licenses
sequencelengths 1
3
| version
stringclasses 677
values | tree_hash
stringlengths 40
40
| path
stringclasses 1
value | type
stringclasses 2
values | size
stringlengths 2
8
| text
stringlengths 25
67.1M
| package_name
stringlengths 2
41
| repo
stringlengths 33
86
|
|---|---|---|---|---|---|---|---|---|
[
"MIT"
] | 0.1.2 | 4217710242df1f4708e58e00763de2569cfbe880 | code | 8210 | """
$(TYPEDEF)
An abstract type whose subtypes denote a specific electric field basis.
"""
abstract type ElectricFieldBasis end
"""
$(TYPEDEF)
The right circular electric field basis, i.e. a right-handed circular feed.
"""
struct RPol <: ElectricFieldBasis end
"""
$(TYPEDEF)
The left circular electric field basis, i.e. a left-handed circular feed.
"""
struct LPol <: ElectricFieldBasis end
"""
$(TYPEDEF)
The horizontal or X electric feed basis, i.e. the horizontal linear feed.
"""
struct XPol <: ElectricFieldBasis end
"""
$(TYPEDEF)
The vertical or Y electric feed basis, i.e. the vertical linear feed.
"""
struct YPol <: ElectricFieldBasis end
abstract type StokesBasis end
struct IPol end
struct QPol end
struct UPol end
struct VPol end
"""
$(TYPEDEF)
Denotes a general polarization basis, with basis vectors (B1,B2) which are typically
`<: Union{ElectricFieldBasis, Missing}`
"""
struct PolBasis{B1<:Union{ElectricFieldBasis, Missing}, B2<:Union{ElectricFieldBasis, Missing}} end
"""
CirBasis <: PolBasis
Measurement uses the circular polarization basis, which is typically used for circular
feed interferometers.
"""
const CirBasis = PolBasis{RPol,LPol}
"""
LinBasis <: PolBasis
Measurement uses the linear polarization basis, which is typically used for linear
feed interferometers.
"""
const LinBasis = PolBasis{XPol,YPol}
"""
$(TYPEDEF)
Static vector that holds the stokes parameters of a polarized
complex visibility
To convert between a `StokesParams` and `CoherencyMatrix` use the `convert`
function
```julia
convert(::CoherencyMatrix, StokesVector(1.0, 0.1, 0.1, 0.4))
```
"""
struct StokesParams{T} <: FieldVector{4,T}
I::T
Q::T
U::T
V::T
end
StokesParams(::SArray) = throw(ArgumentError("argument does not have a basis please wrap it in a `CoherencyMatrix`"))
StaticArraysCore.similar_type(::Type{<:StokesParams}, ::Type{T}, s::Size{(4,)}) where {T} = StokesParams{T}
"""
$(TYPEDEF)
Coherency matrix for a single baseline with bases `B1` and `B2`. The two bases correspond
to the type of feeds used for each telescope and should be subtypes of `PolBasis`. To see which
bases are implemented type `subtypes(Rimes.PolBasis)` in the REPL.
For a circular basis the layout of the coherency matrix is
```
RR* RL*
LR* RR*
```
which can be constructed using
```julia-repl
c = CoherencyMatrix(RR, LR, RL, LL, CirBasis())
```
For a linear basis the layout of the coherency matrix is
```
XX* XY*
YX* YY*
```
which can be constructed using
```julia-repl
c = CoherencyMatrix(XX, YX, XY, YY, CirBasis())
```
For a mixed (e.g., circular and linear basis) the layout of the coherency matrix is
```
RX* RY*
LX* LY*
```
or e.g., linear and circular the layout of the coherency matrix is
```
XR* XL*
YR* YL*
```
These coherency matrices can be constructed using:
```julia-repl
# Circular and linear feeds i.e., |R><X|
c = CoherencyMatrix(RX, LX, RY, LY, LinBasis(), CirBasis())
# Linear and circular feeds i.e., |X><R|
c = CoherencyMatrix(XR, YR, XL, YL, LinBasis(), CirBasis())
```
"""
struct CoherencyMatrix{B1,B2,T} <: StaticArraysCore.FieldMatrix{2,2,T}
e11::T
e21::T
e12::T
e22::T
end
StaticArraysCore.similar_type(::Type{CoherencyMatrix{B1,B2}}, ::Type{T}, s::Size{(2,2)}) where {B1,B2,T} = CoherencyMatrix{B1,B2,T}
"""
CoherencyMatrix(e11, e21, e12, e22, basis::NTuple{2, PolBasis})
Constructs the coherency matrix with components
e11 e12
e21 e22
relative to the tensor product basis, `|basis[1]><basis[2]|`. Note that basis[1] and basis[2]
could be different.
For instance
```julia
c = Coherency(1.0, 0.0, 0.0, 1.0, CirBasis(), LinBasis())
```
elements correspond to
RX* RY*
LX* LY*
"""
@inline function CoherencyMatrix(e11::Number, e21::Number, e12::Number, e22::Number, basis::NTuple{2,PolBasis})
T = promote_type(typeof(e11), typeof(e12), typeof(e21), typeof(e22))
return CoherencyMatrix{typeof(basis[1]), typeof(basis[2]),T}(T(e11), T(e21), T(e12), T(e22))
end
"""
CoherencyMatrix(e11, e21, e12, e22, basis::PolBasis)
Constructs the coherency matrix with components
e11 e12
e21 e22
relative to the tensor product basis, `basis` given by `|basis><basis|`.
For instance
```julia
c = Coherency(1.0, 0.0, 0.0, 1.0, CirBasis())
```
elements correspond to
RR* RL*
LR* LL*
"""
@inline function CoherencyMatrix(e11::Number, e21::Number, e12::Number, e22::Number, basis::PolBasis)
return CoherencyMatrix(e11, e21, e12, e22, (basis, basis))
end
"""
CoherencyMatrix(e11, e21, e12, e22, basis1::PolBasis basis2::PolBasis)
Constructs the coherency matrix with components
e11 e12
e21 e22
relative to the tensor product basis, `basis` given by `|basis1><basis2|`.
For instance
```julia
c = Coherency(1.0, 0.0, 0.0, 1.0, CirBasis(), LinBasis())
```
elements correspond to
RX* RY*
LX* LY*
"""
@inline function CoherencyMatrix(e11::Number, e21::Number, e12::Number, e22::Number, basis1::PolBasis, basis2::PolBasis)
return CoherencyMatrix(e11, e21, e12, e22, (basis1, basis2))
end
@inline function CoherencyMatrix(mat::AbstractMatrix, basis1::PolBasis, basis2::PolBasis)
return CoherencyMatrix(mat[1], mat[2], mat[3], mat[4], basis1, basis2)
end
@inline function CoherencyMatrix(mat::AbstractMatrix, basis::PolBasis)
return CoherencyMatrix(mat[1], mat[2], mat[3], mat[4], basis)
end
"""
CoherencyMatrix(s::StokesParams, basis1::PolBasis)
CoherencyMatrix(s::StokesParams, basis1::PolBasis, basis2::PolBasis)
CoherencyMatrix(s::StokesParams, basis1::PolBasis, basis2::PolBasis, refbasis=CirBasis())
Constructs the coherency matrix from the set of stokes parameters `s`.
This is specialized on `basis1` and `basis2` which form the tensor product basis
`|basis1><basis2|`, or if a single basis is given then by `|basis><basis|`.
For example
```julia
CoherencyMatrix(s, CircBasis())
```
will give the coherency matrix
```
I+V Q+iU
Q-iU I-V
```
while
```julia
CoherencyMatrix(s, LinBasis())
```
will give
```
I+Q U+iV
U-iV I-Q
```
# Notes
Internally this function first converts to a reference basis and then the final basis.
You can select the reference basis used with the optional argument refbasis. By default
we use the circular basis as our reference. Note that this is only important for mixed bases,
e.g., if `basis1` and `basis2` are different. If `basis1==basis2` then the reference basis
is never used.
"""
@inline function CoherencyMatrix(s::StokesParams, b1::PolBasis, b2::PolBasis, refbasis::Union{LinBasis, CirBasis}=CirBasis())
t1 = basis_transform(refbasis=>b1)
# Flip because these are the dual elements
t2 = basis_transform(b2=>refbasis)
# Use circular basis as a reference
c_cir = CoherencyMatrix(s, refbasis)
return CoherencyMatrix(t1*c_cir*t2, b1, b2)
end
function CoherencyMatrix(s::StokesParams, b1::T, b2::T, refbasis=CirBasis()) where {T<:PolBasis}
return CoherencyMatrix(s, b1)
end
@inline function CoherencyMatrix(s::StokesParams, b::PolBasis)
return CoherencyMatrix{typeof(b), typeof(b)}(s)
end
@inline function CoherencyMatrix{CirBasis,CirBasis}(s::StokesParams)
(;I,Q,U,V) = s
RR = complex((I + V))
LR = (Q - 1im*U)
RL = (Q + 1im*U)
LL = complex((I - V))
return CoherencyMatrix(RR, LR, RL, LL, CirBasis(), CirBasis())
end
@inline function CoherencyMatrix{B1, B2}(s::StokesParams) where {B1, B2}
return CoherencyMatrix(s, B1(), B2())
end
@inline function CoherencyMatrix{LinBasis, LinBasis}(s::StokesParams)
(;I,Q,U,V) = s
XX = (I + Q)
YX = (U - 1im*V)
XY = (U + 1im*V)
YY = (I - Q)
return CoherencyMatrix(XX, YX, XY, YY, LinBasis(), LinBasis())
end
@inline function StokesParams(c::CoherencyMatrix{CirBasis, CirBasis})
I = (c.e11 + c.e22)/2
Q = (c.e21 + c.e12)/2
U = 1im*(c.e21 - c.e12)/2
V = (c.e11 - c.e22)/2
return StokesParams(I, Q, U, V)
end
@inline function StokesParams(c::CoherencyMatrix{B1, B2}) where {B1, B2}
t1 = basis_transform(B1()=>CirBasis())
# Flip because these are the dual elements
t2 = basis_transform(CirBasis()=>B2())
c_cir = CoherencyMatrix(t1*c*t2, CirBasis())
return StokesParams(c_cir)
end
| PolarizedTypes | https://github.com/EHTJulia/PolarizedTypes.jl.git |
|
[
"MIT"
] | 0.1.2 | 4217710242df1f4708e58e00763de2569cfbe880 | code | 1227 | ChainRulesCore.ProjectTo(x::CoherencyMatrix{B1, B2, <: Number}) where {B1, B2} = ProjectTo{CoherencyMatrix}(; element = ProjectTo(eltype(x)), basis1=B1(), basis2=B2())
function (project::ProjectTo{CoherencyMatrix})(dx::AbstractMatrix)
@assert size(dx) == (2,2) "Issue in Coherency pullback the matrix is not 2x2"
return CoherencyMatrix(dx, project.basis1, project.basis2)
end
function (project::ProjectTo{CoherencyMatrix})(dx::CoherencyMatrix{B1, B2}) where {B1, B2}
@assert B1() == project.basis1 "First basis does not match in $(typeof(dx)) and $(project.basis1)"
@assert B2() == project.basis2 "Second basis does not match in $(typeof(dx)) and $(project.basis2)"
@assert size(dx) == (2,2) "Issue in Coherency pullback the matrix is not 2x2"
return dx
end
# function ChainRulesCore.rrule(::Type{<:CoherencyMatrix}, e11, e21, e12, e22, basis::NTuple{2, <:PolBasis})
# c = CoherencyMatrix(e11, e21, e12, e22, basis)
# pr = ProjectTo(e11)
# function _CoherencyMatrix_sep_pullback(Δ)
# return NoTangent(), pr(Δ[1,1]), pr(Δ[2,1]), pr(Δ[1,2]), pr(Δ[2,2]), NoTangent()
# end
# return c, _CoherencyMatrix_sep_pullback
# end
# Needed to ensure everything is constructed nicely
| PolarizedTypes | https://github.com/EHTJulia/PolarizedTypes.jl.git |
|
[
"MIT"
] | 0.1.2 | 4217710242df1f4708e58e00763de2569cfbe880 | code | 9269 | using PolarizedTypes
using StaticArrays
using ChainRulesCore
using JET
using Test
@testset "PolarizedTypes.jl" begin
@testset "Basis transform" begin
@test basis_transform(PolBasis{XPol,YPol}()=>PolBasis{RPol,LPol}())*basis_transform(PolBasis{RPol,LPol}()=>PolBasis{XPol,YPol}()) ≈ [1.0 0.0;0.0 1.0]
@test basis_transform(PolBasis{RPol,LPol}()=>PolBasis{XPol,YPol}())*basis_transform(PolBasis{XPol,YPol}()=>PolBasis{RPol,LPol}()) ≈ [1.0 0.0;0.0 1.0]
@test basis_transform(CirBasis(), LinBasis()) == basis_transform(CirBasis()=>LinBasis())
@test basis_components(RPol(), CirBasis()) ≈ basis_transform(CirBasis()=>CirBasis())*SVector(1.0, 0.0)
@test basis_components(LPol(), CirBasis()) ≈ basis_transform(CirBasis()=>CirBasis())*SVector(0.0, 1.0)
@test basis_components(RPol(), LinBasis()) ≈ basis_transform(CirBasis()=>LinBasis())*SVector(1.0, 0.0)
@test basis_components(LPol(), LinBasis()) ≈ basis_transform(CirBasis()=>LinBasis())*SVector(0.0, 1.0)
@test basis_components(XPol(), CirBasis()) ≈ basis_transform(LinBasis()=>CirBasis())*SVector(1.0, 0.0)
@test basis_components(YPol(), CirBasis()) ≈ basis_transform(LinBasis()=>CirBasis())*SVector(0.0, 1.0)
@test basis_components(XPol(), LinBasis()) ≈ basis_transform(LinBasis()=>LinBasis())*SVector(1.0, 0.0)
@test basis_components(YPol(), LinBasis()) ≈ basis_transform(LinBasis()=>LinBasis())*SVector(0.0, 1.0)
for (e1, e2) in [(RPol, LPol), (LPol, RPol),
(XPol, YPol), (XPol, YPol),
]
@test basis_transform(PolBasis{e1,e2}()=>PolBasis{e1,e2}()) ≈ [1.0 0.0; 0.0 1.0]
end
end
@testset "Non-orthogonal" begin
@test_throws AssertionError basis_transform(PolBasis{XPol,YPol}(), PolBasis{RPol,XPol}())
@test_throws AssertionError basis_transform(PolBasis{XPol,YPol}(), PolBasis{RPol,YPol}())
@test_throws AssertionError basis_transform(PolBasis{XPol,YPol}(), PolBasis{XPol,RPol}())
@test_throws AssertionError basis_transform(PolBasis{XPol,YPol}(), PolBasis{XPol,LPol}())
end
@testset "Missing feeds" begin
for E in (XPol,YPol,RPol,LPol)
c1 = basis_components(E(), PolBasis{E, Missing}())
@test c1[1] ≈ 1.0
@test c1[2] isa Missing
c2 = basis_components(E(), PolBasis{Missing, E}())
@test c2[2] ≈ 1.0
@test c2[1] isa Missing
# Test that the compiler realized the Union
@inferred basis_components(E(), PolBasis{Missing, E}())
@inferred basis_components(E(), PolBasis{E, Missing}())
@inferred basis_transform(PolBasis{E, Missing}()=>CirBasis())
@inferred basis_transform(PolBasis{E, Missing}()=>LinBasis())
@inferred basis_transform(CirBasis()=>PolBasis{E, Missing}())
@inferred basis_transform(LinBasis()=>PolBasis{E, Missing}())
end
end
@testset "Simple stokes test" begin
sQ = StokesParams(1.0, 0.5, 0.0, 0.0)
sU = StokesParams(1.0, 0.0, 0.5, 0.0)
sV = StokesParams(1.0, 0.0, 0.0, 0.5)
@test CoherencyMatrix(sQ, LinBasis()) ≈ ([1.5 0.0; 0.0 0.5])
@test CoherencyMatrix(sQ, CirBasis()) ≈ ([1.0 0.5; 0.5 1.0])
@test CoherencyMatrix(sU, LinBasis()) ≈ ([1.0 0.5; 0.5 1.0])
@test CoherencyMatrix(sU, CirBasis()) ≈ ([1.0 0.5im; -0.5im 1.0])
@test CoherencyMatrix(sV, LinBasis()) ≈ ([1.0 0.5im; -0.5im 1.0])
@test CoherencyMatrix(sV, CirBasis()) ≈ ([1.5 0.0; 0.0 0.5])
end
@testset "Simple Coherency test" begin
cRR = CoherencyMatrix(0.5, 0.0, 0.0, 0.0, CirBasis())
cLR = CoherencyMatrix(0.0, 0.5, 0.0, 0.0, CirBasis())
cRL = CoherencyMatrix(0.0, 0.0, 0.5, 0.0, CirBasis())
cLL = CoherencyMatrix(0.0, 0.0, 0.0, 0.5, CirBasis())
@test StokesParams(cRR) ≈ inv(2)*[0.5, 0.0, 0.0, 0.5]
@test StokesParams(cLR) ≈ inv(2)*[0.0, 0.5, 0.5im, 0.0]
@test StokesParams(cRL) ≈ inv(2)*[0.0, 0.5, -0.5im, 0.0]
@test StokesParams(cLL) ≈ inv(2)*[0.5, 0.0, 0.0, -0.5]
cXX = CoherencyMatrix(0.5, 0.0, 0.0, 0.0, LinBasis())
cYX = CoherencyMatrix(0.0, 0.5, 0.0, 0.0, LinBasis())
cXY = CoherencyMatrix(0.0, 0.0, 0.5, 0.0, LinBasis())
cYY = CoherencyMatrix(0.0, 0.0, 0.0, 0.5, LinBasis())
@test StokesParams(cXX) ≈ inv(2)*[0.5, 0.5, 0.0, 0.0]
@test StokesParams(cYX) ≈ inv(2)*[0.0, 0.0, 0.5, 0.5im]
@test StokesParams(cXY) ≈ inv(2)*[0.0, 0.0, 0.5, -0.5im]
@test StokesParams(cYY) ≈ inv(2)*[0.5, -0.5, 0.0, 0.0]
@test StaticArraysCore.similar_type(CoherencyMatrix{CirBasis,LinBasis}, Float64, Size(2,2)) == CoherencyMatrix{CirBasis,LinBasis,Float64}
@test StaticArraysCore.similar_type(StokesParams, Float64, Size(4,)) == StokesParams{Float64}
end
@testset "Conversions back and forward" begin
s = StokesParams(1.0 .+ 0.0im, 0.2 + 0.2im, 0.2 - 0.2im, 0.1+0.05im)
@test s ≈ StokesParams(CoherencyMatrix(s, CirBasis()))
@test s ≈ StokesParams(CoherencyMatrix(s, LinBasis()))
@test s ≈ StokesParams(CoherencyMatrix(s, CirBasis(), LinBasis()))
@test s ≈ StokesParams(CoherencyMatrix(s, LinBasis(), CirBasis()))
@test s ≈ StokesParams(CoherencyMatrix(s, PolBasis{YPol,XPol}(), PolBasis{LPol,RPol}()))
end
@testset "Mixed Pol" begin
I = 2.0 + 0.5im
Q = rand(ComplexF64) - 0.5
U = rand(ComplexF64) - 0.5
V = rand(ComplexF64) - 0.5
s = StokesParams(I, Q, U, V)
c1 = CoherencyMatrix(s, CirBasis(), LinBasis())
c2 = CoherencyMatrix{CirBasis, LinBasis}(s)
c3 = basis_transform(CoherencyMatrix(s, CirBasis()), CirBasis(), LinBasis())
@test c1 ≈ c2
@test c1 ≈ c3
@test StokesParams(c1) ≈ s
@test StokesParams(c2) ≈ s
@test StokesParams(c3) ≈ s
end
@testset "Conversion Consistency" begin
s = StokesParams(1.0 .+ 0.0im, 0.2 + 0.2im, 0.2 - 0.2im, 0.1+0.05im)
c_lin1 = CoherencyMatrix(s, LinBasis())
c_lin2 = CoherencyMatrix(s, PolBasis{XPol,YPol}())
c_lin3 = CoherencyMatrix(s, PolBasis{XPol,YPol}(), PolBasis{XPol,YPol}())
@test c_lin1 ≈ c_lin2 ≈ c_lin3
c_cir1 = CoherencyMatrix(s, CirBasis())
c_cir2 = CoherencyMatrix(s, PolBasis{RPol,LPol}())
c_cir3 = CoherencyMatrix(s, PolBasis{RPol,LPol}(), PolBasis{RPol,LPol}())
@test c_cir1 ≈ c_cir2 ≈ c_cir3
t1 = basis_transform(LinBasis()=>CirBasis())
t2 = basis_transform(CirBasis()=>LinBasis())
@test t2*c_cir1*t1 ≈ c_lin1
@test t1*c_lin1*t2 ≈ c_cir1
@test_throws ArgumentError StokesParams(t1*c_lin1*t2)
# Test the mixed basis
@test c_cir1*t1 ≈ t1*c_lin1
@test c_lin1*t2 ≈ t2*c_cir1
end
@testset "Performance test" begin
s = StokesParams(1.0 .+ 0.0im, 0.2 + 0.2im, 0.2 - 0.2im, 0.1+0.05im)
@test_opt StokesParams(CoherencyMatrix(s, LinBasis()))
@test_opt StokesParams(CoherencyMatrix(s, CirBasis()))
@test_opt StokesParams(CoherencyMatrix(s, LinBasis(), CirBasis()))
@test_opt StokesParams(CoherencyMatrix(s, LinBasis(), CirBasis(), LinBasis()))
end
@testset "Polarized Functions" begin
s = StokesParams(2.0, 0.25, -0.25, 0.25)
@test linearpol(s) == complex(0.25, -0.25)
@test evpa(s) ≈ atan(-0.5, 0.5)/2
@test s[2:end] ≈ polarization(s)
slin = StokesParams(2.0, 0.2, 0.2, 0.0)
fp = fracpolarization(slin)
@test complex(fp[1], fp[2]) ≈ linearpol(slin)/slin.I
@test fp[end] ≈ 0
@testset "ellipse" begin
p = polellipse(s)
@test p.a*p.b ≈ s.V^2/4
@test p.evpa ≈ evpa(s)
plin = polellipse(slin)
@test plin.a ≈ (abs(linearpol(slin)))
@test isapprox(plin.b, 0, atol=1e-8)
@test plin.evpa ≈ evpa(slin)
@test p.sn ≈ sign(s.V)
end
@test mpol(s) ≈ complex(0.25, -0.25)/2
@testset "Complex Vis" begin
I = 2.0 + 0.5im
Q = rand(ComplexF64) - 0.5
U = rand(ComplexF64) - 0.5
V = rand(ComplexF64) - 0.5
s = StokesParams(I, Q, U, V)
@test mbreve(s) == m̆(s)
c = CoherencyMatrix{CirBasis, LinBasis}(s)
@test linearpol(c) ≈ linearpol(s)
@test polarization(c) ≈ polarization(s)
@test fracpolarization(c) ≈ fracpolarization(s)
@test m̆(c) ≈ m̆(s)
@test mbreve(c) ≈ mbreve(s)
@test evpa(c) ≈ evpa(s)
end
end
@testset "ChainRules" begin
I = 2.0 + 0.5im
Q = rand(ComplexF64) - 0.5
U = rand(ComplexF64) - 0.5
V = rand(ComplexF64) - 0.5
s = StokesParams(I, Q, U, V)
c = CoherencyMatrix{CirBasis, LinBasis}(s)
cmat = SMatrix(c)
prc = ChainRulesCore.ProjectTo(c)
@test prc(cmat) == c
@test prc(c) == c
@test_throws AssertionError prc(CoherencyMatrix(cmat, LinBasis()))
@test_throws AssertionError prc(CoherencyMatrix(cmat, CirBasis()))
end
end
| PolarizedTypes | https://github.com/EHTJulia/PolarizedTypes.jl.git |
|
[
"MIT"
] | 0.1.2 | 4217710242df1f4708e58e00763de2569cfbe880 | docs | 1202 | # PolarizedTypes
[](https://ehtjulia.github.io/PolarizedTypes.jl/stable/)
[](https://ehtjulia.github.io/PolarizedTypes.jl/dev/)
[](https://github.com/ehtjulia/PolarizedTypes.jl/actions/workflows/CI.yml?query=branch%3Amain)
[](https://codecov.io/gh/ehtjulia/PolarizedTypes.jl)
[](https://github.com/SciML/ColPrac)
This defines the basic for polarized types for use in VLBI, including:
- `StokesParams` for the stokes parameters
- `CoherencyMatrix` for coherency matrices in arbitrary bases, including a mixed basis.
```julia
using PolarizedTypes
s = StokesParams(1.0, 0.1, 0.1, -0.05)
c = CoherencyMatrix(s, CirBasis(), CirBasis())
l = CoherencyMatrix(s, CirBasis(), LinBasis())
m = CoherencyMatrix(x, LinBasis(), CirBasis())
```
| PolarizedTypes | https://github.com/EHTJulia/PolarizedTypes.jl.git |
|
[
"MIT"
] | 0.1.2 | 4217710242df1f4708e58e00763de2569cfbe880 | docs | 204 | ```@meta
CurrentModule = PolarizedTypes
```
# PolarizedTypes
Documentation for [PolarizedTypes](https://github.com/ptiede/PolarizedTypes.jl).
```@index
```
```@autodocs
Modules = [PolarizedTypes]
```
| PolarizedTypes | https://github.com/EHTJulia/PolarizedTypes.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | code | 413 | using Documenter, AWSTools
makedocs(;
modules=[AWSTools],
format=Documenter.HTML(;
prettyurls=get(ENV, "CI", nothing) == "true", assets=["assets/invenia.css"]
),
pages=["Home" => "index.md"],
repo="https://github.com/JuliaCloud/AWSTools.jl/blob/{commit}{path}#L{line}",
sitename="AWSTools.jl",
authors="Invenia Technical Computing",
checkdocs=:exports,
strict=true,
)
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | code | 1748 | module AWSTools
using AWS
using AWSS3
using Mocking
using Random
using Dates
export assume_role
@service STS
get_caller_identity() = STS.get_caller_identity()["GetCallerIdentityResult"]
account_id() = (@mock get_caller_identity())["GetCallerIdentityResult"]["Account"]
"""
assume_role(role_arn, [role_session_name]) -> AWSConfig
Generate a new `AWSConfig` by assuming a new role. In order to use the assumed role you need
to use this config in the various AWS calls you perform.
# Arguments
- `role_arn::AbstractString`: The ARN of the role to assume.
- `role_session_name::AbstractString`: An optional string which is the unique identifier for
the session name.
# Keywords
- `config::AWSConfig`: The AWS configuration to use when assuming the role.
"""
function assume_role(
role_arn::AbstractString,
role_session_name::AbstractString=randstring(16);
config::AWSConfig=global_aws_config(),
)
function get_role_creds(role_arn, role_session_name, config)
response = @mock STS.assume_role(role_arn, role_session_name; aws_config=config)
response = response["AssumeRoleResult"]
credentials = response["Credentials"]
return AWSCredentials(
credentials["AccessKeyId"],
credentials["SecretAccessKey"],
credentials["SessionToken"];
expiry=DateTime(credentials["Expiration"], dateformat"yyyy-mm-ddTHH:MM:SSZ"),
)
end
renew = () -> get_role_creds(role_arn, role_session_name, config)
creds = renew()
creds.renew = renew
return AWSConfig(; creds=creds)
end
include("timeout.jl")
include("CloudFormation.jl")
include("EC2.jl")
include("ECR.jl")
include("Docker.jl")
Base.@deprecate_binding S3 AWSS3
end # AWSTools
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | code | 3640 | module CloudFormation
using AWS
using AWS: AWSExceptions.AWSException
using EzXML
using MbedTLS: MbedException
using Memento
using Mocking
using OrderedCollections: OrderedDict
using XMLDict
# Improper casing to avoid issues with Module name and AWS.AWSService
@service cloudFormation
const logger = getlogger(@__MODULE__)
# Register the module level logger at runtime so that folks can access the logger via `getlogger(MyModule)`
# NOTE: If this line is not included then the precompiled `MyModule.logger` won't be registered at runtime.
__init__() = Memento.register(logger)
export raw_stack_description, stack_output
function describe_stacks(config::AWSConfig, params::AbstractDict)
dep_msg = """
`describe_stacks(config::AWSConfig, params::AbstractDict)` is deprecated and will be removed.
Use the AWS @service CloudFormation.describe_stacks() functionality instead.
"""
Base.depwarn(dep_msg, :get_authorization_token)
return cloudFormation.describe_stacks(params; aws_config=config)
end
const _NRETRIES = 5
# for APIs we don't want to hammer
function cautious_delays(; kwargs...)
return ExponentialBackOff(; n=_NRETRIES, first_delay=5, max_delay=300, kwargs...)
end
function minimal_delays(; kwargs...)
return ExponentialBackOff(; n=_NRETRIES, first_delay=0.1, max_delay=60, kwargs...)
end
"""
raw_stack_description(stack_name::AbstractString) -> String
Returns the description for the specified stack. Can optionally pass in the aws `config`
as a keyword argument.
"""
function raw_stack_description(
stack_name::AbstractString; config::AWSConfig=global_aws_config()
)
function retry_cond(s, e)
if e isa AWSException
if 500 <= e.cause.status <= 504
debug(logger, "CloudFormation request encountered $(e.code); retrying")
return (s, true)
elseif e.cause.status == 429 || (e.cause.status == 400 && e.code == "Throttling")
debug(logger, "CloudFormation request encountered $(e.code); retrying")
return (s, true)
end
elseif e isa MbedException
debug(logger, "CloudFormation request encountered $e; retrying")
return (s, true)
end
return (s, false)
end
f = retry(; delays=cautious_delays(; jitter=0.2), check=retry_cond) do
@mock describe_stacks(config, Dict("StackName" => stack_name, "return_raw" => true))
end
response = String(f())
return response
end
"""
stack_output(stack_name::AbstractString) -> OrderedDict
The stack's OutputKey and OutputValue values as a dictionary. Can pass in the aws `config`
as a keyword argument.
"""
function stack_output(stack_name::AbstractString; config::AWSConfig=global_aws_config())
outputs = OrderedDict{String,String}()
description = raw_stack_description(stack_name; config=config)
xml = root(parsexml(description))
ns = ["ns" => namespace(xml)]
output_elements = findall("//ns:Outputs/ns:member", xml, ns)
for el in output_elements
key = nodecontent(findfirst("ns:OutputKey", el, ns))
val = nodecontent(findfirst("ns:OutputValue", el, ns))
outputs[key] = val
end
return outputs
end
function output_pair(item::AbstractDict)
key = item["OutputKey"]::String
value = if isa(item["OutputValue"], String)
item["OutputValue"]::String
elseif isa(item["OutputValue"], AbstractDict) && isempty(item["OutputValue"])
""
else
throw(ArgumentError("Unhandled output value: $(repr(item["OutputValue"]))"))
end
return key => value
end
end # CloudFormation
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | code | 1284 | module Docker
using AWS
using ..ECR
export login, pull, push, build
"""
login(registry_id::Union{AbstractString, Integer}="")
Gets the AWS ECR authorization token and runs the corresponding docker login command.
Takes an optional AWS `config` keyword argument or uses the default.
"""
function login(
registry_id::Union{AbstractString,Integer}=""; config::AWSConfig=global_aws_config()
)
return login(ECR.get_login(registry_id; config=config))
end
function login(docker_login::Cmd)
success(pipeline(docker_login, stdout=stdout, stderr=stderr))
end
"""
pull(image::AbstractString, tags::AbstractVector{<:AbstractString}=String[])
Pulls a docker image and tags it if `tags` is specified.
"""
function pull(image::AbstractString, tags::AbstractVector{<:AbstractString}=String[])
run(`docker pull $image`)
for tag in tags
run(`docker tag $image $tag`)
end
end
"""
push(image::AbstractString)
Pushes a docker image.
"""
function push(image::AbstractString)
run(`docker push $image`)
end
"""
build(dir::AbstractString, tag::AbstractString="")
Builds the docker image.
"""
function build(dir::AbstractString, tag::AbstractString="")
opts = isempty(tag) ? `` : `-t $tag`
run(`docker build $opts $dir`)
end
end # Docker
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | code | 1842 | module EC2
using ...AWSTools: timeout
using HTTP: HTTP
using Mocking
export instance_metadata, instance_region, instance_availability_zone
"""
instance_metadata(path) -> Union{String,Nothing}
Retrieve AWS EC2 instance metadata as a string from the provided `path`. If no instance
metadata is available (typically due to not running within an EC2 instance) then `nothing`
will be returned. See the AWS documentation for details on what metadata is available.
https://docs.aws.amazon.com/AWSEC2/latest/UserGuide/ec2-instance-metadata.html
"""
function instance_metadata(path::AbstractString)
# Retrieve details about the instance:
# https://docs.aws.amazon.com/AWSEC2/latest/UserGuide/ec2-instance-metadata.html
#
# Note: When running outside of EC2 the connection to the local-link address will fail
# with a connection timeout (ETIMEDOUT) after a 60 seconds (tested on HTTP.jl v0.8.2)
# See: https://github.com/JuliaWeb/HTTP.jl/issues/114
uri = HTTP.URI(scheme="http", host="169.254.169.254", path=path)
r = timeout(5) do
@mock HTTP.get(uri, status_exception=false)
end
return r !== nothing ? String(something(r).body) : r
end
"""
instance_availability_zone() -> Union{String,Nothing}
Get the availability zone of the host if running inside of an EC2 instance. If not running
within an EC2 instance `nothing` is returned.
"""
function instance_availability_zone()
# Get the availability zone information from the EC2 instance metadata.
return instance_metadata("/latest/meta-data/placement/availability-zone")
end
"""
instance_availability_zone() -> Union{String,Nothing}
Get the region of the host if executed inside of an EC2 instance. If not running within an
EC2 instance `nothing` is returned.
"""
instance_region() = chop(instance_availability_zone())
end
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | code | 1967 | module ECR
using AWS
using Base64
using Mocking
# Improper casing to avoid issues with Module name and AWS.AWSService
@service Ecr
function get_authorization_token(config::AWSConfig, params::AbstractDict=Dict{String,Any}())
dep_msg = """
`get_authorization_token(config::AWSConfig, params::AbstractDict=Dict{String,Any}())` is deprecated and will be removed.
Use the AWS @service ECR.get_authorization_token() functionality instead.
"""
Base.depwarn(dep_msg, :get_authorization_token)
return Ecr.get_authorization_token(params; aws_config=config)
end
export get_login
"""
get_login(registry_ids::Union{AbstractString, Integer}="") -> Cmd
Gets the AWS ECR authorization token and returns the corresponding docker login command.
The AWS `config` keyword parameter is optional (will use the default if it's not passed in).
"""
get_login
function get_login(registry_id::AbstractString=""; config::AWSConfig=global_aws_config())
# Note: Although `get_authorization_token` can take multiple registry IDs at once it
# will only return a "proxyEndpoint" for the first registry. Additionally, the
# `aws ecr get-login` command returns a `docker login` command for each registry ID
# passed in. Because of these factors we'll do our processing on a single registry.
response = if !isempty(registry_id)
@mock get_authorization_token(config, Dict("registryIds" => [registry_id]))
else
@mock get_authorization_token(config)
end
authorization_data = first(response["authorizationData"])
token = String(base64decode(authorization_data["authorizationToken"]))
username, password = split(token, ':')
endpoint = authorization_data["proxyEndpoint"]
return `docker login -u $username -p $password $endpoint`
end
function get_login(registry_id::Integer; config::AWSConfig=global_aws_config())
return get_login(lpad(registry_id, 12, '0'); config=config)
end
end # ECR
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | code | 1662 | """
timeout(f::Function, seconds::Real) -> Union{Some,Nothing}
Executes the provided function `f` and returns the result wrapped in a `Some`. If the given
function takes longer than `seconds` the function is terminated and `nothing` is returned.
Note that the timeout will only work correctly if the called function `f` yields which will
occur automatically if performing an I/O operation.
"""
function timeout(f::Function, seconds::Real)
result = nothing
c = Condition()
try
# Execute the given function as a task so we can interrupt it if necessary
t = @async begin
result = Some(f())
notify(c)
end
# Create a timeout task which aborts the wait early if we hit the timeout.
# Note: We use an async task here to ensure when `f` finishes before the timeout it
# is not blocked by the timer.
@async begin
# Note: The `pollint` will may cause abort events to occur after the specified
# timeout. e.g. if `seconds = 1.5` then the abort would occur at 2 seconds.
timedwait(float(seconds); pollint=1.0) do
istaskdone(t)
end
notify(c)
end
wait(c)
# Kill the function task if it is still executing
istaskdone(t) || @async Base.throwto(t, InterruptException())
wait(t)
catch e
# Unwrap the TaskFailedException
if VERSION >= v"1.3.0-alpha.110" && e isa TaskFailedException
e = e.task.exception
end
# Ignore the kill exception
e isa InterruptException || rethrow(e)
end
return result
end
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | code | 784 | using AWSTools.EC2: instance_metadata, instance_availability_zone, instance_region
@testset "EC2" begin
@testset "instance_metadata" begin
r = HTTP.Response("{}") # Instance identity document is JSON
apply(instance_metadata_patch(r)) do
@test instance_metadata("/latest/dynamic/instance-identity/document") == "{}"
end
end
@testset "instance_availability_zone" begin
r = HTTP.Response("us-east-1a")
apply(instance_metadata_patch(r)) do
@test instance_availability_zone() == "us-east-1a"
end
end
@testset "instance_region" begin
r = HTTP.Response("us-east-1a")
apply(instance_metadata_patch(r)) do
@test instance_region() == "us-east-1"
end
end
end
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | code | 15264 | using AWS
using Base: CmdRedirect
using Base64
using Dates: datetime2unix, now
const invalid_access_key = "ThisIsMyInvalidAccessKey"
const invalid_secret_key = "ThisIsMyInvalidSecretKey"
function get_auth(config::AWSConfig; params::AbstractDict=Dict())
id = lpad(!haskey(params, "registryIds") ? "" : first(params["registryIds"]), 12, '0')
return Dict(
"authorizationData" => [
Dict(
"authorizationToken" => base64encode("AWS:password"),
"proxyEndpoint" => "https://$(id).dkr.ecr.us-east-1.amazonaws.com",
),
],
)
end
get_caller_identity_patch = @patch function AWSTools.get_caller_identity()
account_id = join(rand(0:9, 12), "")
return Dict(
"GetCallerIdentityResult" => Dict(
Dict(
"Account" => account_id,
"Arn" => "arn:aws:iam::$account_id:user/UserName",
"UserId" => join(rand('A':'Z', 21), ""),
),
),
)
end
sts_assume_role = @patch function AWSTools.STS.assume_role(
role_arn, role_session_name; aws_config
)
return Dict(
"AssumeRoleResult" => Dict(
"Credentials" => Dict(
"AccessKeyId" => "TESTACCESSKEYID",
"SecretAccessKey" => "TESTSECRETACEESSKEY",
"SessionToken" => "TestSessionToken",
"Expiration" => "2021-11-03T16:37:10Z",
),
),
)
end
function instance_metadata_patch(result)
@patch HTTP.get(args...; kwargs...) = result
end
get_authorization_token_patch = @patch function AWSTools.ECR.get_authorization_token(
config::AWSConfig, params::AbstractDict
)
return get_auth(config; params=params)
end
get_authorization_token_no_param_patch = @patch function AWSTools.ECR.get_authorization_token(
config::AWSConfig
)
return get_auth(config)
end
describe_stacks_patch = @patch function AWSTools.CloudFormation.describe_stacks(
config, params
)
responses = Dict(
Dict("StackName" => "stackname", "return_raw" => true) => """
<DescribeStacksResponse xmlns="http://cloudformation.amazonaws.com/doc/2010-05-15/">
<DescribeStacksResult>
<Stacks>
<member>
<StackId>Stack Id</StackId>
<StackName>Stack Name</StackName>
<Description>Stack Description</Description>
</member>
</Stacks>
</DescribeStacksResult>
</DescribeStacksResponse>
""",
Dict("StackName" => "1-stack-output-stackname", "return_raw" => true) => """
<DescribeStacksResponse xmlns="http://cloudformation.amazonaws.com/doc/2010-05-15/">
<DescribeStacksResult>
<Stacks>
<member>
<Outputs>
<member>
<OutputKey>TestBucketArn1</OutputKey>
<OutputValue>arn:aws:s3:::test-bucket-1</OutputValue>
</member>
</Outputs>
<StackId>Stack Id</StackId>
<StackName>Stack Name</StackName>
<Description>Stack Description</Description>
</member>
</Stacks>
</DescribeStacksResult>
</DescribeStacksResponse>
""",
Dict("StackName" => "multiple-stack-outputs-stackname", "return_raw" => true) => """
<DescribeStacksResponse xmlns="http://cloudformation.amazonaws.com/doc/2010-05-15/">
<DescribeStacksResult>
<Stacks>
<member>
<Outputs>
<member>
<OutputKey>TestBucketArn1</OutputKey>
<OutputValue>arn:aws:s3:::test-bucket-1</OutputValue>
</member>
<member>
<OutputKey>TestBucketArn2</OutputKey>
<OutputValue>arn:aws:s3:::test-bucket-2</OutputValue>
</member>
</Outputs>
<StackId>Stack Id</StackId>
<StackName>Stack Name</StackName>
<Description>Stack Description</Description>
</member>
</Stacks>
</DescribeStacksResult>
</DescribeStacksResponse>
""",
Dict("StackName" => "empty-value", "return_raw" => true) => """
<DescribeStacksResponse xmlns="http://cloudformation.amazonaws.com/doc/2010-05-15/">
<DescribeStacksResult>
<Stacks>
<member>
<Outputs>
<member>
<OutputKey>ParquetConversionTriggerName</OutputKey>
<OutputValue></OutputValue>
</member>
</Outputs>
<StackId>Stack Id</StackId>
<StackName>Stack Name</StackName>
<Description>Stack Description</Description>
</member>
</Stacks>
</DescribeStacksResult>
</DescribeStacksResponse>
""",
Dict("StackName" => "export", "return_raw" => true) => """
<DescribeStacksResponse xmlns="http://cloudformation.amazonaws.com/doc/2010-05-15/">
<DescribeStacksResult>
<Stacks>
<member>
<Outputs>
<member>
<Description>Exported output for use in other stacks</Description>
<ExportName>ExportedKey</ExportName>
<OutputKey>Key</OutputKey>
<OutputValue>Value</OutputValue>
</member>
</Outputs>
<StackId>Stack Id</StackId>
<StackName>Stack Name</StackName>
<Description>Stack Description</Description>
</member>
</Stacks>
</DescribeStacksResult>
</DescribeStacksResponse>
""",
)
# So we can test that we get an error using the invalid access and secret keys
access_key = config.credentials.access_key_id
secret_key = config.credentials.secret_key
if access_key == invalid_access_key && secret_key == invalid_secret_key
throw(
AWSException(
HTTP.StatusError(
403,
"",
"",
HTTP.Messages.Response(
403,
"""
<ErrorResponse xmlns="http://cloudformation.amazonaws.com/doc/2010-05-15/">
<Error>
<Type>Sender</Type>
<Code>InvalidClientTokenId</Code>
<Message>The security token included in the request is invalid.</Message>
</Error>
<RequestId>cff5beb8-4b7b-11e9-9c2b-43c18f6078dc</RequestId>
</ErrorResponse>
""",
),
),
),
)
else
return responses[params]
end
end
function throttle_patch(allow)
describe_stacks_throttle_count = 0
@patch function AWSTools.CloudFormation.describe_stacks(config, params)
describe_stacks_throttle_count += 1
if !(describe_stacks_throttle_count in allow)
error_message = """
<ErrorResponse xmlns="http://cloudformation.amazonaws.com/doc/2010-05-15/">
<Error>
<Type>Sender</Type>
<Code>Throttling</Code>
<Message>Rate exceeded</Message>
</Error>
<RequestId>d0c477ac-f267-11e8-9d2b-93e3aa6368c5</RequestId>
</ErrorResponse>
"""
response = HTTP.Messages.Response(400, error_message)
http_error = HTTP.ExceptionRequest.StatusError(400, "", "", response)
throw(AWSException(http_error))
end
responses = Dict(
Dict("StackName" => "stackname", "return_raw" => true) => """
<DescribeStacksResponse xmlns="http://cloudformation.amazonaws.com/doc/2010-05-15/">
<DescribeStacksResult>
<Stacks>
<member>
<StackId>Stack Id</StackId>
<StackName>Stack Name</StackName>
<Description>Stack Description</Description>
<ThrottleCount>$describe_stacks_throttle_count</ThrottleCount>
</member>
</Stacks>
</DescribeStacksResult>
</DescribeStacksResponse>
""",
)
return responses[params]
end
end
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | code | 4813 | using AWS
using AWS: AWSExceptions.AWSException
using AWSTools
using Dates
using Documenter
using HTTP
using Memento
using Mocking
using OrderedCollections: OrderedDict
using Test
import AWSTools.Docker
using AWSTools: account_id
using AWSTools.CloudFormation: raw_stack_description, stack_output
using AWSTools.EC2: instance_availability_zone, instance_region
using AWSTools.ECR: get_login
Memento.config!("debug"; fmt="[{level} | {name}]: {msg}")
# Need this so that submodules are able to use the debug log level
setlevel!(getlogger(AWSTools), "debug")
Mocking.activate()
include("patch.jl")
"""
describe_stack_string(throttle_count::Integer=0) -> String
Returns the expected xml string for CloudFormation tests.
