_output_____"
]
],
[
[
"# dictionary comprehension with complex conditional\n# - this means \"pair a string version of the item with its square if it is even, otherwise with the item plus 100, for each item in my_list\"\nsquares = {str(i): i**2 if i%2==0 else i+100 for i in my_list}\n\nsquares",
"_____no_output_____"
]
],
[
[
"## A Practical Example for Dictionaries and Comprehensions",
"_____no_output_____"
],
[
"So how are dictionaries useful? There are countless ways, but let's look at one example. Previously we saw in the section titled \"Lists vs. Tuples\" that a list of tuples with consistent structure is useful because pieces of data have predictable indices. For example, with this definition of `species_data`, the genus of every entry always has index 0:",
"_____no_output_____"
]
],
[
[
"species_data = [\n # genus, species, date named, common name\n ('Aplysia', 'californica', 1863, 'California sea hare'),\n ('Macrocystis', 'pyrifera', 1820, 'Giant kelp'),\n ('Pagurus', 'samuelis', 1857, 'Blueband hermit crab'),\n]\n\n# print every genus\nfor sp in species_data:\n print(sp[0])",
"Aplysia\nMacrocystis\nPagurus\n"
]
],
[
[
"However, this approach requires that we memorize the meaning of each index (0 = genus, 1 = species, etc.). If we use dictionaries instead of tuples, we can access data by name, rather than arbitrary indices. To do this, we could convert every tuple into a dictionary, so that the whole data set is a list of dictionaries with identical keys.\n\nTo demonstrate this, we could write everything out, like this:",
"_____no_output_____"
]
],
[
[
"species_dicts = [\n {'genus': 'Aplysia', 'species': 'californica', 'year': 1863, 'common': 'California sea hare'},\n {'genus': 'Macrocystis', 'species': 'pyrifera', 'year': 1820, 'common': 'Giant kelp'},\n {'genus': 'Pagurus', 'species': 'samuelis', 'year': 1857, 'common': 'Blueband hermit crab'},\n]",
"_____no_output_____"
]
],
[
[
"However, if the `species_data` list already existed and was much longer than it is here, this would be a lot of extra work!\n\nInstead, we could use what we have learned to programmatically construct the list of dictionaries from the existing list of tuples using a `for` loop. Here's one way to do that which uses tuple unpacking for naming each of the four pieces of data in every tuple:",
"_____no_output_____"
]
],
[
[
"# create a new empty list\nspecies_dicts = []\n\n# for each tuple, unpack the 4 pieces of data into 4 temporary variables\nfor genus, species, year, common in species_data:\n\n # build a new dictionary for this species\n d = {'genus': genus, 'species': species, 'year': year, 'common': common}\n\n # append the dictionary to the new list\n species_dicts.append(d)\n\n# display the new list of dictionaries\nspecies_dicts",
"_____no_output_____"
]
],
[
[
"Now the genus of every entry can be accessed using the key `'genus'` rather than index 0:",
"_____no_output_____"
]
],
[
[
"# print every genus\nfor sp in species_dicts:\n print(sp['genus'])",
"Aplysia\nMacrocystis\nPagurus\n"
]
],
[
[
"If we want to be *really* clever, we can do the entire conversion in a single step by *constructing a dictionary inside a list comprehension*. For this, we need to first introduce another built-in function.\n\nThe `zip()` function takes two or more sequences (e.g., lists or tuples) as inputs and groups the elements from each sequence in order. For example:",
"_____no_output_____"
]
],
[
[
"list1 = ['a', 'b', 'c']\nlist2 = [1, 2, 3]\n\n# using list() here to simplify how the result is displayed\nlist(zip(list1, list2))",
"_____no_output_____"
]
],
[
[
"How can we use `zip()` to help us convert our list of tuples into a list of dictionaries? First, define a variable containing the dictionary keys:",
"_____no_output_____"
]
],
[
[
"keys = ('genus', 'species', 'year', 'common')",
"_____no_output_____"
]
],
[
[
"With this, it is possible to pair the values from one of the tuples with these keys. For example, if we just look at the first tuple:",
"_____no_output_____"
]
],
[
[
"values = species_data[0]\nlist(zip(keys, values))",
"_____no_output_____"
]
],
[
[
"Here we have a list of tuples, where each tuple is a key-value pair. This is just like what the `items()` function returns for a dictionary. From this, we could construct a dictionary from this first tuple using dictionary comprehension:",
"_____no_output_____"
]
],
[
[
"{k:v for k,v in zip(keys, values)}",
"_____no_output_____"
]
],
[
[
"Equivalently, because there is no extra manipulation of `k` or `v` here, we could use the built-in function `dict()` to convert the list of key-value pairs directly into a dictionary:",
"_____no_output_____"
]
],
[
[
"dict(zip(keys, values))",
"_____no_output_____"
]
],
[
[
"All we have to do now is generalize this to all tuples in `species_data`. We can use a list comprehension for this, which gives us the final expression that does the entire conversion in one step, from a list of tuples to a list of dictionaries:",
"_____no_output_____"
]
],
[
[
"keys = ('genus', 'species', 'year', 'common')\nspecies_dicts = [dict(zip(keys, values)) for values in species_data]\nspecies_dicts",
"_____no_output_____"
]
],
[
[
"This is much more elegant than writing out the list of dictionaries by hand!\n\nAs a final proof of success, we can once again access the genus of every entry using a key:",
"_____no_output_____"
]
],
[
[
"# print every genus\nfor sp in species_dicts:\n print(sp['genus'])",
"Aplysia\nMacrocystis\nPagurus\n"
]
],
[
[
"For me, the amount of work we get out of this single expression, `[dict(zip(keys, values)) for values in species_data]`, is delightful!",
"_____no_output_____"
],
[
"# Continue to the Next Lesson\n\nReturn to home to continue to the next lession:\n\nhttps://jpgill86.github.io/python-for-neuroscientists/",
"_____no_output_____"
],
[
"# External Resources",
"_____no_output_____"
],
[
"The official language documentation:\n\n* [Python 3 documentation](https://docs.python.org/3/index.html)\n* [Built-in functions](https://docs.python.org/3/library/functions.html)\n* [Standard libraries](https://docs.python.org/3/library/index.html)\n* [Glossary of terms](https://docs.python.org/3/glossary.html)\n* [In-depth tutorial](https://docs.python.org/3/tutorial/index.html)\n\nExtended language documentation:\n* [IPython (Jupyter) vs. Python differences](https://ipython.readthedocs.io/en/stable/interactive/python-ipython-diff.html)\n* [IPython (Jupyter) \"magic\" (`%`) commands](https://ipython.readthedocs.io/en/stable/interactive/magics.html)\n\nFree interactive books created by Jake VanderPlas:\n\n* [A Whirlwind Tour of Python](https://colab.research.google.com/github/jakevdp/WhirlwindTourOfPython/blob/master/Index.ipynb) [[PDF version]](https://www.oreilly.com/programming/free/files/a-whirlwind-tour-of-python.pdf)\n* [Python Data Science Handbook](https://colab.research.google.com/github/jakevdp/PythonDataScienceHandbook/blob/master/notebooks/Index.ipynb)",
"_____no_output_____"
],
[
"# License\n\n[This work](https://github.com/jpgill86/python-for-neuroscientists) is licensed under a [Creative Commons Attribution 4.0 International\nLicense](http://creativecommons.org/licenses/by/4.0/).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0f71c91877cb54f0a1994a0e8eb4dfbd19700df | 145,769 | ipynb | Jupyter Notebook | lecture_code/20_Deep Learning.ipynb | alokranjan1802/ml19-20w | 42b8df929fdc8f91d72b1328daca098266a0b307 | [
"MIT"
] | 108 | 2019-11-26T01:06:51.000Z | 2022-03-20T07:20:55.000Z | lecture_code/20_Deep Learning.ipynb | Lakshita2002/ml19-20w | bd8ef13f7016f1fb3d604b01bc913e18b44868cb | [
"MIT"
] | 4 | 2020-02-10T17:55:52.000Z | 2020-05-14T16:13:14.000Z | lecture_code/20_Deep Learning.ipynb | Lakshita2002/ml19-20w | bd8ef13f7016f1fb3d604b01bc913e18b44868cb | [
"MIT"
] | 225 | 2020-01-03T14:20:41.000Z | 2022-03-23T18:56:31.000Z | 440.389728 | 45,944 | 0.929525 | [
[
[
"# A glimpse into the inner working of a 2 layer Neural network",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import numpy as np\nfrom numpy import random as nprand\nfrom cs771 import plotData as pd, utils, genSyntheticData as gsd\nfrom keras.models import Sequential\nfrom keras.layers import Dense as dense\nfrom keras import optimizers",
"Using TensorFlow backend.\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorflow\\python\\framework\\dtypes.py:516: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorflow\\python\\framework\\dtypes.py:517: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorflow\\python\\framework\\dtypes.py:518: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorflow\\python\\framework\\dtypes.py:519: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorflow\\python\\framework\\dtypes.py:520: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorflow\\python\\framework\\dtypes.py:525: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:541: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:542: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:543: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:544: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:545: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\nC:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:550: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\n"
],
[
"d = 2 \nn = 20\nr = 2\n\ntmp1 = gsd.genSphericalData( d, n, [-5, -5], r )\ntmp2 = gsd.genSphericalData( d, n, [5, 5], r )\nXPos = np.vstack( (tmp1, tmp2) )\nyPos = np.ones( (XPos.shape[0],) )\n\ntmp1 = gsd.genSphericalData( d, n, [-5, 5], r )\ntmp2 = gsd.genSphericalData( d, n, [5, -5], r )\nXNeg = np.vstack( (tmp1, tmp2) )\nyNeg = np.zeros( (XNeg.shape[0],) )\n\nX = np.vstack( (XPos, XNeg) )\ny = np.concatenate( (yPos, yNeg) )\nn = X.shape[0]\nidx = nprand.permutation( n )\n\nX = X[idx]\ny = y[idx]\n\nmu = np.mean( X, axis = 0 )\nsigma = np.std( X, axis = 0 )\n\nX -= mu\nX /= sigma",
"_____no_output_____"
],
[
"# You may get deprecation warnings about tensorflow when you run\n# this cell for the first time. This is okay and not an error\n# It seems TF has disabled several functional API in its new version\n# and keras routines have not (yet) been upgraded to use them and\n# continue to use the old (deprecated) routines hence the warnings\n\nmodel = Sequential()\nmodel.add( dense( units = 2, activation = \"sigmoid\", input_dim = 2, use_bias = True ) )\nmodel.add( dense( units = 1, activation = \"sigmoid\", use_bias = True ) )\n# Setting a very large learning rate lr may make the NN temperamental and cause\n# it to converge to a local optima. Keras supports \"callbacks\" which allow the\n# user to dynamically lower learning rate if progress has stalled\nopt = optimizers.Adam( lr = 0.1, beta_1 = 0.9, beta_2 = 0.999, amsgrad = True )\n# Metrics are just for sake of display, not for sake of training\n# Set verbose = 1 or 2 to see metrics reported for every epoch of training\n# Notice that whereas loss value goes down almost monotonically, the accuracy\n# may fluctuate i.e. go down a bit before finally going up again\nmodel.compile( loss = \"binary_crossentropy\", optimizer = opt, metrics = [\"binary_accuracy\"] )\nhistory = model.fit( X, y, epochs = 50, batch_size = n//8, verbose = 0 )\n\nfig0, ax0 = pd.getFigList( nrows = 1, ncols = 2, sizex = 5, sizey = 4 )\nax0[0].plot(history.history['loss'])\nax0[1].plot(history.history['binary_accuracy'])\nax0[0].set_xlabel( \"Epochs\" )\nax0[0].set_ylabel( \"Binary Cross Entropy Loss\" )\nax0[1].set_xlabel( \"Epochs\" )\nax0[1].set_ylabel( \"Classification Accuracy\" )",
