path
stringlengths 7
265
| concatenated_notebook
stringlengths 46
17M
|
|---|---|
RTE Playground.ipynb | ###Markdown
Bag-of-words Models
###Code
# the simplest model that just tries to classify by linear combination of averages
L0 = nnb.InputLayer(ndim=1)
L1 = nnb.InputLayer(ndim=1)
L2 = nnb.InputLayer(ndim=1)
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=50*3, outsize=1, activation_func=nnb.activation.tanh)
model = (L0 & L1 & L2) | CC | C2
try_model(model, s0a, s1a, s2a, y)
# only slightly more complex model that first merges l0, l1, then tries to learn a linear combination of that with l2
L0 = nnb.InputLayer(ndim=1)
L1 = nnb.InputLayer(ndim=1)
C01 = nnb.ConcatenationModel()
C1 = nnb.PerceptronLayer(insize=2*50, outsize=50)
qajoin = (L0 & L1) | C01 | C1
L2 = nnb.InputLayer(ndim=1)
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=2*50, outsize=1, activation_func=nnb.activation.tanh)
model = (qajoin & L2) | CC | C2
try_model(model, s0a, s1a, s2a, y)
# extension of the above with extra hidden layer
L0 = nnb.InputLayer(ndim=1)
L1 = nnb.InputLayer(ndim=1)
C01 = nnb.ConcatenationModel()
C1a = nnb.PerceptronLayer(insize=2*50, outsize=50)
C1b = nnb.PerceptronLayer(insize=50, outsize=25, activation_func=nnb.activation.tanh)
qajoin = (L0 & L1) | C01 | C1a | C1b
L2 = nnb.InputLayer(ndim=1)
CC = nnb.ConcatenationModel()
C2a = nnb.PerceptronLayer(insize=50+25, outsize=25)
C2b = nnb.PerceptronLayer(insize=25, outsize=1, activation_func=nnb.activation.tanh)
model = (qajoin & L2) | CC | C2a | C2b
try_model(model, s0a, s1a, s2a, y)
###Output
Preprocessing...
Cost function...
Compiling...
Training...
~Epoch 1~
[--------- ]
Finished. Took 0.0217814326286 minutes.
Evaluating...
Error = 0.500357608689
New best!
~Epoch 2~
[--------- ]
Finished. Took 0.0217702666918 minutes.
Evaluating...
Error = 0.500007342751
New best!
~Epoch 3~
[--------- ]
Finished. Took 0.021881600221 minutes.
Evaluating...
Error = 0.500145769065
~Epoch 4~
[--------- ]
Finished. Took 0.0218525330226 minutes.
Evaluating...
Error = 0.500343043051
~Epoch 5~
[--------- ]
Finished. Took 0.0218734184901 minutes.
Evaluating...
Error = 0.500018127542
~Epoch 6~
[--------- ]
Finished. Took 0.0218481183052 minutes.
Evaluating...
Error = 0.500029008664
~Epoch 7~
[--------- ]
Finished. Took 0.0223319689433 minutes.
Evaluating...
Error = 0.501230559823
~Epoch 8~
[--------- ]
Finished. Took 0.0218985676765 minutes.
Evaluating...
Error = 0.500803193592
~Epoch 9~
[--------- ]
Finished. Took 0.0218106150627 minutes.
Evaluating...
Error = 0.500096080358
~Epoch 10~
[--------- ]
Finished. Took 0.0218416015307 minutes.
Evaluating...
Error = 0.500542697909
~Epoch 11~
[--------- ]
Finished. Took 0.0218163013458 minutes.
Evaluating...
Error = 0.503701656726
~Epoch 12~
[--------- ]
Finished. Took 0.0218620498975 minutes.
Evaluating...
Error = 0.499996685223
New best!
~Epoch 13~
[--------- ]
Finished. Took 0.0218279838562 minutes.
Evaluating...
Error = 0.500114915747
~Epoch 14~
[--------- ]
Finished. Took 0.021864883105 minutes.
Evaluating...
Error = 0.50000234246
~Epoch 15~
[--------- ]
Finished. Took 0.0218339165052 minutes.
Evaluating...
Error = 0.501091637636
~Epoch 16~
[--------- ]
Finished. Took 0.0220268646876 minutes.
Evaluating...
Error = 0.500141559823
~Epoch 17~
[--------- ]
Finished. Took 0.0218552192052 minutes.
Evaluating...
Error = 0.501337095868
~Epoch 18~
[--------- ]
Finished. Took 0.0217992186546 minutes.
Evaluating...
Error = 0.500326763224
~Epoch 19~
[--------- ]
Finished. Took 0.0218602180481 minutes.
Evaluating...
Error = 0.500073103204
~Epoch 20~
[--------- ]
Finished. Took 0.0218064347903 minutes.
Evaluating...
Error = 0.500221115396
~Epoch 21~
[--------- ]
Finished. Took 0.0219269990921 minutes.
Evaluating...
Error = 0.500677313315
~Epoch 22~
[--------- ]
Finished. Took 0.0218431830406 minutes.
Evaluating...
Error = 0.499999411133
~Epoch 23~
[--------- ]
Finished. Took 0.0218532681465 minutes.
Evaluating...
Error = 0.500826726601
~Epoch 24~
[--------- ]
Finished. Took 0.0217517852783 minutes.
Evaluating...
Error = 0.500886390623
~Epoch 25~
[--------- ]
Finished. Took 0.0218867341677 minutes.
Evaluating...
Error = 0.500462269425
~Epoch 26~
[--------- ]
Finished. Took 0.0217462658882 minutes.
Evaluating...
Error = 0.501476886195
~Epoch 27~
[--------- ]
Finished. Took 0.0223957339923 minutes.
Evaluating...
Error = 0.500028026309
~Epoch 28~
[--------- ]
Finished. Took 0.021883781751 minutes.
Evaluating...
Error = 0.500362921846
~Epoch 29~
[--------- ]
Finished. Took 0.0218784968058 minutes.
Evaluating...
Error = 0.500223828294
~Epoch 30~
[--------- ]
Finished. Took 0.0222222487132 minutes.
Evaluating...
Error = 0.502864350179
~Epoch 31~
[--------- ]
Finished. Took 0.0218279361725 minutes.
Evaluating...
Error = 0.500619307896
~Epoch 32~
[--------- ]
Finished. Took 0.0218044996262 minutes.
Evaluating...
Error = 0.499987406644
New best!
~Epoch 33~
[--------- ]
Finished. Took 0.0221875667572 minutes.
Evaluating...
Error = 0.500366175275
~Epoch 34~
[--------- ]
Finished. Took 0.0219057679176 minutes.
Evaluating...
Error = 0.500410528936
~Epoch 35~
[--------- ]
Finished. Took 0.0218756159147 minutes.
Evaluating...
Error = 0.500048850828
~Epoch 36~
[--------- ]
Finished. Took 0.0217797160149 minutes.
Evaluating...
Error = 0.500113452477
~Epoch 37~
[--------- ]
Finished. Took 0.0218860348066 minutes.
Evaluating...
Error = 0.500028451843
~Epoch 38~
[--------- ]
Finished. Took 0.0219083189964 minutes.
Evaluating...
Error = 0.499986161081
New best!
~Epoch 39~
[--------- ]
Finished. Took 0.0220331986745 minutes.
Evaluating...
Error = 0.501674818963
~Epoch 40~
[--------- ]
Finished. Took 0.0219795306524 minutes.
Evaluating...
Error = 0.500021788593
~Epoch 41~
[--------- ]
Finished. Took 0.0218236843745 minutes.
Evaluating...
Error = 0.500132441928
~Epoch 42~
[--------- ]
Finished. Took 0.0218075990677 minutes.
Evaluating...
Error = 0.499985138935
New best!
~Epoch 43~
[--------- ]
Finished. Took 0.0219311475754 minutes.
Evaluating...
Error = 0.500253047816
~Epoch 44~
[--------- ]
Finished. Took 0.0223954002062 minutes.
Evaluating...
Error = 0.503078734515
~Epoch 45~
[--------- ]
Finished. Took 0.0218530495962 minutes.
Evaluating...
Error = 0.499995136601
~Epoch 46~
[--------- ]
Finished. Took 0.0223164161046 minutes.
Evaluating...
Error = 0.500738841335
~Epoch 47~
[--------- ]
Finished. Took 0.0219388167063 minutes.
Evaluating...
Error = 0.499974096477
New best!
~Epoch 48~
[--------- ]
Finished. Took 0.0218135674795 minutes.
Evaluating...
Error = 0.500120409531
~Epoch 49~
[--------- ]
Finished. Took 0.0217928806941 minutes.
Evaluating...
Error = 0.500184527706
~Epoch 50~
[--------- ]
Finished. Took 0.0218890508016 minutes.
Evaluating...
Error = 0.500130238553
~Epoch 51~
[--------- ]
Finished. Took 0.0220821499825 minutes.
Evaluating...
Error = 0.50109280612
~Epoch 52~
[--------- ]
Finished. Took 0.0218738516172 minutes.
Evaluating...
Error = 0.500248211362
~Epoch 53~
[--------- ]
Finished. Took 0.0218495686849 minutes.
Evaluating...
Error = 0.50014239258
~Epoch 54~
[--------- ]
Finished. Took 0.0218565146128 minutes.
Evaluating...
Error = 0.50086558956
~Epoch 55~
[--------- ]
Finished. Took 0.0220512986183 minutes.
Evaluating...
Error = 0.499970071088
New best!
~Epoch 56~
[--------- ]
Finished. Took 0.0218093832334 minutes.
Evaluating...
Error = 0.500149102842
~Epoch 57~
[--------- ]
Finished. Took 0.0220838824908 minutes.
Evaluating...
Error = 0.500033025376
~Epoch 58~
[--------- ]
Finished. Took 0.0218762477239 minutes.
Evaluating...
Error = 0.501198219803
~Epoch 59~
[--------- ]
Finished. Took 0.0230452815692 minutes.
Evaluating...
Error = 0.50012025529
~Epoch 60~
[--------- ]
Finished. Took 0.021840496858 minutes.
Evaluating...
Error = 0.500077270832
~Epoch 61~
[--------- ]
Finished. Took 0.0218392491341 minutes.
Evaluating...
Error = 0.50077927355
~Epoch 62~
[--------- ]
Finished. Took 0.0217747489611 minutes.
Evaluating...
Error = 0.499984892315
~Epoch 63~
[--------- ]
Finished. Took 0.0217939813932 minutes.
Evaluating...
Error = 0.499997648061
~Epoch 64~
[--------- ]
Finished. Took 0.0217553496361 minutes.
Evaluating...
Error = 0.500380485899
~Epoch 65~
[--------- ]
Finished. Took 0.0222050984701 minutes.
Evaluating...
Error = 0.499986363764
~Epoch 66~
[--------- ]
Finished. Took 0.0219154993693 minutes.
Evaluating...
Error = 0.499977693214
~Epoch 67~
[--------- ]
Finished. Took 0.0222742160161 minutes.
Evaluating...
Error = 0.50068828969
~Epoch 68~
[--------- ]
Finished. Took 0.0221931139628 minutes.
Evaluating...
Error = 0.499993818277
~Epoch 69~
[--------- ]
Finished. Took 0.0223215699196 minutes.
Evaluating...
Error = 0.500456752386
~Epoch 70~
[--------- ]
Finished. Took 0.0218123356501 minutes.
Evaluating...
Error = 0.501516249963
~Epoch 71~
[--------- ]
Finished. Took 0.022450085481 minutes.
Evaluating...
Error = 0.500085789755
~Epoch 72~
[--------- ]
Finished. Took 0.0222800334295 minutes.
Evaluating...
Error = 0.500401278801
~Epoch 73~
[--------- ]
Finished. Took 0.0218959013621 minutes.
Evaluating...
Error = 0.50036435486
~Epoch 74~
[--------- ]
Finished. Took 0.0217820684115 minutes.
Evaluating...
Error = 0.502541472291
~Epoch 75~
[--------- ]
Finished. Took 0.0224252978961 minutes.
Evaluating...
Error = 0.499963902512
New best!
~Epoch 76~
[--------- ]
Finished. Took 0.0217240651449 minutes.
Evaluating...
Error = 0.500101250946
~Epoch 77~
[--------- ]
Finished. Took 0.0217859864235 minutes.
Evaluating...
Error = 0.50040999318
~Epoch 78~
[--------- ]
Finished. Took 0.0217722177505 minutes.
Evaluating...
Error = 0.500510304056
~Epoch 79~
[--------- ]
Finished. Took 0.0217543999354 minutes.
Evaluating...
Error = 0.500318563833
~Epoch 80~
[--------- ]
Finished. Took 0.0223845322927 minutes.
Evaluating...
Error = 0.499998462164
~Epoch 81~
[--------- ]
Finished. Took 0.0218147158623 minutes.
Evaluating...
Error = 0.502026415632
~Epoch 82~
[--------- ]
Finished. Took 0.0222399671872 minutes.
Evaluating...
Error = 0.502093156353
~Epoch 83~
[--------- ]
Finished. Took 0.0217820326487 minutes.
Evaluating...
Error = 0.500024211545
~Epoch 84~
[--------- ]
Finished. Took 0.0218458970388 minutes.
Evaluating...
Error = 0.502331249226
~Epoch 85~
[--------- ]
Finished. Took 0.0217954158783 minutes.
Evaluating...
Error = 0.499950856299
New best!
~Epoch 86~
[--------- ]
Finished. Took 0.0223637660344 minutes.
Evaluating...
Error = 0.500109465447
~Epoch 87~
[--------- ]
Finished. Took 0.0217625339826 minutes.
Evaluating...
Error = 0.499951075783
~Epoch 88~
[--------- ]
Finished. Took 0.0221950531006 minutes.
Evaluating...
Error = 0.499984625191
~Epoch 89~
[--------- ]
Finished. Took 0.0217854340871 minutes.
Evaluating...
Error = 0.499943631552
New best!
~Epoch 90~
[--------- ]
Finished. Took 0.0223787148794 minutes.
Evaluating...
Error = 0.500429406791
~Epoch 91~
[--------- ]
Finished. Took 0.0217892169952 minutes.
Evaluating...
Error = 0.499942773631
New best!
~Epoch 92~
[--------- ]
Finished. Took 0.0224108338356 minutes.
Evaluating...
Error = 0.500466631022
~Epoch 93~
[--------- ]
Finished. Took 0.0217609643936 minutes.
Evaluating...
Error = 0.500321091069
~Epoch 94~
[--------- ]
Finished. Took 0.0224106987317 minutes.
Evaluating...
Error = 0.500188895129
~Epoch 95~
[--------- ]
Finished. Took 0.0217641671499 minutes.
Evaluating...
Error = 0.501258692385
~Epoch 96~
[--------- ]
Finished. Took 0.0223159313202 minutes.
Evaluating...
Error = 0.500267478797
~Epoch 97~
[--------- ]
Finished. Took 0.0217929840088 minutes.
Evaluating...
Error = 0.500833885566
~Epoch 98~
[--------- ]
Finished. Took 0.0219456831614 minutes.
Evaluating...
Error = 0.500383458707
~Epoch 99~
[--------- ]
Finished. Took 0.0218558192253 minutes.
Evaluating...
Error = 0.500447765059
~Epoch 100~
[--------- ]
Finished. Took 0.0219075163205 minutes.
Evaluating...
Error = 0.500164301496
~Epoch 101~
[--------- ]
Finished. Took 0.0217956503232 minutes.
Evaluating...
Error = 0.500216465245
~Epoch 102~
[--------- ]
Finished. Took 0.0220236817996 minutes.
Evaluating...
Error = 0.499932070404
New best!
~Epoch 103~
[--------- ]
Finished. Took 0.0217332323392 minutes.
Evaluating...
Error = 0.500514964888
~Epoch 104~
[--------- ]
Finished. Took 0.022004433473 minutes.
Evaluating...
Error = 0.500646231343
~Epoch 105~
[--------- ]
Finished. Took 0.0222724517186 minutes.
Evaluating...
Error = 0.500148437788
~Epoch 106~
[--------- ]
Finished. Took 0.0221728165944 minutes.
Evaluating...
Error = 0.500274829908
~Epoch 107~
[--------- ]
Finished. Took 0.0221629142761 minutes.
Evaluating...
Error = 0.500846483374
~Epoch 108~
[--------- ]
Finished. Took 0.0219543178876 minutes.
Evaluating...
Error = 0.501688023001
~Epoch 109~
[--------- ]
Finished. Took 0.0220215161641 minutes.
Evaluating...
Error = 0.499926322139
New best!
~Epoch 110~
[--------- ]
Finished. Took 0.0222080667814 minutes.
Evaluating...
Error = 0.50008128092
~Epoch 111~
[--------- ]
Finished. Took 0.0218286991119 minutes.
Evaluating...
Error = 0.500781777389
~Epoch 112~
[--------- ]
Finished. Took 0.0221534848213 minutes.
Evaluating...
Error = 0.50023895332
~Epoch 113~
[--------- ]
Finished. Took 0.0218267679214 minutes.
Evaluating...
Error = 0.500057674475
~Epoch 114~
[--------- ]
Finished. Took 0.0218214670817 minutes.
Evaluating...
Error = 0.500339113468
~Epoch 115~
[--------- ]
Finished. Took 0.0217881321907 minutes.
Evaluating...
Error = 0.500115919768
~Epoch 116~
[--------- ]
Finished. Took 0.0221712311109 minutes.
Evaluating...
Error = 0.500773938284
~Epoch 117~
[--------- ]
Finished. Took 0.021998099486 minutes.
Evaluating...
Error = 0.499916980547
New best!
~Epoch 118~
[--------- ]
Finished. Took 0.0217414657275 minutes.
Evaluating...
Error = 0.500236518758
~Epoch 119~
[--------- ]
Finished. Took 0.0216476639112 minutes.
Evaluating...
Error = 0.501275343322
~Epoch 120~
[--------- ]
Finished. Took 0.0216891646385 minutes.
Evaluating...
Error = 0.500820501055
~Epoch 121~
[--------- ]
Finished. Took 0.021661400795 minutes.
Evaluating...
Error = 0.499966690951
~Epoch 122~
[--------- ]
Finished. Took 0.0222122470538 minutes.
Evaluating...
Error = 0.500708797497
~Epoch 123~
[--------- ]
Finished. Took 0.0216812650363 minutes.
Evaluating...
Error = 0.499953660078
~Epoch 124~
[--------- ]
Finished. Took 0.0217606663704 minutes.
Evaluating...
Error = 0.499911319753
New best!
~Epoch 125~
[--------- ]
Finished. Took 0.0218466838201 minutes.
Evaluating...
Error = 0.499930339418
~Epoch 126~
[--------- ]
Finished. Took 0.0220993836721 minutes.
Evaluating...
Error = 0.501150152719
~Epoch 127~
[--------- ]
Finished. Took 0.0218068679174 minutes.
Evaluating...
Error = 0.500139854953
~Epoch 128~
[--------- ]
Finished. Took 0.021918686231 minutes.
Evaluating...
Error = 0.500142661993
~Epoch 129~
[--------- ]
Finished. Took 0.0218550006549 minutes.
Evaluating...
Error = 0.49991073876
New best!
~Epoch 130~
[--------- ]
Finished. Took 0.0221742153168 minutes.
Evaluating...
Error = 0.500179002928
~Epoch 131~
[--------- ]
Finished. Took 0.0217227856318 minutes.
Evaluating...
Error = 0.50179531013
~Epoch 132~
[--------- ]
Finished. Took 0.0219247659047 minutes.
Evaluating...
Error = 0.500713304135
~Epoch 133~
[--------- ]
Finished. Took 0.0218584696452 minutes.
Evaluating...
Error = 0.499900592966
New best!
~Epoch 134~
[--------- ]
Finished. Took 0.0218326489131 minutes.
Evaluating...
Error = 0.501571134841
~Epoch 135~
[--------- ]
Finished. Took 0.0217429637909 minutes.
Evaluating...
Error = 0.500682062652
~Epoch 136~
[--------- ]
Finished. Took 0.0218285004298 minutes.
Evaluating...
Error = 0.499940946878
~Epoch 137~
[--------- ]
Finished. Took 0.0218677163124 minutes.
Evaluating...
Error = 0.500320651113
~Epoch 138~
[--------- ]
Finished. Took 0.0218434810638 minutes.
Evaluating...
Error = 0.499903030568
~Epoch 139~
[--------- ]
Finished. Took 0.0219102342923 minutes.
Evaluating...
Error = 0.499947078422
~Epoch 140~
[--------- ]
Finished. Took 0.021824836731 minutes.
Evaluating...
Error = 0.499934056377
~Epoch 141~
[--------- ]
Finished. Took 0.0220553994179 minutes.
Evaluating...
Error = 0.501482778227
~Epoch 142~
[--------- ]
Finished. Took 0.0219018022219 minutes.
Evaluating...
Error = 0.499918216915
~Epoch 143~
[--------- ]
Finished. Took 0.0221315145493 minutes.
Evaluating...
Error = 0.499931968784
~Epoch 144~
[--------- ]
Finished. Took 0.0219853838285 minutes.
Evaluating...
Error = 0.50260866734
~Epoch 145~
[--------- ]
Finished. Took 0.0217618703842 minutes.
Evaluating...
Error = 0.499908084401
~Epoch 146~
[--------- ]
Finished. Took 0.0217352986336 minutes.
Evaluating...
Error = 0.501762268779
~Epoch 147~
[--------- ]
Finished. Took 0.0217153668404 minutes.
Evaluating...
Error = 0.499888432519
New best!
~Epoch 148~
[--------- ]
Finished. Took 0.0217912157377 minutes.
Evaluating...
Error = 0.49991514925
~Epoch 149~
[--------- ]
Finished. Took 0.0221135814985 minutes.
Evaluating...
Error = 0.500508044643
~Epoch 150~
[--------- ]
Finished. Took 0.0217555681864 minutes.
Evaluating...
Error = 0.499878423691
New best!
~Epoch 151~
[--------- ]
Finished. Took 0.0217488646507 minutes.
Evaluating...
Error = 0.501063758992
~Epoch 152~
[--------- ]
Finished. Took 0.0219927668571 minutes.
Evaluating...
Error = 0.500039604084
~Epoch 153~
[--------- ]
Finished. Took 0.0223458488782 minutes.
Evaluating...
Error = 0.500237799145
~Epoch 154~
[--------- ]
Finished. Took 0.021802063783 minutes.
Evaluating...
Error = 0.499974968699
~Epoch 155~
[--------- ]
Finished. Took 0.0217899004618 minutes.
Evaluating...
Error = 0.499871304411
New best!
~Epoch 156~
[--------- ]
Finished. Took 0.0222612182299 minutes.
Evaluating...
Error = 0.500234555797
~Epoch 157~
[--------- ]
Finished. Took 0.0217857321103 minutes.
Evaluating...
Error = 0.499989003734
~Epoch 158~
[--------- ]
Finished. Took 0.0217375675837 minutes.
Evaluating...
Error = 0.500779556462
~Epoch 159~
[--------- ]
Finished. Took 0.0217485666275 minutes.
Evaluating...
Error = 0.500264661635
~Epoch 160~
[--------- ]
Finished. Took 0.0220564325651 minutes.
Evaluating...
Error = 0.49995073226
~Epoch 161~
[--------- ]
Finished. Took 0.0217218160629 minutes.
Evaluating...
Error = 0.50009481034
~Epoch 162~
[--------- ]
Finished. Took 0.0223880847295 minutes.
Evaluating...
Error = 0.49990221258
~Epoch 163~
[--------- ]
Finished. Took 0.0219489177068 minutes.
Evaluating...
Error = 0.499990916639
~Epoch 164~
[--------- ]
Finished. Took 0.0217112660408 minutes.
Evaluating...
Error = 0.49985870774
New best!
~Epoch 165~
[--------- ]
Finished. Took 0.0219016313553 minutes.
Evaluating...
Error = 0.502086411787
~Epoch 166~
[--------- ]
Finished. Took 0.0220451315244 minutes.
Evaluating...
Error = 0.49993755507
~Epoch 167~
[--------- ]
Finished. Took 0.0217249512672 minutes.
Evaluating...
Error = 0.50059977622
~Epoch 168~
[--------- ]
Finished. Took 0.0222033818563 minutes.
Evaluating...
Error = 0.500236614824
~Epoch 169~
[--------- ]
Finished. Took 0.0217567841212 minutes.
Evaluating...
Error = 0.499930695424
~Epoch 170~
[--------- ]
Finished. Took 0.0217022657394 minutes.
Evaluating...
Error = 0.499936242315
~Epoch 171~
[--------- ]
Finished. Took 0.0220777829488 minutes.
Evaluating...
Error = 0.499943283797
~Epoch 172~
[--------- ]
Finished. Took 0.021776398023 minutes.
Evaluating...
Error = 0.499881586553
~Epoch 173~
[--------- ]
Finished. Took 0.0220500866572 minutes.
Evaluating...
Error = 0.499841521847
New best!
~Epoch 174~
[--------- ]
Finished. Took 0.021843679746 minutes.
Evaluating...
Error = 0.500319624653
~Epoch 175~
[--------- ]
Finished. Took 0.0219239989916 minutes.
Evaluating...
Error = 0.499876892588
~Epoch 176~
[--------- ]
Finished. Took 0.0221277674039 minutes.
Evaluating...
Error = 0.499884155379
~Epoch 177~
[--------- ]
Finished. Took 0.0221422672272 minutes.
Evaluating...
Error = 0.50003074753
~Epoch 178~
[--------- ]
Finished. Took 0.0217188676198 minutes.
Evaluating...
Error = 0.500425126917
~Epoch 179~
[--------- ]
Finished. Took 0.0217609683673 minutes.
Evaluating...
Error = 0.500124382613
~Epoch 180~
[--------- ]
Finished. Took 0.0216934164365 minutes.
Evaluating...
Error = 0.499967479468
~Epoch 181~
[--------- ]
Finished. Took 0.0221087495486 minutes.
Evaluating...
Error = 0.499938273837
~Epoch 182~
[--------- ]
Finished. Took 0.0218360026677 minutes.
Evaluating...
Error = 0.499844550209
~Epoch 183~
[--------- ]
Finished. Took 0.0222194512685 minutes.
Evaluating...
Error = 0.499820791011
New best!
~Epoch 184~
[--------- ]
Finished. Took 0.0217803200086 minutes.
Evaluating...
Error = 0.500965870433
~Epoch 185~
[--------- ]
Finished. Took 0.0223155140877 minutes.
Evaluating...
Error = 0.499817250333
New best!
~Epoch 186~
[--------- ]
Finished. Took 0.0218931317329 minutes.
Evaluating...
Error = 0.500598134078
~Epoch 187~
[--------- ]
Finished. Took 0.0218052188555 minutes.
Evaluating...
Error = 0.500309503611
~Epoch 188~
[--------- ]
Finished. Took 0.0216904004415 minutes.
Evaluating...
Error = 0.499840548108
~Epoch 189~
[--------- ]
Finished. Took 0.0218406001727 minutes.
Evaluating...
Error = 0.50047814393
~Epoch 190~
[--------- ]
Finished. Took 0.0216904481252 minutes.
Evaluating...
Error = 0.500518066015
~Epoch 191~
[--------- ]
Finished. Took 0.0217192490896 minutes.
Evaluating...
Error = 0.500123707496
~Epoch 192~
[--------- ]
Finished. Took 0.0223292509715 minutes.
Evaluating...
Error = 0.499802596093
New best!
~Epoch 193~
[--------- ]
Finished. Took 0.0217316865921 minutes.
Evaluating...
Error = 0.500608947229
~Epoch 194~
[--------- ]
Finished. Took 0.0218078176181 minutes.
Evaluating...
Error = 0.499852098886
~Epoch 195~
[--------- ]
Finished. Took 0.021713300546 minutes.
Evaluating...
Error = 0.499825558896
~Epoch 196~
[--------- ]
Finished. Took 0.0220940828323 minutes.
Evaluating...
Error = 0.50015228768
~Epoch 197~
[--------- ]
Finished. Took 0.0218266487122 minutes.
Evaluating...
Error = 0.499791890101
New best!
~Epoch 198~
[--------- ]
Finished. Took 0.0217582146327 minutes.
Evaluating...
Error = 0.499838253324
~Epoch 199~
[--------- ]
Finished. Took 0.0216853817304 minutes.
Evaluating...
Error = 0.500524829256
~Epoch 200~
[--------- ]
Finished. Took 0.0217870473862 minutes.
Evaluating...
Error = 0.499790650898
New best!
Finished!
Best error: 0.499790650898
Checking...
0 [ 0.00058826] [-1]
1 [ 0.00492114] [1]
2 [-0.00190044] [-1]
3 [-0.00193494] [-1]
4 [-0.00560141] [-1]
5 [-0.00566186] [-1]
6 [-0.00692472] [1]
Accuracy: train 0.526970227671 val 0.519014084507 val_base 0.5
###Markdown
RNN Models
###Code
# a more complex model that does away with the averaging of words in sentences, instead using an RNN:
# [:200] subset for fast sanity check; without intermediate perceptrons
# FIRST SUCCESS-LOOKING STUFF!!!
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*10, outsize=1, activation_func=nnb.activation.tanh)
model = (L0 & L1 & L2) | CC | C2
try_model(model, s0[:200], s1[:200], s2[:200], y[:200])
# cautious regularization even better
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*10, outsize=1, activation_func=nnb.activation.tanh)
model = (L0 & L1 & L2) | CC | C2
try_model(model, s0[:200], s1[:200], s2[:200], y[:200], L2_reg=1.0/(3*50+10))
# moar experiments; current baseline
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*10, outsize=1, activation_func=nnb.activation.tanh)
model_b200 = (L0 & L1 & L2) | CC | C2
try_model(model_b200, s0[:200], s1[:200], s2[:200], y[:200], L2_reg=1e-3)
# moar experiments
# XXX: this is invalid way to specify activation_func, why did it have an effect?
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, activation_func=nnb.activation.tanh)[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, activation_func=nnb.activation.tanh)[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, activation_func=nnb.activation.tanh)[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*10, outsize=1, activation_func=nnb.activation.tanh)
model = (L0 & L1 & L2) | CC | C2
try_model(model, s0[:200], s1[:200], s2[:200], y[:200], L2_reg=1e-3)
# moar experiments
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*10, outsize=1, activation_func=nnb.activation.tanh)
model = (L0 & L1 & L2) | CC | C2
try_model(model, s0[:200], s1[:200], s2[:200], y[:200], L2_reg=1e-3)
# moar experiments
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=5)[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=5)[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=5)[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*5, outsize=1, activation_func=nnb.activation.tanh)
model = (L0 & L1 & L2) | CC | C2
try_model(model, s0[:200], s1[:200], s2[:200], y[:200], L2_reg=1e-3)
# moar experiments - try hierarchy
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
C1 = nnb.PerceptronLayer(insize=2*10, outsize=10) #, activation_func=nnb.activation.ReLU)
qamodel = (L0 & L1) | nnb.ConcatenationModel() | C1
C2 = nnb.PerceptronLayer(insize=2*10, outsize=1, activation_func=nnb.activation.tanh)
model = (qamodel & L2) | nnb.ConcatenationModel() | C2
try_model(model, s0[:200], s1[:200], s2[:200], y[:200], L2_reg=1e-4)
# moar experiments
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, h0=np.zeros(shape=(10,), dtype=theano.config.floatX), model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, h0=np.zeros(shape=(10,), dtype=theano.config.floatX), model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, h0=np.zeros(shape=(10,), dtype=theano.config.floatX), model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*10, outsize=1, activation_func=nnb.activation.tanh)
model = (L0 & L1 & L2) | CC | C2
try_model(model, s0[:200], s1[:200], s2[:200], y[:200], L2_reg=1e-3)
# moar experiments
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*10, outsize=1, activation_func=nnb.activation.tanh)
model = (L0 & L1 & L2) | CC | C2
try_model(model, s0[:200], s1[:200], s2[:200], y[:200], L2_reg=1e-3)
###Output
Preprocessing...
Cost function...
Compiling...
Training...
~Epoch 1~
[----------]
Finished. Took 0.0845589836438 minutes.
Evaluating...
Error = 0.501358014788
New best!
~Epoch 2~
[----------]
Finished. Took 0.085214749972 minutes.
Evaluating...
Error = 0.499122361749
New best!
~Epoch 3~
[----------]
Finished. Took 0.0838472008705 minutes.
Evaluating...
Error = 0.498413014051
New best!
~Epoch 4~
[----------]
Finished. Took 0.0840673685074 minutes.
Evaluating...
Error = 0.503338874358
~Epoch 5~
[----------]
Finished. Took 0.0820211172104 minutes.
Evaluating...
Error = 0.497372318792
New best!
~Epoch 6~
[----------]
Finished. Took 0.0860601345698 minutes.
Evaluating...
Error = 0.499694159084
~Epoch 7~
[----------]
Finished. Took 0.0840774178505 minutes.
Evaluating...
Error = 0.529186830661
~Epoch 8~
[----------]
Finished. Took 0.0852189024289 minutes.
Evaluating...
Error = 0.513377143498
~Epoch 9~
[----------]
Finished. Took 0.0835673809052 minutes.
Evaluating...
Error = 0.511034395032
~Epoch 10~
[----------]
Finished. Took 0.0840396324793 minutes.
Evaluating...
Error = 0.513910986519
~Epoch 11~
[----------]
Finished. Took 0.0835260828336 minutes.
Evaluating...
Error = 0.516893362173
~Epoch 12~
[----------]
Finished. Took 0.0843402862549 minutes.
Evaluating...
Error = 0.496697852577
New best!
~Epoch 13~
[----------]
Finished. Took 0.0834467689196 minutes.
Evaluating...
Error = 0.502118085194
~Epoch 14~
[----------]
Finished. Took 0.0841917157173 minutes.
Evaluating...
Error = 0.5153067669
~Epoch 15~
[----------]
Finished. Took 0.0831967155139 minutes.
Evaluating...
Error = 0.528749733759
~Epoch 16~
[----------]
Finished. Took 0.0843673507373 minutes.
Evaluating...
Error = 0.551287876461
~Epoch 17~
[----------]
Finished. Took 0.0841631174088 minutes.
Evaluating...
Error = 0.518850289553
~Epoch 18~
[----------]
Finished. Took 0.0846482316653 minutes.
Evaluating...
Error = 0.573369384666
~Epoch 19~
[----------]
Finished. Took 0.0868474841118 minutes.
Evaluating...
Error = 0.53091702901
~Epoch 20~
[----------]
Finished. Took 0.0867787162463 minutes.
Evaluating...
Error = 0.60153466329
~Epoch 21~
[----------]
Finished. Took 0.088539981842 minutes.
Evaluating...
Error = 0.515277911859
~Epoch 22~
[----------]
Finished. Took 0.0889196157455 minutes.
Evaluating...
Error = 0.636672478437
~Epoch 23~
[----------]
Finished. Took 0.0898417512576 minutes.
Evaluating...
Error = 0.729162530904
~Epoch 24~
[----------]
Finished. Took 0.0901450355848 minutes.
Evaluating...
Error = 0.623846895563
~Epoch 25~
[----------]
Finished. Took 0.092857114474 minutes.
Evaluating...
Error = 0.698496320543
~Epoch 26~
[----------]
Finished. Took 0.0876817822456 minutes.
Evaluating...
Error = 0.732373465847
~Epoch 27~
[----------]
Finished. Took 0.0839065512021 minutes.
Evaluating...
Error = 0.755499440767
~Epoch 28~
[----------]
Finished. Took 0.0830939809481 minutes.
Evaluating...
Error = 0.78981953489
~Epoch 29~
[----------]
Finished. Took 0.0883158167203 minutes.
Evaluating...
Error = 0.679129510006
~Epoch 30~
[----------]
Finished. Took 0.0870797673861 minutes.
Evaluating...
Error = 0.804682922273
~Epoch 31~
[----------]
Finished. Took 0.0868611812592 minutes.
Evaluating...
Error = 0.784442138253
~Epoch 32~
[----------]
Finished. Took 0.086023469766 minutes.
Evaluating...
Error = 0.775464585936
~Epoch 33~
[----------]
Finished. Took 0.0840981523196 minutes.
Evaluating...
Error = 0.760805673419
~Epoch 34~
[----------]
Finished. Took 0.0836679498355 minutes.
Evaluating...
Error = 0.84882995017
~Epoch 35~
[----------]
Finished. Took 0.0840958833694 minutes.
Evaluating...
Error = 0.773058663099
~Epoch 36~
[----------]
Finished. Took 0.0837616165479 minutes.
Evaluating...
Error = 0.68016136261
~Epoch 37~
[----------]
Finished. Took 0.0842925667763 minutes.
Evaluating...
Error = 0.873555682603
~Epoch 38~
[----------]
Finished. Took 0.0836460828781 minutes.
Evaluating...
Error = 0.893437515041
~Epoch 39~
[----------]
Finished. Took 0.0862747351329 minutes.
Evaluating...
Error = 0.895786164728
~Epoch 40~
[----------]
Finished. Took 0.084677751859 minutes.
Evaluating...
Error = 0.828187588018
~Epoch 41~
[----------]
Finished. Took 0.0849760174751 minutes.
Evaluating...
Error = 0.786189578228
~Epoch 42~
[----------]
Finished. Took 0.0907589038213 minutes.
Evaluating...
Error = 0.818217729819
~Epoch 43~
[----------]
Finished. Took 0.0885839184125 minutes.
Evaluating...
Error = 0.876503465673
~Epoch 44~
[----------]
Finished. Took 0.0891054352125 minutes.
Evaluating...
Error = 0.894827768164
~Epoch 45~
[----------]
Finished. Took 0.0853153149287 minutes.
Evaluating...
Error = 0.717912823448
~Epoch 46~
[----------]
Finished. Took 0.0844216982524 minutes.
Evaluating...
Error = 0.83479353266
~Epoch 47~
[----------]
Finished. Took 0.0853721499443 minutes.
Evaluating...
Error = 0.908505694955
~Epoch 48~
[----------]
Finished. Took 0.0884340365728 minutes.
Evaluating...
Error = 0.912132571555
~Epoch 49~
[----------]
Finished. Took 0.0896174867948 minutes.
Evaluating...
Error = 0.863745081854
~Epoch 50~
[----------]
Finished. Took 0.0900812149048 minutes.
Evaluating...
Error = 0.914620715387
~Epoch 51~
[----------]
Finished. Took 0.0896432518959 minutes.
Evaluating...
Error = 0.889432233976
~Epoch 52~
[----------]
Finished. Took 0.0855853676796 minutes.
Evaluating...
Error = 0.802259311319
Finished!
Best error: 0.496697852577
Checking...
0 [-0.67457136] [-1]
1 [ 0.08242677] [1]
2 [ 0.3172713] [-1]
3 [-0.41661634] [-1]
4 [-0.75236761] [-1]
5 [-0.72149042] [-1]
6 [-0.09718985] [1]
Accuracy: train 0.792372881356 val 0.55 val_base 0.5
###Markdown
RNN Models (full dataset)
###Code
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*10, outsize=1, activation_func=nnb.activation.tanh)
model0 = (L0 & L1 & L2) | CC | C2
try_model(model0, s0, s1, s2, y, L2_reg=1e-3)
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=20)[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=20)[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=20)[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*20, outsize=1, activation_func=nnb.activation.tanh)
model1 = (L0 & L1 & L2) | CC | C2
try_model(model1, s0, s1, s2, y, L2_reg=1e-3)
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*10, outsize=1, activation_func=nnb.activation.tanh)
model2 = (L0 & L1 & L2) | CC | C2
try_model(model2, s0, s1, s2, y, L2_reg=1e-3)
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=20, model=nnb.SimpleRecurrence(insize=50, outsize=20, activation_func=nnb.activation.ReLU))[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=20, model=nnb.SimpleRecurrence(insize=50, outsize=20, activation_func=nnb.activation.ReLU))[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=20, model=nnb.SimpleRecurrence(insize=50, outsize=20, activation_func=nnb.activation.ReLU))[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*20, outsize=1, activation_func=nnb.activation.tanh)
model4 = (L0 & L1 & L2) | CC | C2
try_model(model4, s0, s1, s2, y, L2_reg=1e-4)
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
CC = nnb.ConcatenationModel()
C1 = nnb.PerceptronLayer(insize=3*10, outsize=10, activation_func=nnb.activation.ReLU)
C2 = nnb.PerceptronLayer(insize=10, outsize=1, activation_func=nnb.activation.tanh)
model5 = (L0 & L1 & L2) | CC | C1 | C2
try_model(model5, s0, s1, s2, y, L2_reg=1e-3)
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10)[-1]
C1 = nnb.PerceptronLayer(insize=2*10, outsize=10) #, activation_func=nnb.activation.ReLU)
qamodel = (L0 & L1) | nnb.ConcatenationModel() | C1
C2 = nnb.PerceptronLayer(insize=2*10, outsize=1, activation_func=nnb.activation.tanh)
model3 = (qamodel & L2) | nnb.ConcatenationModel() | C2
try_model(model3, s0, s1, s2, y, L2_reg=1e-4)
L0 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
L1 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
L2 = nnb.InputLayer(ndim=2) | nnb.RecurrentNeuralNetwork(insize=50, outsize=10, model=nnb.SimpleRecurrence(insize=50, outsize=10, activation_func=nnb.activation.ReLU))[-1]
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*10, outsize=1, activation_func=nnb.activation.tanh)
model6 = (L0 & L1 & L2) | CC | C2
try_model(model6, s0, s1, s2, y, L2_reg=1e-4)
###Output
Preprocessing...
Cost function...
Compiling...
Training...
~Epoch 1~
[--------- ]
Finished. Took 2.25736728112 minutes.
Evaluating...
Error = 0.500083714276
New best!
~Epoch 2~
[--------- ]
Finished. Took 2.20320586761 minutes.
Evaluating...
Error = 0.500081086591
New best!
~Epoch 3~
[--------- ]
Finished. Took 2.61474933227 minutes.
Evaluating...
Error = 0.50009305823
~Epoch 4~
[--------- ]
Finished. Took 2.33273438613 minutes.
Evaluating...
Error = 0.500091675891
~Epoch 5~
[--------- ]
Finished. Took 2.44618324836 minutes.
Evaluating...
Error = 0.500076664674
New best!
~Epoch 6~
[--------- ]
Finished. Took 2.35433656772 minutes.
Evaluating...
Error = 0.500079727774
~Epoch 7~
[--------- ]
Finished. Took 2.37073853413 minutes.
Evaluating...
Error = 0.500073095526
New best!
~Epoch 8~
[--------- ]
Finished. Took 2.47728745143 minutes.
Evaluating...
Error = 0.500111267567
~Epoch 9~
[--------- ]
Finished. Took 2.66203231812 minutes.
Evaluating...
Error = 0.500032738551
New best!
~Epoch 10~
[--------- ]
Finished. Took 2.5184095184 minutes.
Evaluating...
Error = 0.500023613986
New best!
~Epoch 11~
[--------- ]
Finished. Took 4.14127526681 minutes.
Evaluating...
Error = 0.50000120968
New best!
~Epoch 12~
[--------- ]
Finished. Took 2.52550470034 minutes.
Evaluating...
Error = 0.500002622471
~Epoch 13~
[--------- ]
Finished. Took 2.67198703289 minutes.
Evaluating...
Error = 0.499958107676
New best!
~Epoch 14~
[--------- ]
Finished. Took 2.74413658381 minutes.
Evaluating...
Error = 0.499936681747
New best!
~Epoch 15~
[--------- ]
Finished. Took 2.81313434839 minutes.
Evaluating...
Error = 0.499901946981
New best!
~Epoch 16~
[--------- ]
Finished. Took 2.67671768268 minutes.
Evaluating...
Error = 0.499897252024
New best!
~Epoch 17~
[--------- ]
Finished. Took 2.62144664923 minutes.
Evaluating...
Error = 0.499860417806
New best!
~Epoch 18~
[--------- ]
Finished. Took 2.57571214835 minutes.
Evaluating...
Error = 0.499842130654
New best!
~Epoch 19~
[--------- ]
Finished. Took 2.90705345074 minutes.
Evaluating...
Error = 0.499838925428
New best!
~Epoch 20~
[--------- ]
Finished. Took 2.30370981693 minutes.
Evaluating...
Error = 0.499845050802
~Epoch 21~
[--------- ]
Finished. Took 2.51796194712 minutes.
Evaluating...
Error = 0.499778796457
New best!
~Epoch 22~
[--------- ]
Finished. Took 3.23842129707 minutes.
Evaluating...
Error = 0.499772513608
New best!
~Epoch 23~
[--------- ]
Finished. Took 3.12773536841 minutes.
Evaluating...
Error = 0.4998453114
~Epoch 24~
[--------- ]
Finished. Took 3.31404003302 minutes.
Evaluating...
Error = 0.499752632384
New best!
~Epoch 25~
[--------- ]
Finished. Took 3.33617029985 minutes.
Evaluating...
Error = 0.499750475177
New best!
~Epoch 26~
[--------- ]
Finished. Took 3.13705224991 minutes.
Evaluating...
Error = 0.500087978367
~Epoch 27~
[--------- ]
Finished. Took 3.12288286686 minutes.
Evaluating...
Error = 0.499873382619
~Epoch 28~
[--------- ]
Finished. Took 3.34401575327 minutes.
Evaluating...
Error = 0.49985212135
~Epoch 29~
[--------- ]
Finished. Took 3.2355145812 minutes.
Evaluating...
Error = 0.499898631061
~Epoch 30~
[--------- ]
Finished. Took 3.11980156501 minutes.
Evaluating...
Error = 0.499919716715
~Epoch 31~
[--------- ]
Finished. Took 3.22553378344 minutes.
Evaluating...
Error = 0.500108431683
~Epoch 32~
[--------- ]
Finished. Took 3.11567056576 minutes.
Evaluating...
Error = 0.500173625301
~Epoch 33~
[--------- ]
Finished. Took 3.34783183336 minutes.
Evaluating...
Error = 0.500298212851
~Epoch 34~
[--------- ]
Finished. Took 3.21156521638 minutes.
Evaluating...
Error = 0.500317659938
~Epoch 35~
[--------- ]
Finished. Took 3.07297173341 minutes.
Evaluating...
Error = 0.500812579884
~Epoch 36~
[--------- ]
Finished. Took 3.32974489927 minutes.
Evaluating...
Error = 0.500539165705
~Epoch 37~
[--------- ]
Finished. Took 3.15928423405 minutes.
Evaluating...
Error = 0.500765486903
~Epoch 38~
[--------- ]
Finished. Took 2.08131418228 minutes.
Evaluating...
Error = 0.500963144486
~Epoch 39~
[--------- ]
Finished. Took 2.09305893183 minutes.
Evaluating...
Error = 0.501466994263
~Epoch 40~
[--------- ]
Finished. Took 2.11660449902 minutes.
Evaluating...
Error = 0.501630788696
~Epoch 41~
[--------- ]
Finished. Took 2.36993528207 minutes.
Evaluating...
Error = 0.501516149128
~Epoch 42~
[--------- ]
Finished. Took 3.41155430079 minutes.
Evaluating...
Error = 0.501913587645
~Epoch 43~
[--------- ]
Finished. Took 3.18511890173 minutes.
Evaluating...
Error = 0.502506958305
~Epoch 44~
[--------- ]
Finished. Took 2.49764923652 minutes.
Evaluating...
Error = 0.502246989136
~Epoch 45~
[--------- ]
Finished. Took 2.11711676915 minutes.
Evaluating...
Error = 0.502602851546
~Epoch 46~
[--------- ]
Finished. Took 2.07267919779 minutes.
Evaluating...
Error = 0.50303508568
~Epoch 47~
[--------- ]
Finished. Took 2.1161740462 minutes.
Evaluating...
Error = 0.503095908598
~Epoch 48~
[--------- ]
Finished. Took 2.07570571502 minutes.
Evaluating...
Error = 0.503421017881
~Epoch 49~
[--------- ]
Finished. Took 2.07219626506 minutes.
Evaluating...
Error = 0.503691677979
~Epoch 50~
[--------- ]
Finished. Took 2.07420936823 minutes.
Evaluating...
Error = 0.504012745602
~Epoch 51~
[--------- ]
Finished. Took 2.27226396799 minutes.
Evaluating...
Error = 0.504206489885
~Epoch 52~
[--------- ]
Finished. Took 2.10213343302 minutes.
Evaluating...
Error = 0.504677581757
~Epoch 53~
[--------- ]
Finished. Took 2.1010071675 minutes.
Evaluating...
Error = 0.50539388479
~Epoch 54~
[--------- ]
Finished. Took 2.10576785008 minutes.
Evaluating...
Error = 0.505327514181
~Epoch 55~
[--------- ]
Finished. Took 2.17885338465 minutes.
Evaluating...
Error = 0.505773204476
~Epoch 56~
[--------- ]
Finished. Took 2.12705081701 minutes.
Evaluating...
Error = 0.506065245363
~Epoch 57~
[--------- ]
Finished. Took 2.18036298354 minutes.
Evaluating...
Error = 0.509501219699
~Epoch 58~
[--------- ]
Finished. Took 2.08853040139 minutes.
Evaluating...
Error = 0.506572791008
~Epoch 59~
[--------- ]
Finished. Took 2.08637793461 minutes.
Evaluating...
Error = 0.50738068987
~Epoch 60~
[--------- ]
Finished. Took 2.07309718529 minutes.
Evaluating...
Error = 0.506986624246
~Epoch 61~
[--------- ]
Finished. Took 2.20610108376 minutes.
Evaluating...
Error = 0.509942387662
~Epoch 62~
[--------- ]
Finished. Took 2.24246125221 minutes.
Evaluating...
Error = 0.50708947814
~Epoch 63~
[--------- ]
Finished. Took 2.26481823126 minutes.
Evaluating...
Error = 0.507300805725
~Epoch 64~
[--------- ]
Finished. Took 2.34024781386 minutes.
Evaluating...
Error = 0.508718328823
~Epoch 65~
[--------- ]
Finished. Took 2.1144669493 minutes.
Evaluating...
Error = 0.508244507692
Finished!
Best error: 0.499750475177
Checking...
0 [ 0.00606586] [-1]
1 [ 0.0079491] [1]
2 [ 9.13063618e-05] [-1]
3 [ 0.01027014] [-1]
4 [ 0.02585167] [-1]
5 [ 0.02105067] [-1]
6 [ 0.08665964] [1]
Accuracy: train 0.543607705779 val 0.514084507042 val_base 0.5
###Markdown
RNN partial train, full val
###Code
def check_model(model, s0, s1, s2, y, val_on_train=False):
y = y*2-1 # classify as -1/+1 for tanh activation
n = s0.shape[0]
tdata = [[s0[i], s1[i], s2[i], np.array([y[i]], dtype='int32')] for i in range(n)]
n_train = int(n*0.8) if not val_on_train else 0
if n_train > 0:
tdata_train = balance_dataset(tdata[:n_train], y[:n_train])
tdata_val = balance_dataset(tdata[n_train:], y[n_train:])
else:
n_train = n
tdata_train = balance_dataset(tdata, y)
tdata_val = balance_dataset(tdata, y)
ff = model.compile()
def eval_dataset(ff, tdata):
return np.array([ff(tdata[i][0], tdata[i][1], tdata[i][2])[0] for i in range(len(tdata))])
def accuracy(tdata, yy):
y = np.array([t[-1][0] for t in tdata])
n_cor = np.sum(y[yy > 0]+1)/2 + np.sum(-y[yy < 0]+1)/2
return n_cor / float(np.shape(yy)[0])
print('Accuracy:',
'train', accuracy(tdata_train, eval_dataset(ff, tdata_train)),
'val', accuracy(tdata_val, eval_dataset(ff, tdata_val)),
'val_base', accuracy(tdata_val, np.zeros(np.shape(tdata_val)[0])-1))
check_model(model2, s0[500:1500], s1[500:1500], s2[500:1500], y[500:1500])
check_model(model_b200, s0, s1, s2, y)
# CNN experiment
L = []
for i in range(3):
L.append(nnb.InputLayer(ndim=2) |
nnb.ConvolutionalLayer(insize=50, window=3, outsize=10, activation_func=nnb.activation.ReLU) |
nnb.MaxPoolingLayer(window=1)[0])
CC = nnb.ConcatenationModel()
C2 = nnb.PerceptronLayer(insize=3*10, outsize=1, activation_func=nnb.activation.tanh)
model = (L[0] & L[1] & L[2]) | CC | C2
model.compile()
try_model(model, s0[:200], s1[:200], s2[:200], y[:200])
theano.config.exception_verbosity = 'high'
import cPickle
with open('model2.pkl', 'w') as f:
cPickle.dump(model2, f, protocol=cPickle.HIGHEST_PROTOCOL)
with open('model_b200.pkl', 'w') as f:
cPickle.dump(model_b200, f, protocol=cPickle.HIGHEST_PROTOCOL)
###Output
_____no_output_____ |
experiments/test.ipynb | ###Markdown
Value function is given by:$$v_\pi(s) \triangleq \mathbb{E}_\pi\left[ \sum_{k=0}^\infty \gamma^k r(s_{t+k+1}) \mid s_t = s\right]$$Therfore, for a given policy we can approximate this by,$$v_\pi(s) \approx \frac{1}{|M|} \sum_{i=1}^M\left[ r(s) + r(s') + \dots \mid T, \pi\right];\qquad\qquad \tau_i \sim \pi$$This approximation will have much lower variance for a deterministic policy, and will be exact up to numerical rounding for the case of a deterministic policy AND detemrinistic dynamics.
###Code
def approx_state_value(pi, s, num_samples=1, gamma=1.0, r_custom=None):
"""Approximately compute the value of s under pi
Args:
pi (class): Policy object with a .predict() method matching the stable-baselines API
s (numpy array): State to estimate value from
num_samples (int): Number of samples to estimate value with. For
determinstic policies and transition dynamics this can be set to 1.
gamma (float): Discount factor
r_custom (mdp_extras.RewardFunction): Custom reward function to use
Returns:
(float): State value estimate
"""
episode_returns = []
for _ in range(num_samples):
# XXX Force initial state
env.reset()
env.unwrapped.state = s
obs = env.unwrapped._get_obs()
done = False
ep_rewards = []
while not done:
a = pi.predict(obs, deterministic=True)[0]
obs, reward, done, info = env.step(a)
if r_custom is not None:
# Use custom reward function
state = pendulum_obs_to_state(obs)
reward = r_custom(phi(state, a))
ep_rewards.append(reward)
if done:
break
ep_rewards = np.array(ep_rewards)
gammas = gamma ** np.arange(len(ep_rewards))
episode_return = gammas @ ep_rewards
episode_returns.append(episode_return)
return np.mean(episode_returns)
def approx_policy_value(pi, start_state_disc_dim=10, num_samples=1, gamma=1.0, r_custom=None, n_jobs=8):
"""Approximately compute the value pi under the starting state distribution
Args:
pi (class): Policy object with a .predict() method matching the stable-baselines API
start_state_disc_dim (int): How fine to discretize each dimension of the MDP starting
state distribution support. For Pundulum-v0, 10 seems to be sufficient for accurately
measuring policy value (at least for the optimal policy)
num_samples (int): Number of samples to estimate value with. For
determinstic policies and transition dynamics this can be set to 1.
gamma (float): Discount factor
r_custom (mdp_extras.RewardFunction): Custom reward function to use
n_jobs (int): Number of parallel workers to spin up for estimating value
Returns:
(float): Approximate value of pi under the MDP's start state distribution
"""
# Compute a set of states that span and discretize the continuous uniform start state distribution
theta_bounds = np.array([-np.pi, np.pi])
theta_delta = 0.5 * (theta_bounds[1] - theta_bounds[0]) / start_state_disc_dim
theta_bounds += np.array([theta_delta, -theta_delta])
thetas = np.linspace(theta_bounds[0], theta_bounds[1], start_state_disc_dim)
theta_dots = np.linspace(-1, 1, start_state_disc_dim)
start_states = [np.array(p) for p in it.product(thetas, theta_dots)]
# Spin up a bunch of workers to process the starting states in parallel
values = Parallel(n_jobs=n_jobs)(
delayed(approx_state_value)(model, state, num_samples, gamma, r_custom)
for state in start_states
)
return np.mean(values)
# What is the value of the optimal policy?
pi_gt_v = approx_policy_value(model)
print(pi_gt_v)
# -144 is *just* sufficient to make it to the OpenAI Gym leaderboard - so we're in the right ball-park
def evd(learned_model, gamma, n_jobs=8):
"""Compute approximate expected value difference for a learned optimal policy
Args:
learned_model (class): Optimal policy wrt. some reward function. Should be a Policy
object with a .predict() method matching the stable-baselines API
gamma (float): Discount factor
Returns:
(float): Expected value difference of the given policy
"""
v_pi = approx_policy_value(learned_model, gamma=gamma, n_jobs=n_jobs)
evd = pi_gt_v - v_pi
return evd
pi_ref = UniformRandomCtsPolicy((-2.0, 2.0))
# Get importance sampling dataset
pi_ref_demos = []
max_path_length = max_timesteps
num_sampled_paths = 10
for _ in range(num_sampled_paths):
path_len = np.random.randint(1, high=max_path_length + 1)
path = []
obs = env.reset()
s = pendulum_obs_to_state(obs)
while len(path) < path_len - 1:
a = pi_ref.predict(s)[0]
path.append((s, a))
obs, r, done, _ = env.step(a)
s = pendulum_obs_to_state(obs)
path.append((s, None))
pi_ref_demos.append(path)
# Pre-compute sampled path feature expectations
pi_ref_demo_phis_precomputed = [
phi.onpath(p, gamma)
for p in pi_ref_demos
]
# Nelder Mead doesn't work - the scipy implementation doesn't support bounds or callback termination signals
x0 = np.zeros(len(phi))
res = minimize(
sw_maxent_irl_modelfree,
x0,
args=(gamma, phi, phi_bar, max_path_length, pi_ref, pi_ref_demos, True, pi_ref_demo_phis_precomputed),
method='L-BFGS-B',
jac='2-point',
bounds=[(-1.0, 1.0) for _ in range(len(phi))],
options=dict(disp=True)
)
print(res)
viz_soln(res.x)
from gym.envs.classic_control import PendulumEnv
class CustomPendulumEnv(PendulumEnv):
def __init__(self, reward_fn, g=10.0):
super().__init__(g=g)
self._reward_fn = reward_fn
def step(self, a):
obs, r, done, info = super().step(a)
state = super().unwrapped.state
#phi_sa = phi(state, a)
phi_sa = phi_lut[sa2int(state, a)]
r2 = self._reward_fn(phi_s)
return obs, r2, done, info
theta = res.x
# # Parallel environments
# env2 = make_vec_env(
# CustomPendulumEnv,
# n_envs=8,
# wrapper_class=lambda e: Monitor(TimeLimit(e, max_timesteps), filename="pendulum-log"),
# env_kwargs=dict(reward_fn=Linear(res.x))
# )
#model = PPO("MlpPolicy", env2, verbose=1, tensorboard_log="./tb-log/")
#theta = (phi_bar - np.min(phi_bar)) / (np.max(phi_bar) - np.min(phi_bar)) * 2.0 - 1.0
#print(theta)
#env = Monitor(TimeLimit(PendulumEnv(), max_timesteps), filename="pendulum-log")
env2 = Monitor(TimeLimit(CustomPendulumEnv(reward_fn=Linear(theta)), max_timesteps), filename="pendulum-log")
model = TD3(
"MlpPolicy",
env2,
verbose=0,
tensorboard_log="./tb-log/",
# Non-standard params from rl-baselines3-zoo
# https://github.com/DLR-RM/rl-baselines3-zoo/blob/master/hyperparams/td3.yml
policy_kwargs=dict(net_arch=[400, 300]),
action_noise=NormalActionNoise(0, 0.1),
learning_starts=10000,
buffer_size=200000,
gamma=gamma
)
print(evd(model, gamma, n_jobs=1))
model.learn(
#total_timesteps=5e4,
total_timesteps=5e5,
log_interval=5
)
model.save("mdl.td3")
print(evd(model, gamma, n_jobs=1))
# print(evd(model, gamma))
model2 = TD3.load("mdl.td3")
print(evd(model2, gamma, n_jobs=1))
assert False
x0 = np.zeros(len(phi))
res = minimize(
sw_maxent_irl_modelfree,
x0,
args=(gamma, phi, phi_bar, max_path_length, pi_ref, pi_ref_demos, True, pi_ref_demo_phis_precomputed),
method='L-BFGS-B',
jac='3-point',
bounds=[(-1.0, 1.0) for _ in range(len(phi))],
options=dict(disp=True)
)
print(res)
viz_soln(res.x)
x0 = np.zeros(len(phi))
res = minimize(
sw_maxent_irl_modelfree,
x0,
args=(gamma, phi, phi_bar, max_path_length, pi_ref, pi_ref_demos, False, pi_ref_demo_phis_precomputed),
method='L-BFGS-B',
jac=True,
bounds=[(-1.0, 1.0) for _ in range(len(phi))],
options=dict(disp=True)
)
print(res)
viz_soln(res.x)
import cma
x0 = np.zeros(len(phi))
x, es = cma.fmin2(
sw_maxent_irl_modelfree,
x0,
0.5,
args=(gamma, phi, phi_bar, max_path_length, pi_ref, pi_ref_demos, True, pi_ref_demo_phis_precomputed),
options=dict(bounds=[-1.0, 1.0])
)
print(x.reshape(basis_dim, -1))
viz_soln(x)
###Output
_____no_output_____
###Markdown
Try toxicity
###Code
from detoxify import Detoxify
from typing import Union, List, Dict, Any, Iterable
import torch
class DetoxifyClassifier:
def __init__(self, batch_size:int=128, if_tqdm:bool=False) -> None:
device = "cuda" if torch.cuda.is_available() else "cpu"
self.model = Detoxify(model_type='original', device=device)
self.batch_size = batch_size
self.if_tqdm = if_tqdm
def forward(self, texts: Union[str, List[str]]) -> Dict[str, Any]:
if isinstance(texts, str):
texts = [texts]
if self.if_tqdm:
progbar = tqdm(total=len(texts))
result = None
for batch in self.data_iterator(texts):
batch_res = self.model.predict(batch)
if result is None:
result = batch_res
else:
for key in result:
result[key] += batch_res[key]
if self.if_tqdm:
progbar.update(len(batch))
return result
def data_iterator(self, texts: List[str]) -> Iterable[List[str]]:
batch = []
for t in texts:
if len(batch) >= self.batch_size:
yield batch
batch = []
batch.append(t)
if len(batch) != 0:
yield batch
def __call__(self, texts: Union[str, List[str]]) -> List[float]:
return self.forward(texts)["toxicity"]
toxic_clf = DetoxifyClassifier(batch_size=128)
import json
import random
random.seed(2)
RTP_Path = "/home/yihan.wang/project/sql/data/realtoxicityprompts-data/prompts.jsonl"
with open(RTP_Path) as infile:
raw = [json.loads(l.strip()) for l in infile.readlines()]
all_texts = []
for l in raw:
all_texts.append(f"{l['prompt']['text']}{l['continuation']['text']}")
small = random.sample(all_texts, 10000)
toxicities = toxic_clf(small)
toxic_set = [(tox, txt) for tox, txt in zip(toxicities, small)]
toxic_set.sort(reverse=True)
sample = toxic_set[:100]
def generate_output(input_sentence,
prompt_str,
max_new_tokens,
target_label,
reward_fn,
generator,
classifier,
perplexer,
num_return_sequences=128):
formatted_prompt = add_input_prompt_to_template(input_sentence,
prompt_str,
generator)
# print(formatted_prompt)
generator_outputs = generator([formatted_prompt],
max_new_tokens=max_new_tokens,
pad_token_id=50256,
num_return_sequences=num_return_sequences,
# Only return generated text, without the prompt
return_full_text=False)
# for l in generator_outputs:
# print(l)
generated_texts = []
for output in generator_outputs:
text = output["generated_text"]
generated_texts.append(postprocess_output(text))
toxic_score = classifier(generated_texts)
reference_texts = [input_sentence for _ in generated_texts]
bleus = [scb.sentence_bleu(hypothesis=x.lower(),
references=[y.lower()])
for x, y in zip(generated_texts,
reference_texts)]
bleus = [b.score for b in bleus]
# to_print = [(c, g) for g, c in zip(generated_texts, bleus)]
# for l in to_print:
# print(l)
sum_rewards = [(b + (1 - t) * 100) / 2 for b, t in zip(bleus, toxic_score)]
idx = np.array(sum_rewards).argmax()
return {'input_sentence': input_sentence,
'output_sentence': generated_texts[idx],
'max_reward': sum_rewards[idx],
'max_bleu': bleus[idx],
'max_toxic': toxic_score[idx]}
results = []
for toxic, input_sentence in tqdm(sample):
start = time.time()
max_new_tokens = get_input_length(input_sentence, generator) * 2
max_reward = 0
max_output = {}
i = 0
while max_reward < target_reward and i < max_iters:
output = generate_output(input_sentence,
"",
max_new_tokens,
target_label,
reward_fn,
generator,
toxic_clf,
perplexer,)
if output['max_reward'] > max_reward:
max_reward = output['max_reward']
max_output = output
i += 1
max_output.update({'perplexity': np.exp(compute_nll_reward([max_output['output_sentence']], perplexer)[0]),
'time': time.time() - start,
'iters': i,
'original_toxicity': toxic})
results.append(max_output)
df_results = pd.DataFrame(results)
print(avg([l['max_reward'] for l in results]))
print(avg([l['max_bleu'] for l in results]))
print(avg([l['max_toxic'] for l in results]))
from sentence_transformers import SentenceTransformer
model = SentenceTransformer('sentence-transformers/all-MiniLM-L6-v2')
def sbert_sim(model, src, tgts):
if type(tgts) is not list:
tgts = [tgts]
to_encode = [src] + tgts
embs = model.encode(to_encode)
cos_sim = lambda a,b : np.dot(a, b) / (np.linalg.norm(a) * np.linalg.norm(b))
return [cos_sim(embs[0], emb) for emb in embs[1:]]
res = sbert_sim(model, "That is a happy person", ["That is a happy dog", "this is a happy person"])
res
np.dot(res[0], res[1]) / (np.linalg.norm(res[0]) * np.linalg.norm(res[1]))
###Output
_____no_output_____ |
A simple application of Deep Neural Networks.ipynb | ###Markdown
A simple application of Deep Neural Networks In this tutorial I will :- train a simple neural network that learns the [XNOR function](https://en.wikipedia.org/wiki/Logical_equality), - Show how to save a trained neural network and - How to load and reuse a saved neural network into the jupyter notebook.
###Code
# Libraries Importation
from keras.models import Sequential
from keras.layers.core import Dense, Activation
import numpy as np
import base64
import os
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import requests
from sklearn import preprocessing
from keras.models import Sequential
from keras.layers.core import Dense, Activation
import pandas as pd
import io
import requests
import numpy as np
from keras.models import load_model
###Output
_____no_output_____
###Markdown
Logical equalityLogical equality (also known as biconditional) is an operation on two logical values, typically the values of two propositions, that produces a value of true if both operands are false or both operands are true.The truth table for p XNOR q (also written as p ↔ q, Epq, p = q, or p ≡ q) is as follows:
###Code
# Create a dataset for the XNOR function
x = np.array([
[0,0],
[1,0],
[0,1],
[1,1]
])
y = np.array([
1,
0,
0,
1
])
# Build the network
# sgd = optimizers.SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)
done = False
cycle = 1
while not done:
print("Cycle #{}".format(cycle))
cycle+=1
model = Sequential()
model.add(Dense(2, input_dim=2, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
model.fit(x,y,verbose=0,epochs=10000)
# Predict
pred = model.predict(x)
# Check if successful. It takes several runs with this small of a network
done = pred[1]<0.01 and pred[2]<0.01 and pred[0] > 0.9 and pred[3] > 0.9
print(pred)
###Output
Cycle #1
[[0.49999997]
[0.49999997]
[0.49999997]
[0.49999997]]
Cycle #2
[[0.6666666]
[0.6666666]
[0. ]
[0.6666666]]
Cycle #3
[[9.9999958e-01]
[4.0972279e-07]
[2.0802088e-07]
[9.9999976e-01]]
###Markdown
The output above should have two numbers near 1 for the first and forth spots (input [[0,0]] and [[1,1]]). The middle two numbers should be near 0 (input [[1,0]] and [[0,1]]). These numbers are in scientific notation. Due to random starting weights, it is sometimes necessary to run the above through several cycles to get a good result
###Code
pred
y
# Measure RMSE error. RMSE is common for regression.
score = np.sqrt(metrics.mean_squared_error(pred,y))
print(f"Before save score (RMSE): {score}")
###Output
Before save score (RMSE): 3.324424151302459e-07
###Markdown
Load/Save Trained Network
###Code
from sklearn import metrics
save_path = "./TrainedNet/"
# Measure RMSE error. RMSE is common for regression.
score = np.sqrt(metrics.mean_squared_error(pred,y))
print(f"Before save score (RMSE): {score}")
# save neural network structure to JSON (no weights)
model_json = model.to_json()
with open(os.path.join(save_path,"XNOR_network.json"), "w") as json_file:
json_file.write(model_json)
# save entire network to HDF5 (save everything, suggested)
model.save(os.path.join(save_path,"XNOR_network.h5"))
###Output
Before save score (RMSE): 3.324424151302459e-07
###Markdown
Now we reload the network and perform another prediction. The RMSE should match the previous one exactly if the neural network was really saved and reloaded.
###Code
model2 = load_model(os.path.join(save_path,"XNOR_network.h5"))
pred = model2.predict(x)
# Measure RMSE error. RMSE is common for regression.
score = np.sqrt(metrics.mean_squared_error(pred,y))
print(f"After load score (RMSE): {score}")
###Output
After load score (RMSE): 3.324424151302459e-07
|
26-NeuralNetworks2/26-NeuralNetworks2-solutions.ipynb | ###Markdown
Introduction to Data Science Lecture 25: Neural Networks II*COMP 5360 / MATH 4100, University of Utah, http://datasciencecourse.net/*In this lecture, we'll continue discussing Neural Networks. Recommended Reading:* A. Géron, [Hands-On Machine Learning with Scikit-Learn & TensorFlow](http://proquest.safaribooksonline.com/book/programming/9781491962282) (2017) * I. Goodfellow, Y. Bengio, and A. Courville, [Deep Learning](http://www.deeplearningbook.org/) (2016)* Y. LeCun, Y. Bengio, and G. Hinton, [Deep learning](https://www.nature.com/articles/nature14539), Nature (2015) Recap: Neural NetworksLast time, we introduced *Neural Networks* and discussed how they can be used for classification and regression.There are many different *network architectures* for Neural Networks, but our focus is on **Multi-layer Perceptrons**. Here, there is an *input layer*, typically drawn on the left hand side and an *output layer*, typically drawn on the right hand side. The middle layers are called *hidden layers*. <img src="Colored_neural_network.svg" title="https://en.wikipedia.org/wiki/Artificial_neural_network/media/File:Colored_neural_network.svg" width="300">Given a set of features $X = x^0 = \{x_1, x_2, ..., x_n\}$ and a target $y$, a neural network works as follows. Each layer applies an affine transformation and an [activation function](https://en.wikipedia.org/wiki/Activation_function) (e.g., ReLU, hyperbolic tangent, or logistic) to the output of the previous layer: $$x^{j} = f ( A^{j} x^{j-1} + b^j ). $$At the $j$-th hidden layer, the input is represented as the composition of $j$ such mappings. An additional function, *e.g.* [softmax](https://en.wikipedia.org/wiki/Softmax_function), is applied to the output layer to give the prediction, $\hat y$, for classification or regression. <img src="activationFct.png" title="see Géron, Ch. 10" width="700"> Softmax function for classificaton The *softmax function*, $\sigma:\mathbb{R}^K \to (0,1)^K$ is defined by$$\sigma(\mathbf{z})_j = \frac{e^{z_j}}{\sum_{k=1}^K e^{z_k}}\qquad \qquad \textrm{for } j=1, \ldots, K.$$Note that each component is in the range $(0,1)$ and the values sum to 1. We interpret $\sigma(\mathbf{z})_j$ as the probability that $\mathbf{z}$ is a member of class $j$. Training a neural networkNeural networks uses a loss function of the form $$Loss(\hat{y},y,W) = \frac{1}{2} \sum_{i=1}^n g(\hat{y}_i(W),y_i) + \frac{\alpha}{2} \|W\|_2^2$$Here, + $y_i$ is the label for the $i$-th example, + $\hat{y}_i(W)$ is the predicted label for the $i$-th example, + $g$ is a function that measures the error, typically $L^2$ difference for regression or cross-entropy for classification, and + $\alpha$ is a regularization parameter. Starting from initial random weights, the loss function is minimized by repeatedly updating these weights. Various **optimization methods** can be used, *e.g.*, + gradient descent method + quasi-Newton method,+ stochastic gradient descent, or + ADAM. There are various parameters associated with each method that must be tuned. **Back propagation** is a way of using the chain rule from calculus to compute the gradient of the $Loss$ function for optimization. Neural Networks in scikit-learnIn the previous lecture, we used Neural Network implementations in scikit-learn to do both classification and regression:+ [multi-layer perceptron (MLP) classifier](http://scikit-learn.org/stable/modules/generated/sklearn.neural_network.MLPClassifier.html)+ [multi-layer perceptron (MLP) regressor](http://scikit-learn.org/stable/modules/generated/sklearn.neural_network.MLPRegressor.html)However, there are several limitations to the scikit-learn implementation: - no GPU support- limited network architectures Neural networks with TensorFlowToday, we'll use [TensorFlow](https://github.com/tensorflow/tensorflow) to train a Neural Network. TensorFlow is an open-source library designed for large-scale machine learning. Installing TensorFlowInstructions for installing TensorFlow are available at [the tensorflow install page](https://www.tensorflow.org/versions/r1.0/install/).It is recommended that you use the command: ```pip install tensorflow```
###Code
import tensorflow as tf
print(tf.__version__)
# to make this notebook's output stable across runs
def reset_graph(seed=42):
tf.reset_default_graph()
tf.set_random_seed(seed)
np.random.seed(seed)
###Output
1.13.1
###Markdown
TensorFlow represents computations by connecting op (operation) nodes into a computation graph.A TensorFlow program usually has two components:+ In the *construction phase*, a computational graph is built. During this phase, no computations are performed and the variables are not yet initialized. + In the *execution phase*, the graph is evaluated, typically many times. In this phase, the each operation is given to a CPU or GPU, variables are initialized, and functions can be evaluted.
###Code
# construction phase
x = tf.Variable(3)
y = tf.Variable(4)
f = x*x*y + y + 2
# execution phase
with tf.Session() as sess: # initializes a "session"
x.initializer.run()
y.initializer.run()
print(f.eval())
# alternatively all variables cab be initialized as follows
init = tf.global_variables_initializer()
with tf.Session() as sess: # initializes a "session"
init.run() # initializes all the variables
print(f.eval())
###Output
WARNING:tensorflow:From /Applications/anaconda3/lib/python3.7/site-packages/tensorflow/python/framework/op_def_library.py:263: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.
Instructions for updating:
Colocations handled automatically by placer.
42
42
###Markdown
AutodiffTensorFlow can automatically compute the derivative of functions using [```gradients```](https://www.tensorflow.org/api_docs/python/tf/gradients).
###Code
# construction phase
x = tf.Variable(3.0)
y = tf.Variable(4.0)
f = x + 2*y*y + 2
grads = tf.gradients(f,[x,y])
# execution phase
with tf.Session() as sess:
sess.run(tf.global_variables_initializer()) # initializes all variables
print([g.eval() for g in grads])
###Output
[1.0, 16.0]
###Markdown
This is enormously helpful since training a NN requires the derivate of the loss function with respect to the parameters (and there are a lot of parameters). This is computed using backpropagation (chain rule) and TensorFlow does this work for you. **Exercise:** Use TensorFlow to compute the derivative of $f(x) = e^x$ at $x=2$.
###Code
# your code here
# Reference solution
x = tf.Variable(2.0)
f = tf.exp(x)
grads = tf.gradients(f,[x])
# execution phase
with tf.Session() as sess:
sess.run(tf.global_variables_initializer()) # initializes all variables
print(grads[0].eval())
###Output
7.389056
###Markdown
Optimization methodsTensorflow also has several built-in optimization methods.Other optimization methods in TensorFlow:+ [```tf.train.Optimizer```](https://www.tensorflow.org/api_docs/python/tf/train/Optimizer)+ [```tf.train.GradientDescentOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/GradientDescentOptimizer)+ [```tf.train.AdadeltaOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/AdadeltaOptimizer)+ [```tf.train.AdagradOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/AdagradOptimizer)+ [```tf.train.AdagradDAOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/AdagradDAOptimizer)+ [```tf.train.MomentumOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/MomentumOptimizer)+ [```tf.train.AdamOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/AdamOptimizer)+ [```tf.train.FtrlOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/FtrlOptimizer)+ [```tf.train.ProximalGradientDescentOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/ProximalGradientDescentOptimizer)+ [```tf.train.ProximalAdagradOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/ProximalAdagradOptimizer)+ [```tf.train.RMSPropOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/RMSPropOptimizer)For more information, see the [TensorFlow training webpage](https://www.tensorflow.org/api_guides/python/train). Let's see how to use the [```GradientDescentOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/GradientDescentOptimizer).
###Code
x = tf.Variable(3.0, trainable=True)
y = tf.Variable(2.0, trainable=True)
f = x*x + 100*y*y
opt = tf.train.GradientDescentOptimizer(learning_rate=5e-3).minimize(f)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for i in range(1000):
if i%100 == 0: print(sess.run([x,y,f]))
sess.run(opt)
###Output
[3.0, 2.0, 409.0]
[1.0980968, 0.0, 1.2058167]
[0.40193906, 0.0, 0.161555]
[0.14712274, 0.0, 0.0216451]
[0.053851694, 0.0, 0.0029000049]
[0.019711465, 0.0, 0.00038854184]
[0.0072150305, 0.0, 5.2056665e-05]
[0.0026409342, 0.0, 6.9745333e-06]
[0.00096666755, 0.0, 9.344461e-07]
[0.00035383157, 0.0, 1.2519678e-07]
###Markdown
Using another optimizer, such as the [```MomentumOptimizer```](https://www.tensorflow.org/api_docs/python/tf/train/MomentumOptimizer), has similiar syntax.
###Code
x = tf.Variable(3.0, trainable=True)
y = tf.Variable(2.0, trainable=True)
f = x*x + 100*y*y
opt = tf.train.MomentumOptimizer(learning_rate=1e-2,momentum=.5).minimize(f)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for i in range(1000):
if i%100 == 0: print(sess.run([x,y,f]))
sess.run(opt)
###Output
[3.0, 2.0, 409.0]
[0.043930665, 2.0290405e-15, 0.0019299033]
[0.0006126566, -1.547466e-30, 3.753481e-07]
[8.544106e-06, 0.0, 7.300175e-11]
[1.1915596e-07, 0.0, 1.4198143e-14]
[1.6617479e-09, 0.0, 2.761406e-18]
[2.3174716e-11, 0.0, 5.3706746e-22]
[3.2319424e-13, 0.0, 1.0445451e-25]
[4.5072626e-15, 0.0, 2.0315416e-29]
[6.285822e-17, 0.0, 3.951156e-33]
###Markdown
**Exercise:** Use TensorFlow to find the minimum of the [Rosenbrock function](https://en.wikipedia.org/wiki/Rosenbrock_function): $$f(x,y) = (x-1)^2 + 100*(y-x^2)^2.$$
###Code
# your code here
# Reference solution
x = tf.Variable(3.0, trainable=True)
y = tf.Variable(2.0, trainable=True)
f = tf.pow(x-1,2) + 100*tf.pow(y-tf.pow(x,2),2)
opt = tf.train.MomentumOptimizer(learning_rate=1e-4,momentum=.9).minimize(f)
# execution phase
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for i in range(10000):
if i%1000 == 0: print(sess.run([x,y,f]))
sess.run(opt)
###Output
[3.0, 2.0, 4904.0]
[0.34496623, 0.11588032, 0.43004355]
[0.7261489, 0.5260107, 0.07515866]
[0.84546393, 0.7141316, 0.023927316]
[0.9054483, 0.81943625, 0.008956054]
[0.939964, 0.8832833, 0.0036105204]
[0.96110237, 0.92355835, 0.0015155661]
[0.9744939, 0.94953483, 0.00065163325]
[0.9831504, 0.96651673, 0.0002843708]
[0.9888154, 0.97771096, 0.0001252972]
###Markdown
Classifying the MNIST handwritten digit datasetWe now use TensorFlow to classify the handwritten digits in the MNIST dataset. Using plain TensorFlowWe'll first follow [Géron, Ch. 10](https://github.com/ageron/handson-ml/blob/master/10_introduction_to_artificial_neural_networks.ipynb) to build a NN using plain TensorFlow. Construction phase+ We specify the number of inputs and outputs and the size of each layer. Here the images are 28x28 and there are 10 classes (each corresponding to a digit). We'll choose 2 hidden layers, with 300 and 100 neurons respectively. + Placeholder nodes are used to represent the training data and targets. We use the ```None``` keyword to leave the shape (of the training batch) unspecified. + We add layers to the NN using the ```layers.dense()``` function. In each case, we specify the input, and the size of the layer. We also specify the activation function used in each layer. Here, we choose the ReLU function. + We specify that the output of the NN will be a softmax function. The loss function is cross entropy. + We then specify that we'll use the [GradientDescentOptimizer](https://www.tensorflow.org/api_docs/python/tf/train/GradientDescentOptimizer) with a learning rate of 0.01. + Finally, we specify how the model will be evaluated. The [```in_top_k```](https://www.tensorflow.org/api_docs/python/tf/nn/in_top_k) function checks to see if the targets are in the top k predictions. We then initialize all of the variables and create an object to save the model using the [```saver()```](https://www.tensorflow.org/programmers_guide/saved_model) function. Execution phaseAt each *epoch*, the code breaks the training batch into mini-batches of size 50. Cycling through the mini-batches, it uses gradient descent to train the NN. The accuracy for both the training and test datasets are evaluated.
###Code
import tensorflow as tf
import numpy as np
from sklearn.metrics import confusion_matrix
# to make this notebook's output stable across runs
def reset_graph(seed=42):
tf.reset_default_graph()
tf.set_random_seed(seed)
np.random.seed(seed)
# load the data
(X_train, y_train), (X_test, y_test) = tf.keras.datasets.mnist.load_data()
X_train = X_train.astype(np.float32).reshape(-1, 28*28) / 255.0
X_test = X_test.astype(np.float32).reshape(-1, 28*28) / 255.0
y_train = y_train.astype(np.int32)
y_test = y_test.astype(np.int32)
# helper code
def shuffle_batch(X, y, batch_size):
rnd_idx = np.random.permutation(len(X))
n_batches = len(X) // batch_size
for batch_idx in np.array_split(rnd_idx, n_batches):
X_batch, y_batch = X[batch_idx], y[batch_idx]
yield X_batch, y_batch
# construction phase
n_inputs = 28*28 # MNIST
n_hidden1 = 300
n_hidden2 = 100
n_outputs = 10
reset_graph()
X = tf.placeholder(tf.float32, shape=(None, n_inputs), name="X")
y = tf.placeholder(tf.int32, shape=(None), name="y")
with tf.name_scope("dnn"):
hidden1 = tf.layers.dense(X, n_hidden1, name="hidden1",activation=tf.nn.relu)
hidden2 = tf.layers.dense(hidden1, n_hidden2, name="hidden2",activation=tf.nn.relu)
logits = tf.layers.dense(hidden2, n_outputs, name="outputs")
#y_proba = tf.nn.softmax(logits)
with tf.name_scope("loss"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)
loss = tf.reduce_mean(xentropy, name="loss")
learning_rate = 0.01
with tf.name_scope("train"):
optimizer = tf.train.GradientDescentOptimizer(learning_rate)
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))
# execution phase
init = tf.global_variables_initializer()
saver = tf.train.Saver()
n_epochs = 10
#n_batches = 50
batch_size = 50
with tf.Session() as sess:
init.run()
for epoch in range(n_epochs):
for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
acc_batch = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_valid = accuracy.eval(feed_dict={X: X_test, y: y_test})
print(epoch, "Batch accuracy:", acc_batch, "Validation accuracy:", acc_valid)
save_path = saver.save(sess, "./my_model_final.ckpt")
###Output
0 Batch accuracy: 0.9 Validation accuracy: 0.9055
1 Batch accuracy: 0.9 Validation accuracy: 0.9208
2 Batch accuracy: 0.94 Validation accuracy: 0.9331
3 Batch accuracy: 0.94 Validation accuracy: 0.9406
4 Batch accuracy: 1.0 Validation accuracy: 0.9444
5 Batch accuracy: 0.98 Validation accuracy: 0.9481
6 Batch accuracy: 0.98 Validation accuracy: 0.9537
7 Batch accuracy: 0.98 Validation accuracy: 0.9566
8 Batch accuracy: 0.98 Validation accuracy: 0.9591
9 Batch accuracy: 1.0 Validation accuracy: 0.9597
###Markdown
Since the NN has been saved, we can use it for classification using the [```saver.restore```](https://www.tensorflow.org/programmers_guide/saved_model) function. We can also print the confusion matrix using [```confusion_matrix```](https://www.tensorflow.org/api_docs/python/tf/confusion_matrix).
###Code
with tf.Session() as sess:
saver.restore(sess, save_path)
Z = logits.eval(feed_dict={X: X_test})
y_pred = np.argmax(Z, axis=1)
print(confusion_matrix(y_test,y_pred))
###Output
WARNING:tensorflow:From /Applications/anaconda3/lib/python3.7/site-packages/tensorflow/python/training/saver.py:1266: checkpoint_exists (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.
Instructions for updating:
Use standard file APIs to check for files with this prefix.
INFO:tensorflow:Restoring parameters from ./my_model_final.ckpt
[[ 969 0 1 1 0 3 1 2 2 1]
[ 0 1115 2 2 0 1 4 2 9 0]
[ 7 1 980 12 5 0 6 9 11 1]
[ 1 0 2 984 0 1 0 10 7 5]
[ 1 0 3 1 930 0 7 1 5 34]
[ 10 2 1 25 3 820 10 1 14 6]
[ 8 3 0 2 9 6 925 0 5 0]
[ 0 10 12 4 2 0 0 984 2 14]
[ 3 2 3 13 4 1 8 8 929 3]
[ 5 7 0 13 11 2 1 5 4 961]]
###Markdown
Using TensorFlow's Keras API Next, we'll use TensorFlow's Keras API to build a NN for the MNIST dataset. [Keras](https://keras.io/) is a high-level neural networks API, written in Python and capable of running on top of TensorFlow, CNTK, or Theano. We'll use it with TensorFlow.
###Code
import tensorflow as tf
import numpy as np
from sklearn.metrics import confusion_matrix
(X_train, y_train),(X_test, y_test) = tf.keras.datasets.mnist.load_data()
X_train, X_test = X_train / 255.0, X_test / 255.0
# set the model
model = tf.keras.models.Sequential([
tf.keras.layers.Flatten(input_shape=(28, 28)),
tf.keras.layers.Dense(512, activation=tf.nn.relu),
tf.keras.layers.Dropout(rate=0.2),
tf.keras.layers.Dense(10, activation=tf.nn.softmax)
])
# specifiy optimizer
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# train the model
model.fit(X_train, y_train, epochs=5)
score = model.evaluate(X_test, y_test)
names = model.metrics_names
for ii in np.arange(len(names)):
print(names[ii],score[ii])
model.summary()
y_pred = np.argmax(model.predict(X_test), axis=1)
print(confusion_matrix(y_test,y_pred))
###Output
[[ 974 1 0 0 0 0 3 1 0 1]
[ 0 1124 3 2 0 1 2 0 3 0]
[ 5 0 1018 2 1 0 1 1 3 1]
[ 0 1 3 997 0 0 0 2 2 5]
[ 1 0 1 1 971 0 3 0 0 5]
[ 2 0 0 20 3 852 5 1 6 3]
[ 2 3 0 1 7 3 940 0 2 0]
[ 3 6 10 6 1 0 0 984 4 14]
[ 4 0 7 6 1 3 3 3 945 2]
[ 2 2 0 4 12 1 0 1 1 986]]
###Markdown
Using a pre-trained networkThere are many examples of pre-trained NN that can be accessed [here](https://www.tensorflow.org/api_docs/python/tf/keras/applications). These NN are very large, having been trained on giant computers using massive datasets. It can be very useful to initialize a NN using one of these. This is called [transfer learning](https://en.wikipedia.org/wiki/Transfer_learning). We'll use a NN that was pretrained for image recognition. This NN was trained on the [ImageNet](http://www.image-net.org/) project, which contains > 14 million images belonging to > 20,000 classes (synsets).
###Code
import tensorflow as tf
import numpy as np
from tensorflow.keras.preprocessing import image
from tensorflow.keras.applications import vgg16
vgg_model = tf.keras.applications.VGG16(weights='imagenet',include_top=True)
vgg_model.summary()
img_path = 'images/scout1.jpeg'
img = image.load_img(img_path, target_size=(224, 224))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
x = vgg16.preprocess_input(x)
preds = vgg_model.predict(x)
print('Predicted:', vgg16.decode_predictions(preds, top=5)[0])
###Output
Predicted: [('n02098105', 'soft-coated_wheaten_terrier', 0.3554158), ('n02105641', 'Old_English_sheepdog', 0.23714595), ('n02095314', 'wire-haired_fox_terrier', 0.13490717), ('n02091635', 'otterhound', 0.0611032), ('n02093991', 'Irish_terrier', 0.052789364)]
###Markdown
**Exercise:** Repeat the above steps for an image of your own.**Exercise:** There are several [other pre-trained networks in Keras](https://github.com/keras-team/keras-applications). Try these!
###Code
# your code here
###Output
_____no_output_____ |
Python Tutorial PyTorch/assignments/03_assignment_cifar10_feedforward.ipynb | ###Markdown
Classifying images of everyday objects using a neural networkThe ability to try many different neural network architectures to address a problem is what makes deep learning really powerful, especially compared to shallow learning techniques like linear regression, logistic regression etc. In this assignment, you will:1. Explore the CIFAR10 dataset: https://www.cs.toronto.edu/~kriz/cifar.html2. Set up a training pipeline to train a neural network on a GPU2. Experiment with different network architectures & hyperparametersAs you go through this notebook, you will find a **???** in certain places. Your job is to replace the **???** with appropriate code or values, to ensure that the notebook runs properly end-to-end. Try to experiment with different network structures and hypeparameters to get the lowest loss.You might find these notebooks useful for reference, as you work through this notebook:- https://jovian.ml/aakashns/04-feedforward-nn- https://jovian.ml/aakashns/fashion-feedforward-minimal
###Code
# Uncomment and run the commands below if imports fail
# !conda install numpy pandas pytorch torchvision cpuonly -c pytorch -y
# !pip install matplotlib --upgrade --quiet
import torch
import torchvision
import numpy as np
import matplotlib.pyplot as plt
import torch.nn as nn
import torch.nn.functional as F
from torchvision.datasets import CIFAR10
from torchvision.transforms import ToTensor
from torchvision.utils import make_grid
from torch.utils.data.dataloader import DataLoader
from torch.utils.data import random_split
%matplotlib inline
# Project name used for jovian.commit
project_name = '03-assignment-cifar10-feedforward'
###Output
_____no_output_____
###Markdown
Exploring the CIFAR10 dataset
###Code
dataset = CIFAR10(root='data/', download=True, transform=ToTensor())
test_dataset = CIFAR10(root='data/', train=False, transform=ToTensor())
###Output
Downloading https://www.cs.toronto.edu/~kriz/cifar-10-python.tar.gz to data/cifar-10-python.tar.gz
###Markdown
**Q: How many images does the training dataset contain?**
###Code
dataset_size = len(dataset)
print('The training dataset contains ' + str(dataset_size) + ' images.')
###Output
The training dataset contains 50000 images.
###Markdown
**Q: How many images does the testing dataset contain?**
###Code
test_dataset_size = len(test_dataset)
print('The training dataset contains ' + str(test_dataset_size) + ' images.')
###Output
The training dataset contains 10000 images.
###Markdown
**Q: How many output classes does the dataset contain? Can you list them?**Hint: Use `dataset.classes`
###Code
classes = dataset.classes
print('The classes in the dataset are: ')
print(classes)
num_classes = len(dataset.classes)
print('The number of classes in the dataset is: ', num_classes)
###Output
The number of classes in the dataset is: 10
###Markdown
**Q: What is the shape of an image tensor from the dataset?**
###Code
img, label = dataset[0]
img_shape = img.shape
print('The shape of an image tensor is: ', img_shape)
###Output
The shape of an image tensor is: torch.Size([3, 32, 32])
###Markdown
Note that this dataset consists of 3-channel color images (RGB). Let us look at a sample image from the dataset. `matplotlib` expects channels to be the last dimension of the image tensors (whereas in PyTorch they are the first dimension), so we'll the `.permute` tensor method to shift channels to the last dimension. Let's also print the label for the image.
###Code
img, label = dataset[0]
plt.imshow(img.permute((1, 2, 0)))
print('Label (numeric):', label)
print('Label (textual):', classes[label])
###Output
Label (numeric): 6
Label (textual): frog
###Markdown
**(Optional) Q: Can you determine the number of images belonging to each class?**Hint: Loop through the dataset.
###Code
counter = dict.fromkeys(classes, 0)
for _, l in dataset:
counter[classes[l]] += 1
counter
counter.values()
###Output
_____no_output_____
###Markdown
Let's save our work to Jovian, before continuing.
###Code
!pip install jovian --upgrade --quiet
import jovian
jovian.commit(project=project_name, environment=None)
###Output
_____no_output_____
###Markdown
Preparing the data for trainingWe'll use a validation set with 5000 images (10% of the dataset). To ensure we get the same validation set each time, we'll set PyTorch's random number generator to a seed value of 43.
###Code
torch.manual_seed(43)
val_size = 5000
train_size = len(dataset) - val_size
###Output
_____no_output_____
###Markdown
Let's use the `random_split` method to create the training & validation sets
###Code
train_ds, val_ds = random_split(dataset, [train_size, val_size])
len(train_ds), len(val_ds)
###Output
_____no_output_____
###Markdown
We can now create data loaders to load the data in batches.
###Code
batch_size=128
train_loader = DataLoader(train_ds, batch_size, shuffle=True, num_workers=4, pin_memory=True)
val_loader = DataLoader(val_ds, batch_size*2, num_workers=4, pin_memory=True)
test_loader = DataLoader(test_dataset, batch_size*2, num_workers=4, pin_memory=True)
###Output
_____no_output_____
###Markdown
Let's visualize a batch of data using the `make_grid` helper function from Torchvision.
###Code
for images, _ in train_loader:
print('images.shape:', images.shape)
plt.figure(figsize=(16,8))
plt.axis('off')
plt.imshow(make_grid(images, nrow=16).permute((1, 2, 0)))
break
###Output
images.shape: torch.Size([128, 3, 32, 32])
###Markdown
Can you label all the images by looking at them? Trying to label a random sample of the data manually is a good way to estimate the difficulty of the problem, and identify errors in labeling, if any. Base Model class & Training on GPULet's create a base model class, which contains everything except the model architecture i.e. it wil not contain the `__init__` and `__forward__` methods. We will later extend this class to try out different architectures. In fact, you can extend this model to solve any image classification problem.
###Code
def accuracy(outputs, labels):
_, preds = torch.max(outputs, dim=1)
return torch.tensor(torch.sum(preds == labels).item() / len(preds))
class ImageClassificationBase(nn.Module):
def training_step(self, batch):
images, labels = batch
out = self(images) # Generate predictions
loss = F.cross_entropy(out, labels) # Calculate loss
return loss
def validation_step(self, batch):
images, labels = batch
out = self(images) # Generate predictions
loss = F.cross_entropy(out, labels) # Calculate loss
acc = accuracy(out, labels) # Calculate accuracy
return {'val_loss': loss.detach(), 'val_acc': acc}
def validation_epoch_end(self, outputs):
batch_losses = [x['val_loss'] for x in outputs]
epoch_loss = torch.stack(batch_losses).mean() # Combine losses
batch_accs = [x['val_acc'] for x in outputs]
epoch_acc = torch.stack(batch_accs).mean() # Combine accuracies
return {'val_loss': epoch_loss.item(), 'val_acc': epoch_acc.item()}
def epoch_end(self, epoch, result):
print("Epoch [{}], val_loss: {:.4f}, val_acc: {:.4f}".format(epoch, result['val_loss'], result['val_acc']))
###Output
_____no_output_____
###Markdown
We can also use the exact same training loop as before. I hope you're starting to see the benefits of refactoring our code into reusable functions.
###Code
def evaluate(model, val_loader):
outputs = [model.validation_step(batch) for batch in val_loader]
return model.validation_epoch_end(outputs)
def fit(epochs, lr, model, train_loader, val_loader, opt_func=torch.optim.SGD):
history = []
optimizer = opt_func(model.parameters(), lr)
for epoch in range(epochs):
# Training Phase
for batch in train_loader:
loss = model.training_step(batch)
loss.backward()
optimizer.step()
optimizer.zero_grad()
# Validation phase
result = evaluate(model, val_loader)
model.epoch_end(epoch, result)
history.append(result)
return history
###Output
_____no_output_____
###Markdown
Finally, let's also define some utilities for moving out data & labels to the GPU, if one is available.
###Code
torch.cuda.is_available()
def get_default_device():
"""Pick GPU if available, else CPU"""
if torch.cuda.is_available():
return torch.device('cuda')
else:
return torch.device('cpu')
device = get_default_device()
device
def to_device(data, device):
"""Move tensor(s) to chosen device"""
if isinstance(data, (list,tuple)):
return [to_device(x, device) for x in data]
return data.to(device, non_blocking=True)
class DeviceDataLoader():
"""Wrap a dataloader to move data to a device"""
def __init__(self, dl, device):
self.dl = dl
self.device = device
def __iter__(self):
"""Yield a batch of data after moving it to device"""
for b in self.dl:
yield to_device(b, self.device)
def __len__(self):
"""Number of batches"""
return len(self.dl)
###Output
_____no_output_____
###Markdown
Let us also define a couple of helper functions for plotting the losses & accuracies.
###Code
def plot_losses(history):
losses = [x['val_loss'] for x in history]
plt.plot(losses, '-x')
plt.xlabel('epoch')
plt.ylabel('loss')
plt.title('Loss vs. No. of epochs');
def plot_accuracies(history):
accuracies = [x['val_acc'] for x in history]
plt.plot(accuracies, '-x')
plt.xlabel('epoch')
plt.ylabel('accuracy')
plt.title('Accuracy vs. No. of epochs');
###Output
_____no_output_____
###Markdown
Let's move our data loaders to the appropriate device.
###Code
train_loader = DeviceDataLoader(train_loader, device)
val_loader = DeviceDataLoader(val_loader, device)
test_loader = DeviceDataLoader(test_loader, device)
###Output
_____no_output_____
###Markdown
Training the modelWe will make several attempts at training the model. Each time, try a different architecture and a different set of learning rates. Here are some ideas to try:- Increase or decrease the number of hidden layers- Increase of decrease the size of each hidden layer- Try different activation functions- Try training for different number of epochs- Try different learning rates in every epochWhat's the highest validation accuracy you can get to? **Can you get to 50% accuracy? What about 60%?**
###Code
input_size = 3*32*32
hidden_layer1 = 2048
hidden_layer2 = 1024
hidden_layer3 = 512
hidden_layer4 = 256
hidden_layer5 = 128
hidden_layer6 = 64
hidden_layer7 = 32
output_size = 10
###Output
_____no_output_____
###Markdown
**Q: Extend the `ImageClassificationBase` class to complete the model definition.**Hint: Define the `__init__` and `forward` methods.
###Code
class CIFAR10Model(ImageClassificationBase):
def __init__(self):
super().__init__()
# Input Layer to hidden layer 1
self.linear_input = nn.Linear(input_size, hidden_layer1)
# Hidden layer 1 to hidden layer 2
self.linear_hidden1 = nn.Linear(hidden_layer1, hidden_layer2)
# Hidden layer 2 to hidden layer 3
self.linear_hidden2 = nn.Linear(hidden_layer2, hidden_layer3)
# Hidden layer 3 to hidden layer 4
self.linear_hidden3 = nn.Linear(hidden_layer3, hidden_layer4)
# Hidden layer 4 to hidden layer 5
self.linear_hidden4 = nn.Linear(hidden_layer4, hidden_layer5)
# Hidden layer 5 to hidden layer 6
self.linear_hidden5 = nn.Linear(hidden_layer5, hidden_layer6)
# Hidden layer 6 to Output layer
self.linear_hidden6 = nn.Linear(hidden_layer6, hidden_layer7)
# Hidden layer 7 to Output layer
self.linear_output = nn.Linear(hidden_layer7, output_size)
def forward(self, xb):
# Flatten images into vectors
out = xb.view(xb.size(0), -1)
# Apply input layers
out = self.linear_input(out)
# Apply activation function
out = F.relu(out)
# Get intermediate outputs using hidden layer 1
out = self.linear_hidden1(out)
# Apply activation function
out = F.relu(out)
# Get intermediate outputs using hidden layer 2
out = self.linear_hidden2(out)
# Apply activation function
out = F.relu(out)
# Get intermediate outputs using hidden layer 3
out = self.linear_hidden3(out)
# Apply activation function
out = F.relu(out)
# Get intermediate outputs using hidden layer 4
out = self.linear_hidden4(out)
# Apply activation function
out = F.relu(out)
# Get intermediate outputs using hidden layer 5
out = self.linear_hidden5(out)
# Apply activation function
out = F.relu(out)
out = self.linear_hidden6(out)
# Apply activation function
out = F.relu(out)
# Get predictions using output layer
out = self.linear_output(out)
return out
###Output
_____no_output_____
###Markdown
You can now instantiate the model, and move it the appropriate device.
###Code
model = to_device(CIFAR10Model(), device)
model
###Output
_____no_output_____
###Markdown
Before you train the model, it's a good idea to check the validation loss & accuracy with the initial set of weights.
###Code
history = [evaluate(model, val_loader)]
history
###Output
_____no_output_____
###Markdown
**Q: Train the model using the `fit` function to reduce the validation loss & improve accuracy.**Leverage the interactive nature of Jupyter to train the model in multiple phases, adjusting the no. of epochs & learning rate each time based on the result of the previous training phase.
###Code
history += fit(5, 0.5, model, train_loader, val_loader)
history += fit(5, 0.4, model, train_loader, val_loader)
history += fit(10, 0.2, model, train_loader, val_loader)
history += fit(10, 0.1, model, train_loader, val_loader)
###Output
Epoch [0], val_loss: 1.4829, val_acc: 0.4864
Epoch [1], val_loss: 1.5539, val_acc: 0.4620
Epoch [2], val_loss: 1.4973, val_acc: 0.4888
Epoch [3], val_loss: 1.5712, val_acc: 0.4792
Epoch [4], val_loss: 1.5230, val_acc: 0.4898
Epoch [5], val_loss: 1.4988, val_acc: 0.4951
Epoch [6], val_loss: 1.4691, val_acc: 0.5043
Epoch [7], val_loss: 1.5509, val_acc: 0.4958
Epoch [8], val_loss: 1.5826, val_acc: 0.4837
Epoch [9], val_loss: 1.6598, val_acc: 0.4744
###Markdown
Plot the losses and the accuracies to check if you're starting to hit the limits of how well your model can perform on this dataset. You can train some more if you can see the scope for further improvement.
###Code
plot_losses(history)
plot_accuracies(history)
###Output
_____no_output_____
###Markdown
Finally, evaluate the model on the test dataset report its final performance.
###Code
validation = evaluate(model, test_loader)
validation
###Output
_____no_output_____
###Markdown
Are you happy with the accuracy? Record your results by completing the section below, then you can come back and try a different architecture & hyperparameters. Recoding your resultsAs your perform multiple experiments, it's important to record the results in a systematic fashion, so that you can review them later and identify the best approaches that you might want to reproduce or build upon later. **Q: Describe the model's architecture with a short summary.**E.g. `"3 layers (16,32,10)"` (16, 32 and 10 represent output sizes of each layer)
###Code
arch = [input_size, hidden_layer1, hidden_layer2, hidden_layer3, hidden_layer4, hidden_layer5, hidden_layer6, hidden_layer7, output_size]
###Output
_____no_output_____
###Markdown
**Q: Provide the list of learning rates used while training.**
###Code
lrs = [0.5, 0.4, 0.2, 0.1]
###Output
_____no_output_____
###Markdown
**Q: Provide the list of no. of epochs used while training.**
###Code
epochs = [5, 5, 10, 10]
###Output
_____no_output_____
###Markdown
**Q: What were the final test accuracy & test loss?**
###Code
test_acc = validation['val_acc']
test_loss = validation['val_loss']
test_acc, test_loss
###Output
_____no_output_____
###Markdown
Finally, let's save the trained model weights to disk, so we can use this model later.
###Code
torch.save(model.state_dict(), '03-assignment-cifar10-feedforward.pth')
###Output
_____no_output_____
###Markdown
The `jovian` library provides some utility functions to keep your work organized. With every version of your notebok, you can attach some hyperparameters and metrics from your experiment.
###Code
# Clear previously recorded hyperparams & metrics
jovian.reset()
jovian.log_hyperparams(arch=arch,
lrs=lrs,
epochs=epochs)
jovian.log_metrics(test_loss=test_loss, test_acc=test_acc)
###Output
_____no_output_____
###Markdown
Finally, we can commit the notebook to Jovian, attaching the hypeparameters, metrics and the trained model weights.
###Code
jovian.commit(project=project_name, outputs=['cifar10-feedforward.pth'], environment=None)
###Output
_____no_output_____
###Markdown
Once committed, you can find the recorded metrics & hyperprameters in the "Records" tab on Jovian. You can find the saved model weights in the "Files" tab. Continued experimentationNow go back up to the **"Training the model"** section, and try another network architecture with a different set of hyperparameters. As you try different experiments, you will start to build an undestanding of how the different architectures & hyperparameters affect the final result. Don't worry if you can't get to very high accuracy, we'll make some fundamental changes to our model in the next lecture.Once you have tried multiple experiments, you can compare your results using the **"Compare"** button on Jovian. I could not use the jovian interphase to commit my notebook, but I tracked my changes and are displeyed in the image bellow:neural network comparison (Optional) Write a blog postWriting a blog post is the best way to further improve your understanding of deep learning & model training, because it forces you to articulate your thoughts clearly. Here'are some ideas for a blog post:- Report the results given by different architectures on the CIFAR10 dataset- Apply this training pipeline to a different dataset (it doesn't have to be images, or a classification problem) - Improve upon your model from Assignment 2 using a feedfoward neural network, and write a sequel to your previous blog post- Share some Strategies for picking good hyperparameters for deep learning- Present a summary of the different steps involved in training a deep learning model with PyTorch- Implement the same model using a different deep learning library e.g. Keras ( https://keras.io/ ), and present a comparision.
###Code
###Output
_____no_output_____ |
Intermediate Machine Learning/6 XGBoost/xgboost.ipynb | ###Markdown
In this tutorial, you will learn how to build and optimize models with **gradient boosting**. This method dominates many Kaggle competitions and achieves state-of-the-art results on a variety of datasets. IntroductionFor much of this course, you have made predictions with the random forest method, which achieves better performance than a single decision tree simply by averaging the predictions of many decision trees.We refer to the random forest method as an "ensemble method". By definition, **ensemble methods** combine the predictions of several models (e.g., several trees, in the case of random forests). Next, we'll learn about another ensemble method called gradient boosting. Gradient Boosting**Gradient boosting** is a method that goes through cycles to iteratively add models into an ensemble. It begins by initializing the ensemble with a single model, whose predictions can be pretty naive. (Even if its predictions are wildly inaccurate, subsequent additions to the ensemble will address those errors.)Then, we start the cycle:- First, we use the current ensemble to generate predictions for each observation in the dataset. To make a prediction, we add the predictions from all models in the ensemble. - These predictions are used to calculate a loss function (like [mean squared error](https://en.wikipedia.org/wiki/Mean_squared_error), for instance).- Then, we use the loss function to fit a new model that will be added to the ensemble. Specifically, we determine model parameters so that adding this new model to the ensemble will reduce the loss. (*Side note: The "gradient" in "gradient boosting" refers to the fact that we'll use [gradient descent](https://en.wikipedia.org/wiki/Gradient_descent) on the loss function to determine the parameters in this new model.*)- Finally, we add the new model to ensemble, and ...- ... repeat! ExampleWe begin by loading the training and validation data in `X_train`, `X_valid`, `y_train`, and `y_valid`.
###Code
import pandas as pd
from sklearn.model_selection import train_test_split
# Read the data
data = pd.read_csv('../input/melbourne-housing-snapshot/melb_data.csv')
# Select subset of predictors
cols_to_use = ['Rooms', 'Distance', 'Landsize', 'BuildingArea', 'YearBuilt']
X = data[cols_to_use]
# Select target
y = data.Price
# Separate data into training and validation sets
X_train, X_valid, y_train, y_valid = train_test_split(X, y)
###Output
_____no_output_____
###Markdown
In this example, you'll work with the XGBoost library. **XGBoost** stands for **extreme gradient boosting**, which is an implementation of gradient boosting with several additional features focused on performance and speed. (_Scikit-learn has another version of gradient boosting, but XGBoost has some technical advantages._) In the next code cell, we import the scikit-learn API for XGBoost ([`xgboost.XGBRegressor`](https://xgboost.readthedocs.io/en/latest/python/python_api.htmlmodule-xgboost.sklearn)). This allows us to build and fit a model just as we would in scikit-learn. As you'll see in the output, the `XGBRegressor` class has many tunable parameters -- you'll learn about those soon!
###Code
from xgboost import XGBRegressor
my_model = XGBRegressor()
my_model.fit(X_train, y_train)
###Output
_____no_output_____
###Markdown
We also make predictions and evaluate the model.
###Code
from sklearn.metrics import mean_absolute_error
predictions = my_model.predict(X_valid)
print("Mean Absolute Error: " + str(mean_absolute_error(predictions, y_valid)))
###Output
Mean Absolute Error: 239960.14714193667
###Markdown
Parameter TuningXGBoost has a few parameters that can dramatically affect accuracy and training speed. The first parameters you should understand are: `n_estimators``n_estimators` specifies how many times to go through the modeling cycle described above. It is equal to the number of models that we include in the ensemble. - Too _low_ a value causes _underfitting_, which leads to inaccurate predictions on both training data and test data. - Too _high_ a value causes _overfitting_, which causes accurate predictions on training data, but inaccurate predictions on test data (_which is what we care about_). Typical values range from 100-1000, though this depends a lot on the `learning_rate` parameter discussed below.Here is the code to set the number of models in the ensemble:
###Code
my_model = XGBRegressor(n_estimators=500)
my_model.fit(X_train, y_train)
###Output
_____no_output_____
###Markdown
`early_stopping_rounds``early_stopping_rounds` offers a way to automatically find the ideal value for `n_estimators`. Early stopping causes the model to stop iterating when the validation score stops improving, even if we aren't at the hard stop for `n_estimators`. It's smart to set a high value for `n_estimators` and then use `early_stopping_rounds` to find the optimal time to stop iterating.Since random chance sometimes causes a single round where validation scores don't improve, you need to specify a number for how many rounds of straight deterioration to allow before stopping. Setting `early_stopping_rounds=5` is a reasonable choice. In this case, we stop after 5 straight rounds of deteriorating validation scores.When using `early_stopping_rounds`, you also need to set aside some data for calculating the validation scores - this is done by setting the `eval_set` parameter. We can modify the example above to include early stopping:
###Code
my_model = XGBRegressor(n_estimators=500)
my_model.fit(X_train, y_train,
early_stopping_rounds=5,
eval_set=[(X_valid, y_valid)],
verbose=False)
###Output
_____no_output_____
###Markdown
If you later want to fit a model with all of your data, set `n_estimators` to whatever value you found to be optimal when run with early stopping. `learning_rate`Instead of getting predictions by simply adding up the predictions from each component model, we can multiply the predictions from each model by a small number (known as the **learning rate**) before adding them in. This means each tree we add to the ensemble helps us less. So, we can set a higher value for `n_estimators` without overfitting. If we use early stopping, the appropriate number of trees will be determined automatically.In general, a small learning rate and large number of estimators will yield more accurate XGBoost models, though it will also take the model longer to train since it does more iterations through the cycle. As default, XGBoost sets `learning_rate=0.1`.Modifying the example above to change the learning rate yields the following code:
###Code
my_model = XGBRegressor(n_estimators=1000, learning_rate=0.05)
my_model.fit(X_train, y_train,
early_stopping_rounds=5,
eval_set=[(X_valid, y_valid)],
verbose=False)
###Output
_____no_output_____
###Markdown
`n_jobs`On larger datasets where runtime is a consideration, you can use parallelism to build your models faster. It's common to set the parameter `n_jobs` equal to the number of cores on your machine. On smaller datasets, this won't help. The resulting model won't be any better, so micro-optimizing for fitting time is typically nothing but a distraction. But, it's useful in large datasets where you would otherwise spend a long time waiting during the `fit` command.Here's the modified example:
###Code
my_model = XGBRegressor(n_estimators=1000, learning_rate=0.05, n_jobs=4)
my_model.fit(X_train, y_train,
early_stopping_rounds=5,
eval_set=[(X_valid, y_valid)],
verbose=False)
###Output
_____no_output_____ |
notebooks/DynamicInvariants.ipynb | ###Markdown
Mining Function SpecificationsWhen testing a program, one not only needs to cover its several behaviors; one also needs to _check_ whether the result is as expected. In this chapter, we introduce a technique that allows us to _mine_ function specifications from a set of given executions, resulting in abstract and formal _descriptions_ of what the function expects and what it delivers. These so-called _dynamic invariants_ produce pre- and post-conditions over function arguments and variables from a set of executions. They are useful in a variety of contexts:* Dynamic invariants provide important information for [symbolic fuzzing](SymbolicFuzzer.ipynb), such as types and ranges of function arguments.* Dynamic invariants provide pre- and postconditions for formal program proofs and verification.* Dynamic invariants provide a large number of assertions that can check whether function behavior has changed* Checks provided by dynamic invariants can be very useful as _oracles_ for checking the effects of generated testsTraditionally, dynamic invariants are dependent on the executions they are derived from. However, when paired with comprehensive test generators, they quickly become very precise, as we show in this chapter. **Prerequisites*** You should be familiar with tracing program executions, as in the [chapter on coverage](Coverage.ipynb).* Later in this section, we access the internal _abstract syntax tree_ representations of Python programs and transform them, as in the [chapter on information flow](InformationFlow.ipynb).
###Code
import bookutils
import Coverage
import Intro_Testing
###Output
_____no_output_____
###Markdown
SynopsisTo [use the code provided in this chapter](Importing.ipynb), write```python>>> from fuzzingbook.DynamicInvariants import ```and then make use of the following features.This chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:```python>>> def sum2(a, b):>>> return a + b>>> with TypeAnnotator() as type_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.```python>>> print(type_annotator.typed_functions())def sum2(a: int, b: int) ->int: return a + b```The invariant annotator works in a similar fashion:```python>>> with InvariantAnnotator() as inv_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.```python>>> print(inv_annotator.functions_with_invariants())@precondition(lambda a, b: isinstance(a, int), doc='isinstance(a, int)')@precondition(lambda a, b: isinstance(b, int), doc='isinstance(b, int)')@postcondition(lambda return_value, a, b: a == return_value - b, doc='a == return_value - b')@postcondition(lambda return_value, a, b: b == return_value - a, doc='b == return_value - a')@postcondition(lambda return_value, a, b: isinstance(return_value, int), doc='isinstance(return_value, int)')@postcondition(lambda return_value, a, b: return_value == a + b, doc='return_value == a + b')@postcondition(lambda return_value, a, b: return_value == b + a, doc='return_value == b + a')def sum2(a, b): return a + b```Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs) as well as for all kinds of _symbolic code analyses_. The chapter gives details on how to customize the properties checked for. Specifications and AssertionsWhen implementing a function or program, one usually works against a _specification_ – a set of documented requirements to be satisfied by the code. Such specifications can come in natural language. A formal specification, however, allows the computer to check whether the specification is satisfied.In the [introduction to testing](Intro_Testing.ipynb), we have seen how _preconditions_ and _postconditions_ can describe what a function does. Consider the following (simple) square root function:
###Code
def my_sqrt(x):
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
The assertion `assert p` checks the condition `p`; if it does not hold, execution is aborted. Here, the actual body is not yet written; we use the assertions as a specification of what `my_sqrt()` _expects_, and what it _delivers_.The topmost assertion is the _precondition_, stating the requirements on the function arguments. The assertion at the end is the _postcondition_, stating the properties of the function result (including its relationship with the original arguments). Using these pre- and postconditions as a specification, we can now go and implement a square root function that satisfies them. Once implemented, we can have the assertions check at runtime whether `my_sqrt()` works as expected; a [symbolic](SymbolicFuzzer.ipynb) or [concolic](ConcolicFuzzer.ipynb) test generator will even specifically try to find inputs where the assertions do _not_ hold. (An assertion can be seen as a conditional branch towards aborting the execution, and any technique that tries to cover all code branches will also try to invalidate as many assertions as possible.) However, not every piece of code is developed with explicit specifications in the first place; let alone does most code comes with formal pre- and post-conditions. (Just take a look at the chapters in this book.) This is a pity: As Ken Thompson famously said, "Without specifications, there are no bugs – only surprises". It is also a problem for testing, since, of course, testing needs some specification to test against. This raises the interesting question: Can we somehow _retrofit_ existing code with "specifications" that properly describe their behavior, allowing developers to simply _check_ them rather than having to write them from scratch? This is what we do in this chapter. Why Generic Error Checking is Not EnoughBefore we go into _mining_ specifications, let us first discuss why it could be useful to _have_ them. As a motivating example, consider the full implementation of `my_sqrt()` from the [introduction to testing](Intro_Testing.ipynb):
###Code
import bookutils
def my_sqrt(x):
"""Computes the square root of x, using the Newton-Raphson method"""
approx = None
guess = x / 2
while approx != guess:
approx = guess
guess = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
`my_sqrt()` does not come with any functionality that would check types or values. Hence, it is easy for callers to make mistakes when calling `my_sqrt()`:
###Code
from ExpectError import ExpectError, ExpectTimeout
with ExpectError():
my_sqrt("foo")
with ExpectError():
x = my_sqrt(0.0)
###Output
_____no_output_____
###Markdown
At least, the Python system catches these errors at runtime. The following call, however, simply lets the function enter an infinite loop:
###Code
with ExpectTimeout(1):
x = my_sqrt(-1.0)
###Output
_____no_output_____
###Markdown
Mining Function SpecificationsWhen testing a program, one not only needs to cover its several behaviors; one also needs to _check_ whether the result is as expected. In this chapter, we introduce a technique that allows us to _mine_ function specifications from a set of given executions, resulting in abstract and formal _descriptions_ of what the function expects and what it delivers. These so-called _dynamic invariants_ produce pre- and post-conditions over function arguments and variables from a set of executions. They are useful in a variety of contexts:* Dynamic invariants provide important information for [symbolic fuzzing](SymbolicFuzzer.ipynb), such as types and ranges of function arguments.* Dynamic invariants provide pre- and postconditions for formal program proofs and verification.* Dynamic invariants provide a large number of assertions that can check whether function behavior has changed* Checks provided by dynamic invariants can be very useful as _oracles_ for checking the effects of generated testsTraditionally, dynamic invariants are dependent on the executions they are derived from. However, when paired with comprehensive test generators, they quickly become very precise, as we show in this chapter. **Prerequisites*** You should be familiar with tracing program executions, as in the [chapter on coverage](Coverage.ipynb).* Later in this section, we access the internal _abstract syntax tree_ representations of Python programs and transform them, as in the [chapter on information flow](InformationFlow.ipynb).
###Code
import bookutils
import Coverage
import Intro_Testing
###Output
_____no_output_____
###Markdown
SynopsisTo [use the code provided in this chapter](Importing.ipynb), write```python>>> from fuzzingbook.DynamicInvariants import ```and then make use of the following features.This chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:```python>>> def sum2(a, b):>>> return a + b>>> with TypeAnnotator() as type_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.```python>>> print(type_annotator.typed_functions())def sum2(a: int, b: int) ->int: return a + b```The invariant annotator works in a similar fashion:```python>>> with InvariantAnnotator() as inv_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.```python>>> print(inv_annotator.functions_with_invariants())@precondition(lambda a, b: isinstance(a, int), doc='isinstance(a, int)')@precondition(lambda a, b: isinstance(b, int), doc='isinstance(b, int)')@postcondition(lambda return_value, a, b: a == return_value - b, doc='a == return_value - b')@postcondition(lambda return_value, a, b: b == return_value - a, doc='b == return_value - a')@postcondition(lambda return_value, a, b: isinstance(return_value, int), doc='isinstance(return_value, int)')@postcondition(lambda return_value, a, b: return_value == a + b, doc='return_value == a + b')@postcondition(lambda return_value, a, b: return_value == b + a, doc='return_value == b + a')def sum2(a, b): return a + b```Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs) as well as for all kinds of _symbolic code analyses_. The chapter gives details on how to customize the properties checked for. Specifications and AssertionsWhen implementing a function or program, one usually works against a _specification_ – a set of documented requirements to be satisfied by the code. Such specifications can come in natural language. A formal specification, however, allows the computer to check whether the specification is satisfied.In the [introduction to testing](Intro_Testing.ipynb), we have seen how _preconditions_ and _postconditions_ can describe what a function does. Consider the following (simple) square root function:
###Code
def my_sqrt(x):
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
The assertion `assert p` checks the condition `p`; if it does not hold, execution is aborted. Here, the actual body is not yet written; we use the assertions as a specification of what `my_sqrt()` _expects_, and what it _delivers_.The topmost assertion is the _precondition_, stating the requirements on the function arguments. The assertion at the end is the _postcondition_, stating the properties of the function result (including its relationship with the original arguments). Using these pre- and postconditions as a specification, we can now go and implement a square root function that satisfies them. Once implemented, we can have the assertions check at runtime whether `my_sqrt()` works as expected; a [symbolic](SymbolicFuzzer.ipynb) or [concolic](ConcolicFuzzer.ipynb) test generator will even specifically try to find inputs where the assertions do _not_ hold. (An assertion can be seen as a conditional branch towards aborting the execution, and any technique that tries to cover all code branches will also try to invalidate as many assertions as possible.) However, not every piece of code is developed with explicit specifications in the first place; let alone does most code comes with formal pre- and post-conditions. (Just take a look at the chapters in this book.) This is a pity: As Ken Thompson famously said, "Without specifications, there are no bugs – only surprises". It is also a problem for testing, since, of course, testing needs some specification to test against. This raises the interesting question: Can we somehow _retrofit_ existing code with "specifications" that properly describe their behavior, allowing developers to simply _check_ them rather than having to write them from scratch? This is what we do in this chapter. Why Generic Error Checking is Not EnoughBefore we go into _mining_ specifications, let us first discuss why it could be useful to _have_ them. As a motivating example, consider the full implementation of `my_sqrt()` from the [introduction to testing](Intro_Testing.ipynb):
###Code
import bookutils
def my_sqrt(x):
"""Computes the square root of x, using the Newton-Raphson method"""
approx = None
guess = x / 2
while approx != guess:
approx = guess
guess = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
`my_sqrt()` does not come with any functionality that would check types or values. Hence, it is easy for callers to make mistakes when calling `my_sqrt()`:
###Code
from ExpectError import ExpectError, ExpectTimeout
with ExpectError():
my_sqrt("foo")
with ExpectError():
x = my_sqrt(0.0)
###Output
_____no_output_____
###Markdown
At least, the Python system catches these errors at runtime. The following call, however, simply lets the function enter an infinite loop:
###Code
with ExpectTimeout(1):
x = my_sqrt(-1.0)
###Output
_____no_output_____
###Markdown
Our goal is to avoid such errors by _annotating_ functions with information that prevents errors like the above ones. The idea is to provide a _specification_ of expected properties – a specification that can then be checked at runtime or statically. \todo{Introduce the concept of *contract*.} Specifying and Checking Data TypesFor our Python code, one of the most important "specifications" we need is *types*. Python being a "dynamically" typed language means that all data types are determined at run time; the code itself does not explicitly state whether a variable is an integer, a string, an array, a dictionary – or whatever. As _writer_ of Python code, omitting explicit type declarations may save time (and allows for some fun hacks). It is not sure whether a lack of types helps in _reading_ and _understanding_ code for humans. For a _computer_ trying to analyze code, the lack of explicit types is detrimental. If, say, a constraint solver, sees `if x:` and cannot know whether `x` is supposed to be a number or a string, this introduces an _ambiguity_. Such ambiguities may multiply over the entire analysis in a combinatorial explosion – or in the analysis yielding an overly inaccurate result. Python 3.6 and later allows data types as _annotations_ to function arguments (actually, to all variables) and return values. We can, for instance, state that `my_sqrt()` is a function that accepts a floating-point value and returns one:
###Code
def my_sqrt_with_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
By default, such annotations are ignored by the Python interpreter. Therefore, one can still call `my_sqrt_typed()` with a string as an argument and get the exact same result as above. However, one can make use of special _typechecking_ modules that would check types – _dynamically_ at runtime or _statically_ by analyzing the code without having to execute it. Excursion: Runtime Type Checking(Commented out as `enforce` is not supported by Python 3.9)The Python `enforce` package provides a function decorator that automatically inserts type-checking code that is executed at runtime. Here is how to use it:
###Code
# import enforce
# @enforce.runtime_validation
# def my_sqrt_with_checked_type_annotations(x: float) -> float:
# """Computes the square root of x, using the Newton-Raphson method"""
# return my_sqrt(x)
###Output
_____no_output_____
###Markdown
Now, invoking `my_sqrt_with_checked_type_annotations()` raises an exception when invoked with a type dfferent from the one declared:
###Code
# with ExpectError():
# my_sqrt_with_checked_type_annotations(True)
###Output
_____no_output_____
###Markdown
Note that this error is not caught by the "untyped" variant, where passing a boolean value happily returns $\sqrt{1}$ as result.
###Code
# my_sqrt(True)
###Output
_____no_output_____
###Markdown
In Python (and other languages), the boolean values `True` and `False` can be implicitly converted to the integers 1 and 0; however, it is hard to think of a call to `sqrt()` where this would not be an error. End of Excursion Static Type CheckingType annotations can also be checked _statically_ – that is, without even running the code. Let us create a simple Python file consisting of the above `my_sqrt_typed()` definition and a bad invocation.
###Code
import inspect
import tempfile
f = tempfile.NamedTemporaryFile(mode='w', suffix='.py')
f.name
f.write(inspect.getsource(my_sqrt))
f.write('\n')
f.write(inspect.getsource(my_sqrt_with_type_annotations))
f.write('\n')
f.write("print(my_sqrt_with_type_annotations('123'))\n")
f.flush()
###Output
_____no_output_____
###Markdown
These are the contents of our newly created Python file:
###Code
from bookutils import print_file
print_file(f.name)
###Output
_____no_output_____
###Markdown
[Mypy](http://mypy-lang.org) is a type checker for Python programs. As it checks types statically, types induce no overhead at runtime; plus, a static check can be faster than a lengthy series of tests with runtime type checking enabled. Let us see what `mypy` produces on the above file:
###Code
import subprocess
result = subprocess.run(["mypy", "--strict", f.name], universal_newlines=True, stdout=subprocess.PIPE)
del f # Delete temporary file
print(result.stdout)
###Output
_____no_output_____
###Markdown
We see that `mypy` complains about untyped function definitions such as `my_sqrt()`; most important, however, it finds that the call to `my_sqrt_with_type_annotations()` in the last line has the wrong type. With `mypy`, we can achieve the same type safety with Python as in statically typed languages – provided that we as programmers also produce the necessary type annotations. Is there a simple way to obtain these? Mining Type SpecificationsOur first task will be to mine type annotations (as part of the code) from _values_ we observe at run time. These type annotations would be _mined_ from actual function executions, _learning_ from (normal) runs what the expected argument and return types should be. By observing a series of calls such as these, we could infer that both `x` and the return value are of type `float`:
###Code
y = my_sqrt(25.0)
y
y = my_sqrt(2.0)
y
###Output
_____no_output_____
###Markdown
How can we mine types from executions? The answer is simple: 1. We _observe_ a function during execution2. We track the _types_ of its arguments3. We include these types as _annotations_ into the code.To do so, we can make use of Python's tracing facility we already observed in the [chapter on coverage](Coverage.ipynb). With every call to a function, we retrieve the arguments, their values, and their types. Tracking CallsTo observe argument types at runtime, we define a _tracer function_ that tracks the execution of `my_sqrt()`, checking its arguments and return values. The `Tracker` class is set to trace functions in a `with` block as follows:```pythonwith Tracker() as tracker: function_to_be_tracked(...)info = tracker.collected_information()```As in the [chapter on coverage](Coverage.ipynb), we use the `sys.settrace()` function to trace individual functions during execution. We turn on tracking when the `with` block starts; at this point, the `__enter__()` method is called. When execution of the `with` block ends, `__exit()__` is called.
###Code
import sys
class Tracker(object):
def __init__(self, log=False):
self._log = log
self.reset()
def reset(self):
self._calls = {}
self._stack = []
def traceit(self):
"""Placeholder to be overloaded in subclasses"""
pass
# Start of `with` block
def __enter__(self):
self.original_trace_function = sys.gettrace()
sys.settrace(self.traceit)
return self
# End of `with` block
def __exit__(self, exc_type, exc_value, tb):
sys.settrace(self.original_trace_function)
###Output
_____no_output_____
###Markdown
The `traceit()` method does nothing yet; this is done in specialized subclasses. The `CallTracker` class implements a `traceit()` function that checks for function calls and returns:
###Code
class CallTracker(Tracker):
def traceit(self, frame, event, arg):
"""Tracking function: Record all calls and all args"""
if event == "call":
self.trace_call(frame, event, arg)
elif event == "return":
self.trace_return(frame, event, arg)
return self.traceit
###Output
_____no_output_____
###Markdown
`trace_call()` is called when a function is called; it retrieves the function name and current arguments, and saves them on a stack.
###Code
class CallTracker(CallTracker):
def trace_call(self, frame, event, arg):
"""Save current function name and args on the stack"""
code = frame.f_code
function_name = code.co_name
arguments = get_arguments(frame)
self._stack.append((function_name, arguments))
if self._log:
print(simple_call_string(function_name, arguments))
def get_arguments(frame):
"""Return call arguments in the given frame"""
# When called, all arguments are local variables
local_variables = dict(frame.f_locals) # explicit copy
arguments = [(var, frame.f_locals[var]) for var in local_variables]
arguments.reverse() # Want same order as call
return arguments
###Output
_____no_output_____
###Markdown
When the function returns, `trace_return()` is called. We now also have the return value. We log the whole call with arguments and return value (if desired) and save it in our list of calls.
###Code
class CallTracker(CallTracker):
def trace_return(self, frame, event, arg):
"""Get return value and store complete call with arguments and return value"""
code = frame.f_code
function_name = code.co_name
return_value = arg
# TODO: Could call get_arguments() here to also retrieve _final_ values of argument variables
called_function_name, called_arguments = self._stack.pop()
assert function_name == called_function_name
if self._log:
print(simple_call_string(function_name, called_arguments), "returns", return_value)
self.add_call(function_name, called_arguments, return_value)
###Output
_____no_output_____
###Markdown
`simple_call_string()` is a helper for logging that prints out calls in a user-friendly manner.
###Code
def simple_call_string(function_name, argument_list, return_value=None):
"""Return function_name(arg[0], arg[1], ...) as a string"""
call = function_name + "(" + \
", ".join([var + "=" + repr(value)
for (var, value) in argument_list]) + ")"
if return_value is not None:
call += " = " + repr(return_value)
return call
###Output
_____no_output_____
###Markdown
`add_call()` saves the calls in a list; each function name has its own list.
###Code
class CallTracker(CallTracker):
def add_call(self, function_name, arguments, return_value=None):
"""Add given call to list of calls"""
if function_name not in self._calls:
self._calls[function_name] = []
self._calls[function_name].append((arguments, return_value))
###Output
_____no_output_____
###Markdown
Using `calls()`, we can retrieve the list of calls, either for a given function, or for all functions.
###Code
class CallTracker(CallTracker):
def calls(self, function_name=None):
"""Return list of calls for function_name,
or a mapping function_name -> calls for all functions tracked"""
if function_name is None:
return self._calls
return self._calls[function_name]
###Output
_____no_output_____
###Markdown
Let us now put this to use. We turn on logging to track the individual calls and their return values:
###Code
with CallTracker(log=True) as tracker:
y = my_sqrt(25)
y = my_sqrt(2.0)
###Output
_____no_output_____
###Markdown
After execution, we can retrieve the individual calls:
###Code
calls = tracker.calls('my_sqrt')
calls
###Output
_____no_output_____
###Markdown
Each call is pair (`argument_list`, `return_value`), where `argument_list` is a list of pairs (`parameter_name`, `value`).
###Code
my_sqrt_argument_list, my_sqrt_return_value = calls[0]
simple_call_string('my_sqrt', my_sqrt_argument_list, my_sqrt_return_value)
###Output
_____no_output_____
###Markdown
If the function does not return a value, `return_value` is `None`.
###Code
def hello(name):
print("Hello,", name)
with CallTracker() as tracker:
hello("world")
hello_calls = tracker.calls('hello')
hello_calls
hello_argument_list, hello_return_value = hello_calls[0]
simple_call_string('hello', hello_argument_list, hello_return_value)
###Output
_____no_output_____
###Markdown
Getting TypesDespite what you may have read or heard, Python actually _is_ a typed language. It is just that it is _dynamically typed_ – types are used and checked only at runtime (rather than declared in the code, where they can be _statically checked_ at compile time). We can thus retrieve types of all values within Python:
###Code
type(4)
type(2.0)
type([4])
###Output
_____no_output_____
###Markdown
We can retrieve the type of the first argument to `my_sqrt()`:
###Code
parameter, value = my_sqrt_argument_list[0]
parameter, type(value)
###Output
_____no_output_____
###Markdown
as well as the type of the return value:
###Code
type(my_sqrt_return_value)
###Output
_____no_output_____
###Markdown
Hence, we see that (so far), `my_sqrt()` is a function taking (among others) integers and returning floats. We could declare `my_sqrt()` as:
###Code
def my_sqrt_annotated(x: int) -> float:
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
This is a representation we could place in a static type checker, allowing to check whether calls to `my_sqrt()` actually pass a number. A dynamic type checker could run such checks at runtime. And of course, any [symbolic interpretation](SymbolicFuzzer.ipynb) will greatly profit from the additional annotations. By default, Python does not do anything with such annotations. However, tools can access annotations from functions and other objects:
###Code
my_sqrt_annotated.__annotations__
###Output
_____no_output_____
###Markdown
This is how run-time checkers access the annotations to check against. Accessing Function StructureOur plan is to annotate functions automatically, based on the types we have seen. To do so, we need a few modules that allow us to convert a function into a tree representation (called _abstract syntax trees_, or ASTs) and back; we already have seen these in the chapters on [concolic](ConcolicFuzzer.ipynb) and [symbolic](SymbolicFuzzer.ipynb) testing.
###Code
import ast
import inspect
###Output
_____no_output_____
###Markdown
We can get the source of a Python function using `inspect.getsource()`. (Note that this does not work for functions defined in other notebooks.)
###Code
my_sqrt_source = inspect.getsource(my_sqrt)
my_sqrt_source
###Output
_____no_output_____
###Markdown
To view these in a visually pleasing form, our function `print_content(s, suffix)` formats and highlights the string `s` as if it were a file with ending `suffix`. We can thus view (and highlight) the source as if it were a Python file:
###Code
from bookutils import print_content
print_content(my_sqrt_source, '.py')
###Output
_____no_output_____
###Markdown
Parsing this gives us an abstract syntax tree (AST) – a representation of the program in tree form.
###Code
my_sqrt_ast = ast.parse(my_sqrt_source)
###Output
_____no_output_____
###Markdown
What does this AST look like? The helper functions `ast.dump()` (textual output) and `showast.show_ast()` (graphical output with [showast](https://github.com/hchasestevens/show_ast)) allow us to inspect the structure of the tree. We see that the function starts as a `FunctionDef` with name and arguments, followed by a body, which is a list of statements of type `Expr` (the docstring), type `Assign` (assignments), `While` (while loop with its own body), and finally `Return`.
###Code
print(ast.dump(my_sqrt_ast, indent=4))
###Output
_____no_output_____
###Markdown
Too much text for you? This graphical representation may make things simpler.
###Code
from bookutils import rich_output
if rich_output():
import showast
showast.show_ast(my_sqrt_ast)
###Output
_____no_output_____
###Markdown
The function `ast.unparse()` converts such a tree back into the more familiar textual Python code representation. Comments are gone, and there may be more parentheses than before, but the result has the same semantics:
###Code
print_content(ast.unparse(my_sqrt_ast), '.py')
###Output
_____no_output_____
###Markdown
Annotating Functions with Given TypesLet us now go and transform these trees ti add type annotations. We start with a helper function `parse_type(name)` which parses a type name into an AST.
###Code
def parse_type(name):
class ValueVisitor(ast.NodeVisitor):
def visit_Expr(self, node):
self.value_node = node.value
tree = ast.parse(name)
name_visitor = ValueVisitor()
name_visitor.visit(tree)
return name_visitor.value_node
print(ast.dump(parse_type('int')))
print(ast.dump(parse_type('[object]')))
###Output
_____no_output_____
###Markdown
We now define a helper function that actually adds type annotations to a function AST. The `TypeTransformer` class builds on the Python standard library `ast.NodeTransformer` infrastructure. It would be called as```python TypeTransformer({'x': 'int'}, 'float').visit(ast)```to annotate the arguments of `my_sqrt()`: `x` with `int`, and the return type with `float`. The returned AST can then be unparsed, compiled or analyzed.
###Code
class TypeTransformer(ast.NodeTransformer):
def __init__(self, argument_types, return_type=None):
self.argument_types = argument_types
self.return_type = return_type
super().__init__()
###Output
_____no_output_____
###Markdown
The core of `TypeTransformer` is the method `visit_FunctionDef()`, which is called for every function definition in the AST. Its argument `node` is the subtree of the function definition to be transformed. Our implementation accesses the individual arguments and invokes `annotate_args()` on them; it also sets the return type in the `returns` attribute of the node.
###Code
class TypeTransformer(TypeTransformer):
def visit_FunctionDef(self, node):
"""Add annotation to function"""
# Set argument types
new_args = []
for arg in node.args.args:
new_args.append(self.annotate_arg(arg))
new_arguments = ast.arguments(
node.args.posonlyargs,
new_args,
node.args.vararg,
node.args.kwonlyargs,
node.args.kw_defaults,
node.args.kwarg,
node.args.defaults
)
# Set return type
if self.return_type is not None:
node.returns = parse_type(self.return_type)
return ast.copy_location(
ast.FunctionDef(node.name, new_arguments,
node.body, node.decorator_list,
node.returns), node)
###Output
_____no_output_____
###Markdown
Each argument gets its own annotation, taken from the types originally passed to the class:
###Code
class TypeTransformer(TypeTransformer):
def annotate_arg(self, arg):
"""Add annotation to single function argument"""
arg_name = arg.arg
if arg_name in self.argument_types:
arg.annotation = parse_type(self.argument_types[arg_name])
return arg
###Output
_____no_output_____
###Markdown
Does this work? Let us annotate the AST from `my_sqrt()` with types for the arguments and return types:
###Code
new_ast = TypeTransformer({'x': 'int'}, 'float').visit(my_sqrt_ast)
###Output
_____no_output_____
###Markdown
When we unparse the new AST, we see that the annotations actually are present:
###Code
print_content(ast.unparse(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Similarly, we can annotate the `hello()` function from above:
###Code
hello_source = inspect.getsource(hello)
hello_ast = ast.parse(hello_source)
new_ast = TypeTransformer({'name': 'str'}, 'None').visit(hello_ast)
print_content(ast.unparse(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Annotating Functions with Mined TypesLet us now annotate functions with types mined at runtime. We start with a simple function `type_string()` that determines the appropriate type of a given value (as a string):
###Code
def type_string(value):
return type(value).__name__
type_string(4)
type_string([])
###Output
_____no_output_____
###Markdown
For composite structures, `type_string()` does not examine element types; hence, the type of `[3]` is simply `list` instead of, say, `list[int]`. For now, `list` will do fine.
###Code
type_string([3])
###Output
_____no_output_____
###Markdown
`type_string()` will be used to infer the types of argument values found at runtime, as returned by `CallTracker.calls()`:
###Code
with CallTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(2.0)
tracker.calls()
###Output
_____no_output_____
###Markdown
The function `annotate_types()` takes such a list of calls and annotates each function listed:
###Code
def annotate_types(calls):
annotated_functions = {}
for function_name in calls:
try:
annotated_functions[function_name] = annotate_function_with_types(function_name, calls[function_name])
except KeyError:
continue
return annotated_functions
###Output
_____no_output_____
###Markdown
For each function, we get the source and its AST and then get to the actual annotation in `annotate_function_ast_with_types()`:
###Code
def annotate_function_with_types(function_name, function_calls):
function = globals()[function_name] # May raise KeyError for internal functions
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_types(function_ast, function_calls)
###Output
_____no_output_____
###Markdown
The function `annotate_function_ast_with_types()` invokes the `TypeTransformer` with the calls seen, and for each call, iterate over the arguments, determine their types, and annotate the AST with these. The universal type `Any` is used when we encounter type conflicts, which we will discuss below.
###Code
from typing import Any
def annotate_function_ast_with_types(function_ast, function_calls):
parameter_types = {}
return_type = None
for calls_seen in function_calls:
args, return_value = calls_seen
if return_value is not None:
if return_type is not None and return_type != type_string(return_value):
return_type = 'Any'
else:
return_type = type_string(return_value)
for parameter, value in args:
try:
different_type = parameter_types[parameter] != type_string(value)
except KeyError:
different_type = False
if different_type:
parameter_types[parameter] = 'Any'
else:
parameter_types[parameter] = type_string(value)
annotated_function_ast = TypeTransformer(parameter_types, return_type).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Here is `my_sqrt()` annotated with the types recorded usign the tracker, above.
###Code
print_content(ast.unparse(annotate_types(tracker.calls())['my_sqrt']), '.py')
###Output
_____no_output_____
###Markdown
All-in-one AnnotationLet us bring all of this together in a single class `TypeAnnotator` that first tracks calls of functions and then allows to access the AST (and the source code form) of the tracked functions annotated with types. The method `typed_functions()` returns the annotated functions as a string; `typed_functions_ast()` returns their AST.
###Code
class TypeTracker(CallTracker):
pass
class TypeAnnotator(TypeTracker):
def typed_functions_ast(self, function_name=None):
if function_name is None:
return annotate_types(self.calls())
return annotate_function_with_types(function_name,
self.calls(function_name))
def typed_functions(self, function_name=None):
if function_name is None:
functions = ''
for f_name in self.calls():
try:
f_text = ast.unparse(self.typed_functions_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
return ast.unparse(self.typed_functions_ast(function_name))
###Output
_____no_output_____
###Markdown
Here is how to use `TypeAnnotator`. We first track a series of calls:
###Code
with TypeAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(2.0)
###Output
_____no_output_____
###Markdown
After tracking, we can immediately retrieve an annotated version of the functions tracked:
###Code
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
This also works for multiple and diverse functions. One could go and implement an automatic type annotator for Python files based on the types seen during execution.
###Code
with TypeAnnotator() as annotator:
hello('type annotations')
y = my_sqrt(1.0)
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
A content as above could now be sent to a type checker, which would detect any type inconsistency between callers and callees. Likewise, type annotations such as the ones above greatly benefit symbolic code analysis (as in the chapter on [symbolic fuzzing](SymbolicFuzzer.ipynb)), as they effectively constrain the set of values that arguments and variables can take. Multiple TypesLet us now resolve the role of the magic `Any` type in `annotate_function_ast_with_types()`. If we see multiple types for the same argument, we set its type to `Any`. For `my_sqrt()`, this makes sense, as its arguments can be integers as well as floats:
###Code
with CallTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(4)
print_content(ast.unparse(annotate_types(tracker.calls())['my_sqrt']), '.py')
###Output
_____no_output_____
###Markdown
The following function `sum3()` can be called with floating-point numbers as arguments, resulting in the parameters getting a `float` type:
###Code
def sum3(a, b, c):
return a + b + c
with TypeAnnotator() as annotator:
y = sum3(1.0, 2.0, 3.0)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we call `sum3()` with integers, though, the arguments get an `int` type:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
And we can also call `sum3()` with strings, giving the arguments a `str` type:
###Code
with TypeAnnotator() as annotator:
y = sum3("one", "two", "three")
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we have multiple calls, but with different types, `TypeAnnotator()` will assign an `Any` type to both arguments and return values:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y = sum3("one", "two", "three")
typed_sum3_def = annotator.typed_functions('sum3')
print_content(typed_sum3_def, '.py')
###Output
_____no_output_____
###Markdown
A type `Any` makes it explicit that an object can, indeed, have any type; it will not be typechecked at runtime or statically. To some extent, this defeats the power of type checking; but it also preserves some of the type flexibility that many Python programmers enjoy. Besides `Any`, the `typing` module supports several additional ways to define ambiguous types; we will keep this in mind for a later exercise. Specifying and Checking InvariantsBesides basic data types. we can check several further properties from arguments. We can, for instance, whether an argument can be negative, zero, or positive; or that one argument should be smaller than the second; or that the result should be the sum of two arguments – properties that cannot be expressed in a (Python) type.Such properties are called *invariants*, as they hold across all invocations of a function. Specifically, invariants come as _pre_- and _postconditions_ – conditions that always hold at the beginning and at the end of a function. (There are also _data_ and _object_ invariants that express always-holding properties over the state of data or objects, but we do not consider these in this book.) Annotating Functions with Pre- and PostconditionsThe classical means to specify pre- and postconditions is via _assertions_, which we have introduced in the [chapter on testing](Intro_Testing.ipynb). A precondition checks whether the arguments to a function satisfy the expected properties; a postcondition does the same for the result. We can express and check both using assertions as follows:
###Code
def my_sqrt_with_invariants(x):
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
A nicer way, however, is to syntactically separate invariants from the function at hand. Using appropriate decorators, we could specify pre- and postconditions as follows:```python@precondition lambda x: x >= 0@postcondition lambda return_value, x: return_value * return_value == xdef my_sqrt_with_invariants(x): normal code without assertions ...```The decorators `@precondition` and `@postcondition` would run the given functions (specified as anonymous `lambda` functions) before and after the decorated function, respectively. If the functions return `False`, the condition is violated. `@precondition` gets the function arguments as arguments; `@postcondition` additionally gets the return value as first argument. It turns out that implementing such decorators is not hard at all. Our implementation builds on a [code snippet from StackOverflow](https://stackoverflow.com/questions/12151182/python-precondition-postcondition-for-member-function-how):
###Code
import functools
def condition(precondition=None, postcondition=None):
def decorator(func):
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args, **kwargs):
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated"
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated"
return retval
return wrapper
return decorator
def precondition(check):
return condition(precondition=check)
def postcondition(check):
return condition(postcondition=check)
###Output
_____no_output_____
###Markdown
With these, we can now start decorating `my_sqrt()`:
###Code
@precondition(lambda x: x > 0)
def my_sqrt_with_precondition(x):
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
This catches arguments violating the precondition:
###Code
with ExpectError():
my_sqrt_with_precondition(-1.0)
###Output
_____no_output_____
###Markdown
Likewise, we can provide a postcondition:
###Code
EPSILON = 1e-5
@postcondition(lambda ret, x: ret * ret - x < EPSILON)
def my_sqrt_with_postcondition(x):
return my_sqrt(x)
y = my_sqrt_with_postcondition(2.0)
y
###Output
_____no_output_____
###Markdown
If we have a buggy implementation of $\sqrt{x}$, this gets caught quickly:
###Code
@postcondition(lambda ret, x: ret * ret - x < EPSILON)
def buggy_my_sqrt_with_postcondition(x):
return my_sqrt(x) + 0.1
with ExpectError():
y = buggy_my_sqrt_with_postcondition(2.0)
###Output
_____no_output_____
###Markdown
While checking pre- and postconditions is a great way to catch errors, specifying them can be cumbersome. Let us try to see whether we can (again) _mine_ some of them. Mining InvariantsTo _mine_ invariants, we can use the same tracking functionality as before; instead of saving values for individual variables, though, we now check whether the values satisfy specific _properties_ or not. For instance, if all values of `x` seen satisfy the condition `x > 0`, then we make `x > 0` an invariant of the function. If we see positive, zero, and negative values of `x`, though, then there is no property of `x` left to talk about.The general idea is thus:1. Check all variable values observed against a set of predefined properties; and2. Keep only those properties that hold for all runs observed. Defining PropertiesWhat precisely do we mean by properties? Here is a small collection of value properties that would frequently be used in invariants. All these properties would be evaluated with the _metavariables_ `X`, `Y`, and `Z` (actually, any upper-case identifier) being replaced with the names of function parameters:
###Code
INVARIANT_PROPERTIES = [
"X < 0",
"X <= 0",
"X > 0",
"X >= 0",
"X == 0",
"X != 0",
]
###Output
_____no_output_____
###Markdown
When `my_sqrt(x)` is called as, say `my_sqrt(5.0)`, we see that `x = 5.0` holds. The above properties would then all be checked for `x`. Only the properties `X > 0`, `X >= 0`, and `X != 0` hold for the call seen; and hence `x > 0`, `x >= 0`, and `x != 0` would make potential preconditions for `my_sqrt(x)`. We can check for many more properties such as relations between two arguments:
###Code
INVARIANT_PROPERTIES += [
"X == Y",
"X > Y",
"X < Y",
"X >= Y",
"X <= Y",
]
###Output
_____no_output_____
###Markdown
Types also can be checked using properties. For any function parameter `X`, only one of these will hold:
###Code
INVARIANT_PROPERTIES += [
"isinstance(X, bool)",
"isinstance(X, int)",
"isinstance(X, float)",
"isinstance(X, list)",
"isinstance(X, dict)",
]
###Output
_____no_output_____
###Markdown
We can check for arithmetic properties:
###Code
INVARIANT_PROPERTIES += [
"X == Y + Z",
"X == Y * Z",
"X == Y - Z",
"X == Y / Z",
]
###Output
_____no_output_____
###Markdown
Here's relations over three values, a Python special:
###Code
INVARIANT_PROPERTIES += [
"X < Y < Z",
"X <= Y <= Z",
"X > Y > Z",
"X >= Y >= Z",
]
###Output
_____no_output_____
###Markdown
Finally, we can also check for list or string properties. Again, this is just a tiny selection.
###Code
INVARIANT_PROPERTIES += [
"X == len(Y)",
"X == sum(Y)",
"X.startswith(Y)",
]
###Output
_____no_output_____
###Markdown
Extracting Meta-VariablesLet us first introduce a few _helper functions_ before we can get to the actual mining. `metavars()` extracts the set of meta-variables (`X`, `Y`, `Z`, etc.) from a property. To this end, we parse the property as a Python expression and then visit the identifiers.
###Code
def metavars(prop):
metavar_list = []
class ArgVisitor(ast.NodeVisitor):
def visit_Name(self, node):
if node.id.isupper():
metavar_list.append(node.id)
ArgVisitor().visit(ast.parse(prop))
return metavar_list
assert metavars("X < 0") == ['X']
assert metavars("X.startswith(Y)") == ['X', 'Y']
assert metavars("isinstance(X, str)") == ['X']
###Output
_____no_output_____
###Markdown
Instantiating PropertiesTo produce a property as invariant, we need to be able to _instantiate_ it with variable names. The instantiation of `X > 0` with `X` being instantiated to `a`, for instance, gets us `a > 0`. To this end, the function `instantiate_prop()` takes a property and a collection of variable names and instantiates the meta-variables left-to-right with the corresponding variables names in the collection.
###Code
def instantiate_prop_ast(prop, var_names):
class NameTransformer(ast.NodeTransformer):
def visit_Name(self, node):
if node.id not in mapping:
return node
return ast.Name(id=mapping[node.id], ctx=ast.Load())
meta_variables = metavars(prop)
assert len(meta_variables) == len(var_names)
mapping = {}
for i in range(0, len(meta_variables)):
mapping[meta_variables[i]] = var_names[i]
prop_ast = ast.parse(prop, mode='eval')
new_ast = NameTransformer().visit(prop_ast)
return new_ast
def instantiate_prop(prop, var_names):
prop_ast = instantiate_prop_ast(prop, var_names)
prop_text = ast.unparse(prop_ast).strip()
while prop_text.startswith('(') and prop_text.endswith(')'):
prop_text = prop_text[1:-1]
return prop_text
assert instantiate_prop("X > Y", ['a', 'b']) == 'a > b'
assert instantiate_prop("X.startswith(Y)", ['x', 'y']) == 'x.startswith(y)'
###Output
_____no_output_____
###Markdown
Evaluating PropertiesTo actually _evaluate_ properties, we do not need to instantiate them. Instead, we simply convert them into a boolean function, using `lambda`:
###Code
def prop_function_text(prop):
return "lambda " + ", ".join(metavars(prop)) + ": " + prop
def prop_function(prop):
return eval(prop_function_text(prop))
###Output
_____no_output_____
###Markdown
Here is a simple example:
###Code
prop_function_text("X > Y")
p = prop_function("X > Y")
p(100, 1)
p(1, 100)
###Output
_____no_output_____
###Markdown
Checking InvariantsTo extract invariants from an execution, we need to check them on all possible instantiations of arguments. If the function to be checked has two arguments `a` and `b`, we instantiate the property `X < Y` both as `a < b` and `b < a` and check each of them. To get all combinations, we use the Python `permutations()` function:
###Code
import itertools
for combination in itertools.permutations([1.0, 2.0, 3.0], 2):
print(combination)
###Output
_____no_output_____
###Markdown
The function `true_property_instantiations()` takes a property and a list of tuples (`var_name`, `value`). It then produces all instantiations of the property with the given values and returns those that evaluate to True.
###Code
def true_property_instantiations(prop, vars_and_values, log=False):
instantiations = set()
p = prop_function(prop)
len_metavars = len(metavars(prop))
for combination in itertools.permutations(vars_and_values, len_metavars):
args = [value for var_name, value in combination]
var_names = [var_name for var_name, value in combination]
try:
result = p(*args)
except:
result = None
if log:
print(prop, combination, result)
if result:
instantiations.add((prop, tuple(var_names)))
return instantiations
###Output
_____no_output_____
###Markdown
Here is an example. If `x == -1` and `y == 1`, the property `X < Y` holds for `x < y`, but not for `y < x`:
###Code
invs = true_property_instantiations("X < Y", [('x', -1), ('y', 1)], log=True)
invs
###Output
_____no_output_____
###Markdown
The instantiation retrieves the short form:
###Code
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Likewise, with values for `x` and `y` as above, the property `X < 0` only holds for `x`, but not for `y`:
###Code
invs = true_property_instantiations("X < 0", [('x', -1), ('y', 1)], log=True)
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Extracting InvariantsLet us now run the above invariant extraction on function arguments and return values as observed during a function execution. To this end, we extend the `CallTracker` class into an `InvariantTracker` class, which automatically computes invariants for all functions and all calls observed during tracking. By default, an `InvariantTracker` uses the properties as defined above; however, one can specify alternate sets of properties.
###Code
class InvariantTracker(CallTracker):
def __init__(self, props=None, **kwargs):
if props is None:
props = INVARIANT_PROPERTIES
self.props = props
super().__init__(**kwargs)
###Output
_____no_output_____
###Markdown
The key method of the `InvariantTracker` is the `invariants()` method. This iterates over the calls observed and checks which properties hold. Only the intersection of properties – that is, the set of properties that hold for all calls – is preserved, and eventually returned. The special variable `return_value` is set to hold the return value.
###Code
RETURN_VALUE = 'return_value'
class InvariantTracker(InvariantTracker):
def invariants(self, function_name=None):
if function_name is None:
return {function_name: self.invariants(function_name) for function_name in self.calls()}
invariants = None
for variables, return_value in self.calls(function_name):
vars_and_values = variables + [(RETURN_VALUE, return_value)]
s = set()
for prop in self.props:
s |= true_property_instantiations(prop, vars_and_values, self._log)
if invariants is None:
invariants = s
else:
invariants &= s
return invariants
###Output
_____no_output_____
###Markdown
Here's an example of how to use `invariants()`. We run the tracker on a small set of calls.
###Code
with InvariantTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(10.0)
tracker.calls()
###Output
_____no_output_____
###Markdown
The `invariants()` method produces a set of properties that hold for the observed runs, together with their instantiations over function arguments.
###Code
invs = tracker.invariants('my_sqrt')
invs
###Output
_____no_output_____
###Markdown
As before, the actual instantiations are easier to read:
###Code
def pretty_invariants(invariants):
props = []
for (prop, var_names) in invariants:
props.append(instantiate_prop(prop, var_names))
return sorted(props)
pretty_invariants(invs)
###Output
_____no_output_____
###Markdown
We see that the both `x` and the return value have a `float` type. We also see that both are always greater than zero. These are properties that may make useful pre- and postconditions, notably for symbolic analysis. However, there's also an invariant which does _not_ universally hold, namely `return_value <= x`, as the following example shows:
###Code
my_sqrt(0.01)
###Output
_____no_output_____
###Markdown
Clearly, 0.1 > 0.01 holds. This is a case of us not learning from sufficiently diverse inputs. As soon as we have a call including `x = 0.1`, though, the invariant `return_value <= x` is eliminated:
###Code
with InvariantTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(10.0)
y = my_sqrt(0.01)
pretty_invariants(tracker.invariants('my_sqrt'))
###Output
_____no_output_____
###Markdown
We will discuss later how to ensure sufficient diversity in inputs. (Hint: This involves test generation.) Let us try out our invariant tracker on `sum3()`. We see that all types are well-defined; the properties that all arguments are non-zero, however, is specific to the calls observed.
###Code
with InvariantTracker() as tracker:
y = sum3(1, 2, 3)
y = sum3(-4, -5, -6)
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with strings instead, we get different invariants. Notably, we obtain the postcondition that the return value starts with the value of `a` – a universal postcondition if strings are used.
###Code
with InvariantTracker() as tracker:
y = sum3('a', 'b', 'c')
y = sum3('f', 'e', 'd')
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with both strings and numbers (and zeros, too), there are no properties left that would hold across all calls. That's the price of flexibility.
###Code
with InvariantTracker() as tracker:
y = sum3('a', 'b', 'c')
y = sum3('c', 'b', 'a')
y = sum3(-4, -5, -6)
y = sum3(0, 0, 0)
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
Converting Mined Invariants to AnnotationsAs with types, above, we would like to have some functionality where we can add the mined invariants as annotations to existing functions. To this end, we introduce the `InvariantAnnotator` class, extending `InvariantTracker`. We start with a helper method. `params()` returns a comma-separated list of parameter names as observed during calls.
###Code
class InvariantAnnotator(InvariantTracker):
def params(self, function_name):
arguments, return_value = self.calls(function_name)[0]
return ", ".join(arg_name for (arg_name, arg_value) in arguments)
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = sum3(1, 2, 3)
annotator.params('my_sqrt')
annotator.params('sum3')
###Output
_____no_output_____
###Markdown
Now for the actual annotation. `preconditions()` returns the preconditions from the mined invariants (i.e., those propertes that do not depend on the return value) as a string with annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@precondition(lambda " + self.params(function_name) + ": " + inv + ")"
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
annotator.preconditions('my_sqrt')
###Output
_____no_output_____
###Markdown
`postconditions()` does the same for postconditions:
###Code
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ")")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
annotator.postconditions('my_sqrt')
###Output
_____no_output_____
###Markdown
With these, we can take a function and add both pre- and postconditions as annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def functions_with_invariants(self):
functions = ""
for function_name in self.invariants():
try:
function = self.function_with_invariants(function_name)
except KeyError:
continue
functions += function
return functions
def function_with_invariants(self, function_name):
function = globals()[function_name] # Can throw KeyError
source = inspect.getsource(function)
return "\n".join(self.preconditions(function_name) +
self.postconditions(function_name)) + '\n' + source
###Output
_____no_output_____
###Markdown
Here comes `function_with_invariants()` in all its glory:
###Code
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
print_content(annotator.function_with_invariants('my_sqrt'), '.py')
###Output
_____no_output_____
###Markdown
Quite a lot of invariants, is it? Further below (and in the exercises), we will discuss on how to focus on the most relevant properties. Some ExamplesHere's another example. `list_length()` recursively computes the length of a Python function. Let us see whether we can mine its invariants:
###Code
def list_length(L):
if L == []:
length = 0
else:
length = 1 + list_length(L[1:])
return length
with InvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Almost all these properties (except for the very first) are relevant. Of course, the reason the invariants are so neat is that the return value is equal to `len(L)` is that `X == len(Y)` is part of the list of properties to be checked. The next example is a very simple function:
###Code
def sum2(a, b):
return a + b
with InvariantAnnotator() as annotator:
sum2(31, 45)
sum2(0, 0)
sum2(-1, -5)
###Output
_____no_output_____
###Markdown
The invariants all capture the relationship between `a`, `b`, and the return value as `return_value == a + b` in all its variations.
###Code
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
If we have a function without return value, the return value is `None` and we can only mine preconditions. (Well, we get a "postcondition" that the return value is non-zero, which holds for `None`).
###Code
def print_sum(a, b):
print(a + b)
with InvariantAnnotator() as annotator:
print_sum(31, 45)
print_sum(0, 0)
print_sum(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Checking SpecificationsA function with invariants, as above, can be fed into the Python interpreter, such that all pre- and postconditions are checked. We create a function `my_sqrt_annotated()` which includes all the invariants mined above.
###Code
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
my_sqrt_def = annotator.functions_with_invariants()
my_sqrt_def = my_sqrt_def.replace('my_sqrt', 'my_sqrt_annotated')
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def)
###Output
_____no_output_____
###Markdown
The "annotated" version checks against invalid arguments – or more precisely, against arguments with properties that have not been observed yet:
###Code
with ExpectError():
my_sqrt_annotated(-1.0)
###Output
_____no_output_____
###Markdown
This is in contrast to the original version, which just hangs on negative values:
###Code
with ExpectTimeout(1):
my_sqrt(-1.0)
###Output
_____no_output_____
###Markdown
If we make changes to the function definition such that the properties of the return value change, such _regressions_ are caught as violations of the postconditions. Let us illustrate this by simply inverting the result, and return $-2$ as square root of 4.
###Code
my_sqrt_def = my_sqrt_def.replace('my_sqrt_annotated', 'my_sqrt_negative')
my_sqrt_def = my_sqrt_def.replace('return approx', 'return -approx')
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def)
###Output
_____no_output_____
###Markdown
Technically speaking, $-2$ _is_ a square root of 4, since $(-2)^2 = 4$ holds. Yet, such a change may be unexpected by callers of `my_sqrt()`, and hence, this would be caught with the first call:
###Code
with ExpectError():
my_sqrt_negative(2.0)
###Output
_____no_output_____
###Markdown
We see how pre- and postconditions, as well as types, can serve as *oracles* during testing. In particular, once we have mined them for a set of functions, we can check them again and again with test generators – especially after code changes. The more checks we have, and the more specific they are, the more likely it is we can detect unwanted effects of changes. Mining Specifications from Generated TestsMined specifications can only be as good as the executions they were mined from. If we only see a single call to, say, `sum2()`, we will be faced with several mined pre- and postconditions that _overspecialize_ towards the values seen:
###Code
def sum2(a, b):
return a + b
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
The mined precondition `a == b`, for instance, only holds for the single call observed; the same holds for the mined postcondition `return_value == a * b`. Yet, `sum2()` can obviously be successfully called with other values that do not satisfy these conditions. To get out of this trap, we have to _learn from more and more diverse runs_. If we have a few more calls of `sum2()`, we see how the set of invariants quickly gets smaller:
###Code
with InvariantAnnotator() as annotator:
length = sum2(1, 2)
length = sum2(-1, -2)
length = sum2(0, 0)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
But where to we get such diverse runs from? This is the job of generating software tests. A simple grammar for calls of `sum2()` will easily resolve the problem.
###Code
from GrammarFuzzer import GrammarFuzzer # minor dependency
from Grammars import is_valid_grammar, crange, convert_ebnf_grammar # minor dependency
SUM2_EBNF_GRAMMAR = {
"<start>": ["<sum2>"],
"<sum2>": ["sum2(<int>, <int>)"],
"<int>": ["<_int>"],
"<_int>": ["(-)?<leaddigit><digit>*", "0"],
"<leaddigit>": crange('1', '9'),
"<digit>": crange('0', '9')
}
assert is_valid_grammar(SUM2_EBNF_GRAMMAR)
sum2_grammar = convert_ebnf_grammar(SUM2_EBNF_GRAMMAR)
sum2_fuzzer = GrammarFuzzer(sum2_grammar)
[sum2_fuzzer.fuzz() for i in range(10)]
with InvariantAnnotator() as annotator:
for i in range(10):
eval(sum2_fuzzer.fuzz())
print_content(annotator.function_with_invariants('sum2'), '.py')
###Output
_____no_output_____
###Markdown
But then, writing tests (or a test driver) just to derive a set of pre- and postconditions may possibly be too much effort – in particular, since tests can easily be derived from given pre- and postconditions in the first place. Hence, it would be wiser to first specify invariants and then let test generators or program provers do the job. Also, an API grammar, such as above, will have to be set up such that it actually respects preconditions – in our case, we invoke `sqrt()` with positive numbers only, already assuming its precondition. In some way, one thus needs a specification (a model, a grammar) to mine another specification – a chicken-and-egg problem. However, there is one way out of this problem: If one can automatically generate tests at the system level, then one has an _infinite source of executions_ to learn invariants from. In each of these executions, all functions would be called with values that satisfy the (implicit) precondition, allowing us to mine invariants for these functions. This holds, because at the system level, invalid inputs must be rejected by the system in the first place. The meaningful precondition at the system level, ensuring that only valid inputs get through, thus gets broken down into a multitude of meaningful preconditions (and subsequent postconditions) at the function level. The big requirement for this, though, is that one needs good test generators at the system level. In [the next part](05_Domain-Specific_Fuzzing.ipynb), we will discuss how to automatically generate tests for a variety of domains, from configuration to graphical user interfaces. SynopsisThis chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:
###Code
def sum2(a, b):
return a + b
with TypeAnnotator() as type_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.
###Code
print(type_annotator.typed_functions())
###Output
_____no_output_____
###Markdown
The invariant annotator works in a similar fashion:
###Code
with InvariantAnnotator() as inv_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.
###Code
print(inv_annotator.functions_with_invariants())
###Output
_____no_output_____
###Markdown
Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs) as well as for all kinds of _symbolic code analyses_. The chapter gives details on how to customize the properties checked for. Lessons Learned* Type annotations and explicit invariants allow for _checking_ arguments and results for expected data types and other properties.* One can automatically _mine_ data types and invariants by observing arguments and results at runtime.* The quality of mined invariants depends on the diversity of values observed during executions; this variety can be increased by generating tests. Next StepsThis chapter concludes the [part on semantical fuzzing techniques](04_Semantical_Fuzzing.ipynb). In the next part, we will explore [domain-specific fuzzing techniques](05_Domain-Specific_Fuzzing.ipynb) from configurations and APIs to graphical user interfaces. BackgroundThe [DAIKON dynamic invariant detector](https://plse.cs.washington.edu/daikon/) can be considered the mother of function specification miners. Continuously maintained and extended for more than 20 years, it mines likely invariants in the style of this chapter for a variety of languages, including C, C++, C, Eiffel, F, Java, Perl, and Visual Basic. On top of the functionality discussed above, it holds a rich catalog of patterns for likely invariants, supports data invariants, can eliminate invariants that are implied by others, and determines statistical confidence to disregard unlikely invariants. The corresponding paper \cite{Ernst2001} is one of the seminal and most-cited papers of Software Engineering. A multitude of works have been published based on DAIKON and detecting invariants; see this [curated list](http://plse.cs.washington.edu/daikon/pubs/) for details. The interaction between test generators and invariant detection is already discussed in \cite{Ernst2001} (incidentally also using grammars). The Eclat tool \cite{Pacheco2005} is a model example of tight interaction between a unit-level test generator and DAIKON-style invariant mining, where the mined invariants are used to produce oracles and to systematically guide the test generator towards fault-revealing inputs. Mining specifications is not restricted to pre- and postconditions. The paper "Mining Specifications" \cite{Ammons2002} is another classic in the field, learning state protocols from executions. Grammar mining, as described in [our chapter with the same name](GrammarMiner.ipynb) can also be seen as a specification mining approach, this time learning specifications for input formats. As it comes to adding type annotations to existing code, the blog post ["The state of type hints in Python"](https://www.bernat.tech/the-state-of-type-hints-in-python/) gives a great overview on how Python type hints can be used and checked. To add type annotations, there are two important tools available that also implement our above approach:* [MonkeyType](https://instagram-engineering.com/let-your-code-type-hint-itself-introducing-open-source-monkeytype-a855c7284881) implements the above approach of tracing executions and annotating Python 3 arguments, returns, and variables with type hints.* [PyAnnotate](https://github.com/dropbox/pyannotate) does a similar job, focusing on code in Python 2. It does not produce Python 3-style annotations, but instead produces annotations as comments that can be processed by static type checkers.These tools have been created by engineers at Facebook and Dropbox, respectively, assisting them in checking millions of lines of code for type issues. ExercisesOur code for mining types and invariants is in no way complete. There are dozens of ways to extend our implementations, some of which we discuss in exercises. Exercise 1: Union TypesThe Python `typing` module allows to express that an argument can have multiple types. For `my_sqrt(x)`, this allows to express that `x` can be an `int` or a `float`:
###Code
from typing import Union, Optional
def my_sqrt_with_union_type(x: Union[int, float]) -> float:
...
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it supports union types for arguments and return values. Use `Optional[X]` as a shorthand for `Union[X, None]`. **Solution.** Left to the reader. Hint: extend `type_string()`. Exercise 2: Types for Local VariablesIn Python, one cannot only annotate arguments with types, but actually also local and global variables – for instance, `approx` and `guess` in our `my_sqrt()` implementation:
###Code
def my_sqrt_with_local_types(x: Union[int, float]) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
approx: Optional[float] = None
guess: float = x / 2
while approx != guess:
approx: float = guess
guess: float = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it also annotates local variables with types. Search the function AST for assignments, determine the type of the assigned value, and make it an annotation on the left hand side. **Solution.** Left to the reader. Exercise 3: Verbose Invariant CheckersOur implementation of invariant checkers does not make it clear for the user which pre-/postcondition failed.
###Code
@precondition(lambda s: len(s) > 0)
def remove_first_char(s):
return s[1:]
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
The following implementation adds an optional `doc` keyword argument which is printed if the invariant is violated:
###Code
def condition(precondition=None, postcondition=None, doc='Unknown'):
def decorator(func):
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args, **kwargs):
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated: " + doc
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated: " + doc
return retval
return wrapper
return decorator
def precondition(check, **kwargs):
return condition(precondition=check, doc=kwargs.get('doc', 'Unknown'))
def postcondition(check, **kwargs):
return condition(postcondition=check, doc=kwargs.get('doc', 'Unknown'))
@precondition(lambda s: len(s) > 0, doc="len(s) > 0")
def remove_first_char(s):
return s[1:]
remove_first_char('abc')
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
Extend `InvariantAnnotator` such that it includes the conditions in the generated pre- and postconditions. **Solution.** Here's a simple solution:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@precondition(lambda " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")"
conditions.append(cond)
return conditions
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")")
conditions.append(cond)
return conditions
###Output
_____no_output_____
###Markdown
The resulting annotations are harder to read, but easier to diagnose:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
As an alternative, one may be able to use `inspect.getsource()` on the lambda expression or unparse it. This is left to the reader. Exercise 4: Save Initial ValuesIf the value of an argument changes during function execution, this can easily confuse our implementation: The values are tracked at the beginning of the function, but checked only when it returns. Extend the `InvariantAnnotator` and the infrastructure it uses such that* it saves argument values both at the beginning and at the end of a function invocation;* postconditions can be expressed over both _initial_ values of arguments as well as the _final_ values of arguments;* the mined postconditions refer to both these values as well. **Solution.** To be added. Exercise 5: ImplicationsSeveral mined invariant are actually _implied_ by others: If `x > 0` holds, then this implies `x >= 0` and `x != 0`. Extend the `InvariantAnnotator` such that implications between properties are explicitly encoded, and such that implied properties are no longer listed as invariants. See \cite{Ernst2001} for ideas. **Solution.** Left to the reader. Exercise 6: Local VariablesPostconditions may also refer to the values of local variables. Consider extending `InvariantAnnotator` and its infrastructure such that the values of local variables at the end of the execution are also recorded and made part of the invariant inference mechanism. **Solution.** Left to the reader. Exercise 7: Exploring Invariant AlternativesAfter mining a first set of invariants, have a [concolic fuzzer](ConcolicFuzzer.ipynb) generate tests that systematically attempt to invalidate pre- and postconditions. How far can you generalize? **Solution.** To be added. Exercise 8: Grammar-Generated PropertiesThe larger the set of properties to be checked, the more potential invariants can be discovered. Create a _grammar_ that systematically produces a large set of properties. See \cite{Ernst2001} for possible patterns. **Solution.** Left to the reader. Exercise 9: Embedding Invariants as AssertionsRather than producing invariants as annotations for pre- and postconditions, insert them as `assert` statements into the function code, as in:```pythondef my_sqrt(x): 'Computes the square root of x, using the Newton-Raphson method' assert isinstance(x, int), 'violated precondition' assert (x > 0), 'violated precondition' approx = None guess = (x / 2) while (approx != guess): approx = guess guess = ((approx + (x / approx)) / 2) return_value = approx assert (return_value < x), 'violated postcondition' assert isinstance(return_value, float), 'violated postcondition' return approx```Such a formulation may make it easier for test generators and symbolic analysis to access and interpret pre- and postconditions. **Solution.** Here is a tentative implementation that inserts invariants into function ASTs. Part 1: Embedding Invariants into Functions
###Code
class EmbeddedInvariantAnnotator(InvariantTracker):
def functions_with_invariants_ast(self, function_name=None):
if function_name is None:
return annotate_functions_with_invariants(self.invariants())
return annotate_function_with_invariants(function_name, self.invariants(function_name))
def functions_with_invariants(self, function_name=None):
if function_name is None:
functions = ''
for f_name in self.invariants():
try:
f_text = ast.unparse(self.functions_with_invariants_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
return ast.unparse(self.functions_with_invariants_ast(function_name))
def function_with_invariants(self, function_name):
return self.functions_with_invariants(function_name)
def function_with_invariants_ast(self, function_name):
return self.functions_with_invariants_ast(function_name)
def annotate_invariants(invariants):
annotated_functions = {}
for function_name in invariants:
try:
annotated_functions[function_name] = annotate_function_with_invariants(function_name, invariants[function_name])
except KeyError:
continue
return annotated_functions
def annotate_function_with_invariants(function_name, function_invariants):
function = globals()[function_name]
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_invariants(function_ast, function_invariants)
def annotate_function_ast_with_invariants(function_ast, function_invariants):
annotated_function_ast = EmbeddedInvariantTransformer(function_invariants).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Part 2: Preconditions
###Code
class PreconditionTransformer(ast.NodeTransformer):
def __init__(self, invariants):
self.invariants = invariants
super().__init__()
def preconditions(self):
preconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated precondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) < 0:
preconditions += assertion_ast.body
return preconditions
def insert_assertions(self, body):
preconditions = self.preconditions()
try:
docstring = body[0].value.s
except:
docstring = None
if docstring:
return [body[0]] + preconditions + body[1:]
else:
return preconditions + body
def visit_FunctionDef(self, node):
"""Add invariants to function"""
# print(ast.dump(node))
node.body = self.insert_assertions(node.body)
return node
class EmbeddedInvariantTransformer(PreconditionTransformer):
pass
with EmbeddedInvariantAnnotator() as annotator:
my_sqrt(5)
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Part 3: PostconditionsWe make a few simplifying assumptions: * Variables do not change during execution.* There is a single `return` statement at the end of the function.
###Code
class EmbeddedInvariantTransformer(PreconditionTransformer):
def postconditions(self):
postconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated postcondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) >= 0:
postconditions += assertion_ast.body
return postconditions
def insert_assertions(self, body):
new_body = super().insert_assertions(body)
postconditions = self.postconditions()
body_ends_with_return = isinstance(new_body[-1], ast.Return)
if body_ends_with_return:
saver = RETURN_VALUE + " = " + ast.unparse(new_body[-1].value)
else:
saver = RETURN_VALUE + " = None"
saver_ast = ast.parse(saver)
postconditions = [saver_ast] + postconditions
if body_ends_with_return:
return new_body[:-1] + postconditions + [new_body[-1]]
else:
return new_body + postconditions
with EmbeddedInvariantAnnotator() as annotator:
my_sqrt(5)
my_sqrt_def = annotator.functions_with_invariants()
###Output
_____no_output_____
###Markdown
Here's the full definition with included assertions:
###Code
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def.replace('my_sqrt', 'my_sqrt_annotated'))
with ExpectError():
my_sqrt_annotated(-1)
###Output
_____no_output_____
###Markdown
Here come some more examples:
###Code
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
print_sum(31, 45)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Mining Function SpecificationsIn the [chapter on assertions](Assertions.ipynb), we have seen how important it is to _check_ whether the result is as expected. In this chapter, we introduce a technique that allows us to _mine_ function specifications from a set of given executions, resulting in abstract and formal _descriptions_ of what the function expects and what it delivers.These so-called _dynamic invariants_ produce pre- and post-conditions over function arguments and variables from a set of executions. Within debugging, the resulting _assertions_ can immediately check whether function behavior has changed, but can also be useful to determine the characteristics of _failing_ runs (as opposed to _passing_ runs). Furthermore, the resulting specifications provide pre- and postconditions for formal program proofs, testing, and verification.This chapter is based on [a chapter with the same name in The Fuzzing Book](https://www.fuzzingbook.org/html/DynamicInvariants.html), which focuses on test generation.
###Code
from bookutils import YouTubeVideo
YouTubeVideo("HDu1olXFvv0")
###Output
_____no_output_____
###Markdown
**Prerequisites*** You should be familiar with tracing program executions, as in the [chapter on tracing](Tracer.ipynb).* Later in this section, we access the internal _abstract syntax tree_ representations of Python programs and transform them, as in the [chapter on tracking failure origins](Slicer.ipynb).
###Code
import bookutils
from Tracer import Tracer
# ignore
from typing import Sequence, Any, Callable, Tuple
from typing import Dict, Union, Set, List, cast, Optional
###Output
_____no_output_____
###Markdown
SynopsisTo [use the code provided in this chapter](Importing.ipynb), write```python>>> from debuggingbook.DynamicInvariants import ```and then make use of the following features.This chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:```python>>> def sum2(a, b): type: ignore>>> return a + b>>> with TypeAnnotator() as type_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.```python>>> print(type_annotator.typed_functions())def sum2(a: int, b: int) ->int: return a + b```The invariant annotator works in a similar fashion:```python>>> with InvariantAnnotator() as inv_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.```python>>> print(inv_annotator.functions_with_invariants())@precondition(lambda a, b: isinstance(a, int))@precondition(lambda a, b: isinstance(b, int))@postcondition(lambda return_value, a, b: a == return_value - b)@postcondition(lambda return_value, a, b: b == return_value - a)@postcondition(lambda return_value, a, b: isinstance(return_value, int))@postcondition(lambda return_value, a, b: return_value == a + b)@postcondition(lambda return_value, a, b: return_value == b + a)def sum2(a, b): type: ignore return a + b```Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs). The chapter gives details on how to customize the properties checked for. Specifications and AssertionsWhen implementing a function or program, one usually works against a _specification_ – a set of documented requirements to be satisfied by the code. Such specifications can come in natural language. A formal specification, however, allows the computer to check whether the specification is satisfied.In the [chapter on assertions](Assertions.ipynb), we have seen how _preconditions_ and _postconditions_ can describe what a function does. Consider the following (simple) square root function:
###Code
def square_root(x): # type: ignore
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
The assertion `assert p` checks the condition `p`; if it does not hold, execution is aborted. Here, the actual body is not yet written; we use the assertions as a specification of what `square_root()` _expects_, and what it _delivers_.The topmost assertion is the _precondition_, stating the requirements on the function arguments. The assertion at the end is the _postcondition_, stating the properties of the function result (including its relationship with the original arguments). Using these pre- and postconditions as a specification, we can now go and implement a square root function that satisfies them. Once implemented, we can have the assertions check at runtime whether `square_root()` works as expected. However, not every piece of code is developed with explicit specifications in the first place; let alone does most code comes with formal pre- and post-conditions. (Just take a look at the chapters in this book.) This is a pity: As Ken Thompson famously said, "Without specifications, there are no bugs – only surprises". It is also a problem for debugging, since, of course, debugging needs some specification such that we know what is wrong, and how to fix it. This raises the interesting question: Can we somehow _retrofit_ existing code with "specifications" that properly describe their behavior, allowing developers to simply _check_ them rather than having to write them from scratch? This is what we do in this chapter. Beyond Generic FailuresBefore we go into _mining_ specifications, let us first discuss why it could be useful to _have_ them. As a motivating example, consider the full implementation of `square_root()` from the [chapter on assertions](Assertions.ipynb):
###Code
import bookutils
def square_root(x): # type: ignore
"""Computes the square root of x, using the Newton-Raphson method"""
approx = None
guess = x / 2
while approx != guess:
approx = guess
guess = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
`square_root()` does not come with any functionality that would check types or values. Hence, it is easy for callers to make mistakes when calling `square_root()`:
###Code
from ExpectError import ExpectError, ExpectTimeout
with ExpectError():
square_root("foo")
with ExpectError():
x = square_root(0.0)
###Output
_____no_output_____
###Markdown
At least, the Python system catches these errors at runtime. The following call, however, simply lets the function enter an infinite loop:
###Code
with ExpectTimeout(1):
x = square_root(-1.0)
###Output
_____no_output_____
###Markdown
Our goal is to avoid such errors by _annotating_ functions with information that prevents errors like the above ones. The idea is to provide a _specification_ of expected properties – a specification that can then be checked at runtime or statically. \todo{Introduce the concept of *contract*.} Mining Data TypesFor our Python code, one of the most important "specifications" we need is *types*. Python being a "dynamically" typed language means that all data types are determined at run time; the code itself does not explicitly state whether a variable is an integer, a string, an array, a dictionary – or whatever. As _writer_ of Python code, omitting explicit type declarations may save time (and allows for some fun hacks). It is not sure whether a lack of types helps in _reading_ and _understanding_ code for humans. For a _computer_ trying to analyze code, the lack of explicit types is detrimental. If, say, a constraint solver, sees `if x:` and cannot know whether `x` is supposed to be a number or a string, this introduces an _ambiguity_. Such ambiguities may multiply over the entire analysis in a combinatorial explosion – or in the analysis yielding an overly inaccurate result. Python 3.6 and later allow data types as _annotations_ to function arguments (actually, to all variables) and return values. We can, for instance, state that `square_root()` is a function that accepts a floating-point value and returns one:
###Code
def square_root_with_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return square_root(x)
###Output
_____no_output_____
###Markdown
By default, such annotations are ignored by the Python interpreter. Therefore, one can still call `square_root_typed()` with a string as an argument and get the exact same result as above. However, one can make use of special _typechecking_ modules that would check types – _dynamically_ at runtime or _statically_ by analyzing the code without having to execute it. Excursion: Runtime Type Checking _Please note: `enforce` no longer runs on Python 3.7 and later, so this section is commented out_The Python `enforce` package provides a function decorator that automatically inserts type-checking code that is executed at runtime. Here is how to use it:
###Code
# import enforce
# @enforce.runtime_validation
# def square_root_with_checked_type_annotations(x: float) -> float:
# """Computes the square root of x, using the Newton-Raphson method"""
# return square_root(x)
###Output
_____no_output_____
###Markdown
Now, invoking `square_root_with_checked_type_annotations()` raises an exception when invoked with a type dfferent from the one declared:
###Code
# with ExpectError():
# square_root_with_checked_type_annotations(True)
###Output
_____no_output_____
###Markdown
Note that this error is not caught by the "untyped" variant, where passing a boolean value happily returns $\sqrt{1}$ as result.
###Code
# square_root(True)
###Output
_____no_output_____
###Markdown
In Python (and other languages), the boolean values `True` and `False` can be implicitly converted to the integers 1 and 0; however, it is hard to think of a call to `sqrt()` where this would not be an error.
###Code
from bookutils import quiz
quiz("What happens if we call "
"`square_root_with_checked_type_annotations(1)`?",
[
"`1` is automatically converted to float. It will pass.",
"`1` is a subtype of float. It will pass.",
"`1` is an integer, and no float. The type check will fail.",
"The function will fail for some other reason."
], '37035 // 12345')
###Output
_____no_output_____
###Markdown
"Prediction is very difficult, especially when the future is concerned" (Niels Bohr). We can find out by a simple experiment that `float` actually means `float` – and not `int`:
###Code
# with ExpectError(enforce.exceptions.RuntimeTypeError):
# square_root_with_checked_type_annotations(1)
###Output
_____no_output_____
###Markdown
To allow `int` as a type, we need to specify a _union_ of types.
###Code
# @enforce.runtime_validation
# def square_root_with_union_type(x: Union[int, float]) -> float:
# """Computes the square root of x, using the Newton-Raphson method"""
# return square_root(x)
# square_root_with_union_type(2)
# square_root_with_union_type(2.0)
# with ExpectError(enforce.exceptions.RuntimeTypeError):
# square_root_with_union_type("Two dot zero")
###Output
_____no_output_____
###Markdown
End of Excursion Static Type CheckingType annotations can also be checked _statically_ – that is, without even running the code. Let us create a simple Python file consisting of the above `square_root_typed()` definition and a bad invocation.
###Code
import inspect
import tempfile
f = tempfile.NamedTemporaryFile(mode='w', suffix='.py')
f.name
f.write(inspect.getsource(square_root))
f.write('\n')
f.write(inspect.getsource(square_root_with_type_annotations))
f.write('\n')
f.write("print(square_root_with_type_annotations('123'))\n")
f.flush()
###Output
_____no_output_____
###Markdown
These are the contents of our newly created Python file:
###Code
from bookutils import print_file
print_file(f.name, start_line_number=1)
###Output
_____no_output_____
###Markdown
[Mypy](http://mypy-lang.org) is a type checker for Python programs. As it checks types statically, types induce no overhead at runtime; plus, a static check can be faster than a lengthy series of tests with runtime type checking enabled. Let us see what `mypy` produces on the above file:
###Code
import subprocess
result = subprocess.run(["mypy", "--strict", f.name],
universal_newlines=True, stdout=subprocess.PIPE)
print(result.stdout.replace(f.name + ':', ''))
del f # Delete temporary file
###Output
_____no_output_____
###Markdown
We see that `mypy` complains about untyped function definitions such as `square_root()`; most important, however, it finds that the call to `square_root_with_type_annotations()` in the last line has the wrong type. With `mypy`, we can achieve the same type safety with Python as in statically typed languages – provided that we as programmers also produce the necessary type annotations. Is there a simple way to obtain these? Mining Type SpecificationsOur first task will be to mine type annotations (as part of the code) from _values_ we observe at run time. These type annotations would be _mined_ from actual function executions, _learning_ from (normal) runs what the expected argument and return types should be. By observing a series of calls such as these, we could infer that both `x` and the return value are of type `float`:
###Code
y = square_root(25.0)
y
y = square_root(2.0)
y
###Output
_____no_output_____
###Markdown
How can we mine types from executions? The answer is simple: 1. We _observe_ a function during execution2. We track the _types_ of its arguments3. We include these types as _annotations_ into the code.To do so, we can make use of Python's tracing facility we already observed in the [chapter on tracing executions](Tracer.ipynb). With every call to a function, we retrieve the arguments, their values, and their types. Tracing CallsTo observe argument types at runtime, we define a _tracer function_ that tracks the execution of `square_root()`, checking its arguments and return values. The `CallTracer` class is set to trace functions in a `with` block as follows:```pythonwith CallTracer() as tracer: function_to_be_tracked(...)info = tracer.collected_information()```To create the tracer, we build on the `Tracer` superclass as in the [chapter on tracing executions](Tracer.ipynb).
###Code
from types import FrameType
###Output
_____no_output_____
###Markdown
We start with two helper functions. `get_arguments()` returns a list of call arguments in the given call frame.
###Code
Arguments = List[Tuple[str, Any]]
def get_arguments(frame: FrameType) -> Arguments:
"""Return call arguments in the given frame"""
# When called, all arguments are local variables
local_variables = dict(frame.f_locals) # explicit copy
arguments = [(var, frame.f_locals[var]) for var in local_variables]
arguments.reverse() # Want same order as call
return arguments
###Output
_____no_output_____
###Markdown
`simple_call_string()` is a helper for logging that prints out calls in a user-friendly manner.
###Code
def simple_call_string(function_name: str, argument_list: Arguments,
return_value : Any = None) -> str:
"""Return function_name(arg[0], arg[1], ...) as a string"""
call = function_name + "(" + \
", ".join([var + "=" + repr(value)
for (var, value) in argument_list]) + ")"
if return_value is not None:
call += " = " + repr(return_value)
return call
###Output
_____no_output_____
###Markdown
Now for `CallTracer`. The constructor simply invokes the `Tracer` constructor:
###Code
class CallTracer(Tracer):
def __init__(self, log: bool = False, **kwargs: Any)-> None:
super().__init__(**kwargs)
self._log = log
self.reset()
def reset(self) -> None:
self._calls: Dict[str, List[Tuple[Arguments, Any]]] = {}
self._stack: List[Tuple[str, Arguments]] = []
###Output
_____no_output_____
###Markdown
The `traceit()` method does nothing yet; this is done in specialized subclasses. The `CallTracer` class implements a `traceit()` function that checks for function calls and returns:
###Code
class CallTracer(CallTracer):
def traceit(self, frame: FrameType, event: str, arg: Any) -> None:
"""Tracking function: Record all calls and all args"""
if event == "call":
self.trace_call(frame, event, arg)
elif event == "return":
self.trace_return(frame, event, arg)
###Output
_____no_output_____
###Markdown
`trace_call()` is called when a function is called; it retrieves the function name and current arguments, and saves them on a stack.
###Code
class CallTracer(CallTracer):
def trace_call(self, frame: FrameType, event: str, arg: Any) -> None:
"""Save current function name and args on the stack"""
code = frame.f_code
function_name = code.co_name
arguments = get_arguments(frame)
self._stack.append((function_name, arguments))
if self._log:
print(simple_call_string(function_name, arguments))
###Output
_____no_output_____
###Markdown
When the function returns, `trace_return()` is called. We now also have the return value. We log the whole call with arguments and return value (if desired) and save it in our list of calls.
###Code
class CallTracer(CallTracer):
def trace_return(self, frame: FrameType, event: str, arg: Any) -> None:
"""Get return value and store complete call with arguments and return value"""
code = frame.f_code
function_name = code.co_name
return_value = arg
# TODO: Could call get_arguments() here to also retrieve _final_ values of argument variables
called_function_name, called_arguments = self._stack.pop()
assert function_name == called_function_name
if self._log:
print(simple_call_string(function_name, called_arguments), "returns", return_value)
self.add_call(function_name, called_arguments, return_value)
###Output
_____no_output_____
###Markdown
`add_call()` saves the calls in a list; each function name has its own list.
###Code
class CallTracer(CallTracer):
def add_call(self, function_name: str, arguments: Arguments,
return_value: Any = None) -> None:
"""Add given call to list of calls"""
if function_name not in self._calls:
self._calls[function_name] = []
self._calls[function_name].append((arguments, return_value))
###Output
_____no_output_____
###Markdown
we can retrieve the list of calls, either for a given function name (`calls()`),or for all functions (`all_calls()`).
###Code
class CallTracer(CallTracer):
def calls(self, function_name: str) -> List[Tuple[Arguments, Any]]:
"""Return list of calls for `function_name`."""
return self._calls[function_name]
class CallTracer(CallTracer):
def all_calls(self) -> Dict[str, List[Tuple[Arguments, Any]]]:
"""
Return list of calls for function_name,
or a mapping function_name -> calls for all functions tracked
"""
return self._calls
###Output
_____no_output_____
###Markdown
Let us now put this to use. We turn on logging to track the individual calls and their return values:
###Code
with CallTracer(log=True) as tracer:
y = square_root(25)
y = square_root(2.0)
###Output
_____no_output_____
###Markdown
After execution, we can retrieve the individual calls:
###Code
calls = tracer.calls('square_root')
calls
###Output
_____no_output_____
###Markdown
Each call is a pair (`argument_list`, `return_value`), where `argument_list` is a list of pairs (`parameter_name`, `value`).
###Code
square_root_argument_list, square_root_return_value = calls[0]
simple_call_string('square_root', square_root_argument_list, square_root_return_value)
###Output
_____no_output_____
###Markdown
If the function does not return a value, `return_value` is `None`.
###Code
def hello(name: str) -> None:
print("Hello,", name)
with CallTracer() as tracer:
hello("world")
hello_calls = tracer.calls('hello')
hello_calls
hello_argument_list, hello_return_value = hello_calls[0]
simple_call_string('hello', hello_argument_list, hello_return_value)
###Output
_____no_output_____
###Markdown
Getting TypesDespite what you may have read or heard, Python actually _is_ a typed language. It is just that it is _dynamically typed_ – types are used and checked only at runtime (rather than declared in the code, where they can be _statically checked_ at compile time). We can thus retrieve types of all values within Python:
###Code
type(4)
type(2.0)
type([4])
###Output
_____no_output_____
###Markdown
We can retrieve the type of the first argument to `square_root()`:
###Code
parameter, value = square_root_argument_list[0]
parameter, type(value)
###Output
_____no_output_____
###Markdown
as well as the type of the return value:
###Code
type(square_root_return_value)
###Output
_____no_output_____
###Markdown
Hence, we see that (so far), `square_root()` is a function taking (among others) integers and returning floats. We could declare `square_root()` as:
###Code
def square_root_annotated(x: int) -> float:
return square_root(x)
###Output
_____no_output_____
###Markdown
This is a representation we could place in a static type checker, allowing to check whether calls to `square_root()` actually pass a number. A dynamic type checker could run such checks at runtime. By default, Python does not do anything with such annotations. However, tools can access annotations from functions and other objects:
###Code
square_root_annotated.__annotations__
###Output
_____no_output_____
###Markdown
This is how run-time checkers access the annotations to check against. Annotating Functions with TypesOur plan is to annotate functions automatically, based on the types we have seen. Our aim is to build a class `TypeAnnotator` that can be used as follows. First, it would track some execution:```pythonwith TypeAnnotator() as annotator: some_function_call()```After tracking, `TypeAnnotator` would provide appropriate methods to access (type-)annotated versions of the function seen:```pythonprint(annotator.typed_functions())```Let us put the pieces together to build `TypeAnnotator`. Excursion: Accessing Function Structure To annotate functions, we need to convert a function into a tree representation (called _abstract syntax trees_, or ASTs) and back; we already have seen these in the chapter on [tracking value origins](Slicer.ipynb).
###Code
import ast
import inspect
###Output
_____no_output_____
###Markdown
We can get the source of a Python function using `inspect.getsource()`. (Note that this does not work for functions defined in other notebooks.)
###Code
square_root_source = inspect.getsource(square_root)
square_root_source
###Output
_____no_output_____
###Markdown
To view these in a visually pleasing form, our function `print_content(s, suffix)` formats and highlights the string `s` as if it were a file with ending `suffix`. We can thus view (and highlight) the source as if it were a Python file:
###Code
from bookutils import print_content
print_content(square_root_source, '.py')
###Output
_____no_output_____
###Markdown
Parsing this gives us an abstract syntax tree (AST) – a representation of the program in tree form.
###Code
square_root_ast = ast.parse(square_root_source)
###Output
_____no_output_____
###Markdown
What does this AST look like? The helper functions `ast.dump()` (textual output) and `showast.show_ast()` (graphical output with [showast](https://github.com/hchasestevens/show_ast)) allow us to inspect the structure of the tree. We see that the function starts as a `FunctionDef` with name and arguments, followed by a body, which is a list of statements of type `Expr` (the docstring), type `Assign` (assignments), `While` (while loop with its own body), and finally `Return`.
###Code
print(ast.dump(square_root_ast, indent=4))
###Output
_____no_output_____
###Markdown
Too much text for you? This graphical representation may make things simpler.
###Code
from bookutils import show_ast
show_ast(square_root_ast)
###Output
_____no_output_____
###Markdown
The function `ast.unparse()` converts such a tree back into the more familiar textual Python code representation. Comments are gone, and there may be more (or fewer) parentheses than before, but the result has the same semantics:
###Code
print_content(ast.unparse(square_root_ast), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: Annotating Functions with Given Types Let us now go and transform ASTs to add type annotations. We start with a helper function `parse_type(name)` which parses a type name into an AST.
###Code
def parse_type(name: str) -> ast.expr:
class ValueVisitor(ast.NodeVisitor):
def visit_Expr(self, node: ast.Expr) -> None:
self.value_node = node.value
tree = ast.parse(name)
name_visitor = ValueVisitor()
name_visitor.visit(tree)
return name_visitor.value_node
print(ast.dump(parse_type('int')))
print(ast.dump(parse_type('[object]')))
###Output
_____no_output_____
###Markdown
We now define a helper function that actually adds type annotations to a function AST. The `TypeTransformer` class builds on the Python standard library `ast.NodeTransformer` infrastructure. It would be called as```python TypeTransformer({'x': 'int'}, 'float').visit(ast)```to annotate the arguments of `square_root()`: `x` with `int`, and the return type with `float`. The returned AST can then be unparsed, compiled or analyzed.
###Code
class TypeTransformer(ast.NodeTransformer):
def __init__(self, argument_types: Dict[str, str], return_type: Optional[str] = None):
self.argument_types = argument_types
self.return_type = return_type
super().__init__()
###Output
_____no_output_____
###Markdown
The core of `TypeTransformer` is the method `visit_FunctionDef()`, which is called for every function definition in the AST. Its argument `node` is the subtree of the function definition to be transformed. Our implementation accesses the individual arguments and invokes `annotate_args()` on them; it also sets the return type in the `returns` attribute of the node.
###Code
class TypeTransformer(TypeTransformer):
def visit_FunctionDef(self, node: ast.FunctionDef) -> ast.FunctionDef:
"""Add annotation to function"""
# Set argument types
new_args = []
for arg in node.args.args:
new_args.append(self.annotate_arg(arg))
new_arguments = ast.arguments(
node.args.posonlyargs,
new_args,
node.args.vararg,
node.args.kwonlyargs,
node.args.kw_defaults,
node.args.kwarg,
node.args.defaults
)
# Set return type
if self.return_type is not None:
node.returns = parse_type(self.return_type)
return ast.copy_location(
ast.FunctionDef(node.name, new_arguments,
node.body, node.decorator_list,
node.returns), node)
###Output
_____no_output_____
###Markdown
Each argument gets its own annotation, taken from the types originally passed to the class:
###Code
class TypeTransformer(TypeTransformer):
def annotate_arg(self, arg: ast.arg) -> ast.arg:
"""Add annotation to single function argument"""
arg_name = arg.arg
if arg_name in self.argument_types:
arg.annotation = parse_type(self.argument_types[arg_name])
return arg
###Output
_____no_output_____
###Markdown
Does this work? Let us annotate the AST from `square_root()` with types for the arguments and return types:
###Code
new_ast = TypeTransformer({'x': 'int'}, 'float').visit(square_root_ast)
###Output
_____no_output_____
###Markdown
When we unparse the new AST, we see that the annotations actually are present:
###Code
print_content(ast.unparse(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Similarly, we can annotate the `hello()` function from above:
###Code
hello_source = inspect.getsource(hello)
hello_ast = ast.parse(hello_source)
new_ast = TypeTransformer({'name': 'str'}, 'None').visit(hello_ast)
print_content(ast.unparse(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Mining Function SpecificationsIn the [chapter on assertions](Assertions.ipynb), we have seen how important it is to _check_ whether the result is as expected. In this chapter, we introduce a technique that allows us to _mine_ function specifications from a set of given executions, resulting in abstract and formal _descriptions_ of what the function expects and what it delivers.These so-called _dynamic invariants_ produce pre- and post-conditions over function arguments and variables from a set of executions. Within debugging, the resulting _assertions_ can immediately check whether function behavior has changed, but can also be useful to determine the characteristics of _failing_ runs (as opposed to _passing_ runs). Furthermore, the resulting specifications provide pre- and postconditions for formal program proofs, testing, and verification.This chapter is based on [a chapter with the same name in The Fuzzing Book](https://www.fuzzingbook.org/html/DynamicInvariants.html), which focuses on test generation.
###Code
from bookutils import YouTubeVideo
YouTubeVideo("HDu1olXFvv0")
###Output
_____no_output_____
###Markdown
**Prerequisites*** You should be familiar with tracing program executions, as in the [chapter on tracing](Tracer.ipynb).* Later in this section, we access the internal _abstract syntax tree_ representations of Python programs and transform them, as in the [chapter on tracking failure origins](Slicer.ipynb).
###Code
import bookutils
from Tracer import Tracer
###Output
_____no_output_____
###Markdown
SynopsisTo [use the code provided in this chapter](Importing.ipynb), write```python>>> from debuggingbook.DynamicInvariants import ```and then make use of the following features.This chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:```python>>> def sum2(a, b): type: ignore>>> return a + b>>> with TypeAnnotator() as type_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.```python>>> print(type_annotator.typed_functions())def sum2(a: int, b: int) ->int: return a + b```The invariant annotator works in a similar fashion:```python>>> with InvariantAnnotator() as inv_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.```python>>> print(inv_annotator.functions_with_invariants())@precondition(lambda a, b: isinstance(a, int))@precondition(lambda a, b: isinstance(b, int))@postcondition(lambda return_value, a, b: a == return_value - b)@postcondition(lambda return_value, a, b: b == return_value - a)@postcondition(lambda return_value, a, b: isinstance(return_value, int))@postcondition(lambda return_value, a, b: return_value == a + b)@postcondition(lambda return_value, a, b: return_value == b + a)def sum2(a, b): type: ignore return a + b```Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs). The chapter gives details on how to customize the properties checked for. Specifications and AssertionsWhen implementing a function or program, one usually works against a _specification_ – a set of documented requirements to be satisfied by the code. Such specifications can come in natural language. A formal specification, however, allows the computer to check whether the specification is satisfied.In the [chapter on assertions](Assertions.ipynb), we have seen how _preconditions_ and _postconditions_ can describe what a function does. Consider the following (simple) square root function:
###Code
def square_root(x): # type: ignore
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
The assertion `assert p` checks the condition `p`; if it does not hold, execution is aborted. Here, the actual body is not yet written; we use the assertions as a specification of what `square_root()` _expects_, and what it _delivers_.The topmost assertion is the _precondition_, stating the requirements on the function arguments. The assertion at the end is the _postcondition_, stating the properties of the function result (including its relationship with the original arguments). Using these pre- and postconditions as a specification, we can now go and implement a square root function that satisfies them. Once implemented, we can have the assertions check at runtime whether `square_root()` works as expected. However, not every piece of code is developed with explicit specifications in the first place; let alone does most code comes with formal pre- and post-conditions. (Just take a look at the chapters in this book.) This is a pity: As Ken Thompson famously said, "Without specifications, there are no bugs – only surprises". It is also a problem for debugging, since, of course, debugging needs some specification such that we know what is wrong, and how to fix it. This raises the interesting question: Can we somehow _retrofit_ existing code with "specifications" that properly describe their behavior, allowing developers to simply _check_ them rather than having to write them from scratch? This is what we do in this chapter. Beyond Generic FailuresBefore we go into _mining_ specifications, let us first discuss why it could be useful to _have_ them. As a motivating example, consider the full implementation of `square_root()` from the [chapter on assertions](Assertions.ipynb):
###Code
import bookutils
def square_root(x): # type: ignore
"""Computes the square root of x, using the Newton-Raphson method"""
approx = None
guess = x / 2
while approx != guess:
approx = guess
guess = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
`square_root()` does not come with any functionality that would check types or values. Hence, it is easy for callers to make mistakes when calling `square_root()`:
###Code
from ExpectError import ExpectError, ExpectTimeout
with ExpectError():
square_root("foo")
with ExpectError():
x = square_root(0.0)
###Output
_____no_output_____
###Markdown
At least, the Python system catches these errors at runtime. The following call, however, simply lets the function enter an infinite loop:
###Code
with ExpectTimeout(1):
x = square_root(-1.0)
###Output
_____no_output_____
###Markdown
Our goal is to avoid such errors by _annotating_ functions with information that prevents errors like the above ones. The idea is to provide a _specification_ of expected properties – a specification that can then be checked at runtime or statically. \todo{Introduce the concept of *contract*.} Mining Data TypesFor our Python code, one of the most important "specifications" we need is *types*. Python being a "dynamically" typed language means that all data types are determined at run time; the code itself does not explicitly state whether a variable is an integer, a string, an array, a dictionary – or whatever. As _writer_ of Python code, omitting explicit type declarations may save time (and allows for some fun hacks). It is not sure whether a lack of types helps in _reading_ and _understanding_ code for humans. For a _computer_ trying to analyze code, the lack of explicit types is detrimental. If, say, a constraint solver, sees `if x:` and cannot know whether `x` is supposed to be a number or a string, this introduces an _ambiguity_. Such ambiguities may multiply over the entire analysis in a combinatorial explosion – or in the analysis yielding an overly inaccurate result. Python 3.6 and later allow data types as _annotations_ to function arguments (actually, to all variables) and return values. We can, for instance, state that `square_root()` is a function that accepts a floating-point value and returns one:
###Code
def square_root_with_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return square_root(x)
###Output
_____no_output_____
###Markdown
By default, such annotations are ignored by the Python interpreter. Therefore, one can still call `square_root_typed()` with a string as an argument and get the exact same result as above. However, one can make use of special _typechecking_ modules that would check types – _dynamically_ at runtime or _statically_ by analyzing the code without having to execute it. Runtime Type CheckingThe Python `enforce` package provides a function decorator that automatically inserts type-checking code that is executed at runtime. Here is how to use it:
###Code
import enforce
@enforce.runtime_validation
def square_root_with_checked_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return square_root(x)
###Output
_____no_output_____
###Markdown
Now, invoking `square_root_with_checked_type_annotations()` raises an exception when invoked with a type dfferent from the one declared:
###Code
with ExpectError():
square_root_with_checked_type_annotations(True)
###Output
_____no_output_____
###Markdown
Note that this error is not caught by the "untyped" variant, where passing a boolean value happily returns $\sqrt{1}$ as result.
###Code
square_root(True)
###Output
_____no_output_____
###Markdown
In Python (and other languages), the boolean values `True` and `False` can be implicitly converted to the integers 1 and 0; however, it is hard to think of a call to `sqrt()` where this would not be an error.
###Code
from bookutils import quiz
quiz("What happens if we call "
"`square_root_with_checked_type_annotations(1)`?",
[
"`1` is automatically converted to float. It will pass.",
"`1` is a subtype of float. It will pass.",
"`1` is an integer, and no float. The type check will fail.",
"The function will fail for some other reason."
], '37035 // 12345')
###Output
_____no_output_____
###Markdown
"Prediction is very difficult, especially when the future is concerned" (Niels Bohr). We can find out by a simple experiment that `float` actually means `float` – and not `int`:
###Code
with ExpectError(enforce.exceptions.RuntimeTypeError):
square_root_with_checked_type_annotations(1)
###Output
_____no_output_____
###Markdown
To allow `int` as a type, we need to specify a _union_ of types.
###Code
from typing import Union, Optional
@enforce.runtime_validation
def square_root_with_union_type(x: Union[int, float]) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return square_root(x)
square_root_with_union_type(2)
square_root_with_union_type(2.0)
with ExpectError(enforce.exceptions.RuntimeTypeError):
square_root_with_union_type("Two dot zero")
###Output
_____no_output_____
###Markdown
Static Type CheckingType annotations can also be checked _statically_ – that is, without even running the code. Let us create a simple Python file consisting of the above `square_root_typed()` definition and a bad invocation.
###Code
import inspect
import tempfile
f = tempfile.NamedTemporaryFile(mode='w', suffix='.py')
f.name
f.write(inspect.getsource(square_root))
f.write('\n')
f.write(inspect.getsource(square_root_with_type_annotations))
f.write('\n')
f.write("print(square_root_with_type_annotations('123'))\n")
f.flush()
###Output
_____no_output_____
###Markdown
These are the contents of our newly created Python file:
###Code
from bookutils import print_file
print_file(f.name, start_line_number=1)
###Output
_____no_output_____
###Markdown
[Mypy](http://mypy-lang.org) is a type checker for Python programs. As it checks types statically, types induce no overhead at runtime; plus, a static check can be faster than a lengthy series of tests with runtime type checking enabled. Let us see what `mypy` produces on the above file:
###Code
import subprocess
result = subprocess.run(["mypy", "--strict", f.name],
universal_newlines=True, stdout=subprocess.PIPE)
print(result.stdout.replace(f.name + ':', ''))
del f # Delete temporary file
###Output
_____no_output_____
###Markdown
We see that `mypy` complains about untyped function definitions such as `square_root()`; most important, however, it finds that the call to `square_root_with_type_annotations()` in the last line has the wrong type. With `mypy`, we can achieve the same type safety with Python as in statically typed languages – provided that we as programmers also produce the necessary type annotations. Is there a simple way to obtain these? Mining Type SpecificationsOur first task will be to mine type annotations (as part of the code) from _values_ we observe at run time. These type annotations would be _mined_ from actual function executions, _learning_ from (normal) runs what the expected argument and return types should be. By observing a series of calls such as these, we could infer that both `x` and the return value are of type `float`:
###Code
y = square_root(25.0)
y
y = square_root(2.0)
y
###Output
_____no_output_____
###Markdown
How can we mine types from executions? The answer is simple: 1. We _observe_ a function during execution2. We track the _types_ of its arguments3. We include these types as _annotations_ into the code.To do so, we can make use of Python's tracing facility we already observed in the [chapter on tracing executions](Tracer.ipynb). With every call to a function, we retrieve the arguments, their values, and their types. Tracing CallsTo observe argument types at runtime, we define a _tracer function_ that tracks the execution of `square_root()`, checking its arguments and return values. The `CallTracer` class is set to trace functions in a `with` block as follows:```pythonwith CallTracer() as tracer: function_to_be_tracked(...)info = tracer.collected_information()```To create the tracer, we build on the `Tracer` superclass as in the [chapter on tracing executions](Tracer.ipynb).
###Code
from typing import Sequence, Any, Callable, Optional, Type, Tuple, Any
from typing import Dict, Union, Set, List, cast, TypeVar
from types import FrameType, TracebackType
###Output
_____no_output_____
###Markdown
We start with two helper functions. `get_arguments()` returns a list of call arguments in the given call frame.
###Code
Arguments = List[Tuple[str, Any]]
def get_arguments(frame: FrameType) -> Arguments:
"""Return call arguments in the given frame"""
# When called, all arguments are local variables
arguments = [(var, frame.f_locals[var]) for var in frame.f_locals]
arguments.reverse() # Want same order as call
return arguments
###Output
_____no_output_____
###Markdown
`simple_call_string()` is a helper for logging that prints out calls in a user-friendly manner.
###Code
def simple_call_string(function_name: str, argument_list: Arguments,
return_value : Any = None) -> str:
"""Return function_name(arg[0], arg[1], ...) as a string"""
call = function_name + "(" + \
", ".join([var + "=" + repr(value)
for (var, value) in argument_list]) + ")"
if return_value is not None:
call += " = " + repr(return_value)
return call
###Output
_____no_output_____
###Markdown
Now for `CallTracer`. The constructor simply invokes the `Tracer` constructor:
###Code
class CallTracer(Tracer):
def __init__(self, log: bool = False, **kwargs: Any)-> None:
super().__init__(**kwargs)
self._log = log
self.reset()
def reset(self) -> None:
self._calls: Dict[str, List[Tuple[Arguments, Any]]] = {}
self._stack: List[Tuple[str, Arguments]] = []
###Output
_____no_output_____
###Markdown
The `traceit()` method does nothing yet; this is done in specialized subclasses. The `CallTracer` class implements a `traceit()` function that checks for function calls and returns:
###Code
class CallTracer(CallTracer):
def traceit(self, frame: FrameType, event: str, arg: Any) -> None:
"""Tracking function: Record all calls and all args"""
if event == "call":
self.trace_call(frame, event, arg)
elif event == "return":
self.trace_return(frame, event, arg)
###Output
_____no_output_____
###Markdown
`trace_call()` is called when a function is called; it retrieves the function name and current arguments, and saves them on a stack.
###Code
class CallTracer(CallTracer):
def trace_call(self, frame: FrameType, event: str, arg: Any) -> None:
"""Save current function name and args on the stack"""
code = frame.f_code
function_name = code.co_name
arguments = get_arguments(frame)
self._stack.append((function_name, arguments))
if self._log:
print(simple_call_string(function_name, arguments))
###Output
_____no_output_____
###Markdown
When the function returns, `trace_return()` is called. We now also have the return value. We log the whole call with arguments and return value (if desired) and save it in our list of calls.
###Code
class CallTracer(CallTracer):
def trace_return(self, frame: FrameType, event: str, arg: Any) -> None:
"""Get return value and store complete call with arguments and return value"""
code = frame.f_code
function_name = code.co_name
return_value = arg
# TODO: Could call get_arguments() here to also retrieve _final_ values of argument variables
called_function_name, called_arguments = self._stack.pop()
assert function_name == called_function_name
if self._log:
print(simple_call_string(function_name, called_arguments), "returns", return_value)
self.add_call(function_name, called_arguments, return_value)
###Output
_____no_output_____
###Markdown
`add_call()` saves the calls in a list; each function name has its own list.
###Code
class CallTracer(CallTracer):
def add_call(self, function_name: str, arguments: Arguments,
return_value: Any = None) -> None:
"""Add given call to list of calls"""
if function_name not in self._calls:
self._calls[function_name] = []
self._calls[function_name].append((arguments, return_value))
###Output
_____no_output_____
###Markdown
we can retrieve the list of calls, either for a given function name (`calls()`),or for all functions (`all_calls()`).
###Code
class CallTracer(CallTracer):
def calls(self, function_name: str) -> List[Tuple[Arguments, Any]]:
"""Return list of calls for `function_name`."""
return self._calls[function_name]
class CallTracer(CallTracer):
def all_calls(self) -> Dict[str, List[Tuple[Arguments, Any]]]:
"""
Return list of calls for function_name,
or a mapping function_name -> calls for all functions tracked
"""
return self._calls
###Output
_____no_output_____
###Markdown
Let us now put this to use. We turn on logging to track the individual calls and their return values:
###Code
with CallTracer(log=True) as tracer:
y = square_root(25)
y = square_root(2.0)
###Output
_____no_output_____
###Markdown
After execution, we can retrieve the individual calls:
###Code
calls = tracer.calls('square_root')
calls
###Output
_____no_output_____
###Markdown
Each call is a pair (`argument_list`, `return_value`), where `argument_list` is a list of pairs (`parameter_name`, `value`).
###Code
square_root_argument_list, square_root_return_value = calls[0]
simple_call_string('square_root', square_root_argument_list, square_root_return_value)
###Output
_____no_output_____
###Markdown
If the function does not return a value, `return_value` is `None`.
###Code
def hello(name: str) -> None:
print("Hello,", name)
with CallTracer() as tracer:
hello("world")
hello_calls = tracer.calls('hello')
hello_calls
hello_argument_list, hello_return_value = hello_calls[0]
simple_call_string('hello', hello_argument_list, hello_return_value)
###Output
_____no_output_____
###Markdown
Getting TypesDespite what you may have read or heard, Python actually _is_ a typed language. It is just that it is _dynamically typed_ – types are used and checked only at runtime (rather than declared in the code, where they can be _statically checked_ at compile time). We can thus retrieve types of all values within Python:
###Code
type(4)
type(2.0)
type([4])
###Output
_____no_output_____
###Markdown
We can retrieve the type of the first argument to `square_root()`:
###Code
parameter, value = square_root_argument_list[0]
parameter, type(value)
###Output
_____no_output_____
###Markdown
as well as the type of the return value:
###Code
type(square_root_return_value)
###Output
_____no_output_____
###Markdown
Hence, we see that (so far), `square_root()` is a function taking (among others) integers and returning floats. We could declare `square_root()` as:
###Code
def square_root_annotated(x: int) -> float:
return square_root(x)
###Output
_____no_output_____
###Markdown
This is a representation we could place in a static type checker, allowing to check whether calls to `square_root()` actually pass a number. A dynamic type checker could run such checks at runtime. By default, Python does not do anything with such annotations. However, tools can access annotations from functions and other objects:
###Code
square_root_annotated.__annotations__
###Output
_____no_output_____
###Markdown
This is how run-time checkers access the annotations to check against. Annotating Functions with TypesOur plan is to annotate functions automatically, based on the types we have seen. Our aim is to build a class `TypeAnnotator` that can be used as follows. First, it would track some execution:```pythonwith TypeAnnotator() as annotator: some_function_call()```After tracking, `TypeAnnotator` would provide appropriate methods to access (type-)annotated versions of the function seen:```pythonprint(annotator.typed_functions())```Let us put the pieces together to build `TypeAnnotator`. Excursion: Accessing Function Structure To annotate functions, we need to convert a function into a tree representation (called _abstract syntax trees_, or ASTs) and back; we already have seen these in the chapter on [tracking value origins](Slicer.ipynb).
###Code
import ast
import inspect
import astor
###Output
_____no_output_____
###Markdown
We can get the source of a Python function using `inspect.getsource()`. (Note that this does not work for functions defined in other notebooks.)
###Code
square_root_source = inspect.getsource(square_root)
square_root_source
###Output
_____no_output_____
###Markdown
To view these in a visually pleasing form, our function `print_content(s, suffix)` formats and highlights the string `s` as if it were a file with ending `suffix`. We can thus view (and highlight) the source as if it were a Python file:
###Code
from bookutils import print_content
print_content(square_root_source, '.py')
###Output
_____no_output_____
###Markdown
Parsing this gives us an abstract syntax tree (AST) – a representation of the program in tree form.
###Code
square_root_ast = ast.parse(square_root_source)
###Output
_____no_output_____
###Markdown
What does this AST look like? The helper functions `astor.dump_tree()` (textual output) and `showast.show_ast()` (graphical output with [showast](https://github.com/hchasestevens/show_ast)) allow us to inspect the structure of the tree. We see that the function starts as a `FunctionDef` with name and arguments, followed by a body, which is a list of statements of type `Expr` (the docstring), type `Assign` (assignments), `While` (while loop with its own body), and finally `Return`.
###Code
print(astor.dump_tree(square_root_ast))
###Output
_____no_output_____
###Markdown
Too much text for you? This graphical representation may make things simpler.
###Code
from bookutils import show_ast
show_ast(square_root_ast)
###Output
_____no_output_____
###Markdown
The function `astor.to_source()` converts such a tree back into the more familiar textual Python code representation. Comments are gone, and there may be more parentheses than before, but the result has the same semantics:
###Code
print_content(astor.to_source(square_root_ast), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: Annotating Functions with Given Types Let us now go and transform ASTs to add type annotations. We start with a helper function `parse_type(name)` which parses a type name into an AST.
###Code
def parse_type(name: str) -> ast.expr:
class ValueVisitor(ast.NodeVisitor):
def visit_Expr(self, node: ast.Expr) -> None:
self.value_node = node.value
tree = ast.parse(name)
name_visitor = ValueVisitor()
name_visitor.visit(tree)
return name_visitor.value_node
print(astor.dump_tree(parse_type('int')))
print(astor.dump_tree(parse_type('[object]')))
###Output
_____no_output_____
###Markdown
We now define a helper function that actually adds type annotations to a function AST. The `TypeTransformer` class builds on the Python standard library `ast.NodeTransformer` infrastructure. It would be called as```python TypeTransformer({'x': 'int'}, 'float').visit(ast)```to annotate the arguments of `square_root()`: `x` with `int`, and the return type with `float`. The returned AST can then be unparsed, compiled or analyzed.
###Code
class TypeTransformer(ast.NodeTransformer):
def __init__(self, argument_types: Dict[str, str], return_type: Optional[str] = None):
self.argument_types = argument_types
self.return_type = return_type
super().__init__()
###Output
_____no_output_____
###Markdown
The core of `TypeTransformer` is the method `visit_FunctionDef()`, which is called for every function definition in the AST. Its argument `node` is the subtree of the function definition to be transformed. Our implementation accesses the individual arguments and invokes `annotate_args()` on them; it also sets the return type in the `returns` attribute of the node.
###Code
class TypeTransformer(TypeTransformer):
def visit_FunctionDef(self, node: ast.FunctionDef) -> ast.FunctionDef:
"""Add annotation to function"""
# Set argument types
new_args = []
for arg in node.args.args:
new_args.append(self.annotate_arg(arg))
new_arguments = ast.arguments(
new_args,
node.args.vararg,
node.args.kwonlyargs,
node.args.kw_defaults,
node.args.kwarg,
node.args.defaults
)
# Set return type
if self.return_type is not None:
node.returns = parse_type(self.return_type)
return ast.copy_location(
ast.FunctionDef(node.name, new_arguments,
node.body, node.decorator_list,
node.returns), node)
###Output
_____no_output_____
###Markdown
Each argument gets its own annotation, taken from the types originally passed to the class:
###Code
class TypeTransformer(TypeTransformer):
def annotate_arg(self, arg: ast.arg) -> ast.arg:
"""Add annotation to single function argument"""
arg_name = arg.arg
if arg_name in self.argument_types:
arg.annotation = parse_type(self.argument_types[arg_name])
return arg
###Output
_____no_output_____
###Markdown
Does this work? Let us annotate the AST from `square_root()` with types for the arguments and return types:
###Code
new_ast = TypeTransformer({'x': 'int'}, 'float').visit(square_root_ast)
###Output
_____no_output_____
###Markdown
When we unparse the new AST, we see that the annotations actually are present:
###Code
print_content(astor.to_source(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Similarly, we can annotate the `hello()` function from above:
###Code
hello_source = inspect.getsource(hello)
hello_ast = ast.parse(hello_source)
new_ast = TypeTransformer({'name': 'str'}, 'None').visit(hello_ast)
print_content(astor.to_source(new_ast), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: Annotating Functions with Mined Types Let us now annotate functions with types mined at runtime. We start with a simple function `type_string()` that determines the appropriate type of a given value (as a string):
###Code
def type_string(value: Any) -> str:
return type(value).__name__
type_string(4)
type_string([])
###Output
_____no_output_____
###Markdown
For composite structures, `type_string()` does not examine element types; hence, the type of `[3]` is simply `list` instead of, say, `list[int]`. For now, `list` will do fine.
###Code
type_string([3])
###Output
_____no_output_____
###Markdown
`type_string()` will be used to infer the types of argument values found at runtime, as returned by `CallTracer.all_calls()`:
###Code
with CallTracer() as tracer:
y = square_root(25.0)
y = square_root(2.0)
tracer.all_calls()
###Output
_____no_output_____
###Markdown
The function `annotate_types()` takes such a list of calls and annotates each function listed:
###Code
def annotate_types(calls: Dict[str, List[Tuple[Arguments, Any]]]) -> Dict[str, ast.AST]:
annotated_functions = {}
for function_name in calls:
try:
annotated_functions[function_name] = \
annotate_function_with_types(function_name, calls[function_name])
except KeyError:
continue
return annotated_functions
###Output
_____no_output_____
###Markdown
For each function, we get the source and its AST and then get to the actual annotation in `annotate_function_ast_with_types()`:
###Code
def annotate_function_with_types(function_name: str,
function_calls: List[Tuple[Arguments, Any]]) -> ast.AST:
function = globals()[function_name] # May raise KeyError for internal functions
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_types(function_ast, function_calls)
###Output
_____no_output_____
###Markdown
The function `annotate_function_ast_with_types()` invokes the `TypeTransformer` with the calls seen, and for each call, iterate over the arguments, determine their types, and annotate the AST with these. The universal type `Any` is used when we encounter type conflicts, which we will discuss below.
###Code
from typing import Any
def annotate_function_ast_with_types(function_ast: ast.AST,
function_calls: List[Tuple[Arguments, Any]]) -> ast.AST:
parameter_types: Dict[str, str] = {}
return_type = None
for calls_seen in function_calls:
args, return_value = calls_seen
if return_value:
if return_type and return_type != type_string(return_value):
return_type = 'Any'
else:
return_type = type_string(return_value)
for parameter, value in args:
try:
different_type = (parameter_types[parameter] !=
type_string(value))
except KeyError:
different_type = False
if different_type:
parameter_types[parameter] = 'Any'
else:
parameter_types[parameter] = type_string(value)
annotated_function_ast = \
TypeTransformer(parameter_types, return_type).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Here is `square_root()` annotated with the types recorded usign the tracer, above.
###Code
print_content(astor.to_source(annotate_types(tracer.all_calls())['square_root']), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: A Type Annotator Class Let us bring all of this together in a single class `TypeAnnotator` that first tracks calls of functions and then allows to access the AST (and the source code form) of the tracked functions annotated with types. The method `typed_functions()` returns the annotated functions as a string; `typed_functions_ast()` returns their AST.
###Code
class TypeTracer(CallTracer):
pass
class TypeAnnotator(TypeTracer):
def typed_functions_ast(self) -> Dict[str, ast.AST]:
return annotate_types(self.all_calls())
def typed_function_ast(self, function_name: str) -> ast.AST:
return annotate_function_with_types(function_name, self.calls(function_name))
def typed_functions(self) -> str:
functions = ''
for f_name in self.all_calls():
try:
f_text = astor.to_source(self.typed_function_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
def typed_function(self, function_name: str) -> str:
return astor.to_source(self.typed_function_ast(function_name))
###Output
_____no_output_____
###Markdown
End of Excursion Here is how to use `TypeAnnotator`. We first track a series of calls:
###Code
with TypeAnnotator() as annotator:
y = square_root(25.0)
y = square_root(2.0)
###Output
_____no_output_____
###Markdown
After tracking, we can immediately retrieve an annotated version of the functions tracked:
###Code
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
This also works for multiple and diverse functions. One could go and implement an automatic type annotator for Python files based on the types seen during execution.
###Code
with TypeAnnotator() as annotator:
hello('type annotations')
y = square_root(1.0)
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
A content as above could now be sent to a type checker, which would detect any type inconsistency between callers and callees. Excursion: Handling Multiple TypesLet us now resolve the role of the magic `Any` type in `annotate_function_ast_with_types()`. If we see multiple types for the same argument, we set its type to `Any`. For `square_root()`, this makes sense, as its arguments can be integers as well as floats:
###Code
with CallTracer() as tracer:
y = square_root(25.0)
y = square_root(4)
annotated_square_root_ast = annotate_types(tracer.all_calls())['square_root']
print_content(astor.to_source(annotated_square_root_ast), '.py')
###Output
_____no_output_____
###Markdown
The following function `sum3()` can be called with floating-point numbers as arguments, resulting in the parameters getting a `float` type:
###Code
def sum3(a, b, c): # type: ignore
return a + b + c
with TypeAnnotator() as annotator:
y = sum3(1.0, 2.0, 3.0)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we call `sum3()` with integers, though, the arguments get an `int` type:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
And we can also call `sum3()` with strings, which assigns the arguments a `str` type:
###Code
with TypeAnnotator() as annotator:
y = sum3("one", "two", "three")
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we have multiple calls, but with different types, `TypeAnnotator()` will assign an `Any` type to both arguments and return values:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y = sum3("one", "two", "three")
typed_sum3_def = annotator.typed_function('sum3')
print_content(typed_sum3_def, '.py')
###Output
_____no_output_____
###Markdown
A type `Any` makes it explicit that an object can, indeed, have any type; it will not be typechecked at runtime or statically. To some extent, this defeats the power of type checking; but it also preserves some of the type flexibility that many Python programmers enjoy. Besides `Any`, the `typing` module supports several additional ways to define ambiguous types; we will keep this in mind for a later exercise. End of Excursion Mining InvariantsBesides basic data types. we can check several further properties from arguments. We can, for instance, whether an argument can be negative, zero, or positive; or that one argument should be smaller than the second; or that the result should be the sum of two arguments – properties that cannot be expressed in a (Python) type.Such properties are called *invariants*, as they hold across all invocations of a function. Specifically, invariants come as _pre_- and _postconditions_ – conditions that always hold at the beginning and at the end of a function. (There are also _data_ and _object_ invariants that express always-holding properties over the state of data or objects, but we do not consider these in this book.) Annotating Functions with Pre- and PostconditionsThe classical means to specify pre- and postconditions is via _assertions_, which we have introduced in the [chapter on assertions](Assertions.ipynb). A precondition checks whether the arguments to a function satisfy the expected properties; a postcondition does the same for the result. We can express and check both using assertions as follows:
###Code
def square_root_with_invariants(x): # type: ignore
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
A nicer way, however, is to syntactically separate invariants from the function at hand. Using appropriate decorators, we could specify pre- and postconditions as follows:```python@precondition lambda x: x >= 0@postcondition lambda return_value, x: return_value * return_value == xdef square_root_with_invariants(x): normal code without assertions ...```The decorators `@precondition` and `@postcondition` would run the given functions (specified as anonymous `lambda` functions) before and after the decorated function, respectively. If the functions return `False`, the condition is violated. `@precondition` gets the function arguments as arguments; `@postcondition` additionally gets the return value as first argument. It turns out that implementing such decorators is not hard at all. Our implementation builds on a [code snippet from StackOverflow](https://stackoverflow.com/questions/12151182/python-precondition-postcondition-for-member-function-how):
###Code
import functools
def condition(precondition: Optional[Callable] = None,
postcondition: Optional[Callable] = None) -> Callable:
def decorator(func: Callable) -> Callable:
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args: Any, **kwargs: Any) -> Any:
if precondition is not None:
assert precondition(*args, **kwargs), \
"Precondition violated"
# Call original function or method
retval = func(*args, **kwargs)
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), \
"Postcondition violated"
return retval
return wrapper
return decorator
def precondition(check: Callable) -> Callable:
return condition(precondition=check)
def postcondition(check: Callable) -> Callable:
return condition(postcondition=check)
###Output
_____no_output_____
###Markdown
With these, we can now start decorating `square_root()`:
###Code
@precondition(lambda x: x > 0)
def square_root_with_precondition(x): # type: ignore
return square_root(x)
###Output
_____no_output_____
###Markdown
This catches arguments violating the precondition:
###Code
with ExpectError():
square_root_with_precondition(-1.0)
###Output
_____no_output_____
###Markdown
Likewise, we can provide a postcondition:
###Code
import math
@postcondition(lambda ret, x: math.isclose(ret * ret, x))
def square_root_with_postcondition(x): # type: ignore
return square_root(x)
y = square_root_with_postcondition(2.0)
y
###Output
_____no_output_____
###Markdown
If we have a buggy implementation of $\sqrt{x}$, this gets caught quickly:
###Code
@postcondition(lambda ret, x: math.isclose(ret * ret, x))
def buggy_square_root_with_postcondition(x): # type: ignore
return square_root(x) + 0.1
with ExpectError():
y = buggy_square_root_with_postcondition(2.0)
###Output
_____no_output_____
###Markdown
While checking pre- and postconditions is a great way to catch errors, specifying them can be cumbersome. Let us try to see whether we can (again) _mine_ some of them. Mining InvariantsTo _mine_ invariants, we can use the same tracking functionality as before; instead of saving values for individual variables, though, we now check whether the values satisfy specific _properties_ or not. For instance, if all values of `x` seen satisfy the condition `x > 0`, then we make `x > 0` an invariant of the function. If we see positive, zero, and negative values of `x`, though, then there is no property of `x` left to talk about.The general idea is thus:1. Check all variable values observed against a set of predefined properties; and2. Keep only those properties that hold for all runs observed. Defining PropertiesWhat precisely do we mean by properties? Here is a small collection of value properties that would frequently be used in invariants. All these properties would be evaluated with the _metavariables_ `X`, `Y`, and `Z` (actually, any upper-case identifier) being replaced with the names of function parameters:
###Code
INVARIANT_PROPERTIES = [
"X < 0",
"X <= 0",
"X > 0",
"X >= 0",
# "X == 0", # implied by "X", below
# "X != 0", # implied by "not X", below
]
###Output
_____no_output_____
###Markdown
When `square_root(x)` is called as, say `square_root(5.0)`, we see that `x = 5.0` holds. The above properties would then all be checked for `x`. Only the properties `X > 0`, `X >= 0`, and `not X` hold for the call seen; and hence `x > 0`, `x >= 0`, and `not x` (or better: `x != 0`) would make potential preconditions for `square_root(x)`. We can check for many more properties such as relations between two arguments:
###Code
INVARIANT_PROPERTIES += [
"X == Y",
"X > Y",
"X < Y",
"X >= Y",
"X <= Y",
]
###Output
_____no_output_____
###Markdown
Types also can be checked using properties. For any function parameter `X`, only one of these will hold:
###Code
INVARIANT_PROPERTIES += [
"isinstance(X, bool)",
"isinstance(X, int)",
"isinstance(X, float)",
"isinstance(X, list)",
"isinstance(X, dict)",
]
###Output
_____no_output_____
###Markdown
We can check for arithmetic properties:
###Code
INVARIANT_PROPERTIES += [
"X == Y + Z",
"X == Y * Z",
"X == Y - Z",
"X == Y / Z",
]
###Output
_____no_output_____
###Markdown
Here's relations over three values, a Python special:
###Code
INVARIANT_PROPERTIES += [
"X < Y < Z",
"X <= Y <= Z",
"X > Y > Z",
"X >= Y >= Z",
]
###Output
_____no_output_____
###Markdown
These Boolean properties also check for other types, as in Python, `None`, an empty list, an empty set, an empty string, and the value zero all evaluate to `False`.
###Code
INVARIANT_PROPERTIES += [
"X",
"not X"
]
###Output
_____no_output_____
###Markdown
Finally, we can also check for list or string properties. Again, this is just a tiny selection.
###Code
INVARIANT_PROPERTIES += [
"X == len(Y)",
"X == sum(Y)",
"X in Y",
"X.startswith(Y)",
"X.endswith(Y)",
]
###Output
_____no_output_____
###Markdown
Extracting Meta-VariablesLet us first introduce a few _helper functions_ before we can get to the actual mining. `metavars()` extracts the set of meta-variables (`X`, `Y`, `Z`, etc.) from a property. To this end, we parse the property as a Python expression and then visit the identifiers.