Pass in a throttle count for throttling.
"""
function describe_stack_string(throttle_count::Integer=0)
result = """
<DescribeStacksResponse xmlns="http://cloudformation.amazonaws.com/doc/2010-05-15/">
<DescribeStacksResult>
<Stacks>
<member>
<StackId>Stack Id</StackId>
<StackName>Stack Name</StackName>
<Description>Stack Description</Description>
$(throttle_count > 0 ? "<ThrottleCount>$throttle_count</ThrottleCount>" : "")
</member>
</Stacks>
</DescribeStacksResult>
</DescribeStacksResponse>
"""
return replace(result, r"^\s*\n"m => "")
end
# TODO: Include in Base
function Base.convert(::Type{Vector{String}}, cmd::Cmd)
cmd.exec
end
@testset "AWSTools Tests" begin
include("timeout.jl")
include("EC2.jl")
@testset "account_id" begin
apply(get_caller_identity_patch) do
@test occursin(r"^\d{12}$", account_id())
end
end
@testset "assume_role" begin
apply(sts_assume_role) do
result = AWSTools.assume_role("TestArn")
@test isa(result.credentials, AWSCredentials)
@test isa(result.credentials.renew, Function)
end
end
@testset "CloudFormation" begin
apply(describe_stacks_patch) do
@testset "raw_stack_description" begin
resp = raw_stack_description("stackname")
@test resp == describe_stack_string()
@test_throws AWSException begin
creds = AWSCredentials(invalid_access_key, invalid_secret_key)
raw_stack_description("stackname"; config=AWSConfig(; creds=creds))
end
end
@testset "stack_output" begin
outputs = stack_output("stackname")
@test outputs == Dict()
outputs = stack_output("1-stack-output-stackname")
@test outputs == Dict("TestBucketArn1"=>"arn:aws:s3:::test-bucket-1")
outputs = stack_output("multiple-stack-outputs-stackname")
@test outputs == Dict(
"TestBucketArn1" => "arn:aws:s3:::test-bucket-1",
"TestBucketArn2" => "arn:aws:s3:::test-bucket-2",
)
# Empty output values
outputs = stack_output("empty-value")
@test outputs == Dict(
"ParquetConversionTriggerName" => "",
)
outputs = stack_output("export")
@test outputs == Dict("Key" => "Value")
end
end
end
@testset "raw_stack_description throttling" begin
allow = [1, 3, 5, 7, 8, 11, 13, 14, 15, 16]
apply(throttle_patch(allow)) do
for i in allow
@test_skip raw_stack_description("stackname") == describe_stack_string(i)
end
end
end
@testset "ECR" begin
@testset "Basic login" begin
apply(get_authorization_token_no_param_patch) do
docker_login = get_login()
@test docker_login ==
`docker login -u AWS -p password https://000000000000.dkr.ecr.us-east-1.amazonaws.com`
end
end
@testset "Login specifying registry ID" begin
apply(get_authorization_token_patch) do
docker_login = get_login(1)
@test docker_login == `docker login -u AWS -p password https://000000000001.dkr.ecr.us-east-1.amazonaws.com`
end
end
end
@testset "Online Tests" begin
@testset "ECR" begin
command = convert(Vector{String}, get_login())
@test command[1] == "docker"
@test command[2] == "login"
@test command[3] == "-u"
@test command[4] == "AWS"
@test command[5] == "-p"
@test length(command) == 7
end
end
doctest(AWSTools)
end
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | code | 1345 | using AWSTools: timeout
@testset "timeout" begin
# Ensure that timeout is compiled for elapsed time tests
timeout(() -> 0, 1)
@testset "finish" begin
secs = @elapsed begin
result = timeout(() -> 0, 1)
end
@test result == Some(0)
@test secs < 0.2 # Should execute almost as fast as calling the function directly
end
@testset "abort" begin
secs = @elapsed begin
result = timeout(1) do
sleep(5)
error("unexpected error")
end
end
@test result === nothing
@test 1 <= secs < 5
end
@testset "return nothing" begin
secs = @elapsed begin
result = timeout(() -> nothing, 1)
end
@test result == Some(nothing)
@test secs < 1
@test secs < 0.2 # Should execute almost as fast as calling the function directly
end
@testset "exception" begin
local exception
secs = @elapsed begin
try
timeout(() -> error("function error"), 5)
catch e
exception = e
end
end
@test exception == ErrorException("function error")
@test secs < 5
@test secs < 0.2 # Should execute almost as fast as calling the function directly
end
end
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | docs | 161 | # AWSTools
[](https://github.com/JuliaCloud/AWSTools.jl/actions?query=workflow%3ACI)
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 2.3.0 | 842c0e8cb146524ffa5caf097e3014ab45b441d9 | docs | 1345 | # AWSTools
AWSTools provides several helper methods for working with AWSSDK.jl from julia.
## Installation
```julia
julia> Pkg.add("AWSTools.jl")
```
You will also need to have the proper IAM permissions for the actions you wish to perform.
Currently the permissions AWSTools requires (if run in it's entirety) are:
- cloudformation:DescribeStacks
- ecr:GetAuthorizationToken
- s3:GetObject
- s3:ListBucket
- s3:PutObject
- s3:DeleteObject
## Basic Usage
This example uses the Docker module directly and the ECR module indirectly. See the API for other uses of AWSTools.
```julia
julia> using AWSTools
julia> using AWSTools.Docker
julia> Docker.login()
WARNING! Using --password via the CLI is insecure. Use --password-stdin.
Login Succeeded
true
```
## API
```@docs
AWSTools.assume_role
```
### CloudFormation
```@docs
AWSTools.CloudFormation.raw_stack_description(::AbstractString)
AWSTools.CloudFormation.stack_output(::AbstractString)
```
### Docker
```@docs
AWSTools.Docker.login()
AWSTools.Docker.pull(::AbstractString)
AWSTools.Docker.push(::AbstractString)
AWSTools.Docker.build(::AbstractString, ::AbstractString)
```
### ECR
```@docs
AWSTools.ECR.get_login
```
### EC2
```@docs
AWSTools.EC2.instance_metadata(::AbstractString)
AWSTools.EC2.instance_availability_zone()
AWSTools.EC2.instance_region()
```
| AWSTools | https://github.com/JuliaCloud/AWSTools.jl.git |
|
[
"MIT"
] | 0.2.0 | 5666b8b2ce833fb6502db9ddba352f9e915f44b2 | code | 1360 | module RemoteSemaphores
export RemoteSemaphore, acquire, release
using Base: Semaphore, acquire, release
using Distributed
"""
RemoteSemaphore(n::Int, pid=myid())
A semaphore living on a specific process.
Do not attempt to fetch the future to a different process and use it there, as that will be
an isolated, unsynced copy of the semaphore.
"""
struct RemoteSemaphore
n::Int # stored for printing only
rref::Future
function RemoteSemaphore(n::Integer, pid=myid())
sem = Semaphore(n)
fut = Future(pid)
put!(fut, sem)
return new(n, fut)
end
end
function Base.acquire(rsem::RemoteSemaphore)
fut = rsem.rref
loc = fut.where
remotecall_wait(loc) do
sem = fetch(fut)
acquire(sem)
end
return nothing
end
function Base.release(rsem::RemoteSemaphore)
fut = rsem.rref
loc = fut.where
remotecall_wait(loc) do
sem = fetch(fut)
release(sem)
end
return nothing
end
function Base.show(io::IO, rsem::RemoteSemaphore)
print(io, typeof(rsem), '(', rsem.n, ", pid=", rsem.rref.where, ')')
end
# expensive, easily out of sync, for testing only
function _current_count(rsem::RemoteSemaphore)
fut = rsem.rref
loc = fut.where
return remotecall_fetch(loc) do
sem = fetch(fut)
return sem.curr_cnt
end
end
end
| RemoteSemaphores | https://github.com/invenia/RemoteSemaphores.jl.git |
|
[
"MIT"
] | 0.2.0 | 5666b8b2ce833fb6502db9ddba352f9e915f44b2 | code | 6176 | using RemoteSemaphores
using RemoteSemaphores: _current_count
using Test
using Dates
using Distributed
include("utils.jl")
@testset "RemoteSemaphores.jl" begin
@testset "Single Process" begin
@test_throws ArgumentError RemoteSemaphore(0)
rsem = RemoteSemaphore(2)
@test _current_count(rsem) == 0
@test string(rsem) == "$(typeof(rsem))(2, pid=$(myid()))"
try
asynctimedwait(1.0; kill=true) do
release(rsem)
end
@test "Expected error but no error thrown" == nothing
catch err
@test err isa RemoteException
if VERSION >= v"1.2.0-DEV.28"
expected = ErrorException
@test err.captured.ex isa expected
@test occursin("release count must match acquire count", err.captured.ex.msg)
else
expected = AssertionError
@test err.captured.ex isa expected
end
if !isa(err, RemoteException) || !isa(err.captured.ex, expected)
rethrow(err)
end
end
@test asynctimedwait(1.0; kill=true) do
acquire(rsem)
end
@test _current_count(rsem) == 1
@test asynctimedwait(1.0; kill=true) do
acquire(rsem)
end
@test _current_count(rsem) == 2
acquired = false
@test asynctimedwait(1.0) do
acquire(rsem)
acquired = true
end == false
@test !acquired
@test _current_count(rsem) == 2
@test asynctimedwait(1.0; kill=true) do
release(rsem)
end
@test acquired
@test _current_count(rsem) == 2
@test asynctimedwait(10.0; kill=true) do
@sync for i = 1:100
@async (isodd(i) ? acquire(rsem) : release(rsem))
end
end
@test _current_count(rsem) == 2
end
@testset "Multiple Processes" begin
@testset "Simple remote" begin
worker_pid = addprocs(1)[1]
@everywhere using RemoteSemaphores
@everywhere include("utils.jl")
rsem = RemoteSemaphore(2, worker_pid)
@test _current_count(rsem) == 0
@test string(rsem) == "$(typeof(rsem))(2, pid=$worker_pid)"
try
asynctimedwait(1.0; kill=true) do
release(rsem)
end
@test "Expected error but no error thrown" == nothing
catch err
@test err isa RemoteException
if VERSION >= v"1.2.0-DEV.28"
expected = ErrorException
@test err.captured.ex isa expected
@test occursin("release count must match acquire count", err.captured.ex.msg)
else
expected = AssertionError
@test err.captured.ex isa expected
end
if !isa(err, RemoteException) || !isa(err.captured.ex, expected)
rethrow(err)
end
end
@test asynctimedwait(1.0; kill=true) do
acquire(rsem)
end
@test _current_count(rsem) == 1
@test asynctimedwait(1.0; kill=true) do
acquire(rsem)
end
@test _current_count(rsem) == 2
acquired = false
@test asynctimedwait(1.0) do
acquire(rsem)
acquired = true
end == false
@test !acquired
@test _current_count(rsem) == 2
@test asynctimedwait(1.0; kill=true) do
release(rsem)
end
@test acquired
@test _current_count(rsem) == 2
@test asynctimedwait(10.0; kill=true) do
@sync for i = 1:100
@async (isodd(i) ? acquire(rsem) : release(rsem))
end
end
@test _current_count(rsem) == 2
end
@testset "Multiple processes" begin
addprocs(3 - nprocs())
worker1_pid, worker2_pid = workers()
@everywhere using RemoteSemaphores
@everywhere using RemoteSemaphores: _current_count
@everywhere include("utils.jl")
rsem = RemoteSemaphore(3, worker1_pid)
@test _current_count(rsem) == 0
@test (@fetchfrom worker1_pid _current_count(rsem)) == 0
@test (@fetchfrom worker2_pid _current_count(rsem)) == 0
@test asynctimedwait(10.0; kill=true) do
acquire(rsem)
end
@test _current_count(rsem) == 1
@test (@fetchfrom worker1_pid _current_count(rsem)) == 1
@test (@fetchfrom worker2_pid _current_count(rsem)) == 1
@test @fetchfrom worker2_pid asynctimedwait(1.0; kill=true) do
acquire(rsem)
end
@test _current_count(rsem) == 2
@test (@fetchfrom worker1_pid _current_count(rsem)) == 2
@test (@fetchfrom worker2_pid _current_count(rsem)) == 2
@test @fetchfrom worker1_pid asynctimedwait(1.0; kill=true) do
acquire(rsem)
end
@test _current_count(rsem) == 3
@test (@fetchfrom worker1_pid _current_count(rsem)) == 3
@test (@fetchfrom worker2_pid _current_count(rsem)) == 3
acquired1 = false
@test asynctimedwait(10.0) do
acquire(rsem)
acquired1 = true
end == false
acquired2 = Future()
@test @fetchfrom worker1_pid begin
asynctimedwait(10.0) do
acquire(rsem)
put!(acquired2, true)
end
end == false
acquired3 = Future()
@test @fetchfrom worker2_pid begin
asynctimedwait(10.0) do
acquire(rsem)
put!(acquired3, true)
end
end == false
conditions_hit() = acquired1 + isready(acquired2) + isready(acquired3)
sleep(10)
@test conditions_hit() == 0
@test asynctimedwait(10.0; kill=true) do
release(rsem)
end
sleep(2)
@test conditions_hit() == 1
@test asynctimedwait(10.0; kill=true) do
release(rsem)
end
sleep(2)
@test conditions_hit() == 2
@test asynctimedwait(10.0; kill=true) do
release(rsem)
end
sleep(2)
@test conditions_hit() == 3
@test acquired1
@test isready(acquired2)
@test isready(acquired3)
end
end
end
| RemoteSemaphores | https://github.com/invenia/RemoteSemaphores.jl.git |
|
[
"MIT"
] | 0.2.0 | 5666b8b2ce833fb6502db9ddba352f9e915f44b2 | code | 539 | struct TimeoutException
duration
end
function Base.showerror(io::IO, te::TimeoutException)
print(io, "TimeoutException: Operation did not finish in ", te.duration)
if !isa(te.duration, Period)
print(io, " seconds")
end
end
function asynctimedwait(fn, secs; kill=false)
t = @async fn()
timedwait(() -> istaskdone(t), secs)
if istaskdone(t)
fetch(t)
return true
else
if kill
Base.throwto(t, TimeoutException(secs))
end
return false
end
end
| RemoteSemaphores | https://github.com/invenia/RemoteSemaphores.jl.git |
|
[
"MIT"
] | 0.2.0 | 5666b8b2ce833fb6502db9ddba352f9e915f44b2 | docs | 967 | # RemoteSemaphores
[](https://travis-ci.com/invenia/RemoteSemaphores.jl)
[](https://ci.appveyor.com/project/invenia/RemoteSemaphores-jl)
[](https://codecov.io/gh/invenia/RemoteSemaphores.jl)
## Documentation
```julia
RemoteSemaphore(n::Int, pid=myid())
```
A `RemoteSemaphore` is a [counting semaphore](https://www.quora.com/What-is-a-counting-semaphore) that lives on a particular process in order to control access to a resource from multiple processes.
It is implemented using the unexported `Base.Semaphore` stored inside a `Future` which is only accessed on the process it was initialized on.
Like `Base.Semaphore`, it implements `acquire` and `release`, and is not thread-safe.
| RemoteSemaphores | https://github.com/invenia/RemoteSemaphores.jl.git |
|
[
"MIT"
] | 1.0.1 | 5ed2f06644b4cdc96f25b2f8527cebde1ec77506 | code | 267 | module InteractiveErrorsCthulhuExt
import Cthulhu
import InteractiveErrors
InteractiveErrors.has_cthulhu() = true
InteractiveErrors.ascend(mi::Core.MethodInstance) = Cthulhu.ascend(mi)
InteractiveErrors.descend(mi::Core.MethodInstance) = Cthulhu.descend(mi)
end
| InteractiveErrors | https://github.com/MichaelHatherly/InteractiveErrors.jl.git |
|
[
"MIT"
] | 1.0.1 | 5ed2f06644b4cdc96f25b2f8527cebde1ec77506 | code | 225 | module InteractiveErrorsDebuggerExt
import Debugger
import InteractiveErrors
InteractiveErrors.has_debugger() = true
InteractiveErrors.breakpoint(file::AbstractString, line::Integer) = Debugger.breakpoint(file, line)
end
| InteractiveErrors | https://github.com/MichaelHatherly/InteractiveErrors.jl.git |
|
[
"MIT"
] | 1.0.1 | 5ed2f06644b4cdc96f25b2f8527cebde1ec77506 | code | 389 | module InteractiveErrorsJETExt
import JET
import InteractiveErrors
InteractiveErrors.has_jet() = true
function InteractiveErrors.report_call(mi::Core.MethodInstance)
func = Base.tuple_type_head(mi.specTypes).instance
sig = Base.tuple_type_tail(mi.specTypes)
result = JET.report_call(func, sig)
@info "Press return to continue."
readline()
return result
end
end
| InteractiveErrors | https://github.com/MichaelHatherly/InteractiveErrors.jl.git |
|
[
"MIT"
] | 1.0.1 | 5ed2f06644b4cdc96f25b2f8527cebde1ec77506 | code | 560 | module InteractiveErrorsJuliaFormatterExt
import JuliaFormatter
import InteractiveErrors
import PrecompileTools
InteractiveErrors.has_juliaformatter() = true
function InteractiveErrors.format_julia_source(source::String)
try
return JuliaFormatter.format_text(source)
catch err
@debug "failed to format source" err source
return source
end
end
PrecompileTools.@compile_workload begin
InteractiveErrors.format_julia_source(
read(joinpath(@__DIR__, "..", "src", "InteractiveErrors.jl"), String),
)
end
end
| InteractiveErrors | https://github.com/MichaelHatherly/InteractiveErrors.jl.git |
|
[
"MIT"
] | 1.0.1 | 5ed2f06644b4cdc96f25b2f8527cebde1ec77506 | code | 780 | module InteractiveErrorsOhMyREPLExt
import OhMyREPL
import InteractiveErrors
import PrecompileTools
InteractiveErrors.has_ohmyrepl() = true
function InteractiveErrors.highlight(source::String)
O = OhMyREPL
tokens = collect(O.tokenize(source))
crayons = fill(O.Crayon(), length(tokens))
O.Passes.SyntaxHighlighter.SYNTAX_HIGHLIGHTER_SETTINGS(crayons, tokens, 0, source)
io = IOBuffer()
for (token, crayon) in zip(tokens, crayons)
print(io, crayon)
print(io, O.untokenize(token, source))
print(io, O.Crayon(reset = true))
end
return String(take!(io))
end
PrecompileTools.@compile_workload begin
InteractiveErrors.highlight(
read(joinpath(@__DIR__, "..", "src", "InteractiveErrors.jl"), String),
)
end
end
| InteractiveErrors | https://github.com/MichaelHatherly/InteractiveErrors.jl.git |
|
[
"MIT"
] | 1.0.1 | 5ed2f06644b4cdc96f25b2f8527cebde1ec77506 | code | 13193 | module InteractiveErrors
using FoldingTrees
using REPL, REPL.TerminalMenus, InteractiveUtils, IterTools
import PackageExtensionCompat, PrecompileTools
export toggle, current_theme, set_theme!, reset_theme!, adjust_theme!
#
# Themes.
#
const DEFAULT_THEME = (
function_name = (bold = true,),
directory = (color = :light_black,),
filename = (color = :magenta, bold = true),
line_number = (color = :green, bold = true),
user_stack = (color = :green, bold = true),
system_stack = (color = :red, bold = true),
stdlib_module = (color = :yellow,),
base_module = (color = :blue,),
core_module = (color = :light_black,),
package_module = (color = :cyan, bold = true),
unknown_module = (color = :red,),
inlined_frames = (color = :light_black,),
toplevel_frames = (color = :light_black,),
repeated_frames = (color = :red,),
file_contents = (color = :light_black,),
signature = (color = :light_black, format = true, highlight = true),
source = (color = :normal, bold = true, highlight = true),
line_range = (before = 0, after = 5),
charset = :unicode,
)
const THEME = Ref{Any}(DEFAULT_THEME)
current_theme() = THEME[]
set_theme!(nt::NamedTuple) = THEME[] = nt
set_theme!(; kws...) = set_theme!(_nt(kws))
_nt(kws) = NamedTuple{Tuple(keys(kws))}(values(kws))
reset_theme!() = set_theme!(DEFAULT_THEME)
adjust_theme!(nt::NamedTuple) = set_theme!(merge(current_theme(), nt))
adjust_theme!(; kws...) = adjust_theme!(_nt(kws))
get_theme(key) = get(NamedTuple, current_theme(), key)
get_theme(key, default) = get(current_theme(), key, default)
function style(str; kws...)
sprint(; context = :color => true) do io
printstyled(io, str; bold = get(kws, :bold, false), color = get(kws, :color, :normal))
end
end
style(str, key::Symbol) = style(str; get_theme(key)...)
#
# Stackframe Wrapping.
#
struct StackFrameWrapper
sf::StackTraces.StackFrame
n::Int
StackFrameWrapper(tuple) = new(tuple...)
end
function Base.show(io::IO, s::StackFrameWrapper)
func = style(s.sf.func, :function_name)
file = rewrite_path(s.sf.file)
dir, file = dirname(file), basename(file)
file = style(file, :filename)
dir = style(joinpath(dir, ""), :directory)
line = style(s.sf.line, :line_number)
repeated = s.n > 1 ? style("x $(s.n)", :repeated_frames) : ""
print(io, strip("$func $dir$file:$line $repeated"))
end
function rewrite_path(path)
fn(path, replacer) = replace(String(path), replacer; count = 1)
path = fn(path, normpath(Sys.BUILD_STDLIB_PATH) => "@stdlib")
path = fn(path, normpath(Sys.STDLIB) => "@stdlib")
path = fn(path, homedir() => "~")
return path
end
function find_source(file)
# Binary versions of Julia have the wrong stdlib path, fix it.
file = replace(string(file), normpath(Sys.BUILD_STDLIB_PATH) => Sys.STDLIB; count = 1)
return Base.find_source_file(file)
end
#
# Explorer.
#
struct CapturedError
err::Any
bt::Any
end
Base.show(io::IO, ce::CapturedError) = showerror(io, ce.err, ce.bt)
explore(err::CapturedError) = explore(stdout, err)
function explore(io::IO, err::CapturedError; interactive = true)
# Give a printout of the actual error message prior to launching tree
# explorer since it's probably useful to have.
println(io, sprint(showerror, err.err, context = :color => true))
# Use the default cleaning functionality from Base. No need to reinvent.
clean = Base.process_backtrace(err.bt)
wrapped = StackFrameWrapper.(clean)
toplevel = findfirst(s -> StackTraces.is_top_level_frame(s.sf), wrapped)
toplevel = toplevel === nothing ? length(wrapped) : toplevel
user_frames = wrapped[1:toplevel]
system_frames = wrapped[toplevel+1:end]
root = Node{Any}("(stacktrace)")
function make_nodes(root_node, frames; fold = false)
for (nth, frame_group) in enumerate(aggregate_modules(frames))
m = module_of(first(frame_group))
if m === :unknown
for frame in frame_group
fold!(Node{Any}(frame, root_node))
end
else
name =
m === :inlined ? style("[inlined]", :inlined_frames) :
m === :toplevel ? style("[top-level]", :toplevel_frames) :
is_from_stdlib(m) ? style("$(m)", :stdlib_module) :
is_from_base(m) ? style("$(m)", :base_module) :
is_from_core(m) ? style("$(m)", :core_module) :
is_from_package(m) ? style("$(m)", :package_module) :
style("$(m)", :unknown_module)
node = Node{Any}(name, root_node)
for frame in frame_group
current = Node{Any}(frame, node)
fold!(current)
# Formatted signature for the frame:
if !StackTraces.is_top_level_frame(frame.sf)
let lines = _formatted_signature(frame)
if !isempty(lines)
sig = Node{Any}(style("signature", :signature), current)
fold!(sig)
for line in lines
Node{Any}(line, sig)
end
end
end
end
# Source code for the frame:
let lines = _lines_around(frame)
if !isempty(lines)
src = Node{Any}(style("source", :source), current)
for line in lines
Node{Any}(line, src)
end
end
end
end
# Hide any of the following by default:
if m in (:inlined, :toplevel) ||
is_from_stdlib(m) ||
is_from_base(m) ||
is_from_core(m) ||
fold
fold!(node)
end
# Always open up the very first node, unless it's a toplevel.
if nth === 1 && m !== :toplevel
unfold!(node)
end
end
end
end
user_nodes = Node{Any}(style("(user)", :user_stack), root)
make_nodes(user_nodes, user_frames)
system_nodes = Node{Any}(style("(system)", :system_stack), root)
make_nodes(system_nodes, system_frames; fold = true)
fold!(system_nodes)
menu = TreeMenu(root; dynamic = true, maxsize = 30)
result = interactive ? TerminalMenus.request(menu; cursor = 3) : user_nodes
result === nothing && return
actions = [
"clipboard" =>
() -> (maybe_clipboard(sprint(showerror, err.err, err.bt[1:toplevel])); nothing),
"print" => () -> (showerror(io, err.err, err.bt[1:toplevel]); nothing),
"stacktrace" => () -> clean,
"exception" => () -> err.err,
"backtrace" => () -> err.bt,
]
data = result.data
extras = []
if isa(data, StackFrameWrapper)
file, line = data.sf.file, data.sf.line
file = find_source(file)
if file !== nothing && isfile(file)
file, line
extras = ["edit" => () -> (edit(file, line); nothing), "retry" => () -> true]
has_debugger() && push!(extras, "breakpoint" => () -> breakpoint(file, line))
push!(extras, "less" => () -> (less(file, line); nothing))
actions = vcat(extras, actions)
end
if isdefined(data.sf, :linfo)
mi = data.sf.linfo
if isa(mi, Core.MethodInstance)
extras = []
if has_cthulhu()
push!(extras, "ascend" => () -> ascend(mi))
push!(extras, "descend" => () -> descend(mi))
end
if has_jet()
push!(extras, "JET" => () -> report_call(mi))
end
actions = vcat(extras, actions)
end
end
end
result =
interactive ?
request(MultiSelectMenu(first.(actions); charset = get_theme(:charset, :unicode))) :
collect(1:length(actions))
choice = sort(collect(result))
if !isempty(choice)
output = []
for (name, func) in actions[choice]
out = func()
out === nothing || push!(output, Symbol(name) => out)
end
isempty(output) || return NamedTuple{Tuple(first.(output))}(last.(output))
end
return nothing
end
function _lines_around(s::StackFrameWrapper)
file, line = s.sf.file, s.sf.line
file = find_source(file)
if file !== nothing && isfile(file)
lines = readlines(file)
range = get_theme(:line_range)
above = max(1, line - get(range, :before, 0))
below = min(line + get(range, :after, 5), length(lines))
highlighter =
get(get_theme(:source), :highlight, true) === true ? highlight :
s -> style(s, :file_contents)
return highlighter.(lines[above:below])
else
return String[]
end
end
function _formatted_signature(s::StackFrameWrapper)
str = String(rsplit(string(s.sf), " at "; limit = 2)[1])
str = replace(str, "#unused#" => "")
formatter = get(get_theme(:signature), :format, true) === true ? format_julia_source : identity
highlighter =
get(get_theme(:signature), :highlight, true) === true ? highlight :
s -> style(s, :file_contents)
fmt = highlighter(formatter(str))
return collect(eachline(IOBuffer(fmt)))
end
# Just give up when there is no clipboard available.
function maybe_clipboard(str)
try
clipboard(str)
catch err
@warn "Could not find a clipboard."
end
end
rootmodule(m::Module) = m === Base ? m : m === parentmodule(m) ? m : rootmodule(parentmodule(m))
rootmodule(::Any) = nothing
modulepath(m::Module) = string(pkgdir(m))
modulepath(other) = ""
is_from_stdlib(m) = startswith(modulepath(rootmodule(m)), Sys.STDLIB)
is_from_base(m) = rootmodule(m) === Base
is_from_core(m) = rootmodule(m) === Core
is_from_package(m) = (r = rootmodule(m); !is_from_core(r) && !is_from_base(r) && !is_from_stdlib(r))
module_of(sf) =
sf.sf.inlined ? :inlined :
sf.sf.func === Symbol("top-level scope") ? :toplevel :
isa(sf.sf.linfo, Core.MethodInstance) ? sf.sf.linfo.def.module : :unknown
aggregate_modules(stacktrace) = IterTools.groupby(module_of, stacktrace)
#
# REPL hook.
#
const ENABLED = Ref(true)
"""
Turn interactive errors on or off.
"""
toggle() = ENABLED[] = !ENABLED[]
is_toggle_expr(expr) = Meta.isexpr(expr, :call, 1) && expr.args[1] === :toggle
is_retry(::Nothing) = false
is_retry(nt::NamedTuple) = haskey(nt, :retry) && nt.retry === true
maybe_retry(out, expr) = is_retry(out) ? Core.eval(Main, _ast_transforms(expr)) : out
function _ast_transforms(ast)
if isdefined(Base, :active_repl_backend)
for xf in Base.active_repl_backend.ast_transforms
ast = Base.invokelatest(xf, ast)
end
end
return ast
end
function wrap_errors(expr)
if ENABLED[] && !is_toggle_expr(expr)
quote
try
$(Expr(:toplevel, expr))
catch e
$(maybe_retry)(
$(explore)(($CapturedError)(e, catch_backtrace())),
$(Expr(:quote, expr)),
)
end
end
else
expr
end
end
function setup_repl()
# Skip REPL setup if we are precompiling. Avoids warnings in precompilation.
ccall(:jl_generating_output, Cint, ()) == 1 && return nothing
@async begin
done = false
for _ = 1:10
if isdefined(Base, :active_repl_backend)
backend = Base.active_repl_backend
if isdefined(backend, :ast_transforms)
pushfirst!(backend.ast_transforms, wrap_errors)
done = true
break
end
end
sleep(0.5)
end
done || @warn "Could not start `InteractiveErrors` REPL hook."
end
end
#
# Extensions, these get extended in the `/ext` modules.
#
has_cthulhu(args...) = false
ascend(args...) = @warn "`import Cthulhu` to enable `ascend` action."
descend(args...) = @warn "`import Cthulhu` to enable `descend` action."
has_debugger(args...) = false
breakpoint(args...) = @warn "`import Debugger` to enable `breakpoint` action."
has_jet(args...) = false
report_call(args...) = @warn "`import JET` to enable `report_call` action."
has_juliaformatter(args...) = false
format_julia_source(source) = source
has_ohmyrepl(args...) = false
highlight(source) = style(source, :file_contents)
#
# Module Initialisation.
#
function __init__()
setup_repl()
PackageExtensionCompat.@require_extensions
end
PrecompileTools.@compile_workload begin
try
div(1, 0)
catch error
explore(IOBuffer(), CapturedError(error, catch_backtrace()); interactive = false)
end
end
end # module
| InteractiveErrors | https://github.com/MichaelHatherly/InteractiveErrors.jl.git |
|
[
"MIT"
] | 1.0.1 | 5ed2f06644b4cdc96f25b2f8527cebde1ec77506 | code | 3057 | using Test, InteractiveErrors
@testset "InteractiveErrors" begin
IE = InteractiveErrors
@test IE.rootmodule(Main) === Main
@test IE.rootmodule(Base) === Base
@test IE.rootmodule(Core) === Core
@test IE.rootmodule(InteractiveErrors) === InteractiveErrors
@test IE.rootmodule(Base.Math) === Base
@test IE.is_from_stdlib(Test)
@test !IE.is_from_base(Test)
@test IE.is_from_base(Base)
@test !IE.is_from_package(Base)
@test IE.is_from_package(InteractiveErrors)
@test !IE.is_from_core(InteractiveErrors)
@test IE.is_from_core(Core)
build = joinpath(normpath(Sys.BUILD_STDLIB_PATH), "Test", "src", "Test.jl")
stdlib = joinpath(normpath(Sys.STDLIB), "Test", "src", "Test.jl")
package = @__FILE__
@test isfile(InteractiveErrors.find_source(build))
@test isfile(InteractiveErrors.find_source(stdlib))
@test isfile(InteractiveErrors.find_source(package))
@test startswith(InteractiveErrors.rewrite_path(build), "@stdlib")
@test startswith(InteractiveErrors.rewrite_path(stdlib), "@stdlib")
@test isa(IE.wrap_errors(:(1 + 1)), Expr)
@test isa(IE.wrap_errors(:(toggle())), Expr)
toggle()
@test isa(IE.wrap_errors(:(1 + 1)), Expr)
@test isa(IE.wrap_errors(:(toggle())), Expr)
toggle()
@test IE.style("func", :function_name) == "\e[0m\e[1mfunc\e[22m"
@test isa(IE.adjust_theme!(function_name = (color = :yellow, bold = false)), NamedTuple)
@test IE.style("func", :function_name) == "\e[33mfunc\e[39m"
@test isa(IE.current_theme(), NamedTuple)
@test IE.maybe_retry((; retry = true), :(true))
@test IE.maybe_retry((; retry = false), :(true)) == (; retry = false)
try
div(1, 0)
catch err
ce = IE.CapturedError(err, catch_backtrace())
io = IOBuffer()
nt = IE.explore(io, ce; interactive = false)
str = String(take!(io))
@test !isempty(str)
@test contains(str, "DivideError:")
@test isa(nt, NamedTuple)
@test collect(keys(nt)) == [:stacktrace, :exception, :backtrace]
@test isa(nt.exception, DivideError)
@test !isempty(nt.stacktrace)
@test !isempty(nt.backtrace)
end
@test !IE.has_cthulhu()
@test !IE.has_debugger()
@test !IE.has_jet()
@test !IE.has_ohmyrepl()
@test !IE.has_juliaformatter()
using Cthulhu, Debugger, JET, OhMyREPL, JuliaFormatter
@test IE.has_cthulhu()
@test IE.has_debugger()
@test IE.has_jet()
@test IE.has_ohmyrepl()
@test IE.has_juliaformatter()
try
sqrt(-1)
catch err
ce = IE.CapturedError(err, catch_backtrace())
io = IOBuffer()
nt = IE.explore(io, ce; interactive = false)
str = String(take!(io))
@test !isempty(str)
@test contains(str, "DomainError")
@test isa(nt, NamedTuple)
@test collect(keys(nt)) == [:stacktrace, :exception, :backtrace]
@test isa(nt.exception, DomainError)
@test !isempty(nt.stacktrace)
@test !isempty(nt.backtrace)
end
end
| InteractiveErrors | https://github.com/MichaelHatherly/InteractiveErrors.jl.git |
|
[
"MIT"
] | 1.0.1 | 5ed2f06644b4cdc96f25b2f8527cebde1ec77506 | docs | 5229 | # InteractiveErrors.jl
Interactive error messages for the Julia REPL.

## Installation
Requires Julia `1.6+`.
```
julia> using Pkg
julia> Pkg.add("InteractiveErrors")
```
Add `using InteractiveErrors` to your `startup.jl` file after `using Revise`.
If you don't have that installed yet you should install it.
## Usage
Just start using your REPL normally. Once you hit an error you'll be presented
with an interactive tree representing your stacktrace which you can explore. To
turn interactive errors off and return to using normal stacktraces call
`toggle()`. Call `toggle()` again to turn it back on.
Press `up` and `down` arrows to move through the stacktrace. Press `space` to
fold or unfold the currently selected line. A `+` will appear on folded lines.
Press `enter` once finished. If you are on a line that references a particular
file then that will present additional options in the next menu. `q` can be
pressed to exit back to the REPL.
**Note:** a lot of information is hidden inside some of the folded lines and
some is completely stripped from the display (such as method arguments). The
default choice of information to display is up for discussion. Unfolding a
line containing a file and line number will display the immediate lines
surrounding it.
The second menu offers several actions that can be taken on the selected line.
```
[press: d=done, a=all, n=none]
• [ ] ascend
[ ] descend
[ ] JET
[ ] edit
[ ] retry
[ ] breakpoint
[ ] less
[ ] clipboard
[ ] print
[ ] stacktrace
[ ] exception
[ ] backtrace
```
Press `enter` to choose the currently selected line. More than one can be chosen:
- `ascend` (available if `Cthulhu` is loaded) calls `Cthulhu.ascend` on the selected method.
- `descend` (available if `Cthulhu` is loaded) calls `Cthulhu.descend` on the selected method.
- `JET` (available if `JET` is loaded) calls `JET.report_call` on the selected method.
- `edit` opens default editor on the selected file and line.
- `retry` runs the code entered in the REPL again.
- `breakpoint` (available if `Debugger` is loaded) sets a `Debugger.breakpoint` on the selected file and line.
- `less` opens the pager on the selected file and line.
- `clipboard` copies the normal Julia stacktrace to the clipboard. Useful for
posting bug reports. Don't send the interactive printout as an error
message when reporting issues to packages or Julia.
- `print` prints out the normal Julia stacktrace to `stdout`.
- `stacktrace` returns the stacktrace object.
- `exception` returns the exception object that was caught.
- `backtrace` returns the *raw* backtrace object. Contains `Ptr`s. Not
terribly useful.
More than one action can be selected at once. A common combination is `edit`
and `retry`. Press `d` (for done) once you're finished making your choices.
## Optional Packages
Additional features are available when certain packages are loaded within the REPL session.
- `Cthulhu`, provides `ascend` and `descend` options on selected method.
- `Debugger`, provides `breakpoint` on selected method.
- `JET`, provides `report_call` on selected method.
- `JuliaFormatter`, provides syntax formatting for method signatures.
- `OhMyREPL`, provides syntax highlighting for source code and method signatures.
Depending on the maintenance burden of compatibility with these packages they
may in future versions become direct dependencies instead of using `Requires`.
## Themes
Most of the default coloring in the stack-tree can be adjusted to the user's
liking via a simple theming system.
- `current_theme()` returns the currently active theme: a nested `NamedTuple`
of customisation options.
- `set_theme!` can be used to set your own custom theme that follows the same
naming scheme as the default theme. Takes either keyword arguments, or a
`NamedTuple`.
- `reset_theme!` will reset the theme.
- `adjust_theme!` can be used if you only want to make some minor adjustments
to the `current_theme`. Takes a `NamedTuple` or keyword arguments that will
be `merge`d with the `current_theme`.
The default theme is shown below:
```
pairs(::NamedTuple) with 19 entries:
:function_name => (bold = true,)
:directory => (color = :light_black,)
:filename => (color = :magenta, bold = true)
:line_number => (color = :green, bold = true)
:user_stack => (color = :green, bold = true)
:system_stack => (color = :red, bold = true)
:stdlib_module => (color = :yellow,)
:base_module => (color = :blue,)
:core_module => (color = :light_black,)
:package_module => (color = :cyan, bold = true)
:unknown_module => (color = :red,)
:inlined_frames => (color = :light_black,)
:toplevel_frames => (color = :light_black,)
:repeated_frames => (color = :red,)
:file_contents => (color = :light_black,)
:signature => (color = :light_black, format = true, highlight = true)
:source => (color = :normal, bold = true, highlight = true)
:line_range => (before = 0, after = 5)
:charset => :unicode
```
| InteractiveErrors | https://github.com/MichaelHatherly/InteractiveErrors.jl.git |
|
[
"MIT"
] | 0.2.2 | e2e045a2aba9da8fadb5b14bf28ae96f58d0883e | code | 459 | using Documenter, DailyTreasuryYieldCurve
makedocs(
modules = [DailyTreasuryYieldCurve],
format = Documenter.HTML(; prettyurls = get(ENV, "CI", nothing) == "true"),
authors = "Tyler Beason",
sitename = "DailyTreasuryYieldCurve.jl",
pages = Any["Main"=>"index.md"]
# strict = true,
# clean = true,
# checkdocs = :exports,
)
deploydocs(
repo = "github.com/tbeason/DailyTreasuryYieldCurve.jl.git",
push_preview = true
)
| DailyTreasuryYieldCurve | https://github.com/tbeason/DailyTreasuryYieldCurve.jl.git |
|
[
"MIT"
] | 0.2.2 | e2e045a2aba9da8fadb5b14bf28ae96f58d0883e | code | 4087 | module DailyTreasuryYieldCurve
using Reexport
using EzXML
import HTTP
@reexport using Dates
using Missings
@reexport using DataFrames
using Interpolations
export getyieldcurves
export RateInterpolator, createRateInterpolator
const DATAFEED = "https://data.treasury.gov/feed.svc/DailyTreasuryYieldCurveRateData"
const DATAFEEDREAL = "http://data.treasury.gov/feed.svc/DailyTreasuryRealYieldCurveRateData"
# these fields are provided by the feed (as of 20200413)
# note that we must use :m1 because :1m is not a valid symbol
const COLNAMES = [:id,:date,:m1,:m2,:m3,:m6,:y1,:y2,:y3,:y5,:y7,:y10,:y20,:y30,:y30dup]
const COLNAMESREAL = [:id,:date,:y5,:y7,:y10,:y20,:y30]
"""
getyieldcurves(;real::Bool=false,begdt::Date=Date(1990,1,2),enddt::Date=today())
Download the whole published history of daily US Treasury yield curves from the official data feed.
