code
stringlengths 2.5k
150k
| kind
stringclasses 1
value |
|---|---|
# Word2Vec
**Learning Objectives**
1. Compile all steps into one function
2. Prepare training data for Word2Vec
3. Model and Training
4. Embedding lookup and analysis
## Introduction
Word2Vec is not a singular algorithm, rather, it is a family of model architectures and optimizations that can be used to learn word embeddings from large datasets. Embeddings learned through Word2Vec have proven to be successful on a variety of downstream natural language processing tasks.
Note: This notebook is based on [Efficient Estimation of Word Representations in Vector Space](https://arxiv.org/pdf/1301.3781.pdf) and
[Distributed
Representations of Words and Phrases and their Compositionality](https://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf). It is not an exact implementation of the papers. Rather, it is intended to illustrate the key ideas.
These papers proposed two methods for learning representations of words:
* **Continuous Bag-of-Words Model** which predicts the middle word based on surrounding context words. The context consists of a few words before and after the current (middle) word. This architecture is called a bag-of-words model as the order of words in the context is not important.
* **Continuous Skip-gram Model** which predict words within a certain range before and after the current word in the same sentence. A worked example of this is given below.
You'll use the skip-gram approach in this notebook. First, you'll explore skip-grams and other concepts using a single sentence for illustration. Next, you'll train your own Word2Vec model on a small dataset. This notebook also contains code to export the trained embeddings and visualize them in the [TensorFlow Embedding Projector](http://projector.tensorflow.org/).
Each learning objective will correspond to a __#TODO__ in the [student lab notebook](../labs/word2vec.ipynb) -- try to complete that notebook first before reviewing this solution notebook.
## Skip-gram and Negative Sampling
While a bag-of-words model predicts a word given the neighboring context, a skip-gram model predicts the context (or neighbors) of a word, given the word itself. The model is trained on skip-grams, which are n-grams that allow tokens to be skipped (see the diagram below for an example). The context of a word can be represented through a set of skip-gram pairs of `(target_word, context_word)` where `context_word` appears in the neighboring context of `target_word`.
Consider the following sentence of 8 words.
> The wide road shimmered in the hot sun.
The context words for each of the 8 words of this sentence are defined by a window size. The window size determines the span of words on either side of a `target_word` that can be considered `context word`. Take a look at this table of skip-grams for target words based on different window sizes.
Note: For this tutorial, a window size of *n* implies n words on each side with a total window span of 2*n+1 words across a word.

The training objective of the skip-gram model is to maximize the probability of predicting context words given the target word. For a sequence of words *w<sub>1</sub>, w<sub>2</sub>, ... w<sub>T</sub>*, the objective can be written as the average log probability

where `c` is the size of the training context. The basic skip-gram formulation defines this probability using the softmax function.

where *v* and *v<sup>'<sup>* are target and context vector representations of words and *W* is vocabulary size.
Computing the denominator of this formulation involves performing a full softmax over the entire vocabulary words which is often large (10<sup>5</sup>-10<sup>7</sup>) terms.
The [Noise Contrastive Estimation](https://www.tensorflow.org/api_docs/python/tf/nn/nce_loss) loss function is an efficient approximation for a full softmax. With an objective to learn word embeddings instead of modelling the word distribution, NCE loss can be [simplified](https://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf) to use negative sampling.
The simplified negative sampling objective for a target word is to distinguish the context word from *num_ns* negative samples drawn from noise distribution *P<sub>n</sub>(w)* of words. More precisely, an efficient approximation of full softmax over the vocabulary is, for a skip-gram pair, to pose the loss for a target word as a classification problem between the context word and *num_ns* negative samples.
A negative sample is defined as a (target_word, context_word) pair such that the context_word does not appear in the `window_size` neighborhood of the target_word. For the example sentence, these are few potential negative samples (when `window_size` is 2).
```
(hot, shimmered)
(wide, hot)
(wide, sun)
```
In the next section, you'll generate skip-grams and negative samples for a single sentence. You'll also learn about subsampling techniques and train a classification model for positive and negative training examples later in the tutorial.
## Setup
```
# Use the chown command to change the ownership of repository to user.
!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst
!pip install -q tqdm
# You can use any Python source file as a module by executing an import statement in some other Python source file.
# The import statement combines two operations; it searches for the named module, then it binds the
# results of that search to a name in the local scope.
import io
import itertools
import numpy as np
import os
import re
import string
import tensorflow as tf
import tqdm
from tensorflow.keras import Model, Sequential
from tensorflow.keras.layers import Activation, Dense, Dot, Embedding, Flatten, GlobalAveragePooling1D, Reshape
from tensorflow.keras.layers.experimental.preprocessing import TextVectorization
```
Please check your tensorflow version using the cell below.
```
# Show the currently installed version of TensorFlow
print("TensorFlow version: ",tf.version.VERSION)
SEED = 42
AUTOTUNE = tf.data.experimental.AUTOTUNE
```
### Vectorize an example sentence
Consider the following sentence:
`The wide road shimmered in the hot sun.`
Tokenize the sentence:
```
sentence = "The wide road shimmered in the hot sun"
tokens = list(sentence.lower().split())
print(len(tokens))
```
Create a vocabulary to save mappings from tokens to integer indices.
```
vocab, index = {}, 1 # start indexing from 1
vocab['<pad>'] = 0 # add a padding token
for token in tokens:
if token not in vocab:
vocab[token] = index
index += 1
vocab_size = len(vocab)
print(vocab)
```
Create an inverse vocabulary to save mappings from integer indices to tokens.
```
inverse_vocab = {index: token for token, index in vocab.items()}
print(inverse_vocab)
```
Vectorize your sentence.
```
example_sequence = [vocab[word] for word in tokens]
print(example_sequence)
```
### Generate skip-grams from one sentence
The `tf.keras.preprocessing.sequence` module provides useful functions that simplify data preparation for Word2Vec. You can use the `tf.keras.preprocessing.sequence.skipgrams` to generate skip-gram pairs from the `example_sequence` with a given `window_size` from tokens in the range `[0, vocab_size)`.
Note: `negative_samples` is set to `0` here as batching negative samples generated by this function requires a bit of code. You will use another function to perform negative sampling in the next section.
```
window_size = 2
positive_skip_grams, _ = tf.keras.preprocessing.sequence.skipgrams(
example_sequence,
vocabulary_size=vocab_size,
window_size=window_size,
negative_samples=0)
print(len(positive_skip_grams))
```
Take a look at few positive skip-grams.
```
for target, context in positive_skip_grams[:5]:
print(f"({target}, {context}): ({inverse_vocab[target]}, {inverse_vocab[context]})")
```
### Negative sampling for one skip-gram
The `skipgrams` function returns all positive skip-gram pairs by sliding over a given window span. To produce additional skip-gram pairs that would serve as negative samples for training, you need to sample random words from the vocabulary. Use the `tf.random.log_uniform_candidate_sampler` function to sample `num_ns` number of negative samples for a given target word in a window. You can call the funtion on one skip-grams's target word and pass the context word as true class to exclude it from being sampled.
Key point: *num_ns* (number of negative samples per positive context word) between [5, 20] is [shown to work](https://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf) best for smaller datasets, while *num_ns* between [2,5] suffices for larger datasets.
```
# Get target and context words for one positive skip-gram.
target_word, context_word = positive_skip_grams[0]
# Set the number of negative samples per positive context.
num_ns = 4
context_class = tf.reshape(tf.constant(context_word, dtype="int64"), (1, 1))
negative_sampling_candidates, _, _ = tf.random.log_uniform_candidate_sampler(
true_classes=context_class, # class that should be sampled as 'positive'
num_true=1, # each positive skip-gram has 1 positive context class
num_sampled=num_ns, # number of negative context words to sample
unique=True, # all the negative samples should be unique
range_max=vocab_size, # pick index of the samples from [0, vocab_size]
seed=SEED, # seed for reproducibility
name="negative_sampling" # name of this operation
)
print(negative_sampling_candidates)
print([inverse_vocab[index.numpy()] for index in negative_sampling_candidates])
```
### Construct one training example
For a given positive `(target_word, context_word)` skip-gram, you now also have `num_ns` negative sampled context words that do not appear in the window size neighborhood of `target_word`. Batch the `1` positive `context_word` and `num_ns` negative context words into one tensor. This produces a set of positive skip-grams (labelled as `1`) and negative samples (labelled as `0`) for each target word.
```
# Add a dimension so you can use concatenation (on the next step).
negative_sampling_candidates = tf.expand_dims(negative_sampling_candidates, 1)
# Concat positive context word with negative sampled words.
context = tf.concat([context_class, negative_sampling_candidates], 0)
# Label first context word as 1 (positive) followed by num_ns 0s (negative).
label = tf.constant([1] + [0]*num_ns, dtype="int64")
# Reshape target to shape (1,) and context and label to (num_ns+1,).
target = tf.squeeze(target_word)
context = tf.squeeze(context)
label = tf.squeeze(label)
```
Take a look at the context and the corresponding labels for the target word from the skip-gram example above.
```
print(f"target_index : {target}")
print(f"target_word : {inverse_vocab[target_word]}")
print(f"context_indices : {context}")
print(f"context_words : {[inverse_vocab[c.numpy()] for c in context]}")
print(f"label : {label}")
```
A tuple of `(target, context, label)` tensors constitutes one training example for training your skip-gram negative sampling Word2Vec model. Notice that the target is of shape `(1,)` while the context and label are of shape `(1+num_ns,)`
```
print(f"target :", target)
print(f"context :", context )
print(f"label :", label )
```
### Summary
This picture summarizes the procedure of generating training example from a sentence.

## Lab Task 1: Compile all steps into one function
### Skip-gram Sampling table
A large dataset means larger vocabulary with higher number of more frequent words such as stopwords. Training examples obtained from sampling commonly occuring words (such as `the`, `is`, `on`) don't add much useful information for the model to learn from. [Mikolov et al.](https://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf) suggest subsampling of frequent words as a helpful practice to improve embedding quality.
The `tf.keras.preprocessing.sequence.skipgrams` function accepts a sampling table argument to encode probabilities of sampling any token. You can use the `tf.keras.preprocessing.sequence.make_sampling_table` to generate a word-frequency rank based probabilistic sampling table and pass it to `skipgrams` function. Take a look at the sampling probabilities for a `vocab_size` of 10.
```
sampling_table = tf.keras.preprocessing.sequence.make_sampling_table(size=10)
print(sampling_table)
```
`sampling_table[i]` denotes the probability of sampling the i-th most common word in a dataset. The function assumes a [Zipf's distribution](https://en.wikipedia.org/wiki/Zipf%27s_law) of the word frequencies for sampling.
Key point: The `tf.random.log_uniform_candidate_sampler` already assumes that the vocabulary frequency follows a log-uniform (Zipf's) distribution. Using these distribution weighted sampling also helps approximate the Noise Contrastive Estimation (NCE) loss with simpler loss functions for training a negative sampling objective.
### Generate training data
Compile all the steps described above into a function that can be called on a list of vectorized sentences obtained from any text dataset. Notice that the sampling table is built before sampling skip-gram word pairs. You will use this function in the later sections.