###Code
def metavars(prop: str) -> List[str]:
metavar_list = []
class ArgVisitor(ast.NodeVisitor):
def visit_Name(self, node: ast.Name) -> None:
if node.id.isupper():
metavar_list.append(node.id)
ArgVisitor().visit(ast.parse(prop))
return metavar_list
assert metavars("X < 0") == ['X']
assert metavars("X.startswith(Y)") == ['X', 'Y']
assert metavars("isinstance(X, str)") == ['X']
###Output
_____no_output_____
###Markdown
Instantiating PropertiesTo produce a property as invariant, we need to be able to _instantiate_ it with variable names. The instantiation of `X > 0` with `X` being instantiated to `a`, for instance, gets us `a > 0`. To this end, the function `instantiate_prop()` takes a property and a collection of variable names and instantiates the meta-variables left-to-right with the corresponding variables names in the collection.
###Code
def instantiate_prop_ast(prop: str, var_names: Sequence[str]) -> ast.AST:
class NameTransformer(ast.NodeTransformer):
def visit_Name(self, node: ast.Name) -> ast.Name:
if node.id not in mapping:
return node
return ast.Name(id=mapping[node.id], ctx=ast.Load())
meta_variables = metavars(prop)
assert len(meta_variables) == len(var_names)
mapping = {}
for i in range(0, len(meta_variables)):
mapping[meta_variables[i]] = var_names[i]
prop_ast = ast.parse(prop, mode='eval')
new_ast = NameTransformer().visit(prop_ast)
return new_ast
def instantiate_prop(prop: str, var_names: Sequence[str]) -> str:
prop_ast = instantiate_prop_ast(prop, var_names)
prop_text = astor.to_source(prop_ast).strip()
while prop_text.startswith('(') and prop_text.endswith(')'):
prop_text = prop_text[1:-1]
return prop_text
assert instantiate_prop("X > Y", ['a', 'b']) == 'a > b'
assert instantiate_prop("X.startswith(Y)", ['x', 'y']) == 'x.startswith(y)'
###Output
_____no_output_____
###Markdown
Evaluating PropertiesTo actually _evaluate_ properties, we do not need to instantiate them. Instead, we simply convert them into a boolean function, using `lambda`:
###Code
def prop_function_text(prop: str) -> str:
return "lambda " + ", ".join(metavars(prop)) + ": " + prop
###Output
_____no_output_____
###Markdown
Here is a simple example:
###Code
prop_function_text("X > Y")
###Output
_____no_output_____
###Markdown
We can easily evaluate the function:
###Code
def prop_function(prop: str) -> Callable:
return eval(prop_function_text(prop))
###Output
_____no_output_____
###Markdown
Here is an example:
###Code
p = prop_function("X > Y")
quiz("What is p(100, 1)?",
[
"False",
"True"
], 'p(100, 1) + 1', globals())
p(100, 1)
p(1, 100)
###Output
_____no_output_____
###Markdown
Checking InvariantsTo extract invariants from an execution, we need to check them on all possible instantiations of arguments. If the function to be checked has two arguments `a` and `b`, we instantiate the property `X < Y` both as `a < b` and `b < a` and check each of them. To get all combinations, we use the Python `permutations()` function:
###Code
import itertools
for combination in itertools.permutations([1.0, 2.0, 3.0], 2):
print(combination)
###Output
_____no_output_____
###Markdown
The function `true_property_instantiations()` takes a property and a list of tuples (`var_name`, `value`). It then produces all instantiations of the property with the given values and returns those that evaluate to True.
###Code
Invariants = Set[Tuple[str, Tuple[str, ...]]]
def true_property_instantiations(prop: str, vars_and_values: Arguments,
log: bool = False) -> Invariants:
instantiations = set()
p = prop_function(prop)
len_metavars = len(metavars(prop))
for combination in itertools.permutations(vars_and_values, len_metavars):
args = [value for var_name, value in combination]
var_names = [var_name for var_name, value in combination]
try:
result = p(*args)
except:
result = None
if log:
print(prop, combination, result)
if result:
instantiations.add((prop, tuple(var_names)))
return instantiations
###Output
_____no_output_____
###Markdown
Here is an example. If `x == -1` and `y == 1`, the property `X < Y` holds for `x < y`, but not for `y < x`:
###Code
invs = true_property_instantiations("X < Y", [('x', -1), ('y', 1)], log=True)
invs
###Output
_____no_output_____
###Markdown
The instantiation retrieves the short form:
###Code
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Likewise, with values for `x` and `y` as above, the property `X < 0` only holds for `x`, but not for `y`:
###Code
invs = true_property_instantiations("X < 0", [('x', -1), ('y', 1)], log=True)
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Extracting InvariantsLet us now run the above invariant extraction on function arguments and return values as observed during a function execution. To this end, we extend the `CallTracer` class into an `InvariantTracer` class, which automatically computes invariants for all functions and all calls observed during tracking. By default, an `InvariantTracer` uses the `INVARIANT_PROPERTIES` properties as defined above; however, one can specify alternate sets of properties.
###Code
class InvariantTracer(CallTracer):
def __init__(self, props: Optional[List[str]] = None, **kwargs: Any) -> None:
if props is None:
props = INVARIANT_PROPERTIES
self.props = props
super().__init__(**kwargs)
###Output
_____no_output_____
###Markdown
The key method of the `InvariantTracer` is the `invariants()` method. This iterates over the calls observed and checks which properties hold. Only the intersection of properties – that is, the set of properties that hold for all calls – is preserved, and eventually returned. The special variable `return_value` is set to hold the return value.
###Code
RETURN_VALUE = 'return_value'
class InvariantTracer(InvariantTracer):
def all_invariants(self) -> Dict[str, Invariants]:
return {function_name: self.invariants(function_name)
for function_name in self.all_calls()}
def invariants(self, function_name: str) -> Invariants:
invariants = None
for variables, return_value in self.calls(function_name):
vars_and_values = variables + [(RETURN_VALUE, return_value)]
s = set()
for prop in self.props:
s |= true_property_instantiations(prop, vars_and_values,
self._log)
if invariants is None:
invariants = s
else:
invariants &= s
assert invariants is not None
return invariants
###Output
_____no_output_____
###Markdown
Here's an example of how to use `invariants()`. We run the tracer on a small set of calls.
###Code
with InvariantTracer() as tracer:
y = square_root(25.0)
y = square_root(10.0)
tracer.all_calls()
###Output
_____no_output_____
###Markdown
The `invariants()` method produces a set of properties that hold for the observed runs, together with their instantiations over function arguments.
###Code
invs = tracer.invariants('square_root')
invs
###Output
_____no_output_____
###Markdown
As before, the actual instantiations are easier to read:
###Code
def pretty_invariants(invariants: Invariants) -> List[str]:
props = []
for (prop, var_names) in invariants:
props.append(instantiate_prop(prop, var_names))
return sorted(props)
pretty_invariants(invs)
###Output
_____no_output_____
###Markdown
We see that the both `x` and the return value have a `float` type. We also see that both are always greater than zero. These are properties that may make useful pre- and postconditions, notably for symbolic analysis. However, there's also an invariant which does _not_ universally hold, namely `return_value <= x`, as the following example shows:
###Code
square_root(0.01)
###Output
_____no_output_____
###Markdown
Clearly, 0.1 > 0.01 holds. This is a case of us not learning from sufficiently diverse inputs. As soon as we have a call including `x = 0.1`, though, the invariant `return_value <= x` is eliminated:
###Code
with InvariantTracer() as tracer:
y = square_root(25.0)
y = square_root(10.0)
y = square_root(0.01)
pretty_invariants(tracer.invariants('square_root'))
###Output
_____no_output_____
###Markdown
We will discuss later how to ensure sufficient diversity in inputs. (Hint: This involves test generation.) Let us try out our invariant tracer on `sum3()`. We see that all types are well-defined; the properties that all arguments are non-zero, however, is specific to the calls observed.
###Code
with InvariantTracer() as tracer:
y = sum3(1, 2, 3)
y = sum3(-4, -5, -6)
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with strings instead, we get different invariants. Notably, we obtain the postcondition that the returned string always starts with the string in the first argument `a` – a universal postcondition if strings are used.
###Code
with InvariantTracer() as tracer:
y = sum3('a', 'b', 'c')
y = sum3('f', 'e', 'd')
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with both strings and numbers (and zeros, too), there are no properties left that would hold across all calls. That's the price of flexibility.
###Code
with InvariantTracer() as tracer:
y = sum3('a', 'b', 'c')
y = sum3('c', 'b', 'a')
y = sum3(-4, -5, -6)
y = sum3(0, 0, 0)
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
Converting Mined Invariants to AnnotationsAs with types, above, we would like to have some functionality where we can add the mined invariants as annotations to existing functions. To this end, we introduce the `InvariantAnnotator` class, extending `InvariantTracer`. We start with a helper method. `params()` returns a comma-separated list of parameter names as observed during calls.
###Code
class InvariantAnnotator(InvariantTracer):
def params(self, function_name: str) -> str:
arguments, return_value = self.calls(function_name)[0]
return ", ".join(arg_name for (arg_name, arg_value) in arguments)
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = sum3(1, 2, 3)
annotator.params('square_root')
annotator.params('sum3')
###Output
_____no_output_____
###Markdown
Now for the actual annotation. `preconditions()` returns the preconditions from the mined invariants (i.e., those propertes that do not depend on the return value) as a string with annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = ("@precondition(lambda " + self.params(function_name) +
": " + inv + ")")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
annotator.preconditions('square_root')
###Output
_____no_output_____
###Markdown
`postconditions()` does the same for postconditions:
###Code
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = (f"@postcondition(lambda {RETURN_VALUE},"
f" {self.params(function_name)}: {inv})")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
annotator.postconditions('square_root')
###Output
_____no_output_____
###Markdown
With these, we can take a function and add both pre- and postconditions as annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def functions_with_invariants(self) -> str:
functions = ""
for function_name in self.all_invariants():
try:
function = self.function_with_invariants(function_name)
except KeyError:
continue
functions += function
return functions
def function_with_invariants(self, function_name: str) -> str:
function = globals()[function_name] # Can throw KeyError
source = inspect.getsource(function)
return '\n'.join(self.preconditions(function_name) +
self.postconditions(function_name)) + \
'\n' + source
###Output
_____no_output_____
###Markdown
Here comes `function_with_invariants()` in all its glory:
###Code
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
print_content(annotator.function_with_invariants('square_root'), '.py')
###Output
_____no_output_____
###Markdown
Quite a number of invariants, isn't it? Further below (and in the exercises), we will discuss on how to focus on the most relevant properties. Avoiding OverspecializationMined specifications can only be as good as the executions they were mined from. If we only see a single call, for instance, we will be faced with several mined pre- and postconditions that _overspecialize_ towards the values seen. Let us illustrate this effect on a simple `sum2()` function which adds two numbers.
###Code
def sum2(a, b): # type: ignore
return a + b
###Output
_____no_output_____
###Markdown
If we invoke `sum2()` with a variety of arguments, the invariants all capture the relationship between `a`, `b`, and the return value as `return_value == a + b` in all its variations.
###Code
with InvariantAnnotator() as annotator:
sum2(31, 45)
sum2(0, 0)
sum2(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
If, however, we see only a single call, the invariants will overspecialize to the single call seen:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
The mined precondition `a == b`, for instance, only holds for the single call observed; the same holds for the mined postcondition `return_value == a * b`. Yet, `sum2()` can obviously be successfully called with other values that do not satisfy these conditions. To get out of this trap, we have to _learn from more and more diverse runs_. One way to obtain such runs is by _generating_ inputs. Indeed, a simple test generator for calls of `sum2()` will easily resolve the problem.
###Code
import random
with InvariantAnnotator() as annotator:
for i in range(100):
a = random.randrange(-10, +10)
b = random.randrange(-10, +10)
length = sum2(a, b)
print_content(annotator.function_with_invariants('sum2'), '.py')
###Output
_____no_output_____
###Markdown
Note, though, that an API test generator, such as above, will have to be set up such that it actually respects preconditions – in our case, we invoke `sum2()` with integers only, already assuming its precondition. In some way, one thus needs a specification (a model, a grammar) to mine another specification – a chicken-and-egg problem. However, there is one way out of this problem: If one can automatically generate tests at the system level, then one has an _infinite source of executions_ to learn invariants from. In each of these executions, all functions would be called with values that satisfy the (implicit) precondition, allowing us to mine invariants for these functions. This holds, because at the system level, invalid inputs must be rejected by the system in the first place. The meaningful precondition at the system level, ensuring that only valid inputs get through, thus gets broken down into a multitude of meaningful preconditions (and subsequent postconditions) at the function level. The big requirement for all this, though, is that one needs good test generators. This will be the subject of another book, namely [The Fuzzing Book](https://www.fuzzingbook.org/). Partial InvariantsFor debugging, it can be helpful to focus on invariants produced only by _failing_ runs, thus characterizing the _circumstances under which a function fails_. Let us illustrate this on an example. The `middle()` function from the [chapter on statistical debugging](StatisticalDebugger.ipynb) is supposed to return the middle of three integers `x`, `y`, and `z`.
###Code
from StatisticalDebugger import middle # minor dependency
with InvariantAnnotator() as annotator:
for i in range(100):
x = random.randrange(-10, +10)
y = random.randrange(-10, +10)
z = random.randrange(-10, +10)
mid = middle(x, y, z)
###Output
_____no_output_____
###Markdown
By default, our `InvariantAnnotator()` does not return any particular pre- or postcondition (other than the types observed). That is just fine, as the function indeed imposes no particular precondition; and the postcondition from `middle()` is not covered by the `InvariantAnnotator` patterns.
###Code
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Things get more interesting if we focus on a particular subset of runs only, though - say, a set of inputs where `middle()` fails.
###Code
from StatisticalDebugger import MIDDLE_FAILING_TESTCASES # minor dependency
with InvariantAnnotator() as annotator:
for x, y, z in MIDDLE_FAILING_TESTCASES:
mid = middle(x, y, z)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Now that's an intimidating set of pre- and postconditions. However, almost all of the preconditions are implied by the one precondition```python@precondition(lambda x, y, z: y < x < z, doc='y < x < z')```which characterizes the exact condition under which `middle()` fails (which also happens to be the condition under which the erroneous second `return y` is executed). By checking how _invariants for failing runs_ differ from _invariants for passing runs_, we can identify circumstances for function failures.
###Code
quiz("Could `InvariantAnnotator` also determine a precondition "
"that characterizes _passing_ runs?",
[
"Yes",
"No"
], 'int(math.exp(1))', globals())
###Output
_____no_output_____
###Markdown
Indeed, it cannot – the correct invariant for passing runs would be the _inverse_ of the invariant for failing runs, and `not A < B < C` is not part of our invariant library. We can easily test this:
###Code
from StatisticalDebugger import MIDDLE_PASSING_TESTCASES # minor dependency
with InvariantAnnotator() as annotator:
for x, y, z in MIDDLE_PASSING_TESTCASES:
mid = middle(x, y, z)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Some ExamplesLet us try out the `InvariantAnnotator` on a number of examples. Removing HTML MarkupRunning `InvariantAnnotator` on our ongoing example `remove_html_markup()` does not provide much, as our invariant properties are tailored towards numerical functions.
###Code
from Intro_Debugging import remove_html_markup
with InvariantAnnotator() as annotator:
remove_html_markup("<foo>bar</foo>")
remove_html_markup("bar")
remove_html_markup('"bar"')
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
In the [chapter on DDSet](DDSetDebugger.ipynb), we will see how to express more complex properties for structured inputs. A Recursive FunctionHere's another example. `list_length()` recursively computes the length of a Python function. Let us see whether we can mine its invariants:
###Code
def list_length(elems: List[Any]) -> int:
if elems == []:
length = 0
else:
length = 1 + list_length(elems[1:])
return length
with InvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Almost all these properties are relevant. Of course, the reason the invariants are so neat is that the return value is equal to `len(elems)` is that `X == len(Y)` is part of the list of properties to be checked. Sum of two NumbersThe next example is a very simple function: If we have a function without return value, the return value is `None` and we can only mine preconditions. (Well, we get a "postcondition" `not return_value` that the return value evaluates to False, which holds for `None`).
###Code
def print_sum(a, b): # type: ignore
print(a + b)
with InvariantAnnotator() as annotator:
print_sum(31, 45)
print_sum(0, 0)
print_sum(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Checking SpecificationsA function with invariants, as above, can be fed into the Python interpreter, such that all pre- and postconditions are checked. We create a function `square_root_annotated()` which includes all the invariants mined above.
###Code
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
square_root_def = annotator.functions_with_invariants()
square_root_def = square_root_def.replace('square_root',
'square_root_annotated')
print_content(square_root_def, '.py')
exec(square_root_def)
###Output
_____no_output_____
###Markdown
The "annotated" version checks against invalid arguments – or more precisely, against arguments with properties that have not been observed yet:
###Code
with ExpectError():
square_root_annotated(-1.0) # type: ignore
###Output
_____no_output_____
###Markdown
This is in contrast to the original version, which just hangs on negative values:
###Code
with ExpectTimeout(1):
square_root(-1.0)
###Output
_____no_output_____
###Markdown
If we make changes to the function definition such that the properties of the return value change, such _regressions_ are caught as violations of the postconditions. Let us illustrate this by simply inverting the result, and return $-2$ as square root of 4.
###Code
square_root_def = square_root_def.replace('square_root_annotated',
'square_root_negative')
square_root_def = square_root_def.replace('return approx',
'return -approx')
print_content(square_root_def, '.py')
exec(square_root_def)
###Output
_____no_output_____
###Markdown
Technically speaking, $-2$ _is_ a square root of 4, since $(-2)^2 = 4$ holds. Yet, such a change may be unexpected by callers of `square_root()`, and hence, this would be caught with the first call:
###Code
with ExpectError():
square_root_negative(2.0) # type: ignore
###Output
_____no_output_____
###Markdown
We see how pre- and postconditions, as well as types, can serve as *oracles* during testing. In particular, once we have mined them for a set of functions, we can check them again and again with test generators – especially after code changes. The more checks we have, and the more specific they are, the more likely it is we can detect unwanted effects of changes. SynopsisThis chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:
###Code
def sum2(a, b): # type: ignore
return a + b
with TypeAnnotator() as type_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.
###Code
print(type_annotator.typed_functions())
###Output
_____no_output_____
###Markdown
The invariant annotator works in a similar fashion:
###Code
with InvariantAnnotator() as inv_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.
###Code
print(inv_annotator.functions_with_invariants())
###Output
_____no_output_____
###Markdown
Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs). The chapter gives details on how to customize the properties checked for. Lessons Learned* Type annotations and explicit invariants allow for _checking_ arguments and results for expected data types and other properties.* One can automatically _mine_ data types and invariants by observing arguments and results at runtime.* The quality of mined invariants depends on the diversity of values observed during executions; this variety can be increased by generating tests. Next StepsIn the next chapter, we will explore [abstracting failure conditions](DDSetDebugger.ipynb). BackgroundThe [DAIKON dynamic invariant detector](https://plse.cs.washington.edu/daikon/) can be considered the mother of function specification miners. Continuously maintained and extended for more than 20 years, it mines likely invariants in the style of this chapter for a variety of languages, including C, C++, C, Eiffel, F, Java, Perl, and Visual Basic. On top of the functionality discussed above, it holds a rich catalog of patterns for likely invariants, supports data invariants, can eliminate invariants that are implied by others, and determines statistical confidence to disregard unlikely invariants. The corresponding paper \cite{Ernst2001} is one of the seminal and most-cited papers of Software Engineering. A multitude of works have been published based on DAIKON and detecting invariants; see this [curated list](http://plse.cs.washington.edu/daikon/pubs/) for details. The interaction between test generators and invariant detection is already discussed in \cite{Ernst2001} (incidentally also using grammars). The Eclat tool \cite{Pacheco2005} is a model example of tight interaction between a unit-level test generator and DAIKON-style invariant mining, where the mined invariants are used to produce oracles and to systematically guide the test generator towards fault-revealing inputs. Mining specifications is not restricted to pre- and postconditions. The paper "Mining Specifications" \cite{Ammons2002} is another classic in the field, learning state protocols from executions. Grammar mining \cite{Gopinath2020} can also be seen as a specification mining approach, this time learning specifications for input formats. As it comes to adding type annotations to existing code, the blog post ["The state of type hints in Python"](https://www.bernat.tech/the-state-of-type-hints-in-python/) gives a great overview on how Python type hints can be used and checked. To add type annotations, there are two important tools available that also implement our above approach:* [MonkeyType](https://instagram-engineering.com/let-your-code-type-hint-itself-introducing-open-source-monkeytype-a855c7284881) implements the above approach of tracing executions and annotating Python 3 arguments, returns, and variables with type hints.* [PyAnnotate](https://github.com/dropbox/pyannotate) does a similar job, focusing on code in Python 2. It does not produce Python 3-style annotations, but instead produces annotations as comments that can be processed by static type checkers.These tools have been created by engineers at Facebook and Dropbox, respectively, assisting them in checking millions of lines of code for type issues. ExercisesOur code for mining types and invariants is in no way complete. There are dozens of ways to extend our implementations, some of which we discuss in exercises. Exercise 1: Union TypesThe Python `typing` module allows to express that an argument can have multiple types. For `square_root(x)`, this allows to express that `x` can be an `int` or a `float`:
###Code
def square_root_with_union_type(x: Union[int, float]) -> float: # type: ignore
...
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it supports union types for arguments and return values. Use `Optional[X]` as a shorthand for `Union[X, None]`. **Solution.** Left to the reader. Hint: extend `type_string()`. Exercise 2: Types for Local VariablesIn Python, one cannot only annotate arguments with types, but actually also local and global variables – for instance, `approx` and `guess` in our `square_root()` implementation:
###Code
def square_root_with_local_types(x: Union[int, float]) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
approx: Optional[float] = None
guess: float = x / 2
while approx != guess:
approx: float = guess # type: ignore
guess: float = (approx + x / approx) / 2 # type: ignore
return approx
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it also annotates local variables with types. Search the function AST for assignments, determine the type of the assigned value, and make it an annotation on the left hand side. **Solution.** Left to the reader. Exercise 3: Verbose Invariant CheckersOur implementation of invariant checkers does not make it clear for the user which pre-/postcondition failed.
###Code
@precondition(lambda s: len(s) > 0)
def remove_first_char(s: str) -> str:
return s[1:]
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
The following implementation adds an optional `doc` keyword argument which is printed if the invariant is violated:
###Code
def my_condition(precondition: Optional[Callable] = None,
postcondition: Optional[Callable] = None, doc: str = 'Unknown') -> Callable:
def decorator(func: Callable) -> Callable:
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args: Any, **kwargs: Any) -> Any:
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated: " + doc
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated: " + doc
return retval
return wrapper
return decorator
def my_precondition(check: Callable, **kwargs: Any) -> Callable:
return my_condition(precondition=check, doc=kwargs.get('doc', 'Unknown'))
def my_postcondition(check: Callable, **kwargs: Any) -> Callable:
return my_condition(postcondition=check, doc=kwargs.get('doc', 'Unknown'))
@my_precondition(lambda s: len(s) > 0, doc="len(s) > 0") # type: ignore
def remove_first_char(s: str) -> str:
return s[1:]
remove_first_char('abc')
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
Extend `InvariantAnnotator` such that it includes the conditions in the generated pre- and postconditions. **Solution.** Here's a simple solution:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@my_precondition(lambda " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")"
conditions.append(cond)
return conditions
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@my_postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")")
conditions.append(cond)
return conditions
###Output
_____no_output_____
###Markdown
The resulting annotations are harder to read, but easier to diagnose:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
As an alternative, one may be able to use `inspect.getsource()` on the lambda expression or unparse it. This is left to the reader. Exercise 4: Save Initial ValuesIf the value of an argument changes during function execution, this can easily confuse our implementation: The values are tracked at the beginning of the function, but checked only when it returns. Extend the `InvariantAnnotator` and the infrastructure it uses such that* it saves argument values both at the beginning and at the end of a function invocation;* postconditions can be expressed over both _initial_ values of arguments as well as the _final_ values of arguments;* the mined postconditions refer to both these values as well. **Solution.** To be added. Exercise 5: ImplicationsSeveral mined invariant are actually _implied_ by others: If `x > 0` holds, then this implies `x >= 0` and `x != 0`. Extend the `InvariantAnnotator` such that implications between properties are explicitly encoded, and such that implied properties are no longer listed as invariants. See \cite{Ernst2001} for ideas. **Solution.** Left to the reader. Exercise 6: Local VariablesPostconditions may also refer to the values of local variables. Consider extending `InvariantAnnotator` and its infrastructure such that the values of local variables at the end of the execution are also recorded and made part of the invariant inference mechanism. **Solution.** Left to the reader. Exercise 7: Embedding Invariants as AssertionsRather than producing invariants as annotations for pre- and postconditions, insert them as `assert` statements into the function code, as in:```pythondef square_root(x): 'Computes the square root of x, using the Newton-Raphson method' assert isinstance(x, int), 'violated precondition' assert x > 0, 'violated precondition' approx = None guess = (x / 2) while (approx != guess): approx = guess guess = ((approx + (x / approx)) / 2) return_value = approx assert return_value < x, 'violated postcondition' assert isinstance(return_value, float), 'violated postcondition' return approx```Such a formulation may make it easier for test generators and symbolic analysis to access and interpret pre- and postconditions. **Solution.** Here is a tentative implementation that inserts invariants into function ASTs. Part 1: Embedding Invariants into Functions
###Code
class EmbeddedInvariantAnnotator(InvariantTracer):
def function_with_invariants_ast(self, function_name: str) -> ast.AST:
return annotate_function_with_invariants(function_name, self.invariants(function_name))
def function_with_invariants(self, function_name: str) -> str:
return astor.to_source(self.function_with_invariants_ast(function_name))
def annotate_invariants(invariants: Dict[str, Invariants]) -> Dict[str, ast.AST]:
annotated_functions = {}
for function_name in invariants:
try:
annotated_functions[function_name] = annotate_function_with_invariants(function_name, invariants[function_name])
except KeyError:
continue
return annotated_functions
def annotate_function_with_invariants(function_name: str,
function_invariants: Invariants) -> ast.AST:
function = globals()[function_name]
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_invariants(function_ast, function_invariants)
def annotate_function_ast_with_invariants(function_ast: ast.AST,
function_invariants: Invariants) -> ast.AST:
annotated_function_ast = EmbeddedInvariantTransformer(function_invariants).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Part 2: Preconditions
###Code
class PreconditionTransformer(ast.NodeTransformer):
def __init__(self, invariants: Invariants) -> None:
self.invariants = invariants
super().__init__()
def preconditions(self) -> List[ast.stmt]:
preconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated precondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) < 0:
preconditions += assertion_ast.body
return preconditions
def insert_assertions(self, body: List[ast.stmt]) -> List[ast.stmt]:
preconditions = self.preconditions()
try:
docstring = cast(ast.Constant, body[0]).value.s
except:
docstring = None
if docstring:
return [body[0]] + preconditions + body[1:]
else:
return preconditions + body
def visit_FunctionDef(self, node: ast.FunctionDef) -> ast.FunctionDef:
"""Add invariants to function"""
# print(ast.dump(node))
node.body = self.insert_assertions(node.body)
return node
class EmbeddedInvariantTransformer(PreconditionTransformer):
pass
with EmbeddedInvariantAnnotator() as annotator:
square_root(5)
print_content(annotator.function_with_invariants('square_root'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.function_with_invariants('sum3'), '.py')
###Output
_____no_output_____
###Markdown
Part 3: PostconditionsWe make a few simplifying assumptions: * Variables do not change during execution.* There is a single `return` statement at the end of the function.
###Code
class EmbeddedInvariantTransformer(EmbeddedInvariantTransformer):
def postconditions(self) -> List[ast.stmt]:
postconditions = []
for (prop, var_names) in self.invariants:
assertion = ("assert " + instantiate_prop(prop, var_names) +
', "violated postcondition"')
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) >= 0:
postconditions += assertion_ast.body
return postconditions
def insert_assertions(self, body: List[ast.stmt]) -> List[ast.stmt]:
new_body = super().insert_assertions(body)
postconditions = self.postconditions()
body_ends_with_return = isinstance(new_body[-1], ast.Return)
if body_ends_with_return:
ret_val = cast(ast.Return, new_body[-1]).value
saver = RETURN_VALUE + " = " + astor.to_source(ret_val)
else:
saver = RETURN_VALUE + " = None"
saver_ast = cast(ast.stmt, ast.parse(saver))
postconditions = [saver_ast] + postconditions
if body_ends_with_return:
return new_body[:-1] + postconditions + [new_body[-1]]
else:
return new_body + postconditions
with EmbeddedInvariantAnnotator() as annotator:
square_root(5)
square_root_def = annotator.function_with_invariants('square_root')
###Output
_____no_output_____
###Markdown
Here's the full definition with included assertions:
###Code
print_content(square_root_def, '.py')
exec(square_root_def.replace('square_root', 'square_root_annotated'))
with ExpectError():
square_root_annotated(-1)
###Output
_____no_output_____
###Markdown
Here come some more examples:
###Code
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.function_with_invariants('sum3'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.function_with_invariants('list_length'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
print_sum(31, 45)
print_content(annotator.function_with_invariants('print_sum'), '.py')
###Output
_____no_output_____
###Markdown
Mining Function SpecificationsIn the [chapter on assertions](Assertions.ipynb), we have seen how important it is to _check_ whether the result is as expected. In this chapter, we introduce a technique that allows us to _mine_ function specifications from a set of given executions, resulting in abstract and formal _descriptions_ of what the function expects and what it delivers.These so-called _dynamic invariants_ produce pre- and post-conditions over function arguments and variables from a set of executions. Within debugging, the resulting _assertions_ can immediately check whether function behavior has changed, but can also be useful to determine the characteristics of _failing_ runs (as opposed to _passing_ runs). Furthermore, the resulting specifications provide pre- and postconditions for formal program proofs, testing, and verification.This chapter is based on [a chapter with the same name in The Fuzzing Book](https://www.fuzzingbook.org/html/DynamicInvariants.html), which focuses on test generation.
###Code
from bookutils import YouTubeVideo
YouTubeVideo("HDu1olXFvv0")
###Output
_____no_output_____
###Markdown
**Prerequisites*** You should be familiar with tracing program executions, as in the [chapter on tracing](Tracer.ipynb).* Later in this section, we access the internal _abstract syntax tree_ representations of Python programs and transform them, as in the [chapter on tracking failure origins](Slicer.ipynb).
###Code
import bookutils
from Tracer import Tracer
###Output
_____no_output_____
###Markdown
SynopsisTo [use the code provided in this chapter](Importing.ipynb), write```python>>> from debuggingbook.DynamicInvariants import ```and then make use of the following features.This chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:```python>>> def sum2(a, b):>>> return a + b>>> with TypeAnnotator() as type_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.```python>>> print(type_annotator.typed_functions())def sum2(a: int, b: int) ->int: return a + b```The invariant annotator works in a similar fashion:```python>>> with InvariantAnnotator() as inv_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.```python>>> print(inv_annotator.functions_with_invariants())@precondition(lambda a, b: isinstance(a, int))@precondition(lambda a, b: isinstance(b, int))@postcondition(lambda return_value, a, b: a == return_value - b)@postcondition(lambda return_value, a, b: b == return_value - a)@postcondition(lambda return_value, a, b: isinstance(return_value, int))@postcondition(lambda return_value, a, b: return_value == a + b)@postcondition(lambda return_value, a, b: return_value == b + a)def sum2(a, b): return a + b```Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs). The chapter gives details on how to customize the properties checked for. Specifications and AssertionsWhen implementing a function or program, one usually works against a _specification_ – a set of documented requirements to be satisfied by the code. Such specifications can come in natural language. A formal specification, however, allows the computer to check whether the specification is satisfied.In the [chapter on assertions](Assertions.ipynb), we have seen how _preconditions_ and _postconditions_ can describe what a function does. Consider the following (simple) square root function:
###Code
def square_root(x): # type: ignore
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
The assertion `assert p` checks the condition `p`; if it does not hold, execution is aborted. Here, the actual body is not yet written; we use the assertions as a specification of what `square_root()` _expects_, and what it _delivers_.The topmost assertion is the _precondition_, stating the requirements on the function arguments. The assertion at the end is the _postcondition_, stating the properties of the function result (including its relationship with the original arguments). Using these pre- and postconditions as a specification, we can now go and implement a square root function that satisfies them. Once implemented, we can have the assertions check at runtime whether `square_root()` works as expected. However, not every piece of code is developed with explicit specifications in the first place; let alone does most code comes with formal pre- and post-conditions. (Just take a look at the chapters in this book.) This is a pity: As Ken Thompson famously said, "Without specifications, there are no bugs – only surprises". It is also a problem for debugging, since, of course, debugging needs some specification such that we know what is wrong, and how to fix it. This raises the interesting question: Can we somehow _retrofit_ existing code with "specifications" that properly describe their behavior, allowing developers to simply _check_ them rather than having to write them from scratch? This is what we do in this chapter. Beyond Generic FailuresBefore we go into _mining_ specifications, let us first discuss why it could be useful to _have_ them. As a motivating example, consider the full implementation of `square_root()` from the [chapter on assertions](Assertions.ipynb):
###Code
import bookutils
def square_root(x): # type: ignore
"""Computes the square root of x, using the Newton-Raphson method"""
approx = None
guess = x / 2
while approx != guess:
approx = guess
guess = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
`square_root()` does not come with any functionality that would check types or values. Hence, it is easy for callers to make mistakes when calling `square_root()`:
###Code
from ExpectError import ExpectError, ExpectTimeout
with ExpectError():
square_root("foo")
with ExpectError():
x = square_root(0.0)
###Output
_____no_output_____
###Markdown
At least, the Python system catches these errors at runtime. The following call, however, simply lets the function enter an infinite loop:
###Code
with ExpectTimeout(1):
x = square_root(-1.0)
###Output
_____no_output_____
###Markdown
Our goal is to avoid such errors by _annotating_ functions with information that prevents errors like the above ones. The idea is to provide a _specification_ of expected properties – a specification that can then be checked at runtime or statically. \todo{Introduce the concept of *contract*.} Mining Data TypesFor our Python code, one of the most important "specifications" we need is *types*. Python being a "dynamically" typed language means that all data types are determined at run time; the code itself does not explicitly state whether a variable is an integer, a string, an array, a dictionary – or whatever. As _writer_ of Python code, omitting explicit type declarations may save time (and allows for some fun hacks). It is not sure whether a lack of types helps in _reading_ and _understanding_ code for humans. For a _computer_ trying to analyze code, the lack of explicit types is detrimental. If, say, a constraint solver, sees `if x:` and cannot know whether `x` is supposed to be a number or a string, this introduces an _ambiguity_. Such ambiguities may multiply over the entire analysis in a combinatorial explosion – or in the analysis yielding an overly inaccurate result. Python 3.6 and later allow data types as _annotations_ to function arguments (actually, to all variables) and return values. We can, for instance, state that `square_root()` is a function that accepts a floating-point value and returns one:
###Code
def square_root_with_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return square_root(x)
###Output
_____no_output_____
###Markdown
By default, such annotations are ignored by the Python interpreter. Therefore, one can still call `square_root_typed()` with a string as an argument and get the exact same result as above. However, one can make use of special _typechecking_ modules that would check types – _dynamically_ at runtime or _statically_ by analyzing the code without having to execute it. Runtime Type CheckingThe Python `enforce` package provides a function decorator that automatically inserts type-checking code that is executed at runtime. Here is how to use it:
###Code
import enforce
@enforce.runtime_validation
def square_root_with_checked_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return square_root(x)
###Output
_____no_output_____
###Markdown
Now, invoking `square_root_with_checked_type_annotations()` raises an exception when invoked with a type dfferent from the one declared:
###Code
with ExpectError():
square_root_with_checked_type_annotations(True)
###Output
_____no_output_____
###Markdown
Note that this error is not caught by the "untyped" variant, where passing a boolean value happily returns $\sqrt{1}$ as result.
###Code
square_root(True)
###Output
_____no_output_____
###Markdown
In Python (and other languages), the boolean values `True` and `False` can be implicitly converted to the integers 1 and 0; however, it is hard to think of a call to `sqrt()` where this would not be an error.
###Code
from bookutils import quiz
quiz("What happens if we call "
"`square_root_with_checked_type_annotations(1)`?",
[
"`1` is automatically converted to float. It will pass.",
"`1` is a subtype of float. It will pass.",
"`1` is an integer, and no float. The type check will fail.",
"The function will fail for some other reason."
], '37035 // 12345')
###Output
_____no_output_____
###Markdown
"Prediction is very difficult, especially when the future is concerned" (Niels Bohr). We can find out by a simple experiment that `float` actually means `float` – and not `int`:
###Code
with ExpectError(enforce.exceptions.RuntimeTypeError):
square_root_with_checked_type_annotations(1)
###Output
_____no_output_____
###Markdown
To allow `int` as a type, we need to specify a _union_ of types.
###Code
from typing import Union, Optional
@enforce.runtime_validation
def square_root_with_union_type(x: Union[int, float]) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return square_root(x)
square_root_with_union_type(2)
square_root_with_union_type(2.0)
with ExpectError(enforce.exceptions.RuntimeTypeError):
square_root_with_union_type("Two dot zero")
###Output
_____no_output_____
###Markdown
Static Type CheckingType annotations can also be checked _statically_ – that is, without even running the code. Let us create a simple Python file consisting of the above `square_root_typed()` definition and a bad invocation.
###Code
import inspect
import tempfile
f = tempfile.NamedTemporaryFile(mode='w', suffix='.py')
f.name
f.write(inspect.getsource(square_root))
f.write('\n')
f.write(inspect.getsource(square_root_with_type_annotations))
f.write('\n')
f.write("print(square_root_with_type_annotations('123'))\n")
f.flush()
###Output
_____no_output_____
###Markdown
These are the contents of our newly created Python file:
###Code
from bookutils import print_file
print_file(f.name, start_line_number=1)
###Output
_____no_output_____
###Markdown
[Mypy](http://mypy-lang.org) is a type checker for Python programs. As it checks types statically, types induce no overhead at runtime; plus, a static check can be faster than a lengthy series of tests with runtime type checking enabled. Let us see what `mypy` produces on the above file:
###Code
import subprocess
result = subprocess.run(["mypy", "--strict", f.name],
universal_newlines=True, stdout=subprocess.PIPE)
print(result.stdout.replace(f.name + ':', ''))
del f # Delete temporary file
###Output
_____no_output_____
###Markdown
We see that `mypy` complains about untyped function definitions such as `square_root()`; most important, however, it finds that the call to `square_root_with_type_annotations()` in the last line has the wrong type. With `mypy`, we can achieve the same type safety with Python as in statically typed languages – provided that we as programmers also produce the necessary type annotations. Is there a simple way to obtain these? Mining Type SpecificationsOur first task will be to mine type annotations (as part of the code) from _values_ we observe at run time. These type annotations would be _mined_ from actual function executions, _learning_ from (normal) runs what the expected argument and return types should be. By observing a series of calls such as these, we could infer that both `x` and the return value are of type `float`:
###Code
y = square_root(25.0)
y
y = square_root(2.0)
y
###Output
_____no_output_____
###Markdown
How can we mine types from executions? The answer is simple: 1. We _observe_ a function during execution2. We track the _types_ of its arguments3. We include these types as _annotations_ into the code.To do so, we can make use of Python's tracing facility we already observed in the [chapter on tracing executions](Tracer.ipynb). With every call to a function, we retrieve the arguments, their values, and their types. Tracing CallsTo observe argument types at runtime, we define a _tracer function_ that tracks the execution of `square_root()`, checking its arguments and return values. The `CallTracer` class is set to trace functions in a `with` block as follows:```pythonwith CallTracer() as tracer: function_to_be_tracked(...)info = tracer.collected_information()```To create the tracer, we build on the `Tracer` superclass as in the [chapter on tracing executions](Tracer.ipynb).
###Code
from typing import Sequence, Any, Callable, Optional, Type, Tuple, Any
from typing import Dict, Union, Set, List, cast, TypeVar
from types import FrameType, TracebackType
Arguments = List[Tuple[str, Any]]
class CallTracer(Tracer):
def __init__(self, log: bool = False, **kwargs: Any)-> None:
super().__init__(**kwargs)
self._log = log
self.reset()
def reset(self) -> None:
self._calls: Dict[str, List[Tuple[Arguments, Any]]] = {}
self._stack: List[Tuple[str, Arguments]] = []
###Output
_____no_output_____
###Markdown
The `traceit()` method does nothing yet; this is done in specialized subclasses. The `CallTracer` class implements a `traceit()` function that checks for function calls and returns:
###Code
class CallTracer(CallTracer):
def traceit(self, frame: FrameType, event: str, arg: Any) -> None:
"""Tracking function: Record all calls and all args"""
if event == "call":
self.trace_call(frame, event, arg)
elif event == "return":
self.trace_return(frame, event, arg)
def trace_call(self, frame: FrameType, event: str, arg: Any) -> None:
...
def trace_return(self, frame: FrameType, event: str, arg: Any) -> None:
...
###Output
_____no_output_____
###Markdown
`trace_call()` is called when a function is called; it retrieves the function name and current arguments, and saves them on a stack.
###Code
class CallTracer(CallTracer):
def trace_call(self, frame: FrameType, event: str, arg: Any) -> None:
"""Save current function name and args on the stack"""
code = frame.f_code
function_name = code.co_name
arguments = get_arguments(frame)
self._stack.append((function_name, arguments))
if self._log:
print(simple_call_string(function_name, arguments))
def get_arguments(frame: FrameType) -> Arguments:
"""Return call arguments in the given frame"""
# When called, all arguments are local variables
arguments = [(var, frame.f_locals[var]) for var in frame.f_locals]
arguments.reverse() # Want same order as call
return arguments
###Output
_____no_output_____
###Markdown
When the function returns, `trace_return()` is called. We now also have the return value. We log the whole call with arguments and return value (if desired) and save it in our list of calls.
###Code
class CallTracer(CallTracer):
def trace_return(self, frame: FrameType, event: str, arg: Any) -> None:
"""Get return value and store complete call with arguments and return value"""
code = frame.f_code
function_name = code.co_name
return_value = arg
# TODO: Could call get_arguments() here to also retrieve _final_ values of argument variables
called_function_name, called_arguments = self._stack.pop()
assert function_name == called_function_name
if self._log:
print(simple_call_string(function_name, called_arguments), "returns", return_value)
self.add_call(function_name, called_arguments, return_value)
def add_call(self, function_name: str, arguments: Arguments,
return_value: Any = None) -> None:
...
###Output
_____no_output_____
###Markdown
`simple_call_string()` is a helper for logging that prints out calls in a user-friendly manner.
###Code
def simple_call_string(function_name: str, argument_list: Arguments,
return_value : Any = None) -> str:
"""Return function_name(arg[0], arg[1], ...) as a string"""
call = function_name + "(" + \
", ".join([var + "=" + repr(value)
for (var, value) in argument_list]) + ")"
if return_value is not None:
call += " = " + repr(return_value)
return call
###Output
_____no_output_____
###Markdown
`add_call()` saves the calls in a list; each function name has its own list.
###Code
class CallTracer(CallTracer):
def add_call(self, function_name: str, arguments: Arguments,
return_value: Any = None) -> None:
"""Add given call to list of calls"""
if function_name not in self._calls:
self._calls[function_name] = []
self._calls[function_name].append((arguments, return_value))
###Output
_____no_output_____
###Markdown
we can retrieve the list of calls, either for a given function name (`calls()`),or for all functions (`all_calls()`).
###Code
class CallTracer(CallTracer):
def calls(self, function_name: str) -> List[Tuple[Arguments, Any]]:
"""Return list of calls for `function_name`."""
return self._calls[function_name]
class CallTracer(CallTracer):
def all_calls(self) -> Dict[str, List[Tuple[Arguments, Any]]]:
"""
Return list of calls for function_name,
or a mapping function_name -> calls for all functions tracked
"""
return self._calls
###Output
_____no_output_____
###Markdown
Let us now put this to use. We turn on logging to track the individual calls and their return values:
###Code
with CallTracer(log=True) as tracer:
y = square_root(25)
y = square_root(2.0)
###Output
_____no_output_____
###Markdown
After execution, we can retrieve the individual calls:
###Code
calls = tracer.calls('square_root')
calls
###Output
_____no_output_____
###Markdown
Each call is a pair (`argument_list`, `return_value`), where `argument_list` is a list of pairs (`parameter_name`, `value`).
###Code
square_root_argument_list, square_root_return_value = calls[0]
simple_call_string('square_root', square_root_argument_list, square_root_return_value)
###Output
_____no_output_____
###Markdown
If the function does not return a value, `return_value` is `None`.
###Code
def hello(name: str) -> None:
print("Hello,", name)
with CallTracer() as tracer:
hello("world")
hello_calls = tracer.calls('hello')
hello_calls
hello_argument_list, hello_return_value = hello_calls[0]
simple_call_string('hello', hello_argument_list, hello_return_value)
###Output
_____no_output_____
###Markdown
Getting TypesDespite what you may have read or heard, Python actually _is_ a typed language. It is just that it is _dynamically typed_ – types are used and checked only at runtime (rather than declared in the code, where they can be _statically checked_ at compile time). We can thus retrieve types of all values within Python:
###Code
type(4)
type(2.0)
type([4])
###Output
_____no_output_____
###Markdown
We can retrieve the type of the first argument to `square_root()`:
###Code
parameter, value = square_root_argument_list[0]
parameter, type(value)
###Output
_____no_output_____
###Markdown
as well as the type of the return value:
###Code
type(square_root_return_value)
###Output
_____no_output_____
###Markdown
Hence, we see that (so far), `square_root()` is a function taking (among others) integers and returning floats. We could declare `square_root()` as:
###Code
def square_root_annotated(x: int) -> float:
return square_root(x)
###Output
_____no_output_____
###Markdown
This is a representation we could place in a static type checker, allowing to check whether calls to `square_root()` actually pass a number. A dynamic type checker could run such checks at runtime. By default, Python does not do anything with such annotations. However, tools can access annotations from functions and other objects:
###Code
square_root_annotated.__annotations__
###Output
_____no_output_____
###Markdown
This is how run-time checkers access the annotations to check against. Annotating Functions with TypesOur plan is to annotate functions automatically, based on the types we have seen. Our aim is to build a class `TypeAnnotator` that can be used as follows. First, it would track some execution:```pythonwith TypeAnnotator() as annotator: some_function_call()```After tracking, `TypeAnnotator` would provide appropriate methods to access (type-)annotated versions of the function seen:```pythonprint(annotator.typed_functions())```Let us put the pieces together to build `TypeAnnotator`. Excursion: Accessing Function Structure To annotate functions, we need to convert a function into a tree representation (called _abstract syntax trees_, or ASTs) and back; we already have seen these in the chapter on [tracking value origins](Slicer.ipynb).
###Code
import ast
import inspect
import astor
###Output
_____no_output_____
###Markdown
We can get the source of a Python function using `inspect.getsource()`. (Note that this does not work for functions defined in other notebooks.)
###Code
square_root_source = inspect.getsource(square_root)
square_root_source
###Output
_____no_output_____
###Markdown
To view these in a visually pleasing form, our function `print_content(s, suffix)` formats and highlights the string `s` as if it were a file with ending `suffix`. We can thus view (and highlight) the source as if it were a Python file:
###Code
from bookutils import print_content
print_content(square_root_source, '.py')
###Output
_____no_output_____
###Markdown
Parsing this gives us an abstract syntax tree (AST) – a representation of the program in tree form.
###Code
square_root_ast = ast.parse(square_root_source)
###Output
_____no_output_____
###Markdown
What does this AST look like? The helper functions `astor.dump_tree()` (textual output) and `showast.show_ast()` (graphical output with [showast](https://github.com/hchasestevens/show_ast)) allow us to inspect the structure of the tree. We see that the function starts as a `FunctionDef` with name and arguments, followed by a body, which is a list of statements of type `Expr` (the docstring), type `Assign` (assignments), `While` (while loop with its own body), and finally `Return`.
###Code
print(astor.dump_tree(square_root_ast))
###Output
_____no_output_____
###Markdown
Too much text for you? This graphical representation may make things simpler.
###Code
from bookutils import show_ast
show_ast(square_root_ast)
###Output
_____no_output_____
###Markdown
The function `astor.to_source()` converts such a tree back into the more familiar textual Python code representation. Comments are gone, and there may be more parentheses than before, but the result has the same semantics:
###Code
print_content(astor.to_source(square_root_ast), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: Annotating Functions with Given Types Let us now go and transform ASTs to add type annotations. We start with a helper function `parse_type(name)` which parses a type name into an AST.
###Code
def parse_type(name: str) -> ast.expr:
class ValueVisitor(ast.NodeVisitor):
def visit_Expr(self, node: ast.Expr) -> None:
self.value_node = node.value
tree = ast.parse(name)
name_visitor = ValueVisitor()
name_visitor.visit(tree)
return name_visitor.value_node
print(astor.dump_tree(parse_type('int')))
print(astor.dump_tree(parse_type('[object]')))
###Output
_____no_output_____
###Markdown
We now define a helper function that actually adds type annotations to a function AST. The `TypeTransformer` class builds on the Python standard library `ast.NodeTransformer` infrastructure. It would be called as```python TypeTransformer({'x': 'int'}, 'float').visit(ast)```to annotate the arguments of `square_root()`: `x` with `int`, and the return type with `float`. The returned AST can then be unparsed, compiled or analyzed.
###Code
class TypeTransformer(ast.NodeTransformer):
def __init__(self, argument_types: Dict[str, str], return_type: Optional[str] = None):
self.argument_types = argument_types
self.return_type = return_type
super().__init__()
###Output
_____no_output_____
###Markdown
The core of `TypeTransformer` is the method `visit_FunctionDef()`, which is called for every function definition in the AST. Its argument `node` is the subtree of the function definition to be transformed. Our implementation accesses the individual arguments and invokes `annotate_args()` on them; it also sets the return type in the `returns` attribute of the node.
###Code
class TypeTransformer(TypeTransformer):
def visit_FunctionDef(self, node: ast.FunctionDef) -> ast.FunctionDef:
"""Add annotation to function"""
# Set argument types
new_args = []
for arg in node.args.args:
new_args.append(self.annotate_arg(arg))
new_arguments = ast.arguments(
new_args,
node.args.vararg,
node.args.kwonlyargs,
node.args.kw_defaults,
node.args.kwarg,
node.args.defaults
)
# Set return type
if self.return_type is not None:
node.returns = parse_type(self.return_type)
return ast.copy_location(
ast.FunctionDef(node.name, new_arguments,
node.body, node.decorator_list,
node.returns), node)
def annotate_arg(self, arg: ast.arg) -> ast.arg:
...
###Output
_____no_output_____
###Markdown
Each argument gets its own annotation, taken from the types originally passed to the class:
###Code
class TypeTransformer(TypeTransformer):
def annotate_arg(self, arg: ast.arg) -> ast.arg:
"""Add annotation to single function argument"""
arg_name = arg.arg
if arg_name in self.argument_types:
arg.annotation = parse_type(self.argument_types[arg_name])
return arg
###Output
_____no_output_____
###Markdown
Does this work? Let us annotate the AST from `square_root()` with types for the arguments and return types:
###Code
new_ast = TypeTransformer({'x': 'int'}, 'float').visit(square_root_ast)
###Output
_____no_output_____
###Markdown
When we unparse the new AST, we see that the annotations actually are present:
###Code
print_content(astor.to_source(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Similarly, we can annotate the `hello()` function from above:
###Code
hello_source = inspect.getsource(hello)
hello_ast = ast.parse(hello_source)
new_ast = TypeTransformer({'name': 'str'}, 'None').visit(hello_ast)
print_content(astor.to_source(new_ast), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: Annotating Functions with Mined Types Let us now annotate functions with types mined at runtime. We start with a simple function `type_string()` that determines the appropriate type of a given value (as a string):
###Code
def type_string(value: Any) -> str:
return type(value).__name__
type_string(4)
type_string([])
###Output
_____no_output_____
###Markdown
For composite structures, `type_string()` does not examine element types; hence, the type of `[3]` is simply `list` instead of, say, `list[int]`. For now, `list` will do fine.
###Code
type_string([3])
###Output
_____no_output_____
###Markdown
`type_string()` will be used to infer the types of argument values found at runtime, as returned by `CallTracer.all_calls()`:
###Code
with CallTracer() as tracer:
y = square_root(25.0)
y = square_root(2.0)
tracer.all_calls()
###Output
_____no_output_____
###Markdown
The function `annotate_types()` takes such a list of calls and annotates each function listed:
###Code
def annotate_types(calls: Dict[str, List[Tuple[Arguments, Any]]]) -> Dict[str, ast.AST]:
annotated_functions = {}
for function_name in calls:
try:
annotated_functions[function_name] = \
annotate_function_with_types(function_name, calls[function_name])
except KeyError:
continue
return annotated_functions
###Output
_____no_output_____
###Markdown
For each function, we get the source and its AST and then get to the actual annotation in `annotate_function_ast_with_types()`:
###Code
def annotate_function_with_types(function_name: str,
function_calls: List[Tuple[Arguments, Any]]) -> ast.AST:
function = globals()[function_name] # May raise KeyError for internal functions
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_types(function_ast, function_calls)
###Output
_____no_output_____
###Markdown
The function `annotate_function_ast_with_types()` invokes the `TypeTransformer` with the calls seen, and for each call, iterate over the arguments, determine their types, and annotate the AST with these. The universal type `Any` is used when we encounter type conflicts, which we will discuss below.
###Code
from typing import Any
def annotate_function_ast_with_types(function_ast: ast.AST,
function_calls: List[Tuple[Arguments, Any]]) -> ast.AST:
parameter_types: Dict[str, str] = {}
return_type = None
for calls_seen in function_calls:
args, return_value = calls_seen
if return_value:
if return_type and return_type != type_string(return_value):
return_type = 'Any'
else:
return_type = type_string(return_value)
for parameter, value in args:
try:
different_type = (parameter_types[parameter] !=
type_string(value))
except KeyError:
different_type = False
if different_type:
parameter_types[parameter] = 'Any'
else:
parameter_types[parameter] = type_string(value)
annotated_function_ast = \
TypeTransformer(parameter_types, return_type).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Here is `square_root()` annotated with the types recorded usign the tracer, above.
###Code
print_content(astor.to_source(annotate_types(tracer.all_calls())['square_root']), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: A Type Annotator Class Let us bring all of this together in a single class `TypeAnnotator` that first tracks calls of functions and then allows to access the AST (and the source code form) of the tracked functions annotated with types. The method `typed_functions()` returns the annotated functions as a string; `typed_functions_ast()` returns their AST.
###Code
class TypeTracer(CallTracer):
pass
class TypeAnnotator(TypeTracer):
def typed_functions_ast(self) -> Dict[str, ast.AST]:
return annotate_types(self.all_calls())
def typed_function_ast(self, function_name: str) -> ast.AST:
return annotate_function_with_types(function_name, self.calls(function_name))
def typed_functions(self) -> str:
functions = ''
for f_name in self.all_calls():
try:
f_text = astor.to_source(self.typed_function_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
def typed_function(self, function_name: str) -> str:
return astor.to_source(self.typed_function_ast(function_name))
###Output
_____no_output_____
###Markdown
End of Excursion Here is how to use `TypeAnnotator`. We first track a series of calls:
###Code
with TypeAnnotator() as annotator:
y = square_root(25.0)
y = square_root(2.0)
###Output
_____no_output_____
###Markdown
After tracking, we can immediately retrieve an annotated version of the functions tracked:
###Code
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
This also works for multiple and diverse functions. One could go and implement an automatic type annotator for Python files based on the types seen during execution.
###Code
with TypeAnnotator() as annotator:
hello('type annotations')
y = square_root(1.0)
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
A content as above could now be sent to a type checker, which would detect any type inconsistency between callers and callees. Excursion: Handling Multiple TypesLet us now resolve the role of the magic `Any` type in `annotate_function_ast_with_types()`. If we see multiple types for the same argument, we set its type to `Any`. For `square_root()`, this makes sense, as its arguments can be integers as well as floats:
###Code
with CallTracer() as tracer:
y = square_root(25.0)
y = square_root(4)
annotated_square_root_ast = annotate_types(tracer.all_calls())['square_root']
print_content(astor.to_source(annotated_square_root_ast), '.py')
###Output
_____no_output_____
###Markdown
The following function `sum3()` can be called with floating-point numbers as arguments, resulting in the parameters getting a `float` type:
###Code
def sum3(a, b, c): # type: ignore
return a + b + c
with TypeAnnotator() as annotator:
y = sum3(1.0, 2.0, 3.0)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we call `sum3()` with integers, though, the arguments get an `int` type:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
And we can also call `sum3()` with strings, which assigns the arguments a `str` type:
###Code
with TypeAnnotator() as annotator:
y = sum3("one", "two", "three")
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we have multiple calls, but with different types, `TypeAnnotator()` will assign an `Any` type to both arguments and return values:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y = sum3("one", "two", "three")
typed_sum3_def = annotator.typed_function('sum3')
print_content(typed_sum3_def, '.py')
###Output
_____no_output_____
###Markdown
A type `Any` makes it explicit that an object can, indeed, have any type; it will not be typechecked at runtime or statically. To some extent, this defeats the power of type checking; but it also preserves some of the type flexibility that many Python programmers enjoy. Besides `Any`, the `typing` module supports several additional ways to define ambiguous types; we will keep this in mind for a later exercise. End of Excursion Mining InvariantsBesides basic data types. we can check several further properties from arguments. We can, for instance, whether an argument can be negative, zero, or positive; or that one argument should be smaller than the second; or that the result should be the sum of two arguments – properties that cannot be expressed in a (Python) type.Such properties are called *invariants*, as they hold across all invocations of a function. Specifically, invariants come as _pre_- and _postconditions_ – conditions that always hold at the beginning and at the end of a function. (There are also _data_ and _object_ invariants that express always-holding properties over the state of data or objects, but we do not consider these in this book.) Annotating Functions with Pre- and PostconditionsThe classical means to specify pre- and postconditions is via _assertions_, which we have introduced in the [chapter on assertions](Assertions.ipynb). A precondition checks whether the arguments to a function satisfy the expected properties; a postcondition does the same for the result. We can express and check both using assertions as follows:
###Code
def square_root_with_invariants(x): # type: ignore
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
A nicer way, however, is to syntactically separate invariants from the function at hand. Using appropriate decorators, we could specify pre- and postconditions as follows:```python@precondition lambda x: x >= 0@postcondition lambda return_value, x: return_value * return_value == xdef square_root_with_invariants(x): normal code without assertions ...```The decorators `@precondition` and `@postcondition` would run the given functions (specified as anonymous `lambda` functions) before and after the decorated function, respectively. If the functions return `False`, the condition is violated. `@precondition` gets the function arguments as arguments; `@postcondition` additionally gets the return value as first argument. It turns out that implementing such decorators is not hard at all. Our implementation builds on a [code snippet from StackOverflow](https://stackoverflow.com/questions/12151182/python-precondition-postcondition-for-member-function-how):
###Code
import functools
def condition(precondition: Optional[Callable] = None,
postcondition: Optional[Callable] = None) -> Callable:
def decorator(func: Callable) -> Callable:
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args: Any, **kwargs: Any) -> Any:
if precondition is not None:
assert precondition(*args, **kwargs), \
"Precondition violated"
# Call original function or method
retval = func(*args, **kwargs)
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), \
"Postcondition violated"
return retval
return wrapper
return decorator
def precondition(check: Callable) -> Callable:
return condition(precondition=check)
def postcondition(check: Callable) -> Callable:
return condition(postcondition=check)
###Output
_____no_output_____
###Markdown
With these, we can now start decorating `square_root()`:
###Code
@precondition(lambda x: x > 0)
def square_root_with_precondition(x): # type: ignore
return square_root(x)
###Output
_____no_output_____
###Markdown
This catches arguments violating the precondition:
###Code
with ExpectError():
square_root_with_precondition(-1.0)
###Output
_____no_output_____
###Markdown
Likewise, we can provide a postcondition:
###Code
import math
@postcondition(lambda ret, x: math.isclose(ret * ret, x))
def square_root_with_postcondition(x): # type: ignore
return square_root(x)
y = square_root_with_postcondition(2.0)
y
###Output
_____no_output_____
###Markdown
If we have a buggy implementation of $\sqrt{x}$, this gets caught quickly:
###Code
@postcondition(lambda ret, x: math.isclose(ret * ret, x))
def buggy_square_root_with_postcondition(x): # type: ignore
return square_root(x) + 0.1
with ExpectError():
y = buggy_square_root_with_postcondition(2.0)
###Output
_____no_output_____
###Markdown
While checking pre- and postconditions is a great way to catch errors, specifying them can be cumbersome. Let us try to see whether we can (again) _mine_ some of them. Mining InvariantsTo _mine_ invariants, we can use the same tracking functionality as before; instead of saving values for individual variables, though, we now check whether the values satisfy specific _properties_ or not. For instance, if all values of `x` seen satisfy the condition `x > 0`, then we make `x > 0` an invariant of the function. If we see positive, zero, and negative values of `x`, though, then there is no property of `x` left to talk about.The general idea is thus:1. Check all variable values observed against a set of predefined properties; and2. Keep only those properties that hold for all runs observed. Defining PropertiesWhat precisely do we mean by properties? Here is a small collection of value properties that would frequently be used in invariants. All these properties would be evaluated with the _metavariables_ `X`, `Y`, and `Z` (actually, any upper-case identifier) being replaced with the names of function parameters:
###Code
INVARIANT_PROPERTIES = [
"X < 0",
"X <= 0",
"X > 0",
"X >= 0",
# "X == 0", # implied by "X", below
# "X != 0", # implied by "not X", below
]
###Output
_____no_output_____
###Markdown
When `square_root(x)` is called as, say `square_root(5.0)`, we see that `x = 5.0` holds. The above properties would then all be checked for `x`. Only the properties `X > 0`, `X >= 0`, and `not X` hold for the call seen; and hence `x > 0`, `x >= 0`, and `not x` (or better: `x != 0`) would make potential preconditions for `square_root(x)`. We can check for many more properties such as relations between two arguments:
###Code
INVARIANT_PROPERTIES += [
"X == Y",
"X > Y",
"X < Y",
"X >= Y",
"X <= Y",
]
###Output
_____no_output_____
###Markdown
Types also can be checked using properties. For any function parameter `X`, only one of these will hold:
###Code
INVARIANT_PROPERTIES += [
"isinstance(X, bool)",
"isinstance(X, int)",
"isinstance(X, float)",
"isinstance(X, list)",
"isinstance(X, dict)",
]
###Output
_____no_output_____
###Markdown
We can check for arithmetic properties:
###Code
INVARIANT_PROPERTIES += [
"X == Y + Z",
"X == Y * Z",
"X == Y - Z",
"X == Y / Z",
]
###Output
_____no_output_____
###Markdown
Here's relations over three values, a Python special:
###Code
INVARIANT_PROPERTIES += [
"X < Y < Z",
"X <= Y <= Z",
"X > Y > Z",
"X >= Y >= Z",
]
###Output
_____no_output_____
###Markdown
These Boolean properties also check for other types, as in Python, `None`, an empty list, an empty set, an empty string, and the value zero all evaluate to `False`.
###Code
INVARIANT_PROPERTIES += [
"X",
"not X"
]
###Output
_____no_output_____
###Markdown
Finally, we can also check for list or string properties. Again, this is just a tiny selection.
###Code
INVARIANT_PROPERTIES += [
"X == len(Y)",
"X == sum(Y)",
"X in Y",
"X.startswith(Y)",
"X.endswith(Y)",
]
###Output
_____no_output_____
###Markdown
Extracting Meta-VariablesLet us first introduce a few _helper functions_ before we can get to the actual mining. `metavars()` extracts the set of meta-variables (`X`, `Y`, `Z`, etc.) from a property. To this end, we parse the property as a Python expression and then visit the identifiers.
###Code
def metavars(prop: str) -> List[str]:
metavar_list = []
class ArgVisitor(ast.NodeVisitor):
def visit_Name(self, node: ast.Name) -> None:
if node.id.isupper():
metavar_list.append(node.id)
ArgVisitor().visit(ast.parse(prop))
return metavar_list
assert metavars("X < 0") == ['X']
assert metavars("X.startswith(Y)") == ['X', 'Y']
assert metavars("isinstance(X, str)") == ['X']
###Output
_____no_output_____
###Markdown
Instantiating PropertiesTo produce a property as invariant, we need to be able to _instantiate_ it with variable names. The instantiation of `X > 0` with `X` being instantiated to `a`, for instance, gets us `a > 0`. To this end, the function `instantiate_prop()` takes a property and a collection of variable names and instantiates the meta-variables left-to-right with the corresponding variables names in the collection.
###Code
def instantiate_prop_ast(prop: str, var_names: Sequence[str]) -> ast.AST:
class NameTransformer(ast.NodeTransformer):
def visit_Name(self, node: ast.Name) -> ast.Name:
if node.id not in mapping:
return node
return ast.Name(id=mapping[node.id], ctx=ast.Load())
meta_variables = metavars(prop)
assert len(meta_variables) == len(var_names)
mapping = {}
for i in range(0, len(meta_variables)):
mapping[meta_variables[i]] = var_names[i]
prop_ast = ast.parse(prop, mode='eval')
new_ast = NameTransformer().visit(prop_ast)
return new_ast
def instantiate_prop(prop: str, var_names: Sequence[str]) -> str:
prop_ast = instantiate_prop_ast(prop, var_names)
prop_text = astor.to_source(prop_ast).strip()
while prop_text.startswith('(') and prop_text.endswith(')'):
prop_text = prop_text[1:-1]
return prop_text
assert instantiate_prop("X > Y", ['a', 'b']) == 'a > b'
assert instantiate_prop("X.startswith(Y)", ['x', 'y']) == 'x.startswith(y)'
###Output
_____no_output_____
###Markdown
Evaluating PropertiesTo actually _evaluate_ properties, we do not need to instantiate them. Instead, we simply convert them into a boolean function, using `lambda`:
###Code
def prop_function_text(prop: str) -> str:
return "lambda " + ", ".join(metavars(prop)) + ": " + prop
###Output
_____no_output_____
###Markdown
Here is a simple example:
###Code
prop_function_text("X > Y")
###Output
_____no_output_____
###Markdown
We can easily evaluate the function:
###Code
def prop_function(prop: str) -> Callable:
return eval(prop_function_text(prop))
###Output
_____no_output_____
###Markdown
Here is an example:
###Code
p = prop_function("X > Y")
quiz("What is p(100, 1)?",
[
"False",
"True"
], 'p(100, 1) + 1', globals())
p(100, 1)
p(1, 100)
###Output
_____no_output_____
###Markdown
Checking InvariantsTo extract invariants from an execution, we need to check them on all possible instantiations of arguments. If the function to be checked has two arguments `a` and `b`, we instantiate the property `X < Y` both as `a < b` and `b < a` and check each of them. To get all combinations, we use the Python `permutations()` function:
###Code
import itertools
for combination in itertools.permutations([1.0, 2.0, 3.0], 2):
print(combination)
###Output
_____no_output_____
###Markdown
The function `true_property_instantiations()` takes a property and a list of tuples (`var_name`, `value`). It then produces all instantiations of the property with the given values and returns those that evaluate to True.
###Code
Invariants = Set[Tuple[str, Tuple[str, ...]]]
def true_property_instantiations(prop: str, vars_and_values: Arguments,
log: bool = False) -> Invariants:
instantiations = set()
p = prop_function(prop)
len_metavars = len(metavars(prop))
for combination in itertools.permutations(vars_and_values, len_metavars):
args = [value for var_name, value in combination]
var_names = [var_name for var_name, value in combination]
try:
result = p(*args)
except:
result = None
if log:
print(prop, combination, result)
if result:
instantiations.add((prop, tuple(var_names)))
return instantiations
###Output
_____no_output_____
###Markdown
Here is an example. If `x == -1` and `y == 1`, the property `X < Y` holds for `x < y`, but not for `y < x`:
###Code
invs = true_property_instantiations("X < Y", [('x', -1), ('y', 1)], log=True)
invs
###Output
_____no_output_____
###Markdown
The instantiation retrieves the short form:
###Code
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Likewise, with values for `x` and `y` as above, the property `X < 0` only holds for `x`, but not for `y`:
###Code
invs = true_property_instantiations("X < 0", [('x', -1), ('y', 1)], log=True)
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Extracting InvariantsLet us now run the above invariant extraction on function arguments and return values as observed during a function execution. To this end, we extend the `CallTracer` class into an `InvariantTracer` class, which automatically computes invariants for all functions and all calls observed during tracking. By default, an `InvariantTracer` uses the `INVARIANT_PROPERTIES` properties as defined above; however, one can specify alternate sets of properties.
###Code
class InvariantTracer(CallTracer):
def __init__(self, props: Optional[List[str]] = None, **kwargs: Any) -> None:
if props is None:
props = INVARIANT_PROPERTIES
self.props = props
super().__init__(**kwargs)
###Output
_____no_output_____
###Markdown
The key method of the `InvariantTracer` is the `invariants()` method. This iterates over the calls observed and checks which properties hold. Only the intersection of properties – that is, the set of properties that hold for all calls – is preserved, and eventually returned. The special variable `return_value` is set to hold the return value.
###Code
RETURN_VALUE = 'return_value'
class InvariantTracer(InvariantTracer):
def all_invariants(self) -> Dict[str, Invariants]:
return {function_name: self.invariants(function_name)
for function_name in self.all_calls()}
def invariants(self, function_name: str) -> Invariants:
invariants = None
for variables, return_value in self.calls(function_name):
vars_and_values = variables + [(RETURN_VALUE, return_value)]
s = set()
for prop in self.props:
s |= true_property_instantiations(prop, vars_and_values,
self._log)
if invariants is None:
invariants = s
else:
invariants &= s
assert invariants is not None
return invariants
###Output
_____no_output_____
###Markdown
Here's an example of how to use `invariants()`. We run the tracer on a small set of calls.
###Code
with InvariantTracer() as tracer:
y = square_root(25.0)
y = square_root(10.0)
tracer.all_calls()
###Output
_____no_output_____
###Markdown
The `invariants()` method produces a set of properties that hold for the observed runs, together with their instantiations over function arguments.
###Code
invs = tracer.invariants('square_root')
invs
###Output
_____no_output_____
###Markdown
As before, the actual instantiations are easier to read:
###Code
def pretty_invariants(invariants: Invariants) -> List[str]:
props = []
for (prop, var_names) in invariants:
props.append(instantiate_prop(prop, var_names))
return sorted(props)
pretty_invariants(invs)
###Output
_____no_output_____
###Markdown
We see that the both `x` and the return value have a `float` type. We also see that both are always greater than zero. These are properties that may make useful pre- and postconditions, notably for symbolic analysis. However, there's also an invariant which does _not_ universally hold, namely `return_value <= x`, as the following example shows:
###Code
square_root(0.01)
###Output
_____no_output_____
###Markdown
Clearly, 0.1 > 0.01 holds. This is a case of us not learning from sufficiently diverse inputs. As soon as we have a call including `x = 0.1`, though, the invariant `return_value <= x` is eliminated:
###Code
with InvariantTracer() as tracer:
y = square_root(25.0)
y = square_root(10.0)
y = square_root(0.01)
pretty_invariants(tracer.invariants('square_root'))
###Output
_____no_output_____
###Markdown
We will discuss later how to ensure sufficient diversity in inputs. (Hint: This involves test generation.) Let us try out our invariant tracer on `sum3()`. We see that all types are well-defined; the properties that all arguments are non-zero, however, is specific to the calls observed.
###Code
with InvariantTracer() as tracer:
y = sum3(1, 2, 3)
y = sum3(-4, -5, -6)
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with strings instead, we get different invariants. Notably, we obtain the postcondition that the returned string always starts with the string in the first argument `a` – a universal postcondition if strings are used.
###Code
with InvariantTracer() as tracer:
y = sum3('a', 'b', 'c')
y = sum3('f', 'e', 'd')
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with both strings and numbers (and zeros, too), there are no properties left that would hold across all calls. That's the price of flexibility.
###Code
with InvariantTracer() as tracer:
y = sum3('a', 'b', 'c')
y = sum3('c', 'b', 'a')
y = sum3(-4, -5, -6)
y = sum3(0, 0, 0)
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
Converting Mined Invariants to AnnotationsAs with types, above, we would like to have some functionality where we can add the mined invariants as annotations to existing functions. To this end, we introduce the `InvariantAnnotator` class, extending `InvariantTracer`. We start with a helper method. `params()` returns a comma-separated list of parameter names as observed during calls.
###Code
class InvariantAnnotator(InvariantTracer):
def params(self, function_name: str) -> str:
arguments, return_value = self.calls(function_name)[0]
return ", ".join(arg_name for (arg_name, arg_value) in arguments)
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = sum3(1, 2, 3)
annotator.params('square_root')
annotator.params('sum3')
###Output
_____no_output_____
###Markdown
Now for the actual annotation. `preconditions()` returns the preconditions from the mined invariants (i.e., those propertes that do not depend on the return value) as a string with annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = ("@precondition(lambda " + self.params(function_name) +
": " + inv + ")")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
annotator.preconditions('square_root')
###Output
_____no_output_____
###Markdown
`postconditions()` does the same for postconditions:
###Code
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = (f"@postcondition(lambda {RETURN_VALUE},"
f" {self.params(function_name)}: {inv})")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
annotator.postconditions('square_root')
###Output
_____no_output_____
###Markdown
With these, we can take a function and add both pre- and postconditions as annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def functions_with_invariants(self) -> str:
functions = ""
for function_name in self.all_invariants():
try:
function = self.function_with_invariants(function_name)
except KeyError:
continue
functions += function
return functions
def function_with_invariants(self, function_name: str) -> str:
function = globals()[function_name] # Can throw KeyError
source = inspect.getsource(function)
return '\n'.join(self.preconditions(function_name) +
self.postconditions(function_name)) + \
'\n' + source
###Output
_____no_output_____
###Markdown
Here comes `function_with_invariants()` in all its glory:
###Code
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
print_content(annotator.function_with_invariants('square_root'), '.py')
###Output
_____no_output_____
###Markdown
Quite a number of invariants, isn't it? Further below (and in the exercises), we will discuss on how to focus on the most relevant properties. Avoiding OverspecializationMined specifications can only be as good as the executions they were mined from. If we only see a single call, for instance, we will be faced with several mined pre- and postconditions that _overspecialize_ towards the values seen. Let us illustrate this effect on a simple `sum2()` function which adds two numbers.
###Code
def sum2(a, b): # type: ignore
return a + b
###Output
_____no_output_____
###Markdown
If we invoke `sum2()` with a variety of arguments, the invariants all capture the relationship between `a`, `b`, and the return value as `return_value == a + b` in all its variations.
###Code
with InvariantAnnotator() as annotator:
sum2(31, 45)
sum2(0, 0)
sum2(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
If, however, we see only a single call, the invariants will overspecialize to the single call seen:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
The mined precondition `a == b`, for instance, only holds for the single call observed; the same holds for the mined postcondition `return_value == a * b`. Yet, `sum2()` can obviously be successfully called with other values that do not satisfy these conditions. To get out of this trap, we have to _learn from more and more diverse runs_. One way to obtain such runs is by _generating_ inputs. Indeed, a simple test generator for calls of `sum2()` will easily resolve the problem.
###Code
import random
with InvariantAnnotator() as annotator:
for i in range(100):
a = random.randrange(-10, +10)
b = random.randrange(-10, +10)
length = sum2(a, b)
print_content(annotator.function_with_invariants('sum2'), '.py')
###Output
_____no_output_____
###Markdown
Note, though, that an API test generator, such as above, will have to be set up such that it actually respects preconditions – in our case, we invoke `sum2()` with integers only, already assuming its precondition. In some way, one thus needs a specification (a model, a grammar) to mine another specification – a chicken-and-egg problem. However, there is one way out of this problem: If one can automatically generate tests at the system level, then one has an _infinite source of executions_ to learn invariants from. In each of these executions, all functions would be called with values that satisfy the (implicit) precondition, allowing us to mine invariants for these functions. This holds, because at the system level, invalid inputs must be rejected by the system in the first place. The meaningful precondition at the system level, ensuring that only valid inputs get through, thus gets broken down into a multitude of meaningful preconditions (and subsequent postconditions) at the function level. The big requirement for all this, though, is that one needs good test generators. This will be the subject of another book, namely [The Fuzzing Book](https://www.fuzzingbook.org/). Partial InvariantsFor debugging, it can be helpful to focus on invariants produced only by _failing_ runs, thus characterizing the _circumstances under which a function fails_. Let us illustrate this on an example. The `middle()` function from the [chapter on statistical debugging](StatisticalDebugger.ipynb) is supposed to return the middle of three integers `x`, `y`, and `z`.
###Code
from StatisticalDebugger import middle # minor dependency
with InvariantAnnotator() as annotator:
for i in range(100):
x = random.randrange(-10, +10)
y = random.randrange(-10, +10)
z = random.randrange(-10, +10)
mid = middle(x, y, z)
###Output
_____no_output_____
###Markdown
By default, our `InvariantAnnotator()` does not return any particular pre- or postcondition (other than the types observed). That is just fine, as the function indeed imposes no particular precondition; and the postcondition from `middle()` is not covered by the `InvariantAnnotator` patterns.
###Code
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Things get more interesting if we focus on a particular subset of runs only, though - say, a set of inputs where `middle()` fails. (TODO: Move these from `Repairer` to here, or to statistical debugging)
###Code
from Repairer import MIDDLE_FAILING_TESTCASES # minor dependency
with InvariantAnnotator() as annotator:
for x, y, z in MIDDLE_FAILING_TESTCASES:
mid = middle(x, y, z)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Now that's an intimidating set of pre- and postconditions. However, almost all of the preconditions are implied by the one precondition```python@precondition(lambda x, y, z: y < x < z, doc='y < x < z')```which characterizes the exact condition under which `middle()` fails (which also happens to be the condition under which the erroneous second `return y` is executed). By checking how _invariants for failing runs_ differ from _invariants for passing runs_, we can identify circumstances for function failures.
###Code
quiz("Could `InvariantAnnotator` also determine a precondition "
"that characterizes _passing_ runs?",
[
"Yes",
"No"
], 'int(math.exp(1))', globals())
###Output
_____no_output_____
###Markdown
Indeed, it cannot – the correct invariant for passing runs would be the _inverse_ of the invariant for failing runs, and `not A < B < C` is not part of our invariant library. We can easily test this:
###Code
from Repairer import MIDDLE_PASSING_TESTCASES # minor dependency
with InvariantAnnotator() as annotator:
for x, y, z in MIDDLE_PASSING_TESTCASES:
mid = middle(x, y, z)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Some ExamplesLet us try out the `InvariantAnnotator` on a number of examples. Removing HTML MarkupRunning `InvariantAnnotator` on our ongoing example `remove_html_markup()` does not provide much, as our invariant properties are tailored towards numerical functions.
###Code
from Intro_Debugging import remove_html_markup
with InvariantAnnotator() as annotator:
remove_html_markup("<foo>bar</foo>")
remove_html_markup("bar")
remove_html_markup('"bar"')
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
We will see in the chapter on [Alhazen](Alhazen.ipynb) on how to express more complex properties for structured inputs. A Recursive FunctionHere's another example. `list_length()` recursively computes the length of a Python function. Let us see whether we can mine its invariants:
###Code
def list_length(elems: List[Any]) -> int:
if elems == []:
length = 0
else:
length = 1 + list_length(elems[1:])
return length
with InvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Almost all these properties are relevant. Of course, the reason the invariants are so neat is that the return value is equal to `len(elems)` is that `X == len(Y)` is part of the list of properties to be checked. Sum of two NumbersThe next example is a very simple function: If we have a function without return value, the return value is `None` and we can only mine preconditions. (Well, we get a "postcondition" `not return_value` that the return value evaluates to False, which holds for `None`).
###Code
def print_sum(a, b): # type: ignore
print(a + b)
with InvariantAnnotator() as annotator:
print_sum(31, 45)
print_sum(0, 0)
print_sum(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Checking SpecificationsA function with invariants, as above, can be fed into the Python interpreter, such that all pre- and postconditions are checked. We create a function `square_root_annotated()` which includes all the invariants mined above.
###Code
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
square_root_def = annotator.functions_with_invariants()
square_root_def = square_root_def.replace('square_root',
'square_root_annotated')
print_content(square_root_def, '.py')
exec(square_root_def)
###Output
_____no_output_____
###Markdown
The "annotated" version checks against invalid arguments – or more precisely, against arguments with properties that have not been observed yet:
###Code
with ExpectError():
square_root_annotated(-1.0) # type: ignore
###Output
_____no_output_____
###Markdown
This is in contrast to the original version, which just hangs on negative values:
###Code
with ExpectTimeout(1):
square_root(-1.0)
###Output
_____no_output_____
###Markdown
If we make changes to the function definition such that the properties of the return value change, such _regressions_ are caught as violations of the postconditions. Let us illustrate this by simply inverting the result, and return $-2$ as square root of 4.
###Code
square_root_def = square_root_def.replace('square_root_annotated',
'square_root_negative')
square_root_def = square_root_def.replace('return approx',
'return -approx')
print_content(square_root_def, '.py')
exec(square_root_def)
###Output
_____no_output_____
###Markdown
Technically speaking, $-2$ _is_ a square root of 4, since $(-2)^2 = 4$ holds. Yet, such a change may be unexpected by callers of `square_root()`, and hence, this would be caught with the first call:
###Code
with ExpectError():
square_root_negative(2.0) # type: ignore
###Output
_____no_output_____
###Markdown
We see how pre- and postconditions, as well as types, can serve as *oracles* during testing. In particular, once we have mined them for a set of functions, we can check them again and again with test generators – especially after code changes. The more checks we have, and the more specific they are, the more likely it is we can detect unwanted effects of changes. SynopsisThis chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:
###Code
def sum2(a, b): # type: ignore
return a + b
with TypeAnnotator() as type_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.
###Code
print(type_annotator.typed_functions())
###Output
_____no_output_____
###Markdown
The invariant annotator works in a similar fashion:
###Code
with InvariantAnnotator() as inv_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.
###Code
print(inv_annotator.functions_with_invariants())
###Output
_____no_output_____
###Markdown
Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs). The chapter gives details on how to customize the properties checked for. Lessons Learned* Type annotations and explicit invariants allow for _checking_ arguments and results for expected data types and other properties.* One can automatically _mine_ data types and invariants by observing arguments and results at runtime.* The quality of mined invariants depends on the diversity of values observed during executions; this variety can be increased by generating tests. Next StepsIn the next chapter, we will explore [abstracting failure conditions](DDSetDebugger.ipynb). BackgroundThe [DAIKON dynamic invariant detector](https://plse.cs.washington.edu/daikon/) can be considered the mother of function specification miners. Continuously maintained and extended for more than 20 years, it mines likely invariants in the style of this chapter for a variety of languages, including C, C++, C, Eiffel, F, Java, Perl, and Visual Basic. On top of the functionality discussed above, it holds a rich catalog of patterns for likely invariants, supports data invariants, can eliminate invariants that are implied by others, and determines statistical confidence to disregard unlikely invariants. The corresponding paper \cite{Ernst2001} is one of the seminal and most-cited papers of Software Engineering. A multitude of works have been published based on DAIKON and detecting invariants; see this [curated list](http://plse.cs.washington.edu/daikon/pubs/) for details. The interaction between test generators and invariant detection is already discussed in \cite{Ernst2001} (incidentally also using grammars). The Eclat tool \cite{Pacheco2005} is a model example of tight interaction between a unit-level test generator and DAIKON-style invariant mining, where the mined invariants are used to produce oracles and to systematically guide the test generator towards fault-revealing inputs. Mining specifications is not restricted to pre- and postconditions. The paper "Mining Specifications" \cite{Ammons2002} is another classic in the field, learning state protocols from executions. Grammar mining \cite{Gopinath2020} can also be seen as a specification mining approach, this time learning specifications for input formats. As it comes to adding type annotations to existing code, the blog post ["The state of type hints in Python"](https://www.bernat.tech/the-state-of-type-hints-in-python/) gives a great overview on how Python type hints can be used and checked. To add type annotations, there are two important tools available that also implement our above approach:* [MonkeyType](https://instagram-engineering.com/let-your-code-type-hint-itself-introducing-open-source-monkeytype-a855c7284881) implements the above approach of tracing executions and annotating Python 3 arguments, returns, and variables with type hints.* [PyAnnotate](https://github.com/dropbox/pyannotate) does a similar job, focusing on code in Python 2. It does not produce Python 3-style annotations, but instead produces annotations as comments that can be processed by static type checkers.These tools have been created by engineers at Facebook and Dropbox, respectively, assisting them in checking millions of lines of code for type issues. ExercisesOur code for mining types and invariants is in no way complete. There are dozens of ways to extend our implementations, some of which we discuss in exercises. Exercise 1: Union TypesThe Python `typing` module allows to express that an argument can have multiple types. For `square_root(x)`, this allows to express that `x` can be an `int` or a `float`:
###Code
def square_root_with_union_type(x: Union[int, float]) -> float: # type: ignore
...
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it supports union types for arguments and return values. Use `Optional[X]` as a shorthand for `Union[X, None]`. **Solution.** Left to the reader. Hint: extend `type_string()`. Exercise 2: Types for Local VariablesIn Python, one cannot only annotate arguments with types, but actually also local and global variables – for instance, `approx` and `guess` in our `square_root()` implementation:
###Code
def square_root_with_local_types(x: Union[int, float]) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
approx: Optional[float] = None
guess: float = x / 2
while approx != guess:
approx: float = guess # type: ignore
guess: float = (approx + x / approx) / 2 # type: ignore
return approx
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it also annotates local variables with types. Search the function AST for assignments, determine the type of the assigned value, and make it an annotation on the left hand side. **Solution.** Left to the reader. Exercise 3: Verbose Invariant CheckersOur implementation of invariant checkers does not make it clear for the user which pre-/postcondition failed.
###Code
@precondition(lambda s: len(s) > 0)
def remove_first_char(s: str) -> str:
return s[1:]
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
The following implementation adds an optional `doc` keyword argument which is printed if the invariant is violated:
###Code
def my_condition(precondition: Optional[Callable] = None,
postcondition: Optional[Callable] = None, doc: str = 'Unknown') -> Callable:
def decorator(func: Callable) -> Callable:
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args: Any, **kwargs: Any) -> Any:
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated: " + doc
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated: " + doc
return retval
return wrapper
return decorator
def my_precondition(check: Callable, **kwargs: Any) -> Callable:
return my_condition(precondition=check, doc=kwargs.get('doc', 'Unknown'))
def my_postcondition(check: Callable, **kwargs: Any) -> Callable:
return my_condition(postcondition=check, doc=kwargs.get('doc', 'Unknown'))
@my_precondition(lambda s: len(s) > 0, doc="len(s) > 0") # type: ignore
def remove_first_char(s: str) -> str:
return s[1:]
remove_first_char('abc')
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
Extend `InvariantAnnotator` such that it includes the conditions in the generated pre- and postconditions. **Solution.** Here's a simple solution:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@my_precondition(lambda " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")"
conditions.append(cond)
return conditions
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@my_postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")")
conditions.append(cond)
return conditions
###Output
_____no_output_____
###Markdown
The resulting annotations are harder to read, but easier to diagnose:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
As an alternative, one may be able to use `inspect.getsource()` on the lambda expression or unparse it. This is left to the reader. Exercise 4: Save Initial ValuesIf the value of an argument changes during function execution, this can easily confuse our implementation: The values are tracked at the beginning of the function, but checked only when it returns. Extend the `InvariantAnnotator` and the infrastructure it uses such that* it saves argument values both at the beginning and at the end of a function invocation;* postconditions can be expressed over both _initial_ values of arguments as well as the _final_ values of arguments;* the mined postconditions refer to both these values as well. **Solution.** To be added. Exercise 5: ImplicationsSeveral mined invariant are actually _implied_ by others: If `x > 0` holds, then this implies `x >= 0` and `x != 0`. Extend the `InvariantAnnotator` such that implications between properties are explicitly encoded, and such that implied properties are no longer listed as invariants. See \cite{Ernst2001} for ideas. **Solution.** Left to the reader. Exercise 6: Local VariablesPostconditions may also refer to the values of local variables. Consider extending `InvariantAnnotator` and its infrastructure such that the values of local variables at the end of the execution are also recorded and made part of the invariant inference mechanism. **Solution.** Left to the reader. Exercise 7: Embedding Invariants as AssertionsRather than producing invariants as annotations for pre- and postconditions, insert them as `assert` statements into the function code, as in:```pythondef square_root(x): 'Computes the square root of x, using the Newton-Raphson method' assert isinstance(x, int), 'violated precondition' assert x > 0, 'violated precondition' approx = None guess = (x / 2) while (approx != guess): approx = guess guess = ((approx + (x / approx)) / 2) return_value = approx assert return_value < x, 'violated postcondition' assert isinstance(return_value, float), 'violated postcondition' return approx```Such a formulation may make it easier for test generators and symbolic analysis to access and interpret pre- and postconditions. **Solution.** Here is a tentative implementation that inserts invariants into function ASTs. Part 1: Embedding Invariants into Functions
###Code
class EmbeddedInvariantAnnotator(InvariantTracer):
def function_with_invariants_ast(self, function_name: str) -> ast.AST:
return annotate_function_with_invariants(function_name, self.invariants(function_name))
def function_with_invariants(self, function_name: str) -> str:
return astor.to_source(self.function_with_invariants_ast(function_name))
def annotate_invariants(invariants: Dict[str, Invariants]) -> Dict[str, ast.AST]:
annotated_functions = {}
for function_name in invariants:
try:
annotated_functions[function_name] = annotate_function_with_invariants(function_name, invariants[function_name])
except KeyError:
continue
return annotated_functions
def annotate_function_with_invariants(function_name: str,
function_invariants: Invariants) -> ast.AST:
function = globals()[function_name]
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_invariants(function_ast, function_invariants)
def annotate_function_ast_with_invariants(function_ast: ast.AST,
function_invariants: Invariants) -> ast.AST:
annotated_function_ast = EmbeddedInvariantTransformer(function_invariants).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Part 2: Preconditions
###Code
class PreconditionTransformer(ast.NodeTransformer):
def __init__(self, invariants: Invariants) -> None:
self.invariants = invariants
super().__init__()
def preconditions(self) -> List[ast.stmt]:
preconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated precondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) < 0:
preconditions += assertion_ast.body
return preconditions
def insert_assertions(self, body: List[ast.stmt]) -> List[ast.stmt]:
preconditions = self.preconditions()
try:
docstring = cast(ast.Constant, body[0]).value.s
except:
docstring = None
if docstring:
return [body[0]] + preconditions + body[1:]
else:
return preconditions + body
def visit_FunctionDef(self, node: ast.FunctionDef) -> ast.FunctionDef:
"""Add invariants to function"""
# print(ast.dump(node))
node.body = self.insert_assertions(node.body)
return node
class EmbeddedInvariantTransformer(PreconditionTransformer):
pass
with EmbeddedInvariantAnnotator() as annotator:
square_root(5)
print_content(annotator.function_with_invariants('square_root'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.function_with_invariants('sum3'), '.py')
###Output
_____no_output_____
###Markdown
Part 3: PostconditionsWe make a few simplifying assumptions: * Variables do not change during execution.* There is a single `return` statement at the end of the function.
###Code
class EmbeddedInvariantTransformer(EmbeddedInvariantTransformer):
def postconditions(self) -> List[ast.stmt]:
postconditions = []
for (prop, var_names) in self.invariants:
assertion = ("assert " + instantiate_prop(prop, var_names) +
', "violated postcondition"')
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) >= 0:
postconditions += assertion_ast.body
return postconditions
def insert_assertions(self, body: List[ast.stmt]) -> List[ast.stmt]:
new_body = super().insert_assertions(body)
postconditions = self.postconditions()
body_ends_with_return = isinstance(new_body[-1], ast.Return)
if body_ends_with_return:
ret_val = cast(ast.Return, new_body[-1]).value
saver = RETURN_VALUE + " = " + astor.to_source(ret_val)
else:
saver = RETURN_VALUE + " = None"
saver_ast = cast(ast.stmt, ast.parse(saver))
postconditions = [saver_ast] + postconditions
if body_ends_with_return:
return new_body[:-1] + postconditions + [new_body[-1]]
else:
return new_body + postconditions
with EmbeddedInvariantAnnotator() as annotator:
square_root(5)
square_root_def = annotator.function_with_invariants('square_root')
###Output
_____no_output_____
###Markdown
Here's the full definition with included assertions:
###Code
print_content(square_root_def, '.py')
exec(square_root_def.replace('square_root', 'square_root_annotated'))
with ExpectError():
square_root_annotated(-1)
###Output
_____no_output_____
###Markdown
Here come some more examples:
###Code
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.function_with_invariants('sum3'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.function_with_invariants('list_length'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
print_sum(31, 45)
print_content(annotator.function_with_invariants('print_sum'), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: Annotating Functions with Mined Types Let us now annotate functions with types mined at runtime. We start with a simple function `type_string()` that determines the appropriate type of a given value (as a string):
###Code
def type_string(value: Any) -> str:
return type(value).__name__
type_string(4)
type_string([])
###Output
_____no_output_____
###Markdown
For composite structures, `type_string()` does not examine element types; hence, the type of `[3]` is simply `list` instead of, say, `list[int]`. For now, `list` will do fine.
###Code
type_string([3])
###Output
_____no_output_____
###Markdown
`type_string()` will be used to infer the types of argument values found at runtime, as returned by `CallTracer.all_calls()`:
###Code
with CallTracer() as tracer:
y = square_root(25.0)
y = square_root(2.0)
tracer.all_calls()
###Output
_____no_output_____
###Markdown
The function `annotate_types()` takes such a list of calls and annotates each function listed:
###Code
def annotate_types(calls: Dict[str, List[Tuple[Arguments, Any]]]) -> Dict[str, ast.AST]:
annotated_functions = {}
for function_name in calls:
try:
annotated_functions[function_name] = \
annotate_function_with_types(function_name, calls[function_name])
except KeyError:
continue
return annotated_functions
###Output
_____no_output_____
###Markdown
For each function, we get the source and its AST and then get to the actual annotation in `annotate_function_ast_with_types()`:
###Code
def annotate_function_with_types(function_name: str,
function_calls: List[Tuple[Arguments, Any]]) -> ast.AST:
function = globals()[function_name] # May raise KeyError for internal functions
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_types(function_ast, function_calls)
###Output
_____no_output_____
###Markdown
The function `annotate_function_ast_with_types()` invokes the `TypeTransformer` with the calls seen, and for each call, iterate over the arguments, determine their types, and annotate the AST with these. The universal type `Any` is used when we encounter type conflicts, which we will discuss below.
###Code
def annotate_function_ast_with_types(function_ast: ast.AST,
function_calls: List[Tuple[Arguments, Any]]) -> ast.AST:
parameter_types: Dict[str, str] = {}
return_type = None
for calls_seen in function_calls:
args, return_value = calls_seen
if return_value:
if return_type and return_type != type_string(return_value):
return_type = 'Any'
else:
return_type = type_string(return_value)
for parameter, value in args:
try:
different_type = (parameter_types[parameter] !=
type_string(value))
except KeyError:
different_type = False
if different_type:
parameter_types[parameter] = 'Any'
else:
parameter_types[parameter] = type_string(value)
annotated_function_ast = \
TypeTransformer(parameter_types, return_type).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Here is `square_root()` annotated with the types recorded usign the tracer, above.
###Code
print_content(ast.unparse(annotate_types(tracer.all_calls())['square_root']), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: A Type Annotator Class Let us bring all of this together in a single class `TypeAnnotator` that first tracks calls of functions and then allows to access the AST (and the source code form) of the tracked functions annotated with types. The method `typed_functions()` returns the annotated functions as a string; `typed_functions_ast()` returns their AST.
###Code
class TypeTracer(CallTracer):
pass
class TypeAnnotator(TypeTracer):
def typed_functions_ast(self) -> Dict[str, ast.AST]:
return annotate_types(self.all_calls())
def typed_function_ast(self, function_name: str) -> ast.AST:
return annotate_function_with_types(function_name, self.calls(function_name))
def typed_functions(self) -> str:
functions = ''
for f_name in self.all_calls():
try:
f_text = ast.unparse(self.typed_function_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
def typed_function(self, function_name: str) -> str:
return ast.unparse(self.typed_function_ast(function_name))
###Output
_____no_output_____
###Markdown
End of Excursion Here is how to use `TypeAnnotator`. We first track a series of calls:
###Code
with TypeAnnotator() as annotator:
y = square_root(25.0)
y = square_root(2.0)
###Output
_____no_output_____
###Markdown
After tracking, we can immediately retrieve an annotated version of the functions tracked:
###Code
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
This also works for multiple and diverse functions. One could go and implement an automatic type annotator for Python files based on the types seen during execution.
###Code
with TypeAnnotator() as annotator:
hello('type annotations')
y = square_root(1.0)
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
A content as above could now be sent to a type checker, which would detect any type inconsistency between callers and callees. Excursion: Handling Multiple TypesLet us now resolve the role of the magic `Any` type in `annotate_function_ast_with_types()`. If we see multiple types for the same argument, we set its type to `Any`. For `square_root()`, this makes sense, as its arguments can be integers as well as floats:
###Code
with CallTracer() as tracer:
y = square_root(25.0)
y = square_root(4)
annotated_square_root_ast = annotate_types(tracer.all_calls())['square_root']
print_content(ast.unparse(annotated_square_root_ast), '.py')
###Output
_____no_output_____
###Markdown
The following function `sum3()` can be called with floating-point numbers as arguments, resulting in the parameters getting a `float` type:
###Code
def sum3(a, b, c): # type: ignore
return a + b + c
with TypeAnnotator() as annotator:
y = sum3(1.0, 2.0, 3.0)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we call `sum3()` with integers, though, the arguments get an `int` type:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
And we can also call `sum3()` with strings, which assigns the arguments a `str` type:
###Code
with TypeAnnotator() as annotator:
y = sum3("one", "two", "three")
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we have multiple calls, but with different types, `TypeAnnotator()` will assign an `Any` type to both arguments and return values:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y = sum3("one", "two", "three")
typed_sum3_def = annotator.typed_function('sum3')
print_content(typed_sum3_def, '.py')
###Output
_____no_output_____
###Markdown
A type `Any` makes it explicit that an object can, indeed, have any type; it will not be typechecked at runtime or statically. To some extent, this defeats the power of type checking; but it also preserves some of the type flexibility that many Python programmers enjoy. Besides `Any`, the `typing` module supports several additional ways to define ambiguous types; we will keep this in mind for a later exercise. End of Excursion Mining InvariantsBesides basic data types. we can check several further properties from arguments. We can, for instance, whether an argument can be negative, zero, or positive; or that one argument should be smaller than the second; or that the result should be the sum of two arguments – properties that cannot be expressed in a (Python) type.Such properties are called *invariants*, as they hold across all invocations of a function. Specifically, invariants come as _pre_- and _postconditions_ – conditions that always hold at the beginning and at the end of a function. (There are also _data_ and _object_ invariants that express always-holding properties over the state of data or objects, but we do not consider these in this book.) Annotating Functions with Pre- and PostconditionsThe classical means to specify pre- and postconditions is via _assertions_, which we have introduced in the [chapter on assertions](Assertions.ipynb). A precondition checks whether the arguments to a function satisfy the expected properties; a postcondition does the same for the result. We can express and check both using assertions as follows:
###Code
def square_root_with_invariants(x): # type: ignore
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
A nicer way, however, is to syntactically separate invariants from the function at hand. Using appropriate decorators, we could specify pre- and postconditions as follows:```python@precondition lambda x: x >= 0@postcondition lambda return_value, x: return_value * return_value == xdef square_root_with_invariants(x): normal code without assertions ...```The decorators `@precondition` and `@postcondition` would run the given functions (specified as anonymous `lambda` functions) before and after the decorated function, respectively. If the functions return `False`, the condition is violated. `@precondition` gets the function arguments as arguments; `@postcondition` additionally gets the return value as first argument. It turns out that implementing such decorators is not hard at all. Our implementation builds on a [code snippet from StackOverflow](https://stackoverflow.com/questions/12151182/python-precondition-postcondition-for-member-function-how):
###Code
import functools
def condition(precondition: Optional[Callable] = None,
postcondition: Optional[Callable] = None) -> Callable:
def decorator(func: Callable) -> Callable:
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args: Any, **kwargs: Any) -> Any:
if precondition is not None:
assert precondition(*args, **kwargs), \
"Precondition violated"
# Call original function or method
retval = func(*args, **kwargs)
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), \
"Postcondition violated"
return retval
return wrapper
return decorator
def precondition(check: Callable) -> Callable:
return condition(precondition=check)
def postcondition(check: Callable) -> Callable:
return condition(postcondition=check)
###Output
_____no_output_____
###Markdown
With these, we can now start decorating `square_root()`:
###Code
@precondition(lambda x: x > 0)
def square_root_with_precondition(x): # type: ignore
return square_root(x)
###Output
_____no_output_____
###Markdown
This catches arguments violating the precondition:
###Code
with ExpectError():
square_root_with_precondition(-1.0)
###Output
_____no_output_____
###Markdown
Likewise, we can provide a postcondition:
###Code
import math
@postcondition(lambda ret, x: math.isclose(ret * ret, x))
def square_root_with_postcondition(x): # type: ignore
return square_root(x)
y = square_root_with_postcondition(2.0)
y
###Output
_____no_output_____
###Markdown
If we have a buggy implementation of $\sqrt{x}$, this gets caught quickly:
###Code
@postcondition(lambda ret, x: math.isclose(ret * ret, x))
def buggy_square_root_with_postcondition(x): # type: ignore
return square_root(x) + 0.1
with ExpectError():
y = buggy_square_root_with_postcondition(2.0)
###Output
_____no_output_____
###Markdown
While checking pre- and postconditions is a great way to catch errors, specifying them can be cumbersome. Let us try to see whether we can (again) _mine_ some of them. Mining InvariantsTo _mine_ invariants, we can use the same tracking functionality as before; instead of saving values for individual variables, though, we now check whether the values satisfy specific _properties_ or not. For instance, if all values of `x` seen satisfy the condition `x > 0`, then we make `x > 0` an invariant of the function. If we see positive, zero, and negative values of `x`, though, then there is no property of `x` left to talk about.The general idea is thus:1. Check all variable values observed against a set of predefined properties; and2. Keep only those properties that hold for all runs observed. Defining PropertiesWhat precisely do we mean by properties? Here is a small collection of value properties that would frequently be used in invariants. All these properties would be evaluated with the _metavariables_ `X`, `Y`, and `Z` (actually, any upper-case identifier) being replaced with the names of function parameters:
###Code
INVARIANT_PROPERTIES = [
"X < 0",
"X <= 0",
"X > 0",
"X >= 0",
# "X == 0", # implied by "X", below
# "X != 0", # implied by "not X", below
]
###Output
_____no_output_____
###Markdown
When `square_root(x)` is called as, say `square_root(5.0)`, we see that `x = 5.0` holds. The above properties would then all be checked for `x`. Only the properties `X > 0`, `X >= 0`, and `not X` hold for the call seen; and hence `x > 0`, `x >= 0`, and `not x` (or better: `x != 0`) would make potential preconditions for `square_root(x)`. We can check for many more properties such as relations between two arguments:
###Code
INVARIANT_PROPERTIES += [
"X == Y",
"X > Y",
"X < Y",
"X >= Y",
"X <= Y",
]
###Output
_____no_output_____
###Markdown
Types also can be checked using properties. For any function parameter `X`, only one of these will hold:
###Code
INVARIANT_PROPERTIES += [
"isinstance(X, bool)",
"isinstance(X, int)",
"isinstance(X, float)",
"isinstance(X, list)",
"isinstance(X, dict)",
]
###Output
_____no_output_____
###Markdown
We can check for arithmetic properties:
###Code
INVARIANT_PROPERTIES += [
"X == Y + Z",
"X == Y * Z",
"X == Y - Z",
"X == Y / Z",
]
###Output
_____no_output_____
###Markdown
Here's relations over three values, a Python special:
###Code
INVARIANT_PROPERTIES += [
"X < Y < Z",
"X <= Y <= Z",
"X > Y > Z",
"X >= Y >= Z",
]
###Output
_____no_output_____
###Markdown
These Boolean properties also check for other types, as in Python, `None`, an empty list, an empty set, an empty string, and the value zero all evaluate to `False`.
###Code
INVARIANT_PROPERTIES += [
"X",
"not X"
]
###Output
_____no_output_____
###Markdown
Finally, we can also check for list or string properties. Again, this is just a tiny selection.
###Code
INVARIANT_PROPERTIES += [
"X == len(Y)",
"X == sum(Y)",
"X in Y",
"X.startswith(Y)",
"X.endswith(Y)",
]
###Output
_____no_output_____
###Markdown
Extracting Meta-VariablesLet us first introduce a few _helper functions_ before we can get to the actual mining. `metavars()` extracts the set of meta-variables (`X`, `Y`, `Z`, etc.) from a property. To this end, we parse the property as a Python expression and then visit the identifiers.
###Code
def metavars(prop: str) -> List[str]:
metavar_list = []
class ArgVisitor(ast.NodeVisitor):
def visit_Name(self, node: ast.Name) -> None:
if node.id.isupper():
metavar_list.append(node.id)
ArgVisitor().visit(ast.parse(prop))
return metavar_list
assert metavars("X < 0") == ['X']
assert metavars("X.startswith(Y)") == ['X', 'Y']
assert metavars("isinstance(X, str)") == ['X']
###Output
_____no_output_____
###Markdown
Instantiating PropertiesTo produce a property as invariant, we need to be able to _instantiate_ it with variable names. The instantiation of `X > 0` with `X` being instantiated to `a`, for instance, gets us `a > 0`. To this end, the function `instantiate_prop()` takes a property and a collection of variable names and instantiates the meta-variables left-to-right with the corresponding variables names in the collection.
###Code
def instantiate_prop_ast(prop: str, var_names: Sequence[str]) -> ast.AST:
class NameTransformer(ast.NodeTransformer):
def visit_Name(self, node: ast.Name) -> ast.Name:
if node.id not in mapping:
return node
return ast.Name(id=mapping[node.id], ctx=ast.Load())
meta_variables = metavars(prop)
assert len(meta_variables) == len(var_names)
mapping = {}
for i in range(0, len(meta_variables)):
mapping[meta_variables[i]] = var_names[i]
prop_ast = ast.parse(prop, mode='eval')
new_ast = NameTransformer().visit(prop_ast)
return new_ast
def instantiate_prop(prop: str, var_names: Sequence[str]) -> str:
prop_ast = instantiate_prop_ast(prop, var_names)
prop_text = ast.unparse(prop_ast).strip()
while prop_text.startswith('(') and prop_text.endswith(')'):
prop_text = prop_text[1:-1]
return prop_text
assert instantiate_prop("X > Y", ['a', 'b']) == 'a > b'
assert instantiate_prop("X.startswith(Y)", ['x', 'y']) == 'x.startswith(y)'
###Output
_____no_output_____
###Markdown
Evaluating PropertiesTo actually _evaluate_ properties, we do not need to instantiate them. Instead, we simply convert them into a boolean function, using `lambda`:
###Code
def prop_function_text(prop: str) -> str:
return "lambda " + ", ".join(metavars(prop)) + ": " + prop
###Output
_____no_output_____
###Markdown
Here is a simple example:
###Code
prop_function_text("X > Y")
###Output
_____no_output_____
###Markdown
We can easily evaluate the function:
###Code
def prop_function(prop: str) -> Callable:
return eval(prop_function_text(prop))
###Output
_____no_output_____
###Markdown
Here is an example:
###Code
p = prop_function("X > Y")
quiz("What is p(100, 1)?",
[
"False",
"True"
], 'p(100, 1) + 1', globals())
p(100, 1)
p(1, 100)
###Output
_____no_output_____
###Markdown
Checking InvariantsTo extract invariants from an execution, we need to check them on all possible instantiations of arguments. If the function to be checked has two arguments `a` and `b`, we instantiate the property `X < Y` both as `a < b` and `b < a` and check each of them. To get all combinations, we use the Python `permutations()` function:
###Code
import itertools
for combination in itertools.permutations([1.0, 2.0, 3.0], 2):
print(combination)
###Output
_____no_output_____
###Markdown
The function `true_property_instantiations()` takes a property and a list of tuples (`var_name`, `value`). It then produces all instantiations of the property with the given values and returns those that evaluate to True.
###Code
Invariants = Set[Tuple[str, Tuple[str, ...]]]
def true_property_instantiations(prop: str, vars_and_values: Arguments,
log: bool = False) -> Invariants:
instantiations = set()
p = prop_function(prop)
len_metavars = len(metavars(prop))
for combination in itertools.permutations(vars_and_values, len_metavars):
args = [value for var_name, value in combination]
var_names = [var_name for var_name, value in combination]
try:
result = p(*args)
except:
result = None
if log:
print(prop, combination, result)
if result:
instantiations.add((prop, tuple(var_names)))
return instantiations
###Output
_____no_output_____
###Markdown
Here is an example. If `x == -1` and `y == 1`, the property `X < Y` holds for `x < y`, but not for `y < x`:
###Code
invs = true_property_instantiations("X < Y", [('x', -1), ('y', 1)], log=True)
invs
###Output
_____no_output_____
###Markdown
The instantiation retrieves the short form:
###Code
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Likewise, with values for `x` and `y` as above, the property `X < 0` only holds for `x`, but not for `y`:
###Code
invs = true_property_instantiations("X < 0", [('x', -1), ('y', 1)], log=True)
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Extracting InvariantsLet us now run the above invariant extraction on function arguments and return values as observed during a function execution. To this end, we extend the `CallTracer` class into an `InvariantTracer` class, which automatically computes invariants for all functions and all calls observed during tracking. By default, an `InvariantTracer` uses the `INVARIANT_PROPERTIES` properties as defined above; however, one can specify alternate sets of properties.
###Code
class InvariantTracer(CallTracer):
def __init__(self, props: Optional[List[str]] = None, **kwargs: Any) -> None:
if props is None:
props = INVARIANT_PROPERTIES
self.props = props
super().__init__(**kwargs)
###Output
_____no_output_____
###Markdown
The key method of the `InvariantTracer` is the `invariants()` method. This iterates over the calls observed and checks which properties hold. Only the intersection of properties – that is, the set of properties that hold for all calls – is preserved, and eventually returned. The special variable `return_value` is set to hold the return value.
###Code
RETURN_VALUE = 'return_value'
class InvariantTracer(InvariantTracer):
def all_invariants(self) -> Dict[str, Invariants]:
return {function_name: self.invariants(function_name)
for function_name in self.all_calls()}
def invariants(self, function_name: str) -> Invariants:
invariants = None
for variables, return_value in self.calls(function_name):
vars_and_values = variables + [(RETURN_VALUE, return_value)]
s = set()
for prop in self.props:
s |= true_property_instantiations(prop, vars_and_values,
self._log)
if invariants is None:
invariants = s
else:
invariants &= s
assert invariants is not None
return invariants
###Output
_____no_output_____
###Markdown
Here's an example of how to use `invariants()`. We run the tracer on a small set of calls.
###Code
with InvariantTracer() as tracer:
y = square_root(25.0)
y = square_root(10.0)
tracer.all_calls()
###Output
_____no_output_____
###Markdown
The `invariants()` method produces a set of properties that hold for the observed runs, together with their instantiations over function arguments.
###Code
invs = tracer.invariants('square_root')
invs
###Output
_____no_output_____
###Markdown
As before, the actual instantiations are easier to read:
###Code
def pretty_invariants(invariants: Invariants) -> List[str]:
props = []
for (prop, var_names) in invariants:
props.append(instantiate_prop(prop, var_names))
return sorted(props)
pretty_invariants(invs)
###Output
_____no_output_____
###Markdown
We see that the both `x` and the return value have a `float` type. We also see that both are always greater than zero. These are properties that may make useful pre- and postconditions, notably for symbolic analysis. However, there's also an invariant which does _not_ universally hold, namely `return_value <= x`, as the following example shows:
###Code
square_root(0.01)
###Output
_____no_output_____
###Markdown
Clearly, 0.1 > 0.01 holds. This is a case of us not learning from sufficiently diverse inputs. As soon as we have a call including `x = 0.1`, though, the invariant `return_value <= x` is eliminated:
###Code
with InvariantTracer() as tracer:
y = square_root(25.0)
y = square_root(10.0)
y = square_root(0.01)
pretty_invariants(tracer.invariants('square_root'))
###Output
_____no_output_____
###Markdown
We will discuss later how to ensure sufficient diversity in inputs. (Hint: This involves test generation.) Let us try out our invariant tracer on `sum3()`. We see that all types are well-defined; the properties that all arguments are non-zero, however, is specific to the calls observed.
###Code
with InvariantTracer() as tracer:
y = sum3(1, 2, 3)
y = sum3(-4, -5, -6)
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with strings instead, we get different invariants. Notably, we obtain the postcondition that the returned string always starts with the string in the first argument `a` – a universal postcondition if strings are used.
###Code
with InvariantTracer() as tracer:
y = sum3('a', 'b', 'c')
y = sum3('f', 'e', 'd')
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with both strings and numbers (and zeros, too), there are no properties left that would hold across all calls. That's the price of flexibility.
###Code
with InvariantTracer() as tracer:
y = sum3('a', 'b', 'c')
y = sum3('c', 'b', 'a')
y = sum3(-4, -5, -6)
y = sum3(0, 0, 0)
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
Converting Mined Invariants to AnnotationsAs with types, above, we would like to have some functionality where we can add the mined invariants as annotations to existing functions. To this end, we introduce the `InvariantAnnotator` class, extending `InvariantTracer`. We start with a helper method. `params()` returns a comma-separated list of parameter names as observed during calls.
###Code
class InvariantAnnotator(InvariantTracer):
def params(self, function_name: str) -> str:
arguments, return_value = self.calls(function_name)[0]
return ", ".join(arg_name for (arg_name, arg_value) in arguments)
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = sum3(1, 2, 3)
annotator.params('square_root')
annotator.params('sum3')
###Output
_____no_output_____
###Markdown
Now for the actual annotation. `preconditions()` returns the preconditions from the mined invariants (i.e., those propertes that do not depend on the return value) as a string with annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = ("@precondition(lambda " + self.params(function_name) +
": " + inv + ")")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
annotator.preconditions('square_root')
###Output
_____no_output_____
###Markdown
`postconditions()` does the same for postconditions:
###Code
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = (f"@postcondition(lambda {RETURN_VALUE},"
f" {self.params(function_name)}: {inv})")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
annotator.postconditions('square_root')
###Output
_____no_output_____
###Markdown
With these, we can take a function and add both pre- and postconditions as annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def functions_with_invariants(self) -> str:
functions = ""
for function_name in self.all_invariants():
try:
function = self.function_with_invariants(function_name)
except KeyError:
continue
functions += function
return functions
def function_with_invariants(self, function_name: str) -> str:
function = globals()[function_name] # Can throw KeyError
source = inspect.getsource(function)
return '\n'.join(self.preconditions(function_name) +
self.postconditions(function_name)) + \
'\n' + source
###Output
_____no_output_____
###Markdown
Here comes `function_with_invariants()` in all its glory:
###Code
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
print_content(annotator.function_with_invariants('square_root'), '.py')
###Output
_____no_output_____
###Markdown
Quite a number of invariants, isn't it? Further below (and in the exercises), we will discuss on how to focus on the most relevant properties. Avoiding OverspecializationMined specifications can only be as good as the executions they were mined from. If we only see a single call, for instance, we will be faced with several mined pre- and postconditions that _overspecialize_ towards the values seen. Let us illustrate this effect on a simple `sum2()` function which adds two numbers.
###Code
def sum2(a, b): # type: ignore
return a + b
###Output
_____no_output_____
###Markdown
If we invoke `sum2()` with a variety of arguments, the invariants all capture the relationship between `a`, `b`, and the return value as `return_value == a + b` in all its variations.
###Code
with InvariantAnnotator() as annotator:
sum2(31, 45)
sum2(0, 0)
sum2(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
If, however, we see only a single call, the invariants will overspecialize to the single call seen:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
The mined precondition `a == b`, for instance, only holds for the single call observed; the same holds for the mined postcondition `return_value == a * b`. Yet, `sum2()` can obviously be successfully called with other values that do not satisfy these conditions. To get out of this trap, we have to _learn from more and more diverse runs_. One way to obtain such runs is by _generating_ inputs. Indeed, a simple test generator for calls of `sum2()` will easily resolve the problem.
###Code
import random
with InvariantAnnotator() as annotator:
for i in range(100):
a = random.randrange(-10, +10)
b = random.randrange(-10, +10)
length = sum2(a, b)
print_content(annotator.function_with_invariants('sum2'), '.py')
###Output
_____no_output_____
###Markdown
Note, though, that an API test generator, such as above, will have to be set up such that it actually respects preconditions – in our case, we invoke `sum2()` with integers only, already assuming its precondition. In some way, one thus needs a specification (a model, a grammar) to mine another specification – a chicken-and-egg problem. However, there is one way out of this problem: If one can automatically generate tests at the system level, then one has an _infinite source of executions_ to learn invariants from. In each of these executions, all functions would be called with values that satisfy the (implicit) precondition, allowing us to mine invariants for these functions. This holds, because at the system level, invalid inputs must be rejected by the system in the first place. The meaningful precondition at the system level, ensuring that only valid inputs get through, thus gets broken down into a multitude of meaningful preconditions (and subsequent postconditions) at the function level. The big requirement for all this, though, is that one needs good test generators. This will be the subject of another book, namely [The Fuzzing Book](https://www.fuzzingbook.org/). Partial InvariantsFor debugging, it can be helpful to focus on invariants produced only by _failing_ runs, thus characterizing the _circumstances under which a function fails_. Let us illustrate this on an example. The `middle()` function from the [chapter on statistical debugging](StatisticalDebugger.ipynb) is supposed to return the middle of three integers `x`, `y`, and `z`.
###Code
from StatisticalDebugger import middle # minor dependency
with InvariantAnnotator() as annotator:
for i in range(100):
x = random.randrange(-10, +10)
y = random.randrange(-10, +10)
z = random.randrange(-10, +10)
mid = middle(x, y, z)
###Output
_____no_output_____
###Markdown
By default, our `InvariantAnnotator()` does not return any particular pre- or postcondition (other than the types observed). That is just fine, as the function indeed imposes no particular precondition; and the postcondition from `middle()` is not covered by the `InvariantAnnotator` patterns.
###Code
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Things get more interesting if we focus on a particular subset of runs only, though - say, a set of inputs where `middle()` fails.
###Code
from StatisticalDebugger import MIDDLE_FAILING_TESTCASES # minor dependency
with InvariantAnnotator() as annotator:
for x, y, z in MIDDLE_FAILING_TESTCASES:
mid = middle(x, y, z)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Now that's an intimidating set of pre- and postconditions. However, almost all of the preconditions are implied by the one precondition```python@precondition(lambda x, y, z: y < x < z, doc='y < x < z')```which characterizes the exact condition under which `middle()` fails (which also happens to be the condition under which the erroneous second `return y` is executed). By checking how _invariants for failing runs_ differ from _invariants for passing runs_, we can identify circumstances for function failures.
###Code
quiz("Could `InvariantAnnotator` also determine a precondition "
"that characterizes _passing_ runs?",
[
"Yes",
"No"
], 'int(math.exp(1))', globals())
###Output
_____no_output_____
###Markdown
Indeed, it cannot – the correct invariant for passing runs would be the _inverse_ of the invariant for failing runs, and `not A < B < C` is not part of our invariant library. We can easily test this:
###Code
from StatisticalDebugger import MIDDLE_PASSING_TESTCASES # minor dependency
with InvariantAnnotator() as annotator:
for x, y, z in MIDDLE_PASSING_TESTCASES:
mid = middle(x, y, z)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Some ExamplesLet us try out the `InvariantAnnotator` on a number of examples. Removing HTML MarkupRunning `InvariantAnnotator` on our ongoing example `remove_html_markup()` does not provide much, as our invariant properties are tailored towards numerical functions.
###Code
from Intro_Debugging import remove_html_markup
with InvariantAnnotator() as annotator:
remove_html_markup("<foo>bar</foo>")
remove_html_markup("bar")
remove_html_markup('"bar"')
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
In the [chapter on DDSet](DDSetDebugger.ipynb), we will see how to express more complex properties for structured inputs. A Recursive FunctionHere's another example. `list_length()` recursively computes the length of a Python function. Let us see whether we can mine its invariants:
###Code
def list_length(elems: List[Any]) -> int:
if elems == []:
length = 0
else:
length = 1 + list_length(elems[1:])
return length
with InvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Almost all these properties are relevant. Of course, the reason the invariants are so neat is that the return value is equal to `len(elems)` is that `X == len(Y)` is part of the list of properties to be checked. Sum of two NumbersThe next example is a very simple function: If we have a function without return value, the return value is `None` and we can only mine preconditions. (Well, we get a "postcondition" `not return_value` that the return value evaluates to False, which holds for `None`).
###Code
def print_sum(a, b): # type: ignore
print(a + b)
with InvariantAnnotator() as annotator:
print_sum(31, 45)
print_sum(0, 0)
print_sum(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Checking SpecificationsA function with invariants, as above, can be fed into the Python interpreter, such that all pre- and postconditions are checked. We create a function `square_root_annotated()` which includes all the invariants mined above.
###Code
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
square_root_def = annotator.functions_with_invariants()
square_root_def = square_root_def.replace('square_root',
'square_root_annotated')
print_content(square_root_def, '.py')
exec(square_root_def)
###Output
_____no_output_____
###Markdown
The "annotated" version checks against invalid arguments – or more precisely, against arguments with properties that have not been observed yet:
###Code
with ExpectError():
square_root_annotated(-1.0) # type: ignore
###Output
_____no_output_____
###Markdown
This is in contrast to the original version, which just hangs on negative values:
###Code
with ExpectTimeout(1):
square_root(-1.0)
###Output
_____no_output_____
###Markdown
If we make changes to the function definition such that the properties of the return value change, such _regressions_ are caught as violations of the postconditions. Let us illustrate this by simply inverting the result, and return $-2$ as square root of 4.
###Code
square_root_def = square_root_def.replace('square_root_annotated',
'square_root_negative')
square_root_def = square_root_def.replace('return approx',
'return -approx')
print_content(square_root_def, '.py')
exec(square_root_def)
###Output
_____no_output_____
###Markdown
Technically speaking, $-2$ _is_ a square root of 4, since $(-2)^2 = 4$ holds. Yet, such a change may be unexpected by callers of `square_root()`, and hence, this would be caught with the first call:
###Code
with ExpectError():
square_root_negative(2.0) # type: ignore
###Output
_____no_output_____
###Markdown
We see how pre- and postconditions, as well as types, can serve as *oracles* during testing. In particular, once we have mined them for a set of functions, we can check them again and again with test generators – especially after code changes. The more checks we have, and the more specific they are, the more likely it is we can detect unwanted effects of changes. SynopsisThis chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:
###Code
def sum2(a, b): # type: ignore
return a + b
with TypeAnnotator() as type_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.
###Code
print(type_annotator.typed_functions())
###Output
_____no_output_____
###Markdown
The invariant annotator works in a similar fashion:
###Code
with InvariantAnnotator() as inv_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.
###Code
print(inv_annotator.functions_with_invariants())
###Output
_____no_output_____
###Markdown
Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs). The chapter gives details on how to customize the properties checked for. Lessons Learned* Type annotations and explicit invariants allow for _checking_ arguments and results for expected data types and other properties.* One can automatically _mine_ data types and invariants by observing arguments and results at runtime.* The quality of mined invariants depends on the diversity of values observed during executions; this variety can be increased by generating tests. Next StepsIn the next chapter, we will explore [abstracting failure conditions](DDSetDebugger.ipynb). BackgroundThe [DAIKON dynamic invariant detector](https://plse.cs.washington.edu/daikon/) can be considered the mother of function specification miners. Continuously maintained and extended for more than 20 years, it mines likely invariants in the style of this chapter for a variety of languages, including C, C++, C, Eiffel, F, Java, Perl, and Visual Basic. On top of the functionality discussed above, it holds a rich catalog of patterns for likely invariants, supports data invariants, can eliminate invariants that are implied by others, and determines statistical confidence to disregard unlikely invariants. The corresponding paper \cite{Ernst2001} is one of the seminal and most-cited papers of Software Engineering. A multitude of works have been published based on DAIKON and detecting invariants; see this [curated list](http://plse.cs.washington.edu/daikon/pubs/) for details. The interaction between test generators and invariant detection is already discussed in \cite{Ernst2001} (incidentally also using grammars). The Eclat tool \cite{Pacheco2005} is a model example of tight interaction between a unit-level test generator and DAIKON-style invariant mining, where the mined invariants are used to produce oracles and to systematically guide the test generator towards fault-revealing inputs. Mining specifications is not restricted to pre- and postconditions. The paper "Mining Specifications" \cite{Ammons2002} is another classic in the field, learning state protocols from executions. Grammar mining \cite{Gopinath2020} can also be seen as a specification mining approach, this time learning specifications for input formats. As it comes to adding type annotations to existing code, the blog post ["The state of type hints in Python"](https://www.bernat.tech/the-state-of-type-hints-in-python/) gives a great overview on how Python type hints can be used and checked. To add type annotations, there are two important tools available that also implement our above approach:* [MonkeyType](https://instagram-engineering.com/let-your-code-type-hint-itself-introducing-open-source-monkeytype-a855c7284881) implements the above approach of tracing executions and annotating Python 3 arguments, returns, and variables with type hints.* [PyAnnotate](https://github.com/dropbox/pyannotate) does a similar job, focusing on code in Python 2. It does not produce Python 3-style annotations, but instead produces annotations as comments that can be processed by static type checkers.These tools have been created by engineers at Facebook and Dropbox, respectively, assisting them in checking millions of lines of code for type issues. ExercisesOur code for mining types and invariants is in no way complete. There are dozens of ways to extend our implementations, some of which we discuss in exercises. Exercise 1: Union TypesThe Python `typing` module allows to express that an argument can have multiple types. For `square_root(x)`, this allows to express that `x` can be an `int` or a `float`:
###Code
def square_root_with_union_type(x: Union[int, float]) -> float: # type: ignore
...
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it supports union types for arguments and return values. Use `Optional[X]` as a shorthand for `Union[X, None]`. **Solution.** Left to the reader. Hint: extend `type_string()`. Exercise 2: Types for Local VariablesIn Python, one cannot only annotate arguments with types, but actually also local and global variables – for instance, `approx` and `guess` in our `square_root()` implementation:
###Code
def square_root_with_local_types(x: Union[int, float]) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
approx: Optional[float] = None
guess: float = x / 2
while approx != guess:
approx: float = guess # type: ignore
guess: float = (approx + x / approx) / 2 # type: ignore
return approx
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it also annotates local variables with types. Search the function AST for assignments, determine the type of the assigned value, and make it an annotation on the left hand side. **Solution.** Left to the reader. Exercise 3: Verbose Invariant CheckersOur implementation of invariant checkers does not make it clear for the user which pre-/postcondition failed.
###Code
@precondition(lambda s: len(s) > 0)
def remove_first_char(s: str) -> str:
return s[1:]
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
The following implementation adds an optional `doc` keyword argument which is printed if the invariant is violated:
###Code
def my_condition(precondition: Optional[Callable] = None,
postcondition: Optional[Callable] = None, doc: str = 'Unknown') -> Callable:
def decorator(func: Callable) -> Callable:
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args: Any, **kwargs: Any) -> Any:
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated: " + doc
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated: " + doc
return retval
return wrapper
return decorator
def my_precondition(check: Callable, **kwargs: Any) -> Callable:
return my_condition(precondition=check, doc=kwargs.get('doc', 'Unknown'))
def my_postcondition(check: Callable, **kwargs: Any) -> Callable:
return my_condition(postcondition=check, doc=kwargs.get('doc', 'Unknown'))
@my_precondition(lambda s: len(s) > 0, doc="len(s) > 0") # type: ignore
def remove_first_char(s: str) -> str:
return s[1:]
remove_first_char('abc')
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
Extend `InvariantAnnotator` such that it includes the conditions in the generated pre- and postconditions. **Solution.** Here's a simple solution:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@my_precondition(lambda " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")"
conditions.append(cond)
return conditions
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@my_postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")")
conditions.append(cond)
return conditions
###Output
_____no_output_____
###Markdown
The resulting annotations are harder to read, but easier to diagnose:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
As an alternative, one may be able to use `inspect.getsource()` on the lambda expression or unparse it. This is left to the reader. Exercise 4: Save Initial ValuesIf the value of an argument changes during function execution, this can easily confuse our implementation: The values are tracked at the beginning of the function, but checked only when it returns. Extend the `InvariantAnnotator` and the infrastructure it uses such that* it saves argument values both at the beginning and at the end of a function invocation;* postconditions can be expressed over both _initial_ values of arguments as well as the _final_ values of arguments;* the mined postconditions refer to both these values as well. **Solution.** To be added. Exercise 5: ImplicationsSeveral mined invariant are actually _implied_ by others: If `x > 0` holds, then this implies `x >= 0` and `x != 0`. Extend the `InvariantAnnotator` such that implications between properties are explicitly encoded, and such that implied properties are no longer listed as invariants. See \cite{Ernst2001} for ideas. **Solution.** Left to the reader. Exercise 6: Local VariablesPostconditions may also refer to the values of local variables. Consider extending `InvariantAnnotator` and its infrastructure such that the values of local variables at the end of the execution are also recorded and made part of the invariant inference mechanism. **Solution.** Left to the reader. Exercise 7: Embedding Invariants as AssertionsRather than producing invariants as annotations for pre- and postconditions, insert them as `assert` statements into the function code, as in:```pythondef square_root(x): 'Computes the square root of x, using the Newton-Raphson method' assert isinstance(x, int), 'violated precondition' assert x > 0, 'violated precondition' approx = None guess = (x / 2) while (approx != guess): approx = guess guess = ((approx + (x / approx)) / 2) return_value = approx assert return_value < x, 'violated postcondition' assert isinstance(return_value, float), 'violated postcondition' return approx```Such a formulation may make it easier for test generators and symbolic analysis to access and interpret pre- and postconditions. **Solution.** Here is a tentative implementation that inserts invariants into function ASTs. Part 1: Embedding Invariants into Functions
###Code
class EmbeddedInvariantAnnotator(InvariantTracer):
def function_with_invariants_ast(self, function_name: str) -> ast.AST:
return annotate_function_with_invariants(function_name, self.invariants(function_name))
def function_with_invariants(self, function_name: str) -> str:
return ast.unparse(self.function_with_invariants_ast(function_name))
def annotate_invariants(invariants: Dict[str, Invariants]) -> Dict[str, ast.AST]:
annotated_functions = {}
for function_name in invariants:
try:
annotated_functions[function_name] = annotate_function_with_invariants(function_name, invariants[function_name])
except KeyError:
continue
return annotated_functions
def annotate_function_with_invariants(function_name: str,
function_invariants: Invariants) -> ast.AST:
function = globals()[function_name]
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_invariants(function_ast, function_invariants)
def annotate_function_ast_with_invariants(function_ast: ast.AST,
function_invariants: Invariants) -> ast.AST:
annotated_function_ast = EmbeddedInvariantTransformer(function_invariants).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Part 2: Preconditions
###Code
class PreconditionTransformer(ast.NodeTransformer):
def __init__(self, invariants: Invariants) -> None:
self.invariants = invariants
super().__init__()
def preconditions(self) -> List[ast.stmt]:
preconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated precondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) < 0:
preconditions += assertion_ast.body
return preconditions
def insert_assertions(self, body: List[ast.stmt]) -> List[ast.stmt]:
preconditions = self.preconditions()
try:
docstring = cast(ast.Constant, body[0]).value.s
except:
docstring = None
if docstring:
return [body[0]] + preconditions + body[1:]
else:
return preconditions + body
def visit_FunctionDef(self, node: ast.FunctionDef) -> ast.FunctionDef:
"""Add invariants to function"""
# print(ast.dump(node))
node.body = self.insert_assertions(node.body)
return node
class EmbeddedInvariantTransformer(PreconditionTransformer):
pass
with EmbeddedInvariantAnnotator() as annotator:
square_root(5)
print_content(annotator.function_with_invariants('square_root'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.function_with_invariants('sum3'), '.py')
###Output
_____no_output_____
###Markdown
Part 3: PostconditionsWe make a few simplifying assumptions: * Variables do not change during execution.* There is a single `return` statement at the end of the function.
###Code
class EmbeddedInvariantTransformer(EmbeddedInvariantTransformer):
def postconditions(self) -> List[ast.stmt]:
postconditions = []
for (prop, var_names) in self.invariants:
assertion = ("assert " + instantiate_prop(prop, var_names) +
', "violated postcondition"')
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) >= 0:
postconditions += assertion_ast.body
return postconditions
def insert_assertions(self, body: List[ast.stmt]) -> List[ast.stmt]:
new_body = super().insert_assertions(body)
postconditions = self.postconditions()
body_ends_with_return = isinstance(new_body[-1], ast.Return)
if body_ends_with_return:
ret_val = cast(ast.Return, new_body[-1]).value
saver = RETURN_VALUE + " = " + ast.unparse(cast(ast.AST, ret_val))
else:
saver = RETURN_VALUE + " = None"
saver_ast = cast(ast.stmt, ast.parse(saver))
postconditions = [saver_ast] + postconditions
if body_ends_with_return:
return new_body[:-1] + postconditions + [new_body[-1]]
else:
return new_body + postconditions
with EmbeddedInvariantAnnotator() as annotator:
square_root(5)
square_root_def = annotator.function_with_invariants('square_root')
###Output
_____no_output_____
###Markdown
Here's the full definition with included assertions:
###Code
print_content(square_root_def, '.py')
exec(square_root_def.replace('square_root', 'square_root_annotated'))
with ExpectError():
square_root_annotated(-1)
###Output
_____no_output_____
###Markdown
Here come some more examples:
###Code
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.function_with_invariants('sum3'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.function_with_invariants('list_length'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
print_sum(31, 45)
print_content(annotator.function_with_invariants('print_sum'), '.py')
###Output
_____no_output_____
###Markdown
Mining Function SpecificationsWhen testing a program, one not only needs to cover its several behaviors; one also needs to _check_ whether the result is as expected. In this chapter, we introduce a technique that allows us to _mine_ function specifications from a set of given executions, resulting in abstract and formal _descriptions_ of what the function expects and what it delivers. These so-called _dynamic invariants_ produce pre- and post-conditions over function arguments and variables from a set of executions. They are useful in a variety of contexts:* Dynamic invariants provide important information for [symbolic fuzzing](SymbolicFuzzer.ipynb), such as types and ranges of function arguments.* Dynamic invariants provide pre- and postconditions for formal program proofs and verification.* Dynamic invariants provide a large number of assertions that can check whether function behavior has changed* Checks provided by dynamic invariants can be very useful as _oracles_ for checking the effects of generated testsTraditionally, dynamic invariants are dependent on the executions they are derived from. However, when paired with comprehensive test generators, they quickly become very precise, as we show in this chapter. **Prerequisites*** You should be familiar with tracing program executions, as in the [chapter on coverage](Coverage.ipynb).* Later in this section, we access the internal _abstract syntax tree_ representations of Python programs and transform them, as in the [chapter on information flow](InformationFlow.ipynb).
###Code
import fuzzingbook_utils
import Coverage
import Intro_Testing
###Output
_____no_output_____
###Markdown
SynopsisTo [use the code provided in this chapter](Importing.ipynb), write```python>>> from fuzzingbook.DynamicInvariants import ```and then make use of the following features.This chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:```python>>> def sum2(a, b):>>> return a + b>>> with TypeAnnotator() as type_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.```python>>> print(type_annotator.typed_functions())def sum2(a: int, b: int) ->int: return a + b```The invariant annotator works in a similar fashion:```python>>> with InvariantAnnotator() as inv_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.```python>>> print(inv_annotator.functions_with_invariants())@precondition(lambda a, b: isinstance(a, int), doc='isinstance(a, int)')@precondition(lambda a, b: isinstance(b, int), doc='isinstance(b, int)')@postcondition(lambda return_value, a, b: a == return_value - b, doc='a == return_value - b')@postcondition(lambda return_value, a, b: b == return_value - a, doc='b == return_value - a')@postcondition(lambda return_value, a, b: isinstance(return_value, int), doc='isinstance(return_value, int)')@postcondition(lambda return_value, a, b: return_value == a + b, doc='return_value == a + b')@postcondition(lambda return_value, a, b: return_value == b + a, doc='return_value == b + a')def sum2(a, b): return a + b```Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs) as well as for all kinds of _symbolic code analyses_. The chapter gives details on how to customize the properties checked for. Specifications and AssertionsWhen implementing a function or program, one usually works against a _specification_ – a set of documented requirements to be satisfied by the code. Such specifications can come in natural language. A formal specification, however, allows the computer to check whether the specification is satisfied.In the [introduction to testing](Intro_Testing.ipynb), we have seen how _preconditions_ and _postconditions_ can describe what a function does. Consider the following (simple) square root function:
###Code
def my_sqrt(x):
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
The assertion `assert p` checks the condition `p`; if it does not hold, execution is aborted. Here, the actual body is not yet written; we use the assertions as a specification of what `my_sqrt()` _expects_, and what it _delivers_.The topmost assertion is the _precondition_, stating the requirements on the function arguments. The assertion at the end is the _postcondition_, stating the properties of the function result (including its relationship with the original arguments). Using these pre- and postconditions as a specification, we can now go and implement a square root function that satisfies them. Once implemented, we can have the assertions check at runtime whether `my_sqrt()` works as expected; a [symbolic](SymbolicFuzzer.ipynb) or [concolic](ConcolicFuzzer.ipynb) test generator will even specifically try to find inputs where the assertions do _not_ hold. (An assertion can be seen as a conditional branch towards aborting the execution, and any technique that tries to cover all code branches will also try to invalidate as many assertions as possible.) However, not every piece of code is developed with explicit specifications in the first place; let alone does most code comes with formal pre- and post-conditions. (Just take a look at the chapters in this book.) This is a pity: As Ken Thompson famously said, "Without specifications, there are no bugs – only surprises". It is also a problem for testing, since, of course, testing needs some specification to test against. This raises the interesting question: Can we somehow _retrofit_ existing code with "specifications" that properly describe their behavior, allowing developers to simply _check_ them rather than having to write them from scratch? This is what we do in this chapter. Why Generic Error Checking is Not EnoughBefore we go into _mining_ specifications, let us first discuss why it could be useful to _have_ them. As a motivating example, consider the full implementation of `my_sqrt()` from the [introduction to testing](Intro_Testing.ipynb):
###Code
import fuzzingbook_utils
def my_sqrt(x):
"""Computes the square root of x, using the Newton-Raphson method"""
approx = None
guess = x / 2
while approx != guess:
approx = guess
guess = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
`my_sqrt()` does not come with any functionality that would check types or values. Hence, it is easy for callers to make mistakes when calling `my_sqrt()`:
###Code
from ExpectError import ExpectError, ExpectTimeout
with ExpectError():
my_sqrt("foo")
with ExpectError():
x = my_sqrt(0.0)
###Output
_____no_output_____
###Markdown
At least, the Python system catches these errors at runtime. The following call, however, simply lets the function enter an infinite loop:
###Code
with ExpectTimeout(1):
x = my_sqrt(-1.0)
###Output
_____no_output_____
###Markdown
Our goal is to avoid such errors by _annotating_ functions with information that prevents errors like the above ones. The idea is to provide a _specification_ of expected properties – a specification that can then be checked at runtime or statically. \todo{Introduce the concept of *contract*.} Specifying and Checking Data TypesFor our Python code, one of the most important "specifications" we need is *types*. Python being a "dynamically" typed language means that all data types are determined at run time; the code itself does not explicitly state whether a variable is an integer, a string, an array, a dictionary – or whatever. As _writer_ of Python code, omitting explicit type declarations may save time (and allows for some fun hacks). It is not sure whether a lack of types helps in _reading_ and _understanding_ code for humans. For a _computer_ trying to analyze code, the lack of explicit types is detrimental. If, say, a constraint solver, sees `if x:` and cannot know whether `x` is supposed to be a number or a string, this introduces an _ambiguity_. Such ambiguities may multiply over the entire analysis in a combinatorial explosion – or in the analysis yielding an overly inaccurate result. Python 3.6 and later allows data types as _annotations_ to function arguments (actually, to all variables) and return values. We can, for instance, state that `my_sqrt()` is a function that accepts a floating-point value and returns one:
###Code
def my_sqrt_with_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
By default, such annotations are ignored by the Python interpreter. Therefore, one can still call `my_sqrt_typed()` with a string as an argument and get the exact same result as above. However, one can make use of special _typechecking_ modules that would check types – _dynamically_ at runtime or _statically_ by analyzing the code without having to execute it. Runtime Type CheckingThe Python `enforce` package provides a function decorator that automatically inserts type-checking code that is executed at runtime. Here is how to use it:
###Code
import enforce
@enforce.runtime_validation
def my_sqrt_with_checked_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
Now, invoking `my_sqrt_with_checked_type_annotations()` raises an exception when invoked with a type dfferent from the one declared:
###Code
with ExpectError():
my_sqrt_with_checked_type_annotations(True)
###Output
_____no_output_____
###Markdown
Note that this error is not caught by the "untyped" variant, where passing a boolean value happily returns $\sqrt{1}$ as result.
###Code
my_sqrt(True)
###Output
_____no_output_____
###Markdown
In Python (and other languages), the boolean values `True` and `False` can be implicitly converted to the integers 1 and 0; however, it is hard to think of a call to `sqrt()` where this would not be an error. Static Type CheckingType annotations can also be checked _statically_ – that is, without even running the code. Let us create a simple Python file consisting of the above `my_sqrt_typed()` definition and a bad invocation.
###Code
import inspect
import tempfile
f = tempfile.NamedTemporaryFile(mode='w', suffix='.py')
f.name
f.write(inspect.getsource(my_sqrt))
f.write('\n')
f.write(inspect.getsource(my_sqrt_with_type_annotations))
f.write('\n')
f.write("print(my_sqrt_with_type_annotations('123'))\n")
f.flush()
###Output
_____no_output_____
###Markdown
These are the contents of our newly created Python file:
###Code
from fuzzingbook_utils import print_file
print_file(f.name)
###Output
_____no_output_____
###Markdown
[Mypy](http://mypy-lang.org) is a type checker for Python programs. As it checks types statically, types induce no overhead at runtime; plus, a static check can be faster than a lengthy series of tests with runtime type checking enabled. Let us see what `mypy` produces on the above file:
###Code
import subprocess
result = subprocess.run(["mypy", "--strict", f.name], universal_newlines=True, stdout=subprocess.PIPE)
del f # Delete temporary file
print(result.stdout)
###Output
_____no_output_____
###Markdown
We see that `mypy` complains about untyped function definitions such as `my_sqrt()`; most important, however, it finds that the call to `my_sqrt_with_type_annotations()` in the last line has the wrong type. With `mypy`, we can achieve the same type safety with Python as in statically typed languages – provided that we as programmers also produce the necessary type annotations. Is there a simple way to obtain these? Mining Type SpecificationsOur first task will be to mine type annotations (as part of the code) from _values_ we observe at run time. These type annotations would be _mined_ from actual function executions, _learning_ from (normal) runs what the expected argument and return types should be. By observing a series of calls such as these, we could infer that both `x` and the return value are of type `float`:
###Code
y = my_sqrt(25.0)
y
y = my_sqrt(2.0)
y
###Output
_____no_output_____
###Markdown
How can we mine types from executions? The answer is simple: 1. We _observe_ a function during execution2. We track the _types_ of its arguments3. We include these types as _annotations_ into the code.To do so, we can make use of Python's tracing facility we already observed in the [chapter on coverage](Coverage.ipynb). With every call to a function, we retrieve the arguments, their values, and their types. Tracking CallsTo observe argument types at runtime, we define a _tracer function_ that tracks the execution of `my_sqrt()`, checking its arguments and return values. The `Tracker` class is set to trace functions in a `with` block as follows:```pythonwith Tracker() as tracker: function_to_be_tracked(...)info = tracker.collected_information()```As in the [chapter on coverage](Coverage.ipynb), we use the `sys.settrace()` function to trace individual functions during execution. We turn on tracking when the `with` block starts; at this point, the `__enter__()` method is called. When execution of the `with` block ends, `__exit()__` is called.
###Code
import sys
class Tracker(object):
def __init__(self, log=False):
self._log = log
self.reset()
def reset(self):
self._calls = {}
self._stack = []
def traceit(self):
"""Placeholder to be overloaded in subclasses"""
pass
# Start of `with` block
def __enter__(self):
self.original_trace_function = sys.gettrace()
sys.settrace(self.traceit)
return self
# End of `with` block
def __exit__(self, exc_type, exc_value, tb):
sys.settrace(self.original_trace_function)
###Output
_____no_output_____
###Markdown
The `traceit()` method does nothing yet; this is done in specialized subclasses. The `CallTracker` class implements a `traceit()` function that checks for function calls and returns:
###Code
class CallTracker(Tracker):
def traceit(self, frame, event, arg):
"""Tracking function: Record all calls and all args"""
if event == "call":
self.trace_call(frame, event, arg)
elif event == "return":
self.trace_return(frame, event, arg)
return self.traceit
###Output
_____no_output_____
###Markdown
`trace_call()` is called when a function is called; it retrieves the function name and current arguments, and saves them on a stack.
###Code
class CallTracker(CallTracker):
def trace_call(self, frame, event, arg):
"""Save current function name and args on the stack"""
code = frame.f_code
function_name = code.co_name
arguments = get_arguments(frame)
self._stack.append((function_name, arguments))
if self._log:
print(simple_call_string(function_name, arguments))
def get_arguments(frame):
"""Return call arguments in the given frame"""
# When called, all arguments are local variables
arguments = [(var, frame.f_locals[var]) for var in frame.f_locals]
arguments.reverse() # Want same order as call
return arguments
###Output
_____no_output_____
###Markdown
When the function returns, `trace_return()` is called. We now also have the return value. We log the whole call with arguments and return value (if desired) and save it in our list of calls.
###Code
class CallTracker(CallTracker):
def trace_return(self, frame, event, arg):
"""Get return value and store complete call with arguments and return value"""
code = frame.f_code
function_name = code.co_name
return_value = arg
# TODO: Could call get_arguments() here to also retrieve _final_ values of argument variables
called_function_name, called_arguments = self._stack.pop()
assert function_name == called_function_name
if self._log:
print(simple_call_string(function_name, called_arguments), "returns", return_value)
self.add_call(function_name, called_arguments, return_value)
###Output
_____no_output_____
###Markdown
`simple_call_string()` is a helper for logging that prints out calls in a user-friendly manner.
###Code
def simple_call_string(function_name, argument_list, return_value=None):
"""Return function_name(arg[0], arg[1], ...) as a string"""
call = function_name + "(" + \
", ".join([var + "=" + repr(value)
for (var, value) in argument_list]) + ")"
if return_value is not None:
call += " = " + repr(return_value)
return call
###Output
_____no_output_____
###Markdown
`add_call()` saves the calls in a list; each function name has its own list.
###Code
class CallTracker(CallTracker):
def add_call(self, function_name, arguments, return_value=None):
"""Add given call to list of calls"""
if function_name not in self._calls:
self._calls[function_name] = []
self._calls[function_name].append((arguments, return_value))
###Output
_____no_output_____
###Markdown
Using `calls()`, we can retrieve the list of calls, either for a given function, or for all functions.
###Code
class CallTracker(CallTracker):
def calls(self, function_name=None):
"""Return list of calls for function_name,
or a mapping function_name -> calls for all functions tracked"""
if function_name is None:
return self._calls
return self._calls[function_name]
###Output
_____no_output_____
###Markdown
Let us now put this to use. We turn on logging to track the individual calls and their return values:
###Code
with CallTracker(log=True) as tracker:
y = my_sqrt(25)
y = my_sqrt(2.0)
###Output
_____no_output_____
###Markdown
After execution, we can retrieve the individual calls:
###Code
calls = tracker.calls('my_sqrt')
calls
###Output
_____no_output_____
###Markdown
Each call is pair (`argument_list`, `return_value`), where `argument_list` is a list of pairs (`parameter_name`, `value`).
###Code
my_sqrt_argument_list, my_sqrt_return_value = calls[0]
simple_call_string('my_sqrt', my_sqrt_argument_list, my_sqrt_return_value)
###Output
_____no_output_____
###Markdown
If the function does not return a value, `return_value` is `None`.
###Code
def hello(name):
print("Hello,", name)
with CallTracker() as tracker:
hello("world")
hello_calls = tracker.calls('hello')
hello_calls
hello_argument_list, hello_return_value = hello_calls[0]
simple_call_string('hello', hello_argument_list, hello_return_value)
###Output
_____no_output_____
###Markdown
Getting TypesDespite what you may have read or heard, Python actually _is_ a typed language. It is just that it is _dynamically typed_ – types are used and checked only at runtime (rather than declared in the code, where they can be _statically checked_ at compile time). We can thus retrieve types of all values within Python:
###Code
type(4)
type(2.0)
type([4])
###Output
_____no_output_____
###Markdown
We can retrieve the type of the first argument to `my_sqrt()`:
###Code
parameter, value = my_sqrt_argument_list[0]
parameter, type(value)
###Output
_____no_output_____
###Markdown
as well as the type of the return value:
###Code
type(my_sqrt_return_value)
###Output
_____no_output_____
###Markdown
Hence, we see that (so far), `my_sqrt()` is a function taking (among others) integers and returning floats. We could declare `my_sqrt()` as:
###Code
def my_sqrt_annotated(x: int) -> float:
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
This is a representation we could place in a static type checker, allowing to check whether calls to `my_sqrt()` actually pass a number. A dynamic type checker could run such checks at runtime. And of course, any [symbolic interpretation](SymbolicFuzzer.ipynb) will greatly profit from the additional annotations. By default, Python does not do anything with such annotations. However, tools can access annotations from functions and other objects:
###Code
my_sqrt_annotated.__annotations__
###Output
_____no_output_____
###Markdown
This is how run-time checkers access the annotations to check against. Accessing Function StructureOur plan is to annotate functions automatically, based on the types we have seen. To do so, we need a few modules that allow us to convert a function into a tree representation (called _abstract syntax trees_, or ASTs) and back; we already have seen these in the chapters on [concolic](ConcolicFuzzer.ipynb) and [symbolic](SymbolicFuzzer.ipynb) testing.
###Code
import ast
import inspect
import astor
###Output
_____no_output_____
###Markdown
We can get the source of a Python function using `inspect.getsource()`. (Note that this does not work for functions defined in other notebooks.)
###Code
my_sqrt_source = inspect.getsource(my_sqrt)
my_sqrt_source
###Output
_____no_output_____
###Markdown
To view these in a visually pleasing form, our function `print_content(s, suffix)` formats and highlights the string `s` as if it were a file with ending `suffix`. We can thus view (and highlight) the source as if it were a Python file:
###Code
from fuzzingbook_utils import print_content
print_content(my_sqrt_source, '.py')
###Output
_____no_output_____
###Markdown
Parsing this gives us an abstract syntax tree (AST) – a representation of the program in tree form.
###Code
my_sqrt_ast = ast.parse(my_sqrt_source)
###Output
_____no_output_____
###Markdown
What does this AST look like? The helper functions `astor.dump_tree()` (textual output) and `showast.show_ast()` (graphical output with [showast](https://github.com/hchasestevens/show_ast)) allow us to inspect the structure of the tree. We see that the function starts as a `FunctionDef` with name and arguments, followed by a body, which is a list of statements of type `Expr` (the docstring), type `Assign` (assignments), `While` (while loop with its own body), and finally `Return`.
###Code
print(astor.dump_tree(my_sqrt_ast))
###Output
_____no_output_____
###Markdown
Too much text for you? This graphical representation may make things simpler.
###Code
from fuzzingbook_utils import rich_output
if rich_output():
import showast
showast.show_ast(my_sqrt_ast)
###Output
_____no_output_____
###Markdown
The function `astor.to_source()` converts such a tree back into the more familiar textual Python code representation. Comments are gone, and there may be more parentheses than before, but the result has the same semantics:
###Code
print_content(astor.to_source(my_sqrt_ast), '.py')
###Output
_____no_output_____
###Markdown
Annotating Functions with Given TypesLet us now go and transform these trees ti add type annotations. We start with a helper function `parse_type(name)` which parses a type name into an AST.
###Code
def parse_type(name):
class ValueVisitor(ast.NodeVisitor):
def visit_Expr(self, node):
self.value_node = node.value
tree = ast.parse(name)
name_visitor = ValueVisitor()
name_visitor.visit(tree)
return name_visitor.value_node
print(astor.dump_tree(parse_type('int')))
print(astor.dump_tree(parse_type('[object]')))
###Output
_____no_output_____
###Markdown
We now define a helper function that actually adds type annotations to a function AST. The `TypeTransformer` class builds on the Python standard library `ast.NodeTransformer` infrastructure. It would be called as```python TypeTransformer({'x': 'int'}, 'float').visit(ast)```to annotate the arguments of `my_sqrt()`: `x` with `int`, and the return type with `float`. The returned AST can then be unparsed, compiled or analyzed.
###Code
class TypeTransformer(ast.NodeTransformer):
def __init__(self, argument_types, return_type=None):
self.argument_types = argument_types
self.return_type = return_type
super().__init__()
###Output
_____no_output_____
###Markdown
The core of `TypeTransformer` is the method `visit_FunctionDef()`, which is called for every function definition in the AST. Its argument `node` is the subtree of the function definition to be transformed. Our implementation accesses the individual arguments and invokes `annotate_args()` on them; it also sets the return type in the `returns` attribute of the node.
###Code
class TypeTransformer(TypeTransformer):
def visit_FunctionDef(self, node):
"""Add annotation to function"""
# Set argument types
new_args = []
for arg in node.args.args:
new_args.append(self.annotate_arg(arg))
new_arguments = ast.arguments(
new_args,
node.args.vararg,
node.args.kwonlyargs,
node.args.kw_defaults,
node.args.kwarg,
node.args.defaults
)
# Set return type
if self.return_type is not None:
node.returns = parse_type(self.return_type)
return ast.copy_location(ast.FunctionDef(node.name, new_arguments,
node.body, node.decorator_list,
node.returns), node)
###Output
_____no_output_____
###Markdown
Each argument gets its own annotation, taken from the types originally passed to the class:
###Code
class TypeTransformer(TypeTransformer):
def annotate_arg(self, arg):
"""Add annotation to single function argument"""
arg_name = arg.arg
if arg_name in self.argument_types:
arg.annotation = parse_type(self.argument_types[arg_name])
return arg
###Output
_____no_output_____
###Markdown
Does this work? Let us annotate the AST from `my_sqrt()` with types for the arguments and return types:
###Code
new_ast = TypeTransformer({'x': 'int'}, 'float').visit(my_sqrt_ast)
###Output
_____no_output_____
###Markdown
When we unparse the new AST, we see that the annotations actually are present:
###Code
print_content(astor.to_source(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Similarly, we can annotate the `hello()` function from above:
###Code
hello_source = inspect.getsource(hello)
hello_ast = ast.parse(hello_source)
new_ast = TypeTransformer({'name': 'str'}, 'None').visit(hello_ast)
print_content(astor.to_source(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Annotating Functions with Mined TypesLet us now annotate functions with types mined at runtime. We start with a simple function `type_string()` that determines the appropriate type of a given value (as a string):
###Code
def type_string(value):
return type(value).__name__
type_string(4)
type_string([])
###Output
_____no_output_____
###Markdown
For composite structures, `type_string()` does not examine element types; hence, the type of `[3]` is simply `list` instead of, say, `list[int]`. For now, `list` will do fine.
###Code
type_string([3])
###Output
_____no_output_____
###Markdown
`type_string()` will be used to infer the types of argument values found at runtime, as returned by `CallTracker.calls()`:
###Code
with CallTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(2.0)
tracker.calls()
###Output
_____no_output_____
###Markdown
The function `annotate_types()` takes such a list of calls and annotates each function listed:
###Code
def annotate_types(calls):
annotated_functions = {}
for function_name in calls:
try:
annotated_functions[function_name] = annotate_function_with_types(function_name, calls[function_name])
except KeyError:
continue
return annotated_functions
###Output
_____no_output_____
###Markdown
For each function, we get the source and its AST and then get to the actual annotation in `annotate_function_ast_with_types()`:
###Code
def annotate_function_with_types(function_name, function_calls):
function = globals()[function_name] # May raise KeyError for internal functions
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_types(function_ast, function_calls)
###Output
_____no_output_____
###Markdown
The function `annotate_function_ast_with_types()` invokes the `TypeTransformer` with the calls seen, and for each call, iterate over the arguments, determine their types, and annotate the AST with these. The universal type `Any` is used when we encounter type conflicts, which we will discuss below.
###Code
from typing import Any
def annotate_function_ast_with_types(function_ast, function_calls):
parameter_types = {}
return_type = None
for calls_seen in function_calls:
args, return_value = calls_seen
if return_value is not None:
if return_type is not None and return_type != type_string(return_value):
return_type = 'Any'
else:
return_type = type_string(return_value)
for parameter, value in args:
try:
different_type = parameter_types[parameter] != type_string(value)
except KeyError:
different_type = False
if different_type:
parameter_types[parameter] = 'Any'
else:
parameter_types[parameter] = type_string(value)
annotated_function_ast = TypeTransformer(parameter_types, return_type).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Here is `my_sqrt()` annotated with the types recorded usign the tracker, above.
###Code
print_content(astor.to_source(annotate_types(tracker.calls())['my_sqrt']), '.py')
###Output
_____no_output_____
###Markdown
All-in-one AnnotationLet us bring all of this together in a single class `TypeAnnotator` that first tracks calls of functions and then allows to access the AST (and the source code form) of the tracked functions annotated with types. The method `typed_functions()` returns the annotated functions as a string; `typed_functions_ast()` returns their AST.
###Code
class TypeTracker(CallTracker):
pass
class TypeAnnotator(TypeTracker):
def typed_functions_ast(self, function_name=None):
if function_name is None:
return annotate_types(self.calls())
return annotate_function_with_types(function_name, self.calls(function_name))
def typed_functions(self, function_name=None):
if function_name is None:
functions = ''
for f_name in self.calls():
try:
f_text = astor.to_source(self.typed_functions_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
return astor.to_source(self.typed_functions_ast(function_name))
###Output
_____no_output_____
###Markdown
Here is how to use `TypeAnnotator`. We first track a series of calls:
###Code
with TypeAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(2.0)
###Output
_____no_output_____
###Markdown
After tracking, we can immediately retrieve an annotated version of the functions tracked:
###Code
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
This also works for multiple and diverse functions. One could go and implement an automatic type annotator for Python files based on the types seen during execution.
###Code
with TypeAnnotator() as annotator:
hello('type annotations')
y = my_sqrt(1.0)
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
A content as above could now be sent to a type checker, which would detect any type inconsistency between callers and callees. Likewise, type annotations such as the ones above greatly benefit symbolic code analysis (as in the chapter on [symbolic fuzzing](SymbolicFuzzer.ipynb)), as they effectively constrain the set of values that arguments and variables can take. Multiple TypesLet us now resolve the role of the magic `Any` type in `annotate_function_ast_with_types()`. If we see multiple types for the same argument, we set its type to `Any`. For `my_sqrt()`, this makes sense, as its arguments can be integers as well as floats:
###Code
with CallTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(4)
print_content(astor.to_source(annotate_types(tracker.calls())['my_sqrt']), '.py')
###Output
_____no_output_____
###Markdown
The following function `sum3()` can be called with floating-point numbers as arguments, resulting in the parameters getting a `float` type:
###Code
def sum3(a, b, c):
return a + b + c
with TypeAnnotator() as annotator:
y = sum3(1.0, 2.0, 3.0)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we call `sum3()` with integers, though, the arguments get an `int` type:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
And we can also call `sum3()` with strings, giving the arguments a `str` type:
###Code
with TypeAnnotator() as annotator:
y = sum3("one", "two", "three")
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we have multiple calls, but with different types, `TypeAnnotator()` will assign an `Any` type to both arguments and return values:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y = sum3("one", "two", "three")
typed_sum3_def = annotator.typed_functions('sum3')
print_content(typed_sum3_def, '.py')
###Output
_____no_output_____
###Markdown
A type `Any` makes it explicit that an object can, indeed, have any type; it will not be typechecked at runtime or statically. To some extent, this defeats the power of type checking; but it also preserves some of the type flexibility that many Python programmers enjoy. Besides `Any`, the `typing` module supports several additional ways to define ambiguous types; we will keep this in mind for a later exercise. Specifying and Checking InvariantsBesides basic data types. we can check several further properties from arguments. We can, for instance, whether an argument can be negative, zero, or positive; or that one argument should be smaller than the second; or that the result should be the sum of two arguments – properties that cannot be expressed in a (Python) type.Such properties are called *invariants*, as they hold across all invocations of a function. Specifically, invariants come as _pre_- and _postconditions_ – conditions that always hold at the beginning and at the end of a function. (There are also _data_ and _object_ invariants that express always-holding properties over the state of data or objects, but we do not consider these in this book.) Annotating Functions with Pre- and PostconditionsThe classical means to specify pre- and postconditions is via _assertions_, which we have introduced in the [chapter on testing](Intro_Testing.ipynb). A precondition checks whether the arguments to a function satisfy the expected properties; a postcondition does the same for the result. We can express and check both using assertions as follows:
###Code
def my_sqrt_with_invariants(x):
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
A nicer way, however, is to syntactically separate invariants from the function at hand. Using appropriate decorators, we could specify pre- and postconditions as follows:```python@precondition lambda x: x >= 0@postcondition lambda return_value, x: return_value * return_value == xdef my_sqrt_with_invariants(x): normal code without assertions ...```The decorators `@precondition` and `@postcondition` would run the given functions (specified as anonymous `lambda` functions) before and after the decorated function, respectively. If the functions return `False`, the condition is violated. `@precondition` gets the function arguments as arguments; `@postcondition` additionally gets the return value as first argument. It turns out that implementing such decorators is not hard at all. Our implementation builds on a [code snippet from StackOverflow](https://stackoverflow.com/questions/12151182/python-precondition-postcondition-for-member-function-how):
###Code
import functools
def condition(precondition=None, postcondition=None):
def decorator(func):
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args, **kwargs):
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated"
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated"
return retval
return wrapper
return decorator
def precondition(check):
return condition(precondition=check)
def postcondition(check):
return condition(postcondition=check)
###Output
_____no_output_____
###Markdown
With these, we can now start decorating `my_sqrt()`:
###Code
@precondition(lambda x: x > 0)
def my_sqrt_with_precondition(x):
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
This catches arguments violating the precondition:
###Code
with ExpectError():
my_sqrt_with_precondition(-1.0)
###Output
_____no_output_____
###Markdown
Likewise, we can provide a postcondition:
###Code
EPSILON = 1e-5
@postcondition(lambda ret, x: ret * ret - x < EPSILON)
def my_sqrt_with_postcondition(x):
return my_sqrt(x)
y = my_sqrt_with_postcondition(2.0)
y
###Output
_____no_output_____
###Markdown
If we have a buggy implementation of $\sqrt{x}$, this gets caught quickly:
###Code
@postcondition(lambda ret, x: ret * ret - x < EPSILON)
def buggy_my_sqrt_with_postcondition(x):
return my_sqrt(x) + 0.1
with ExpectError():
y = buggy_my_sqrt_with_postcondition(2.0)
###Output
_____no_output_____
###Markdown
While checking pre- and postconditions is a great way to catch errors, specifying them can be cumbersome. Let us try to see whether we can (again) _mine_ some of them. Mining InvariantsTo _mine_ invariants, we can use the same tracking functionality as before; instead of saving values for individual variables, though, we now check whether the values satisfy specific _properties_ or not. For instance, if all values of `x` seen satisfy the condition `x > 0`, then we make `x > 0` an invariant of the function. If we see positive, zero, and negative values of `x`, though, then there is no property of `x` left to talk about.The general idea is thus:1. Check all variable values observed against a set of predefined properties; and2. Keep only those properties that hold for all runs observed. Defining PropertiesWhat precisely do we mean by properties? Here is a small collection of value properties that would frequently be used in invariants. All these properties would be evaluated with the _metavariables_ `X`, `Y`, and `Z` (actually, any upper-case identifier) being replaced with the names of function parameters:
###Code
INVARIANT_PROPERTIES = [
"X < 0",
"X <= 0",
"X > 0",
"X >= 0",
"X == 0",
"X != 0",
]
###Output
_____no_output_____
###Markdown
When `my_sqrt(x)` is called as, say `my_sqrt(5.0)`, we see that `x = 5.0` holds. The above properties would then all be checked for `x`. Only the properties `X > 0`, `X >= 0`, and `X != 0` hold for the call seen; and hence `x > 0`, `x >= 0`, and `x != 0` would make potential preconditions for `my_sqrt(x)`. We can check for many more properties such as relations between two arguments:
###Code
INVARIANT_PROPERTIES += [
"X == Y",
"X > Y",
"X < Y",
"X >= Y",
"X <= Y",
]
###Output
_____no_output_____
###Markdown
Types also can be checked using properties. For any function parameter `X`, only one of these will hold:
###Code
INVARIANT_PROPERTIES += [
"isinstance(X, bool)",
"isinstance(X, int)",
"isinstance(X, float)",
"isinstance(X, list)",
"isinstance(X, dict)",
]
###Output
_____no_output_____
###Markdown
We can check for arithmetic properties:
###Code
INVARIANT_PROPERTIES += [
"X == Y + Z",
"X == Y * Z",
"X == Y - Z",
"X == Y / Z",
]
###Output
_____no_output_____
###Markdown
Here's relations over three values, a Python special:
###Code
INVARIANT_PROPERTIES += [
"X < Y < Z",
"X <= Y <= Z",
"X > Y > Z",
"X >= Y >= Z",
]
###Output
_____no_output_____
###Markdown
Finally, we can also check for list or string properties. Again, this is just a tiny selection.
###Code
INVARIANT_PROPERTIES += [
"X == len(Y)",
"X == sum(Y)",
"X.startswith(Y)",
]
###Output
_____no_output_____
###Markdown
Extracting Meta-VariablesLet us first introduce a few _helper functions_ before we can get to the actual mining. `metavars()` extracts the set of meta-variables (`X`, `Y`, `Z`, etc.) from a property. To this end, we parse the property as a Python expression and then visit the identifiers.
###Code
def metavars(prop):
metavar_list = []
class ArgVisitor(ast.NodeVisitor):
def visit_Name(self, node):
if node.id.isupper():
metavar_list.append(node.id)
ArgVisitor().visit(ast.parse(prop))
return metavar_list
assert metavars("X < 0") == ['X']
assert metavars("X.startswith(Y)") == ['X', 'Y']
assert metavars("isinstance(X, str)") == ['X']
###Output
_____no_output_____
###Markdown
Instantiating PropertiesTo produce a property as invariant, we need to be able to _instantiate_ it with variable names. The instantiation of `X > 0` with `X` being instantiated to `a`, for instance, gets us `a > 0`. To this end, the function `instantiate_prop()` takes a property and a collection of variable names and instantiates the meta-variables left-to-right with the corresponding variables names in the collection.
###Code
def instantiate_prop_ast(prop, var_names):
class NameTransformer(ast.NodeTransformer):
def visit_Name(self, node):
if node.id not in mapping:
return node
return ast.Name(id=mapping[node.id], ctx=ast.Load())
meta_variables = metavars(prop)
assert len(meta_variables) == len(var_names)
mapping = {}
for i in range(0, len(meta_variables)):
mapping[meta_variables[i]] = var_names[i]
prop_ast = ast.parse(prop, mode='eval')
new_ast = NameTransformer().visit(prop_ast)
return new_ast
def instantiate_prop(prop, var_names):
prop_ast = instantiate_prop_ast(prop, var_names)
prop_text = astor.to_source(prop_ast).strip()
while prop_text.startswith('(') and prop_text.endswith(')'):
prop_text = prop_text[1:-1]
return prop_text
assert instantiate_prop("X > Y", ['a', 'b']) == 'a > b'
assert instantiate_prop("X.startswith(Y)", ['x', 'y']) == 'x.startswith(y)'
###Output
_____no_output_____
###Markdown
Evaluating PropertiesTo actually _evaluate_ properties, we do not need to instantiate them. Instead, we simply convert them into a boolean function, using `lambda`:
###Code
def prop_function_text(prop):
return "lambda " + ", ".join(metavars(prop)) + ": " + prop
def prop_function(prop):
return eval(prop_function_text(prop))
###Output
_____no_output_____
###Markdown
Here is a simple example:
###Code
prop_function_text("X > Y")
p = prop_function("X > Y")
p(100, 1)
p(1, 100)
###Output
_____no_output_____
###Markdown
Checking InvariantsTo extract invariants from an execution, we need to check them on all possible instantiations of arguments. If the function to be checked has two arguments `a` and `b`, we instantiate the property `X < Y` both as `a < b` and `b < a` and check each of them. To get all combinations, we use the Python `permutations()` function:
###Code
import itertools
for combination in itertools.permutations([1.0, 2.0, 3.0], 2):
print(combination)
###Output
_____no_output_____
###Markdown
The function `true_property_instantiations()` takes a property and a list of tuples (`var_name`, `value`). It then produces all instantiations of the property with the given values and returns those that evaluate to True.
###Code
def true_property_instantiations(prop, vars_and_values, log=False):
instantiations = set()
p = prop_function(prop)
len_metavars = len(metavars(prop))
for combination in itertools.permutations(vars_and_values, len_metavars):
args = [value for var_name, value in combination]
var_names = [var_name for var_name, value in combination]
try:
result = p(*args)
except:
result = None
if log:
print(prop, combination, result)
if result:
instantiations.add((prop, tuple(var_names)))
return instantiations
###Output
_____no_output_____
###Markdown
Here is an example. If `x == -1` and `y == 1`, the property `X < Y` holds for `x < y`, but not for `y < x`:
###Code
invs = true_property_instantiations("X < Y", [('x', -1), ('y', 1)], log=True)
invs
###Output
_____no_output_____
###Markdown
The instantiation retrieves the short form:
###Code
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Likewise, with values for `x` and `y` as above, the property `X < 0` only holds for `x`, but not for `y`:
###Code
invs = true_property_instantiations("X < 0", [('x', -1), ('y', 1)], log=True)
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Extracting InvariantsLet us now run the above invariant extraction on function arguments and return values as observed during a function execution. To this end, we extend the `CallTracker` class into an `InvariantTracker` class, which automatically computes invariants for all functions and all calls observed during tracking. By default, an `InvariantTracker` uses the properties as defined above; however, one can specify alternate sets of properties.
###Code
class InvariantTracker(CallTracker):
def __init__(self, props=None, **kwargs):
if props is None:
props = INVARIANT_PROPERTIES
self.props = props
super().__init__(**kwargs)
###Output
_____no_output_____
###Markdown
The key method of the `InvariantTracker` is the `invariants()` method. This iterates over the calls observed and checks which properties hold. Only the intersection of properties – that is, the set of properties that hold for all calls – is preserved, and eventually returned. The special variable `return_value` is set to hold the return value.
###Code
RETURN_VALUE = 'return_value'
class InvariantTracker(InvariantTracker):
def invariants(self, function_name=None):
if function_name is None:
return {function_name: self.invariants(function_name) for function_name in self.calls()}
invariants = None
for variables, return_value in self.calls(function_name):
vars_and_values = variables + [(RETURN_VALUE, return_value)]
s = set()
for prop in self.props:
s |= true_property_instantiations(prop, vars_and_values, self._log)
if invariants is None:
invariants = s
else:
invariants &= s
return invariants
###Output
_____no_output_____
###Markdown
Here's an example of how to use `invariants()`. We run the tracker on a small set of calls.
###Code
with InvariantTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(10.0)
tracker.calls()
###Output
_____no_output_____
###Markdown
The `invariants()` method produces a set of properties that hold for the observed runs, together with their instantiations over function arguments.
###Code
invs = tracker.invariants('my_sqrt')
invs
###Output
_____no_output_____
###Markdown
As before, the actual instantiations are easier to read:
###Code
def pretty_invariants(invariants):
props = []
for (prop, var_names) in invariants:
props.append(instantiate_prop(prop, var_names))
return sorted(props)
pretty_invariants(invs)
###Output
_____no_output_____
###Markdown
We see that the both `x` and the return value have a `float` type. We also see that both are always greater than zero. These are properties that may make useful pre- and postconditions, notably for symbolic analysis. However, there's also an invariant which does _not_ universally hold, namely `return_value <= x`, as the following example shows:
###Code
my_sqrt(0.01)
###Output
_____no_output_____
###Markdown
Clearly, 0.1 > 0.01 holds. This is a case of us not learning from sufficiently diverse inputs. As soon as we have a call including `x = 0.1`, though, the invariant `return_value <= x` is eliminated:
###Code
with InvariantTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(10.0)
y = my_sqrt(0.01)
pretty_invariants(tracker.invariants('my_sqrt'))
###Output
_____no_output_____
###Markdown
We will discuss later how to ensure sufficient diversity in inputs. (Hint: This involves test generation.) Let us try out our invariant tracker on `sum3()`. We see that all types are well-defined; the properties that all arguments are non-zero, however, is specific to the calls observed.
###Code
with InvariantTracker() as tracker:
y = sum3(1, 2, 3)
y = sum3(-4, -5, -6)
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with strings instead, we get different invariants. Notably, we obtain the postcondition that the return value starts with the value of `a` – a universal postcondition if strings are used.
###Code
with InvariantTracker() as tracker:
y = sum3('a', 'b', 'c')
y = sum3('f', 'e', 'd')
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with both strings and numbers (and zeros, too), there are no properties left that would hold across all calls. That's the price of flexibility.
###Code
with InvariantTracker() as tracker:
y = sum3('a', 'b', 'c')
y = sum3('c', 'b', 'a')
y = sum3(-4, -5, -6)
y = sum3(0, 0, 0)
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
Converting Mined Invariants to AnnotationsAs with types, above, we would like to have some functionality where we can add the mined invariants as annotations to existing functions. To this end, we introduce the `InvariantAnnotator` class, extending `InvariantTracker`. We start with a helper method. `params()` returns a comma-separated list of parameter names as observed during calls.
###Code
class InvariantAnnotator(InvariantTracker):
def params(self, function_name):
arguments, return_value = self.calls(function_name)[0]
return ", ".join(arg_name for (arg_name, arg_value) in arguments)
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = sum3(1, 2, 3)
annotator.params('my_sqrt')
annotator.params('sum3')
###Output
_____no_output_____
###Markdown
Now for the actual annotation. `preconditions()` returns the preconditions from the mined invariants (i.e., those propertes that do not depend on the return value) as a string with annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@precondition(lambda " + self.params(function_name) + ": " + inv + ")"
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
annotator.preconditions('my_sqrt')
###Output
_____no_output_____
###Markdown
`postconditions()` does the same for postconditions:
###Code
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ")")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
annotator.postconditions('my_sqrt')
###Output
_____no_output_____
###Markdown
With these, we can take a function and add both pre- and postconditions as annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def functions_with_invariants(self):
functions = ""
for function_name in self.invariants():
try:
function = self.function_with_invariants(function_name)
except KeyError:
continue
functions += function
return functions
def function_with_invariants(self, function_name):
function = globals()[function_name] # Can throw KeyError
source = inspect.getsource(function)
return "\n".join(self.preconditions(function_name) +
self.postconditions(function_name)) + '\n' + source
###Output
_____no_output_____
###Markdown
Here comes `function_with_invariants()` in all its glory:
###Code
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
print_content(annotator.function_with_invariants('my_sqrt'), '.py')
###Output
_____no_output_____
###Markdown
Quite a lot of invariants, is it? Further below (and in the exercises), we will discuss on how to focus on the most relevant properties. Some ExamplesHere's another example. `list_length()` recursively computes the length of a Python function. Let us see whether we can mine its invariants:
###Code
def list_length(L):
if L == []:
length = 0
else:
length = 1 + list_length(L[1:])
return length
with InvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Almost all these properties (except for the very first) are relevant. Of course, the reason the invariants are so neat is that the return value is equal to `len(L)` is that `X == len(Y)` is part of the list of properties to be checked. The next example is a very simple function:
###Code
def sum2(a, b):
return a + b
with InvariantAnnotator() as annotator:
sum2(31, 45)
sum2(0, 0)
sum2(-1, -5)
###Output
_____no_output_____
###Markdown
The invariants all capture the relationship between `a`, `b`, and the return value as `return_value == a + b` in all its variations.
###Code
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
If we have a function without return value, the return value is `None` and we can only mine preconditions. (Well, we get a "postcondition" that the return value is non-zero, which holds for `None`).
###Code
def print_sum(a, b):
print(a + b)
with InvariantAnnotator() as annotator:
print_sum(31, 45)
print_sum(0, 0)
print_sum(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Checking SpecificationsA function with invariants, as above, can be fed into the Python interpreter, such that all pre- and postconditions are checked. We create a function `my_sqrt_annotated()` which includes all the invariants mined above.
###Code
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
my_sqrt_def = annotator.functions_with_invariants()
my_sqrt_def = my_sqrt_def.replace('my_sqrt', 'my_sqrt_annotated')
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def)
###Output
_____no_output_____
###Markdown
The "annotated" version checks against invalid arguments – or more precisely, against arguments with properties that have not been observed yet:
###Code
with ExpectError():
my_sqrt_annotated(-1.0)
###Output
_____no_output_____
###Markdown
This is in contrast to the original version, which just hangs on negative values:
###Code
with ExpectTimeout(1):
my_sqrt(-1.0)
###Output
_____no_output_____
###Markdown
If we make changes to the function definition such that the properties of the return value change, such _regressions_ are caught as violations of the postconditions. Let us illustrate this by simply inverting the result, and return $-2$ as square root of 4.
###Code
my_sqrt_def = my_sqrt_def.replace('my_sqrt_annotated', 'my_sqrt_negative')
my_sqrt_def = my_sqrt_def.replace('return approx', 'return -approx')
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def)
###Output
_____no_output_____
###Markdown
Technically speaking, $-2$ _is_ a square root of 4, since $(-2)^2 = 4$ holds. Yet, such a change may be unexpected by callers of `my_sqrt()`, and hence, this would be caught with the first call:
###Code
with ExpectError():
my_sqrt_negative(2.0)
###Output
_____no_output_____
###Markdown
We see how pre- and postconditions, as well as types, can serve as *oracles* during testing. In particular, once we have mined them for a set of functions, we can check them again and again with test generators – especially after code changes. The more checks we have, and the more specific they are, the more likely it is we can detect unwanted effects of changes. Mining Specifications from Generated TestsMined specifications can only be as good as the executions they were mined from. If we only see a single call to, say, `sum2()`, we will be faced with several mined pre- and postconditions that _overspecialize_ towards the values seen:
###Code
def sum2(a, b):
return a + b
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
The mined precondition `a == b`, for instance, only holds for the single call observed; the same holds for the mined postcondition `return_value == a * b`. Yet, `sum2()` can obviously be successfully called with other values that do not satisfy these conditions. To get out of this trap, we have to _learn from more and more diverse runs_. If we have a few more calls of `sum2()`, we see how the set of invariants quickly gets smaller:
###Code
with InvariantAnnotator() as annotator:
length = sum2(1, 2)
length = sum2(-1, -2)
length = sum2(0, 0)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
But where to we get such diverse runs from? This is the job of generating software tests. A simple grammar for calls of `sum2()` will easily resolve the problem.
###Code
from GrammarFuzzer import GrammarFuzzer # minor dependency
from Grammars import is_valid_grammar, crange, convert_ebnf_grammar # minor dependency
SUM2_EBNF_GRAMMAR = {
"<start>": ["<sum2>"],
"<sum2>": ["sum2(<int>, <int>)"],
"<int>": ["<_int>"],
"<_int>": ["(-)?<leaddigit><digit>*", "0"],
"<leaddigit>": crange('1', '9'),
"<digit>": crange('0', '9')
}
assert is_valid_grammar(SUM2_EBNF_GRAMMAR)
sum2_grammar = convert_ebnf_grammar(SUM2_EBNF_GRAMMAR)
sum2_fuzzer = GrammarFuzzer(sum2_grammar)
[sum2_fuzzer.fuzz() for i in range(10)]
with InvariantAnnotator() as annotator:
for i in range(10):
eval(sum2_fuzzer.fuzz())
print_content(annotator.function_with_invariants('sum2'), '.py')
###Output
_____no_output_____
###Markdown
But then, writing tests (or a test driver) just to derive a set of pre- and postconditions may possibly be too much effort – in particular, since tests can easily be derived from given pre- and postconditions in the first place. Hence, it would be wiser to first specify invariants and then let test generators or program provers do the job. Also, an API grammar, such as above, will have to be set up such that it actually respects preconditions – in our case, we invoke `sqrt()` with positive numbers only, already assuming its precondition. In some way, one thus needs a specification (a model, a grammar) to mine another specification – a chicken-and-egg problem. However, there is one way out of this problem: If one can automatically generate tests at the system level, then one has an _infinite source of executions_ to learn invariants from. In each of these executions, all functions would be called with values that satisfy the (implicit) precondition, allowing us to mine invariants for these functions. This holds, because at the system level, invalid inputs must be rejected by the system in the first place. The meaningful precondition at the system level, ensuring that only valid inputs get through, thus gets broken down into a multitude of meaningful preconditions (and subsequent postconditions) at the function level. The big requirement for this, though, is that one needs good test generators at the system level. In [the next part](05_Domain-Specific_Fuzzing.ipynb), we will discuss how to automatically generate tests for a variety of domains, from configuration to graphical user interfaces. SynopsisThis chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:
###Code
def sum2(a, b):
return a + b
with TypeAnnotator() as type_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.
###Code
print(type_annotator.typed_functions())
###Output
_____no_output_____
###Markdown
The invariant annotator works in a similar fashion:
###Code
with InvariantAnnotator() as inv_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.
###Code
print(inv_annotator.functions_with_invariants())
###Output
_____no_output_____
###Markdown
Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs) as well as for all kinds of _symbolic code analyses_. The chapter gives details on how to customize the properties checked for. Lessons Learned* Type annotations and explicit invariants allow for _checking_ arguments and results for expected data types and other properties.* One can automatically _mine_ data types and invariants by observing arguments and results at runtime.* The quality of mined invariants depends on the diversity of values observed during executions; this variety can be increased by generating tests. Next StepsThis chapter concludes the [part on semantical fuzzing techniques](04_Semantical_Fuzzing.ipynb). In the next part, we will explore [domain-specific fuzzing techniques](05_Domain-Specific_Fuzzing.ipynb) from configurations and APIs to graphical user interfaces. BackgroundThe [DAIKON dynamic invariant detector](https://plse.cs.washington.edu/daikon/) can be considered the mother of function specification miners. Continuously maintained and extended for more than 20 years, it mines likely invariants in the style of this chapter for a variety of languages, including C, C++, C, Eiffel, F, Java, Perl, and Visual Basic. On top of the functionality discussed above, it holds a rich catalog of patterns for likely invariants, supports data invariants, can eliminate invariants that are implied by others, and determines statistical confidence to disregard unlikely invariants. The corresponding paper \cite{Ernst2001} is one of the seminal and most-cited papers of Software Engineering. A multitude of works have been published based on DAIKON and detecting invariants; see this [curated list](http://plse.cs.washington.edu/daikon/pubs/) for details. The interaction between test generators and invariant detection is already discussed in \cite{Ernst2001} (incidentally also using grammars). The Eclat tool \cite{Pacheco2005} is a model example of tight interaction between a unit-level test generator and DAIKON-style invariant mining, where the mined invariants are used to produce oracles and to systematically guide the test generator towards fault-revealing inputs. Mining specifications is not restricted to pre- and postconditions. The paper "Mining Specifications" \cite{Ammons2002} is another classic in the field, learning state protocols from executions. Grammar mining, as described in [our chapter with the same name](GrammarMiner.ipynb) can also be seen as a specification mining approach, this time learning specifications for input formats. As it comes to adding type annotations to existing code, the blog post ["The state of type hints in Python"](https://www.bernat.tech/the-state-of-type-hints-in-python/) gives a great overview on how Python type hints can be used and checked. To add type annotations, there are two important tools available that also implement our above approach:* [MonkeyType](https://instagram-engineering.com/let-your-code-type-hint-itself-introducing-open-source-monkeytype-a855c7284881) implements the above approach of tracing executions and annotating Python 3 arguments, returns, and variables with type hints.* [PyAnnotate](https://github.com/dropbox/pyannotate) does a similar job, focusing on code in Python 2. It does not produce Python 3-style annotations, but instead produces annotations as comments that can be processed by static type checkers.These tools have been created by engineers at Facebook and Dropbox, respectively, assisting them in checking millions of lines of code for type issues. ExercisesOur code for mining types and invariants is in no way complete. There are dozens of ways to extend our implementations, some of which we discuss in exercises. Exercise 1: Union TypesThe Python `typing` module allows to express that an argument can have multiple types. For `my_sqrt(x)`, this allows to express that `x` can be an `int` or a `float`:
###Code
from typing import Union, Optional
def my_sqrt_with_union_type(x: Union[int, float]) -> float:
...
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it supports union types for arguments and return values. Use `Optional[X]` as a shorthand for `Union[X, None]`. **Solution.** Left to the reader. Hint: extend `type_string()`. Exercise 2: Types for Local VariablesIn Python, one cannot only annotate arguments with types, but actually also local and global variables – for instance, `approx` and `guess` in our `my_sqrt()` implementation:
###Code
def my_sqrt_with_local_types(x: Union[int, float]) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
approx: Optional[float] = None
guess: float = x / 2
while approx != guess:
approx: float = guess
guess: float = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it also annotates local variables with types. Search the function AST for assignments, determine the type of the assigned value, and make it an annotation on the left hand side. **Solution.** Left to the reader. Exercise 3: Verbose Invariant CheckersOur implementation of invariant checkers does not make it clear for the user which pre-/postcondition failed.
###Code
@precondition(lambda s: len(s) > 0)
def remove_first_char(s):
return s[1:]
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
The following implementation adds an optional `doc` keyword argument which is printed if the invariant is violated:
###Code
def condition(precondition=None, postcondition=None, doc='Unknown'):
def decorator(func):
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args, **kwargs):
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated: " + doc
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated: " + doc
return retval
return wrapper
return decorator
def precondition(check, **kwargs):
return condition(precondition=check, doc=kwargs.get('doc', 'Unknown'))
def postcondition(check, **kwargs):
return condition(postcondition=check, doc=kwargs.get('doc', 'Unknown'))
@precondition(lambda s: len(s) > 0, doc="len(s) > 0")
def remove_first_char(s):
return s[1:]
remove_first_char('abc')
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
Extend `InvariantAnnotator` such that it includes the conditions in the generated pre- and postconditions. **Solution.** Here's a simple solution:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@precondition(lambda " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")"
conditions.append(cond)
return conditions
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")")
conditions.append(cond)
return conditions
###Output
_____no_output_____
###Markdown
The resulting annotations are harder to read, but easier to diagnose:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
As an alternative, one may be able to use `inspect.getsource()` on the lambda expression or unparse it. This is left to the reader. Exercise 4: Save Initial ValuesIf the value of an argument changes during function execution, this can easily confuse our implementation: The values are tracked at the beginning of the function, but checked only when it returns. Extend the `InvariantAnnotator` and the infrastructure it uses such that* it saves argument values both at the beginning and at the end of a function invocation;* postconditions can be expressed over both _initial_ values of arguments as well as the _final_ values of arguments;* the mined postconditions refer to both these values as well. **Solution.** To be added. Exercise 5: ImplicationsSeveral mined invariant are actually _implied_ by others: If `x > 0` holds, then this implies `x >= 0` and `x != 0`. Extend the `InvariantAnnotator` such that implications between properties are explicitly encoded, and such that implied properties are no longer listed as invariants. See \cite{Ernst2001} for ideas. **Solution.** Left to the reader. Exercise 6: Local VariablesPostconditions may also refer to the values of local variables. Consider extending `InvariantAnnotator` and its infrastructure such that the values of local variables at the end of the execution are also recorded and made part of the invariant inference mechanism. **Solution.** Left to the reader. Exercise 7: Exploring Invariant AlternativesAfter mining a first set of invariants, have a [concolic fuzzer](ConcolicFuzzer.ipynb) generate tests that systematically attempt to invalidate pre- and postconditions. How far can you generalize? **Solution.** To be added. Exercise 8: Grammar-Generated PropertiesThe larger the set of properties to be checked, the more potential invariants can be discovered. Create a _grammar_ that systematically produces a large set of properties. See \cite{Ernst2001} for possible patterns. **Solution.** Left to the reader. Exercise 9: Embedding Invariants as AssertionsRather than producing invariants as annotations for pre- and postconditions, insert them as `assert` statements into the function code, as in:```pythondef my_sqrt(x): 'Computes the square root of x, using the Newton-Raphson method' assert isinstance(x, int), 'violated precondition' assert (x > 0), 'violated precondition' approx = None guess = (x / 2) while (approx != guess): approx = guess guess = ((approx + (x / approx)) / 2) return_value = approx assert (return_value < x), 'violated postcondition' assert isinstance(return_value, float), 'violated postcondition' return approx```Such a formulation may make it easier for test generators and symbolic analysis to access and interpret pre- and postconditions. **Solution.** Here is a tentative implementation that inserts invariants into function ASTs. Part 1: Embedding Invariants into Functions
###Code
class EmbeddedInvariantAnnotator(InvariantTracker):
def functions_with_invariants_ast(self, function_name=None):
if function_name is None:
return annotate_functions_with_invariants(self.invariants())
return annotate_function_with_invariants(function_name, self.invariants(function_name))
def functions_with_invariants(self, function_name=None):
if function_name is None:
functions = ''
for f_name in self.invariants():
try:
f_text = astor.to_source(self.functions_with_invariants_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
return astor.to_source(self.functions_with_invariants_ast(function_name))
def function_with_invariants(self, function_name):
return self.functions_with_invariants(function_name)
def function_with_invariants_ast(self, function_name):
return self.functions_with_invariants_ast(function_name)
def annotate_invariants(invariants):
annotated_functions = {}
for function_name in invariants:
try:
annotated_functions[function_name] = annotate_function_with_invariants(function_name, invariants[function_name])
except KeyError:
continue
return annotated_functions
def annotate_function_with_invariants(function_name, function_invariants):
function = globals()[function_name]
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_invariants(function_ast, function_invariants)
def annotate_function_ast_with_invariants(function_ast, function_invariants):
annotated_function_ast = EmbeddedInvariantTransformer(function_invariants).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Part 2: Preconditions
###Code
class PreconditionTransformer(ast.NodeTransformer):
def __init__(self, invariants):
self.invariants = invariants
super().__init__()
def preconditions(self):
preconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated precondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) < 0:
preconditions += assertion_ast.body
return preconditions
def insert_assertions(self, body):
preconditions = self.preconditions()
try:
docstring = body[0].value.s
except:
docstring = None
if docstring:
return [body[0]] + preconditions + body[1:]
else:
return preconditions + body
def visit_FunctionDef(self, node):
"""Add invariants to function"""
# print(ast.dump(node))
node.body = self.insert_assertions(node.body)
return node
class EmbeddedInvariantTransformer(PreconditionTransformer):
pass
with EmbeddedInvariantAnnotator() as annotator:
my_sqrt(5)
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Part 3: PostconditionsWe make a few simplifying assumptions: * Variables do not change during execution.* There is a single `return` statement at the end of the function.
###Code
class EmbeddedInvariantTransformer(PreconditionTransformer):
def postconditions(self):
postconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated postcondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) >= 0:
postconditions += assertion_ast.body
return postconditions
def insert_assertions(self, body):
new_body = super().insert_assertions(body)
postconditions = self.postconditions()
body_ends_with_return = isinstance(new_body[-1], ast.Return)
if body_ends_with_return:
saver = RETURN_VALUE + " = " + astor.to_source(new_body[-1].value)
else:
saver = RETURN_VALUE + " = None"
saver_ast = ast.parse(saver)
postconditions = [saver_ast] + postconditions
if body_ends_with_return:
return new_body[:-1] + postconditions + [new_body[-1]]
else:
return new_body + postconditions
with EmbeddedInvariantAnnotator() as annotator:
my_sqrt(5)
my_sqrt_def = annotator.functions_with_invariants()
###Output
_____no_output_____
###Markdown
Here's the full definition with included assertions:
###Code
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def.replace('my_sqrt', 'my_sqrt_annotated'))
with ExpectError():
my_sqrt_annotated(-1)
###Output
_____no_output_____
###Markdown
Here come some more examples:
###Code
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
print_sum(31, 45)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Mining Function SpecificationsIn the [chapter on assertions](Assertions.ipynb), we have seen how important it is to _check_ whether the result is as expected. In this chapter, we introduce a technique that allows us to _mine_ function specifications from a set of given executions, resulting in abstract and formal _descriptions_ of what the function expects and what it delivers.These so-called _dynamic invariants_ produce pre- and post-conditions over function arguments and variables from a set of executions. Within debugging, the resulting _assertions_ can immediately check whether function behavior has changed, but can also be useful to determine the characteristics of _failing_ runs (as opposed to _passing_ runs). Furthermore, the resulting specifications provide pre- and postconditions for formal program proofs, testing, and verification.This chapter is based on [a chapter with the same name in The Fuzzing Book](https://www.fuzzingbook.org/html/DynamicInvariants.html), which focuses on test generation.
###Code
from bookutils import YouTubeVideo
YouTubeVideo("HDu1olXFvv0")
###Output
_____no_output_____
###Markdown
**Prerequisites*** You should be familiar with tracing program executions, as in the [chapter on tracing](Tracer.ipynb).* Later in this section, we access the internal _abstract syntax tree_ representations of Python programs and transform them, as in the [chapter on tracking failure origins](Slicer.ipynb).
###Code
import bookutils
from Tracer import Tracer
# ignore
from typing import Sequence, Any, Callable, Tuple
from typing import Dict, Union, Set, List, cast, Optional
###Output
_____no_output_____
###Markdown
SynopsisTo [use the code provided in this chapter](Importing.ipynb), write```python>>> from debuggingbook.DynamicInvariants import ```and then make use of the following features.This chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:```python>>> def sum2(a, b): type: ignore>>> return a + b>>> with TypeAnnotator() as type_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.```python>>> print(type_annotator.typed_functions())def sum2(a: int, b: int) ->int: return a + b```The invariant annotator works in a similar fashion:```python>>> with InvariantAnnotator() as inv_annotator:>>> sum2(1, 2)>>> sum2(-4, -5)>>> sum2(0, 0)```The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.```python>>> print(inv_annotator.functions_with_invariants())@precondition(lambda a, b: isinstance(a, int))@precondition(lambda a, b: isinstance(b, int))@postcondition(lambda return_value, a, b: a == return_value - b)@postcondition(lambda return_value, a, b: b == return_value - a)@postcondition(lambda return_value, a, b: isinstance(return_value, int))@postcondition(lambda return_value, a, b: return_value == a + b)@postcondition(lambda return_value, a, b: return_value == b + a)def sum2(a, b): type: ignore return a + b```Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs). The chapter gives details on how to customize the properties checked for. Specifications and AssertionsWhen implementing a function or program, one usually works against a _specification_ – a set of documented requirements to be satisfied by the code. Such specifications can come in natural language. A formal specification, however, allows the computer to check whether the specification is satisfied.In the [chapter on assertions](Assertions.ipynb), we have seen how _preconditions_ and _postconditions_ can describe what a function does. Consider the following (simple) square root function:
###Code
def square_root(x): # type: ignore
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
The assertion `assert p` checks the condition `p`; if it does not hold, execution is aborted. Here, the actual body is not yet written; we use the assertions as a specification of what `square_root()` _expects_, and what it _delivers_.The topmost assertion is the _precondition_, stating the requirements on the function arguments. The assertion at the end is the _postcondition_, stating the properties of the function result (including its relationship with the original arguments). Using these pre- and postconditions as a specification, we can now go and implement a square root function that satisfies them. Once implemented, we can have the assertions check at runtime whether `square_root()` works as expected. However, not every piece of code is developed with explicit specifications in the first place; let alone does most code comes with formal pre- and post-conditions. (Just take a look at the chapters in this book.) This is a pity: As Ken Thompson famously said, "Without specifications, there are no bugs – only surprises". It is also a problem for debugging, since, of course, debugging needs some specification such that we know what is wrong, and how to fix it. This raises the interesting question: Can we somehow _retrofit_ existing code with "specifications" that properly describe their behavior, allowing developers to simply _check_ them rather than having to write them from scratch? This is what we do in this chapter. Beyond Generic FailuresBefore we go into _mining_ specifications, let us first discuss why it could be useful to _have_ them. As a motivating example, consider the full implementation of `square_root()` from the [chapter on assertions](Assertions.ipynb):
###Code
import bookutils
def square_root(x): # type: ignore
"""Computes the square root of x, using the Newton-Raphson method"""
approx = None
guess = x / 2
while approx != guess:
approx = guess
guess = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
`square_root()` does not come with any functionality that would check types or values. Hence, it is easy for callers to make mistakes when calling `square_root()`:
###Code
from ExpectError import ExpectError, ExpectTimeout
with ExpectError():
square_root("foo")
with ExpectError():
x = square_root(0.0)
###Output
_____no_output_____
###Markdown
At least, the Python system catches these errors at runtime. The following call, however, simply lets the function enter an infinite loop:
###Code
with ExpectTimeout(1):
x = square_root(-1.0)
###Output
_____no_output_____
###Markdown
Our goal is to avoid such errors by _annotating_ functions with information that prevents errors like the above ones. The idea is to provide a _specification_ of expected properties – a specification that can then be checked at runtime or statically. \todo{Introduce the concept of *contract*.} Mining Data TypesFor our Python code, one of the most important "specifications" we need is *types*. Python being a "dynamically" typed language means that all data types are determined at run time; the code itself does not explicitly state whether a variable is an integer, a string, an array, a dictionary – or whatever. As _writer_ of Python code, omitting explicit type declarations may save time (and allows for some fun hacks). It is not sure whether a lack of types helps in _reading_ and _understanding_ code for humans. For a _computer_ trying to analyze code, the lack of explicit types is detrimental. If, say, a constraint solver, sees `if x:` and cannot know whether `x` is supposed to be a number or a string, this introduces an _ambiguity_. Such ambiguities may multiply over the entire analysis in a combinatorial explosion – or in the analysis yielding an overly inaccurate result. Python 3.6 and later allow data types as _annotations_ to function arguments (actually, to all variables) and return values. We can, for instance, state that `square_root()` is a function that accepts a floating-point value and returns one:
###Code
def square_root_with_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return square_root(x)
###Output
_____no_output_____
###Markdown
By default, such annotations are ignored by the Python interpreter. Therefore, one can still call `square_root_typed()` with a string as an argument and get the exact same result as above. However, one can make use of special _typechecking_ modules that would check types – _dynamically_ at runtime or _statically_ by analyzing the code without having to execute it. Runtime Type CheckingThe Python `enforce` package provides a function decorator that automatically inserts type-checking code that is executed at runtime. Here is how to use it:
###Code
import enforce
@enforce.runtime_validation
def square_root_with_checked_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return square_root(x)
###Output
_____no_output_____
###Markdown
Now, invoking `square_root_with_checked_type_annotations()` raises an exception when invoked with a type dfferent from the one declared:
###Code
with ExpectError():
square_root_with_checked_type_annotations(True)
###Output
_____no_output_____
###Markdown
Note that this error is not caught by the "untyped" variant, where passing a boolean value happily returns $\sqrt{1}$ as result.
###Code
square_root(True)
###Output
_____no_output_____
###Markdown
In Python (and other languages), the boolean values `True` and `False` can be implicitly converted to the integers 1 and 0; however, it is hard to think of a call to `sqrt()` where this would not be an error.
###Code
from bookutils import quiz
quiz("What happens if we call "
"`square_root_with_checked_type_annotations(1)`?",
[
"`1` is automatically converted to float. It will pass.",
"`1` is a subtype of float. It will pass.",
"`1` is an integer, and no float. The type check will fail.",
"The function will fail for some other reason."
], '37035 // 12345')
###Output
_____no_output_____
###Markdown
"Prediction is very difficult, especially when the future is concerned" (Niels Bohr). We can find out by a simple experiment that `float` actually means `float` – and not `int`:
###Code
with ExpectError(enforce.exceptions.RuntimeTypeError):
square_root_with_checked_type_annotations(1)
###Output
_____no_output_____
###Markdown
To allow `int` as a type, we need to specify a _union_ of types.
###Code
@enforce.runtime_validation
def square_root_with_union_type(x: Union[int, float]) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return square_root(x)
square_root_with_union_type(2)
square_root_with_union_type(2.0)
with ExpectError(enforce.exceptions.RuntimeTypeError):
square_root_with_union_type("Two dot zero")
###Output
_____no_output_____
###Markdown
Static Type CheckingType annotations can also be checked _statically_ – that is, without even running the code. Let us create a simple Python file consisting of the above `square_root_typed()` definition and a bad invocation.
###Code
import inspect
import tempfile
f = tempfile.NamedTemporaryFile(mode='w', suffix='.py')
f.name
f.write(inspect.getsource(square_root))
f.write('\n')
f.write(inspect.getsource(square_root_with_type_annotations))
f.write('\n')
f.write("print(square_root_with_type_annotations('123'))\n")
f.flush()
###Output
_____no_output_____
###Markdown
These are the contents of our newly created Python file:
###Code
from bookutils import print_file
print_file(f.name, start_line_number=1)
###Output
_____no_output_____
###Markdown
[Mypy](http://mypy-lang.org) is a type checker for Python programs. As it checks types statically, types induce no overhead at runtime; plus, a static check can be faster than a lengthy series of tests with runtime type checking enabled. Let us see what `mypy` produces on the above file:
###Code
import subprocess
result = subprocess.run(["mypy", "--strict", f.name],
universal_newlines=True, stdout=subprocess.PIPE)
print(result.stdout.replace(f.name + ':', ''))
del f # Delete temporary file
###Output
_____no_output_____
###Markdown
We see that `mypy` complains about untyped function definitions such as `square_root()`; most important, however, it finds that the call to `square_root_with_type_annotations()` in the last line has the wrong type. With `mypy`, we can achieve the same type safety with Python as in statically typed languages – provided that we as programmers also produce the necessary type annotations. Is there a simple way to obtain these? Mining Type SpecificationsOur first task will be to mine type annotations (as part of the code) from _values_ we observe at run time. These type annotations would be _mined_ from actual function executions, _learning_ from (normal) runs what the expected argument and return types should be. By observing a series of calls such as these, we could infer that both `x` and the return value are of type `float`:
###Code
y = square_root(25.0)
y
y = square_root(2.0)
y
###Output
_____no_output_____
###Markdown
How can we mine types from executions? The answer is simple: 1. We _observe_ a function during execution2. We track the _types_ of its arguments3. We include these types as _annotations_ into the code.To do so, we can make use of Python's tracing facility we already observed in the [chapter on tracing executions](Tracer.ipynb). With every call to a function, we retrieve the arguments, their values, and their types. Tracing CallsTo observe argument types at runtime, we define a _tracer function_ that tracks the execution of `square_root()`, checking its arguments and return values. The `CallTracer` class is set to trace functions in a `with` block as follows:```pythonwith CallTracer() as tracer: function_to_be_tracked(...)info = tracer.collected_information()```To create the tracer, we build on the `Tracer` superclass as in the [chapter on tracing executions](Tracer.ipynb).
###Code
from types import FrameType
###Output
_____no_output_____
###Markdown
We start with two helper functions. `get_arguments()` returns a list of call arguments in the given call frame.
###Code
Arguments = List[Tuple[str, Any]]
def get_arguments(frame: FrameType) -> Arguments:
"""Return call arguments in the given frame"""
# When called, all arguments are local variables
arguments = [(var, frame.f_locals[var]) for var in frame.f_locals]
arguments.reverse() # Want same order as call
return arguments
###Output
_____no_output_____
###Markdown
`simple_call_string()` is a helper for logging that prints out calls in a user-friendly manner.
###Code
def simple_call_string(function_name: str, argument_list: Arguments,
return_value : Any = None) -> str:
"""Return function_name(arg[0], arg[1], ...) as a string"""
call = function_name + "(" + \
", ".join([var + "=" + repr(value)
for (var, value) in argument_list]) + ")"
if return_value is not None:
call += " = " + repr(return_value)
return call
###Output
_____no_output_____
###Markdown
Now for `CallTracer`. The constructor simply invokes the `Tracer` constructor:
###Code
class CallTracer(Tracer):
def __init__(self, log: bool = False, **kwargs: Any)-> None:
super().__init__(**kwargs)
self._log = log
self.reset()
def reset(self) -> None:
self._calls: Dict[str, List[Tuple[Arguments, Any]]] = {}
self._stack: List[Tuple[str, Arguments]] = []
###Output
_____no_output_____
###Markdown
The `traceit()` method does nothing yet; this is done in specialized subclasses. The `CallTracer` class implements a `traceit()` function that checks for function calls and returns:
###Code
class CallTracer(CallTracer):
def traceit(self, frame: FrameType, event: str, arg: Any) -> None:
"""Tracking function: Record all calls and all args"""
if event == "call":
self.trace_call(frame, event, arg)
elif event == "return":
self.trace_return(frame, event, arg)
###Output
_____no_output_____
###Markdown
`trace_call()` is called when a function is called; it retrieves the function name and current arguments, and saves them on a stack.
###Code
class CallTracer(CallTracer):
def trace_call(self, frame: FrameType, event: str, arg: Any) -> None:
"""Save current function name and args on the stack"""
code = frame.f_code
function_name = code.co_name
arguments = get_arguments(frame)
self._stack.append((function_name, arguments))
if self._log:
print(simple_call_string(function_name, arguments))
###Output
_____no_output_____
###Markdown
When the function returns, `trace_return()` is called. We now also have the return value. We log the whole call with arguments and return value (if desired) and save it in our list of calls.
###Code
class CallTracer(CallTracer):
def trace_return(self, frame: FrameType, event: str, arg: Any) -> None:
"""Get return value and store complete call with arguments and return value"""
code = frame.f_code
function_name = code.co_name
return_value = arg
# TODO: Could call get_arguments() here to also retrieve _final_ values of argument variables
called_function_name, called_arguments = self._stack.pop()
assert function_name == called_function_name
if self._log:
print(simple_call_string(function_name, called_arguments), "returns", return_value)
self.add_call(function_name, called_arguments, return_value)
###Output
_____no_output_____
###Markdown
`add_call()` saves the calls in a list; each function name has its own list.
###Code
class CallTracer(CallTracer):
def add_call(self, function_name: str, arguments: Arguments,
return_value: Any = None) -> None:
"""Add given call to list of calls"""
if function_name not in self._calls:
self._calls[function_name] = []
self._calls[function_name].append((arguments, return_value))
###Output
_____no_output_____
###Markdown
we can retrieve the list of calls, either for a given function name (`calls()`),or for all functions (`all_calls()`).
###Code
class CallTracer(CallTracer):
def calls(self, function_name: str) -> List[Tuple[Arguments, Any]]:
"""Return list of calls for `function_name`."""
return self._calls[function_name]
class CallTracer(CallTracer):
def all_calls(self) -> Dict[str, List[Tuple[Arguments, Any]]]:
"""
Return list of calls for function_name,
or a mapping function_name -> calls for all functions tracked
"""
return self._calls
###Output
_____no_output_____
###Markdown
Let us now put this to use. We turn on logging to track the individual calls and their return values:
###Code
with CallTracer(log=True) as tracer:
y = square_root(25)
y = square_root(2.0)
###Output
_____no_output_____
###Markdown
After execution, we can retrieve the individual calls:
###Code
calls = tracer.calls('square_root')
calls
###Output
_____no_output_____
###Markdown
Each call is a pair (`argument_list`, `return_value`), where `argument_list` is a list of pairs (`parameter_name`, `value`).
###Code
square_root_argument_list, square_root_return_value = calls[0]
simple_call_string('square_root', square_root_argument_list, square_root_return_value)
###Output
_____no_output_____
###Markdown
If the function does not return a value, `return_value` is `None`.
###Code
def hello(name: str) -> None:
print("Hello,", name)
with CallTracer() as tracer:
hello("world")
hello_calls = tracer.calls('hello')
hello_calls
hello_argument_list, hello_return_value = hello_calls[0]
simple_call_string('hello', hello_argument_list, hello_return_value)
###Output
_____no_output_____
###Markdown
Getting TypesDespite what you may have read or heard, Python actually _is_ a typed language. It is just that it is _dynamically typed_ – types are used and checked only at runtime (rather than declared in the code, where they can be _statically checked_ at compile time). We can thus retrieve types of all values within Python:
###Code
type(4)
type(2.0)
type([4])
###Output
_____no_output_____
###Markdown
We can retrieve the type of the first argument to `square_root()`:
###Code
parameter, value = square_root_argument_list[0]
parameter, type(value)
###Output
_____no_output_____
###Markdown
as well as the type of the return value:
###Code
type(square_root_return_value)
###Output
_____no_output_____
###Markdown
Hence, we see that (so far), `square_root()` is a function taking (among others) integers and returning floats. We could declare `square_root()` as:
###Code
def square_root_annotated(x: int) -> float:
return square_root(x)
###Output
_____no_output_____
###Markdown
This is a representation we could place in a static type checker, allowing to check whether calls to `square_root()` actually pass a number. A dynamic type checker could run such checks at runtime. By default, Python does not do anything with such annotations. However, tools can access annotations from functions and other objects:
###Code
square_root_annotated.__annotations__
###Output
_____no_output_____
###Markdown
This is how run-time checkers access the annotations to check against. Annotating Functions with TypesOur plan is to annotate functions automatically, based on the types we have seen. Our aim is to build a class `TypeAnnotator` that can be used as follows. First, it would track some execution:```pythonwith TypeAnnotator() as annotator: some_function_call()```After tracking, `TypeAnnotator` would provide appropriate methods to access (type-)annotated versions of the function seen:```pythonprint(annotator.typed_functions())```Let us put the pieces together to build `TypeAnnotator`. Excursion: Accessing Function Structure To annotate functions, we need to convert a function into a tree representation (called _abstract syntax trees_, or ASTs) and back; we already have seen these in the chapter on [tracking value origins](Slicer.ipynb).
###Code
import ast
import inspect
import astor
###Output
_____no_output_____
###Markdown
We can get the source of a Python function using `inspect.getsource()`. (Note that this does not work for functions defined in other notebooks.)
###Code
square_root_source = inspect.getsource(square_root)
square_root_source
###Output
_____no_output_____
###Markdown
To view these in a visually pleasing form, our function `print_content(s, suffix)` formats and highlights the string `s` as if it were a file with ending `suffix`. We can thus view (and highlight) the source as if it were a Python file:
###Code
from bookutils import print_content
print_content(square_root_source, '.py')
###Output
_____no_output_____
###Markdown
Parsing this gives us an abstract syntax tree (AST) – a representation of the program in tree form.
###Code
square_root_ast = ast.parse(square_root_source)
###Output
_____no_output_____
###Markdown
What does this AST look like? The helper functions `astor.dump_tree()` (textual output) and `showast.show_ast()` (graphical output with [showast](https://github.com/hchasestevens/show_ast)) allow us to inspect the structure of the tree. We see that the function starts as a `FunctionDef` with name and arguments, followed by a body, which is a list of statements of type `Expr` (the docstring), type `Assign` (assignments), `While` (while loop with its own body), and finally `Return`.
###Code
print(astor.dump_tree(square_root_ast))
###Output
_____no_output_____
###Markdown
Too much text for you? This graphical representation may make things simpler.
###Code
from bookutils import show_ast
show_ast(square_root_ast)
###Output
_____no_output_____
###Markdown
The function `astor.to_source()` converts such a tree back into the more familiar textual Python code representation. Comments are gone, and there may be more parentheses than before, but the result has the same semantics:
###Code
print_content(astor.to_source(square_root_ast), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: Annotating Functions with Given Types Let us now go and transform ASTs to add type annotations. We start with a helper function `parse_type(name)` which parses a type name into an AST.
###Code
def parse_type(name: str) -> ast.expr:
class ValueVisitor(ast.NodeVisitor):
def visit_Expr(self, node: ast.Expr) -> None:
self.value_node = node.value
tree = ast.parse(name)
name_visitor = ValueVisitor()
name_visitor.visit(tree)
return name_visitor.value_node
print(astor.dump_tree(parse_type('int')))
print(astor.dump_tree(parse_type('[object]')))
###Output
_____no_output_____
###Markdown
We now define a helper function that actually adds type annotations to a function AST. The `TypeTransformer` class builds on the Python standard library `ast.NodeTransformer` infrastructure. It would be called as```python TypeTransformer({'x': 'int'}, 'float').visit(ast)```to annotate the arguments of `square_root()`: `x` with `int`, and the return type with `float`. The returned AST can then be unparsed, compiled or analyzed.
###Code
class TypeTransformer(ast.NodeTransformer):
def __init__(self, argument_types: Dict[str, str], return_type: Optional[str] = None):
self.argument_types = argument_types
self.return_type = return_type
super().__init__()
###Output
_____no_output_____
###Markdown
The core of `TypeTransformer` is the method `visit_FunctionDef()`, which is called for every function definition in the AST. Its argument `node` is the subtree of the function definition to be transformed. Our implementation accesses the individual arguments and invokes `annotate_args()` on them; it also sets the return type in the `returns` attribute of the node.
###Code
class TypeTransformer(TypeTransformer):
def visit_FunctionDef(self, node: ast.FunctionDef) -> ast.FunctionDef:
"""Add annotation to function"""
# Set argument types
new_args = []
for arg in node.args.args:
new_args.append(self.annotate_arg(arg))
new_arguments = ast.arguments(
new_args,
node.args.vararg,
node.args.kwonlyargs,
node.args.kw_defaults,
node.args.kwarg,
node.args.defaults
)
# Set return type
if self.return_type is not None:
node.returns = parse_type(self.return_type)
return ast.copy_location(
ast.FunctionDef(node.name, new_arguments,
node.body, node.decorator_list,
node.returns), node)
###Output
_____no_output_____
###Markdown
Each argument gets its own annotation, taken from the types originally passed to the class:
###Code
class TypeTransformer(TypeTransformer):
def annotate_arg(self, arg: ast.arg) -> ast.arg:
"""Add annotation to single function argument"""
arg_name = arg.arg
if arg_name in self.argument_types:
arg.annotation = parse_type(self.argument_types[arg_name])
return arg
###Output
_____no_output_____
###Markdown
Does this work? Let us annotate the AST from `square_root()` with types for the arguments and return types:
###Code
new_ast = TypeTransformer({'x': 'int'}, 'float').visit(square_root_ast)
###Output
_____no_output_____
###Markdown
When we unparse the new AST, we see that the annotations actually are present:
###Code
print_content(astor.to_source(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Similarly, we can annotate the `hello()` function from above:
###Code
hello_source = inspect.getsource(hello)
hello_ast = ast.parse(hello_source)
new_ast = TypeTransformer({'name': 'str'}, 'None').visit(hello_ast)
print_content(astor.to_source(new_ast), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: Annotating Functions with Mined Types Let us now annotate functions with types mined at runtime. We start with a simple function `type_string()` that determines the appropriate type of a given value (as a string):
###Code
def type_string(value: Any) -> str:
return type(value).__name__
type_string(4)
type_string([])
###Output
_____no_output_____
###Markdown
For composite structures, `type_string()` does not examine element types; hence, the type of `[3]` is simply `list` instead of, say, `list[int]`. For now, `list` will do fine.
###Code
type_string([3])
###Output
_____no_output_____
###Markdown
`type_string()` will be used to infer the types of argument values found at runtime, as returned by `CallTracer.all_calls()`:
###Code
with CallTracer() as tracer:
y = square_root(25.0)
y = square_root(2.0)
tracer.all_calls()
###Output
_____no_output_____
###Markdown
The function `annotate_types()` takes such a list of calls and annotates each function listed:
###Code
def annotate_types(calls: Dict[str, List[Tuple[Arguments, Any]]]) -> Dict[str, ast.AST]:
annotated_functions = {}
for function_name in calls:
try:
annotated_functions[function_name] = \
annotate_function_with_types(function_name, calls[function_name])
except KeyError:
continue
return annotated_functions
###Output
_____no_output_____
###Markdown
For each function, we get the source and its AST and then get to the actual annotation in `annotate_function_ast_with_types()`:
###Code
def annotate_function_with_types(function_name: str,
function_calls: List[Tuple[Arguments, Any]]) -> ast.AST:
function = globals()[function_name] # May raise KeyError for internal functions
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_types(function_ast, function_calls)
###Output
_____no_output_____
###Markdown
The function `annotate_function_ast_with_types()` invokes the `TypeTransformer` with the calls seen, and for each call, iterate over the arguments, determine their types, and annotate the AST with these. The universal type `Any` is used when we encounter type conflicts, which we will discuss below.
###Code
def annotate_function_ast_with_types(function_ast: ast.AST,
function_calls: List[Tuple[Arguments, Any]]) -> ast.AST:
parameter_types: Dict[str, str] = {}
return_type = None
for calls_seen in function_calls:
args, return_value = calls_seen
if return_value:
if return_type and return_type != type_string(return_value):
return_type = 'Any'
else:
return_type = type_string(return_value)
for parameter, value in args:
try:
different_type = (parameter_types[parameter] !=
type_string(value))
except KeyError:
different_type = False
if different_type:
parameter_types[parameter] = 'Any'
else:
parameter_types[parameter] = type_string(value)
annotated_function_ast = \
TypeTransformer(parameter_types, return_type).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Here is `square_root()` annotated with the types recorded usign the tracer, above.
###Code
print_content(astor.to_source(annotate_types(tracer.all_calls())['square_root']), '.py')
###Output
_____no_output_____
###Markdown
End of Excursion Excursion: A Type Annotator Class Let us bring all of this together in a single class `TypeAnnotator` that first tracks calls of functions and then allows to access the AST (and the source code form) of the tracked functions annotated with types. The method `typed_functions()` returns the annotated functions as a string; `typed_functions_ast()` returns their AST.
###Code
class TypeTracer(CallTracer):
pass
class TypeAnnotator(TypeTracer):
def typed_functions_ast(self) -> Dict[str, ast.AST]:
return annotate_types(self.all_calls())
def typed_function_ast(self, function_name: str) -> ast.AST:
return annotate_function_with_types(function_name, self.calls(function_name))
def typed_functions(self) -> str:
functions = ''
for f_name in self.all_calls():
try:
f_text = astor.to_source(self.typed_function_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
def typed_function(self, function_name: str) -> str:
return astor.to_source(self.typed_function_ast(function_name))
###Output
_____no_output_____
###Markdown
End of Excursion Here is how to use `TypeAnnotator`. We first track a series of calls:
###Code
with TypeAnnotator() as annotator:
y = square_root(25.0)
y = square_root(2.0)
###Output
_____no_output_____
###Markdown
After tracking, we can immediately retrieve an annotated version of the functions tracked:
###Code
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
This also works for multiple and diverse functions. One could go and implement an automatic type annotator for Python files based on the types seen during execution.
###Code
with TypeAnnotator() as annotator:
hello('type annotations')
y = square_root(1.0)
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
A content as above could now be sent to a type checker, which would detect any type inconsistency between callers and callees. Excursion: Handling Multiple TypesLet us now resolve the role of the magic `Any` type in `annotate_function_ast_with_types()`. If we see multiple types for the same argument, we set its type to `Any`. For `square_root()`, this makes sense, as its arguments can be integers as well as floats:
###Code
with CallTracer() as tracer:
y = square_root(25.0)
y = square_root(4)
annotated_square_root_ast = annotate_types(tracer.all_calls())['square_root']
print_content(astor.to_source(annotated_square_root_ast), '.py')
###Output
_____no_output_____
###Markdown
The following function `sum3()` can be called with floating-point numbers as arguments, resulting in the parameters getting a `float` type:
###Code
def sum3(a, b, c): # type: ignore
return a + b + c
with TypeAnnotator() as annotator:
y = sum3(1.0, 2.0, 3.0)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we call `sum3()` with integers, though, the arguments get an `int` type:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
And we can also call `sum3()` with strings, which assigns the arguments a `str` type:
###Code
with TypeAnnotator() as annotator:
y = sum3("one", "two", "three")
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we have multiple calls, but with different types, `TypeAnnotator()` will assign an `Any` type to both arguments and return values:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y = sum3("one", "two", "three")
typed_sum3_def = annotator.typed_function('sum3')
print_content(typed_sum3_def, '.py')
###Output
_____no_output_____
###Markdown
A type `Any` makes it explicit that an object can, indeed, have any type; it will not be typechecked at runtime or statically. To some extent, this defeats the power of type checking; but it also preserves some of the type flexibility that many Python programmers enjoy. Besides `Any`, the `typing` module supports several additional ways to define ambiguous types; we will keep this in mind for a later exercise. End of Excursion Mining InvariantsBesides basic data types. we can check several further properties from arguments. We can, for instance, whether an argument can be negative, zero, or positive; or that one argument should be smaller than the second; or that the result should be the sum of two arguments – properties that cannot be expressed in a (Python) type.Such properties are called *invariants*, as they hold across all invocations of a function. Specifically, invariants come as _pre_- and _postconditions_ – conditions that always hold at the beginning and at the end of a function. (There are also _data_ and _object_ invariants that express always-holding properties over the state of data or objects, but we do not consider these in this book.) Annotating Functions with Pre- and PostconditionsThe classical means to specify pre- and postconditions is via _assertions_, which we have introduced in the [chapter on assertions](Assertions.ipynb). A precondition checks whether the arguments to a function satisfy the expected properties; a postcondition does the same for the result. We can express and check both using assertions as follows:
###Code
def square_root_with_invariants(x): # type: ignore
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
A nicer way, however, is to syntactically separate invariants from the function at hand. Using appropriate decorators, we could specify pre- and postconditions as follows:```python@precondition lambda x: x >= 0@postcondition lambda return_value, x: return_value * return_value == xdef square_root_with_invariants(x): normal code without assertions ...```The decorators `@precondition` and `@postcondition` would run the given functions (specified as anonymous `lambda` functions) before and after the decorated function, respectively. If the functions return `False`, the condition is violated. `@precondition` gets the function arguments as arguments; `@postcondition` additionally gets the return value as first argument. It turns out that implementing such decorators is not hard at all. Our implementation builds on a [code snippet from StackOverflow](https://stackoverflow.com/questions/12151182/python-precondition-postcondition-for-member-function-how):
###Code
import functools
def condition(precondition: Optional[Callable] = None,
postcondition: Optional[Callable] = None) -> Callable:
def decorator(func: Callable) -> Callable:
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args: Any, **kwargs: Any) -> Any:
if precondition is not None:
assert precondition(*args, **kwargs), \
"Precondition violated"
# Call original function or method
retval = func(*args, **kwargs)
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), \
"Postcondition violated"
return retval
return wrapper
return decorator
def precondition(check: Callable) -> Callable:
return condition(precondition=check)
def postcondition(check: Callable) -> Callable:
return condition(postcondition=check)
###Output
_____no_output_____
###Markdown
With these, we can now start decorating `square_root()`:
###Code
@precondition(lambda x: x > 0)
def square_root_with_precondition(x): # type: ignore
return square_root(x)
###Output
_____no_output_____
###Markdown
This catches arguments violating the precondition:
###Code
with ExpectError():
square_root_with_precondition(-1.0)
###Output
_____no_output_____
###Markdown
Likewise, we can provide a postcondition:
###Code
import math
@postcondition(lambda ret, x: math.isclose(ret * ret, x))
def square_root_with_postcondition(x): # type: ignore
return square_root(x)
y = square_root_with_postcondition(2.0)
y
###Output
_____no_output_____
###Markdown
If we have a buggy implementation of $\sqrt{x}$, this gets caught quickly:
###Code
@postcondition(lambda ret, x: math.isclose(ret * ret, x))
def buggy_square_root_with_postcondition(x): # type: ignore
return square_root(x) + 0.1
with ExpectError():
y = buggy_square_root_with_postcondition(2.0)
###Output
_____no_output_____
###Markdown
While checking pre- and postconditions is a great way to catch errors, specifying them can be cumbersome. Let us try to see whether we can (again) _mine_ some of them. Mining InvariantsTo _mine_ invariants, we can use the same tracking functionality as before; instead of saving values for individual variables, though, we now check whether the values satisfy specific _properties_ or not. For instance, if all values of `x` seen satisfy the condition `x > 0`, then we make `x > 0` an invariant of the function. If we see positive, zero, and negative values of `x`, though, then there is no property of `x` left to talk about.The general idea is thus:1. Check all variable values observed against a set of predefined properties; and2. Keep only those properties that hold for all runs observed. Defining PropertiesWhat precisely do we mean by properties? Here is a small collection of value properties that would frequently be used in invariants. All these properties would be evaluated with the _metavariables_ `X`, `Y`, and `Z` (actually, any upper-case identifier) being replaced with the names of function parameters:
###Code
INVARIANT_PROPERTIES = [
"X < 0",
"X <= 0",
"X > 0",
"X >= 0",
# "X == 0", # implied by "X", below
# "X != 0", # implied by "not X", below
]
###Output
_____no_output_____
###Markdown
When `square_root(x)` is called as, say `square_root(5.0)`, we see that `x = 5.0` holds. The above properties would then all be checked for `x`. Only the properties `X > 0`, `X >= 0`, and `not X` hold for the call seen; and hence `x > 0`, `x >= 0`, and `not x` (or better: `x != 0`) would make potential preconditions for `square_root(x)`. We can check for many more properties such as relations between two arguments:
###Code
INVARIANT_PROPERTIES += [
"X == Y",
"X > Y",
"X < Y",
"X >= Y",
"X <= Y",
]
###Output
_____no_output_____
###Markdown
Types also can be checked using properties. For any function parameter `X`, only one of these will hold:
###Code
INVARIANT_PROPERTIES += [
"isinstance(X, bool)",
"isinstance(X, int)",
"isinstance(X, float)",
"isinstance(X, list)",
"isinstance(X, dict)",
]
###Output
_____no_output_____
###Markdown
We can check for arithmetic properties:
###Code
INVARIANT_PROPERTIES += [
"X == Y + Z",
"X == Y * Z",
"X == Y - Z",
"X == Y / Z",
]
###Output
_____no_output_____
###Markdown
Here's relations over three values, a Python special:
###Code
INVARIANT_PROPERTIES += [
"X < Y < Z",
"X <= Y <= Z",
"X > Y > Z",
"X >= Y >= Z",
]
###Output
_____no_output_____
###Markdown
These Boolean properties also check for other types, as in Python, `None`, an empty list, an empty set, an empty string, and the value zero all evaluate to `False`.
###Code
INVARIANT_PROPERTIES += [
"X",
"not X"
]
###Output
_____no_output_____
###Markdown
Finally, we can also check for list or string properties. Again, this is just a tiny selection.
###Code
INVARIANT_PROPERTIES += [
"X == len(Y)",
"X == sum(Y)",
"X in Y",
"X.startswith(Y)",
"X.endswith(Y)",
]
###Output
_____no_output_____
###Markdown
Extracting Meta-VariablesLet us first introduce a few _helper functions_ before we can get to the actual mining. `metavars()` extracts the set of meta-variables (`X`, `Y`, `Z`, etc.) from a property. To this end, we parse the property as a Python expression and then visit the identifiers.
###Code
def metavars(prop: str) -> List[str]:
metavar_list = []
class ArgVisitor(ast.NodeVisitor):
def visit_Name(self, node: ast.Name) -> None:
if node.id.isupper():
metavar_list.append(node.id)
ArgVisitor().visit(ast.parse(prop))
return metavar_list
assert metavars("X < 0") == ['X']
assert metavars("X.startswith(Y)") == ['X', 'Y']
assert metavars("isinstance(X, str)") == ['X']
###Output
_____no_output_____
###Markdown
Instantiating PropertiesTo produce a property as invariant, we need to be able to _instantiate_ it with variable names. The instantiation of `X > 0` with `X` being instantiated to `a`, for instance, gets us `a > 0`. To this end, the function `instantiate_prop()` takes a property and a collection of variable names and instantiates the meta-variables left-to-right with the corresponding variables names in the collection.
###Code
def instantiate_prop_ast(prop: str, var_names: Sequence[str]) -> ast.AST:
class NameTransformer(ast.NodeTransformer):
def visit_Name(self, node: ast.Name) -> ast.Name:
if node.id not in mapping:
return node
return ast.Name(id=mapping[node.id], ctx=ast.Load())
meta_variables = metavars(prop)
assert len(meta_variables) == len(var_names)
mapping = {}
for i in range(0, len(meta_variables)):
mapping[meta_variables[i]] = var_names[i]
prop_ast = ast.parse(prop, mode='eval')
new_ast = NameTransformer().visit(prop_ast)
return new_ast
def instantiate_prop(prop: str, var_names: Sequence[str]) -> str:
prop_ast = instantiate_prop_ast(prop, var_names)
prop_text = astor.to_source(prop_ast).strip()
while prop_text.startswith('(') and prop_text.endswith(')'):
prop_text = prop_text[1:-1]
return prop_text
assert instantiate_prop("X > Y", ['a', 'b']) == 'a > b'
assert instantiate_prop("X.startswith(Y)", ['x', 'y']) == 'x.startswith(y)'
###Output
_____no_output_____
###Markdown
Evaluating PropertiesTo actually _evaluate_ properties, we do not need to instantiate them. Instead, we simply convert them into a boolean function, using `lambda`:
###Code
def prop_function_text(prop: str) -> str:
return "lambda " + ", ".join(metavars(prop)) + ": " + prop
###Output
_____no_output_____
###Markdown
Here is a simple example:
###Code
prop_function_text("X > Y")
###Output
_____no_output_____
###Markdown
We can easily evaluate the function:
###Code
def prop_function(prop: str) -> Callable:
return eval(prop_function_text(prop))
###Output
_____no_output_____
###Markdown
Here is an example:
###Code
p = prop_function("X > Y")
quiz("What is p(100, 1)?",
[
"False",
"True"
], 'p(100, 1) + 1', globals())
p(100, 1)
p(1, 100)
###Output
_____no_output_____
###Markdown
Checking InvariantsTo extract invariants from an execution, we need to check them on all possible instantiations of arguments. If the function to be checked has two arguments `a` and `b`, we instantiate the property `X < Y` both as `a < b` and `b < a` and check each of them. To get all combinations, we use the Python `permutations()` function:
###Code
import itertools
for combination in itertools.permutations([1.0, 2.0, 3.0], 2):
print(combination)
###Output
_____no_output_____
###Markdown
The function `true_property_instantiations()` takes a property and a list of tuples (`var_name`, `value`). It then produces all instantiations of the property with the given values and returns those that evaluate to True.
###Code
Invariants = Set[Tuple[str, Tuple[str, ...]]]
def true_property_instantiations(prop: str, vars_and_values: Arguments,
log: bool = False) -> Invariants:
instantiations = set()
p = prop_function(prop)
len_metavars = len(metavars(prop))
for combination in itertools.permutations(vars_and_values, len_metavars):
args = [value for var_name, value in combination]
var_names = [var_name for var_name, value in combination]
try:
result = p(*args)
except:
result = None
if log:
print(prop, combination, result)
if result:
instantiations.add((prop, tuple(var_names)))
return instantiations
###Output
_____no_output_____
###Markdown
Here is an example. If `x == -1` and `y == 1`, the property `X < Y` holds for `x < y`, but not for `y < x`:
###Code
invs = true_property_instantiations("X < Y", [('x', -1), ('y', 1)], log=True)
invs
###Output
_____no_output_____
###Markdown
The instantiation retrieves the short form:
###Code
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Likewise, with values for `x` and `y` as above, the property `X < 0` only holds for `x`, but not for `y`:
###Code
invs = true_property_instantiations("X < 0", [('x', -1), ('y', 1)], log=True)
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Extracting InvariantsLet us now run the above invariant extraction on function arguments and return values as observed during a function execution. To this end, we extend the `CallTracer` class into an `InvariantTracer` class, which automatically computes invariants for all functions and all calls observed during tracking. By default, an `InvariantTracer` uses the `INVARIANT_PROPERTIES` properties as defined above; however, one can specify alternate sets of properties.
###Code
class InvariantTracer(CallTracer):
def __init__(self, props: Optional[List[str]] = None, **kwargs: Any) -> None:
if props is None:
props = INVARIANT_PROPERTIES
self.props = props
super().__init__(**kwargs)
###Output
_____no_output_____
###Markdown
The key method of the `InvariantTracer` is the `invariants()` method. This iterates over the calls observed and checks which properties hold. Only the intersection of properties – that is, the set of properties that hold for all calls – is preserved, and eventually returned. The special variable `return_value` is set to hold the return value.
###Code
RETURN_VALUE = 'return_value'
class InvariantTracer(InvariantTracer):
def all_invariants(self) -> Dict[str, Invariants]:
return {function_name: self.invariants(function_name)
for function_name in self.all_calls()}
def invariants(self, function_name: str) -> Invariants:
invariants = None
for variables, return_value in self.calls(function_name):
vars_and_values = variables + [(RETURN_VALUE, return_value)]
s = set()
for prop in self.props:
s |= true_property_instantiations(prop, vars_and_values,
self._log)
if invariants is None:
invariants = s
else:
invariants &= s
assert invariants is not None
return invariants
###Output
_____no_output_____
###Markdown
Here's an example of how to use `invariants()`. We run the tracer on a small set of calls.
###Code
with InvariantTracer() as tracer:
y = square_root(25.0)
y = square_root(10.0)
tracer.all_calls()
###Output
_____no_output_____
###Markdown
The `invariants()` method produces a set of properties that hold for the observed runs, together with their instantiations over function arguments.
###Code
invs = tracer.invariants('square_root')
invs
###Output
_____no_output_____
###Markdown
As before, the actual instantiations are easier to read:
###Code
def pretty_invariants(invariants: Invariants) -> List[str]:
props = []
for (prop, var_names) in invariants:
props.append(instantiate_prop(prop, var_names))
return sorted(props)
pretty_invariants(invs)
###Output
_____no_output_____
###Markdown
We see that the both `x` and the return value have a `float` type. We also see that both are always greater than zero. These are properties that may make useful pre- and postconditions, notably for symbolic analysis. However, there's also an invariant which does _not_ universally hold, namely `return_value <= x`, as the following example shows:
###Code
square_root(0.01)
###Output
_____no_output_____
###Markdown
Clearly, 0.1 > 0.01 holds. This is a case of us not learning from sufficiently diverse inputs. As soon as we have a call including `x = 0.1`, though, the invariant `return_value <= x` is eliminated:
###Code
with InvariantTracer() as tracer:
y = square_root(25.0)
y = square_root(10.0)
y = square_root(0.01)
pretty_invariants(tracer.invariants('square_root'))
###Output
_____no_output_____
###Markdown
We will discuss later how to ensure sufficient diversity in inputs. (Hint: This involves test generation.) Let us try out our invariant tracer on `sum3()`. We see that all types are well-defined; the properties that all arguments are non-zero, however, is specific to the calls observed.