Optionally, pass a filename if you have already downloaded the data, eg. `getyieldcurves(fn::AbstractString)`.
By default, gets the nominal yield curve. Pass `realrates=true` to get the real yield curve. The nominal series starts in 1990, while the real series starts in 2003.
Returns a `DataFrame`.
"""
function getyieldcurves(;realrates::Bool=false,begdt::Date=Date(1990,1,2),enddt::Date=today())
startyr=max(year(begdt),1990)-1
endyr=min(year(enddt),year(today()))+1
datefilter = string("?\$filter=","year(NEW_DATE)%20gt%20",startyr,"%20and%20","year(NEW_DATE)%20lt%20",endyr)
if realrates
h = HTTP.get(string(DATAFEEDREAL,datefilter))
else
h = HTTP.get(string(DATAFEED,datefilter))
end
thexml = parsexml(String(h.body))
df =_parseyieldcurves(thexml,realrates)
filter!(row -> begdt <= row.date <= enddt,df)
return df
end
function getyieldcurves(fn::AbstractString;realrates::Bool=false,begdt::Date=Date(1990,1,2),enddt::Date=today())
thexml = readxml(fn)
df =_parseyieldcurves(thexml,realrates)
filter!(row -> begdt <= row.date <= enddt,df)
return df
end
"""
_parseyieldcurves(thexml,realrates)
Parser function for Treasury yield curve data. (unexported)
"""
function _parseyieldcurves(thexml,realrates)
if realrates
return _parserealcurves(thexml)
else
return _parsenominalcurves(thexml)
end
end
function _parsenominalcurves(thexml)
r = thexml.root
cnod=findall("/*/*/*[position()=7]",r)
@assert length(cnod) >= 1 "Rate XML parse error likely."
stripsplitstrip(s) = strip.(split(strip(s),"\n"))
strarr = map(stripsplitstrip,nodecontent.(cnod))
df=DataFrame(Tuple.(strarr))
@assert nrow(df) >= 1 "Rate XML parse error likely."
rename!(df,COLNAMES)
select!(df,Not([:id; :y30dup]))
for c in COLNAMES[2:end-1]
if c == :date
df[!,c] = Date.(parse.(DateTime,df[!,c]))
else
df[!,c] = passmissing(x->parse(Float64,x)).(replace(df[!,c],""=>missing))
end
end
sort!(df,:date)
return df
end
function _parserealcurves(thexml)
r = thexml.root
cnod=findall("/*/*/*[position()=7]",r)
@assert length(cnod) >= 1 "Rate XML parse error likely."
nodestr = nodecontent.(cnod)
function splitandfill(s)
spl = split(s)
L = length(spl)
L == length(COLNAMESREAL) && return spl
strvec = Vector{eltype(spl)}(undef,length(COLNAMESREAL))
for i in 1:length(COLNAMESREAL)
if i <= L
strvec[i] = spl[i]
else
strvec[i] = ""
end
end
return strvec
end
strarr = splitandfill.(nodestr)
df=DataFrame(Tuple.(strarr))
@assert nrow(df) >= 1 "Rate XML parse error likely."
rename!(df,COLNAMESREAL)
select!(df,Not(:id))
for c in COLNAMESREAL[2:end]
if c == :date
df[!,c] = Date.(parse.(DateTime,df[!,c]))
else
df[!,c] = passmissing(x->parse(Float64,x)).(replace(df[!,c],""=>missing))
end
end
sort!(df,:date)
return df
end
# include additional files
include("interp.jl")
end # module
| DailyTreasuryYieldCurve | https://github.com/tbeason/DailyTreasuryYieldCurve.jl.git |
|
[
"MIT"
] | 0.2.2 | e2e045a2aba9da8fadb5b14bf28ae96f58d0883e | code | 2730 |
# Treasury uses Act/Act (ICMA) day count convention
# unclear how this translates to constant maturities
# I use months = 30 days, years = 365 days
const DAYSTOMATURITY = [30,60,90,180,365,730,1095,1825,2555,3650,7300,10950]
const DAYSTOMATURITYREAL = [1825,2555,3650,7300,10950]
const TTM = (;Iterators.zip(DailyTreasuryYieldCurve.COLNAMES[3:end-1],DAYSTOMATURITY)...)
const TTMREAL = (;Iterators.zip(DailyTreasuryYieldCurve.COLNAMESREAL[3:end],DAYSTOMATURITYREAL)...)
"""
RateInterpolator
A container for the series of daily yield curve interpolators.
Fields: `dates` and `interpolators`, both `Vector`.
To interpolate, just call it with the value to interpolate (days to maturity) and the date of the curve.
```
(ri::RateInterpolator)(d::Real,dt::Date)
```
Currently supports only linear interpolation and extrapolation. See [`createRateInterpolator`](@ref) for construction.
"""
struct RateInterpolator{T}
dates::Vector{Date}
interpolators::Vector{T}
realrates::Bool
end
function (ri::RateInterpolator)(d::Real,dt::Date)
idx = searchsortedlast(ri.dates,dt)
return ri.interpolators[idx](d)
end
"""
createRateInterpolator(df;realrates::Bool=false)
Preferred method to construct a [`RateInterpolator`](@ref), just pass the `df` that you get from [`getyieldcurves`](@ref).
"""
function createRateInterpolator(df0;realrates::Bool=false)
loc = [!all(ismissing(x) for x in r) for r in eachrow(select(df0,Not(:date)))] # 0 for rows with only missing data
df = df0[loc,:]
dates = unique(df.date)
dfs = DataFrames.stack(df,Not(:date); variable_eltype=Symbol)
if realrates
dfs.DTM = [TTMREAL[k] for k in dfs.variable]
else
dfs.DTM = [TTM[k] for k in dfs.variable]
end
gd = groupby(dfs,:date)
rateinterps = [buildsingleinterpolator(g) for g in gd]
@assert length(dates) == length(rateinterps) "Number of dates is different than number of daily interpolators."
return RateInterpolator(dates,rateinterps,realrates)
end
"""
buildsingleinterpolator(df::AbstractDataFrame)
Builds a single (one day) interpolation of the yield curve. Used in `createRateInterpolator`.
"""
function buildsingleinterpolator(df::AbstractDataFrame)
dfdm = dropmissing(df)
x = dfdm.DTM
y = dfdm.value
itp = LinearInterpolation(x,y,extrapolation_bc=Line())
return itp
end
function Base.show(io::IO, ri::RateInterpolator)
itpT = Interpolations.itptype(first(ri.interpolators))
begdt = first(ri.dates)
enddt = last(ri.dates)
ndays = length(ri.dates)
kind = ri.realrates ? "Real" : "Nominal"
print(io,"$kind RateInterpolator{$itpT interpolators} starting $begdt and ending $enddt ($ndays days)")
end
| DailyTreasuryYieldCurve | https://github.com/tbeason/DailyTreasuryYieldCurve.jl.git |
|
[
"MIT"
] | 0.2.2 | e2e045a2aba9da8fadb5b14bf28ae96f58d0883e | code | 999 | using DailyTreasuryYieldCurve, Dates
using Test
testdf = getyieldcurves()
@testset "Nominal" begin
@test Symbol.(names(testdf)) == DailyTreasuryYieldCurve.COLNAMES[2:end-1]
@test testdf[1,:date] == Date(1990,1,2)
@test isapprox(testdf[1,:y30],8.0)
end
@testset "Real" begin
testdfr = getyieldcurves(;realrates=true)
@test Symbol.(names(testdfr)) == DailyTreasuryYieldCurve.COLNAMESREAL[2:end]
@test testdfr[1,:date] == Date(2003,1,2)
@test isapprox(testdfr[1,:y5],1.752231)
end
@testset "Interpolation" begin
# testdf = getyieldcurves()
testri = createRateInterpolator(testdf)
@test isapprox(testri(45,Date(2020,4,13)),0.23)
@test isapprox(testri.([30;60],Date(2020,4,13)),[0.17;0.29])
end
@testset "Filters" begin
# testdf = getyieldcurves()
smalltestdf = getyieldcurves(;begdt=Date(2019,1,1),enddt=Date(2019,12,31))
filter!(row->Date(2019,1,1) <= row.date <= Date(2019,12,31),testdf)
@test testdf == smalltestdf
end
| DailyTreasuryYieldCurve | https://github.com/tbeason/DailyTreasuryYieldCurve.jl.git |
|
[
"MIT"
] | 0.2.2 | e2e045a2aba9da8fadb5b14bf28ae96f58d0883e | code | 363 | ####
#### Coverage summary, printed as "(percentage) covered".
####
#### Useful for CI environments that just want a summary (eg a Gitlab setup).
####
using Coverage
cd(joinpath(@__DIR__, "..", "..")) do
covered_lines, total_lines = get_summary(process_folder())
percentage = covered_lines / total_lines * 100
println("($(percentage)%) covered")
end
| DailyTreasuryYieldCurve | https://github.com/tbeason/DailyTreasuryYieldCurve.jl.git |
|
[
"MIT"
] | 0.2.2 | e2e045a2aba9da8fadb5b14bf28ae96f58d0883e | code | 266 | # only push coverage from one bot
get(ENV, "TRAVIS_OS_NAME", nothing) == "linux" || exit(0)
get(ENV, "TRAVIS_JULIA_VERSION", nothing) == "1.3" || exit(0)
using Coverage
cd(joinpath(@__DIR__, "..", "..")) do
Codecov.submit(Codecov.process_folder())
end
| DailyTreasuryYieldCurve | https://github.com/tbeason/DailyTreasuryYieldCurve.jl.git |
|
[
"MIT"
] | 0.2.2 | e2e045a2aba9da8fadb5b14bf28ae96f58d0883e | docs | 2164 | # DailyTreasuryYieldCurve.jl
<!--




 -->
[](https://travis-ci.com/tbeason/DailyTreasuryYieldCurve.jl)
[](http://codecov.io/github/tbeason/DailyTreasuryYieldCurve.jl?branch=master)
[](https://tbeason.github.io/DailyTreasuryYieldCurve.jl/stable)
<!---[](https://tbeason.github.io/DailyTreasuryYieldCurve.jl/dev)-->
This Julia package does one thing: gets you daily yield curves from the [US Treasury](https://www.treasury.gov/resource-center/data-chart-center/interest-rates/Pages/TextView.aspx?data=yield). The data is served via an XML feed, but this package cleans it up into a `DataFrame` so that you can use it.
# Example
Add the package via the Julia Package Manager.
```julia
] add DailyTreasuryYieldCurve
```
It is easy to get the historical yield curves:
```julia
using DailyTreasuryYieldCurve
df_rates = getyieldcurves()
df_realrates = getyieldcurves(;realrates=true)
```
You can also build a `RateInterpolator` which helps you interpolate/extrapolate using the data:
```julia
using Dates
nominal_itp = createRateInterpolator(df_rates)
nominal_itp(45,Date(2020,4,13)) # gets the 45 day rate on 2020-4-13
```
For more information check the [documentation](https://tbeason.github.io/DailyTreasuryYieldCurve.jl/stable).
## Disclaimer
This package is provided as-is and without guarantees. I am not affiliated with the US Treasury. Please cite the original source when using this data.
| DailyTreasuryYieldCurve | https://github.com/tbeason/DailyTreasuryYieldCurve.jl.git |
|
[
"MIT"
] | 0.2.2 | e2e045a2aba9da8fadb5b14bf28ae96f58d0883e | docs | 2271 | # DailyTreasuryYieldCurve
[DailyTreasuryYieldCurve.jl](https://github.com/tbeason/DailyTreasuryYieldCurve.jl) is a Julia package for downloading and working with historical daily yield curve data from the [US Treasury](https://www.treasury.gov/resource-center/data-chart-center/interest-rates/Pages/TextView.aspx?data=yield).
## Getting Daily Yield Curves
```@docs
getyieldcurves
```
Structure of returned data for *nominal* curve:
| Column Name | Description |
| ---- | ---- |
| date | Date of yield curve |
| m1 | 1 month constant maturity rate |
| m2 | 2 month constant maturity rate |
| m3 | 3 month constant maturity rate |
| m6 | 6 month constant maturity rate |
| y1 | 1 year constant maturity rate |
| y2 | 2 year constant maturity rate |
| y3 | 3 year constant maturity rate |
| y5 | 5 year constant maturity rate |
| y7 | 7 year constant maturity rate |
| y10 | 10 year constant maturity rate |
| y20 | 20 year constant maturity rate |
| y30 | 30 year constant maturity rate |
Structure of returned data for *real* curve:
| Column Name | Description |
| ---- | ---- |
| date | Date of yield curve |
| y5 | 5 year constant maturity real rate |
| y7 | 7 year constant maturity real rate |
| y10 | 10 year constant maturity real rate |
| y20 | 20 year constant maturity real rate |
| y30 | 30 year constant maturity real rate |
Not all maturities were reported on every day.
## Interpolation
The package contains some convenience utilities for interpolating/extrapolating with the yield curve data.
```@docs
RateInterpolator
createRateInterpolator
```
!!! warning "Last curve carried forward"
If you request a date that does not exist in `RateInterpolator.dates`, it will (blindly) carry forward the curve from the previous available date. It is your responsibility to double-check your inputs.
## Day Count Convention
The standard day count convention for valuing US Treasuries is [Actual/Actual (ICMA)](https://en.wikipedia.org/wiki/Day_count_convention#Actual/Actual_ICMA) (as opposed to something like 30/360). However, it is unclear (to me) exactly how to match this to constant maturities. Therefore, I use the convention that months are 30 days (for maturities less than 1 year) and years are 365 days (for maturities 1 year or more). | DailyTreasuryYieldCurve | https://github.com/tbeason/DailyTreasuryYieldCurve.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 765 | using BioMASS
using Documenter
makedocs(;
modules=[BioMASS],
authors="Hiroaki Imoto <[email protected]>",
repo="https://github.com/biomass-dev/BioMASS.jl/blob/{commit}{path}#L{line}",
sitename="BioMASS.jl",
format=Documenter.HTML(;
prettyurls=get(ENV, "CI", "false") == "true",
canonical="https://biomass-dev.github.io/BioMASS.jl",
assets=String[],
),
pages=[
"Home" => "index.md",
"Getting started with BioMASS.jl" => [
"Parameter Estimation" => "usage/parameter_estimation.md",
"Bifurcation Analysis" => "usage/bifurcation_analysis.md",
],
"References" => "references.md"
],
)
deploydocs(;
repo="github.com/biomass-dev/BioMASS.jl",
)
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 2596 | """
Barr, A. R., Heldt, F. S., Zhang, T., Bakal, C. & Novák, B. A Dynamical
Framework for the All-or-None G1/S Transition. Cell Syst. 2, 27–37 (2016).
https://doi.org/10.1016/j.cels.2016.01.001
"""
include("./name2idx/parameters.jl")
include("./name2idx/species.jl")
include("./set_model.jl")
include("./forwarddiff.jl")
const BP = C.CycElevel # name(index) of bifurcation parameter
const SN = V.NUM # num of state variables
const PN = 1 # num of parameters
const VN = SN + PN # num of variables
function calc_fixed_point_vec(model_path::String)::Tuple{Array,Array}
fp::Array = []
ev::Array = []
br::Array = []
for i in 1:6
global p = param_values()
if i == 1
p[C.Emi1T] = 0.0
elseif i == 2
p[C.Emi1T] = 0.5
elseif i == 3
p[C.Emi1T] = 0.75
elseif i == 4
p[C.Emi1T] = 1.0
elseif i == 5
p[C.Emi1T] = 1.25
elseif i == 6
p[C.Emi1T] = 2.0
end
new_curve!(
model_path, p, diffeq, get_derivatives, get_steady_state,
direction=false, bifparam=BP, n_state=SN
)
push!(fp, readdlm(joinpath(model_path, "data", "fp.dat"), '\t', Float64, '\n'))
push!(ev, readdlm(joinpath(model_path, "data", "ev.dat"), '\t', Float64, '\n'))
push!(br, get_bistable_regime(ev[i], SN))
end
return fp, br
end
function bifurcation_diagram(model_path::String, fp::Array, br::Array)
rc("figure", figsize=(9, 6))
rc("font", family="Arial")
rc("font", size=20)
rc("axes", linewidth=1.2)
rc("xtick.major", width=1.2)
rc("ytick.major", width=1.2)
rc("lines", linewidth=2)
for (i, (fixed_point, unstable_ss)) in enumerate(zip(fp, br))
if i == 1
color = "red"
else
color = "silver"
end
plot(
fixed_point[1:unstable_ss[1]-1, VN+1],
fixed_point[1:unstable_ss[1]-1, V.p27T+1],
"-", color=color
)
plot(
fixed_point[unstable_ss, VN+1],
fixed_point[unstable_ss, V.p27T+1],
"--", color=color
)
plot(
fixed_point[unstable_ss[end]+1:end, VN+1],
fixed_point[unstable_ss[end]+1:end, V.p27T+1],
"-", color=color
)
end
xlabel("CycE level")
ylabel("p27 level")
xlim(0.0, 1.0)
xticks([0, 0.5, 1])
ylim(0.0, 2.05)
yticks([0, 1, 2])
savefig(joinpath(model_path, "bifurcation_diagram.pdf"), bbox_inches="tight")
close()
end
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 2551 | function diffeq!(du,u,p,t)
CycE = p[C.CycElevel] - u[V.CycEp27]
Vdp27 = p[C.kd27] + (p[C.kd27e]*CycE)*u[V.Skp2]
Vdcyce = p[C.kdcyce] + p[C.kdcycee]*CycE
Vdskp2 = p[C.kdskp2] + p[C.kdskp2c1]*u[V.Cdh1]
Vicdh1 = p[C.kicdh1e]*CycE
du[V.p27T] = p[C.ks27] - Vdp27*u[V.p27T]
du[V.Skp2] = p[C.ksskp2] - Vdskp2*u[V.Skp2]
du[V.CycEp27] = p[C.kasse]*(p[C.CycElevel]-u[V.CycEp27])*(u[V.p27T]-u[V.CycEp27])-(p[C.kdise]+Vdp27+Vdcyce)*u[V.CycEp27]
du[V.EmiC] = p[C.kasec]*(p[C.Cdh1T]-u[V.EmiC])*(p[C.Emi1T]-u[V.EmiC]) - (p[C.kdiec]+p[C.kdemi1])*u[V.EmiC]
du[V.Cdh1dp] = p[C.kacdh1]*(p[C.Cdh1T]-u[V.Cdh1dp]) - Vicdh1*u[V.Cdh1dp]
du[V.Cdh1] = (p[C.kdiec]+p[C.kdemi1])*(u[V.Cdh1dp]-u[V.Cdh1]) - p[C.kasec]*u[V.Cdh1]*(p[C.Emi1T]-u[V.EmiC])+p[C.kacdh1]*(p[C.Cdh1T]-u[V.EmiC]-u[V.Cdh1])-Vicdh1*u[V.Cdh1]
end
function param_values()::Vector{Float64}
p::Vector{Float64} = zeros(C.NUM)
p[C.kscyce] = 0.003
p[C.kdcyce] = 0.001
p[C.kdcycee] = 0.0001
p[C.kdcycea] = 0.03
p[C.kasse] = 1
p[C.kdise] = 0.02
## CYCA SYNTHESISp[C.DEGRADATION AND P27 BINDING/DISSOCIATION:
p[C.kscyca] = 0.0025
p[C.kdcyca] = 0.002
p[C.kdcycac1] = 0.4
p[C.kassa] = 1
p[C.kdisa] = 0.02
## P27 SYNTHESIS AND DEGRADATION:
p[C.ks27] = 0.008
p[C.kd27] = 0.004
p[C.kd27e] = 2
p[C.kd27a] = 2
## EMI1 SYNTHESIS AND DEGRADATION:
p[C.ksemi1] = 0.003
p[C.kdemi1] = 0.001
## CDH1 REGULATION:
p[C.Cdh1T] = 1
p[C.kacdh1] = 0.02
p[C.kicdh1e] = 0.07
p[C.kicdh1a] = 0.2
p[C.kasec] = 2
p[C.kdiec] = 0.02
## SKP2 SYNTHESIS AND DEGRADATION:
p[C.ksskp2] = 0.004
p[C.kdskp2] = 0.002
p[C.kdskp2c1] = 0.2
## CDK INHIBITOR
p[C.Inhibitor] = 0.0
p[C.Emi1T] = 0.0
p[C.CycElevel] = 1.0
return p
end
function get_derivatives(u::Vector{Float64},p::Vector{Float64})
# derivatives: dF/d[bifurcation_param]
dFdp::Vector{Float64} = zeros(V.NUM)
dFdp[V.p27T] = -p[C.kd27e]*u[V.Skp2]*u[V.p27T]
dFdp[V.CycEp27] = p[C.kasse]*(1.0-u[V.CycEp27])*(u[V.p27T]-u[V.CycEp27])-(p[C.kd27e]*u[V.Skp2]+p[C.kdcycee])*u[V.CycEp27]
dFdp[V.Cdh1dp] = -p[C.kicdh1e]*u[V.Cdh1dp]
dFdp[V.Cdh1] = -p[C.kicdh1e]*u[V.Cdh1]
return dFdp
end
function get_steady_state(p::Vector{Float64})
tspan::Tuple{Float64,Float64} = (0.0,Inf)
u0::Vector{Float64} = zeros(V.NUM)
prob = ODEProblem(diffeq!,u0,tspan,p)
prob = SteadyStateProblem(prob)
sol = solve(prob,DynamicSS(CVODE_BDF()),dt=1.0)
return sol.u
end | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 538 | module C
const NAMES = [
"kscyce"
"kdcyce"
"kdcycee"
"kdcycea"
"kasse"
"kdise"
"kscyca"
"kdcyca"
"kdcycac1"
"kassa"
"kdisa"
"ks27"
"kd27"
"kd27e"
"kd27a"
"ksemi1"
"kdemi1"
"Cdh1T"
"kacdh1"
"kicdh1e"
"kicdh1a"
"kasec"
"kdiec"
"ksskp2"
"kdskp2"
"kdskp2c1"
"Inhibitor"
"CycElevel"
"Emi1T"
]
#name2idx
for (idx,name) in enumerate(NAMES)
eval(Meta.parse("const $name = $idx"))
end
const NUM = length(NAMES)
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 232 | module V
const NAMES = [
"p27T"
"CycEp27"
"Cdh1dp"
"EmiC"
"Cdh1"
"Skp2"
]
#name2idx
for (idx,name) in enumerate(NAMES)
eval(Meta.parse("const $name = $idx"))
end
const NUM = length(NAMES)
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 5280 | """
Rata, S. et al. Two Interlinked Bistable Switches Govern Mitotic Control in
Mammalian Cells. Curr. Biol. 28, 3824-3832.e6 (2018).
https://doi.org/10.1016/j.cub.2018.09.059
"""
include("./name2idx/parameters.jl")
include("./name2idx/species.jl")
include("./set_model.jl")
include("./forwarddiff.jl")
const BP = C.InhCDK # name(index) of bifurcation parameter
const SN = V.NUM # num of state variables
const PN = 1 # num of parameters
const VN = SN + PN # num of variables
function calc_fixed_point_vec(model_path::String)::Tuple{Array,Array}
fp::Array = []
ev::Array = []
br::Array = []
for i in 1:4
global p = param_values()
# i==1 -> Control
if i == 2 # Wee1 inhibition
p[C.kweeS] = 0.0
p[C.kweeF] = 0.0
elseif i == 3 # Gwl siRNA
p[C.Gwtot] = 0.0
elseif i == 4 # Wee1 inhibition & Gwl siRNA
p[C.kweeS] = 0.0
p[C.kweeF] = 0.0
p[C.Gwtot] = 0.0
end
new_curve!(
model_path, p, diffeq, get_derivatives, get_steady_state,
direction=false, bifparam=BP, n_state=SN
)
push!(fp, readdlm(joinpath(model_path, "data", "fp.dat"), '\t', Float64, '\n'))
push!(ev, readdlm(joinpath(model_path, "data", "ev.dat"), '\t', Float64, '\n'))
push!(br, get_bistable_regime(ev[i], SN))
end
return fp, br
end
function bifurcation_diagram(model_path::String, fp::Array, br::Array)
rc("figure", figsize=(20, 3))
rc("font", family="Arial")
rc("font", size=14)
rc("axes", linewidth=1.2)
rc("xtick.major", width=1.2)
rc("ytick.major", width=1.2)
rc("lines", linewidth=2)
for (i, (fixed_point, unstable_ss)) in enumerate(zip(fp, br))
if length(unstable_ss) > 0
intermediate_ss = []
for j = 2:length(unstable_ss)
if unstable_ss[j] - unstable_ss[j-1] != 1
intermediate_ss = unstable_ss[j-1]:unstable_ss[j]
end
end
end
subplot(1, 4, i)
if i == 1
plot(
fixed_point[1:unstable_ss[1]-1, VN+1],
fixed_point[1:unstable_ss[1]-1, V.Subp+1],
color="royalblue"
)
plot(
fixed_point[unstable_ss[1]:intermediate_ss[1]-1, VN+1],
fixed_point[unstable_ss[1]:intermediate_ss[1]-1, V.Subp+1],
color="darkgray", "--"
)
plot(
fixed_point[intermediate_ss, VN+1],
fixed_point[intermediate_ss, V.Subp+1],
color="darkorange"
)
plot(
fixed_point[intermediate_ss[end]+1:unstable_ss[end], VN+1],
fixed_point[intermediate_ss[end]+1:unstable_ss[end], V.Subp+1],
color="darkgray", "--"
)
plot(
fixed_point[unstable_ss[end]+1:end, VN+1],
fixed_point[unstable_ss[end]+1:end, V.Subp+1],
color="crimson"
)
xlim(0, 0.81)
xlabel("1NMPP1 (μM)")
ylim(-0.05, 1.05)
yticks([0, 0.5, 1], [0, 50, 100])
ylabel("Sub-p (%)")
title("Control", fontsize=18)
elseif i == 2
plot(
fixed_point[1:unstable_ss[1]-1, VN+1],
fixed_point[1:unstable_ss[1]-1, V.Subp+1],
color="royalblue"
)
plot(
fixed_point[unstable_ss, VN+1],
fixed_point[unstable_ss, V.Subp+1],
color="darkgray", "--"
)
plot(
fixed_point[unstable_ss[end]+1:end, VN+1],
fixed_point[unstable_ss[end]+1:end, V.Subp+1],
color="crimson"
)
xlim(0, 0.81)
xlabel("1NMPP1 (μM)")
ylim(-0.05, 1.05)
yticks([0, 0.5, 1], [0, 50, 100])
title("Wee1 inhibition", fontsize=18)
elseif i == 3
plot(
fixed_point[1:unstable_ss[1]-1, VN+1],
fixed_point[1:unstable_ss[1]-1, V.Subp+1],
color="royalblue"
)
plot(
fixed_point[unstable_ss, VN+1],
fixed_point[unstable_ss, V.Subp+1],
color="darkgray", "--"
)
plot(
fixed_point[unstable_ss[end]+1:end, VN+1],
fixed_point[unstable_ss[end]+1:end, V.Subp+1],
color="crimson"
)
xlim(0, 0.81)
xlabel("1NMPP1 (μM)")
ylim(-0.05, 1.05)
yticks([0, 0.5, 1], [0, 50, 100])
title("Gwl siRNA", fontsize=18)
elseif i == 4
plot(
fixed_point[:, VN+1],
fixed_point[:, V.Subp+1],
color="darkorange"
)
xlim(0, 0.81)
xlabel("1NMPP1 (μM)")
ylim(-0.05, 1.05)
yticks([0, 0.5, 1], [0, 50, 100])
title("Wee1 inhibition & Gwl siRNA", fontsize=18)
end
end
savefig(joinpath(model_path, "bifurcation_diagram.pdf"), bbox_inches="tight")
close()
end
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 4106 | function diffeq!(du,u,p,t)
Complex = p[C.B55tot] - u[V.PP2AB55]
Wee1p = 1 - u[V.Wee1] - u[V.Wee1pp]
Cdc25p = 1 - u[V.Cdc25] - u[V.Cdc25pp]
Vwee = (p[C.kweeS]*(1-u[V.Wee1]) + p[C.kweeF]*u[V.Wee1])
V25 = p[C.k25S]*(1-u[V.Cdc25pp]) + p[C.k25F]*u[V.Cdc25pp]
VGwl = p[C.kGwENSA]*u[V.Gwlp]
du[V.Subp] = p[C.kcBc1Sub]*u[V.CycBCdk1]/((1 + (p[C.InhCDK]/p[C.Kd])))*(p[C.SubT]-u[V.Subp]) - p[C.kB55Sub]*u[V.PP2AB55]*u[V.Subp]
du[V.CycBCdk1] = V25*(p[C.CycBCdk1T] - u[V.CycBCdk1]) - Vwee*u[V.CycBCdk1]
du[V.PP1] = (p[C.kapp1] + p[C.kapp1a]*u[V.PP1])*(p[C.PP1T] - u[V.PP1]) - (p[C.kipp1] + p[C.kipp1C]*u[V.CycBCdk1]/((1 + (p[C.InhCDK]/p[C.Kd]))))*u[V.PP1]
du[V.pENSAt] = VGwl*(p[C.ENSAtot] - u[V.pENSAt]) - p[C.kcatB55]*Complex
du[V.Gwlp] = (p[C.kcBc1G]*u[V.CycBCdk1]/((1 + (p[C.InhCDK]/p[C.Kd]))) + p[C.kcAc2G]*p[C.CycACdk2T])*(p[C.Gwtot] - u[V.Gwlp]) - (p[C.kB55G]*u[V.PP2AB55] + p[C.kppxGwl] + p[C.kPP1Gw]*u[V.PP1])*u[V.Gwlp]
du[V.PP2AB55] = p[C.kdis]*Complex + p[C.kcatB55]*Complex - p[C.kass]*u[V.PP2AB55]*(u[V.pENSAt] - Complex)
du[V.Wee1] = (p[C.kppxY15] + p[C.kB55W1]*u[V.PP2AB55])*Wee1p - (p[C.kcBc1W1]*u[V.CycBCdk1]/((1 + (p[C.InhCDK]/p[C.Kd]))) + p[C.kcAc2W1]*p[C.CycACdk2T])*u[V.Wee1]
du[V.Wee1pp] = (p[C.kcBc1W1]*u[V.CycBCdk1]/((1 + (p[C.InhCDK]/p[C.Kd]))) + p[C.kcAc2W1]*p[C.CycACdk2T])*Wee1p - (p[C.kppxY15] + p[C.kB55W1]*u[V.PP2AB55])*u[V.Wee1pp]
du[V.Cdc25] = (p[C.kppxY15] + p[C.kB5525]*u[V.PP2AB55])*Cdc25p - (p[C.kcBc125]*u[V.CycBCdk1]/((1 + (p[C.InhCDK]/p[C.Kd]))) + p[C.kcAc225]*p[C.CycACdk2T])*u[V.Cdc25]
du[V.Cdc25pp] = (p[C.kcBc125]*u[V.CycBCdk1]/((1 + (p[C.InhCDK]/p[C.Kd]))) + p[C.kcAc225]*p[C.CycACdk2T])*(Cdc25p) - (p[C.kppxY15] + p[C.kB5525]*u[V.PP2AB55])*u[V.Cdc25pp]
end
function param_values()::Vector{Float64}
p::Vector{Float64} = zeros(C.NUM)
p[C.InhCDK] = 2.0
p[C.CycBCdk1T] = 8.1808
p[C.CycACdk2T] = 1.0000
p[C.PP1T] = 1.0000
p[C.kapp1] = 0.0115
p[C.kapp1a] = 0.7054
p[C.kipp1] = 0.0018
p[C.kipp1C] = 0.7549
p[C.kPP1Gw] = 18.4724
p[C.ENSAtot] = 1.0000
p[C.B55tot] = 0.2500
p[C.SubT] = 1.0000
p[C.kass] = 617.2807
p[C.kdis] = 0.0088
p[C.kcatB55] = 1.0338
p[C.kGwENSA] = 20.8811
p[C.kppxGwl] = 0.1560
p[C.kcBc1Sub] = 0.0080
p[C.kcBc1G] = 0.2393
p[C.Gwtot] = 1.0000
p[C.kB55G] = 496.5636
p[C.kB55Sub] = 0.0593
p[C.kcAc2G] = 0.1916
p[C.k25S] = 0.0050
p[C.k25F] = 0.9411
p[C.kweeS] = 0.0050
p[C.kweeF] = 47.2937
p[C.kcBc1W1] = 1.3132
p[C.kcBc125] = 1.3132
p[C.kppxY15] = 0.0050
p[C.kcAc2W1] = 0.1096
p[C.kcAc225] = 0.1096
p[C.kB55W1] = 0.5511
p[C.kB5525] = 0.5511
p[C.Kd] = 0.025
return p
end
function get_derivatives(u::Vector{Float64},p::Vector{Float64})
# derivatives: dF/d[bifurcation_param]
dFdp::Vector{Float64} = zeros(V.NUM)
Wee1p = 1 - u[V.Wee1] - u[V.Wee1pp]
Cdc25p = 1 - u[V.Cdc25] - u[V.Cdc25pp]
dFdp[V.Subp] = - (1/p[C.Kd])*(p[C.kcBc1Sub]*u[V.CycBCdk1]/(1 + (p[C.InhCDK]/p[C.Kd]))^2)*(p[C.SubT]-u[V.Subp])
dFdp[V.PP1] = (1/p[C.Kd])*(p[C.kipp1] + p[C.kipp1C]*u[V.CycBCdk1]/(1 + (p[C.InhCDK]/p[C.Kd]))^2)*u[V.PP1]
dFdp[V.Gwlp] = - (1/p[C.Kd])*(p[C.kcBc1G]*u[V.CycBCdk1]/(1 + (p[C.InhCDK]/p[C.Kd]))^2)*(p[C.Gwtot] - u[V.Gwlp])
dFdp[V.Wee1] = (1/p[C.Kd])*(p[C.kcBc1W1]*u[V.CycBCdk1]/(1 + (p[C.InhCDK]/p[C.Kd]))^2)*u[V.Wee1]
dFdp[V.Wee1pp] = - (1/p[C.Kd])*(p[C.kcBc1W1]*u[V.CycBCdk1]/(1 + (p[C.InhCDK]/p[C.Kd]))^2)*Wee1p
dFdp[V.Cdc25] = (1/p[C.Kd])*(p[C.kcBc125]*u[V.CycBCdk1]/(1 + (p[C.InhCDK]/p[C.Kd]))^2)*u[V.Cdc25]
dFdp[V.Cdc25pp] = - (1/p[C.Kd])*(p[C.kcBc125]*u[V.CycBCdk1]/(1 + (p[C.InhCDK]/p[C.Kd]))^2)*(Cdc25p)
return dFdp
end
function get_steady_state(p::Vector{Float64})
tspan::Tuple{Float64,Float64} = (0.0,Inf)
u0::Vector{Float64} = zeros(V.NUM)
u0[V.PP1] = 1.0
u0[V.PP2AB55] = 0.25
u0[V.Wee1] = 1.0
u0[V.Cdc25] = 1.0
prob = ODEProblem(diffeq!,u0,tspan,p)
prob = SteadyStateProblem(prob)
sol = solve(prob,DynamicSS(CVODE_BDF()),dt=1.0)
return sol.u
end | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 612 | module C
const NAMES = [
"InhCDK"
"CycBCdk1T"
"CycACdk2T"
"PP1T"
"kapp1"
"kapp1a"
"kipp1"
"kipp1C"
"kPP1Gw"
"ENSAtot"
"B55tot"
"SubT"
"kass"
"kdis"
"kcatB55"
"kGwENSA"
"kppxGwl"
"kcBc1Sub"
"kcBc1G"
"Gwtot"
"kB55G"
"kB55Sub"
"kcAc2G"
"k25S"
"k25F"
"kweeS"
"kweeF"
"kcBc1W1"
"kcBc125"
"kppxY15"
"kcAc2W1"
"kcAc225"
"kB55W1"
"kB5525"
"Kd"
]
#name2idx
for (idx,name) in enumerate(NAMES)
eval(Meta.parse("const $name = $idx"))
end
const NUM = length(NAMES)
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 285 | module V
const NAMES = [
"Subp"
"CycBCdk1"
"PP1"
"pENSAt"
"Gwlp"
"PP2AB55"
"Wee1"
"Wee1pp"
"Cdc25"
"Cdc25pp"
]
#name2idx
for (idx,name) in enumerate(NAMES)
eval(Meta.parse("const $name = $idx"))
end
const NUM = length(NAMES)
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 1699 | """
Yao, G., Lee, T. J., Mori, S., Nevins, J. R. & You, L. A bistable Rb-E2F switch
underlies the restriction point. Nat. Cell Biol. 10, 476–482 (2008).
