licenses
sequencelengths 1
3
| version
stringclasses 677
values | tree_hash
stringlengths 40
40
| path
stringclasses 1
value | type
stringclasses 2
values | size
stringlengths 2
8
| text
stringlengths 25
67.1M
| package_name
stringlengths 2
41
| repo
stringlengths 33
86
|
|---|---|---|---|---|---|---|---|---|
[
"MIT"
] | 0.3.0 | e7f8a3e4bafabc4154558c1f3cd6f3e5915a534f | code | 2367 | # Author: Mathias Louboutin
# Date: June 2021
#
using JUDI, LinearAlgebra, Images, PyPlot, DSP, ImageGather, SlimPlotting
# Set up model structure
n = (601, 333) # (x,y,z) or (x,z)
d = (15., 15.)
o = (0., 0.)
# Velocity [km/s]
v = ones(Float32,n) .+ 0.5f0
for i=1:12
v[:,25*i+1:end] .= 1.5f0 + i*.25f0
end
v0 = imfilter(v, Kernel.gaussian(5))
# Slowness squared [s^2/km^2]
m = (1f0 ./ v).^2
m0 = (1f0 ./ v0).^2
# Setup info and model structure
nsrc = 1 # number of sources
model = Model(n, d, o, m; nb=40)
model0 = Model(n, d, o, m0; nb=40)
# Set up receiver geometry
nxrec = 401
xrec = range(0f0, stop=(n[1] -1)*d[1], length=nxrec)
yrec = 0f0
zrec = range(20f0, stop=20f0, length=nxrec)
# receiver sampling and recording time
timeR = 4000f0 # receiver recording time [ms]
dtR = 4f0 # receiver sampling interval [ms]
# Set up receiver structure
recGeometry = Geometry(xrec, yrec, zrec; dt=dtR, t=timeR, nsrc=nsrc)
# Set up source geometry (cell array with source locations for each shot)
xsrc = 4500f0
ysrc = 0f0
zsrc = 20f0
# source sampling and number of time steps
timeS = 4000f0 # ms
dtS = 4f0 # ms
# Set up source structure
srcGeometry = Geometry(xsrc, ysrc, zsrc; dt=dtS, t=timeS)
# setup wavelet
f0 = 0.015f0 # kHz
wavelet = ricker_wavelet(timeS, dtS, f0)
q = judiVector(srcGeometry, wavelet)
###################################################################################################
opt = Options(space_order=16, IC="as")
# Setup operators
F = judiModeling(model, srcGeometry, recGeometry; options=opt)
F0 = judiModeling(model0, srcGeometry, recGeometry; options=opt)
# Nonlinear modeling
dD = F*q
# Make rtms
J = judiJacobian(F0, q)
# Get offsets and mute data
offs = abs.(xrec .- xsrc)
res = deepcopy(dD)
mute!(res.data[1], offs)
reso = deepcopy(res)
I = inv(judiIllumination(J))
rtm = I*J'*res
omap = Array{Any}(undef, 2)
i = 1
# try a bunch of weighting functions
for (wf, iwf) = zip([x-> x .+ 5f3, x-> log.(x .+ 10)], [x -> x .- 5f3, x-> exp.(x) .- 10])
reso.data[1] .= res.data[1] .* wf(offs)'
rtmo = I*J'*reso
omap[i] = iwf(offset_map(rtm.data, rtmo.data))
global i+=1
end
figure()
for (i, name)=enumerate(["shift", "log"])
subplot(1,2,i)
plot_velocity(omap[i]', (1,1); cmap="jet", aspect="auto", perc=98, new_fig=false, vmax=5000)
colorbar()
title(name)
end
tight_layout()
| ImageGather | https://github.com/slimgroup/ImageGather.jl.git |
|
[
"MIT"
] | 0.3.0 | e7f8a3e4bafabc4154558c1f3cd6f3e5915a534f | code | 640 | module ImageGather
using JUDI
using JUDI.DSP, JUDI.PyCall
import Base: getindex, *
import JUDI: judiAbstractJacobian, judiMultiSourceVector, judiComposedPropagator, judiJacobian, make_input, propagate
import JUDI.LinearAlgebra: adjoint
const impl = PyNULL()
function __init__()
pushfirst!(PyVector(pyimport("sys")."path"),dirname(pathof(ImageGather)))
copy!(impl, pyimport("implementation"))
end
# Utility functions
include("utils.jl")
# Surface offset gathers
include("surface_gather.jl")
# Subsurface offset gathers
include("subsurface_gather.jl")
end # module
| ImageGather | https://github.com/slimgroup/ImageGather.jl.git |
|
[
"MIT"
] | 0.3.0 | e7f8a3e4bafabc4154558c1f3cd6f3e5915a534f | code | 6023 | export judiExtendedJacobian
struct judiExtendedJacobian{D, O, FT} <: judiAbstractJacobian{D, O, FT}
m::AbstractSize
n::AbstractSize
F::FT
q::judiMultiSourceVector
offsets::Vector{D}
dims::Vector{Symbol}
end
"""
J = judiExtendedJacobian(F, q, offsets; options::JUDIOptions, omni=false, dims=nothing)
Extended jacobian (extended Born modeling operator) for subsurface horsizontal offsets `offsets`. Its adjoint
comput the subsurface common offset volume. In succint way, the extened born modeling Operator can summarized in a linear algebra frmaework as:
Options structure for seismic modeling.
`omni`: If `true`, the extended jacobian will be computed for all dimensions.
`dims`: If `omni` is `false`, the extended jacobian will be computed for the dimension(s) specified in `dims`.
"""
function judiExtendedJacobian(F::judiComposedPropagator{D, O}, q::judiMultiSourceVector, offsets;
options=nothing, omni=false, dims=nothing) where {D, O}
JUDI.update!(F.options, options)
offsets = Vector{D}(offsets)
ndim = length(F.model.n)
if omni
dims = [:x, :y, :z][1:ndim]
else
if isnothing(dims)
dims = [:x]
else
dims = symvec(dims)
if ndim == 2
dims[dims .== :z] .= :y
end
end
end
return judiExtendedJacobian{D, :born, typeof(F)}(F.m, space(F.model.n), F, q, offsets, dims)
end
symvec(s::Symbol) = [s]
symvec(s::Tuple) = [symvec(ss)[1] for ss in s]::Vector{Symbol}
symvec(s::Vector) = [symvec(ss)[1] for ss in s]::Vector{Symbol}
adjoint(J::judiExtendedJacobian{D, O, FT}) where {D, O, FT} = judiExtendedJacobian{D, adjoint(O), FT}(J.n, J.m, J.F, J.q, J.offsets, J.dims)
getindex(J::judiExtendedJacobian{D, O, FT}, i) where {D, O, FT} = judiExtendedJacobian{D, O, FT}(J.m[i], J.n[i], J.F[i], J.q[i], J.offsets, J.dims)
function make_input(J::judiExtendedJacobian{D, :adjoint_born, FT}, q) where {D, FT}
srcGeom, srcData = JUDI.make_src(J.q, J.F.qInjection)
recGeom, recData = JUDI.make_src(q, J.F.rInterpolation)
return srcGeom, srcData, recGeom, recData, nothing
end
function make_input(J::judiExtendedJacobian{D, :born, FT}, dm) where {D<:Number, FT}
srcGeom, srcData = JUDI.make_src(J.q, J.F.qInjection)
return srcGeom, srcData, J.F.rInterpolation.data[1], nothing, dm
end
*(J::judiExtendedJacobian{T, :born, O}, dm::Array{T, 3}) where {T, O} = J*vec(dm)
*(J::judiExtendedJacobian{T, :born, O}, dm::Array{T, 4}) where {T, O} = J*vec(dm)
JUDI.process_input_data(::judiExtendedJacobian{D, :born, FT}, q::Vector{D}) where {D<:Number, FT} = q
############################################################
function propagate(J::judiExtendedJacobian{T, :born, O}, q::AbstractArray{T}, illum::Bool) where {T, O}
srcGeometry, srcData, recGeometry, _, dm = make_input(J, q)
# Load full geometry for out-of-core geometry containers
recGeometry = Geometry(recGeometry)
srcGeometry = Geometry(srcGeometry)
# Avoid useless propage without perturbation
if minimum(dm) == 0 && maximum(dm) == 0
return judiVector(recGeometry, zeros(Float32, recGeometry.nt[1], length(recGeometry.xloc[1])))
end
# Set up Python model structure
modelPy = devito_model(J.model, J.options)
nh = [length(J.offsets) for _=1:length(J.dims)]
dmd = reshape(dm, nh..., J.model.n...)
dtComp = convert(Float32, modelPy."critical_dt")
# Extrapolate input data to computational grid
qIn = time_resample(srcData, srcGeometry, dtComp)
# Set up coordinates
src_coords = setup_grid(srcGeometry, J.model.n) # shifts source coordinates by origin
rec_coords = setup_grid(recGeometry, J.model.n) # shifts rec coordinates by origin
# Devito interface
dD = JUDI.wrapcall_data(impl."cig_lin", modelPy, src_coords, qIn, rec_coords,
dmd, J.offsets, ic=J.options.IC, space_order=J.options.space_order, dims=J.dims)
dD = time_resample(dD, dtComp, recGeometry)
# Output shot record as judiVector
return judiVector{Float32, Matrix{Float32}}(1, recGeometry, [dD])
end
function propagate(J::judiExtendedJacobian{T, :adjoint_born, O}, q::AbstractArray{T}, illum::Bool) where {T, O}
srcGeometry, srcData, recGeometry, recData, _ = make_input(J, q)
# Load full geometry for out-of-core geometry containers
recGeometry = Geometry(recGeometry)
srcGeometry = Geometry(srcGeometry)
# Set up Python model
modelPy = devito_model(J.model, J.options)
dtComp = convert(Float32, modelPy."critical_dt")
# Extrapolate input data to computational grid
qIn = time_resample(srcData, srcGeometry, dtComp)
dObserved = time_resample(recData, recGeometry, dtComp)
# Set up coordinates
src_coords = setup_grid(srcGeometry, J.model.n) # shifts source coordinates by origin
rec_coords = setup_grid(recGeometry, J.model.n) # shifts rec coordinates by origin
# Devito
g = JUDI.pylock() do
pycall(impl."cig_grad", PyArray, modelPy, src_coords, qIn, rec_coords, dObserved, J.offsets,
illum=false, ic=J.options.IC, space_order=J.options.space_order, dims=J.dims)
end
g = remove_padding_cig(g, modelPy.padsizes; true_adjoint=J.options.sum_padding)
return g
end
function remove_padding_cig(gradient::AbstractArray{DT}, nb::NTuple{Nd, NTuple{2, Int64}}; true_adjoint::Bool=false) where {DT, Nd}
no = ndims(gradient) - length(nb)
N = size(gradient)[no+1:end]
hd = tuple([Colon() for _=1:no]...)
if true_adjoint
for (dim, (nbl, nbr)) in enumerate(nb)
diml = dim+no
selectdim(gradient, diml, nbl+1) .+= dropdims(sum(selectdim(gradient, diml, 1:nbl), dims=diml), dims=diml)
selectdim(gradient, diml, N[dim]-nbr) .+= dropdims(sum(selectdim(gradient, diml, N[dim]-nbr+1:N[dim]), dims=diml), dims=diml)
end
end
out = gradient[hd..., [nbl+1:nn-nbr for ((nbl, nbr), nn) in zip(nb, N)]...]
return out
end
| ImageGather | https://github.com/slimgroup/ImageGather.jl.git |
|
[
"MIT"
] | 0.3.0 | e7f8a3e4bafabc4154558c1f3cd6f3e5915a534f | code | 4124 | import JUDI: AbstractModel, rlock_pycall, devito
export surface_gather, double_rtm_cig
"""
surface_gather(model, q, data; offsets=nothing, options=Options())
Compute the surface offset gathers volume (nx (X ny) X nz X no) for via the double rtm method whith `no` offsets.
Parameters
* `model`: JUDI Model structure.
* `q`: Source, judiVector.
* `data`: Obeserved data, judiVector.
* `offsets`: List of offsets to compute the gather at. Optional (defaults to 0:10*model.d:model.extent)
* `options`: JUDI Options structure.
"""
function surface_gather(model::AbstractModel, q::judiVector, data::judiVector; offsets=nothing, mute=true, options=Options())
isnothing(offsets) && (offsets = 0f0:10*model.d[1]:(model.n[1]-1)*model.d[1])
offsets = collect(offsets)
pool = JUDI._worker_pool()
# Distribute source
arg_func = i -> (model, q[i], data[i], offsets, options[i], mute)
# Distribute source
ncig = (model.n..., length(offsets))
out = PhysicalParameter(ncig, (model.d..., 1f0), (model.o..., minimum(offsets)), zeros(Float32, ncig...))
out = out + JUDI.run_and_reduce(double_rtm_cig, pool, q.nsrc, arg_func)
return out.data
end
"""
double_rtm_cig(model, q, data, offsets, options)
Compute the single shot contribution to the surface offset gather via double rtm. This single source contribution consists of the following steps:
1. Mute direct arrival in the data.
2. Compute standard RTM ``R``.
3. Compute the offset RTM ``R_o`` for the the weighted data where each trace is weighted by its offset `(rec_x - src_x)`.
4. Compute the envelope ``R_e = \\mathcal{E}(R)`` and ``R_{oe} = \\mathcal{E}(R_o)``.
5. Compute the offset map ``\\frac{R_e \\odot R_{oe}}{R_e \\odot R_e + \\epsilon}``.
6. Apply illumination correction and laplace filter ``R_l = \\mathcal{D} \\Delta R``.
7. Compute each offset contribution ``\\mathcal{I}[:, h] = R_l \\odot \\delta[ha - h]_{tol}`` [`delta_h`](@ref).
8. Return ``\\mathcal{I}``.
"""
function double_rtm_cig(model_full, q::judiVector, data::judiVector, offs, options, mute)
GC.gc(true)
devito.clear_cache()
# Load full geometry for out-of-core geometry containers
data.geometry = Geometry(data.geometry)
q.geometry = Geometry(q.geometry)
# Limit model to area with sources/receivers
if options.limit_m == true
model = deepcopy(model_full)
model, _ = limit_model_to_receiver_area(q.geometry, data.geometry, model, options.buffer_size)
else
model = model_full
end
# Set up Python model
modelPy = devito_model(model, options)
dtComp = convert(Float32, modelPy."critical_dt")
# Extrapolate input data to computational grid
qIn = time_resample(make_input(q), q.geometry, dtComp)
res = time_resample(make_input(data), data.geometry, dtComp)
# Set up coordinates
src_coords = setup_grid(q.geometry, model.n) # shifts source coordinates by origin
rec_coords = setup_grid(data.geometry, model.n) # shifts rec coordinates by origin
# Src-rec offsets
scale = 1f1
off_r = log.(abs.(data.geometry.xloc[1] .- q.geometry.xloc[1]) .+ scale)
inv_off(x) = exp.(x) .- scale
# mute
if mute
mute!(res, off_r .- scale; dt=dtComp/1f3, t0=.25)
end
res_o = res .* off_r'
# Double rtm
rtm, rtmo, illum = rlock_pycall(impl."double_rtm", Tuple{PyArray, PyArray, PyArray},
modelPy, qIn, src_coords, res, res_o, rec_coords,
ic=options.IC)
rtm = remove_padding(rtm, modelPy.padsizes)
rtmo = remove_padding(rtmo, modelPy.padsizes)
illum = remove_padding(illum, modelPy.padsizes)
# offset map
h_map = inv_off(offset_map(rtm, rtmo))
rtm = laplacian(rtm)
rtm[illum .> 0] ./= illum[illum .> 0]
soffs = zeros(Float32, size(model)..., length(offs))
for (i, h) in enumerate(offs)
soffs[:, :, i] .+= rtm .* delta_h(h_map, h, 2*diff(offs)[1])
end
d = (spacing(model)..., 1f0)
n = size(soffs)
o = (origin(model)..., minimum(offs))
return PhysicalParameter(n, d, o, soffs)
end
| ImageGather | https://github.com/slimgroup/ImageGather.jl.git |
|
[
"MIT"
] | 0.3.0 | e7f8a3e4bafabc4154558c1f3cd6f3e5915a534f | code | 2857 | export mv_avg_2d, delta_h, envelope, mute, mute!, laplacian, offset_map
"""
mv_avg_2d(x; k=5)
2D moving average with a square window of width k
"""
function mv_avg_2d(x::AbstractArray{T, 2}; k=5) where T
out = 1f0 * x
nx, ny = size(x)
kl, kr = k÷2 + 1, k÷2
for i=kl:nx-kr, j=kl:ny-kr
out[i, j] = sum(x[i+k, j+l] for k=-kr:kr, l=-kr:kr)/k^2
end
out
end
"""
envelope(x)
Envelope of a vector or a 2D matrix. The envelope over the first dimension is taken for a 2D matrix (see DSP `hilbert`)
"""
envelope(x::AbstractArray{T, 1}) where T = abs.(hilbert(x))
"""
envelope(x)
Envelope of a vector or a 1D vector (see DSP `hilbert`)
"""
envelope(x::AbstractArray{T, 2}) where T = abs.(hilbert(x))
"""
delta_h(ha, h, tol)
Compute the binary mask where `ha` is within `tol` of `h`.
"""
delta_h(ha::AbstractArray{T, 2}, h::Number, tol::Number) where T = Float32.(abs.(h .- ha) .<= tol)
"""
mute!(shot, offsets;vp=1500, t0=1/10, dt=0.004)
In place direct wave muting of a shot record with water sound speed `vp`, time sampling `dt` and firing time `t0`.
"""
function mute!(shot::AbstractArray{Ts, 2}, offsets::Vector{To}; vp=1500, t0=1/10, dt=.004) where {To, Ts}
length(offsets) == size(shot, 2) || throw(DimensionMismatch("Number of offsets has to match the number of traces"))
inds = trunc.(Integer, (offsets ./ vp .+ t0) ./ dt)
inds = min.(max.(1, inds), size(shot, 1))
for (rx, i) = enumerate(inds)
shot[1:i, rx] .= 0f0
end
end
"""
mute(shot, offsets;vp=1500, t0=1/10, dt=0.004)
Direct wave muting of a shot record with water sound speed `vp`, time sampling `dt` and firing time `t0`.
"""
function mute(shot::AbstractArray{Ts, 2}, offsets::Vector{To}; vp=1500, t0=1/10, dt=.004) where {To, Ts}
out = Ts(1) .* shot
mute!(out, offsets; vp=vp, t0=t0, dt=dt)
out
end
"""
laplacian(image; hx=1, hy=1)
2D laplacian of an image with grid spacings (hx, hy)
"""
function laplacian(image::AbstractArray{T, 2}; hx=1, hy=1) where T
scale = 1/(hx*hy)
out = 1 .* image
@views begin
out[2:end-1, 2:end-1] .= -4 .* image[2:end-1, 2:end-1]
out[2:end-1, 2:end-1] .+= image[1:end-2, 2:end-1] + image[2:end-1, 1:end-2]
out[2:end-1, 2:end-1] .+= image[3:end, 2:end-1] + image[2:end-1, 3:end]
end
return scale .* out
end
"""
offset_map(rtm, rtmo; scale=0)
Return the regularized least-square division of rtm and rtmo. The regularization consists of the envelope and moving average
followed by the least-square division [`surface_gather`](@ref)
"""
function offset_map(rtm::AbstractArray{T, 2}, rtmo::AbstractArray{T, 2}; scale=0) where T
rtmn = mv_avg_2d(envelope(rtm))
rtmo = mv_avg_2d(envelope(rtmo))
offset_map = rtmn .* rtmo ./ (rtmn .* rtmn .+ eps(Float32)) .- scale
return offset_map
end
| ImageGather | https://github.com/slimgroup/ImageGather.jl.git |
|
[
"MIT"
] | 0.3.0 | e7f8a3e4bafabc4154558c1f3cd6f3e5915a534f | code | 2340 | using ImageGather, Test
using JUDI, LinearAlgebra
# Set up model structure
n = (301, 151) # (x,y,z) or (x,z)
d = (10., 10.)
o = (0., 0.)
# Velocity [km/s]
v = 1.5f0 .* ones(Float32,n)
v[:, 76:end] .= 2.5f0
v0 = 1.5f0 .* ones(Float32,n)
# Slowness squared [s^2/km^2]
m = (1f0 ./ v).^2
m0 = (1f0 ./ v0).^2
# Setup info and model structure
nsrc = 1 # number of sources
model = Model(n, d, o, m; nb=40)
model0 = Model(n, d, o, m0; nb=40)
dm = model.m - model0.m
# Set up receiver geometry
nxrec = 151
xrec = range(0f0, stop=(n[1] -1)*d[1], length=nxrec)
yrec = 0f0
zrec = range(20f0, stop=20f0, length=nxrec)
# receiver sampling and recording time
timeD = 2000f0 # receiver recording time [ms]
dtD = 4f0 # receiver sampling interval [ms]
# Set up receiver structure
recGeometry = Geometry(xrec, yrec, zrec; dt=dtD, t=timeD, nsrc=nsrc)
# Set up source geometry (cell array with source locations for each shot)
xsrc = 1500f0
ysrc = 0f0
zsrc = 20f0
# Set up source structure
srcGeometry = Geometry(xsrc, ysrc, zsrc; dt=dtD, t=timeD)
# setup wavelet
f0 = 0.015f0 # kHz
wavelet = ricker_wavelet(timeD, dtD, f0)
q = diff(judiVector(srcGeometry, wavelet))
###################################################################################################
opt = Options()
# Setup operators
F = judiModeling(model, srcGeometry, recGeometry; options=opt)
J0 = judiJacobian(F(model0), q)
# Nonlinear modeling
dD = J0*dm
rtm = J0'*dD
# Common surface offset image gather
offsets = -40f0:model.d[1]:40f0
nh = length(offsets)
for dims in ((:x, :z), :z, :x)
J = judiExtendedJacobian(F(model0), q, offsets, dims=dims)
ssodm = J'*dD
@show size(ssodm)
@test size(ssodm, 1) == nh
ssor = zeros(Float32, size(ssodm)...)
for h=1:size(ssor, 1)
if dims == (:x, :z)
for h2=1:size(ssor, 2)
ssor[h, h2, :, :] .= dm.data
end
else
ssor[h, :, :] .= dm.data
end
end
dDe = J*ssor
# @show norm(dDe - dD), norm(ssor[:] - dm[:])
a, b = dot(dD, dDe), dot(ssodm[:], ssor[:])
@test (a-b)/(a+b) ≈ 0 atol=1f-3 rtol=0
# Make sure zero offset is the rtm, remove the sumpadding
ih = div(nh, 2)+1
rtmc = dims == (:x, :z) ? ssodm[ih, ih, :, :] : ssodm[ih, :, :]
@test norm(rtm.data - rtmc, Inf) ≈ 0f0 atol=1f-4 rtol=0
end | ImageGather | https://github.com/slimgroup/ImageGather.jl.git |
|
[
"MIT"
] | 0.3.0 | e7f8a3e4bafabc4154558c1f3cd6f3e5915a534f | docs | 2381 |
[](https://slimgroup.github.io/ImageGather.jl/dev)
[](https://zenodo.org/badge/latestdoi/376881077)
# ImageGather.jl
This package implements image gather functions for seismic inversion and QC. We currently offer surface gathers via the double RTM method and subsurface offset image gather. The subsurface offset common image gather are implemented via the extended born modeling operator and its adjoint allowing for extended LSRTM.
# Example
A simple example of a surface image gather for a layered model can be found in `examples/layers_cig.jl`. This example produces the following image gathers:

: Offset gather for a good and bad background velocity model at a different position along X.
This first plot shows the expected behavior with respect to the offset. We clearly see the flat events with a correct velocity while we obtain the predicted upwards and downwards parabolic events for a respectively low and high velocity at large offset.

: Stack of offset gather along the X direction showing the difference in flatness and alignment for a good and bad background velocity model.
This second plot shows the stack along X of different gathers that shows the focusing onto the reflectors with a correct velocity while the high and low-velocity models produce unfocused and misplaced events.
# Contributions
Contributions are welcome.
# References
This work is inspired by the original [double RTM](https://library.seg.org/doi/pdfplus/10.1190/segam2012-1007.1) paper for the surface gather and the [Extended Born]() paper for extended jacobian for subsurface gathers.
- Matteo Giboli, Reda Baina, Laurence Nicoletis, and Bertrand Duquet, "Reverse Time Migration surface offset gathers part 1: a new method to produce ‘classical’ common image gathers", SEG Technical Program Expanded Abstracts 2012.
- Raanan Dafni, William W Symes, "Generalized reflection tomography formulation based on subsurface offset extended imaging",
Geophysical Journal International, Volume 216, Issue 2, February 2019, Pages 1025–1042, https://doi.org/10.1093/gji/ggy478
# Authors
This package is developed and maintained by Mathias Louboutin <[email protected]>
| ImageGather | https://github.com/slimgroup/ImageGather.jl.git |
|
[
"MIT"
] | 0.3.0 | e7f8a3e4bafabc4154558c1f3cd6f3e5915a534f | docs | 877 | # ImageGather.jl documentation
ImageGather.jl provides computational QC tools for wave-equation based inversion. Namely we provides two main widely used workflows:
- Surface offset gathers (also called surface common image gather). Surface gather compute images of (RTMs) for different offset to verify the accuracy of the background velocity. The method we implement here is based on the double-rtm method [Giboli](@cite) that allows the computation of the gather with two RTMs only instead of one per offset (or offset bin).
- Subsurface offset gathers (also called subsurface common image gather) [sscig](@cite).
## Surface offset gathers
```@docs
surface_gather
double_rtm_cig
```
## Subsurface offset gathers
```@docs
judiExtendedJacobian
```
## Utility functions
```@autodocs
Modules = [ImageGather]
Pages = ["utils.jl"]
```
# References
```@bibliography
```
| ImageGather | https://github.com/slimgroup/ImageGather.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 1357 | using ClusterDepth
using Documenter
using Glob
using Literate
DocMeta.setdocmeta!(ClusterDepth, :DocTestSetup, :(using ClusterDepth); recursive=true)
GENERATED = joinpath(@__DIR__, "src")
for subfolder ∈ ["explanations", "HowTo", "tutorials", "reference"]
local SOURCE_FILES = Glob.glob(subfolder * "/*.jl", GENERATED)
#config=Dict(:repo_root_path=>"https://github.com/unfoldtoolbox/UnfoldSim")
foreach(fn -> Literate.markdown(fn, GENERATED * "/" * subfolder), SOURCE_FILES)
end
makedocs(;
modules=[ClusterDepth],
authors="Benedikt V. Ehinger, Maanik Marathe",
repo="https://github.com/s-ccs/ClusterDepth.jl/blob/{commit}{path}#{line}",
sitename="ClusterDepth.jl",
format=Documenter.HTML(;
prettyurls=get(ENV, "CI", "false") == "true",
canonical="https://s-ccs.github.io/ClusterDepth.jl",
edit_link="main",
assets=String[],
),
pages=[
"Home" => "index.md",
"Tutorials" => [
"An EEG Example" => "tutorials/eeg.md",
"EEG Example - Multichannel data" => "tutorials/eeg-multichannel.md",
],
"Reference" => [
"Clusterdepth FWER" => "reference/type1.md",
"Troendle FWER" => "reference/type1_troendle.md",
],
],
)
deploydocs(;
repo="github.com/s-ccs/ClusterDepth.jl",
devbranch="main",
)
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 4439 | using ClusterDepth
using Random
using CairoMakie
using UnfoldSim
using StatsBase
using Distributions
using DataFrames
# # Family Wise Error of ClusterDepth
# Here we calculate the Family Wise Error the ClusterDepth Correct
# That is, we want to check that pvalues we get, indeed return us a type-1 of 5% for all time-points
# The point being, that if you do tests on 113 timepoints, the chance that one is significant is not 5% but
(1 - (1 - 0.05)^113) * 100 ##%
# ## Setup Simulation
# Let's setup a simulation using UnfoldSim.jl. We simulate a simple 1x2 design with 20 subjects
n_subjects = 20
design = MultiSubjectDesign(n_subjects=n_subjects, n_items=2, items_between=Dict(:condition => ["small", "large"]))
first(generate_events(design), 5)
#
# Next we define a ground-truth signal + relation to events/design with Wilkinson Formulas.
# we want no condition effect, therefore β for the condition should be 0. We further add some inter-subject variability with the mixed models.
# We will use a simulated P300 signal, which at 250Hz has 113 samples.
signal = MixedModelComponent(;
basis=UnfoldSim.p300(; sfreq=250),
formula=@formula(0 ~ 1 + condition + (1 | subject)),
β=[1.0, 0.0],
σs=Dict(:subject => [1]),
);
#
# Let's move the actual simulation into a function, so we can call it many times.
# Note that we use (`RedNoise`)[https://unfoldtoolbox.github.io/UnfoldSim.jl/dev/literate/reference/noisetypes/] which has lot's of Autocorrelation between timepoints. nice!
function run_fun(r)
data, events = simulate(MersenneTwister(r), design, signal, UniformOnset(; offset=5, width=4), RedNoise(noiselevel=1); return_epoched=true)
data = reshape(data, size(data, 1), :)
data = data[:, events.condition.=="small"] .- data[:, events.condition.=="large"]
return data, clusterdepth(data'; τ=quantile(TDist(n_subjects - 1), 0.95), nperm=1000)
end;
# ## Understanding the simulation
# let's have a look at the actual data by running it once, plotting condition wise trials, the ERP and histograms of uncorrected and corrected p-values
data, pval = run_fun(5)
conditionSmall = data[:, 1:2:end]
conditionLarge = data[:, 2:2:end]
pval_uncorrected = 1 .- cdf.(TDist(n_subjects - 1), abs.(ClusterDepth.studentt(conditionSmall .- conditionLarge)))
sig = pval_uncorrected .<= 0.025;
# For the uncorrected p-values based on the t-distribution, we get a type1 error over "time":
mean(sig)
# this is the type 1 error of 5% we expected.
# !!! note
# Type-I error is not the FWER (family wise error rate). FWER is the property of a set of tests (in this case tests per time-point), we can calculate it by repeating such tests,
# and checking for each repetition whether any sample of a repetition is significant (e.g. `any(sig)` followed by a `mean(repetitions_anysig)`).
f = Figure();
series!(Axis(f[1, 1], title="condition==small"), conditionSmall', solid_color=:red)
series!(Axis(f[1, 2], title="condition==large"), conditionLarge', solid_color=:blue)
ax = Axis(f[2, 1:2], title="ERP (mean over trials)")
sig = allowmissing(sig)
sig[sig.==0] .= missing
@show sum(skipmissing(sig))
lines!(sig, color=:gray, linewidth=4)
lines!(ax, mean(conditionSmall, dims=2)[:, 1], solid_color=:red)
lines!(ax, mean(conditionLarge, dims=2)[:, 1], solid_color=:blue)
hist!(Axis(f[3, 1], title="uncorrected pvalues"), pval_uncorrected, bins=0:0.01:1.1)
hist!(Axis(f[3, 2], title="clusterdepth corrected pvalues"), pval, bins=0:0.01:1.1)
f
#----
# ## Run simulations
# This takes some seconds (depending on your infrastructure)
reps = 500
res = fill(NaN, reps, 2)
Threads.@threads for r = 1:reps
data, pvals = run_fun(r)
res[r, 1] = mean(pvals .<= 0.05)
res[r, 2] = mean(abs.(ClusterDepth.studentt(data)) .>= quantile(TDist(n_subjects - 1), 0.975))
end;
# Finally, let's calculate the percentage of simulations where we find a significant effect somewhere
mean(res .> 0, dims=1) |> x -> (:clusterdepth => x[1], :uncorrected => x[2])
# Nice, correction seems to work in principle :) Clusterdepth is not exactly 5%, but with more repetitions we should get there (e.g. with 5000 repetitions, we get 0.051%).
# !!! info
# if you look closely, the `:uncorrected` value (around 60%) is not as bad as the 99% promised in the introduction. This is due to the correlation between the tests introduced by the noise. Indeed, a good exercise is to repeat everything, but put `RedNoise` to `WhiteNoise` | ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 2456 | using ClusterDepth
using Random
using CairoMakie
using UnfoldSim
using StatsBase
using ProgressMeter
using Distributions
# ### Family Wise Error of Troendle
# Here we calculate the Family Wise Error of doing `ntests` at the same time.
# That is, we want to check that Troendle indeed returns us a type-1 of 5% for a _set_ of tests.
#
# The point being, that if you do 30 tests, the chance that one is significant is not 5% but actually
(1 - (1 - 0.05)^30) * 100 ##%
# Let's setup some simulation parameters
reps = 1000
perms = 1000
ntests = 30;
# we will use the student-t in it's 2-sided variant (abs of it)
fun = x -> abs.(ClusterDepth.studentt(x));
# this function simulates data without any effect (H0), then the permutations, and finally calls troendle
function run_fun(r, perms, fun, ntests)
rng = MersenneTwister(r)
data = randn(rng, ntests, 50)
perm = Matrix{Float64}(undef, size(data, 1), perms)
stat = fun(data)
for p = 1:perms
ClusterDepth.sign_permute!(rng, data)
perm[:, p] = fun(data)
end
return data, stat, troendle(perm, stat)
end;
# let's test it once
data, stats_t, pvals = run_fun(1, perms, fun, ntests);
println("data:", size(data), " t-stats:", size(stats_t), " pvals:", size(pvals))
# run the above function `reps=1000`` times - we also save the uncorrected t-based pvalue
pvals_all = fill(NaN, reps, 2, ntests)
Threads.@threads for r = 1:reps
data, stat, pvals = run_fun(r, perms, fun, ntests)
pvals_all[r, 1, :] = pvals
pvals_all[r, 2, :] = (1 .- cdf.(TDist(size(data, 2)), abs.(stat))) .* 2 # * 2 becaue of twosided. Troendle takes this into account already
end;
# Let's check in how many of our simlations we have a significant p-value =<0.05
res = any(pvals_all[:, :, :] .<= 0.05, dims=3)[:, :, 1]
mean(res .> 0, dims=1) |> x -> (:troendle => x[1], :uncorrected => x[2])
# Nice. Troendle fits perfectly and the uncorrected is pretty close to what we calculated above!
# Finally we end this with a short figure to get a better idea of how this data looks like and a histogram of the p-values
f = Figure()
ax = f[1, 1] = Axis(f)
lines!(ax, abs.(ClusterDepth.studentt(data)))
heatmap!(Axis(f[2, 1]), data)
series!(Axis(f[2, 2]), data[:, 1:7]')
h1 = scatter!(Axis(f[1, 2]; yscale=log10), pvals, label="troendle")
hlines!([0.05, 0.01])
hist!(Axis(f[3, 1]), pvals_all[:, 1, :][:], bins=0:0.01:1.1)
hist!(Axis(f[3, 2]), pvals_all[:, 2, :][:], bins=0:0.01:1.1)
f
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 746 | using ClusterDepth
using Random
using CairoMakie
n_t =40 # timepoints
n_sub = 50
n_perm = 5000
snr = 0.5 # signal to nois
## add a signal to the middle
signal = vcat(zeros(n_t÷4), sin.(range(0,π,length=n_t÷2)), zeros(n_t÷4))
## same signal for all subs
signal = repeat(signal,1,n_sub)
## add noise
data = randn(MersenneTwister(123),n_t,n_sub).+ snr .* signal
## by default assumes τ=2.3 (~alpha=0.05), and one-sample ttest
@time pvals = clusterdepth(data);
f = Figure()
ax = f[1,1] = Axis(f)
lines!(abs.(ClusterDepth.studentt(data)))
h1 = scatter(f[1,2],pvals;axis=(;yscale=log10),label="troendle")
pvals2 = clusterdepth(data;pval_type=:naive)
h2 = scatter!(1.2:40.2,pvals2,color="red",label="naive")
#hlines!(([0.05]))
axislegend()
f | ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 3245 | using ClusterDepth
using Random
using CairoMakie
using UnfoldSim
using Unfold
using UnfoldMakie
using Statistics
# ## How to use clusterDepth multiple comparison correction on multichannel data
# This tutorial is adapted from the first EEG example and uses the HArtMuT NYhead model (https://github.com/harmening/HArtMuT) to simulate multiple channels.
# First set up the EEG simulation as before, with one subject and 40 trials:
design = SingleSubjectDesign(conditions=Dict(:condition => ["car", "face"])) |> x -> RepeatDesign(x, 40);
p1 = LinearModelComponent(;
basis=p100(; sfreq=250),
formula=@formula(0 ~ 1),
β=[1.0]
);
n170 = LinearModelComponent(;
basis=UnfoldSim.n170(; sfreq=250),
formula=@formula(0 ~ 1 + condition),
β=[1.0, 0.5], # condition effect - faces are more negative than cars
);
p300 = LinearModelComponent(;
basis=UnfoldSim.p300(; sfreq=250),
formula=@formula(0 ~ 1 + condition),
β=[1.0, 0], # no p300 condition effect
);
# Now choose some source coordinates for each of the p100, n170, p300 that we want to simulate, and use the helper function closest_srcs to get the HArtMuT sources that are closest to these coordinates:
src_coords = [
[20, -78, -10], #p100
[-20, -78, -10], #p100
[50, -40, -25], #n170
[0, -50, 40], #p300
[0, 5, 20], #p300
];
headmodel_HArtMuT = headmodel()
get_closest = coord -> UnfoldSim.closest_src(coord, headmodel_HArtMuT.cortical["pos"]) |> pi -> magnitude(headmodel_HArtMuT; type="perpendicular")[:, pi]
p1_l = p1 |> c -> MultichannelComponent(c, get_closest([-20, -78, -10]))
p1_r = p1 |> c -> MultichannelComponent(c, get_closest([20, -78, -10]))
n170_r = n170 |> c -> MultichannelComponent(c, get_closest([50, -40, -25]))
p300_do = p300 |> c -> MultichannelComponent(c, get_closest([0, -50, -40]))
p300_up = p300 |> c -> MultichannelComponent(c, get_closest([0, 5, 20]))
data, events = simulate(MersenneTwister(1), design, [p1_l, p1_r, n170_r, p300_do, p300_up],
UniformOnset(; offset=0.5 * 250, width=100),
RedNoise(noiselevel=1); return_epoched=true);
# ## Plotting
# This is what the data looks like, for one channel/trial respectively:
f = Figure()
Axis(f[1, 1], title="Single channel, all trials", xlabel="time", ylabel="y")
series!(data[1, :, :]', solid_color=:black)
lines!(mean(data[1, :, :], dims=2)[:, 1], color=:red)
hlines!([0], color=:gray)
Axis(f[2, 1], title="All channels, average over trials", xlabel="time", ylabel="y")
series!(mean(data, dims=3)[:, :, 1], solid_color=:black)
hlines!([0], color=:gray)
f
# And some topoplots:
positions = [Point2f(p[1] + 0.5, p[2] + 0.5) for p in to_positions(headmodel_HArtMuT.electrodes["pos"]')]
df = UnfoldMakie.eeg_matrix_to_dataframe(mean(data, dims=3)[:, :, 1], string.(1:length(positions)));
Δbin = 20 # 20 samples / bin
plot_topoplotseries(df, Δbin; positions=positions, visual=(; enlarge=1, label_scatter=false))
# ## ClusterDepth
# Now that the simulation is done, let's try out ClusterDepth and plot our results
# Note that this is a simple test of "activity" vs. 0
pvals = clusterdepth(data; τ=1.6, nperm=100);
fig, ax, hm = heatmap(transpose(pvals))
ax.title = "pvals";
ax.xlabel = "time";
ax.ylabel = "channel";
Colorbar(fig[:, end+1], hm);
fig
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 3918 | using ClusterDepth
using Random
using CairoMakie
using UnfoldSim
using StatsBase
using Distributions
using DataFrames
using Unfold
using UnfoldMakie
# ## How to use clusterDepth multiple comparison correction
# !!! info
# This tutorial focuses on single-channel data. For multichannel data, see the tutorial "Further EEG Example".
# Let's setup an EEG simulation using UnfoldSim.jl. We simulate a simple 1x2 design with 20 subjects, each with 40 trials
n_subjects = 20
design = MultiSubjectDesign(n_subjects=n_subjects, n_items=40, items_between=Dict(:condition => ["car", "face"]))
first(generate_events(design), 5)
# next we define a ground-truth signal + relation to events/design with Wilkinson Formulas
# let's simulate a P100, a N170 and a P300 - but an effect only on the N170
p1 = MixedModelComponent(;
basis=UnfoldSim.p100(; sfreq=250),
formula=@formula(0 ~ 1 + (1 | subject)),
β=[1.0],
σs=Dict(:subject => [1]),
);
n170 = MixedModelComponent(;
basis=UnfoldSim.n170(; sfreq=250),
formula=@formula(0 ~ 1 + condition + (1 + condition | subject)),
β=[1.0, -0.5], # condition effect - faces are more negative than cars
σs=Dict(:subject => [1, 0.2]), # random slope yes please!
);
p300 = MixedModelComponent(;
basis=UnfoldSim.p300(; sfreq=250),
formula=@formula(0 ~ 1 + condition + (1 + condition | subject)),
β=[1.0, 0], ## no p300 condition effect
σs=Dict(:subject => [1, 1.0]), # but a random slope for condition
);
## Start the simulation
data, events = simulate(MersenneTwister(1), design, [p1, n170, p300], UniformOnset(; offset=500, width=100), RedNoise(noiselevel=1); return_epoched=true)
times = range(0, stop=size(data, 1) / 250, length=size(data, 1));
# let's fit an Unfold Model for each subject
# !!! note
# In principle, we do not need Unfold here - we could simply calculate (subjectwise) means of the conditions, and their time-resolved difference. Using Unfold.jl here simply generalizes it to more complex design, e.g. with continuous predictors etc.
models = map((d, ev) -> (fit(UnfoldModel, @formula(0 ~ 1 + condition), DataFrame(ev), d, times), ev.subject[1]),
eachslice(data; dims=3),
groupby(events, :subject))
# now we can inspect the data easily, and extract the face-effect
function add_subject!(df, s)
df[!, :subject] .= s
return df
end
allEffects = map((x) -> (effects(Dict(:condition => ["car", "face"]), x[1]), x[2]) |> (x) -> add_subject!(x[1], x[2]), models) |> e -> reduce(vcat, e)
plot_erp(allEffects; mapping=(color=:condition, group=:subject))
# extract the face-coefficient from the linear model
allCoefs = map(m -> (coeftable(m[1]), m[2]) |> (x) -> add_subject!(x[1], x[2]), models) |> e -> reduce(vcat, e)
plot_erp(allCoefs; mapping=(group=:subject, col=:coefname))
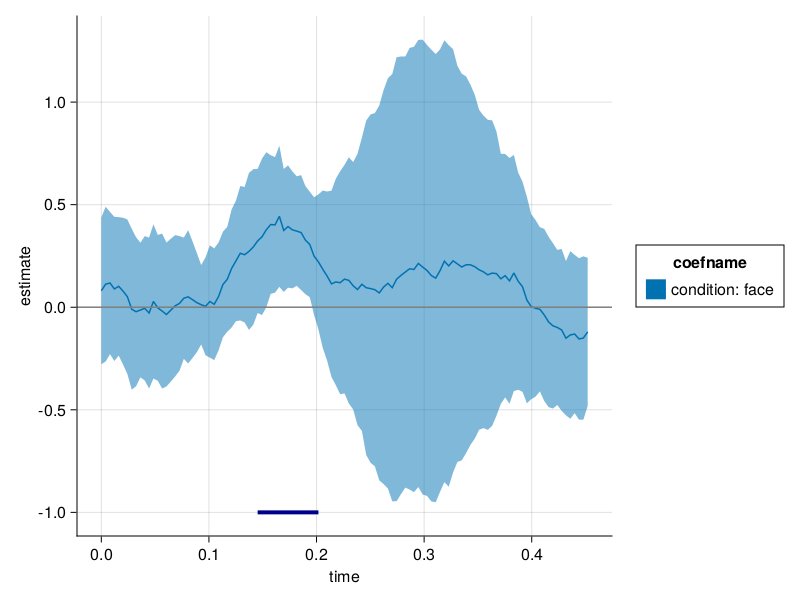
# let's unstack the tidy-coef table into a matrix and put it to clusterdepth for clusterpermutation testing
faceCoefs = allCoefs |> x -> subset(x, :coefname => x -> x .== "condition: face")
erpMatrix = unstack(faceCoefs, :subject, :time, :estimate) |> x -> Matrix(x[:, 2:end])' |> collect
summary(erpMatrix)
# ## Clusterdepth
pvals = clusterdepth(erpMatrix; τ=quantile(TDist(n_subjects - 1), 0.95), nperm=5000);
# well - that was fast, less than a second for a cluster permutation test. not bad at all!
# ## Plotting
# Some plotting, and we add the identified cluster
# first calculate the ERP
faceERP = groupby(faceCoefs, [:time, :coefname]) |>
x -> combine(x, :estimate => mean => :estimate,
:estimate => std => :stderror);
# put the pvalues into a nicer format
pvalDF = ClusterDepth.cluster(pvals .<= 0.05) |> x -> DataFrame(:from => x[1] ./ 250, :to => (x[1] .+ x[2]) ./ 250, :coefname => "condition: face")
plot_erp(faceERP; stderror=true, pvalue=pvalDF)
# Looks good to me! We identified the cluster :-)
# old unused code to use extra=(;pvalue=pvalDF) in the plotting function, but didnt work.
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 569 | module ClusterDepth
using Random
using Images
using SparseArrays
#using ExtendableSparse
using StatsBase
import Base.show
struct ClusterDepthMatrix{T} <: AbstractMatrix{T}
J::Any
ClusterDepthMatrix(x) = new{SparseMatrixCSC}(sparse(x))
end
Base.show(io::IO, x::ClusterDepthMatrix) = show(io, x.J)
Base.show(io::IO, m::MIME"text/plain", x::ClusterDepthMatrix) = show(io, m, x.J)
struct test{T} <: AbstractArray{T,2}
J::Any
end
include("cluster.jl")
include("pvals.jl")
include("utils.jl")
include("troendle.jl")
export troendle
export clusterdepth
end
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 7523 | """
clusterdepth(rng,data::AbstractArray;τ=2.3, statfun=x->abs.(studentt(x)),permfun=sign_permute!,nperm=5000,pval_type=:troendle)
calculate clusterdepth of given datamatrix.
- `data`: `statfun` will be applied on last dimension of data (typically this will be subjects)
Optional
- `τ`: Cluster-forming threshold
- `nperm`: number of permutations, default 5000
- `stat_type`: default the one-sample `t-test`, custom function can be specified (see `statfun!` and `statfun`)
- `side_type`: default: `:abs` - what function should be applied after the `statfun`? could be `:abs`, `:square`, `:positive` to test positive clusters, `:negative` to test negative clusters. Custom function can be provided, see `sidefun``
- `perm_type`: default `:sign` for one-sample data (e.g. differences), performs sign flips. custom function can be provided, see `permfun`
- `pval_type`: how to calculate pvalues within each cluster, default `:troendle`, see `?pvals`
- `statfun` / `statfun!` a function that either takes one or two arguments and aggregates over last dimension. in the two argument case we expect the first argument to be modified inplace and provide a suitable Vector/Matrix.
- `sidefun`: default `abs`. Provide a function to be applied on each element of the output of `statfun`.