###Code
with InvariantTracer() as tracer:
y = sum3(1, 2, 3)
y = sum3(-4, -5, -6)
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with strings instead, we get different invariants. Notably, we obtain the postcondition that the returned string always starts with the string in the first argument `a` – a universal postcondition if strings are used.
###Code
with InvariantTracer() as tracer:
y = sum3('a', 'b', 'c')
y = sum3('f', 'e', 'd')
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with both strings and numbers (and zeros, too), there are no properties left that would hold across all calls. That's the price of flexibility.
###Code
with InvariantTracer() as tracer:
y = sum3('a', 'b', 'c')
y = sum3('c', 'b', 'a')
y = sum3(-4, -5, -6)
y = sum3(0, 0, 0)
pretty_invariants(tracer.invariants('sum3'))
###Output
_____no_output_____
###Markdown
Converting Mined Invariants to AnnotationsAs with types, above, we would like to have some functionality where we can add the mined invariants as annotations to existing functions. To this end, we introduce the `InvariantAnnotator` class, extending `InvariantTracer`. We start with a helper method. `params()` returns a comma-separated list of parameter names as observed during calls.
###Code
class InvariantAnnotator(InvariantTracer):
def params(self, function_name: str) -> str:
arguments, return_value = self.calls(function_name)[0]
return ", ".join(arg_name for (arg_name, arg_value) in arguments)
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = sum3(1, 2, 3)
annotator.params('square_root')
annotator.params('sum3')
###Output
_____no_output_____
###Markdown
Now for the actual annotation. `preconditions()` returns the preconditions from the mined invariants (i.e., those propertes that do not depend on the return value) as a string with annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = ("@precondition(lambda " + self.params(function_name) +
": " + inv + ")")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
annotator.preconditions('square_root')
###Output
_____no_output_____
###Markdown
`postconditions()` does the same for postconditions:
###Code
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = (f"@postcondition(lambda {RETURN_VALUE},"
f" {self.params(function_name)}: {inv})")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
annotator.postconditions('square_root')
###Output
_____no_output_____
###Markdown
With these, we can take a function and add both pre- and postconditions as annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def functions_with_invariants(self) -> str:
functions = ""
for function_name in self.all_invariants():
try:
function = self.function_with_invariants(function_name)
except KeyError:
continue
functions += function
return functions
def function_with_invariants(self, function_name: str) -> str:
function = globals()[function_name] # Can throw KeyError
source = inspect.getsource(function)
return '\n'.join(self.preconditions(function_name) +
self.postconditions(function_name)) + \
'\n' + source
###Output
_____no_output_____
###Markdown
Here comes `function_with_invariants()` in all its glory:
###Code
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
y = sum3(1, 2, 3)
print_content(annotator.function_with_invariants('square_root'), '.py')
###Output
_____no_output_____
###Markdown
Quite a number of invariants, isn't it? Further below (and in the exercises), we will discuss on how to focus on the most relevant properties. Avoiding OverspecializationMined specifications can only be as good as the executions they were mined from. If we only see a single call, for instance, we will be faced with several mined pre- and postconditions that _overspecialize_ towards the values seen. Let us illustrate this effect on a simple `sum2()` function which adds two numbers.
###Code
def sum2(a, b): # type: ignore
return a + b
###Output
_____no_output_____
###Markdown
If we invoke `sum2()` with a variety of arguments, the invariants all capture the relationship between `a`, `b`, and the return value as `return_value == a + b` in all its variations.
###Code
with InvariantAnnotator() as annotator:
sum2(31, 45)
sum2(0, 0)
sum2(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
If, however, we see only a single call, the invariants will overspecialize to the single call seen:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
The mined precondition `a == b`, for instance, only holds for the single call observed; the same holds for the mined postcondition `return_value == a * b`. Yet, `sum2()` can obviously be successfully called with other values that do not satisfy these conditions. To get out of this trap, we have to _learn from more and more diverse runs_. One way to obtain such runs is by _generating_ inputs. Indeed, a simple test generator for calls of `sum2()` will easily resolve the problem.
###Code
import random
with InvariantAnnotator() as annotator:
for i in range(100):
a = random.randrange(-10, +10)
b = random.randrange(-10, +10)
length = sum2(a, b)
print_content(annotator.function_with_invariants('sum2'), '.py')
###Output
_____no_output_____
###Markdown
Note, though, that an API test generator, such as above, will have to be set up such that it actually respects preconditions – in our case, we invoke `sum2()` with integers only, already assuming its precondition. In some way, one thus needs a specification (a model, a grammar) to mine another specification – a chicken-and-egg problem. However, there is one way out of this problem: If one can automatically generate tests at the system level, then one has an _infinite source of executions_ to learn invariants from. In each of these executions, all functions would be called with values that satisfy the (implicit) precondition, allowing us to mine invariants for these functions. This holds, because at the system level, invalid inputs must be rejected by the system in the first place. The meaningful precondition at the system level, ensuring that only valid inputs get through, thus gets broken down into a multitude of meaningful preconditions (and subsequent postconditions) at the function level. The big requirement for all this, though, is that one needs good test generators. This will be the subject of another book, namely [The Fuzzing Book](https://www.fuzzingbook.org/). Partial InvariantsFor debugging, it can be helpful to focus on invariants produced only by _failing_ runs, thus characterizing the _circumstances under which a function fails_. Let us illustrate this on an example. The `middle()` function from the [chapter on statistical debugging](StatisticalDebugger.ipynb) is supposed to return the middle of three integers `x`, `y`, and `z`.
###Code
from StatisticalDebugger import middle # minor dependency
with InvariantAnnotator() as annotator:
for i in range(100):
x = random.randrange(-10, +10)
y = random.randrange(-10, +10)
z = random.randrange(-10, +10)
mid = middle(x, y, z)
###Output
_____no_output_____
###Markdown
By default, our `InvariantAnnotator()` does not return any particular pre- or postcondition (other than the types observed). That is just fine, as the function indeed imposes no particular precondition; and the postcondition from `middle()` is not covered by the `InvariantAnnotator` patterns.
###Code
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Things get more interesting if we focus on a particular subset of runs only, though - say, a set of inputs where `middle()` fails.
###Code
from StatisticalDebugger import MIDDLE_FAILING_TESTCASES # minor dependency
with InvariantAnnotator() as annotator:
for x, y, z in MIDDLE_FAILING_TESTCASES:
mid = middle(x, y, z)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Now that's an intimidating set of pre- and postconditions. However, almost all of the preconditions are implied by the one precondition```python@precondition(lambda x, y, z: y < x < z, doc='y < x < z')```which characterizes the exact condition under which `middle()` fails (which also happens to be the condition under which the erroneous second `return y` is executed). By checking how _invariants for failing runs_ differ from _invariants for passing runs_, we can identify circumstances for function failures.
###Code
quiz("Could `InvariantAnnotator` also determine a precondition "
"that characterizes _passing_ runs?",
[
"Yes",
"No"
], 'int(math.exp(1))', globals())
###Output
_____no_output_____
###Markdown
Indeed, it cannot – the correct invariant for passing runs would be the _inverse_ of the invariant for failing runs, and `not A < B < C` is not part of our invariant library. We can easily test this:
###Code
from StatisticalDebugger import MIDDLE_PASSING_TESTCASES # minor dependency
with InvariantAnnotator() as annotator:
for x, y, z in MIDDLE_PASSING_TESTCASES:
mid = middle(x, y, z)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Some ExamplesLet us try out the `InvariantAnnotator` on a number of examples. Removing HTML MarkupRunning `InvariantAnnotator` on our ongoing example `remove_html_markup()` does not provide much, as our invariant properties are tailored towards numerical functions.
###Code
from Intro_Debugging import remove_html_markup
with InvariantAnnotator() as annotator:
remove_html_markup("<foo>bar</foo>")
remove_html_markup("bar")
remove_html_markup('"bar"')
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
In the [chapter on DDSet](DDSetDebugger.ipynb), we will see how to express more complex properties for structured inputs. A Recursive FunctionHere's another example. `list_length()` recursively computes the length of a Python function. Let us see whether we can mine its invariants:
###Code
def list_length(elems: List[Any]) -> int:
if elems == []:
length = 0
else:
length = 1 + list_length(elems[1:])
return length
with InvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Almost all these properties are relevant. Of course, the reason the invariants are so neat is that the return value is equal to `len(elems)` is that `X == len(Y)` is part of the list of properties to be checked. Sum of two NumbersThe next example is a very simple function: If we have a function without return value, the return value is `None` and we can only mine preconditions. (Well, we get a "postcondition" `not return_value` that the return value evaluates to False, which holds for `None`).
###Code
def print_sum(a, b): # type: ignore
print(a + b)
with InvariantAnnotator() as annotator:
print_sum(31, 45)
print_sum(0, 0)
print_sum(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Checking SpecificationsA function with invariants, as above, can be fed into the Python interpreter, such that all pre- and postconditions are checked. We create a function `square_root_annotated()` which includes all the invariants mined above.
###Code
with InvariantAnnotator() as annotator:
y = square_root(25.0)
y = square_root(0.01)
square_root_def = annotator.functions_with_invariants()
square_root_def = square_root_def.replace('square_root',
'square_root_annotated')
print_content(square_root_def, '.py')
exec(square_root_def)
###Output
_____no_output_____
###Markdown
The "annotated" version checks against invalid arguments – or more precisely, against arguments with properties that have not been observed yet:
###Code
with ExpectError():
square_root_annotated(-1.0) # type: ignore
###Output
_____no_output_____
###Markdown
This is in contrast to the original version, which just hangs on negative values:
###Code
with ExpectTimeout(1):
square_root(-1.0)
###Output
_____no_output_____
###Markdown
If we make changes to the function definition such that the properties of the return value change, such _regressions_ are caught as violations of the postconditions. Let us illustrate this by simply inverting the result, and return $-2$ as square root of 4.
###Code
square_root_def = square_root_def.replace('square_root_annotated',
'square_root_negative')
square_root_def = square_root_def.replace('return approx',
'return -approx')
print_content(square_root_def, '.py')
exec(square_root_def)
###Output
_____no_output_____
###Markdown
Technically speaking, $-2$ _is_ a square root of 4, since $(-2)^2 = 4$ holds. Yet, such a change may be unexpected by callers of `square_root()`, and hence, this would be caught with the first call:
###Code
with ExpectError():
square_root_negative(2.0) # type: ignore
###Output
_____no_output_____
###Markdown
We see how pre- and postconditions, as well as types, can serve as *oracles* during testing. In particular, once we have mined them for a set of functions, we can check them again and again with test generators – especially after code changes. The more checks we have, and the more specific they are, the more likely it is we can detect unwanted effects of changes. SynopsisThis chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:
###Code
def sum2(a, b): # type: ignore
return a + b
with TypeAnnotator() as type_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.
###Code
print(type_annotator.typed_functions())
###Output
_____no_output_____
###Markdown
The invariant annotator works in a similar fashion:
###Code
with InvariantAnnotator() as inv_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.
###Code
print(inv_annotator.functions_with_invariants())
###Output
_____no_output_____
###Markdown
Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs). The chapter gives details on how to customize the properties checked for. Lessons Learned* Type annotations and explicit invariants allow for _checking_ arguments and results for expected data types and other properties.* One can automatically _mine_ data types and invariants by observing arguments and results at runtime.* The quality of mined invariants depends on the diversity of values observed during executions; this variety can be increased by generating tests. Next StepsIn the next chapter, we will explore [abstracting failure conditions](DDSetDebugger.ipynb). BackgroundThe [DAIKON dynamic invariant detector](https://plse.cs.washington.edu/daikon/) can be considered the mother of function specification miners. Continuously maintained and extended for more than 20 years, it mines likely invariants in the style of this chapter for a variety of languages, including C, C++, C, Eiffel, F, Java, Perl, and Visual Basic. On top of the functionality discussed above, it holds a rich catalog of patterns for likely invariants, supports data invariants, can eliminate invariants that are implied by others, and determines statistical confidence to disregard unlikely invariants. The corresponding paper \cite{Ernst2001} is one of the seminal and most-cited papers of Software Engineering. A multitude of works have been published based on DAIKON and detecting invariants; see this [curated list](http://plse.cs.washington.edu/daikon/pubs/) for details. The interaction between test generators and invariant detection is already discussed in \cite{Ernst2001} (incidentally also using grammars). The Eclat tool \cite{Pacheco2005} is a model example of tight interaction between a unit-level test generator and DAIKON-style invariant mining, where the mined invariants are used to produce oracles and to systematically guide the test generator towards fault-revealing inputs. Mining specifications is not restricted to pre- and postconditions. The paper "Mining Specifications" \cite{Ammons2002} is another classic in the field, learning state protocols from executions. Grammar mining \cite{Gopinath2020} can also be seen as a specification mining approach, this time learning specifications for input formats. As it comes to adding type annotations to existing code, the blog post ["The state of type hints in Python"](https://www.bernat.tech/the-state-of-type-hints-in-python/) gives a great overview on how Python type hints can be used and checked. To add type annotations, there are two important tools available that also implement our above approach:* [MonkeyType](https://instagram-engineering.com/let-your-code-type-hint-itself-introducing-open-source-monkeytype-a855c7284881) implements the above approach of tracing executions and annotating Python 3 arguments, returns, and variables with type hints.* [PyAnnotate](https://github.com/dropbox/pyannotate) does a similar job, focusing on code in Python 2. It does not produce Python 3-style annotations, but instead produces annotations as comments that can be processed by static type checkers.These tools have been created by engineers at Facebook and Dropbox, respectively, assisting them in checking millions of lines of code for type issues. ExercisesOur code for mining types and invariants is in no way complete. There are dozens of ways to extend our implementations, some of which we discuss in exercises. Exercise 1: Union TypesThe Python `typing` module allows to express that an argument can have multiple types. For `square_root(x)`, this allows to express that `x` can be an `int` or a `float`:
###Code
def square_root_with_union_type(x: Union[int, float]) -> float: # type: ignore
...
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it supports union types for arguments and return values. Use `Optional[X]` as a shorthand for `Union[X, None]`. **Solution.** Left to the reader. Hint: extend `type_string()`. Exercise 2: Types for Local VariablesIn Python, one cannot only annotate arguments with types, but actually also local and global variables – for instance, `approx` and `guess` in our `square_root()` implementation:
###Code
def square_root_with_local_types(x: Union[int, float]) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
approx: Optional[float] = None
guess: float = x / 2
while approx != guess:
approx: float = guess # type: ignore
guess: float = (approx + x / approx) / 2 # type: ignore
return approx
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it also annotates local variables with types. Search the function AST for assignments, determine the type of the assigned value, and make it an annotation on the left hand side. **Solution.** Left to the reader. Exercise 3: Verbose Invariant CheckersOur implementation of invariant checkers does not make it clear for the user which pre-/postcondition failed.
###Code
@precondition(lambda s: len(s) > 0)
def remove_first_char(s: str) -> str:
return s[1:]
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
The following implementation adds an optional `doc` keyword argument which is printed if the invariant is violated:
###Code
def my_condition(precondition: Optional[Callable] = None,
postcondition: Optional[Callable] = None, doc: str = 'Unknown') -> Callable:
def decorator(func: Callable) -> Callable:
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args: Any, **kwargs: Any) -> Any:
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated: " + doc
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated: " + doc
return retval
return wrapper
return decorator
def my_precondition(check: Callable, **kwargs: Any) -> Callable:
return my_condition(precondition=check, doc=kwargs.get('doc', 'Unknown'))
def my_postcondition(check: Callable, **kwargs: Any) -> Callable:
return my_condition(postcondition=check, doc=kwargs.get('doc', 'Unknown'))
@my_precondition(lambda s: len(s) > 0, doc="len(s) > 0") # type: ignore
def remove_first_char(s: str) -> str:
return s[1:]
remove_first_char('abc')
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
Extend `InvariantAnnotator` such that it includes the conditions in the generated pre- and postconditions. **Solution.** Here's a simple solution:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@my_precondition(lambda " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")"
conditions.append(cond)
return conditions
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name: str) -> List[str]:
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@my_postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")")
conditions.append(cond)
return conditions
###Output
_____no_output_____
###Markdown
The resulting annotations are harder to read, but easier to diagnose:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
As an alternative, one may be able to use `inspect.getsource()` on the lambda expression or unparse it. This is left to the reader. Exercise 4: Save Initial ValuesIf the value of an argument changes during function execution, this can easily confuse our implementation: The values are tracked at the beginning of the function, but checked only when it returns. Extend the `InvariantAnnotator` and the infrastructure it uses such that* it saves argument values both at the beginning and at the end of a function invocation;* postconditions can be expressed over both _initial_ values of arguments as well as the _final_ values of arguments;* the mined postconditions refer to both these values as well. **Solution.** To be added. Exercise 5: ImplicationsSeveral mined invariant are actually _implied_ by others: If `x > 0` holds, then this implies `x >= 0` and `x != 0`. Extend the `InvariantAnnotator` such that implications between properties are explicitly encoded, and such that implied properties are no longer listed as invariants. See \cite{Ernst2001} for ideas. **Solution.** Left to the reader. Exercise 6: Local VariablesPostconditions may also refer to the values of local variables. Consider extending `InvariantAnnotator` and its infrastructure such that the values of local variables at the end of the execution are also recorded and made part of the invariant inference mechanism. **Solution.** Left to the reader. Exercise 7: Embedding Invariants as AssertionsRather than producing invariants as annotations for pre- and postconditions, insert them as `assert` statements into the function code, as in:```pythondef square_root(x): 'Computes the square root of x, using the Newton-Raphson method' assert isinstance(x, int), 'violated precondition' assert x > 0, 'violated precondition' approx = None guess = (x / 2) while (approx != guess): approx = guess guess = ((approx + (x / approx)) / 2) return_value = approx assert return_value < x, 'violated postcondition' assert isinstance(return_value, float), 'violated postcondition' return approx```Such a formulation may make it easier for test generators and symbolic analysis to access and interpret pre- and postconditions. **Solution.** Here is a tentative implementation that inserts invariants into function ASTs. Part 1: Embedding Invariants into Functions
###Code
class EmbeddedInvariantAnnotator(InvariantTracer):
def function_with_invariants_ast(self, function_name: str) -> ast.AST:
return annotate_function_with_invariants(function_name, self.invariants(function_name))
def function_with_invariants(self, function_name: str) -> str:
return astor.to_source(self.function_with_invariants_ast(function_name))
def annotate_invariants(invariants: Dict[str, Invariants]) -> Dict[str, ast.AST]:
annotated_functions = {}
for function_name in invariants:
try:
annotated_functions[function_name] = annotate_function_with_invariants(function_name, invariants[function_name])
except KeyError:
continue
return annotated_functions
def annotate_function_with_invariants(function_name: str,
function_invariants: Invariants) -> ast.AST:
function = globals()[function_name]
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_invariants(function_ast, function_invariants)
def annotate_function_ast_with_invariants(function_ast: ast.AST,
function_invariants: Invariants) -> ast.AST:
annotated_function_ast = EmbeddedInvariantTransformer(function_invariants).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Part 2: Preconditions
###Code
class PreconditionTransformer(ast.NodeTransformer):
def __init__(self, invariants: Invariants) -> None:
self.invariants = invariants
super().__init__()
def preconditions(self) -> List[ast.stmt]:
preconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated precondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) < 0:
preconditions += assertion_ast.body
return preconditions
def insert_assertions(self, body: List[ast.stmt]) -> List[ast.stmt]:
preconditions = self.preconditions()
try:
docstring = cast(ast.Constant, body[0]).value.s
except:
docstring = None
if docstring:
return [body[0]] + preconditions + body[1:]
else:
return preconditions + body
def visit_FunctionDef(self, node: ast.FunctionDef) -> ast.FunctionDef:
"""Add invariants to function"""
# print(ast.dump(node))
node.body = self.insert_assertions(node.body)
return node
class EmbeddedInvariantTransformer(PreconditionTransformer):
pass
with EmbeddedInvariantAnnotator() as annotator:
square_root(5)
print_content(annotator.function_with_invariants('square_root'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.function_with_invariants('sum3'), '.py')
###Output
_____no_output_____
###Markdown
Part 3: PostconditionsWe make a few simplifying assumptions: * Variables do not change during execution.* There is a single `return` statement at the end of the function.
###Code
class EmbeddedInvariantTransformer(EmbeddedInvariantTransformer):
def postconditions(self) -> List[ast.stmt]:
postconditions = []
for (prop, var_names) in self.invariants:
assertion = ("assert " + instantiate_prop(prop, var_names) +
', "violated postcondition"')
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) >= 0:
postconditions += assertion_ast.body
return postconditions
def insert_assertions(self, body: List[ast.stmt]) -> List[ast.stmt]:
new_body = super().insert_assertions(body)
postconditions = self.postconditions()
body_ends_with_return = isinstance(new_body[-1], ast.Return)
if body_ends_with_return:
ret_val = cast(ast.Return, new_body[-1]).value
saver = RETURN_VALUE + " = " + astor.to_source(ret_val)
else:
saver = RETURN_VALUE + " = None"
saver_ast = cast(ast.stmt, ast.parse(saver))
postconditions = [saver_ast] + postconditions
if body_ends_with_return:
return new_body[:-1] + postconditions + [new_body[-1]]
else:
return new_body + postconditions
with EmbeddedInvariantAnnotator() as annotator:
square_root(5)
square_root_def = annotator.function_with_invariants('square_root')
###Output
_____no_output_____
###Markdown
Here's the full definition with included assertions:
###Code
print_content(square_root_def, '.py')
exec(square_root_def.replace('square_root', 'square_root_annotated'))
with ExpectError():
square_root_annotated(-1)
###Output
_____no_output_____
###Markdown
Here come some more examples:
###Code
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.function_with_invariants('sum3'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.function_with_invariants('list_length'), '.py')
with EmbeddedInvariantAnnotator() as annotator:
print_sum(31, 45)
print_content(annotator.function_with_invariants('print_sum'), '.py')
###Output
_____no_output_____
###Markdown
Our goal is to avoid such errors by _annotating_ functions with information that prevents errors like the above ones. The idea is to provide a _specification_ of expected properties – a specification that can then be checked at runtime or statically. \todo{Introduce the concept of *contract*.} Specifying and Checking Data TypesFor our Python code, one of the most important "specifications" we need is *types*. Python being a "dynamically" typed language means that all data types are determined at run time; the code itself does not explicitly state whether a variable is an integer, a string, an array, a dictionary – or whatever. As _writer_ of Python code, omitting explicit type declarations may save time (and allows for some fun hacks). It is not sure whether a lack of types helps in _reading_ and _understanding_ code for humans. For a _computer_ trying to analyze code, the lack of explicit types is detrimental. If, say, a constraint solver, sees `if x:` and cannot know whether `x` is supposed to be a number or a string, this introduces an _ambiguity_. Such ambiguities may multiply over the entire analysis in a combinatorial explosion – or in the analysis yielding an overly inaccurate result. Python 3.6 and later allows data types as _annotations_ to function arguments (actually, to all variables) and return values. We can, for instance, state that `my_sqrt()` is a function that accepts a floating-point value and returns one:
###Code
def my_sqrt_with_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
By default, such annotations are ignored by the Python interpreter. Therefore, one can still call `my_sqrt_typed()` with a string as an argument and get the exact same result as above. However, one can make use of special _typechecking_ modules that would check types – _dynamically_ at runtime or _statically_ by analyzing the code without having to execute it. Runtime Type CheckingThe Python `enforce` package provides a function decorator that automatically inserts type-checking code that is executed at runtime. Here is how to use it:
###Code
import enforce
@enforce.runtime_validation
def my_sqrt_with_checked_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
Now, invoking `my_sqrt_with_checked_type_annotations()` raises an exception when invoked with a type dfferent from the one declared:
###Code
with ExpectError():
my_sqrt_with_checked_type_annotations(True)
###Output
_____no_output_____
###Markdown
Note that this error is not caught by the "untyped" variant, where passing a boolean value happily returns $\sqrt{1}$ as result.
###Code
my_sqrt(True)
###Output
_____no_output_____
###Markdown
In Python (and other languages), the boolean values `True` and `False` can be implicitly converted to the integers 1 and 0; however, it is hard to think of a call to `sqrt()` where this would not be an error. Static Type CheckingType annotations can also be checked _statically_ – that is, without even running the code. Let us create a simple Python file consisting of the above `my_sqrt_typed()` definition and a bad invocation.
###Code
import inspect
import tempfile
f = tempfile.NamedTemporaryFile(mode='w', suffix='.py')
f.name
f.write(inspect.getsource(my_sqrt))
f.write('\n')
f.write(inspect.getsource(my_sqrt_with_type_annotations))
f.write('\n')
f.write("print(my_sqrt_with_type_annotations('123'))\n")
f.flush()
###Output
_____no_output_____
###Markdown
These are the contents of our newly created Python file:
###Code
from bookutils import print_file
print_file(f.name)
###Output
_____no_output_____
###Markdown
[Mypy](http://mypy-lang.org) is a type checker for Python programs. As it checks types statically, types induce no overhead at runtime; plus, a static check can be faster than a lengthy series of tests with runtime type checking enabled. Let us see what `mypy` produces on the above file:
###Code
import subprocess
result = subprocess.run(["mypy", "--strict", f.name], universal_newlines=True, stdout=subprocess.PIPE)
del f # Delete temporary file
print(result.stdout)
###Output
_____no_output_____
###Markdown
We see that `mypy` complains about untyped function definitions such as `my_sqrt()`; most important, however, it finds that the call to `my_sqrt_with_type_annotations()` in the last line has the wrong type. With `mypy`, we can achieve the same type safety with Python as in statically typed languages – provided that we as programmers also produce the necessary type annotations. Is there a simple way to obtain these? Mining Type SpecificationsOur first task will be to mine type annotations (as part of the code) from _values_ we observe at run time. These type annotations would be _mined_ from actual function executions, _learning_ from (normal) runs what the expected argument and return types should be. By observing a series of calls such as these, we could infer that both `x` and the return value are of type `float`:
###Code
y = my_sqrt(25.0)
y
y = my_sqrt(2.0)
y
###Output
_____no_output_____
###Markdown
How can we mine types from executions? The answer is simple: 1. We _observe_ a function during execution2. We track the _types_ of its arguments3. We include these types as _annotations_ into the code.To do so, we can make use of Python's tracing facility we already observed in the [chapter on coverage](Coverage.ipynb). With every call to a function, we retrieve the arguments, their values, and their types. Tracking CallsTo observe argument types at runtime, we define a _tracer function_ that tracks the execution of `my_sqrt()`, checking its arguments and return values. The `Tracker` class is set to trace functions in a `with` block as follows:```pythonwith Tracker() as tracker: function_to_be_tracked(...)info = tracker.collected_information()```As in the [chapter on coverage](Coverage.ipynb), we use the `sys.settrace()` function to trace individual functions during execution. We turn on tracking when the `with` block starts; at this point, the `__enter__()` method is called. When execution of the `with` block ends, `__exit()__` is called.
###Code
import sys
class Tracker(object):
def __init__(self, log=False):
self._log = log
self.reset()
def reset(self):
self._calls = {}
self._stack = []
def traceit(self):
"""Placeholder to be overloaded in subclasses"""
pass
# Start of `with` block
def __enter__(self):
self.original_trace_function = sys.gettrace()
sys.settrace(self.traceit)
return self
# End of `with` block
def __exit__(self, exc_type, exc_value, tb):
sys.settrace(self.original_trace_function)
###Output
_____no_output_____
###Markdown
The `traceit()` method does nothing yet; this is done in specialized subclasses. The `CallTracker` class implements a `traceit()` function that checks for function calls and returns:
###Code
class CallTracker(Tracker):
def traceit(self, frame, event, arg):
"""Tracking function: Record all calls and all args"""
if event == "call":
self.trace_call(frame, event, arg)
elif event == "return":
self.trace_return(frame, event, arg)
return self.traceit
###Output
_____no_output_____
###Markdown
`trace_call()` is called when a function is called; it retrieves the function name and current arguments, and saves them on a stack.
###Code
class CallTracker(CallTracker):
def trace_call(self, frame, event, arg):
"""Save current function name and args on the stack"""
code = frame.f_code
function_name = code.co_name
arguments = get_arguments(frame)
self._stack.append((function_name, arguments))
if self._log:
print(simple_call_string(function_name, arguments))
def get_arguments(frame):
"""Return call arguments in the given frame"""
# When called, all arguments are local variables
arguments = [(var, frame.f_locals[var]) for var in frame.f_locals]
arguments.reverse() # Want same order as call
return arguments
###Output
_____no_output_____
###Markdown
When the function returns, `trace_return()` is called. We now also have the return value. We log the whole call with arguments and return value (if desired) and save it in our list of calls.
###Code
class CallTracker(CallTracker):
def trace_return(self, frame, event, arg):
"""Get return value and store complete call with arguments and return value"""
code = frame.f_code
function_name = code.co_name
return_value = arg
# TODO: Could call get_arguments() here to also retrieve _final_ values of argument variables
called_function_name, called_arguments = self._stack.pop()
assert function_name == called_function_name
if self._log:
print(simple_call_string(function_name, called_arguments), "returns", return_value)
self.add_call(function_name, called_arguments, return_value)
###Output
_____no_output_____
###Markdown
`simple_call_string()` is a helper for logging that prints out calls in a user-friendly manner.
###Code
def simple_call_string(function_name, argument_list, return_value=None):
"""Return function_name(arg[0], arg[1], ...) as a string"""
call = function_name + "(" + \
", ".join([var + "=" + repr(value)
for (var, value) in argument_list]) + ")"
if return_value is not None:
call += " = " + repr(return_value)
return call
###Output
_____no_output_____
###Markdown
`add_call()` saves the calls in a list; each function name has its own list.
###Code
class CallTracker(CallTracker):
def add_call(self, function_name, arguments, return_value=None):
"""Add given call to list of calls"""
if function_name not in self._calls:
self._calls[function_name] = []
self._calls[function_name].append((arguments, return_value))
###Output
_____no_output_____
###Markdown
Using `calls()`, we can retrieve the list of calls, either for a given function, or for all functions.
###Code
class CallTracker(CallTracker):
def calls(self, function_name=None):
"""Return list of calls for function_name,
or a mapping function_name -> calls for all functions tracked"""
if function_name is None:
return self._calls
return self._calls[function_name]
###Output
_____no_output_____
###Markdown
Let us now put this to use. We turn on logging to track the individual calls and their return values:
###Code
with CallTracker(log=True) as tracker:
y = my_sqrt(25)
y = my_sqrt(2.0)
###Output
_____no_output_____
###Markdown
After execution, we can retrieve the individual calls:
###Code
calls = tracker.calls('my_sqrt')
calls
###Output
_____no_output_____
###Markdown
Each call is pair (`argument_list`, `return_value`), where `argument_list` is a list of pairs (`parameter_name`, `value`).
###Code
my_sqrt_argument_list, my_sqrt_return_value = calls[0]
simple_call_string('my_sqrt', my_sqrt_argument_list, my_sqrt_return_value)
###Output
_____no_output_____
###Markdown
If the function does not return a value, `return_value` is `None`.
###Code
def hello(name):
print("Hello,", name)
with CallTracker() as tracker:
hello("world")
hello_calls = tracker.calls('hello')
hello_calls
hello_argument_list, hello_return_value = hello_calls[0]
simple_call_string('hello', hello_argument_list, hello_return_value)
###Output
_____no_output_____
###Markdown
Getting TypesDespite what you may have read or heard, Python actually _is_ a typed language. It is just that it is _dynamically typed_ – types are used and checked only at runtime (rather than declared in the code, where they can be _statically checked_ at compile time). We can thus retrieve types of all values within Python:
###Code
type(4)
type(2.0)
type([4])
###Output
_____no_output_____
###Markdown
We can retrieve the type of the first argument to `my_sqrt()`:
###Code
parameter, value = my_sqrt_argument_list[0]
parameter, type(value)
###Output
_____no_output_____
###Markdown
as well as the type of the return value:
###Code
type(my_sqrt_return_value)
###Output
_____no_output_____
###Markdown
Hence, we see that (so far), `my_sqrt()` is a function taking (among others) integers and returning floats. We could declare `my_sqrt()` as:
###Code
def my_sqrt_annotated(x: int) -> float:
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
This is a representation we could place in a static type checker, allowing to check whether calls to `my_sqrt()` actually pass a number. A dynamic type checker could run such checks at runtime. And of course, any [symbolic interpretation](SymbolicFuzzer.ipynb) will greatly profit from the additional annotations. By default, Python does not do anything with such annotations. However, tools can access annotations from functions and other objects:
###Code
my_sqrt_annotated.__annotations__
###Output
_____no_output_____
###Markdown
This is how run-time checkers access the annotations to check against. Accessing Function StructureOur plan is to annotate functions automatically, based on the types we have seen. To do so, we need a few modules that allow us to convert a function into a tree representation (called _abstract syntax trees_, or ASTs) and back; we already have seen these in the chapters on [concolic](ConcolicFuzzer.ipynb) and [symbolic](SymbolicFuzzer.ipynb) testing.
###Code
import ast
import inspect
import astor
###Output
_____no_output_____
###Markdown
We can get the source of a Python function using `inspect.getsource()`. (Note that this does not work for functions defined in other notebooks.)
###Code
my_sqrt_source = inspect.getsource(my_sqrt)
my_sqrt_source
###Output
_____no_output_____
###Markdown
To view these in a visually pleasing form, our function `print_content(s, suffix)` formats and highlights the string `s` as if it were a file with ending `suffix`. We can thus view (and highlight) the source as if it were a Python file:
###Code
from bookutils import print_content
print_content(my_sqrt_source, '.py')
###Output
_____no_output_____
###Markdown
Parsing this gives us an abstract syntax tree (AST) – a representation of the program in tree form.
###Code
my_sqrt_ast = ast.parse(my_sqrt_source)
###Output
_____no_output_____
###Markdown
What does this AST look like? The helper functions `astor.dump_tree()` (textual output) and `showast.show_ast()` (graphical output with [showast](https://github.com/hchasestevens/show_ast)) allow us to inspect the structure of the tree. We see that the function starts as a `FunctionDef` with name and arguments, followed by a body, which is a list of statements of type `Expr` (the docstring), type `Assign` (assignments), `While` (while loop with its own body), and finally `Return`.
###Code
print(astor.dump_tree(my_sqrt_ast))
###Output
_____no_output_____
###Markdown
Too much text for you? This graphical representation may make things simpler.
###Code
from bookutils import rich_output
if rich_output():
import showast
showast.show_ast(my_sqrt_ast)
###Output
_____no_output_____
###Markdown
The function `astor.to_source()` converts such a tree back into the more familiar textual Python code representation. Comments are gone, and there may be more parentheses than before, but the result has the same semantics:
###Code
print_content(astor.to_source(my_sqrt_ast), '.py')
###Output
_____no_output_____
###Markdown
Annotating Functions with Given TypesLet us now go and transform these trees ti add type annotations. We start with a helper function `parse_type(name)` which parses a type name into an AST.
###Code
def parse_type(name):
class ValueVisitor(ast.NodeVisitor):
def visit_Expr(self, node):
self.value_node = node.value
tree = ast.parse(name)
name_visitor = ValueVisitor()
name_visitor.visit(tree)
return name_visitor.value_node
print(astor.dump_tree(parse_type('int')))
print(astor.dump_tree(parse_type('[object]')))
###Output
_____no_output_____
###Markdown
We now define a helper function that actually adds type annotations to a function AST. The `TypeTransformer` class builds on the Python standard library `ast.NodeTransformer` infrastructure. It would be called as```python TypeTransformer({'x': 'int'}, 'float').visit(ast)```to annotate the arguments of `my_sqrt()`: `x` with `int`, and the return type with `float`. The returned AST can then be unparsed, compiled or analyzed.
###Code
class TypeTransformer(ast.NodeTransformer):
def __init__(self, argument_types, return_type=None):
self.argument_types = argument_types
self.return_type = return_type
super().__init__()
###Output
_____no_output_____
###Markdown
The core of `TypeTransformer` is the method `visit_FunctionDef()`, which is called for every function definition in the AST. Its argument `node` is the subtree of the function definition to be transformed. Our implementation accesses the individual arguments and invokes `annotate_args()` on them; it also sets the return type in the `returns` attribute of the node.
###Code
class TypeTransformer(TypeTransformer):
def visit_FunctionDef(self, node):
"""Add annotation to function"""
# Set argument types
new_args = []
for arg in node.args.args:
new_args.append(self.annotate_arg(arg))
new_arguments = ast.arguments(
new_args,
node.args.vararg,
node.args.kwonlyargs,
node.args.kw_defaults,
node.args.kwarg,
node.args.defaults
)
# Set return type
if self.return_type is not None:
node.returns = parse_type(self.return_type)
return ast.copy_location(ast.FunctionDef(node.name, new_arguments,
node.body, node.decorator_list,
node.returns), node)
###Output
_____no_output_____
###Markdown
Each argument gets its own annotation, taken from the types originally passed to the class:
###Code
class TypeTransformer(TypeTransformer):
def annotate_arg(self, arg):
"""Add annotation to single function argument"""
arg_name = arg.arg
if arg_name in self.argument_types:
arg.annotation = parse_type(self.argument_types[arg_name])
return arg
###Output
_____no_output_____
###Markdown
Does this work? Let us annotate the AST from `my_sqrt()` with types for the arguments and return types:
###Code
new_ast = TypeTransformer({'x': 'int'}, 'float').visit(my_sqrt_ast)
###Output
_____no_output_____
###Markdown
When we unparse the new AST, we see that the annotations actually are present:
###Code
print_content(astor.to_source(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Similarly, we can annotate the `hello()` function from above:
###Code
hello_source = inspect.getsource(hello)
hello_ast = ast.parse(hello_source)
new_ast = TypeTransformer({'name': 'str'}, 'None').visit(hello_ast)
print_content(astor.to_source(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Annotating Functions with Mined TypesLet us now annotate functions with types mined at runtime. We start with a simple function `type_string()` that determines the appropriate type of a given value (as a string):
###Code
def type_string(value):
return type(value).__name__
type_string(4)
type_string([])
###Output
_____no_output_____
###Markdown
For composite structures, `type_string()` does not examine element types; hence, the type of `[3]` is simply `list` instead of, say, `list[int]`. For now, `list` will do fine.
###Code
type_string([3])
###Output
_____no_output_____
###Markdown
`type_string()` will be used to infer the types of argument values found at runtime, as returned by `CallTracker.calls()`:
###Code
with CallTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(2.0)
tracker.calls()
###Output
_____no_output_____
###Markdown
The function `annotate_types()` takes such a list of calls and annotates each function listed:
###Code
def annotate_types(calls):
annotated_functions = {}
for function_name in calls:
try:
annotated_functions[function_name] = annotate_function_with_types(function_name, calls[function_name])
except KeyError:
continue
return annotated_functions
###Output
_____no_output_____
###Markdown
For each function, we get the source and its AST and then get to the actual annotation in `annotate_function_ast_with_types()`:
###Code
def annotate_function_with_types(function_name, function_calls):
function = globals()[function_name] # May raise KeyError for internal functions
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_types(function_ast, function_calls)
###Output
_____no_output_____
###Markdown
The function `annotate_function_ast_with_types()` invokes the `TypeTransformer` with the calls seen, and for each call, iterate over the arguments, determine their types, and annotate the AST with these. The universal type `Any` is used when we encounter type conflicts, which we will discuss below.
###Code
from typing import Any
def annotate_function_ast_with_types(function_ast, function_calls):
parameter_types = {}
return_type = None
for calls_seen in function_calls:
args, return_value = calls_seen
if return_value is not None:
if return_type is not None and return_type != type_string(return_value):
return_type = 'Any'
else:
return_type = type_string(return_value)
for parameter, value in args:
try:
different_type = parameter_types[parameter] != type_string(value)
except KeyError:
different_type = False
if different_type:
parameter_types[parameter] = 'Any'
else:
parameter_types[parameter] = type_string(value)
annotated_function_ast = TypeTransformer(parameter_types, return_type).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Here is `my_sqrt()` annotated with the types recorded usign the tracker, above.
###Code
print_content(astor.to_source(annotate_types(tracker.calls())['my_sqrt']), '.py')
###Output
_____no_output_____
###Markdown
All-in-one AnnotationLet us bring all of this together in a single class `TypeAnnotator` that first tracks calls of functions and then allows to access the AST (and the source code form) of the tracked functions annotated with types. The method `typed_functions()` returns the annotated functions as a string; `typed_functions_ast()` returns their AST.
###Code
class TypeTracker(CallTracker):
pass
class TypeAnnotator(TypeTracker):
def typed_functions_ast(self, function_name=None):
if function_name is None:
return annotate_types(self.calls())
return annotate_function_with_types(function_name, self.calls(function_name))
def typed_functions(self, function_name=None):
if function_name is None:
functions = ''
for f_name in self.calls():
try:
f_text = astor.to_source(self.typed_functions_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
return astor.to_source(self.typed_functions_ast(function_name))
###Output
_____no_output_____
###Markdown
Here is how to use `TypeAnnotator`. We first track a series of calls:
###Code
with TypeAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(2.0)
###Output
_____no_output_____
###Markdown
After tracking, we can immediately retrieve an annotated version of the functions tracked:
###Code
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
This also works for multiple and diverse functions. One could go and implement an automatic type annotator for Python files based on the types seen during execution.
###Code
with TypeAnnotator() as annotator:
hello('type annotations')
y = my_sqrt(1.0)
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
A content as above could now be sent to a type checker, which would detect any type inconsistency between callers and callees. Likewise, type annotations such as the ones above greatly benefit symbolic code analysis (as in the chapter on [symbolic fuzzing](SymbolicFuzzer.ipynb)), as they effectively constrain the set of values that arguments and variables can take. Multiple TypesLet us now resolve the role of the magic `Any` type in `annotate_function_ast_with_types()`. If we see multiple types for the same argument, we set its type to `Any`. For `my_sqrt()`, this makes sense, as its arguments can be integers as well as floats:
###Code
with CallTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(4)
print_content(astor.to_source(annotate_types(tracker.calls())['my_sqrt']), '.py')
###Output
_____no_output_____
###Markdown
The following function `sum3()` can be called with floating-point numbers as arguments, resulting in the parameters getting a `float` type:
###Code
def sum3(a, b, c):
return a + b + c
with TypeAnnotator() as annotator:
y = sum3(1.0, 2.0, 3.0)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we call `sum3()` with integers, though, the arguments get an `int` type:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
And we can also call `sum3()` with strings, giving the arguments a `str` type:
###Code
with TypeAnnotator() as annotator:
y = sum3("one", "two", "three")
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we have multiple calls, but with different types, `TypeAnnotator()` will assign an `Any` type to both arguments and return values:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y = sum3("one", "two", "three")
typed_sum3_def = annotator.typed_functions('sum3')
print_content(typed_sum3_def, '.py')
###Output
_____no_output_____
###Markdown
A type `Any` makes it explicit that an object can, indeed, have any type; it will not be typechecked at runtime or statically. To some extent, this defeats the power of type checking; but it also preserves some of the type flexibility that many Python programmers enjoy. Besides `Any`, the `typing` module supports several additional ways to define ambiguous types; we will keep this in mind for a later exercise. Specifying and Checking InvariantsBesides basic data types. we can check several further properties from arguments. We can, for instance, whether an argument can be negative, zero, or positive; or that one argument should be smaller than the second; or that the result should be the sum of two arguments – properties that cannot be expressed in a (Python) type.Such properties are called *invariants*, as they hold across all invocations of a function. Specifically, invariants come as _pre_- and _postconditions_ – conditions that always hold at the beginning and at the end of a function. (There are also _data_ and _object_ invariants that express always-holding properties over the state of data or objects, but we do not consider these in this book.) Annotating Functions with Pre- and PostconditionsThe classical means to specify pre- and postconditions is via _assertions_, which we have introduced in the [chapter on testing](Intro_Testing.ipynb). A precondition checks whether the arguments to a function satisfy the expected properties; a postcondition does the same for the result. We can express and check both using assertions as follows:
###Code
def my_sqrt_with_invariants(x):
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
A nicer way, however, is to syntactically separate invariants from the function at hand. Using appropriate decorators, we could specify pre- and postconditions as follows:```python@precondition lambda x: x >= 0@postcondition lambda return_value, x: return_value * return_value == xdef my_sqrt_with_invariants(x): normal code without assertions ...```The decorators `@precondition` and `@postcondition` would run the given functions (specified as anonymous `lambda` functions) before and after the decorated function, respectively. If the functions return `False`, the condition is violated. `@precondition` gets the function arguments as arguments; `@postcondition` additionally gets the return value as first argument. It turns out that implementing such decorators is not hard at all. Our implementation builds on a [code snippet from StackOverflow](https://stackoverflow.com/questions/12151182/python-precondition-postcondition-for-member-function-how):
###Code
import functools
def condition(precondition=None, postcondition=None):
def decorator(func):
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args, **kwargs):
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated"
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated"
return retval
return wrapper
return decorator
def precondition(check):
return condition(precondition=check)
def postcondition(check):
return condition(postcondition=check)
###Output
_____no_output_____
###Markdown
With these, we can now start decorating `my_sqrt()`:
###Code
@precondition(lambda x: x > 0)
def my_sqrt_with_precondition(x):
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
This catches arguments violating the precondition:
###Code
with ExpectError():
my_sqrt_with_precondition(-1.0)
###Output
_____no_output_____
###Markdown
Likewise, we can provide a postcondition:
###Code
EPSILON = 1e-5
@postcondition(lambda ret, x: ret * ret - x < EPSILON)
def my_sqrt_with_postcondition(x):
return my_sqrt(x)
y = my_sqrt_with_postcondition(2.0)
y
###Output
_____no_output_____
###Markdown
If we have a buggy implementation of $\sqrt{x}$, this gets caught quickly:
###Code
@postcondition(lambda ret, x: ret * ret - x < EPSILON)
def buggy_my_sqrt_with_postcondition(x):
return my_sqrt(x) + 0.1
with ExpectError():
y = buggy_my_sqrt_with_postcondition(2.0)
###Output
_____no_output_____
###Markdown
While checking pre- and postconditions is a great way to catch errors, specifying them can be cumbersome. Let us try to see whether we can (again) _mine_ some of them. Mining InvariantsTo _mine_ invariants, we can use the same tracking functionality as before; instead of saving values for individual variables, though, we now check whether the values satisfy specific _properties_ or not. For instance, if all values of `x` seen satisfy the condition `x > 0`, then we make `x > 0` an invariant of the function. If we see positive, zero, and negative values of `x`, though, then there is no property of `x` left to talk about.The general idea is thus:1. Check all variable values observed against a set of predefined properties; and2. Keep only those properties that hold for all runs observed. Defining PropertiesWhat precisely do we mean by properties? Here is a small collection of value properties that would frequently be used in invariants. All these properties would be evaluated with the _metavariables_ `X`, `Y`, and `Z` (actually, any upper-case identifier) being replaced with the names of function parameters:
###Code
INVARIANT_PROPERTIES = [
"X < 0",
"X <= 0",
"X > 0",
"X >= 0",
"X == 0",
"X != 0",
]
###Output
_____no_output_____
###Markdown
When `my_sqrt(x)` is called as, say `my_sqrt(5.0)`, we see that `x = 5.0` holds. The above properties would then all be checked for `x`. Only the properties `X > 0`, `X >= 0`, and `X != 0` hold for the call seen; and hence `x > 0`, `x >= 0`, and `x != 0` would make potential preconditions for `my_sqrt(x)`. We can check for many more properties such as relations between two arguments:
###Code
INVARIANT_PROPERTIES += [
"X == Y",
"X > Y",
"X < Y",
"X >= Y",
"X <= Y",
]
###Output
_____no_output_____
###Markdown
Types also can be checked using properties. For any function parameter `X`, only one of these will hold:
###Code
INVARIANT_PROPERTIES += [
"isinstance(X, bool)",
"isinstance(X, int)",
"isinstance(X, float)",
"isinstance(X, list)",
"isinstance(X, dict)",
]
###Output
_____no_output_____
###Markdown
We can check for arithmetic properties:
###Code
INVARIANT_PROPERTIES += [
"X == Y + Z",
"X == Y * Z",
"X == Y - Z",
"X == Y / Z",
]
###Output
_____no_output_____
###Markdown
Here's relations over three values, a Python special:
###Code
INVARIANT_PROPERTIES += [
"X < Y < Z",
"X <= Y <= Z",
"X > Y > Z",
"X >= Y >= Z",
]
###Output
_____no_output_____
###Markdown
Finally, we can also check for list or string properties. Again, this is just a tiny selection.
###Code
INVARIANT_PROPERTIES += [
"X == len(Y)",
"X == sum(Y)",
"X.startswith(Y)",
]
###Output
_____no_output_____
###Markdown
Extracting Meta-VariablesLet us first introduce a few _helper functions_ before we can get to the actual mining. `metavars()` extracts the set of meta-variables (`X`, `Y`, `Z`, etc.) from a property. To this end, we parse the property as a Python expression and then visit the identifiers.
###Code
def metavars(prop):
metavar_list = []
class ArgVisitor(ast.NodeVisitor):
def visit_Name(self, node):
if node.id.isupper():
metavar_list.append(node.id)
ArgVisitor().visit(ast.parse(prop))
return metavar_list
assert metavars("X < 0") == ['X']
assert metavars("X.startswith(Y)") == ['X', 'Y']
assert metavars("isinstance(X, str)") == ['X']
###Output
_____no_output_____
###Markdown
Instantiating PropertiesTo produce a property as invariant, we need to be able to _instantiate_ it with variable names. The instantiation of `X > 0` with `X` being instantiated to `a`, for instance, gets us `a > 0`. To this end, the function `instantiate_prop()` takes a property and a collection of variable names and instantiates the meta-variables left-to-right with the corresponding variables names in the collection.
###Code
def instantiate_prop_ast(prop, var_names):
class NameTransformer(ast.NodeTransformer):
def visit_Name(self, node):
if node.id not in mapping:
return node
return ast.Name(id=mapping[node.id], ctx=ast.Load())
meta_variables = metavars(prop)
assert len(meta_variables) == len(var_names)
mapping = {}
for i in range(0, len(meta_variables)):
mapping[meta_variables[i]] = var_names[i]
prop_ast = ast.parse(prop, mode='eval')
new_ast = NameTransformer().visit(prop_ast)
return new_ast
def instantiate_prop(prop, var_names):
prop_ast = instantiate_prop_ast(prop, var_names)
prop_text = astor.to_source(prop_ast).strip()
while prop_text.startswith('(') and prop_text.endswith(')'):
prop_text = prop_text[1:-1]
return prop_text
assert instantiate_prop("X > Y", ['a', 'b']) == 'a > b'
assert instantiate_prop("X.startswith(Y)", ['x', 'y']) == 'x.startswith(y)'
###Output
_____no_output_____
###Markdown
Evaluating PropertiesTo actually _evaluate_ properties, we do not need to instantiate them. Instead, we simply convert them into a boolean function, using `lambda`:
###Code
def prop_function_text(prop):
return "lambda " + ", ".join(metavars(prop)) + ": " + prop
def prop_function(prop):
return eval(prop_function_text(prop))
###Output
_____no_output_____
###Markdown
Here is a simple example:
###Code
prop_function_text("X > Y")
p = prop_function("X > Y")
p(100, 1)
p(1, 100)
###Output
_____no_output_____
###Markdown
Checking InvariantsTo extract invariants from an execution, we need to check them on all possible instantiations of arguments. If the function to be checked has two arguments `a` and `b`, we instantiate the property `X < Y` both as `a < b` and `b < a` and check each of them. To get all combinations, we use the Python `permutations()` function:
###Code
import itertools
for combination in itertools.permutations([1.0, 2.0, 3.0], 2):
print(combination)
###Output
_____no_output_____
###Markdown
The function `true_property_instantiations()` takes a property and a list of tuples (`var_name`, `value`). It then produces all instantiations of the property with the given values and returns those that evaluate to True.
###Code
def true_property_instantiations(prop, vars_and_values, log=False):
instantiations = set()
p = prop_function(prop)
len_metavars = len(metavars(prop))
for combination in itertools.permutations(vars_and_values, len_metavars):
args = [value for var_name, value in combination]
var_names = [var_name for var_name, value in combination]
try:
result = p(*args)
except:
result = None
if log:
print(prop, combination, result)
if result:
instantiations.add((prop, tuple(var_names)))
return instantiations
###Output
_____no_output_____
###Markdown
Here is an example. If `x == -1` and `y == 1`, the property `X < Y` holds for `x < y`, but not for `y < x`:
###Code
invs = true_property_instantiations("X < Y", [('x', -1), ('y', 1)], log=True)
invs
###Output
_____no_output_____
###Markdown
The instantiation retrieves the short form:
###Code
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Likewise, with values for `x` and `y` as above, the property `X < 0` only holds for `x`, but not for `y`:
###Code
invs = true_property_instantiations("X < 0", [('x', -1), ('y', 1)], log=True)
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Extracting InvariantsLet us now run the above invariant extraction on function arguments and return values as observed during a function execution. To this end, we extend the `CallTracker` class into an `InvariantTracker` class, which automatically computes invariants for all functions and all calls observed during tracking. By default, an `InvariantTracker` uses the properties as defined above; however, one can specify alternate sets of properties.
###Code
class InvariantTracker(CallTracker):
def __init__(self, props=None, **kwargs):
if props is None:
props = INVARIANT_PROPERTIES
self.props = props
super().__init__(**kwargs)
###Output
_____no_output_____
###Markdown
The key method of the `InvariantTracker` is the `invariants()` method. This iterates over the calls observed and checks which properties hold. Only the intersection of properties – that is, the set of properties that hold for all calls – is preserved, and eventually returned. The special variable `return_value` is set to hold the return value.
###Code
RETURN_VALUE = 'return_value'
class InvariantTracker(InvariantTracker):
def invariants(self, function_name=None):
if function_name is None:
return {function_name: self.invariants(function_name) for function_name in self.calls()}
invariants = None
for variables, return_value in self.calls(function_name):
vars_and_values = variables + [(RETURN_VALUE, return_value)]
s = set()
for prop in self.props:
s |= true_property_instantiations(prop, vars_and_values, self._log)
if invariants is None:
invariants = s
else:
invariants &= s
return invariants
###Output
_____no_output_____
###Markdown
Here's an example of how to use `invariants()`. We run the tracker on a small set of calls.
###Code
with InvariantTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(10.0)
tracker.calls()
###Output
_____no_output_____
###Markdown
The `invariants()` method produces a set of properties that hold for the observed runs, together with their instantiations over function arguments.
###Code
invs = tracker.invariants('my_sqrt')
invs
###Output
_____no_output_____
###Markdown
As before, the actual instantiations are easier to read:
###Code
def pretty_invariants(invariants):
props = []
for (prop, var_names) in invariants:
props.append(instantiate_prop(prop, var_names))
return sorted(props)
pretty_invariants(invs)
###Output
_____no_output_____
###Markdown
We see that the both `x` and the return value have a `float` type. We also see that both are always greater than zero. These are properties that may make useful pre- and postconditions, notably for symbolic analysis. However, there's also an invariant which does _not_ universally hold, namely `return_value <= x`, as the following example shows:
###Code
my_sqrt(0.01)
###Output
_____no_output_____
###Markdown
Clearly, 0.1 > 0.01 holds. This is a case of us not learning from sufficiently diverse inputs. As soon as we have a call including `x = 0.1`, though, the invariant `return_value <= x` is eliminated:
###Code
with InvariantTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(10.0)
y = my_sqrt(0.01)
pretty_invariants(tracker.invariants('my_sqrt'))
###Output
_____no_output_____
###Markdown
We will discuss later how to ensure sufficient diversity in inputs. (Hint: This involves test generation.) Let us try out our invariant tracker on `sum3()`. We see that all types are well-defined; the properties that all arguments are non-zero, however, is specific to the calls observed.
###Code
with InvariantTracker() as tracker:
y = sum3(1, 2, 3)
y = sum3(-4, -5, -6)
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with strings instead, we get different invariants. Notably, we obtain the postcondition that the return value starts with the value of `a` – a universal postcondition if strings are used.
###Code
with InvariantTracker() as tracker:
y = sum3('a', 'b', 'c')
y = sum3('f', 'e', 'd')
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with both strings and numbers (and zeros, too), there are no properties left that would hold across all calls. That's the price of flexibility.
###Code
with InvariantTracker() as tracker:
y = sum3('a', 'b', 'c')
y = sum3('c', 'b', 'a')
y = sum3(-4, -5, -6)
y = sum3(0, 0, 0)
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
Converting Mined Invariants to AnnotationsAs with types, above, we would like to have some functionality where we can add the mined invariants as annotations to existing functions. To this end, we introduce the `InvariantAnnotator` class, extending `InvariantTracker`. We start with a helper method. `params()` returns a comma-separated list of parameter names as observed during calls.
###Code
class InvariantAnnotator(InvariantTracker):
def params(self, function_name):
arguments, return_value = self.calls(function_name)[0]
return ", ".join(arg_name for (arg_name, arg_value) in arguments)
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = sum3(1, 2, 3)
annotator.params('my_sqrt')
annotator.params('sum3')
###Output
_____no_output_____
###Markdown
Now for the actual annotation. `preconditions()` returns the preconditions from the mined invariants (i.e., those propertes that do not depend on the return value) as a string with annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@precondition(lambda " + self.params(function_name) + ": " + inv + ")"
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
annotator.preconditions('my_sqrt')
###Output
_____no_output_____
###Markdown
`postconditions()` does the same for postconditions:
###Code
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ")")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
annotator.postconditions('my_sqrt')
###Output
_____no_output_____
###Markdown
With these, we can take a function and add both pre- and postconditions as annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def functions_with_invariants(self):
functions = ""
for function_name in self.invariants():
try:
function = self.function_with_invariants(function_name)
except KeyError:
continue
functions += function
return functions
def function_with_invariants(self, function_name):
function = globals()[function_name] # Can throw KeyError
source = inspect.getsource(function)
return "\n".join(self.preconditions(function_name) +
self.postconditions(function_name)) + '\n' + source
###Output
_____no_output_____
###Markdown
Here comes `function_with_invariants()` in all its glory:
###Code
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
print_content(annotator.function_with_invariants('my_sqrt'), '.py')
###Output
_____no_output_____
###Markdown
Quite a lot of invariants, is it? Further below (and in the exercises), we will discuss on how to focus on the most relevant properties. Some ExamplesHere's another example. `list_length()` recursively computes the length of a Python function. Let us see whether we can mine its invariants:
###Code
def list_length(L):
if L == []:
length = 0
else:
length = 1 + list_length(L[1:])
return length
with InvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Almost all these properties (except for the very first) are relevant. Of course, the reason the invariants are so neat is that the return value is equal to `len(L)` is that `X == len(Y)` is part of the list of properties to be checked. The next example is a very simple function:
###Code
def sum2(a, b):
return a + b
with InvariantAnnotator() as annotator:
sum2(31, 45)
sum2(0, 0)
sum2(-1, -5)
###Output
_____no_output_____
###Markdown
The invariants all capture the relationship between `a`, `b`, and the return value as `return_value == a + b` in all its variations.
###Code
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
If we have a function without return value, the return value is `None` and we can only mine preconditions. (Well, we get a "postcondition" that the return value is non-zero, which holds for `None`).
###Code
def print_sum(a, b):
print(a + b)
with InvariantAnnotator() as annotator:
print_sum(31, 45)
print_sum(0, 0)
print_sum(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Checking SpecificationsA function with invariants, as above, can be fed into the Python interpreter, such that all pre- and postconditions are checked. We create a function `my_sqrt_annotated()` which includes all the invariants mined above.
###Code
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
my_sqrt_def = annotator.functions_with_invariants()
my_sqrt_def = my_sqrt_def.replace('my_sqrt', 'my_sqrt_annotated')
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def)
###Output
_____no_output_____
###Markdown
The "annotated" version checks against invalid arguments – or more precisely, against arguments with properties that have not been observed yet:
###Code
with ExpectError():
my_sqrt_annotated(-1.0)
###Output
_____no_output_____
###Markdown
This is in contrast to the original version, which just hangs on negative values:
###Code
with ExpectTimeout(1):
my_sqrt(-1.0)
###Output
_____no_output_____
###Markdown
If we make changes to the function definition such that the properties of the return value change, such _regressions_ are caught as violations of the postconditions. Let us illustrate this by simply inverting the result, and return $-2$ as square root of 4.
###Code
my_sqrt_def = my_sqrt_def.replace('my_sqrt_annotated', 'my_sqrt_negative')
my_sqrt_def = my_sqrt_def.replace('return approx', 'return -approx')
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def)
###Output
_____no_output_____
###Markdown
Technically speaking, $-2$ _is_ a square root of 4, since $(-2)^2 = 4$ holds. Yet, such a change may be unexpected by callers of `my_sqrt()`, and hence, this would be caught with the first call:
###Code
with ExpectError():
my_sqrt_negative(2.0)
###Output
_____no_output_____
###Markdown
We see how pre- and postconditions, as well as types, can serve as *oracles* during testing. In particular, once we have mined them for a set of functions, we can check them again and again with test generators – especially after code changes. The more checks we have, and the more specific they are, the more likely it is we can detect unwanted effects of changes. Mining Specifications from Generated TestsMined specifications can only be as good as the executions they were mined from. If we only see a single call to, say, `sum2()`, we will be faced with several mined pre- and postconditions that _overspecialize_ towards the values seen:
###Code
def sum2(a, b):
return a + b
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
The mined precondition `a == b`, for instance, only holds for the single call observed; the same holds for the mined postcondition `return_value == a * b`. Yet, `sum2()` can obviously be successfully called with other values that do not satisfy these conditions. To get out of this trap, we have to _learn from more and more diverse runs_. If we have a few more calls of `sum2()`, we see how the set of invariants quickly gets smaller:
###Code
with InvariantAnnotator() as annotator:
length = sum2(1, 2)
length = sum2(-1, -2)
length = sum2(0, 0)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
But where to we get such diverse runs from? This is the job of generating software tests. A simple grammar for calls of `sum2()` will easily resolve the problem.
###Code
from GrammarFuzzer import GrammarFuzzer # minor dependency
from Grammars import is_valid_grammar, crange, convert_ebnf_grammar # minor dependency
SUM2_EBNF_GRAMMAR = {
"<start>": ["<sum2>"],
"<sum2>": ["sum2(<int>, <int>)"],
"<int>": ["<_int>"],
"<_int>": ["(-)?<leaddigit><digit>*", "0"],
"<leaddigit>": crange('1', '9'),
"<digit>": crange('0', '9')
}
assert is_valid_grammar(SUM2_EBNF_GRAMMAR)
sum2_grammar = convert_ebnf_grammar(SUM2_EBNF_GRAMMAR)
sum2_fuzzer = GrammarFuzzer(sum2_grammar)
[sum2_fuzzer.fuzz() for i in range(10)]
with InvariantAnnotator() as annotator:
for i in range(10):
eval(sum2_fuzzer.fuzz())
print_content(annotator.function_with_invariants('sum2'), '.py')
###Output
_____no_output_____
###Markdown
But then, writing tests (or a test driver) just to derive a set of pre- and postconditions may possibly be too much effort – in particular, since tests can easily be derived from given pre- and postconditions in the first place. Hence, it would be wiser to first specify invariants and then let test generators or program provers do the job. Also, an API grammar, such as above, will have to be set up such that it actually respects preconditions – in our case, we invoke `sqrt()` with positive numbers only, already assuming its precondition. In some way, one thus needs a specification (a model, a grammar) to mine another specification – a chicken-and-egg problem. However, there is one way out of this problem: If one can automatically generate tests at the system level, then one has an _infinite source of executions_ to learn invariants from. In each of these executions, all functions would be called with values that satisfy the (implicit) precondition, allowing us to mine invariants for these functions. This holds, because at the system level, invalid inputs must be rejected by the system in the first place. The meaningful precondition at the system level, ensuring that only valid inputs get through, thus gets broken down into a multitude of meaningful preconditions (and subsequent postconditions) at the function level. The big requirement for this, though, is that one needs good test generators at the system level. In [the next part](05_Domain-Specific_Fuzzing.ipynb), we will discuss how to automatically generate tests for a variety of domains, from configuration to graphical user interfaces. SynopsisThis chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:
###Code
def sum2(a, b):
return a + b
with TypeAnnotator() as type_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.
###Code
print(type_annotator.typed_functions())
###Output
_____no_output_____
###Markdown
The invariant annotator works in a similar fashion:
###Code
with InvariantAnnotator() as inv_annotator:
sum2(1, 2)
sum2(-4, -5)
sum2(0, 0)
###Output
_____no_output_____
###Markdown
The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.
###Code
print(inv_annotator.functions_with_invariants())
###Output
_____no_output_____
###Markdown
Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs) as well as for all kinds of _symbolic code analyses_. The chapter gives details on how to customize the properties checked for. Lessons Learned* Type annotations and explicit invariants allow for _checking_ arguments and results for expected data types and other properties.* One can automatically _mine_ data types and invariants by observing arguments and results at runtime.* The quality of mined invariants depends on the diversity of values observed during executions; this variety can be increased by generating tests. Next StepsThis chapter concludes the [part on semantical fuzzing techniques](04_Semantical_Fuzzing.ipynb). In the next part, we will explore [domain-specific fuzzing techniques](05_Domain-Specific_Fuzzing.ipynb) from configurations and APIs to graphical user interfaces. BackgroundThe [DAIKON dynamic invariant detector](https://plse.cs.washington.edu/daikon/) can be considered the mother of function specification miners. Continuously maintained and extended for more than 20 years, it mines likely invariants in the style of this chapter for a variety of languages, including C, C++, C, Eiffel, F, Java, Perl, and Visual Basic. On top of the functionality discussed above, it holds a rich catalog of patterns for likely invariants, supports data invariants, can eliminate invariants that are implied by others, and determines statistical confidence to disregard unlikely invariants. The corresponding paper \cite{Ernst2001} is one of the seminal and most-cited papers of Software Engineering. A multitude of works have been published based on DAIKON and detecting invariants; see this [curated list](http://plse.cs.washington.edu/daikon/pubs/) for details. The interaction between test generators and invariant detection is already discussed in \cite{Ernst2001} (incidentally also using grammars). The Eclat tool \cite{Pacheco2005} is a model example of tight interaction between a unit-level test generator and DAIKON-style invariant mining, where the mined invariants are used to produce oracles and to systematically guide the test generator towards fault-revealing inputs. Mining specifications is not restricted to pre- and postconditions. The paper "Mining Specifications" \cite{Ammons2002} is another classic in the field, learning state protocols from executions. Grammar mining, as described in [our chapter with the same name](GrammarMiner.ipynb) can also be seen as a specification mining approach, this time learning specifications for input formats. As it comes to adding type annotations to existing code, the blog post ["The state of type hints in Python"](https://www.bernat.tech/the-state-of-type-hints-in-python/) gives a great overview on how Python type hints can be used and checked. To add type annotations, there are two important tools available that also implement our above approach:* [MonkeyType](https://instagram-engineering.com/let-your-code-type-hint-itself-introducing-open-source-monkeytype-a855c7284881) implements the above approach of tracing executions and annotating Python 3 arguments, returns, and variables with type hints.* [PyAnnotate](https://github.com/dropbox/pyannotate) does a similar job, focusing on code in Python 2. It does not produce Python 3-style annotations, but instead produces annotations as comments that can be processed by static type checkers.These tools have been created by engineers at Facebook and Dropbox, respectively, assisting them in checking millions of lines of code for type issues. ExercisesOur code for mining types and invariants is in no way complete. There are dozens of ways to extend our implementations, some of which we discuss in exercises. Exercise 1: Union TypesThe Python `typing` module allows to express that an argument can have multiple types. For `my_sqrt(x)`, this allows to express that `x` can be an `int` or a `float`:
###Code
from typing import Union, Optional
def my_sqrt_with_union_type(x: Union[int, float]) -> float:
...
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it supports union types for arguments and return values. Use `Optional[X]` as a shorthand for `Union[X, None]`. **Solution.** Left to the reader. Hint: extend `type_string()`. Exercise 2: Types for Local VariablesIn Python, one cannot only annotate arguments with types, but actually also local and global variables – for instance, `approx` and `guess` in our `my_sqrt()` implementation:
###Code
def my_sqrt_with_local_types(x: Union[int, float]) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
approx: Optional[float] = None
guess: float = x / 2
while approx != guess:
approx: float = guess
guess: float = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it also annotates local variables with types. Search the function AST for assignments, determine the type of the assigned value, and make it an annotation on the left hand side. **Solution.** Left to the reader. Exercise 3: Verbose Invariant CheckersOur implementation of invariant checkers does not make it clear for the user which pre-/postcondition failed.
###Code
@precondition(lambda s: len(s) > 0)
def remove_first_char(s):
return s[1:]
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
The following implementation adds an optional `doc` keyword argument which is printed if the invariant is violated:
###Code
def condition(precondition=None, postcondition=None, doc='Unknown'):
def decorator(func):
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args, **kwargs):
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated: " + doc
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated: " + doc
return retval
return wrapper
return decorator
def precondition(check, **kwargs):
return condition(precondition=check, doc=kwargs.get('doc', 'Unknown'))
def postcondition(check, **kwargs):
return condition(postcondition=check, doc=kwargs.get('doc', 'Unknown'))
@precondition(lambda s: len(s) > 0, doc="len(s) > 0")
def remove_first_char(s):
return s[1:]
remove_first_char('abc')
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
Extend `InvariantAnnotator` such that it includes the conditions in the generated pre- and postconditions. **Solution.** Here's a simple solution:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@precondition(lambda " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")"
conditions.append(cond)
return conditions
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")")
conditions.append(cond)
return conditions
###Output
_____no_output_____
###Markdown
The resulting annotations are harder to read, but easier to diagnose:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
As an alternative, one may be able to use `inspect.getsource()` on the lambda expression or unparse it. This is left to the reader. Exercise 4: Save Initial ValuesIf the value of an argument changes during function execution, this can easily confuse our implementation: The values are tracked at the beginning of the function, but checked only when it returns. Extend the `InvariantAnnotator` and the infrastructure it uses such that* it saves argument values both at the beginning and at the end of a function invocation;* postconditions can be expressed over both _initial_ values of arguments as well as the _final_ values of arguments;* the mined postconditions refer to both these values as well. **Solution.** To be added. Exercise 5: ImplicationsSeveral mined invariant are actually _implied_ by others: If `x > 0` holds, then this implies `x >= 0` and `x != 0`. Extend the `InvariantAnnotator` such that implications between properties are explicitly encoded, and such that implied properties are no longer listed as invariants. See \cite{Ernst2001} for ideas. **Solution.** Left to the reader. Exercise 6: Local VariablesPostconditions may also refer to the values of local variables. Consider extending `InvariantAnnotator` and its infrastructure such that the values of local variables at the end of the execution are also recorded and made part of the invariant inference mechanism. **Solution.** Left to the reader. Exercise 7: Exploring Invariant AlternativesAfter mining a first set of invariants, have a [concolic fuzzer](ConcolicFuzzer.ipynb) generate tests that systematically attempt to invalidate pre- and postconditions. How far can you generalize? **Solution.** To be added. Exercise 8: Grammar-Generated PropertiesThe larger the set of properties to be checked, the more potential invariants can be discovered. Create a _grammar_ that systematically produces a large set of properties. See \cite{Ernst2001} for possible patterns. **Solution.** Left to the reader. Exercise 9: Embedding Invariants as AssertionsRather than producing invariants as annotations for pre- and postconditions, insert them as `assert` statements into the function code, as in:```pythondef my_sqrt(x): 'Computes the square root of x, using the Newton-Raphson method' assert isinstance(x, int), 'violated precondition' assert (x > 0), 'violated precondition' approx = None guess = (x / 2) while (approx != guess): approx = guess guess = ((approx + (x / approx)) / 2) return_value = approx assert (return_value < x), 'violated postcondition' assert isinstance(return_value, float), 'violated postcondition' return approx```Such a formulation may make it easier for test generators and symbolic analysis to access and interpret pre- and postconditions. **Solution.** Here is a tentative implementation that inserts invariants into function ASTs. Part 1: Embedding Invariants into Functions
###Code
class EmbeddedInvariantAnnotator(InvariantTracker):
def functions_with_invariants_ast(self, function_name=None):
if function_name is None:
return annotate_functions_with_invariants(self.invariants())
return annotate_function_with_invariants(function_name, self.invariants(function_name))
def functions_with_invariants(self, function_name=None):
if function_name is None:
functions = ''
for f_name in self.invariants():
try:
f_text = astor.to_source(self.functions_with_invariants_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
return astor.to_source(self.functions_with_invariants_ast(function_name))
def function_with_invariants(self, function_name):
return self.functions_with_invariants(function_name)
def function_with_invariants_ast(self, function_name):
return self.functions_with_invariants_ast(function_name)
def annotate_invariants(invariants):
annotated_functions = {}
for function_name in invariants:
try:
annotated_functions[function_name] = annotate_function_with_invariants(function_name, invariants[function_name])
except KeyError:
continue
return annotated_functions
def annotate_function_with_invariants(function_name, function_invariants):
function = globals()[function_name]
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_invariants(function_ast, function_invariants)
def annotate_function_ast_with_invariants(function_ast, function_invariants):
annotated_function_ast = EmbeddedInvariantTransformer(function_invariants).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Part 2: Preconditions
###Code
class PreconditionTransformer(ast.NodeTransformer):
def __init__(self, invariants):
self.invariants = invariants
super().__init__()
def preconditions(self):
preconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated precondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) < 0:
preconditions += assertion_ast.body
return preconditions
def insert_assertions(self, body):
preconditions = self.preconditions()
try:
docstring = body[0].value.s
except:
docstring = None
if docstring:
return [body[0]] + preconditions + body[1:]
else:
return preconditions + body
def visit_FunctionDef(self, node):
"""Add invariants to function"""
# print(ast.dump(node))
node.body = self.insert_assertions(node.body)
return node
class EmbeddedInvariantTransformer(PreconditionTransformer):
pass
with EmbeddedInvariantAnnotator() as annotator:
my_sqrt(5)
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Part 3: PostconditionsWe make a few simplifying assumptions: * Variables do not change during execution.* There is a single `return` statement at the end of the function.
###Code
class EmbeddedInvariantTransformer(PreconditionTransformer):
def postconditions(self):
postconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated postcondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) >= 0:
postconditions += assertion_ast.body
return postconditions
def insert_assertions(self, body):
new_body = super().insert_assertions(body)
postconditions = self.postconditions()
body_ends_with_return = isinstance(new_body[-1], ast.Return)
if body_ends_with_return:
saver = RETURN_VALUE + " = " + astor.to_source(new_body[-1].value)
else:
saver = RETURN_VALUE + " = None"
saver_ast = ast.parse(saver)
postconditions = [saver_ast] + postconditions
if body_ends_with_return:
return new_body[:-1] + postconditions + [new_body[-1]]
else:
return new_body + postconditions
with EmbeddedInvariantAnnotator() as annotator:
my_sqrt(5)
my_sqrt_def = annotator.functions_with_invariants()
###Output
_____no_output_____
###Markdown
Here's the full definition with included assertions:
###Code
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def.replace('my_sqrt', 'my_sqrt_annotated'))
with ExpectError():
my_sqrt_annotated(-1)
###Output
_____no_output_____
###Markdown
Here come some more examples:
###Code
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
print_sum(31, 45)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Mining Function SpecificationsWhen testing a program, one not only needs to cover its several behaviors; one also needs to _check_ whether the result is as expected. In this chapter, we introduce a technique that allows us to _mine_ function specifications from a set of given executions, resulting in abstract and formal _descriptions_ of what the function expects and what it delivers. These so-called _dynamic invariants_ produce pre- and post-conditions over function arguments and variables from a set of executions. They are useful in a variety of contexts:* Dynamic invariants provide important information for [symbolic fuzzing](SymbolicFuzzer.ipynb), such as types and ranges of function arguments.* Dynamic invariants provide pre- and postconditions for formal program proofs and verification.* Dynamic invariants provide a large number of assertions that can check whether function behavior has changed* Checks provided by dynamic invariants can be very useful as _oracles_ for checking the effects of generated testsTraditionally, dynamic invariants are dependent on the executions they are derived from. However, when paired with comprehensive test generators, they quickly become very precise, as we show in this chapter. **Prerequisites*** You should be familiar with tracing program executions, as in the [chapter on coverage](Coverage.ipynb).* Later in this section, we access the internal _abstract syntax tree_ representations of Python programs and transform them, as in the [chapter on information flow](InformationFlow.ipynb).
###Code
import bookutils
import Coverage
import Intro_Testing
###Output
_____no_output_____
###Markdown
SynopsisTo [use the code provided in this chapter](Importing.ipynb), write```python>>> from fuzzingbook.DynamicInvariants import ```and then make use of the following features.This chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:```python>>> def sum(a, b):>>> return a + b>>> with TypeAnnotator() as type_annotator:>>> sum(1, 2)>>> sum(-4, -5)>>> sum(0, 0)```The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.```python>>> print(type_annotator.typed_functions())def sum(a: int, b: int) -> int: return a + b```The invariant annotator works in a similar fashion:```python>>> with InvariantAnnotator() as inv_annotator:>>> sum(1, 2)>>> sum(-4, -5)>>> sum(0, 0)```The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.```python>>> print(inv_annotator.functions_with_invariants())@precondition(lambda b, a: isinstance(a, int))@precondition(lambda b, a: isinstance(b, int))@postcondition(lambda return_value, b, a: a == return_value - b)@postcondition(lambda return_value, b, a: b == return_value - a)@postcondition(lambda return_value, b, a: isinstance(return_value, int))@postcondition(lambda return_value, b, a: return_value == a + b)@postcondition(lambda return_value, b, a: return_value == b + a)def sum(a, b): return a + b```Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs) as well as for all kinds of _symbolic code analyses_. The chapter gives details on how to customize the properties checked for. Specifications and AssertionsWhen implementing a function or program, one usually works against a _specification_ – a set of documented requirements to be satisfied by the code. Such specifications can come in natural language. A formal specification, however, allows the computer to check whether the specification is satisfied.In the [introduction to testing](Intro_Testing.ipynb), we have seen how _preconditions_ and _postconditions_ can describe what a function does. Consider the following (simple) square root function:
###Code
def any_sqrt(x):
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
The assertion `assert p` checks the condition `p`; if it does not hold, execution is aborted. Here, the actual body is not yet written; we use the assertions as a specification of what `any_sqrt()` _expects_, and what it _delivers_.The topmost assertion is the _precondition_, stating the requirements on the function arguments. The assertion at the end is the _postcondition_, stating the properties of the function result (including its relationship with the original arguments). Using these pre- and postconditions as a specification, we can now go and implement a square root function that satisfies them. Once implemented, we can have the assertions check at runtime whether `any_sqrt()` works as expected; a [symbolic](SymbolicFuzzer.ipynb) or [concolic](ConcolicFuzzer.ipynb) test generator will even specifically try to find inputs where the assertions do _not_ hold. (An assertion can be seen as a conditional branch towards aborting the execution, and any technique that tries to cover all code branches will also try to invalidate as many assertions as possible.) However, not every piece of code is developed with explicit specifications in the first place; let alone does most code comes with formal pre- and post-conditions. (Just take a look at the chapters in this book.) This is a pity: As Ken Thompson famously said, "Without specifications, there are no bugs – only surprises". It is also a problem for testing, since, of course, testing needs some specification to test against. This raises the interesting question: Can we somehow _retrofit_ existing code with "specifications" that properly describe their behavior, allowing developers to simply _check_ them rather than having to write them from scratch? This is what we do in this chapter. Why Generic Error Checking is Not EnoughBefore we go into _mining_ specifications, let us first discuss why it could be useful to _have_ them. As a motivating example, consider the full implementation of a square root function from the [introduction to testing](Intro_Testing.ipynb):
###Code
import bookutils
def my_sqrt(x):
"""Computes the square root of x, using the Newton-Raphson method"""
approx = None
guess = x / 2
while approx != guess:
approx = guess
guess = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
`my_sqrt()` does not come with any functionality that would check types or values. Hence, it is easy for callers to make mistakes when calling `my_sqrt()`:
###Code
from ExpectError import ExpectError, ExpectTimeout
with ExpectError():
my_sqrt("foo")
with ExpectError():
x = my_sqrt(0.0)
###Output
_____no_output_____
###Markdown
At least, the Python system catches these errors at runtime. The following call, however, simply lets the function enter an infinite loop:
###Code
with ExpectTimeout(1):
x = my_sqrt(-1.0)
###Output
_____no_output_____
###Markdown
Our goal is to avoid such errors by _annotating_ functions with information that prevents errors like the above ones. The idea is to provide a _specification_ of expected properties – a specification that can then be checked at runtime or statically. \todo{Introduce the concept of *contract*.} Specifying and Checking Data TypesFor our Python code, one of the most important "specifications" we need is *types*. Python being a "dynamically" typed language means that all data types are determined at run time; the code itself does not explicitly state whether a variable is an integer, a string, an array, a dictionary – or whatever. As _writer_ of Python code, omitting explicit type declarations may save time (and allows for some fun hacks). It is not sure whether a lack of types helps in _reading_ and _understanding_ code for humans. For a _computer_ trying to analyze code, the lack of explicit types is detrimental. If, say, a constraint solver, sees `if x:` and cannot know whether `x` is supposed to be a number or a string, this introduces an _ambiguity_. Such ambiguities may multiply over the entire analysis in a combinatorial explosion – or in the analysis yielding an overly inaccurate result. Python 3.6 and later allows data types as _annotations_ to function arguments (actually, to all variables) and return values. We can, for instance, state that `my_sqrt()` is a function that accepts a floating-point value and returns one:
###Code
def my_sqrt_with_type_annotations(x: float) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
By default, such annotations are ignored by the Python interpreter. Therefore, one can still call `my_sqrt_typed()` with a string as an argument and get the exact same result as above. However, one can make use of special _typechecking_ modules that would check types – _dynamically_ at runtime or _statically_ by analyzing the code without having to execute it. Excursion: Runtime Type Checking(Commented out as `enforce` is not supported by Python 3.9)The Python `enforce` package provides a function decorator that automatically inserts type-checking code that is executed at runtime. Here is how to use it:
###Code
# import enforce
# @enforce.runtime_validation
# def my_sqrt_with_checked_type_annotations(x: float) -> float:
# """Computes the square root of x, using the Newton-Raphson method"""
# return my_sqrt(x)
###Output
_____no_output_____
###Markdown
Now, invoking `my_sqrt_with_checked_type_annotations()` raises an exception when invoked with a type dfferent from the one declared:
###Code
# with ExpectError():
# my_sqrt_with_checked_type_annotations(True)
###Output
_____no_output_____
###Markdown
Note that this error is not caught by the "untyped" variant, where passing a boolean value happily returns $\sqrt{1}$ as result.
###Code
# my_sqrt(True)
###Output
_____no_output_____
###Markdown
In Python (and other languages), the boolean values `True` and `False` can be implicitly converted to the integers 1 and 0; however, it is hard to think of a call to `sqrt()` where this would not be an error. End of Excursion Static Type CheckingType annotations can also be checked _statically_ – that is, without even running the code. Let us create a simple Python file consisting of the above `my_sqrt_typed()` definition and a bad invocation.
###Code
import inspect
import tempfile
f = tempfile.NamedTemporaryFile(mode='w', suffix='.py')
f.name
f.write(inspect.getsource(my_sqrt))
f.write('\n')
f.write(inspect.getsource(my_sqrt_with_type_annotations))
f.write('\n')
f.write("print(my_sqrt_with_type_annotations('123'))\n")
f.flush()
###Output
_____no_output_____
###Markdown
These are the contents of our newly created Python file:
###Code
from bookutils import print_file
print_file(f.name)
###Output
_____no_output_____
###Markdown
[Mypy](http://mypy-lang.org) is a type checker for Python programs. As it checks types statically, types induce no overhead at runtime; plus, a static check can be faster than a lengthy series of tests with runtime type checking enabled. Let us see what `mypy` produces on the above file:
###Code
import subprocess
result = subprocess.run(["mypy", "--strict", f.name], universal_newlines=True, stdout=subprocess.PIPE)
del f # Delete temporary file
print(result.stdout)
###Output
_____no_output_____
###Markdown
We see that `mypy` complains about untyped function definitions such as `my_sqrt()`; most important, however, it finds that the call to `my_sqrt_with_type_annotations()` in the last line has the wrong type. With `mypy`, we can achieve the same type safety with Python as in statically typed languages – provided that we as programmers also produce the necessary type annotations. Is there a simple way to obtain these? Mining Type SpecificationsOur first task will be to mine type annotations (as part of the code) from _values_ we observe at run time. These type annotations would be _mined_ from actual function executions, _learning_ from (normal) runs what the expected argument and return types should be. By observing a series of calls such as these, we could infer that both `x` and the return value are of type `float`:
###Code
y = my_sqrt(25.0)
y
y = my_sqrt(2.0)
y
###Output
_____no_output_____
###Markdown
How can we mine types from executions? The answer is simple: 1. We _observe_ a function during execution2. We track the _types_ of its arguments3. We include these types as _annotations_ into the code.To do so, we can make use of Python's tracing facility we already observed in the [chapter on coverage](Coverage.ipynb). With every call to a function, we retrieve the arguments, their values, and their types. Tracking CallsTo observe argument types at runtime, we define a _tracer function_ that tracks the execution of `my_sqrt()`, checking its arguments and return values. The `Tracker` class is set to trace functions in a `with` block as follows:```pythonwith Tracker() as tracker: function_to_be_tracked(...)info = tracker.collected_information()```As in the [chapter on coverage](Coverage.ipynb), we use the `sys.settrace()` function to trace individual functions during execution. We turn on tracking when the `with` block starts; at this point, the `__enter__()` method is called. When execution of the `with` block ends, `__exit()__` is called.
###Code
import sys
class Tracker:
def __init__(self, log=False):
self._log = log
self.reset()
def reset(self):
self._calls = {}
self._stack = []
def traceit(self):
"""Placeholder to be overloaded in subclasses"""
pass
# Start of `with` block
def __enter__(self):
self.original_trace_function = sys.gettrace()
sys.settrace(self.traceit)
return self
# End of `with` block
def __exit__(self, exc_type, exc_value, tb):
sys.settrace(self.original_trace_function)
###Output
_____no_output_____
###Markdown
The `traceit()` method does nothing yet; this is done in specialized subclasses. The `CallTracker` class implements a `traceit()` function that checks for function calls and returns:
###Code
class CallTracker(Tracker):
def traceit(self, frame, event, arg):
"""Tracking function: Record all calls and all args"""
if event == "call":
self.trace_call(frame, event, arg)
elif event == "return":
self.trace_return(frame, event, arg)
return self.traceit
###Output
_____no_output_____
###Markdown
`trace_call()` is called when a function is called; it retrieves the function name and current arguments, and saves them on a stack.
###Code
class CallTracker(CallTracker):
def trace_call(self, frame, event, arg):
"""Save current function name and args on the stack"""
code = frame.f_code
function_name = code.co_name
arguments = get_arguments(frame)
self._stack.append((function_name, arguments))
if self._log:
print(simple_call_string(function_name, arguments))
def get_arguments(frame):
"""Return call arguments in the given frame"""
# When called, all arguments are local variables
local_variables = dict(frame.f_locals) # explicit copy
arguments = [(var, frame.f_locals[var]) for var in local_variables]
arguments.reverse() # Want same order as call
return arguments
###Output
_____no_output_____
###Markdown
When the function returns, `trace_return()` is called. We now also have the return value. We log the whole call with arguments and return value (if desired) and save it in our list of calls.
###Code
class CallTracker(CallTracker):
def trace_return(self, frame, event, arg):
"""Get return value and store complete call with arguments and return value"""
code = frame.f_code
function_name = code.co_name
return_value = arg
# TODO: Could call get_arguments() here to also retrieve _final_ values of argument variables
called_function_name, called_arguments = self._stack.pop()
assert function_name == called_function_name
if self._log:
print(simple_call_string(function_name, called_arguments), "returns", return_value)
self.add_call(function_name, called_arguments, return_value)
###Output
_____no_output_____
###Markdown
`simple_call_string()` is a helper for logging that prints out calls in a user-friendly manner.
###Code
def simple_call_string(function_name, argument_list, return_value=None):
"""Return function_name(arg[0], arg[1], ...) as a string"""
call = function_name + "(" + \
", ".join([var + "=" + repr(value)
for (var, value) in argument_list]) + ")"
if return_value is not None:
call += " = " + repr(return_value)
return call
###Output
_____no_output_____
###Markdown
`add_call()` saves the calls in a list; each function name has its own list.
###Code
class CallTracker(CallTracker):
def add_call(self, function_name, arguments, return_value=None):
"""Add given call to list of calls"""
if function_name not in self._calls:
self._calls[function_name] = []
self._calls[function_name].append((arguments, return_value))
###Output
_____no_output_____
###Markdown
Using `calls()`, we can retrieve the list of calls, either for a given function, or for all functions.
###Code
class CallTracker(CallTracker):
def calls(self, function_name=None):
"""Return list of calls for function_name,
or a mapping function_name -> calls for all functions tracked"""
if function_name is None:
return self._calls
return self._calls[function_name]
###Output
_____no_output_____
###Markdown
Let us now put this to use. We turn on logging to track the individual calls and their return values:
###Code
with CallTracker(log=True) as tracker:
y = my_sqrt(25)
y = my_sqrt(2.0)
###Output
_____no_output_____
###Markdown
After execution, we can retrieve the individual calls:
###Code
calls = tracker.calls('my_sqrt')
calls
###Output
_____no_output_____
###Markdown
Each call is pair (`argument_list`, `return_value`), where `argument_list` is a list of pairs (`parameter_name`, `value`).
###Code
my_sqrt_argument_list, my_sqrt_return_value = calls[0]
simple_call_string('my_sqrt', my_sqrt_argument_list, my_sqrt_return_value)
###Output
_____no_output_____
###Markdown
If the function does not return a value, `return_value` is `None`.
###Code
def hello(name):
print("Hello,", name)
with CallTracker() as tracker:
hello("world")
hello_calls = tracker.calls('hello')
hello_calls
hello_argument_list, hello_return_value = hello_calls[0]
simple_call_string('hello', hello_argument_list, hello_return_value)
###Output
_____no_output_____
###Markdown
Getting TypesDespite what you may have read or heard, Python actually _is_ a typed language. It is just that it is _dynamically typed_ – types are used and checked only at runtime (rather than declared in the code, where they can be _statically checked_ at compile time). We can thus retrieve types of all values within Python:
###Code
type(4)
type(2.0)
type([4])
###Output
_____no_output_____
###Markdown
We can retrieve the type of the first argument to `my_sqrt()`:
###Code
parameter, value = my_sqrt_argument_list[0]
parameter, type(value)
###Output
_____no_output_____
###Markdown
as well as the type of the return value:
###Code
type(my_sqrt_return_value)
###Output
_____no_output_____
###Markdown
Hence, we see that (so far), `my_sqrt()` is a function taking (among others) integers and floats and returning floats. We could declare `my_sqrt()` as:
###Code
def my_sqrt_annotated(x: float) -> float:
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
This is a representation we could place in a static type checker, allowing to check whether calls to `my_sqrt()` actually pass a number. A dynamic type checker could run such checks at runtime. And of course, any [symbolic interpretation](SymbolicFuzzer.ipynb) will greatly profit from the additional annotations. By default, Python does not do anything with such annotations. However, tools can access annotations from functions and other objects:
###Code
my_sqrt_annotated.__annotations__
###Output
_____no_output_____
###Markdown
This is how run-time checkers access the annotations to check against. Accessing Function StructureOur plan is to annotate functions automatically, based on the types we have seen. To do so, we need a few modules that allow us to convert a function into a tree representation (called _abstract syntax trees_, or ASTs) and back; we already have seen these in the chapters on [concolic](ConcolicFuzzer.ipynb) and [symbolic](SymbolicFuzzer.ipynb) testing.
###Code
import ast
import inspect
###Output
_____no_output_____
###Markdown
We can get the source of a Python function using `inspect.getsource()`. (Note that this does not work for functions defined in other notebooks.)
###Code
my_sqrt_source = inspect.getsource(my_sqrt)
my_sqrt_source
###Output
_____no_output_____
###Markdown
To view these in a visually pleasing form, our function `print_content(s, suffix)` formats and highlights the string `s` as if it were a file with ending `suffix`. We can thus view (and highlight) the source as if it were a Python file:
###Code
from bookutils import print_content
print_content(my_sqrt_source, '.py')
###Output
_____no_output_____
###Markdown
Parsing this gives us an abstract syntax tree (AST) – a representation of the program in tree form.
###Code
my_sqrt_ast = ast.parse(my_sqrt_source)
###Output
_____no_output_____
###Markdown
What does this AST look like? The helper functions `ast.dump()` (textual output) and `showast.show_ast()` (graphical output with [showast](https://github.com/hchasestevens/show_ast)) allow us to inspect the structure of the tree. We see that the function starts as a `FunctionDef` with name and arguments, followed by a body, which is a list of statements of type `Expr` (the docstring), type `Assign` (assignments), `While` (while loop with its own body), and finally `Return`.
###Code
print(ast.dump(my_sqrt_ast, indent=4))
###Output
_____no_output_____
###Markdown
Too much text for you? This graphical representation may make things simpler.
###Code
from bookutils import rich_output
if rich_output():
import showast
showast.show_ast(my_sqrt_ast)
###Output
_____no_output_____
###Markdown
The function `ast.unparse()` converts such a tree back into the more familiar textual Python code representation. Comments are gone, and there may be more parentheses than before, but the result has the same semantics:
###Code
print_content(ast.unparse(my_sqrt_ast), '.py')
###Output
_____no_output_____
###Markdown
Annotating Functions with Given TypesLet us now go and transform these trees ti add type annotations. We start with a helper function `parse_type(name)` which parses a type name into an AST.
###Code
def parse_type(name):
class ValueVisitor(ast.NodeVisitor):
def visit_Expr(self, node):
self.value_node = node.value
tree = ast.parse(name)
name_visitor = ValueVisitor()
name_visitor.visit(tree)
return name_visitor.value_node
print(ast.dump(parse_type('int')))
print(ast.dump(parse_type('[object]')))
###Output
_____no_output_____
###Markdown
We now define a helper function that actually adds type annotations to a function AST. The `TypeTransformer` class builds on the Python standard library `ast.NodeTransformer` infrastructure. It would be called as```python TypeTransformer({'x': 'int'}, 'float').visit(ast)```to annotate the arguments of `my_sqrt()`: `x` with `int`, and the return type with `float`. The returned AST can then be unparsed, compiled or analyzed.
###Code
class TypeTransformer(ast.NodeTransformer):
def __init__(self, argument_types, return_type=None):
self.argument_types = argument_types
self.return_type = return_type
super().__init__()
###Output
_____no_output_____
###Markdown
The core of `TypeTransformer` is the method `visit_FunctionDef()`, which is called for every function definition in the AST. Its argument `node` is the subtree of the function definition to be transformed. Our implementation accesses the individual arguments and invokes `annotate_args()` on them; it also sets the return type in the `returns` attribute of the node.
###Code
class TypeTransformer(TypeTransformer):
def visit_FunctionDef(self, node):
"""Add annotation to function"""
# Set argument types
new_args = []
for arg in node.args.args:
new_args.append(self.annotate_arg(arg))
new_arguments = ast.arguments(
node.args.posonlyargs,
new_args,
node.args.vararg,
node.args.kwonlyargs,
node.args.kw_defaults,
node.args.kwarg,
node.args.defaults
)
# Set return type
if self.return_type is not None:
node.returns = parse_type(self.return_type)
return ast.copy_location(
ast.FunctionDef(node.name, new_arguments,
node.body, node.decorator_list,
node.returns), node)
###Output
_____no_output_____
###Markdown
Each argument gets its own annotation, taken from the types originally passed to the class:
###Code
class TypeTransformer(TypeTransformer):
def annotate_arg(self, arg):
"""Add annotation to single function argument"""
arg_name = arg.arg
if arg_name in self.argument_types:
arg.annotation = parse_type(self.argument_types[arg_name])
return arg
###Output
_____no_output_____
###Markdown
Does this work? Let us annotate the AST from `my_sqrt()` with types for the arguments and return types:
###Code
new_ast = TypeTransformer({'x': 'int'}, 'float').visit(my_sqrt_ast)
###Output
_____no_output_____
###Markdown
When we unparse the new AST, we see that the annotations actually are present:
###Code
print_content(ast.unparse(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Similarly, we can annotate the `hello()` function from above:
###Code
hello_source = inspect.getsource(hello)
hello_ast = ast.parse(hello_source)
new_ast = TypeTransformer({'name': 'str'}, 'None').visit(hello_ast)
print_content(ast.unparse(new_ast), '.py')
###Output
_____no_output_____
###Markdown
Annotating Functions with Mined TypesLet us now annotate functions with types mined at runtime. We start with a simple function `type_string()` that determines the appropriate type of a given value (as a string):
###Code
def type_string(value):
return type(value).__name__
type_string(4)
type_string([])
###Output
_____no_output_____
###Markdown
For composite structures, `type_string()` does not examine element types; hence, the type of `[3]` is simply `list` instead of, say, `list[int]`. For now, `list` will do fine.
###Code
type_string([3])
###Output
_____no_output_____
###Markdown
`type_string()` will be used to infer the types of argument values found at runtime, as returned by `CallTracker.calls()`:
###Code
with CallTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(2.0)
tracker.calls()
###Output
_____no_output_____
###Markdown
The function `annotate_types()` takes such a list of calls and annotates each function listed:
###Code
def annotate_types(calls):
annotated_functions = {}
for function_name in calls:
try:
annotated_functions[function_name] = annotate_function_with_types(function_name, calls[function_name])
except KeyError:
continue
return annotated_functions
###Output
_____no_output_____
###Markdown
For each function, we get the source and its AST and then get to the actual annotation in `annotate_function_ast_with_types()`:
###Code
def annotate_function_with_types(function_name, function_calls):
function = globals()[function_name] # May raise KeyError for internal functions
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_types(function_ast, function_calls)
###Output
_____no_output_____
###Markdown
The function `annotate_function_ast_with_types()` invokes the `TypeTransformer` with the calls seen, and for each call, iterate over the arguments, determine their types, and annotate the AST with these. The universal type `Any` is used when we encounter type conflicts, which we will discuss below.
###Code
from typing import Any
def annotate_function_ast_with_types(function_ast, function_calls):
parameter_types = {}
return_type = None
for calls_seen in function_calls:
args, return_value = calls_seen
if return_value is not None:
if return_type is not None and return_type != type_string(return_value):
return_type = 'Any'
else:
return_type = type_string(return_value)
for parameter, value in args:
try:
different_type = parameter_types[parameter] != type_string(value)
except KeyError:
different_type = False
if different_type:
parameter_types[parameter] = 'Any'
else:
parameter_types[parameter] = type_string(value)
annotated_function_ast = TypeTransformer(parameter_types, return_type).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Here is `my_sqrt()` annotated with the types recorded usign the tracker, above.
###Code
print_content(ast.unparse(annotate_types(tracker.calls())['my_sqrt']), '.py')
###Output
_____no_output_____
###Markdown
All-in-one AnnotationLet us bring all of this together in a single class `TypeAnnotator` that first tracks calls of functions and then allows to access the AST (and the source code form) of the tracked functions annotated with types. The method `typed_functions()` returns the annotated functions as a string; `typed_functions_ast()` returns their AST.
###Code
class TypeTracker(CallTracker):
pass
class TypeAnnotator(TypeTracker):
def typed_functions_ast(self, function_name=None):
if function_name is None:
return annotate_types(self.calls())
return annotate_function_with_types(function_name,
self.calls(function_name))
def typed_functions(self, function_name=None):
if function_name is None:
functions = ''
for f_name in self.calls():
try:
f_text = ast.unparse(self.typed_functions_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
return ast.unparse(self.typed_functions_ast(function_name))
###Output
_____no_output_____
###Markdown
Here is how to use `TypeAnnotator`. We first track a series of calls:
###Code
with TypeAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(2.0)
###Output
_____no_output_____
###Markdown
After tracking, we can immediately retrieve an annotated version of the functions tracked:
###Code
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
This also works for multiple and diverse functions. One could go and implement an automatic type annotator for Python files based on the types seen during execution.
###Code
with TypeAnnotator() as annotator:
hello('type annotations')
y = my_sqrt(1.0)
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
A content as above could now be sent to a type checker, which would detect any type inconsistency between callers and callees. Likewise, type annotations such as the ones above greatly benefit symbolic code analysis (as in the chapter on [symbolic fuzzing](SymbolicFuzzer.ipynb)), as they effectively constrain the set of values that arguments and variables can take. Multiple TypesLet us now resolve the role of the magic `Any` type in `annotate_function_ast_with_types()`. If we see multiple types for the same argument, we set its type to `Any`. For `my_sqrt()`, this makes sense, as its arguments can be integers as well as floats:
###Code
with CallTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(4)
print_content(ast.unparse(annotate_types(tracker.calls())['my_sqrt']), '.py')
###Output
_____no_output_____
###Markdown
The following function `sum3()` can be called with floating-point numbers as arguments, resulting in the parameters getting a `float` type:
###Code
def sum3(a, b, c):
return a + b + c
with TypeAnnotator() as annotator:
y = sum3(1.0, 2.0, 3.0)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we call `sum3()` with integers, though, the arguments get an `int` type:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
And we can also call `sum3()` with strings, giving the arguments a `str` type:
###Code
with TypeAnnotator() as annotator:
y = sum3("one", "two", "three")
y
print_content(annotator.typed_functions(), '.py')
###Output
_____no_output_____
###Markdown
If we have multiple calls, but with different types, `TypeAnnotator()` will assign an `Any` type to both arguments and return values:
###Code
with TypeAnnotator() as annotator:
y = sum3(1, 2, 3)
y = sum3("one", "two", "three")
typed_sum3_def = annotator.typed_functions('sum3')
print_content(typed_sum3_def, '.py')
###Output
_____no_output_____
###Markdown
A type `Any` makes it explicit that an object can, indeed, have any type; it will not be typechecked at runtime or statically. To some extent, this defeats the power of type checking; but it also preserves some of the type flexibility that many Python programmers enjoy. Besides `Any`, the `typing` module supports several additional ways to define ambiguous types; we will keep this in mind for a later exercise. Specifying and Checking InvariantsBesides basic data types. we can check several further properties from arguments. We can, for instance, whether an argument can be negative, zero, or positive; or that one argument should be smaller than the second; or that the result should be the sum of two arguments – properties that cannot be expressed in a (Python) type.Such properties are called *invariants*, as they hold across all invocations of a function. Specifically, invariants come as _pre_- and _postconditions_ – conditions that always hold at the beginning and at the end of a function. (There are also _data_ and _object_ invariants that express always-holding properties over the state of data or objects, but we do not consider these in this book.) Annotating Functions with Pre- and PostconditionsThe classical means to specify pre- and postconditions is via _assertions_, which we have introduced in the [chapter on testing](Intro_Testing.ipynb). A precondition checks whether the arguments to a function satisfy the expected properties; a postcondition does the same for the result. We can express and check both using assertions as follows:
###Code
def my_sqrt_with_invariants(x):
assert x >= 0 # Precondition
...
assert result * result == x # Postcondition
return result
###Output
_____no_output_____
###Markdown
A nicer way, however, is to syntactically separate invariants from the function at hand. Using appropriate decorators, we could specify pre- and postconditions as follows:```python@precondition lambda x: x >= 0@postcondition lambda return_value, x: return_value * return_value == xdef my_sqrt_with_invariants(x): normal code without assertions ...```The decorators `@precondition` and `@postcondition` would run the given functions (specified as anonymous `lambda` functions) before and after the decorated function, respectively. If the functions return `False`, the condition is violated. `@precondition` gets the function arguments as arguments; `@postcondition` additionally gets the return value as first argument. It turns out that implementing such decorators is not hard at all. Our implementation builds on a [code snippet from StackOverflow](https://stackoverflow.com/questions/12151182/python-precondition-postcondition-for-member-function-how):
###Code
import functools
def condition(precondition=None, postcondition=None):
def decorator(func):
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args, **kwargs):
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated"
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated"
return retval
return wrapper
return decorator
def precondition(check):
return condition(precondition=check)
def postcondition(check):
return condition(postcondition=check)
###Output
_____no_output_____
###Markdown
With these, we can now start decorating `my_sqrt()`:
###Code
@precondition(lambda x: x > 0)
def my_sqrt_with_precondition(x):
return my_sqrt(x)
###Output
_____no_output_____
###Markdown
This catches arguments violating the precondition:
###Code
with ExpectError():
my_sqrt_with_precondition(-1.0)
###Output
_____no_output_____
###Markdown
Likewise, we can provide a postcondition:
###Code
EPSILON = 1e-5
@postcondition(lambda ret, x: ret * ret - x < EPSILON)
def my_sqrt_with_postcondition(x):
return my_sqrt(x)
y = my_sqrt_with_postcondition(2.0)
y
###Output
_____no_output_____
###Markdown
If we have a buggy implementation of $\sqrt{x}$, this gets caught quickly:
###Code
@postcondition(lambda ret, x: ret * ret - x < EPSILON)
def buggy_my_sqrt_with_postcondition(x):
return my_sqrt(x) + 0.1
with ExpectError():
y = buggy_my_sqrt_with_postcondition(2.0)
###Output
_____no_output_____
###Markdown
While checking pre- and postconditions is a great way to catch errors, specifying them can be cumbersome. Let us try to see whether we can (again) _mine_ some of them. Mining InvariantsTo _mine_ invariants, we can use the same tracking functionality as before; instead of saving values for individual variables, though, we now check whether the values satisfy specific _properties_ or not. For instance, if all values of `x` seen satisfy the condition `x > 0`, then we make `x > 0` an invariant of the function. If we see positive, zero, and negative values of `x`, though, then there is no property of `x` left to talk about.The general idea is thus:1. Check all variable values observed against a set of predefined properties; and2. Keep only those properties that hold for all runs observed. Defining PropertiesWhat precisely do we mean by properties? Here is a small collection of value properties that would frequently be used in invariants. All these properties would be evaluated with the _metavariables_ `X`, `Y`, and `Z` (actually, any upper-case identifier) being replaced with the names of function parameters:
###Code
INVARIANT_PROPERTIES = [
"X < 0",
"X <= 0",
"X > 0",
"X >= 0",
"X == 0",
"X != 0",
]
###Output
_____no_output_____
###Markdown
When `my_sqrt(x)` is called as, say `my_sqrt(5.0)`, we see that `x = 5.0` holds. The above properties would then all be checked for `x`. Only the properties `X > 0`, `X >= 0`, and `X != 0` hold for the call seen; and hence `x > 0`, `x >= 0`, and `x != 0` would make potential preconditions for `my_sqrt(x)`. We can check for many more properties such as relations between two arguments:
###Code
INVARIANT_PROPERTIES += [
"X == Y",
"X > Y",
"X < Y",
"X >= Y",
"X <= Y",
]
###Output
_____no_output_____
###Markdown
Types also can be checked using properties. For any function parameter `X`, only one of these will hold:
###Code
INVARIANT_PROPERTIES += [
"isinstance(X, bool)",
"isinstance(X, int)",
"isinstance(X, float)",
"isinstance(X, list)",
"isinstance(X, dict)",
]
###Output
_____no_output_____
###Markdown
We can check for arithmetic properties:
###Code
INVARIANT_PROPERTIES += [
"X == Y + Z",
"X == Y * Z",
"X == Y - Z",
"X == Y / Z",
]
###Output
_____no_output_____
###Markdown
Here's relations over three values, a Python special:
###Code
INVARIANT_PROPERTIES += [
"X < Y < Z",
"X <= Y <= Z",
"X > Y > Z",
"X >= Y >= Z",
]
###Output
_____no_output_____
###Markdown
Finally, we can also check for list or string properties. Again, this is just a tiny selection.
###Code
INVARIANT_PROPERTIES += [
"X == len(Y)",
"X == sum(Y)",
"X.startswith(Y)",
]
###Output
_____no_output_____
###Markdown
Extracting Meta-VariablesLet us first introduce a few _helper functions_ before we can get to the actual mining. `metavars()` extracts the set of meta-variables (`X`, `Y`, `Z`, etc.) from a property. To this end, we parse the property as a Python expression and then visit the identifiers.
###Code
def metavars(prop):
metavar_list = []
class ArgVisitor(ast.NodeVisitor):
def visit_Name(self, node):
if node.id.isupper():
metavar_list.append(node.id)
ArgVisitor().visit(ast.parse(prop))
return metavar_list
assert metavars("X < 0") == ['X']
assert metavars("X.startswith(Y)") == ['X', 'Y']
assert metavars("isinstance(X, str)") == ['X']
###Output
_____no_output_____
###Markdown
Instantiating PropertiesTo produce a property as invariant, we need to be able to _instantiate_ it with variable names. The instantiation of `X > 0` with `X` being instantiated to `a`, for instance, gets us `a > 0`. To this end, the function `instantiate_prop()` takes a property and a collection of variable names and instantiates the meta-variables left-to-right with the corresponding variables names in the collection.
###Code
def instantiate_prop_ast(prop, var_names):
class NameTransformer(ast.NodeTransformer):
def visit_Name(self, node):
if node.id not in mapping:
return node
return ast.Name(id=mapping[node.id], ctx=ast.Load())
meta_variables = metavars(prop)
assert len(meta_variables) == len(var_names)
mapping = {}
for i in range(0, len(meta_variables)):
mapping[meta_variables[i]] = var_names[i]
prop_ast = ast.parse(prop, mode='eval')
new_ast = NameTransformer().visit(prop_ast)
return new_ast
def instantiate_prop(prop, var_names):
prop_ast = instantiate_prop_ast(prop, var_names)
prop_text = ast.unparse(prop_ast).strip()
while prop_text.startswith('(') and prop_text.endswith(')'):
prop_text = prop_text[1:-1]
return prop_text
assert instantiate_prop("X > Y", ['a', 'b']) == 'a > b'
assert instantiate_prop("X.startswith(Y)", ['x', 'y']) == 'x.startswith(y)'
###Output
_____no_output_____
###Markdown
Evaluating PropertiesTo actually _evaluate_ properties, we do not need to instantiate them. Instead, we simply convert them into a boolean function, using `lambda`:
###Code
def prop_function_text(prop):
return "lambda " + ", ".join(metavars(prop)) + ": " + prop
def prop_function(prop):
return eval(prop_function_text(prop))
###Output
_____no_output_____
###Markdown
Here is a simple example:
###Code
prop_function_text("X > Y")
p = prop_function("X > Y")
p(100, 1)
p(1, 100)
###Output
_____no_output_____
###Markdown
Checking InvariantsTo extract invariants from an execution, we need to check them on all possible instantiations of arguments. If the function to be checked has two arguments `a` and `b`, we instantiate the property `X < Y` both as `a < b` and `b < a` and check each of them. To get all combinations, we use the Python `permutations()` function:
###Code
import itertools
for combination in itertools.permutations([1.0, 2.0, 3.0], 2):
print(combination)
###Output
_____no_output_____
###Markdown
The function `true_property_instantiations()` takes a property and a list of tuples (`var_name`, `value`). It then produces all instantiations of the property with the given values and returns those that evaluate to True.
###Code
def true_property_instantiations(prop, vars_and_values, log=False):
instantiations = set()
p = prop_function(prop)
len_metavars = len(metavars(prop))
for combination in itertools.permutations(vars_and_values, len_metavars):
args = [value for var_name, value in combination]
var_names = [var_name for var_name, value in combination]
try:
result = p(*args)
except:
result = None
if log:
print(prop, combination, result)
if result:
instantiations.add((prop, tuple(var_names)))
return instantiations
###Output
_____no_output_____
###Markdown
Here is an example. If `x == -1` and `y == 1`, the property `X < Y` holds for `x < y`, but not for `y < x`:
###Code
invs = true_property_instantiations("X < Y", [('x', -1), ('y', 1)], log=True)
invs
###Output
_____no_output_____
###Markdown
The instantiation retrieves the short form:
###Code
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Likewise, with values for `x` and `y` as above, the property `X < 0` only holds for `x`, but not for `y`:
###Code
invs = true_property_instantiations("X < 0", [('x', -1), ('y', 1)], log=True)
for prop, var_names in invs:
print(instantiate_prop(prop, var_names))
###Output
_____no_output_____
###Markdown
Extracting InvariantsLet us now run the above invariant extraction on function arguments and return values as observed during a function execution. To this end, we extend the `CallTracker` class into an `InvariantTracker` class, which automatically computes invariants for all functions and all calls observed during tracking. By default, an `InvariantTracker` uses the properties as defined above; however, one can specify alternate sets of properties.
###Code
class InvariantTracker(CallTracker):
def __init__(self, props=None, **kwargs):
if props is None:
props = INVARIANT_PROPERTIES
self.props = props
super().__init__(**kwargs)
###Output
_____no_output_____
###Markdown
The key method of the `InvariantTracker` is the `invariants()` method. This iterates over the calls observed and checks which properties hold. Only the intersection of properties – that is, the set of properties that hold for all calls – is preserved, and eventually returned. The special variable `return_value` is set to hold the return value.
###Code
RETURN_VALUE = 'return_value'
class InvariantTracker(InvariantTracker):
def invariants(self, function_name=None):
if function_name is None:
return {function_name: self.invariants(function_name) for function_name in self.calls()}
invariants = None
for variables, return_value in self.calls(function_name):
vars_and_values = variables + [(RETURN_VALUE, return_value)]
s = set()
for prop in self.props:
s |= true_property_instantiations(prop, vars_and_values, self._log)
if invariants is None:
invariants = s
else:
invariants &= s
return invariants
###Output
_____no_output_____
###Markdown
Here's an example of how to use `invariants()`. We run the tracker on a small set of calls.
###Code
with InvariantTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(10.0)
tracker.calls()
###Output
_____no_output_____
###Markdown
The `invariants()` method produces a set of properties that hold for the observed runs, together with their instantiations over function arguments.
###Code
invs = tracker.invariants('my_sqrt')
invs
###Output
_____no_output_____
###Markdown
As before, the actual instantiations are easier to read:
###Code
def pretty_invariants(invariants):
props = []
for (prop, var_names) in invariants:
props.append(instantiate_prop(prop, var_names))
return sorted(props)
pretty_invariants(invs)
###Output
_____no_output_____
###Markdown
We see that the both `x` and the return value have a `float` type. We also see that both are always greater than zero. These are properties that may make useful pre- and postconditions, notably for symbolic analysis. However, there's also an invariant which does _not_ universally hold, namely `return_value <= x`, as the following example shows:
###Code
my_sqrt(0.01)
###Output
_____no_output_____
###Markdown
Clearly, 0.1 > 0.01 holds. This is a case of us not learning from sufficiently diverse inputs. As soon as we have a call including `x = 0.1`, though, the invariant `return_value <= x` is eliminated:
###Code
with InvariantTracker() as tracker:
y = my_sqrt(25.0)
y = my_sqrt(10.0)
y = my_sqrt(0.01)
pretty_invariants(tracker.invariants('my_sqrt'))
###Output
_____no_output_____
###Markdown
We will discuss later how to ensure sufficient diversity in inputs. (Hint: This involves test generation.) Let us try out our invariant tracker on `sum3()`. We see that all types are well-defined; the properties that all arguments are non-zero, however, is specific to the calls observed.
###Code
with InvariantTracker() as tracker:
y = sum3(1, 2, 3)
y = sum3(-4, -5, -6)
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with strings instead, we get different invariants. Notably, we obtain the postcondition that the return value starts with the value of `a` – a universal postcondition if strings are used.
###Code
with InvariantTracker() as tracker:
y = sum3('a', 'b', 'c')
y = sum3('f', 'e', 'd')
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
If we invoke `sum3()` with both strings and numbers (and zeros, too), there are no properties left that would hold across all calls. That's the price of flexibility.
###Code
with InvariantTracker() as tracker:
y = sum3('a', 'b', 'c')
y = sum3('c', 'b', 'a')
y = sum3(-4, -5, -6)
y = sum3(0, 0, 0)
pretty_invariants(tracker.invariants('sum3'))
###Output
_____no_output_____
###Markdown
Converting Mined Invariants to AnnotationsAs with types, above, we would like to have some functionality where we can add the mined invariants as annotations to existing functions. To this end, we introduce the `InvariantAnnotator` class, extending `InvariantTracker`. We start with a helper method. `params()` returns a comma-separated list of parameter names as observed during calls.
###Code
class InvariantAnnotator(InvariantTracker):
def params(self, function_name):
arguments, return_value = self.calls(function_name)[0]
return ", ".join(arg_name for (arg_name, arg_value) in arguments)
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = sum3(1, 2, 3)
annotator.params('my_sqrt')
annotator.params('sum3')
###Output
_____no_output_____
###Markdown
Now for the actual annotation. `preconditions()` returns the preconditions from the mined invariants (i.e., those propertes that do not depend on the return value) as a string with annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@precondition(lambda " + self.params(function_name) + ": " + inv + ")"
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
annotator.preconditions('my_sqrt')
###Output
_____no_output_____
###Markdown
`postconditions()` does the same for postconditions:
###Code
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ")")
conditions.append(cond)
return conditions
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
annotator.postconditions('my_sqrt')
###Output
_____no_output_____
###Markdown
With these, we can take a function and add both pre- and postconditions as annotations:
###Code
class InvariantAnnotator(InvariantAnnotator):
def functions_with_invariants(self):
functions = ""
for function_name in self.invariants():
try:
function = self.function_with_invariants(function_name)
except KeyError:
continue
functions += function
return functions
def function_with_invariants(self, function_name):
function = globals()[function_name] # Can throw KeyError
source = inspect.getsource(function)
return "\n".join(self.preconditions(function_name) +
self.postconditions(function_name)) + '\n' + source
###Output
_____no_output_____
###Markdown
Here comes `function_with_invariants()` in all its glory:
###Code
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
y = sum3(1, 2, 3)
print_content(annotator.function_with_invariants('my_sqrt'), '.py')
###Output
_____no_output_____
###Markdown
Quite a lot of invariants, is it? Further below (and in the exercises), we will discuss on how to focus on the most relevant properties. Some ExamplesHere's another example. `list_length()` recursively computes the length of a Python function. Let us see whether we can mine its invariants:
###Code
def list_length(L):
if L == []:
length = 0
else:
length = 1 + list_length(L[1:])
return length
with InvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Almost all these properties (except for the very first) are relevant. Of course, the reason the invariants are so neat is that the return value is equal to `len(L)` is that `X == len(Y)` is part of the list of properties to be checked. The next example is a very simple function:
###Code
def sum2(a, b):
return a + b
with InvariantAnnotator() as annotator:
sum2(31, 45)
sum2(0, 0)
sum2(-1, -5)
###Output
_____no_output_____
###Markdown
The invariants all capture the relationship between `a`, `b`, and the return value as `return_value == a + b` in all its variations.
###Code
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
If we have a function without return value, the return value is `None` and we can only mine preconditions. (Well, we get a "postcondition" that the return value is non-zero, which holds for `None`).
###Code
def print_sum(a, b):
print(a + b)
with InvariantAnnotator() as annotator:
print_sum(31, 45)
print_sum(0, 0)
print_sum(-1, -5)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Checking SpecificationsA function with invariants, as above, can be fed into the Python interpreter, such that all pre- and postconditions are checked. We create a function `my_sqrt_annotated()` which includes all the invariants mined above.
###Code
with InvariantAnnotator() as annotator:
y = my_sqrt(25.0)
y = my_sqrt(0.01)
my_sqrt_def = annotator.functions_with_invariants()
my_sqrt_def = my_sqrt_def.replace('my_sqrt', 'my_sqrt_annotated')
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def)
###Output
_____no_output_____
###Markdown
The "annotated" version checks against invalid arguments – or more precisely, against arguments with properties that have not been observed yet:
###Code
with ExpectError():
my_sqrt_annotated(-1.0)
###Output
_____no_output_____
###Markdown
This is in contrast to the original version, which just hangs on negative values:
###Code
with ExpectTimeout(1):
my_sqrt(-1.0)
###Output
_____no_output_____
###Markdown
If we make changes to the function definition such that the properties of the return value change, such _regressions_ are caught as violations of the postconditions. Let us illustrate this by simply inverting the result, and return $-2$ as square root of 4.
###Code
my_sqrt_def = my_sqrt_def.replace('my_sqrt_annotated', 'my_sqrt_negative')
my_sqrt_def = my_sqrt_def.replace('return approx', 'return -approx')
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def)
###Output
_____no_output_____
###Markdown
Technically speaking, $-2$ _is_ a square root of 4, since $(-2)^2 = 4$ holds. Yet, such a change may be unexpected by callers of `my_sqrt()`, and hence, this would be caught with the first call:
###Code
with ExpectError():
my_sqrt_negative(2.0) # type: ignore
###Output
_____no_output_____
###Markdown
We see how pre- and postconditions, as well as types, can serve as *oracles* during testing. In particular, once we have mined them for a set of functions, we can check them again and again with test generators – especially after code changes. The more checks we have, and the more specific they are, the more likely it is we can detect unwanted effects of changes. Mining Specifications from Generated TestsMined specifications can only be as good as the executions they were mined from. If we only see a single call to, say, `sum2()` as defined above, we will be faced with several mined pre- and postconditions that _overspecialize_ towards the values seen:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
The mined precondition `a == b`, for instance, only holds for the single call observed; the same holds for the mined postcondition `return_value == a * b`. Yet, `sum2()` can obviously be successfully called with other values that do not satisfy these conditions. To get out of this trap, we have to _learn from more and more diverse runs_. If we have a few more calls of `sum2()`, we see how the set of invariants quickly gets smaller:
###Code
with InvariantAnnotator() as annotator:
length = sum2(1, 2)
length = sum2(-1, -2)
length = sum2(0, 0)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
But where to we get such diverse runs from? This is the job of generating software tests. A simple grammar for calls of `sum2()` will easily resolve the problem.
###Code
from GrammarFuzzer import GrammarFuzzer # minor dependency
from Grammars import is_valid_grammar, crange # minor dependency
from Grammars import convert_ebnf_grammar, Grammar # minor dependency
SUM2_EBNF_GRAMMAR: Grammar = {
"<start>": ["<sum2>"],
"<sum2>": ["sum2(<int>, <int>)"],
"<int>": ["<_int>"],
"<_int>": ["(-)?<leaddigit><digit>*", "0"],
"<leaddigit>": crange('1', '9'),
"<digit>": crange('0', '9')
}
assert is_valid_grammar(SUM2_EBNF_GRAMMAR)
sum2_grammar = convert_ebnf_grammar(SUM2_EBNF_GRAMMAR)
sum2_fuzzer = GrammarFuzzer(sum2_grammar)
[sum2_fuzzer.fuzz() for i in range(10)]
with InvariantAnnotator() as annotator:
for i in range(10):
eval(sum2_fuzzer.fuzz())
print_content(annotator.function_with_invariants('sum2'), '.py')
###Output
_____no_output_____
###Markdown
But then, writing tests (or a test driver) just to derive a set of pre- and postconditions may possibly be too much effort – in particular, since tests can easily be derived from given pre- and postconditions in the first place. Hence, it would be wiser to first specify invariants and then let test generators or program provers do the job. Also, an API grammar, such as above, will have to be set up such that it actually respects preconditions – in our case, we invoke `sqrt()` with positive numbers only, already assuming its precondition. In some way, one thus needs a specification (a model, a grammar) to mine another specification – a chicken-and-egg problem. However, there is one way out of this problem: If one can automatically generate tests at the system level, then one has an _infinite source of executions_ to learn invariants from. In each of these executions, all functions would be called with values that satisfy the (implicit) precondition, allowing us to mine invariants for these functions. This holds, because at the system level, invalid inputs must be rejected by the system in the first place. The meaningful precondition at the system level, ensuring that only valid inputs get through, thus gets broken down into a multitude of meaningful preconditions (and subsequent postconditions) at the function level. The big requirement for this, though, is that one needs good test generators at the system level. In [the next part](05_Domain-Specific_Fuzzing.ipynb), we will discuss how to automatically generate tests for a variety of domains, from configuration to graphical user interfaces. SynopsisThis chapter provides two classes that automatically extract specifications from a function and a set of inputs:* `TypeAnnotator` for _types_, and* `InvariantAnnotator` for _pre-_ and _postconditions_.Both work by _observing_ a function and its invocations within a `with` clause. Here is an example for the type annotator:
###Code
def sum(a, b):
return a + b
with TypeAnnotator() as type_annotator:
sum(1, 2)
sum(-4, -5)
sum(0, 0)
###Output
_____no_output_____
###Markdown
The `typed_functions()` method will return a representation of `sum2()` annotated with types observed during execution.
###Code
print(type_annotator.typed_functions())
###Output
_____no_output_____
###Markdown
The invariant annotator works in a similar fashion:
###Code
with InvariantAnnotator() as inv_annotator:
sum(1, 2)
sum(-4, -5)
sum(0, 0)
###Output
_____no_output_____
###Markdown
The `functions_with_invariants()` method will return a representation of `sum2()` annotated with inferred pre- and postconditions that all hold for the observed values.
###Code
print(inv_annotator.functions_with_invariants())
###Output
_____no_output_____
###Markdown
Such type specifications and invariants can be helpful as _oracles_ (to detect deviations from a given set of runs) as well as for all kinds of _symbolic code analyses_. The chapter gives details on how to customize the properties checked for. Lessons Learned* Type annotations and explicit invariants allow for _checking_ arguments and results for expected data types and other properties.* One can automatically _mine_ data types and invariants by observing arguments and results at runtime.* The quality of mined invariants depends on the diversity of values observed during executions; this variety can be increased by generating tests. Next StepsThis chapter concludes the [part on semantical fuzzing techniques](04_Semantical_Fuzzing.ipynb). In the next part, we will explore [domain-specific fuzzing techniques](05_Domain-Specific_Fuzzing.ipynb) from configurations and APIs to graphical user interfaces. BackgroundThe [DAIKON dynamic invariant detector](https://plse.cs.washington.edu/daikon/) can be considered the mother of function specification miners. Continuously maintained and extended for more than 20 years, it mines likely invariants in the style of this chapter for a variety of languages, including C, C++, C, Eiffel, F, Java, Perl, and Visual Basic. On top of the functionality discussed above, it holds a rich catalog of patterns for likely invariants, supports data invariants, can eliminate invariants that are implied by others, and determines statistical confidence to disregard unlikely invariants. The corresponding paper \cite{Ernst2001} is one of the seminal and most-cited papers of Software Engineering. A multitude of works have been published based on DAIKON and detecting invariants; see this [curated list](http://plse.cs.washington.edu/daikon/pubs/) for details. The interaction between test generators and invariant detection is already discussed in \cite{Ernst2001} (incidentally also using grammars). The Eclat tool \cite{Pacheco2005} is a model example of tight interaction between a unit-level test generator and DAIKON-style invariant mining, where the mined invariants are used to produce oracles and to systematically guide the test generator towards fault-revealing inputs. Mining specifications is not restricted to pre- and postconditions. The paper "Mining Specifications" \cite{Ammons2002} is another classic in the field, learning state protocols from executions. Grammar mining, as described in [our chapter with the same name](GrammarMiner.ipynb) can also be seen as a specification mining approach, this time learning specifications for input formats. As it comes to adding type annotations to existing code, the blog post ["The state of type hints in Python"](https://www.bernat.tech/the-state-of-type-hints-in-python/) gives a great overview on how Python type hints can be used and checked. To add type annotations, there are two important tools available that also implement our above approach:* [MonkeyType](https://instagram-engineering.com/let-your-code-type-hint-itself-introducing-open-source-monkeytype-a855c7284881) implements the above approach of tracing executions and annotating Python 3 arguments, returns, and variables with type hints.* [PyAnnotate](https://github.com/dropbox/pyannotate) does a similar job, focusing on code in Python 2. It does not produce Python 3-style annotations, but instead produces annotations as comments that can be processed by static type checkers.These tools have been created by engineers at Facebook and Dropbox, respectively, assisting them in checking millions of lines of code for type issues. ExercisesOur code for mining types and invariants is in no way complete. There are dozens of ways to extend our implementations, some of which we discuss in exercises. Exercise 1: Union TypesThe Python `typing` module allows to express that an argument can have multiple types. For `my_sqrt(x)`, this allows to express that `x` can be an `int` or a `float`:
###Code
from typing import Union, Optional
def my_sqrt_with_union_type(x: Union[int, float]) -> float:
...
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it supports union types for arguments and return values. Use `Optional[X]` as a shorthand for `Union[X, None]`. **Solution.** Left to the reader. Hint: extend `type_string()`. Exercise 2: Types for Local VariablesIn Python, one cannot only annotate arguments with types, but actually also local and global variables – for instance, `approx` and `guess` in our `my_sqrt()` implementation:
###Code
def my_sqrt_with_local_types(x: Union[int, float]) -> float:
"""Computes the square root of x, using the Newton-Raphson method"""
approx: Optional[float] = None
guess: float = x / 2
while approx != guess:
approx = guess
guess = (approx + x / approx) / 2
return approx
###Output
_____no_output_____
###Markdown
Extend the `TypeAnnotator` such that it also annotates local variables with types. Search the function AST for assignments, determine the type of the assigned value, and make it an annotation on the left hand side. **Solution.** Left to the reader. Exercise 3: Verbose Invariant CheckersOur implementation of invariant checkers does not make it clear for the user which pre-/postcondition failed.
###Code
@precondition(lambda s: len(s) > 0)
def remove_first_char(s):
return s[1:]
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
The following implementation adds an optional `doc` keyword argument which is printed if the invariant is violated:
###Code
def verbose_condition(precondition=None, postcondition=None, doc='Unknown'):
def decorator(func):
@functools.wraps(func) # preserves name, docstring, etc
def wrapper(*args, **kwargs):
if precondition is not None:
assert precondition(*args, **kwargs), "Precondition violated: " + doc
retval = func(*args, **kwargs) # call original function or method
if postcondition is not None:
assert postcondition(retval, *args, **kwargs), "Postcondition violated: " + doc
return retval
return wrapper
return decorator
def verbose_precondition(check, **kwargs): # type: ignore
return verbose_condition(precondition=check, doc=kwargs.get('doc', 'Unknown'))
def verbose_postcondition(check, **kwargs): # type: ignore
return verbose_condition(postcondition=check, doc=kwargs.get('doc', 'Unknown'))
@verbose_precondition(lambda s: len(s) > 0, doc="len(s) > 0") # type: ignore
def remove_first_char(s):
return s[1:]
remove_first_char('abc')
with ExpectError():
remove_first_char('')
###Output
_____no_output_____
###Markdown
Extend `InvariantAnnotator` such that it includes the conditions in the generated pre- and postconditions. **Solution.** Here's a simple solution:
###Code
class InvariantAnnotator(InvariantAnnotator):
def preconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) >= 0:
continue # Postcondition
cond = "@verbose_precondition(lambda " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")"
conditions.append(cond)
return conditions
class InvariantAnnotator(InvariantAnnotator):
def postconditions(self, function_name):
conditions = []
for inv in pretty_invariants(self.invariants(function_name)):
if inv.find(RETURN_VALUE) < 0:
continue # Precondition
cond = ("@verbose_postcondition(lambda " +
RETURN_VALUE + ", " + self.params(function_name) + ": " + inv + ', doc=' + repr(inv) + ")")
conditions.append(cond)
return conditions
###Output
_____no_output_____
###Markdown
The resulting annotations are harder to read, but easier to diagnose:
###Code
with InvariantAnnotator() as annotator:
y = sum2(2, 2)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
As an alternative, one may be able to use `inspect.getsource()` on the lambda expression or unparse it. This is left to the reader. Exercise 4: Save Initial ValuesIf the value of an argument changes during function execution, this can easily confuse our implementation: The values are tracked at the beginning of the function, but checked only when it returns. Extend the `InvariantAnnotator` and the infrastructure it uses such that* it saves argument values both at the beginning and at the end of a function invocation;* postconditions can be expressed over both _initial_ values of arguments as well as the _final_ values of arguments;* the mined postconditions refer to both these values as well. **Solution.** To be added. Exercise 5: ImplicationsSeveral mined invariant are actually _implied_ by others: If `x > 0` holds, then this implies `x >= 0` and `x != 0`. Extend the `InvariantAnnotator` such that implications between properties are explicitly encoded, and such that implied properties are no longer listed as invariants. See \cite{Ernst2001} for ideas. **Solution.** Left to the reader. Exercise 6: Local VariablesPostconditions may also refer to the values of local variables. Consider extending `InvariantAnnotator` and its infrastructure such that the values of local variables at the end of the execution are also recorded and made part of the invariant inference mechanism. **Solution.** Left to the reader. Exercise 7: Exploring Invariant AlternativesAfter mining a first set of invariants, have a [concolic fuzzer](ConcolicFuzzer.ipynb) generate tests that systematically attempt to invalidate pre- and postconditions. How far can you generalize? **Solution.** To be added. Exercise 8: Grammar-Generated PropertiesThe larger the set of properties to be checked, the more potential invariants can be discovered. Create a _grammar_ that systematically produces a large set of properties. See \cite{Ernst2001} for possible patterns. **Solution.** Left to the reader. Exercise 9: Embedding Invariants as AssertionsRather than producing invariants as annotations for pre- and postconditions, insert them as `assert` statements into the function code, as in:```pythondef my_sqrt(x): 'Computes the square root of x, using the Newton-Raphson method' assert isinstance(x, int), 'violated precondition' assert (x > 0), 'violated precondition' approx = None guess = (x / 2) while (approx != guess): approx = guess guess = ((approx + (x / approx)) / 2) return_value = approx assert (return_value < x), 'violated postcondition' assert isinstance(return_value, float), 'violated postcondition' return approx```Such a formulation may make it easier for test generators and symbolic analysis to access and interpret pre- and postconditions. **Solution.** Here is a tentative implementation that inserts invariants into function ASTs. Part 1: Embedding Invariants into Functions
###Code
class EmbeddedInvariantAnnotator(InvariantTracker):
def functions_with_invariants_ast(self, function_name=None):
if function_name is None:
return annotate_functions_with_invariants(self.invariants())
return annotate_function_with_invariants(function_name, self.invariants(function_name))
def functions_with_invariants(self, function_name=None):
if function_name is None:
functions = ''
for f_name in self.invariants():
try:
f_text = ast.unparse(self.functions_with_invariants_ast(f_name))
except KeyError:
f_text = ''
functions += f_text
return functions
return ast.unparse(self.functions_with_invariants_ast(function_name))
def function_with_invariants(self, function_name):
return self.functions_with_invariants(function_name)
def function_with_invariants_ast(self, function_name):
return self.functions_with_invariants_ast(function_name)
def annotate_invariants(invariants):
annotated_functions = {}
for function_name in invariants:
try:
annotated_functions[function_name] = annotate_function_with_invariants(function_name, invariants[function_name])
except KeyError:
continue
return annotated_functions
def annotate_function_with_invariants(function_name, function_invariants):
function = globals()[function_name]
function_code = inspect.getsource(function)
function_ast = ast.parse(function_code)
return annotate_function_ast_with_invariants(function_ast, function_invariants)
def annotate_function_ast_with_invariants(function_ast, function_invariants):
annotated_function_ast = EmbeddedInvariantTransformer(function_invariants).visit(function_ast)
return annotated_function_ast
###Output
_____no_output_____
###Markdown
Part 2: Preconditions
###Code
class PreconditionTransformer(ast.NodeTransformer):
def __init__(self, invariants):
self.invariants = invariants
super().__init__()
def preconditions(self):
preconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated precondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) < 0:
preconditions += assertion_ast.body
return preconditions
def insert_assertions(self, body):
preconditions = self.preconditions()
try:
docstring = body[0].value.s
except:
docstring = None
if docstring:
return [body[0]] + preconditions + body[1:]
else:
return preconditions + body
def visit_FunctionDef(self, node):
"""Add invariants to function"""
# print(ast.dump(node))
node.body = self.insert_assertions(node.body)
return node
class EmbeddedInvariantTransformer(PreconditionTransformer):
pass
with EmbeddedInvariantAnnotator() as annotator:
my_sqrt(5)
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____
###Markdown
Part 3: PostconditionsWe make a few simplifying assumptions: * Variables do not change during execution.* There is a single `return` statement at the end of the function.
###Code
class EmbeddedInvariantTransformer(PreconditionTransformer):
def postconditions(self):
postconditions = []
for (prop, var_names) in self.invariants:
assertion = "assert " + instantiate_prop(prop, var_names) + ', "violated postcondition"'
assertion_ast = ast.parse(assertion)
if assertion.find(RETURN_VALUE) >= 0:
postconditions += assertion_ast.body
return postconditions
def insert_assertions(self, body):
new_body = super().insert_assertions(body)
postconditions = self.postconditions()
body_ends_with_return = isinstance(new_body[-1], ast.Return)
if body_ends_with_return:
saver = RETURN_VALUE + " = " + ast.unparse(new_body[-1].value)
else:
saver = RETURN_VALUE + " = None"
saver_ast = ast.parse(saver)
postconditions = [saver_ast] + postconditions
if body_ends_with_return:
return new_body[:-1] + postconditions + [new_body[-1]]
else:
return new_body + postconditions
with EmbeddedInvariantAnnotator() as annotator:
my_sqrt(5)
my_sqrt_def = annotator.functions_with_invariants()
###Output
_____no_output_____
###Markdown
Here's the full definition with included assertions:
###Code
print_content(my_sqrt_def, '.py')
exec(my_sqrt_def.replace('my_sqrt', 'my_sqrt_annotated'))
with ExpectError():
my_sqrt_annotated(-1)
###Output
_____no_output_____
###Markdown
Here come some more examples:
###Code
with EmbeddedInvariantAnnotator() as annotator:
y = sum3(3, 4, 5)
y = sum3(-3, -4, -5)
y = sum3(0, 0, 0)
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
length = list_length([1, 2, 3])
print_content(annotator.functions_with_invariants(), '.py')
with EmbeddedInvariantAnnotator() as annotator:
print_sum(31, 45)
print_content(annotator.functions_with_invariants(), '.py')
###Output
_____no_output_____ |
ch01/Chapter1.ipynb | ###Markdown
And with the fact that conditions are also propagated to individual elements, we gain a very convenient way to access our data:
###Code
a>4
a[a>4]
###Output
_____no_output_____
###Markdown
We can use this feature to trim outliers:
###Code
a[a>4] = 4
a
###Output
_____no_output_____
###Markdown
This is a common use case, so there is a special clip function for it, clipping values at both ends of an interval with one function call:
###Code
a.clip(0, 4)
###Output
_____no_output_____
###Markdown
Handling nonexisting valuesThe power of NumPy's indexing capabilities comes in handy when preprocessing data that we have just read in from a text file. Most likely, that will contain invalid values that we will mark as not being a real number using numpy.NAN
###Code
c = np.array([1,2,np.NAN,3,4]) # fake data
np.isnan(c)
c[~np.isnan(c)]
np.mean(c[~np.isnan(c)])
###Output
_____no_output_____
###Markdown
Comparing the runtimeLet's compare the runtime behavior of NumPy compared with normal Python lists. In the following code, we will calculate the sum of all squared numbers from 1 to 1000 and see how much time it will take.
###Code
import timeit
normal_py_sec = timeit.timeit('sum(x*x for x in range(1000))', number=10000)
naive_np_sec = timeit.timeit('sum(na*na)', setup="import numpy as np; na=np.arange(1000)",
number=10000)
good_np_sec = timeit.timeit('na.dot(na)', setup="import numpy as np; na=np.arange(1000)",
number=10000)
print("Normal Python: %f sec" % normal_py_sec)
print("Naive NumPy: %f sec" %naive_np_sec)
print("Good NumPy: %f sec" % good_np_sec)
###Output
Normal Python: 3.576253 sec
Naive NumPy: 4.167665 sec
Good NumPy: 0.069332 sec
###Markdown
There are some interesting observations:* Using NumPy as data storage takes more time, which is surprising since we believe it must be much faster as it is written as a C extension. But the explanation is on the access to individual elements from Python itself is rather costly. * The dot() function does exactly the same thing However we no longer have the incredible flexibility of Python lists, wich can hold basically anything. NumPy arrays always have only one data type.
###Code
a = np.array([1,2,3])
a.dtype
###Output
_____no_output_____
###Markdown
If we try to use elements of different types, shuch as the ones shown in the following code, NumPy will do its best to coerce them to be the most reasonable common data type:
###Code
np.array([1, "stringy"])
np.array([1, "stringy", set([1,2,3])])
###Output
_____no_output_____
###Markdown
Learning SciPyOn top of the efficient data structures of NumPy, SciPy offers a magnitude of algorithms working on those arrays. Whatever numerical heavy algorithm you take from current books, on numerical recipes, most likely you will find support for them in SciPy on one way or the other. For convenience, the complete namespace of NumPy is also accessible via SciPy. You can check this easily comparing the function references of any base function, such as:
###Code
import scipy, numpy
scipy.version.full_version
scipy.dot is numpy.dot
###Output
_____no_output_____
###Markdown
Our first (tiny) application of machine learningLet's get our hands dirty and take a look at our hypothetical web start-up, MLaaS,which sells the service of providing machine learning algorithms via HTTP. Withincreasing success of our company, the demand for better infrastructure increasesto serve all incoming web requests successfully. We don't want to allocate toomany resources as that would be too costly. On the other side, we will lose money,if we have not reserved enough resources to serve all incoming requests. Now,the question is, when will we hit the limit of our current infrastructure, which weestimated to be at 100,000 requests per hour. We would like to know in advancewhen we have to request additional servers in the cloud to serve all the incomingrequests successfully without paying for unused ones. Reading in the dataWe have collected the web stats for the last month and aggregated them in ch01/data/web_traffic.tsv ( .tsv because it contains tab-separated values). They arestored as the number of hits per hour. Each line contains the hour consecutively andthe number of web hits in that hour.Using gentfromtxt(), we can easily read in the data using the following code:
###Code
import scipy as sp
data = sp.genfromtxt("data/web_traffic.tsv", delimiter="\t")
###Output
_____no_output_____
###Markdown
We have to specify tab as the delimiter so that columns are correctly determined. A quick check shows that we have correctly read in the data:
###Code
print(data[:10])
print(data.shape)
###Output
(743, 2)
###Markdown
As you can see, we have 743 data points with two dimensions. Preprocessing and cleaning the dataIt is more convenient for SciPy to separate the dimensions into two vectors, eachof size 743. The first vector, x , will contain the hours, and the other, y , will containthe Web hits in that particular hour. This splitting is done using the special indexnotation of SciPy, by which we can choose the columns individually:
###Code
x = data[:,0]
y = data[:,1]
###Output
_____no_output_____
###Markdown
Let's check how many hours contain invaliddata, by running the following code:
###Code
sp.sum(sp.isnan(y))
###Output
_____no_output_____
###Markdown
As you can see, we are missing only 8 out of 743 entries, so we can afford to removethem.
###Code
x = x[~sp.isnan(y)]
y = y[~sp.isnan(y)]
###Output
_____no_output_____
###Markdown
We can plot our data using matplotlib which contains the pyplot package, wich tries to mimic MATLAB's interface, which is very convenient and easy to use as you can see in the following code:
###Code
%matplotlib inline
import matplotlib.pyplot as plt
# plot the (x,y) points with dots of size 10
plt.scatter(x,y, s=10)
plt.title("Web traffic over the last month")
plt.xlabel("Time")
plt.ylabel("Hits/hour")
plt.xticks([w*7*24 for w in range(10)],
['week %i' % w for w in range(10)])
plt.autoscale(tight=True)
# draw a slightly opaque, dashed grid
plt.grid(True, linestyle='-', color='0.75')
plt.show()
###Output
_____no_output_____
###Markdown
Choosing the right model and learning algorithmNow that we have a first impression of the data, we return to the initial question:How long will our server handle the incoming web traffic? To answer this we haveto do the following:1. Find the real model behind the noisy data points.2. Following this, use the model to extrapolate into the future to find the point in time where our infrastructure has to be extended. Before building our first model...When we talk about models, you can think of them as simplified theoreticalapproximations of complex reality. As such there is always some inferiorityinvolved, also called the approximation error. This error will guide us in choosingthe right model among the myriad of choices we have. And this error will becalculated as the squared distance of the model's prediction to the real data; forexample, for a learned model function f , the error is calculated as follows:
###Code
def error(f, x, y):
return sp.sum((f(x)-y)**2)
###Output
_____no_output_____
###Markdown
The vectors x and y contain the web stats data that we have extracted earlier. It isthe beauty of SciPy's vectorized functions that we exploit here with f(x) . The trainedmodel is assumed to take a vector and return the results again as a vector of the samesize so that we can use it to calculate the difference to y . Starting with a simple straight line Let's assume for a second that the underlying model is a straight line. Then thechallenge is how to best put that line into the chart so that it results in the smallestapproximation error. SciPy's polyfit() function does exactly that. Given data x andy and the desired order of the polynomial (a straight line has order 1), it finds themodel function that minimizes the error function defined earlier:
###Code
fp1, residuals, rank, sv, rcond = sp.polyfit(x, y, 1, full=True)
###Output
_____no_output_____
###Markdown
The polyfit() function returns the parameters of the fitted model function, fp1 .And by setting full=True , we also get additional background information on thefitting process. Of this, only residuals are of interest, which is exactly the error ofthe approximation:
###Code
print("Model parameters: %s" % fp1)
print(residuals)
###Output
[ 3.17389767e+08]
###Markdown
This means the best straight line fit is the following functionf(x) = 2.59619213 * x + 989.02487106We then use poly1d() to create a model function from the model parameters:
###Code
f1 = sp.poly1d(fp1)
print(error(f1, x, y))
###Output
317389767.34
###Markdown
We have used full=True to retrieve more details on the fitting process. Normally,we would not need it, in which case only the model parameters would be returned.We can now use f1() to plot our first trained model. In addition to the precedingplotting instructions, we simply add the following code:
###Code
import matplotlib.pyplot as plt
# plot the (x,y) points with dots of size 10
plt.scatter(x,y, s=10)
plt.title("Web traffic over the last month")
plt.xlabel("Time")
plt.ylabel("Hits/hour")
plt.xticks([w*7*24 for w in range(10)],
['week %i' % w for w in range(10)])
plt.autoscale(tight=True)
# draw a slightly opaque, dashed grid
plt.grid(True, linestyle='-', color='0.75')
fx = sp.linspace(0,x[-1], 1000) # generate values for plotting
plt.plot(fx, f1(fx), linewidth=4)
plt.legend(["d=%i" % f1.order], loc="upper left")
plt.show()
###Output
_____no_output_____
###Markdown
It seems like the first 4 weeks are not that far off, although we clearly see that there issomething wrong with our initial assumption that the underlying model is a straightline. The absolute value of the error is seldom of use in isolation. However, whencomparing two competing models, we can use their errors to judge which one ofthem is better. Although our first model clearly is not the one we would use, it servesa very important purpose in the workflow. We will use it as our baseline until wefind a better one. Whatever model we come up with in the future, we will compare itagainst the current baseline. Towards some advanced stuffLet's now fit a more complex model, a polynomial of degree 2, to see whether itbetter understands our data:
###Code
f2p = sp.polyfit(x, y, 2)
print(f2p)
f2 = sp.poly1d(f2p)
print(error(f2, x, y))
import matplotlib.pyplot as plt
# plot the (x,y) points with dots of size 10
plt.scatter(x,y, s=10)
plt.title("Web traffic over the last month")
plt.xlabel("Time")
plt.ylabel("Hits/hour")
plt.xticks([w*7*24 for w in range(10)],
['week %i' % w for w in range(10)])
plt.autoscale(tight=True)
# draw a slightly opaque, dashed grid
plt.grid(True, linestyle='-', color='0.75')
fx = sp.linspace(0,x[-1], 1000) # generate values for plotting
plt.plot(fx, f2(fx), linewidth=4)
plt.legend(["d=%i" % f2.order], loc="upper left")
plt.show()
###Output
_____no_output_____
###Markdown
The error is 179,983,507.878, which is almost half the error of the straight line model.This is good but unfortunately this comes with a price: We now have a more complexfunction, meaning that we have one parameter more to tune inside polyfit() . Thefitted polynomial is as follows: f(x) = 0.0105322215 * x**2 - 5.26545650 * x + 1974.76082 So, if more complexity gives better results, why not increase the complexity evenmore? Let's try it for degrees 3, 10, and 100.
###Code
f3p = sp.polyfit(x, y, 3)
print(f3p)
f3 = sp.poly1d(f3p)
f10p = sp.polyfit(x, y, 10)
print(f10p)
f10 = sp.poly1d(f10p)
f100p = sp.polyfit(x, y, 100)
print(f100p)
f100 = sp.poly1d(f100p)
import matplotlib.pyplot as plt
# plot the (x,y) points with dots of size 10
plt.scatter(x,y, s=10)
plt.title("Web traffic over the last month")
plt.xlabel("Time")
plt.ylabel("Hits/hour")
plt.xticks([w*7*24 for w in range(10)],
['week %i' % w for w in range(10)])
plt.autoscale(tight=True)
# draw a slightly opaque, dashed grid
plt.grid(True, linestyle='-', color='0.75')
fx = sp.linspace(0,x[-1], 1000) # generate values for plotting
# plotting with a 1 order
plt.plot(fx, f1(fx), linewidth=4, label="d1=%i" % f1.order)
# plotting with a 2 order
plt.plot(fx, f2(fx), linewidth=4, label="d2=%i" % f2.order)
# plotting with a 3 order
plt.plot(fx, f3(fx), linewidth=4, label="d3=%i" % f3.order)
# plotting with a 10 order
plt.plot(fx, f10(fx), linewidth=4, label="d10=%i" % f10.order)
# plotting with a 100 order
plt.plot(fx, f100(fx), linewidth=4, label="d100=%i" % f100.order)
plt.legend(loc='upper left')
plt.show()
###Output
_____no_output_____
###Markdown
Interestingly, when you have plotted your graph that d100=53 this means that polyfit cannot determine a good fit with 100 degrees. Instead, it figured that 53 must be good enough. It seems like the curves capture and better the fitted data the more complex they get.And also, the errors seem to tell the same story:
###Code
print("Error d=1: %f", error(f1, x, y))
print("Error d=2: %f", error(f2, x, y))
print("Error d=3: %f", error(f3, x, y))
print("Error d=10: %f", error(f10, x, y))
print("Error d=100: %f", error(f100, x, y))
###Output
Error d=1: %f 317389767.34
Error d=2: %f 179983507.878
Error d=3: %f 139350144.032
Error d=10: %f 121942326.364
Error d=100: %f 109452401.18
###Markdown
However, taking a closer look at the fitted curves, we start to wonder whether they alsocapture the true process that generated that data. Framed differently, do our modelscorrectly represent the underlying mass behavior of customers visiting our website?Looking at the polynomial of degree 10 and 53, we see wildly oscillating behavior. Itseems that the models are fitted too much to the data. So much that it is now capturingnot only the underlying process but also the noise. This is called overfitting. So we have the following choices:* Choosing one of the fitted polynomial models.* Switching to another more complex model class.* Thinking differently about the data and start again. Out of the five fitted models, the first order model clearly is too simple, and themodels of order 10 and 53 are clearly overfitting. Only the second and third ordermodels seem to somehow match the data. However, if we extrapolate them at bothborders, we see them going berserk.Switching to a more complex class seems also not to be the right way to go. Whatarguments would back which class? At this point, we realize that we probably havenot fully understood our data. Stepping back to go forward – another look at our dataSo, we step back and take another look at the data. It seems that there is an inflection point between weeks 3 and 4. So let's separate the data and train two lines using week 3.5 as a separation point:
###Code
inflection = 3.5*7*25 #calculate the inflection point in hours
int_inflection = round(inflection)
xa = x[:int_inflection] # data before the inflection point
ya = y[:int_inflection]
xb = x[int_inflection:] # data after
yb = y[int_inflection:]
fa = sp.poly1d(sp.polyfit(xa, ya, 1))
fb = sp.poly1d(sp.polyfit(xb, yb, 1))
fa_error = error(fa, xa, ya)
fb_error = error(fb, xb, yb)
print("Error inflection=%f" %(fa_error + fb_error))
import matplotlib.pyplot as plt
# plot the (x,y) points with dots of size 10
plt.scatter(x,y, s=10)
plt.title("Web traffic over the last month")
plt.xlabel("Time")
plt.ylabel("Hits/hour")
plt.xticks([w*7*24 for w in range(10)],
['week %i' % w for w in range(10)])
plt.autoscale(tight=True)
# draw a slightly opaque, dashed grid
plt.grid(True, linestyle='-', color='0.75')
fx = sp.linspace(0,x[-1], 1000) # generate values for plotting
# plotting with a 1 order
plt.plot(fx, fa(fx), linewidth=4, label="d1=%i" % fa.order)
# plotting with a 2 order
plt.plot(fx, fb(fx), linewidth=4, label="d1=%i" % fb.order)
plt.legend(loc='upper left')
plt.show()
###Output
_____no_output_____
###Markdown
Clearly, the combination of these two lines seems to be a much better fit to the datathan anything we have modeled before. But still, the combined error is higher thanthe higher order polynomials. Can we trust the error at the end? Asked differently, why do we trust the straight line fitted only at the last week of ourdata more than any of the more complex models? It is because we assume that it willcapture future data better. If we plot the models into the future, we see how right weare (d=1 is again our initial straight line). Training and testingIf we only had some data from the future that we could use to measure our modelsagainst, then we should be able to judge our model choice only on the resultingapproximation error.Although we cannot look into the future, we can and should simulate a similar effectby holding out a part of our data. Let's remove, for instance, a certain percentage ofthe data and train on the remaining one. Then we used the held-out data to calculatethe error. As the model has been trained not knowing the held-out data, we shouldget a more realistic picture of how the model will behave in the future.The test errors for the models trained only on the time after inflection point nowshow a completely different picture:
###Code
inflection = 3.5*7*25 #calculate the inflection point in hours
int_inflection = inflection
xb = x[int_inflection:] # data after
yb = y[int_inflection:]
fb1 = sp.poly1d(sp.polyfit(xb, yb, 1))
fb2 = sp.poly1d(sp.polyfit(xb, yb, 2))
fb3 = sp.poly1d(sp.polyfit(xb, yb, 3))
fb10 = sp.poly1d(sp.polyfit(xb, yb, 10))
fb53 = sp.poly1d(sp.polyfit(xb, yb, 53))
fb1_error = error(fb1, xb, yb)
fb2_error = error(fb2, xb, yb)
fb3_error = error(fb3, xb, yb)
fb10_error = error(fb10, xb, yb)
fb53_error = error(fb53, xb, yb)
frac = 0.3
split_idx = int(frac * len(xb))
shuffled = sp.random.permutation(list(range(len(xb))))
test = sorted(shuffled[:split_idx])
train = sorted(shuffled[split_idx:])
fbt1 = sp.poly1d(sp.polyfit(xb[train], yb[train], 1))
fbt2 = sp.poly1d(sp.polyfit(xb[train], yb[train], 2))
fbt3 = sp.poly1d(sp.polyfit(xb[train], yb[train], 3))
fbt10 = sp.poly1d(sp.polyfit(xb[train], yb[train], 10))
fbt100 = sp.poly1d(sp.polyfit(xb[train], yb[train], 53))
print("Test errors for only the time after inflection point")
for f in [fbt1, fbt2, fbt3, fbt10, fbt100]:
print("Error d=%i: %f" % (f.order, error(f, xb[test], yb[test])))
###Output
Test errors for only the time after inflection point
Error d=1: 3946251.726017
Error d=2: 3873979.021186
Error d=3: 3883897.384542
Error d=10: 4038394.035316
Error d=53: 4437453.980905
###Markdown
It seems that we finally have a clear winner: The model with degree 2 has the lowesttest error, which is the error when measured using data that the model did not seeduring training. And this gives us hope that we won't get bad surprises when futuredata arrives Answering our initial questionFinally we have arrived at a model which we think represents the underlyingprocess best; it is now a simple task of finding out when our infrastructure willreach 100,000 requests per hour. We have to calculate when our model functionreaches the value 100,000.Having a polynomial of degree 2, we could simply compute the inverse of thefunction and calculate its value at 100,000. Of course, we would like to have anapproach that is applicable to any model function easily.This can be done by subtracting 100,000 from the polynomial, which results inanother polynomial, and finding its root. SciPy's optimize module has the functionfsolve that achieves this, when providing an initial starting position with parameterx0 . As every entry in our input data file corresponds to one hour, and we have 743 ofthem, we set the starting position to some value after that. Let fbt2 be the winningpolynomial of degree 2.
###Code
fbt2 = sp.poly1d(sp.polyfit(xb[train], yb[train], 2))
print("fbt2(x) = \n%s" %fbt2)
print("fbt2(x)-100,000 = \n%s" %(fbt2-100000))
from scipy.optimize import fsolve
reached_max = fsolve(fbt2-100000, x0=800)/(7*24)
print("100,000 hits/hour expected at week %f" % reached_max[0])
###Output
100,000 hits/hour expected at week 9.806430
###Markdown
Learning NumPy
###Code
import numpy as np
a = np.array([0, 1, 2, 3, 4, 5])
a
a.ndim
a.shape
###Output
_____no_output_____
###Markdown
We just created an array like we would create a list in Python. However, the NumPy arrays have additional information about the shape. In this case, it is a one-dimensional array of six elements. We can now transform this array to a two-dimensional matrix:
###Code
b = a.reshape((3, 2))
b
b.ndim
b.shape
###Output
_____no_output_____
###Markdown
We have a trouble if we wan't to make a real copy, this shows how much the NumPy package is optimized.
###Code
b[1][0] = 77
b
###Output
_____no_output_____
###Markdown
But now if we see the values of a:
###Code
a
###Output
_____no_output_____
###Markdown
We see immediately the same change reflected in "a" as well. If we need a true copy we can perform:
###Code
c = a.reshape((3, 2)).copy()
c
c[0][0] = -99
a
c
###Output
_____no_output_____
###Markdown
Another big advantage of NumPy arrays is that operations are propagated to the individual elements.
###Code
d = np.array([1,2,3,4,5])
d*2
###Output
_____no_output_____
###Markdown
Similarly, for other operations:
###Code
d**2
###Output
_____no_output_____
###Markdown
What it's not the case with Python lists.
###Code
[1,2,3,4,5]*2
[1,2,3,4,5]**2
###Output
_____no_output_____
###Markdown
When using NumPy arrays, we sacrafice the agility Python lists offer. Simple operations such as adding or removing are bit complex for NumPy arrays. But we can choose the right tool for the task. IndexingNumpy allows you to use the arrays themselves as indices by performing:
###Code
a[np.array([2,3,4])]
###Output
_____no_output_____ |
content/en/wuhan-sir/wuhan-sir.ipynb | ###Markdown
When Will Coronavirous (COVID-19) End?by [Frank Zheng](http://frankzheng.me) *This notebook is licensed under the MIT License. If you use the code or data visualization designs contained within this notebook, it would be greatly appreciated if proper attribution is given back to this notebook and/or myself. Thanks! :)*Coronavirus Disease 2019 ([COVID-19](https://www.who.int/emergencies/diseases/novel-coronavirus-2019)) was identified as the cause of a cluster of pneumonia cases in Wuhan, a city in the Hubei Province of China, at the end of 2019. It subsequently spread throughout China and elsewhere, becoming a global health emergency. In February 2020, the World Health Organization (WHO) designated the disease COVID-19, which stands for coronavirus disease 2019. This post will use current data with SIR model and (try to) forcast when the number of coronavirous infected patients reaches its peak and eventually declines. SIR and SIRS ModelIn 1927, the SIR model for sperad of disease was first propsed in in a collection of three articles in the Proceedings of the Royal Society by [Anderson Gray](https://en.wikipedia.org/wiki/Anderson_Gray_McKendrick) McKendrick and [William Ogilvy Kermack](https://en.wikipedia.org/wiki/William_Ogilvy_Kermack); the resulting theory is known as [Kermack–McKendrick theory](https://en.wikipedia.org/wiki/Kermack%E2%80%93McKendrick_theory); now considered a subclass of a more general theory known as [compartmental models](https://en.wikipedia.org/wiki/Compartmental_models_in_epidemiology) in epidemiology. The three original articles were republished in 1991, in a special issue of the Bulletin of [Mathematical Biology](https://link.springer.com/journal/11538/53/1/page/1).SIR model stands for a (suspectible - infectious - recovered) model, while SIRS represents a (suspectible - infectious - recovered - susceptible) model. The main difference between these two is the latter does not confer lifelong immunity, so individual may become susceptible again. The SIR/SIRS diagram from [IDM](https://idmod.org/docs/hiv/model-si.html) below shows how individuals move through each compartment in the model.  - $\beta$, the transmission rate, controls the rate of spread which represents the probability of transmitting disease between a susceptible and an infectious individual. - $\gamma$ = 1/D, recovery rate, is determined by the average duration, D, of infection. - $\xi$, only used for the SIRS model, is the rate which recovered individuals return to the susceptible statue due to loss of immunity.Besides the immunity issue, SIR model also considers the population changes. If the course of the infection is short (emergent outbreak) compared with the lifetime of an individual and the disease is non-fatal, vital dynamics (birth and death) can be ignored. SIR model with vital dynamics considers new births that can provide more susceptible individuals to the population, sustaining an epidemic or allowing new introductions to spread throughout the population. In our COVID-19 case, becuase the disease is such a outbreak, it's not likely to be fatal (3.4% death rate reported by [WHO](https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---3-march-2020)), and most patients who recoved from conronavirus gain immunity, therefore, SIR model without vital dynamics is more situable. In the deterministic form, the SIR model can be written as the following there ordinary differential equations (ODE), parameterized by two growth factors β and γ:$$\frac{dS}{dt} = - \frac{\beta S I}{N}$$$$\frac{dI}{dt} = \frac{\beta S I}{N} - \gamma I$$$$\frac{dR}{dt} = \gamma I$$where N is the population, and since each of S, I and R represent the number of people in mutually exclusive sets, we should have $$S+I+R=N$$Note that the right hand sides of the equations sum to zero, hence$$\frac{dS}{dt} + \frac{dI}{dt} + \frac{dR}{dt} = \frac{dN}{dt}=0$$which indicates that the population is constant.In a closed population with no vital dynamics, an epidemic will eventually die out due to an insufficient number of susceptible individuals to sustain the disease. Infected individuals who are added later will not start another epidemic due to the lifelong immunity of the existing population.A discrete model can be derived from the above equations for the evolution of the disease.$${S}_{n+1} = S_n − \frac{\beta}{N} S_n I_n$$$${I}_{n+1} = I_n + \frac{\beta}{N} S_n I_n - \gamma I_n$$$${R}_{n+1} = R_n + \gamma I_n$$- New infecteds, $I_{n+1}$, result from contact between the susceptibles, $S_n$, and infecteds, $I_n$, with rate of infection $\frac{\beta}{N}$.- Infecteds are cured at a rate proportional to the number of infecteds, $\gamma I_n$, which become recovered, $R_n$- The term $\frac{\beta}{N} S_n$ represents the proportion of contacts by an infected individual that result in the infection of a susceptible individual.- The ratio $1/\gamma$ is the average length of the infectious period of the disease.While these may look very confusing, once explained they make a lot of sense. For example, ${S}_{n+1} = S_n − \frac{\beta}{N} S_n I_n$the equation simply means that the number of susceptible people today(${S}_{n+1}$) would be the number of susceptible people yesterday $S_n$ deduct the number of people that got infected yesterday and today. We would calculate the number of people infected yesterday by multiplying by the rate of infection. Similarly, ${I}_{n+1} = I_n + \frac{\beta}{N} S_n I_n - \gamma I_n$ this simply means that the number of people that number of people infected today would be the number of people that were infected yesterday plus the number of people that got infected yesterday and today, then deduct the number of people that recovered yesterday. The final number is the number of people in the infected category today. Lastly, ${R}_{n+1} = R_n + \gamma I_n$ this equation simply means that the number of people recovered today are the number of people that were recovered yesterday as well as the number of infected people that recovered today.This model is clearly not applicable to all possible epidemics: there may be births and deaths, people may be re-infected, and so on. More complex models take these and other factors into account. However, based on we've learnt from coronavirous so far, I believe the **SIR model without vital dynamics** is the simplist but adequate model to use. Load packages
###Code
import numpy as np
import matplotlib.pyplot as plt
import math
###Output
_____no_output_____
###Markdown
Setup Parameters for SIR ModelUse current data to find {\gamma}(recovery rate) and {\beta}(transmission rate). On 01/24/2020, Wuhan has 258 cumulative total confirmed cases, 25 recovered case and 60 new confirmed cases.
###Code
### based on the data until 01/24/2020 for Wuhan
# infected
inf = 233
# recovered
rec = 25
# recovery rate
gamma = 25.0/258
# transmission rate
beta = (60+25)/(258-25)
# population of Wuhan
n = 1100 * 10000
###Output
_____no_output_____
###Markdown
Build SIR Model
###Code
class SIR:
def __init__(self, inff, recc, suss, time):
self.inff = inff
self.recc = recc
self.suss = suss
self.time = time
def sir(beta, gamma, n, inf, rec):
sus = n - inf - rec
t = 1
inff = []
recc = []
suss = []
time = []
inff.append(inf)
recc.append(rec)
suss.append(sus)
time.append(t)
while True:
if t > 200:
break
t += 1
a = inf
b = rec
c = sus
rec = b + gamma * a
inf = (a + beta * a * c / n - gamma * a)
sus = n - rec - inf
inff.append(round(inf))
recc.append(rec)
suss.append(round(sus))
time.append(t)
return SIR(inff, recc, suss, time)
###Output
_____no_output_____
###Markdown
Plot results
###Code
p = sir(beta, gamma, n, inf, rec)
fig = plt.figure(figsize=(12,4))
plt.plot(p.time, p.suss, 'y-', label = 'Suspectible')
plt.plot(p.time, p.inff, 'r-', label = 'Infected')
plt.plot(p.time, p.recc, 'g-', label = 'Recovered')
plt.axvline(p.time[p.inff.index(max(p.inff))], ls = '--')
plt.title('SIR Model Prediction on COVID-19 in Wuhan \n with 01/24/2020 as Day 1')
plt.xlabel('Time(day) \n by Frank Zheng — frankzheng.me')
plt.ylabel('Population')
plt.xlim(0, 201)
plt.legend(loc = 'best')
plt.show()
###Output
_____no_output_____ |
Lessons/Lesson06-opencv.ipynb | ###Markdown
Lesson 06: OpenCV OpenCV is a library that contains algorithms and functions that are related to and perform image processing and computer vision tasks.
###Code
import cv2
###Output
_____no_output_____
###Markdown
Opening Images
###Code
img = cv2.imread('../images/test-image.png')
print(img.shape)
cv2.imshow('img', img)
# press 'q' to close image or else it will not be properly closed.
cv2.waitKey(0)
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Changing Color Space
###Code
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
cv2.imwrite('../images/gray.jpg', gray)
cv2.imshow('img', gray)
# press 'q' to close image or else it will not be properly closed.
cv2.waitKey(0)
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Resizing
###Code
height, width = img.shape[:-1]
big_image = cv2.resize(img, (2*height, 2*width))
cv2.imshow('img', big_image)
cv2.waitKey(0)
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Cropping
###Code
img_crop = img[0:500, 300:500]
cv2.imshow('img', img_crop)
cv2.waitKey(0)
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Thresholding Thresholding only takes grayscale image.The first argument is the grayscale image, the second argument is the threshold value which is used to classify the pixel values. The third argument is the maximum value which is assigned to pixel values exceeding the threshold. OpenCV provides different types of thresholding which is given by the fourth parameter of the function.The method returns two outputs. The first is the threshold that was used and the second output is the thresholded image.
###Code
thresholded = cv2.threshold(gray, 120, 255, cv2.THRESH_BINARY)
cv2.imshow('img', thresholded[1])
cv2.waitKey(0)
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Filters
###Code
import numpy as np
kernel = np.array([
[1, 1, 1],
[1, 1, 1],
[1, 1, 1]
])
# here second argument means the depth of the output image.
# If you set this argument to -1 then the output image will have the same depth as input image.
filtered = cv2.filter2D(img, -1, kernel)
cv2.imshow('img', filtered)
cv2.waitKey(0)
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Gaussian Blur Takes img, kernel size, standard deviation. If std is 0 then it is calculated from kernel size.
###Code
gaussian = cv2.GaussianBlur(img, (5, 5), 0)
cv2.imshow('img', gaussian)
cv2.waitKey(0)
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Edge Detection Canny Edge Detector First argument is grayscale image, then lower threshold, upper threshold and then kernel size which is by default 3.The edge pixels above the upper threshold are considered in an edge map and edge pixels below the threshold are discarded. The pixel in between the thresholds are considered only if they are connected to pixels in upper threshold. Thus we get a clean edge map.
###Code
edges = cv2.Canny(gray, 100, 200, 3)
cv2.imshow('img', edges)
cv2.waitKey(0)
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Feature Detection SIFT
###Code
sift_img = cv2.imread('../images/test-image.png')
sift_obj = cv2.xfeatures2d.SIFT_create()
# you can also pass a mask in place of None if you want to find features in a specific region.
keypoints = sift_obj.detect(gray)
drawn = cv2.drawKeypoints(gray, keypoints, sift_img)
cv2.imshow('img', sift_img)
cv2.waitKey(0)
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Feature Matching
###Code
img_rot = cv2.imread('../images/test-image-rot.png')
gray_rot = cv2.cvtColor(img_rot, cv2.COLOR_BGR2GRAY)
import random
sift = cv2.xfeatures2d.SIFT_create()
# detectAndCompute will find keypoints as well as compute the descriptors
# descriptors store information about keypoints that make them distinguishable
kp, desc = sift.detectAndCompute(gray, None)
kp_rot, desc_rot = sift.detectAndCompute(gray_rot, None)
# this is will match the descriptors and find the ones that are similar
bf = cv2.BFMatcher()
matches = bf.knnMatch(desc, desc_rot, k=2)
good = []
for m, n in matches:
# applying the ratio test to filter out the bad matches due to noise
# since we set k=2 in bf.knnMatch() we have two matches for each keypoint
# the first one is the best and the second one of the second best
# we are decreasing the value of the worst match and checking whether its
# value becomes smaller then the best match. If it does then that match is discarded
if m.distance < 0.4 * n.distance:
good.append([m])
random.shuffle(good)
image_match = cv2.drawMatchesKnn(img, kp, img_rot, kp_rot, good[:10], outImg=None)
cv2.imshow('img', image_match)
cv2.waitKey(0)
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Opening Videos
###Code
cam = cv2.VideoCapture(0)
while (cam.isOpened()):
ret, frame = cam.read()
cv2.imshow('frame', frame)
if cv2.waitKey(1) & 0xFF == ord('q'):
break
cam.release()
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Converting to Grayscale
###Code
cam = cv2.VideoCapture(0)
while (cam.isOpened()):
ret, frame = cam.read()
gray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)
cv2.imshow('gray', gray)
cv2.imshow('frame', frame)
if cv2.waitKey(1) & 0xFF == ord('q'):
break
cam.release()
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Saving Video
###Code
cam = cv2.VideoCapture(0)
ret, frame = cam.read()
h, w = frame.shape[:2]
fourcc = cv2.VideoWriter_fourcc(*'DIVX')
# 1st argument: where to save video, 2nd compression format, 3rd fps, 4th size of video
video_writer = cv2.VideoWriter('../images/gray_vid.mp4', fourcc, 25.0, (w, h))
while (cam.isOpened()):
ret, frame = cam.read()
gray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)
# gray = cv2.Canny(gray, 30, 35, 3)
# have to convert back to BGR format because video needs 3 channels
# this will not bring back the colors
# gray = cv2.cvtColor(gray, cv2.COLOR_GRAY2BGR)
video_writer.write(gray)
cv2.imshow('frame', frame)
cv2.imshow('gray', gray)
if cv2.waitKey(1) & 0xFF == ord('q'):
break
cam.release()
video_writer.release()
cv2.destroyAllWindows()
###Output
_____no_output_____ |
examples/DKT.ipynb | ###Markdown
Udacity - Machine Learning Engineer Nanodegree Capstone Project Title: Development of a LSTM Network to Predict Students’ Answers on Exam Questions Implementation of DKT: Part 1: Define constants
###Code
fn = "data/ASSISTments_skill_builder_data.csv" # Dataset path
verbose = 1 # Verbose = {0,1,2}
best_model_weights = "weights/bestmodel" # File to save the model.
log_dir = "logs" # Path to save the logs.
optimizer = "adam" # Optimizer to use
lstm_units = 100 # Number of LSTM units
batch_size = 32 # Batch size
epochs = 10 # Number of epochs to train
dropout_rate = 0.3 # Dropout rate
test_fraction = 0.2 # Portion of data to be used for testing
validation_fraction = 0.2 # Portion of training data to be used for validation
###Output
_____no_output_____
###Markdown
Part 2: Pre-processing
###Code
from deepkt import deepkt, data_util, metrics
dataset, length, nb_features, nb_skills = data_util.load_dataset(fn=fn,
batch_size=batch_size,
shuffle=True)
train_set, test_set, val_set = data_util.split_dataset(dataset=dataset,
total_size=length,
test_fraction=test_fraction,
val_fraction=validation_fraction)
set_sz = length * batch_size
test_set_sz = (set_sz * test_fraction)
val_set_sz = (set_sz - test_set_sz) * validation_fraction
train_set_sz = set_sz - test_set_sz - val_set_sz
print("============= Data Summary =============")
print("Total number of students: %d" % set_sz)
print("Training set size: %d" % train_set_sz)
print("Validation set size: %d" % val_set_sz)
print("Testing set size: %d" % test_set_sz)
print("Number of skills: %d" % nb_skills)
print("Number of features in the input: %d" % nb_features)
print("========================================")
###Output
============= Data Summary =============
Total number of students: 4160
Training set size: 2662
Validation set size: 665
Testing set size: 832
Number of skills: 123
Number of features in the input: 246
========================================
###Markdown
Part 3: Building the model
###Code
student_model = deepkt.DKTModel(
nb_features=nb_features,
nb_skills=nb_skills,
hidden_units=lstm_units,
dropout_rate=dropout_rate)
student_model.compile(
optimizer=optimizer,
metrics=[
metrics.BinaryAccuracy(),
metrics.AUC(),
metrics.Precision(),
metrics.Recall()
])
student_model.summary()
###Output
Model: "DKTModel"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
inputs (InputLayer) [(None, None, 246)] 0
_________________________________________________________________
masking (Masking) (None, None, 246) 0
_________________________________________________________________
lstm (LSTM) (None, None, 100) 138800
_________________________________________________________________
outputs (TimeDistributed) (None, None, 123) 12423
=================================================================
Total params: 151,223
Trainable params: 151,223
Non-trainable params: 0
_________________________________________________________________
###Markdown
Part 4: Train the Model
###Code
import tensorflow as tf
history = student_model.fit(dataset=train_set,
epochs=epochs,
verbose=verbose,
validation_data=val_set,
callbacks=[
tf.keras.callbacks.CSVLogger(f"{log_dir}/train.log"),
tf.keras.callbacks.ModelCheckpoint(best_model_weights,
save_best_only=True,
save_weights_only=True),
tf.keras.callbacks.TensorBoard(log_dir=log_dir)
])
###Output
Epoch 1/10
83/83 [==============================] - 12s 146ms/step - loss: 0.0657 - binary_accuracy: 0.7288 - auc: 0.7076 - precision: 0.7478 - recall: 0.9222 - val_loss: 0.0000e+00 - val_binary_accuracy: 0.0000e+00 - val_auc: 0.0000e+00 - val_precision: 0.0000e+00 - val_recall: 0.0000e+00
Epoch 2/10
83/83 [==============================] - 9s 108ms/step - loss: 0.0557 - binary_accuracy: 0.7720 - auc: 0.8120 - precision: 0.7765 - recall: 0.9440 - val_loss: 0.0666 - val_binary_accuracy: 0.7878 - val_auc: 0.8344 - val_precision: 0.7908 - val_recall: 0.9487
Epoch 3/10
83/83 [==============================] - 9s 108ms/step - loss: 0.0612 - binary_accuracy: 0.7788 - auc: 0.8233 - precision: 0.7862 - recall: 0.9348 - val_loss: 0.0607 - val_binary_accuracy: 0.7715 - val_auc: 0.8289 - val_precision: 0.7733 - val_recall: 0.9310
Epoch 4/10
83/83 [==============================] - 9s 103ms/step - loss: 0.0596 - binary_accuracy: 0.7877 - auc: 0.8315 - precision: 0.7991 - recall: 0.9319 - val_loss: 0.0618 - val_binary_accuracy: 0.7871 - val_auc: 0.8322 - val_precision: 0.7917 - val_recall: 0.9436
Epoch 5/10
83/83 [==============================] - 9s 109ms/step - loss: 0.0532 - binary_accuracy: 0.7817 - auc: 0.8291 - precision: 0.7958 - recall: 0.9199 - val_loss: 0.0518 - val_binary_accuracy: 0.8046 - val_auc: 0.8671 - val_precision: 0.8093 - val_recall: 0.9352
Epoch 6/10
83/83 [==============================] - 8s 98ms/step - loss: 0.0584 - binary_accuracy: 0.7768 - auc: 0.8201 - precision: 0.7889 - recall: 0.9232 - val_loss: 0.0522 - val_binary_accuracy: 0.7863 - val_auc: 0.8385 - val_precision: 0.8047 - val_recall: 0.9141
Epoch 7/10
83/83 [==============================] - 9s 104ms/step - loss: 0.0536 - binary_accuracy: 0.7839 - auc: 0.8295 - precision: 0.7970 - recall: 0.9238 - val_loss: 0.0603 - val_binary_accuracy: 0.7838 - val_auc: 0.8345 - val_precision: 0.7932 - val_recall: 0.9277
Epoch 8/10
83/83 [==============================] - 9s 107ms/step - loss: 0.0546 - binary_accuracy: 0.7958 - auc: 0.8498 - precision: 0.8080 - recall: 0.9248 - val_loss: 0.0607 - val_binary_accuracy: 0.8053 - val_auc: 0.8586 - val_precision: 0.8169 - val_recall: 0.9346
Epoch 9/10
83/83 [==============================] - 9s 106ms/step - loss: 0.0543 - binary_accuracy: 0.7795 - auc: 0.8309 - precision: 0.7928 - recall: 0.9147 - val_loss: 0.0509 - val_binary_accuracy: 0.7806 - val_auc: 0.8216 - val_precision: 0.7891 - val_recall: 0.9329
Epoch 10/10
83/83 [==============================] - 9s 105ms/step - loss: 0.0547 - binary_accuracy: 0.7950 - auc: 0.8464 - precision: 0.8063 - recall: 0.9248 - val_loss: 0.0603 - val_binary_accuracy: 0.7717 - val_auc: 0.8144 - val_precision: 0.7823 - val_recall: 0.9200
###Markdown
Part 5: Load the Model with the Best Validation Loss
###Code
student_model.load_weights(best_model_weights)
###Output
_____no_output_____
###Markdown
Part 6: Test the Model
###Code
result = student_model.evaluate(test_set, verbose=verbose)
###Output
26/Unknown - 2s 67ms/step - loss: 0.0529 - binary_accuracy: 0.8071 - auc: 0.8599 - precision: 0.8176 - recall: 0.9329
###Markdown
Confirm we can use a GPU to run the model
###Code
import tensorflow as tf
import pandas as pd
import numpy as np
###Output
_____no_output_____
###Markdown
Define constants
###Code
data = "data/kuze_data/evaluations_per_ans_with_taxonomy_ids_PPL.csv"
factorized_taxonomies = "data/kuze_data/factorized_math_taxonomies.csv"
factorized_students = "data/kuze_data/factorized_student_ids.csv"
verbose = 1
best_model_weights = "weights/bestmodel"
log_dir = "logs"
optimizer = "adam"
lstm_units = 200
batch_size = 64
epochs = 1
dropout_rate = 0.3
test_fraction = 0.2
validation_fraction = 0.2
###Output
_____no_output_____
###Markdown
Pre-processing
###Code
import sys
sys.path.append('/home/grenouille/Documents/jenga/final_project/code/kuze_dkt_imp')
from deepkt import deepkt, data_util, metrics
dataset, length, nb_features, nb_taxonomies = data_util.load_dataset(data, factorized_taxonomies, factorized_students, batch_size=batch_size, shuffle=True)
train_set, test_set, val_set = data_util.split_dataset(dataset=dataset, total_size=length, test_fraction=test_fraction, val_fraction=validation_fraction)
set_size = length * batch_size
test_set_size = (set_size * test_fraction)
val_set_size = (set_size - test_set_size) * validation_fraction
train_set_size = set_size - test_set_size - val_set_size
print("============== Data Summary ==============")
print("Total number of students: %d" % set_size)
print("Training set size: %d" % train_set_size)
print("Validation set size: %d" % val_set_size)
print("Testing set size: %d" % test_set_size)
print("Number of skills: %d" % nb_taxonomies)
print("Number of features in the input: %d" % nb_features)
print("========================================= ")
###Output
_____no_output_____
###Markdown
Building the model
###Code
student_model = deepkt.DKTModel(
nb_features=nb_features,
nb_taxonomies=nb_taxonomies,
hidden_units=lstm_units,
dropout_rate=dropout_rate)
student_model.compile(
optimizer=optimizer,
metrics=[
metrics.BinaryAccuracy(),
metrics.AUC(),
metrics.Precision(),
metrics.Recall()
])
student_model.summary()
###Output
_____no_output_____
###Markdown
Train the model
###Code
history = student_model.fit(
dataset=train_set,
epochs=epochs,
verbose=verbose,
validation_data=val_set,
callbacks=[
tf.keras.callbacks.CSVLogger(f"{log_dir}/train.log"),
tf.keras.callbacks.ModelCheckpoint(best_model_weights, save_best_only=True, save_weights_only=True),
tf.keras.callbacks.TensorBoard(log_dir=log_dir)
]
)
###Output
_____no_output_____
###Markdown
Load the model with the best validation loss
###Code
student_model.load_weights(best_model_weights)
###Output
_____no_output_____
###Markdown
Test the model
###Code
result = student_model.evaluate(test_set, verbose=verbose)
result
student_model.save('student_prediction')
student_model.input_shape
student_model.output_shape
###Output
_____no_output_____
###Markdown
Prediction
###Code
def preprocess_for_prediction(dataframe):
seq = dataframe.groupby('student_id').apply(
lambda r: (
r['factorized_student_id'],
r['factorized_taxonomy_id']
)
)
dataset = tf.data.Dataset.from_generator(
generator=lambda: seq,
output_types=(tf.int32, tf.int32)
)
# Add 1 since indexing starts from 0
student_depth = int(students['factorized_student_id'].max() + 1)
taxonomy_depth = int(taxonomies['factorized_taxonomy_code'].max() + 1)
dataset = dataset.map(
lambda factorized_student_id, factorized_taxonomy_code: (
tf.one_hot(factorized_student_id, depth=student_depth),
tf.one_hot(factorized_taxonomy_code, depth=taxonomy_depth)
)
)
dataset = dataset.padded_batch(
batch_size=64,
padding_values=(
tf.constant(-1, dtype=tf.float32),
tf.constant(-1, dtype=tf.float32)),
padded_shapes=([None, None], [None, None])
)
return dataset
def process_student_data(dataset):
"""Preprocess the tensorflow Dataset type used for prediction.
The first item in the dataset corresponds to the student information.
Dimensions:
-> batch size
-> number of elements per batch
-> one-hot encoded data (number of students)
We want to get the categorical student_id from the one-hot encoding.
Return a list containing the categorical student_id
"""
student_id_list = []
student_val_list = []
for i in range(len(dataset[0][0])):
for j in range(len(dataset[0][0][i])):
array = dataset[0][0][i][j]
idx = np.argmax(array)
student_id_list.append(idx)
student_val_list.append(array[idx].numpy())
return student_id_list, student_val_list
def process_taxonomy_data(dataset):
"""Preprocess the tensorflow Dataset type used for prediction.
The second item in the dataset corresponds to the taxonomy information.
Dimensions:
-> batch size
-> number of elements per batch
-> one-hot encoded data (number of students)
We want to get the categorical student_id from the one-hot encoding.
Return a list containing the categorical student_id
"""
taxonomy_id_list = []
taxonomy_val_list = []
for i in range(len(dataset[0][1])):
for j in range(len(dataset[0][1][i])):
array = dataset[0][1][i][j]
idx = np.argmax(array)
taxonomy_id_list.append(idx)
taxonomy_val_list.append(array[idx].numpy())
return taxonomy_id_list, taxonomy_val_list
def preprocess_prediction_data(predictions):
"""Expose relevant predictions from the predictions array.
Dimensions:
-> batch size
-> number of elements per batch
-> one-hot encoded data (number of taxonomies)
Return one-hot encoded arrays sequentially ordered.
"""
prediction_array_list = []
for i in range(len(predictions)):
for j in range(len(predictions[i])):
prediction_array_list.append(predictions[i][j])
return prediction_array_list
def process_prediction_data(predictions, taxonomy_id_list):
"""Get the predicted value for a taxonomy.
Predictions is a list of arrays containing predictions for all
taxonomies.
The arrays within the list are sequentially ordered.
To get the relevant array we index into the list of arrays
with the index of the taxonomy_id of current interest within
the taxonomy_id_list
To get the prediction for the taxonomy of interest, we index
into the array with the taxonomy_id.
Return a list of predicted values.
Length should be equal to that of taxonomy_id_list.
"""
taxonomy_predictions = []
for idx, taxonomy_code in enumerate(taxonomy_id_list):
prediction_array = predictions[idx]
taxonomy_predictions.append(prediction_array[taxonomy_code])
assert len(taxonomy_predictions) == len(taxonomy_id_list)
return taxonomy_predictions
def post_prediction_preprocessing(dataset, predictions):
"""Process the dataset and predictions into a pandas DataFrame.
We want to take the input dataset and match it to the corresponding
predictions.
The dataset has paddings in order to conform to expected dimensions.
Padding value is -1 and that is where the student_val_list and
taxonomy_val_list come in handy.
Any values with -1 in those 2 lists corresponds to a padding value
and can therefore be dropped"""
# convert the dataset into a list for easy access and manipulation
dataset = list(dataset)
student_id_list, student_val_list = process_student_data(dataset)
taxonomy_id_list, taxonomy_val_list = process_taxonomy_data(dataset)
preprocessed_prediction_list = preprocess_prediction_data(predictions)
taxonomy_predictions = process_prediction_data(
preprocessed_prediction_list, taxonomy_id_list)
# round off all values in taxonomy_predictions to 2 decimal places
# for readability
taxonomy_predictions = [round(i, 4) for i in taxonomy_predictions]
column_names = ['factorized_student_id', 'one-hot_student_value', 'factorized_taxonomy_id',
'one-hot_taxonomy_value', 'prediction']
prediction_df = pd.DataFrame(list(zip(student_id_list,
student_val_list,
taxonomy_id_list,
taxonomy_val_list,
taxonomy_predictions)),
columns=column_names)
# remove padding values from students and taxonomies
prediction_df = prediction_df[prediction_df['one-hot_student_value'] != -1]
prediction_df = prediction_df[prediction_df['one-hot_taxonomy_value'] != -1]
# if the value of the prediction is greater than or equal to 0.5
# the predicted answer should be 1 else 0
# astype('int') converts a boolean value to an integer True == 1, False == 0
prediction_df['predicted_answer'] = prediction_df['prediction'].ge(0.5).astype('int')
prediction_df['predicted_answer'] = prediction_df['predicted_answer'].astype('int')
return prediction_df
data = pd.read_csv('data/kuze_data/predictor_evaluations.csv')
prediction_data = data[data['subject'] == 'math']
taxonomies = pd.read_csv(factorized_taxonomies)
students = pd.read_csv(factorized_students)
prediction_data['factorized_taxonomy_id'] = prediction_data['taxonomy_id_0'].map(
taxonomies.set_index('taxonomy_id_0')['factorized_taxonomy_code'])
prediction_data['factorized_student_id'] = prediction_data['student_id'].map(
students.set_index('student_id')['factorized_student_id'])
shape = prediction_data.shape[0]
# due to limitations in dimensionality we want each dataframe we predict on to have
# 95 items
no_of_dataframes = shape // 95
# split the data into n number of dataframes each with at least 95 rows
partitions = np.array_split(prediction_data, no_of_dataframes)
# carry out prediction on a partition of the predicted data and append
# the returned dataframe to a list
predicted_partitions = []
for df in partitions:
dataset = preprocess_for_prediction(df)
predictions = student_model.predict(dataset)
prediction_data = post_prediction_preprocessing(dataset, predictions)
predicted_partitions.append(prediction_data)
prediction_data = pd.concat(partitions, ignore_index=True)
predictions = pd.concat(predicted_partitions, ignore_index=True)
assert prediction_data.shape[0] == predictions.shape[0]
rows = prediction_data.shape[0]
answer_predictions = predictions['predicted_answer'].values
for i in range(rows):
prediction_data.at[i, 'answer_selection_prediction'] = answer_predictions[i]
def get_aggregated_evaluation_performance(dataframe, with_preds=True):
# Group data by student and evaluation id and calculate actual and predicted
# performance on questions
eval_id = []
student = []
total_questions = []
actual_performance = []
predicted_performance = []
date_of_evaluation = []
subject = []
student_full_name = []
class_name = []
class_grade = []
school_name = []
grouped_data = dataframe.groupby(['evaluation_id', 'student_id'])
for item in grouped_data:
evaluation_id, student_id = item[0]
data = item[1]
actual = data['answer_selection_correct'].value_counts()
total_nu_questions = actual.sum()
first_name = data['student_first_name'].unique()[0]
last_name = data['student_last_name'].unique()[0]
if first_name is np.nan:
first_name = ''
if last_name is np.nan:
last_name = ''
full_name = first_name + ' ' + last_name
try:
actual_correct = actual[1]
except KeyError:
# if a KeyError occurs it means the student got all of the
# questions in that evaluation wrong
actual_correct = 0
actual_perc = int((actual_correct / total_nu_questions) * 100)
if with_preds: # if prediction data is included
predicted = data['answer_selection_prediction'].astype('int').value_counts()
# ensure acual no of questions done matches no of questions predicted
assert actual.sum() == predicted.sum()
predicted_correct = predicted[1]
predicted_perc = int((predicted_correct/ total_nu_questions) * 100)
predicted_performance.append(predicted_perc)
else:
predicted_performance.append(0)
eval_id.append(evaluation_id)
student.append(student_id)
total_questions.append(total_nu_questions)
actual_performance.append(actual_perc)
date_of_evaluation.append(data['date_of_evaluation'].unique()[0].date())
subject.append(data['subject'].unique()[0])
student_full_name.append(full_name)
class_name.append(data['class_name'].unique()[0])
class_grade.append(data['class_grade'].unique()[0])
school_name.append(data['school_name'].unique()[0])
column_names = ['evaluation_id', 'student_id', 'total_number_of_questions', 'actual_performance (%)',
'predicted_performance (%)', 'date_of_evaluation', 'subject', 'student_full_name',
'class_name', 'class_grade', 'school_name']
performance_df = pd.DataFrame(list(zip(eval_id,
student,
total_questions,
actual_performance,
predicted_performance,
date_of_evaluation,
subject,
student_full_name,
class_name,
class_grade,
school_name)),
columns=column_names)
return performance_df
mask = (ds_evaluation_per_ans_sci_prediction_df['date_of_evaluation'] < '2021-07-01')
training_data = ds_evaluation_per_ans_sci_prediction_df.loc[mask]
# training_data.dropna(subset=['answer_selection_correct'], inplace=True)
aggregated_training_data = get_aggregated_evaluation_performance(training_data, with_preds=False)
performance_data = get_aggregated_evaluation_performance(prediction_data)
aggregated_performance_data = pd.concat([aggregated_training_data, performance_data], ignore_index=True)
###Output
_____no_output_____ |
lab/breast-cancer-classification.ipynb | ###Markdown
Predicting breast cancer tumor type: Benign or malignantApply a:- linear classifier, `SVC(kernel='linear')` - non-linear classifier, `RandomForestClassifier(random_state=82)` to the Wisconsin breast cancer dataset.The data is hosted on UCI https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+(Diagnostic)we will need `wdbc.names` and `wdbc.data` from the data folder on UCI.
###Code
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
###Output
_____no_output_____
###Markdown
1. Download the data TODO: add base url and filenames.
###Code
import os
import requests
#TODO: add base url and filenames
base_url = ...
file_names = [..., ...] # .names then .data
for file_name in file_names:
if not os.path.isfile(file_name):
print(f"Downloading {file_name} from {base_url}")
response = requests.get(base_url+file_name)
with open(file_name, 'wb') as f:
f.write(response.content)
else:
print(f"{file_name} found on disk")
###Output
wdbc.names found on disk
wdbc.data found on disk
###Markdown
2. Prepare the data 2.1 A look at the data files
###Code
with open(file_names[0]) as f:
print(f.read())
###Output
1. Title: Wisconsin Diagnostic Breast Cancer (WDBC)
2. Source Information
a) Creators:
Dr. William H. Wolberg, General Surgery Dept., University of
Wisconsin, Clinical Sciences Center, Madison, WI 53792
[email protected]
W. Nick Street, Computer Sciences Dept., University of
Wisconsin, 1210 West Dayton St., Madison, WI 53706
[email protected] 608-262-6619
Olvi L. Mangasarian, Computer Sciences Dept., University of
Wisconsin, 1210 West Dayton St., Madison, WI 53706
[email protected]
b) Donor: Nick Street
c) Date: November 1995
3. Past Usage:
first usage:
W.N. Street, W.H. Wolberg and O.L. Mangasarian
Nuclear feature extraction for breast tumor diagnosis.
IS&T/SPIE 1993 International Symposium on Electronic Imaging: Science
and Technology, volume 1905, pages 861-870, San Jose, CA, 1993.
OR literature:
O.L. Mangasarian, W.N. Street and W.H. Wolberg.
Breast cancer diagnosis and prognosis via linear programming.
Operations Research, 43(4), pages 570-577, July-August 1995.
Medical literature:
W.H. Wolberg, W.N. Street, and O.L. Mangasarian.
Machine learning techniques to diagnose breast cancer from
fine-needle aspirates.
Cancer Letters 77 (1994) 163-171.
W.H. Wolberg, W.N. Street, and O.L. Mangasarian.
Image analysis and machine learning applied to breast cancer
diagnosis and prognosis.
Analytical and Quantitative Cytology and Histology, Vol. 17
No. 2, pages 77-87, April 1995.
W.H. Wolberg, W.N. Street, D.M. Heisey, and O.L. Mangasarian.
Computerized breast cancer diagnosis and prognosis from fine
needle aspirates.
Archives of Surgery 1995;130:511-516.
W.H. Wolberg, W.N. Street, D.M. Heisey, and O.L. Mangasarian.
Computer-derived nuclear features distinguish malignant from
benign breast cytology.
Human Pathology, 26:792--796, 1995.
See also:
http://www.cs.wisc.edu/~olvi/uwmp/mpml.html
http://www.cs.wisc.edu/~olvi/uwmp/cancer.html
Results:
- predicting field 2, diagnosis: B = benign, M = malignant
- sets are linearly separable using all 30 input features
- best predictive accuracy obtained using one separating plane
in the 3-D space of Worst Area, Worst Smoothness and
Mean Texture. Estimated accuracy 97.5% using repeated
10-fold crossvalidations. Classifier has correctly
diagnosed 176 consecutive new patients as of November
1995.
4. Relevant information
Features are computed from a digitized image of a fine needle
aspirate (FNA) of a breast mass. They describe
characteristics of the cell nuclei present in the image.
A few of the images can be found at
http://www.cs.wisc.edu/~street/images/
Separating plane described above was obtained using
Multisurface Method-Tree (MSM-T) [K. P. Bennett, "Decision Tree
Construction Via Linear Programming." Proceedings of the 4th
Midwest Artificial Intelligence and Cognitive Science Society,
pp. 97-101, 1992], a classification method which uses linear
programming to construct a decision tree. Relevant features
were selected using an exhaustive search in the space of 1-4
features and 1-3 separating planes.
The actual linear program used to obtain the separating plane
in the 3-dimensional space is that described in:
[K. P. Bennett and O. L. Mangasarian: "Robust Linear
Programming Discrimination of Two Linearly Inseparable Sets",
Optimization Methods and Software 1, 1992, 23-34].
This database is also available through the UW CS ftp server:
ftp ftp.cs.wisc.edu
cd math-prog/cpo-dataset/machine-learn/WDBC/
5. Number of instances: 569
6. Number of attributes: 32 (ID, diagnosis, 30 real-valued input features)
7. Attribute information
1) ID number
2) Diagnosis (M = malignant, B = benign)
3-32)
Ten real-valued features are computed for each cell nucleus:
a) radius (mean of distances from center to points on the perimeter)
b) texture (standard deviation of gray-scale values)
c) perimeter
d) area
e) smoothness (local variation in radius lengths)
f) compactness (perimeter^2 / area - 1.0)
g) concavity (severity of concave portions of the contour)
h) concave points (number of concave portions of the contour)
i) symmetry
j) fractal dimension ("coastline approximation" - 1)
Several of the papers listed above contain detailed descriptions of
how these features are computed.
The mean, standard error, and "worst" or largest (mean of the three
largest values) of these features were computed for each image,
resulting in 30 features. For instance, field 3 is Mean Radius, field
13 is Radius SE, field 23 is Worst Radius.
All feature values are recoded with four significant digits.
8. Missing attribute values: none
9. Class distribution: 357 benign, 212 malignant
###Markdown
Data set summary Question 1: In the dataset, which columns are features, which column is the target?From the dataset desciption above, summarize how many features we have and what their column names are. State which column is the target, the values we would like to predict.Note, we will ignore the patient ID column and only consider columns 1-12. TODO: answer question below. *ADD dataset summary* The data
###Code
with open(file_names[1]) as f:
for _ in range(10):
print(f.readline(), end="")
###Output
842302,M,17.99,10.38,122.8,1001,0.1184,0.2776,0.3001,0.1471,0.2419,0.07871,1.095,0.9053,8.589,153.4,0.006399,0.04904,0.05373,0.01587,0.03003,0.006193,25.38,17.33,184.6,2019,0.1622,0.6656,0.7119,0.2654,0.4601,0.1189
842517,M,20.57,17.77,132.9,1326,0.08474,0.07864,0.0869,0.07017,0.1812,0.05667,0.5435,0.7339,3.398,74.08,0.005225,0.01308,0.0186,0.0134,0.01389,0.003532,24.99,23.41,158.8,1956,0.1238,0.1866,0.2416,0.186,0.275,0.08902
84300903,M,19.69,21.25,130,1203,0.1096,0.1599,0.1974,0.1279,0.2069,0.05999,0.7456,0.7869,4.585,94.03,0.00615,0.04006,0.03832,0.02058,0.0225,0.004571,23.57,25.53,152.5,1709,0.1444,0.4245,0.4504,0.243,0.3613,0.08758
84348301,M,11.42,20.38,77.58,386.1,0.1425,0.2839,0.2414,0.1052,0.2597,0.09744,0.4956,1.156,3.445,27.23,0.00911,0.07458,0.05661,0.01867,0.05963,0.009208,14.91,26.5,98.87,567.7,0.2098,0.8663,0.6869,0.2575,0.6638,0.173
84358402,M,20.29,14.34,135.1,1297,0.1003,0.1328,0.198,0.1043,0.1809,0.05883,0.7572,0.7813,5.438,94.44,0.01149,0.02461,0.05688,0.01885,0.01756,0.005115,22.54,16.67,152.2,1575,0.1374,0.205,0.4,0.1625,0.2364,0.07678
843786,M,12.45,15.7,82.57,477.1,0.1278,0.17,0.1578,0.08089,0.2087,0.07613,0.3345,0.8902,2.217,27.19,0.00751,0.03345,0.03672,0.01137,0.02165,0.005082,15.47,23.75,103.4,741.6,0.1791,0.5249,0.5355,0.1741,0.3985,0.1244
844359,M,18.25,19.98,119.6,1040,0.09463,0.109,0.1127,0.074,0.1794,0.05742,0.4467,0.7732,3.18,53.91,0.004314,0.01382,0.02254,0.01039,0.01369,0.002179,22.88,27.66,153.2,1606,0.1442,0.2576,0.3784,0.1932,0.3063,0.08368
84458202,M,13.71,20.83,90.2,577.9,0.1189,0.1645,0.09366,0.05985,0.2196,0.07451,0.5835,1.377,3.856,50.96,0.008805,0.03029,0.02488,0.01448,0.01486,0.005412,17.06,28.14,110.6,897,0.1654,0.3682,0.2678,0.1556,0.3196,0.1151
844981,M,13,21.82,87.5,519.8,0.1273,0.1932,0.1859,0.09353,0.235,0.07389,0.3063,1.002,2.406,24.32,0.005731,0.03502,0.03553,0.01226,0.02143,0.003749,15.49,30.73,106.2,739.3,0.1703,0.5401,0.539,0.206,0.4378,0.1072
84501001,M,12.46,24.04,83.97,475.9,0.1186,0.2396,0.2273,0.08543,0.203,0.08243,0.2976,1.599,2.039,23.94,0.007149,0.07217,0.07743,0.01432,0.01789,0.01008,15.09,40.68,97.65,711.4,0.1853,1.058,1.105,0.221,0.4366,0.2075
###Markdown
2.2 Loading the data with PandasSee `pandas.ipynb` for more information on the pandas python library. TODO: add the names of the 12 columns as a list to the names parameter.
###Code
#TODO: add the names of the 12 columns as a list to the names parameter
data = pd.read_csv( 'wdbc.data',
index_col=False,
names=[ ... ])
data.head()
data.info()
data.describe()
data['diagnosis'].value_counts()
###Output
_____no_output_____
###Markdown
renaming values in `'diagnosis'` column TODO: change 'B' to 'benign' and 'M' to 'malginant'.
###Code
#TODO: change 'B' to 'benign' and 'M' to 'malginant'
...
data['diagnosis'].value_counts()
###Output
_____no_output_____
###Markdown
2.3 Getting an overview by visualizing the dataSee `visualization.ipynb` for more information on plotting in python. Comparing healthy and diseased feature values TODO: create sns.histplot() for all columns in data except id and diagnosis.
###Code
#TODO: create sns.histplot() for all columns in data except id and diagnosis
plt.tight_layout()
###Output
_____no_output_____
###Markdown
Pair-wise correlations of numerical features TODO: Create sns.pairplot() for columns 'radius', 'area', 'concavity', 'symmetry' in data using 'diagnosis' as hue.
###Code
#TODO: Create sns.pairplot() for columns 'radius', 'area', 'concavity', 'symmetry' in data using 'diagnosis' as hue
###Output
_____no_output_____
###Markdown
2.4 Create feature matrix and target vector The feature matrix $X$ is the input to the model. The target vector $y$ contains the values the model should produce, the desired output. See [Python Data Science Handbook 05.02-Introducing-Scikit-Learn](https://github.com/jakevdp/PythonDataScienceHandbook/blob/8a34a4f653bdbdc01415a94dc20d4e9b97438965/notebooks/05.02-Introducing-Scikit-Learn.ipynb) for more information. TODO: use all columns except id and diagnosis for feature matrix X. assign diagnosis column to target vector y
###Code
#TODO: use all columns except id and diagnosis for feature matrix X
X = ...
#TODO: assign diagnosis column to target vector y
y = ...
print(f"feature matrix X shape={X.shape}")
print(f"target vector y shape={y.shape}")
###Output
feature matrix X shape=(569, 10)
target vector y shape=(569,)
###Markdown
2.5 Create training and validation setsWe split the data into two sets:1. Training set - used to create the machine learning model1. Validation set - used to evaluate model performance
###Code
from sklearn.model_selection import train_test_split
X_train, X_val, y_train, y_val = train_test_split(X, y,
test_size=0.1,
stratify=y,
random_state=31)
print(f"train shape={X_train.shape}")
print(f"val shape={X_val.shape}")
print("training samples:")
y_train.value_counts()
print("validation samples:")
y_val.value_counts()
###Output
validation samples:
###Markdown
3. Machine learning: Supervised classification 3.1 Train a linear support vector classifier `SVC(kernel='linear')` See [Python Data Science Handbook 05.07-Support-Vector-Machines](https://github.com/jakevdp/PythonDataScienceHandbook/blob/master/notebooks/05.07-Support-Vector-Machines.ipynb) for more information on linear support vector classifiers.
###Code
from sklearn.svm import SVC
model_svc = SVC(kernel='linear')
model_svc.fit(X_train, y_train)
###Output
_____no_output_____
###Markdown
3.2 Compare training and validation accuracy**Important:** We assess the performance of the model on data the model has not seen yet. This set of data is called the *validation* set. The data used to create the model is used the *training* set.See [Python Data Science Handbook 05.03-Hyperparameters-and-Model-Validation](https://github.com/jakevdp/PythonDataScienceHandbook/blob/master/notebooks/05.03-Hyperparameters-and-Model-Validation.ipynb) for more information on interpreting model performance
###Code
print(f"{model_svc} training accuracy={model_svc.score(X_train, y_train):.2f}")
print(f"{model_svc} validation accuracy={model_svc.score(X_val, y_val):.2f}")
###Output
SVC(kernel='linear') validation accuracy=0.89
###Markdown
Question 2: SVC - Are we over or underfitting? We are over-/under-fitting. We need more/less regularization and a higher/lower `C`. We will try `C=0.01`/`C=100` next. TODO: answer question below. *ANSWER QUESTION HERE* TODO: Re-train the model based on your answer.
###Code
#TODO Re-train the model based on your answer
print(f"{model_svc} training accuracy={model_svc.score(X_train, y_train):.2f}")
print(f"{model_svc} validation accuracy={model_svc.score(X_val, y_val):.2f}")
###Output
SVC(C=100, kernel='linear') training accuracy=0.93
SVC(C=100, kernel='linear') validation accuracy=0.93
###Markdown
Nice increase in accuracy. 3.3 Train a non-linear random forest classifier `RandomForestClassifier(random_state=82)` See [Python Data Science Handbook 05.08-Random-Forests](https://github.com/jakevdp/PythonDataScienceHandbook/blob/master/notebooks/05.08-Random-Forests.ipynb) for more information on random forest classifiers.
###Code
from sklearn.ensemble import RandomForestClassifier
model_rf = RandomForestClassifier(random_state=82)
model_rf.fit(X_train, y_train)
###Output
_____no_output_____
###Markdown
3.4 Compare training and validation accuracy**Important:** We assess the performance of the model on data the model has not seen yet. This set of data is called the *validation* set. The data used to create the model is used the *training* set.See [Python Data Science Handbook 05.03-Hyperparameters-and-Model-Validation](https://github.com/jakevdp/PythonDataScienceHandbook/blob/master/notebooks/05.03-Hyperparameters-and-Model-Validation.ipynb) for more information on interpreting model performance
###Code
print(f"{model_rf} training accuracy={model_rf.score(X_train, y_train):.2f}")
print(f"{model_rf} validation accuracy={model_rf.score(X_val, y_val):.2f}")
###Output
RandomForestClassifier(random_state=82) validation accuracy=0.89
###Markdown
Question 3: RandomForest - Are we over or underfitting? We are over-/under-fitting. We need more/less regularization and adjust/not adjust `max_depth` or adjust/not adjust `max_features`. We will try `max_depth=3`/`max_features=None` next. TODO: answer question below. *ANSWER QUESTION HERE* TODO: Re-train the model based on your answer.
###Code
#TODO: Re-train the model based on your answer
print(f"{model_rf} training accuracy={model_rf.score(X_train, y_train):.2f}")
print(f"{model_rf} validation accuracy={model_rf.score(X_val, y_val):.2f}")
###Output
RandomForestClassifier(max_depth=3, random_state=82) training accuracy=0.96
RandomForestClassifier(max_depth=3, random_state=82) validation accuracy=0.91
###Markdown
This helped a little bit. 3.5 The confusion matrix More information on [wikipedia confusion matrix](https://en.wikipedia.org/wiki/Confusion_matrix) Using the svc model from step 3.2
###Code
from sklearn.metrics import confusion_matrix
mat = confusion_matrix(y_val, model_svc.predict(X_val))
labels = ['benign', 'malignant']
sns.heatmap(mat, square=True, annot=True, cbar=False,
xticklabels=labels,
yticklabels=labels)
plt.xlabel('predicted value')
plt.ylabel('true value')
plt.title(f'{model_svc}');
###Output
_____no_output_____
###Markdown
Using the random forest model from step 3.4
###Code
from sklearn.metrics import confusion_matrix
mat = confusion_matrix(y_val, model_rf.predict(X_val))
labels = ['benign', 'malignant']
sns.heatmap(mat, square=True, annot=True, cbar=False,
xticklabels=labels,
yticklabels=labels)
plt.xlabel('predicted value')
plt.ylabel('true value')
plt.title(f'{model_rf}');
###Output
_____no_output_____
###Markdown
Question 4: What kind of errors do SVC and RandomForest make?Based on the confusion matrices above, summarize false positives and false negatives of SVC and RandomForest make? TODO: answer question below. *ANSWER QUESTION HERE* 3.6 Summarize the different scores
###Code
print(f'--- {model_svc} ---')
print('training accuracy (all data) {:.3f}'.format(model_svc.score(X_train, y_train)))
print('validation accuracy (new data) {:.3f}'.format(model_svc.score(X_val, y_val)))
print(f'--- {model_rf} ---')
print('training accuracy (all data) {:.3f}'.format(model_rf.score(X_train, y_train)))
print('validation accuracy (new data) {:.3f}'.format(model_rf.score(X_val, y_val)))
###Output
--- SVC(C=100, kernel='linear') ---
training accuracy (all data) 0.926
validation accuracy (new data) 0.930
--- RandomForestClassifier(max_depth=3, random_state=82) ---
training accuracy (all data) 0.965
validation accuracy (new data) 0.912
|
course1/.ipynb_checkpoints/Mnist1-LearningRateScheduler-checkpoint.ipynb | ###Markdown
Fashion Mnist: example with a Learning Rate (LR) scheduler
###Code
import numpy as np
import pandas as pd
import tensorflow as tf
import matplotlib.pyplot as plt
# check TensorFlow version
print(tf.__version__)
data = tf.keras.datasets.fashion_mnist
# globals
EPOCHS = 60
BATCH_SIZE = 256
(train_images, train_labels), (test_images, test_labels) = data.load_data()
print('Number of training images:', train_images.shape[0])
print('Number of test images:', test_images.shape[0])
# how many distinct classes
np.unique(train_labels)
# ok 10 distinct classes labeled as 0..9
# images are greyscale images
train_images.shape
###Output
_____no_output_____
###Markdown
first of all, normalize imagestrain_images /= 255.test_images /= 255.
###Code
# let's sess one image
plt.imshow(train_images[0], cmap = 'Greys');
# let's build a first classification model
def build_model():
model = tf.keras.models.Sequential([
tf.keras.layers.Flatten(input_shape=(28,28)),
tf.keras.layers.Dense(128, activation = 'relu'),
# adding these two intermediate layers I go from 0.85 to 0.87 validation accuracy
tf.keras.layers.Dense(64, activation = 'relu'),
tf.keras.layers.Dense(32, activation = 'relu'),
tf.keras.layers.Dense(10, activation = 'softmax')
])
model.compile(optimizer='adam', loss = 'sparse_categorical_crossentropy',
metrics = ['accuracy'])
return model
# let's see how many parameters, etc
model = build_model()
model.summary()
# add a Callback that changes the learning rate during the training
def lr_schedule(epoch):
lr_initial = 0.001
ep_start_decay = 20
decay_rate = 0.9
if epoch > ep_start_decay:
lr = lr_initial * decay_rate ** (epoch - ep_start_decay)
else:
lr = lr_initial
return lr
# let's plot it
def plot_ls_scheduler(fn):
vet_epochs = np.arange(EPOCHS)
vet_lr = [fn(epoch) for epoch in vet_epochs]
plt.plot(vet_epochs, vet_lr, 'b*')
plt.grid()
# here we plot
plot_ls_scheduler(lr_schedule)
# the callback for the LR cheduler
cbk1 = tf.keras.callbacks.LearningRateScheduler(lr_schedule, verbose = 1)
history = model.fit(train_images, train_labels, epochs = EPOCHS, batch_size = BATCH_SIZE, validation_split = 0.1, callbacks = [cbk1])
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
plt.xlabel('epoch')
plt.ylabel('loss')
plt.plot(loss, label = 'loss')
plt.plot(val_loss, label = 'validation loss')
plt.legend(loc = 'upper right')
plt.grid();
# wants to see if it starts overfitting
START = 20
plt.xlabel('epoch')
plt.ylabel('loss')
plt.plot(loss[START:], label = 'loss')
plt.plot(val_loss[START:], label = 'validation loss')
plt.legend(loc = 'upper right')
plt.grid();
# ok it has slightly started overfitting.. as we can see from epochs 15 validation loss becomes higher and training loss is still decreasing
plt.xlabel('epoch')
plt.ylabel('accuracy')
plt.plot(accuracy, label = 'accuracy')
plt.plot(val_accuracy, label = 'validation accuracy')
plt.legend(loc = 'lower right')
plt.grid();
model.evaluate(test_images, test_labels)
###Output
313/313 [==============================] - 0s 925us/step - loss: 0.4801 - accuracy: 0.8766
|
.ipynb_checkpoints/Europeandataporta-checkpoint.ipynb | ###Markdown
Europeandataportal.eu Test access SPARQL endpoint
###Code
import sys,json
import pandas as pd
from SPARQLWrapper import SPARQLWrapper, JSON
endpoint_url = "https://www.europeandataportal.eu/sparql"
query = """SELECT * WHERE {
?s ?p ?o
}
limit 100"""
def get_sparql_dataframe(endpoint_url, query):
# print (query)
user_agent = "user: salgo60/%s.%s" % (sys.version_info[0], sys.version_info[1])
sparql = SPARQLWrapper(endpoint_url, agent=user_agent)
sparql.setQuery(query)
sparql.setReturnFormat(JSON)
result = sparql.query().convert()
print(type(result))
#print(results)
column_names = ["s", "p", "q"]
cols = results["head"]
out = []
for row in processed_results['head']:
item = []
print (cols)
for c in cols:
print(row)
item.append(row.get(c, {}).get('value'))
out.append(item)
return pd.DataFrame(out, columns=cols)
df = get_sparql_dataframe(endpoint_url, query)
###Output
<class 'dict'>
{'link': [], 'vars': ['s', 'p', 'o']}
link
|
keras/keras_multiclass_classification.ipynb | ###Markdown
AbstractThis notebook is an example that shows how to build a multiclass classification model with the Keras API. References: - [Multiclass Classification with Keras](http://machinelearningmastery.com/multi-class-classification-tutorial-keras-deep-learning-library/)- [Display deep learning model training history with Keras](http://machinelearningmastery.com/display-deep-learning-model-training-history-in-keras/) Build Model
###Code
import keras
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation
from keras.optimizers import SGD
# Generate dummy data
import numpy as np
import pandas as pd
x_train = np.random.random((1000, 20))
y_train = keras.utils.to_categorical(np.random.randint(10, size=(1000, 1)), num_classes=10)
x_test = np.random.random((100, 20))
y_test = keras.utils.to_categorical(np.random.randint(10, size=(100, 1)), num_classes=10)
model = Sequential()
# Dense(64) is a fully-connected layer with 64 hidden units.
# in the first layer, you must specify the expected input data shape:
# here, 20-dimensional vectors.
model.add(Dense(64, activation='relu', input_dim=20))
model.add(Dropout(0.5))
model.add(Dense(64, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(10, activation='softmax'))
sgd = SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)
model.compile(loss='categorical_crossentropy',
optimizer=sgd,
metrics=['accuracy'])
model.summary()
print("stateful:", model.stateful)
###Output
Using TensorFlow backend.
###Markdown
Training
###Code
# prevent the application from occupying all the memory in the GPU.
import tensorflow as tf
from keras.backend.tensorflow_backend import set_session
config = tf.ConfigProto()
#config.gpu_options.per_process_gpu_memory_fraction = 0.2
config.gpu_options.allow_growth=True
session = tf.Session(config=config)
set_session(session)
history = model.fit(x_train, y_train, epochs=100, batch_size=128, verbose=0)
history.history.keys()
import matplotlib.pyplot as plt
# configure notebook to display plots
%matplotlib inline
from matplotlib.pylab import rcParams
rcParams['figure.figsize'] = 15,6
metrics = pd.DataFrame(history.history)
def plot_metrics(metrics):
'''
'''
ax = plt.subplot(211)
# plot the evolution of loss and accurary during the training peroid
ax.plot(metrics['loss'], color='r')
#plt.plot(metrics['acc'])
plt.ylabel('loss')
plt.xlabel('epochs')
ax = plt.subplot(212)
ax.plot(metrics['acc'])
plt.xlabel('epochs')
_ = plt.ylabel('accuracy')
plot_metrics(metrics)
###Output
_____no_output_____
###Markdown
The evolution of `loss` and `accurary` metrics show that the model is constantly approaching to a more optimal values. Yet, there still certain potential to get to a more optimal point, if one adds more `epochs`.Indeed, as we see from the following experiment, on top of the model trained after `100 epochs`, we added another 100 epochs, the model continues to evolve torwards a more optimal position, i.e. loss decreases and accuracy increases.
###Code
history = model.fit(x_train, y_train, epochs=100, batch_size=128, verbose=0)
plot_metrics(pd.DataFrame(history.history))
###Output
_____no_output_____
###Markdown
Evaluation
###Code
score = model.evaluate(x_test, y_test, batch_size=128)
print(score)
# as long as the Notebook is alive, the application would keep occupying the GPU memory.
session.close()
###Output
_____no_output_____ |
assignments/.ipynb_checkpoints/assignment-3-checkpoint.ipynb | ###Markdown
Assignment 3 - Handling arrays with NumPy Loan Pham and Brandan Owens Exercise 1 Import the data set “Boston_Housing.csv” Extract ['PRICE'] into an arrayPlot a histogram of housing priceFind the mean, max, 75th percentile of the housing price.Create an array of two rows, with the first row from [‘’RM”], and the second row from [“PRICE”]Find the number of houses with “RM” Find the mean of the housing price, with “RM” > 5 Plot a scatter plot to show the relationship between number of rooms and housing price (use plt.scatter())
###Code
# import the data set
import os
import pandas as pd
import numpy as np
data = pd.read_csv("../dataFiles/Boston_Housing.csv")
data
# extract price
price = np.array(data["PRICE"])
price
#plot histogram
import matplotlib.pyplot as plt
plt.hist(price)
# find the mean, max, 75th percentile of the housing price
print(price.max())
print(price.mean())
print(np.percentile(price,75))
# create an array of two rows from "RM" and "PRICE"
rm = np.array(data["RM"])
rm
first_row = rm[0:2]
print(first_row)
second_row = price[0:2]
print(second_row)
# Find the number of houses with “RM” < 5
# print(np.sum(rm < 5))
np.count_nonzero(rm < 5)
# find the mean housing price where "RM" is greater than 5
np.mean(rm > 5)
# plot scatterplot to show the relationship between number of rooms and housing price
plt.scatter(rm, price)
###Output
_____no_output_____
###Markdown
Exercise 2Create a 1000x1 array of numbers, x which divides the interval from -10 to 10 into equal widths.Reshape the array x into 20x50 array, then: (a) Find the shape, dimension, and data type of the array. (b) Access the last element of each row (c) Access first element and then every other elements of each row (d) Access the subarray 7th to 10th rows and 5th to 11th columns (e) find the sum of the 7th column (f) Print the elements in each column which is greater 0. (g) Replace all the negative numbers of the array with 0. (h) Sort each column of the array in descending order.
###Code
# Create a 1000x1 array of numbers, x which divides the interval from -10 to 10 into equal widths.
x = np.linspace(-10,10, num =1000)
x.reshape((1000,1))
# reshape the array into 20x50
x_reshaped = x.reshape(20,50)
x_reshaped
# (a) Find the shape, dimension, and data type of the array.
print(x_reshaped.shape)
print(x_reshaped.ndim)
print(x_reshaped.dtype)
# (b) Access the last element of each row
x_reshaped[:,-1]
#(c) Access first element and then every other elements of each row
x_reshaped[::,::2]
# (d) Access the subarray 7th to 10th rows and 5th to 11th columns
acc = x_reshaped[6:10, 4:11]
acc
# (e) find the sum of the 7th column
sum_col = x_reshaped[:, 6].sum()
sum_col
# (f) print the elements in each column which is greater 0.
for row in x_reshaped:
for element in row:
if element > 0:
print(element)
# (g) Replace all the negative numbers of the array with 0.
num = x_reshaped
print(np.where(num < 0, 0, num))
# (h) sort each column of the array in descending order.
arrg = (-np.sort(-x_reshaped))
arrg
###Output
_____no_output_____
###Markdown
Exercise 3 This exercise illustrates a simple machine learning algorithm. Suppose you have two arrays of numbers. You are going to teach the machine to learn the relationship between the two arrays.
###Code
# - Create an array of 100 random numbers, x. Each number is between 0 and 1.
x = np.random.uniform(0, 1, 100)
x
# Create an array, y, and y = 3x + 2
y = 3*x + 2
y
# - Create two numbers, a and b. Initialize them to be 0.
a = 0
b = 0
# - Create 3 empty lists.
list1 = []
list2 = []
list3 = []
# - the machine predicts the value of y, y_pred: y_pred = a*x+b
# calculate the cost value = sum of the square of difference between y_pred and actual y
# Iterate the above optimization steps 1000 times.
y_pred = a*x +b
for i in range(1000):
cost = np.dot((y_pred - y), (y_pred - y))
# update the values of a and b
da = 2*np.dot((y_pred - y),(x))
db = 2*np.sum(y_pred - y)
a = a - 0.001*da
b = b - 0.001*db
y_pred = a*x + b
# - Store the values of a, b and cost in the 3 lists you created.
list1.append(a)
list2.append(b)
list3.append(cost)
# Plot a graph to show how the values of a and b change over iteration.
plt.plot(list1, label='a')
plt.plot(list2, label='b')
plt.legend()
plt.plot(list3, label='list3')
plt.legend()
###Output
_____no_output_____ |
notebooks/lstm-gait-classifier.ipynb | ###Markdown
Preparing dataset:
###Code
def prepare_dataset(features: str = 'upcv1/train/coordinates'):
print(f"Getting {features} features...")
with open(file=f'{FEATURES_FOLDER}{features}.pickle', mode='rb+') as file:
X, y = pickle.load(file=file)
S, W, C = X.shape
assert S == y.shape[0]
print(f"""
{X.shape}, {y.shape}
Selected sequences: {S}
Sequence window size: {W}
Feature vector size: {C}
Unique subjects: {len(np.unique(y))}""")
return X, y
"""
-- dataset: upcv1, ks20, oumvlp
-- augmentation: rotation and flipping
-- feature combinations: coordinates, velocity, distance, orientation
-- dimentionality reduction: PCA w/ varying dimension
"""
FEATURES = [
('upcv1/train/coordinates', 'upcv1/test/coordinates'),
('upcv1/train/coordinates-augmented', 'upcv1/test/coordinates'),
('upcv1/train/velocity', 'upcv1/test/velocity'),
('upcv1/train/distance', 'upcv1/test/distance'),
('upcv1/train/orientation', 'upcv1/test/orientation'),
('upcv1/train/c-v', 'upcv1/test/c-v'),
('upcv1/train/d-o', 'upcv1/test/d-o'),
('upcv1/train/c-d-o', 'upcv1/test/c-d-o'),
('upcv1/train/c-v-d-o', 'upcv1/test/c-v-d-o'),
('ks20/train/coordinates', 'ks20/test/coordinates'),
('ks20/train/coordinates-augmented', 'ks20/test/coordinates'),
('ks20/train/velocity', 'ks20/test/velocity'),
('ks20/train/distance', 'ks20/test/distance'),
('ks20/train/orientation', 'ks20/test/orientation'),
('ks20/train/c-v', 'ks20/test/c-v'),
('ks20/train/d-o', 'ks20/test/d-o'),
('ks20/train/c-d-o', 'ks20/test/c-d-o'),
('ks20/train/c-v-d-o', 'ks20/test/c-v-d-o')
]
train_features, test_features = FEATURES[7]
X_train, y_train = prepare_dataset(features=train_features)
X_test, y_test = prepare_dataset(features=test_features)
def get_pca(X, R=None):
"""
R - desired reduced dimension size
"""
C, F, L = X.shape
if R is None:
R = L
X = X.reshape(C*F, L)
pca = PCA(n_components=R).fit(X)
return pca
def transform_pca(pca, X):
C, F, L = X.shape
X = X.reshape(C*F, -1)
return pca.transform(X).reshape(C, F, -1)
pca = get_pca(X_train, R=None)
X_train = transform_pca(pca, X_train)
X_test = transform_pca(pca, X_test)
X_train.shape
###Output
_____no_output_____
###Markdown
Set Parameters:
###Code
n_classes = len(np.unique(y_train))
# n_steps - timesteps per series
# n_input - num input parameters per timestep
training_data_count, n_steps, n_input = X_train.shape
test_data_count, _, _ = X_test.shape
global_step = tf.Variable(0, trainable=False)
learning_rate = tf.convert_to_tensor(0.001)
# Hidden layer num of features
n_hidden = 64
batch_size = 128
epochs = 3000
###Output
_____no_output_____
###Markdown
Utility functions for training:
###Code
def LSTM(_X, _weights, _biases):
# model architecture based on "guillaume-chevalier" and "aymericdamien" under the MIT license.
_X = tf.transpose(_X, [1, 0, 2]) # permute n_steps and batch_size
_X = tf.reshape(_X, [-1, n_input])
# Rectified Linear Unit activation function used
_X = tf.nn.relu(tf.matmul(_X, _weights['hidden']) + _biases['hidden'])
# Split data because rnn cell need a list of inputs for the RNN inner loop
_X = tf.split(_X, n_steps, 0)
# Define two stacked LSTM cells (two recurrent layers deep) with tensorflow
lstm_cell_1 = tf.nn.rnn_cell.LSTMCell(n_hidden, forget_bias=1.0, state_is_tuple=True)
lstm_cell_2 = tf.nn.rnn_cell.LSTMCell(n_hidden, forget_bias=1.0, state_is_tuple=True)
lstm_cells = tf.nn.rnn_cell.MultiRNNCell([lstm_cell_1, lstm_cell_2], state_is_tuple=True)
outputs, states = tf.nn.static_rnn(lstm_cells, _X, dtype=tf.float32)
# A single output is produced, in style of "many to one" classifier, refer to http://karpathy.github.io/2015/05/21/rnn-effectiveness/ for details
lstm_last_output = outputs[-1]
# Linear activation
return tf.matmul(lstm_last_output, _weights['out']) + _biases['out']
def extract_batch_size(_train, _labels, _unsampled, batch_size):
# Fetch a "batch_size" amount of data and labels from "(X|y)_train" data.
# Elements of each batch are chosen randomly, without replacement, from X_train with corresponding label from Y_train
# unsampled_indices keeps track of sampled data ensuring non-replacement. Resets when remaining datapoints < batch_size
shape = list(_train.shape)
shape[0] = batch_size
batch_s = np.empty(shape)
batch_labels = np.empty((batch_size,1))
for i in range(batch_size):
# Loop index
# index = random sample from _unsampled (indices)
index = random.choice(_unsampled)
batch_s[i] = _train[index]
batch_labels[i] = _labels[index]
_unsampled.remove(index)
return batch_s, batch_labels, _unsampled
def one_hot(y_):
# One hot encoding of the network outputs
# e.g.: [[5], [0], [3]] --> [[0, 0, 0, 0, 0, 1], [1, 0, 0, 0, 0, 0], [0, 0, 0, 1, 0, 0]]
y_ = y_.reshape(len(y_))
n_values = int(np.max(y_)) + 1
return np.eye(n_values)[np.array(y_, dtype=np.int32)] # Returns FLOATS
###Output
_____no_output_____
###Markdown
Build the network:
###Code
# Graph input/output
x = tf.placeholder(tf.float32, [None, n_steps, n_input])
y = tf.placeholder(tf.float32, [None, n_classes])
# Graph weights
weights = {
'hidden': tf.Variable(tf.random_normal([n_input, n_hidden])), # Hidden layer weights
'out': tf.Variable(tf.random_normal([n_hidden, n_classes], mean=1.0))
}
biases = {
'hidden': tf.Variable(tf.random_normal([n_hidden])),
'out': tf.Variable(tf.random_normal([n_classes]))
}
pred = LSTM(x, weights, biases)
# Loss, optimizer and evaluation
l2 = 0.001 * sum(
tf.nn.l2_loss(tf_var) for tf_var in tf.trainable_variables()
)
# L2 loss prevents this overkill neural network to overfit the data
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(labels=y, logits=pred)) + l2 # Softmax loss
# if decaying_learning_rate:
# learning_rate = tf.train.exponential_decay(init_learning_rate, global_step*batch_size, decay_steps, decay_rate, staircase=True)
#decayed_learning_rate = learning_rate * decay_rate ^ (global_step / decay_steps) # exponentially decayed learning rate
optimizer = tf.train.AdamOptimizer(learning_rate=learning_rate).minimize(cost,global_step=global_step) # Adam Optimizer
correct_pred = tf.equal(tf.argmax(pred,1), tf.argmax(y,1))
accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
###Output
WARNING:tensorflow:`tf.nn.rnn_cell.MultiRNNCell` is deprecated. This class is equivalent as `tf.keras.layers.StackedRNNCells`, and will be replaced by that in Tensorflow 2.0.
WARNING:tensorflow:From <ipython-input-9-5a77964f3916>:15: static_rnn (from tensorflow.python.ops.rnn) is deprecated and will be removed in a future version.
Instructions for updating:
Please use `keras.layers.RNN(cell, unroll=True)`, which is equivalent to this API
WARNING:tensorflow:From /home/user/.local/lib/python3.8/site-packages/keras/layers/legacy_rnn/rnn_cell_impl.py:979: calling Zeros.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.
Instructions for updating:
Call initializer instance with the dtype argument instead of passing it to the constructor
###Markdown
Train the network:
###Code
print(f"Training on {train_features}")
test_losses = []
test_accuracies = []
train_losses = []
train_accuracies = []
sess = tf.InteractiveSession(config=tf.ConfigProto(log_device_placement=True))
init = tf.global_variables_initializer()
sess.run(init)
# Perform Training steps with "batch_size" amount of data at each loop.
# Elements of each batch are chosen randomly, without replacement, from X_train,
# restarting when remaining datapoints < batch_size
step = 1
time_start = time.time()
unsampled_indices = list(range(0,len(X_train)))
for i in range(epochs):
if len(unsampled_indices) < batch_size:
unsampled_indices = list(range(0,len(X_train)))
batch_xs, raw_labels, unsampled_indicies = extract_batch_size(X_train, y_train, unsampled_indices, batch_size)
batch_ys = one_hot(raw_labels)
# check that encoded output is same length as num_classes, if not, pad it
if len(batch_ys[0]) < n_classes:
temp_ys = np.zeros((batch_size, n_classes))
temp_ys[:batch_ys.shape[0],:batch_ys.shape[1]] = batch_ys
batch_ys = temp_ys
# Fit training using batch data
_, loss, acc = sess.run(
[optimizer, cost, accuracy],
feed_dict={
x: batch_xs,
y: batch_ys
}
)
train_losses.append(loss)
train_accuracies.append(acc)
# Evaluation on the test set (no learning made here - just evaluation for diagnosis)
test_loss, test_acc = sess.run(
[cost, accuracy],
feed_dict={
x: X_test,
y: one_hot(y_test)
}
)
test_losses.append(test_loss)
test_accuracies.append(test_acc)
# print(f"Epoch #{i} Loss = {loss:.2f} Accuracy = {acc:.2f}")
if i % 100 == 0:
print(f"Epoch {i}/{epochs} Loss={loss:.2f} Accuracy={acc:.2f} Test Loss={test_loss:.2f} Test Accuracy={test_acc:.2f}")
# Performance on test data
one_hot_predictions, accuracy, final_loss = sess.run(
[pred, accuracy, cost],
feed_dict={
x: X_test,
y: one_hot(y_test)
}
)
# test_losses.append(final_loss)
# test_accuracies.append(accuracy)
print(f"Training finished. Loss = {final_loss} Accuracy = {accuracy}")
print(f"Duration: {time.time() - time_start} sec")
###Output
Training on upcv1/train/c-d-o
Device mapping: no known devices.