"WARNING:tensorflow:From C:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\keras\\backend\\tensorflow_backend.py:66: The name tf.get_default_graph is deprecated. Please use tf.compat.v1.get_default_graph instead.\n\nWARNING:tensorflow:From C:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\keras\\backend\\tensorflow_backend.py:541: The name tf.placeholder is deprecated. Please use tf.compat.v1.placeholder instead.\n\nWARNING:tensorflow:From C:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\keras\\backend\\tensorflow_backend.py:4432: The name tf.random_uniform is deprecated. Please use tf.random.uniform instead.\n\nWARNING:tensorflow:From C:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\keras\\optimizers.py:793: The name tf.train.Optimizer is deprecated. Please use tf.compat.v1.train.Optimizer instead.\n\nWARNING:tensorflow:From C:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\keras\\backend\\tensorflow_backend.py:3657: The name tf.log is deprecated. Please use tf.math.log instead.\n\nWARNING:tensorflow:From C:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\tensorflow\\python\\ops\\nn_impl.py:180: add_dispatch_support.<locals>.wrapper (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\nWARNING:tensorflow:From C:\\Users\\purushot\\AppData\\Roaming\\Python\\Python37\\site-packages\\keras\\backend\\tensorflow_backend.py:1033: The name tf.assign_add is deprecated. Please use tf.compat.v1.assign_add instead.\n\n"
],
[
"def ffpredict( X ):\n # Our shading code anyway converts predictions to [0,1] scores\n return model.predict_classes( X )\n\nfig = pd.getFigure( 10, 10 )\n(xlim, ylim) = np.max( np.abs( X ), axis = 0 ) * 1.1\npd.shade2D( ffpredict, fig, mode = \"batch\", xlim = xlim, ylim = ylim )\npd.plot2D( X[y == 1], fig, color = 'g', marker = '+' )\npd.plot2D( X[y == 0], fig, color = 'r', marker = 'x' )",
"_____no_output_____"
],
[
"def sigmoid( a ):\n return 1/(1 + np.exp( -a ))\n\ndef getHiddenLayerActivations( X ):\n return sigmoid( X.dot( w ) + b )\n\n# Our network learns a function of the form (s = sigmoid function)\n# s( u.T * s( P.T * x + q ) + v )\n\n# Weights that go to the hidden layer\nP = model.layers[0].get_weights()[0]\nq = model.layers[0].get_weights()[1]\n\n# Weights that go to the output layer\nu = model.layers[1].get_weights()[0]\nv = model.layers[1].get_weights()[1]\n\n# Get the post activations of the first hidden layer neuron\n# The multiplication with sign(u[0]) is just to make sure\n# that the colors turn out nicely in the plots\nw = P[:,0] * np.sign( u[0] ) \nb = q[0] * np.sign( u[0] )\n\nfig2 = pd.getFigure( 10, 10 )\npd.shade2DProb( getHiddenLayerActivations, fig2, mode = \"batch\", xlim = xlim, ylim = ylim )\npd.plot2D( X[y == 1], fig2, color = 'g', marker = '+' )\npd.plot2D( X[y == 0], fig2, color = 'r', marker = 'x' )\n\n# Get the post activations of the second hidden layer neuron\n# The multiplication with sign(u[1]) is yet again just to make\n# sure that the colors turn out nicely in the plots\nw = P[:,1] * np.sign( u[1] ) \nb = q[1] * np.sign( u[1] )\n\nfig3 = pd.getFigure( 10, 10 )\npd.shade2DProb( getHiddenLayerActivations, fig3, mode = \"batch\", xlim = xlim, ylim = ylim )\npd.plot2D( X[y == 1], fig3, color = 'g', marker = '+' )\npd.plot2D( X[y == 0], fig3, color = 'r', marker = 'x' )\n\n# Note that the two nodes in the hidden layer cooperate to learn the classifier\n# Neither node can fully classify the red points from the green points on its own\n# so they share the burden. Each node takes up the responsibility of isolating\n# one red clump from the rest of the data. Together they make a perfect classifier :)\n# One can interpret these two nodes as learning two useful features such that the\n# learning problem become linearly separable when given these two new features\nprint( model.layers[0].get_weights() )\nprint( model.layers[1].get_weights() )\n\n# See the value of the weights below and verify that they indeed are of the form\n# that we saw in the toy code (that demonstrated universality of NN)",
"[array([[ 8.140034 , 6.2257695],\n [-8.022345 , -6.2070103]], dtype=float32), array([ 7.6521316, -6.4985967], dtype=float32)]\n[array([[ 7.7717934],\n [-8.765444 ]], dtype=float32), array([-3.4945343], dtype=float32)]\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0f7325ac4b7763597326f536b78ad1dadda2a70 | 20,318 | ipynb | Jupyter Notebook | webgraph-course/week-1/GraphDiameter.ipynb | menshikh-iv/hw | 71d674e580cfe9f9eb19d3c2f96b4dc5f2ea9070 | [
"MIT"
] | null | null | null | webgraph-course/week-1/GraphDiameter.ipynb | menshikh-iv/hw | 71d674e580cfe9f9eb19d3c2f96b4dc5f2ea9070 | [
"MIT"
] | null | null | null | webgraph-course/week-1/GraphDiameter.ipynb | menshikh-iv/hw | 71d674e580cfe9f9eb19d3c2f96b4dc5f2ea9070 | [
"MIT"
] | null | null | null | 197.262136 | 18,032 | 0.905207 | [
[
[
"import networkx as nx\nimport itertools as it\nfrom collections import Counter\n\ng = nx.read_edgelist(\"1.graph.txt\", nodetype=int)\n\n%time max_comp = max(nx.connected_component_subgraphs(g), key=lambda _: _.number_of_nodes())\n%time pathes = nx.networkx.shortest_path_length(max_comp)\n\n%time cnt = Counter(pathes[u][v] for (u, v) in it.combinations(max_comp.nodes(), 2))\nprint(\"diameter(G): {}\".format(max(cnt.keys())))",
"CPU times: user 340 ms, sys: 16 ms, total: 356 ms\nWall time: 355 ms\nCPU times: user 58.1 s, sys: 584 ms, total: 58.6 s\nWall time: 58 s\nCPU times: user 2.26 s, sys: 16 ms, total: 2.28 s\nWall time: 2.25 s\ndiameter(G): 11\n"
],
[
"data = []\ndet = float(sum(cnt.values()))\nfor i in range(1, 12):\n data.append((i, sum(cnt[j] for j in range(1, 12) if j <= i) / det))",
"_____no_output_____"
],
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\n\nx, y = zip(*data)\nplt.plot(x, y)\nplt.grid()\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d0f73884d2f621d6ae5f9ba6a04281fa6d755ae7 | 60,755 | ipynb | Jupyter Notebook | bronze/B39_Controlled_Operations.ipynb | HasanIjaz-HB/Quantum-Computing | 53c2df99cd2efbfb827857125991342f336a3097 | [
"MIT"
] | null | null | null | bronze/B39_Controlled_Operations.ipynb | HasanIjaz-HB/Quantum-Computing | 53c2df99cd2efbfb827857125991342f336a3097 | [
"MIT"
] | null | null | null | bronze/B39_Controlled_Operations.ipynb | HasanIjaz-HB/Quantum-Computing | 53c2df99cd2efbfb827857125991342f336a3097 | [
"MIT"
] | null | null | null | 48.103721 | 9,080 | 0.645412 | [
[
[
"<table>\n <tr><td align=\"right\" style=\"background-color:#ffffff;\">\n <img src=\"../images/logo.jpg\" width=\"20%\" align=\"right\">\n </td></tr>\n <tr><td align=\"right\" style=\"color.:#777777;background-color:#ffffff;font-size:12px;\">\n Prepared by Abuzer Yakaryilmaz and Maksim Dimitrijev<br>\n Özlem Salehi | December 4, 2019 (updated)\n </td></tr>\n <tr><td align=\"right\" style=\"color:#bbbbbb;background-color:#ffffff;font-size:11px;font-style:italic;\">\n This cell contains some macros. If there is a problem with displaying mathematical formulas, please run this cell to load these macros.\n </td></tr>\n</table>\n$ \\newcommand{\\bra}[1]{\\langle #1|} $\n$ \\newcommand{\\ket}[1]{|#1\\rangle} $\n$ \\newcommand{\\braket}[2]{\\langle #1|#2\\rangle} $\n$ \\newcommand{\\dot}[2]{ #1 \\cdot #2} $\n$ \\newcommand{\\biginner}[2]{\\left\\langle #1,#2\\right\\rangle} $\n$ \\newcommand{\\mymatrix}[2]{\\left( \\begin{array}{#1} #2\\end{array} \\right)} $\n$ \\newcommand{\\myvector}[1]{\\mymatrix{c}{#1}} $\n$ \\newcommand{\\myrvector}[1]{\\mymatrix{r}{#1}} $\n$ \\newcommand{\\mypar}[1]{\\left( #1 \\right)} $\n$ \\newcommand{\\mybigpar}[1]{ \\Big( #1 \\Big)} $\n$ \\newcommand{\\sqrttwo}{\\frac{1}{\\sqrt{2}}} $\n$ \\newcommand{\\dsqrttwo}{\\dfrac{1}{\\sqrt{2}}} $\n$ \\newcommand{\\onehalf}{\\frac{1}{2}} $\n$ \\newcommand{\\donehalf}{\\dfrac{1}{2}} $\n$ \\newcommand{\\hadamard}{ \\mymatrix{rr}{ \\sqrttwo & \\sqrttwo \\\\ \\sqrttwo & -\\sqrttwo }} $\n$ \\newcommand{\\vzero}{\\myvector{1\\\\0}} $\n$ \\newcommand{\\vone}{\\myvector{0\\\\1}} $\n$ \\newcommand{\\vhadamardzero}{\\myvector{ \\sqrttwo \\\\ \\sqrttwo } } $\n$ \\newcommand{\\vhadamardone}{ \\myrvector{ \\sqrttwo \\\\ -\\sqrttwo } } $\n$ \\newcommand{\\myarray}[2]{ \\begin{array}{#1}#2\\end{array}} $\n$ \\newcommand{\\X}{ \\mymatrix{cc}{0 & 1 \\\\ 1 & 0} } $\n$ \\newcommand{\\Z}{ \\mymatrix{rr}{1 & 0 \\\\ 0 & -1} } $\n$ \\newcommand{\\Htwo}{ \\mymatrix{rrrr}{ \\frac{1}{2} & \\frac{1}{2} & \\frac{1}{2} & \\frac{1}{2} \\\\ \\frac{1}{2} & -\\frac{1}{2} & \\frac{1}{2} & -\\frac{1}{2} \\\\ \\frac{1}{2} & \\frac{1}{2} & -\\frac{1}{2} & -\\frac{1}{2} \\\\ \\frac{1}{2} & -\\frac{1}{2} & -\\frac{1}{2} & \\frac{1}{2} } } $\n$ \\newcommand{\\CNOT}{ \\mymatrix{cccc}{1 & 0 & 0 & 0 \\\\ 0 & 1 & 0 & 0 \\\\ 0 & 0 & 0 & 1 \\\\ 0 & 0 & 1 & 0} } $\n$ \\newcommand{\\norm}[1]{ \\left\\lVert #1 \\right\\rVert } $",
"_____no_output_____"
],
[
"<h2>Controlled Operations</h2>",
"_____no_output_____"
],
[
"We are going to look at controlled operators acting on multiple qubits. ",
"_____no_output_____"
],
[
"<h3> CNOT operator </h3>",
"_____no_output_____"
],
[
"CNOT is an operator defined on two qubits:\n\n$$\n CNOT = \\mymatrix{cccc}{1 & 0 & 0 & 0 \\\\ 0 & 1 & 0 & 0 \\\\ 0 & 0 & 0 & 1 \\\\ 0 & 0 & 1 & 0} .\n$$\n\nIts effect is very simple: if the state of the first qubit is one, then the state of the second qubit is flipped. \n\nIf the state of the first bit is zero, then the state of the second qubit remains the same. \n\nIn summary:\n<ul>\n <li>$ CNOT \\ket{00} = \\ket{00} $, </li>\n <li>$ CNOT \\ket{01} = \\ket{01} $, </li>\n <li>$ CNOT \\ket{10} = \\ket{11} $, </li>\n <li>$ CNOT \\ket{11} = \\ket{10} $. </li>\n</ul>\n\nCNOT refers to as Controlled-NOT: NOT operator is applied in a controlled way.",
"_____no_output_____"
],
[
"<h3> cx-gate </h3>\n\nIn Qiskit, CNOT operator is represented as cx-gate.\n\nIt takes two arguments: controller-qubit and target-qubit.\n\nIts implementation is as follows:\n\n<i> <b>x-gate</b> (NOT operator) is applied to <u>the target qubit</u> that is <b>CONTROLLED</b> by <u>the controller qubit</u>.</i> \n\n",