https://doi.org/10.1038/ncb1711
"""
include("./name2idx/parameters.jl")
include("./name2idx/species.jl")
include("./set_model.jl")
include("./forwarddiff.jl")
const BP = C.S # name(index) of bifurcation parameter
const SN = V.NUM # num of state variables
const PN = 1 # num of parameters
const VN = SN + PN # num of variables
function calc_fixed_point_vec(model_path::String)::Tuple{Array,Array}
global p = param_values()
new_curve!(
model_path, p, diffeq, get_derivatives, get_steady_state,
direction=false, bifparam=BP, n_state=SN
)
fp::Array = readdlm(joinpath(model_path, "data", "fp.dat"), '\t', Float64, '\n')
ev::Array = readdlm(joinpath(model_path, "data", "ev.dat"), '\t', Float64, '\n')
br::Array = get_bistable_regime(ev, SN)
return fp, br
end
function bifurcation_diagram(model_path::String, fp::Array, br::Array)
rc("figure", figsize=(8, 6))
rc("font", family="Arial")
rc("font", size=24)
rc("axes", linewidth=1)
rc("xtick.major", width=1)
rc("ytick.major", width=1)
rc("lines", linewidth=3)
plot(fp[1:br[1]-1, VN+1], fp[1:br[1]-1, V.E+1], "k-")
plot(fp[br, VN+1], fp[br, V.E+1], lw=1.5, "k--")
plot(fp[br[end]+1:end, VN+1], fp[br[end]+1:end, V.E+1], "k-")
xlabel("Serum (percentage)")
ylabel("E2F (μM)")
xlim(0, 2)
xticks([0, 0.5, 1, 1.5, 2])
yscale("log")
ylim(1e-4, 2)
yticks([1e-4, 1e-2, 1])
savefig(joinpath(model_path, "bifurcation_diagram.pdf"), bbox_inches="tight")
close()
end
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 2469 | function diffeq!(du, u, p, t)
du[V.M] = p[C.kM] * p[C.S] / (p[C.KS] + p[C.S]) - p[C.dM] * u[V.M]
du[V.E] = p[C.kE] * (u[V.M] / (p[C.KM] + u[V.M])) * (u[V.E] / (p[C.KE] + u[V.E])) + p[C.kb] * u[V.M] / (p[C.KM] + u[V.M]) +
p[C.kP1] * u[V.CD] * u[V.RE] / (p[C.KCD] + u[V.RE]) + p[C.kP2] * u[V.CE] * u[V.RE] / (p[C.KCE] + u[V.RE]) - p[C.dE] * u[V.E] - p[C.kRE] * u[V.R] * u[V.E]
du[V.CD] = p[C.kCD] * u[V.M] / (p[C.KM] + u[V.M]) + p[C.kCDS] * p[C.S] / (p[C.KS] + p[C.S]) - p[C.dCD] * u[V.CD]
du[V.CE] = p[C.kCE] * u[V.E] / (p[C.KE] + u[V.E]) - p[C.dCE] * u[V.CE]
du[V.R] = p[C.kR] + p[C.kDP] * u[V.RP] / (p[C.KRP] + u[V.RP]) - p[C.kRE] * u[V.R] * u[V.E] - p[C.kP1] * u[V.CD] * u[V.R] / (p[C.KCD] + u[V.R]) -
p[C.kP2] * u[V.CE] * u[V.R] / (p[C.KCE] + u[V.R]) - p[C.dR] * u[V.R]
du[V.RP] = p[C.kP1] * u[V.CD] * u[V.R] / (p[C.KCD] + u[V.R]) + p[C.kP2] * u[V.CE] * u[V.R] / (p[C.KCE] + u[V.R]) + p[C.kP1] * u[V.CD] * u[V.RE] / (p[C.KCD] + u[V.RE]) +
p[C.kP2] * u[V.CE] * u[V.RE] / (p[C.KCE] + u[V.RE]) - p[C.kDP] * u[V.RP] / (p[C.KRP] + u[V.RP]) - p[C.dRP] * u[V.RP]
du[V.RE] = p[C.kRE] * u[V.R] * u[V.E] - p[C.kP1] * u[V.CD] * u[V.RE] / (p[C.KCD] + u[V.RE]) - p[C.kP2] * u[V.CE] * u[V.RE] / (p[C.KCE] + u[V.RE]) - p[C.dRE] * u[V.RE]
end
function param_values()::Vector{Float64}
p::Vector{Float64} = zeros(C.NUM)
p[C.S] = 2.0
p[C.kE] = 0.4
p[C.kM] = 1.0
p[C.kCD] = 0.03
p[C.kCDS] = 0.45
p[C.kR] = 0.18
p[C.kRE] = 180
p[C.kb] = 0.003
p[C.KS] = 0.5
p[C.kCE] = 0.35
p[C.dM] = 0.7
p[C.dE] = 0.25
p[C.dCD] = 1.5
p[C.dCE] = 1.5
p[C.dR] = 0.06
p[C.dRP] = 0.06
p[C.dRE] = 0.03
p[C.kP1] = 18.0
p[C.kP2] = 18.0
p[C.kDP] = 3.6
p[C.KM] = 0.15
p[C.KE] = 0.15
p[C.KCD] = 0.92
p[C.KCE] = 0.92
p[C.KRP] = 0.01
return p
end
function get_derivatives(u::Vector{Float64}, p::Vector{Float64})
# derivatives: dF/d[bifurcation_param]
dFdp::Vector{Float64} = zeros(V.NUM)
dFdp[V.M] = p[C.kM] * p[C.KS] / (p[C.KS] + p[C.S])^2
dFdp[V.CD] = p[C.kCDS] * p[C.KS] / (p[C.KS] + p[C.S])^2
return dFdp
end
function get_steady_state(p::Vector{Float64})
tspan::Tuple{Float64,Float64} = (0.0, Inf)
u0::Vector{Float64} = zeros(V.NUM)
prob = ODEProblem(diffeq!, u0, tspan, p)
prob = SteadyStateProblem(prob)
sol = solve(prob, DynamicSS(CVODE_BDF()), dt=1.0)
return sol.u
end | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 399 | module C
const NAMES = [
"S"
"kE"
"kM"
"kCD"
"kCDS"
"kR"
"kRE"
"kb"
"KS"
"kCE"
"dM"
"dE"
"dCD"
"dCE"
"dR"
"dRP"
"dRE"
"kP1"
"kP2"
"kDP"
"KM"
"KE"
"KCD"
"KCE"
"KRP"
]
#name2idx
for (idx,name) in enumerate(NAMES)
eval(Meta.parse("const $name = $idx"))
end
const NUM = length(NAMES)
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 210 | module V
const NAMES = [
"M"
"E"
"CD"
"CE"
"R"
"RP"
"RE"
]
for (idx,name) in enumerate(NAMES)
eval(Meta.parse("const $name = $idx"))
end
const NUM = length(NAMES)
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 3698 | module Exp
include("./observable.jl")
experiments = Array{Dict{String,Array{Float64,1}},1}(undef, length(observables))
error_bars = Array{Dict{String,Array{Float64,1}},1}(undef, length(observables))
experiments[observables_index("Phosphorylated_MEKc")] = Dict(
"EGF" => [0.000,0.773,0.439,0.252,0.130,0.087,0.080,0.066],
"HRG" => [0.000,0.865,1.000,0.837,0.884,0.920,0.875,0.789],
)
error_bars[observables_index("Phosphorylated_MEKc")] = Dict(
"EGF" => [0.000,0.030,0.048,0.009,0.009,0.017,0.012,0.008] ./ sqrt(3),
"HRG" => [0.000,0.041,0.000,0.051,0.058,0.097,0.157,0.136] ./ sqrt(3),
)
experiments[observables_index("Phosphorylated_ERKc")] = Dict(
"EGF" => [0.000,0.867,0.799,0.494,0.313,0.266,0.200,0.194],
"HRG" => [0.000,0.848,1.000,0.971,0.950,0.812,0.747,0.595],
)
error_bars[observables_index("Phosphorylated_ERKc")] = Dict(
"EGF" => [0.000,0.137,0.188,0.126,0.096,0.087,0.056,0.012] ./ sqrt(3),
"HRG" => [0.000,0.120,0.000,0.037,0.088,0.019,0.093,0.075] ./ sqrt(3),
)
experiments[observables_index("Phosphorylated_RSKw")] = Dict(
"EGF" => [0,0.814,0.812,0.450,0.151,0.059,0.038,0.030],
"HRG" => [0,0.953,1.000,0.844,0.935,0.868,0.779,0.558],
)
error_bars[observables_index("Phosphorylated_RSKw")] = Dict(
"EGF" => [0,0.064,0.194,0.030,0.027,0.031,0.043,0.051] ./ sqrt(3),
"HRG" => [0,0.230,0.118,0.058,0.041,0.076,0.090,0.077] ./ sqrt(3),
)
experiments[observables_index("Phosphorylated_cFos")] = Dict(
"EGF" => [0,0.060,0.109,0.083,0.068,0.049,0.027,0.017],
"HRG" => [0,0.145,0.177,0.158,0.598,1.000,0.852,0.431],
)
error_bars[observables_index("Phosphorylated_cFos")] = Dict(
"EGF" => [0,0.003,0.021,0.013,0.016,0.007,0.003,0.002] ./ sqrt(3),
"HRG" => [0,0.010,0.013,0.001,0.014,0.000,0.077,0.047] ./ sqrt(3),
)
experiments[observables_index("Phosphorylated_CREBw")] = Dict(
"EGF" => [0,0.446,0.030,0.000,0.000],
"HRG" => [0,1.000,0.668,0.460,0.340],
)
error_bars[observables_index("Phosphorylated_CREBw")] = Dict(
"EGF" => [0,0.0,0.0,0.0,0.0] ./ sqrt(3),
"HRG" => [0,0.0,0.0,0.0,0.0] ./ sqrt(3),
)
experiments[observables_index("cfos_mRNA")] = Dict(
"EGF" => [0,0.181,0.476,0.518,0.174,0.026,0.000],
"HRG" => [0,0.353,0.861,1.000,0.637,0.300,0.059],
)
error_bars[observables_index("cfos_mRNA")] = Dict(
"EGF" => [0.017,0.004,0.044,0.004,0.023,0.007,0.008] ./ sqrt(3),
"HRG" => [0.017,0.006,0.065,0.044,0.087,0.023,0.001] ./ sqrt(3),
)
experiments[observables_index("cFos_Protein")] = Dict(
"EGF" => [0,0.078,0.216,0.240,0.320,0.235],
"HRG" => [0,0.089,0.552,0.861,1.000,0.698],
)
error_bars[observables_index("cFos_Protein")] = Dict(
"EGF" => [0,0.036,0.028,0.056,0.071,0.048] ./ sqrt(3),
"HRG" => [0,0.021,0.042,0.063,0.000,0.047] ./ sqrt(3),
)
experiments[observables_index("dusp_mRNA")] = Dict(
"EGF" => [0.000,0.177,0.331,0.214,0.177,0.231],
"HRG" => [0.000,0.221,0.750,1.000,0.960,0.934],
)
error_bars[observables_index("dusp_mRNA")] = Dict(
"EGF" => [0.033,0.060,0.061,0.032,0.068,0.050] ./ sqrt(3),
"HRG" => [0.027,0.059,0.094,0.124,0.113,0.108] ./ sqrt(3),
)
function get_timepoint(obs_name::String)::Vector{Float64}
if obs_name in ["Phosphorylated_MEKc", "Phosphorylated_ERKc",
"Phosphorylated_RSKw", "Phosphorylated_cFos"]
return [0., 300., 600., 900., 1800., 2700., 3600., 5400.]
elseif obs_name == "Phosphorylated_CREBw"
return [0., 600., 1800., 3600., 5400.]
elseif obs_name == "cfos_mRNA"
return [0.,600.,1200.,1800.,2700.,3600.,5400.]
elseif obs_name in ["cFos_Protein", "dusp_mRNA"]
return [0.,900.,1800.,2700.,3600.,5400.]
end
end
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 440 | const observables = [
"Phosphorylated_MEKc"
"Phosphorylated_ERKc"
"Phosphorylated_RSKw"
"Phosphorylated_CREBw"
"dusp_mRNA"
"cfos_mRNA"
"cFos_Protein"
"Phosphorylated_cFos"
]
function observables_index(observable_name::String)::Int
if !(observable_name in observables)
error("$observable_name is not defined in observables.")
end
return findfirst(isequal(observable_name),observables)
end | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 10340 | function get_ppMEK_slope(t, ligand)::Float64
timepoints = [0, 300, 600, 900, 1800, 2700, 3600, 5400]
ppMEK_data = Dict(
"EGF" => [0.000, 0.773, 0.439, 0.252, 0.130, 0.087, 0.080, 0.066],
"HRG" => [0.000, 0.865, 1.000, 0.837, 0.884, 0.920, 0.875, 0.789],
)
slope = [
(ppMEK_data[ligand][i+1] - activity) / (timepoints[i+1] - timepoint)
for (i, (timepoint, activity)) in enumerate(zip(timepoints, ppMEK_data[ligand]))
if i + 1 <= length(timepoints)
]
for (i, timepoint) in enumerate(timepoints)
if timepoint <= t <= timepoints[i+1]
return slope[i]
end
end
end
function diffeq!(du, u, p, t)
#=
for (i, ui) in enumerate(u)
if ui < 0 && abs(ui) < eps()
u[i] = 0.0
end
end
=#
v = Dict{Int64,Float64}()
v[1] = p[C.V1] * p[C.a] * u[V.ppMEKc] * u[V.ERKc] / (p[C.Km1] * (1 + u[V.pERKc] / p[C.Km2]) + u[V.ERKc])
v[2] = p[C.V2] * p[C.a] * u[V.ppMEKc] * u[V.pERKc] / (p[C.Km2] * (1 + u[V.ERKc] / p[C.Km1]) + u[V.pERKc])
v[3] = p[C.V3] * u[V.pERKc] / (p[C.Km3] * (1 + u[V.ppERKc] / p[C.Km4]) + u[V.pERKc])
v[4] = p[C.V4] * u[V.ppERKc] / (p[C.Km4] * (1 + u[V.pERKc] / p[C.Km3]) + u[V.ppERKc])
v[5] = p[C.V5] * u[V.pERKn] / (p[C.Km5] * (1 + u[V.ppERKn] / p[C.Km6]) + u[V.pERKn])
v[6] = p[C.V6] * u[V.ppERKn] / (p[C.Km6] * (1 + u[V.pERKn] / p[C.Km5]) + u[V.ppERKn])
v[7] = p[C.KimERK] * u[V.ERKc] - p[C.KexERK] * (p[C.Vn] / p[C.Vc]) * u[V.ERKn]
v[8] = p[C.KimpERK] * u[V.pERKc] - p[C.KexpERK] * (p[C.Vn] / p[C.Vc]) * u[V.pERKn]
v[9] = p[C.KimppERK] * u[V.ppERKc] - p[C.KexppERK] * (p[C.Vn] / p[C.Vc]) * u[V.ppERKn]
v[10] = p[C.V10] * u[V.ppERKn]^p[C.n10] / (p[C.Km10]^p[C.n10] + u[V.ppERKn]^p[C.n10])
v[11] = p[C.p11] * u[V.PreduspmRNAn]
v[12] = p[C.p12] * u[V.duspmRNAc]
v[13] = p[C.p13] * u[V.duspmRNAc]
v[14] = p[C.V14] * u[V.ppERKc] * u[V.DUSPc] / (p[C.Km14] + u[V.DUSPc])
v[15] = p[C.V15] * u[V.pDUSPc] / (p[C.Km15] + u[V.pDUSPc])
v[16] = p[C.p16] * u[V.DUSPc]
v[17] = p[C.p17] * u[V.pDUSPc]
v[18] = p[C.KimDUSP] * u[V.DUSPc] - p[C.KexDUSP] * (p[C.Vn] / p[C.Vc]) * u[V.DUSPn]
v[19] = p[C.KimpDUSP] * u[V.pDUSPc] - p[C.KexpDUSP] * (p[C.Vn] / p[C.Vc]) * u[V.pDUSPn]
v[20] = p[C.V20] * u[V.ppERKn] * u[V.DUSPn] / (p[C.Km20] + u[V.DUSPn])
v[21] = p[C.V21] * u[V.pDUSPn] / (p[C.Km21] + u[V.pDUSPn])
v[22] = p[C.p22] * u[V.DUSPn]
v[23] = p[C.p23] * u[V.pDUSPn]
v[24] = p[C.V24] * u[V.ppERKc] * u[V.RSKc] / (p[C.Km24] + u[V.RSKc])
v[25] = p[C.V25] * u[V.pRSKc] / (p[C.Km25] + u[V.pRSKc])
v[26] = p[C.KimRSK] * u[V.pRSKc] - p[C.KexRSK] * (p[C.Vn] / p[C.Vc]) * u[V.pRSKn]
v[27] = p[C.V27] * u[V.pRSKn] * u[V.CREBn] / (p[C.Km27] + u[V.CREBn])
v[28] = p[C.V28] * u[V.pCREBn] / (p[C.Km28] + u[V.pCREBn])
v[29] = p[C.V29] * u[V.ppERKn] * u[V.Elk1n] / (p[C.Km29] + u[V.Elk1n])
v[30] = p[C.V30] * u[V.pElk1n] / (p[C.Km30] + u[V.pElk1n])
v[31] = p[C.V31] * (u[V.pCREBn] * u[V.pElk1n])^p[C.n31] / (p[C.Km31]^p[C.n31] + (u[V.pCREBn] * u[V.pElk1n])^p[C.n31] + (u[V.Fn] / p[C.KF31])^p[C.nF31])
v[32] = p[C.p32] * u[V.PrecfosmRNAn]
v[33] = p[C.p33] * u[V.cfosmRNAc]
v[34] = p[C.p34] * u[V.cfosmRNAc]
v[35] = p[C.V35] * u[V.ppERKc] * u[V.cFOSc] / (p[C.Km35] + u[V.cFOSc])
v[36] = p[C.V36] * u[V.pRSKc] * u[V.cFOSc] / (p[C.Km36] + u[V.cFOSc])
v[37] = p[C.V37] * u[V.pcFOSc] / (p[C.Km37] + u[V.pcFOSc])
v[38] = p[C.p38] * u[V.cFOSc]
v[39] = p[C.p39] * u[V.pcFOSc]
v[40] = p[C.KimFOS] * u[V.cFOSc] - p[C.KexFOS] * (p[C.Vn] / p[C.Vc]) * u[V.cFOSn]
v[41] = p[C.KimpcFOS] * u[V.pcFOSc] - p[C.KexpcFOS] * (p[C.Vn] / p[C.Vc]) * u[V.pcFOSn]
v[42] = p[C.V42] * u[V.ppERKn] * u[V.cFOSn] / (p[C.Km42] + u[V.cFOSn])
v[43] = p[C.V43] * u[V.pRSKn] * u[V.cFOSn] / (p[C.Km43] + u[V.cFOSn])
v[44] = p[C.V44] * u[V.pcFOSn] / (p[C.Km44] + u[V.pcFOSn])
v[45] = p[C.p45] * u[V.cFOSn]
v[46] = p[C.p46] * u[V.pcFOSn]
v[47] = p[C.p47] * u[V.DUSPn] * u[V.ppERKn] - p[C.m47] * u[V.DUSPn_ppERKn]
v[48] = p[C.p48] * u[V.DUSPn_ppERKn]
v[49] = p[C.p49] * u[V.DUSPn] * u[V.pERKn] - p[C.m49] * u[V.DUSPn_pERKn]
v[50] = p[C.p50] * u[V.DUSPn_pERKn]
v[51] = p[C.p51] * u[V.DUSPn] * u[V.ERKn] - p[C.m51] * u[V.DUSPn_ERKn]
v[52] = p[C.p52] * u[V.pDUSPn] * u[V.ppERKn] - p[C.m52] * u[V.pDUSPn_ppERKn]
v[53] = p[C.p53] * u[V.pDUSPn_ppERKn]
v[54] = p[C.p54] * u[V.pDUSPn] * u[V.pERKn] - p[C.m54] * u[V.pDUSPn_pERKn]
v[55] = p[C.p55] * u[V.pDUSPn_pERKn]
v[56] = p[C.p56] * u[V.pDUSPn] * u[V.ERKn] - p[C.m56] * u[V.pDUSPn_ERKn]
v[57] = p[C.V57] * u[V.pcFOSn]^p[C.n57] / (p[C.Km57]^p[C.n57] + u[V.pcFOSn]^p[C.n57])
v[58] = p[C.p58] * u[V.PreFmRNAn]
v[59] = p[C.p59] * u[V.FmRNAc]
v[60] = p[C.p60] * u[V.FmRNAc]
v[61] = p[C.p61] * u[V.Fc]
v[62] = p[C.KimF] * u[V.Fc] - p[C.KexF] * (p[C.Vn] / p[C.Vc]) * u[V.Fn]
v[63] = p[C.p63] * u[V.Fn]
if p[C.Ligand] == p[C.EGF] # EGF=10nM
du[V.ppMEKc] = get_ppMEK_slope(t, "EGF")
elseif p[C.Ligand] == p[C.HRG] # HRG=10nM
du[V.ppMEKc] = get_ppMEK_slope(t, "HRG")
else
du[V.ppMEKc] = 0.0
end
du[V.CREBn] = -v[27] + v[28]
du[V.pCREBn] = v[27] - v[28]
du[V.ERKc] = -v[1] + v[3] - v[7]
du[V.ERKn] = v[5] + v[7] * (p[C.Vc] / p[C.Vn]) + v[50] - v[51] + v[55] - v[56]
du[V.pERKc] = v[1] - v[2] - v[3] + v[4] - v[8]
du[V.pERKn] = -v[5] + v[6] + v[8] * (p[C.Vc] / p[C.Vn]) + v[48] - v[49] + v[53] - v[54]
du[V.ppERKc] = v[2] - v[4] - v[9]
du[V.ppERKn] = -v[6] + v[9] * (p[C.Vc] / p[C.Vn]) - v[47] - v[52]
du[V.Elk1n] = -v[29] + v[30]
du[V.pElk1n] = v[29] - v[30]
du[V.cFOSc] = v[34] - v[35] - v[36] + v[37] - v[38] - v[40]
du[V.cFOSn] = v[40] * (p[C.Vc] / p[C.Vn]) - v[42] - v[43] + v[44] - v[45]
du[V.pcFOSc] = v[35] + v[36] - v[37] - v[39] - v[41]
du[V.pcFOSn] = v[41] * (p[C.Vc] / p[C.Vn]) + v[42] + v[43] - v[44] - v[46]
du[V.DUSPc] = v[13] - v[14] + v[15] - v[16] - v[18]
du[V.DUSPn] = v[18] * (p[C.Vc] / p[C.Vn]) - v[20] + v[21] - v[22] - v[47] + v[48] - v[49] + v[50] - v[51]
du[V.pDUSPc] = v[14] - v[15] - v[17] - v[19]
du[V.pDUSPn] = v[19] * (p[C.Vc] / p[C.Vn]) + v[20] - v[21] - v[23] - v[52] + v[53] - v[54] + v[55] - v[56]
du[V.DUSPn_ERKn] = v[51]
du[V.DUSPn_pERKn] = v[49] - v[50]
du[V.DUSPn_ppERKn] = v[47] - v[48]
du[V.pDUSPn_ERKn] = v[56]
du[V.pDUSPn_pERKn] = v[54] - v[55]
du[V.pDUSPn_ppERKn] = v[52] - v[53]
du[V.RSKc] = -v[24] + v[25]
du[V.pRSKc] = v[24] - v[25] - v[26]
du[V.pRSKn] = v[26] * (p[C.Vc] / p[C.Vn])
du[V.PrecfosmRNAn] = v[31] - v[32]
du[V.PreduspmRNAn] = v[10] - v[11]
du[V.cfosmRNAc] = v[32] * (p[C.Vn] / p[C.Vc]) - v[33]
du[V.duspmRNAc] = v[11] * (p[C.Vn] / p[C.Vc]) - v[12]
du[V.Fc] = v[60] - v[61] - v[62]
du[V.Fn] = v[62] * (p[C.Vc] / p[C.Vn]) - v[63]
du[V.FmRNAc] = v[58] * (p[C.Vn] / p[C.Vc]) - v[59]
du[V.PreFmRNAn] = v[57] - v[58]
end
function param_values()::Vector{Float64}
p::Vector{Float64} = zeros(C.NUM)
p[C.V1] = 0.34284837
p[C.Km1] = 307.0415253
p[C.V2] = 2.20e-01
p[C.Km2] = 3.50e+02
p[C.V3] = 7.20e-01
p[C.Km3] = 1.60e+02
p[C.V4] = 6.48e-01
p[C.Km4] = 6.00e+01
p[C.V5] = 19.49872346
p[C.Km5] = 29.94073716
p[C.V6] = p[C.V5]
p[C.Km6] = p[C.Km5]
p[C.KimERK] = 1.20e-02
p[C.KexERK] = 1.80e-02
p[C.KimpERK] = 1.20e-02
p[C.KexpERK] = 1.80e-02
p[C.KimppERK] = 1.10e-02
p[C.KexppERK] = 1.30e-02
p[C.V10] = 29.24109258
p[C.Km10] = 169.0473748
p[C.n10] = 3.970849295
p[C.p11] = 0.000126129
p[C.p12] = 0.007875765
p[C.p13] = 0.001245747
p[C.V14] = 5.636949216
p[C.Km14] = 34180.48
p[C.V15] = 2.992346912
p[C.Km15] = 0.001172165
p[C.p16] = 2.57e-04
p[C.p17] = 9.63e-05
p[C.KimDUSP] = 0.024269764
p[C.KexDUSP] = 0.070467899
p[C.KimpDUSP] = p[C.KimDUSP]
p[C.KexpDUSP] = p[C.KexDUSP]
p[C.V20] = 0.157678678
p[C.Km20] = 735598.6967
p[C.V21] = 0.005648117
p[C.Km21] = 387.8377182
p[C.p22] = 2.57e-04
p[C.p23] = 9.63e-05
p[C.V24] = 0.550346114
p[C.Km24] = 29516.06587
p[C.V25] = 10.09063736
p[C.Km25] = 0.913939859
p[C.KimRSK] = 0.025925065
p[C.KexRSK] = 0.129803956
p[C.V27] = 19.23118154
p[C.Km27] = 441.5834425
p[C.V28] = 6.574759504
p[C.Km28] = 14.99180922
p[C.V29] = 0.518529841
p[C.Km29] = 21312.69109
p[C.V30] = 13.79479021
p[C.Km30] = 15.04396629
p[C.V31] = 0.655214248
p[C.Km31] = 185.9760682
p[C.n31] = 1.988003164
p[C.KF31] = 0.013844393
p[C.nF31] = 2.800340453
p[C.p32] = 0.003284434
p[C.p33] = 0.000601234
p[C.p34] = 7.65E-05
p[C.V35] = 8.907637012
p[C.Km35] = 8562.744184
p[C.V36] = 0.000597315
p[C.Km36] = 528.552427
p[C.V37] = 1.745848179
p[C.Km37] = 0.070379236
p[C.p38] = 2.57e-04
p[C.p39] = 9.63e-05
p[C.KimFOS] = 0.54528521
p[C.KexFOS] = 0.133249762
p[C.KimpcFOS] = p[C.KimFOS]
p[C.KexpcFOS] = p[C.KexFOS]
p[C.V42] = 0.909968714
p[C.Km42] = 3992.061328
p[C.V43] = 0.076717457
p[C.Km43] = 1157.116021
p[C.V44] = 0.078344305
p[C.Km44] = 0.051168202
p[C.p45] = 2.57e-04
p[C.p46] = 9.63e-05
p[C.p47] = 0.001670815
p[C.m47] = 15.80783969
p[C.p48] = 0.686020478
p[C.p49] = 0.314470502
p[C.m49] = 2.335459127
p[C.p50] = 26.59483436
p[C.p51] = 0.01646825
p[C.m51] = 9.544308421
p[C.p52] = p[C.p47]
p[C.m52] = p[C.m47]
p[C.p53] = p[C.p48]
p[C.p54] = p[C.p49]
p[C.m54] = p[C.m49]
p[C.p55] = p[C.p50]
p[C.p56] = p[C.p51]
p[C.m56] = p[C.m51]
p[C.V57] = 1.026834758
p[C.Km57] = 0.637490056
p[C.n57] = 3.584464176
p[C.p58] = 0.000270488
p[C.p59] = 0.001443889
p[C.p60] = 0.002448164
p[C.p61] = 3.50E-05
p[C.KimF] = 0.019898797
p[C.KexF] = 0.396950616
p[C.p63] = 4.13E-05
p[C.a] = 218.6276381
p[C.Vn] = 0.22
p[C.Vc] = 0.94
p[C.EGF] = 0.0
p[C.HRG] = 1.0
p[C.no_ligand] = 2.0
return p
end
function initial_values()::Vector{Float64}
u0::Vector{Float64} = zeros(V.NUM)
u0[V.ERKc] = 9.60e+02
u0[V.RSKc] = 3.53e+02
u0[V.CREBn] = 1.00e+03
u0[V.Elk1n] = 1.51e+03
return u0
end
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 3118 | # Residual Sum of Squares
function compute_objval_rss(
sim_data::Vector{Float64},
exp_data::Vector{Float64})::Float64
error::Float64 = 0.0
for i in eachindex(exp_data)
@inbounds error += (sim_data[i] - exp_data[i])^2
end
return error
end
# Cosine similarity
function compute_objval_cos(
sim_data::Vector{Float64},
exp_data::Vector{Float64})::Float64
error::Float64 = 1.0 - dot(sim_data, exp_data) / (norm(sim_data) * norm(exp_data))
return error
end
function conditions_index(condition_name::String)::Int
if !(condition_name in Sim.conditions)
error("$condition_name is not defined in Sim.conditions")
end
return findfirst(isequal(condition_name), Sim.conditions)
end
function diff_sim_and_exp(
sim_matrix::Matrix{Float64},
exp_dict::Dict{String,Array{Float64,1}},
exp_timepoint::Vector{Float64},
conditions::Vector{String};
sim_norm_max::Float64)::Tuple{Vector{Float64},Vector{Float64}}
sim_result::Vector{Float64} = []
exp_result::Vector{Float64} = []
for (idx, condition) in enumerate(conditions)
if condition in keys(exp_dict)
append!(sim_result, sim_matrix[idx, Int.(exp_timepoint .+ 1)])
append!(exp_result, exp_dict[condition])
end
end
return (sim_result ./ sim_norm_max, exp_result)
end
# Define an objective function to be minimized.
function objective(indiv_gene)::Float64
indiv::Vector{Float64} = decode_gene2val(indiv_gene)
(p, u0) = update_param(indiv)
if Sim.simulate!(p, u0) === nothing
error::Vector{Float64} = zeros(length(observables))
for (i, obs_name) in enumerate(observables)
if isassigned(Exp.experiments, i)
if length(Sim.normalization) > 0
norm_max::Float64 = (
Sim.normalization[obs_name]["timepoint"] !== nothing ? maximum(
Sim.simulations[
i,
[conditions_index(c) for c in Sim.normalization[obs_name]["condition"]],
Sim.normalization[obs_name]["timepoint"]
]
) : maximum(
Sim.simulations[
i,
[conditions_index(c) for c in Sim.normalization[obs_name]["condition"]],
:,
]
)
)
end
error[i] = compute_objval_rss(
diff_sim_and_exp(
Sim.simulations[i, :, :],
Exp.experiments[i],
Exp.get_timepoint(obs_name),
Sim.conditions,
sim_norm_max=ifelse(
length(Sim.normalization) == 0, 1.0, norm_max
)
)...
)
end
end
return sum(error) # < 1e12
else
return 1e12
end
end
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 11992 | # Specify model parameters and/or initial values to optimize
function get_search_index()::Tuple{Array{Int64,1},Array{Int64,1}}
# parameters
search_idx_params::Vector{Int} = [
C.V1
C.Km1
C.V5
C.Km5
C.V10
C.Km10
C.n10
C.p11
C.p12
C.p13
C.V14
C.Km14
C.V15
C.Km15
C.KimDUSP
C.KexDUSP
C.V20
C.Km20
C.V21
C.Km21
C.V24
C.Km24
C.V25
C.Km25
C.KimRSK
C.KexRSK
C.V27
C.Km27
C.V28
C.Km28
C.V29
C.Km29
C.V30
C.Km30
C.V31
C.Km31
C.n31
C.p32
C.p33
C.p34
C.V35
C.Km35
C.V36
C.Km36
C.V37
C.Km37
C.KimFOS
C.KexFOS
C.V42
C.Km42
C.V43
C.Km43
C.V44
C.Km44
C.p47
C.m47
C.p48
C.p49
C.m49
C.p50
C.p51
C.m51
C.V57
C.Km57
C.n57
C.p58
C.p59
C.p60
C.p61
C.KimF
C.KexF
C.p63
C.KF31
C.nF31
C.a
]
# initial values
search_idx_initials::Vector{Int} = [
# V.(variableName)
]
return search_idx_params, search_idx_initials
end
function get_search_region()::Matrix{Float64}
p::Vector{Float64} = param_values()
u0::Vector{Float64} = initial_values()
search_idx::Tuple{Array{Int64,1},Array{Int64,1}} = get_search_index()
search_param::Vector{Float64} = initialize_search_param(search_idx, p, u0)
search_rgn::Matrix{Float64} = zeros(2, length(p) + length(u0))
# Default: 0.1 ~ 10x
for (i, j) in enumerate(search_idx[1])
search_rgn[1, j] = search_param[i] * 0.1 # lower bound
search_rgn[2, j] = search_param[i] * 10.0 # upper bound
end
# Default: 0.5 ~ 2x
for (i, j) in enumerate(search_idx[2])
search_rgn[1, j+length(p)] = search_param[i+length(search_idx[1])] * 0.5 # lower bound
search_rgn[2, j+length(p)] = search_param[i+length(search_idx[1])] * 2.0 # upper bound
end
# search_rgn[:, C.param_name] = [lower_bound, upper_bound]
# search_rgn[:, V.var_name+length(p)] = [lower_bound, upper_bound]
search_rgn[:, C.V1] = [7.33e-2, 6.60e-01]
search_rgn[:, C.Km1] = [1.83e+2, 8.50e+2]
search_rgn[:, C.V5] = [6.48e-3, 7.20e+1]
search_rgn[:, C.Km5] = [6.00e-1, 1.60e+04]
search_rgn[:, C.V10] = [exp(-10), exp(10)]
search_rgn[:, C.Km10] = [exp(-10), exp(10)]
search_rgn[:, C.n10] = [1.00, 4.00]
search_rgn[:, C.p11] = [8.30e-13, 1.44e-2]
search_rgn[:, C.p12] = [8.00e-8, 5.17e-2]
search_rgn[:, C.p13] = [1.38e-7, 4.84e-1]
search_rgn[:, C.V14] = [4.77e-3, 4.77e+1]
search_rgn[:, C.Km14] = [2.00e+2, 2.00e+6]
search_rgn[:, C.V15] = [exp(-10), exp(10)]
search_rgn[:, C.Km15] = [exp(-10), exp(10)]
search_rgn[:, C.KimDUSP] = [2.20e-4, 5.50e-1]
search_rgn[:, C.KexDUSP] = [2.60e-4, 6.50e-1]
search_rgn[:, C.V20] = [4.77e-3, 4.77e+1]
search_rgn[:, C.Km20] = [2.00e+2, 2.00e+6]
search_rgn[:, C.V21] = [exp(-10), exp(10)]
search_rgn[:, C.Km21] = [exp(-10), exp(10)]
search_rgn[:, C.V24] = [4.77e-2, 4.77e+0]
search_rgn[:, C.Km24] = [2.00e+3, 2.00e+5]
search_rgn[:, C.V25] = [exp(-10), exp(10)]
search_rgn[:, C.Km25] = [exp(-10), exp(10)]
search_rgn[:, C.KimRSK] = [2.20e-4, 5.50e-1]
search_rgn[:, C.KexRSK] = [2.60e-4, 6.50e-1]
search_rgn[:, C.V27] = [exp(-10), exp(10)]
search_rgn[:, C.Km27] = [1.00e+2, 1.00e+4]
search_rgn[:, C.V28] = [exp(-10), exp(10)]
search_rgn[:, C.Km28] = [exp(-10), exp(10)]
search_rgn[:, C.V29] = [4.77e-2, 4.77e+0]
search_rgn[:, C.Km29] = [2.93e+3, 2.93e+5]
search_rgn[:, C.V30] = [exp(-10), exp(10)]
search_rgn[:, C.Km30] = [exp(-10), exp(10)]
search_rgn[:, C.V31] = [exp(-10), exp(10)]
search_rgn[:, C.Km31] = [exp(-10), exp(10)]
search_rgn[:, C.n31] = [1.00, 4.00]
search_rgn[:, C.p32] = [8.30e-13, 1.44e-2]
search_rgn[:, C.p33] = [8.00e-8, 5.17e-2]
search_rgn[:, C.p34] = [1.38e-7, 4.84e-1]
search_rgn[:, C.V35] = [4.77e-3, 4.77e+1]
search_rgn[:, C.Km35] = [2.00e+2, 2.00e+6]
search_rgn[:, C.V36] = [exp(-10), exp(10)]
search_rgn[:, C.Km36] = [1.00e+2, 1.00e+4]
search_rgn[:, C.V37] = [exp(-10), exp(10)]
search_rgn[:, C.Km37] = [exp(-10), exp(10)]
search_rgn[:, C.KimFOS] = [2.20e-4, 5.50e-1]
search_rgn[:, C.KexFOS] = [2.60e-4, 6.50e-1]
search_rgn[:, C.V42] = [4.77e-3, 4.77e+1]
search_rgn[:, C.Km42] = [2.00e+2, 2.00e+6]
search_rgn[:, C.V43] = [exp(-10), exp(10)]
search_rgn[:, C.Km43] = [1.00e+2, 1.00e+4]
search_rgn[:, C.V44] = [exp(-10), exp(10)]
search_rgn[:, C.Km44] = [exp(-10), exp(10)]
search_rgn[:, C.p47] = [1.45e-4, 1.45e+0]
search_rgn[:, C.m47] = [6.00e-3, 6.00e+1]
search_rgn[:, C.p48] = [2.70e-3, 2.70e+1]
search_rgn[:, C.p49] = [5.00e-5, 5.00e-1]
search_rgn[:, C.m49] = [5.00e-3, 5.00e+1]
search_rgn[:, C.p50] = [3.00e-3, 3.00e+1]
search_rgn[:, C.p51] = [exp(-10), exp(10)]
search_rgn[:, C.m51] = [exp(-10), exp(10)]
search_rgn[:, C.V57] = [exp(-10), exp(10)]
search_rgn[:, C.Km57] = [exp(-10), exp(10)]
search_rgn[:, C.n57] = [1.00, 4.00]
search_rgn[:, C.p58] = [8.30e-13, 1.44e-2]
search_rgn[:, C.p59] = [8.00e-8, 5.17e-2]
search_rgn[:, C.p60] = [1.38e-7, 4.84e-1]
search_rgn[:, C.p61] = [exp(-10), exp(10)]
search_rgn[:, C.KimF] = [2.20e-4, 5.50e-1]
search_rgn[:, C.KexF] = [2.60e-4, 6.50e-1]
search_rgn[:, C.p63] = [exp(-10), exp(10)]
search_rgn[:, C.KF31] = [exp(-10), exp(10)]
search_rgn[:, C.nF31] = [1.00, 4.00]
search_rgn[:, C.a] = [1.00e+2, 5.00e+2]
search_rgn = convert_scale!(search_rgn, search_idx)
return search_rgn
end
function update_param(indiv::Vector{Float64})::Tuple{Array{Float64,1},Array{Float64,1}}
p::Vector{Float64} = param_values()
u0::Vector{Float64} = initial_values()
search_idx::Tuple{Array{Int64,1},Array{Int64,1}} = get_search_index()
for (i, j) in enumerate(search_idx[1])
@inbounds p[j] = indiv[i]
end
for (i, j) in enumerate(search_idx[2])
@inbounds u0[j] = indiv[i+length(search_idx[1])]
end
# constraints --------------------------------------------------------------
p[C.V6] = p[C.V5]
p[C.Km6] = p[C.Km5]
p[C.KimpDUSP] = p[C.KimDUSP]
p[C.KexpDUSP] = p[C.KexDUSP]
p[C.KimpcFOS] = p[C.KimFOS]
p[C.KexpcFOS] = p[C.KexFOS]
p[C.p52] = p[C.p47]
p[C.m52] = p[C.m47]
p[C.p53] = p[C.p48]
p[C.p54] = p[C.p49]
p[C.m54] = p[C.m49]
p[C.p55] = p[C.p50]
p[C.p56] = p[C.p51]
p[C.m56] = p[C.m51]
# --------------------------------------------------------------------------
return p, u0
end
function decode_gene2val(indiv_gene)::Vector{Float64}
search_rgn::Matrix{Float64} = get_search_region()
indiv::Vector{Float64} = zeros(length(indiv_gene))
for (i, g) in enumerate(indiv_gene)
indiv[i] = 10^(
g * (
search_rgn[2, i] - search_rgn[1, i]
) + search_rgn[1, i]
)
end
# return round.(indiv, sigdigits=7)
return indiv
end
function encode_val2gene(indiv::Vector{Float64})
search_rgn::Matrix{Float64} = get_search_region()
indiv_gene::Vector{Float64} = zeros(length(indiv))
for i in eachindex(indiv)
indiv_gene[i] = (
log10(indiv[i]) - search_rgn[1, i]
) / (
search_rgn[2, i] - search_rgn[1, i]
)
end
return indiv_gene
end
function encode_bestIndivVal2randGene(
gene_idx::Int64,
best_indiv::Vector{Float64},
p0_bounds::Vector{Float64})::Float64
search_rgn::Matrix{Float64} = get_search_region()
rand_gene::Float64 = (
log10(
best_indiv[gene_idx] * 10^(
rand() * log10(p0_bounds[2] / p0_bounds[1]) + log10(p0_bounds[1])
)
) - search_rgn[1, gene_idx]
) / (
search_rgn[2, gene_idx] - search_rgn[1, gene_idx]
)
return rand_gene
end
function initialize_search_param(
search_idx::Tuple{Array{Int64,1},Array{Int64,1}},
p::Vector{Float64},
u0::Vector{Float64})::Vector{Float64}
duplicate::Vector{String} = []
if length(search_idx[1]) != length(unique(search_idx[1]))
for idx in findall(
[count(x -> x == i, search_idx[1]) for i in unique(search_idx[1])] .!= 1
)
push!(duplicate, C.NAMES[search_idx[1][idx]])
end
error(
"Duplicate parameters (C.): $duplicate"
)
elseif length(search_idx[2]) != length(unique(search_idx[2]))
for idx in findall(
[count(x -> x == i, search_idx[2]) for i in unique(search_idx[2])] .!= 1
)
push!(duplicate, V.NAMES[search_idx[2][idx]])
end
error(
"Duplicate initial conditions (V.): $duplicate"
)
end
search_param = zeros(
length(search_idx[1]) + length(search_idx[2])
)
for (i, j) in enumerate(search_idx[1])
@inbounds search_param[i] = p[j]
end
for (i, j) in enumerate(search_idx[2])
@inbounds search_param[i+length(search_idx[1])] = u0[j]
end
if any(x -> x == 0.0, search_param)
msg::String = "search_param must not contain zero."
for idx in search_idx[1]
if p[idx] == 0.0
error(
@sprintf(
"`C.%s` in search_idx_params: ", C.NAMES[idx]
) * msg
)
end
end
for idx in search_idx[2]
if u0[idx] == 0.0
error(
@sprintf(
"`V.%s` in search_idx_initials: ", V.NAMES[idx]
) * msg
)
end
end
end
return search_param
end
function convert_scale!(
search_rgn::Matrix{Float64},
search_idx::Tuple{Array{Int64,1},Array{Int64,1}})::Matrix{Float64}
for i = 1:size(search_rgn, 2)
if minimum(search_rgn[:, i]) < 0.0
msg = "search_rgn[lower_bound,upper_bound] must be positive.\n"
if i <= C.NUM
error(@sprintf("`C.%s` ", C.NAMES[i]) * msg)
else
error(@sprintf("`V.%s` ", V.NAMES[i-C.NUM]) * msg)
end
elseif minimum(search_rgn[:, i]) == 0.0 && maximum(search_rgn[:, i]) != 0.0
msg = "lower_bound must be larger than 0.\n"
if i <= C.NUM
error(@sprintf("`C.%s` ", C.NAMES[i]) * msg)
else
error(@sprintf("`V.%s` ", V.NAMES[i-C.NUM]) * msg)
end
elseif search_rgn[2, i] - search_rgn[1, i] < 0.0
msg = "lower_bound must be smaller than upper_bound.\n"
if i <= C.NUM
error(@sprintf("`C.%s` ", C.NAMES[i]) * msg)
else
error(@sprintf("`V.%s` ", V.NAMES[i-C.NUM]) * msg)
end
end
end
nonzero_idx::Vector{Int} = []
for i = 1:size(search_rgn, 2)
if search_rgn[:, i] != [0.0, 0.0]
push!(nonzero_idx, i)
end
end
difference::Vector{Int} = collect(
symdiff(
Set(nonzero_idx),
Set(append!(search_idx[1], C.NUM .+ search_idx[2]))
)
)
if length(difference) > 0
for idx in difference
if idx <= C.NUM
println(@sprintf("`C.%s`", C.NAMES[Int(idx)]))
else
println(@sprintf("`V.%s`", V.NAMES[Int(idx)-C.NUM]))
end
end
error(
"Set these search_params in both search_idx and search_rgn."