Epoch 0/3000 Loss=45.71 Accuracy=0.01 Test Loss=45.23 Test Accuracy=0.05
Epoch 100/3000 Loss=34.90 Accuracy=1.00 Test Loss=35.30 Test Accuracy=0.86
Epoch 200/3000 Loss=30.04 Accuracy=1.00 Test Loss=30.42 Test Accuracy=0.88
Epoch 300/3000 Loss=25.87 Accuracy=1.00 Test Loss=26.25 Test Accuracy=0.88
Epoch 400/3000 Loss=22.29 Accuracy=1.00 Test Loss=22.67 Test Accuracy=0.88
Epoch 500/3000 Loss=19.21 Accuracy=1.00 Test Loss=19.59 Test Accuracy=0.89
Epoch 600/3000 Loss=16.56 Accuracy=1.00 Test Loss=16.94 Test Accuracy=0.88
Epoch 700/3000 Loss=14.27 Accuracy=1.00 Test Loss=14.65 Test Accuracy=0.89
Epoch 800/3000 Loss=12.29 Accuracy=1.00 Test Loss=12.66 Test Accuracy=0.90
Epoch 900/3000 Loss=10.58 Accuracy=1.00 Test Loss=10.93 Test Accuracy=0.90
Epoch 1000/3000 Loss=9.11 Accuracy=1.00 Test Loss=9.44 Test Accuracy=0.91
Epoch 1100/3000 Loss=7.83 Accuracy=1.00 Test Loss=8.15 Test Accuracy=0.92
Epoch 1200/3000 Loss=6.73 Accuracy=1.00 Test Loss=7.04 Test Accuracy=0.92
Epoch 1300/3000 Loss=5.88 Accuracy=1.00 Test Loss=6.10 Test Accuracy=0.92
Epoch 1400/3000 Loss=5.06 Accuracy=1.00 Test Loss=5.27 Test Accuracy=0.92
Epoch 1500/3000 Loss=4.37 Accuracy=1.00 Test Loss=4.60 Test Accuracy=0.92
Epoch 1600/3000 Loss=3.77 Accuracy=1.00 Test Loss=4.02 Test Accuracy=0.92
Epoch 1700/3000 Loss=3.26 Accuracy=1.00 Test Loss=3.51 Test Accuracy=0.92
Epoch 1800/3000 Loss=2.81 Accuracy=1.00 Test Loss=3.07 Test Accuracy=0.92
Epoch 1900/3000 Loss=2.42 Accuracy=1.00 Test Loss=2.69 Test Accuracy=0.92
Epoch 2000/3000 Loss=2.09 Accuracy=1.00 Test Loss=2.36 Test Accuracy=0.92
Epoch 2100/3000 Loss=1.80 Accuracy=1.00 Test Loss=2.08 Test Accuracy=0.91
Epoch 2200/3000 Loss=1.55 Accuracy=1.00 Test Loss=1.84 Test Accuracy=0.91
Epoch 2300/3000 Loss=1.33 Accuracy=1.00 Test Loss=1.65 Test Accuracy=0.91
Epoch 2400/3000 Loss=1.15 Accuracy=1.00 Test Loss=1.49 Test Accuracy=0.91
Epoch 2500/3000 Loss=0.99 Accuracy=1.00 Test Loss=1.33 Test Accuracy=0.90
Epoch 2600/3000 Loss=0.85 Accuracy=1.00 Test Loss=1.20 Test Accuracy=0.91
Epoch 2700/3000 Loss=0.89 Accuracy=0.97 Test Loss=1.21 Test Accuracy=0.88
Epoch 2800/3000 Loss=0.69 Accuracy=1.00 Test Loss=0.89 Test Accuracy=0.94
Epoch 2900/3000 Loss=0.60 Accuracy=1.00 Test Loss=0.81 Test Accuracy=0.94
Training finished. Loss = 0.7445202469825745 Accuracy = 0.9446367025375366
Duration: 610.8951778411865 sec
###Markdown
Results:
###Code
# summarize history for accuracy
plt.plot(train_accuracies)
plt.plot(test_accuracies)
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='lower right')
plt.show()
# summarize history for loss
plt.plot(train_losses)
plt.plot(test_losses)
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper right')
plt.show()
predictions = one_hot_predictions.argmax(1)
print("Testing Accuracy: {}%".format(100*accuracy))
print("Precision: {}%".format(100*metrics.precision_score(y_test, predictions, average="weighted")))
print("Recall: {}%".format(100*metrics.recall_score(y_test, predictions, average="weighted")))
print("f1_score: {}%".format(100*metrics.f1_score(y_test, predictions, average="weighted")))
print("Confusion Matrix: Created using test set of {} datapoints, normalised to % of each class in the test dataset".format(len(y_test)))
confusion_matrix = metrics.confusion_matrix(y_test, predictions)
normalised_confusion_matrix = np.array(confusion_matrix, dtype=np.float32)/np.sum(confusion_matrix)*100
# Plot Results:
plt.figure(figsize=(12, 12))
plt.imshow(
normalised_confusion_matrix,
interpolation='nearest',
cmap=plt.cm.Blues
)
plt.title("Confusion matrix \n(normalised to % of total test data)")
plt.colorbar()
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
plt.show()
###Output
Testing Accuracy: 94.46367025375366%
Precision: 95.23200383772824%
Recall: 94.4636678200692%
f1_score: 94.3922567745365%
Confusion Matrix: Created using test set of 578 datapoints, normalised to % of each class in the test dataset
|
Notebooks/Implementations/JUP_SURGP_GLO_B_D__LEI_Evaluation.ipynb | ###Markdown
Calculate LEI
###Code
for ghsl_file in GHSL_files:
print(f'{ghsl_file}')
out_file = ghsl_file.replace(".tif", "new_LEI_90_00.csv")
if not os.path.exists(out_file):
lei = calculate_LEI(ghsl_file, old_list = [5,6], new_list=[4])
xx = pd.DataFrame(lei, columns=['geometry', 'old', 'total'])
xx['LEI'] = xx['old'] / xx['total']
xx.to_csv(out_file)
# Process LEI results
base_folder = '/home/wb411133/data/Projects/LEI'
all_results_files = []
for root, folders, files in os.walk(base_folder):
for f in files:
if "GHSLnew_LEI_90_00" in f:
all_results_files.append(os.path.join(root, f))
summarized_results = {}
for res_file in all_results_files:
res = summarize_LEI(res_file)
baseName = os.path.basename(os.path.dirname(res_file))
summarized_results[baseName] = res
all_results = pd.DataFrame(summarized_results).transpose()
# Old test to determine which files were not processed correctly
#bas_res = all_results[all_results['Expansion'] == 123282000.0].index
all_results.head()
all_results.to_csv(os.path.join(LEI_folder, "Summarized_LEI_Results_90_00.csv"))
###Output
_____no_output_____
###Markdown
Summarize total built per city
###Code
all_res = {}
for g_file in GHSL_files:
city = os.path.basename(os.path.dirname(g_file))
inR = rasterio.open(g_file)
inD = inR.read()
built2014 = (inD >= 3).sum() * (30 * 30)
built2000 = (inD >= 4).sum() * (30 * 30)
built1990 = (inD >= 5).sum() * (30 * 30)
built1975 = (inD >= 6).sum() * (30 * 30)
all_res[city] = [built1975, built1990, built2000, built2014]
print(city)
xx = pd.DataFrame(all_res).head().transpose()
xx.columns = ['built75', 'built90', 'built00', 'built14']
#xx[xx.index.isin(['1'])]
xx.head()
xx.to_csv("/home/wb411133/temp/LEI_cities_built.csv")
###Output
_____no_output_____
###Markdown
Combining results
###Code
csv_files = [x for x in os.listdir(LEI_folder) if x[-4:] == ".csv"]
lei0014 = pd.read_csv(os.path.join(LEI_folder, 'Summarized_LEI_Results.csv'),index_col=0)
lei0014.columns = ["%s_0014" % x for x in lei0014.columns]
lei9014 = pd.read_csv(os.path.join(LEI_folder, 'Summarized_LEI_Results_90_0014.csv'),index_col=0)
lei9014.columns = ["%s_9014" % x for x in lei9014.columns]
lei9000 = pd.read_csv(os.path.join(LEI_folder, 'Summarized_LEI_Results_90_00.csv'),index_col=0)
lei9000.columns = ["%s_9000" % x for x in lei9000.columns]
built_area = pd.read_csv("/home/wb411133/temp/LEI_cities_built.csv",index_col=0)
built_area.columns = ["%s_BUILT" % x for x in built_area.columns]
combined_results = lei0014.join(lei9014).join(lei9000).join(built_area)
combined_results.to_csv(os.path.join(LEI_folder, 'LEI_COMBINED.csv'))
combined_results['Expansion_0014'] + combined_results['Infill_0014'] + combined_results['Leapfrog_0014'] - (combined_results['built14_BUILT'] - combined_results['built00_BUILT'])
built_area.head()
###Output
_____no_output_____
###Markdown
Summarizing methods
###Code
in_ghsl = "/home/wb411133/data/Projects/LEI/1/GHSL.tif"
inR = rasterio.open(in_ghsl)
inD = inR.read()
# Get cell counts of each built category
built2014 = (inD >= 3).sum()
built2000 = (inD >= 4).sum()
built1990 = (inD >= 5).sum()
built1975 = (inD >= 6).sum()
print("%s\n%s\n%s\n%s" % (built2014, built2000, built1990, built1975))
lei_2000_2014 = calculate_LEI(in_ghsl, old_list = [4,5,6], new_list=[3])
lei_1990_2000 = calculate_LEI(in_ghsl, old_list = [5,6], new_list=[4])
xx = pd.DataFrame(lei, columns=['geometry', 'old', 'total'])
xx['LEI'] = xx['old'] / xx['total']
in_file = "/home/wb411133/data/Projects/LEI/1/GHSLnew_LEI_90_00.csv"
inD = pd.read_csv(in_file, index_col=0)
inD.head()
summarize_LEI(in_file)
###Output
_____no_output_____
###Markdown
DEBUGGING
###Code
bad_files = []
for root, dirs, files in os.walk('/home/wb411133/data/Projects/LEI/'):
for f in files:
if "90_00.csv" in f:
bad_files.append(os.path.join(root, f))
bad_files
import shutil
for b in bad_files:
new_file = b.replace("_90_00", "_90_0014")
shutil.move(b, new_file)
###Output
_____no_output_____
###Markdown
Calculating LEIThis script is used for exploring LEI methods - in order to calculate LEI proper, look for the LEIFast.py script in GOST_Rocks/Urban; this implements multi-threading.
###Code
import os, sys, logging
import geojson, rasterio
import rasterio.features
import pandas as pd
import numpy as np
from shapely.geometry import shape, GeometryCollection
from shapely.wkt import loads
from matplotlib import pyplot
from rasterio.plot import show, show_hist
#Import GOST urban functions
sys.path.append("../../")
from src.LEI import *
calculate_LEI?
LEI_folder = '/home/wb411133/data/Projects/LEI'
results = {}
GHSL_files = []
for root, dirs, files in os.walk(LEI_folder):
if os.path.exists(os.path.join(root, "GHSL.tif")):
GHSL_files.append(os.path.join(root, "GHSL.tif"))
try:
results[os.path.basename(root)] = [len(files), os.stat(os.path.join(root, "GHSL.tif")).st_size]
if len(files) != 6:
print("%s - %s" % (os.path.basename(root), os.stat(os.path.join(root, "GHSL.tif")).st_size))
except:
pass
###Output
_____no_output_____
###Markdown
Vizualize raster data - GHSL
###Code
root = '/home/wb411133/data/Projects/LEI/634/'
inputGHSL = os.path.join(root, "GHSL.tif")
inRaster = rasterio.open(inputGHSL)
inR = inRaster.read()
newR = (inR == 3).astype('int')
oldR = (np.isin(inR, [4,5,6])).astype('int')
fig, (axr, axg) = pyplot.subplots(1, 2, figsize=(20,20))
show(oldR, ax=axr, title='OLD')
show(newR, ax=axg, title='NEW')
#write out raster to file
outProperties = inRaster.profile
outRaster = outRaster.astype('int32')
outProperties['dtype'] = 'int32'
with rasterio.open(inputGHSL.replace(".tif", "_LEI.tif"), 'w', **outProperties) as out:
out.write(outRaster)
###Output
_____no_output_____ |
assets/sample/model-logger.ipynb | ###Markdown
Hey !! Let's get started
###Code
!pip install modellogger
from modellogger.modellogger import ModelLogger
###Output
_____no_output_____
###Markdown
Setup model-logger
###Code
# create an instance of modellogger ,It will automatically setup the db for you :)
mlog = ModelLogger('mydb.db')
import numpy as np
import pandas as pd
from matplotlib import rcParams
from sklearn.datasets import load_boston
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
boston = load_boston()
bos = pd.DataFrame(boston.data)
bos.columns = boston.feature_names
bos['PRICE'] = boston.target
###Output
_____no_output_____
###Markdown
Train-->Predict-->Store-->Repeat
###Code
#boston house pricing dataset
X = bos.drop('PRICE', axis = 1)
Y = bos['PRICE']
X_train, X_test, Y_train, Y_test =train_test_split(X, Y, test_size = 0.33, random_state = 5)
#scoring
def score(Y_test,Y_pred):
return mean_squared_error(Y_test,Y_pred)
#train the model as normal
lr1 = LinearRegression()
lr1.fit(X_train, Y_train)
Y_pred = lr1.predict(X_test)
#Store the model
mlog.store_model('logistic_v1', lr1 , X_train , score(Y_test,Y_pred))
mlog.store_model('linear_v2', lr1 , X_train , score(Y_test,Y_pred))
# let's train another regression model with different set of columns
X_train =X_train[['RM', 'AGE', 'DIS', 'RAD', 'TAX','PTRATIO', 'B', 'LSTAT']]
X_test = X_test[['RM', 'AGE', 'DIS', 'RAD', 'TAX','PTRATIO', 'B', 'LSTAT']]
lr2 = LinearRegression(n_jobs=11)
lr2.fit(X_train,Y_train)
Y_pred = lr2.predict(X_test)
mlog.store_model( 'logistic_v2' , lr2 , X_train , score(Y_test,Y_pred))
#mlog.store_model( 'logistic_v3' , lr2 , X_train , score(Y_test,Y_pred))
mlog.store_model( 'linear_v1' , lr2 , X_train , score(Y_test,Y_pred))
###Output
_____no_output_____
###Markdown
Quick Sneak - peek
###Code
mlog.view_results()
###Output
_____no_output_____
###Markdown
Delete a model entry by 'model name'
###Code
mlog.view_results()
mlog.delete_model(Model_Name='logistic_v1')
mlog.delete_model(Model_id= 4)
mlog.delete_all()
###Output
_____no_output_____
###Markdown
Model profiling - The fun part begins !!
###Code
#sample database can be found in github modellogger.github.io/assests/sample
run it local machine not in colab
mlog = ModelLogger('financial.db')
mlog.model_profiles()
#Please refer to the documentaion if you face any issue with parameters
mlog.info()
###Output
_____no_output_____ |
Comparison_2015_2020_Notebook.ipynb | ###Markdown
Has the programming world changed over the past 5 years? Business understanding In order to investigate the changes occured in the programming world over the past 5 years, I would like to answer following questions:Is the programming industry showing more gender diversity in 2020 compared to 2015?Which programming languages are most common in 2020 and are they the same ones as in 2015?Are programmers nowadays having a formal computer science background and has this changed since 2015? Is programming a rewarding profession and has this changed since 2015? Data understanding The data used to answer the questions are the Stack Overflow Developer survey results from the years 2015 and 2020.First the relevant libraries and the datasets have been imported and examined to identify the columns needed for the analysis.Since the survey results are assumed to be a representative sample of the population, some demographics (age and location of respondents) have been evaluated and compared, in order to ensure that the data sets are similarly distributed.
###Code
#Libraries import
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
%pylab inline
#Import of Stack Overflow 2015 survey data
df_2015 = pd.read_csv('2015 Stack Overflow Developer Survey Responses.csv', header=1)
print(df_2015.shape)
print(df_2015.head())
#Import of Stack Overflow 2020 survey data
df_2020 = pd.read_csv('survey_results_public_2020.csv', header=0)
print(df_2020.shape)
print(df_2020.head())
#Check of age distribution of respondents in 2015
df_age_2015 = pd.DataFrame(df_2015.Age.value_counts().reset_index())
df_age_2015.columns = ('Age', 'Count')
df_age_2015['Age'] = df_age_2015['Age'].str.replace('< 20','0-20')
df_age_2015.sort_values(by=['Age']).plot.bar(x='Age', y='Count', legend=False)
plt.show()
#Check of geographical distribution of respondents in 2015
df_coun_2015 = pd.DataFrame(df_2015.Country.dropna().value_counts() / df_2015.Country.dropna().shape)
df_coun_2015.reset_index(inplace = True)
#For simplification purposes the countries having a percentage below 2% as well as the NaN values are aggregated as 'Other'
df_coun_2015.rename(columns={'index': 'Country', 'Country': 'Count'}, inplace = True)
df_coun_2015['Country_new'] = np.where((df_coun_2015['Count'] < 0.02) | (df_coun_2015['Count'].isna()), 'Other',df_coun_2015['Country'])
df_coun_2015 = pd.DataFrame(df_coun_2015.groupby('Country_new')['Count'].sum().reset_index()).sort_values(by=['Count'], ascending=False)
df_coun_2015.plot.pie(y='Count', labels=df_coun_2015['Country_new'], legend = False, autopct='%1.1f%%', radius = 2)
plt.show()
#Check of age distribution of respondents in 2020; ages have been grouped in the same bins as in 2015 data to allow comparison
bins=[0,20,24,29,34,39,50,60,100]
df_age_2020 = pd.DataFrame(df_2020.groupby(pd.cut(df_2020['Age'], bins=bins, include_lowest=False)).size())
df_age_2020.reset_index(inplace = True)
df_age_2020.columns = ('Age', 'Count')
df_age_2020.plot.bar(x='Age', y='Count', legend=False)
plt.show()
#Check of geographical distribution of respondents in 2020
df_coun_2020 = pd.DataFrame(df_2020.Country.dropna().value_counts() / df_2020.Country.dropna().shape)
df_coun_2020.reset_index(inplace = True)
#For simplification purposes the countries having a percentage below 2% are aggregated as 'Other'
df_coun_2020.rename(columns={'index': 'Country', 'Country': 'Count'}, inplace = True)
df_coun_2020['Country_new'] = np.where((df_coun_2020['Count'] < 0.02) | (df_coun_2020['Count'].isna()), 'Other',df_coun_2020['Country'])
df_coun_2020 = pd.DataFrame(df_coun_2020.groupby('Country_new')['Count'].sum().reset_index()).sort_values(by=['Count'], ascending=False)
df_coun_2020.plot.pie(y='Count', labels=df_coun_2020['Country_new'], legend = False, autopct='%1.1f%%', radius = 2)
pylab.ylabel('')
plt.show()
###Output
_____no_output_____
###Markdown
Prepare and Model data First step for data preparation is to check the error message shown upon import of the 2015 csv file, hinting at mixed data types:
###Code
df_2015.iloc[:, [5,108,121,196,197,198]]
###Output
_____no_output_____
###Markdown
Among the columns having mixed type the only column that will be used for the analysis is occupation, which will be formatted as string:
###Code
df_2015['Occupation'] = df_2015['Occupation'].astype('str')
###Output
_____no_output_____
###Markdown
1. Question: Is the programming industry showing more gender diversity in 2020 compared to 2015?
###Code
#Check for the percentage of NaN values in the column Gender in 2015
df_2015['Gender'].isna().sum() / df_2015.shape[0] * 100
#Since NaN represents around 1% of the total I will not consider them in the analysis
df_gen_2015 = pd.DataFrame(df_2015.Gender.dropna().value_counts() / df_2015.Gender.dropna().shape)
df_gen_2015.reset_index(inplace = True)
df_gen_2015.rename(columns={'index': 'Gender', 'Gender': 'Count'}, inplace = True)
#The occurences 'Other' and 'Prefer not to disclose' are aggregated as 'Other' and considered as non-binary genders
df_gen_2015['Gender_agg'] = np.where((~df_gen_2015['Gender'].isin(('Male','Female'))), 'Other',df_gen_2015['Gender'])
df_gen_2015 = pd.DataFrame(df_gen_2015.groupby('Gender_agg')['Count'].sum().reset_index()).sort_values(by=['Count'], ascending=False)
df_gen_2015.plot.pie(y='Count', labels=df_gen_2015['Gender_agg'], legend=None, autopct='%1.1f%%', radius = 2)
pylab.ylabel('')
plt.show()
#Check for the percentage of NaN values in the column Gender in 2020
df_2020['Gender'].isna().sum() / df_2020.shape[0] * 100
#In this cases NaN values are around 21% but they cannot be interpretable and will therefore be dropped.
#For transparency this will be declared in the results.
df_gen_2020 = pd.DataFrame(df_2020.Gender.dropna().value_counts() /df_2020.Gender.dropna().shape)
df_gen_2020.reset_index(inplace = True)
df_gen_2020.rename(columns={'index': 'Gender', 'Gender': 'Count'}, inplace = True)
df_gen_2020['Gender_agg'] = np.where((~df_gen_2020['Gender'].isin(('Man','Woman'))), 'Other',df_gen_2020['Gender'])
df_gen_2020 = pd.DataFrame(df_gen_2020.groupby('Gender_agg')['Count'].sum().reset_index()).sort_values(by=['Count'], ascending=False)
df_gen_2020.plot.pie(y='Count', labels=df_gen_2020['Gender_agg'], legend = False, autopct='%1.1f%%', radius = 2)
pylab.ylabel('')
plt.show()
###Output
_____no_output_____
###Markdown
2. Question: Which programming languages are most common in 2020 and are they the same ones as in 2015?
###Code
#Evaluation of the top 10 most common languages in the 2015 dataset
df_lan_2015 = pd.DataFrame(df_2015.filter(like='Current Lang & Tech', axis=1).count().rename('Count').reset_index()).sort_values(by=['Count'], ascending=False)
df_lan_2015.rename(columns={'index': 'Lang&Tech'}, inplace = True)
df_lan_2015['Lang&Tech'] = df_lan_2015['Lang&Tech'].str.replace(r'Current Lang & Tech: ','')
df_lan_2015.head(10).plot.barh(x='Lang&Tech',figsize=(15,5), legend=False)
pylab.ylabel('')
plt.show()
df_lan_2015.plot.pie(y='Count', labels=df_lan_2015['Lang&Tech'], legend = False, autopct='%1.1f%%', radius = 2)
pylab.ylabel('')
plt.show()
#Evaluation of the top 10 languages to be learned in the future in the 2015 dataset
df_lan_2015 = pd.DataFrame(df_2015.filter(like='Future Lang & Tech', axis=1).count().rename('Count').reset_index()).sort_values(by=['Count'], ascending=False)
df_lan_2015.rename(columns={'index': 'Future_Lang&Tech'}, inplace = True)
df_lan_2015['Future_Lang&Tech'] = df_lan_2015['Future_Lang&Tech'].str.replace(r'Future Lang & Tech: ','')
df_lan_2015.head(10).plot.barh(x='Future_Lang&Tech',figsize=(15,5), legend=False)
pylab.ylabel('')
plt.show()
df_lan_2015.plot.pie(y='Count', labels=df_lan_2015['Future_Lang&Tech'], legend = False, autopct='%1.1f%%', radius = 2)
pylab.ylabel('')
plt.show()
#Evaluation of the top 10 most common languages in the 2020 dataset
df_lan_2020 = pd.DataFrame(df_2020.LanguageWorkedWith.str.split(';').explode('LanguageWorkedWith').reset_index(drop=True))
df_lan_2020 = pd.DataFrame(df_lan_2020['LanguageWorkedWith'].value_counts().rename('Count').reset_index())
df_lan_2020.rename(columns={'index': 'Language'}, inplace = True)
df_lan_2020.head(10).plot.barh(x='Language',figsize=(15,5), legend=False)
pylab.ylabel('')
plt.show()
df_lan_2020.plot.pie(y='Count', labels=df_lan_2020['Language'], legend = False, autopct='%1.1f%%', radius = 2)
pylab.ylabel('')
plt.show()
#Evaluation of the top 10 languages to be learned in the future in the 2020 dataset
df_lan_2020 = df_2020[['LanguageDesireNextYear']]
df_lan_2020 = pd.DataFrame(df_lan_2020.LanguageDesireNextYear.str.split(';').explode('LanguageDesireNextYear').reset_index(drop=True))
df_lan_2020 = pd.DataFrame(df_lan_2020['LanguageDesireNextYear'].value_counts().rename('Count').reset_index())
df_lan_2020.rename(columns={'index': 'Language'}, inplace = True)
df_lan_2020.head(10).plot.barh(x='Language',figsize=(15,5), legend=False)
pylab.ylabel('')
plt.show()
###Output
_____no_output_____
###Markdown
Question 3: Are coders having a formal computer science background and has this changed since 2015?
###Code
#Computation of the percentage of respondents having no computer science background by counting the rows with all three fields related to computer science education left blank (NaN)
df_educ_2015 = pd.DataFrame(df_2015[['Training & Education: BS in CS','Training & Education: Masters in CS', 'Training & Education: PhD in CS']].isna().sum(axis=1).rename('Count'))
df_educ_2015[df_educ_2015.Count == 3].count()/df_2015.shape[0]
#Visual representation of the academic background of the respondents in 2020
df_educ_2020 = pd.DataFrame(df_2020['UndergradMajor'].value_counts().rename('Count').reset_index())
df_educ_2020.rename(columns={'index': 'Major'}, inplace = True)
df_educ_2020.plot.pie(y='Count', labels=df_educ_2020['Major'], legend = False, autopct='%1.1f%%', radius = 2)
pylab.ylabel('')
plt.show()
###Output
_____no_output_____
###Markdown
Question 4: Is the job satisfaction of programmers increasing as the technology advances?
###Code
#Evaluation of the level of job satisfaction of programmers in 2015
df_satisf_2015 = df_2015[['Job Satisfaction']]
df_satisf_2015 = pd.DataFrame(df_satisf_2015['Job Satisfaction'].value_counts().rename('Count').reset_index())
df_satisf_2015.rename(columns={'index': 'Job Satisfaction'}, inplace = True)
df_satisf_2015.plot.pie(y='Count', labels=df_satisf_2015['Job Satisfaction'], legend = False, autopct='%1.1f%%', radius = 2)
pylab.ylabel('')
plt.show()
#Evaluation of the level of job satisfaction of programmers in 2020
df_satisf_2020 = df_2020[['JobSat']]
df_satisf_2020 = pd.DataFrame(df_satisf_2020['JobSat'].value_counts().rename('Count').reset_index())
df_satisf_2020.rename(columns={'index': 'Job Satisfaction'}, inplace = True)
df_satisf_2020.plot.pie(y='Count', labels=df_satisf_2020['Job Satisfaction'], legend = False, autopct='%1.1f%%', radius = 2)
pylab.ylabel('')
plt.show()
###Output
_____no_output_____ |
examples/Engineering Metrics Example.ipynb | ###Markdown
Engineering Metrics Example In order to authenticate with the current Jira server we are required to use oauth with karhoo google apps credentials. To set this up you require the private and public keys of the Jira application for engineering metrics. Once you have these, set up your access as outlined here. Once config files are in place we can simple pass the path to the oauth config to an instance of the Engineering Metrics engine to gain access to all the juicy data stored in our Jira.
###Code
import sys
sys.path.append("..")
from engineeringmetrics.engine import EngineeringMetrics
from pathlib import Path
config_dict = {
'jira_oauth_config_path': Path.home()
}
kem = EngineeringMetrics(config_dict)
projects_data = kem.jirametrics.populate_projects(['INT'], max_results=110)
import pandas as pd
df = pd.DataFrame(projects_data['INT'].issues)
df.shape
df.head()
oso_query = kem.jirametrics.populate_from_jql('project = "OSO" ORDER BY Rank ASC')
oso_df = pd.DataFrame(oso_query.resolved_issues())
oso_df.shape
oso_df.head()
oso_df.set_index('created',drop=False,inplace=True)
oso_df.cycle_time.plot(kind='line',style='ko-').set_title('Time between creation and resolution of OSO issues')
closed_df = pd.DataFrame(projects_data['INT'].resolved_issues())
closed_df.shape
closed_df.set_index('created',drop=False,inplace=True)
closed_df.cycle_time.plot(kind='line',style='ko-').set_title('Time between creation and resolution of INT issues')
###Output
_____no_output_____ |
Brandon_Rhodes_PyCon15/Exercises-5.ipynb | ###Markdown
Make a bar plot of the months in which movies with "Christmas" in their title tend to be released in the USA.
###Code
r = release_dates
r = r[r['title'].apply(lambda x:x.lower()).str.contains("christmas")]
r = r[r['country']=='USA']['date']
r.dt.month.value_counts().sort_index().plot(kind='bar')
###Output
_____no_output_____
###Markdown
Make a bar plot of the months in which movies whose titles start with "The Hobbit" are released in the USA.
###Code
r = release_dates
r = r[r['title'].str.startswith("The Hobbit")]
r = r[r['country']=='USA']['date']
r.dt.month.value_counts().sort_index().plot(kind='bar')
###Output
_____no_output_____
###Markdown
Make a bar plot of the day of the week on which movies with "Romance" in their title tend to be released in the USA.
###Code
r = release_dates
r = r[r['title'].apply(lambda x:x.lower()).str.contains("romance")]
r = r[r['country']=='USA']['date']
r.dt.dayofweek.value_counts().sort_index().plot(kind='bar')
###Output
_____no_output_____
###Markdown
Make a bar plot of the day of the week on which movies with "Action" in their title tend to be released in the USA.
###Code
r = release_dates
r = r[r['title'].apply(lambda x:x.lower()).str.contains("action")]
r = r[r['country']=='USA']['date']
r.dt.dayofweek.value_counts().sort_index().plot(kind='bar')
###Output
_____no_output_____
###Markdown
On which date was each Judi Dench movie from the 1990s released in the USA?
###Code
c = cast
r = release_dates
r = r[r['country']=='USA']
c = c[(c['name']=='Judi Dench') & (c['year']//10*10== 1990)]
c = c.merge(r)
c.date.dt.day
###Output
_____no_output_____
###Markdown
In which months do films with Judi Dench tend to be released in the USA?
###Code
c = cast
r = release_dates
r = r[r['country']=='USA']
c = c[(c['name']=='Judi Dench')]
c = c.merge(r)
c.date.dt.month.value_counts().sort_index().plot(kind='bar')
usa = release_dates[release_dates.country == 'USA']
c = cast
c = c[c.name == 'Judi Dench']
m = c.merge(usa).sort_values('date')
m.date.dt.month.value_counts().sort_index().plot(kind='bar')
###Output
_____no_output_____
###Markdown
In which months do films with Tom Cruise tend to be released in the USA?
###Code
c = cast
r = release_dates
r = r[r['country']=='USA']
c = c[(c['name']=='Tom Cruise') ]
c = c.merge(r)
c.date.dt.month.value_counts().sort_index().plot(kind='bar')
###Output
_____no_output_____ |
4_4_Kalman_Filters/2_1. New Mean and Variance, exercise.ipynb | ###Markdown
New Mean and VarianceNow let's take the formulas from the example below and use them to write a program that takes in two means and variances, and returns a *new*, updated mean and variance for a gaussian. This step is called the parameter or **measurement update** because it is the update that happens when an initial belief (represented by the blue Gaussian, below) is merged with a new piece of information, a measurement with some uncertainty (the orange Gaussian). As you've seen in the previous quizzes, the updated Gaussian will be a combination of these two Gaussians with a new mean that is in between both of theirs and a variance that is less than the smallest of the two given variances; this means that after a measurement, our new mean is more certain than that of the initial belief! Below is our usual Gaussian equation and imports.
###Code
# import math functions
from math import *
import matplotlib.pyplot as plt
import numpy as np
# gaussian function
def f(mu, sigma2, x):
''' f takes in a mean and squared variance, and an input x
and returns the gaussian value.'''
coefficient = 1.0 / sqrt(2.0 * pi *sigma2)
exponential = exp(-0.5 * (x-mu) ** 2 / sigma2)
return coefficient * exponential
###Output
_____no_output_____
###Markdown
QUIZ: Write an `update` function that performs the measurement update.This function should combine the given Gaussian parameters and return new values for the mean and squared variance.This function does not have to perform any exponential math, it simply has to follow the equations for the measurement update as seen in the image at the top of this notebook. You may assume that the given variances `var1` and `var2` are squared terms.
###Code
# the update function
def update(mean1, var1, mean2, var2):
''' This function takes in two means and two squared variance terms,
and returns updated gaussian parameters.'''
## TODO: Calculate the new parameters
new_mean = (var2 * mean1 + var1 * mean2) / (var2 + var1)
new_var = 1 / (1/var2 + 1/var1)
return [new_mean, new_var]
# test your implementation
new_params = update(10, 4, 12, 4)
print(new_params)
###Output
[11.0, 2.0]
###Markdown
Plot a GaussianPlot a Gaussian by looping through a range of x values and creating a resulting list of Gaussian values, `g`, as shown below. You're encouraged to see what happens if you change the values of `mu` and `sigma2`.
###Code
# display a gaussian over a range of x values
# define the parameters
mu = new_params[0]
sigma2 = new_params[1]
# define a range of x values
x_axis = np.arange(0, 20, 0.1)
# create a corresponding list of gaussian values
g = []
for x in x_axis:
g.append(f(mu, sigma2, x))
# plot the result
plt.plot(x_axis, g)
###Output
_____no_output_____
###Markdown
New Mean and VarianceNow let's take the formulas from the example below and use them to write a program that takes in two means and variances, and returns a *new*, updated mean and variance for a gaussian. This step is called the parameter or **measurement update** because it is the update that happens when an initial belief (represented by the blue Gaussian, below) is merged with a new piece of information, a measurement with some uncertainty (the orange Gaussian). As you've seen in the previous quizzes, the updated Gaussian will be a combination of these two Gaussians with a new mean that is in between both of theirs and a variance that is less than the smallest of the two given variances; this means that after a measurement, our new mean is more certain than that of the initial belief! Below is our usual Gaussian equation and imports.
###Code
# import math functions
from math import *
import matplotlib.pyplot as plt
import numpy as np
# gaussian function
def f(mu, sigma2, x):
''' f takes in a mean and squared variance, and an input x
and returns the gaussian value.'''
coefficient = 1.0 / sqrt(2.0 * pi *sigma2)
exponential = exp(-0.5 * (x-mu) ** 2 / sigma2)
return coefficient * exponential
###Output
_____no_output_____
###Markdown
QUIZ: Write an `update` function that performs the measurement update.This function should combine the given Gaussian parameters and return new values for the mean and squared variance.This function does not have to perform any exponential math, it simply has to follow the equations for the measurement update as seen in the image at the top of this notebook. You may assume that the given variances `var1` and `var2` are squared terms.
###Code
# the update function
def update(mean1, var1, mean2, var2):
''' This function takes in two means and two squared variance terms,
and returns updated gaussian parameters.'''
## TODO: Calculate the new parameters
new_mean = None
new_var = None
return [new_mean, new_var]
# test your implementation
new_params = update(10, 4, 12, 4)
print(new_params)
###Output
_____no_output_____
###Markdown
Plot a GaussianPlot a Gaussian by looping through a range of x values and creating a resulting list of Gaussian values, `g`, as shown below. You're encouraged to see what happens if you change the values of `mu` and `sigma2`.
###Code
# display a gaussian over a range of x values
# define the parameters
mu = new_params[0]
sigma2 = new_params[1]
# define a range of x values
x_axis = np.arange(0, 20, 0.1)
# create a corresponding list of gaussian values
g = []
for x in x_axis:
g.append(f(mu, sigma2, x))
# plot the result
plt.plot(x_axis, g)
###Output
_____no_output_____
###Markdown
New Mean and VarianceNow let's take the formulas from the example below and use them to write a program that takes in two means and variances, and returns a *new*, updated mean and variance for a gaussian. This step is called the parameter or **measurement update** because it is the update that happens when an initial belief (represented by the blue Gaussian, below) is merged with a new piece of information, a measurement with some uncertainty (the orange Gaussian). As you've seen in the previous quizzes, the updated Gaussian will be a combination of these two Gaussians with a new mean that is in between both of theirs and a variance that is less than the smallest of the two given variances; this means that after a measurement, our new mean is more certain than that of the initial belief! Below is our usual Gaussian equation and imports.
###Code
# import math functions
from math import *
import matplotlib.pyplot as plt
import numpy as np
# gaussian function
def f(mu, sigma2, x):
''' f takes in a mean and squared variance, and an input x
and returns the gaussian value.'''
coefficient = 1.0 / sqrt(2.0 * pi *sigma2)
exponential = exp(-0.5 * (x-mu) ** 2 / sigma2)
return coefficient * exponential
###Output
_____no_output_____
###Markdown
QUIZ: Write an `update` function that performs the measurement update.This function should combine the given Gaussian parameters and return new values for the mean and squared variance.This function does not have to perform any exponential math, it simply has to follow the equations for the measurement update as seen in the image at the top of this notebook. You may assume that the given variances `var1` and `var2` are squared terms.
###Code
# the update function
def update(mean1, var1, mean2, var2):
''' This function takes in two means and two squared variance terms,
and returns updated gaussian parameters.'''
## TODO: Calculate the new parameters
new_mean = (var2*mean1 + var1*mean2) / (var1 + var2)
new_var = 1 / (1/var1 + 1/var2)
return [new_mean, new_var]
# test your implementation
new_params = update(10, 4, 12, 4)
print(new_params)
###Output
[11.0, 2.0]
###Markdown
Plot a GaussianPlot a Gaussian by looping through a range of x values and creating a resulting list of Gaussian values, `g`, as shown below. You're encouraged to see what happens if you change the values of `mu` and `sigma2`.
###Code
# display a gaussian over a range of x values
# define the parameters
mu = new_params[0]
sigma2 = new_params[1]
# define a range of x values
x_axis = np.arange(0, 20, 0.1)
# create a corresponding list of gaussian values
g = []
for x in x_axis:
g.append(f(mu, sigma2, x))
# plot the result
plt.plot(x_axis, g)
###Output
_____no_output_____
###Markdown
New Mean and VarianceNow let's take the formulas from the example below and use them to write a program that takes in two means and variances, and returns a *new*, updated mean and variance for a gaussian. This step is called the parameter or **measurement update** because it is the update that happens when an initial belief (represented by the blue Gaussian, below) is merged with a new piece of information, a measurement with some uncertainty (the orange Gaussian). As you've seen in the previous quizzes, the updated Gaussian will be a combination of these two Gaussians with a new mean that is in between both of theirs and a variance that is less than the smallest of the two given variances; this means that after a measurement, our new mean is more certain than that of the initial belief! Below is our usual Gaussian equation and imports.
###Code
# import math functions
from math import *
import matplotlib.pyplot as plt
import numpy as np
# gaussian function
def f(mu, sigma2, x):
''' f takes in a mean and squared variance, and an input x
and returns the gaussian value.'''
coefficient = 1.0 / sqrt(2.0 * pi *sigma2)
exponential = exp(-0.5 * (x-mu) ** 2 / sigma2)
return coefficient * exponential
###Output
_____no_output_____
###Markdown
QUIZ: Write an `update` function that performs the measurement update.This function should combine the given Gaussian parameters and return new values for the mean and squared variance.This function does not have to perform any exponential math, it simply has to follow the equations for the measurement update as seen in the image at the top of this notebook. You may assume that the given variances `var1` and `var2` are squared terms.
###Code
# the update function
def update(mean1, var1, mean2, var2):
''' This function takes in two means and two squared variance terms,
and returns updated gaussian parameters.'''
## TODO: Calculate the new parameters
new_mean = (var2*mean1+var1*mean2)/(var1+var2)
new_var = 1./(1/var1+1/var2)
return [new_mean, new_var]
# test your implementation
new_params = update(10, 4, 12, 4)
print(new_params)
###Output
[11.0, 2.0]
###Markdown
Plot a GaussianPlot a Gaussian by looping through a range of x values and creating a resulting list of Gaussian values, `g`, as shown below. You're encouraged to see what happens if you change the values of `mu` and `sigma2`.
###Code
# display a gaussian over a range of x values
# define the parameters
mu = new_params[0]
sigma2 = new_params[1]
# define a range of x values
x_axis = np.arange(0, 20, 0.1)
# create a corresponding list of gaussian values
g = []
for x in x_axis:
#print(mu, sigma2, x)
g.append(f(mu, sigma2, x))
#print(g)
# plot the result
plt.plot(x_axis, g)
###Output
_____no_output_____
###Markdown
New Mean and VarianceNow let's take the formulas from the example below and use them to write a program that takes in two means and variances, and returns a *new*, updated mean and variance for a gaussian. This step is called the parameter or **measurement update** because it is the update that happens when an initial belief (represented by the blue Gaussian, below) is merged with a new piece of information, a measurement with some uncertainty (the orange Gaussian). As you've seen in the previous quizzes, the updated Gaussian will be a combination of these two Gaussians with a new mean that is in between both of theirs and a variance that is less than the smallest of the two given variances; this means that after a measurement, our new mean is more certain than that of the initial belief! Below is our usual Gaussian equation and imports.
###Code
# import math functions
from math import *
import matplotlib.pyplot as plt
import numpy as np
# gaussian function
def f(mu, sigma2, x):
''' f takes in a mean and squared variance, and an input x
and returns the gaussian value.'''
coefficient = 1.0 / sqrt(2.0 * pi *sigma2)
exponential = exp(-0.5 * (x-mu) ** 2 / sigma2)
return coefficient * exponential
###Output
_____no_output_____
###Markdown
QUIZ: Write an `update` function that performs the measurement update.This function should combine the given Gaussian parameters and return new values for the mean and squared variance.This function does not have to perform any exponential math, it simply has to follow the equations for the measurement update as seen in the image at the top of this notebook. You may assume that the given variances `var1` and `var2` are squared terms.
###Code
# the update function
def update(mean1, var1, mean2, var2):
''' This function takes in two means and two squared variance terms,
and returns updated gaussian parameters.'''
## TODO: Calculate the new parameters
new_mean = ((var1*mean1) + (var2*mean2)) / (var1 + var2)
new_var = 1 / (1/var1 + 1/var2)
return [new_mean, new_var]
# test your implementation
new_params = update(10, 4, 12, 4)
print(new_params)
###Output
[11.0, 2.0]
###Markdown
Plot a GaussianPlot a Gaussian by looping through a range of x values and creating a resulting list of Gaussian values, `g`, as shown below. You're encouraged to see what happens if you change the values of `mu` and `sigma2`.
###Code
# display a gaussian over a range of x values
# define the parameters
mu = new_params[0]
sigma2 = new_params[1]
# define a range of x values
x_axis = np.arange(0, 20, 0.1)
# create a corresponding list of gaussian values
g = []
for x in x_axis:
g.append(f(mu, sigma2, x))
# plot the result
plt.plot(x_axis, g)
###Output
_____no_output_____
###Markdown
New Mean and VarianceNow let's take the formulas from the example below and use them to write a program that takes in two means and variances, and returns a *new*, updated mean and variance for a gaussian. This step is called the parameter or **measurement update** because it is the update that happens when an initial belief (represented by the blue Gaussian, below) is merged with a new piece of information, a measurement with some uncertainty (the orange Gaussian). As you've seen in the previous quizzes, the updated Gaussian will be a combination of these two Gaussians with a new mean that is in between both of theirs and a variance that is less than the smallest of the two given variances; this means that after a measurement, our new mean is more certain than that of the initial belief! Below is our usual Gaussian equation and imports.
###Code
# import math functions
from math import *
import matplotlib.pyplot as plt
import numpy as np
# gaussian function
def f(mu, sigma2, x):
''' f takes in a mean and squared variance, and an input x
and returns the gaussian value.'''
coefficient = 1.0 / sqrt(2.0 * pi *sigma2)
exponential = exp(-0.5 * (x-mu) ** 2 / sigma2)
return coefficient * exponential
###Output
_____no_output_____
###Markdown
QUIZ: Write an `update` function that performs the measurement update.This function should combine the given Gaussian parameters and return new values for the mean and squared variance.This function does not have to perform any exponential math, it simply has to follow the equations for the measurement update as seen in the image at the top of this notebook. You may assume that the given variances `var1` and `var2` are squared terms.
###Code
# the update function
def update(mean1, var1, mean2, var2):
''' This function takes in two means and two squared variance terms,
and returns updated gaussian parameters.'''
## TODO: Calculate the new parameters
new_mean = None
new_var = None
return [new_mean, new_var]
# test your implementation
new_params = update(10, 4, 12, 4)
print(new_params)
###Output
_____no_output_____
###Markdown
Plot a GaussianPlot a Gaussian by looping through a range of x values and creating a resulting list of Gaussian values, `g`, as shown below. You're encouraged to see what happens if you change the values of `mu` and `sigma2`.
###Code
# display a gaussian over a range of x values
# define the parameters
mu = new_params[0]
sigma2 = new_params[1]
# define a range of x values
x_axis = np.arange(0, 20, 0.1)
# create a corresponding list of gaussian values
g = []
for x in x_axis:
g.append(f(mu, sigma2, x))
# plot the result
plt.plot(x_axis, g)
###Output
_____no_output_____
###Markdown
New Mean and VarianceNow let's take the formulas from the example below and use them to write a program that takes in two means and variances, and returns a *new*, updated mean and variance for a gaussian. This step is called the parameter or **measurement update** because it is the update that happens when an initial belief (represented by the blue Gaussian, below) is merged with a new piece of information, a measurement with some uncertainty (the orange Gaussian). As you've seen in the previous quizzes, the updated Gaussian will be a combination of these two Gaussians with a new mean that is in between both of theirs and a variance that is less than the smallest of the two given variances; this means that after a measurement, our new mean is more certain than that of the initial belief! Below is our usual Gaussian equation and imports.
###Code
# import math functions
from math import *
import matplotlib.pyplot as plt
import numpy as np
# gaussian function
def f(mu, sigma2, x):
''' f takes in a mean and squared variance, and an input x
and returns the gaussian value.'''
coefficient = 1.0 / sqrt(2.0 * pi *sigma2)
exponential = exp(-0.5 * (x-mu) ** 2 / sigma2)
return coefficient * exponential
###Output
_____no_output_____
###Markdown
QUIZ: Write an `update` function that performs the measurement update.This function should combine the given Gaussian parameters and return new values for the mean and squared variance.This function does not have to perform any exponential math, it simply has to follow the equations for the measurement update as seen in the image at the top of this notebook. You may assume that the given variances `var1` and `var2` are squared terms.
###Code
# the update function
def update(mean1, var1, mean2, var2):
''' This function takes in two means and two squared variance terms,
and returns updated gaussian parameters.'''
## TODO: Calculate the new parameters
new_mean = (mean1 * var2 + mean2 * var1) / (var1 + var2)
new_var = 1 / (1/var1 + 1/var2)
return [new_mean, new_var]
# test your implementation
new_params = update(10, 4, 12, 4)
print(new_params)
###Output
[11.0, 2.0]
###Markdown
Plot a GaussianPlot a Gaussian by looping through a range of x values and creating a resulting list of Gaussian values, `g`, as shown below. You're encouraged to see what happens if you change the values of `mu` and `sigma2`.
###Code
# display a gaussian over a range of x values
# define the parameters
mu = new_params[0]
sigma2 = new_params[1]
# define a range of x values
x_axis = np.arange(0, 20, 0.1)
# create a corresponding list of gaussian values
g = []
for x in x_axis:
g.append(f(mu, sigma2, x))
# plot the result
plt.plot(x_axis, g)
###Output
_____no_output_____
###Markdown
New Mean and VarianceNow let's take the formulas from the example below and use them to write a program that takes in two means and variances, and returns a *new*, updated mean and variance for a gaussian. This step is called the parameter or **measurement update** because it is the update that happens when an initial belief (represented by the blue Gaussian, below) is merged with a new piece of information, a measurement with some uncertainty (the orange Gaussian). As you've seen in the previous quizzes, the updated Gaussian will be a combination of these two Gaussians with a new mean that is in between both of theirs and a variance that is less than the smallest of the two given variances; this means that after a measurement, our new mean is more certain than that of the initial belief! Below is our usual Gaussian equation and imports.
###Code
# import math functions
from math import *
import matplotlib.pyplot as plt
import numpy as np
# gaussian function
def f(mu, sigma2, x):
''' f takes in a mean and squared variance, and an input x
and returns the gaussian value.'''
coefficient = 1.0 / sqrt(2.0 * pi *sigma2)
exponential = exp(-0.5 * (x-mu) ** 2 / sigma2)
return coefficient * exponential
###Output
_____no_output_____
###Markdown
QUIZ: Write an `update` function that performs the measurement update.This function should combine the given Gaussian parameters and return new values for the mean and squared variance.This function does not have to perform any exponential math, it simply has to follow the equations for the measurement update as seen in the image at the top of this notebook. You may assume that the given variances `var1` and `var2` are squared terms.
###Code
# the update function
def update(mean1, var1, mean2, var2):
''' This function takes in two means and two squared variance terms,
and returns updated gaussian parameters.'''
## TODO: Calculate the new parameters
new_mean = (var1*mean2+var2*mean1)/(var1+var2)
new_var = 1/((1/var1)+(1/var2))
return [new_mean, new_var]
# test your implementation
new_params = update(10, 4, 12, 4)
print(new_params)
###Output
[11.0, 2.0]
###Markdown
Plot a GaussianPlot a Gaussian by looping through a range of x values and creating a resulting list of Gaussian values, `g`, as shown below. You're encouraged to see what happens if you change the values of `mu` and `sigma2`.
###Code
# display a gaussian over a range of x values
# define the parameters
mu = new_params[0]
sigma2 = new_params[1]
# define a range of x values
x_axis = np.arange(0, 20, 0.1)
# create a corresponding list of gaussian values
g = []
for x in x_axis:
g.append(f(mu, sigma2, x))
# plot the result
plt.plot(x_axis, g)
###Output
_____no_output_____
###Markdown
New Mean and VarianceNow let's take the formulas from the example below and use them to write a program that takes in two means and variances, and returns a *new*, updated mean and variance for a gaussian. This step is called the parameter or **measurement update** because it is the update that happens when an initial belief (represented by the blue Gaussian, below) is merged with a new piece of information, a measurement with some uncertainty (the orange Gaussian). As you've seen in the previous quizzes, the updated Gaussian will be a combination of these two Gaussians with a new mean that is in between both of theirs and a variance that is less than the smallest of the two given variances; this means that after a measurement, our new mean is more certain than that of the initial belief! Below is our usual Gaussian equation and imports.
###Code
# import math functions
from math import *
import matplotlib.pyplot as plt
import numpy as np
# gaussian function
def f(mu, sigma2, x):
''' f takes in a mean and squared variance, and an input x
and returns the gaussian value.'''
coefficient = 1.0 / sqrt(2.0 * pi *sigma2)
exponential = exp(-0.5 * (x-mu) ** 2 / sigma2)
return coefficient * exponential
###Output
_____no_output_____
###Markdown
QUIZ: Write an `update` function that performs the measurement update.This function should combine the given Gaussian parameters and return new values for the mean and squared variance.This function does not have to perform any exponential math, it simply has to follow the equations for the measurement update as seen in the image at the top of this notebook. You may assume that the given variances `var1` and `var2` are squared terms.
###Code
# the update function
def update(mean1, var1, mean2, var2):
''' This function takes in two means and two squared variance terms,
and returns updated gaussian parameters.'''
## TODO: Calculate the new parameters
new_mean = ((var1 * mean1) + (var2 * mean2)) / (var1 + var2)
new_var = 1 / ((1/var1) + (1/var2))
return [new_mean, new_var]
# test your implementation
new_params = update(10, 4, 12, 4)
print(new_params)
###Output
[11.0, 2.0]
###Markdown
Plot a GaussianPlot a Gaussian by looping through a range of x values and creating a resulting list of Gaussian values, `g`, as shown below. You're encouraged to see what happens if you change the values of `mu` and `sigma2`.
###Code
# display a gaussian over a range of x values
# define the parameters
mu = new_params[0]
sigma2 = new_params[1]
# define a range of x values
x_axis = np.arange(0, 20, 0.1)
# create a corresponding list of gaussian values
g = []
for x in x_axis:
g.append(f(mu, sigma2, x))
# plot the result
plt.plot(x_axis, g)
###Output
_____no_output_____
###Markdown
New Mean and VarianceNow let's take the formulas from the example below and use them to write a program that takes in two means and variances, and returns a *new*, updated mean and variance for a gaussian. This step is called the parameter or **measurement update** because it is the update that happens when an initial belief (represented by the blue Gaussian, below) is merged with a new piece of information, a measurement with some uncertainty (the orange Gaussian). As you've seen in the previous quizzes, the updated Gaussian will be a combination of these two Gaussians with a new mean that is in between both of theirs and a variance that is less than the smallest of the two given variances; this means that after a measurement, our new mean is more certain than that of the initial belief! Below is our usual Gaussian equation and imports.
###Code
# import math functions
from math import *
import matplotlib.pyplot as plt
import numpy as np
# gaussian function
def f(mu, sigma2, x):
''' f takes in a mean and squared variance, and an input x
and returns the gaussian value.'''
coefficient = 1.0 / sqrt(2.0 * pi *sigma2)
exponential = exp(-0.5 * (x-mu) ** 2 / sigma2)
return coefficient * exponential
###Output
_____no_output_____
###Markdown
QUIZ: Write an `update` function that performs the measurement update.This function should combine the given Gaussian parameters and return new values for the mean and squared variance.This function does not have to perform any exponential math, it simply has to follow the equations for the measurement update as seen in the image at the top of this notebook. You may assume that the given variances `var1` and `var2` are squared terms.
###Code
# the update function
def update(mean1, var1, mean2, var2):
''' This function takes in two means and two squared variance terms,
and returns updated gaussian parameters.'''
## Calculate the new parameters
new_mean = (var2*mean1 + var1*mean2)/(var1+var2)
new_var = 1/(1/var2 + 1/var1)
return [new_mean, new_var]
# test your implementation
new_params = update(10, 4, 12, 4)
print(new_params)
###Output
[11.0, 2.0]
###Markdown
Plot a GaussianPlot a Gaussian by looping through a range of x values and creating a resulting list of Gaussian values, `g`, as shown below. You're encouraged to see what happens if you change the values of `mu` and `sigma2`.
###Code
# display a gaussian over a range of x values
# define the parameters
mu = new_params[0]
sigma2 = new_params[1]
# define a range of x values
x_axis = np.arange(0, 20, 0.1)
# create a corresponding list of gaussian values
g = []
for x in x_axis:
g.append(f(mu, sigma2, x))
# plot the result
plt.plot(x_axis, g)
###Output
_____no_output_____ |
01_startup.ipynb | ###Markdown
Dataset made using **abalajiaus/oct_ca:latest-fastai-skl-ski-fire-onnx-mlflow**
###Code
#exports
from fastai.vision import *
import mlflow
import model
import pandas as pd
import mlflow.pytorch as MYPY
#exports
def saveDictToConfigJSON(dictiontary, name):
with open('/workspace/oct_ca_seg/oct/configs/'+ name, 'w') as file:
json.dump(dictiontary, file)
#exports
def loadConfigJSONToDict(fn):
with open('/workspace/oct_ca_seg/oct/configs/'+ fn, 'r') as file:
config = json.load(file)
return config
#exports
class DeepConfig():
def __init__(self, config_dict):
self.config_dict = config_dict
for k,v in self.config_dict.items():
setattr(self, k, DeepConfig(v) if isinstance(v, dict) else v)
config_test = loadConfigJSONToDict('init_config.json')
config = DeepConfig(config_test)
config.DATSET.bs = 1
config.MODEL.dims1 = 8
config.MODEL.dims2 = 12
config.MODEL.dims3 = 16
config.MODEL.maps1 = 2
config.MODEL.maps2 = 8
config.MODEL.maps3 = 12
config.MODEL.f1dims = 8
config.MODEL.f2dims = 12
config.MODEL.f1maps = 2
config.MODEL.f2maps = 2 #this should be number of classes. background, lumen => 2 etc.
config.LEARNER.lr = 0.01
cocodata_path = Path('/workspace/oct_ca_seg/COCOdata/')
train_path = cocodata_path/'train/images'
valid_path = cocodata_path/'valid/images'
test_path = cocodata_path/'test/images'
anno_name = 'medium_set_annotations.json' #or 'annotations.json' for full set
train_anno = train_path/anno_name
valid_anno = valid_path/anno_name
test_anno = test_path/anno_name
trainData = ImageList.from_folder(train_path)
validData = ImageList.from_folder(valid_path)
testData = ImageList.from_folder(test_path)
#export
binify = lambda x : x.point(lambda p: float(p>100.))#.point(lambda p: float(p))
fn = get_image_files(cocodata_path/'train/labels', recurse=True)[10]
labal = open_mask(fn, after_open=binify, convert_mode='L')
labal
labal.data
plt.hist(labal.data.flatten())
#export
class SegCustomLabelList(SegmentationLabelList):
def open(self, fn): return open_mask(fn, after_open=binify, convert_mode='L')
#export
class SegCustomItemList(SegmentationItemList):
_label_cls = SegCustomLabelList
src = SegCustomItemList.from_folder(cocodata_path, recurse=True, extensions='.jpg').filter_by_func(lambda fname: Path(fname).parent.name == 'images', ).split_by_folder('train', 'valid')
codes = np.loadtxt(cocodata_path/'codes.txt', dtype=str)
fn_get_y = lambda image_name: Path(image_name).parent.parent/('labels/'+Path(image_name).name)
src.label_from_func(fn_get_y, classes=codes)
tfms=get_transforms()[0].append(TfmPixel(to_float))
src.transform(tfms, size=256, tfm_y=True)
src.train.y[0].data
bs = config.DATSET.bs
data = src.databunch(bs=bs, val_bs=bs*2)
#data.transform(get_transforms()[0], tfm_y=True)
stats = [torch.tensor([0.2190, 0.1984, 0.1928]), torch.tensor([0.0645, 0.0473, 0.0434])]
data.normalize(stats)
src.train.y[0].data
torch.argmax(src.train.x[0].data, 0, keepdim=True).size()
#data.transform(tfms[0], tfm_y=True, size=(256, 256))
data.show_batch(10, ds_type=DatasetType.Train)
data.one_batch()
###Output
_____no_output_____
###Markdown
Metrics
###Code
#export
def sens(c, l):
n_targs=l.size()[0]
c = c.argmax(dim=1).view(n_targs,-1).float()
l = l.view(n_targs, -1).float()
inter = torch.sum(c*l, dim=(1))
union = torch.sum(c, dim=(1)) + torch.sum(l, dim=1) - inter
return (inter/union).mean()
def spec(c, l):
n_targs=l.size()[0]
c = c.argmax(dim=1).view(n_targs,-1).float()
l = l.view(n_targs, -1).float()
c=1-c
l=1-l
inter = torch.sum(c*l, dim=(1))
union = torch.sum(c, dim=(1)) + torch.sum(l, dim=1) - inter
return (inter/union).mean()
'''def acc(c, l):
n_targs=l.size()[0]
c = c.argmax(dim=1).view(n_targs,-1)
l = l.view(n_targs, -1).float()
return torch.sum(c,dim=(1)) / l.size()[-1]'''
def acc(c, l):
n_targs=l.size()[0]
c = c.argmax(dim=1).view(n_targs,-1)
l = l.argmax(dim=1).view(n_targs,-1)
c = torch.sum(torch.eq(c,l).float(),dim=1)
return (c/l.size()[-1]).mean()
class Dice_Loss(torch.nn.Module):
"""This is a custom Dice Similarity Coefficient loss function that we use
to the accuracy of the segmentation. it is defined as ;
DSC = 2 * (pred /intersect label) / (pred /union label) for the losss we use
1- DSC so gradient descent leads to better outputs."""
def __init__(self, weight=None, size_average=False):
super(Dice_Loss, self).__init__()
def forward(self, pred, label):
label = label.float()
smooth = 1. #helps with backprop
intersection = torch.sum(pred * label)
union = torch.sum(pred) + torch.sum(label)
loss = (2. * intersection + smooth) / (union + smooth)
#return 1-loss because we want to minimise dissimilarity
return 1 - (loss)
def my_Dice_Loss(pred, label):
pred = torch.argmax(pred, dim=0,keepdim=True)
label = label.float()
smooth = 1. #helps with backprop
intersection = torch.sum(pred * label)
union = torch.sum(pred) + torch.sum(label)
loss = (2. * intersection + smooth) / (union + smooth)
#return 1-loss because we want to minimise dissimilarity
return 1 - (loss)
###Output
_____no_output_____
###Markdown
Model Deep Cap the way configs were originally set up needs some help to work here. just made a little recursive config class
###Code
from model import CapsNet
###Output
_____no_output_____
###Markdown
note these functions have the absolute path in the docker image to teh configs save dir
###Code
torch.cuda.is_available()
#saveDictToConfigJSON(config.config_dict, 'config_test_save.json')
capsnet = CapsNet(config.MODEL).cuda()
capsnet(data.one_batch()[0].cuda()).shape
metrics = [sens, spec, acc, my_Dice_Loss, dice]
learner = Learner(data, model=capsnet, metrics=metrics)
lr = config.LEARNER.lr; lr
###Output
_____no_output_____
###Markdown
Fastai UNET
###Code
fast_unet_learner = unet_learner(data, models.resnet18, pretrained=False, y_range=[0,1], metrics=dice)
fast_unet_learner.summary();
###Output
_____no_output_____
###Markdown
Fastai UNET with a resnet 18 backbone has 31,113,008 parameters. Let's make a capsnet with around that many parameters, train them both for an epoch and see which is better. MLFLOW Callback
###Code
import mlflow
#export
clean_tensor_lists = lambda l : [x.item() for x in l]
def saveResultsJSON(json, run_dir, name): #name = exp_name+'item name+ .json'
with open(run_dir +'/'+ name, 'w') as file:
file.write(json)
def save_all_results(learner, run_dir, exp_name): #only call this function after training
train_losses = pd.DataFrame(clean_tensor_lists(learner.recorder.losses)).to_json()
valid_losses = pd.DataFrame(learner.recorder.val_losses).to_json()
metrics = np.array([clean_tensor_lists(x) for x in learner.recorder.metrics])
metric_names = learner.recorder.metrics_names
metrics = pd.DataFrame(metrics, columns=metric_names).to_json()
saveResultsJSON(train_losses, run_dir,'trainL.json')
saveResultsJSON(valid_losses, run_dir,'validL.json')
saveResultsJSON(metrics, run_dir,'metrics.json')
#export
## Tracking Class
from mlflow.tracking import MlflowClient
from mlflow.entities.run import Run
class MLFlowTracker(LearnerCallback):
"A `TrackerCallback` that tracks the loss and metrics into MLFlow"
def __init__(self, learn:Learner, exp_name: str, params: dict, nb_path: str, log_model: bool,uri: str = "http://localhost:5000"):
super().__init__(learn)
self.learn = learn
self.exp_name = exp_name
self.params = params
self.nb_path = nb_path
self.log_model = log_model
self.uri = uri
self.metrics_names = ['train_loss', 'valid_loss'] + [o.__name__ for o in learn.metrics]
def on_train_begin(self, **kwargs: Any) -> None:
"Prepare MLflow experiment and log params"
self.client = mlflow.tracking.MlflowClient(self.uri)
exp = self.client.get_experiment_by_name(self.exp_name)
if exp is None:
self.exp_id = self.client.create_experiment(self.exp_name)
else:
self.exp_id = exp.experiment_id
run = self.client.create_run(experiment_id=self.exp_id)
self.artifact_uri = run.info.artifact_uri
self.run = run.info.run_uuid
for k,v in self.params.items():
self.client.log_param(run_id=self.run, key=k, value=v)
def on_epoch_end(self, epoch, **kwargs:Any)->None:
"Send loss and metrics values to MLFlow after each epoch"
if kwargs['smooth_loss'] is None or kwargs["last_metrics"] is None:
return
metrics = [kwargs['smooth_loss']] + kwargs["last_metrics"]
for name, val in zip(self.metrics_names, metrics):
self.client.log_metric(self.run, name, np.float(val))
def on_train_end(self, **kwargs: Any) -> None:
"Store the notebook and stop run"
#self.client.log_artifact(run_id=self.run, local_path=self.nb_path)
#if self.log_model: MYPY.log_model(self.learn.model, self.exp_name)
self.client.set_terminated(run_id=self.run)
!nbdev_install_git_hooks
from nbdev.export import *
notebook2script()
!nbdev_build_lib
###Output
Converted 00_core.ipynb.
Converted 01_startup.ipynb.
Converted 01a_COCOnutte.ipynb.
Converted 02_runs.ipynb.
Converted index.ipynb.
|
colabs/intriguing_properties/generalized_contrastive_loss.ipynb | ###Markdown
Copyright 2020 Google LLC.Licensed under the Apache License, Version 2.0 (the "License");you may not use this file except in compliance with the License.You may obtain a copy of the License athttps://www.apache.org/licenses/LICENSE-2.0Unless required by applicable law or agreed to in writing, softwaredistributed under the License is distributed on an "AS IS" BASIS,WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.See the License for the specific language governing permissions andlimitations under the License. Generalized contrastive lossThis notebook contains implementation of generalized contrastive loss from ***Intriguing Properties of Contrastive Losses***.
###Code
def generalized_contrastive_loss(
hidden1,
hidden2,
lambda_weight=1.0,
temperature=1.0,
dist='normal',
hidden_norm=True,
loss_scaling=1.0):
"""Generalized contrastive loss.
Both hidden1 and hidden2 should have shape of (n, d).
Configurations to get following losses:
* decoupled NT-Xent loss: set dist='logsumexp', hidden_norm=True
* SWD with normal distribution: set dist='normal', hidden_norm=False
* SWD with uniform hypersphere: set dist='normal', hidden_norm=True
* SWD with uniform hypercube: set dist='uniform', hidden_norm=False
"""
hidden_dim = hidden1.shape[-1] # get hidden dimension
if hidden_norm:
hidden1 = tf.math.l2_normalize(hidden1, -1)
hidden2 = tf.math.l2_normalize(hidden2, -1)
loss_align = tf.reduce_mean((hidden1 - hidden2)**2) / 2.
hiddens = tf.concat([hidden1, hidden2], 0)
if dist == 'logsumexp':
loss_dist_match = get_logsumexp_loss(hiddens, temperature)
else:
initializer = tf.keras.initializers.Orthogonal()
rand_w = initializer([hidden_dim, hidden_dim])
loss_dist_match = get_swd_loss(hiddens, rand_w,
prior=dist,
hidden_norm=hidden_norm)
return loss_scaling * (loss_align + lambda_weight * loss_dist_match)
# Utilities for loss implementation.
def get_logsumexp_loss(states, temperature):
scores = tf.matmul(states, states, transpose_b=True) # (bsz, bsz)
bias = tf.math.log(tf.cast(tf.shape(states)[1], tf.float32)) # a constant
return tf.reduce_mean(
tf.math.reduce_logsumexp(scores / temperature, 1) - bias)
def sort(x):
"""Returns the matrix x where each row is sorted (ascending)."""
xshape = tf.shape(x)
rank = tf.reduce_sum(
tf.cast(tf.expand_dims(x, 2) > tf.expand_dims(x, 1), tf.int32), axis=2)
rank_inv = tf.einsum(
'dbc,c->db',
tf.transpose(tf.cast(tf.one_hot(rank, xshape[1]), tf.float32), [0, 2, 1]),
tf.range(xshape[1], dtype='float32')) # (dim, bsz)
x = tf.gather(x, tf.cast(rank_inv, tf.int32), axis=-1, batch_dims=-1)
return x
def get_swd_loss(states, rand_w, prior='normal', stddev=1., hidden_norm=True):
states_shape = tf.shape(states)
states = tf.matmul(states, rand_w)
states_t = sort(tf.transpose(states)) # (dim, bsz)
if prior == 'normal':
states_prior = tf.random.normal(states_shape, mean=0, stddev=stddev)
elif prior == 'uniform':
states_prior = tf.random.uniform(states_shape, -stddev, stddev)
else:
raise ValueError('Unknown prior {}'.format(prior))
if hidden_norm:
states_prior = tf.math.l2_normalize(states_prior, -1)
states_prior = tf.matmul(states_prior, rand_w)
states_prior_t = sort(tf.transpose(states_prior)) # (dim, bsz)
return tf.reduce_mean((states_prior_t - states_t)**2)
###Output
_____no_output_____ |
EA_FORM1_ImpedanciaRadiacao.ipynb | ###Markdown
**Circuito aproximado para impedância de radiação**
###Code
rho = 1.21
c = 343
pi = 3.141592653589793
###Output
_____no_output_____
###Markdown
 Baffle infinito$R_{A1} = \large\frac{0.1404\rho_{0}c_{0}}{\pi a^{2}}$$R_{A2} = \large\frac{\rho_{0}c_{0}}{\pi a^{2}}$$C_{A1} = \large\frac{5.94a^{3}}{\rho_{0}c_{0}^{2}}$$M_{A1} = \large\frac{0.27\rho_{0}}{a}$
###Code
def zrad_baffle_infto(a):
Ra1 = (0.1404 * rho * c) / (pi * a * a)
Ra2 = (rho * c) / (pi * a * a)
Ca1 = (5.94 * a * a * a) / (rho * c * c)
Ma1 = (0.27 * rho) / a
return Ra1, Ra2, Ca1, Ma1
###Output
_____no_output_____
###Markdown
Final de duto longo$R_{A1} = \large\frac{0.504\rho_{0}c_{0}}{\pi a^{2}}$$R_{A2} = \large\frac{\rho_{0}c_{0}}{\pi a^{2}}$$C_{A1} = \large\frac{5.44a^{3}}{\rho_{0}c_{0}^{2}}$$M_{A1} = \large\frac{0.1952\rho_{0}}{a}$
###Code
def zrad_duto_longo(a):
Ra1 = (0.504 * rho * c) / (pi * a * a)
Ra2 = (rho * c) / (pi * a * a)
Ca1 = (5.44 * a * a * a) / (rho * c * c)
Ma1 = (0.1952 * rho) / a
return Ra1, Ra2, Ca1, Ma1
a12pol = 0.126
zrad_baffle_infto(a12pol)
a6pol = 0.0133/2
zrad_baffle_infto(a6pol)
###Output
_____no_output_____ |
d2l/tensorflow/chapter_convolutional-neural-networks/conv-layer.ipynb | ###Markdown
图像卷积:label:`sec_conv_layer`上节我们解析了卷积层的原理,现在我们看看它的实际应用。由于卷积神经网络的设计是用于探索图像数据,本节我们将以图像为例。 互相关运算严格来说,卷积层是个错误的叫法,因为它所表达的运算其实是*互相关运算*(cross-correlation),而不是卷积运算。根据 :numref:`sec_why-conv`中的描述,在卷积层中,输入张量和核张量通过(**互相关运算**)产生输出张量。首先,我们暂时忽略通道(第三维)这一情况,看看如何处理二维图像数据和隐藏表示。在 :numref:`fig_correlation`中,输入是高度为$3$、宽度为$3$的二维张量(即形状为$3 \times 3$)。卷积核的高度和宽度都是$2$,而卷积核窗口(或卷积窗口)的形状由内核的高度和宽度决定(即$2 \times 2$)。:label:`fig_correlation`在二维互相关运算中,卷积窗口从输入张量的左上角开始,从左到右、从上到下滑动。当卷积窗口滑动到新一个位置时,包含在该窗口中的部分张量与卷积核张量进行按元素相乘,得到的张量再求和得到一个单一的标量值,由此我们得出了这一位置的输出张量值。在如上例子中,输出张量的四个元素由二维互相关运算得到,这个输出高度为$2$、宽度为$2$,如下所示:$$0\times0+1\times1+3\times2+4\times3=19,\\1\times0+2\times1+4\times2+5\times3=25,\\3\times0+4\times1+6\times2+7\times3=37,\\4\times0+5\times1+7\times2+8\times3=43.$$注意,输出大小略小于输入大小。这是因为卷积核的宽度和高度大于1,而卷积核只与图像中每个大小完全适合的位置进行互相关运算。所以,输出大小等于输入大小$n_h \times n_w$减去卷积核大小$k_h \times k_w$,即:$$(n_h-k_h+1) \times (n_w-k_w+1).$$这是因为我们需要足够的空间在图像上“移动”卷积核。稍后,我们将看到如何通过在图像边界周围填充零来保证有足够的空间移动内核,从而保持输出大小不变。接下来,我们在`corr2d`函数中实现如上过程,该函数接受输入张量`X`和卷积核张量`K`,并返回输出张量`Y`。
###Code
import tensorflow as tf
from d2l import tensorflow as d2l
def corr2d(X, K): #@save
"""计算二维互相关运算"""
h, w = K.shape
Y = tf.Variable(tf.zeros((X.shape[0] - h + 1, X.shape[1] - w + 1)))
for i in range(Y.shape[0]):
for j in range(Y.shape[1]):
Y[i, j].assign(tf.reduce_sum(
X[i: i + h, j: j + w] * K))
return Y
###Output
_____no_output_____
###Markdown
通过 :numref:`fig_correlation`的输入张量`X`和卷积核张量`K`,我们来[**验证上述二维互相关运算的输出**]。
###Code
X = tf.constant([[0.0, 1.0, 2.0], [3.0, 4.0, 5.0], [6.0, 7.0, 8.0]])
K = tf.constant([[0.0, 1.0], [2.0, 3.0]])
corr2d(X, K)
###Output
_____no_output_____
###Markdown
卷积层卷积层对输入和卷积核权重进行互相关运算,并在添加标量偏置之后产生输出。所以,卷积层中的两个被训练的参数是卷积核权重和标量偏置。就像我们之前随机初始化全连接层一样,在训练基于卷积层的模型时,我们也随机初始化卷积核权重。基于上面定义的`corr2d`函数[**实现二维卷积层**]。在`__init__`构造函数中,将`weight`和`bias`声明为两个模型参数。前向传播函数调用`corr2d`函数并添加偏置。
###Code
class Conv2D(tf.keras.layers.Layer):
def __init__(self):
super().__init__()
def build(self, kernel_size):
initializer = tf.random_normal_initializer()
self.weight = self.add_weight(name='w', shape=kernel_size,
initializer=initializer)
self.bias = self.add_weight(name='b', shape=(1, ),
initializer=initializer)
def call(self, inputs):
return corr2d(inputs, self.weight) + self.bias
###Output
_____no_output_____
###Markdown
高度和宽度分别为$h$和$w$的卷积核可以被称为$h \times w$卷积或$h \times w$卷积核。我们也将带有$h \times w$卷积核的卷积层称为$h \times w$卷积层。 图像中目标的边缘检测如下是[**卷积层的一个简单应用:**]通过找到像素变化的位置,来(**检测图像中不同颜色的边缘**)。首先,我们构造一个$6\times 8$像素的黑白图像。中间四列为黑色($0$),其余像素为白色($1$)。
###Code
X = tf.Variable(tf.ones((6, 8)))
X[:, 2:6].assign(tf.zeros(X[:, 2:6].shape))
X
###Output
_____no_output_____
###Markdown
接下来,我们构造一个高度为$1$、宽度为$2$的卷积核`K`。当进行互相关运算时,如果水平相邻的两元素相同,则输出为零,否则输出为非零。
###Code
K = tf.constant([[1.0, -1.0]])
###Output
_____no_output_____
###Markdown
现在,我们对参数`X`(输入)和`K`(卷积核)执行互相关运算。如下所示,[**输出`Y`中的1代表从白色到黑色的边缘,-1代表从黑色到白色的边缘**],其他情况的输出为$0$。
###Code
Y = corr2d(X, K)
Y
###Output
_____no_output_____
###Markdown
现在我们将输入的二维图像转置,再进行如上的互相关运算。其输出如下,之前检测到的垂直边缘消失了。不出所料,这个[**卷积核`K`只可以检测垂直边缘**],无法检测水平边缘。
###Code
corr2d(tf.transpose(X), K)
###Output
_____no_output_____
###Markdown
学习卷积核如果我们只需寻找黑白边缘,那么以上`[1, -1]`的边缘检测器足以。然而,当有了更复杂数值的卷积核,或者连续的卷积层时,我们不可能手动设计过滤器。那么我们是否可以[**学习由`X`生成`Y`的卷积核**]呢?现在让我们看看是否可以通过仅查看“输入-输出”对来学习由`X`生成`Y`的卷积核。我们先构造一个卷积层,并将其卷积核初始化为随机张量。接下来,在每次迭代中,我们比较`Y`与卷积层输出的平方误差,然后计算梯度来更新卷积核。为了简单起见,我们在此使用内置的二维卷积层,并忽略偏置。
###Code
# 构造一个二维卷积层,它具有1个输出通道和形状为(1,2)的卷积核
conv2d = tf.keras.layers.Conv2D(1, (1, 2), use_bias=False)
# 这个二维卷积层使用四维输入和输出格式(批量大小、高度、宽度、通道),
# 其中批量大小和通道数都为1
X = tf.reshape(X, (1, 6, 8, 1))
Y = tf.reshape(Y, (1, 6, 7, 1))
lr = 3e-2 # 学习率
Y_hat = conv2d(X)
for i in range(10):
with tf.GradientTape(watch_accessed_variables=False) as g:
g.watch(conv2d.weights[0])
Y_hat = conv2d(X)
l = (abs(Y_hat - Y)) ** 2
# 迭代卷积核
update = tf.multiply(lr, g.gradient(l, conv2d.weights[0]))
weights = conv2d.get_weights()
weights[0] = conv2d.weights[0] - update
conv2d.set_weights(weights)
if (i + 1) % 2 == 0:
print(f'epoch {i+1}, loss {tf.reduce_sum(l):.3f}')
###Output
epoch 2, loss 16.792
epoch 4, loss 5.780
epoch 6, loss 2.183
epoch 8, loss 0.863
epoch 10, loss 0.348
###Markdown
在$10$次迭代之后,误差已经降到足够低。现在我们来看看我们[**所学的卷积核的权重张量**]。
###Code
tf.reshape(conv2d.get_weights()[0], (1, 2))
###Output
_____no_output_____
###Markdown
Convolutions for Images:label:`sec_conv_layer`Now that we understand how convolutional layers work in theory,we are ready to see how they work in practice.Building on our motivation of convolutional neural networksas efficient architectures for exploring structure in image data,we stick with images as our running example. The Cross-Correlation OperationRecall that strictly speaking, convolutional layersare a misnomer, since the operations they expressare more accurately described as cross-correlations.Based on our descriptions of convolutional layers in :numref:`sec_why-conv`,in such a layer, an input tensorand a kernel tensor are combinedto produce an output tensor through a (**cross-correlation operation.**)Let us ignore channels for now and see how this workswith two-dimensional data and hidden representations.In :numref:`fig_correlation`,the input is a two-dimensional tensorwith a height of 3 and width of 3.We mark the shape of the tensor as $3 \times 3$ or ($3$, $3$).The height and width of the kernel are both 2.The shape of the *kernel window* (or *convolution window*)is given by the height and width of the kernel(here it is $2 \times 2$).:label:`fig_correlation`In the two-dimensional cross-correlation operation,we begin with the convolution window positionedat the upper-left corner of the input tensorand slide it across the input tensor,both from left to right and top to bottom.When the convolution window slides to a certain position,the input subtensor contained in that windowand the kernel tensor are multiplied elementwiseand the resulting tensor is summed upyielding a single scalar value.This result gives the value of the output tensorat the corresponding location.Here, the output tensor has a height of 2 and width of 2and the four elements are derived fromthe two-dimensional cross-correlation operation:$$0\times0+1\times1+3\times2+4\times3=19,\\1\times0+2\times1+4\times2+5\times3=25,\\3\times0+4\times1+6\times2+7\times3=37,\\4\times0+5\times1+7\times2+8\times3=43.$$Note that along each axis, the output sizeis slightly smaller than the input size.Because the kernel has width and height greater than one,we can only properly compute the cross-correlationfor locations where the kernel fits wholly within the image,the output size is given by the input size $n_h \times n_w$minus the size of the convolution kernel $k_h \times k_w$via$$(n_h-k_h+1) \times (n_w-k_w+1).$$This is the case since we need enough spaceto "shift" the convolution kernel across the image.Later we will see how to keep the size unchangedby padding the image with zeros around its boundaryso that there is enough space to shift the kernel.Next, we implement this process in the `corr2d` function,which accepts an input tensor `X` and a kernel tensor `K`and returns an output tensor `Y`.
###Code
import tensorflow as tf
from d2l import tensorflow as d2l
def corr2d(X, K): #@save
"""Compute 2D cross-correlation."""
h, w = K.shape
Y = tf.Variable(tf.zeros((X.shape[0] - h + 1, X.shape[1] - w + 1)))
for i in range(Y.shape[0]):
for j in range(Y.shape[1]):
Y[i, j].assign(tf.reduce_sum(
X[i: i + h, j: j + w] * K))
return Y
###Output
_____no_output_____
###Markdown
We can construct the input tensor `X` and the kernel tensor `K`from :numref:`fig_correlation`to [**validate the output of the above implementation**]of the two-dimensional cross-correlation operation.
###Code
X = tf.constant([[0.0, 1.0, 2.0], [3.0, 4.0, 5.0], [6.0, 7.0, 8.0]])
K = tf.constant([[0.0, 1.0], [2.0, 3.0]])
corr2d(X, K)
###Output
_____no_output_____
###Markdown
Convolutional LayersA convolutional layer cross-correlates the input and kerneland adds a scalar bias to produce an output.The two parameters of a convolutional layerare the kernel and the scalar bias.When training models based on convolutional layers,we typically initialize the kernels randomly,just as we would with a fully-connected layer.We are now ready to [**implement a two-dimensional convolutional layer**]based on the `corr2d` function defined above.In the `__init__` constructor function,we declare `weight` and `bias` as the two model parameters.The forward propagation functioncalls the `corr2d` function and adds the bias.
###Code
class Conv2D(tf.keras.layers.Layer):
def __init__(self):
super().__init__()
def build(self, kernel_size):
initializer = tf.random_normal_initializer()
self.weight = self.add_weight(name='w', shape=kernel_size,
initializer=initializer)
self.bias = self.add_weight(name='b', shape=(1, ),
initializer=initializer)
def call(self, inputs):
return corr2d(inputs, self.weight) + self.bias
###Output
_____no_output_____
###Markdown
In$h \times w$ convolutionor a $h \times w$ convolution kernel,the height and width of the convolution kernel are $h$ and $w$, respectively.We also refer toa convolutional layer with a $h \times w$convolution kernel simply as a $h \times w$ convolutional layer. Object Edge Detection in ImagesLet us take a moment to parse [**a simple application of a convolutional layer:detecting the edge of an object in an image**]by finding the location of the pixel change.First, we construct an "image" of $6\times 8$ pixels.The middle four columns are black (0) and the rest are white (1).
###Code
X = tf.Variable(tf.ones((6, 8)))
X[:, 2:6].assign(tf.zeros(X[:, 2:6].shape))
X
###Output
_____no_output_____
###Markdown
Next, we construct a kernel `K` with a height of 1 and a width of 2.When we perform the cross-correlation operation with the input,if the horizontally adjacent elements are the same,the output is 0. Otherwise, the output is non-zero.
###Code
K = tf.constant([[1.0, -1.0]])
###Output
_____no_output_____
###Markdown
We are ready to perform the cross-correlation operationwith arguments `X` (our input) and `K` (our kernel).As you can see, [**we detect 1 for the edge from white to blackand -1 for the edge from black to white.**]All other outputs take value 0.
###Code
Y = corr2d(X, K)
Y
###Output
_____no_output_____
###Markdown
We can now apply the kernel to the transposed image.As expected, it vanishes. [**The kernel `K` only detects vertical edges.**]
###Code
corr2d(tf.transpose(X), K)
###Output
_____no_output_____
###Markdown
Learning a KernelDesigning an edge detector by finite differences `[1, -1]` is neatif we know this is precisely what we are looking for.However, as we look at larger kernels,and consider successive layers of convolutions,it might be impossible to specifyprecisely what each filter should be doing manually.Now let us see whether we can [**learn the kernel that generated `Y` from `X`**]by looking at the input--output pairs only.We first construct a convolutional layerand initialize its kernel as a random tensor.Next, in each iteration, we will use the squared errorto compare `Y` with the output of the convolutional layer.We can then calculate the gradient to update the kernel.For the sake of simplicity,in the followingwe use the built-in classfor two-dimensional convolutional layersand ignore the bias.
###Code
# Construct a two-dimensional convolutional layer with 1 output channel and a
# kernel of shape (1, 2). For the sake of simplicity, we ignore the bias here
conv2d = tf.keras.layers.Conv2D(1, (1, 2), use_bias=False)
# The two-dimensional convolutional layer uses four-dimensional input and
# output in the format of (example, height, width, channel), where the batch
# size (number of examples in the batch) and the number of channels are both 1
X = tf.reshape(X, (1, 6, 8, 1))
Y = tf.reshape(Y, (1, 6, 7, 1))
lr = 3e-2 # Learning rate
Y_hat = conv2d(X)
for i in range(10):
with tf.GradientTape(watch_accessed_variables=False) as g:
g.watch(conv2d.weights[0])
Y_hat = conv2d(X)
l = (abs(Y_hat - Y)) ** 2
# Update the kernel
update = tf.multiply(lr, g.gradient(l, conv2d.weights[0]))
weights = conv2d.get_weights()
weights[0] = conv2d.weights[0] - update
conv2d.set_weights(weights)
if (i + 1) % 2 == 0:
print(f'epoch {i + 1}, loss {tf.reduce_sum(l):.3f}')
###Output
epoch 2, loss 34.684
epoch 4, loss 11.937
epoch 6, loss 4.509
epoch 8, loss 1.783
epoch 10, loss 0.720
###Markdown
Note that the error has dropped to a small value after 10 iterations. Now we will [**take a look at the kernel tensor we learned.**]
###Code
tf.reshape(conv2d.get_weights()[0], (1, 2))
###Output
_____no_output_____ |
tutorials/tutorial.ipynb | ###Markdown
Introduction Key Problems with ML in Chemistry1. Translational Variance2. Rotational Variance3. Permutation VarianceTraditional ML algorithms struggle with making molecular predictions because of the nature of how we represent molecules. Typically, a molecule in space is represented as a 3D cartesian array, typically of shape (points, 3). Something like a Neural Network (NN) cannot operate on an array like this, because each position in the array doesn't mean something on it's own. For example, a molecule could have one cartesian array centered at (0, 0, 0), and another centered at (15, 15, 15). Both arrays represent the same molecule, but numerically they are very different. This is an example of **translational variance**. A similar example exists for **rotational variance**, but imagine rotating the molecule by $90^{\circ}$ instead of translating the molecule. In both of these examples, inputting the two different arrays which represent a single molecule would result in the NN seeing the arrays as representing **different** molecules, which they are not. Much work has gone into creating novel **input representations** for molecules in an attempt to resolve problems with translational and rotational variance. Translational Variance FixWe can solve the translational variance problem by computing the distance matrix of a molecule to obtain a representation of the molecule that is **invariant** to translation. However, in practice, a distance matrix array of shape (N, N) has a **single number** to characterize the distance between two points. Simply put, there is not enough **explicit information** contained within a distance matrix array to be useful to a NN. However, expanding that single number into a vector which probes various distances provides a more sparse representation, but one which is ultimately more **explicit**. An implementation of this idea, known as Atom-centered symmetry functions ([Behler, 2011](https://aip.scitation.org/doi/10.1063/1.3553717)) achieve this representation. These symmetry functions probe continuous space with gaussian functions centered on a grid, descretizing the provided molecular distance matrix. As each position on the grid **explicitly** means something (e.g. two points positioned exactly $1.42e^{-10}m$ apart), the network is capable of making inferences from each position in the array.Grid visualization: [desmos](https://www.desmos.com/calculator/4bncqy2bol) Basis Function Demonstration
###Code
# Package written by Trevor Profitt, modified for TF 2.0 by Riley Jackson
from tfn.layers.atomic_images import DistanceMatrix, GaussianBasis
import numpy as np
from matplotlib import pyplot as plt
# batch, points, 3
np.random.seed(0)
cartesians = np.random.randint(1, 5, size=(5, 15, 3)).astype('float32')
print('cartesians shape: {}'.format(cartesians.shape))
distance_matrix = DistanceMatrix()(cartesians)
print('distance_matrix shape: {}'.format(distance_matrix.shape))
image = GaussianBasis(
width=0.2,
spacing=0.2,
min_value=-1.0,
max_value=15.0
)(distance_matrix)
print('image shape: {}'.format(image.shape))
# (batch, points, points, functions)
plt.plot(image[0, 0, 1, :30])
plt.axis(xlim=16.0, ylim=1.0)
plt.show()
###Output
cartesians shape: (5, 15, 3)
distance_matrix shape: (5, 15, 15)
image shape: (5, 15, 15, 80)
###Markdown
Permutation Variance FixThe ordering of points in an input tensor is meaningless in a molecular context, so we must be careful to avoid ever depending on this ordering. We can achieve this by only ever operating the network atom-wise to obtain **atomic property predictions**. Whenever we want to predict **molecular properties**, we must use cummulative combinations of these atomic predictions (e.g. summation). Rotational Variance FixAtom-centered symmetry functions are rotationally **invariant** because the distance matrix is invariant. The is fine for networks which predict scalar values from molecules, e.g. energy. However, for problems that are direction dependent, we want rotational **equivariance**, where the output of the network rotations *the same* as the input, e.g. force prediction, molecular dynamics, etc.Need a network that can encode directional information and feed it forward to influence outputs. Enter the **Tensor Field Networks**. Tensor Field NetworksTensor Field Networks (TFN) are **Rotationally Equivariant Continuous Convolution Neural Networks** which are capable of inputing continuous 3D point-clouds (e.g. molecules) and making scalar, vector, and higher order tensor predictions which rotate with the original input point-cloud ([Thomas et. al. 2018](https://arxiv.org/abs/1802.08219)).Ignoring the **continuous convolution** part, this means that TFNs are capable of knowing when an image has been rotated, something vanilla convolution nets are not capable of.For example, a traditional conv. net trained to recognize cats on **non-rotated images** would not identify a cat in the second picture: While TFNs will still identify a cat in the rotated image, trained only on images in a single orientation. We can convince ourselves this is true by visualizing TFNs outputing a vector upon input of a molecule: Equivariance Demonstration
###Code
import tensorflow as tf
import numpy as np
import math
from matplotlib import pyplot as plt
from tensorflow.python.keras.models import Model
print('tensorflow version: {}\neager mode on: {}'.format(tf.__version__, tf.executing_eagerly()))
def rotation_matrix(axis_matrix=None, theta=math.pi/2):
"""
Return the 3D rotation matrix associated with counterclockwise rotation about
the given `axis` by `theta` radians.
:param axis_matrix: np.ndarray. Defaults to [1, 0, 0], the x-axis.
:param theta: float. Defaults to pi/2. Rotation in radians.
"""
axis_matrix = axis_matrix or [1, 0, 0]
axis_matrix = np.asarray(axis_matrix)
axis_matrix = axis_matrix / math.sqrt(np.dot(axis_matrix, axis_matrix))
a = math.cos(theta / 2.0)
b, c, d = -axis_matrix * math.sin(theta / 2.0)
aa, bb, cc, dd = a * a, b * b, c * c, d * d
bc, ad, ac, ab, bd, cd = b * c, a * d, a * c, a * b, b * d, c * d
return np.array([[aa + bb - cc - dd, 2 * (bc + ad), 2 * (bd - ac)],
[2 * (bc - ad), aa + cc - bb - dd, 2 * (cd + ab)],
[2 * (bd + ac), 2 * (cd - ab), aa + dd - bb - cc]])
class MyModel(Model):
def __init__(self, max_z: int = 5):
super().__init__(dynamic=True)
self.max_z = max_z
self.embedding = SelfInteraction(16)
self.conv_0 = MolecularConvolution()
self.conv_1 = MolecularConvolution(si_units=1, output_orders=[1])
def call(self, inputs, training=None, mask=None):
r, z = inputs
one_hot, image, vectors = Preprocessing(self.max_z)([r, z])
features = self.embedding(K.expand_dims(one_hot, axis=-1))
output = self.conv_0([one_hot, image, vectors] + features)
output = self.conv_1([one_hot, image, vectors] + output)[0]
output = K.squeeze(output, axis=-2)
return K.mean(output, axis=1) # (batch, 3)
def compute_output_shape(self, input_shape):
r, z = input_shape
batch, *_ = r
return tf.TensorShape([batch, 3])
# Set up random input/target data, instantiate model
seed = 1
np.random.seed(seed)
tf.random.set_seed(seed)
cartesians = np.random.rand(5, 29, 3)
atomic_nums = np.random.randint(0, 5, size=[5, 29])
vectors = np.random.rand(5, 3)
model = MyModel()
model.compile(optimizer='adam', loss='mae', run_eagerly=True)
model.fit(x=[cartesians, atomic_nums], y=vectors, epochs=2, batch_size=5)
# Make predictions on normal/rotated vectors
predicted_vectors = model.predict([cartesians, atomic_nums])
R = rotation_matrix([1, 0, 0], theta=math.pi/4)
rotated_cartesians = np.dot(cartesians, R)
rotated_predicted_vectors = model.predict([rotated_cartesians, atomic_nums])
print('equivariance perserved: {}'.format(
np.alltrue(
np.isclose(
np.dot(predicted_vectors, R), rotated_predicted_vectors,
rtol=0.,
atol=1e-5
)
)
))
# Matplotlib display stuff
fig = plt.figure()
ax = fig.gca(projection='3d')
ax.set_xlim3d(-1, 1)
ax.set_ylim3d(-1, 1)
ax.set_zlim3d(-1, 1)
pv, rpv = predicted_vectors[0], rotated_predicted_vectors[0]
ax.quiver(
[0, 0], [0, 0], [0, 0],
[pv[0], rpv[0]], [pv[1], rpv[1]], [pv[2], rpv[2]],
length =2, normalize = True
)
cosang = np.dot(pv, rpv)
sinang = np.linalg.norm(np.cross(pv, rpv))
print('angle between predicted vectors: {}'.format(np.rad2deg(np.arctan2(sinang, cosang))))
plt.show()
###Output
tensorflow version: 2.0.0
eager mode on: True
Train on 5 samples
Epoch 1/2
5/5 [==============================] - 3s 629ms/sample - loss: 0.4732
Epoch 2/2
5/5 [==============================] - 0s 72ms/sample - loss: 0.4692
equivariance perserved: True
angle between predicted vectors: 44.04207229614258
###Markdown
Tensor Field Networks are powerful... How do they work?TFNs combine continuous convolution (i.e. a filter generating neural network) with the spherical harmonics to produce **spherically symmetric filters** (dubbed Harmonic Filters) which are point-convolved with feature tensors associated with the 3D point-cloud. These Harmonic Filters are capable of convolving with features associated with the point-cloud (e.g. atom type information for molecules, mass, velocity, acceleration, etc.) to produce outputs which rotate **equivariantly** with the input point-cloud. Spherical HarmonicsThe Spherical Harmonics are a set of orthogonal basis functions defined on the surface of a sphere. They can be utilized to represent any function which exists on a sphere. They have many special properties, however, for our purposes, we are interested in their **spherical symmetry**. The filters of our network are composed of a directionless **Radial** function and the real component of the spherical harmonic function centered on the unit-vectors of our point-cloud.We're not mathematicians, so the important thing to know is that the spherical symmetry of the spherical harmonics provide the filters of our network with **rotational equivariance**. The implementation of these basis functions is quite simple due to a few shortcuts, which we'll see soon. Tensor Rotation OrderFeature tensors that are part of the group $S(O3)$ (i.e. tensors that rotate normally in 3D), require $2l + 1$ dimensions to be represented. "$l$" is referred to as the **rotation order** of that tensor. e.g. an array which represents a scalar value has rotation order $l=0$, and an array representing a vector has rotation order $l=1$.e.g. an *energy* (scalar) array for a batch of molecules would have the shape **(molecules, points, 1)**, while a *force* (vector) array for a batch of molecules would have shape **(molecules, points, 3)**.Once again, we're not mathematicians, so all we need to know is that rotation order refers to the **rank** of what our feature tensor *represents*, and that we can only combine tensors of varying rotation order in **specific ways** if we hope to conserve **rotational equivariance**. Equivariant Combination TypesThe combination rules are essentially the traditional ways of combining scalars, vectors, and higher order tensors. With notation $A \cdot B \rightarrow O$, the combinations are:$ L \cdot 0 \rightarrow L $; scalar multiplication$ 1 \cdot 1 \rightarrow 1 $; element-wise multiplication$ 1 \cdot 1 \rightarrow 0 $; dot product Equations of Tensor Field Networks The Harmonic Filter$$\Large{F^{(l_{f}l_{i})}_{cm}(\vec{r}) = R^{(l_{f}l_{i})}_{c}(r)Y^{l_{f}}_{m'}(\hat{r})}$$Where:$\vec{r}$ are the distance vectors for all points in the point-cloud,$F^{(l_{f}l_{i})}_{cm}$ is the Harmonic Filter of rotation order $l_{f}$ to be applied to feature of rotation order $l_{i}$$Y^{l_{f}}_{m'}(\hat{r})$ is the output of the spherical harmonic function applied to the unit vector $\hat{r}$ $R^{(l_{f}l_{i})}_{c}$ is the filter generating network, know as the **Radial** function.Some lucky shortcuts:$Y^{0}(\vec{r}) \propto 1$; since $Y^{0}$ is a sphere,$Y^{1}(\vec{r}) \propto \hat{r}$; since $Y^{1}$ is centered on the unit vectors, $\vec{r}$Some notes:the Radial takes as input the directionless discretized distance matrix, *r*, and outputs some learned representation of that distance matrix. The Radial function is isolated from either vectors or feature tensors, and as such is **general to any function**, so long as the output's shape matches with that of the feature tensor it will eventually be convolved with. The current default implementation is a multi-layer dense neural network, but could just as easily be replaced with a convolutional network, or some traditional architecture.A **Harmonic Filter** is created upon multiplying the output of the Radial with the output of the spherical harmonic function. Point ConvolutionCore function of the TFNs, contains 90%+ of the networks weights:$$\Large{ L^{l_{o}}_{acm_{o}}(\vec{r}_{a}, V^{(l_{\textit{i}})}_{acm_{i}}) := \sum_{m_{\textit{f}}, m_{\textit{i}}}C^{(l_{o}m_{o})}_{(l_{f},m_{f})(l_{i},m_{i})} \sum_{b \in S} F^{(l_{f},l_{i})}_{cm_{f}}(\vec{r}_{ab})V^{l_{i}}_{bcm_{i}}}$$Where:$\vec{r}_{a}$ is the image (discretized distance matrix),$V^{(l_{\textit{i}})}_{acm_{i}}$ is the collection of feature vectors, indexed by *a*, the point; *c*, the *channel* or *feature* of point *a*; *m*, the representation order of that *feature*.$F^{(l_{f},l_{i})}_{cm_{f}}(\vec{r}_{ab})$ is the Harmonic Filter$C^{(l_{o}m_{o})}_{(l_{f},m_{f})(l_{i},m_{i})}$ is the Clebsch-Gordan Coefficient for the convolution of Harmonic Filter and feature tensorSome Notes:The three indices $l_{i}$, $l_{f}$, and $l_{o}$ denote rotation order for input tensor (feature), Harmonic Filter, and output tensor, respectively. Based on the rules for combining tensors of varying rotation order (RO) described earlier, it may now be clear that there are **multiple ways** of combining filter and feature to obtain an output tensor. As such, usage of increasingly higher RO inputs and filters results in an explosion of total output tensors. For example, if you have a point-cloud with feature tensors representing mass and velocity for each point in the cloud, i.e. two feature tensors, one of RO 0 and the other RO 1, and convolve those feature tensors with Harmonic Filters of RO0 and RO1, there will be a total of **5 output tensors**. Once tensors higher than vectors are introduced the space explodes. However, there are very few instances where tensors of RO higher than 1 are required.It is also import to note that each Harmonic Filter is indexed by $l_{i}$ and $l_{f}$. This means that for every convolution operation in a TFN, each input tensor is convolved with two full-fledged neural networks, both potentially quite large.On the surface, this is not much different from a traditional convolutional network. Typically, filters are small 3x3 weight arrays. The power of a conv. net comes from layering many of these filters, which serve as basic 'feature' detectors, on top of each other to build larger structure detection in data, typically images. TFNs are quite similar, only the filters are much larger than 3x3 arrays and they leverage symmetry from the Spherical Harmonics to gain rotational equivariance to input data. ConcatenationAs we saw in the last section, a point convolution can output many tensors of varying RO, depending on our choice of Harmonic Filter, and our input tensors. In the last example, we had **5 tensors**, **3** of RO *0*, **2** of RO *1*. Only tensors of different RO must be stored seperately, so we can combine tensors of like RO by concatenating along the second last axis, the channels/feature dimension. So, following the same example, our list of **5** examples condenses to **2** tensors, one for RO *0* and another for RO *1*. Self Interaction$$\Large{ \sum_{c'}W^{(l)}_{cc'}V^{(l)}_{ac'm}}$$Where:$W^{(l)}_{cc'}$ refers to a kernel matrix$V^{(l)}_{ac'm}$ refers to an input feature tensor indexed by representation index, *m**c'* refers to the old channels dimension of the input tensor *V*, while *c* refers to the new channels dimensionSome Notes:This operation is analagous to a typical dense layer without biases. This operation can be thought of as a scalar transform, and is thus also rotationally equivariant. Equivariant Activation$$\Large{ \eta^{(0)}(V^{(0)}_{ac} + b^{(0)}_{c})}$$$$\Large{ \eta^{(l)}(||V||^{(l)}_{ac} + b^{(l)}_{c})V^{(l)}_{acm}}$$Where:$\eta$ refers to the nonlinear activation function applied to the tensors. This is typically a shifted softplus.$V^{(l)}_{ac'm}$ refers to the input tensors, indexed by the representation index, *m*.Some Notes:Much like the Self Interaction layer, this is a scalar transform operation, and is thus rotationally equivariant. Tensor Field Networks Implemented using Keras & TF 2.0All of these operations are contained nicely with the Keras Layer pattern. convenience layers, `tfn.layers.Convolution` and `tfn.layers.MolecularConvolution`, encapsulate all of the above operations with a very simple interface. Also contained in the `tfn.layers` package is `Preprocessing` layer, which takes as input cartesian and atomic number tensors and outputs the tensors: `one_hot`, which is used to identify the type of each atom; `image`, which is the discretized distance matrix of the cartesian tensor; `vectors`, which is the set of unit vectors between every point in the cartesian tensor. Below is an example script for setting up and training a TFN to predict molecular energies and atomic forces concurrently:
###Code
import numpy as np
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Input, Lambda
from tensorflow.keras import backend as K
# Package written by Riley Jackson
from tfn.layers import MolecularConvolution, Preprocessing, SelfInteraction
### MODEL ###
# Define inputs
r = Input(shape=(29, 3), dtype='float32', name='r')
z = Input(shape=(29,), dtype='int32', name='z')
# Set up point-cloud/feature tensors
one_hot, image, vectors = Preprocessing(max_z=6)([r, z])
expanded_onehot = Lambda(lambda x: K.expand_dims(x, axis=-1))(one_hot)
feature_tensor = SelfInteraction(units=32)(expanded_onehot)
# Neural Network
output = MolecularConvolution(name='conv_0')([one_hot, image, vectors] + feature_tensor)
output = MolecularConvolution(name='conv_1')([one_hot, image, vectors] + output)
output = MolecularConvolution(name='conv_2', si_units=1)([one_hot, image, vectors] + output)
# Reshape output tensors
atomic_energies = Lambda(lambda x: K.squeeze(x, axis=-1))(output[0])
molecular_energies = Lambda(lambda x: K.sum(x, axis=-2), name='molecular_energies')(atomic_energies) # (batch, 1)
forces = Lambda(lambda x: K.squeeze(x, axis=-2), name='forces')(output[1]) # (batch, points, 3)
model = Model(inputs=[r, z], outputs=[molecular_energies, forces])
# Generate some random inputs/targets
cartesians = np.random.rand(5, 29, 3)
atomic_nums = np.random.randint(0, 5, size=(5, 29))
e = np.random.rand(5, 1)
f = np.random.rand(5, 29, 3)
# Compile and fit
model.compile(loss='mae', optimizer='adam')
model.fit(x=[cartesians, atomic_nums], y=[e, f], epochs=2)
###Output
Train on 5 samples
Epoch 1/2
5/5 [==============================] - 19s 4s/sample - loss: 6.6132 - molecular_energies_loss: 6.0510 - forces_loss: 0.5622
Epoch 2/2
5/5 [==============================] - 0s 27ms/sample - loss: 6.1962 - molecular_energies_loss: 5.6547 - forces_loss: 0.5415
###Markdown
Remember to install the package: pip install git+https://github.com/jrob93/nice_orbsAnd install dependencies! (see nice_orbs_env.yml)
###Code
# If not pip installing add import path
import sys
import os
print(os.getcwd())
sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), '..')))
from nice_orbs.orb_class import BodyOrb
import nice_orbs.orb_funcs as orb_funcs
###Output
_____no_output_____
###Markdown
Create a blank body to add orbit values to
###Code
bod = BodyOrb()
###Output
_____no_output_____
###Markdown
Set some values manually. Note that by default nice_orbs accepts distances in units of semimajor axis, angles in radians. By default G=1 and M_sun=1 therefore time is in units of 1yr/2pi
###Code
bod.a = 2.5
bod.e = 0.1
bod.inc = np.radians(5)
bod.print_orb()
###Output
a=2.5,e=0.1,inc=0.08726646259971647,peri=None,node=None,f=None
###Markdown
An orbit can also be passed from a dictionary
###Code
orb_dict = {"a":2.5,
"e":0.1,
"inc":np.radians(5),
"peri":np.radians(10),
"node":np.radians(15)
}
bod.load_dict(orb_dict)
bod.print_orb()
###Output
a=2.5,e=0.1,inc=0.08726646259971647,peri=0.17453292519943295,node=0.2617993877991494,f=None
###Markdown
Note that we have only passed the 5 elements required to describe the shape and orientation of the elliptical orbit in 3d space. Also we need to call calc_orb_vectors in order to find the unit vectors of this ellipse from the elements
###Code
bod.calc_orb_vectors()
bod.calc_values()
bod.ep,bod.eQ
###Output
_____no_output_____
###Markdown
Now we can find the xyz positions that describe the elliptical orbit in space
###Code
df_pos = bod.planet_orbit()
fig = plt.figure()
ax = plt.axes(projection='3d')
ax.scatter3D(0,0,0,marker="+",c="k") # plot origin (sun)
ax.plot3D(df_pos["x"],df_pos["y"],df_pos["z"],label = "bod")
ax.set_box_aspect((np.ptp(df_pos["x"]), np.ptp(df_pos["y"]), np.ptp(df_pos["z"]))) # aspect ratio is 1:1:1 in data space
ax.legend()
ax.set_xlabel("x")
ax.set_ylabel("y")
ax.set_zlabel("z")
plt.show()
###Output
_____no_output_____
###Markdown
We can also calculate the positions of an object from elements such as true_anomaly
###Code
bod.f = np.radians(30)
bod.n
df_rv = bod.pos_vel_from_orbit(bod.f)
fig = plt.figure()
ax = plt.axes(projection='3d')
ax.scatter3D(0,0,0,marker="+",c="k") # plot origin (sun)
ax.plot3D(df_pos["x"],df_pos["y"],df_pos["z"],label = "bod")
ax.scatter3D(df_rv["x"],df_rv["y"],df_rv["z"],label = "f_true = {:.2f} deg".format(np.degrees(bod.f)))
ax.set_box_aspect((np.ptp(df_pos["x"]), np.ptp(df_pos["y"]), np.ptp(df_pos["z"]))) # aspect ratio is 1:1:1 in data space
ax.legend()
ax.set_xlabel("x")
ax.set_ylabel("y")
ax.set_zlabel("z")
plt.show()
###Output
_____no_output_____
###Markdown
At time = 0 the object should be at its perihelion. Be wary of the time units, Kepler's second law (a^3 = T^2) gives the period from the semimajor axis in years (remember that 1 year = 2pi in these default units).
###Code
t_list = np.linspace(0,2*np.pi*(bod.a**(3.0/2.0)))
fig = plt.figure()
ax = plt.axes(projection='3d')
ax.scatter3D(0,0,0,marker="+",c="k") # plot origin (sun)
ax.plot3D(df_pos["x"],df_pos["y"],df_pos["z"],label = "bod")
for t in t_list:
bod.f = orb_funcs.f_from_t(t,bod.e,bod.n,0,0)
df_rv = bod.pos_vel_from_orbit(bod.f)
s1 = ax.scatter3D(df_rv["x"],df_rv["y"],df_rv["z"],c=t,vmin = np.amin(t_list), vmax = np.amax(t_list))
cbar1=fig.colorbar(s1)
cbar1.set_label("time")
ax.set_box_aspect((np.ptp(df_pos["x"]), np.ptp(df_pos["y"]), np.ptp(df_pos["z"]))) # aspect ratio is 1:1:1 in data space
ax.legend()
ax.set_xlabel("x")
ax.set_ylabel("y")
ax.set_zlabel("z")
plt.show()
###Output
_____no_output_____
###Markdown
*synthtorch* tutorialTrain a network for a synthesis task with synthtorchAuthor: Jacob ReinholdDate: Mar 1, 2019 Setup notebook
###Code
import logging
import sys
import matplotlib.pyplot as plt
import nibabel as nib
import numpy as np
from PIL import Image
import synthtorch
###Output
_____no_output_____
###Markdown
Setup a logger (change the level here to control output logging information)
###Code
logging.basicConfig(format='%(asctime)s - %(name)s - %(levelname)s - %(message)s',
level=logging.getLevelName('INFO'))
logger = logging.getLogger(__name__)
###Output
_____no_output_____
###Markdown
Support in-notebook plotting
###Code
%matplotlib inline
###Output
_____no_output_____
###Markdown
Report versions
###Code
print('numpy version: {}'.format(np.__version__))
from matplotlib import __version__ as mplver
print('matplotlib version: {}'.format(mplver))
pv = sys.version_info
print('python version: {}.{}.{}'.format(pv.major, pv.minor, pv.micro))
###Output
python version: 3.7.2
###Markdown
Reload packages where content for package development
###Code
%load_ext autoreload
%autoreload 2
###Output
_____no_output_____
###Markdown
Setup learnerIn this section, we instantiate a `learner` (inspired by the [fastai](https://github.com/fastai/fastai) library) which holds all the functions relevant to training and prediction; however, the library is tailored for experimentation with *synthesis* tasks, which is why we do not use fastai by itself.The easiest way to interface with the library is to create a configuration file (see [here](https://gist.github.com/jcreinhold/793d26387f6a8b9f6966b59c6705f249) for an example with instructions on how to fill out the file at the bottom) which holds the experimental parameters and setup.Below we import a configuration file for a T1-to-T1 (MPRAGE) task, i.e., a (denoising) autoencoder using a U-net architecture. In this case, I will actually load a pretrained model and continue training it (for one epoch). `synthtorch` knows to continue training if the configuration file field of `trained_model` is already an existing file, otherwise it will train a new network from scratch (and save it to the location in `trained_model`).
###Code
config = synthtorch.ExperimentConfig.load_json('config.json')
###Output
_____no_output_____
###Markdown
Now we create the model in a wrapper called `Learner` with the specified configuration.
###Code
learner = synthtorch.Learner.train_setup(config)
###Output
2019-03-01 17:27:25,516 - synthnn.learn.learner - INFO - Number of trainable parameters in model: 531052
2019-03-01 17:27:25,544 - synthnn.learn.learner - INFO - Loaded checkpoint: tutorial.pth (epoch 30)
2019-03-01 17:27:25,546 - synthnn.learn.learner - INFO - Adding data augmentation transforms
2019-03-01 17:27:25,821 - synthnn.learn.learner - INFO - Number of training images: 57275
2019-03-01 17:27:25,856 - synthnn.learn.learner - INFO - Number of validation images: 7250
2019-03-01 17:27:25,857 - synthnn.learn.learner - INFO - LR: 0.00300
###Markdown
I can now train the model with the following command (I am only training for one epoch for this tutorial):
###Code
learner.fit(1)
###Output
2019-03-01 13:12:28,931 - synthnn.learn.learner - INFO - Epoch: 1 - Training Loss: 4.70e+00, Validation Loss: 4.65e+00
###Markdown
Now we can save the model to `tutorial.pth` with the following command:
###Code
learner.save('tutorial.pth')
###Output
_____no_output_____
###Markdown
You can run the commands below without the following command; however, I will load a model here which produces ok results.
###Code
learner.load('good.pth')
###Output
_____no_output_____
###Markdown
Now we can do a prediction with either a `.tif` file (since this example is a 2D network) or a `.nii` file (or `.png`, if that is what was used for training). Note that the filename(s) must be in a list because `synthtorch` supports multiple modalities for input/output. In this case I use a `.tif` file for the convenience of displaying the result.
###Code
test_fn = 'test_img.tif'
out_img = learner.predict([test_fn])
orig = np.asarray(Image.open(test_fn))
synth = np.asarray(out_img)
f, (ax1,ax2) = plt.subplots(1,2,figsize=(8,8))
ax1.imshow(orig,cmap='gray',vmax=synth.max()); ax1.axis('off'); ax1.set_title('Original')
ax2.imshow(synth,cmap='gray'); ax2.axis('off'); ax2.set_title('Synthesized');
###Output
_____no_output_____
###Markdown
Example 1: Finding CRISPR-Cas systems in a cyanobacteria genome In this example, we will annotate and visualize CRISPR-Cas systems in the cyanobacteria species Rippkaea orientalis. CRISPR-Cas is a widespread bacterial defense system, found in at least 50% of all known prokaryotic species. This system is significant in that it can be leveraged as a precision gene editing tool, an advancement that was awarded the 2020 Nobel Prize in Chemistry. The genome of R. orientalis harbors two complete CRISPR-Cas loci (one chromosomal, and one extrachromosomal/plasmid).You can download the complete assembled genome [here](https://www.ncbi.nlm.nih.gov/assembly/GCF_000024045.1/); it is also available at https://github.com/wilkelab/Opfi in the `tutorials` folder (`tutorials/GCF_000024045.1_ASM2404v1_genomic.fna.gz`). This tutorial assumes the user has already installed Opfi and all dependencies (if installing with conda, this is done automatically). Some familiarity with BLAST and the basic homology search algorithm may also be helpful, but is not required. **Note:** This tutorial uses several input data files, all of which are provided in the `tutorials` directory. If running this notebook in another working directory, be sure copy over all three data files as well. **1. Use the makeblastdb utility to convert a Cas protein database to BLAST format** We start by converting a Cas sequence database to a format that BLAST can recognize, using the command line utility `makeblastdb`, which is part of the core NCBI BLAST+ distribution. A set of ~20,000 non-redundant Cas sequences, downloaded from [Uniprot](https://www.uniprot.org/uniref/) is available as a tar archive (`tutorials/cas_database.tar.gz`). We'll make a new directory, "blastdb", and extract sequences there:
###Code
! mkdir blastdb
! cd blastdb && tar -xzf ../cas_database.tar.gz && cd ..
###Output
_____no_output_____
###Markdown
Next, create two BLAST databases for the sequence data: one containing Cas1 sequences only, and another that contains the remaining Cas sequences.
###Code
! cd blastdb && cat cas1.fasta | makeblastdb -dbtype prot -title cas1 -hash_index -out cas1_db && cd ..
! cd blastdb && cat cas[2-9].fasta cas1[0-2].fasta casphi.fasta | makeblastdb -dbtype prot -title cas_all -hash_index -out cas_all_but_1_db
###Output
_____no_output_____
###Markdown
`-dbtype prot` simply tells `makeblastdb` to expect amino acid sequences. We use `-title` and `-out` to name the database (required by BLAST) and to prefix the database files, respectively. `-hash_index` directs `makeblastdb` to generate a hash index of protein sequences, which can speed up computation time. **2. Use Opfi's Gene Finder module to search for CRISPR-Cas loci** CRISPR-Cas systems are extremely diverse. The most recent [classification effort](https://www.nature.com/articles/s41579-019-0299-x) identifies 6 major types, and over 40 subtypes, of compositionally destinct systems. Although there is sufficent sequence similarity between subtypes to infer the existence of a common ancestor, the only protein family present in the majority of CRISPR-cas subtypes is the conserved endonuclease Cas1. For our search, we will define candidate CRISPR-cas loci as having, minimally, a cas1 gene. First, create another directory for output:
###Code
! mkdir example_1_output
###Output
_____no_output_____
###Markdown
The following bit of code uses Opfi's Gene Finder module to search for CRISPR-Cas systems:
###Code
from gene_finder.pipeline import Pipeline
import os
genomic_data = "GCF_000024045.1_ASM2404v1_genomic.fna.gz"
output_directory = "example_1_output"
p = Pipeline()
p.add_seed_step(db="blastdb/cas1_db", name="cas1", e_val=0.001, blast_type="PROT", num_threads=1)
p.add_filter_step(db="blastdb/cas_all_but_1_db", name="cas_all", e_val=0.001, blast_type="PROT", num_threads=1)
p.add_crispr_step()
# use the input filename as the job id
# results will be written to the file <job id>_results.csv
job_id = os.path.basename(genomic_data)
results = p.run(job_id=job_id, data=genomic_data, output_directory=output_directory, min_prot_len=90, span=10000, gzip=True)
###Output
_____no_output_____
###Markdown
First, we initialize a `Pipeline` object, which keeps track of all search parameters, as well as a running list of systems that meet search criteria. Next, we add three search steps to the pipeline:1. `add_seed_step`: BLAST is used to search the input genome against a database of Cas1 sequences. Regions around putative Cas1 hits become the intial candidates, and the rest of the genome is ignored.2. `add_filter_step`: Candidate regions are searched for any additional Cas genes. Candidates without at least one additional putative Cas gene are also discarded.3. `add_crispr_step`: Remaining candidates are annotated for CRISPR repeat sequences using PILER-CR. Finally, we run the pipeline, executing steps in the order they we added. `min_prot_len` sets the minimum length (in amino acid residues) of hits to keep (really short hits are unlikely real protein encoding genes). `span` is the region directly up- and downstream of initial hits. So, each candidate system will be about 20 kbp in length. Results are written to a single CSV file. Final candidate loci contain at least one putative Cas1 gene and one additional Cas gene. As we will see, this relatively permissive criteria captures some non-CRISPR-Cas loci. Opfi has additional modules for reducing unlikely systems after the gene finding stage. **3. Visualize annotated CRISPR-Cas gene clusters using Opfi's Operon Analyzer module** It is sometimes useful to visualize candidate systems, especially during the exploratory phase of a genomics survey. Opfi provides a few functions for visualizing candidate systems as gene diagrams. We'll use these to visualize the CRISPR-Cas gene clusters in R. orientalis:
###Code
import csv
import sys
from operon_analyzer import load, visualize
feature_colors = { "cas1": "lightblue",
"cas2": "seagreen",
"cas3": "gold",
"cas4": "springgreen",
"cas5": "darkred",
"cas6": "thistle",
"cas7": "coral",
"cas8": "red",
"cas9": "palegreen",
"cas10": "yellow",
"cas11": "tan",
"cas12": "orange",
"cas13": "saddlebrown",
"casphi": "olive",
"CRISPR array": "purple"
}
# read in the output from Gene Finder and create a gene diagram for each cluster (operon)
with open("example_1_output/GCF_000024045.1_ASM2404v1_genomic.fna.gz_results.csv", "r") as operon_data:
operons = load.load_operons(operon_data)
visualize.plot_operons(operons=operons, output_directory="example_1_output", feature_colors=feature_colors, nucl_per_line=25000)
###Output
_____no_output_____
###Markdown
Looking at the gene diagrams, it is clear that we identified both CRISPR-Cas systems in this genome. We also see some systems that don't resemble functional CRISPR-Cas operons. Because we used a relatively permissive e-value threshhold of 0.001 when running BLAST, Opfi retained regions with very low sequence similarity to true CRISPR-Cas genes. In fact, these regions are likely not CRISPR-Cas loci at all. Using a lower e-value would likely eliminate these "false positive" systems, but Opfi also has additional functions for filtering out unlikely candidates _after_ the intial BLAST search. In general, we have found that using permissive BLAST parameters intially, and then filtering or eliminating candidates during the downstream analysis, is an effective way to search for gene clusters in large amounts of genomic/metagenomic data. In this toy example, we could re-run BLAST many times without significant cost. But on a more realistic dataset, needing to re-do the computationally expensive homology search could detrail a project. Since the optimal search parameters may not be known _a priori_, it can be better to do a permissive homology search initially, and then narrow down results later. Finally, clean up the temporary directories, if desired:
###Code
! rm -r example_1_output blastdb
###Output
_____no_output_____
###Markdown
Example 2: Filter and classify CRISPR-Cas systems based on genomic composition As mentioned in the previous example, known CRISPR-Cas systems fall into 6 broad categories, based on the presence of particular "signature" genes, as well as overall composition and genomic architecture. In this example, we will use Opfi to search for and classify CRISPR-Cas systems in ~300 strains of fusobacteria. This dataset was chosen because it is more representative (in magnitude) of what would be encountered in a real genomics study. Additionally, the fusobacteria phylum contains a variety of CRISPR-Cas subtypes. Given that the homology search portion of the analysis takes several hours (using a single core) to complete, we have pre-run Gene Finder using the same setup as the previous example. **1. Make another temporary directory for output:**
###Code
! mkdir example_2_output
###Output
_____no_output_____
###Markdown
**2. Filter Gene Finder output and extract high-confidence CRISPR-Cas systems** The following code reads in unfiltered Gene Finder output and applies a set of conditions ("rules") to accomplish two things:1. Select (and bin) systems according to type, and,2. Eliminate candidates that likely do not represent true CRISPR-Cas systems
###Code
from operon_analyzer import analyze, rules
fs = rules.FilterSet().pick_overlapping_features_by_bit_score(0.9)
cas_types = ["I", "II", "III", "V"]
rulesets = []
# type I rules
rulesets.append(rules.RuleSet().contains_group(feature_names = ["cas5", "cas7"], max_gap_distance_bp = 1000, require_same_orientation = True) \
.require("cas3"))
# type II rules
rulesets.append(rules.RuleSet().contains_at_least_n_features(feature_names = ["cas1", "cas2", "cas9"], feature_count = 3) \
.minimum_size("cas9", 3000))
# type III rules
rulesets.append(rules.RuleSet().contains_group(feature_names = ["cas5", "cas7"], max_gap_distance_bp = 1000, require_same_orientation = True) \
.require("cas10"))
# type V rules
rulesets.append(rules.RuleSet().contains_at_least_n_features(feature_names = ["cas1", "cas2", "cas12"], feature_count = 3))
for rs, cas_type in zip(rulesets, cas_types):
with open("refseq_fusobacteria.csv", "r") as input_csv:
with open(f"example_2_output/refseq_fuso_filtered_type{cas_type}.csv", "w") as output_csv:
analyze.evaluate_rules_and_reserialize(input_csv, rs, fs, output_csv)
###Output
_____no_output_____
###Markdown
The rule sets are informed by an established CRISPR-Cas classification system, which you can read more about [here](https://www.nature.com/articles/s41579-019-0299-x). The most recent system recognizes 6 major CRISPR-Cas types, but since fusobacteria doesn't contain type IV or VI systems that can be identified with our protein dataset, we didn't define the corresponding rule sets. **3. Verify results with additional visualizations** Altogther, this analysis will identify several hundred systems. We won't look at each system individually (but you are free to do so!). For the sake of confirming that the code ran as expected, we'll create gene diagrams for just the type V systems, since there are only two:
###Code
import csv
import sys
from operon_analyzer import load, visualize
feature_colors = { "cas1": "lightblue",
"cas2": "seagreen",
"cas3": "gold",
"cas4": "springgreen",
"cas5": "darkred",
"cas6": "thistle",
"cas7": "coral",
"cas8": "red",
"cas9": "palegreen",
"cas10": "yellow",
"cas11": "tan",
"cas12": "orange",
"cas13": "saddlebrown",
"casphi": "olive",
"CRISPR array": "purple"
}
# read in the output from Gene Finder and create a gene diagram for each cluster (operon)
with open("example_2_output/refseq_fuso_filtered_typeV.csv", "r") as operon_data:
operons = load.load_operons(operon_data)
visualize.plot_operons(operons=operons, output_directory="example_2_output", feature_colors=feature_colors, nucl_per_line=25000)
###Output
_____no_output_____
###Markdown
Finally, clean up the temporary output directory, if desired:
###Code
! rm -r example_2_output
###Output
_____no_output_____
###Markdown
Example 1: Finding CRISPR-Cas systems in a cyanobacteria genome In this example, we will annotate and visualize CRISPR-Cas systems in the cyanobacteria species Rippkaea orientalis. CRISPR-Cas is a widespread bacterial defense system, found in at least 50% of all known prokaryotic species. This system is significant in that it can be leveraged as a precision gene editing tool, an advancement that was awarded the 2020 Nobel Prize in Chemistry. The genome of R. orientalis harbors two complete CRISPR-Cas loci (one chromosomal, and one extrachromosomal/plasmid).You can download the complete assembled genome [here](https://www.ncbi.nlm.nih.gov/assembly/GCF_000024045.1/); it is also available at https://github.com/wilkelab/Opfi in the `tutorials` folder (`data/GCF_000024045.1_ASM2404v1_genomic.fna.gz`). This tutorial assumes the user has already cloned and built Opfi, installed NCBI BLAST+ **and** PILER-CR, and has set `tutorials` as the working directory. Some familiarity with BLAST and the basic homology search algorithm may also be helpful, but is not required. **1. Use the makeblastdb utility to convert a Cas protein database to BLAST format** We start by converting a Cas sequence database to a format that BLAST can recognize, using the command line utility `makeblastdb`, which is part of the core NCBI BLAST+ distribution. A set of ~20,000 non-redundant Cas sequences, downloaded from [Uniprot](https://www.uniprot.org/uniref/) is available as a tar archive (`tutorials/data/cas_database.tar.gz`). We'll make a new directory, "blastdb", and extract sequences there:
###Code
! mkdir blastdb
! cd blastdb && tar -xzf ../data/cas_database.tar.gz && cd ..
###Output
_____no_output_____
###Markdown
Next, create two BLAST databases for the sequence data: one containing Cas1 sequences only, and another that contains the remaining Cas sequences.
###Code
! cd blastdb && cat cas1.fasta | makeblastdb -dbtype prot -title cas1 -hash_index -out cas1_db && cd ..
! cd blastdb && cat cas[2-9].fasta cas1[0-2].fasta casphi.fasta | makeblastdb -dbtype prot -title cas_all -hash_index -out cas_all_but_1_db
###Output
_____no_output_____
###Markdown
`-dbtype prot` simply tells `makeblastdb` to expect amino acid sequences. We use `-title` and `-out` to name the database (required by BLAST) and to prefix the database files, respectively. `-hash_index` directs `makeblastdb` to generate a hash index of protein sequences, which can speed up computation time. **2. Use Opfi's Gene Finder module to search for CRISPR-Cas loci** CRISPR-Cas systems are extremely diverse. The most recent [classification effort](https://www.nature.com/articles/s41579-019-0299-x) identifies 6 major types, and over 40 subtypes, of compositionally destinct systems. Although there is sufficent sequence similarity between subtypes to infer the existence of a common ancestor, the only protein family present in the majority of CRISPR-cas subtypes is the conserved endonuclease Cas1. For our search, we will define candidate CRISPR-cas loci as having, minimally, a cas1 gene. First, create another directory for output:
###Code
! mkdir example_1_output
###Output
_____no_output_____
###Markdown
The following bit of code uses Opfi's Gene Finder module to search for CRISPR-Cas systems:
###Code
from gene_finder.pipeline import Pipeline
import os
genomic_data = "data/GCF_000024045.1_ASM2404v1_genomic.fna.gz"
output_directory = "example_1_output"
p = Pipeline()
p.add_seed_step(db="blastdb/cas1_db", name="cas1", e_val=0.001, blast_type="PROT", num_threads=1)
p.add_filter_step(db="blastdb/cas_all_but_1_db", name="cas_all", e_val=0.001, blast_type="PROT", num_threads=1)
p.add_crispr_step()
# use the input filename as the job id
# results will be written to the file <job id>_results.csv
job_id = os.path.basename(genomic_data)
results = p.run(job_id=job_id, data=genomic_data, output_directory=output_directory, min_prot_len=90, span=10000, gzip=True)
###Output
_____no_output_____
###Markdown
First, we initialize a `Pipeline` object, which keeps track of all search parameters, as well as a running list of systems that meet search criteria. Next, we add three search steps to the pipeline:1. `add_seed_step`: BLAST is used to search the input genome against a database of Cas1 sequences. Regions around putative Cas1 hits become the intial candidates, and the rest of the genome is ignored.2. `add_filter_step`: Candidate regions are searched for any additional Cas genes. Candidates without at least one additional putative Cas gene are also discarded.3. `add_crispr_step`: Remaining candidates are annotated for CRISPR repeat sequences using PILER-CR. Finally, we run the pipeline, executing steps in the order they we added. `min_prot_len` sets the minimum length (in amino acid residues) of hits to keep (really short hits are unlikely real protein encoding genes). `span` is the region directly up- and downstream of initial hits. So, each candidate system will be about 20 kbp in length. Results are written to a single CSV file. Final candidate loci contain at least one putative Cas1 gene and one additional Cas gene. As we will see, this relatively permissive criteria captures some non-CRISPR-Cas loci. Opfi has additional modules for reducing unlikely systems after the gene finding stage. **3. Visualize annotated CRISPR-Cas gene clusters using Opfi's Operon Analyzer module** It is sometimes useful to visualize candidate systems, especially during the exploratory phase of a genomics survey. Opfi provides a few functions for visualizing candidate systems as gene diagrams. We'll use these to visualize the CRISPR-Cas gene clusters in R. orientalis:
###Code
import csv
import sys
from operon_analyzer import load, visualize
feature_colors = { "cas1": "lightblue",
"cas2": "seagreen",
"cas3": "gold",
"cas4": "springgreen",
"cas5": "darkred",
"cas6": "thistle",
"cas7": "coral",
"cas8": "red",
"cas9": "palegreen",
"cas10": "yellow",
"cas11": "tan",
"cas12": "orange",
"cas13": "saddlebrown",
"casphi": "olive",
"CRISPR array": "purple"
}
# read in the output from Gene Finder and create a gene diagram for each cluster (operon)
with open("example_1_output/GCF_000024045.1_ASM2404v1_genomic.fna.gz_results.csv", "r") as operon_data:
operons = load.load_operons(operon_data)
visualize.plot_operons(operons=operons, output_directory="example_1_output", feature_colors=feature_colors, nucl_per_line=25000)
###Output
_____no_output_____
###Markdown
Looking at the gene diagrams, it is clear that we identified both CRISPR-Cas systems in this genome. We also see some systems that don't resemble functional CRISPR-Cas operons. Because we used a relatively permissive e-value threshhold of 0.001 when running BLAST, Opfi retained regions with very low sequence similarity to true CRISPR-Cas genes. In fact, these regions are likely not CRISPR-Cas loci at all. Using a lower e-value would likely eliminate these "false positive" systems, but Opfi also has additional functions for filtering out unlikely candidates _after_ the intial BLAST search. In general, we have found that using permissive BLAST parameters intially, and then filtering or eliminating candidates during the downstream analysis, is an effective way to search for gene clusters in large amounts of genomic/metagenomic data. In this toy example, we could re-run BLAST many times without significant cost. But on a more realistic dataset, needing to re-do the computationally expensive homology search could detrail a project. Since the optimal search parameters may not be known _a priori_, it can be better to do a permissive homology search initially, and then narrow down results later. Finally, clean up the temporary directories, if desired:
###Code
! rm -r example_1_output blastdb
###Output
_____no_output_____
###Markdown
Example 2: Filter and classify CRISPR-Cas systems based on genomic composition As mentioned in the previous example, known CRISPR-Cas systems fall into 6 broad categories, based on the presence of particular "signature" genes, as well as overall composition and genomic architecture. In this example, we will use Opfi to search for and classify CRISPR-Cas systems in ~300 strains of fusobacteria. This dataset was chosen because it is more representative (in magnitude) of what would be encountered in a real genomics study. Additionally, the fusobacteria phylum contains a variety of CRISPR-Cas subtypes. Given that the homology search portion of the analysis takes several hours (using a single core) to complete, we have pre-run Gene Finder using the same setup as the previous example. **1. Make another temporary directory for output:**
###Code
! mkdir example_2_output
###Output
_____no_output_____
###Markdown
**2. Filter Gene Finder output and extract high-confidence CRISPR-Cas systems** The following code reads in unfiltered Gene Finder output and applies a set of conditions ("rules") to accomplish two things:1. Select (and bin) systems according to type, and,2. Eliminate candidates that likely do not represent true CRISPR-Cas systems
###Code
from operon_analyzer import analyze, rules
fs = rules.FilterSet().pick_overlapping_features_by_bit_score(0.9)
cas_types = ["I", "II", "III", "V"]
rulesets = []
# type I rules
rulesets.append(rules.RuleSet().contains_group(feature_names = ["cas5", "cas7"], max_gap_distance_bp = 1000, require_same_orientation = True) \
.require("cas3"))
# type II rules
rulesets.append(rules.RuleSet().contains_at_least_n_features(feature_names = ["cas1", "cas2", "cas9"], feature_count = 3) \
.minimum_size("cas9", 3000))
# type III rules
rulesets.append(rules.RuleSet().contains_group(feature_names = ["cas5", "cas7"], max_gap_distance_bp = 1000, require_same_orientation = True) \
.require("cas10"))
# type V rules
rulesets.append(rules.RuleSet().contains_at_least_n_features(feature_names = ["cas1", "cas2", "cas12"], feature_count = 3))
for rs, cas_type in zip(rulesets, cas_types):
with open("data/refseq_fusobacteria.csv", "r") as input_csv:
with open(f"example_2_output/refseq_fuso_filtered_type{cas_type}.csv", "w") as output_csv:
analyze.evaluate_rules_and_reserialize(input_csv, rs, fs, output_csv)
###Output
_____no_output_____
###Markdown
The rule sets are informed by an established CRISPR-Cas classification system, which you can read more about [here](https://www.nature.com/articles/s41579-019-0299-x). The most recent system recognizes 6 major CRISPR-Cas types, but since fusobacteria doesn't contain type IV or VI systems that can be identified with our protein dataset, we didn't define the corresponding rule sets. **3. Verify results with additional visualizations** Altogther, this analysis will identify several hundred systems. We won't look at each system individually (but you are free to do so!). For the sake of confirming that the code ran as expected, we'll create gene diagrams for just the type V systems, since there are only two:
###Code
import csv
import sys
from operon_analyzer import load, visualize
feature_colors = { "cas1": "lightblue",
"cas2": "seagreen",
"cas3": "gold",
"cas4": "springgreen",
"cas5": "darkred",
"cas6": "thistle",
"cas7": "coral",
"cas8": "red",
"cas9": "palegreen",
"cas10": "yellow",
"cas11": "tan",
"cas12": "orange",
"cas13": "saddlebrown",
"casphi": "olive",
"CRISPR array": "purple"
}
# read in the output from Gene Finder and create a gene diagram for each cluster (operon)
with open("example_2_output/refseq_fuso_filtered_typeV.csv", "r") as operon_data:
operons = load.load_operons(operon_data)
visualize.plot_operons(operons=operons, output_directory="example_2_output", feature_colors=feature_colors, nucl_per_line=25000)
###Output
_____no_output_____
###Markdown
Finally, clean up the temporary output directory, if desired:
###Code
! rm -r example_2_output
###Output
_____no_output_____
###Markdown
Note: restart runtime after this import before running the augmentations```>>> pip install dist/arizona-0.0.1-py3-none-any.whl```
###Code
import arizona
arizona.__version__
###Output
_____no_output_____
###Markdown
Using by each input text
###Code
from arizona.textmentations.functions import keyboard_func
text = "giới thiệu về công ty ftech"
results = keyboard_func(
text,
num_samples=5,
intent='faq_company',
tags='O O O B-work_unit I-word_unit I-work_unit',
aug_char_percent=0.2,
aug_word_percent=0.1,
unikey_percent=0.5,
config_file='../configs/unikey.json',
)
from pprint import pprint
pprint(results)
from arizona.textmentations.functions import remove_accent_func
text = 'giới thiệu về công ty ftech'
results = remove_accent_func(
text,
num_samples=5,
intent='faq_company',
tags='O O O B-work_unit I-word_unit I-work_unit'
)
from pprint import pprint
pprint(results)
from arizona.textmentations.functions import abbreviates_func
text = "giới thiệu về công ty ftech"
results = abbreviates_func(
text,
num_samples=10,
intent='faq_company',
tags='O O O B-work_unit I-word_unit I-work_unit',
config_file='../configs/abbreviations.json'
)
from pprint import pprint
pprint(results)
###Output
{'intent': ['faq_company',
'faq_company',
'faq_company',
'faq_company',
'faq_company',
'faq_company',
'faq_company',
'faq_company',
'faq_company',
'faq_company'],
'tags': ['O O B-work_unit I-work_unit',
'O O B-work_unit I-work_unit',
'O O B-work_unit I-word_unit I-work_unit',
'O O B-work_unit I-word_unit I-work_unit',
'O O B-work_unit I-work_unit',
'O O O B-work_unit I-work_unit',
'O O B-work_unit I-work_unit',
'O O B-work_unit I-work_unit',
'O O B-work_unit I-word_unit I-work_unit',
'O O B-work_unit I-word_unit I-work_unit'],
'text': ['gt về cty ftech',
'gthieu về cty ftech',
'gthieu về công ty ftech',
'gthieu về công ty ftech',
'gthieu về cty ftech',
'giới thiệu về cty ftech',
'gthieu về cty ftech',
'gthieu về cty ftech',
'gthieu về công ty ftech',
'gthieu về công ty ftech']}
###Markdown
Using by a .csv file
###Code
from arizona.textmentations import TextAugmentation
data_path = '../data/nlu.csv'
text_augs = TextAugmentation(
data=data_path,
text_col='text',
intent_col='intent',
tags_col='tags'
)
df = text_augs.augment(
methods=['remove_accent', 'abbreviation', 'keyboard'],
without_origin_data=True,
write_file=False
)
df.head(10)
###Output
100%|██████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 10/10 [00:05<00:00, 1.92it/s]
###Markdown
Reasonable Crowd Dataset Tutorial
###Code
import json
import geopandas as gpd
import matplotlib.pyplot as plt
import numpy as np
from shapely.geometry import Polygon
%matplotlib inline
###Output
_____no_output_____
###Markdown
Simple manipulations Plotting a footprint In this section, we show a simple code snippet for plotting a footprint
###Code
# Enter the path to a trajectory
trajectory_path = "<INSERT_PATH_TO_DOWNLOADED_DATA>/reasonable_crowd_data/trajectories/U_27-a.json"
# load the trajectory
with open(trajectory_path, "r") as j_file:
traj_states = json.load(j_file)
# let's plot the footprint of the first state
state = traj_states[0]
fig = plt.figure()
ax = fig.add_subplot(111)
ax.set_aspect("equal")
plt.axis("off")
footprint = Polygon(state["footprint"])
ax.plot(*footprint.exterior.xy)
plt.show()
###Output
_____no_output_____
###Markdown
Plotting a layer of the map Let's plot a layer of one of the maps.
###Code
# Path to a gpkg file representing one of the layers
map_path = "<INSERT_PATH_TO_DOWNLOADED_DATA>/reasonable_crowd_data/maps/S_boundaries.gpkg"
# thanks to the wide support of the gpkg format, plotting is only a couple of lines
map_df = gpd.read_file(map_path)
fig, ax = plt.subplots(1, 1)
map_df.plot(ax = ax)
ax.set_aspect('equal')
plt.axis("off")
plt.show()
###Output
_____no_output_____
###Markdown
More advanced manipulations To get better acquainted with the data, we wrote a small script: `visualize_realization.py` that visualizes a realization by making use of the map files and the trajectory files. The output of this script is something like `movie.mp4` in this directory. (Legend for the movie: the black polygon is ego, blue polygons are other cars, red polygons are pedestrians.)We highly recommend that you go through the code to become really familiar with the data.
###Code
from visualize_realization import visualize_realization
trajectory_path = "<INSERT_PATH_TO_DOWNLOADED_DATA>/reasonable_crowd_data/trajectories/S_1-a.json"
map_path = "<INSERT_PATH_TO_DOWNLOADED_DATA>/reasonable_crowd_data/maps/S_boundaries.gpkg"
# directory where to save the ouputs of this script (a video and an image for each frame)
save_dir = "<INSERT_PATH_TO_DIRECTORY_WHERE_SAVE_PLOTS>"
# NOTE: This might take a couple mins to run;
visualize_realization(trajectory_path, map_path, save_dir)
###Output
_____no_output_____
###Markdown
sconascona is a tool to perform network analysis over correlation networks of brain regions. This tutorial will go through the basic functionality of scona, taking us from our inputs (a matrix of structural regional measures over subjects) to a report of local network measures for each brain region, and network level comparisons to a cohort of random graphs of the same degree.
###Code
import numpy as np
import networkx as nx
import scona as scn
import scona.datasets as datasets
###Output
_____no_output_____
###Markdown
Importing dataA scona analysis starts with four inputs.* __regional_measures__ A pandas DataFrame with subjects as rows. The columns should include structural measures for each brain region, as well as any subject-wise covariates. * __names__ A list of names of the brain regions. This will be used to specify which columns of the __regional_measures__ matrix to want to correlate over.* __covars__ _(optional)_ A list of your covariates. This will be used to specify which columns of __regional_measure__ you wish to correct for. * __centroids__ A list of tuples representing the cartesian coordinates of brain regions. This list should be in the same order as the list of brain regions to accurately assign coordinates to regions. The coordinates are expected to obey the convention the the x=0 plane is the same plane that separates the left and right hemispheres of the brain.
###Code
# Read in sample data from the NSPN WhitakerVertes PNAS 2016 paper.
df, names, covars, centroids = datasets.NSPN_WhitakerVertes_PNAS2016.import_data()
df.head()
###Output
_____no_output_____
###Markdown
Create a correlation matrixWe calculate residuals of the matrix df for the columns of names, correcting for the columns in covars.
###Code
df_res = scn.create_residuals_df(df, names, covars)
df_res
###Output
_____no_output_____
###Markdown
Now we create a correlation matrix over the columns of df_res
###Code
M = scn.create_corrmat(df_res, method='pearson')
###Output
_____no_output_____
###Markdown
Create a weighted graphA short sidenote on the BrainNetwork class: This is a very lightweight subclass of the [`Networkx.Graph`](https://networkx.github.io/documentation/stable/reference/classes/graph.html) class. This means that any methods you can use on a `Networkx.Graph` object can also be used on a `BrainNetwork` object, although the reverse is not true. We have added various methods which allow us to keep track of measures that have already been calculated, which, especially later on when one is dealing with 10^3 random graphs, saves a lot of time. All scona measures are implemented in such a way that they can be used on a regular `Networkx.Graph` object. For example, instead of `G.threshold(10)` you can use `scn.threshold_graph(G, 10)`. Also you can create a `BrainNetwork` from a `Networkx.Graph` `G`, using `scn.BrainNetwork(network=G)` Initialise a weighted graph `G` from the correlation matrix `M`. The `parcellation` and `centroids` arguments are used to label nodes with names and coordinates respectively.
###Code
G = scn.BrainNetwork(network=M, parcellation=names, centroids=centroids)
###Output
_____no_output_____
###Markdown
Threshold to create a binary graphWe threshold G at cost 10 to create a binary graph with 10% as many edges as the complete graph G. Ordinarily when thresholding one takes the 10% of edges with the highest weight. In our case, because we want the resulting graph to be connected, we calculate a minimum spanning tree first. If you want to omit this step, you can pass the argument `mst=False` to `threshold`.The threshold method does not edit objects inplace
###Code
H = G.threshold(10)
###Output
_____no_output_____
###Markdown
Calculate nodal summary. `calculate_nodal_measures` will compute and record the following nodal measures * average_dist (if centroids available)* total_dist (if centroids available)* betweenness* closeness* clustering coefficient* degree* interhem (if centroids are available)* interhem_proportion (if centroids are available)* nodal partition* participation coefficient under partition calculated above* shortest_path_length `export_nodal_measure` returns nodal attributes in a DataFrame. Let's try it now.
###Code
H.report_nodal_measures().head()
###Output
_____no_output_____
###Markdown
Use `calculate_nodal_measures` to fill in a bunch of nodal measures
###Code
H.calculate_nodal_measures()
H.report_nodal_measures().head()
###Output
_____no_output_____
###Markdown
We can also add measures as one might normally add nodal attributes to a networkx graph
###Code
nx.set_node_attributes(H, name="hat", values={x: x**2 for x in H.nodes})
###Output
_____no_output_____
###Markdown
These show up in our DataFrame too
###Code
H.report_nodal_measures(columns=['name', 'degree', 'hat']).head()
###Output
_____no_output_____
###Markdown
Calculate Global measures
###Code
H.calculate_global_measures()
H.rich_club();
###Output
_____no_output_____
###Markdown
Create a GraphBundleThe `GraphBundle` object is the scona way to handle across network comparisons. What is it? Essentially it's a python dictionary with `BrainNetwork` objects as values.
###Code
brain_bundle = scn.GraphBundle([H], ['NSPN_cost=10'])
###Output
_____no_output_____
###Markdown
This creates a dictionary-like object with BrainNetwork `H` keyed by `'NSPN_cost=10'`
###Code
brain_bundle
###Output
_____no_output_____
###Markdown
Now add a series of random_graphs created by edge swap randomisation of H (keyed by `'NSPN_cost=10'`)
###Code
# Note that 10 is not usually a sufficient number of random graphs to do meaningful analysis,
# it is used here for time considerations
brain_bundle.create_random_graphs('NSPN_cost=10', 10)
brain_bundle
###Output
_____no_output_____
###Markdown
Report on a GraphBundleThe following method will calculate global measures ( if they have not already been calculated) for all of the graphs in `graph_bundle` and report the results in a DataFrame. We can do the same for rich club coefficients below.
###Code
brain_bundle.report_global_measures()
brain_bundle.report_rich_club()
###Output
_____no_output_____
###Markdown
TutorialIn this little tutorial, we will show you how to use Moniker to sample from categorical distributions in constant time. We will also compare this to the famous `numpy.random.choice` function for sampling categorical distributions to get a sense of how much faster Alias sampling is!First, let's import our packages.
###Code
import sys
sys.path.append("../src/")
import numpy as np
import matplotlib.pyplot as plt
import time
from moniker import Sampler
###Output
_____no_output_____
###Markdown
Let's use an array of four weights (unnormalized probabilities) to show that Moniker's samples and the samples from `numpy.random.choice` match the actual distribution.
###Code
weights = np.array([4, 1, 5, 2])
sampler = Sampler(weights)
num_samples = 100000
samples = sampler.sample(num_samples = num_samples)
np_samples = np.zeros(num_samples)
for s in range(num_samples):
np_samples[s] = np.random.choice(
range(weights.shape[0]),
p=weights/np.sum(weights)
)
my_uniques, my_counts = np.unique(samples, return_counts=True)
np_uniques, np_counts = np.unique(np_samples, return_counts=True)
plt.figure()
plt.bar(my_uniques+0.25, my_counts/num_samples, color='blue',
label="Moniker", align='center', width=0.25)
plt.bar(range(weights.shape[0]), weights/np.sum(weights), color='black',
label="True dist.", align='center', width=0.25)
plt.bar(np_uniques-0.25, np_counts/num_samples, color='red',
label="np.random.choice", align='center', width=0.25)
plt.xlabel('Sample index')
plt.ylabel('Sample frequency')
plt.xticks(range(4))
plt.legend()
plt.show()
###Output
_____no_output_____
###Markdown
So, we know Moniker works! The distribution of its generated samples match with the true distribution and `numpy.random.choice`.Now, let's see how much faster Moniker is compared to `numpy.random.choice`. We will sample from randomly initialized distributions (`weights = np.random.uniform(size=num_weights`) of varying length (`num_weights`), and plot the time it takes to generate a varying number of samples (`num_samples`) using `numpy.random.choice` and Moniker.
###Code
def colors_from_array(arr, c): # nice for plotting
cmap = plt.get_cmap(c)
colors = [cmap(i) for i in np.linspace(0.3,1,len(arr))]
return colors, cmap
num_samples_range = np.arange(10000,15000,1000)
num_weights_range = np.arange(5,11)
colors, my_cmap = colors_from_array(num_weights_range, "Blues")
plt.figure()
for w,num_weights in enumerate(num_weights_range):
weights = np.random.uniform(size=num_weights)
Δt = np.zeros_like(num_samples_range, dtype=np.float64)
sampler = Sampler(weights)
for n,num_samples in enumerate(num_samples_range):
ti_moniker = time.time()
samples = sampler.sample(num_samples=num_samples)
t_moniker = time.time() - ti_moniker
np_samples = np.zeros(num_samples)
ti_numpy = time.time()
for i in range(num_samples):
np_samples[i] = np.random.choice(
num_weights,
p=weights/sum(weights)
)
t_numpy = time.time() - ti_numpy
Δt[n] = t_numpy - t_moniker
plt.plot(
num_samples_range, Δt,
color=colors[w],
label=r'$N = {}$'.format(num_weights)
)
sm = plt.cm.ScalarMappable(
cmap=my_cmap,
norm=plt.Normalize(
vmin=min(num_weights_range),
vmax=max(num_weights_range)
)
)
cbar = plt.colorbar(sm)
cbar.set_label(r'$N$', rotation=0)
plt.xlabel('Number of samples')
plt.ylabel(r'$\Delta t$', rotation=0)
plt.show()
###Output
_____no_output_____
###Markdown
This module helps solve systems of linear equations. There are several ways of doing this. The first is to just pass the coefficients as a list of lists. Say we want to solve the system of equations:$$\begin{array}{c|c|c}x - y = 5\\x + y = -1\end{array}$$This is done with a simple call to linear_solver.solve_linear_system(), like so
###Code
import linear_solver as ls
xs = ls.solve_linear_system(
[[1, -1, 5],
[1, 1, -1]])
print(xs)
###Output
[[ 2.]
[-3.]]
###Markdown
Clearly, the solution set $(2, -3)$ satisfies the two equations above.If a system of equations that has no unique solutions is given, a warning is printed and ```None``` is returned.
###Code
xs = ls.solve_linear_system(
[[1, 1, 0],
[2, 2, 0]])
print(xs)
xs = ls.solve_linear_system(
[[1, 1, 0],
[2, 2, 1]])
print(xs)
###Output
Determinant of coefficients matrix is 0. No unique solution.
None
###Markdown
Additionally, the coefficients of the equation can be read from a text file, where expressions are evaluated before they are read. For example, consider the following system of equations:$$\begin{array}{c|c|c|c}22m_1 + 22m_2 - m_3 = 0\\(0.1)(22)m_1 + (0.9)(22)m_2 - 0.6m_3 = 0\\\frac{22}{0.68} m_1 + \frac{22}{0.78} m_2 = (500)(3.785)\end{array}$$We can put these coefficients into a text file, ```'coefficients.txt'```, which has the contents\ contents of coefficients.txt22 22 -1 00.1\*22 0.9\*22 -0.6 022/0.68 22/0.78 0 500\*3.785and then pass that file to the solver function.
###Code
sol = ls.solve_linear_system('coefficients.txt')
for i, row in enumerate(sol):
print('m_{0} = {1:.2f}'.format(i, row[0,0]))
###Output
m_0 = 23.85
m_1 = 39.74
m_2 = 1399.00
###Markdown
Simulate some source activity and EEG data
###Code
sources_sim = run_simulations(pth_fwd, durOfTrial=0)
eeg_sim = create_eeg(sources_sim, pth_fwd)
###Output
_____no_output_____
###Markdown
Plot a simulated sample and corresponding EEG
###Code
%matplotlib qt
sample = 0 # index of the simulation
title = f'Simulation {sample}'
# Topographic plot
eeg_sim[sample].average().plot_topomap([0.5])
# Source plot
sources_sim.plot(hemi='both', initial_time=sample, surface='white', colormap='inferno', title=title, time_viewer=False)
###Output
_____no_output_____
###Markdown
Load and train ConvDip with simulated data
###Code
# Find out input and output dimensions based on the shape of the leadfield
input_dim, output_dim = load_leadfield(pth_fwd).shape
# Initialize the artificial neural network model
model = get_model(input_dim, output_dim)
# Train the model
model, history = train_model(model, sources_sim, eeg_sim, delta=1)
###Output
_____no_output_____
###Markdown
Evaluate ConvDipLet's evaluate our model!
###Code
%matplotlib qt
# Load some files from the forward model
leadfield = load_leadfield(pth_fwd)
info = load_info(pth_fwd)
# Simulate a brand new sample:
sources_eval = run_simulations(pth_fwd, 1, durOfTrial=0)
eeg_eval = create_eeg(sources_eval, pth_fwd)
# Calculate the ERP (average across trials):
eeg_sample = np.squeeze( eeg_eval )
# Predict
source_predicted = predict(model, eeg_sample, pth_fwd)
# Visualize ground truth...
title = f'Ground Truth'
sources_eval.plot(hemi='both', initial_time=0.5, surface='white', colormap='inferno', title=title, time_viewer=False)
# ... and prediction
title = f'ConvDip Prediction'
source_predicted.plot(hemi='both', initial_time=0.5, surface='white', colormap='inferno', title=title, time_viewer=False)
# ... and the 'True' EEG topography
title = f'Simulated EEG'
eeg_eval[0].average().plot_topomap([0], title=title)
###Output
_____no_output_____
###Markdown
sconascona is a tool to perform network analysis over correlation networks of brain regions. This tutorial will go through the basic functionality of scona, taking us from our inputs (a matrix of structural regional measures over subjects) to a report of local network measures for each brain region, and network level comparisons to a cohort of random graphs of the same degree.
###Code
import numpy as np
import networkx as nx
import scona as scn
import scona.datasets as datasets
###Output
_____no_output_____
###Markdown
Importing dataA scona analysis starts with four inputs.* __regional_measures__ A pandas DataFrame with subjects as rows. The columns should include structural measures for each brain region, as well as any subject-wise covariates. * __names__ A list of names of the brain regions. This will be used to specify which columns of the __regional_measures__ matrix to want to correlate over.* __covars__ _(optional)_ A list of your covariates. This will be used to specify which columns of __regional_measure__ you wish to correct for. * __centroids__ A list of tuples representing the cartesian coordinates of brain regions. This list should be in the same order as the list of brain regions to accurately assign coordinates to regions. The coordinates are expected to obey the convention the the x=0 plane is the same plane that separates the left and right hemispheres of the brain.
###Code
# Read in sample data from the NSPN WhitakerVertes PNAS 2016 paper.
df, names, covars, centroids = datasets.NSPN_WhitakerVertes_PNAS2016.import_data()
df.head()
###Output
_____no_output_____
###Markdown
Create a correlation matrixWe calculate residuals of the matrix df for the columns of names, correcting for the columns in covars.
###Code
df_res = scn.create_residuals_df(df, names, covars)
df_res
###Output
_____no_output_____
###Markdown
Now we create a correlation matrix over the columns of df_res
###Code
M = scn.create_corrmat(df_res, method='pearson')
###Output
_____no_output_____
###Markdown
Create a weighted graphA short sidenote on the BrainNetwork class: This is a very lightweight subclass of the [`Networkx.Graph`](https://networkx.github.io/documentation/stable/reference/classes/graph.html) class. This means that any methods you can use on a `Networkx.Graph` object can also be used on a `BrainNetwork` object, although the reverse is not true. We have added various methods which allow us to keep track of measures that have already been calculated, which, especially later on when one is dealing with 10^3 random graphs, saves a lot of time. All scona measures are implemented in such a way that they can be used on a regular `Networkx.Graph` object. For example, instead of `G.threshold(10)` you can use `scn.threshold_graph(G, 10)`. Also you can create a `BrainNetwork` from a `Networkx.Graph` `G`, using `scn.BrainNetwork(network=G)` Initialise a weighted graph `G` from the correlation matrix `M`. The `parcellation` and `centroids` arguments are used to label nodes with names and coordinates respectively.
###Code
G = scn.BrainNetwork(network=M, parcellation=names, centroids=centroids)
###Output
_____no_output_____
###Markdown
Threshold to create a binary graphWe threshold G at cost 10 to create a binary graph with 10% as many edges as the complete graph G. Ordinarily when thresholding one takes the 10% of edges with the highest weight. In our case, because we want the resulting graph to be connected, we calculate a minimum spanning tree first. If you want to omit this step, you can pass the argument `mst=False` to `threshold`.The threshold method does not edit objects inplace
###Code
H = G.threshold(10)
###Output
_____no_output_____
###Markdown
Calculate nodal summary. `calculate_nodal_measures` will compute and record the following nodal measures * average_dist (if centroids available)* total_dist (if centroids available)* betweenness* closeness* clustering coefficient* degree* interhem (if centroids are available)* interhem_proportion (if centroids are available)* nodal partition* participation coefficient under partition calculated above* shortest_path_length `export_nodal_measure` returns nodal attributes in a DataFrame. Let's try it now.
###Code
H.report_nodal_measures().head()
###Output
_____no_output_____
###Markdown
Use `calculate_nodal_measures` to fill in a bunch of nodal measures
###Code
H.calculate_nodal_measures()
H.report_nodal_measures().head()
###Output
_____no_output_____
###Markdown
We can also add measures as one might normally add nodal attributes to a networkx graph
###Code
nx.set_node_attributes(H, name="hat", values={x: x**2 for x in H.nodes})
###Output
_____no_output_____
###Markdown
These show up in our DataFrame too
###Code
H.report_nodal_measures(columns=['name', 'degree', 'hat']).head()
###Output
_____no_output_____
###Markdown
Calculate Global measures
###Code
H.calculate_global_measures()
H.rich_club();
###Output
_____no_output_____
###Markdown
Create a GraphBundleThe `GraphBundle` object is the scona way to handle across network comparisons. What is it? Essentially it's a python dictionary with `BrainNetwork` objects as values.
###Code
brain_bundle = scn.GraphBundle([H], ['NSPN_cost=10'])
###Output
_____no_output_____
###Markdown
This creates a dictionary-like object with BrainNetwork `H` keyed by `'NSPN_cost=10'`
###Code
brain_bundle
###Output
_____no_output_____
###Markdown
Now add a series of random_graphs created by edge swap randomisation of H (keyed by `'NSPN_cost=10'`)
###Code
# Note that 10 is not usually a sufficient number of random graphs to do meaningful analysis,
# it is used here for time considerations
brain_bundle.create_random_graphs('NSPN_cost=10', 10)
brain_bundle
###Output
_____no_output_____
###Markdown
Report on a GraphBundleThe following method will calculate global measures ( if they have not already been calculated) for all of the graphs in `graph_bundle` and report the results in a DataFrame. We can do the same for rich club coefficients below.
###Code
brain_bundle.report_global_measures()
brain_bundle.report_rich_club()
###Output
_____no_output_____
###Markdown
Convert Calcium Imaging data from .mat to NWB fileMore details on [NWB Calcium imaging data](https://pynwb.readthedocs.io/en/stable/tutorials/domain/ophys.htmlcalcium-imaging-data).**0.** We start importing the relevant modules to read from .mat file and to manipulate NWB file groups and datasets
###Code
from datetime import datetime
from dateutil.tz import tzlocal
from pynwb import NWBFile, NWBHDF5IO, ProcessingModule
from pynwb.ophys import TwoPhotonSeries, OpticalChannel, ImageSegmentation, Fluorescence, DfOverF, MotionCorrection
from pynwb.device import Device
from pynwb.base import TimeSeries
import scipy.io
import numpy as np
import h5py
import os
###Output
_____no_output_____
###Markdown
**1.** Load the .mat files containing calcium imaging data
###Code
path_to_files = '/Users/bendichter/Desktop/Axel Lab/data' #r'C:\Users\Luiz\Desktop\Axel'
# Open info file
fname0 = 'fly2_run1_info.mat'
fpath0 = os.path.join(path_to_files, fname0)
f_info = scipy.io.loadmat(fpath0, struct_as_record=False, squeeze_me=True)
info = f_info['info']
# Open .mat file containing Calcium Imaging data
fname1 = '2019_04_18_Nsyb_NLS6s_Su_walk_G_fly2_run1_8401reg.mat'
fpath1 = os.path.join(path_to_files, fname1)
file = h5py.File(fpath1, 'r')
options = file['options']
landmarkThreshold = file['landmarkThreshold']
templates = file['templates']
R = file['R']
Y = file['Y']
###Output
_____no_output_____
###Markdown
**2.** Create a new [NWB file instance](https://pynwb.readthedocs.io/en/stable/pynwb.file.htmlpynwb.file.NWBFile), fill it with all the relevant information
###Code
#Create new NWB file
nwb = NWBFile(session_description='my CaIm recording',
identifier='EXAMPLE_ID',
session_start_time=datetime.now(tzlocal()),
experimenter='Dr. ABC',
lab='My Lab',
institution='My University',
experiment_description='Some description.',
session_id='IDX')
print(nwb)
###Output
_____no_output_____
###Markdown
**3.** Create [Device](https://pynwb.readthedocs.io/en/stable/pynwb.device.htmlpynwb.device.Device) and [OpticalChannel](https://pynwb.readthedocs.io/en/stable/pynwb.ophys.htmlpynwb.ophys.OpticalChannel) containers to be used by a specific [ImagingPlane](https://pynwb.readthedocs.io/en/stable/pynwb.ophys.htmlpynwb.ophys.ImagingPlane).
###Code
#Create and add device
device = Device(info.objective.replace('/','_'))
nwb.add_device(device)
# Create an Imaging Plane for Yellow
optical_channel_Y = OpticalChannel(name='optical_channel_Y',
description='2P Optical Channel',
emission_lambda=510.)
imaging_plane_Y = nwb.create_imaging_plane(name='imaging_plane_Y',
optical_channel=optical_channel_Y,
description='Imaging plane',
device=device,
excitation_lambda=488.,
imaging_rate=info.daq.scanRate,
indicator='NLS-GCaMP6s',
location='whole central brain')
# Create an Imaging Plane for Red
optical_channel_R = OpticalChannel(name='optical_channel_R',
description='2P Optical Channel',
emission_lambda=633.)
imaging_plane_R = nwb.create_imaging_plane(name='imaging_plane_R',
optical_channel=optical_channel_R,
description='Imaging plane',
device=device,
excitation_lambda=488.,
imaging_rate=info.daq.scanRate,
indicator='redStinger',
location='whole central brain')
print(nwb)
###Output
_____no_output_____
###Markdown
**4.** Create a [TwoPhotonSeries](https://pynwb.readthedocs.io/en/stable/pynwb.ophys.htmlpynwb.ophys.TwoPhotonSeries) container to store the raw data. Raw data usually goes on the `acquisition` group of NWB files.
###Code
#Stores raw data in acquisition group - dims=(X,Y,Z,T)
raw_image_series_Y = TwoPhotonSeries(name='TwoPhotonSeries_Y',
imaging_plane=imaging_plane_Y,
rate=info.daq.scanRate,
dimension=[36, 167, 257],
data=Y[:,:,:,:])
raw_image_series_R = TwoPhotonSeries(name='TwoPhotonSeries_R',
imaging_plane=imaging_plane_R,
rate=info.daq.scanRate,
dimension=[36, 167, 257],
data=R[:,:,:,:])
nwb.add_acquisition(raw_image_series_Y)
nwb.add_acquisition(raw_image_series_R)
print(nwb.acquisition['TwoPhotonSeries'])
###Output
_____no_output_____
###Markdown
**5.** A very important data preprocessing step for calcium signals is motion correction. We can store the processed result data in the [MotionCorrection](https://pynwb.readthedocs.io/en/stable/pynwb.ophys.htmlpynwb.ophys.MotionCorrection) container, inside the `processing` group of NWB files.
###Code
#Creates ophys ProcessingModule and add to file
ophys_module = ProcessingModule(name='ophys',
description='contains optical physiology processed data.')
nwb.add_processing_module(ophys_module)
#Stores corrected data in TwoPhotonSeries container
corrected_image_series = TwoPhotonSeries(name='TwoPhotonSeries_corrected',
imaging_plane=imaging_plane,
rate=info.daq.scanRate,
dimension=[36, 167, 257],
data=Y[:,:,:,0])
#TimeSeries XY translation correction values
xy_translation = TimeSeries(name='xy_translation',
data=np.zeros((257,2)),
rate=info.daq.scanRate)
#Adds the corrected image stack to MotionCorrection container
motion_correction = MotionCorrection()
motion_correction.create_corrected_image_stack(corrected=corrected_image_series,
original=raw_image_series,
xy_translation=xy_translation)
#Add MotionCorrection to processing group
ophys_module.add_data_interface(motion_correction)
###Output
_____no_output_____
###Markdown
**6.** Any processed data should be stored in the `processing` group of NWB files. A list of available containers can be found [here](https://pynwb.readthedocs.io/en/stable/overview_nwbfile.htmlprocessing-modules). These include, for example, [DfOverF](https://pynwb.readthedocs.io/en/stable/pynwb.ophys.htmlpynwb.ophys.DfOverF), [ImageSegmentation](https://pynwb.readthedocs.io/en/stable/pynwb.ophys.htmlpynwb.ophys.ImageSegmentation), [Fluorescence](https://pynwb.readthedocs.io/en/stable/pynwb.ophys.htmlpynwb.ophys.Fluorescence) and others.
###Code
#Stores processed data of different types in ProcessingModule group
#Image segmentation
img_seg = ImageSegmentation()
ophys_module.add_data_interface(img_seg)
#Fluorescence
fl = Fluorescence()
ophys_module.add_data_interface(fl)
#DfOverF
dfoverf = DfOverF()
ophys_module.add_data_interface(dfoverf)
print(nwb.processing['ophys'])
###Output
_____no_output_____
###Markdown
**7.** The NWB structure is is place, but we still need to save it to file:
###Code
#Saves to NWB file
fname_nwb = 'file_1.nwb'
fpath_nwb = os.path.join(path_to_files, fname_nwb)
with NWBHDF5IO(fpath_nwb, mode='w') as io:
io.write(nwb)
print('File saved with size: ', os.stat(fpath_nwb).st_size/1e6, ' mb')
###Output
_____no_output_____
###Markdown
**8.** Finally, let's load it and check the file contents:
###Code
#Loads NWB file
with NWBHDF5IO(fpath_nwb, mode='r') as io:
nwb = io.read()
print(nwb)
print(nwb.processing['ophys'].data_interfaces['MotionCorrection'])
###Output
_____no_output_____
###Markdown
Evan Schaffer datafrom `.npz` files to NWB.
###Code
from datetime import datetime
from dateutil.tz import tzlocal
from pynwb import NWBFile, NWBHDF5IO, ProcessingModule
from pynwb.ophys import TwoPhotonSeries, OpticalChannel, ImageSegmentation, Fluorescence, DfOverF, MotionCorrection
from pynwb.device import Device
from pynwb.base import TimeSeries
import scipy.io
import numpy as np
import h5py
import os
import matplotlib.pyplot as plt
a = np.load('2019_07_01_Nsyb_NLS6s_walk_fly2.npz')
print('First file:')
print('Groups:', a.files)
print('Dims (height,width,depth):', a['dims'])
print('dFF shape: ', a['dFF'].shape)
b = np.load('2019_07_01_Nsyb_NLS6s_walk_fly2_A.npz')
print(' ')
print('Second file - Sparse Matrix:')
print('Groups:', b.files)
print('Indices: ', b['indices'], ' | Shape: ',b['indices'].shape)
print('Indptr: ', b['indptr'], ' | Shape: ',b['indptr'].shape)
print('Format: ', b['format'])
print('Shape: ', b['shape'])
print('Data: ', b['data'], ' | Shape: ',b['data'].shape)
###Output
_____no_output_____
###Markdown
Start creating a new NWB file instance and populating it with fake raw data
###Code
#Create new NWB file
nwb = NWBFile(session_description='my CaIm recording',
identifier='EXAMPLE_ID',
session_start_time=datetime.now(tzlocal()),
experimenter='Dr. ABC',
lab='My Lab',
institution='My University',
experiment_description='Some description.',
session_id='IDX')
#Create and add device
device = Device('MyDevice')
nwb.add_device(device)
# Create an Imaging Plane for Yellow
optical_channel = OpticalChannel(name='optical_channel',
description='2P Optical Channel',
emission_lambda=510.)
imaging_plane = nwb.create_imaging_plane(name='imaging_plane',
optical_channel=optical_channel,
description='Imaging plane',
device=device,
excitation_lambda=488.,
imaging_rate=1000.,
indicator='NLS-GCaMP6s',
location='whole central brain',
conversion=1.0)
#Stores raw data in acquisition group - dims=(X,Y,Z,T)
Xp = a['dims'][0][0]
Yp = a['dims'][0][1]
Zp = a['dims'][0][2]
T = a['dFF'].shape[1]
nCells = a['dFF'].shape[0]
fake_data = np.random.randn(Xp,Yp,Zp,100)
raw_image_series = TwoPhotonSeries(name='TwoPhotonSeries',
imaging_plane=imaging_plane,
rate=1000.,
dimension=[Xp,Yp,Zp],
data=fake_data)
nwb.add_acquisition(raw_image_series)
#Creates ophys ProcessingModule and add to file
ophys_module = ProcessingModule(name='ophys',
description='contains optical physiology processed data.')
nwb.add_processing_module(ophys_module)
###Output
_____no_output_____
###Markdown
Now transform the lists of indices into (xp,yp,zp) masks. With the masks created, we can add them to a plane segmentation class.
###Code
def make_voxel_mask(indices, dims):
"""
indices - List with voxels indices, e.g. [64371, 89300, 89301, ..., 3763753, 3763842, 3763843]
dims - (height, width, depth) in pixels
"""
voxel_mask = []
for ind in indices:
zp = np.floor(ind/(dims[0]*dims[1])).astype('int')
rest = ind%(dims[0]*dims[1])
yp = np.floor(rest/dims[0]).astype('int')
xp = rest%dims[0]
voxel_mask.append((xp,yp,zp,1))
return voxel_mask
#Call function
indices = b['indices']
indptr = indices[b['indptr'][0:-1]]
dims = np.squeeze(a['dims'])
voxel_mask = make_voxel_mask(indptr, dims)
#Create Image Segmentation compartment
img_seg = ImageSegmentation()
ophys_module.add_data_interface(img_seg)
#Create plane segmentation and add ROIs
ps = img_seg.create_plane_segmentation(description='plane segmentation',
imaging_plane=imaging_plane,
reference_images=raw_image_series)
ps.add_roi(voxel_mask=voxel_mask)
###Output
_____no_output_____
###Markdown
With the ROIs created, we can add the dF/F data
###Code
#DFF measures
dff = DfOverF(name='dff_interface')
ophys_module.add_data_interface(dff)
#create ROI regions
roi_region = ps.create_roi_table_region(description='ROI table region',
region=[0])
#create ROI response series
dff_data = a['dFF']
dFF_series = dff.create_roi_response_series(name='df_over_f',
data=dff_data,
unit='NA',
rois=roi_region,
rate=1000.)
###Output
_____no_output_____
###Markdown
The file contains two arrays with pixel indexing: one with all pixels at cells and one with only one reference pixel per cell. Let's see how it looks like in a 3D plot:
###Code
#3D scatter with masks points
from mpl_toolkits.mplot3d import Axes3D
#Reference points
xptr = [p[0] for p in voxel_mask]
yptr = [p[1] for p in voxel_mask]
zptr = [p[2] for p in voxel_mask]
#All points in mask
all_pt_mask = make_voxel_mask(indices, dims)
x = [p[0] for p in all_pt_mask]
y = [p[1] for p in all_pt_mask]
z = [p[2] for p in all_pt_mask]
%matplotlib notebook
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
ax.scatter(x, y, z, c='k', marker='.', s=.5)
ax.scatter(xptr, yptr, zptr, c='r', marker='o', s=20)
#Saves to NWB file
path_to_files = ''
fname_nwb = 'file_1.nwb'
fpath_nwb = os.path.join(path_to_files, fname_nwb)
with NWBHDF5IO(fpath_nwb, mode='w') as io:
io.write(nwb)
print('File saved with size: ', os.stat(fpath_nwb).st_size/1e6, ' mb')
#Loads NWB file
with NWBHDF5IO(fpath_nwb, mode='r') as io:
nwb = io.read()
print(nwb)
print(nwb.processing['ophys'].data_interfaces['dff_interface'].roi_response_series['df_over_f'])
###Output
_____no_output_____
###Markdown
데이터셋데이터는 영화 '라라랜드'의 네이버영화의 평점에 기록되어 있는 커멘트 15,603개 입니다.
###Code
from pprint import pprint
with open('./data/134963_norm.txt', encoding='utf-8') as f:
docs = [doc.strip() for doc in f]
print('n docs = %d' % len(docs))
pprint(docs[:5])
###Output
n docs = 15603
['시사회에서 보고왔습니다동화와 재즈뮤지컬의 만남 지루하지않고 재밌습니다',
'사랑과 꿈 그 흐름의 아름다움을 음악과 영상으로 최대한 담아놓았다 배우들 연기는 두말할것없고',
'지금껏 영화 평가 해본 적이 없는데 진짜 최고네요 색감 스토리 음악 연기 모두ㅜㅜ최고입니다',
'방금 시사회 보고 왔어요 배우들 매력이 눈을 뗄 수가 없게 만드네요 한편의 그림 같은 장면들도 많고 음악과 춤이 눈과 귀를 사로 잡았어요 '
'한번 더 보고 싶네요',
'초반부터 끝까지 재미있게 잘보다가 결말에서 고국마 왕창먹음 힐링 받는 느낌들다가 막판에 기분 잡쳤습니다 마치 감독이 하고싶은 말은 '
'너희들이 원하는 결말은 이거지 하지만 현실은 이거다 라고 말하고 싶었나보군요']
###Markdown
PyCRFSuite spacing 학습데이터 만들기
###Code
import sys
sys.path.append('../')
from pycrfsuite_spacing import TemplateGenerator
from pycrfsuite_spacing import CharacterFeatureTransformer
from pycrfsuite_spacing import sent_to_chartags
from pycrfsuite_spacing import sent_to_xy
###Output
_____no_output_____
###Markdown
sent_to_chars는 띄어쓰기가 있는 문장을 글자와 띄어쓰기 태그로 변환해줍니다. 띄어쓰기/붙여쓰기의 태그를 지정할 수 있습니다. default는 space = 1, nonspace = 0으로 int 입니다.
###Code
chars, tags = sent_to_chartags('이것도 너프해 보시지', space='1', nonspace='0')
print(chars)
print(tags)
###Output
이것도너프해보시지
['0', '0', '1', '0', '0', '1', '0', '0', '1']
###Markdown
빈 문자가 입력되면 빈 글자와 empty list 가 변환됩니다.
###Code
sent_to_chartags('')
###Output
_____no_output_____
###Markdown
TemplateGenerator는 앞 2 글자부터 뒤 2 글짜까지 길이가 3인 templates를 만든다는 의미입니다. 반드시 현재 글자 (index 0)가 포함된 부분만 template으로 만듭니다.
###Code
TemplateGenerator(begin=-2, end=2, min_range_length=3,max_range_length=3).tolist()
###Output
_____no_output_____
###Markdown
to_feature()는 template을 이용하여 문장에서 features를 만드는 함수입니다. CharacterFeatureTransformer의 instance는 callable 입니다.
###Code
to_feature = CharacterFeatureTransformer(
TemplateGenerator(begin=-2,
end=2,
min_range_length=3,
max_range_length=3)
)
x, y = sent_to_xy('이것도 너프해 보시지', to_feature)
pprint(x)
print(y)
###Output
[['X[0,2]=이것도'],
['X[-1,1]=이것도', 'X[0,2]=것도너'],
['X[-2,0]=이것도', 'X[-1,1]=것도너', 'X[0,2]=도너프'],
['X[-2,0]=것도너', 'X[-1,1]=도너프', 'X[0,2]=너프해'],
['X[-2,0]=도너프', 'X[-1,1]=너프해', 'X[0,2]=프해보'],
['X[-2,0]=너프해', 'X[-1,1]=프해보', 'X[0,2]=해보시'],
['X[-2,0]=프해보', 'X[-1,1]=해보시', 'X[0,2]=보시지'],
['X[-2,0]=해보시', 'X[-1,1]=보시지'],
['X[-2,0]=보시지']]
['0', '0', '1', '0', '0', '1', '0', '0', '1']
###Markdown
PyCRFSuite spacing 모델 학습하기모델을 학습합니다. 사용할 수 있는 arguments 입니다. - feature_minfreq - 기본값은 0 입니다. 이 값이 지나치게 작으면 features 개수가 커집니다. 메모리가 많이 필요하니 적절한 수준으로 잡아줍니다. - max_iterations - 기본값은 100 입니다. - l1_cost, l2_cost - 기본값은 각각 l1_cost = 0, l2_cost=1.0 입니다. Lasso CRF 를 이용하려면 l2_cost = 0, l1_cost > 0 으로 설정합니다. - L1, L2 regularization 을 모두 이용하려면 l1_cost > 0 & l2_cost > 0 으로 설정합니다. default parameter 를 이용하려면 아래처럼 to_feature 만 입력합니다. correct = PyCRFSuiteSpacing(to_feature)
###Code
from pycrfsuite_spacing import PyCRFSuiteSpacing
correct = PyCRFSuiteSpacing(
to_feature = to_feature,
feature_minfreq=3, # default = 0
max_iterations=100,
l1_cost=1.0,
l2_cost=1.0
)
correct.train(docs, 'demo_model.crfsuite')
###Output
_____no_output_____
###Markdown
학습된 모델을 불러올 수도 있습니다.
###Code
correct.load_tagger('demo_model.crfsuite')
###Output
_____no_output_____
###Markdown
PyCRFSuite spacing 으로 띄어쓰기 오류 교정하기
###Code
correct('이건진짜좋은영화라라랜드진짜좋은영화')
###Output
_____no_output_____ |
P09-RootFinding.ipynb | ###Markdown
Root Finding============This week we're exploring algorithms for finding roots of arbitrary functions.Any time you try to solve an algebraic problem and end up with a transcendental equation you can find yourself with root finding as the only viable means of extracting answers.As an example there's a nice quantum mechanical system (The finite square well , you don't need to follow this podcast, it's just an example for which the result of a transcendental equation is important) for which the bound energy states can be found by solving the two transcendental equations:$$\sin(z)=z/z_0$$and $$\cos(z)=z/z_0$$Where $z_0$ is a unitless real number that characterizes the depth and width of the potential well and $z$ is a unitless real number (less that $z_0$) that characterizes the energy level.Since the $\cos(z)$ version always has at least one solution, let's look at it first.
###Code
%matplotlib inline
import numpy as np
import matplotlib.pyplot as pl
N=100
z0=2.0
z=np.linspace(0,1.5,N)
def leftS(z):
return np.cos(z)
def rightS(z,z0=z0):
return z/z0
def f(z,z0=z0):
return leftS(z)-rightS(z,z0)
pl.grid()
pl.title("Investigating $\cos(z)=z/z_0$")
pl.ylabel("left, right and difference")
pl.xlabel("$z$")
pl.plot(z,leftS(z),'r-',label='$\cos(z)$')
pl.plot(z, rightS(z),'b-',label='$z/z_0$')
pl.plot(z, f(z),'g-', label='$\cos(z)-z/z_0$')
pl.legend(loc=3)
def fp(z):
"""
We need a function to evaluate the derivative of f(z) for Newton's method.
"""
return -np.sin(z)-1.0/z0
def newtonsMethod(f, fp, zi, eps=1e-15, Nmax=100, showCount=False):
"""
Very simple implementation of Newton's Method.
Try to find a zero of 'f' near zi to within eps.
Don't use up over Nmax iterations
"""
z=zi # start at zi
y=f(z) # evaluate y
count=0 # start count at zero
while (abs(y)>eps) and count<Nmax:
dz=y/fp(z) # evaluate dz
z=z-dz # update z
y=f(z) # update y
count += 1 # update count
if count>=Nmax:
raise RuntimeError("Ack! I can't find a zero.")
elif showCount:
print( "Found root", z, "in", count, "iterations. y=", y)
return z
z = newtonsMethod(f, fp, 1.0, showCount=True)
from scipy.optimize import brentq
print (brentq(f, 0.9, 1.1))
###Output
1.0298665293222566
###Markdown
Suppose we have some potential function and we want to find a "bound state" wavefunction that satisfies the boundary conditions of the potential. There are of course many different possible potentials that could be considered. Let's focus on a class that goes to infinity for $xL$. Between those limits the potential is defined by a function $V(x)$.We can ues RK4 to integrate from $x=0$ to $x=L$. What shall we integrate? The Schrodinger Wave Equation of course!$$-\frac{\hbar^2}{2m} \psi''(x) + V(x)\psi(x) = E\psi(x)$$$$\psi'' = \frac{2m}{\hbar^2}\left(V(x)-E\right)\psi(x)$$
###Code
def V(x, a=3.0):
"""
Here's an example potential V(x)=0.0
"""
return 0.0
psi0, psip0 = 0.0, 1.0 # start psi and psi' at $x=0$.
s=np.array([psi0, psip0])
hbar=1.0 # pick convenient units
m=1.0
L=1.0
x=0.0
dx=L/20
E=0.90*(hbar**2/(2*m))*(np.pi/L)**2 # start at 90% of known ground state energy.
xList=[x] # put in the first value
psiList=[psi0]
def RK4Step(s, x, derivs, dx, E):
"""
Take a single RK4 step. (our old friend)
But this time we're integrating in 'x', not 't'.
"""
dxh=dx/2.0
f1 = derivs(s, x, E)
f2 = derivs(s+f1*dxh, x+dxh, E)
f3 = derivs(s+f2*dxh, x+dxh, E)
f4 = derivs(s+f3*dx, x+dx, E)
return s + (f1+2*f2+2*f3+f4)*dx/6.0
def SWE_derivs(s, x, E):
psi=s[0]
psip=s[1]
psipp =(2*m/hbar**2)*(V(x)-E)*psi
return np.array([psip, psipp])
while x<=L:
s=RK4Step(s, x, SWE_derivs, dx, E)
x+=dx
xList.append(x)
psiList.append(s[0])
pl.title("Test Wave Function at Energy %3.2f" % E)
pl.ylabel("$\psi(x)$ (un-normalized)")
pl.xlabel("$x$")
pl.plot(xList, psiList, 'b-')
def calcBC(E):
"""
Compute the value of psi(x) at x=L for a given value of E
assuming psi(0) is zero.
"""
s=np.array([psi0, psip0])
x=0.0
while x<L:
s=RK4Step(s, x, SWE_derivs, dx, E)
x+=dx
return s[0]
print ("BC at E=4.4:",calcBC(4.4))
print ("BC at E=5.4:",calcBC(5.4))
Ezero = brentq(calcBC, 4.4, 5.4) # find "root" with brentq
Ezero
print( ((hbar**2)/(2*m))*(np.pi/L)**2 )# exact result
def calcBC_wPsi(E):
"""
Compute the value of psi(x) at x=L for a given value of E
assuming psi(0) is zero.
"""
s=np.array([psi0, psip0])
x=0.0
xList=[x]
psiList=[psi0]
while x<L:
s=RK4Step(s, x, SWE_derivs, dx, E)
x+=dx
xList.append(x)
psiList.append(s[0])
return xList, psiList
xList, psiList = calcBC_wPsi(Ezero)
pl.plot(xList, psiList, 'b-')
pl.grid()
###Output
_____no_output_____
###Markdown
Root Finding============This week we're exploring algorithms for finding roots of arbitrary functions.Any time you try to solve an algebraic problem and end up with a transcendental equation you can find yourself with root finding as the only viable means of extracting answers.As an example there's a nice quantum mechanical system (The finite square well , you don't need to follow this podcast, it's just an example for which the result of a transcendental equation is important) for which the bound energy states can be found by solving the two transcendental equations:$$\sin(z)=z/z_0$$and $$\cos(z)=z/z_0$$Where $z_0$ is a unitless real number that characterizes the depth and width of the potential well and $z$ is a unitless real number (less that $z_0$) that characterizes the energy level.Since the $\cos(z)$ version always has at least one solution, let's look at it first.
###Code
%pylab inline
N=100
z0=2.0
z=linspace(0,1.5,N)
def leftS(z):
return cos(z)
def rightS(z,z0=z0):
return z/z0
def f(z,z0=z0):
return leftS(z)-rightS(z,z0)
grid()
title("Investigating $\cos(z)=z/z_0$")
ylabel("left, right and difference")
xlabel("$z$")
plot(z,leftS(z),'r-',label='$\cos(z)$')
plot(z, rightS(z),'b-',label='$z/z_0$')
plot(z, f(z),'g-', label='$\cos(z)-z/z_0$')
legend(loc=3)
def fp(z):
"""
We need a function to evaluate the derivative of f(z) for Newton's method.
"""
return -sin(z)-1.0/z0
def newtonsMethod(f, fp, zi, eps=1e-15, Nmax=100, showCount=False):
"""
Very simple implementation of Newton's Method.
Try to find a zero of 'f' near zi to within eps.
Don't use up over Nmax iterations
"""
z=zi # start at zi
y=f(z) # evaluate y
count=0 # start count at zero
while (abs(y)>eps) and count<Nmax:
dz=y/fp(z) # evaluate dz
z=z-dz # update z
y=f(z) # update y
count += 1 # update count
if count>=Nmax:
raise RuntimeError("Ack! I can't find a zero.")
elif showCount:
print( "Found root", z, "in", count, "iterations. y=", y)
return z
z = newtonsMethod(f, fp, 1.0, showCount=True)
from scipy.optimize import brentq
print (brentq(f, 0.9, 1.1))
###Output
1.0298665293222566
###Markdown
Suppose we have some potential function and we want to find a "bound state" wavefunction that satisfies the boundary conditions of the potential. There are of course many different possible potentials that could be considered. Let's focus on a class that goes to infinity for $xL$. Between those limits the potential is defined by a function $V(x)$.We can ues RK4 to integrate from $x=0$ to $x=L$. What shall we integrate? The Schrodinger Wave Equation of course!$$-\frac{\hbar^2}{2m} \psi''(x) + V(x)\psi(x) = E\psi(x)$$$$\psi'' = \frac{2m}{\hbar^2}\left(V(x)-E\right)\psi(x)$$
###Code
def V(x, a=3.0):
"""
Here's an example potential V(x)=0.0
"""
return 0.0
psi0, psip0 = 0.0, 1.0 # start psi and psi' at $x=0$.
s=array([psi0, psip0])
hbar=1.0 # pick convenient units
m=1.0
L=1.0
x=0.0
dx=L/20
E=0.90*(hbar**2/(2*m))*(pi/L)**2 # start at 90% of known ground state energy.
xList=[x] # put in the first value
psiList=[psi0]
def RK4Step(s, x, derivs, dx, E):
"""
Take a single RK4 step. (our old friend)
But this time we're integrating in 'x', not 't'.
"""
dxh=dx/2.0
f1 = derivs(s, x, E)
f2 = derivs(s+f1*dxh, x+dxh, E)
f3 = derivs(s+f2*dxh, x+dxh, E)
f4 = derivs(s+f3*dx, x+dx, E)
return s + (f1+2*f2+2*f3+f4)*dx/6.0
def SWE_derivs(s, x, E):
psi=s[0]
psip=s[1]
psipp =(2*m/hbar**2)*(V(x)-E)*psi
return array([psip, psipp])
while x<=L:
s=RK4Step(s, x, SWE_derivs, dx, E)
x+=dx
xList.append(x)
psiList.append(s[0])
title("Test Wave Function at Energy %3.2f" % E)
ylabel("$\psi(x)$ (un-normalized)")
xlabel("$x$")
plot(xList, psiList, 'b-')
def calcBC(E):
"""
Compute the value of psi(x) at x=L for a given value of E
assuming psi(0) is zero.
"""
s=array([psi0, psip0])
x=0.0
while x<L:
s=RK4Step(s, x, SWE_derivs, dx, E)
x+=dx
return s[0]
print ("BC at E=4.4:",calcBC(4.4))
print ("BC at E=5.4:",calcBC(5.4))
Ezero = brentq(calcBC, 4.4, 5.4) # find "root" with brentq
Ezero
print( ((hbar**2)/(2*m))*(pi/L)**2 )# exact result
def calcBC_wPsi(E):
"""
Compute the value of psi(x) at x=L for a given value of E
assuming psi(0) is zero.
"""
s=array([psi0, psip0])
x=0.0
xList=[x]
psiList=[psi0]
while x<L:
s=RK4Step(s, x, SWE_derivs, dx, E)
x+=dx
xList.append(x)
psiList.append(s[0])
return xList, psiList
xList, psiList = calcBC_wPsi(Ezero)
plot(xList, psiList, 'b-')
###Output
_____no_output_____ |
plot_history.ipynb | ###Markdown
Notebook to plot the history
###Code
import numpy as np
import json
import os
import matplotlib.pyplot as plt
from matplotlib import rcParams
class PlotStyler():
def __init__(self, n, cmap="Set1", change_linestyle=True):
assert cmap in ["hsv", "Set1", "Accent"]
self.cmap = cmap
self.i = -1
self.n = n
self.cm = plt.get_cmap(cmap)
if change_linestyle:
self.linestyles = [
"solid", "dotted", "dashed", "dashdot",
(0, (3, 1, 1, 1, 1, 1)), (0, (5, 1)), (0, (3, 5, 1, 5, 1, 5))
]
else:
self.linestyles = ["solid"]
def get(self):
self.i += 1
ls = self.linestyles[self.i % len(self.linestyles)]
if self.cmap in ["hsv", "Accent"]:
c = self.cm(self.i/self.n)
else:
c = self.cm(self.i)
return c, ls
def plot(d, xlim=None, ylim=None, title=None, plot_limbs=False, replace_reward_by_success_rate=False):
generations = d["generations"]
best_rewards = d["best_rewards"]
mean_rewards = d["mean_rewards"]
min_rewards = d["min_rewards"]
species_mean_rewards = d["species_mean_rewards"]
species_eval_rewards = d["species_eval_rewards"]
num_limbs = d["num_limbs"]
num_species = len(species_eval_rewards[0])
if replace_reward_by_success_rate:
species_eval_rewards = d["success_rates"]
ylim = (0, 1)
plt.rcParams["font.size"] = 30
ps = PlotStyler(16)
fig=plt.figure(figsize=(16, 10), dpi= 80, facecolor="w", edgecolor="k")
if title is not None:
fig.suptitle(title)
ax1 = fig.add_subplot(111)
# Decide the color and line style first
styles = []
for i in range(num_species):
styles.append(ps.get())
if plot_limbs:
# rigid parts graph
c, ls = ps.get()
ax2 = ax1.twinx()
ax2.plot(generations, num_limbs[:, 0], color=c, linestyle = ls, label="number of rigid parts")
# reward graph
for i in range(num_species):
c, ls = styles[i]
label = f"reward in worker {i+1}"
if num_species == 1:
label = "reward"
ax1.plot(generations, species_mean_rewards[:, i], color=c, alpha=0.2)
ax1.plot(generations, species_eval_rewards[:, i], color=c, linestyle = ls, label=label)
h, l = ax1.get_legend_handles_labels()
if plot_limbs:
h2, l2 = ax2.get_legend_handles_labels()
h += h2
l += l2
ax2.set_ylabel("num_limbs")
plt.rcParams["font.size"] = 24
if "ax2" in locals():
# To eliminate overlap with ax2 graph
ax2.legend(h, l, loc="upper left")
else:
ax1.legend(h, l, loc="upper left")
ax1.set_xlabel("generation")
ax1.set_ylabel("reward")
if ylim is not None:
ax1.set_ylim(ylim)
if xlim is not None:
ax1.set_xlim(xlim)
if plot_limbs:
ax2.set_xlim(xlim)
def extract_data(d):
generations = np.array([h["generation"] for h in d])
elapseds = np.array([h["elapsed"] for h in d])
best_species_eval_rewards = np.array([h["best_reward"] for h in d])
best_rewards = np.array([h["current_best_reward"] for h in d])
mean_rewards = np.array([h["current_mean_reward"] for h in d])
min_rewards = np.array([h["current_min_reward"] for h in d])
species_mean_rewards = np.array([h["current_mean_rewards"] for h in d])
species_eval_rewards = np.array([h["current_eval_rewards"] for h in d])
success_rates = np.mean(np.array([h["success_rate"] for h in d]), axis=2)
num_limbs = np.array([h["num_limbs"] for h in d])
return {
"generations": generations,
"elapseds": elapseds,
"best_species_eval_rewards": best_species_eval_rewards,
"best_rewards": best_rewards,
"mean_rewards": mean_rewards,
"min_rewards": min_rewards,
"species_mean_rewards": species_mean_rewards,
"species_eval_rewards": species_eval_rewards,
"success_rates": success_rates,
"num_limbs": num_limbs
}
simname = "old/0.8.8_20220203_225723"
# simname = "old/0.8.8_20220205_213427"
# Data collection
filename = os.path.join("log", simname, "history.json")
with open(filename, "r") as f:
d = extract_data(json.load(f))
plot(d)
###Output
_____no_output_____
###Markdown
History
###Code
experiments = !ls -1 ./checkpoints
names = [
'loss',
# SSD
'conf_loss', 'loc_loss',
#'pos_conf_loss', 'neg_conf_loss', 'pos_loc_loss',
'precision', 'recall', 'fmeasure',
# SegLink
'seg_precision', 'seg_recall', 'seg_fmeasure',
#'link_precision', 'link_recall', 'link_fmeasure',
#'pos_seg_conf_loss', 'neg_seg_conf_loss', 'seg_loc_loss', 'pos_link_conf_loss', 'neg_link_conf_loss',
#'num_pos_seg', 'num_neg_seg', 'num_pos_link', 'num_neg_link',
#'seg_conf_loss', 'seg_loc_loss', 'link_conf_loss',
]
#names = None
plot_history(experiments[-4:], names)
###Output
_____no_output_____
###Markdown
History
###Code
d = experiments[1]
checkpath = os.path.join('.', 'checkpoints', d)
df = pd.read_csv(os.path.join(checkpath, 'history.csv'))
epochs = np.arange(len(df)) + 1
ticks = epochs
#print(list(df.keys()))
plt.figure(figsize=(12,6))
plt.plot(epochs, df['loss'])
plt.plot(epochs, df['val_loss'])
plt.title('loss')
plt.legend(['training','validation'])
ax = plt.gca()
ax.set_xticks(ticks)
ax.set_xticklabels(ticks)
plt.grid()
#plt.ylim([0.0, 5.0])
#plt.ylim([0.0, 20.0])
if False:
plotpath = os.path.join(checkpath, 'plots')
os.makedirs(plotpath, exist_ok=True)
plt.savefig(os.path.join(plotpath, 'history_loss.png'), bbox_inches='tight') # jpg/png/pgf
plt.show()
signals = [
# SSD
#'conf_loss', 'loc_loss',
#'pos_conf_loss', 'neg_conf_loss', 'pos_loc_loss',
#'precision', 'recall', 'fmeasure',
# SegLink
'seg_precision', 'seg_recall', 'seg_fmeasure',
#'link_precision', 'link_recall', 'link_fmeasure',
#'pos_seg_conf_loss', 'neg_seg_conf_loss', 'seg_loc_loss', 'pos_link_conf_loss', 'neg_link_conf_loss',
#'num_pos_seg', 'num_neg_seg', 'num_pos_link', 'num_neg_link',
#'seg_conf_loss', 'seg_loc_loss', 'link_conf_loss',
]
fig, axs = plt.subplots(1, len(signals), figsize=(20,4))
for i, s in enumerate(signals):
if s not in df.keys():
print('missing %s' %(s))
continue
axs[i].plot(epochs, df[s])
axs[i].plot(epochs, df['val_'+s])
axs[i].set_title(s)
if s.split('_')[-1] in ['precision', 'recall', 'fmeasure']:
axs[i].set_ylim([0,1])
axs[i].set_xticks(ticks)
axs[i].set_xticklabels(ticks)
axs[i].grid()
if False:
plotpath = os.path.join(checkpath, 'plots')
os.makedirs(plotpath, exist_ok=True)
plt.savefig(os.path.join(plotpath, 'history_metrics.png'), bbox_inches='tight') # jpg/png/pgf
plt.show()
###Output
_____no_output_____
###Markdown
Compare History
###Code
d1 = experiments[0]
d2 = experiments[1]
signals = [
#'loss',
#'precision',
#'recall',
#'fmeasure',
'seg_fmeasure',
'link_fmeasure',
]
df1 = pd.read_csv(os.path.join('.', 'checkpoints', d1, 'history.csv'))
df2 = pd.read_csv(os.path.join('.', 'checkpoints', d2, 'history.csv'))
epochs1 = np.arange(len(df1)) + 1
epochs2 = np.arange(len(df2)) + 1
ticks = np.arange(max(len(df1),len(df2))) + 1
fig, axs = plt.subplots(1, len(signals), figsize=(20,6))
for i, s in enumerate(signals):
if s not in df1.keys() or s not in df2.keys():
print('missing %s' %(s))
continue
ax = axs[i] if len(signals) > 1 else axs
ax.plot(epochs1, df1[s])
ax.plot(epochs1, df1['val_'+s])
ax.plot(epochs2, df2[s])
ax.plot(epochs2, df2['val_'+s])
ax.set_title(s)
if s.split('_')[-1] in ['precision', 'recall', 'fmeasure']:
ax.set_ylim([0,1])
ax.set_xlim([0, ticks[-1]+1])
ax.set_xticks(ticks)
ax.set_xticklabels(ticks)
ax.grid()
plt.show()
###Output
_____no_output_____
###Markdown
History
###Code
checkdir = experiments[-1]
checkpath = os.path.join('.', 'checkpoints', checkdir)
hist = pd.DataFrame.from_csv(os.path.join(checkpath, 'history.csv'))
epochs = np.arange(len(hist)) + 1
ticks = epochs
#print(list(hist.keys()))
plt.figure(figsize=(12,6))
plt.plot(epochs, hist['loss'])
plt.plot(epochs, hist['val_loss'])
plt.title('loss')
plt.legend(['training','validation'])
ax = plt.gca()
ax.set_xticks(ticks)
ax.set_xticklabels(ticks)
plt.grid()
#plt.ylim([0.0, 5.0])
#plt.ylim([0.0, 20.0])
plotpath = os.path.join(checkpath, 'plots')
os.makedirs(plotpath, exist_ok=True)
plt.savefig(os.path.join(plotpath, 'history_loss.png'), bbox_inches='tight') # jpg/png/pgf
plt.show()
signals = [
# SSD
#'conf_loss', 'loc_loss',
#'pos_conf_loss', 'neg_conf_loss', 'pos_loc_loss',
#'precision', 'recall', 'fmeasure',
# SegLink
'seg_precision', 'seg_recall', 'seg_fmeasure',
#'link_precision', 'link_recall', 'link_fmeasure',
#'pos_seg_conf_loss', 'neg_seg_conf_loss', 'seg_loc_loss', 'pos_link_conf_loss', 'neg_link_conf_loss',
#'num_pos_seg', 'num_neg_seg', 'num_pos_link', 'num_neg_link',
#'seg_conf_loss', 'seg_loc_loss', 'link_conf_loss',
]
fig, axs = plt.subplots(1, len(signals), figsize=(20,4))
for i, s in enumerate(signals):
if s not in hist.keys():
print('missing %s' %(s))
continue
axs[i].plot(epochs, hist[s])
axs[i].plot(epochs, hist['val_'+s])
axs[i].set_title(s)
if s.split('_')[-1] in ['precision', 'recall', 'fmeasure']:
axs[i].set_ylim([0,1])
axs[i].set_xticks(ticks)
axs[i].set_xticklabels(ticks)
axs[i].grid()
plotpath = os.path.join(checkpath, 'plots')
os.makedirs(plotpath, exist_ok=True)
plt.savefig(os.path.join(plotpath, 'history_metrics.png'), bbox_inches='tight') # jpg/png/pgf
plt.show()
###Output
_____no_output_____
###Markdown
Compare History
###Code
checkdir1 = experiments[-5]
checkdir2 = experiments[-1]
signals = [
#'loss',
#'precision',
#'recall',
#'fmeasure',
'predictions_seg_fmeasure',
'predictions_inter_link_fmeasure',
'predictions_cross_link_fmeasure',
]
hist1 = pd.DataFrame.from_csv(os.path.join('.', 'checkpoints', checkdir1, 'history.csv'))
hist2 = pd.DataFrame.from_csv(os.path.join('.', 'checkpoints', checkdir2, 'history.csv'))
epochs1 = np.arange(len(hist1)) + 1
epochs2 = np.arange(len(hist2)) + 1
ticks = np.arange(max(len(hist1),len(hist1))) + 1
fig, axs = plt.subplots(1, len(signals), figsize=(20,6))
for i, s in enumerate(signals):
if s not in hist1.keys() or s not in hist2.keys():
print('missing %s' %(s))
continue
ax = axs[i] if len(signals) > 1 else axs
ax.plot(epochs1, hist1[s])
ax.plot(epochs1, hist1['val_'+s])
ax.plot(epochs2, hist2[s])
ax.plot(epochs2, hist2['val_'+s])
ax.set_title(s)
if s.split('_')[-1] in ['precision', 'recall', 'fmeasure']:
ax.set_ylim([0,1])
ax.set_xlim([0, ticks[-1]])
ax.set_xticks(ticks)
ax.set_xticklabels(ticks)
ax.grid()
plt.show()
###Output
_____no_output_____
###Markdown
History
###Code
experiments = !ls -1 ./checkpoints
names = [
'loss',
# SSD
'conf_loss', 'loc_loss',
#'pos_conf_loss', 'neg_conf_loss', 'pos_loc_loss',
'precision', 'recall', 'fmeasure',
# SegLink
'seg_precision', 'seg_recall', 'seg_fmeasure',
#'link_precision', 'link_recall', 'link_fmeasure',
#'pos_seg_conf_loss', 'neg_seg_conf_loss', 'seg_loc_loss', 'pos_link_conf_loss', 'neg_link_conf_loss',
#'num_pos_seg', 'num_neg_seg', 'num_pos_link', 'num_neg_link',
#'seg_conf_loss', 'seg_loc_loss', 'link_conf_loss',
]
#names = None
plot_history(experiments[-4:], names)
###Output
_____no_output_____ |
examples/notebooks/generators/5_rest_api.ipynb | ###Markdown
Using the xcube generator REST API This notebook demonstrates direct use of the xcube generator REST API,without the higher-level Python client library. It performs the following steps:1. Use the xcube gen credentials to obtain an access token from the gen server.2. Issue a gen server request to generate a cube and store it in an S3 bucket.3. Monitor the status of the request until it has completed.4. Open the newly generated cube from the S3 bucket and plot an image using its data.This notebook reads the remote generator service configuration from the configuration file `edc_service.yml`, which may in turn contain references to environment variables. The notebook also requires the following environment variables to be set in order to write the generated cube into an S3 bucket:```AWS_ACCESS_KEY_IDAWS_SECRET_ACCESS_KEY``` First, import some necessary libraries.
###Code
import os
import requests
import time
import string
import yaml
from xcube.core.store import new_data_store
###Output
_____no_output_____
###Markdown
Set the endpoint URL, client ID, and client secret for the generator service, reading the values from the `edc_service.yml` configuration file.
###Code
with open('edc-service.yml', 'r') as fh:
yaml_content = fh.read()
yaml_substituted = string.Template(yaml_content).safe_substitute(os.environ)
service_params = yaml.safe_load(yaml_substituted)
client_id = service_params['client_id']
client_secret = service_params['client_secret']
endpoint = service_params['endpoint_url']
###Output
_____no_output_____
###Markdown
Post a request to the generator's authorization service to get an access token authorized by the client credentials.
###Code
token_response = requests.post(
endpoint + 'oauth/token',
json = {
'audience': endpoint,
'client_id': client_id,
'client_secret': client_secret,
'grant_type': 'client-credentials',
},
headers = {'Accept': 'application/json'},
)
access_token = token_response.json()['access_token']
###Output
_____no_output_____
###Markdown
Create and issue a request to generate a cube containing soil moisture data for the European region. Note the use of the access token to authorize the request.
###Code
cubegen_request = {
'input_config': {
'store_id': '@cds',
'data_id': 'satellite-soil-moisture:saturation:monthly',
'open_params': {}
},
'cube_config': {
'variable_names': ['soil_moisture_saturation'],
'crs': 'WGS84',
# 'bbox': [-13.00, 41.15, 21.79, 57.82],
'spatial_res': 0.25,
'time_range': ['2000-01-01', '2001-01-01'],
'time_period': '1M',
'chunks': {'lat': 65, 'lon': 139, 'time': 12},
},
'output_config': {
'store_id': 's3',
'data_id': 'cds-soil-moisture.zarr',
'store_params': {'root': 'eurodatacube-scratch'},
'replace': True,
},
}
cubegen_response = requests.put(
endpoint + 'cubegens',
json=cubegen_request,
headers={
'Accept': 'application/json',
'Authorization': 'Bearer ' + access_token
},
)
###Output
_____no_output_____
###Markdown
Get the ID of the cube generation request so that we can monitor its progress.
###Code
cubegen_id = cubegen_response.json()['job_id']
cubegen_id
cubegen_response.json()
###Output
_____no_output_____
###Markdown
Check the request status repeatedly at five-second intervals until the request completes, then show the status.
###Code
while True:
status_response = requests.get(
endpoint + 'cubegens/' + cubegen_id,
headers={
'Accept': 'application/json',
'Authorization': 'Bearer ' + access_token
}
)
json_response = status_response.json()
if json_response['job_status']['succeeded'] or json_response['job_status']['failed']:
break
time.sleep(5)
json_response['job_status']
###Output
_____no_output_____
###Markdown
If the request was successful, the cube has now been written into an S3 bucket. Create an xcube data store to read the data from this bucket and list its contents.
###Code
data_store = new_data_store(
's3',
root='eurodatacube-scratch',
storage_options={
'key': os.environ['AWS_ACCESS_KEY_ID'],
'secret': os.environ['AWS_SECRET_ACCESS_KEY']
}
)
list(data_store.get_data_ids())
###Output
_____no_output_____
###Markdown
Open our newly-created dataset from this store and show a summary of its properties and contents.
###Code
dataset = data_store.open_data('cds-soil-moisture.zarr')
dataset
###Output
_____no_output_____
###Markdown
Plot a map showing the generated data at a particular point in time.
###Code
dataset.sm.isel(time=6).plot.imshow()
###Output
_____no_output_____ |
kudo-kubeflow/Anomaly-Detection-with-Serving.ipynb | ###Markdown
Function arguments specified with `InputPath` and `OutputPath` are the key to defining dependencies.For now, it suffices to think of them as the input and output of each step.How we can define dependencies is explained in the [next section](How-to-Combine-the-Components-into-a-Pipeline). Component 1: Download & Prepare the Data Set
###Code
%%writefile minio_secret.yaml
apiVersion: v1
kind: Secret
metadata:
name: minio-s3-secret
annotations:
serving.kubeflow.org/s3-endpoint: minio-service.kubeflow:9000
serving.kubeflow.org/s3-usehttps: "0" # Default: 1. Must be 0 when testing with MinIO!
type: Opaque
data:
awsAccessKeyID: bWluaW8=
awsSecretAccessKey: bWluaW8xMjM=
---
apiVersion: v1
kind: ServiceAccount
metadata:
name: default
secrets:
- name: minio-s3-secret
! kubectl apply -f minio_secret.yaml
def download_and_process_dataset(data_dir: OutputPath(str)):
import pandas as pd
import numpy as np
import os
if not os.path.exists(data_dir):
os.makedirs(data_dir)
# Generated training sequences for use in the model.
def create_sequences(values, time_steps=288):
output = []
for i in range(len(values) - time_steps):
output.append(values[i : (i + time_steps)])
return np.stack(output)
master_url_root = "https://raw.githubusercontent.com/numenta/NAB/master/data/"
df_small_noise_url_suffix = "artificialNoAnomaly/art_daily_small_noise.csv"
df_small_noise_url = master_url_root + df_small_noise_url_suffix
df_small_noise = pd.read_csv(
df_small_noise_url, parse_dates=True, index_col="timestamp"
)
df_daily_jumpsup_url_suffix = "artificialWithAnomaly/art_daily_jumpsup.csv"
df_daily_jumpsup_url = master_url_root + df_daily_jumpsup_url_suffix
df_daily_jumpsup = pd.read_csv(
df_daily_jumpsup_url, parse_dates=True, index_col="timestamp"
)
# Normalize and save the mean and std we get,
# for normalizing test data.
training_mean = df_small_noise.mean()
training_std = df_small_noise.std()
df_training_value = (df_small_noise - training_mean) / training_std
print("Number of training samples:", len(df_training_value))
x_train = create_sequences(df_training_value.values)
print("Training input shape: ", x_train.shape)
np.save(data_dir + "/x_train.npy", x_train)
###Output
_____no_output_____
###Markdown
Component 2: Build the Model
###Code
def build_and_train_model(data_dir: InputPath(str), model_dir: OutputPath(str)):
import os
import numpy as np
from tensorflow import keras
from keras import layers
import tensorflow as tf
x_train = np.load(data_dir + "/x_train.npy")
print("Training input shape: ", x_train.shape)
# TODO: make x_train into a layer so it can be used as input layer below
class Conv1DTranspose(tf.keras.layers.Layer):
def __init__(self, filters, kernel_size, strides=1, padding="valid"):
super().__init__()
self.conv2dtranspose = tf.keras.layers.Conv2DTranspose(
filters, (kernel_size, 1), (strides, 1), padding
)
def call(self, x):
x = tf.expand_dims(x, axis=2)
x = self.conv2dtranspose(x)
x = tf.squeeze(x, axis=2)
return x
model = keras.Sequential(
[
layers.Input(shape=(x_train.shape[1], x_train.shape[2])),
layers.Conv1D(
filters=32, kernel_size=7, padding="same", strides=2, activation="relu"
),
layers.Dropout(rate=0.2),
layers.Conv1D(
filters=16, kernel_size=7, padding="same", strides=2, activation="relu"
),
Conv1DTranspose(filters=16, kernel_size=7, padding="same", strides=2),
layers.Dropout(rate=0.2),
Conv1DTranspose(filters=32, kernel_size=7, padding="same", strides=2),
Conv1DTranspose(filters=1, kernel_size=7, padding="same"),
]
)
model.compile(optimizer=keras.optimizers.Adam(learning_rate=0.001), loss="mse")
model.summary()
history = model.fit(
x_train,
x_train,
epochs=50,
batch_size=128,
validation_split=0.1,
callbacks=[
keras.callbacks.EarlyStopping(monitor="val_loss", patience=5, mode="min")
],
)
model.save(model_dir)
print(f"Model saved {model_dir}")
print(os.listdir(model_dir))
###Output
_____no_output_____
###Markdown
Combine into pipeline
###Code
def train_and_serve(
data_dir: str,
model_dir: str,
export_bucket: str,
model_name: str,
model_version: int,
):
# For GPU support, please add the "-gpu" suffix to the base image
BASE_IMAGE = "mesosphere/kubeflow:1.0.1-0.3.1-tensorflow-2.2.0"
downloadOp = components.func_to_container_op(
download_and_process_dataset, base_image=BASE_IMAGE
)()
trainOp = components.func_to_container_op(
build_and_train_model, base_image=BASE_IMAGE
)(downloadOp.output)
# evaluateOp = components.func_to_container_op(evaluate_model, base_image=BASE_IMAGE)(
# downloadOp.output, trainOp.output
# )
# exportOp = components.func_to_container_op(export_model, base_image=BASE_IMAGE)(
# trainOp.output, evaluateOp.output, export_bucket, model_name, model_version
# )
# # Create an inference server from an external component
# kfserving_op = components.load_component_from_url(
# "https://raw.githubusercontent.com/kubeflow/pipelines/8d738ea7ddc350e9b78719910982abcd8885f93f/components/kubeflow/kfserving/component.yaml"
# )
# kfserving = kfserving_op(
# action="create",
# default_model_uri=f"s3://{export_bucket}/{model_name}",
# model_name="mnist",
# namespace=NAMESPACE,
# framework="tensorflow",
# )
# kfserving.after(exportOp)
# See: https://github.com/kubeflow/kfserving/blob/master/docs/DEVELOPER_GUIDE.md#troubleshooting
def op_transformer(op):
op.add_pod_annotation(name="sidecar.istio.io/inject", value="false")
return op
@dsl.pipeline(name="anomaly-pipeline")
def anomaly_pipeline(
model_dir: str = "/train/model",
data_dir: str = "/train/data",
export_bucket: str = "airframe-anomaly",
model_name: str = "airframe-anomaly",
model_version: int = 1,
):
train_and_serve(
data_dir=data_dir,
model_dir=model_dir,
export_bucket=export_bucket,
model_name=model_name,
model_version=model_version,
)
dsl.get_pipeline_conf().add_op_transformer(op_transformer)
pipeline_func = anomaly_pipeline
run_name = pipeline_func.__name__ + " run"
experiment_name = "airframe-anomaly-detection"
arguments = {
"model_dir": "/train/model",
"data_dir": "/train/data",
# "export_bucket": "airframe-anomaly",
"model_name": "airframe-anomaly",
"model_version": "1",
}
client = kfp.Client()
run_result = client.create_run_from_pipeline_func(
pipeline_func,
experiment_name=experiment_name,
run_name=run_name,
arguments=arguments,
)
###Output
_____no_output_____
###Markdown
Component 3: Evaluate the ModelWith the following Python function the model is evaluated.The metrics [metadata](https://www.kubeflow.org/docs/pipelines/sdk/pipelines-metrics/) (loss and accuracy) is available to the Kubeflow Pipelines UI.Metadata can automatically be visualized with output viewer(s).Please go [here](https://www.kubeflow.org/docs/pipelines/sdk/output-viewer/) to see how to do that.
###Code
def evaluate_model(
data_dir: InputPath(str), model_dir: InputPath(str), metrics_path: OutputPath(str)
) -> NamedTuple("EvaluationOutput", [("mlpipeline_metrics", "Metrics")]):
"""Loads a saved model from file and uses a pre-downloaded dataset for evaluation.
Model metrics are persisted to `/mlpipeline-metrics.json` for Kubeflow Pipelines
metadata."""
import json
import tensorflow as tf
import tensorflow_datasets as tfds
from collections import namedtuple
def normalize_test(values, mean, std):
values -= mean
values /= std
return values
df_test_value = (df_daily_jumpsup - training_mean) / training_std
# Create sequences from test values.
x_test = create_sequences(df_test_value.values)
print("Test input shape: ", x_test.shape)
# Get test MAE loss.
x_test_pred = model.predict(x_test)
test_mae_loss = np.mean(np.abs(x_test_pred - x_test), axis=1)
test_mae_loss = test_mae_loss.reshape((-1))
# Detect all the samples which are anomalies.
anomalies = test_mae_loss > threshold
print("Number of anomaly samples: ", np.sum(anomalies))
print("Indices of anomaly samples: ", np.where(anomalies))
ds_test, ds_info = tfds.load(
"mnist",
split="test",
shuffle_files=True,
as_supervised=True,
withj_info=True,
download=False,
data_dir=data_dir,
)
# See: https://www.tensorflow.org/datasets/keras_example#build_training_pipeline
ds_test = ds_test.map(
normalize_image, num_parallel_calls=tf.data.experimental.AUTOTUNE
)
ds_test = ds_test.batch(128)
ds_test = ds_test.cache()
ds_test = ds_test.prefetch(tf.data.experimental.AUTOTUNE)
model = tf.keras.models.load_model(model_dir)
(loss, accuracy) = model.evaluate(ds_test)
metrics = {
"metrics": [
{"name": "loss", "numberValue": str(loss), "format": "PERCENTAGE"},
{"name": "accuracy", "numberValue": str(accuracy), "format": "PERCENTAGE"},
]
}
with open(metrics_path, "w") as f:
json.dump(metrics, f)
out_tuple = namedtuple("EvaluationOutput", ["mlpipeline_metrics"])
return out_tuple(json.dumps(metrics))
###Output
_____no_output_____
###Markdown
Component 4: Export the Model
###Code
def export_model(
model_dir: InputPath(str),
metrics: InputPath(str),
export_bucket: str,
model_name: str,
model_version: int,
):
import os
import boto3
from botocore.client import Config
s3 = boto3.client(
"s3",
endpoint_url="http://minio-service.kubeflow:9000",
aws_access_key_id="minio",
aws_secret_access_key="minio123",
config=Config(signature_version="s3v4"),
)
# Create export bucket if it does not yet exist
response = s3.list_buckets()
export_bucket_exists = False
for bucket in response["Buckets"]:
if bucket["Name"] == export_bucket:
export_bucket_exists = True
if not export_bucket_exists:
s3.create_bucket(ACL="public-read-write", Bucket=export_bucket)
# Save model files to S3
for root, dirs, files in os.walk(model_dir):
for filename in files:
local_path = os.path.join(root, filename)
s3_path = os.path.relpath(local_path, model_dir)
s3.upload_file(
local_path,
export_bucket,
f"{model_name}/{model_version}/{s3_path}",
ExtraArgs={"ACL": "public-read"},
)
response = s3.list_objects(Bucket=export_bucket)
print(f"All objects in {export_bucket}:")
for file in response["Contents"]:
print("{}/{}".format(export_bucket, file["Key"]))
###Output
_____no_output_____
###Markdown
How to Combine the Components into a PipelineNote that up to this point we have not yet used the Kubeflow Pipelines SDK!With our four components (i.e. self-contained funtions) defined, we can wire up the dependencies with Kubeflow Pipelines.The call [`components.func_to_container_op(f, base_image=img)(*args)`](https://www.kubeflow.org/docs/pipelines/sdk/sdk-overview/) has the following ingredients:- `f` is the Python function that defines a component- `img` is the base (Docker) image used to package the function- `*args` lists the arguments to `f`What the `*args` mean is best explained by going forward through the graph:- `downloadOp` is the very first step and has no dependencies; it therefore has no `InputPath`. Its output (i.e. `OutputPath`) is stored in `data_dir`.- `trainOp` needs the data downloaded from `downloadOp` and its signature lists `data_dir` (input) and `model_dir` (output). So, it _depends on_ `downloadOp.output` (i.e. the previous step's output) and stores its own outputs in `model_dir`, which can be used by another step. `downloadOp` is the parent of `trainOp`, as required.- `evaluateOp`'s function takes three arguments: `data_dir` (i.e. `downloadOp.output`), `model_dir` (i.e. `trainOp.output`), and `metrics_path`, which is where the function stores its evaluation metrics. That way, `evaluateOp` can only run after the successful completion of both `downloadOp` and `trainOp`.- `exportOp` runs the function `export_model`, which accepts five parameters: `model_dir`, `metrics`, `export_bucket`, `model_name`, and `model_version`. From where do we get the `model_dir`? It is nothing but `trainOp.output`. Similarly, `metrics` is `evaluateOp.output`. The remaining three arguments are regular Python arguments that are static for the pipeline: they do not depend on any step's output being available. Hence, they are defined without using `InputPath`. Since it is the last step of the pipeline, we also do not list any `OutputPath` for use in another step.
###Code
def train_and_serve(
data_dir: str,
model_dir: str,
export_bucket: str,
model_name: str,
model_version: int,
):
# For GPU support, please add the "-gpu" suffix to the base image
BASE_IMAGE = "mesosphere/kubeflow:1.0.1-0.3.1-tensorflow-2.2.0"
downloadOp = components.func_to_container_op(
download_dataset, base_image=BASE_IMAGE
)()
trainOp = components.func_to_container_op(train_model, base_image=BASE_IMAGE)(
downloadOp.output
)
evaluateOp = components.func_to_container_op(evaluate_model, base_image=BASE_IMAGE)(
downloadOp.output, trainOp.output
)
exportOp = components.func_to_container_op(export_model, base_image=BASE_IMAGE)(
trainOp.output, evaluateOp.output, export_bucket, model_name, model_version
)
# Create an inference server from an external component
kfserving_op = components.load_component_from_url(
"https://raw.githubusercontent.com/kubeflow/pipelines/8d738ea7ddc350e9b78719910982abcd8885f93f/components/kubeflow/kfserving/component.yaml"
)
kfserving = kfserving_op(
action="create",
default_model_uri=f"s3://{export_bucket}/{model_name}",
model_name="mnist",
namespace=NAMESPACE,
framework="tensorflow",
)
kfserving.after(exportOp)
###Output
_____no_output_____
###Markdown
Just in case it isn't obvious: this will build the Docker images for you.Each image is based on `BASE_IMAGE` and includes the Python functions as executable files.Each component _can_ use a different base image though.This may come in handy if you want to have reusable components for automatic data and/or model analysis (e.g. to investigate bias).Note that you did not have to use [Kubeflow Fairing](../fairing/Kubeflow%20Fairing.ipynb) or `docker build` locally at all! Remember when we said all dependencies have to be included in the base image? Well, that was not quite accurate. It's a good idea to have everything included and tested before you define and use your pipeline components to make sure that there are not dependency conflicts. There is, however, a way to add packages (packages_to_install) and additional code to execute before the function code (extra_code).Is that it?Not quite!We still have to define the pipeline itself.Our `train_and_serve` function defines dependencies but we must use the KFP domain-specific language (DSL) to register the pipeline with its four components:
###Code
# See: https://github.com/kubeflow/kfserving/blob/master/docs/DEVELOPER_GUIDE.md#troubleshooting
def op_transformer(op):
op.add_pod_annotation(name="sidecar.istio.io/inject", value="false")
return op
@dsl.pipeline(
name="End-to-End MNIST Pipeline",
description="A sample pipeline to demonstrate multi-step model training, evaluation, export, and serving",
)
def mnist_pipeline(
model_dir: str = "/train/model",
data_dir: str = "/train/data",
export_bucket: str = "mnist",
model_name: str = "mnist",
model_version: int = 1,
):
train_and_serve(
data_dir=data_dir,
model_dir=model_dir,
export_bucket=export_bucket,
model_name=model_name,
model_version=model_version,
)
dsl.get_pipeline_conf().add_op_transformer(op_transformer)
###Output
_____no_output_____
###Markdown
With that in place, let's submit the pipeline directly from our notebook:
###Code
pipeline_func = mnist_pipeline
run_name = pipeline_func.__name__ + " run"
experiment_name = "End-to-End MNIST Pipeline"
arguments = {
"model_dir": "/train/model",
"data_dir": "/train/data",
"export_bucket": "mnist",
"model_name": "mnist",
"model_version": "1",
}
client = kfp.Client()
run_result = client.create_run_from_pipeline_func(
pipeline_func,
experiment_name=experiment_name,
run_name=run_name,
arguments=arguments,
)
###Output
[I 200902 12:10:38 _client:267] Creating experiment End-to-End MNIST Pipeline.
###Markdown
The graph will look like this:If there are any issues with our pipeline definition, this is where they would flare up.So, until you submit it, you won't know if your pipeline definition is correct. We have so far claimed that Kubeflow Pipelines is for automation of multi-step (ad hoc) workflows and usage in CI/CD. You may have wondered why that is. After all, it is possible to set up recurring runs of pipelines. The reason is that these pipeline steps are one-offs. Even though you can parameterize each step, including the ones that kick off an entire pipeline, there is no orchestration of workflows. Stated differently, if a step fails, there is no mechanism for automatic retries. Nor is there any support for marking success: if the step is scheduled to be run again, it will be run again, whether or not the previous execution was successful, obviating any subsequent runs (except in cases where it may be warranted). Kubeflow Pipelines allows retries but it is not configurable out of the box. If you want Airflow- or Luigi-like behaviour for dependency management of workflows, Kubeflow Pipelines is not the tool. How to Predict with the Inference ServerThe simplest way to check that our inference server is up and running is to check it with `curl` ( pre-installed on the cluster).To do so, let's define a few helper functions for plotting and displaying images:
###Code
import matplotlib.pyplot as plt
def display_image(x_test, image_index):
plt.imshow(x_test[image_index].reshape(28, 28), cmap="binary")
def predict_number(model, x_test, image_index):
pred = model.predict(x_test[image_index : image_index + 1])
print(pred.argmax())
import tensorflow as tf
(x_train, y_train), (x_test, y_test) = tf.keras.datasets.mnist.load_data()
x_test = x_test / 255.0 # We must transform the data in the same way as before!
image_index = 1005
display_image(x_test, image_index)
###Output
Downloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/mnist.npz
11493376/11490434 [==============================] - 0s 0us/step
###Markdown
The inference server expects a JSON payload:
###Code
import codecs, json
tf_serving_req = {"instances": x_test[image_index : image_index + 1].tolist()}
with open("input.json", "w") as json_file:
json.dump(tf_serving_req, json_file)
model = "mnist"
url = f"http://{model}-predictor-default.{NAMESPACE}.svc.cluster.local/v1/models/{model}:predict"
! curl -L $url [email protected]
###Output
{
"predictions": [[1.40836079e-07, 5.1750153e-06, 2.76334504e-06, 0.00259963516, 0.00124566338, 4.93358129e-06, 1.92605313e-07, 0.000128401283, 0.00166672678, 0.994346321]
]
} |
twitter_analysis2.ipynb | ###Markdown
snatwitter-searchhashtag.py
###Code
from twitter import *
import networkx as nx
import json
import os
# Clear screen
os.system('cls' if os.name=='nt' else 'clear')
graph=nx.DiGraph()
print("")
print(".....................................................")
print ("RT NETWORK OF AN HASHTAG")
print("")
hashtag = "#priyans49767484"
# Log in
OAUTH_TOKEN = "793203628429438978-o1Gs0rjBN3TesBu6cV5muiic15XeZpl"
OAUTH_SECRET = "GVyujMi5n0gE0QE2R2fWK6R7wwxE3R2olAr4s0xd1NCnh"
CONSUMER_KEY = "hCsfln5TQNt3p39CDC3dCJf6d"
CONSUMER_SECRET = "7G3Q5a7Q33D8ENP2oI4ef1Earv4WfEldaPA1eaeR69NtblDQYY"
auth = OAuth(OAUTH_TOKEN, OAUTH_SECRET, CONSUMER_KEY, CONSUMER_SECRET)
twitter = Twitter(auth = auth)
# search
# https://dev.twitter.com/docs/api/1.1/get/search/tweets
query = twitter.search.tweets(q=hashtag, count=100)
# Debug line
#print json.dumps(query, sort_keys=True, indent=4)
# Print results
print("Search complete (%f seconds)" % (query["search_metadata"]["completed_in"]))
print("Found",len(query["statuses"]),"results.")
# Get results and find retweets and mentions
for result in query["statuses"]:
print ("")
print ("Tweet:",result["text"])
print ("By user:",result["user"]["name"])
if len(result["entities"]["user_mentions"]) != 0:
print ("Mentions:")
for i in result["entities"]["user_mentions"]:
print (" - by",i["screen_name"])
graph.add_edge(i["screen_name"],result["user"]["name"])
if "retweeted_status" in result:
if len(result["retweeted_status"]["entities"]["user_mentions"]) != 0:
print( "Retweets:")
for i in result["retweeted_status"]["entities"]["user_mentions"]:
print (" - by",i["screen_name"])
graph.add_edge(i["screen_name"],result["user"]["name"])
else:
pass
# Save graph
print( "")
print ("The network of RT of the hashtag was analyzed succesfully.")
print( "")
print ("Saving the file as "+hashtag+"-rt-network.gexf...")
nx.write_gexf(graph,"/home/"+ hashtag+"-rt-network.gexf")
nx.write_gexf(graph,"/home/"+ hashtag+"-rt-network.txt")
###Output
_____no_output_____ |
playbooks/windows/02_execution/T1035_service_execution/remote_service_creation.ipynb | ###Markdown
Remote Service Creation Playbook Tags**ID:** WINEXEC190815181010**Author:** Roberto Rodriguez [@Cyb3rWard0g](https://twitter.com/Cyb3rWard0g)**References:** WINEXEC190813181010 ATT&CK Tags**Tactic:** Execution, Lateral Movement**Technique:** Service Execution (T1035) Applies To Technical DescriptionAdversaries may execute a binary, command, or script via a method that interacts with Windows services, such as the Service Control Manager. This can be done by by adversaries creating a new service. Adversaries can create services remotely to execute code and move lateraly across the environment. Permission RequiredAdministrator HypothesisAdversaries might be creating new services remotely to execute code and move laterally in my environment Attack Simulation Dataset| Environment| Name | Description ||--------|---------|---------|| [Shire](https://github.com/Cyb3rWard0g/mordor/tree/acf9f6be6a386783a20139ceb2faf8146378d603/environment/shire) | [empire_invoke_psexec](https://github.com/Cyb3rWard0g/mordor/blob/master/small_datasets/windows/execution/service_execution_T1035/empire_invoke_psexec.md) | A mordor dataset to simulate an adversary creating a service | Recommended Data Sources| Event ID | Event Name | Log Provider | Audit Category | Audit Sub-Category | ATT&CK Data Source ||---------|---------|----------|----------|---------|---------|| [4697](https://docs.microsoft.com/en-us/windows/security/threat-protection/auditing/event-4697) | A service was installed in the system | Microsoft-Windows-Security-Auditing | | | Windows Services || [4624](https://github.com/Cyb3rWard0g/OSSEM/blob/master/data_dictionaries/windows/security/events/event-4624.md) | An account was successfully logged on | Microsoft-Windows-Security-Auditing | Audit Logon/Logoff | Audit Logon | Windows Event Logs | Data Analytics Initialize Analytics Engine
###Code
from openhunt.logparser import winlogbeat
from pyspark.sql import SparkSession
win = winlogbeat()
spark = SparkSession.builder.appName("Mordor").config("spark.sql.caseSensitive", "True").getOrCreate()
print(spark)
###Output
<pyspark.sql.session.SparkSession object at 0x7fe65115b198>
###Markdown
Prepare & Process Mordor File
###Code
mordor_file = win.extract_nested_fields("mordor/small_datasets/empire_invoke_psexec_2019-05-18210652.json",spark)
###Output
[+] Processing a Spark DataFrame..
[+] Reading Mordor file..
[+] Processing Data from Winlogbeat version 6..
[+] DataFrame Returned !
###Markdown
Register Mordor DataFrame as a SQL temporary view
###Code
mordor_file.createOrReplaceTempView("mordor_file")
###Output
_____no_output_____
###Markdown
Validate Analytic I| FP Rate | Source | Analytic Logic | Description ||--------|---------|---------|---------|| Low | Sysmon | SELECT o.`@timestamp`, o.computer_name, o.SubjectUserName, o.SubjectUserName, o.ServiceName, a.IpAddress FROM mordor_file o INNER JOIN (SELECT computer_name,TargetUserName,TargetLogonId,IpAddress FROM mordor_file WHERE channel = "Security" AND LogonType = 3 AND IpAddress is not null AND NOT TargetUserName LIKE "%$") a ON o.SubjectLogonId = a.TargetLogonId WHERE o.channel = "Security" AND o.event_id = 4697 | Look for new services being created in your environment under a network logon session (3). That is a sign that the service creation was performed from another endpoint in the environment |
###Code
security_4697_4624_df = spark.sql(
'''
SELECT o.`@timestamp`, o.computer_name, o.SubjectUserName, o.SubjectUserName, o.ServiceName, a.IpAddress
FROM mordor_file o
INNER JOIN (
SELECT computer_name,TargetUserName,TargetLogonId,IpAddress
FROM mordor_file
WHERE channel = "Security"
AND LogonType = 3
AND IpAddress is not null
AND NOT TargetUserName LIKE "%$"
) a
ON o.SubjectLogonId = a.TargetLogonId
WHERE o.channel = "Security"
AND o.event_id = 4697
'''
)
security_4697_4624_df.show(20,False)
###Output
+------------------------+---------------+---------------+---------------+-----------+-------------+
|@timestamp |computer_name |SubjectUserName|SubjectUserName|ServiceName|IpAddress |
+------------------------+---------------+---------------+---------------+-----------+-------------+
|2019-05-18T21:07:23.035Z|IT001.shire.com|pgustavo |pgustavo |Updater |172.18.39.106|
+------------------------+---------------+---------------+---------------+-----------+-------------+
|
gpt-pandas-code-generation.ipynb | ###Markdown
Adding Examples for GPT Model
###Code
gpt.add_example(Example('How many unique values in Division Column?',
'df["Division"].nunique()'))
gpt.add_example(Example('Find the Division of boy who scored 55 marks',
'df.loc[(df.loc[:, "Gender"] == "boy") & (df.loc[:, "Marks"] == 55), "Division"]'))
gpt.add_example(Example('Find the average Marks scored by Girls',
'np.mean(df.loc[(df.loc[:, "Gender"] == "girl"), "Marks"])'))
###Output
_____no_output_____
###Markdown
Example 1
###Code
prompt = "Display Division of girl who scored maximum marks"
print(gpt.get_top_reply(prompt))
df.loc[(df.loc[:, "Gender"] == "girl") &
(df.loc[:, "Marks"] == max(df.loc[:, "Marks"])),
"Division"]
###Output
_____no_output_____
###Markdown
Example 2
###Code
prompt = "Find the median Marks scored by Boys"
print(gpt.get_top_reply(prompt))
np.median(df.loc[(df.loc[:, "Gender"] == "boy"), "Marks"])
###Output
_____no_output_____ |
US Stocks Performance Analysis/StockDataAnalysis.ipynb | ###Markdown
Extracting Stock Data Using a Python Library 1. Importing libraries
###Code
!pip install yfinance==0.1.67
#!pip install pandas==1.3.3
import yfinance as yf
import pandas as pd
###Output
_____no_output_____
###Markdown
2.1. Using the yfinance Library to Extract Stock Data of Apple Using the `Ticker` module we can create an object that will allow us to access functions to extract data. To do this we need to provide the ticker symbol for the stock, here the company is Apple and the ticker symbol is `AAPL`.
###Code
apple = yf.Ticker("AAPL")
###Output
_____no_output_____
###Markdown
Now we can access functions and variables to extract the type of data we need. You can view them and what they represent here [https://aroussi.com/post/python-yahoo-finance](https://aroussi.com/post/python-yahoo-finance?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0220ENSkillsNetwork23455606-2021-01-01). Stock Info Using the attribute info we can extract information about the stock as a Python dictionary.
###Code
apple_info=apple.info
apple_info
###Output
_____no_output_____
###Markdown
We can get the 'country' using the key country
###Code
apple_info['country']
###Output
_____no_output_____
###Markdown
Extracting Share Price A share is the single smallest part of a company's stock that you can buy, the prices of these shares fluctuate over time. Using the history() method we can get the share price of the stock over a certain period of time. Using the `period` parameter we can set how far back from the present to get data. The options for `period` are 1 day (1d), 5d, 1 month (1mo) , 3mo, 6mo, 1 year (1y), 2y, 5y, 10y, ytd, and max.
###Code
apple_share_price_data = apple.history(period="max")
###Output
_____no_output_____
###Markdown
The format that the data is returned in is a Pandas DataFrame. With the `Date` as the index the share `Open`, `High`, `Low`, `Close`, `Volume`, and `Stock Splits` are given for each day.
###Code
apple_share_price_data.head()
###Output
_____no_output_____
###Markdown
We can reset the index of the DataFrame with the `reset_index` function. We also set the `inplace` paramter to `True` so the change takes place to the DataFrame itself.
###Code
apple_share_price_data.reset_index(inplace=True)
###Output
_____no_output_____
###Markdown
We can plot the `Open` price against the `Date`:
###Code
apple_share_price_data.plot(x="Date", y="Open")
###Output
_____no_output_____
###Markdown
Extracting Dividends Dividends are the distribution of a companys profits to shareholders. In this case they are defined as an amount of money returned per share an investor owns. Using the variable `dividends` we can get a dataframe of the data. The period of the data is given by the period defined in the 'history\` function.
###Code
apple.dividends
###Output
_____no_output_____
###Markdown
We can plot the dividends overtime:
###Code
apple.dividends.plot()
###Output
_____no_output_____
###Markdown
2.2. Using the yfinance Library to Extract Stock Data of AMD Now using the `Ticker` module create an object for AMD (Advanced Micro Devices) with the ticker symbol is `AMD` called; name the object amd.
###Code
AMD = yf.Ticker("amd")
###Output
_____no_output_____
###Markdown
Question 1 Use the key 'country' to find the country the stock belongs to, remember it as it will be a quiz question.
###Code
AMD_info=AMD.info
AMD_info
AMD_info['country']
###Output
_____no_output_____
###Markdown
Question 2 Use the key 'sector' to find the sector the stock belongs to, remember it as it will be a quiz question.
###Code
AMD_info['sector']
###Output
_____no_output_____
###Markdown
Question 3 Obtain stock data for AMD using the `history` function, set the `period` to max. Find the `Volume` traded on the first day (first row).