"_____no_output_____"
],
[
"<h3> Unitary backend</h3>\n\nUnitary_simulator gives a unitary representation of all gates in the circuit until this point.\n\n``` python\n job = execute(circuit, Aer.get_backend('unitary_simulator'))\n current_unitary = job.result().get_unitary(circuit, decimals=3)\n print(current_unitary)\n```",
"_____no_output_____"
],
[
"\nLet's check the unitary operator corresponding to the CNOT. We follow the qiskit order.",
"_____no_output_____"
]
],
[
[
"# draw the circuit\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\nqreg1 = QuantumRegister(2)\ncreg1 = ClassicalRegister(2)\n\nmycircuit1 = QuantumCircuit(qreg1,creg1)\n\nmycircuit1.cx(qreg1[1],qreg1[0])\n\n\njob = execute(mycircuit1,Aer.get_backend('unitary_simulator'))\nu=job.result().get_unitary(mycircuit1,decimals=3)\nfor i in range(len(u)):\n s=\"\"\n for j in range(len(u)):\n val = str(u[i][j].real)\n while(len(val)<5): val = \" \"+val\n s = s + val\n print(s)\n\nmycircuit1.draw(output=\"mpl\")\n",
" 1.0 0.0 0.0 0.0\n 0.0 1.0 0.0 0.0\n 0.0 0.0 0.0 1.0\n 0.0 0.0 1.0 0.0\n"
]
],
[
[
"Now, let's apply CNOT to the states $ \\ket{00}, \\ket{01}, \\ket{10}, \\ket{11} $ iteratively where qreg[1] is the control and qreg[0] is the target.",
"_____no_output_____"
]
],
[
[
"# import all necessary objects and methods for quantum circuits\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\nall_inputs=['00','01','10','11']\n\nfor input in all_inputs:\n qreg2 = QuantumRegister(2) # quantum register with 2 qubits\n creg2 = ClassicalRegister(2) # classical register with 2 bits\n mycircuit2 = QuantumCircuit(qreg2,creg2) # quantum circuit with quantum and classical registers\n \n #initialize the inputs\n if input[0]=='1':\n mycircuit2.x(qreg2[1]) # set the state of qreg[1] to |1>\n if input[1]=='1':\n mycircuit2.x(qreg2[0]) # set the state of qreg[0] to |1>\n\n # apply cx(first-qubit,second-qubit)\n mycircuit2.cx(qreg2[1],qreg2[0])\n\n # measure both qubits\n mycircuit2.measure(qreg2,creg2)\n \n # execute the circuit 100 times in the local simulator\n job = execute(mycircuit2,Aer.get_backend('qasm_simulator'),shots=100)\n counts = job.result().get_counts(mycircuit2)\n for outcome in counts: # print the reverse of the outcomes\n print(\"our input is\",input,\": \",outcome,\"is observed\",counts[outcome],\"times\")",
"our input is 00 : 00 is observed 100 times\nour input is 01 : 01 is observed 100 times\nour input is 10 : 11 is observed 100 times\nour input is 11 : 10 is observed 100 times\n"
]
],
[
[
"<h3>Task 1</h3>\n\nOur task is to learn the behavior of the following quantum circuit by doing experiments.\n\nOur circuit has two qubits. \n<ul>\n <li> Apply Hadamard to both qubits.\n <li> Apply CNOT(qreg[1] is the control,qreg[0] is the target).\n <li> Apply Hadamard to both qubits.\n <li> Measure the circuit.\n</ul>\n\nIteratively initialize the qubits to $ \\ket{00} $, $ \\ket{01} $, $ \\ket{10} $, and $ \\ket{11} $.\n\nExecute your program 100 times for each iteration, and then check the outcomes for each iteration. \n\nObserve that the overall circuit implements CNOT(qreg[0] is the control, qreg[1] is the target).",
"_____no_output_____"
]
],
[
[
"# import all necessary objects and methods for quantum circuits\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\n#\n# your code is here\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\nall_inputs=['00','01','10','11']\n\nfor input in all_inputs:\n qreg1 = QuantumRegister(2) # quantum register with 2 qubits\n creg1 = ClassicalRegister(2) # classical register with 2 bits\n mycircuit1 = QuantumCircuit(qreg1,creg1) # quantum circuit with quantum and classical registers\n \n #initialize the inputs\n if input[0]=='1':\n mycircuit1.x(qreg1[1]) # set the state of the qubit to |1>\n if input[1]=='1':\n mycircuit1.x(qreg1[0]) # set the state of the qubit to |1>\n\n # apply h-gate to both qubits\n mycircuit1.h(qreg1[0])\n mycircuit1.h(qreg1[1])\n\n # apply cx\n mycircuit1.cx(qreg1[1],qreg1[0])\n\n # apply h-gate to both qubits\n mycircuit1.h(qreg1[0])\n mycircuit1.h(qreg1[1])\n\n # measure both qubits\n mycircuit1.measure(qreg1,creg1)\n \n # execute the circuit 100 times in the local simulator\n job = execute(mycircuit1,Aer.get_backend('qasm_simulator'),shots=100)\n counts = job.result().get_counts(mycircuit1)\n for outcome in counts: # print the reverse of the outcomes\n print(\"our input is\",input,\": \",outcome,\"is observed\",counts[outcome],\"times\")\n#\n",
"our input is 00 : 00 is observed 100 times\nour input is 01 : 11 is observed 100 times\nour input is 10 : 10 is observed 100 times\nour input is 11 : 01 is observed 100 times\n"
]
],
[
[
"<a href=\"B39_Controlled_Operations_Solutions.ipynb#task1\">click for our solution</a>",
"_____no_output_____"
],
[
"<h3>Task 2</h3>\n\nOur task is to learn the behavior of the following quantum circuit by doing experiments.\n\nOur circuit has two qubits. \n<ul>\n <li> Apply CNOT(qreg[1] is the control, qreg[0] is the target).\n <li> Apply CNOT(qreg[0] is the control, qreg[1] is the target).\n <li> Apply CNOT(qreg[0] is the control, qreg[1] is the target).\n</ul>\n\nIteratively initialize the qubits to $ \\ket{00} $, $ \\ket{01} $, $ \\ket{10} $, and $ \\ket{11} $.\n\nExecute your program 100 times for each iteration, and then check the outcomes for each iteration. \n\nObserve that the overall circuit swaps the values of the two qubits:\n<ul>\n <li> $\\ket{00} \\rightarrow \\ket{00} $ </li>\n <li> $\\ket{01} \\rightarrow \\ket{10} $ </li>\n <li> $\\ket{10} \\rightarrow \\ket{01} $ </li>\n <li> $\\ket{11} \\rightarrow \\ket{11} $ </li>\n</ul>",
"_____no_output_____"
]
],
[
[
"# import all necessary objects and methods for quantum circuits\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\n#\n# your code is here\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\nall_inputs=['00','01','10','11']\n\nfor input in all_inputs:\n qreg2 = QuantumRegister(2) # quantum register with 2 qubits\n creg2 = ClassicalRegister(2) # classical register with 2 bits\n mycircuit2 = QuantumCircuit(qreg2,creg2) # quantum circuit with quantum and classical registers\n \n #initialize the inputs\n if input[0]=='1':\n mycircuit2.x(qreg2[1]) # set the value of the qubit to |1>\n if input[1]=='1':\n mycircuit2.x(qreg2[0]) # set the value of the qubit to |1>\n\n # apply cx(qreg2[0] is the target)\n mycircuit2.cx(qreg2[1],qreg2[0])\n # apply cx(qreg2[1] is the target)\n mycircuit2.cx(qreg2[0],qreg2[1])\n # apply cx(qreg2[1] is the target)\n mycircuit2.cx(qreg2[0],qreg2[1])\n \n mycircuit2.measure(qreg2,creg2)\n \n # execute the circuit 100 times in the local simulator\n job = execute(mycircuit2,Aer.get_backend('qasm_simulator'),shots=100)\n counts = job.result().get_counts(mycircuit2)\n for outcome in counts: # print the reverse of the outcomes\n print(\"our input is\",input,\": \",outcome,\"is observed\",counts[outcome],\"times\")\n#\n",
"our input is 00 : 00 is observed 100 times\nour input is 01 : 01 is observed 100 times\nour input is 10 : 11 is observed 100 times\nour input is 11 : 10 is observed 100 times\n"
]
],
[
[
"<a href=\"B39_Controlled_Operations_Solutions.ipynb#task2\">click for our solution</a>",
"_____no_output_____"
],
[
"<h3> Task 3 [Extra] </h3>\n\nCreate a quantum curcuit with $ n=5 $ qubits.\n\nSet each qubit to $ \\ket{1} $.\n\nRepeat 4 times:\n<ul>\n <li>Randomly pick a pair of qubits, and apply cx-gate (CNOT operator) on the pair.</li>\n</ul>\n\nDraw your circuit, and execute your program 100 times.\n\nVerify your measurement results by checking the diagram of the circuit. ",
"_____no_output_____"
]
],
[
[
"# import all necessary objects and methods for quantum circuits\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n# import randrange for random choices\nfrom random import randrange\n\n#\n# your code is here\n#\n",
"_____no_output_____"
]
],
[
[
"<a href=\"B39_Controlled_Operations_Solutions.ipynb#task3\">click for our solution</a>",
"_____no_output_____"
],
[
"<h3> Task 4 </h3>\n\nIn this task, our aim is to create an operator which will apply the NOT operator to the target qubit qreg[0] when the control qubit qreg[1] is in state $\\ket{0}$. In other words, we want to obtain the following operator:\n\n$\\mymatrix{cccc}{0 & 1 & 0 & 0 \\\\ 1 & 0 & 0 & 0 \\\\ 0 & 0 & 1 & 0 \\\\ 0 & 0 & 0 & 1}$.\n\nWe can summarize its effect as follows:\n<ul>\n <li>$ \\ket{00} \\rightarrow \\ket{01} $, </li>\n <li>$ \\ket{01} \\rightarrow \\ket{00} $, </li>\n <li>$ \\ket{10} \\rightarrow \\ket{10} $, </li>\n <li>$ \\ket{11} \\rightarrow \\ket{11} $. </li>\n</ul>\n\nWrite a function named c0x which takes the circuit name and the register as parameters and implements the operation. Check the corresponding unitary matrix using the code given below.\n<ul>\n <li>Apply NOT operator to qreg[1];</li>\n <li>Apply CNOT operator, where qreg[1] is control and qreg[0] is target;</li>\n <li>Apply NOT operator to qreg[1] - to revert it to the initial state.</li>\n</ul>",
"_____no_output_____"
],
[
"<b>Idea:</b> We can use our regular CNOT operator, and to change the condition for the control qubit we can apply NOT operator to it before the CNOT - this way the NOT operator will be applied to the target qubit when initially the state of the control qubit was $\\ket{0}$.\n\nAlthough this trick is quite simple, this approach is important and will be very useful in our following implementations.",
"_____no_output_____"
]
],
[
[
"def c0x(mycircuit,qreg):\n\n#\n# Your code here\n#",
"_____no_output_____"
],
[
"from qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\nqreg4 = QuantumRegister(2)\ncreg4 = ClassicalRegister(2)\n\nmycircuit4 = QuantumCircuit(qreg4,creg4)\n\n#We apply the operator c0x by calling the function\nc0x(mycircuit4,qreg4)\n\njob = execute(mycircuit1,Aer.get_backend('unitary_simulator'))\nu=job.result().get_unitary(mycircuit1,decimals=3)\nfor i in range(len(u)):\n s=\"\"\n for j in range(len(u)):\n val = str(u[i][j].real)\n while(len(val)<5): val = \" \"+val\n s = s + val\n print(s)\n\nmycircuit1.draw(output=\"mpl\")",
"_____no_output_____"
]
],
[
[
"<a href=\"B39_Controlled_Operations_Solutions.ipynb#task4\">click for our solution</a>",
"_____no_output_____"
],
[