- `permfun` function to permute the data, should accept an RNG-object and the data. can be inplace, the data is copied, but the same array is shared between permutations
"""
clusterdepth(data::AbstractArray, args...; kwargs...) =
clusterdepth(MersenneTwister(1), data, args...; kwargs...)
function clusterdepth(
rng,
data::AbstractArray;
τ = 2.3,
stat_type = :onesample_ttest,
perm_type = :sign,
side_type = :abs,
nperm = 5000,
pval_type = :troendle,
(statfun!) = nothing,
statfun = nothing,
)
if stat_type == :onesample_ttest
statfun! = studentt!
statfun = studentt
end
if perm_type == :sign
permfun = sign_permute!
end
if side_type == :abs
sidefun = abs
elseif side_type == :square
sidefun = x -> x^2
elseif side_type == :negative
sidefun = x -> -x
elseif side_type == :positive
sidefun = nothing # the default :)
else
@assert isnothing(side_type) "unknown side_type ($side_type) specified. Check your spelling and ?clusterdepth"
end
cdmTuple = perm_clusterdepths_both(
rng,
data,
permfun,
τ;
nₚ = nperm,
(statfun!) = statfun!,
statfun = statfun,
sidefun = sidefun,
)
return pvals(statfun(data), cdmTuple, τ; type = pval_type)
end
function perm_clusterdepths_both(
rng,
data,
permfun,
τ;
statfun = nothing,
(statfun!) = nothing,
nₚ = 1000,
sidefun = nothing,
)
@assert !(isnothing(statfun) && isnothing(statfun!)) "either statfun or statfun! has to be defined"
data_perm = deepcopy(data)
rows_h = Int[]
cols_h = Int[]
vals_h = Float64[]
rows_t = Int[]
cols_t = Int[]
vals_t = Float64[]
if ndims(data_perm) == 2
d0 = Array{Float64}(undef, size(data_perm, 1))
else
d0 = Array{Float64}(undef, size(data_perm)[[1, 2]])
end
#@debug size(d0)
#@debug size(data_perm)
for i = 1:nₚ
# permute
d_perm = permfun(rng, data_perm)
if isnothing(statfun!)
d0 = statfun(d_perm)
else
# inplace!
statfun!(d0, d_perm)
end
if !isnothing(sidefun)
d0 .= sidefun.(d0)
end
# get clusterdepth
(fromTo, head, tail) = calc_clusterdepth(d0, τ)
# save it
if !isempty(head)
append!(rows_h, fromTo)
append!(cols_h, fill(i, length(fromTo)))
append!(vals_h, head)
#Jₖ_head[fromTo,i] .+=head
end
if !isempty(tail)
append!(rows_t, fromTo)
append!(cols_t, fill(i, length(fromTo)))
append!(vals_t, tail)
#Jₖ_tail[fromTo,i] .+=tail
end
end
Jₖ_head = sparse(rows_h, cols_h, vals_h)#SparseMatrixCSC(nₚ,maximum(rows_h), cols_h,rows_h,vals_h)
Jₖ_tail = sparse(rows_t, cols_t, vals_t)#SparseMatrixCSC(nₚ,maximum(rows_t), cols_t,rows_t,vals_t)
return ClusterDepthMatrix((Jₖ_head)), ClusterDepthMatrix((Jₖ_tail))
end
"""
calc_clusterdepth(data,τ)
returns tuple with three entries:
1:maxLength, maximal clustervalue per clusterdepth head, same for tail
We assume data and τ have already been transformed for one/two sided testing, so that we can do d0.>τ for finding clusters
"""
function calc_clusterdepth(d0::AbstractArray{<:Real,2}, τ)
nchan = size(d0, 1)
# save all the results from calling calc_clusterdepth on individual channels
(allFromTo, allHead, allTail) = (
Array{Vector{Integer}}(undef, nchan),
Array{Vector{Float64}}(undef, nchan),
Array{Vector{Float64}}(undef, nchan),
)
fromTo = []
for i = 1:nchan
(a, b, c) = calc_clusterdepth(d0[i, :], τ)
allFromTo[i] = a
allHead[i] = b
allTail[i] = c
if (length(a) > length(fromTo)) # running check to find the length ('fromTo') of the largest cluster
fromTo = a
end
end
# for each clusterdepth value, select the largest cluster value found across all channels
(head, tail) = (zeros(length(fromTo)), zeros(length(fromTo)))
for i = 1:nchan
for j in allFromTo[i]
if allHead[i][j] > head[j]
head[j] = allHead[i][j]
end
if allTail[i][j] > tail[j]
tail[j] = allTail[i][j]
end
end
end
return fromTo, head, tail
end
function calc_clusterdepth(d0, τ)
startIX, len = cluster(d0 .> τ)
if isempty(len) # if nothing above threshold, just go on
return [], [], []
end
maxL = 1 + maximum(len) # go up only to max-depth
valCol_head = Vector{Float64}(undef, maxL)
valCol_tail = Vector{Float64}(undef, maxL)
fromTo = 1:maxL
for j in fromTo
# go over clusters implicitly
# select clusters that are larger (at least one)
selIX = len .>= (j - 1)
if !isempty(selIX)
ix = startIX[selIX] .+ (j - 1)
valCol_head[j] = maximum(d0[ix])
# potential optimization is that for j = 0 and maxL = 0, tail and head are identical
ix = startIX[selIX] .+ (len[selIX]) .- (j - 1)
valCol_tail[j] = maximum(d0[ix])
end
end
return fromTo, valCol_head, valCol_tail
end
"""
finds neighbouring clusters in the vector and returns start + length vectors
if the first and last cluster start on the first/last sample, we dont know their real depth
Input is assumed to be a thresholded Array with only 0/1
"""
function cluster(data)
label = label_components(data)
K = maximum(label)
start = fill(0, K)
stop = fill(0, K)
for k = 1:K
#length[k] = sum(label.==k)
start[k] = findfirst(==(k), label)
stop[k] = findlast(==(k), label)
end
len = stop .- start
# if the first and last cluster start on the first/last sample, we dont know their real depth
if length(start) > 0 && start[end] + len[end] == length(data)
start = start[1:end-1]
len = len[1:end-1]
end
if length(start) > 0 && start[1] == 1
start = start[2:end]
len = len[2:end]
end
return start, len
end
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 1071 | """
permute data and returns clusterdepth
returns for each cluster-depth, the maximum stat-value over all detected clusters, from both sides at the same time
Conversely, perm_clusterdepths_both, returns the maximum stat-value from head and tail separately (combined after calcuating pvalues)
"""
function perm_clusterdepths_combined(rng, data, statFun)
Jₖ = spzeros(m - 1, nₚ)
for i = 1:nₚ
d0 = perm(rng, data, statFun)
startIX, len = get_clusterdepths(d0, τ)
maxL = maximum(len)
# go up to max-depth
valCol = Vector{Float64}(undef, maxL)
fromTo = 1:maxL
for j in fromTo
# go over clusters implicitly
# select clusters that are larger (at least one)
selIX = len .>= j
if !isempty(selIX)
ix_head = startIX[selIX] .+ j
ix_tail = len[selIX] .+ startIX[selIX] .- j
valCol[j] = maximum(d0[vcat(ix_head, ix_tail)])
end
end
Jₖ[fromTo, i] = valCol
end
return Jₖ
end;
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 2697 |
"""
pvals(data;kwargs...) = pvals(data[2:end],data[1];kwargs...)
pvals(data::AbstractVector,stat::Real;type=:twosided)
calculates pvalues based on permutation results
if called with `stat`, first entry is assumed to be the observation
"""
pvals(data; kwargs...) = pvals(data[2:end], data[1]; kwargs...)
function pvals(data::AbstractVector, stat::Real; type = :twosided)
data = vcat(stat, data)
if type == :greater || type == :twosided
comp = >=
if type == :twosided
data = abs.(data)
stat = abs.(stat)
end
elseif type == :lesser
comp = <=
else
error("not implemented")
end
pvals = (sum(comp(stat[1]), data)) / (length(data))
return pvals
end
"""
Calculate pvals from cluster-depth permutation matrices
"""
pvals(stat::Matrix, args...; kwargs...) =
mapslices(x -> pvals(x, args...; kwargs...), stat; dims = (2))
pvals(stat, J::ClusterDepthMatrix, args...; kwargs...) =
pvals(stat, (J,), args...; kwargs...)
function pvals(
stat::AbstractVector,
Jₖ::NTuple{T,ClusterDepthMatrix},
τ;
type = :troendle,
) where {T}
start, len = cluster(stat .> τ) # get observed clusters
p = fill(1.0, size(stat, 1))
if type == :troendle
for k = 1:length(start) # go over clusters
s = start[k]
l = len[k]
forwardIX = s:(s+l)
@views t_head = troendle(Jₖ[1], sparsevec(1:(l+1), stat[forwardIX], l + 1))
@views p[forwardIX] = t_head[1:(l+1)]
if length(Jₖ) == 2
backwardIX = (s+l):-1:s
@views t_tail = troendle(Jₖ[2], sparsevec(1:(l+1), stat[backwardIX], l + 1))
@views p[backwardIX] = max.(p[backwardIX], t_tail[1:(l+1)])
end
end
elseif type == :naive
function getJVal(Jₖ, l)
if l >= size(Jₖ, 1)
valsNull = 0
else
valsNull = @view Jₖ[l+1, :]
end
return valsNull
end
for k = 1:length(start) # go over clusters
for ix = start[k]:(start[k]+len[k])
p[ix] =
(1 + sum(stat[ix] .<= getJVal(Jₖ[1].J, len[k]))) /
(size(Jₖ[1].J, 2) + 1)
if length(Jₖ) == 2
tail_p =
(1 + sum((stat[ix] .<= getJVal(Jₖ[2].J, len[k])))) ./
(size(Jₖ[2].J, 2) + 1)
p[ix] = max(p[ix], tail_p)
end
end
end
#p = p .+ 1/size(Jₖ[1].J,2)
else
error("unknown type")
end
# add conservative permutation fix
return p
end
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 5098 | """
pvals_rankbased(perm::AbstractMatrix,stat::AbstractVector;kwargs...)
pvals_rankbased(cdm::ClusterDepthMatrix,stat::AbstractSparseVector;kwargs...)
Takes a matrix filled with permutation results and a observed matrix (size ntests) and calculates the p-values for each entry along
the permutation dimension
`perm`: Matrix of permutations with dimensions `(ntests x permutations)`
`stat`: Vector of observed statistics size `ntests`
For `cdm::ClusterDepthMatrix` we can easily trim the permutation matrix towards the end (as it is actually a ragged Matrix).
That is, the permutation matrix might look like:
perm = [
x x x x x x x;
x x . x . x x;
x x . x . . x;
x x . . . . x;
. x . . . . .;
. . . . . . .;
. . . . . . .;
. . . . . . .;
]
Then the last three rows can simply be removed. rowwise-Ranks/pvalues would be identical anyway
The same will be checked for the stat-vector, if the stat vector is only "3" depths long, but the permutation has been calculated for "10" depths, we do not need to check the last 7 depths
of the permutation matrix.
**Output** will always be Dense Matrix (length(stat),nperm+1) with the first column being the pvalues of the observations
"""
function pvals_rankbased(cdm::ClusterDepthMatrix, stat::AbstractSparseVector; kwargs...)
perm = cdm.J
if length(stat) > size(perm, 1) # larger cluster in stat than perms
perm = sparse(findnz(perm)..., length(stat), size(perm, 2))
elseif length(stat) < size(perm, 1)
#stat = sparsevec(findnz(stat)...,size(perm,1))
i, j, v = findnz(perm)
ix = i .<= length(stat)
perm = sparse(i[ix], j[ix], v[ix], length(stat), size(perm, 2))
end
pvals = pvals_rankbased(perm, stat; kwargs...)
return pvals
end
function pvals_rankbased(perm::AbstractMatrix, stat::AbstractVector; type = :twosided)
# add stat to perm
d = hcat(stat, perm)
# fix specific testing
if type == :twosided
d = .-abs.(d)
elseif type == :greater
d = .-d
elseif type == :lesser
else
error("unknown type")
end
d = Matrix(d)
# potential improvement, competerank 1224 -> but should be modified competerank 1334, then we could skip the expensive ceil below
d = mapslices(tiedrank, d, dims = 2)
# rank & calc p-val
#@show(d[1:10,1:10])
d .= ceil.(d) ./ (size(d, 2))
return d
end
"""
in some sense: `argsort(argunique(x))`, returns the indices to get a sorted unique of x
"""
function ix_sortUnique(x)
uniqueidx(v) = unique(i -> v[i], eachindex(v))
un_ix = uniqueidx(x)
sort_ix = sortperm(x[un_ix])
sortUn_ix = un_ix[sort_ix]
return sortUn_ix
end
"""
calculates the minimum in `X` along `dims=2` in the columns specified by àrrayOfIndicearrays` which could be e.g. `[[1,2],[5,6],[3,4,7]]`
"""
function multicol_minimum(x::AbstractMatrix, arrayOfIndicearrays::AbstractVector)
min = fill(NaN, size(x, 2), length(arrayOfIndicearrays))
for to = 1:length(arrayOfIndicearrays)
@views min[:, to] = minimum(x[arrayOfIndicearrays[to], :], dims = 1)
end
return min
end
"""
function troendle(perm::AbstractMatrix,stat::AbstractVector;type=:twosided)
Multiple Comparison Correction as in Troendle 1995
`perm` with size ntests x nperms
`stat` with size ntests
`type` can be :twosided (default), :lesser, :greater
Heavily inspired by the R implementation in permuco from Jaromil Frossard
Note: While permuco is released under BSD, the author Jaromil Frossard gave us an MIT license for the troendle and the clusterdepth R-functions.
"""
function troendle(perm::AbstractMatrix, stat::AbstractVector; type = :twosided)
pAll = pvals_rankbased(perm, stat; type = type)
# get uncorrected pvalues of data
#@show size(pAll)
pD = pAll[:, 1] # get first observation
# rank the pvalues
pD_rank = tiedrank(pD)
# test in ascending order, same p-vals will be combined later
# the following two lines are
sortUn_ix = ix_sortUnique(pD_rank)
# these two lines would be identical
#testOrder = pD_rank[sortUn_ix]
#testOrder = sort(unique(pD_rank))
# as ranks can be tied, we have to take those columns, and run a "min" on them
testOrder_all = [findall(x .== pD_rank) for x in pD_rank[sortUn_ix]]
minPermPArray = multicol_minimum(pAll, testOrder_all)
# the magic happens here, per permutation
resortPermPArray = similar(minPermPArray)
# the following line can be made much faster by using views & reverse the arrays
#resortPermPArray = reverse(accumulate(min,reverse(minPermPArray),dims=2))
@views accumulate!(
min,
resortPermPArray[:, end:-1:1],
minPermPArray[:, end:-1:1],
dims = 2,
)
pval_testOrder = pvals.(eachcol(resortPermPArray); type = :lesser)
pval_testOrderMax = accumulate(max, pval_testOrder) # no idea why though
uniqueToNonUnique = vcat([fill(x, length(v)) for (x, v) in enumerate(testOrder_all)]...)
return pval_testOrderMax[uniqueToNonUnique][invperm(vcat(testOrder_all...))]
end
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 1827 | function studentt!(out::AbstractMatrix, x::AbstractArray{<:Real,3}; kwargs...)
for (x_ch, o_ch) in zip(eachslice(x, dims = 1), eachslice(out, dims = 1))
#@debug size(x_ch),size(o_ch)
studentt!(o_ch, x_ch; kwargs...)
end
return out
end
"""
studentt_test!(out,x;type=:abs)
strongly optimized one-sample t-test function.
Implements: t = mean(x) / ( sqrt(var(x))) / sqrt(size(x,2)-1)
Accepts 2D or 3D matrices, always aggregates over the last dimension
"""
function studentt_test!(out, x::AbstractMatrix)
mean!(out, x)
df = 1 ./ sqrt(size(x, 2) - 1)
#@debug size(out),size(x)
tmp = [1.0]
for k in eachindex(out)
std!(tmp, @view(x[k, :]), out[k])
@views out[k] /= (sqrt(tmp[1]) * df)
end
return out
end
function std!(tmp, x_slice, μ)
@views x_slice .= (x_slice .- μ) .^ 2
sum!(tmp, x_slice)
tmp .= sqrt.(tmp ./ (length(x_slice) - 1))
end
function studentt!(out, x)
#@debug size(out),size(x)
mean!(out, x)
out .= out ./ (std(x, mean = out, dims = 2)[:, 1] ./ sqrt(size(x, 2) - 1))
end
function studentt(x::AbstractMatrix)
# more efficient than this one liner
# studentt(x::AbstractMatrix) = (mean(x,dims=2)[:,1])./(std(x,dims=2)[:,1]./sqrt(size(x,2)-1))
μ = mean(x, dims = 2)[:, 1]
μ .= μ ./ (std(x, mean = μ, dims = 2)[:, 1] ./ sqrt(size(x, 2) - 1))
end
studentt(x::AbstractArray{<:Real,3}) =
dropdims(mapslices(studentt, x, dims = (2, 3)), dims = 3)
"""
Permutation via random sign-flip
Flips signs along the last dimension
"""
function sign_permute!(rng, x::AbstractArray)
n = ndims(x)
@assert n > 1 "vectors cannot be permuted"
fl = rand(rng, [-1, 1], size(x, n))
for (flip, xslice) in zip(fl, eachslice(x; dims = n))
xslice .= xslice .* flip
end
return x
end
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 562 | @testset "cluster" begin
s, l = ClusterDepth.cluster(
[4.0, 0.0, 10.0, 0.0, 3.0, 4.0, 0, 4.0, 4.0, 0.0, 0.0, 5.0] .> 0.9,
)
@test s == [3, 5, 8]
@test l == [0, 1, 1]
s, l = ClusterDepth.cluster([0.0, 0.0, 0.0, 0.0] .> 0.9)
@test s == []
@test l == []
end
@testset "Tests for 2D data" begin
data = randn(StableRNG(1), 4, 5)
@show ClusterDepth.calc_clusterdepth(data, 0)
end
@testset "Tests for 3D data" begin
data = randn(StableRNG(1), 3, 20, 5)
@show ClusterDepth.clusterdepth(data; τ = 0.4, nperm = 5)
end
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 1960 | @testset "pvals" begin
@testset "pvals-onePerm" begin
@test ClusterDepth.pvals(zeros(100), 1) == 1 / 101
@test ClusterDepth.pvals(vcat(1, zeros(100))) == 1 / 101
@test ClusterDepth.pvals(vcat(0, ones(100))) == 101 / 101
@test ClusterDepth.pvals(vcat(80, 0:99)) == 21 / 101
end
@testset "pvals-ClusterDepthMatrix" begin
cdm = ClusterDepth.ClusterDepthMatrix(sparse(ones(10, 1000)))
p = ClusterDepth.pvals([0.0, 0.0, 0.0, 0.0, 0.0, 1.0, 2.0, 1.0, 0.0, 0.0], cdm, 0.1)
@test p[7] ≈ 1 / 1001
p = ClusterDepth.pvals([0.0, 0.0, 0.0, 0.0, 0.0, 1.0, 2.0, 1.0, 0.0, 0.0], cdm, 2.1)
p .≈ 1000 / 1000
p = ClusterDepth.pvals(
[0.0, 0.0, 0.0, 0.0, 0.0, 1.0, 2.0, 1.0, 0.0, 0.0],
cdm,
0.1,
type = :naive,
)
@test p[7] ≈ 1 / 1001
J = zeros(10, 1000)
J[1, :] .= 5
J[2, :] .= 3
J[3, :] .= 1
cdm = ClusterDepth.ClusterDepthMatrix(sparse(J))
p = ClusterDepth.pvals(
[4.0, 0.0, 10.0, 0.0, 3.0, 4.0, 0.0, 4.0, 4.0, 0.0, 0.0],
cdm,
0.9,
)
@test all((p .> 0.05) .== [1, 1, 0, 1, 1, 0, 1, 1, 0, 1, 1])
#two tailed
p = ClusterDepth.pvals(
[4.0, 0.0, 10.0, 0.0, 3.0, 4.0, 0.0, 4.0, 4.0, 0.0, 0.0],
(cdm, cdm),
0.9,
)
@test all((p .> 0.05) .== [1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1])
p = ClusterDepth.pvals([0.0, 1.0, 2.0, 3.0, 4.0, 5.1, 2.0, 6.0], (cdm, cdm), 0.9)
@test all((p .> 0.05) .== [1, 1, 1, 1, 1, 1, 1, 1])
p = ClusterDepth.pvals([0.0, 1.0, 2.0, 3.0, 4.0, 5.1, 2.0, 0], (cdm, cdm), 0.9)
@test all((p .> 0.05) .== [1, 1, 1, 0, 0, 0, 1, 1])
p = ClusterDepth.pvals([0.0, 1.0, 2.0, 3.0, 4.0, 2.0, 1.0, 0.0, 6], (cdm, cdm), 0.9)
@test all((p .> 0.05) .== [1, 1, 1, 0, 0, 1, 1, 1, 1])
end
end
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 179 | using ClusterDepth
include("setup.jl")
@testset "ClusterDepth.jl" begin
include("troendle.jl")
include("cluster.jl")
include("utils.jl")
include("pvals.jl")
end
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 96 | using Test
using Random
using LinearAlgebra
using StableRNGs
using StatsBase
using SparseArrays
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 2542 | @testset "troendle" begin
@testset "troendle()" begin
nperm = 900
ntests = 9
rng = StableRNG(1)
perm = randn(rng, ntests, nperm)
stat = [0.0, 0.0, -1, 1, -3.5, 3, 10, 10, -10]
p_cor = troendle(perm, stat)
@test length(p_cor) == ntests
@test all(p_cor[1:2] .== 1.0)
@test all(p_cor[7:9] .< 0.008)
#
statSig = [100.0, 100, 100, 100, 100, 100, 100, 100, 100]
p_cor = troendle(perm, statSig)
@test length(unique(p_cor)) == 1
@test all(p_cor .== (1 / (1 + nperm)))
# sidedness
h_cor = troendle(perm, stat; type = :greater) .< 0.05
@test h_cor == [0, 0, 0, 0, 0, 1, 1, 1, 0]
h_cor = troendle(perm, stat; type = :lesser) .< 0.05
@test h_cor == [0, 0, 0, 0, 1, 0, 0, 0, 1]
end
@testset "pvals_rankbased" begin
nperm = 9
ntests = 5
perm = randn(StableRNG(1), ntests, nperm)
perm[4:5, :] .= 0
J = sparse(perm)
stat_short = sparsevec(1:2, [3, 3])
stat_longer = sparsevec(1:2, [3, 3], ntests + 2)
stat = sparsevec(1:2, [3, 3], ntests)
cdm = ClusterDepth.ClusterDepthMatrix(J)
p = ClusterDepth.pvals_rankbased(perm, stat)
@test ClusterDepth.pvals_rankbased(cdm, stat) == p
# trimming shorter vec
p_short = ClusterDepth.pvals_rankbased(cdm, stat_short)
@test size(p_short) == (2, nperm + 1)
@test p_short == p[1:2, :]
# extending longer vec
p_long = ClusterDepth.pvals_rankbased(cdm, stat_longer)
@test size(p_long) == (ntests + 2, nperm + 1)
p = ClusterDepth.pvals_rankbased([1 2 3 4 5; 1 2 3 4 5], [3.5, 3])
@test p[:, 1] ≈ [0.5, 2.0 / 3]
@test p[1, 2:end] ≈ p[2, 2:end]
p = ClusterDepth.pvals_rankbased([1 2 3 4 5; 1 2 3 4 5], [0, 6])
@test p[:, 1] ≈ [1.0, 1.0 / 6]
p = ClusterDepth.pvals_rankbased(
ClusterDepth.ClusterDepthMatrix([1 2 3 4 5; 1 2 3 4 5]),
sparse([-6, -6, -6, -6, -6]),
)
@test all(p[:, 1] .== [1 / 6.0])
# test sidedness
p = ClusterDepth.pvals_rankbased([1 2 3 4 5; 1 2 3 4 5], [-6, 6])
@test p[:, 1] ≈ [1.0 / 6, 1.0 / 6]
p = ClusterDepth.pvals_rankbased([1 2 3 4 5; 1 2 3 4 5], [-6, 6]; type = :lesser)
@test p[:, 1] ≈ [1.0 / 6, 1.0]
p = ClusterDepth.pvals_rankbased([1 2 3 4 5; 1 2 3 4 5], [-6, 6]; type = :greater)
@test p[:, 1] ≈ [1.0, 1.0 / 6]
end
end
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | code | 1587 | @testset "sign_permute" begin
m = [1 1 1; 2 2 2; 3 3 3; 4 4 4]
p = ClusterDepth.sign_permute!(StableRNG(2), deepcopy(m))
@test p[1, :] == [1, -1, 1]
# different seeds are different
@test p != ClusterDepth.sign_permute!(StableRNG(3), deepcopy(m))
# same seeds are the same
@test p == ClusterDepth.sign_permute!(StableRNG(2), deepcopy(m))
m = ones(1, 1000000)
@test abs(mean(ClusterDepth.sign_permute!(StableRNG(1), deepcopy(m)))) < 0.001
m = ones(1, 2, 3, 4, 5, 6, 7, 100)
o = ClusterDepth.sign_permute!(StableRNG(1), deepcopy(m))
@test sort(unique(mean(o, dims = 1:ndims(o)-1))) == [-1.0, 1.0]
end
@testset "studentt" begin
x = randn(StableRNG(1), 10000, 50)
t = ClusterDepth.studentt(x)
@test length(t) == 10000
@test maximum(abs.(t)) < 10 # we'd need to be super lucky ;)
@test mean(abs.(t) .> 2) < 0.06
#2D input data
data = randn(StableRNG(1), 4, 5)
@test size(ClusterDepth.studentt(data)) == (4,)
#3D input data
data = randn(StableRNG(1), 3, 4, 5)
@test size(ClusterDepth.studentt(data)) == (3, 4)
#
t = rand(10000)
ClusterDepth.studentt!(t, x)
@test t ≈ ClusterDepth.studentt(x)
@test length(t) == 10000
@test maximum(abs.(t)) < 10 # we'd need to be super lucky ;)
@test mean(abs.(t) .> 2) < 0.06
#2D input data
data = randn(StableRNG(1), 4, 5)
t = rand(4)
ClusterDepth.studentt!(t, data)
@test size(t) == (4,)
#3D input data
data = randn(StableRNG(1), 3, 4, 5)
@test size(ClusterDepth.studentt(data)) == (3, 4)
end
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | docs | 2277 | # ClusterDepth
[](https://s-ccs.github.io/ClusterDepth.jl/dev/)
[](https://github.com/s-ccs/ClusterDepth.jl/actions/workflows/CI.yml?query=branch%3Amain)
[](https://codecov.io/gh/s-ccs/ClusterDepth.jl)
Fast implementation (~>100x faster than R/C++) of the ClusterDepth multiple comparison algorithm from Frossard and Renaud [Neuroimage 2022](https://doi.org/10.1016/j.neuroimage.2021.118824)
This is especially interesting to EEG signals. Currently only acts on a single channel/timeseries. Multichannel as discussed in the paper is the next step.
## Quickstart
```julia
using ClusterDepth
pval_corrected = clusterdepth(erpMatrix; τ=2.3,nperm=5000)
```
## FWER check
We checked FWER for `troendle(...)` and `clusterdepth(...)` [(link to docs)](https://www.s-ccs.de/ClusterDepth.jl/dev/reference/type1/)
For clustedepth we used 5000 repetitions, 5000 permutations, 200 tests.
|simulation|noise|uncorrected|type|
|---|---|---|---|
|clusterdepth|white|1.0|0.0554|
|clusterdepth|red*|0.835|0.0394|
|troendle|white|XX|XX|
|troendle|red*|XX|XX|
Uncorrected should be 1 - it is very improbable, than none of the 200 tests in one repetition is not significant (we expect 5% to be).
Corrected should be 0.05 (CI-95 [0.043,0.0564])
\* red noise introduces strong correlations between individual trials, thus makes the tests correlated while following the H0.
## Citing
Algorithm published in https://doi.org/10.1016/j.neuroimage.2021.118824 - Frossard & Renaud 2022, Neuroimage
Please also cite this toolbox: [](https://zenodo.org/badge/latestdoi/593411464)
Some functions are inspired by [R::permuco](https://cran.r-project.org/web/packages/permuco/index.html), written by Jaromil Frossard. Note: Permuco is GPL licensed, but Jaromil Frossard released the relevant clusteterdepth functions to me under MIT. Thereby, this repository can be licensed under MIT.
1
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 06fdba84326a461861f836d352308d4a17f3370b | docs | 627 | ```@meta
CurrentModule = ClusterDepth
```
# ClusterDepth
## Comparison to permuco R implementation
The implementation to Permuco is similar, but ClusterDepth.jl is more barebone - that is, we dont offer many permutation schemes, focus on the ClusterDepth Algorithm, and don't provide the nice wrappers like `clusterLM`.
Timing wise, a simple test on 50 subjects, 100 repetitions, 5000 permutations shows the following results:
|timepoints|ClusterDepth.jl|permuco|julia-speedup|
|---|---|---|---|
|40|0.03s|2.9s|~100x|
|400|0.14s|22s|~160x|
|4000|1.88s|240s|~120x|
```@index
```
```@autodocs
Modules = [ClusterDepth]
```
| ClusterDepth | https://github.com/s-ccs/ClusterDepth.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 4059 | module Bench
using IncompleteLU
using BenchmarkTools
using SparseArrays
using LinearAlgebra: axpy!, I
using Random: seed!
using Profile
function go()
seed!(1)
A = sprand(10_000, 10_000, 10 / 10_000) + 15I
LU = ilu(A)
Profile.clear_malloc_data()
@profile ilu(A)
end
function axpy_perf()
A = sprand(1_000, 1_000, 10 / 1_000) + 15I
y = SparseVectorAccumulator{Float64}(1_000)
axpy!(1.0, A, 1, A.colptr[1], y)
axpy!(1.0, A, 2, A.colptr[2], y)
axpy!(1.0, A, 3, A.colptr[3], y)
Profile.clear_malloc_data()
axpy!(1.0, A, 1, A.colptr[1], y)
axpy!(1.0, A, 2, A.colptr[2], y)
axpy!(1.0, A, 3, A.colptr[3], y)
end
function sum_values_row_wise(A::SparseMatrixCSC)
n = size(A, 1)
reader = RowReader(A)
sum = 0.0
for row = 1 : n
column = first_in_row(reader, row)
while is_column(column)
sum += nzval(reader, column)
column = next_column!(reader, column)
end
end
sum
end
function sum_values_column_wise(A::SparseMatrixCSC)
n = size(A, 1)
sum = 0.0
for col = 1 : n
for idx = A.colptr[col] : A.colptr[col + 1] - 1
sum += A.nzval[idx]
end
end
sum
end
function bench_alloc()
A = sprand(1_000, 1_000, 10 / 1_000) + 15I
sum_values_row_wise(A)
Profile.clear_malloc_data()
sum_values_row_wise(A)
end
function bench_perf()
A = sprand(10_000, 10_000, 10 / 10_000) + 15I
@show sum_values_row_wise(A)
@show sum_values_column_wise(A)
fst = @benchmark Bench.sum_values_row_wise($A)
snd = @benchmark Bench.sum_values_column_wise($A)
fst, snd
end
function bench_ILU()
seed!(1)
A = sprand(10_000, 10_000, 10 / 10_000) + 15I
LU = ilu(A, τ = 0.1)
@show nnz(LU.L) nnz(LU.U)
# nnz(LU.L) = 44836
# nnz(LU.U) = 54827
result = @benchmark ilu($A, τ = 0.1)
# BenchmarkTools.Trial:
# memory estimate: 16.24 MiB
# allocs estimate: 545238
# --------------
# minimum time: 116.923 ms (0.00% GC)
# median time: 127.514 ms (2.18% GC)
# mean time: 130.932 ms (1.75% GC)
# maximum time: 166.202 ms (3.05% GC)
# --------------
# samples: 39
# evals/sample: 1
# After switching to row reader.
# BenchmarkTools.Trial:
# memory estimate: 15.96 MiB
# allocs estimate: 545222
# --------------
# minimum time: 55.264 ms (0.00% GC)
# median time: 61.872 ms (4.73% GC)
# mean time: 61.906 ms (3.72% GC)
# maximum time: 74.615 ms (4.12% GC)
# --------------
# samples: 81
# evals/sample: 1
# After skipping off-diagonal elements in A
# BenchmarkTools.Trial:
# memory estimate: 15.96 MiB
# allocs estimate: 545222
# --------------
# minimum time: 51.187 ms (0.00% GC)
# median time: 55.767 ms (4.27% GC)
# mean time: 56.586 ms (3.50% GC)
# maximum time: 72.987 ms (7.53% GC)
# --------------
# samples: 89
# evals/sample: 1
# After moving L and U to Row Reader structs
# BenchmarkTools.Trial:
# memory estimate: 13.03 MiB
# allocs estimate: 495823
# --------------
# minimum time: 43.062 ms (0.00% GC)
# median time: 46.205 ms (2.83% GC)
# mean time: 47.076 ms (1.76% GC)
# maximum time: 65.956 ms (1.96% GC)
# --------------
# samples: 107
# evals/sample: 1
# After emptying the fill-in vecs during copy.
# BenchmarkTools.Trial:
# memory estimate: 13.03 MiB
# allocs estimate: 495823
# --------------
# minimum time: 41.930 ms (0.00% GC)
# median time: 44.583 ms (2.25% GC)
# mean time: 45.785 ms (1.38% GC)
# maximum time: 66.683 ms (1.59% GC)
# --------------
# samples: 110
# evals/sample: 1
end
end
# Bench.go() | IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 1026 | module BenchInsertion
using IncompleteLU
using BenchmarkTools
function do_stuff(A)
n = size(A, 1)
U_row = IncompleteLU.InsertableSparseVector{Float64}(n);
A_reader = IncompleteLU.RowReader(A)
for k = 1 : n
col = first_in_row(A_reader, k)
while is_column(col)
IncompleteLU.add!(U_row, nzval(A_reader, col), col, n + 1)
next_col = next_column(A_reader, col)
next_row!(A_reader, col)
if has_next_nonzero(A_reader, col) && nzrow(A_reader, col) ≤ col
enqueue_next_nonzero!(A_reader, col)
end
col = next_col
end
U_row.indices[n + 1] = n + 1
end
U_row
end
function wut(n = 1_000)
A = sprand(n, n, 10 / n) + 15I
@benchmark BenchInsertion.do_stuff($A)
end
function check_allocs(n = 100_000)
srand(1)
A = sprand(n, n, 10 / n) + 15I
do_stuff(A)
Profile.clear()
Profile.clear_malloc_data()
@profile do_stuff(A)
end
end
# BenchInsertion.check_allocs() | IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 357 | module IncompleteLU
using SparseArrays
using Base: @propagate_inbounds
struct ILUFactorization{Tv,Ti}
L::SparseMatrixCSC{Tv,Ti}
U::SparseMatrixCSC{Tv,Ti}
end
include("sorted_set.jl")
include("linked_list.jl")
include("sparse_vector_accumulator.jl")
include("insertion_sort_update_vector.jl")
include("application.jl")
include("crout_ilu.jl")
end | IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 1943 | import SparseArrays: nnz
import LinearAlgebra: ldiv!
import Base.\
export forward_substitution!, backward_substitution!
"""
Returns the number of nonzeros of the `L` and `U`
factor combined.
Excludes the unit diagonal of the `L` factor,
which is not stored.
"""
nnz(F::ILUFactorization) = nnz(F.L) + nnz(F.U)
function ldiv!(F::ILUFactorization, y::AbstractVector)
forward_substitution!(F, y)
backward_substitution!(F, y)
end
function ldiv!(y::AbstractVector, F::ILUFactorization, x::AbstractVector)
y .= x
ldiv!(F, y)
end
(\)(F::ILUFactorization, y::AbstractVector) = ldiv!(F, copy(y))
"""
Applies in-place backward substitution with the U factor of F, under the assumptions:
1. U is stored transposed / row-wise
2. U has no lower-triangular elements stored
3. U has (nonzero) diagonal elements stored.
"""
function backward_substitution!(F::ILUFactorization, y::AbstractVector)
U = F.U
@inbounds for col = U.n : -1 : 1
# Substitutions
for idx = U.colptr[col + 1] - 1 : -1 : U.colptr[col] + 1
y[col] -= U.nzval[idx] * y[U.rowval[idx]]
end
# Final answer for y[col]
y[col] /= U.nzval[U.colptr[col]]
end
y
end
function backward_substitution!(v::AbstractVector, F::ILUFactorization, y::AbstractVector)
v .= y
backward_substitution!(F, v)
end
"""
Applies in-place forward substitution with the L factor of F, under the assumptions:
1. L is stored column-wise (unlike U)
2. L has no upper triangular elements
3. L has *no* diagonal elements
"""
function forward_substitution!(F::ILUFactorization, y::AbstractVector)
L = F.L
@inbounds for col = 1 : L.n - 1
for idx = L.colptr[col] : L.colptr[col + 1] - 1
y[L.rowval[idx]] -= L.nzval[idx] * y[col]
end
end
y
end
function forward_substitution!(v::AbstractVector, F::ILUFactorization, y::AbstractVector)
v .= y
forward_substitution!(F, v)
end
| IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 3193 | export ilu
function lutype(T::Type)
UT = typeof(oneunit(T) - oneunit(T) * (oneunit(T) / (oneunit(T) + zero(T))))
LT = typeof(oneunit(UT) / oneunit(UT))
S = promote_type(T, LT, UT)
end
function ilu(A::SparseMatrixCSC{ATv,Ti}; τ = 1e-3) where {ATv,Ti}
n = size(A, 1)
Tv = lutype(ATv)
L = spzeros(Tv, Ti, n, n)
U = spzeros(Tv, Ti, n, n)
U_row = SparseVectorAccumulator{Tv,Ti}(n)
L_col = SparseVectorAccumulator{Tv,Ti}(n)
A_reader = RowReader(A)
L_reader = RowReader(L, Val{false})
U_reader = RowReader(U, Val{false})
@inbounds for k = Ti(1) : Ti(n)
##
## Copy the new row into U_row and the new column into L_col
##
col::Int = first_in_row(A_reader, k)
while is_column(col)
add!(U_row, nzval(A_reader, col), col)
next_col = next_column(A_reader, col)
next_row!(A_reader, col)
# Check if the next nonzero in this column
# is still above the diagonal
if has_next_nonzero(A_reader, col) && nzrow(A_reader, col) ≤ col
enqueue_next_nonzero!(A_reader, col)
end
col = next_col
end
# Copy the remaining part of the column into L_col
axpy!(one(Tv), A, k, nzidx(A_reader, k), L_col)
##
## Combine the vectors:
##
# U_row[k:n] -= L[k,i] * U[i,k:n] for i = 1 : k - 1
col = first_in_row(L_reader, k)
while is_column(col)
axpy!(-nzval(L_reader, col), U, col, nzidx(U_reader, col), U_row)
next_col = next_column(L_reader, col)
next_row!(L_reader, col)
if has_next_nonzero(L_reader, col)
enqueue_next_nonzero!(L_reader, col)
end
col = next_col
end
# Nothing is happening here when k = n, maybe remove?
# L_col[k+1:n] -= U[i,k] * L[i,k+1:n] for i = 1 : k - 1
if k < n
col = first_in_row(U_reader, k)
while is_column(col)
axpy!(-nzval(U_reader, col), L, col, nzidx(L_reader, col), L_col)
next_col = next_column(U_reader, col)
next_row!(U_reader, col)
if has_next_nonzero(U_reader, col)
enqueue_next_nonzero!(U_reader, col)
end
col = next_col
end
end
##
## Apply a drop rule
##
U_diag_element = U_row.nzval[k]
# U_diag_element = U_row.values[k]
# Append the columns
append_col!(U, U_row, k, τ)
append_col!(L, L_col, k, τ, inv(U_diag_element))
# Add the new row and column to U_nonzero_col, L_nonzero_row, U_first, L_first
# (First index *after* the diagonal)
U_reader.next_in_column[k] = U.colptr[k] + 1
if U.colptr[k] < U.colptr[k + 1] - 1
enqueue_next_nonzero!(U_reader, k)
end
L_reader.next_in_column[k] = L.colptr[k]
if L.colptr[k] < L.colptr[k + 1]
enqueue_next_nonzero!(L_reader, k)
end
end
return ILUFactorization(L, U)
end
| IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 2608 | import Base: getindex, setindex!, empty!, Vector
import LinearAlgebra: axpy!
"""
`InsertableSparseVector` accumulates the sparse vector
result from SpMV. Initialization requires O(N) work,
therefore the data structure is reused. Insertion
requires O(nnz) at worst, as insertion sort is used.
"""
struct InsertableSparseVector{Tv}
values::Vector{Tv}
indices::SortedSet
InsertableSparseVector{Tv}(n::Int) where {Tv} = new(Vector{Tv}(undef, n), SortedSet(n))
end
@propagate_inbounds getindex(v::InsertableSparseVector{Tv}, idx::Int) where {Tv} = v.values[idx]
@propagate_inbounds setindex!(v::InsertableSparseVector{Tv}, value::Tv, idx::Int) where {Tv} = v.values[idx] = value
@inline indices(v::InsertableSparseVector) = Vector(v.indices)
function Vector(v::InsertableSparseVector{Tv}) where {Tv}
vals = zeros(Tv, v.indices.N - 1)
for index in v.indices
@inbounds vals[index] = v.values[index]
end
return vals
end
"""
Sets `v[idx] += a` when `idx` is occupied, or sets `v[idx] = a`.
Complexity is O(nnz). The `prev_idx` can be used to start the linear
search at `prev_idx`, useful when multiple already sorted values
are added.
"""
function add!(v::InsertableSparseVector, a, idx::Integer, prev_idx::Integer)
if push!(v.indices, idx, prev_idx)
@inbounds v[idx] = a
else
@inbounds v[idx] += a
end
v
end
"""
Add without providing a previous index.
"""
@propagate_inbounds add!(v::InsertableSparseVector, a, idx::Integer) = add!(v, a, idx, v.indices.N)
function axpy!(a, A::SparseMatrixCSC, column::Integer, start::Integer, y::InsertableSparseVector)
prev_index = y.indices.N
@inbounds for idx = start : A.colptr[column + 1] - 1
add!(y, a * A.nzval[idx], A.rowval[idx], prev_index)
prev_index = A.rowval[idx]
end
y
end
"""
Empties the InsterableSparseVector in O(1) operations.
"""
@inline empty!(v::InsertableSparseVector) = empty!(v.indices)
"""
Basically `A[:, j] = scale * drop(y)`, where drop removes
values less than `drop`.
Resets the `InsertableSparseVector`.
Note: does *not* update `A.colptr` for columns > j + 1,
as that is done during the steps.
"""
function append_col!(A::SparseMatrixCSC{Tv}, y::InsertableSparseVector{Tv}, j::Int, drop::Tv, scale::Tv = one(Tv)) where {Tv}
total = 0
@inbounds for row = y.indices
if abs(y[row]) ≥ drop || row == j
push!(A.rowval, row)
push!(A.nzval, scale * y[row])
total += 1
end
end
@inbounds A.colptr[j + 1] = A.colptr[j] + total
empty!(y)
nothing
end | IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 2353 | import Base: push!
"""
The factor L is stored column-wise, but we need
all nonzeros in row `row`. We already keep track of
the first nonzero in each column (at most `n` indices).
Take `l = LinkedLists(n)`. Let `l.head[row]` be the column
of some nonzero in row `row`. Then we can store the column
of the next nonzero of row `row` in `l.next[l.head[row]]`, etc.
That "spot" is empty and there will never be a conflict
because as long as we only store the first nonzero per column:
the column is then a unique identifier.
"""
struct LinkedLists{Ti}
head::Vector{Ti}
next::Vector{Ti}
end
LinkedLists{Ti}(n::Integer) where {Ti} = LinkedLists(zeros(Ti, n), zeros(Ti, n))
"""
For the L-factor: insert in row `head` column `value`
For the U-factor: insert in column `head` row `value`
"""
@propagate_inbounds function push!(l::LinkedLists, head::Integer, value::Integer)
l.head[head], l.next[value] = value, l.head[head]
return l
end
struct RowReader{Tv,Ti}
A::SparseMatrixCSC{Tv,Ti}
next_in_column::Vector{Ti}
rows::LinkedLists{Ti}
end
function RowReader(A::SparseMatrixCSC{Tv,Ti}) where {Tv,Ti}
n = size(A, 2)
@inbounds next_in_column = [A.colptr[i] for i = 1 : n]
rows = LinkedLists{Ti}(n)
@inbounds for i = Ti(1) : Ti(n)
push!(rows, A.rowval[A.colptr[i]], i)
end
return RowReader(A, next_in_column, rows)
end
function RowReader(A::SparseMatrixCSC{Tv,Ti}, initialize::Type{Val{false}}) where {Tv,Ti}
n = size(A, 2)
return RowReader(A, zeros(Ti, n), LinkedLists{Ti}(n))
end
@propagate_inbounds nzidx(r::RowReader, column::Integer) = r.next_in_column[column]
@propagate_inbounds nzrow(r::RowReader, column::Integer) = r.A.rowval[nzidx(r, column)]
@propagate_inbounds nzval(r::RowReader, column::Integer) = r.A.nzval[nzidx(r, column)]
@propagate_inbounds has_next_nonzero(r::RowReader, column::Integer) = nzidx(r, column) < r.A.colptr[column + 1]
@propagate_inbounds enqueue_next_nonzero!(r::RowReader, column::Integer) = push!(r.rows, nzrow(r, column), column)
@propagate_inbounds next_column(r::RowReader, column::Integer) = r.rows.next[column]
@propagate_inbounds first_in_row(r::RowReader, row::Integer) = r.rows.head[row]
@propagate_inbounds is_column(column::Integer) = column != 0
@propagate_inbounds next_row!(r::RowReader, column::Integer) = r.next_in_column[column] += 1
| IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 1917 | import Base: iterate, push!, Vector, getindex, setindex!, show, empty!
"""
SortedSet keeps track of a sorted set of integers ≤ N
using insertion sort with a linked list structure in a pre-allocated
vector. Requires O(N + 1) memory. Insertion goes via a linear scan in O(n)
where `n` is the number of stored elements, but can be accelerated
by passing along a known value in the set (which is useful when pushing
in an already sorted list). The insertion itself requires O(1) operations
due to the linked list structure. Provides iterators:
```julia
ints = SortedSet(10)
push!(ints, 5)
push!(ints, 3)
for value in ints
println(value)
end
```
"""
struct SortedSet
next::Vector{Int}
N::Int
function SortedSet(N::Int)
next = Vector{Int}(undef, N + 1)
@inbounds next[N + 1] = N + 1
new(next, N + 1)
end
end
# Convenience wrappers for indexing
@propagate_inbounds getindex(s::SortedSet, i::Int) = s.next[i]
@propagate_inbounds setindex!(s::SortedSet, value::Int, i::Int) = s.next[i] = value
# Iterate in
@inline function iterate(s::SortedSet, p::Int = s.N)
@inbounds nxt = s[p]
return nxt == s.N ? nothing : (nxt, nxt)
end
show(io::IO, s::SortedSet) = print(io, typeof(s), " with values ", Vector(s))
"""
For debugging and testing
"""
function Vector(s::SortedSet)
v = Int[]
for index in s
push!(v, index)
end
return v
end
"""
Insert `index` after a known value `after`
"""
function push!(s::SortedSet, value::Int, after::Int)
@inbounds begin
while s[after] < value
after = s[after]
end
if s[after] == value
return false
end
s[after], s[value] = value, s[after]
return true
end
end
"""
Make the head pointer do a self-loop.
"""
@inline empty!(s::SortedSet) = s[s.N] = s.N
@inline push!(s::SortedSet, index::Int) = push!(s, index, s.N)
| IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 3386 | import Base: setindex!, empty!, Vector
import LinearAlgebra: axpy!
"""
`SparseVectorAccumulator` accumulates the sparse vector
resulting from SpMV. Initialization requires O(N) work,
therefore the data structure is reused. Insertion is O(1).
Note that `nzind` is unordered. Also note that there is
wasted space: `nzind` could be a growing list. Pre-allocation
seems faster though.
SparseVectorAccumulator incorporates the multiple switch technique
by Gustavson (1976), which makes resetting an O(1) operation rather
than O(nnz): the `curr` value is used to flag the occupied indices,
and `curr` is increased at each reset.
occupied = [0, 1, 0, 1, 0, 0, 0]
nzind = [2, 4, 0, 0, 0, 0]
nzval = [0., .1234, 0., .435, 0., 0., 0.]
nnz = 2
length = 7
curr = 1
"""
mutable struct SparseVectorAccumulator{Tv,Ti}
occupied::Vector{Ti}
nzind::Vector{Ti}
nzval::Vector{Tv}
nnz::Ti
length::Ti
curr::Ti
return SparseVectorAccumulator{Tv,Ti}(N::Integer) where {Tv,Ti} = new(
zeros(Ti, N),
Vector{Ti}(undef, N),
Vector{Tv}(undef, N),
0,
N,
1
)
end
function Vector(v::SparseVectorAccumulator{T}) where {T}
x = zeros(T, v.length)
@inbounds x[v.nzind[1 : v.nnz]] = v.nzval[v.nzind[1 : v.nnz]]
return x
end
"""
Add a part of a SparseMatrixCSC column to a SparseVectorAccumulator,
starting at a given index until the end.
"""
function axpy!(a, A::SparseMatrixCSC, column, start, y::SparseVectorAccumulator)
# Loop over the whole column of A
@inbounds for idx = start : A.colptr[column + 1] - 1
add!(y, a * A.nzval[idx], A.rowval[idx])
end
return y
end
"""
Sets `v[idx] += a` when `idx` is occupied, or sets `v[idx] = a`.
Complexity is O(1).
"""
function add!(v::SparseVectorAccumulator, a, idx)
@inbounds begin
if isoccupied(v, idx)
v.nzval[idx] += a
else
v.nnz += 1
v.occupied[idx] = v.curr
v.nzval[idx] = a
v.nzind[v.nnz] = idx
end
end
return nothing
end
"""
Check whether `idx` is nonzero.
"""
@propagate_inbounds isoccupied(v::SparseVectorAccumulator, idx::Integer) = v.occupied[idx] == v.curr
"""
Empty the SparseVectorAccumulator in O(1) operations.
"""
@inline function empty!(v::SparseVectorAccumulator)
v.curr += 1
v.nnz = 0
end
"""
Basically `A[:, j] = scale * drop(y)`, where drop removes
values less than `drop`. Note: sorts the `nzind`'s of `y`,
so that the column can be appended to a SparseMatrixCSC.
Resets the `SparseVectorAccumulator`.
Note: does *not* update `A.colptr` for columns > j + 1,
as that is done during the steps.