###Code
AMD_share_price_data = AMD.history(period="max")
AMD_share_price_data.head()
AMD_share_price_data.reset_index(inplace=True)
AMD_share_price_data.plot(x="Date", y="Open")
AMD.dividends
#AMD.dividends.plot()
###Output
_____no_output_____ |
04_HowTos/FileService/How_to_use_the_File_Services.ipynb | ###Markdown
How to Use the Data Lab Public File Services_Mike Fitzpatrick and Glenn Eychaner_*Revised: Jan 03, 2019*Files in the virtual storage are usually identified via the prefix "_vos://_". This shorthand identifier is resolved to a user's home directory of the storage space in the service. If the "_vos://_" prefix is instead the name of another user (e.g. "_geychaner://_", and the remainder of the path grants public or group read/write access, then the other user's spaces will be accessed. Most user spaces have a "_/public_" directory to facilitate file sharing (e.g. '_geychaner://public/foo.fits_' will access the '_foo.fits_' file from user '_geychaner_'). Users can make any file (or directory) public by moving it to (or creating a link in) their "/public" directory._Public file services_ are specially created areas where all files are world-readable, and are used for serving files from Data Lab datasets.
###Code
# Make matplotlib plot inline
%matplotlib inline
# Standard DL imports, note we only need storeClient
from dl import storeClient as sc
# 3rd Party Imports
import io
import numpy as np
from matplotlib import pyplot as p
from astropy.io import fits
###Output
_____no_output_____
###Markdown
Listing another user's 'public/' folder in their vospaceThe user in our example is 'demo00'
###Code
print(sc.ls ('demo00://public', format='short'))
###Output
test2.csv test3.csv test6.csv
###Markdown
An example using the **SDSS DR14** public file service.A 'file service' is a _public_ vospace, readable by all users. Set base directory and plate number
###Code
# Set the base directory and plate number
# These can be found by explring the SDSS DR14 space using 'sc.ls()'
print(sc.ls ('sdss_dr14://'))
print(sc.ls ('sdss_dr14://eboss'))
print(sc.ls ('sdss_dr14://eboss/spectro'))
print(sc.ls ('sdss_dr14://eboss/spectro/redux'))
base = 'sdss_dr14://eboss/spectro/redux/v5_10_0/'
plate = '3615'
###Output
apo,apogee,eboss,env,manga,marvels,sdss
calib,elg,lss,lya,photo,photoObj,qso,resolve,spectro,spiders,sweeps,target
data,firefly,redux
images,platelist-mjdsort.html,platelist-mjdsort.txt,platelist.fits,platelist.html,platelist.txt,platequality-mjdsort.html,platequality-mjdsort.txt,platequality.html,platequality.txt,redmonster,v5_10_0
###Markdown
List all available FITS plate files in the plate directory
###Code
# Construct the vospace path to the plate directory
spPlate = base + plate + '/spPlate-' + plate
print(sc.ls (spPlate + '*.fits', format='short'))
###Output
spPlate-3615-55179.fits spPlate-3615-55208.fits spPlate-3615-55445.fits
spPlate-3615-55856.fits spPlate-3615-56219.fits spPlate-3615-56544.fits
###Markdown
Pick a modified Julian date and fiber
###Code
mjd = '56544'
fiber = 39
# Construct the vospace path to the plate file and verify
spfile = spPlate + '-' + mjd + '.fits'
print ('File: ' + spfile)
print (sc.ls (spfile))
###Output
File: sdss_dr14://eboss/spectro/redux/v5_10_0/3615/spPlate-3615-56544.fits
spPlate-3615-56544.fits
###Markdown
Now read the spectrum from the file and construct the wavelength array
###Code
try:
with fits.open(sc.get(spfile, mode='fileobj')) as hdulist:
hdr = hdulist[0].header
flux = hdulist[0].data[fiber-1, :]
ivar = hdulist[1].data[fiber-1, :]
sky = hdulist[6].data[fiber-1, :]
except Exception as e:
raise ValueError("Could not find spPlate file for plate={0:s}, mjd={1:s}!".format(plate, mjd))
loglam = hdr['COEFF0'] + hdr['COEFF1']*np.arange(hdr['NAXIS1'], dtype=flux.dtype)
wavelength = 10.0**loglam
print ("{} {} {}".format(len(flux),len(ivar),len(wavelength)))
###Output
4645 4645 4645
###Markdown
Make a plot of the spectrum
###Code
p.plot(wavelength, flux * (ivar > 0), 'k')
p.xlabel('Angstroms')
p.ylabel('Flux')
###Output
_____no_output_____
###Markdown
List all available public file spacesThe '_sc.services()_' function allows a user to list all the available file services.
###Code
print(sc.services())
###Output
name svc description
-------- ---- --------
chandra vos ChaMPlane: Measuring the Faint X-ray Bin ...
cosmic_dawn vos Cosmic DAWN survey
deeprange vos Deeprange Survey
deep_ecliptic vos Depp Ecliptic Survey
dls vos Deep Lens Survey
flamex vos FLAMINGOS Extragalactic Survey
fls vos First Look Survey
fsvs vos Faint Sky Variability Survey
ir_bootes vos Infrared Bootes Imaging Survey
lgs vos Local Group Survey
lmc vos SuperMACHO Survey
ls_dr1 vos DECam Legacy Survey DR1
ls_dr2 vos DECam Legacy Survey DR2
ls_dr3 vos DECam Legacy Survey DR3
ls_dr4 vos DECam Legacy Survey DR4
ls_dr5 vos DECam Legacy Survey DR5
ls_dr6 vos DECam Legacy Survey DR6
ls_dr7 vos DECam Legacy Survey DR7
ls_dr8 vos DECam Legacy Survey DR8
m31_newfirm vos M31 NEWFIRM Survey
ndwfs vos NOAO Deep-Wide Survey
nfp vos NOAO Fundamental Plane Survey
nmbs vos NEWFIRM Medium Band Survey
nmbs_2 vos NEWFIRM Medium Band Survey II
nsc vos NOAO Source Catalog
sdss_dr14 vos SDSS DR14
sdss_dr16 vos SDSS DR16
singg vos Survey for Ionization in Neutral-Gas Gal ...
smash_dr1 vos SMASH DR1
smash_dr2 vos SMASH DR2
sze vos SZE+Optical Studies of the Cosmic Accele ...
w_project vos The w Project
zbootes vos z-band Photometry of the NOAO Deep-Wide ...
###Markdown
How to Use the Data Lab Public File Services_Mike Fitzpatrick and Glenn Eychaner_*Revised: Jan 03, 2019*Files in the virtual storage are usually identified via the prefix "_vos://_". This shorthand identifier is resolved to a user's home directory of the storage space in the service. If the "_vos://_" prefix is instead the name of another user (e.g. "_geychaner://_", and the remainder of the path grants public or group read/write access, then the other user's spaces will be accessed. Most user spaces have a "_/public_" directory to facilitate file sharing (e.g. '_geychaner://public/foo.fits_' will access the '_foo.fits_' file from user '_geychaner_'). Users can make any file (or directory) public by moving it to (or creating a link in) their "/public" directory._Public file services_ are specially created areas where all files are world-readable, and are used for serving files from Data Lab datasets.
###Code
# Make matplotlib plot inline
%matplotlib inline
# Standard DL imports, note we only need storeClient
from dl import storeClient as sc
# 3rd Party Imports
import io
import numpy as np
from matplotlib import pyplot as p
from astropy.io import fits
###Output
_____no_output_____
###Markdown
Listing another user's public file space
###Code
print(sc.ls ('geychaner://public', format='short'))
###Output
grzw1_3sn10_29M.jpg grzw1_sn10_15M.jpg
###Markdown
An example using the **SDSS DR14** public file service. Set base directory and plate number
###Code
# Set the base directory and plate number
# These can be found by explring the SDSS DR14 space using 'sc.ls()'
print(sc.ls ('sdss_dr14://'))
print(sc.ls ('sdss_dr14://eboss'))
print(sc.ls ('sdss_dr14://eboss/spectro'))
print(sc.ls ('sdss_dr14://eboss/spectro/redux'))
base = 'sdss_dr14://eboss/spectro/redux/v5_10_0/'
plate = '3615'
###Output
apo,apogee,eboss,env,manga,marvels,sdss
calib,elg,lss,lya,photo,photoObj,qso,resolve,spectro,spiders,sweeps,target
data,firefly,redux
images,platelist-mjdsort.html,platelist-mjdsort.txt,platelist.fits,platelist.html,platelist.txt,platequality-mjdsort.html,platequality-mjdsort.txt,platequality.html,platequality.txt,redmonster,v5_10_0
###Markdown
List all available FITS plate files in the plate directory
###Code
# Construct the vospace path to the plate directory
spPlate = base + plate + '/spPlate-' + plate
print(sc.ls (spPlate + '*.fits', format='short'))
###Output
spPlate-3615-55179.fits spPlate-3615-55208.fits spPlate-3615-55445.fits
spPlate-3615-55856.fits spPlate-3615-56219.fits spPlate-3615-56544.fits
###Markdown
Pick a modified Julian date and fiber
###Code
mjd = '56544'
fiber = 39
# Construct the vospace path to the plate file and verify
spfile = spPlate + '-' + mjd + '.fits'
print ('File: ' + spfile)
print (sc.ls (spfile))
###Output
File: sdss_dr14://eboss/spectro/redux/v5_10_0/3615/spPlate-3615-56544.fits
spPlate-3615-56544.fits
###Markdown
Now read the spectrum from the file and construct the wavelength array
###Code
try:
with fits.open(sc.get(spfile, mode='fileobj')) as hdulist:
hdr = hdulist[0].header
flux = hdulist[0].data[fiber-1, :]
ivar = hdulist[1].data[fiber-1, :]
sky = hdulist[6].data[fiber-1, :]
except Exception as e:
raise ValueError("Could not find spPlate file for plate={0:s}, mjd={1:s}!".format(plate, mjd))
loglam = hdr['COEFF0'] + hdr['COEFF1']*np.arange(hdr['NAXIS1'], dtype=flux.dtype)
wavelength = 10.0**loglam
print ("{} {} {}".format(len(flux),len(ivar),len(wavelength)))
###Output
4645 4645 4645
###Markdown
Make a plot of the spectrum
###Code
p.plot(wavelength, flux * (ivar > 0), 'k')
p.xlabel('Angstroms')
p.ylabel('Flux')
###Output
_____no_output_____
###Markdown
List all available public file spacesThe '_sc.services()_' function allows a user to list all the available file services.
###Code
print(sc.services())
###Output
name svc description
-------- ---- --------
chandra vos ChaMPlane: Measuring the Faint X-ray Bin ...
cosmic_dawn vos Cosmic DAWN survey
deeprange vos Deeprange Survey
deep_ecliptic vos Depp Ecliptic Survey
dls vos Deep Lens Survey
flamex vos FLAMINGOS Extragalactic Survey
fls vos First Look Survey
fsvs vos Faint Sky Variability Survey
ir_bootes vos Infrared Bootes Imaging Survey
lgs vos Local Group Survey
lmc vos SuperMACHO Survey
ls_dr1 vos DECam Legacy Survey DR1
ls_dr2 vos DECam Legacy Survey DR2
ls_dr3 vos DECam Legacy Survey DR3
ls_dr4 vos DECam Legacy Survey DR4
ls_dr5 vos DECam Legacy Survey DR5
ls_dr6 vos DECam Legacy Survey DR6
ls_dr7 vos DECam Legacy Survey DR7
m31_newfirm vos M31 NEWFIRM Survey
ndwfs vos NOAO Deep-Wide Survey
nfp vos NOAO Fundamental Plane Survey
nmbs vos NEWFIRM Medium Band Survey
nmbs_2 vos NEWFIRM Medium Band Survey II
sdss_dr14 vos SDSS DR14
singg vos Survey for Ionization in Neutral-Gas Gal ...
smash_dr1 vos SMASH DR1
sze vos SZE+Optical Studies of the Cosmic Accele ...
w_project vos The w Project
###Markdown
How to Use the Data Lab Public File Services_Mike Fitzpatrick, Glenn Eychaner, Robert Nikutta_Files in the virtual storage are usually identified via the prefix "_vos://_". This shorthand identifier is resolved to a user's home directory of the storage space in the service. If the "_vos://_" prefix is instead the name of another user (e.g. "_demo01://_", and the remainder of the path grants public or group read/write access, then the other user's spaces will be accessed. Most user spaces have a "_/public_" directory to facilitate file sharing (e.g. '_demo01://public/foo.fits_' will access the '_foo.fits_' file from user '_demo01_'). Users can make any file (or directory) public by moving it to (or creating a link in) their "/public" directory._Public file services_ are specially created areas where all files are world-readable, and are used for serving files from Data Lab datasets.
###Code
# Make matplotlib plot inline
%matplotlib inline
# Standard DL imports, note we only need storeClient
from dl import storeClient as sc
# 3rd Party Imports
import io
import numpy as np
from matplotlib import pyplot as p
from astropy.io import fits
###Output
_____no_output_____
###Markdown
Listing another user's 'public/' folder in their vospaceThe user in our example is 'demo00'
###Code
print(sc.ls ('demo00://public', format='short'))
###Output
test2.csv test3.csv test6.csv
###Markdown
An example using the **SDSS DR14** public file service.A 'file service' is a _public_ vospace, readable by all users. Set base directory and plate number
###Code
# Set the base directory and plate number
# These can be found by explring the SDSS DR14 space using 'sc.ls()'
print(sc.ls ('sdss_dr14://'))
print(sc.ls ('sdss_dr14://eboss'))
print(sc.ls ('sdss_dr14://eboss/spectro'))
print(sc.ls ('sdss_dr14://eboss/spectro/redux'))
base = 'sdss_dr14://eboss/spectro/redux/v5_10_0/'
plate = '3615'
###Output
apo,apogee,eboss,env,manga,marvels,sdss
calib,elg,lss,lya,photo,photoObj,qso,resolve,spectro,spiders,sweeps,target
data,firefly,redux
images,platelist-mjdsort.html,platelist-mjdsort.txt,platelist.fits,platelist.html,platelist.txt,platequality-mjdsort.html,platequality-mjdsort.txt,platequality.html,platequality.txt,redmonster,v5_10_0
###Markdown
List all available FITS plate files in the plate directory
###Code
# Construct the vospace path to the plate directory
spPlate = base + plate + '/spPlate-' + plate
print(sc.ls (spPlate + '*.fits', format='short'))
###Output
spPlate-3615-55179.fits spPlate-3615-55208.fits spPlate-3615-55445.fits
spPlate-3615-55856.fits spPlate-3615-56219.fits spPlate-3615-56544.fits
###Markdown
Pick a modified Julian date and fiber
###Code
mjd = '56544'
fiber = 39
# Construct the vospace path to the plate file and verify
spfile = spPlate + '-' + mjd + '.fits'
print ('File: ' + spfile)
print (sc.ls (spfile))
###Output
File: sdss_dr14://eboss/spectro/redux/v5_10_0/3615/spPlate-3615-56544.fits
spPlate-3615-56544.fits
###Markdown
Now read the spectrum from the file and construct the wavelength array
###Code
try:
with fits.open(sc.get(spfile, mode='fileobj')) as hdulist:
hdr = hdulist[0].header
flux = hdulist[0].data[fiber-1, :]
ivar = hdulist[1].data[fiber-1, :]
sky = hdulist[6].data[fiber-1, :]
except Exception as e:
raise ValueError("Could not find spPlate file for plate={0:s}, mjd={1:s}!".format(plate, mjd))
loglam = hdr['COEFF0'] + hdr['COEFF1']*np.arange(hdr['NAXIS1'], dtype=flux.dtype)
wavelength = 10.0**loglam
print ("{} {} {}".format(len(flux),len(ivar),len(wavelength)))
###Output
4645 4645 4645
###Markdown
Make a plot of the spectrum
###Code
p.plot(wavelength, flux * (ivar > 0), 'k')
p.xlabel('Angstroms')
p.ylabel('Flux')
###Output
_____no_output_____
###Markdown
List all available public file spacesThe '_sc.services()_' function allows a user to list all the available file services.
###Code
print(sc.services())
###Output
name svc description
-------- ---- --------
nfp vos NOAO Fundamental Plane Survey
chandra vos ChaMPlane: Measuring the Faint X-ray Bin ...
cosmic_dawn vos Cosmic DAWN survey
deeprange vos Deeprange Survey
deep_ecliptic vos Deep Ecliptic Survey
des_dr2 vos Dark Energy Survey DR2
desi_ets vos DESI Early Target Selection
dls vos Deep Lens Survey
flamex vos FLAMINGOS Extragalactic Survey
fls vos First Look Survey
fsvs vos Faint Sky Variability Survey
ir_bootes vos Infrared Bootes Imaging Survey
lgs vos Local Group Survey
gogreen_dr1 vos GOGREEN DR1 Survey
lmc vos SuperMACHO Survey
ls_dr1 vos DECam Legacy Survey DR1
ls_dr2 vos DECam Legacy Survey DR2
ls_dr3 vos DECam Legacy Survey DR3
ls_dr4 vos DECam Legacy Survey DR4
ls_dr5 vos DECam Legacy Survey DR5
ls_dr6 vos DECam Legacy Survey DR6
ls_dr7 vos DECam Legacy Survey DR7
ls_dr8 vos DECam Legacy Survey DR8
ls_dr9 vos DECam Legacy Survey DR9
ls_dr9sv vos DECam Legacy Survey DR9sv
m31_newfirm vos M31 NEWFIRM Survey
ndwfs vos NOAO Deep-Wide Survey
nmbs vos NEWFIRM Medium Band Survey
nmbs_2 vos NEWFIRM Medium Band Survey II
nsc vos NOAO Source Catalog
sdss_dr8 vos SDSS DR8
sdss_dr9 vos SDSS DR9
sdss_dr10 vos SDSS DR10
sdss_dr11 vos SDSS DR11
sdss_dr12 vos SDSS DR12
sdss_dr13 vos SDSS DR13
sdss_dr14 vos SDSS DR14
sdss_dr15 vos SDSS DR15
sdss_dr16 vos SDSS DR16
singg vos Survey for Ionization in Neutral-Gas Gal ...
smash_dr1 vos SMASH DR1
smash_dr2 vos SMASH DR2
sze vos SZE+Optical Studies of the Cosmic Accele ...
w_project vos The w Project
zbootes vos z-band Photometry of the NOAO Deep-Wide ...
|
notebooks/ISIS Serial Numbers.ipynb | ###Markdown
Generate the dataThe next three cells take a directory of cleaned `.trn` files (extra values like AUTO or OPTIONAL removed) and generate a `data.db` database of ISIS serial number translations.
###Code
# Database population and a declared class
Base = declarative.declarative_base()
class Translations(Base):
__tablename__ = 'isis_translations'
id = sqlalchemy.Column(sqlalchemy.INTEGER, primary_key=True)
mission = sqlalchemy.Column(sqlalchemy.String)
instrument = sqlalchemy.Column(sqlalchemy.String)
translation = sqlalchemy.Column(NestedJsonObject)
def __init__(self, mission, instrument, translation):
self.mission = mission
self.instrument = instrument
self.translation = translation
class StringToMission(Base):
__tablename__ = 'isis_mission_to_standard'
id = sqlalchemy.Column(sqlalchemy.INTEGER, primary_key=True)
key = sqlalchemy.Column(sqlalchemy.String)
value = sqlalchemy.Column(sqlalchemy.String)
def __init__(self, key, value):
self.key = key
self.value = value
engine = sqlalchemy.create_engine('sqlite:///data.db')
Base.metadata.bind = engine
Base.metadata.create_all()
session = orm.sessionmaker(bind=engine)()
files = glob.glob('../autocnet/examples/serial_number_translations/*.trn')
for f in files:
p = pvl.load(f)
name = os.path.basename(f[:-16])
try:
v = re.findall("([a-z, 0-9]*)([A-Z, a-z, 0-9]*)", name)[0]
mission, instrument = v
except:
v = re.findall("[a-z, 0-9]*", name)
mission = v[0]
instrument = None
r = Translations(mission, instrument, p)
session.add(r)
session.commit()
# Build the mission names lookup table
v = """
Group = MissionName
InputKey = SpacecraftName
InputGroup = "IsisCube,Instrument"
InputPosition = (IsisCube, Instrument)
Translation = (Aircraft, "Aircraft")
Translation = (Apollo15, "APOLLO 15")
Translation = (Apollo15, "APOLLO15")
Translation = (Apollo16, "APOLLO 16")
Translation = (Apollo16, "APOLLO16")
Translation = (Apollo17, "APOLLO 17")
Translation = (Apollo17, "APOLLO17")
Translation = (Cassini, Cassini-Huygens)
# Translation = (Chan1, "CHANDRAYAAN-1 ORBITER")
# Translation = (Chan1, CHANDRAYAAN1_ORBITER)
# Translation = (Chan1, CHANDRAYAAN-1)
Translation = (Chandrayaan1, "CHANDRAYAAN-1 ORBITER")
Translation = (Chandrayaan1, CHANDRAYAAN1_ORBITER)
Translation = (Chandrayaan1, CHANDRAYAAN-1)
Translation = (Clementine1, CLEMENTINE_1)
Translation = (Clementine1, "CLEMENTINE 1")
Translation = (Dawn, "DAWN")
Translation = (Galileo, "Galileo Orbiter")
Translation = (Hayabusa, HAYABUSA)
Translation = (Ideal, IdealSpacecraft)
Translation = (Kaguya, KAGUYA)
Translation = (Kaguya, SELENE-M)
Translation = (Lo, "Lunar Orbiter 3")
Translation = (Lo, "Lunar Orbiter 4")
Translation = (Lo, "Lunar Orbiter 5")
Translation = (Lro, "LUNAR RECONNAISSANCE ORBITER")
Translation = (Lro, "Lunar Reconnaissance Orbiter")
Translation = (Mariner10, Mariner_10)
Translation = (Mariner10, MARINER_10)
Translation = (Mer, "MARS EXPLORATION ROVER 1")
Translation = (Mer, MARS_EXPLORATION_ROVER_1)
Translation = (Mer, "MARS EXPLORATION ROVER 2")
Translation = (Mer, "SIMULATED MARS EXPLORATION ROVER 1")
Translation = (Mer, "SIMULATED MARS EXPLORATION ROVER 2")
Translation = (Messenger, MESSENGER)
Translation = (Messenger, Messenger)
Translation = (Mex, "MARS EXPRESS")
Translation = (Mex, "Mars Express")
Translation = (Mgs, MARSGLOBALSURVEYOR)
Translation = (Mgs, "MARS GLOBAL SURVEYOR")
Translation = (Mro, "MARS RECONNAISSANCE ORBITER")
Translation = (Mro, Mars_Reconnaissance_Orbiter)
Translation = (NewHorizons, "NEW HORIZONS")
Translation = (Near, NEAR)
Translation = (Near, "NEAR EARTH ASTEROID RENDEZVOUS")
Translation = (Odyssey, MARS_ODYSSEY)
Translation = (OsirisRex, OSIRIS-REX)
Translation = (Smart1, SMART1)
Translation = (Viking1, VIKING_ORBITER_1)
Translation = (Viking2, VIKING_ORBITER_2)
Translation = (Voyager1, VOYAGER_1)
Translation = (Voyager2, VOYAGER_2)
End_Group
End
"""
p = pvl.loads(v)
for k, v in p['MissionName'].items():
if k == 'Translation':
r = StringToMission(v[1], v[0])
session.add(r)
session.commit()
###Output
_____no_output_____
###Markdown
Sample Query
###Code
# Sample querying the database
for i, j, t in session.query(Translations.mission, Translations.instrument, Translations.translation):
print(i,j)
d = PVLModule(t)
print(d)
break
###Output
_____no_output_____
###Markdown
Testing code to prototype the functionality now in autocnet
###Code
class SerialNumberDecoder(pvl.decoder.PVLDecoder):
"""
A PVL Decoder class to handle cube label parsing for the purpose of creating a valid ISIS
serial number. Inherits from the PVLDecoder in planetarypy's pvl module.
"""
def cast_unquoated_string(self, value):
"""
Overrides the parent class's method so that any un-quoted string type value found in the
parsed pvl will just return the original value. This is needed so that keyword values
are not re-formatted from what is originally in the ISIS cube label.
Note: This affects value types that are recognized as null, boolean, number, datetime,
et at.
"""
return value.decode('utf-8')
def get_isis_translation(label):
"""
Compute the ISIS serial number for a given image using
the input cube or the label extracted from the cube.
Parameters
----------
label : dict or str
A PVL dict object or file name to extract
the PVL object from
Returns
-------
translation : dict
A PVLModule object containing the extracted
translation file
"""
if not isinstance(label, PVLModule):
label = pvl.load(label)
cube_obj = find_in_dict(label, 'Instrument')
# Grab the spacecraft name and run it through the ISIS lookup
spacecraft_name = find_in_dict(cube_obj, 'SpacecraftName')
for row in session.query(StringToMission).filter(StringToMission.key==spacecraft_name):
spacecraft_name = row.value.lower()
#Try and pull an instrument identifier
try:
instrumentid = find_in_dict(cube_obj, 'InstrumentId').capitalize()
except:
instrumentid = None
# Grab the translation PVL object using the lookup
for row in session.query(Translations).filter(Translations.mission==spacecraft_name,
Translations.instrument==instrumentid):
# Convert the JSON back to a PVL object
translation = PVLModule(row.translation)
return translation
def extract_subgroup(data, key_list):
return reduce(lambda d, k: d[k], key_list, data)
def generate_serial_number(label):
if not isinstance(label, PVLModule):
label = pvl.load(label, cls=SerialNumberDecoder)
# Get the translation information
translation = get_isis_translation(label)
serial_number = []
# Sort the keys to ensure proper iteration order
keys = sorted(translation.keys())
for k in keys:
group = translation[k]
search_key = group['InputKey']
search_position = group['InputPosition']
search_translation = {group['Translation'][1]:group['Translation'][0]}
print(search_key, search_position, search_translation)
sub_group = extract_subgroup(label, search_position)
serial_entry = sub_group[search_key]
if serial_entry in search_translation.keys():
serial_entry = search_translation[serial_entry]
elif '*' in search_translation.keys() and search_translation['*'] != '*':
serial_entry = search_translation['*']
serial_number.append(serial_entry)
return '/'.join(serial_number)
###Output
SpacecraftName ['IsisCube', 'Instrument'] {'*': 'APOLLO15'}
InstrumentId ['IsisCube', 'Instrument'] {'*': '*'}
StartTime ['IsisCube', 'Instrument'] {'*': '*'}
APOLLO15/METRIC/1971-07-31T01:24:36.970
###Markdown
Edit below code for the correct path to an Apollo cube and uncomment to test. Must remain commented out for a non-failing build server-side.
###Code
#serial = generate_serial_number('/Users/jlaura/Desktop/Apollo15/AS15-M-0296_sub4.cub')
#print(serial)
###Output
_____no_output_____ |
Lab_1/Matplotlib_Lab_1.ipynb | ###Markdown
Introduction to Numerical Computing with Numpy and Matplotlib What is MatplotlibMatplotlib is a Python 2D plotting library which produces publication quality figures in a variety of hardcopy formats and interactive environments across platforms. Matplotlib can be used in Python scripts, the Python and IPython shells, the Jupyter notebook, web application servers, and four graphical user interface toolkits. Matplotlib tries to make easy things easy and hard things possible. You can generate plots, histograms, power spectra, bar charts, errorcharts, scatterplots, etc., with just a few lines of code.For more infomration: [Matplotlib Webpage](https://matplotlib.org/) Malplotlib Object Model Matplotlib State's MachineMatplotlib behaves like a state machine. Any command is applied to the current plotting area.
###Code
import matplotlib.pyplot as plt
import numpy as np
t = np.linspace(0, 2*np.pi, 50)
x = np.sin(t)
y = np.cos(t)
# Now create a figure
plt.figure()
# and plot x inside it
plt.plot(x)
# Now create a new figure
plt.figure()
# and plot y inside it...
plt.plot(y)
# Add a title
plt.title("Cos")
###Output
_____no_output_____
###Markdown
Line Plot
###Code
x = np.linspace(0, 2*np.pi, 50)
y1 = np.sin(x)
y2 = np.sin(2*x)
plt.figure() # Create figure
plt.plot(y1)
plt.plot(x, y1)
# red dot-dash circle
plt.plot(x, y1, 'r')
# red marker only circle
plt.plot(x, y1, 'r-o')
# clear figure then plot 2 curves
plt.clf()
plt.plot(x, y1, 'g-o', x, y2, 'b-+')
plt.legend(['sin(x)','sin(2x)'])
###Output
_____no_output_____
###Markdown
Scatter Plot
###Code
N = 50 # no. of points
x = np.linspace(0, 10, N)
#print(x)
from numpy.random import rand
e = rand(N)*5.0 # noise
y1 = x + e
areas = rand(N)*300
plt.scatter(x, y1, s=areas)
colors = rand(N)
plt.scatter(x, y1, s=areas,c=colors)
plt.colorbar()
plt.title("Random scatter")
###Output
_____no_output_____
###Markdown
Image Plot: Correlation Plot
###Code
# Create some data
e1 = rand(100)
e2 = rand(100)*2
e3 = rand(100)*50
e4 = rand(100)*100
corrmatrix = np.corrcoef([e1, e2, e3, e4])
print(corrmatrix)
# Plot corr matrix as image
plt.imshow(corrmatrix, cmap='GnBu')
plt.colorbar()
###Output
[[ 1. 0.11927474 0.049266 0.02849791]
[ 0.11927474 1. 0.02444701 0.0252238 ]
[ 0.049266 0.02444701 1. -0.19692395]
[ 0.02849791 0.0252238 -0.19692395 1. ]]
###Markdown
Multiple plots using subplot
###Code
t = np.linspace(0, 2*np.pi)
x = np.sin(t)
y = np.cos(t)
# To divide the plotting area
plt.subplot(2, 1, 1)
plt.plot(x)
# Now activate a new plot
# area.
plt.subplot(2, 1, 2)
plt.plot(y)
###Output
_____no_output_____
###Markdown
Histogram plot
###Code
# Create array of data
from numpy.random import randint
data = randint(10000, size=(10,1000))
# Approx norm distribution
x = np.sum(data, axis=0)
# Set up for stacked plots
plt.subplot(2,1,1)
plt.hist(x, color='r')
# Plot cumulative dist
plt.subplot(2,1,2)
plt.hist(x, cumulative=True)
###Output
_____no_output_____
###Markdown
Legend, Title, and Label Axis
###Code
# Add labels in plot command.
plt.plot(np.sin(t), label='sin')
plt.plot(np.cos(t), label='cos')
plt.legend()
plt.plot(t, np.sin(t))
plt.xlabel('radians')
# Keywords set text properties.
plt.ylabel('amplitude',fontsize='large')
plt.title('Sin(x)')
###Output
_____no_output_____
###Markdown
Other Visualization Libraries: **Seaborn**: Better looking, high-level, plot-based on matplotlib [Seaborn](https://seaborn.pydata.org/)**Bokeh**: D3 like visulization in web browser [Bokeh](https://github.com/bokeh/bokeh) NumPy ArrayNumpy is the core library for scientific computing in Python. It provides a high-performance multidimensional array object, and tools for working with these arrays. A numpy array is a grid of values, all of the same type, and is indexed by a tuple of nonnegative integers. The number of dimensions is the rank of the array; the shape of an array is a tuple of integers giving the size of the array along each dimension.The Python core library provided Lists. A list is the Python equivalent of an array, but is resizeable and can contain elements of different types.**Why NumPy array**?Size - Numpy data structures take up less spacePerformance - they have a need for speed and are faster than listsFunctionality - SciPy and NumPy have optimized functions such as linear algebra operations built in. Introducing NumPy Array
###Code
import numpy as np
# Simple array creation
a = np.array([0, 1, 2, 3, 4, 5])
print(a)
# Checking the type and type of elements
print(type(a))
print(a.dtype)
# Check dimensions and shape
print(a.ndim)
print(a.shape)
# Check Bytes per element and Bytes of memory use
print(a.itemsize)
print(a.nbytes)
###Output
8
48
###Markdown
Array Operations
###Code
a = np.array([1, 2, 3, 4])
b = np.array([2, 3, 4, 5])
print(a*b)
print(a**b)
print(a+b)
print(a/b)
# Setting array elements
print(a[0])
a[0] = 10
print(a[0])
print(a)
# Multi-dimensional array
a = np.array([[ 0, 1, 2, 3],
[10,11,12,13]])
print(a)
print(a.shape)
print(a.size)
print(a.ndim)
# Get and set elements in Multi-dimensional array
print(a[1, 3])
a[1, 3] = -1
print(a[1, 3])
print(a[1])
###Output
[10 11 12 -1]
###Markdown
Slicing array
###Code
# Slicing array
print(a)
# print all row and 3rd column
print(a[:,2])
# print first row and all column
print(a[0,:])
# negative indices work also
print(a)
print(a[:,:-2])
###Output
[[ 0 1 2 3]
[10 11 12 -1]]
[ 2 12]
[0 1 2 3]
[[ 0 1 2 3]
[10 11 12 -1]]
[[ 0 1]
[10 11]]
|
tarea_02/tarea_02.ipynb | ###Markdown
Tarea N°02 Instrucciones1.- Completa tus datos personales (nombre y rol USM) en siguiente celda.**Nombre**: Pedro Paillalef**Rol**: 201304686-k2.- Debes pushear este archivo con tus cambios a tu repositorio personal del curso, incluyendo datos, imágenes, scripts, etc.3.- Se evaluará:- Soluciones- Código- Que Binder esté bien configurado.- Al presionar `Kernel -> Restart Kernel and Run All Cells` deben ejecutarse todas las celdas sin error. I.- Clasificación de dígitosEn este laboratorio realizaremos el trabajo de reconocer un dígito a partir de una imagen.  El objetivo es a partir de los datos, hacer la mejor predicción de cada imagen. Para ellos es necesario realizar los pasos clásicos de un proyecto de _Machine Learning_, como estadística descriptiva, visualización y preprocesamiento. * Se solicita ajustar al menos tres modelos de clasificación: * Regresión logística * K-Nearest Neighbours * Uno o más algoritmos a su elección [link](https://scikit-learn.org/stable/supervised_learning.htmlsupervised-learning) (es obligación escoger un _estimator_ que tenga por lo menos un hiperparámetro). * En los modelos que posean hiperparámetros es mandatorio buscar el/los mejores con alguna técnica disponible en `scikit-learn` ([ver más](https://scikit-learn.org/stable/modules/grid_search.htmltuning-the-hyper-parameters-of-an-estimator)).* Para cada modelo, se debe realizar _Cross Validation_ con 10 _folds_ utilizando los datos de entrenamiento con tal de determinar un intervalo de confianza para el _score_ del modelo.* Realizar una predicción con cada uno de los tres modelos con los datos _test_ y obtener el _score_. * Analizar sus métricas de error (**accuracy**, **precision**, **recall**, **f-score**) Exploración de los datosA continuación se carga el conjunto de datos a utilizar, a través del sub-módulo `datasets` de `sklearn`.
###Code
import numpy as np
import pandas as pd
from sklearn import datasets
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import confusion_matrix
import time
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import f1_score
from sklearn.svm import SVC
from sklearn.model_selection import validation_curve
from itertools import cycle
from sklearn import svm, datasets
from sklearn.metrics import roc_curve, auc
from sklearn.preprocessing import label_binarize
from sklearn.multiclass import OneVsRestClassifier
from scipy import interp
from sklearn.metrics import roc_auc_score
from sklearn import linear_model
from sklearn.model_selection import cross_validate
from sklearn.metrics import make_scorer
from sklearn.feature_selection import SelectKBest, f_classif
from sklearn.feature_selection import chi2
%matplotlib inline
digits_dict = datasets.load_digits()
print(digits_dict["DESCR"])
digits_dict
digits_dict.keys()
digits_dict["target"]
###Output
_____no_output_____
###Markdown
A continuación se crea dataframe declarado como `digits` con los datos de `digits_dict` tal que tenga 65 columnas, las 6 primeras a la representación de la imagen en escala de grises (0-blanco, 255-negro) y la última correspondiente al dígito (`target`) con el nombre _target_.
###Code
digits = (
pd.DataFrame(
digits_dict["data"],
)
.rename(columns=lambda x: f"c{x:02d}")
.assign(target=digits_dict["target"])
.astype(int)
)
digits.head()
digits.describe()
###Output
_____no_output_____
###Markdown
Ejercicio 1**Análisis exploratorio:** Realiza tu análisis exploratorio, no debes olvidar nada! Recuerda, cada análisis debe responder una pregunta.Algunas sugerencias:* ¿Cómo se distribuyen los datos?* ¿Cuánta memoria estoy utilizando?* ¿Qué tipo de datos son?* ¿Cuántos registros por clase hay?* ¿Hay registros que no se correspondan con tu conocimiento previo de los datos? ¿Cómo se distribuyen los datos? Los datos se distribuyen en un dataframe llamado digits, donde las columnas son string c+dígito desde 0 a 63. con un target que sirve para agrupar la base de datos y finalmente, estos últimos se dejan en formato int. DISCRIBUCUPIB BOMAL ¿Cuánta memoria estoy utilizando?
###Code
digits.info
###Output
_____no_output_____
###Markdown
En este data frame hay 456.4 KB de uso. ¿Qué tipo de datos son? Los datos son enteros ¿Cuántos registros por clase hay? 1797 ¿Hay registros que no se correspondan con tu conocimiento previo de los datos? no hay datos raros porque son int32 Ejercicio 2**Visualización:** Para visualizar los datos utilizaremos el método `imshow` de `matplotlib`. Resulta necesario convertir el arreglo desde las dimensiones (1,64) a (8,8) para que la imagen sea cuadrada y pueda distinguirse el dígito. Superpondremos además el label correspondiente al dígito, mediante el método `text`. Esto nos permitirá comparar la imagen generada con la etiqueta asociada a los valores. Realizaremos lo anterior para los primeros 25 datos del archivo.
###Code
digits_dict["images"][0]
###Output
_____no_output_____
###Markdown
Visualiza imágenes de los dígitos utilizando la llave `images` de `digits_dict`. Sugerencia: Utiliza `plt.subplots` y el método `imshow`. Puedes hacer una grilla de varias imágenes al mismo tiempo!
###Code
nx, ny = 5, 5
fig, axs = plt.subplots(nx,ny, figsize=(12, 12))
for i in range(1, nx*ny +1):
img = digits_dict["images"][i]
fig.add_subplot(nx, ny, i)
plt.imshow(img)
plt.show()
###Output
_____no_output_____
###Markdown
Ejercicio 3**Machine Learning**: En esta parte usted debe entrenar los distintos modelos escogidos desde la librería de `skelearn`. Para cada modelo, debe realizar los siguientes pasos:* **train-test** * Crear conjunto de entrenamiento y testeo (usted determine las proporciones adecuadas). * Imprimir por pantalla el largo del conjunto de entrenamiento y de testeo. * **modelo**: * Instanciar el modelo objetivo desde la librería sklearn. * *Hiper-parámetros*: Utiliza `sklearn.model_selection.GridSearchCV` para obtener la mejor estimación de los parámetros del modelo objetivo.* **Métricas**: * Graficar matriz de confusión. * Analizar métricas de error.__Preguntas a responder:__* ¿Cuál modelo es mejor basado en sus métricas?* ¿Cuál modelo demora menos tiempo en ajustarse?* ¿Qué modelo escoges?
###Code
X = digits.drop(columns="target").values
y = digits["target"].values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.4, random_state=42)
###Output
_____no_output_____
###Markdown
Regresión logistica
###Code
tiempos_ejecucion = []
lista_nombres = ["Logistic Regresor","Random Forest","knn","Logistic Regresor -","Random Forest-","knn-"]
tiempo_inicial = time.time()
clf = LogisticRegression()
clf.fit(X_train,y_train)
tiempo_final = time.time()
tiempos_ejecucion.append(tiempo_final-tiempo_inicial)
###Output
C:\Users\pedro\miniconda3\envs\mat281\lib\site-packages\sklearn\linear_model\_logistic.py:764: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG)
###Markdown
Random Forest
###Code
tiempo_inicial = time.time()
rf = RandomForestClassifier(max_depth=12, random_state=0)
rf.fit(X_train,y_train)
tiempo_final = time.time()
tiempos_ejecucion.append(tiempo_final-tiempo_inicial)
###Output
_____no_output_____
###Markdown
KNN
###Code
tiempo_inicial = time.time()
knn = KNeighborsClassifier(n_neighbors=7)
knn.fit(X_train,y_train)
tiempo_final = time.time()
tiempos_ejecucion.append(tiempo_final-tiempo_inicial)
###Output
_____no_output_____
###Markdown
Score
###Code
### regresión logistica
clf.score( X_test, y_test)
#### matriz de contución regresión logistica
y_pred = clf.predict(X_test)
confusion_matrix(y_test, y_pred)
### Random Forest
rf.score( X_test, y_test)
#### Matriz de contusión random foret
y_pred = rf.predict(X_test)
confusion_matrix(y_test, y_pred)
### KNN
knn.score( X_test, y_test)
### Matriz de contusión knn
y_pred = knn.predict(X_test)
confusion_matrix(y_test, y_pred)
###Output
_____no_output_____
###Markdown
¿Cuál modelo es mejor basado en sus métricas? El mejor modelo corresponde al KNN. ¿Cuál modelo demora menos tiempo en ajustarse?
###Code
print(tiempos_ejecucion)
###Output
[1.7783284187316895, 0.4449596405029297, 0.06295633316040039]
###Markdown
El modelo que demora menos en ajustarse es el KNN con delta inicial 0.093 y luego con la reducción de dimensionalidad queda en 0.015. A modo complementario, el modelo cuyo ajusto no efecta practicamente nada corresponde al Random Forest con delta inicial 0.45 y luego obtuvo 0.42. ¿Qué modelo escoges? El modelo que escogo es KNN, por presentar mejores métricas. Ejercicio 4__Comprensión del modelo:__ Tomando en cuenta el mejor modelo entontrado en el `Ejercicio 3`, debe comprender e interpretar minuciosamente los resultados y gráficos asocados al modelo en estudio, para ello debe resolver los siguientes puntos: * **Cross validation**: usando **cv** (con n_fold = 10), sacar una especie de "intervalo de confianza" sobre alguna de las métricas estudiadas en clases: * $\mu \pm \sigma$ = promedio $\pm$ desviación estandar * **Curva de Validación**: Replica el ejemplo del siguiente [link](https://scikit-learn.org/stable/auto_examples/model_selection/plot_validation_curve.htmlsphx-glr-auto-examples-model-selection-plot-validation-curve-py) pero con el modelo, parámetros y métrica adecuada. Saque conclusiones del gráfico. * **Curva AUC–ROC**: Replica el ejemplo del siguiente [link](https://scikit-learn.org/stable/auto_examples/model_selection/plot_roc.htmlsphx-glr-auto-examples-model-selection-plot-roc-py) pero con el modelo, parámetros y métrica adecuada. Saque conclusiones del gráfico. Cross validation
###Code
cv_results = cross_validate(knn, X, y, cv=10)
cv_results['test_score']
###Output
_____no_output_____
###Markdown
Curva de validación
###Code
param_range = np.logspace(-6, -1, 5)
train_scores, test_scores = validation_curve(
SVC(), X, y, param_name="gamma", param_range=param_range,
scoring="accuracy", n_jobs=1)
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
plt.title("Validation Curve with SVM")
plt.xlabel(r"$\gamma$")
plt.ylabel("Score")
plt.ylim(0.0, 1.1)
lw = 2
plt.semilogx(param_range, train_scores_mean, label="Training score",
color="darkorange", lw=lw)
plt.fill_between(param_range, train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.2,
color="darkorange", lw=lw)
plt.semilogx(param_range, test_scores_mean, label="Cross-validation score",
color="navy", lw=lw)
plt.fill_between(param_range, test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.2,
color="navy", lw=lw)
plt.legend(loc="best")
plt.show()
###Output
_____no_output_____
###Markdown
Curva AUC–ROC
###Code
# Binarize the output
y = label_binarize(y, classes=[0, 1, 2,3,4,5,6,7,8,9])
n_classes = y.shape[1]
# Add noisy features to make the problem harder
random_state = np.random.RandomState(0)
n_samples, n_features = X.shape
X = np.c_[X, random_state.randn(n_samples, 200 * n_features)]
# shuffle and split training and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=.5,
random_state=0)
# Learn to predict each class against the other
classifier = OneVsRestClassifier(svm.SVC(kernel='linear', probability=True,
random_state=random_state))
y_score = classifier.fit(X_train, y_train).decision_function(X_test)
# Compute ROC curve and ROC area for each class
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], y_score[:, i])
roc_auc[i] = auc(fpr[i], tpr[i])
# Compute micro-average ROC curve and ROC area
fpr["micro"], tpr["micro"], _ = roc_curve(y_test.ravel(), y_score.ravel())
roc_auc["micro"] = auc(fpr["micro"], tpr["micro"])
plt.figure()
lw = 2
plt.plot(fpr[2], tpr[2], color='darkorange',
lw=lw, label='ROC curve (area = %0.2f)' % roc_auc[2])
plt.plot([0, 1], [0, 1], color='navy', lw=lw, linestyle='--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic example')
plt.legend(loc="lower right")
plt.show()
###Output
_____no_output_____
###Markdown
La curva roc obtenida es típica y refleja buenos resultados (que se verán más adelante) Ejercicio 5__Reducción de la dimensión:__ Tomando en cuenta el mejor modelo encontrado en el `Ejercicio 3`, debe realizar una redcción de dimensionalidad del conjunto de datos. Para ello debe abordar el problema ocupando los dos criterios visto en clases: * **Selección de atributos*** **Extracción de atributos**__Preguntas a responder:__Una vez realizado la reducción de dimensionalidad, debe sacar algunas estadísticas y gráficas comparativas entre el conjunto de datos original y el nuevo conjunto de datos (tamaño del dataset, tiempo de ejecución del modelo, etc.)
###Code
X = digits.drop(columns="target").values
y = digits["target"].values
X_new = SelectKBest(chi2, k=18).fit_transform(X, y)
X = X_new
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.4, random_state=42)
###Output
_____no_output_____
###Markdown
Regresión logística
###Code
tiempo_inicial = time.time()
clf = LogisticRegression()
clf.fit(X_test,y_test)
tiempo_final = time.time()
tiempos_ejecucion.append(tiempo_final-tiempo_inicial)
###Output
C:\Users\pedro\miniconda3\envs\mat281\lib\site-packages\sklearn\linear_model\_logistic.py:764: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG)
###Markdown
Random forest
###Code
tiempo_inicial = time.time()
rf = RandomForestClassifier(max_depth=12, random_state=0)
rf.fit(X_train,y_train)
tiempo_final = time.time()
tiempos_ejecucion.append(tiempo_final-tiempo_inicial)
###Output
_____no_output_____
###Markdown
KNN
###Code
tiempo_inicial = time.time()
knn = KNeighborsClassifier(n_neighbors=7)
knn.fit(X_train,y_train)
tiempo_final = time.time()
tiempos_ejecucion.append(tiempo_final-tiempo_inicial)
###Output
_____no_output_____
###Markdown
Ejercicio 6__Visualizando Resultados:__ A continuación se provee código para comparar las etiquetas predichas vs las etiquetas reales del conjunto de _test_.
###Code
def mostar_resultados(digits,model,nx=5, ny=5,label = "correctos"):
"""
Muestra los resultados de las prediciones de un modelo
de clasificacion en particular. Se toman aleatoriamente los valores
de los resultados.
- label == 'correcto': retorna los valores en que el modelo acierta.
- label == 'incorrecto': retorna los valores en que el modelo no acierta.
Observacion: El modelo que recibe como argumento debe NO encontrarse
'entrenado'.
:param digits: dataset 'digits'
:param model: modelo de sklearn
:param nx: numero de filas (subplots)
:param ny: numero de columnas (subplots)
:param label: datos correctos o incorrectos
:return: graficos matplotlib
"""
X = digits.drop(columns="target").values
y = digits["target"].values
X_train, X_test, Y_train, Y_test = train_test_split(X, y, test_size=0.2, random_state = 42)
model.fit(X_train, Y_train) # ajustando el modelo
y_pred = list(model.predict(X_test))
# Mostrar los datos correctos
if label=="correctos":
mask = (y_pred == y_test)
color = "green"
# Mostrar los datos correctos
elif label=="incorrectos":
mask = (y_pred != y_test)
color = "red"
else:
raise ValueError("Valor incorrecto")
X_aux = X_test
y_aux_true = y_test
y_aux_pred = y_pred
# We'll plot the first 100 examples, randomly choosen
fig, ax = plt.subplots(nx, ny, figsize=(12,12))
for i in range(nx):
for j in range(ny):
index = j + ny * i
data = X_aux[index, :].reshape(8,8)
label_pred = str(int(y_aux_pred[index]))
label_true = str(int(y_aux_true[index]))
ax[i][j].imshow(data, interpolation='nearest', cmap='gray_r')
ax[i][j].text(0, 0, label_pred, horizontalalignment='center', verticalalignment='center', fontsize=10, color=color)
ax[i][j].text(7, 0, label_true, horizontalalignment='center', verticalalignment='center', fontsize=10, color='blue')
ax[i][j].get_xaxis().set_visible(False)
ax[i][j].get_yaxis().set_visible(False)
plt.show()
model=KNeighborsClassifier(n_neighbors=7)
mostar_resultados(digits,model,nx=5, ny=5,label = "correctos")
###Output
C:\Users\pedro\miniconda3\envs\mat281\lib\site-packages\ipykernel_launcher.py:33: DeprecationWarning: elementwise comparison failed; this will raise an error in the future.
###Markdown
**Pregunta*** Tomando en cuenta el mejor modelo entontrado en el `Ejercicio 3`, grafique los resultados cuando: * el valor predicho y original son iguales * el valor predicho y original son distintos * Cuando el valor predicho y original son distintos , ¿Por qué ocurren estas fallas?
###Code
#mostar_resultados(digits,knn,nx=5, ny=5,label = "correctos")
model=KNeighborsClassifier(n_neighbors=7)
mostar_resultados(digits,model,nx=3, ny=3,label = "incorrectos")
###Output
C:\Users\pedro\miniconda3\envs\mat281\lib\site-packages\ipykernel_launcher.py:38: DeprecationWarning: elementwise comparison failed; this will raise an error in the future.
###Markdown
Tarea N°02 Instrucciones1.- Completa tus datos personales (nombre y rol USM) en siguiente celda.**Nombre**: Gabriel Vergara Schifferli**Rol**: 201510519-72.- Debes pushear este archivo con tus cambios a tu repositorio personal del curso, incluyendo datos, imágenes, scripts, etc.3.- Se evaluará:- Soluciones- Código- Que Binder esté bien configurado.- Al presionar `Kernel -> Restart Kernel and Run All Cells` deben ejecutarse todas las celdas sin error. I.- Clasificación de dígitosEn este laboratorio realizaremos el trabajo de reconocer un dígito a partir de una imagen.  El objetivo es a partir de los datos, hacer la mejor predicción de cada imagen. Para ellos es necesario realizar los pasos clásicos de un proyecto de _Machine Learning_, como estadística descriptiva, visualización y preprocesamiento. * Se solicita ajustar al menos tres modelos de clasificación: * Regresión logística * K-Nearest Neighbours * Uno o más algoritmos a su elección [link](https://scikit-learn.org/stable/supervised_learning.htmlsupervised-learning) (es obligación escoger un _estimator_ que tenga por lo menos un hiperparámetro). * En los modelos que posean hiperparámetros es mandatorio buscar el/los mejores con alguna técnica disponible en `scikit-learn` ([ver más](https://scikit-learn.org/stable/modules/grid_search.htmltuning-the-hyper-parameters-of-an-estimator)).* Para cada modelo, se debe realizar _Cross Validation_ con 10 _folds_ utilizando los datos de entrenamiento con tal de determinar un intervalo de confianza para el _score_ del modelo.* Realizar una predicción con cada uno de los tres modelos con los datos _test_ y obtener el _score_. * Analizar sus métricas de error (**accuracy**, **precision**, **recall**, **f-score**) Exploración de los datosA continuación se carga el conjunto de datos a utilizar, a través del sub-módulo `datasets` de `sklearn`.
###Code
import numpy as np
import pandas as pd
from sklearn import datasets
import matplotlib.pyplot as plt
%matplotlib inline
digits_dict = datasets.load_digits()
print(digits_dict["DESCR"])
digits_dict.keys()
digits_dict["target"]
digits_dict["data"]
###Output
_____no_output_____
###Markdown
A continuación se crea dataframe declarado como `digits` con los datos de `digits_dict` tal que tenga 65 columnas, las 6 primeras a la representación de la imagen en escala de grises (0-blanco, 255-negro) y la última correspondiente al dígito (`target`) con el nombre _target_.
###Code
digits = (
pd.DataFrame(
digits_dict["data"],
)
.rename(columns=lambda x: f"c{x:02d}")
.assign(target=digits_dict["target"])
.astype(int)
)
digits.head()
###Output
_____no_output_____
###Markdown
Ejercicio 1**Análisis exploratorio:** Realiza tu análisis exploratorio, no debes olvidar nada! Recuerda, cada análisis debe responder una pregunta.Algunas sugerencias:* ¿Cómo se distribuyen los datos?* ¿Cuánta memoria estoy utilizando?* ¿Qué tipo de datos son?* ¿Cuántos registros por clase hay?* ¿Hay registros que no se correspondan con tu conocimiento previo de los datos?
###Code
numeros = []
for i in range(0,10):
numeros.append( digits.loc[ digits["target"] == i ])
numeros[0].describe()
numeros[0].describe().iloc[2]
fig, ax = plt.subplots(2, 5, sharex='col', sharey='row')
print("graficos de frecuencio número 0")
for i in range(0,5):
numeros[i].describe().iloc[2].hist(ax=ax[0,i]),
numeros[i +5].describe().iloc[2].hist(ax=ax[1,i])
###Output
graficos de frecuencio número 0
###Markdown
Estos gráficos muestran la frecuencia que hay en la media para cada casilla de la imagen para los números 0 hasta 9. Entonces esto muestra el patron medio que tiene un número asociado a su dibujo.
###Code
###
# LA MEMORIA UTILIZADA
import sys
Memoria = digits.memory_usage()
total = (Memoria[1]*65)/1000
print(total, 'kilobytes')
for i in range(0,10):
print("hay %f resgistros para la clase %s" % ( numeros[i].shape[0] , i ) )
###Output
hay 178.000000 resgistros para la clase 0
hay 182.000000 resgistros para la clase 1
hay 177.000000 resgistros para la clase 2
hay 183.000000 resgistros para la clase 3
hay 181.000000 resgistros para la clase 4
hay 182.000000 resgistros para la clase 5
hay 181.000000 resgistros para la clase 6
hay 179.000000 resgistros para la clase 7
hay 174.000000 resgistros para la clase 8
hay 180.000000 resgistros para la clase 9
###Markdown
Ejercicio 2**Visualización:** Para visualizar los datos utilizaremos el método `imshow` de `matplotlib`. Resulta necesario convertir el arreglo desde las dimensiones (1,64) a (8,8) para que la imagen sea cuadrada y pueda distinguirse el dígito. Superpondremos además el label correspondiente al dígito, mediante el método `text`. Esto nos permitirá comparar la imagen generada con la etiqueta asociada a los valores. Realizaremos lo anterior para los primeros 25 datos del archivo.
###Code
digits_dict["images"][0]
###Output
_____no_output_____
###Markdown
Visualiza imágenes de los dígitos utilizando la llave `images` de `digits_dict`. Sugerencia: Utiliza `plt.subplots` y el método `imshow`. Puedes hacer una grilla de varias imágenes al mismo tiempo!
###Code
fig, axs = plt.subplots(4, 4, figsize=(15, 15))
for i in range(1, 17):
img = digits_dict["images"][i]
fig.add_subplot(4, 4, i)
plt.imshow(img)
plt.axis('off')
plt.show()
###Output
_____no_output_____
###Markdown
Ejercicio 3**Machine Learning**: En esta parte usted debe entrenar los distintos modelos escogidos desde la librería de `skelearn`. Para cada modelo, debe realizar los siguientes pasos:* **train-test** * Crear conjunto de entrenamiento y testeo (usted determine las proporciones adecuadas). * Imprimir por pantalla el largo del conjunto de entrenamiento y de testeo. * **modelo**: * Instanciar el modelo objetivo desde la librería sklearn. * *Hiper-parámetros*: Utiliza `sklearn.model_selection.GridSearchCV` para obtener la mejor estimación de los parámetros del modelo objetivo.* **Métricas**: * Graficar matriz de confusión. * Analizar métricas de error.__Preguntas a responder:__* ¿Cuál modelo es mejor basado en sus métricas?* ¿Cuál modelo demora menos tiempo en ajustarse?* ¿Qué modelo escoges?
###Code
x = digits.drop(columns="target").values
y = digits["target"].values
###Output
_____no_output_____
###Markdown
**TRAIN-TEST SET**
###Code
from metrics_classification import summary_metrics as sm
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.20,
train_size=0.80,
random_state=2020)
print('cantidad de datos para entrenamiento : ',len(x_train))
print('cantidad de datos para testeo : ',len(x_test))
###Output
cantidad de datos para entrenamiento : 1437
cantidad de datos para testeo : 360
###Markdown
**MODELO**
###Code
#REGRESIÓN LOGÍSTICA
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV
import time
#Selección de hiperparámetros
parameters = {
'penalty' : ['l2', None],
'class_weight' : ['balanced', None],
'solver' : ['liblinear', 'lbfgs'],
'random_state':[2020]
}
lr = LogisticRegression()
lr_gridsearchcv = GridSearchCV(estimator = lr,
param_grid = parameters,
cv = 10)
lr_grid_result = lr_gridsearchcv.fit(x_train, y_train)
#KNN
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier()
knnparameters = {
'metric' : ['euclidean', 'manhattan'],
'weights' : ['uniform','distance'],
'algorithm' : ['auto', 'ball_tree', 'kd_tree']
}
knn_gridsearchcv = GridSearchCV(estimator = knn,
param_grid = knnparameters,
cv = 10)
knn_grid_result = knn_gridsearchcv.fit(x_train, y_train)
#PERCEPTRON
from sklearn.linear_model import Perceptron
per = Perceptron()
Pparameters ={
'penalty' : ['l2','l1','elasticnet', None],
'tol':[1e-3,1e-5,1e-1],
'alpha' : [0.0001, 0.00001,0.000001]
}
scores = ['precision', 'recall']
P_gridsearchcv = GridSearchCV(estimator = per,
param_grid = Pparameters,
cv = 10)
p_grid_result = P_gridsearchcv.fit(x_train, y_train)
print("LOGISTIC REGRESSION")
print("El mejor tiempo es de LR: %f usando %s" % (lr_grid_result.best_score_, lr_grid_result.best_params_))
#Calculo de métricas con matriz de confusión
lr_predict = lr_gridsearchcv.predict(x_test)
d = dict( y=y_test, yhat = lr_predict)
df= pd.DataFrame.from_dict(d, orient='index').transpose()
print(confusion_matrix(y_test,lr_predict))
print(sm(df))
print('')
########################################
print("KNN")
print("El mejor tiempo es de KNN: %f usando %s" % (knn_grid_result.best_score_, knn_grid_result.best_params_))
knn_predict = knn_gridsearchcv.predict(x_test)
d = dict( y=y_test, yhat = knn_predict)
df= pd.DataFrame.from_dict(d, orient='index').transpose()
print(confusion_matrix(y_test,knn_predict))
print(sm(df))
print('')
#########################################
print("SGDR")
print("El mejor tiempo es de SGDR: %f usando %s" % (p_grid_result.best_score_, p_grid_result.best_params_))
p_predict = P_gridsearchcv.predict(x_test)
d = dict( y=y_test, yhat = p_predict)
df= pd.DataFrame.from_dict(d, orient='index').transpose()
print(confusion_matrix(y_test,p_predict))
print(sm(df))
print('')
###Output
LOGISTIC REGRESSION
El mejor tiempo es de LR: 0.959630 usando {'class_weight': 'balanced', 'penalty': 'l2', 'random_state': 2020, 'solver': 'lbfgs'}
[[37 0 0 0 1 0 0 0 0 0]
[ 0 34 0 0 0 0 1 0 0 0]
[ 0 0 35 0 0 0 0 0 0 0]
[ 0 0 0 35 0 0 0 0 0 1]
[ 1 0 0 0 40 0 0 0 0 1]
[ 0 0 0 0 1 35 0 0 0 1]
[ 0 0 0 0 0 0 36 0 0 0]
[ 0 0 0 0 0 0 0 37 0 0]
[ 1 0 0 0 1 0 0 0 27 0]
[ 0 0 0 0 1 0 0 0 1 33]]
accuracy recall precision fscore
0 0.9694 0.969 0.9712 0.9698
KNN
El mejor tiempo es de KNN: 0.985383 usando {'algorithm': 'auto', 'metric': 'euclidean', 'weights': 'uniform'}
[[38 0 0 0 0 0 0 0 0 0]
[ 0 35 0 0 0 0 0 0 0 0]
[ 0 0 35 0 0 0 0 0 0 0]
[ 0 0 0 36 0 0 0 0 0 0]
[ 0 0 0 0 42 0 0 0 0 0]
[ 0 0 0 0 0 36 0 0 0 1]
[ 0 0 0 0 0 0 36 0 0 0]
[ 0 0 0 0 0 0 0 37 0 0]
[ 0 0 0 0 0 0 0 0 29 0]
[ 0 0 0 0 0 0 0 0 0 35]]
accuracy recall precision fscore
0 0.9972 0.9973 0.9972 0.9972
SGDR
El mejor tiempo es de SGDR: 0.938068 usando {'alpha': 0.0001, 'penalty': 'l1', 'tol': 0.001}
[[38 0 0 0 0 0 0 0 0 0]
[ 0 32 0 2 1 0 0 0 0 0]
[ 0 0 35 0 0 0 0 0 0 0]
[ 0 0 0 32 0 0 0 0 3 1]
[ 0 0 0 0 41 0 0 0 1 0]
[ 0 2 0 0 0 35 0 0 0 0]
[ 0 0 0 0 0 0 35 0 1 0]
[ 0 0 0 0 0 0 0 37 0 0]
[ 0 1 0 0 0 0 0 0 28 0]
[ 1 0 0 0 0 0 0 1 9 24]]
accuracy recall precision fscore
0 0.9361 0.9349 0.9406 0.9325
###Markdown
**RESUMEN:** el método knn y lr tienen un tiempo de ejecución similar, siendo lr el más rápido y perceptron el más lento.En cuanto a la cantidad de aciertos y errores, el método knn tiene la accuracy más alta y perceptron la peor. Esto se mantiene esta misma relación en cuanto a recall, presición y fscore.Por lo tanto el método que mejor se adapta en este caso es el KNN. Ejercicio 4__Comprensión del modelo:__ Tomando en cuenta el mejor modelo entontrado en el `Ejercicio 3`, debe comprender e interpretar minuciosamente los resultados y gráficos asocados al modelo en estudio, para ello debe resolver los siguientes puntos: * **Cross validation**: usando **cv** (con n_fold = 10), sacar una especie de "intervalo de confianza" sobre alguna de las métricas estudiadas en clases: * $\mu \pm \sigma$ = promedio $\pm$ desviación estandar * **Curva de Validación**: Replica el ejemplo del siguiente [link](https://scikit-learn.org/stable/auto_examples/model_selection/plot_validation_curve.htmlsphx-glr-auto-examples-model-selection-plot-validation-curve-py) pero con el modelo, parámetros y métrica adecuada. Saque conclusiones del gráfico. * **Curva AUC–ROC**: Replica el ejemplo del siguiente [link](https://scikit-learn.org/stable/auto_examples/model_selection/plot_roc.htmlsphx-glr-auto-examples-model-selection-plot-roc-py) pero con el modelo, parámetros y métrica adecuada. Saque conclusiones del gráfico.
###Code
#Cross Validation usando KNN
from sklearn.model_selection import cross_val_score
precision = cross_val_score(estimator=knn_gridsearchcv,
X=x_train,
y=y_train,
cv=10)
precision = [round(x,2) for x in precision]
print('Precisiones: {} '.format(precision))
print('Precision promedio: {0: .3f} +/- {1: .3f}'.format(np.mean(precision),
np.std(precision)))
#curva de validación
from sklearn.model_selection import validation_curve
param_range = np.array([i for i in range(1, 10)])
#Validation curve usando los mejores hiperparámetros
train_scores, test_scores = validation_curve(
KNeighborsClassifier(weights = 'distance',
metric = 'euclidean'),
x_train, y_train,
param_name="n_neighbors",
param_range=param_range,
scoring="accuracy", n_jobs=1)
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
plt.title("Validation Curve with SVM")
plt.xlabel(r"$\gamma$")
plt.ylabel("Score")
plt.ylim(0.0, 1.1)
lw = 2
plt.semilogx(param_range, train_scores_mean, label="Training score",
color="darkorange", lw=lw)
plt.fill_between(param_range, train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.2,
color="darkorange", lw=lw)
plt.semilogx(param_range, test_scores_mean, label="Cross-validation score",
color="navy", lw=lw)
plt.fill_between(param_range, test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.2,
color="navy", lw=lw)
plt.legend(loc="best")
plt.show()
###Output
_____no_output_____
###Markdown
la curva de cross validation es practicamente constante, por lo tanto, se tiene que el modelo de knn se comporta de buena forma independiente de la cantidad de clusters dado que hay un leve decresimiento.
###Code
from itertools import cycle
from sklearn.metrics import roc_curve, auc
from sklearn.preprocessing import label_binarize
from sklearn.multiclass import OneVsRestClassifier
from scipy import interp
from sklearn.metrics import roc_auc_score
index = np.argmax(test_scores_mean)
param_range[index]
y = label_binarize(y, classes=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
n_classes = y.shape[1]
n_samples, n_features = x.shape
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.20,
train_size=0.80,
random_state=2020)
classifier = KNeighborsClassifier(weights = 'distance',metric = 'euclidean', n_neighbors = param_range[index])
y_score = classifier.fit(x_train, y_train).predict(x_test)
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], y_score[:, i])
roc_auc[i] = auc(fpr[i], tpr[i])
# Compute micro-average ROC curve and ROC area
fpr["micro"], tpr["micro"], _ = roc_curve(y_test.ravel(), y_score.ravel())
roc_auc["micro"] = auc(fpr["micro"], tpr["micro"])
#AOC-ROC para multiples clases (código también obtenido del link)
all_fpr = np.unique(np.concatenate([fpr[i] for i in range(n_classes)]))
# Then interpolate all ROC curves at this points
mean_tpr = np.zeros_like(all_fpr)
for i in range(n_classes):
mean_tpr += np.interp(all_fpr, fpr[i], tpr[i])
# Finally average it and compute AUC
mean_tpr /= n_classes
fpr["macro"] = all_fpr
tpr["macro"] = mean_tpr
roc_auc["macro"] = auc(fpr["macro"], tpr["macro"])
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(n_classes), colors):
plt.plot(fpr[i], tpr[i], color=color, lw=lw,
label='ROC curve of class {0} (area = {1:0.2f})'
''.format(i, roc_auc[i]))
plt.plot([0, 1], [0, 1], 'k--', lw=lw)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Some extension of Receiver operating characteristic to multi-class')
plt.legend(loc="lower right")
plt.show()
#Curva promedio de las multi-clases
import sys
# First aggregate all false positive rates
all_fpr = np.unique(np.concatenate([fpr[i] for i in range(n_classes)]))
# Then interpolate all ROC curves at this points
mean_tpr = np.zeros_like(all_fpr)
for i in range(n_classes):
mean_tpr += np.interp(all_fpr, fpr[i], tpr[i])
# Finally average it and compute AUC
mean_tpr /= n_classes
fpr["macro"] = all_fpr
tpr["macro"] = mean_tpr
roc_auc["macro"] = auc(fpr["macro"], tpr["macro"])
plt.plot(fpr["macro"], tpr["macro"],
label='macro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["macro"]),
color='navy', linestyle='-', linewidth=4)
plt.plot([0, 1], [0, 1], 'k--', lw=lw)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Some extension of Receiver operating characteristic to multi-class')
plt.legend(loc="lower right")
plt.show()
###Output
_____no_output_____
###Markdown
Ejercicio 5__Reducción de la dimensión:__ Tomando en cuenta el mejor modelo encontrado en el `Ejercicio 3`, debe realizar una redcción de dimensionalidad del conjunto de datos. Para ello debe abordar el problema ocupando los dos criterios visto en clases: * **Selección de atributos*** **Extracción de atributos**__Preguntas a responder:__Una vez realizado la reducción de dimensionalidad, debe sacar algunas estadísticas y gráficas comparativas entre el conjunto de datos original y el nuevo conjunto de datos (tamaño del dataset, tiempo de ejecución del modelo, etc.)
###Code
#SELECCIÓN DE ATRIBUTOS
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_classif
x_training = digits.drop(columns="target")
y_training = digits["target"]
x_training = x_training.drop(['c00','c32','c39'],axis=1)
k = 30 # número de atributos a seleccionar
columnas = list(x_training.columns.values)
seleccionadas = SelectKBest(f_classif, k=k).fit(x_training, y_training)
catrib = seleccionadas.get_support()
atributos = [columnas[i] for i in list(catrib.nonzero()[0])]
X_a=x_training[atributos]
start_time = time.time()
knn_grid_result = knn_gridsearchcv.fit(x_training, y_training)
print("%s segundos, que demora antes de seleccionar atributos" % (time.time() - start_time))
start_time = time.time()
knn_grid_result = knn_gridsearchcv.fit(X_a, y_training)
print('%s segundos, que demora despues de seleccionar atributos' % (time.time() - start_time))
###Output
tiempo de 4.0168678760528564 segundos, que demora antes de seleccionar atributos
tiempo de 2.3130300045013428 segundos, que demora despues de seleccionar atributos
###Markdown
Ejercicio 6__Visualizando Resultados:__ A continuación se provee código para comparar las etiquetas predichas vs las etiquetas reales del conjunto de _test_.
###Code
def mostar_resultados(digits,model,nx=5, ny=5,label = "correctos"):
"""
Muestra los resultados de las prediciones de un modelo
de clasificacion en particular. Se toman aleatoriamente los valores
de los resultados.
- label == 'correcto': retorna los valores en que el modelo acierta.
- label == 'incorrecto': retorna los valores en que el modelo no acierta.
Observacion: El modelo que recibe como argumento debe NO encontrarse
'entrenado'.
:param digits: dataset 'digits'
:param model: modelo de sklearn
:param nx: numero de filas (subplots)
:param ny: numero de columnas (subplots)
:param label: datos correctos o incorrectos
:return: graficos matplotlib
"""
X = digits.drop(columns="target").values
y = digits["target"].values
X_train, X_test, Y_train, Y_test = train_test_split(X, y, test_size=0.2, random_state = 42)
model.fit(X_train, Y_train) # ajustando el modelo
y_pred = list(model.predict(X_test))
# Mostrar los datos correctos
if label=="correctos":
mask = (y_pred == Y_test)
color = "green"
# Mostrar los datos correctos
elif label=="incorrectos":
mask = (y_pred != Y_test)
color = "red"
else:
raise ValueError("Valor incorrecto")
X_aux = X_test[mask]
y_aux_true = Y_test[mask]
y_aux_pred = np.array(y_pred)[mask]
# We'll plot the first 100 examples, randomly choosen
fig, ax = plt.subplots(nx, ny, figsize=(12,12))
fix = X_aux.shape[0]
for i in range(nx):
for j in range(ny):
index = j + ny * i
if index < fix:
data = X_aux[index, :].reshape(8,8)
label_pred = str(int(y_aux_pred[index]))
label_true = str(int(y_aux_true[index]))
ax[i][j].imshow(data, interpolation='nearest', cmap='gray_r')
ax[i][j].text(0, 0, label_pred,
horizontalalignment='center',
verticalalignment='center',
fontsize=10,
color=color)
ax[i][j].text(7, 0, label_true,
horizontalalignment='center',
verticalalignment='center',
fontsize=10,
color='blue')
ax[i][j].get_xaxis().set_visible(False)
ax[i][j].get_yaxis().set_visible(False)
plt.show()
###Output
_____no_output_____
###Markdown
**Pregunta*** Tomando en cuenta el mejor modelo entontrado en el `Ejercicio 3`, grafique los resultados cuando: * el valor predicho y original son iguales * el valor predicho y original son distintos * Cuando el valor predicho y original son distintos , ¿Por qué ocurren estas fallas?
###Code
mostar_resultados(digits,KNeighborsClassifier(),nx=5, ny=5,label = "correctos")
mostar_resultados(digits,KNeighborsClassifier(),nx=5, ny=5,label = "incorrectos")
###Output
_____no_output_____
###Markdown
Tarea N°02 Instrucciones1.- Completa tus datos personales (nombre y rol USM) en siguiente celda.**Nombre**: Lucas Montero**Rol**: 201473588-k2.- Debes pushear este archivo con tus cambios a tu repositorio personal del curso, incluyendo datos, imágenes, scripts, etc.3.- Se evaluará:- Soluciones- Código- Que Binder esté bien configurado.- Al presionar `Kernel -> Restart Kernel and Run All Cells` deben ejecutarse todas las celdas sin error. I.- Clasificación de dígitosEn este laboratorio realizaremos el trabajo de reconocer un dígito a partir de una imagen. 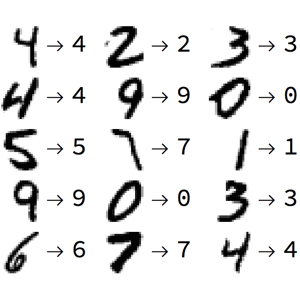 El objetivo es a partir de los datos, hacer la mejor predicción de cada imagen. Para ellos es necesario realizar los pasos clásicos de un proyecto de _Machine Learning_, como estadística descriptiva, visualización y preprocesamiento. * Se solicita ajustar al menos tres modelos de clasificación: * Regresión logística * K-Nearest Neighbours * Uno o más algoritmos a su elección [link](https://scikit-learn.org/stable/supervised_learning.htmlsupervised-learning) (es obligación escoger un _estimator_ que tenga por lo menos un hiperparámetro). * En los modelos que posean hiperparámetros es mandatorio buscar el/los mejores con alguna técnica disponible en `scikit-learn` ([ver más](https://scikit-learn.org/stable/modules/grid_search.htmltuning-the-hyper-parameters-of-an-estimator)).* Para cada modelo, se debe realizar _Cross Validation_ con 10 _folds_ utilizando los datos de entrenamiento con tal de determinar un intervalo de confianza para el _score_ del modelo.* Realizar una predicción con cada uno de los tres modelos con los datos _test_ y obtener el _score_. * Analizar sus métricas de error (**accuracy**, **precision**, **recall**, **f-score**) Exploración de los datosA continuación se carga el conjunto de datos a utilizar, a través del sub-módulo `datasets` de `sklearn`.
###Code
import numpy as np
import pandas as pd
from sklearn import datasets
import matplotlib.pyplot as plt
%matplotlib inline
digits_dict = datasets.load_digits()
print(digits_dict["DESCR"])
digits_dict.keys()
digits_dict["target"]
###Output
_____no_output_____
###Markdown
A continuación se crea dataframe declarado como `digits` con los datos de `digits_dict` tal que tenga 65 columnas, las 6 primeras a la representación de la imagen en escala de grises (0-blanco, 255-negro) y la última correspondiente al dígito (`target`) con el nombre _target_.
###Code
digits = (
pd.DataFrame(
digits_dict["data"],
)
.rename(columns=lambda x: f"c{x:02d}")
.assign(target=digits_dict["target"])
.astype(int)
)
digits.head()
###Output
_____no_output_____
###Markdown
Ejercicio 1**Análisis exploratorio:** Realiza tu análisis exploratorio, no debes olvidar nada! Recuerda, cada análisis debe responder una pregunta.Algunas sugerencias:* ¿Cómo se distribuyen los datos?* ¿Cuánta memoria estoy utilizando?* ¿Qué tipo de datos son?* ¿Cuántos registros por clase hay?* ¿Hay registros que no se correspondan con tu conocimiento previo de los datos?
###Code
#¿Cómo se distribuyen los datos?
# Los datos se encuentran en el dataframe llamado "digits"
# Se encuentran en columnas de string c+(dígito entre 00-63)
# La ultima columna es un "target" que se utiliza para agrupar datos.
#¿Cuánta memoria estoy utilizando?
digits.info()
#¿Qué tipo de datos son?
# La información de la tabla es del formato int.
#¿Cuántos registros por clase hay?
#1797
#¿Hay registros que no se correspondan con tu conocimiento previo de los datos?
#no
###Output
_____no_output_____
###Markdown
Ejercicio 2**Visualización:** Para visualizar los datos utilizaremos el método `imshow` de `matplotlib`. Resulta necesario convertir el arreglo desde las dimensiones (1,64) a (8,8) para que la imagen sea cuadrada y pueda distinguirse el dígito. Superpondremos además el label correspondiente al dígito, mediante el método `text`. Esto nos permitirá comparar la imagen generada con la etiqueta asociada a los valores. Realizaremos lo anterior para los primeros 25 datos del archivo.
###Code
digits_dict["images"][0]
###Output
_____no_output_____
###Markdown
Visualiza imágenes de los dígitos utilizando la llave `images` de `digits_dict`. Sugerencia: Utiliza `plt.subplots` y el método `imshow`. Puedes hacer una grilla de varias imágenes al mismo tiempo!
###Code
nx, ny = 5, 5
fig, axs = plt.subplots(nx,ny, figsize=(12, 12))
for i in range(1, nx*ny +1):
img = digits_dict["images"][i]
fig.add_subplot(nx, ny, i)
plt.imshow(img)
plt.show()
###Output
_____no_output_____
###Markdown
Ejercicio 3**Machine Learning**: En esta parte usted debe entrenar los distintos modelos escogidos desde la librería de `skelearn`. Para cada modelo, debe realizar los siguientes pasos:* **train-test** * Crear conjunto de entrenamiento y testeo (usted determine las proporciones adecuadas). * Imprimir por pantalla el largo del conjunto de entrenamiento y de testeo. * **modelo**: * Instanciar el modelo objetivo desde la librería sklearn. * *Hiper-parámetros*: Utiliza `sklearn.model_selection.GridSearchCV` para obtener la mejor estimación de los parámetros del modelo objetivo.* **Métricas**: * Graficar matriz de confusión. * Analizar métricas de error.__Preguntas a responder:__* ¿Cuál modelo es mejor basado en sus métricas?* ¿Cuál modelo demora menos tiempo en ajustarse?* ¿Qué modelo escoges?
###Code
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import confusion_matrix
import time
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import f1_score
from sklearn.svm import SVC
from sklearn.model_selection import validation_curve
from itertools import cycle
from sklearn import svm, datasets
from sklearn.metrics import roc_curve, auc
from sklearn.preprocessing import label_binarize
from sklearn.multiclass import OneVsRestClassifier
from scipy import interp
from sklearn.metrics import roc_auc_score
from sklearn import linear_model
from sklearn.model_selection import cross_validate
from sklearn.metrics import make_scorer
from sklearn.feature_selection import SelectKBest, f_classif
from sklearn.feature_selection import chi2
X = digits.drop(columns="target").values
y = digits["target"].values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.4, random_state=42)
tiempos = [] #Creamos una lista vacía para guardar los tiempos
########## Logistic Regresor ##########
tiempo_i = time.time() #Guardamos el tiempo inicial
LR = LogisticRegression() #Guardamos LogisticRegression() en LR
LR.fit(X_train,y_train) #Usamos regresión logistica
tiempo_f = time.time() #Guardamos el tiempo final
tiempos.append(tiempo_f-tiempo_i) #Guardamos el tiempo que se demoró
LR.score( X_test, y_test) #Vemos el score
prediccion = LR.predict(X_test) #Guardamos la predicción
confusion_matrix(y_test, prediccion) #Creamos la matriz de confusión
f1_score(y_test, prediccion,average='micro') #Vemos el score final
########## Random Forest ##########
tiempo_i = time.time() #Guardamos el tiempo inicial
RF = RandomForestClassifier(max_depth=12, random_state=0) #Guardamos RandomForest en RF
RF.fit(X_train,y_train) #Usamos random forest
tiempo_f = time.time() #Guardamos el tiempo inicial
tiempos.append(tiempo_f-tiempo_i) #Guardamos el tiempo que se demoró
RF.score( X_test, y_test) #Vemos el score
prediccion = RF.predict(X_test) #Guardamos la predicción
confusion_matrix(y_test, prediccion) #Creamos la matriz de confusión
f1_score(y_test, prediccion,average='micro') #Vemos el score final
########## KNN ##########
tiempo_i = time.time() #Guardamos el tiempo inicial
KNN = KNeighborsClassifier(n_neighbors=7) #Guardamos KNeighborsClassifier en KNN
KNN.fit(X_train,y_train) #Usamos random forest
tiempo_f = time.time() #Guardamos el tiempo inicial
tiempos.append(tiempo_f-tiempo_i) #Guardamos el tiempo que se demoró
KNN.score( X_test, y_test) #Vemos el score
prediccion = KNN.predict(X_test) #Guardamos la predicción
confusion_matrix(y_test, prediccion) #Creamos la matriz de confusión
f1_score(y_test, prediccion,average='micro') #Vemos el score final
###Output
_____no_output_____
###Markdown
Ejercicio 4__Comprensión del modelo:__ Tomando en cuenta el mejor modelo entontrado en el `Ejercicio 3`, debe comprender e interpretar minuciosamente los resultados y gráficos asocados al modelo en estudio, para ello debe resolver los siguientes puntos: * **Cross validation**: usando **cv** (con n_fold = 10), sacar una especie de "intervalo de confianza" sobre alguna de las métricas estudiadas en clases: * $\mu \pm \sigma$ = promedio $\pm$ desviación estandar * **Curva de Validación**: Replica el ejemplo del siguiente [link](https://scikit-learn.org/stable/auto_examples/model_selection/plot_validation_curve.htmlsphx-glr-auto-examples-model-selection-plot-validation-curve-py) pero con el modelo, parámetros y métrica adecuada. Saque conclusiones del gráfico. * **Curva AUC–ROC**: Replica el ejemplo del siguiente [link](https://scikit-learn.org/stable/auto_examples/model_selection/plot_roc.htmlsphx-glr-auto-examples-model-selection-plot-roc-py) pero con el modelo, parámetros y métrica adecuada. Saque conclusiones del gráfico.
###Code
cv_results = cross_validate(LR, X, y, cv=10) #Vemos la cross validation de regresión logistica
cv_results['test_score']
cv_results = cross_validate(RF, X, y, cv=10) #Vemos la cross validation de random forest
cv_results['test_score']
cv_results = cross_validate(KNN, X, y, cv=10) #Vemos la cross validation de knn
cv_results['test_score']
###Output
_____no_output_____
###Markdown
Ejercicio 5__Reducción de la dimensión:__ Tomando en cuenta el mejor modelo encontrado en el `Ejercicio 3`, debe realizar una redcción de dimensionalidad del conjunto de datos. Para ello debe abordar el problema ocupando los dos criterios visto en clases: * **Selección de atributos*** **Extracción de atributos**__Preguntas a responder:__Una vez realizado la reducción de dimensionalidad, debe sacar algunas estadísticas y gráficas comparativas entre el conjunto de datos original y el nuevo conjunto de datos (tamaño del dataset, tiempo de ejecución del modelo, etc.)
###Code
## FIX ME PLEASE
###Output
_____no_output_____
###Markdown
Ejercicio 6__Visualizando Resultados:__ A continuación se provee código para comparar las etiquetas predichas vs las etiquetas reales del conjunto de _test_.
###Code
def mostar_resultados(digits,model,nx=5, ny=5,label = "correctos"):
"""
Muestra los resultados de las prediciones de un modelo
de clasificacion en particular. Se toman aleatoriamente los valores
de los resultados.
- label == 'correcto': retorna los valores en que el modelo acierta.
- label == 'incorrecto': retorna los valores en que el modelo no acierta.
Observacion: El modelo que recibe como argumento debe NO encontrarse
'entrenado'.
:param digits: dataset 'digits'
:param model: modelo de sklearn
:param nx: numero de filas (subplots)
:param ny: numero de columnas (subplots)
:param label: datos correctos o incorrectos
:return: graficos matplotlib
"""
X = digits.drop(columns="target").values
y = digits["target"].values
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2, random_state = 42)
model.fit(X_train, Y_train) # ajustando el modelo
y_pred = list(model.predict(X_test))
# Mostrar los datos correctos
if label=="correctos":
mask = (y_pred == y_test)
color = "green"
# Mostrar los datos correctos
elif label=="incorrectos":
mask = (y_pred != y_test)
color = "red"
else:
raise ValueError("Valor incorrecto")
X_aux = X_test[mask]
y_aux_true = y_test[mask]
y_aux_pred = y_pred[mask]
# We'll plot the first 100 examples, randomly choosen
fig, ax = plt.subplots(nx, ny, figsize=(12,12))
for i in range(nx):
for j in range(ny):
index = j + ny * i
data = X_aux[index, :].reshape(8,8)
label_pred = str(int(y_aux_pred[index]))
label_true = str(int(y_aux_true[index]))
ax[i][j].imshow(data, interpolation='nearest', cmap='gray_r')
ax[i][j].text(0, 0, label_pred, horizontalalignment='center', verticalalignment='center', fontsize=10, color=color)
ax[i][j].text(7, 0, label_true, horizontalalignment='center', verticalalignment='center', fontsize=10, color='blue')
ax[i][j].get_xaxis().set_visible(False)
ax[i][j].get_yaxis().set_visible(False)
plt.show()
###Output
_____no_output_____
###Markdown
**Pregunta*** Tomando en cuenta el mejor modelo entontrado en el `Ejercicio 3`, grafique los resultados cuando: * el valor predicho y original son iguales * el valor predicho y original son distintos * Cuando el valor predicho y original son distintos , ¿Por qué ocurren estas fallas?
###Code
## FIX ME PLEASE
###Output
_____no_output_____
###Markdown
Tarea N°02 Instrucciones1.- Completa tus datos personales (nombre y rol USM) en siguiente celda.**Nombre**:**Rol**:2.- Debes pushear este archivo con tus cambios a tu repositorio personal del curso, incluyendo datos, imágenes, scripts, etc.3.- Se evaluará:- Soluciones- Código- Que Binder esté bien configurado.- Al presionar `Kernel -> Restart Kernel and Run All Cells` deben ejecutarse todas las celdas sin error. I.- Clasificación de dígitosEn este laboratorio realizaremos el trabajo de reconocer un dígito a partir de una imagen. 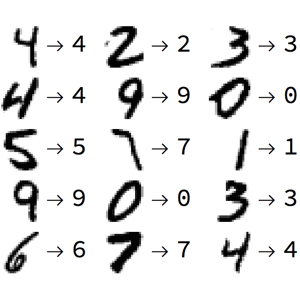 El objetivo es a partir de los datos, hacer la mejor predicción de cada imagen. Para ellos es necesario realizar los pasos clásicos de un proyecto de _Machine Learning_, como estadística descriptiva, visualización y preprocesamiento. * Se solicita ajustar al menos tres modelos de clasificación: * Regresión logística * K-Nearest Neighbours * Uno o más algoritmos a su elección [link](https://scikit-learn.org/stable/supervised_learning.htmlsupervised-learning) (es obligación escoger un _estimator_ que tenga por lo menos un hiperparámetro). * En los modelos que posean hiperparámetros es mandatorio buscar el/los mejores con alguna técnica disponible en `scikit-learn` ([ver más](https://scikit-learn.org/stable/modules/grid_search.htmltuning-the-hyper-parameters-of-an-estimator)).* Para cada modelo, se debe realizar _Cross Validation_ con 10 _folds_ utilizando los datos de entrenamiento con tal de determinar un intervalo de confianza para el _score_ del modelo.* Realizar una predicción con cada uno de los tres modelos con los datos _test_ y obtener el _score_. * Analizar sus métricas de error (**accuracy**, **precision**, **recall**, **f-score**) Exploración de los datosA continuación se carga el conjunto de datos a utilizar, a través del sub-módulo `datasets` de `sklearn`.
###Code
import numpy as np
import pandas as pd
from sklearn import datasets
import matplotlib.pyplot as plt
%matplotlib inline
digits_dict = datasets.load_digits()
print(digits_dict["DESCR"])
digits_dict.keys()
digits_dict["target"]
###Output
_____no_output_____
###Markdown
A continuación se crea dataframe declarado como `digits` con los datos de `digits_dict` tal que tenga 65 columnas, las 6 primeras a la representación de la imagen en escala de grises (0-blanco, 255-negro) y la última correspondiente al dígito (`target`) con el nombre _target_.
###Code
digits = (
pd.DataFrame(
digits_dict["data"],
)
.rename(columns=lambda x: f"c{x:02d}")
.assign(target=digits_dict["target"])
.astype(int)
)
digits.head()
###Output
_____no_output_____
###Markdown
Ejercicio 1**Análisis exploratorio:** Realiza tu análisis exploratorio, no debes olvidar nada! Recuerda, cada análisis debe responder una pregunta.Algunas sugerencias:* ¿Cómo se distribuyen los datos?* ¿Cuánta memoria estoy utilizando?* ¿Qué tipo de datos son?* ¿Cuántos registros por clase hay?* ¿Hay registros que no se correspondan con tu conocimiento previo de los datos?
###Code
## FIX ME PLEASE
digits.describe()
digits.memory_usage()
digits.dtypes
len(digits.columns)
###Output
_____no_output_____
###Markdown
Ejercicio 2**Visualización:** Para visualizar los datos utilizaremos el método `imshow` de `matplotlib`. Resulta necesario convertir el arreglo desde las dimensiones (1,64) a (8,8) para que la imagen sea cuadrada y pueda distinguirse el dígito. Superpondremos además el label correspondiente al dígito, mediante el método `text`. Esto nos permitirá comparar la imagen generada con la etiqueta asociada a los valores. Realizaremos lo anterior para los primeros 25 datos del archivo.
###Code
digits_dict["images"][0]
###Output
_____no_output_____
###Markdown
Visualiza imágenes de los dígitos utilizando la llave `images` de `digits_dict`. Sugerencia: Utiliza `plt.subplots` y el método `imshow`. Puedes hacer una grilla de varias imágenes al mismo tiempo!
###Code
nx, ny = 5, 5
fig, axs = plt.subplots(nx, ny, figsize=(12, 12))
## FIX ME PLEASE
plt.imshow(digits_dict['images'][0])
###Output
_____no_output_____
###Markdown
Ejercicio 3**Machine Learning**: En esta parte usted debe entrenar los distintos modelos escogidos desde la librería de `skelearn`. Para cada modelo, debe realizar los siguientes pasos:* **train-test** * Crear conjunto de entrenamiento y testeo (usted determine las proporciones adecuadas). * Imprimir por pantalla el largo del conjunto de entrenamiento y de testeo. * **modelo**: * Instanciar el modelo objetivo desde la librería sklearn. * *Hiper-parámetros*: Utiliza `sklearn.model_selection.GridSearchCV` para obtener la mejor estimación de los parámetros del modelo objetivo.* **Métricas**: * Graficar matriz de confusión. * Analizar métricas de error.__Preguntas a responder:__* ¿Cuál modelo es mejor basado en sus métricas?* ¿Cuál modelo demora menos tiempo en ajustarse?* ¿Qué modelo escoges?
###Code
X = digits.drop(columns="target").values
y = digits["target"].values
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state = 2)
model=LogisticRegression()
len(X_train)
len(X_test)
len(y_train)
len(y_test)
from sklearn.linear_model import LogisticRegression
rlog = LogisticRegression()
rlog.fit(X_train, y_train)
import sklearn as sk
sk.model_selection.GridSearchCV
import matplotlib.pyplot as plt # doctest: +SKIP
from sklearn.datasets import make_classification
from sklearn.metrics import plot_confusion_matrix
from sklearn.model_selection import train_test_split
from sklearn.svm import SVC
X, y = make_classification(random_state=0)
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
clf = SVC(random_state=0)
clf.fit(X_train, y_train)
SVC(random_state=0)
plot_confusion_matrix(clf, X_test, y_test) # doctest: +SKIP
plt.show() # doctest: +SKIP
# metrics
from metrics_classification import *
from sklearn.metrics import confusion_matrix
y_true = list(y_test)
y_pred = list(rlog.predict(X_test))
print('Valores:\n')
print('originales: ', y_true)
print('predicho: ', y_pred)
print('\nMatriz de confusion:\n ')
print(confusion_matrix(y_true,y_pred))
# ejemplo
df_temp = pd.DataFrame(
{
'y':y_true,
'yhat':y_pred
}
)
df_metrics = summary_metrics(df_temp)
print("\nMetricas para los regresores : 'sepal length (cm)' y 'sepal width (cm)'")
print("")
print(df_metrics)
#k-means
# librerias
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.datasets.samples_generator import make_blobs
pd.set_option('display.max_columns', 500) # Ver más columnas de los dataframes
# Ver gráficos de matplotlib en jupyter notebook/lab
%matplotlib inline
def init_blobs(N, k, seed=1):
X, y = make_blobs(n_samples=N, centers=k,
random_state=seed, cluster_std=0.60)
return X
digits.head()
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
columns = [i for i in digits.columns]
digits[columns] = scaler.fit_transform(digits[columns])
digits.head()
digits.describe()
# graficar
#un ejemplo
sns.set(rc={'figure.figsize':(11.7,8.27)})
ax = sns.scatterplot( data=digits,x="c00", y="c01")
# ajustar modelo: k-means
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters=4)
kmeans.fit(X)
centroids = kmeans.cluster_centers_ # centros
clusters = kmeans.labels_ # clusters
print(digits.columns)
len(clusters)
len(centroids)
# etiquetar los datos con los clusters encontrados
clusters=[clusters[i] for i in range(0,1797)]
clusters=np.array(clusters)
digits["cluster"] = clusters
digits["cluster"] = digits["cluster"].astype('category')
centroids_df = pd.DataFrame(centroids, columns=['c00','c01'])
centroids_df["cluster"] = [0,1,2,3]
# graficar los datos etiquetados con k-means
fig, ax = plt.subplots(figsize=(11, 8.5))
sns.scatterplot( data=digits,
x="c00",
y="c01",
hue="cluster",
legend='full')
sns.scatterplot(x="c00", y="c01",
s=100, color="black", marker="x",
data=centroids_df)
#regresion logistica
# librerias
import os
import numpy as np
import pandas as pd
from sklearn import datasets
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import seaborn as sns
pd.set_option('display.max_columns', 500) # Ver más columnas de los dataframes
# Ver gráficos de matplotlib en jupyter notebook/lab
%matplotlib inline
# datos
from sklearn.linear_model import LogisticRegression
X = digits[['c00', 'c01']]
Y = digits['c02']
# split dataset
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.1, random_state = 1)
# print rows train and test sets
print('Separando informacion:\n')
print('numero de filas data original : ',len(X))
print('numero de filas train set : ',len(X_train))
print('numero de filas test set : ',len(X_test))
# Creando el modelo
rlog = LogisticRegression()
rlog.fit(X_train, Y_train) # ajustando el modelo
# grafica de la regresion logistica
plt.figure(figsize=(12,4))
# dataframe a matriz
X = X.values
Y = Y.values
x_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5
y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5
h = .02 # step size in the mesh
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
Z = rlog.predict(np.c_[xx.ravel(), yy.ravel()])
# Put the result into a color plot
Z = Z.reshape(xx.shape)
plt.figure(1, figsize=(4, 3))
plt.pcolormesh(xx, yy, Z, cmap=plt.cm.Paired)
# Plot also the training points
plt.scatter(X[:, 0], X[:, 1], c=Y, edgecolors='k', cmap=plt.cm.Paired)
plt.xlabel('Sepal length')
plt.ylabel('Sepal width')
plt.show()
###Output
_____no_output_____
###Markdown
Ejercicio 4__Comprensión del modelo:__ Tomando en cuenta el mejor modelo entontrado en el `Ejercicio 3`, debe comprender e interpretar minuciosamente los resultados y gráficos asocados al modelo en estudio, para ello debe resolver los siguientes puntos: * **Cross validation**: usando **cv** (con n_fold = 10), sacar una especie de "intervalo de confianza" sobre alguna de las métricas estudiadas en clases: * $\mu \pm \sigma$ = promedio $\pm$ desviación estandar * **Curva de Validación**: Replica el ejemplo del siguiente [link](https://scikit-learn.org/stable/auto_examples/model_selection/plot_validation_curve.htmlsphx-glr-auto-examples-model-selection-plot-validation-curve-py) pero con el modelo, parámetros y métrica adecuada. Saque conclusiones del gráfico. * **Curva AUC–ROC**: Replica el ejemplo del siguiente [link](https://scikit-learn.org/stable/auto_examples/model_selection/plot_roc.htmlsphx-glr-auto-examples-model-selection-plot-roc-py) pero con el modelo, parámetros y métrica adecuada. Saque conclusiones del gráfico.
###Code
from sklearn.model_selection import cross_val_score
from sklearn.tree import DecisionTreeClassifier,DecisionTreeRegressor
# separ los datos en train y eval
X_train, X_eval, y_train, y_eval = train_test_split(X, y, test_size=0.35,
train_size=0.65,
random_state=1982)
model = DecisionTreeClassifier(criterion='entropy',
max_depth=5)
precision = cross_val_score(estimator=model,
X=X_train,
y=y_train,
cv=10)
precision = [round(x,2) for x in precision]
print('Precisiones: {} '.format(precision))
print('Precision promedio: {0: .3f} +/- {1: .3f}'.format(np.mean(precision),np.std(precision)))
print(__doc__)
import matplotlib.pyplot as plt
import numpy as np
import sklearn as sk
from sklearn.datasets import load_digits
from sklearn.svm import SVC
from sklearn.model_selection import validation_curve
param_range = np.logspace(-6, -1, 5)
train_scores, test_scores = validation_curve(
SVC(), X, y, "gamma", param_range,
param_name='accuracy')
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
plt.title("Validation Curve with SVM")
plt.xlabel(r"$\gamma$")
plt.ylabel("Score")
plt.ylim(0.0, 1.1)
lw = 2
plt.semilogx(param_range, train_scores_mean, label="Training score",
color="darkorange", lw=lw)
plt.fill_between(param_range, train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.2,
color="darkorange", lw=lw)
plt.semilogx(param_range, test_scores_mean, label="Cross-validation score",
color="navy", lw=lw)
plt.fill_between(param_range, test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.2,
color="navy", lw=lw)
plt.legend(loc="best")
plt.show()
sk.metrics.SCORERS.keys()
###Output
_____no_output_____
###Markdown
Ejercicio 5__Reducción de la dimensión:__ Tomando en cuenta el mejor modelo encontrado en el `Ejercicio 3`, debe realizar una redcción de dimensionalidad del conjunto de datos. Para ello debe abordar el problema ocupando los dos criterios visto en clases: * **Selección de atributos*** **Extracción de atributos**__Preguntas a responder:__Una vez realizado la reducción de dimensionalidad, debe sacar algunas estadísticas y gráficas comparativas entre el conjunto de datos original y el nuevo conjunto de datos (tamaño del dataset, tiempo de ejecución del modelo, etc.)
###Code
digits.columns = [f'V{k}' for k in range(1,digits.shape[1]+1)]
digits['y']=y
digits.head()
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_classif
# Separamos las columnas objetivo
x_training = digits.drop(['y'], axis=1)
y_training = digits['y']
# Aplicando el algoritmo univariante de prueba F.
k = 15 # número de atributos a seleccionar
columnas = list(x_training.columns.values)
seleccionadas = SelectKBest(f_classif, k=k).fit(x_training, y_training)
catrib = digits['y'].get_support()
atributos = [columnas[i] for i in list(catrib.nonzero()[0])]
atributos
###Output
_____no_output_____
###Markdown
Ejercicio 6__Visualizando Resultados:__ A continuación se provee código para comparar las etiquetas predichas vs las etiquetas reales del conjunto de _test_.
###Code
def mostar_resultados(digits,model,nx=5, ny=5,label = "correctos"):
"""
Muestra los resultados de las prediciones de un modelo
de clasificacion en particular. Se toman aleatoriamente los valores
de los resultados.
- label == 'correcto': retorna los valores en que el modelo acierta.
- label == 'incorrecto': retorna los valores en que el modelo no acierta.
Observacion: El modelo que recibe como argumento debe NO encontrarse
'entrenado'.
:param digits: dataset 'digits'
:param model: modelo de sklearn
:param nx: numero de filas (subplots)
:param ny: numero de columnas (subplots)
:param label: datos correctos o incorrectos
:return: graficos matplotlib
"""
X = digits.drop(columns="target").values
y = digits["target"].values
X_train, X_test, Y_train, Y_test = train_test_split(X, y, test_size=0.2, random_state = 42)
model.fit(X_train, Y_train) # ajustando el modelo
y_pred = list(model.predict(X_test))
# Mostrar los datos correctos
if label=="correctos":
mask = (y_pred == y_test)
color = "green"
# Mostrar los datos correctos
elif label=="incorrectos":
mask = (y_pred != y_test)
color = "red"
else:
raise ValueError("Valor incorrecto")
X_aux = X_test[mask]
y_aux_true = y_test[mask]
y_aux_pred = y_pred[mask]
# We'll plot the first 100 examples, randomly choosen
fig, ax = plt.subplots(nx, ny, figsize=(12,12))
for i in range(nx):
for j in range(ny):
index = j + ny * i
data = X_aux[index, :].reshape(8,8)
label_pred = str(int(y_aux_pred[index]))
label_true = str(int(y_aux_true[index]))
ax[i][j].imshow(data, interpolation='nearest', cmap='gray_r')
ax[i][j].text(0, 0, label_pred, horizontalalignment='center', verticalalignment='center', fontsize=10, color=color)
ax[i][j].text(7, 0, label_true, horizontalalignment='center', verticalalignment='center', fontsize=10, color='blue')
ax[i][j].get_xaxis().set_visible(False)
ax[i][j].get_yaxis().set_visible(False)
plt.show()
###Output
_____no_output_____
###Markdown
**Pregunta*** Tomando en cuenta el mejor modelo entontrado en el `Ejercicio 3`, grafique los resultados cuando: * el valor predicho y original son iguales * el valor predicho y original son distintos * Cuando el valor predicho y original son distintos , ¿Por qué ocurren estas fallas?
###Code
mostar_resultados(digits,model,nx=5, ny=5,label = "correctos")
###Output
_____no_output_____
###Markdown
Tarea N°02 Instrucciones1.- Completa tus datos personales (nombre y rol USM) en siguiente celda.**Nombre**:Andrés Montecinos López**Rol**:201204515-02.- Debes pushear este archivo con tus cambios a tu repositorio personal del curso, incluyendo datos, imágenes, scripts, etc.3.- Se evaluará:- Soluciones- Código- Que Binder esté bien configurado.- Al presionar `Kernel -> Restart Kernel and Run All Cells` deben ejecutarse todas las celdas sin error. I.- Clasificación de dígitosEn este laboratorio realizaremos el trabajo de reconocer un dígito a partir de una imagen. 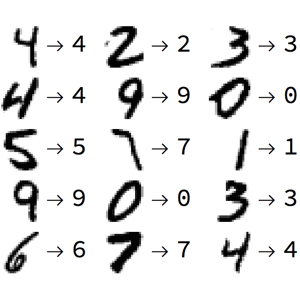 El objetivo es a partir de los datos, hacer la mejor predicción de cada imagen. Para ellos es necesario realizar los pasos clásicos de un proyecto de _Machine Learning_, como estadística descriptiva, visualización y preprocesamiento. * Se solicita ajustar al menos tres modelos de clasificación: * Regresión logística * K-Nearest Neighbours * Uno o más algoritmos a su elección [link](https://scikit-learn.org/stable/supervised_learning.htmlsupervised-learning) (es obligación escoger un _estimator_ que tenga por lo menos un hiperparámetro). * En los modelos que posean hiperparámetros es mandatorio buscar el/los mejores con alguna técnica disponible en `scikit-learn` ([ver más](https://scikit-learn.org/stable/modules/grid_search.htmltuning-the-hyper-parameters-of-an-estimator)).* Para cada modelo, se debe realizar _Cross Validation_ con 10 _folds_ utilizando los datos de entrenamiento con tal de determinar un intervalo de confianza para el _score_ del modelo.* Realizar una predicción con cada uno de los tres modelos con los datos _test_ y obtener el _score_. * Analizar sus métricas de error (**accuracy**, **precision**, **recall**, **f-score**) Exploración de los datosA continuación se carga el conjunto de datos a utilizar, a través del sub-módulo `datasets` de `sklearn`.
###Code
import numpy as np
import pandas as pd
from sklearn import datasets
import matplotlib.pyplot as plt
%matplotlib inline
digits_dict = datasets.load_digits()
print(digits_dict["DESCR"])
digits_dict.keys()
digits_dict["target"]
###Output
_____no_output_____
###Markdown
A continuación se crea dataframe declarado como `digits` con los datos de `digits_dict` tal que tenga 65 columnas, las 6 primeras a la representación de la imagen en escala de grises (0-blanco, 255-negro) y la última correspondiente al dígito (`target`) con el nombre _target_.
###Code
digits = (
pd.DataFrame(
digits_dict["data"],
)
.rename(columns=lambda x: f"c{x:02d}")
.assign(target=digits_dict["target"])
.astype(int)
)
digits.head()
###Output
_____no_output_____
###Markdown
Ejercicio 1**Análisis exploratorio:** Realiza tu análisis exploratorio, no debes olvidar nada! Recuerda, cada análisis debe responder una pregunta.Algunas sugerencias:* ¿Cómo se distribuyen los datos?* ¿Cuánta memoria estoy utilizando?* ¿Qué tipo de datos son?* ¿Cuántos registros por clase hay?* ¿Hay registros que no se correspondan con tu conocimiento previo de los datos? ¿Qué pregunta (s) estás tratando de resolver (o probar que estás equivocado)? * Se intenta desarrollar un clasificador de digitos a partir de imagenes escritas a mano, empleando diferentes algoritmos de clasificación pertenecientes a la rama de Machine Learnig y que fueron vistos en clases. Para ello se provee de una matriz de datos previamente manipulada y normalizada, donde se reduce a una entrada de una matriz de 8x8 con datos que van del rango de 0 a 16 que representan el dígito entregado, además del valor objetivo. ¿Qué tipos de Datos son? * Inicialmente se observa que los datos a analizar son de tipo númerico, conformando un total de 1797 datos. De estos se consideran 64 variables de entrada, correspondiente a los pixeles de una imagen, con el objetivo de predecir un digito.* Por otro lado, cabe señalar que para elaborar la base de datos se consideraron digitos escritos por 43 personas donde 30 de ellas participaron generando digitos para la etapa de entrenamiento y el resto para la etapa de prueba.
###Code
digits.info()
digits.shape # En base a esto se y la información de la celda anterior se obtiene que la base de datos
# no presenta valores nulos
###Output
_____no_output_____
###Markdown
¿Cuántos registros por clase hay?* En este caso se analizarán al menos 10 clases con digitos de 0 a 10.
###Code
digits['target'].unique()
###Output
_____no_output_____
###Markdown
Ejercicio 2**Visualización:** Para visualizar los datos utilizaremos el método `imshow` de `matplotlib`. Resulta necesario convertir el arreglo desde las dimensiones (1,64) a (8,8) para que la imagen sea cuadrada y pueda distinguirse el dígito. Superpondremos además el label correspondiente al dígito, mediante el método `text`. Esto nos permitirá comparar la imagen generada con la etiqueta asociada a los valores. Realizaremos lo anterior para los primeros 25 datos del archivo.
###Code
#Ejemplo de digitos
a=digits_dict["images"][0]
plt.imshow(a, cmap=plt.cm.gray_r, interpolation='nearest')
###Output
_____no_output_____
###Markdown
Visualiza imágenes de los dígitos utilizando la llave `images` de `digits_dict`. Sugerencia: Utiliza `plt.subplots` y el método `imshow`. Puedes hacer una grilla de varias imágenes al mismo tiempo!
###Code
nx, ny = 5, 5
fig, ax = plt.subplots(nx, ny, figsize=(12, 12))
#plt.imshow(a, cmap=plt.cm.gray_r, interpolation='nearest')
axes = []
for i in range(nx*ny):
axes.append(fig.add_subplot(nx,ny,i+1))
a=digits_dict["images"][i]
plt.imshow(a, cmap=plt.cm.gray_r, interpolation='nearest')
plt.subplots_adjust(top = 0.9)
fig.suptitle('Distribución variables numéricas', fontsize = 10, fontweight = "bold")
###Output
_____no_output_____
###Markdown
Ejercicio 3**Machine Learning**: En esta parte usted debe entrenar los distintos modelos escogidos desde la librería de `skelearn`. Para cada modelo, debe realizar los siguientes pasos:* **train-test** * Crear conjunto de entrenamiento y testeo (usted determine las proporciones adecuadas). * Imprimir por pantalla el largo del conjunto de entrenamiento y de testeo. * **modelo**: * Instanciar el modelo objetivo desde la librería sklearn. * *Hiper-parámetros*: Utiliza `sklearn.model_selection.GridSearchCV` para obtener la mejor estimación de los parámetros del modelo objetivo.* **Métricas**: * Graficar matriz de confusión. * Analizar métricas de error.__Preguntas a responder:__* ¿Cuál modelo es mejor basado en sus métricas?* ¿Cuál modelo demora menos tiempo en ajustarse?* ¿Qué modelo escoges? train-test
###Code
X = digits.drop(columns="target").values
y = digits["target"].values
#División de Datos
# Reparto de datos en train y test
# ==============================================================================
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
digits.drop('target', axis = 'columns'),
digits['target'],
train_size = 0.8,
random_state = 1234,
shuffle = True
)
print('El conjuto de entrenamiento se conforma de:',y_train.size,'ejemplos')
print('El conjuto de prueba se conforma de:',y_test.size,'ejemplos')
print("Partición de entrenamento")
print("-----------------------")
print(y_train.describe())
print("Partición de test")
print("-----------------------")
print(y_test.describe())
###Output
Partición de test
-----------------------
count 360.000000
mean 4.452778
std 2.774334
min 0.000000
25% 2.000000
50% 4.000000
75% 7.000000
max 9.000000
Name: target, dtype: float64
###Markdown
Metricas modelos analizados
###Code
from sklearn.model_selection import GridSearchCV
from sklearn import tree
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
# nombre modelos
names_models = [
"RBF SVM",
"Decision Tree",
"Random Forest"
]
# modelos
classifiers = [
SVC(kernel='poly',C=1),
DecisionTreeClassifier(),
RandomForestClassifier(),
]
list_models = list(zip(names_models,classifiers))
hiperparametros={
'RBF SVM': {'gamma': [0.01, 0.1, 2,10]},
'Decision Tree' : {'max_depth':[2,3,4,5,6]},
'Random Forest' : {'max_depth':[2,3,4,5,6]}
}
hiperparametros['RBF SVM']
from metrics_classification import*
from sklearn.metrics import confusion_matrix
from timeit import default_timer
import seaborn as sns
frames_metrics = pd.DataFrame([])
for nombre,modelo in list_models:
inicio=default_timer()
grid = GridSearchCV(
estimator = modelo, #modelo a ajustar
param_grid = hiperparametros[nombre], #función segúnel nombre del modelo arroje los hiperametros cargados anteriormente
scoring = 'neg_root_mean_squared_error',
n_jobs = - 1,
cv = 10,
verbose = 0,
return_train_score = True)
model_fit = grid.fit(X = X_train, y = y_train)
modelo_final = grid.best_estimator_
print(modelo_final)
preds = modelo_final.predict(X = X_test)
df_temp = pd.DataFrame(
{
'y':y_test,
'yhat': preds
}
)
a=summary_metrics(df_temp)
fin=default_timer()
a['name models']=nombre
a['tiempo']=fin-inicio
frames_metrics=pd.concat([frames_metrics,a],axis=0)
matr=confusion_matrix(y_test,preds)
ind=['0','1','2','3','4','5','6','7','8','9']
df_cn=pd.DataFrame(matr,index=ind,columns=ind)
ax = plt.axes()
grafica=sns.heatmap(df_cn, cmap="YlGnBu",annot=True)
plt.xlabel='Digito'
plt.ylabel='Digito'
ax.set_title(nombre)
plt.show()
frames_metrics
###Output
SVC(C=1, gamma=0.01, kernel='poly')
###Markdown
* el mejor modelo basado en sus métricas es RBF SVM. Además respesto a random forest que tuvo buenas métricas, SVM es más rápido. Ejercicio 4__Comprensión del modelo:__ Tomando en cuenta el mejor modelo entontrado en el `Ejercicio 3`, debe comprender e interpretar minuciosamente los resultados y gráficos asocados al modelo en estudio, para ello debe resolver los siguientes puntos: * **Cross validation**: usando **cv** (con n_fold = 10), sacar una especie de "intervalo de confianza" sobre alguna de las métricas estudiadas en clases: * $\mu \pm \sigma$ = promedio $\pm$ desviación estandar * **Curva de Validación**: Replica el ejemplo del siguiente [link](https://scikit-learn.org/stable/auto_examples/model_selection/plot_validation_curve.htmlsphx-glr-auto-examples-model-selection-plot-validation-curve-py) pero con el modelo, parámetros y métrica adecuada. Saque conclusiones del gráfico. * **Curva AUC–ROC**: Replica el ejemplo del siguiente [link](https://scikit-learn.org/stable/auto_examples/model_selection/plot_roc.htmlsphx-glr-auto-examples-model-selection-plot-roc-py) pero con el modelo, parámetros y métrica adecuada. Saque conclusiones del gráfico. Curva de Validación
###Code
import matplotlib.pyplot as plt
import numpy as np
from matplotlib.pyplot import subplots, show
from sklearn.datasets import load_digits
from sklearn.svm import SVC
from sklearn.model_selection import validation_curve
X, y = load_digits(return_X_y=True)
param_range = np.logspace(-6, 3, 5)
train_scores, test_scores = validation_curve(
SVC(kernel='poly',C=1), X, y, param_name="gamma", param_range=param_range,
scoring="accuracy", n_jobs=1,cv=10)
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
fig, ax = subplots()
plt.title('Validation Curve with SVM')
plt.ylim(0.0, 1.1)
lw = 2
ax.semilogx(param_range, train_scores_mean, label="Training score",
color="darkorange", lw=lw)
ax.fill_between(param_range, train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.2,
color="darkorange", lw=lw)
ax.semilogx(param_range, test_scores_mean, label="Cross-validation score",
color="navy", lw=lw)
ax.fill_between(param_range, test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.2,
color="navy", lw=lw)
ax.legend(loc="best")
ax.set_xlabel('gamma')
ax.set_ylabel('Score')
show()
###Output
_____no_output_____
###Markdown
* se aprecia que del gráfico que para entrenamiento y cross-validation el mejor ajuste del parámetro gama es 0.01 Curva AUC–ROC
###Code
import numpy as np
import matplotlib.pyplot as plt
from itertools import cycle
from sklearn import svm, datasets
from sklearn.metrics import roc_curve, auc
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import label_binarize
from sklearn.multiclass import OneVsRestClassifier
from scipy import interp
from sklearn.metrics import roc_auc_score
# Import some data to play with
X = digits.drop(columns="target").values
y=digits["target"].values
# Binarize the output
y = label_binarize(y, classes=[0, 1, 2,3,4,5,6,7,8,9])
n_classes = y.shape[1]
# shuffle and split training and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=.2,
random_state=0)
# Learn to predict each class against the other
classifier = OneVsRestClassifier(svm.SVC(kernel='linear', probability=True,
random_state=0))
y_score = classifier.fit(X_train, y_train).decision_function(X_test)
# Compute ROC curve and ROC area for each class
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], y_score[:, i])
roc_auc[i] = auc(fpr[i], tpr[i])
# Compute micro-average ROC curve and ROC area
fpr["micro"], tpr["micro"], _ = roc_curve(y_test.ravel(), y_score.ravel())
roc_auc["micro"] = auc(fpr["micro"], tpr["micro"])
# First aggregate all false positive rates
all_fpr = np.unique(np.concatenate([fpr[i] for i in range(n_classes)]))
# Then interpolate all ROC curves at this points
mean_tpr = np.zeros_like(all_fpr)
for i in range(n_classes):
mean_tpr += interp(all_fpr, fpr[i], tpr[i])
# Finally average it and compute AUC
mean_tpr /= n_classes
fpr["macro"] = all_fpr
tpr["macro"] = mean_tpr
roc_auc["macro"] = auc(fpr["macro"], tpr["macro"])
# Plot all ROC curves
fig, ax = subplots( figsize=(8,6))
ax.plot(fpr["micro"], tpr["micro"],
label='micro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["micro"]),
color='deeppink', linestyle=':', linewidth=4)
ax.plot(fpr["macro"], tpr["macro"],
label='macro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["macro"]),
color='navy', linestyle=':', linewidth=4)
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(n_classes), colors):
ax.plot(fpr[i], tpr[i], color=color, lw=lw,
label='ROC curve of class {0} (area = {1:0.2f})'
''.format(i, roc_auc[i]))
ax.plot([0, 1], [0, 1], 'k--', lw=lw)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
ax.set_xlabel('False Positive Rate')
ax.set_ylabel('True Positive Rate')
plt.title('Some extension of Receiver operating characteristic to multi-class')
ax.legend(bbox_to_anchor=(1.05, 1))
show()
###Output
<ipython-input-20-4836a1f385bd>:7: DeprecationWarning: scipy.interp is deprecated and will be removed in SciPy 2.0.0, use numpy.interp instead
mean_tpr += interp(all_fpr, fpr[i], tpr[i])
###Markdown
* Las mejores clasificaciones son hechas para la curva de clase 1-7-9.* Por otro lado existe tendencia a clasificar bien, esto dado que la razon de verdadero positivo es alta y se aleja de la diagonal. Ejercicio 5__Reducción de la dimensión:__ Tomando en cuenta el mejor modelo encontrado en el `Ejercicio 3`, debe realizar una redcción de dimensionalidad del conjunto de datos. Para ello debe abordar el problema ocupando los dos criterios visto en clases: * **Selección de atributos*** **Extracción de atributos**__Preguntas a responder:__Una vez realizado la reducción de dimensionalidad, debe sacar algunas estadísticas y gráficas comparativas entre el conjunto de datos original y el nuevo conjunto de datos (tamaño del dataset, tiempo de ejecución del modelo, etc.)
###Code
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_regression, mutual_info_regression
from sklearn.feature_selection import chi2,mutual_info_classif
X1 = digits.drop(columns="target")
y1 = digits["target"]
# Aplicando el algoritmo univariante de prueba F.
k = 18 # número de atributos a seleccionar
#columnas = list(X_train1.columns.values)
seleccionadas = SelectKBest(mutual_info_classif, k=k).fit(X1, y1)
selected_features_df = pd.DataFrame({'Feature':list(X1.columns),
'Scores MI':seleccionadas.scores_})
selected_features_df.sort_values(by='Scores MI', ascending=False).head(9)
###Output
_____no_output_____
###Markdown
* aqui se muestran las mejores variables usando selección de atributos
###Code
from sklearn.svm import SVC
from sklearn.datasets import load_digits
from sklearn.feature_selection import RFE
import matplotlib.pyplot as plt
# Load the digits dataset
X2 = digits.drop(columns="target")
y2 = digits["target"]
# Create the RFE object and rank each pixel
def error_rfe(n):
svc = SVC(kernel="linear", C=1,gamma=2)
rfe = RFE(estimator=svc, n_features_to_select=n, step=1)
selec=rfe.fit(X2, y2)
y_pr=selec.predict(X2)
fina=pd.DataFrame({'pixel':list(range(0,64,1)),'Ranking':selec.ranking_})
df_temp = pd.DataFrame(
{
'y':list(y2),
'yhat': list(y_pr)
}
)
a=summary_metrics(df_temp)
return a
error_rfe(2)
error_rfe(4)
error_rfe(6)
error_rfe(8)
error_rfe(10)
error_rfe(12)
error_rfe(14)
error_rfe(18)
###Output
_____no_output_____
###Markdown
Sobre 15 variables es suficiente para estimar. Ejercicio 6__Visualizando Resultados:__ A continuación se provee código para comparar las etiquetas predichas vs las etiquetas reales del conjunto de _test_.
###Code
def mostar_resultados(digits,model,nx=5, ny=5,label = "correctos"):
"""
Muestra los resultados de las prediciones de un modelo
de clasificacion en particular. Se toman aleatoriamente los valores
de los resultados.
- label == 'correcto': retorna los valores en que el modelo acierta.
- label == 'incorrecto': retorna los valores en que el modelo no acierta.
Observacion: El modelo que recibe como argumento debe NO encontrarse
'entrenado'.
:param digits: dataset 'digits'
:param model: modelo de sklearn
:param nx: numero de filas (subplots)
:param ny: numero de columnas (subplots)
:param label: datos correctos o incorrectos
:return: graficos matplotlib
"""
X = digits.drop(columns="target").values
y = digits["target"].values
X_train, X_test, Y_train, y_test = train_test_split(X, y, test_size=0.2, random_state = 42)
model.fit(X_train, Y_train) # ajustando el modelo
y_pred = list(model.predict(X_test))
# Mostrar los datos correctos
if label=="correctos":
mask = (y_pred == y_test)
color = "green"
# Mostrar los datos correctos
elif label=="incorrectos":
mask = (y_pred != y_test)
color = "red"
else:
raise ValueError("Valor incorrecto")
X_aux = X_test
y_aux_true = y_test
y_aux_pred = y_pred
# We'll plot the first 100 examples, randomly choosen
fig, ax = plt.subplots(nx, ny, figsize=(12,12))
for i in range(nx):
for j in range(ny):
index = j + ny * i
data = X_aux[index, :].reshape(8,8)
label_pred = str(int(y_aux_pred[index]))
label_true = str(int(y_aux_true[index]))
ax[i][j].imshow(data, interpolation='nearest', cmap='gray_r')
ax[i][j].text(0, 0, label_pred, horizontalalignment='center', verticalalignment='center', fontsize=10, color=color)
ax[i][j].text(7, 0, label_true, horizontalalignment='center', verticalalignment='center', fontsize=10, color='blue')
ax[i][j].get_xaxis().set_visible(False)
ax[i][j].get_yaxis().set_visible(False)
plt.show()
model=svm.SVC(C=1, gamma=0.01, kernel='poly')
mostar_resultados(digits,model,nx=5, ny=5,label = "correctos")
model=svm.SVC(C=1, gamma=0.01, kernel='poly')
mostar_resultados(digits,model,nx=5, ny=5,label="incorrectos")
###Output
_____no_output_____ |
tutorials/1_basis/1_basis.ipynb | ###Markdown
1 - 机器学习基础``` 描述:机器学习基础 作者:Chenyyx 时间:2019-12-23``````目录: 1 - 机器学习是什么? 2 - 为什么使用机器学习? 3 - 机器学习的类型 4 - 小实例 5 - 总结``` 1 - 机器学习是什么?机器学习是计算机编程的科学,它使得计算机可以从数据中学习。一个更加面向工程的定义:如果一个计算机在任务 T 上的性能由 P 这个指标来度量,并且随着经验 E 的提升而提升,那么我们就可以说这个计算机程序在任务 T 上使用 P 度量指标从经验 E 中学习。最简单的一个小例子,您的垃圾邮件过滤器就是一个机器学习程序,可以通过给出的垃圾邮件样本以及常规邮件样本来学习标记垃圾邮件。在这种情况下,任务 T 是为新的电子邮件标记垃圾邮件,经验 E 是训练数据集,并且需要定义性能度量 P。例如,您可以使用正确分类的电子邮件的比例。这种特殊的性能度量被称为准确性,经常用于分类任务。 2 - 为什么使用机器学习? - 传统方法传统方法,我们会针对每一个模式写一个检测算法,如果检测到这些模式中的一些,我们的程序才会把目标检测出来。由于每次遇到的问题不会是很小的问题,我们的程序会发展为一个复杂的规则很长的列表,这在后期往往是很难进行维护的。 - 机器学习方法相比之下,基于机器学习所编写的程序会更短,更加容易维护,而且可能会更加准确。就拿垃圾邮件分类任务举例来说,如果垃圾邮件发送者将垃圾邮件之前的规则调整一下,比如将 `4` 换成 `For` 。我们的传统方法则需要更新来标记 `For` 电子邮件,这样我们就需要不断写入新的规则。相比之下,基于机器学习技术的垃圾邮件过滤器会自动注意到 `For` 在用户标记的垃圾邮件中变得异常频繁,并且在没有我们干预的情况下开始对其进行标记。机器学习还对传统方法过于复杂或者没有已知算法的问题有些优势。例如,考虑语音识别:假如你想简单地开始写一个能区分单词 “one” 和 “two” 的程序。你可能会注意到 “two” 这个单词开头是一个高音 (“T”),所以你可以硬编码一个测量高音强度的算法,并用它来区分 “one” 和 “two” 。很明显,这种技术不会在嘈杂的环境和数十种语言中扩展成千上万非常不同的人所说的话。最好的解决方案(至少是现在最好的)是编写一个自学习的算法,给出每个单词的许多示例记录。最后,机器学习可以帮助人类学习(图 1-4):可以检查 ML 算法,看看他们已经学到了什么(尽管对于一些算法来说这可能很困难)。例如,一旦垃圾邮件过滤器已经接受足够的垃圾邮件训练,就可以很容易地检查垃圾邮件过滤器是否能够预测垃圾邮件的最佳预测效果。有时候,这将揭示出未知的相关性或新的趋势,从而导致对问题有更好的理解。应用 ML 技术挖掘大量的数据可以帮助发现那些并不明显的模式。这被称为数据挖掘。总结一下,机器学习对于下面的问题有较好的表现: - 现有解决方案需要大量手动调整或长规则列表的问题:机器学习算法通常可以简化代码并且得到更好的效果。 - 使用传统的方法没有好的解决方案的复杂的问题:最好的机器学习技术可以找到一个解决方案。 - 不是很稳定,时常波动的环境:机器学习系统可以适应新的数据。 - 需要深入了解复杂的问题和数据量比较大的问题。 3 - 机器学习系统的类型 3.1 - 有监督学习在有监督学习中,提供给算法的训练数据包括其所需的分类或解决方案,称之为 **label(标签)** 。典型的监督学习任务是分类,垃圾邮件过滤器就是一个很好的例子:它与许多示例电子邮件和它们的类(垃圾邮件和常规邮件)一起训练,并且它必须学会如何分类新的电子邮件。另一个典型的任务是预测一个目标数值,如给定一组特征(里程,年限,品牌等),来预测一辆汽车的价格,称为预测。这种任务被称为回归。下面列出了一些比较重要的监督学习算法: - k-Nearest Neighbors(k近邻) - Linear Regression(线性回归) - Logistic Regression(Logistic 回归) - Support Vector Machines (SVMs)(支持向量机) - Decision Trees and Random Forests(决策树和随机森林) - Neural network(神经网络) 3.2 - 无监督学习在无监督学习中,训练数据是没有 label(标签)的。下面列出了一些重要的无监督学习算法: - 聚类 - k-Means(k均值) - Hierarchical Cluster Analysis (HCA)(分层聚类分析) - Expectation Maximization(期望最大化) - 可视化和降维 - Principal Component Analysis (PCA)(主成分分析) - Kernel PCA— Locally-Linear Embedding (LLE)(内核 PCA - 局部线性嵌入) - t-distributed Stochastic Neighbor Embedding (t-SNE)(t 分布的随机相邻嵌入) - 关联规则学习 - Apriori - Eclat 3.3 - 半监督学习还有一些算法可以处理部分标记的训练数据,通常是很多未标记的数据和一些标记的数据,这被称为半监督学习。大多数半监督学习算法是无监督和监督算法的组合。以无监督的方式训练模型,然后使用监督学习技术对整个系统进行微调。 3.4 - 强化学习在强化学习中,一个被称为 agent 的学习系统可以观察环境,选择和执行行动,并获取回报(或负面回报的惩罚)。然后,它必须自己学习什么是最好的策略,称为 policy,以获取最大的回报。一项 policy 规定了 agent 在特定情况下应该选择的行动。 4 - 小实例下面举个小例子,使用 scikit-learn 中的 k近邻算法 进行分类。
###Code
from sklearn.neighbors import (NeighborhoodComponentsAnalysis, KNeighborsClassifier)
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
X, y = load_iris(return_X_y=True)
X_train, X_test, y_train, y_test = train_test_split(X, y, stratify=y, test_size=0.7, random_state=42)
nca = NeighborhoodComponentsAnalysis(random_state=42)
knn = KNeighborsClassifier(n_neighbors=3)
nca_pipe = Pipeline([('nca', nca), ('knn', knn)])
nca_pipe.fit(X_train, y_train)
print(nca_pipe.score(X_test, y_test))
###Output
0.9619047619047619
|
run_locaNMF_notebooks/run_locaNMF_and_yongxu_original_code.ipynb | ###Markdown
User-defined Parameters
###Code
##################################################################################################################
## PARAMETERS THAT YOU SHOULD CHANGE
##################################################################################################################
# Path to data and atlas
mouse_name = 'IJ1'
session_name = 'Mar3'
root_dir = '/media/cat/4TBSSD/yuki/yongxu/data/'
data_folder = root_dir+mouse_name+'/'+session_name+'/'
# spatial_data_filename = "IA1_spatial.npy" #
spatial_data_filename =mouse_name+'pm_'+session_name+'_30Hz_code_04_trial_ROItimeCourses_30sec_pca_0.95_spatial.npy'
# temporal_data_filename = "IA1_temporal.npy" #
temporal_data_filename = mouse_name+'pm_'+session_name+'_30Hz_code_04_trial_ROItimeCourses_30sec_pca_0.95.npy'
# atlas_filename = "maskwarp_1.npy" # contains 'atlas'
random_spatial_data_filename =mouse_name+'pm_'+session_name+'_30Hz_code_04_random_ROItimeCourses_30sec_pca_0.95_spatial.npy'
# temporal_data_filename = "IA1_temporal.npy" #
random_temporal_data_filename = mouse_name+'pm_'+session_name+'_30Hz_code_04_random_ROItimeCourses_30sec_pca_0.95.npy'
atlas_filename = "atlas_split.npy"
# maxrank = how many max components per brain region. Set maxrank to around 4 for regular dataset.
maxrank = 1
# min_pixels = minimum number of pixels in Allen map for it to be considered a brain region
# default min_pixels = 100
min_pixels = 200
# loc_thresh = Localization threshold, i.e. percentage of area restricted to be inside the 'Allen boundary'
# default loc_thresh = 80
loc_thresh = 75
# r2_thresh = Fraction of variance in the data to capture with LocaNMF
# default r2_thresh = 0.99
r2_thresh = 0.96
# Do you want nonnegative temporal components? The data itself should also be nonnegative in this case.
# default nonnegative_temporal = False
nonnegative_temporal = False
##################################################################################################################
## PARAMETERS THAT YOU SHOULD PROBABLY NOT CHANGE (unless you know what you're doing)
##################################################################################################################
# maxiter_hals = Number of iterations in innermost loop (HALS). Keeping this low provides a sort of regularization.
# default maxiter_hals = 20
maxiter_hals = 20
# maxiter_lambda = Number of iterations for the lambda loop. Keep this high for finding a good solution.
# default maxiter_lambda = 100
maxiter_lambda = 150
# lambda_step = Amount to multiply lambda after every lambda iteration.
# lambda_init = initial value of lambda. Keep this low. default lambda_init = 0.000001
# lambda_{i+1}=lambda_i*lambda_step. lambda_0=lambda_init. default lambda_step = 1.35
lambda_step = 1.25
lambda_init = 1e-4
# # spatial_data_filename.shape
# a=np.load(data_folder+spatial_data_filename)
# print(a.shape)
# b=np.load(data_folder+temporal_data_filename)
# print(b.shape)
###Output
_____no_output_____
###Markdown
Load & Format Data
###Code
spatial = np.load(data_folder+spatial_data_filename)
# spatial_random = np.load(data_folder+random_spatial_data_filename)
# spatial=np.concatenate(spatial_trial,spatial_random,axis=0)
spatial = np.transpose(spatial,[1,0])
denoised_spatial_name = np.reshape(spatial,[128,128,-1])
temporal_trial = np.load(data_folder+temporal_data_filename)
temporal_random = np.load(data_folder+random_temporal_data_filename)
temporal=np.concatenate((temporal_trial,temporal_random),axis=0)
temporal = np.transpose(temporal,[1,0,2])
# denoised_temporal_name = np.reshape(temporal,[-1,42*601])
denoised_temporal_name = np.reshape(temporal,[-1,temporal.shape[1]*temporal.shape[2]])
print('loaded data\n',flush=True)
atlas = np.load('/home/cat/code/widefieldPredict/locanmf/atlas_fixed_pixel.npy')#['atlas'].astype(float)
#areanames = sio.loadmat(data_folder+atlas_filename)['areanames']
# atlas.shape
fig = plt.figure()
plt.imshow(atlas)
# fig=plt.figure(figsize=(20,15))
# for it in np.unique(atlas):
# plotmap=np.zeros((atlas.shape)); plotmap.fill(np.nan); plotmap[atlas==it]=atlas[atlas==it]
# # plt.subplot(5,6,it+1)
# plt.imshow(plotmap,cmap='Spectral'); plt.axis('off'); plt.title('Allen region map'); plt.show();
# plt.show()
print(denoised_temporal_name.shape)
print(denoised_spatial_name.shape)
print(atlas.shape)
fig = plt.figure()
plt.plot(denoised_temporal_name[:,:601].T); plt.show()
fig=plt.figure()
for i in np.arange(7):
plt.imshow(denoised_spatial_name[:,:,i]); plt.show()
# Get data in the correct format
V=denoised_temporal_name
U=denoised_spatial_name
#
brainmask = np.ones(U.shape[:2],dtype=bool)
# Load true areas if simulated data
simulation=0
# Include nan values of U in brainmask, and put those values to 0 in U
brainmask[np.isnan(np.sum(U,axis=2))]=False
U[np.isnan(U)]=0
# Preprocess V: flatten and remove nans
dimsV=V.shape
keepinds=np.nonzero(np.sum(np.isfinite(V),axis=0))[0]
V=V[:,keepinds]
# del arrays
# U.shape
# Check that data has the correct shapes. V [K_d x T], U [X x Y x K_d], brainmask [X x Y]
if V.shape[0]!=U.shape[-1]:
print('Wrong dimensions of U and V!')
print("Rank of video : %d" % V.shape[0]); print("Number of timepoints : %d" % V.shape[1]);
# Plot the maximum of U
# plotmap=np.zeros((atlas.shape)); plotmap.fill(np.nan); plotmap[brainmask]=atlas[brainmask]
# fig=plt.figure()
# plt.imshow(plotmap,cmap='Spectral'); plt.axis('off'); plt.title('Allen region map'); plt.show();
# fig=plt.figure()
# plt.imshow(np.max(U,axis=2)); plt.axis('off'); plt.title('Max U'); plt.show()
# Perform the LQ decomposition. Time everything.
t0_global = time.time()
t0 = time.time()
if nonnegative_temporal:
r = V.T
else:
q, r = np.linalg.qr(V.T)
time_ests={'qr_decomp':time.time() - t0}
###Output
_____no_output_____
###Markdown
Initialize LocaNMF
###Code
# Put in data structure for LocaNMF
video_mats = (np.copy(U[brainmask]), r.T)
rank_range = (1, maxrank, 1)
del U
# region_mats[0] = [unique regions x pixels] the mask of each region
# region_mats[1] = [unique regions x pixels] the distance penalty of each region
# region_mats[2] = [unique regions] area code
region_mats = LocaNMF.extract_region_metadata(brainmask, atlas, min_size=min_pixels)
region_metadata = LocaNMF.RegionMetadata(region_mats[0].shape[0],
region_mats[0].shape[1:],
device=device)
region_metadata.set(torch.from_numpy(region_mats[0].astype(np.uint8)),
torch.from_numpy(region_mats[1]),
torch.from_numpy(region_mats[2].astype(np.int64)))
# print (region_mats[1].shape)
# print (region_mats[2])
# print (region_mats[2].shape)
# grab region names
rois=np.load('/home/cat/code/widefieldPredict/locanmf/rois_50.npz')
rois_name=rois['names']
# rois_name
rois_ids=rois['ids']
# rois_ids
# Do SVD as initialization
if device=='cuda':
torch.cuda.synchronize()
#
print('v SVD Initialization')
t0 = time.time()
region_videos = LocaNMF.factor_region_videos(video_mats,
region_mats[0],
rank_range[1],
device=device)
#
if device=='cuda':
torch.cuda.synchronize()
print("\'-total : %f" % (time.time() - t0))
time_ests['svd_init'] = time.time() - t0
#
low_rank_video = LocaNMF.LowRankVideo(
(int(np.sum(brainmask)),) + video_mats[1].shape, device=device
)
low_rank_video.set(torch.from_numpy(video_mats[0].T),
torch.from_numpy(video_mats[1]))
###Output
_____no_output_____
###Markdown
LocaNMF
###Code
#
if device=='cuda':
torch.cuda.synchronize()
#
print('v Rank Line Search')
t0 = time.time()
#locanmf_comps,loc_save
(nmf_factors,
loc_save,
save_lam,
save_scale,
save_per,
save_spa,
save_scratch) = LocaNMF.rank_linesearch(low_rank_video,
region_metadata,
region_videos,
maxiter_rank=maxrank,
maxiter_lambda=maxiter_lambda, # main param to tweak
maxiter_hals=maxiter_hals,
lambda_step=lambda_step,
lambda_init=lambda_init,
loc_thresh=loc_thresh,
r2_thresh=r2_thresh,
rank_range=rank_range,
# nnt=nonnegative_temporal,
verbose=[True, False, False],
sample_prop=(1,1),
device=device
)
#
if device=='cuda':
torch.cuda.synchronize()
#
print("\'-total : %f" % (time.time() - t0))
time_ests['rank_linesearch'] = time.time() - t0
# print (loc_save.scale.data)
# print (loc_save.spatial.data.shape)
# print (loc_save.spatial.scratch.shape)
# how much of spatial components is in each region
ratio=torch.norm(loc_save.spatial.scratch,p=2,dim=-1)/torch.norm(loc_save.spatial.data,p=2,dim=-1)
print ("ratio: ", ratio)
# how much of the spatial component is inside the ROI
per=100*(ratio**2)
print ('percentage; ', per)
#
print ("The threshold is 75%. all copmonents should be above, otherwise increase lambda iterations ")
# #
# print("Number of components : %d" % len(locanmf_comps))
# computing the variance in each component;
print ("Compute the variance explained by each component ")
mov = torch.matmul(low_rank_video.spatial.data.t(),
low_rank_video.temporal.data)
#
var = torch.mean(torch.var(mov, dim=1, unbiased=False)) # TODO: Precompute this
var_ests=np.zeros((len(nmf_factors)))
for i in np.arange(len(nmf_factors)):
mov = torch.matmul(torch.index_select(nmf_factors.spatial.data,0,torch.tensor([i])).t(),
torch.index_select(nmf_factors.temporal.data,0,torch.tensor([i])))
var_i = torch.mean(torch.var(mov, dim=1, unbiased=False))
# mean(var(dataest))/mean(var(data))
var_ests[i] = var_i.item() / var.item()
# to return to this; should sum to 1, perhaps
plt.plot(var_ests);
plt.show()
#
print (np.argsort(var_ests))
print (var_ests[np.argsort(var_ests)])
print (var_ests.sum())
print ("TODO: verify that locaNMF svm decoding is similar to pca")
# compute the r-squared again
mov = torch.matmul(low_rank_video.spatial.data.t(),low_rank_video.temporal.data)
var = torch.mean(torch.var(mov, dim=1, unbiased=False)) # TODO: Precompute this
torch.addmm(beta=1,
input=mov,
alpha=-1,
mat1=nmf_factors.spatial.data.t(),
mat2=nmf_factors.temporal.data,
out=mov)
r2_est = 1 - (torch.mean(mov.pow_(2)).item() / var.item())
print(r2_est);
print ("TODO: Save rsquared components")
# # for each component what is the rsquared - OPTIONAL - for checks
# r2_ests=np.zeros((len(locanmf_comps)))
# for i in np.arange(len(locanmf_comps)):
# mov = torch.matmul(low_rank_video.spatial.data.t(),low_rank_video.temporal.data)
# var = torch.mean(torch.var(mov, dim=1, unbiased=False)) # TODO: Precompute this
# # mov = data-dataest = data-mat1*mat2
# torch.addmm(beta=1,
# input=mov,
# alpha=-1,
# mat1=torch.index_select(locanmf_comps.spatial.data,
# 0,
# torch.tensor([i])).t(),
# mat2=torch.index_select(locanmf_comps.temporal.data,
# 0,
# torch.tensor([i])),
# out=mov)
# # r2_ests = 1-mean(var(data-dataest))/mean(var(data))
# r2_ests[i] = 1 - (torch.mean(mov.pow_(2)).item() / var.item())
# # for each component what is the rsquared - OPTIONAL - for checks
# mov = torch.matmul(low_rank_video.spatial.data.t(),low_rank_video.temporal.data)
# var = torch.mean(torch.var(mov, dim=1, unbiased=False)) # TODO: Precompute this
# r2_ests_2=np.zeros((len(locanmf_comps)))
# for i in np.arange(len(locanmf_comps)):
# mov_i = torch.matmul(torch.index_select(locanmf_comps.spatial.data,0,torch.tensor([i])).t(),
# torch.index_select(locanmf_comps.temporal.data,0,torch.tensor([i])))
# var_i = torch.mean(torch.var(mov-mov_i, dim=1, unbiased=False))
# # 1 - mean(var(data-dataest))/mean(var(data))
# r2_ests_2[i] = 1 - (var_i.item() / var.item())
# r2_ests_loo=np.zeros((len(locanmf_comps)))
# for i in np.arange(len(locanmf_comps)):
# mov = torch.matmul(low_rank_video.spatial.data.t(),low_rank_video.temporal.data)
# var = torch.mean(torch.var(mov, dim=1, unbiased=False)) # TODO: Precompute this
# torch.addmm(beta=1,
# input=mov,
# alpha=-1,
# mat1=torch.index_select(locanmf_comps.spatial.data,
# 0,
# torch.tensor(np.concatenate((np.arange(0,i),np.arange(i+1,len(locanmf_comps)))))).t(),
# mat2=torch.index_select(locanmf_comps.temporal.data,
# 0,
# torch.tensor(np.concatenate((np.arange(0,i),np.arange(i+1,len(locanmf_comps)))))),
# out=mov)
# r2_ests_loo[i] = 1 - (torch.mean(mov.pow_(2)).item() / var.item())
# _,r2_fit=LocaNMF.evaluate_fit_to_region(low_rank_video,
# locanmf_comps,
# region_metadata.support.data.sum(0),
# sample_prop=(1, 1))
# Evaluate R^2
_,r2_fit=LocaNMF.evaluate_fit_to_region(low_rank_video,
nmf_factors,
region_metadata.support.data.sum(0),
sample_prop=(1, 1)
)
#
print("R^2 fit on all data : %f" % r2_fit)
time_ests['global_time'] = time.time()-t0_global
# C is the temporal components
C = np.matmul(q,nmf_factors.temporal.data.cpu().numpy().T).T
print ("n_comps, n_time pts x n_trials: ", C.shape)
qc, rc = np.linalg.qr(C.T)
# back to visualizing variance
print(np.sum(var_ests))
plt.bar(np.arange(len(nmf_factors)),var_ests); plt.show()
#
var_est=np.zeros((len(nmf_factors)))
for i in np.arange(len(nmf_factors)):
var_est[i]=np.var(C[i,:])/np.var(C)
# locanmf_comps.regions.data
###Output
_____no_output_____
###Markdown
Reformat spatial and temporal matrices, and save
###Code
# Assigning regions to components
region_ranks = []; region_idx = []
locanmf_comps = nmf_factors
for rdx in torch.unique(locanmf_comps.regions.data, sorted=True):
region_ranks.append(torch.sum(rdx == locanmf_comps.regions.data).item())
region_idx.append(rdx.item())
areas=region_metadata.labels.data[locanmf_comps.regions.data].cpu().numpy()
# Get LocaNMF spatial and temporal components
A=locanmf_comps.spatial.data.cpu().numpy().T
A_reshape=np.zeros((brainmask.shape[0],brainmask.shape[1],A.shape[1]));
A_reshape.fill(np.nan)
A_reshape[brainmask,:]=A
# C is already computed above delete above
if nonnegative_temporal:
C=locanmf_comps.temporal.data.cpu().numpy()
else:
C=np.matmul(q,locanmf_comps.temporal.data.cpu().numpy().T).T
# Add back removed columns from C as nans
C_reshape=np.full((C.shape[0],dimsV[1]),np.nan)
C_reshape[:,keepinds]=C
C_reshape=np.reshape(C_reshape,[C.shape[0],dimsV[1]])
# Get lambdas
lambdas=np.squeeze(locanmf_comps.lambdas.data.cpu().numpy())
# A_reshape.shape
# c_p is the trial sturcutre
c_p=C_reshape.reshape(A_reshape.shape[2],int(C_reshape.shape[1]/1801),1801)
#
c_plot=c_p.transpose((1,0,2))
c_plot.shape
# save LocaNMF data
data_folder = root_dir+mouse_name+'/'+session_name+'/'
areas_saved = []
for area in areas:
idx = np.where(rois_ids==np.abs(area))[0]
temp_name = str(rois_name[idx].squeeze())
if area <0:
temp_name += " - right"
else:
temp_name += " - left"
areas_saved.append(temp_name)
np.savez(os.path.join(root_dir,mouse_name,session_name,'locanmf_trial.npz'),
temporal = c_plot[:int(c_plot.shape[0]/2),:,:],
areas = areas,
names = areas_saved)
np.savez(os.path.join(root_dir,mouse_name,session_name,'locanmf_random.npz'),
temporal = c_plot[int(c_plot.shape[0]/2):,:,:],
areas = areas,
names = areas_saved)
d = np.load('/media/cat/4TBSSD/yuki/yongxu/data/IJ1/Mar3/locanmf_trial.npz')
temporal = d['temporal']
areas = d['areas']
names = d['names']
print (temporal.shape)
print (areas)
print (names)
# from scipy.signal import savgol_filter
# t = np.arange(temporal.shape[2])/30 - 30
# max_vals = []
# for k in range(temporal.shape[1]):
# temp = temporal[:,k].mean(0)
# #plt.plot(temp)
# temp2 = savgol_filter(temp, 15, 2)
# plt.subplot(121)
# plt.xlim(-15,0)
# plt.plot(t,temp2)
# m = np.max(temp2[:temp2.shape[0]//2])
# max_vals.append(m)
# plt.subplot(122)
# plt.xlim(-15,0)
# temp2 = temp2/np.max(temp2)
# plt.plot(t,temp2)
# #break
# plt.show()
# max_vals = np.array(max_vals)
# args = np.argsort(max_vals)[::-1]
# print (max_vals[args])
# print (names[args])
# temp componetns of behavior + control data vstacked
t_plot=temporal.transpose((1,0,2))
print (temporal.shape)
# Plot the distribution of lambdas. OPTIONAL
# If lots of values close to the minimum, decrease lambda_init.
# If lots of values close to the maximum, increase maxiter_lambda or lambda_step.
plt.hist(locanmf_comps.lambdas.data.cpu(),
bins=torch.unique(locanmf_comps.lambdas.data).shape[0])
plt.show()
print(locanmf_comps.lambdas.data.cpu())
region_name=region_mats[2]
region_name.shape
region_name
def parse_areanames_new(region_name,rois_name):
areainds=[]; areanames=[];
for i,area in enumerate(region_name):
areainds.append(area)
areanames.append(rois_name[np.where(rois_ids==np.abs(area))][0])
sortvec=np.argsort(np.abs(areainds))
areanames=[areanames[i] for i in sortvec]
areainds=[areainds[i] for i in sortvec]
return areainds,areanames
#
region_name=region_mats[2]
# Get area names for all components
areainds,areanames_all = parse_areanames_new(region_name,rois_name)
areanames_area=[]
for i,area in enumerate(areas):
areanames_area.append(areanames_all[areainds.index(area)])
# # Get area names for all components
areainds,areanames_all =parse_areanames_new(region_name,rois_name)
areanames_area=[]
for i,area in enumerate(areas):
areanames_area.append(areanames_all[areainds.index(area)])
# Save results! - USE .NPZ File
print("LocaNMF completed successfully in "+ str(time.time()-t0_global) + "\n")
print("Results saved in "+data_folder+'locanmf_decomp_loc'+str(loc_thresh)+'.mat')
# Prefer to save c_p which is already converted
sio.savemat(data_folder+'locanmf_decomp_loc'+str(loc_thresh)+'.mat',
{'C':C_reshape,
'A':A_reshape,
'lambdas':lambdas,
'areas':areas,
'r2_fit':r2_fit,
'time_ests':time_ests,
'areanames':areanames_area
})
torch.cuda.empty_cache()
atlas_split=atlas
itt=0
fig=plt.figure() #figsize=(10,10))
b_=[]
for it in np.unique(atlas_split):
if np.abs(it) !=0:
plotmap=np.zeros((atlas_split.shape)); plotmap.fill(np.nan); plotmap[atlas_split==it]=atlas_split[atlas_split==it]
plt.subplot(5,8,itt+1)
plt.imshow(plotmap,cmap='Spectral'); plt.axis('off');
plt.title(rois_name[np.where(rois_ids==np.abs(it))][0],fontsize=6);
b_.append(plotmap)
# plt.show()
itt=itt+1
plt.tight_layout(h_pad=0.5,w_pad=0.5)
# how much of the spatial component is inside the ROI
# per
######################################################
##### PLACE TO LOOK FOR large spatial components #####
######################################################
fig=plt.figure()
for i in range(A_reshape.shape[2]):
plt.subplot(5,8,i+1)
plt.imshow(A_reshape[:,:,i])
plt.title(areanames_area[i],fontsize=6)
plt.tight_layout(h_pad=0.5,w_pad=0.5)
plt.show()
# calculate ROI data
roi_spatial=np.zeros((A_reshape.shape[2],denoised_spatial_name.shape[2]))
for i in range(denoised_spatial_name.shape[2]):
for j in range(A_reshape.shape[2]):
A_masking=np.zeros((A_reshape[:,:,j].shape))
A_masking[A_reshape[:,:,j]!=0]=1
A_multiply=A_masking*denoised_spatial_name[:,:,i]
roi_spatial[j,i]=np.sum(A_multiply)/np.sum(A_masking)
roi_data=[]
for s in range(temporal_trial.shape[0]):
roi_each=roi_spatial@temporal_trial[s]
roi_data.append(roi_each)
roi_save_trial=np.array(roi_data)
roi_save_trial.shape
# roi_spatial=np.zeros((A_reshape.shape[2],denoised_spatial_name.shape[2]))
# for i in range(denoised_spatial_name.shape[2]):
# for j in range(A_reshape.shape[2]):
# A_masking=np.zeros((A_reshape[:,:,j].shape))
# A_masking[A_reshape[:,:,j]!=0]=1
# A_multiply=A_masking*denoised_spatial_name[:,:,i]
# roi_spatial[j,i]=np.sum(A_multiply)/np.sum(A_masking)
# roi_data=[]
# for s in range(temporal_random.shape[0]):
# roi_each=roi_spatial@temporal_random[s]
# roi_data.append(roi_each)
# roi_save_random=np.array(roi_data)
# roi_save_random.shape
# save ROI data
# np.save(save_folder+save_name+'trial.npy',roi_save_trial)
# np.save(save_folder+save_name+'random.npy',roi_save_random)
###Output
_____no_output_____
###Markdown
Visualization of components
###Code
# CAT: Use actual averages not random data
C_area_rois = c_plot[:int(c_plot.shape[0]/2),:,:].mean(0)
print (C_area_rois.shape)
t = np.arange(C_area_rois.shape[1])/30.-30
clrs_local = ['magenta','brown','pink','lightblue','darkblue']
fig = plt.figure(figsize=(12,16))
############### SHREYA"S CODE ####################
# Spatial and Temporal Components: Summary
atlascolor=np.zeros((atlas.shape[0],atlas.shape[1],4))
A_color=np.zeros((A_reshape.shape[0],A_reshape.shape[1],4))
cmap=plt.cm.get_cmap('jet')
colors=cmap(np.arange(len(areainds))/len(areainds))
#
for i,area_i in enumerate(areainds):
if area_i not in areas:
continue
atlascolor[atlas==area_i,:]=colors[i,:]
C_area=C[np.where(areas==area_i)[0],:]
#
for z in range(len(names_plot)):
if names_plot[z] in names_plot[z]:
clr = clrs_local[z]
break
#
for j in np.arange(colors.shape[1]):
A_color[:,:,j]=A_color[:,:,j]+colors[i,j]*A_reshape[:,:,np.where(areas==area_i)[0][0]]
#fig=plt.figure(figsize=(15,8))
ax1=fig.add_subplot(2,2,1)
ax1.imshow(atlascolor)
#ax1.set_title('Atlas Regions')
ax1.axis('off')
ax2=fig.add_subplot(2,2,3)
ax2.imshow(A_color)
#ax2.set_title('Spatial Components (One per region)')
ax2.axis('off')
ax3=fig.add_subplot(1,2,2)
axvar=0
print (areas)
names_plot = ['Retrosplenial','barrel','limb','visual','motor']
for i,area_i in enumerate(areainds):
if area_i not in areas:
continue
# C_area=C[np.where(areas==area_i)[0][0],:min(1000,C.shape[1])]
C_area=C_area_rois[np.where(areas==area_i)[0][0]]
#ax3.plot(1.5*axvar+C_area/np.nanmax(np.abs(C_area)),color=colors[i,:])
for z in range(len(names_plot)):
if names_plot[z] in names_plot[z]:
clr = clrs_local[z]
break
ax3.plot(t, 1.5*axvar+C_area*23,
#color=colors[i,:],
color = clr,
linewidth=3)
print (C_area.shape)
axvar+=1
ax3.set_xlim(-15,0)
#ax3.set_title('Temporal Components (One per region)')
ax3.axis('off')
if True:
plt.savefig('/home/cat/locanmf.svg',dpi=300)
plt.close()
else:
plt.show()
data1 = np.load('/media/cat/4TBSSD/yuki/AQ2/tif_files/AQ2am_Dec17_30Hz/AQ2am_Dec17_30Hz_locanmf.npz',
allow_pickle=True)
names_rois = data1['names']
print (names_rois)
ids = data1['areas']
print (areas)
# Plotting all the regions' components
for i,area in enumerate(areas):
try:
fig=plt.figure(figsize=(20,4))
ax1 = fig.add_subplot(1,3,1)
plotmap_area = np.zeros((atlas.shape));
plotmap_area.fill(np.nan);
plotmap_area[brainmask] = atlas[brainmask]==area
ax1.imshow(plotmap_area);
ax1.set_title('Atlas '+areanames_area[i])
ax1.axis('off')
ax2 = fig.add_subplot(1,3,2)
ax2.imshow(A_reshape[:,:,i])
ax2.set_title('LocaNMF A [%s]'%(i+1))
ax2.axis('off')
ax3 = fig.add_subplot(1,3,3)
ax3.plot(C[i,:min(1000,C.shape[1])],'k')
if simulation:
ax3.plot(V[np.where(area==trueareas)[0][0],:min(1000,V.shape[1])],'r');
if i==0: ax3.legend(('LocaNMF','True'))
ax3.set_title('LocaNMF C [%s]'%(i+1))
ax3.axis('off')
plt.show()
except:
pass
# Calculate Canonical Correlations between components in each pair of regions
corrmat=np.zeros((len(areainds),len(areainds)))
skipinds=[]
for i,area_i in enumerate(areainds):
for j,area_j in enumerate(areainds):
if i==0 and area_j not in areas:
skipinds.append(j)
C_i=C[np.where(areas==area_i)[0],:].T
C_j=C[np.where(areas==area_j)[0],:].T
if i not in skipinds and j not in skipinds:
cca=CCA(n_components=1)
cca.fit(C_i,C_j)
C_i_cca,C_j_cca=cca.transform(C_i,C_j)
try: C_i_cca=C_i_cca[:,0]
except: pass
try: C_j_cca=C_j_cca[:,0]
except: pass
corrmat[i,j]=np.corrcoef(C_i_cca,C_j_cca)[0,1]
corrmat=np.delete(corrmat,skipinds,axis=0);
corrmat=np.delete(corrmat,skipinds,axis=1);
corr_areanames=np.delete(areanames_all,skipinds)
# Plot correlations
fig=plt.figure()
plt.imshow(corrmat,cmap=plt.cm.get_cmap('jet')); plt.clim(-1,1); plt.colorbar(shrink=0.8)
plt.get_cmap('jet')
plt.xticks(ticks=np.arange(len(areainds)-len(skipinds)),labels=corr_areanames,rotation=90);
plt.yticks(ticks=np.arange(len(areainds)-len(skipinds)),labels=corr_areanames);
plt.title('CCA between all regions',fontsize=12)
plt.xlabel('Region i',fontsize=12)
plt.ylabel('Region j',fontsize=12)
plt.show()
# Save visualized components and correlations
# print('Saving postprocessing results!')
# postprocess.plot_components(A_reshape,C,areas,atlas,areanames,data_folder)
# postprocess.plot_correlations(A_reshape,C,areas,atlas,areanames,data_folder)
import numpy as np
data = np.load('/media/cat/4TBSSD/yuki/IA1/tif_files/IA1pm_Feb2_30Hz/IA1pm_Feb2_30Hz_locanmf.npz',
allow_pickle=True)
trials = data['temporal_trial']
random = data['temporal_random']
print (trials.shape, random.shape)
trials = np.load('/media/cat/4TBSSD/yuki/IA1/tif_files/IA1am_Mar4_30Hz/IA1am_Mar4_30Hz_code_04_trial_ROItimeCourses_30sec_pca_0.95.npy')
print (trials.shape)
data = np.load('/media/cat/4TBSSD/yuki/IA1/tif_files/IA1am_Mar4_30Hz/IA1am_Mar4_30Hz_whole_stack_trial_ROItimeCourses_15sec_pca30components.npy')
print (data.shape)
spatial = np.load('/media/cat/4TBSSD/yuki/IA1/tif_files/IA1am_Mar4_30Hz/IA1am_Mar4_30Hz_whole_stack_trial_ROItimeCourses_15sec_pca30components_spatial.npy')
print (spatial.shape)
###Output
(40000, 30)
(30, 16384)
|
Chapter_4/Chapter_4-5.ipynb | ###Markdown
Chapter4: 実践的なアプリケーションを作ってみよう 4.5 テキストエディッタを作ってみよう①
###Code
# リスト4.5.1: サブウインドウの作成例1
# tkinterのインポート
import tkinter as tk
### GUI ###
# ウインドウの作成
root = tk.Tk()
# サブウインドウの作成
mini_window = tk.Toplevel(root)
# ウインドウ状態の維持
root.mainloop()
# リスト4.5.2: サブウインドウの作成例2
# tkinterのインポート
import tkinter as tk
### 関数 ###
def sub_window():
# サブウインドウの作成
mini_window = tk.Toplevel(root)
# ボタン2
button2 = tk.Button(mini_window, text = "Push2")
button2.pack(padx = 50, pady = 25)
### GUI ###
# ウインドウの作成
root = tk.Tk()
# ボタン1
button1 = tk.Button(root, text = "Push1", command = sub_window)
button1.pack(padx = 100, pady = 50)
# ウインドウ状態の維持
root.mainloop()
###Output
_____no_output_____
###Markdown
Chapter4: 実践的なアプリケーションを作ってみよう 4.5 テキストエディッタを作ってみよう①
###Code
# リスト4.5.1: サブウインドウの作成例1
# tkinterのインポート
import tkinter as tk
### GUI ###
# ウインドウの作成
root = tk.Tk()
# サブウインドウの作成
mini_window = tk.Toplevel(root)
# ウインドウ状態の維持
root.mainloop()
# リスト4.5.2: サブウインドウの作成例2
# tkinterのインポート
import tkinter as tk
### 関数 ###
def sub_window():
# サブウインドウの作成
mini_window = tk.Toplevel(root)
# ボタン2
button2 = tk.Button(mini_window, text = "Push2")
button2.pack(padx = 50, pady = 25)
### GUI ###
# ウインドウの作成
root = tk.Tk()
# ボタン1
button1 = tk.Button(root, text = "Push1", command = sub_window)
button1.pack(padx = 100, pady = 50)
# ウインドウ状態の維持
root.mainloop()
###Output
_____no_output_____ |
python/example/dictionary-sentiment/sentiment.ipynb | ###Markdown
Rule-based Sentiment AnalysisIn the following example, we walk-through a simple use case for our straight forward SentimentDetector annotator.This annotator will work on top of a list of labeled sentences which can have any of the following features positive negative revert increment decrementEach of these sentences will be used for giving a score to text Spark `2.4` and Spark NLP `2.0.1` 1. Call necessary imports and set the resource path to read local data files
###Code
#Imports
import sys
sys.path.append('../../')
from pyspark.sql import SparkSession
from pyspark.ml import Pipeline
from pyspark.sql.functions import array_contains
from sparknlp.annotator import *
from sparknlp.common import RegexRule
from sparknlp.base import DocumentAssembler, Finisher
#Setting location of resource Directory
resource_path= "../../../src/test/resources/"
###Output
_____no_output_____
###Markdown
2. Load SparkSession if not already there
###Code
spark = SparkSession.builder \
.appName("SentimentDetector")\
.master("local[*]")\
.config("spark.driver.memory","8G")\
.config("spark.driver.maxResultSize", "2G")\
.config("spark.jars.packages", "JohnSnowLabs:spark-nlp:2.0.1")\
.config("spark.kryoserializer.buffer.max", "500m")\
.getOrCreate()
data = spark. \
read. \
parquet(resource_path+"sentiment.parquet"). \
limit(10000).cache()
data.show()
###Output
_____no_output_____
###Markdown
3. Create appropriate annotators. We are using Sentence Detection, Tokenizing the sentences, and find the lemmas of those tokens. The Finisher will only output the Sentiment.
###Code
document_assembler = DocumentAssembler() \
.setInputCol("text")
sentence_detector = SentenceDetector() \
.setInputCols(["document"]) \
.setOutputCol("sentence")
tokenizer = Tokenizer() \
.setInputCols(["sentence"]) \
.setOutputCol("token")
lemmatizer = Lemmatizer() \
.setInputCols(["token"]) \
.setOutputCol("lemma") \
.setDictionary(resource_path+"lemma-corpus-small/lemmas_small.txt", key_delimiter="->", value_delimiter="\t")
sentiment_detector = SentimentDetector() \
.setInputCols(["lemma", "sentence"]) \
.setOutputCol("sentiment_score") \
.setDictionary(resource_path+"sentiment-corpus/default-sentiment-dict.txt", ",")
finisher = Finisher() \
.setInputCols(["sentiment_score"]) \
.setOutputCols(["sentiment"])
###Output
_____no_output_____
###Markdown
4. Train the pipeline, which is only being trained from external resources, not from the dataset we pass on. The prediction runs on the target dataset
###Code
pipeline = Pipeline(stages=[document_assembler, sentence_detector, tokenizer, lemmatizer, sentiment_detector, finisher])
model = pipeline.fit(data)
result = model.transform(data)
###Output
_____no_output_____
###Markdown
5. filter the finisher output, to find the positive sentiment lines
###Code
result.where(array_contains(result.sentiment, "positive")).show(10,False)
###Output
_____no_output_____
###Markdown
Necessary imports
###Code
#Imports
import sys
sys.path.append('../../')
from pyspark.sql import SparkSession
from pyspark.ml import Pipeline
from sparknlp.annotator import *
from sparknlp.common import RegexRule
from sparknlp.base import DocumentAssembler, Finisher
###Output
_____no_output_____
###Markdown
Create a spark dataset
###Code
data = spark. \
read. \
parquet("../../../src/test/resources/sentiment.parquet"). \
limit(10000)
data.cache()
data.count()
###Output
_____no_output_____
###Markdown
Create appropriate annotators. We are using Sentence Detection, Tokenizing the sentences, and find the lemmas of those tokensThe Finisher will only output the Sentiment.
###Code
document_assembler = DocumentAssembler() \
.setInputCol("text")
sentence_detector = SentenceDetector() \
.setInputCols(["document"]) \
.setOutputCol("sentence")
tokenizer = Tokenizer() \
.setInputCols(["sentence"]) \
.setOutputCol("token")
lemmatizer = Lemmatizer() \
.setInputCols(["token"]) \
.setOutputCol("lemma") \
.setDictionary("../../../src/test/resources/lemma-corpus-small/lemmas_small.txt", key_delimiter="->", value_delimiter="\t")
sentiment_detector = SentimentDetector() \
.setInputCols(["lemma", "sentence"]) \
.setOutputCol("sentiment_score") \
.setDictionary("../../../src/test/resources/sentiment-corpus/default-sentiment-dict.txt", ",")
finisher = Finisher() \
.setInputCols(["sentiment_score"]) \
.setOutputCols(["sentiment"])
###Output
_____no_output_____
###Markdown
Train the pipeline, which is only being trained from external resources, not from the dataset we pass on.The prediction runs on the target dataset
###Code
pipeline = Pipeline(stages=[document_assembler, sentence_detector, tokenizer, lemmatizer, sentiment_detector, finisher])
model = pipeline.fit(data)
result = model.transform(data)
###Output
_____no_output_____
###Markdown
We filter the finisher output, to find the positive sentiment lines
###Code
result.filter("sentiment != 'positive'").show()
###Output
_____no_output_____ |
python/08_keras_functional_api.ipynb | ###Markdown
Utilizando a API funcional do keras para criar uma rede neural MLPNesse notebook é utilizado o dataset [Concrete Compressive Strength Data Set](https://archive.ics.uci.edu/ml/datasets/concrete+compressive+strength), onde as features são:* Aglomerante: Cement (kg in a m3 mixture)* Escória de Alto Forno: Blast Furnace Slag (kg in a m3 mixture)* Cinzas volantes: Fly Ash (kg in a m3 mixture)* Água: Water (kg in a m3 mixture)* Superplastificante: Superplasticizer (kg in a m3 mixture)* Agregado graúdo: Coarse Aggregate (kg in a m3 mixture)* Agregado fino: Fine Aggregate (kg in a m3 mixture)* Idade: Age (Day (1~365))O objetivo é com essas features obter a resistência à compressão do concreto, em inglês: Concrete compressive strength (MPa).Primeiro são importadas as bibliotecas.
###Code
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from keras.models import Model
from keras.layers import Input
from keras.layers import Dense
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import cross_val_score
import numpy as np
import pandas as pd
np.random.seed(0)
tf.random.set_seed(0)
###Output
_____no_output_____
###Markdown
O dataset é aberto usando o pandas. O formato original é em xls, então também é especificada a planilha aser aberta, a primeira. As features são renomeadas para facilitar as referências.São definidos os dataframes com features e variável de saída.
###Code
url = "https://archive.ics.uci.edu/ml/machine-learning-databases/concrete/compressive/Concrete_Data.xls"
df = pd.read_excel(url, sheet_name=0)
df.columns = ['cement', 'slag', 'ash', 'water', 'superplasticizer', 'coarse_aggregate', 'fine_aggregate', 'age', 'compressive_strength']
X = (df.drop(['compressive_strength'], axis=1))
y = (df['compressive_strength'])
###Output
_____no_output_____
###Markdown
O dataset é dividido em treino e teste.
###Code
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
from sklearn.preprocessing import StandardScaler
scl = StandardScaler()
X_train = scl.fit_transform(X_train)
X_test = scl.transform(X_test)
###Output
_____no_output_____
###Markdown
São definidas duas variáveis para a média e o desvio padrão da variável alvo.
###Code
mean = y_train.mean()
std = y_train.std()
###Output
_____no_output_____
###Markdown
Uma função para ciração da rede é definida utilizando a API funcional do Keras. Isso permite com que o resultado final da rede seja multiplicado pelo desvio padrão e somado à média. Dessa forma os pesos são ajustados à variàvel objetivo padronizada.
###Code
def create_model():
x = Input(shape=(X_train.shape[1],))
hidden1 = Dense(20, activation='relu')(x)
hidden2 = Dense(100, activation='relu')(hidden1)
hidden3 = Dense(20, activation='relu')(hidden2)
hidden4 = Dense(1)(hidden3)
output = hidden4 * std + mean
model = Model(inputs=x, outputs=output)
opt = keras.optimizers.Adam(learning_rate=1e-3)
model.compile(loss="mean_absolute_error", optimizer=opt)
return model
###Output
_____no_output_____
###Markdown
O modelo é criado e as camadas são mostradas.
###Code
model = create_model()
model.summary()
###Output
Model: "model"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) [(None, 8)] 0
_________________________________________________________________
dense (Dense) (None, 20) 180
_________________________________________________________________
dense_1 (Dense) (None, 100) 2100
_________________________________________________________________
dense_2 (Dense) (None, 20) 2020
_________________________________________________________________
dense_3 (Dense) (None, 1) 21
_________________________________________________________________
tf.math.multiply (TFOpLambda (None, 1) 0
_________________________________________________________________
tf.__operators__.add (TFOpLa (None, 1) 0
=================================================================
Total params: 4,321
Trainable params: 4,321
Non-trainable params: 0
_________________________________________________________________
###Markdown
Um wraper é criado para utilizar a função de validação cruzado do Scikit-learn.
###Code
from keras.wrappers.scikit_learn import KerasRegressor
estimator = KerasRegressor(build_fn=create_model, epochs=300, batch_size=300, verbose=0)
###Output
_____no_output_____
###Markdown
O score da validação cruzada é calculado.
###Code
print(-cross_val_score(estimator, X_train, y_train, cv=5, scoring='neg_mean_absolute_error').mean())
###Output
WARNING:tensorflow:5 out of the last 5 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7f5d4b7fc710> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
4.075838752319069
###Markdown
O score da validação cruzada no conjunto de treino é de 4.08 MPa. Por fim, o modelo é treinado utilizando todo o conjunto de treino.
###Code
model.fit(x=X_train, y=y_train.values, epochs=300, batch_size=300)
###Output
Epoch 1/300
3/3 [==============================] - 1s 8ms/step - loss: 13.0678
Epoch 2/300
3/3 [==============================] - 0s 6ms/step - loss: 12.3589
Epoch 3/300
3/3 [==============================] - 0s 3ms/step - loss: 11.7695
Epoch 4/300
3/3 [==============================] - 0s 4ms/step - loss: 11.4504
Epoch 5/300
3/3 [==============================] - 0s 3ms/step - loss: 10.9317
Epoch 6/300
3/3 [==============================] - 0s 5ms/step - loss: 10.5465
Epoch 7/300
3/3 [==============================] - 0s 5ms/step - loss: 10.2404
Epoch 8/300
3/3 [==============================] - 0s 5ms/step - loss: 9.9319
Epoch 9/300
3/3 [==============================] - 0s 4ms/step - loss: 9.5963
Epoch 10/300
3/3 [==============================] - 0s 4ms/step - loss: 9.3915
Epoch 11/300
3/3 [==============================] - 0s 5ms/step - loss: 9.4176
Epoch 12/300
3/3 [==============================] - 0s 4ms/step - loss: 9.1122
Epoch 13/300
3/3 [==============================] - 0s 5ms/step - loss: 8.9046
Epoch 14/300
3/3 [==============================] - 0s 4ms/step - loss: 8.8182
Epoch 15/300
3/3 [==============================] - 0s 7ms/step - loss: 8.8467
Epoch 16/300
3/3 [==============================] - 0s 5ms/step - loss: 8.5716
Epoch 17/300
3/3 [==============================] - 0s 4ms/step - loss: 8.1219
Epoch 18/300
3/3 [==============================] - 0s 6ms/step - loss: 7.9176
Epoch 19/300
3/3 [==============================] - 0s 4ms/step - loss: 7.9593
Epoch 20/300
3/3 [==============================] - 0s 8ms/step - loss: 7.7760
Epoch 21/300
3/3 [==============================] - 0s 5ms/step - loss: 7.5013
Epoch 22/300
3/3 [==============================] - 0s 6ms/step - loss: 7.2442
Epoch 23/300
3/3 [==============================] - 0s 3ms/step - loss: 7.1579
Epoch 24/300
3/3 [==============================] - 0s 7ms/step - loss: 6.9899
Epoch 25/300
3/3 [==============================] - 0s 6ms/step - loss: 6.9342
Epoch 26/300
3/3 [==============================] - 0s 5ms/step - loss: 6.6572
Epoch 27/300
3/3 [==============================] - 0s 5ms/step - loss: 6.5209
Epoch 28/300
3/3 [==============================] - 0s 6ms/step - loss: 6.5546
Epoch 29/300
3/3 [==============================] - 0s 5ms/step - loss: 6.3050
Epoch 30/300
3/3 [==============================] - 0s 4ms/step - loss: 6.2221
Epoch 31/300
3/3 [==============================] - 0s 5ms/step - loss: 6.0136
Epoch 32/300
3/3 [==============================] - 0s 3ms/step - loss: 5.8539
Epoch 33/300
3/3 [==============================] - 0s 3ms/step - loss: 5.7161
Epoch 34/300
3/3 [==============================] - 0s 3ms/step - loss: 5.6015
Epoch 35/300
3/3 [==============================] - 0s 4ms/step - loss: 5.4892
Epoch 36/300
3/3 [==============================] - 0s 4ms/step - loss: 5.5415
Epoch 37/300
3/3 [==============================] - 0s 4ms/step - loss: 5.5744
Epoch 38/300
3/3 [==============================] - 0s 4ms/step - loss: 5.4764
Epoch 39/300
3/3 [==============================] - 0s 4ms/step - loss: 5.2958
Epoch 40/300
3/3 [==============================] - 0s 8ms/step - loss: 5.2382
Epoch 41/300
3/3 [==============================] - 0s 5ms/step - loss: 5.1268
Epoch 42/300
3/3 [==============================] - 0s 6ms/step - loss: 5.0339
Epoch 43/300
3/3 [==============================] - 0s 4ms/step - loss: 5.1394
Epoch 44/300
3/3 [==============================] - 0s 5ms/step - loss: 4.8920
Epoch 45/300
3/3 [==============================] - 0s 4ms/step - loss: 4.7908
Epoch 46/300
3/3 [==============================] - 0s 4ms/step - loss: 4.7065
Epoch 47/300
3/3 [==============================] - 0s 6ms/step - loss: 4.7412
Epoch 48/300
3/3 [==============================] - 0s 5ms/step - loss: 4.6897
Epoch 49/300
3/3 [==============================] - 0s 5ms/step - loss: 4.7332
Epoch 50/300
3/3 [==============================] - 0s 5ms/step - loss: 4.6011
Epoch 51/300
3/3 [==============================] - 0s 5ms/step - loss: 4.5975
Epoch 52/300
3/3 [==============================] - 0s 5ms/step - loss: 4.5720
Epoch 53/300
3/3 [==============================] - 0s 4ms/step - loss: 4.3383
Epoch 54/300
3/3 [==============================] - 0s 4ms/step - loss: 4.4053
Epoch 55/300
3/3 [==============================] - 0s 4ms/step - loss: 4.2993
Epoch 56/300
3/3 [==============================] - 0s 4ms/step - loss: 4.2008
Epoch 57/300
3/3 [==============================] - 0s 4ms/step - loss: 4.0486
Epoch 58/300
3/3 [==============================] - 0s 4ms/step - loss: 4.1533
Epoch 59/300
3/3 [==============================] - 0s 4ms/step - loss: 4.1466
Epoch 60/300
3/3 [==============================] - 0s 4ms/step - loss: 4.0242
Epoch 61/300
3/3 [==============================] - 0s 6ms/step - loss: 4.0413
Epoch 62/300
3/3 [==============================] - 0s 4ms/step - loss: 4.0967
Epoch 63/300
3/3 [==============================] - 0s 11ms/step - loss: 3.9375
Epoch 64/300
3/3 [==============================] - 0s 5ms/step - loss: 3.8908
Epoch 65/300
3/3 [==============================] - 0s 5ms/step - loss: 3.8731
Epoch 66/300
3/3 [==============================] - 0s 4ms/step - loss: 3.9600
Epoch 67/300
3/3 [==============================] - 0s 5ms/step - loss: 3.8554
Epoch 68/300
3/3 [==============================] - 0s 5ms/step - loss: 3.9097
Epoch 69/300
3/3 [==============================] - 0s 7ms/step - loss: 3.8020
Epoch 70/300
3/3 [==============================] - 0s 5ms/step - loss: 3.7046
Epoch 71/300
3/3 [==============================] - 0s 4ms/step - loss: 3.6340
Epoch 72/300
3/3 [==============================] - 0s 4ms/step - loss: 3.6736
Epoch 73/300
3/3 [==============================] - 0s 5ms/step - loss: 3.6278
Epoch 74/300
3/3 [==============================] - 0s 4ms/step - loss: 3.5951
Epoch 75/300
3/3 [==============================] - 0s 5ms/step - loss: 3.5178
Epoch 76/300
3/3 [==============================] - 0s 4ms/step - loss: 3.6078
Epoch 77/300
3/3 [==============================] - 0s 7ms/step - loss: 3.4314
Epoch 78/300
3/3 [==============================] - 0s 5ms/step - loss: 3.5365
Epoch 79/300
3/3 [==============================] - 0s 7ms/step - loss: 3.4496
Epoch 80/300
3/3 [==============================] - 0s 3ms/step - loss: 3.4851
Epoch 81/300
3/3 [==============================] - 0s 3ms/step - loss: 3.5547
Epoch 82/300
3/3 [==============================] - 0s 4ms/step - loss: 3.4007
Epoch 83/300
3/3 [==============================] - 0s 4ms/step - loss: 3.3138
Epoch 84/300
3/3 [==============================] - 0s 3ms/step - loss: 3.4823
Epoch 85/300
3/3 [==============================] - 0s 4ms/step - loss: 3.4060
Epoch 86/300
3/3 [==============================] - 0s 5ms/step - loss: 3.2788
Epoch 87/300
3/3 [==============================] - 0s 4ms/step - loss: 3.3946
Epoch 88/300
3/3 [==============================] - 0s 9ms/step - loss: 3.1992
Epoch 89/300
3/3 [==============================] - 0s 4ms/step - loss: 3.3129
Epoch 90/300
3/3 [==============================] - 0s 3ms/step - loss: 3.1984
Epoch 91/300
3/3 [==============================] - 0s 5ms/step - loss: 3.1731
Epoch 92/300
3/3 [==============================] - 0s 4ms/step - loss: 3.1287
Epoch 93/300
3/3 [==============================] - 0s 6ms/step - loss: 3.2374
Epoch 94/300
3/3 [==============================] - 0s 6ms/step - loss: 3.1677
Epoch 95/300
3/3 [==============================] - 0s 3ms/step - loss: 3.1069
Epoch 96/300
3/3 [==============================] - 0s 6ms/step - loss: 3.3899
Epoch 97/300
3/3 [==============================] - 0s 3ms/step - loss: 3.2320
Epoch 98/300
3/3 [==============================] - 0s 8ms/step - loss: 3.1388
Epoch 99/300
3/3 [==============================] - 0s 6ms/step - loss: 3.0814
Epoch 100/300
3/3 [==============================] - 0s 8ms/step - loss: 3.1803
Epoch 101/300
3/3 [==============================] - 0s 6ms/step - loss: 3.1019
Epoch 102/300
3/3 [==============================] - 0s 6ms/step - loss: 3.0957
Epoch 103/300
3/3 [==============================] - 0s 4ms/step - loss: 3.0721
Epoch 104/300
3/3 [==============================] - 0s 4ms/step - loss: 3.0754
Epoch 105/300
3/3 [==============================] - 0s 5ms/step - loss: 2.9981
Epoch 106/300
3/3 [==============================] - 0s 5ms/step - loss: 2.9634
Epoch 107/300
3/3 [==============================] - 0s 5ms/step - loss: 2.8686
Epoch 108/300
3/3 [==============================] - 0s 6ms/step - loss: 2.8122
Epoch 109/300
3/3 [==============================] - 0s 4ms/step - loss: 2.8525
Epoch 110/300
3/3 [==============================] - 0s 4ms/step - loss: 2.9299
Epoch 111/300
3/3 [==============================] - 0s 10ms/step - loss: 2.7503
Epoch 112/300
3/3 [==============================] - 0s 6ms/step - loss: 2.8197
Epoch 113/300
3/3 [==============================] - 0s 7ms/step - loss: 2.8883
Epoch 114/300
3/3 [==============================] - 0s 7ms/step - loss: 2.9060
Epoch 115/300
3/3 [==============================] - 0s 4ms/step - loss: 2.9429
Epoch 116/300
3/3 [==============================] - 0s 5ms/step - loss: 2.8969
Epoch 117/300
3/3 [==============================] - 0s 6ms/step - loss: 2.8524
Epoch 118/300
3/3 [==============================] - 0s 5ms/step - loss: 2.8659
Epoch 119/300
3/3 [==============================] - 0s 9ms/step - loss: 2.7554
Epoch 120/300
3/3 [==============================] - 0s 7ms/step - loss: 2.7084
Epoch 121/300
3/3 [==============================] - 0s 5ms/step - loss: 2.7586
Epoch 122/300
3/3 [==============================] - 0s 7ms/step - loss: 2.8650
Epoch 123/300
3/3 [==============================] - 0s 5ms/step - loss: 2.8447
Epoch 124/300
3/3 [==============================] - 0s 5ms/step - loss: 2.7594
Epoch 125/300
3/3 [==============================] - 0s 5ms/step - loss: 2.7502
Epoch 126/300
3/3 [==============================] - 0s 3ms/step - loss: 2.6893
Epoch 127/300
3/3 [==============================] - 0s 5ms/step - loss: 2.6585
Epoch 128/300
3/3 [==============================] - 0s 4ms/step - loss: 2.7050
Epoch 129/300
3/3 [==============================] - 0s 9ms/step - loss: 2.7045
Epoch 130/300
3/3 [==============================] - 0s 5ms/step - loss: 2.6622
Epoch 131/300
3/3 [==============================] - 0s 4ms/step - loss: 2.6670
Epoch 132/300
3/3 [==============================] - 0s 5ms/step - loss: 2.6024
Epoch 133/300
3/3 [==============================] - 0s 5ms/step - loss: 2.5943
Epoch 134/300
3/3 [==============================] - 0s 6ms/step - loss: 2.6538
Epoch 135/300
3/3 [==============================] - 0s 5ms/step - loss: 2.6637
Epoch 136/300
3/3 [==============================] - 0s 5ms/step - loss: 2.6373
Epoch 137/300
3/3 [==============================] - 0s 4ms/step - loss: 2.6858
Epoch 138/300
3/3 [==============================] - 0s 4ms/step - loss: 2.5966
Epoch 139/300
3/3 [==============================] - 0s 6ms/step - loss: 2.6709
Epoch 140/300
3/3 [==============================] - 0s 5ms/step - loss: 2.6183
Epoch 141/300
3/3 [==============================] - 0s 6ms/step - loss: 2.6100
Epoch 142/300
3/3 [==============================] - 0s 6ms/step - loss: 2.5553
Epoch 143/300
3/3 [==============================] - 0s 5ms/step - loss: 2.5414
Epoch 144/300
3/3 [==============================] - 0s 9ms/step - loss: 2.5682
Epoch 145/300
3/3 [==============================] - 0s 6ms/step - loss: 2.5174
Epoch 146/300
3/3 [==============================] - 0s 6ms/step - loss: 2.5529
Epoch 147/300
3/3 [==============================] - 0s 4ms/step - loss: 2.5442
Epoch 148/300
3/3 [==============================] - 0s 4ms/step - loss: 2.4639
Epoch 149/300
3/3 [==============================] - 0s 5ms/step - loss: 2.4506
Epoch 150/300
3/3 [==============================] - 0s 8ms/step - loss: 2.4646
Epoch 151/300
3/3 [==============================] - 0s 4ms/step - loss: 2.4134
Epoch 152/300
3/3 [==============================] - 0s 5ms/step - loss: 2.4788
Epoch 153/300
3/3 [==============================] - 0s 5ms/step - loss: 2.5309
Epoch 154/300
3/3 [==============================] - 0s 5ms/step - loss: 2.4614
Epoch 155/300
3/3 [==============================] - 0s 5ms/step - loss: 2.4905
Epoch 156/300
3/3 [==============================] - 0s 6ms/step - loss: 2.3991
Epoch 157/300
3/3 [==============================] - 0s 5ms/step - loss: 2.4559
Epoch 158/300
3/3 [==============================] - 0s 8ms/step - loss: 2.4049
Epoch 159/300
3/3 [==============================] - 0s 4ms/step - loss: 2.3743
Epoch 160/300
3/3 [==============================] - 0s 5ms/step - loss: 2.4227
Epoch 161/300
3/3 [==============================] - 0s 7ms/step - loss: 2.4768
Epoch 162/300
3/3 [==============================] - 0s 5ms/step - loss: 2.4126
Epoch 163/300
3/3 [==============================] - 0s 6ms/step - loss: 2.4278
Epoch 164/300
3/3 [==============================] - 0s 5ms/step - loss: 2.5466
Epoch 165/300
3/3 [==============================] - 0s 7ms/step - loss: 2.4127
Epoch 166/300
3/3 [==============================] - 0s 5ms/step - loss: 2.3923
Epoch 167/300
3/3 [==============================] - 0s 6ms/step - loss: 2.3465
Epoch 168/300
3/3 [==============================] - 0s 5ms/step - loss: 2.3937
Epoch 169/300
3/3 [==============================] - 0s 6ms/step - loss: 2.3169
Epoch 170/300
3/3 [==============================] - 0s 5ms/step - loss: 2.3969
Epoch 171/300
3/3 [==============================] - 0s 9ms/step - loss: 2.4319
Epoch 172/300
3/3 [==============================] - 0s 6ms/step - loss: 2.4004
Epoch 173/300
3/3 [==============================] - 0s 5ms/step - loss: 2.3470
Epoch 174/300
3/3 [==============================] - 0s 8ms/step - loss: 2.3041
Epoch 175/300
3/3 [==============================] - 0s 6ms/step - loss: 2.3253
Epoch 176/300
3/3 [==============================] - 0s 5ms/step - loss: 2.3492
Epoch 177/300
3/3 [==============================] - 0s 5ms/step - loss: 2.3213
Epoch 178/300
3/3 [==============================] - 0s 4ms/step - loss: 2.3783
Epoch 179/300
3/3 [==============================] - 0s 6ms/step - loss: 2.3139
Epoch 180/300
3/3 [==============================] - 0s 6ms/step - loss: 2.3099
Epoch 181/300
3/3 [==============================] - 0s 5ms/step - loss: 2.3086
Epoch 182/300
3/3 [==============================] - 0s 5ms/step - loss: 2.2976
Epoch 183/300
3/3 [==============================] - 0s 6ms/step - loss: 2.2649
Epoch 184/300
3/3 [==============================] - 0s 5ms/step - loss: 2.1898
Epoch 185/300
3/3 [==============================] - 0s 6ms/step - loss: 2.2692
Epoch 186/300
3/3 [==============================] - 0s 3ms/step - loss: 2.2780
Epoch 187/300
3/3 [==============================] - 0s 6ms/step - loss: 2.3409
Epoch 188/300
3/3 [==============================] - 0s 4ms/step - loss: 2.2484
Epoch 189/300
3/3 [==============================] - 0s 4ms/step - loss: 2.2649
Epoch 190/300
3/3 [==============================] - 0s 5ms/step - loss: 2.3110
Epoch 191/300
3/3 [==============================] - 0s 7ms/step - loss: 2.3460
Epoch 192/300
3/3 [==============================] - 0s 8ms/step - loss: 2.3201
Epoch 193/300
3/3 [==============================] - 0s 6ms/step - loss: 2.2863
Epoch 194/300
3/3 [==============================] - 0s 6ms/step - loss: 2.2222
Epoch 195/300
3/3 [==============================] - 0s 7ms/step - loss: 2.2940
Epoch 196/300
3/3 [==============================] - 0s 5ms/step - loss: 2.2871
Epoch 197/300
3/3 [==============================] - 0s 5ms/step - loss: 2.2805
Epoch 198/300
3/3 [==============================] - 0s 6ms/step - loss: 2.3587
Epoch 199/300
3/3 [==============================] - 0s 6ms/step - loss: 2.1920
Epoch 200/300
3/3 [==============================] - 0s 4ms/step - loss: 2.3573
Epoch 201/300
3/3 [==============================] - 0s 4ms/step - loss: 2.3062
Epoch 202/300
3/3 [==============================] - 0s 6ms/step - loss: 2.3282
Epoch 203/300
3/3 [==============================] - 0s 7ms/step - loss: 2.2466
Epoch 204/300
3/3 [==============================] - 0s 7ms/step - loss: 2.2842
Epoch 205/300
3/3 [==============================] - 0s 8ms/step - loss: 2.2419
Epoch 206/300
3/3 [==============================] - 0s 11ms/step - loss: 2.2574
Epoch 207/300
3/3 [==============================] - 0s 6ms/step - loss: 2.1632
Epoch 208/300
3/3 [==============================] - 0s 9ms/step - loss: 2.1888
Epoch 209/300
3/3 [==============================] - 0s 5ms/step - loss: 2.1692
Epoch 210/300
3/3 [==============================] - 0s 5ms/step - loss: 2.1626
Epoch 211/300
3/3 [==============================] - 0s 5ms/step - loss: 2.1501
Epoch 212/300
3/3 [==============================] - 0s 4ms/step - loss: 2.1647
Epoch 213/300
3/3 [==============================] - 0s 4ms/step - loss: 2.1637
Epoch 214/300
3/3 [==============================] - 0s 5ms/step - loss: 2.1203
Epoch 215/300
3/3 [==============================] - 0s 4ms/step - loss: 2.1552
Epoch 216/300
3/3 [==============================] - 0s 8ms/step - loss: 2.0658
Epoch 217/300
3/3 [==============================] - 0s 5ms/step - loss: 2.1176
Epoch 218/300
3/3 [==============================] - 0s 7ms/step - loss: 2.1204
Epoch 219/300
3/3 [==============================] - 0s 5ms/step - loss: 2.2285
Epoch 220/300
3/3 [==============================] - 0s 4ms/step - loss: 2.1378
Epoch 221/300
3/3 [==============================] - 0s 6ms/step - loss: 2.0813
Epoch 222/300
3/3 [==============================] - 0s 11ms/step - loss: 2.1350
Epoch 223/300
3/3 [==============================] - 0s 7ms/step - loss: 2.0436
Epoch 224/300
3/3 [==============================] - 0s 7ms/step - loss: 2.1569
Epoch 225/300
3/3 [==============================] - 0s 7ms/step - loss: 2.0411
Epoch 226/300
3/3 [==============================] - 0s 5ms/step - loss: 2.0955
Epoch 227/300
3/3 [==============================] - 0s 5ms/step - loss: 2.1530
Epoch 228/300
3/3 [==============================] - 0s 4ms/step - loss: 2.1007
Epoch 229/300
3/3 [==============================] - 0s 6ms/step - loss: 2.0824
Epoch 230/300
3/3 [==============================] - 0s 6ms/step - loss: 2.1998
Epoch 231/300
3/3 [==============================] - 0s 4ms/step - loss: 2.1093
Epoch 232/300
3/3 [==============================] - 0s 4ms/step - loss: 2.1621
Epoch 233/300
3/3 [==============================] - 0s 4ms/step - loss: 1.9906
Epoch 234/300
3/3 [==============================] - 0s 6ms/step - loss: 2.0186
Epoch 235/300
3/3 [==============================] - 0s 4ms/step - loss: 2.0769
Epoch 236/300
3/3 [==============================] - 0s 7ms/step - loss: 1.9972
Epoch 237/300
3/3 [==============================] - 0s 5ms/step - loss: 2.1243
Epoch 238/300
3/3 [==============================] - 0s 5ms/step - loss: 2.0725
Epoch 239/300
3/3 [==============================] - 0s 3ms/step - loss: 2.0143
Epoch 240/300
3/3 [==============================] - 0s 4ms/step - loss: 2.2119
Epoch 241/300
3/3 [==============================] - 0s 6ms/step - loss: 2.0125
Epoch 242/300
3/3 [==============================] - 0s 7ms/step - loss: 2.0594
Epoch 243/300
3/3 [==============================] - 0s 5ms/step - loss: 2.0224
Epoch 244/300
3/3 [==============================] - 0s 6ms/step - loss: 2.0041
Epoch 245/300
3/3 [==============================] - 0s 6ms/step - loss: 1.9769
Epoch 246/300
3/3 [==============================] - 0s 7ms/step - loss: 1.9534
Epoch 247/300
3/3 [==============================] - 0s 7ms/step - loss: 1.9638
Epoch 248/300
3/3 [==============================] - 0s 7ms/step - loss: 2.1759
Epoch 249/300
3/3 [==============================] - 0s 5ms/step - loss: 2.0925
Epoch 250/300
3/3 [==============================] - 0s 3ms/step - loss: 2.1773
Epoch 251/300
3/3 [==============================] - 0s 4ms/step - loss: 2.0704
Epoch 252/300
3/3 [==============================] - 0s 6ms/step - loss: 2.0607
Epoch 253/300
3/3 [==============================] - 0s 4ms/step - loss: 2.0396
Epoch 254/300
3/3 [==============================] - 0s 6ms/step - loss: 1.9907
Epoch 255/300
3/3 [==============================] - 0s 4ms/step - loss: 1.9852
Epoch 256/300
3/3 [==============================] - 0s 4ms/step - loss: 2.0055
Epoch 257/300
3/3 [==============================] - 0s 5ms/step - loss: 1.9374
Epoch 258/300
3/3 [==============================] - 0s 6ms/step - loss: 2.0446
Epoch 259/300
3/3 [==============================] - 0s 3ms/step - loss: 1.9537
Epoch 260/300
3/3 [==============================] - 0s 5ms/step - loss: 2.0097
Epoch 261/300
3/3 [==============================] - 0s 4ms/step - loss: 2.0104
Epoch 262/300
3/3 [==============================] - 0s 4ms/step - loss: 2.0684
Epoch 263/300
3/3 [==============================] - 0s 4ms/step - loss: 2.0035
Epoch 264/300
3/3 [==============================] - 0s 5ms/step - loss: 1.9062
Epoch 265/300
3/3 [==============================] - 0s 4ms/step - loss: 2.0256
Epoch 266/300
3/3 [==============================] - 0s 6ms/step - loss: 1.9907
Epoch 267/300
3/3 [==============================] - 0s 6ms/step - loss: 1.9669
Epoch 268/300
3/3 [==============================] - 0s 6ms/step - loss: 1.9967
Epoch 269/300
3/3 [==============================] - 0s 5ms/step - loss: 1.8749
Epoch 270/300
3/3 [==============================] - 0s 5ms/step - loss: 1.8846
Epoch 271/300
3/3 [==============================] - 0s 3ms/step - loss: 1.9608
Epoch 272/300
3/3 [==============================] - 0s 5ms/step - loss: 1.8159
Epoch 273/300
3/3 [==============================] - 0s 5ms/step - loss: 1.8787
Epoch 274/300
3/3 [==============================] - 0s 11ms/step - loss: 1.8671
Epoch 275/300
3/3 [==============================] - 0s 6ms/step - loss: 1.9226
Epoch 276/300
3/3 [==============================] - 0s 6ms/step - loss: 2.0416
Epoch 277/300
3/3 [==============================] - 0s 6ms/step - loss: 1.9772
Epoch 278/300
3/3 [==============================] - 0s 5ms/step - loss: 1.9869
Epoch 279/300
3/3 [==============================] - 0s 6ms/step - loss: 1.9468
Epoch 280/300
3/3 [==============================] - 0s 8ms/step - loss: 1.9844
Epoch 281/300
3/3 [==============================] - 0s 5ms/step - loss: 1.9181
Epoch 282/300
3/3 [==============================] - 0s 5ms/step - loss: 1.9181
Epoch 283/300
3/3 [==============================] - 0s 5ms/step - loss: 2.0166
Epoch 284/300
3/3 [==============================] - 0s 4ms/step - loss: 1.9712
Epoch 285/300
3/3 [==============================] - 0s 5ms/step - loss: 1.9028
Epoch 286/300
3/3 [==============================] - 0s 5ms/step - loss: 1.9395
Epoch 287/300
3/3 [==============================] - 0s 4ms/step - loss: 1.9845
Epoch 288/300
3/3 [==============================] - 0s 5ms/step - loss: 1.8723
Epoch 289/300
3/3 [==============================] - 0s 3ms/step - loss: 1.8071
Epoch 290/300
3/3 [==============================] - 0s 4ms/step - loss: 1.8252
Epoch 291/300
3/3 [==============================] - 0s 7ms/step - loss: 1.8454
Epoch 292/300
3/3 [==============================] - 0s 8ms/step - loss: 1.9126
Epoch 293/300
3/3 [==============================] - 0s 6ms/step - loss: 1.8903
Epoch 294/300
3/3 [==============================] - 0s 6ms/step - loss: 1.9082
Epoch 295/300
3/3 [==============================] - 0s 6ms/step - loss: 1.8252
Epoch 296/300
3/3 [==============================] - 0s 6ms/step - loss: 1.8035
Epoch 297/300
3/3 [==============================] - 0s 6ms/step - loss: 1.8343
Epoch 298/300
3/3 [==============================] - 0s 5ms/step - loss: 1.8999
Epoch 299/300
3/3 [==============================] - 0s 4ms/step - loss: 1.9006
Epoch 300/300
3/3 [==============================] - 0s 6ms/step - loss: 1.8484
###Markdown
O erro médio absoluto final é então calculado.
###Code
mean_absolute_error(y_test, model(X_test))
###Output
_____no_output_____
###Markdown
O modelo prevê a resistência à compressão do concreto com um erro médio absoluto de 3.845 MPa para o conunto de teste.
###Code
###Output
_____no_output_____ |
statistics/nonparametric_methods/Distribution_Free_Methods.ipynb | ###Markdown
Creating Empirical Cumulative Distribution Function
###Code
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
plt.style.use('ggplot')
data = np.array([
5.9, 6.0, 6.4, 6.4, 6.5, 6.5, 6.6, 6.7, 6.9, 7.0, 7.1, 7.2, 7.5, 7.5, 7.8, 7.9, 8.1, 8.1, 8.2, 8.9, 9.3, 9.3,
9.6, 10.4, 10.6, 11.8, 11.8, 12.6, 12.9, 14.3, 15.0, 16.2, 16.3, 17.0, 17.2, 22.8, 23.1, 33.0, 40.0, 42.8,
43.0, 44.8, 45.0, 45.8
])
data
len(data)
###Output
_____no_output_____
###Markdown
The ECDF is generated with this function: $\large{\hat{F}(x) = \frac{\x_i \leq x}{n}}$ The above basically means for each value of x in your data set consisting of only unique values of x, the function is equal to the number of Xs that are less than or equal to the current X value. F(5.9) = 1/44 = 0.023F(6) = 2/44 = 0.045F(6.4) = 4/44 = 0.091...F(45.8) = 44/44 = 1.0
###Code
def ecdf(x):
"""Return empirical CDF of x."""
sx = np.sort(x)
cdf = (1.0 + np.arange(len(sx)))/len(sx)
return sx, cdf
sx, y = ecdf(data)
plt.step(sx, y)
plt.title("Empirical CDF", fontsize=14, weight='bold')
plt.show()
###Output
_____no_output_____
###Markdown
Creating 95% confidence band based on [Dvoretzky–Kiefer–Wolfowitz](http://stats.stackexchange.com/questions/55500/confidence-intervals-for-empirical-cdf) inequality: $\large{ecdf\pm\sqrt{\frac{1}{2n}log(\frac{2}{\alpha})}}$
###Code
alpha = 0.05
nobs = 44
interval = np.sqrt(np.log(2./alpha) / (2 * nobs))
print("Confidence band width: " + str(interval))
lower = np.clip(y - interval, 0, 1)
upper = np.clip(y + interval, 0, 1)
plt.step(sx, y, sx, upper, sx, lower)
plt.fill_between(sx, y, upper, color='grey', alpha=0.5, step='pre')
plt.fill_between(sx, y, lower, color='grey',alpha=0.5, step='pre')
plt.title("Emprical CDF with 95% Confidence Interval", fontsize=12, weight='bold')
plt.show()
###Output
Confidence band width: 0.204741507042
###Markdown
Found out that the [statsmodels](http://statsmodels.sourceforge.net/devel/_modules/statsmodels/distributions/empirical_distribution.html) package has ECDF function and function for making confidence interval.
###Code
%matplotlib inline
import numpy as np
from statsmodels.distributions import ECDF
from statsmodels.distributions.empirical_distribution import _conf_set
import matplotlib.pyplot as plt
data = np.array([
5.9, 6.0, 6.4, 6.4, 6.5, 6.5, 6.6, 6.7, 6.9, 7.0, 7.1, 7.2, 7.5, 7.5, 7.8, 7.9, 8.1, 8.1, 8.2, 8.9, 9.3, 9.3,
9.6, 10.4, 10.6, 11.8, 11.8, 12.6, 12.9, 14.3, 15.0, 16.2, 16.3, 17.0, 17.2, 22.8, 23.1, 33.0, 40.0, 42.8,
43.0, 44.8, 45.0, 45.8
])
cdf = ECDF(data)
y = cdf.y
x = cdf.x
plt.step(x, y)
lower, upper = _conf_set(y, alpha=0.05)
plt.step(x, lower)
plt.step(x, upper)
plt.fill_between(x, y, upper, color='grey', alpha=0.5, interpolate=False, step='pre')
plt.fill_between(x, y, lower, color='grey',alpha=0.5, interpolate=False, step='pre')
plt.vlines(x, 0, .05) # show location of steps
plt.title("Empirical CDF with 95% Confidence Interval", fontsize=12, weight='bold')
plt.show()
###Output
_____no_output_____ |
4-Condition.ipynb | ###Markdown
4. Conditional Statements, If...Else Conditions are used a lot in programming. Sometimes, requirements could be broken down into some if-else statements. Let us try running some if-else statements.
###Code
#compare strings, try replacing the name.
name = "tom"
if name == "tom":
print("You are Tom.")
else:
print("You are not Tom.")
###Output
_____no_output_____
###Markdown
You could compare numbers too. The following conditions check if a number is larger than 10 or smaller than 10, and also if the number is 10.
###Code
a = 10
if a > 10:
print("a is larger than 10")
elif a < 10:
print("a is lesser than 10")
else:
print("a is 10")
###Output
_____no_output_____
###Markdown
We can also have multiple conditions such as the following, where we want to find a number larger than 10 and is an even number.
###Code
a = 2
if a >= 10 or a % 2 == 0:
print('a is either larger than or equals to 10 or it is an even number.')
else:
print('a is smaller than 10 and is not even.')
###Output
_____no_output_____
###Markdown
Try it yourselfInstead of comparing with a fixed number, you can compare both variables. Try rewriting the above and replace 10 with a variable named "b".
###Code
# Type in your code below
###Output
_____no_output_____ |
InitialDataCleaning02.ipynb | ###Markdown
Initial Data CleaningThis script is designed to fill in nulls in important missing fields and to format and select the data which will be required to create a model later on. Import Statements
###Code
import geopandas as gpd
import folium
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import sqlite3
from shapely.geometry import Point
###Output
_____no_output_____
###Markdown
Filling in NullsTo fill in nulls in this process we need some background on the data itself and how I intend to use it. First off, the dataset of interest contains over 2 million rows - each of which pertains to a specific wildfire. For each fire there is a Latitude and Longitude point for where the fire originated. Also, in most rows there are county and state code identifiers, however, in many rows (approx. 600K) there are no county identifiers. The lack of those county identifiers is an issue since we intend to analyze and predict wildfires on a county level. To address the issue of the missing county identifiers we take the following actions:1. From an online source, get a data set which has Geofences (Polygons) for all the counties in the U.S. 2. From the U.S. Census Bureau get a correlation of all the counties and states with their correct code 3. Merge the county polygons to the county/state codes based on the county name and state abbreviation4. In the wildfire data create spatial Points out of the longitude and latitude columns5. Execute a Spatial merge between the wildfire Points and the county Polygons based on whether the Point is in the Polygon6. Extract the county/state codes from the merged dataframe and then subset to the desired columns for further cleaning Read in the .shp file which has all of the Polygons for the U.S. counties. Subset to just the three needed columns: county name, country/state/county abbreviation (which we extract state abbreviation from), and the Polygon for the county
###Code
geodf = gpd.read_file("./Data/CountyGeoFences/gadm36_USA_2.shp")[["NAME_2", "HASC_2", 'geometry']]
geodf.head(1)
###Output
_____no_output_____
###Markdown
Create a new GeoDataFrame which reformats the county and state into the correct format to join on later
###Code
new_df = gpd.GeoDataFrame()
new_df['COUNTY'] = geodf['NAME_2'].str.upper().str.strip()
new_df['STATE'] = geodf['HASC_2'].str.slice(3,5).str.strip()
new_df['geometry'] = geodf['geometry']
new_df.head(1)
###Output
_____no_output_____
###Markdown
Read in the Census data which correlates the county to the county/state code. In this data the *whole* states and the whole *US* are included, but are formatted so that their names are in all uppercase - whereas individual counties are in title case. So we filter out those rows which are already in upper case - as they are the whole states and whole US - which we are not interested in.
###Code
lnd_data = pd.read_csv("./Data/CountyLandArea.csv")[['Areaname', 'STCOU']]
lnd_data = lnd_data.loc[lnd_data['Areaname'] != lnd_data['Areaname'].str.upper()]
lnd_data.head(1)
###Output
_____no_output_____
###Markdown
Now format the data by separating the county name and the state abbreviation and by turning the code into a zero-padded five digit long value. For reference the code is built in a way such that the first two number indicate the state and the last three numbers indicate the county.
###Code
new_lnd_data = pd.DataFrame()
new_lnd_data['COUNTY'] = lnd_data['Areaname'].str.split(',').apply(lambda x: x[0].upper()).str.strip()
new_lnd_data['STATE'] = lnd_data['Areaname'].str.split(',').apply(lambda x: x[1].upper()).str.strip()
new_lnd_data['FIPS_CODE'] = lnd_data['STCOU'].astype(str).str.zfill(5)
new_lnd_data.head(1)
###Output
_____no_output_____
###Markdown
Merge the county Polygon data with the Census data to get the county/state code with the Polygons. Then ensure that the object is a Spatial DataFrame with the correct geospatial reference.
###Code
comb = gpd.GeoDataFrame(pd.merge(new_lnd_data, new_df, on=['COUNTY', "STATE"], how='inner'), crs="EPSG:4326")
###Output
_____no_output_____
###Markdown
Set that dataset aside. Read in the wildfire data from the .sqlite table, selecting (in SQL) only the few columns which we want
###Code
cursor = sqlite3.connect('./Data/Fire/FPA_FOD_20210617.sqlite')
query = "SELECT f.FOD_ID, f.DISCOVERY_DATE, f.FIRE_SIZE, f.LATITUDE, f.LONGITUDE, f.FIPS_CODE from Fires f"
all_data = pd.read_sql_query(query, cursor)
all_data.head(1)
###Output
_____no_output_____
###Markdown
Take the wildfire data and turn it into a Spatial DataFrame and create the geometry columns to be the Lat/Long points and ensure that the Spatial DataFrame has the correct reference
###Code
geo_fire_df = gpd.GeoDataFrame(all_data, geometry=gpd.points_from_xy(all_data.LONGITUDE, all_data.LATITUDE), crs="EPSG:4326")
geo_fire_df.head(1)
###Output
_____no_output_____
###Markdown
Now take both DataFrames - with the points (wildfire data) and with the polygons (county/state data)We will execute a spatial join such that we are joining the rows from the county/state data to the wildfire data wherever the Point in the wildfire data is *within* the Polygon of county/state data.
###Code
merged = geo_fire_df.sjoin(comb, how='inner', predicate='within')
###Output
_____no_output_____
###Markdown
There may be some duplicates because of overlap in counties or some edge cases within the data. So we remove those duplicates and retain only the first instance of the row.
###Code
mgd = merged.drop_duplicates(subset=["DISCOVERY_DATE", "FIRE_SIZE", "FIPS_CODE_right"])
###Output
_____no_output_____
###Markdown
Formatting Now that we have the county/state codes filled in for all of our wildfire instances we can get to formatting the data. To format the data we will select just the few columns which we want and set the names correctly. Then we'll turn the discovery date into an actual date column and then finally calculate the discovery month of the fire - as we will be focused on monthly predictions of wildfires.
###Code
final_mgd = mgd[["DISCOVERY_DATE", "FIRE_SIZE", "FIPS_CODE_right"]].rename({'FIPS_CODE_right': "FIPS_CODE"}, axis=1)
final_mgd.loc[:, 'DISCOVERY_DATE'] = pd.to_datetime(final_mgd.loc[:, 'DISCOVERY_DATE'])
final_mgd['MONTH'] = final_mgd['DISCOVERY_DATE'].to_numpy().astype('datetime64[M]')
final_mgd.head(1)
final_mgd.shape
###Output
_____no_output_____
###Markdown
From this data we sum up all of the fire sizes for each month and county/state code (FIPS)
###Code
agg_data = final_mgd.groupby(by=['MONTH', 'FIPS_CODE']).agg({'FIRE_SIZE': sum}).reset_index()
agg_data.rename({'FIRE_SIZE': 'TOTAL_BURN_AREA'}, axis=1, inplace=True)
###Output
_____no_output_____
###Markdown
Now - we want to standardize the area burned by the size of the county itself. This way we can better assess the actual impact of the fires on the county since some small fires may be very impactful in certain counties (i.e. small ones) but large fires may not matter at all since it's really only a small portion of the county (i.e. in Alaska where the counties are larger than most states)To do this standardization we turn back to the census bureau and we can find the land size (in Sq. Miles) for all of the counties and then merge it to our aggregated data and then divide the Fire Size by the county size. We will convert the Sq. Miles into Acres for a better apples-apples comparison of total area to area burned. Also note that some fires may extend beyond one county - or start in one county and then blow directly into the neighnoring county - and while this may lead some of our numbers to be decieving, as a whole the concept is still strong. However, from a data perspective this does mean that total area burned may be greater than county area.
###Code
land_area_data = pd.read_csv("./Data/CountyLandArea.csv")[["STCOU", "LND010190D"]]
land_area_data.rename({'STCOU': "FIPS_CODE", "LND010190D": "COUNTY_AREA"}, axis=1, inplace=True)
land_area_data['FIPS_CODE'] = land_area_data["FIPS_CODE"].astype(str).str.zfill(5)
land_area_data['COUNTY_AREA'] = land_area_data['COUNTY_AREA'] * 640
land_area_data = land_area_data.iloc[np.where(land_area_data['COUNTY_AREA'] != 0)]
land_area_data.head(1)
###Output
_____no_output_____
###Markdown
Now merge the aggregated data with the land area data and divide to get the burned to toal area ratio
###Code
agg_and_area = pd.merge(agg_data, land_area_data, how='inner', on=['FIPS_CODE'])
agg_and_area.head(1)
###Output
_____no_output_____
###Markdown
We will keep the burn area and county data so that we can aggregate separately for state and national geographic areas. So output the data.
###Code
agg_and_area.to_csv("./Data/clean_fire_data.csv", index=False)
###Output
_____no_output_____
###Markdown
Our last step is to pull in some weather data, clean it and then join in based on the state for which the weather data pertains to. This data is kept separate for now but will be converted into a format where it can be easily joined into a state level aggregation of the wildfire data First read in the weather data csv file
###Code
files = ["California", "Arizona", "Colorado", "Montana", "Nevada", "Oregon", "Washington", "Idaho", "Utah", "NewMexico",
"Wyoming"]
data = []
for f in files:
df = pd.read_csv(f"./Data/Weather/{f}.csv", parse_dates=['DATE'])[["NAME", "DATE", "PRCP", "TMAX", "TMIN"]]
df["STATE"] = df['NAME'].str.split(',').apply(lambda x: x[1]).str.strip().str.split(' ').apply(lambda x: x[0]).str.strip()
df.drop('NAME', axis=1, inplace=True)
data.append(df)
weather_data = pd.concat(data, axis=0)
###Output
_____no_output_____
###Markdown
Now, we will average out the measurements which we have for multiple stats down into measurements on a state level
###Code
grped_weather = weather_data.groupby(by=["STATE", "DATE"]).mean().reset_index()
grped_weather.rename({'DATE': 'MONTH'}, axis=1, inplace=True)
###Output
_____no_output_____
###Markdown
Now, once again, read in the Census data to get state codes - there is a bit of logic to extract the state codes but not too bad since we can use work we have already done to format the file correctly. In the end we can see that we do indeed end up with codes for our 50 states
###Code
census_data = pd.read_csv("./Data/CountyLandArea.csv")[["Areaname", "STCOU"]]
census_data = census_data.loc[census_data['Areaname'] != census_data['Areaname'].str.upper()]
census_data['STATE'] = census_data['Areaname'].str.split(',').apply(lambda x: x[1].upper()).str.strip()
census_data['STATE_CODE'] = census_data['STCOU'].astype(str).str.zfill(5).str.slice(0,2)
census_data = census_data.drop_duplicates(subset=['STATE_CODE', 'STATE'])[["STATE", "STATE_CODE"]].reset_index(drop=True)
###Output
_____no_output_____
###Markdown
Now we will join the weather and census data together to get the state codes for our states
###Code
weather_w_codes = pd.merge(grped_weather, census_data, how='inner', on='STATE')
###Output
_____no_output_____
###Markdown
Now export the data to a csv file where we can pull it into our modelling efforts later on
###Code
weather_w_codes.to_csv("./Data/cleaned_weather_data.csv", index=False)
###Output
_____no_output_____ |
chapter_3/YELP_Dataset_full.ipynb | ###Markdown
Preprocessing Yelp Dataset (Full version)
###Code
import re
import collections
import numpy as np
import pandas as pd
from tqdm.notebook import tqdm
from argparse import Namespace
tqdm.pandas(desc="Preprocessing Reviews")
###Output
/opt/miniconda3/envs/notebook_env/lib/python3.7/site-packages/tqdm/std.py:668: FutureWarning: The Panel class is removed from pandas. Accessing it from the top-level namespace will also be removed in the next version
from pandas import Panel
###Markdown
Define configurations
###Code
args = Namespace(
raw_train_dataset_csv = "data/yelp/raw_train.csv",
raw_test_dataset_csv = "data/yelp/raw_test.csv",
output_munged_csv="data/yelp/reviews_with_splits_full.csv",
seed=1337,
)
###Output
_____no_output_____
###Markdown
Read Data
###Code
train_reviews = pd.read_csv(
args.raw_train_dataset_csv,
header = None,
names = ["rating", "review"]
)
train_reviews = train_reviews[~pd.isnull(train_reviews.review)]
test_reviews = pd.read_csv(
args.raw_test_dataset_csv,
header=None,
names=["rating", "review"]
)
test_reviews = test_reviews[~pd.isnull(test_reviews.review)]
train_reviews.head()
###Output
_____no_output_____
###Markdown
Split data into Train/Dev/Test
###Code
from sklearn.model_selection import train_test_split
train_reviews.rating.value_counts(), test_reviews.rating.value_counts()
train, val = train_test_split(train_reviews, train_size=0.7,
stratify=train_reviews.rating.values)
train = train.copy()
val = val.copy()
train.rating.value_counts() # Have the same distribution
val.rating.value_counts() # Have the same distribution
test_reviews.rating.value_counts() # Have the same distribution
train.reset_index(drop=True, inplace=True)
val.reset_index(drop=True, inplace=True)
test_reviews.reset_index(drop=True, inplace=True)
train["split"] = "train"
val["split"] = "val"
test_reviews["split"] = "test"
###Output
_____no_output_____
###Markdown
Final reviews
###Code
final_reviews = pd.concat([train, val, test_reviews], axis=0, copy=True)
final_reviews.reset_index(drop=True, inplace=True)
final_reviews.split.value_counts()
final_reviews.review.head()
final_reviews[(final_reviews.review.isnull())]
###Output
_____no_output_____
###Markdown
Preprocess Reviews
###Code
# Preprocess the reviews
def preprocess_text(text):
text = text.lower()
text = re.sub(r"([.,!?])", r" \1 ", text)
text = re.sub(r"[^a-zA-Z.,!?]+", r" ", text)
return text
final_reviews.review = final_reviews.review.progress_apply(
preprocess_text)
final_reviews['rating'] = final_reviews.rating.progress_apply(
{1: 'negative', 2: 'positive'}.get)
final_reviews.head()
final_reviews.to_csv(args.output_munged_csv, index=False)
###Output
_____no_output_____ |
Reasearch Code - Jupyter Notebooks/Step2-Feature-Engineering.ipynb | ###Markdown
Machine Learning Model Building Pipeline: Feature Engineering1. Data Analysis2. Feature Engineering3. Feature Selection4. Model Building**This is the notebook for step 2: Feature Engineering**The dataset used is the house price dataset available on [Kaggle.com](https://www.kaggle.com/c/house-prices-advanced-regression-techniques/data). See below for more details.=================================================================================================== Predicting Sale Price of HousesThe aim of the project is to build a machine learning model to predict the sale price of homes based on different explanatory variables describing aspects of residential houses. How do I download the dataset?To download the House Price dataset go this website:https://www.kaggle.com/c/house-prices-advanced-regression-techniques/dataDownload 'train.csv'. Rename the file as 'houseprice.csv' and save where you saved this jupyter notebook. House Prices dataset: Feature EngineeringIn the following cells, we will engineer / pre-process the variables of the House Price Dataset from Kaggle. We will engineer the variables so that we tackle:1. Missing values2. Temporal variables3. Non-Gaussian distributed variables4. Categorical variables: remove rare labels5. Categorical variables: convert strings to numbers5. Standarise the values of the variables to the same range Setting the seedIt is important to note that we are engineering variables and pre-processing data with the idea of deploying the model. Therefore, from now on, for each step that includes some element of randomness, it is extremely important that we **set the seed**. This way, we can obtain reproducibility between our research and our development code.**Always set the seeds**.
###Code
# to handle datasets
import pandas as pd
import numpy as np
# for plotting
import matplotlib.pyplot as plt
# to divide train and test set
from sklearn.model_selection import train_test_split
# feature scaling
from sklearn.preprocessing import MinMaxScaler
# to visualise al the columns in the dataframe
pd.pandas.set_option('display.max_columns', None)
import warnings
warnings.simplefilter(action='ignore')
# load dataset
data = pd.read_csv('houseprice.csv')
print(data.shape)
data.head()
###Output
(1460, 81)
###Markdown
Separate dataset into train and testBefore beginning to engineer our features, it is important to separate our data intro training and testing set. When we engineer features, some techniques learn parameters from data. It is important to learn this parameters only from the train set. This is to avoid over-fitting. **Separating the data into train and test involves randomness, therefore, we need to set the seed.**
###Code
# Let's separate into train and test set
# Remember to set the seed (random_state for this sklearn function)
X_train, X_test, y_train, y_test = train_test_split(data,
data['SalePrice'],
test_size=0.1,
# we are setting the seed here:
random_state=0)
X_train.shape, X_test.shape
###Output
_____no_output_____
###Markdown
Missing values Categorical variablesFor categorical variables, we will replace missing values with the string "missing".
###Code
# make a list of the categorical variables that contain missing values
vars_with_na = [
var for var in data.columns
if X_train[var].isnull().sum() > 0 and X_train[var].dtypes == 'O'
]
# print percentage of missing values per variable
X_train[vars_with_na].isnull().mean()
# replace missing values with new label: "Missing"
X_train[vars_with_na] = X_train[vars_with_na].fillna('Missing')
X_test[vars_with_na] = X_test[vars_with_na].fillna('Missing')
# check that we have no missing information in the engineered variables
X_train[vars_with_na].isnull().sum()
# check that test set does not contain null values in the engineered variables
[var for var in vars_with_na if X_test[var].isnull().sum() > 0]
###Output
_____no_output_____
###Markdown
Numerical variablesTo engineer missing values in numerical variables, we will:- add a binary missing value indicator variable- and then replace the missing values in the original variable with the mode
###Code
# make a list with the numerical variables that contain missing values
vars_with_na = [
var for var in data.columns
if X_train[var].isnull().sum() > 0 and X_train[var].dtypes != 'O'
]
# print percentage of missing values per variable
X_train[vars_with_na].isnull().mean()
# replace engineer missing values as we described above
for var in vars_with_na:
# calculate the mode using the train set
mode_val = X_train[var].mode()[0]
# add binary missing indicator (in train and test)
X_train[var+'_na'] = np.where(X_train[var].isnull(), 1, 0)
X_test[var+'_na'] = np.where(X_test[var].isnull(), 1, 0)
# replace missing values by the mode
# (in train and test)
X_train[var] = X_train[var].fillna(mode_val)
X_test[var] = X_test[var].fillna(mode_val)
# check that we have no more missing values in the engineered variables
X_train[vars_with_na].isnull().sum()
# check that test set does not contain null values in the engineered variables
[vr for var in vars_with_na if X_test[var].isnull().sum() > 0]
# check the binary missing indicator variables
X_train[['LotFrontage_na', 'MasVnrArea_na', 'GarageYrBlt_na']].head()
###Output
_____no_output_____
###Markdown
Temporal variables Capture elapsed timeThere are 4 variables that refer to the years in which the house or the garage were built or remodeled. We will capture the time elapsed between those variables and the year in which the house was sold:
###Code
def elapsed_years(df, var):
# capture difference between the year variable
# and the year in which the house was sold
df[var] = df['YrSold'] - df[var]
return df
for var in ['YearBuilt', 'YearRemodAdd', 'GarageYrBlt']:
X_train = elapsed_years(X_train, var)
X_test = elapsed_years(X_test, var)
###Output
_____no_output_____
###Markdown
Numerical variable transformationIn the previous Jupyter notebook, we observed that the numerical variables are not normally distributed.We will log transform the positive numerical variables in order to get a more Gaussian-like distribution. This tends to help Linear machine learning models.
###Code
for var in ['LotFrontage', 'LotArea', '1stFlrSF', 'GrLivArea', 'SalePrice']:
X_train[var] = np.log(X_train[var])
X_test[var] = np.log(X_test[var])
# check that test set does not contain null values in the engineered variables
[var for var in ['LotFrontage', 'LotArea', '1stFlrSF',
'GrLivArea', 'SalePrice'] if X_test[var].isnull().sum() > 0]
# same for train set
[var for var in ['LotFrontage', 'LotArea', '1stFlrSF',
'GrLivArea', 'SalePrice'] if X_train[var].isnull().sum() > 0]
###Output
_____no_output_____
###Markdown
Categorical variables Removing rare labelsFirst, we will group those categories within variables that are present in less than 1% of the observations. That is, all values of categorical variables that are shared by less than 1% of houses, well be replaced by the string "Rare".
###Code
# let's capture the categorical variables in a list
cat_vars = [var for var in X_train.columns if X_train[var].dtype == 'O']
def find_frequent_labels(df, var, rare_perc):
# function finds the labels that are shared by more than
# a certain % of the houses in the dataset
df = df.copy()
tmp = df.groupby(var)['SalePrice'].count() / len(df)
return tmp[tmp > rare_perc].index
for var in cat_vars:
# find the frequent categories
frequent_ls = find_frequent_labels(X_train, var, 0.01)
# replace rare categories by the string "Rare"
X_train[var] = np.where(X_train[var].isin(
frequent_ls), X_train[var], 'Rare')
X_test[var] = np.where(X_test[var].isin(
frequent_ls), X_test[var], 'Rare')
###Output
_____no_output_____
###Markdown
Encoding of categorical variablesNext, we need to transform the strings of the categorical variables into numbers. We will do it so that we capture the monotonic relationship between the label and the target.
###Code
# this function will assign discrete values to the strings of the variables,
# so that the smaller value corresponds to the category that shows the smaller
# mean house sale price
def replace_categories(train, test, var, target):
# order the categories in a variable from that with the lowest
# house sale price, to that with the highest
ordered_labels = train.groupby([var])[target].mean().sort_values().index
# create a dictionary of ordered categories to integer values
ordinal_label = {k: i for i, k in enumerate(ordered_labels, 0)}
# use the dictionary to replace the categorical strings by integers
train[var] = train[var].map(ordinal_label)
test[var] = test[var].map(ordinal_label)
for var in cat_vars:
replace_categories(X_train, X_test, var, 'SalePrice')
# check absence of na in the train set
[var for var in X_train.columns if X_train[var].isnull().sum() > 0]
# check absence of na in the test set
[var for var in X_test.columns if X_test[var].isnull().sum() > 0]
# let me show you what I mean by monotonic relationship
# between labels and target
def analyse_vars(df, var):
# function plots median house sale price per encoded
# category
df = df.copy()
df.groupby(var)['SalePrice'].median().plot.bar()
plt.title(var)
plt.ylabel('SalePrice')
plt.show()
for var in cat_vars:
analyse_vars(X_train, var)
###Output
_____no_output_____
###Markdown
The monotonic relationship is particularly clear for the variables MSZoning, Neighborhood, and ExterQual. Note how, the higher the integer that now represents the category, the higher the mean house sale price.(remember that the target is log-transformed, that is why the differences seem so small). Feature ScalingFor use in linear models, features need to be either scaled or normalised. In the next section, I will scale features to the minimum and maximum values:
###Code
# capture all variables in a list
# except the target and the ID
train_vars = [var for var in X_train.columns if var not in ['Id', 'SalePrice']]
# count number of variables
len(train_vars)
# create scaler
scaler = MinMaxScaler()
# fit the scaler to the train set
scaler.fit(X_train[train_vars])
# transform the train and test set
X_train[train_vars] = scaler.transform(X_train[train_vars])
X_test[train_vars] = scaler.transform(X_test[train_vars])
X_train.head()
# let's now save the train and test sets for the next notebook!
X_train.to_csv('xtrain.csv', index=False)
X_test.to_csv('xtest.csv', index=False)
###Output
_____no_output_____ |
is_square.ipynb | ###Markdown
TaskGiven an integral number, determine if it's a square number:
###Code
def is_square(n):
if (n * 1/2) * (n * 1/2) == n:
return True
return False
###Output
_____no_output_____ |
old/Model-01.ipynb | ###Markdown
UCI Epileptic Seizure Recognition Data Set Link to the dataset: https://archive.ics.uci.edu/ml/datasets/Epileptic+Seizure+Recognition
###Code
import os
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torchvision import transforms
from torch.utils.data import Dataset, DataLoader
from torch.utils.data.sampler import SubsetRandomSampler
###Output
_____no_output_____
###Markdown
Hyperparamters
###Code
batch_size = 64
test_split = .2
shuffle_dataset = True
random_seed = 42
epochs = 100
learning_rate = 1e-3
###Output
_____no_output_____
###Markdown
Dataset
###Code
class UCIEpilepsy(Dataset):
def __init__(self, data_path):
self.data = pd.read_csv(data_path).iloc[:, 1:].to_numpy().astype(np.single)
def __len__(self):
return len(self.data)
def __getitem__(self, idx):
data = torch.from_numpy(self.data[idx,2:178])
label = torch.from_numpy(self.data[idx,178:])
return data, label
dataset = UCIEpilepsy(data_path='data/data.csv')
dataset_size = len(dataset)
indices = list(range(dataset_size))
split = int(np.floor(test_split * dataset_size))
if shuffle_dataset:
np.random.seed(random_seed)
np.random.shuffle(indices)
test_indices, train_indices = indices[:split], indices[split:]
train_sampler = SubsetRandomSampler(train_indices)
test_sampler = SubsetRandomSampler(test_indices)
train_dataloader = DataLoader(dataset, batch_size=batch_size, sampler=train_sampler)
test_dataloader = DataLoader(dataset, batch_size=batch_size, sampler=test_sampler)
###Output
_____no_output_____
###Markdown
Neural Network
###Code
class NeuralNetwork(nn.Module):
def __init__(self):
super(NeuralNetwork, self).__init__()
self.linear_relu_stack = nn.Sequential(
nn.Linear(176, 128),
nn.ReLU(),
nn.Linear(128, 128),
nn.ReLU(),
nn.Linear(128, 5)
)
def forward(self, x):
return self.linear_relu_stack(x)
###Output
_____no_output_____
###Markdown
Training Loop
###Code
def train_loop(X,y, model, loss_fn, optimizer):
size = len(X)
# for batch, (X, y) in enumerate(dataloader, 0):
outputs = model(X)
loss = loss_fn(outputs, y.squeeze(1).long()-1)
optimizer.zero_grad()
loss.backward()
optimizer.step()
# if batch % 10 == 0:
# loss, current = loss.item(), batch * len(X)
print(f"loss: {loss:>7f}")
###Output
_____no_output_____
###Markdown
Test Loop
###Code
def test_loop(X, y, model, criterion):
size = len(X)
num_correct = 0
num_samples = 0
with torch.no_grad():
#for X, y in dataloader:
outputs = model(X)
_, predictions = outputs.max(1)
num_correct += (predictions == y).sum()
num_samples += predictions.size(0)
print(f"Got {num_correct} / {num_samples} with {float(num_correct) / float(num_samples) * 100:.2f}")
###Output
_____no_output_____
###Markdown
Execution
###Code
model = NeuralNetwork()
X, y = next(iter(train_dataloader))
textX, testy = next(iter(test_dataloader))
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(model.parameters(), lr=learning_rate, momentum=0.9)
for epoch in range(epochs):
print(f"Epoch [{epoch+1}/{epochs}]")
train_loop(X, y, model, criterion, optimizer)
test_loop(X, y, model, criterion)
###Output
Epoch [1/100]
loss: 18.408686
Got 9 / 64 with 14.06
Epoch [2/100]
loss: 13.081454
Got 15 / 64 with 23.44
Epoch [3/100]
loss: 13.784674
Got 345 / 64 with 539.06
Epoch [4/100]
loss: 8.900457
Got 735 / 64 with 1148.44
Epoch [5/100]
loss: 9.482249
Got 582 / 64 with 909.38
Epoch [6/100]
loss: 6.394184
Got 543 / 64 with 848.44
Epoch [7/100]
loss: 4.073208
Got 486 / 64 with 759.38
Epoch [8/100]
loss: 2.911988
Got 495 / 64 with 773.44
Epoch [9/100]
loss: 2.571485
Got 432 / 64 with 675.00
Epoch [10/100]
loss: 1.705727
Got 414 / 64 with 646.88
Epoch [11/100]
loss: 1.073463
Got 444 / 64 with 693.75
Epoch [12/100]
loss: 0.786391
Got 510 / 64 with 796.88
Epoch [13/100]
loss: 0.661567
Got 546 / 64 with 853.12
Epoch [14/100]
loss: 0.446909
Got 567 / 64 with 885.94
Epoch [15/100]
loss: 0.332226
Got 585 / 64 with 914.06
Epoch [16/100]
loss: 0.268042
Got 597 / 64 with 932.81
Epoch [17/100]
loss: 0.234971
Got 594 / 64 with 928.12
Epoch [18/100]
loss: 0.207456
Got 591 / 64 with 923.44
Epoch [19/100]
loss: 0.183676
Got 591 / 64 with 923.44
Epoch [20/100]
loss: 0.157857
Got 585 / 64 with 914.06
Epoch [21/100]
loss: 0.132126
Got 579 / 64 with 904.69
Epoch [22/100]
loss: 0.107050
Got 579 / 64 with 904.69
Epoch [23/100]
loss: 0.084840
Got 579 / 64 with 904.69
Epoch [24/100]
loss: 0.067431
Got 579 / 64 with 904.69
Epoch [25/100]
loss: 0.054594
Got 579 / 64 with 904.69
Epoch [26/100]
loss: 0.045662
Got 579 / 64 with 904.69
Epoch [27/100]
loss: 0.039722
Got 579 / 64 with 904.69
Epoch [28/100]
loss: 0.035914
Got 579 / 64 with 904.69
Epoch [29/100]
loss: 0.032929
Got 579 / 64 with 904.69
Epoch [30/100]
loss: 0.029694
Got 579 / 64 with 904.69
Epoch [31/100]
loss: 0.026613
Got 579 / 64 with 904.69
Epoch [32/100]
loss: 0.024110
Got 579 / 64 with 904.69
Epoch [33/100]
loss: 0.022154
Got 579 / 64 with 904.69
Epoch [34/100]
loss: 0.020560
Got 579 / 64 with 904.69
Epoch [35/100]
loss: 0.019147
Got 579 / 64 with 904.69
Epoch [36/100]
loss: 0.017855
Got 579 / 64 with 904.69
Epoch [37/100]
loss: 0.016668
Got 579 / 64 with 904.69
Epoch [38/100]
loss: 0.015573
Got 579 / 64 with 904.69
Epoch [39/100]
loss: 0.014584
Got 579 / 64 with 904.69
Epoch [40/100]
loss: 0.013697
Got 579 / 64 with 904.69
Epoch [41/100]
loss: 0.012875
Got 579 / 64 with 904.69
Epoch [42/100]
loss: 0.012111
Got 579 / 64 with 904.69
Epoch [43/100]
loss: 0.011405
Got 579 / 64 with 904.69
Epoch [44/100]
loss: 0.010749
Got 579 / 64 with 904.69
Epoch [45/100]
loss: 0.010141
Got 579 / 64 with 904.69
Epoch [46/100]
loss: 0.009581
Got 579 / 64 with 904.69
Epoch [47/100]
loss: 0.009067
Got 579 / 64 with 904.69
Epoch [48/100]
loss: 0.008596
Got 579 / 64 with 904.69
Epoch [49/100]
loss: 0.008165
Got 579 / 64 with 904.69
Epoch [50/100]
loss: 0.007770
Got 579 / 64 with 904.69
Epoch [51/100]
loss: 0.007410
Got 579 / 64 with 904.69
Epoch [52/100]
loss: 0.007082
Got 579 / 64 with 904.69
Epoch [53/100]
loss: 0.006781
Got 579 / 64 with 904.69
Epoch [54/100]
loss: 0.006506
Got 579 / 64 with 904.69
Epoch [55/100]
loss: 0.006254
Got 579 / 64 with 904.69
Epoch [56/100]
loss: 0.006022
Got 579 / 64 with 904.69
Epoch [57/100]
loss: 0.005809
Got 579 / 64 with 904.69
Epoch [58/100]
loss: 0.005612
Got 579 / 64 with 904.69
Epoch [59/100]
loss: 0.005430
Got 579 / 64 with 904.69
Epoch [60/100]
loss: 0.005261
Got 579 / 64 with 904.69
Epoch [61/100]
loss: 0.005104
Got 579 / 64 with 904.69
Epoch [62/100]
loss: 0.004958
Got 579 / 64 with 904.69
Epoch [63/100]
loss: 0.004822
Got 579 / 64 with 904.69
Epoch [64/100]
loss: 0.004695
Got 579 / 64 with 904.69
Epoch [65/100]
loss: 0.004576
Got 579 / 64 with 904.69
Epoch [66/100]
loss: 0.004464
Got 579 / 64 with 904.69
Epoch [67/100]
loss: 0.004359
Got 579 / 64 with 904.69
Epoch [68/100]
loss: 0.004260
Got 579 / 64 with 904.69
Epoch [69/100]
loss: 0.004166
Got 579 / 64 with 904.69
Epoch [70/100]
loss: 0.004077
Got 579 / 64 with 904.69
Epoch [71/100]
loss: 0.003993
Got 579 / 64 with 904.69
Epoch [72/100]
loss: 0.003913
Got 579 / 64 with 904.69
Epoch [73/100]
loss: 0.003837
Got 579 / 64 with 904.69
Epoch [74/100]
loss: 0.003765
Got 579 / 64 with 904.69
Epoch [75/100]
loss: 0.003696
Got 579 / 64 with 904.69
Epoch [76/100]
loss: 0.003631
Got 579 / 64 with 904.69
Epoch [77/100]
loss: 0.003568
Got 579 / 64 with 904.69
Epoch [78/100]
loss: 0.003508
Got 579 / 64 with 904.69
Epoch [79/100]
loss: 0.003451
Got 579 / 64 with 904.69
Epoch [80/100]
loss: 0.003396
Got 579 / 64 with 904.69
Epoch [81/100]
loss: 0.003343
Got 579 / 64 with 904.69
Epoch [82/100]
loss: 0.003293
Got 579 / 64 with 904.69
Epoch [83/100]
loss: 0.003244
Got 579 / 64 with 904.69
Epoch [84/100]
loss: 0.003197
Got 579 / 64 with 904.69
Epoch [85/100]
loss: 0.003152
Got 579 / 64 with 904.69
Epoch [86/100]
loss: 0.003109
Got 579 / 64 with 904.69
Epoch [87/100]
loss: 0.003067
Got 579 / 64 with 904.69
Epoch [88/100]
loss: 0.003026
Got 579 / 64 with 904.69
Epoch [89/100]
loss: 0.002987
Got 579 / 64 with 904.69
Epoch [90/100]
loss: 0.002950
Got 579 / 64 with 904.69
Epoch [91/100]
loss: 0.002913
Got 579 / 64 with 904.69
Epoch [92/100]
loss: 0.002878
Got 579 / 64 with 904.69
Epoch [93/100]
loss: 0.002844
Got 579 / 64 with 904.69
Epoch [94/100]
loss: 0.002811
Got 579 / 64 with 904.69
Epoch [95/100]
loss: 0.002779
Got 579 / 64 with 904.69
Epoch [96/100]
loss: 0.002747
Got 579 / 64 with 904.69
Epoch [97/100]
loss: 0.002717
Got 579 / 64 with 904.69
Epoch [98/100]
loss: 0.002688
Got 579 / 64 with 904.69
Epoch [99/100]
loss: 0.002659
Got 579 / 64 with 904.69
Epoch [100/100]
loss: 0.002631
Got 579 / 64 with 904.69
|
doc/source/notebooks/ordinal.ipynb | ###Markdown
Ordinal Regression with GPflow Clement Tiennot, Camille Tiennot 2016 edits by James Hensman 2017
###Code
import gpflow
import numpy as np
import matplotlib
%matplotlib inline
matplotlib.rcParams['figure.figsize'] = (12, 6)
plt = matplotlib.pyplot
#make a one dimensional ordinal regression problem
np.random.seed(1)
num_data = 20
X = np.random.rand(num_data, 1)
K = np.exp(-0.5*np.square(X - X.T)/0.01) + np.eye(num_data)*1e-6
f = np.dot(np.linalg.cholesky(K), np.random.randn(num_data, 1))
plt.plot(X, f, '.')
plt.ylabel('latent function value')
Y = np.round((f + f.min())*3)
Y = Y - Y.min()
Y = np.asarray(Y, np.float64)
plt.twinx()
plt.plot(X, Y, 'kx', mew=1.5)
plt.ylabel('observed data value')
# construct ordinal likelihood
bin_edges = np.array(np.arange(np.unique(Y).size + 1), dtype=float)
bin_edges = bin_edges - bin_edges.mean()
likelihood=gpflow.likelihoods.Ordinal(bin_edges)
# build a model with this likelihood
m = gpflow.models.VGP(X, Y,
kern=gpflow.kernels.Matern32(1),
likelihood=likelihood)
#fit the model
gpflow.train.ScipyOptimizer().minimize(m)
# here we'll plot the expected value of Y +- 2 std deviations, as if the distribution were Gaussian
plt.figure(figsize=(14, 6))
Xtest = np.linspace(m.X.read_value().min(), m.X.read_value().max(), 100).reshape(-1, 1)
mu, var = m.predict_y(Xtest)
line, = plt.plot(Xtest, mu, lw=2)
col=line.get_color()
plt.plot(Xtest, mu+2*np.sqrt(var), '--', lw=2, color=col)
plt.plot(Xtest, mu-2*np.sqrt(var), '--', lw=2, color=col)
plt.plot(m.X.read_value(), m.Y.read_value(), 'kx', mew=2)
# to see the predictive density, try predicting every possible value.
def pred_density(m):
Xtest = np.linspace(m.X.read_value().min(), m.X.read_value().max(), 100).reshape(-1, 1)
ys = np.arange(m.Y.read_value().max()+1)
densities = []
for y in ys:
Ytest = np.ones_like(Xtest) * y
densities.append(m.predict_density(Xtest, Ytest))
return np.hstack(densities).T
plt.imshow(np.exp(pred_density(m)), interpolation='nearest',
extent=[m.X.read_value().min(), m.X.read_value().max(), -0.5, m.Y.read_value().max()+0.5],
origin='lower', aspect='auto', cmap=plt.cm.viridis)
plt.colorbar()
plt.plot(X, Y, 'kx', mew=2, scalex=False, scaley=False)
###Output
_____no_output_____
###Markdown
Ordinal Regression with GPflow Clement Tiennot, Camille Tiennot 2016 edits by James Hensman 2017
###Code
import gpflow
import numpy as np
import matplotlib
%matplotlib inline
matplotlib.rcParams['figure.figsize'] = (12, 6)
plt = matplotlib.pyplot
#make a one dimensional ordinal regression problem
np.random.seed(1)
num_data = 20
X = np.random.rand(num_data, 1)
K = np.exp(-0.5*np.square(X - X.T)/0.01) + np.eye(num_data)*1e-6
f = np.dot(np.linalg.cholesky(K), np.random.randn(num_data, 1))
plt.plot(X, f, '.')
plt.ylabel('latent function value')
Y = np.round((f + f.min())*3)
Y = Y - Y.min()
Y = np.asarray(Y, np.float64)
plt.twinx()
plt.plot(X, Y, 'kx', mew=1.5)
plt.ylabel('observed data value')
# construct ordinal likelihood
bin_edges = np.array(np.arange(np.unique(Y).size + 1), dtype=float)
bin_edges = bin_edges - bin_edges.mean()
likelihood=gpflow.likelihoods.Ordinal(bin_edges)
# build a model with this likelihood
m = gpflow.models.VGP(X, Y,
kern=gpflow.kernels.Matern32(1),
likelihood=likelihood)
#fit the model
gpflow.train.ScipyOptimizer().minimize(m)
# here we'll plot the expected value of Y +- 2 std deviations, as if the distribution were Gaussian
plt.figure(figsize=(14, 6))
Xtest = np.linspace(m.X.read_value().min(), m.X.read_value().max(), 100).reshape(-1, 1)
mu, var = m.predict_y(Xtest)
line, = plt.plot(Xtest, mu, lw=2)
col=line.get_color()
plt.plot(Xtest, mu+2*np.sqrt(var), '--', lw=2, color=col)
plt.plot(Xtest, mu-2*np.sqrt(var), '--', lw=2, color=col)
plt.plot(m.X.read_value(), m.Y.read_value(), 'kx', mew=2)
# to see the predictive density, try predicting every possible value.
def pred_density(m):
Xtest = np.linspace(m.X.read_value().min(), m.X.read_value().max(), 100).reshape(-1, 1)
ys = np.arange(m.Y.read_value().max()+1)
densities = []
for y in ys:
Ytest = np.ones_like(Xtest) * y
densities.append(m.predict_density(Xtest, Ytest))
return np.hstack(densities).T
plt.imshow(np.exp(pred_density(m)), interpolation='nearest',
extent=[m.X.read_value().min(), m.X.read_value().max(), -0.5, m.Y.read_value().max()+0.5],
origin='lower', aspect='auto', cmap=plt.cm.viridis)
plt.colorbar()
plt.plot(X, Y, 'kx', mew=2, scalex=False, scaley=False)
###Output
_____no_output_____
###Markdown
Ordinal Regression with GPflow
###Code
import GPflow
import tensorflow as tf
import matplotlib
import numpy as np
%matplotlib inline
matplotlib.style.use('ggplot')
plt = matplotlib.pyplot
#make a one dimensional ordinal regression problem
np.random.seed(1)
X = np.random.rand(100,1)
K = np.exp(-0.5*np.square(X - X.T)/0.01) + np.eye(100)*1e-6
f = np.dot(np.linalg.cholesky(K), np.random.randn(100,1))
plt.plot(X, f, '.')