)
end
search_rgn = search_rgn[:, nonzero_idx]
return log10.(search_rgn)
end
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 4146 | module Sim
include("./name2idx/parameters.jl")
include("./name2idx/species.jl")
include("./ode.jl")
include("./observable.jl")
using .C
using .V
using Sundials
using SteadyStateDiffEq
# Options for ODE solver
const ABSTOL = 1e-8
const RELTOL = 1e-8
normalization = Dict{String,Dict{}}()
for observable in observables
normalization[observable] = Dict(
"timepoint" => nothing,
"condition" => ["EGF", "HRG"]
)
end
const dt = 1.0
const t = collect(0.0:dt:5400.0) # 0, 1, 2, ..., 5400 [sec.]
const conditions = ["EGF", "HRG"]
simulations = Array{Float64,3}(
undef, length(observables), length(conditions), length(t)
)
function solveode(
f::Function,
u0::Vector{Float64},
t::Vector{Float64},
p::Vector{Float64})::Union{ODESolution{},Nothing}
local sol::ODESolution{}, is_successful::Bool
prob = ODEProblem(f, u0, (t[1], t[end]), p)
try
prob = ODEProblem(f, u0, (t[1], t[end]), p)
sol = solve(
prob, CVODE_BDF(),
abstol=ABSTOL,
reltol=RELTOL,
saveat=dt,
dtmin=eps(),
verbose=false
)
is_successful = ifelse(sol.t[end] == t[end], true, false)
catch
is_successful = false
#finally
# if !is_successful
# GC.gc()
# end
end
return is_successful ? sol : nothing
end
function get_steady_state(
f::Function,
u0::Vector{Float64},
p::Vector{Float64})::Vector{Float64}
local sol::SteadyStateSolution{}
try
prob = ODEProblem(f, u0, (0.0, Inf), p)
prob = SteadyStateProblem(prob)
sol = solve(
prob,
DynamicSS(
CVODE_BDF();
abstol=ABSTOL,
reltol=RELTOL
),
dt=dt,
dtmin=eps(),
verbose=false
)
#is_successful = ifelse(sol.retcode === :Success, true, false)
return sol.u
catch
#is_successful = false
# GC.gc()
#finally
# if !is_successful
# GC.gc()
# end
return []
end
#return is_successful ? sol.u : []
end
function simulate!(p::Vector{Float64}, u0::Vector{Float64})::Union{Bool,Nothing}
# get steady state
p[C.Ligand] = p[C.no_ligand]
u0 = get_steady_state(diffeq!, u0, p)
if isempty(u0)
return false
end
# add ligand
for (i, condition) in enumerate(conditions)
if condition == "EGF"
p[C.Ligand] = p[C.EGF]
elseif condition == "HRG"
p[C.Ligand] = p[C.HRG]
end
sol = solveode(diffeq!, u0, t, p)
if sol === nothing
return false
else
@inbounds @simd for j in eachindex(t)
simulations[observables_index("Phosphorylated_MEKc"), i, j] = (
sol.u[j][V.ppMEKc]
)
simulations[observables_index("Phosphorylated_ERKc"), i, j] = (
sol.u[j][V.pERKc] + sol.u[j][V.ppERKc]
)
simulations[observables_index("Phosphorylated_RSKw"), i, j] = (
sol.u[j][V.pRSKc] + sol.u[j][V.pRSKn] * (p[C.Vn] / p[C.Vc])
)
simulations[observables_index("Phosphorylated_CREBw"), i, j] = (
sol.u[j][V.pCREBn] * (p[C.Vn] / p[C.Vc])
)
simulations[observables_index("dusp_mRNA"), i, j] = (
sol.u[j][V.duspmRNAc]
)
simulations[observables_index("cfos_mRNA"), i, j] = (
sol.u[j][V.cfosmRNAc]
)
simulations[observables_index("cFos_Protein"), i, j] = (
(sol.u[j][V.pcFOSn] + sol.u[j][V.cFOSn]) * (p[C.Vn] / p[C.Vc])
+ sol.u[j][V.cFOSc] + sol.u[j][V.pcFOSc]
)
simulations[observables_index("Phosphorylated_cFos"), i, j] = (
sol.u[j][V.pcFOSn] * (p[C.Vn] / p[C.Vc]) + sol.u[j][V.pcFOSc]
)
end
end
end
end
end # module
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 1394 | module C
const NAMES = [
"V1"
"Km1"
"V2"
"Km2"
"V3"
"Km3"
"V4"
"Km4"
"V5"
"Km5"
"V6"
"Km6"
"KimERK"
"KexERK"
"KimpERK"
"KexpERK"
"KimppERK"
"KexppERK"
"V10"
"Km10"
"n10"
"p11"
"p12"
"p13"
"V14"
"Km14"
"V15"
"Km15"
"p16"
"p17"
"KimDUSP"
"KexDUSP"
"KimpDUSP"
"KexpDUSP"
"V20"
"Km20"
"V21"
"Km21"
"p22"
"p23"
"V24"
"Km24"
"V25"
"Km25"
"KimRSK"
"KexRSK"
"V27"
"Km27"
"V28"
"Km28"
"V29"
"Km29"
"V30"
"Km30"
"V31"
"Km31"
"n31"
"p32"
"p33"
"p34"
"V35"
"Km35"
"V36"
"Km36"
"V37"
"Km37"
"p38"
"p39"
"KimFOS"
"KexFOS"
"KimpcFOS"
"KexpcFOS"
"V42"
"Km42"
"V43"
"Km43"
"V44"
"Km44"
"p45"
"p46"
"p47"
"m47"
"p48"
"p49"
"m49"
"p50"
"p51"
"m51"
"p52"
"m52"
"p53"
"p54"
"m54"
"p55"
"p56"
"m56"
"V57"
"Km57"
"n57"
"p58"
"p59"
"p60"
"p61"
"KimF"
"KexF"
"p63"
"KF31"
"nF31"
#
"a"
"Vn"
"Vc"
"Ligand"
"EGF"
"HRG"
"no_ligand"
]
for (idx,name) in enumerate(NAMES)
eval(Meta.parse("const $name = $idx"))
end
const NUM = length(NAMES)
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 650 | module V
const NAMES = [
"ppMEKc"
"CREBn"
"pCREBn"
"ERKc"
"ERKn"
"pERKc"
"pERKn"
"ppERKc"
"ppERKn"
"Elk1n"
"pElk1n"
"cFOSc"
"cFOSn"
"pcFOSc"
"pcFOSn"
"DUSPc"
"DUSPn"
"pDUSPc"
"pDUSPn"
"DUSPn_ERKn"
"DUSPn_pERKn"
"DUSPn_ppERKn"
"pDUSPn_ERKn"
"pDUSPn_pERKn"
"pDUSPn_ppERKn"
"RSKc"
"pRSKc"
"pRSKn"
"PrecfosmRNAn"
"PreduspmRNAn"
"cfosmRNAc"
"duspmRNAc"
"Fc"
"Fn"
"FmRNAc"
"PreFmRNAn"
]
for (idx,name) in enumerate(NAMES)
eval(Meta.parse("const $name = $idx"))
end
const NUM = length(NAMES)
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 906 | module Exp
include("./observable.jl")
experiments = Array{Dict{String,Array{Float64,1}},1}(undef, length(observables))
error_bars = Array{Dict{String,Array{Float64,1}},1}(undef, length(observables))
experiments[observables_index("Nuclear_NFkB")] = Dict(
"WT" => [
11.505
51.52262857
59.289
83.68132857
64.46
45.027
34.62007143
36.81375
53.31616
49.223625
38.17835714
39.38095714
44.80163333
37.23936667
30.32244
36.295
35.46456
31.04
34.05333333
32.175
29.0548
30.96566
35.023
28.58318571
22.54135
] ./ 83.68132857
)
function get_timepoint(obs_name::String)::Vector{Float64}
if obs_name == "Nuclear_NFkB"
[15.0*i for i in 0:24] # 0, 15, 30, ..., 360 [min.]
end
end
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 164 | const observables = [
"Nuclear_NFkB"
]
function observables_index(observable_name::String)::Int
return findfirst(isequal(observable_name),observables)
end | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 32354 | function MA(k,S)
return k*S
end
function MAcom2(k,S1,S2)
return k*S1*S2
end
function MM(k,Km,S)
return k*S/(Km+S)
end
function Hill2(A,n)
return A^n
end
function libPulseDelay(t,X,term)
# signal parameter
sinput = 1.0 - (1.0 - 0.01)*X
sbase = 0.01
slate = 0.05 - (0.05 - 0.01)*X
tpulse = 0.25
traise = 0.5
tdecay = 0.5
tdelay = 0.0
if t <= tdelay || term == 1.0
return sbase
elseif tdelay < t <= (traise + tdelay)
return (t - tdelay)*(sinput - sbase)/traise + sbase
elseif (traise + tdelay) < t <= (tpulse + traise + tdelay)
return sinput
elseif (tpulse + traise + tdelay) < t
return (sinput - slate)*exp(-(t - tpulse - traise - tdelay)/tdecay) + slate
else
return 0.0
end
end
function diffeq!(du,u,h,p,t)
#IKK activity
IKKa = u[V.IKKp] + u[V.IKKpC] + u[V.IKKppC] + u[V.IKKpp]
if IKKa == 0.0
IKKa = 1.0
end
##### time delay
NFKBnDelay = h(p,t-p[C.delayrnae])[V.NFKBn]
###################----CBM module----#######################
signal = libPulseDelay(t,p[C.X],p[C.term])
v = Dict{Int64,Float64}()
v[1] = MA(p[C.kCp0], u[V.C]) # (*basal C phosphorylation*)
v[2] = signal * MM(p[C.kCpS], p[C.kmCpS], u[V.C]) # (*signal-dependent C phosphorylation*)
v[3] = MM(p[C.kCpu], p[C.kmCpu], u[V.Cp]) # (* Cp dephosphorylation*)
v[4] = MA(p[C.kCpB0], u[V.CB]) # (*basal CB phosphorylation*)
v[5] = signal * MM(p[C.kCpBS], p[C.kmCpBS], u[V.CB]) # (*signal-dependent CB phosphorylation*)
v[6] = MM(p[C.kCpBu], p[C.kmCpBu], u[V.CpB]) # (* CpB dephosphorylation*)
v[7] = MA(p[C.kCpM0], u[V.CM]) # (*basal CM phosphorylation*)
v[8] = signal * MM(p[C.kCpMS], p[C.kmCpMS], u[V.CM]) # (*signal-dependent CM phosphorylation*)
v[9] = MM(p[C.kCpMu], p[C.kmCpMu], u[V.CpM]) # (* CpM dephosphorylation*)
v[10] = MA(p[C.kCpBM0], u[V.CBM]) # (*basal CBM phosphorylation*)
v[11] = signal * MM(p[C.kCpBMS], p[C.kmCpBMS], u[V.CBM]) # (*signal-dependent CBM phosphorylation*)
v[12] = MM(p[C.kCpBMu], p[C.kmCpBMu], u[V.CpBM]) # (* CpBM dephosphorylation*)
v[13] = MAcom2(p[C.kBaM], u[V.B], u[V.M]) # (*association between B and M*)
v[14] = MA(p[C.kBdM], u[V.BM]) # (*disassociation between B and M*)
v[15] = MAcom2(p[C.kCaB], u[V.C], u[V.B]) # (*association between C and B*)
v[16] = MA(p[C.kCdB], u[V.CB]) # (*disassociation between C and B*)
v[17] = MAcom2(p[C.kCpaB], u[V.Cp], u[V.B]) # (*association between Cp and B*)
v[18] = MA(p[C.kCpdB], u[V.CpB]) # (*disassociation between Cp and B*)
v[19] = MAcom2(p[C.kCaM], u[V.C], u[V.M]) # (*association between C and M*)
v[20] = MA(p[C.kCdM], u[V.CM]) # (*disassociation between C and M*)
v[21] = MAcom2(p[C.kCpaM], u[V.Cp], u[V.M]) # (*association between Cp and M*)
v[22] = MA(p[C.kCpdM], u[V.CpM]) # (*disassociation between Cp and M*)
v[23] = MAcom2(p[C.kCBaM], u[V.CB], u[V.M]) # (*association between CB and M*)
v[24] = MA(p[C.kCBdM], u[V.CBM]) # (*disassociation between CB and M*)
v[25] = MAcom2(p[C.kCpBaM], u[V.CpB], u[V.M]) # (*association between CpB and M*)
v[26] = MA(p[C.kCpBdM], u[V.CpBM]) # (*disassociation between CpB and M*)
v[27] = MAcom2(p[C.kCMaB], u[V.CM], u[V.B]) # (*association between CM and B*)
v[28] = MA(p[C.kCMdB], u[V.CBM]) # (*disassociation between CM and B*)
v[29] = MAcom2(p[C.kCpMaB], u[V.CpM], u[V.B]) # (*association between CpM and B*)
v[30] = MA(p[C.kCpMdB], u[V.CpBM]) # (*disassociation between CpM and B*)
v[31] = MAcom2(p[C.kCaBM], u[V.C], u[V.BM]) # (*association between C and BM*)
v[32] = MA(p[C.kCdBM], u[V.CBM]) # (*disassociation between C and BM*)
v[33] = MAcom2(p[C.kCpaBM], u[V.Cp], u[V.BM]) # (*association between Cp and BM*)
v[34] = MA(p[C.kCpdBM], u[V.CpBM]) # (*disassociation between Cp and BM*)
###################----CBM module----#######################
###################----TAK1 module----######################
v[35] = MA(p[C.kTp0], u[V.TAK1]) # (*basal TAK1 phosphorylation*)
v[36] = u[V.Cp] * MM(p[C.kCTpS], p[C.kmCTpS], u[V.TAK1]) # (*Cp-dependent TAK1 phosphorylation*)
v[37] = u[V.CpB] * MM(p[C.kCBTpS], p[C.kmCBTpS], u[V.TAK1]) # (*CpB-dependent TAK1 phosphorylation*)
v[38] = u[V.CpM] * MM(p[C.kCMTpS], p[C.kmCMTpS], u[V.TAK1]) # (*CpM-dependent TAK1 phosphorylation*)
v[39] = u[V.CpBM] * MM(p[C.kCBMTpS], p[C.kmCBMTpS], u[V.TAK1]) # (*CpBM-dependent TAK1 phosphorylation*)
v[40] = u[V.IKKp] * MM(p[C.kTpIKK1], p[C.kmTpIKK1], u[V.TAK1]) # (*IKKp-dependent TAK1 phosphorylation*)
v[41] = MM(p[C.kTpu], p[C.kmTpu], u[V.TAK1p]) # (*TAK1 dephosphorylation*)
v[42] = MA(p[C.kTpC0], u[V.TAK1C]) # (*basal TAK1C phosphorylation*)
v[43] = u[V.Cp] * MM(p[C.kCTpCS], p[C.kmCTpCS], u[V.TAK1C]) # (*Cp-dependent TAK1C phosphorylation*)
v[44] = u[V.CpB] * MM(p[C.kCBTpCS], p[C.kmCBTpCS], u[V.TAK1C]) # (*CpB-dependent TAK1C phosphorylation*)
v[45] = u[V.CpM] * MM(p[C.kCMTpCS], p[C.kmCMTpCS], u[V.TAK1C]) # (*CpM-dependent TAK1C phosphorylation*)
v[46] = u[V.CpBM] * MM(p[C.kCBMTpCS], p[C.kmCBMTpCS], u[V.TAK1C]) # (*CpBM-dependent TAK1C phosphorylation*)
v[47] = u[V.IKKp] * MM(p[C.kTpCIKK1], p[C.kmTpCIKK1], u[V.TAK1C]) # (*IKKp-dependent TAK1C phosphorylation*)
v[48] = u[V.IKKpC] * MM(p[C.kTpCIKK2], p[C.kmTpCIKK2], u[V.TAK1C]) # (*IKKpC-dependent TAK1C phosphorylation*)
v[49] = u[V.IKKppC] * MM(p[C.kTpCIKK3], p[C.kmTpCIKK3], u[V.TAK1C]) # (*IKKppC-dependent TAK1C phosphorylation*)
v[50] = MM(p[C.kTpCu], p[C.kmTpCu], u[V.TAK1pC]) # (*basal TAK1C dephosphorylation*)
v[51] = MAcom2(p[C.kTaB], (u[V.CB] + u[V.CpB]), u[V.TAK1]) # (*association between TAK1 and CB*)
v[52] = MAcom2(p[C.kTaM], (u[V.CM] + u[V.CpM]), u[V.TAK1]) # (*association between TAK1 and CM*)
v[53] = MAcom2(p[C.kTaBM], (u[V.CBM] + u[V.CpBM]), u[V.TAK1]) # (*association between TAK1 and CBM*)
v[54] = MA(p[C.kTCd], u[V.TAK1C]) # (*dissociation between TAK1 and CBM*)
v[55] = MAcom2(p[C.kTpaB], (u[V.CB] + u[V.CpB]), u[V.TAK1p]) # (*association between TAK1p and CBM*)
v[56] = MAcom2(p[C.kTpaM], (u[V.CM] + u[V.CpM]), u[V.TAK1p]) # (*association between TAK1p and CBM*)
v[57] = MAcom2(p[C.kTpaBM], (u[V.CBM] + u[V.CpBM]), u[V.TAK1p]) # (*association between TAK1p and CBM*)
v[58] = MA(p[C.kTpCd], u[V.TAK1pC]) # (*dissociation between TAK1 and CBM*)
###################----TAK1 module----######################
###################----IKK module----#######################
v[59] = MA(p[C.kIp0], u[V.IKK]) # (*basal IKK phosphorylation*)
v[60] = u[V.TAK1p] * MM(p[C.kIpTAKp], p[C.kmIpTAKp], u[V.IKK]) # (*TAK1p-dependent IKK phosphorylation*)
v[61] = u[V.TAK1pC] * MM(p[C.kIpTAKpC], p[C.kmIpTAKpC], u[V.IKK]) # (*TAKpC-dependent IKK phosphorylation*)
v[62] = MA(p[C.kICp0], u[V.IKKC]) # (*basal IKKC phosphorylation*)
v[63] = u[V.TAK1p] * MM(p[C.kICpTAKp], p[C.kmICpTAKp], u[V.IKKC]) # (*TAK1p-dependent IKKC phosphorylation*)
v[64] = u[V.TAK1pC] * MM(p[C.kICpTAKpC], p[C.kmICpTAKpC], u[V.IKKC]) # (*TAK1pC-dependent IKKC phosphorylation*)
v[65] = MM(p[C.kIpu], p[C.kmIpu], u[V.IKKp]) # (*IKKp dephosphorylation*)
v[66] = MM(p[C.kIpCu], p[C.kmIpCu], u[V.IKKpC]) # (*IKKpC dephosphorylation*)
v[67] = MAcom2(p[C.kIaB], (u[V.CB] + u[V.CpB]), u[V.IKK]) # (*association between IKK and CB*)
v[68] = MAcom2(p[C.kIaM], (u[V.CM] + u[V.CpM]), u[V.IKK]) # (*association between IKK and CM*)
v[69] = MAcom2(p[C.kIaBM], (u[V.CBM] + u[V.CpBM]), u[V.IKK]) # (*association between IKK and CBM*)
v[70] = MA(p[C.kICd], u[V.IKKC]) # (*dissociation between IKK and CBM*)
v[71] = MAcom2(p[C.kIpaB], (u[V.CB] + u[V.CpB]), u[V.IKKp]) # (*association between IKKp and CB*)
v[72] = MAcom2(p[C.kIpaM], (u[V.CM] + u[V.CpM]), u[V.IKKp]) # (*association between IKKp and CM*)
v[73] = MAcom2(p[C.kIpaBM], (u[V.CBM] + u[V.CpBM]), u[V.IKKp]) # (*association between IKKp and CBM*)
v[74] = MA(p[C.kIpCd], u[V.IKKpC]) # (*dissociation between IKKp and CBM*)
v[75] = MM(p[C.kIpCfaIKKpC], p[C.kmIpCfaIKKpC], u[V.IKKpC]) # (*basal IKKpC phosphorylation*)
v[76] = u[V.IKKppC] * MM(p[C.kIpCfaIKKppC], p[C.kmIpCfaIKKppC], u[V.IKKpC]) # (*IKKppC-dependent IKKpC phosphorylation*)
v[77] = MM(p[C.kIppCu], p[C.kmIppCu], u[V.IKKppC]) # (*basal IKKppC dephosphorylation*)
v[78] = MA(p[C.kIppCd], u[V.IKKppC]) # (*dissociation between IKKpp and CBM (IKKppC->IKKpp*)
v[79] = MM(p[C.kIpphf], p[C.kmIpphf], u[V.IKKpp]) # (*basal IKKpp inactivation (IKKpp->IKKi)*)
v[80] = MM(p[C.kIppChf], p[C.kmIppChf], u[V.IKKppC]) # (*basal IKKppC inactivation (IKKppC->IKKi)*)
v[81] = MM(p[C.kIir], p[C.kmIir], u[V.IKKi]) # (* recycling (IKKi->IKK)*)
v[82] = u[V.A20c] * MM(p[C.kIpA20], p[C.kmIpA20], u[V.IKKp]) # (* A20-dependent IKKp phosphorylation inhibition *)
v[83] = u[V.A20c] * MM(p[C.kIpCA20], p[C.kmIpCA20], u[V.IKKpC]) # (* A20-dependent IKKpC phosphorylation inhibition *)
###################----IKK module----#######################
########----IKKNFkBIkB module----################
v[84] = MAcom2(p[C.kassanfkbikk], IKKa, u[V.NFKBIKBac]) # (*association between IKKa and NFkBIkBac*)
v[85] = MAcom2(p[C.kassbnfkbikk], IKKa, u[V.NFKBIKBbc]) # (*association between IKKa and NFkBIkBbc*)
v[86] = MAcom2(p[C.kassenfkbikk], IKKa, u[V.NFKBIKBec]) # (*association between IKKa and NFkBIkBec*)
v[87] = MA(p[C.kdisanfkbikk], u[V.IKKNFKBIKBac]) # (*dissociation between IKKa and NFkBIkBac*)
v[88] = MA(p[C.kdisbnfkbikk], u[V.IKKNFKBIKBbc]) # (*dissociation between IKKa and NFkBIkBbc*)
v[89] = MA(p[C.kdisenfkbikk], u[V.IKKNFKBIKBec]) # (*dissociation between IKKa and NFkBIkBec*)
v[90] = MA(p[C.kdegboundaIKK], u[V.IKKNFKBIKBac]) # (*dissociation between IKKa and NFkBc and degradation of IkBac*)
v[91] = MA(p[C.kdegboundbIKK], u[V.IKKNFKBIKBbc]) # (*dissociation between IKKa and NFkBc and degradation of IkBbc*)
v[92] = MA(p[C.kdegboundeIKK], u[V.IKKNFKBIKBec]) # (*dissociation between IKKa and NFkBc and degradation of IkBec*)
v[93] = MAcom2(p[C.kassaikk], u[V.IKBac], IKKa) # (*association between IKKa and IkBac*)
v[94] = MAcom2(p[C.kassbikk], u[V.IKBbc], IKKa) # (*association between IKKa and IkBbc*)
v[95] = MAcom2(p[C.kasseikk], u[V.IKBec], IKKa) # (*association between IKKa and IkBec*)
v[96] = MA(p[C.kdisaikk], u[V.IKKIKBac]) # (*dissociation between IKKa and IkBac*)
v[97] = MA(p[C.kdisbikk], u[V.IKKIKBbc]) # (*dissociation between IKKa and IkBbc*)
v[98] = MA(p[C.kdiseikk], u[V.IKKIKBec]) # (*dissociation between IKKa and IkBec*)
v[99] = MA(p[C.kdegfreeaIKK], u[V.IKKIKBac]) # (*dissociation of IKKa and degradation of IkBac*)
v[100] = MA(p[C.kdegfreebIKK], u[V.IKKIKBbc]) # (*dissociation of IKKa and degradation of IkBbc*)
v[101] = MA(p[C.kdegfreeeIKK], u[V.IKKIKBec]) # (*dissociation of IKKa and degradation of IkBec*)
########----IKKNFkBIkB module----################
###################----NFkB module----######################
v[102] = MAcom2(p[C.kassa], u[V.IKBac], u[V.NFKBc]) # (*association between NFkBc and IkBac*)
v[103] = MA(p[C.kdisa], u[V.NFKBIKBac]) # (*dissociation between NFkBc and IkBac*)
v[104] = MA(p[C.kdegbounda], u[V.NFKBIKBac]) # (*dissociation of NFkBc and degradation of IkBac*)
v[105] = MAcom2(p[C.kassaikknfkb], u[V.IKKIKBac], u[V.NFKBc]) # (*association between NFkBc and IKKaIkBac*)
v[106] = MA(p[C.kdisaikknfkb], u[V.IKKNFKBIKBac]) # (*dissociation between NFkBc and IKKaIkBac*)
v[107] = MAcom2(p[C.kassa], u[V.IKBan], u[V.NFKBn]) # (*association between NFkBn and IkBan*)
v[108] = MA(p[C.kdisa], u[V.NFKBIKBan]) # (*dissociation between NFkBn and IkBan*)
v[109] = MA(p[C.kdegbounda], u[V.NFKBIKBan]) # (*dissociation of NFkBn and degradation of IkBan*)
v[110] = MAcom2(p[C.kassb], u[V.IKBbc], u[V.NFKBc]) # (*association between NFkBc and IkBbc*)
v[111] = MA(p[C.kdisb], u[V.NFKBIKBbc]) # (*dissociation between NFkBc and IkBbc*)
v[112] = MA(p[C.kdegboundb], u[V.NFKBIKBbc]) # (*dissociation of NFkBc and degradation of IkBbc*)
v[113] = MAcom2(p[C.kassbikknfkb], u[V.IKKIKBbc], u[V.NFKBc]) # (*association between NFkBc and IKKaIkBbc*)
v[114] = MA(p[C.kdisbikknfkb], u[V.IKKNFKBIKBbc]) # (*dissociation between NFkBc and IKKaIkBbc*)
v[115] = MAcom2(p[C.kassb], u[V.IKBbn], u[V.NFKBn]) # (*association between NFkBn and IkBbn*)
v[116] = MA(p[C.kdisb], u[V.NFKBIKBbn]) # (*dissociation between NFkBn and IkBbn*)
v[117] = MA(p[C.kdegboundb], u[V.NFKBIKBbn]) # (*dissociation of NFkBn and degradation of IkBbn*)
v[118] = MAcom2(p[C.kasse], u[V.IKBec], u[V.NFKBc]) # (*association between NFkBc and IkBec*)
v[119] = MA(p[C.kdise], u[V.NFKBIKBec]) # (*dissociation between NFkBc and IkBec*)
v[120] = MA(p[C.kdegbounde], u[V.NFKBIKBec]) # (*dissociation of NFkBc and degradation of IkBec*)
v[121] = MAcom2(p[C.kasseikknfkb], u[V.IKKIKBec], u[V.NFKBc]) # (*association between NFkBc and IKKaIkBec*)
v[122] = MA(p[C.kdiseikknfkb], u[V.IKKNFKBIKBec]) # (*dissociation between NFkBc and IKKaIkBec*)
v[123] = MAcom2(p[C.kasse], u[V.IKBen], u[V.NFKBn]) # (*association between NFkBn and IkBen*)
v[124] = MA(p[C.kdise], u[V.NFKBIKBen]) # (*dissociation between NFkBn and IkBen*)
v[125] = MA(p[C.kdegbounde], u[V.NFKBIKBen]) # (*dissociation of NFkBn and degradation of IkBen*)
v[126] = MA(p[C.kshutboundikbain], u[V.NFKBIKBac]) # (*transport NFkBIkBac into nucleos*)
v[127] = MA(p[C.kshutboundikbaout], u[V.NFKBIKBan]) # (*transport NFkBIkBan into cytoplasm*)
v[128] = MA(p[C.kshutboundikbbin], u[V.NFKBIKBbc]) # (*transport NFkBIkBbc into nucleos*)
v[129] = MA(p[C.kshutboundikbbout], u[V.NFKBIKBbn]) # (*transport NFkBIkBbn into cytoplasm*)
v[130] = MA(p[C.kshutboundikbein], u[V.NFKBIKBec]) # (*transport NFkBIkBec into nucleos*)
v[131] = MA(p[C.kshutboundikbeout], u[V.NFKBIKBen]) # (*transport NFkBIkBen into cytoplasm*)
v[132] = MA(p[C.kshutfreeikbain], u[V.IKBac]) # (*transport IkBac into nucleos*)
v[133] = MA(p[C.kshutfreeikbaout], u[V.IKBan]) # (*transport IkBan into cytoplasm*)
v[134] = MA(p[C.kshutfreeikbbin], u[V.IKBbc]) # (*transport IkBbc into nucleos*)
v[135] = MA(p[C.kshutfreeikbbout], u[V.IKBbn]) # (*transport IkBbn into cytoplasm*)
v[136] = MA(p[C.kshutfreeikbein], u[V.IKBec]) # (*transport IkBec into nucleos*)
v[137] = MA(p[C.kshutfreeikbeout], u[V.IKBen]) # (*transport IkBen into cytoplasm*)
v[138] = MA(p[C.kshutnfkbin], u[V.NFKBc]) # (*transport NFkBc into nucleos*)
v[139] = MA(p[C.kshutnfkbout], u[V.NFKBn]) # (*transport NFkBn into cytoplasm*)
v[140] = p[C.k0mrnaikba] # (*basal mRNA(IkBa) transcription*)
v[141] = p[C.kprodmrnaikba] * Hill2(u[V.NFKBn], p[C.khillprodmrnaikba]) # (*NFkB-induced IkBa transcription*)
v[142] = MA(p[C.kdegmrnaikba], u[V.mRNAac]) # (*mRNA(IkBa) degradation*)
v[143] = MA(p[C.kpikba], u[V.mRNAac]) # (*mRNA(IkBa) translation*)
v[144] = MA(p[C.kdegfreea], u[V.IKBac]) # (*IkBac degradation*)
v[145] = MA(p[C.kdegfreea], u[V.IKBan]) # (*IkBan degradation*)
v[146] = p[C.k0mrnaikbb] # (*basal mRNA(IkBb) transcription*)
v[147] = MA(p[C.kdegmrnaikbb], u[V.mRNAbc]) # (*mRNA(IkBb) degradation*)
v[148] = MA(p[C.kpikbb], u[V.mRNAbc]) # (*mRNA(IkBb) translation*)
v[149] = MA(p[C.kdegfreeb], u[V.IKBbc]) # (*IkBbc degradation*)
v[150] = MA(p[C.kdegfreeb], u[V.IKBbn]) # (*IkBbn degradation*)
v[151] = p[C.k0mrnaikbe] # (*basal mRNA(IkBe) transcription*)
v[152] = p[C.kprodmrnaikbe] * Hill2(NFKBnDelay, p[C.khillprodmrnaikbe]) # (*NFkB-induced IkBe transcription*)
v[153] = MA(p[C.kdegmrnaikbe], u[V.mRNAec]) # (*mRNA(IkBe) degradation*)
v[154] = MA(p[C.kpikbe], u[V.mRNAec]) # (*mRNA(IkBe) translation*)
v[155] = MA(p[C.kdegfreee], u[V.IKBec]) # (*IkBec degradation*)
v[156] = MA(p[C.kdegfreee], u[V.IKBen]) # (*IkBen degradation*)
v[157] = p[C.k0mrnaa20] # (*basal mRNA(A20) transcription*)
v[158] = p[C.kprodmrnaa20] * Hill2(u[V.NFKBn], p[C.khillprodmrnaa20]) # (*NFkB-induced A20 transcription*)
v[159] = MA(p[C.kdegmrnaa20], u[V.mRNAa20c]) # (*mRNA(A20) degradation*)
v[160] = MA(p[C.kpa20], u[V.mRNAa20c]) # (*mRNA(A20) translation*)
v[161] = MA(p[C.kdega20], u[V.A20c]) # (*A20 degradation*)
IKKaNFKBIKB = - v[93] - v[94] - v[95] + v[99] + v[96] + v[100] + v[97] + v[101] + v[98] + v[90] + v[87] +
v[91] + v[88] + v[92] + v[89] - v[84] - v[85] - v[86]
###################----CBM module----######################
du[V.B] = (- v[15] + v[16]) + (- v[17] + v[18]) + (- v[27] + v[28]) + (- v[29] + v[30]) + (- v[13] + v[14])
du[V.M] = (- v[19] + v[20]) + (- v[21] + v[22]) + (- v[23] + v[24]) + (- v[25] + v[26]) + (- v[13] + v[14])
du[V.BM] = ( v[13] - v[14]) + (- v[31] + v[32]) + (- v[33] + v[34])
du[V.C] = (- v[15] + v[16]) + (- v[19] + v[20]) + (- v[31] + v[32]) + ( v[3] -(v[1] + v[2]) )
du[V.CB] = (v[15] - v[16]) + (- v[23] + v[24]) + ( v[6] -(v[4] + v[5]) )
du[V.CM] = (v[19] - v[20]) + (- v[27] + v[28]) + ( v[9] -(v[7] + v[8]) )
du[V.Cp] = (- v[17] + v[18]) + (- v[21] + v[22]) + (- v[33] + v[34]) + (- v[3] +(v[1] + v[2]) )
du[V.CpB] = (v[17] - v[18]) + (- v[25] + v[26]) + (- v[6] +(v[4] + v[5]) )
du[V.CpM] = (v[21] - v[22]) + (- v[29] + v[30]) + (- v[9] +(v[7] + v[8]) )
du[V.CBM] = (v[27] - v[28]) + (v[23] - v[24]) + ( v[31] - v[32]) + ( v[12] -(v[10] + v[11]) )
du[V.CpBM] = (v[29] - v[30]) + (v[25] - v[26]) + ( v[33] - v[34]) + ( - v[12] +(v[10] + v[11]) )
###################----CBM module----######################
###################----TAK1 module---######################
du[V.TAK1] = -(v[35] + v[36] + v[37] + v[38] + v[39] + v[40]) + v[41] - (v[51] + v[52] + v[53]) + v[54]
du[V.TAK1p] = (v[35] + v[36] + v[37] + v[38] + v[39] + v[40]) - v[41] - (v[55] + v[56] + v[57]) + v[58]
du[V.TAK1C] = -(v[42] + v[43] + v[44] + v[45] + v[46] + v[47] + v[48] + v[49] ) + v[50] + (v[51] + v[52] + v[53]) - v[54]
du[V.TAK1pC] = (v[42] + v[43] + v[44] + v[45] + v[46] + v[47] + v[48] + v[49] ) - v[50] + (v[55] + v[56] + v[57]) - v[58]
###################----TAK1 module---######################
###################----IKK module----######################
du[V.IKK] = -(v[59] + v[60] + v[61]) + v[65] + v[81] - (v[67] + v[68] + v[69]) + v[70] + v[82]
du[V.IKKC] = -(v[62] + v[63] + v[64]) + v[66] + (v[67] + v[68] + v[69]) - v[70] + v[83]
du[V.IKKp] = (v[59] + v[60] + v[61]) - v[65] - (v[71] + v[72] + v[73]) + v[74] - v[82] + u[V.IKKp]*IKKaNFKBIKB/IKKa
du[V.IKKpC] = (v[71] + v[72] + v[73]) - v[74] - ( v[75] + v[76]) + v[77]+ (v[62] + v[63] + v[64]) - v[66] - v[83] + u[V.IKKpC]*IKKaNFKBIKB/IKKa
du[V.IKKppC] = ( v[75] + v[76]) - v[77] - v[78] - v[80] + u[V.IKKppC]*IKKaNFKBIKB/IKKa
du[V.IKKpp] = v[78] - v[79] + u[V.IKKpp]*IKKaNFKBIKB/IKKa
du[V.IKKi] = v[79] + v[80] - v[81]
###################----IKK module----######################
###################----NFkB module----######################
du[V.NFKBc] = v[90] + v[91] + v[92] - v[102] + v[103]+ v[104] - v[105] + v[106] - v[110] + v[111] +
v[112] - v[113] + v[114] - v[118] + v[119] + v[120] - v[121] + v[122] - v[138] + v[139]
du[V.NFKBn] = - v[107] + v[108] + v[109] - v[115] + v[116] + v[117] - v[123] + v[124] + v[125] + v[138] - v[139]
du[V.IKBac] = - v[93] + v[96] - v[102] + v[103] - v[132] + v[133] + v[143] - v[144]
du[V.IKBbc] = - v[94] + v[97] - v[110] + v[111] - v[134] + v[135] + v[148] - v[149]
du[V.IKBec] = - v[95] + v[98] - v[118] + v[119] - v[136] + v[137] + v[154] - v[155]
du[V.IKBan] = - v[107] + v[108] + v[132] - v[133] - v[145]
du[V.IKBbn] = - v[115] + v[116] + v[134] - v[135] - v[150]
du[V.IKBen] = - v[123] + v[124] + v[136] - v[137] - v[156]
du[V.NFKBIKBac] = - v[84] + v[87] + v[102] - v[103] - v[104] - v[126] + v[127]
du[V.NFKBIKBbc] = - v[85] + v[88] + v[110] - v[111] - v[112] - v[128] + v[129]
du[V.NFKBIKBec] = - v[86] + v[89] + v[118] - v[119] - v[120] - v[130] + v[131]
du[V.NFKBIKBan] = v[107] - v[108] - v[109] + v[126] - v[127]
du[V.NFKBIKBbn] = v[115] - v[116] - v[117] + v[128] - v[129]
du[V.NFKBIKBen] = v[123] - v[124] - v[125] + v[130] - v[131]
du[V.IKKIKBac] = v[93] - v[96] - v[99] - v[105] + v[106]
du[V.IKKIKBbc] = v[94] - v[97] - v[100] - v[113] + v[114]
du[V.IKKIKBec] = v[95] - v[98] - v[101] - v[121] + v[122]
du[V.IKKNFKBIKBac] = v[84] - v[87] - v[90] + v[105] - v[106]
du[V.IKKNFKBIKBbc] = v[85] - v[88] - v[91] + v[113] - v[114]
du[V.IKKNFKBIKBec] = v[86] - v[89] - v[92] + v[121] - v[122]
du[V.mRNAac] = v[140] + v[141] - v[142]
du[V.mRNAbc] = v[146] - v[147]
du[V.mRNAec] = v[151] + v[152] - v[153]
du[V.mRNAa20c] = v[157] + v[158] - v[159]
du[V.A20c] = v[160] - v[161]
###################----NFkB module----######################
end
function param_values()
p = zeros(C.NUM)
p[C.kCp0] = 1.000000e-04
p[C.kCpB0] = 1.000000e-04
p[C.kCpM0] = 1.000000e-04
p[C.kCpBM0] = 1.000000e-04
p[C.kCpS] = 7.000000e+00
p[C.kCpBS] = 2.000000e+01
p[C.kCpMS] = 1.700000e+01
p[C.kCpBMS] = 1.000000e+00
p[C.kmCpS] = 1.000000e+00
p[C.kmCpBS] = 1.000000e+00
p[C.kmCpMS] = 1.000000e+00
p[C.kmCpBMS] = 5.000000e+00
p[C.kCpu] = 7.500000e-01
p[C.kCpBu] = 1.950000e+00
p[C.kCpMu] = 3.750000e+00
p[C.kCpBMu] = 2.500000e-01
p[C.kmCpu] = 1.000000e+00
p[C.kmCpBu] = 1.000000e+00
p[C.kmCpMu] = 1.000000e+00
p[C.kmCpBMu] = 2.500000e+00
p[C.kBaM] = 5.000000e-01
p[C.kCaB] = 5.000000e-01
p[C.kCaM] = 5.000000e-01
p[C.kCBaM] = 1.500000e+00
p[C.kCMaB] = 5.000000e-01
p[C.kCaBM] = 5.000000e-01
p[C.kCpaBM] = 1.885000e+00
p[C.kCpaB] = 7.885000e+00
p[C.kCpaM] = 4.585000e+00
p[C.kCpBaM] = 2.500000e+00
p[C.kCpMaB] = 1.500000e+00
p[C.kBdM] = 2.500000e+00
p[C.kCdB] = 1.000000e+00
p[C.kCdM] = 2.500000e+00
p[C.kCBdM] = 1.500000e+00
p[C.kCMdB] = 2.500000e+00
p[C.kCpdB] = 3.000000e-01
p[C.kCpdM] = 3.000000e-01
p[C.kCpBdM] = 1.500000e-01
p[C.kCpMdB] = 4.500000e-01
p[C.kCdBM] = 2.000000e-01
p[C.kCpdBM] = 2.500000e-01
p[C.kTp0] = 2.700000e-01
p[C.kCTpS] = 3.200000e-01
p[C.kCBTpS] = 1.320000e+00
p[C.kCMTpS] = 1.320000e+00
p[C.kCBMTpS] = 1.320000e+00
p[C.kmCTpS] = 2.100000e+00
p[C.kmCBTpS] = 2.100000e+00
p[C.kmCMTpS] = 2.100000e+00
p[C.kmCBMTpS] = 2.100000e+00
p[C.kTpIKK1] = 1.670000e+01
p[C.kmTpIKK1] = 7.800000e-01
p[C.kTpu] = 3.400000e+00
p[C.kmTpu] = 1.790000e+00
p[C.kTpC0] = 3.300000e-01
p[C.kCTpCS] = 5.500000e+00
p[C.kCBTpCS] = 1.750000e+01
p[C.kCMTpCS] = 1.650000e+01
p[C.kCBMTpCS] = 2.450000e+01
p[C.kmCTpCS] = 5.200000e-01
p[C.kmCBTpCS] = 5.200000e-01
p[C.kmCMTpCS] = 5.200000e-01
p[C.kmCBMTpCS] = 5.200000e-01
p[C.kTpCIKK1] = 1.000000e+01
p[C.kmTpCIKK1] = 7.800000e-01
p[C.kTpCIKK2] = 1.710000e+01
p[C.kmTpCIKK2] = 1.300000e-01
p[C.kTpCIKK3] = 8.000000e+01
p[C.kmTpCIKK3] = 7.000000e-01
p[C.kTpCu] = 1.533000e+01
p[C.kmTpCu] = 8.000000e-01
p[C.kTaB] = 1.790000e+02
p[C.kTaM] = 1.690000e+02
p[C.kTaBM] = 3.690000e+02
p[C.kTpaB] = 3.790000e+00
p[C.kTpaM] = 3.790000e+00
p[C.kTpaBM] = 3.790000e+00
p[C.kTCd] = 4.000000e+02
p[C.kTpCd] = 3.210000e+00
p[C.kIp0] = 5.500000e-02
p[C.kIpTAKp] = 1.930000e+00
p[C.kmIpTAKp] = 4.000000e-01
p[C.kIpTAKpC] = 3.800000e+00
p[C.kmIpTAKpC] = 5.800000e-01
p[C.kIpu] = 1.500000e+01
p[C.kmIpu] = 6.700000e-01
p[C.kICp0] = 5.100000e-02
p[C.kICpTAKp] = 8.640000e+00
p[C.kmICpTAKp] = 7.100000e+00
p[C.kICpTAKpC] = 1.230000e+01
p[C.kmICpTAKpC] = 7.177000e+00
p[C.kIpCu] = 1.250000e+01
p[C.kmIpCu] = 5.410000e+00
p[C.kIaB] = 3.030000e+01
p[C.kIaM] = 2.230000e+01
p[C.kIaBM] = 5.030000e+01
p[C.kIpaB] = 3.130000e+01
p[C.kIpaM] = 2.330000e+01
p[C.kIpaBM] = 6.030000e+01
p[C.kICd] = 3.280000e+01
p[C.kIpCd] = 1.320000e+00
p[C.kIpCfaIKKpC] = 1.000000e-05
p[C.kmIpCfaIKKpC] = 2.560000e+00
p[C.kIpCfaIKKppC] = 1.085700e+02
p[C.kmIpCfaIKKppC] = 2.010000e+00
p[C.kIppCu] = 1.530000e+00
p[C.kmIppCu] = 2.000000e-01
p[C.kIppCd] = 4.500000e-01
p[C.kIppChf] = 1.000000e-01
p[C.kmIppChf] = 5.000000e-01
p[C.kIpphf] = 9.000000e-03
p[C.kmIpphf] = 1.520000e+00
p[C.kIir] = 1.440000e+00
p[C.kmIir] = 3.440000e+00
p[C.kIpA20] = 2.000000e+01
p[C.kmIpA20] = 1.000000e-03
p[C.kIpCA20] = 2.000000e+01
p[C.kmIpCA20] = 1.000000e-03
p[C.kassaikk] = 1.631051e-03
p[C.kassaikknfkb] = 1.305073e+01
p[C.kassanfkbikk] = 4.812297e-02
p[C.kdisaikk] = 1.375438e-01
p[C.kdisaikknfkb] = 2.296074e-05
p[C.kdisanfkbikk] = 3.030804e-02
p[C.kassa] = 3.655315e+01
p[C.kdisa] = 3.495116e-04
p[C.kdegbounda] = 2.089360e-03
p[C.kdegboundaIKK] = 1.774837e-01
p[C.kdegfreea] = 2.010515e-02
p[C.kdegfreeaIKK] = 2.265038e-03
p[C.kshutboundikbain] = 4.102701e-01
p[C.kshutboundikbaout] = 1.632918e-01
p[C.kshutfreeikbain] = 1.011242e-01
p[C.kshutfreeikbaout] = 1.158262e-03
p[C.k0mrnaikba] = 3.450000e-05
p[C.kdegmrnaikba] = 8.326217e-02
p[C.khillprodmrnaikba] = 1.620503e+00
p[C.kpikba] = 8.484488e+00
p[C.kprodmrnaikba] = 1.290500e-01
p[C.kassbikk] = 5.609029e-03
p[C.kassbikknfkb] = 1.423728e+02
p[C.kassbnfkbikk] = 1.541680e-02
p[C.kdisbikk] = 2.771705e-02
p[C.kdisbikknfkb] = 1.742842e-03
p[C.kdisbnfkbikk] = 1.068148e-01
p[C.kassb] = 2.437209e+01
p[C.kdegboundb] = 2.804468e-04
p[C.kdegboundbIKK] = 3.645044e-01
p[C.kdegfreeb] = 1.486087e-01
p[C.kdegfreebIKK] = 2.818781e-04
p[C.kdisb] = 1.856372e-04
p[C.kshutboundikbbin] = 2.155431e-01
p[C.kshutboundikbbout] = 7.783843e-01
p[C.kshutfreeikbbin] = 1.505364e-03
p[C.kshutfreeikbbout] = 8.350086e-04
p[C.k0mrnaikbb] = 9.336099e-05
p[C.kdegmrnaikbb] = 4.101465e-03
p[C.khillprodmrnaikbb] = 0.000000e+00
p[C.kpikbb] = 4.171048e-02
p[C.kprodmrnaikbb] = 0.000000e+00
p[C.kasseikk] = 4.423523e-03
p[C.kasseikknfkb] = 1.485352e+02
p[C.kassenfkbikk] = 1.531672e-02
p[C.kdiseikk] = 3.827432e-01
p[C.kdiseikknfkb] = 2.124084e-04
p[C.kdisenfkbikk] = 2.691468e-01
p[C.kasse] = 2.413478e+02
p[C.kdegbounde] = 1.712910e-02
p[C.kdegboundeIKK] = 6.934859e-02
p[C.kdegfreee] = 2.201754e-01
p[C.kdegfreeeIKK] = 4.819481e-05
p[C.kdise] = 2.215707e-02
p[C.kshutboundikbein] = 8.666062e-03
p[C.kshutboundikbeout] = 4.006261e-02
p[C.kshutfreeikbein] = 1.364442e-03
p[C.kshutfreeikbeout] = 1.014252e-04
p[C.k0mrnaikbe] = 1.232627e-04
p[C.kdegmrnaikbe] = 9.773320e-03
p[C.khillprodmrnaikbe] = 1.944545e+00
p[C.kpikbe] = 2.384772e-01
p[C.kprodmrnaikbe] = 1.052365e-01
p[C.kshutnfkbin] = 2.072176e-01
p[C.kshutnfkbout] = 5.537618e-04
p[C.k0mrnaa20] = 1.118559e-03
p[C.kdegmrnaa20] = 2.809754e-01
p[C.kprodmrnaa20] = 1.788303e+00
p[C.kpa20] = 2.445338e-01
p[C.kdega20] = 6.217866e+00
p[C.khillprodmrnaa20] = 1.243583e+00
p[C.delayrnae] = 4.500000e+01
p[C.X] = 0.0
p[C.term] = 1.0
return p
end
function initial_values()
u0 = zeros(V.NUM)
u0[V.B] = 1.0 # B
u0[V.M] = 1.0 # M
u0[V.C] = 1.0 # C
u0[V.TAK1] = 1.0 # TAK1
u0[V.IKK] = 1.0 # IKK
u0[V.NFKBc] = 1.0 # NFKBc
return u0
end | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 3117 | # Residual Sum of Squares
function compute_objval_rss(
sim_data::Vector{Float64},
exp_data::Vector{Float64})::Float64
error::Float64 = 0.0
for i in eachindex(exp_data)
@inbounds error += (sim_data[i] - exp_data[i])^2
end
return error
end
# Cosine similarity
function compute_objval_cos(
sim_data::Vector{Float64},
exp_data::Vector{Float64})::Float64
error::Float64 = 1.0 - dot(sim_data, exp_data) / (norm(sim_data) * norm(exp_data))
return error
end
function conditions_index(condition_name::String)::Int
if !(condition_name in Sim.conditions)
error("$condition_name is not defined in Sim.conditions")
end
return findfirst(isequal(condition_name), Sim.conditions)
end
function diff_sim_and_exp(
sim_matrix::Matrix{Float64},
exp_dict::Dict{String,Array{Float64,1}},
exp_timepoint::Vector{Float64},
conditions::Vector{String};
sim_norm_max::Float64)::Tuple{Vector{Float64},Vector{Float64}}
sim_result::Vector{Float64} = []
exp_result::Vector{Float64} = []
for (idx, condition) in enumerate(conditions)
if condition in keys(exp_dict)
append!(sim_result, sim_matrix[idx, Int.(exp_timepoint .+ 1)])
append!(exp_result, exp_dict[condition])
end
end
return (sim_result ./ sim_norm_max, exp_result)