"""
function append_col!(A::SparseMatrixCSC, y::SparseVectorAccumulator, j::Integer, drop, scale = one(eltype(A)))
# Move the indices of interest up front
total = 0
@inbounds for idx = 1 : y.nnz
row = y.nzind[idx]
value = y.nzval[row]
if abs(value) ≥ drop || row == j
total += 1
y.nzind[total] = row
end
end
# Sort the retained values.
sort!(y.nzind, 1, total, Base.Sort.QuickSort, Base.Order.Forward)
@inbounds for idx = 1 : total
row = y.nzind[idx]
push!(A.rowval, row)
push!(A.nzval, scale * y.nzval[row])
end
@inbounds A.colptr[j + 1] = A.colptr[j] + total
empty!(y)
return nothing
end
| IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 1973 | using Test
using IncompleteLU: ILUFactorization, forward_substitution!, backward_substitution!
using LinearAlgebra
@testset "Forward and backward substitutions" begin
function test_fw_substitution(F::ILUFactorization)
A = F.L
n = size(A, 1)
x = rand(n)
y = copy(x)
v = zeros(n)
forward_substitution!(v, F, x)
forward_substitution!(F, x)
ldiv!(LowerTriangular(A + I), y)
@test v ≈ y
@test x ≈ y
end
function test_bw_substitution(F::ILUFactorization)
A = F.U
n = size(A, 1)
x = rand(n)
y = copy(x)
v = zeros(n)
backward_substitution!(v, F, x)
backward_substitution!(F, x)
ldiv!(UpperTriangular(transpose(A)), y)
@test v ≈ y
@test x ≈ y
end
L = sparse(tril(rand(10, 10), -1))
U = sparse(tril(rand(10, 10)) + 10I)
F = ILUFactorization(L, U)
test_fw_substitution(F)
test_bw_substitution(F)
L = sparse(tril(tril(sprand(10, 10, .5), -1)))
U = sparse(tril(sprand(10, 10, .5) + 10I))
F = ILUFactorization(L, U)
test_fw_substitution(F)
test_bw_substitution(F)
L = spzeros(10, 10)
U = spzeros(10, 10) + 10I
F = ILUFactorization(L, U)
test_fw_substitution(F)
test_bw_substitution(F)
end
@testset "ldiv!" begin
function test_ldiv!(L, U)
LU = ILUFactorization(L, U)
x = rand(size(LU.L, 1))
y = copy(x)
z = copy(x)
w = copy(x)
ldiv!(LU, x)
ldiv!(LowerTriangular(LU.L + I), y)
ldiv!(UpperTriangular(transpose(LU.U)), y)
@test x ≈ y
@test LU \ z == x
ldiv!(w, LU, z)
@test w == x
end
test_ldiv!(tril(sprand(10, 10, .5), -1), tril(sprand(10, 10, .5) + 10I))
end
@testset "nnz" begin
L = tril(sprand(10, 10, .5), -1)
U = tril(sprand(10, 10, .5)) + 10I
LU = ILUFactorization(L, U)
@test nnz(LU) == nnz(L) + nnz(U)
end | IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 1126 | using Test
using IncompleteLU
using SparseArrays
using LinearAlgebra
@testset "Crout ILU" for Tv in (Float64, Float32, ComplexF64, ComplexF32), Ti in (Int64, Int32)
let
# Test if it performs full LU if droptol is zero
A = convert(SparseMatrixCSC{Tv, Ti}, sprand(Tv, 10, 10, .5) + 10I)
ilu = IncompleteLU.ilu(A, τ = 0)
flu = lu(Matrix(A), Val(false))
@test typeof(ilu) == IncompleteLU.ILUFactorization{Tv,Ti}
@test Matrix(ilu.L + I) ≈ flu.L
@test Matrix(transpose(ilu.U)) ≈ flu.U
end
let
# Test if L = I and U = diag(A) when the droptol is large.
A = convert(SparseMatrixCSC{Tv, Ti}, sprand(10, 10, .5) + 10I)
ilu = IncompleteLU.ilu(A, τ = 1.0)
@test nnz(ilu.L) == 0
@test nnz(ilu.U) == 10
@test diag(ilu.U) == diag(A)
end
end
@testset "Crout ILU with integer matrix" begin
A = sparse(Int32(1):Int32(10), Int32(1):Int32(10), 1)
ilu = IncompleteLU.ilu(A, τ = 0)
@test typeof(ilu) == IncompleteLU.ILUFactorization{Float64,Int32}
@test nnz(ilu.L) == 0
@test diag(ilu.U) == diag(A)
end | IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 1715 | using Test
using IncompleteLU: InsertableSparseVector, add!, axpy!, append_col!, indices
@testset "InsertableSparseVector" begin
@testset "Insertion sorted sparse vector" begin
v = InsertableSparseVector{Float64}(10)
add!(v, 3.0, 6, 11)
add!(v, 3.0, 3, 11)
add!(v, 3.0, 3, 11)
@test v[6] == 3.0
@test v[3] == 6.0
@test indices(v) == [3, 6]
end
@testset "Add column of SparseMatrixCSC" begin
v = InsertableSparseVector{Float64}(5)
A = sprand(5, 5, 1.0)
axpy!(2., A, 3, A.colptr[3], v)
axpy!(3., A, 4, A.colptr[4], v)
@test Vector(v) == 2 * A[:, 3] + 3 * A[:, 4]
end
@testset "Append column to SparseMatrixCSC" begin
A = spzeros(5, 5)
v = InsertableSparseVector{Float64}(5)
add!(v, 0.3, 1)
add!(v, 0.009, 3)
add!(v, 0.12, 4)
add!(v, 0.007, 5)
append_col!(A, v, 1, 0.1)
# Test whether the column is copied correctly
# and the dropping rule is applied
@test A[1, 1] == 0.3
@test A[2, 1] == 0.0 # zero
@test A[3, 1] == 0.0 # dropped
@test A[4, 1] == 0.12
@test A[5, 1] == 0.0 # dropped
# Test whether the InsertableSparseVector is reset
# when reusing it for the second column. Also do
# scaling with a factor of 10.
add!(v, 0.5, 2)
add!(v, 0.009, 3)
add!(v, 0.5, 4)
add!(v, 0.007, 5)
append_col!(A, v, 2, 0.1, 10.0)
@test A[1, 2] == 0.0 # zero
@test A[2, 2] == 5.0 # scaled
@test A[3, 2] == 0.0 # dropped
@test A[4, 2] == 5.0 # scaled
@test A[5, 2] == 0.0 # dropped
end
end | IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 1170 | using Test
using IncompleteLU: LinkedLists, RowReader, first_in_row, is_column, nzval, next_column,
next_row!, has_next_nonzero, enqueue_next_nonzero!
using SparseArrays
@testset "Linked List" begin
n = 5
let
lists = LinkedLists{Int}(n)
# head[2] -> 5 -> nil
# head[5] -> 4 -> 3 -> nil
push!(lists, 5, 3)
push!(lists, 5, 4)
push!(lists, 2, 5)
@test lists.head[5] == 4
@test lists.next[4] == 3
@test lists.next[3] == 0
@test lists.head[2] == 5
@test lists.next[5] == 0
end
end
@testset "Read SparseMatrixCSC row by row" begin
# Read a sparse matrix row by row.
n = 10
A = sprand(n, n, .5)
reader = RowReader(A)
for row = 1 : n
column = first_in_row(reader, row)
while is_column(column)
@test nzval(reader, column) == A[row, column]
next_col = next_column(reader, column)
next_row!(reader, column)
if has_next_nonzero(reader, column)
enqueue_next_nonzero!(reader, column)
end
column = next_col
end
end
end | IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 216 | using Test
using IncompleteLU
include("sorted_set.jl")
include("linked_list.jl")
include("sparse_vector_accumulator.jl")
include("insertion_sort_update_vector.jl")
include("application.jl")
include("crout_ilu.jl")
| IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 1097 | using Test
import IncompleteLU: SortedSet, push!
@testset "Sorted indices" begin
@testset "New values" begin
indices = SortedSet(10)
@test push!(indices, 5)
@test push!(indices, 7)
@test push!(indices, 4)
@test push!(indices, 6)
@test push!(indices, 8)
as_vec = Vector(indices)
@test as_vec == [4, 5, 6, 7, 8]
end
@testset "Duplicate values" begin
indices = SortedSet(10)
@test push!(indices, 3)
@test push!(indices, 3) == false
@test push!(indices, 8)
@test push!(indices, 8) == false
@test Vector(indices) == [3, 8]
end
@testset "Quick insertion with known previous index" begin
indices = SortedSet(10)
@test push!(indices, 3)
@test push!(indices, 4, 3)
@test push!(indices, 8, 4)
@test Vector(indices) == [3, 4, 8]
end
@testset "Pretty printing" begin
indices = SortedSet(10)
push!(indices, 3)
push!(indices, 2)
@test occursin("with values", sprint(show, indices))
end
end | IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | code | 2225 | using IncompleteLU: SparseVectorAccumulator, add!, append_col!, isoccupied
using LinearAlgebra
@testset "SparseVectorAccumulator" for Ti in (Int32, Int64), Tv in (Float64, Float32)
@testset "Initialization" begin
v = SparseVectorAccumulator{Tv,Ti}(10)
@test iszero(v.nnz)
@test iszero(v.occupied)
end
@testset "Add to SparseVectorAccumulator" begin
v = SparseVectorAccumulator{Tv,Ti}(3)
add!(v, Tv(1.0), Ti(3))
add!(v, Tv(1.0), Ti(3))
add!(v, Tv(3.0), Ti(2))
@test v.nnz == 2
@test isoccupied(v, 1) == false
@test isoccupied(v, 2)
@test isoccupied(v, 3)
@test Vector(v) == Tv[0.; 3.0; 2.0]
end
@testset "Add column of SparseMatrixCSC" begin
# Copy all columns of a
v = SparseVectorAccumulator{Tv,Ti}(5)
A = convert(SparseMatrixCSC{Tv,Ti}, sprand(Tv, 5, 5, 1.0))
axpy!(Tv(2), A, Ti(3), A.colptr[3], v)
axpy!(Tv(3), A, Ti(4), A.colptr[4], v)
@test Vector(v) == 2 * A[:, 3] + 3 * A[:, 4]
end
@testset "Append column to SparseMatrixCSC" begin
A = spzeros(Tv, Ti, 5, 5)
v = SparseVectorAccumulator{Tv,Ti}(5)
add!(v, Tv(0.3), Ti(1))
add!(v, Tv(0.009), Ti(3))
add!(v, Tv(0.12), Ti(4))
add!(v, Tv(0.007), Ti(5))
append_col!(A, v, Ti(1), Tv(0.1))
# Test whether the column is copied correctly
# and the dropping rule is applied
@test A[1, 1] == Tv(0.3)
@test A[2, 1] == Tv(0.0) # zero
@test A[3, 1] == Tv(0.0) # dropped
@test A[4, 1] == Tv(0.12)
@test A[5, 1] == Tv(0.0) # dropped
# Test whether the InsertableSparseVector is reset
# when reusing it for the second column. Also do
# scaling with a factor of 10.
add!(v, Tv(0.5), Ti(2))
add!(v, Tv(0.009), Ti(3))
add!(v, Tv(0.5), Ti(4))
add!(v, Tv(0.007), Ti(5))
append_col!(A, v, Ti(2), Tv(0.1), Tv(10.0))
@test A[1, 2] == Tv(0.0) # zero
@test A[2, 2] == Tv(5.0) # scaled
@test A[3, 2] == Tv(0.0) # dropped
@test A[4, 2] == Tv(5.0) # scaled
@test A[5, 2] == Tv(0.0) # dropped
end
end | IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 0.2.1 | 6c676e79f98abb6d33fa28122cad099f1e464afe | docs | 6488 | [](https://travis-ci.org/haampie/IncompleteLU.jl) [](https://codecov.io/gh/haampie/IncompleteLU.jl)
# ILU for SparseMatrixCSC
This package implements the left-looking or Crout version of ILU for
the `SparseMatrixCSC` type. It exports the function `ilu`.
## How to install
`IncompleteLU` can be installed through the Julia package manager:
```julia
julia> ]
pkg> add IncompleteLU
```
The package is then available via
```julia
julia> using IncompleteLU
```
## When to use this package
Whenever you need an incomplete factorization of a sparse and _non-symmetric_ matrix.
The package also provides means to apply the factorization in-place via `ldiv!`, `forward_substitution!` and `backward_substitution!`.
This is useful in the context of left, right or split preconditioning. See the example below.
## Example
Using a drop tolerance of `0.01`, we get a reasonable preconditioner with a bit of fill-in.
```julia
> using IncompleteLU, LinearAlgebra, SparseArrays
> using BenchmarkTools
> A = sprand(1000, 1000, 5 / 1000) + 10I
> fact = @btime ilu($A, τ = 0.001)
2.894 ms (90 allocations: 1.18 MiB)
> norm((fact.L + I) * fact.U' - A)
0.05736313452207798
> nnz(fact) / nnz(A)
3.6793806030969844
```
Full LU is obtained when the drop tolerance is `0.0`.
```julia
> fact = @btime ilu($A, τ = 0.)
209.293 ms (106 allocations: 12.18 MiB)
> norm((fact.L + I) * fact.U' - A)
1.5262736852530086e-13
> nnz(fact) / nnz(A)
69.34213528932355
```
## Preconditioner
ILU is typically used as preconditioner for iterative methods. For instance
```julia
using IterativeSolvers, IncompleteLU
using SparseArrays, LinearAlgebra
using BenchmarkTools
using Plots
"""
Benchmarks a non-symmetric n × n × n problem
with and without the ILU preconditioner.
"""
function mytest(n = 64)
N = n^3
A = spdiagm(
-1 => fill(-1.0, n - 1),
0 => fill(3.0, n),
1 => fill(-2.0, n - 1)
)
Id = sparse(1.0I, n, n)
A = kron(A, Id) + kron(Id, A)
A = kron(A, Id) + kron(Id, A)
x = ones(N)
b = A * x
LU = ilu(A, τ = 0.1)
@show nnz(LU) / nnz(A)
# Benchmarks
prec = @benchmark ilu($A, τ = 0.1)
@show prec
with = @benchmark bicgstabl($A, $b, 2, Pl = $LU, max_mv_products = 2000)
@show with
without = @benchmark bicgstabl($A, $b, 2, max_mv_products = 2000)
@show without
# Result
x_with, hist_with = bicgstabl(A, b, 2, Pl = LU, max_mv_products = 2000, log = true)
x_without, hist_without = bicgstabl(A, b, 2, max_mv_products = 2000, log = true)
@show norm(b - A * x_with) / norm(b)
@show norm(b - A * x_without) / norm(b)
plot(hist_with[:resnorm], yscale = :log10, label = "With ILU preconditioning", xlabel = "Iteration", ylabel = "Residual norm (preconditioned)", mark = :x)
plot!(hist_without[:resnorm], label = "Without preconditioning", mark = :x)
end
mytest()
```
Outputs
```julia
nnz(LU) / nnz(A) = 2.1180353639352374
prec = Trial(443.781 ms)
with = Trial(766.141 ms)
without = Trial(2.595 s)
norm(b - A * x_with) / norm(b) = 2.619046427010899e-9
norm(b - A * x_without) / norm(b) = 1.2501603557459283e-8
```
## The algorithm
The basic algorithm loops roughly as follows:
```
for k = 1 : n
row = zeros(n); row[k:n] = A[k,k:n]
col = zeros(n); col[k+1:n] = A[k+1:n,k]
for i = 1 : k - 1 where L[k,i] != 0
row -= L[k,i] * U[i,k:n]
end
for i = 1 : k - 1 where U[i,k] != 0
col -= U[i,k] * L[k+1:n,i]
end
# Apply a dropping rule in row and col
U[k,:] = row
L[:,k] = col / U[k,k]
L[k,k] = 1
end
```
which means that at each step `k` a complete row and column are computed based on the previous rows and columns:
```
k
+---+---+---+---+---+---+---+---+
| \ | | x | x | x | x | x | x |
+---+---+---+---+---+---+---+---+
| | \ | x | x | x | x | x | x |
+---+---+---+---+---+---+---+---+
| | | . | . | . | . | . | . | k
+---+---+---+---+---+---+---+---+
| x | x | . | \ | | | | |
+---+---+---+---+---+---+---+---+
| x | x | . | | \ | | | |
+---+---+---+---+---+---+---+---+
| x | x | . | | | \ | | |
+---+---+---+---+---+---+---+---+
| x | x | . | | | | \ | |
+---+---+---+---+---+---+---+---+
| x | x | . | | | | | \ |
+---+---+---+---+---+---+---+---+
col and row are the .'s, updated by the x's.
```
At step `k` we load (part of) a row and column of the matrix `A`, and subtract the previous rows and columns times a scalar (basically a SpMV product). The problem is that our matrix is column-major, so that loading a row is not cheap. Secondly, it makes sense to store the `L` factor column-wise and the `U` factor row-wise (so that we can append columns and rows without data movement), yet we need access to a row of `L` and a column of `U`.
The latter problem can be worked around without expensive searches. It's basically smart bookkeeping: going from step `k` to `k+1` requires updating indices to the next nonzero of each row of `U` after column `k`. If you now store for each column of `U` a list of nonzero indices, this is the moment you can update it. Similarly for the `L` factor.
The matrix `A` can be read row by row as well with the same trick.
## Accumulating a new sparse row or column
Throughout the steps two temporary row and column accumulators are used to store the linear combinations of previous sparse rows and columns. There are two implementations of this accumulator: the `SparseVectorAccumulator` performs insertion in `O(1)`, but stores the indices unordered; therefore a sort is required when appending to the `SparseMatrixCSC`. The `InsertableSparseVector` performs insertion sort, which can be slow, but turns out to be fast in practice. The latter is a result of insertion itself being an `O(1)` operation due to a linked list structure, and the fact that sorted vectors are added, so that the linear scan does not have to restart at each insertion.
The advantage of `SparseVectorAccumulator` over `InsertableSparseVector` is that the former postpones sorting until after dropping, while `InsertableSparseVector` also performs insertion sort on dropped values.
## Todo
The method does not implement scaling techniques, so the `τ` parameter is really an
absolute dropping tolerance parameter.
| IncompleteLU | https://github.com/haampie/IncompleteLU.jl.git |
|
[
"MIT"
] | 1.0.2 | 7e3de57fa445342a33b6f0350c32e32943de75e2 | code | 725 | using RamseySpectroscopyCore
using Documenter
DocMeta.setdocmeta!(RamseySpectroscopyCore, :DocTestSetup, :(using RamseySpectroscopyCore); recursive=true)
makedocs(;
modules=[RamseySpectroscopyCore],
authors="Maksim Radchenko",
repo="https://github.com/m0Cey/RamseySpectroscopyCore.jl/blob/{commit}{path}#{line}",
sitename="RamseySpectroscopyCore.jl",
format=Documenter.HTML(;
prettyurls=get(ENV, "CI", "false") == "true",
canonical="https://m0Cey.github.io/RamseySpectroscopyCore.jl",
edit_link="main",
assets=String[],
),
pages=[
"Home" => "index.md",
],
)
deploydocs(;
repo="github.com/m0Cey/RamseySpectroscopyCore.jl",
devbranch="main",
)
| RamseySpectroscopyCore | https://github.com/m0Cey/RamseySpectroscopyCore.jl.git |
|
[
"MIT"
] | 1.0.2 | 7e3de57fa445342a33b6f0350c32e32943de75e2 | code | 369 | """
RamseySpectroscopyCore.jl provides core toolset for simulating Rabi/Ramsey spectroscopy
experiments. It consists of controling functions `pump()`, `rest()` and an interrogating
function `probe()`.
"""
module RamseySpectroscopyCore
export StateVector, PerturbEvol, FreeEvol
include("structs.jl")
export pump, pump!, rest, rest!, probe
include("methods.jl")
end
| RamseySpectroscopyCore | https://github.com/m0Cey/RamseySpectroscopyCore.jl.git |
|
[
"MIT"
] | 1.0.2 | 7e3de57fa445342a33b6f0350c32e32943de75e2 | code | 3415 | """
pump!(self::StateVector, pulse::PerturbEvol) -> StateVector
Transform the state of a quantum system according to system's evolution in
a presence of the laser pulse.
# Arguments
* `self` :
The current state vector of a quantum system.
* `pulse` :
The laser pulse which is applied on the quantum system.
# Returns
* `StateVector` :
Composite type instance with transformed `state` object.
# See also
* [`StateVector`](@ref)
* [`PerturbEvol`](@ref)
# References
* Wikipedia: https://en.wikipedia.org/wiki/Ramsey_interferometry
"""
function pump!(self::StateVector, pulse::PerturbEvol)
local Ψ⃗ = self.state
local Ŵ = pulse.operator
self.state = Ŵ * Ψ⃗
return self
end
"""
pump(state_vector::StateVector, pulse::PerturbEvol) -> StateVector
Transform the state of a quantum system according to system's evolution in
a presence of the laser pulse.
# Arguments
* `state_vector` :
The current state vector of a quantum system.
* `pulse` :
The laser pulse which is applied on the quantum system.
# Returns
* `StateVector` :
Composite type instance with transformed `state` object.
# See also
* [`StateVector`](@ref)
* [`PerturbEvol`](@ref)
# References
* Wikipedia: https://en.wikipedia.org/wiki/Ramsey_interferometry
"""
function pump(state_vector::StateVector, pulse::PerturbEvol)
local Ψ⃗ = state_vector.state
local Ŵ = pulse.operator
return StateVector(Ŵ * Ψ⃗)
end
"""
rest!(self::StateVector, duration::FreeEvol) -> StateVector
Transform the state of a quantum system according to system's evolution in
a presence of no perturbations.
# Arguments
* `self` :
The current state vector of a quantum system.
* `duration` :
The time of free evolution of the quantum system.
# Returns
* `StateVector` :
Composite type instance with transformed `state` object.
# See also
* [`StateVector`](@ref)
* [`FreeEvol`](@ref)
# References
* Wikipedia: https://en.wikipedia.org/wiki/Ramsey_interferometry
"""
function rest!(self::StateVector, duration::FreeEvol)
local Ψ⃗ = self.state
local V̂ = duration.operator
self.state = V̂ * Ψ⃗
return self
end
"""
rest(state_vector::StateVector, duration::FreeEvol) -> StateVector
Transform the state of a quantum system according to system's evolution in
a presence of no perturbations.
# Arguments
* `state_vector` :
The current state vector of a quantum system.
* `duration` :
The time of free evolution of the quantum system.
# Returns
* `StateVector` :
Composite type instance with transformed `state` object.
# See also
* [`StateVector`](@ref)
* [`FreeEvol`](@ref)
# References
* Wikipedia: https://en.wikipedia.org/wiki/Ramsey_interferometry
"""
function rest(state_vector::StateVector, duration::FreeEvol)
local Ψ⃗ = state_vector.state
local V̂ = duration.operator
return StateVector(V̂ * Ψ⃗)
end
"""
probe(state_vector::StateVector) -> Real
Method that computes the value of probability `nₑ` for a quantum system to be in
excited state and can be interpret as population of excited state.
# Arguments
* `self` :
The current state vector of a quantum system.
# Returns
* `nₑ` :
Real number.
# See also
* [`StateVector`](@ref)
# References
* Wikipedia: https://en.wikipedia.org/wiki/Ramsey_interferometry
"""
function probe(state_vector::StateVector)::Real
local Ψ⃗ = state_vector.state
return abs2(Ψ⃗[1])
end
| RamseySpectroscopyCore | https://github.com/m0Cey/RamseySpectroscopyCore.jl.git |
|
[
"MIT"
] | 1.0.2 | 7e3de57fa445342a33b6f0350c32e32943de75e2 | code | 3074 | """
StateVector
Struct that describes a state of the quantum two-level system via ket vector
in Dirac notation.
# Fields
* `state` :
Vector that represents the state of the system in (|e>, |g>) basis.
# Arguments
* `e` :
Numerical value that implies the probability of a quantum system to be in excited state
or the amount of particles in the ground state.
Value of |e> ket vector.
* `g` :
Numerical value that implies the probability of a quantum system to be in ground state
or the amount of particles in the ground state.
Value of |g> ket vector.
# Returns
* `StateVector` :
Composite type instance.
# References
* Wikipedia: https://en.wikipedia.org/wiki/Two-state_quantum_system
* Wikipedia: https://en.wikipedia.org/wiki/Bra%E2%80%93ket_notation
"""
mutable struct StateVector
state::Vector{ComplexF64}
function StateVector(e::Number = 0, g::Number = 1)
norm::Number = hypot(e, g)
return new(complex([e/norm, g/norm]))
end
function StateVector(state::Vector{T} where T<:Number)
return new(state)
end
end
"""
PerturbEvol
Struct for a quantum physics operator that describes evolution of quantum two-level system
in a presence of some perturbation field.
# Fields
* `operator`:
Represents operator itself in a form of a matrix.
# Arguments
* `δ` :
The `detuning`, a measure of how far the perturbation field oscillation frequency is
off-resonance relative to the transition
(Hz).
* `τ` :
The `duration` of Rabi/Ramsey pulse
(s).
* `Ω₀` :
The `Rabi frequency` at which the probability amplitudes of two energy levels fluctuate
in an oscillating perturbation field
(Hz).
# Returns
* `PerturbEvol` :
Composite type instance.
# See also
* [`StateVector`](@ref)
# References
* Wikipedia: https://en.wikipedia.org/wiki/Two-state_quantum_system
* Wikipedia: https://en.wikipedia.org/wiki/Rabi_frequency
"""
struct PerturbEvol
operator::Matrix{ComplexF64}
function PerturbEvol(δ::Real, τ::Real, Ω₀::Real)
Ω = hypot(δ, Ω₀)
Ŵ = [
cos(0.5*Ω*τ) + 1im*(δ/Ω)*sin(0.5*Ω*τ) 1im*(Ω₀/Ω)*sin(0.5*Ω*τ);
1im*(Ω₀/Ω)*sin(0.5*Ω*τ) cos(0.5*Ω*τ) - 1im*(δ/Ω)*sin(0.5*Ω*τ)
]
return new(Ŵ)
end
end
"""
FreeEvol
Struct for a quantum physics operator that describes evolution of unperturbed
quantum two-level system. This evolution can be described in the way of changing
phase difference between `|e>` and `|g>` states.
# Fields
* `operator`:
Represents operator itself in a form of a matrix.
# Arguments
* `x` :
The `phase difference` between `|e>` and `|g>` states
as a result of free evolution.
# Returns
* `FreeEvol` :
Composite type instance.
# See also
* [`StateVector`](@ref)
# References
* Wikipedia: https://en.wikipedia.org/wiki/Two-state_quantum_system
"""
struct FreeEvol
operator::Matrix{ComplexF64}
function FreeEvol(x::Real)
V̂ = [
exp(1im*x) 0;
0 1
]
return new(V̂)
end
end
| RamseySpectroscopyCore | https://github.com/m0Cey/RamseySpectroscopyCore.jl.git |
|
[
"MIT"
] | 1.0.2 | 7e3de57fa445342a33b6f0350c32e32943de75e2 | code | 551 | using RamseySpectroscopyCore
using Test
@testset "RamseySpectroscopyCore.jl" begin
@test rest!(StateVector(0, 1), FreeEvol(0)).state == [0; 1]
@test rest(StateVector(0, 1), FreeEvol(0)).state == [0; 1]
@test pump!(StateVector(0, 1), PerturbEvol(0, 0, 1)).state == [0; 1]
@test pump(StateVector(0, 1), PerturbEvol(0, 0, 1)).state == [0; 1]
@test probe(StateVector(rand(), rand())) ≤ 1
@test probe(StateVector(rand(ComplexF64), rand(ComplexF64))) ≤ 1
@test probe(StateVector(rand(Complex{Int}), rand(Complex{Int}))) ≤ 1
end
| RamseySpectroscopyCore | https://github.com/m0Cey/RamseySpectroscopyCore.jl.git |
|
[
"MIT"
] | 1.0.2 | 7e3de57fa445342a33b6f0350c32e32943de75e2 | docs | 2416 | # RamseySpectroscopyCore.jl
[](https://m0Cey.github.io/RamseySpectroscopyCore.jl/stable/)
[](https://m0Cey.github.io/RamseySpectroscopyCore.jl/dev/)
[](https://github.com/m0Cey/RamseySpectroscopyCore.jl/actions/workflows/CI.yml?query=branch%3Amain)
[](https://codecov.io/gh/m0Cey/RamseySpectroscopyCore.jl)
[](https://github.com/invenia/BlueStyle)
(This section is WIP)
RamseySpectroscopyCore.jl provides core toolset for simulating Rabi/Ramsey spectroscopy experiments.
The main goal is to have ready-to-use, simple-to-understand modeling toolkit with experimenters in mind.
Furthermore, it can be used as introduction to spectroscopy for people with little physics background or
students.
## Installation
To install RamseySpectroscopyCore.jl, use the Julia package manager:
```julia
julia> using Pkg
julia> Pkg.add("RabiRamseySpectroscopy")
```
## Example
(This section is WIP)
Lets start by creating our experement setup:
1. Gas cell or a single atom - research object;
2. Tunable laser - research instrument.
```julia
julia> using RabiRamseySpectroscopy
julia> atom = StateVector(0, 1)
julia> laser = PerturbEvol(0, 3.14, 1)
```
Now we want to pump our atom with a laser to excited energy level:
```julia
julia> pump!(atom, laser)
```
After that, atom needs to be probed with a second laser pulse to check a populaton of the excited
state (or how many atoms in a gas cell are excited).
```julia
julia> probe(atom)
0.9999
```
We'll get some value approximately close to 1 which means that laser frequency is resonant with
transition frequency of an atom.
Congrats, you performed your first spectroscopy experement in Julia!
## Development
(This section is WIP)
If you want to help develop this package, you can do it via GitHub default instruments (pull requests,
issues etc.) and/or contact me: [email protected].
In additon, it is highly recommended to read or
modify code of RamseySpectroscopyCore.jl with JuliaMono font installed. That way UTF-8 symbols will be
displayed correctly. | RamseySpectroscopyCore | https://github.com/m0Cey/RamseySpectroscopyCore.jl.git |
|
[
"MIT"
] | 1.0.2 | 7e3de57fa445342a33b6f0350c32e32943de75e2 | docs | 1695 | # RamseySpectroscopyCore.jl
(This section is WIP)
RamseySpectroscopyCore.jl provides core toolset for simulating Rabi/Ramsey spectroscopy experiments.
The main goal is to have ready-to-use, simple-to-understand modeling toolkit with experimenters in mind.
Furthermore, it can be used as introduction to spectroscopy for people with little physics background or
students.
## Installation
To install RamseySpectroscopyCore.jl, use the Julia package manager:
```julia
julia> using Pkg
julia> Pkg.add("RamseySpectroscopyCore")
```
## Example
(This section is WIP)
Lets start by creating our experement setup:
1. Gas cell or a single atom - research object;
2. Tunable laser - research instrument.
```julia
julia> using RabiRamseySpectroscopy
julia> atom = StateVector(0, 1)
julia> laser = PerturbEvol(0, 3.14, 1)
```
Now we want to pump our atom with a laser to excited energy level:
```julia
julia> pump!(atom, laser)
```
After that, atom needs to be probed with a second laser pulse to check a populaton of the excited
state (or how many atoms in a gas cell are excited).
```julia
julia> probe(atom)
0.9999
```
We'll get some value approximately close to 1, which means that laser frequency is resonant with
transition frequency of an atom.
Congrats, you performed your first spectroscopy experement in Julia!
## Development
(This section is WIP)
If you want to help develop this package, you can do it via GitHub default instruments(pull requests,
issues etc.) and/or contact me: [email protected]. In additon, it is highly recommended to read or
modify code of RamseySpectroscopyCore.jl with JuliaMono font installed. That way UTF-8 symbols will be
displayed correctly.
| RamseySpectroscopyCore | https://github.com/m0Cey/RamseySpectroscopyCore.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 6558 | using BenchmarkTools
using ParticleDA
using MPI
using Random
using Base.Threads
using HDF5
include(joinpath(joinpath(@__DIR__, "..", "test"), "models", "llw2d.jl"))
using .LLW2d
if !MPI.Initialized()
MPI.Init()
end
const SUITE = BenchmarkGroup()
const my_rank = MPI.Comm_rank(MPI.COMM_WORLD)
const my_size = MPI.Comm_size(MPI.COMM_WORLD)
const params = Dict(
"filter" => Dict(
"nprt" => 32,
"enable_timers" => true,
"verbose" => true,
),
"model" => Dict(
"llw2d" => Dict(
"nx" => 200,
"ny" => 200,
"n_stations_x" => 8,
"n_stations_y" => 8,
"padding" => 0,
),
),
)
const n_time_step = 20
const nprt_per_rank = Int(params["filter"]["nprt"] / my_size)
const rng = Random.TaskLocalRNG()
Random.seed!(rng, 1234)
const filter_params = ParticleDA.get_params(ParticleDA.FilterParameters, params["filter"])
const n_tasks = (
filter_params.n_tasks > 0
? filter_params.n_tasks
: Threads.nthreads() * abs(filter_params.n_tasks)
)
const model = LLW2d.init(params["model"], n_tasks)
const state = Vector{ParticleDA.get_state_eltype(model)}(
undef, ParticleDA.get_state_dimension(model)
)
const observation = Vector{ParticleDA.get_observation_eltype(model)}(
undef, ParticleDA.get_observation_dimension(model)
)
const observation_sequence = ParticleDA.simulate_observations_from_model(
model, n_time_step
)
SUITE["Model interface"] = BenchmarkGroup()
SUITE["Model interface"]["sample_initial_state!"] = @benchmarkable ParticleDA.sample_initial_state!(
local_state, $(model), local_rng
) setup=(
local_state = copy($(state));
local_rng = copy($(rng));
)
SUITE["Model interface"]["update_state_deterministic!"] = @benchmarkable ParticleDA.update_state_deterministic!(
local_state, $(model), 0
) setup=(
local_state = copy($(state));
local_rng = copy($(rng));
ParticleDA.sample_initial_state!(local_state, $(model), local_rng);
)
SUITE["Model interface"]["update_state_stochastic!"] = @benchmarkable ParticleDA.update_state_stochastic!(
local_state, $(model), local_rng
) setup=(
local_state = copy($(state));
local_rng = copy($(rng));
ParticleDA.sample_initial_state!(local_state, $(model), local_rng);
ParticleDA.update_state_deterministic!(local_state, $(model), 0);
)
SUITE["Model interface"]["sample_observation_given_state!"] = @benchmarkable ParticleDA.sample_observation_given_state!(
local_observation, local_state, $(model), local_rng
) setup=(
local_state = copy($(state));
local_observation = copy($(observation));
local_rng = copy($(rng));
ParticleDA.sample_initial_state!(local_state, $(model), local_rng);
)
SUITE["Model interface"]["get_log_density_observation_given_state!"] = @benchmarkable ParticleDA.get_log_density_observation_given_state(
local_observation, local_state, $(model)
) setup=(
local_state = copy($(state));
local_observation = copy($(observation));
local_rng = copy($(rng));
ParticleDA.sample_initial_state!(local_state, $(model), local_rng);
ParticleDA.sample_observation_given_state!(
local_observation, local_state, $(model), local_rng
);
)
SUITE["Model interface"]["get_observation_mean_given_state!"] = @benchmarkable ParticleDA.get_observation_mean_given_state!(
observation_mean, local_state, $(model)
) setup=(
local_state = copy($(state));
observation_mean = copy($(observation));
local_rng = copy($(rng));
ParticleDA.sample_initial_state!(local_state, $(model), local_rng);
)
SUITE["Model interface"]["get_covariance_observation_noise"] = (
@benchmarkable ParticleDA.get_covariance_observation_noise($(model))
)
SUITE["Model interface"]["get_covariance_state_observation_given_previous_state"] = (
@benchmarkable ParticleDA.get_covariance_state_observation_given_previous_state(
$(model)
)
)
SUITE["Model interface"]["get_covariance_observation_observation_given_previous_state"] = (
@benchmarkable ParticleDA.get_covariance_observation_observation_given_previous_state(
$(model)
)
)
SUITE["Model interface"]["simulate_observations_from_model"] = @benchmarkable (
ParticleDA.simulate_observations_from_model($(model), $(n_time_step))
)
for filter_type in (BootstrapFilter, OptimalFilter), statistics_type in (
ParticleDA.NaiveMeanSummaryStat,
ParticleDA.NaiveMeanAndVarSummaryStat,
ParticleDA.MeanSummaryStat,
ParticleDA.MeanAndVarSummaryStat
)
group = SUITE["Filtering ($(filter_type), $(statistics_type))"] = BenchmarkGroup()
group["init_filter"] = @benchmarkable (
ParticleDA.init_filter(
$(filter_params),
$(model),
$(nprt_per_rank),
$(n_tasks),
$(filter_type),
$(statistics_type)
)
)
group["sample_proposal_and_compute_log_weights!"] = @benchmarkable (
ParticleDA.sample_proposal_and_compute_log_weights!(
states,
log_weights,
local_observation,
0,
$(model),
filter_data,
$(filter_type),
local_rng
)
) setup=(
local_rng=copy($(rng));
states=ParticleDA.init_states($(model), $(nprt_per_rank), 1, local_rng);
log_weights=Vector{Float64}(undef, $(nprt_per_rank));
local_state = copy($(state));
local_observation = copy($(observation));
ParticleDA.sample_initial_state!(local_state, $(model), local_rng);
ParticleDA.sample_observation_given_state!(
local_observation, local_state, $(model), local_rng
);
filter_data = ParticleDA.init_filter(
$(filter_params),
$(model),
$(nprt_per_rank),
$(n_tasks),
$(filter_type),
$(statistics_type)
)
)
group["run_particle_filter"] = @benchmarkable (
ParticleDA.run_particle_filter(
LLW2d.init,
local_filter_params,
$(params["model"]),
$(observation_sequence),
$(filter_type),
$(statistics_type);
rng=local_rng
);
rm(output_filename);
) seconds=30 evals=1 setup=(
local_rng=copy($(rng));
output_filename = tempname();
local_filter_params = ParticleDA.FilterParameters(;
output_filename=output_filename,
(; (Symbol(k) => v for (k, v) in $(params["filter"]))...)...
);
)
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 449 | using Documenter, ParticleDA
# Load the `LLW2d` module from the test/models to show the docstring of the parameters
test_dir = joinpath(dirname(pathof(ParticleDA)), "..", "test")
include(joinpath(test_dir, "models", "llw2d.jl"))
using .LLW2d
makedocs(
modules = [ParticleDA, LLW2d],
sitename = "ParticleDA",
)
deploydocs(
repo = "github.com/Team-RADDISH/ParticleDA.jl",
target = "build",
deps = nothing,
make = nothing,
)
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 5791 | ### A Pluto.jl notebook ###
# v0.14.5
using Markdown
using InteractiveUtils
# This Pluto notebook uses @bind for interactivity. When running this notebook outside of Pluto, the following 'mock version' of @bind gives bound variables a default value (instead of an error).
macro bind(def, element)
quote
local el = $(esc(element))
global $(esc(def)) = Core.applicable(Base.get, el) ? Base.get(el) : missing
el
end
end
# ╔═╡ c7383ff4-ba4c-11eb-1977-b31b330b20d0
begin
import Pkg
Pkg.activate(@__DIR__)
Pkg.instantiate()
using Plots
using HDF5
using Unitful
using UnitfulRecipes
using PlutoUI
end
# ╔═╡ 3e85f1c4-32ca-4e8e-ab9f-2fefaaabffb7
md"## Load HDF5 output file"
# ╔═╡ 72c703c9-3c32-45ab-b910-25a3c6652bdc
filename = "particle_da.h5"
# ╔═╡ 7cbbad5a-3767-422c-9d6b-f32524d4bf04
fh = h5open(filename, "r")
# ╔═╡ 7c316637-defe-4f60-a7e5-2e10511f7044
md"## Set these parameters to choose what to plot"
# ╔═╡ e1acfe11-0ead-4ada-946e-eff674e6d44e
begin
timestamps = keys(fh["data_syn"])
md"""
Select the timestamp
$(@bind timestamp_idx Slider(1:length(timestamps)))
"""
end
# ╔═╡ 775fbb1e-a760-4862-829d-455051942255
timestamp = timestamps[timestamp_idx]
# ╔═╡ d442ef52-1566-484d-af31-ba3565307502
md"""
Select the field
$(@bind field Select([f => f for f in keys(fh["data_syn"]["t0000"])]))
"""
# ╔═╡ 384116c1-ed19-4a8c-85a7-6c0dfd9c164f
md"## Contour plots of surface height"
# ╔═╡ 8f67e2b3-9a01-42a3-a56a-11b84776a5e1
md"## Scatter plot of particle weights"
# ╔═╡ a35f5895-5066-4e1d-b252-6172888aa92d
begin
weights = read(fh["weights"][timestamp])
p1 = scatter(weights, marker=:star)
p2 = scatter(weights, marker=:star, yscale=:log10)
for plt in (p1, p2)
plot!(plt; xlabel="Particle ID", ylabel="Weight")
end
plot(p1, p2, label="")
end
# ╔═╡ e5335e14-bdb6-432c-94ab-c666c304efc6
md"## Time series of Estimated Sample Size"
# ╔═╡ 343a1d50-38f8-4457-81dc-5d962a2acb4a
plot([1 / sum(read(w) .^ 2) for w in fh["weights"]];
label="", marker=:o, xlabel="Time step", ylabel="Estimated Sample Size (1 / sum(weight^2))")
# ╔═╡ a52a3f7e-1d8e-4153-b0c2-2cb47584c447
md"## Animation"
# ╔═╡ 8520dcbb-0bd8-4020-aea3-009e24df2099
md"## Collect data from the output file"
# ╔═╡ cff1a64f-03ba-4150-9501-fa4803901808
# All time-independent quantities
begin
field_unit = read(fh["data_syn"]["t0000"][field]["Unit"])
var_unit = read(fh["data_var"]["t0000"][field]["Unit"])
field_desc = read(fh["data_syn"]["t0000"][field]["Description"])
x_unit = read(fh["grid"]["x"]["Unit"])
y_unit = read(fh["grid"]["y"]["Unit"])
x_st_unit = read(fh["stations"]["x"]["Unit"])
y_st_unit = read(fh["stations"]["y"]["Unit"])
x = read(fh["grid"]["x"]) .* uparse(x_unit) .|> u"km"
y = read(fh["grid"]["y"]) .* uparse(y_unit) .|> u"km"
x_st = read(fh["stations"]["x"]) .* uparse(x_st_unit) .|> u"km"
y_st = read(fh["stations"]["y"]) .* uparse(y_st_unit) .|> u"km"
end
# ╔═╡ e17da3a7-39e4-4326-aa2e-08f70b574878
function plot_data(x, y, z_t, z_avg, z_std, field_desc)
n_contours = 100
zmax = max(maximum(z_t), maximum(z_avg))
zmin = min(minimum(z_t), minimum(z_avg))
levels = range(zmin, zmax; length=n_contours)
# Note that for heatmaps we need to permute the dimensions of the z matrix
p1 = heatmap(x, y, z_t'; title="True $(lowercase(field_desc))")
p2 = heatmap(x, y, z_avg'; title="Assimilated $(lowercase(field_desc))")
p3 = heatmap(x, y, z_std'; title="Std of assimilated $(lowercase(field_desc))")
for (i, plt) in enumerate((p1, p2, p3))
# Set labels
plot!(plt; xlabel="x", ylabel="y")
# Set range of color bar for first two plots
i ∈ (1, 2) && plot!(plt; clims=(ustrip(zmin), ustrip(zmax)))
# Add the positions of the stations
scatter!(plt, x_st, y_st, color=:red, marker=:star, label="")
end
plot(p1, p2, p3; titlefontsize=8, guidefontsize=8)
end
# ╔═╡ ba33b9a1-7d73-4247-b298-ccf30acc8859
function animate_data(fh, field, field_unit, var_unit, x, y)
animation = @animate for timestamp ∈ keys(fh["data_syn"])
z_t = read(fh["data_syn"][timestamp][field]) .* uparse(field_unit)
z_avg = read(fh["data_avg"][timestamp][field]) .* uparse(field_unit)
z_std = sqrt.(read(fh["data_var"][timestamp][field]) .* uparse(var_unit))
plot_data(x, y, z_t, z_avg, z_std, field_desc)
end
return mp4(animation, "animation_jl.mp4"; fps=5)
end
# ╔═╡ a9343779-de40-4d33-8487-27d53ec095c0
animate_data(fh, field, field_unit, var_unit, x, y)
# ╔═╡ da1315e0-71de-4df6-9d74-259979571e1e
# Quantities specific to the current timestamp
begin
z_t = read(fh["data_syn"][timestamp][field]) .* uparse(field_unit)
z_avg = read(fh["data_avg"][timestamp][field]) .* uparse(field_unit)
z_std = sqrt.(read(fh["data_var"][timestamp][field]) .* uparse(var_unit))
end
# ╔═╡ 1d230245-5f29-4895-b0cb-4e49f6c125ff
plot_data(x, y, z_t, z_avg, z_std, field_desc)
# ╔═╡ Cell order:
# ╠═c7383ff4-ba4c-11eb-1977-b31b330b20d0
# ╟─3e85f1c4-32ca-4e8e-ab9f-2fefaaabffb7
# ╠═72c703c9-3c32-45ab-b910-25a3c6652bdc
# ╟─7cbbad5a-3767-422c-9d6b-f32524d4bf04
# ╟─7c316637-defe-4f60-a7e5-2e10511f7044
# ╟─e1acfe11-0ead-4ada-946e-eff674e6d44e
# ╟─775fbb1e-a760-4862-829d-455051942255
# ╟─d442ef52-1566-484d-af31-ba3565307502
# ╟─384116c1-ed19-4a8c-85a7-6c0dfd9c164f
# ╟─e17da3a7-39e4-4326-aa2e-08f70b574878
# ╟─1d230245-5f29-4895-b0cb-4e49f6c125ff
# ╟─8f67e2b3-9a01-42a3-a56a-11b84776a5e1
# ╟─a35f5895-5066-4e1d-b252-6172888aa92d
# ╟─e5335e14-bdb6-432c-94ab-c666c304efc6
# ╟─343a1d50-38f8-4457-81dc-5d962a2acb4a
# ╟─a52a3f7e-1d8e-4153-b0c2-2cb47584c447
# ╟─ba33b9a1-7d73-4247-b298-ccf30acc8859
# ╟─a9343779-de40-4d33-8487-27d53ec095c0
# ╟─8520dcbb-0bd8-4020-aea3-009e24df2099
# ╟─cff1a64f-03ba-4150-9501-fa4803901808
# ╟─da1315e0-71de-4df6-9d74-259979571e1e
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 8131 | ### A Pluto.jl notebook ###
# v0.19.22
using Markdown
using InteractiveUtils
# ╔═╡ 7eeee6d4-b299-11ed-22e4-0dcb77cafa96
begin
import Pkg
Pkg.activate("../Project.toml")
using ParticleDA
using LinearAlgebra
using PDMats
using FillArrays
using Random
using HDF5
using Plots
using Statistics
end
# ╔═╡ 116a8654-c619-4683-8d9a-073aa548fe37
include("../test/models/lineargaussian.jl")
# ╔═╡ 4d2656ca-eacb-4d2b-91cb-bc82fdb49520
include("../test/kalman.jl")
# ╔═╡ a64762bb-3a9f-4b1c-83db-f1a366f282eb
function plot_filtering_distribution_comparison(
n_time_step,
n_particle,
filter_type,
init_model,
model_parameters_dict,
seed,
)
output_filename = tempname()
rng = Random.TaskLocalRNG()
Random.seed!(rng, seed)
model = init_model(model_parameters_dict)
observation_seq = ParticleDA.simulate_observations_from_model(
model, n_time_step; rng=rng
)
true_state_mean_seq, true_state_var_seq = Kalman.run_kalman_filter(
model, observation_seq
)
filter_parameters = ParticleDA.FilterParameters(
nprt=n_particle, verbose=true, output_filename=output_filename
)
isfile(output_filename) && rm(output_filename)
states, statistics = ParticleDA.run_particle_filter(
init_model,
filter_parameters,
model_parameters_dict,
observation_seq,
filter_type,
ParticleDA.MeanAndVarSummaryStat;
rng=rng
)
state_mean_seq = Matrix{ParticleDA.get_state_eltype(model)}(
undef, ParticleDA.get_state_dimension(model), n_time_step
)
state_var_seq = Matrix{ParticleDA.get_state_eltype(model)}(
undef, ParticleDA.get_state_dimension(model), n_time_step
)
weights_seq = Matrix{Float64}(undef, n_particle, n_time_step)
h5open(output_filename, "r") do file
for t in 1:n_time_step
key = ParticleDA.time_index_to_hdf5_key(t)
state_mean_seq[:, t] = read(file["state_avg"][key])
state_var_seq[:, t] = read(file["state_var"][key])
weights_seq[:, t] = read(file["weights"][key])
end
end
plots = Array{Plots.Plot}(undef, 1 + ParticleDA.get_state_dimension(model))
plots[1] = plot(
1:n_time_step,
1 ./ sum(x -> x.^2, weights_seq; dims=1)[1, :],
xlabel="Time index",
label="Estimated ESS",
legend=:outerright,
)
for (i, (m, v, tm, tv)) in enumerate(zip(
eachrow(state_mean_seq),
eachrow(state_var_seq),
eachrow(true_state_mean_seq),
eachrow(true_state_var_seq),
))
plots[i + 1] = plot(
1:n_time_step,
m,
xlabel="Time index",
ylabel="\$x_$i\$",
label="Filtering estimate",
ribbon=3 * v.^0.5,
fillalpha=0.25,
legend=:outerright,
)
plots[i + 1] = plot(
plots[i + 1],
1:n_time_step,
tm,
label="Truth",
ribbon=3 * tv.^0.5,
fillalpha=0.25,
)
end
plot(
plots...,
layout=(size(plots, 1), 1),
size=(800, 800),
left_margin=20Plots.px,
)
end
# ╔═╡ 2ad564f3-48a2-4c2a-8d7d-384a84f7d6d2
function plot_filter_estimate_rmse_vs_n_particles(
n_time_step,
n_particles,
init_model,
model_parameters_dict,
seed
)
rng = Random.TaskLocalRNG()
Random.seed!(rng, seed)
model = init_model(model_parameters_dict)
observation_seq = ParticleDA.simulate_observations_from_model(
model, n_time_step; rng=rng
)
true_state_mean_seq, true_state_var_seq = Kalman.run_kalman_filter(
model, observation_seq
)
plots = Array{Plots.Plot}(undef, 2)
for (j, (filter_type, label)) in enumerate(
zip(
(BootstrapFilter, OptimalFilter),
("Bootstrap proposal", "Locally optimal proposal")
)
)
mean_rmses = Vector{Float64}(undef, length(n_particles))
log_var_rmses = Vector{Float64}(undef, length(n_particles))
for (i, n_particle) in enumerate(n_particles)
output_filename = tempname()
filter_parameters = ParticleDA.FilterParameters(
nprt=n_particle, verbose=true, output_filename=output_filename
)
states, statistics = ParticleDA.run_particle_filter(
LinearGaussian.init,
filter_parameters,
model_parameters_dict,
observation_seq,
filter_type,
ParticleDA.MeanAndVarSummaryStat;
rng=rng
)
state_mean_seq = Matrix{ParticleDA.get_state_eltype(model)}(
undef, ParticleDA.get_state_dimension(model), n_time_step
)
state_var_seq = Matrix{ParticleDA.get_state_eltype(model)}(
undef, ParticleDA.get_state_dimension(model), n_time_step
)
weights_seq = Matrix{Float64}(undef, n_particle, n_time_step)
h5open(output_filename, "r") do file
for t in 1:n_time_step
key = ParticleDA.time_index_to_hdf5_key(t)
state_mean_seq[:, t] = read(file["state_avg"][key])
state_var_seq[:, t] = read(file["state_var"][key])
weights_seq[:, t] = read(file["weights"][key])
end
end
mean_rmses[i] = sqrt(
mean(x -> x.^2, state_mean_seq .- true_state_mean_seq)
)
log_var_rmses[i] = sqrt(
mean(x -> x.^2, log.(state_var_seq) .- log.(true_state_var_seq))
)
end
plots[j] = plot(
n_particles,
[mean_rmses, log_var_rmses],
labels=["mean" "log(variance)"],
xlabel="Number of particles",
ylabel="RMSE(truth, estimate)",
xaxis=:log,
yaxis=:log,
xticks=n_particles,
title=label,
)
end
plot(
plots...,
layout=(1, 2),
size=(1000, 400),
left_margin=20Plots.px,
bottom_margin=20Plots.px,
)
end
# ╔═╡ 89dae12b-0010-4ea1-ae69-490137196662
let
n_time_step = 200
n_particle = 100
filter_type = BootstrapFilter
seed = 20230222
plot_filtering_distribution_comparison(
n_time_step,
n_particle,
filter_type,
LinearGaussian.init,
LinearGaussian.diagonal_linear_gaussian_model_parameters(),
seed
)
end
# ╔═╡ 3e0abdfc-8668-431c-8ad3-61802e21d34e
let
n_particles = [10, 100, 1000, 10_000, 100_000]
n_time_step = 200
seed = 20230222
figure = plot_filter_estimate_rmse_vs_n_particles(
n_time_step,
n_particles,
LinearGaussian.init,
LinearGaussian.diagonal_linear_gaussian_model_parameters(),
seed
)
figure
end
# ╔═╡ 64a289be-75ce-42e2-9e43-8e0286f70a35
let
n_time_step = 200
n_particle = 100
filter_type = BootstrapFilter
seed = 20230222
plot_filtering_distribution_comparison(
n_time_step,
n_particle,
filter_type,
LinearGaussian.init,
LinearGaussian.stochastically_driven_dsho_model_parameters(),
seed
)
end
# ╔═╡ b396f776-885b-437a-94c3-693f318d7ed2
let
n_time_step = 200
n_particles = [10, 100, 1000, 10_000, 100_000]
n_time_step = 200
seed = 20230222
figure = plot_filter_estimate_rmse_vs_n_particles(
n_time_step,
n_particles,
LinearGaussian.init,
LinearGaussian.stochastically_driven_dsho_model_parameters(),
seed
)
figure
end
# ╔═╡ Cell order:
# ╠═7eeee6d4-b299-11ed-22e4-0dcb77cafa96
# ╠═116a8654-c619-4683-8d9a-073aa548fe37
# ╠═4d2656ca-eacb-4d2b-91cb-bc82fdb49520
# ╠═a64762bb-3a9f-4b1c-83db-f1a366f282eb
# ╠═2ad564f3-48a2-4c2a-8d7d-384a84f7d6d2
# ╠═89dae12b-0010-4ea1-ae69-490137196662
# ╠═3e0abdfc-8668-431c-8ad3-61802e21d34e
# ╠═64a289be-75ce-42e2-9e43-8e0286f70a35
# ╠═b396f776-885b-437a-94c3-693f318d7ed2
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 1066 | using ParticleDA
using TimerOutputs
using MPI
# Initialise MPI
MPI.Init()
mpi_size = MPI.Comm_size(MPI.COMM_WORLD)
# Save some variables for later use
test_dir = joinpath(dirname(pathof(ParticleDA)), "..", "test")
module_src = joinpath(test_dir, "model", "model.jl")
input_file = joinpath(test_dir, "integration_test_1.yaml")
truth_file = "test_observations.h5"
# Instantiate the test environment
using Pkg
Pkg.activate(test_dir)
Pkg.instantiate()
# Include the sample model source code and load it
include(module_src)
using .Model
input_dict = ParticleDA.read_input_file("parametersW1.yaml")
run_custom_params = Dict(input_dict)
# Real run
TimerOutputs.enable_debug_timings(ParticleDA)
run_custom_params["model"]["llw2d"]["padding"]=0
run_custom_params["filter"]["verbose"]=true
run_custom_params["filter"]["enable_timers"]=true
run_custom_params["filter"]["output_filename"]=string("weak_scaling_r",mpi_size,".h5")
run_custom_params["filter"]["nprt"]=mpi_size * 64
ParticleDA.run_particle_filter(Model.init, run_custom_params, BootstrapFilter(), truth_file)
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 13314 | module ParticleDA
using Random
using Statistics
using MPI
using Base.Threads
using YAML
using HDF5
using TimerOutputs
using LinearAlgebra
using PDMats
using StructArrays
using ChunkSplitters
export run_particle_filter, simulate_observations_from_model
export BootstrapFilter, OptimalFilter
export MeanSummaryStat, MeanAndVarSummaryStat, NaiveMeanSummaryStat, NaiveMeanAndVarSummaryStat
include("params.jl")
include("io.jl")
include("models.jl")
include("statistics.jl")
include("filters.jl")
include("utils.jl")
"""
simulate_observations_from_model(
init_model, input_file_path, output_file_path; rng=Random.TaskLocalRNG()
) -> Matrix
Simulate observations from the state space model initialised by the `init_model`
function with parameters specified by the `model` key in the input YAML file at
`input_file_path` and save the simulated observation and state sequences to a HDF5 file
at `output_file_path`. `rng` is a random number generator to use to generate random
variates while simulating from the model - a seeded random number generator may be
specified to ensure reproducible results.