Y = np.round((f + f.min())*3)
Y = Y - Y.min()
Y = np.asarray(Y, np.int32)
plt.twinx()
plt.plot(X, Y, 'kx')
# construct ordinal likelihood
bin_edges = np.arange(np.unique(Y).size)
bin_edges = bin_edges - bin_edges.mean()
likelihood=GPflow.likelihoods.Ordinal(bin_edges)
# build a model with this likelihood
m = GPflow.vgp.VGP(X, Y,
kern=GPflow.kernels.Matern32(1),
likelihood=likelihood)
#fit the model
_ = m.optimize()
# here we'll plot the expected value of Y +- 2 std deviations, as if the distribution were Gaussian
plt.figure(figsize=(14, 6))
Xtest = np.linspace(m.X.value.min(), m.X.value.max(), 100).reshape(-1, 1)
mu, var = m.predict_y(Xtest)
line, = plt.plot(Xtest, mu, lw=2)
col=line.get_color()
plt.plot(Xtest, mu+2*np.sqrt(var), '--', lw=2, color=col)
plt.plot(Xtest, mu-2*np.sqrt(var), '--', lw=2, color=col)
plt.plot(m.X.value, m.Y.value, 'kx', mew=2)
# to see the predictive density, try predicting every possible value.
def pred_density(m):
Xtest = np.linspace(m.X.value.min(), m.X.value.max(), 100).reshape(-1, 1)
ys = np.arange(m.Y.value.max()+1)
densities = []
for y in ys:
Ytest = np.ones_like(Xtest) * y
densities.append(m.predict_density(Xtest, Ytest))
return np.hstack(densities).T
plt.figure(figsize=(16,6))
plt.imshow(np.exp(pred_density(m)), interpolation='nearest',
extent=[m.X.value.min(), m.X.value.max(), -0.5, m.Y.value.max()+0.5],
origin='lower', aspect='auto', cmap=plt.cm.viridis)
plt.colorbar()
plt.plot(X, Y, 'kx', mew=2, scalex=False, scaley=False)
###Output
_____no_output_____ |
Weatherpy/HM6 APi OpenWeather-Final.ipynb | ###Markdown
temperature(F) vs Latitude
###Code
#temperature vs Latitude
temp_lat_df = cities_df[['temperature','latitude']]
fig,ax = plt.subplots()
ax.scatter(temp_lat_df['latitude'],temp_lat_df['temperature'],marker='.')
ax.set_xlabel('Latitude')
ax.set_ylabel('Temperature(F)')
plt.title('City Latitude vs Temperature(F)')
ax.grid()
fig.savefig('City Latitude vs Average Temperature(F).png')
plt.show()
###Output
_____no_output_____
###Markdown
First Oberservation: based on the graph above we can see that the tendency is that cities located at lower latitudes tend to have higher temperatures and cities located at higher latitudes tend to have lower temperatures. Humidity (%) vs. Latitude
###Code
#Humidity (%) vs. Latitude
hum_lat_df = cities_df[['humidity','latitude']]
fig1,ax1 = plt.subplots()
ax1.scatter(hum_lat_df['latitude'],hum_lat_df['humidity'],marker='.',color='g')
ax1.set_xlabel('Latitude')
ax1.set_ylabel('humidity(%)')
plt.title('City Latitude vs Humidity(%)')
ax1.grid()
fig1.savefig('City Latitude vs Humidity(%).png')
plt.show()
###Output
_____no_output_____
###Markdown
Second Oberservation: Based on the graph above we can see that most of the cities located from equator up to a latitude of ~50 degrees have the highest humidity while those cities located below the equator tend to have less humidity. Cloudiness (%) vs. Latitude
###Code
#Cloudiness (%) vs. Latitude
cloud_lat_df = cities_df[['cloudiness','latitude']]
fig2,ax2 = plt.subplots()
ax2.scatter(cloud_lat_df['latitude'],cloud_lat_df['cloudiness'],marker='.',color='r')
ax2.set_xlabel('Latitude')
ax2.set_ylabel('cloudiness(%)')
plt.title('City Latitude vs Cloudiness(%)')
ax2.grid()
fig2.savefig('City Latitude vs Cloudiness(%).png')
plt.show()
###Output
_____no_output_____
###Markdown
Third Observation: Based on the sample we have most of the cities where located from latitude from range 40 to 50 degrees. We can determine that at latitude 50 degrees we tend to have more cloudiness. Wind Speed (mph) vs. Latitude
###Code
#Wind Speed (mph) vs. Latitude
wind_lat_df = cities_df[['windspeed','latitude']]
fig3,ax3 = plt.subplots()
ax3.scatter(wind_lat_df['latitude'],wind_lat_df['windspeed'],marker='.',color='b')
ax3.set_xlabel('Latitude')
ax3.set_ylabel('windspeed(mph)')
plt.title('City Latitude vs Windspeed(MPH)')
ax3.grid()
fig3.savefig('City Latitude vs Windspeed(MPH).png')
plt.show()
###Output
_____no_output_____ |
DSC 540 - Data Preparation/Week 3 & 4.ipynb | ###Markdown
Week 3 & 4TitleTitle -->H1 -->H2 -->H3 -->H4 -->Patrick Weatherford[Green]: (bce35b)[Purple]: (ae8bd5)[Coral]: (9c4957)[Grey]: (8c8c8c) ****** Importing libraries
###Code
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import os
import requests
from collections import deque as deq
from itertools import permutations, dropwhile, zip_longest
import re
import math
import io
## import custom config file with keys and change working directory
class_path = os.path.expanduser('~') + '\\OneDrive - Bellevue University\\Bellevue_University\\DSC 540 - Data Preparation'
bu_path = os.path.expanduser('~') + '\\OneDrive - Bellevue University\\Bellevue_University'
os.chdir(bu_path); import config; os.chdir(class_path)
## matplotlib default settings
plt.style.use('dark_background')
mpl.rcParams.update({'lines.linewidth':3})
mpl.rcParams.update({'axes.labelsize':14})
mpl.rcParams.update({'axes.titlesize':16})
mpl.rcParams.update({'axes.titleweight':'bold'})
mpl.rcParams.update({'figure.autolayout':True})
mpl.rcParams.update({'axes.grid':True, 'grid.color':'#424242', 'grid.linestyle':'--'})
## see list of current settings
# plt.rcParams.keys()
###Output
_____no_output_____
###Markdown
Instantiating random number generator
###Code
rng_seed = 777
rng = np.random.default_rng(rng_seed)
###Output
_____no_output_____
###Markdown
*** Book ActivitiesTitleTitle -->H1 -->H2 -->H3 -->H4 -->Patrick Weatherford -->[Green]: (bce35b)[Purple]: (ae8bd5)[Coral]: (9c4957)[Grey]: (8c8c8c) Activity 5TitleTitle -->H1 -->H2 -->H3 -->H4 -->Patrick Weatherford -->[Green]: (bce35b)[Purple]: (ae8bd5)[Coral]: (9c4957)[Grey]: (8c8c8c)Boston Housing data set.
###Code
## get variable info
def skip_logic(x):
if 7 <= x <= 20: # get specific rows from website which hold variable info
return False
else:
return True
data_url = "http://lib.stat.cmu.edu/datasets/boston"
var_df = pd.read_csv(data_url, sep=' +', skiprows=lambda x: skip_logic(x), header=None, engine='python', names=['VARIABLE','DESC']) # create DataFrame of variables
var_df['VARIABLE'] = var_df['VARIABLE'].replace(' ', '') # remove whiate space from variable name
var_df_disp = var_df.style.set_table_styles([dict(selector='th', props=[('text-align', 'left')])]) # lef-align headers
var_df_disp.set_properties(subset=['DESC'], **{'width': '400px'}) # widen description column
var_df_disp.set_properties(**{'text-align': 'left'}) # left align all values
var_df_disp
## get Boston Housing data
raw_df = pd.read_csv(data_url, sep="\s+", skiprows=22, header=None) # start data feed from 23 row
data = np.hstack([raw_df.values[::2, :], raw_df.values[1::2, :3]]) # data in rows on site is not on single row. Must skip every other row, hstack, then exclude NaN values from overlapping row
labels = var_df['VARIABLE'].values # assign variable names to list
bost_df = pd.DataFrame(data, columns=labels) # create df
## first 10 rows
bost_df.head(10)
## drop specified columns
bost_df.drop(['CHAS','NOX','B','LSTAT'], inplace=True, axis=1)
bost_df
## last 7 rows in df
bost_df.tail(7)
###Output
_____no_output_____
###Markdown
Looping through df variables using pandas implementation.
###Code
## plot histogram of each variable
bost_df.hist(figsize=(15,13), bins=30)
plt.show()
###Output
_____no_output_____
###Markdown
Custom function to loop through df and create histogram of each variable.
###Code
def df_plot(df, plot_type, hist_bins=10, n_plot_cols=3, figsize=None):
cols = df.columns # get list of column names
n_vars = len(cols) # get number of variables in df
n_plot_cols = n_plot_cols # get number of columns that will be in the subplot figure
n_plot_rows = math.ceil(n_vars / n_plot_cols) # based on the number of columns in the subplot figure & number of df columns, calculate how many rows should be in the figure
plt.figure(figsize=figsize) # set figure size
graph_cnt = 1 # initiate counter for variable plots
for r in range(1, n_plot_rows+1): # for row in subplot figure
for c in range(1, n_plot_cols+1): # for col in subplot figure
if graph_cnt > n_vars: # if variable counter is greater than the number of variables in the df, break the loop
break
else: # else plot each variable in the correct subplot location going from left->right, top->bottom
plt.subplot(n_plot_rows, n_plot_cols, graph_cnt)
if plot_type.lower() == 'hist':
plt.hist(df.iloc[:, graph_cnt-1], bins=hist_bins)
plt.title(cols[graph_cnt-1])
graph_cnt += 1
plt.show()
df_plot(bost_df, plot_type='hist', hist_bins=30, figsize=(15,13))
###Output
_____no_output_____
###Markdown
Scatter plot
###Code
plt.scatter(
x=bost_df['RM']
, y=bost_df['MEDV']
, alpha=.7
)
plt.title('Median Home Value vs. Number of Rooms')
plt.xlabel('Number of Rooms')
plt.ylabel('Median Home Value ($1k)')
plt.show()
plt.scatter(
x=np.log10(bost_df['CRIM'])
, y=bost_df['MEDV']
, alpha=.7
)
plt.title('Median Home Value vs. Crime Rate')
plt.xlabel('Crime Rate (log10)')
plt.ylabel('Median Home Value ($1k)')
plt.show()
###Output
_____no_output_____
###Markdown
Summary statistics
###Code
bost_df.describe()
###Output
_____no_output_____
###Markdown
Percent (%) of houses < $20k
###Code
p = round(100 * sum(bost_df['MEDV'] < 20) / len(bost_df), 2)
print(f"{p}% of homes are less thatn $20k")
###Output
41.5% of homes are less thatn $20k
###Markdown
Activity 6TitleTitle -->H1 -->H2 -->H3 -->H4 -->Patrick Weatherford -->[Green]: (bce35b)[Purple]: (ae8bd5)[Coral]: (9c4957)[Grey]: (8c8c8c)Adult Income Dataset (UCI)
###Code
var_url = 'https://raw.githubusercontent.com/TrainingByPackt/Data-Wrangling-with-Python/master/Lesson04/Activity06/adult_income_names.txt'
var_df = pd.read_csv(var_url, sep=':', engine='python', header=None, names=['VARIABLE','DESC'])
var_df['VARIABLE'] = var_df['VARIABLE'].str.strip()
var_df_disp = var_df.style.set_table_styles([dict(selector='th', props=[('text-align', 'left')])]) # lef-align headers
var_df_disp.set_properties(subset=['DESC'], **{'width': '800px'}) # widen description column
var_df_disp.set_properties(**{'text-align': 'left'}) # left align all values
var_df_disp
cols = [i for i in var_df['VARIABLE']]
cols.append('income')
data_url = 'https://raw.githubusercontent.com/TrainingByPackt/Data-Wrangling-with-Python/master/Lesson04/Activity06/adult_income_data.csv'
a6_df = pd.read_csv(data_url, header=None, names=cols)
a6_df
###Output
_____no_output_____
###Markdown
Read text file line-by-line
###Code
def ReadText(file_path):
with open(file_path, 'r') as TextFile:
for line in TextFile:
line = line.rstrip('\n')
print(line)
file_path = 'RandomTextFile.txt'
ReadText(file_path)
###Output
This is line 1
This is line 2
This is line 3
This is line 4 yo yo
This is line 5 blahblah
.-.
|_:_|
/(_Y_)\
. ( \/M\/ )
'. _.'-/'-'\-'._
': _/.--'[[[[]'--.\_
': /_' : |::"| : '.\
': // ./ |oUU| \.' :\
': _:'..' \_|___|_/ : :|
':. .' |_[___]_| :.':\
[::\ | : | | : ; : \
'-' \/'.| |.' \ .;.' |
|\_ \ '-' : |
| \ \ .: : | |
| \ | '. : \ |
/ \ :. .; |
/ | | :__/ : \\
| | | \: | \ | ||
/ \ : : |: / |__| /|
| : : :_/_| /'._\ '--|_\
/___.-/_|-' \ \
'-'
###Markdown
Find missing values row index locations.
###Code
a6_df.isnull().sum()
###Output
_____no_output_____
###Markdown
Create df with only age, education, and occupation
###Code
df_sub = a6_df[['age','education','occupation']]
df_sub
plt.hist(a6_df['age'], bins=20)
plt.title('Histogram of Age')
plt.xlabel('Age (bins = 20)')
plt.ylabel('Frequency')
plt.show()
###Output
_____no_output_____
###Markdown
Using apply(), strip white space from string columns
###Code
def df_str_strip(df):
for c in df.columns:
if df[c].dtype == 'O':
c_new = f"{c}_new"
df[c_new] = df[c].apply(lambda x: x.strip())
df[c] = df[c_new]
df.drop(labels=c_new, axis=1, inplace=True)
else:
continue
df_str_strip(a6_df)
a6_df
###Output
_____no_output_____
###Markdown
Find people aged between 30 and 50.
###Code
len((a6_df[(a6_df['age'] >=30) & (a6_df['age'] <= 50)]).index)
###Output
_____no_output_____
###Markdown
Group records based on age and education to find how the mean age is distributed.
###Code
pd.DataFrame(a6_df[['age','education']].groupby('education')['age'].mean())
occ_summ_df = a6_df.groupby('occupation')['age'].describe()
occ_summ_df
###Output
_____no_output_____
###Markdown
occupation with oldest workers
###Code
occ_summ_df['mean'].max()
###Output
_____no_output_____
###Markdown
occupation with highest share of 75% percentile
###Code
occ_summ_df[occ_summ_df['75%'] == occ_summ_df['75%'].max()].index[0]
###Output
_____no_output_____
###Markdown
Use subset and groupby to find outliers.- Are there classes with very few observations?- Are there extreme numerical values?
###Code
for c in a6_df.columns:
n_unique_vals = a6_df[[c]].nunique()[0]
if n_unique_vals <= 20:
pd.DataFrame(a6_df[[c]].groupby(c).size(), columns=[c]).plot.bar()
else:
pd.DataFrame(a6_df[[c]].groupby(c).size(), columns=[f"{c} (frequency)"]).plot.box()
###Output
_____no_output_____
###Markdown
Joining based on values
###Code
initial_df = a6_df[['age','workclass','education']]
agg_df = a6_df[a6_df['income'] == '>50K'].loc[:,['workclass']]
agg_df_grp = agg_df.groupby('workclass')
agg_df = pd.DataFrame(agg_df_grp.size(), columns=['workclass >50K count'])
pd.merge(left=initial_df, right=agg_df, left_on='workclass', right_index=True)
###Output
_____no_output_____
###Markdown
*** Series and Basic ArithmeticTitleTitle -->H1 -->H2 -->H3 -->H4 -->Patrick Weatherford -->[Green]: (bce35b)[Purple]: (ae8bd5)[Coral]: (9c4957)[Grey]: (8c8c8c)
###Code
series1 = [7.3, -2.5, 3.4, 1.5]
s1_index = ['a','c','d','e']
series2 = [-2.1, 3.6, -1.5, 4, 3.1]
s2_index = ['a','c','e','f','g']
s1 = pd.Series(series1, index=s1_index)
s2 = pd.Series(series2, index=s2_index)
###Output
_____no_output_____
###Markdown
Add series 1 to series 2
###Code
s1 + s2
###Output
_____no_output_____
###Markdown
Subtract series 1 from series 2
###Code
s1 - s2
###Output
_____no_output_____
###Markdown
***Below code is for practice only
###Code
np.arange(start=1, stop=20, step=2)
np.arange(start=30, stop=-1, step=-2)
np.linspace(start=1, stop=10, num=5)
np.linspace(start=30, stop=0, num=20)
a1 = rng.integers(1,100,30)
a1
a1_re = a1.reshape(2,3,5)
a1_re
a1_re[1,0,3]
## index[1] in axis 0
## index[0] in axis 1
## index[3] in axis 2
a1_re.size, a1_re.shape, a1_re.ndim
a1_rav = a1_re.ravel()
a1_rav
a2 = rng.integers(1,10,20).reshape(4,5)
a2
df = pd.DataFrame(a2)
df
df1 = df.drop(0, axis=1)
df1
rng.binomial(n=8, p=.6, size=10)
url = "https://github.com/TrainingByPackt/Data-Wrangling-with-Python/raw/master/Lesson04/Exercise48-51/Sample%20-%20Superstore.xls" # Make sure the url is the raw version of the file on GitHub
download = requests.get(url).content
ss_df = pd.read_excel(io.BytesIO(download))
ss_df.drop(['Row ID'], inplace=True, axis=1)
print(f"""
{ss_df.shape}
{ss_df.columns}
""")
ss_df_sub = ss_df.loc[
[i for i in range(5,10)]
, ['Customer ID','Customer Name','City','Postal Code','Sales']
]
ss_df_sub
ss_df.loc[
[i for i in range(100,200)] # axis 0 range(100,200)
,['Sales','Profit'] # axis 1 columns to include
].describe()
unique_states = ss_df['State'].unique()
n_unique_states = ss_df['State'].nunique()
print(f"""
States in DataFrame:
{unique_states}
Number of Distinct States in DataFrame:
{n_unique_states}
""")
matrix_data = np.matrix([[22,66,140],[42,70,148],[30,62,125],[35,68,160],[25,62,152]])
row_labels = ['A','B','C','D','E']
column_headings = ['Age','Height','Weight']
matrix_data
matrix_df = pd.DataFrame(data=matrix_data
,index=row_labels
,columns=column_headings)
matrix_df
matrix_df.reset_index(inplace=True, drop=True)
matrix_df
matrix_df['Profession'] = 'Student Teacher Engineer Doctor Nurse'.split()
matrix_df
matrix_df = matrix_df.set_index('Profession')
matrix_df
df_sub = ss_df.loc[
[i for i in range(10)]
,['Ship Mode','State','Sales']
]
df_sub
by_state = df_sub.groupby('State')
by_state.mean()
pd.DataFrame(by_state.describe().loc['California'])
df_sub.groupby('Ship Mode').describe().loc[['Second Class','Standard Class']]
byStateCity = ss_df.groupby(['State','City'])['Sales']
byStateCity.describe()
url = "https://github.com/TrainingByPackt/Data-Wrangling-with-Python/raw/master/Lesson04/Exercise48-51/Sample%20-%20Superstore.xls" # Make sure the url is the raw version of the file on GitHub
download = requests.get(url).content
miss_df = pd.read_excel(io.BytesIO(download), sheet_name='Missing')
miss_df
miss_df.isnull()
for c in miss_df.columns:
miss = miss_df[c].isnull().sum()
if miss > 0:
print(f"{c} has {miss} missing value(s)")
else:
print(f"{c} has NO missing value(s)!")
###Output
Customer has 1 missing value(s)
Product has 2 missing value(s)
Sales has 1 missing value(s)
Quantity has 1 missing value(s)
Discount has NO missing value(s)!
Profit has 1 missing value(s)
|
even-more-python-for-beginners-data-tools/15 - Visualizing data with Matplotlib/15 - Visualizing correlations.ipynb | ###Markdown
Visualizing data with matplotlib Somtimes graphs provide the best way to visualize dataThe **matplotlib** library allows you to draw graphs to help with visualizationIf we want to visualize data, we will need to load some data into a DataFrame
###Code
import pandas as pd
# Load our data from the csv file
delays_df = pd.read_csv('Data/Lots_of_flight_data.csv')
###Output
_____no_output_____
###Markdown
In order to display plots we need to import the **matplotlib** library
###Code
import matplotlib.pyplot as plt
###Output
_____no_output_____
###Markdown
A common plot used in data science is the scatter plot for checking the relationship between two columnsIf you see dots scattered everywhere, there is no correlation between the two columnsIf you see somethign resembling a line, there is a correlation between the two columnsYou can use the plot method of the DataFrame to draw the scatter plot* kind - the type of graph to draw* x - value to plot as x* y - value to plot as y* color - color to use for the graph points* alpha - opacity - useful to show density of points in a scatter plot* title - title of the graph
###Code
#Check if there is a relationship between the distance of a flight and how late the flight arrives
delays_df.plot(
kind='scatter',
x='DISTANCE',
y='ARR_DELAY',
color='blue',
alpha=0.3,
title='Correlation of arrival and distance'
)
plt.show()
#Check if there is a relationship between the how late the flight leaves and how late the flight arrives
delays_df.plot(
kind='scatter',
x='DEP_DELAY',
y='ARR_DELAY',
color='blue',
alpha=0.3,
title='Correlation of arrival and departure delay'
)
plt.show()
###Output
_____no_output_____
###Markdown
Visualizing data with matplotlib Somtimes graphs provide the best way to visualize dataThe **matplotlib** library allows you to draw graphs to help with visualizationIf we want to visualize data, we will need to load some data into a DataFrame
###Code
import pandas as pd
# Load our data from the csv file
delays_df = pd.read_csv('Data/Lots_of_flight_data.csv')
###Output
_____no_output_____
###Markdown
In order to display plots we need to import the **matplotlib** library
###Code
import matplotlib.pyplot as plt
###Output
_____no_output_____
###Markdown
A common plot used in data science is the scatter plot for checking the relationship between two columnsIf you see dots scattered everywhere, there is no correlation between the two columnsIf you see somethign resembling a line, there is a correlation between the two columnsYou can use the plot method of the DataFrame to draw the scatter plot* kind - the type of graph to draw* x - value to plot as x* y - value to plot as y* color - color to use for the graph points* alpha - opacity - useful to show density of points in a scatter plot* title - title of the graph
###Code
#Check if there is a relationship between the distance of a flight and how late the flight arrives
delays_df.plot(
kind='scatter',
x='DISTANCE',
y='ARR_DELAY',
color='blue',
alpha=0.3,
title='Correlation of arrival and distance'
)
plt.show()
#Check if there is a relationship between the how late the flight leaves and how late the flight arrives
delays_df.plot(
kind='scatter',
x='DEP_DELAY',
y='ARR_DELAY',
color='blue',
alpha=0.3,
title='Correlation of arrival and departure delay'
)
plt.show()
###Output
_____no_output_____
###Markdown
Visualizing data with matplotlib Somtimes graphs provide the best way to visualize dataThe **matplotlib** library allows you to draw graphs to help with visualizationIf we want to visualize data, we will need to load some data into a DataFrame
###Code
import pandas as pd
# Load our data from the csv file
delays_df = pd.read_csv('Lots_of_flight_data.csv')
###Output
_____no_output_____
###Markdown
In order to display plots we need to import the **matplotlib** library
###Code
import matplotlib.pyplot as plt
###Output
_____no_output_____
###Markdown
A common plot used in data science is the scatter plot for checking the relationship between two columnsIf you see dots scattered everywhere, there is no correlation between the two columnsIf you see somethign resembling a line, there is a correlation between the two columnsYou can use the plot method of the DataFrame to draw the scatter plot* kind - the type of graph to draw* x - value to plot as x* y - value to plot as y* color - color to use for the graph points* alpha - opacity - useful to show density of points in a scatter plot* title - title of the graph
###Code
#Check if there is a relationship between the distance of a flight and how late the flight arrives
delays_df.plot(
kind='scatter',
x='DISTANCE',
y='ARR_DELAY',
color='blue',
alpha=0.3,
title='Correlation of arrival and distance'
)
plt.show()
#Check if there is a relationship between the how late the flight leaves and how late the flight arrives
delays_df.plot(
kind='scatter',
x='DEP_DELAY',
y='ARR_DELAY',
color='blue',
alpha=0.3,
title='Correlation of arrival and departure delay'
)
plt.show()
###Output
_____no_output_____
###Markdown
Visualizing data with matplotlib Somtimes graphs provide the best way to visualize dataThe **matplotlib** library allows you to draw graphs to help with visualizationIf we want to visualize data, we will need to load some data into a DataFrame
###Code
import pandas as pd
# Load our data from the csv file
delays_df = pd.read_csv('Lots_of_flight_data.csv')
###Output
_____no_output_____
###Markdown
In order to display plots we need to import the **matplotlib** library
###Code
import matplotlib.pyplot as plt
###Output
_____no_output_____
###Markdown
A common plot used in data science is the scatter plot for checking the relationship between two columnsIf you see dots scattered everywhere, there is no correlation between the two columnsIf you see somethign resembling a line, there is a correlation between the two columnsYou can use the plot method of the DataFrame to draw the scatter plot* kind - the type of graph to draw* x - value to plot as x* y - value to plot as y* color - color to use for the graph points* alpha - opacity - useful to show density of points in a scatter plot* title - title of the graph
###Code
#Check if there is a relationship between the distance of a flight and how late the flight arrives
delays_df.plot(
kind='scatter',
x='DISTANCE',
y='ARR_DELAY',
color='blue',
alpha=0.3,
title='Correlation of arrival and distance'
)
plt.show()
#Check if there is a relationship between the how late the flight leaves and how late the flight arrives
delays_df.plot(
kind='scatter',
x='DEP_DELAY',
y='ARR_DELAY',
color='blue',
alpha=0.3,
title='Correlation of arrival and departure delay'
)
plt.show()
###Output
_____no_output_____
###Markdown
The scatter plot allows us to see there is no correlation between distance and arrival delay but there is a strong correlation between departure delay and arrival delay.
###Code
plt.xlabel('Departure delay (minutes)')
plt.ylabel('Arrival delay (minutes)')
plt.title('Correlation Departure and Arrival Delay')
plt.scatter(x=delays_df['DEP_DELAY'],
y=delays_df['ARR_DELAY'],
color='blue', alpha=0.3)
plt.show()
# Grab our libaries
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
# Remove rows with null values since those will crash our linear regression model training
delays_df.dropna(inplace=True)
# Move our features into the X DataFrame
X = delays_df.loc[:,['DEP_DELAY']]
# Move our labels into the y DataFrame
y = delays_df.loc[:,['ARR_DELAY']]
# Split our data into test and training DataFrames
X_train, X_test, y_train, y_test = train_test_split(
X,
y,
test_size=0.3,
random_state=42
)
regressor = LinearRegression() # Create a scikit learn LinearRegression object
regressor.fit(X_train, y_train) # Use the fit method to train the model using your training data
y_pred = regressor.predict(X_test)
plt.xlabel('Departure delay (minutes)')
plt.ylabel('Arrival delay (minutes)')
plt.title('Predicted Arrival delays')
plt.plot(
X_test,
y_pred,
color='red',
linewidth=1)
plt.scatter(x=delays_df['DEP_DELAY'],
y=delays_df['ARR_DELAY'],
color='blue', alpha=0.3)
plt.show()
###Output
_____no_output_____
###Markdown
Matplotlib를 이용한 데이터 시각화 그래프는 데이터를 시각화하는 가장 좋은 방법입니다.**matplotlib** 라이브러리는 시각화를 돕는 그래프를 그리는 것을 제공합니다.데이터를 시각화하기 원한다면 DataFrame으로 데이터를 불러오는 것이 필요합니다.
###Code
import pandas as pd
# csv 파일로부터 데이터를 불러오기
delays_df = pd.read_csv('Data/Lots_of_flight_data.csv')
###Output
_____no_output_____
###Markdown
plot을 표시하려면 ** matplotlib ** 라이브러리를 가져와야합니다.
###Code
import matplotlib.pyplot as plt
###Output
_____no_output_____
###Markdown
데이터 과학에서 사용되는 일반적인 플롯은 두 열 간의 관계를 확인하기위한 산점도입니다.사방에 점이 흩어져있는 경우 두 열간에 상관 관계가없는 것입니다.선과 닮은 부분이 보이면 두 열 사이에 상관 관계가있는 것입니다.DataFrame의 plot 메서드를 사용하여 산점도를 그릴 수 있습니다.* kind - 그릴 그래프의 타입* x - x로 표시할 값* y - y로 표시할 값* color - 그래프 점에 사용할 색상* alpha - 불투명도 - 산점도에서 점의 밀도를 표시하는 데 유용합니다.* title - 그래프의 제목
###Code
# 항공편 거리와 항공편 도착 늦게 사이에 관계가 있는지 확인
delays_df.plot(
kind='scatter',
x='DISTANCE',
y='ARR_DELAY',
color='blue',
alpha=0.3,
title='Correlation of arrival and distance'
)
plt.show()
# 항공편 출발 시간과 도착 시간 사이에 관계가 있는지 확인하세요.
delays_df.plot(
kind='scatter',
x='DEP_DELAY',
y='ARR_DELAY',
color='blue',
alpha=0.3,
title='Correlation of arrival and departure delay'
)
plt.show()
###Output
_____no_output_____ |
shemi_piran_1990.ipynb | ###Markdown
[ADS Entry](http://adsabs.harvard.edu/abs/1990ApJ...365L..55S)
###Code
import sympy
sympy.init_printing()
###Output
_____no_output_____
###Markdown
One difficulty that I had with equation 1 is that is assumes that the pair production cross section decreases quadratically with the photon energy. The cross section for pair production can be found in equation 32 in [Weaver 1976](https://journals.aps.org/pra/abstract/10.1103/PhysRevA.13.1563). The full calculation can be found in [Theory of Photons and Electrons by Jauch and Rohrlich (1955)](https://archive.org/details/TheoryOfPhotonsElectrons). Equation 1
###Code
sigma_T = sympy.Symbol('sigma_T')
epsilon = sympy.Symbol('epsilon')
n_gamma = sympy.Symbol('n_gamma')
mathscrE = sympy.Symbol(r'\mathscr{E}')
mathscrR = sympy.Symbol(r'\mathscr{R}')
tau = sympy.Symbol('tau')
hbar = sympy.Symbol('hbar', positive=True)
c = sympy.Symbol('c',positive=True)
m_e = sympy.Symbol('m_e', positive=True)
R = sympy.Symbol('R', positive=True)
alpha = sympy.Symbol('alpha')
compton_wavelength = hbar/(m_e*c)
_ = sigma_T*n_gamma*R/epsilon**2
_ = _.subs(n_gamma,mathscrE/epsilon/R**3)
_ = _.subs(R,mathscrR*compton_wavelength)
_ = _.subs(sigma_T,alpha**2*compton_wavelength**2)
eqn_1 = sympy.Eq(tau,_)
eqn_1
###Output
_____no_output_____
###Markdown
To derive equation 2, we begin with the Fermi distribution function for electrons$n = \int_0^{\infty} \frac{p^2 dp}{\hbar^3} \left[1+\exp \left(\frac{\mu + \sqrt{p^2 c^2 + m_e^2 c^4}}{k T} \right)\right]^{-1} =$Since the electrons are in equilibrium with photons, the chemical potential vanishes $\mu = 0$$ = \int_0^{\infty} \frac{p^2 dp}{\hbar^3} \left[1+\exp \left(\frac{\sqrt{p^2 c^2 + m_e^2 c^4}}{k T} \right)\right]^{-1} \approx$Since we are dealing with temperatures much smaller than the electron rest mass energy, the exponential term in the denominator is much larger than unity$ \approx \int_0^{\infty} \frac{p^2 dp}{\hbar^3} \exp \left(-\frac{\sqrt{p^2 c^2 + m_e^2 c^4}}{k T} \right) \approx$Most of the contribution to the integral comes from the $p \ll m_e c$, so$ \approx \exp \left(-\frac{m_e c^2}{k T} \right) \int_0^{\infty} \frac{p^2 dp}{\hbar^3} \exp \left(-\frac{p^2}{m_e k T} \right) \approx \exp \left(-\frac{m_e c^2}{k T} \right) \left(\frac{m_e k T}{\hbar^2} \right)^{3/2}$
###Code
k = sympy.Symbol('k')
T = sympy.Symbol('T')
lambda_e = sympy.Symbol('lambda_e')
mathscrT = sympy.Symbol(r'\mathscr{T}')
n_pm = sympy.Symbol(r'n_{\pm}')
_ = sympy.exp(-m_e*c**2/k/T)*(m_e*k*T/hbar**2)**sympy.Rational(3,2)
_ = _.subs(T,mathscrT*m_e*c**2/k)
_ = _.subs(hbar,sympy.solve(sympy.Eq(lambda_e,compton_wavelength),hbar)[0])
eqn_2 = sympy.Eq(n_pm,_)
eqn_2
###Output
_____no_output_____
###Markdown
Equation 3
###Code
_ = sympy.Eq(mathscrE,R**3*(k*T)**4/(hbar*c)**3/(m_e*c**2))
_ = _.subs(T,mathscrT*m_e*c**2/k)
_ = _.subs(R, mathscrR*compton_wavelength)
eqn_3 = _
eqn_3
###Output
_____no_output_____
###Markdown
The energy density in the fireball is comparable to the pressure $p$. The total energy in the fireball is therefore $R^3 p$. In order to escape, a photon has to travel a distance $R$. If the mean free path is $l$, then the number of scatterings it takes for a photon to reach the same is $\left(R/l \right)^2$, and so the actual path the photon travels before escaping is larger $R^2/l$. The photon moves all the while at the speed of light, so the escape time is $\tau \approx \frac{R^2}{c l} \approx \frac{R}{c} \tau$. The luminosity is therefore $L \approx \frac{p R^2 c}{\tau}$.The ratio between the luminosity and mechanical power is$\frac{L}{p dV/dt} \approx {p R^2 c}{p R^2 dR/dt} \frac{1}{\tau} \approx \frac{1}{\tau}$ Equation 5
###Code
V = sympy.Symbol('V')
gamma = sympy.Symbol('gamma')
_ = V**(gamma-1)*T
_ = _.subs(gamma,sympy.Rational(4,3))
_ = _.subs(V,R**3)
_.simplify()
###Output
_____no_output_____ |
notebooks/rna_graph_tutorial.ipynb | ###Markdown
Graphein RNA Graph Construction Tutorial[API Reference](https://graphein.ai/modules/graphein.rna.html)In this notebook we construct graphs of RNA secondary structures.The inputs we require are a sequence (optional) and a [dotbracket](https://www.tbi.univie.ac.at/RNA/ViennaRNA/doc/html/rna_structure_notations.html) specification of the secondary structure.The workflow we follow is similar to the other data modalities. The desired edge constructions are passed as a list of functions to the construction function.[](https://colab.research.google.com/github/a-r-j/graphein/blob/master/notebooks/rna_graph_tutorial.ipynb)
###Code
# Install Graphein if necessary
# !pip install graphein
import logging
logging.getLogger("matplotlib").setLevel(logging.WARNING)
logging.getLogger("graphein").setLevel(logging.WARNING)
from typing import List, Callable
import networkx as nx
from graphein.rna.graphs import construct_rna_graph
from graphein.rna.edges import (
add_all_dotbracket_edges,
add_pseudoknots,
add_phosphodiester_bonds,
add_base_pairing_interactions
)
###Output
_____no_output_____
###Markdown
Construction with a DotbracketGraph construction is handled by the [`construct_rna_graph()`](https://graphein.ai/modules/graphein.rna.htmlgraphein.rna.graphs.construct_rna_graph) function.
###Code
from graphein.rna.visualisation import plot_rna_graph
edge_funcs_1: List[Callable] = [
add_base_pairing_interactions,
add_phosphodiester_bonds,
add_pseudoknots,
]
edge_funcs_2: List[Callable] = [add_all_dotbracket_edges]
g = construct_rna_graph(
"......((((((......[[[))))))......]]]....",
sequence=None,
edge_construction_funcs=edge_funcs_1,
)
nx.draw(g)
plot_rna_graph(g, layout=nx.layout.spring_layout)
###Output
WARNING:graphein.rna.visualisation:No sequence data found in graph. Skipping base type labelling.
###Markdown
Construction with a dotbracket and sequence
###Code
g = construct_rna_graph(
sequence="CGUCUUAAACUCAUCACCGUGUGGAGCUGCGACCCUUCCCUAGAUUCGAAGACGAG",
dotbracket="((((((...(((..(((...))))))...(((..((.....))..)))))))))..",
edge_construction_funcs=edge_funcs_1,
)
plot_rna_graph(g, layout=nx.layout.spring_layout)
g = construct_rna_graph(
sequence="CGUCUUAAACUCAUCACCGUGUGGAGCUGCGACCCUUCCCUAGAUUCGAAGACGAG",
dotbracket="((((((...(((..(((...))))))...(((..((.....))..)))))))))..",
edge_construction_funcs=edge_funcs_1,
)
plot_rna_graph(g, layout=nx.layout.circular_layout)
###Output
_____no_output_____
###Markdown
Graphein RNA Graph Construction TutorialIn this notebook we construct graphs of RNA secondary structures.The inputs we require are a sequence (optional) and a [dotbracket](https://www.tbi.univie.ac.at/RNA/ViennaRNA/doc/html/rna_structure_notations.html) specification of the secondary structure. The workflow we follow is similar to the other data modalities. The desired edge constructions are passed as a list of functions to the construction function. Construction with a dotbracket
###Code
from graphein.rna.visualisation import plot_rna_graph
edge_funcs_1: List[Callable] = [
add_base_pairing_interactions,
add_phosphodiester_bonds,
add_pseudoknots,
]
edge_funcs_2: List[Callable] = [add_all_dotbracket_edges]
g = construct_rna_graph(
"......((((((......[[[))))))......]]]....",
sequence=None,
edge_construction_funcs=edge_funcs_1,
)
nx.draw(g)
###Output
_____no_output_____
###Markdown
Construction with a dotbracket and sequence
###Code
g = construct_rna_graph(
sequence="CGUCUUAAACUCAUCACCGUGUGGAGCUGCGACCCUUCCCUAGAUUCGAAGACGAG",
dotbracket="((((((...(((..(((...))))))...(((..((.....))..)))))))))..",
edge_construction_funcs=edge_funcs_1,
)
plot_rna_graph(g)
###Output
_____no_output_____ |
Scalable-Machine-Learning/spark_tutorial_student.ipynb | ###Markdown
 +  **Spark Tutorial: Learning Apache Spark** This tutorial will teach you how to use [Apache Spark](http://spark.apache.org/), a framework for large-scale data processing, within a notebook. Many traditional frameworks were designed to be run on a single computer. However, many datasets today are too large to be stored on a single computer, and even when a dataset can be stored on one computer (such as the datasets in this tutorial), the dataset can often be processed much more quickly using multiple computers. Spark has efficient implementations of a number of transformations and actions that can be composed together to perform data processing and analysis. Spark excels at distributing these operations across a cluster while abstracting away many of the underlying implementation details. Spark has been designed with a focus on scalability and efficiency. With Spark you can begin developing your solution on your laptop, using a small dataset, and then use that same code to process terabytes or even petabytes across a distributed cluster. **During this tutorial we will cover:** *Part 1:* Basic notebook usage and [Python](https://docs.python.org/2/) integration *Part 2:* An introduction to using [Apache Spark](https://spark.apache.org/) with the Python [pySpark API](https://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD) running in the browser *Part 3:* Using RDDs and chaining together transformations and actions *Part 4:* Lambda functions *Part 5:* Additional RDD actions *Part 6:* Additional RDD transformations *Part 7:* Caching RDDs and storage options *Part 8:* Debugging Spark applications and lazy evaluation The following transformations will be covered:* `map()`, `mapPartitions()`, `mapPartitionsWithIndex()`, `filter()`, `flatMap()`, `reduceByKey()`, `groupByKey()` The following actions will be covered:* `first()`, `take()`, `takeSample()`, `takeOrdered()`, `collect()`, `count()`, `countByValue()`, `reduce()`, `top()` Also covered:* `cache()`, `unpersist()`, `id()`, `setName()` Note that, for reference, you can look up the details of these methods in [Spark's Python API](https://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD) **Part 1: Basic notebook usage and [Python](https://docs.python.org/2/) integration ** **(1a) Notebook usage** A notebook is comprised of a linear sequence of cells. These cells can contain either markdown or code, but we won't mix both in one cell. When a markdown cell is executed it renders formatted text, images, and links just like HTML in a normal webpage. The text you are reading right now is part of a markdown cell. Python code cells allow you to execute arbitrary Python commands just like in any Python shell. Place your cursor inside the cell below, and press "Shift" + "Enter" to execute the code and advance to the next cell. You can also press "Ctrl" + "Enter" to execute the code and remain in the cell. These commands work the same in both markdown and code cells.
###Code
# This is a Python cell. You can run normal Python code here...
print 'The sum of 1 and 1 is {0}'.format(1+1)
# Here is another Python cell, this time with a variable (x) declaration and an if statement:
x = 42
if x > 40:
print 'The sum of 1 and 2 is {0}'.format(1+2)
###Output
The sum of 1 and 2 is 3
###Markdown
**(1b) Notebook state** As you work through a notebook it is important that you run all of the code cells. The notebook is stateful, which means that variables and their values are retained until the notebook is detached (in Databricks Cloud) or the kernel is restarted (in IPython notebooks). If you do not run all of the code cells as you proceed through the notebook, your variables will not be properly initialized and later code might fail. You will also need to rerun any cells that you have modified in order for the changes to be available to other cells.
###Code
# This cell relies on x being defined already.
# If we didn't run the cells from part (1a) this code would fail.
print x * 2
###Output
84
###Markdown
**(1c) Library imports** We can import standard Python libraries ([modules](https://docs.python.org/2/tutorial/modules.html)) the usual way. An `import` statement will import the specified module. In this tutorial and future labs, we will provide any imports that are necessary.
###Code
# Import the regular expression library
import re
m = re.search('(?<=abc)def', 'abcdef')
m.group(0)
# Import the datetime library
import datetime
print 'This was last run on: {0}'.format(datetime.datetime.now())
###Output
This was last run on: 2018-03-14 08:31:43.498335
###Markdown
**Part 2: An introduction to using [Apache Spark](https://spark.apache.org/) with the Python [pySpark API](https://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD) running in the browser** **Spark Context** In Spark, communication occurs between a driver and executors. The driver has Spark jobs that it needs to run and these jobs are split into tasks that are submitted to the executors for completion. The results from these tasks are delivered back to the driver. In part 1, we saw that normal python code can be executed via cells. When using Databricks Cloud this code gets executed in the Spark driver's Java Virtual Machine (JVM) and not in an executor's JVM, and when using an IPython notebook it is executed within the kernel associated with the notebook. Since no Spark functionality is actually being used, no tasks are launched on the executors. In order to use Spark and its API we will need to use a `SparkContext`. When running Spark, you start a new Spark application by creating a [SparkContext](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.SparkContext). When the `SparkContext` is created, it asks the master for some cores to use to do work. The master sets these cores aside just for you; they won't be used for other applications. When using Databricks Cloud or the virtual machine provisioned for this class, the `SparkContext` is created for you automatically as `sc`. **(2a) Example Cluster** The diagram below shows an example cluster, where the cores allocated for an application are outlined in purple.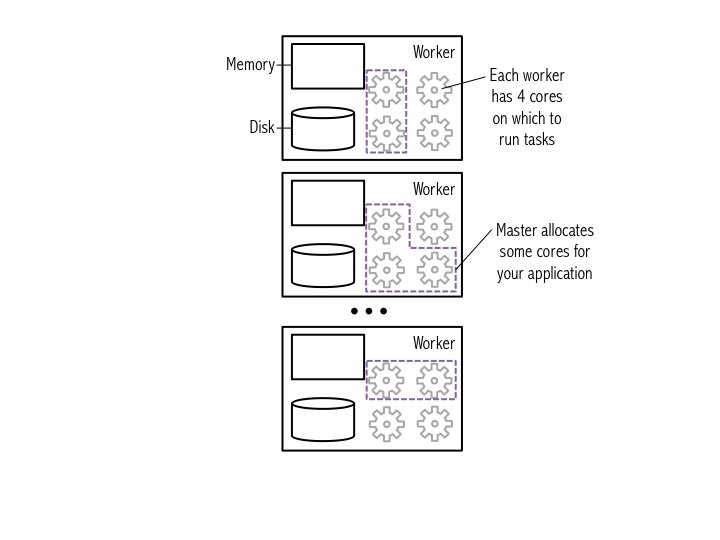 You can view the details of your Spark application in the Spark web UI. The web UI is accessible in Databricks cloud by going to "Clusters" and then clicking on the "View Spark UI" link for your cluster. When running locally you'll find it at [localhost:4040](http://localhost:4040). In the web UI, under the "Jobs" tab, you can see a list of jobs that have been scheduled or run. It's likely there isn't any thing interesting here yet because we haven't run any jobs, but we'll return to this page later. At a high level, every Spark application consists of a driver program that launches various parallel operations on executor Java Virtual Machines (JVMs) running either in a cluster or locally on the same machine. In Databricks Cloud, "Databricks Shell" is the driver program. When running locally, "PySparkShell" is the driver program. In all cases, this driver program contains the main loop for the program and creates distributed datasets on the cluster, then applies operations (transformations & actions) to those datasets. Driver programs access Spark through a SparkContext object, which represents a connection to a computing cluster. A Spark context object (`sc`) is the main entry point for Spark functionality. A Spark context can be used to create Resilient Distributed Datasets (RDDs) on a cluster. Try printing out `sc` to see its type.
###Code
# Display the type of the Spark Context sc
type(sc)
###Output
_____no_output_____
###Markdown
**(2b) `SparkContext` attributes** You can use Python's [dir()](https://docs.python.org/2/library/functions.html?highlight=dirdir) function to get a list of all the attributes (including methods) accessible through the `sc` object.
###Code
# List sc's attributes
dir(sc)
###Output
_____no_output_____
###Markdown
**(2c) Getting help** Alternatively, you can use Python's [help()](https://docs.python.org/2/library/functions.html?highlight=helphelp) function to get an easier to read list of all the attributes, including examples, that the `sc` object has.
###Code
# Use help to obtain more detailed information
help(sc)
# After reading the help we've decided we want to use sc.version to see what version of Spark we are running
sc.version
# Help can be used on any Python object
help(map)
###Output
Help on built-in function map in module __builtin__:
map(...)
map(function, sequence[, sequence, ...]) -> list
Return a list of the results of applying the function to the items of
the argument sequence(s). If more than one sequence is given, the
function is called with an argument list consisting of the corresponding
item of each sequence, substituting None for missing values when not all
sequences have the same length. If the function is None, return a list of
the items of the sequence (or a list of tuples if more than one sequence).
###Markdown
**Part 3: Using RDDs and chaining together transformations and actions** **Working with your first RDD** In Spark, we first create a base [Resilient Distributed Dataset](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD) (RDD). We can then apply one or more transformations to that base RDD. *An RDD is immutable, so once it is created, it cannot be changed.* As a result, each transformation creates a new RDD. Finally, we can apply one or more actions to the RDDs. Note that Spark uses lazy evaluation, so transformations are not actually executed until an action occurs. We will perform several exercises to obtain a better understanding of RDDs:* Create a Python collection of 10,000 integers* Create a Spark base RDD from that collection* Subtract one from each value using `map`* Perform action `collect` to view results* Perform action `count` to view counts* Apply transformation `filter` and view results with `collect`* Learn about lambda functions* Explore how lazy evaluation works and the debugging challenges that it introduces **(3a) Create a Python collection of integers in the range of 1 .. 10000** We will use the [xrange()](https://docs.python.org/2/library/functions.html?highlight=xrangexrange) function to create a [list()](https://docs.python.org/2/library/functions.html?highlight=listlist) of integers. `xrange()` only generates values as they are needed. This is different from the behavior of [range()](https://docs.python.org/2/library/functions.html?highlight=rangerange) which generates the complete list upon execution. Because of this `xrange()` is more memory efficient than `range()`, especially for large ranges.
###Code
data = xrange(1, 10001)
# Data is just a normal Python list
# Obtain data's first element
data[0]
# We can check the size of the list using the len() function
len(data)
###Output
_____no_output_____
###Markdown
**(3b) Distributed data and using a collection to create an RDD** In Spark, datasets are represented as a list of entries, where the list is broken up into many different partitions that are each stored on a different machine. Each partition holds a unique subset of the entries in the list. Spark calls datasets that it stores "Resilient Distributed Datasets" (RDDs). One of the defining features of Spark, compared to other data analytics frameworks (e.g., Hadoop), is that it stores data in memory rather than on disk. This allows Spark applications to run much more quickly, because they are not slowed down by needing to read data from disk. The figure below illustrates how Spark breaks a list of data entries into partitions that are each stored in memory on a worker.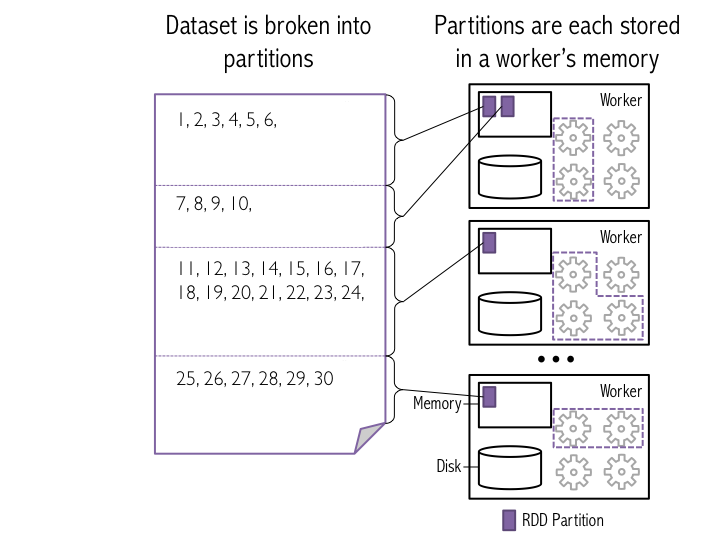 To create the RDD, we use `sc.parallelize()`, which tells Spark to create a new set of input data based on data that is passed in. In this example, we will provide an `xrange`. The second argument to the [sc.parallelize()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.SparkContext.parallelize) method tells Spark how many partitions to break the data into when it stores the data in memory (we'll talk more about this later in this tutorial). Note that for better performance when using `parallelize`, `xrange()` is recommended if the input represents a range. This is the reason why we used `xrange()` in 3a. There are many different types of RDDs. The base class for RDDs is [pyspark.RDD](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD) and other RDDs subclass `pyspark.RDD`. Since the other RDD types inherit from `pyspark.RDD` they have the same APIs and are functionally identical. We'll see that `sc.parallelize()` generates a `pyspark.rdd.PipelinedRDD` when its input is an `xrange`, and a `pyspark.RDD` when its input is a `range`. After we generate RDDs, we can view them in the "Storage" tab of the web UI. You'll notice that new datasets are not listed until Spark needs to return a result due to an action being executed. This feature of Spark is called "lazy evaluation". This allows Spark to avoid performing unnecessary calculations.
###Code
# Parallelize data using 8 partitions
# This operation is a transformation of data into an RDD
# Spark uses lazy evaluation, so no Spark jobs are run at this point
xrangeRDD = sc.parallelize(data, 8)
# Let's view help on parallelize
help(sc.parallelize)
# Let's see what type sc.parallelize() returned
print 'type of xrangeRDD: {0}'.format(type(xrangeRDD))
# How about if we use a range
dataRange = range(1, 10001)
rangeRDD = sc.parallelize(dataRange, 8)
print 'type of dataRangeRDD: {0}'.format(type(rangeRDD))
# Each RDD gets a unique ID
print 'xrangeRDD id: {0}'.format(xrangeRDD.id())
print 'rangeRDD id: {0}'.format(rangeRDD.id())
# We can name each newly created RDD using the setName() method
xrangeRDD.setName('My first RDD')
# Let's view the lineage (the set of transformations) of the RDD using toDebugString()
print xrangeRDD.toDebugString()
# Let's use help to see what methods we can call on this RDD
help(xrangeRDD)
# Let's see how many partitions the RDD will be split into by using the getNumPartitions()
xrangeRDD.getNumPartitions()
###Output
_____no_output_____
###Markdown
**(3c): Subtract one from each value using `map`** So far, we've created a distributed dataset that is split into many partitions, where each partition is stored on a single machine in our cluster. Let's look at what happens when we do a basic operation on the dataset. Many useful data analysis operations can be specified as "do something to each item in the dataset". These data-parallel operations are convenient because each item in the dataset can be processed individually: the operation on one entry doesn't effect the operations on any of the other entries. Therefore, Spark can parallelize the operation. `map(f)`, the most common Spark transformation, is one such example: it applies a function `f` to each item in the dataset, and outputs the resulting dataset. When you run `map()` on a dataset, a single *stage* of tasks is launched. A *stage* is a group of tasks that all perform the same computation, but on different input data. One task is launched for each partitition, as shown in the example below. A task is a unit of execution that runs on a single machine. When we run `map(f)` within a partition, a new *task* applies `f` to all of the entries in a particular partition, and outputs a new partition. In this example figure, the dataset is broken into four partitions, so four `map()` tasks are launched.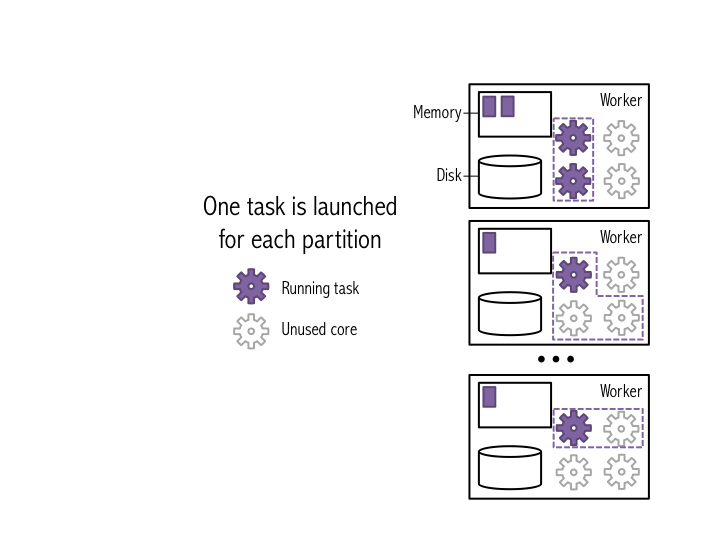 The figure below shows how this would work on the smaller data set from the earlier figures. Note that one task is launched for each partition.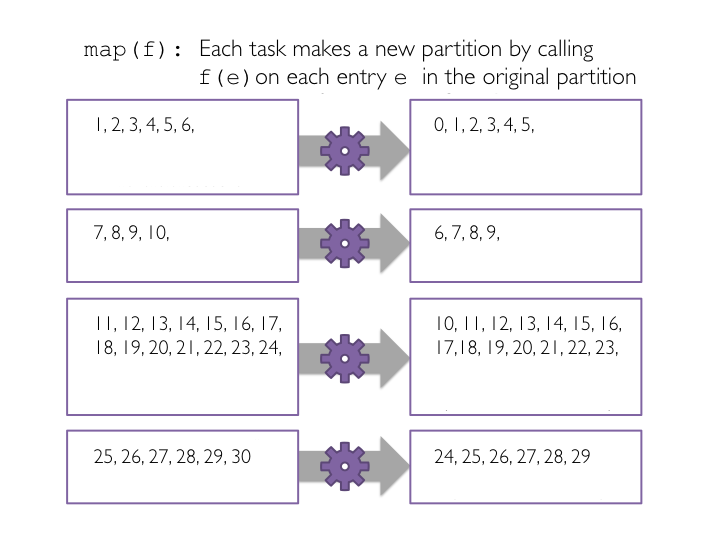 When applying the `map()` transformation, each item in the parent RDD will map to one element in the new RDD. So, if the parent RDD has twenty elements, the new RDD will also have twenty items. Now we will use `map()` to subtract one from each value in the base RDD we just created. First, we define a Python function called `sub()` that will subtract one from the input integer. Second, we will pass each item in the base RDD into a `map()` transformation that applies the `sub()` function to each element. And finally, we print out the RDD transformation hierarchy using `toDebugString()`.
###Code
# Create sub function to subtract 1
def sub(value):
""""Subtracts one from `value`.
Args:
value (int): A number.
Returns:
int: `value` minus one.
"""
return (value - 1)
# Transform xrangeRDD through map transformation using sub function
# Because map is a transformation and Spark uses lazy evaluation, no jobs, stages,
# or tasks will be launched when we run this code.
subRDD = xrangeRDD.map(sub)
# Let's see the RDD transformation hierarchy
print subRDD.toDebugString()
###Output
(8) PythonRDD[3] at RDD at PythonRDD.scala:43 []
| ParallelCollectionRDD[0] at parallelize at PythonRDD.scala:392 []
###Markdown
** (3d) Perform action `collect` to view results ** To see a list of elements decremented by one, we need to create a new list on the driver from the the data distributed in the executor nodes. To do this we call the `collect()` method on our RDD. `collect()` is often used after a filter or other operation to ensure that we are only returning a *small* amount of data to the driver. This is done because the data returned to the driver must fit into the driver's available memory. If not, the driver will crash. The `collect()` method is the first action operation that we have encountered. Action operations cause Spark to perform the (lazy) transformation operations that are required to compute the RDD returned by the action. In our example, this means that tasks will now be launched to perform the `parallelize`, `map`, and `collect` operations. In this example, the dataset is broken into four partitions, so four `collect()` tasks are launched. Each task collects the entries in its partition and sends the result to the SparkContext, which creates a list of the values, as shown in the figure below.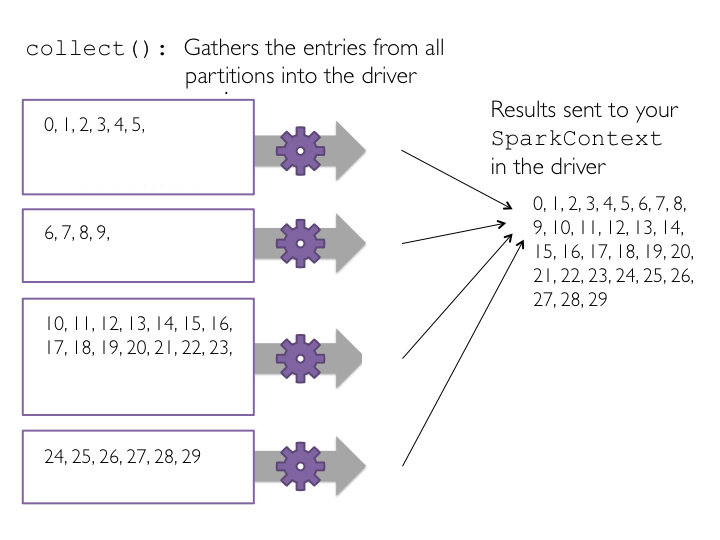 The above figures showed what would happen if we ran `collect()` on a small example dataset with just four partitions. Now let's run `collect()` on `subRDD`.
###Code
# Let's collect the data
print subRDD.collect()
###Output
[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210, 211, 212, 213, 214, 215, 216, 217, 218, 219, 220, 221, 222, 223, 224, 225, 226, 227, 228, 229, 230, 231, 232, 233, 234, 235, 236, 237, 238, 239, 240, 241, 242, 243, 244, 245, 246, 247, 248, 249, 250, 251, 252, 253, 254, 255, 256, 257, 258, 259, 260, 261, 262, 263, 264, 265, 266, 267, 268, 269, 270, 271, 272, 273, 274, 275, 276, 277, 278, 279, 280, 281, 282, 283, 284, 285, 286, 287, 288, 289, 290, 291, 292, 293, 294, 295, 296, 297, 298, 299, 300, 301, 302, 303, 304, 305, 306, 307, 308, 309, 310, 311, 312, 313, 314, 315, 316, 317, 318, 319, 320, 321, 322, 323, 324, 325, 326, 327, 328, 329, 330, 331, 332, 333, 334, 335, 336, 337, 338, 339, 340, 341, 342, 343, 344, 345, 346, 347, 348, 349, 350, 351, 352, 353, 354, 355, 356, 357, 358, 359, 360, 361, 362, 363, 364, 365, 366, 367, 368, 369, 370, 371, 372, 373, 374, 375, 376, 377, 378, 379, 380, 381, 382, 383, 384, 385, 386, 387, 388, 389, 390, 391, 392, 393, 394, 395, 396, 397, 398, 399, 400, 401, 402, 403, 404, 405, 406, 407, 408, 409, 410, 411, 412, 413, 414, 415, 416, 417, 418, 419, 420, 421, 422, 423, 424, 425, 426, 427, 428, 429, 430, 431, 432, 433, 434, 435, 436, 437, 438, 439, 440, 441, 442, 443, 444, 445, 446, 447, 448, 449, 450, 451, 452, 453, 454, 455, 456, 457, 458, 459, 460, 461, 462, 463, 464, 465, 466, 467, 468, 469, 470, 471, 472, 473, 474, 475, 476, 477, 478, 479, 480, 481, 482, 483, 484, 485, 486, 487, 488, 489, 490, 491, 492, 493, 494, 495, 496, 497, 498, 499, 500, 501, 502, 503, 504, 505, 506, 507, 508, 509, 510, 511, 512, 513, 514, 515, 516, 517, 518, 519, 520, 521, 522, 523, 524, 525, 526, 527, 528, 529, 530, 531, 532, 533, 534, 535, 536, 537, 538, 539, 540, 541, 542, 543, 544, 545, 546, 547, 548, 549, 550, 551, 552, 553, 554, 555, 556, 557, 558, 559, 560, 561, 562, 563, 564, 565, 566, 567, 568, 569, 570, 571, 572, 573, 574, 575, 576, 577, 578, 579, 580, 581, 582, 583, 584, 585, 586, 587, 588, 589, 590, 591, 592, 593, 594, 595, 596, 597, 598, 599, 600, 601, 602, 603, 604, 605, 606, 607, 608, 609, 610, 611, 612, 613, 614, 615, 616, 617, 618, 619, 620, 621, 622, 623, 624, 625, 626, 627, 628, 629, 630, 631, 632, 633, 634, 635, 636, 637, 638, 639, 640, 641, 642, 643, 644, 645, 646, 647, 648, 649, 650, 651, 652, 653, 654, 655, 656, 657, 658, 659, 660, 661, 662, 663, 664, 665, 666, 667, 668, 669, 670, 671, 672, 673, 674, 675, 676, 677, 678, 679, 680, 681, 682, 683, 684, 685, 686, 687, 688, 689, 690, 691, 692, 693, 694, 695, 696, 697, 698, 699, 700, 701, 702, 703, 704, 705, 706, 707, 708, 709, 710, 711, 712, 713, 714, 715, 716, 717, 718, 719, 720, 721, 722, 723, 724, 725, 726, 727, 728, 729, 730, 731, 732, 733, 734, 735, 736, 737, 738, 739, 740, 741, 742, 743, 744, 745, 746, 747, 748, 749, 750, 751, 752, 753, 754, 755, 756, 757, 758, 759, 760, 761, 762, 763, 764, 765, 766, 767, 768, 769, 770, 771, 772, 773, 774, 775, 776, 777, 778, 779, 780, 781, 782, 783, 784, 785, 786, 787, 788, 789, 790, 791, 792, 793, 794, 795, 796, 797, 798, 799, 800, 801, 802, 803, 804, 805, 806, 807, 808, 809, 810, 811, 812, 813, 814, 815, 816, 817, 818, 819, 820, 821, 822, 823, 824, 825, 826, 827, 828, 829, 830, 831, 832, 833, 834, 835, 836, 837, 838, 839, 840, 841, 842, 843, 844, 845, 846, 847, 848, 849, 850, 851, 852, 853, 854, 855, 856, 857, 858, 859, 860, 861, 862, 863, 864, 865, 866, 867, 868, 869, 870, 871, 872, 873, 874, 875, 876, 877, 878, 879, 880, 881, 882, 883, 884, 885, 886, 887, 888, 889, 890, 891, 892, 893, 894, 895, 896, 897, 898, 899, 900, 901, 902, 903, 904, 905, 906, 907, 908, 909, 910, 911, 912, 913, 914, 915, 916, 917, 918, 919, 920, 921, 922, 923, 924, 925, 926, 927, 928, 929, 930, 931, 932, 933, 934, 935, 936, 937, 938, 939, 940, 941, 942, 943, 944, 945, 946, 947, 948, 949, 950, 951, 952, 953, 954, 955, 956, 957, 958, 959, 960, 961, 962, 963, 964, 965, 966, 967, 968, 969, 970, 971, 972, 973, 974, 975, 976, 977, 978, 979, 980, 981, 982, 983, 984, 985, 986, 987, 988, 989, 990, 991, 992, 993, 994, 995, 996, 997, 998, 999, 1000, 1001, 1002, 1003, 1004, 1005, 1006, 1007, 1008, 1009, 1010, 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1018, 1019, 1020, 1021, 1022, 1023, 1024, 1025, 1026, 1027, 1028, 1029, 1030, 1031, 1032, 1033, 1034, 1035, 1036, 1037, 1038, 1039, 1040, 1041, 1042, 1043, 1044, 1045, 1046, 1047, 1048, 1049, 1050, 1051, 1052, 1053, 1054, 1055, 1056, 1057, 1058, 1059, 1060, 1061, 1062, 1063, 1064, 1065, 1066, 1067, 1068, 1069, 1070, 1071, 1072, 1073, 1074, 1075, 1076, 1077, 1078, 1079, 1080, 1081, 1082, 1083, 1084, 1085, 1086, 1087, 1088, 1089, 1090, 1091, 1092, 1093, 1094, 1095, 1096, 1097, 1098, 1099, 1100, 1101, 1102, 1103, 1104, 1105, 1106, 1107, 1108, 1109, 1110, 1111, 1112, 1113, 1114, 1115, 1116, 1117, 1118, 1119, 1120, 1121, 1122, 1123, 1124, 1125, 1126, 1127, 1128, 1129, 1130, 1131, 1132, 1133, 1134, 1135, 1136, 1137, 1138, 1139, 1140, 1141, 1142, 1143, 1144, 1145, 1146, 1147, 1148, 1149, 1150, 1151, 1152, 1153, 1154, 1155, 1156, 1157, 1158, 1159, 1160, 1161, 1162, 1163, 1164, 1165, 1166, 1167, 1168, 1169, 1170, 1171, 1172, 1173, 1174, 1175, 1176, 1177, 1178, 1179, 1180, 1181, 1182, 1183, 1184, 1185, 1186, 1187, 1188, 1189, 1190, 1191, 1192, 1193, 1194, 1195, 1196, 1197, 1198, 1199, 1200, 1201, 1202, 1203, 1204, 1205, 1206, 1207, 1208, 1209, 1210, 1211, 1212, 1213, 1214, 1215, 1216, 1217, 1218, 1219, 1220, 1221, 1222, 1223, 1224, 1225, 1226, 1227, 1228, 1229, 1230, 1231, 1232, 1233, 1234, 1235, 1236, 1237, 1238, 1239, 1240, 1241, 1242, 1243, 1244, 1245, 1246, 1247, 1248, 1249, 1250, 1251, 1252, 1253, 1254, 1255, 1256, 1257, 1258, 1259, 1260, 1261, 1262, 1263, 1264, 1265, 1266, 1267, 1268, 1269, 1270, 1271, 1272, 1273, 1274, 1275, 1276, 1277, 1278, 1279, 1280, 1281, 1282, 1283, 1284, 1285, 1286, 1287, 1288, 1289, 1290, 1291, 1292, 1293, 1294, 1295, 1296, 1297, 1298, 1299, 1300, 1301, 1302, 1303, 1304, 1305, 1306, 1307, 1308, 1309, 1310, 1311, 1312, 1313, 1314, 1315, 1316, 1317, 1318, 1319, 1320, 1321, 1322, 1323, 1324, 1325, 1326, 1327, 1328, 1329, 1330, 1331, 1332, 1333, 1334, 1335, 1336, 1337, 1338, 1339, 1340, 1341, 1342, 1343, 1344, 1345, 1346, 1347, 1348, 1349, 1350, 1351, 1352, 1353, 1354, 1355, 1356, 1357, 1358, 1359, 1360, 1361, 1362, 1363, 1364, 1365, 1366, 1367, 1368, 1369, 1370, 1371, 1372, 1373, 1374, 1375, 1376, 1377, 1378, 1379, 1380, 1381, 1382, 1383, 1384, 1385, 1386, 1387, 1388, 1389, 1390, 1391, 1392, 1393, 1394, 1395, 1396, 1397, 1398, 1399, 1400, 1401, 1402, 1403, 1404, 1405, 1406, 1407, 1408, 1409, 1410, 1411, 1412, 1413, 1414, 1415, 1416, 1417, 1418, 1419, 1420, 1421, 1422, 1423, 1424, 1425, 1426, 1427, 1428, 1429, 1430, 1431, 1432, 1433, 1434, 1435, 1436, 1437, 1438, 1439, 1440, 1441, 1442, 1443, 1444, 1445, 1446, 1447, 1448, 1449, 1450, 1451, 1452, 1453, 1454, 1455, 1456, 1457, 1458, 1459, 1460, 1461, 1462, 1463, 1464, 1465, 1466, 1467, 1468, 1469, 1470, 1471, 1472, 1473, 1474, 1475, 1476, 1477, 1478, 1479, 1480, 1481, 1482, 1483, 1484, 1485, 1486, 1487, 1488, 1489, 1490, 1491, 1492, 1493, 1494, 1495, 1496, 1497, 1498, 1499, 1500, 1501, 1502, 1503, 1504, 1505, 1506, 1507, 1508, 1509, 1510, 1511, 1512, 1513, 1514, 1515, 1516, 1517, 1518, 1519, 1520, 1521, 1522, 1523, 1524, 1525, 1526, 1527, 1528, 1529, 1530, 1531, 1532, 1533, 1534, 1535, 1536, 1537, 1538, 1539, 1540, 1541, 1542, 1543, 1544, 1545, 1546, 1547, 1548, 1549, 1550, 1551, 1552, 1553, 1554, 1555, 1556, 1557, 1558, 1559, 1560, 1561, 1562, 1563, 1564, 1565, 1566, 1567, 1568, 1569, 1570, 1571, 1572, 1573, 1574, 1575, 1576, 1577, 1578, 1579, 1580, 1581, 1582, 1583, 1584, 1585, 1586, 1587, 1588, 1589, 1590, 1591, 1592, 1593, 1594, 1595, 1596, 1597, 1598, 1599, 1600, 1601, 1602, 1603, 1604, 1605, 1606, 1607, 1608, 1609, 1610, 1611, 1612, 1613, 1614, 1615, 1616, 1617, 1618, 1619, 1620, 1621, 1622, 1623, 1624, 1625, 1626, 1627, 1628, 1629, 1630, 1631, 1632, 1633, 1634, 1635, 1636, 1637, 1638, 1639, 1640, 1641, 1642, 1643, 1644, 1645, 1646, 1647, 1648, 1649, 1650, 1651, 1652, 1653, 1654, 1655, 1656, 1657, 1658, 1659, 1660, 1661, 1662, 1663, 1664, 1665, 1666, 1667, 1668, 1669, 1670, 1671, 1672, 1673, 1674, 1675, 1676, 1677, 1678, 1679, 1680, 1681, 1682, 1683, 1684, 1685, 1686, 1687, 1688, 1689, 1690, 1691, 1692, 1693, 1694, 1695, 1696, 1697, 1698, 1699, 1700, 1701, 1702, 1703, 1704, 1705, 1706, 1707, 1708, 1709, 1710, 1711, 1712, 1713, 1714, 1715, 1716, 1717, 1718, 1719, 1720, 1721, 1722, 1723, 1724, 1725, 1726, 1727, 1728, 1729, 1730, 1731, 1732, 1733, 1734, 1735, 1736, 1737, 1738, 1739, 1740, 1741, 1742, 1743, 1744, 1745, 1746, 1747, 1748, 1749, 1750, 1751, 1752, 1753, 1754, 1755, 1756, 1757, 1758, 1759, 1760, 1761, 1762, 1763, 1764, 1765, 1766, 1767, 1768, 1769, 1770, 1771, 1772, 1773, 1774, 1775, 1776, 1777, 1778, 1779, 1780, 1781, 1782, 1783, 1784, 1785, 1786, 1787, 1788, 1789, 1790, 1791, 1792, 1793, 1794, 1795, 1796, 1797, 1798, 1799, 1800, 1801, 1802, 1803, 1804, 1805, 1806, 1807, 1808, 1809, 1810, 1811, 1812, 1813, 1814, 1815, 1816, 1817, 1818, 1819, 1820, 1821, 1822, 1823, 1824, 1825, 1826, 1827, 1828, 1829, 1830, 1831, 1832, 1833, 1834, 1835, 1836, 1837, 1838, 1839, 1840, 1841, 1842, 1843, 1844, 1845, 1846, 1847, 1848, 1849, 1850, 1851, 1852, 1853, 1854, 1855, 1856, 1857, 1858, 1859, 1860, 1861, 1862, 1863, 1864, 1865, 1866, 1867, 1868, 1869, 1870, 1871, 1872, 1873, 1874, 1875, 1876, 1877, 1878, 1879, 1880, 1881, 1882, 1883, 1884, 1885, 1886, 1887, 1888, 1889, 1890, 1891, 1892, 1893, 1894, 1895, 1896, 1897, 1898, 1899, 1900, 1901, 1902, 1903, 1904, 1905, 1906, 1907, 1908, 1909, 1910, 1911, 1912, 1913, 1914, 1915, 1916, 1917, 1918, 1919, 1920, 1921, 1922, 1923, 1924, 1925, 1926, 1927, 1928, 1929, 1930, 1931, 1932, 1933, 1934, 1935, 1936, 1937, 1938, 1939, 1940, 1941, 1942, 1943, 1944, 1945, 1946, 1947, 1948, 1949, 1950, 1951, 1952, 1953, 1954, 1955, 1956, 1957, 1958, 1959, 1960, 1961, 1962, 1963, 1964, 1965, 1966, 1967, 1968, 1969, 1970, 1971, 1972, 1973, 1974, 1975, 1976, 1977, 1978, 1979, 1980, 1981, 1982, 1983, 1984, 1985, 1986, 1987, 1988, 1989, 1990, 1991, 1992, 1993, 1994, 1995, 1996, 1997, 1998, 1999, 2000, 2001, 2002, 2003, 2004, 2005, 2006, 2007, 2008, 2009, 2010, 2011, 2012, 2013, 2014, 2015, 2016, 2017, 2018, 2019, 2020, 2021, 2022, 2023, 2024, 2025, 2026, 2027, 2028, 2029, 2030, 2031, 2032, 2033, 2034, 2035, 2036, 2037, 2038, 2039, 2040, 2041, 2042, 2043, 2044, 2045, 2046, 2047, 2048, 2049, 2050, 2051, 2052, 2053, 2054, 2055, 2056, 2057, 2058, 2059, 2060, 2061, 2062, 2063, 2064, 2065, 2066, 2067, 2068, 2069, 2070, 2071, 2072, 2073, 2074, 2075, 2076, 2077, 2078, 2079, 2080, 2081, 2082, 2083, 2084, 2085, 2086, 2087, 2088, 2089, 2090, 2091, 2092, 2093, 2094, 2095, 2096, 2097, 2098, 2099, 2100, 2101, 2102, 2103, 2104, 2105, 2106, 2107, 2108, 2109, 2110, 2111, 2112, 2113, 2114, 2115, 2116, 2117, 2118, 2119, 2120, 2121, 2122, 2123, 2124, 2125, 2126, 2127, 2128, 2129, 2130, 2131, 2132, 2133, 2134, 2135, 2136, 2137, 2138, 2139, 2140, 2141, 2142, 2143, 2144, 2145, 2146, 2147, 2148, 2149, 2150, 2151, 2152, 2153, 2154, 2155, 2156, 2157, 2158, 2159, 2160, 2161, 2162, 2163, 2164, 2165, 2166, 2167, 2168, 2169, 2170, 2171, 2172, 2173, 2174, 2175, 2176, 2177, 2178, 2179, 2180, 2181, 2182, 2183, 2184, 2185, 2186, 2187, 2188, 2189, 2190, 2191, 2192, 2193, 2194, 2195, 2196, 2197, 2198, 2199, 2200, 2201, 2202, 2203, 2204, 2205, 2206, 2207, 2208, 2209, 2210, 2211, 2212, 2213, 2214, 2215, 2216, 2217, 2218, 2219, 2220, 2221, 2222, 2223, 2224, 2225, 2226, 2227, 2228, 2229, 2230, 2231, 2232, 2233, 2234, 2235, 2236, 2237, 2238, 2239, 2240, 2241, 2242, 2243, 2244, 2245, 2246, 2247, 2248, 2249, 2250, 2251, 2252, 2253, 2254, 2255, 2256, 2257, 2258, 2259, 2260, 2261, 2262, 2263, 2264, 2265, 2266, 2267, 2268, 2269, 2270, 2271, 2272, 2273, 2274, 2275, 2276, 2277, 2278, 2279, 2280, 2281, 2282, 2283, 2284, 2285, 2286, 2287, 2288, 2289, 2290, 2291, 2292, 2293, 2294, 2295, 2296, 2297, 2298, 2299, 2300, 2301, 2302, 2303, 2304, 2305, 2306, 2307, 2308, 2309, 2310, 2311, 2312, 2313, 2314, 2315, 2316, 2317, 2318, 2319, 2320, 2321, 2322, 2323, 2324, 2325, 2326, 2327, 2328, 2329, 2330, 2331, 2332, 2333, 2334, 2335, 2336, 2337, 2338, 2339, 2340, 2341, 2342, 2343, 2344, 2345, 2346, 2347, 2348, 2349, 2350, 2351, 2352, 2353, 2354, 2355, 2356, 2357, 2358, 2359, 2360, 2361, 2362, 2363, 2364, 2365, 2366, 2367, 2368, 2369, 2370, 2371, 2372, 2373, 2374, 2375, 2376, 2377, 2378, 2379, 2380, 2381, 2382, 2383, 2384, 2385, 2386, 2387, 2388, 2389, 2390, 2391, 2392, 2393, 2394, 2395, 2396, 2397, 2398, 2399, 2400, 2401, 2402, 2403, 2404, 2405, 2406, 2407, 2408, 2409, 2410, 2411, 2412, 2413, 2414, 2415, 2416, 2417, 2418, 2419, 2420, 2421, 2422, 2423, 2424, 2425, 2426, 2427, 2428, 2429, 2430, 2431, 2432, 2433, 2434, 2435, 2436, 2437, 2438, 2439, 2440, 2441, 2442, 2443, 2444, 2445, 2446, 2447, 2448, 2449, 2450, 2451, 2452, 2453, 2454, 2455, 2456, 2457, 2458, 2459, 2460, 2461, 2462, 2463, 2464, 2465, 2466, 2467, 2468, 2469, 2470, 2471, 2472, 2473, 2474, 2475, 2476, 2477, 2478, 2479, 2480, 2481, 2482, 2483, 2484, 2485, 2486, 2487, 2488, 2489, 2490, 2491, 2492, 2493, 2494, 2495, 2496, 2497, 2498, 2499, 2500, 2501, 2502, 2503, 2504, 2505, 2506, 2507, 2508, 2509, 2510, 2511, 2512, 2513, 2514, 2515, 2516, 2517, 2518, 2519, 2520, 2521, 2522, 2523, 2524, 2525, 2526, 2527, 2528, 2529, 2530, 2531, 2532, 2533, 2534, 2535, 2536, 2537, 2538, 2539, 2540, 2541, 2542, 2543, 2544, 2545, 2546, 2547, 2548, 2549, 2550, 2551, 2552, 2553, 2554, 2555, 2556, 2557, 2558, 2559, 2560, 2561, 2562, 2563, 2564, 2565, 2566, 2567, 2568, 2569, 2570, 2571, 2572, 2573, 2574, 2575, 2576, 2577, 2578, 2579, 2580, 2581, 2582, 2583, 2584, 2585, 2586, 2587, 2588, 2589, 2590, 2591, 2592, 2593, 2594, 2595, 2596, 2597, 2598, 2599, 2600, 2601, 2602, 2603, 2604, 2605, 2606, 2607, 2608, 2609, 2610, 2611, 2612, 2613, 2614, 2615, 2616, 2617, 2618, 2619, 2620, 2621, 2622, 2623, 2624, 2625, 2626, 2627, 2628, 2629, 2630, 2631, 2632, 2633, 2634, 2635, 2636, 2637, 2638, 2639, 2640, 2641, 2642, 2643, 2644, 2645, 2646, 2647, 2648, 2649, 2650, 2651, 2652, 2653, 2654, 2655, 2656, 2657, 2658, 2659, 2660, 2661, 2662, 2663, 2664, 2665, 2666, 2667, 2668, 2669, 2670, 2671, 2672, 2673, 2674, 2675, 2676, 2677, 2678, 2679, 2680, 2681, 2682, 2683, 2684, 2685, 2686, 2687, 2688, 2689, 2690, 2691, 2692, 2693, 2694, 2695, 2696, 2697, 2698, 2699, 2700, 2701, 2702, 2703, 2704, 2705, 2706, 2707, 2708, 2709, 2710, 2711, 2712, 2713, 2714, 2715, 2716, 2717, 2718, 2719, 2720, 2721, 2722, 2723, 2724, 2725, 2726, 2727, 2728, 2729, 2730, 2731, 2732, 2733, 2734, 2735, 2736, 2737, 2738, 2739, 2740, 2741, 2742, 2743, 2744, 2745, 2746, 2747, 2748, 2749, 2750, 2751, 2752, 2753, 2754, 2755, 2756, 2757, 2758, 2759, 2760, 2761, 2762, 2763, 2764, 2765, 2766, 2767, 2768, 2769, 2770, 2771, 2772, 2773, 2774, 2775, 2776, 2777, 2778, 2779, 2780, 2781, 2782, 2783, 2784, 2785, 2786, 2787, 2788, 2789, 2790, 2791, 2792, 2793, 2794, 2795, 2796, 2797, 2798, 2799, 2800, 2801, 2802, 2803, 2804, 2805, 2806, 2807, 2808, 2809, 2810, 2811, 2812, 2813, 2814, 2815, 2816, 2817, 2818, 2819, 2820, 2821, 2822, 2823, 2824, 2825, 2826, 2827, 2828, 2829, 2830, 2831, 2832, 2833, 2834, 2835, 2836, 2837, 2838, 2839, 2840, 2841, 2842, 2843, 2844, 2845, 2846, 2847, 2848, 2849, 2850, 2851, 2852, 2853, 2854, 2855, 2856, 2857, 2858, 2859, 2860, 2861, 2862, 2863, 2864, 2865, 2866, 2867, 2868, 2869, 2870, 2871, 2872, 2873, 2874, 2875, 2876, 2877, 2878, 2879, 2880, 2881, 2882, 2883, 2884, 2885, 2886, 2887, 2888, 2889, 2890, 2891, 2892, 2893, 2894, 2895, 2896, 2897, 2898, 2899, 2900, 2901, 2902, 2903, 2904, 2905, 2906, 2907, 2908, 2909, 2910, 2911, 2912, 2913, 2914, 2915, 2916, 2917, 2918, 2919, 2920, 2921, 2922, 2923, 2924, 2925, 2926, 2927, 2928, 2929, 2930, 2931, 2932, 2933, 2934, 2935, 2936, 2937, 2938, 2939, 2940, 2941, 2942, 2943, 2944, 2945, 2946, 2947, 2948, 2949, 2950, 2951, 2952, 2953, 2954, 2955, 2956, 2957, 2958, 2959, 2960, 2961, 2962, 2963, 2964, 2965, 2966, 2967, 2968, 2969, 2970, 2971, 2972, 2973, 2974, 2975, 2976, 2977, 2978, 2979, 2980, 2981, 2982, 2983, 2984, 2985, 2986, 2987, 2988, 2989, 2990, 2991, 2992, 2993, 2994, 2995, 2996, 2997, 2998, 2999, 3000, 3001, 3002, 3003, 3004, 3005, 3006, 3007, 3008, 3009, 3010, 3011, 3012, 3013, 3014, 3015, 3016, 3017, 3018, 3019, 3020, 3021, 3022, 3023, 3024, 3025, 3026, 3027, 3028, 3029, 3030, 3031, 3032, 3033, 3034, 3035, 3036, 3037, 3038, 3039, 3040, 3041, 3042, 3043, 3044, 3045, 3046, 3047, 3048, 3049, 3050, 3051, 3052, 3053, 3054, 3055, 3056, 3057, 3058, 3059, 3060, 3061, 3062, 3063, 3064, 3065, 3066, 3067, 3068, 3069, 3070, 3071, 3072, 3073, 3074, 3075, 3076, 3077, 3078, 3079, 3080, 3081, 3082, 3083, 3084, 3085, 3086, 3087, 3088, 3089, 3090, 3091, 3092, 3093, 3094, 3095, 3096, 3097, 3098, 3099, 3100, 3101, 3102, 3103, 3104, 3105, 3106, 3107, 3108, 3109, 3110, 3111, 3112, 3113, 3114, 3115, 3116, 3117, 3118, 3119, 3120, 3121, 3122, 3123, 3124, 3125, 3126, 3127, 3128, 3129, 3130, 3131, 3132, 3133, 3134, 3135, 3136, 3137, 3138, 3139, 3140, 3141, 3142, 3143, 3144, 3145, 3146, 3147, 3148, 3149, 3150, 3151, 3152, 3153, 3154, 3155, 3156, 3157, 3158, 3159, 3160, 3161, 3162, 3163, 3164, 3165, 3166, 3167, 3168, 3169, 3170, 3171, 3172, 3173, 3174, 3175, 3176, 3177, 3178, 3179, 3180, 3181, 3182, 3183, 3184, 3185, 3186, 3187, 3188, 3189, 3190, 3191, 3192, 3193, 3194, 3195, 3196, 3197, 3198, 3199, 3200, 3201, 3202, 3203, 3204, 3205, 3206, 3207, 3208, 3209, 3210, 3211, 3212, 3213, 3214, 3215, 3216, 3217, 3218, 3219, 3220, 3221, 3222, 3223, 3224, 3225, 3226, 3227, 3228, 3229, 3230, 3231, 3232, 3233, 3234, 3235, 3236, 3237, 3238, 3239, 3240, 3241, 3242, 3243, 3244, 3245, 3246, 3247, 3248, 3249, 3250, 3251, 3252, 3253, 3254, 3255, 3256, 3257, 3258, 3259, 3260, 3261, 3262, 3263, 3264, 3265, 3266, 3267, 3268, 3269, 3270, 3271, 3272, 3273, 3274, 3275, 3276, 3277, 3278, 3279, 3280, 3281, 3282, 3283, 3284, 3285, 3286, 3287, 3288, 3289, 3290, 3291, 3292, 3293, 3294, 3295, 3296, 3297, 3298, 3299, 3300, 3301, 3302, 3303, 3304, 3305, 3306, 3307, 3308, 3309, 3310, 3311, 3312, 3313, 3314, 3315, 3316, 3317, 3318, 3319, 3320, 3321, 3322, 3323, 3324, 3325, 3326, 3327, 3328, 3329, 3330, 3331, 3332, 3333, 3334, 3335, 3336, 3337, 3338, 3339, 3340, 3341, 3342, 3343, 3344, 3345, 3346, 3347, 3348, 3349, 3350, 3351, 3352, 3353, 3354, 3355, 3356, 3357, 3358, 3359, 3360, 3361, 3362, 3363, 3364, 3365, 3366, 3367, 3368, 3369, 3370, 3371, 3372, 3373, 3374, 3375, 3376, 3377, 3378, 3379, 3380, 3381, 3382, 3383, 3384, 3385, 3386, 3387, 3388, 3389, 3390, 3391, 3392, 3393, 3394, 3395, 3396, 3397, 3398, 3399, 3400, 3401, 3402, 3403, 3404, 3405, 3406, 3407, 3408, 3409, 3410, 3411, 3412, 3413, 3414, 3415, 3416, 3417, 3418, 3419, 3420, 3421, 3422, 3423, 3424, 3425, 3426, 3427, 3428, 3429, 3430, 3431, 3432, 3433, 3434, 3435, 3436, 3437, 3438, 3439, 3440, 3441, 3442, 3443, 3444, 3445, 3446, 3447, 3448, 3449, 3450, 3451, 3452, 3453, 3454, 3455, 3456, 3457, 3458, 3459, 3460, 3461, 3462, 3463, 3464, 3465, 3466, 3467, 3468, 3469, 3470, 3471, 3472, 3473, 3474, 3475, 3476, 3477, 3478, 3479, 3480, 3481, 3482, 3483, 3484, 3485, 3486, 3487, 3488, 3489, 3490, 3491, 3492, 3493, 3494, 3495, 3496, 3497, 3498, 3499, 3500, 3501, 3502, 3503, 3504, 3505, 3506, 3507, 3508, 3509, 3510, 3511, 3512, 3513, 3514, 3515, 3516, 3517, 3518, 3519, 3520, 3521, 3522, 3523, 3524, 3525, 3526, 3527, 3528, 3529, 3530, 3531, 3532, 3533, 3534, 3535, 3536, 3537, 3538, 3539, 3540, 3541, 3542, 3543, 3544, 3545, 3546, 3547, 3548, 3549, 3550, 3551, 3552, 3553, 3554, 3555, 3556, 3557, 3558, 3559, 3560, 3561, 3562, 3563, 3564, 3565, 3566, 3567, 3568, 3569, 3570, 3571, 3572, 3573, 3574, 3575, 3576, 3577, 3578, 3579, 3580, 3581, 3582, 3583, 3584, 3585, 3586, 3587, 3588, 3589, 3590, 3591, 3592, 3593, 3594, 3595, 3596, 3597, 3598, 3599, 3600, 3601, 3602, 3603, 3604, 3605, 3606, 3607, 3608, 3609, 3610, 3611, 3612, 3613, 3614, 3615, 3616, 3617, 3618, 3619, 3620, 3621, 3622, 3623, 3624, 3625, 3626, 3627, 3628, 3629, 3630, 3631, 3632, 3633, 3634, 3635, 3636, 3637, 3638, 3639, 3640, 3641, 3642, 3643, 3644, 3645, 3646, 3647, 3648, 3649, 3650, 3651, 3652, 3653, 3654, 3655, 3656, 3657, 3658, 3659, 3660, 3661, 3662, 3663, 3664, 3665, 3666, 3667, 3668, 3669, 3670, 3671, 3672, 3673, 3674, 3675, 3676, 3677, 3678, 3679, 3680, 3681, 3682, 3683, 3684, 3685, 3686, 3687, 3688, 3689, 3690, 3691, 3692, 3693, 3694, 3695, 3696, 3697, 3698, 3699, 3700, 3701, 3702, 3703, 3704, 3705, 3706, 3707, 3708, 3709, 3710, 3711, 3712, 3713, 3714, 3715, 3716, 3717, 3718, 3719, 3720, 3721, 3722, 3723, 3724, 3725, 3726, 3727, 3728, 3729, 3730, 3731, 3732, 3733, 3734, 3735, 3736, 3737, 3738, 3739, 3740, 3741, 3742, 3743, 3744, 3745, 3746, 3747, 3748, 3749, 3750, 3751, 3752, 3753, 3754, 3755, 3756, 3757, 3758, 3759, 3760, 3761, 3762, 3763, 3764, 3765, 3766, 3767, 3768, 3769, 3770, 3771, 3772, 3773, 3774, 3775, 3776, 3777, 3778, 3779, 3780, 3781, 3782, 3783, 3784, 3785, 3786, 3787, 3788, 3789, 3790, 3791, 3792, 3793, 3794, 3795, 3796, 3797, 3798, 3799, 3800, 3801, 3802, 3803, 3804, 3805, 3806, 3807, 3808, 3809, 3810, 3811, 3812, 3813, 3814, 3815, 3816, 3817, 3818, 3819, 3820, 3821, 3822, 3823, 3824, 3825, 3826, 3827, 3828, 3829, 3830, 3831, 3832, 3833, 3834, 3835, 3836, 3837, 3838, 3839, 3840, 3841, 3842, 3843, 3844, 3845, 3846, 3847, 3848, 3849, 3850, 3851, 3852, 3853, 3854, 3855, 3856, 3857, 3858, 3859, 3860, 3861, 3862, 3863, 3864, 3865, 3866, 3867, 3868, 3869, 3870, 3871, 3872, 3873, 3874, 3875, 3876, 3877, 3878, 3879, 3880, 3881, 3882, 3883, 3884, 3885, 3886, 3887, 3888, 3889, 3890, 3891, 3892, 3893, 3894, 3895, 3896, 3897, 3898, 3899, 3900, 3901, 3902, 3903, 3904, 3905, 3906, 3907, 3908, 3909, 3910, 3911, 3912, 3913, 3914, 3915, 3916, 3917, 3918, 3919, 3920, 3921, 3922, 3923, 3924, 3925, 3926, 3927, 3928, 3929, 3930, 3931, 3932, 3933, 3934, 3935, 3936, 3937, 3938, 3939, 3940, 3941, 3942, 3943, 3944, 3945, 3946, 3947, 3948, 3949, 3950, 3951, 3952, 3953, 3954, 3955, 3956, 3957, 3958, 3959, 3960, 3961, 3962, 3963, 3964, 3965, 3966, 3967, 3968, 3969, 3970, 3971, 3972, 3973, 3974, 3975, 3976, 3977, 3978, 3979, 3980, 3981, 3982, 3983, 3984, 3985, 3986, 3987, 3988, 3989, 3990, 3991, 3992, 3993, 3994, 3995, 3996, 3997, 3998, 3999, 4000, 4001, 4002, 4003, 4004, 4005, 4006, 4007, 4008, 4009, 4010, 4011, 4012, 4013, 4014, 4015, 4016, 4017, 4018, 4019, 4020, 4021, 4022, 4023, 4024, 4025, 4026, 4027, 4028, 4029, 4030, 4031, 4032, 4033, 4034, 4035, 4036, 4037, 4038, 4039, 4040, 4041, 4042, 4043, 4044, 4045, 4046, 4047, 4048, 4049, 4050, 4051, 4052, 4053, 4054, 4055, 4056, 4057, 4058, 4059, 4060, 4061, 4062, 4063, 4064, 4065, 4066, 4067, 4068, 4069, 4070, 4071, 4072, 4073, 4074, 4075, 4076, 4077, 4078, 4079, 4080, 4081, 4082, 4083, 4084, 4085, 4086, 4087, 4088, 4089, 4090, 4091, 4092, 4093, 4094, 4095, 4096, 4097, 4098, 4099, 4100, 4101, 4102, 4103, 4104, 4105, 4106, 4107, 4108, 4109, 4110, 4111, 4112, 4113, 4114, 4115, 4116, 4117, 4118, 4119, 4120, 4121, 4122, 4123, 4124, 4125, 4126, 4127, 4128, 4129, 4130, 4131, 4132, 4133, 4134, 4135, 4136, 4137, 4138, 4139, 4140, 4141, 4142, 4143, 4144, 4145, 4146, 4147, 4148, 4149, 4150, 4151, 4152, 4153, 4154, 4155, 4156, 4157, 4158, 4159, 4160, 4161, 4162, 4163, 4164, 4165, 4166, 4167, 4168, 4169, 4170, 4171, 4172, 4173, 4174, 4175, 4176, 4177, 4178, 4179, 4180, 4181, 4182, 4183, 4184, 4185, 4186, 4187, 4188, 4189, 4190, 4191, 4192, 4193, 4194, 4195, 4196, 4197, 4198, 4199, 4200, 4201, 4202, 4203, 4204, 4205, 4206, 4207, 4208, 4209, 4210, 4211, 4212, 4213, 4214, 4215, 4216, 4217, 4218, 4219, 4220, 4221, 4222, 4223, 4224, 4225, 4226, 4227, 4228, 4229, 4230, 4231, 4232, 4233, 4234, 4235, 4236, 4237, 4238, 4239, 4240, 4241, 4242, 4243, 4244, 4245, 4246, 4247, 4248, 4249, 4250, 4251, 4252, 4253, 4254, 4255, 4256, 4257, 4258, 4259, 4260, 4261, 4262, 4263, 4264, 4265, 4266, 4267, 4268, 4269, 4270, 4271, 4272, 4273, 4274, 4275, 4276, 4277, 4278, 4279, 4280, 4281, 4282, 4283, 4284, 4285, 4286, 4287, 4288, 4289, 4290, 4291, 4292, 4293, 4294, 4295, 4296, 4297, 4298, 4299, 4300, 4301, 4302, 4303, 4304, 4305, 4306, 4307, 4308, 4309, 4310, 4311, 4312, 4313, 4314, 4315, 4316, 4317, 4318, 4319, 4320, 4321, 4322, 4323, 4324, 4325, 4326, 4327, 4328, 4329, 4330, 4331, 4332, 4333, 4334, 4335, 4336, 4337, 4338, 4339, 4340, 4341, 4342, 4343, 4344, 4345, 4346, 4347, 4348, 4349, 4350, 4351, 4352, 4353, 4354, 4355, 4356, 4357, 4358, 4359, 4360, 4361, 4362, 4363, 4364, 4365, 4366, 4367, 4368, 4369, 4370, 4371, 4372, 4373, 4374, 4375, 4376, 4377, 4378, 4379, 4380, 4381, 4382, 4383, 4384, 4385, 4386, 4387, 4388, 4389, 4390, 4391, 4392, 4393, 4394, 4395, 4396, 4397, 4398, 4399, 4400, 4401, 4402, 4403, 4404, 4405, 4406, 4407, 4408, 4409, 4410, 4411, 4412, 4413, 4414, 4415, 4416, 4417, 4418, 4419, 4420, 4421, 4422, 4423, 4424, 4425, 4426, 4427, 4428, 4429, 4430, 4431, 4432, 4433, 4434, 4435, 4436, 4437, 4438, 4439, 4440, 4441, 4442, 4443, 4444, 4445, 4446, 4447, 4448, 4449, 4450, 4451, 4452, 4453, 4454, 4455, 4456, 4457, 4458, 4459, 4460, 4461, 4462, 4463, 4464, 4465, 4466, 4467, 4468, 4469, 4470, 4471, 4472, 4473, 4474, 4475, 4476, 4477, 4478, 4479, 4480, 4481, 4482, 4483, 4484, 4485, 4486, 4487, 4488, 4489, 4490, 4491, 4492, 4493, 4494, 4495, 4496, 4497, 4498, 4499, 4500, 4501, 4502, 4503, 4504, 4505, 4506, 4507, 4508, 4509, 4510, 4511, 4512, 4513, 4514, 4515, 4516, 4517, 4518, 4519, 4520, 4521, 4522, 4523, 4524, 4525, 4526, 4527, 4528, 4529, 4530, 4531, 4532, 4533, 4534, 4535, 4536, 4537, 4538, 4539, 4540, 4541, 4542, 4543, 4544, 4545, 4546, 4547, 4548, 4549, 4550, 4551, 4552, 4553, 4554, 4555, 4556, 4557, 4558, 4559, 4560, 4561, 4562, 4563, 4564, 4565, 4566, 4567, 4568, 4569, 4570, 4571, 4572, 4573, 4574, 4575, 4576, 4577, 4578, 4579, 4580, 4581, 4582, 4583, 4584, 4585, 4586, 4587, 4588, 4589, 4590, 4591, 4592, 4593, 4594, 4595, 4596, 4597, 4598, 4599, 4600, 4601, 4602, 4603, 4604, 4605, 4606, 4607, 4608, 4609, 4610, 4611, 4612, 4613, 4614, 4615, 4616, 4617, 4618, 4619, 4620, 4621, 4622, 4623, 4624, 4625, 4626, 4627, 4628, 4629, 4630, 4631, 4632, 4633, 4634, 4635, 4636, 4637, 4638, 4639, 4640, 4641, 4642, 4643, 4644, 4645, 4646, 4647, 4648, 4649, 4650, 4651, 4652, 4653, 4654, 4655, 4656, 4657, 4658, 4659, 4660, 4661, 4662, 4663, 4664, 4665, 4666, 4667, 4668, 4669, 4670, 4671, 4672, 4673, 4674, 4675, 4676, 4677, 4678, 4679, 4680, 4681, 4682, 4683, 4684, 4685, 4686, 4687, 4688, 4689, 4690, 4691, 4692, 4693, 4694, 4695, 4696, 4697, 4698, 4699, 4700, 4701, 4702, 4703, 4704, 4705, 4706, 4707, 4708, 4709, 4710, 4711, 4712, 4713, 4714, 4715, 4716, 4717, 4718, 4719, 4720, 4721, 4722, 4723, 4724, 4725, 4726, 4727, 4728, 4729, 4730, 4731, 4732, 4733, 4734, 4735, 4736, 4737, 4738, 4739, 4740, 4741, 4742, 4743, 4744, 4745, 4746, 4747, 4748, 4749, 4750, 4751, 4752, 4753, 4754, 4755, 4756, 4757, 4758, 4759, 4760, 4761, 4762, 4763, 4764, 4765, 4766, 4767, 4768, 4769, 4770, 4771, 4772, 4773, 4774, 4775, 4776, 4777, 4778, 4779, 4780, 4781, 4782, 4783, 4784, 4785, 4786, 4787, 4788, 4789, 4790, 4791, 4792, 4793, 4794, 4795, 4796, 4797, 4798, 4799, 4800, 4801, 4802, 4803, 4804, 4805, 4806, 4807, 4808, 4809, 4810, 4811, 4812, 4813, 4814, 4815, 4816, 4817, 4818, 4819, 4820, 4821, 4822, 4823, 4824, 4825, 4826, 4827, 4828, 4829, 4830, 4831, 4832, 4833, 4834, 4835, 4836, 4837, 4838, 4839, 4840, 4841, 4842, 4843, 4844, 4845, 4846, 4847, 4848, 4849, 4850, 4851, 4852, 4853, 4854, 4855, 4856, 4857, 4858, 4859, 4860, 4861, 4862, 4863, 4864, 4865, 4866, 4867, 4868, 4869, 4870, 4871, 4872, 4873, 4874, 4875, 4876, 4877, 4878, 4879, 4880, 4881, 4882, 4883, 4884, 4885, 4886, 4887, 4888, 4889, 4890, 4891, 4892, 4893, 4894, 4895, 4896, 4897, 4898, 4899, 4900, 4901, 4902, 4903, 4904, 4905, 4906, 4907, 4908, 4909, 4910, 4911, 4912, 4913, 4914, 4915, 4916, 4917, 4918, 4919, 4920, 4921, 4922, 4923, 4924, 4925, 4926, 4927, 4928, 4929, 4930, 4931, 4932, 4933, 4934, 4935, 4936, 4937, 4938, 4939, 4940, 4941, 4942, 4943, 4944, 4945, 4946, 4947, 4948, 4949, 4950, 4951, 4952, 4953, 4954, 4955, 4956, 4957, 4958, 4959, 4960, 4961, 4962, 4963, 4964, 4965, 4966, 4967, 4968, 4969, 4970, 4971, 4972, 4973, 4974, 4975, 4976, 4977, 4978, 4979, 4980, 4981, 4982, 4983, 4984, 4985, 4986, 4987, 4988, 4989, 4990, 4991, 4992, 4993, 4994, 4995, 4996, 4997, 4998, 4999, 5000, 5001, 5002, 5003, 5004, 5005, 5006, 5007, 5008, 5009, 5010, 5011, 5012, 5013, 5014, 5015, 5016, 5017, 5018, 5019, 5020, 5021, 5022, 5023, 5024, 5025, 5026, 5027, 5028, 5029, 5030, 5031, 5032, 5033, 5034, 5035, 5036, 5037, 5038, 5039, 5040, 5041, 5042, 5043, 5044, 5045, 5046, 5047, 5048, 5049, 5050, 5051, 5052, 5053, 5054, 5055, 5056, 5057, 5058, 5059, 5060, 5061, 5062, 5063, 5064, 5065, 5066, 5067, 5068, 5069, 5070, 5071, 5072, 5073, 5074, 5075, 5076, 5077, 5078, 5079, 5080, 5081, 5082, 5083, 5084, 5085, 5086, 5087, 5088, 5089, 5090, 5091, 5092, 5093, 5094, 5095, 5096, 5097, 5098, 5099, 5100, 5101, 5102, 5103, 5104, 5105, 5106, 5107, 5108, 5109, 5110, 5111, 5112, 5113, 5114, 5115, 5116, 5117, 5118, 5119, 5120, 5121, 5122, 5123, 5124, 5125, 5126, 5127, 5128, 5129, 5130, 5131, 5132, 5133, 5134, 5135, 5136, 5137, 5138, 5139, 5140, 5141, 5142, 5143, 5144, 5145, 5146, 5147, 5148, 5149, 5150, 5151, 5152, 5153, 5154, 5155, 5156, 5157, 5158, 5159, 5160, 5161, 5162, 5163, 5164, 5165, 5166, 5167, 5168, 5169, 5170, 5171, 5172, 5173, 5174, 5175, 5176, 5177, 5178, 5179, 5180, 5181, 5182, 5183, 5184, 5185, 5186, 5187, 5188, 5189, 5190, 5191, 5192, 5193, 5194, 5195, 5196, 5197, 5198, 5199, 5200, 5201, 5202, 5203, 5204, 5205, 5206, 5207, 5208, 5209, 5210, 5211, 5212, 5213, 5214, 5215, 5216, 5217, 5218, 5219, 5220, 5221, 5222, 5223, 5224, 5225, 5226, 5227, 5228, 5229, 5230, 5231, 5232, 5233, 5234, 5235, 5236, 5237, 5238, 5239, 5240, 5241, 5242, 5243, 5244, 5245, 5246, 5247, 5248, 5249, 5250, 5251, 5252, 5253, 5254, 5255, 5256, 5257, 5258, 5259, 5260, 5261, 5262, 5263, 5264, 5265, 5266, 5267, 5268, 5269, 5270, 5271, 5272, 5273, 5274, 5275, 5276, 5277, 5278, 5279, 5280, 5281, 5282, 5283, 5284, 5285, 5286, 5287, 5288, 5289, 5290, 5291, 5292, 5293, 5294, 5295, 5296, 5297, 5298, 5299, 5300, 5301, 5302, 5303, 5304, 5305, 5306, 5307, 5308, 5309, 5310, 5311, 5312, 5313, 5314, 5315, 5316, 5317, 5318, 5319, 5320, 5321, 5322, 5323, 5324, 5325, 5326, 5327, 5328, 5329, 5330, 5331, 5332, 5333, 5334, 5335, 5336, 5337, 5338, 5339, 5340, 5341, 5342, 5343, 5344, 5345, 5346, 5347, 5348, 5349, 5350, 5351, 5352, 5353, 5354, 5355, 5356, 5357, 5358, 5359, 5360, 5361, 5362, 5363, 5364, 5365, 5366, 5367, 5368, 5369, 5370, 5371, 5372, 5373, 5374, 5375, 5376, 5377, 5378, 5379, 5380, 5381, 5382, 5383, 5384, 5385, 5386, 5387, 5388, 5389, 5390, 5391, 5392, 5393, 5394, 5395, 5396, 5397, 5398, 5399, 5400, 5401, 5402, 5403, 5404, 5405, 5406, 5407, 5408, 5409, 5410, 5411, 5412, 5413, 5414, 5415, 5416, 5417, 5418, 5419, 5420, 5421, 5422, 5423, 5424, 5425, 5426, 5427, 5428, 5429, 5430, 5431, 5432, 5433, 5434, 5435, 5436, 5437, 5438, 5439, 5440, 5441, 5442, 5443, 5444, 5445, 5446, 5447, 5448, 5449, 5450, 5451, 5452, 5453, 5454, 5455, 5456, 5457, 5458, 5459, 5460, 5461, 5462, 5463, 5464, 5465, 5466, 5467, 5468, 5469, 5470, 5471, 5472, 5473, 5474, 5475, 5476, 5477, 5478, 5479, 5480, 5481, 5482, 5483, 5484, 5485, 5486, 5487, 5488, 5489, 5490, 5491, 5492, 5493, 5494, 5495, 5496, 5497, 5498, 5499, 5500, 5501, 5502, 5503, 5504, 5505, 5506, 5507, 5508, 5509, 5510, 5511, 5512, 5513, 5514, 5515, 5516, 5517, 5518, 5519, 5520, 5521, 5522, 5523, 5524, 5525, 5526, 5527, 5528, 5529, 5530, 5531, 5532, 5533, 5534, 5535, 5536, 5537, 5538, 5539, 5540, 5541, 5542, 5543, 5544, 5545, 5546, 5547, 5548, 5549, 5550, 5551, 5552, 5553, 5554, 5555, 5556, 5557, 5558, 5559, 5560, 5561, 5562, 5563, 5564, 5565, 5566, 5567, 5568, 5569, 5570, 5571, 5572, 5573, 5574, 5575, 5576, 5577, 5578, 5579, 5580, 5581, 5582, 5583, 5584, 5585, 5586, 5587, 5588, 5589, 5590, 5591, 5592, 5593, 5594, 5595, 5596, 5597, 5598, 5599, 5600, 5601, 5602, 5603, 5604, 5605, 5606, 5607, 5608, 5609, 5610, 5611, 5612, 5613, 5614, 5615, 5616, 5617, 5618, 5619, 5620, 5621, 5622, 5623, 5624, 5625, 5626, 5627, 5628, 5629, 5630, 5631, 5632, 5633, 5634, 5635, 5636, 5637, 5638, 5639, 5640, 5641, 5642, 5643, 5644, 5645, 5646, 5647, 5648, 5649, 5650, 5651, 5652, 5653, 5654, 5655, 5656, 5657, 5658, 5659, 5660, 5661, 5662, 5663, 5664, 5665, 5666, 5667, 5668, 5669, 5670, 5671, 5672, 5673, 5674, 5675, 5676, 5677, 5678, 5679, 5680, 5681, 5682, 5683, 5684, 5685, 5686, 5687, 5688, 5689, 5690, 5691, 5692, 5693, 5694, 5695, 5696, 5697, 5698, 5699, 5700, 5701, 5702, 5703, 5704, 5705, 5706, 5707, 5708, 5709, 5710, 5711, 5712, 5713, 5714, 5715, 5716, 5717, 5718, 5719, 5720, 5721, 5722, 5723, 5724, 5725, 5726, 5727, 5728, 5729, 5730, 5731, 5732, 5733, 5734, 5735, 5736, 5737, 5738, 5739, 5740, 5741, 5742, 5743, 5744, 5745, 5746, 5747, 5748, 5749, 5750, 5751, 5752, 5753, 5754, 5755, 5756, 5757, 5758, 5759, 5760, 5761, 5762, 5763, 5764, 5765, 5766, 5767, 5768, 5769, 5770, 5771, 5772, 5773, 5774, 5775, 5776, 5777, 5778, 5779, 5780, 5781, 5782, 5783, 5784, 5785, 5786, 5787, 5788, 5789, 5790, 5791, 5792, 5793, 5794, 5795, 5796, 5797, 5798, 5799, 5800, 5801, 5802, 5803, 5804, 5805, 5806, 5807, 5808, 5809, 5810, 5811, 5812, 5813, 5814, 5815, 5816, 5817, 5818, 5819, 5820, 5821, 5822, 5823, 5824, 5825, 5826, 5827, 5828, 5829, 5830, 5831, 5832, 5833, 5834, 5835, 5836, 5837, 5838, 5839, 5840, 5841, 5842, 5843, 5844, 5845, 5846, 5847, 5848, 5849, 5850, 5851, 5852, 5853, 5854, 5855, 5856, 5857, 5858, 5859, 5860, 5861, 5862, 5863, 5864, 5865, 5866, 5867, 5868, 5869, 5870, 5871, 5872, 5873, 5874, 5875, 5876, 5877, 5878, 5879, 5880, 5881, 5882, 5883, 5884, 5885, 5886, 5887, 5888, 5889, 5890, 5891, 5892, 5893, 5894, 5895, 5896, 5897, 5898, 5899, 5900, 5901, 5902, 5903, 5904, 5905, 5906, 5907, 5908, 5909, 5910, 5911, 5912, 5913, 5914, 5915, 5916, 5917, 5918, 5919, 5920, 5921, 5922, 5923, 5924, 5925, 5926, 5927, 5928, 5929, 5930, 5931, 5932, 5933, 5934, 5935, 5936, 5937, 5938, 5939, 5940, 5941, 5942, 5943, 5944, 5945, 5946, 5947, 5948, 5949, 5950, 5951, 5952, 5953, 5954, 5955, 5956, 5957, 5958, 5959, 5960, 5961, 5962, 5963, 5964, 5965, 5966, 5967, 5968, 5969, 5970, 5971, 5972, 5973, 5974, 5975, 5976, 5977, 5978, 5979, 5980, 5981, 5982, 5983, 5984, 5985, 5986, 5987, 5988, 5989, 5990, 5991, 5992, 5993, 5994, 5995, 5996, 5997, 5998, 5999, 6000, 6001, 6002, 6003, 6004, 6005, 6006, 6007, 6008, 6009, 6010, 6011, 6012, 6013, 6014, 6015, 6016, 6017, 6018, 6019, 6020, 6021, 6022, 6023, 6024, 6025, 6026, 6027, 6028, 6029, 6030, 6031, 6032, 6033, 6034, 6035, 6036, 6037, 6038, 6039, 6040, 6041, 6042, 6043, 6044, 6045, 6046, 6047, 6048, 6049, 6050, 6051, 6052, 6053, 6054, 6055, 6056, 6057, 6058, 6059, 6060, 6061, 6062, 6063, 6064, 6065, 6066, 6067, 6068, 6069, 6070, 6071, 6072, 6073, 6074, 6075, 6076, 6077, 6078, 6079, 6080, 6081, 6082, 6083, 6084, 6085, 6086, 6087, 6088, 6089, 6090, 6091, 6092, 6093, 6094, 6095, 6096, 6097, 6098, 6099, 6100, 6101, 6102, 6103, 6104, 6105, 6106, 6107, 6108, 6109, 6110, 6111, 6112, 6113, 6114, 6115, 6116, 6117, 6118, 6119, 6120, 6121, 6122, 6123, 6124, 6125, 6126, 6127, 6128, 6129, 6130, 6131, 6132, 6133, 6134, 6135, 6136, 6137, 6138, 6139, 6140, 6141, 6142, 6143, 6144, 6145, 6146, 6147, 6148, 6149, 6150, 6151, 6152, 6153, 6154, 6155, 6156, 6157, 6158, 6159, 6160, 6161, 6162, 6163, 6164, 6165, 6166, 6167, 6168, 6169, 6170, 6171, 6172, 6173, 6174, 6175, 6176, 6177, 6178, 6179, 6180, 6181, 6182, 6183, 6184, 6185, 6186, 6187, 6188, 6189, 6190, 6191, 6192, 6193, 6194, 6195, 6196, 6197, 6198, 6199, 6200, 6201, 6202, 6203, 6204, 6205, 6206, 6207, 6208, 6209, 6210, 6211, 6212, 6213, 6214, 6215, 6216, 6217, 6218, 6219, 6220, 6221, 6222, 6223, 6224, 6225, 6226, 6227, 6228, 6229, 6230, 6231, 6232, 6233, 6234, 6235, 6236, 6237, 6238, 6239, 6240, 6241, 6242, 6243, 6244, 6245, 6246, 6247, 6248, 6249, 6250, 6251, 6252, 6253, 6254, 6255, 6256, 6257, 6258, 6259, 6260, 6261, 6262, 6263, 6264, 6265, 6266, 6267, 6268, 6269, 6270, 6271, 6272, 6273, 6274, 6275, 6276, 6277, 6278, 6279, 6280, 6281, 6282, 6283, 6284, 6285, 6286, 6287, 6288, 6289, 6290, 6291, 6292, 6293, 6294, 6295, 6296, 6297, 6298, 6299, 6300, 6301, 6302, 6303, 6304, 6305, 6306, 6307, 6308, 6309, 6310, 6311, 6312, 6313, 6314, 6315, 6316, 6317, 6318, 6319, 6320, 6321, 6322, 6323, 6324, 6325, 6326, 6327, 6328, 6329, 6330, 6331, 6332, 6333, 6334, 6335, 6336, 6337, 6338, 6339, 6340, 6341, 6342, 6343, 6344, 6345, 6346, 6347, 6348, 6349, 6350, 6351, 6352, 6353, 6354, 6355, 6356, 6357, 6358, 6359, 6360, 6361, 6362, 6363, 6364, 6365, 6366, 6367, 6368, 6369, 6370, 6371, 6372, 6373, 6374, 6375, 6376, 6377, 6378, 6379, 6380, 6381, 6382, 6383, 6384, 6385, 6386, 6387, 6388, 6389, 6390, 6391, 6392, 6393, 6394, 6395, 6396, 6397, 6398, 6399, 6400, 6401, 6402, 6403, 6404, 6405, 6406, 6407, 6408, 6409, 6410, 6411, 6412, 6413, 6414, 6415, 6416, 6417, 6418, 6419, 6420, 6421, 6422, 6423, 6424, 6425, 6426, 6427, 6428, 6429, 6430, 6431, 6432, 6433, 6434, 6435, 6436, 6437, 6438, 6439, 6440, 6441, 6442, 6443, 6444, 6445, 6446, 6447, 6448, 6449, 6450, 6451, 6452, 6453, 6454, 6455, 6456, 6457, 6458, 6459, 6460, 6461, 6462, 6463, 6464, 6465, 6466, 6467, 6468, 6469, 6470, 6471, 6472, 6473, 6474, 6475, 6476, 6477, 6478, 6479, 6480, 6481, 6482, 6483, 6484, 6485, 6486, 6487, 6488, 6489, 6490, 6491, 6492, 6493, 6494, 6495, 6496, 6497, 6498, 6499, 6500, 6501, 6502, 6503, 6504, 6505, 6506, 6507, 6508, 6509, 6510, 6511, 6512, 6513, 6514, 6515, 6516, 6517, 6518, 6519, 6520, 6521, 6522, 6523, 6524, 6525, 6526, 6527, 6528, 6529, 6530, 6531, 6532, 6533, 6534, 6535, 6536, 6537, 6538, 6539, 6540, 6541, 6542, 6543, 6544, 6545, 6546, 6547, 6548, 6549, 6550, 6551, 6552, 6553, 6554, 6555, 6556, 6557, 6558, 6559, 6560, 6561, 6562, 6563, 6564, 6565, 6566, 6567, 6568, 6569, 6570, 6571, 6572, 6573, 6574, 6575, 6576, 6577, 6578, 6579, 6580, 6581, 6582, 6583, 6584, 6585, 6586, 6587, 6588, 6589, 6590, 6591, 6592, 6593, 6594, 6595, 6596, 6597, 6598, 6599, 6600, 6601, 6602, 6603, 6604, 6605, 6606, 6607, 6608, 6609, 6610, 6611, 6612, 6613, 6614, 6615, 6616, 6617, 6618, 6619, 6620, 6621, 6622, 6623, 6624, 6625, 6626, 6627, 6628, 6629, 6630, 6631, 6632, 6633, 6634, 6635, 6636, 6637, 6638, 6639, 6640, 6641, 6642, 6643, 6644, 6645, 6646, 6647, 6648, 6649, 6650, 6651, 6652, 6653, 6654, 6655, 6656, 6657, 6658, 6659, 6660, 6661, 6662, 6663, 6664, 6665, 6666, 6667, 6668, 6669, 6670, 6671, 6672, 6673, 6674, 6675, 6676, 6677, 6678, 6679, 6680, 6681, 6682, 6683, 6684, 6685, 6686, 6687, 6688, 6689, 6690, 6691, 6692, 6693, 6694, 6695, 6696, 6697, 6698, 6699, 6700, 6701, 6702, 6703, 6704, 6705, 6706, 6707, 6708, 6709, 6710, 6711, 6712, 6713, 6714, 6715, 6716, 6717, 6718, 6719, 6720, 6721, 6722, 6723, 6724, 6725, 6726, 6727, 6728, 6729, 6730, 6731, 6732, 6733, 6734, 6735, 6736, 6737, 6738, 6739, 6740, 6741, 6742, 6743, 6744, 6745, 6746, 6747, 6748, 6749, 6750, 6751, 6752, 6753, 6754, 6755, 6756, 6757, 6758, 6759, 6760, 6761, 6762, 6763, 6764, 6765, 6766, 6767, 6768, 6769, 6770, 6771, 6772, 6773, 6774, 6775, 6776, 6777, 6778, 6779, 6780, 6781, 6782, 6783, 6784, 6785, 6786, 6787, 6788, 6789, 6790, 6791, 6792, 6793, 6794, 6795, 6796, 6797, 6798, 6799, 6800, 6801, 6802, 6803, 6804, 6805, 6806, 6807, 6808, 6809, 6810, 6811, 6812, 6813, 6814, 6815, 6816, 6817, 6818, 6819, 6820, 6821, 6822, 6823, 6824, 6825, 6826, 6827, 6828, 6829, 6830, 6831, 6832, 6833, 6834, 6835, 6836, 6837, 6838, 6839, 6840, 6841, 6842, 6843, 6844, 6845, 6846, 6847, 6848, 6849, 6850, 6851, 6852, 6853, 6854, 6855, 6856, 6857, 6858, 6859, 6860, 6861, 6862, 6863, 6864, 6865, 6866, 6867, 6868, 6869, 6870, 6871, 6872, 6873, 6874, 6875, 6876, 6877, 6878, 6879, 6880, 6881, 6882, 6883, 6884, 6885, 6886, 6887, 6888, 6889, 6890, 6891, 6892, 6893, 6894, 6895, 6896, 6897, 6898, 6899, 6900, 6901, 6902, 6903, 6904, 6905, 6906, 6907, 6908, 6909, 6910, 6911, 6912, 6913, 6914, 6915, 6916, 6917, 6918, 6919, 6920, 6921, 6922, 6923, 6924, 6925, 6926, 6927, 6928, 6929, 6930, 6931, 6932, 6933, 6934, 6935, 6936, 6937, 6938, 6939, 6940, 6941, 6942, 6943, 6944, 6945, 6946, 6947, 6948, 6949, 6950, 6951, 6952, 6953, 6954, 6955, 6956, 6957, 6958, 6959, 6960, 6961, 6962, 6963, 6964, 6965, 6966, 6967, 6968, 6969, 6970, 6971, 6972, 6973, 6974, 6975, 6976, 6977, 6978, 6979, 6980, 6981, 6982, 6983, 6984, 6985, 6986, 6987, 6988, 6989, 6990, 6991, 6992, 6993, 6994, 6995, 6996, 6997, 6998, 6999, 7000, 7001, 7002, 7003, 7004, 7005, 7006, 7007, 7008, 7009, 7010, 7011, 7012, 7013, 7014, 7015, 7016, 7017, 7018, 7019, 7020, 7021, 7022, 7023, 7024, 7025, 7026, 7027, 7028, 7029, 7030, 7031, 7032, 7033, 7034, 7035, 7036, 7037, 7038, 7039, 7040, 7041, 7042, 7043, 7044, 7045, 7046, 7047, 7048, 7049, 7050, 7051, 7052, 7053, 7054, 7055, 7056, 7057, 7058, 7059, 7060, 7061, 7062, 7063, 7064, 7065, 7066, 7067, 7068, 7069, 7070, 7071, 7072, 7073, 7074, 7075, 7076, 7077, 7078, 7079, 7080, 7081, 7082, 7083, 7084, 7085, 7086, 7087, 7088, 7089, 7090, 7091, 7092, 7093, 7094, 7095, 7096, 7097, 7098, 7099, 7100, 7101, 7102, 7103, 7104, 7105, 7106, 7107, 7108, 7109, 7110, 7111, 7112, 7113, 7114, 7115, 7116, 7117, 7118, 7119, 7120, 7121, 7122, 7123, 7124, 7125, 7126, 7127, 7128, 7129, 7130, 7131, 7132, 7133, 7134, 7135, 7136, 7137, 7138, 7139, 7140, 7141, 7142, 7143, 7144, 7145, 7146, 7147, 7148, 7149, 7150, 7151, 7152, 7153, 7154, 7155, 7156, 7157, 7158, 7159, 7160, 7161, 7162, 7163, 7164, 7165, 7166, 7167, 7168, 7169, 7170, 7171, 7172, 7173, 7174, 7175, 7176, 7177, 7178, 7179, 7180, 7181, 7182, 7183, 7184, 7185, 7186, 7187, 7188, 7189, 7190, 7191, 7192, 7193, 7194, 7195, 7196, 7197, 7198, 7199, 7200, 7201, 7202, 7203, 7204, 7205, 7206, 7207, 7208, 7209, 7210, 7211, 7212, 7213, 7214, 7215, 7216, 7217, 7218, 7219, 7220, 7221, 7222, 7223, 7224, 7225, 7226, 7227, 7228, 7229, 7230, 7231, 7232, 7233, 7234, 7235, 7236, 7237, 7238, 7239, 7240, 7241, 7242, 7243, 7244, 7245, 7246, 7247, 7248, 7249, 7250, 7251, 7252, 7253, 7254, 7255, 7256, 7257, 7258, 7259, 7260, 7261, 7262, 7263, 7264, 7265, 7266, 7267, 7268, 7269, 7270, 7271, 7272, 7273, 7274, 7275, 7276, 7277, 7278, 7279, 7280, 7281, 7282, 7283, 7284, 7285, 7286, 7287, 7288, 7289, 7290, 7291, 7292, 7293, 7294, 7295, 7296, 7297, 7298, 7299, 7300, 7301, 7302, 7303, 7304, 7305, 7306, 7307, 7308, 7309, 7310, 7311, 7312, 7313, 7314, 7315, 7316, 7317, 7318, 7319, 7320, 7321, 7322, 7323, 7324, 7325, 7326, 7327, 7328, 7329, 7330, 7331, 7332, 7333, 7334, 7335, 7336, 7337, 7338, 7339, 7340, 7341, 7342, 7343, 7344, 7345, 7346, 7347, 7348, 7349, 7350, 7351, 7352, 7353, 7354, 7355, 7356, 7357, 7358, 7359, 7360, 7361, 7362, 7363, 7364, 7365, 7366, 7367, 7368, 7369, 7370, 7371, 7372, 7373, 7374, 7375, 7376, 7377, 7378, 7379, 7380, 7381, 7382, 7383, 7384, 7385, 7386, 7387, 7388, 7389, 7390, 7391, 7392, 7393, 7394, 7395, 7396, 7397, 7398, 7399, 7400, 7401, 7402, 7403, 7404, 7405, 7406, 7407, 7408, 7409, 7410, 7411, 7412, 7413, 7414, 7415, 7416, 7417, 7418, 7419, 7420, 7421, 7422, 7423, 7424, 7425, 7426, 7427, 7428, 7429, 7430, 7431, 7432, 7433, 7434, 7435, 7436, 7437, 7438, 7439, 7440, 7441, 7442, 7443, 7444, 7445, 7446, 7447, 7448, 7449, 7450, 7451, 7452, 7453, 7454, 7455, 7456, 7457, 7458, 7459, 7460, 7461, 7462, 7463, 7464, 7465, 7466, 7467, 7468, 7469, 7470, 7471, 7472, 7473, 7474, 7475, 7476, 7477, 7478, 7479, 7480, 7481, 7482, 7483, 7484, 7485, 7486, 7487, 7488, 7489, 7490, 7491, 7492, 7493, 7494, 7495, 7496, 7497, 7498, 7499, 7500, 7501, 7502, 7503, 7504, 7505, 7506, 7507, 7508, 7509, 7510, 7511, 7512, 7513, 7514, 7515, 7516, 7517, 7518, 7519, 7520, 7521, 7522, 7523, 7524, 7525, 7526, 7527, 7528, 7529, 7530, 7531, 7532, 7533, 7534, 7535, 7536, 7537, 7538, 7539, 7540, 7541, 7542, 7543, 7544, 7545, 7546, 7547, 7548, 7549, 7550, 7551, 7552, 7553, 7554, 7555, 7556, 7557, 7558, 7559, 7560, 7561, 7562, 7563, 7564, 7565, 7566, 7567, 7568, 7569, 7570, 7571, 7572, 7573, 7574, 7575, 7576, 7577, 7578, 7579, 7580, 7581, 7582, 7583, 7584, 7585, 7586, 7587, 7588, 7589, 7590, 7591, 7592, 7593, 7594, 7595, 7596, 7597, 7598, 7599, 7600, 7601, 7602, 7603, 7604, 7605, 7606, 7607, 7608, 7609, 7610, 7611, 7612, 7613, 7614, 7615, 7616, 7617, 7618, 7619, 7620, 7621, 7622, 7623, 7624, 7625, 7626, 7627, 7628, 7629, 7630, 7631, 7632, 7633, 7634, 7635, 7636, 7637, 7638, 7639, 7640, 7641, 7642, 7643, 7644, 7645, 7646, 7647, 7648, 7649, 7650, 7651, 7652, 7653, 7654, 7655, 7656, 7657, 7658, 7659, 7660, 7661, 7662, 7663, 7664, 7665, 7666, 7667, 7668, 7669, 7670, 7671, 7672, 7673, 7674, 7675, 7676, 7677, 7678, 7679, 7680, 7681, 7682, 7683, 7684, 7685, 7686, 7687, 7688, 7689, 7690, 7691, 7692, 7693, 7694, 7695, 7696, 7697, 7698, 7699, 7700, 7701, 7702, 7703, 7704, 7705, 7706, 7707, 7708, 7709, 7710, 7711, 7712, 7713, 7714, 7715, 7716, 7717, 7718, 7719, 7720, 7721, 7722, 7723, 7724, 7725, 7726, 7727, 7728, 7729, 7730, 7731, 7732, 7733, 7734, 7735, 7736, 7737, 7738, 7739, 7740, 7741, 7742, 7743, 7744, 7745, 7746, 7747, 7748, 7749, 7750, 7751, 7752, 7753, 7754, 7755, 7756, 7757, 7758, 7759, 7760, 7761, 7762, 7763, 7764, 7765, 7766, 7767, 7768, 7769, 7770, 7771, 7772, 7773, 7774, 7775, 7776, 7777, 7778, 7779, 7780, 7781, 7782, 7783, 7784, 7785, 7786, 7787, 7788, 7789, 7790, 7791, 7792, 7793, 7794, 7795, 7796, 7797, 7798, 7799, 7800, 7801, 7802, 7803, 7804, 7805, 7806, 7807, 7808, 7809, 7810, 7811, 7812, 7813, 7814, 7815, 7816, 7817, 7818, 7819, 7820, 7821, 7822, 7823, 7824, 7825, 7826, 7827, 7828, 7829, 7830, 7831, 7832, 7833, 7834, 7835, 7836, 7837, 7838, 7839, 7840, 7841, 7842, 7843, 7844, 7845, 7846, 7847, 7848, 7849, 7850, 7851, 7852, 7853, 7854, 7855, 7856, 7857, 7858, 7859, 7860, 7861, 7862, 7863, 7864, 7865, 7866, 7867, 7868, 7869, 7870, 7871, 7872, 7873, 7874, 7875, 7876, 7877, 7878, 7879, 7880, 7881, 7882, 7883, 7884, 7885, 7886, 7887, 7888, 7889, 7890, 7891, 7892, 7893, 7894, 7895, 7896, 7897, 7898, 7899, 7900, 7901, 7902, 7903, 7904, 7905, 7906, 7907, 7908, 7909, 7910, 7911, 7912, 7913, 7914, 7915, 7916, 7917, 7918, 7919, 7920, 7921, 7922, 7923, 7924, 7925, 7926, 7927, 7928, 7929, 7930, 7931, 7932, 7933, 7934, 7935, 7936, 7937, 7938, 7939, 7940, 7941, 7942, 7943, 7944, 7945, 7946, 7947, 7948, 7949, 7950, 7951, 7952, 7953, 7954, 7955, 7956, 7957, 7958, 7959, 7960, 7961, 7962, 7963, 7964, 7965, 7966, 7967, 7968, 7969, 7970, 7971, 7972, 7973, 7974, 7975, 7976, 7977, 7978, 7979, 7980, 7981, 7982, 7983, 7984, 7985, 7986, 7987, 7988, 7989, 7990, 7991, 7992, 7993, 7994, 7995, 7996, 7997, 7998, 7999, 8000, 8001, 8002, 8003, 8004, 8005, 8006, 8007, 8008, 8009, 8010, 8011, 8012, 8013, 8014, 8015, 8016, 8017, 8018, 8019, 8020, 8021, 8022, 8023, 8024, 8025, 8026, 8027, 8028, 8029, 8030, 8031, 8032, 8033, 8034, 8035, 8036, 8037, 8038, 8039, 8040, 8041, 8042, 8043, 8044, 8045, 8046, 8047, 8048, 8049, 8050, 8051, 8052, 8053, 8054, 8055, 8056, 8057, 8058, 8059, 8060, 8061, 8062, 8063, 8064, 8065, 8066, 8067, 8068, 8069, 8070, 8071, 8072, 8073, 8074, 8075, 8076, 8077, 8078, 8079, 8080, 8081, 8082, 8083, 8084, 8085, 8086, 8087, 8088, 8089, 8090, 8091, 8092, 8093, 8094, 8095, 8096, 8097, 8098, 8099, 8100, 8101, 8102, 8103, 8104, 8105, 8106, 8107, 8108, 8109, 8110, 8111, 8112, 8113, 8114, 8115, 8116, 8117, 8118, 8119, 8120, 8121, 8122, 8123, 8124, 8125, 8126, 8127, 8128, 8129, 8130, 8131, 8132, 8133, 8134, 8135, 8136, 8137, 8138, 8139, 8140, 8141, 8142, 8143, 8144, 8145, 8146, 8147, 8148, 8149, 8150, 8151, 8152, 8153, 8154, 8155, 8156, 8157, 8158, 8159, 8160, 8161, 8162, 8163, 8164, 8165, 8166, 8167, 8168, 8169, 8170, 8171, 8172, 8173, 8174, 8175, 8176, 8177, 8178, 8179, 8180, 8181, 8182, 8183, 8184, 8185, 8186, 8187, 8188, 8189, 8190, 8191, 8192, 8193, 8194, 8195, 8196, 8197, 8198, 8199, 8200, 8201, 8202, 8203, 8204, 8205, 8206, 8207, 8208, 8209, 8210, 8211, 8212, 8213, 8214, 8215, 8216, 8217, 8218, 8219, 8220, 8221, 8222, 8223, 8224, 8225, 8226, 8227, 8228, 8229, 8230, 8231, 8232, 8233, 8234, 8235, 8236, 8237, 8238, 8239, 8240, 8241, 8242, 8243, 8244, 8245, 8246, 8247, 8248, 8249, 8250, 8251, 8252, 8253, 8254, 8255, 8256, 8257, 8258, 8259, 8260, 8261, 8262, 8263, 8264, 8265, 8266, 8267, 8268, 8269, 8270, 8271, 8272, 8273, 8274, 8275, 8276, 8277, 8278, 8279, 8280, 8281, 8282, 8283, 8284, 8285, 8286, 8287, 8288, 8289, 8290, 8291, 8292, 8293, 8294, 8295, 8296, 8297, 8298, 8299, 8300, 8301, 8302, 8303, 8304, 8305, 8306, 8307, 8308, 8309, 8310, 8311, 8312, 8313, 8314, 8315, 8316, 8317, 8318, 8319, 8320, 8321, 8322, 8323, 8324, 8325, 8326, 8327, 8328, 8329, 8330, 8331, 8332, 8333, 8334, 8335, 8336, 8337, 8338, 8339, 8340, 8341, 8342, 8343, 8344, 8345, 8346, 8347, 8348, 8349, 8350, 8351, 8352, 8353, 8354, 8355, 8356, 8357, 8358, 8359, 8360, 8361, 8362, 8363, 8364, 8365, 8366, 8367, 8368, 8369, 8370, 8371, 8372, 8373, 8374, 8375, 8376, 8377, 8378, 8379, 8380, 8381, 8382, 8383, 8384, 8385, 8386, 8387, 8388, 8389, 8390, 8391, 8392, 8393, 8394, 8395, 8396, 8397, 8398, 8399, 8400, 8401, 8402, 8403, 8404, 8405, 8406, 8407, 8408, 8409, 8410, 8411, 8412, 8413, 8414, 8415, 8416, 8417, 8418, 8419, 8420, 8421, 8422, 8423, 8424, 8425, 8426, 8427, 8428, 8429, 8430, 8431, 8432, 8433, 8434, 8435, 8436, 8437, 8438, 8439, 8440, 8441, 8442, 8443, 8444, 8445, 8446, 8447, 8448, 8449, 8450, 8451, 8452, 8453, 8454, 8455, 8456, 8457, 8458, 8459, 8460, 8461, 8462, 8463, 8464, 8465, 8466, 8467, 8468, 8469, 8470, 8471, 8472, 8473, 8474, 8475, 8476, 8477, 8478, 8479, 8480, 8481, 8482, 8483, 8484, 8485, 8486, 8487, 8488, 8489, 8490, 8491, 8492, 8493, 8494, 8495, 8496, 8497, 8498, 8499, 8500, 8501, 8502, 8503, 8504, 8505, 8506, 8507, 8508, 8509, 8510, 8511, 8512, 8513, 8514, 8515, 8516, 8517, 8518, 8519, 8520, 8521, 8522, 8523, 8524, 8525, 8526, 8527, 8528, 8529, 8530, 8531, 8532, 8533, 8534, 8535, 8536, 8537, 8538, 8539, 8540, 8541, 8542, 8543, 8544, 8545, 8546, 8547, 8548, 8549, 8550, 8551, 8552, 8553, 8554, 8555, 8556, 8557, 8558, 8559, 8560, 8561, 8562, 8563, 8564, 8565, 8566, 8567, 8568, 8569, 8570, 8571, 8572, 8573, 8574, 8575, 8576, 8577, 8578, 8579, 8580, 8581, 8582, 8583, 8584, 8585, 8586, 8587, 8588, 8589, 8590, 8591, 8592, 8593, 8594, 8595, 8596, 8597, 8598, 8599, 8600, 8601, 8602, 8603, 8604, 8605, 8606, 8607, 8608, 8609, 8610, 8611, 8612, 8613, 8614, 8615, 8616, 8617, 8618, 8619, 8620, 8621, 8622, 8623, 8624, 8625, 8626, 8627, 8628, 8629, 8630, 8631, 8632, 8633, 8634, 8635, 8636, 8637, 8638, 8639, 8640, 8641, 8642, 8643, 8644, 8645, 8646, 8647, 8648, 8649, 8650, 8651, 8652, 8653, 8654, 8655, 8656, 8657, 8658, 8659, 8660, 8661, 8662, 8663, 8664, 8665, 8666, 8667, 8668, 8669, 8670, 8671, 8672, 8673, 8674, 8675, 8676, 8677, 8678, 8679, 8680, 8681, 8682, 8683, 8684, 8685, 8686, 8687, 8688, 8689, 8690, 8691, 8692, 8693, 8694, 8695, 8696, 8697, 8698, 8699, 8700, 8701, 8702, 8703, 8704, 8705, 8706, 8707, 8708, 8709, 8710, 8711, 8712, 8713, 8714, 8715, 8716, 8717, 8718, 8719, 8720, 8721, 8722, 8723, 8724, 8725, 8726, 8727, 8728, 8729, 8730, 8731, 8732, 8733, 8734, 8735, 8736, 8737, 8738, 8739, 8740, 8741, 8742, 8743, 8744, 8745, 8746, 8747, 8748, 8749, 8750, 8751, 8752, 8753, 8754, 8755, 8756, 8757, 8758, 8759, 8760, 8761, 8762, 8763, 8764, 8765, 8766, 8767, 8768, 8769, 8770, 8771, 8772, 8773, 8774, 8775, 8776, 8777, 8778, 8779, 8780, 8781, 8782, 8783, 8784, 8785, 8786, 8787, 8788, 8789, 8790, 8791, 8792, 8793, 8794, 8795, 8796, 8797, 8798, 8799, 8800, 8801, 8802, 8803, 8804, 8805, 8806, 8807, 8808, 8809, 8810, 8811, 8812, 8813, 8814, 8815, 8816, 8817, 8818, 8819, 8820, 8821, 8822, 8823, 8824, 8825, 8826, 8827, 8828, 8829, 8830, 8831, 8832, 8833, 8834, 8835, 8836, 8837, 8838, 8839, 8840, 8841, 8842, 8843, 8844, 8845, 8846, 8847, 8848, 8849, 8850, 8851, 8852, 8853, 8854, 8855, 8856, 8857, 8858, 8859, 8860, 8861, 8862, 8863, 8864, 8865, 8866, 8867, 8868, 8869, 8870, 8871, 8872, 8873, 8874, 8875, 8876, 8877, 8878, 8879, 8880, 8881, 8882, 8883, 8884, 8885, 8886, 8887, 8888, 8889, 8890, 8891, 8892, 8893, 8894, 8895, 8896, 8897, 8898, 8899, 8900, 8901, 8902, 8903, 8904, 8905, 8906, 8907, 8908, 8909, 8910, 8911, 8912, 8913, 8914, 8915, 8916, 8917, 8918, 8919, 8920, 8921, 8922, 8923, 8924, 8925, 8926, 8927, 8928, 8929, 8930, 8931, 8932, 8933, 8934, 8935, 8936, 8937, 8938, 8939, 8940, 8941, 8942, 8943, 8944, 8945, 8946, 8947, 8948, 8949, 8950, 8951, 8952, 8953, 8954, 8955, 8956, 8957, 8958, 8959, 8960, 8961, 8962, 8963, 8964, 8965, 8966, 8967, 8968, 8969, 8970, 8971, 8972, 8973, 8974, 8975, 8976, 8977, 8978, 8979, 8980, 8981, 8982, 8983, 8984, 8985, 8986, 8987, 8988, 8989, 8990, 8991, 8992, 8993, 8994, 8995, 8996, 8997, 8998, 8999, 9000, 9001, 9002, 9003, 9004, 9005, 9006, 9007, 9008, 9009, 9010, 9011, 9012, 9013, 9014, 9015, 9016, 9017, 9018, 9019, 9020, 9021, 9022, 9023, 9024, 9025, 9026, 9027, 9028, 9029, 9030, 9031, 9032, 9033, 9034, 9035, 9036, 9037, 9038, 9039, 9040, 9041, 9042, 9043, 9044, 9045, 9046, 9047, 9048, 9049, 9050, 9051, 9052, 9053, 9054, 9055, 9056, 9057, 9058, 9059, 9060, 9061, 9062, 9063, 9064, 9065, 9066, 9067, 9068, 9069, 9070, 9071, 9072, 9073, 9074, 9075, 9076, 9077, 9078, 9079, 9080, 9081, 9082, 9083, 9084, 9085, 9086, 9087, 9088, 9089, 9090, 9091, 9092, 9093, 9094, 9095, 9096, 9097, 9098, 9099, 9100, 9101, 9102, 9103, 9104, 9105, 9106, 9107, 9108, 9109, 9110, 9111, 9112, 9113, 9114, 9115, 9116, 9117, 9118, 9119, 9120, 9121, 9122, 9123, 9124, 9125, 9126, 9127, 9128, 9129, 9130, 9131, 9132, 9133, 9134, 9135, 9136, 9137, 9138, 9139, 9140, 9141, 9142, 9143, 9144, 9145, 9146, 9147, 9148, 9149, 9150, 9151, 9152, 9153, 9154, 9155, 9156, 9157, 9158, 9159, 9160, 9161, 9162, 9163, 9164, 9165, 9166, 9167, 9168, 9169, 9170, 9171, 9172, 9173, 9174, 9175, 9176, 9177, 9178, 9179, 9180, 9181, 9182, 9183, 9184, 9185, 9186, 9187, 9188, 9189, 9190, 9191, 9192, 9193, 9194, 9195, 9196, 9197, 9198, 9199, 9200, 9201, 9202, 9203, 9204, 9205, 9206, 9207, 9208, 9209, 9210, 9211, 9212, 9213, 9214, 9215, 9216, 9217, 9218, 9219, 9220, 9221, 9222, 9223, 9224, 9225, 9226, 9227, 9228, 9229, 9230, 9231, 9232, 9233, 9234, 9235, 9236, 9237, 9238, 9239, 9240, 9241, 9242, 9243, 9244, 9245, 9246, 9247, 9248, 9249, 9250, 9251, 9252, 9253, 9254, 9255, 9256, 9257, 9258, 9259, 9260, 9261, 9262, 9263, 9264, 9265, 9266, 9267, 9268, 9269, 9270, 9271, 9272, 9273, 9274, 9275, 9276, 9277, 9278, 9279, 9280, 9281, 9282, 9283, 9284, 9285, 9286, 9287, 9288, 9289, 9290, 9291, 9292, 9293, 9294, 9295, 9296, 9297, 9298, 9299, 9300, 9301, 9302, 9303, 9304, 9305, 9306, 9307, 9308, 9309, 9310, 9311, 9312, 9313, 9314, 9315, 9316, 9317, 9318, 9319, 9320, 9321, 9322, 9323, 9324, 9325, 9326, 9327, 9328, 9329, 9330, 9331, 9332, 9333, 9334, 9335, 9336, 9337, 9338, 9339, 9340, 9341, 9342, 9343, 9344, 9345, 9346, 9347, 9348, 9349, 9350, 9351, 9352, 9353, 9354, 9355, 9356, 9357, 9358, 9359, 9360, 9361, 9362, 9363, 9364, 9365, 9366, 9367, 9368, 9369, 9370, 9371, 9372, 9373, 9374, 9375, 9376, 9377, 9378, 9379, 9380, 9381, 9382, 9383, 9384, 9385, 9386, 9387, 9388, 9389, 9390, 9391, 9392, 9393, 9394, 9395, 9396, 9397, 9398, 9399, 9400, 9401, 9402, 9403, 9404, 9405, 9406, 9407, 9408, 9409, 9410, 9411, 9412, 9413, 9414, 9415, 9416, 9417, 9418, 9419, 9420, 9421, 9422, 9423, 9424, 9425, 9426, 9427, 9428, 9429, 9430, 9431, 9432, 9433, 9434, 9435, 9436, 9437, 9438, 9439, 9440, 9441, 9442, 9443, 9444, 9445, 9446, 9447, 9448, 9449, 9450, 9451, 9452, 9453, 9454, 9455, 9456, 9457, 9458, 9459, 9460, 9461, 9462, 9463, 9464, 9465, 9466, 9467, 9468, 9469, 9470, 9471, 9472, 9473, 9474, 9475, 9476, 9477, 9478, 9479, 9480, 9481, 9482, 9483, 9484, 9485, 9486, 9487, 9488, 9489, 9490, 9491, 9492, 9493, 9494, 9495, 9496, 9497, 9498, 9499, 9500, 9501, 9502, 9503, 9504, 9505, 9506, 9507, 9508, 9509, 9510, 9511, 9512, 9513, 9514, 9515, 9516, 9517, 9518, 9519, 9520, 9521, 9522, 9523, 9524, 9525, 9526, 9527, 9528, 9529, 9530, 9531, 9532, 9533, 9534, 9535, 9536, 9537, 9538, 9539, 9540, 9541, 9542, 9543, 9544, 9545, 9546, 9547, 9548, 9549, 9550, 9551, 9552, 9553, 9554, 9555, 9556, 9557, 9558, 9559, 9560, 9561, 9562, 9563, 9564, 9565, 9566, 9567, 9568, 9569, 9570, 9571, 9572, 9573, 9574, 9575, 9576, 9577, 9578, 9579, 9580, 9581, 9582, 9583, 9584, 9585, 9586, 9587, 9588, 9589, 9590, 9591, 9592, 9593, 9594, 9595, 9596, 9597, 9598, 9599, 9600, 9601, 9602, 9603, 9604, 9605, 9606, 9607, 9608, 9609, 9610, 9611, 9612, 9613, 9614, 9615, 9616, 9617, 9618, 9619, 9620, 9621, 9622, 9623, 9624, 9625, 9626, 9627, 9628, 9629, 9630, 9631, 9632, 9633, 9634, 9635, 9636, 9637, 9638, 9639, 9640, 9641, 9642, 9643, 9644, 9645, 9646, 9647, 9648, 9649, 9650, 9651, 9652, 9653, 9654, 9655, 9656, 9657, 9658, 9659, 9660, 9661, 9662, 9663, 9664, 9665, 9666, 9667, 9668, 9669, 9670, 9671, 9672, 9673, 9674, 9675, 9676, 9677, 9678, 9679, 9680, 9681, 9682, 9683, 9684, 9685, 9686, 9687, 9688, 9689, 9690, 9691, 9692, 9693, 9694, 9695, 9696, 9697, 9698, 9699, 9700, 9701, 9702, 9703, 9704, 9705, 9706, 9707, 9708, 9709, 9710, 9711, 9712, 9713, 9714, 9715, 9716, 9717, 9718, 9719, 9720, 9721, 9722, 9723, 9724, 9725, 9726, 9727, 9728, 9729, 9730, 9731, 9732, 9733, 9734, 9735, 9736, 9737, 9738, 9739, 9740, 9741, 9742, 9743, 9744, 9745, 9746, 9747, 9748, 9749, 9750, 9751, 9752, 9753, 9754, 9755, 9756, 9757, 9758, 9759, 9760, 9761, 9762, 9763, 9764, 9765, 9766, 9767, 9768, 9769, 9770, 9771, 9772, 9773, 9774, 9775, 9776, 9777, 9778, 9779, 9780, 9781, 9782, 9783, 9784, 9785, 9786, 9787, 9788, 9789, 9790, 9791, 9792, 9793, 9794, 9795, 9796, 9797, 9798, 9799, 9800, 9801, 9802, 9803, 9804, 9805, 9806, 9807, 9808, 9809, 9810, 9811, 9812, 9813, 9814, 9815, 9816, 9817, 9818, 9819, 9820, 9821, 9822, 9823, 9824, 9825, 9826, 9827, 9828, 9829, 9830, 9831, 9832, 9833, 9834, 9835, 9836, 9837, 9838, 9839, 9840, 9841, 9842, 9843, 9844, 9845, 9846, 9847, 9848, 9849, 9850, 9851, 9852, 9853, 9854, 9855, 9856, 9857, 9858, 9859, 9860, 9861, 9862, 9863, 9864, 9865, 9866, 9867, 9868, 9869, 9870, 9871, 9872, 9873, 9874, 9875, 9876, 9877, 9878, 9879, 9880, 9881, 9882, 9883, 9884, 9885, 9886, 9887, 9888, 9889, 9890, 9891, 9892, 9893, 9894, 9895, 9896, 9897, 9898, 9899, 9900, 9901, 9902, 9903, 9904, 9905, 9906, 9907, 9908, 9909, 9910, 9911, 9912, 9913, 9914, 9915, 9916, 9917, 9918, 9919, 9920, 9921, 9922, 9923, 9924, 9925, 9926, 9927, 9928, 9929, 9930, 9931, 9932, 9933, 9934, 9935, 9936, 9937, 9938, 9939, 9940, 9941, 9942, 9943, 9944, 9945, 9946, 9947, 9948, 9949, 9950, 9951, 9952, 9953, 9954, 9955, 9956, 9957, 9958, 9959, 9960, 9961, 9962, 9963, 9964, 9965, 9966, 9967, 9968, 9969, 9970, 9971, 9972, 9973, 9974, 9975, 9976, 9977, 9978, 9979, 9980, 9981, 9982, 9983, 9984, 9985, 9986, 9987, 9988, 9989, 9990, 9991, 9992, 9993, 9994, 9995, 9996, 9997, 9998, 9999]
###Markdown
** (3d) Perform action `count` to view counts ** One of the most basic jobs that we can run is the `count()` job which will count the number of elements in an RDD using the `count()` action. Since `map()` creates a new RDD with the same number of elements as the starting RDD, we expect that applying `count()` to each RDD will return the same result. Note that because `count()` is an action operation, if we had not already performed an action with `collect()`, then Spark would now perform the transformation operations when we executed `count()`. Each task counts the entries in its partition and sends the result to your SparkContext, which adds up all of the counts. The figure below shows what would happen if we ran `count()` on a small example dataset with just four partitions.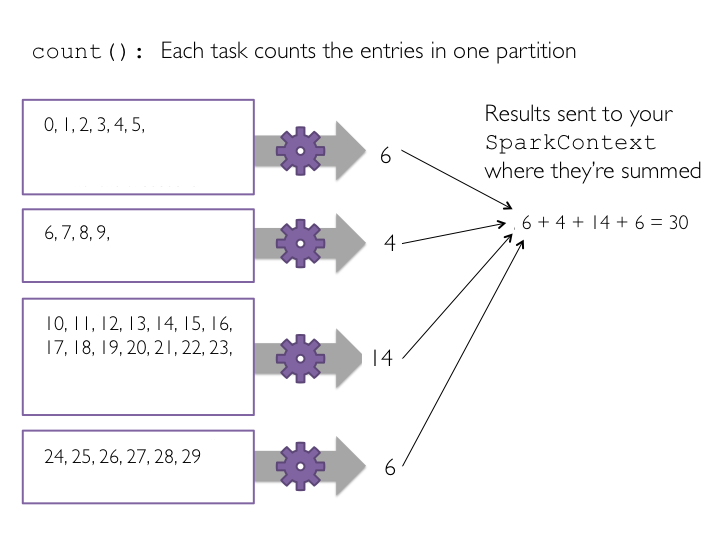
###Code
print xrangeRDD.count()
print subRDD.count()
###Output
10000
10000
###Markdown
** (3e) Apply transformation `filter` and view results with `collect` ** Next, we'll create a new RDD that only contains the values less than ten by using the `filter(f)` data-parallel operation. The `filter(f)` method is a transformation operation that creates a new RDD from the input RDD by applying filter function `f` to each item in the parent RDD and only passing those elements where the filter function returns `True`. Elements that do not return `True` will be dropped. Like `map()`, filter can be applied individually to each entry in the dataset, so is easily parallelized using Spark. The figure below shows how this would work on the small four-partition dataset.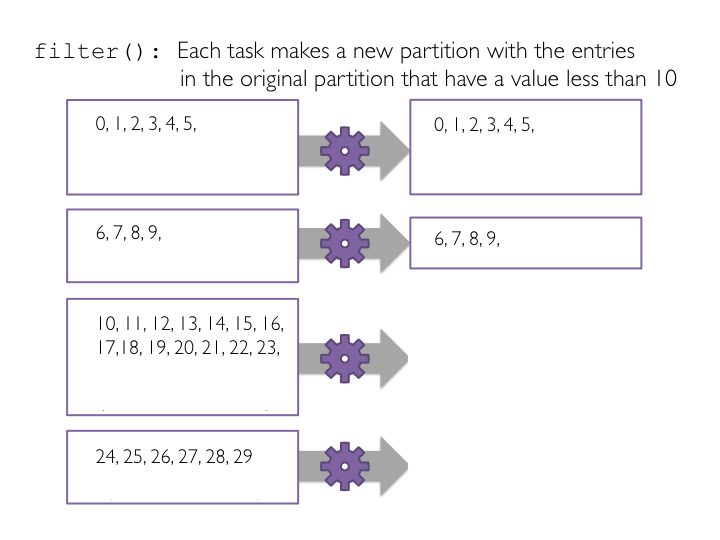 To filter this dataset, we'll define a function called `ten()`, which returns `True` if the input is less than 10 and `False` otherwise. This function will be passed to the `filter()` transformation as the filter function `f`. To view the filtered list of elements less than ten, we need to create a new list on the driver from the distributed data on the executor nodes. We use the `collect()` method to return a list that contains all of the elements in this filtered RDD to the driver program.
###Code
# Define a function to filter a single value
def ten(value):
"""Return whether value is below ten.
Args:
value (int): A number.
Returns:
bool: Whether `value` is less than ten.
"""
if (value < 10):
return True
else:
return False
# The ten function could also be written concisely as: def ten(value): return value < 10
# Pass the function ten to the filter transformation
# Filter is a transformation so no tasks are run
filteredRDD = subRDD.filter(ten)
# View the results using collect()
# Collect is an action and triggers the filter transformation to run
print filteredRDD.collect()
###Output
[0, 1, 2, 3, 4, 5, 6, 7, 8, 9]
###Markdown
** Part 4: Lambda Functions ** ** (4a) Using Python `lambda()` functions ** Python supports the use of small one-line anonymous functions that are not bound to a name at runtime. Borrowed from LISP, these `lambda` functions can be used wherever function objects are required. They are syntactically restricted to a single expression. Remember that `lambda` functions are a matter of style and using them is never required - semantically, they are just syntactic sugar for a normal function definition. You can always define a separate normal function instead, but using a `lambda()` function is an equivalent and more compact form of coding. Ideally you should consider using `lambda` functions where you want to encapsulate non-reusable code without littering your code with one-line functions. Here, instead of defining a separate function for the `filter()` transformation, we will use an inline `lambda()` function.
###Code
lambdaRDD = subRDD.filter(lambda x: x < 10)
lambdaRDD.collect()
# Let's collect the even values less than 10
evenRDD = lambdaRDD.filter(lambda x: x % 2 == 0)
evenRDD.collect()
###Output
_____no_output_____
###Markdown
** Part 5: Additional RDD actions ** ** (5a) Other common actions ** Let's investigate the additional actions: [first()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.first), [take()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.take), [top()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.top), [takeOrdered()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.takeOrdered), and [reduce()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.reduce) One useful thing to do when we have a new dataset is to look at the first few entries to obtain a rough idea of what information is available. In Spark, we can do that using the `first()`, `take()`, `top()`, and `takeOrdered()` actions. Note that for the `first()` and `take()` actions, the elements that are returned depend on how the RDD is *partitioned*. Instead of using the `collect()` action, we can use the `take(n)` action to return the first n elements of the RDD. The `first()` action returns the first element of an RDD, and is equivalent to `take(1)`. The `takeOrdered()` action returns the first n elements of the RDD, using either their natural order or a custom comparator. The key advantage of using `takeOrdered()` instead of `first()` or `take()` is that `takeOrdered()` returns a deterministic result, while the other two actions may return differing results, depending on the number of partions or execution environment. `takeOrdered()` returns the list sorted in *ascending order*. The `top()` action is similar to `takeOrdered()` except that it returns the list in *descending order.* The `reduce()` action reduces the elements of a RDD to a single value by applying a function that takes two parameters and returns a single value. The function should be commutative and associative, as `reduce()` is applied at the partition level and then again to aggregate results from partitions. If these rules don't hold, the results from `reduce()` will be inconsistent. Reducing locally at partitions makes `reduce()` very efficient.
###Code
# Let's get the first element
print filteredRDD.first()
# The first 4
print filteredRDD.take(4)
# Note that it is ok to take more elements than the RDD has
print filteredRDD.take(12)
# Retrieve the three smallest elements
print filteredRDD.takeOrdered(3)
# Retrieve the five largest elements
print filteredRDD.top(5)
# Pass a lambda function to takeOrdered to reverse the order
filteredRDD.takeOrdered(4, lambda s: -s)
# Obtain Python's add function
from operator import add
# Efficiently sum the RDD using reduce
print filteredRDD.reduce(add)
# Sum using reduce with a lambda function
print filteredRDD.reduce(lambda a, b: a + b)
# Note that subtraction is not both associative and commutative
print filteredRDD.reduce(lambda a, b: a - b)
print filteredRDD.repartition(4).reduce(lambda a, b: a - b)
# While addition is
print filteredRDD.repartition(4).reduce(lambda a, b: a + b)
###Output
45
45
-45
21
45
###Markdown
** (5b) Advanced actions ** Here are two additional actions that are useful for retrieving information from an RDD: [takeSample()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.takeSample) and [countByValue()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.countByValue) The `takeSample()` action returns an array with a random sample of elements from the dataset. It takes in a `withReplacement` argument, which specifies whether it is okay to randomly pick the same item multiple times from the parent RDD (so when `withReplacement=True`, you can get the same item back multiple times). It also takes an optional `seed` parameter that allows you to specify a seed value for the random number generator, so that reproducible results can be obtained. The `countByValue()` action returns the count of each unique value in the RDD as a dictionary that maps values to counts.
###Code
# takeSample reusing elements
print filteredRDD.takeSample(withReplacement=True, num=6)
# takeSample without reuse
print filteredRDD.takeSample(withReplacement=False, num=6)
# Set seed for predictability
print filteredRDD.takeSample(withReplacement=False, num=6, seed=500)
# Try reruning this cell and the cell above -- the results from this cell will remain constant
# Use ctrl-enter to run without moving to the next cell
# Create new base RDD to show countByValue
repetitiveRDD = sc.parallelize([1, 2, 3, 1, 2, 3, 1, 2, 1, 2, 3, 3, 3, 4, 5, 4, 6])
print repetitiveRDD.countByValue()
###Output
defaultdict(<type 'int'>, {1: 4, 2: 4, 3: 5, 4: 2, 5: 1, 6: 1})
###Markdown
** Part 6: Additional RDD transformations ** ** (6a) `flatMap` ** When performing a `map()` transformation using a function, sometimes the function will return more (or less) than one element. We would like the newly created RDD to consist of the elements outputted by the function. Simply applying a `map()` transformation would yield a new RDD made up of iterators. Each iterator could have zero or more elements. Instead, we often want an RDD consisting of the values contained in those iterators. The solution is to use a [flatMap()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.flatMap) transformation, `flatMap()` is similar to `map()`, except that with `flatMap()` each input item can be mapped to zero or more output elements. To demonstrate `flatMap()`, we will first emit a word along with its plural, and then a range that grows in length with each subsequent operation.
###Code
# Let's create a new base RDD to work from
wordsList = ['cat', 'elephant', 'rat', 'rat', 'cat']
wordsRDD = sc.parallelize(wordsList, 4)
# Use map
singularAndPluralWordsRDDMap = wordsRDD.map(lambda x: (x, x + 's'))
# Use flatMap
singularAndPluralWordsRDD = wordsRDD.flatMap(lambda x: (x, x + 's'))
# View the results
print singularAndPluralWordsRDDMap.collect()
print singularAndPluralWordsRDD.collect()
# View the number of elements in the RDD
print singularAndPluralWordsRDDMap.count()
print singularAndPluralWordsRDD.count()
simpleRDD = sc.parallelize([2, 3, 4])
print simpleRDD.map(lambda x: range(1, x)).collect()
print simpleRDD.flatMap(lambda x: range(1, x)).collect()
###Output
[[1], [1, 2], [1, 2, 3]]
[1, 1, 2, 1, 2, 3]
###Markdown
** (6b) `groupByKey` and `reduceByKey` ** Let's investigate the additional transformations: [groupByKey()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.groupByKey) and [reduceByKey()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.reduceByKey). Both of these transformations operate on pair RDDs. A pair RDD is an RDD where each element is a pair tuple (key, value). For example, `sc.parallelize([('a', 1), ('a', 2), ('b', 1)])` would create a pair RDD where the keys are 'a', 'a', 'b' and the values are 1, 2, 1. The `reduceByKey()` transformation gathers together pairs that have the same key and applies a function to two associated values at a time. `reduceByKey()` operates by applying the function first within each partition on a per-key basis and then across the partitions. While both the `groupByKey()` and `reduceByKey()` transformations can often be used to solve the same problem and will produce the same answer, the `reduceByKey()` transformation works much better for large distributed datasets. This is because Spark knows it can combine output with a common key on each partition *before* shuffling (redistributing) the data across nodes. Only use `groupByKey()` if the operation would not benefit from reducing the data before the shuffle occurs. Look at the diagram below to understand how `reduceByKey` works. Notice how pairs on the same machine with the same key are combined (by using the lamdba function passed into reduceByKey) before the data is shuffled. Then the lamdba function is called again to reduce all the values from each partition to produce one final result.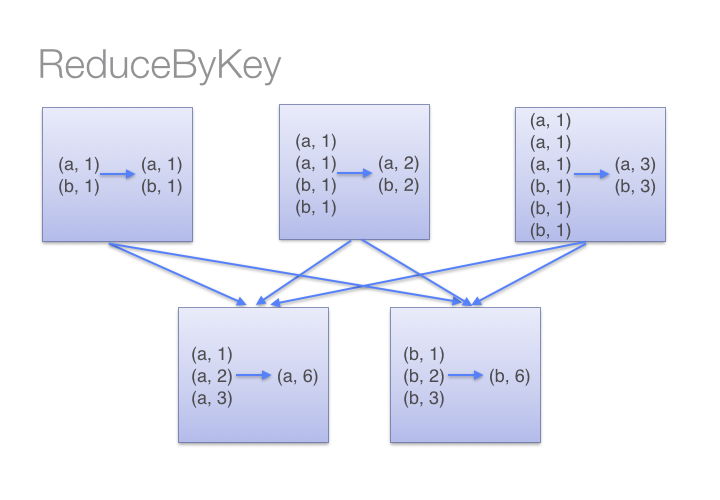 On the other hand, when using the `groupByKey()` transformation - all the key-value pairs are shuffled around, causing a lot of unnecessary data to being transferred over the network. To determine which machine to shuffle a pair to, Spark calls a partitioning function on the key of the pair. Spark spills data to disk when there is more data shuffled onto a single executor machine than can fit in memory. However, it flushes out the data to disk one key at a time, so if a single key has more key-value pairs than can fit in memory an out of memory exception occurs. This will be more gracefully handled in a later release of Spark so that the job can still proceed, but should still be avoided. When Spark needs to spill to disk, performance is severely impacted.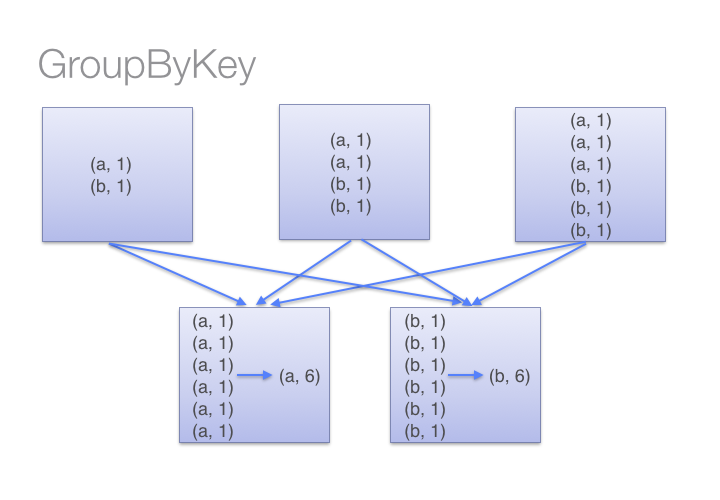 As your dataset grows, the difference in the amount of data that needs to be shuffled, between the `reduceByKey()` and `groupByKey()` transformations, becomes increasingly exaggerated. Here are more transformations to prefer over `groupByKey()`: + [combineByKey()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.combineByKey) can be used when you are combining elements but your return type differs from your input value type. + [foldByKey()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.foldByKey) merges the values for each key using an associative function and a neutral "zero value". Now let's go through a simple `groupByKey()` and `reduceByKey()` example.
###Code
pairRDD = sc.parallelize([('a', 1), ('a', 2), ('b', 1)])
# mapValues only used to improve format for printing
print pairRDD.groupByKey().mapValues(lambda x: list(x)).collect()
# Different ways to sum by key
print pairRDD.groupByKey().map(lambda (k, v): (k, sum(v))).collect()
# Using mapValues, which is recommended when they key doesn't change
print pairRDD.groupByKey().mapValues(lambda x: sum(x)).collect()
# reduceByKey is more efficient / scalable
print pairRDD.reduceByKey(add).collect()
###Output
[('a', [1, 2]), ('b', [1])]
[('a', 3), ('b', 1)]
[('a', 3), ('b', 1)]
[('a', 3), ('b', 1)]
###Markdown
** (6c) Advanced transformations ** [Optional] Let's investigate the advanced transformations: [mapPartitions()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.mapPartitions) and [mapPartitionsWithIndex()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.mapPartitionsWithIndex) The `mapPartitions()` transformation uses a function that takes in an iterator (to the items in that specific partition) and returns an iterator. The function is applied on a partition by partition basis. The `mapPartitionsWithIndex()` transformation uses a function that takes in a partition index (think of this like the partition number) and an iterator (to the items in that specific partition). For every partition (index, iterator) pair, the function returns a tuple of the same partition index number and an iterator of the transformed items in that partition.
###Code
# mapPartitions takes a function that takes an iterator and returns an iterator
print wordsRDD.collect()
itemsRDD = wordsRDD.mapPartitions(lambda iterator: [','.join(iterator)])
print itemsRDD.collect()
itemsByPartRDD = wordsRDD.mapPartitionsWithIndex(lambda index, iterator: [(index, list(iterator))])
# We can see that three of the (partitions) workers have one element and the fourth worker has two
# elements, although things may not bode well for the rat...
print itemsByPartRDD.collect()
# Rerun without returning a list (acts more like flatMap)
itemsByPartRDD = wordsRDD.mapPartitionsWithIndex(lambda index, iterator: (index, list(iterator)))
print itemsByPartRDD.collect()
###Output
[(0, ['cat']), (1, ['elephant']), (2, ['rat']), (3, ['rat', 'cat'])]
[0, ['cat'], 1, ['elephant'], 2, ['rat'], 3, ['rat', 'cat']]
###Markdown
** Part 7: Caching RDDs and storage options ** ** (7a) Caching RDDs ** For efficiency Spark keeps your RDDs in memory. By keeping the contents in memory, Spark can quickly access the data. However, memory is limited, so if you try to keep too many RDDs in memory, Spark will automatically delete RDDs from memory to make space for new RDDs. If you later refer to one of the RDDs, Spark will automatically recreate the RDD for you, but that takes time. So, if you plan to use an RDD more than once, then you should tell Spark to cache that RDD. You can use the `cache()` operation to keep the RDD in memory. However, if you cache too many RDDs and Spark runs out of memory, it will delete the least recently used (LRU) RDD first. Again, the RDD will be automatically recreated when accessed. You can check if an RDD is cached by using the `is_cached` attribute, and you can see your cached RDD in the "Storage" section of the Spark web UI. If you click on the RDD's name, you can see more information about where the RDD is stored.
###Code
# Name the RDD
filteredRDD.setName('My Filtered RDD')
# Cache the RDD
filteredRDD.cache()
# Is it cached
print filteredRDD.is_cached
###Output
True
###Markdown
** (7b) Unpersist and storage options ** Spark automatically manages the RDDs cached in memory and will save them to disk if it runs out of memory. For efficiency, once you are finished using an RDD, you can optionally tell Spark to stop caching it in memory by using the RDD's `unpersist()` method to inform Spark that you no longer need the RDD in memory. You can see the set of transformations that were applied to create an RDD by using the `toDebugString()` method, which will provide storage information, and you can directly query the current storage information for an RDD using the `getStorageLevel()` operation. ** Advanced: ** Spark provides many more options for managing how RDDs are stored in memory or even saved to disk. You can explore the API for RDD's [persist()](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.RDD.persist) operation using Python's [help()](https://docs.python.org/2/library/functions.html?highlight=helphelp) command. The `persist()` operation, optionally, takes a pySpark [StorageLevel](http://spark.apache.org/docs/latest/api/python/pyspark.htmlpyspark.StorageLevel) object.
###Code
# Note that toDebugString also provides storage information
print filteredRDD.toDebugString()
# If we are done with the RDD we can unpersist it so that its memory can be reclaimed
filteredRDD.unpersist()
# Storage level for a non cached RDD
print filteredRDD.getStorageLevel()
filteredRDD.cache()
# Storage level for a cached RDD
print filteredRDD.getStorageLevel()
###Output
Serialized 1x Replicated
Memory Serialized 1x Replicated
###Markdown
** Part 8: Debugging Spark applications and lazy evaluation ** ** How Python is Executed in Spark ** Internally, Spark executes using a Java Virtual Machine (JVM). pySpark runs Python code in a JVM using [Py4J](http://py4j.sourceforge.net). Py4J enables Python programs running in a Python interpreter to dynamically access Java objects in a Java Virtual Machine. Methods are called as if the Java objects resided in the Python interpreter and Java collections can be accessed through standard Python collection methods. Py4J also enables Java programs to call back Python objects. Because pySpark uses Py4J, coding errors often result in a complicated, confusing stack trace that can be difficult to understand. In the following section, we'll explore how to understand stack traces. ** (8a) Challenges with lazy evaluation using transformations and actions ** Spark's use of lazy evaluation can make debugging more difficult because code is not always executed immediately. To see an example of how this can happen, let's first define a broken filter function. Next we perform a `filter()` operation using the broken filtering function. No error will occur at this point due to Spark's use of lazy evaluation. The `filter()` method will not be executed *until* an action operation is invoked on the RDD. We will perform an action by using the `collect()` method to return a list that contains all of the elements in this RDD.
###Code
def brokenTen(value):
"""Incorrect implementation of the ten function.
Note:
The `if` statement checks an undefined variable `val` instead of `value`.
Args:
value (int): A number.
Returns:
bool: Whether `value` is less than ten.
Raises:
NameError: The function references `val`, which is not available in the local or global
namespace, so a `NameError` is raised.
"""
if (val < 10):
return True
else:
return False
brokenRDD = subRDD.filter(brokenTen)
# Now we'll see the error
brokenRDD.collect()
###Output
_____no_output_____
###Markdown
** (8b) Finding the bug ** When the `filter()` method is executed, Spark evaluates the RDD by executing the `parallelize()` and `filter()` methods. Since our `filter()` method has an error in the filtering function `brokenTen()`, an error occurs. Scroll through the output "Py4JJavaError Traceback (most recent call last)" part of the cell and first you will see that the line that generated the error is the `collect()` method line. There is *nothing wrong with this line*. However, it is an action and that caused other methods to be executed. Continue scrolling through the Traceback and you will see the following error line: NameError: global name 'val' is not defined Looking at this error line, we can see that we used the wrong variable name in our filtering function `brokenTen()`. ** (8c) Moving toward expert style ** As you are learning Spark, I recommend that you write your code in the form: RDD.transformation1() RDD.action1() RDD.transformation2() RDD.action2() Using this style will make debugging your code much easier as it makes errors easier to localize - errors in your transformations will occur when the next action is executed. Once you become more experienced with Spark, you can write your code with the form: RDD.transformation1().transformation2().action() We can also use `lambda()` functions instead of separately defined functions when their use improves readability and conciseness.
###Code
# Cleaner code through lambda use
subRDD.filter(lambda x: x < 10).collect()
# Even better by moving our chain of operators into a single line.
sc.parallelize(data).map(lambda y: y - 1).filter(lambda x: x < 10).collect()
###Output
_____no_output_____
###Markdown
** (8d) Readability and code style ** To make the expert coding style more readable, enclose the statement in parentheses and put each method, transformation, or action on a separate line.
###Code
# Final version
(sc
.parallelize(data)
.map(lambda y: y - 1)
.filter(lambda x: x < 10)
.collect())
###Output
_____no_output_____ |
Insee/notebooks/02.Filtering_data.ipynb | ###Markdown
2 Filtering dataIn chapter 1, we have seen how to retrieve data with select. Now you may want to know how can we only retrieve the data that interest us,but not all the rows. We can use the **where** statement to filter data that satisfied certain conditions.
###Code
%load_ext sql
%config SqlMagic.autocommit=False
%config SqlMagic.autolimit=20
%config SqlMagic.displaylimit=20
%sql postgresql://user-pengfei:gv8eba5xmsw4kt2uk1mn@postgresql-124499/test
%%sql
SELECT * from orders limit 5;
###Output
* postgresql://user-pengfei:***@postgresql-124499/test
5 rows affected.
###Markdown
2.1 Filtering digit columnsTo filter digit column, we need to build a boolean expression such as column_name comparator valuePossible comparators:- equality : =- inequality : !=, or - greater than: >- less than : <- Greater than equal to : >=- Less than equal to : <=Below query is an example, in the boolean expression column name is order_date (we extract the year from the date), comparator is =, value is 1996This query should only return records where the order_date equals to 1996
###Code
%%sql
select * from orders where extract(year from order_date)=1996 limit 5
###Output
* postgresql://user-pengfei:***@postgresql-124499/test
5 rows affected.
###Markdown
What if I want to get all records where year is not equals to 2010. There are two possible ways (!=, or ) to express inequality. Most database servers such as Mysql, Postgresql, SQLite, etc. support both. However, some database server such as **Microsoft Access and IBM DB2 only support **.
###Code
%%sql
select * from orders where extract(year from order_date)!=1996 limit 5
%%sql
select * from orders where extract(year from order_date)<>1996 limit 5
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
5 rows affected.
###Markdown
These two queries do the same thing. You should see the same output. We can also qualify inclusive ranges using a BETWEEN statement, as shown here(“inclusive” means that 1996 and 1997 are included in the range):
###Code
%%sql
select *
from orders
where extract(year from order_date) between 1996 and 1997
limit 5
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
5 rows affected.
###Markdown
2.2 Combining multiple filtering condition 2.2.1 And operatorWe can express the "between and" range with another expression. For instance the year must be greater than or equal to 2005 and less than or equal to 2010. We need to add two filter and combine the result of the two filter with an **and**.Below query should return exactly the same result as the above query
###Code
%%sql
select *
from orders
where extract(year from order_date) >= 1996 and extract(year from order_date)<= 1997
limit 5
###Output
* postgresql://user-pengfei:***@postgresql-124499/test
5 rows affected.
###Markdown
Note the "between and" express an inclusive range, for not inclusive range, we can not use it. As a result, evaluate the and of two filters become very useful. Below query returns orders that order_date > 1996 and < 1998.
###Code
%%sql
select *
from orders
where extract(year from order_date) > 1996 and extract(year from order_date)< 1998
limit 5
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
5 rows affected.
###Markdown
2.2.2 Or operatorThe OR operator will return the record if at least one of the criteria is true for the record. For instance, if we wanted only orders with months 3, 6, 9, or 12, we can use below query:
###Code
%%sql
select * from orders
where extract(month from order_date)=3
or extract(month from order_date)=6
or extract(month from order_date)=9
or extract(month from order_date)=12
limit 5;
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
5 rows affected.
###Markdown
2.2.3 In operatorIn the above example, we tested column month with a list of possible value. In this kind of situation, we can use the **In** operator.
###Code
%%sql
select * from orders
where extract(month from order_date) in (3,6,9,12)
limit 5;
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
5 rows affected.
###Markdown
We can also express the negation of the in operator by adding not in front of **in** operator.
###Code
%%sql
select * from orders
where extract(month from order_date) not in (3,6,9,12)
limit 5;
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
5 rows affected.
###Markdown
2.2.4 Arithmetic operatorsWe can notice 3,6,9,12 can all be divided by 3. So we can also use another way to get the same records as above examples.**Note some database such as Oracle does not support the modulus operator. It instead uses the MOD() function.**
###Code
%%sql
select * from orders
where cast(extract(month from order_date) as integer ) %3=0
limit 5;
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
5 rows affected.
###Markdown
2.3 Filtering text columnsThe rules for qualifying text fields follow the same structure, although there are subtle(small) differences. You can**use =, AND, OR, and IN statements with text**. However, when using text, you must wrap literals (or text values you specify) **in single quotes**.
###Code
%%sql
SELECT * FROM orders
WHERE ship_city = 'Paris'
limit 5;
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
4 rows affected.
###Markdown
Note **postgresql does not support double quote** on string value. So below query will return error in postgresql. It works for Mysql or Sqlite
###Code
%%sql
SELECT * FROM orders
WHERE ship_city = "Paris"
limit 5;
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
(psycopg2.errors.UndefinedColumn) column "Paris" does not exist
LINE 2: WHERE ship_city = "Paris"
^
[SQL: SELECT * FROM orders
WHERE ship_city = "Paris"
limit 5;]
(Background on this error at: https://sqlalche.me/e/14/f405)
###Markdown
If we do not add " or ' on Paris, the database server will get confused and think Paris is a column name rather than a text value. This single-quote rule applies to all text operations. For example, below query will return error.
###Code
%%sql
SELECT * FROM orders
WHERE ship_city = Paris
limit 5;
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
(psycopg2.errors.UndefinedColumn) column "paris" does not exist
LINE 2: WHERE ship_city = Paris
^
[SQL: SELECT * FROM orders
WHERE ship_city = Paris
limit 5;]
(Background on this error at: https://sqlalche.me/e/14/f405)
###Markdown
We can also use string value in other filter operations. Below query will return all rows that the ship_city is Paris, London, or Madrid.
###Code
%%sql
SELECT * FROM orders
WHERE ship_city in ('Paris','London','Madrid')
limit 5;
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
5 rows affected.
###Markdown
2.3.1 Other useful functions filter by using length**length** operator can get the length of a text field. For instance, below query will filter all rows that ship_country length is <=2 (e.g. UK).
###Code
%%sql
SELECT * FROM orders
WHERE length(ship_country) <= 2
limit 5
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
5 rows affected.
###Markdown
Wild card text filtering with like operatorAnother common operation is to use wildcards with LIKE followed by a regular expression, in the regular expression:- % : means any number of characters an- _ : means any single character.- Any other character is interpreted literally.So, if you wanted to find all orders that have ship_country start with the letter “C” you would run below query to find all text start with “C” and followed by any characters
###Code
%%sql
select * from orders
where ship_country like 'C%'
limit 5;
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
5 rows affected.
###Markdown
If you want find all text that have a “C” as the first character and a “n” as the third character, you can run below query.**Note the character inside '' is case-sensitive unlike the query command**. Try change below query to 'C_N' and see what happens.
###Code
%%sql
select * from orders
where ship_country like 'C_n%'
limit 5;
###Output
* postgresql://pliu:***@127.0.0.1:5432/north_wind
5 rows affected.
###Markdown
2.4 Handling nullYou may have noticed that in table orders the ship_region column have null values. **A null is a value that has no value. It is the complete absence of any content. It is a vacuous state**.In sql, **Null values cannot be determined with an = . You need to use the IS NULL or IS NOT NULL statements to identify null values**Below query returns all rows that snow_depth is null
###Code
%%sql
select * from orders
where ship_region is null
limit 5;
###Output
* postgresql://user-pengfei:***@postgresql-124499/test
5 rows affected.
###Markdown
2.4.1 Why we even have null values in the database? The null values is useful for some use case. For example, for a table of weather station, the column such as snow_depth or precipitation, it does make sense. Not because it was a sunny day (in this case, it is better to record the values as 0), but rather because some stations might not have the necessary instruments to take those measurements. Can we replace null by 0 for snow_depth?No, It might be misleading to set those values to 0 (which implies data was recorded), so those measurements should be left null.In some columns, we can not have null values. For example, the **order_id** column should be designed that it never allows nulls. Because if it's null, all the rest columns of this row become orphan data that belongs to no order.We can see that nulls values are ambiguous and it can be difficult to determine their business meaning. It is important that nullable columns (columns that are allowed to have null values) **have documented what a null value means from a business perspective**. **Otherwise, nulls should be banned from those table columns**.Do not confuse nulls with empty text(i.e., '' ). This also applies to whitespace text (i.e., ' ') . These will be treated as values andnever will be considered null. A null is definitely not the same as 0 either, because 0 is a value, whereas null is an absence of a value. 2.4.2 Problems caused by null valuesWe know that freight column has null values, try to run the following query. You can notice that the returned rows do not contain any null values. Because null is not 0 or any number, it will not qualify to any condition. So the **freight <= 6** filtered out all rows that contain null value.
###Code
%%sql
SELECT order_id, freight FROM orders
WHERE freight <= 6
limit 5
###Output
* postgresql://user-pengfei:***@postgresql-124499/test
5 rows affected.
###Markdown
But having a null value does not mean the freight > 6, so we may want to keep those rows that have null values. We can have them by using the following query
###Code
%%sql
SELECT order_id, freight FROM orders
WHERE freight is null
or freight <= 6
limit 5
###Output
* postgresql://user-pengfei:***@postgresql-124499/test
5 rows affected.
###Markdown
The above query works, but we have a more elegant way of handling null values. We can use the **coalesce() function**- coalesce(col_name,replacement_value): It takes a column name that may have null value, if the row value is null, then replace it with the given replacement_value.Below query use coalesce to replace all null value of column freight by 0 than compare them with <=6. This will not modify the origin table.
###Code
%%sql
SELECT order_id, freight FROM orders
WHERE coalesce(freight,0) <= 6
limit 5
###Output
* postgresql://user-pengfei:***@postgresql-124499/test
5 rows affected.
###Markdown
If we want the other user can use the replaced value, we can create a new column by using below query
###Code
%%sql
SELECT order_id, coalesce(freight, 0) as freight_without_null
FROM orders
limit 5;
###Output
* postgresql://user-pengfei:***@postgresql-124499/test
5 rows affected.
###Markdown
2.5 Grouping ConditionsWhen you start chaining "AND" and "OR" together, you need to make sure that you organize each set of conditions between each OR in a way that groups related conditions.For example, we need to find all orders that shipped by company 2 and freight is not bigger than 6 or the . We could write below query.
###Code
%%sql
select * from orders
where ship_via = 2 and freight <=6
or ship_city = 'Paris'
limit 5;
###Output
* postgresql://user-pengfei:***@postgresql-124499/test
5 rows affected.
###Markdown
We are lucky this works, because AND, OR have the same priority, sql resolve from left to right. So **ship_via = 2 and freight <=6** resolved to a value, then this value get resolved with **or ship_city = 'Paris'**. But for a begginer, this can be confusing, he will wonder if AND condition get resolved first or the OR condition get resolved first.To avoid this, we can explicitly group conditions in parentheses. This makes not only makes the semantics clearer, but also the execution safer.
###Code
%%sql
select * from orders
where (ship_via = 2 and freight <=6)
or ship_city = 'Paris'
limit 5;
###Output
* postgresql://user-pengfei:***@postgresql-124499/test
5 rows affected.
|
Notebooks/plots_for_midpoint_presentation.ipynb | ###Markdown
Some Prior Draws
###Code
lh = gpytorch.likelihoods.GaussianLikelihood()
model = Surrogate(test_x, true_y, lh, RBFKernel)
model.train()
lh.train();
n_samples = 4
samples = model(test_x).sample(sample_shape=torch.Size((n_samples,))).squeeze()
label_fs = 18
title_fs = 20
leg_fs = 16
lwd = 3.
plt.figure(figsize=(10, 6))
sns.set_style("white")
colors = sns.color_palette("muted")
for smp in range(n_samples):
plt.plot(test_x, samples[smp, :].detach(), color=colors[0],
linewidth=lwd)
# plt.plot(train_x, train_y.detach(), marker="o", linestyle="None",
# label="Observations", color=colors[1], markersize=5)
# plt.plot(test_x, pred_mean.detach(), label="Predicted Mean", color=colors[0],
# linewidth=lwd)
# plt.fill_between(test_x, lower.detach(), upper.detach(),
# alpha=0.2)
plt.title("Prior Samples From Gaussian Process", fontsize=title_fs)
plt.xlabel("Input", fontsize=label_fs)
plt.ylabel("Response", fontsize=label_fs)
plt.xticks([])
plt.yticks([])
sns.despine()
plt.legend( fontsize=leg_fs)
plt.show()
###Output
_____no_output_____
###Markdown
Train and Get Posterior Samples
###Code
lh = gpytorch.likelihoods.GaussianLikelihood()
model = Surrogate(train_x, train_y, lh, RBFKernel)
model.train()
lh.train()
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
mll = gpytorch.mlls.ExactMarginalLogLikelihood(lh, model)
for i in range(500):
# Zero gradients from previous iteration
optimizer.zero_grad()
# Output from model
output = model(train_x)
# Calc loss and backprop gradients
loss = -mll(output, train_y)
loss.backward()
optimizer.step()
# print(loss.item())
model.eval()
lh.eval()
pred_dist = lh(model(test_x))
pred_mean = pred_dist.mean.detach()
lower, upper = pred_dist.confidence_region()
import seaborn as sns
label_fs = 18
title_fs = 20
leg_fs = 16
lwd = 3.
plt.figure(figsize=(10, 6))
sns.set_style("white")
colors = sns.color_palette("muted")
plt.plot(test_x, true_y.detach(), label="Ground Truth", color=colors[2],
linewidth=lwd)
plt.plot(train_x, train_y.detach(), marker="o", linestyle="None",
label="Observations", color=colors[1], markersize=5)
plt.plot(test_x, pred_mean.detach(), label="Predicted Mean", color=colors[0],
linewidth=lwd)
plt.fill_between(test_x, lower.detach(), upper.detach(),
alpha=0.2)
plt.xlabel("Input", fontsize=label_fs)
plt.ylabel("Response", fontsize=label_fs)
plt.title("Posterior Gaussian Process Distribution",
fontsize=title_fs)
plt.xticks([])
plt.yticks([])
sns.despine()
plt.legend(fontsize=leg_fs)
plt.show()
###Output
_____no_output_____
###Markdown
Bayesian Optimization
###Code
train_x = torch.rand(5) * 5
train_y = true_func(train_x)
bayesopt = BayesOpt(train_x, train_y, normalize=False, normalize_y=False, max_x=5.)
# bayesopt.surrogate.train()
# bayesopt.surrogate_lh.train()
# optimizer = torch.optim.Adam(bayesopt.surrogate.parameters(), lr=0.01)
# mll = gpytorch.mlls.ExactMarginalLogLikelihood(bayesopt.surrogate_lh,
# bayesopt.surrogate)
# for i in range(200):
# # Zero gradients from previous iteration
# optimizer.zero_grad()
# # Output from model
# output = bayesopt.surrogate(bayesopt.train_x)
# # Calc loss and backprop gradients
# loss = -mll(output, bayesopt.train_y)
# loss.backward()
# optimizer.step()
# print(loss.item())
bayesopt.train_surrogate(200)
bayesopt.surrogate
next_query = bayesopt.acquire()
bayesopt.surrogate.eval();
bayesopt.surrogate_lh.eval();
pred_dist = bayesopt.surrogate_lh(bayesopt.surrogate(test_x))
pred_mean = pred_dist.mean.detach()
lower, upper = pred_dist.confidence_region()
label_fs = 18
title_fs = 20
leg_fs = 16
lwd = 3.
plt.figure(figsize=(10, 6))
sns.set_style("white")
colors = sns.color_palette("muted")
plt.plot(test_x, true_y.detach(), label="Ground Truth", color=colors[2],
linewidth=lwd, linestyle="--")
plt.plot(train_x, train_y.detach(), marker="o", linestyle="None",
label="Observations", color=colors[1], markersize=8)
plt.plot(test_x, pred_mean.detach(), label="Predictive Distribution", color=colors[0],
linewidth=lwd)
plt.fill_between(test_x, lower.detach(), upper.detach(),
alpha=0.2)
plt.scatter(next_query, true_func(next_query, 0.).detach(), marker="P", s=200,
label="Next Query")
plt.xlabel("Input", fontsize=label_fs)
plt.ylabel("Response", fontsize=label_fs)
plt.title("Optimization",
fontsize=title_fs)
plt.xticks([])
plt.yticks([])
plt.ylim(-2.5, 2.5)
sns.despine()
plt.legend(fontsize=leg_fs)
plt.show()
bayesopt.update_obs(next_query.unsqueeze(0), true_func(next_query).unsqueeze(0))
bayesopt.train_surrogate(200);
next_query = bayesopt.acquire()
bayesopt.surrogate.eval();
bayesopt.surrogate_lh.eval();
pred_dist = bayesopt.surrogate_lh(bayesopt.surrogate(test_x))
pred_mean = pred_dist.mean.detach()
lower, upper = pred_dist.confidence_region()
label_fs = 18
title_fs = 20
leg_fs = 16
lwd = 3.
plt.figure(figsize=(10, 6))
sns.set_style("white")
colors = sns.color_palette("muted")
plt.plot(test_x, true_y.detach(), label="Ground Truth", color=colors[2],
linewidth=lwd, linestyle="--")
plt.plot(bayesopt.train_x, bayesopt.train_y.detach(), marker="o", linestyle="None",
label="Observations", color=colors[1], markersize=8)
plt.plot(test_x, pred_mean.detach(), label="Predictive Distribution", color=colors[0],
linewidth=lwd)
plt.fill_between(test_x, lower.detach(), upper.detach(),
alpha=0.2)
plt.scatter(next_query, true_func(next_query, 0.).detach(), marker="P", s=200,
label="Next Query")
plt.xlabel("Input", fontsize=label_fs)
plt.ylabel("Response", fontsize=label_fs)
plt.title("Optimization",
fontsize=title_fs)
plt.xticks([])
plt.yticks([])
plt.ylim(-2.5, 2.5)
sns.despine()
plt.legend(fontsize=leg_fs)
plt.show()
bayesopt.update_obs(next_query.unsqueeze(0), true_func(next_query).unsqueeze(0))
bayesopt.train_surrogate(200);
next_query = bayesopt.acquire()
bayesopt.surrogate.eval();
bayesopt.surrogate_lh.eval();
pred_dist = bayesopt.surrogate_lh(bayesopt.surrogate(test_x))
pred_mean = pred_dist.mean.detach()
lower, upper = pred_dist.confidence_region()
label_fs = 18
title_fs = 20
leg_fs = 16
lwd = 3.
plt.figure(figsize=(10, 6))
sns.set_style("white")
colors = sns.color_palette("muted")
plt.plot(test_x, true_y.detach(), label="Ground Truth", color=colors[2],
linewidth=lwd, linestyle="--")
plt.plot(bayesopt.train_x, bayesopt.train_y.detach(), marker="o", linestyle="None",
label="Observations", color=colors[1], markersize=8)
plt.plot(test_x, pred_mean.detach(), label="Predictive Distribution", color=colors[0],
linewidth=lwd)
plt.fill_between(test_x, lower.detach(), upper.detach(),
alpha=0.2)
plt.scatter(next_query, true_func(next_query, 0.).detach(), marker="P", s=200,
label="Next Query")
plt.xlabel("Input", fontsize=label_fs)
plt.ylabel("Response", fontsize=label_fs)
plt.title("Optimization",
fontsize=title_fs)
plt.xticks([])
plt.yticks([])
plt.ylim(-2.5, 2.5)
sns.despine()
plt.legend(fontsize=leg_fs)
plt.show()
###Output
_____no_output_____
###Markdown
Network Plots
###Code
import sys
sys.path.append("../ntwrk/gym/")
import network_sim
import gym
env = gym.make("PccNs-v0")
env.reset()
send_rates = torch.arange(1, 1000, 25)
n_trial = 100
rwrds = torch.zeros(send_rates.numel(), n_trial)
for ind, sr in enumerate(send_rates):
for tt in range(n_trial):
rwrds[ind, tt] = env.step(sr.unsqueeze(0))[1]
env.reset()
mean_rwrd = rwrds.mean(1).unsqueeze(-1)
std_rwrd = rwrds.std(1).unsqueeze(-1)
mean_rwrd.shape
sns.set_style("white")
sns.set_palette("muted")
label_fs = 20
title_fs = 24
tick_fs = 16
plt.figure(figsize=(10, 6))
plt.errorbar(x=send_rates, y=mean_rwrd, yerr=std_rwrd,
linestyle="None", marker='o',
capsize=3)
plt.tick_params(labelsize=tick_fs)
plt.title("Reward At Initialization", fontsize=title_fs)
plt.xlabel("Send Rate (packets/sec)", fontsize=label_fs)
plt.ylabel("Scaled Reward", fontsize=label_fs)
sns.despine()
plt.show()
# init_send_rate = torch.tensor(850).unsqueeze(0)
# n_init_send = 20
# n_trial = 100
# rwrds = torch.zeros(send_rates.numel(), n_trial)
# for ind, sr in enumerate(send_rates):
# for tt in range(n_trial):
# for snd in range(n_init_send):
# env.step(init_send_rate)
# rwrds[ind, tt] = env.step(sr.unsqueeze(0))[1]
# env.reset()
# mean_rwrd = rwrds.mean(1).unsqueeze(-1)
# std_rwrd = rwrds.std(1).unsqueeze(-1)
# sns.set_style("white")
# sns.set_palette("muted")
# label_fs = 20
# title_fs = 24
# tick_fs = 16
# plt.figure(figsize=(10, 6))
# plt.errorbar(x=send_rates, y=mean_rwrd, yerr=std_rwrd,
# linestyle="None", marker='o',
# capsize=3)
# plt.tick_params(labelsize=tick_fs)
# plt.title("Reward After 20 Time Steps", fontsize=title_fs)
# plt.xlabel("Send Rate (packets/sec)", fontsize=label_fs)
# plt.ylabel("Scaled Reward", fontsize=label_fs)
# sns.despine()
# plt.show()
###Output
_____no_output_____
###Markdown
Bayesian Optimization for Network
###Code
env = gym.make("PccNs-v0")
env.reset()
env.senders[0].rate = env.senders[0].starting_rate
rate = torch.tensor(env.senders[0].rate).unsqueeze(0)
rwrd = torch.tensor(env.step(rate)[1]).unsqueeze(0)
max_x = 1000.
bayes_opt = BayesOpt(rate, rwrd, normalize=True, normalize_y=False, max_x=max_x,
max_jump=400)
bayes_opt.surrogate_lh.noise.data = torch.tensor([-1.])
rnds = 10
max_obs = 3
test_points = torch.arange(1, 1000).float().div(max_x)
rate_history = torch.zeros(rnds, max_obs + 1)
rwrd_history = torch.zeros(rnds, max_obs + 1)
pred_means = torch.zeros(rnds, test_points.numel())
pred_lower = torch.zeros(rnds, test_points.numel())
pred_upper = torch.zeros(rnds, test_points.numel())
for ii in range(rnds):
bayes_opt.train_surrogate(iters=200, overwrite=True)
next_rate = bayes_opt.acquire(explore=0.5).unsqueeze(0)
# next_rate = torch.rand(1)
rwrd = torch.tensor(env.step(next_rate.mul(bayes_opt.max_x))[1]).unsqueeze(0)
print(next_rate, rwrd)
rate_history[ii, :bayes_opt.train_x.numel()] = bayes_opt.train_x.clone()
rwrd_history[ii, :bayes_opt.train_y.numel()] = bayes_opt.train_y.clone()
bayes_opt.update_obs(next_rate, rwrd, max_obs=max_obs)
print("train x = ", bayes_opt.train_x)
bayes_opt.surrogate_lh.eval()
bayes_opt.surrogate.eval()
test_dist = bayes_opt.surrogate_lh(bayes_opt.surrogate(test_points))
pred_means[ii, :] = test_dist.mean.detach()
pred_lower[ii, :], pred_upper[ii, :] = test_dist.confidence_region()
rate_history[ii, -1] = bayes_opt.train_x[-1].clone()
rwrd_history[ii, -1] = bayes_opt.train_y[-1].clone()
# plt.plot(bayes_opt.train_x[:-1], bayes_opt.train_y[:-1], marker='.', linestyle="None")
# plt.plot(bayes_opt.train_x[-1], bayes_opt.train_y[-1], marker='*', linestyle="None")
# plt.plot(test_points, test_dist.mean.detach())
# plt.show()
# sns.set_style("white")
# colors = sns.color_palette("muted")
label_fs = 20
title_fs = 24
tick_fs = 16
leg_fs = 18
marksize=15
lwd = 2.5
for rnd in range(rnds):
plt.figure(figsize=(10, 6))
plt.plot(rate_history[rnd, :-1], rwrd_history[rnd, :-1], marker=".", linestyle="None",
label="Observations", color=colors[1], markersize=marksize)
plt.plot(test_points, pred_means[rnd, :], label="Predictive Distribution",
linewidth=lwd)
plt.scatter(rate_history[rnd, -1], rwrd_history[rnd, -1], marker="P",s=200,
label="Next Query", linestyle="None", color=colors[2])
plt.tick_params(labelsize=tick_fs)
plt.title("Reward vs Rate Optimization", fontsize=title_fs)
plt.xlabel("Send Rate (normalized packets/sec)", fontsize=label_fs)
plt.ylabel("Scaled Reward", fontsize=label_fs)
plt.legend(loc='upper left')
sns.despine()
plt.ylim(0, 0.6)
plt.show()
###Output
_____no_output_____ |
EDA/EDA_items.ipynb | ###Markdown
Análisis exploratorio de *items_ordered_2years* En este cuadeerno se detalla el proceso de análisis de datos de los pedidos realizados Índice de Contenidos 1. Importación de paquetes2. Carga de datos3. Análisis de los datos 3.1. Variables 3.2. Duplicados 3.3. Clientes 3.4. Ventas totales 3.5. Ventas por localización 4. Conclusiones Importación de paquetes Cargamos las librerías a usar.
###Code
import pandas as pd
import plotly.express as px
import random
seed = 124
random.seed(seed)
###Output
_____no_output_____
###Markdown
Carga de datos Abrimos el fichero y lo cargamos para poder manejar la información
###Code
df_items = pd.read_csv('../Data/items_ordered_2years.txt', sep='|', on_bad_lines='skip',parse_dates=['created_at'])
df_items.sample(5, random_state=seed)
###Output
_____no_output_____
###Markdown
Análisis de los datos En este apartado se analizarán todas las características de las variables
###Code
print("Dimensiones de los datos:", df_items.shape)
df_items.isna().sum()
###Output
_____no_output_____
###Markdown
Existen datos perdidos o no proporcionados de 3 variables distintas
###Code
percent_missing = df_items.isnull().sum() * 100 / df_items.shape[0]
df_missing_values = pd.DataFrame({
'column_name': df_items.columns,
'percent_missing': percent_missing
})
df_missing_values.sort_values(by='percent_missing',ascending=False,inplace=True)
print(df_missing_values.to_string(index=False))
###Output
column_name percent_missing
city 0.323019
zipcode 0.322697
base_cost 0.258028
num_order 0.000000
item_id 0.000000
created_at 0.000000
product_id 0.000000
qty_ordered 0.000000
price 0.000000
discount_percent 0.000000
customer_id 0.000000
###Markdown
Aunque haya datos perdidos, estos no representan una gran proporción respecto al dataset
###Code
df_items.info()
###Output
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 930905 entries, 0 to 930904
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 num_order 930905 non-null object
1 item_id 930905 non-null object
2 created_at 930905 non-null datetime64[ns]
3 product_id 930905 non-null int64
4 qty_ordered 930905 non-null int64
5 base_cost 928503 non-null float64
6 price 930905 non-null float64
7 discount_percent 930905 non-null float64
8 customer_id 930905 non-null object
9 city 927898 non-null object
10 zipcode 927901 non-null object
dtypes: datetime64[ns](1), float64(3), int64(2), object(5)
memory usage: 60.4+ MB
###Markdown
Podemos observar que tenemos 1 variable de tipo fecha, 2 variables que son números enteros (el identificador de producto y la cantidad pedida), 3 variables son números decimales (precio base, precio de venta y porcentaje de descuento) y las demás variables que son clásificadas como tipo **object** también las podemos considerar de tipo *string*
###Code
df_items.describe()
###Output
_____no_output_____
###Markdown
Al echar un vistazo por encima nos podemos dar cuenta de un par de cosas un tanto *extrañas*:* El mínimo precio base es negativo* El mínimo precio de venta es 0* El mínimo porcentaje de descuento es 1, por lo tanto no hay ningún pedido que no tenga aplicado un porcentaje de descuento
###Code
df_items.describe(include=object)
###Output
_____no_output_____
###Markdown
Vemos que el número de pedido se repite y esto se debe a que por cada producto distinto que existen dentro de un pedido necesitamos un registro nuevo, y por eso se repide el identificador de pedido Variables Ahora realizaremos un análisis más específico de cada variable *num_order* Esta variable es el identificador de los pedidos y la podemos ver repetida ya que en un mismo pedido puede haber diferentes productos, y para cada producto distinto dentro de un mismo pedido se usan distintas entradas.
###Code
orders = df_items['num_order'].nunique()
print(f'Hay {orders} pedidos registrados')
df_items['num_order'].value_counts()
###Output
_____no_output_____
###Markdown
El máximo de líneas de pedidos son 60, mientras que hay pedidos con tan solo una línea. *item_id* La variable *item_id* se usa para identificar el producto dentro del pedido, por lo tanto un mismo producto que este en pedidos distintos tendrá distintos *item_id*
###Code
items = df_items['item_id'].nunique()
print(f'Identificadores : {items}')
print(f'Registros : {df_items.shape[0]}')
###Output
Identificadores : 906266
Registros : 930905
###Markdown
Como se ve no tenemos identificadores de items como registros del dataset lo que nos lleva a pensar que podría haber registros repetidos
###Code
df_items['item_id'].value_counts()
df_items[df_items['item_id'] == '642b9b87df5b13e91ce86962684c2613']
###Output
_____no_output_____
###Markdown
Nuestras sospechas se confirman, ya que vemos que el mismo pedido tiene las tres mismas líneas. Por tanto, se trata de datos duplicados *created_at* Instante en el que se realizó el pedido
###Code
df_items['created_at'].sample(1,random_state=seed)
###Output
_____no_output_____
###Markdown
El formato de la fecha y tiempo es *YYYY-MM-DD HH:MM:SS* *product_id* Identificador del producto
###Code
products = df_items['product_id'].nunique()
print(f'Hay {products} identificadores de productos')
# Productos en más frecuentas en los pedidos
df_items['product_id'].value_counts()
###Output
_____no_output_____
###Markdown
Los productos más comunes en los pedidos, no son exactamente los mismos que los que más cantidad se piden.
###Code
# Productos más vendidos (cantidad)
df_best_products = df_items.groupby("product_id",as_index=False).agg({"num_order":"count", "qty_ordered":"sum"}).sort_values("qty_ordered", ascending=False)
df_best_products.head()
###Output
_____no_output_____
###Markdown
*qty_ordered* Cantidad pedida de cierto producto para un cierto pedido
###Code
df_q = df_items.groupby('num_order',as_index=False).agg(total_qty=('qty_ordered','sum'),n_items=('item_id','count')).sort_values('total_qty',ascending=False)
df_q.head(10)
df_q.tail(10)
###Output
_____no_output_____
###Markdown
En el resumen de la descripción del conjunto de datos, ya veíamos que los mínimos y máximos de las cantidades que se piden eran bastante dispares. Con esta tabla vemos como se reparten las cantidades pedidas respecto a los productos dentro del pedido.
###Code
df_q['total_qty'].value_counts().head(10)
df_q[df_q['total_qty'] > 10].shape[0]
###Output
_____no_output_____
###Markdown
Vemos que lo más común es que en los pedidos haya entre *2* y *10* unidades de productos. Aun así hay un número sustancia del pedidos que tienen cantidades superiores a las *10* unidades. *base_cost* Precio de coste de una unidad de producto sin IVA Como vimos al principio esta columna tiene valores perdidos. Veamos si sería posible recuperarlos mirando *base_cost* del mismo producto en otros pedidos.
###Code
products = df_items[df_items['base_cost'].isna()]['product_id'].drop_duplicates().to_list()
print(f'Nº productos con precios nulos: {len(products)}')
df_pr = df_items[(df_items['product_id'].isin(products)) & (df_items['base_cost'].notna())][['product_id','base_cost']]
print(f"Nº productos que se puede recuperar base_cost: {df_pr['product_id'].nunique()}")
df_pr.groupby('product_id',as_index=False).agg(n_cost=('base_cost','count'),mean_cost=('base_cost','mean')).sort_values('n_cost',ascending=False)
###Output
Nº productos con precios nulos: 564
Nº productos que se puede recuperar base_cost: 499
###Markdown
Parece ser que se puede recuperar bastantes valores, eso sí hay que decidir que valores imputar porque los productos varían de precio en los diferentes pedidos. Lo más extraño a mencionar es que el precio de coste medio de uno de los productos sea directamente un cero.
###Code
neg_bcost = df_items[df_items['base_cost'] < 0].shape[0]
print(f'Registros con base_cost negativo: {neg_bcost}')
###Output
Registros con base_cost negativo: 46
###Markdown
*price* Precio unitario de los productos.
###Code
pr_zero = df_items[df_items['price'] == 0].shape[0]
null_benefits = df_items[(df_items['price'] < df_items['base_cost'])][['product_id','base_cost','price','discount_percent']]
print(f'Productos con precio cero: {pr_zero}')
print(f'Líneas pedido sin beneficios: {null_benefits.shape[0]}')
###Output
Productos con precio cero: 61
Líneas pedido sin beneficios: 10729
###Markdown
Existen registros con precios nulos y también bastantes líneas de pedidos en los que nos se generan beneficios. Puede que estas últimas se traten de descuentos aplicados.
###Code
null_benefits.corr()
###Output
_____no_output_____
###Markdown
Donde nos surge la gran duda es en que no hay correspondencia entre los costes, precios y descuentos de los productos. Por tanto, podemos intuir que los precios sobre los que se aplican los descuentos no son estos, sino otros, ya que nosotros tenemos los precios finales. *discount_percent* Porcentaje de descuento aplicado, pero no al precio con el que contamos nosotros.
###Code
print(f"Líneas con descuento: {df_items[df_items['discount_percent'] > 0].shape[0]}")
print(f'Lineas total: {df_items.shape[0]}')
###Output
Líneas con descuento: 930905
Lineas total: 930905
###Markdown
Algo que llama la atención es que todos las líneas de pedidos tienen descuentos
###Code
print(f"Cantidad de porc. desc.: {df_items['discount_percent'].nunique()}")
df_items['discount_percent'].value_counts().sort_values(ascending=False).head(10)
df_items['discount_percent'].value_counts().sort_values(ascending=False).tail(10)
###Output
_____no_output_____
###Markdown
Los porcentajes de descuento más comunes son por debajo de *25%*, mientras que los menos comunes oscilan entre el *30% y 50%*. *customer_id* El identificador del cliente que ha realizado el pedido
###Code
clients = df_items['customer_id'].nunique()
print(f'{clients} clientes han realizado pedidos')
df_items['customer_id'].sample(5,random_state=seed)
###Output
_____no_output_____
###Markdown
Este campo es un *hash* con el fin de anonimizar los datos personales. *city* y *zipcode* Estas variables indican la ciudad en la cuál se han realizado el pedido
###Code
print(f"Nº de ciudades: {df_items['city'].nunique()}")
print(f"Nº de ciudades: {df_items['zipcode'].nunique()}")
###Output
Nº de ciudades: 20993
Nº de ciudades: 11116
###Markdown
Vimos antes que estas dos columnas tienen un número similar de registros nulos, lo que da a entender que puede que coincidan.
###Code
df_items[(df_items.city.isna()) & (df_items.zipcode.isna())].shape[0]
###Output
_____no_output_____
###Markdown
Tenemos 2910 registros donde no tenemos ninguna información de la dirección, ni la ciudad ni el código postal. A pesar de que no son demasiados registros, es información que no podremos recuperar de ninguna manera, ya que los datos están anonimizados
###Code
cities = df_items[(df_items['city'].notna()) & (df_items['city'].str.contains('alba'))]['city']
cities.value_counts()
###Output
_____no_output_____
###Markdown
Haciendo una búsqueda sencilla de cadenas, vemos que el campo no está normalizado y hay nombres de ciudades escritos de diferentes maneras.
###Code
# Normalizamos los nombres de ciudades
indexes = df_items['city'].notna().index
df_items.loc[indexes, 'city'] = df_items.loc[indexes, 'city'].apply(lambda x: str(x).lower().strip())
print(f"Nº de ciudades: {df_items['city'].nunique()}")
###Output
Nº de ciudades: 11569
###Markdown
Cuando normalizamos vemos que el número de ciudades disminuye. Aun así, faltarían muchos más métodos que aplicar para llegar a normalizar el campo. **NOTA** Por otro lado, mientras que estabamos limpiando datos, nos dimos cuenta que hay valores númericos para ciudades y códigos postales imputados como nombres.
###Code
df_city = df_items[(df_items['city'].notna()) & (df_items['city'].str.isnumeric())]
print(f"Nº ciudad con dígitos: {df_city.shape[0]}")
df_city[['num_order','city','zipcode']].sample(5,random_state=seed)
df_zip = df_items[(df_items['zipcode'].notna()) & (df_items['zipcode'].notna())]
df_zip = df_zip[(df_zip['city'].str.isnumeric()) & (~df_zip['zipcode'].str.isnumeric())]
print(f"Nº códigos postales y ciudad invertidos: {df_zip.shape[0]}")
df_zip[['city','zipcode']].sample(10,random_state=seed)
###Output
Nº códigos postales y ciudad invertidos: 357
###Markdown
Duplicados En apartados anteriores intuimos la presencia de registros duplicados, veamos a que se debe.
###Code
print(f"Regitros duplicados: {df_items.duplicated().sum()}")
df_items[df_items.duplicated()].head(5)
###Output
Regitros duplicados: 17599
###Markdown
Clientes que más compran Clientes que más pedidos hacen
###Code
df_clients = df_items.groupby(['customer_id', 'num_order'],as_index=False).agg({'qty_ordered':'sum'}) # num_order se repite, por tanto no se puede agregar por el de primeras
df_clients = df_clients.groupby(['customer_id'],as_index=False).agg({'num_order':'count', 'qty_ordered':'sum'})
df_clients = df_clients.sort_values('num_order',ascending=False)
df_clients.head()
###Output
_____no_output_____
###Markdown
Clientes que han comprado una mayor cantidad de productos
###Code
df_clientes = df_clients.sort_values('qty_ordered',ascending=False)
df_clientes.head()
###Output
_____no_output_____
###Markdown
Ventas totales En este apartado veremos las fechas en las cuáles se producen más ventas
###Code
# Modificamos la columna para que solo muestre la fecha sin la hora
df_items.loc[:, 'date'] = df_items['created_at'].dt.date
df_items[['created_at','date']].sample(5,random_state=seed)
###Output
_____no_output_____
###Markdown
Agrupamos por fecha para saber la cantidad de productos y el número de pedidos realizados por cada día
###Code
df_temp = df_items.groupby(['date', 'num_order']).agg({'qty_ordered':'sum'}).reset_index() # num_order se repite, por tanto no se puede agregar por el de primeras
df_temp = df_temp.groupby(['date']).agg({'num_order':'count', 'qty_ordered':'sum'}).reset_index()
df_temp.sample(5,random_state=seed)
fig = px.area(df_temp, x="date", y="num_order", title='Pedidos realizados por día',)
fig.show()
###Output
_____no_output_____
###Markdown
Vemos los picos de pedidos en los meses como Noviembre (BlackFriday) y picos en las rebajas tanto de invierno como de verano
###Code
fig = px.area(df_temp, x="date", y="qty_ordered", title='Cantidad de productos pedidos por día')
fig.show()
###Output
_____no_output_____
###Markdown
La cantidad de productos pedidos por día es muy similar en forma a la anterior gráfica, por lo que tienen cierta correlación. Aun así, nos debemos fijar en la escala del eje Y.
###Code
df_temp.corr()
###Output
_____no_output_____
###Markdown
Ventas por localización Puesto que hemos sacado las coordenadas de cada localización podemos mostrar visualmente donde se han producido más ventas
###Code
df_temp = df_items.groupby(['city', 'num_order'],as_index=False).agg({'qty_ordered':'sum'}) # num_order se repite, por tanto no se puede agregar por el de primeras
df_temp = df_temp.groupby(['city'],as_index=False).agg({'num_order':'count', 'qty_ordered':'sum'})
df_temp.sort_values('num_order',ascending=False).head(10)
###Output
_____no_output_____ |
conditionals.ipynb | ###Markdown
Conditionals A very simple program might just be a sequence of statements: a = 1 print(a) a = 2 print(a) But virtually nothing interesting can be written this way -- because what happens can never change. The program cannot change depending on what button you clicked on screen, what packets have come in over the network, what bytes you have read from a file or upon calculations done in advance.We can use simple diagrams to show how branching affects programs. Consider the sequence of statements above. We can draw that as:This is pretty boring, so we can simplify all the statements to just:where the arrow is assumed to include a sequence of statements without branches.If we add a **branch** to our program, we now have two possible exits at some block in the code:Here, **true_path** is only executed if some condition was true at the `if` block. Learning to see the structure of programs as graphs like this is a key part of understanding how programs will run. We'll see several of these graphs in this lecture. ConditionalsA conditional is an expression that evaluates to True or False (i.e. a **boolean** expression). `if` takes a conditional to decide which branch of code to execute.The comparison operators are used to test values and the boolean operators combine sub-expressions together.Valid conditional expressions include: x == 0 [equality] x > 5 [comparison] status == "alive" or status == "dead" [boolean operator] The expression following an if statement must be a conditional. It must evaluate to either `True` or `False`. Block syntax: indentationIn Python, we denote the section of code following `if` by **indenting** it (putting spaces in front of the following lines). The `if` continues until the code indentation moves back to where it was before. An indented block always starts with a colon `:`.**All blocks of code in Python are denoted this way. Python is whitespace-sensitive. **There are no block markers like braces `{ }` or `begin` and `end` in Python. The block of code is defined by the colon followed by the indentation (spacing) **alone**. The code that "belongs" to a `if` statement is everything which has the matching indentation. All other language features which deal with blocks of code (i.e. need to work with multiple statements) use indentation to mark the blocks of code.
###Code
# this is fine
x = False
if x:
print("hello")
print("there")
print("after")
# this is an indentation error -- it will not run!
x = True
if x:
print ("hello")
print ("there")
# ^
# | does not match any possible indentation!
# this is also an indentation error -- it will not run!
x = True
if x:
print ("hello")
print ("there")
###Output
_____no_output_____
###Markdown
ifHow do we introduce such branches in our programs. The simplest branching statement is `if`. `if` takes an expression and executes one sequence of code only if the given expression is `True`. if : ... elseWe can have a branch with two different paths using `else`. One of the two paths is always executed; the `if` path if the expression is True or the `else` path if it is False. if : else:
###Code
#If the if condition is true, the if statement is executed.
#The program then continues after the else statement.
exam = 28
if exam < 40:
print("You failed", end = " ")
else:
print("You passed", end = " ")
print("the exams")
###Output
You failed the exams
###Markdown
if....elif....elseSometimes we need to have more than two branches. For example, we might have a temperature scale in a recipe in Celsius, and we need to work out what Gas Mark to put our oven at: We can use `elif` to combine an `else` and an `if` together. We can still use a final `else` statement.
###Code
temp = 150
if temp<135:
gas_mark=1
elif temp<149:
gas_mark =2
elif temp<163:
gas_mark = 3
else:
# if it's hotter than that, just turn the burner on full blast!
gas_mark = 100
print(gas_mark)
###Output
3
###Markdown
5. Compound Boolean ExpressionsThe can have more than one condition on an if statement. The conditions need to be joined together with `and` , `or` or `not` operators to create a large boolean expression. `and` All the conditions need to be true for the expression to be true. `or` Any of the conditions need to be true for the expression to be true.
###Code
#Both conditions of the or expression are true.
weather = "wet"
temp = 24
if weather == "wet" or temp < 0:
print("I'm not going outside today")
elif weather == "sunny" and temp > 20:
print("I'm going to need suncream today")
else:
print("Not sure what I'll do today")
###Output
I'm not going outside today
|
Lesson_5/convolution_visualization/conv_visualization.ipynb | ###Markdown
Convolutional LayerIn this notebook, we visualize four filtered outputs (a.k.a. activation maps) of a convolutional layer. In this example, *we* are defining four filters that are applied to an input image by initializing the **weights** of a convolutional layer, but a trained CNN will learn the values of these weights. Import the image
###Code
import cv2
import matplotlib.pyplot as plt
%matplotlib inline
# TODO: Feel free to try out your own images here by changing img_path
# to a file path to another image on your computer!
img_path = 'data/udacity_sdc.png'
# load color image
bgr_img = cv2.imread(img_path)
# convert to grayscale
gray_img = cv2.cvtColor(bgr_img, cv2.COLOR_BGR2GRAY)
# normalize, rescale entries to lie in [0,1]
gray_img = gray_img.astype("float32")/255
# plot image
plt.imshow(gray_img, cmap='gray')
plt.show()
###Output
_____no_output_____
###Markdown
Define and visualize the filters
###Code
import numpy as np
## TODO: Feel free to modify the numbers here, to try out another filter!
filter_vals = np.array([[-1, -1, 1, 1], [-1, -1, 1, 1], [-1, -1, 1, 1], [-1, -1, 1, 1]])
print('Filter shape: ', filter_vals.shape)
# Defining four different filters,
# all of which are linear combinations of the `filter_vals` defined above
# define four filters
filter_1 = filter_vals
filter_2 = -filter_1
filter_3 = filter_1.T
filter_4 = -filter_3
filters = np.array([filter_1, filter_2, filter_3, filter_4])
# For an example, print out the values of filter 1
print('Filter 1: \n', filter_1)
#filter 3
print('Filter 3: \n', filter_3)
# visualize all four filters
fig = plt.figure(figsize=(10, 5))
for i in range(4):
ax = fig.add_subplot(1, 4, i+1, xticks=[], yticks=[])
ax.imshow(filters[i], cmap='gray')
ax.set_title('Filter %s' % str(i+1))
width, height = filters[i].shape
for x in range(width):
for y in range(height):
ax.annotate(str(filters[i][x][y]), xy=(y,x),
horizontalalignment='center',
verticalalignment='center',
color='white' if filters[i][x][y]<0 else 'black')
###Output
_____no_output_____
###Markdown
Define a convolutional layer The various layers that make up any neural network are documented, [here](http://pytorch.org/docs/stable/nn.html). For a convolutional neural network, we'll start by defining a:* Convolutional layerInitialize a single convolutional layer so that it contains all your created filters. Note that you are not training this network; you are initializing the weights in a convolutional layer so that you can visualize what happens after a forward pass through this network! `__init__` and `forward`To define a neural network in PyTorch, you define the layers of a model in the function `__init__` and define the forward behavior of a network that applyies those initialized layers to an input (`x`) in the function `forward`. In PyTorch we convert all inputs into the Tensor datatype, which is similar to a list data type in Python. Below, I define the structure of a class called `Net` that has a convolutional layer that can contain four 3x3 grayscale filters.
###Code
import torch
import torch.nn as nn
import torch.nn.functional as F
# define a neural network with a single convolutional layer with four filters
class Net(nn.Module):
def __init__(self, weight):
super(Net, self).__init__()
# initializes the weights of the convolutional layer to be the weights of the 4 defined filters
k_height, k_width = weight.shape[2:]
# assumes there are 4 grayscale filters
self.conv = nn.Conv2d(1, 4, kernel_size=(k_height, k_width), bias=False)
self.conv.weight = torch.nn.Parameter(weight)
def forward(self, x):
# calculates the output of a convolutional layer
# pre- and post-activation
conv_x = self.conv(x)
activated_x = F.relu(conv_x)
# returns both layers
return conv_x, activated_x
# instantiate the model and set the weights
weight = torch.from_numpy(filters).unsqueeze(1).type(torch.FloatTensor)
model = Net(weight)
# print out the layer in the network
print(model)
###Output
Net(
(conv): Conv2d(1, 4, kernel_size=(4, 4), stride=(1, 1), bias=False)
)
###Markdown
Visualize the output of each filterFirst, we'll define a helper function, `viz_layer` that takes in a specific layer and number of filters (optional argument), and displays the output of that layer once an image has been passed through.
###Code
# helper function for visualizing the output of a given layer
# default number of filters is 4
def viz_layer(layer, n_filters= 4):
fig = plt.figure(figsize=(20, 20))
for i in range(n_filters):
ax = fig.add_subplot(1, n_filters, i+1, xticks=[], yticks=[])
# grab layer outputs
ax.imshow(np.squeeze(layer[0,i].data.numpy()), cmap='gray')
ax.set_title('Output %s' % str(i+1))
###Output
_____no_output_____
###Markdown
Let's look at the output of a convolutional layer, before and after a ReLu activation function is applied.
###Code
# plot original image
plt.imshow(gray_img, cmap='gray')
# visualize all filters
fig = plt.figure(figsize=(12, 6))
fig.subplots_adjust(left=0, right=1.5, bottom=0.8, top=1, hspace=0.05, wspace=0.05)
for i in range(4):
ax = fig.add_subplot(1, 4, i+1, xticks=[], yticks=[])
ax.imshow(filters[i], cmap='gray')
ax.set_title('Filter %s' % str(i+1))
# convert the image into an input Tensor
gray_img_tensor = torch.from_numpy(gray_img).unsqueeze(0).unsqueeze(1)
# get the convolutional layer (pre and post activation)
conv_layer, activated_layer = model(gray_img_tensor)
# visualize the output of a conv layer
viz_layer(conv_layer)
###Output
_____no_output_____
###Markdown
ReLu activationIn this model, we've used an activation function that scales the output of the convolutional layer. We've chose a ReLu function to do this, and this function simply turns all negative pixel values in 0's (black). See the equation pictured below for input pixel values, `x`.
###Code
# after a ReLu is applied
# visualize the output of an activated conv layer
viz_layer(activated_layer)
###Output
_____no_output_____ |
Basic/PythonNegativeIndexing.ipynb | ###Markdown
We can index single characters in strings using the bracket notation. The first character has index 0, the second index 1, and so on. Did you ever want to access the last element in a string? Counting the indices can be a real pain for long strings with more than 8-10 characters.But no worries, Python has a language feature for this.Instead of starting counting from the left, you can also start from the right. Access the last character with the negative index -1, the second last with the index -2, and so on.In summary, there are two ways to index sequence positions, from the left and from the right:
###Code
x = 'cool'
print(x[-1] + x[-2] + x[-4] + x[-3])
###Output
_____no_output_____ |
Exercise py/07-Sets and Booleans.ipynb | ###Markdown
Set and BooleansThere are two other object types in Python that we should quickly cover: Sets and Booleans. SetsSets are an unordered collection of *unique* elements. We can construct them by using the set() function. Let's go ahead and make a set to see how it works
###Code
x = set()
# We add to sets with the add() method
x.add(1)
#Show
x
###Output
_____no_output_____
###Markdown
Note the curly brackets. This does not indicate a dictionary! Although you can draw analogies as a set being a dictionary with only keys.We know that a set has only unique entries. So what happens when we try to add something that is already in a set?