"<h3>CCNOT</h3>\n\nNow we will discuss CNOT gate controlled by two qubits (also called the Toffoli gate). \n\nThe idea behind this gate is simple - NOT operator is applied to the target qubit when both control qubits are in state $\\ket{1}$. Below you can see its matrix representation:\n\n$\\mymatrix{cccccccc}{1 & 0 & 0 & 0 & 0 & 0 & 0 & 0 \\\\ 0 & 1 & 0 & 0 & 0 & 0 & 0 & 0 \\\\ 0 & 0 & 1 & 0 & 0 & 0 & 0 & 0 \\\\ 0 & 0 & 0 & 1 & 0 & 0 & 0 & 0 \\\\ 0 & 0 & 0 & 0 & 1 & 0 & 0 & 0 \\\\ 0 & 0 & 0 & 0 & 0 & 1 & 0 & 0 \\\\ 0 & 0 & 0 & 0 & 0 & 0 & 0 & 1 \\\\ 0 & 0 & 0 & 0 & 0 & 0 & 1 & 0}$.\n\n\nIn summary:\n<ul>\n <li>$ CCNOT \\ket{000} = \\ket{000} $, </li>\n <li>$ CCNOT \\ket{001} = \\ket{001} $, </li>\n <li>$ CCNOT \\ket{010} = \\ket{010} $, </li>\n <li>$ CCNOT \\ket{011} = \\ket{011} $. </li>\n <li>$ CCNOT \\ket{100} = \\ket{100} $, </li>\n <li>$ CCNOT \\ket{101} = \\ket{101} $, </li>\n <li>$ CCNOT \\ket{110} = \\ket{111} $, </li>\n <li>$ CCNOT \\ket{111} = \\ket{110} $. </li>\n</ul>",
"_____no_output_____"
],
[
"<h3> ccx-gate </h3>\n\nIn Qiskit, CCNOT operator is represented as ccx-gate.\n\nIt takes three arguments: two controller-qubits and target-qubit.\n\n circuit.ccx(control-qubit1,control-qubit2,target-qubit)\n\n<i> <b>x-gate</b> (NOT operator) is applied to <u>the target qubit</u> that is <b>CONTROLLED</b> by <u>the controller qubits</u>.</i> \n",
"_____no_output_____"
],
[
"Now, let's apply CCNOT iteratively to see its effect. (Note that we follow the qiskit order.)",
"_____no_output_____"
]
],
[
[
"# import all necessary objects and methods for quantum circuits\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\nall_inputs=['000','001','010','011','100','101','110','111']\n\nfor input in all_inputs:\n qreg3 = QuantumRegister(3) # quantum register with 3 qubits\n creg3 = ClassicalRegister(3) # classical register with 3 bits\n mycircuit3 = QuantumCircuit(qreg3,creg3) # quantum circuit with quantum and classical registers\n \n #initialize the inputs\n if input[0]=='1':\n mycircuit3.x(qreg3[2]) # set the state of the first qubit to |1>\n if input[1]=='1':\n mycircuit3.x(qreg3[1]) # set the state of the second qubit to |1>\n if input[2]=='1':\n mycircuit3.x(qreg3[0]) # set the state of the third qubit to |1>\n\n # apply ccx(first-qubit,second-qubit,third-qubit)\n mycircuit3.ccx(qreg3[2],qreg3[1],qreg3[0])\n\n # measure the qubits\n mycircuit3.measure(qreg3,creg3)\n \n # execute the circuit 100 times in the local simulator\n job = execute(mycircuit3,Aer.get_backend('qasm_simulator'),shots=100)\n counts = job.result().get_counts(mycircuit3)\n for outcome in counts: # print the reverse of the outcomes\n print(\"our input is\",input,\": \",outcome,\"is observed\",counts[outcome],\"times\")",
"our input is 000 : 000 is observed 100 times\nour input is 001 : 001 is observed 100 times\nour input is 010 : 010 is observed 100 times\nour input is 011 : 011 is observed 100 times\nour input is 100 : 100 is observed 100 times\nour input is 101 : 101 is observed 100 times\nour input is 110 : 111 is observed 100 times\nour input is 111 : 110 is observed 100 times\n"
]
],
[
[
"<hr>\nRecall Task 4. Similarly, we can create an operator which applies NOT operator to a target qubit, controlled by two qubits which are in states different than 1. For example, the following implementation allows to apply NOT operator to the target qubit if both control qubits are in state $\\ket{0}$. \n\nThe matrix form of the operator is given as follows:\n\n$\\mymatrix{cccc}{0 & 1 & 0 & 0 & 0 & 0 & 0 & 0 \\\\ 1 & 0 & 0 & 0& 0 & 0 & 0 & 0 \\\\ 0 & 0 & 1 & 0& 0 & 0 & 0 & 0 \\\\ 0 & 0 & 0 & 1& 0 & 0 & 0 & 0 \\\\ 0 & 0 & 0 & 0& 1 & 0 & 0 & 0 \\\\ 0 & 0 & 0 & 0& 0 & 1 & 0 & 0 \\\\ 0 & 0 & 0 & 0& 0 & 0 & 1 & 0 \\\\ 0 & 0 & 0 & 0& 0 & 0 & 0 & 1}$.\n",
"_____no_output_____"
]
],
[
[
"def cc0x(mycircuit,qreg):\n\n mycircuit.x(qreg[2])\n mycircuit.x(qreg[1])\n\n mycircuit.ccx(qreg[2],qreg[1],qreg[0])\n\n # Returning control qubits to the initial state\n mycircuit.x(qreg[1])\n mycircuit.x(qreg[2])",
"_____no_output_____"
],
[
"from qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\nqreg4 = QuantumRegister(3)\ncreg4 = ClassicalRegister(3)\n\nmycircuit4 = QuantumCircuit(qreg4,creg4)\n\ncc0x(mycircuit4,qreg4)\n\njob = execute(mycircuit4,Aer.get_backend('unitary_simulator'))\nu=job.result().get_unitary(mycircuit4,decimals=3)\nfor i in range(len(u)):\n s=\"\"\n for j in range(len(u)):\n val = str(u[i][j].real)\n while(len(val)<5): val = \" \"+val\n s = s + val\n print(s)\n\nmycircuit4.draw(output=\"mpl\")",
" 0.0 1.0 0.0 0.0 0.0 0.0 0.0 0.0\n 1.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0\n 0.0 0.0 1.0 0.0 0.0 0.0 0.0 0.0\n 0.0 0.0 0.0 1.0 0.0 0.0 0.0 0.0\n 0.0 0.0 0.0 0.0 1.0 0.0 0.0 0.0\n 0.0 0.0 0.0 0.0 0.0 1.0 0.0 0.0\n 0.0 0.0 0.0 0.0 0.0 0.0 1.0 0.0\n 0.0 0.0 0.0 0.0 0.0 0.0 0.0 1.0\n"
]
],
[
[
"<h3>Task 5</h3>\n\nYou have a circuit with three qubits. Apply NOT operator to qreg[1] if qreg[0] is in state 0 and qreg[2] is in state 1. Check its efffect on different inputs.",
"_____no_output_____"
]
],
[
[
"from qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\nall_inputs=['000','001','010','011','100','101','110','111']\n\nfor input in all_inputs:\n qreg5 = QuantumRegister(3) # quantum register with 3 qubits\n creg5 = ClassicalRegister(3) # classical register with 3 bits\n mycircuit5 = QuantumCircuit(qreg5,creg5) # quantum circuit with quantum and classical registers\n \n #initialize the inputs\n if input[0]=='1':\n mycircuit5.x(qreg5[2]) # set the state of the first qubit to |1>\n if input[1]=='1':\n mycircuit5.x(qreg5[1]) # set the state of the second qubit to |1>\n if input[2]=='1':\n mycircuit5.x(qreg5[0]) # set the state of the third qubit to |1>\n\n#\n# You code here\n mycircuit5.x(qreg5[0])\n\n mycircuit5.ccx(qreg5[2],qreg5[0],qreg5[1])\n\n #Set back to initial value\n mycircuit5.x(qreg5[0])\n#\n\n\n# measure the qubits\n mycircuit5.measure(qreg5,creg5)\n \n # execute the circuit 100 times in the local simulator\n job = execute(mycircuit5,Aer.get_backend('qasm_simulator'),shots=100)\n counts = job.result().get_counts(mycircuit5)\n for outcome in counts: # print the reverse of the outcomes\n print(\"our input is\",input,\": \",outcome,\"is observed\",counts[outcome],\"times\")",
"our input is 000 : 000 is observed 100 times\nour input is 001 : 001 is observed 100 times\nour input is 010 : 010 is observed 100 times\nour input is 011 : 011 is observed 100 times\nour input is 100 : 110 is observed 100 times\nour input is 101 : 101 is observed 100 times\nour input is 110 : 100 is observed 100 times\nour input is 111 : 111 is observed 100 times\n"
]
],
[
[
"<a href=\"B39_Controlled_Operations_Solutions.ipynb#task5\">click for our solution</a>",
"_____no_output_____"
],
[
"<h3>More controls</h3>\n\nSuppose that you are given ccx operator which applies a not operator controlled by two qubits. You can use additional qubits to implement a not operator controlled by more than two qubits.\n\n\nThe following code implements a NOT operator controlled by the three qubits qreg[1], qreg[2] and qreg[3], qreg[4] is used as the additional qubit and qreg[0] is the target. We apply it iteratively. Note that the first qubit in the output is due to additional qubit.",
"_____no_output_____"
]
],
[
[
"def cccx(mycircuit,qreg):\n #qreg[4] is set to 1 if qreg[1] and qreg[2] are 1 \n mycircuit.ccx(qreg[1],qreg[2],qreg[4])\n \n #NOT operator is applied to qreg[0] if all three qubits are 1\n mycircuit5.ccx(qreg[3],qreg[4],qreg[0])\n \n #We set back qreg[4] back to its initial value\n mycircuit5.ccx(qreg[1],qreg[2],qreg[4]) ",
"_____no_output_____"
],
[
"# import all necessary objects and methods for quantum circuits\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\nall_inputs=['0000','0001','0010','0011','0100','0101','0110','0111','0000','1001','1010','1011','1100',\n '1101','1110','1111']\n\nfor input in all_inputs:\n qreg5 = QuantumRegister(5) # quantum register with 5 qubits\n creg5 = ClassicalRegister(5) # classical register with 5 bits\n mycircuit5 = QuantumCircuit(qreg5,creg5) # quantum circuit with quantum and classical registers\n \n #initialize the inputs\n if input[0]=='1':\n mycircuit5.x(qreg5[3]) # set the state of the qubit to |1>\n if input[1]=='1':\n mycircuit5.x(qreg5[2]) # set the state of the qubit to |1>\n if input[2]=='1':\n mycircuit5.x(qreg5[1]) # set the state of the qubit to |1>\n if input[3]=='1':\n mycircuit5.x(qreg5[0]) # set the state of the qubit to |1>\n\n cccx(mycircuit5,qreg5) \n \n # measure the qubits\n mycircuit5.measure(qreg5,creg5)\n \n # execute the circuit 100 times in the local simulator\n job = execute(mycircuit5,Aer.get_backend('qasm_simulator'),shots=100)\n counts = job.result().get_counts(mycircuit5)\n for outcome in counts: # print the reverse of the outcomes\n print(\"our input is\",input,\": \",outcome,\"is observed\",counts[outcome],\"times\")",
"our input is 0000 : 00000 is observed 100 times\nour input is 0001 : 00001 is observed 100 times\nour input is 0010 : 00010 is observed 100 times\nour input is 0011 : 00011 is observed 100 times\nour input is 0100 : 00100 is observed 100 times\nour input is 0101 : 00101 is observed 100 times\nour input is 0110 : 00110 is observed 100 times\nour input is 0111 : 00111 is observed 100 times\nour input is 0000 : 00000 is observed 100 times\nour input is 1001 : 01001 is observed 100 times\nour input is 1010 : 01010 is observed 100 times\nour input is 1011 : 01011 is observed 100 times\nour input is 1100 : 01100 is observed 100 times\nour input is 1101 : 01101 is observed 100 times\nour input is 1110 : 01111 is observed 100 times\nour input is 1111 : 01110 is observed 100 times\n"
]
],
[
[
"<h3>Task 6</h3>\n\nImplement the NOT operator controlled by 4 qubits where qreg[0] is the target and apply it iteratively to all possible states. Note that you will need additional qubits.",
"_____no_output_____"
]
],
[
[
"def ccccx(mycircuit,qreg):\n \n #\n #Your code here\n mycircuit.ccx(qreg[4],qreg[3],qreg[5])\n mycircuit.ccx(qreg[2],qreg[1],qreg[6])\n\n mycircuit.ccx(qreg[5],qreg[6],qreg[0])\n\n # Returning additional qubits to the initial state\n mycircuit.ccx(qreg[2],qreg[1],qreg[6])\n mycircuit.ccx(qreg[4],qreg[3],qreg[5])",
"_____no_output_____"
],
[
"# import all necessary objects and methods for quantum circuits\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\n\nall_inputs=['00000','00001','00010','00011','00100','00101','00110','00111','00000',\n '01001','01010','01011','01100','01101','01110','01111','10000','10001',\n '10010','10011','10100','10101','10110','10111','10000','11001','11010',\n '11011','11100','11101','11110','11111']\n\nfor input in all_inputs:\n qreg6 = QuantumRegister(7) # quantum register with 7 qubits\n creg6 = ClassicalRegister(7) # classical register with 7 bits\n mycircuit6 = QuantumCircuit(qreg6,creg6) # quantum circuit with quantum and classical registers\n \n #initialize the inputs\n if input[0]=='1':\n mycircuit6.x(qreg6[4]) # set the state of the first qubit to |1>\n if input[1]=='1':\n mycircuit6.x(qreg6[3]) # set the state of the second qubit to |1>\n if input[2]=='1':\n mycircuit6.x(qreg6[2]) # set the state of the third qubit to |1>\n if input[3]=='1':\n mycircuit6.x(qreg6[1]) # set the state of the fourth qubit to |1>\n if input[4]=='1':\n mycircuit6.x(qreg6[0]) # set the state of the fifth qubit to |1> \n \n ccccx(mycircuit6,qreg6)\n\n mycircuit6.measure(qreg6,creg6)\n\n job = execute(mycircuit6,Aer.get_backend('qasm_simulator'),shots=10000)\n counts = job.result().get_counts(mycircuit6)\n for outcome in counts: # print the reverse of the outcomes\n print(\"our input is\",input,\": \",outcome,\"is observed\",counts[outcome],\"times\")",