The input YAML file at `input_file_path` should have a `simulate_observations` key
with value a dictionary with keys `seed` and `n_time_step` corresponding to respectively
the number of time steps to generate observations for from the model and the seed to
use to initialise the state of the random number generator used to simulate the
observations.
The simulated observation sequence is returned as a matrix with columns corresponding to
the observation vectors at each time step.
"""
function simulate_observations_from_model(
init_model,
input_file_path::String,
output_file_path::String;
rng::Random.AbstractRNG=Random.TaskLocalRNG()
)
input_dict = read_input_file(input_file_path)
model_dict = get(input_dict, "model", Dict())
model = init_model(model_dict)
simulate_observations_dict = get(input_dict, "simulate_observations", Dict())
n_time_step = get(simulate_observations_dict, "n_time_step", 1)
seed = get(simulate_observations_dict, "seed", nothing)
Random.seed!(rng, seed)
h5open(output_file_path, "cw") do output_file
return simulate_observations_from_model(
model, n_time_step; output_file, rng
)
end
end
function simulate_observations_from_model(
model,
num_time_step::Integer;
output_file::Union{Nothing, HDF5.File}=nothing,
rng::Random.AbstractRNG=Random.TaskLocalRNG()
)
state = Vector{get_state_eltype(model)}(undef, get_state_dimension(model))
observation_sequence = Matrix{get_observation_eltype(model)}(
undef, get_observation_dimension(model), num_time_step
)
sample_initial_state!(state, model, rng)
if !isnothing(output_file)
write_state(output_file, state, 0, "state", model)
end
for (time_index, observation) in enumerate(eachcol(observation_sequence))
update_state_deterministic!(state, model, time_index)
update_state_stochastic!(state, model, rng)
sample_observation_given_state!(observation, state, model, rng)
if !isnothing(output_file)
write_state(output_file, state, time_index, "state", model)
write_observation(output_file, observation, time_index, model)
end
end
return observation_sequence
end
"""
run_particle_filter(
init_model,
input_file_path,
observation_file_path,
filter_type=BootstrapFilter,
summary_stat_type=MeanAndVarSummaryStat;
rng=Random.TaskLocalRNG()
) -> Tuple{Matrix, Union{NamedTuple, Nothing}}
Run particle filter. `init_model` is the function which initialise the model,
`input_file_path` is the path to the YAML file with the input parameters.
`observation_file_path` is the path to the HDF5 file containing the observation
sequence to perform filtering for. `filter_type` is the particle filter type to use.
See [`ParticleFilter`](@ref) for the possible values. `summary_stat_type` is a type
specifying the summary statistics of the particles to compute at each time step. See
[`AbstractSummaryStat`](@ref) for the possible values. `rng` is a random number
generator to use to generate random variates while filtering - a seeded random
number generator may be specified to ensure reproducible results. If running with
multiple threads a thread-safe generator such as `Random.TaskLocalRNG` (the default)
must be used.
Returns a tuple containing the state particles representing an estimate of the filtering
distribution at the final observation time (with each particle a column of the returned
matrix) and a named tuple containing the estimated summary statistics of this final
filtering distribution. If running on multiple ranks using MPI, the returned states
array will correspond only to the particles local to this rank and the summary
statistics will be returned only on the master rank with all other ranks returning
`nothing` for their second return value.
"""
function run_particle_filter(
init_model,
input_file_path::String,
observation_file_path::String,
filter_type::Type{<:ParticleFilter}=BootstrapFilter,
summary_stat_type::Type{<:AbstractSummaryStat}=MeanAndVarSummaryStat;
rng::Random.AbstractRNG=Random.TaskLocalRNG()
)
MPI.Init()
# Do I/O on rank 0 only and then broadcast
my_rank = MPI.Comm_rank(MPI.COMM_WORLD)
if my_rank == 0
input_dict = read_input_file(input_file_path)
observation_sequence = h5open(
read_observation_sequence, observation_file_path, "r"
)
else
input_dict = nothing
observation_sequence = nothing
end
input_dict = MPI.bcast(input_dict, 0, MPI.COMM_WORLD)
observation_sequence = MPI.bcast(observation_sequence, 0, MPI.COMM_WORLD)
filter_params = get_params(FilterParameters, get(input_dict, "filter", Dict()))
if !isnothing(filter_params.seed)
# Use a linear congruential generator to generate different seeds for each rank
seed = UInt64(filter_params.seed)
multiplier, increment = 0x5851f42d4c957f2d, 0x14057b7ef767814f
for _ in 1:my_rank
# As seed is UInt64 operations will be modulo 2^64
seed = multiplier * seed + increment
end
# Seed per-rank random number generator
Random.seed!(rng, seed)
end
model_params_dict = get(input_dict, "model", Dict())
return run_particle_filter(
init_model,
filter_params,
model_params_dict,
observation_sequence,
filter_type,
summary_stat_type;
rng
)
end
function run_particle_filter(
init_model,
filter_params::FilterParameters,
model_params_dict::Dict,
observation_sequence::AbstractMatrix,
filter_type::Type{<:ParticleFilter},
summary_stat_type::Type{<:AbstractSummaryStat};
rng::Random.AbstractRNG=Random.TaskLocalRNG()
)
MPI.Init()
my_rank = MPI.Comm_rank(MPI.COMM_WORLD)
my_size = MPI.Comm_size(MPI.COMM_WORLD)
# For now, assume that the particles can be evenly divided between ranks
@assert mod(filter_params.nprt, my_size) == 0
nprt_per_rank = Int(filter_params.nprt / my_size)
if filter_params.enable_timers
TimerOutputs.enable_debug_timings(ParticleDA)
end
timer = TimerOutput()
# Number of tasks to schedule operations which can be parallelized acoss particles
# over - negative n_tasks filter parameter values are assumed to correspond to
# number of tasks per thread and we force the maximum number of tasks that can be
# set to the number of particles on each rank so that there is at least one
# particle per task
n_tasks = min(
filter_params.n_tasks > 0
? filter_params.n_tasks
: Threads.nthreads() * abs(filter_params.n_tasks),
nprt_per_rank
)
# Do memory allocations
@timeit_debug timer "Model initialization" model = init_model(
model_params_dict, n_tasks
)
@timeit_debug timer "State initialization" states = init_states(
model, nprt_per_rank, n_tasks, rng
)
@timeit_debug timer "Filter initialization" filter_data = init_filter(
filter_params, model, nprt_per_rank, n_tasks, filter_type, summary_stat_type
)
@timeit_debug timer "Summary statistics" update_statistics!(
filter_data.statistics, states, filter_params.master_rank
)
# Write initial state (time = 0) + metadata
if(filter_params.verbose && my_rank == filter_params.master_rank)
@timeit_debug timer "Unpack statistics" unpack_statistics!(
filter_data.unpacked_statistics, filter_data.statistics
)
@timeit_debug timer "Write snapshot" write_snapshot(
filter_params.output_filename,
model,
filter_data,
states,
0,
0 in filter_params.particle_save_time_indices,
)
end
for (time_index, observation) in enumerate(eachcol(observation_sequence))
# Sample updated values for particles from proposal distribution and compute
# unnormalized log weights for each particle in ensemble given observations
# for current time step
@timeit_debug timer "Proposals and weights" sample_proposal_and_compute_log_weights!(
states,
@view(filter_data.weights[1:nprt_per_rank]),
observation,
time_index,
model,
filter_data,
filter_type,
rng
)
# Gather weights to master rank and resample particles, doing MPI collectives
# inplace to save memory allocations.
# Note that only master_rank allocates memory for all particles. Other ranks
# only allocate for their chunk of state.
if my_rank == filter_params.master_rank
@timeit_debug timer "Gather weights" MPI.Gather!(
MPI.IN_PLACE,
UBuffer(filter_data.weights, nprt_per_rank),
filter_params.master_rank,
MPI.COMM_WORLD
)
@timeit_debug timer "Normalize weights" normalized_exp!(filter_data.weights)
@timeit_debug timer "Resample" resample!(
filter_data.resampling_indices, filter_data.weights, rng
)
else
@timeit_debug timer "Gather weights" MPI.Gather!(
filter_data.weights, nothing, filter_params.master_rank, MPI.COMM_WORLD
)
end
# Broadcast resampled particle indices to all ranks
MPI.Bcast!(filter_data.resampling_indices, filter_params.master_rank, MPI.COMM_WORLD)
@timeit_debug timer "Copy states" copy_states!(
states,
filter_data.copy_buffer,
filter_data.resampling_indices,
my_rank,
nprt_per_rank
)
if filter_params.verbose
@timeit_debug timer "Update statistics" update_statistics!(
filter_data.statistics, states, filter_params.master_rank
)
end
if my_rank == filter_params.master_rank && filter_params.verbose
@timeit_debug timer "Unpack statistics" unpack_statistics!(
filter_data.unpacked_statistics, filter_data.statistics
)
@timeit_debug timer "Write snapshot" write_snapshot(
filter_params.output_filename,
model,
filter_data,
states,
time_index,
time_index in filter_params.particle_save_time_indices,
)
end
end
if !filter_params.verbose
# Do final update and unpack of statistics if not performed in filtering loop
@timeit_debug timer "Update statistics" update_statistics!(
filter_data.statistics, states, filter_params.master_rank
)
if my_rank == filter_params.master_rank
@timeit_debug timer "Unpack statistics" unpack_statistics!(
filter_data.unpacked_statistics, filter_data.statistics
)
end
end
if filter_params.enable_timers
if my_rank == filter_params.master_rank
print_timer(timer)
end
if filter_params.verbose
# Gather string representations of timers from all ranks and write them on master
str_timer = string(timer)
timer_lengths = MPI.Gather(
sizeof(str_timer), filter_params.master_rank, MPI.COMM_WORLD
)
if my_rank == filter_params.master_rank
timer_chars = MPI.Gatherv!(
str_timer,
MPI.VBuffer(Vector{UInt8}(undef, sum(timer_lengths)), timer_lengths),
filter_params.master_rank,
MPI.COMM_WORLD
)
write_timers(timer_lengths, my_size, timer_chars, filter_params)
else
MPI.Gatherv!(str_timer, nothing, filter_params.master_rank, MPI.COMM_WORLD)
end
end
end
return states, filter_data.unpacked_statistics
end
end # module
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 10100 | """
ParticleFilter
Abstract type for particle filters. Currently implemented subtypes are:
* [`BootstrapFilter`](@ref)
* [`OptimalFilter`](@ref)
"""
abstract type ParticleFilter end
"""
ParticleDA.init_filter(filter_params, model, nprt_per_rank, ::T) -> NamedTuple
Initialise any data structures required by filter of type `T`, with filtering specific
parameters specified by `filter_params`, state space model to perform filtering with
described by `model` and `nprt_per_rank` particles per MPI rank.
New filter implementations should extend this method specifying `T` as the appropriate
singleton type for the new filter.
"""
function init_filter end
"""
ParticleDA.sample_proposal_and_compute_log_weights!(
states, log_weights, observation, model, filter_data, ::T, rng
)
Sample new values for two-dimensional array of state vectors `states` from proposal
distribution writing in-place to `states` array and compute logarithm of unnormalized
particle weights writing to `log_weights` given observation vector `observation`, with
state space model described by `model`, named tuple of filter specific data
structures `filter_data`, filter type `T` and random number generator `rng` used to
generate any random draws.
New filter implementations should extend this method specifying `T` as the appropriate
singleton type for the new filter.
"""
function sample_proposal_and_compute_log_weights! end
"""
BootstrapFilter <: ParticleFilter
Singleton type `BootstrapFilter`. This can be used as argument of
[`run_particle_filter`](@ref) to select the bootstrap filter.
"""
struct BootstrapFilter <: ParticleFilter end
"""
OptimalFilter <: ParticleFilter
Singleton type `OptimalFilter`. This can be used as argument of
[`run_particle_filter`](@ref) to select the optimal proposal filter (for conditionally
linear-Gaussian models).
"""
struct OptimalFilter <: ParticleFilter end
# Initialize arrays used by the filter
function init_filter(
filter_params::FilterParameters,
model,
nprt_per_rank::Int,
n_tasks::Int,
::Type{BootstrapFilter},
summary_stat_type::Type{<:AbstractSummaryStat}
)
state_dimension = get_state_dimension(model)
state_eltype = get_state_eltype(model)
if MPI.Comm_rank(MPI.COMM_WORLD) == filter_params.master_rank
weights = Vector{state_eltype}(undef, filter_params.nprt)
unpacked_statistics = init_unpacked_statistics(
summary_stat_type, state_eltype, state_dimension
)
else
weights = Vector{state_eltype}(undef, nprt_per_rank)
unpacked_statistics = nothing
end
resampling_indices = Vector{Int}(undef, filter_params.nprt)
statistics = init_statistics(summary_stat_type, state_eltype, state_dimension)
# Memory buffer used during copy of the states
copy_buffer = Array{state_eltype, 2}(undef, state_dimension, nprt_per_rank)
return (;
weights,
resampling_indices,
statistics,
unpacked_statistics,
copy_buffer,
n_tasks,
)
end
struct OfflineMatrices{R<:Real, M<:AbstractMatrix{R}, F<:AbstractPDMat{R}}
# Covariance between state X and observations Y given previous state x
cov_X_Y::M
# Covariance between observations Y given previous state x
cov_Y_Y::F
end
struct OnlineMatrices{T<:AbstractMatrix}
# Buffer of size (observation dimension, number of particles per rank) for holding
# intermediate values in computation of optimal proposal update
observation_buffer::T
# Buffer of size (state dimension, number of particles per rank) for holding
# intermediate values in computation of optimal proposal update
state_buffer::T
end
# Allocate and compute matrices that do not depend on time-dependent variables
function init_offline_matrices(model)
return OfflineMatrices(
get_covariance_state_observation_given_previous_state(model),
get_covariance_observation_observation_given_previous_state(model),
)
end
# Allocate memory for matrices that will be updated during the time stepping loop.
function init_online_matrices(model, nprt_per_rank::Int)
observation_dimension = get_observation_dimension(model)
updated_state_dimension = length(
get_state_indices_correlated_to_observations(model)
)
return OnlineMatrices(
Matrix{get_observation_eltype(model)}(
undef, observation_dimension, nprt_per_rank
),
Matrix{get_state_eltype(model)}(
undef, updated_state_dimension, nprt_per_rank
),
)
end
# Initialize arrays used by the filter
function init_filter(
filter_params::FilterParameters,
model,
nprt_per_rank::Int,
n_tasks::Int,
::Type{OptimalFilter},
summary_stat_type::Type{<:AbstractSummaryStat}
)
filter_data = init_filter(
filter_params, model, nprt_per_rank, n_tasks, BootstrapFilter, summary_stat_type
)
offline_matrices = init_offline_matrices(model)
online_matrices = init_online_matrices(model, nprt_per_rank)
observation_dimension = get_observation_dimension(model)
observation_eltype = get_observation_eltype(model)
observation_mean_buffer = Array{observation_eltype, 2}(
undef, observation_dimension, filter_data.n_tasks
)
return (; filter_data..., offline_matrices, online_matrices, observation_mean_buffer)
end
function sample_proposal_and_compute_log_weights!(
states::AbstractMatrix,
log_weights::AbstractVector,
observation::AbstractVector,
time_index::Integer,
model,
filter_data::NamedTuple,
::Type{BootstrapFilter},
rng::Random.AbstractRNG,
)
n_particle = size(states, 2)
@sync for (particle_indices, task_index) in chunks(1:n_particle, filter_data.n_tasks)
Threads.@spawn for particle_index in particle_indices
state = selectdim(states, 2, particle_index)
update_state_deterministic!(state, model, time_index, task_index)
update_state_stochastic!(state, model, rng, task_index)
log_weights[particle_index] = get_log_density_observation_given_state(
observation, state, model, task_index
)
end
end
end
function get_log_density_observation_given_previous_state(
observation::AbstractVector{T},
pre_noise_state::AbstractVector{S},
model,
filter_data::NamedTuple,
task_index::Integer=1
) where {S, T}
observation_mean = selectdim(filter_data.observation_mean_buffer, 2, task_index)
get_observation_mean_given_state!(observation_mean, pre_noise_state, model, task_index)
return -invquad(
filter_data.offline_matrices.cov_Y_Y, observation - observation_mean
) / 2
end
# ldiv! not currently defined for PDMat so define here
LinearAlgebra.ldiv!(A::PDMat, B::AbstractMatrix) = ldiv!(A.chol, B)
function update_states_given_observations!(
states::AbstractMatrix,
observation::AbstractVector,
model,
filter_data,
rng::Random.AbstractRNG
)
observation_buffer = filter_data.online_matrices.observation_buffer
state_buffer = filter_data.online_matrices.state_buffer
cov_X_Y = filter_data.offline_matrices.cov_X_Y
cov_Y_Y = filter_data.offline_matrices.cov_Y_Y
# Compute Y ~ Normal(HX, R) for each particle X
n_particle = size(states, 2)
@sync for (particle_indices, task_index) in chunks(1:n_particle, filter_data.n_tasks)
Threads.@spawn for particle_index in particle_indices
sample_observation_given_state!(
selectdim(observation_buffer, 2, particle_index),
selectdim(states, 2, particle_index),
model,
rng,
task_index
)
end
end
# To allow for only a subset of state components being correlated to observations
# (given previous state) and so needing to be updated as part of optimal proposal
# the model can specify the relevant indices to update. This avoids computing a
# zero update for such state components
update_indices = get_state_indices_correlated_to_observations(model)
# Update particles to account for observations, X = X - QHᵀ(HQHᵀ + R)⁻¹(Y − y)
# The following lines are equivalent to the single statement version
# states[update_indices..., :] .-= (
# cov_X_Y * (cov_Y_Y \ (observation_buffer .- observation))
# )
# but we stage across multiple statements to allow using in-place operations to
# avoid unnecessary allocations.
observation_buffer .-= observation
ldiv!(cov_Y_Y, observation_buffer)
mul!(state_buffer, cov_X_Y, observation_buffer)
@view(states[update_indices, :]) .-= state_buffer
end
function sample_proposal_and_compute_log_weights!(
states::AbstractMatrix,
log_weights::AbstractVector,
observation::AbstractVector,
time_index::Integer,
model,
filter_data::NamedTuple,
::Type{OptimalFilter},
rng::Random.AbstractRNG,
)
n_particle = size(states, 2)
@sync for (particle_indices, task_index) in chunks(1:n_particle, filter_data.n_tasks)
Threads.@spawn for particle_index in particle_indices
state = selectdim(states, 2, particle_index)
update_state_deterministic!(state, model, time_index, task_index)
# Particle weights for optimal proposal _do not_ depend on state noise values
# therefore we calculate them using states after applying deterministic part of
# time update but before adding state noise
log_weights[particle_index] = get_log_density_observation_given_previous_state(
observation, state, model, filter_data, task_index
)
update_state_stochastic!(state, model, rng, task_index)
end
end
# Update to account for conditioning on observations can be performed using matrix-
# matrix level 3 BLAS operations therefore perform outside of threaded loop over
# particles
update_states_given_observations!(states, observation, model, filter_data, rng)
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 3208 | function create_or_open_group(
file::HDF5.File, group_name::String, subgroup_name::Union{Nothing, String}=nothing
)
if !haskey(file, group_name)
group = create_group(file, group_name)
else
group = open_group(file, group_name)
end
if !isnothing(subgroup_name)
if !haskey(group, subgroup_name)
subgroup = create_group(group, subgroup_name)
else
subgroup = open_group(group, subgroup_name)
end
else
subgroup = nothing
end
return group, subgroup
end
function write_array(
group::HDF5.Group,
dataset_name::String,
array::AbstractArray,
dataset_attributes::Union{Dict{String, Any}, Nothing}=nothing
)
if !haskey(group, dataset_name)
group[dataset_name] = array
if !isnothing(dataset_attributes)
for (key, value) in pairs(dataset_attributes)
attributes(group[dataset_name])[key] = value
end
end
else
@warn "Write failed, dataset $dataset_name already exists in $group"
end
end
function write_timers(
lengths::Vector{Int},
size::Int,
chars::AbstractVector{UInt8},
params::FilterParameters
)
write_timers(lengths, size, chars, params.output_filename)
end
function write_timers(
lengths::Vector{Int}, size::Int, chars::AbstractVector{UInt8}, filename::String
)
group_name = "timer"
h5open(filename, "cw") do file
group, _ = create_or_open_group(file, group_name)
sum_lengths = cumsum(lengths)
for i in 1:size
timer_string = String(
chars[1 + (i > 1 ? sum_lengths[i - 1] : 0):sum_lengths[i]]
)
dataset_name = "rank" * string(i-1)
if !haskey(group, dataset_name)
group[dataset_name] = timer_string
else
@warn "Write failed, dataset $dataset_name already exists in $group"
end
end
end
end
function read_input_file(path_to_input_file::String)
# Read input provided in a yaml file. Overwrite default input parameters with the values provided.
if isfile(path_to_input_file)
user_input_dict = YAML.load_file(path_to_input_file)
else
@warn "Input file " * path_to_input_file * " not found, using default parameters"
user_input_dict = Dict()
end
return user_input_dict
end
function read_observation_sequence(observation_file::HDF5.File)
observation_group = observation_file["observations"]
time_keys = sort(keys(observation_group), by=hdf5_key_to_time_index)
@assert Set(map(hdf5_key_to_time_index, time_keys)) == Set(
hdf5_key_to_time_index(time_keys[1]):hdf5_key_to_time_index(time_keys[end])
) "Observations in $observation_file_path are at non-contiguous time indices"
observation = observation_group[time_keys[1]]
observation_dimension = length(observation)
observation_sequence = Matrix{eltype(observation)}(
undef, observation_dimension, length(time_keys)
)
for (time_index, key) in enumerate(time_keys)
observation_sequence[:, time_index] .= read(observation_group[key])
end
return observation_sequence
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 17739 | # Functions to extend in the model - required for all filters
"""
ParticleDA.get_state_dimension(model) -> Integer
Return the positive integer dimension of the state vector which is assumed to be fixed
for all time steps.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`.
"""
function get_state_dimension end
"""
ParticleDA.get_observation_dimension(model) -> Integer
Return the positive integer dimension of the observation vector which is assumed to be
fixed for all time steps.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`.
"""
function get_observation_dimension end
"""
ParticleDA.get_state_eltype(model) -> Type
Return the element type of the state vector which is assumed to be fixed for all time
steps.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`.
"""
function get_state_eltype end
"""
ParticleDA.get_observation_eltype(model) -> Type
Return the element type of the observation vector which is assumed to be fixed for all
time steps.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`.
"""
function get_observation_eltype end
"""
ParticleDA.sample_initial_state!(state, model, rng, task_index=1)
Sample value for state vector from its initial distribution for model described by
`model` using random number generator `rng` to generate random draws and writing
to `state` argument.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`.
"""
function sample_initial_state! end
"""
ParticleDA.update_state_deterministic!(state, model, time_index, task_index=1)
Apply the deterministic component of the state time update at discrete time index
`time_index` for the model described by `model` for the state vector `state`
writing the updated state back to the `state` argument.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`.
"""
function update_state_deterministic! end
"""
ParticleDA.update_state_stochastic!(state, model, rng, task_index=1)
Apply the stochastic component of the state time update for the model described by
`model` for the state vector `state`, using random number generator `rng` to
generate random draws and writing the updated state back to the `state` argument.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`.
"""
function update_state_stochastic! end
"""
ParticleDA.sample_observation_given_state!(
observation, state, model, rng, task_index=1
)
Simulate noisy observations of the state `state` of model described by `model`
and write to `observation` array using `rng` to generate any random draws.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`.
"""
function sample_observation_given_state! end
"""
ParticleDA.get_log_density_observation_given_state(
observation, state, model, task_index=1
) -> Real
Return the logarithm of the probability density of an observation vector `observation`
given a state vector `state` for the model associated with `model`. Any additive
terms that are constant with respect to the state may be neglected.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`.
"""
function get_log_density_observation_given_state end
"""
ParticleDA.write_model_metadata(file::HDF5.File, model)
Write metadata for with the model described by `model` to the HDF5 file `file`.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`.
"""
function write_model_metadata end
# Functions to extend in the model - required only for optimal proposal filter
"""
ParticleDA.get_observation_mean_given_state!(
observation_mean, state, model, task_index=1
)
Compute the mean of the multivariate normal distribution on the observations given
the current state and write to the first argument.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`. Only required for filtering conditionally Gaussian models with
the optimal proposal filter implementation in [`OptimalFilter`](@ref).
"""
function get_observation_mean_given_state! end
"""
ParticleDA.get_covariance_state_noise(model, i, j) -> Real
Return covariance `cov(U[i], U[j])` between components of the zero-mean Gaussian state
noise vector `U`.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`. Only required for filtering conditionally Gaussian models with
the optimal proposal filter implementation in [`OptimalFilter`](@ref).
"""
function get_covariance_state_noise end
"""
ParticleDA.get_covariance_observation_noise(model, i, j) -> Real
Return covariance `cov(V[i], V[j])` between components of the zero-mean Gaussian
observation noise vector `V`.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`. Only required for filtering conditionally Gaussian models with
the optimal proposal filter implementation in [`OptimalFilter`](@ref).
"""
function get_covariance_observation_noise end
"""
ParticleDA.get_covariance_state_observation_given_previous_state(
model, i, j
) -> Real
Return the covariance `cov(X[i], Y[j])` between components of the state vector
`X = F(x) + U` and observation vector `Y = H * X + V` where `H` is the linear
observation operator, `F` the (potentially non-linear) forward operator describing the
deterministic state dynamics, `U` is a zero-mean Gaussian state noise vector, `V` is a
zero-mean Gaussian observation noise vector and `x` is the state at the previous
observation time.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`. Only required for filtering conditionally Gaussian models with
the optimal proposal filter implementation in [`OptimalFilter`](@ref).
"""
function get_covariance_state_observation_given_previous_state end
"""
ParticleDA.get_covariance_observation_observation_given_previous_state(
model, i, j
) -> Real
Return covariance `cov(Y[i], Y[j])` between components of the observation vector
`Y = H * (F(x) + U) + V` where `H` is the linear observation operator, `F` the
(potentially non-linear) forward operator describing the deterministic state dynamics,
`U` is a zero-mean Gaussian state noise vector, `V` is a zero-mean Gaussian observation
noise vector and `x` is the state at the previous observation time.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`. Only required for filtering conditionally Gaussian models with
the optimal proposal filter implementation in [`OptimalFilter`](@ref).
"""
function get_covariance_observation_observation_given_previous_state end
# Functions to extend in the model - required only for Kalman filter tests
"""
ParticleDA.get_initial_state_mean!(state_mean, model)
Compute the mean of the multivariate normal distribution on the initial state and write
to the first argument.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`. Only required for filtering linear-Gaussian models with the
Kalman filter for testing.
"""
function get_initial_state_mean! end
"""
ParticleDA.get_covariance_initial_state(model, i, j) -> Real
Return covariance `cov(X[i], X[j])` between components of the initial state vector `X`.
This method is intended to be extended by the user with the above signature, specifying
the type of `model`. Only required for filtering linear-Gaussian models with the
Kalman filter for testing.
"""
function get_covariance_initial_state end
# Additional methods for models. These may be optionally extended by the user for a
# specific `model` type, for example to provide more efficient implementations,
# however the versions below will work providing methods for the generic functions
# described above are implemented by the model.
"""
ParticleDA.get_state_indices_correlated_to_observations(
model
) -> AbstractVector{Int}
Return the vector containing the indices of the state vector `X` which at least one of
the observations `Y`` are correlated to, that is `i ∈ 1:get_state_dimension(model)`
such that `cov(X[i], Y[j]) > 0` for at least one
`j ∈ 1:get_observation_dimension(model)`. This is used to avoid needing to compute
and store zero covariance terms. Defaults to returning all state indices which will
always give correct results but will be inefficient if there are zero blocks in
`cov(X, Y)`.
"""
function get_state_indices_correlated_to_observations(model)
return 1:get_state_dimension(model)
end
function construct_dense_covariance_matrix(
model, get_covariance, dimension::Int, eltype::Type{<:Real}
)
cov = Matrix{eltype}(undef, dimension, dimension)
for i in 1:dimension
for j in 1:i
cov[i, j] = get_covariance(model, i, j)
end
end
return PDMat(Symmetric(cov, :L))
end
"""
ParticleDA.get_covariance_state_noise(model) -> AbstractPDMat
Return covariance matrix `cov(U, U)` of zero-mean Gaussian state noise vector `U`.
Defaults to computing dense matrix using index-based
`ParticleDA.get_covariance_state_noise` method. Models may extend to exploit any
sparsity structure in covariance matrix.
"""
function get_covariance_state_noise(model)
return construct_dense_covariance_matrix(
model,
get_covariance_state_noise,
get_state_dimension(model),
get_state_eltype(model)
)
end
"""
ParticleDA.get_covariance_observation_noise(model) -> AbstractMatrix
Return covariance matrix `cov(V, V)` of zero-mean Gaussian observation noise vector `V`.
Defaults to computing dense matrix using index-based
`ParticleDA.get_covariance_observation_noise` method. Models may extend to exploit any
sparsity structure in covariance matrix.
"""
function get_covariance_observation_noise(model)
return construct_dense_covariance_matrix(
model,
get_covariance_observation_noise,
get_observation_dimension(model),
get_observation_eltype(model)
)
end
"""
ParticleDA.get_covariance_state_observation_given_previous_state(
model
) -> AbstractMatrix
Return the covariance matrix `cov(X, Y)` between the state vector `X = F(x) + U` and
observation vector `Y = H * X + V` where `H` is the linear observation operator, `F`
the (potentially non-linear) forward operator describing the deterministic
state dynamics, `U` is a zero-mean Gaussian state noise vector, `V` is a
zero-mean Gaussian observation noise vector and `x` is the state at the previous
observation time. The indices `i` here are those returned by
[`get_state_indices_correlated_to_observations`](@ref) which can be used to avoid
computing and storing blocks of `cov(X, Y)` which will always be zero.
"""
function get_covariance_state_observation_given_previous_state(model)
state_indices = get_state_indices_correlated_to_observations(model)
observation_dimension = get_observation_dimension(model)
cov = Matrix{get_state_eltype(model)}(
undef, length(state_indices), observation_dimension
)
for (i, state_index) in enumerate(state_indices)
for j in 1:observation_dimension
cov[i, j] = get_covariance_state_observation_given_previous_state(
model, state_index, j
)
end
end
return cov
end
"""
ParticleDA.get_covariance_observation_observation_given_previous_state(
model
) -> AbstractPDMat
Return covariance matrix `cov(Y, Y)` of the observation vector `Y = H * (F(x) + U) + V`
where `H` is the linear observation operator, `F` the (potentially non-linear) forward
operator describing the deterministic state dynamics, `U` is a zero-mean Gaussian state
noise vector, `V` is a zero-mean Gaussian observation noise vector and `x` is the state
at the previous observation time. Defaults to computing a dense matrix. Models may
extend to exploit any sparsity structure in covariance matrix.
"""
function get_covariance_observation_observation_given_previous_state(
model
)
return construct_dense_covariance_matrix(
model,
get_covariance_observation_observation_given_previous_state,
get_observation_dimension(model),
get_observation_eltype(model)
)
end
"""
ParticleDA.get_initial_state_mean(model)
Compute the mean of the multivariate normal distribution on the initial state.
"""
function get_initial_state_mean(model)
state_mean = Vector{get_state_eltype(model)}(undef, get_state_dimension(model))
get_initial_state_mean!(state_mean, model)
return state_mean
end
"""
ParticleDA.get_covariance_initial_state(model) -> AbstractPDMat
Return covariance matrix `cov(X, X)` of the initial state vector `X`.
Defaults to computing dense matrix using index-based
`ParticleDA.get_covariance_initial_state` method. Models may extend to exploit any
sparsity structure in covariance matrix.
"""
function get_covariance_initial_state(model)
return construct_dense_covariance_matrix(
model,
get_covariance_initial_state,
get_state_dimension(model),
get_state_eltype(model)
)
end
# Additional IO methods for models. These may be optionally extended by the user for a
# specific `model` type, for example to write out arrays in a more useful format.
time_index_to_hdf5_key(time_index::Int) = "t" * lpad(string(time_index), 4, "0")
hdf5_key_to_time_index(key::String) = parse(Int, key[2:end])
"""
ParticleDA.write_observation(
file::HDF5.File,
observation::AbstractVector,
time_index::Int,
model
)
Write the observations at time index `time_index` represented by the vector
`observation` to the HDF5 file `file` for the model represented by `model`.
"""
function write_observation(
file::HDF5.File, observation::AbstractVector, time_index::Int, model
)
time_stamp = time_index_to_hdf5_key(time_index)
group, _ = create_or_open_group(file, "observations")
attributes = Dict("Description" => "Observations", "Time index" => time_index)
write_array(group, time_stamp, observation, attributes)
end
"""
ParticleDA.write_state(
file::HDF5.File,
state::AbstractVector{T},
time_index::Int,
group_name::String,
model
)
Write the model state at time index `time_index` represented by the vector `state` to
the HDF5 file `file` with `group_name` for the model represented by `model`.
"""
function write_state(
file::HDF5.File,
state::AbstractVector,
time_index::Int,
group_name::String,
model
)
time_stamp = time_index_to_hdf5_key(time_index)
group, _ = create_or_open_group(file, group_name)
attributes = Dict("Description" => "Model state", "Time index" => time_index)
write_array(group, time_stamp, state, attributes)
end
"""
ParticleDA.write_weights(
file::HDF5.File,
weights::AbstractVector{T},
time_index::Int,
model
)
Write the particle weights at time index `time_index` represented by the vector
`weights` to the HDF5 file `file` for the model represented by `model`.
"""
function write_weights(
file::HDF5.File,
weights::AbstractVector,
time_index::Int,
model
)
time_stamp = time_index_to_hdf5_key(time_index)
group, _ = create_or_open_group(file, "weights")
attributes = Dict("Description" => "Particle weights", "Time index" => time_index)
write_array(group, time_stamp, weights, attributes)
end
"""
ParticleDA.write_snapshot(
output_filename, model, filter_data, states, time_index, save_states
)
Write a snapshot of the model and filter states to the HDF5 file `output_filename` for
the model and filters described by `model` and `filter_data` respectively at time
index `time_index`, optionally saving the current ensemble of state particles
represented by the two-dimensional array `states` (first axis state component, second
particle index) if `save_states == true`. `time_index == 0` corresponds to the initial
model and filter states before any updates and non-time dependent model data will be
written out when called with this value of `time_index`.
"""
function write_snapshot(
output_filename::AbstractString,
model,
filter_data::NamedTuple,
states::AbstractMatrix{T},
time_index::Int,
save_states::Bool,
) where T
println("Writing output at timestep = ", time_index)
h5open(output_filename, "cw") do file
time_index == 0 && write_model_metadata(file, model)
for name in keys(filter_data.unpacked_statistics)
write_state(
file,
filter_data.unpacked_statistics[name],
time_index,
"state_$name",
model
)
end
write_weights(file, filter_data.weights, time_index, model)
if save_states
println("Writing particle states at timestep = ", time_index)
for (index, state) in enumerate(eachcol(states))
group_name = "state_particle_$index"
write_state(file, state, time_index, group_name, model)
end
end
end
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 1827 | """
FilterParameters()
Parameters for ParticleDA run. Keyword arguments:
* `master_rank` : Id of MPI rank that performs serial computations
* `verbose::Bool` : Flag to control whether to write output
* `output_filename::String` : Name of output file
* `nprt::Int` : Number of particles for particle filter
* `enable_timers::Bool` : Flag to control run time measurements
* `particle_save_time_indices`: Set of time indices to save particles at
* `seed`: Seed to initialise state of random number generator used for filtering
* `n_tasks`: Number of tasks to use for running parallelisable operations. Positive
integers indicate the number of tasks directly, while the absolute value of negative
integers indicate the number of task to use per-thread (as reported by
`Threads.nthreads()`). Using multiple tasks per thread will improve the ability of
the scheduler to balance load across threads but potentially increase overheads.
If simulation of the model being filtered use multiple threads then it may be
beneficial to set the `n_tasks = 1` to avoid too much contention between threads.
"""
Base.@kwdef struct FilterParameters{V<:Union{AbstractSet, AbstractVector}}
master_rank::Int = 0
verbose::Bool = false
output_filename::String = "particle_da.h5"
nprt::Int = 4
enable_timers::Bool = false
particle_save_time_indices::V = []
seed::Union{Nothing, Int} = nothing
n_tasks::Int = -1
end
# Initialise params struct with user-defined dict of values.
function get_params(T, user_input_dict::Dict)
user_input = (; (Symbol(k) => v for (k,v) in user_input_dict)...)
params = T(;user_input...)
end
get_params(user_input_dict::Dict) = get_params(FilterParameters, user_input_dict)
# Initialise params struct with default values
get_params() = FilterParameters()
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 7198 | """
AbstractSummaryStat
Abstract type for summary statistics of particle ensemble. Concrete subtypes can be
passed as the `filter_type` argument to [`run_particle_filter`](@ref) to specify which
summary statistics to record and how they are computed.
See also: [`AbstractSumReductionSummaryStat`](@ref),
[`AbstractCustomReductionSummaryStat`](@ref).
"""
abstract type AbstractSummaryStat{T} end
"""
AbstractSumReductionSummaryStat <: AbstractSummaryStat
Abstract type for summary statistics computed using standard MPI sum reductions.
Compatible with a wider range of CPU architectures but may require less numerically
stable implementations.
"""
abstract type AbstractSumReductionSummaryStat{T} <: AbstractSummaryStat{T} end
"""
AbstractCustomReductionSummaryStat <: AbstractSummaryStat
Abstract type for summary statistics computed using custom MPI reductions. Allows
greater flexibility in computing statistics which can support more numerically stable
implementations, but at a cost of not being compatible with all CPU architectures. In
particular, `MPI.jl` does not currently support custom operators
[on Power PC and ARM architecures](https://github.com/JuliaParallel/MPI.jl/issues/404).
"""
abstract type AbstractCustomReductionSummaryStat{T} <: AbstractSummaryStat{T} end
"""
NaiveMeanSummaryStat <: AbstractSumReductionSummaryStat
Sum reduction based summary statistic type which computes the means of the particle
ensemble for each state dimension. The mean is computed by directly accumulating the
sums of the particle values and number of particles on each rank. If custom reductions
are supported by the CPU architecture in use the more numerically stable
[`MeanSummaryStat`](@ref) should be used instead.
"""
struct NaiveMeanSummaryStat{T} <: AbstractSumReductionSummaryStat{T}
sum::T
n::Int
end
compute_statistic(::Type{<:NaiveMeanSummaryStat}, x::AbstractVector) = (
NaiveMeanSummaryStat(sum(x), length(x))
)
statistic_names(::Type{<:NaiveMeanSummaryStat}) = (:avg,)
unpack(S::NaiveMeanSummaryStat) = (; avg=S.sum / S.n)
"""
NaiveMeanAndVarSummaryStat <: AbstractSumReductionSummaryStat
Sum reduction based summary statistic type which computes the means and variances of the
particle ensemble for each state dimension. The mean and variance are computed by
directly accumulating the sums of the particle values, the squared particle values and
number of particles on each rank, with the variance computed as the scaled difference
between the sum of the squares and square of the sums. This 'naive' implementation
avoids custom MPI reductions but can be numerically unstable for large ensembles or
state components with large values. If custom reductions are supported by the CPU
architecture in use the more numerically stable [`MeanAndVarSummaryStat`](@ref) should
be used instead.
"""
struct NaiveMeanAndVarSummaryStat{T} <: AbstractSumReductionSummaryStat{T}
sum::T
sum_sq::T
n::Int
end
compute_statistic(::Type{<:NaiveMeanAndVarSummaryStat}, x::AbstractVector) = (
NaiveMeanAndVarSummaryStat(sum(x), sum(abs2, x), length(x))
)
statistic_names(::Type{<:NaiveMeanAndVarSummaryStat}) = (:avg, :var)
unpack(S::NaiveMeanAndVarSummaryStat) = (
avg=S.sum / S.n, var=(S.sum_sq - S.sum^2 / S.n) / (S.n - 1)
)
function init_statistics(
S::Type{<:AbstractSumReductionSummaryStat}, T::Type, dimension::Int
)
return StructVector{S{T}}(undef, dimension)
end
function update_statistics!(
statistics::StructVector{S}, states::AbstractMatrix{T}, master_rank::Int,
) where {T, S <: AbstractSumReductionSummaryStat{T}}
Threads.@threads for i in eachindex(statistics)
statistics[i] = compute_statistic(S, selectdim(states, 1, i))
end
for name in fieldnames(S)
MPI.Reduce!(getproperty(statistics, name), +, master_rank, MPI.COMM_WORLD)
end
end
function unpack_statistics!(
unpacked_statistics::NamedTuple, statistics::StructVector{S}
) where {T, S <: AbstractSumReductionSummaryStat{T}}
Threads.@threads for i in eachindex(statistics)
for (name, val) in pairs(unpack(statistics[i]))
unpacked_statistics[name][i] = val
end
end
end
"""
MeanSummaryStat <: AbstractCustomReductionSummaryStat
Custom reduction based summary statistic type which computes the means of the particle
ensemble for each state dimension. On CPU architectures which do not support custom
reductions [`NaiveMeanSummaryStat`](@ref) can be used instead.
"""
struct MeanSummaryStat{T} <: AbstractCustomReductionSummaryStat{T}
avg::T
n::Int
end
compute_statistic(::Type{<:MeanSummaryStat}, x::AbstractVector) = (
MeanSummaryStat(mean(x), length(x))
)
statistic_names(::Type{<:MeanSummaryStat}) = (:avg,)
function combine_statistics(s1::MeanSummaryStat, s2::MeanSummaryStat)
n = s1.n + s2.n
m = (s1.avg * s1.n + s2.avg * s2.n) / n
MeanSummaryStat(m, n)
end
"""
MeanAndVarSummaryStat <: AbstractCustomReductionSummaryStat
Custom reduction based summary statistic type which computes the means and variances o
the particle ensemble for each state dimension. On CPU architectures which do not
support custom reductions [`NaiveMeanAndVarSummaryStat`](@ref) can be used instead.
"""
struct MeanAndVarSummaryStat{T} <: AbstractCustomReductionSummaryStat{T}
avg::T
var::T
n::Int
end
function compute_statistic(::Type{<:MeanAndVarSummaryStat}, x::AbstractVector)
m = mean(x)
v = varm(x, m, corrected=true)
n = length(x)
MeanAndVarSummaryStat(m, v, n)
end
statistic_names(::Type{<:MeanAndVarSummaryStat}) = (:avg, :var)
function combine_statistics(s1::MeanAndVarSummaryStat, s2::MeanAndVarSummaryStat)
n = s1.n + s2.n
m = (s1.avg * s1.n + s2.avg * s2.n) / n
# Calculate pooled unbiased sample variance of two groups.
# From https://stats.stackexchange.com/q/384951
# Can be found in https://www.tandfonline.com/doi/abs/10.1080/00031305.2014.966589
v = (
(s1.n - 1) * s1.var
+ (s2.n - 1) * s2.var
+ s1.n * s2.n / n * (s2.avg - s1.avg)^2
) / (n - 1)
MeanAndVarSummaryStat(m, v, n)
end
function init_statistics(
S::Type{<:AbstractCustomReductionSummaryStat}, T::Type, dimension::Int
)
return Array{S{T}}(undef, dimension)
end
function update_statistics!(
statistics::AbstractVector{S}, states::AbstractMatrix{T}, master_rank::Int,
) where {T, S <: AbstractCustomReductionSummaryStat{T}}
Threads.@threads for i in eachindex(statistics)
statistics[i] = compute_statistic(S, selectdim(states, 1, i))
end
MPI.Reduce!(statistics, combine_statistics, master_rank, MPI.COMM_WORLD)
end
function unpack_statistics!(
unpacked_statistics::NamedTuple, statistics::AbstractVector{S}
) where {T, S <: AbstractCustomReductionSummaryStat{T}}
Threads.@threads for i in eachindex(statistics)
for name in statistic_names(S)
unpacked_statistics[name][i] = getfield(statistics[i], name)
end
end
end
init_unpacked_statistics(S::Type{<:AbstractSummaryStat}, T::Type, dimension::Int) = (;
(name => Array{T}(undef, dimension) for name in statistic_names(S))...