```
# Generates skip-gram pairs with negative sampling for a list of sequences
# (int-encoded sentences) based on window size, number of negative samples
# and vocabulary size.
def generate_training_data(sequences, window_size, num_ns, vocab_size, seed):
# Elements of each training example are appended to these lists.
targets, contexts, labels = [], [], []
# Build the sampling table for vocab_size tokens.
# TODO 1a
sampling_table = tf.keras.preprocessing.sequence.make_sampling_table(vocab_size)
# Iterate over all sequences (sentences) in dataset.
for sequence in tqdm.tqdm(sequences):
# Generate positive skip-gram pairs for a sequence (sentence).
positive_skip_grams, _ = tf.keras.preprocessing.sequence.skipgrams(
sequence,
vocabulary_size=vocab_size,
sampling_table=sampling_table,
window_size=window_size,
negative_samples=0)
# Iterate over each positive skip-gram pair to produce training examples
# with positive context word and negative samples.
# TODO 1b
for target_word, context_word in positive_skip_grams:
context_class = tf.expand_dims(
tf.constant([context_word], dtype="int64"), 1)
negative_sampling_candidates, _, _ = tf.random.log_uniform_candidate_sampler(
true_classes=context_class,
num_true=1,
num_sampled=num_ns,
unique=True,
range_max=vocab_size,
seed=SEED,
name="negative_sampling")
# Build context and label vectors (for one target word)
negative_sampling_candidates = tf.expand_dims(
negative_sampling_candidates, 1)
context = tf.concat([context_class, negative_sampling_candidates], 0)
label = tf.constant([1] + [0]*num_ns, dtype="int64")
# Append each element from the training example to global lists.
targets.append(target_word)
contexts.append(context)
labels.append(label)
return targets, contexts, labels
```
## Lab Task 2: Prepare training data for Word2Vec
With an understanding of how to work with one sentence for a skip-gram negative sampling based Word2Vec model, you can proceed to generate training examples from a larger list of sentences!
### Download text corpus
You will use a text file of Shakespeare's writing for this tutorial. Change the following line to run this code on your own data.
```
path_to_file = tf.keras.utils.get_file('shakespeare.txt', 'https://storage.googleapis.com/download.tensorflow.org/data/shakespeare.txt')
```
Read text from the file and take a look at the first few lines.
```
with open(path_to_file) as f:
lines = f.read().splitlines()
for line in lines[:20]:
print(line)
```
Use the non empty lines to construct a `tf.data.TextLineDataset` object for next steps.
```
# TODO 2a
text_ds = tf.data.TextLineDataset(path_to_file).filter(lambda x: tf.cast(tf.strings.length(x), bool))
```
### Vectorize sentences from the corpus
You can use the `TextVectorization` layer to vectorize sentences from the corpus. Learn more about using this layer in this [Text Classification](https://www.tensorflow.org/tutorials/keras/text_classification) tutorial. Notice from the first few sentences above that the text needs to be in one case and punctuation needs to be removed. To do this, define a `custom_standardization function` that can be used in the TextVectorization layer.
```
# We create a custom standardization function to lowercase the text and
# remove punctuation.
def custom_standardization(input_data):
lowercase = tf.strings.lower(input_data)
return tf.strings.regex_replace(lowercase,
'[%s]' % re.escape(string.punctuation), '')
# Define the vocabulary size and number of words in a sequence.
vocab_size = 4096
sequence_length = 10
# Use the text vectorization layer to normalize, split, and map strings to
# integers. Set output_sequence_length length to pad all samples to same length.
vectorize_layer = TextVectorization(
standardize=custom_standardization,
max_tokens=vocab_size,
output_mode='int',
output_sequence_length=sequence_length)
```
Call `adapt` on the text dataset to create vocabulary.
```
vectorize_layer.adapt(text_ds.batch(1024))
```
Once the state of the layer has been adapted to represent the text corpus, the vocabulary can be accessed with `get_vocabulary()`. This function returns a list of all vocabulary tokens sorted (descending) by their frequency.
```
# Save the created vocabulary for reference.
inverse_vocab = vectorize_layer.get_vocabulary()
print(inverse_vocab[:20])
```
The vectorize_layer can now be used to generate vectors for each element in the `text_ds`.
```
def vectorize_text(text):
text = tf.expand_dims(text, -1)
return tf.squeeze(vectorize_layer(text))
# Vectorize the data in text_ds.
text_vector_ds = text_ds.batch(1024).prefetch(AUTOTUNE).map(vectorize_layer).unbatch()
```
### Obtain sequences from the dataset
You now have a `tf.data.Dataset` of integer encoded sentences. To prepare the dataset for training a Word2Vec model, flatten the dataset into a list of sentence vector sequences. This step is required as you would iterate over each sentence in the dataset to produce positive and negative examples.
Note: Since the `generate_training_data()` defined earlier uses non-TF python/numpy functions, you could also use a `tf.py_function` or `tf.numpy_function` with `tf.data.Dataset.map()`.
```
sequences = list(text_vector_ds.as_numpy_iterator())
print(len(sequences))
```
Take a look at few examples from `sequences`.
```
for seq in sequences[:5]:
print(f"{seq} => {[inverse_vocab[i] for i in seq]}")
```
### Generate training examples from sequences
`sequences` is now a list of int encoded sentences. Just call the `generate_training_data()` function defined earlier to generate training examples for the Word2Vec model. To recap, the function iterates over each word from each sequence to collect positive and negative context words. Length of target, contexts and labels should be same, representing the total number of training examples.
```
targets, contexts, labels = generate_training_data(
sequences=sequences,
window_size=2,
num_ns=4,
vocab_size=vocab_size,
seed=SEED)
print(len(targets), len(contexts), len(labels))
```
### Configure the dataset for performance
To perform efficient batching for the potentially large number of training examples, use the `tf.data.Dataset` API. After this step, you would have a `tf.data.Dataset` object of `(target_word, context_word), (label)` elements to train your Word2Vec model!
```
BATCH_SIZE = 1024
BUFFER_SIZE = 10000
dataset = tf.data.Dataset.from_tensor_slices(((targets, contexts), labels))
dataset = dataset.shuffle(BUFFER_SIZE).batch(BATCH_SIZE, drop_remainder=True)
print(dataset)
```
Add `cache()` and `prefetch()` to improve performance.
```
dataset = dataset.cache().prefetch(buffer_size=AUTOTUNE)
print(dataset)
```
## Lab Task 3: Model and Training
The Word2Vec model can be implemented as a classifier to distinguish between true context words from skip-grams and false context words obtained through negative sampling. You can perform a dot product between the embeddings of target and context words to obtain predictions for labels and compute loss against true labels in the dataset.
### Subclassed Word2Vec Model
Use the [Keras Subclassing API](https://www.tensorflow.org/guide/keras/custom_layers_and_models) to define your Word2Vec model with the following layers:
* `target_embedding`: A `tf.keras.layers.Embedding` layer which looks up the embedding of a word when it appears as a target word. The number of parameters in this layer are `(vocab_size * embedding_dim)`.
* `context_embedding`: Another `tf.keras.layers.Embedding` layer which looks up the embedding of a word when it appears as a context word. The number of parameters in this layer are the same as those in `target_embedding`, i.e. `(vocab_size * embedding_dim)`.
* `dots`: A `tf.keras.layers.Dot` layer that computes the dot product of target and context embeddings from a training pair.
* `flatten`: A `tf.keras.layers.Flatten` layer to flatten the results of `dots` layer into logits.
With the sublassed model, you can define the `call()` function that accepts `(target, context)` pairs which can then be passed into their corresponding embedding layer. Reshape the `context_embedding` to perform a dot product with `target_embedding` and return the flattened result.
Key point: The `target_embedding` and `context_embedding` layers can be shared as well. You could also use a concatenation of both embeddings as the final Word2Vec embedding.
```
class Word2Vec(Model):
def __init__(self, vocab_size, embedding_dim):
super(Word2Vec, self).__init__()
self.target_embedding = Embedding(vocab_size,
embedding_dim,
input_length=1,
name="w2v_embedding", )
self.context_embedding = Embedding(vocab_size,
embedding_dim,
input_length=num_ns+1)
self.dots = Dot(axes=(3,2))
self.flatten = Flatten()
def call(self, pair):
target, context = pair
we = self.target_embedding(target)
ce = self.context_embedding(context)
dots = self.dots([ce, we])
return self.flatten(dots)
```
### Define loss function and compile model
For simplicity, you can use `tf.keras.losses.CategoricalCrossEntropy` as an alternative to the negative sampling loss. If you would like to write your own custom loss function, you can also do so as follows:
``` python
def custom_loss(x_logit, y_true):
return tf.nn.sigmoid_cross_entropy_with_logits(logits=x_logit, labels=y_true)
```
It's time to build your model! Instantiate your Word2Vec class with an embedding dimension of 128 (you could experiment with different values). Compile the model with the `tf.keras.optimizers.Adam` optimizer.
```
# TODO 3a
embedding_dim = 128
word2vec = Word2Vec(vocab_size, embedding_dim)
word2vec.compile(optimizer='adam',
loss=tf.keras.losses.CategoricalCrossentropy(from_logits=True),
metrics=['accuracy'])
```
Also define a callback to log training statistics for tensorboard.
```
tensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir="logs")
```
Train the model with `dataset` prepared above for some number of epochs.
```
word2vec.fit(dataset, epochs=20, callbacks=[tensorboard_callback])
```
Tensorboard now shows the Word2Vec model's accuracy and loss.
```
!tensorboard --bind_all --port=8081 --load_fast=false --logdir logs
```
Run the following command in **Cloud Shell:**
<code>gcloud beta compute ssh --zone <instance-zone> <notebook-instance-name> --project <project-id> -- -L 8081:localhost:8081</code>
Make sure to replace <instance-zone>, <notebook-instance-name> and <project-id>.
In Cloud Shell, click *Web Preview* > *Change Port* and insert port number *8081*. Click *Change and Preview* to open the TensorBoard.

**To quit the TensorBoard, click Kernel > Interrupt kernel**.
## Lab Task 4: Embedding lookup and analysis
Obtain the weights from the model using `get_layer()` and `get_weights()`. The `get_vocabulary()` function provides the vocabulary to build a metadata file with one token per line.
```
# TODO 4a
weights = word2vec.get_layer('w2v_embedding').get_weights()[0]
vocab = vectorize_layer.get_vocabulary()
```
Create and save the vectors and metadata file.
```
out_v = io.open('vectors.tsv', 'w', encoding='utf-8')
out_m = io.open('metadata.tsv', 'w', encoding='utf-8')
for index, word in enumerate(vocab):
if index == 0: continue # skip 0, it's padding.
vec = weights[index]
out_v.write('\t'.join([str(x) for x in vec]) + "\n")
out_m.write(word + "\n")
out_v.close()
out_m.close()
```
Download the `vectors.tsv` and `metadata.tsv` to analyze the obtained embeddings in the [Embedding Projector](https://projector.tensorflow.org/).
```
try:
from google.colab import files
files.download('vectors.tsv')
files.download('metadata.tsv')
except Exception as e:
pass
```
## Next steps
This tutorial has shown you how to implement a skip-gram Word2Vec model with negative sampling from scratch and visualize the obtained word embeddings.
* To learn more about word vectors and their mathematical representations, refer to these [notes](https://web.stanford.edu/class/cs224n/readings/cs224n-2019-notes01-wordvecs1.pdf).
* To learn more about advanced text processing, read the [Transformer model for language understanding](https://www.tensorflow.org/tutorials/text/transformer) tutorial.