end
# Define an objective function to be minimized.
function objective(indiv_gene)::Float64
indiv::Vector{Float64} = decode_gene2val(indiv_gene)
(p, u0) = update_param(indiv)
if Sim.simulate!(p, u0) isa Nothing
error::Vector{Float64} = zeros(length(observables))
for (i, obs_name) in enumerate(observables)
if isassigned(Exp.experiments, i)
if length(Sim.normalization) > 0
norm_max::Float64 = (
Sim.normalization[obs_name]["timepoint"] !== nothing ? maximum(
Sim.simulations[
i,
[conditions_index(c) for c in Sim.normalization[obs_name]["condition"]],
Sim.normalization[obs_name]["timepoint"]
]
) : maximum(
Sim.simulations[
i,
[conditions_index(c) for c in Sim.normalization[obs_name]["condition"]],
:,
]
)
)
end
error[i] = compute_objval_rss(
diff_sim_and_exp(
Sim.simulations[i, :, :],
Exp.experiments[i],
Exp.get_timepoint(obs_name),
Sim.conditions,
sim_norm_max=ifelse(
length(Sim.normalization) == 0, 1.0, norm_max
)
)...
)
end
end
return sum(error) # < 1e12
else
return 1e12
end
end | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 8644 | # Specify model parameters and/or initial values to optimize
function get_search_index()::Tuple{Array{Int64,1},Array{Int64,1}}
# parameters
search_idx_params::Vector{Int} = [
C.kassaikk
C.kassaikknfkb
C.kassanfkbikk
C.kdisaikk
C.kdisaikknfkb
C.kdisanfkbikk
C.kassa
C.kdisa
C.kdegbounda
C.kdegboundaIKK
C.kdegfreea
C.kdegfreeaIKK
C.kshutboundikbain
C.kshutboundikbaout
C.kshutfreeikbain
C.kshutfreeikbaout
C.k0mrnaikba
C.kdegmrnaikba
C.khillprodmrnaikba
C.kpikba
C.kprodmrnaikba
C.kassbikk
C.kassbikknfkb
C.kassbnfkbikk
C.kdisbikk
C.kdisbikknfkb
C.kdisbnfkbikk
C.kassb
C.kdegboundb
C.kdegboundbIKK
C.kdegfreeb
C.kdegfreebIKK
C.kdisb
C.kshutboundikbbin
C.kshutboundikbbout
C.kshutfreeikbbin
C.kshutfreeikbbout
C.k0mrnaikbb
C.kdegmrnaikbb
# C.khillprodmrnaikbb
C.kpikbb
# C.kprodmrnaikbb
C.kasseikk
C.kasseikknfkb
C.kassenfkbikk
C.kdiseikk
C.kdiseikknfkb
C.kdisenfkbikk
C.kasse
C.kdegbounde
C.kdegboundeIKK
C.kdegfreee
C.kdegfreeeIKK
C.kdise
C.kshutboundikbein
C.kshutboundikbeout
C.kshutfreeikbein
C.kshutfreeikbeout
C.k0mrnaikbe
C.kdegmrnaikbe
C.khillprodmrnaikbe
C.kpikbe
C.kprodmrnaikbe
C.kshutnfkbin
C.kshutnfkbout
C.k0mrnaa20
C.kdegmrnaa20
C.kprodmrnaa20
C.kpa20
C.kdega20
C.khillprodmrnaa20
]
# initial values
search_idx_initials::Vector{Int} = [
# V.(variableName)
]
return search_idx_params, search_idx_initials
end
function get_search_region()::Matrix{Float64}
p::Vector{Float64} = param_values()
u0::Vector{Float64} = initial_values()
search_idx::Tuple{Array{Int64,1},Array{Int64,1}} = get_search_index()
search_param::Vector{Float64} = initialize_search_param(search_idx, p, u0)
search_rgn::Matrix{Float64} = zeros(2, length(p) + length(u0))
# Default: 0.1 ~ 10x
for (i, j) in enumerate(search_idx[1])
search_rgn[1,j] = search_param[i] * 0.1 # lower bound
search_rgn[2,j] = search_param[i] * 10.0 # upper bound
end
# Default: 0.5 ~ 2x
for (i, j) in enumerate(search_idx[2])
search_rgn[1,j + length(p)] = search_param[i + length(search_idx[1])] * 0.5 # lower bound
search_rgn[2,j + length(p)] = search_param[i + length(search_idx[1])] * 2.0 # upper bound
end
# search_rgn[:, C.param_name] = [lower_bound, upper_bound]
# search_rgn[:, V.var_name+length(p)] = [lower_bound, upper_bound]
search_rgn[:, C.khillprodmrnaikba] = [1.0, 2.0]
search_rgn[:, C.khillprodmrnaikbe] = [1.0, 2.0]
search_rgn[:, C.khillprodmrnaa20] = [1.0, 2.0]
search_rgn = convert_scale!(search_rgn, search_idx)
return search_rgn
end
function update_param(indiv::Vector{Float64})::Tuple{Array{Float64,1},Array{Float64,1}}
p::Vector{Float64} = param_values()
u0::Vector{Float64} = initial_values()
search_idx::Tuple{Array{Int64,1},Array{Int64,1}} = get_search_index()
for (i, j) in enumerate(search_idx[1])
@inbounds p[j] = indiv[i]
end
for (i, j) in enumerate(search_idx[2])
@inbounds u0[j] = indiv[i + length(search_idx[1])]
end
return p, u0
end
function decode_gene2val(indiv_gene)::Vector{Float64}
search_rgn::Matrix{Float64} = get_search_region()
indiv::Vector{Float64} = zeros(length(indiv_gene))
for i in eachindex(indiv_gene)
indiv[i] = 10^(
indiv_gene[i] * (
search_rgn[2,i] - search_rgn[1,i]
) + search_rgn[1,i]
)
end
return round.(indiv, sigdigits=7)
end
function encode_val2gene(indiv::Vector{Float64})
search_rgn::Matrix{Float64} = get_search_region()
indiv_gene::Vector{Float64} = zeros(length(indiv))
for i in eachindex(indiv)
indiv_gene[i] = (
log10(indiv[i]) - search_rgn[1,i]
) / (
search_rgn[2,i] - search_rgn[1,i]
)
end
return indiv_gene
end
function encode_bestIndivVal2randGene(
gene_idx::Int64,
best_indiv::Vector{Float64},
p0_bounds::Vector{Float64})::Float64
search_rgn::Matrix{Float64} = get_search_region()
rand_gene::Float64 = (
log10(
best_indiv[gene_idx] * 10^(
rand() * log10(p0_bounds[2] / p0_bounds[1]) + log10(p0_bounds[1])
)
) - search_rgn[1,gene_idx]
) / (
search_rgn[2,gene_idx] - search_rgn[1,gene_idx]
)
return rand_gene
end
function initialize_search_param(
search_idx::Tuple{Array{Int64,1},Array{Int64,1}},
p::Vector{Float64},
u0::Vector{Float64})::Vector{Float64}
duplicate::Vector{String} = []
if length(search_idx[1]) != length(unique(search_idx[1]))
for idx in findall(
[count(x -> x == i, search_idx[1]) for i in unique(search_idx[1])] .!= 1
)
push!(duplicate, C.NAMES[search_idx[1][idx]])
end
error(
"Duplicate parameters (C.): $duplicate"
)
elseif length(search_idx[2]) != length(unique(search_idx[2]))
for idx in findall(
[count(x -> x == i, search_idx[2]) for i in unique(search_idx[2])] .!= 1
)
push!(duplicate, V.NAMES[search_idx[2][idx]])
end
error(
"Duplicate initial conditions (V.): $duplicate"
)
end
search_param = zeros(
length(search_idx[1]) + length(search_idx[2])
)
for (i, j) in enumerate(search_idx[1])
@inbounds search_param[i] = p[j]
end
for (i, j) in enumerate(search_idx[2])
@inbounds search_param[i + length(search_idx[1])] = u0[j]
end
if any(x -> x == 0.0, search_param)
msg::String = "search_param must not contain zero."
for idx in search_idx[1]
if p[idx] == 0.0
error(
@sprintf(
"`C.%s` in search_idx_params: ", C.NAMES[idx]
) * msg
)
end
end
for idx in search_idx[2]
if u0[idx] == 0.0
error(
@sprintf(
"`V.%s` in search_idx_initials: ", V.NAMES[idx]
) * msg
)
end
end
end
return search_param
end
function convert_scale!(
search_rgn::Matrix{Float64},
search_idx::Tuple{Array{Int64,1},Array{Int64,1}})::Matrix{Float64}
for i = 1:size(search_rgn, 2)
if minimum(search_rgn[:,i]) < 0.0
msg = "search_rgn[lower_bound,upper_bound] must be positive.\n"
if i <= C.NUM
error(@sprintf("`C.%s` ", C.NAMES[i]) * msg)
else
error(@sprintf("`V.%s` ", V.NAMES[i - C.NUM]) * msg)
end
elseif minimum(search_rgn[:,i]) == 0.0 && maximum(search_rgn[:,i]) != 0.0
msg = "lower_bound must be larger than 0.\n"
if i <= C.NUM
error(@sprintf("`C.%s` ", C.NAMES[i]) * msg)
else
error(@sprintf("`V.%s` ", V.NAMES[i - C.NUM]) * msg)
end
elseif search_rgn[2,i] - search_rgn[1,i] < 0.0
msg = "lower_bound must be smaller than upper_bound.\n"
if i <= C.NUM
error(@sprintf("`C.%s` ", C.NAMES[i]) * msg)
else
error(@sprintf("`V.%s` ", V.NAMES[i - C.NUM]) * msg)
end
end
end
nonzero_idx::Vector{Int} = []
for i = 1:size(search_rgn, 2)
if search_rgn[:,i] != [0.0,0.0]
push!(nonzero_idx, i)
end
end
difference::Vector{Int} = collect(
symdiff(
Set(nonzero_idx),
Set(append!(search_idx[1], C.NUM .+ search_idx[2]))
)
)
if length(difference) > 0
for idx in difference
if j <= C.NUM
println(@sprintf("`C.%s`", C.NAMES[Int(idx)]))
else
println(@sprintf("`V.%s`", V.NAMES[Int(idx) - C.NUM]))
end
end
error(
"Set these search_params in both search_idx and search_rgn."
)
end
search_rgn = search_rgn[:,nonzero_idx]
return log10.(search_rgn)
end | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 2567 | module Sim
include("./name2idx/parameters.jl")
include("./name2idx/species.jl")
include("./ode.jl")
include("./observable.jl")
using .C
using .V
using DelayDiffEq
normalization = Dict{String,Dict{}}()
for observable in observables
normalization[observable] = Dict(
"timepoint" => nothing,
"condition" => ["WT"]
)
end
const dt = 1.0
t = collect(0.0:1.0:360.0) # 0, 1, 2, ..., 360 [min.]
const sstime = 1000.0 # time to reach steady state
const conditions = ["WT"]
simulations = Array{Float64,3}(
undef, length(observables), length(conditions), length(t)
)
function solvedde(
f::Function, u0::Vector{Float64}, history::Vector{Float64},
tspan::Tuple{Float64,Float64}, p::Vector{Float64}, tau::Float64)
h(p, t) = history
lags = [tau]
prob = DDEProblem(f, u0, h, tspan, p; constant_lags=lags)
alg = MethodOfSteps(BS3())
sol = solve(
prob, alg, saveat=dt, abstol=1e-8, reltol=1e-8, verbose=false
)
return sol
end
function get_steady_state(
p::Vector{Float64}, u0::Vector{Float64},
sstime::Float64, tau::Float64)::Vector{Float64}
# get steady state (t<0)
p[C.term] = 1.0
history::Vector{Float64} = u0
tspan::Tuple{Float64,Float64} = (0.0, sstime)
try
sol = solvedde(diffeq!, u0, history, tspan, p, tau)
if sol.retcode === :Success
return sol[:, end]
else
return []
end
catch
return []
end
end
function get_time_course(
p::Vector{Float64}, u0::Vector{Float64},
sstime::Float64, tau::Float64)
p1::Vector{Float64} = copy(p)
p1[C.term] = 0.0
u1::Vector{Float64} = get_steady_state(p, u0, sstime, tau)
if isempty(u1)
return nothing
end
history::Vector{Float64} = u1
tspan::Tuple{Float64,Float64} = (0.0, t[end])
try
sol = solvedde(diffeq!, u1, history, tspan, p1, tau)
return ifelse(sol.retcode === :Success, sol, nothing)
catch
return nothing
end
end
function simulate!(
p::Vector{Float64},
u0::Vector{Float64})::Union{Bool,Nothing}
for (i, condition) in enumerate(conditions)
# if condition == "WT"
# pass
# end
sol = get_time_course(p, u0, sstime, p[C.delayrnae])
if sol === nothing
return false
else
@inbounds @simd for j in eachindex(t)
simulations[observables_index("Nuclear_NFkB"), i, j] = (
sol.u[j][V.NFKBn]
)
end
end
end
end
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 3058 | module C
const NAMES = [
"kCp0"
"kCpB0"
"kCpM0"
"kCpBM0"
"kCpS"
"kCpBS"
"kCpMS"
"kCpBMS"
"kmCpS"
"kmCpBS"
"kmCpMS"
"kmCpBMS"
"kCpu"
"kCpBu"
"kCpMu"
"kCpBMu"
"kmCpu"
"kmCpBu"
"kmCpMu"
"kmCpBMu"
"kBaM"
"kCaB"
"kCaM"
"kCBaM"
"kCMaB"
"kCaBM"
"kCpaBM"
"kCpaB"
"kCpaM"
"kCpBaM"
"kCpMaB"
"kBdM"
"kCdB"
"kCdM"
"kCBdM"
"kCMdB"
"kCpdB"
"kCpdM"
"kCpBdM"
"kCpMdB"
"kCdBM"
"kCpdBM"
"kTp0"
"kCTpS"
"kCBTpS"
"kCMTpS"
"kCBMTpS"
"kmCTpS"
"kmCBTpS"
"kmCMTpS"
"kmCBMTpS"
"kTpIKK1"
"kmTpIKK1"
"kTpu"
"kmTpu"
"kTpC0"
"kCTpCS"
"kCBTpCS"
"kCMTpCS"
"kCBMTpCS"
"kmCTpCS"
"kmCBTpCS"
"kmCMTpCS"
"kmCBMTpCS"
"kTpCIKK1"
"kmTpCIKK1"
"kTpCIKK2"
"kmTpCIKK2"
"kTpCIKK3"
"kmTpCIKK3"
"kTpCu"
"kmTpCu"
"kTaB"
"kTaM"
"kTaBM"
"kTpaB"
"kTpaM"
"kTpaBM"
"kTCd"
"kTpCd"
"kIp0"
"kIpTAKp"
"kmIpTAKp"
"kIpTAKpC"
"kmIpTAKpC"
"kIpu"
"kmIpu"
"kICp0"
"kICpTAKp"
"kmICpTAKp"
"kICpTAKpC"
"kmICpTAKpC"
"kIpCu"
"kmIpCu"
"kIaB"
"kIaM"
"kIaBM"
"kIpaB"
"kIpaM"
"kIpaBM"
"kICd"
"kIpCd"
"kIpCfaIKKpC"
"kmIpCfaIKKpC"
"kIpCfaIKKppC"
"kmIpCfaIKKppC"
"kIppCu"
"kmIppCu"
"kIppCd"
"kIppChf"
"kmIppChf"
"kIpphf"
"kmIpphf"
"kIir"
"kmIir"
"kIpA20"
"kmIpA20"
"kIpCA20"
"kmIpCA20"
"kassaikk"
"kassaikknfkb"
"kassanfkbikk"
"kdisaikk"
"kdisaikknfkb"
"kdisanfkbikk"
"kassa"
"kdisa"
"kdegbounda"
"kdegboundaIKK"
"kdegfreea"
"kdegfreeaIKK"
"kshutboundikbain"
"kshutboundikbaout"
"kshutfreeikbain"
"kshutfreeikbaout"
"k0mrnaikba"
"kdegmrnaikba"
"khillprodmrnaikba"
"kpikba"
"kprodmrnaikba"
"kassbikk"
"kassbikknfkb"
"kassbnfkbikk"
"kdisbikk"
"kdisbikknfkb"
"kdisbnfkbikk"
"kassb"
"kdegboundb"
"kdegboundbIKK"
"kdegfreeb"
"kdegfreebIKK"
"kdisb"
"kshutboundikbbin"
"kshutboundikbbout"
"kshutfreeikbbin"
"kshutfreeikbbout"
"k0mrnaikbb"
"kdegmrnaikbb"
"khillprodmrnaikbb"
"kpikbb"
"kprodmrnaikbb"
"kasseikk"
"kasseikknfkb"
"kassenfkbikk"
"kdiseikk"
"kdiseikknfkb"
"kdisenfkbikk"
"kasse"
"kdegbounde"
"kdegboundeIKK"
"kdegfreee"
"kdegfreeeIKK"
"kdise"
"kshutboundikbein"
"kshutboundikbeout"
"kshutfreeikbein"
"kshutfreeikbeout"
"k0mrnaikbe"
"kdegmrnaikbe"
"khillprodmrnaikbe"
"kpikbe"
"kprodmrnaikbe"
"kshutnfkbin"
"kshutnfkbout"
"k0mrnaa20"
"kdegmrnaa20"
"kprodmrnaa20"
"kpa20"
"kdega20"
"khillprodmrnaa20"
"delayrnae"
"X"
"term"
]
for (idx,name) in enumerate(NAMES)
eval(Meta.parse("const $name = $idx"))
end
const NUM = length(NAMES)
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 726 | module V
const NAMES = [
"B"
"M"
"BM"
"C"
"CB"
"CM"
"Cp"
"CpB"
"CpM"
"CBM"
"CpBM"
#(* "TAK1 module*)
"TAK1"
"TAK1p"
"TAK1C"
"TAK1pC"
#(* "IKK module*)
"IKK"
"IKKC"
"IKKp"
"IKKpC"
"IKKppC"
"IKKpp"
"IKKi"
#(* NFkB module*) c:cytoplasm n:nucleus
"NFKBc"
"NFKBn"
"IKBac"
"IKBbc"
"IKBec"
"IKBan"
"IKBbn"
"IKBen"
"NFKBIKBac"
"NFKBIKBbc"
"NFKBIKBec"
"NFKBIKBan"
"NFKBIKBbn"
"NFKBIKBen"
"IKKIKBac"
"IKKIKBbc"
"IKKIKBec"
"IKKNFKBIKBac"
"IKKNFKBIKBbc"
"IKKNFKBIKBec"
"mRNAac"
"mRNAbc"
"mRNAec"
"mRNAa20c"
"A20c"
]
for (idx,name) in enumerate(NAMES)
eval(Meta.parse("const $name = $idx"))
end
const NUM = length(NAMES)
end # module | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 1867 | module BioMASS
using Printf
using LinearAlgebra
using StatsBase
using Statistics
using DelimitedFiles
export Model,
numpy_load,
generate_initial_population,
scipy_differential_evolution,
run_simulation,
create_diffeq,
new_curve!,
get_bistable_regime
const requirements = [
joinpath("name2idx", "parameters.jl"),
joinpath("name2idx", "species.jl"),
"ode.jl",
"observable.jl",
"simulation.jl",
"experimental_data.jl",
"search_param.jl",
"problem.jl",
]
struct Model
path::String
parameters::Module
species::Module
observables::Vector{String}
sim::Module
exp::Module
obj_func::Function
cond2idx::Function
search_idx::Function
search_region::Function
update_param::Function
gene2val::Function
val2gene::Function
bestIndivVal2randGene::Function
end
function Model(model_path::String, show_info::Bool=false)
for req in requirements
include(joinpath(model_path, req))
end
if show_info
print(
"Model information\n"
* "-----------------\n"
* @sprintf(
"%d species\n%d parameters, of which %d to be estimated",
length(V.NAMES), length(C.NAMES), length(get_search_index()[1])
)
)
end
Model(
model_path,
C,
V,
observables,
Sim,
Exp,
objective,
conditions_index,
get_search_index,
get_search_region,
update_param,
decode_gene2val,
encode_val2gene,
encode_bestIndivVal2randGene,
)
end
function isinstalled(pymodule::String)::Bool
try
pyimport(pymodule)
return true
catch
return false
end
end
include("pyproject.jl")
include("visulalize.jl")
include("continuation.jl")
end # module
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 9785 | using ForwardDiff: jacobian
function create_diffeq(model_path::String)
lines::Vector{String} = []
open(joinpath(model_path, "set_model.jl"), "r") do f
append!(lines, readlines(f))
end
for (i, line) in enumerate(lines)
if occursin("function diffeq!", line)
lines[i] = "function diffeq(u::Vector)\n du = similar(u)\n\n"
elseif line == "end"
lines[i] = " return du\nend"
lines = lines[1:i]
break
end
end
open(joinpath(model_path, "forwarddiff.jl"), "w") do f
for line in lines
write(f, line * "\n")
end
end
end
# matrix transformation (large diagonal elements move to upper)
function pivoting!(s::Matrix{Float64}, pivot::Int, dim_newton::Int)
v0::Vector{Float64} = zeros(dim_newton + 1)
v1::Vector{Float64} = zeros(dim_newton + 1)
possess::Int = 0
max_element::Float64 = 0.0
for i in pivot:size(s, 1)
current_element = abs(s[i, pivot])
if max_element <= current_element
max_element = current_element
possess = i
end
end
for j in 1:size(s, 2)
v0[j] = s[possess, j]
v1[j] = s[pivot, j]
end
for j in 1:size(s, 2)
s[possess, j] = v1[j]
s[pivot, j] = v0[j]
end
end
# Gaussian elimination (row reduction)
function gaussian_elimination!(s::Matrix{Float64}, e::Vector{Float64}, dim_newton::Int)
for i in 1:dim_newton
pivoting!(s, i, dim_newton)
end
# forward
for k in 1:size(s, 1)
w = (s[k, k] != 0.0) ? 1.0 / s[k, k] : 1.0
for j in k:size(s, 2)
s[k, j] *= w
for i in k:size(s, 1)
s[i, j] -= s[i, k] * s[k, j]
end
end
end
# backward
for i in size(s, 1):-1:1
sum = 0.0
for j in i:length(e)
sum += s[i, j] * e[j]
end
e[i] = s[i, end] - sum
end
end
# Newton's method
function newtons_method!(
diffeq::Function,
get_derivatives::Function,
x::Vector{Float64},
real_part::Vector{Float64},
imaginary_part::Vector{Float64},
fix_num::Int,
p::Vector{Float64},
successful::Bool,
bifparam::Int,
n_state::Int,
dim_newton::Int,
n_variable::Int;
NEPS::Float64)
u::Vector{Float64} = zeros(n_state)
vx::Vector{Float64} = zeros(dim_newton)
s::Matrix{Float64} = zeros(dim_newton, dim_newton + 1)
for i in eachindex(x)
if fix_num == i
for j in eachindex(vx)
idx = i + j
if idx > length(x)
idx -= length(x)
end
vx[j] = x[idx]
end
break
else
continue
end
end
# initial error
e::Vector{Float64} = zeros(dim_newton)
error::Float64 = 1.0
while error > NEPS
for i in 1:n_variable
if fix_num == i
idx_param = n_variable - i
p[bifparam] = (idx_param == 0) ? x[fix_num] : vx[idx_param]
for j in eachindex(u)
idx = j - i
if idx == 0
u[j] = x[fix_num]
elseif idx < 0
u[j] = vx[n_variable+idx]
else
u[j] = vx[idx]
end
end
break
else
continue
end
end
# initialization
dFdx::Matrix{Float64} = jacobian(diffeq, u)
dFdp::Vector{Float64} = get_derivatives(u, p)
F::Vector{Float64} = diffeq(u)
eigenvalues::Array{Complex{Float64},1} = eigvals(dFdx)
for (i, eigenvalue) in enumerate(eigenvalues)
real_part[i] = real(eigenvalue)
imaginary_part[i] = imag(eigenvalue)
end
# s = [dF-F]
for i in 1:n_variable
if fix_num == i
for k in 1:n_state
for j in 1:n_state
idx = i + j
if idx == n_variable
s[k, j] = dFdp[k]
elseif idx > n_variable
s[k, j] = dFdx[k, idx-n_variable]
else
s[k, j] = dFdx[k, idx]
end
end
s[k, n_variable] = -F[k]
end
break
else
continue
end
end
gaussian_elimination!(s, e, dim_newton)
# update error
error = 0.0
@inbounds for i in eachindex(e)
vx[i] += e[i]
error += e[i] * e[i]
end
error = sqrt(error)
if isnan(error) || isinf(error)
successful = false
break
end
end
for i in eachindex(x)
if fix_num == i
for j in eachindex(vx)
idx = i + j
if idx > length(x)
idx -= length(x)
end
x[idx] = vx[j]
end
break
else
continue
end
end
end
function new_curve!(
model_path::Union{String,SubString{String}},
p::Vector{Float64},
diffeq::Function,
get_derivatives::Function,
get_steady_state::Function;
direction::Bool=false,
bifparam::Int,
n_state::Int,
n_param::Int=1,
n_variable::Int=n_state + 1,
dim_newton::Int=n_state,
MC::Int=100000,
IVAL::Float64=1e-2,
RATE::Float64=1e-3,
NEPS::Float64=1e-12)
# direction : Set to true to +IVAL, false to -IVAL
# bifparam : name(index) of bifurcation parameter
# n_state : num of state variables
# n_param : num of parameters
# n_variable : num of variables
# dim_newton : dim of Newton's method
# MC : maximum of counts
# IVAL : first variation
# RATE : variation rate
# NEPS : eps of Newton's method
count::Int = 1
x::Vector{Float64} = zeros(n_variable)
dx::Vector{Float64} = zeros(n_variable)
real_part::Vector{Float64} = zeros(n_state)
imaginary_part::Vector{Float64} = zeros(n_state)
# file
if !isdir(joinpath(model_path, "data"))
mkdir(joinpath(model_path, "data"))
else
files::Vector{String} = readdir(joinpath(model_path, "data"))
for file in files
rm(joinpath(model_path, "data", "$file"))
end
end
FOUT1 = open(joinpath(model_path, "data", "fp.dat"), "w") # file for fixed point
FOUT2 = open(joinpath(model_path, "data", "ev.dat"), "w") # file for eigenvalues
# initial condition
x[1:n_state] = get_steady_state(p)
x[end] = p[bifparam] # x-axis
# initial fixed
fix_val::Float64 = x[end]
fix_num::Int = n_variable
x[fix_num] = fix_val
# first Newton's method
successful::Bool = true
newtons_method!(
diffeq,
get_derivatives,
x,
real_part,
imaginary_part,
fix_num,
p,
successful,
bifparam,
n_state,
dim_newton,
n_variable,
NEPS=NEPS,
)
write(FOUT1, @sprintf("%d\t", count))
for i in eachindex(x)
write(FOUT1, @sprintf("%10.8e\t", x[i]))
end
write(FOUT1, @sprintf("%d\n", fix_num))
write(FOUT2, @sprintf("%d\t", count))
for i in 1:n_state
write(
FOUT2, @sprintf(
"%10.8e\t%10.8e\t", real_part[i], imaginary_part[i]
)
)
end
write(FOUT2, @sprintf("%10.8e\t%d\n", p[bifparam], fix_num))
count += 1
# keep optimums
px::Vector{Float64} = copy(x)
# variation
fix_val += ifelse(direction, +IVAL, -IVAL)
# same fixed variable
x[fix_num] = fix_val
while count <= MC && successful
newtons_method!(
diffeq,
get_derivatives,
x,
real_part,
imaginary_part,
fix_num,
p,
successful,
bifparam,
n_state,
dim_newton,
n_variable,
NEPS=NEPS,
)
# maximum variation
for (i, prev) in enumerate(px)
@inbounds dx[i] = x[i] - prev
end
sum::Float64 = 0.0
for i in eachindex(dx)
@inbounds sum += dx[i] * dx[i]
end
ave::Float64 = sqrt(sum)
for i in eachindex(dx)
@inbounds dx[i] /= ave
end
px = copy(x)
for (i, diff) in enumerate(dx)
@inbounds x[i] += abs(RATE) * diff
end
# fix variable with maximum variation
fix_num = 1
for i in 2:length(dx)
if abs(dx[fix_num]) < abs(dx[i])
fix_num = i
end
end
# Stop calc.
if x[end] <= 0.0
successful = false
end
write(FOUT1, @sprintf("%d\t", count))
for i in eachindex(x)
write(FOUT1, @sprintf("%10.8e\t", x[i]))
end
write(FOUT1, @sprintf("%d\n", fix_num))
write(FOUT2, @sprintf("%d\t", count))
for i in 1:n_state
write(
FOUT2, @sprintf(
"%10.8e\t%10.8e\t", real_part[i], imaginary_part[i]
)
)
end
write(FOUT2, @sprintf("%10.8e\t%d\n", p[bifparam], fix_num))
count += 1
end
close(FOUT1)
close(FOUT2)
end
function get_bistable_regime(ev::Matrix{Float64}, n_state::Int)
br::Vector{Int} = []
for i in 1:size(ev, 1)
if maximum(ev[i, [2j for j in 1:n_state]]) > 0.0
push!(br, i)
end
end
return br
end
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 5985 | using PyCall
function __init__()
py"""
import os
import shutil
import re
import sys
import warnings
from typing import Callable, Optional, List, Union
try:
import numpy as np
except ImportError:
print("numpy: Not installed")
DIRNAME = "_tmp"
class _Logger(object):
def __init__(self, model_path: str, x_id: int):
self.log = open(
os.path.join(model_path, "fitparam", DIRNAME + str(x_id), "optimization.log"),
mode="w",
encoding="utf-8",
)
def write(self, message: str):
self.log.write(message)
class Optimizer(object):
def __init__(
self,
model_path,
model_objective,
model_gene2val,
x_id,
):
self.model_path = model_path
self.model_objective = model_objective
self.model_gene2val = model_gene2val
self.x_id = x_id
self.savedir = os.path.join(self.model_path, "fitparam", f"{self.x_id}")
if os.path.isdir(self.savedir):
raise ValueError(
f"out{os.sep}{self.x_id} already exists in {self.model_path}. "
"Use another parameter id."
)
else:
os.makedirs(self.savedir)
os.makedirs(os.path.join(self.model_path, "fitparam", DIRNAME + str(self.x_id)), exist_ok=True)
self.default_stdout = sys.stdout
def minimize(self, *args, **kwargs):
try:
from scipy.optimize import differential_evolution
except ImportError:
print("scipy: Not installed.")
os.makedirs(os.path.join(self.model_path, "fitparam", DIRNAME + str(self.x_id)), exist_ok=True)
try:
sys.stdout = _Logger(self.model_path, self.x_id)
with warnings.catch_warnings():
warnings.simplefilter("ignore")
res = differential_evolution(*args, **kwargs)
return res
finally:
sys.stdout = self.default_stdout
def _get_n_iter(self) -> int:
n_iter: int = 0
path_to_log = os.path.join(self.savedir, "optimization.log")
with open(path_to_log, mode="r", encoding="utf-8") as f:
log_file = f.readlines()
for message in log_file:
if len(message.strip()) > 0:
n_iter += 1
return n_iter
def import_solution(self, x: Union[np.ndarray, List[float]], cleanup: bool = True) -> None:
shutil.move(
os.path.join(self.model_path, "fitparam", DIRNAME + str(self.x_id), "optimization.log"),
self.savedir,
)
best_fitness: float = self.model_objective(x)
n_iter = self._get_n_iter()
np.save(os.path.join(self.savedir, "best_fitness"), best_fitness)
np.save(os.path.join(self.savedir, "count_num"), n_iter)
np.save(os.path.join(self.savedir, "generation"), n_iter)
np.save(os.path.join(self.savedir, f"fit_param{n_iter}"), x)
if cleanup:
shutil.rmtree(os.path.join(self.model_path, "fitparam", DIRNAME + str(self.x_id)))
def optimize(
model_path,
model_objective,
model_gene2val,
n_search_param,
x_id,
**kwargs,
) -> None:
optimizer = Optimizer(
model_path,
model_objective,
model_gene2val,
x_id,
)
res = optimizer.minimize(
model_objective,
[(0.0, 1.0) for _ in range(n_search_param)],
**kwargs,
)
param_values = model_gene2val(res.x)
optimizer.import_solution(param_values)
"""
end
param2biomass(model_path::String) = py"convert"(model_path)
numpy_load(path::String) = py"np.load"(path)
function scipy_differential_evolution(
model::Model,
x_id::Int;
strategy::String="best1bin",
maxiter::Int=100,
popsize::Int=3,
tol::Float64=1e-4,
mutation::Union{Float64,Tuple{Float64,Float64}}=0.1,
recombination::Float64=0.5,
seed::Union{Nothing,Int}=nothing,
disp::Bool=true,
polish::Bool=false,
init::Union{String,Matrix{Float64}}="latinhypercube",
atol::Float64=0.0,
updating::String="immediate"
)::Nothing
search_bounds::Matrix{Float64} = model.search_region()
n_search_param::Int = size(search_bounds)[2]
if !disp
error("Set 'disp' to true.")
end
if polish
error("Set 'polish' to false.")
end
return py"optimize"(
model.path,
model.obj_func,
model.gene2val,
n_search_param,
x_id,
strategy=strategy,
maxiter=maxiter,
popsize=popsize,
tol=tol,
mutation=mutation,
recombination=recombination,
seed=seed,
disp=disp,
polish=polish,
init=init,
atol=atol,
updating=updating,
)
end
function generate_initial_population(
model::Model;
popsize::Int=3,
threshold::Float64=1e12,
show_progress::Bool=true
)::Matrix{Float64}
search_bounds::Matrix{Float64} = model.search_region()
n_gene::Int = size(search_bounds)[2]
n_population::Int = popsize * n_gene
population::Matrix{Float64} = fill(
Inf, (n_population, n_gene + 1)
)
for i = 1:n_population
while threshold <= population[i, end]
for j = 1:n_gene
population[i, j] = rand()
end
population[i, end] = model.obj_func(population[i, 1:n_gene])
end
if show_progress
print("\r$i / $n_population")
end
end
population = sortslices(population, dims=1, by=x -> x[end])
return population[:, 1:n_gene]
end
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 15650 | using PyPlot
function get_indiv(model::Model, paramset::Int)::Vector{Float64}
dirname = joinpath(
model.path,
"fitparam",
"$paramset",
)
if isfile(
local path_to_generation = joinpath(dirname, "generation.dat")
)
best_generation::Int64 = readdlm(
path_to_generation
)[1, 1]
else
best_generation = numpy_load(
joinpath(dirname, "generation.npy")
)[1, 1]
end
if isfile(
local path_to_fitparam = joinpath(dirname, "fit_param$best_generation.dat")
)
best_indiv::Vector{Float64} = readdlm(
path_to_fitparam
)[:, 1]
else
best_indiv = numpy_load(
joinpath(dirname, "fit_param$best_generation.npy")
)[:, 1]
end
return best_indiv
end
function load_param(
model::Model,
paramset::Int)::Tuple{Array{Float64,1},Array{Float64,1}}
best_indiv::Vector{Float64} = get_indiv(model, paramset)
(p, u0) = model.update_param(best_indiv)
return p, u0
end
function get_executable(model::Model)::Vector{Int}
n_file::Vector{Int} = []
fitparam_files::Vector{String} = readdir(
joinpath(
model.path,
"fitparam"
)
)
for file in fitparam_files
if occursin(r"\d", file)
push!(n_file, parse(Int64, file))
end
end
empty_folder::Vector{Int} = []
for (i, nth_param_set) in enumerate(n_file)
local filepath = joinpath(
model.path,
"fitparam",
"$nth_param_set",
"generation",
)
if !isfile(filepath * ".dat") && !isfile(filepath * ".npy")
push!(empty_folder, i)
end
end
for i in sort(empty_folder, rev=true)
deleteat!(n_file, i)
end
return n_file
end
function validate!(model::Model, nth_param_set::Int64)
(p, u0) = load_param(model, nth_param_set)
if model.sim.simulate!(p, u0) isa Nothing
return model, true
else
print("Simulation failed. #$nth_param_set\n")
return model, false
end
return model
end
function get_norm_max(
model::Model, i::Int, j::Int, obs_name::String, simulations_all::Array{Float64,4})::Float64
if length(model.sim.normalization) > 0
norm_max::Float64 = (
model.sim.normalization[obs_name]["timepoint"] !== nothing ? maximum(
simulations_all[
i,
j,
[model.cond2idx(c) for c in model.sim.normalization[obs_name]["condition"]],
model.sim.normalization[obs_name]["timepoint"],
]
) : maximum(
simulations_all[
i,
j,
[model.cond2idx(c) for c in model.sim.normalization[obs_name]["condition"]],
:,
]
)
)
return norm_max
else
return 0.0
end
end
function plot_timecourse(
model::Model,
n_file::Vector{Int},
viz_type::String,
show_all::Bool,
stdev::Bool,
simulations_all::Array{Float64,4},
save_format::String)
if !isdir(
joinpath(
model.path,
"figure",
"simulation",
"$viz_type"
)
)
mkpath(
joinpath(
model.path,
"figure",
"simulation",
"$viz_type"
)
)
end
cmap = [
"#1f77b4", "#ff7f0e", "#2ca02c", "#d62728", "#9467bd",
"#8c564b", "#e377c2", "#7f7f7f", "#bcbd22", "#17becf"
]
shape = [
"o", "v", "^", "<", ">", "8", "s", "p", "*", "h", "H", "D", "d", "P", "X"
]
# rcParams
rc("figure", figsize=(4, 3))
rc("font", size=18)
rc("axes", linewidth=1.5)
rc("xtick.major", width=1.5)
rc("ytick.major", width=1.5)
rc("lines", linewidth=1.8)
rc("lines", markersize=12)
for (i, obs_name) in enumerate(model.observables)
gca().spines["right"].set_visible(false)
gca().spines["top"].set_visible(false)
gca().yaxis.set_ticks_position("left")
gca().xaxis.set_ticks_position("bottom")
if viz_type != "experiment"
if show_all
for j in eachindex(n_file)
if length(model.sim.normalization) > 0
norm_max = get_norm_max(model, i, j, obs_name, simulations_all)
end
for (k, condition) in enumerate(model.sim.conditions)
plot(
model.sim.t,
simulations_all[i, j, k, :] ./ ifelse(
length(model.sim.normalization) == 0 || maximum(simulations_all[i, j, k, :]) == 0.0,
1.0,
norm_max
),
color=cmap[k],
lw=0.5, alpha=0.35
)
end
end
end
if viz_type == "average"
normalized = Array{Float64,4}(
undef,
length(model.observables), length(n_file), length(model.sim.t), length(model.sim.conditions)
)
@inbounds for j in eachindex(n_file)
if length(model.sim.normalization) > 0
norm_max = get_norm_max(model, i, j, obs_name, simulations_all)
end
@simd for k in eachindex(model.sim.conditions)
normalized[i, j, k, :] = (
simulations_all[i, j, k, :] ./ ifelse(
length(model.sim.normalization) == 0 || maximum(simulations_all[i, j, k, :]) == 0.0,
1.0,
norm_max
)
)
end
end
if length(model.sim.normalization) > 0 && model.sim.normalization[obs_name]["timepoint"] === nothing
mean_norm_max::Float64 = maximum(
vcat(
[
[
mean(
filter(
!isnan, normalized[i, :, k, l]
)
) for l in eachindex(model.sim.t)
] for k in eachindex(model.sim.normalization[obs_name]["condition"])
]...