)
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 3150 |
function normalized_exp!(weight::AbstractVector)
weight .-= maximum(weight)
@. weight = exp(weight)
weight ./= sum(weight)
end
# Resample particles from given weights using Stochastic Universal Sampling
function resample!(
resampled_indices::AbstractVector{Int},
weights::AbstractVector{T},
rng::Random.AbstractRNG=Random.TaskLocalRNG()
) where T
nprt = length(weights)
nprt_inv = 1.0 / nprt
k = 1
weight_cdf = cumsum(weights)
u0 = nprt_inv * rand(rng, T)
# Note: To parallelise this loop, updates to k and u have to be atomic.
# TODO: search for better parallel implementations
for ip in 1:nprt
u = u0 + (ip - 1) * nprt_inv
while(u > weight_cdf[k])
k += 1
end
resampled_indices[ip] = k
end
end
function init_states(model, nprt_per_rank::Int, n_tasks::Int, rng::AbstractRNG)
state_el_type = ParticleDA.get_state_eltype(model)
state_dimension = ParticleDA.get_state_dimension(model)
states = Matrix{state_el_type}(undef, state_dimension, nprt_per_rank)
@sync for (particle_indices, task_index) in chunks(1:nprt_per_rank, n_tasks)
Threads.@spawn for particle_index in particle_indices
sample_initial_state!(
selectdim(states, 2, particle_index), model, rng, task_index
)
end
end
return states
end
function copy_states!(
particles::AbstractMatrix{T},
buffer::AbstractMatrix{T},
resampling_indices::Vector{Int},
my_rank::Int,
nprt_per_rank::Int
) where T
# These are the particle indices stored on this rank
particles_have = my_rank * nprt_per_rank + 1:(my_rank + 1) * nprt_per_rank
# These are the particle indices this rank should have after resampling
particles_want = resampling_indices[particles_have]
# These are the ranks that have the particles this rank should have
rank_has = floor.(Int, (particles_want .- 1) / nprt_per_rank)
# We could work out how many sends and receives we have to do and allocate
# this appropriately but, lazy
reqs = Vector{MPI.Request}(undef, 0)
# Send particles to processes that want them
for (k,id) in enumerate(resampling_indices)
rank_wants = floor(Int, (k - 1) / nprt_per_rank)
if id in particles_have && rank_wants != my_rank
local_id = id - my_rank * nprt_per_rank
req = MPI.Isend(view(particles, :, local_id), rank_wants, id, MPI.COMM_WORLD)
push!(reqs, req)
end
end
# Receive particles this rank wants from ranks that have them
# If I already have them, just do a local copy
# Receive into a buffer so we dont accidentally overwrite stuff
for (k,proc,id) in zip(1:nprt_per_rank, rank_has, particles_want)
if proc == my_rank
local_id = id - my_rank * nprt_per_rank
buffer[:, k] .= view(particles, :, local_id)
else
req = MPI.Irecv!(view(buffer, :, k), proc, id, MPI.COMM_WORLD)
push!(reqs,req)
end
end
# Wait for all comms to complete
MPI.Waitall(reqs)
particles .= buffer
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 6412 | module Kalman
using LinearAlgebra
using PDMats
using ParticleDA
"""Compute `matrix = state_transition_matrix * matrix`."""
function lmult_by_state_transition_matrix!(
matrix::AbstractMatrix, model, time_index
)
for col in eachcol(matrix)
ParticleDA.update_state_deterministic!(
col, model, time_index
)
end
end
"""Compute `output_matrix = observation_matrix * rhs_matrix`."""
function lmult_by_observation_matrix!(
output_matrix::AbstractMatrix, rhs_matrix::AbstractMatrix, model
)
for (observation_vector, state_vector) in zip(
eachcol(output_matrix), eachcol(rhs_matrix)
)
ParticleDA.get_observation_mean_given_state!(
observation_vector, state_vector, model
)
end
end
abstract type AbstractKalmanFilter end
"""Kalman filter for linear Gaussian state space models using matrix-free updates.
Applies model state transition and observation functions directly to perform covariance
updates. This is more memory efficient and allows for time varying state transitions but
may be slower compared to using explicit matrix-matrix multiplies.
"""
struct MatrixFreeKalmanFilter <: AbstractKalmanFilter
model
end
function pre_and_postmultiply_by_state_transition_matrix!(
state_covar::Matrix, filter::MatrixFreeKalmanFilter, time_index::Int
)
lmult_by_state_transition_matrix!(state_covar, filter.model, time_index)
lmult_by_state_transition_matrix!(state_covar', filter.model, time_index)
end
function pre_and_postmultiply_by_observation_matrix!(
state_observation_covar::Matrix,
observation_covar::Matrix,
state_covar::Matrix,
filter::MatrixFreeKalmanFilter
)
lmult_by_observation_matrix!(
state_observation_covar', state_covar', filter.model
)
lmult_by_observation_matrix!(
observation_covar, state_observation_covar, filter.model
)
end
"""Kalman filter for linear Gaussian state space models.
Explicitly constructs state transition and observation matrices. Assumes state
transition matrix is time-invariant.
"""
struct KalmanFilter <: AbstractKalmanFilter
transition_matrix::Matrix
observation_matrix::Matrix
temp_matrix::Matrix
end
function KalmanFilter(model)
transition_matrix = Matrix{ParticleDA.get_state_eltype(model)}(
I,
ParticleDA.get_state_dimension(model),
ParticleDA.get_state_dimension(model)
)
observation_matrix = Matrix{ParticleDA.get_state_eltype(model)}(
undef,
ParticleDA.get_observation_dimension(model),
ParticleDA.get_state_dimension(model),
)
temp_matrix = Matrix{ParticleDA.get_state_eltype(model)}(
undef,
ParticleDA.get_state_dimension(model),
ParticleDA.get_state_dimension(model)
)
lmult_by_observation_matrix!(
observation_matrix, transition_matrix, model
)
lmult_by_state_transition_matrix!(transition_matrix, model, 0)
return KalmanFilter(transition_matrix, observation_matrix, temp_matrix)
end
function pre_and_postmultiply_by_state_transition_matrix!(
state_covar::Matrix, filter::KalmanFilter, time_index::Int
)
mul!(filter.temp_matrix, state_covar, filter.transition_matrix')
mul!(state_covar, filter.transition_matrix, filter.temp_matrix)
end
function pre_and_postmultiply_by_observation_matrix!(
state_observation_covar::Matrix,
observation_covar::Matrix,
state_covar::Matrix,
filter::KalmanFilter
)
mul!(state_observation_covar, state_covar, filter.observation_matrix')
mul!(observation_covar, filter.observation_matrix, state_observation_covar)
end
"""
run_kalman_filter(
model, observation_sequence[, filter_type=KalmanFilter]
)
Run Kalman filter on a linear-Gaussian state space model `model` with observations
`observation_sequence`. The `filter_type` argument can be used to set the implementation
used for the filtering updates.
"""
function run_kalman_filter(
model,
observation_sequence::Matrix ,
filter_type::Type{<:AbstractKalmanFilter}=KalmanFilter
)
state_mean = ParticleDA.get_initial_state_mean(model)
state_covar = Matrix(ParticleDA.get_covariance_initial_state(model))
state_mean_sequence = Matrix{ParticleDA.get_state_eltype(model)}(
undef,
ParticleDA.get_state_dimension(model),
size(observation_sequence, 2),
)
state_var_sequence = Matrix{ParticleDA.get_state_eltype(model)}(
undef,
ParticleDA.get_state_dimension(model),
size(observation_sequence, 2),
)
observation_mean = Vector{ParticleDA.get_observation_eltype(model)}(
undef, ParticleDA.get_observation_dimension(model)
)
state_observation_covar = Matrix{ParticleDA.get_state_eltype(model)}(
undef,
ParticleDA.get_state_dimension(model),
ParticleDA.get_observation_dimension(model)
)
observation_covar = Matrix{ParticleDA.get_observation_eltype(model)}(
undef,
ParticleDA.get_observation_dimension(model),
ParticleDA.get_observation_dimension(model)
)
observation_noise_covar = ParticleDA.get_covariance_observation_noise(model)
state_noise_covar = ParticleDA.get_covariance_state_noise(model)
kalman_filter = filter_type(model)
for (time_index, observation) in enumerate(eachcol(observation_sequence))
ParticleDA.update_state_deterministic!(
state_mean, model, time_index
)
ParticleDA.get_observation_mean_given_state!(
observation_mean, state_mean, model
)
pre_and_postmultiply_by_state_transition_matrix!(
state_covar, kalman_filter, time_index
)
pdadd!(state_covar, state_noise_covar)
pre_and_postmultiply_by_observation_matrix!(
state_observation_covar, observation_covar, state_covar, kalman_filter
)
pdadd!(observation_covar, observation_noise_covar)
chol_observation_covar = cholesky!(Symmetric(observation_covar))
state_mean .+= state_observation_covar * (
chol_observation_covar \ (observation - observation_mean)
)
state_covar .-= (
state_observation_covar * (
chol_observation_covar \ state_observation_covar'
)
)
state_mean_sequence[:, time_index] = state_mean
state_var_sequence[:, time_index] = diag(state_covar)
end
return state_mean_sequence, state_var_sequence
end
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 1509 | using Test, ParticleDA, MPI, Random
MPI.Init()
my_rank = MPI.Comm_rank(MPI.COMM_WORLD)
my_size = MPI.Comm_size(MPI.COMM_WORLD)
n_particle_per_rank = 3
n_particle = n_particle_per_rank * my_size
verbose = "-v" in ARGS || "--verbose" in ARGS
local_states = float(
collect(
my_rank * n_particle_per_rank + 1 : (my_rank + 1) * n_particle_per_rank
)
)
if verbose
for i = 1:my_size
if i == my_rank + 1
println("rank ", my_rank, ": local states: ", local_states)
end
MPI.Barrier(MPI.COMM_WORLD)
end
end
buffer = zeros((1, n_particle_per_rank))
Random.seed!(1234)
indices = rand(1:n_particle, n_particle)
if verbose && my_rank == 0
println()
println("Resampling particles to indices ", indices)
println()
end
ParticleDA.copy_states!(
reshape(local_states, (1, n_particle_per_rank)),
buffer,
indices,
my_rank,
n_particle_per_rank
)
if verbose
for i = 1:my_size
if i == my_rank + 1
test = (
local_states
== float(
indices[
my_rank * n_particle_per_rank + 1 : (my_rank + 1) * n_particle_per_rank
]
)
)
println("rank ", my_rank, ": local states: ", local_states, " -- ", test)
end
MPI.Barrier(MPI.COMM_WORLD)
end
end
@test local_states == float(
indices[my_rank * n_particle_per_rank + 1 : (my_rank + 1) * n_particle_per_rank]
)
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 2361 | using ParticleDA, MPI, Statistics, Test, YAML
include(joinpath(@__DIR__, "models", "llw2d.jl"))
using .LLW2d
# Initialise MPI
MPI.Init()
# Get the number or ranks, so that we can set a number of particle as an integer
# multiple of them.
my_size = MPI.Comm_size(MPI.COMM_WORLD)
n_particle_per_rank = 5
n_particle = n_particle_per_rank * my_size
my_rank = MPI.Comm_rank(MPI.COMM_WORLD)
master_rank = 0
input_file_path = tempname()
output_file_path = tempname()
observation_file_path = tempname()
input_params = Dict(
"filter" => Dict(
"nprt" => n_particle,
"enable_timers" => true,
"verbose"=> false,
"output_filename" => output_file_path,
"seed" => 456,
),
"model" => Dict(
"llw2d" => Dict(
"nx" => 21,
"ny" => 21,
"n_stations_x" => 2,
"n_stations_y" => 2,
"padding" => 0,
"obs_noise_std" => [10.],
),
),
"simulate_observations" => Dict(
"n_time_step" => 100,
"seed" => 123,
)
)
if my_rank == master_rank
YAML.write_file(input_file_path, input_params)
simulate_observations_from_model(LLW2d.init, input_file_path, observation_file_path)
end
for filter_type in (ParticleDA.BootstrapFilter, ParticleDA.OptimalFilter),
stat_type in (ParticleDA.NaiveMeanSummaryStat, ParticleDA.MeanAndVarSummaryStat)
states, statistics = run_particle_filter(
LLW2d.init, input_file_path, observation_file_path, filter_type, stat_type
)
@test !any(isnan.(states))
# Gather final states from all MPI ranks to master
flat_global_states = MPI.Gather(vec(states), master_rank, MPI.COMM_WORLD)
if my_rank == master_rank
global_states = reshape(flat_global_states, (size(states, 1), n_particle))
reference_statistics = (
avg=mean(global_states; dims=2),
var=var(global_states, corrected=true; dims=2)
)
for name in ParticleDA.statistic_names(stat_type)
@test size(statistics[name]) == size(states[:, 1])
@test !any(isnan.(statistics[name]))
@test all(
(statistics[name] .≈ reference_statistics[name])
.| isapprox.(reference_statistics[name], 0; atol=1e-15)
)
end
else
@test isnothing(statistics)
end
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 2121 | using Test, ParticleDA, MPI, Statistics, Random
MPI.Init()
my_rank = MPI.Comm_rank(MPI.COMM_WORLD)
my_size = MPI.Comm_size(MPI.COMM_WORLD)
rng = Random.TaskLocalRNG()
Random.seed!(rng, 1234 + my_rank)
master_rank = 0
n_particle_per_rank = 5
n_particle = n_particle_per_rank * my_size
state_eltype = Float64
dimension = 2
verbose = "-v" in ARGS || "--verbose" in ARGS
local_states = rand(rng, state_eltype, (dimension, n_particle_per_rank))
if verbose
for i = 1:my_size
if i == my_rank + 1
println("rank ", my_rank, " local states")
for state in eachcol(local_states)
println(state)
end
end
MPI.Barrier(MPI.COMM_WORLD)
end
end
flat_global_states = MPI.Gather(vec(local_states), master_rank, MPI.COMM_WORLD)
if my_rank == master_rank
global_states = reshape(flat_global_states, (dimension, n_particle))
if verbose
println("global states")
for state in eachcol(global_states)
println(state)
end
end
reference_statistics = (;
avg=mean(global_states; dims=2), var=var(global_states, corrected=true; dims=2)
)
end
for stats_type in (
ParticleDA.NaiveMeanSummaryStat,
ParticleDA.NaiveMeanAndVarSummaryStat,
ParticleDA.MeanSummaryStat,
ParticleDA.MeanAndVarSummaryStat
)
statistics = ParticleDA.init_statistics(stats_type, state_eltype, dimension)
ParticleDA.update_statistics!(statistics, local_states, master_rank)
if my_rank == master_rank
unpacked_statistics = ParticleDA.init_unpacked_statistics(
stats_type, state_eltype, dimension
)
ParticleDA.unpack_statistics!(unpacked_statistics, statistics)
for name in ParticleDA.statistic_names(stats_type)
verbose && println(
name,
", locally computed: ",
unpacked_statistics[name],
", globally computed: ",
reference_statistics[name]
)
@test all(unpacked_statistics[name] .≈ reference_statistics[name])
end
end
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 35683 | using ParticleDA
using HDF5, LinearAlgebra, MPI, PDMats, Random, StableRNGs, Statistics, Test, YAML
include(joinpath(@__DIR__, "models", "llw2d.jl"))
include(joinpath(@__DIR__, "models", "lorenz63.jl"))
include(joinpath(@__DIR__, "models", "lineargaussian.jl"))
include(joinpath(@__DIR__, "kalman.jl"))
using .LLW2d
using .Lorenz63
@testset "LLW2d model unit tests" begin
dx = dy = 2e3
### get_station_grid_indices
station_grid_indices = LLW2d.get_station_grid_indices(
2, 2, 20e3, 20e3, 150e3, 150e3, dx, dy
)
@test station_grid_indices[:, 1] == [76, 76, 86, 86]
@test station_grid_indices[:, 2] == [76, 86, 76, 86]
station_grid_indices = LLW2d.get_station_grid_indices(
3, 3, 20e3, 20e3, 150e3, 150e3, dx, dy
)
@test station_grid_indices[:, 1] == [76, 76, 76, 86, 86, 86, 96, 96, 96]
@test station_grid_indices[:, 2] == [76, 86, 96, 76, 86, 96, 76, 86, 96]
### initheight!
nx = 2
ny = 2
eta = ones(nx, ny)
ocean_depth = ones(nx, ny)
peak_position = [floor(Int, nx/4) * dx, floor(Int, ny/4) * dy]
LLW2d.initheight!(eta, ocean_depth, dx, dy, 3e4, 1.0, peak_position)
@test eta ≈ [0.978266982572228 0.9463188389826958;
0.9463188389826958 0.9154140546161575]
eta = ones(2, 2)
ocean_depth = zeros(2, 2)
LLW2d.initheight!(eta, ocean_depth, dx, dy, 3e4, 1.0, peak_position)
@test eta ≈ zeros(2, 2)
# timestep. TODO: add real tests. So far we're just making sure code won't
# crash
n = 200
eta1 = rand(n, n)
mm1 = rand(n, n)
nn1 = rand(n, n)
eta0 = rand(n, n)
mm0 = rand(n, n)
nn0 = rand(n, n)
y_averaged_depth = rand(n, n)
x_averaged_depth = rand(n, n)
land_filter_m = rand(n, n)
land_filter_n = rand(n, n)
land_filter_e = rand(n,n)
absorbing_boundary = rand(n,n)
model_matrices = LLW2d.Matrices(absorbing_boundary, ocean_depth,
x_averaged_depth, y_averaged_depth,
land_filter_m, land_filter_n, land_filter_e)
dxeta = Matrix{Float64}(undef, n, n)
dyeta = Matrix{Float64}(undef, n, n)
LLW2d.timestep!(dxeta, dyeta, eta1, mm1, nn1, eta0, mm0, nn0, model_matrices, dx, dy, 1)
# setup. TODO: add real tests. So far we're just making sure code won't
# crash
LLW2d.setup(n, n, 3e4, 0.1, 0.015, 10.0)
end
function check_hdf5_group_valid(parent, group, group_name)
@test haskey(parent, group_name)
@test isa(group, HDF5.Group)
@test occursin(group_name, HDF5.name(group))
end
@testset "File IO unit tests" begin
output_filename = tempname()
group_name = "test_group"
subgroup_name = "test_subgroup"
dataset_name = "test_dataset"
test_array = [1, 2, 3]
test_attributes = Dict("string_attribute" => "value", "int_attribute" => 1)
# test create group in empty file and write array
h5open(output_filename, "cw") do file
group, subgroup = ParticleDA.create_or_open_group(file, group_name)
check_hdf5_group_valid(file, group, group_name)
@test isnothing(subgroup)
ParticleDA.write_array(group, dataset_name, test_array, test_attributes)
end
h5open(output_filename, "cw") do file
@test haskey(file, group_name)
# test opening existing group in file and array written previously matches
group, _ = ParticleDA.create_or_open_group(file, group_name)
check_hdf5_group_valid(file, group, group_name)
@test read(group, dataset_name) == test_array
@test all([
read_attribute(group[dataset_name], k) == test_attributes[k]
for k in keys(test_attributes)
])
# test writing to existing dataset name results in warning and does not update
@test_logs (:warn, r"already exists") ParticleDA.write_array(
group, dataset_name, []
)
@test read(group, dataset_name) == test_array
# test opening subgroup
_, subgroup = ParticleDA.create_or_open_group(file, group_name, subgroup_name)
check_hdf5_group_valid(group, subgroup, subgroup_name)
end
# test writing timer data
timer_strings = ["ab", "cde", "fg", "hij"]
ParticleDA.write_timers(
map(length, timer_strings),
length(timer_strings),
codeunits(join(timer_strings)),
output_filename
)
h5open(output_filename, "cw") do file
@test haskey(file, "timer")
for (i, timer_string) in enumerate(timer_strings)
timer_dataset_name = "rank$(i-1)"
@test haskey(file["timer"], timer_dataset_name)
@test read(file["timer"], timer_dataset_name) == timer_string
end
end
end
function run_unit_tests_for_generic_model_interface(model, seed)
state_dimension = ParticleDA.get_state_dimension(model)
@test isa(state_dimension, Integer)
@test state_dimension > 0
observation_dimension = ParticleDA.get_observation_dimension(model)
@test isa(observation_dimension, Integer)
@test observation_dimension > 0
state_eltype = ParticleDA.get_state_eltype(model)
@test isa(state_eltype, DataType)
observation_eltype = ParticleDA.get_observation_eltype(model)
@test isa(observation_eltype, DataType)
state = Vector{state_eltype}(undef, state_dimension)
state .= NaN
ParticleDA.sample_initial_state!(state, model, StableRNG(seed))
@test !any(isnan.(state))
# sample_initial_state! should generate same state when passed random number
# generator with same state / seed
state_copy = copy(state)
ParticleDA.sample_initial_state!(state, model, StableRNG(seed))
@test all(state .== state_copy)
# update_state_deterministic! should give same updated state for same input state
ParticleDA.update_state_deterministic!(state, model, 1)
ParticleDA.update_state_deterministic!(state_copy, model, 1)
@test !any(isnan.(state))
@test all(state .== state_copy)
# update_state_stochastic! should give same updated state for same input + rng state
ParticleDA.update_state_stochastic!(state, model, StableRNG(seed))
ParticleDA.update_state_stochastic!(state_copy, model, StableRNG(seed))
@test !any(isnan.(state))
@test all(state == state_copy)
observation = Vector{observation_eltype}(undef, observation_dimension)
observation .= NaN
observation_copy = copy(observation)
# sample_observation_given_state! should give same observation for same input +
# rng state
ParticleDA.sample_observation_given_state!(
observation, state, model, StableRNG(seed)
)
ParticleDA.sample_observation_given_state!(
observation_copy, state, model, StableRNG(seed)
)
@test !any(isnan.(observation))
@test all(observation .== observation_copy)
log_density = ParticleDA.get_log_density_observation_given_state(
observation, state, model
)
@test isa(log_density, Real)
@test !isnan(log_density)
# get_log_density_observation_given_state should give same output for same inputs
@test log_density == ParticleDA.get_log_density_observation_given_state(
observation, state, model
)
# Tests for model IO functions
output_filename = tempname()
h5open(output_filename, "cw") do file
# As write_model_metadata could be a no-op we just test it runs without error
ParticleDA.write_model_metadata(file, model)
ParticleDA.write_observation(file, observation, 0, model)
@test haskey(file, "observations")
state_group_name = "state"
ParticleDA.write_state(file, state, 0, state_group_name, model)
@test haskey(file, state_group_name)
ParticleDA.write_weights(file, [1, 1, 1], 0, model)
@test haskey(file, "weights")
end
n_particle = 2
filter_data = (
weights=ones(n_particle),
unpacked_statistics=Dict(
"avg" => zeros(state_dimension), "var" => zeros(state_dimension)
)
)
states = zeros(state_dimension, n_particle)
for save_states in (true, false)
output_filename = tempname()
ParticleDA.write_snapshot(
output_filename, model, filter_data, states, 0, save_states
)
h5open(output_filename, "r") do file
for key in keys(filter_data.unpacked_statistics)
@test haskey(file, "state_$key")
end
@test haskey(file, "weights")
for i in 1:n_particle
key = "state_particle_$i"
@test save_states ? haskey(file, key) : !haskey(file, key)
end
end
end
end
@testset (
"Generic model interface unit tests - $(parentmodule(typeof(model)))"
) for model in (
LLW2d.init(Dict()),
Lorenz63.init(Dict()),
LinearGaussian.init(LinearGaussian.stochastically_driven_dsho_model_parameters())
)
seed = 1234
run_unit_tests_for_generic_model_interface(model, seed)
end
function check_mean_function(
set_mean!,
set_sample!,
rng,
bound_constant,
estimate_n_samples,
dimension,
el_type
)
mean = Vector{el_type}(undef, dimension)
mean .= NaN
mean_copy = copy(mean)
set_mean!(mean)
@test !any(isnan.(mean))
set_mean!(mean_copy)
@test all(mean .== mean_copy)
sample = Vector{el_type}(undef, dimension)
for n_sample in estimate_n_samples
empirical_mean = zeros(el_type, dimension)
for _ in 1:n_sample
set_sample!(sample, rng)
empirical_mean .+= sample ./ n_sample
end
# Monte Carlo estimate of mean should have O(sqrt(n_sample)) convergence to
# true mean
@test (
norm(empirical_mean - mean, Inf)
< bound_constant / sqrt(n_sample)
)
end
end
function check_covariance_function(
get_covariance_ij,
set_mean!,
set_sample!,
rng,
bound_constant,
estimate_n_samples,
dimension,
el_type
)
cov = Matrix{Float64}(undef, dimension, dimension)
all_entries_valid = true
for i in 1:dimension
for j in 1:i
cov_ij = get_covariance_ij(i, j)
all_entries_valid &= isa(cov_ij, Real)
all_entries_valid &= !isnan(cov_ij)
if i == j
all_entries_valid &= cov_ij > 0
else
all_entries_valid &= (cov_ij == get_covariance_ij(j, i))
end
cov[i, j] = cov_ij
end
end
@test all_entries_valid
cov = Symmetric(cov, :L)
@test isposdef(cov)
function get_variances_and_correlations(covariance_matrix)
variances = diag(covariance_matrix)
inv_scale_matrix = Diagonal(1 ./ sqrt.(variances))
correlation_matrix = inv_scale_matrix * covariance_matrix * inv_scale_matrix
return variances, correlation_matrix
end
# Get vector of variances and correlation matrix
var, corr = get_variances_and_correlations(cov)
mean = Vector{el_type}(undef, dimension)
set_mean!(mean)
sample = Vector{el_type}(undef, dimension)
for n_sample in estimate_n_samples
empirical_cov = zeros(dimension, dimension)
for _ in 1:n_sample
set_sample!(sample, rng)
sample .-= mean
empirical_cov .+= (sample * sample') ./ (n_sample - 1)
end
# Monte Carlo estimates of variances and correlations should have roughly
# O(sqrt(n_sample)) convergence to true values
empirical_var, empirical_corr = get_variances_and_correlations(empirical_cov)
@test (
norm(empirical_var - var, Inf) < bound_constant / sqrt(n_sample)
)
@test (
norm(empirical_corr - corr, Inf) < bound_constant / sqrt(n_sample)
)
end
end
function check_cross_covariance_function(
get_cross_covariance_ij,
set_means!,
set_samples!,
rng,
bound_constant,
estimate_n_samples,
dimensions,
el_types
)
cross_cov = Matrix{Float64}(undef, dimensions...)
all_entries_valid = true
for i in 1:dimensions[1]
for j in 1:dimensions[2]
cross_cov_ij = get_cross_covariance_ij(i, j)
all_entries_valid &= isa(cross_cov_ij, Real)
all_entries_valid &= !isnan(cross_cov_ij)
cross_cov[i, j] = cross_cov_ij
end
end
@test all_entries_valid
means = Tuple(
Vector{el_type}(undef, dimension)
for (el_type, dimension) in zip(el_types, dimensions)
)
set_means!(means...)
samples = Tuple(
Vector{el_type}(undef, dimension)
for (el_type, dimension) in zip(el_types, dimensions)
)
for n_sample in estimate_n_samples
empirical_cross_cov = zeros(dimensions...)
for _ in 1:n_sample
set_samples!(samples..., rng)
samples[1] .-= means[1]
samples[2] .-= means[2]
empirical_cross_cov .+= (samples[1] * samples[2]') ./ (n_sample - 1)
end
# Monte Carlo estimates of cross_covariances should have roughly
# O(sqrt(n_sample)) convergence to true values
@test (
norm(empirical_cross_cov - cross_cov, Inf)
< bound_constant / sqrt(n_sample)
)
end
end
function run_tests_for_optimal_proposal_model_interface(
model, seed, estimate_bound_constant, estimate_n_samples
)
state_dimension = ParticleDA.get_state_dimension(model)
observation_dimension = ParticleDA.get_observation_dimension(model)
state_eltype = ParticleDA.get_state_eltype(model)
observation_eltype = ParticleDA.get_observation_eltype(model)
state = Vector{state_eltype}(undef, state_dimension)
ParticleDA.sample_initial_state!(state, model, StableRNG(seed))
check_mean_function(
m -> ParticleDA.get_observation_mean_given_state!(m, state, model),
(s, r) -> ParticleDA.sample_observation_given_state!(s, state, model, r),
StableRNG(seed),
estimate_bound_constant,
estimate_n_samples,
observation_dimension,
observation_eltype
)
function state_transition_mean!(mean)
mean[:] = state
ParticleDA.update_state_deterministic!(mean, model, 0)
end
function sample_state_transition(next_state, rng)
next_state[:] = state
ParticleDA.update_state_deterministic!(next_state, model, 0)
ParticleDA.update_state_stochastic!(next_state, model, rng)
end
check_mean_function(
state_transition_mean!,
sample_state_transition,
StableRNG(seed),
estimate_bound_constant,
estimate_n_samples,
state_dimension,
state_eltype
)
check_covariance_function(
(i, j) -> ParticleDA.get_covariance_observation_noise(model, i, j),
m -> ParticleDA.get_observation_mean_given_state!(m, state, model),
(s, r) -> ParticleDA.sample_observation_given_state!(s, state, model, r),
StableRNG(seed),
estimate_bound_constant,
estimate_n_samples,
observation_dimension,
observation_eltype
)
check_covariance_function(
(i, j) -> ParticleDA.get_covariance_state_noise(model, i, j),
state_transition_mean!,
sample_state_transition,
StableRNG(seed),
estimate_bound_constant * 10,
estimate_n_samples,
state_dimension,
state_eltype
)
state_buffer = Vector{state_eltype}(undef, state_dimension)
function sample_observation_given_previous_state!(observation, rng)
state_buffer[:] = state
ParticleDA.update_state_deterministic!(state_buffer, model, 0)
ParticleDA.update_state_stochastic!(state_buffer, model, rng)
ParticleDA.sample_observation_given_state!(
observation, state_buffer, model, rng
)
end
function observation_given_previous_state_mean!(mean)
state_buffer[:] = state
ParticleDA.update_state_deterministic!(state_buffer, model, 0)
ParticleDA.get_observation_mean_given_state!(mean, state_buffer, model)
end
check_covariance_function(
(i, j) -> ParticleDA.get_covariance_observation_observation_given_previous_state(
model, i, j
),
observation_given_previous_state_mean!,
sample_observation_given_previous_state!,
StableRNG(seed),
estimate_bound_constant,
estimate_n_samples,
observation_dimension,
observation_eltype
)
function sample_state_observation_given_previous_state!(state_, observation, rng)
state_[:] = state
ParticleDA.update_state_deterministic!(state_, model, 0)
ParticleDA.update_state_stochastic!(state_, model, rng)
ParticleDA.sample_observation_given_state!(
observation, state_, model, rng
)
end
function state_observation_given_previous_state_mean!(state_mean, observation_mean)
state_mean[:] = state
ParticleDA.update_state_deterministic!(state_mean, model, 0)
ParticleDA.get_observation_mean_given_state!(
observation_mean, state_mean, model
)
end
check_cross_covariance_function(
(i, j) -> ParticleDA.get_covariance_state_observation_given_previous_state(
model, i, j
),
state_observation_given_previous_state_mean!,
sample_state_observation_given_previous_state!,
StableRNG(seed),
estimate_bound_constant,
estimate_n_samples,
(state_dimension, observation_dimension),
(state_eltype, observation_eltype)
)
end
@testset (
"Optimal proposal model interface unit tests - $(parentmodule(typeof(config.model)))"
) for config in (
(;
# Use sigma != 1. to test if covariance is being scaled by sigma correctly
# Reduce mesh dimensions to keep test run time reasonable
model = LLW2d.init(
Dict(
"llw2d" => Dict(
"sigma" => [0.5, 1.5, 1.5],
"nx" => 11,
"ny" => 11,
"x_length" => 100e3,
"y_length" => 100e3,
"station_boundary_x" => 30e3,
"station_boundary_y" => 30e3,
)
)
),
estimate_n_samples = [10, 100],
),
(; model = Lorenz63.init(Dict()), estimate_n_samples = [10, 100, 1000]),
(;
model = LinearGaussian.init(
LinearGaussian.stochastically_driven_dsho_model_parameters()
),
estimate_n_samples = [10, 100, 1000]
)
)
seed = 1234
# Constant factor used in Monte Carlo estimate convergence tests. Set based on some
# trial and error to keep tests relatively sensitive while avoiding too high
# probability of false failures but may require tweaking for each model
estimate_bound_constant = 12.5
run_tests_for_optimal_proposal_model_interface(
config.model, seed, estimate_bound_constant, config.estimate_n_samples
)
end
function run_tests_for_convergence_of_filter_estimates_against_kalman_filter(
filter_type,
init_model,
model_parameters_dict,
seed,
n_time_step,
n_particles,
mean_rmse_bound_constant,
log_var_rmse_bound_constant,
)
rng = Random.TaskLocalRNG()
Random.seed!(rng, seed)
model = init_model(model_parameters_dict)
observation_seq = ParticleDA.simulate_observations_from_model(
model, n_time_step; rng=rng
)
true_state_mean_seq, true_state_var_seq = Kalman.run_kalman_filter(
model, observation_seq
)
for n_particle in n_particles
output_filename = tempname()
filter_parameters = ParticleDA.FilterParameters(
nprt=n_particle, verbose=true, output_filename=output_filename
)
states, statistics = ParticleDA.run_particle_filter(
init_model,
filter_parameters,
model_parameters_dict,
observation_seq,
filter_type,
ParticleDA.MeanAndVarSummaryStat;
rng=rng
)
state_mean_seq = Matrix{ParticleDA.get_state_eltype(model)}(
undef, ParticleDA.get_state_dimension(model), n_time_step
)
state_var_seq = Matrix{ParticleDA.get_state_eltype(model)}(
undef, ParticleDA.get_state_dimension(model), n_time_step
)
weights_seq = Matrix{Float64}(undef, n_particle, n_time_step)
h5open(output_filename, "r") do file
for t in 1:n_time_step
key = ParticleDA.time_index_to_hdf5_key(t)
state_mean_seq[:, t] = read(file["state_avg"][key])
state_var_seq[:, t] = read(file["state_var"][key])
weights_seq[:, t] = read(file["weights"][key])
end
end
mean_rmse = sqrt(
mean(x -> x.^2, state_mean_seq .- true_state_mean_seq)
)
log_var_rmse = sqrt(
mean(x -> x.^2, log.(state_var_seq) .- log.(true_state_var_seq))
)
# Monte Carlo estimates of mean and log variance should have O(sqrt(n_particle))
# convergence to true values
@test mean_rmse < mean_rmse_bound_constant / sqrt(n_particle)
@test log_var_rmse < log_var_rmse_bound_constant / sqrt(n_particle)
end
end
@testset (
"Filter estimate validation against Kalman filter - $(filter_type)"
) for filter_type in (BootstrapFilter, OptimalFilter)
seed = 1234
n_time_step = 100
n_particles = [30, 100, 300, 1000]
# Constant factora used in Monte Carlo estimate convergence tests. Set based on some
# trial and error to keep tests relatively sensitive while avoiding too high
# probability of false failures
mean_rmse_bound_constant = 1.
log_var_rmse_bound_constant = 5.
run_tests_for_convergence_of_filter_estimates_against_kalman_filter(
filter_type,
LinearGaussian.init,
LinearGaussian.stochastically_driven_dsho_model_parameters(),
seed,
n_time_step,
n_particles,
mean_rmse_bound_constant,
log_var_rmse_bound_constant,
)
end
@testset "Summary statistics unit tests" begin
MPI.Init()
seed = 5678
dimension = 100
state_eltype = Float64
n_particle = 5
master_rank = 0
rng = StableRNG(seed)
states = randn(rng, (dimension, n_particle))
reference_statistics = (
avg=mean(states; dims=2), var=var(states, corrected=true; dims=2)
)
for statistics_type in (
ParticleDA.NaiveMeanSummaryStat,
ParticleDA.NaiveMeanAndVarSummaryStat,
ParticleDA.MeanSummaryStat,
ParticleDA.MeanAndVarSummaryStat
)
names = ParticleDA.statistic_names(statistics_type)
@test isa(names, Tuple)
@test eltype(names) == Symbol
statistics = ParticleDA.init_statistics(
statistics_type, state_eltype, dimension
)
ParticleDA.update_statistics!(statistics, states, master_rank)
unpacked_statistics = ParticleDA.init_unpacked_statistics(
statistics_type, state_eltype, dimension
)
@test keys(unpacked_statistics) == names
ParticleDA.unpack_statistics!(unpacked_statistics, statistics)
for name in names
@test all(unpacked_statistics[name] .≈ reference_statistics[name])
end
end
end
@testset "Generic filter unit tests" begin
MPI.Init()
seed = 1357
rng = Random.TaskLocalRNG()
Random.seed!(rng, seed)
summary_stat_type = ParticleDA.MeanAndVarSummaryStat
model = LLW2d.init(Dict())
filter_params = ParticleDA.get_params()
nprt_per_rank = filter_params.nprt
n_tasks = 1
states = ParticleDA.init_states(model, nprt_per_rank, n_tasks, rng)
@test size(states) == (ParticleDA.get_state_dimension(model), nprt_per_rank)
@test eltype(states) == ParticleDA.get_state_eltype(model)
# Sample an observation from model to use for testing filter update
time_index = 0
particle_index = 1
state = copy(states[:, 1])
ParticleDA.update_state_deterministic!(state, model, time_index)
ParticleDA.update_state_stochastic!(state, model, rng)
observation = Vector{ParticleDA.get_observation_eltype(model)}(
undef, ParticleDA.get_observation_dimension(model)
)
ParticleDA.sample_observation_given_state!(observation, state, model, rng)
log_weights = Vector{Float64}(undef, nprt_per_rank)
log_weights .= NaN
for filter_type in (BootstrapFilter, OptimalFilter)
filter_data = ParticleDA.init_filter(
filter_params, model, nprt_per_rank, n_tasks, filter_type, summary_stat_type
)
@test isa(filter_data, NamedTuple)
new_states = copy(states)
Random.seed!(rng, seed)
ParticleDA.sample_proposal_and_compute_log_weights!(
new_states,
log_weights,
observation,
time_index,
model,
filter_data,
filter_type,
rng
)
@test all(new_states != states)
@test !any(isnan.(log_weights))
# Test that log weight for particle used to simulate observation is greater
# than for other particles: this is not guaranteed to be the case, but should be
# with high probability if the initial particles are widely dispersed
@test all(log_weights[1] .> log_weights[2:end])
new_states_2 = copy(states)
log_weights_2 = Vector{Float64}(undef, nprt_per_rank)
# Check filter update gives deterministic updates when rng state is fixed
Random.seed!(rng, seed)
ParticleDA.sample_proposal_and_compute_log_weights!(
new_states_2,
log_weights_2,
observation,
time_index,
model,
filter_data,
filter_type,
rng
)
@test all(log_weights .== log_weights_2)
@test all(new_states .== new_states_2)
end
end
@testset "Optimal proposal filter specific unit tests" begin
rng = StableRNG(2468)
n_task = 1
model_params_dict = Dict(
"llw2d" => Dict(
"nx" => 32,
"ny" => 32,
"n_stations_x" => 4,
"n_stations_y" => 4,
"padding" => 0
)
)
model = LLW2d.init(model_params_dict)
# offline_matrices struct fields should all be matrix-like objects (either subtypes
# of AbstractMatrix or Factorization) and should all be initialised to finite values
# by init_offline_matrices
offline_matrices = ParticleDA.init_offline_matrices(model)
for f in nfields(offline_matrices)
matrix = getfield(offline_matrices, f)
@test isa(matrix, AbstractMatrix) || isa(matrix, Factorization)
@test !isa(matrix, AbstractMatrix) || all(isfinite, matrix)
end
# online_matrices struct fields should all be AbstractMatrix subtypes but may be
# unintialised so cannot say anything about values
online_matrices = ParticleDA.init_online_matrices(model, 1)
for f in nfields(online_matrices)
matrix = getfield(online_matrices, f)
@test isa(matrix, AbstractMatrix)
end
state_dimension = ParticleDA.get_state_dimension(model)
updated_indices = ParticleDA.get_state_indices_correlated_to_observations(
model
)
cov_X_X = ParticleDA.get_covariance_state_noise(model)
# State noise covariance should be positive definite and so symmetric and
# satisfying the trace inequality tr(C) > 0
@test all(isfinite, cov_X_X) && issymmetric(cov_X_X) && tr(cov_X_X) > 0
cov_X_Y = ParticleDA.get_covariance_state_observation_given_previous_state(
model
)
@test all(isfinite, cov_X_Y)
cov_Y_Y = ParticleDA.get_covariance_observation_observation_given_previous_state(
model
)
@test all(isfinite, cov_X_Y) && issymmetric(cov_Y_Y) && tr(cov_Y_Y) > 0
# Generate simulated observation
obs_state = Vector{ParticleDA.get_state_eltype(model)}(undef, state_dimension)
ParticleDA.sample_initial_state!(obs_state, model, rng)
ParticleDA.update_state_deterministic!(obs_state, model, 0)
observation = Vector{ParticleDA.get_observation_eltype(model)}(
undef, ParticleDA.get_observation_dimension(model)
)
ParticleDA.sample_observation_given_state!(observation, obs_state, model, rng)
# Sample new initial state and apply deterministic state update
state = Vector{ParticleDA.get_state_eltype(model)}(undef, state_dimension)
ParticleDA.sample_initial_state!(state, model, rng)
ParticleDA.update_state_deterministic!(state, model, 0)
# Get observation mean given updated state
observation_mean_given_state = Vector{
ParticleDA.get_observation_eltype(model)
}(undef, ParticleDA.get_observation_dimension(model))
ParticleDA.get_observation_mean_given_state!(
observation_mean_given_state, state, model
)
# Optimal proposal for conditionally Gaussian state-space model with updates
# X = F(x) + U and Y = HX + V where x is the previous state value, F the forward
# operator for the deterministic state dynamics, U ~ Normal(0, Q) the additive
# state noise, X the state at the next time step, H the linear observation
# operator, V ~ Normal(0, R) the additive observation noise and Y the modelled
# observations, is Normal(m, C) where
# m = F(x) + QHᵀ(HQHᵀ + R)⁻¹(y − HF(x))
# = F(x) + cov(X, Y) @ cov(Y, Y)⁻¹ (y − HF(x))
# and C = Q − QHᵀ(HQHᵀ + R)⁻¹HQ = cov(X, X) - cov(X, Y) cov(Y, Y)⁻¹ cov(X, Y)ᵀ
analytic_mean = copy(state)
@view(analytic_mean[updated_indices]) .+= (
cov_X_Y * (cov_Y_Y \ (observation .- observation_mean_given_state))
)
analytic_cov = copy(cov_X_X)
analytic_cov[updated_indices, updated_indices] .-= cov_X_Y * (cov_Y_Y \ cov_X_Y')
# init_filter assumes MPI.Init() has been called
MPI.Init()
for nprt in [25, 100, 400, 2500, 10000]
filter_params = ParticleDA.get_params(
ParticleDA.FilterParameters, Dict("nprt" => nprt)
)
filter_data = ParticleDA.init_filter(
filter_params, model, nprt, n_task, OptimalFilter, ParticleDA.MeanSummaryStat
)
# Create set of state 'particles' all equal to propagated state
states = Matrix{ParticleDA.get_state_eltype(model)}(
undef, (state_dimension, nprt)
)
states .= state
updated_states = copy(states)
for state in eachcol(updated_states)
ParticleDA.update_state_stochastic!(state, model, rng)
end
noise = updated_states .- states
# Mean of noise added by update_particle_noise! should be zero in all components
# and empirical mean should therefore be zero to within Monte Carlo error. The
# constant in the tolerance below was set by looking at scale of typical
# deviation, the point of check is that errors scale at expected O(1/√N) rate.
@test maximum(abs.(mean(noise, dims=2))) < (10. / sqrt(nprt))
# Covariance of noise added by update_particle_noise! to observed state
# components should be cov_X_X as computed above and empirical covariance of
# these components should therefore be within Monte Carlo error of cov_X_X. The
# constant in tolerance below was set by looking at scale of typical deviations,
# the point of check is that errors scale at expected O(1/√N) rate.
noise_cov = cov(noise, dims=2)
@test maximum(abs.(noise_cov .- cov_X_X)) < (10. / sqrt(nprt))
ParticleDA.update_states_given_observations!(
updated_states, observation, model, filter_data, rng
)
updated_mean = mean(updated_states, dims=2)
updated_cov = cov(updated_states, dims=2)
# Mean and covariance of updated particles should be within O(1/√N) Monte Carlo
# error of analytic values - constants in tolerances were set by looking at
# scale of typical deviations, main point of checks are that errors scale at
# expected O(1/√N) rate.
@test maximum(abs.(updated_mean .- analytic_mean)) < (10. / sqrt(nprt))
@test maximum(abs.(updated_cov .- analytic_cov)) < (10. / sqrt(nprt))
end
end
@testset "Resampling unit tests" begin
rng = StableRNG(4321)
for n_particle in (4, 8, 20, 50)
log_weights = randn(rng, n_particle)
weights = copy(log_weights)
ParticleDA.normalized_exp!(weights)
@test all(weights .>= 0)
@test all(weights .<= 1)
@test sum(weights) ≈ 1
@test all(weights .≈ (exp.(log_weights) ./ sum(exp.(log_weights))))
for n_sample in (100, 10000)
counts = zeros(Int64, n_particle)
for _ in 1:n_sample
resampled_indices = Vector{Int64}(undef, n_particle)
ParticleDA.resample!(resampled_indices, weights, rng)
for i in resampled_indices
counts[i] = counts[i] + 1
end
end
proportions = counts ./ (n_sample * n_particle)
@test norm(proportions - weights, Inf) < 0.5 / sqrt(n_sample)
end
end
# weight of 1.0 on first particle returns only copies of that particle
weights = [1., 0., 0., 0., 0.]
resampled_indices = Vector{Int64}(undef, length(weights))
ParticleDA.resample!(resampled_indices, weights)
@test all(resampled_indices .== 1)
# weight of 1.0 on last particle returns only copies of that particle
weights = [0., 0., 0., 0., 1.]
resampled_indices = Vector{Int64}(undef, length(weights))
ParticleDA.resample!(resampled_indices, weights)
@test all(resampled_indices .== 5)
end
@testset "Integration test -- $(input_file) with $(filter_type) and $(stat_type)" for
filter_type in (ParticleDA.BootstrapFilter, ParticleDA.OptimalFilter),
stat_type in (ParticleDA.MeanSummaryStat, ParticleDA.MeanAndVarSummaryStat),
input_file in ["integration_test_$i.yaml" for i in 1:6]
observation_file_path = tempname()
ParticleDA.simulate_observations_from_model(
LLW2d.init,
joinpath(@__DIR__, input_file),
observation_file_path
)
observation_sequence = h5open(
ParticleDA.read_observation_sequence, observation_file_path, "r"
)
@test !any(isnan.(observation_sequence))
states, statistics = ParticleDA.run_particle_filter(
LLW2d.init,
joinpath(@__DIR__, input_file),
observation_file_path,
filter_type,
stat_type,
)
@test !any(isnan.(states))
reference_statistics = (
avg=mean(states; dims=2), var=var(states, corrected=true; dims=2)
)
for name in ParticleDA.statistic_names(stat_type)
@test size(statistics[name]) == size(states[:, 1])
@test !any(isnan.(statistics[name]))
@test all(statistics[name] .≈ reference_statistics[name])
end
end
@testset "MPI test -- $(file)" for file in (
"mpi_filtering.jl", "mpi_copy_states.jl", "mpi_summary_statistics.jl"
)
julia = Base.julia_cmd()
flags = ["--startup-file=no", "-q", "-t$(Base.Threads.nthreads())"]
script = joinpath(@__DIR__, file)
mpiexec() do mpiexec
@test success(run(ignorestatus(`$(mpiexec) -n 3 $(julia) $(flags) $(script)`)))
end
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 7910 | module LinearGaussian
using Distributions
using HDF5
using Random
using PDMats
using ParticleDA
Base.@kwdef struct LinearGaussianModelParameters{
S <: Real,
T <: Real,
TM <: AbstractMatrix{S},
OM <: AbstractMatrix{T},
IV <: AbstractVector{S},
ICM <: AbstractMatrix{S},
SCM <: AbstractMatrix{S},
OCM <: AbstractMatrix{T}
}
state_transition_matrix::TM
observation_matrix::OM
initial_state_mean::IV
initial_state_covar::ICM
state_noise_covar::SCM
observation_noise_covar::OCM
end
struct LinearGaussianModel{S <: Real, T <: Real}
state_dimension::Int
observation_dimension::Int
parameters::LinearGaussianModelParameters{S, T}
initial_state_distribution::MvNormal{S}
state_noise_distribution::MvNormal{S}
observation_noise_distribution::MvNormal{T}
end
function diagonal_linear_gaussian_model_parameters(
state_dimension=3,
state_transition_coefficient=0.8,
observation_coefficient=1.0,
initial_state_std=1.0,
state_noise_std=0.6,
observation_noise_std=0.5,
)
return Dict(
:state_transition_matrix => ScalMat(
state_dimension, state_transition_coefficient
),
:observation_matrix => ScalMat(
state_dimension, observation_coefficient
),
:initial_state_mean => zeros(state_dimension),
:initial_state_covar => ScalMat(
state_dimension, initial_state_std^2
),
:state_noise_covar => ScalMat(
state_dimension, state_noise_std^2
),
:observation_noise_covar => ScalMat(
state_dimension, observation_noise_std^2
),
)
end
function stochastically_driven_dsho_model_parameters(
δ=0.2,
ω=1.,
Q=2.,
σ=0.5,
)
β = sqrt(Q^2 - 1 / 4)
return Dict(
:state_transition_matrix => exp(-ω * δ / 2Q) * [
[
cos(ω * β * δ / Q) + sin(ω * β * δ / Q) / 2β,
Q * sin(ω * β * δ / Q) / (ω * β)
]';
[
-Q * ω * sin(ω * δ * β / Q) / β,
cos(ω * δ * β / Q) - sin(ω * δ * β / Q) / 2β
]'
],
:observation_matrix => ScalMat(2, 1.),
:initial_state_mean => zeros(2),
:initial_state_covar => ScalMat(2, 1.),
:state_noise_covar => PDMat(
Q * exp(-ω * δ / Q) * [
[
(
(cos(2ω * δ * β / Q) - 1)
- 2β * sin(2ω * δ * β / Q)
+ 4β^2 * (exp(ω * δ / Q) - 1)
) / (8ω^3 * β^2),
Q * sin(ω * δ * β / Q)^2 / (2ω^2 * β^2)
]';
[
Q * sin(ω * δ * β / Q)^2 / (2ω^2 * β^2),
(
(cos(2ω * δ * β / Q) - 1)
+ 2β * sin(2ω * δ * β / Q)
+ 4β^2 * (exp(ω * δ / Q) - 1)
) / (8ω * β^2),
]'
]
),
:observation_noise_covar => ScalMat(2, σ^2)
)
end
function init(parameters_dict::Dict, n_tasks::Int=1)
parameters = LinearGaussianModelParameters(; parameters_dict...)