* If you’re interested in pre-trained embedding models, you may also be interested in [Exploring the TF-Hub CORD-19 Swivel Embeddings](https://www.tensorflow.org/hub/tutorials/cord_19_embeddings_keras), or the [Multilingual Universal Sentence Encoder](https://www.tensorflow.org/hub/tutorials/cross_lingual_similarity_with_tf_hub_multilingual_universal_encoder)
* You may also like to train the model on a new dataset (there are many available in [TensorFlow Datasets](https://www.tensorflow.org/datasets)).
| github_jupyter |
# Numbers and Integer Math
Watch the full [C# 101 video](https://www.youtube.com/watch?v=jEE0pWTq54U&list=PLdo4fOcmZ0oVxKLQCHpiUWun7vlJJvUiN&index=5) for this module.
## Integer Math
You have a few `integers` defined below. An `integer` is a positive or negative whole number.
> Before you run the code, what should c be?
## Addition
```
int a = 18;
int b = 6;
int c = a + b;
Console.WriteLine(c);
```
## Subtraction
```
int c = a - b;
Console.WriteLine(c);
```
## Multiplication
```
int c = a * b;
Console.WriteLine(c);
```
## Division
```
int c = a / b;
Console.WriteLine(c);
```
# Order of operations
C# follows the order of operation when it comes to math. That is, it does multiplication and division first, then addition and subtraction.
> What would the math be if C# didn't follow the order of operation, and instead just did math left to right?
```
int a = 5;
int b = 4;
int c = 2;
int d = a + b * c;
Console.WriteLine(d);
```
## Using parenthesis
You can also force different orders by putting parentheses around whatever you want done first
> Try it out
```
int d = (a + b) * c;
Console.WriteLine(d);
```
You can make math as long and complicated as you want.
> Can you make this line even more complicated?
```
int d = (a + b) - 6 * c + (12 * 4) / 3 + 12;
Console.WriteLine(d);
```
## Integers: Whole numbers no matter what
Integer math will always produce integers. What that means is that even when math should result in a decimal or fraction, the answer will be truncated to a whole number.
> Check it out. WHat should the answer truly be?
```
int a = 7;
int b = 4;
int c = 3;
int d = (a + b) / c;
Console.WriteLine(d);
```
# Playground
Play around with what you've learned! Here's some starting ideas:
> Do you have any homework or projects that need math? Try using code in place of a calculator!
>
> How do integers round? Do they always round up? down? to the nearest integer?
>
> How do the Order of Operations work? Play around with parentheses.
```
Console.WriteLine("Playground");
```
# Continue learning
There are plenty more resources out there to learn!
> [⏩ Next Module - Numbers and Integer Precision](http://tinyurl.com/csharp-notebook05)
>
> [⏪ Last Module - Searching Strings](http://tinyurl.com/csharp-notebook03)
>
> [Watch the video](https://www.youtube.com/watch?v=jEE0pWTq54U&list=PLdo4fOcmZ0oVxKLQCHpiUWun7vlJJvUiN&index=5)
>
> [Documentation: Numbers in C#](https://docs.microsoft.com/dotnet/csharp/tour-of-csharp/tutorials/numbers-in-csharp?WT.mc_id=Educationalcsharp-c9-scottha)
>
> [Start at the beginning: What is C#?](https://www.youtube.com/watch?v=BM4CHBmAPh4&list=PLdo4fOcmZ0oVxKLQCHpiUWun7vlJJvUiN&index=1)
# Other resources
Here's some more places to explore:
> [Other 101 Videos](https://dotnet.microsoft.com/learn/videos?WT.mc_id=csharpnotebook-35129-website)
>
> [Microsoft Learn](https://docs.microsoft.com/learn/dotnet/?WT.mc_id=csharpnotebook-35129-website)
>
> [C# Documentation](https://docs.microsoft.com/dotnet/csharp/?WT.mc_id=csharpnotebook-35129-website)
| github_jupyter |
## **Nigerian Music scraped from Spotify - an analysis**
Clustering is a type of [Unsupervised Learning](https://wikipedia.org/wiki/Unsupervised_learning) that presumes that a dataset is unlabelled or that its inputs are not matched with predefined outputs. It uses various algorithms to sort through unlabeled data and provide groupings according to patterns it discerns in the data.
[**Pre-lecture quiz**](https://white-water-09ec41f0f.azurestaticapps.net/quiz/27/)
### **Introduction**
[Clustering](https://link.springer.com/referenceworkentry/10.1007%2F978-0-387-30164-8_124) is very useful for data exploration. Let's see if it can help discover trends and patterns in the way Nigerian audiences consume music.
> ✅ Take a minute to think about the uses of clustering. In real life, clustering happens whenever you have a pile of laundry and need to sort out your family members' clothes 🧦👕👖🩲. In data science, clustering happens when trying to analyze a user's preferences, or determine the characteristics of any unlabeled dataset. Clustering, in a way, helps make sense of chaos, like a sock drawer.
In a professional setting, clustering can be used to determine things like market segmentation, determining what age groups buy what items, for example. Another use would be anomaly detection, perhaps to detect fraud from a dataset of credit card transactions. Or you might use clustering to determine tumors in a batch of medical scans.
✅ Think a minute about how you might have encountered clustering 'in the wild', in a banking, e-commerce, or business setting.
> 🎓 Interestingly, cluster analysis originated in the fields of Anthropology and Psychology in the 1930s. Can you imagine how it might have been used?
Alternately, you could use it for grouping search results - by shopping links, images, or reviews, for example. Clustering is useful when you have a large dataset that you want to reduce and on which you want to perform more granular analysis, so the technique can be used to learn about data before other models are constructed.
✅ Once your data is organized in clusters, you assign it a cluster Id, and this technique can be useful when preserving a dataset's privacy; you can instead refer to a data point by its cluster id, rather than by more revealing identifiable data. Can you think of other reasons why you'd refer to a cluster Id rather than other elements of the cluster to identify it?
### Getting started with clustering
> 🎓 How we create clusters has a lot to do with how we gather up the data points into groups. Let's unpack some vocabulary:
>
> 🎓 ['Transductive' vs. 'inductive'](https://wikipedia.org/wiki/Transduction_(machine_learning))
>
> Transductive inference is derived from observed training cases that map to specific test cases. Inductive inference is derived from training cases that map to general rules which are only then applied to test cases.
>
> An example: Imagine you have a dataset that is only partially labelled. Some things are 'records', some 'cds', and some are blank. Your job is to provide labels for the blanks. If you choose an inductive approach, you'd train a model looking for 'records' and 'cds', and apply those labels to your unlabeled data. This approach will have trouble classifying things that are actually 'cassettes'. A transductive approach, on the other hand, handles this unknown data more effectively as it works to group similar items together and then applies a label to a group. In this case, clusters might reflect 'round musical things' and 'square musical things'.
>
> 🎓 ['Non-flat' vs. 'flat' geometry](https://datascience.stackexchange.com/questions/52260/terminology-flat-geometry-in-the-context-of-clustering)
>
> Derived from mathematical terminology, non-flat vs. flat geometry refers to the measure of distances between points by either 'flat' ([Euclidean](https://wikipedia.org/wiki/Euclidean_geometry)) or 'non-flat' (non-Euclidean) geometrical methods.
>
> 'Flat' in this context refers to Euclidean geometry (parts of which are taught as 'plane' geometry), and non-flat refers to non-Euclidean geometry. What does geometry have to do with machine learning? Well, as two fields that are rooted in mathematics, there must be a common way to measure distances between points in clusters, and that can be done in a 'flat' or 'non-flat' way, depending on the nature of the data. [Euclidean distances](https://wikipedia.org/wiki/Euclidean_distance) are measured as the length of a line segment between two points. [Non-Euclidean distances](https://wikipedia.org/wiki/Non-Euclidean_geometry) are measured along a curve. If your data, visualized, seems to not exist on a plane, you might need to use a specialized algorithm to handle it.
<p >
<img src="../../images/flat-nonflat.png"
width="600"/>
<figcaption>Infographic by Dasani Madipalli</figcaption>
> 🎓 ['Distances'](https://web.stanford.edu/class/cs345a/slides/12-clustering.pdf)
>
> Clusters are defined by their distance matrix, e.g. the distances between points. This distance can be measured a few ways. Euclidean clusters are defined by the average of the point values, and contain a 'centroid' or center point. Distances are thus measured by the distance to that centroid. Non-Euclidean distances refer to 'clustroids', the point closest to other points. Clustroids in turn can be defined in various ways.
>
> 🎓 ['Constrained'](https://wikipedia.org/wiki/Constrained_clustering)
>
> [Constrained Clustering](https://web.cs.ucdavis.edu/~davidson/Publications/ICDMTutorial.pdf) introduces 'semi-supervised' learning into this unsupervised method. The relationships between points are flagged as 'cannot link' or 'must-link' so some rules are forced on the dataset.
>
> An example: If an algorithm is set free on a batch of unlabelled or semi-labelled data, the clusters it produces may be of poor quality. In the example above, the clusters might group 'round music things' and 'square music things' and 'triangular things' and 'cookies'. If given some constraints, or rules to follow ("the item must be made of plastic", "the item needs to be able to produce music") this can help 'constrain' the algorithm to make better choices.
>
> 🎓 'Density'
>
> Data that is 'noisy' is considered to be 'dense'. The distances between points in each of its clusters may prove, on examination, to be more or less dense, or 'crowded' and thus this data needs to be analyzed with the appropriate clustering method. [This article](https://www.kdnuggets.com/2020/02/understanding-density-based-clustering.html) demonstrates the difference between using K-Means clustering vs. HDBSCAN algorithms to explore a noisy dataset with uneven cluster density.
Deepen your understanding of clustering techniques in this [Learn module](https://docs.microsoft.com/learn/modules/train-evaluate-cluster-models?WT.mc_id=academic-15963-cxa)
### **Clustering algorithms**
There are over 100 clustering algorithms, and their use depends on the nature of the data at hand. Let's discuss some of the major ones:
- **Hierarchical clustering**. If an object is classified by its proximity to a nearby object, rather than to one farther away, clusters are formed based on their members' distance to and from other objects. Hierarchical clustering is characterized by repeatedly combining two clusters.
<p >
<img src="../../images/hierarchical.png"
width="600"/>
<figcaption>Infographic by Dasani Madipalli</figcaption>
- **Centroid clustering**. This popular algorithm requires the choice of 'k', or the number of clusters to form, after which the algorithm determines the center point of a cluster and gathers data around that point. [K-means clustering](https://wikipedia.org/wiki/K-means_clustering) is a popular version of centroid clustering which separates a data set into pre-defined K groups. The center is determined by the nearest mean, thus the name. The squared distance from the cluster is minimized.
<p >
<img src="../../images/centroid.png"
width="600"/>
<figcaption>Infographic by Dasani Madipalli</figcaption>
- **Distribution-based clustering**. Based in statistical modeling, distribution-based clustering centers on determining the probability that a data point belongs to a cluster, and assigning it accordingly. Gaussian mixture methods belong to this type.
- **Density-based clustering**. Data points are assigned to clusters based on their density, or their grouping around each other. Data points far from the group are considered outliers or noise. DBSCAN, Mean-shift and OPTICS belong to this type of clustering.
- **Grid-based clustering**. For multi-dimensional datasets, a grid is created and the data is divided amongst the grid's cells, thereby creating clusters.
The best way to learn about clustering is to try it for yourself, so that's what you'll do in this exercise.