"our input is 00000 : 0000000 is observed 10000 times\nour input is 00001 : 0000001 is observed 10000 times\nour input is 00010 : 0000010 is observed 10000 times\nour input is 00011 : 0000011 is observed 10000 times\nour input is 00100 : 0000100 is observed 10000 times\nour input is 00101 : 0000101 is observed 10000 times\nour input is 00110 : 0000110 is observed 10000 times\nour input is 00111 : 0000111 is observed 10000 times\nour input is 00000 : 0000000 is observed 10000 times\nour input is 01001 : 0001001 is observed 10000 times\nour input is 01010 : 0001010 is observed 10000 times\nour input is 01011 : 0001011 is observed 10000 times\nour input is 01100 : 0001100 is observed 10000 times\nour input is 01101 : 0001101 is observed 10000 times\nour input is 01110 : 0001110 is observed 10000 times\nour input is 01111 : 0001111 is observed 10000 times\nour input is 10000 : 0010000 is observed 10000 times\nour input is 10001 : 0010001 is observed 10000 times\nour input is 10010 : 0010010 is observed 10000 times\nour input is 10011 : 0010011 is observed 10000 times\nour input is 10100 : 0010100 is observed 10000 times\nour input is 10101 : 0010101 is observed 10000 times\nour input is 10110 : 0010110 is observed 10000 times\nour input is 10111 : 0010111 is observed 10000 times\nour input is 10000 : 0010000 is observed 10000 times\nour input is 11001 : 0011001 is observed 10000 times\nour input is 11010 : 0011010 is observed 10000 times\nour input is 11011 : 0011011 is observed 10000 times\nour input is 11100 : 0011100 is observed 10000 times\nour input is 11101 : 0011101 is observed 10000 times\nour input is 11110 : 0011111 is observed 10000 times\nour input is 11111 : 0011110 is observed 10000 times\n"
]
],
[
[
"<a href=\"B39_Controlled_Operations_Solutions.ipynb#task6\">click for our solution</a>",
"_____no_output_____"
],
[
"<h3>Task 7</h3>\n\nImplement the following control: the NOT operator is applied to the target qubit qreg[0] if 5 control qubits qreg[5] to qreg[1] are initially in the state $\\ket{10101}$. Check your operator by trying different initial states. You may define a function or write your code directly.",
"_____no_output_____"
]
],
[
[
"# import all necessary objects and methods for quantum circuits\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\n#Try different initial states\nall_inputs=['101010','101011','100000','111111']\n\nfor input in all_inputs:\n qreg7 = QuantumRegister(9) # quantum register with 9 qubits\n creg7 = ClassicalRegister(9) # classical register with 9 bits\n mycircuit7 = QuantumCircuit(qreg7,creg7) # quantum circuit with quantum and classical registers\n \n #initialize the inputs\n if input[0]=='1':\n mycircuit7.x(qreg7[5]) # set the state of the first qubit to |1>\n if input[1]=='1':\n mycircuit7.x(qreg7[4]) # set the state of the second qubit to |1>\n if input[2]=='1':\n mycircuit7.x(qreg7[3]) # set the state of the third qubit to |1>\n if input[3]=='1':\n mycircuit7.x(qreg7[2]) # set the state of the fourth qubit to |1>\n if input[4]=='1':\n mycircuit7.x(qreg7[1]) # set the state of the fifth qubit to |1>\n if input[5]=='1':\n mycircuit7.x(qreg7[0]) # set the state of the fifth qubit to |1>\n\n #\n # Your code here\n #\n \n mycircuit7.measure(qreg7,creg7)\n\n job = execute(mycircuit7,Aer.get_backend('qasm_simulator'),shots=10000)\n counts = job.result().get_counts(mycircuit7)\n for outcome in counts: # print the reverse of the outcomes\n print(\"our input is\",input,\": \",outcome,\"is observed\",counts[outcome],\"times\")",
"_____no_output_____"
]
],
[
[
"<a href=\"B39_Controlled_Operations_Solutions.ipynb#task7\">click for our solution</a>",
"_____no_output_____"
],
[
"<h3>Task 8 (Optional)</h3>\n\nImplement the parametrized controlled NOT operator with 4 control qubits, where parameter will be the state of control qubits for which NOT operator will be applied to the target qubit.\n\nAs a result you need to define the following function: <i>control(circuit,quantum_reg,number)</i>, where:\n<ul>\n <li><i>circuit</i> allows to pass the quantum circuit;</li>\n <li><i>quantum_reg</i> allows to pass the quantum register;</li>\n <li><i>state</i> is the state of control qubits, between 0 and 15, where 0 corresponds to 0000 and 15 corresponds to 1111 (like binary numbers :) ).</li>\n</ul>",
"_____no_output_____"
]
],
[
[
"#state - the state of control qubits, between 0 and 15.\ndef control(circuit,quantum_reg,state):\n\n#\n# your code is here\n#",
"_____no_output_____"
]
],
[
[
"You can try different inputs to see that your function is implementing the mentioned control operation.",
"_____no_output_____"
]
],
[
[
"#Try different initial states\nall_inputs=['01010','01011','10000','11111']\n\nfor input in all_inputs:\n qreg8 = QuantumRegister(7) # quantum register with 7 qubits\n creg8 = ClassicalRegister(7) # classical register with 7 bits\n mycircuit8 = QuantumCircuit(qreg8,creg8) # quantum circuit with quantum and classical registers\n \n #initialize the inputs\n if input[0]=='1':\n mycircuit8.x(qreg8[4]) # set the state of the first qubit to |1>\n if input[1]=='1':\n mycircuit8.x(qreg8[3]) # set the state of the second qubit to |1>\n if input[2]=='1':\n mycircuit8.x(qreg8[2]) # set the state of the third qubit to |1>\n if input[3]=='1':\n mycircuit8.x(qreg8[1]) # set the state of the fourth qubit to |1>\n if input[4]=='1':\n mycircuit8.x(qreg8[0]) # set the state of the fifth qubit to |1>\n\n\n control(mycircuit8,qreg8,5)\n mycircuit8.measure(qreg8,creg8)\n\n job = execute(mycircuit8,Aer.get_backend('qasm_simulator'),shots=10000)\n counts = job.result().get_counts(mycircuit8)\n for outcome in counts: # print the reverse of the outcomes\n print(\"our input is\",input,\": \",outcome,\"is observed\",counts[outcome],\"times\")",
"_____no_output_____"
]
],
[
[
"<a href=\"B39_Controlled_Operations_Solutions.ipynb#task8\">click for our solution</a>",
"_____no_output_____"
],
[
"<h3> Multi-controlled Not Gate </h3>",
"_____no_output_____"
],
[
"In Qiskit there is a multi-controlled not gate, known as the multi controlled Toffoli gate. It is represented by mct.\n\n circuit.mct(control_list,target_qubit,ancilla_list)\n\n<i> <b>x-gate</b> (NOT operator) is applied to <u>the target qubit</u> <b>CONTROLLED</b> by <u>the list of control qubits</u> using the <u>ancilla list</u> as the additional qubits</u>.</i> \n",
"_____no_output_____"
],
[
"If there are $n$ control qubits, how many additional qubits do you need?",
"_____no_output_____"
],
[
"Let's apply a NOT operator controlled by the four qubits qreg[1], qreg[2], qreg[3] and qreg[4]. Let qreg[5] and qreg[6] be the additional qubits and let qreg[0] be the target. Let's check the inputs 11111 and 11110.",
"_____no_output_____"
]
],
[
[
"# import all necessary objects and methods for quantum circuits\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\nall_inputs=['11110','11111']\n\nfor input in all_inputs:\n qreg = QuantumRegister(7) # quantum register with 7 qubits\n creg = ClassicalRegister(7) # classical register with 7 bits\n mycircuit = QuantumCircuit(qreg,creg) # quantum circuit with quantum and classical registers\n \n #initialize the inputs\n if input[0]=='1':\n mycircuit.x(qreg[4]) # set the state of the qubit to |1>\n if input[1]=='1':\n mycircuit.x(qreg[3]) # set the state of the qubit to |1>\n if input[2]=='1':\n mycircuit.x(qreg[2]) # set the state of the qubit to |1>\n if input[3]=='1':\n mycircuit.x(qreg[1]) # set the state of the qubit to |1>\n if input[4]=='1':\n mycircuit.x(qreg[0]) # set the state of the qubit to |1>\n\n control_list=[]\n for i in range(1,5):\n control_list.append(qreg[i])\n mycircuit.mct(control_list,qreg[0],[qreg[5],qreg[6]]) \n \n # measure the qubits\n mycircuit.measure(qreg,creg)\n \n # execute the circuit 100 times in the local simulator\n job = execute(mycircuit,Aer.get_backend('qasm_simulator'),shots=100)\n counts = job.result().get_counts(mycircuit)\n for outcome in counts: # print the reverse of the outcomes\n print(\"our input is\",input,\": \",outcome,\"is observed\",counts[outcome],\"times\")",
"our input is 11110 : 0011111 is observed 100 times\nour input is 11111 : 0011110 is observed 100 times\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
d0f76209c655dbcedaae554b16787e4594ee0f14 | 472,032 | ipynb | Jupyter Notebook | tutorials/W3D2_DynamicNetworks/W3D2_Tutorial1.ipynb | sanchobarriga/course-content | a7cbe0fa40dee200bd964b349e685513bb9f71c4 | [
"CC-BY-4.0"
] | 2 | 2020-07-03T04:39:09.000Z | 2020-07-12T02:08:31.000Z | tutorials/W3D2_DynamicNetworks/W3D2_Tutorial1.ipynb | sanchobarriga/course-content | a7cbe0fa40dee200bd964b349e685513bb9f71c4 | [
"CC-BY-4.0"
] | 1 | 2020-06-22T22:57:03.000Z | 2020-06-22T22:57:03.000Z | tutorials/W3D2_DynamicNetworks/W3D2_Tutorial1.ipynb | sanchobarriga/course-content | a7cbe0fa40dee200bd964b349e685513bb9f71c4 | [
"CC-BY-4.0"
] | 1 | 2021-03-29T21:08:26.000Z | 2021-03-29T21:08:26.000Z | 155.632047 | 45,008 | 0.881358 | [
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W3D2_DynamicNetworks/W3D2_Tutorial1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Neuromatch Academy: Week 3, Day 2, Tutorial 1\n\n# Neuronal Network Dynamics: Neural Rate Models\n\n",
"_____no_output_____"
],
[
"## Background\n\nThe brain is a complex system, not because it is composed of a large number of diverse types of neurons, but mainly because of how neurons are connected to each other. The brain is a very large network of densely interconnected neurons. \n\nThe activity of neurons is constantly evolving in time. For this reason, neurons can be modeled as dynamical systems. The dynamical system approach is only one of the many modeling approaches that computational neuroscientists have developed (other points of views include information processing, network science, and statistical models). How the dynamics of neuronal networks affect the representation and processing of information in the brain is an open question. However, signatures of altered brain dynamics present in many brain diseases (e.g., in epilepsy or Parkinson's disease) tell us that it is crucial to study neuronal dynamics if we want to understand the brain.\n\nIn this tutorial, we will simulate and study one of the simplest models of biological neuronal networks. Instead of modeling and simulating individual excitatory neurons (e.g., LIF models that you implemented yesterday), we will treat them as a single homogeneous population and approximate their dynamics using a single one-dimensional equation describing the evolution of their average spiking rate in time.\n\n## Objectives\nIn this tutorial we will learn how to build a firing rate model of a single population of excitatory neurons. \n\nSteps:\n- Write the equation for the firing rate dynamics of a 1D excitatory population.\n- Visualize the response of the population as a function of parameters such as threshold level and gain, using the frequency-current (F-I) curve.\n- Numerically simulate the dynamics of the excitatory population and find the fixed points of the system. \n- Investigate the stability of the fixed points by linearizing the dynamics around them.\n \n",