(observation_dimension, state_dimension) = size(
parameters.observation_matrix
)
return LinearGaussianModel(
state_dimension,
observation_dimension,
parameters,
(
MvNormal(m, c)
for (m, c) in (
(parameters.initial_state_mean, parameters.initial_state_covar),
(zeros(state_dimension), parameters.state_noise_covar),
(zeros(observation_dimension), parameters.observation_noise_covar),
)
)...
)
end
ParticleDA.get_state_dimension(model::LinearGaussianModel) = model.state_dimension
ParticleDA.get_observation_dimension(model::LinearGaussianModel) = model.observation_dimension
ParticleDA.get_state_eltype(::LinearGaussianModel{S, T}) where {S, T} = S
ParticleDA.get_observation_eltype(::LinearGaussianModel{S, T}) where {S, T} = T
function ParticleDA.sample_initial_state!(
state::AbstractVector{T},
model::LinearGaussianModel{S, T},
rng::Random.AbstractRNG,
task_index::Integer=1,
) where {S, T}
rand!(rng, model.initial_state_distribution, state)
end
function ParticleDA.update_state_deterministic!(
state::AbstractVector{T},
model::LinearGaussianModel{S, T},
time_index::Int,
task_index::Integer=1,
) where {S, T}
state .= model.parameters.state_transition_matrix * state
end
function ParticleDA.update_state_stochastic!(
state::AbstractVector{T},
model::LinearGaussianModel{S, T},
rng::Random.AbstractRNG,
task_index::Integer=1,
) where {S, T}
rand!(rng, state + model.state_noise_distribution, state)
end
function ParticleDA.sample_observation_given_state!(
observation::AbstractVector{T},
state::AbstractVector{S},
model::LinearGaussianModel{S, T},
rng::Random.AbstractRNG,
task_index::Integer=1,
) where {S <: Real, T <: Real}
rand!(
rng,
(model.parameters.observation_matrix * state)
+ model.observation_noise_distribution,
observation
)
end
function ParticleDA.get_log_density_observation_given_state(
observation::AbstractVector{T},
state::AbstractVector{S},
model::LinearGaussianModel{S, T},
task_index::Integer=1,
) where {S <: Real, T <: Real}
return logpdf(
(model.parameters.observation_matrix * state)
+ model.observation_noise_distribution,
observation
)
end
function ParticleDA.write_model_metadata(file::HDF5.File, model::LinearGaussianModel)
group_name = "parameters"
if !haskey(file, group_name)
group = create_group(file, group_name)
for field in fieldnames(typeof(model.parameters))
value = getfield(model.parameters, field)
attributes(group)[string(field)] = (
isa(value, AbstractArray) ? collect(value) : value
)
end
else
@warn "Write failed, group $group_name already exists in $(file.filename)!"
end
end
function ParticleDA.get_observation_mean_given_state!(
observation_mean::AbstractVector{T},
state::AbstractVector{S},
model::LinearGaussianModel{S, T},
task_index::Integer=1,
) where {S <: Real, T <: Real}
observation_mean .= model.parameters.observation_matrix * state
end
function ParticleDA.get_initial_state_mean(model::LinearGaussianModel)
return collect(model.initial_state_distribution.μ)
end
function ParticleDA.get_covariance_initial_state(model::LinearGaussianModel)
return model.initial_state_distribution.Σ
end
function ParticleDA.get_covariance_state_noise(model::LinearGaussianModel)
return model.state_noise_distribution.Σ
end
function ParticleDA.get_covariance_state_noise(model::LinearGaussianModel, i::Int, j::Int)
return model.state_noise_distribution.Σ[i, j]
end
function ParticleDA.get_covariance_observation_noise(model::LinearGaussianModel)
return model.observation_noise_distribution.Σ
end
function ParticleDA.get_covariance_observation_noise(
model::LinearGaussianModel, i::Int, j::Int
)
return model.observation_noise_distribution.Σ[i, j]
end
function ParticleDA.get_covariance_state_observation_given_previous_state(
model::LinearGaussianModel, i::Int, j::Int
)
return model.state_noise_distribution.Σ[i, j]
end
function ParticleDA.get_covariance_observation_observation_given_previous_state(
model::LinearGaussianModel, i::Int, j::Int
)
return (
model.state_noise_distribution.Σ[i, j]
+ model.observation_noise_distribution.Σ[i, j]
)
end
function ParticleDA.get_state_indices_correlated_to_observations(model::LinearGaussianModel)
return 1:ParticleDA.get_state_dimension(model)
end
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 25172 | module LLW2d
using ParticleDA
using LinearAlgebra, Random, Distributions, Base.Threads, GaussianRandomFields, HDF5
using DelimitedFiles
using PDMats
include("llw2d_timestepping.jl")
"""
LLW2dModelParameters()
Parameters for the linear long wave two-dimensional (LLW2d) model. Keyword arguments:
* `nx::Int` : Number of grid points in the x direction
* `ny::Int` : Number of grid points in the y direction
* `x_length::AbstractFloat` : Domain size in metres in the x direction
* `y_length::AbstractFloat` : Domain size in metres in the y direction
* `dx::AbstractFloat` : Distance in metres between grid points in the x direction
* `dy::AbstractFloat` : Distance in metrtes between grid points in the y direction
* `station_filename::String` : Name of input file for station coordinates
* `n_stations_x::Int` : Number of observation stations in the x direction (if using regular grid)
* `n_stations_y::Int` : Number of observation stations in the y direction (if using regular grid)
* `station_distance_x::Float` : Distance in metres between stations in the x direction (if using regular grid)
* `station_distance_y::Float` : Distance in metres between stations in the y direction (if using regular grid)
* `station_boundary_x::Float` : Distance in metres between bottom left edge of box and first station in the x direction (if using regular grid)
* `station_boundary_y::Float` : Distance in metres between bottom left edge of box and first station in the y direction (if using regular grid)
* `n_integration_step::Int` : Number of sub-steps to integrate the forward model per time step
* `time_step::AbstractFloat` : Time step length in seconds
* `peak_position::Vector{AbstractFloat}` : The `[x, y] coordinates in metres of the initial wave peak
* `peak_height::AbstractFloat` : The height in metres of the initial wave peak
* `source_size::AbstractFloat` : Cutoff distance in metres from the peak for the initial wave
* `bathymetry_setup::AbstractFloat` : Bathymetry set-up
* `lambda::AbstractFloat` : Length scale for Matérn covariance kernel in background noise
* `nu::AbstractFloat` : Smoothess parameter for Matérn covariance kernel in background noise
* `sigma::AbstractFloat` : Marginal standard deviation for Matérn covariance kernel in background noise
* `lambda_initial_state::AbstractFloat` : Length scale for Matérn covariance kernel in initial state of particles
* `nu_initial_state::AbstractFloat` : Smoothess parameter for Matérn covariance kernel in initial state of particles
* `sigma_initial_state::AbstractFloat` : Marginal standard deviation for Matérn covariance kernel in initial state of particles
* `padding::Int` : Min padding for circulant embedding gaussian random field generator
* `primes::Int`: Whether the size of the minimum circulant embedding of the covariance matrix can be written as a product of small primes (2, 3, 5 and 7). Default is `true`.
* `use_peak_initial_state_mean::Bool`: Whether to set mean of initial height field to a wave peak (true) or to all zeros (false).
In both cases the initial mean of the other state variables is zero.
* `absorber_thickness_fraction::Float` : Thickness of absorber for sponge absorbing boundary conditions, fraction of grid size
* `boundary_damping::Float` : Damping for boundaries
* `cutoff_depth::Float` : Shallowest water depth
* `obs_noise_std::Vector`: Standard deviations of noise added to observations of the true state
* `observed_state_var_indices::Vector`: Vector containing the indices of the observed state variables (1: height, 2: velocity x-component, 3: velocity y-component)
"""
Base.@kwdef struct LLW2dModelParameters{T<:AbstractFloat}
nx::Int = 41
ny::Int = 41
x_length::T = 400.0e3
y_length::T = 400.0e3
dx::T = x_length / (nx - 1)
dy::T = y_length / (ny - 1)
time_step::T = 50.0
n_integration_step::Int = 50
station_filename::String = ""
n_stations_x::Int = 4
n_stations_y::Int = 4
station_distance_x::T = 20.0e3
station_distance_y::T = 20.0e3
station_boundary_x::T = 150.0e3
station_boundary_y::T = 150.0e3
obs_noise_std::Vector{T} = [1.0]
# Observed indices
observed_state_var_indices::Vector{Int} = [1]
source_size::T = 3.0e4
bathymetry_setup::T = 3.0e3
peak_height = 1.0
peak_position = [floor(Int, nx / 4) * dx, floor(Int, ny / 4) * dy]
lambda::Vector{T} = [1.0e4, 1.0e4, 1.0e4]
nu::Vector{T} = [2.5, 2.5, 2.5]
sigma::Vector{T} = [1.0, 1.0, 1.0]
lambda_initial_state::Vector{T} = [1.0e4, 1.0e4, 1.0e4]
nu_initial_state::Vector{T} = [2.5, 2.5, 2.5]
sigma_initial_state::Vector{T} = [10.0, 10.0, 10.0]
padding::Int = 100
primes::Bool = true
use_peak_initial_state_mean::Bool = false
absorber_thickness_fraction::T = 0.1
boundary_damping::T = 0.015
cutoff_depth::T = 10.0
end
# Number of state variables in model
const n_state_var = 3
get_float_eltype(::Type{<:LLW2dModelParameters{T}}) where {T} = T
get_float_eltype(p::LLW2dModelParameters) = get_float_eltype(typeof(p))
struct RandomField{T<:Real, F<:GaussianRandomField}
grf::F
xi::Array{T, 3}
w::Array{Complex{T}, 3}
z::Array{T, 3}
end
struct LLW2dModel{T <: Real, U <: Real, G <: GaussianRandomField}
parameters::LLW2dModelParameters{T}
station_grid_indices::Matrix{Int}
field_buffer::Array{T, 4}
observation_buffer::Matrix{U}
initial_state_grf::Vector{RandomField{T, G}}
state_noise_grf::Vector{RandomField{T, G}}
model_matrices::Matrices{T}
end
function ParticleDA.get_params(T::Type{LLW2dModelParameters}, user_input_dict::Dict)
for key in ("lambda", "nu", "sigma", "lambda_initial_state", "nu_initial_state", "sigma_initial_state")
if haskey(user_input_dict, key) && !isa(user_input_dict[key], Vector)
user_input_dict[key] = fill(user_input_dict[key], 3)
end
end
user_input = (; (Symbol(k) => v for (k,v) in user_input_dict)...)
params = T(;user_input...)
end
function flat_state_to_fields(state::AbstractArray, params::LLW2dModelParameters)
if ndims(state) == 1
return reshape(state, (params.nx, params.ny, n_state_var))
else
return reshape(state, (params.nx, params.ny, n_state_var, :))
end
end
function get_grid_axes(params::LLW2dModelParameters)
x = range(0, length=params.nx, step=params.dx)
y = range(0, length=params.ny, step=params.dy)
return x, y
end
# Initialize a gaussian random field generating function using the Matern covariance kernel
# and circulant embedding generation method
# TODO: Could generalise this
function init_gaussian_random_field_generator(
lambda::Vector{T},
nu::Vector{T},
sigma::Vector{T},
x::AbstractVector{T},
y::AbstractVector{T},
pad::Int,
primes::Bool,
n_tasks::Int,
) where T
# Let's limit ourselves to two-dimensional fields
dim = 2
function _generate(l, n, s)
cov = CovarianceFunction(dim, Matern(l, n, σ = s))
grf = GaussianRandomField(cov, CirculantEmbedding(), x, y, minpadding=pad, primes=primes)
v = grf.data[1]
xi = Array{eltype(grf.cov)}(undef, size(v)..., n_tasks)
w = Array{complex(float(eltype(v)))}(undef, size(v)..., n_tasks)
z = Array{eltype(grf.cov)}(undef, length.(grf.pts)..., n_tasks)
RandomField(grf, xi, w, z)
end
return [_generate(l, n, s) for (l, n, s) in zip(lambda, nu, sigma)]
end
# Get a random sample from random_field_generator using random number generator rng
function sample_gaussian_random_field!(
field::AbstractMatrix{T},
random_field_generator::RandomField,
rng::Random.AbstractRNG,
task_index::Integer=1
) where T
randn!(rng, selectdim(random_field_generator.xi, 3, task_index))
sample_gaussian_random_field!(
field,
random_field_generator,
selectdim(random_field_generator.xi, 3, task_index),
task_index
)
end
# Get a random sample from random_field_generator using random_numbers
function sample_gaussian_random_field!(
field::AbstractMatrix{T},
random_field_generator::RandomField,
random_numbers::AbstractArray{T},
task_index::Integer=1
) where T
field .= GaussianRandomFields._sample!(
selectdim(random_field_generator.w, 3, task_index),
selectdim(random_field_generator.z, 3, task_index),
random_field_generator.grf,
random_numbers
)
end
function add_random_field!(
state_fields::AbstractArray{T, 3},
field_buffer::AbstractMatrix{T},
generators::Vector{<:RandomField},
rng::Random.AbstractRNG,
task_index::Integer
) where T
for (field, generator) in zip(eachslice(state_fields, dims=3), generators)
sample_gaussian_random_field!(field_buffer, generator, rng, task_index)
field .+= field_buffer
end
end
function ParticleDA.get_initial_state_mean!(
state_mean::AbstractVector{T},
model::LLW2dModel,
) where {T <: Real}
state_mean_fields = flat_state_to_fields(state_mean, model.parameters)
if model.parameters.use_peak_initial_state_mean
initheight!(
selectdim(state_mean_fields, 3, 1),
model.model_matrices,
model.parameters.dx,
model.parameters.dy,
model.parameters.source_size,
model.parameters.peak_height,
model.parameters.peak_position
)
state_mean_fields[:, :, 2:3] .= 0
else
state_mean_fields .= 0
end
return state_mean
end
function ParticleDA.sample_initial_state!(
state::AbstractVector{T},
model::LLW2dModel,
rng::Random.AbstractRNG,
task_index::Integer=1
) where T
ParticleDA.get_initial_state_mean!(state, model)
# Add samples of the initial random field to all particles
add_random_field!(
flat_state_to_fields(state, model.parameters),
view(model.field_buffer, :, :, 1, task_index),
model.initial_state_grf,
rng,
task_index
)
return state
end
function get_station_grid_indices(params::LLW2dModelParameters)
if params.station_filename != ""
return get_station_grid_indices(
params.station_filename,
params.dx,
params.dy,
)
else
return get_station_grid_indices(
params.n_stations_x,
params.n_stations_y,
params.station_distance_x,
params.station_distance_y,
params.station_boundary_x,
params.station_boundary_y,
params.dx,
params.dy,
)
end
end
function get_station_grid_indices(
filename::String,
dx::T,
dy::T,
) where T
coords = readdlm(filename, ',', Float64, '\n'; comments=true, comment_char='#')
return floor.(Int, coords ./ [dx dy]) .+ 1
end
function get_station_grid_indices(
n_stations_x::Integer,
n_stations_y::Integer,
station_distance_x::T,
station_distance_y::T,
station_boundary_x::T,
station_boundary_y::T,
dx::T,
dy::T
) where T
# synthetic station locations
station_grid_indices = Matrix{Int}(undef, n_stations_x * n_stations_y, 2)
n = 0
@inbounds for i in 1:n_stations_x, j in 1:n_stations_y
n += 1
station_grid_indices[n, 1] = round(
Int, (station_boundary_x + (i - 1) * station_distance_x) / dx + 1
)
station_grid_indices[n, 2] = round(
Int, (station_boundary_y + (j - 1) * station_distance_y) / dy + 1
)
end
return station_grid_indices
end
ParticleDA.get_state_dimension(model::LLW2dModel) = (
model.parameters.nx * model.parameters.ny * n_state_var
)
ParticleDA.get_observation_dimension(model::LLW2dModel) = (
size(model.station_grid_indices, 1) * length(model.parameters.observed_state_var_indices)
)
ParticleDA.get_state_eltype(::Type{<:LLW2dModel{T, U, G}}) where {T, U, G} = T
ParticleDA.get_state_eltype(model::LLW2dModel) = ParticleDA.get_state_eltype(typeof(model))
ParticleDA.get_observation_eltype(::Type{<:LLW2dModel{T, U, G}}) where {T, U, G} = U
ParticleDA.get_observation_eltype(model::LLW2dModel) = ParticleDA.get_observation_eltype(typeof(model))
function ParticleDA.get_covariance_observation_noise(
model::LLW2dModel, state_index_1::CartesianIndex, state_index_2::CartesianIndex
)
x_index_1, y_index_1, var_index_1 = state_index_1.I
x_index_2, y_index_2, var_index_2 = state_index_2.I
if (x_index_1 == x_index_2 && y_index_1 == y_index_2 && var_index_1 == var_index_2)
return (model.parameters.obs_noise_std[var_index_1]^2)
else
return 0.
end
end
function ParticleDA.get_covariance_observation_noise(
model::LLW2dModel, state_index_1::Int, state_index_2::Int
)
return ParticleDA.get_covariance_observation_noise(
model,
flat_state_index_to_cartesian_index(model.parameters, state_index_1),
flat_state_index_to_cartesian_index(model.parameters, state_index_2),
)
end
function ParticleDA.get_covariance_observation_noise(model::LLW2dModel)
observation_dimension = ParticleDA.get_observation_dimension(model)
return PDiagMat(
observation_dimension,
[
ParticleDA.get_covariance_observation_noise(model, i, i)
for i in 1:observation_dimension
]
)
end
function flat_state_index_to_cartesian_index(
parameters::LLW2dModelParameters, flat_index::Integer
)
n_grid = parameters.nx * parameters.ny
state_var_index, flat_grid_index = fldmod1(flat_index, n_grid)
grid_y_index, grid_x_index = fldmod1(flat_grid_index, parameters.nx)
return CartesianIndex(grid_x_index, grid_y_index, state_var_index)
end
function grid_index_to_grid_point(
parameters::LLW2dModelParameters, grid_index::Tuple{T, T}
) where {T <: Integer}
return [
(grid_index[1] - 1) * parameters.dx, (grid_index[2] - 1) * parameters.dy
]
end
function observation_index_to_cartesian_state_index(
parameters::LLW2dModelParameters, station_grid_indices::AbstractMatrix, observation_index::Integer
)
n_station = size(station_grid_indices,1)
state_var_index, station_index = fldmod1(observation_index, n_station)
return CartesianIndex(
station_grid_indices[station_index, :]..., state_var_index
)
end
function get_covariance_gaussian_random_fields(
gaussian_random_fields::Vector{RandomField{T, G}},
model_parameters::LLW2dModelParameters,
state_index_1::CartesianIndex,
state_index_2::CartesianIndex
) where {T <: Real, G <: GaussianRandomField}
x_index_1, y_index_1, var_index_1 = state_index_1.I
x_index_2, y_index_2, var_index_2 = state_index_2.I
if var_index_1 == var_index_2
grid_point_1 = grid_index_to_grid_point(
model_parameters, (x_index_1, y_index_1)
)
grid_point_2 = grid_index_to_grid_point(
model_parameters, (x_index_2, y_index_2)
)
covariance_structure = gaussian_random_fields[var_index_1].grf.cov.cov
return apply(
covariance_structure, abs.(grid_point_1 .- grid_point_2)
)
else
return 0.
end
end
function ParticleDA.get_covariance_state_noise(
model::LLW2dModel, state_index_1::CartesianIndex, state_index_2::CartesianIndex
)
return get_covariance_gaussian_random_fields(
model.state_noise_grf, model.parameters, state_index_1, state_index_2,
)
end
function ParticleDA.get_covariance_state_noise(
model::LLW2dModel, state_index_1::Integer, state_index_2::Integer
)
return get_covariance_gaussian_random_fields(
model.state_noise_grf,
model.parameters,
flat_state_index_to_cartesian_index(model.parameters, state_index_1),
flat_state_index_to_cartesian_index(model.parameters, state_index_2),
)
end
function ParticleDA.get_covariance_initial_state(
model::LLW2dModel, state_index_1::Integer, state_index_2::Integer
)
return get_covariance_gaussian_random_fields(
model.initial_state_grf,
model.parameters,
flat_state_index_to_cartesian_index(model.parameters, state_index_1),
flat_state_index_to_cartesian_index(model.parameters, state_index_2),
)
end
function ParticleDA.get_covariance_observation_observation_given_previous_state(
model::LLW2dModel, observation_index_1::Integer, observation_index_2::Integer
)
observation_1 = observation_index_to_cartesian_state_index(
model.parameters,
model.station_grid_indices,
observation_index_1
)
observation_2 = observation_index_to_cartesian_state_index(
model.parameters,
model.station_grid_indices,
observation_index_2
)
return ParticleDA.get_covariance_state_noise(
model,
observation_1,
observation_2,
) + ParticleDA.get_covariance_observation_noise(
model, observation_1, observation_2
)
end
function ParticleDA.get_covariance_state_observation_given_previous_state(
model::LLW2dModel, state_index::Integer, observation_index::Integer
)
return ParticleDA.get_covariance_state_noise(
model,
flat_state_index_to_cartesian_index(model.parameters, state_index),
observation_index_to_cartesian_state_index(
model.parameters, model.station_grid_indices, observation_index
),
)
end
function ParticleDA.get_state_indices_correlated_to_observations(model::LLW2dModel)
n_grid = model.parameters.nx * model.parameters.ny
return vcat(
(
(i - 1) * n_grid + 1 : i * n_grid
for i in model.parameters.observed_state_var_indices
)...
)
end
function init(parameters_dict::Dict, n_tasks::Int=1)
parameters = ParticleDA.get_params(
LLW2dModelParameters, get(parameters_dict, "llw2d", Dict())
)
station_grid_indices = get_station_grid_indices(parameters)
T = get_float_eltype(parameters)
n_stations = size(station_grid_indices, 1)
n_observations = n_stations * length(parameters.observed_state_var_indices)
# Buffer array to be used in the tsunami update
field_buffer = Array{T}(undef, parameters.nx, parameters.ny, 2, n_tasks)
# Buffer array to be used in computing observation mean
observation_buffer = Array{T}(undef, n_observations, n_tasks)
# Gaussian random fields for generating intial state and state noise
x, y = get_grid_axes(parameters)
initial_state_grf = init_gaussian_random_field_generator(
parameters.lambda_initial_state,
parameters.nu_initial_state,
parameters.sigma_initial_state,
x,
y,
parameters.padding,
parameters.primes,
n_tasks,
)
state_noise_grf = init_gaussian_random_field_generator(
parameters.lambda,
parameters.nu,
parameters.sigma,
x,
y,
parameters.padding,
parameters.primes,
n_tasks
)
# Set up tsunami model
model_matrices = setup(
parameters.nx,
parameters.ny,
parameters.bathymetry_setup,
parameters.absorber_thickness_fraction,
parameters.boundary_damping,
parameters.cutoff_depth
)
return LLW2dModel(
parameters,
station_grid_indices,
field_buffer,
observation_buffer,
initial_state_grf,
state_noise_grf,
model_matrices
)
end
function ParticleDA.get_observation_mean_given_state!(
observation_mean::AbstractVector,
state::AbstractVector,
model::LLW2dModel,
task_index::Integer=1
)
state_fields = flat_state_to_fields(state, model.parameters)
n = 1
for k in model.parameters.observed_state_var_indices
for (i, j) in eachrow(model.station_grid_indices)
observation_mean[n] = state_fields[i, j, k]
n += 1
end
end
end
function ParticleDA.sample_observation_given_state!(
observation::AbstractVector{S},
state::AbstractVector{T},
model::LLW2dModel,
rng::AbstractRNG,
task_index::Integer=1,
) where{S, T}
ParticleDA.get_observation_mean_given_state!(observation, state, model, task_index)
observation .+= rand(
rng, MvNormal(ParticleDA.get_covariance_observation_noise(model))
)
return observation
end
function ParticleDA.get_log_density_observation_given_state(
observation::AbstractVector,
state::AbstractVector,
model::LLW2dModel,
task_index::Integer=1
)
observation_mean = selectdim(model.observation_buffer, 2, task_index)
ParticleDA.get_observation_mean_given_state!(observation_mean, state, model, task_index)
return -invquad(
ParticleDA.get_covariance_observation_noise(model),
observation - observation_mean
) / 2
end
function ParticleDA.update_state_deterministic!(
state::AbstractVector, model::LLW2dModel, time_index::Integer, task_index::Integer=1
)
# Parts of state vector are aliased to tsunami height and velocity component fields
state_fields = flat_state_to_fields(state, model.parameters)
height_field = selectdim(state_fields, 3, 1)
velocity_x_field = selectdim(state_fields, 3, 2)
velocity_y_field = selectdim(state_fields, 3, 3)
dx_buffer = view(model.field_buffer, :, :, 1, task_index)
dy_buffer = view(model.field_buffer, :, :, 2, task_index)
dt = model.parameters.time_step / model.parameters.n_integration_step
for _ in 1:model.parameters.n_integration_step
# Update tsunami wavefield with LLW2d.timestep in-place
timestep!(
dx_buffer,
dy_buffer,
height_field,
velocity_x_field,
velocity_y_field,
height_field,
velocity_x_field,
velocity_y_field,
model.model_matrices,
model.parameters.dx,
model.parameters.dy,
dt
)
end
end
function ParticleDA.update_state_stochastic!(
state::AbstractVector, model::LLW2dModel, rng::AbstractRNG, task_index::Integer=1
)
# Add state noise
add_random_field!(
flat_state_to_fields(state, model.parameters),
view(model.field_buffer, :, :, 1, task_index),
model.state_noise_grf,
rng,
task_index
)
end
### Model IO
function write_parameters(group::HDF5.Group, params::LLW2dModelParameters)
fields = fieldnames(typeof(params))
for field in fields
attributes(group)[string(field)] = getfield(params, field)
end
end
function write_coordinates(group::HDF5.Group, x::AbstractVector, y::AbstractVector)
for (dataset_name, val) in zip(("x", "y"), (x, y))
dataset, _ = create_dataset(group, dataset_name, val)
dataset[:] = val
attributes(dataset)["Description"] = "$dataset_name coordinate"
attributes(dataset)["Unit"] = "m"
end
end
function ParticleDA.write_model_metadata(file::HDF5.File, model::LLW2dModel)
parameters = model.parameters
grid_x, grid_y = map(collect, get_grid_axes(parameters))
stations_x = (model.station_grid_indices[:, 1] .- 1) .* parameters.dx
stations_y = (model.station_grid_indices[:, 2] .- 1) .* parameters.dy
for (group_name, write_group) in [
("parameters", group -> write_parameters(group, parameters)),
("grid_coordinates", group -> write_coordinates(group, grid_x, grid_y)),
("station_coordinates", group -> write_coordinates(group, stations_x, stations_y)),
]
if !haskey(file, group_name)
group = create_group(file, group_name)
write_group(group)
else
@warn "Write failed, group $group_name already exists in $(file.filename)!"
end
end
end
function ParticleDA.write_state(
file::HDF5.File,
state::AbstractVector{T},
time_index::Int,
group_name::String,
model::LLW2dModel
) where T
parameters = model.parameters
subgroup_name = ParticleDA.time_index_to_hdf5_key(time_index)
_, subgroup = ParticleDA.create_or_open_group(file, group_name, subgroup_name)
state_fields = flat_state_to_fields(state, parameters)
state_fields_metadata = [
(name="height", unit="m", description="Ocean surface height"),
(name="vx", unit="m/s", description="Ocean surface velocity x-component"),
(name="vy", unit="m/s", description="Ocean surface velocity y-component")
]
for (field, metadata) in zip(eachslice(state_fields, dims=3), state_fields_metadata)
if !haskey(subgroup, metadata.name)
subgroup[metadata.name] = field
dataset_attributes = attributes(subgroup[metadata.name])
dataset_attributes["Description"] = metadata.description
dataset_attributes["Unit"] = metadata.unit
dataset_attributes["Time index"] = time_index
dataset_attributes["Time (s)"] = time_index * parameters.time_step
else
@warn "Write failed, dataset $(metadata.name) already exists in $(subgroup)!"
end
end
end
end # module
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 7849 | # Linear Long Wave (LLW) tsunami in 2D Cartesian Coordinate
struct Matrices{T,M<:AbstractMatrix{T}}
absorbing_boundary::M
ocean_depth::M
x_averaged_depth::M
y_averaged_depth::M
land_filter_m::M
land_filter_n::M
land_filter_e::M
end
const g_n = 9.80665
function timestep!(dx_buffer::AbstractMatrix{T},
dy_buffer::AbstractMatrix{T},
eta1::AbstractMatrix{T},
mm1::AbstractMatrix{T},
nn1::AbstractMatrix{T},
eta0::AbstractMatrix{T},
mm0::AbstractMatrix{T},
nn0::AbstractMatrix{T},
absorbing_boundary::AbstractMatrix{T},
x_averaged_depth::AbstractMatrix{T},
y_averaged_depth::AbstractMatrix{T},
land_filter_m::AbstractMatrix{T},
land_filter_n::AbstractMatrix{T},
land_filter_e::AbstractMatrix{T},
dx::Real,dy::Real,dt::Real) where T
nx, ny = size(eta1)
@assert (nx, ny) == size(mm1) == size(nn1) == size(eta0) == size(mm0) ==
size(nn0) == size(x_averaged_depth) == size(y_averaged_depth) ==
size(land_filter_m) == size(land_filter_n) == size(land_filter_e) ==
size(absorbing_boundary)
# diffs
for j in 1:ny
for i in 2:nx
@inbounds dx_buffer[i,j] = (eta0[i,j] - eta0[i - 1,j]) / dx
end
@inbounds dx_buffer[1,j] = (eta0[1,j] - 0) / dx
end
for i in 1:nx
for j in 2:ny
@inbounds dy_buffer[i,j] = (eta0[i,j] - eta0[i, j - 1]) / dy
end
@inbounds dy_buffer[i,1] = (eta0[i,1] - 0) / dy
end
# Update Velocity
for j in 1:ny, i in 1:nx
@inbounds mm1[i,j] = mm0[i, j] - g_n * x_averaged_depth[i, j] * dx_buffer[i, j] * dt
@inbounds nn1[i,j] = nn0[i, j] - g_n * y_averaged_depth[i, j] * dy_buffer[i, j] * dt
end
# boundary condition
for j in 1:ny, i in 1:nx
@inbounds mm1[i, j] = mm1[i, j] * land_filter_m[i, j] * absorbing_boundary[i, j]
@inbounds nn1[i, j] = nn1[i, j] * land_filter_n[i, j] * absorbing_boundary[i, j]
end
# diffs
for j in 1:ny
@inbounds dx_buffer[nx, j] = (-mm1[nx, j]) / dx
for i in 1:(nx-1)
@inbounds dx_buffer[i, j] = (mm1[i + 1, j] - mm1[i, j]) / dx
end
end
for i in 1:nx
@inbounds dy_buffer[i, ny] = (-nn1[i, ny]) / dy
for j in 1:(ny-1)
@inbounds dy_buffer[i, j] = (nn1[i,j + 1] - nn1[i, j]) / dy
end
end
# Update Wave Heigt
for j in 1:ny, i in 1:nx
@inbounds eta1[i, j] = eta0[i, j] - (dx_buffer[i, j] + dy_buffer[i, j]) * dt
end
# boundary condition
for j in 1:ny, i in 1:nx
@inbounds eta1[i, j] = eta1[i, j] * land_filter_e[i, j] * absorbing_boundary[i, j]
end
return eta1, mm1, nn1
end
function timestep!(dx_buffer::AbstractMatrix{T},
dy_buffer::AbstractMatrix{T},
eta1::AbstractMatrix{T},
mm1::AbstractMatrix{T},
nn1::AbstractMatrix{T},
eta0::AbstractMatrix{T},
mm0::AbstractMatrix{T},
nn0::AbstractMatrix{T},
matrices::Matrices{T},
dx::Real,dy::Real,dt::Real) where T
# Unpack the relevant fields of `matrices`
return timestep!(dx_buffer, dy_buffer, eta1, mm1, nn1, eta0, mm0, nn0,
matrices.absorbing_boundary, matrices.x_averaged_depth,
matrices.y_averaged_depth, matrices.land_filter_m,
matrices.land_filter_n, matrices.land_filter_e, dx, dy, dt)
end
function setup(nx::Int,
ny::Int,
bathymetry_val::Real,
absorber_thickness_fraction::Real,
apara::Real,
cutoff_depth::Real,
T::DataType = Float64)
# Memory allocation
ocean_depth = Matrix{T}(undef, nx, ny)
x_averaged_depth = Matrix{T}(undef, nx, ny)
y_averaged_depth = Matrix{T}(undef, nx, ny)
absorbing_boundary = ones(T, nx, ny)
land_filter_m = ones(T, nx, ny) # land filters
land_filter_n = ones(T, nx, ny) # "
land_filter_e = ones(T, nx, ny) # "
nxa = floor(Int, nx * absorber_thickness_fraction)
nya = floor(Int, nx * absorber_thickness_fraction)
# Bathymetry set-up. Users may need to modify it
fill!(ocean_depth, bathymetry_val)
@inbounds for j in 1:ny, i in 1:nx
if ocean_depth[i,j] < 0
ocean_depth[i,j] = 0
elseif ocean_depth[i,j] < cutoff_depth
ocean_depth[i,j] = cutoff_depth
end
end
# average bathymetry for staabsorbing_boundaryered-grid computation
for j in 1:ny
for i in 2:nx
x_averaged_depth[i, j] = (ocean_depth[i, j] + ocean_depth[i - 1, j]) / 2
if ocean_depth[i, j] <= 0 || ocean_depth[i - 1, j] <= 0
x_averaged_depth[i, j] = 0
end
end
x_averaged_depth[1, j] = ocean_depth[1, j]
end
for i in 1:nx
for j in 2:ny
y_averaged_depth[i, j] = (ocean_depth[i, j] + ocean_depth[i, j - 1]) / 2
if ocean_depth[i, j] <= 0 || ocean_depth[i, j - 1] <= 0
y_averaged_depth[i, j] = 0
end
end
y_averaged_depth[i, 1] = ocean_depth[i, 1]
end
# Land filter
@inbounds for j in 1:ny,i in 1:nx
(x_averaged_depth[i, j] < 0) && (land_filter_m[i, j] = 0)
(y_averaged_depth[i, j] < 0) && (land_filter_n[i, j] = 0)
(ocean_depth[i, j] < 0) && (land_filter_e[i, j] = 0)
end
# Sponge absorbing boundary condition by Cerjan (1985)
@inbounds for j in 1:ny, i in 1:nx
if i <= nxa
absorbing_boundary[i, j] *= exp(-((apara * (nxa - i)) ^ 2))
end
if i >= nx - nxa + 1
absorbing_boundary[i, j] *= exp(-((apara * (i - nx + nxa - 1)) ^ 2))
end
if j <= nya
absorbing_boundary[i, j] *= exp(-((apara * (nya - j)) ^ 2))
end
if j >= ny - nya + 1
absorbing_boundary[i, j] *= exp(-((apara * (j - ny + nya - 1)) ^ 2))
end
end
return Matrices(absorbing_boundary, ocean_depth, x_averaged_depth, y_averaged_depth,
land_filter_m, land_filter_n, land_filter_e)
end
# Calculates the initial surface height at point x,y
function get_initial_height(x::T, y::T, peak_x::T, peak_y::T, peak_height::T, cutoff::T) where T
dx = peak_x - x
dy = peak_y - y
distance_to_peak = sqrt(dx^2 + dy^2)
if distance_to_peak <= cutoff
return peak_height / 4.0 * ((1 + cospi(dx / cutoff)) * (1 + cospi(dy / cutoff)))
else
return 0.0
end
end
# Initializes the surface height on all points of the grid
function initheight!(height::AbstractMatrix{T},
ocean_depth::AbstractMatrix{T},
dx::Real,dy::Real,cutoff_distance::Real,
peak_height::Real, peak_position::AbstractVector{T}) where T
@assert size(height) == size(ocean_depth)
nx, ny = size(height)
x = (1:nx) .* dx
y = (1:ny) .* dy
height .= get_initial_height.(x', y, peak_position[1], peak_position[2], peak_height, cutoff_distance)
height[ocean_depth .< eps(T)] .= 0.0
end
# Initializes the surface height on all points of the grid
function initheight!(height::AbstractMatrix{T},
matrices::Matrices{T},
dx::Real,dy::Real,cutoff_distance::Real,
peak_height::Real, peak_position::AbstractVector{T}) where T
# Unpack the relevant field of `matrices`
initheight!(height, matrices.ocean_depth, dx, dy, cutoff_distance, peak_height, peak_position)
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | code | 6297 | module Lorenz63
using Base.Threads
using Distributions
using HDF5
using Random
using PDMats
using OrdinaryDiffEq
using ParticleDA
Base.@kwdef struct Lorenz63ModelParameters{S <: Real, T <: Real}
σ::S = 10.
ρ::S = 28.
β::S = 8. / 3.
time_step::S = 0.1
observed_indices::Union{UnitRange{Int}, StepRange{Int, Int}, Vector{Int}} = 1:3
initial_state_std::Union{S, Vector{S}} = 0.05
state_noise_std::Union{S, Vector{S}} = 0.05
observation_noise_std::Union{T, Vector{T}} = 2.
end
function get_params(
P::Type{Lorenz63ModelParameters{S, T}}, model_params_dict::Dict
) where {S <: Real, T <: Real}
return P(; (; (Symbol(k) => v for (k, v) in model_params_dict)...)...)
end
struct Lorenz63Model{S <: Real, T <: Real, I}
parameters::Lorenz63ModelParameters{S, T}
integrators::Vector{I}
initial_state_distribution::MvNormal{S}
state_noise_distribution::MvNormal{S}
observation_noise_distribution::MvNormal{T}
end
function update_time_derivative!(
du_dt::Vector{S}, u::Vector{S}, parameters::Lorenz63ModelParameters{S, T}, t::U
) where {S <: Real, T <: Real, U <: Real}
du_dt[1] = parameters.σ * (u[2] - u[1])
du_dt[2] = u[1] * (parameters.ρ - u[3]) - u[2]
du_dt[3] = u[1] * u[2] - parameters.β * u[3]
end
function init(
parameters_dict::Dict,
n_tasks::Int=1;
S::Type{<:Real}=Float64,
T::Type{<:Real}=Float64
)
parameters = get_params(Lorenz63ModelParameters{S, T}, parameters_dict)
time_span = (0, parameters.time_step)
integrators = [
OrdinaryDiffEq.init(
ODEProblem(update_time_derivative!, u, time_span, parameters),
Tsit5();
save_everystep=false
)
for u in eachcol(zeros(S, 3, n_tasks))
]
state_dimension = 3
observation_dimension = length(parameters.observed_indices)
return Lorenz63Model(
parameters,
integrators,
(
MvNormal(m, isa(s, Vector) ? PDiagMat(s.^2) : ScalMat(length(m), s.^2))
for (m, s) in (
(ones(S, state_dimension), parameters.initial_state_std),
(zeros(S, state_dimension), parameters.state_noise_std),
(zeros(T, observation_dimension), parameters.observation_noise_std),
)
)...
)
end
ParticleDA.get_state_dimension(::Lorenz63Model) = 3
ParticleDA.get_observation_dimension(model::Lorenz63Model) = length(
model.parameters.observed_indices
)
ParticleDA.get_state_eltype(::Lorenz63Model{S, T}) where {S, T} = S
ParticleDA.get_observation_eltype(::Lorenz63Model{S, T}) where {S, T} = T
function ParticleDA.sample_initial_state!(
state::AbstractVector{T},
model::Lorenz63Model{S, T},
rng::Random.AbstractRNG,
task_index::Integer=1
) where {S, T}
rand!(rng, model.initial_state_distribution, state)
end
function ParticleDA.update_state_deterministic!(
state::AbstractVector{T},
model::Lorenz63Model{S, T},
time_index::Integer,
task_index::Integer=1
) where {S, T}
reinit!(model.integrators[task_index], state)
step!(model.integrators[task_index], model.parameters.time_step, true)
state .= model.integrators[task_index].u
end
function ParticleDA.update_state_stochastic!(
state::AbstractVector{T},
model::Lorenz63Model{S, T},
rng::Random.AbstractRNG,
task_index::Integer=1
) where {S, T}
rand!(rng, state + model.state_noise_distribution, state)
end
function ParticleDA.sample_observation_given_state!(
observation::AbstractVector{T},
state::AbstractVector{S},
model::Lorenz63Model{S, T},
rng::Random.AbstractRNG,
task_index::Integer=1
) where {S <: Real, T <: Real}
rand!(
rng,
view(state, model.parameters.observed_indices)
+ model.observation_noise_distribution,
observation
)
end
function ParticleDA.get_log_density_observation_given_state(
observation::AbstractVector{T},
state::AbstractVector{S},
model::Lorenz63Model{S, T},
task_index::Integer=1
) where {S <: Real, T <: Real}
return logpdf(
view(state, model.parameters.observed_indices)
+ model.observation_noise_distribution,
observation
)
end
function ParticleDA.write_model_metadata(file::HDF5.File, model::Lorenz63Model)
group_name = "parameters"
if !haskey(file, group_name)
group = create_group(file, group_name)
for field in fieldnames(typeof(model.parameters))
value = getfield(model.parameters, field)
attributes(group)[string(field)] = (
isa(value, AbstractVector) ? collect(value) : value
)
end
else
@warn "Write failed, group $group_name already exists in $(file.filename)!"
end
end
function ParticleDA.get_observation_mean_given_state!(
observation_mean::AbstractVector{T},
state::AbstractVector{S},
model::Lorenz63Model{S, T},
task_index::Integer=1
) where {S <: Real, T <: Real}
observation_mean .= view(state, model.parameters.observed_indices)
end
function ParticleDA.get_covariance_state_noise(model::Lorenz63Model)
return model.state_noise_distribution.Σ
end
function ParticleDA.get_covariance_state_noise(model::Lorenz63Model, i::Int, j::Int)
return model.state_noise_distribution.Σ[i, j]
end
function ParticleDA.get_covariance_observation_noise(model::Lorenz63Model)
return model.observation_noise_distribution.Σ
end
function ParticleDA.get_covariance_observation_noise(
model::Lorenz63Model, i::Int, j::Int
)
return model.observation_noise_distribution.Σ[i, j]
end
function ParticleDA.get_covariance_state_observation_given_previous_state(
model::Lorenz63Model, i::Int, j::Int
)
return model.state_noise_distribution.Σ[i, model.parameters.observed_indices[j]]
end
function ParticleDA.get_covariance_observation_observation_given_previous_state(
model::Lorenz63Model, i::Int, j::Int
)
return (
model.state_noise_distribution.Σ[
model.parameters.observed_indices[i], model.parameters.observed_indices[j]
]
+ model.observation_noise_distribution.Σ[i, j]
)
end
function ParticleDA.get_state_indices_correlated_to_observations(model::Lorenz63Model)
return model.parameters.observed_indices
end
end
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | docs | 1926 | # ParticleDA
[](https://team-raddish.github.io/ParticleDA.jl/stable/)
[](https://team-raddish.github.io/ParticleDA.jl/dev/)
[](https://github.com/Team-RADDISH/ParticleDA.jl/actions/workflows/ci.yml?query=branch%3Amain)
[](https://codecov.io/gh/Team-RADDISH/ParticleDA.jl)
[](https://zenodo.org/badge/latestdoi/232626497)
[](https://doi.org/10.5194/gmd-2023-38)
`ParticleDA.jl` is a Julia package to perform data assimilation with particle filters,
supporting both thread-based parallelism and distributed processing using MPI.
This project is developed in collaboration with the
[Centre for Advanced Research Computing](https://ucl.ac.uk/arc), University College London.
## Installation
To install the latest stable version of the package, open the [Julia
REPL](https://docs.julialang.org/en/v1/stdlib/REPL/), enter the package manager
with `]`, then run the command
```
add ParticleDA
```
If you plan to develop the package (make changes, submit pull requests, etc), in
the package manager mode run this command
```
dev ParticleDA
```
This will automatically clone the repository to your local directory
`~/.julia/dev/ParticleDA`.
You can exit from the package manager mode by pressing `CTRL + C` or,
alternatively, the backspace key when there is no input in the prompt.
## Documentation
[Documentation Website](https://team-raddish.github.io/ParticleDA.jl/dev/)
## License
The `ParticleDA.jl` package is licensed under the MIT "Expat" License.
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | docs | 2430 | # Running benchmarks
We use [`PkgBenchmark.jl`](https://github.com/JuliaCI/PkgBenchmark.jl) and
[`BenchmarkCI.jl`](https://github.com/tkf/BenchmarkCI.jl) to run benchmarks for
this package. Benchmarks can be run in two different modes:
* only on the current branch;
* on the current branch and the default one, comparing the results between the
two to highlight performance improvements and regressions. This is
automatically run in all pull requests opened in the GitHub repository.
## Run benchmarks on the current branch
Before running the benchmarks, activate the environment of the `benchmark/`
directory with:
```julia
using Pkg, ParticleDA
Pkg.activate(joinpath(pkgdir(ParticleDA), "benchmark"))
Pkg.instantiate()
```
To run the benchmarks, execute the commands
```julia
using PkgBenchmark, ParticleDA
benchmarkpkg(ParticleDA)
```
See the docstring of
[`PkgBenchmark.benchmarkpkg`](https://juliaci.github.io/PkgBenchmark.jl/stable/run_benchmarks/#PkgBenchmark.benchmarkpkg)
for more details about its usage, but note you can pass a configuration like
```
benchmarkpkg(ParticleDA, BenchmarkConfig(; env = Dict("JULIA_NUM_THREADS" => 2)))
```
to specify the number of threads.
Remember you can go back to the top-level environment in Julia with
```julia
Pkg.activate()
```
or in the Pkg REPL mode with:
```julia
]activate
```
## Compare benchmarks on the current branch and the default one
In addition to activating the `benchmark/` environment as shown above:
```julia
using Pkg, ParticleDA
Pkg.activate(joinpath(pkgdir(ParticleDA), "benchmark"))
Pkg.instantiate()
```
you need to change the working directory to the root folder of `ParticleDA`:
```julia
cd(pkgdir(ParticleDA))
```
You can run the benchmarks with
```julia
using PkgBenchmark, BenchmarkCI
BenchmarkCI.judge() # run the benchmarks
BenchmarkCI.displayjudgement() # show the results in the terminal
```
Note that you can pass a
[`PkgBenchmark.BenchmarkConfig`](https://juliaci.github.io/PkgBenchmark.jl/stable/run_benchmarks/#PkgBenchmark.BenchmarkConfig)
also to
[`BenchmarkCI.judge`](https://tkf.github.io/BenchmarkCI.jl/dev/#BenchmarkCI.judge)
```julia
BenchmarkCI.judge(BenchmarkConfig(; env = Dict("JULIA_NUM_THREADS" => 2)))
```
Note: remember that you should not have uncommitted changes in the local
repository in order to run benchmarks with `BenchmarkCI`, because it needs to
automatically switch to the default branch.
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | docs | 10242 | # ParticleDA.jl
[`ParticleDA.jl`](https://github.com/Team-RADDISH/ParticleDA.jl) is a Julia
package to perform data assimilation with particle filters, distributed using MPI.
## Installation
To install the latest stable version of the package, open the [Julia
REPL](https://docs.julialang.org/en/v1/stdlib/REPL/), enter the package manager
with `]`, then run the command
```
add ParticleDA.jl
```
If you plan to develop the package (make changes, submit pull requests, etc), in
the package manager mode run this command
```
dev ParticleDA
```
This will automatically clone the repository to your local directory
`~/.julia/dev/ParticleDA`.
You can exit from the package manager mode by pressing `CTRL + C` or,
alternatively, the backspace key when there is no input in the prompt.
## Usage
After installing the package, you can start using it in Julia's REPL with
```julia
using ParticleDA
```
The run the particle filter you can use the function `run_particle_filter`:
```@docs
run_particle_filter
```
To simulate observations from the model (which can be used to for example test the
filtering algorithms) you can use the function `simulate_observations_from_model`
```@docs
simulate_observations_from_model
```
The `filter_type` argument to [`run_particle_filter`](@ref) should be a concrete subtype
of the `ParticleFilter` abstract type.
```@docs
ParticleDA.ParticleFilter
BootstrapFilter
OptimalFilter
```
The `summary_stat_type` argument to [`run_particle_filter`](@ref) should be a concrete
subtype of the `AbstractSummaryStat` abstract type.
```@docs
ParticleDA.AbstractSummaryStat
ParticleDA.AbstractCustomReductionSummaryStat
ParticleDA.AbstractSumReductionSummaryStat
ParticleDA.MeanAndVarSummaryStat
ParticleDA.MeanSummaryStat
ParticleDA.NaiveMeanAndVarSummaryStat
ParticleDA.NaiveMeanSummaryStat
```
The next section details how to write the interface between the model and the
particle filter.