We'll require some packages to knock-off this module. You can have them installed as: `install.packages(c('tidyverse', 'tidymodels', 'DataExplorer', 'summarytools', 'plotly', 'paletteer', 'corrplot', 'patchwork'))`
Alternatively, the script below checks whether you have the packages required to complete this module and installs them for you in case some are missing.
```
suppressWarnings(if(!require("pacman")) install.packages("pacman"))
pacman::p_load('tidyverse', 'tidymodels', 'DataExplorer', 'summarytools', 'plotly', 'paletteer', 'corrplot', 'patchwork')
```
## Exercise - cluster your data
Clustering as a technique is greatly aided by proper visualization, so let's get started by visualizing our music data. This exercise will help us decide which of the methods of clustering we should most effectively use for the nature of this data.
Let's hit the ground running by importing the data.
```
# Load the core tidyverse and make it available in your current R session
library(tidyverse)
# Import the data into a tibble
df <- read_csv(file = "https://raw.githubusercontent.com/microsoft/ML-For-Beginners/main/5-Clustering/data/nigerian-songs.csv")
# View the first 5 rows of the data set
df %>%
slice_head(n = 5)
```
Sometimes, we may want some little more information on our data. We can have a look at the `data` and `its structure` by using the [*glimpse()*](https://pillar.r-lib.org/reference/glimpse.html) function:
```
# Glimpse into the data set
df %>%
glimpse()
```
Good job!💪
We can observe that `glimpse()` will give you the total number of rows (observations) and columns (variables), then, the first few entries of each variable in a row after the variable name. In addition, the *data type* of the variable is given immediately after each variable's name inside `< >`.
`DataExplorer::introduce()` can summarize this information neatly:
```
# Describe basic information for our data
df %>%
introduce()
# A visual display of the same
df %>%
plot_intro()
```
Awesome! We have just learnt that our data has no missing values.
While we are at it, we can explore common central tendency statistics (e.g [mean](https://en.wikipedia.org/wiki/Arithmetic_mean) and [median](https://en.wikipedia.org/wiki/Median)) and measures of dispersion (e.g [standard deviation](https://en.wikipedia.org/wiki/Standard_deviation)) using `summarytools::descr()`
```
# Describe common statistics
df %>%
descr(stats = "common")
```
Let's look at the general values of the data. Note that popularity can be `0`, which show songs that have no ranking. We'll remove those shortly.
> 🤔 If we are working with clustering, an unsupervised method that does not require labeled data, why are we showing this data with labels? In the data exploration phase, they come in handy, but they are not necessary for the clustering algorithms to work.
### 1. Explore popular genres
Let's go ahead and find out the most popular genres 🎶 by making a count of the instances it appears.
```
# Popular genres
top_genres <- df %>%
count(artist_top_genre, sort = TRUE) %>%
# Encode to categorical and reorder the according to count
mutate(artist_top_genre = factor(artist_top_genre) %>% fct_inorder())
# Print the top genres
top_genres
```
That went well! They say a picture is worth a thousand rows of a data frame (actually nobody ever says that 😅). But you get the gist of it, right?
One way to visualize categorical data (character or factor variables) is using barplots. Let's make a barplot of the top 10 genres:
```
# Change the default gray theme
theme_set(theme_light())
# Visualize popular genres
top_genres %>%
slice(1:10) %>%
ggplot(mapping = aes(x = artist_top_genre, y = n,
fill = artist_top_genre)) +
geom_col(alpha = 0.8) +
paletteer::scale_fill_paletteer_d("rcartocolor::Vivid") +
ggtitle("Top genres") +
theme(plot.title = element_text(hjust = 0.5),
# Rotates the X markers (so we can read them)
axis.text.x = element_text(angle = 90))
```
Now it's way easier to identify that we have `missing` genres 🧐!
> A good visualisation will show you things that you did not expect, or raise new questions about the data - Hadley Wickham and Garrett Grolemund, [R For Data Science](https://r4ds.had.co.nz/introduction.html)
Note, when the top genre is described as `Missing`, that means that Spotify did not classify it, so let's get rid of it.
```
# Visualize popular genres
top_genres %>%
filter(artist_top_genre != "Missing") %>%
slice(1:10) %>%
ggplot(mapping = aes(x = artist_top_genre, y = n,
fill = artist_top_genre)) +
geom_col(alpha = 0.8) +
paletteer::scale_fill_paletteer_d("rcartocolor::Vivid") +
ggtitle("Top genres") +
theme(plot.title = element_text(hjust = 0.5),
# Rotates the X markers (so we can read them)
axis.text.x = element_text(angle = 90))
```
From the little data exploration, we learn that the top three genres dominate this dataset. Let's concentrate on `afro dancehall`, `afropop`, and `nigerian pop`, additionally filter the dataset to remove anything with a 0 popularity value (meaning it was not classified with a popularity in the dataset and can be considered noise for our purposes):
```
nigerian_songs <- df %>%
# Concentrate on top 3 genres
filter(artist_top_genre %in% c("afro dancehall", "afropop","nigerian pop")) %>%
# Remove unclassified observations
filter(popularity != 0)
# Visualize popular genres
nigerian_songs %>%
count(artist_top_genre) %>%
ggplot(mapping = aes(x = artist_top_genre, y = n,
fill = artist_top_genre)) +
geom_col(alpha = 0.8) +
paletteer::scale_fill_paletteer_d("ggsci::category10_d3") +
ggtitle("Top genres") +
theme(plot.title = element_text(hjust = 0.5))
```
Let's see whether there is any apparent linear relationship among the numerical variables in our data set. This relationship is quantified mathematically by the [correlation statistic](https://en.wikipedia.org/wiki/Correlation).
The correlation statistic is a value between -1 and 1 that indicates the strength of a relationship. Values above 0 indicate a *positive* correlation (high values of one variable tend to coincide with high values of the other), while values below 0 indicate a *negative* correlation (high values of one variable tend to coincide with low values of the other).
```
# Narrow down to numeric variables and fid correlation
corr_mat <- nigerian_songs %>%
select(where(is.numeric)) %>%
cor()
# Visualize correlation matrix
corrplot(corr_mat, order = 'AOE', col = c('white', 'black'), bg = 'gold2')
```
The data is not strongly correlated except between `energy` and `loudness`, which makes sense, given that loud music is usually pretty energetic. `Popularity` has a correspondence to `release date`, which also makes sense, as more recent songs are probably more popular. Length and energy seem to have a correlation too.
It will be interesting to see what a clustering algorithm can make of this data!
> 🎓 Note that correlation does not imply causation! We have proof of correlation but no proof of causation. An [amusing web site](https://tylervigen.com/spurious-correlations) has some visuals that emphasize this point.
### 2. Explore data distribution
Let's ask some more subtle questions. Are the genres significantly different in the perception of their danceability, based on their popularity? Let's examine our top three genres data distribution for popularity and danceability along a given x and y axis using [density plots](https://www.khanacademy.org/math/ap-statistics/density-curves-normal-distribution-ap/density-curves/v/density-curves).
```
# Perform 2D kernel density estimation
density_estimate_2d <- nigerian_songs %>%
ggplot(mapping = aes(x = popularity, y = danceability, color = artist_top_genre)) +
geom_density_2d(bins = 5, size = 1) +
paletteer::scale_color_paletteer_d("RSkittleBrewer::wildberry") +
xlim(-20, 80) +
ylim(0, 1.2)
# Density plot based on the popularity
density_estimate_pop <- nigerian_songs %>%
ggplot(mapping = aes(x = popularity, fill = artist_top_genre, color = artist_top_genre)) +
geom_density(size = 1, alpha = 0.5) +
paletteer::scale_fill_paletteer_d("RSkittleBrewer::wildberry") +
paletteer::scale_color_paletteer_d("RSkittleBrewer::wildberry") +
theme(legend.position = "none")
# Density plot based on the danceability
density_estimate_dance <- nigerian_songs %>%
ggplot(mapping = aes(x = danceability, fill = artist_top_genre, color = artist_top_genre)) +
geom_density(size = 1, alpha = 0.5) +
paletteer::scale_fill_paletteer_d("RSkittleBrewer::wildberry") +
paletteer::scale_color_paletteer_d("RSkittleBrewer::wildberry")
# Patch everything together
library(patchwork)
density_estimate_2d / (density_estimate_pop + density_estimate_dance)
```
We see that there are concentric circles that line up, regardless of genre. Could it be that Nigerian tastes converge at a certain level of danceability for this genre?
In general, the three genres align in terms of their popularity and danceability. Determining clusters in this loosely-aligned data will be a challenge. Let's see whether a scatter plot can support this.
```
# A scatter plot of popularity and danceability
scatter_plot <- nigerian_songs %>%
ggplot(mapping = aes(x = popularity, y = danceability, color = artist_top_genre, shape = artist_top_genre)) +
geom_point(size = 2, alpha = 0.8) +
paletteer::scale_color_paletteer_d("futurevisions::mars")
# Add a touch of interactivity
ggplotly(scatter_plot)
```
A scatterplot of the same axes shows a similar pattern of convergence.
In general, for clustering, you can use scatterplots to show clusters of data, so mastering this type of visualization is very useful. In the next lesson, we will take this filtered data and use k-means clustering to discover groups in this data that see to overlap in interesting ways.
## **🚀 Challenge**
In preparation for the next lesson, make a chart about the various clustering algorithms you might discover and use in a production environment. What kinds of problems is the clustering trying to address?
## [**Post-lecture quiz**](https://white-water-09ec41f0f.azurestaticapps.net/quiz/28/)
## **Review & Self Study**
Before you apply clustering algorithms, as we have learned, it's a good idea to understand the nature of your dataset. Read more on this topic [here](https://www.kdnuggets.com/2019/10/right-clustering-algorithm.html)
Deepen your understanding of clustering techniques:
- [Train and Evaluate Clustering Models using Tidymodels and friends](https://rpubs.com/eR_ic/clustering)
- Bradley Boehmke & Brandon Greenwell, [*Hands-On Machine Learning with R*](https://bradleyboehmke.github.io/HOML/)*.*
## **Assignment**
[Research other visualizations for clustering](https://github.com/microsoft/ML-For-Beginners/blob/main/5-Clustering/1-Visualize/assignment.md)
## THANK YOU TO:
[Jen Looper](https://www.twitter.com/jenlooper) for creating the original Python version of this module ♥️
[`Dasani Madipalli`](https://twitter.com/dasani_decoded) for creating the amazing illustrations that make machine learning concepts more interpretable and easier to understand.
Happy Learning,
[Eric](https://twitter.com/ericntay), Gold Microsoft Learn Student Ambassador.
| github_jupyter |
# ADMM Optimizer
## Introduction
The ADMM Optimizer can solve classes of mixed-binary constrained optimization problems, hereafter (MBCO), which often appear in logistic, finance, and operation research. In particular, the ADMM Optimizer here designed can tackle the following optimization problem $(P)$:
$$
\min_{x \in \mathcal{X},u\in\mathcal{U} \subseteq \mathbb{R}^l } \quad q(x) + \varphi(u),
$$
subject to the constraints:
$$
\mathrm{s.t.:~} \quad G x = b, \quad g(x) \leq 0, \quad \ell(x, u) \leq 0,
$$
with the corresponding functional assumptions.