###Code
# Add a different element
x.add(2)
#Show
x
# Try to add the same element
x.add(1)
#Show
x
###Output
_____no_output_____
###Markdown
Notice how it won't place another 1 there. That's because a set is only concerned with unique elements! We can cast a list with multiple repeat elements to a set to get the unique elements. For example:
###Code
# Create a list with repeats
list1 = [1,1,2,2,3,4,5,6,1,1]
# Cast as set to get unique values
set(list1)
###Output
_____no_output_____
###Markdown
BooleansPython comes with Booleans (with predefined True and False displays that are basically just the integers 1 and 0). It also has a placeholder object called None. Let's walk through a few quick examples of Booleans (we will dive deeper into them later in this course).
###Code
# Set object to be a boolean
a = True
#Show
a
###Output
_____no_output_____
###Markdown
We can also use comparison operators to create booleans. We will go over all the comparison operators later on in the course.
###Code
# Output is boolean
1 > 2
###Output
_____no_output_____
###Markdown
We can use None as a placeholder for an object that we don't want to reassign yet:
###Code
# None placeholder
b = None
# Show
print(b)
###Output
None
|
files/IO/06.Protobuf.ipynb | ###Markdown
[Protobuf](https://developers.google.com/protocol-buffers/docs/pythontutorial?hl=rucompiling-your-protocol-buffers)
###Code
40*10000/1024
!sudo apt-get install protobuf-compiler
!pip install protobuf
!mkdir proto_example
!touch proto_example/__init__.py
!protoc --python_out=proto_example example.proto
from proto_example.example_pb2 import ParticleList
particle_list = ParticleList()
with open("build/example.bin", "rb") as fin:
particle_list.ParseFromString(fin.read())
for indx, particle in enumerate(particle_list.particle):
if indx > 100:
md = particle.momentum_direction
print(particle.id, particle.energy)
print(md.x, md.y, md.z)
break
###Output
101 317.14
-1.0 1.0 1.0
|
docs/memo/notebooks/data/zacks.broker_ratings/notebook.ipynb | ###Markdown
Zack's Broker Ratings Revision HistoryIn this notebook, we'll take a look at Zack's *Broker Ratings Revision History* dataset, available on [Quantopian](https://www.quantopian.com/store). This dataset spans 2002 through the current day and provides Analyst Ratings History for Securities.Update time: Zacks updates this dataset sometime during the first week of each month. In pipeline, your data will be updated close to midnight. So on the 27th, you will have data with a maximum asof_date of the 26th. Notebook ContentsThere are two ways to access the data and you'll find both of them listed below. Just click on the section you'd like to read through.- Interactive overview: This is only available on Research and uses blaze to give you access to large amounts of data. Recommended for exploration and plotting.- Pipeline overview: Data is made available through pipeline which is available on both the Research & Backtesting environment. Recommended for custom factor development and moving back & forth between research/backtesting. Free samples and limitsOne key caveat: we limit the number of results returned from any given expression to 10,000 to protect against runaway memory usage. To be clear, you have access to all the data server side. We are limiting the size of the responses back from Blaze.There is a *free* version of this dataset as well as a paid one. The free sample includes data until 2 months prior to the current date.To access the most up-to-date values for this data set for trading a live algorithm (as with other partner sets), you need to purchase acess to the full set.With preamble in place, let's get started:Interactive Overview Accessing the data with Blaze and Interactive on ResearchPartner datasets are available on Quantopian Research through an API service known as [Blaze](http://blaze.pydata.org). Blaze provides the Quantopian user with a convenient interface to access very large datasets, in an interactive, generic manner.Blaze provides an important function for accessing these datasets. Some of these sets are many millions of records. Bringing that data directly into Quantopian Research directly just is not viable. So Blaze allows us to provide a simple querying interface and shift the burden over to the server side.It is common to use Blaze to reduce your dataset in size, convert it over to Pandas and then to use Pandas for further computation, manipulation and visualization.Helpful links:* [Query building for Blaze](http://blaze.readthedocs.io/en/latest/queries.html)* [Pandas-to-Blaze dictionary](http://blaze.readthedocs.io/en/latest/rosetta-pandas.html)* [SQL-to-Blaze dictionary](http://blaze.readthedocs.io/en/latest/rosetta-sql.html).Once you've limited the size of your Blaze object, you can convert it to a Pandas DataFrames using:> `from odo import odo` > `odo(expr, pandas.DataFrame)`To see how this data can be used in your algorithm, search for the `Pipeline Overview` section of this notebook or head straight to Pipeline Overview
###Code
# import the free sample of the dataset
from quantopian.interactive.data.zacks import broker_ratings as dataset
# import data operations
from odo import odo
# import other libraries we will use
import pandas as pd
import matplotlib.pyplot as plt
# Let's use blaze to understand the data a bit using Blaze dshape()
dataset.timestamp.min()
# And how many rows are there?
# N.B. we're using a Blaze function to do this, not len()
dataset.count()
# Let's see what the data looks like. We'll grab three rows
dataset.tail(3)
###Output
_____no_output_____
###Markdown
Let's go over the columns:- **file_prod_date**: File production date- **m_ticker**: Master ticker or trading symbol- **symbol**: Ticker- **comp_name**: Company name- **comp_name_2**: Company name 2- **exchange**: Exchange traded- **currency_code**: Currency code- **rating_cnt_strong_buys**: Number of analysts with a strong buy rating- **rating_cnt_mod_buys**: Number of analysts with a moderate buy rating- **rating_cnt_holds**: Number of analysts with a hold rating- **rating_cnt_mod_sells**: Number of analysts with a moderate sell rating- **rating_cnt_strong_sells**: Number of analysts with a strong sell rating- **rating_mean_recom**: Average rating recommendation- **rating_cnt_with**: Number of analysts with a rating- **rating_cnt_without**: Number of analysts with no rating- **asof_date**: Observation date- **timestamp**: This is our timestamp on when we registered the data.We've done much of the data processing for you. Fields like `timestamp` and `sid` are standardized across all our Store Datasets, so the datasets are easy to combine. We have standardized the `sid` across all our equity databases.We can select columns and rows with ease. Below, we'll fetch all rows for Apple (sid 24) and explore the scores a bit with a chart.
###Code
aapl_data = dataset_100[dataset_100.symbol == 'AAPL']
aapl = odo(aapl_data, pd.DataFrame)
# suppose we want the rows to be indexed by timestamp.
aapl.index = list(aapl['asof_date'])
aapl.drop('asof_date',1,inplace=True)
aapl[-3:]
###Output
_____no_output_____
###Markdown
Pipeline Overview Accessing the data in your algorithms & researchThe only method for accessing partner data within algorithms running on Quantopian is via the pipeline API. Different data sets work differently but in the case of this data, you can add this data to your pipeline as follows:
###Code
# Import necessary Pipeline modules
from quantopian.pipeline import Pipeline
from quantopian.research import run_pipeline
from quantopian.pipeline.factors import AverageDollarVolume
# For use in your algorithms
# Using the full/sample paid dataset in your pipeline algo
from quantopian.pipeline.data.zacks import broker_ratings
###Output
_____no_output_____
###Markdown
Now that we've imported the data, let's take a look at which fields are available for each dataset.You'll find the dataset, the available fields, and the datatypes for each of those fields.
###Code
print "Here are the list of available fields per dataset:"
print "---------------------------------------------------\n"
def _print_fields(dataset):
print "Dataset: %s\n" % dataset.__name__
print "Fields:"
for field in list(dataset.columns):
print "%s - %s" % (field.name, field.dtype)
print "\n"
for data in (broker_ratings,):
_print_fields(data)
print "---------------------------------------------------\n"
###Output
Here are the list of available fields per dataset:
---------------------------------------------------
Dataset: broker_ratings
Fields:
comp_name - object
exchange - object
currency_code - object
symbol - object
m_ticker - object
rating_mean_recom - float64
rating_cnt_without - float64
rating_cnt_strong_buys - float64
asof_date - datetime64[ns]
rating_cnt_mod_buys - float64
rating_cnt_holds - float64
file_prod_date - datetime64[ns]
rating_cnt_mod_sells - float64
comp_name_2 - object
rating_cnt_strong_sells - float64
rating_cnt_with - float64
---------------------------------------------------
###Markdown
Now that we know what fields we have access to, let's see what this data looks like when we run it through Pipeline.This is constructed the same way as you would in the backtester. For more information on using Pipeline in Research view this thread:https://www.quantopian.com/posts/pipeline-in-research-build-test-and-visualize-your-factors-and-filters
###Code
# Let's see what this data looks like when we run it through Pipeline
# This is constructed the same way as you would in the backtester. For more information
# on using Pipeline in Research view this thread:
# https://www.quantopian.com/posts/pipeline-in-research-build-test-and-visualize-your-factors-and-filters
columns = {'Strong Sells': broker_ratings.rating_cnt_strong_sells.latest,
'Strong Buys': broker_ratings.rating_cnt_strong_buys.latest}
pipe = Pipeline(columns=columns, screen=broker_ratings.rating_cnt_mod_sells.latest.notnan())
# The show_graph() method of pipeline objects produces a graph to show how it is being calculated.
pipe.show_graph(format='png')
# run_pipeline will show the output of your pipeline
pipe_output = run_pipeline(pipe, start_date='2013-11-01', end_date='2013-11-25')
pipe_output
###Output
_____no_output_____
###Markdown
Zack's Broker Ratings Revision HistoryIn this notebook, we'll take a look at Zack's *Broker Ratings Revision History* dataset, available on [Quantopian](https://www.quantopian.com/store). This dataset spans 2002 through the current day and provides Analyst Ratings History for Securities.Update time: Zacks updates this dataset sometime during the first week of each month. In pipeline, your data will be updated close to midnight. So on the 27th, you will have data with a maximum asof_date of the 26th. Notebook ContentsThere are two ways to access the data and you'll find both of them listed below. Just click on the section you'd like to read through.- Interactive overview: This is only available on Research and uses blaze to give you access to large amounts of data. Recommended for exploration and plotting.- Pipeline overview: Data is made available through pipeline which is available on both the Research & Backtesting environment. Recommended for custom factor development and moving back & forth between research/backtesting. Free samples and limitsOne key caveat: we limit the number of results returned from any given expression to 10,000 to protect against runaway memory usage. To be clear, you have access to all the data server side. We are limiting the size of the responses back from Blaze.There is a *free* version of this dataset as well as a paid one. The free sample includes data until 2 months prior to the current date.To access the most up-to-date values for this data set for trading a live algorithm (as with other partner sets), you need to purchase acess to the full set.With preamble in place, let's get started:Interactive Overview Accessing the data with Blaze and Interactive on ResearchPartner datasets are available on Quantopian Research through an API service known as [Blaze](http://blaze.pydata.org). Blaze provides the Quantopian user with a convenient interface to access very large datasets, in an interactive, generic manner.Blaze provides an important function for accessing these datasets. Some of these sets are many millions of records. Bringing that data directly into Quantopian Research directly just is not viable. So Blaze allows us to provide a simple querying interface and shift the burden over to the server side.It is common to use Blaze to reduce your dataset in size, convert it over to Pandas and then to use Pandas for further computation, manipulation and visualization.Helpful links:* [Query building for Blaze](http://blaze.readthedocs.io/en/latest/queries.html)* [Pandas-to-Blaze dictionary](http://blaze.readthedocs.io/en/latest/rosetta-pandas.html)* [SQL-to-Blaze dictionary](http://blaze.readthedocs.io/en/latest/rosetta-sql.html).Once you've limited the size of your Blaze object, you can convert it to a Pandas DataFrames using:> `from odo import odo` > `odo(expr, pandas.DataFrame)`To see how this data can be used in your algorithm, search for the `Pipeline Overview` section of this notebook or head straight to Pipeline Overview
###Code
# import the free sample of the dataset
from quantopian.interactive.data.zacks import broker_ratings as dataset
# import data operations
from odo import odo
# import other libraries we will use
import pandas as pd
import matplotlib.pyplot as plt
# Let's use blaze to understand the data a bit using Blaze dshape()
dataset.timestamp.min()
# And how many rows are there?
# N.B. we're using a Blaze function to do this, not len()
dataset.count()
# Let's see what the data looks like. We'll grab three rows
dataset.tail(3)
###Output
_____no_output_____
###Markdown
Let's go over the columns:- **file_prod_date**: File production date- **m_ticker**: Master ticker or trading symbol- **symbol**: Ticker- **comp_name**: Company name- **comp_name_2**: Company name 2- **exchange**: Exchange traded- **currency_code**: Currency code- **rating_cnt_strong_buys**: Number of analysts with a strong buy rating- **rating_cnt_mod_buys**: Number of analysts with a moderate buy rating- **rating_cnt_holds**: Number of analysts with a hold rating- **rating_cnt_mod_sells**: Number of analysts with a moderate sell rating- **rating_cnt_strong_sells**: Number of analysts with a strong sell rating- **rating_mean_recom**: Average rating recommendation- **rating_cnt_with**: Number of analysts with a rating- **rating_cnt_without**: Number of analysts with no rating- **asof_date**: Observation date- **timestamp**: This is our timestamp on when we registered the data.We've done much of the data processing for you. Fields like `timestamp` and `sid` are standardized across all our Store Datasets, so the datasets are easy to combine. We have standardized the `sid` across all our equity databases.We can select columns and rows with ease. Below, we'll fetch all rows for Apple (sid 24) and explore the scores a bit with a chart.
###Code
aapl_data = dataset_100[dataset_100.symbol == 'AAPL']
aapl = odo(aapl_data, pd.DataFrame)
# suppose we want the rows to be indexed by timestamp.
aapl.index = list(aapl['asof_date'])
aapl.drop('asof_date',1,inplace=True)
aapl[-3:]
###Output
_____no_output_____
###Markdown
Pipeline Overview Accessing the data in your algorithms & researchThe only method for accessing partner data within algorithms running on Quantopian is via the pipeline API. Different data sets work differently but in the case of this data, you can add this data to your pipeline as follows:
###Code
# Import necessary Pipeline modules
from quantopian.pipeline import Pipeline
from quantopian.research import run_pipeline
from quantopian.pipeline.factors import AverageDollarVolume
# For use in your algorithms
# Using the full/sample paid dataset in your pipeline algo
from quantopian.pipeline.data.zacks import broker_ratings
###Output
_____no_output_____
###Markdown
Now that we've imported the data, let's take a look at which fields are available for each dataset.You'll find the dataset, the available fields, and the datatypes for each of those fields.
###Code
print "Here are the list of available fields per dataset:"
print "---------------------------------------------------\n"
def _print_fields(dataset):
print "Dataset: %s\n" % dataset.__name__
print "Fields:"
for field in list(dataset.columns):
print "%s - %s" % (field.name, field.dtype)
print "\n"
for data in (broker_ratings,):
_print_fields(data)
print "---------------------------------------------------\n"
###Output
Here are the list of available fields per dataset:
---------------------------------------------------
Dataset: broker_ratings
Fields:
comp_name - object
exchange - object
currency_code - object
symbol - object
m_ticker - object
rating_mean_recom - float64
rating_cnt_without - float64
rating_cnt_strong_buys - float64
asof_date - datetime64[ns]
rating_cnt_mod_buys - float64
rating_cnt_holds - float64
file_prod_date - datetime64[ns]
rating_cnt_mod_sells - float64
comp_name_2 - object
rating_cnt_strong_sells - float64
rating_cnt_with - float64
---------------------------------------------------
###Markdown
Now that we know what fields we have access to, let's see what this data looks like when we run it through Pipeline.This is constructed the same way as you would in the backtester. For more information on using Pipeline in Research view this thread:https://www.quantopian.com/posts/pipeline-in-research-build-test-and-visualize-your-factors-and-filters
###Code
# Let's see what this data looks like when we run it through Pipeline
# This is constructed the same way as you would in the backtester. For more information
# on using Pipeline in Research view this thread:
# https://www.quantopian.com/posts/pipeline-in-research-build-test-and-visualize-your-factors-and-filters
columns = {'Strong Sells': broker_ratings.rating_cnt_strong_sells.latest,
'Strong Buys': broker_ratings.rating_cnt_strong_buys.latest}
pipe = Pipeline(columns=columns, screen=broker_ratings.rating_cnt_mod_sells.latest.notnan())
# The show_graph() method of pipeline objects produces a graph to show how it is being calculated.
pipe.show_graph(format='png')
# run_pipeline will show the output of your pipeline
pipe_output = run_pipeline(pipe, start_date='2013-11-01', end_date='2013-11-25')
pipe_output
###Output
_____no_output_____ |
RankingIntermediateChanges.ipynb | ###Markdown
Application of ML in Ranking Application of ML in Ranking of Search Engines Downloading Data From Kaggle Might Take a while
###Code
from google.colab import drive
drive.mount('/content/drive', force_remount=True)
# Download Data
!pip install kaggle
import json
import os
from pprint import pprint
with open('/content/drive/My Drive/kaggle.json') as kc:
kaggle_config = json.load(kc)
os.environ['KAGGLE_USERNAME'] = kaggle_config['username']
os.environ['KAGGLE_KEY'] = kaggle_config['key']
!kaggle competitions download -c text-relevance-competition-ir-2-itmo-fall-2019
!unzip docs.tsv.zip
!rm docs.tsv.zip
###Output
Requirement already satisfied: kaggle in /usr/local/lib/python3.6/dist-packages (1.5.6)
Requirement already satisfied: six>=1.10 in /usr/local/lib/python3.6/dist-packages (from kaggle) (1.12.0)
Requirement already satisfied: urllib3<1.25,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from kaggle) (1.24.3)
Requirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from kaggle) (2.21.0)
Requirement already satisfied: certifi in /usr/local/lib/python3.6/dist-packages (from kaggle) (2019.11.28)
Requirement already satisfied: python-dateutil in /usr/local/lib/python3.6/dist-packages (from kaggle) (2.6.1)
Requirement already satisfied: python-slugify in /usr/local/lib/python3.6/dist-packages (from kaggle) (4.0.0)
Requirement already satisfied: tqdm in /usr/local/lib/python3.6/dist-packages (from kaggle) (4.41.1)
Requirement already satisfied: idna<2.9,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->kaggle) (2.8)
Requirement already satisfied: chardet<3.1.0,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->kaggle) (3.0.4)
Requirement already satisfied: text-unidecode>=1.3 in /usr/local/lib/python3.6/dist-packages (from python-slugify->kaggle) (1.3)
Warning: Looks like you're using an outdated API Version, please consider updating (server 1.5.6 / client 1.5.4)
Downloading docs.tsv.zip to /content
100% 1.17G/1.17G [00:31<00:00, 39.1MB/s]
100% 1.17G/1.17G [00:31<00:00, 39.7MB/s]
Downloading sample.submission.txt to /content
0% 0.00/434k [00:00<?, ?B/s]
100% 434k/434k [00:00<00:00, 59.8MB/s]
Downloading queries.numerate.txt to /content
0% 0.00/31.9k [00:00<?, ?B/s]
100% 31.9k/31.9k [00:00<00:00, 32.2MB/s]
Archive: docs.tsv.zip
inflating: docs.tsv
###Markdown
Code
###Code
!pip install -U tqdm
!pip install rank_bm25
!pip install keyw
import os
import gzip
import nltk
import random
import pickle
import pandas as pd
import logging
import keyw
import itertools
import re
import time
import numpy as np
import pandas as pd
from rank_bm25 import BM25Okapi
from nltk.corpus import stopwords
nltk.download("stopwords")
from string import punctuation
from collections import defaultdict
from collections import OrderedDict
from tqdm.notebook import tqdm
logging.basicConfig(level=logging.DEBUG)
logging.debug('Test Logger')
indexing_data_location = '/content/docs.tsv'
queries_list = '/content/queries.numerate.txt'
db_directory_name = 'database'
###Output
_____no_output_____
###Markdown
Convert Docs from memory load to IO operations
###Code
# database = pd.read_csv(indexing_data_location, sep='\t', header=None,
# names=['id', 'subject', 'content'],
# dtype = {'id': int,'subject': str,
# 'content': str})
os.mkdir(db_directory_name)
with open(indexing_data_location, "rb") as f:
for line in tqdm(f):
line = line.decode().split('\t')
file_number = line[0]
subject = line[1]
text = line[2]
line = subject + ' ' + text
with open(os.path.join(db_directory_name, file_number), 'w') as output:
output.write(line)
###Output
_____no_output_____
###Markdown
Building Inverted Index Inverted Indexer Class
###Code
class InvertedIndexer:
"""
This class makes inverted index
"""
def __init__(self, filename=False):
self.filename = filename
self.stemmer_ru = nltk.SnowballStemmer("russian")
self.stopwords = set(stopwords.words("russian")) | set(stopwords.words("english"))
self.punctuation = punctuation # from string import punctuation
if filename:
self.inverted_index = self._build_index(self.filename)
else:
self.inverted_index = defaultdict(set)
def preprocess(self, sentence):
"""
Method to remove stop words and punctuations return tokens
"""
NONTEXT = re.compile('[^0-9 a-z#+_а-яё]')
sentence = sentence.lower()
sentence = sentence.translate(str.maketrans('', '', punctuation))
sentence = re.sub(NONTEXT,'',sentence)
# Heavy Operation Taking lot of time will move it outside
# tokens = [self.stemmer_ru.stem(word) for word in sentence.split()]
tokens = [token for token in sentence.split() if token not in self.stopwords]
return tokens
def stem_keys(self, inverted_index):
"""
Called after index is built to stem all the keys and normalize them
"""
logging.debug('Indexing Complete will not Stem keys and remap indexes')
temp_dict = defaultdict(set)
i = 0
for word in tqdm(inverted_index):
stemmed_key = keyw.engrus(word)
stemmed_key = self.stemmer_ru.stem(stemmed_key)
temp_dict[stemmed_key].update(inverted_index[word])
inverted_index[word] = None
inverted_index = temp_dict
logging.debug('Done Stemmping Indexes')
return inverted_index
def _build_index(self, indexing_data_location):
"""
This method builds the inverted index and returns the invrted index dictionary
"""
inverted_index = defaultdict(set)
with open(indexing_data_location, "rb") as f:
for line in tqdm(f):
line = line.decode().split('\t')
file_number = line[0]
subject = line[1]
text = line[2]
line = subject + ' ' + text
for word in self.preprocess(line):
inverted_index[word].add(int(file_number))
inverted_index = self.stem_keys(inverted_index)
return inverted_index
def save(self, filename_to_save):
"""
Save method to save the inverted indexes
"""
with open(filename_to_save, mode='wb') as f:
pickle.dump(self.inverted_index, f)
def load(self, filelocation_to_load):
"""
Load method to load the inverted indexes
"""
with open(filelocation_to_load, mode='rb') as f:
self.inverted_index = pickle.load(f)
###Output
_____no_output_____
###Markdown
SolutionPredictor Class
###Code
class SolutionPredictor:
"""
This classes uses object of Hw1SolutionIndexer
to make boolean search
"""
def __init__(self, indexer):
"""
indexer : object of class Hw1SolutionIndexer
"""
self.indexer = indexer
def find_docs(self, query):
"""
This method provides booleaen search
query : string with text of query
Returns Python set with documents which contain query words
Should return maximum 100 docs
"""
query = keyw.engrus(query)
tokens = self.indexer.preprocess(query)
tokens = [self.indexer.stemmer_ru.stem(word) for word in tokens]
docs_list = set()
for word in tokens:
if len(docs_list) > 0:
docs_list.intersection_update(self.indexer.inverted_index[word]) # changed from intersection_update
else:
docs_list.update(self.indexer.inverted_index[word])
# Handling case when no set is returned by intersection
# if len(docs_list) < 10:
# for word in tokens:
# docs_list.update(self.indexer.inverted_index[word]) # changed from intersection_update
return docs_list
###Output
_____no_output_____
###Markdown
Generate Index Run Only when Index is not generated Otherwise use the Loading code block
###Code
logging.debug('Index is creating...')
start = time.time()
new_index = InvertedIndexer(indexing_data_location)
end = time.time()
logging.debug('Index has been created and saved as inverted_index.pickle in {:.4f}s'.format(end-start))
len(new_index.inverted_index)
new_index.save('inverted_index_all_rus.pickle')
!cp inverted_index.pickle '/content/drive/My Drive/Homeworks/InformationRetrieval/inverted_index_temp.pickle'
###Output
_____no_output_____
###Markdown
Load IndexRun when Index is not generated
###Code
index_location = '/content/drive/My Drive/Homeworks/InformationRetrieval/inverted_index_4.pickle'
logging.debug('Loading Index...')
start = time.time()
new_index = InvertedIndexer()
new_index.load(index_location)
end = time.time()
logging.debug('Index has been loaded from inverted_index.pickle in {:.4f}s'.format(end-start))
len(new_index.inverted_index)
###Output
_____no_output_____
###Markdown
Testing Block
###Code
test_sentence = 'бандиты боятся ли ментов'
boolean_model = SolutionPredictor(new_index)
start = time.time()
print(boolean_model.find_docs(test_sentence))
logging.debug('Search Time: {}'.format(time.time() - start))
test_docs = boolean_model.find_docs(test_sentence)
test_dict = {}
for doc in test_docs:
with open(os.path.join(db_directory_name, str(doc))) as f:
test_dict[doc] = f.readlines()[0]
###Output
_____no_output_____
###Markdown
Loading File to DataFrameWe are doing this for faster search based on index numbers
###Code
# %%time
# database = pd.read_csv(indexing_data_location, sep='\t', header=None,
# names=['id', 'subject', 'content'],
# dtype = {'id': int,'subject': str,
# 'content': str})
# database.head()
###Output
_____no_output_____
###Markdown
Implementing BM25 (When all data in memory)
###Code
# # boolean_model = SolutionPredictor(new_index)
# output=open('submission.csv', 'w')
# with open(queries_list) as queries:
# output.write('QueryId,DocumentId\n')
# for line in tqdm(queries.readlines()):
# line = line.split('\t')
# id = int(line[0])
# query = line[1]
# value_present = boolean_model.find_docs(query)
# data = database[database['id'].isin(list(value_present))].copy()
# data.loc[:, 'content'] = data[['subject', 'content']].astype(str).apply(' '.join, axis=1)
# del data['subject']
# data = OrderedDict(data.set_index('id')['content'].to_dict())
# for key in data:
# data[key] = new_index.preprocess(data[key])
# if data:
# bm25 = BM25Okapi(data.values())
# rankings = sorted(list(zip(data.keys(), bm25.get_scores(new_index.preprocess(query)))), key=lambda x: x[1], reverse=True)[:10]
# for rank in rankings:
# output.write('{},{}\n'.format(id, rank[0]))
# if (not data) or len(rankings) < 10:
# if not data:
# rankings = []
# logging.debug('Found Some values with no query results or less than 10 {} ranking'.format(id, len(rankings)))
# print(id, query)
# for i in range(10 - len(rankings)):
# output.write('{},{}\n'.format(id, random.randrange(50000)))
# output.close()
# !cp submission.csv '/content/drive/My Drive/Homeworks/InformationRetrieval/submission.csv'
###Output
_____no_output_____
###Markdown
Implementing BM25 when all data on disk
###Code
corpus = {
1: "Hello there good man!",
2: "It is quite windy in London",
3: "How is the weather today?"
}
tokenized_corpus = [doc.split(" ") for doc in corpus.values()]
print(tokenized_corpus)
bm25 = BM25Okapi(tokenized_corpus)
query = "windy London"
tokenized_query = query.split(" ")
doc_scores = bm25.get_scores(tokenized_query)
print(sorted(list(zip(corpus.keys(),doc_scores)), key=lambda x: x[1], reverse=True))
output=open('submission.csv', 'w')
with open(queries_list) as queries:
output.write('QueryId,DocumentId\n')
for line in tqdm(queries.readlines()):
line = line.split('\t')
id = int(line[0])
query = line[1]
value_present = boolean_model.find_docs(query)
logging.debug('Found Value {}'.format(len(value_present)))
data = OrderedDict()
for doc in value_present:
with open(os.path.join(db_directory_name, str(doc))) as f:
data[doc] = new_index.preprocess(f.readlines()[0])
logging.debug('Loaded Data')
if data:
bm25 = BM25Okapi(data.values())
start = time.time()
rankings = sorted(list(zip(data.keys(), bm25.get_scores(new_index.preprocess(query)))), key=lambda x: x[1], reverse=True)[:100]
for rank in rankings:
output.write('{},{}\n'.format(id, rank[0]))
logging.debug('Calculated ranking in {}'.format(time.time()- start))
if (not data) or len(rankings) < 10:
if not data:
rankings = []
logging.debug('Found Some values with no query results or less than 10 {} ranking'.format(id, len(rankings)))
print(id, query)
for i in range(10 - len(rankings)):
output.write('{},{}\n'.format(id, random.randrange(50000)))
output.close()
data = OrderedDict()
with open(queries_list) as queries:
for line in tqdm(queries.readlines()):
line = line.split('\t')
id = int(line[0])
query = line[1]
value_present = boolean_model.find_docs(query)
data[id] = value_present
i = 0
for k in data:
print(k, data[k])
if i == 5:
break
i += 1
for key in data:
if len(data[key]) < 10:
print(key, data[key])
###Output
_____no_output_____ |
lessons/NLP Pipelines/tokenization_practice.ipynb | ###Markdown
Note on NLTK data downloadRun the cell below to download the necessary nltk data packages. Note, because we are working in classroom workspaces, we will be downloading specific packages in each notebook throughout the lesson. However, you can download all packages by entering `nltk.download()` on your computer. Keep in mind this does take up a bit more space. You can learn more about nltk data installation [here](https://www.nltk.org/data.html).
###Code
import nltk
nltk.download('punkt')
###Output
[nltk_data] Downloading package punkt to /Users/zacks/nltk_data...
[nltk_data] Package punkt is already up-to-date!
###Markdown
TokenizationTry out the tokenization methods in nltk to split the following text into words and then sentences.**Note: All solution notebooks can be found by clicking on the Jupyter icon on the top left of this workspace.**
###Code
# import statements
from nltk.tokenize import word_tokenize
from nltk.tokenize import sent_tokenize
text = "Dr. Smith graduated from the University of Washington. He later started an analytics firm called Lux, which catered to enterprise customers."
print(text)
# Split text into words using NLTK
word_tokenize(text)
# Split text into sentences
sent_tokenize(text)
###Output
_____no_output_____
###Markdown
Table of Contents1 Note on NLTK data download2 Tokenization Note on NLTK data downloadRun the cell below to download the necessary nltk data packages. Note, because we are working in classroom workspaces, we will be downloading specific packages in each notebook throughout the lesson. However, you can download all packages by entering `nltk.download()` on your computer. Keep in mind this does take up a bit more space. You can learn more about nltk data installation [here](https://www.nltk.org/data.html).
###Code
import nltk
###Output
_____no_output_____
###Markdown
TokenizationTry out the tokenization methods in nltk to split the following text into words and then sentences.**Note: All solution notebooks can be found by clicking on the Jupyter icon on the top left of this workspace.**
###Code
# import statements
from nltk.tokenize import word_tokenize
from nltk.tokenize import sent_tokenize
text = "Dr. Smith graduated from the University of Washington. He later started an analytics firm called Lux, which catered to enterprise customers."
print(text)
# Split text into words using NLTK
word_tokens = word_tokenize(text)
#word_tokens
# Split text into sentences
sent_tokenize = sent_tokenize(text)
sent_tokenize
###Output
_____no_output_____
###Markdown
Note on NLTK data downloadRun the cell below to download the necessary nltk data packages. Note, because we are working in classroom workspaces, we will be downloading specific packages in each notebook throughout the lesson. However, you can download all packages by entering `nltk.download()` on your computer. Keep in mind this does take up a bit more space. You can learn more about nltk data installation [here](https://www.nltk.org/data.html).
###Code
import nltk
nltk.download('punkt')
###Output
[nltk_data] Downloading package punkt to
[nltk_data] C:\Users\amira\AppData\Roaming\nltk_data...
[nltk_data] Unzipping tokenizers\punkt.zip.
###Markdown
TokenizationTry out the tokenization methods in nltk to split the following text into words and then sentences.**Note: All solution notebooks can be found by clicking on the Jupyter icon on the top left of this workspace.**
###Code
# import statements
from nltk.tokenize import word_tokenize
from nltk.tokenize import sent_tokenize
text = "Dr. Smith graduated from the University of Washington. He later started an analytics firm called Lux, which catered to enterprise customers."
print(text)
# Split text into words using NLTK
words = word_tokenize(text)
print(words)
# Split text into sentences
sentences = sent_tokenize(text)
print(sentences)
###Output
['Dr. Smith graduated from the University of Washington.', 'He later started an analytics firm called Lux, which catered to enterprise customers.']
###Markdown
Note on NLTK data downloadRun the cell below to download the necessary nltk data packages. Note, because we are working in classroom workspaces, we will be downloading specific packages in each notebook throughout the lesson. However, you can download all packages by entering `nltk.download()` on your computer. Keep in mind this does take up a bit more space. You can learn more about nltk data installation [here](https://www.nltk.org/data.html).
###Code
import nltk
nltk.download('punkt')
###Output
_____no_output_____
###Markdown
TokenizationTry out the tokenization methods in nltk to split the following text into words and then sentences.**Note: All solution notebooks can be found by clicking on the Jupyter icon on the top left of this workspace.**
###Code
# import statements
text = "Dr. Smith graduated from the University of Washington. He later started an analytics firm called Lux, which catered to enterprise customers."
print(text)
# Split text into words using NLTK
# Split text into sentences
###Output
_____no_output_____
###Markdown
Note on NLTK data downloadRun the cell below to download the necessary nltk data packages. Note, because we are working in classroom workspaces, we will be downloading specific packages in each notebook throughout the lesson. However, you can download all packages by entering `nltk.download()` on your computer. Keep in mind this does take up a bit more space. You can learn more about nltk data installation [here](https://www.nltk.org/data.html).
###Code
import nltk
nltk.download('punkt')
###Output
[nltk_data] Downloading package punkt to /home/jovyan/nltk_data...
[nltk_data] Unzipping tokenizers/punkt.zip.
###Markdown
TokenizationTry out the tokenization methods in nltk to split the following text into words and then sentences.**Note: All solution notebooks can be found by clicking on the Jupyter icon on the top left of this workspace.**
###Code
# import statements
from nltk.tokenize import word_tokenize
text = "Dr. Smith graduated from the University of Washington. He later started an analytics firm called Lux, which catered to enterprise customers."
print(text)
# Split text into words using NLTK
word_tokenize(text)
# Split text into sentences
text.split()
###Output
_____no_output_____ |
Final Problem 1 - Double Pendulum(1).ipynb | ###Markdown
Problem 1: Double Pendulum Solved Using Lagrangian Method Physics 5700 Final, Spring 2021 Alex Bumgarner.187 Part 1: Python Setup: Here, I import the packages needed for the rest of the notebook:
###Code
%matplotlib inline
from IPython.display import Image #Allows us to display images from the web
import numpy as np #For our more complicated math
from scipy.integrate import solve_ivp #Allows us to solve first-order ODE's
import matplotlib.pyplot as plt #For plotting
###Output
_____no_output_____
###Markdown
Part 2: Problem Setup:
###Code
Image(url = 'https://upload.wikimedia.org/wikipedia/commons/7/78/Double-Pendulum.svg')
###Output
_____no_output_____
###Markdown
**Fig. 1: Double Pendulum***Source: Wikimedia Commons* A mass $m_1$ is attached to the ceiling by a massless rope of length $L_1$. A second mass $m_2$ is attached to the first mass by a massless string of length $m_2$, forming the double pendulum shown in Fig. 1. Our goal is to predict the motion of these two masses given a set of initial conditions. We define our Cartesian axes such that $\hat{x}$ points to the *right* and $\hat{y}$ points *up*. Part 3: Solving the Euler-Lagrange Equations The Lagrangian is defined as $\mathscr{L} = T - U$, where $T$ is the kinetic energy and $U$ is the potential energy of the system. We will define these energies in terms of our general coordinates $\theta_1$ and $\theta_2$ Assuming $m_1$ is $h_1$ high off the ground when $\theta_1 = 0$, the height of $m_1$ is equal to $h_1 + L_1 - L_1\cos{\theta_1}$. Thus, the potential energy of $m_1$ is given by: $$U_1 = m_1 g [L_1 (1-\cos{\theta_1})+ h_1]$$ Assuming $m_2$ is $h_2$ high ff the ground with $\theta_1 = \theta_2 = 0$, the height of $m_2$ is given by $h_1 + L_1 - L_1\cos{\theta_1} + h_2 + L_2 - L_2\cos{\theta_2}$. Thus, the potential energy of $m_2$ is given by: $$U_2 = m_2 g [L_1 (1-\cos{\theta_1}) + h_1 + L_2 (1-\cos{\theta_2})+h_2]$$ Summing these two potential energies (and omitting the $h$'s, as they will "disappear" when differentiating), we get an expression for the total potential energy: $$U = (m_1 + m_2) g L_1 (1-\cos{\theta_1}) + m_2 g L_2 (1-\cos{\theta_2})$$ The kinetic energy of $m_1$ is given by $\frac{1}{2} m_1 v_1^2$, where $v_1$ is the *magnitdue* of the mass's velocity. $v_1 = L_1 \dot{\theta_1}$, so the kinetic energy is: $$ T_1 = \frac{1}{2} m_1 L_1^2 \dot{\theta_1}^2 $$ The kinetic energy of $m_2$ is given by $\frac{1}{2} m_2 v_2^2$. Like $m_1$, $m_2$ moves with a tangential velocity $L_2 \dot{\theta_2}$. However, it is also affected by the velocity of $m_1$. As such, we must find the velocity of $m_2$ in each Carteian direction and add them in quadreture to get the magnitude. In the x-direction, the velocity is $v_2,x = L_1 \dot{\theta_1} \cos{\theta_1} + L_2 \dot{\theta_2} \cos{\theta_2}$ In the y-direction, the velocity is $v_2,y = L_1 \dot{\theta_1} \sin{\theta_1} + L_2 \dot{\theta_2} \sin{\theta_2}$ Thus, the kinetic energy of $m_2$ is given by: $$T_2 = \frac{1}{2} m_2 [(L_1 \dot\theta_1 \cos{\theta_1} + L_2 \dot\theta_2 \cos{\theta_2})^2 + (L_1 \dot\theta_1 \sin{\theta_1} + L_2 \dot\theta_2 \sin{\theta_2})^2]$$ This can be expanded and simplified: $$T_2 = \frac{1}{2} m_2 (L_1^2 \dot\theta_1^2 +L_2^2 \dot\theta_2^2 + 2 L_1 L_2 \dot\theta_1 \dot\theta_2 (\sin{\theta_1} \sin{\theta_2} + \cos{\theta_1} \cos{\theta_2}))$$ $$T_2 = \frac{1}{2} m_2 (L_1^2 \dot\theta_1^2 +L_2^2 \dot\theta_2^2 + 2 L_1 L_2 \dot\theta_1 \dot\theta_2 \cos{(\theta_1-\theta_2))}$$ Summing these energies gives us the total kinetic energy of the system: $$ T = \frac{1}{2}(m_1 + m_2) L_1^2 \dot\theta_1^2 + \frac{1}{2} m_2 L_2^2 \dot\theta_2^2 + m_2 L_1 L_2 \dot\theta_1 \dot\theta_2 \cos{(\theta_1 - \theta_2)}$$ Thus, the Lagrangian $\mathscr{L} = T - U$ is given by: $$\mathscr{L} = \frac{1}{2}(m_1 + m_2) L_1^2 \dot\theta_1^2 + \frac{1}{2} m_2 L_2^2 \dot\theta_2^2 + m_2 L_1 L_2 \dot\theta_1 \dot\theta_2 \cos{(\theta_1 - \theta_2)} - (m_1 + m_2) g L_1 (1-\cos{\theta_1}) - m_2 g L_2 (1-\cos{\theta_2})$$ The Euler-Lagrange equations for our generalized variables, $\theta_1$ and $\theta_2$, are: $$\frac{d}{dt} (\frac{\partial\mathscr{L}}{\partial \dot\theta_1}) = \frac{\partial\mathscr{L}}{\partial\theta_1} $$ $$\frac{d}{dt} (\frac{\partial\mathscr{L}}{\partial \dot\theta_2}) = \frac{\partial\mathscr{L}}{\partial\theta_2} $$ Plugging in our Lagrangian and simplifying gives us: $$ \ddot\theta_1 (m_1+m_2) L_1 + m_2 L_2 \ddot\theta_2 \cos(\theta_1 - \theta_2) + \dot\theta_2^2 m_2 L_2 \sin(\theta_1 - \theta_2) = -(m_1 + m_2) g \sin(\theta_1)$$ $$ \ddot\theta_2 m_2 L_2 + m_2 L_1 \ddot\theta_1 \cos(\theta_1-\theta_2) -m_2 L_1 \dot\theta_1^2 \sin(\theta_1 - \theta_2) = -m_2 g \sin(\theta_2) $$ To solve with scipy, we need these as first-order ordinary differential equations. We rewrite the above equations using $z_i = \dot\theta_i$ and $\dot{z_i} = \ddot\theta_i$. Solving for $\dot z_1$ and $\dot z_2$, we get: $$ \dot{z_1} = \frac{-(m_1+m2)g\sin(\theta_1) - m_2 \sin(\theta_1 - \theta_2) (L_2 z_2^2 + L_1 z_1^2 \cos(\theta_1 - \theta_2)) + g m_2 \cos(\theta_1 - \theta_2) \sin(\theta_2)}{L_1 (m_1 + m_2 \sin^2(\theta_1 - \theta_2))}$$ $$ \dot{z_2} = \frac{(m_1 + m_2) [g \cos(\theta_1 - \theta_2) \sin(\theta_1) + L_1 z_1^2 \sin(\theta_1 - \theta_2) - g \sin(\theta_2)] + L_2 m_2 z_2^2 \cos(\theta_1-\theta_2)\sin(\theta_1-\theta_2)}{L_2 (m_1 + m_2 \sin^2(\theta_1 - \theta_2))}$$ Part 4: Solving for the Motion *The following is largely adapted from the Lagrangian_pendulum.ipynb notebook provided in class* We now want to solve our system of differntial equations and plot the course of the masses given initial conditions:
###Code
class Pendulum():
"""
This class creates and solves for the motion of a pendulum of two masses using Lagrange's equations
Parameters
------------
L1: float
length of first pendulum
L2: float
length of second pendulum
m1: float
mass of first object
m2: float
mass of second object
g: float
gravitational acceleration at the Earth's surface
"""
def __init__(self, L1 = 1., L2 = 1., m1 = 1., m2 = 1., g = 1.):
"""
Initializes the pendulum and provides default values if none are provided by user
"""
self.L1 = L1
self.L2 = L2
self.m1 = m1
self.m2 = m2
self.g = g
def dy_dt(self,t,y):
"""
Inputs a four component vector y[theta1,theta1dot,theta2,thetadot2] and
outputs the time derivative of each component
"""
theta1 = y[0]
theta1dot = y[1]
theta2 = y[2]
theta2dot = y[3]
z1 = theta1dot #We defined these in the last section to get 1st order ODE's
z2 = theta2dot
#Below, a handful of common functions to simplify formula input (thanks for the idea!)
c = np.cos(theta1-theta2)
s = np.sin(theta1-theta2)
denom = (self.m1 + self.m2*s**2)
#Now, the equations from above:
z1_dot = (-(self.m1 + self.m2)*g*np.sin(theta1)-self.m2*s*(self.L2*z2**2 + self.L1*z1**2 * c)+self.g*self.m2*c*np.sin(theta2))/(self.L1*denom)
z2_dot = ((self.m1+self.m2)*(self.g*c*np.sin(theta1)+self.L1*z1**2 *s - g*np.sin(theta2))+self.L2*self.m2*z2**2 *c*s)/(self.L2*denom)
return(z1,z1_dot,z2,z2_dot)
def solve_ode(self, t_pts, theta1_0, theta1dot_0, theta2_0, theta2dot_0, abserr=1.0e-10,relerr = 1.0e-10):
"""
As the name suggests, this function inputs initial values for each theta and theta dot and solves the ODE along the specified t_pts
"""
#initial position y-vector:
y = [theta1_0, theta1dot_0, theta2_0, theta2dot_0]
#Below, we use the solve_ivp function to solve for the motion over our set of t_pts
solution = solve_ivp(self.dy_dt, (t_pts[0], t_pts[-1]), y, t_eval=t_pts, atol = abserr, rtol = relerr)
theta1, theta1dot, theta2, theta2dot = solution.y
return(theta1,theta1dot,theta2,theta2dot)
###Output
_____no_output_____
###Markdown
Now, a few plotting functions:
###Code
def plot_y_vs_x(x, y, axis_labels=None, label=None, title=None,
color=None, linestyle=None, semilogy=False, loglog=False,
ax=None):
"""
Simple plot of points y vs. points x, found in Lagrangian_pendulum.ipynb
"""
if ax is None:
ax = plt.gca()
if (semilogy):
line, = ax.semilogy(x, y, label=label,
color=color, linestyle=linestyle)
elif (loglog):
line, = ax.loglog(x, y, label=label,
color=color, linestyle=linestyle)
else:
line, = ax.plot(x, y, label=label,
color=color, linestyle=linestyle)
if label is not None:
ax.legend()
if title is not None:
ax.set_title(title)
if axis_labels is not None:
ax.set_xlabel(axis_labels[0])
ax.set_ylabel(axis_labels[1])
return ax, line
def start_stop_indices(t_pts, plot_start, plot_stop):
start_index = (np.fabs(t_pts-plot_start)).argmin()
stop_index = (np.fabs(t_pts-plot_stop)).argmin()
return start_index, stop_index
###Output
_____no_output_____
###Markdown
Finally, some labels for our plots:
###Code
theta_vs_time_labels = (r'$t$', r'$\theta(t)$')
###Output
_____no_output_____
###Markdown
Part 5: Plotting the Motion Below, we'll make some representative plots of the motion for various parameters and initial conditions:
###Code
# Define plotting time
t_start = 0.
t_end = 50.
delta_t = 0.001
t_pts = np.arange(t_start, t_end+delta_t, delta_t)
###Output
_____no_output_____
###Markdown
Pendulum 1: Basic parameters, initially at rest
###Code
#Parameters:
L1 = 1.0
L2 = 1.0
m1 = 1.0
m2 = 1.0
g = 1.0
#Initial conditions:
theta1_0 = np.pi/2
theta1dot_0 = 0.0
theta2_0 = np.pi
theta2dot_0 = 0.0
#Create the pendulum:
p1 = Pendulum(L1,L2,m1,m2,g)
theta1, theta1dot, theta2, theta2dot = p1.solve_ode(t_pts,theta1_0,theta1dot_0,theta2_0,theta2dot_0)
fig = plt.figure(figsize = (10,5))
overall_title = 'Double Pendulum: ' + \
rf' $\theta_1(0) = {theta1_0:.2f},$' + \
rf' $\dot\theta_1(0) = {theta1dot_0:.2f},$' + \
rf' $\theta_2(0) = {theta2_0:.2f},$' + \
rf' $\dot\theta_2(0) = {theta2dot_0:.2f}$'
fig.suptitle(overall_title, va='baseline')
ax_a = fig.add_subplot(1,1,1)
start,stop = start_stop_indices(t_pts,t_start,t_end)
plot_y_vs_x(t_pts[start:stop],theta1[start:stop],
axis_labels = theta_vs_time_labels,
label = r'$\theta_1(t)$',
ax = ax_a)
plot_y_vs_x(t_pts[start:stop],theta2[start:stop],
axis_labels = theta_vs_time_labels,
label = r'$\theta_2(t)$',
ax = ax_a)
###Output
_____no_output_____
###Markdown
Here, we see the first mass stays fairly close to its initial conditions, while the second mass moves around for a bit then abruptly makes multiple full rotations before settling down, again. Pendulum 2: Basic parameters, begin by pulling back and releasing bottom mass
###Code
#Parameters:
L1 = 1.0
L2 = 1.0
m1 = 1.0
m2 = 1.0
g = 1.0
#Initial conditions:
theta1_0 = 0
theta1dot_0 = 0.0
theta2_0 = np.pi/4
theta2dot_0 = 0.0
#Create the pendulum:
p2 = Pendulum(L1,L2,m1,m2,g)
theta1, theta1dot, theta2, theta2dot = p2.solve_ode(t_pts,theta1_0,theta1dot_0,theta2_0,theta2dot_0)
fig = plt.figure(figsize = (10,5))
overall_title = 'Double Pendulum: ' + \
rf' $\theta_1(0) = {theta1_0:.2f},$' + \
rf' $\dot\theta_1(0) = {theta1dot_0:.2f},$' + \
rf' $\theta_2(0) = {theta2_0:.2f},$' + \
rf' $\dot\theta_2(0) = {theta2dot_0:.2f}$'
fig.suptitle(overall_title, va='baseline')
ax_a = fig.add_subplot(1,1,1)
start,stop = start_stop_indices(t_pts,t_start,t_end)
plot_y_vs_x(t_pts[start:stop],theta1[start:stop],
axis_labels = theta_vs_time_labels,
label = r'$\theta_1(t)$',
ax = ax_a)
plot_y_vs_x(t_pts[start:stop],theta2[start:stop],
axis_labels = theta_vs_time_labels,
label = r'$\theta_2(t)$',
ax = ax_a)
###Output
_____no_output_____
###Markdown
While neither mass appears periodic, they both stay quite close to their initial starting positions, never making full rotations. Pendulum 3: $m_2 > m_1$
###Code
#Parameters:
L1 = 1.0
L2 = 1.0
m1 = 1.0
m2 = 10.0
g = 1.0
#Initial conditions (same as Pendulum 1):
theta1_0 = np.pi/2
theta1dot_0 = 0.0
theta2_0 = np.pi
theta2dot_0 = 0.0
#Create the pendulum:
p3 = Pendulum(L1,L2,m1,m2,g)
theta1, theta1dot, theta2, theta2dot = p3.solve_ode(t_pts,theta1_0,theta1dot_0,theta2_0,theta2dot_0)
fig = plt.figure(figsize = (10,5))
overall_title = 'Double Pendulum: ' + \
rf' $\theta_1(0) = {theta1_0:.2f},$' + \
rf' $\dot\theta_1(0) = {theta1dot_0:.2f},$' + \
rf' $\theta_2(0) = {theta2_0:.2f},$' + \
rf' $\dot\theta_2(0) = {theta2dot_0:.2f}$'
fig.suptitle(overall_title, va='baseline')
ax_a = fig.add_subplot(1,1,1)
start,stop = start_stop_indices(t_pts,t_start,t_end)
plot_y_vs_x(t_pts[start:stop],theta1[start:stop],
axis_labels = theta_vs_time_labels,
label = r'$\theta_1(t)$',
ax = ax_a)
plot_y_vs_x(t_pts[start:stop],theta2[start:stop],
axis_labels = theta_vs_time_labels,
label = r'$\theta_2(t)$',
ax = ax_a)
###Output
_____no_output_____
###Markdown
In this case, both masses make full rotations, with the first mass making multiple rotations before finally settling down. Part 6: Investigating Chaos Some of the signature features of chaotic system include a lack of periodicity and an exponential sensitivity to initial conditions. In this section, I show that the double pendulum is chaotic for initial conditions outside of the small-angle approximation. We'll start by initializing a pendulum with fairly large initial angles:
###Code
#Parameters:
L1 = 1.0
L2 = 1.0
m1 = 1.0
m2 = 1.0
g = 1.0
#Initial conditions (same as Pendulum 1):
theta1_0 = np.pi/4
theta1dot_0 = 0
theta2_0 = np.pi
theta2dot_0 = 0.0
#Create the pendulum:
p4 = Pendulum(L1,L2,m1,m2,g)
theta1, theta1dot, theta2, theta2dot = p4.solve_ode(t_pts,theta1_0,theta1dot_0,theta2_0,theta2dot_0)
fig = plt.figure(figsize = (10,5))
overall_title = 'Double Pendulum: ' + \
rf' $\theta_1(0) = {theta1_0:.2f},$' + \
rf' $\dot\theta_1(0) = {theta1dot_0:.2f},$' + \
rf' $\theta_2(0) = {theta2_0:.2f},$' + \
rf' $\dot\theta_2(0) = {theta2dot_0:.2f}$'
fig.suptitle(overall_title, va='baseline')
ax_a = fig.add_subplot(1,1,1)
start,stop = start_stop_indices(t_pts,t_start,t_end)
plot_y_vs_x(t_pts[start:stop],theta1[start:stop],
axis_labels = theta_vs_time_labels,
label = r'$\theta_1(t)$',
ax = ax_a)
plot_y_vs_x(t_pts[start:stop],theta2[start:stop],
axis_labels = theta_vs_time_labels,
label = r'$\theta_2(t)$',
ax = ax_a)
###Output
_____no_output_____
###Markdown
Already, it appears that neither mass is periodic with time, indicating chaos. However, we need a bit more evidence. Below, we'll plot the state space plot of each mass with respect to time. If either plot closes on itself, the mass's motion is periodic.
###Code
state_space_labels = (r'$\theta$', r'$d\theta/dt$') #New labels for comparing thetadot to theta
fig = plt.figure(figsize = (10,5))
overall_title = 'Double Pendulum: ' + \
rf' $\theta_1(0) = {theta1_0:.2f},$' + \
rf' $\dot\theta_1(0) = {theta1dot_0:.2f},$' + \
rf' $\theta_2(0) = {theta2_0:.2f},$' + \
rf' $\dot\theta_2(0) = {theta2dot_0:.2f}$'
fig.suptitle(overall_title, va='baseline')
ax_a = fig.add_subplot(1,2,1) #1 row, 2 columns, position 1
ax_b = fig.add_subplot(1,2,2) #1 row, 2 columns, position 2
start,stop = start_stop_indices(t_pts,t_start,t_end)
plot_y_vs_x(theta1[start:stop],theta1dot[start:stop],
axis_labels = state_space_labels,
label = r'$\theta_1(t)$',
ax = ax_a)
plot_y_vs_x(theta2[start:stop],theta2dot[start:stop],
axis_labels = state_space_labels,
label = r'$\theta_2(t)$',
ax = ax_b)
###Output
_____no_output_____
###Markdown
As expected, neither plot closes on itself, indicating that the motion is chaotic. B As a final check for chaos, we'll plot the difference between each $\theta$ for two pendulums with *almost* identical initial positions. In other words, we will create a second pendulum with initial conditions only slighly different than the first. If the difference between respective $\theta$ increases exponentially with time, we have a chaotic system.
###Code
# Our first pendulum is p4, created above. Here, we create the slightly different pendulum:
#Parameters:
L1 = 1.0
L2 = 1.0
m1 = 1.0
m2 = 1.0
g = 1.0
#Initial conditions (slightly different than Pendulum 4):
theta1_0 = np.pi/4 + 0.001
theta1dot_0 = 0
theta2_0 = np.pi + 0.001
theta2dot_0 = 0.0
#Create the pendulum:
p5 = Pendulum(L1,L2,m1,m2,g)
theta1_diff, theta1dot_diff, theta2_diff, theta2dot_diff = p5.solve_ode(t_pts,theta1_0,theta1dot_0,theta2_0,theta2dot_0)
#Labels for our new plots:
delta_labels = (r'$t$', r'$\Delta\theta$')
#Define variables to plot, the difference between each theta
diff1 = np.abs(theta1-theta1_diff)
diff2 = np.abs(theta2-theta2_diff)
#Plot our functions:
fig = plt.figure(figsize = (10,5))
overall_title = 'Double Pendulum: ' + \
rf' $\theta_1(0) = {theta1_0:.2f},$' + \
rf' $\dot\theta_1(0) = {theta1dot_0:.2f},$' + \
rf' $\theta_2(0) = {theta2_0:.2f},$' + \
rf' $\dot\theta_2(0) = {theta2dot_0:.2f}$'
fig.suptitle(overall_title, va='baseline')
ax_a = fig.add_subplot(1,1,1) #1 row, 1 columns, position 1
start,stop = start_stop_indices(t_pts,t_start,t_end)
plot_y_vs_x(t_pts[start:stop],diff1[start:stop],
axis_labels = delta_labels,
label = r'$\Delta\theta_1(t)$',
semilogy = True, #Semi log plot
ax = ax_a)
plot_y_vs_x(t_pts[start:stop],diff2[start:stop],
axis_labels = delta_labels,
label = r'$\Delta\theta_2(t)$',
semilogy = True, #Semi log plot
ax = ax_a)
###Output
_____no_output_____ |
tutorials/regression/03-CurveFitting-TensorFlow.ipynb | ###Markdown
3 Curve Fitting (TensorFlow)Two methods are used to fit a curve in this tutorial, using [TensorFlow](https://www.tensorflow.org/):- Direct solution using least-squares method - this is the same method used in the previous tutorial that uses NumPy - Iterative optimisation using stochastic gradient descent 3.1 DataFirst, we sample $n$ observed data from the underlying polynomial defined by weights $w$:
###Code
import random
import numpy as np
# get ground-truth data from the "true" model
n = 100
w = [4, 3, 2, 1]
deg = len(w)-1
x = np.linspace(-1,1,n)[:,np.newaxis]
t = np.matmul(np.power(np.reshape(x,[-1,1]),
np.linspace(deg,0,deg+1)), w)
std_noise = 0.2
t_observed = np.reshape(
[t[idx]+random.gauss(0,std_noise) for idx in range(n)],
[-1,1])
###Output
_____no_output_____
###Markdown
3.2 Least-Squares SolutionThis is mathematcally the same method used in previous NumPy tutorial. The advantage using TensorFlow here is not particularly obvious.
###Code
import tensorflow as tf
X = tf.pow(x, tf.linspace(deg,0,deg+1))
w_lstsq = tf.linalg.lstsq(X, t_observed)
print(w_lstsq)
###Output
tf.Tensor(
[[4.05248568]
[2.95396288]
[1.956563 ]
[1.00741483]], shape=(4, 1), dtype=float64)
###Markdown
3.3 Stochastic Gradient Descend MethodInstead of least-squares, weights can be optimised by minimising a loss function between the predicted- and observed target values, using [SGD](https://en.wikipedia.org/wiki/Stochastic_gradient_descent). It is not an efficient method for this curve fitting problem, is only for the purpose of demonstrating how an iterative method can be implemented in TensorFlow.
###Code
w_sgd = tf.Variable(initial_value=tf.zeros([deg+1,1],tf.float64))
polynomial = lambda x_input : tf.matmul(x_input, w_sgd)
optimizer = tf.optimizers.SGD(5e-3)
total_iter = int(2e4)
for step in range(total_iter):
index = step % n
with tf.GradientTape() as g:
loss = tf.reduce_mean((polynomial(X[None,index,:])-t_observed[index])**2) #MSE
gradients = g.gradient(loss, [w_sgd])
optimizer.apply_gradients(zip(gradients, [w_sgd]))
if (step%1000)==0:
print('Step %d: Loss=%f' % (step, loss))
print(w_sgd)
###Output
Step 0: Loss=4.267816
Step 1000: Loss=0.037259
Step 2000: Loss=0.042375
Step 3000: Loss=0.133096
Step 4000: Loss=0.154306
Step 5000: Loss=0.136392
Step 6000: Loss=0.108063
Step 7000: Loss=0.081463
Step 8000: Loss=0.060006
Step 9000: Loss=0.043758
Step 10000: Loss=0.031807
Step 11000: Loss=0.023136
Step 12000: Loss=0.016880
Step 13000: Loss=0.012372
Step 14000: Loss=0.009123
Step 15000: Loss=0.006774
Step 16000: Loss=0.005071
Step 17000: Loss=0.003832
Step 18000: Loss=0.002925
Step 19000: Loss=0.002258
<tf.Variable 'Variable:0' shape=(4, 1) dtype=float64, numpy=
array([[3.99621521],
[2.96221558],
[1.99494511],
[1.00508842]])>
|
tests/test_vae_ts.ipynb | ###Markdown
Identifiability Test of Linear VAE on Synthetic Dataset
###Code
%load_ext autoreload
%autoreload 2
import torch
import torch.nn.functional as F
from torch.utils.data import DataLoader, random_split
import ltcl
import numpy as np
from ltcl.datasets.sim_dataset import SimulationDatasetTSTwoSample
from ltcl.modules.srnn import SRNNSynthetic
from ltcl.tools.utils import load_yaml
import random
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
use_cuda = True
device = torch.device("cuda:0" if use_cuda else "cpu")
latent_size = 8
data = SimulationDatasetTSTwoSample(directory = '/srv/data/ltcl/data/',
transition='linear_nongaussian_ts')
num_validation_samples = 2500
train_data, val_data = random_split(data, [len(data)-num_validation_samples, num_validation_samples])
train_loader = DataLoader(train_data, batch_size=12800, shuffle=True, pin_memory=True)
val_loader = DataLoader(val_data, batch_size=16, shuffle=False, pin_memory=True)
cfg = load_yaml('../ltcl/configs/toy_linear_ts.yaml')
model = SRNNSynthetic.load_from_checkpoint(checkpoint_path="/srv/data/ltcl/log/weiran/toy_linear_ts/lightning_logs/version_1/checkpoints/epoch=299-step=228599.ckpt",
input_dim=cfg['VAE']['INPUT_DIM'],
length=cfg['VAE']['LENGTH'],
z_dim=cfg['VAE']['LATENT_DIM'],
lag=cfg['VAE']['LAG'],
hidden_dim=cfg['VAE']['ENC']['HIDDEN_DIM'],
trans_prior=cfg['VAE']['TRANS_PRIOR'],
bound=cfg['SPLINE']['BOUND'],
count_bins=cfg['SPLINE']['BINS'],
order=cfg['SPLINE']['ORDER'],
beta=cfg['VAE']['BETA'],
gamma=cfg['VAE']['GAMMA'],
sigma=cfg['VAE']['SIGMA'],
lr=cfg['VAE']['LR'],
bias=cfg['VAE']['BIAS'],
use_warm_start=cfg['SPLINE']['USE_WARM_START'],
spline_pth=cfg['SPLINE']['PATH'],
decoder_dist=cfg['VAE']['DEC']['DIST'],
correlation=cfg['MCC']['CORR'])
###Output
Load pretrained spline flow
###Markdown
Load model checkpoint
###Code
model.eval()
model.to('cpu')
###Output
_____no_output_____
###Markdown
Compute permutation and sign flip
###Code
for batch in train_loader:
break
batch_size = batch['s1']['xt'].shape[0]
zs.shape
zs, mu, logvar = model.forward(batch['s1'])
mu = mu.view(batch_size, -1, latent_size)
A = mu[:,0,:].detach().cpu().numpy()
B = batch['s1']['yt'][:,0,:].detach().cpu().numpy()
C = np.zeros((latent_size,latent_size))
for i in range(latent_size):
C[i] = -np.abs(np.corrcoef(B, A, rowvar=False)[i,latent_size:])
from scipy.optimize import linear_sum_assignment
row_ind, col_ind = linear_sum_assignment(C)
A = A[:, col_ind]
mask = np.ones(latent_size)
for i in range(latent_size):
if np.corrcoef(B, A, rowvar=False)[i,latent_size:][i] > 0:
mask[i] = -1
print("Permutation:",col_ind)
print("Sign Flip:", mask)
fig = plt.figure(figsize=(4,4))
sns.heatmap(-C, vmin=0, vmax=1, annot=True, fmt=".2f", linewidths=.5, cbar=False, cmap='Greens')
plt.xlabel("Estimated latents ")
plt.ylabel("True latents ")
plt.title("MCC=%.3f"%np.abs(C[row_ind, col_ind]).mean());
figure_path = '/home/weiran/figs/'
from matplotlib.backends.backend_pdf import PdfPages
with PdfPages(figure_path + '/mcc_var.pdf') as pdf:
fig = plt.figure(figsize=(4,4))
sns.heatmap(-C, vmin=0, vmax=1, annot=True, fmt=".2f", linewidths=.5, cbar=False, cmap='Greens')
plt.xlabel("Estimated latents ")
plt.ylabel("True latents ")
plt.title("MCC=%.3f"%np.abs(C[row_ind, col_ind]).mean());
pdf.savefig(fig, bbox_inches="tight")
# Permute column here
mu = mu[:,:,col_ind]
# Flip sign here
mu = mu * torch.Tensor(mask, device=mu.device).view(1,1,latent_size)
mu = -mu
fig = plt.figure(figsize=(8,2))
col = 0
plt.plot(mu[:250,-1,col].detach().cpu().numpy(), color='b', label='True', alpha=0.75)
plt.plot(batch['yt_'].squeeze()[:250,col].detach().cpu().numpy(), color='r', label="Estimated", alpha=0.75)
plt.legend()
plt.title("Current latent variable $z_t$")
fig = plt.figure(figsize=(8,2))
col = 3
l = 1
plt.plot(batch['yt'].squeeze()[:250,l,col].detach().cpu().numpy(), color='b', label='True')
plt.plot(mu[:,:-1,:][:250,l,col].detach().cpu().numpy(), color='r', label="Estimated")
plt.xlabel("Sample index")
plt.ylabel("Latent variable value")
plt.legend()
plt.title("Past latent variable $z_l$")
fig = plt.figure(figsize=(2,2))
eps = model.sample(batch["xt"].cpu())
eps = eps.detach().cpu().numpy()
component_idx = 4
sns.distplot(eps[:,component_idx], hist=False, kde=True,
bins=None,
hist_kws={'edgecolor':'black'},
kde_kws={'linewidth': 2});
plt.title("Learned noise prior")
###Output
/home/cmu_wyao/anaconda3/envs/py37/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `kdeplot` (an axes-level function for kernel density plots).
warnings.warn(msg, FutureWarning)
###Markdown
System identification (causal discovery)
###Code
from ltcl.modules.components.base import GroupLinearLayer
trans_func = GroupLinearLayer(din = 8,
dout = 8,
num_blocks = 2,
diagonal = False)
b = torch.nn.Parameter(0.001 * torch.randn(1, 8))
opt = torch.optim.Adam(trans_func.parameters(),lr=0.01)
lossfunc = torch.nn.L1Loss()
max_iters = 2
counter = 0
for step in range(max_iters):
for batch in train_loader:
batch_size = batch['yt'].shape[0]
x_recon, mu, logvar, z = model.forward(batch)
mu = mu.view(batch_size, -1, 8)
# Fix permutation before training
mu = mu[:,:,col_ind]
# Fix sign flip before training
mu = mu * torch.Tensor(mask, device=mu.device).view(1,1,8)
mu = -mu
pred = trans_func(mu[:,:-1,:]).sum(dim=1) + b
true = mu[:,-1,:]
loss = lossfunc(pred, true) #+ torch.mean(adaptive.lossfun((pred - true)))
opt.zero_grad()
loss.backward()
opt.step()
if counter % 100 == 0:
print(loss.item())
counter += 1
###Output
1.0767319202423096
0.213593527674675
###Markdown
Visualize causal matrix
###Code
B2 = model.transition_prior.transition.w[0][col_ind][:, col_ind].detach().cpu().numpy()
B1 = model.transition_prior.transition.w[1][col_ind][:, col_ind].detach().cpu().numpy()
B1 = B1 * mask.reshape(1,-1) * (mask).reshape(-1,1)
B2 = B2 * mask.reshape(1,-1) * (mask).reshape(-1,1)
BB2 = np.load("/srv/data/ltcl/data/linear_nongaussian_ts/W2.npy")
BB1 = np.load("/srv/data/ltcl/data/linear_nongaussian_ts/W1.npy")
# b = np.concatenate((B1,B2), axis=0)
# bb = np.concatenate((BB1,BB2), axis=0)
# b = b / np.linalg.norm(b, axis=0).reshape(1, -1)
# bb = bb / np.linalg.norm(bb, axis=0).reshape(1, -1)
# pred = (b / np.linalg.norm(b, axis=0).reshape(1, -1)).reshape(-1)
# true = (bb / np.linalg.norm(bb, axis=0).reshape(1, -1)).reshape(-1)
bs = [B1, B2]
bbs = [BB1, BB2]
with PdfPages(figure_path + '/entries.pdf') as pdf:
fig, axs = plt.subplots(1,2, figsize=(4,2))
for tau in range(2):
ax = axs[tau]
b = bs[tau]
bb = bbs[tau]
b = b / np.linalg.norm(b, axis=0).reshape(1, -1)
bb = bb / np.linalg.norm(bb, axis=0).reshape(1, -1)
pred = (b / np.linalg.norm(b, axis=0).reshape(1, -1)).reshape(-1)
true = (bb / np.linalg.norm(bb, axis=0).reshape(1, -1)).reshape(-1)
ax.scatter(pred, true, s=10, cmap=plt.cm.coolwarm, zorder=10, color='b')
lims = [-0.75,0.75
]
# now plot both limits against eachother
ax.plot(lims, lims, '-.', alpha=0.75, zorder=0)
# ax.set_xlim(lims)
# ax.set_ylim(lims)
ax.set_xlabel("Estimated weight")
ax.set_ylabel("Truth weight")
ax.set_title(r"Entries of $\mathbf{B}_%d$"%(tau+1))
plt.tight_layout()
pdf.savefig(fig, bbox_inches="tight")
fig, axs = plt.subplots(2,4, figsize=(4,2))
for i in range(8):
row = i // 4
col = i % 4
ax = axs[row,col]
ax.scatter(B[:,i], A[:,i], s=4, color='b', alpha=0.25)
ax.axis('off')
# ax.set_xlabel('Ground truth latent')
# ax.set_ylabel('Estimated latent')
# ax.grid('..')
fig.tight_layout()
import numpy as numx
def calculate_amari_distance(matrix_one,
matrix_two,
version=1):
""" Calculate the Amari distance between two input matrices.
:param matrix_one: the first matrix
:type matrix_one: numpy array
:param matrix_two: the second matrix
:type matrix_two: numpy array
:param version: Variant to use.
:type version: int
:return: The amari distance between two input matrices.
:rtype: float
"""
if matrix_one.shape != matrix_two.shape:
return "Two matrices must have the same shape."
product_matrix = numx.abs(numx.dot(matrix_one,
numx.linalg.inv(matrix_two)))
product_matrix_max_col = numx.array(product_matrix.max(0))
product_matrix_max_row = numx.array(product_matrix.max(1))
n = product_matrix.shape[0]
""" Formula from ESLII
Here they refered to as "amari error"
The value is in [0, N-1].
reference:
Bach, F. R.; Jordan, M. I. Kernel Independent Component
Analysis, J MACH LEARN RES, 2002, 3, 1--48
"""
amari_distance = product_matrix / numx.tile(product_matrix_max_col, (n, 1))
amari_distance += product_matrix / numx.tile(product_matrix_max_row, (n, 1)).T
amari_distance = amari_distance.sum() / (2 * n) - 1
amari_distance = amari_distance / (n-1)
return amari_distance
print("Amari distance for B1:", calculate_amari_distance(B1, BB1))
print("Amari distance for B2:", calculate_amari_distance(B2, BB2))
###Output
Amari distance for B1: 0.16492316394918433
Amari distance for B2: 0.352378050478662
###Markdown
Identifiability Test of Linear VAE on Synthetic Dataset
###Code
%load_ext autoreload
%autoreload 2
import torch
import torch.nn.functional as F
from torch.utils.data import DataLoader, random_split
import leap
import numpy as np
from leap.datasets.sim_dataset import SimulationDatasetTSTwoSample
from leap.modules.srnn import SRNNSynthetic
from leap.tools.utils import load_yaml
import random
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
use_cuda = True
device = torch.device("cuda:0" if use_cuda else "cpu")
latent_size = 8
data = SimulationDatasetTSTwoSample(directory = '/srv/data/leap/data/',
transition='linear_nongaussian_ts')
num_validation_samples = 2500
train_data, val_data = random_split(data, [len(data)-num_validation_samples, num_validation_samples])
train_loader = DataLoader(train_data, batch_size=12800, shuffle=True, pin_memory=True)
val_loader = DataLoader(val_data, batch_size=16, shuffle=False, pin_memory=True)
cfg = load_yaml('../leap/configs/toy_linear_ts.yaml')
model = SRNNSynthetic.load_from_checkpoint(checkpoint_path="/srv/data/ltcl/log/weiran/toy_linear_ts/lightning_logs/version_1/checkpoints/epoch=299-step=228599.ckpt",
input_dim=cfg['VAE']['INPUT_DIM'],
length=cfg['VAE']['LENGTH'],
z_dim=cfg['VAE']['LATENT_DIM'],
lag=cfg['VAE']['LAG'],
hidden_dim=cfg['VAE']['ENC']['HIDDEN_DIM'],
trans_prior=cfg['VAE']['TRANS_PRIOR'],
bound=cfg['SPLINE']['BOUND'],
count_bins=cfg['SPLINE']['BINS'],
order=cfg['SPLINE']['ORDER'],
beta=cfg['VAE']['BETA'],
gamma=cfg['VAE']['GAMMA'],
sigma=cfg['VAE']['SIGMA'],
lr=cfg['VAE']['LR'],
bias=cfg['VAE']['BIAS'],
use_warm_start=cfg['SPLINE']['USE_WARM_START'],
spline_pth=cfg['SPLINE']['PATH'],
decoder_dist=cfg['VAE']['DEC']['DIST'],
correlation=cfg['MCC']['CORR'])
###Output
Load pretrained spline flow
###Markdown
Load model checkpoint
###Code
model.eval()
model.to('cpu')
###Output
_____no_output_____
###Markdown
Compute permutation and sign flip
###Code
for batch in train_loader:
break
batch_size = batch['s1']['xt'].shape[0]
zs.shape
zs, mu, logvar = model.forward(batch['s1'])
mu = mu.view(batch_size, -1, latent_size)
A = mu[:,0,:].detach().cpu().numpy()
B = batch['s1']['yt'][:,0,:].detach().cpu().numpy()
C = np.zeros((latent_size,latent_size))
for i in range(latent_size):
C[i] = -np.abs(np.corrcoef(B, A, rowvar=False)[i,latent_size:])
from scipy.optimize import linear_sum_assignment
row_ind, col_ind = linear_sum_assignment(C)
A = A[:, col_ind]
mask = np.ones(latent_size)
for i in range(latent_size):
if np.corrcoef(B, A, rowvar=False)[i,latent_size:][i] > 0:
mask[i] = -1
print("Permutation:",col_ind)
print("Sign Flip:", mask)
fig = plt.figure(figsize=(4,4))
sns.heatmap(-C, vmin=0, vmax=1, annot=True, fmt=".2f", linewidths=.5, cbar=False, cmap='Greens')
plt.xlabel("Estimated latents ")
plt.ylabel("True latents ")
plt.title("MCC=%.3f"%np.abs(C[row_ind, col_ind]).mean());
figure_path = '/home/weiran/figs/'
from matplotlib.backends.backend_pdf import PdfPages
with PdfPages(figure_path + '/mcc_var.pdf') as pdf:
fig = plt.figure(figsize=(4,4))
sns.heatmap(-C, vmin=0, vmax=1, annot=True, fmt=".2f", linewidths=.5, cbar=False, cmap='Greens')
plt.xlabel("Estimated latents ")
plt.ylabel("True latents ")
plt.title("MCC=%.3f"%np.abs(C[row_ind, col_ind]).mean());
pdf.savefig(fig, bbox_inches="tight")
# Permute column here
mu = mu[:,:,col_ind]
# Flip sign here
mu = mu * torch.Tensor(mask, device=mu.device).view(1,1,latent_size)
mu = -mu
fig = plt.figure(figsize=(8,2))
col = 0
plt.plot(mu[:250,-1,col].detach().cpu().numpy(), color='b', label='True', alpha=0.75)
plt.plot(batch['yt_'].squeeze()[:250,col].detach().cpu().numpy(), color='r', label="Estimated", alpha=0.75)
plt.legend()
plt.title("Current latent variable $z_t$")
fig = plt.figure(figsize=(8,2))
col = 3
l = 1
plt.plot(batch['yt'].squeeze()[:250,l,col].detach().cpu().numpy(), color='b', label='True')
plt.plot(mu[:,:-1,:][:250,l,col].detach().cpu().numpy(), color='r', label="Estimated")
plt.xlabel("Sample index")
plt.ylabel("Latent variable value")
plt.legend()
plt.title("Past latent variable $z_l$")
fig = plt.figure(figsize=(2,2))
eps = model.sample(batch["xt"].cpu())
eps = eps.detach().cpu().numpy()
component_idx = 4
sns.distplot(eps[:,component_idx], hist=False, kde=True,
bins=None,
hist_kws={'edgecolor':'black'},
kde_kws={'linewidth': 2});
plt.title("Learned noise prior")
###Output
/home/cmu_wyao/anaconda3/envs/py37/lib/python3.7/site-packages/seaborn/distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `kdeplot` (an axes-level function for kernel density plots).
warnings.warn(msg, FutureWarning)
###Markdown
System identification (causal discovery)
###Code
from leap.modules.components.base import GroupLinearLayer
trans_func = GroupLinearLayer(din = 8,
dout = 8,
num_blocks = 2,
diagonal = False)
b = torch.nn.Parameter(0.001 * torch.randn(1, 8))
opt = torch.optim.Adam(trans_func.parameters(),lr=0.01)
lossfunc = torch.nn.L1Loss()
max_iters = 2
counter = 0
for step in range(max_iters):
for batch in train_loader:
batch_size = batch['yt'].shape[0]
x_recon, mu, logvar, z = model.forward(batch)
mu = mu.view(batch_size, -1, 8)
# Fix permutation before training
mu = mu[:,:,col_ind]
# Fix sign flip before training
mu = mu * torch.Tensor(mask, device=mu.device).view(1,1,8)
mu = -mu
pred = trans_func(mu[:,:-1,:]).sum(dim=1) + b
true = mu[:,-1,:]
loss = lossfunc(pred, true) #+ torch.mean(adaptive.lossfun((pred - true)))
opt.zero_grad()
loss.backward()
opt.step()
if counter % 100 == 0:
print(loss.item())
counter += 1
###Output
1.0767319202423096
0.213593527674675
###Markdown
Visualize causal matrix
###Code
B2 = model.transition_prior.transition.w[0][col_ind][:, col_ind].detach().cpu().numpy()
B1 = model.transition_prior.transition.w[1][col_ind][:, col_ind].detach().cpu().numpy()
B1 = B1 * mask.reshape(1,-1) * (mask).reshape(-1,1)
B2 = B2 * mask.reshape(1,-1) * (mask).reshape(-1,1)
BB2 = np.load("/srv/data/ltcl/data/linear_nongaussian_ts/W2.npy")
BB1 = np.load("/srv/data/ltcl/data/linear_nongaussian_ts/W1.npy")
# b = np.concatenate((B1,B2), axis=0)
# bb = np.concatenate((BB1,BB2), axis=0)
# b = b / np.linalg.norm(b, axis=0).reshape(1, -1)
# bb = bb / np.linalg.norm(bb, axis=0).reshape(1, -1)
# pred = (b / np.linalg.norm(b, axis=0).reshape(1, -1)).reshape(-1)
# true = (bb / np.linalg.norm(bb, axis=0).reshape(1, -1)).reshape(-1)
bs = [B1, B2]
bbs = [BB1, BB2]
with PdfPages(figure_path + '/entries.pdf') as pdf:
fig, axs = plt.subplots(1,2, figsize=(4,2))
for tau in range(2):
ax = axs[tau]
b = bs[tau]
bb = bbs[tau]
b = b / np.linalg.norm(b, axis=0).reshape(1, -1)
bb = bb / np.linalg.norm(bb, axis=0).reshape(1, -1)
pred = (b / np.linalg.norm(b, axis=0).reshape(1, -1)).reshape(-1)
true = (bb / np.linalg.norm(bb, axis=0).reshape(1, -1)).reshape(-1)
ax.scatter(pred, true, s=10, cmap=plt.cm.coolwarm, zorder=10, color='b')
lims = [-0.75,0.75
]
# now plot both limits against eachother
ax.plot(lims, lims, '-.', alpha=0.75, zorder=0)
# ax.set_xlim(lims)
# ax.set_ylim(lims)
ax.set_xlabel("Estimated weight")
ax.set_ylabel("Truth weight")
ax.set_title(r"Entries of $\mathbf{B}_%d$"%(tau+1))
plt.tight_layout()
pdf.savefig(fig, bbox_inches="tight")
fig, axs = plt.subplots(2,4, figsize=(4,2))
for i in range(8):
row = i // 4
col = i % 4
ax = axs[row,col]
ax.scatter(B[:,i], A[:,i], s=4, color='b', alpha=0.25)
ax.axis('off')
# ax.set_xlabel('Ground truth latent')
# ax.set_ylabel('Estimated latent')
# ax.grid('..')
fig.tight_layout()
import numpy as numx
def calculate_amari_distance(matrix_one,
matrix_two,
version=1):
""" Calculate the Amari distance between two input matrices.
:param matrix_one: the first matrix
:type matrix_one: numpy array
:param matrix_two: the second matrix
:type matrix_two: numpy array
:param version: Variant to use.
:type version: int
:return: The amari distance between two input matrices.
:rtype: float
"""
if matrix_one.shape != matrix_two.shape:
return "Two matrices must have the same shape."
product_matrix = numx.abs(numx.dot(matrix_one,
numx.linalg.inv(matrix_two)))
product_matrix_max_col = numx.array(product_matrix.max(0))
product_matrix_max_row = numx.array(product_matrix.max(1))
n = product_matrix.shape[0]
""" Formula from ESLII
Here they refered to as "amari error"
The value is in [0, N-1].
reference:
Bach, F. R.; Jordan, M. I. Kernel Independent Component
Analysis, J MACH LEARN RES, 2002, 3, 1--48
"""
amari_distance = product_matrix / numx.tile(product_matrix_max_col, (n, 1))
amari_distance += product_matrix / numx.tile(product_matrix_max_row, (n, 1)).T
amari_distance = amari_distance.sum() / (2 * n) - 1
amari_distance = amari_distance / (n-1)
return amari_distance
print("Amari distance for B1:", calculate_amari_distance(B1, BB1))
print("Amari distance for B2:", calculate_amari_distance(B2, BB2))
###Output
Amari distance for B1: 0.16492316394918433
Amari distance for B2: 0.352378050478662
|
Semana1-Projeto.ipynb | ###Markdown
Descrição: As a data scientist working for an investment firm, you will extract the revenue data for Tesla and GameStop and build a dashboard to compare the price of the stock vs the revenue. Tarefas:- Question 1 - Extracting Tesla Stock Data Using yfinance - 2 Points- Question 2 - Extracting Tesla Revenue Data Using Webscraping - 1 Points- Question 3 - Extracting GameStop Stock Data Using yfinance - 2 Points- Question 4 - Extracting GameStop Revenue Data Using Webscraping - 1 Points- Question 5 - Tesla Stock and Revenue Dashboard - 2 Points- Question 6 - GameStop Stock and Revenue Dashboard- 2 Points- Question 7 - Sharing your Assignment Notebook - 2 Points
###Code
# installing dependencies
!pip install yfinance pandas bs4
# importing modules
import yfinance as yf
import pandas as pd
import requests
from bs4 import BeautifulSoup
# needed to cast datetime values
from datetime import datetime
###Output
Requirement already satisfied: yfinance in /opt/conda/lib/python3.8/site-packages (0.1.63)
Requirement already satisfied: pandas in /opt/conda/lib/python3.8/site-packages (1.2.4)
Requirement already satisfied: bs4 in /opt/conda/lib/python3.8/site-packages (0.0.1)
Requirement already satisfied: beautifulsoup4 in /opt/conda/lib/python3.8/site-packages (from bs4) (4.9.3)
Requirement already satisfied: python-dateutil>=2.7.3 in /opt/conda/lib/python3.8/site-packages (from pandas) (2.8.1)
Requirement already satisfied: pytz>=2017.3 in /opt/conda/lib/python3.8/site-packages (from pandas) (2021.1)
Requirement already satisfied: numpy>=1.16.5 in /opt/conda/lib/python3.8/site-packages (from pandas) (1.20.2)
Requirement already satisfied: six>=1.5 in /opt/conda/lib/python3.8/site-packages (from python-dateutil>=2.7.3->pandas) (1.15.0)
Requirement already satisfied: multitasking>=0.0.7 in /opt/conda/lib/python3.8/site-packages (from yfinance) (0.0.9)
Requirement already satisfied: requests>=2.20 in /opt/conda/lib/python3.8/site-packages (from yfinance) (2.25.1)
Requirement already satisfied: lxml>=4.5.1 in /opt/conda/lib/python3.8/site-packages (from yfinance) (4.6.3)
Requirement already satisfied: urllib3<1.27,>=1.21.1 in /opt/conda/lib/python3.8/site-packages (from requests>=2.20->yfinance) (1.26.4)
Requirement already satisfied: chardet<5,>=3.0.2 in /opt/conda/lib/python3.8/site-packages (from requests>=2.20->yfinance) (4.0.0)
Requirement already satisfied: idna<3,>=2.5 in /opt/conda/lib/python3.8/site-packages (from requests>=2.20->yfinance) (2.10)
Requirement already satisfied: certifi>=2017.4.17 in /opt/conda/lib/python3.8/site-packages (from requests>=2.20->yfinance) (2020.12.5)
Requirement already satisfied: soupsieve>1.2 in /opt/conda/lib/python3.8/site-packages (from beautifulsoup4->bs4) (2.0.1)
###Markdown
Question 1 - Extracting Tesla Stock Data Using yfinance
###Code
# making Ticker object
tesla_ticker = yf.Ticker("TSLA")
# creating a dataframe with history values
tesla_data = tesla_ticker.history(period="max")
# now we need to reset dataframe index and show first values
tesla_data.reset_index(inplace=True)
tesla_data.head()
###Output
_____no_output_____
###Markdown
Question 2 - Extracting Tesla Revenue Data Using Webscraping
###Code
# getting TSLA revenue data from https://www.macrotrends.net/stocks/charts/TSLA/tesla/revenue
tesla_revenue_url="https://www.macrotrends.net/stocks/charts/TSLA/tesla/revenue"
html_data=requests.get(tesla_revenue_url).text
# parsing html data
soup = BeautifulSoup(html_data,"html.parser")
# now we need to create a dataframe with columns "date" and "revenue", using data scrapped from soup
tesla_revenue_table = soup.find("table", class_="historical_data_table")
# now we will create an empty dataframe
tesla_revenue_data = pd.DataFrame(columns=["Date", "Revenue"])
# now we will loop through table and populate the dataframe
for row in tesla_revenue_table.tbody.find_all("tr"):
col = row.find_all("td")
if (col != []):
row_date = col[0].text
row_date = datetime.strptime(row_date, '%Y')
row_revenue = col[1].text
# we need to strip "," and "$" chars from revenue value
row_revenue = row_revenue.replace(",","").replace("$","")
row_revenue = int(row_revenue)
# printing var types
# print(type(row_date), type(row_revenue) )
tesla_revenue_data = tesla_revenue_data.append(
{
'Date': row_date,
'Revenue': row_revenue
}, ignore_index=True)
tesla_revenue_data.head()
###Output
_____no_output_____
###Markdown
Question 3 - Extracting GameStop Stock Data Using yfinance
###Code
# making Ticker object
gme_ticker = yf.Ticker("GME")
# creating a dataframe with history values
gme_data = gme_ticker.history(period="max")
# now we need to reset dataframe index and show first values
gme_data.reset_index(inplace=True)
gme_data.head()
###Output
_____no_output_____
###Markdown
Question 4 - Extracting GameStop Revenue Data Using Webscraping
###Code
# getting GME revenue data from https://www.macrotrends.net/stocks/charts/GME/gamestop/revenue
gme_revenue_url="https://www.macrotrends.net/stocks/charts/GME/gamestop/revenue"
html_data=requests.get(gme_revenue_url).text
# parsing html data
soup = BeautifulSoup(html_data,"html.parser")
# now we need to create a dataframe with columns "date" and "revenue", using data scrapped from soup
gme_revenue_table = soup.find("table", class_="historical_data_table")
# now we will create an empty dataframe
gme_revenue_data = pd.DataFrame(columns=["Date", "Revenue"])
# now we will loop through table and populate the dataframe
for row in gme_revenue_table.tbody.find_all("tr"):
col = row.find_all("td")
if (col != []):
row_date = col[0].text
row_date = datetime.strptime(row_date, '%Y')
row_revenue = col[1].text
# we need to strip "," and "$" chars from revenue value
row_revenue = row_revenue.replace(",","").replace("$","")
row_revenue = int(row_revenue)
# printing var types
# print(type(row_date), type(row_revenue) )
gme_revenue_data = gme_revenue_data.append(
{
'Date': row_date,
'Revenue': row_revenue
}, ignore_index=True)
gme_revenue_data.head()
###Output
_____no_output_____
###Markdown
Question 5 - Tesla Stock and Revenue Dashboard
###Code
# plotting tesla stock data
tesla_data.plot(x="Date", y="Open")
# plotting tesla revenue data
tesla_revenue_data.plot(x="Date", y="Revenue")
###Output
_____no_output_____
###Markdown
Question 6 - GameStop Stock and Revenue Dashboard
###Code
# plotting gamestop stock data
gme_data.plot(x="Date", y="Open")
# plotting gamestop revenue data
gme_revenue_data.plot(x="Date", y="Revenue")
###Output
_____no_output_____ |
program/.ipynb_checkpoints/2_6_Qlearning-checkpoint.ipynb | ###Markdown
2.6 Q学習で迷路を攻略
###Code
# 使用するパッケージの宣言
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# 初期位置での迷路の様子
# 図を描く大きさと、図の変数名を宣言
fig = plt.figure(figsize=(5, 5))
ax = plt.gca()
# 赤い壁を描く
plt.plot([1, 1], [0, 1], color='red', linewidth=2)
plt.plot([1, 2], [2, 2], color='red', linewidth=2)
plt.plot([2, 2], [2, 1], color='red', linewidth=2)
plt.plot([2, 3], [1, 1], color='red', linewidth=2)
# 状態を示す文字S0~S8を描く
plt.text(0.5, 2.5, 'S0', size=14, ha='center')
plt.text(1.5, 2.5, 'S1', size=14, ha='center')
plt.text(2.5, 2.5, 'S2', size=14, ha='center')
plt.text(0.5, 1.5, 'S3', size=14, ha='center')
plt.text(1.5, 1.5, 'S4', size=14, ha='center')
plt.text(2.5, 1.5, 'S5', size=14, ha='center')
plt.text(0.5, 0.5, 'S6', size=14, ha='center')
plt.text(1.5, 0.5, 'S7', size=14, ha='center')
plt.text(2.5, 0.5, 'S8', size=14, ha='center')
plt.text(0.5, 2.3, 'START', ha='center')
plt.text(2.5, 0.3, 'GOAL', ha='center')
# 描画範囲の設定と目盛りを消す設定
ax.set_xlim(0, 3)
ax.set_ylim(0, 3)
plt.tick_params(axis='both', which='both', bottom='off', top='off',
labelbottom='off', right='off', left='off', labelleft='off')
# 現在地S0に緑丸を描画する
line, = ax.plot([0.5], [2.5], marker="o", color='g', markersize=60)
# 初期の方策を決定するパラメータtheta_0を設定
# 行は状態0~7、列は移動方向で↑、→、↓、←を表す
theta_0 = np.array([[np.nan, 1, 1, np.nan], # s0
[np.nan, 1, np.nan, 1], # s1
[np.nan, np.nan, 1, 1], # s2
[1, 1, 1, np.nan], # s3
[np.nan, np.nan, 1, 1], # s4
[1, np.nan, np.nan, np.nan], # s5
[1, np.nan, np.nan, np.nan], # s6
[1, 1, np.nan, np.nan], # s7、※s8はゴールなので、方策はなし
])
# 方策パラメータtheta_0をランダム方策piに変換する関数の定義
def simple_convert_into_pi_from_theta(theta):
'''単純に割合を計算する'''
[m, n] = theta.shape # thetaの行列サイズを取得
pi = np.zeros((m, n))
for i in range(0, m):
pi[i, :] = theta[i, :] / np.nansum(theta[i, :]) # 割合の計算
pi = np.nan_to_num(pi) # nanを0に変換
return pi
# ランダム行動方策pi_0を求める
pi_0 = simple_convert_into_pi_from_theta(theta_0)
# 初期の行動価値関数Qを設定
[a, b] = theta_0.shape # 行と列の数をa, bに格納
Q = np.random.rand(a, b) * theta_0 * 0.1
# *theta0をすることで要素ごとに掛け算をし、Qの壁方向の値がnanになる
# ε-greedy法を実装
def get_action(s, Q, epsilon, pi_0):
direction = ["up", "right", "down", "left"]
# 行動を決める
if np.random.rand() < epsilon:
# εの確率でランダムに動く
next_direction = np.random.choice(direction, p=pi_0[s, :])
else:
# Qの最大値の行動を採用する
next_direction = direction[np.nanargmax(Q[s, :])]
# 行動をindexに
if next_direction == "up":
action = 0
elif next_direction == "right":
action = 1
elif next_direction == "down":
action = 2
elif next_direction == "left":
action = 3
return action
def get_s_next(s, a, Q, epsilon, pi_0):
direction = ["up", "right", "down", "left"]
next_direction = direction[a] # 行動aの方向
# 行動から次の状態を決める
if next_direction == "up":
s_next = s - 3 # 上に移動するときは状態の数字が3小さくなる
elif next_direction == "right":
s_next = s + 1 # 右に移動するときは状態の数字が1大きくなる
elif next_direction == "down":
s_next = s + 3 # 下に移動するときは状態の数字が3大きくなる
elif next_direction == "left":
s_next = s - 1 # 左に移動するときは状態の数字が1小さくなる
return s_next
# Q学習による行動価値関数Qの更新
def Q_learning(s, a, r, s_next, Q, eta, gamma):
if s_next == 8: # ゴールした場合
Q[s, a] = Q[s, a] + eta * (r - Q[s, a])
else:
Q[s, a] = Q[s, a] + eta * (r + gamma * np.nanmax(Q[s_next,: ]) - Q[s, a])
return Q
# Q学習で迷路を解く関数の定義、状態と行動の履歴および更新したQを出力
def goal_maze_ret_s_a_Q(Q, epsilon, eta, gamma, pi):
s = 0 # スタート地点
a = a_next = get_action(s, Q, epsilon, pi) # 初期の行動
s_a_history = [[0, np.nan]] # エージェントの移動を記録するリスト
while (1): # ゴールするまでループ
a = a_next # 行動更新
s_a_history[-1][1] = a
# 現在の状態(つまり一番最後なのでindex=-1)に行動を代入
s_next = get_s_next(s, a, Q, epsilon, pi)
# 次の状態を格納
s_a_history.append([s_next, np.nan])
# 次の状態を代入。行動はまだ分からないのでnanにしておく
# 報酬を与え, 次の行動を求めます
if s_next == 8:
r = 1 # ゴールにたどり着いたなら報酬を与える
a_next = np.nan
else:
r = 0
a_next = get_action(s_next, Q, epsilon, pi)
# 次の行動a_nextを求めます。
# 価値関数を更新
Q = Q_learning(s, a, r, s_next, Q, eta, gamma)
# 終了判定
if s_next == 8: # ゴール地点なら終了
break
else:
s = s_next
return [s_a_history, Q]
# Q学習で迷路を解く
eta = 0.1 # 学習率
gamma = 0.9 # 時間割引率
epsilon = 0.5 # ε-greedy法の初期値
v = np.nanmax(Q, axis=1) # 状態ごとに価値の最大値を求める
is_continue = True
episode = 1
V = [] # エピソードごとの状態価値を格納する
V.append(np.nanmax(Q, axis=1)) # 状態ごとに行動価値の最大値を求める
while is_continue: # is_continueがFalseになるまで繰り返す
print("エピソード:" + str(episode))
# ε-greedyの値を少しずつ小さくする
epsilon = epsilon / 2
# Q学習で迷路を解き、移動した履歴と更新したQを求める
[s_a_history, Q] = goal_maze_ret_s_a_Q(Q, epsilon, eta, gamma, pi_0)
# 状態価値の変化
new_v = np.nanmax(Q, axis=1) # 状態ごとに行動価値の最大値を求める
print(np.sum(np.abs(new_v - v))) # 状態価値関数の変化を出力
v = new_v
V.append(v) # このエピソード終了時の状態価値関数を追加
print("迷路を解くのにかかったステップ数は" + str(len(s_a_history) - 1) + "です")
# 100エピソード繰り返す
episode = episode + 1
if episode > 100:
break
# 状態価値の変化を可視化します
# 参考URL http://louistiao.me/posts/notebooks/embedding-matplotlib-animations-in-jupyter-notebooks/
from matplotlib import animation
from IPython.display import HTML
import matplotlib.cm as cm # color map
def init():
# 背景画像の初期化
line.set_data([], [])
return (line,)
def animate(i):
# フレームごとの描画内容
# 各マスに状態価値の大きさに基づく色付きの四角を描画
line, = ax.plot([0.5], [2.5], marker="s",
color=cm.jet(V[i][0]), markersize=85) # S0
line, = ax.plot([1.5], [2.5], marker="s",
color=cm.jet(V[i][1]), markersize=85) # S1
line, = ax.plot([2.5], [2.5], marker="s",
color=cm.jet(V[i][2]), markersize=85) # S2
line, = ax.plot([0.5], [1.5], marker="s",
color=cm.jet(V[i][3]), markersize=85) # S3
line, = ax.plot([1.5], [1.5], marker="s",
color=cm.jet(V[i][4]), markersize=85) # S4
line, = ax.plot([2.5], [1.5], marker="s",
color=cm.jet(V[i][5]), markersize=85) # S5
line, = ax.plot([0.5], [0.5], marker="s",
color=cm.jet(V[i][6]), markersize=85) # S6
line, = ax.plot([1.5], [0.5], marker="s",
color=cm.jet(V[i][7]), markersize=85) # S7
line, = ax.plot([2.5], [0.5], marker="s",
color=cm.jet(1.0), markersize=85) # S8
return (line,)
# 初期化関数とフレームごとの描画関数を用いて動画を作成
anim = animation.FuncAnimation(
fig, animate, init_func=init, frames=len(V), interval=200, repeat=False)
HTML(anim.to_jshtml())
###Output
_____no_output_____ |
doc/examples/Simple Live plotting example with DDH5.ipynb | ###Markdown
The idea is pretty simple:We first define the structure of the datadict (you can also use a datadict that is already populated); this is equivalent to the idea of registering parameters in qcodes.You can then use the DDH5 writer to start saving data -- it'll determine file location automatically, within the base directory that is the first argument.To look at the data, you can use the `autoplot_ddh5` app. The easiest way might be to copy the file `apps/templates/autoplot_ddh5.bat` to some location of your choice, and edit the pathname variable to the correct folder in which `autoplot_ddh5.py` is located, such as:`` @set "APPPATH=c:\code\plottr\apps"``(note: this is the apps directory in the plottr base repository, not in the package). You can then associate opening `.ddh5` files with that batch file.
###Code
data = dd.DataDict(
x = dict(unit='A'),
y = dict(unit='B'),
z = dict(axes=['x', 'y']),
)
data.validate()
nrows = 100
with dds.DDH5Writer(r"d:\data", data) as writer:
for n in range(nrows):
writer.add_data(x=[n],
y=np.linspace(0,1,11).reshape(1,-1),
z=np.random.rand(11).reshape(1,-1)
)
time.sleep(1)
###Output
_____no_output_____ |
practice/cython-demo.ipynb | ###Markdown
Compare speed between native and cythonized math functions
###Code
%load_ext Cython
%%cython
from libc.math cimport log, sqrt
def log_c(float x):
return log(x)/2.302585092994046
def sqrt_c(float x):
return sqrt(x)
import os
from os.path import expanduser
import pandas as pd
import pandas.io.sql as pd_sql
import math
from functions.auth.connections import postgres_connection
connection_uri = postgres_connection('mountain_project')
def transform_features(df):
""" Add log and sqrt values
"""
# add log values for ols linear regression
df['log_star_ratings'] = df['star_ratings'].apply(lambda x: math.log(x+1, 10))
df['log_ticks'] = df['ticks'].apply(lambda x: math.log(x+1, 10))
df['log_avg_stars'] = df['avg_stars'].apply(lambda x: math.log(x+1, 10))
df['log_length'] = df['length_'].apply(lambda x: math.log(x+1, 10))
df['log_grade'] = df['grade'].apply(lambda x: math.log(x+2, 10))
df['log_on_to_do_lists'] = df['on_to_do_lists'].apply(lambda x: math.log(x+1, 10)) # Target
# add sqrt values for Poisson regression
df['sqrt_star_ratings'] = df['star_ratings'].apply(lambda x: math.sqrt(x))
df['sqrt_ticks'] = df['ticks'].apply(lambda x: math.sqrt(x))
df['sqrt_avg_stars'] = df['avg_stars'].apply(lambda x: math.sqrt(x))
df['sqrt_length'] = df['length_'].apply(lambda x: math.sqrt(x))
df['sqrt_grade'] = df['grade'].apply(lambda x: math.sqrt(x+1))
return df
def transform_features_cythonized(df):
""" Add log and sqrt values using cythonized math functions
"""
# add log values for ols linear regression
df['log_star_ratings'] = df.star_ratings.apply(lambda x: log_c(x+1))
df['log_ticks'] = df.ticks.apply(lambda x: log_c(x+1))
df['log_avg_stars'] = df.avg_stars.apply(lambda x: log_c(x+1))
df['log_length'] = df.length_.apply(lambda x: log_c(x+1))
df['log_grade'] = df.grade.apply(lambda x: log_c(x+2))
df['log_on_to_do_lists'] = df.on_to_do_lists.apply(lambda x: log_c(x+1))
# add sqrt values for Poisson regression
df['sqrt_star_ratings'] = df.star_ratings.apply(lambda x: sqrt_c(x))
df['sqrt_ticks'] = df.ticks.apply(lambda x: sqrt_c(x))
df['sqrt_avg_stars'] = df.avg_stars.apply(lambda x: sqrt_c(x))
df['sqrt_length'] = df.length_.apply(lambda x: sqrt_c(x))
df['sqrt_grade'] = df.grade.apply(lambda x: sqrt_c(x+1))
return df
query = """
SELECT b.avg_stars, b.your_stars, b.length_, b.grade,
r.star_ratings, r.suggested_ratings, r.on_to_do_lists, r.ticks
FROM routes b
LEFT JOIN ratings r ON b.url_id = r.url_id
WHERE b.area_name = 'buttermilks'
AND length_ IS NOT NULL
"""
df = pd_sql.read_sql(query, connection_uri) # grab data as a dataframe
df.head()
%%timeit
transform_features(df)
%%timeit
transform_features_cythonized(df) # Cythonized math functions are only barely faster
###Output
2.98 ms ± 31.3 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
|
cleared-demos/ivp_odes/Dissipation in Runge-Kutta Methods.ipynb | ###Markdown
Dissipation in Runge-Kutta MethodsCopyright (C) 2020 Andreas KloecknerMIT LicensePermission is hereby granted, free of charge, to any person obtaining a copyof this software and associated documentation files (the "Software"), to dealin the Software without restriction, including without limitation the rightsto use, copy, modify, merge, publish, distribute, sublicense, and/or sellcopies of the Software, and to permit persons to whom the Software isfurnished to do so, subject to the following conditions:The above copyright notice and this permission notice shall be included inall copies or substantial portions of the Software.THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS ORIMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THEAUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHERLIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS INTHE SOFTWARE.
###Code
import numpy as np
import matplotlib.pyplot as pt
def rk4_step(y, t, h, f):
k1 = f(t, y)
k2 = f(t+h/2, y + h/2*k1)
k3 = f(t+h/2, y + h/2*k2)
k4 = f(t+h, y + h*k3)
return y + h/6*(k1 + 2*k2 + 2*k3 + k4)
###Output
_____no_output_____
###Markdown
Consider the second-order harmonic oscillator$$u''=-u$$with initial conditions $u(0) = 1$ and $u'(0)=0$.$\cos(x)$ is a solution to this problem.`f` below gives the right-hand side for this ODE converted to first-order form:
###Code
def f(t, y):
u, up = y
return np.array([up, -u])
###Output
_____no_output_____
###Markdown
Now, we use 4th-order Runge Kutta to integrate this system over "many" time steps:
###Code
times = [0]
y_values = [np.array([1,0])]
h = 0.5
t_end = 800
while times[-1] < t_end:
y_values.append(rk4_step(y_values[-1], times[-1], h, f))
times.append(times[-1]+h)
###Output
_____no_output_____
###Markdown
Lastly, plot the computed solution:
###Code
y_values = np.array(y_values)
pt.figure(figsize=(15,5))
pt.plot(times, y_values[:, 0])
###Output
_____no_output_____ |
assignments/assignment9/Assignment9_Model1_NarrativeQA_dataset.ipynb | ###Markdown
2 - Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine TranslationIn this second notebook on sequence-to-sequence models using PyTorch and TorchText, we'll be implementing the model from [Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine Translation](https://arxiv.org/abs/1406.1078). This model will achieve improved test perplexity whilst only using a single layer RNN in both the encoder and the decoder. IntroductionLet's remind ourselves of the general encoder-decoder model.We use our encoder (green) over the embedded source sequence (yellow) to create a context vector (red). We then use that context vector with the decoder (blue) and a linear layer (purple) to generate the target sentence.In the previous model, we used an multi-layered LSTM as the encoder and decoder.One downside of the previous model is that the decoder is trying to cram lots of information into the hidden states. Whilst decoding, the hidden state will need to contain information about the whole of the source sequence, as well as all of the tokens have been decoded so far. By alleviating some of this information compression, we can create a better model!We'll also be using a GRU (Gated Recurrent Unit) instead of an LSTM (Long Short-Term Memory). Why? Mainly because that's what they did in the paper (this paper also introduced GRUs) and also because we used LSTMs last time. To understand how GRUs (and LSTMs) differ from standard RNNS, check out [this](https://colah.github.io/posts/2015-08-Understanding-LSTMs/) link. Is a GRU better than an LSTM? [Research](https://arxiv.org/abs/1412.3555) has shown they're pretty much the same, and both are better than standard RNNs. Preparing DataAll of the data preparation will be (almost) the same as last time, so we'll very briefly detail what each code block does. See the previous notebook for a recap.We'll import PyTorch, TorchText, spaCy and a few standard modules.
###Code
import torch
import torch.nn as nn
import torch.optim as optim
from torchtext.datasets import Multi30k
from torchtext.data import Field, BucketIterator, Example, Dataset
import spacy
import numpy as np
import random
import math
import time
###Output
_____no_output_____
###Markdown
Then set a random seed for deterministic results/reproducability.
###Code
SEED = 1234
random.seed(SEED)
np.random.seed(SEED)
torch.manual_seed(SEED)
torch.cuda.manual_seed(SEED)
torch.backends.cudnn.deterministic = True
###Output
_____no_output_____
###Markdown
Instantiate our German and English spaCy models. Previously we reversed the source (German) sentence, however in the paper we are implementing they don't do this, so neither will we. Create our fields to process our data. This will append the "start of sentence" and "end of sentence" tokens as well as converting all words to lowercase.
###Code
SRC = Field(tokenize='spacy',
init_token='<sos>',
eos_token='<eos>',
lower=True)
TRG = Field(tokenize = 'spacy',
init_token='<sos>',
eos_token='<eos>',
lower=True)
###Output
_____no_output_____
###Markdown
Load our data.
###Code
import pandas as pd
#Read and preprocess the data
docs_index = pd.read_csv('https://raw.githubusercontent.com/deepmind/narrativeqa/master/documents.csv')
qapair = pd.read_csv('https://raw.githubusercontent.com/deepmind/narrativeqa/master/qaps.csv')
wiki_summary = pd.read_csv("https://raw.githubusercontent.com/deepmind/narrativeqa/master/third_party/wikipedia/summaries.csv")
wiki_summary.head(2)
docs_index.head()
qapair
docs_index.shape, qapair.shape, wiki_summary.shape
qapair.columns, wiki_summary.columns
mapped_ads = qapair.merge(wiki_summary[['document_id', 'summary']], on='document_id', how='left')
mapped_ads_filtered = mapped_ads[['question', 'answer1', 'summary', 'set']]
mapped_ads_filtered['question'][0]
mapped_ads_filtered['summary'][0][:400]
MAX_LEN = 400
mapped_ads_filtered['summary_question'] = mapped_ads_filtered['summary'].apply(lambda x: x[:MAX_LEN]) + mapped_ads_filtered['question']
mapped_ads_filtered['answer'] = mapped_ads_filtered['answer1']
len(mapped_ads_filtered['summary_question'][0])
mapped_ads_filtered[mapped_ads_filtered['summary'].isna()]
df= mapped_ads_filtered[['summary_question', 'answer', 'set']]
test_df, train_df, valid_df = [group for _, group in df.groupby('set')]
test_df.reset_index(drop=True, inplace=True)
train_df.reset_index(drop=True, inplace=True)
valid_df.reset_index(drop=True, inplace=True)
len(train_df['summary_question'][10])
# This pandas dataframe should be mapped to the list of fields object
# formed the list of fields object
fields = [('src',SRC), ('trg', TRG)]
# map operation to map the rows of pandas dataframe to torchdataset field
train_mapped_rows_to_torchtext = [Example.fromlist([train_df['summary_question'][i], train_df['answer'][i]], fields) for i in range(train_df.shape[0])]
test_mapped_rows_to_torchtext = [Example.fromlist([test_df['summary_question'][i], test_df['answer'][i]], fields) for i in range(test_df.shape[0])]
valid_mapped_rows_to_torchtext = [Example.fromlist([valid_df['summary_question'][i], valid_df['answer'][i]], fields) for i in range(valid_df.shape[0])]
train_data = Dataset(train_mapped_rows_to_torchtext, fields)
valid_data = Dataset(valid_mapped_rows_to_torchtext, fields)
test_data = Dataset(test_mapped_rows_to_torchtext, fields)
###Output
_____no_output_____
###Markdown
We'll also print out an example just to double check they're not reversed.
###Code
print(vars(valid_data.examples[0]))
print(vars(train_data.examples[0]))
###Output
{'src': [' ', 'at', 'madeline', 'hall', ',', 'an', 'old', 'mansion', '-', 'house', 'near', 'southampton', 'belonging', 'to', 'the', 'wealthy', 'de', 'versely', 'family', ',', 'lives', 'an', 'elderly', 'spinster', 'miss', 'delmar', ',', 'the', 'aunt', 'of', 'the', 'earl', 'de', 'versely', 'and', 'captain', 'delmar', '.', 'miss', 'delmar', 'invites', 'arabella', 'mason', ',', 'the', 'daughter', 'of', 'a', 'deceased', ',', 'well', '-', 'liked', 'steward', 'to', 'stay', 'with', 'her', 'as', 'a', 'lower', '-', 'class', 'guest', 'in', 'the', 'house', '.', 'captain', 'delmar', 'is', 'known', 'to', 'visit', 'his', 'aunt', 'at', 'madeline', 'hall', 'frequently', ',', 'who', 'is', 'miss', 'delmer', '?'], 'trg': ['the', 'elderly', 'spinster', 'aunt', 'of', 'the', 'earl', 'de', 'verseley', 'and', 'captain', 'delmar']}
###Markdown
Then create our vocabulary, converting all tokens appearing less than twice into `` tokens.
###Code
SRC.build_vocab(train_data, min_freq = 2)
TRG.build_vocab(train_data, min_freq = 2)
###Output
_____no_output_____
###Markdown
Finally, define the `device` and create our iterators.
###Code
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
BATCH_SIZE = 128
train_iterator, valid_iterator, test_iterator = BucketIterator.splits(
(train_data, valid_data, test_data),
batch_size = BATCH_SIZE,
device = device, sort=False)
print("dataload complete")
###Output
dataload complete
###Markdown
Building the Seq2Seq Model EncoderThe encoder is similar to the previous one, with the multi-layer LSTM swapped for a single-layer GRU. We also don't pass the dropout as an argument to the GRU as that dropout is used between each layer of a multi-layered RNN. As we only have a single layer, PyTorch will display a warning if we try and use pass a dropout value to it.Another thing to note about the GRU is that it only requires and returns a hidden state, there is no cell state like in the LSTM.$$\begin{align*}h_t &= \text{GRU}(e(x_t), h_{t-1})\\(h_t, c_t) &= \text{LSTM}(e(x_t), h_{t-1}, c_{t-1})\\h_t &= \text{RNN}(e(x_t), h_{t-1})\end{align*}$$From the equations above, it looks like the RNN and the GRU are identical. Inside the GRU, however, is a number of *gating mechanisms* that control the information flow in to and out of the hidden state (similar to an LSTM). Again, for more info, check out [this](https://colah.github.io/posts/2015-08-Understanding-LSTMs/) excellent post. The rest of the encoder should be very familar from the last session, it takes in a sequence, $X = \{x_1, x_2, ... , x_T\}$, passes it through the embedding layer, recurrently calculates hidden states, $H = \{h_1, h_2, ..., h_T\}$, and returns a context vector (the final hidden state), $z=h_T$.$$h_t = \text{EncoderGRU}(e(x_t), h_{t-1})$$This is identical to the encoder of the general seq2seq model, with all the "magic" happening inside the GRU (green).
###Code
class Encoder(nn.Module):
def __init__(self, input_dim, emb_dim, hid_dim, dropout):
super().__init__()
self.hid_dim = hid_dim
self.embedding = nn.Embedding(input_dim, emb_dim) #no dropout as only one layer!
self.rnn = nn.GRU(emb_dim, hid_dim)
self.dropout = nn.Dropout(dropout)
def forward(self, src):
#src = [src len, batch size]
embedded = self.dropout(self.embedding(src))
#embedded = [src len, batch size, emb dim]
outputs, hidden = self.rnn(embedded) #no cell state!
#outputs = [src len, batch size, hid dim * n directions]
#hidden = [n layers * n directions, batch size, hid dim]
#outputs are always from the top hidden layer
return hidden
###Output
_____no_output_____
###Markdown
DecoderThe decoder is where the implementation differs significantly from the previous model and we alleviate some of the information compression.Instead of the GRU in the decoder taking just the embedded target token, $d(y_t)$ and the previous hidden state $s_{t-1}$ as inputs, it also takes the context vector $z$. $$s_t = \text{DecoderGRU}(d(y_t), s_{t-1}, z)$$Note how this context vector, $z$, does not have a $t$ subscript, meaning we re-use the same context vector returned by the encoder for every time-step in the decoder. Before, we predicted the next token, $\hat{y}_{t+1}$, with the linear layer, $f$, only using the top-layer decoder hidden state at that time-step, $s_t$, as $\hat{y}_{t+1}=f(s_t^L)$. Now, we also pass the embedding of current token, $d(y_t)$ and the context vector, $z$ to the linear layer.$$\hat{y}_{t+1} = f(d(y_t), s_t, z)$$Thus, our decoder now looks something like this:Note, the initial hidden state, $s_0$, is still the context vector, $z$, so when generating the first token we are actually inputting two identical context vectors into the GRU.How do these two changes reduce the information compression? Well, hypothetically the decoder hidden states, $s_t$, no longer need to contain information about the source sequence as it is always available as an input. Thus, it only needs to contain information about what tokens it has generated so far. The addition of $y_t$ to the linear layer also means this layer can directly see what the token is, without having to get this information from the hidden state. However, this hypothesis is just a hypothesis, it is impossible to determine how the model actually uses the information provided to it (don't listen to anyone that says differently). Nevertheless, it is a solid intuition and the results seem to indicate that this modifications are a good idea!Within the implementation, we will pass $d(y_t)$ and $z$ to the GRU by concatenating them together, so the input dimensions to the GRU are now `emb_dim + hid_dim` (as context vector will be of size `hid_dim`). The linear layer will take $d(y_t), s_t$ and $z$ also by concatenating them together, hence the input dimensions are now `emb_dim + hid_dim*2`. We also don't pass a value of dropout to the GRU as it only uses a single layer.`forward` now takes a `context` argument. Inside of `forward`, we concatenate $y_t$ and $z$ as `emb_con` before feeding to the GRU, and we concatenate $d(y_t)$, $s_t$ and $z$ together as `output` before feeding it through the linear layer to receive our predictions, $\hat{y}_{t+1}$.
###Code
class Decoder(nn.Module):
def __init__(self, output_dim, emb_dim, hid_dim, dropout):
super().__init__()
self.hid_dim = hid_dim
self.output_dim = output_dim
self.embedding = nn.Embedding(output_dim, emb_dim)
self.rnn = nn.GRU(emb_dim + hid_dim, hid_dim)
self.fc_out = nn.Linear(emb_dim + hid_dim * 2, output_dim)
self.dropout = nn.Dropout(dropout)
def forward(self, input, hidden, context):
#input = [batch size]
#hidden = [n layers * n directions, batch size, hid dim]
#context = [n layers * n directions, batch size, hid dim]
#n layers and n directions in the decoder will both always be 1, therefore:
#hidden = [1, batch size, hid dim]
#context = [1, batch size, hid dim]
input = input.unsqueeze(0)
#input = [1, batch size]
embedded = self.dropout(self.embedding(input))
#embedded = [1, batch size, emb dim]
emb_con = torch.cat((embedded, context), dim = 2)
#emb_con = [1, batch size, emb dim + hid dim]
output, hidden = self.rnn(emb_con, hidden)
#output = [seq len, batch size, hid dim * n directions]
#hidden = [n layers * n directions, batch size, hid dim]
#seq len, n layers and n directions will always be 1 in the decoder, therefore:
#output = [1, batch size, hid dim]
#hidden = [1, batch size, hid dim]
output = torch.cat((embedded.squeeze(0), hidden.squeeze(0), context.squeeze(0)),
dim = 1)
#output = [batch size, emb dim + hid dim * 2]
prediction = self.fc_out(output)
#prediction = [batch size, output dim]
return prediction, hidden
###Output
_____no_output_____
###Markdown
Seq2Seq ModelPutting the encoder and decoder together, we get:Again, in this implementation we need to ensure the hidden dimensions in both the encoder and the decoder are the same.Briefly going over all of the steps:- the `outputs` tensor is created to hold all predictions, $\hat{Y}$- the source sequence, $X$, is fed into the encoder to receive a `context` vector- the initial decoder hidden state is set to be the `context` vector, $s_0 = z = h_T$- we use a batch of `` tokens as the first `input`, $y_1$- we then decode within a loop: - inserting the input token $y_t$, previous hidden state, $s_{t-1}$, and the context vector, $z$, into the decoder - receiving a prediction, $\hat{y}_{t+1}$, and a new hidden state, $s_t$ - we then decide if we are going to teacher force or not, setting the next input as appropriate (either the ground truth next token in the target sequence or the highest predicted next token)
###Code
class Seq2Seq(nn.Module):
def __init__(self, encoder, decoder, device):
super().__init__()
self.encoder = encoder
self.decoder = decoder
self.device = device
assert encoder.hid_dim == decoder.hid_dim, \
"Hidden dimensions of encoder and decoder must be equal!"
def forward(self, src, trg, teacher_forcing_ratio = 0.5):
#src = [src len, batch size]
#trg = [trg len, batch size]
#teacher_forcing_ratio is probability to use teacher forcing
#e.g. if teacher_forcing_ratio is 0.75 we use ground-truth inputs 75% of the time
batch_size = trg.shape[1]
trg_len = trg.shape[0]
trg_vocab_size = self.decoder.output_dim
#tensor to store decoder outputs
outputs = torch.zeros(trg_len, batch_size, trg_vocab_size).to(self.device)
#last hidden state of the encoder is the context
context = self.encoder(src)
#context also used as the initial hidden state of the decoder
hidden = context
#first input to the decoder is the <sos> tokens
input = trg[0,:]
for t in range(1, trg_len):
#insert input token embedding, previous hidden state and the context state
#receive output tensor (predictions) and new hidden state
output, hidden = self.decoder(input, hidden, context)
#place predictions in a tensor holding predictions for each token
outputs[t] = output
#decide if we are going to use teacher forcing or not
teacher_force = random.random() < teacher_forcing_ratio
#get the highest predicted token from our predictions
top1 = output.argmax(1)
#if teacher forcing, use actual next token as next input
#if not, use predicted token
input = trg[t] if teacher_force else top1
return outputs
###Output
_____no_output_____
###Markdown
Training the Seq2Seq ModelThe rest of this session is very similar to the previous one. We initialise our encoder, decoder and seq2seq model (placing it on the GPU if we have one). As before, the embedding dimensions and the amount of dropout used can be different between the encoder and the decoder, but the hidden dimensions must remain the same.
###Code
INPUT_DIM = len(SRC.vocab)
OUTPUT_DIM = len(TRG.vocab)
ENC_EMB_DIM = 256
DEC_EMB_DIM = 256
HID_DIM = 512
ENC_DROPOUT = 0.5
DEC_DROPOUT = 0.5
enc = Encoder(INPUT_DIM, ENC_EMB_DIM, HID_DIM, ENC_DROPOUT)
dec = Decoder(OUTPUT_DIM, DEC_EMB_DIM, HID_DIM, DEC_DROPOUT)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model = Seq2Seq(enc, dec, device).to(device)
###Output
_____no_output_____
###Markdown
Next, we initialize our parameters. The paper states the parameters are initialized from a normal distribution with a mean of 0 and a standard deviation of 0.01, i.e. $\mathcal{N}(0, 0.01)$. It also states we should initialize the recurrent parameters to a special initialization, however to keep things simple we'll also initialize them to $\mathcal{N}(0, 0.01)$.
###Code
def init_weights(m):
for name, param in m.named_parameters():
nn.init.normal_(param.data, mean=0, std=0.01)
model.apply(init_weights)
###Output
_____no_output_____
###Markdown
We print out the number of parameters.Even though we only have a single layer RNN for our encoder and decoder we actually have **more** parameters than the last model. This is due to the increased size of the inputs to the GRU and the linear layer. However, it is not a significant amount of parameters and causes a minimal amount of increase in training time (~3 seconds per epoch extra).
###Code
def count_parameters(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
print(f'The model has {count_parameters(model):,} trainable parameters')
###Output
The model has 24,638,663 trainable parameters
###Markdown
We initiaize our optimizer.
###Code
optimizer = optim.Adam(model.parameters())
###Output
_____no_output_____
###Markdown
We also initialize the loss function, making sure to ignore the loss on `` tokens.
###Code
TRG_PAD_IDX = TRG.vocab.stoi[TRG.pad_token]
criterion = nn.CrossEntropyLoss(ignore_index = TRG_PAD_IDX)
###Output
_____no_output_____
###Markdown
We then create the training loop...
###Code
def train(model, iterator, optimizer, criterion, clip):
model.train()
epoch_loss = 0
for i, batch in enumerate(iterator):
src = batch.src
trg = batch.trg
optimizer.zero_grad()
output = model(src, trg)
#trg = [trg len, batch size]
#output = [trg len, batch size, output dim]
output_dim = output.shape[-1]
output = output[1:].view(-1, output_dim)
trg = trg[1:].view(-1)
#trg = [(trg len - 1) * batch size]
#output = [(trg len - 1) * batch size, output dim]
loss = criterion(output, trg)
loss.backward()
torch.nn.utils.clip_grad_norm_(model.parameters(), clip)
optimizer.step()
epoch_loss += loss.item()
return epoch_loss / len(iterator)
###Output
_____no_output_____
###Markdown
...and the evaluation loop, remembering to set the model to `eval` mode and turn off teaching forcing.
###Code
def evaluate(model, iterator, criterion):
model.eval()
epoch_loss = 0
with torch.no_grad():
for i, batch in enumerate(iterator):
src = batch.src
trg = batch.trg
output = model(src, trg, 0) #turn off teacher forcing
#trg = [trg len, batch size]
#output = [trg len, batch size, output dim]
output_dim = output.shape[-1]
output = output[1:].view(-1, output_dim)
trg = trg[1:].view(-1)
#trg = [(trg len - 1) * batch size]
#output = [(trg len - 1) * batch size, output dim]
loss = criterion(output, trg)
epoch_loss += loss.item()
return epoch_loss / len(iterator)
###Output
_____no_output_____
###Markdown
We'll also define the function that calculates how long an epoch takes.
###Code
def epoch_time(start_time, end_time):
elapsed_time = end_time - start_time
elapsed_mins = int(elapsed_time / 60)
elapsed_secs = int(elapsed_time - (elapsed_mins * 60))
return elapsed_mins, elapsed_secs
###Output
_____no_output_____
###Markdown
Then, we train our model, saving the parameters that give us the best validation loss.
###Code
N_EPOCHS = 10
CLIP = 1
best_valid_loss = float('inf')
for epoch in range(N_EPOCHS):
start_time = time.time()
train_loss = train(model, train_iterator, optimizer, criterion, CLIP)
valid_loss = evaluate(model, test_iterator, criterion)
end_time = time.time()
epoch_mins, epoch_secs = epoch_time(start_time, end_time)
if valid_loss < best_valid_loss:
best_valid_loss = valid_loss
torch.save(model.state_dict(), 'tut2-model.pt')
print(f'Epoch: {epoch+1:02} | Time: {epoch_mins}m {epoch_secs}s')
print(f'\tTrain Loss: {train_loss:.3f} | Train PPL: {math.exp(train_loss):7.3f}')
print(f'\t Val. Loss: {valid_loss:.3f} | Val. PPL: {math.exp(valid_loss):7.3f}')
###Output
Epoch: 01 | Time: 1m 4s
Train Loss: 6.010 | Train PPL: 407.627
Val. Loss: 5.360 | Val. PPL: 212.814
Epoch: 02 | Time: 1m 5s
Train Loss: 5.555 | Train PPL: 258.535
Val. Loss: 5.364 | Val. PPL: 213.547
Epoch: 03 | Time: 1m 5s
Train Loss: 5.425 | Train PPL: 227.036
Val. Loss: 5.355 | Val. PPL: 211.694
Epoch: 04 | Time: 1m 6s
Train Loss: 5.304 | Train PPL: 201.061
Val. Loss: 5.408 | Val. PPL: 223.160
Epoch: 05 | Time: 1m 7s
Train Loss: 5.162 | Train PPL: 174.467
Val. Loss: 5.323 | Val. PPL: 205.019
Epoch: 06 | Time: 1m 6s
Train Loss: 5.028 | Train PPL: 152.612
Val. Loss: 5.317 | Val. PPL: 203.845
Epoch: 07 | Time: 1m 7s
Train Loss: 4.871 | Train PPL: 130.504
Val. Loss: 5.354 | Val. PPL: 211.498
Epoch: 08 | Time: 1m 7s
Train Loss: 4.684 | Train PPL: 108.236
Val. Loss: 5.360 | Val. PPL: 212.657
Epoch: 09 | Time: 1m 7s
Train Loss: 4.406 | Train PPL: 81.967
Val. Loss: 5.458 | Val. PPL: 234.543
Epoch: 10 | Time: 1m 6s
Train Loss: 4.103 | Train PPL: 60.502
Val. Loss: 5.606 | Val. PPL: 271.940
###Markdown
Finally, we test the model on the test set using these "best" parameters.
###Code
model.load_state_dict(torch.load('tut2-model.pt'))
test_loss = evaluate(model, test_iterator, criterion)
print(f'| Test Loss: {test_loss:.3f} | Test PPL: {math.exp(test_loss):7.3f} |')
###Output
| Test Loss: 5.317 | Test PPL: 203.845 |
###Markdown
2 - Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine TranslationIn this second notebook on sequence-to-sequence models using PyTorch and TorchText, we'll be implementing the model from [Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine Translation](https://arxiv.org/abs/1406.1078). This model will achieve improved test perplexity whilst only using a single layer RNN in both the encoder and the decoder. IntroductionLet's remind ourselves of the general encoder-decoder model.We use our encoder (green) over the embedded source sequence (yellow) to create a context vector (red). We then use that context vector with the decoder (blue) and a linear layer (purple) to generate the target sentence.In the previous model, we used an multi-layered LSTM as the encoder and decoder.One downside of the previous model is that the decoder is trying to cram lots of information into the hidden states. Whilst decoding, the hidden state will need to contain information about the whole of the source sequence, as well as all of the tokens have been decoded so far. By alleviating some of this information compression, we can create a better model!We'll also be using a GRU (Gated Recurrent Unit) instead of an LSTM (Long Short-Term Memory). Why? Mainly because that's what they did in the paper (this paper also introduced GRUs) and also because we used LSTMs last time. To understand how GRUs (and LSTMs) differ from standard RNNS, check out [this](https://colah.github.io/posts/2015-08-Understanding-LSTMs/) link. Is a GRU better than an LSTM? [Research](https://arxiv.org/abs/1412.3555) has shown they're pretty much the same, and both are better than standard RNNs. Preparing DataAll of the data preparation will be (almost) the same as last time, so we'll very briefly detail what each code block does. See the previous notebook for a recap.We'll import PyTorch, TorchText, spaCy and a few standard modules.
###Code
import torch
import torch.nn as nn
import torch.optim as optim
from torchtext.datasets import Multi30k
from torchtext.data import Field, BucketIterator, Example, Dataset
import spacy
import numpy as np
import random
import math
import time
###Output
_____no_output_____
###Markdown
Then set a random seed for deterministic results/reproducability.
###Code
SEED = 1234
random.seed(SEED)
np.random.seed(SEED)
torch.manual_seed(SEED)
torch.cuda.manual_seed(SEED)
torch.backends.cudnn.deterministic = True
###Output
_____no_output_____
###Markdown
Instantiate our German and English spaCy models. Previously we reversed the source (German) sentence, however in the paper we are implementing they don't do this, so neither will we. Create our fields to process our data. This will append the "start of sentence" and "end of sentence" tokens as well as converting all words to lowercase.
###Code
SRC = Field(tokenize='spacy',
init_token='<sos>',
eos_token='<eos>',
lower=True)
TRG = Field(tokenize = 'spacy',
init_token='<sos>',
eos_token='<eos>',
lower=True)
###Output
_____no_output_____
###Markdown
Load our data.
###Code
import pandas as pd
#Read and preprocess the data
docs_index = pd.read_csv('https://raw.githubusercontent.com/deepmind/narrativeqa/master/documents.csv')
qapair = pd.read_csv('https://raw.githubusercontent.com/deepmind/narrativeqa/master/qaps.csv')
wiki_summary = pd.read_csv("https://raw.githubusercontent.com/deepmind/narrativeqa/master/third_party/wikipedia/summaries.csv")
wiki_summary.head(2)
docs_index.head()
qapair
docs_index.shape, qapair.shape, wiki_summary.shape
qapair.columns, wiki_summary.columns
mapped_ads = qapair.merge(wiki_summary[['document_id', 'summary']], on='document_id', how='left')
mapped_ads_filtered = mapped_ads[['question', 'answer1', 'summary', 'set']]
mapped_ads_filtered['question'][0]
mapped_ads_filtered['summary'][0][:400]
MAX_LEN = 400
mapped_ads_filtered['summary_question'] = mapped_ads_filtered['summary'].apply(lambda x: x[:MAX_LEN]) + mapped_ads_filtered['question']
mapped_ads_filtered['answer'] = mapped_ads_filtered['answer1']
len(mapped_ads_filtered['summary_question'][0])
mapped_ads_filtered[mapped_ads_filtered['summary'].isna()]
df= mapped_ads_filtered[['summary_question', 'answer', 'set']]
test_df, train_df, valid_df = [group for _, group in df.groupby('set')]
test_df.reset_index(drop=True, inplace=True)
train_df.reset_index(drop=True, inplace=True)
valid_df.reset_index(drop=True, inplace=True)
len(train_df['summary_question'][10])
# This pandas dataframe should be mapped to the list of fields object
# formed the list of fields object
fields = [('src',SRC), ('trg', TRG)]
# map operation to map the rows of pandas dataframe to torchdataset field
train_mapped_rows_to_torchtext = [Example.fromlist([train_df['summary_question'][i], train_df['answer'][i]], fields) for i in range(train_df.shape[0])]
test_mapped_rows_to_torchtext = [Example.fromlist([test_df['summary_question'][i], test_df['answer'][i]], fields) for i in range(test_df.shape[0])]
valid_mapped_rows_to_torchtext = [Example.fromlist([valid_df['summary_question'][i], valid_df['answer'][i]], fields) for i in range(valid_df.shape[0])]
train_data = Dataset(train_mapped_rows_to_torchtext, fields)
valid_data = Dataset(valid_mapped_rows_to_torchtext, fields)
test_data = Dataset(test_mapped_rows_to_torchtext, fields)
###Output
_____no_output_____
###Markdown
We'll also print out an example just to double check they're not reversed.
###Code
print(vars(valid_data.examples[0]))
print(vars(train_data.examples[0]))
###Output
{'src': [' ', 'at', 'madeline', 'hall', ',', 'an', 'old', 'mansion', '-', 'house', 'near', 'southampton', 'belonging', 'to', 'the', 'wealthy', 'de', 'versely', 'family', ',', 'lives', 'an', 'elderly', 'spinster', 'miss', 'delmar', ',', 'the', 'aunt', 'of', 'the', 'earl', 'de', 'versely', 'and', 'captain', 'delmar', '.', 'miss', 'delmar', 'invites', 'arabella', 'mason', ',', 'the', 'daughter', 'of', 'a', 'deceased', ',', 'well', '-', 'liked', 'steward', 'to', 'stay', 'with', 'her', 'as', 'a', 'lower', '-', 'class', 'guest', 'in', 'the', 'house', '.', 'captain', 'delmar', 'is', 'known', 'to', 'visit', 'his', 'aunt', 'at', 'madeline', 'hall', 'frequently', ',', 'who', 'is', 'miss', 'delmer', '?'], 'trg': ['the', 'elderly', 'spinster', 'aunt', 'of', 'the', 'earl', 'de', 'verseley', 'and', 'captain', 'delmar']}
###Markdown
Then create our vocabulary, converting all tokens appearing less than twice into `` tokens.
###Code
SRC.build_vocab(train_data, min_freq = 2)
TRG.build_vocab(train_data, min_freq = 2)
###Output
_____no_output_____
###Markdown
Finally, define the `device` and create our iterators.
###Code
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
BATCH_SIZE = 128
train_iterator, valid_iterator, test_iterator = BucketIterator.splits(
(train_data, valid_data, test_data),
batch_size = BATCH_SIZE,
device = device, sort=False)
print("dataload complete")
###Output
dataload complete
###Markdown
Building the Seq2Seq Model EncoderThe encoder is similar to the previous one, with the multi-layer LSTM swapped for a single-layer GRU. We also don't pass the dropout as an argument to the GRU as that dropout is used between each layer of a multi-layered RNN. As we only have a single layer, PyTorch will display a warning if we try and use pass a dropout value to it.Another thing to note about the GRU is that it only requires and returns a hidden state, there is no cell state like in the LSTM.$$\begin{align*}h_t &= \text{GRU}(e(x_t), h_{t-1})\\(h_t, c_t) &= \text{LSTM}(e(x_t), h_{t-1}, c_{t-1})\\h_t &= \text{RNN}(e(x_t), h_{t-1})\end{align*}$$From the equations above, it looks like the RNN and the GRU are identical. Inside the GRU, however, is a number of *gating mechanisms* that control the information flow in to and out of the hidden state (similar to an LSTM). Again, for more info, check out [this](https://colah.github.io/posts/2015-08-Understanding-LSTMs/) excellent post. The rest of the encoder should be very familar from the last session, it takes in a sequence, $X = \{x_1, x_2, ... , x_T\}$, passes it through the embedding layer, recurrently calculates hidden states, $H = \{h_1, h_2, ..., h_T\}$, and returns a context vector (the final hidden state), $z=h_T$.$$h_t = \text{EncoderGRU}(e(x_t), h_{t-1})$$This is identical to the encoder of the general seq2seq model, with all the "magic" happening inside the GRU (green).
###Code
class Encoder(nn.Module):
def __init__(self, input_dim, emb_dim, hid_dim, dropout):
super().__init__()
self.hid_dim = hid_dim
self.embedding = nn.Embedding(input_dim, emb_dim) #no dropout as only one layer!
self.rnn = nn.GRU(emb_dim, hid_dim)
self.dropout = nn.Dropout(dropout)
def forward(self, src):
#src = [src len, batch size]
embedded = self.dropout(self.embedding(src))
#embedded = [src len, batch size, emb dim]
outputs, hidden = self.rnn(embedded) #no cell state!
#outputs = [src len, batch size, hid dim * n directions]
#hidden = [n layers * n directions, batch size, hid dim]
#outputs are always from the top hidden layer
return hidden
###Output
_____no_output_____
###Markdown
DecoderThe decoder is where the implementation differs significantly from the previous model and we alleviate some of the information compression.Instead of the GRU in the decoder taking just the embedded target token, $d(y_t)$ and the previous hidden state $s_{t-1}$ as inputs, it also takes the context vector $z$. $$s_t = \text{DecoderGRU}(d(y_t), s_{t-1}, z)$$Note how this context vector, $z$, does not have a $t$ subscript, meaning we re-use the same context vector returned by the encoder for every time-step in the decoder. Before, we predicted the next token, $\hat{y}_{t+1}$, with the linear layer, $f$, only using the top-layer decoder hidden state at that time-step, $s_t$, as $\hat{y}_{t+1}=f(s_t^L)$. Now, we also pass the embedding of current token, $d(y_t)$ and the context vector, $z$ to the linear layer.$$\hat{y}_{t+1} = f(d(y_t), s_t, z)$$Thus, our decoder now looks something like this:Note, the initial hidden state, $s_0$, is still the context vector, $z$, so when generating the first token we are actually inputting two identical context vectors into the GRU.How do these two changes reduce the information compression? Well, hypothetically the decoder hidden states, $s_t$, no longer need to contain information about the source sequence as it is always available as an input. Thus, it only needs to contain information about what tokens it has generated so far. The addition of $y_t$ to the linear layer also means this layer can directly see what the token is, without having to get this information from the hidden state. However, this hypothesis is just a hypothesis, it is impossible to determine how the model actually uses the information provided to it (don't listen to anyone that says differently). Nevertheless, it is a solid intuition and the results seem to indicate that this modifications are a good idea!Within the implementation, we will pass $d(y_t)$ and $z$ to the GRU by concatenating them together, so the input dimensions to the GRU are now `emb_dim + hid_dim` (as context vector will be of size `hid_dim`). The linear layer will take $d(y_t), s_t$ and $z$ also by concatenating them together, hence the input dimensions are now `emb_dim + hid_dim*2`. We also don't pass a value of dropout to the GRU as it only uses a single layer.`forward` now takes a `context` argument. Inside of `forward`, we concatenate $y_t$ and $z$ as `emb_con` before feeding to the GRU, and we concatenate $d(y_t)$, $s_t$ and $z$ together as `output` before feeding it through the linear layer to receive our predictions, $\hat{y}_{t+1}$.
###Code
class Decoder(nn.Module):
def __init__(self, output_dim, emb_dim, hid_dim, dropout):
super().__init__()
self.hid_dim = hid_dim
self.output_dim = output_dim
self.embedding = nn.Embedding(output_dim, emb_dim)
self.rnn = nn.GRU(emb_dim + hid_dim, hid_dim)
self.fc_out = nn.Linear(emb_dim + hid_dim * 2, output_dim)
self.dropout = nn.Dropout(dropout)
def forward(self, input, hidden, context):
#input = [batch size]
#hidden = [n layers * n directions, batch size, hid dim]
#context = [n layers * n directions, batch size, hid dim]
#n layers and n directions in the decoder will both always be 1, therefore:
#hidden = [1, batch size, hid dim]
#context = [1, batch size, hid dim]
input = input.unsqueeze(0)
#input = [1, batch size]
embedded = self.dropout(self.embedding(input))
#embedded = [1, batch size, emb dim]
emb_con = torch.cat((embedded, context), dim = 2)
#emb_con = [1, batch size, emb dim + hid dim]
output, hidden = self.rnn(emb_con, hidden)
#output = [seq len, batch size, hid dim * n directions]
#hidden = [n layers * n directions, batch size, hid dim]
#seq len, n layers and n directions will always be 1 in the decoder, therefore:
#output = [1, batch size, hid dim]
#hidden = [1, batch size, hid dim]
output = torch.cat((embedded.squeeze(0), hidden.squeeze(0), context.squeeze(0)),
dim = 1)
#output = [batch size, emb dim + hid dim * 2]
prediction = self.fc_out(output)
#prediction = [batch size, output dim]
return prediction, hidden
###Output
_____no_output_____
###Markdown
Seq2Seq ModelPutting the encoder and decoder together, we get:Again, in this implementation we need to ensure the hidden dimensions in both the encoder and the decoder are the same.Briefly going over all of the steps:- the `outputs` tensor is created to hold all predictions, $\hat{Y}$- the source sequence, $X$, is fed into the encoder to receive a `context` vector- the initial decoder hidden state is set to be the `context` vector, $s_0 = z = h_T$- we use a batch of `` tokens as the first `input`, $y_1$- we then decode within a loop: - inserting the input token $y_t$, previous hidden state, $s_{t-1}$, and the context vector, $z$, into the decoder - receiving a prediction, $\hat{y}_{t+1}$, and a new hidden state, $s_t$ - we then decide if we are going to teacher force or not, setting the next input as appropriate (either the ground truth next token in the target sequence or the highest predicted next token)
###Code
class Seq2Seq(nn.Module):
def __init__(self, encoder, decoder, device):
super().__init__()
self.encoder = encoder
self.decoder = decoder
self.device = device
assert encoder.hid_dim == decoder.hid_dim, \
"Hidden dimensions of encoder and decoder must be equal!"
def forward(self, src, trg, teacher_forcing_ratio = 0.5):
#src = [src len, batch size]
#trg = [trg len, batch size]
#teacher_forcing_ratio is probability to use teacher forcing
#e.g. if teacher_forcing_ratio is 0.75 we use ground-truth inputs 75% of the time
batch_size = trg.shape[1]
trg_len = trg.shape[0]
trg_vocab_size = self.decoder.output_dim
#tensor to store decoder outputs
outputs = torch.zeros(trg_len, batch_size, trg_vocab_size).to(self.device)
#last hidden state of the encoder is the context
context = self.encoder(src)
#context also used as the initial hidden state of the decoder
hidden = context
#first input to the decoder is the <sos> tokens
input = trg[0,:]
for t in range(1, trg_len):
#insert input token embedding, previous hidden state and the context state
#receive output tensor (predictions) and new hidden state
output, hidden = self.decoder(input, hidden, context)
#place predictions in a tensor holding predictions for each token
outputs[t] = output
#decide if we are going to use teacher forcing or not
teacher_force = random.random() < teacher_forcing_ratio
#get the highest predicted token from our predictions
top1 = output.argmax(1)
#if teacher forcing, use actual next token as next input
#if not, use predicted token
input = trg[t] if teacher_force else top1
return outputs
###Output
_____no_output_____
###Markdown
Training the Seq2Seq ModelThe rest of this session is very similar to the previous one. We initialise our encoder, decoder and seq2seq model (placing it on the GPU if we have one). As before, the embedding dimensions and the amount of dropout used can be different between the encoder and the decoder, but the hidden dimensions must remain the same.
###Code
INPUT_DIM = len(SRC.vocab)
OUTPUT_DIM = len(TRG.vocab)
ENC_EMB_DIM = 256
DEC_EMB_DIM = 256
HID_DIM = 512
ENC_DROPOUT = 0.5
DEC_DROPOUT = 0.5
enc = Encoder(INPUT_DIM, ENC_EMB_DIM, HID_DIM, ENC_DROPOUT)
dec = Decoder(OUTPUT_DIM, DEC_EMB_DIM, HID_DIM, DEC_DROPOUT)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model = Seq2Seq(enc, dec, device).to(device)
###Output
_____no_output_____
###Markdown
Next, we initialize our parameters. The paper states the parameters are initialized from a normal distribution with a mean of 0 and a standard deviation of 0.01, i.e. $\mathcal{N}(0, 0.01)$. It also states we should initialize the recurrent parameters to a special initialization, however to keep things simple we'll also initialize them to $\mathcal{N}(0, 0.01)$.
###Code
def init_weights(m):
for name, param in m.named_parameters():
nn.init.normal_(param.data, mean=0, std=0.01)
model.apply(init_weights)
###Output
_____no_output_____
###Markdown
We print out the number of parameters.Even though we only have a single layer RNN for our encoder and decoder we actually have **more** parameters than the last model. This is due to the increased size of the inputs to the GRU and the linear layer. However, it is not a significant amount of parameters and causes a minimal amount of increase in training time (~3 seconds per epoch extra).
###Code
def count_parameters(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
print(f'The model has {count_parameters(model):,} trainable parameters')
###Output
The model has 24,638,663 trainable parameters
###Markdown
We initiaize our optimizer.
###Code
optimizer = optim.Adam(model.parameters())
###Output
_____no_output_____
###Markdown
We also initialize the loss function, making sure to ignore the loss on `` tokens.
###Code
TRG_PAD_IDX = TRG.vocab.stoi[TRG.pad_token]
criterion = nn.CrossEntropyLoss(ignore_index = TRG_PAD_IDX)
###Output
_____no_output_____
###Markdown
We then create the training loop...
###Code
def train(model, iterator, optimizer, criterion, clip):
model.train()
epoch_loss = 0
for i, batch in enumerate(iterator):
src = batch.src
trg = batch.trg
optimizer.zero_grad()
output = model(src, trg)
#trg = [trg len, batch size]
#output = [trg len, batch size, output dim]
output_dim = output.shape[-1]
output = output[1:].view(-1, output_dim)
trg = trg[1:].view(-1)
#trg = [(trg len - 1) * batch size]
#output = [(trg len - 1) * batch size, output dim]
loss = criterion(output, trg)
loss.backward()
torch.nn.utils.clip_grad_norm_(model.parameters(), clip)
optimizer.step()
epoch_loss += loss.item()
return epoch_loss / len(iterator)
###Output
_____no_output_____
###Markdown
...and the evaluation loop, remembering to set the model to `eval` mode and turn off teaching forcing.
###Code
def evaluate(model, iterator, criterion):
model.eval()
epoch_loss = 0
with torch.no_grad():
for i, batch in enumerate(iterator):
src = batch.src
trg = batch.trg
output = model(src, trg, 0) #turn off teacher forcing
#trg = [trg len, batch size]
#output = [trg len, batch size, output dim]
output_dim = output.shape[-1]
output = output[1:].view(-1, output_dim)
trg = trg[1:].view(-1)
#trg = [(trg len - 1) * batch size]
#output = [(trg len - 1) * batch size, output dim]
loss = criterion(output, trg)
epoch_loss += loss.item()
return epoch_loss / len(iterator)
###Output
_____no_output_____
###Markdown
We'll also define the function that calculates how long an epoch takes.
###Code
def epoch_time(start_time, end_time):
elapsed_time = end_time - start_time
elapsed_mins = int(elapsed_time / 60)
elapsed_secs = int(elapsed_time - (elapsed_mins * 60))
return elapsed_mins, elapsed_secs
###Output
_____no_output_____
###Markdown
Then, we train our model, saving the parameters that give us the best validation loss.
###Code
N_EPOCHS = 10
CLIP = 1
best_valid_loss = float('inf')
for epoch in range(N_EPOCHS):
start_time = time.time()
train_loss = train(model, train_iterator, optimizer, criterion, CLIP)
valid_loss = evaluate(model, test_iterator, criterion)
end_time = time.time()
epoch_mins, epoch_secs = epoch_time(start_time, end_time)
if valid_loss < best_valid_loss:
best_valid_loss = valid_loss
torch.save(model.state_dict(), 'tut2-model.pt')
print(f'Epoch: {epoch+1:02} | Time: {epoch_mins}m {epoch_secs}s')
print(f'\tTrain Loss: {train_loss:.3f} | Train PPL: {math.exp(train_loss):7.3f}')
print(f'\t Val. Loss: {valid_loss:.3f} | Val. PPL: {math.exp(valid_loss):7.3f}')
###Output
Epoch: 01 | Time: 1m 4s
Train Loss: 6.010 | Train PPL: 407.627
Val. Loss: 5.360 | Val. PPL: 212.814
Epoch: 02 | Time: 1m 5s
Train Loss: 5.555 | Train PPL: 258.535
Val. Loss: 5.364 | Val. PPL: 213.547
Epoch: 03 | Time: 1m 5s
Train Loss: 5.425 | Train PPL: 227.036
Val. Loss: 5.355 | Val. PPL: 211.694
Epoch: 04 | Time: 1m 6s
Train Loss: 5.304 | Train PPL: 201.061
Val. Loss: 5.408 | Val. PPL: 223.160
Epoch: 05 | Time: 1m 7s
Train Loss: 5.162 | Train PPL: 174.467
Val. Loss: 5.323 | Val. PPL: 205.019
Epoch: 06 | Time: 1m 6s
Train Loss: 5.028 | Train PPL: 152.612
Val. Loss: 5.317 | Val. PPL: 203.845
Epoch: 07 | Time: 1m 7s
Train Loss: 4.871 | Train PPL: 130.504
Val. Loss: 5.354 | Val. PPL: 211.498
Epoch: 08 | Time: 1m 7s
Train Loss: 4.684 | Train PPL: 108.236
Val. Loss: 5.360 | Val. PPL: 212.657
Epoch: 09 | Time: 1m 7s
Train Loss: 4.406 | Train PPL: 81.967
Val. Loss: 5.458 | Val. PPL: 234.543
Epoch: 10 | Time: 1m 6s
Train Loss: 4.103 | Train PPL: 60.502
Val. Loss: 5.606 | Val. PPL: 271.940
###Markdown
Finally, we test the model on the test set using these "best" parameters.
###Code
model.load_state_dict(torch.load('tut2-model.pt'))
test_loss = evaluate(model, test_iterator, criterion)
print(f'| Test Loss: {test_loss:.3f} | Test PPL: {math.exp(test_loss):7.3f} |')
###Output
| Test Loss: 5.317 | Test PPL: 203.845 |
|
urls.ipynb | ###Markdown
Extract already-prepared urls dataData overview [in this py notebook](https://github.com/sisudata/coloring/blob/master/coloring.ipynb)
###Code
%%bash
cd /tmp
test -e coloring || git clone https://github.com/sisudata/coloring
cd coloring
if ! [ -d url_svmlight ] ; then
wget --quiet "http://www.sysnet.ucsd.edu/projects/url/url_svmlight.tar.gz"
tar xzf url_svmlight.tar.gz
fi
if ! [ -f parallelSort.o ] || ! [ -f u4_sort.so ] || ! [ -f u8_sort.so ]; then
./build.sh 2>/dev/null
fi
import os
orig_wd = os.getcwd()
os.chdir('/tmp/coloring')
import numpy as np
import random
np.random.seed(1234)
random.seed(1234)
import utils_graph_coloring as urlutils
Xcontinuous, Xcsr, _, y, nrows, ncols = urlutils.get_all_data()
os.chdir(orig_wd)
from svmlight_loader_install import dump_svmlight_file
ixs = np.arange(nrows, dtype=int)
# malicious urls is temporally correlated, only 1 split to make
cut = int(.7 * nrows)
train_ixs, test_ixs = ixs[:cut], ixs[cut:]
Xcontinuous_train = Xcontinuous[:cut, :]
Xcontinuous_test = Xcontinuous[cut:, :]
Xcsr_train = Xcsr[:cut, :]
Xcsr_test = Xcsr[cut:, :]
y_train = y[:cut]
y_test = y[cut:]
print('urls-data/cont00.svm')
dump_svmlight_file(Xcontinuous_train, y_train, 'urls-data/cont00.svm')
print('urls-data/sps00.svm')
dump_svmlight_file(Xcsr_train, y_train, 'urls-data/sps00.svm')
print('urls-data/cont01.svm')
dump_svmlight_file(Xcontinuous_test, y_test, 'urls-data/cont01.svm')
print('urls-data/sps01.svm')
dump_svmlight_file(Xcsr_test, y_test, 'urls-data/sps01.svm')
Xcsr_train.shape # (1677291, 3381345)
Xcontinuous_train.shape # (1677291, 64)
###Output
_____no_output_____
###Markdown
From [Feature Engineering and Analysis on kaggle.com](https://www.kaggle.com/br0kej/feature-engineering-and-analysis/notebook). Adapted to save the split and feature engineered dataset as csv and to only use xgboost.
###Code
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import re
from tld import get_tld
from typing import Tuple, Union, Any
from sklearn.preprocessing import OrdinalEncoder
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
df = pd.read_csv('data/malicious_phish.csv')
df.head()
df.shape
df.type.value_counts()
sns.countplot(x = 'type', data = df, order = df['type'].value_counts().index)
def is_url_ip_address(url: str) -> bool:
return re.search(
# IPv4
'(([01]?\\d\\d?|2[0-4]\\d|25[0-5])\\.([01]?\\d\\d?|2[0-4]\\d|25[0-5])\\.([01]?\\d\\d?|2[0-4]\\d|25[0-5])\\.'
'([01]?\\d\\d?|2[0-4]\\d|25[0-5])\\/)|'
# IPv4 with port
'(([01]?\\d\\d?|2[0-4]\\d|25[0-5])\\.([01]?\\d\\d?|2[0-4]\\d|25[0-5])\\.([01]?\\d\\d?|2[0-4]\\d|25[0-5])\\.'
'([01]?\\d\\d?|2[0-4]\\d|25[0-5])\\/)|'
# IPv4 in hexadecimal
'((0x[0-9a-fA-F]{1,2})\\.(0x[0-9a-fA-F]{1,2})\\.(0x[0-9a-fA-F]{1,2})\\.(0x[0-9a-fA-F]{1,2})\\/)'
# Ipv6
'(?:[a-fA-F0-9]{1,4}:){7}[a-fA-F0-9]{1,4}|'
'([0-9]+(?:\.[0-9]+){3}:[0-9]+)|'
'((?:(?:\d|[01]?\d\d|2[0-4]\d|25[0-5])\.){3}(?:25[0-5]|2[0-4]\d|[01]?\d\d|\d)(?:\/\d{1,2})?)', url) is not None
df['is_ip'] = df['url'].apply(lambda i: is_url_ip_address(i))
df['is_ip'].value_counts()
def process_tld(url: str, fix_protos: bool = False) -> Tuple[str, str, str, str]:
"""
Takes a URL string and uses the tld library to extract subdomain, domain, top
level domain and full length domain
"""
res = get_tld(url, as_object=True, fail_silently=False,
fix_protocol=fix_protos)
subdomain = res.subdomain
domain = res.domain
tld = res.tld
fld = res.fld
return subdomain, domain, tld, fld
def process_url_with_tld(row: pd.Series) -> Tuple[str, str, str, str]:
"""
Takes in a dataframe row, checks to see if rows `is_ip` column is
False. If it is false, continues to process the URL and extract the
features, otherwise sets four features to None before returning.
This processing is wrapped in a try/except block to enable debugging
and it prints out the inputs that caused a failure as well as a
failure counter.
"""
try:
if not row['is_ip']:
if str(row['url']).startswith('http:'):
return process_tld(row['url'])
else:
return process_tld(row['url'], fix_protos=True)
else:
subdomain = None
domain = None
tld = None
fld = None
return subdomain, domain, tld, fld
except:
idx = row.name
url = row['url']
type = row['type']
print(f'Failed - {idx}: {url} is a {type} example')
return None, None, None, None
df[['subdomain', 'domain', 'tld', 'fld']] = df.apply(lambda x: process_url_with_tld(x), axis=1, result_type="expand")
df[['subdomain', 'domain', 'tld', 'fld']].value_counts()
df[['subdomain', 'domain', 'tld', 'fld']].isna().sum(), df['is_ip'].value_counts()
df[['subdomain', 'domain', 'tld', 'fld']].isna().sum()[0] - \
df['is_ip'].value_counts()[1]
df.head()
def get_url_path(url: str) -> Union[str, None]:
"""
Get's the path from a URL
For example:
If the URL was "www.google.co.uk/my/great/path"
The path returned would be "my/great/path"
"""
try:
res = get_tld(url, as_object=True,
fail_silently=False, fix_protocol=True)
if res.parsed_url.query:
joined = res.parsed_url.path + res.parsed_url.query
return joined
else:
return res.parsed_url.path
except:
return None
def alpha_count(url: str) -> int:
"""
Counts the number of alpha characters in a URL
"""
alpha = 0
for i in url:
if i.isalpha():
alpha += 1
return alpha
def digit_count(url: str) -> int:
"""
Counts the number of digit characters in a URL
"""
digits = 0
for i in url:
if i.isnumeric():
digits += 1
return digits
def count_dir_in_url_path(url_path: Union[str, None]) -> int:
"""
Counts number of / in url path to count number of
sub directories
"""
if url_path:
n_dirs = url_path.count('/')
return n_dirs
else:
return 0
def get_first_dir_len(url_path: Union[str, None]) -> int:
"""
Counts the length of the first directory within
the URL provided
"""
if url_path:
if len(url_path.split('/')) > 1:
first_dir_len = len(url_path.split('/')[1])
return first_dir_len
else:
return 0
def contains_shortening_service(url: str) -> int:
"""
Checks to see whether URL contains a shortening service
"""
return re.search('bit\.ly|goo\.gl|shorte\.st|go2l\.ink|x\.co|ow\.ly|t\.co|tinyurl|tr\.im|is\.gd|cli\.gs|'
'yfrog\.com|migre\.me|ff\.im|tiny\.cc|url4\.eu|twit\.ac|su\.pr|twurl\.nl|snipurl\.com|'
'short\.to|BudURL\.com|ping\.fm|post\.ly|Just\.as|bkite\.com|snipr\.com|fic\.kr|loopt\.us|'
'doiop\.com|short\.ie|kl\.am|wp\.me|rubyurl\.com|om\.ly|to\.ly|bit\.do|t\.co|lnkd\.in|'
'db\.tt|qr\.ae|adf\.ly|goo\.gl|bitly\.com|cur\.lv|tinyurl\.com|ow\.ly|bit\.ly|ity\.im|'
'q\.gs|is\.gd|po\.st|bc\.vc|twitthis\.com|u\.to|j\.mp|buzurl\.com|cutt\.us|u\.bb|yourls\.org|'
'x\.co|prettylinkpro\.com|scrnch\.me|filoops\.info|vzturl\.com|qr\.net|1url\.com|tweez\.me|v\.gd|'
'tr\.im|link\.zip\.net',
url) is not None
# General Features
df['url_path'] = df['url'].apply(lambda x: get_url_path(x))
df['contains_shortener'] = df['url'].apply(
lambda x: contains_shortening_service(x))
# URL component length
df['url_len'] = df['url'].apply(lambda x: len(str(x)))
df['subdomain_len'] = df['subdomain'].apply(lambda x: len(str(x)))
df['tld_len'] = df['tld'].apply(lambda x: len(str(x)))
df['fld_len'] = df['fld'].apply(lambda x: len(str(x)))
df['url_path_len'] = df['url_path'].apply(lambda x: len(str(x)))
# Simple count features
df['url_alphas'] = df['url'].apply(lambda i: alpha_count(i))
df['url_digits'] = df['url'].apply(lambda i: digit_count(i))
df['url_puncs'] = (df['url_len'] - (df['url_alphas'] + df['url_digits']))
df['count.'] = df['url'].apply(lambda x: x.count('.'))
df['count@'] = df['url'].apply(lambda x: x.count('@'))
df['count-'] = df['url'].apply(lambda x: x.count('-'))
df['count%'] = df['url'].apply(lambda x: x.count('%'))
df['count?'] = df['url'].apply(lambda x: x.count('?'))
df['count='] = df['url'].apply(lambda x: x.count('='))
df['count_dirs'] = df['url_path'].apply(lambda x: count_dir_in_url_path(x))
df['first_dir_len'] = df['url_path'].apply(lambda x: get_first_dir_len(x))
# Binary Label
df['binary_label'] = df['type'].apply(lambda x: 0 if x == 'benign' else 1)
# Binned Features
groups = ['Short', 'Medium', 'Long', 'Very Long']
# URL Lengths in 4 bins
df['url_len_q'] = pd.qcut(df['url_len'], q=4, labels=groups)
# FLD Lengths in 4 bins
df['fld_len_q'] = pd.qcut(df['fld_len'], q=4, labels=groups)
# Percentage Features
df['pc_alphas'] = df['url_alphas'] / df['url_len']
df['pc_digits'] = df['url_digits'] / df['url_len']
df['pc_puncs'] = df['url_puncs'] / df['url_len']
df[["url_len_q", "fld_len_q"]] = OrdinalEncoder().fit_transform(
df[["url_len_q", "fld_len_q"]])
df.head()
df.describe()
fig, axes = plt.subplots(2, 2, figsize=(20, 10), sharey=False)
sns.boxplot(ax=axes[0, 0], x='type', y='url_len', data=df)
axes[0, 0].set_title('URL Length')
sns.boxplot(ax=axes[0, 1], x='type', y='subdomain_len', data=df)
axes[0, 1].set_title('Subdomain Length')
sns.boxplot(ax=axes[1, 0], x='type', y='tld_len', data=df)
axes[1, 0].set_title('Top-Level Domain Length')
sns.boxplot(ax=axes[1, 1], x='type', y='fld_len', data=df)
axes[1, 1].set_title('Full Length Domain Length')
fig, axes = plt.subplots(1, 3, figsize=(20, 5), sharey=True)
sns.boxplot(ax=axes[0], x='type', y='pc_alphas', data=df)
axes[0].set_title('Ratio of Alpha Characeters to URL')
sns.boxplot(ax=axes[1], x='type', y='pc_puncs', data=df)
axes[1].set_title('Ratio of Puncutation to URL')
sns.boxplot(ax=axes[2], x='type', y='pc_digits', data=df)
axes[2].set_title('Ratio of Digits to URL')
fig, axes = plt.subplots(1, 3, figsize=(20, 5), sharey=True)
fig.suptitle('% of Character Type in URL - with length quartiles')
sns.boxplot(ax=axes[0], x='type', y='pc_alphas', hue='url_len_q', data=df)
axes[0].set_title('Ratio of Alpha Characeters to URL')
sns.boxplot(ax=axes[1], x='type', y='pc_puncs', hue='url_len_q', data=df)
axes[1].set_title('Ratio of Puncutation to URL')
sns.boxplot(ax=axes[2], x='type', y='pc_digits', hue='url_len_q', data=df)
axes[2].set_title('Ratio of Digits to URL')
fig, axes = plt.subplots(3, 1, figsize=(20, 15), sharey=True)
sns.kdeplot(ax=axes[0], data=df, x="pc_alphas", hue='type')
sns.kdeplot(ax=axes[1], data=df, x="pc_puncs", hue='type')
sns.kdeplot(ax=axes[2], data=df, x="pc_digits", hue='type')
X = df[['is_ip', 'url_len', 'subdomain_len', 'tld_len', 'fld_len', 'url_path_len',
'url_alphas', 'url_digits', 'url_puncs', 'count.', 'count@', 'count-',
'count%', 'count?', 'count=',
'pc_alphas', 'pc_digits', 'pc_puncs', 'count_dirs',
'contains_shortener', 'first_dir_len',
'url_len_q', 'fld_len_q']]
y = df['binary_label']
X.isna().sum()
X[X.isnull().any(axis=1)]
df[df['first_dir_len'].isnull()]
X['first_dir_len'].value_counts()
X = X.fillna(0)
X.isna().sum()
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, shuffle=True, random_state=5)
def make_binary(X):
df = X
df['is_ip'] = df['is_ip'] \
.apply(lambda x: 1 if x else 0)
df['contains_shortener'] = df['contains_shortener'] \
.apply(lambda x: 1 if x else 0)
return df
make_binary(X_train).to_csv(
'data/malicious_phish_X_train.csv', index=False)
make_binary(X_test).to_csv('data/malicious_phish_X_test.csv', index=False)
y_train.to_csv('data/malicious_phish_y_train.csv', index=False)
y_test.to_csv('data/malicious_phish_y_test.csv', index=False)
from xgboost import XGBClassifier
model = XGBClassifier(n_estimators=100, use_label_encoder=False)
model.fit(X_train, y_train)
print(model)
y_pred = model.predict(X_test)
print(classification_report(y_test, y_pred))
score = accuracy_score(y_test, y_pred)
print("accuracy: %0.3f" % score)
sns.heatmap(confusion_matrix(y_test, y_pred),
annot=True, fmt='g', cmap='Blues')
xgb_features = model.feature_importances_.tolist()
cols = X.columns
feature_importances = pd.DataFrame({'features': cols, 'xgb': xgb_features})
feature_importances['mean_importance'] = feature_importances.mean(axis=1)
feature_importances = feature_importances.sort_values(
by='mean_importance', ascending=False)
feature_importances
fig, axes = plt.subplots(1, figsize=(20, 10))
sns.barplot(data=feature_importances, x="mean_importance", y='features', ax=axes)
###Output
_____no_output_____ |
Machine Learning/tasks/1 term/HW_2_Texts/homework2_texts.ipynb | ###Markdown
Homework 2. Simple text processing.
###Code
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
###Output
_____no_output_____
###Markdown
Prohibited Comment ClassificationThis part of assigment is fully based on YSDA NLP_course homework. Special thanks to YSDA team for making it available on github.__In this part__ you will build an algorithm that classifies social media comments into normal or toxic.Like in many real-world cases, you only have a small (10^3) dataset of hand-labeled examples to work with. We'll tackle this problem using both classical nlp methods and embedding-based approach.
###Code
# In colab uncomment this cell
# ! wget https://raw.githubusercontent.com/ml-mipt/ml-mipt/basic/homeworks/homework2_texts/comments.tsv
import pandas as pd
data = pd.read_csv("comments.tsv", sep='\t')
texts = data['comment_text'].values
target = data['should_ban'].values
data[50::200]
from sklearn.model_selection import train_test_split
texts_train, texts_test, y_train, y_test = train_test_split(texts, target, test_size=0.5, random_state=42)
###Output
_____no_output_____
###Markdown
__Note:__ it is generally a good idea to split data into train/test before anything is done to them.It guards you against possible data leakage in the preprocessing stage. For example, should you decide to select words present in obscene tweets as features, you should only count those words over the training set. Otherwise your algoritm can cheat evaluation. Preprocessing and tokenizationComments contain raw text with punctuation, upper/lowercase letters and even newline symbols.To simplify all further steps, we'll split text into space-separated tokens using one of nltk tokenizers.Generally, library `nltk` [link](https://www.nltk.org) is widely used in NLP. It is not necessary in here, but mentioned to intoduce it to you.
###Code
from nltk.tokenize import TweetTokenizer
tokenizer = TweetTokenizer()
preprocess = lambda text: ' '.join(tokenizer.tokenize(text.lower()))
text = 'How to be a grown-up at work: replace "I don\'t want to do that" with "Ok, great!".'
print("before:", text,)
print("after:", preprocess(text),)
# task: preprocess each comment in train and test
texts_train = pd.DataFrame(texts_train)[0].apply(preprocess).values
texts_test = pd.DataFrame(texts_test)[0].apply(preprocess).values
# Small check that everything is done properly
assert texts_train[5] == 'who cares anymore . they attack with impunity .'
assert texts_test[89] == 'hey todds ! quick q ? why are you so gay'
assert len(texts_test) == len(y_test)
###Output
_____no_output_____
###Markdown
Solving it: bag of wordsOne traditional approach to such problem is to use bag of words features:1. build a vocabulary of frequent words (use train data only)2. for each training sample, count the number of times a word occurs in it (for each word in vocabulary).3. consider this count a feature for some classifier__Note:__ in practice, you can compute such features using sklearn. __Please don't do that in the current assignment, though.__* `from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer`
###Code
import re
import operator
all_words_train = ' '.join(texts_train).split()
word_frequency = {word: all_words_train.count(word) for word in set(all_words_train)}
word_frequency_sorted = dict(sorted(word_frequency.items(), key = operator.itemgetter(1), reverse = True))
# task: find up to k most frequent tokens in texts_train,
# sort them by number of occurences (highest first)
k = min(10000, len(set(' '.join(texts_train).split())))
bow_vocabulary = list(word_frequency_sorted)[:k]
print('example features:', sorted(bow_vocabulary)[::100])
def text_to_bow(text):
""" convert text string to an array of token counts. Use bow_vocabulary. """
word_freq_text = [text.split().count(word) for word in bow_vocabulary]
return np.array(word_freq_text, 'float32')
X_train_bow = np.stack(list(map(text_to_bow, texts_train)))
X_test_bow = np.stack(list(map(text_to_bow, texts_test)))
# Small check that everything is done properly
k_max = len(set(' '.join(texts_train).split()))
assert X_train_bow.shape == (len(texts_train), min(k, k_max))
assert X_test_bow.shape == (len(texts_test), min(k, k_max))
assert np.all(X_train_bow[5:10].sum(-1) == np.array([len(s.split()) for s in texts_train[5:10]]))
assert len(bow_vocabulary) <= min(k, k_max)
assert X_train_bow[6, bow_vocabulary.index('.')] == texts_train[6].split().count('.')
###Output
_____no_output_____
###Markdown
Machine learning stuff: fit, predict, evaluate. You know the drill.
###Code
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV
bow_model = LogisticRegression(solver = 'liblinear', max_iter=1000).fit(X_train_bow, y_train)
from sklearn.metrics import roc_auc_score, roc_curve
def plotting(X_train_bow, X_test_bow, y_train, y_test, bow_model):
for name, X, y, model in [
('train', X_train_bow, y_train, bow_model),
('test ', X_test_bow, y_test, bow_model)
]:
proba = model.predict_proba(X)[:, 1]
auc = roc_auc_score(y, proba)
plt.plot(*roc_curve(y, proba)[:2], label='%s AUC=%.4f' % (name, auc))
plt.plot([0, 1], [0, 1], '--', color='black',)
plt.legend(fontsize='large')
plt.grid()
plotting(X_train_bow, X_test_bow, y_train, y_test, bow_model)
###Output
_____no_output_____
###Markdown
Try to vary the number of tokens `k` and check how the model performance changes. Show it on a plot.
###Code
import tqdm
aucs = []
tokens = np.arange(4500, 6000, 100)
for token in tqdm.tqdm_notebook(tokens):
bow_vocabulary = list(word_frequency_sorted)[:token]
X_train_bow = np.stack(list(map(text_to_bow, texts_train)))
X_test_bow = np.stack(list(map(text_to_bow, texts_test)))
bow_model = LogisticRegression(solver = 'liblinear', max_iter=1000).fit(X_train_bow, y_train)
proba = bow_model.predict_proba(X_test_bow)[:, 1]
aucs.append(roc_auc_score(y_test, proba))
plt.plot(tokens, aucs, label = 'Test')
plt.xlabel('Number of tokens')
plt.ylabel('AUC Score')
plt.title('AUC')
plt.legend()
plt.grid()
###Output
_____no_output_____
###Markdown
Task: implement TF-IDF featuresNot all words are equally useful. One can prioritize rare words and downscale words like "and"/"or" by using __tf-idf features__. This abbreviation stands for __text frequency/inverse document frequence__ and means exactly that:$$ feature_i = { Count(word_i \in x) \times { log {N \over Count(word_i \in D) + \alpha} }}, $$where x is a single text, D is your dataset (a collection of texts), N is a total number of documents and $\alpha$ is a smoothing hyperparameter (typically 1). And $Count(word_i \in D)$ is the number of documents where $word_i$ appears.It may also be a good idea to normalize each data sample after computing tf-idf features.__Your task:__ implement tf-idf features, train a model and evaluate ROC curve. Compare it with basic BagOfWords model from above.__Please don't use sklearn/nltk builtin tf-idf vectorizers in your solution :)__ You can still use 'em for debugging though. Blog post about implementing the TF-IDF features from scratch: https://triton.ml/blog/tf-idf-from-scratch
###Code
# Your beautiful code here
alpha = 0.95
def idf(word):
freq = sum(1 for text in texts_train if word in text.split())
return np.log(len(texts_train) / (freq + alpha))
def tf_idf(text):
features = [text.split().count(word) * idf(word) if word in text.split() else 0 for word in bow_vocabulary]
return np.array(features)
X_train_feat = np.stack(list(map(tf_idf, texts_train)))
X_test_feat = np.stack(list(map(tf_idf, texts_test)))
feat_model = LogisticRegression(solver = 'liblinear', max_iter=1000).fit(X_train_feat, y_train)
plotting(X_train_feat, X_test_feat, y_train, y_test, feat_model)
###Output
_____no_output_____ |
winner.ipynb | ###Markdown
for i in range(len(y)): if y[i]==X[i][0]: y[i]=0 else: y[i]=1 imbalanced dataset so it is going to print(np.unique(y, return_counts=True))
###Code
zeros = 0
for i in range(len(X)):
if y[i] == X[i][0]:
if zeros <= 250:
y[i] = 0
zeros = zeros + 1
else:
y[i] = 1
t = X[i][0]
X[i][0] = X[i][1]
X[i][1] = t
else:
y[i] = 1
for i in range(len(X)):
if X[i][3]==X[i][0]:
X[i][3]=0
else:
X[i][3]=1
X = np.array(X , dtype='int32')
y = np.array(y , dtype='int32')
y = y.ravel()
print(np.unique(y, return_counts=True))
# now balanced dataset
from sklearn.model_selection import train_test_split
X_train, X_test, y_train,y_test = train_test_split(X, y, test_size=0.2 , random_state=0)
alg1 = LogisticRegression(solver='liblinear')
start = time.time()
alg1.fit(X_train , y_train)
end = time.time()
total_time1 = end - start
y_pred1 = alg1.predict(X_test)
print('accuracy : ', alg1.score(X_test , y_test))
print('time : ' , total_time1)
print(classification_report(y_test , y_pred1))
print(confusion_matrix(y_test , y_pred1))
alg2 = RandomForestClassifier(n_estimators=60)
start = time.time()
alg2.fit(X_train , y_train)
end = time.time()
total_time2 = end - start
y_pred2 = alg2.predict(X_test)
print('accuracy : ', alg2.score(X_test , y_test))
print('time : ' , total_time2)
print(classification_report(y_test , y_pred2))
print(confusion_matrix(y_test , y_pred2))
alg3 = DecisionTreeClassifier(max_depth=1 , criterion='gini')
start = time.time()
alg3.fit(X_train , y_train)
end = time.time()
total_time3 = end - start
y_pred3 = alg3.predict(X_test)
print('accuracy : ', alg3.score(X_test , y_test))
print('time : ' , total_time3)
print(classification_report(y_test , y_pred3))
print(confusion_matrix(y_test , y_pred3))
# Printing tree alongwith class names
dot_data = StringIO()
export_graphviz(alg3, out_file=dot_data, filled=True, rounded=True, special_characters=True, feature_names = ['team1', 'team2', 'venue','toss_winner', 'toss_decision'])
graph = pydotplus.graph_from_dot_data(dot_data.getvalue())
Image(graph.create_png())
dot_data = export_graphviz(alg3, out_file=None,
feature_names=['team1', 'team2', 'venue', 'toss_winner', 'toss_decision'])
graph = pydotplus.graph_from_dot_data(dot_data)
graph.write_pdf("ipl_winner_decision_tree.pdf")
alg4 = BernoulliNB()
start = time.time()
alg4.fit(X_train,y_train)
end = time.time()
total_time4 = end - start
y_pred4 = alg4.predict(X_test)
print('accuracy : ', alg4.score(X_test , y_test))
print('time : ' , total_time4)
print(classification_report(y_test , y_pred4))
print(confusion_matrix(y_test , y_pred4))
alg5 = GaussianNB()
start = time.time()
alg5.fit(X_train,y_train)
end = time.time()
total_time5 = end - start
y_pred5 = alg5.predict(X_test)
print('accuracy : ', alg5.score(X_test , y_test))
print('time : ' , total_time5)
print(classification_report(y_test , y_pred5))
print(confusion_matrix(y_test , y_pred5))
alg6 = MultinomialNB()
start = time.time()
alg6.fit(X_train,y_train)
end = time.time()
total_time6 = end - start
y_pred6 = alg6.predict(X_test)
print('accuracy : ', alg6.score(X_test , y_test))
print('time : ' , total_time6)
print(classification_report(y_test , y_pred6))
print(confusion_matrix(y_test , y_pred6))
x_axis = []
y_axis = []
for k in range(1, 26, 2):
clf = KNeighborsClassifier(n_neighbors = k)
score = cross_val_score(clf, X_train, y_train, cv = KFold(n_splits=5, shuffle=True, random_state=0))
x_axis.append(k)
y_axis.append(score.mean())
import matplotlib.pyplot as plt
plt.plot(x_axis, y_axis)
plt.xlabel("k")
plt.ylabel("cross_val_score")
plt.title("variation of score on different values of k")
plt.show()
alg7 = KNeighborsClassifier(n_neighbors=19, weights='distance', algorithm='auto', p=2, metric='minkowski')
start = time.time()
alg7.fit(X_train, y_train)
end = time.time()
total_time7 = end - start
y_pred7 = alg7.predict(X_test)
print('accuracy : ', alg7.score(X_test , y_test))
print('time : ' , total_time7)
print(classification_report(y_test , y_pred7))
print(confusion_matrix(y_test , y_pred7))
clf = SVC(kernel='rbf')
grid = {'C': [1e2,1e3, 5e3, 1e4, 5e4, 1e5],
'gamma': [1e-3, 5e-4, 1e-4, 5e-3]}
alg8 = GridSearchCV(clf, grid)
start = time.time()
alg8.fit(X_train, y_train)
end = time.time()
total_time8 = end - start
y_pred8 = alg8.predict(X_test)
print(alg8.best_estimator_)
print('accuracy : ', alg8.score(X_test , y_test))
print('time : ' , total_time8)
print(classification_report(y_test , y_pred8))
print(confusion_matrix(y_test , y_pred8))
alg9 = LinearSVC(multi_class='crammer_singer')
start = time.time()
alg9.fit(X_train, y_train)
end = time.time()
total_time9 = end - start
y_pred9 = alg9.predict(X_test)
print('accuracy : ', alg9.score(X_test , y_test))
print('time : ' , total_time9)
print(classification_report(y_test , y_pred9))
print(confusion_matrix(y_test , y_pred9))
ridge = RidgeClassifier()
parameters={'alpha':[1e-15,1e-10,1e-8,1e-3,1e-2,1,5,10,20,30,35,40]}
alg10=GridSearchCV(ridge,parameters)
start = time.time()
alg10.fit(X_train, y_train)
end = time.time()
total_time10 = end - start
y_pred10 = alg10.predict(X_test)
print('accuracy : ', alg10.score(X_test , y_test))
print('time : ' , total_time10)
print(classification_report(y_test , y_pred10))
print(confusion_matrix(y_test , y_pred10))
test = np.array([2, 4, 1, 1, 1]).reshape(1,-1)
print('alg1 : ' , alg1.predict(test))
print('alg2 : ' , alg2.predict(test))
print('alg3 : ' , alg3.predict(test))
print('alg4 : ' , alg4.predict(test))
print('alg5 : ' , alg5.predict(test))
print('alg6 : ' , alg6.predict(test))
print('alg7 : ' , alg7.predict(test))
print('alg8 : ' , alg8.predict(test))
print('alg9 : ' , alg9.predict(test))
print('alg10 :' , alg10.predict(test))
test = np.array([4, 2, 1, 0, 1]).reshape(1,-1)
print('alg1 : ' , alg1.predict(test))
print('alg2 : ' , alg2.predict(test))
print('alg3 : ' , alg3.predict(test))
print('alg4 : ' , alg4.predict(test))
print('alg5 : ' , alg5.predict(test))
print('alg6 : ' , alg6.predict(test))
print('alg7 : ' , alg7.predict(test))
print('alg8 : ' , alg8.predict(test))
print('alg9 : ' , alg9.predict(test))
print('alg10 :' , alg10.predict(test))
df_model=pd.DataFrame({
'Model_Applied':['Logistic_Regression', 'Random_Forest', 'Decision_tree', 'BernoulliNB', 'GausianNB', 'MultinomialNB', 'KNN', 'SVC', 'Linear_SVC', 'Ridge_Classifier'],
'Accuracy':[alg1.score(X_test,y_test), alg2.score(X_test,y_test), alg3.score(X_test,y_test), alg4.score(X_test,y_test),
alg5.score(X_test,y_test), alg6.score(X_test,y_test), alg7.score(X_test,y_test), alg8.score(X_test,y_test),
alg9.score(X_test,y_test), alg10.score(X_test,y_test)],
'Training_Time':[total_time1, total_time2, total_time3, total_time4, total_time5, total_time6, total_time7, total_time8,
total_time9, total_time10]})
df_model
df_model.plot(kind='bar',x='Model_Applied', ylim=[0,1] , y='Accuracy', figsize=(10,10) , ylabel='Accuracy', title='Accurcy comparison of different Models')
df_model.plot(kind='bar',x='Model_Applied', ylim=[0,0.14] , y='Training_Time', figsize=(10,10), ylabel='Training Time', title='Training time comparison of different Models')
import pickle as pkl
with open('winner.pkl', 'wb') as f:
pkl.dump(alg3, f)
with open('winner.pkl', 'rb') as f:
model = pkl.load(f)
model.predict(test)
###Output
_____no_output_____ |
pytorch3d_render.ipynb | ###Markdown
Attempt 2 (Using PyTorch3D)
###Code
import sys
import torch
pyt_version_str=torch.__version__.split("+")[0].replace(".", "")
# print(pyt_version_str)
version_str="".join([
f"py3{sys.version_info.minor}_cu",
torch.version.cuda.replace(".",""),
f"_pyt{pyt_version_str}"
])
!pip install --no-index --no-cache-dir pytorch3d -f https://dl.fbaipublicfiles.com/pytorch3d/packaging/wheels/{version_str}/download.html
import pytorch3d as p3d
print(p3d.__version__)
!pip uninstall pytorch3d
import os
import torch
import numpy as np
from tqdm.notebook import tqdm
import imageio
import torch.nn as nn
import torch.nn.functional as F
import matplotlib.pyplot as plt
from skimage import img_as_ubyte
# io utils
from pytorch3d.io import load_obj,load_objs_as_meshes
# datastructures
from pytorch3d.structures import Meshes
# 3D transformations functions
from pytorch3d.transforms import Rotate, Translate
# rendering components
from pytorch3d.renderer import (
FoVPerspectiveCameras, look_at_view_transform, look_at_rotation,
RasterizationSettings, MeshRenderer, MeshRasterizer, BlendParams,
SoftSilhouetteShader, HardPhongShader, PointLights, TexturesVertex,
)
# verts_l, faces_l, aux_l = load_obj(f'{data_path}/{fname}/{fname}_lower.obj')
# verts_u, faces_u, aux_u = load_obj(f'{data_path}/{fname}/{fname}_upper.obj')
# verts_u, faces_u, _= load_obj('/content/016KWDMV_upper.obj')
!wget https://dl.fbaipublicfiles.com/pytorch3d/data/teapot/teapot.obj
# verts_u, faces_u, _ = load_obj("/content/teapot.obj")
verts_u, faces_u, _= load_obj('/content/016KWDMV_upper.obj')
print(verts_u.shape)
print(faces_u.textures_idx.shape)
# print(aux_u)
# !rm -rf /content/data
# device = torch.device("cuda:0")
# verts_u, faces_u, _= load_obj('/content/016KWDMV_upper.obj')
faces = faces_u.verts_idx
# Initialize each vertex to be white in color.
verts_rgb = torch.ones_like(verts_u)[None] # (1, V, 3)
textures = TexturesVertex(verts_features=verts_rgb.to('cuda'))
# Create a Meshes object for the teapot. Here we have only one mesh in the batch.
mesh = Meshes(
verts=[verts_u.to('cuda')],
faces=[faces.to('cuda')],
textures=textures
)
print(verts_u)
print(verts_rgb)
# plt.figure(figsize=(7,7))
# print(mesh.textures)
# # texturesuv_image_matplotlib(mesh.textures, subsample=None)
# plt.axis("off");
# Initialize a perspective camera.
cameras = FoVPerspectiveCameras(device='cuda')
# To blend the 100 faces we set a few parameters which control the opacity and the sharpness of
# edges. Refer to blending.py for more details.
blend_params = BlendParams(sigma=1e-4, gamma=1e-4)
# Define the settings for rasterization and shading. Here we set the output image to be of size
# 256x256. To form the blended image we use 100 faces for each pixel. We also set bin_size and max_faces_per_bin to None which ensure that
# the faster coarse-to-fine rasterization method is used. Refer to rasterize_meshes.py for
# explanations of these parameters. Refer to docs/notes/renderer.md for an explanation of
# the difference between naive and coarse-to-fine rasterization.
raster_settings = RasterizationSettings(
image_size=256,
blur_radius=np.log(1. / 1e-4 - 1.) * blend_params.sigma,
faces_per_pixel=100,
)
# Create a silhouette mesh renderer by composing a rasterizer and a shader.
silhouette_renderer = MeshRenderer(
rasterizer=MeshRasterizer(
cameras=cameras,
raster_settings=raster_settings
),
shader=SoftSilhouetteShader(blend_params=blend_params)
)
# We will also create a Phong renderer. This is simpler and only needs to render one face per pixel.
raster_settings = RasterizationSettings(
image_size=256,
blur_radius=0.0,
faces_per_pixel=1,
)
# We can add a point light in front of the object.
lights = PointLights(device='cuda', location=((2.0, 2.0, -2.0),))
phong_renderer = MeshRenderer(
rasterizer=MeshRasterizer(
cameras=cameras,
raster_settings=raster_settings
),
shader=HardPhongShader(device='cuda', cameras=cameras, lights=lights)
)
# Select the viewpoint using spherical angles
distance = 10 # distance from camera to the object
elevation = 0.0 # angle of elevation in degrees
azimuth = 0.0 # No rotation so the camera is positioned on the +Z axis.
# Get the position of the camera based on the spherical angles
R, T = look_at_view_transform(distance, elevation, azimuth, device='cuda')
# Render the teapot providing the values of R and T.
silhouette = silhouette_renderer(meshes_world=mesh, R=R, T=T)
image_ref = phong_renderer(meshes_world=mesh, R=R, T=T)
silhouette = silhouette.cpu().numpy()
image_ref = image_ref.cpu().numpy()
plt.figure(figsize=(16, 16))
plt.subplot(1, 2, 1)
plt.imshow(silhouette.squeeze()[..., 3]) # only plot the alpha channel of the RGBA image
plt.grid(False)
plt.subplot(1, 2, 2)
plt.imshow(image_ref.squeeze())
plt.grid(False)
###Output
_____no_output_____ |
noteboooks/dispersion.ipynb | ###Markdown
FODO dispersion
###Code
cirumference = 1000 # meters
proton_energy = 15 # GeV
dipole_length = 5 # meters
dipole_B_max = 2 # T
n_cells = 8 # ??
dipole_angle = np.pi / 16 # ??
quad_length = 3
quad_strength = 8.89e-3 / quad_length
dipole_length = 5
dipole_angle = np.pi / 16
dipole_bending_radius = dipole_length / dipole_angle
# reduce the drift lengths to compensate for the now thick elements
drift_length = (cirumference / n_cells - (2 * quad_length) - (4 * dipole_length)) / 6
half_quad_f = Quadrupole(quad_strength, quad_length/2, name="quad_f")
quad_d = Quadrupole(-quad_strength, quad_length, name="quad_d")
dipole = Dipole(dipole_bending_radius, dipole_angle)
drift = Drift(drift_length)
# We take the same FODO as exercise 1 and add some quadupoles
FODO_thick = Lattice([half_quad_f, drift, dipole, drift, dipole, drift,
quad_d, drift, dipole, drift, dipole, drift, half_quad_f])
FODO_thick.plot()
FODO_thick.m
FODO_thick.dispersion_solution()
(FODO_thick * 8).plot.top_down()
tracked = FODO_thick.dispersion()
plt.plot(tracked.s, tracked.x, label="dispersion")
plt.legend()
###Output
_____no_output_____
###Markdown
particles with energy spread
###Code
beam = Beam(n_particles=50, sigma_energy=1e-6)
beam
transported = FODO_thick.transport(beam.match(FODO_thick.twiss_solution()))
plt.plot(transported.s, transported.x.T);
s, disp, *_ = FODO_thick.dispersion()
plt.plot(s, disp)
FODO_thick.plot()
###Output
_____no_output_____
###Markdown
dp/p orbit
###Code
lat = Lattice([Dipole(1, np.pi/2)])
tracked = lat.slice(Dipole, 100).transport([[0, 0.1], [0, 0], [0, 0], [0, 0], [0, 0.1]])
plt.plot(tracked.s, tracked.x.T)
# top down view, projecting around the bend
x_circle = np.cos(tracked.s) + tracked.x * np.cos(tracked.s)
y_circle = np.sin(tracked.s) + tracked.x * np.sin(tracked.s)
fig, ax = plt.subplots(1, 1)
ax.plot(x_circle.T, y_circle.T)
ax.set_aspect("equal")
###Output
_____no_output_____ |
.ipynb_checkpoints/hypothesis-checkpoint.ipynb | ###Markdown
Hypothesis Testing==================Copyright 2016 Allen DowneyLicense: [Creative Commons Attribution 4.0 International](http://creativecommons.org/licenses/by/4.0/)
###Code
from __future__ import print_function, division
import numpy
import scipy.stats
import matplotlib.pyplot as pyplot
from ipywidgets import interact, interactive, fixed
import ipywidgets as widgets
import first
# seed the random number generator so we all get the same results
numpy.random.seed(19)
# some nicer colors from http://colorbrewer2.org/
COLOR1 = '#7fc97f'
COLOR2 = '#beaed4'
COLOR3 = '#fdc086'
COLOR4 = '#ffff99'
COLOR5 = '#386cb0'
%matplotlib inline
###Output
_____no_output_____
###Markdown
Part One Suppose you observe an apparent difference between two groups and you want to check whether it might be due to chance.As an example, we'll look at differences between first babies and others. The `first` module provides code to read data from the National Survey of Family Growth (NSFG).
###Code
live, firsts, others = first.MakeFrames()
###Output
_____no_output_____
###Markdown
We'll look at a couple of variables, including pregnancy length and birth weight. The effect size we'll consider is the difference in the means.Other examples might include a correlation between variables or a coefficient in a linear regression. The number that quantifies the size of the effect is called the "test statistic".
###Code
def TestStatistic(data):
group1, group2 = data
test_stat = abs(group1.mean() - group2.mean())
return test_stat
###Output
_____no_output_____
###Markdown
For the first example, I extract the pregnancy length for first babies and others. The results are pandas Series objects.
###Code
group1 = firsts.prglngth
group2 = others.prglngth
###Output
_____no_output_____
###Markdown
The actual difference in the means is 0.078 weeks, which is only 13 hours.
###Code
actual = TestStatistic((group1, group2))
actual
###Output
_____no_output_____
###Markdown
The null hypothesis is that there is no difference between the groups. We can model that by forming a pooled sample that includes first babies and others.
###Code
n, m = len(group1), len(group2)
pool = numpy.hstack((group1, group2))
###Output
_____no_output_____
###Markdown
Then we can simulate the null hypothesis by shuffling the pool and dividing it into two groups, using the same sizes as the actual sample.
###Code
def RunModel():
numpy.random.shuffle(pool)
data = pool[:n], pool[n:]
return data
###Output
_____no_output_____
###Markdown
The result of running the model is two NumPy arrays with the shuffled pregnancy lengths:
###Code
RunModel()
###Output
_____no_output_____
###Markdown
Then we compute the same test statistic using the simulated data:
###Code
TestStatistic(RunModel())
###Output
_____no_output_____
###Markdown
If we run the model 1000 times and compute the test statistic, we can see how much the test statistic varies under the null hypothesis.
###Code
test_stats = numpy.array([TestStatistic(RunModel()) for i in range(1000)])
test_stats.shape
###Output
_____no_output_____
###Markdown
Here's the sampling distribution of the test statistic under the null hypothesis, with the actual difference in means indicated by a gray line.
###Code
pyplot.vlines(actual, 0, 300, linewidth=3, color='0.8')
pyplot.hist(test_stats, color=COLOR5)
pyplot.xlabel('difference in means')
pyplot.ylabel('count')
None
###Output
_____no_output_____
###Markdown
The p-value is the probability that the test statistic under the null hypothesis exceeds the actual value.
###Code
pvalue = sum(test_stats >= actual) / len(test_stats)
pvalue
###Output
_____no_output_____
###Markdown
In this case the result is about 15%, which means that even if there is no difference between the groups, it is plausible that we could see a sample difference as big as 0.078 weeks.We conclude that the apparent effect might be due to chance, so we are not confident that it would appear in the general population, or in another sample from the same population.STOP HERE--------- Part Two========We can take the pieces from the previous section and organize them in a class that represents the structure of a hypothesis test.
###Code
class HypothesisTest(object):
"""Represents a hypothesis test."""
def __init__(self, data):
"""Initializes.
data: data in whatever form is relevant
"""
self.data = data
self.MakeModel()
self.actual = self.TestStatistic(data)
self.test_stats = None
def PValue(self, iters=1000):
"""Computes the distribution of the test statistic and p-value.
iters: number of iterations
returns: float p-value
"""
self.test_stats = numpy.array([self.TestStatistic(self.RunModel())
for _ in range(iters)])
count = sum(self.test_stats >= self.actual)
return count / iters
def MaxTestStat(self):
"""Returns the largest test statistic seen during simulations.
"""
return max(self.test_stats)
def PlotHist(self, label=None):
"""Draws a Cdf with vertical lines at the observed test stat.
"""
ys, xs, patches = pyplot.hist(ht.test_stats, color=COLOR4)
pyplot.vlines(self.actual, 0, max(ys), linewidth=3, color='0.8')
pyplot.xlabel('test statistic')
pyplot.ylabel('count')
def TestStatistic(self, data):
"""Computes the test statistic.
data: data in whatever form is relevant
"""
raise UnimplementedMethodException()
def MakeModel(self):
"""Build a model of the null hypothesis.
"""
pass
def RunModel(self):
"""Run the model of the null hypothesis.
returns: simulated data
"""
raise UnimplementedMethodException()
###Output
_____no_output_____
###Markdown
`HypothesisTest` is an abstract parent class that encodes the template. Child classes fill in the missing methods. For example, here's the test from the previous section.
###Code
class DiffMeansPermute(HypothesisTest):
"""Tests a difference in means by permutation."""
def TestStatistic(self, data):
"""Computes the test statistic.
data: data in whatever form is relevant
"""
group1, group2 = data
test_stat = abs(group1.mean() - group2.mean())
return test_stat
def MakeModel(self):
"""Build a model of the null hypothesis.
"""
group1, group2 = self.data
self.n, self.m = len(group1), len(group2)
self.pool = numpy.hstack((group1, group2))
def RunModel(self):
"""Run the model of the null hypothesis.
returns: simulated data
"""
numpy.random.shuffle(self.pool)
data = self.pool[:self.n], self.pool[self.n:]
return data
###Output
_____no_output_____
###Markdown
Now we can run the test by instantiating a DiffMeansPermute object:
###Code
data = (firsts.prglngth, others.prglngth)
ht = DiffMeansPermute(data)
p_value = ht.PValue(iters=1000)
print('\nmeans permute pregnancy length')
print('p-value =', p_value)
print('actual =', ht.actual)
print('ts max =', ht.MaxTestStat())
###Output
means permute pregnancy length
p-value = 0.169
actual = 0.07803726677754952
ts max = 0.20696822841
###Markdown
And we can plot the sampling distribution of the test statistic under the null hypothesis.
###Code
ht.PlotHist()
###Output
_____no_output_____
###Markdown
Difference in standard deviation**Exercize 1**: Write a class named `DiffStdPermute` that extends `DiffMeansPermute` and overrides `TestStatistic` to compute the difference in standard deviations. Is the difference in standard deviations statistically significant?
###Code
# Solution goes here
###Output
_____no_output_____
###Markdown
Here's the code to test your solution to the previous exercise.
###Code
data = (firsts.prglngth, others.prglngth)
ht = DiffStdPermute(data)
p_value = ht.PValue(iters=1000)
print('\nstd permute pregnancy length')
print('p-value =', p_value)
print('actual =', ht.actual)
print('ts max =', ht.MaxTestStat())
###Output
std permute pregnancy length
p-value = 0.156
actual = 0.1760490642294399
ts max = 0.503028294469
###Markdown
Difference in birth weightsNow let's run DiffMeansPermute again to see if there is a difference in birth weight between first babies and others.
###Code
data = (firsts.totalwgt_lb.dropna(), others.totalwgt_lb.dropna())
ht = DiffMeansPermute(data)
p_value = ht.PValue(iters=1000)
print('\nmeans permute birthweight')
print('p-value =', p_value)
print('actual =', ht.actual)
print('ts max =', ht.MaxTestStat())
###Output
means permute birthweight
p-value = 0.0
actual = 0.12476118453549034
ts max = 0.0924976865499
|
Inferential_Statistical_Analysis_with_Python-master/week1/week1_assessment.ipynb | ###Markdown
You will use the values of what you find in this assignment to answer questions in the quiz that follows. You may want to open this notebook to be displayed side-by-side on screen with this next quiz. 1. Write a function that inputs an integers and returns the negative
###Code
# Write your function here
def some_function(Int_input):
return -Int_input
# Test your function with input x
x = 4
Output = some_function(x)
print(Output)
###Output
-4
###Markdown
2. Write a function that inputs a list of integers and returns the minimum value
###Code
# Write your function here
def ListFun(lst):
return min(lst)
# Test your function with input lst
lst = [-3, 0, 2, 100, -1, 2]
print(ListFun(lst))
# Create you own input list to test with
my_lst = [324,-234,284,34975,0,-23,34978,-213]
print(ListFun(my_lst))
###Output
-3
-234
###Markdown
Challenge problem: Write a function that take in four arguments: lst1, lst2, str1, str2, and returns a pandas DataFrame that has the first column labeled str1 and the second column labaled str2, that have values lst1 and lst2 scaled to be between 0 and 1.For example```lst1 = [1, 2, 3]lst2 = [2, 4, 5]str1 = 'one'str2 = 'two'my_function(lst1, lst2, str1, str2)``` should return a DataFrame that looks like:| | one | two || --- | --- | --- || 0 | 0 | 0 || 1 | .5 | .666 || 2 | 1 | 1 |
###Code
import pandas as pd
def Challenge(lst1, lst2, str1, str2):
df = pd.DataFrame({str1:lst1, str2:lst2})
return df
# test your challenge problem function
import numpy as np
lst1 = np.random.randint(-234, 938, 100)
lst2 = np.random.randint(-522, 123, 100)
str1 = 'one'
str2 = 'alpha'
print(Challenge(lst1, lst2, str1, str2))
###Output
one alpha
0 795 -52
1 637 -241
2 665 -46
3 655 -104
4 417 -486
5 497 57
6 773 -370
7 824 -128
8 111 -170
9 479 102
10 -103 -450
11 439 -385
12 151 -266
13 621 -104
14 582 41
15 -116 -431
16 -94 -24
17 332 -239
18 184 -262
19 232 4
20 344 -349
21 634 -35
22 920 -107
23 -117 -275
24 871 -380
25 828 -252
26 371 -496
27 893 -224
28 687 -103
29 734 -223
.. ... ...
70 355 -194
71 743 -142
72 464 -73
73 536 -219
74 323 -21
75 714 36
76 -84 -37
77 -111 -180
78 -155 -143
79 702 54
80 -125 67
81 -180 -35
82 339 -119
83 325 -368
84 127 -488
85 116 -78
86 265 -190
87 138 -315
88 763 -123
89 754 -286
90 595 61
91 769 -49
92 -228 -501
93 76 -110
94 811 8
95 149 -415
96 850 -60
97 258 25
98 774 -426
99 393 -440
[100 rows x 2 columns]
|
notebooks/firefox_p2/tc_br_tracing/bm25-model.ipynb | ###Markdown
IntroductionIn this notebook we demonstrate the use of **BM25 (Best Matching 25)** Information Retrieval technique to make trace link recovery between Test Cases and Bug Reports.We model our study as follows:* Each bug report title, summary and description compose a single query.* We use each test case content as an entire document that must be returned to the query made Import Libraries
###Code
%load_ext autoreload
%autoreload 2
from mod_finder_util import mod_finder_util
mod_finder_util.add_modules_origin_search_path()
import pandas as pd
from modules.models_runner.tc_br_models_runner import TC_BR_Runner
from modules.models_runner.tc_br_models_runner import TC_BR_Models_Hyperp
from modules.utils import aux_functions
from modules.utils import firefox_dataset_p2 as fd
from modules.utils import tokenizers as tok
from modules.models.bm25 import BM_25
from IPython.display import display
import warnings; warnings.simplefilter('ignore')
###Output
_____no_output_____
###Markdown
Load Datasets
###Code
tcs = [x for x in range(37,59)]
orc = fd.Tc_BR_Oracles.read_oracle_expert_df()
orc_subset = orc[orc.index.isin(tcs)]
#aux_functions.highlight_df(orc_subset)
tcs = [13,37,60]
brs = [1267501]
testcases = fd.Datasets.read_testcases_df()
testcases = testcases[testcases.TC_Number.isin(tcs)]
bugreports = fd.Datasets.read_selected_bugreports_df()
bugreports = bugreports[bugreports.Bug_Number.isin(brs)]
print('tc.shape: {}'.format(testcases.shape))
print('br.shape: {}'.format(bugreports.shape))
###Output
TestCases.shape: (195, 12)
SelectedBugReports.shape: (91, 18)
tc.shape: (3, 12)
br.shape: (1, 18)
###Markdown
Running BM25 Model
###Code
corpus = testcases.tc_desc
query = bugreports.br_desc
test_cases_names = testcases.tc_name
bug_reports_names = bugreports.br_name
bm25_hyperp = TC_BR_Models_Hyperp.get_bm25_model_hyperp()
bm25_model = BM_25(**bm25_hyperp)
bm25_model.set_name('BM25_Model_TC_BR')
bm25_model.recover_links(corpus, query, test_cases_names, bug_reports_names)
bm25_model.get_sim_matrix().shape
sim_matrix_normalized = bm25_model.get_sim_matrix()
aux_functions.highlight_df(sim_matrix_normalized)
sim_matrix_origin = bm25_model._sim_matrix_origin
aux_functions.highlight_df(sim_matrix_origin)
df = pd.DataFrame()
df['tc'] = corpus
df.index = test_cases_names
df.index.name = ''
#df = df.T
df.head(10)
###Output
_____no_output_____
###Markdown
Query Vector
###Code
tokenizer = tok.PorterStemmerBased_Tokenizer()
query_vec = [tokenizer.__call__(doc) for doc in query]
df_q = pd.DataFrame(query_vec)
df_q.index = bug_reports_names
df_q.index.name = ''
df_q = df_q.T
df_q.head(10)
###Output
_____no_output_____
###Markdown
Average Document Length
###Code
bm25_model.bm25.avgdl
###Output
_____no_output_____
###Markdown
Number of documents
###Code
bm25_model.bm25.corpus_size
###Output
_____no_output_____
###Markdown
Term frequency by document
###Code
bm25_model.bm25.df['apz']
###Output
_____no_output_____
###Markdown
Most Relevant Words
###Code
bm25_model.mrw_tcs
bm25_model.docs_feats_df
###Output
_____no_output_____ |
ICCT_en/examples/01/M-01_Complex_numbers_Cartesian_form.ipynb | ###Markdown
Complex numbers in Cartesian formFeel free to use this interactive example to visualize complex numbers in a complex plane, utilizing the Cartesian form. Also, you can test basic mathematical operators while working with complex numbers: addition, subtraction, multiplying, and dividing. All results are presented in the respective plot, as well as in the typical mathematical notation.You can manipulate complex numbers directly on the plot (by simple clicking), or/and use input fields at the same time. In order to provide better visibility of the respective vectors in the plot widget, the complex number coefficients are limited to $\pm10$.
###Code
%matplotlib notebook
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import numpy as np
import ipywidgets as widgets
from IPython.display import display
red_patch = mpatches.Patch(color='red', label='z1')
blue_patch = mpatches.Patch(color='blue', label='z2')
green_patch = mpatches.Patch(color='green', label='z1 + z2')
yellow_patch = mpatches.Patch(color='yellow', label='z1 - z2')
black_patch = mpatches.Patch(color='black', label='z1 * z2')
magenta_patch = mpatches.Patch(color='magenta', label='z1 / z2')
# Init values
XLIM = 5
YLIM = 5
vectors_index_first = False;
V = [None, None]
V_complex = [None, None]
# Complex plane
fig = plt.figure(num='Complex numbers in Cartesian form')
ax = fig.add_subplot(1, 1, 1)
def get_interval(lim):
if lim <= 10:
return 1
if lim < 75:
return 5
if lim > 100:
return 25
return 10
def set_ticks():
XLIMc = int((XLIM / 10) + 1) * 10
YLIMc = int((YLIM / 10) + 1) * 10
if XLIMc > 150:
XLIMc += 10
if YLIMc > 150:
YLIMc += 10
xstep = get_interval(XLIMc)
ystep = get_interval(YLIMc)
#print(stepx, stepy)
major_ticks = np.arange(-XLIMc, XLIMc, xstep)
major_ticks_y = np.arange(-YLIMc, YLIMc, ystep)
ax.set_xticks(major_ticks)
ax.set_yticks(major_ticks_y)
ax.grid(which='both')
def clear_plot():
plt.cla()
set_ticks()
ax.set_xlabel('Re')
ax.set_ylabel('Im')
plt.ylim([-YLIM, YLIM])
plt.xlim([-XLIM, XLIM])
plt.legend(handles=[red_patch, blue_patch, green_patch, yellow_patch, black_patch, magenta_patch])
clear_plot()
set_ticks()
plt.show()
set_ticks()
# Set a complex number using direct manipulation on the plot
def set_vector(i, data_x, data_y):
clear_plot()
V.pop(i)
V.insert(i, (0, 0, round(data_x, 2), round(data_y, 2)))
V_complex.pop(i)
V_complex.insert(i, complex(round(data_x, 2), round(data_y, 2)))
if i == 0:
ax.arrow(*V[0], head_width=0.25, head_length=0.5, color="r", length_includes_head=True)
a1.value = round(data_x, 2)
b1.value = round(data_y, 2)
if V[1] != None:
ax.arrow(*V[1], head_width=0.25, head_length=0.5, color="b", length_includes_head=True)
elif i == 1:
ax.arrow(*V[1], head_width=0.25, head_length=0.5, color="b", length_includes_head=True)
a2.value = round(data_x, 2)
b2.value = round(data_y, 2)
if V[0] != None:
ax.arrow(*V[0], head_width=0.25, head_length=0.5, color="r", length_includes_head=True)
max_bound()
def onclick(event):
global vectors_index_first
vectors_index_first = not vectors_index_first
x = event.xdata
y = event.ydata
if (x > 10):
x = 10.0
if (x < - 10):
x = -10.0
if (y > 10):
y = 10.0
if (y < - 10):
y = -10.0
if vectors_index_first:
set_vector(0, x, y)
else:
set_vector(1, x, y)
fig.canvas.mpl_connect('button_press_event', onclick)
# Widgets
a1 = widgets.BoundedFloatText(layout=widgets.Layout(width='10%'), min = -10, max = 10, step = 0.5)
b1 = widgets.BoundedFloatText(layout=widgets.Layout(width='10%'), min = -10, max = 10, step = 0.5)
button_set_z1 = widgets.Button(description="Plot z1")
a2 = widgets.BoundedFloatText(layout=widgets.Layout(width='10%'), min = -10, max = 10, step = 0.5)
b2 = widgets.BoundedFloatText(layout=widgets.Layout(width='10%'), min = -10, max = 10, step = 0.5)
button_set_z2 = widgets.Button(description="Plot z2")
box_layout_z1 = widgets.Layout(border='solid red', padding='10px')
box_layout_z2 = widgets.Layout(border='solid blue', padding='10px')
box_layout_opers = widgets.Layout(border='solid black', padding='10px')
items_z1 = [widgets.Label("z1 = "), a1, widgets.Label("+ j * "), b1, button_set_z1]
items_z2 = [widgets.Label("z2 = "), a2, widgets.Label("+ j * "), b2, button_set_z2]
display(widgets.Box(children=items_z1, layout=box_layout_z1))
display(widgets.Box(children=items_z2, layout=box_layout_z2))
button_add = widgets.Button(description="Add")
button_substract = widgets.Button(description="Subtract")
button_multiply = widgets.Button(description="Multiply")
button_divide = widgets.Button(description="Divide")
button_reset = widgets.Button(description="Reset")
output = widgets.Output()
print('Complex number operations:')
items_operations = [button_add, button_substract, button_multiply, button_divide, button_reset]
display(widgets.Box(children=items_operations))
display(output)
# Set complex number using input widgets (Text and Button)
def on_button_set_z1_clicked(b):
z1_old = V[0];
z1_new = (0, 0, a1.value, b1.value)
if z1_old != z1_new:
set_vector(0, a1.value, b1.value)
change_lims()
def on_button_set_z2_clicked(b):
z2_old = V[1];
z2_new = (0, 0, a2.value, b2.value)
if z2_old != z2_new:
set_vector(1, a2.value, b2.value)
change_lims()
# Complex number operations:
def perform_operation(oper):
global XLIM, YLIM
if (V_complex[0] != None) and (V_complex[1] != None):
if (oper == '+'):
result = V_complex[0] + V_complex[1]
v_color = "g"
elif (oper == '-'):
result = V_complex[0] - V_complex[1]
v_color = "y"
elif (oper == '*'):
result = V_complex[0] * V_complex[1]
v_color = "black"
elif (oper == '/'):
result = V_complex[0] / V_complex[1]
v_color = "magenta"
result = complex(round(result.real, 2), round(result.imag, 2))
ax.arrow(0, 0, result.real, result.imag, head_width=0.25, head_length=0.5, color=v_color, length_includes_head=True)
if abs(result.real) > XLIM:
XLIM = round(abs(result.real) + 1)
if abs(result.imag) > YLIM:
YLIM = round(abs(result.imag) + 1)
change_lims()
with output:
print(V_complex[0], oper, V_complex[1], "=", result)
def on_button_add_clicked(b):
perform_operation("+")
def on_button_substract_clicked(b):
perform_operation("-")
def on_button_multiply_clicked(b):
perform_operation("*")
def on_button_divide_clicked(b):
perform_operation("/")
# Plot init methods
def on_button_reset_clicked(b):
global V, V_complex, XLIM, YLIM
with output:
output.clear_output()
clear_plot()
vectors_index_first = False;
V = [None, None]
V_complex = [None, None]
a1.value = 0
b1.value = 0
a2.value = 0
b2.value = 0
XLIM = 5
YLIM = 5
change_lims()
def clear_plot():
plt.cla()
set_ticks()
ax.set_xlabel('Re')
ax.set_ylabel('Im')
plt.ylim([-YLIM, YLIM])
plt.xlim([-XLIM, XLIM])
plt.legend(handles=[red_patch, blue_patch, green_patch, yellow_patch, black_patch, magenta_patch])
def change_lims():
set_ticks()
plt.ylim([-YLIM, YLIM])
plt.xlim([-XLIM, XLIM])
set_ticks()
def max_bound():
global XLIM, YLIM
mx = 0
my = 0
if V_complex[0] != None:
z = V_complex[0]
if abs(z.real) > mx:
mx = abs(z.real)
if abs(z.imag) > my:
my = abs(z.imag)
if V_complex[1] != None:
z = V_complex[1]
if abs(z.real) > mx:
mx = abs(z.real)
if abs(z.imag) > my:
my = abs(z.imag)
if mx > XLIM:
XLIM = round(mx + 1)
elif mx <=5:
XLIM = 5
if my > YLIM:
YLIM = round(my + 1)
elif my <=5:
YLIM = 5
change_lims()
# Button events
button_set_z1.on_click(on_button_set_z1_clicked)
button_set_z2.on_click(on_button_set_z2_clicked)
button_add.on_click(on_button_add_clicked)
button_substract.on_click(on_button_substract_clicked)
button_multiply.on_click(on_button_multiply_clicked)
button_divide.on_click(on_button_divide_clicked)
button_reset.on_click(on_button_reset_clicked)
###Output
_____no_output_____ |
backend.ipynb | ###Markdown
Backend Data Generation---This notebook generates the backend data needed for the app to fetch latest stat of a fighter
###Code
import pandas as pd
import numpy as np
pd.set_option('display.max_columns', 100)
df = pd.read_csv("data/UFC_processed.csv")
df["date"] = pd.to_datetime(df["date"]) # date as datetime
features = ["date","fighter"]
for name in df.columns[4:25]: # slice string to separate prefix (e.g: R_)
features.append(name[2:]) # add more to features
# separate fighters
blueFighter = pd.concat([df.iloc[:,[0,1]],df.iloc[:,4:25]],axis=1)
redFighter = pd.concat([df.iloc[:,[0,2]],df.iloc[:,25:]],axis=1)
# rename columns
blueFighter.columns = features
redFighter.columns = features
# join them in one table
fighters = pd.concat([redFighter,blueFighter],axis=0,).reset_index(drop=True)
# as each fighter has fought multiple matches, in order to get the latest stat for each fighter, we have to group them by name
# and get the details of their latest match:
l = fighters.groupby("fighter")
fighters_detail = []
# for each unique fighter:
# 1- groupby fighter's name to get all their fights
# 2- sort the values by data in newest to oldest format
# 3- get the first element (i.e: iloc0) which is the newest
for fighter in fighters["fighter"].unique():
fighters_detail.append(l.get_group(fighter).sort_values(by=["date"],ascending=False).iloc[0])
fighter_stat = pd.DataFrame(fighters_detail).sort_values(by="fighter")
fighter_stat.insert(0, 'ID', np.arange(1,len(fighter_stat.index)+1))
fighter_stat.reset_index(drop=True, inplace=True)
# export dataset
fighter_stat.to_csv("data/FIGHTER_STAT.csv",index=False)
###Output
_____no_output_____ |
1.- Fundamentos de Pandas/1.1 Colab/Creando_el_primer_DataFrame_desde_un_csv.ipynb | ###Markdown
Descargando y consumiendo un csv
###Code
# Descargamos el Dataset
! wget http://srea64.github.io/msan622/project/pokemon.csv
"""
Importamos la librería pandas
"""
import pandas
"""
pd: responde al alias
read_csv: Función de Pandas que permite trabajar con csv
"""
pandas.read_csv('pokemon.csv')
"""
Importamos la librería pandas y le colocamos el alias pd para ser llamada
más adelante
"""
import pandas as pd
"""
pd: responde al alias
read_csv: Función de Pandas que permite trabajar con csv
"""
df_pokemon = pd.read_csv('pokemon.csv')
df_pokemon
###Output
_____no_output_____
###Markdown
Leemos directamente de la URL origen
###Code
import pandas as pd
url = "http://srea64.github.io/msan622/project/pokemon.csv"
pd.read_csv(url)
###Output
_____no_output_____
###Markdown
Columna con valores NA
###Code
# Saber que columna del dataframe tiene valores NA
df_pokemon.isnull().sum()
###Output
_____no_output_____
###Markdown
Nombre de las columnas
###Code
# Saber los nombres de las columnas que conforman mi dataframe
list(df_pokemon)
###Output
_____no_output_____
###Markdown
Consultar datos en base al nombre de la columna
###Code
# Extraer información de la columna con su nombre
df_pokemon['id']
# En vase al nombre de la columna extraemos el mismo elemento
df_pokemon['Name'][1]
###Output
_____no_output_____
###Markdown
Consultar datos en base a la posición de la columna
###Code
# Extraer el nombre de la columna por su posición
df_pokemon.columns[1]
"""
1.- Extraemos el nombre de la columna por su posición
2.- Usamos ese nombre para extraer la información
"""
df_pokemon[df_pokemon.columns[1]]
"""
1.- Extraemos el nombre de la columna por su posición
2.- Usamos ese nombre para extraer la información
3.- Tomamos el primer elemento de la información extraída
"""
df_pokemon[df_pokemon.columns[1]][1]
###Output
_____no_output_____
###Markdown
Extraer información en base a los datos de una fila
###Code
# Extraer información en base a la primera columna en este caso 'id'
df_pokemon.loc[0]
# Extraer información en base a la columna seleccionada por el usuario en este caso 'Name'
df_pokemon.loc[df_pokemon['Name'] == 'Bulbasaur']
# Extraer información en base a la posición de columna en este caso 'Name'
df_pokemon.loc[df_pokemon[df_pokemon.columns[1]] == 'Bulbasaur']
###Output
_____no_output_____
###Markdown
Ejemplo de uso para filtrar datos de un dataframe
###Code
"""
Otro ejemplo
Extraemos los pokemons que tengan un ataque mayor o igual a 50
"""
df_pokemon.loc[df_pokemon['Attack'] >= 50]
###Output
_____no_output_____ |
LHS_PRCC.ipynb | ###Markdown
Latin Hypercube Sampling & Partial Rank Correlation Coefficients *~ a method for analyzing model sensitivity to parameters ~* Importing packages that will be used.
###Code
import numpy as np
from scipy import special
import random
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
from IPython.display import display
import pandas as pd
import matplotlib.pyplot as plt
###Output
_____no_output_____
###Markdown
Specify the number of parameters to sample and the number of samples to draw from each parameter distribution. *Do not include any parameters that should be left fixed in parameterCount - those will be specified later. When choosing number of samples to draw, note that more samples (~ 1000) yields better results while fewer (~50) is faster for testing, since it requires fewer model solves.*
###Code
# Number of parameters to sample
parameterCount = 2;
# Number of samples to draw for each parameter
sampleCount = 100;
###Output
_____no_output_____
###Markdown
This defines functions for specifying sampled parameters' names and distributions as well as drawing samples from a user-specified parameter distribution. Does not need any user edits.
###Code
def parNameDist(Name,Distribution):
paramTemp = {}
paramTemp['Name']=Name
paramTemp['Dist']=Distribution
return paramTemp
def sampleDistrib(modelParamName,distrib,distribSpecs):
if distrib == 'uniform':
mmin = distribSpecs[0].value
mmax = distribSpecs[1].value
intervalwidth = (mmax - mmin) / sampleCount # width of each
# sampling interval
samples = []
for sample in range(sampleCount):
lower = mmin + intervalwidth * (sample-1) # lb of interval
upper = mmin + intervalwidth * (sample) # ub of interval
sampleVal = np.random.uniform(lower, upper) # draw a random sample
# within the interval
samples.append(sampleVal)
elif distrib == 'normal':
mmean= distribSpecs[0].value
mvar = distribSpecs[1].value
lower = mvar*np.sqrt(2)*special.erfinv(-0.9999)+mmean # set lb of 1st
# sample interval
samples = []
for sample in range(sampleCount):
n = sample + 1
if n != sampleCount:
upper = (np.sqrt(2*mvar)*special.erfinv(2*n/sampleCount-1)
+ mmean) # ub of sample interval
else:
upper = np.sqrt(2*mvar)*special.erfinv(0.9999) + mmean
sampleVal = np.random.uniform(lower, upper) # draw a random sample
# within the interval
samples.append(sampleVal)
lower = upper # set current ub as the lb for next interval
elif distrib == 'triangle':
mmin = distribSpecs[0].value
mmax = distribSpecs[1].value
mmode= distribSpecs[2].value
samples = []
for sample in range(sampleCount):
n = sample + 1
intervalarea = 1/sampleCount
ylower = intervalarea*(n-1) # use cdf to read off area as y's &
yupper = intervalarea*(n) # get corresponding x's for the pdf
# Check to see if y values = cdf(x <= mmode)
# for calculating correxponding x values:
if ylower <= ((mmode - mmin)/(mmax - mmin)):
lower = np.sqrt(ylower*(mmax - mmin)*(mmode - mmin)) + mmin
else:
lower = mmax-np.sqrt((1 - ylower)*(mmax - mmin)*(mmax - mmode))
if yupper <= ((mmode - mmin)/(mmax - mmin)):
upper = np.sqrt(yupper*(mmax - mmin)*(mmode - mmin)) + mmin;
else:
upper = mmax-np.sqrt((1 - yupper)*(mmax - mmin)*(mmax - mmode))
sampleVal = np.random.uniform(lower, upper)
samples.append(sampleVal)
b = int(np.ceil(sampleCount/10))
plt.hist(samples, density = 1, bins = b)
B=str(b)
plt.title('Histogram of ' + modelParamName
+ ' parameter samples for ' + B + ' bins')
plt.ylabel('proportion of samples');
plt.xlabel(modelParamName + ' value')
plt.show()
return samples
###Output
_____no_output_____
###Markdown
Calls the function to ask for user input to name parameters and specify distributions. Type these in text input boxes and dropdowns that will appear below after running the cell.
###Code
params = {}
for i in range(parameterCount):
s=str(i)
params[i] = interactive(parNameDist,
Name='Type parameter ' + s + ' name',
Distribution=['uniform','normal','triangle'])
display(params[i])
###Output
_____no_output_____
###Markdown
Input parameter distribution specifics in the interactive boxes that appear below after running this cell.
###Code
distribSpecs={}
for i in range(parameterCount):
parName = params[i].result['Name']
print('Enter distribution specifics for parameter ' + parName + ':')
if params[i].result['Dist'] == 'normal':
distribSpecs[parName] = {}
distribSpecs[parName][0] = widgets.FloatText(
value=2,
description='Mean:'
)
distribSpecs[parName][1] = widgets.FloatText(
value=1,
description='Variance:'
)
display(distribSpecs[parName][0], distribSpecs[parName][1])
elif params[i].result['Dist'] == 'uniform':
distribSpecs[parName] = {}
distribSpecs[parName][0] = widgets.FloatText(
value=0,
description='Minimum:'
)
distribSpecs[parName][1] = widgets.FloatText(
value=2,
description='Maximum:'
)
display(distribSpecs[parName][0], distribSpecs[parName][1])
elif params[i].result['Dist'] == 'triangle':
distribSpecs[parName] = {}
distribSpecs[parName][0] = widgets.FloatText(
value=0,
description='Minimum:'
)
distribSpecs[parName][1] = widgets.FloatText(
value=2,
description='Maximum:'
)
distribSpecs[parName][2] = widgets.FloatText(
value=1,
description='Mode:'
)
display(distribSpecs[parName][0], distribSpecs[parName][1], distribSpecs[parName][2])
###Output
_____no_output_____
###Markdown
This passes the distributions to the code for generating parameter samples, and histogram plots of samples for each parameter will appear below.
###Code
parameters = {}
for j in range(parameterCount):
parameters[params[j].result['Name']] = sampleDistrib(params[j].result['Name'],
params[j].result['Dist'],
distribSpecs[params[j].result['Name']])
###Output
_____no_output_____
###Markdown
Randomly permute each set of parameter samples in order to randomly pair the samples to more fully sample the parameter space for the Monte Carlo simulations.
###Code
LHSparams=[]
for p in parameters:
temp = parameters[p]
random.shuffle(temp)
LHSparams.append(temp)
###Output
_____no_output_____
###Markdown
Define your model function. Two examples have been provided below: (1) a linear function with two sampled parameters: slope and intercept, and (2) a Lotka-Volterra predator - prey model.Note that the order and number of the parameters needs to match the order and number of parameters speficied above to ensure accuracy when the model is solved below.
###Code
def testlinear(x,sampledParams,unsampledParams):
m = sampledParams[0]
b = sampledParams[1]
a = unsampledParams
y = m * x + b + a;
return y
def myodes(y, t, sampledParams, unsampledParams):
q, r = y # unpack current values of y
alpha, beta = sampledParams # unpack sampled parameters
delta, lambdaa, gamma = unsampledParams # unpack unsampled parameters
derivs = [alpha*q*r - lambdaa*q, # list of dy/dt=f functions
beta*r - gamma*q*r - delta*r]
return derivs
###Output
_____no_output_____
###Markdown
Run Monte Carlo simulations for each parameter sample set. *Be sure to specify a call to your model function and any necessary arguments below.*
###Code
# EDIT THE FOLLOWING VARIABLES, UNSAMPLED PARAMETERS, & ANY OTHER ARGS HERE,
# AS WELL AS THE CALL TO YOUR OWN MODEL FUNCTION INSIDE THE FOR LOOP BELOW
x = np.linspace(0, 10, num=101)
unsampledParams = 2;
Output = []
for j in range(sampleCount):
sampledParams=[i[j] for i in LHSparams]
sol = testlinear(x,sampledParams,unsampledParams)
Output.append(sol)
# EDIT THE STRING TO NAME YOUR SIM OUTPUT (for fig labels, filenames):
labelstring = 'y'
# # EXAMPLE CODE FOR A COUPLED ODE MODEL:
# import scipy.integrate as spi
# t = np.linspace(0,17,num=171) # time domain for myodes
# # odesic = [q0, r0]
# odesic = [500,1000]
# lambdaa = np.log(2)/7
# delta = 0.5
# gamma = 1
# unsampledParams = [lambdaa, delta, gamma]
# Simdata={}
# Output = []
# for i in range(sampleCount):
# Simdata[i]={}
# Simdata[i]['q']=[]
# Simdata[i]['r']=[]
# for j in range(sampleCount):
# sampledParams=[i[j] for i in LHSparams]
# sol=spi.odeint(myodes, odesic, t, args=(sampledParams,unsampledParams))
# Simdata[j]['q'] = sol[:,0] # solution to the equation for variable r
# Simdata[j]['r'] = sol[:,1] # solution to the equation for variable s
# Ratio = np.divide(sol[:,0],sol[:,1]) # compute ratio to compare w/ param samples
# Output.append(Ratio)
# labelstring = 'predator to prey ratio (q/r)'; # id for fig labels, filenames
###Output
_____no_output_____
###Markdown
Plot the range of simulation output generated by the all of the Monte Carlo simulations using errorbars.
###Code
yavg = np.mean(Output, axis=0)
yerr = np.std(Output, axis=0)
plt.errorbar(t,yavg,yerr)
plt.xlabel('x')
# plt.xlabel('time (days)') # for myodes
plt.ylabel(labelstring)
plt.title('Error bar plot of ' + labelstring + ' from LHS simulations')
plt.show()
###Output
_____no_output_____
###Markdown
Compute partial rank correlation coefficients to compare simulation outputs with parameters
###Code
SampleResult=[]
x_idx = 11 # time or location index of sim results
x_idx2= x_idx+1 # to compare w/ param sample vals
LHS=[*zip(*LHSparams)]
LHSarray=np.array(LHS)
Outputarray=np.array(Output)
subOut=Outputarray[0:,x_idx:x_idx2]
LHSout = np.hstack((LHSarray,subOut))
SampleResult = LHSout.tolist()
Ranks=[]
for s in range(sampleCount):
indices = list(range(len(SampleResult[s])))
indices.sort(key=lambda k: SampleResult[s][k])
r = [0] * len(indices)
for i, k in enumerate(indices):
r[k] = i
Ranks.append(r)
C=np.corrcoef(Ranks);
if np.linalg.det(C) < 1e-16: # determine if singular
Cinv = np.linalg.pinv(C) # may need to use pseudo inverse
else:
Cinv = np.linalg.inv(C)
resultIdx = parameterCount+1
prcc=np.zeros(resultIdx)
for w in range(parameterCount): # compute PRCC btwn each param & sim result
prcc[w]=-Cinv[w,resultIdx]/np.sqrt(Cinv[w,w]*Cinv[resultIdx,resultIdx])
###Output
_____no_output_____
###Markdown
Plot the PRCCs for each parameter
###Code
xp=[i for i in range(parameterCount)]
plt.bar(xp,prcc[0:parameterCount], align='center')
bLabels=list(parameters.keys())
plt.xticks(xp, bLabels)
plt.ylabel('PRCC value');
N=str(sampleCount)
loc=str(x_idx)
plt.title('Partial rank correlation of params with ' + labelstring
+ ' results \n from ' + N + ' LHS sims, at x = ' +loc);
plt.show()
###Output
_____no_output_____
###Markdown
Can also do PRCCs over time...
###Code
SampleResult=[]
resultIdx = parameterCount+1
prcc=np.zeros((resultIdx,len(x)))
LHS=[*zip(*LHSparams)]
LHSarray=np.array(LHS)
Outputarray=np.array(Output)
for xi in range(len(x)): # loop through time or location of sim results
xi2 = xi+1 # to compare w/ parameter sample vals
subOut = Outputarray[0:,xi:xi2]
LHSout = np.hstack((LHSarray,subOut))
SampleResult = LHSout.tolist()
Ranks=[]
for s in range(sampleCount):
indices = list(range(len(SampleResult[s])))
indices.sort(key=lambda k: SampleResult[s][k])
r = [0] * len(indices)
for i, k in enumerate(indices):
r[k] = i
Ranks.append(r)
C=np.corrcoef(Ranks);
if np.linalg.det(C) < 1e-16: # determine if singular
Cinv = np.linalg.pinv(C) # may need to use pseudo inverse
else:
Cinv = np.linalg.inv(C)
for w in range(parameterCount): # compute PRCC btwn each param & sim result
prcc[w,xi]=-Cinv[w,resultIdx]/np.sqrt(Cinv[w,w]*Cinv[resultIdx,resultIdx])
###Output
_____no_output_____
###Markdown
Plot PRCC values as they vary over time or space. *Notice PRCC can change with respect to the independent variable (x-axis). This may be helpful for certain applications, as opposed to only looking at a "snapshot."*
###Code
for p in range(parameterCount):
plt.plot(x,prcc[p,])
labels=list(parameters.keys())
plt.legend(labels)
plt.ylabel('PRCC value');
plt.xlabel('x')
N=str(sampleCount)
plt.title('Partial rank correlation of params with ' + labelstring
+ ' results \n from ' + N + ' LHS sims');
plt.show()
###Output
_____no_output_____
###Markdown
Latin Hypercube Sampling & Partial Rank Correlation Coefficients *~ a method for analyzing model sensitivity to parameters ~* Importing packages that will be used.
###Code
import numpy as np
from scipy import special
import random
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
from IPython.display import display
import pandas as pd
import matplotlib.pyplot as plt
###Output
_____no_output_____
###Markdown
Specify the number of parameters to sample and the number of samples to draw from each parameter distribution. *Do not include any parameters that should be left fixed in parameterCount - those will be specified later. When choosing number of samples to draw, note that more samples (~ 1000) yields better results while fewer (~50) is faster for testing, since it requires fewer model solves.*
###Code
# Number of parameters to sample
parameterCount = 2;
# Number of samples to draw for each parameter
sampleCount = 100;
###Output
_____no_output_____
###Markdown
This defines functions for specifying sampled parameters' names and distributions as well as drawing samples from a user-specified parameter distribution. Does not need any user edits.
###Code
def parNameDist(Name,Distribution):
paramTemp = {}
paramTemp['Name']=Name
paramTemp['Dist']=Distribution
return paramTemp
def sampleDistrib(modelParamName,distrib,distribSpecs):
if distrib == 'uniform':
mmin = distribSpecs[0].value
mmax = distribSpecs[1].value
intervalwidth = (mmax - mmin) / sampleCount # width of each
# sampling interval
samples = []
for sample in range(sampleCount):
lower = mmin + intervalwidth * (sample-1) # lb of interval
upper = mmin + intervalwidth * (sample) # ub of interval
sampleVal = np.random.uniform(lower, upper) # draw a random sample
# within the interval
samples.append(sampleVal)
elif distrib == 'normal':
mmean= distribSpecs[0].value
mvar = distribSpecs[1].value
lower = mvar*np.sqrt(2)*special.erfinv(-0.9999)+mmean # set lb of 1st
# sample interval
samples = []
for sample in range(sampleCount):
n = sample + 1
if n != sampleCount:
upper = (np.sqrt(2*mvar)*special.erfinv(2*n/sampleCount-1)
+ mmean) # ub of sample interval
else:
upper = np.sqrt(2*mvar)*special.erfinv(0.9999) + mmean
sampleVal = np.random.uniform(lower, upper) # draw a random sample
# within the interval
samples.append(sampleVal)
lower = upper # set current ub as the lb for next interval
elif distrib == 'triangle':
mmin = distribSpecs[0].value
mmax = distribSpecs[1].value
mmode= distribSpecs[2].value
samples = []
for sample in range(sampleCount):
n = sample + 1
intervalarea = 1/sampleCount
ylower = intervalarea*(n-1) # use cdf to read off area as y's &
yupper = intervalarea*(n) # get corresponding x's for the pdf
# Check to see if y values = cdf(x <= mmode)
# for calculating correxponding x values:
if ylower <= ((mmode - mmin)/(mmax - mmin)):
lower = np.sqrt(ylower*(mmax - mmin)*(mmode - mmin)) + mmin
else:
lower = mmax-np.sqrt((1 - ylower)*(mmax - mmin)*(mmax - mmode))
if yupper <= ((mmode - mmin)/(mmax - mmin)):
upper = np.sqrt(yupper*(mmax - mmin)*(mmode - mmin)) + mmin;
else:
upper = mmax-np.sqrt((1 - yupper)*(mmax - mmin)*(mmax - mmode))
sampleVal = np.random.uniform(lower, upper)
samples.append(sampleVal)
b = int(np.ceil(sampleCount/10))
plt.hist(samples, density = 1, bins = b)
B=str(b)
plt.title('Histogram of ' + modelParamName
+ ' parameter samples for ' + B + ' bins')
plt.ylabel('proportion of samples');
plt.xlabel(modelParamName + ' value')
plt.show()
return samples
###Output
_____no_output_____
###Markdown
Calls the function to ask for user input to name parameters and specify distributions. Type these in text input boxes and dropdowns that will appear below after running the cell.
###Code
params = {}
for i in range(parameterCount):
s=str(i)
params[i] = interactive(parNameDist,
Name='Type parameter ' + s + ' name',
Distribution=['uniform','normal','triangle'])
display(params[i])
###Output
_____no_output_____
###Markdown
Input parameter distribution specifics in the interactive boxes that appear below after running this cell.
###Code
distribSpecs={}
for i in range(parameterCount):
parName = params[i].result['Name']
print('Enter distribution specifics for parameter ' + parName + ':')
if params[i].result['Dist'] == 'normal':
distribSpecs[parName] = {}
distribSpecs[parName][0] = widgets.FloatText(
value=2,
description='Mean:'
)
distribSpecs[parName][1] = widgets.FloatText(
value=1,
description='Variance:'
)
display(distribSpecs[parName][0], distribSpecs[parName][1])
elif params[i].result['Dist'] == 'uniform':
distribSpecs[parName] = {}
distribSpecs[parName][0] = widgets.FloatText(
value=0,
description='Minimum:'
)
distribSpecs[parName][1] = widgets.FloatText(
value=2,
description='Maximum:'
)
display(distribSpecs[parName][0], distribSpecs[parName][1])
elif params[i].result['Dist'] == 'triangle':
distribSpecs[parName] = {}
distribSpecs[parName][0] = widgets.FloatText(
value=0,
description='Minimum:'
)
distribSpecs[parName][1] = widgets.FloatText(
value=2,
description='Maximum:'
)
distribSpecs[parName][2] = widgets.FloatText(
value=1,
description='Mode:'
)
display(distribSpecs[parName][0], distribSpecs[parName][1], distribSpecs[parName][2])
###Output
Enter distribution specifics for parameter Type parameter 0 name:
###Markdown
This passes the distributions to the code for generating parameter samples, and histogram plots of samples for each parameter will appear below.