### Interfacing the model
The model needs to define a custom data structure and a few functions, that will
be used by [`run_particle_filter`](@ref):
* a custom structure which holds the data about the model. This will be used to
dispatch the methods to be defined, listed below;
* an initialisation function with the following signature:
```julia
init(params_dict::Dict, n_tasks::Integer) -> model
```
with `params_dict` a dictionary with the parameters of the model and `n_tasks` an
integer specifying the maximum number tasks (coroutines) parallelisable operations
will be scheduled over. This initialisation function should create an instance of the
model data structure and return it. The value of `n_tasks` can be used to create
task-specific buffers for writing to when computing the model updates to avoid
reallocating data structures on each function call. As tasks may be run in parallel
over multiple threads, any buffers used in functions called within tasks should be
unique to the task; to facilitate this functions in the model interface (see below)
which may be called within tasks scheduled in parallel are passed a `task_index`
argument which is a integer index in `1:n_tasks` which is guaranteed to be unique to a
particular task and so can be used to index in to task specific buffers.
* The model needs to extend the following methods, using the model type for dispatch:
```@docs
ParticleDA.get_state_dimension
ParticleDA.get_observation_dimension
ParticleDA.get_state_eltype
ParticleDA.get_observation_eltype
ParticleDA.sample_initial_state!
ParticleDA.update_state_deterministic!
ParticleDA.update_state_stochastic!
ParticleDA.sample_observation_given_state!
ParticleDA.get_log_density_observation_given_state
ParticleDA.write_model_metadata
```
* Optionally, if the model has additive Gaussian observation and state noise, it may
also extend the following methods, again using the model type for dispatch, to allow
using the more statistically efficient `OptimalFilter` for filtering
```@docs
ParticleDA.get_observation_mean_given_state!
ParticleDA.get_covariance_state_noise
ParticleDA.get_covariance_observation_noise
ParticleDA.get_covariance_state_observation_given_previous_state
ParticleDA.get_covariance_observation_observation_given_previous_state
```
### Input parameters
You can store the input parameters in an YAML file with the following structure
```yaml
filter:
key1: value1
model:
model_name1:
key2: value2
key3: value3
model_name2:
key4: value4
key5: value5
```
The parameters under `filter` are related to the particle filter, under `model`
you can specify the parameters for different models.
The particle filter parameters are saved in the following data structure:
```@docs
ParticleDA.FilterParameters
```
## Example: estimating the state in a tsunami model
A full example of a model interfacing `ParticleDA` is available in `test/models/llw2d.jl`.
This model represents a simple two dimensional simulation of tsunami dynamics and is
partly based on the [tsunami data assimilation code](https://github.com/tktmyd/tdac) by
Takuto Maeda. The particle filter can be run with observations simulated from the model
as follows
```julia
# Load ParticleDA
using ParticleDA
# Save some variables for later use
test_dir = joinpath(dirname(pathof(ParticleDA)), "..", "test")
llw2d_src = joinpath(test_dir, "models", "llw2d.jl")
input_file = joinpath(test_dir, "integration_test_1.yaml")
observation_file = tempname()
# Instantiate the test environment
using Pkg
Pkg.activate(test_dir)
Pkg.instantiate()
# Include the sample model source code and load it
include(llw2d_src)
using .LLW2d
# Simulate observations from the model to use
simulate_observations_from_model(LLW2d.init, input_file, observation_file)
# Run the (optimal proposal) particle filter with simulated observations computing the
# mean and variance of the particle ensemble. On non-Intel architectures you may need
# to use NaiveMeanAndVarSummaryStat instead
final_states, final_statistics = run_particle_filter(
LLW2d.init, input_file, observation_file, OptimalFilter, MeanAndVarSummaryStat
)
```
### Parameters of the test model
The [Input parameters](@ref) section shows how to pass the input parameters for
the filter and the model. For the model included in the test suite, called
`llw2d`, you can set the following parameters:
```@docs
LLW2d.LLW2dModelParameters
```
### Observation station coordinates
The coordinates of the observation stations can be set in two different ways.
1. Provide the coordinates in an input file. Set the parameter `station_filename` to the name of your input file.
The input file is in plain text, the format is one row per station containing x, y - coordinates in metres. Here is
a simple example with two stations
```julia
# Comment lines starting with '#' will be ignored by the code
# This file contains two stations: at [1km, 1km] and [2km, 2km]
1.0e3, 1.0e3
2.0e3, 2.0e3
```
2. Provide parameters for an equispaced rectilinear grid of observation stations. The values of these parameters should then be set:
* `n_stations_x` : Number of observation stations in the x direction
* `n_stations_y` : Number of observation stations in the y direction
* `station_distance_x` : Distance between stations in the x direction (m)
* `station_distance_y` : Distance between stations in the y direction (m)
* `station_boundary_x` : Distance between bottom left edge of box and first station in the x direction (m)
* `station_boundary_y` : Distance between bottom left edge of box and first station in the y direction (m)
As an example, one could set
```julia
n_stations_x=2,
n_stations_y=2,
station_distance_x=1.0e3,
station_distance_y=1.0e3,
station_boundary_x=10.0e3,
station_boundary_y=10.0e3,
```
to generate 4 stations at `[10km, 10km]`, `[10km, 11km]`, `[11km, 10km]` and `[11km, 11km]`.
## Output
If the filter parameter `verbose` is set to `true`, [`run_particle_filter`](@ref) will produce an HDF5 file in the run directory. The file name is `particle_da.h5` by default (this is configurable using the `output_filename` filter parameter). The file contains the summary statistics of the estimate state distribution (by default the mean and variance), particle weights, parameters used, and other metadata at each time step observation were assimilated. To read the output file, use the [HDF5 library](https://www.hdfgroup.org/solutions/hdf5/).
A basic plotting tool is provided in a [Jupyter notebook](https://github.com/Team-RADDISH/ParticleDA.jl/blob/main/extra/Plot_tdac_output.ipynb). This is intended as a template to build more sophisticated postprocessing tools, but can be used for some simple analysis. Set the variable `timestamp` in the third cell to plot different time slices from the output file. More functionality may be added as the package develops.
## Running in parallel
The particle state update is parallelised using both MPI and threading. According to our preliminary tests both methods work well at small scale. To use the threading, set the environment variable `JULIA_NUM_THREADS` to the number of threads you want to use before starting Julia and then call the [`run_particle_filter`](@ref) function normally. You can check the number of threads julia has available by calling in Julia's REPL
```julia
Threads.nthreads()
```
To use the MPI parallelisation, write a Julia script that calls the [`run_particle_filter`](@ref) function for the relevant model and observations and run it in an Unix shell with
```bash
mpirun -np <your_number_of_processes> julia <your_julia_script>
```
Note that the parallel performance may vary depending on the performance of the algorithm. In general, a degeneracy of the particle weights will lead to poor load balance and parallel performance. See [this issue](https://github.com/Team-RADDISH/ParticleDA.jl/issues/115#issuecomment-675468511) for more details.
## Testing
We have a basic test suite for `ParticleDA.jl`. You can run the tests by entering the
package manager mode in Julia's REPL with `]` and running the command
```
test ParticleDA
```
## License
The `ParticleDA.jl` package is licensed under the MIT "Expat" License.
| ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | docs | 313 | # Weak Scaling
This directory contains a batch script and a julia script that can be used
to run a weak scaling experiment on a HPC system. This is an updated version
to what was used to run on CSD3 in the past (look for the old files in git
history if interested). So far only tested on a small number of ranks. | ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 1.1.0 | 445106e67dc21688691ed762446d393965e1d982 | docs | 338 | # List of tracked input files
## Alex May21
These inputs were developed by Alex Beskos in May21 during development of the Optimal Filter. They were found to give good performance of the filter in the test case with the TDAC model and the results have been shown in several talks in 2021.
### Files
- parametersW1.yaml
- stationsW1.txt | ParticleDA | https://github.com/Team-RADDISH/ParticleDA.jl.git |
|
[
"MIT"
] | 0.4.0 | d49e35f413186528f1d7cc675e67d0ed16fd7800 | code | 136 | module ScanByte
using Libdl: Libdl
using SIMD: SIMD
include("byteset.jl")
include("codegen.jl")
export ByteSet, memchr
end # module
| ScanByte | https://github.com/jakobnissen/ScanByte.jl.git |
|
[
"MIT"
] | 0.4.0 | d49e35f413186528f1d7cc675e67d0ed16fd7800 | code | 2189 | struct ByteSet <: AbstractSet{UInt8}
data::NTuple{4, UInt64}
ByteSet(x::NTuple{4, UInt64}) = new(x)
end
ByteSet() = ByteSet((UInt64(0), UInt64(0), UInt64(0), UInt64(0)))
Base.length(s::ByteSet) = mapreduce(count_ones, +, s.data)
Base.isempty(s::ByteSet) = s === ByteSet()
function ByteSet(it)
a = b = c = d = UInt64(0)
for i in it
vi = convert(UInt8, i)
if vi < 0x40
a |= UInt(1) << ((vi - 0x00) & 0x3f)
elseif vi < 0x80
b |= UInt(1) << ((vi - 0x40) & 0x3f)
elseif vi < 0xc0
c |= UInt(1) << ((vi - 0x80) & 0x3f)
else
d |= UInt(1) << ((vi - 0xc0) & 0x3f)
end
end
ByteSet((a, b, c, d))
end
function Base.minimum(s::ByteSet)
y = iterate(s)
y === nothing ? Base._empty_reduce_error() : first(y)
end
function Base.maximum(s::ByteSet)
offset = 0x03 * UInt8(64)
for i in 0:3
@inbounds bits = s.data[4 - i]
iszero(bits) && continue
return ((3-i)*64 + (64 - leading_zeros(bits)) - 1) % UInt8
end
Base._empty_reduce_error()
end
function Base.in(byte::UInt8, s::ByteSet)
i, o = divrem(byte, UInt8(64))
@inbounds !(iszero(s.data[i & 0x03 + 0x01] >>> (o & 0x3f) & UInt(1)))
end
@inline function Base.iterate(s::ByteSet, state=UInt(0))
ioffset, offset = divrem(state, UInt(64))
n = UInt(0)
while iszero(n)
ioffset > 3 && return nothing
n = s.data[ioffset + 1] >>> offset
offset *= !iszero(n)
ioffset += 1
end
tz = trailing_zeros(n)
result = (64 * (ioffset - 1) + offset + tz) % UInt8
(result, UInt(result) + UInt(1))
end
function Base.:~(s::ByteSet)
a, b, c, d = s.data
ByteSet((~a, ~b, ~c, ~d))
end
is_contiguous(s::ByteSet) = isempty(s) || (maximum(s) - minimum(s) + 1 == length(s))
Base.union(a::ByteSet, b::ByteSet) = ByteSet(map(|, a.data, b.data))
Base.intersect(a::ByteSet, b::ByteSet) = ByteSet(map(&, a.data, b.data))
Base.symdiff(a::ByteSet, b::ByteSet) = ByteSet(map(⊻, a.data, b.data))
Base.setdiff(a::ByteSet, b::ByteSet) = ByteSet(map((i,j) -> i & ~j, a.data, b.data))
Base.isdisjoint(a::ByteSet, b::ByteSet) = isempty(intersect(a, b))
| ScanByte | https://github.com/jakobnissen/ScanByte.jl.git |
|
[
"MIT"
] | 0.4.0 | d49e35f413186528f1d7cc675e67d0ed16fd7800 | code | 17539 | # The overall goal of the code in this file is to create code which can
# use SIMD to find the first byte in a bytevector not contained in a ByteSet.
# Due to historical reasons, this code finds the first byte that is NOT part
# of a byteset. To make it work, I simply invert the input byteset in the
# _gen_function_content method.
# The code proceeds in the following steps in a loop:
# * load 16 (SSSE3) or 32 (AVX2) bytes at a time to a SIMD vector V
# * Use zerovec_* function to zero out all bytes which are in the byteset
# * Use haszerolayout to check if all bytes are zero. If so, skip 16/32 bytes.
# * Else, use leading_zero_bytes to skip that amount ahead.
const v256 = SIMD.Vec{32, UInt8}
const v128 = SIMD.Vec{16, UInt8}
const BVec = Union{v128, v256}
const _ZERO_v256 = v256(ntuple(i -> VecElement{UInt8}(0x00), 32))
# Discover if the system CPU has SSSE or AVX2 instruction sets
let
llvmpath = if VERSION ≥ v"1.6.0-DEV.1429"
Base.libllvm_path()
else
only(filter(lib->occursin(r"LLVM\b", basename(lib)), Libdl.dllist()))
end
libllvm = Libdl.dlopen(llvmpath)
gethostcpufeatures = Libdl.dlsym(libllvm, :LLVMGetHostCPUFeatures)
features_cstring = ccall(gethostcpufeatures, Cstring, ())
features = split(unsafe_string(features_cstring), ',')
Libc.free(features_cstring)
# Need both SSE2 and SSSE3 to process v128 vectors.
@eval const SSSE3 = $(any(isequal("+ssse3"), features) & any(isequal("+sse2"), features))
@eval const AVX2 = $(any(isequal("+avx2"), features))
# Prefer 32-byte vectors because larger vectors = higher speed
@eval const DEFVEC = if get(ENV, "JULIA_CPU_TARGET", "native") != "native"
nothing
elseif AVX2
v256
elseif SSSE3
v128
else
nothing
end
end
"""
vpcmpeqb(a::BVec, b::BVec) -> BVec
Compare vectors `a` and `b` element wise and return a vector with `0x00`
where elements are not equal, and `0xff` where they are. Maps to the `vpcmpeqb`
AVX2 CPU instruction, or the `pcmpeqb` SSE2 instruction.
"""
function vpcmpeqb end
"""
vpshufb(a::BVec, b::BVec) -> BVec
Maps to the AVX2 `vpshufb` instruction or the SSSE3 `pshufb` instruction depending
on the width of the BVec.
"""
function vpshufb end
"""
vec_uge(a::BVec, b::BVec) -> BVec
Compare vectors `a` and `b` element wise and return a vector with `0xff`
where `a[i] ≥ b[i]``, and `0x00` otherwise. Implemented efficiently for CPUs
with the `vpcmpeqb` and `vpmaxub` instructions.
See also: [`vpcmpeqb`](@ref)
"""
function vec_uge end
# In this statement, define some functions for either 16-byte or 32-byte vectors
let
# icmp eq instruction yields bool (i1) values. We extend with sext to 0x00/0xff.
# since that's the native output of vcmpeqb instruction, LLVM will optimize it
# to just that.
vpcmpeqb_template = """%res = icmp eq <N x i8> %0, %1
%resb = sext <N x i1> %res to <N x i8>
ret <N x i8> %resb
"""
uge_template = """%res = icmp uge <N x i8> %0, %1
%resb = sext <N x i1> %res to <N x i8>
ret <N x i8> %resb
"""
for N in (16, 32)
T = NTuple{N, VecElement{UInt8}}
ST = SIMD.Vec{N, UInt8}
instruction_set = N == 16 ? "ssse3" : "avx2"
instruction_tail = N == 16 ? ".128" : ""
intrinsic = "llvm.x86.$(instruction_set).pshuf.b$(instruction_tail)"
vpcmpeqb_code = replace(vpcmpeqb_template, "<N x" => "<$(sizeof(T)) x")
@eval @inline function vpcmpeqb(a::$ST, b::$ST)
$(ST)(Base.llvmcall($vpcmpeqb_code, $T, Tuple{$T, $T}, a.data, b.data))
end
@eval @inline function vpshufb(a::$ST, b::$ST)
$(ST)(ccall($intrinsic, llvmcall, $T, ($T, $T), a.data, b.data))
end
@eval const $(Symbol("_SHIFT", string(8N))) = $(ST)(ntuple(i -> 0x01 << ((i-1)%8), $N))
@eval @inline bitshift_ones(shift::$ST) = vpshufb($(Symbol("_SHIFT", string(8N))), shift)
uge_code = replace(uge_template, "<N x" => "<$(sizeof(T)) x")
@eval @inline function vec_uge(a::$ST, b::$ST)
$(ST)(Base.llvmcall($uge_code, $T, Tuple{$T, $T}, a.data, b.data))
end
end
end
"""
haszerolayout(x::BVec) -> Bool
Test if the vector consists of all zeros.
"""
@inline haszerolayout(v::BVec) = iszero(mapreduce(i -> i.value, |, v.data))
"Count the number of 0x00 bytes in a vector"
@inline function leading_zero_bytes(v::v256)
# First compare to zero to get vector of 0xff where is zero, else 0x00
# Then use vpcmpeqb to extract top bits of each byte to a single UInt32,
# which is a bitvector, where the 1's were 0x00 in the original vector
# Then use trailing/leading ones to count the number
eqzero = vpcmpeqb(v, _ZERO_v256).data
packed = ccall("llvm.x86.avx2.pmovmskb", llvmcall, UInt32, (NTuple{32, VecElement{UInt8}},), eqzero)
@static if ENDIAN_BOM == 0x04030201
return trailing_ones(packed)
else
return leading_ones(packed)
end
end
# vpmovmskb requires AVX2, so we fall back to this.
@inline function leading_zero_bytes(v::v128)
lzbits = @static if ENDIAN_BOM == 0x04030201
trailing_zeros(reinterpret(UInt128, v))
else
leading_zeros(reinterpret(UInt128, v))
end
div(lzbits % UInt, 8)
end
@inline loadvector(::Type{T}, p::Ptr) where {T <: BVec} = unsafe_load(Ptr{T}(p))
# We have this as a separate function to keep the same constant mask in memory.
@inline shrl4(x) = x >>> 0x04
### ---- ZEROVEC_ FUNCTIONS
# The zerovec functions takes a single vector x of type BVec, and some more arguments
# which are supposed to be constant folded. The constant folded arguments are computed
# using a ByteSet. The resulting zerovec code will highly efficiently zero out the
# bytes which are contained in the original byteset.
# This is the generic fallback. For each of the 16 possible values of the lower 4
# bytes, we use vpshufb to get a byte B. That byte encodes which of the 8 values of
# H = bits 5-7 of the input bytes are not allowed. Let's say the byte is 0b11010011.
# Then values 3, 4, 6 are allowed. So we compute if B & (0x01 << H), that will be
# zero only if the byte is allowed.
# For the highest 8th bit, we exploit the fact that if that bit is set, vpshufb
# always returns 0x00. So either `upper` or `lower` will be 0x00, and we can or it
# together to get a combined table.
# See also http://0x80.pl/articles/simd-byte-lookup.html for explanation.
@inline function zerovec_generic(x::T, topzero::T, topone::T) where {T <: BVec}
lower = vpshufb(topzero, x)
upper = vpshufb(topone, x ⊻ 0b10000000)
bitmap = lower | upper
return bitmap & bitshift_ones(shrl4(x))
end
# Essentially like above, except in the special case of all accepted values being
# within 128 of each other, we can shift the acceptable values down in 0x00:0x7f,
# and then only use one table. if f == ~, and input top bit is set, the bitmap
# will be 0xff, and it will fail no matter the shift.
# By changing the offset and/or setting f == identity, this function can also cover
# cases where all REJECTED values are within 128 of each other.
@inline function zerovec_128(x::T, lut::T, offset::UInt8, f::Function) where {T <: BVec}
y = x - offset
bitmap = f(vpshufb(lut, y))
return bitmap & bitshift_ones(shrl4(y))
end
# If there are only 8 accepted elements, we use a vpshufb to get the bitmask
# of the accepted top 4 bits directly.
@inline function zerovec_8elem(x::T, lut1::T, lut2::T) where {T <: BVec}
# Get a 8-bit bitarray of the possible ones
mask = vpshufb(lut1, x & 0b00001111)
shifted = vpshufb(lut2, shrl4(x))
return vpcmpeqb(shifted, mask & shifted)
end
# One where it's a single range. After subtracting low, all accepted values end
# up in 0x00:len-1, and so a >= (aka. uge) check will zero out accepted bytes.
@inline function zerovec_range(x::BVec, low::UInt8, len::UInt8)
vec_uge((x - low), typeof(x)(len))
end
# One where, in all the disallowed values, the lower nibble is unique.
# This one is surprisingly common and very efficient.
# If all 0x80:0xff are allowed, the mask can be 0xff, and is compiled away
@inline function zerovec_inv_nibble(x::T, lut::T, mask::UInt8) where {T <: BVec}
# If upper bit is set, vpshufb yields 0x00. 0x00 is not equal to any bytes with the
# upper biset set, so the comparison will return 0x00, allowing it.
return vpcmpeqb(x, vpshufb(lut, x & mask))
end
# Same as above, but inverted. Even better!
@inline function zerovec_nibble(x::T, lut::T, mask::UInt8) where {T <: BVec}
return x ⊻ vpshufb(lut, x & mask)
end
# Simplest of all - and fastest!
@inline zerovec_same(x::BVec, y::UInt8) = x ⊻ y
function load_lut(::Type{T}, v::Vector{UInt8}) where {T <: BVec}
T === v256 && (v = repeat(v, 2))
return unsafe_load(Ptr{T}(pointer(v)))
end
# Compute the table (aka. LUT) used to generate the bitmap in zerovec_generic.
function generic_luts(::Type{T}, ::Val{byteset}, offset::UInt8, invert::Bool) where {
byteset, T <: BVec}
# If ascii, we set each allowed bit, but invert after vpshufb. Hence, if top bit
# is set, it returns 0x00 and is inverted to 0xff, guaranteeing failure
topzero = fill(invert ? 0xff : 0x00, 16)
topone = copy(topzero)
for byte in byteset
byte -= offset
# Lower 4 bits is used in vpshufb, so it's the index into the LUT
index = (byte & 0x0f) + 0x01
# Upper bit sets which of the two bitmaps we use.
bitmap = (byte & 0x80) == 0x80 ? topone : topzero
# Bits 5,6,7 from lowest control the shift. If, after a shift, the bit
# aligns with a zero, it's in the bitmask
shift = (byte >> 0x04) & 0x07
bitmap[index] ⊻= 0x01 << shift
end
return load_lut(T, topzero), load_lut(T, topone)
end
# Compute the LUT for use in zerovec_8elem.
function elem8_luts(::Type{T}, ::Val{byteset}) where {byteset, T <: BVec}
allowed_mask = fill(0xff, 16)
bitindices = fill(0x00, 16)
for (i, byte) in enumerate(byteset)
bitindex = 0x01 << (i - 1)
allowed_mask[(byte & 0x0f) + 0x01] ⊻= bitindex
bitindices[(byte >>> 0x04) + 0x01] ⊻= bitindex
end
return load_lut(T, allowed_mask), load_lut(T, bitindices)
end
# Compute LUT for zerovec_unique_nibble.
function unique_lut(::Type{T}, ::Val{byteset}, invert::Bool) where {byteset, T <: BVec}
# The default, unset value of the vector v must be one where v[x & 0x0f + 1] ⊻ x
# is never accidentally zero.
allowed = collect(0x01:0x10)
for byte in (invert ? ~byteset : byteset)
allowed[(byte & 0b00001111) + 1] = byte
end
return load_lut(T, allowed)
end
### ---- GEN_ZERO_ FUNCTIONS
# These will take a symbol sym and a byteset x. They will then compute all the
# relevant values for use in the zerovec_* functions, and produce code that
# will zero out the accepted bytes. Notably, the calculated values will all
# be compile-time constants, EXCEPT the symbol, which represents the input vector.
@inline @generated function zero_generic(x::BVec, ::Val{bs}) where bs
lut1, lut2 = generic_luts(x, Val{bs}(), 0x00, true)
:(zerovec_generic(x, $lut1, $lut2))
end
@inline @generated function zero_8elem(x::BVec, ::Val{bs}) where bs
lut1, lut2 = elem8_luts(x, Val{bs}())
:(zerovec_8elem(x, $lut1, $lut2))
end
@inline @generated function zero_128(x::BVec, ::Val{bs}, ::Val{ascii}, ::Val{inverted}) where {
bs, ascii, inverted
}
if ascii && !inverted
offset, f, invert = 0x00, ~, false
elseif ascii && inverted
offset, f, invert = 0x80, ~, false
elseif !ascii && !inverted
offset, f, invert = minimum(bs), ~, false
else
offset, f, invert = minimum(~bs), identity, true
end
(lut, _) = generic_luts(x, Val{bs}(), offset, invert)
:(zerovec_128(x, $lut, $offset, $f))
end
@inline @generated function zero_range(x::BVec, ::Val{bs}) where bs
:(zerovec_range(x, $(minimum(bs)), $(UInt8(length(bs)))))
end
@inline @generated function zero_inv_range(x::BVec, ::Val{bs}) where bs
# An inverted range is the same as a shifted range, because UInt8 arithmetic
# is circular. So we can simply adjust the shift, and return regular vec_range
:(zerovec_range(x, $(maximum(~bs) + 0x01), $(UInt8(length(bs)))))
end
@inline @generated function zero_nibble(x::BVec, ::Val{bs}, ::Val{invert}) where {bs, invert}
lut = unique_lut(x, Val{bs}(), invert)
mask = maximum(invert ? ~bs : bs) > 0x7f ? 0x0f : 0xff
if invert
:(zerovec_inv_nibble(x, $lut, $mask))
else
:(zerovec_nibble(x, $lut, $mask))
end
end
@inline @generated function zero_same(x::BVec, ::Val{bs}) where bs
:(zerovec_same(x, $(minimum(bs))))
end
### ----- GEN ZERO CODE
# This is the main function of this file. Given a BVec type T and a byteset B,
# it will produce the most optimal code which will zero out all bytes in input
# vector of type T, which are present in B. This function tests for the most
# efficient special cases, in order, until finally defaulting to generics.
@inline @generated function zero_code(x::BVec, valbs::Val{bs}) where bs
# These special cases are handled completely differently
# and dispatch should not reach here if this is the case
@assert !in(length(bs), (0, 255, 256))
if length(bs) == 1
:(zero_same(x, valbs))
elseif length(bs) == length(Set([i & 0x0f for i in bs]))
:(zero_nibble(x, valbs, Val(false)))
elseif length(~bs) == length(Set([i & 0x0f for i in ~bs]))
:(zero_nibble(x, valbs, Val(true)))
elseif is_contiguous(bs)
:(zero_range(x, valbs))
elseif is_contiguous(~bs)
:(zero_inv_range(x, valbs))
elseif minimum(bs) > 127
:(zero_128(x, valbs, $(Val(true)), $(Val(true))))
elseif maximum(bs) < 128
:(zero_128(x, valbs, $(Val(true)), $(Val(false))))
elseif maximum(~bs) - minimum(~bs) < 128
:(zero_128(x, valbs, $(Val(false)), $(Val(true))))
elseif maximum(bs) - minimum(bs) < 128
:(zero_128(x, valbs, $(Val(false)), $(Val(false))))
elseif length(bs) < 9
:(zero_8elem(x, valbs))
else
:(zero_generic(x, valbs))
end
end
"""
SizedMemory
This is an internal type, and is considered unstable.
Construct a `SizedMemory` from a string, or something that implements `pointer` and
`sizeof`. This struct simply wraps a pointer and a length and is completely unsafe.
Care must be taken to ensure the underlying memory isn't garbage collected or moved.
"""
struct SizedMemory
ptr::Ptr{UInt8}
len::UInt
end
Base.pointer(mem::SizedMemory) = mem.ptr
Base.length(mem::SizedMemory) = mem.len
Base.isempty(mem::SizedMemory) = iszero(length(mem))
SizedMemory(x) = SizedMemory(pointer(x), sizeof(x))
"""
memchr(x, bytes)
memchr(ptr::Ptr, len::UInt, bytes)
Return first position of any byte in `bytes`, in memory `mem`, or
`nothing` if no bytes were found
`bytes` can be a `Val{::ByteSet}`, in which case this function specializes
to the byteset, or a single `UInt8`, in which case it does not.
`x` can be any type that implements `pointer` and `sizeof`, or alternatively
a pointer and a memory length can be passed.
"""
function memchr end
@inline function _memchr_nonempty(::Type{T}, mem::SizedMemory, bs::Val) where {T <: BVec}
local zeroed
local nscanned
nscanned = zero(UInt)
while true
vector = loadvector(T, pointer(mem) + nscanned)
zeroed = zero_code(vector, bs)
haszerolayout(zeroed) || break
nscanned += sizeof(T)
nscanned > length(mem) && break
end
nscanned += leading_zero_bytes(zeroed) + 1
nscanned > length(mem) ? nothing : nscanned % Int
end
@inline function _memchr_nonempty(::Nothing, mem::SizedMemory, valbs::Val{byteset}) where byteset
for i in Base.OneTo(mem.len)
in(unsafe_load(mem.ptr + i - 1), byteset) || return i % Int
end
nothing
end
@inline @generated function _memchr(T, mem::SizedMemory, ::Val{byteset}) where byteset
byteset isa ByteSet || error("memchr must be passed a Val of a ByteSet instance")
len = length(byteset)
# Three special cases:
# 1) Empty byteset: The byte is never found and always returns nothing
# 2) Byteset is all bytes: Byte is found at pos 1 iff mem is not empty
# 3) One-element byteset: Just use the non-byteset memchr method
if iszero(len)
quote nothing end
elseif isone(len)
return :(memchr(mem, $(first(byteset))))
elseif len == 256
:(ifelse(isempty(mem), nothing, 1))
else
# We invert the byteset, because for historical reasons, this package
# was originally written to find the first byte NOT in a byteset
inverted = ~byteset
:(_memchr_nonempty(T, mem, Val{$(inverted)}()))
end
end
# Default method given a byteset, dispatches to default vector type
@inline memchr(mem::SizedMemory, valbs::Val) = _memchr(DEFVEC, mem, valbs)
# TODO: We might eventually replace this with a pure Julia function, but
# for now, libc's memchr is 2x faster.
@inline function memchr(mem::SizedMemory, byte::UInt8)
pos = @ccall memchr(pointer(mem)::Ptr{UInt8}, byte::Cint, length(mem)::Csize_t)::Ptr{Cchar}
pos == C_NULL ? nothing : ((pos - pointer(mem)) + 1) % Int
end
@inline function memchr(ptr::Ptr, len::UInt, byte_s::Union{UInt8, Val})
memchr(SizedMemory(Ptr{UInt8}(ptr), len), byte_s)
end
@inline function memchr(x, byte_s::Union{UInt8, Val})
GC.@preserve x memchr(SizedMemory(x), byte_s)
end
| ScanByte | https://github.com/jakobnissen/ScanByte.jl.git |
|
[
"MIT"
] | 0.4.0 | d49e35f413186528f1d7cc675e67d0ed16fd7800 | code | 2041 | function test_membership(members)
bs = ByteSet(members)
refset = Set{UInt8}([UInt8(i) for i in members])
@test refset == Set{UInt8}(collect(bs))
@test all(i -> in(i, bs), refset)
end
function test_inversion(bs)
inv = ~bs
all = true
for i in 0x00:0xff
all &= (in(i, bs) ⊻ in(i, inv))
end
@test all
end
@testset "Instantiation" begin
@test isempty(ByteSet())
@test iszero(length(ByteSet()))
for set in ["hello", "kdjy82zxxcbnpw", [0x00, 0x1a, 0xff, 0xf8, 0xd2]]
test_membership(set)
end
end
@testset "Min/max" begin
@test_throws ArgumentError maximum(ByteSet())
@test_throws ArgumentError minimum(ByteSet())
@test minimum(ByteSet("xylophone")) == UInt8('e')
@test maximum(ByteSet([0xa1, 0x0f, 0x4e, 0xf1, 0x40, 0x39])) == 0xf1
end
@testset "Contiguity" begin
@test ScanByte.is_contiguous(ByteSet(0x03:0x41))
@test ScanByte.is_contiguous(ByteSet())
@test ScanByte.is_contiguous(ByteSet(0x51))
@test ScanByte.is_contiguous(ByteSet(0xc1:0xd2))
@test ScanByte.is_contiguous(ByteSet(0x00:0xff))
@test !ScanByte.is_contiguous(ByteSet([0x12:0x3a; 0x3c:0x4a]))
@test !ScanByte.is_contiguous(ByteSet([0x01, 0x02, 0x04, 0x05]))
end
@testset "Inversion" begin
test_inversion(ByteSet())
test_inversion(ByteSet(0x00:0xff))
test_inversion(ByteSet([0x04, 0x06, 0x91, 0x92]))
test_inversion(ByteSet(0x54:0x71))
test_inversion(ByteSet(0x12:0x11))
test_inversion(ByteSet("abracadabra"))
end
@testset "Set operations" begin
sets = map(ByteSet, [
[],
[0x00:0xff;],
[0x00:0x02; 0x04; 0x19],
[0x01; 0x03; 0x09; 0xa1; 0xa1],
[0x41:0x8f; 0xd1:0xe1; 0xa0:0xf0],
[0x81:0x89; 0xd0:0xd0]
])
ssets = map(Set, sets)
for (s1, ss1) in zip(sets, ssets), (s2, ss2) in zip(sets, ssets)
for f in [union, intersect, symdiff, setdiff]
@test Set(f(s1, s2)) == f(ss1, ss2)
end
@test isdisjoint(s1, s2) == isdisjoint(ss1, ss2)
end
end
| ScanByte | https://github.com/jakobnissen/ScanByte.jl.git |
|
[
"MIT"
] | 0.4.0 | d49e35f413186528f1d7cc675e67d0ed16fd7800 | code | 71 | using ScanByte
using Test
include("byteset.jl")
include("scanbyte.jl") | ScanByte | https://github.com/jakobnissen/ScanByte.jl.git |
|
[
"MIT"
] | 0.4.0 | d49e35f413186528f1d7cc675e67d0ed16fd7800 | code | 4589 | SizedMemory = ScanByte.SizedMemory
@testset "SizedMemory" begin
for str in ["Hello, world!", "", "αβγ", " "]
mem = SizedMemory(str)
@test pointer(mem) == pointer(str)
@test length(mem) == ncodeunits(str)
arr = collect(codeunits(str))
mem = SizedMemory(arr)
@test pointer(mem) == pointer(arr)
@test length(mem) == length(arr)
end
for str in ["Hello, world!", "αβγδ", " "]
for start in (1, 3), len in (0, 3, 5)
substr = SubString(str, start:start + len - 1)
mem = SizedMemory(substr)
@test pointer(mem) == pointer(substr)
@test length(mem) == ncodeunits(substr)
end
end
end
@testset "memchr of byte" begin
@test memchr(SizedMemory([]), 0x01) === nothing
@test memchr(SizedMemory([1,2,3]), 0x02) === 9
@test memchr(SizedMemory([1,2,3]), 0x04) === nothing
for byte in [0x00, 0xa0, 0xda, 0xff]
@test memchr(SizedMemory(UInt8[]), byte) === nothing
@test memchr(UInt8[], byte) === nothing
not_bytes = [i for i in 0x00:0xff if i != byte]
for (len, ind) in [(2, 2), (20, 14), (500, 431)]
v = rand(not_bytes, len)
@test memchr(SizedMemory(v), byte) === nothing
@test memchr(v, byte) === nothing
v[ind] = byte
@test memchr(SizedMemory(v), byte) == ind
@test memchr(v, byte) == ind
end
end
# Test the memchr method with pointer
bytes = [1,2,3]
@test memchr(pointer(bytes), UInt(11), 0x02) === 9
@test memchr(pointer(bytes), UInt(11), 0x03) === nothing
end
@testset "Scanning" begin
# We test the inverted bytesets, because the codegen is built to look for
# the bytes its NOT scanning for. Just to make the test easier
for T in (ScanByte.v128, ScanByte.v256, nothing)
for inv_byteset in [
ByteSet(), # empty
ByteSet(0x00:0xff), # full
ByteSet(0x41), # one member
~ByteSet(0x6a), # all but one member
ByteSet([0xa2, 0xb4, 0xc8, 0xf9, 0xf6]), # unique lower nibble
~ByteSet([0x38, 0x40, 0x51, 0x79, 0x94]), # inv uniq nibble
ByteSet(0x49:0x71), # Contiguous
~ByteSet(0x02:0x31), # Inverted contiguous
ByteSet(rand(0x80:0xff, 20)), # 128 - 255
ByteSet(rand(0x00:0x7f, 20)), # 0 - 127
ByteSet(rand(0x5a:0x94, 20)), # within 127 of each other
~ByteSet(rand(0x5a:0x94, 20)), # inv within 127
ByteSet([0x38, 0x40, 0x90, 0xba, 0xc5, 0xc8, 0xe7]), # at most 8 elements
ByteSet([i for i in 0x00:0xff if rand(Bool)]) # fallback
]
byteset = ~inv_byteset
# Empty
@test ScanByte._memchr(T, SizedMemory(UInt8[]), Val(byteset)) === nothing
# 1000 bytes not in the set
if length(byteset) != 256
bytes = rand(collect(~byteset), 1000)
@test ScanByte._memchr(T, SizedMemory(bytes), Val(byteset)) === nothing
if !isempty(byteset)
bytes[500] = first(iterate(byteset))
@test ScanByte._memchr(T, SizedMemory(bytes), Val(byteset)) == 500
bytes[25] = first(iterate(byteset))
@test ScanByte._memchr(T, SizedMemory(bytes), Val(byteset)) == 25
end
resize!(bytes, 20)
@test ScanByte._memchr(T, SizedMemory(bytes), Val(byteset)) === nothing
end
# Short vector of bytes in the set
if !isempty(byteset)
bytes = rand(collect(byteset), 128)
@test ScanByte._memchr(T, SizedMemory(bytes), Val(byteset)) == 1
end
end
end
# Also test gen_scan_function directly (instead of _gen_scan_function)
byteset = ByteSet([0x02, 0x09, 0x11])
@test memchr(SizedMemory([1,2,3]), Val(byteset)) == 9
@test memchr(SizedMemory([1,3,4]), Val(byteset)) === nothing
@test memchr(SizedMemory([1,5,6,7,4,17,13]), Val(byteset)) == 41
# Test the memchr method with pointer
bytes = [1,5,6,7,4,17,13]
@test memchr(pointer(bytes), UInt(45), Val(byteset)) === 41
@test memchr(pointer(bytes), UInt(33), Val(byteset)) === nothing
# Test the memchr method with array/string directly
bytes = [1,5,6,7,4,17,13]
@test memchr(UInt8.(bytes), Val(byteset)) === 6
@test memchr(String(UInt8.(bytes)), Val(byteset)) === 6
@test memchr("\x03\x02", Val(byteset)) === 2
end
| ScanByte | https://github.com/jakobnissen/ScanByte.jl.git |
|
[
"MIT"
] | 0.4.0 | d49e35f413186528f1d7cc675e67d0ed16fd7800 | docs | 937 | ## Release 0.4.0
__Breaking changes__
* Breaking: `SizedMemory` is now internal
__Other_
_
* Now falls back to generic code if `ENV["JULIA_CPU_TARGET"]` is set.
* Use libc memchr when byteset length is one
* Add more set ops to `ByteSet`
## Release 0.3.2
* Optimise ByteSet
* Optimise `memchr(x, ::Byte)` by calling libc's memchr
* Add generic method `memchr(::Any, x)`, automatically creating `SizedMemory` from x
## Release 0.3.1
Fix issue #3 - SIMD fallback warning only emitted at package precompile time.
## Release 0.3
__Breaking changes__
* `gen_scan_function` has been removed. Instead, you should now scan for multiple bytes by calling `memchr(::SizedMemory, Val(byteset))`.
## Release 0.2
__Breaking changes__
* `SizedMemory(x)` now requires that `x` implements `sizeof`
__New features__
* New function `memchr(x::SizedMemory, b::UInt8)`, which finds the first position of `b` in `x`.
## Release 0.1
Initial release
| ScanByte | https://github.com/jakobnissen/ScanByte.jl.git |
|
[
"MIT"
] | 0.4.0 | d49e35f413186528f1d7cc675e67d0ed16fd7800 | docs | 3362 | # ScanByte.jl

[](https://codecov.io/gh/jakobnissen/ScanByte.jl)
_Find your bytes. Fast._
ScanByte is a package to solve a simple problem: Find the first occurrence of a byte or any byte from a set of bytes in a chunk of memory. Think of it like a much faster version of `findfirst` that only iterates over bytes in memory.
ScanByte is micro-optimized for speed. On my laptop it can hit the RAM bandwidth limit of around 20 GB/s. This speed makes it a suitable building block for string search engines, Regex implementations, parsers and similar use cases.
### Usage
The central function of interest in this package is `memchr`.
The function takes a chunk of memory and some bytes, and returns the first position (1-indexed) in the chunk of memory where any byte from the byte set is found, or `nothing` if no bytes are found.
The chunk of memory can be any type which implements methods for `pointer` and `sizeof`, or alternatively you can input a raw pointer and a size.
The byte set can be passed in in two different ways:
* As a single `UInt8`, in which case ScanByte will simply dispatch to libc's memchr function
* A `Val{bs}`, where `bs` is an instance of the type `ByteSet <: AbstractSet{UInt8}` from this package.
In the latter case, ScanByte will, at compile time, pick an efficient SIMD algorithm based on the content of the byteset.
Currently ScanByte only has SIMD algorithms for SSSE3 and AVX2 instruction sets (found in all x86-based PCs), and uses a slow fallback for CPUs without these instructions.
### Example usage
In all these examples, the input data can be a `String`, a `codeunits` object, an `Array{UInt8}`, or a pointer+length.
Any type that implements `pointer` and `sizeof` will do.
Search for a single byte:
```julia
julia> memchr("Julia", UInt8('i'))
4
julia> memchr(codeunits("Julia"), UInt8('z')) === nothing
true
julia> str = "Julia";
julia> GC.@preserve str memchr(pointer(str), sizeof(str) % UInt, UInt8('i'))
4
```
Search for a byteset. Here, `Val` must be used to force specialization on the byteset:
```julia
julia> bs = ByteSet([0x01, 0x6a, 0xf1]);
julia> memchr([0x4a, 0xf1], Val(bs))
3
```
Search using a function. To do this, you must construct a `ByteSet` using the predicate on `0x00:0xff`:
```julia
julia> f(x) = in(x, 0x1a:0x4c) || in(x, 0xd1:0xf1); # some function
julia> bs = ByteSet(filter(f, 0x00:0xff));
julia> memchr("hello, Bob", Val(bs))
6
```
## Drawbacks
At the moment, ScanByte has three major drawbacks:
* If you are search for a predicate/byteset and not a single byte, tt relies on generated functions to compute the optimal Julia code to create the scanning function. This means the byte set must be known at compile time.
* It relies on explicit SIMD instructions. To be fast, it must run on computers with ideally the `AVX2` instruction set, or with the `SSE2` and `SSSE3` sets. Also, if you create the scanning function on a computer with `AVX2` but runs it on a computer without, LLVM will probably crash. Currently, the fallback methods are fairly slow.
* There is no guaranteed stable version of detecting which SIMD instructions your Julia supports. So this package tries to guess by parsing some output from LLVM.
| ScanByte | https://github.com/jakobnissen/ScanByte.jl.git |
|
[
"MIT"
] | 0.1.0 | 5eca57672b04c93780e07a3363fbbac6fc822324 | code | 439 | module AltInplaceOpsInterface
add!(a::A, b::B) where {A,B} = error("add!(a::$A, b::$B) is not implemented")
minus!(a::A, b::B) where {A,B} = error("minus!(a::$A, b::$B) is not implemented")
pow!(a::A, b::B) where {A,B} = error("pow!(a::$A, b::$B) is not implemented")
max!(a::A, b::B) where {A,B} = error("max!(a::$A, b::$B) is not implemented")
min!(a::A, b::B) where {A,B} = error("min!(a::$A, b::$B) is not implemented")
end # module
| AltInplaceOpsInterface | https://github.com/baxmittens/AltInplaceOpsInterface.jl.git |
|
[
"MIT"
] | 0.1.0 | 5eca57672b04c93780e07a3363fbbac6fc822324 | code | 46 | using Test
@testset begin
@test 1 == 1
end | AltInplaceOpsInterface | https://github.com/baxmittens/AltInplaceOpsInterface.jl.git |
|
[
"MIT"
] | 0.1.0 | 5eca57672b04c93780e07a3363fbbac6fc822324 | docs | 108 | # AltInplaceOpsInterface.jl
This is a package for interfacing some inplace operations due to old julia code
| AltInplaceOpsInterface | https://github.com/baxmittens/AltInplaceOpsInterface.jl.git |
|
[
"MIT"
] | 0.1.2 | cd110f79356a53100ea1ca281eceb7b7fe39462c | code | 391 | import Pkg; Pkg.activate("."); Pkg.instantiate()
using Documenter
#include("../src/Pitches.jl")
push!(LOAD_PATH, "../src/")
using Pitches
makedocs(
sitename="Pitches.jl",
pages = [
"index.md",
"tutorial.md",
"interface.md",
"Pitch and Interval Types" => [
"spelled.md",
"midi.md",
"logfreq.md",
]
]
)
| Pitches | https://github.com/DCMLab/Pitches.jl.git |
|
[
"MIT"
] | 0.1.2 | cd110f79356a53100ea1ca281eceb7b7fe39462c | code | 105 | include("make.jl")
deploydocs(
repo = "github.com/DCMLab/Pitches.jl.git",
devbranch = "main",
)
| Pitches | https://github.com/DCMLab/Pitches.jl.git |
|
[
"MIT"
] | 0.1.2 | cd110f79356a53100ea1ca281eceb7b7fe39462c | code | 4365 | module Pitches
import Base: show, +, -, *, convert, zero, isless, isequal, isapprox
export Interval, IntervalClass, Pitch
export octave, unison, ic, isstep, chromsemi, embed
export intervaltype, intervalclasstype
export topitch, tointerval, pc
# Pitch: basic Types and Interfaces
# =================================
"""
abstract type Interval end
Any interval type should be a subtype of `Interval`.
Intervals should implement the following operations as far as possible:
- `ic`
- `octave(T)`
- `isstep`
- `chromsemi(T)`
- `intervalclasstype(T)`
- `Base.+`
- `Base.-` (negation and substraction)
- `Base.*` (with integers, both sides)
- `Base.zero(T)`
- `Base.sign`
Where `(T)` marks operations on the type itself.
"""
abstract type Interval end
"""
abstract type IntervalClass <: Interval end
Any interval class type should be a subtype of `IntervalClass`.
In addition to the methods on intervals, interval classes should implement:
- `embed`
- `intervaltype(T)`
`intervalclasstype(T)` and `ic` should be identities.
"""
abstract type IntervalClass <: Interval end
# interfaces
# ----------
"""
unison(T)
Returns the interval of a unison for interval type `T`.
Alias for `Base.zero`.
New interval types should implement `Base.zero`,
but user code should call `unison` for better interpretability.
"""
const unison = Base.zero
"""
octave(T, [n=1])
Returns the interval corresponding to an octave for interval type `T`.
For interval classes, this should return `zero(T)`
(a default method is provided).
If `n` is specified, the octave is multiplied by `n` first.
This is equivalent to `octave(T) * n`.
For convenience, a fallback for `octave(p::T, [n])` is provided.
Only `octave(T)` needs to be implemented.
"""
function octave end
octave(T::Type{PC}) where {PC<:IntervalClass} = zero(T)
octave(T, n::Int) = octave(T) * n
octave(p::Interval) = octave(typeof(p))
"""
ic(i)
Returns the interval class of an interval, removing the octave
"""
function ic end
"""
embed(ic, [oct=0])
embed(pc, [oct=0])
Converts an interval class to an interval in the canonical octave,
adding `oct` octaves, if supplied.
Also works for pitches.
"""
function embed end
embed(ic, oct) = embed(ic) + octave(intervaltype(typeof(ic)), oct)
"""
intervaltype(IC::Type)
Returns for an interval class type `IC` the corresponding interval type.
For convenience, `intervaltype(ic::IC)` is also provided.
"""
function intervaltype end
intervaltype(::Any) = nothing
intervaltype(::IC) where {IC<:IntervalClass} = intervaltype(IC)
"""
intervalclasstype(I::Type)
Returns for an interval type `I` the corresponding interval class type.
For convenience, `intervalclasstype(p::P)` is also provided.
"""
function intervalclasstype end
intervalclasstype(::Any) = nothing
intervalclasstype(::I) where {I<:Interval} = intervalclasstype(I)
"""
isstep(p)
For diatonic intervals, indicates whether `p` is a step.
"""
function isstep end
"""
chromsemi(I::Type)
Returns a chromatic semitone of type `I`.
"""
function chromsemi end
# pitches
# =======
"""
Pitch{I}
Represents a pitch for the interval type `I`.
The interval is interpreted as an absolute pitch
by assuming a reference pitch.
The reference pitch is type dependent and known from context.
"""
struct Pitch{I<:Interval}
pitch :: I
end
topitch(i::I) where {I<:Interval} = Pitch(i)
tointerval(p::Pitch{I}) where {I<:Interval} = p.pitch
+(p::Pitch{I}, i::I) where {I<:Interval} = Pitch(p.pitch + i)
+(i::I, p::Pitch{I}) where {I<:Interval} = Pitch(p.pitch + i)
-(p::Pitch{I}, i::I) where {I<:Interval} = Pitch(p.pitch - i)
-(p1::Pitch{I}, p2::Pitch{I}) where {I<:Interval} = p1.pitch - p2.pitch
"""
pc(p)
Return the pitch class that corresponds to `p`.
"""
pc(p::Pitch{I}) where {I<:Interval} = Pitch(ic(p.pitch))
embed(p::Pitch{I}, octs::Int=0) where {I<:Interval} = Pitch(embed(p.pitch, octs))
Base.isequal(p1::Pitch{I}, p2::Pitch{I}) where {I<:Interval} = p1.pitch == p2.pitch
Base.isapprox(p1::Pitch, p2::Pitch; kwargs...) =
Base.isapprox(p1.pitch, p2.pitch; kwargs...)
Base.isless(p1::Pitch{I}, p2::Pitch{I}) where {I<:Interval} = p1.pitch < p2.pitch
Base.hash(p::Pitch, x::UInt) = hash(p.pitch, x)
# specific interval types
# =======================
include("pitches/midi.jl")
include("pitches/spelled.jl")
include("pitches/logfreq.jl")
end # module
| Pitches | https://github.com/DCMLab/Pitches.jl.git |
|
[
"MIT"
] | 0.1.2 | cd110f79356a53100ea1ca281eceb7b7fe39462c | code | 4921 | export FreqInterval, logfreqi, logfreqp, freqi, freqp
export FreqIC, logfreqic, logfreqpc, freqic, freqpc
import Base: +, -, *, ==
# some helper constants
const oct = log(2)
const halfoct = oct/2
# FreqInterval
# ============
"""
FreqInterval <: Interval
A frequency ratio interval.