1. Function $q: \mathbb{R}^n \to \mathbb{R}$ is quadratic, i.e., $q(x) = x^{\intercal} Q x + a^{\intercal} x$ for a given symmetric squared matrix $Q \in \mathbb{R}^n \times \mathbb{R}^n, Q = Q^{\intercal}$, and vector $a \in \mathbb{R}^n$;
2. The set $\mathcal{X} = \{0,1\}^n = \{x_{(i)} (1-x_{(i)}) = 0, \forall i\}$ enforces the binary constraints;
3. Matrix $G\in\mathbb{R}^n \times \mathbb{R}^{n'}$, vector $b \in \mathbb{R}^{n'}$, and function $g: \mathbb{R}^n \to \mathbb{R}$ is convex;
4. Function $\varphi: \mathbb{R}^l \to \mathbb{R}$ is convex and $\mathcal{U}$ is a convex set;
5. Function $\ell: \mathbb{R}^n\times \mathbb{R}^l \to \mathbb{R}$ is *jointly* convex in $x, u$.
In order to solve MBO problems, [1] proposed heuristics for $(P)$ based on the Alternating Direction Method of Multipliers (ADMM) [2]. ADMM is an operator splitting algorithm with a long history in convex optimization, and it is known to have residual, objective and dual variable convergence properties, provided that convexity assumptions are holding.
The method of [1] (referred to as 3-ADMM-H) leverages the ADMM operator-splitting procedure to devise a decomposition for certain classes of MBOs into:
- a QUBO subproblem to be solved by on the quantum device via variational algorithms, such as VQE or QAOA;
- continuous convex constrained subproblem, which can be efficiently solved with classical optimization solvers.
The algorithm 3-ADMM-H works as follows:
0. Initialization phase (set the parameters and the QUBO and convex solvers);
1. For each ADMM iterations ($k = 1, 2, \ldots, $) untill termination:
- Solve a properly defined QUBO subproblem (with a classical or quantum solver);
- Solve properly defined convex problems (with a classical solver);
- Update the dual variables.
2. Return optimizers and cost.
A comprehensive discussion on the conditions for convergence, feasibility and optimality of the algorithm can be found in [1]. A variant with 2 ADMM blocks, namely a QUBO subproblem, and a continuous convex constrained subproblem, is also introduced in [1].
## References
[1] [C. Gambella and A. Simonetto, *Multi-block ADMM heuristics for mixed-binary optimization, on classical and quantum computers,* arXiv preprint arXiv:2001.02069 (2020).](https://arxiv.org/abs/2001.02069)
[2] [S. Boyd, N. Parikh, E. Chu, B. Peleato, and J. Eckstein, *Distributed optimization and statistical learning via the alternating direction method of multipliers,* Foundations and Trends in Machine learning, 3, 1–122 (2011).](https://web.stanford.edu/~boyd/papers/pdf/admm_distr_stats.pdf)
## Initialization
First of all we load all the packages that we need.
```
import time
from typing import List, Optional, Any
import numpy as np
import matplotlib.pyplot as plt
from docplex.mp.model import Model
from qiskit import BasicAer
from qiskit.aqua.algorithms import QAOA, NumPyMinimumEigensolver
from qiskit.optimization.algorithms import CobylaOptimizer, MinimumEigenOptimizer
from qiskit.optimization.problems import QuadraticProgram
from qiskit.optimization.algorithms.admm_optimizer import ADMMParameters, ADMMOptimizer
# If CPLEX is installed, you can uncomment this line to import the CplexOptimizer.
# CPLEX can be used in this tutorial to solve the convex continuous problem,
# but also as a reference to solve the QUBO, or even the full problem.
#
# from qiskit.optimization.algorithms import CplexOptimizer
```
We first initialize all the algorithms we plan to use later in this tutorial.
To solve the QUBO problems we can choose between
- `MinimumEigenOptimizer` using different `MinimumEigensolver`, such as `VQE`, `QAOA` or `NumpyMinimumEigensolver` (classical)
- `GroverOptimizer`
- `CplexOptimizer` (classical, if CPLEX is installed)
and to solve the convex continuous problems we can choose between the following classical solvers:
- `CplexOptimizer` (if CPLEX is installed)
- `CobylaOptimizer`
In case CPLEX is not available, the `CobylaOptimizer` (for convex continuous problems) and the `MinimumEigenOptimizer` using the `NumpyMinimumEigensolver` (for QUBOs) can be used as classical alternatives to CPLEX for testing, validation, and benchmarking.
```
# define COBYLA optimizer to handle convex continuous problems.
cobyla = CobylaOptimizer()
# define QAOA via the minimum eigen optimizer
qaoa = MinimumEigenOptimizer(QAOA(quantum_instance=BasicAer.get_backend('statevector_simulator')))
# exact QUBO solver as classical benchmark
exact = MinimumEigenOptimizer(NumPyMinimumEigensolver()) # to solve QUBOs
# in case CPLEX is installed it can also be used for the convex problems, the QUBO,
# or as a benchmark for the full problem.
#
# cplex = CplexOptimizer()
```
## Example
We test 3-ADMM-H algorithm on a simple Mixed-Binary Quadratic Problem with equality and inequality constraints (Example 6 reported in [1]). We first construct a docplex problem and then load it into a `QuadraticProgram`.
```
# construct model using docplex
mdl = Model('ex6')
v = mdl.binary_var(name='v')
w = mdl.binary_var(name='w')
t = mdl.binary_var(name='t')
u = mdl.continuous_var(name='u')
mdl.minimize(v + w + t + 5 * (u-2)**2)
mdl.add_constraint(v + 2 * w + t + u <= 3, "cons1")
mdl.add_constraint(v + w + t >= 1, "cons2")
mdl.add_constraint(v + w == 1, "cons3")
# load quadratic program from docplex model
qp = QuadraticProgram()
qp.from_docplex(mdl)
print(qp.export_as_lp_string())
```
## Classical Solution
3-ADMM-H needs a QUBO optimizer to solve the QUBO subproblem, and a continuous optimizer to solve the continuous convex constrained subproblem. We first solve the problem classically: we use the `MinimumEigenOptimizer` with the `NumPyMinimumEigenSolver` as a classical and exact QUBO solver and we use the `CobylaOptimizer` as a continuous convex solver. 3-ADMM-H supports any other suitable solver available in Qiskit. For instance, VQE, QAOA, and GroverOptimizer can be invoked as quantum solvers, as demonstrated later.
If CPLEX is installed, the `CplexOptimizer` can also be used as both, a QUBO and convex solver.
### Parameters
The 3-ADMM-H are wrapped in class `ADMMParameters`. Customized parameter values can be set as arguments of the class. In this example, parameters $\rho, \beta$ are initialized to $1001$ and $1000$, respectively. The penalization `factor_c` of equality constraints $Gx = b$ is set to $900$. The tolerance `tol` for primal residual convergence is set to `1.e-6`.
In this case, the 3-block implementation is guaranteed to converge for Theorem 4 of [1], because the inequality constraint with the continuous variable is always active. The 2-block implementation can be run by setting `three_block=False`, and practically converges to a feasible not optimal solution.
```
admm_params = ADMMParameters(
rho_initial=1001,
beta=1000,
factor_c=900,
max_iter=100,
three_block=True, tol=1.e-6
)
```
### Calling 3-ADMM-H algorithm
To invoke the 3-ADMM-H algorithm, an instance of the `ADMMOptimizer` class needs to be created. This takes ADMM-specific parameters and the subproblem optimizers separately into the constructor. The solution returned is an instance of `OptimizationResult` class.
```
# define QUBO optimizer
qubo_optimizer = exact
# qubo_optimizer = cplex # uncomment to use CPLEX instead
# define classical optimizer
convex_optimizer = cobyla
# convex_optimizer = cplex # uncomment to use CPLEX instead
# initialize ADMM with classical QUBO and convex optimizer
admm = ADMMOptimizer(params=admm_params,
qubo_optimizer=qubo_optimizer,
continuous_optimizer=convex_optimizer)
# run ADMM to solve problem
result = admm.solve(qp)
```
### Classical Solver Result
The 3-ADMM-H solution can be then printed and visualized. The `x` attribute of the solution contains respectively, the
values of the binary decision variables and the values of the continuous decision variables. The `fval` is the objective
value of the solution.
```
print("x={}".format(result.x))
print("fval={:.2f}".format(result.fval))
```
Solution statistics can be accessed in the `state` field and visualized. We here display the convergence of 3-ADMM-H, in terms of primal residuals.
```
plt.plot(result.state.residuals)
plt.xlabel("Iterations")
plt.ylabel("Residuals")
plt.show()
```
## Quantum Solution
We now solve the same optimization problem with QAOA as QUBO optimizer, running on simulated quantum device.
First, one need to select the classical optimizer of the eigensolver QAOA. Then, the simulation backened is set. Finally,
the eigensolver is wrapped into the `MinimumEigenOptimizer` class. A new instance of `ADMMOptimizer` is populated with QAOA as QUBO optimizer.
```
# define QUBO optimizer
qubo_optimizer = qaoa
# define classical optimizer
convex_optimizer = cobyla
# convex_optimizer = cplex # uncomment to use CPLEX instead
# initialize ADMM with quantum QUBO optimizer and classical convex optimizer
admm_q = ADMMOptimizer(params=admm_params,
qubo_optimizer=qubo_optimizer,
continuous_optimizer=convex_optimizer)
# run ADMM to solve problem
result_q = admm_q.solve(qp)
```
### Quantum Solver Results
Here we present the results obtained from the quantum solver. As in the example above `x` stands for the solution, the `fval` is for objective value.
```
print("x={}".format(result_q.x))
print("fval={:.2f}".format(result_q.fval))
plt.clf()
plt.plot(result_q.state.residuals)
plt.xlabel("Iterations")
plt.ylabel("Residuals")
plt.show()
import qiskit.tools.jupyter
%qiskit_version_table
%qiskit_copyright
```
| github_jupyter |
# 4 Setting the initial SoC
Setting the initial SoC for your pack is performed with an argument passed to the solve algorithm. Currently the same value is applied to each battery but in future it will be possible to vary the SoC across the pack.