)
)
for j in eachindex(n_file)
for k in eachindex(model.sim.conditions)
for l in eachindex(model.sim.t)
if !isnan(mean_norm_max) && mean_norm_max != 0.0
@inbounds normalized[i, j, k, l] /= mean_norm_max
end
end
end
end
end
for (k, condition) in enumerate(model.sim.conditions)
plot(
model.sim.t, [
mean(
filter(
!isnan, normalized[i, :, k, l]
)
) for l in eachindex(model.sim.t)
],
color=cmap[k],
label=condition
)
end
if stdev
for (k, condition) in enumerate(model.sim.conditions)
y_mean = [
mean(
filter(
!isnan, normalized[i, :, k, l]
)
) for l in eachindex(model.sim.t)
]
y_std = [
std(
filter(
!isnan, normalized[i, :, k, l]
)
) for l in eachindex(model.sim.t)
]
fill_between(
model.sim.t,
y_mean - y_std, y_mean + y_std,
color=cmap[k],
lw=0,
alpha=0.1,
)
end
end
else
norm_max = length(model.sim.normalization) > 0 ? (
model.sim.normalization[obs_name]["timepoint"] !== nothing ? maximum(
model.sim.simulations[
i,
[model.cond2idx(c) for c in model.sim.normalization[obs_name]["condition"]],
model.sim.normalization[obs_name]["timepoint"],
]
) : maximum(
model.sim.simulations[
i,
[model.cond2idx(c) for c in model.sim.normalization[obs_name]["condition"]],
:,
]
)
) : 1.0
for (j, condition) in enumerate(model.sim.conditions)
plot(
model.sim.t,
model.sim.simulations[i, j, :] ./ ifelse(
length(model.sim.normalization) == 0 || maximum(model.sim.simulations[i, j, :]) == 0.0,
1.0,
norm_max
),
color=cmap[j],
label=condition
)
end
end
end
if isassigned(model.exp.experiments, i)
exp_t = model.exp.get_timepoint(obs_name)
if isassigned(model.exp.error_bars, i)
for (k, condition) in enumerate(model.sim.conditions)
if condition in keys(model.exp.experiments[i])
exp_data = errorbar(
exp_t,
model.exp.experiments[i][condition],
yerr=model.exp.error_bars[i][condition],
lw=1, markerfacecolor="None",
color=cmap[k],
markeredgecolor=cmap[k],
ecolor=cmap[k],
fmt=shape[k],
capsize=8,
clip_on=false
)
for capline in exp_data[2]
capline.set_clip_on(false)
end
for barlinecol in exp_data[3]
barlinecol.set_clip_on(false)
end
end
end
else
for (k, condition) in enumerate(model.sim.conditions)
if condition in keys(model.exp.experiments[i])
plot(
exp_t,
model.exp.experiments[i][condition],
shape[k],
color=cmap[k],
markerfacecolor="None",
markeredgecolor=cmap[k],
clip_on=false
)
end
end
end
end
xlabel("Time")
ylabel(replace(obs_name, "_" => " "))
savefig(
joinpath(
model.path,
"figure",
"simulation",
"$viz_type",
"$obs_name." * "$save_format"
),
dpi=save_format == "pdf" ? nothing : 600,
bbox_inches="tight"
)
close()
end
end
function run_simulation(
model::Model;
viz_type::String="original",
show_all::Bool=false,
stdev::Bool=false,
save_format::String="pdf")
if !isdir(
joinpath(
model.path,
"figure"
)
)
mkdir(
joinpath(
model.path,
"figure"
)
)
end
if !(viz_type in ["best", "average", "original", "experiment"])
try
parse(Int64, viz_type)
catch
error(
"Avairable viz_type are: 'best','average','original','experiment','n(=1,2,...)'"
)
end
end
n_file::Vector{Int} = viz_type in ["original", "experiment"] ? [] : get_executable(model)
simulaitons_all::Array{Float64,4} = fill(
NaN,
(
length(model.observables),
length(n_file),
length(model.sim.conditions),
length(model.sim.t),
)
)
if viz_type != "experiment"
if length(n_file) > 0
if length(n_file) == 1 && viz_type == "average"
error("viz_type should be best, not $viz_type")
end
for (j, nth_param_set) in enumerate(n_file)
(model, is_successful) = validate!(model, nth_param_set)
if is_successful
for i in eachindex(model.observables)
@inbounds simulaitons_all[i, j, :, :] = model.sim.simulations[i, :, :]
end
end
end
best_fitness_all::Vector{Float64} = fill(Inf, length(n_file))
for (i, nth_param_set) in enumerate(n_file)
local filepath = joinpath(
model.path,
"fitparam",
"$nth_param_set",
"best_fitness",
)
if isfile(filepath * ".dat")
best_fitness_all[i] = readdlm(
filepath * ".dat"
)[1, 1]
elseif isfile(filepath * ".npy")
best_fitness_all[i] = numpy_load(
filepath * ".npy"
)[1, 1]
end
end
best_param_set::Int = n_file[argmin(best_fitness_all)]
if viz_type == "best"
model, _ = validate!(model, best_param_set)
elseif viz_type != "average" && parse(Int64, viz_type) <= length(n_file)
model, _ = validate!(model, parse(Int64, viz_type))
elseif viz_type != "average" && parse(Int64, viz_type) > length(n_file)
error(
@sprintf(
"n (%d) must be smaller than n_fitparam (%d)",
parse(Int64, viz_type), length(n_file)
)
)
end
else
p::Vector{Float64} = param_values()
u0::Vector{Float64} = initial_values()
if model.sim.simulate!(p, u0) !== nothing
error(
"Simulation failed."
)
end
end
end
plot_timecourse(
model, n_file, viz_type, show_all, stdev, simulaitons_all, save_format
)
end | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 904 | using DelimitedFiles
using Sundials
using SteadyStateDiffEq
import BioMASS: isinstalled
@testset "Bifurcation analysis" begin
for model in ["restriction_point", "g1s_transition", "mitotic_control"]
@testset "$model" begin
MODEL_PATH = "../examples/bifurcation/" * model
create_diffeq(MODEL_PATH)
include(MODEL_PATH * "/diagram.jl")
fp, br = calc_fixed_point_vec(MODEL_PATH)
for file in ["/data/fp.dat", "/data/ev.dat"]
@test isfile(MODEL_PATH * file)
end
if isinstalled("matplotlib")
using PyPlot
bifurcation_diagram(MODEL_PATH, fp, br)
@test isfile(MODEL_PATH * "/bifurcation_diagram.pdf")
end
rm(MODEL_PATH * "/forwarddiff.jl")
rm(MODEL_PATH * "/data", recursive=true, force=true)
end
end
end
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 1228 | @testset "Parameter Estimation" begin
let model_ode = Model("../examples/fos_model")
output = []
@testset "optimization" begin
initpop = generate_initial_population(model_ode)
@test size(initpop) == (225, 75)
scipy_differential_evolution(model_ode, 1, maxiter=10, init=initpop)
lines = open(joinpath(model_ode.path, "fitparam", "1", "optimization.log"), "r") do f
readlines(f)
end
@test startswith(lines[end], "differential_evolution step 10:")
push!(output, "logs")
push!(output, "fitparam")
end
@testset "visualization" begin
@test run_simulation(model_ode, viz_type="best") === nothing
files = readdir(joinpath(model_ode.path, "figure", "simulation", "best"))
n_pdf = 0
for file in files
if occursin(".pdf", file)
n_pdf += 1
end
end
@test n_pdf == length(model_ode.observables)
push!(output, "figure")
end
for dir in output
rm(joinpath(model_ode.path, "$dir"), recursive=true, force=true)
end
end
end
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | code | 168 | using BioMASS
using Test
@time begin
@testset "BioMASS.jl" begin
include("parameter_estimation.jl")
include("bifurcation_analysis.jl")
end
end
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | docs | 3389 | # The BioMASS module for Julia
[](https://biomass-dev.github.io/BioMASS.jl/stable)
[](https://biomass-dev.github.io/BioMASS.jl/dev)
[](https://github.com/biomass-dev/BioMASS.jl/actions)
[](https://opensource.org/licenses/MIT)
[](https://doi.org/10.3390/cancers12102878)
This module provides a Julia interface to the [BioMASS](https://github.com/biomass-dev/biomass) parameter estimation.
## Installation
The package is a registered package, and can be installed with `Pkg.add`.
```julia
julia> using Pkg; Pkg.add("BioMASS")
```
or through the `pkg` REPL mode by typing
```
] add BioMASS
```
### Python package requirements:
- numpy - https://numpy.org
- scipy - https://scipy.org
- matplotlib - https://matplotlib.org
## Example
### Model development
This example shows you how to build a simple Michaelis-Menten two-step enzyme catalysis model.
> E + S ⇄ ES → E + P
[`pasmopy.Text2Model`](https://pasmopy.readthedocs.io/en/latest/model_development.html) allows you to build a BioMASS model from text. You simply describe biochemical reactions and the molecular mechanisms extracted from text are converted into an executable model.
Prepare a text file describing the biochemical reactions (e.g., `michaelis_menten.txt`)
```
E + S ⇄ ES | kf=0.003, kr=0.001 | E=100, S=50
ES → E + P | kf=0.002
@obs Substrate: u[S]
@obs E_free: u[E]
@obs E_total: u[E] + u[ES]
@obs Product: u[P]
@obs Complex: u[ES]
@sim tspan: [0, 100]
```
Convert the text into an executable model
```shell
$ python # pasmopy requires Python 3.7+
```
```python
>>> from pasmopy import Text2Model
>>> description = Text2Model("michaelis_menten.txt", lang="julia")
>>> description.convert() # generate 'michaelis_menten_jl/'
```
Simulate the model using BioMASS.jl
```shell
$ julia
```
```julia
using BioMASS
model = Model("./michaelis_menten_jl");
run_simulation(model)
```
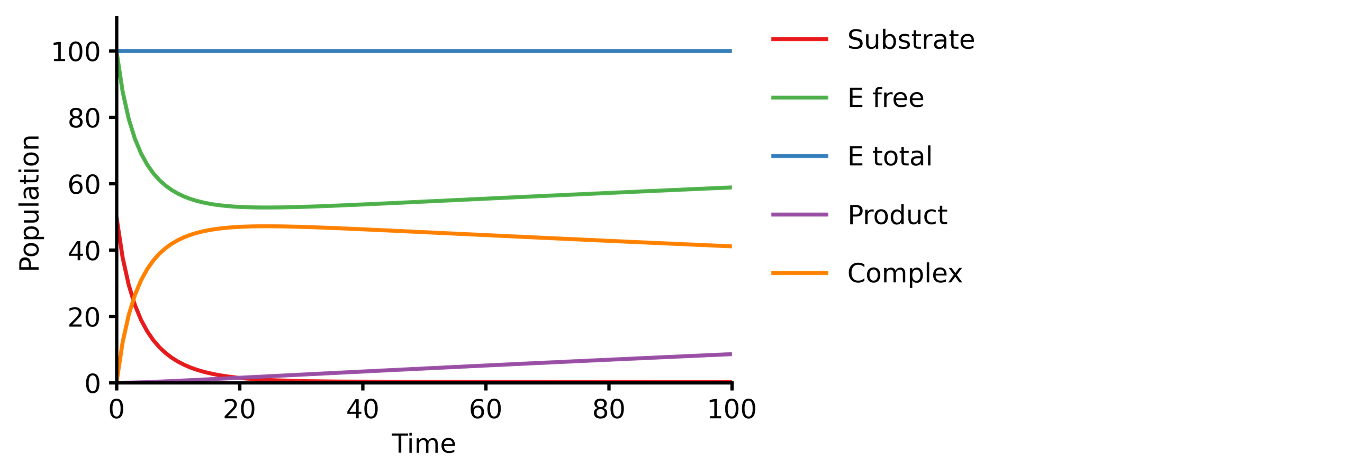
### Parameter estimation
```julia
using BioMASS
model = Model("./examples/fos_model");
# Estimate unknown model parameters from experimental observations
scipy_differential_evolution(model, 1) # requires scipy package
# Save simulation results to figure/ in the model folder
run_simulation(model, viz_type="best", show_all=true)
```
## References
- Imoto, H., Zhang, S. & Okada, M. A Computational Framework for Prediction and Analysis of Cancer Signaling Dynamics from RNA Sequencing Data—Application to the ErbB Receptor Signaling Pathway. _Cancers_ **12**, 2878 (2020). https://doi.org/10.3390/cancers12102878
- Imoto, H., Yamashiro, S. & Okada, M. A text-based computational framework for patient -specific modeling for classification of cancers. _iScience_ **25**, 103944 (2022). https://doi.org/10.1016/j.isci.2022.103944
## License
[MIT](https://github.com/biomass-dev/BioMASS.jl/blob/master/LICENSE)
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | docs | 1113 | # BioMASS.jl
[](https://opensource.org/licenses/MIT)
[](https://github.com/biomass-dev/BioMASS.jl)
[](https://github.com/biomass-dev/BioMASS.jl/actions)
[](https://doi.org/10.3390/cancers12102878)
This module provides a Julia interface to the [BioMASS](https://github.com/biomass-dev/biomass) parameter estimation.
The open access publication describing BioMASS is available here:
- Imoto, H., Zhang, S. & Okada, M. A Computational Framework for Prediction and Analysis of Cancer Signaling Dynamics from RNA Sequencing Data—Application to the ErbB Receptor Signaling Pathway. _Cancers_ **12**, 2878 (2020). https://doi.org/10.3390/cancers12102878
## Installation
```
] add BioMASS
```
```@contents
Pages = [
"usage/parameter_estimation.md",
"usage/bifurcation_analysis.md",
]
Depth = 3
```
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | docs | 1253 | - Imoto, H., Zhang, S. & Okada, M. A Computational Framework for Prediction and Analysis of Cancer Signaling Dynamics from RNA Sequencing Data—Application to the ErbB Receptor Signaling Pathway. _Cancers_ **12**, 2878 (2020). https://doi.org/10.3390/cancers12102878
- Imoto, H., Yamashiro, S. & Okada, M. A text-based computational framework for patient -specific modeling for classification of cancers. _iScience_ **25**, 103944 (2022). https://doi.org/10.1016/j.isci.2022.103944
- Nakakuki, T. *et al.* Ligand-specific c-Fos expression emerges from the spatiotemporal control of ErbB network dynamics. *Cell* **141**, 884–896 (2010). https://doi.org/10.1016/j.cell.2010.03.054
- Yao, G., Lee, T. J., Mori, S., Nevins, J. R. & You, L. A bistable Rb-E2F switch underlies the restriction point. *Nat. Cell Biol.* **10**, 476–482 (2008). https://doi.org/10.1038/ncb1711
- Barr, A. R., Heldt, F. S., Zhang, T., Bakal, C. & Novák, B. A Dynamical Framework for the All-or-None G1/S Transition. *Cell Syst.* **2**, 27–37 (2016). https://doi.org/10.1016/j.cels.2016.01.001
- Rata, S. *et al.* Two Interlinked Bistable Switches Govern Mitotic Control in Mammalian Cells. *Curr. Biol.* **28**, 3824-3832.e6 (2018). https://doi.org/10.1016/j.cub.2018.09.059
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | docs | 4137 | # Bifurcation Analysis
A numerical study of the changes in the dynamics and stability of a system upon variations in its parameters.

## Procedure for stability analysis at fixed points
Consider the following system of ordinary differential equations:
```math
\dfrac{dx}{dt} = F(x)
```
1. Determine the fixed point vector, ``x^*``, solving ``F(x^*) = 0``
1. Construct the Jacobian matrix, ``J(x) = \dfrac{\partial F(x)}{\partial x}``
1. Compute eigenvalues of ``J(x^*)``: ``|J(x^*) − λE| = 0``
1. Conclude on stability or instability of ``x^*`` based on the real parts of eigenvalues
- All eigenvalues have real parts less than zero → ``x^*`` is stable
- At least one of the eigenvalues has a real part greater than zero → ``x^*`` is unstable
## Usage
Here I would like to use a mathematical model of Rb–E2F pathway ([Yao et al., 2008](https://www.nature.com/articles/ncb1711)) to show you how to perform bifurcation analysis with [`BioMASS.jl`](https://github.com/biomass-dev/BioMASS.jl).
### Prepare `name2idx/` to define model species and parameters
- See [examples](https://github.com/biomass-dev/BioMASS.jl/tree/master/examples/bifurcation/restriction_point/name2idx).
### Create [`set_model.jl`](https://github.com/biomass-dev/BioMASS.jl/blob/master/examples/bifurcation/restriction_point/set_model.jl)
In this file, you will need to prepare four functions:
- `diffeq!`: Ordinary differential equations of the model.
- `param_values`: Model parameters.
- `get_derivatives`: ``\dfrac{\partial F(x)}{\partial bp}``, where ``bp`` is the bifurcation parameter (x-axis).
- `get_steady_state`: Function to equilibrate the system.
### Run `create_diffeq` function
```julia
create_diffeq(".")
```
Then you will get `forwarddiff.jl` file in your model folder.
### Load requirements
```julia
using DelimitedFiles
using Sundials
using SteadyStateDiffEq
using PyPlot
include("./name2idx/parameters.jl")
include("./name2idx/species.jl")
include("./set_model.jl")
include("./forwarddiff.jl")
const BP = C.S # name(index) of bifurcation parameter (x-axis)
const SN = V.NUM # num of state variables
const PN = 1 # num of parameters
const VN = SN + PN # num of variables
```
### Calculate fixed points and analyze their stability
After executing `new_curve!` function, you will get `data/fp.dat` and `data/ev.dat` files, where they contain fixed points and eigenvalues, respectively.
```julia
function calc_fixed_point_vec(model_path::String)::Tuple{Array,Array}
global p = param_values()
new_curve!(
model_path, p, diffeq, get_derivatives, get_steady_state,
direction=false, bifparam=BP, n_state=SN
)
fp::Array = readdlm(joinpath(model_path, "data", "fp.dat"), '\t', Float64, '\n')
ev::Array = readdlm(joinpath(model_path, "data", "ev.dat"), '\t', Float64, '\n')
br::Array = get_bistable_regime(ev, SN)
return fp, br
end
```
### Plot results
```julia
function bifurcation_diagram(model_path::String, fp::Array, br::Array)
rc("figure", figsize=(8, 6))
rc("font", family="Arial")
rc("font", size=24)
rc("axes", linewidth=1)
rc("xtick.major", width=1)
rc("ytick.major", width=1)
rc("lines", linewidth=3)
plot(fp[1:br[1]-1, VN+1], fp[1:br[1]-1, V.E+1], "k-")
plot(fp[br, VN+1], fp[br, V.E+1], lw=1.5, "k--")
plot(fp[br[end]+1:end, VN+1], fp[br[end]+1:end, V.E+1], "k-")
xlabel("Serum (percentage)")
ylabel("E2F (μM)")
xlim(0, 2)
xticks([0, 0.5, 1, 1.5, 2])
yscale("log")
ylim(1e-4, 2)
yticks([1e-4, 1e-2, 1])
savefig(joinpath(model_path, "bifurcation_diagram.pdf"), bbox_inches="tight")
close()
end
```
### Run all functions defined above
```julia
const MODEL_PATH = "."
fp, br = calc_fixed_point_vec(MODEL_PATH);
bifurcation_diagram(MODEL_PATH, fp, br);
```

*Stable (solid) and unstable (dashed) steady states of E2F activity with respect to serum stimulation.*
For more examples, please refer to [examples/bifurcation](https://github.com/biomass-dev/BioMASS.jl/tree/master/examples/bifurcation).
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | docs | 1586 | # Model Construction
[`pasmopy.Text2Model`](https://pasmopy.readthedocs.io/en/latest/model_development.html) allows you to build a BioMASS model from text. You simply describe biochemical reactions and the molecular mechanisms extracted from text are converted into an executable model.
## Example
This example shows you how to build a simple Michaelis-Menten two-step enzyme catalysis model with Pasmopy.
> E + S ⇄ ES → E + P
_An enzyme, E, binding to a substrate, S, to form a complex, ES, which in turn releases a product, P, regenerating the original enzyme._
1. Prepare a text file describing biochemical reactions (e.g., `michaelis_menten.txt`)
```
E binds S <--> ES | kf=0.003, kr=0.001 | E=100, S=50
ES dissociates to E and P | kf=0.002, kr=0
@obs Substrate: u[S]
@obs E_free: u[E]
@obs E_total: u[E] + u[ES]
@obs Product: u[P]
@obs Complex: u[ES]
@sim tspan: [0, 100]
```
1. Convert the text into an executable model
```shell
$ python
```
```python
>>> from pasmopy import Text2Model
>>> description = Text2Model("michaelis_menten.txt", lang="julia")
>>> description.convert() # generate 'michaelis_menten_jl/'
Model information
-----------------
2 reactions
4 species
4 parameters
```
1. Run simulation
```shell
$ julia
```
```julia
using BioMASS
model = Model("./michaelis_menten_jl");
run_simulation(model)
```
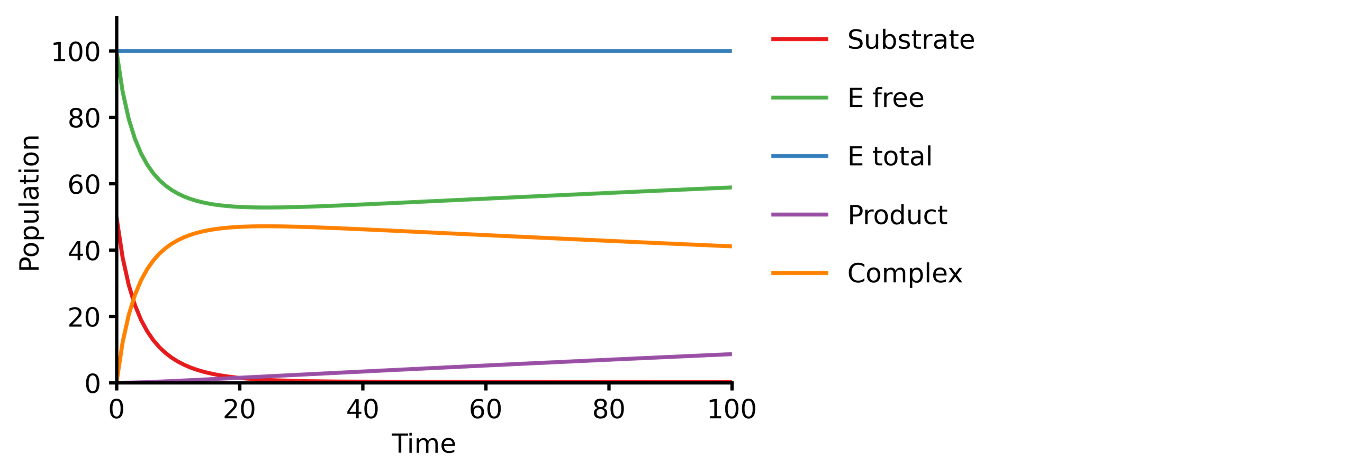 | BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.8.2 | 398e9ae41e9e5898e13166a251d542056f36e54c | docs | 4495 | # Parameter Estimation

## Core functions
---
### Model(`path_to_model`::String)
Load a BioMASS model. The model must include the following files:
| Name | Content |
| :--------------------- | :------------------------------------------------------------------------------------------------------- |
| `name2idx/` | Names of model parameters and species |
| `ode.jl` | Differential equation, parameters and initial condition |
| `observalbe.jl` | Model observables for correlating simulation results with experimental observations |
| `simulation.jl` | Simulation condition |
| `experimental_data.jl` | Experimental measurements |
| `search_param.jl` | Lower and upper bounds of model parameters to be estimated |
| `problem.jl` | An objective function to be minimized, i.e., the distance between model simulation and experimental data |
- **Parameters**
- `path_to_model`::String
- The model folder to read.
- **Returns**
- `model`::Model
- The executable model in BioMASS.
!!! note
[`pasmopy.Text2Model`](https://pasmopy.readthedocs.io/en/latest/model_development.html) allows you to build a BioMASS model from text [[Imoto et al., 2022](https://www.cell.com/iscience/fulltext/S2589-0042(22)00214-0)]. You simply describe biochemical reactions and the molecular mechanisms extracted from text are converted into an executable model. To build a model for BioMASS.jl, please set `lang="julia"`.
---
### scipy\_differential\_evolution(`model`::Model, `ix_id`::Int, `kwargs`...)
Estimate model parameters from experimental data.
- **Parameters**
- `model`::Model
- Model for parameter estimation.
- `x_id`::Int
- Index of parameter set to estimate.
- `kwargs`...
- Keyword arguments to pass to [`scipy.optimize.differential_evolution`](https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.differential_evolution.html.).
---
### run\_simulation(`model`::Model, `viz_type`::String, `show_all`::Bool=false, `stdev`::Bool=false)
Save simulation results with optimized parameter values.
- **Parameters**
- `viz_type`::String
- `"average"`
- `"best"`
- `"original"`
- `"experiment"`
- `show_all`::Bool (default: `false`)
- Whether to show all simulation results.
- `stdev`::Bool (default: `false`)
- If True, the standard deviation of simulated values will be shown
(only available for `"average"` visualization type).
- `save_format`::String (default: `"pdf"`)
- Either "png" or "pdf", indicating whether to save figures as png or pdf format.
## Estimate unknown model parameters
```julia
using BioMASS
model = Model("./examples/fos_model");
initpop = generate_initial_population(model)
scipy_differential_evolution(model, 1, init=initpop)
```
## Simultaneous parameter optimization
### Using module `Distributed`
```julia
using Distributed
addprocs(); # add worker processes
@everywhere using BioMASS
@everywhere begin
model = Model("./examples/fos_model")
function optimize_parallel(i)
scipy_differential_evolution(model, i)
end
end
pmap(optimize_parallel, 1:10)
```
### Calling multiple bash scripts
- main.jl
```julia
using BioMASS
model = Model("./examples/fos_model")
if abspath(PROGRAM_FILE) == @__FILE__
scipy_differential_evolution(model, parse(Int64, ARGS[1]))
end
```
- optimize_parallel.sh
```bash
#!/bin/sh
for i in $(seq 1 10); do
nohup julia main.jl $i >> errout/$i.log 2>&1 &
done
# To terminate the process,
# $ pgrep -f main.jl | xargs kill -9
```
Run optimize_parallel.sh
```bash
$ mkdir errout
$ sh optimize_parallel.sh
```
## How to track optimization process
The temporary result will be saved in `path_to_model/fitparam/n/optimization.log`.
```bash
$ tail examples/fos_model/fitparam/1/optimization.log
```
## Visualization of simulation results
The simulation results will be saved in `figure/`.
```julia
run_simulation(model, viz_type="best", show_all=true)
```
| BioMASS | https://github.com/biomass-dev/BioMASS.jl.git |
|
[
"MIT"
] | 0.5.2 | 7979027234c190955d3aa25ca5d9fbdf94e896c6 | code | 329 | using Documenter, HTMLTables
Documenter.makedocs(
modules=[HTMLTables],
format=Documenter.HTML(),
pages=["Home" => "index.md", "Reading" => "reading.md", "Writing" => "writing.md"],
sitename="HTMLTables.jl",
authors="Ceco Elijah Maples",
)
Documenter.deploydocs(repo="github.com/cecoeco/HTMLTables.jl.git")
| HTMLTables | https://github.com/cecoeco/HTMLTables.jl.git |
|
[
"MIT"
] | 0.5.2 | 7979027234c190955d3aa25ca5d9fbdf94e896c6 | code | 11643 | """
# HTMLTables
Julia package for reading and writing HTML tables using the Tables.jl interface
## exported functions:
- `HTMLTables.readtable`: reads an HTML table into a Julia table such as a `DataFrame`.
- `HTMLTables.writetable`: converts a Julia table to an HTML table and writes it to a file or IO stream.
"""
module HTMLTables
using Cascadia, Colors, ColorSchemes, Gumbo, HTTP, Tables
export readtable, writetable
include("themes.jl")
function isurl(source::String)::Bool
return Base.startswith(source, r"^(http://|https://|ftp://)")
end
function ishtmlfile(source::String)::Bool
return Base.Filesystem.splitext(source)[2] == ".html"
end
function extract_row_data(row::Gumbo.HTMLNode)::Vector
cells::Vector{Gumbo.HTMLNode} = Base.eachmatch(Cascadia.Selector("td,th"), row)
return [Cascadia.nodeText(cell) for cell in cells]
end
"""
readtable(
source,
sink;
id::String="",
class::Union{String,Vector{String}}="",
index::Int=1
)
Reads an HTML table into a sink function such as `DataFrame`.
## Arguments
- `source`: URL or path to the HTML table.
- `sink`: the function that materializes the table data.
## Keyword Arguments
- `id::String`: the id of the HTML table in the HTML document.
- `class::Union{String,Vector{String}}`: the class of the HTML table.
- `index::Int`: the index of the HTML table in the HTML document.
## Returns
- `sink`: the sink function such as `DataFrame` with the HTML table data.
## Examples
reading an HTML table from a website into a DataFrame:
```julia
using HTMLTables, DataFrames
url = "https://www.w3schools.com/html/html_tables.asp"
df = HTMLTables.readtable(url, DataFrame, index=1)
println(df)
```
output:
```
6×3 DataFrame
Row │ Company Contact Country
│ String String String
─────┼─────────────────────────────────────────────────────────
1 │ Alfreds Futterkiste Maria Anders Germany
2 │ Centro comercial Moctezuma Francisco Chang Mexico
3 │ Ernst Handel Roland Mendel Austria
4 │ Island Trading Helen Bennett UK
5 │ Laughing Bacchus Winecellars Yoshi Tannamuri Canada
6 │ Magazzini Alimentari Riuniti Giovanni Rovelli Italy
```
"""
function readtable(
source, sink; id::String="", class::Union{String,Vector{String}}="", index::Int=1
)
if isa(source, IO)
source = Base.read(source, String)
end
html_content::String = ""
if isurl(source) == true
html_content *= Base.String(HTTP.get(source).body)
elseif ishtmlfile(source) == true
html_content *= Base.read(source, String)
else
html_content *= source
end
html_document::Gumbo.HTMLDocument = Gumbo.parsehtml(html_content)
selector::String = ""
if Base.isempty(id)
if Base.isempty(class)
selector *= "table"
elseif !Base.isempty(class) && Base.isa(class, String)
selector *= "table.$class"
elseif !Base.isempty(class) && Base.isa(class, Vector{String})
selector *= "table." * Base.join(class, ".")
end
elseif !Base.isempty(id)
selector *= "#$id"
end
tables::Vector{Gumbo.HTMLNode} = Base.eachmatch(
Cascadia.Selector(selector), html_document.root
)
table::Gumbo.HTMLNode = tables[index]
rows::Vector{Gumbo.HTMLNode} = Base.eachmatch(Cascadia.Selector("tr"), table)
headers::Vector = []
data::Vector{Vector} = []
for (i, row) in Base.enumerate(rows)
rowdata = extract_row_data(row)
if (i == 1 && Base.isempty(headers)) == true
headers = rowdata
else
Base.push!(data, rowdata)
end
end
tuples::Vector = [Base.Tuple(row) for row in data]
return sink(tuples, headers)
end
function writetheme(theme::Symbol; styles::Bool)::String
if theme == "" || !styles
return ""
end
theme_dictionary::Dict{Symbol,String} = Base.Dict(
:default => DEFAULT,
:red => RED,
:orange => ORANGE,
:yellow => YELLOW,
:green => GREEN,
:blue => BLUE,
:violet => VIOLET,
:magenta => MAGENTA,
:brown => BROWN,
:gray => GRAY,
:black => BLACK,
:gold => GOLD,
:silver => SILVER,
:bronze => BRONZE,
:julia => JULIA,
:sunstone => SUNSTONE,
:moonstone => MOONSTONE,
:dracula => DRACULA,
:solarized => SOLARIZED
)
if Base.haskey(theme_dictionary, theme)
theme = theme_dictionary[theme]
else
Base.throw(Base.ArgumentError("$(theme) is not a valid theme"))
end
return """<style>\n$theme\n</style>\n"""
end
function writetheme(theme::String; styles::Bool)::String
return writetheme(Symbol(theme); styles=styles)
end
function iscssfile(file::String)::Bool
if file == ""
return false
end
return Base.splitext(file)[end] == ".css"
end
function writestyle(css::String; styles::Bool)::String
if css == "" || !styles
return ""
end
if iscssfile(css)
css::String = Base.read(css, String)
end
return """<style>\n$css\n</style>\n"""
end
function writeid(id::String)::String
if id == ""
return ""
else
return " id=\"$id\""
end
end
function writeclass(class::String)::String
if class == ""
return ""
else
return " class=\"$class\""
end
end
function writeclass(class::Vector)::String
if class == []
return ""
else
return " class=\"" * Base.join(class, " ") * "\""
end
end
function writecaption(caption::String)::String
if caption == ""
return ""
else
return "<caption>$caption</caption>\n"
end
end
function writetooltip(tooltips::Bool, cell_value)::String
if tooltips
return " title=\"$cell_value\""
else
return ""
end
end
function writethead(tbl; header::Bool, editable::Bool)::String
if !header
return ""
end
contenteditable::String = ""
if editable
contenteditable *= " contenteditable=\"true\""
else
contenteditable *= ""
end
thead::String = "<thead$contenteditable>\n<tr>\n"
for col in Base.names(tbl)
thead *= "<td$contenteditable>$col</td>\n"
end
thead *= "</tr>\n</thead>\n"
return thead
end
function getnumbers(tbl)::Vector{Float64}
numbers::Vector{Float64} = Float64[]
for col in Base.names(tbl)
for val in tbl[!, col]
if Base.isa(val, Number)
Base.push!(numbers, val)
end
end
end
return numbers
end
function css_rgb(color::Colors.Colorant)::String
r::Float64 = Colors.red(color) * 255
g::Float64 = Colors.green(color) * 255
b::Float64 = Colors.blue(color) * 255
return "rgb(" * Base.join(["$r", "$g", "$b"], ",") * ")"
end
function cellcolor(tbl; colorscale::String, cell_value, styles::Bool)::String
numbers::Vector{Number} = getnumbers(tbl)
if colorscale == "" || Base.ismissing(cell_value) || !(cell_value in numbers) || !styles
return ""
end
colorscheme::ColorSchemes.ColorScheme = Base.getfield(ColorSchemes, Symbol(colorscale))
cell_position::Float64 =
(cell_value - Base.minimum(numbers)) /
(Base.maximum(numbers) - Base.minimum(numbers))
color::Colors.Colorant = ColorSchemes.get(colorscheme, cell_position)
css_color::String = css_rgb(color)
return " style=\"background-color: $css_color;\""
end
function writetbody(
tbl; colorscale::String, tooltips::Bool, styles::Bool, editable::Bool
)::String
contenteditable::String = ""
if editable
contenteditable *= " contenteditable=\"true\""
else
contenteditable *= ""
end
tbody::String = "<tbody $contenteditable>\n"
for row in Tables.rows(tbl)
tbody *= "<tr $contenteditable>\n"
for col in Base.names(tbl)
cell_value = row[Base.Symbol(col)]
cell::String = ""
cell *= "<td $contenteditable"
cell *= writetooltip(tooltips, cell_value)
cell *= cellcolor(
tbl; colorscale=colorscale, cell_value=cell_value, styles=styles
)
cell *= ">$cell_value</td>\n"
tbody *= cell
end
tbody *= "</tr>\n"
end
tbody *= "</tbody>\n"
return tbody
end
function writetfoot(tbl; footer::Bool, editable::Bool)::String
if !footer
return ""
end
contenteditable::String = ""
if editable
contenteditable *= " contenteditable=\"true\""
else
contenteditable *= ""
end
tfoot::String = "<tfoot $contenteditable>\n<tr>\n"
for _ in Base.names(tbl)
tfoot *= "<td $contenteditable></td>\n"
end
tfoot *= "</tr>\n</tfoot>\n"
return tfoot
end
"""
writetable(
out,
tbl;
header::Bool=true,
footer::Bool=true,
id::String="",
class::Union{String,Vector{String}}="",
caption::String="",
editable::Bool=false,
tooltips::Bool=true,
styles::Bool=true,
css::String="",
theme::Union{String,Symbol}=:default,
colorscale::Union{String,Symbol}=""
)
Uses the Tables.jl interface to write an HTML table.
## Arguments
- `out`: accepts the same types as [`Base.write`](https://docs.julialang.org/en/v1/base/io-network/#Base.write).
- `tbl`: the table to write.
## Keyword Arguments
- `header::Bool`: whether to include the table header.
- `footer::Bool`: whether to include the table footer.
- `id::String`: the id of the HTML table.
- `class::Union{String,Vector{String}}`: the class of the HTML table.
- `caption::String`: the caption of the HTML table.
- `editable::Bool`: whether to enable table editing.
- `tooltips::Bool`: whether to include tooltips.
- `styles::Bool`: whether to include the CSS. If false `css`, `theme` and `colorscale` are ignored.
- `css::String`: the path to the CSS file to include.
- `theme::Union{Symbol,String}`: the theme of the HTML table.
- `colorscale::Union{Symbol,String}`: the colorscale of the HTML table.