"_____no_output_____"
],
[
"# Setup",
"_____no_output_____"
]
],
[
[
"# Imports\nimport matplotlib.pyplot as plt # import matplotlib\nimport numpy as np # import numpy\nimport scipy.optimize as opt # import root-finding algorithm\nimport ipywidgets as widgets # interactive display",
"_____no_output_____"
],
[
"#@title Figure Settings\n%matplotlib inline\n\nfig_w, fig_h = 6, 4\nmy_fontsize = 16\nmy_params = {'axes.labelsize': my_fontsize,\n 'axes.titlesize': my_fontsize,\n 'figure.figsize': [fig_w, fig_h],\n 'font.size': my_fontsize,\n 'legend.fontsize': my_fontsize-4,\n 'lines.markersize': 8.,\n 'lines.linewidth': 2.,\n 'xtick.labelsize': my_fontsize-2,\n 'ytick.labelsize': my_fontsize-2}\n\nplt.rcParams.update(my_params)",
"_____no_output_____"
],
[
"# @title Helper functions\n\ndef plot_fI(x, f):\n plt.figure(figsize=(6,4)) # plot the figure\n plt.plot(x, f, 'k')\n plt.xlabel('x (a.u.)', fontsize=14.)\n plt.ylabel('F(x)', fontsize=14.)\n plt.show()",
"_____no_output_____"
],
[
"#@title Helper functions\ndef plot_dE_E(E, dEdt):\n plt.figure()\n plt.plot(E_grid, dEdt, 'k')\n plt.plot(E_grid, 0.*E_grid, 'k--')\n plt.xlabel('E activity')\n plt.ylabel(r'$\\frac{dE}{dt}$', fontsize=20)\n plt.ylim(-0.1, 0.1)\n\ndef plot_dFdt(x,dFdt):\n plt.figure()\n plt.plot(x, dFdt, 'r')\n plt.xlabel('x (a.u.)', fontsize=14.)\n plt.ylabel('dF(x)', fontsize=14.)\n plt.show()\n",
"_____no_output_____"
]
],
[
[
"# Neuronal network dynamics",
"_____no_output_____"
]
],
[
[
"#@title Video: Dynamic networks\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"ZSsAaeaG9ZM\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"Video available at https://youtube.com/watch?v=ZSsAaeaG9ZM\n"
]
],
[
[
"## Dynamics of a single excitatory population\n\nIndividual neurons respond by spiking. When we average the spikes of neurons in a population, we can define the average firing activity of the population. In this model, we are interested in how the population-averaged firing varies as a function of different network parameters.\n\n\\begin{align}\n\\tau_E \\frac{dE}{dt} &= -E + F(w_{EE}E + I^{\\text{ext}}_E) \\quad\\qquad (1)\n\\end{align}\n\n$E(t)$ represents the average firing rate of the excitatory population at time $t$, $\\tau_E$ controls the timescale of the evolution of the average firing rate, $w_{EE}$ denotes the strength (synaptic weight) of the recurrent excitatory input to the population, $I^{\\text{ext}}_E$ represents the external input, and the transfer function $F(\\cdot)$ (which can be related to f-I curve of individual neurons described in the next sections) represents the population activation function in response to all received inputs.\n\n\nTo start building the model, please execute the cell below to initialize the simulation parameters.",
"_____no_output_____"
]
],
[
[
"#@title Default parameters for a single excitatory population model\ndef default_parsE( **kwargs):\n pars = {}\n \n ### Excitatory parameters ###\n pars['tau_E'] = 1. # Timescale of the E population [ms]\n pars['a_E'] = 1.2 # Gain of the E population\n pars['theta_E'] = 2.8 # Threshold of the E population\n\n ### Connection strength ###\n pars['wEE'] = 0. # E to E, we first set it to 0\n\n ### External input ###\n pars['I_ext_E'] = 0.\n\n ### simulation parameters ###\n pars['T'] = 20. # Total duration of simulation [ms]\n pars['dt'] = .1 # Simulation time step [ms]\n pars['E_init'] = 0.2 # Initial value of E\n \n ### External parameters if any ###\n for k in kwargs:\n pars[k] = kwargs[k]\n \n pars['range_t'] = np.arange(0, pars['T'], pars['dt']) # Vector of discretized time points [ms]\n \n return pars",
"_____no_output_____"
]
],
[
[
"You can use:\n- `pars = default_parsE()` to get all the parameters, and then you can execute `print(pars)` to check these parameters. \n- `pars = default_parsE(T=T_sim, dt=time_step)` to set new simulation time and time step\n- After `pars = default_parsE()`, use `pars['New_para'] = value` to add an new parameter with its value",
"_____no_output_____"
],
[
"## F-I curves\nIn electrophysiology, a neuron is often characterized by its spike rate output in response to input currents. This is often called the **F-I** curve, denoting the spike frequency (**F**) in response to different injected currents (**I**). We estimated this for an LIF neuron in yesterday's tutorial.\n\nThe transfer function $F(\\cdot)$ in Equation (1) represents the gain of the population as a function of the total input. The gain is often modeled as a sigmoidal function, i.e., more input drive leads to a nonlinear increase in the population firing rate. The output firing rate will eventually saturate for high input values. \n\nA sigmoidal $F(\\cdot)$ is parameterized by its gain $a$ and threshold $\\theta$.\n\n$$ F(x;a,\\theta) = \\frac{1}{1+\\text{e}^{-a(x-\\theta)}} - \\frac{1}{1+\\text{e}^{a\\theta}} \\quad(2)$$\n\nThe argument $x$ represents the input to the population. Note that the second term is chosen so that $F(0;a,\\theta)=0$.\n\nMany other transfer functions (generally monotonic) can be also used. Examples are the rectified linear function $ReLU(x)$ or the hyperbolic tangent $tanh(x)$.",
"_____no_output_____"
],
[
"### Exercise 1: Implement F-I curve \n\nLet's first investigate the activation functions before simulating the dynamics of the entire population. \n\nIn this exercise, you will implement a sigmoidal **F-I** curve or transfer function $F(x)$, with gain $a$ and threshold level $\\theta$ as parameters.",
"_____no_output_____"
]
],
[
[
"# Excercise 1\ndef F(x,a,theta): \n \"\"\"\n Population activation function.\n\n Args:\n x (float): the population input\n a (float): the gain of the function\n theta (float): the threshold of the function\n \n Returns:\n float: the population activation response F(x) for input x\n \"\"\"\n\n #################################################################################\n ## TODO for students: compute f = F(x), remove the NotImplementedError once done# \n #################################################################################\n # the exponential function: np.exp(.)\n # f = ...\n raise NotImplementedError(\"Student excercise: implement the f-I function\")\n\n return f\n\n# Uncomment these lines when you've filled the function, then run the cell again \n# to plot the f-I curve.\npars = default_parsE() # get default parameters \n# print(pars) # print out pars to get familiar with parameters\nx = np.arange(0,10,.1) # set the range of input\n\n# Uncomment this when you fill the exercise, and call the function\n# plot_fI(x, F(x,pars['a_E'],pars['theta_E']))",
"_____no_output_____"
],
[
"# to_remove solution\ndef F(x,a,theta): \n \"\"\"\n Population activation function.\n\n Args:\n x : the population input\n a : the gain of the function\n theta : the threshold of the function\n \n Returns:\n the population activation response F(x) for input x\n \"\"\"\n\n # add the expression of f = F(x)\n f = (1+np.exp(-a*(x-theta)))**-1 - (1+np.exp(a*theta))**-1\n\n return f\n\npars = default_parsE() # get default parameters\nx = np.arange(0,10,.1) # set the range of input\n\nwith plt.xkcd():\n plot_fI(x, F(x,pars['a_E'],pars['theta_E']))",
"findfont: Font family ['xkcd', 'xkcd Script', 'Humor Sans', 'Comic Neue', 'Comic Sans MS'] not found. Falling back to DejaVu Sans.\n"
]
],
[
[
"### Interactive Demo: Parameter exploration of F-I curve\nHere's an interactive demo that shows how the F-I curve is changing for different values of the gain and threshold parameters.\n\n\n**Remember to enable the demo by running the cell.**",
"_____no_output_____"
]
],
[
[
"#@title F-I curve Explorer\ndef interactive_plot_FI(a, theta):\n '''\n Population activation function.\n\n Expecxts:\n a : the gain of the function\n theta : the threshold of the function\n \n Returns:\n plot the F-I curve with give parameters\n '''\n # set the range of input\n x = np.arange(0,10,.1)\n plt.figure()\n plt.plot(x, F(x, a, theta), 'k')\n plt.xlabel('x (a.u.)', fontsize=14.)\n plt.ylabel('F(x)', fontsize=14.)\n plt.show()\n\n\n\n_ = widgets.interact(interactive_plot_FI, a = (0.3, 3., 0.3), \\\n theta = (2., 4., 0.2)) ",
"_____no_output_____"
]
],
[
[
"## Simulation scheme of E dynamics\n\nBecause $F(\\cdot)$ is a nonlinear function, the exact solution of Equation $1$ can not be determined via analytical methods. Therefore, numerical methods must be used to find the solution. In practice, the derivative on the left-hand side of Equation (1) can be approximated using the Euler method on a time-grid of stepsize $\\Delta t$:\n\n\\begin{align}\n&\\frac{dE}{dt} \\approx \\frac{E[k+1]-E[k]}{\\Delta t} \n\\end{align}\nwhere $E[k] = E(k\\Delta t)$. \n\nThus,\n\n$$\\Delta E[k] = \\frac{\\Delta t}{\\tau_E}[-E[k] + F(w_{EE}E[k] + I^{\\text{ext}}_E(k;a_E,\\theta_E)]$$\n\n\nHence, Equation (1) is updated at each time step by:\n\n$$E[k+1] = E[k] + \\Delta E[k]$$\n\n**_Please execute the following cell to enable the WC simulator_**",
"_____no_output_____"
]
],
[
[
"#@title E population simulator: `simulate_E`\ndef simulate_E(pars):\n \n \"\"\"\n Simulate an excitatory population of neurons \n \n Args:\n pars : Parameter dictionary\n \n Returns:\n E : Activity of excitatory population (array)\n \"\"\"\n \n # Set parameters\n tau_E, a_E, theta_E = pars['tau_E'], pars['a_E'], pars['theta_E']\n wEE = pars['wEE']\n I_ext_E = pars['I_ext_E']\n E_init = pars['E_init'] \n dt, range_t = pars['dt'], pars['range_t'] \n Lt = range_t.size \n \n # Initialize activity\n E = np.zeros(Lt)\n E[0] = E_init\n I_ext_E = I_ext_E*np.ones(Lt)\n\n # Update the E activity \n for k in range(Lt-1):\n dE = dt/tau_E * (-E[k] + F(wEE*E[k]+I_ext_E[k], a_E, theta_E))\n E[k+1] = E[k] + dE\n \n return E\n\nprint(help(simulate_E))\n",
"Help on function simulate_E in module __main__:\n\nsimulate_E(pars)\n Simulate an excitatory population of neurons \n \n Args:\n pars : Parameter dictionary\n \n Returns:\n E : Activity of excitatory population (array)\n\nNone\n"