```
import liionpack as lp
import pybamm
import numpy as np
import matplotlib.pyplot as plt
```
Lets set up the most simple pack possible with 1 battery and very low busbar resistance to compare to a pure PyBaMM simulation
```
Rsmall = 1e-6
netlist = lp.setup_circuit(Np=1, Ns=1, Rb=Rsmall, Rc=Rsmall, Ri=5e-2, V=4.0, I=1.0)
# Heat transfer coefficients
htc = np.ones(1) * 10
# PyBaMM parameters
chemistry = pybamm.parameter_sets.Chen2020
parameter_values = pybamm.ParameterValues(chemistry=chemistry)
# Cycling experiment
experiment = pybamm.Experiment(
[
(
"Discharge at 1 A for 1000 s or until 3.3 V",
"Rest for 1000 s",
"Charge at 1 A for 1000 s or until 4.0 V",
"Rest for 1000 s",
)
]
* 3, period="10 s"
)
SoC = 0.5
# Solve pack
output = lp.solve(netlist=netlist,
parameter_values=parameter_values,
experiment=experiment,
htc=htc, initial_soc=SoC)
```
Let's compare to the PyBaMM simulation
```
parameter_values = pybamm.ParameterValues(chemistry=chemistry)
parameter_values.update({"Total heat transfer coefficient [W.m-2.K-1]": 10.0})
sim = lp.create_simulation(parameter_values, experiment, make_inputs=False)
sol = sim.solve(initial_soc=SoC)
def compare(sol, output):
# Get pack level results
time = sol["Time [s]"].entries
v_pack = output["Pack terminal voltage [V]"]
i_pack = output["Pack current [A]"]
v_batt = sol["Terminal voltage [V]"].entries
i_batt = sol["Current [A]"].entries
# Plot pack voltage and current
_, (axl, axr) = plt.subplots(1, 2, tight_layout=True, figsize=(15, 10), sharex=True, sharey=True)
axl.plot(time[1:], v_pack, color="green", label="simulation")
axl.set_xlabel("Time [s]")
axl.set_ylabel("Pack terminal voltage [V]", color="green")
axl2 = axl.twinx()
axl2.plot(time[1:], i_pack, color="black", label="simulation")
axl2.set_ylabel("Pack current [A]", color="black")
axl2.set_title("Liionpack Simulation")
axr.plot(time, v_batt, color="red", label="simulation")
axr.set_xlabel("Time [s]")
axr.set_ylabel("Battery terminal voltage [V]", color="red")
axr2 = axr.twinx()
axr2.plot(time, i_batt, color="blue", label="simulation")
axr2.set_ylabel("Battery current [A]", color="blue")
axr2.set_title("Single PyBaMM Simulation")
compare(sol, output)
```
Now lets start the simulation from a different state of charge
```
SoC = 0.25
# Solve pack
output = lp.solve(netlist=netlist,
parameter_values=parameter_values,
experiment=experiment,
htc=htc, initial_soc=SoC)
compare(sol, output)
```
Here we are still comparing to the PyBaMM simulation at 0.5 SoC and we can see that liionpack started at a lower voltage corresponding to a lower SoC.
```
parameter_values = pybamm.ParameterValues(chemistry=chemistry)
parameter_values.update({"Total heat transfer coefficient [W.m-2.K-1]": 10.0})
sim = lp.create_simulation(parameter_values, experiment, make_inputs=False)
sol = sim.solve(initial_soc=SoC)
```
Now we can re-run the PyBaMM simulation and compare again
```
compare(sol, output)
lp.draw_circuit(netlist)
```
| github_jupyter |
# Découverte du format CSV - *Comma-Separated values*
**Plan du document**
- Le format **CSV**
- Représenter des données CSV avec Python
- Première solution: un tableau de tuples
- **Deuxième solution**: un tableau de *tuples nommés* (dictionnaires)
- l'*unpacking*,
- l'opération *zip*
- syntaxe en *compréhension des dictionnaires*
- **synthèse**: CSV -> tableau de tuples nommés
## Le format CSV
*Comma*: virgule; *CSV*: valeurs séparées par des virgule.
CSV est un format **textuel** (par opposition à *binaire*) qui sert à représenter des **données en tables**; voici à quoi cela ressemble:
```
nom,prenom,date_naissance
Durand,Jean-Pierre,23/05/1985
Dupont,Christophe,15/12/1967
Terta,Henry,12/06/1978
```
On devine qu'il s'agit d'informations à propos d'individus: Jean pierre Durand né le 23 mai 1985, etc. En informatique on parle de **collection de données**.
La première ligne précise le sens des valeurs trouvées aux lignes suivantes; ses valeurs `nom`, `prenom`, `date_naissance` sont appelées **descripteurs** ou encore **attributs**.
Les lignes suivantes correspondent à des individus différents; en informatique on parle souvent d'**objets** ou d'**entités**.
Chaque «*objet*» (ici individu) correspond à une ligne: les **valeurs** qu'on y trouve sont associées aux *descripteurs* de même position.
On peut (re)présenter la même information plus agréablement avec un rendu table:
| nom | prenom | date_naissance |
| ------------- |:-------------:| -----:|
| Durand | Jean-Pierre | 23/05/1985 |
| Dupont | Christophe | 15/12/1967 |
| Tertra | Henry | 12/06/1978 |
## Représenter des données CSV avec Python
### Première solution: une liste de tuples
Cela donnerait `[('Durand', 'Jean-Pierre', '23/05/1985'), ('Dupont',..),(..)]`
On y parvient assez simplement à l'aide de `str.split(..)`:
```
donnees_CSV = """nom,prenom,date_naissance
Durand,Jean-Pierre,23/05/1985
Dupont,Christophe,15/12/1967
Terta,Henry,12/06/1978"""
etape1 = donnees_CSV.split('\n')
etape1
etape2 = [obj.split(',') for obj in etape1]
etape2 # une liste de liste
etape3 = [tuple(obj) for obj in etape2]
etape3 # une liste de tuple
fin = etape3[1:] # un petit slice
fin # sans l'en-tête
```
#### À faire toi-même
On peut parvenir à `fin` à partir de `donnees_CSV` en **une seule fois** par *composition* ... essais!
```
# deux_en_un = [ obj.split(',') for obj in donnees_CSV.split('\n') ]
# trois_en_un = [ tuple( obj.split(',') ) for obj in donnees_CSV.split('\n') ]
# tu peux essayer de faire deux_en_un, puis trois_en_un avant.
quatre_en_un = [ tuple( obj.split(',') ) for obj in donnees_CSV.split('\n') ][1:]
# pour tester
assert quatre_en_un == fin
```
___
L'inconvénient de cette représentation c'est qu'elle «oublie» les descripteurs.
Pourquoi ne pas les conserver comme à l'étape3? Pour éviter d'avoir un tableau *hétérogène*: le premier élément ne serait pas un «objet». De tels tableaux sont plus difficile à manipuler.
### Deuxième solution: un tableau de *tuples nommés*
**n-uplet (ou tuples) nommés**: tuple dont chaque valeur est associée à un descripteur.
Malheureusement Python ne possède pas un tel type par défaut (il existe toutefois dans la bibliothèque standard).
Pour représenter ce type, nous utiliserons un dictionnaire dont les clés sont les descripteurs; voici un exemple:
```python
{'nom': 'Durand', 'prenom': 'Jean-Pierre', 'date_naissance': '23/05/1985'}
```
Pour y parvenir, nous partons de:
```
donnees_CSV = """nom,prenom,date_naissance
Durand,Jean-Pierre,23/05/1985
Dupont,Christophe,15/12/1967
Terta,Henry,12/06/1978"""
```
Les étapes qui suivent servent à séparer les descripteurs et les objets:
```
tmp = donnees_CSV.split('\n')
tmp
descripteurs_str = tmp[0]
descripteurs = tuple(descripteurs_str.split(','))
print(f"le tuple des descripteurs: {descripteurs}")
donnees_str = tmp[1:]
donnees_str
objets = [tuple(obj.split(',')) for obj in donnees_str]
print(f"la liste des objets (des personnes ici):\n {objets}")
```
#### À faire toi-même
Peux-tu compléter les parties manquantes pour obtenir le même résultat plus rapidement?
```
descripteurs = tuple( donnees_CSV.split('\n')[0].split(',') )
objets = [ tuple( ligne.split(',') ) for ligne in donnees_CSV.split('\n')[1:] ]
print(f"- les descripteurs:\n\t {descripteurs}\n- les objets:\n\t {objets}")
```
______
#### À faire toi-même - *découverte de l'**unpacking** (déballage)*
Peux-tu réaliser le traitement précédent en **vraiment** une seule ligne? Pour cela observe les trois exemples qui suivent:
```
# exemple1 d'unpacking
tete, *queue = [1, 2, 3, 4]
print(f"La tête: {tete} et la queue: {queue}")
# exemple2 d'unpacking
un, deux, *reste = [1, 2, 3, 4]
print(f"un: {un}\ndeux: {deux}\nreste: {reste}")
# exemple3 d'unpacking
tete, *corps, pied = [1,2,3,4]
print(f"tete: {tete}\ncorps: {corps}\npied: {pied}")
# À toi de jouer!
descripteurs, *objets = [tuple(d.split(',')) for d in donnees_CSV.split('\n')]
print(f"les descripteurs:\n\t {descripteurs}\nles objets:\n\t {objets}")
```
____
Arrivé à ce stade nous voudrions combiner:
- `('descr1', 'descr2', ...)` et `('v1', 'v2', ...)` en ...
- `{'descr1': 'v1', 'descr2': 'v2', ..}` (n-uplet nommé)
#### Appareiller deux séquences - `zip`
On a souvent besoin de grouper par paires deux séquences de même longueur `len`.
*Ex*: je **dispose** de `['a', 'b', 'c']` et `[3, 2, 1]`
j'ai **besoin de** `[('a', 3), ('b', 2), ('c', 1)]`.
#### À faire toi-même
La fonction `appareiller(t1, t2)` prend deux tableaux de même taille en argument et renvoie un tableau obtenue en appararillant les éléments de `t1` et `t2` de même index.
Compléter le code qui suit pour résoudre ce problème
```
def appareiller(t1, t2):
assert len(t1) == len(t2)
t = []
for i in range(len(t1)):
couple = (t1[i], t2[i])
t.append( couple )
return t
# autre solution avec la syntaxe en compréhension
def appareiller2(t1, t2):
assert len(t1) == len(t2)
return [
(t1[i], t2[i])
for i in range(len(t1))
]
# vérifier votre solution
tab1 = ['a', 'b', 'c']
tab2 = [3, 2, 1]
assert appareiller(tab1, tab2) == [('a', 3), ('b', 2), ('c', 1)]
assert appareiller2(tab1, tab2) == [('a', 3), ('b', 2), ('c', 1)]
```
___
Un cas d'utilisation fréquent de l'apparaillement est la lecture dans une boucle des paires
```
# tester moi
tab1 = ['a', 'b', 'c']
tab2 = [3, 2, 1]
for a, b in appareiller(tab1, tab2):
print(f'a vaut "{a}" et b vaut "{b}"')
```
en fait, Python dispose d'une fonction prédéfinie `zip(seq1, seq2, ...)` qui fait la même chose avec des «séquences» (`list` est un cas particulier de séquence).
*note*: `zip`?? penser à la «fermeture-éclair» d'un blouson ...
```
z = zip(tab1, tab2)
print(z)
print(list(z))
```
*note*: elle renvoie un objet spécial de type `zip` car on l'utilise souvent dans une boucle directement c'est-à-dire sans mémoriser le zip (un peu comme avec `range`)
```
# tester moi
tab1 = ['a', 'b', 'c']
tab2 = [3, 2, 1]
for a, b in zip(tab1, tab):
print(f'a vaut "{a}" et b vaut "{b}"')
```
#### Découverte: la syntaxe en compréhension est aussi valable pour les `dict`
Voici un exemple simple:
```
modele_tuple_nomme = {desc: None for desc in descripteurs}
modele_tuple_nomme
```
Bien Noter que la partie avant `for` est de la forme `<cle>: <val>`.
On utilise généralement cela avec `zip`:
```
cles = ("cle1", "cle2", "cle3")
valeurs = ("ah", "oh", "hein")
{c: v for c, v in zip(cles, valeurs)} # zip fonctionne aussi avec des tuples de même longueur!
```
Voici encore un exemple bien utile pour réaliser un tableau à partir de données CSV.
```
cles = ("cle1", "cle2", "cle3")
objets = [("ah", "oh", "hein"), ('riri', 'fifi', 'loulou')]
# on veut un tableau de tuples nommés
[ {desc: val for desc, val in zip(cles, objet)} for objet in objets ]
```
### Synthèse: retour au problème des données au format CSV
En combinant tout ce que tu as appris et les exemples précédents, tu devrais être capable d'obtenir notre liste de n-uplets nommés en quelques lignes ... Non?