###Code
parameters = {}
for j in range(parameterCount):
parameters[params[j].result['Name']] = sampleDistrib(params[j].result['Name'],
params[j].result['Dist'],
distribSpecs[params[j].result['Name']])
###Output
_____no_output_____
###Markdown
Randomly permute each set of parameter samples in order to randomly pair the samples to more fully sample the parameter space for the Monte Carlo simulations.
###Code
LHSparams=[]
for p in parameters:
temp = parameters[p]
random.shuffle(temp)
LHSparams.append(temp)
###Output
_____no_output_____
###Markdown
Define your model function. Two examples have been provided below: (1) a linear function with two sampled parameters: slope and intercept, and (2) a Lotka-Volterra predator - prey model.Note that the order and number of the parameters needs to match the order and number of parameters speficied above to ensure accuracy when the model is solved below.
###Code
def testlinear(x,sampledParams,unsampledParams):
m = sampledParams[0]
b = sampledParams[1]
a = unsampledParams
y = m * x + b + a;
return y
def myodes(y, t, sampledParams, unsampledParams):
q, r = y # unpack current values of y
alpha, beta = sampledParams # unpack sampled parameters
delta, lambdaa, gamma = unsampledParams # unpack unsampled parameters
derivs = [alpha*q*r - lambdaa*q, # list of dy/dt=f functions
beta*r - gamma*q*r - delta*r]
return derivs
###Output
_____no_output_____
###Markdown
Run Monte Carlo simulations for each parameter sample set. *Be sure to specify a call to your model function and any necessary arguments below.*
###Code
# EDIT THE FOLLOWING VARIABLES, UNSAMPLED PARAMETERS, & ANY OTHER ARGS HERE,
# AS WELL AS THE CALL TO YOUR OWN MODEL FUNCTION INSIDE THE FOR LOOP BELOW
x = np.linspace(0, 10, num=101)
unsampledParams = 2;
Output = []
for j in range(sampleCount):
sampledParams=[i[j] for i in LHSparams]
sol = testlinear(x,sampledParams,unsampledParams)
Output.append(sol)
# EDIT THE STRING TO NAME YOUR SIM OUTPUT (for fig labels, filenames):
labelstring = 'y'
# # EXAMPLE CODE FOR A COUPLED ODE MODEL:
# import scipy.integrate as spi
# t = np.linspace(0,17,num=171) # time domain for myodes
# # odesic = [q0, r0]
# odesic = [500,1000]
# lambdaa = np.log(2)/7
# delta = 0.5
# gamma = 1
# unsampledParams = [lambdaa, delta, gamma]
# Simdata={}
# Output = []
# for i in range(sampleCount):
# Simdata[i]={}
# Simdata[i]['q']=[]
# Simdata[i]['r']=[]
# for j in range(sampleCount):
# sampledParams=[i[j] for i in LHSparams]
# sol=spi.odeint(myodes, odesic, t, args=(sampledParams,unsampledParams))
# Simdata[j]['q'] = sol[:,0] # solution to the equation for variable r
# Simdata[j]['r'] = sol[:,1] # solution to the equation for variable s
# Ratio = np.divide(sol[:,0],sol[:,1]) # compute ratio to compare w/ param samples
# Output.append(Ratio)
# labelstring = 'predator to prey ratio (q/r)'; # id for fig labels, filenames
###Output
_____no_output_____
###Markdown
Plot the range of simulation output generated by the all of the Monte Carlo simulations using errorbars.
###Code
yavg = np.mean(Output, axis=0)
yerr = np.std(Output, axis=0)
plt.errorbar(t,yavg,yerr)
plt.xlabel('x')
# plt.xlabel('time (days)') # for myodes
plt.ylabel(labelstring)
plt.title('Error bar plot of ' + labelstring + ' from LHS simulations')
plt.show()
###Output
_____no_output_____
###Markdown
Compute partial rank correlation coefficients to compare simulation outputs with parameters
###Code
SampleResult=[]
x_idx = 11 # time or location index of sim results
x_idx2= x_idx+1 # to compare w/ param sample vals
LHS=[*zip(*LHSparams)]
LHSarray=np.array(LHS)
Outputarray=np.array(Output)
subOut=Outputarray[0:,x_idx:x_idx2]
LHSout = np.hstack((LHSarray,subOut))
SampleResult = LHSout.tolist()
Ranks=[]
for s in range(sampleCount):
indices = list(range(len(SampleResult[s])))
indices.sort(key=lambda k: SampleResult[s][k])
r = [0] * len(indices)
for i, k in enumerate(indices):
r[k] = i
Ranks.append(r)
C=np.corrcoef(Ranks);
if np.linalg.det(C) < 1e-16: # determine if singular
Cinv = np.linalg.pinv(C) # may need to use pseudo inverse
else:
Cinv = np.linalg.inv(C)
resultIdx = parameterCount+1
prcc=np.zeros(resultIdx)
for w in range(parameterCount): # compute PRCC btwn each param & sim result
prcc[w]=-Cinv[w,resultIdx]/np.sqrt(Cinv[w,w]*Cinv[resultIdx,resultIdx])
###Output
_____no_output_____
###Markdown
Plot the PRCCs for each parameter
###Code
xp=[i for i in range(parameterCount)]
plt.bar(xp,prcc[0:parameterCount], align='center')
bLabels=list(parameters.keys())
plt.xticks(xp, bLabels)
plt.ylabel('PRCC value');
N=str(sampleCount)
loc=str(x_idx)
plt.title('Partial rank correlation of params with ' + labelstring
+ ' results \n from ' + N + ' LHS sims, at x = ' +loc);
plt.show()
###Output
_____no_output_____
###Markdown
Can also do PRCCs over time...
###Code
SampleResult=[]
resultIdx = parameterCount+1
prcc=np.zeros((resultIdx,len(x)))
LHS=[*zip(*LHSparams)]
LHSarray=np.array(LHS)
Outputarray=np.array(Output)
for xi in range(len(x)): # loop through time or location of sim results
xi2 = xi+1 # to compare w/ parameter sample vals
subOut = Outputarray[0:,xi:xi2]
LHSout = np.hstack((LHSarray,subOut))
SampleResult = LHSout.tolist()
Ranks=[]
for s in range(sampleCount):
indices = list(range(len(SampleResult[s])))
indices.sort(key=lambda k: SampleResult[s][k])
r = [0] * len(indices)
for i, k in enumerate(indices):
r[k] = i
Ranks.append(r)
C=np.corrcoef(Ranks);
if np.linalg.det(C) < 1e-16: # determine if singular
Cinv = np.linalg.pinv(C) # may need to use pseudo inverse
else:
Cinv = np.linalg.inv(C)
for w in range(parameterCount): # compute PRCC btwn each param & sim result
prcc[w,xi]=-Cinv[w,resultIdx]/np.sqrt(Cinv[w,w]*Cinv[resultIdx,resultIdx])
###Output
_____no_output_____
###Markdown
Plot PRCC values as they vary over time or space. *Notice PRCC can change with respect to the independent variable (x-axis). This may be helpful for certain applications, as opposed to only looking at a "snapshot."*
###Code
for p in range(parameterCount):
plt.plot(x,prcc[p,])
labels=list(parameters.keys())
plt.legend(labels)
plt.ylabel('PRCC value');
plt.xlabel('x')
N=str(sampleCount)
plt.title('Partial rank correlation of params with ' + labelstring
+ ' results \n from ' + N + ' LHS sims');
plt.show()
###Output
_____no_output_____
###Markdown
Latin Hypercube Sampling & Partial Rank Correlation Coefficients *~ a method for analyzing model sensitivity to parameters ~* Importing packages that will be used.
###Code
import numpy as np
from scipy import special
import random
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
from IPython.display import display
import pandas as pd
import matplotlib.pyplot as plt
###Output
_____no_output_____
###Markdown
Specify the number of parameters to sample and the number of samples to draw from each parameter distribution. *Do not include any parameters that should be left fixed in parameterCount - those will be specified later. When choosing number of samples to draw, note that more samples (~ 1000) yields better results while fewer (~50) is faster for testing, since it requires fewer model solves.*
###Code
# Number of parameters to sample
parameterCount = 4;
# Number of samples to draw for each parameter
sampleCount = 100; # 1000 plus
###Output
_____no_output_____
###Markdown
This defines functions for specifying sampled parameters' names and distributions as well as drawing samples from a user-specified parameter distribution. Does not need any user edits.
###Code
def parNameDist(Name,Distribution):
paramTemp = {}
paramTemp['Name']=Name
paramTemp['Dist']=Distribution
return paramTemp
def sampleDistrib(modelParamName,distrib,distribSpecs):
if distrib == 'uniform':
mmin = distribSpecs[0].value
mmax = distribSpecs[1].value
intervalwidth = (mmax - mmin) / sampleCount # width of each
# sampling interval
samples = []
for sample in range(sampleCount):
lower = mmin + intervalwidth * (sample-1) # lb of interval
upper = mmin + intervalwidth * (sample) # ub of interval
sampleVal = np.random.uniform(lower, upper) # draw a random sample
# within the interval
samples.append(sampleVal)
elif distrib == 'normal':
mmean= distribSpecs[0].value
mvar = distribSpecs[1].value
lower = mvar*np.sqrt(2)*special.erfinv(-0.9999)+mmean # set lb of 1st
# sample interval
samples = []
for sample in range(sampleCount):
n = sample + 1
if n != sampleCount:
upper = (np.sqrt(2*mvar)*special.erfinv(2*n/sampleCount-1)
+ mmean) # ub of sample interval
else:
upper = np.sqrt(2*mvar)*special.erfinv(0.9999) + mmean
sampleVal = np.random.uniform(lower, upper) # draw a random sample
# within the interval
samples.append(sampleVal)
lower = upper # set current ub as the lb for next interval
elif distrib == 'triangle':
mmin = distribSpecs[0].value
mmax = distribSpecs[1].value
mmode= distribSpecs[2].value
samples = []
for sample in range(sampleCount):
n = sample + 1
intervalarea = 1/sampleCount
ylower = intervalarea*(n-1) # use cdf to read off area as y's &
yupper = intervalarea*(n) # get corresponding x's for the pdf
# Check to see if y values = cdf(x <= mmode)
# for calculating correxponding x values:
if ylower <= ((mmode - mmin)/(mmax - mmin)):
lower = np.sqrt(ylower*(mmax - mmin)*(mmode - mmin)) + mmin
else:
lower = mmax-np.sqrt((1 - ylower)*(mmax - mmin)*(mmax - mmode))
if yupper <= ((mmode - mmin)/(mmax - mmin)):
upper = np.sqrt(yupper*(mmax - mmin)*(mmode - mmin)) + mmin;
else:
upper = mmax-np.sqrt((1 - yupper)*(mmax - mmin)*(mmax - mmode))
sampleVal = np.random.uniform(lower, upper)
samples.append(sampleVal)
b = int(np.ceil(sampleCount/10))
plt.hist(samples, density = 1, bins = b)
B=str(b)
plt.title('Histogram of ' + modelParamName
+ ' parameter samples for ' + B + ' bins')
plt.ylabel('proportion of samples');
plt.xlabel(modelParamName + ' value')
plt.show()
return samples
###Output
_____no_output_____
###Markdown
Calls the function to ask for user input to name parameters and specify distributions. Type these in text input boxes and dropdowns that will appear below after running the cell.
###Code
params = {}
for i in range(parameterCount):
s=str(i)
params[i] = interactive(parNameDist,
Name='Type parameter ' + s + ' name',
Distribution=['uniform','normal','triangle'])
display(params[i])
params
###Output
_____no_output_____
###Markdown
Input parameter distribution specifics in the interactive boxes that appear below after running this cell.
###Code
distribSpecs={}
for i in range(parameterCount):
parName = params[i].result['Name']
print('Enter distribution specifics for parameter ' + parName + ':')
if params[i].result['Dist'] == 'normal':
distribSpecs[parName] = {}
distribSpecs[parName][0] = widgets.FloatText(
value=2,
description='Mean:'
)
distribSpecs[parName][1] = widgets.FloatText(
value=1,
description='Variance:'
)
display(distribSpecs[parName][0], distribSpecs[parName][1])
elif params[i].result['Dist'] == 'uniform':
distribSpecs[parName] = {}
distribSpecs[parName][0] = widgets.FloatText(
value=0,
description='Minimum:'
)
distribSpecs[parName][1] = widgets.FloatText(
value=2,
description='Maximum:'
)
display(distribSpecs[parName][0], distribSpecs[parName][1])
elif params[i].result['Dist'] == 'triangle':
distribSpecs[parName] = {}
distribSpecs[parName][0] = widgets.FloatText(
value=0,
description='Minimum:'
)
distribSpecs[parName][1] = widgets.FloatText(
value=2,
description='Maximum:'
)
distribSpecs[parName][2] = widgets.FloatText(
value=1,
description='Mode:'
)
display(distribSpecs[parName][0], distribSpecs[parName][1], distribSpecs[parName][2])
# parameter of interest
a=1.5
b = 1.64
delta = 4.0
beta = 2
lf = 0.50
uf = 2
varbound=np.array([[a*lf,a*uf],[b*lf,b*uf],[lf*delta,uf*delta],[lf*beta,uf*beta]])
varbound
###Output
_____no_output_____
###Markdown
This passes the distributions to the code for generating parameter samples, and histogram plots of samples for each parameter will appear below.
###Code
parameters = {}
for j in range(parameterCount):
parameters[params[j].result['Name']] = sampleDistrib(params[j].result['Name'],
params[j].result['Dist'],
distribSpecs[params[j].result['Name']])
###Output
_____no_output_____
###Markdown
Randomly permute each set of parameter samples in order to randomly pair the samples to more fully sample the parameter space for the Monte Carlo simulations.
###Code
LHSparams=[]
for p in parameters:
temp = parameters[p]
random.shuffle(temp)
LHSparams.append(temp)
# parameters
# LHSparams
###Output
_____no_output_____
###Markdown
Our IDM/FIDM model packages
###Code
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Mon Jan 24 15:17:39 2022
@author: rafiul
"""
# import scipy.integrate as integrate
# from scipy.integrate import odeint
import sys
import os
# from geneticalgorithm import geneticalgorithm as ga
#from geneticalgorithm_pronto import geneticalgorithm as ga
# from ga import ga
import numpy as np
import scipy.integrate as integrate
from scipy import special
from scipy.interpolate import interp1d
import pandas as pd
###Output
_____no_output_____
###Markdown
Functions
###Code
def RK4(func, X0, ts):
"""
Runge Kutta 4 solver.
"""
dt = ts[1] - ts[0]
nt = len(ts)
X = np.zeros((nt, X0.shape[0]),dtype=np.float64)
X[0] = X0
for i in range(nt-1):
k1 = func(X[i], ts[i])
k2 = func(X[i] + dt/2. * k1, ts[i] + dt/2.)
k3 = func(X[i] + dt/2. * k2, ts[i] + dt/2.)
k4 = func(X[i] + dt * k3, ts[i] + dt)
X[i+1] = X[i] + dt / 6. * (k1 + 2. * k2 + 2. * k3 + k4)
return X
# see this link for model and paramterts https://en.wikipedia.org/wiki/Intelligent_driver_model
# DOI: 10.1098/rsta.2010.0084
# @jit(nopython=True)
def idm_model(x,t):
X,V = x[0],x[1]
dX,dV = np.zeros(1,dtype=np.float64), np.zeros(1,dtype=np.float64)
dX = V # Differtial Equation 1
###
s = position_LV(t) - X - 5 # 5 = length of the car
deltaV = V - speed_LV(t)
sstar = s0+V*T + (V*deltaV)/(2*np.sqrt(a*b))
# ###
dV = a*(1-(V/V_0)**delta - (sstar/s)**2) # Differtial Equation 2
return np.array([dX,dV],dtype=np.float64)
# @jit(nopython=True)
def speed_LV(t):
return interp1d(nth_car_data['time'],nth_car_data['speed'],bounds_error=False)(t)
def position_LV(t):
return interp1d(nth_car_data['time'],postion_of_the_LV,bounds_error=False)(t)
def fractional_idm_model_1d(V,t,X):
# index = round(t) #convert into integer number
current_position_of_follower = X
###
s = position_LV(t) - current_position_of_follower - 5 # 5 = length of the car
deltaV = V - speed_LV(t)
sstar = s0+V*T + (V*deltaV)/(2*np.sqrt(a*b))
# ###
dV = a*(1-(V/V_0)**delta - (sstar/s)**2) # Differtial Equation 2
return dV
def speed_error(sol,nth_car_speed):
return np.sum((sol[1,:-1]-nth_car_speed[1:])**2)
def gap_error(sol,postion_of_the_LV):
return np.sum((sol[0,:]-postion_of_the_LV)**2)
def caputoEuler_1d(a, f, y0, tspan, x0_f):
"""Use one-step Adams-Bashforth (Euler) method to integrate Caputo equation
D^a y(t) = f(y,t)
Args:
a: fractional exponent in the range (0,1)
f: callable(y,t) returning a numpy array of shape (d,)
Vector-valued function to define the right hand side of the system
y0: array of shape (d,) giving the initial state vector y(t==0)
tspan (array): The sequence of time points for which to solve for y.
These must be equally spaced, e.g. np.arange(0,10,0.005)
tspan[0] is the intial time corresponding to the initial state y0.
Returns:
y: array, with shape (len(tspan), len(y0))
With the initial value y0 in the first row
Raises:
FODEValueError
See also:
K. Diethelm et al. (2004) Detailed error analysis for a fractional Adams
method
C. Li and F. Zeng (2012) Finite Difference Methods for Fractional
Differential Equations
"""
#(d, a, f, y0, tspan) = _check_args(a, f, y0, tspan)
N = len(tspan)
h = (tspan[N-1] - tspan[0])/(N - 1)
c = special.rgamma(a) * np.power(h, a) / a
w = c * np.diff(np.power(np.arange(N), a))
fhistory = np.zeros(N - 1, dtype=np.float64)
y = np.zeros(N, dtype=np.float64)
x = np.zeros(N, dtype=np.float64)
y[0] = y0;
x[0] = x0_f;
for n in range(0, N - 1):
tn = tspan[n]
yn = y[n]
fhistory[n] = f(yn, tn, x[n])
y[n+1] = y0 + np.dot(w[0:n+1], fhistory[n::-1])
x[n+1] = x[n] + y[n+1] * h
return np.array([x,y])
def error_func_idm(variable_X):
# varbound=np.array([[a*lf,a*uf],[lf*delta,uf*delta],[lf*beta,uf*beta]])
a = variable_X[0]
delta = variable_X[1]
beta = variable_X[2]
x0 = np.array([initial_position,initial_velocity],dtype=np.float64) #initial position and velocity
# Classical ODE
# sol = integrate.odeint(idm_model, x0, time_span)
sol = RK4(idm_model, x0, time_span)
sol = sol.transpose(1,0)
# print(np.sum((sol[1,:-1]-nth_car_speed[1:])**2))
return np.sum((sol[1,1:]-nth_car_speed[:-1])**2)
def error_func_fidm(variable_X):
# varbound=np.array([[a*lf,a*uf],[lf*delta,uf*delta],[lf*beta,uf*beta]])
a = variable_X[0]
delta = variable_X[1]
beta = variable_X[2]
alpha = variable_X[3]
if alpha > .99999:
alpha = .99999
sol = caputoEuler_1d(alpha,fractional_idm_model_1d, initial_velocity, time_span, initial_position) #, args=(number_groups,beta_P,beta_C,beta_A,v,w,mu_E,mu_A,mu_P,mu_C,p,q,contact_by_group))
return np.sum((sol[1,1:]-nth_car_speed[:-1])**2)
# np.array(Output).reshape(len(Output),1)
###Output
_____no_output_____
###Markdown
Run Monte Carlo simulations for each parameter sample set. *Be sure to specify a call to your model function and any necessary arguments below.*
###Code
######################################
# Global variables
# see this link for model and paramterts https://en.wikipedia.org/wiki/Intelligent_driver_model
V_0 = 20 # desired speed m/s
s0 = 30
T = 1.5
nth_car = 3
# a=1.5
# b = 1.67
# delta = 4.0
# beta = 2
# find best values for our model
# a_alpha = 1.2
# ######################################
# Actual data
# df = pd.read_csv('RAllCarDataTime350.csv')
git_raw_url = 'https://raw.githubusercontent.com/m-rafiul-islam/driver-behavior-model/main/RAllCarDataTime350.csv'
df = pd.read_csv(git_raw_url)
nth_car_data = df.loc[df['nthcar'] == nth_car, :]
nth_car_speed = np.array(df.loc[df['nthcar'] == nth_car,'speed'])
# leader vehicle profile
# 7 m/s - 25.2 km/h 11 m/s - 39.6 km/h 18 m/s - 64.8 km/h 22 m/s - 79.2 km/h
# 25 km/h -- 6.95 m/s 40 km/h -- 11.11 m/s 60 km/h -- 16.67 m/s
# dt=1 #time step -- 1 sec
time_span = np.array(nth_car_data['time'])
dt = time_span[1]-time_span[0]
# speed_of_the_LV = 15*np.ones(600+1) # we will need data
# speed_of_the_LV = np.concatenate((np.linspace(0,7,60),7*np.ones(120),np.linspace(7,11,60), 11*np.ones(120), np.linspace(11,0,60) ))# we will need data
speed_of_the_LV = nth_car_speed
num_points = len(speed_of_the_LV)
postion_of_the_LV = np.zeros(num_points)
initla_position_of_the_LV = 18.45 # 113
postion_of_the_LV[0] = initla_position_of_the_LV
for i in range(1,num_points):
postion_of_the_LV[i] = postion_of_the_LV[i-1] + dt*(speed_of_the_LV[i]+speed_of_the_LV[i-1])/2
initial_position = 0.
initial_velocity = 6.72
x0 = np.array([initial_position,initial_velocity],dtype=np.float64) #initial position and velocity
alpha =1
# a=1.5
b = 1.67
# delta = 4.0
# beta = 2
Output = []
for j in range(sampleCount):
sampledParams=[i[j] for i in LHSparams]
SSE = error_func_idm(sampledParams)
Output.append(SSE)
###Output
_____no_output_____
###Markdown
Plot the range of simulation output generated by the all of the Monte Carlo simulations using errorbars.
###Code
# yavg = np.mean(Output, axis=0)
# yerr = np.std(Output, axis=0)
# plt.errorbar(t,yavg,yerr)
# plt.xlabel('x')
# # plt.xlabel('time (days)') # for myodes
# plt.ylabel(labelstring)
# plt.title('Error bar plot of ' + labelstring + ' from LHS simulations')
# plt.show()
###Output
_____no_output_____
###Markdown
Compute partial rank correlation coefficients to compare simulation outputs with parameters
###Code
#
LHSout = np.hstack((LHSarray,np.array(Output).reshape(len(Output),1)))
SampleResult = LHSout.tolist()
Ranks=[]
for s in range(sampleCount):
indices = list(range(len(SampleResult[s])))
indices.sort(key=lambda k: SampleResult[s][k])
r = [0] * len(indices)
for i, k in enumerate(indices):
r[k] = i
Ranks.append(r)
C=np.corrcoef(Ranks);
if np.linalg.det(C) < 1e-16: # determine if singular
Cinv = np.linalg.pinv(C) # may need to use pseudo inverse
else:
Cinv = np.linalg.inv(C)
resultIdx = parameterCount+1
prcc=np.zeros(resultIdx)
for w in range(parameterCount): # compute PRCC btwn each param & sim result
prcc[w]=-Cinv[w,resultIdx]/np.sqrt(Cinv[w,w]*Cinv[resultIdx,resultIdx])
xp=[i for i in range(parameterCount)]
plt.bar(xp,prcc[0:parameterCount], align='center')
bLabels=list(parameters.keys())
plt.xticks(xp, bLabels)
plt.ylabel('PRCC value');
plt.show()
###Output
_____no_output_____ |
notebooks/tutorials/landscape_evolution/smooth_threshold_eroder/stream_power_smooth_threshold_eroder.ipynb | ###Markdown
The `StreamPowerSmoothThresholdEroder` componentLandlab's `StreamPowerSmoothThresholdEroder` (here SPSTE for short) is a fluvial landscape evolution component that uses a thresholded form of the stream power erosion law. The novel aspect is that the threshold takes a smoothed form rather than an abrupt mathematical discontinuity: as long as slope and drainage area are greater than zero, there is always *some* erosion rate even if the erosive potential function is below the nominal threshold value. This approach is motivated by the finding that mathematically discontinuous functions in numerical models can lead to "numerical daemons": non-smooth functional behavior that can greatly complicate optimization (Clark & Kavetski, 2010; Kavetski & Clark, 2010, 2011). The SPSTE is one of the fluvial erosion components used in the *terrainBento* collection of landscape evolution models (Barnhart et al., 2019).This tutorial provides a brief overview of how to use the SPSTE component.*(G.E. Tucker, 2021)* TheoryThe SPSTE formulation is as follows. Consider a location on a stream channel that has local downstream slope gradient $S$ and drainage area $A$. We define an erosion potential function $\omega$ as$$\omega = KA^mS^n$$where $K$ is an erodibility coefficient with dimensions of $[L^{(1-2m)}/T]$. The erosion potential function has dimensions of erosion (lowering) rate, [L/T], and it represents the rate of erosion that would occur if there were no threshold term. The expression takes the form of the familiar area-slope erosion law, also known as the "stream power law" because the exponents can be configured to represent an erosion law that depends on stream power per unit bed area (Whipple & Tucker, 1999). A common choice of exponents is $m=1/2$, $n=1$, but other combinations are possible depending on one's assumptions about process, hydrology, channel geometry, and other factors (e.g., Howard et al., 1994; Whipple et al., 2000).We also define a threshold erosion potential function, $\omega_c$, below which erosion rate declines precipitously. Given these definitions, a mathematically discontinuous threshold erosion function would look like this:$$E = \max (\omega - \omega_c, 0)$$This kind of formulation is mathematically simple, and given data on $E$ and $\omega$, one could easily find $K$ and $\omega_c$ empirically by fitting a line. Yet even in the case of sediment transport, where the initial motion of grains is usually represented by a threshold shear stress (often referred to as the *critical shear stress* for initiation of sediment motion), we know that *some* transport still occurs below the nominal threshold (e.g, Wilcock & McArdell, 1997). Although it is undeniably true that the rate of sediment transport declines rapidly when the average shear stress drops below a critical value, the strictly linear-with-threshold formulation is really more of convenient mathematical fiction than an accurate reflection of geophysical reality. In bed-load sediment transport, reality seems to be smoother than this mathematical fiction, if one transport rates averaged over a suitably long time period. The same is likely true for the hydraulic detachment and removal of cohesive/rocky material as well. Furthermore, as alluded to above, a strict threshold expression for transport or erosion can create numerical daemons that complicate model analysis.To avoid the mathematical discontinuity at $\omega=\omega_c$, SPSTE uses a smoothed version of the above function:$$E = \omega - \omega_c \left( 1 - e^{-\omega / \omega_c} \right)$$The code below generates a plot that compares the strict threshold and smooth threshold erosion laws.
###Code
import numpy as np
import matplotlib.pyplot as plt
from landlab import RasterModelGrid, imshow_grid
from landlab.components import FlowAccumulator, StreamPowerSmoothThresholdEroder
omega = np.arange(0, 5.01, 0.01)
omegac = 1.0
Eabrupt = np.maximum(omega - omegac, 0.0)
Esmooth = omega - omegac * (1.0 - np.exp(-omega / omegac))
plt.plot(omega, Esmooth, "k", label="Smoothed threshold")
plt.plot(omega, Eabrupt, "k--", label="Hard threshold")
plt.plot([1.0, 1.0], [0.0, 4.0], "g:", label=r"$\omega=\omega_c$")
plt.xlabel(r"Erosion potential function ($\omega$)")
plt.ylabel("Erosion rate")
plt.legend()
###Output
_____no_output_____
###Markdown
Notice that the SPSTE formulation effectively smooths over the sharp discontinuity at $\omega = \omega_c$. EquilibriumConsider a case of steady, uniform fluvial erosion. Let the ratio of the erosion potential function to its threshold value be a constant, as$$\beta = \omega / \omega_c$$This allows us to replace instances of $\omega_c$ with $(1/\beta) \omega$,$$E = KA^m S^n - \frac{1}{\beta} KA^m S^n \left( 1 - e^{-\beta} \right)$$$$ = K A^m S^n \left( 1 - \frac{1}{\beta} \left( 1 - e^{-\beta} \right)\right)$$Let$$\alpha = \left( 1 - \frac{1}{\beta} \left( 1 - e^{-\beta} \right)\right)$$Then we can solve for the steady-state slope as$$\boxed{S = \left( \frac{E}{\alpha K A^m} \right)^{1/n}}$$We can relate $\beta$ and $\omega_c$ via$$\omega_c = E / (1-\beta (1 - e^{-\beta} ))$$ UsageHere we get a summary of the component's usage and input parameters by printing out the component's header docstring:
###Code
print(StreamPowerSmoothThresholdEroder.__doc__)
###Output
_____no_output_____
###Markdown
ExampleHere we'll run a steady-state example with $\beta = 1$. To do this, we'll start with a slightly inclined surface with some superimposed random noise, and subject it to a steady rate of rock uplift relative to baselevel, $U$, until it reaches a steady state.
###Code
# Parameters
K = 0.0001 # erodibility coefficient, 1/yr
m = 0.5 # drainage area exponent
beta = 1.0 # ratio of w / wc [-]
uplift_rate = 0.001 # rate of uplift relative to baselevel, m/yr
nrows = 16 # number of grid rows (small for speed)
ncols = 25 # number of grid columns (")
dx = 100.0 # grid spacing, m
dt = 1000.0 # time-step duration, yr
run_duration = 2.5e5 # duration of run, yr
init_slope = 0.001 # initial slope gradient of topography, m/m
noise_amplitude = 0.1 # amplitude of random noise on init. topo.
# Derived parameters
omega_c = uplift_rate / (beta - (1 - np.exp(-beta)))
nsteps = int(run_duration / dt)
# Create grid and elevation field with initial ramp
grid = RasterModelGrid((nrows, ncols), xy_spacing=dx)
grid.set_closed_boundaries_at_grid_edges(True, True, True, False)
elev = grid.add_zeros("topographic__elevation", at="node")
elev[:] = init_slope * grid.y_of_node
np.random.seed(0)
elev[grid.core_nodes] += noise_amplitude * np.random.rand(grid.number_of_core_nodes)
# Display starting topography
imshow_grid(grid, elev)
# Instantiate the two components
# (note that m=0.5, n=1 are the defaults for SPSTE)
fa = FlowAccumulator(grid, flow_director="D8")
spste = StreamPowerSmoothThresholdEroder(grid, K_sp=K, threshold_sp=omega_c)
# Run the model
for i in range(nsteps):
# flow accumulation
fa.run_one_step()
# uplift / baselevel
elev[grid.core_nodes] += uplift_rate * dt
# erosion
spste.run_one_step(dt)
# Display the final topopgraphy
imshow_grid(grid, elev)
# Calculate the analytical solution in slope-area space
alpha = 1.0 - (1.0 / beta) * (1.0 - np.exp(-beta))
area_pred = np.array([1.0e4, 1.0e6])
slope_pred = uplift_rate / (alpha * K * area_pred ** m)
# Plot the slope-area relation and compare with analytical
area = grid.at_node["drainage_area"]
slope = grid.at_node["topographic__steepest_slope"]
cores = grid.core_nodes
plt.loglog(area[cores], slope[cores], "k.")
plt.plot(area_pred, slope_pred)
plt.legend(["Numerical", "Analytical"])
plt.title("Equilibrium slope-area relation")
plt.xlabel(r"Drainage area (m$^2$)")
_ = plt.ylabel("Slope (m/m)")
###Output
_____no_output_____
###Markdown
The above plot shows that the simulation has reached steady state, and that the slope-area relation matches the analytical solution.We can also inspect the erosion potential function, which should be uniform in space, and (because $\beta = 1$ in this example) equal to the threshold $\omega_c$. We can also compare this with the uplift rate and the erosion-rate function:
###Code
# Plot the erosion potential function
omega = K * area[cores] ** m * slope[cores]
plt.plot([0.0, 1.0e6], [omega_c, omega_c], "g", label=r"$\omega_c$")
plt.plot(area[cores], omega, ".", label=r"$\omega$")
plt.plot([0.0, 1.0e6], [uplift_rate, uplift_rate], "r", label=r"$U$")
erorate = omega - omega_c * (1.0 - np.exp(-omega / omega_c))
plt.plot(
area[cores], erorate, "k+", label=r"$\omega - \omega_c (1 - e^{-\omega/\omega_c})$"
)
plt.ylim([0.0, 2 * omega_c])
plt.legend()
plt.title("Erosion potential function vs. threshold term")
plt.xlabel(r"Drainage area (m$^2$)")
_ = plt.ylabel("Erosion potential function (m/yr)")
###Output
_____no_output_____
###Markdown
The `StreamPowerSmoothThresholdEroder` componentLandlab's `StreamPowerSmoothThresholdEroder` (here SPSTE for short) is a fluvial landscape evolution component that uses a thresholded form of the stream power erosion law. The novel aspect is that the threshold takes a smoothed form rather than an abrupt mathematical discontinuity: as long as slope and drainage area are greater than zero, there is always *some* erosion rate even if the erosive potential function is below the nominal threshold value. This approach is motivated by the finding that mathematically discontinuous functions in numerical models can lead to "numerical daemons": non-smooth functional behavior that can greatly complicate optimization (Clark & Kavetski, 2010; Kavetski & Clark, 2010, 2011). The SPSTE is one of the fluvial erosion components used in the *terrainBento* collection of landscape evolution models (Barnhart et al., 2019).This tutorial provides a brief overview of how to use the SPSTE component.*(G.E. Tucker, 2021)* TheoryThe SPSTE formulation is as follows. Consider a location on a stream channel that has local downstream slope gradient $S$ and drainage area $A$. We define an erosion potential function $\omega$ as$$\omega = KA^mS^n$$where $K$ is an erodibility coefficient with dimensions of $[L^{(1-2m)}/T]$. The erosion potential function has dimensions of erosion (lowering) rate, [L/T], and it represents the rate of erosion that would occur if there were no threshold term. The expression takes the form of the familiar area-slope erosion law, also known as the "stream power law" because the exponents can be configured to represent an erosion law that depends on stream power per unit bed area (Whipple & Tucker, 1999). A common choice of exponents is $m=1/2$, $n=1$, but other combinations are possible depending on one's assumptions about process, hydrology, channel geometry, and other factors (e.g., Howard et al., 1994; Whipple et al., 2000).We also define a threshold erosion potential function, $\omega_c$, below which erosion rate declines precipitously. Given these definitions, a mathematically discontinuous threshold erosion function would look like this:$$E = \max (\omega - \omega_c, 0)$$This kind of formulation is mathematically simple, and given data on $E$ and $\omega$, one could easily find $K$ and $\omega_c$ empirically by fitting a line. Yet even in the case of sediment transport, where the initial motion of grains is usually represented by a threshold shear stress (often referred to as the *critical shear stress* for initiation of sediment motion), we know that *some* transport still occurs below the nominal threshold (e.g, Wilcock & McArdell, 1997). Although it is undeniably true that the rate of sediment transport declines rapidly when the average shear stress drops below a critical value, the strictly linear-with-threshold formulation is really more of convenient mathematical fiction than an accurate reflection of geophysical reality. In bed-load sediment transport, reality seems to be smoother than this mathematical fiction, if one transport rates averaged over a suitably long time period. The same is likely true for the hydraulic detachment and removal of cohesive/rocky material as well. Furthermore, as alluded to above, a strict threshold expression for transport or erosion can create numerical daemons that complicate model analysis.To avoid the mathematical discontinuity at $\omega=\omega_c$, SPSTE uses a smoothed version of the above function:$$E = \omega - \omega_c \left( 1 - e^{-\omega / \omega_c} \right)$$The code below generates a plot that compares the strict threshold and smooth threshold erosion laws.
###Code
import numpy as np
import matplotlib.pyplot as plt
from landlab import RasterModelGrid, imshow_grid
from landlab.components import (FlowAccumulator,
StreamPowerSmoothThresholdEroder)
omega = np.arange(0, 5.01, 0.01)
omegac = 1.0
Eabrupt = np.maximum(omega - omegac, 0.0)
Esmooth = omega - omegac * (1.0 - np.exp(-omega / omegac))
plt.plot(omega, Esmooth, 'k', label='Smoothed threshold')
plt.plot(omega, Eabrupt, 'k--', label='Hard threshold')
plt.plot([1., 1.], [0., 4.], 'g:', label=r'$\omega=\omega_c$')
plt.xlabel(r'Erosion potential function ($\omega$)')
plt.ylabel('Erosion rate')
plt.legend()
###Output
_____no_output_____
###Markdown
Notice that the SPSTE formulation effectively smooths over the sharp discontinuity at $\omega = \omega_c$. EquilibriumConsider a case of steady, uniform fluvial erosion. Let the ratio of the erosion potential function to its threshold value be a constant, as$$\beta = \omega / \omega_c$$This allows us to replace instances of $\omega_c$ with $(1/\beta) \omega$,$$E = KA^m S^n - \frac{1}{\beta} KA^m S^n \left( 1 - e^{-\beta} \right)$$$$ = K A^m S^n \left( 1 - \frac{1}{\beta} \left( 1 - e^{-\beta} \right)\right)$$Let$$\alpha = \left( 1 - \frac{1}{\beta} \left( 1 - e^{-\beta} \right)\right)$$Then we can solve for the steady-state slope as$$\boxed{S = \left( \frac{E}{\alpha K A^m} \right)^{1/n}}$$We can relate $\beta$ and $\omega_c$ via$$\omega_c = E / (1-\beta (1 - e^{-\beta} ))$$ UsageHere we get a summary of the component's usage and input parameters by printing out the component's header docstring:
###Code
print(StreamPowerSmoothThresholdEroder.__doc__)
###Output
_____no_output_____
###Markdown
ExampleHere we'll run a steady-state example with $\beta = 1$. To do this, we'll start with a slightly inclined surface with some superimposed random noise, and subject it to a steady rate of rock uplift relative to baselevel, $U$, until it reaches a steady state.
###Code
# Parameters
K = 0.0001 # erodibility coefficient, 1/yr
m = 0.5 # drainage area exponent
beta = 1.0 # ratio of w / wc [-]
uplift_rate = 0.001 # rate of uplift relative to baselevel, m/yr
nrows = 16 # number of grid rows (small for speed)
ncols = 25 # number of grid columns (")
dx = 100.0 # grid spacing, m
dt = 1000.0 # time-step duration, yr
run_duration = 2.5e5 # duration of run, yr
init_slope = 0.001 # initial slope gradient of topography, m/m
noise_amplitude = 0.1 # amplitude of random noise on init. topo.
# Derived parameters
omega_c = uplift_rate / (beta - (1 - np.exp(-beta)))
nsteps = int(run_duration / dt)
# Create grid and elevation field with initial ramp
grid = RasterModelGrid((nrows, ncols), xy_spacing=dx)
grid.set_closed_boundaries_at_grid_edges(True, True, True, False)
elev = grid.add_zeros('topographic__elevation', at='node')
elev[:] = init_slope * grid.y_of_node
np.random.seed(0)
elev[grid.core_nodes] += (noise_amplitude
* np.random.rand(grid.number_of_core_nodes))
# Display starting topography
imshow_grid(grid, elev)
# Instantiate the two components
# (note that m=0.5, n=1 are the defaults for SPSTE)
fa = FlowAccumulator(grid, flow_director='D8')
spste = StreamPowerSmoothThresholdEroder(
grid,
K_sp=K,
threshold_sp=omega_c
)
# Run the model
for i in range(nsteps):
# flow accumulation
fa.run_one_step()
# uplift / baselevel
elev[grid.core_nodes] += uplift_rate * dt
# erosion
spste.run_one_step(dt)
# Display the final topopgraphy
imshow_grid(grid, elev)
# Calculate the analytical solution in slope-area space
alpha = (1.0 - (1.0 / beta) * (1.0 - np.exp(-beta)))
area_pred = np.array([1.0e4, 1.0e6])
slope_pred = (uplift_rate / (alpha * K * area_pred**m))
# Plot the slope-area relation and compare with analytical
area = grid.at_node['drainage_area']
slope = grid.at_node['topographic__steepest_slope']
cores = grid.core_nodes
plt.loglog(area[cores], slope[cores], 'k.')
plt.plot(area_pred, slope_pred)
plt.legend(['Numerical', 'Analytical'])
plt.title('Equilibrium slope-area relation')
plt.xlabel(r'Drainage area (m$^2$)')
_ = plt.ylabel('Slope (m/m)')
###Output
_____no_output_____
###Markdown
The above plot shows that the simulation has reached steady state, and that the slope-area relation matches the analytical solution.We can also inspect the erosion potential function, which should be uniform in space, and (because $\beta = 1$ in this example) equal to the threshold $\omega_c$. We can also compare this with the uplift rate and the erosion-rate function:
###Code
# Plot the erosion potential function
omega = K * area[cores]**m * slope[cores]
plt.plot([0.0, 1.0e6], [omega_c, omega_c], 'g', label=r'$\omega_c$')
plt.plot(area[cores], omega, '.', label=r'$\omega$')
plt.plot([0.0, 1.0e6],
[uplift_rate, uplift_rate],
'r',
label=r'$U$'
)
erorate = omega - omega_c * (1.0 - np.exp(-omega / omega_c))
plt.plot(area[cores],
erorate,
'k+',
label=r'$\omega - \omega_c (1 - e^{-\omega/\omega_c})$')
plt.ylim([0.0, 2 * omega_c])
plt.legend()
plt.title('Erosion potential function vs. threshold term')
plt.xlabel(r'Drainage area (m$^2$)')
_ = plt.ylabel('Erosion potential function (m/yr)')
###Output
_____no_output_____ |
YgroupChallenge.ipynb | ###Markdown
Librerias utilizadas
###Code
import pandas as pd
import matplotlib.pyplot as plt
import ipywidgets as widgets
from ipywidgets import interact
from area import area
import ipydatetime
###Output
_____no_output_____
###Markdown
Histograma de tiempos de viaje para un año dado
###Code
def histograma_por_año(año):
df = pd.read_csv("Data/OD_{}.csv".format(año))
duration = df["duration_sec"]
plt.hist(duration, bins=20 ,histtype='bar', edgecolor='k')
plt.xlabel('Duracion del viaje(segundos)')
plt.ylabel('Numero de viajes')
plt.title('Histograma de tiempos de viaje año {}'.format(año))
return plt.show()
interact(histograma_por_año, año=[2014,2015,2016,2017])
###Output
_____no_output_____
###Markdown
Listado del Top N de estaciones más utilizadas para un año dado. Dividirlo en:- Estaciones de salida- Estaciones de llegada- En general
###Code
def listado_top_estaciones_año(año,N):
df = pd.read_csv("Data/OD_{}.csv".format(año))
start_st = pd.DataFrame({'station_code': df['start_station_code']})
end_st = pd.DataFrame({'station_code': df['end_station_code']})
general_st = start_st.append(end_st)
start_st = start_st.groupby('station_code').size().reset_index(name='counts')
start_st = start_st.sort_values(by=['counts'],ascending=False)
start_st = start_st.head(int(N))
start_st.index = range(1, int(N)+1)
end_st = end_st.groupby('station_code').size().reset_index(name='counts')
end_st = end_st.sort_values(by=['counts'],ascending=False)
end_st = end_st.head(int(N))
end_st.index = range(1, int(N)+1)
general_st = general_st.groupby('station_code').size().reset_index(name='counts')
general_st = general_st.sort_values(by=['counts'],ascending=False)
general_st = general_st.head(int(N))
general_st.index = range(1, int(N)+1)
informe = pd.concat([start_st,end_st,general_st],axis=1)
informe.columns = ["start_station_code","start_count","end_station_code","end_count","general_station_code","general_count"]
informe.to_csv("listado_top{}_estaciones_{}.csv".format(int(N),año),index=False)
return informe
interact(listado_top_estaciones_año, año=[2014,2015,2016,2017],N=(0.0,200.0,1))
###Output
_____no_output_____
###Markdown
Listado del Top N de viajes más comunes para un año dado. Donde un viaje se define por su estación de salida y de llegada
###Code
def listado_top_viajes_año(año,N):
df = pd.read_csv("Data/OD_{}.csv".format(año))
viajes = pd.DataFrame({'start_station_code': df['start_station_code'],
'end_station_code': df['end_station_code']})
viajes["viaje"] = viajes["start_station_code"].astype(str) + '-' + viajes["end_station_code"].astype(str)
viajes = viajes.groupby('viaje').size().reset_index(name='counts')
viajes = viajes.sort_values(by=['counts'],ascending=False)
viajes = viajes.head(int(N))
viajes.index = range(1, int(N)+1)
viajes.to_csv("listado_top{}_viajes_{}.csv".format(int(N),año),index=False)
return viajes
interact(listado_top_viajes_año, año=[2014,2015,2016,2017],N=(0.0,200.0,1))
###Output
_____no_output_____
###Markdown
Identificación de horas punta para un año determinado sin tener en cuenta el día. Es decir, si es día de semana, fin de semana, festivo o temporada del año.
###Code
def histograma_horas_por_año(año):
df = pd.read_csv("Data/OD_{}.csv".format(año))
hours = pd.to_datetime(df["start_date"]).dt.hour
plt.hist(hours, bins=24 ,histtype='bar', edgecolor='k')
plt.xlabel('Hora del viage')
plt.xticks(range(0,24))
plt.xlabel('Hora')
plt.ylabel('Numero de viajes')
plt.title('Histograma de horas puntas de viaje año {}'.format(año))
return plt.show()
interact(histograma_horas_por_año, año=[2014,2015,2016,2017])
###Output
_____no_output_____
###Markdown
Comparación de utilización del sistema entre dos años cualesquiera. La utilización del sistema se puede medir como:- Cantidad de viajes totales- Tiempo total de utilización del sistema- Cantidad de viajes por estaciones/bicicletas disponibles
###Code
def comparacion_sistema_año(año1,año2,medida):
df1 = pd.read_csv("Data/OD_{}.csv".format(año1))
df2 = pd.read_csv("Data/OD_{}.csv".format(año2))
if medida=="viajes totales":
plt.bar([str(año1),str(año2)],[len(df1),len(df2)])
plt.xlabel('Año')
plt.ylabel('Numero de viajes')
plt.title('Comparacion Nº viajes entre el {} y el {}'.format(año1,año2))
return plt.show()
if medida=="tiempo de uso":
uso1 = str(pd.Timedelta(df1["duration_sec"].sum(), unit ='s'))
uso2 = str(pd.Timedelta(df2["duration_sec"].sum(), unit ='s'))
usoTotal = pd.DataFrame({str(año1):uso1,
str(año2):uso2},index=[0])
return usoTotal
else:
start_st1 = pd.DataFrame({'station_code': df1['start_station_code']})
end_st1 = pd.DataFrame({'station_code': df1['end_station_code']})
general_st1 = start_st1.append(end_st1)
start_st2 = pd.DataFrame({'station_code': df2['start_station_code']})
end_st2 = pd.DataFrame({'station_code': df2['end_station_code']})
general_st2 = start_st2.append(end_st2)
st1 = general_st1.groupby('station_code').size().reset_index(name='counts')
st1.index = range(1, len(st1)+1)
st2 = general_st2.groupby('station_code').size().reset_index(name='counts')
st2.index = range(1, len(st2)+1)
comp_est = pd.concat([st1,st2],axis=1)
comp_est.columns = ["station_code_"+str(año1),"count_"+str(año1),"station_code_"+str(año2),"count_"+str(año2)]
comp_est.to_csv("comparacion_{}_{}_{}.csv".format(año1,año2,medida),index=False)
return comp_est
interact(comparacion_sistema_año, año1=[2014,2015,2016,2017],año2=[2014,2015,2016,2017],
medida=["viajes totales","tiempo de uso", "cantidad de viajes por estacion"])
###Output
_____no_output_____
###Markdown
Capacidad instalada total (suma de la capacidad total de cada estación)
###Code
stations = pd.read_json("Data/stations.json")
capacity = pd.json_normalize(stations["stations"])
print("\nCapacidad total : {}".format(capacity['ba'].sum()))
###Output
Capacidad total : 4925
###Markdown
Cambio en la capacidad instalada entre dos años puntuales
###Code
def comparacion_capacidad_año(año1,año2):
df1 = pd.read_csv("Data/Stations_{}.csv".format(año1))
df2 = pd.read_csv("Data/Stations_{}.csv".format(año2))
stations = pd.read_json("Data/stations.json")
capacity = pd.json_normalize(stations["stations"])
sum1 = capacity[capacity.n.astype(int).isin(df1["code"])]
sum2 = capacity[capacity.n.astype(int).isin(df2["code"])]
change = sum2['ba'].sum() - sum1['ba'].sum()
print("El cambio de capacidad entre los años {} y {} ha sido de : {} unidades".format(año1,año2,change))
interact(comparacion_capacidad_año, año1=[2014,2015,2016,2017],año2=[2014,2015,2016,2017])
###Output
_____no_output_____
###Markdown
Ampliación de la cobertura de la red entre dos años puntuales. La misma se puede medir como el área total que generan las estaciones.
###Code
def ampliacion_red_año(año1,año2):
df1 = pd.read_csv("Data/Stations_{}.csv".format(año1))
df2 = pd.read_csv("Data/Stations_{}.csv".format(año2))
coordinates1 = []
for i in range(len(df1)):
coordinates1.append([df1["latitude"][i],df1["longitude"][i]])
coordinates2 = []
for i in range(len(df2)):
coordinates2.append([df2["latitude"][i],df2["longitude"][i]])
coordinates1.sort(key = lambda x: (-x[0], x[1]))
coordinates2.sort(key = lambda x: (-x[0], x[1]))
obj1 = {'type':'Polygon','coordinates':[coordinates1]}
obj2 = {'type':'Polygon','coordinates':[coordinates2]}
area1 = area(obj1)
area2 = area(obj2)
print("La diferencia entre la cobertura de la red entre el {} y el {} es: {} m".format(año1,año2,area2-area1))
interact(ampliacion_red_año, año1=[2014,2015,2016,2017],año2=[2014,2015,2016,2017])
###Output
_____no_output_____
###Markdown
Comparación de densidad de la red para un par de años puntuales. La densidad de la red se mide como el área que abarcan todas las estaciones,dividida la cantidad de estaciones.
###Code
def densidad_red_año(año1,año2):
df1 = pd.read_csv("Data/Stations_{}.csv".format(año1))
df2 = pd.read_csv("Data/Stations_{}.csv".format(año2))
coordinates1 = []
for i in range(len(df1)):
coordinates1.append([df1["latitude"][i],df1["longitude"][i]])
coordinates2 = []
for i in range(len(df2)):
coordinates2.append([df2["latitude"][i],df2["longitude"][i]])
coordinates1.sort(key = lambda x: (-x[0], x[1]))
coordinates2.sort(key = lambda x: (-x[0], x[1]))
obj1 = {'type':'Polygon','coordinates':[coordinates1]}
obj2 = {'type':'Polygon','coordinates':[coordinates2]}
densidad1 = area(obj1) / len(df1)
densidad2 = area(obj2) / len(df2)
print("La diferencia entre la densidad de la red entre el {} y el {} es: {}".format(año1,año2,densidad2-densidad1))
interact(densidad_red_año, año1=[2014,2015,2016,2017],año2=[2014,2015,2016,2017])
###Output
_____no_output_____
###Markdown
Velocidad promedio de los ciclistas para un año determinado
###Code
from math import sin, cos, sqrt, atan2, radians
def velocidad_año(año):
df = pd.read_csv("Data/OD_{}.csv".format(año))
stations = pd.read_json("Data/stations.json")
stations = pd.json_normalize(stations["stations"])
stations.n = stations.n.astype(int)
start = pd.DataFrame({'n': df["start_station_code"]})
result_start = pd.merge(start,stations,on='n',how="left")
end = pd.DataFrame({'n': df["end_station_code"]})
result_end = pd.merge(end,stations,on='n',how="left")
combine = pd.concat([result_start[["n","id","la","lo"]],result_end[["n","id","la","lo"]],df["duration_sec"]],axis=1)
combine.columns = ["start_station_code","id","start_station_la","start_station_lo",
"end_station_code","end_station_id","end_station_la","end_station_lo","duration_sec"]
R = 6373.0
lat1 = combine["start_station_la"].map(radians)
lon1 = combine["start_station_lo"].map(radians)
lat2 = combine["end_station_la"].map(radians)
lon2 = combine["end_station_lo"].map(radians)
dlon = lon2 - lon1
dlat = lat2 - lat1
a = (dlat/2).map(sin)**2 + lat1.map(cos) * lat2.map(cos) * (dlon/2).map(sin)**2
c = 2 * a.map(sqrt).map(asin)
distance = R * c
combine["speed"] = distance / (combine["duration_sec"]/3600)
print("La velocidad media de los ciclistas en el año {} es: {} km/h".format(año,combine["speed"].mean()))
interact(velocidad_año, año=[2014,2015,2016,2017])
###Output
_____no_output_____
###Markdown
Cantidad de bicicletas totales para un momento dado. Considerando la misma como la cantidad de bicicletas que hay en todas las estaciones activas para ese momento, más todos los viajes que se estén realizando.
###Code
from datetime import datetime
import pytz
datetime_picker = ipydatetime.DatetimePicker()
def total_bikes_at_date(date):
if date!=None:
df = pd.read_csv("Data/OD_{}.csv".format(date.year))
df.start_date = pd.to_datetime(df.start_date)
df.end_date = pd.to_datetime(df.end_date)
date = pd.to_datetime(date).tz_localize(tz=None)
total = len(df[(date>df.start_date) & (date<df.end_date)])
print("En el momento {} hay {} bicicletas".format(str(date),total))
else:
return 0
interact(total_bikes_at_date,date=datetime_picker)
###Output
_____no_output_____ |
docs/_static/notebooks/old_quickstart.ipynb | ###Markdown
Quickstart ===================================In this notebook, we will explore `visual`, a companion package to `torchbearer` that enables complex feature visualisation with minimal work. We shall first show how easy it is to get a feature visualisation going using visual.
###Code
!pip install -q torchbearer-visual
###Output
_____no_output_____
###Markdown
We import a couple things from visual and then print the layer names of squeezenet v1.1 so that we can decide which layer to ascend on.
###Code
from visual.models import inception_v3
from visual import BasicAscent, Channel, image
from visual.transforms import RandomRotate, RandomScale, SpatialJitter, Compose
from visual.models.utils import IntermediateLayerGetter
return_layers = {'Mixed_5b': 'feat1'}
model = IntermediateLayerGetter(inception_v3(True, False), return_layers)
print(model.layer_names)
###Output
['Conv2d_1a_3x3', 'Conv2d_1a_3x3_conv', 'Conv2d_1a_3x3_bn', 'Conv2d_1a_3x3_relu', 'Conv2d_2a_3x3', 'Conv2d_2a_3x3_conv', 'Conv2d_2a_3x3_bn', 'Conv2d_2a_3x3_relu', 'Conv2d_2b_3x3', 'Conv2d_2b_3x3_conv', 'Conv2d_2b_3x3_bn', 'Conv2d_2b_3x3_relu', 'MaxPool2b', 'Conv2d_3b_1x1', 'Conv2d_3b_1x1_conv', 'Conv2d_3b_1x1_bn', 'Conv2d_3b_1x1_relu', 'Conv2d_4a_3x3', 'Conv2d_4a_3x3_conv', 'Conv2d_4a_3x3_bn', 'Conv2d_4a_3x3_relu', 'MaxPool4a', 'Mixed_5b', 'Mixed_5b_branch1x1', 'Mixed_5b_branch1x1_conv', 'Mixed_5b_branch1x1_bn', 'Mixed_5b_branch1x1_relu', 'Mixed_5b_branch5x5_1', 'Mixed_5b_branch5x5_1_conv', 'Mixed_5b_branch5x5_1_bn', 'Mixed_5b_branch5x5_1_relu', 'Mixed_5b_branch5x5_2', 'Mixed_5b_branch5x5_2_conv', 'Mixed_5b_branch5x5_2_bn', 'Mixed_5b_branch5x5_2_relu', 'Mixed_5b_branch3x3dbl_1', 'Mixed_5b_branch3x3dbl_1_conv', 'Mixed_5b_branch3x3dbl_1_bn', 'Mixed_5b_branch3x3dbl_1_relu', 'Mixed_5b_branch3x3dbl_2', 'Mixed_5b_branch3x3dbl_2_conv', 'Mixed_5b_branch3x3dbl_2_bn', 'Mixed_5b_branch3x3dbl_2_relu', 'Mixed_5b_branch3x3dbl_3', 'Mixed_5b_branch3x3dbl_3_conv', 'Mixed_5b_branch3x3dbl_3_bn', 'Mixed_5b_branch3x3dbl_3_relu', 'Mixed_5b_AvgPool', 'Mixed_5b_branch_pool', 'Mixed_5b_branch_pool_conv', 'Mixed_5b_branch_pool_bn', 'Mixed_5b_branch_pool_relu', 'Mixed_5c', 'Mixed_5c_branch1x1', 'Mixed_5c_branch1x1_conv', 'Mixed_5c_branch1x1_bn', 'Mixed_5c_branch1x1_relu', 'Mixed_5c_branch5x5_1', 'Mixed_5c_branch5x5_1_conv', 'Mixed_5c_branch5x5_1_bn', 'Mixed_5c_branch5x5_1_relu', 'Mixed_5c_branch5x5_2', 'Mixed_5c_branch5x5_2_conv', 'Mixed_5c_branch5x5_2_bn', 'Mixed_5c_branch5x5_2_relu', 'Mixed_5c_branch3x3dbl_1', 'Mixed_5c_branch3x3dbl_1_conv', 'Mixed_5c_branch3x3dbl_1_bn', 'Mixed_5c_branch3x3dbl_1_relu', 'Mixed_5c_branch3x3dbl_2', 'Mixed_5c_branch3x3dbl_2_conv', 'Mixed_5c_branch3x3dbl_2_bn', 'Mixed_5c_branch3x3dbl_2_relu', 'Mixed_5c_branch3x3dbl_3', 'Mixed_5c_branch3x3dbl_3_conv', 'Mixed_5c_branch3x3dbl_3_bn', 'Mixed_5c_branch3x3dbl_3_relu', 'Mixed_5c_AvgPool', 'Mixed_5c_branch_pool', 'Mixed_5c_branch_pool_conv', 'Mixed_5c_branch_pool_bn', 'Mixed_5c_branch_pool_relu', 'Mixed_5d', 'Mixed_5d_branch1x1', 'Mixed_5d_branch1x1_conv', 'Mixed_5d_branch1x1_bn', 'Mixed_5d_branch1x1_relu', 'Mixed_5d_branch5x5_1', 'Mixed_5d_branch5x5_1_conv', 'Mixed_5d_branch5x5_1_bn', 'Mixed_5d_branch5x5_1_relu', 'Mixed_5d_branch5x5_2', 'Mixed_5d_branch5x5_2_conv', 'Mixed_5d_branch5x5_2_bn', 'Mixed_5d_branch5x5_2_relu', 'Mixed_5d_branch3x3dbl_1', 'Mixed_5d_branch3x3dbl_1_conv', 'Mixed_5d_branch3x3dbl_1_bn', 'Mixed_5d_branch3x3dbl_1_relu', 'Mixed_5d_branch3x3dbl_2', 'Mixed_5d_branch3x3dbl_2_conv', 'Mixed_5d_branch3x3dbl_2_bn', 'Mixed_5d_branch3x3dbl_2_relu', 'Mixed_5d_branch3x3dbl_3', 'Mixed_5d_branch3x3dbl_3_conv', 'Mixed_5d_branch3x3dbl_3_bn', 'Mixed_5d_branch3x3dbl_3_relu', 'Mixed_5d_AvgPool', 'Mixed_5d_branch_pool', 'Mixed_5d_branch_pool_conv', 'Mixed_5d_branch_pool_bn', 'Mixed_5d_branch_pool_relu', 'Mixed_6a', 'Mixed_6a_branch3x3', 'Mixed_6a_branch3x3_conv', 'Mixed_6a_branch3x3_bn', 'Mixed_6a_branch3x3_relu', 'Mixed_6a_branch3x3dbl_1', 'Mixed_6a_branch3x3dbl_1_conv', 'Mixed_6a_branch3x3dbl_1_bn', 'Mixed_6a_branch3x3dbl_1_relu', 'Mixed_6a_branch3x3dbl_2', 'Mixed_6a_branch3x3dbl_2_conv', 'Mixed_6a_branch3x3dbl_2_bn', 'Mixed_6a_branch3x3dbl_2_relu', 'Mixed_6a_branch3x3dbl_3', 'Mixed_6a_branch3x3dbl_3_conv', 'Mixed_6a_branch3x3dbl_3_bn', 'Mixed_6a_branch3x3dbl_3_relu', 'Mixed_6a_MaxPool', 'Mixed_6b', 'Mixed_6b_branch1x1', 'Mixed_6b_branch1x1_conv', 'Mixed_6b_branch1x1_bn', 'Mixed_6b_branch1x1_relu', 'Mixed_6b_branch7x7_1', 'Mixed_6b_branch7x7_1_conv', 'Mixed_6b_branch7x7_1_bn', 'Mixed_6b_branch7x7_1_relu', 'Mixed_6b_branch7x7_2', 'Mixed_6b_branch7x7_2_conv', 'Mixed_6b_branch7x7_2_bn', 'Mixed_6b_branch7x7_2_relu', 'Mixed_6b_branch7x7_3', 'Mixed_6b_branch7x7_3_conv', 'Mixed_6b_branch7x7_3_bn', 'Mixed_6b_branch7x7_3_relu', 'Mixed_6b_branch7x7dbl_1', 'Mixed_6b_branch7x7dbl_1_conv', 'Mixed_6b_branch7x7dbl_1_bn', 'Mixed_6b_branch7x7dbl_1_relu', 'Mixed_6b_branch7x7dbl_2', 'Mixed_6b_branch7x7dbl_2_conv', 'Mixed_6b_branch7x7dbl_2_bn', 'Mixed_6b_branch7x7dbl_2_relu', 'Mixed_6b_branch7x7dbl_3', 'Mixed_6b_branch7x7dbl_3_conv', 'Mixed_6b_branch7x7dbl_3_bn', 'Mixed_6b_branch7x7dbl_3_relu', 'Mixed_6b_branch7x7dbl_4', 'Mixed_6b_branch7x7dbl_4_conv', 'Mixed_6b_branch7x7dbl_4_bn', 'Mixed_6b_branch7x7dbl_4_relu', 'Mixed_6b_branch7x7dbl_5', 'Mixed_6b_branch7x7dbl_5_conv', 'Mixed_6b_branch7x7dbl_5_bn', 'Mixed_6b_branch7x7dbl_5_relu', 'Mixed_6b_AvgPool', 'Mixed_6b_branch_pool', 'Mixed_6b_branch_pool_conv', 'Mixed_6b_branch_pool_bn', 'Mixed_6b_branch_pool_relu', 'Mixed_6c', 'Mixed_6c_branch1x1', 'Mixed_6c_branch1x1_conv', 'Mixed_6c_branch1x1_bn', 'Mixed_6c_branch1x1_relu', 'Mixed_6c_branch7x7_1', 'Mixed_6c_branch7x7_1_conv', 'Mixed_6c_branch7x7_1_bn', 'Mixed_6c_branch7x7_1_relu', 'Mixed_6c_branch7x7_2', 'Mixed_6c_branch7x7_2_conv', 'Mixed_6c_branch7x7_2_bn', 'Mixed_6c_branch7x7_2_relu', 'Mixed_6c_branch7x7_3', 'Mixed_6c_branch7x7_3_conv', 'Mixed_6c_branch7x7_3_bn', 'Mixed_6c_branch7x7_3_relu', 'Mixed_6c_branch7x7dbl_1', 'Mixed_6c_branch7x7dbl_1_conv', 'Mixed_6c_branch7x7dbl_1_bn', 'Mixed_6c_branch7x7dbl_1_relu', 'Mixed_6c_branch7x7dbl_2', 'Mixed_6c_branch7x7dbl_2_conv', 'Mixed_6c_branch7x7dbl_2_bn', 'Mixed_6c_branch7x7dbl_2_relu', 'Mixed_6c_branch7x7dbl_3', 'Mixed_6c_branch7x7dbl_3_conv', 'Mixed_6c_branch7x7dbl_3_bn', 'Mixed_6c_branch7x7dbl_3_relu', 'Mixed_6c_branch7x7dbl_4', 'Mixed_6c_branch7x7dbl_4_conv', 'Mixed_6c_branch7x7dbl_4_bn', 'Mixed_6c_branch7x7dbl_4_relu', 'Mixed_6c_branch7x7dbl_5', 'Mixed_6c_branch7x7dbl_5_conv', 'Mixed_6c_branch7x7dbl_5_bn', 'Mixed_6c_branch7x7dbl_5_relu', 'Mixed_6c_AvgPool', 'Mixed_6c_branch_pool', 'Mixed_6c_branch_pool_conv', 'Mixed_6c_branch_pool_bn', 'Mixed_6c_branch_pool_relu', 'Mixed_6d', 'Mixed_6d_branch1x1', 'Mixed_6d_branch1x1_conv', 'Mixed_6d_branch1x1_bn', 'Mixed_6d_branch1x1_relu', 'Mixed_6d_branch7x7_1', 'Mixed_6d_branch7x7_1_conv', 'Mixed_6d_branch7x7_1_bn', 'Mixed_6d_branch7x7_1_relu', 'Mixed_6d_branch7x7_2', 'Mixed_6d_branch7x7_2_conv', 'Mixed_6d_branch7x7_2_bn', 'Mixed_6d_branch7x7_2_relu', 'Mixed_6d_branch7x7_3', 'Mixed_6d_branch7x7_3_conv', 'Mixed_6d_branch7x7_3_bn', 'Mixed_6d_branch7x7_3_relu', 'Mixed_6d_branch7x7dbl_1', 'Mixed_6d_branch7x7dbl_1_conv', 'Mixed_6d_branch7x7dbl_1_bn', 'Mixed_6d_branch7x7dbl_1_relu', 'Mixed_6d_branch7x7dbl_2', 'Mixed_6d_branch7x7dbl_2_conv', 'Mixed_6d_branch7x7dbl_2_bn', 'Mixed_6d_branch7x7dbl_2_relu', 'Mixed_6d_branch7x7dbl_3', 'Mixed_6d_branch7x7dbl_3_conv', 'Mixed_6d_branch7x7dbl_3_bn', 'Mixed_6d_branch7x7dbl_3_relu', 'Mixed_6d_branch7x7dbl_4', 'Mixed_6d_branch7x7dbl_4_conv', 'Mixed_6d_branch7x7dbl_4_bn', 'Mixed_6d_branch7x7dbl_4_relu', 'Mixed_6d_branch7x7dbl_5', 'Mixed_6d_branch7x7dbl_5_conv', 'Mixed_6d_branch7x7dbl_5_bn', 'Mixed_6d_branch7x7dbl_5_relu', 'Mixed_6d_AvgPool', 'Mixed_6d_branch_pool', 'Mixed_6d_branch_pool_conv', 'Mixed_6d_branch_pool_bn', 'Mixed_6d_branch_pool_relu', 'Mixed_6e', 'Mixed_6e_branch1x1', 'Mixed_6e_branch1x1_conv', 'Mixed_6e_branch1x1_bn', 'Mixed_6e_branch1x1_relu', 'Mixed_6e_branch7x7_1', 'Mixed_6e_branch7x7_1_conv', 'Mixed_6e_branch7x7_1_bn', 'Mixed_6e_branch7x7_1_relu', 'Mixed_6e_branch7x7_2', 'Mixed_6e_branch7x7_2_conv', 'Mixed_6e_branch7x7_2_bn', 'Mixed_6e_branch7x7_2_relu', 'Mixed_6e_branch7x7_3', 'Mixed_6e_branch7x7_3_conv', 'Mixed_6e_branch7x7_3_bn', 'Mixed_6e_branch7x7_3_relu', 'Mixed_6e_branch7x7dbl_1', 'Mixed_6e_branch7x7dbl_1_conv', 'Mixed_6e_branch7x7dbl_1_bn', 'Mixed_6e_branch7x7dbl_1_relu', 'Mixed_6e_branch7x7dbl_2', 'Mixed_6e_branch7x7dbl_2_conv', 'Mixed_6e_branch7x7dbl_2_bn', 'Mixed_6e_branch7x7dbl_2_relu', 'Mixed_6e_branch7x7dbl_3', 'Mixed_6e_branch7x7dbl_3_conv', 'Mixed_6e_branch7x7dbl_3_bn', 'Mixed_6e_branch7x7dbl_3_relu', 'Mixed_6e_branch7x7dbl_4', 'Mixed_6e_branch7x7dbl_4_conv', 'Mixed_6e_branch7x7dbl_4_bn', 'Mixed_6e_branch7x7dbl_4_relu', 'Mixed_6e_branch7x7dbl_5', 'Mixed_6e_branch7x7dbl_5_conv', 'Mixed_6e_branch7x7dbl_5_bn', 'Mixed_6e_branch7x7dbl_5_relu', 'Mixed_6e_AvgPool', 'Mixed_6e_branch_pool', 'Mixed_6e_branch_pool_conv', 'Mixed_6e_branch_pool_bn', 'Mixed_6e_branch_pool_relu', 'AuxLogits', 'AuxLogits_AvgPool0', 'AuxLogits_conv0', 'AuxLogits_conv0_conv', 'AuxLogits_conv0_bn', 'AuxLogits_conv0_relu', 'AuxLogits_conv1', 'AuxLogits_conv1_conv', 'AuxLogits_conv1_bn', 'AuxLogits_conv1_relu', 'AuxLogits_AvgPool1', 'AuxLogits_fc', 'Mixed_7a', 'Mixed_7a_branch3x3_1', 'Mixed_7a_branch3x3_1_conv', 'Mixed_7a_branch3x3_1_bn', 'Mixed_7a_branch3x3_1_relu', 'Mixed_7a_branch3x3_2', 'Mixed_7a_branch3x3_2_conv', 'Mixed_7a_branch3x3_2_bn', 'Mixed_7a_branch3x3_2_relu', 'Mixed_7a_branch7x7x3_1', 'Mixed_7a_branch7x7x3_1_conv', 'Mixed_7a_branch7x7x3_1_bn', 'Mixed_7a_branch7x7x3_1_relu', 'Mixed_7a_branch7x7x3_2', 'Mixed_7a_branch7x7x3_2_conv', 'Mixed_7a_branch7x7x3_2_bn', 'Mixed_7a_branch7x7x3_2_relu', 'Mixed_7a_branch7x7x3_3', 'Mixed_7a_branch7x7x3_3_conv', 'Mixed_7a_branch7x7x3_3_bn', 'Mixed_7a_branch7x7x3_3_relu', 'Mixed_7a_branch7x7x3_4', 'Mixed_7a_branch7x7x3_4_conv', 'Mixed_7a_branch7x7x3_4_bn', 'Mixed_7a_branch7x7x3_4_relu', 'Mixed_7a_MaxPool', 'Mixed_7b', 'Mixed_7b_branch1x1', 'Mixed_7b_branch1x1_conv', 'Mixed_7b_branch1x1_bn', 'Mixed_7b_branch1x1_relu', 'Mixed_7b_branch3x3_1', 'Mixed_7b_branch3x3_1_conv', 'Mixed_7b_branch3x3_1_bn', 'Mixed_7b_branch3x3_1_relu', 'Mixed_7b_branch3x3_2a', 'Mixed_7b_branch3x3_2a_conv', 'Mixed_7b_branch3x3_2a_bn', 'Mixed_7b_branch3x3_2a_relu', 'Mixed_7b_branch3x3_2b', 'Mixed_7b_branch3x3_2b_conv', 'Mixed_7b_branch3x3_2b_bn', 'Mixed_7b_branch3x3_2b_relu', 'Mixed_7b_branch3x3dbl_1', 'Mixed_7b_branch3x3dbl_1_conv', 'Mixed_7b_branch3x3dbl_1_bn', 'Mixed_7b_branch3x3dbl_1_relu', 'Mixed_7b_branch3x3dbl_2', 'Mixed_7b_branch3x3dbl_2_conv', 'Mixed_7b_branch3x3dbl_2_bn', 'Mixed_7b_branch3x3dbl_2_relu', 'Mixed_7b_branch3x3dbl_3a', 'Mixed_7b_branch3x3dbl_3a_conv', 'Mixed_7b_branch3x3dbl_3a_bn', 'Mixed_7b_branch3x3dbl_3a_relu', 'Mixed_7b_branch3x3dbl_3b', 'Mixed_7b_branch3x3dbl_3b_conv', 'Mixed_7b_branch3x3dbl_3b_bn', 'Mixed_7b_branch3x3dbl_3b_relu', 'Mixed_7b_AvgPool', 'Mixed_7b_branch_pool', 'Mixed_7b_branch_pool_conv', 'Mixed_7b_branch_pool_bn', 'Mixed_7b_branch_pool_relu', 'Mixed_7c', 'Mixed_7c_branch1x1', 'Mixed_7c_branch1x1_conv', 'Mixed_7c_branch1x1_bn', 'Mixed_7c_branch1x1_relu', 'Mixed_7c_branch3x3_1', 'Mixed_7c_branch3x3_1_conv', 'Mixed_7c_branch3x3_1_bn', 'Mixed_7c_branch3x3_1_relu', 'Mixed_7c_branch3x3_2a', 'Mixed_7c_branch3x3_2a_conv', 'Mixed_7c_branch3x3_2a_bn', 'Mixed_7c_branch3x3_2a_relu', 'Mixed_7c_branch3x3_2b', 'Mixed_7c_branch3x3_2b_conv', 'Mixed_7c_branch3x3_2b_bn', 'Mixed_7c_branch3x3_2b_relu', 'Mixed_7c_branch3x3dbl_1', 'Mixed_7c_branch3x3dbl_1_conv', 'Mixed_7c_branch3x3dbl_1_bn', 'Mixed_7c_branch3x3dbl_1_relu', 'Mixed_7c_branch3x3dbl_2', 'Mixed_7c_branch3x3dbl_2_conv', 'Mixed_7c_branch3x3dbl_2_bn', 'Mixed_7c_branch3x3dbl_2_relu', 'Mixed_7c_branch3x3dbl_3a', 'Mixed_7c_branch3x3dbl_3a_conv', 'Mixed_7c_branch3x3dbl_3a_bn', 'Mixed_7c_branch3x3dbl_3a_relu', 'Mixed_7c_branch3x3dbl_3b', 'Mixed_7c_branch3x3dbl_3b_conv', 'Mixed_7c_branch3x3dbl_3b_bn', 'Mixed_7c_branch3x3dbl_3b_relu', 'Mixed_7c_AvgPool', 'Mixed_7c_branch_pool', 'Mixed_7c_branch_pool_conv', 'Mixed_7c_branch_pool_bn', 'Mixed_7c_branch_pool_relu', 'AvgPool', 'fc']
###Markdown
Let's choose the final layer, where we have 1000 channels, each corresponding to 1 imagenet class. We'll choose the 256th feature, which is the class "Newfoundland Dog". After running a `BasicAscent` and viewing it in pyplot, you can confirm for yourself that we indeed see things that look like dogs.
###Code
transforms = Compose([
RandomRotate(list(range(-30, 30, 5))),
RandomScale([0.9, 0.95, 1.0, 1.05, 1.1, 1.15, 1.2]),
])
crit = Channel(10, 'feat1')
img = image((3, 256, 256), transform=transforms, correlate=True, fft=True)
a = BasicAscent(img, crit, verbose=2).to_pyplot().run(model, device='cuda')
###Output
_____no_output_____
###Markdown
So, we have seen how quick it is to get a simple visualisation running, but what were did we actually do to get there? We'll now introduce, one at a time, the abstractions that visual uses:- Images- Transforms- Criterions- Ascenders- Models
###Code
###Output
_____no_output_____ |
trial.ipynb | ###Markdown
Define Muskingum coefficients Parameters
###Code
K1 = 7.722
X1 = 0.3
delta_t = 15*60
alpha1 = (delta_t-2*K1*X1)/(2*K1*(1-X1)+delta_t)
beta1 = (delta_t+2*K1*X1)/(2*K1*(1-X1)+delta_t)
xi1 = (2*K1*(1-X1)-delta_t)/(2*K1*(1-X1)+delta_t)
K2 = 7.3317
X2 = 0.5
delta_t = 15*60
alpha2 = (delta_t-2*K2*X2)/(2*K2*(1-X2)+delta_t)
beta2 = (delta_t+2*K1*X1)/(2*K2*(1-X2)+delta_t)
xi2 = (2*K2*(1-X1)-delta_t)/(2*K2*(1-X2)+delta_t)
K3 = 7.722
X3 = 0.2
delta_t = 15*60
alpha3 = (delta_t-2*K2*X2)/(2*K2*(1-X2)+delta_t)
beta3 = (delta_t+2*K2*X2)/(2*K2*(1-X2)+delta_t)
xi3 = (2*K2*(1-X2)-delta_t)/(2*K2*(1-X2)+delta_t)
###Output
_____no_output_____
###Markdown
Define state space system Matrices
###Code
#Define the matrices
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.dates import DateFormatter
B_I = np.array([0,0,0,0,0,0,beta1,beta2,0]).reshape(3,3)
X_I = np.array([0,0,0,0,0,0,xi1,xi2,0]).reshape(3,3)
B_O = np.array([beta1,0,0,0,beta2,0,alpha3*beta1,alpha3*beta2,beta3]).reshape(3,3)
X_O = np.array([xi1,0,0,0,xi2,0,alpha3*xi1,alpha3*xi2,xi3]).reshape(3,3)
P_I = np.array([1,0,0,0,1,0,alpha1,alpha2,1]).reshape(3,3)
P_O = np.array([alpha1,0,0,0,alpha2,0,alpha3*alpha1,alpha3*alpha2,alpha3]).reshape(3,3)
###Output
_____no_output_____
###Markdown
Create block Matrices
###Code
df = pd.read_csv('Total_inflow.csv')
mat_6x6 = np.block([[B_I,X_I],[B_O,X_O]]) # Creating 6x6 Matrix
mat_6x3 = np.block([[P_I],[P_O]]) # Creating 6 * 3 matrix
I_intial=np.array([0,0,0]).reshape(3,1) # Intializing I matrix
O_intial=np.array([0,0,0]).reshape(3,1) # Initializing O matrix
IO_mat_6x1 = np.block([[I_intial],[O_intial]]) # Creating a block matrix
eig = np.linalg.eig(mat_6x6)
eig
###Output
_____no_output_____
###Markdown
Finding the State Space System Solution
###Code
P1 = np.array(df['Inflow_1'])
P2 = np.array(df['Inflow_2'])
P3 = np.array(df['Inflow_3'])
time = df['Time']
# P_t = np.array([[P1[0]],[P2[0]],[P3[0]]])
result = IO_mat_6x1.reshape(1,6) # Initialing result with initial value for Input and output
for idx in range(len(P1)): # running till the length of P1 i.e. the number of time steps
P_t = np.array([[P1[idx]],[P2[idx]],[P3[idx]]]) # extracting value of each arrary for each time step
val = mat_6x6@result[-1,:].reshape(6,1)+mat_6x3@P_t # selecting the last value of result and reshaping it to 6x1
val = val.reshape(1,6)
result = np.concatenate((result,val),axis=0) # each row result for each time step
df_nwm = pd.read_csv('Total_Discharge_nwm.csv')
# plot the discharge time series
import matplotlib.pyplot as plt
from matplotlib.dates import DateFormatter
time= pd.to_datetime(time)
plt.figure(facecolor='white')
plt.plot(time, result[1:,3])
plt.plot(df_nwm['Outflow1'],label = 'NWM')
date_form = DateFormatter("%b %d")
plt.legend()
plt.figure(facecolor = 'white')
plt.plot(result[1:,4],label = 'Model')
plt.plot(df_nwm['Outflow2'],label = 'NWM')
plt.legend()
plt.figure(facecolor = 'white')
plt.plot(result[1:,5],label = 'Model')
plt.plot(df_nwm['Outflow3'],label = 'NWM')
plt.legend()
# # plt.rc('font', size=14)
# fig, ax = plt.subplots(figsize=(10, 6))
# ax.plot(result[:,3], time, color='tab:blue', label='Q')
type(time)
#Load the sensor data and model data
sensor = pd.read_csv('D:/Sujana/Project/ce397/DD6_sensor.csv')
Y = np.array(sensor['DD6_discharge'])
X = pd.read_csv('D:/Sujana/Project/ce397/result_outflow_kalman.csv')
I1 = np.array(X['I1'])
I2 = np.array(X['I2'])
I3 = np.array(X['I3'])
O1 = np.array(X['O1'])
O2 = np.array(X['O2'])
O3 = np.array(X['O3'])
C = np.array([1/4,0,0,3/4,0,0]).reshape(1,6)
#Define noise parameters
sigma_v = 0.05 # Measurement noise std. dev
sigma_w = 0.5 # Process noise std. dev
add_v = 0 # Add measurement noise by setting to 1
add_w = 0 # Add process noise by setting to 1
#Generate y_hat values
# for i in range(len(I1)):
# x_h = np.array([[I1[i]],[I2[i]],[I3[i]],[O1[i]],[O2[i]],[O3[i]]])
# value = C@x_h.reshape(6,1)
# y_hat = np.concatenate((result,val),axis=0)
# y_hat += add_v * sigma_v * np.random.randn(*y_hat.shape)
###Output
_____no_output_____
###Markdown
Kalman Filter
###Code
A = np.block([[B_I,X_I],[B_O,X_O]])
B = np.block([[P_I],[P_O]])
V = sigma_v**2 * np.eye(len(C)) # Measurement noise covariance
W = sigma_w**2 * np.eye(len(A)) # Process noise covariance
S = 1e-2 * np.eye(len(A)) # Initial estimate of error covariance
x_hat = np.zeros(6).reshape(1,6) # Estimate of initial state
# X_hat = [x_hat]
for k in range(len(I1)):
y = Y[k]
u = np.array([[P1[k]],[P2[k]],[P3[k]]])
# u += add_w * sigma_w * np.random.randn(B.shape[1])
S = A @ (S - S @ C.T @ np.linalg.inv(C @ S @ C.T + V) @ C @ S) @ A.T + W
L = S @ C.T @ np.linalg.inv(C @ S @ C.T + V)
y_hat = C @ (A @ x_hat[-1,:].reshape(6,1) + B @ u)
x = A @ x_hat[-1,:].reshape(6,1) + B @ u + L @ (y - y_hat)
x = x.reshape(1,6)
x_hat = np.concatenate((x_hat,x),axis=0)
X_hat = x_hat
# X_hat = np.vstack(X_hat)
# for idx in range(len(P1)): # running till the length of P1 i.e. the number of time steps
# P_t = np.array([[P1[idx]],[P2[idx]],[P3[idx]]]) # extracting value of each arrary for each time step
# val = mat_6x6@result[-1,:].reshape(6,1)+mat_6x3@P_t # selecting the last value of result and reshaping it to 6x1
# val = val.reshape(1,6)
# result = np.concatenate((result,val),axis=0)
# plot the discharge time series
# Outflow1
plt.figure(facecolor='white')
plt.plot(O1)
plt.plot(Y, label = 'sensor')
plt.plot(X_hat[:,3])
# plt.plot(df_nwm['Outflow1'],label = 'NWM')
date_form = DateFormatter("%b %d")
plt.legend()
# Outflow3
plt.figure(facecolor = 'white')
plt.plot(result[1:,5],label = 'Model')
plt.plot(df_nwm['Outflow3'],label = 'NWM')
plt.legend()
# # plt.rc('font', size=14)
# fig, ax = plt.subplots(figsize=(10, 6))
# ax.plot(result[:,3], time, color='tab:blue', label='Q')
# plot the discharge time series
import matplotlib.pyplot as plt
from matplotlib.dates import DateFormatter
plt.figure(facecolor='white')
plt.plot(time, result[1:,3])
plt.plot(df_nwm['Outflow1'],label = 'NWM')
date_form = DateFormatter("%b %d")
ax.xaxis.set_major_formatter(date_form)
plt.legend()
import matplotlib.pyplot as plt
from matplotlib.dates import DateFormatter
plt.figure(facecolor='white')
plt.rc('font', size=14)
fig, ax = plt.subplots(figsize=(10, 6))
ax.plot(X['O1'], color='tab:blue', label='Q')
# ax.line(X.index.values,
# X['O1'],
# color='purple')
ax.set(xlabel='Date',
ylabel='Discharge [cms]',
title='Outflow 1')
date_form = DateFormatter("%b %d")
ax.xaxis.set_major_formatter(date_form)
ax.legend()
ax.grid(True)
plt.show()
X = X.set_index('Time')
m =
type(X.index.values)
###Output
_____no_output_____
###Markdown
Check the path of your csv files before loading the data
###Code
col_types = {'Id': 'int',
'OwnerUserId': 'float',
'CreationDate': 'str',
'ParentId': 'int',
'Score': 'int',
'Title': 'str',
'Body':'str'}
questions = pd.read_csv('/pythonquestions/Questions.csv', encoding = "ISO-8859-1", dtype=col_types)
answers = pd.read_csv('/pythonquestions/Answers.csv', encoding = "ISO-8859-1", dtype=col_types)
# question to answer: (ans_id, score, owner_id)
q_to_a = load_obj('obj/q_to_a.pkl')
if not q_to_a:
q_to_a = dict()
for _, row in answers[['Id', 'ParentId', 'Score', 'OwnerUserId']].iterrows():
q_id = row['ParentId']
a_id = row['Id']
a_score = row['Score']
a_owner_id = row['OwnerUserId'] if not np.isnan(row['OwnerUserId']) else None
if q_id not in q_to_a:
q_to_a[q_id] = [(a_id, a_score, a_owner_id)]
else:
q_to_a[q_id].append((a_id, a_score, a_owner_id))
save_obj(q_to_a, 'obj/q_to_a.pkl')
# Keep only the questions with 4-10 answers and a distinguishable answer
q_to_a = {k:v for k, v in q_to_a.items() if len(v)>3 and len(v)<11 and max(v, key=lambda x: x[1])[1]>0}
# keep only qualified questions
questions = questions[questions['Id'].isin(q_to_a)]
# Keep only answers related to a qualified question
def answer_in_use(answer):
if answer['ParentId'] in q_to_a:
for item in q_to_a[answer['ParentId']]:
if answer['Id'] == item[0]:
return True
return False
answers = answers[answers.apply(lambda x: answer_in_use(x), axis=1)]
questions.info()
answers.info()
###Output
<class 'pandas.core.frame.DataFrame'>
Int64Index: 215203 entries, 0 to 987106
Data columns (total 6 columns):
Id 215203 non-null int32
OwnerUserId 213419 non-null float64
CreationDate 215203 non-null object
ParentId 215203 non-null int32
Score 215203 non-null int32
Body 215203 non-null object
dtypes: float64(1), int32(3), object(2)
memory usage: 9.0+ MB
###Markdown
Semantic features: Question-Ans similarity and Ans-Ans similarity (It took 1.5 hr to run on my Core-i7-7700 / 32GB RAM laptop)
###Code
#t_start = time.time()
def compute_sim(q_to_a, df_questions, df_answers):
a_sim = dict()
tokenizer = RegexpTokenizer(r'\w+')
print(len(q_to_a))
c = 0
for q_id, a_list in q_to_a.items():
c+=1
print(str(c) + ' ' + str(len(a_list)), end='\r')
# get split body text for a question and the answers
q_body = df_questions[df_questions['Id']==q_id].iloc[0]['Body']
q_body = BeautifulSoup(q_body, 'html.parser').get_text()#.split()
q_body = tokenizer.tokenize(q_body.lower())
q_body = [w for w in q_body if w not in stopwords.words('english')]
#print(q_body)
a_bodies = list()
for a_id, _, _ in a_list:
a_body = df_answers[df_answers['Id']==a_id].iloc[0]['Body']
a_body = BeautifulSoup(a_body, 'html.parser').get_text()#.split()
a_body = tokenizer.tokenize(a_body.lower())
a_body = [w for w in a_body if w not in stopwords.words('english')]
a_bodies.append(a_body)
#print(a_bodies)
# apply a series of transformations to the answers: bag-of-word, tf-idf, and lsi
dictionary = corpora.Dictionary(a_bodies)
corpus = [dictionary.doc2bow(a) for a in a_bodies]
#print(a_bodies[0], len(a_bodies[0]))
#print(corpus[0], len(corpus[0]))
tfidf = models.TfidfModel(corpus)
corpus_tfidf = tfidf[corpus]
#print(corpus_tfidf[0], len(corpus_tfidf[0]))
lsi = models.LsiModel(corpus_tfidf, id2word=dictionary, num_topics=4)
corpus_lsi = lsi[corpus_tfidf]
#print(corpus_lsi[0], len(corpus_lsi[0]))
index = similarities.MatrixSimilarity(corpus_lsi)
# question-to-answer similarity
q_bow = dictionary.doc2bow(q_body)
q_lsi = lsi[q_bow]
q_to_a_sim = index[q_lsi]
#print(q_to_a_sim)
# ans-to-ans similarity, excluding the answer itself
for idx, a_lsi in enumerate(corpus_lsi):
a_to_a_sim = index[a_lsi]
a_to_a_sim = [a_to_a_sim[i] for i in range(len(a_to_a_sim)) if i != idx] # exclude itself
# construct the dictionary a_sim
a_id = a_list[idx][0]
sim_to_q = q_to_a_sim[idx]
max_sim_to_a = max(a_to_a_sim)
min_sim_to_a = min(a_to_a_sim)
a_sim[a_id] = (sim_to_q, max_sim_to_a, min_sim_to_a)
return a_sim
# a_sim: a_id -> (sim_to_question, max_sim_to_other_ans, min_sim_to_other_ans)
a_sim = load_obj('obj/a_sim.pkl')
if not a_sim:
a_sim = compute_sim(q_to_a, questions, answers)
save_obj(a_sim, 'obj/a_sim.pkl')
#t_end = time.time()
#print('time elapsed: %f minutes', ((t_end-t_start)/60.0))
answers = answers.assign(SimToQ = answers['Id'].apply(lambda a_id: a_sim[a_id][0]))
answers = answers.assign(MaxSimToA = answers['Id'].apply(lambda a_id: a_sim[a_id][1]))
answers = answers.assign(MinSimToA = answers['Id'].apply(lambda a_id: a_sim[a_id][2]))
answers.head(3)
###Output
_____no_output_____
###Markdown
Shallow Features
###Code
def strip_code(html):
bs = BeautifulSoup(html, 'html.parser')
[s.extract() for s in bs('code')]
return bs.get_text()
answers = answers.assign(BodyEnglishText = answers['Body'].apply(strip_code))
answers = answers.assign(EnglishCount = answers['BodyEnglishText'].apply(lambda x: len(x.split())))
answers.head(3)
def count_links(html):
bs = BeautifulSoup(html, 'html.parser')
return len(bs.find_all('a'))
answers = answers.assign(LinkCount = answers['Body'].apply(count_links))
answers.head(3)
def has_code(html):
bs = BeautifulSoup(html, 'html.parser')
return 1 if bs.find('code') else 0
answers = answers.assign(HasCode = answers['Body'].apply(has_code))
answers.head(3)
def code_length(html):
bs = BeautifulSoup(html, 'html.parser')
contents = [tag.text.split() for tag in bs.find_all('code')]
return sum(len(item) for item in contents)
answers = answers.assign(CodeLength = answers['Body'].apply(code_length))
answers = answers.assign(TotalLength = answers.apply(lambda row: row['EnglishCount'] + row['CodeLength'], axis=1))
answers.head(3)
def get_order(qid, aid):
ids = [i[0] for i in q_to_a[qid]]
return ids.index(aid)+1
answers = answers.assign(PostOrder = answers.apply(lambda row: get_order(row['ParentId'], row['Id']), axis=1))
answers.head(3)
def is_answer(qid, aid):
if aid == max(q_to_a[qid], key=lambda item: item[1])[0]:
return 1
else:
return 0
answers = answers.assign(IsAnswer = answers.apply(lambda row: is_answer(row['ParentId'], row['Id']), axis=1))
answers.head(3)
# calculate reputation scores for users
user_rep = dict()
for ans_list in q_to_a.values():
for _, score, owner_id in ans_list:
if owner_id:
if owner_id in user_rep:
user_rep[owner_id] += score
else:
user_rep[owner_id] = score
def get_reputation(userId):
if not np.isnan(userId) and userId in user_rep:
return user_rep[userId]
else:
return 0
answers = answers.assign(Reputation = answers['OwnerUserId'].apply(get_reputation))
answers.head(3)
###Output
_____no_output_____
###Markdown
Training: RandomForest
###Code
q_train, q_validation = model_selection.train_test_split(questions, test_size=0.2, random_state=42)
a_train = answers[answers['ParentId'].isin(q_train['Id'])]
a_validation = answers[answers['ParentId'].isin(q_validation['Id'])]
x_train = a_train[['TotalLength','LinkCount', 'CodeLength', 'PostOrder', 'Reputation', 'SimToQ', 'MaxSimToA', 'MinSimToA']]
y_train = a_train[['IsAnswer']]
x_val = a_validation[['Id', 'TotalLength','LinkCount', 'CodeLength', 'PostOrder', 'Reputation', 'SimToQ', 'MaxSimToA', 'MinSimToA']]
y_val = a_validation[['IsAnswer']]
rf_classifier = RandomForestClassifier(
n_estimators=1000, min_samples_leaf=4, n_jobs=-1, oob_score=True, random_state=42)
rf_classifier.fit(x_train, y_train.values.ravel())
y_pred = rf_classifier.predict_proba(x_val.iloc[:, 1:])
def get_accuracy(q_ids, a_ids, prob_pred):
a_to_prob = dict()
for idx, a_id in enumerate(a_ids):
prob = prob_pred[:, 1][idx]
a_to_prob[a_id] = prob
count = 0
for q_id in q_ids:
right_answer = max(q_to_a[q_id], key=lambda item: item[1])[0]
predict_answer = 0
highest_score = 0
for a_id, score, _ in q_to_a[q_id]:
pred_score = a_to_prob[a_id]
if pred_score > highest_score:
predict_answer = a_id
highest_score = pred_score
if right_answer==predict_answer:
count += 1
return count/len(q_ids)
print('RF model accuracy:', get_accuracy(q_validation['Id'].tolist(), a_validation['Id'].tolist(), y_pred))
###Output
RF model accuracy: 0.4722190788729207
###Markdown
Baseline
###Code
def baseline(q_ids, a_val):
count_first = 0
count_last = 0
count_long = 0
for q_id in q_ids:
right_answer = max(q_to_a[q_id], key=lambda item: item[1])[0]
first_answer = q_to_a[q_id][0][0]
if right_answer==first_answer:
count_first += 1
last_answer = q_to_a[q_id][-1][0]
if right_answer==last_answer:
count_last += 1
longest_answer = -1
max_length = 0
for a_id, score, _ in q_to_a[q_id]:
leng = a_val[a_val.Id==a_id].iloc[0]['TotalLength']
if leng > max_length:
longest_answer = a_id
max_length = leng
if right_answer==longest_answer:
count_long += 1
print('Baseline for the first answer:', count_first/len(q_ids))
print('Baseline for the last answer:', count_last/len(q_ids))
print('Baseline for the longest answer:', count_long/len(q_ids))
baseline(q_validation['Id'].tolist(), a_validation)
###Output
Baseline for the first answer: 0.3969672965938667
Baseline for the last answer: 0.07649654860246691
Baseline for the longest answer: 0.29885707819395724
###Markdown
Explaintion of important features
###Code
x_val = x_val.assign(PredictProba = y_pred[:, 1])
x_val.head(10)
importances = rf_classifier.feature_importances_
std = np.std([tree.feature_importances_ for tree in rf_classifier.estimators_], axis=0)
indices = np.argsort(importances)[::-1]
# Print the feature ranking
print("Feature ranking:")
for f in range(x_val.shape[1]-2):
print("%d. feature %d (%f)" % (f + 1, indices[f], importances[indices[f]]))
import matplotlib.pyplot as plt
plt.figure()
plt.title("Feature importances")
plt.bar(range(x_val.shape[1]-2), importances[indices],
color="r", yerr=std[indices], align="center")
plt.xticks(range(x_val.shape[1]-2), indices)
plt.xlim([-1, x_val.shape[1]-2])
plt.show()
###Output
_____no_output_____
###Markdown
Checking installation.
###Code
import numpy as np
from scipy import special as sp
import matplotlib.pyplot as plt
import pandas as pd
%matplotlib inline
###Output
_____no_output_____
###Markdown
Parabola
###Code
x = np.array([0,1,2,3,4,5,15,25,35,50])
y = np.sqrt(4*x)
plt.plot(x, y, label='up')
plt.plot(x, -y, label='down')
plt.xlabel('X')
plt.ylabel('Y')
plt.legend()
###Output
_____no_output_____
###Markdown
Factorial square
###Code
x = np.array([0,1,2,3,4,5,15,25,35,50])
y = (x/sp.factorial(5))**2
plt.plot(x, y, label='up')
plt.plot(x, -y, label='down')
plt.xlabel('X')
plt.ylabel('Y')
plt.legend()
###Output
_____no_output_____
###Markdown
Use of pandas
###Code
pd.DataFrame.from_dict({'students':['a','b', 'c'],
'marks':[95,30,40]})
###Output
_____no_output_____
###Markdown
###Code
print('mont ra mad an traoù ?')
###Output
mont ra mad an traoù ?
###Markdown
Import Packages
###Code
# Importing Packages
import numpy as np
import pandas as pd
#Importing packages for cross_validation
from sklearn.model_selection import cross_val_score
#for modeling
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
import xgboost as xgb
from sklearn.model_selection import train_test_split
from collections import Counter
#for feature selection
from sklearn.feature_selection import SelectKBest, f_classif
## Grid search cross validation
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import StratifiedKFold
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib
from scipy import stats
import pickle
###Output
_____no_output_____
###Markdown
Read Test and Train Data
###Code
train = pd.read_csv('./data/train.csv')
test = pd.read_csv('./data/test.csv')
###Output
_____no_output_____
###Markdown
Exploratory Data Analysis
###Code
train.info()
train.describe()
train.tail()
test.info()
train.isnull().sum().sum()
test.isnull().sum().sum()
%config InlineBackend.figure_format = 'retina'
features = ['var_0', 'var_1','var_2','var_3', 'var_4', 'var_5']
train_sub = train[features]
sns.pairplot(train_sub)
%config InlineBackend.figure_format = 'retina'
sns.countplot(x= "target", data=train)
t0 = train.loc[train['target'] == 0].drop(['target'], axis=1)
t1 = train.loc[train['target'] == 1].drop(['target'], axis=1)
features = t0.columns.values[1:51]
t0.columns.values[1]
x = features.shape[0]
rows = np.int((np.ceil(x/5.0)))
%config InlineBackend.figure_format = 'retina'
f, axes = plt.subplots(rows, 5, figsize=(20, rows * 3), sharex=True)
font = {'family' : 'Times new roman',
'weight' : 'bold',
'size' : 18}
matplotlib.rc('font', **font)
for i, ax in enumerate(axes.flatten()):
sns.kdeplot(t0.iloc[:, i+1], Label='0', ax=ax)
sns.kdeplot(t1.iloc[:, i+1], Label='1', ax=ax)
ax.set_xlabel(features[i])
# ax.legend(fontsize=18)
plt.tight_layout()
%config InlineBackend.figure_format = 'retina'
font = {'family' : 'Times new roman',
'weight' : 'bold',
'size' : 22}
matplotlib.rc('font', **font)
f, axes = plt.subplots(1, 2, figsize=(20, 8))
#features = train.columns.values[2:202]
sns.distplot(t0.mean(axis=1),color="green", kde=True,bins=120, label='target 0', hist_kws={"alpha" : 0.4}, ax=axes[0])
sns.distplot(t1.mean(axis=1),color="blue", kde=True,bins=120, label='target 1', hist_kws={"alpha" : 0.4}, ax=axes[0])
axes[0].set_title("Distribution of mean values per row in the \n training set for target 0 and target 1", fontsize=20)
axes[0].legend()
sns.distplot(t0.mean(axis=0),color="green", kde=True,bins=120, label='target 0', hist_kws={"alpha" : 0.4}, ax=axes[1])
sns.distplot(t1.mean(axis=0),color="blue", kde=True,bins=120, label='target 1', hist_kws={"alpha" : 0.4}, ax=axes[1])
axes[1].set_title("Distribution of mean values per column in the \n training set for target 0 and target 1", fontsize=20)
axes[1].legend()
plt.show()
plt.tight_layout()
%config InlineBackend.figure_format = 'retina'
font = {'family' : 'Times new roman',
'weight' : 'bold',
'size' : 22}
matplotlib.rc('font', **font)
f, axes = plt.subplots(1, 2, figsize=(20, 8))
#features = train.columns.values[2:202]
sns.distplot(t0.std(axis=1),color="green", kde=True,bins=120, label='target 0', hist_kws={"alpha" : 0.4}, ax=axes[0])
sns.distplot(t1.std(axis=1),color="blue", kde=True,bins=120, label='target 1', hist_kws={"alpha" : 0.4}, ax=axes[0])
axes[0].set_title("Distribution of standard deviations per row in the \n training set for target 0 and target 1", fontsize=20)
axes[0].legend()
sns.distplot(t0.std(axis=0),color="green", kde=True,bins=120, label='target 0', hist_kws={"alpha" : 0.4}, ax=axes[1])
sns.distplot(t1.std(axis=0),color="blue", kde=True,bins=120, label='target 1', hist_kws={"alpha" : 0.4}, ax=axes[1])
axes[1].set_title("Distribution of standard deviations per column in the \n training set for target 0 and target 1", fontsize=20)
axes[1].legend()
plt.show()
plt.tight_layout()
%config InlineBackend.figure_format = 'retina'
font = {'family' : 'Times new roman',
'weight' : 'bold',
'size' : 22}
matplotlib.rc('font', **font)
f, axes = plt.subplots(1, 2, figsize=(20, 8))
#features = train.columns.values[2:202]
sns.distplot(t0.min(axis=1),color="green", kde=True,bins=120, label='target 0', hist_kws={"alpha" : 0.4}, ax=axes[0])
sns.distplot(t1.min(axis=1),color="blue", kde=True,bins=120, label='target 1', hist_kws={"alpha" : 0.4}, ax=axes[0])
axes[0].set_title("Distribution of minimum values per row in the \n training set for target 0 and target 1", fontsize=20)
axes[0].legend()
sns.distplot(t0.max(axis=1),color="green", kde=True,bins=120, label='target 0', hist_kws={"alpha" : 0.4}, ax=axes[1])
sns.distplot(t1.max(axis=1),color="blue", kde=True,bins=120, label='target 1', hist_kws={"alpha" : 0.4}, ax=axes[1])
axes[1].set_title("Distribution of maximum values per row in the \n training set for target 0 and target 1", fontsize=20)
axes[1].legend()
plt.show()
plt.tight_layout()
%%time
correlations = train[features].corr().abs().unstack().sort_values(kind="quicksort").reset_index()
correlations = correlations[correlations['level_0'] != correlations['level_1']]
correlation = correlations[correlations.index % 2 != 0].reset_index()
correlation.tail()
features = t0.columns.values[1:202]
duplicate = []
# dups_shape = train['var_68'].nunique()
for fea in features:
dups_shape = train.pivot_table(index=[fea], aggfunc='size')
duplicate.append([fea, dups_shape.max(), dups_shape.idxmax()])
duplicate_df = pd.DataFrame(duplicate,columns=['Features', 'Max_duplicates', 'Value'])
np.transpose(duplicate_df.sort_values(by = 'Max_duplicates', ascending=False).head(20))
###Output
_____no_output_____
###Markdown
4. Feature Selection* [Feature selection algorithms](https://www.analyticsvidhya.com/blog/2016/12/introduction-to-feature-selection-methods-with-an-example-or-how-to-select-the-right-variables/) help to determine which features significantly influence the output variable and then eliminate the remaining features from consideration. This step was useful as this dataset has large number of features, being able to eliminate features that have little to no impact on the analysis reduced the complexity of the model and enabled machine learning algortihms to train faster. * The scikit module provides [several ways](https://scikit-learn.org/stable/modules/feature_selection.html) for identifying significant features either by evaluating the statistical correlation with the outcome variable (e.g. univariate feature selection method) or by measuring the usefulness of a subset of feature by actually training a machine learning model on it (e.g. recursive feature elimination). * Considering the size of this particular dataset and the computational time usually required for recursive feature elimination, the universal feature seleciton method was adopted to select the top 25 features that are critical for modeling. Later, submissions were also made by selecting top 50, 100 and 150 features to check the effect of feature selection on accuracy and time required for computation. * **Define Target Variable and Test Variables for Modeling**
###Code
import numpy as np
from sklearn.feature_selection import SelectKBest, f_classif
feature_data = train.drop(columns=['target','ID_code'],axis=1)
target_data = train['target']
X_train, X_test, y_train, y_test = train_test_split(
feature_data, target_data, test_size=0.3, random_state=40)
X_train
###Output
_____no_output_____
###Markdown
* **Select Features to be Considered for Modeling**
###Code
selector = SelectKBest(f_classif, k=50)
X_new=selector.fit_transform(X_train, y_train)
mask = selector.get_support(indices=True)
colname = X_train.columns[mask]# Names of the selected columns
scores = -np.log10(selector.pvalues_)
# def plot_joint_plot(df, feature, target):
# j = sns.jointplot(feature, target, data = df, kind = 'reg')
# j.annotate(stats.pearsonr)
# return plt.show()
pearson_r = []
for column in X_train.columns:
corr_tuple = stats.pearsonr(X_train[column], y_train)
pearson_r.append([column, corr_tuple[0], corr_tuple[1]])
corr_df = pd.DataFrame(pearson_r, columns = ['Features', 'Correlation', 'P_value' ])
corr_df.head(5)
corr_df.sort_values(by = ['P_value'], inplace=True)
corr_df.head(5)
###Output
_____no_output_____
###Markdown
* **Redifining Train and Test Data Using the Selected Features**
###Code
X_train_new = X_train[colname]
X_test_new = X_test[colname]
###Output
_____no_output_____
###Markdown
5. Train Different Classification Models using Selected Features* All models (KNN, Logistic Regresson, Decision Tree,XGBoost and Random Forest) were trained using the train dataset with selected features. * To estimate the accuracy of these models when applied to unseen data, cross validation was performed for each model by splitting the train dataset (80%/20% for train/test split), fitting the model and computing the score 5 consecutive times (with different splits each time) based on ROC_AUC score. The mean of the accuracies for the 5 trials were then reported as the CV score for each model. * **Defining models**
###Code
#Random Forest, Decision Tree, Logistic Regression and K Nearest Neighbours, and XGBoost models
RFmodel=RandomForestClassifier(random_state=1)
DTmodel=DecisionTreeClassifier(random_state=1)
logreg = LogisticRegression(solver='newton-cg', max_iter=1000)
knn = KNeighborsClassifier(n_neighbors = 3)
xgbm = xgb.XGBClassifier(silent=True, scale_pos_weight=1,learning_rate=0.01,
colsample_bytree = 0.4,
subsample = 0.8,
objective='binary:logistic',
n_estimators=1000,
reg_alpha = 0.3,
max_depth=4,
gamma=10)
###Output
_____no_output_____
###Markdown
* **Training the Models**
###Code
# RFmodel.fit(X_train_new, y_train)
# print("RF done")
# filename = 'RF_model.sav'
# pickle.dump(RFmodel, open(filename, 'wb'))
# DTmodel.fit(X_train_new, y_train)
# print("DT done")
# filename = 'DT_model.sav'
# pickle.dump(DTmodel, open(filename, 'wb'))
# logreg.fit(X_train_new, y_train)
# print("logreg done")
# filename = 'logreg_model.sav'
# pickle.dump(logreg, open(filename, 'wb'))
# knn.fit(X_train_new, y_train)
# print("knn done")
# filename = 'knn_model.sav'
# pickle.dump(knn, open(filename, 'wb'))
# xgbm.fit(X_train_new, y_train)# print("xgbm done")
# filename = 'xgbm_model.sav'
# pickle.dump(xgbm, open(filename, 'wb'))
from sklearn import metrics
from sklearn.metrics import roc_curve
def model_performance(model_name, file_name, test_X, test_Y):
loaded_model = pickle.load(open(file_name, 'rb'))
y_pred = loaded_model.predict(test_X)
y_pred_prob = loaded_model.predict_proba(test_X)[:,1]
fpr, tpr, thresholds = roc_curve(test_Y, y_pred_prob)
cnf_matrix = metrics.confusion_matrix(test_Y, y_pred)
print("Accuracy:",metrics.accuracy_score(y_test, y_pred))
print("Precision:",metrics.precision_score(y_test, y_pred))
print("Recall:",metrics.recall_score(y_test, y_pred))
%config InlineBackend.figure_format = 'retina'
fig, axes = plt.subplots(1, 2, figsize=(20, 8))
sns.set(font_scale=2)
sns.heatmap(pd.DataFrame(cnf_matrix), annot=True, annot_kws={"size": 20},
cmap="viridis", fmt='g', ax = axes[0])
axes[0].xaxis.set_label_position("top")
axes[0].set_title('Confusion matrix for {}'.format(model), y=1.1, fontsize=20)
axes[0].set_ylabel('Actual label', fontsize=20)
axes[0].set_xlabel('Predicted label', fontsize=20)
axes[0].xaxis.set_tick_params(labelsize=20)
axes[0].yaxis.set_tick_params(labelsize=20)
axes[1].plot([0, 1], [0, 1], 'k--', linewidth=3.0)
axes[1].plot(fpr,tpr, label='Logistic Regression', linewidth=3.0)
axes[1].set_xlabel('False Positive Rate')
axes[1].set_ylabel('True Positive Rate')
axes[1].set_title('ROC Curve')
plt.tight_layout()
model = 'Random Forest'
file_name = 'RF_model.sav'
model_performance(model, file_name, X_test_new, y_test)
model = 'Logistic Regression'
file_name = 'logreg_model.sav'
model_performance(model, file_name, X_test_new, y_test)
###Output
Accuracy: 0.9066833333333333
Precision: 0.6394453004622496
Recall: 0.13923838282167422
###Markdown
* **Calculating Cross Validation Scores**
###Code
#define a function that can be called to calculate cross validation score for each model
def cross_val(X,x,y):
scores = cross_val_score(X, x, y, cv=5, scoring = "roc_auc")
return scores.mean()
#Calculate Cross Validation Score based on ROC_AUC by calling th ecross_val function defined before
RF_score = round(cross_val(RFmodel, X_train_new, y_train) * 100, 2)
print("RF done")
DT_score =round(cross_val(DTmodel, X_train_new, y_train) * 100, 2)
print("DT done")
logreg_score = round(cross_val(logreg, X_train_new, y_train) * 100, 2)
print("logreg done")
knn_score = round(cross_val(knn, X_train_new, y_train) * 100, 2)
print("knn done")
xgb_score = round(cross_val(xgbm,X_train_new, y_train) * 100, 2)
print("xgb done")
RF_score
###Output
_____no_output_____
###Markdown
* **Tabulating the CV Scores for Different Models**
###Code
results = pd.DataFrame({'Model': ['XGB', 'Logistic Regression', 'Random_Forest','KNN','DT'
],'Score': [xgb_score , logreg_score,RF_score,knn_score,DT_score]})
results = results.sort_values(['Score'], ascending=[False])
results.head()
###Output
_____no_output_____
###Markdown
6. Hyperparameter Tuning for Logistic RegressionResults from the previous step shows that XGBoost and Logistic Regression have the best CV scores (this was also found to be true when the iteration was repeated with 50,100 and 150 features). To reduce the possibility of overfitting, the GridSearchCV method from scikit is used to perform a hyper-paramter optimization on these two models to check if it improves the accuracy of prediction. Below is the code used for a hyperparamter search for logistic regression. * **Grid Search to Optimize the Parameters**
###Code
grid={"C":np.logspace(-2,3,7), "penalty":["l1","l2"]}# l1 lasso l2 ridge
logreg1=LogisticRegression()
logreg_hp=GridSearchCV(logreg1,grid,cv=3,verbose=0)
logreg_hp.fit(train_X_new,train_Y)
print("tuned hpyerparameters :(best parameters) ",logreg_hp.best_params_)
print("accuracy :",logreg_hp.best_score_)
###Output
_____no_output_____
###Markdown
* **Modeling with the Best Parameters from GridSearch**
###Code
logreg2=LogisticRegression(C=1000,penalty="l2")#Grid Search Result C:1000 l2 Accuracy :0.902095
logreg2.fit(train_X_new,train_Y)
logreg2_score = round(cross_val(logreg2,train_X_new, train_Y) * 100, 2)
print("Accuracy for logical regression after doing a GridSearchCV: ", logreg2_score)
###Output
_____no_output_____
###Markdown
7. Hyperparameter Tuning for XGBoost *The code below is commented out as Kaggle timed out before the grid search is complete for these paramters.
###Code
#xgbm_h = xgb.XGBClassifier(silent=False, scale_pos_weight=1,learning_rate=0.01, colsample_bytree = 0.4,subsample = 0.8,objective='binary:logistic',
n_estimators=1000,
reg_alpha = 0.3,
max_depth=4,
gamma=10)
#params = {
# 'min_child_weight': [1, 5, 10],
# 'gamma': [0.5, 1, 1.5, 2, 5],
# 'subsample': [0.6, 0.8, 1.0],
# 'colsample_bytree': [0.6, 0.8, 1.0],
# 'max_depth': [3, 4, 5]
}
#xgbm_hp= GridSearchCV(estimator = xgbm, param_grid = params, scoring='roc_auc',n_jobs=1,iid=False, cv=3)
#xgbm_hp.fit(train_X_new,train_Y)
#xgbm_hp.best_params_, xgbm_hp.best_score_
###Output
_____no_output_____
###Markdown
* **Training XGB Model with Best Parameters from GridSearch **
###Code
#xgbm2 = xgb.XGBClassifier(silent=1,
# scale_pos_weight=1,
# learning_rate=0.01,
# colsample_bytree = 0.4,
# subsample = 0.8,
# objective='binary:logistic',
# n_estimators=1000,
# reg_alpha = 0.3,
# max_depth=4,
# gamma=10)
#xgbm2.fit(train_X_new,train_Y)
#xgb2_score = round(cross_val(xgbm2,train_X_new, train_Y) * 100, 2)
###Output
_____no_output_____
###Markdown
8. Prediction on Test Data using the Best Performing Model
###Code
#Comment out the models you wont use in this iteration
#logreg_pred = logreg.predict_proba(test_X_new)[:,1]
#logreg2_pred = logreg2.predict_proba(test_X_new)[:,1]
xgb_pred =xgbm.predict_proba (test_X_new)[:,1]
#xgb2_pred =xgbm2.predict_proba (test_X_new)[:,1]
###Output
_____no_output_____
###Markdown
8. Generate Output File for Submission
###Code
#The lines below shows you how to save your data in the format needed to score it in the competition
output = pd.DataFrame({'ID_code': test.ID_code,
'target': xgb_pred})
output.to_csv('submission.csv', index=False)
###Output
_____no_output_____
###Markdown
不完備情報2社参入ゲームのサンプルデータを作る。利得関数に入るのはPop, Dist, sigma(相手の参入確率)で、それぞれにつくパラメータとサンプルマーケットの数を指定してオブジェクトを作ることを考える。
###Code
import numpy as np
import matplotlib.pyplot as plt
% matplotlib inline
class Seim:
def __init__(self, alpha, beta, delta, Market_num):
self.alpha, self.beta, self.delta, self.Market_num = alpha, beta, delta, Market_num
def pop(self):
Population = np.random.uniform(size = (self.Merket_num, 1))
return Population
def dist(self):
Distance = np.random.uniform(size = (self.Merket_num, 2))
return Distance
def fixedpoint(self, error):
# ここで初期値に依存して到達する均衡が異なる。→今のモデルであれば具体的にそのthresholdが求められる。
###Output
_____no_output_____
###Markdown
このモデルにおいて別々の均衡に到達する初期値の場所を探す
###Code
# PopとDistを固定した一つの市場に注目する。
def update(sigma1, sigma2, pop, dist, alpha, beta, delta):
updatesigma1 = np.exp(alpha*pop + beta*dist[0] + delta*sigma2)/(1 + np.exp(alpha*pop + beta*dist[0] + delta*sigma2))
updatesigma2 = np.exp(alpha*pop + beta*dist[1] + delta*sigma1)/(1 +np.exp(alpha*pop + beta*dist[1] + delta*sigma1))
return [updatesigma1, updatesigma2]
def fixpoint(sigma1, sigma2, pop, dist, alpha, beta, delta, error):
diff = (update(sigma1, sigma2, pop, dist, alpha, beta, delta)[0] - sigma1)**2 + (update(sigma1, sigma2, pop, dist, alpha, beta, delta)[1] - sigma2)**2
while diff > error:
sigma1 = update(sigma1, sigma2, pop, dist, alpha, beta, delta)[0]
sigma2 = update(sigma1, sigma2, pop, dist, alpha, beta, delta)[1]
diff = (update(sigma1, sigma2, pop, dist, alpha, beta, delta)[0] - sigma1)**2 + (update(sigma1, sigma2, pop, dist, alpha, beta, delta)[1] - sigma2)**2
return [sigma1, sigma2]
# 設定
pop = 1
dist = [1, 2]
alpha = 0.5
beta = -0.7
delta = -0.3
error = 1.0e-20
fixpoint(0, 0, pop, dist, alpha, beta, delta, error)
fixpoint(1,1, pop, dist, alpha, beta, delta, error)
fixpoint(0, 1, pop, dist, alpha, beta, delta, error)
fixpoint(1, 0, pop, dist, alpha, beta, delta, error)
###Output
_____no_output_____
###Markdown
単一っぽい。updateの過程を描く
###Code
point_num = 100
sigmas = np.zeros((point_num, 2))
sigma1 = 0
sigma2 = 0
initial = [sigma1, sigma2]
sigmas[0, :] = initial
for i in range(point_num-1):
sigmas[i+1 , :] = update(sigmas[i, 0], sigmas[i, 1], pop, dist, alpha, beta, delta)
sigmas[0:5, :]
###Output
_____no_output_____
###Markdown
一瞬で収束しちゃうね。 複数均衡が現れる過程を描く。そのためにbest response functionをお互いについて書く。設定はpop = 1dist = [1, 2]alpha = 0.5beta = -0.7delta = -0.3error = 1.0e-20
###Code
# 設定1
pop = 1
dist = [1, 1]
alpha = 0
beta = 0
delta = 0.3
error = 1.0e-20
def br1(sigma2, pop, dist, alpha, beta, delta):
return np.exp(alpha*pop + beta*dist[0] + delta*sigma2)/(1 + np.exp(alpha*pop + beta*dist[0] + delta*sigma2))
def br2(sigma1, pop, dist, alpha, beta, delta):
return np.exp(alpha*pop + beta*dist[1] + delta*sigma1)/(1 +np.exp(alpha*pop + beta*dist[1] + delta*sigma1))
grid_num = 200
grid = np.linspace(0,1,grid_num)
BRs = np.zeros((grid_num, 2))
for i in range(grid_num):
BRs[i, :] = [br1(grid[i], pop, dist, alpha, beta, delta), br2(grid[i], pop, dist, alpha, beta, delta)]
plt.plot(grid, BRs[:, 1], label = "player 2's BR function")
plt.plot(BRs[:, 0], grid, label = "player 1's BR function")
plt.legend()
plt.xlabel("player 1's entry probability")
plt.ylabel("player 2's entry probability")
# 設定2 複数均衡を出す
pop = 1
dist = [1, 1]
alpha = 0
beta = 3
delta = -6
for i in range(grid_num):
BRs[i, :] = [br1(grid[i], pop, dist, alpha, beta, delta), br2(grid[i], pop, dist, alpha, beta, delta)]
plt.plot(grid, BRs[:, 1], label = "player 2's BR function")
plt.plot(BRs[:, 0], grid, label = "player 1's BR function")
plt.legend()
plt.xlabel("player 1's entry probability")
plt.ylabel("player 2's entry probability")
# 設定3 deltaが正、つまりstrategic somplemetaryな時
pop = 1
dist = [1, 1]
alpha = 0
beta = -3
delta = 6
for i in range(grid_num):
BRs[i, :] = [br1(grid[i], pop, dist, alpha, beta, delta), br2(grid[i], pop, dist, alpha, beta, delta)]
plt.plot(grid, BRs[:, 1], label = "player 2's BR function")
plt.plot(BRs[:, 0], grid, label = "player 1's BR function")
plt.legend()
plt.xlabel("player 1's entry probability")
plt.ylabel("player 2's entry probability")
###Output
_____no_output_____
###Markdown
若干の知見- alphaは均衡の数にほぼ関係ない。(BRを上下にシフトさせるだけ)- betaは逆にかなり重要。betaを0のままdeltaを弄って複数の均衡を存在させるにはdeltaがかなり大きくないといけない。しかも、それで均衡が出せるのはdeltaが負の時、すなわちstarategic substituteの時だけっぽい。(証明はなし)- betaの値は重要っぽいが、別にplayerごとにdistが異なってなくても構わない。
###Code
# 設定4 betaを0のままdeltaだけを弄って複数の均衡を生み出す
pop = 1
dist = [1, 1]
alpha = 0
beta = 0
delta = -10
for i in range(grid_num):
BRs[i, :] = [br1(grid[i], pop, dist, alpha, beta, delta), br2(grid[i], pop, dist, alpha, beta, delta)]
plt.plot(grid, BRs[:, 1], label = "player 2's BR function")
plt.plot(BRs[:, 0], grid, label = "player 1's BR function")
plt.legend()
plt.xlabel("player 1's entry probability")
plt.ylabel("player 2's entry probability")
###Output
_____no_output_____ |
notebooks/rotating tetrahedron.ipynb | ###Markdown
Adapted from https://www.tutorialspoint.com/webgl/webgl_sample_application.htm
###Code
import feedWebGL2.feedback as fd
from ipywidgets import interact, interactive, fixed, interact_manual
import numpy as np
fd.widen_notebook()
np.set_printoptions(precision=4)
corners = 0.5 * np.array([
[1, 1, 1],
[1, -1, -1],
[-1, -1, 1],
[-1, 1, -1],
])
colors = corners + 0.5
def tetrahedron_triangles(corners):
triangles = np.zeros([4, 3, 3], dtype=np.float)
for i in range(4):
triangles[i, :i] = corners[:i]
triangles[i, i:] = corners[i+1:]
return triangles
triangles = tetrahedron_triangles(corners)
tcolors = tetrahedron_triangles(colors)
# make faces "flat colored"
if 0:
for i in range(4):
for j in range(3):
tcolors[i,j] = colors[i]
def matrix1(phi, i=0):
result = np.eye(4)
result[0,0] = np.cos(phi)
result[i,i] = np.cos(phi)
result[0,i] = np.sin(phi)
result[i,0] = -np.sin(phi)
return result
def matrix(phi=0.0, theta=0.0, xt=0, yt=0, zt=0):
M1 = matrix1(phi, 1)
#print(M1)
M2 = matrix1(theta, 2)
#print(M2)
M12 = M1.dot(M2)
Mt = np.eye(4)
Mt[3,0] = xt
Mt[3,1] = yt
Mt[3,2] = zt
return M12.dot(Mt)
M = matrix(1.0, 2.0, 0.1, -0.1, 0.2)
M
vertices = triangles.ravel()
def rotate(phi=-0.5, theta=0.0, xt=0.0, yt=0.0, zt=0.0):
M = matrix(phi * np.pi, theta * np.pi, xt, yt, zt)
assert np.abs(np.linalg.det(M) - 1.0) < 0.0001
feedback_program.change_uniform_vector("rotation_matrix", M.ravel())
feedback_program.run()
return M
vertex_shader = """#version 300 es
uniform mat4 rotation_matrix;
in vec3 coordinates;
in vec3 vcolor;
out vec3 output_vertex;
out vec3 coord_color;
void main() {
coord_color = vcolor;
gl_Position = vec4(coordinates, 1.0);
gl_Position = rotation_matrix * gl_Position;
gl_Position[3] = 1.0;
output_vertex = gl_Position.xyz;
}
"""
fragment_shader = """#version 300 es
// For some reason it is required to specify precision, otherwise error.
precision highp float;
in vec3 coord_color;
//out vec4 color;
out vec4 fragmentColor;
void main() {
fragmentColor = vec4(coord_color, 1.0);
}
"""
feedback_program = fd.FeedbackProgram(
program = fd.Program(
vertex_shader = vertex_shader,
fragment_shader = fragment_shader,
feedbacks = fd.Feedbacks(
output_vertex = fd.Feedback(num_components=3),
),
),
runner = fd.Runner(
vertices_per_instance = 3 * len(triangles),
run_type = "TRIANGLES",
uniforms = fd.Uniforms(
rotation_matrix = fd.Uniform(
default_value = list(M.ravel()),
vtype = "4fv",
is_matrix = True,
),
),
inputs = fd.Inputs(
coordinates = fd.Input(
num_components = 3,
from_buffer = fd.BufferLocation(
name = "coordinates_buffer", # start at the beginning, don't skip any values...
),
),
vcolor = fd.Input(
num_components = 3,
from_buffer = fd.BufferLocation(
name = "colors_buffer", # start at the beginning, don't skip any values...
),
),
),
),
context = fd.Context(
buffers = fd.Buffers(
coordinates_buffer = fd.Buffer(
array=list(vertices),
),
colors_buffer = fd.Buffer(
array=list(tcolors.ravel()),
)
),
width = 600,
show = True,
),
)
# display the widget and debugging information
feedback_program.debugging_display()
#feedback_program
#feedback_program.run()
interact(rotate, phi=(-1.0, 1.0), theta=(-1.0, 1.0), xt=(-1.0, 1.0), yt=(-1.0, 1.0), zt=(-1.0, 1.0))
#move_corner(x=-0.1)
A1 = np.array(feedback_program.get_feedback("output_vertex"))
A1
A2 = np.array(feedback_program.get_feedback("output_vertex"))
A2
A1 - A2
colors
np.abs(np.linalg.det(M))
###Output
_____no_output_____ |
itertools.ipynb | ###Markdown
exploring itertools- itertools https://docs.python.org/3/library/itertools.html- more itertools https://more-itertools.readthedocs.io/en/stable/index.html itertools
###Code
import itertools as it
inf=['count', 'cycle', 'repeat']
iter_short=['accumulate', 'chain', 'chain.from_iterable', 'compress', 'dropwhile', 'filterfalse', 'groupby', 'islice', 'pairwise', 'starmap', 'takewhile', 'tee', 'zip_longest']
combinatoric = ['product', 'permutations', 'combinations', 'combinations_with_replacement']
notes="pairwise is new in vesrion 3.10 but exists in more-itertools for earlier versions"
recipes=['take', 'prepend', 'tabulate', 'tail', 'consume', 'nth', 'all_equal', 'quantify', 'pad_none', 'ncycles', 'flatten', 'repeatfunc', 'grouper', 'triplewise', 'sliding_window', 'roundrobin', 'partition', 'before_and_after', 'powerset', 'unique_everseen', 'unique_justseen', 'iter_except', 'first_true', 'random_product', 'random_combination_with_replacement', 'nth_combination']
for x in combinatoric:
print(x)
help(it.__dict__[x])
for x in inf:
print(x)
help(it.__dict__[x])
for x in iter_short:
print(x)
help(it.__dict__[x])
help(it.chain.from_iterable)
for x in combinatoric:
print(x)
help(it.__dict__[x])
###Output
product
Help on class product in module itertools:
class product(builtins.object)
| product(*iterables, repeat=1) --> product object
|
| Cartesian product of input iterables. Equivalent to nested for-loops.
|
| For example, product(A, B) returns the same as: ((x,y) for x in A for y in B).
| The leftmost iterators are in the outermost for-loop, so the output tuples
| cycle in a manner similar to an odometer (with the rightmost element changing
| on every iteration).
|
| To compute the product of an iterable with itself, specify the number
| of repetitions with the optional repeat keyword argument. For example,
| product(A, repeat=4) means the same as product(A, A, A, A).
|
| product('ab', range(3)) --> ('a',0) ('a',1) ('a',2) ('b',0) ('b',1) ('b',2)
| product((0,1), (0,1), (0,1)) --> (0,0,0) (0,0,1) (0,1,0) (0,1,1) (1,0,0) ...
|
| Methods defined here:
|
| __getattribute__(self, name, /)
| Return getattr(self, name).
|
| __iter__(self, /)
| Implement iter(self).
|
| __next__(self, /)
| Implement next(self).
|
| __reduce__(...)
| Return state information for pickling.
|
| __setstate__(...)
| Set state information for unpickling.
|
| __sizeof__(...)
| Returns size in memory, in bytes.
|
| ----------------------------------------------------------------------
| Static methods defined here:
|
| __new__(*args, **kwargs) from builtins.type
| Create and return a new object. See help(type) for accurate signature.
permutations
Help on class permutations in module itertools:
class permutations(builtins.object)
| permutations(iterable, r=None)
|
| Return successive r-length permutations of elements in the iterable.
|
| permutations(range(3), 2) --> (0,1), (0,2), (1,0), (1,2), (2,0), (2,1)
|
| Methods defined here:
|
| __getattribute__(self, name, /)
| Return getattr(self, name).
|
| __iter__(self, /)
| Implement iter(self).
|
| __next__(self, /)
| Implement next(self).
|
| __reduce__(...)
| Return state information for pickling.
|
| __setstate__(...)
| Set state information for unpickling.
|
| __sizeof__(...)
| Returns size in memory, in bytes.
|
| ----------------------------------------------------------------------
| Static methods defined here:
|
| __new__(*args, **kwargs) from builtins.type
| Create and return a new object. See help(type) for accurate signature.
combinations
Help on class combinations in module itertools:
class combinations(builtins.object)
| combinations(iterable, r)
|
| Return successive r-length combinations of elements in the iterable.
|
| combinations(range(4), 3) --> (0,1,2), (0,1,3), (0,2,3), (1,2,3)
|
| Methods defined here:
|
| __getattribute__(self, name, /)
| Return getattr(self, name).
|
| __iter__(self, /)
| Implement iter(self).
|
| __next__(self, /)
| Implement next(self).
|
| __reduce__(...)
| Return state information for pickling.
|
| __setstate__(...)
| Set state information for unpickling.
|
| __sizeof__(...)
| Returns size in memory, in bytes.
|
| ----------------------------------------------------------------------
| Static methods defined here:
|
| __new__(*args, **kwargs) from builtins.type
| Create and return a new object. See help(type) for accurate signature.
combinations_with_replacement
Help on class combinations_with_replacement in module itertools:
class combinations_with_replacement(builtins.object)
| combinations_with_replacement(iterable, r)
|
| Return successive r-length combinations of elements in the iterable allowing individual elements to have successive repeats.
|
| combinations_with_replacement('ABC', 2) --> AA AB AC BB BC CC"
|
| Methods defined here:
|
| __getattribute__(self, name, /)
| Return getattr(self, name).
|
| __iter__(self, /)
| Implement iter(self).
|
| __next__(self, /)
| Implement next(self).
|
| __reduce__(...)
| Return state information for pickling.
|
| __setstate__(...)
| Set state information for unpickling.
|
| __sizeof__(...)
| Returns size in memory, in bytes.
|
| ----------------------------------------------------------------------
| Static methods defined here:
|
| __new__(*args, **kwargs) from builtins.type
| Create and return a new object. See help(type) for accurate signature.
###Markdown
more_itertools
###Code
import more_itertools as mi
grouping="chunked, ichunked, sliced, distribute, divide, split_at, split_before, split_after, split_into, split_when, bucket, unzip, grouper, partition"
look_ahead_behind="spy, peekable, seekable"
windowing="windowed, substrings, substrings_indexes, stagger, windowed_complete, pairwise, triplewise, sliding_window"
Augmenting="count_cycle, intersperse, padded, mark_ends, repeat_last, adjacent, groupby_transform, pad_none, ncycles"
Combining="collapse, sort_together, interleave, interleave_longest, interleave_evenly, zip_offset, zip_equal, zip_broadcast, dotproduct, convolve, flatten, roundrobin, prepend, value_chain"
Summarizing="ilen, unique_to_each, sample, consecutive_groups, run_length, map_reduce, exactly_n, is_sorted, all_equal, all_unique, minmax, first_true, quantify"
Selecting="islice_extended, first, last, one, only, strictly_n, strip, lstrip, rstrip, filter_except, map_except, nth_or_last, unique_in_window, before_and_after, nth, take, tail, unique_everseen, unique_justseen, duplicates_everseen, duplicates_justseen"
Combinatorics="distinct_permutations, distinct_combinations, circular_shifts, partitions, set_partitions, product_index, combination_index, permutation_index, powerset, random_product, random_permutation, random_combination, random_combination_with_replacement, nth_product, nth_permutation, nth_combination"
Wrapping="always_iterable, always_reversible, countable, consumer, with_iter, iter_except"
Others = "locate, rlocate, replace, numeric_range, side_effect, iterate, difference, make_decorator, SequenceView, time_limited, consume, tabulate, repeatfunc"
names = 'grouping, look_ahead_behind, windowing, Augmenting, Combining, Summarizing, Selecting, Combinatorics, Wrapping, Others'.replace(',','').split()
names
recipes_in_more = []
recipes_in_more.extend([x for x in grouping.replace(",","").split() if x in recipes])
recipes_in_more.extend([x for x in look_ahead_behind.replace(",","").split() if x in recipes])
recipes_in_more.extend([x for x in windowing.replace(",","").split() if x in recipes])
recipes_in_more.extend([x for x in Augmenting.replace(",","").split() if x in recipes])
recipes_in_more.extend([x for x in Combining.replace(",","").split() if x in recipes])
recipes_in_more.extend([x for x in Summarizing.replace(",","").split() if x in recipes])
recipes_in_more.extend([x for x in Selecting.replace(",","").split() if x in recipes])
recipes_in_more.extend([x for x in Combinatorics.replace(",","").split() if x in recipes])
recipes_in_more.extend([x for x in Wrapping.replace(",","").split() if x in recipes])
recipes_in_more.extend([x for x in Others.replace(",","").split() if x in recipes])
recipes_in_more
set(recipes)-set(recipes_in_more)
for n in [x.strip() for x in grouping.split(',')]:
print(n)
help(mi.__dict__[n])
for n in [x.strip() for x in look_ahead_behind.split(',')]:
print(n)
help(mi.__dict__[n])
for n in [x.strip() for x in windowing.split(',')]:
print(n)
help(mi.__dict__[n])
for n in [x.strip() for x in Augmenting.split(',')]:
print(n)
help(mi.__dict__[n])
for n in [x.strip() for x in Combining.split(',')]:
print(n)
help(mi.__dict__[n])
for n in [x.strip() for x in Summarizing.split(',')]:
print(n)
help(mi.__dict__[n])
for n in [x.strip() for x in Selecting.split(',')]:
print(n)
help(mi.__dict__[n])
for n in [x.strip() for x in Combinatorics.split(',')]:
print(n)
help(mi.__dict__[n])
re.sub?
import re
worditer = re.sub(r'[\*,\.]','', "Slices with negative values require some caching of *iterable*, but this").lower().split()
print(repr(worditer))
import random
random.shuffle(worditer)
print(repr(worditer))
from queue import
i = [1,2,78, 'fred', ['nested', 'list', 1], 22.56]
mi.random_combination_with_replacement(i, 5)
for n in [x.strip() for x in Wrapping.split(',')]:
print(n)
help(mi.__dict__[n])
for n in [x.strip() for x in Others.split(',')]:
print(n)
help(mi.__dict__[n])
###Output
locate
Help on function locate in module more_itertools.more:
locate(iterable, pred=<class 'bool'>, window_size=None)
Yield the index of each item in *iterable* for which *pred* returns
``True``.
*pred* defaults to :func:`bool`, which will select truthy items:
>>> list(locate([0, 1, 1, 0, 1, 0, 0]))
[1, 2, 4]
Set *pred* to a custom function to, e.g., find the indexes for a particular
item.
>>> list(locate(['a', 'b', 'c', 'b'], lambda x: x == 'b'))
[1, 3]
If *window_size* is given, then the *pred* function will be called with
that many items. This enables searching for sub-sequences:
>>> iterable = [0, 1, 2, 3, 0, 1, 2, 3, 0, 1, 2, 3]
>>> pred = lambda *args: args == (1, 2, 3)
>>> list(locate(iterable, pred=pred, window_size=3))
[1, 5, 9]
Use with :func:`seekable` to find indexes and then retrieve the associated
items:
>>> from itertools import count
>>> from more_itertools import seekable
>>> source = (3 * n + 1 if (n % 2) else n // 2 for n in count())
>>> it = seekable(source)
>>> pred = lambda x: x > 100
>>> indexes = locate(it, pred=pred)
>>> i = next(indexes)
>>> it.seek(i)
>>> next(it)
106
rlocate
Help on function rlocate in module more_itertools.more:
rlocate(iterable, pred=<class 'bool'>, window_size=None)
Yield the index of each item in *iterable* for which *pred* returns
``True``, starting from the right and moving left.
*pred* defaults to :func:`bool`, which will select truthy items:
>>> list(rlocate([0, 1, 1, 0, 1, 0, 0])) # Truthy at 1, 2, and 4
[4, 2, 1]
Set *pred* to a custom function to, e.g., find the indexes for a particular
item:
>>> iterable = iter('abcb')
>>> pred = lambda x: x == 'b'
>>> list(rlocate(iterable, pred))
[3, 1]
If *window_size* is given, then the *pred* function will be called with
that many items. This enables searching for sub-sequences:
>>> iterable = [0, 1, 2, 3, 0, 1, 2, 3, 0, 1, 2, 3]
>>> pred = lambda *args: args == (1, 2, 3)
>>> list(rlocate(iterable, pred=pred, window_size=3))
[9, 5, 1]
Beware, this function won't return anything for infinite iterables.
If *iterable* is reversible, ``rlocate`` will reverse it and search from
the right. Otherwise, it will search from the left and return the results
in reverse order.
See :func:`locate` to for other example applications.
replace
Help on function replace in module more_itertools.more:
replace(iterable, pred, substitutes, count=None, window_size=1)
Yield the items from *iterable*, replacing the items for which *pred*
returns ``True`` with the items from the iterable *substitutes*.
>>> iterable = [1, 1, 0, 1, 1, 0, 1, 1]
>>> pred = lambda x: x == 0
>>> substitutes = (2, 3)
>>> list(replace(iterable, pred, substitutes))
[1, 1, 2, 3, 1, 1, 2, 3, 1, 1]
If *count* is given, the number of replacements will be limited:
>>> iterable = [1, 1, 0, 1, 1, 0, 1, 1, 0]
>>> pred = lambda x: x == 0
>>> substitutes = [None]
>>> list(replace(iterable, pred, substitutes, count=2))
[1, 1, None, 1, 1, None, 1, 1, 0]
Use *window_size* to control the number of items passed as arguments to
*pred*. This allows for locating and replacing subsequences.
>>> iterable = [0, 1, 2, 5, 0, 1, 2, 5]
>>> window_size = 3
>>> pred = lambda *args: args == (0, 1, 2) # 3 items passed to pred
>>> substitutes = [3, 4] # Splice in these items
>>> list(replace(iterable, pred, substitutes, window_size=window_size))
[3, 4, 5, 3, 4, 5]
numeric_range
Help on class numeric_range in module more_itertools.more:
class numeric_range(collections.abc.Sequence, collections.abc.Hashable)
| numeric_range(*args)
|
| An extension of the built-in ``range()`` function whose arguments can
| be any orderable numeric type.
|
| With only *stop* specified, *start* defaults to ``0`` and *step*
| defaults to ``1``. The output items will match the type of *stop*:
|
| >>> list(numeric_range(3.5))
| [0.0, 1.0, 2.0, 3.0]
|
| With only *start* and *stop* specified, *step* defaults to ``1``. The
| output items will match the type of *start*:
|
| >>> from decimal import Decimal
| >>> start = Decimal('2.1')
| >>> stop = Decimal('5.1')
| >>> list(numeric_range(start, stop))
| [Decimal('2.1'), Decimal('3.1'), Decimal('4.1')]
|
| With *start*, *stop*, and *step* specified the output items will match
| the type of ``start + step``:
|
| >>> from fractions import Fraction
| >>> start = Fraction(1, 2) # Start at 1/2
| >>> stop = Fraction(5, 2) # End at 5/2
| >>> step = Fraction(1, 2) # Count by 1/2
| >>> list(numeric_range(start, stop, step))
| [Fraction(1, 2), Fraction(1, 1), Fraction(3, 2), Fraction(2, 1)]
|
| If *step* is zero, ``ValueError`` is raised. Negative steps are supported:
|
| >>> list(numeric_range(3, -1, -1.0))
| [3.0, 2.0, 1.0, 0.0]
|
| Be aware of the limitations of floating point numbers; the representation
| of the yielded numbers may be surprising.
|
| ``datetime.datetime`` objects can be used for *start* and *stop*, if *step*
| is a ``datetime.timedelta`` object:
|
| >>> import datetime
| >>> start = datetime.datetime(2019, 1, 1)
| >>> stop = datetime.datetime(2019, 1, 3)
| >>> step = datetime.timedelta(days=1)
| >>> items = iter(numeric_range(start, stop, step))
| >>> next(items)
| datetime.datetime(2019, 1, 1, 0, 0)
| >>> next(items)
| datetime.datetime(2019, 1, 2, 0, 0)
|
| Method resolution order:
| numeric_range
| collections.abc.Sequence
| collections.abc.Reversible
| collections.abc.Collection
| collections.abc.Sized
| collections.abc.Iterable
| collections.abc.Container
| collections.abc.Hashable
| builtins.object
|
| Methods defined here:
|
| __bool__(self)
|
| __contains__(self, elem)
|
| __eq__(self, other)
| Return self==value.
|
| __getitem__(self, key)
|
| __hash__(self)
| Return hash(self).
|
| __init__(self, *args)
| Initialize self. See help(type(self)) for accurate signature.
|
| __iter__(self)
|
| __len__(self)
|
| __reduce__(self)
| Helper for pickle.
|
| __repr__(self)
| Return repr(self).
|
| __reversed__(self)
|
| count(self, value)
| S.count(value) -> integer -- return number of occurrences of value
|
| index(self, value)
| S.index(value, [start, [stop]]) -> integer -- return first index of value.
| Raises ValueError if the value is not present.
|
| Supporting start and stop arguments is optional, but
| recommended.
|
| ----------------------------------------------------------------------
| Data descriptors defined here:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __abstractmethods__ = frozenset()
|
| ----------------------------------------------------------------------
| Class methods inherited from collections.abc.Reversible:
|
| __subclasshook__(C) from abc.ABCMeta
| Abstract classes can override this to customize issubclass().
|
| This is invoked early on by abc.ABCMeta.__subclasscheck__().
| It should return True, False or NotImplemented. If it returns
| NotImplemented, the normal algorithm is used. Otherwise, it
| overrides the normal algorithm (and the outcome is cached).
side_effect
Help on function side_effect in module more_itertools.more:
side_effect(func, iterable, chunk_size=None, before=None, after=None)
Invoke *func* on each item in *iterable* (or on each *chunk_size* group
of items) before yielding the item.
`func` must be a function that takes a single argument. Its return value
will be discarded.
*before* and *after* are optional functions that take no arguments. They
will be executed before iteration starts and after it ends, respectively.
`side_effect` can be used for logging, updating progress bars, or anything
that is not functionally "pure."
Emitting a status message:
>>> from more_itertools import consume
>>> func = lambda item: print('Received {}'.format(item))
>>> consume(side_effect(func, range(2)))
Received 0
Received 1
Operating on chunks of items:
>>> pair_sums = []
>>> func = lambda chunk: pair_sums.append(sum(chunk))
>>> list(side_effect(func, [0, 1, 2, 3, 4, 5], 2))
[0, 1, 2, 3, 4, 5]
>>> list(pair_sums)
[1, 5, 9]
Writing to a file-like object:
>>> from io import StringIO
>>> from more_itertools import consume
>>> f = StringIO()
>>> func = lambda x: print(x, file=f)
>>> before = lambda: print(u'HEADER', file=f)
>>> after = f.close
>>> it = [u'a', u'b', u'c']
>>> consume(side_effect(func, it, before=before, after=after))
>>> f.closed
True
iterate
Help on function iterate in module more_itertools.more:
iterate(func, start)
Return ``start``, ``func(start)``, ``func(func(start))``, ...
>>> from itertools import islice
>>> list(islice(iterate(lambda x: 2*x, 1), 10))
[1, 2, 4, 8, 16, 32, 64, 128, 256, 512]
difference
Help on function difference in module more_itertools.more:
difference(iterable, func=<built-in function sub>, *, initial=None)
This function is the inverse of :func:`itertools.accumulate`. By default
it will compute the first difference of *iterable* using
:func:`operator.sub`:
>>> from itertools import accumulate
>>> iterable = accumulate([0, 1, 2, 3, 4]) # produces 0, 1, 3, 6, 10
>>> list(difference(iterable))
[0, 1, 2, 3, 4]
*func* defaults to :func:`operator.sub`, but other functions can be
specified. They will be applied as follows::
A, B, C, D, ... --> A, func(B, A), func(C, B), func(D, C), ...
For example, to do progressive division:
>>> iterable = [1, 2, 6, 24, 120]
>>> func = lambda x, y: x // y
>>> list(difference(iterable, func))
[1, 2, 3, 4, 5]
If the *initial* keyword is set, the first element will be skipped when
computing successive differences.
>>> it = [10, 11, 13, 16] # from accumulate([1, 2, 3], initial=10)
>>> list(difference(it, initial=10))
[1, 2, 3]
make_decorator
Help on function make_decorator in module more_itertools.more:
make_decorator(wrapping_func, result_index=0)
Return a decorator version of *wrapping_func*, which is a function that
modifies an iterable. *result_index* is the position in that function's
signature where the iterable goes.
This lets you use itertools on the "production end," i.e. at function
definition. This can augment what the function returns without changing the
function's code.
For example, to produce a decorator version of :func:`chunked`:
>>> from more_itertools import chunked
>>> chunker = make_decorator(chunked, result_index=0)
>>> @chunker(3)
... def iter_range(n):
... return iter(range(n))
...
>>> list(iter_range(9))
[[0, 1, 2], [3, 4, 5], [6, 7, 8]]
To only allow truthy items to be returned:
>>> truth_serum = make_decorator(filter, result_index=1)
>>> @truth_serum(bool)
... def boolean_test():
... return [0, 1, '', ' ', False, True]
...
>>> list(boolean_test())
[1, ' ', True]
The :func:`peekable` and :func:`seekable` wrappers make for practical
decorators:
>>> from more_itertools import peekable
>>> peekable_function = make_decorator(peekable)
>>> @peekable_function()
... def str_range(*args):
... return (str(x) for x in range(*args))
...
>>> it = str_range(1, 20, 2)
>>> next(it), next(it), next(it)
('1', '3', '5')
>>> it.peek()
'7'
>>> next(it)
'7'
SequenceView
Help on class SequenceView in module more_itertools.more:
class SequenceView(collections.abc.Sequence)
| SequenceView(target)
|
| Return a read-only view of the sequence object *target*.
|
| :class:`SequenceView` objects are analogous to Python's built-in
| "dictionary view" types. They provide a dynamic view of a sequence's items,
| meaning that when the sequence updates, so does the view.
|
| >>> seq = ['0', '1', '2']
| >>> view = SequenceView(seq)
| >>> view
| SequenceView(['0', '1', '2'])
| >>> seq.append('3')
| >>> view
| SequenceView(['0', '1', '2', '3'])
|
| Sequence views support indexing, slicing, and length queries. They act
| like the underlying sequence, except they don't allow assignment:
|
| >>> view[1]
| '1'
| >>> view[1:-1]
| ['1', '2']
| >>> len(view)
| 4
|
| Sequence views are useful as an alternative to copying, as they don't
| require (much) extra storage.
|
| Method resolution order:
| SequenceView
| collections.abc.Sequence
| collections.abc.Reversible
| collections.abc.Collection
| collections.abc.Sized
| collections.abc.Iterable
| collections.abc.Container
| builtins.object
|
| Methods defined here:
|
| __getitem__(self, index)
|
| __init__(self, target)
| Initialize self. See help(type(self)) for accurate signature.
|
| __len__(self)
|
| __repr__(self)
| Return repr(self).
|
| ----------------------------------------------------------------------
| Data descriptors defined here:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __abstractmethods__ = frozenset()
|
| ----------------------------------------------------------------------
| Methods inherited from collections.abc.Sequence:
|
| __contains__(self, value)
|
| __iter__(self)
|
| __reversed__(self)
|
| count(self, value)
| S.count(value) -> integer -- return number of occurrences of value
|
| index(self, value, start=0, stop=None)
| S.index(value, [start, [stop]]) -> integer -- return first index of value.
| Raises ValueError if the value is not present.
|
| Supporting start and stop arguments is optional, but
| recommended.
|
| ----------------------------------------------------------------------
| Class methods inherited from collections.abc.Reversible:
|
| __subclasshook__(C) from abc.ABCMeta
| Abstract classes can override this to customize issubclass().
|
| This is invoked early on by abc.ABCMeta.__subclasscheck__().
| It should return True, False or NotImplemented. If it returns
| NotImplemented, the normal algorithm is used. Otherwise, it
| overrides the normal algorithm (and the outcome is cached).
time_limited
Help on class time_limited in module more_itertools.more:
class time_limited(builtins.object)
| time_limited(limit_seconds, iterable)
|
| Yield items from *iterable* until *limit_seconds* have passed.
| If the time limit expires before all items have been yielded, the
| ``timed_out`` parameter will be set to ``True``.
|
| >>> from time import sleep
| >>> def generator():
| ... yield 1
| ... yield 2
| ... sleep(0.2)
| ... yield 3
| >>> iterable = time_limited(0.1, generator())
| >>> list(iterable)
| [1, 2]
| >>> iterable.timed_out
| True
|
| Note that the time is checked before each item is yielded, and iteration
| stops if the time elapsed is greater than *limit_seconds*. If your time
| limit is 1 second, but it takes 2 seconds to generate the first item from
| the iterable, the function will run for 2 seconds and not yield anything.
|
| Methods defined here:
|
| __init__(self, limit_seconds, iterable)
| Initialize self. See help(type(self)) for accurate signature.
|
| __iter__(self)
|
| __next__(self)
|
| ----------------------------------------------------------------------
| Data descriptors defined here:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
consume
Help on function consume in module more_itertools.recipes:
consume(iterator, n=None)
Advance *iterable* by *n* steps. If *n* is ``None``, consume it
entirely.
Efficiently exhausts an iterator without returning values. Defaults to
consuming the whole iterator, but an optional second argument may be
provided to limit consumption.
>>> i = (x for x in range(10))
>>> next(i)
0
>>> consume(i, 3)
>>> next(i)
4
>>> consume(i)
>>> next(i)
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
StopIteration
If the iterator has fewer items remaining than the provided limit, the
whole iterator will be consumed.
>>> i = (x for x in range(3))
>>> consume(i, 5)
>>> next(i)
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
StopIteration
tabulate
Help on function tabulate in module more_itertools.recipes:
tabulate(function, start=0)
Return an iterator over the results of ``func(start)``,
``func(start + 1)``, ``func(start + 2)``...
*func* should be a function that accepts one integer argument.
If *start* is not specified it defaults to 0. It will be incremented each
time the iterator is advanced.
>>> square = lambda x: x ** 2
>>> iterator = tabulate(square, -3)
>>> take(4, iterator)
[9, 4, 1, 0]
repeatfunc
Help on function repeatfunc in module more_itertools.recipes:
repeatfunc(func, times=None, *args)
Call *func* with *args* repeatedly, returning an iterable over the
results.
If *times* is specified, the iterable will terminate after that many
repetitions:
>>> from operator import add
>>> times = 4
>>> args = 3, 5
>>> list(repeatfunc(add, times, *args))
[8, 8, 8, 8]
If *times* is ``None`` the iterable will not terminate:
>>> from random import randrange
>>> times = None
>>> args = 1, 11
>>> take(6, repeatfunc(randrange, times, *args)) # doctest:+SKIP
[2, 4, 8, 1, 8, 4]
###Markdown
itertools> This module implements a number of iterator building blocks inspired by constructs from APL, Haskell, and SML. Each has been recast in a form suitable for Python. The module standardizes a core set of fast, memory efficient tools that are useful by themselves or in combination. Together, they form an “iterator algebra” making it possible to construct specialized tools succinctly and efficiently in pure Python.[Python 3 Standard Library - itertools](https://docs.python.org/3/library/itertools.html)Infinite iterators - count() - cycle() - repeat()Iterators terminating on the shortest input sequence: - accumulate() - chain() - chain.from_iterable() - compress() - dropwhile() - filterfalse() - groupby() - islice() - starmap() - takewhile() - tee() - zip_longest()Combinatoric iterators: - product() - permutations() - combinations() - combinations_with_replacement() accumulate()Return series of accumulated sums (or other binary function results).[Python 3 Standard Library - itertools.accumulate](https://docs.python.org/3/library/itertools.htmlitertools.accumulate)
###Code
from itertools import accumulate
accumulations = accumulate([1, 2, 3])
list(accumulations)
###Output
_____no_output_____
###Markdown
chain()Return a chain object whose __next__() method returns elements from the first iterable until it is exhausted, then elements from the next iterable, until all of the iterables are exhausted.[Python 3 Standard Library - itertools.chain](https://docs.python.org/3/library/itertools.htmlitertools.chain)
###Code
# Example (slightly modified) from:
# https://realpython.com/python-itertools/#section-recap_2
from itertools import chain
chained_iterator = chain('abc', [1, 2, 3])
chained_iterator
list(chained_iterator)
###Output
_____no_output_____
###Markdown
chain.from_iterable()Alternate constructor for chain(). Gets chained inputs from a single iterable argument.[Python 3 Standard Library - itertools.chain.from_iterable](https://docs.python.org/3/library/itertools.htmlitertools.chain.from_iterable)
###Code
from itertools import chain
chained_iterator = chain.from_iterable(['ABC', 'DEF'])
chained_iterator
list(chained_iterator)
###Output
_____no_output_____
###Markdown
combinations()Return successive n-length combinations of elements in the iterable.[Python 3 Standard Library - itertools.combinations](https://docs.python.org/3/library/itertools.htmlitertools.combinations)
###Code
# Example (slightly modified) from:
# https://realpython.com/python-itertools/#section-recap
from itertools import combinations
combinations = combinations([1, 2, 3], 2)
list(combinations)
###Output
_____no_output_____
###Markdown
combinations_with_replacement()Return successive n-length combinations of elements in the iterable allowing individual elements to have successive repeats.[Python 3 Standard Library - itertools.combinations_with_replacement](https://docs.python.org/3/library/itertools.htmlitertools.combinations_with_replacement)
###Code
# Example (slightly modified) from:
# https://realpython.com/python-itertools/#section-recap
from itertools import combinations_with_replacement
combinations = combinations_with_replacement([1, 2, 3], 2)
list(combinations)
###Output
_____no_output_____
###Markdown
compress()Make an iterator that filters elements from data returning only those that have a corresponding element in selectors that evaluates to True.[Python 3 Standard Library - itertools.compress](https://docs.python.org/3/library/itertools.htmlitertools.compress)
###Code
from itertools import compress
filtered_values = compress('ABCDEF', [1,0,1,0,1,1])
filtered_values
list(filtered_values)
###Output
_____no_output_____
###Markdown
count()Return a count object whose .__next__() method returns consecutive values.[Python 3 Standard Library - itertools.count](https://docs.python.org/3/library/itertools.htmlitertools.count)``` Example from: https://realpython.com/python-itertools/section-recap>>> from itertools import count>>> count(start=1, step=2)1, 3, 5, 7, 9, ...``` cycle()Return elements from the iterable until it is exhausted. Then repeat the sequence indefinitely.[Python 3 Standard Library - itertools.cycle](https://docs.python.org/3/library/itertools.htmlitertools.cycle)```>>> from itertools import cycle>>> cycle('ABCD')A B C D A B C D A B C D ...``` dropwhile()Drop items from the iterable while pred(item) is true. Afterwards, return every element until the iterable is exhausted.[Python 3 Standard Library - itertools.dropwhile](https://docs.python.org/3/library/itertools.htmlitertools.dropwhile)
###Code
from itertools import dropwhile
filtered_values = dropwhile(lambda x: x<5, [1,4,6,4,1])
filtered_values
list(filtered_values)
###Output
_____no_output_____
###Markdown
islice()Return an iterator whose __next__() method returns selected values from an iterable. Works like a slice() on a list but returns an iterator.[Python 3 Standard Library - itertools.islice](https://docs.python.org/3/library/itertools.htmlitertools.islice)
###Code
from itertools import islice
iterable = [0, 1, 2, 3, 4, 5, 6, 7, 8, 9]
sliced_iterator = islice(iterable, 1, 7, 2) # iterable, start, stop, step
sliced_iterator
list(sliced_iterator)
###Output
_____no_output_____
###Markdown
filterfalse()Return those items of sequence for which pred(item) is false. If pred is None, return the items that are false.[Python 3 Standard Library - itertools.filterfalse](https://docs.python.org/3/library/itertools.htmlitertools.filterfalse)
###Code
# Example from:
# https://docs.python.org/3/library/itertools.html#itertools.filterfalse
from itertools import filterfalse
even_numbers = filterfalse(lambda x: x%2, range(10))
even_numbers
list(even_numbers)
###Output
_____no_output_____
###Markdown
groupby()Make an iterator that returns consecutive keys and groups from the iterable.[Python 3 Standard Library - itertools.groupby](https://docs.python.org/3/library/itertools.htmlitertools.groupby)
###Code
from itertools import groupby
grouped_values = (k for k, g in groupby('AAAABBBCCDAABBB'))
grouped_values
list(grouped_values)
###Output
_____no_output_____
###Markdown
permutations()Return successive n-length permutations of elements in the iterable.[Python 3 Standard Library - itertools.permutations](https://docs.python.org/3/library/itertools.htmlitertools.permutations)
###Code
# Example (slightly modified) from:
# https://realpython.com/python-itertools/#section-recap
from itertools import permutations
permutations = permutations('abc')
list(permutations)
###Output
_____no_output_____
###Markdown
product()Cartesian product of input iterables. Equivalent to nested for-loops. ((x,y) for x in A for y in B)[Python 3 Standard Library - itertools.product](https://docs.python.org/3/library/itertools.htmlitertools.product)
###Code
# Example (slightly modified) from:
# https://realpython.com/python-itertools/#section-recap_2
from itertools import product
product = product([1, 2], ['a', 'b'])
product
list(product)
# Example (slightly modified) from:
# https://docs.python.org/3/library/itertools.html#itertools.product
same_product = ((x,y) for x in [1, 2] for y in ['a', 'b'])
same_product
list(same_product)
###Output
_____no_output_____
###Markdown
repeat()Create an iterator which returns the object for the specified number of times. If not specified, returns the object endlessly.[Python 3 Standard Library - itertools.repeat](https://docs.python.org/3/library/itertools.htmlitertools.repeat)
###Code
# Example (slightly modified) from:
# https://realpython.com/python-itertools/#section-recap
from itertools import repeat
repetitions = repeat(2, 5)
list(repetitions)
###Output
_____no_output_____
###Markdown
starmap()Make an iterator that computes the function using arguments obtained from the iterable. Used instead of [map()](https://docs.python.org/3/library/functions.htmlmap) when argument parameters are already grouped in tuples from a single iterable (the data has been “pre-zipped”)[Python 3 Standard Library - itertools.starmap](https://docs.python.org/3/library/itertools.htmlitertools.starmap)
###Code
from itertools import starmap
powers = starmap(pow, [(2,5), (3,2), (10,3)])
powers
list(powers)
###Output
_____no_output_____
###Markdown
takewhile()Make an iterator that returns elements from the iterable as long as the predicate is true.[Python 3 Standard Library - itertools.takewhile](https://docs.python.org/3/library/itertools.htmlitertools.takewhile)
###Code
# Example from:
# https://docs.python.org/3/library/itertools.html#itertools.takewhile
from itertools import takewhile
filtered_values = takewhile(lambda x: x<5, [1,4,6,4,1])
filtered_values
list(filtered_values)
###Output
_____no_output_____
###Markdown
tee()Create any number of independent iterators from a single input iterable.[Python 3 Standard Library - itertools.tee](https://docs.python.org/3/library/itertools.htmlitertools.tee)
###Code
# Example (slightly modified) from:
# https://realpython.com/python-itertools/#section-recap_2
from itertools import tee
iterable = ['a', 'b', 'c']
clone_1, clone_2 = tee(iterable, 2)
list(clone_1)
list(clone_2)
###Output
_____no_output_____
###Markdown
zip_longest()Make an iterator that aggregates elements from each of the iterables.[Python 3 Standard Library - itertools.zip_longest](https://docs.python.org/3/library/itertools.htmlitertools.zip_longest)
###Code
from itertools import zip_longest
zipped = zip_longest('ABCD', 'xy', fillvalue='-')
zipped
list(zipped)
###Output
_____no_output_____
###Markdown
How do we loop over the elements on a list?
###Code
numbers = [1, 2, 3, 4, 5]
for n in numbers:
print(n)
###Output
1
2
3
4
5
###Markdown
We all agree that's much better than:
###Code
for index in range(0, len(numbers)):
print(numbers[index])
###Output
1
2
3
4
5
###Markdown
Every time somebody loops over the indexes, a kitten dies. ... or, even worse:
###Code
index = 0
while index < len(numbers):
print(numbers[index])
index += 1
###Output
1
2
3
###Markdown
This is going to get at least *two* kittens dead. Python's for loop- Known as 'for-each' in other programming languages.- **Higher abstraction** level $\rightarrow$ more **readable** and clear.- We don't care about indexes or implementation details.- We just say *"loop over this"* (whatever it is) Recommended talk: [Loop Like a Native](https://www.youtube.com/watch?v=EnSu9hHGq5o), by Ned Batchelder. It's safer- Not manipulating indexes $\rightarrow$ fewer places were we can make mistakes.- Actually, we can arguably err nowhere using a for-each loop.
###Code
index = 0
while index <= len(numbers): # Ooops!
print(numbers[index])
index += 1
###Output
1
2
3
###Markdown
The for loop not only works with lists Strings
###Code
for letter in "aeiou":
print(letter)
###Output
a
e
i
o
u
###Markdown
Sets
###Code
numbers = {"one", "two", "three"}
for n in numbers:
print(n)
###Output
two
three
one
###Markdown
Note: no guarantees as for the order. See [All Your Ducks In A Row](https://www.youtube.com/watch?v=fYlnfvKVDoM), by Brandon Rhodes Dictionaries
###Code
numbers = {1 : "one", 2 : "two", 3 : "three"}
for key, value in numbers.items():
print(key, "->", value)
###Output
1 -> one
2 -> two
3 -> three
###Markdown
Files
###Code
with open("quixote.txt") as fd:
for line in fd:
print(line, end="") # line already includes a newline
###Output
In a village of La Mancha, the name of which I have no desire to call
to mind, there lived not long since one of those gentlemen that keep a
lance in the lance-rack, an old buckler, a lean hack, and a greyhound
for coursing.
###Markdown
We can use the for loop with anything that's *iterable* Advanced tip: we can use [Abstract Base Classes](https://docs.python.org/3/library/abc.html).[collections.abc.Iterable](https://docs.python.org/3/library/collections.abc.htmlcollections.abc.Iterable) is an ABC for classes that are iterable.
###Code
import collections.abc
print(issubclass(list, collections.abc.Iterable))
print(issubclass(str, collections.abc.Iterable))
isinstance({1, 2}, collections.Iterable)
isinstance("abcd", collections.Iterable)
###Output
_____no_output_____
###Markdown
Detour: What's an iterable? From the [Glossary](https://docs.python.org/3/glossary.htmlterm-iterable):> An object capable of returning its members one at a time. Examples of iterables include all sequence types (such as `list`, `str`, and `tuple`) and some non-sequence types like dict, file objects, and objects of any classes you define with an `__iter__()` or `__getitem__()` method. So our object will be iterable if it implements `__iter__()` [`*`] [`*`] ... or `__getitem__()`, but let's focus on `__iter__()`. What's `__iter__()`? Again, [from the docs](https://docs.python.org/3/reference/datamodel.htmlobject.__iter__):> This method is called when an iterator is required for a container. This method should return a new iteratorobject that can iterate over all the objects in the container. For mappings, it should iterate over the keys of the container.- So we need to implement `__iter__()` and make it return an *iterator*- This method is called implicitly by the built-in `iter()` I don't believe you
###Code
numbers = [1, 2, 3]
print(numbers.__iter__)
###Output
<method-wrapper '__iter__' of list object at 0x7fdbbc074788>
###Markdown
OK, `__iter__()` exists.
###Code
it = iter(numbers) # calls __iter__()
print(it)
###Output
<list_iterator object at 0x7fdb9e410860>
###Markdown
... and it returns an iterator. But... what's an *iterator*?It's an object that conforms to the [**Iterator Protocol**](https://docs.python.org/3/library/stdtypes.htmliterator-types), implementing two methods. `iterator.__iter__()`> Return the iterator object itself. This is required to allow both containers and iterators to be used with the for and in statements. This method corresponds to the `tp_iter` slot of the type structure for Python objects in the Python/C API.- TL;DR: implement `__iter__()`, make it return `self`.- We need to do this, for... whatever reasons. I'll follow the instructions, then.
###Code
class MyIterator(object):
def __iter__(self):
return self
###Output
_____no_output_____
###Markdown
This feels pointless. `iterator.__next__()` > Return the next item from the container. If there are no further items, raise the `StopIteration` exception. This method corresponds to the `tp_iternext` slot of the type structure for Python objects in the Python/C API.- No, I did not understand the second sentence either.- This was `.next()` in the now long-forgotten, barely remembered Python 2.
###Code
class MyIterator(object):
def __init__(self, elements):
self.elements = elements
self.index = 0
def __iter__(self):
return self
def __next__(self):
# No elements left, so raise exception.
if self.index >= len(self.elements):
raise StopIteration
result = self.elements[self.index]
self.index += 1
return result
it = MyIterator([1, 2, 3])
print(next(it)) # calls __next__()
print(next(it))
print(next(it))
###Output
1
2
3
###Markdown
- We've built our own ugly, awkward iterator object. Yay!- Recommended reading: [Understanding Python Iterables and Iterators](http://www.shutupandship.com/2012/01/understanding-python-iterables-and.html) The fourth time we call `next()`, `StopIteration` happens:
###Code
print(next(it))
###Output
_____no_output_____
###Markdown
`next()` must be used on iteratorsEven if our class is iterable, we need an iterator to loop over it:
###Code
numbers = [1, 2, 3]
next(numbers)
numbers = [1, 2, 3]
it = iter(numbers) # calls __iter__(), returns iterator
print("Iterator:", it) # it is an iterator indeed
print(next(it)) # returns 1
print(next(it)) # returns 2
print(next(it)) # returns 3
print(next(it)) # raises StopIteration
###Output
1
2
3
###Markdown
This is exactly what the `for` loop does under the hood- Uses `iter()` to get an iterator.- Repeatedly calls `next()`.- Stops when `StopIteration` is raised.
###Code
numbers = [1, 2, 3]
def awkward_for(iterable):
it = iter(iterable)
while True:
try:
print(next(it))
except StopIteration:
break
awkward_for(numbers)
###Output
1
2
3
###Markdown
I'm so happy we have the built-in `for` instead of this monster. `for` and `StopIteration`- Although the iterator *will* raise StopIteration, we don't have to worry about it.- It turns out that the for loop listens for StopIteration explicitly.- It doesn't catch other exceptions raised by the iterator...- ... or `StopIteration` raised within the body of the loop.
###Code
numbers = [1, 2, 3]
for n in numbers:
raise StopIteration # break stuff
###Output
_____no_output_____
###Markdown
Detour: GeneratorsGenerators are a special type of iterator.How a normal function works:- The execution starts at the function's first line.- It continues until we reach a `return` statement...- ... or the end of the function — that returns `None` implicitly.- Whatever result we return, must be returned at once. Recommended readings:- ['yield' and Generators Explained](https://jeffknupp.com/blog/2013/04/07/improve-your-python-yield-and-generators-explained/)- [Generator Tricks for Systems Programmers](http://www.dabeaz.com/generators/) Single exit point
###Code
def foo(x):
"""Add one, then square."""
y = x + 1 # we first do this
z = y ** 2 # then we do this
return z # exit the function here
print(foo(4))
###Output
25
###Markdown
Multiple exit points
###Code
import random
def spam(x):
"""Square if even, halve if odd."""
if x % 2 == 0:
return x ** 2 # we may exit here...
return x / 2 # ... or here.
print(spam(4)) # 4 ** 2 = 16
print(spam(7)) # 7 / 2 = 3.5
###Output
16
3.5
###Markdown
Implicit `None`
###Code
def foobar(x):
y = x + 1 # do nothing with 'y'
# implicit `return None`
print(foobar(2))
###Output
None
###Markdown
Let's say that we want to work with even numbers.
###Code
def get_even(stop):
"""Return all the even numbers <= stop."""
numbers = []
n = 0
while n <= stop:
numbers.append(n)
n += 2
return numbers
###Output
_____no_output_____
###Markdown
... or, of course, simply:
###Code
def get_even(stop):
return list(range(0, stop + 1, 2))
print(get_even(10)) # [2 ... 10]
print("Sum:", sum(get_even(10)))
###Output
[0, 2, 4, 6, 8, 10]
Sum: 30
###Markdown
When problems arise- The problem is that we're building the list...- ... and returning all the elements **at once**.- Working with large intervals becomes impractical, to say the least.
###Code
sum(get_even(int(1e18))) # not enough RAM modules
###Output
_____no_output_____
###Markdown
Generators to the rescue- Generators are functions that are able to return values **one by one**.- The state of the function is frozen until the next value is requested.- They use they `yield` keyword instead of `return`.- If there's $\gt 1$ `yield` in our function, it becomes a `generator function`.- Generator functions return generator iterators (generators, for short)
###Code
def simple_generator():
yield 1 # will return this the first time
yield 2 # this the second time
yield 3 # and this the third time
g = simple_generator()
print(g)
print(next(g)) # returns 1
print(next(g)) # returns 2
print(next(g)) # returns 3
print(next(g)) # raises StopIteration
###Output
<generator object simple_generator at 0x7fdbe0916a20>
1
2
3
###Markdown
Let's say that we want a function that returns **all** natural numbers:
###Code
def count():
n = 1
while True:
yield n
n += 1
numbers = count()
print(numbers) # again, it's a generator
print(next(numbers)) # 1
print(next(numbers)) # 2
print(next(numbers)) # 3
###Output
<generator object count at 0x7fdbe0930828>
1
2
3
###Markdown
... and so on, up to infinity and beyond. This is not possible with a normal function — we can't return *all* the natural numbers.
###Code
def count():
"""Try to return all natural numbers, and fail."""
result = []
n = 1
while True:
result.append(n)
n += 1
###Output
_____no_output_____
###Markdown
Don't run this code. Extra points: generator expressions- Like comprehension lists, but with **lazy** evaluation.- They generate the elements one by one, as needed.
###Code
g = (x ** 2 for x in range(1, 1000000))
print(g) # it's a generator indeed
print(next(g)) # computes the square of 1
print(next(g)) # computes the square of 2
print(next(g)) # computes the square of 3
###Output
<generator object <genexpr> at 0x7fdb9e428ab0>
1
4
9
###Markdown
Even more extra points:- `range()` is **not** a generator- In the old world of Python 2 it returned a list.- But now, in Python3, generates elements one by one.
###Code
numbers = range(1, 100)
print(numbers) # range(1, 100)
print(type(numbers)) # it's a 'range' object
it = iter(numbers)
print(next(it)) # returns the first element
print(next(it)) # returns the second element
###Output
1
2
###Markdown
So...- `range` is a `sequence object` that produces numbers on demand.- `sequences` in Python are object that support random access.- It can also contain if the number is part of the range in $\mathcal{O}(1)$...- ... instead of the $\mathcal{O}(n)$ that it would take to scan through all of them.
###Code
odd = range(1, int(1e50), 2)
print(odd) # a really large range indeed
###Output
range(1, 100000000000000007629769841091887003294964970946560, 2)
###Markdown
print(odd[9]) tell me what's the 10th odd number
###Code
print(int(1e48) in odd) # completes before the heat death of the Universe
###Output
False
###Markdown
Summary- Iterators are objects on which we can call `next()`- Generators generate elements one by one.- Every generator is an iterator, but not vice versa.- We could achieve the same with a custom iterator — but this is nicer.Recommended reading: [this answer by Alex Martelli on Stack Owerflow](http://stackoverflow.com/a/2776865). Extra: Measuring a generatorHow do we calculate the number of elements of a generator?
###Code
g = (x ** 2 for x in range(1, 101))
###Output
_____no_output_____
###Markdown
This doesn't work.
###Code
len(g) # TypeError
###Output
_____no_output_____
###Markdown
Idea: cast to list
###Code
g = (x ** 2 for x in range(1, 101))
len(list(g))
###Output
_____no_output_____
###Markdown
But we need a temporary list. Better: use `sum()`
###Code
g = (x ** 2 for x in range(1, 101))
sum(1 for x in g) # use another generator
###Output
_____no_output_____
###Markdown
Or more idiomatic, as we don't care about the vale of the numbers:
###Code
g = (x ** 2 for x in range(1, 101))
sum(1 for _ in g)
###Output
_____no_output_____
###Markdown
Use `_` as a throwaway variable. Finally, time for some Kung Fu[Photo](https://www.flickr.com/photos/kurt-b/9453209945/) by Kurt Bauschardt / [CC BY-SA 2.0](https://creativecommons.org/licenses/by-sa/2.0/) Prime numbersHow do we determine whether a number is prime?
###Code
def is_prime(n):
"""Checks whether a number is prime."""
divisor = 2
while divisor < n:
if n % divisor == 0:
return False
divisor += 1
return True
print(is_prime(2)) # True
print(is_prime(3)) # True
print(is_prime(8)) # False
###Output
True
True
False
###Markdown
But we already now there's a much better way with `range()`!
###Code
def is_prime(n):
"""Checks whether a number is prime."""
for divisor in range(2, n):
if n % divisor == 0:
return False
divisor += 1
return True
print(is_prime(5)) # True
print(is_prime(6)) # False
print(is_prime(7)) # True
###Output
True
False
True
###Markdown
Mandatory optimization: check divisors only until $\sqrt{n}$- Because a non-prime number $n$ will have a divisor $\le \sqrt{n}$- Brings us down from $\mathcal{O}(n)$ to $\mathcal{O}(\sqrt{n})$. Not bad.- Recommended reading, with proof for humans: [this answer on Stack Overflow](http://stackoverflow.com/a/5811176)
###Code
import math
def is_prime(n):
"""Checks whether a number is prime."""
stop = int(math.sqrt(n)) + 1
for divisor in range(2, stop):
if n % divisor == 0:
return False
divisor += 1
return True
print(is_prime(10))
print(is_prime(13))
###Output
False
True
###Markdown
Integer factorization- Now we want to decompose a number into its prime factors.- For example, $8 = 2 \times 2 \times 2 = 2^3$, and $30 = 2 \times 3 \times 5$- Simplest method: [trial division](https://en.wikipedia.org/wiki/Trial_division), one by one- For this we'll need to loop over prime numbers Getting the first n prime numbers Attempt 1
###Code
def get_primes(how_many):
"""Return the first 'how_many' prime numbers."""
primes = []
n = 2
while len(primes) < how_many:
if is_prime(n):
primes.append(n)
n += 1
return primes
print(get_primes(5))
###Output
[2, 3, 5, 7, 11]
###Markdown
**Problem:** we need to generate all the prime numbers at once.Let's use a generator instead! Attempt 2
###Code
def get_primes(how_many):
"""Return the first 'how_many' prime numbers."""
counter = 0
n = 2
while counter < how_many:
if is_prime(n):
yield n
counter += 1
n += 1
###Output
<generator object get_primes at 0x7fdbe0941120>
[2, 3, 5, 7, 11]
###Markdown
Twice as many places where we can commit a mistake, as we're keeping track of:- How many prime numbers we have generated so far.- What's the next number we need to evaluate.`range()` is not an option because we don't know what the stop is. But we can use... itertools.count()- Makes an iterator that returns evenly spaced values starting with $n$.- By default, we start counting from zero.- The default step is one.
###Code
import itertools
numbers = itertools.count()
print(numbers) # tells us what the next element will be
print(next(numbers)) # 0
print(next(numbers)) # 1
print(next(numbers)) # 2
odd = itertools.count(1, step=2)
print(next(odd)) # first odd number
print(next(odd)) # second odd number
###Output
1
3
###Markdown
Attempt 3We can thus simplify our code a little bit:
###Code
def get_primes(how_many):
"""Return the first 'how_many' prime numbers."""
counter = 0
for n in itertools.count(2):
if is_prime(n):
yield n
counter += 1
if counter == how_many:
break
###Output
_____no_output_____
###Markdown
However, this is still awkward, as our function does two things:- Generates prime numbers- Keeps track if how many we have generated so far. Let's split it in two different steps. Attempt 4
###Code
def primes():
"""An endless generator of prime numbers."""
for n in itertools.count(2):
if is_prime(n):
yield n
def get_primes(how_many):
"""Return the first 'how_many' prime numbers."""
counter = 0
all_primes = primes()
while counter < how_many:
yield next(all_primes)
counter += 1
###Output
_____no_output_____
###Markdown
Good! But this can be simplified even further! To get all the prime numbers we can use a generator expression
###Code
primes = (n for n in itertools.count(2) if is_prime(n))
###Output
_____no_output_____
###Markdown
... or `filter()`:
###Code
primes = filter(is_prime, itertools.count(2))
###Output
_____no_output_____
###Markdown
Notes:- generator expressions are usually shorter and more readable- But in this case `filter()` is arguably better — we're taking the value as it is.- `filter()` was `itertools.ifilter()` before...- ... but nobody uses Python 2 anymore. Attempt 5
###Code
def get_primes(how_many):
"""Return the first 'how_many' prime numbers."""
counter = 0
primes = filter(is_prime, itertools.count(2))
while counter < how_many:
yield next(all_primes)
counter += 1
###Output
_____no_output_____
###Markdown
Take how many?- This thing that we're doing in `get_primes()` is a common pattern.- *"Give me the first n elements of this iterable"*- We cannot use slice notation:
###Code
numbers = [1, 2, 3, 4, 5]
print(numbers[:3]) # the first three
print(get_primes(10)[:5]) # slicing doesn't work with generators
###Output
[1, 2, 3]
###Markdown
itertools.islice()- Like slice notation, but works with all things iterable.- Lazy evaluation, of course, asking for elements once at a time.
###Code
for n in itertools.islice(itertools.count(), 5):
print(n)
word = "abcde"
print(list(itertools.islice(word, 3))) # the first three letters
# If start != 0, skip elements until it's reached
squares = (x ** 2 for x in itertools.count(1))
print(list(itertools.islice(squares, 10, 15))) # start=10, stop=15
# We can use a step other than one.
numbers = range(100000)
print(list(itertools.islice(numbers, 0, 20, 2))) # step=2
###Output
[0, 2, 4, 6, 8, 10, 12, 14, 16, 18]
###Markdown
So if we want the first $n$ prime numbers:
###Code
primes = filter(is_prime, itertools.count(2))
for n in itertools.islice(primes, 10):
print(n)
###Output
2
3
5
7
11
13
17
19
23
29
###Markdown
Attempt 6Let's rewrite our function:
###Code
def get_primes(how_many):
"""Return the first 'how_many' prime numbers."""
primes = filter(is_prime, itertools.count(2))
for n in itertools.islice(primes, how_many):
yield n
for n in get_primes(5):
print(n)
###Output
2
3
5
7
11
###Markdown
We're just looping over the elements of a iterator and in turn `yield`ing them. This is what `yield from` was invented for! yield from- Allows a generator to delegate part of its operations to another generator.- That is, each `next()` which ask the sub-generator for another value.- Usually we delegate to subgenerators, but it works with anything *iterable*.- `yield from iterable` $\approx$ `for item in iterable: yield item`- It is also a transparent two-way channel from the caller to the sub-generator.
###Code
def foo():
yield 1
yield 2
yield 3
def spam():
yield from foo() # for x in foo(): yield x
print(list(spam()))
def vowels():
# Works with anything iterable
yield from "aeiou"
print(list(vowels()))
###Output
['a', 'e', 'i', 'o', 'u']
###Markdown
Recommended reading:[A Curious Course on Coroutines and Concurrency](http://dabeaz.com/coroutines/) Attempt 7So we can rewrite our function as:
###Code
def get_primes(how_many):
"""Return the first 'how_many' prime numbers."""
primes = filter(is_prime, itertools.count(2))
yield from itertools.islice(primes, how_many)
for n in get_primes(5):
print(n)
###Output
2
3
5
7
11
###Markdown
Yet we can make this even shorter.- We're just defining a generator that delegates all the work to another generator.- The first argument to `islice()` is always the same: `primes`.- The only thing that changes is the second one, `how_many`. This is what `functools.partial()` was invented for! functools.partial()- Used for partial function application. That's formal language for...- ... *"freezing some portion of a function's arguments"*.- It also supports freezing keyword arguments.- Allows us to simplifying a function signature.- You don't know you need it in your life until you come across it.
###Code
import functools
power_two = functools.partial(math.pow, 2) # freeze 'math.pow(2, ...'
print(power_two)
print(power_two(3)) # math.pow(2, 3)
print(power_two(5)) # math.pow(2, 5)
print(power_two(9)) # math.pow(2, 9)
###Output
8.0
32.0
512.0
###Markdown
Used in real life to create handy shortcuts for a function we'll **call repeatedly**, and where some arguments are always the same:
###Code
import os.path
src_dir = "/home/vterron/src/"
print(os.path.join(src_dir, "file1.py"))
print(os.path.join(src_dir, "file2.py"))
print(os.path.join(src_dir, "file3.py"))
# This is getting a little tedious...
###Output
/home/vterron/src/file1.py
/home/vterron/src/file2.py
/home/vterron/src/file3.py
###Markdown
We can do instead:
###Code
get_src = functools.partial(os.path.join, "/home/vterron/src/")
print(get_src("file1.py")) # os.path.join("/home/vterron/src/", "file1.py")
print(get_src("file2.py"))
print(get_src("file3.py"))
###Output
/home/vterron/src/file1.py
/home/vterron/src/file2.py
/home/vterron/src/file3.py
###Markdown
... or as a shorcut to define a function:
###Code
import random
def bonoloto():
"""Get a random ticket to play BonoLoto."""
return random.sample(range(1, 50), 6)
bonoloto()
import functools
import random
bonoloto = functools.partial(random.sample, range(1, 50), 6)
bonoloto()
###Output
_____no_output_____
###Markdown
Attempt 8So going back to our function...
###Code
def get_primes(how_many):
"""Return the first 'how_many' prime numbers."""
primes = filter(is_prime, itertools.count(2))
yield from itertools.islice(primes, how_many)
###Output
_____no_output_____
###Markdown
We can do instead:
###Code
primes = filter(is_prime, itertools.count(2))
get_primes = functools.partial(itertools.islice, primes)
for n in get_primes(5):
print(n)
###Output
2
3
5
7
11
###Markdown
Integer factorization, Part II- This is all neat and good, but not exactly what we need here.- We don't want the first whatever prime numbers...- ... but all the prime numbers $\le \sqrt{n}$.- For example, to factorize 15 we need to test all the primes until $\left \lceil{\sqrt{15}}\right \rceil = 3$: Generating all the divisors $\le x$ Naive attempt
###Code
def primes_until(stop):
"""Return all the prime numbers <= stop."""
primes = filter(is_prime, itertools.count(2))
for p in primes:
if p <= stop:
yield p
else:
break
print(list(primes_until(15)))
###Output
[2, 3, 5, 7, 11, 13]
###Markdown
This is also a common pattern: loop over the elements of an iterable, stopping as soon as some condition is no longer satisfied. And this is what `takewhile()` was invented for! `itertools.takewhile()`- Make an iterator that returns elements.- Stop when the predicate is no longer `True`.- Frequently used in conjunction with lambda functions.
###Code
numbers = range(1, 10000)
# Take numbers as long as they're < 4
it = itertools.takewhile(lambda x: x < 4, numbers)
print(next(it)) # 1 is < 4, so we return it
print(next(it)) # 2 is < 4, so we return it
print(next(it)) # 3 is < 4, so we return it
print(next(it)) # 4 is *not* < 4, so stop
###Output
1
2
3
###Markdown
Lambda functions are not mandatory
###Code
def smaller_than_ten(x):
"""Checks whether the number is < 10."""
return x < 10
numbers = range(0, int(1e12), 2)
list(itertools.takewhile(smaller_than_ten, numbers))
###Output
_____no_output_____
###Markdown
But since we're at it...
###Code
def is_smaller(x, than=float("inf")):
return x < than
numbers = range(0, int(1e18), 2)
# functools.partial() + keyword argument = more readable!
for n in itertools.takewhile(functools.partial(is_smaller, than=10), numbers):
print(n)
###Output
0
2
4
6
8
###Markdown
`operator`This [must-know module](https://docs.python.org/3/library/operator.html) exports all standard operators as functions.
###Code
import operator
print(operator.lt(3, 5)) # 3 < 5 -> True
print(operator.not_(True)) # not True -> False
print(operator.add(2, 1)) # 2 + 1 -> 3
print(operator.neg(-1)) # -(-1) -> 1
# In-place addition
x = [1, 2, 3]
operator.iadd(x, [4, 5]) # x += [4, 5]
print(x) # [1, 2, 3, 4, 5]
###Output
True
False
3
1
[1, 2, 3, 4, 5]
###Markdown
So we can rewrite our solution as...
###Code
numbers = range(0, int(1e24), 2)
smaller_than_ten = functools.partial(operator.gt, 10)
for n in itertools.takewhile(smaller_than_ten, numbers):
print(n)
###Output
0
2
4
6
8
###Markdown
- Note that with `partial()` we have to bind arguments left to right.- Instead of testing whether $x < 10$...- ... we're rewriting it as $10 > x$Anyway. Generating all the divisors $\le x$ Kung Fu version
###Code
def primes_until(stop):
"""Return all the prime numbers <= stop."""
primes = filter(is_prime, itertools.count(2))
yield from itertools.takewhile(lambda p: p < stop, primes)
for x in primes_until(15):
print(x)
###Output
2
3
5
7
11
13
###Markdown
Woo-hoo! And what about the *opposite*? `itertools.dropwhile()`- Make an iterator that returns elements.- Ignore elements as long as the predicate is `True`.- As soon as it's not anymore, return everything.
###Code
numbers = range(1, 10000)
# Ignore numbers as long as they're < 4
it = itertools.dropwhile(lambda x: x < 4, numbers)
print(next(it)) # 'it' returns 1 -> 1 < 4, so we drop it, so ...
# 'it' returns 2 -> 2 < 4, so we drop it, so ...
# 'it' returns 3 -> 3 < 4, so we drop it, so ...
# returns 4
print(next(it)) # returns 5
print(next(it)) # returns 6
print(next(it)) # returns 7
print(next(it)) # returns 8
# ... and so on until we exhaust 'numbers'
###Output
4
5
6
7
8
###Markdown
Prime numbers within a rangeUsing both `takewhile()` and `dropwhile()` gets us this:
###Code
def primes_range(start=2, stop=float("inf")):
"""Return all the prime numbers start <= x < stop."""
primes = filter(is_prime, itertools.count(2))
it = itertools.dropwhile(lambda x: x < start, primes)
yield from itertools.takewhile(lambda y: y < stop, it)
print(list(primes_range(100, 125))) # 100 <= x < 125
print(list(primes_range(stop=20))) # all primes < 20
it = primes_range(start=1000) # all primes >= 1000
print(next(it)) # 1009
print(next(it)) # 1013
print(next(it)) # 1019
print(next(it)) # 1021
# ... and so on until infinity.
###Output
1009
1013
1019
1021
###Markdown
Integer factorization: The Algorithm- Iterate over the prime numbers $p \le \sqrt{n}$- If $n$ is divisible by $p$, then $p$ is a factor.- Divide $n$ by $p$ and factorize the quotient, recursively.- Base case: we ran out of prime numbers, so $n$ is prime too.- The factorization is all found factors. Example: factorize $30$- Is $30$ divisible by $2$? Yes.- Therefore, $2$ is a factor.Factorize now the quotient, $30 \div 2 = 15$:- Is $15$ divisible by $2$? No.- Is $15$ divisible by $3$? Yes.- Therefore, $3$ is a factor.Factorize now the quotient, $15 \div 3 = 5$:- There're no primes $\le \sqrt{5}$, so $5$ is prime too.- $30 = 2 \times 3 \times 5$ Show me the code
###Code
import math
def factorize(n):
"""Decompose n into prime numbers."""
for p in primes_until(int(math.sqrt(n))):
quotient, remainder = divmod(n, p)
if remainder == 0:
# 'p' is a prime factor
return [p] + factorize(quotient)
# Base case, reached only if we run out of primes
return [n] # n is prime
print(factorize(30)) # [2, 3, 5]
print(factorize(70)) # [2, 3, 7]
print(factorize(78)) # [2, 3, 13]
print(factorize(11)) # we hit the base case directly
###Output
[2, 3, 5]
[2, 5, 7]
[2, 3, 13]
[11]
###Markdown
The `uniq` commandA Unix command that collapses adjacent identical lines into one.
###Code
%%bash
cat ~/letters.txt
%%bash
uniq ~/letters.txt
###Output
a
b
a
###Markdown
Note that we're only removing **consecutive** duplicates. Our own version Padawan attemptFor simplicity's sake, let's accept only strings as input.
###Code
def uniq(word):
"""Remove consecutive duplicates letters."""
# Don't break if string is empty
if not word:
return ''
result = [word[0]]
for letter in word[1:]:
if letter != result[-1]:
result.append(letter)
return ''.join(result)
print(uniq("a")) # 'a'
print(uniq("bbb")) # 'b'
print(uniq("abc")) # 'abc'
print(uniq("aabbbccccdee")) # 'abcde'
###Output
a
b
abc
abcde
###Markdown
itertools.groupby()- Make an iterator that returns consecutive `keys` and `groups` from the iterable.- A new `group` starts every time the value of the key changes.- The `group` objects are a sub-iterator over the elements in the group.
###Code
word = "aabccc"
groups = itertools.groupby(word)
print(groups) # a 'groupby' object — an iterable
key1, group1 = next(groups) # give me the first group
print("Key:", key1) # 'a'
print("Group:", group1) # a '_grouper' object... another iterable
print("Next:", next(group1)) # the first 'a'
print("Next:", next(group1)) # the second 'a'
print("Next:", next(group1)) # StopIteration
###Output
Next: a
Next: a
###Markdown
- Each call to `next()` will return two elements: the `key` and the `group`.- The `key` is what this group contains — e.g., the letter *a*. - The `group` are the actual occurrences of the key — e.g., *"aa"*.Let's keep going on...
###Code
print("Word", word) # 'aabcc'
key2, group2 = next(groups) # give me the second group
print("Key:", key2) # ok, so this group contains 'b'
# And how many occurrences of 'b' are there?
print("Group:", group2) # the occurrences of 'b'
print(list(group2)) # ['b'] — oh, only one.
# The third and last group
key3, group3 = next(groups)
print("Key:", key3) # 'c'
print(list(group3)) # ['c', 'c', 'c']
###Output
Key: c
['c', 'c', 'c']
###Markdown
Using a `for` loop instead
###Code
word = "aabccc"
for key, group in itertools.groupby(word):
print(key, "->", list(group))
###Output
a -> ['a', 'a']
b -> ['b']
c -> ['c', 'c', 'c']
###Markdown
This might seem silly, but it's very powerful indeed. Our `uniq` command Kung Fu version
###Code
def uniq(word):
result = []
# We don't need the group for all, hence the '_'
for key, _ in itertools.groupby(word):
result.append(key)
return ''.join(result)
word = "aaabbcccccdee"
print(uniq(word))
###Output
abcde
###Markdown
... or even more succinct:
###Code
def uniq(word):
return ''.join(key for key, _ in itertools.groupby(word))
print(uniq(word))
###Output
abcde
###Markdown
A (rudimentary) compression algorithm- Let's implement a very basic string compression function...- ... using the counts of repeated characters.- For example, "aaabbcccccdee" returns "a3b2c5d1e2" Without `itertools`, the solution brings us pain and misery.
###Code
def compress(word):
"""Return a compressed version of the string."""
result = []
current = word[0]
counter = 1
for letter in word[1:]:
if letter == current:
# We're still in the same group
counter += 1
else:
# We need to start a new group
result += [current, str(counter)]
current = letter # start a new group
counter = 1
result += [current, str(counter)]
return ''.join(result)
word = "aaabbcccccdee"
print(compress(word)) # 'a3b2c5d1e2'
###Output
a3b2c5d1e2
###Markdown
Kung Fu version
###Code
def compress(word):
"""Return a compressed version of the string."""
result = []
for key, group in itertools.groupby(word):
result.append("{}{}".format(key, len(list(group))))
return ''.join(result)
word = "aaabbcccccdee"
print(compress(word)) # 'a3b2c5d1e2'
###Output
a3b2c5d1e2
###Markdown
Making it case case-insensitiveA problem is that letters in different case are considered to be different.
###Code
print(compress("aaAAAA")) # 'a2A4'
###Output
a2A4
###Markdown
This might make sense, but we're into compressing our strings so much that we decide to ignore the case... *respecting the case of the first occurrence* — otherwise, we would just `.upper()` or `.lower()` the entire string. That is:- "aaAAAA" $\rightarrow$ "a6"- "AAaaaa" $\rightarrow$ "A6"- "BbBccC" $\rightarrow$ "B3c3".How do we do this? The `key` function- By default, `groupby()` compares the elements as they are seen.- The keyword argument `key` allows us to specify a different criteria.- It is a function that transforms each elements before it's compared.- This is known as the *"key value for each element"* How is this useful? Let's do this:
###Code
def compress_more(word):
"""Return an even more compressed version of the string."""
result = []
# Make the comparison between letters case-insensitive.
for _, group in itertools.groupby(word, key=lambda x: x.upper()):
group = list(group)
# Take first element to respect case of the first occurrence.
result.append("{}{}".format(group[0], len(group)))
return ''.join(result)
print(compress_more("aaAAAA")) # 'a6'
print(compress_more("AAaaaa")) # 'A6'
print(compress_more("BbBccC")) # 'B3c3'
###Output
a6
A6
B3c3
###Markdown
- We can't use the key, as they will always be in upper case.- 'group' is an iterator, so we store it as a list to use slices and `len()`. Grouping letters by lengthLet's say that we have a series of words, and we want to group them by their length.
###Code
words = "red orange yellow green blue indigo violet gray".split()
###Output
_____no_output_____
###Markdown
Rookie version
###Code
# Map each length to a list of words
lengths = collections.defaultdict(list)
for w in words:
lengths[len(w)].append(w)
for key, value in lengths.items():
print(key, '->', value)
###Output
3 -> ['red']
4 -> ['blue', 'gray']
5 -> ['green']
6 -> ['orange', 'yellow', 'indigo', 'violet']
###Markdown
Kung Fu version
###Code
words.sort(key=len)
for key, group in itertools.groupby(words, key=len):
print(key, '->', list(group))
###Output
3 -> ['red']
4 -> ['blue', 'gray']
5 -> ['green']
6 -> ['orange', 'yellow', 'indigo', 'violet']
###Markdown
Two things to note:- We have to sort the list first; otherwise it only groups consecutive items.- The first solution is $\mathcal{O}(n)$ vs $\mathcal{O}(n \log n)$, because we have to sort the list. But `groupby()` is arguably better here as it's shorter and clearly conveys what we're doing: grouping the words according to a criteria. AnagramsHow do we find anagrams in a list of wors?
###Code
words = ['listen', 'meteor', 'dawn', 'remote', 'silent']
# If two words are an anagram, sorted() returns the same
print(sorted('listen')) # ['e', 'i', 'l', 'n', 's', 't']
print(sorted('silent')) # ['e', 'i', 'l', 'n', 's', 't']
###Output
['e', 'i', 'l', 'n', 's', 't']
['e', 'i', 'l', 'n', 's', 't']
###Markdown
Neophyte version
###Code
anagrams = collections.defaultdict(list)
for w in words:
# Because lists are not hashable
anagrams[tuple(sorted(w))].append(w)
for words_ in anagrams.values():
# 'dawn' doesn't match anything else
if len(words_) >= 2:
print(words_)
###Output
['listen', 'silent']
['meteor', 'remote']
###Markdown
Kung Fu version
###Code
words.sort(key=sorted)
for _, group in itertools.groupby(words, key=sorted):
words_ = list(group)
if len(words_) >= 2:
print(words_)
%matplotlib inline
###Output
_____no_output_____
###Markdown
Consecutive values over a thresholdLet's say we have this function:$\frac{x^3}{14} + \frac{x^2}{14} - \frac{13 x}{14} - \frac{1}{14}$
###Code
import numpy
# Build polynomial from coefficients
coeffs = [1/14, 1/14, -13/14, -1/14]
func = numpy.poly1d(coeffs)
func(0.5) # evaluate function at 0.5
###Output
_____no_output_____
###Markdown
Maths! Run for your lives! It looks like this
###Code
from matplotlib import pyplot
xp = numpy.linspace(-4, 4.5, 100)
pyplot.plot(xp, func(xp), 'b', alpha = 0.7)
pyplot.grid(True)
###Output
_____no_output_____
###Markdown
Let's say it's time versus some value we're monitoring We want to **trigger an alarm** if we spent $>= 3$ seconds below zero.
###Code
import numpy
from matplotlib import pyplot
coeffs = [1/14, 1/14, -13/14, -1/14]
func = numpy.poly1d(coeffs)
print(numpy.roots(coeffs))
xp = numpy.linspace(-4, 4.5, 100)
xzero = numpy.linspace(-0.077, 3.183, 100)
yzero = func(xzero)
pyplot.plot(xp, func(xp), 'b', alpha = 0.7)
pyplot.grid(True)
pyplot.xlabel("Time")
pyplot.ylabel("Important Thing")
pyplot.fill_between(xzero, 0, func(xzero), facecolor='red')
pyplot.savefig("images/curve-three.svg")
###Output
[-4.106 3.183 -0.077]
###Markdown
We know that the answer is yes...How do we do it... in code?
###Code
# Use three decimal digits
numpy.set_printoptions(precision=3)
###Output
_____no_output_____
###Markdown
For simplicity, evaluate it only at integers.
###Code
xp = range(-4, 5)
yp = func(xp)
for x, y in zip(xp, yp):
print("f({}) -> {:.3f}".format(x, y))
###Output
f(-4) -> 0.214
f(-3) -> 1.429
f(-2) -> 1.500
f(-1) -> 0.857
f(0) -> -0.071
f(1) -> -0.857
f(2) -> -1.071
f(3) -> -0.286
f(4) -> 1.929
###Markdown
Are there three consecutive points here with negative sign? It's as simple as this
###Code
# Group the func(x) values by whether they're negative or not.
for is_negative, group in itertools.groupby(yp, key=lambda x: x < 0):
values = numpy.array(list(group))
print(is_negative, "->", values)
if is_negative and len(values) >= 3:
print("Trigger alarm!!")
break
###Output
False -> [ 0.214 1.429 1.5 0.857]
True -> [-0.071 -0.857 -1.071 -0.286]
Trigger alarm!!
###Markdown
Don't even try to implement this without `groupby()`. [Photo](https://en.wikipedia.org/wiki/File:Wushu_dao.jpg) by AEMHZ~commonswiki / [CC BY-SA 3.0](https://creativecommons.org/licenses/by-sa/3.0/deed.en) / Cropped from original [Photo](https://en.wikipedia.org/wiki/File:Wushu_dao.jpg) by Alexandre Ferreira / [CC BY-SA 2.0](https://creativecommons.org/licenses/by-sa/2.0/) / Cropped from original Iterators are a one-way roadIn order to check how many elements there are in a iterator we need to consume it.
###Code
g = (x ** 2 for x in range(1, 101))
sum(1 for _ in g)
###Output
_____no_output_____
###Markdown
Which... works, but the generator is now exhausted, so we can't use it anymore.
###Code
list(g) # iterator is empty, so empty list
###Output
_____no_output_____
###Markdown
There is no way we can go backwards in the traversal of the iterator. There's no going backEvery time we call `next()` the iterator moves one step forward — forever.
###Code
it = iter("abcdefghijk")
next(it) # 'a'
next(it) # now b, we'll never see 'a' again
###Output
_____no_output_____
###Markdown
That's why, if we'll need each element more than once, we need to store them in a list. `itertools.tee()`- Returns $n$ independent iterators from a single iterable.- This allows us to move forward using of these iterators...- ... while the rest still point at the same place.
###Code
even = itertools.count(2, step=2)
a, b = itertools.tee(even)
print(next(a)) # 2
print(next(a)) # 4
###Output
2
4
###Markdown
We haven't yet called `next()` with `b`, to it's still at $2$.
###Code
print(next(b)) # 2
###Output
4
###Markdown
Measuring an iterator, Part IIWe could be tempted to do something like this:
###Code
g = (x ** 2 for x in range(1, 101))
# Fork the iterator, rebind name 'g'
g, tmp = itertools.tee(g)
length = sum(1 for _ in tmp)
print("Length:", length)
###Output
Length: 100
###Markdown
- We have exahusted the original iterator `g`...- ... but we use the same name for one of the copies.- The other copy, `tmp`, is the one we use for counting. However...This is a bad idea. From [the documentation](https://docs.python.org/3/library/itertools.htmlitertools.tee):> This itertool may require significant auxiliary storage (depending on how much temporary data needs to be stored). In general, if one iterator uses most or all of the data before another iterator starts, it is faster to use `list()` instead of `tee()`.These independent iterators need to store the elements in memory until they've been consumed by all the elements, so if we are going to exahust one of the iterators we're better off by simply using a list.
###Code
g = (x ** 2 for x in range(1, 11))
g = list(g)
print(len(g))
print(g)
###Output
[1, 4, 9, 16, 25, 36, 49, 64, 81, 100]
###Markdown
When is this iseful, then?When the independent generators are going to be used more or less **at the same time**.For example, if we need to compute the difference between consecutive elements in list:
###Code
numbers = [1, 5, 7]
# The output will be [4, 2], because 5 - 1 = 4 and 7 - 5 = 2
def diff(sequence):
# Pair element with the previous one
pairs = zip(sequence, sequence[1:])
return [p[1] - p[0] for p in pairs]
diff(numbers)
###Output
_____no_output_____
###Markdown
However, we can't use this with a generator, because **we can't use slicing**.
###Code
numbers = (x ** 2 for x in [1, 5, 7])
diff(numbers)
###Output
_____no_output_____
###Markdown
`tee()` to the rescueThis is a job `tee()` was designed for:
###Code
def diff(iterable):
a, b = itertools.tee(iterable)
next(b)
return [p[1] - p[0] for p in zip(a, b)]
numbers = (x for x in [1, 5, 7])
diff(numbers)
###Output
_____no_output_____
###Markdown
NumPy already did itBy the way, if we're working with sequences NumPy already includes [a function for this](http://docs.scipy.org/doc/numpy/reference/generated/numpy.diff.html):
###Code
import numpy
numpy.diff([1, 5, 7])
###Output
_____no_output_____
###Markdown
Don't reinvent the wheel! A NeverEnding StoryHow do we alternate indefinitely between -1 and 1? Absolute n00b version
###Code
x = 1
while True:
print(x)
if x == 1:
x = -1
else:
x = 1
###Output
_____no_output_____
###Markdown
An infinite loop, of catastrophic consequences if executed. A little-less-beginner version
###Code
x = 1
while True:
print(x)
x *= -1
###Output
_____no_output_____
###Markdown
We're all still going to die if we run this. The FIFO versionUsing a [first in, first out](https://en.wikipedia.org/wiki/FIFO_(computing_and_electronics) queue.
###Code
import collections
class Alternate(object):
"""Alternate between 1 and -1 endlessly, via pop()."""
def __init__(self):
self.values = collections.deque()
self._refill()
def _refill(self):
"""Insert some 1's and -1's in the queue."""
self.values.extend([1, -1] * 10)
def __len__(self):
return len(self.values)
def pop(self):
"""Return the next 1 or -1 value."""
# We ran out of values, so generate a few more.
if not len(self):
self._refill()
return self.values.popleft()
###Output
_____no_output_____
###Markdown
The [`collections`](https://docs.python.org/3/library/collections.htmlcollections.deque) module is awesome, but this is totally overkill. The pr0 versionThe *"I was paying attention, so I know we have to use a generator"* approach:
###Code
def alternate():
"""Alternate between 1 and -1 endlessly."""
x = 1
while True:
yield x
x *= -1
ones = alternate()
print(next(ones)) # 1
print(next(ones)) # -1
print(next(ones)) # 1
# ...
###Output
1
-1
1
###Markdown
Kung Fu version
###Code
ones = itertools.cycle([1, -1])
print(next(ones)) # 1
print(next(ones)) # -1
print(next(ones)) # 1
print(next(ones)) # -1
print(next(ones)) # 1
# ...
###Output
1
-1
1
-1
1
###Markdown
`itertools.cycle()`- Make an iterator returning elements from the iterable...- ... and save a copy of each of the returned elements.- When the iterable is exhausted, iterate over the saved copy.- Repeat indefinitely, until the end of time.
###Code
vowels = itertools.cycle("aeiou")
# Endless generator, so we need islice()
for v in itertools.islice(vowels, 10):
print(v)
g = (x ** 2 for x in range(1, 4))
squares = itertools.cycle(g)
print(list(itertools.islice(squares, 11)))
# We've exahusted the generator...
next(g) # StopIteration
# ... but 'cycle' still keeps a copy.
next(squares) # 9
###Output
_____no_output_____
###Markdown
Waiting patientlyA real-life case where `cycle()` proves useful:
###Code
import itertools
import sys
import time
def sleep(seconds):
"""Show an animated spinner while we sleep."""
symbols = itertools.cycle('-\|/')
tend = time.time() + seconds
while time.time() < tend:
# '\r' is carriage return: return cursor to the start of the line.
sys.stdout.write('\rPlease wait... ' + next(symbols)) # no newline
sys.stdout.flush()
time.sleep(0.1)
print() # newline
if __name__ == "__main__":
sleep(20)
###Output
_____no_output_____
###Markdown
[](https://asciinema.org/a/8ocljajktav6nkm3lbtrv7i8l?size=medium&t=3&autoplay=1) Iterating over *two* (or more) iterablesLet's count from $1$ to $10$, then down to $1$ again, and so on.
###Code
up = list(range(1, 11))
down = list(reversed(range(2, 10)))
print("Up :", up)
print("Down:", down)
itertools.cycle(numbers, vowels)
###Output
_____no_output_____
###Markdown
We have a problem — `cycle()` only accepts an iterable. And, by extension, what do we do if we want to loop over two iterables? This?
###Code
numbers = [1, 2, 3]
vowels = "aeiou"
for item in numbers:
print(item)
for item in vowels:
print(item)
###Output
1
2
3
a
e
i
o
u
###Markdown
There must be a better way — as [Raymond Hettinger always reminds us](https://youtu.be/wf-BqAjZb8M?t=23m7s). `itertools.chain()`- Make an iterator returning elements from the first iterable...- ... then proceed to the next iterable...- ... and so on until all of the iterables are exhausted.
###Code
numbers = [1, 2, 3]
vowels = "aeiou"
for item in itertools.chain(numbers, vowels):
print(item)
###Output
1
2
3
a
e
i
o
u
###Markdown
So going back to what we wanted to do...
###Code
up = range(1, 11)
down = reversed(range(2, 10))
numbers = itertools.cycle(itertools.chain(up, down))
list(itertools.islice(numbers, 25))
###Output
_____no_output_____
###Markdown
Doing it lazilyAnd the other way around? What it we have our iterables already in a list?
###Code
words = "Past glories are poor feeding".split() # ― Isaac Asimov
print(words)
###Output
['Past', 'glories', 'are', 'poor', 'feeding']
###Markdown
We want to loop other the letters of all the words. I'll use `chain()` then
###Code
for letter in itertools.chain(words):
print(letter)
###Output
Past
glories
are
poor
feeding
###Markdown
It doesn't work! `chain()` iterates over the strings of the single iterable (the list) it receives. What we want is to chain the *letters* in each word — so each word will need to be an input argument. Unpacking Arguments- Here's where we would normally use **argument unpacking**.- From list or tuple to separate positional arguments...- ... using the `*` operator to unpack the arguments out.
###Code
def add(x, y):
return x + y
values = [2, 3]
# Works, but terribly ugly
add(values[0], values[1])
# values[0] becomes the first positional argument...
# ... and values[1] becomes the second one.
add(*values)
###Output
_____no_output_____
###Markdown
Unpacking our words
###Code
words = "Past glories are poor feeding".split() # ― Isaac Asimov
letters = itertools.chain(*words) # notice the *
list(itertools.islice(letters, 15))
###Output
_____no_output_____
###Markdown
Works! But — Intertwining iterators
###Code
squares = (x ** 2 for x in range(2, 100000000)) # 1e8
sep = itertools.cycle(["->"])
powers = zip(squares, sep)
print(next(powers)) # (4, '->')
print(next(powers)) # (9, '->')
print(next(powers)) # (16, '->')
###Output
(4, '->')
(9, '->')
(16, '->')
###Markdown
How do we chain these values? That is: $4 \rightarrow 9 \rightarrow 16 \rightarrow$... Do we... unpack it, as we just said?
###Code
squares = (x ** 2 for x in range(2, 100000000)) # 1e8
sep = itertools.cycle(["->"])
powers = zip(squares, sep)
it = itertools.chain(*powers) # unpacks 1e8 arguments
###Output
_____no_output_____
###Markdown
Blood, toil, tears, and sweat ensue. `itertools.chain.from_iterable()`- Alternate constructor for `chain()`- Chained inputs from a **single iterable argument**...- ... that is evaluated lazily.
###Code
numbers = [1, 2, 3]
vowels = "aeiou"
things = [numbers, vowels]
# Instead of chain() + argument unpacking
for item in itertools.chain.from_iterable(things):
print(item)
###Output
1
2
3
a
e
i
o
u
###Markdown
Since the evaluation is lazy, we could even chain an *infinite* number of iterables.
###Code
squares = (x ** 2 for x in range(2, 100000000)) # 1e8
sep = itertools.cycle(["->"])
powers = zip(squares, sep)
it = itertools.chain.from_iterable(powers) # doesn't explode
list(itertools.islice(it, 10)) # just the first ten elements
###Output
_____no_output_____
###Markdown
[Photo](https://commons.wikimedia.org/wiki/File:Steven_ho_about.jpg) by Jane Davees / [CC BY 3.0](https://creativecommons.org/licenses/by/3.0/) Rolling dicesIf we roll two six-sided dices, how many unique combinations are there?
###Code
def product(first, second):
"""A generator of the Cartesian product of two iterables."""
for x in first:
for y in second:
yield (x, y)
dice = range(1, 7)
pairs = list(product(dice, dice))
pairs[:8]
# set() needed because (1, 3) is the same result as (3, 1)
len(set(pairs))
###Output
_____no_output_____
###Markdown
What if it's *three* dices? Three (or more) dicesWe need to generalize our solution. For example, via recursion:
###Code
def product(*args):
"""A generator of the Cartesian product of multiple iterables."""
# Base case
if not args:
yield ()
return
for element in args[0]:
for subproduct in product(*args[1:]):
yield (element,) + subproduct
dice = range(1, 7)
triplets = product(dice, dice, dice)
for x in itertools.islice(triplets, 7):
print(x)
###Output
(1, 1, 1)
(1, 1, 2)
(1, 1, 3)
(1, 1, 4)
(1, 1, 5)
(1, 1, 6)
(1, 2, 1)
###Markdown
`itertools.product()`- Make an iterator returning the Cartesian product of the input iterables.- Cycles with the rightmost element advancing on every iteration.- To compute the product of an iterable with itself, we use `repeat`.
###Code
g = itertools.product([1, 2], "ab")
print(g)
for pair in g:
print(pair)
###Output
(1, 'a')
(1, 'b')
(2, 'a')
(2, 'b')
###Markdown
`repeat` keyword argumentInstead of writing several times the same input iterable:
###Code
dice = range(1, 7)
pairs = itertools.product(dice, repeat=2)
for pair in itertools.islice(pairs, 10):
print(pair)
###Output
(1, 1)
(1, 2)
(1, 3)
(1, 4)
(1, 5)
(1, 6)
(2, 1)
(2, 2)
(2, 3)
(2, 4)
###Markdown
An easy questionIf we roll four six-sided dices, in how many outcomes they add up to 5?
###Code
results = itertools.product(range(1, 7), repeat=4)
results = filter(lambda x: sum(x) == 5, results)
for x in results:
print(x)
###Output
(1, 1, 1, 2)
(1, 1, 2, 1)
(1, 2, 1, 1)
(2, 1, 1, 1)
###Markdown
That is — we need a $2$ in one dice and $1$ in the other three. PixelsLet's say we need to work with pixels — lots of them!
###Code
class Pixel(object):
def __init__(self, row, column):
self.row = row
self.column = column
def __repr__(self):
"""Return a nicely formatted representation string."""
return "Pixel(row={}, column={})".format(self.row, self.column)
p = Pixel(2, 3)
print(p)
###Output
Pixel(row=2, column=3)
###Markdown
[`collections.namedtuple()`](https://docs.python.org/3/library/collections.htmlcollections.namedtuple)- Factory function for creating tuple subclasses with named fields.- Makes our live much easier if we don't need our class to be mutable...- ... because we get `__init__()`, `__repr__()` and others for free.- We can add our own methods via inheritance.
###Code
Pixel = collections.namedtuple("Pixel", "row, column")
###Output
_____no_output_____
###Markdown
That's all.
###Code
p = Pixel(5, 1)
print(p)
###Output
Pixel(row=5, column=1)
###Markdown
Adding a methodTo compute the distance between two points:
###Code
class Pixel(collections.namedtuple("Pixel", "row, column")):
def distance(self, other):
"""Return the Euclidean distance between two picels."""
delta_x = self.row - other.row
delta_y = self.column - other.column
return math.sqrt(delta_x ** 2 + delta_y ** 2)
p1 = Pixel(2, 3)
p2 = Pixel(5, 1)
print("Distance:", p1.distance(p2), "pixels")
###Output
Distance: 3.605551275463989 pixels
###Markdown
Getting the neighborsLet's add a method to get the eight neighbors, lateral and diagonal: Naive version:
###Code
class Pixel(collections.namedtuple("Pixel", "row, column")):
def neighbours(self):
"""Return a generator of the eight neighbours of the Pixel."""
yield Pixel(self.row - 1, self.column - 1)
yield Pixel(self.row - 1, self.column )
yield Pixel(self.row - 1, self.column + 1)
yield Pixel(self.row , self.column - 1)
yield Pixel(self.row , self.column + 1)
yield Pixel(self.row + 1, self.column - 1)
yield Pixel(self.row + 1, self.column )
yield Pixel(self.row + 1, self.column + 1)
###Output
_____no_output_____
###Markdown
That's a lot of code. Kung Fu version
###Code
class Pixel(collections.namedtuple("Pixel", "row column")):
def neighbours(self):
"""Return a generator of the eight neighbours of the Pixel."""
for x_delta, y_delta in itertools.product([-1, 0, 1], repeat=2):
if x_delta or y_delta: # ignore delta (0, 0)
yield Pixel(self.row + x_delta, self.column + y_delta)
for p in Pixel(3, 4).neighbours():
print(p)
###Output
Pixel(row=2, column=3)
Pixel(row=2, column=4)
Pixel(row=2, column=5)
Pixel(row=3, column=3)
Pixel(row=3, column=5)
Pixel(row=4, column=3)
Pixel(row=4, column=4)
Pixel(row=4, column=5)
###Markdown
Kung Fu Master versionDon't hardcode the class name.
###Code
class Pixel(collections.namedtuple("Pixel", "row column")):
def neighbours(self):
"""Return a generator of the eight neighbours of the Pixel."""
cls = type(self) # we can use now 'cls' to create new objects
for x_delta, y_delta in itertools.product([-1, 0, 1], repeat=2):
if x_delta or y_delta: # ignore delta (0, 0)
yield cls(self.row + x_delta, self.column + y_delta)
for p in Pixel(4, 7).neighbours():
print(p)
###Output
Pixel(row=3, column=6)
Pixel(row=3, column=7)
Pixel(row=3, column=8)
Pixel(row=4, column=6)
Pixel(row=4, column=8)
Pixel(row=5, column=6)
Pixel(row=5, column=7)
Pixel(row=5, column=8)
|
provider/docs/tutorial/notebooks/01-register-features.ipynb | ###Markdown
Copyright (c) Microsoft Corporation.Licensed under the MIT license. Feast Azure Provider Tutorial: Register FeaturesIn this notebook you will connect to your feature store and register features into a central repository hosted on Azure Blob Storage. It should be noted that best practice for registering features would be through a CI/CD process e.g. GitHub Actions, or Azure DevOps. Configure Feature RepoThe cell below connects to your feature store. __You need to update the feature_repo/feature_store.yaml file so that the registry path points to your blob location__
###Code
import os
from feast import FeatureStore
from azureml.core import Workspace
# access key vault to get secrets
ws = Workspace.from_config()
kv = ws.get_default_keyvault()
# update with your connection string
os.environ['SQL_CONN']=kv.get_secret("FEAST-SQL-CONN")
os.environ['REDIS_CONN']=kv.get_secret("FEAST-REDIS-CONN")
# connect to feature store
fs = FeatureStore("./feature_repo")
###Output
_____no_output_____
###Markdown
Define the data source (offline store)The data source refers to raw underlying data (a table in Azure SQL DB or Synapse SQL). Feast uses a time-series data model to represent data. This data model is used to interpret feature data in data sources in order to build training datasets or when materializing features into an online store.
###Code
from feast_azure_provider.mssqlserver_source import MsSqlServerSource
orders_table = "orders"
driver_hourly_table = "driver_hourly"
customer_profile_table = "customer_profile"
driver_source = MsSqlServerSource(
table_ref=driver_hourly_table,
event_timestamp_column="datetime",
created_timestamp_column="created",
)
customer_source = MsSqlServerSource(
table_ref=customer_profile_table,
event_timestamp_column="datetime",
created_timestamp_column="",
)
###Output
_____no_output_____
###Markdown
Define Feature ViewsA feature view is an object that represents a logical group of time-series feature data as it is found in a data source. Feature views consist of one or more entities, features, and a data source. Feature views allow Feast to model your existing feature data in a consistent way in both an offline (training) and online (serving) environment.Feature views are used during:- The generation of training datasets by querying the data source of feature views in order to find historical feature values. A single training dataset may consist of features from multiple feature views. - Loading of feature values into an online store. Feature views determine the storage schema in the online store.- Retrieval of features from the online store. Feature views provide the schema definition to Feast in order to look up features from the online store.__NOTE: Feast does not generate feature values. It acts as the ingestion and serving system. The data sources described within feature views should reference feature values in their already computed form.__
###Code
from feast import Feature, FeatureView, ValueType
from datetime import timedelta
driver_fv = FeatureView(
name="driver_stats",
entities=["driver"],
features=[
Feature(name="conv_rate", dtype=ValueType.FLOAT),
Feature(name="acc_rate", dtype=ValueType.FLOAT),
Feature(name="avg_daily_trips", dtype=ValueType.INT32),
],
batch_source=driver_source,
ttl=timedelta(hours=2),
)
customer_fv = FeatureView(
name="customer_profile",
entities=["customer_id"],
features=[
Feature(name="current_balance", dtype=ValueType.FLOAT),
Feature(name="avg_passenger_count", dtype=ValueType.FLOAT),
Feature(name="lifetime_trip_count", dtype=ValueType.INT32),
],
batch_source=customer_source,
ttl=timedelta(days=2),
)
###Output
_____no_output_____
###Markdown
Define entitiesAn entity is a collection of semantically related features. Users define entities to map to the domain of their use case. For example, a ride-hailing service could have customers and drivers as their entities, which group related features that correspond to these customers and drivers.Entities are defined as part of feature views. Entities are used to identify the primary key on which feature values should be stored and retrieved. These keys are used during the lookup of feature values from the online store and the join process in point-in-time joins. It is possible to define composite entities (more than one entity object) in a feature view.Entities should be reused across feature views. Entity keyA related concept is an entity key. These are one or more entity values that uniquely describe a feature view record. In the case of an entity (like a driver) that only has a single entity field, the entity is an entity key. However, it is also possible for an entity key to consist of multiple entity values. For example, a feature view with the composite entity of (customer, country) might have an entity key of (1001, 5).Entity keys act as primary keys. They are used during the lookup of features from the online store, and they are also used to match feature rows across feature views during point-in-time joins.
###Code
from feast import Entity
driver = Entity(name="driver", join_key="driver_id", value_type=ValueType.INT64)
customer = Entity(name="customer_id", value_type=ValueType.INT64)
###Output
_____no_output_____
###Markdown
Feast `apply()`Feast `apply` will:1. Feast will scan Python files in your feature repository and find all Feast object definitions, such as feature views, entities, and data sources.1. Feast will validate your feature definitions1. Feast will sync the metadata about Feast objects to the registry. If a registry does not exist, then it will be instantiated. The standard registry is a simple protobuf binary file that is stored on Azure Blob Storage.1. Feast CLI will create all necessary feature store infrastructure. The exact infrastructure that is deployed or configured depends on the provider configuration that you have set in feature_store.yaml.
###Code
fs.apply([driver, driver_fv, customer, customer_fv])
###Output
_____no_output_____
###Markdown
Copyright (c) Microsoft Corporation.Licensed under the MIT license. Feast Azure Provider Tutorial: Register FeaturesIn this notebook you will connect to your feature store and register features into a central repository hosted on Azure Blob Storage. It should be noted that best practice for registering features would be through a CI/CD process e.g. GitHub Actions, or Azure DevOps. Configure Feature RepoThe cell below connects to your feature store. The `registry_blob_url` should point to the location on blob where you want your feature repsository to be stored.
###Code
from feast import FeatureStore, RepoConfig
from feast.registry import RegistryConfig
from feast_azure_provider.mssqlserver import MsSqlServerOfflineStoreConfig
from feast.infra.online_stores.redis import RedisOnlineStoreConfig
from azureml.core import Workspace
# update this to your location on blob
registry_blob_url = "https://<ACCOUNT_NAME>.blob.core.windows.net/<CONTAINER>/<PATH>/registry.db"
# access key vault to get secrets
ws = Workspace.from_config()
kv = ws.get_default_keyvault()
# update with your connection string
offline_conn_str = kv.get_secret("FEAST-SQL-CONN")
online_conn_str = kv.get_secret("FEAST-REDIS-CONN")
# set RegistryConfig
reg_config = RegistryConfig(
registry_store_type="feast_azure_provider.registry_store.AzBlobRegistryStore",
path=registry_blob_url,
)
# set RepoConfig
repo_cfg = RepoConfig(
registry=reg_config,
project="production",
provider="feast_azure_provider.azure_provider.AzureProvider",
offline_store=MsSqlServerOfflineStoreConfig(connection_string=offline_conn_str),
online_store=RedisOnlineStoreConfig(connection_string=online_conn_str),
)
# connect to feature store
store = FeatureStore(config=repo_cfg)
###Output
_____no_output_____
###Markdown
Define the data source (offline store)The data source refers to raw underlying data (a table in Azure SQL DB or Synapse SQL). Feast uses a time-series data model to represent data. This data model is used to interpret feature data in data sources in order to build training datasets or when materializing features into an online store.
###Code
from feast_azure_provider.mssqlserver_source import MsSqlServerSource
orders_table = "orders"
driver_hourly_table = "driver_hourly"
customer_profile_table = "customer_profile"
driver_source = MsSqlServerSource(
table_ref=driver_hourly_table,
event_timestamp_column="datetime",
created_timestamp_column="created",
)
customer_source = MsSqlServerSource(
table_ref=customer_profile_table,
event_timestamp_column="datetime",
created_timestamp_column="",
)
###Output
_____no_output_____
###Markdown
Define Feature ViewsA feature view is an object that represents a logical group of time-series feature data as it is found in a data source. Feature views consist of one or more entities, features, and a data source. Feature views allow Feast to model your existing feature data in a consistent way in both an offline (training) and online (serving) environment.Feature views are used during:- The generation of training datasets by querying the data source of feature views in order to find historical feature values. A single training dataset may consist of features from multiple feature views. - Loading of feature values into an online store. Feature views determine the storage schema in the online store.- Retrieval of features from the online store. Feature views provide the schema definition to Feast in order to look up features from the online store.__NOTE: Feast does not generate feature values. It acts as the ingestion and serving system. The data sources described within feature views should reference feature values in their already computed form.__
###Code
from feast import Feature, FeatureView, ValueType
from datetime import timedelta
driver_fv = FeatureView(
name="driver_stats",
entities=["driver"],
features=[
Feature(name="conv_rate", dtype=ValueType.FLOAT),
Feature(name="acc_rate", dtype=ValueType.FLOAT),
Feature(name="avg_daily_trips", dtype=ValueType.INT32),
],
batch_source=driver_source,
ttl=timedelta(hours=2),
)
customer_fv = FeatureView(
name="customer_profile",
entities=["customer_id"],
features=[
Feature(name="current_balance", dtype=ValueType.FLOAT),
Feature(name="avg_passenger_count", dtype=ValueType.FLOAT),
Feature(name="lifetime_trip_count", dtype=ValueType.INT32),
],
batch_source=customer_source,
ttl=timedelta(days=2),
)
###Output
_____no_output_____
###Markdown
Define entitiesAn entity is a collection of semantically related features. Users define entities to map to the domain of their use case. For example, a ride-hailing service could have customers and drivers as their entities, which group related features that correspond to these customers and drivers.Entities are defined as part of feature views. Entities are used to identify the primary key on which feature values should be stored and retrieved. These keys are used during the lookup of feature values from the online store and the join process in point-in-time joins. It is possible to define composite entities (more than one entity object) in a feature view.Entities should be reused across feature views. Entity keyA related concept is an entity key. These are one or more entity values that uniquely describe a feature view record. In the case of an entity (like a driver) that only has a single entity field, the entity is an entity key. However, it is also possible for an entity key to consist of multiple entity values. For example, a feature view with the composite entity of (customer, country) might have an entity key of (1001, 5).Entity keys act as primary keys. They are used during the lookup of features from the online store, and they are also used to match feature rows across feature views during point-in-time joins.
###Code
from feast import Entity
driver = Entity(name="driver", join_key="driver_id", value_type=ValueType.INT64)
customer = Entity(name="customer_id", value_type=ValueType.INT64)
###Output
_____no_output_____
###Markdown
Feast `apply()`Feast `apply` will:1. Feast will scan Python files in your feature repository and find all Feast object definitions, such as feature views, entities, and data sources.1. Feast will validate your feature definitions1. Feast will sync the metadata about Feast objects to the registry. If a registry does not exist, then it will be instantiated. The standard registry is a simple protobuf binary file that is stored on Azure Blob Storage.1. Feast CLI will create all necessary feature store infrastructure. The exact infrastructure that is deployed or configured depends on the provider configuration that you have set in feature_store.yaml.
###Code
store.apply([driver, driver_fv, customer, customer_fv])
###Output
_____no_output_____ |
TypeDataStructures/Nesting Dictionary.ipynb | ###Markdown
Nesting Dictionary
###Code
Y1 ={ 'bb':11}
Y10 = {'hhh':555}
Y7 = { 'ff':44, 'gg': Y10}
Y = {'a': Y1, 'c':2, 'd':3, 'e': Y7 }
print(Y)
###Output
{'a': {'bb': 11}, 'd': 3, 'e': {'gg': {'hhh': 555}, 'ff': 44}, 'c': 2}
|
misc/day44_querying_database.ipynb | ###Markdown
Questions to answer- What kind of projects are popular on Kickstarter?- How much are people asking for?- What kind of projects tend to be more funded? Connect to database
###Code
dbname="kick"
tblname="info"
engine = create_engine(
'postgresql://localhost:5432/{dbname}'.format(dbname=dbname))
# Connect to database
conn = psycopg2.connect(dbname=dbname)
cur = conn.cursor()
###Output
_____no_output_____
###Markdown
Remind myself of the columns in the table:
###Code
cur.execute("SELECT column_name,data_type FROM information_schema.columns WHERE table_name = '{table}';".format(table=tblname))
rows = cur.fetchall()
pd.DataFrame(rows, columns=["column_name", "data_type"])
###Output
_____no_output_____
###Markdown
Number of records in table:
###Code
cur.execute("SELECT COUNT(*) from {table}".format(table=tblname))
cur.fetchone()
###Output
_____no_output_____
###Markdown
--- Question 1: Project topics- How many different types of projects are on Kickstarter?- What is most popular?- What is most rare?
###Code
cur.execute("SELECT topic, COUNT(*) from {table} GROUP BY topic ORDER BY count DESC;".format(table=tblname))
rows = cur.fetchall()
df = pd.DataFrame(rows, columns=["topic", "count"])
# Plot findings
plt.rcParams["figure.figsize"] = [17,5]
df.plot(kind="bar", x="topic", y="count", legend=False)
plt.ylabel("Kickstarter projects")
plt.xlabel("Topic")
plt.title("Kickstarter projects by topic")
plt.tick_params(axis='x', labelsize=7)
"There are {num_topics} different types of Kickstarter projects".format(num_topics=df.shape[0])
# Most popular project topic is
df[df["count"] == df["count"].max()]
# Most rare project topic is
df[df["count"] == df["count"].min()]
###Output
_____no_output_____
###Markdown
What are the rare projects?
###Code
cur.execute("SELECT id, blurb, goal*static_usd_rate as goal_usd FROM {table} WHERE topic = '{topic}'".format(table=tblname, topic="Taxidermy"))
rows = cur.fetchall()
for row in rows:
row_id, blurb, goal = row
print(">>> $%d | id: %s" % (goal, row_id),
blurb, sep="\n")
###Output
>>> $8319 | id: 2009417734
Master prop maker and creator of the dead fairy hoax that captured the world’s imagination invites you into his studio to reveal all!
>>> $4300 | id: 874566085
Gallery | Public Dissections | Events |
A space in downtown San Francisco to reflect upon the less considered means of living & dying.
>>> $5000 | id: 304809544
TaxiClear transforms biology into brilliant works of art that illustrate the beauty and complexity of life
>>> $500 | id: 1111529080
Insects: the most abundant and intriguing organisms on Earth. May be seen as pests but are works of art that I will add to wreaths.
>>> $3000 | id: 48529158
I take my passion for leather working and create hand carved notebook covers to honor a person's legacy and career accomplishments.
>>> $2000 | id: 588593322
Casting skulls from rare and unique species of animal. To include miniature skulls in the future.
>>> $705 | id: 1539840572
The "Meteor Grip" ergonomic craft, taxidermy knife fro precise accurate cutting. This hand specific design is a leader in grip design.
>>> $456 | id: 1450117214
Creating beautiful and unique statement pieces with the use of taxidermy, skulls and ornate frames.
>>> $15450 | id: 1370425060
Bringing art and science together through hand made ethically sourced bone jewellery and art sculptures
>>> $5464 | id: 1052896826
A new and creative way to display the beauty of ocean,swamp and land creatures of the world by electroless silver plating.
>>> $7208 | id: 1208011665
Hollow Earth is slated to be Perth's 1st true Oddity/Collectables store.
Selling anything from hand-made trinkets to occult items.
>>> $250000 | id: 1984141754
I want to go into the wilds to find and subsequently kill sasquatch. Donors will be sent a small piece of his hide if I succeed.
###Markdown
Question 2: Project funding goals- How much are people asking for in general? by topics?
###Code
sql = "SELECT id, topic, goal*static_usd_rate as goal_usd FROM {table}".format(table=tblname)
cur.execute(sql)
rows = cur.fetchall()
df = pd.DataFrame(rows, columns=["id", "topic", "goal_usd"])
# Asking average
np.log10(df.goal_usd).plot.kde()
plt.xlabel("log(funding goal in USD)")
"Most projects are asking for: $%d - $%d" % (10**2.5, 10**5)
sns.barplot(x="topic", y="goal_usd",
data=df.groupby("topic").mean().reset_index().sort_values(by="goal_usd", ascending=False))
_ = plt.xticks(rotation='vertical')
plt.ylabel("Average goal (USD)")
plt.xlabel("Kickstarter project topic")
plt.title("Funding goals on Kickstarter by topic")
plt.tick_params(axis='x', labelsize=7)
###Output
_____no_output_____
###Markdown
"Movie Theaters" and "Space exploration" have the average higest funding goals Question 3: Funding successWhat tends to get funded?
###Code
sql = "SELECT id, topic, goal, pledged, pledged/goal as progress FROM info ORDER BY progress DESC;"
cur.execute(sql)
rows = cur.fetchall()
df = pd.DataFrame(rows, columns=["id", "topic", "goal", "pledged", "progress"])
df["well_funded"] = df.progress >= 1
plt.rcParams["figure.figsize"] = [17,5]
sns.boxplot(x="topic", y="progress", data=df[df.well_funded].sort_values(by="topic"))
_ = plt.xticks(rotation='vertical')
plt.yscale('log')
plt.ylabel("Percent of funding goal")
plt.xlabel("Topic")
plt.title("Projects that were successfully funded by Topic")
plt.tick_params(axis='x', labelsize=7)
sns.barplot(x="topic", y="progress",
data=df[df.well_funded].groupby("topic").count().reset_index().sort_values(by="progress", ascending=False))
_ = plt.xticks(rotation='vertical')
plt.ylabel("Project that were successfully funded")
plt.xlabel("Topic")
plt.title("Projects that were successfully funded by Topic")
plt.tick_params(axis='x', labelsize=7)
plt.rcParams["figure.figsize"] = [17,5]
sns.boxplot(x="topic", y="progress",
data=df[np.invert(df.well_funded)].sort_values(by="topic"))
_ = plt.xticks(rotation='vertical')
plt.ylabel("Percent of funding goal met")
plt.xlabel("Topic")
plt.title("Pojects that have yet to meet their funding goals")
plt.tick_params(axis='x', labelsize=7)
sns.barplot(x="topic", y="progress",
data=df[np.invert(df.well_funded)].groupby("topic").count().reset_index().sort_values(by="progress", ascending=False))
_ = plt.xticks(rotation='vertical')
plt.ylabel("Project that were not yet successfully funded")
plt.xlabel("Topic")
plt.title("Pojects that have yet to meet their funding goals")
plt.tick_params(axis='x', labelsize=7)
###Output
_____no_output_____
###Markdown
Close connection
###Code
# close communication with the PostgreSQL database server
cur.close()
# commit the changes
conn.commit()
# close connection
conn.close()
###Output
_____no_output_____ |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.