]
],
[
[
"#### Interactive Demo: Parameter Exploration of single population dynamics\n\nNote that $w_{EE}=0$, as in the default setting, means no recurrent input to the excitatory population in Equation (1). Hence, the dynamics is entirely determined by the external input $I_{E}^{\\text{ext}}$. Try to explore how $E_{sim}(t)$ changes with different $I_{E}^{\\text{ext}}$ and $\\tau_E$ parameter values, and investigate the relationship between $F(I_{E}^{\\text{ext}}; a_E, \\theta_E)$ and the steady value of E. Note that, $E_{ana}(t)$ denotes the analytical solution.",
"_____no_output_____"
]
],
[
[
"#@title Mean-field model Explorer\n\n# get default parameters\npars = default_parsE(T=20.)\n\ndef Myplot_E_diffI_difftau(I_ext, tau_E):\n # set external input and time constant\n pars['I_ext_E'] = I_ext\n pars['tau_E'] = tau_E\n\n # simulation\n E = simulate_E(pars)\n \n # Analytical Solution\n E_ana = pars['E_init'] + (F(I_ext,pars['a_E'],pars['theta_E'])-pars['E_init'])*\\\n (1.-np.exp(-pars['range_t']/pars['tau_E']))\n\n # plot\n plt.figure()\n plt.plot(pars['range_t'], E, 'b', label=r'$E_{\\mathrm{sim}}$(t)', alpha=0.5, zorder=1)\n plt.plot(pars['range_t'], E_ana, 'b--', lw=5, dashes=(2,2),\\\n label=r'$E_{\\mathrm{ana}}$(t)', zorder=2)\n plt.plot(pars['range_t'], F(I_ext,pars['a_E'],pars['theta_E'])\\\n *np.ones(pars['range_t'].size), 'k--', label=r'$F(I_E^{\\mathrm{ext}})$')\n plt.xlabel('t (ms)', fontsize=16.)\n plt.ylabel('E activity', fontsize=16.)\n plt.legend(loc='best', fontsize=14.)\n plt.show()\n\n_ = widgets.interact(Myplot_E_diffI_difftau, I_ext = (0.0, 10., 1.),\\\n tau_E = (1., 5., 0.2)) \n",
"_____no_output_____"
]
],
[
[
"### Think!\nAbove, we have numerically solved a system driven by a positive input and that, if $w_{EE} \\neq 0$, receives an excitatory recurrent input (**try changing the value of $w_{EE}$ to a positive number**). Yet, $E(t)$ either decays to zero or reaches a fixed non-zero value.\n- Why doesn't the solution of the system \"explode\" in a finite time? In other words, what guarantees that E(t) stays finite? \n- Which parameter would you change in order to increase the maximum value of the response? ",
"_____no_output_____"
],
[
"## Fixed points of the E system\n",
"_____no_output_____"
]
],
[
[
"#@title Video: Fixed point\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"B31fX6V0PZ4\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"Video available at https://youtube.com/watch?v=B31fX6V0PZ4\n"
]
],
[
[
"As you varied the two parameters in the last Interactive Demo, you noticed that, while at first the system output quickly changes, with time, it reaches its maximum/minimum value and does not change anymore. The value eventually reached by the system is called the **steady state** of the system, or the **fixed point**. Essentially, in the steady states the derivative with respect to time of the activity ($E$) is zero, i.e. $\\frac{dE}{dt}=0$. \n\nWe can find that the steady state of the Equation $1$ by setting $\\displaystyle{\\frac{dE}{dt}=0}$ and solve for $E$:\n\n$$E_{\\text{steady}} = F(w_{EE}E_{\\text{steady}} + I^{\\text{ext}}_E;a_E,\\theta_E) = 0, \\qquad (3)$$\n\nWhen it exists, the solution of Equation $3$ defines a **fixed point** of the dynamics which satisfies $\\displaystyle{\\frac{dE}{dt}=0}$ (and determines steady state of the system). Notice that the right-hand side of the last equation depends itself on $E_{steady}$. If $F(x)$ is nonlinear it is not always possible to find an analytical solution that can instead be found via numerical simulations, as we will do later.\n\nFrom the Interactive Demo one could also notice that the value of $\\tau_E$ influences how quickly the activity will converge to the steady state from its initial value. \n\nIn the specific case of $w_{EE}=0$, we can also analytically compute the analytical solution of Equation $1$ (i.e., the thick blue dashed line) and deduce the role of $\\tau_E$ in determining the convergence to the fixed point: \n\n$$\\displaystyle{E(t) = \\big{[}F(I^{\\text{ext}}_E;a_E,\\theta_E) -E(t=0)\\big{]} (1-\\text{e}^{-\\frac{t}{\\tau_E}})} + E(t=0)$$ \\\\\n\nWe can now numerically calculate the fixed point with the `scipy.optimize.root` function.\n\n<font size=3><font color='gray'>_(note that at the very beginning, we `import scipy.optimize as opt` )_</font></font>.\n\n\\\\\n\nPlease execute the cell below to define the functions `my_fpE`, `check_fpE`, and `plot_fpE`",
"_____no_output_____"
]
],
[
[
"#@title Function of calculating the fixed point\ndef my_fpE(pars, E_init):\n\n # get the parameters\n a_E, theta_E = pars['a_E'], pars['theta_E']\n wEE = pars['wEE']\n I_ext_E = pars['I_ext_E']\n \n # define the right hand of E dynamics\n def my_WCr(x):\n E = x[0]\n dEdt=(-E + F(wEE*E+I_ext_E,a_E,theta_E))\n y = np.array(dEdt)\n \n return y\n \n x0 = np.array(E_init)\n x_fp = opt.root(my_WCr, x0).x\n \n return x_fp\n\ndef check_fpE(pars, x_fp):\n\n a_E, theta_E = pars['a_E'], pars['theta_E']\n wEE = pars['wEE']\n I_ext_E = pars['I_ext_E']\n\n # calculate Equation(3)\n y = x_fp- F(wEE*x_fp+I_ext_E, a_E, theta_E)\n\n return np.abs(y)<1e-4\n\ndef plot_fpE(pars, x_fp, mycolor):\n \n wEE = pars['wEE']\n I_ext_E = pars['I_ext_E']\n\n plt.plot(wEE*x_fp+I_ext_E, x_fp, 'o', color=mycolor)\n",
"_____no_output_____"
]
],
[
[
"#### Exercise 2: Visualization of the fixed point\n\nWhen no analytical solution of Equation $3$ can be found, it is often useful to plot $\\displaystyle{\\frac{dE}{dt}=0}$ as a function of $E$. The values of E for which the plotted function crosses zero on the y axis correspond to fixed points. \n\nHere, let us, for example, set $w_{EE}=5.0$ and $I^{\\text{ext}}_E=0.5$. Define $\\displaystyle{\\frac{dE}{dt}}$ using Equation $1$, plot the result, and check for the presence of fixed points. \n\nWe will now try to find the fixed points using the previously defined function `my_fpE(pars, E_init)` with different initial values ($E_{\\text{init}}$). Use the previously defined function `check_fpE(pars, x_fp)` to verify that the values of $E$ for which $\\displaystyle{\\frac{dE}{dt}} = 0$ are the true fixed points.",
"_____no_output_____"
]
],
[
[
"# Exercise 2\npars = default_parsE() # get default parameters\n\n# set your external input and wEE\npars['I_ext_E'] = 0.5\npars['wEE'] = 5.0\n\nE_grid = np.linspace(0, 1., 1000)# give E_grid\n\n#figure, line (E, dEdt)\n###############################\n## TODO for students: #\n## Calculate dEdt = -E + F(.) #\n## Then plot the lines #\n###############################\n# Calculate dEdt\n# dEdt = ...\n\n# Uncomment this to plot the dEdt across E\n# plot_dE_E(E_grid, dEdt)\n\n# Add fixed point\n#####################################################\n## TODO for students: #\n# Calculate the fixed point with your initial value #\n# verify your fixed point and plot the corret ones #\n#####################################################\n\n# Calculate the fixed point with your initial value\nx_fp_1 = my_fpE(pars, 1)\n\n#check if x_fp is the intersection of the lines with the given function check_fpE(pars, x_fp)\n#vary different initial values to find the correct fixed point (Should be 3)\n# Use blue, red and yellow colors, respectively ('b', 'r', 'y' codenames)\n\n# if check_fpE(pars, x_fp_1):\n# plt.plot(x_fp_1, 0, 'bo', ms=8)\n\n# Replicate the code above (lines 35-36) for all fixed points.\n",
"_____no_output_____"
],
[
"# to_remove solution\npars = default_parsE() # get default parameters\n\n#set your external input and wEE\npars['I_ext_E'] = 0.5\npars['wEE'] = 5.0\n \n# give E_grid\nE_grid = np.linspace(0, 1., 1000)\n\n# Calculate dEdt\ndEdt = -E_grid + F(pars['wEE']*E_grid+pars['I_ext_E'], pars['a_E'], pars['theta_E'])\n\n\nwith plt.xkcd():\n plot_dE_E(E_grid, dEdt)\n #Calculate the fixed point with your initial value\n \n x_fp_1 = my_fpE(pars, 0.)\n if check_fpE(pars, x_fp_1):\n plt.plot(x_fp_1, 0, 'bo', ms=8)\n\n x_fp_2 = my_fpE(pars, 0.4)\n if check_fpE(pars, x_fp_2):\n plt.plot(x_fp_2, 0, 'ro', ms=8)\n\n x_fp_3 = my_fpE(pars, 0.9)\n if check_fpE(pars, x_fp_3):\n plt.plot(x_fp_3, 0, 'yo', ms=8)\n\n plt.show()",
"findfont: Font family ['xkcd', 'xkcd Script', 'Humor Sans', 'Comic Neue', 'Comic Sans MS'] not found. Falling back to DejaVu Sans.\n"
]
],
[
[
"#### Interactive Demo: fixed points as a function of recurrent and external inputs.\n\nYou can now explore how the previous plot changes when the recurrent coupling $w_{\\text{EE}}$ and the external input $I_E^{\\text{ext}}$ take different values.",
"_____no_output_____"
]
],
[
[
"#@title Fixed point Explorer\n\ndef plot_intersection_E(wEE, I_ext_E):\n #set your parameters\n pars['wEE'] = wEE\n pars['I_ext_E'] = I_ext_E\n\n #note that wEE !=0\n if wEE>0:\n # find fixed point\n x_fp_1 = my_fpE(pars, 0.)\n x_fp_2 = my_fpE(pars, 0.4)\n x_fp_3 = my_fpE(pars, 0.9)\n\n plt.figure()\n E_grid = np.linspace(0, 1., 1000)\n dEdt = -E_grid + F(wEE*E_grid+I_ext_E, pars['a_E'], pars['theta_E'])\n\n plt.plot(E_grid, dEdt, 'k')\n plt.plot(E_grid, 0.*E_grid, 'k--')\n \n if check_fpE(pars, x_fp_1):\n plt.plot(x_fp_1, 0, 'bo', ms=8)\n if check_fpE(pars, x_fp_2):\n plt.plot(x_fp_2, 0, 'bo', ms=8)\n if check_fpE(pars, x_fp_3):\n plt.plot(x_fp_3, 0, 'bo', ms=8)\n\n plt.xlabel('E activity', fontsize=14.)\n plt.ylabel(r'$\\frac{dE}{dt}$', fontsize=18.)\n\n plt.show()\n\n_ = widgets.interact(plot_intersection_E, wEE = (1., 7., 0.2), \\\n I_ext_E = (0., 3., 0.1)) ",
"_____no_output_____"
]
],
[
[
"## Summary\n\nIn this tutorial, we have investigated the dynamics of a rate-based single excitatory population of neurons.\n\nWe learned about:\n- The effect of the input parameters and the time constant of the network on the dynamics of the population.\n- How to find the fixed point(s) of the system.\n\nNext, we have two Bonus, but important concepts in dynamical system analysis and simulation. If you have time left, watch the next video and proceed to solve the exercises. You will learn:\n\n- How to determine the stability of a fixed point by linearizing the system.\n- How to add realistic inputs to our model.",
"_____no_output_____"
],
[
"## Bonus 1: Stability of a fixed point",
"_____no_output_____"
]
],
[
[
"#@title Video: Stability of fixed points\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"nvxxf59w2EA\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"Video available at https://youtube.com/watch?v=nvxxf59w2EA\n"
]
],
[
[
"#### Initial values and trajectories\n\nHere, let us first set $w_{EE}=5.0$ and $I^{\\text{ext}}_E=0.5$, and investigate the dynamics of $E(t)$ starting with different initial values $E(0) \\equiv E_{\\text{init}}$. We will plot the trajectories of $E(t)$ with $E_{\\text{init}} = 0.0, 0.1, 0.2,..., 0.9$.",