*rappel*: au départ, on **dispose de**
```python
donnees_CSV = """nom,prenom,date_naissance
Durand,Jean-Pierre,23/05/1985
Dupont,Christophe,15/12/1967
Terta,Henry,12/06/1978"""
```
au final, on veut **produire** une liste de *tuples nommés*:
```python
[
{'nom': 'Durand', 'prenom': 'Jean-Pierre', 'date_naissance': '23/05/1985'},
{'nom': 'Dupont', 'prenom': 'Christophe', 'date_naissance': '15/12/1967'},
{'nom': 'Terta', 'prenom': 'Henry', 'date_naissance': '12/06/1978'}
]
```
Voici comment y parvenir en deux «compréhensions»
```
descripteurs, *objets = [tuple(ligne.split(',')) for ligne in donnees_CSV.split('\n')]
objets = [ # sur plusieurs ligne pour plus de clarté.
{
desc: val for desc, val in zip(descripteurs, obj)
}
for obj in objets
]
objets
```
#### À faire toi-même
La syntaxe en compréhension des listes et des dictionnaires est utile et puissante mais nécessite pas mal d'investissement pour être bien maîtrisée.
Pour cette raison, reprend le problème en écrivant une fonction `csv_vers_objets(csv_str)` qui prend en argument la chaîne au format csv et renvoie le tableau de n-uplets nommés correspondant.
Nous la réutiliserons dans le 05_applications...
```
def csv_vers_objets(csv_str):
descripteurs, *objets = [tuple(ligne.split(',')) for ligne in csv_str.split('\n')]
objets = [ # sur plusieurs ligne pour plus de clarté.
{
desc: val for desc, val in zip(descripteurs, obj)
}
for obj in objets
]
return objets
csv_vers_objets(donnees_CSV)
assert csv_vers_objets(donnees_CSV) == objets
```
| github_jupyter |
# TF neural net with normalized ISO spectra
```
# TensorFlow and tf.keras
import tensorflow as tf
from tensorflow import keras
# Helper libraries
import glob
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from concurrent.futures import ProcessPoolExecutor
from IPython.core.debugger import set_trace as st
from sklearn.model_selection import train_test_split
from time import time
# My modules
from swsnet import helpers
print(tf.__version__)
```
## Dataset: ISO-SWS (normalized, culled)
```
# Needed directories
base_dir = '../data/isosws_atlas/'
# Pickles containing our spectra in the form of pandas dataframes:
spec_dir = base_dir + 'spectra_normalized/'
spec_files = np.sort(glob.glob(spec_dir + '*.pkl'))
# Metadata pickle (pd.dataframe). Note each entry contains a pointer to the corresponding spectrum pickle.
metadata = base_dir + 'metadata_sorted_normalized_culled.pkl'
```
#### Labels ('group'):
1. Naked stars
2. Stars with dust
3. Warm, dusty objects
4. Cool, dusty objects
5. Very red objects
6. Continuum-free objects but having emission lines
7. Flux-free and/or fatally flawed spectra
### Subset 1: all data included
```
features, labels = helpers.load_data(base_dir=base_dir, metadata=metadata,
only_ok_data=False, clean=False, verbose=False)
print(features.shape)
print(labels.shape)
```
### Subset 2: exclude group 7
```
features_clean, labels_clean = \
helpers.load_data(base_dir=base_dir, metadata=metadata,
only_ok_data=False, clean=True, verbose=False)
print(features_clean.shape)
print(labels_clean.shape)
```
### Subset 3: exclude group 7, uncertain data
```
features_certain, labels_certain = \
helpers.load_data(base_dir=base_dir, metadata=metadata,
only_ok_data=True, clean=False, verbose=False)
print(features_certain.shape)
print(labels_certain.shape)
```
# Testing l2norms
```
def neural(features, labels, test_size=0.3, l2norm=0.01):
X_train, X_test, y_train, y_test = \
train_test_split(features, labels, test_size=test_size, random_state = 42)
# Sequential model, 7 classes of output.
model = keras.Sequential()
model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm), input_dim=359))
model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))
model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))
model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))
model.add(keras.layers.Dense(7, activation='softmax'))
# Early stopping condition.
callback = [tf.keras.callbacks.EarlyStopping(monitor='acc', patience=5, verbose=0)]
# Recompile model and fit.
model.compile(optimizer=keras.optimizers.Adam(0.0005),
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# model.fit(X_train, y_train, epochs=50, batch_size=32, verbose=False)
model.fit(X_train, y_train, epochs=100, batch_size=32, callbacks=callback, verbose=False)
# Check accuracy.
score = model.evaluate(X_test, y_test, verbose=0)
accuracy = score[1]
print("L2 norm, accuracy: ", l2norm, accuracy)
return model, test_size, accuracy
# for l2norm in (0.1, 0.01, 0.001, 0.0001, 0.00001):
# model, test_size, accuracy = neural(features, labels, l2norm=l2norm)
# for l2norm in (0.1, 0.01, 0.001, 0.0001, 0.00001):
# model, test_size, accuracy = neural(features_clean, labels_clean, l2norm=l2norm)
# for l2norm in (0.1, 0.01, 0.001, 0.0001, 0.00001):
# model, test_size, accuracy = neural(features_certain, labels_certain, l2norm=l2norm)
# for l2norm in (0.001, 0.0001, 0.00001, 0.000001):
# model, test_size, accuracy = neural(features_certain, labels_certain, l2norm=l2norm)
```
***
# Testing training size vs. accuracy
Model:
```
def run_NN(input_tuple):
"""Run a Keras NN for the purpose of examining the effect of training set size.
Args:
features (ndarray): Array containing the spectra (fluxes).
labels (ndarray): Array containing the group labels for the spectra.
test_size (float): Fraction of test size relative to (test + training).
Returns:
test_size (float): Input test_size, just a sanity check!
accuracy (float): Accuracy of this neural net when applied to the test set.
"""
features, labels, test_size = input_tuple
l2norm = 0.001
X_train, X_test, y_train, y_test = \
train_test_split(features, labels, test_size=test_size, random_state = 42)
# Sequential model, 7 classes of output.
model = keras.Sequential()
model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm), input_dim=359))
model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))
model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))
model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))
model.add(keras.layers.Dense(7, activation='softmax'))
# Early stopping condition.
callback = [tf.keras.callbacks.EarlyStopping(monitor='acc', patience=5, verbose=0)]
# Recompile model and fit.
model.compile(optimizer=keras.optimizers.Adam(0.0005),
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# model.fit(X_train, y_train, epochs=50, batch_size=32, verbose=False)
model.fit(X_train, y_train, epochs=100, batch_size=32, callbacks=callback, verbose=False)
# Check accuracy.
score = model.evaluate(X_test, y_test, verbose=0)
accuracy = score[1]
# print("Test size, accuracy: ", test_size, accuracy)
return test_size, accuracy
def run_networks(search_map):
# Run the networks in parallel.
start = time()
pool = ProcessPoolExecutor(max_workers=14)
results = list(pool.map(run_NN, search_map))
end = time()
print('Took %.3f seconds' % (end - start))
run_matrix = np.array(results)
return run_matrix
def plot_results(run_matrix):
# Examine results.
plt.plot(run_matrix.T[0], run_matrix.T[1], 's', mfc='w', ms=5, mew=2, mec='r');
plt.xlabel('Test size (fraction)');
plt.ylabel('Test accuracy');
plt.minorticks_on();
# plt.xlim(left=0);
return
```
Search space (training size):
```
# Values of test_size to probe.
search_space = np.arange(0.14, 0.60, 0.02)
print('Size of test set considered: ', search_space)
# Number of iterations for each test_size value.
n_iterations = 20
# Create a vector to iterate over.
rx = np.array([search_space] * n_iterations).T
search_space_full = rx.flatten()
print('Number of iterations per test_size: ', n_iterations)
print('Total number of NN iterations required: ', n_iterations * len(search_space))
# Wrap up tuple inputs for running in parallel.
search_map = [(features, labels, x) for x in search_space_full]
search_map_clean = [(features_clean, labels_clean, x) for x in search_space_full]
search_map_certain = [(features_certain, labels_certain, x) for x in search_space_full]
run_matrix = run_networks(search_map)
run_matrix_clean = run_networks(search_map_clean)
run_matrix_certain = run_networks(search_map_certain)
```
## Full set:
```
plot_results(run_matrix)
```
## Clean set:
```
plot_results(run_matrix_clean)
```
## Certain set:
```
plot_results(run_matrix_certain)
```
***
Based on the above, probably need to do more data preprocessing:
- e.g., remove untrustworthy data
```
# save_path = '../models/nn_sorted_normalized_culled.h5'
# model.save(save_path)
```
| github_jupyter |
Filename: MNIST_data.ipynb
From <a href="http://neuralnetworksanddeeplearning.com/chap1.html"> this </a> book
Abbreviation: MNIST = Modified (handwritten digits data set from the U.S.) National Institute of Standards and Technology
Purpose: Explore the MNIST digits data to get familiar with the content and quality of the data.
```
import mnist_loader
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# training_data, validation_data, test_data = mnist_loader.load_data_wrapper()
#training_data, validation_data, test_data = mnist_loader.load_data()
training, validation, test = mnist_loader.load_data()
struct = [{'name': 'training', 'data': training[0], 'label': training[1]},
{'name': 'validation', 'data': validation[0], 'label': validation[1]},
{'name': 'test', 'data': test[0], 'label': test[1]}]
```
Training, validation, and test data structures are 2 element tuples having the following structure:
* <pre>[[p,i,x,e,l,s, , i,n, i,m,a,g,e, ,1], [...]]</pre>
* <pre>[num_represented_by_image1, ...]
```
fig, axes = plt.subplots(1, 3)
train = pd.Series(training_data[1])
train.hist(ax=axes[0])
axes[0].set_title("Training Data")
#display(train.describe())
validate = pd.Series(validation_data[1])
validate.hist(ax=axes[1])
axes[1].set_title("Validation Data")
#display(validate.describe())
test = pd.Series(test_data[1])
test.hist(ax=axes[2])
axes[2].set_title("Test Data")
#display(test.describe())
display("Distribution of validation data values")
values = pd.Series(validation_data[1])
values.hist()
values.describe()
display("Distribution of validation data values")
values = pd.Series(test_data[1])
values.hist()
values.describe()
```
<h3>Training Data</h3>
```
pixels = pd.DataFrame(training_data[0])
display("Images: {} Pixels-per-image: {}".format(*pixels.shape))
pixels.head()
#pixels.T.describe() # takes FOREVER ...