Is internally represented by the logarithm of the frequency ratio,
so conversion to Float64 will return a log-frequency ratio!
"""
struct FreqInterval <: Interval
lfr :: Float64
end
# constructors
"""
logfreqi(logratio)
Creates a frequency ratio interval from a log-ratio.
"""
logfreqi(lfr) = FreqInterval(lfr)
"""
logfreqp(logfrequency)
Creates a frequency pitch from a log-frequency.
"""
logfreqp(lf) = Pitch(FreqInterval(lf))
"""
freqi(ratio)
Creates a frequency ratio interval from a frequency ratio.
"""
freqi(ratio) = FreqInterval(log(ratio))
"""
freqp(frequency)
Creates a frequency pitch from a frequency.
"""
freqp(freq) = Pitch(FreqInterval(log(freq)))
# Base interface
Base.show(io::IO, i::FreqInterval) = print(io, string("fr", exp(i.lfr)))
Base.show(io::IO, p::Pitch{FreqInterval}) = print(io, string(exp(p.pitch.lfr), "Hz"))
Base.isless(i1::FreqInterval, i2::FreqInterval) =
i1.lfr < i2.lfr
Base.isequal(i1::FreqInterval, i2::FreqInterval) =
i1.lfr == i2.lfr
Base.isapprox(a::FreqInterval, b::FreqInterval; kwargs...) =
Base.isapprox(a.lfr, b.lfr; kwargs...)
Base.hash(i::FreqInterval, x::UInt64) = hash(i.lfr, x)
Base.Float64(i::FreqInterval) = i.lfr
Base.Float64(p::Pitch{FreqInterval}) = p.pitch.lfr
convert(::Type{FreqInterval}, x::N) where {N<:Real} = logfreqi(Float64(x))
convert(::Type{N}, i::FreqInterval) where {N<:Real} = convert(N, i.lfr)
# TODO: conversion to other pitch types, in particular tomidi
# interval interface
+(i1::FreqInterval, i2::FreqInterval) = logfreqi(i1.lfr + i2.lfr)
-(i1::FreqInterval, i2::FreqInterval) = logfreqi(i1.lfr - i2.lfr)
-(i::FreqInterval) = logfreqi(-i.lfr)
*(i::FreqInterval, n::Integer) = logfreqi(n*i.lfr)
*(n::Integer, i::FreqInterval) = logfreqi(n*i.lfr)
Base.sign(i::FreqInterval) = sign(i.lfr)
Base.abs(i::FreqInterval) = logfreqi(abs(i.lfr))
Base.zero(::Type{FreqInterval}) = logfreqi(0.)
Base.zero(::FreqInterval) = logfreqi(0.)
octave(::Type{FreqInterval}) = logfreqi(oct)
chromsemi(::Type{FreqInterval}) = logfreqi(oct/12)
# isstep is left unimplemented
ic(i::FreqInterval) = logfreqic(i.lfr)
embed(i::FreqInterval) = i
intervaltype(::Type{FreqInterval}) = FreqInterval
intervalclasstype(::Type{FreqInterval}) = FreqIC
# FreqIC
# ======
"""
FreqIC <: Interval
A frequency ratio interval class.
Is internally represented by the logarithm of the frequency ratio,
so conversion to Float64 will return a log-frequency ratio!
Maintains octave equivalence.
"""
struct FreqIC <: IntervalClass
lfr :: Float64
end
# constructors
"""
logfreqic(logratio)
Creates a frequency ratio interval class from a log-ratio.
"""
logfreqic(lfr) = FreqIC(mod(lfr, oct))
"""
logfreqpc(logfrequency)
Creates a frequency pitch class from a log-frequency.
"""
logfreqpc(lf) = Pitch(logfreqic(lf))
"""
freqic(ratio)
Creates a frequency ratio interval class from a frequency ratio.
"""
freqic(ratio) = logfreqic(log(ratio))
"""
freqpc(frequency)
Creates a frequency pitch class from a frequency.
"""
freqpc(freq) = Pitch(freqic(freq))
# Base interface
Base.show(io::IO, i::FreqIC) = print(io, string("fr[", exp(i.lfr), "]"))
Base.show(io::IO, p::Pitch{FreqIC}) = print(io, string("[", exp(p.pitch.lfr), "]Hz"))
Base.isless(i1::FreqIC, i2::FreqIC) =
i1.lfr < i2.lfr
Base.isequal(i1::FreqIC, i2::FreqIC) =
i1.lfr == i2.lfr
Base.isapprox(a::FreqIC, b::FreqIC; kwargs...) =
Base.isapprox(a.lfr, b.lfr; kwargs...) ||
Base.isapprox(a.lfr + log(2), b.lfr; kwargs...) ||
Base.isapprox(a.lfr, b.lfr + log(2); kwargs...)
Base.hash(i::FreqIC, x::UInt64) = hash(i.lfr, x)
Base.Float64(i::FreqIC) = i.lfr
Base.Float64(p::Pitch{FreqIC}) = p.pitch.lfr
convert(::Type{FreqIC}, x::N) where {N<:Real} = logfreqic(Float64(x))
convert(::Type{N}, i::FreqIC) where {N<:Real} = convert(N, i.lfr)
# TODO: conversion to other pitch types, in particular tomidi
# interval interface
+(i1::FreqIC, i2::FreqIC) = logfreqic(i1.lfr + i2.lfr)
-(i1::FreqIC, i2::FreqIC) = logfreqic(i1.lfr - i2.lfr)
-(i::FreqIC) = logfreqic(-i.lfr)
*(i::FreqIC, n::Integer) = logfreqic(n*i.lfr)
*(n::Integer, i::FreqIC) = logfreqic(n*i.lfr)
Base.sign(i::FreqIC) = i.lfr == 0 ? 0 : -sign(i.lfr - halfoct)
Base.abs(i::FreqIC) = logfreqic(abs(mod(i.lfr + halfoct, oct) - halfoct))
Base.zero(::Type{FreqIC}) = logfreqic(0)
Base.zero(::FreqIC) = logfreqic(0)
octave(::Type{FreqIC}) = logfreqic(0)
chromsemi(::Type{FreqIC}) = logfreqic(oct/12)
# isstep is left unimplemented
ic(i::FreqIC) = i
embed(i::FreqIC) = logfreqi(i.lfr)
intervaltype(::Type{FreqIC}) = FreqInterval
intervalclasstype(::Type{FreqIC}) = FreqIC
| Pitches | https://github.com/DCMLab/Pitches.jl.git |
|
[
"MIT"
] | 0.1.2 | cd110f79356a53100ea1ca281eceb7b7fe39462c | code | 6120 | # midi intervals and classes
# ==========================
export MidiInterval, midi, midis, @midi, midip, midips, @midip
export MidiIC, midic, midics, @midic, midipc, midipcs, @midipc
export tomidi
# special interface
"""
tomidi(i [, ...])
tomidi(p [, ...])
Converts a pitch or interval to the corresponding midi type.
Depending on the input type, this might require additional parameters.
"""
function tomidi end
# types
"""
MidiInterval <: Interval
Intervals represented as chromatic integers.
`60` is Middle C.
"""
struct MidiInterval <: Interval
interval :: Int
end
"""
MidiIC <: IntervalClass
Interval classes represented as cromatic integers in Z_12, where `0` is C.
"""
struct MidiIC <: IntervalClass
ic :: Int
MidiIC(ic) = new(mod(ic,12))
end
# constructors
"""
midi(interval)
Creates a `MidiInterval` from an integer.
"""
midi(interval::Int) = MidiInterval(interval)
"""
midip(n)
Creates a midi pitch (`Pitch{MidiInterval}`) from an integer.
"""
midip(n::Int) = Pitch(midi(n))
"""
midic(interval)
Creates a `MidiIC` from an integer.
"""
midic(interval::Int) = MidiIC(interval)
"""
midipc(n)
Creates a midi pitch class (`Pitch{MidiIC}`) from an integer.
"""
midipc(n::Int) = Pitch(midic(n))
# constructor macros
"""
@midi expr
Replaces all `Int`s in `expr` with a call to `midi(::Int)`.
This allows the user to write integers where midi intervals are required.
Does not work when `expr` contains integers that should not be converted
or intervals that are not written as literal integers.
"""
macro midi(expr)
mkmidi(x) = x
mkmidi(e::Expr) = Expr(e.head, map(mkmidi, e.args)...)
mkmidi(n::Int) = :(Pitches.midi($n))
return esc(mkmidi(expr))
end
"""
@midic expr
Replaces all `Int`s in `expr` with a call to `midi(::Int)`.
This allows the user to write integers where midi intervals are required.
Does not work when `expr` contains integers that should not be converted
or intervals that are not written as literal integers.
"""
macro midic(expr)
mkmidi(x) = x
mkmidi(e::Expr) = Expr(e.head, map(mkmidi, e.args)...)
mkmidi(n::Int) = :(Pitches.midic($n))
return esc(mkmidi(expr))
end
"""
@midip expr
Replaces all `Int`s in `expr` with a call to `midip(::Int)`.
This allows the user to write integers where midi intervals are required.
Does not work when `expr` contains integers that should not be converted
or intervals that are not written as literal integers.
"""
macro midip(expr)
mkmidi(x) = x
mkmidi(e::Expr) = Expr(e.head, map(mkmidi, e.args)...)
mkmidi(n::Int) = :(Pitches.midip($n))
return esc(mkmidi(expr))
end
"""
@midipc expr
Replaces all `Int`s in `expr` with a call to `midipc(::Int)`.
This allows the user to write integers where midi intervals are required.
Does not work when `expr` contains integers that should not be converted
or intervals that are not written as literal integers.
"""
macro midipc(expr)
mkmidi(x) = x
mkmidi(e::Expr) = Expr(e.head, map(mkmidi, e.args)...)
mkmidi(n::Int) = :(Pitches.midipc($n))
return esc(mkmidi(expr))
end
# Base interface
show(io::IO, p::MidiInterval) = print(io, string("i", p.interval))
show(io::IO, p::MidiIC) = print(io, string("ic", p.ic))
show(io::IO, p::Pitch{MidiInterval}) = print(io, string("p", p.pitch.interval))
show(io::IO, p::Pitch{MidiIC}) = print(io, string("pc", p.pitch.ic))
Base.isless(p1::MidiInterval, p2::MidiInterval) = isless(p1.interval, p2.interval)
Base.isless(p1::MidiIC, p2::MidiIC) = isless(p1.ic, p2.ic)
Base.isequal(p1::MidiInterval, p2::MidiInterval) = p1.interval == p2.interval
Base.isequal(p1::MidiIC, p2::MidiIC) = p1.ic == p2.ic
Base.hash(p::MidiInterval, x::UInt) = hash(p.interval, x)
Base.hash(p::MidiIC, x::UInt) = hash(p.ic, x)
Base.Int64(p::MidiInterval) = p.interval
Base.Int64(p::MidiIC) = p.ic
Base.Int64(p::Pitch{MidiInterval}) = p.pitch.interval
Base.Int64(p::Pitch{MidiIC}) = p.pitch.ic
# conversion
convert(::Type{MidiInterval}, x::N) where {N<:Number} = midi(convert(Int, x))
convert(::Type{Interval}, x::N) where {N<:Number} = midi(convert(Int, x))
convert(::Type{Int}, p::MidiInterval) = p.interval
convert(::Type{N}, p::MidiInterval) where {N<:Number} = convert(N, p.interval)
convert(::Type{MidiIC}, x::N) where {N<:Number} = midic(convert(Int, x))
convert(::Type{IntervalClass}, x::N) where {N<:Number} = midic(convert(Int, x))
convert(::Type{Int}, p::MidiIC) = p.ic
convert(::Type{N}, p::MidiIC) where {N<:Number} = convert(N, p.ic)
## tomidi (identities)
tomidi(i::MidiInterval) = i
tomidi(i::MidiIC) = i
tomidi(p::Pitch{MidiInterval}) = p
tomidi(p::Pitch{MidiIC}) = p
# interval interface (midi interval)
+(p1::MidiInterval, p2::MidiInterval) = midi(p1.interval + p2.interval)
-(p1::MidiInterval, p2::MidiInterval) = midi(p1.interval - p2.interval)
-(p::MidiInterval) = midi(-p.interval)
zero(::Type{MidiInterval}) = midi(0)
zero(::MidiInterval) = midi(0)
*(p::MidiInterval, n::Integer) = midi(p.interval*n)
*(n::Integer, p::MidiInterval) = midi(p.interval*n)
octave(::Type{MidiInterval}) = midi(12)
Base.sign(p::MidiInterval) = sign(p.interval)
Base.abs(p::MidiInterval) = midi(abs(p.interval))
ic(p::MidiInterval) = midic(p.interval)
embed(p::MidiInterval) = p
intervaltype(::Type{MidiInterval}) = MidiInterval
intervalclasstype(::Type{MidiInterval}) = MidiIC
isstep(p::MidiInterval) = abs(p.interval) <= 2
chromsemi(::Type{MidiInterval}) = midi(1)
# interval interface (midi interval class)
+(p1::MidiIC, p2::MidiIC) = midic(p1.ic + p2.ic)
-(p1::MidiIC, p2::MidiIC) = midic(p1.ic - p2.ic)
-(p::MidiIC) = midic(-p.ic)
zero(::Type{MidiIC}) = midic(0)
zero(::MidiIC) = midic(0)
*(p::MidiIC, n::Integer) = midic(p.ic*n)
*(n::Integer, p::MidiIC) = midic(p.ic*n)
octave(::Type{MidiIC}) = midic(0)
Base.sign(p::MidiIC) = p.ic == 0 ? 0 : -sign(p.ic-6)
Base.abs(p::MidiIC) = midic(abs(mod(p.ic + 6, 12) - 6))
ic(p::MidiIC) = p
embed(p::MidiIC) = midi(p.ic)
intervaltype(::Type{MidiIC}) = MidiInterval
intervalclasstype(::Type{MidiIC}) = MidiIC
isstep(p::MidiIC) = p.ic <= 2 || p.ic >= 10
chromsemi(::Type{MidiIC}) = midic(1)
| Pitches | https://github.com/DCMLab/Pitches.jl.git |
|
[
"MIT"
] | 0.1.2 | cd110f79356a53100ea1ca281eceb7b7fe39462c | code | 11846 | export SpelledInterval, spelled, spelledp
export SpelledIC, sic, spc
export octaves, internalocts, fifths
export degree, generic, diasteps, alteration, letter
export parsespelled, parsespelledpitch, @i_str, @p_str
import Base: +, -, *, ==
# helpers
const perfectints = Set{Int}([0,3,4])
accstr(n, u, d) =
if n > 0; repeat(u,n) elseif n < 0; repeat(d,-n) else "" end
qualpf(n, a, p, d) =
if n > 0; repeat(a,n) elseif n < 0; repeat(d,-n) else p end
qualimpf(n, a, mj, mn, d) =
if n > 0; repeat(a,n) elseif n < -1; repeat(d,-n-1) elseif n == -1; mn else mj end
# common functions (special interface)
"""
degree(i)
degree(p)
Return the "relative scale degree" (0-6) to which the interval points
(unison=`0`, 2nd=`1`, octave=`0`, 2nd down=`6`, etc.).
For pitches, return the integer that corresponds to the letter (C=`0`, D=`1`, ...).
See also: [`generic`](@ref), [`diasteps`](@ref), [`letter`](@ref)
"""
function degree end
degree(fifths::Int) = mod(fifths*4,7)
degree(p::Pitch) = degree(p.pitch)
"""
generic(i)
Return the generic interval, i.e. the number of diatonic steps modulo octave.
Unlike [`degree`](@ref), the result respects the sign of the interval
(unison=`0`, 2nd up=`1`, 2nd down=`-1`).
For pitches, use [`degree`](@ref).
See also: [`degree`](@ref), [`diasteps`](@ref)
"""
function generic end
"""
letter(p)
Returns the letter of a pitch as a character.
See also: [`degree`](@ref).
"""
letter(p::Pitch) = 'A' + mod(degree(p)+2, 7)
"""
diasteps(i)
Return the diatonic steps of the interval (unison=`0`, 2nd=`1`, ..., octave=`7`).
Respects both direction and octaves.
See also [`degree`](@ref) and [`generic`](@ref).
"""
function diasteps end
"""
alteration(i)
alteration(p)
Return the number of semitones by which the interval is altered from its the perfect or major variant.
Positive alteration always indicates augmentation,
negative alteration indicates diminution (minor or smaller) of the interval.
For interval classes, the alteration always corresponds to the upward version of the interval.
For pitches, return the accidentals (positive=sharps, negative=flats, `0`=natural).
"""
function alteration end
alteration(p::Pitch) = alteration(ic(p.pitch))
"""
octaves(i)
octaves(p)
Return the number of octaves the interval spans.
Positive intervals start at 0 octaves, increasing.
Negative intervals start at -1 octaves, decreasing.
(You might want to use `octaves(abs(i))` instead).
For a pitch, return its octave.
"""
function octaves end
octaves(p::Pitch) = octaves(p.pitch)
"""
internalocts(i)
Return the internal octaves (i.e. dependent on the 5ths dimension) of an interval.
"""
function internalocts end
"""
fifths(i)
fifths(p)
Return the octave-invariant part of the interval in fifths
(unison=`0`, 5th up=`1`, 4th up/5th down=`-1`).
For a pitch, return the pitch class on the line of fifths.
"""
function fifths end
fifths(p::Pitch) = fifths(p.pitch)
# spelled interval
# ----------------
"""
SpelledInterval <: Interval
Spelled intervals represented as pairs of fifths and octaves.
E.g., `SpelledInterval(-3, 2)` represents a minor 3rd upwards
(3 fifths down, 2 octaves up).
"""
struct SpelledInterval <: Interval
fifths :: Int
octaves :: Int
end
"""
spelled(fifths, octaves)
Creates a spelled interval from `fifths` and `octaves`.
"""
spelled(f, o) = SpelledInterval(f, o)
"""
spelledp(fifths, octaves)
Creates a spelled pitch from `fifths` and `octaves`.
"""
spelledp(f, o) = Pitch(spelled(f, o))
# accessors
degree(i::SpelledInterval) = degree(i.fifths)
generic(i::SpelledInterval) = if sign(i) < 0; -degree(-i.fifths) else degree(i.fifths) end
diasteps(i::SpelledInterval) = i.fifths*4 + i.octaves*7
alteration(i::SpelledInterval) = let absi = abs(i); fld(absi.fifths+1, 7) end
octaves(i::SpelledInterval) = i.octaves + fld(i.fifths*4, 7)
internalocts(i::SpelledInterval) = i.octaves
fifths(i::SpelledInterval) = i.fifths
# general interface
function Base.show(io::IO, i::SpelledInterval)
# negative? print as -abs(i)
if sign(i) == -1
print(io, "-")
print(io, abs(i))
return
end
dia = degree(i)
diff = alteration(i) # interval is always positive, so direction is correct
qual = if dia ∈ perfectints
qualpf(diff, 'a', 'P', 'd')
else
qualimpf(diff, 'a', 'M', 'm', 'd')
end
octs = octaves(i)
print(io, qual * string(dia+1) * ':' * string(octs))
end
function Base.show(io::IO, p::Pitch{SpelledInterval})
dia = degree(p)
alter = alteration(p)
print(io, string('A' + mod(dia + 2, 7), accstr(alter, '♯', '♭'), string(octaves(p))))
end
Base.isless(i1::SpelledInterval, i2::SpelledInterval) = sign(i1 - i2) == -1
Base.isequal(i1::SpelledInterval, i2::SpelledInterval) =
i1.octaves == i2.octaves && i1.fifths == i2.fifths
Base.hash(i::SpelledInterval, x::UInt) = hash(i.octaves, hash(i.fifths, x))
+(i1::SpelledInterval, i2::SpelledInterval) =
spelled(i1.fifths + i2.fifths, i1.octaves + i2.octaves)
-(i1::SpelledInterval, i2::SpelledInterval) =
spelled(i1.fifths - i2.fifths, i1.octaves - i2.octaves)
-(i::SpelledInterval) = spelled(-i.fifths, -i.octaves)
Base.zero(::Type{SpelledInterval}) = spelled(0,0)
Base.zero(::SpelledInterval) = spelled(0,0)
*(i::SpelledInterval, n::Integer) = spelled(i.fifths*n, i.octaves*n)
*(n::Integer,i::SpelledInterval) = spelled(i.fifths*n, i.octaves*n)
octave(::Type{SpelledInterval}) = spelled(0,1)
Base.sign(i::SpelledInterval) = cmp((diasteps(i), fld(i.fifths + 1, 7)), (0,0))
Base.abs(i::SpelledInterval) = if sign(i) < 0; -i else i end
ic(i::SpelledInterval) = sic(i.fifths)
embed(i::SpelledInterval) = i
intervaltype(::Type{SpelledInterval}) = SpelledInterval
intervalclasstype(::Type{SpelledInterval}) = SpelledIC
isstep(i::SpelledInterval) = abs(diasteps(i)) <= 1
chromsemi(::Type{SpelledInterval}) = spelled(7,-4)
# conversion
tomidi(i::SpelledInterval) = midi(7*i.fifths + 12*i.octaves)
tomidi(p::Pitch{SpelledInterval}) = Pitch(tomidi(p.pitch) + midi(12)) # C4 = 48 semitones above C0 = midi(60)
# spelled interval class
# ----------------------
"""
SpelledIC <: IntervalClass
Spelled interval class represented on the line of 5ths with `0 = C`.
E.g., `SpelledIC(3)` represents a major 6th upwards or minor 3rd downwards
(i.e., three 5ths up modulo octave).
"""
struct SpelledIC <: IntervalClass
fifths :: Int
end
"""
sic(fifths)
Creates a spelled interval class going `fifths` 5ths upwards.
"""
sic(fs) = SpelledIC(fs)
"""
spc(fifths)
Creates a spelled pitch class.
In analogy to `sic`, this function takes a number of 5ths.
"""
spc(fs) = Pitch(sic(fs))
# accessors
degree(i::SpelledIC) = degree(i.fifths)
generic(i::SpelledIC) = degree(i.fifths)
diasteps(i::SpelledIC) = degree(i.fifths)
alteration(i::SpelledIC) = fld(i.fifths + 1, 7)
octaves(i::SpelledIC) = 0
internalocts(i::SpelledIC) = 0
fifths(i::SpelledIC) = i.fifths
# interface functions
function Base.show(io::IO, ic::SpelledIC)
dia = degree(ic)
diff = alteration(ic)
qual = if dia ∈ perfectints
qualpf(diff, 'a', 'P', 'd')
else
qualimpf(diff, 'a', 'M', 'm', 'd')
end
print(io, qual * string(dia+1))
end
function Base.show(io::IO, p::Pitch{SpelledIC})
i = p.pitch
dia = degree(i)
alter = alteration(i)
print(io, ('A' + mod(dia+2, 7)) * accstr(alter, '♯', '♭'))
end
Base.isless(i1::SpelledIC, i2::SpelledIC) = isless(i1.fifths,i2.fifths)
Base.isequal(i1::SpelledIC, i2::SpelledIC) = isequal(i1.fifths,i2.fifths)
Base.hash(i::SpelledIC, x::UInt) = hash(i.fifths, x)
+(i1::SpelledIC, i2::SpelledIC) = sic(i1.fifths + i2.fifths)
-(i1::SpelledIC, i2::SpelledIC) = sic(i1.fifths - i2.fifths)
-(i::SpelledIC) = sic(-i.fifths)
Base.zero(::Type{SpelledIC}) = sic(0)
Base.zero(::SpelledIC) = sic(0)
*(i::SpelledIC, n::Integer) = sic(i.fifths * n)
*(n::Integer,i::SpelledIC) = sic(i.fifths * n)
octave(::Type{SpelledIC}) = sic(0)
function Base.sign(i::SpelledIC)
dia = degree(i)
if dia == 0
cmp(fld(i.fifths + 1, 4), 0)
elseif dia > 3
-1
else
1
end
end
Base.abs(i::SpelledIC) = if sign(i) < 0; -i else i end
ic(i::SpelledIC) = i
embed(i::SpelledIC) = spelled(i.fifths, -fld(i.fifths*4, 7))
intervaltype(::Type{SpelledIC}) = SpelledInterval
intervalclasstype(::Type{SpelledIC}) = SpelledIC
isstep(i::SpelledIC) = degree(i) ∈ [0,1,6]
chromsemi(::Type{SpelledIC}) = sic(7)
# conversion
tomidi(i::SpelledIC) = midic(i.fifths * 7)
tomidi(p::Pitch{SpelledIC}) = midipc(p.pitch.fifths * 7)
# parsing
# -------
const rgsic = r"^(-?)(a+|d+|[MPm])([1-7])$"
const rgspelled = r"^(-?)(a+|d+|[MPm])([1-7]):(-?)(\d+)$"
function matchinterval(modifier, num)
dia = parse(Int, num) - 1
perfect = dia ∈ perfectints
alt = if modifier == "M" && !perfect
0
elseif modifier == "m" && !perfect
-1
elseif lowercase(modifier) == "p" && perfect
0
elseif occursin(r"^a+$", modifier)
length(modifier)
elseif occursin(r"^d+$", modifier)
-length(modifier) - (perfect ? 0 : 1)
else
error("cannot parse interval \"$modifier$num\"")
end
mod(dia*2+1, 7) - 1 + 7*alt
end
"""
parsespelled(str)
Parse a spelled interval or interval class string.
The type is determined from the string,
so `i"M3:0"` returns an interval while `i"M3"` returns an interval class.
See also: [`@i_str`](@ref), [`parsespelledpitch`](@ref).
"""
function parsespelled(str)
m = match(rgsic, str)
if m != nothing
int = sic(matchinterval(m[2], m[3]))
else
m = match(rgspelled, str)
if m != nothing
fifths = matchinterval(m[2], m[3])
octs = parse(Int, m[4]*m[5])
int = spelled(fifths, octs - fld(fifths*4, 7))
else
error("cannot parse interval \"$str\"")
end
end
# invert if necessary
if m[1] == "-"
-int
else
int
end
end
"""
i"str"
Parse a spelled interval or interval class string.
The type is determined from the string,
so `i"M3:0"` returns an interval while `i"M3"` returns an interval class.
See also: [`@p_str`](@ref), [`parsespelled`](@ref), [`parsespelledpitch`](@ref).
"""
macro i_str(str)
parsespelled(str)
end
const rgspelledpc = r"^([a-g])(♭+|♯+|b+|#+)?$"i
const rgspelledp = r"^([a-g])(♭+|♯+|b+|#+)?(-?\d+)$"i
function matchpitch(letter, accs)
letter = uppercase(letter)[1]
if letter >= 'A' && letter <= 'G'
dia = mod(letter - 'A' - 2, 7)
else
error("cannot parse pitch letter \"$letter\"")
end
alt = if accs == nothing || accs == ""
0
elseif occursin(r"^♭+|b+$"i, accs)
-length(accs)
elseif occursin(r"^♯+|#+$"i, accs)
length(accs)
else
error("cannot parse accidentals \"$accs\"")
end
mod(dia*2 + 1, 7) - 1 + 7*alt
end
"""
parsespelledpitch(str)
Parse a spelled pitch or pitch class string.
The type is determined from the string,
so `p"G4"` returns a pitch while `p"G"` returns a pitch class.
See also: [`@p_str`](@ref), [`parsespelled`](@ref).
"""
function parsespelledpitch(str)
m = match(rgspelledpc, str)
if m != nothing
spc(matchpitch(m[1], m[2]))
else
m = match(rgspelledp, str)
if m != nothing
octs = parse(Int, m[3])
fifths = matchpitch(m[1], m[2])
spelledp(fifths, octs - fld(fifths*4, 7))
else
error("cannot parse pitch \"$str\"")
end
end
end
"""
p"str"
Parse a spelled pitch or pitch class string.
The type is determined from the string,
so `p"G4"` returns a pitch while `p"G"` returns a pitch class.
See also: [`@i_str`](@ref), [`parsespelledpitch`](@ref), [`parsespelled`](@ref).
"""
macro p_str(str)
parsespelledpitch(str)
end
| Pitches | https://github.com/DCMLab/Pitches.jl.git |
|
[
"MIT"
] | 0.1.2 | cd110f79356a53100ea1ca281eceb7b7fe39462c | code | 104 | using Test
using Pitches
include("test_midi.jl")
include("test_spelled.jl")
include("test_logfreq.jl")
| Pitches | https://github.com/DCMLab/Pitches.jl.git |
|
[
"MIT"
] | 0.1.2 | cd110f79356a53100ea1ca281eceb7b7fe39462c | code | 4648 | @testset "Frequency pitches" begin
@testset "types ok and compact" begin
@test isbitstype(FreqInterval)
@test isbitstype(FreqIC)
@test isbitstype(Pitch{FreqInterval})
@test isbitstype(Pitch{FreqIC})
end
@testset "constructors" begin
@test logfreqi(log(3.8)) == freqi(3.8)
@test logfreqic(log(3.8)) == freqic(3.8)
@test logfreqic(log(3.8) + log(2)) ≈ freqic(3.8)
@test logfreqp(log(441)) == freqp(441)
@test logfreqpc(log(441)) == freqpc(441)
@test logfreqpc(log(441) + log(2)) ≈ freqpc(441)
end
@testset "printing" begin
#@test string(freqi(3.8)) == "fr3.8"
#@test string(freqic(3.8)) == "fr[1.9]"
#@test string(freqp(441)) == "441.0Hz"
#@test string(freqpc(441)) == "[1.7226562500000004]Hz"
end
@testset "hashing" begin
@test hash(logfreqi(log(3.8))) == hash(freqi(3.8))
@test hash(logfreqic(log(3.8))) == hash(freqic(3.8))
@test hash(logfreqi(log(441))) == hash(freqi(441))
@test hash(logfreqic(log(441))) == hash(freqic(441))
end
@testset "interval interface" begin
@test freqi(3/2) + freqi(4/3) ≈ freqi(2)
@test freqi(2/3) + freqi(4/3) ≈ freqi(8/9)
@test (-freqi(3/2)) + freqi(4/3) ≈ freqi(8/9)
@test freqi(4/3) - freqi(3/2) ≈ -freqi(9/8)
@test -freqi(3/2) ≈ freqi(2/3)
@test -freqi(4/5) ≈ unison(FreqInterval) - freqi(4/5)
@test unison(FreqInterval) == freqi(1)
@test unison(freqi(0.42)) == freqi(1)
@test freqi(3/2) * 2 == freqi(9/4)
@test freqi(3/2) * -2 == freqi(4/9)
@test 2 * freqi(3/2) == freqi(9/4)
@test -2 * freqi(3/2) == freqi(4/9)
@test sign(freqi(3/2)) == 1
@test sign(freqi(2/3)) == -1
@test sign(freqi(1)) == 0
@test abs(freqi(3/2)) == freqi(3/2)
@test abs(freqi(2/3)) ≈ freqi(3/2)
@test abs(freqi(1)) == freqi(1)
@test octave(FreqInterval) == freqi(2)
@test chromsemi(FreqInterval) == logfreqi(log(2)/12)
@test ic(freqi(3)) ≈ freqic(3)
@test ic(freqi(3)) ≈ freqic(3/2)
@test ic(freqi(3/2)) ≈ freqic(3)
@test embed(freqi(3/2)) == freqi(3/2)
@test intervaltype(FreqInterval) == FreqInterval
@test intervalclasstype(FreqInterval) == FreqIC
end
@testset "interval class interface" begin
@test freqic(3/2) + freqic(4/3) ≈ freqic(2)
@test freqic(2/3) + freqic(4/3) ≈ freqic(8/9)
@test (-freqic(3/2)) + freqic(4/3) ≈ freqic(8/9)
@test freqic(4/3) - freqic(3/2) ≈ -freqic(9/8)
@test -freqic(3/2) ≈ freqic(2/3)
@test -freqic(4/5) ≈ unison(FreqIC) - freqic(4/5)
@test unison(FreqIC) == freqic(1)
@test unison(freqic(0.42)) == freqic(1)
@test freqic(3/2) * 2 == freqic(9/4)
@test freqic(3/2) * -2 == freqic(4/9)
@test 2 * freqic(3/2) == freqic(9/4)
@test -2 * freqic(3/2) == freqic(4/9)
@test sign(freqic(3/2)) == -1
@test sign(freqic(2/3)) == 1
@test sign(freqic(1)) == 0
@test abs(freqic(3/2)) == freqic(2/3)
@test abs(freqic(2/3)) ≈ freqic(2/3)
@test abs(freqic(1)) == freqic(1)
@test octave(FreqIC) == freqic(2)
@test chromsemi(FreqIC) == logfreqic(log(2)/12)
@test ic(freqic(3)) ≈ freqic(3)
@test ic(freqic(3)) ≈ freqic(3/2)
@test ic(freqic(3/2)) ≈ freqic(3)
@test embed(freqic(3/2)) == freqi(3/2)
@test intervaltype(FreqIC) == FreqInterval
@test intervalclasstype(FreqIC) == FreqIC
end
@testset "pitch interface" begin
@test topitch(freqi(441)) == freqp(441)
@test tointerval(freqp(441)) == freqi(441)
@test freqp(441) + freqi(2) ≈ freqp(882)
@test freqp(441) + freqi(1/2) ≈ freqp(220.5)
@test freqp(441) - freqi(2) ≈ freqp(220.5)
@test freqp(441) - freqp(431.5) ≈ freqi(441/431.5)
@test pc(freqp(441)) == freqpc(441)
@test embed(freqp(441)) == freqp(441)
end
@testset "pitch class interface" begin
@test topitch(freqic(441)) == freqpc(441)
@test tointerval(freqpc(441)) == freqic(441)
@test freqpc(441) + freqic(3/2) ≈ freqpc(661.5)
@test freqpc(441) + freqic(2/3) ≈ freqpc(294)
@test freqpc(441) - freqic(3/2) ≈ freqpc(294)
@test freqpc(441) - freqpc(431.5) ≈ freqic(441/431.5)
@test pc(freqpc(441)) == freqpc(441)
@test embed(freqpc(441)) == logfreqp(mod(log(441), log(2)))
@test embed(freqpc(441), 8) == freqp(441)
end
end
| Pitches | https://github.com/DCMLab/Pitches.jl.git |
|
[
"MIT"
] | 0.1.2 | cd110f79356a53100ea1ca281eceb7b7fe39462c | code | 4954 | @testset "MIDI pitches" begin
@testset "types ok and compact" begin
@test isbitstype(MidiInterval)
@test isbitstype(MidiIC)
@test isbitstype(Pitch{MidiInterval})
@test isbitstype(Pitch{MidiIC})
end
@testset "constructors" begin
@test midi(3) == midi(3)
@test midip(60) == midip(60)
@test midic(15) == midic(3)
@test midipc(63) == midipc(3)
end
@testset "macro constructors" begin
@test (@midi [1,2,3]) == midi.([1,2,3])
@test (@midip [1,2,3]) == midip.([1,2,3])
@test (@midic [1,2,3]) == midic.([1,2,3])
@test (@midipc [1,2,3]) == midipc.([1,2,3])
end
@testset "String conversion" begin
@test string(midi(3)) == "i3"
@test string(midip(63)) == "p63"
@test string(midic(15)) == "ic3"
@test string(midipc(63)) == "pc3"
end
@testset "Int conversion" begin
@test Int(midi(3)) == 3
@test Int(midip(63)) == 63
@test Int(midic(15)) == 3
@test Int(midipc(63)) == 3
end
@testset "hashing" begin
@test hash(midi(3)) == hash(midi(3))
@test hash(midip(3)) == hash(midip(3))
@test hash(midic(3)) == hash(midic(3))
@test hash(midipc(3)) == hash(midipc(3))
end
@testset "interval interface" begin
@test midi(3) + midi(10) == midi(13)
@test midi(-3) + midi(4) == midi(1)
@test midi(3) + midi(-4) == midi(-1)
@test midi(3) - midi(4) == midi(-1)
@test -midi(5) == midi(-5)
@test -midi(7) == midi(0) - midi(7)
@test zero(MidiInterval) == midi(0)
@test zero(midi(3)) == midi(0)
@test midi(2) * 7 == midi(14)
@test midi(-3) * 4 == midi(-12)
@test midi(4) * -3 == midi(-12)
@test 6 * midi(2) == midi(12)
@test 4 * midi(-4) == midi(-16)
@test 5 * midi(4) == midi(20)
@test tomidi(midi(42)) == midi(42)
@test octave(MidiInterval) == midi(12)
@test sign(midi(3)) == 1
@test sign(midi(0)) == 0
@test sign(midi(-3)) == -1
@test abs(midi(-3)) == midi(3)
@test ic(midi(15)) == midic(3)
@test ic(midi(-3)) == midic(9)
@test embed(midi(42)) == midi(42)
@test intervaltype(MidiInterval) == MidiInterval
@test intervalclasstype(MidiInterval) == MidiIC
for i in -2:2
@test isstep(midi(i)) == true
end
for i in -13:-3
@test isstep(midi(i)) == false
end
for i in 3:13
@test isstep(midi(i)) == false
end
@test chromsemi(MidiInterval) == midi(1)
end
@testset "interval class interface" begin
@test midic(3) + midic(10) == midic(1)
@test midic(-3) + midic(4) == midic(1)
@test midic(3) + midic(-4) == midic(11)
@test midic(3) - midic(4) == midic(11)
@test -midic(5) == midic(-5)
@test -midic(7) == midic(0) - midic(7)
@test zero(MidiIC) == midic(0)
@test zero(midic(3)) == midic(0)
@test midic(2) * 7 == midic(14)
@test midic(-3) * 4 == midic(-12)
@test midic(4) * -3 == midic(-12)
@test 6 * midic(2) == midic(12)
@test 4 * midic(-4) == midic(-16)
@test 5 * midic(4) == midic(20)
@test tomidi(midic(42)) == midic(42)
@test octave(MidiIC) == midic(0)
@test sign(midic(3)) == 1
@test sign(midic(0)) == 0
@test sign(midic(-3)) == -1
@test abs(midic(-3)) == midic(3)
@test ic(midic(15)) == midic(3)
@test ic(midic(-3)) == midic(9)
@test embed(midic(3)) == midi(3)
@test embed(midic(-3)) == midi(9)
@test intervaltype(MidiIC) == MidiInterval
@test intervalclasstype(MidiIC) == MidiIC
for i in -2:2
@test isstep(midic(i)) == true
end
for i in -9:-3
@test isstep(midic(i)) == false
end
for i in 3:9
@test isstep(midic(i)) == false
end
@test chromsemi(MidiIC) == midic(1)
end
@testset "pitch interface" begin
@test topitch(midi(3)) == midip(3)
@test tointerval(midip(42)) == midi(42)
@test midip(63) + midi(7) == midip(70)
@test midip(63) + midi(-3) == midip(60)
@test midip(63) - midi(7) == midip(56)
@test midip(67) - midip(61) == midi(6)
@test pc(midip(63)) == midipc(3)
@test embed(midip(8)) == midip(8)
end
@testset "pitch class interface" begin
@test topitch(midic(3)) == midipc(3)
@test tointerval(midipc(42)) == midic(42)
@test midipc(63) + midic(7) == midipc(70)
@test midipc(63) + midic(-3) == midipc(60)
@test midipc(63) - midic(7) == midipc(56)
@test midipc(67) - midipc(61) == midic(6)
@test pc(midipc(63)) == midipc(3)
@test embed(midipc(8)) == midip(8)
end
end
| Pitches | https://github.com/DCMLab/Pitches.jl.git |
|
[
"MIT"
] | 0.1.2 | cd110f79356a53100ea1ca281eceb7b7fe39462c | code | 8243 | @testset "Spelled pitches" begin
@testset "types ok and compact" begin
@test isbitstype(SpelledInterval)
@test isbitstype(SpelledIC)
@test isbitstype(Pitch{SpelledInterval})
@test isbitstype(Pitch{SpelledIC})
end
@testset "constructors" begin
@test spelled(-3, 0) == spelled(-3, 0)
@test spelledp(0, 4) == spelledp(0, 4)
@test sic(-3) == sic(-3)
@test spc(3) == spc(3)
end
@testset "string macros" begin
@test i"M3:1" == spelled(4, -1)
@test i"-M3:0" == spelled(-4, 2)
@test p"C♭4" == spelledp(-7, 8)
@test p"Cb4" == spelledp(-7, 8)
@test p"Cb-1" == spelledp(-7, 3)
@test p"C#-1" == spelledp(7, -5)
@test i"m3" == sic(-3)
@test i"-m3" == sic(3)
@test p"C♯" == spc(7)
@test p"C#" == spc(7)
# TODO: test more complex parsing cases
end
@testset "accessors" begin
@test octaves(i"M3:1") == 1
@test internalocts(i"M3:1") == -1
@test fifths(i"M3:1") == 4
@test degree(i"M3:1") == 2
@test generic(i"M3:1") == 2
@test diasteps(i"M3:1") == 9
@test alteration(i"M3:1") == 0
@test octaves(i"-M3:1") == -2
@test internalocts(i"-M3:1") == 1
@test fifths(i"-M3:1") == -4
@test degree(i"-M3:1") == 5
@test generic(i"-M3:1") == -2
@test diasteps(i"-M3:1") == -9
@test alteration(i"-M3:1") == 0
@test octaves(i"a5") == 0
@test internalocts(i"a5") == 0
@test fifths(i"a5") == 8
@test degree(i"a5") == 4
@test generic(i"a5") == 4
@test diasteps(i"a5") == 4
@test alteration(i"a5") == 1
@test octaves(p"Ebb5") == 5
@test fifths(p"Ebb5") == -10
@test degree(p"Ebb5") == 2
@test alteration(p"Ebb5") == -2
@test letter(p"Ebb5") == 'E'
@test octaves(p"F#") == 0
@test fifths(p"F#") == 6
@test degree(p"F#") == 3
@test alteration(p"F#") == 1
@test letter(p"F#") == 'F'
# edge cases
@test alteration(i"-P4:0") == 0
@test alteration(i"-M7:0") == 0
@test alteration(i"a4:0") == 1
@test alteration(i"m7:0") == -1
@test alteration(i"-a4:0") == 1
@test alteration(i"-m7:0") == -1
@test alteration(i"d1:0") == 1 # d1:0 == -a1:0
@test alteration(i"P4") == 0
@test alteration(i"M7") == 0
@test alteration(i"d1") == -1
@test alteration(p"C#-1") == 1
@test alteration(p"Cb-1") == -1
@test alteration(p"F") == 0
@test alteration(p"B") == 0
end
@testset "printing" begin
@test string(i"m3:1") == "m3:1"
@test string(p"Eb4") == "E♭4"
@test string(i"m3") == "m3"
@test string(p"E#") == "E♯"
end
@testset "hashing" begin
@test hash(spelled(-3,1)) == hash(spelled(-3,1))
@test hash(spelledp(3,0)) == hash(spelledp(3,0))
@test hash(sic(3)) == hash(sic(3))
@test hash(spc(3)) == hash(spc(3))
end
@testset "interval interface" begin
@test i"m3:0" + i"M3:0" == i"P5:0"
@test i"m3:0" + i"M7:0" == i"M2:1"
@test i"P5:0" + i"P5:0" == i"M2:1"
@test i"-m3:0" + i"M3:0" == i"a1:0"
@test i"m3:0" + i"-M3:0" == i"-a1:0"
@test i"m3:0" - i"M3:0" == i"-a1:0"
@test i"m3:0" - i"M6:0" == i"-a4:0"
@test -i"P4:0" == i"-P4:0"
@test -i"P4:0" == i"P5:-1"
@test -i"P5:0" == zero(SpelledInterval) - i"P5:0"
@test zero(SpelledInterval) == i"P1:0"
@test zero(i"m3:0") == i"P1:0"
@test i"P5:0" * 2 == i"M2:1"
@test i"M2:0" * 4 == i"a5:0"
@test i"-m3:0" * 4 == i"-d2:1"
@test i"M3:0" * -3 == i"-a7:0"
@test 4 * i"M2:0" == i"a5:0"
@test 4 * i"-M3:0" == i"-aa2:1"
@test 5 * i"M3:0" == i"aaa4:1"
@test tomidi(i"aaa4:1") == midi(20)
@test tomidi(i"-P5:0") == midi(-7)
@test octave(SpelledInterval) == i"P1:1"
@test sign(i"m2:0") == 1
@test sign(i"P1:0") == 0
@test sign(i"d1:0") == -1
@test sign(i"a1:0") == 1
@test sign(i"-m3:0") == -1
@test sign(i"P4:0") == 1
@test sign(i"-M7:0") == -1
@test abs(i"-m3:0") == i"m3:0"
@test i"m2:0" < i"M2:0"
@test i"-m2:0" > i"-M2:0"
@test ic(i"M3:3") == i"M3"
@test ic(i"-M3:1") == i"m6"
@test embed(i"M3:3") == i"M3:3"
@test intervaltype(SpelledInterval) == SpelledInterval
@test intervalclasstype(SpelledInterval) == SpelledIC
@test isstep(i"d1:0")
@test isstep(i"P1:0")
@test isstep(i"a1:0")
@test isstep(i"d2:0")
@test isstep(i"m2:0")
@test isstep(i"M2:0")
@test isstep(i"a2:0")
@test isstep(i"-d2:0")
@test isstep(i"-m2:0")
@test isstep(i"-M2:0")
@test isstep(i"-a2:0")
@test !isstep(i"d3:0")
@test !isstep(i"-d3:0")
@test !isstep(i"M7:0")
@test !isstep(i"-M7:0")
@test !isstep(i"P1:1")
@test !isstep(i"-P1:1")
@test !isstep(i"m2:1")
@test !isstep(i"-m2:1")
@test chromsemi(SpelledInterval) == i"a1:0"
end
@testset "interval class interface" begin
@test i"m3" + i"M3" == i"P5"
@test i"m3" + i"M7" == i"M2"
@test i"P5" + i"P5" == i"M2"
@test i"-m3" + i"M3" == i"a1"
@test i"m3" + i"-M3" == i"-a1"
@test i"m3" - i"M3" == i"-a1"
@test i"m3" - i"M6" == i"-a4"
@test -i"P4" == i"-P4"
@test -i"P4" == i"P5"
@test -i"P5" == zero(SpelledIC) - i"P5"
@test zero(SpelledIC) == i"P1"
@test zero(i"m3") == i"P1"
@test i"P5" * 2 == i"M2"
@test i"M2" * 4 == i"a5"
@test i"-m3" * 4 == i"-d2"
@test i"M3" * -3 == i"-a7"
@test 4 * i"M2" == i"a5"
@test 4 * i"-M3" == i"-aa2"
@test 5 * i"M3" == i"aaa4"
@test tomidi(i"aaa4") == midic(8)
@test tomidi(i"-P5") == midic(-7)
@test octave(SpelledIC) == i"P1"
@test sign(i"m2") == 1
@test sign(i"P1") == 0
@test sign(i"d1") == -1
@test sign(i"a1") == 1
@test sign(i"-m3") == -1
@test sign(i"P4") == 1
@test sign(i"-M7") == 1
@test abs(i"-m3") == i"m3"
@test ic(i"-M3") == i"m6"
@test embed(i"M3") == i"M3:0"
@test embed(i"M7") == i"M7:0"
@test embed(i"P4") == i"P4:0"
@test embed(i"a1") == i"a1:0"
@test intervaltype(SpelledIC) == SpelledInterval
@test intervalclasstype(SpelledIC) == SpelledIC
@test isstep(i"d1")
@test isstep(i"P1")
@test isstep(i"a1")
@test isstep(i"d2")
@test isstep(i"m2")
@test isstep(i"M2")
@test isstep(i"a2")
@test isstep(i"-d2")
@test isstep(i"-m2")
@test isstep(i"-M2")
@test isstep(i"-a2")
@test !isstep(i"d3")
@test !isstep(i"-d3")
@test chromsemi(SpelledIC) == i"a1"
end
@testset "pitch interface" begin
@test topitch(i"m3:4") == p"Eb4"
@test tointerval(p"C#3") == i"a1:3"
@test p"Eb4" + i"P5:0" == p"Bb4"
@test p"Eb4" + i"-m3:0" == p"C4"
@test p"Eb4" - i"P5:0" == p"Ab3"
@test p"G4" - p"C#4" == i"d5:0"
@test alteration(p"Ab-1") == -1
@test alteration(p"A#-1") == 1
@test p"C-1" > p"Cb-1"
@test pc(p"Eb4") == p"Eb"
@test embed(p"Eb4") == p"Eb4"
@test tomidi(p"C#3") == midip(49)
@test tomidi(p"Db3") == midip(49)
end
@testset "pitch class interface" begin
@test topitch(i"m3") == p"Eb"
@test tointerval(p"E") == i"M3"
@test p"Eb" + i"P5" == p"Bb"
@test p"Eb" + i"-m3" == p"C"
@test p"Eb" - i"P5" == p"Ab"
@test p"G" - p"C#" == i"d5"
@test pc(p"Eb") == p"Eb"
@test embed(p"Eb") == p"Eb0"
@test embed(p"Eb", 4) == p"Eb4"
@test tomidi(p"C#") == midipc(1)
@test tomidi(p"Db") == midipc(1)
end
end
| Pitches | https://github.com/DCMLab/Pitches.jl.git |
|
[
"MIT"
] | 0.1.2 | cd110f79356a53100ea1ca281eceb7b7fe39462c | docs | 493 | # Changes
## 0.1.2
- The definition of `sign` has been updated for spelled pitches:
- Unisons now have a direction (e.g., `sign(a1:0) == 1`).
- As a consequence, the `alteration` of `d1:0` (= `-a1:0`) is now 1
because it is considered downward.
- The interval class `d1` is considered downward:
- For interval classes, `sign` takes the shortest realization (as before)
but `alteration` always considers the upward interval (as in printing).-
## 0.1.0/0.1.1
- first version
| Pitches | https://github.com/DCMLab/Pitches.jl.git |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.