## Examples
creates a simple HTML table from a DataFrame and writes it to the standard output:
```julia
using HTMLTables, DataFrames
df = DataFrame(x=[1, 2, 3], y=[45, 67, 89])
HTMLTables.writetable(stdout, df)
```
creates a simple HTML table from a DataFrame and writes it to a file:
```julia
using HTMLTables, DataFrames
df = DataFrame(x=[1, 2, 3], y=[4, 11, 28])
HTMLTables.writetable("table.html", df)
```
"""
function writetable(
out,
tbl;
header::Bool=true,
footer::Bool=true,
id::String="",
class::Union{String,Vector{String}}="",
caption::String="",
editable::Bool=false,
tooltips::Bool=true,
styles::Bool=true,
css::String="",
theme::Union{String,Symbol}=:default,
colorscale::Union{String,Symbol}="",
)
html_table::String = ""
html_table *= writetheme(theme; styles=styles)
html_table *= writestyle(css; styles=styles)
html_table *= "<table$(writeid(id))$(writeclass(class))>\n"
html_table *= writecaption(caption)
html_table *= writethead(tbl; header=header, editable=editable)
html_table *= writetbody(
tbl; colorscale=colorscale, tooltips=tooltips, styles=styles, editable=editable
)
html_table *= writetfoot(tbl; footer=footer, editable=editable)
html_table *= "</table>"
Base.write(out, html_table)
return nothing
end
end
| HTMLTables | https://github.com/cecoeco/HTMLTables.jl.git |
|
[
"MIT"
] | 0.5.2 | 7979027234c190955d3aa25ca5d9fbdf94e896c6 | code | 7588 | const NORMALIZE::String = """
* {
margin: 0;
padding: 0;
box-sizing: border-box;
}
html, body {
display: flex;
flex-direction: column;
justify-content: center;
align-items: center;
width: 100%;
height: 100%;
}
"""
const BASIC_STYLING::String = """
table {
font-family: Helvetica;
font-size: 15px;
color: black;
border: 0;
border-spacing: 0;
border-collapse: collapse;
border-top: 1px solid black;
border-bottom: 1px solid black;
cursor: default;
}
caption {
caption-side: top;
padding: 10px;
}
thead {
font-weight: bold;
border-bottom: 1px solid black;
}
tbody td:hover {
text-decoration: underline;
}
th, td {
padding: 5px 10px;
text-align: center;
}
tfoot {
height: 20px;
border-top: 1px solid black;
}
"""
const DEFAULT::String = """
$NORMALIZE
$BASIC_STYLING
table {
background-color: white;
}
tfoot {
display: none;
}
"""
const RED::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background-color: hsl(0, 100%, 60%);
}
tbody tr:nth-child(odd) {
background-color: hsl(0, 100%, 75%);
}
tbody tr:nth-child(even) {
background-color: hsl(0, 100%, 85%);
}
"""
const ORANGE::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background-color: hsl(25, 100%, 60%);
}
tbody tr:nth-child(odd) {
background-color: hsl(25, 100%, 75%);
}
tbody tr:nth-child(even) {
background-color: hsl(25, 100%, 85%);
}
"""
const YELLOW::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background-color: hsl(60, 100%, 60%);
}
tbody tr:nth-child(odd) {
background-color: hsl(60, 100%, 75%);
}
tbody tr:nth-child(even) {
background-color: hsl(60, 100%, 85%);
}
"""
const GREEN::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background-color: hsl(115, 100%, 60%);
}
tbody tr:nth-child(odd) {
background-color: hsl(115, 100%, 75%);
}
tbody tr:nth-child(even) {
background-color: hsl(115, 100%, 85%);
}
"""
const BLUE::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background-color: hsl(205, 100%, 60%);
}
tbody tr:nth-child(odd) {
background-color: hsl(205, 100%, 75%);
}
tbody tr:nth-child(even) {
background-color: hsl(205, 100%, 85%);
}
"""
const VIOLET::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background-color: hsl(260, 100%, 60%);
}
tbody tr:nth-child(odd) {
background-color: hsl(260, 100%, 75%);
}
tbody tr:nth-child(even) {
background-color: hsl(260, 100%, 85%);
}
"""
const MAGENTA::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background-color: hsl(320, 100%, 60%);
}
tbody tr:nth-child(odd) {
background-color: hsl(320, 100%, 75%);
}
tbody tr:nth-child(even) {
background-color: hsl(320, 100%, 85%);
}
"""
const BROWN::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background-color: hsl(20, 40%, 50%);
}
tbody tr:nth-child(odd) {
background-color: hsl(20, 40%, 65%);
}
tbody tr:nth-child(even) {
background-color: hsl(20, 40%, 75%);
}
"""
const GRAY::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background-color: hsl(0, 0%, 60%);
}
tbody tr:nth-child(odd) {
background-color: hsl(0, 0%, 75%);
}
tbody tr:nth-child(even) {
background-color: hsl(0, 0%, 85%);
}
"""
const BLACK::String = """
$NORMALIZE
table {
font-family: Helvetica;
font-size: 15px;
color: white;
border: 0;
border-spacing: 0;
border-collapse: collapse;
border-top: 1px solid white;
border-bottom: 1px solid white;
cursor: default;
}
caption {
caption-side: top;
padding: 10px;
}
thead {
font-weight: bold;
border-bottom: 1px solid white;
}
tbody td:hover {
text-decoration: underline;
}
th, td {
padding: 5px 10px;
text-align: center;
}
tfoot {
height: 20px;
border-top: 1px solid white;
}
thead, tfoot {
background-color: hsl(0, 0%, 0%);
}
tbody tr:nth-child(odd) {
background-color: hsl(0, 0%, 10%);
}
tbody tr:nth-child(even) {
background-color: hsl(0, 0%, 20%);
}
"""
const GOLD::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background: linear-gradient(135deg, #FFD700 0%, #FFFACD 100%);
}
tbody tr:nth-child(odd) {
background: linear-gradient(135deg, #FFF8DC 0%, #FFD700 100%);
}
tbody tr:nth-child(even) {
background: linear-gradient(135deg, #FFFACD 0%, #FFD700 100%);
}
"""
const SILVER::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background: linear-gradient(135deg, #C0C0C0 0%, #E0E0E0 100%);
}
tbody tr:nth-child(odd) {
background: linear-gradient(135deg, #D3D3D3 0%, #C0C0C0 100%);
}
tbody tr:nth-child(even) {
background: linear-gradient(135deg, #E0E0E0 0%, #C0C0C0 100%);
}
"""
const BRONZE::String = """
$NORMALIZE
$BASIC_STYLING
thead, tfoot {
background: linear-gradient(135deg, #CD7F32 0%, #F4A460 100%);
}
tbody tr:nth-child(odd) {
background: linear-gradient(135deg, #D2B48C 0%, #CD7F32 100%);
}
tbody tr:nth-child(even) {
background: linear-gradient(135deg, #F4A460 0%, #CD7F32 100%);
}
"""
const JULIA::String = """
@import url("https://fonts.googleapis.com/css2?family=Arya:wght@400;700");
$NORMALIZE
$BASIC_STYLING
table {
font-size: 15px;
color: #222222;
background-color: white;
}
thead, tfoot {
font-family: "Arya";
background-color: #a270ba;
}
"""
const SUNSTONE::String = """
@import url('https://fonts.googleapis.com/css2?family=Caladea:ital,wght@0,400;0,700;1,400;1,700&display=swap');
$NORMALIZE
table {
font-family: "Arial";
font-size: 15px;
color: hsl(20, 100%, 15%);
border: 0;
border-spacing: 0;
border-collapse: collapse;
border-top: 2px solid #ff8850;
border-bottom: 2px solid #ff8850;
cursor: default;
}
caption {
caption-side: top;
padding: 10px;
}
thead {
font-family: "Caladea";
font-weight: bold;
border-bottom: 2px solid #ff8850;
color: #fffdce
}
thead,
tfoot {
height: 25px;
min-height: 25px;
background-color: #e16c52;
}
tbody td:hover {
text-decoration: underline;
}
tbody tr:nth-child(odd) {
background-color: #ffaf91;
}
tbody tr:nth-child(even) {
background-color: #ee8c76;
}
th, td {
padding: 5px 10px;
text-align: center;
}
tfoot {
border-top: 2px solid #ff8850;
}
"""
const MOONSTONE::String = """
@import url('https://fonts.googleapis.com/css2?family=Caladea:ital,wght@0,400;0,700;1,400;1,700&display=swap');
$NORMALIZE
table {
font-family: "Arial";
font-size: 15px;
color: hsl(220, 100%, 20%);
border: 0;
border-spacing: 0;
border-collapse: collapse;
border-top: 2px solid #477edf;
border-bottom: 2px solid #477edf;
cursor: default;
}
caption {
caption-side: top;
padding: 10px;
}
thead {
font-family: "Caladea";
font-weight: bold;
border-bottom: 2px solid #477edf;
color: #fffdcc
}
thead,
tfoot {
height: 25px;
min-height: 25px;
background-color: #88c3ff;
}
tbody td:hover {
text-decoration: underline;
}
tbody tr:nth-child(odd) {
background-color: #f5faff;
}
tbody tr:nth-child(even) {
background-color: #c8e4ff;
}
th, td {
padding: 5px 10px;
text-align: center;
}
tfoot {
border-top: 2px solid #477edf;
}
"""
# https://draculatheme.com/
const DRACULA::String = """
$NORMALIZE
$BASIC_STYLING
thead {
color: #bd93f9;
}
thead, tfoot {
background-color: hsl(232, 15%, 18%);
}
tbody tr:nth-child(odd) {
background-color: hsl(232, 15%, 31%);
}
tbody tr:nth-child(even) {
background-color: hsl(232, 15%, 25%);
}
"""
# https://ethanschoonover.com/solarized/
const SOLARIZED::String = """
$NORMALIZE
$BASIC_STYLING
thead {
color: #2aa198;
}
thead, tfoot, tbody tr:nth-child(odd) {
background-color: hsl(45, 81%, 95%);
}
tbody tr:nth-child(even) {
background-color: hsl(45, 41%, 90%);
}
"""
| HTMLTables | https://github.com/cecoeco/HTMLTables.jl.git |
|
[
"MIT"
] | 0.5.2 | 7979027234c190955d3aa25ca5d9fbdf94e896c6 | code | 4815 | @testset "read HTML table from HTML documents using DataFrame" begin
df_01::DataFrame = HTMLTables.readtable(HTML_FILE_01, DataFrame)
df_01[!, :Age] = parse.(Int, df_01[!, :Age])
@test size(df_01) == (3, 3)
@test names(df_01) == ["Name", "Age", "Occupation"]
@test df_01[1, 1] == "Alice"
@test df_01[1, 2] == 30
@test df_01[1, 3] == "Engineer"
@test df_01[2, 1] == "Bob"
@test df_01[2, 2] == 25
@test df_01[2, 3] == "Designer"
@test df_01[3, 1] == "Charlie"
@test df_01[3, 2] == 35
@test df_01[3, 3] == "Teacher"
@test isa(df_01, DataFrame)
@test isa(df_01[1, 1], String)
@test isa(df_01[1, 2], Int)
@test isa(df_01[1, 3], String)
@test isa(df_01[2, 1], String)
@test isa(df_01[2, 2], Int)
@test isa(df_01[2, 3], String)
@test isa(df_01[3, 1], String)
@test isa(df_01[3, 2], Int)
@test isa(df_01[3, 3], String)
df_02::DataFrame = HTMLTables.readtable(HTML_FILE_02, DataFrame)
@test size(df_02) == (3, 3)
@test names(df_02) == ["Name", "Age", "Occupation"]
@test isa(df_02, DataFrame)
@test all(x -> x == "" || ismissing(x), df_02[!, :Name])
@test all(x -> x == "" || ismissing(x), df_02[!, :Age])
@test all(x -> x == "" || ismissing(x), df_02[!, :Occupation])
for row in eachindex(df_02[:, 1])
@test df_02[row, :Name] == "" || ismissing(df_02[row, :Name])
@test df_02[row, :Age] == "" || ismissing(df_02[row, :Age])
@test df_02[row, :Occupation] == "" || ismissing(df_02[row, :Occupation])
end
end
@testset "read HTML table from HTML string using DataFrame" begin
df_01::DataFrame = HTMLTables.readtable(Base.read(HTML_FILE_01, String), DataFrame)
df_01[!, :Age] = parse.(Int, df_01[!, :Age])
@test size(df_01) == (3, 3)
@test names(df_01) == ["Name", "Age", "Occupation"]
@test df_01[1, 1] == "Alice"
@test df_01[1, 2] == 30
@test df_01[1, 3] == "Engineer"
@test df_01[2, 1] == "Bob"
@test df_01[2, 2] == 25
@test df_01[2, 3] == "Designer"
@test df_01[3, 1] == "Charlie"
@test df_01[3, 2] == 35
@test df_01[3, 3] == "Teacher"
@test isa(df_01, DataFrame)
@test isa(df_01[1, 1], String)
@test isa(df_01[1, 2], Int)
@test isa(df_01[1, 3], String)
@test isa(df_01[2, 1], String)
@test isa(df_01[2, 2], Int)
@test isa(df_01[2, 3], String)
@test isa(df_01[3, 1], String)
@test isa(df_01[3, 2], Int)
@test isa(df_01[3, 3], String)
df_02::DataFrame = HTMLTables.readtable(Base.read(HTML_FILE_02, String), DataFrame)
@test size(df_02) == (3, 3)
@test names(df_02) == ["Name", "Age", "Occupation"]
@test isa(df_02, DataFrame)
@test all(x -> x == "" || ismissing(x), df_02[!, :Name])
@test all(x -> x == "" || ismissing(x), df_02[!, :Age])
@test all(x -> x == "" || ismissing(x), df_02[!, :Occupation])
for row in eachindex(df_02[:, 1])
@test df_02[row, :Name] == "" || ismissing(df_02[row, :Name])
@test df_02[row, :Age] == "" || ismissing(df_02[row, :Age])
@test df_02[row, :Occupation] == "" || ismissing(df_02[row, :Occupation])
end
end
@testset "read HTML table from IO using DataFrame" begin
html_io_01::IO = open(HTML_FILE_01, "r")
df_01::DataFrame = HTMLTables.readtable(html_io_01, DataFrame)
df_01[!, :Age] = parse.(Int, df_01[!, :Age])
@test size(df_01) == (3, 3)
@test names(df_01) == ["Name", "Age", "Occupation"]
@test df_01[1, 1] == "Alice"
@test df_01[1, 2] == 30
@test df_01[1, 3] == "Engineer"
@test df_01[2, 1] == "Bob"
@test df_01[2, 2] == 25
@test df_01[2, 3] == "Designer"
@test df_01[3, 1] == "Charlie"
@test df_01[3, 2] == 35
@test df_01[3, 3] == "Teacher"
@test isa(df_01, DataFrame)
@test isa(df_01[1, 1], String)
@test isa(df_01[1, 2], Int)
@test isa(df_01[1, 3], String)
@test isa(df_01[2, 1], String)
@test isa(df_01[2, 2], Int)
@test isa(df_01[2, 3], String)
@test isa(df_01[3, 1], String)
@test isa(df_01[3, 2], Int)
@test isa(df_01[3, 3], String)
html_io_02::IO = Base.open(HTML_FILE_02, "r")
df_02::DataFrame = HTMLTables.readtable(html_io_02, DataFrame)
@test size(df_02) == (3, 3)
@test names(df_02) == ["Name", "Age", "Occupation"]
@test isa(df_02, DataFrame)
@test all(x -> x == "" || ismissing(x), df_02[!, :Name])
@test all(x -> x == "" || ismissing(x), df_02[!, :Age])
@test all(x -> x == "" || ismissing(x), df_02[!, :Occupation])
for row in eachindex(df_02[:, 1])
@test df_02[row, :Name] == "" || ismissing(df_02[row, :Name])
@test df_02[row, :Age] == "" || ismissing(df_02[row, :Age])
@test df_02[row, :Occupation] == "" || ismissing(df_02[row, :Occupation])
end
end | HTMLTables | https://github.com/cecoeco/HTMLTables.jl.git |
|
[
"MIT"
] | 0.5.2 | 7979027234c190955d3aa25ca5d9fbdf94e896c6 | code | 1103 | module TestHTMLTables
using DataFrames, HTMLTables, Test
const DIR::String = Base.Filesystem.dirname(Base.@__FILE__)
const CSS_DIR::String = Base.Filesystem.joinpath(DIR, "css")
const CSS_FILE_01::String = Base.Filesystem.joinpath(CSS_DIR, "example-01.css")
const CSS_FILE_02::String = Base.Filesystem.joinpath(CSS_DIR, "example-02.css")
const CSS_STRING_01::String = Base.read(CSS_FILE_01, String)
const CSS_STRING_02::String = Base.read(CSS_FILE_02, String)
const HTML_DIR::String = Base.Filesystem.joinpath(DIR, "html")
const HTML_FILE_01::String = Base.Filesystem.joinpath(HTML_DIR, "example-01.html")
const HTML_FILE_02::String = Base.Filesystem.joinpath(HTML_DIR, "example-02.html")
const HTML_STRING_01::String = Base.read(HTML_FILE_01, String)
const HTML_STRING_02::String = Base.read(HTML_FILE_02, String)
const URL_01::String = "https://www.w3schools.com/html/html_tables.asp"
const GLOBAL_DF_01::DataFrame = HTMLTables.readtable(HTML_FILE_01, DataFrame)
const GLOBAL_DF_02::DataFrame = HTMLTables.readtable(HTML_FILE_02, DataFrame)
include("readtests.jl")
include("writetests.jl")
end | HTMLTables | https://github.com/cecoeco/HTMLTables.jl.git |
|
[
"MIT"
] | 0.5.2 | 7979027234c190955d3aa25ca5d9fbdf94e896c6 | code | 7173 | @testset "built-in CSS theme" begin
themes::Vector{Symbol} = [
:default,
:red,
:orange,
:yellow,
:green,
:blue,
:violet,
:magenta,
:brown,
:gray,
:black,
:gold,
:silver,
:bronze,
:julia,
:sunstone,
:moonstone,
:dracula,
:solarized
]
for theme in themes
io_01::IO = IOBuffer()
HTMLTables.writetable(io_01, GLOBAL_DF_01; theme=theme)
table_01::String = String(Base.take!(io_01))
Base.close(io_01)
@test occursin("<style>", table_01)
io_02::IO = IOBuffer()
HTMLTables.writetable(io_02, GLOBAL_DF_01; theme=string(theme))
table_02::String = String(Base.take!(io_02))
Base.close(io_02)
@test occursin("<style>", table_02)
@test table_01 == table_02
end
end
@testset "CSS file" begin
io_01::IO = IOBuffer()
HTMLTables.writetable(io_01, GLOBAL_DF_01; css=CSS_FILE_01)
table_01::String = String(Base.take!(io_01))
Base.close(io_01)
@test occursin("<style>", table_01)
io_02::IO = IOBuffer()
HTMLTables.writetable(io_02, GLOBAL_DF_01; css=CSS_FILE_02)
table_02::String = String(Base.take!(io_02))
Base.close(io_02)
@test occursin("<style>", table_02)
end
@testset "CSS string" begin
io_01::IO = IOBuffer()
HTMLTables.writetable(io_01, GLOBAL_DF_01; css=CSS_STRING_01)
table_01::String = String(Base.take!(io_01))
Base.close(io_01)
@test occursin("<style>", table_01)
io_02::IO = IOBuffer()
HTMLTables.writetable(io_02, GLOBAL_DF_01; css=CSS_STRING_02)
table_02::String = String(Base.take!(io_02))
Base.close(io_02)
@test occursin("<style>", table_02)
end
@testset "header" begin
io_01::IO = IOBuffer()
HTMLTables.writetable(io_01, GLOBAL_DF_01; header=true)
table_01::String = String(Base.take!(io_01))
Base.close(io_01)
@test occursin("<thead", table_01)
io_02::IO = IOBuffer()
HTMLTables.writetable(io_02, GLOBAL_DF_01; header=false)
table_02::String = String(Base.take!(io_02))
Base.close(io_02)
@test !occursin("<thead", table_02)
io_03::IO = IOBuffer()
HTMLTables.writetable(io_03, GLOBAL_DF_01)
table_03::String = String(Base.take!(io_03))
Base.close(io_03)
@test occursin("<thead", table_03)
end
@testset "footer" begin
io_01::IO = IOBuffer()
HTMLTables.writetable(io_01, GLOBAL_DF_01; footer=true)
table_01::String = String(Base.take!(io_01))
Base.close(io_01)
@test occursin("<tfoot", table_01)
io_02::IO = IOBuffer()
HTMLTables.writetable(io_02, GLOBAL_DF_01; footer=false)
table_02::String = String(Base.take!(io_02))
Base.close(io_02)
@test !occursin("<tfoot", table_02)
io_03::IO = IOBuffer()
HTMLTables.writetable(io_03, GLOBAL_DF_01)
table_03::String = String(Base.take!(io_03))
Base.close(io_03)
@test occursin("<tfoot", table_03)
end
@testset "id" begin
io_01::IO = IOBuffer()
HTMLTables.writetable(io_01, GLOBAL_DF_01; id="table_01")
table_01::String = String(Base.take!(io_01))
Base.close(io_01)
@test occursin("id=\"table_01\"", table_01)
io_02::IO = IOBuffer()
HTMLTables.writetable(io_02, GLOBAL_DF_01; id="table_02")
table_02::String = String(Base.take!(io_02))
Base.close(io_02)
@test occursin("id=\"table_02\"", table_02)
end
@testset "class" begin
io_01::IO = IOBuffer()
HTMLTables.writetable(io_01, GLOBAL_DF_01; class="table_01")
table_01::String = String(Base.take!(io_01))
Base.close(io_01)
@test occursin("class=\"table_01\"", table_01)
io_02::IO = IOBuffer()
HTMLTables.writetable(io_02, GLOBAL_DF_01; class="table_02")
table_02::String = String(Base.take!(io_02))
Base.close(io_02)
@test occursin("class=\"table_02\"", table_02)
io_03::IO = IOBuffer()
HTMLTables.writetable(io_03, GLOBAL_DF_01; class="table_01 table_02")
table_03::String = String(Base.take!(io_03))
Base.close(io_03)
@test occursin("class=\"table_01 table_02\"", table_03)
io_04::IO = IOBuffer()
HTMLTables.writetable(io_04, GLOBAL_DF_01; class="table_02 table_01")
table_04::String = String(Base.take!(io_04))
Base.close(io_04)
@test occursin("class=\"table_02 table_01\"", table_04)
io_05::IO = IOBuffer()
HTMLTables.writetable(io_05, GLOBAL_DF_01; class=["table_01", "table_02"])
table_05::String = String(Base.take!(io_05))
Base.close(io_05)
@test occursin("class=\"table_01 table_02\"", table_05)
io_06::IO = IOBuffer()
HTMLTables.writetable(io_06, GLOBAL_DF_01; class=["table_02", "table_01"])
table_06::String = String(Base.take!(io_06))
Base.close(io_06)
@test occursin("class=\"table_02 table_01\"", table_06)
end
@testset "caption" begin
io_01::IO = IOBuffer()
HTMLTables.writetable(io_01, GLOBAL_DF_01; caption="Table 1")
table_01::String = String(Base.take!(io_01))
Base.close(io_01)
@test occursin("<caption", table_01)
io_02::IO = IOBuffer()
HTMLTables.writetable(io_02, GLOBAL_DF_01)
table_02::String = String(Base.take!(io_02))
Base.close(io_02)
@test !occursin("<caption", table_02)
end
@testset "tooltips" begin
io_01::IO = IOBuffer()
HTMLTables.writetable(io_01, GLOBAL_DF_01; tooltips=true)
table_01::String = String(Base.take!(io_01))
Base.close(io_01)
@test occursin("title", table_01)
io_02::IO = IOBuffer()
HTMLTables.writetable(io_02, GLOBAL_DF_01; tooltips=false)
table_02::String = String(Base.take!(io_02))
Base.close(io_02)
@test !occursin("title", table_02)
io_03::IO = IOBuffer()
HTMLTables.writetable(io_03, GLOBAL_DF_01)
table_03::String = String(Base.take!(io_03))
Base.close(io_03)
@test occursin("title", table_03)
end
@testset "editable" begin
io_01::IO = IOBuffer()
HTMLTables.writetable(io_01, GLOBAL_DF_01; editable=true)
table_01::String = String(Base.take!(io_01))
Base.close(io_01)
@test occursin("contenteditable", table_01)
io_02::IO = IOBuffer()
HTMLTables.writetable(io_02, GLOBAL_DF_01; editable=false)
table_02::String = String(Base.take!(io_02))
Base.close(io_02)
@test !occursin("contenteditable", table_02)
io_03::IO = IOBuffer()
HTMLTables.writetable(io_03, GLOBAL_DF_01)
table_03::String = String(Base.take!(io_03))
Base.close(io_03)
@test !occursin("contenteditable", table_03)
end
@testset "disabled CSS" begin
io_01::IO = IOBuffer()
HTMLTables.writetable(
io_01,
GLOBAL_DF_01;
styles=false,
css=CSS_STRING_01,
theme=:default,
colorscale="Reds",
)
table_01::String = String(Base.take!(io_01))
@test !occursin("<style>", table_01)
io_02::IO = IOBuffer()
HTMLTables.writetable(
io_02,
GLOBAL_DF_01;
styles=false,
css=CSS_FILE_01,
theme=:gold,
colorscale="Viridis",
)
table_02::String = String(Base.take!(io_02))
@test !occursin("<style>", table_02)
@test table_01 == table_02
end | HTMLTables | https://github.com/cecoeco/HTMLTables.jl.git |
|
[
"MIT"
] | 0.5.2 | 7979027234c190955d3aa25ca5d9fbdf94e896c6 | docs | 2306 | <div>
<a href="https://github.com/cecoeco/HTMLTables.jl/actions/workflows/CI.yml"><img alt="CI test" src="https://github.com/cecoeco/HTMLTables.jl/actions/workflows/CI.yml/badge.svg"></a>
<a href="https://zenodo.org/doi/10.5281/zenodo.11253769"><img src="https://zenodo.org/badge/779591300.svg" alt="DOI"></a>
<a href="https://juliapkgstats.com/pkg/HTMLTables"><img src="https://img.shields.io/badge/dynamic/json?url=http%3A%2F%2Fjuliapkgstats.com%2Fapi%2Fv1%2Fmonthly_downloads%2FHTMLTables&query=total_requests&suffix=%2Fmonth&label=Downloads" alt="Package Statistics"></a>
<a href="https://github.com/JuliaDiff/BlueStyle"><img alt="Style: Blue" src="https://img.shields.io/badge/code%20style-blue-4495d1.svg"></a>
</div>
# HTMLTables
<i>Julia package for reading and writing HTML tables.</i>
:book: **Documentation:** <a href="https://cecoeco.github.io/HTMLTables.jl/stable/"><img src="https://img.shields.io/badge/docs-stable-aquamarine.svg" alt="Documentation Stable" /></a> <a href="https://cecoeco.github.io/HTMLTables.jl/dev/"><img src="https://img.shields.io/badge/docs-dev-aquamarine.svg" alt="Documentation Dev"></a>
:arrow_down: **Installation:** use this command in the Julia REPL: `using Pkg; Pkg.add("HTMLTables")`
**Examples:**
create an HTML table using a `DataFrame`:
```julia
using HTMLTables, DataFrames
df = DataFrame([i:i+19 for i in 1:20:501], Symbol.('a':'z'))
HTMLTables.writetable("viridis.html", df, colorscale="viridis")
```
output:

create a `DataFrame` from parsing HTML:
```julia
using HTMLTables, DataFrames
url = "https://www.w3schools.com/html/html_tables.asp"
df = HTMLTables.readtable(url, DataFrame)
println(df)
```
output:
```
6×3 DataFrame
Row │ Company Contact Country
│ String String String
─────┼─────────────────────────────────────────────────────────
1 │ Alfreds Futterkiste Maria Anders Germany
2 │ Centro comercial Moctezuma Francisco Chang Mexico
3 │ Ernst Handel Roland Mendel Austria
4 │ Island Trading Helen Bennett UK
5 │ Laughing Bacchus Winecellars Yoshi Tannamuri Canada
6 │ Magazzini Alimentari Riuniti Giovanni Rovelli Italy
```
| HTMLTables | https://github.com/cecoeco/HTMLTables.jl.git |
|
[
"MIT"
] | 0.5.2 | 7979027234c190955d3aa25ca5d9fbdf94e896c6 | docs | 49 | # HTMLTables
```@docs
HTMLTables.HTMLTables
```
| HTMLTables | https://github.com/cecoeco/HTMLTables.jl.git |
|
[
"MIT"
] | 0.5.2 | 7979027234c190955d3aa25ca5d9fbdf94e896c6 | docs | 45 | ## Reading
```@docs
HTMLTables.readtable
```
| HTMLTables | https://github.com/cecoeco/HTMLTables.jl.git |
|
[
"MIT"
] | 0.5.2 | 7979027234c190955d3aa25ca5d9fbdf94e896c6 | docs | 46 | ## Writing
```@docs
HTMLTables.writetable
```
| HTMLTables | https://github.com/cecoeco/HTMLTables.jl.git |
|
[
"MIT"
] | 1.0.2 | 377585d2d44d9717448ddb079abe61272ada26b4 | code | 289 | using Documenter, DungBase
makedocs(
modules = [DungBase],
format = :html,
sitename = "DungBase.jl",
pages = Any["index.md"]
)
deploydocs(
repo = "github.com/yakir12/DungBase.jl.git",
target = "build",
julia = "1.0",
deps = nothing,
make = nothing,
)
| DungBase | https://github.com/yakir12/DungBase.jl.git |
|
[
"MIT"
] | 1.0.2 | 377585d2d44d9717448ddb079abe61272ada26b4 | code | 1425 | module DungBase
using Dates, UUIDs, StaticArrays
import IntervalSets: width, (..), AbstractInterval, leftendpoint
using StatsBase, StructArrays, Dierckx, AngleBetweenVectors, LinearAlgebra
export VideoFile, WholeVideo, FragmentedVideo, DisjointVideo, AbstractTimeLine, AbstractPeriod, Instantaneous, Prolonged, Temporal, Board, Calibration, POI, Metadata, Run, Experiment, Track, Common, Point, PointCollection
export start, duration, stop, files, filenames, point, pointcollection, homing, searching, searchcenter, turningpoint
__init__() = @warn "This package is deprecated"
stop(x) = start(x) + duration(x)
include("videos.jl")
include("temporals.jl")
include("calibrations.jl")
include("pois.jl")
include("track.jl")
# include(joinpath(@__DIR__, "experimental_setup.jl"))
struct Metadata
setup::Dict{Symbol, Any}
comment::String
date::DateTime
end
# Base.getproperty(x::Metadata, level::Symbol) = get(x.setup, level, missing)
#=getcoord(x::Run, poi::Symbol) = get(x.pois, poi, missing)
_getlevel(x::Metadata, level::Symbol) = get(x, level, missing)
getlevel(x::Run, level::Symbol) = _getlevel(x.metadata, level)=#
struct Run{T, M}
data::T # pois::Dict{Symbol, T} or Track
metadata::M
end
#=struct Run
setup::Dict{Symbol, Any}
pois::Dict{Symbol, POI}
comment::String
date::DateTime
end=#
struct Experiment
runs::Vector{Run}
description::String
end
end # module
| DungBase | https://github.com/yakir12/DungBase.jl.git |
|
[
"MIT"
] | 1.0.2 | 377585d2d44d9717448ddb079abe61272ada26b4 | code | 1216 | struct Board
designation::String
checker_width_cm::Float64
dimensions::Tuple{Int, Int}
description::String
function Board(designation, checker_width_cm, dimensions, description)
@assert !isempty(designation) "empty board designation is not allowed"
@assert checker_width_cm > 0 "checker width must be larger zero"
w, h = dimensions
@assert w > 1 "number of checkers must be larger one"
@assert h > 1 "number of checkers must be larger one"
@assert !isempty(description) "empty board description is not allowed"
new(designation, checker_width_cm, dimensions, description)
end
end
Board() = Board("_", 1.0, (2,2), "_")
struct Calibration{T, S}
intrinsic::T # Missing or Temporal{Prolonged}
extrinsic::S # Temporal{Instantaneous}
board::Board
comment::String
end
Calibration() = Calibration(missing, Temporal(), Board(), "")
Calibration(c::Calibration, args) = Calibration(get(args, :intrinsic, c.intrinsic), get(args, :extrinsic, c.extrinsic), get(args, :board, c.board), get(args, :comment, c.comment))
filenames(c::Calibration) = first(filenames(c.extrinsic.video))
start(c::Calibration) = start(c.extrinsic.time)
| DungBase | https://github.com/yakir12/DungBase.jl.git |
|
[
"MIT"
] | 1.0.2 | 377585d2d44d9717448ddb079abe61272ada26b4 | code | 792 | #=using DataStructures
abstract type AbstractFactor end
struct Nominal <: AbstractFactor
levels::Set{String}
end
struct Ordinal <: AbstractFactor
levels::OrderedSet{String}
end
struct Discrete <: AbstractFactor
levels::OrderedSet{Int}
end
struct Continuous <: AbstractFactor
levels::Vector{Float64}
end
struct Run
setup::Int
start::DateTime
comment::String
pois::Vector{POI}
end
struct Experiment{N}
description::String
factors::NTuple{N, String}
levels::NTuple{N, AbstractFactor}
setups::Vector{NTuple{N, Int}}
runs::Vector{Run}
end=#
struct Run
setup::Vector{Union{Missing, String}}
start::DateTime
comment::String
pois::Dict{Symbol, POI}
end
struct Experiment
description::String
runs::Vector{Run}
end
| DungBase | https://github.com/yakir12/DungBase.jl.git |
|
[
"MIT"
] | 1.0.2 | 377585d2d44d9717448ddb079abe61272ada26b4 | code | 370 | struct POI{T, U}
calib::T # Calibration -> name for the matlab calibration file or maybe the transformation matrix itself
data::U # Temporal -> pixel coordinates
end
POI{T, U}() where {T, U} = POI(T(), U())
# struct POI{V <: AbstractTimeLine, T <: AbstractPeriod}
# calib::Dict{UUID, <:Calibration}
# temporal::Temporal{V, T}
# spatial::UUID
# end
| DungBase | https://github.com/yakir12/DungBase.jl.git |
|
[
"MIT"
] | 1.0.2 | 377585d2d44d9717448ddb079abe61272ada26b4 | code | 2015 | abstract type AbstractPeriod end
function checkInstantaneous(x::T) where {T <: TimePeriod}
@assert x ≥ zero(T) "period start cannot be negative"
x
end
checkInstantaneous(x) = x
struct Instantaneous{T} <: AbstractPeriod
data::T
Instantaneous{T}(x) where {T} = new(checkInstantaneous(x))
end
Instantaneous(x::T) where {T} = Instantaneous{T}(x)
Instantaneous() = Instantaneous(Millisecond(0))
function checkProlonged(x::I) where {T <: TimePeriod, I <: AbstractInterval{T}}
@assert leftendpoint(x) ≥ zero(T) "period start cannot be negative"
@assert width(x) > zero(T) "period duration must be larger than zero"
x
end
checkProlonged(x) = x
struct Prolonged{T} <: AbstractPeriod
data::T
Prolonged{T}(x) where {T} = new(checkProlonged(x))
end
Prolonged(x::T) where {T} = Prolonged{T}(x)
Prolonged() = Prolonged(Millisecond(0)..Millisecond(1))
AbstractPeriod(start::T) where {T <: TimePeriod} = Instantaneous(start)
AbstractPeriod(x::I) where {T <: TimePeriod, I <: AbstractInterval{T}} = Prolonged(x)
AbstractPeriod(start::T1, stop::T2) where {T1 <: TimePeriod, T2 <: TimePeriod} = Prolonged(start..stop)
AbstractPeriod(start::T, _::Nothing) where {T <: TimePeriod} = Instantaneous(start)
start(x::Instantaneous) = x.data
start(x::Prolonged) = leftendpoint(x.data)
duration(x::Instantaneous{T}) where {T <: TimePeriod} = zero(T)
duration(x::Prolonged) = width(x.data)
struct Temporal{V <: AbstractTimeLine, T <: AbstractPeriod}
video::V
time::T
comment::String
function Temporal{V, T}(video, time, comment) where {V <: AbstractTimeLine, T <: AbstractPeriod}
@assert stop(time) ≤ duration(video) "POIs are outside the video's timeline (stop: $(stop(time)); duration: $(duration(video)); file: $(files(video))"
new(video, time, comment)
end
end
Temporal(video::V, time::T, comment::String) where {V <: AbstractTimeLine, T <: AbstractPeriod} = Temporal{V, T}(video, time, comment)
Temporal() = Temporal(WholeVideo(), Instantaneous(), "")
| DungBase | https://github.com/yakir12/DungBase.jl.git |
|
[
"MIT"
] | 1.0.2 | 377585d2d44d9717448ddb079abe61272ada26b4 | code | 2577 |
const ignorefirst = 10 # cm
const bigturn = π/3 # 60°
# const smallturn = π/93 # 60°
const s = 500
const Point = SVector{2, Float64}
point(x::Missing) = x
point(x::Instantaneous)= Point(x.data[1], x.data[2])
_getv(spl, k) = SVector{2, Float64}(derivative(spl, k))
function gettpindex(spl, ks)
tp = ks[1]
vlast = _getv(spl, ks[1])
for k in Iterators.drop(ks, 1)
v = _getv(spl, k)
Δ = angle(vlast, v)
tp = k
Δ > bigturn && break
vlast = v
end
return tp
end
function gettpknot(spl)
ks = Dierckx.get_knots(spl)
filter!(k -> norm(spl(k) - spl(0)) > ignorefirst, ks)
tp2 = gettpindex(spl, ks)
# return tp2
tp1 = copy(tp2)
for k in ks
k == tp2 && break
tp1 = k
end
tp1 += 0.1
if tp1 < tp2
main = _getv(spl, tp1)
for t in tp2:-0.3:tp1
v = _getv(spl, t)
Δ = angle(main, v)
Δ < bigturn && return t
end
end
return tp2
end
mutable struct TimedPoint
xy::Point
t::Float64
end
const PointCollection = StructVector{TimedPoint}
pointcollection(x::Missing, t₀) = StructVector{TimedPoint}(undef, 0)
pointcollection(x, t₀) = StructVector(TimedPoint(Point(i[1], i[2]), i[3] - t₀) for i in eachrow(x.data))
struct Track
coords::Vector{Point}
t::StepRangeLen{Float64,Base.TwicePrecision{Float64},Base.TwicePrecision{Float64}}
tp::Int
rawcoords::StructArray{TimedPoint}
end
function filterdance(xy, Δt)
xy2 = [xy[1,:]]
t = [0.0]
for p in eachrow(xy)
if norm(p - xy2[end]) > 4
push!(xy2, p)
push!(t, t[end] + Δt)
else
t[end] += Δt
end
end
t .-= t[1]
return t, hcat(xy2...)
end
function Track(x::Prolonged)
xyt = !issorted(x.data[:, 3]) ? sortslices(x.data, dims = 1, lt = (x, y) -> isless(x[3], y[3])) : x.data
Δt = mean(trim(diff(xyt[:, 3]), prop = 0.1))
t, xy = filterdance(xyt[:,1:2], Δt)
spl = ParametricSpline(t, xy; s = s, k = 2)
tl = range(0.0, step = Δt, stop = t[end])
xyl = Point.(spl.(tl))
tp = gettpknot(spl)
i = findfirst(≥(tp), tl)
if isnothing(i)
i = length(tl)
end
raw = pointcollection((data = xyt, ), xyt[1,3])
Track(xyl, tl, i, raw)
end
homing(t::Track) = t.coords[1:t.tp]
searching(t::Track) = t.coords[t.tp:end]
searchcenter(t::Track) = mean(searching(t))
turningpoint(t::Track) = t.coords[t.tp]
mutable struct Common{N}
feeder::Point
nest::Point
track::Track
pellet::PointCollection
originalnest::N
end
| DungBase | https://github.com/yakir12/DungBase.jl.git |
|
[
"MIT"
] | 1.0.2 | 377585d2d44d9717448ddb079abe61272ada26b4 | code | 4388 | struct VideoFile
name::String
start::DateTime
duration::Millisecond
function VideoFile(name, start, duration::Millisecond)
@assert !isempty(name) "file name is empty"
@assert start > DateTime(0) "starting date & time must be larger than zero"
@assert duration > Millisecond(0) "zero duration is not supported"
new(name, start, duration)
end
end
function VideoFile(name, start, duration::T) where T <: TimePeriod
ms = duration/convert(T, Millisecond(1))
VideoFile(name, start, Millisecond(round(Int, ms)))
end
VideoFile() = VideoFile("_", DateTime(1), Millisecond(1))
start(x::VideoFile) = x.start
duration(x::VideoFile) = x.duration
VideoFile(vf::VideoFile, args) = VideoFile(get(args, :name, vf.name), get(args, :start, start(vf)), get(args, :duration, duration(vf)))
abstract type AbstractTimeLine end
struct WholeVideo <: AbstractTimeLine
file::VideoFile
comment::String
end
WholeVideo() = WholeVideo(VideoFile(), "")
WholeVideo(wv::WholeVideo, args) = WholeVideo(get(args, :files, VideoFile(wv.file, args)), get(args, :comment, wv.comment))
files(x::WholeVideo) = SVector{1, VideoFile}(x.file)
struct FragmentedVideo{N, SV <: SVector{N, VideoFile}} <: AbstractTimeLine
files::SV
comment::String
function FragmentedVideo{N, SV}(files, comment) where {N, SV <: SVector{N, VideoFile}}
@assert N > 1 "video collection must include more than one file"
last = stop(files[1])
for file in files[2:N]
@assert start(file) == last "there is a gap between two adjacent videos: $(start(file) - last)"
last = stop(file)
end
@assert allunique(getfield.(files, :name)) "all file names must be unique"
new(files, comment)
end
end
FragmentedVideo(files::SV, comment::String) where {N, SV <: SVector{N, VideoFile}} = FragmentedVideo{N, SV}(files, comment)
function FragmentedVideo(files::T, comment::String) where {T <: AbstractVector{VideoFile}}
n = length(files)
FragmentedVideo(SVector{n, VideoFile}(files), comment)
end
function FragmentedVideo()
v1 = VideoFile()
v2 = VideoFile("__", stop(v1), Millisecond(1))
FragmentedVideo([v1, v2], "")
end
FragmentedVideo(fv::FragmentedVideo, args) = FragmentedVideo(get(args, :files, fv.files), get(args, :comment, fv.comment))
struct DisjointVideo{N, SV <: SVector{N, VideoFile}} <: AbstractTimeLine
files::SV
comment::String
function DisjointVideo{N, SV}(files, comment) where {N, SV <: SVector{N, VideoFile}}
n = length(files)
@assert n > 1 "video collection must include more than one file"
last = stop(files[1])
for file in files[2:end]
@assert start(file) ≥ last "one file starts before the next one ends"
last = stop(file)
end
@assert allunique(getfield.(files, :name)) "all file names must be unique"
new(SVector{n}(files), comment)
end
end
DisjointVideo(files::SV, comment::String) where {N, SV <: SVector{N, VideoFile}} = DisjointVideo{N, SV}(files, comment)
function DisjointVideo(files::T, comment::String) where {T <: AbstractVector{VideoFile}}
n = length(files)
DisjointVideo(SVector{n, VideoFile}(files), comment)
end
function DisjointVideo()
v1 = VideoFile()
v2 = VideoFile("__", stop(v1) + Millisecond(1), Millisecond(1))
DisjointVideo([v1, v2], "")
end
DisjointVideo(fv::DisjointVideo, args) = DisjointVideo(get(args, :files, fv.files), get(args, :comment, fv.comment))
AbstractTimeLine(file::VideoFile, comment::String) = WholeVideo(file, comment)
function AbstractTimeLine(files::Vector{VideoFile}, comment::String)
@assert !isempty(files) "must have some files"
if length(files) == 1
WholeVideo(files[], comment)
else
if all(stop(prev) == start(next) for (prev, next) in zip(files[1:end-1], files[2:end]))
FragmentedVideo(files, comment)
else
DisjointVideo(files, comment)
end
end
end
files(x::AbstractTimeLine) = x.files
filenames(x::AbstractTimeLine) = getfield.(files(x), :name)
start(x::WholeVideo) = start(x.file)
start(x::AbstractTimeLine) = start(first(x.files))
duration(x::WholeVideo) = duration(x.file)
duration(x::FragmentedVideo) = sum(duration, x.files)
duration(x::DisjointVideo) = stop(last(x.files)) - start(first(x.files))
| DungBase | https://github.com/yakir12/DungBase.jl.git |
|
[
"MIT"
] | 1.0.2 | 377585d2d44d9717448ddb079abe61272ada26b4 | code | 26 | using DungBase
using Test
| DungBase | https://github.com/yakir12/DungBase.jl.git |
|
[
"MIT"
] | 1.0.2 | 377585d2d44d9717448ddb079abe61272ada26b4 | code | 266 | # only push coverage from one bot
get(ENV, "TRAVIS_OS_NAME", nothing) == "linux" || exit(0)
get(ENV, "TRAVIS_JULIA_VERSION", nothing) == "1.0" || exit(0)
using Coverage
cd(joinpath(@__DIR__, "..", "..")) do
Codecov.submit(Codecov.process_folder())
end
| DungBase | https://github.com/yakir12/DungBase.jl.git |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.