"_____no_output_____"
]
],
[
[
"#@title Initial values\npars = default_parsE()\npars['wEE'] = 5.0\npars['I_ext_E'] = 0.5\n\nplt.figure(figsize=(10,6))\nfor ie in range(10): \n pars['E_init'] = 0.1*ie # set the initial value\n E = simulate_E(pars) # run the simulation\n # plot the activity with given initial\n plt.plot(pars['range_t'], E, 'b', alpha=0.1 + 0.1*ie, label= r'E$_{\\mathrm{init}}$=%.1f' % (0.1*ie))\n\nplt.xlabel('t (ms)')\nplt.title('Two steady states?')\nplt.ylabel('E(t)')\nplt.legend(loc=[0.72, 0.13], fontsize=14)\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"#### Interactive Demo: dynamics as a function of the initial value.\n\n\nLet's now set $E_{init}$ to a value of your choice in this demo. How does the solution change? What do you observe?",
"_____no_output_____"
]
],
[
[
"#@title Initial value Explorer\npars = default_parsE()\npars['wEE'] = 5.0\npars['I_ext_E'] = 0.5\n\ndef plot_E_diffEinit(E_init):\n pars['E_init'] = E_init\n E = simulate_E(pars)\n \n plt.figure()\n plt.plot(pars['range_t'], E, 'b', label='E(t)')\n plt.xlabel('t (ms)', fontsize=16.)\n plt.ylabel('E activity', fontsize=16.)\n plt.show()\n\n_ = widgets.interact(plot_E_diffEinit, E_init = (0., 1., 0.02)) ",
"_____no_output_____"
]
],
[
[
"### Stability analysis via linearization of the dynamics\n\nJust like Equation $1$ in the case ($w_{EE}=0$) discussed above, a generic linear system \n$$\\frac{dx}{dt} = \\lambda (x - b),$$ \nhas a fixed point for $x=b$. The analytical solution of such a system can be found to be:\n$$x(t) = b + \\big{(} x(0) - b \\big{)} \\text{e}^{\\lambda t}.$$ \nNow consider a small perturbation of the activity around the fixed point: $x(0) = b+ \\epsilon$, where $|\\epsilon| \\ll 1$. Will the perturbation $\\epsilon(t)$ grow with time or will it decay to the fixed point? The evolution of the perturbation with time can be written, using the analytical solution for $x(t)$, as:\n $$\\epsilon (t) = x(t) - b = \\epsilon \\text{e}^{\\lambda t}$$\n\n- if $\\lambda < 0$, $\\epsilon(t)$ decays to zero, $x(t)$ will still converge to $b$ and the fixed point is \"**stable**\".\n\n- if $\\lambda > 0$, $\\epsilon(t)$ grows with time, $x(t)$ will leave the fixed point $b$ exponentially and the fixed point is, therefore, \"**unstable**\" .",
"_____no_output_____"
],
[
"### Compute the stability of Equation (1)\n\nSimilar to what we did in the linear system above, in order to determine the stability of a fixed point $E_{\\rm fp}$ of the excitatory population dynamics, we perturb Equation $1$ around $E_{\\rm fp}$ by $\\epsilon$, i.e. $E = E_{\\rm fp} + \\epsilon$. We can plug in Equation $1$ and obtain the equation determining the time evolution of the perturbation $\\epsilon(t)$:\n\n\\begin{align}\n\\tau_E \\frac{d\\epsilon}{dt} \\approx -\\epsilon + w_{EE} F'(w_{EE}E_{\\text{fp}} + I^{\\text{ext}}_E;a_E,\\theta_E) \\epsilon \n\\end{align}\nwhere $F'(\\cdot)$ is the derivative of the transfer function $F(\\cdot)$. We can rewrite the above equation as:\n\\begin{align}\n\\frac{d\\epsilon}{dt} \\approx \\frac{\\epsilon}{\\tau_E }[-1 + w_{EE} F'(w_{EE}E_{\\text{fp}} + I^{\\text{ext}}_E;a_E,\\theta_E)] \n\\end{align}\n\nThat is, as in the linear system above, the value of $\\lambda = [-1+ w_{EE}F'(w_{EE}E_{\\text{fp}} + I^{\\text{ext}}_E;a_E,\\theta_E)]/\\tau_E$ determines whether the perturbation will grow or decay to zero, i.e., $\\lambda$ defines the stability of the fixed point. This value is called the **eigenvalue** of the dynamical system.",
"_____no_output_____"
],
[
"### Exercise 4: Compute $dF$ and Eigenvalue\n\nThe derivative of the sigmoid transfer function is:\n\\begin{align} \n\\frac{dF}{dx} & = \\frac{d}{dx} (1+\\exp\\{-a(x-\\theta)\\})^{-1} \\\\\n& = a\\exp\\{-a(x-\\theta)\\} (1+\\exp\\{-a(x-\\theta)\\})^{-2}. \n\\end{align}\n\nLet's now find the expression for the derivative $\\displaystyle{\\frac{dF}{dx}}$ in the following cell and plot it.",
"_____no_output_____"
]
],
[
[
"# Exercise 4\n\ndef dF(x,a,theta): \n \"\"\"\n Population activation function.\n\n Args:\n x : the population input\n a : the gain of the function\n theta : the threshold of the function\n \n Returns:\n dFdx : the population activation response F(x) for input x\n \"\"\"\n\n #####################################################################\n ## TODO for students: compute dFdx, then remove NotImplementedError #\n #####################################################################\n # dFdx = ...\n raise NotImplementedError(\"Student excercise: compute the deravitive of F(x)\")\n\n return dFdx\n\npars = default_parsE() # get default parameters\nx = np.arange(0,10,.1) # set the range of input\n\n# Uncomment below lines after completing the dF function\n# plot_dFdt(x,dF(x,pars['a_E'],pars['theta_E']))\n",
"_____no_output_____"
],
[
"# to_remove solution\ndef dF(x,a,theta): \n \"\"\"\n Population activation function.\n\n Args:\n x : the population input\n a : the gain of the function\n theta : the threshold of the function\n \n Returns:\n dFdx : the population activation response F(x) for input x\n \"\"\"\n\n dFdx = a*np.exp(-a*(x-theta))*(1+np.exp(-a*(x-theta)))**-2\n\n return dFdx\n\n# get default parameters\npars = default_parsE()\n\n# set the range of input\nx = np.arange(0,10,.1)\n\n# plot figure\nwith plt.xkcd():\n plot_dFdt(x,dF(x,pars['a_E'],pars['theta_E']))\n\n",
"_____no_output_____"
]
],
[
[
"### Exercise 5: Compute eigenvalues \n\nAs discussed above, for the case with $w_{EE}=5.0$ and $I^{\\text{ext}}_E=0.5$, the system displays **3** fixed points. However, when we simulated the dynamics and varied the initial conditions $E_{\\rm init}$, we could only obtain **two** steady states. In this exercise, we will now check the stability of each of the $3$ fixed points by calculating the corresponding eigenvalues with the function `eig_E` defined above. Check the sign of each eigenvalue (i.e., stability of each fixed point). How many of the fixed points are stable?",
"_____no_output_____"
]
],
[
[
"# Exercise 5\npars = default_parsE()\npars['wEE'] = 5.0\npars['I_ext_E'] = 0.5\n\ndef eig_E(pars, fp):\n \"\"\"\n Args:\n pars : Parameter dictionary\n fp : fixed point E\n \n Returns:\n eig : eigevalue of the linearized system\n \"\"\"\n \n #get the parameters\n tau_E, a_E, theta_E = pars['tau_E'], pars['a_E'], pars['theta_E']\n wEE, I_ext_E = pars['wEE'], pars['I_ext_E']\n # fixed point\n E = fp\n\n #######################################################################\n ## TODO for students: compute eigenvalue, remove NotImplementedError #\n #######################################################################\n # eig = ...\n raise NotImplementedError(\"Student excercise: compute the eigenvalue\")\n\n return eig\n\n# Uncomment below lines after completing the eigE function.\n# x_fp_1 = fpE(pars, 0.)\n# eig_fp_1 = eig_E(pars, x_fp_1)\n# print('Fixed point1=%.3f, Eigenvalue=%.3f' % (x_fp_1, eig_E1))\n\n# Continue by finding the eigenvalues for all fixed points of Exercise 2",
"_____no_output_____"
],
[
"# to_remove solution\npars = default_parsE()\npars['wEE'] = 5.0\npars['I_ext_E'] = 0.5\n\ndef eig_E(pars, fp):\n \"\"\"\n Args:\n pars : Parameter dictionary\n fp : fixed point E\n \n Returns:\n eig : eigevalue of the linearized system\n \"\"\"\n \n #get the parameters\n tau_E, a_E, theta_E = pars['tau_E'], pars['a_E'], pars['theta_E']\n wEE, I_ext_E = pars['wEE'], pars['I_ext_E']\n # fixed point\n E = fp\n\n eig = (-1. + wEE*dF(wEE*E + I_ext_E, a_E, theta_E)) / tau_E \n\n return eig\n\n# Uncomment below lines after completing the eigE function\nx_fp_1 = my_fpE(pars, 0.)\neig_E1 = eig_E(pars, x_fp_1)\nprint('Fixed point1=%.3f, Eigenvalue=%.3f' % (x_fp_1, eig_E1))\n\n# Continue by finding the eigenvalues for all fixed points of Exercise 2\nx_fp_2 = my_fpE(pars, 0.4)\neig_E2 = eig_E(pars, x_fp_2)\nprint('Fixed point2=%.3f, Eigenvalue=%.3f' % (x_fp_2, eig_E2))\n\nx_fp_3 = my_fpE(pars, 0.9)\neig_E3 = eig_E(pars, x_fp_3)\nprint('Fixed point3=%.3f, Eigenvalue=%.3f' % (x_fp_3, eig_E3))",
"Fixed point1=0.042, Eigenvalue=-0.583\nFixed point2=0.447, Eigenvalue=0.498\nFixed point3=0.900, Eigenvalue=-0.626\n"
]
],
[
[
"### Think! \nThroughout the tutorial, we have assumed $w_{\\rm EE}> 0 $, i.e., we considered a single population of **excitatory** neurons. What do you think will be the behavior of a population of inhibitory neurons, i.e., where $w_{\\rm EE}> 0$ is replaced by $w_{\\rm II}< 0$? ",
"_____no_output_____"
],
[
"## Bonus 2: Noisy input drives transition between two stable states\n\n",
"_____no_output_____"
],
[
"### Ornstein-Uhlenbeck (OU) process\n\nAs discussed in several previous tutorials, the OU process is usually used to generate a noisy input into the neuron. The OU input $\\eta(t)$ follows: \n\n$$\\tau_\\eta \\frac{d}{dt}\\eta(t) = -\\eta (t) + \\sigma_\\eta\\sqrt{2\\tau_\\eta}\\xi(t)$$\n\nExecute the following function `my_OU(pars, sig, myseed=False)` to generate an OU process.",
"_____no_output_____"
]
],
[
[
"#@title OU process `my_OU(pars, sig, myseed=False)`\ndef my_OU(pars, sig, myseed=False):\n \"\"\"\n A functions that generates Ornstein-Uhlenback process\n\n Args:\n pars : parameter dictionary\n sig : noise amplitute\n myseed : random seed. int or boolean\n \n Returns:\n I : Ornstein-Uhlenbeck input current\n \"\"\"\n \n # Retrieve simulation parameters\n dt, range_t = pars['dt'], pars['range_t']\n Lt = range_t.size\n tau_ou = pars['tau_ou'] # [ms]\n \n # set random seed\n if myseed:\n np.random.seed(seed=myseed) \n else:\n np.random.seed()\n \n # Initialize\n noise = np.random.randn(Lt)\n I = np.zeros(Lt)\n I[0] = noise[0] * sig\n \n #generate OU\n for it in range(Lt-1):\n I[it+1] = I[it] + dt/tau_ou*(0.-I[it]) + np.sqrt(2.*dt/tau_ou) * sig * noise[it+1]\n \n return I\n\npars = default_parsE(T=100)\npars['tau_ou'] = 1. #[ms]\nsig_ou = 0.1\nI_ou = my_OU(pars, sig=sig_ou, myseed=1998)\nplt.figure(figsize=(10, 4))\nplt.plot(pars['range_t'], I_ou, 'b')\nplt.xlabel('Time (ms)')\nplt.ylabel(r'$I_{\\mathrm{OU}}$');",
"_____no_output_____"
]
],
[
[
"### Bonus Example: Up-Down transition\n\nIn the presence of two or more fixed points, noisy inputs can drive a transition between the fixed points! Here, we stimulate an E population for 1,000 ms applying OU inputs.",
"_____no_output_____"
]
],
[
[
"#@title Simulation of an E population with OU inputs\npars = default_parsE(T = 1000)\npars['wEE'] = 5.0\nsig_ou = 0.7\npars['tau_ou'] = 1. #[ms]\npars['I_ext_E'] = 0.56 + my_OU(pars, sig=sig_ou, myseed=2020)\n\nE = simulate_E(pars)\n\nplt.figure(figsize=(10, 4))\nplt.plot(pars['range_t'], E, 'r', alpha=0.8)\nplt.xlabel('t (ms)')\nplt.ylabel('E activity')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.