print('\033[1m'+"validation_data:"+'\033[0m')
display(validation_data)
print('{1:32s}{0}'.format(type(validation_data),'\033[1m'+"validation_data type:"+'\033[0m'))
print('{1:32s}{0}'.format(len(validation_data),'\033[1m'+"num of components:"+'\033[0m'))
print('')
print('{1:32s}{0}'.format(type(validation_data[0]),'\033[1m'+"first component type:"+'\033[0m'))
print('{1:32s}{0}'.format(len(validation_data[0]),'\033[1m'+"num of sub-components:"+'\033[0m'))
print('')
print('{1:32s}{0}'.format(type(validation_data[1]),'\033[1m'+"second component type:"+'\033[0m'))
print('{1:32s}{0}'.format(len(validation_data[1]),'\033[1m'+"num of sub-components:"+'\033[0m'))
print('\033[1m'+"test_data:"+'\033[0m')
display(test_data)
print('{1:32s}{0}'.format(type(test_data),'\033[1m'+"test_data type:"+'\033[0m'))
print('{1:32s}{0}'.format(len(test_data),'\033[1m'+"num of components:"+'\033[0m'))
print('')
print('{1:32s}{0}'.format(type(test_data[0]),'\033[1m'+"first component type:"+'\033[0m'))
print('{1:32s}{0}'.format(len(test_data[0]),'\033[1m'+"num of sub-components:"+'\033[0m'))
print('')
print('{1:32s}{0}'.format(type(test_data[1]),'\033[1m'+"second component type:"+'\033[0m'))
print('{1:32s}{0}'.format(len(test_data[1]),'\033[1m'+"num of sub-components:"+'\033[0m'))
import numpy as np
import matplotlib.pyplot as plt
print(type(training_data[0][0]))
print(len(training_data[0][0]))
print(28*28)
# break data into 28 x 28 square array (from 1 x 784 array)
plottable_image = np.reshape(training_data[0][0], (28, 28))
#display(plottable_image)
# plot
plt.imshow(plottable_image, cmap='gray_r')
plt.show()
import matplotlib.pyplot as plt
%matplotlib inline
table_cols = 18
table_rows = 12
rand_grid = np.random.rand(28, 28)
# plottable_image = np.reshape(training_data[0][0], (28, 28))
k = 0
for i in range(0,table_cols) :
for j in range(0,table_rows) :
if i==0 and j==0 :
plottable_images = [rand_grid]
else :
plottable_images.append( np.reshape(training_data[0][k], (28, 28)) )
k += 1
print(len(plottable_images))
fig, axes = plt.subplots(table_rows, table_cols, figsize=(15, 12),
subplot_kw={'xticks': [], 'yticks': []})
fig.subplots_adjust(hspace=0.5, wspace=0)
for i, ax in enumerate(axes.flat):
ax.imshow(plottable_images[i], cmap='gray_r')
if i == 0 :
ax.set_title("rand")
else :
digit = str(training_data[1][i-1])
index = str(i-1)
ax.set_title("({}) {}".format(index, digit))
plt.show()
print(len(list(training_data)))
print(len(list(validation_data)))
print(len(list(test_data)))
training_data, validation_data, test_data = mnist_loader.load_data_wrapper()
training_data = list(training_data)
validation_data = list(validation_data)
test_data = list(test_data)
print(len(training_data))
print(len(validation_data))
print(len(test_data))
print(len(training_data))
print(len(validation_data))
print(len(test_data))
print(training_data)
print(training_data[0][1])
training_data.shape
```
| github_jupyter |
# `asyncio` Beispiel
Ab IPython≥7.0 könnt ihr `asyncio` direkt in Jupyter Notebooks verwenden; seht auch [IPython 7.0, Async REPL](https://blog.jupyter.org/ipython-7-0-async-repl-a35ce050f7f7).
Wenn ihr die Fehlermeldung `RuntimeError: This event loop is already running` erhaltet, hilft euch vielleicht [nest-asyncio] weiter.
Ihr könnt das Paket in eurer Jupyter- oder JupyterHub-Umgebung installieren mit
```bash
$ pipenv install nest-asyncio
```
Ihr könnt es dann in euer Notebook importieren und verwenden mit:
```
import nest_asyncio
nest_asyncio.apply()
```
## Einfaches *Hello world*-Beispiel
```
import asyncio
async def hello():
print('Hello')
await asyncio.sleep(1)
print('world')
await hello()
```
## Ein bisschen näher an einem realen Beispiel
```
import asyncio
import random
async def produce(queue, n):
for x in range(1, n + 1):
# produce an item
print('producing {}/{}'.format(x, n))
# simulate i/o operation using sleep
await asyncio.sleep(random.random())
item = str(x)
# put the item in the queue
await queue.put(item)
# indicate the producer is done
await queue.put(None)
async def consume(queue):
while True:
# wait for an item from the producer
item = await queue.get()
if item is None:
# the producer emits None to indicate that it is done
break
# process the item
print('consuming {}'.format(item))
# simulate i/o operation using sleep
await asyncio.sleep(random.random())
loop = asyncio.get_event_loop()
queue = asyncio.Queue(loop=loop)
asyncio.ensure_future(produce(queue, 10), loop=loop)
loop.run_until_complete(consume(queue))
```
## Ausnahmebehandlung
> **Siehe auch:** [set_exception_handler](https://docs.python.org/3/library/asyncio-eventloop.html#asyncio.loop.set_exception_handler)
```
def main():
loop = asyncio.get_event_loop()
# May want to catch other signals too
signals = (signal.SIGHUP, signal.SIGTERM, signal.SIGINT)
for s in signals:
loop.add_signal_handler(
s, lambda s=s: asyncio.create_task(shutdown(loop, signal=s)))
loop.set_exception_handler(handle_exception)
queue = asyncio.Queue()
```
## Testen mit `pytest`
### Beispiel:
```
import pytest
@pytest.mark.asyncio
async def test_consume(mock_get, mock_queue, message, create_mock_coro):
mock_get.side_effect = [message, Exception("break while loop")]
with pytest.raises(Exception, match="break while loop"):
await consume(mock_queue)
```
### Bibliotheken von Drittanbietern
* [pytest-asyncio](https://github.com/pytest-dev/pytest-asyncio) hat hilfreiche Dinge wie Test-Fixtures für `event_loop`, `unused_tcp_port`, und `unused_tcp_port_factory`; und die Möglichkeit zum Erstellen eurer eigenen [asynchronen Fixtures](https://github.com/pytest-dev/pytest-asyncio/#async-fixtures).
* [asynctest](https://asynctest.readthedocs.io/en/latest/index.html) verfügt über hilfreiche Werkzeuge, einschließlich Coroutine-Mocks und [exhaust_callbacks](https://asynctest.readthedocs.io/en/latest/asynctest.helpers.html#asynctest.helpers.exhaust_callbacks) so dass wir `await task` nicht manuell erstellen müssen.
* [aiohttp](https://docs.aiohttp.org/en/stable/) hat ein paar wirklich nette eingebaute Test-Utilities.
## Debugging
`asyncio` hat bereits einen [debug mode](https://docs.python.org/3.6/library/asyncio-dev.html#debug-mode-of-asyncio) in der Standardbibliothek. Ihr könnt ihn einfach mit der Umgebungsvariablen `PYTHONASYNCIODEBUG` oder im Code mit `loop.set_debug(True)` aktivieren.
### Verwendet den Debug-Modus zum Identifizieren langsamer asynchroner Aufrufe
Der Debug-Modus von `asyncio` hat einen kleinen eingebauten Profiler. Wenn der Debug-Modus aktiviert ist, protokolliert `asyncio` alle asynchronen Aufrufe, die länger als 100 Millisekunden dauern.
### Debugging im Produktivbetrieb mit `aiodebug`
[aiodebug](https://github.com/qntln/aiodebug) ist eine kleine Bibliothek zum Überwachen und Testen von Asyncio-Programmen.
#### Beispiel
```
from aiodebug import log_slow_callbacks
def main():
loop = asyncio.get_event_loop()
log_slow_callbacks.enable(0.05)
```
## Logging
[aiologger](https://github.com/b2wdigital/aiologger) ermöglicht eine nicht-blockierendes Logging.
## Asynchrone Widgets
> **Seht auch:** [Asynchronous Widgets](https://ipywidgets.readthedocs.io/en/stable/examples/Widget%20Asynchronous.html)
```
def wait_for_change(widget, value):
future = asyncio.Future()
def getvalue(change):
# make the new value available
future.set_result(change.new)
widget.unobserve(getvalue, value)
widget.observe(getvalue, value)
return future
from ipywidgets import IntSlider
slider = IntSlider()
async def f():
for i in range(10):
print('did work %s'%i)
x = await wait_for_change(slider, 'value')
print('async function continued with value %s'%x)
asyncio.ensure_future(f())
slider
```
## Weiterlesen
* Lynn Root: [asyncio: We Did It Wrong](https://www.roguelynn.com/words/asyncio-we-did-it-wrong/)
* Mike Driscoll: [An Intro to asyncio](https://www.blog.pythonlibrary.org/2016/07/26/python-3-an-intro-to-asyncio/)
* Yeray Diaz: [Asyncio Coroutine Patterns: Beyond await](https://medium.com/python-pandemonium/asyncio-coroutine-patterns-beyond-await-a6121486656f)
| github_jupyter |
# B - A Closer Look at Word Embeddings
We have very briefly covered how word embeddings (also known as word vectors) are used in the tutorials. In this appendix we'll have a closer look at these embeddings and find some (hopefully) interesting results.
Embeddings transform a one-hot encoded vector (a vector that is 0 in elements except one, which is 1) into a much smaller dimension vector of real numbers. The one-hot encoded vector is also known as a *sparse vector*, whilst the real valued vector is known as a *dense vector*.
The key concept in these word embeddings is that words that appear in similar _contexts_ appear nearby in the vector space, i.e. the Euclidean distance between these two word vectors is small. By context here, we mean the surrounding words. For example in the sentences "I purchased some items at the shop" and "I purchased some items at the store" the words 'shop' and 'store' appear in the same context and thus should be close together in vector space.
You may have also heard about *word2vec*. *word2vec* is an algorithm (actually a bunch of algorithms) that calculates word vectors from a corpus. In this appendix we use *GloVe* vectors, *GloVe* being another algorithm to calculate word vectors. If you want to know how *word2vec* works, check out a two part series [here](http://mccormickml.com/2016/04/19/word2vec-tutorial-the-skip-gram-model/) and [here](http://mccormickml.com/2017/01/11/word2vec-tutorial-part-2-negative-sampling/), and if you want to find out more about *GloVe*, check the website [here](https://nlp.stanford.edu/projects/glove/).
In PyTorch, we use word vectors with the `nn.Embedding` layer, which takes a _**[sentence length, batch size]**_ tensor and transforms it into a _**[sentence length, batch size, embedding dimensions]**_ tensor.
In tutorial 2 onwards, we also used pre-trained word embeddings (specifically the GloVe vectors) provided by TorchText. These embeddings have been trained on a gigantic corpus. We can use these pre-trained vectors within any of our models, with the idea that as they have already learned the context of each word they will give us a better starting point for our word vectors. This usually leads to faster training time and/or improved accuracy.
In this appendix we won't be training any models, instead we'll be looking at the word embeddings and finding a few interesting things about them.
A lot of the code from the first half of this appendix is taken from [here](https://github.com/spro/practical-pytorch/blob/master/glove-word-vectors/glove-word-vectors.ipynb). For more information about word embeddings, go [here](https://monkeylearn.com/blog/word-embeddings-transform-text-numbers/).
## Loading the GloVe vectors
First, we'll load the GloVe vectors. The `name` field specifies what the vectors have been trained on, here the `6B` means a corpus of 6 billion words. The `dim` argument specifies the dimensionality of the word vectors. GloVe vectors are available in 50, 100, 200 and 300 dimensions. There is also a `42B` and `840B` glove vectors, however they are only available at 300 dimensions.
```
import torchtext.vocab
glove = torchtext.vocab.GloVe(name = '6B', dim = 100)
print(f'There are {len(glove.itos)} words in the vocabulary')
```
As shown above, there are 400,000 unique words in the GloVe vocabulary. These are the most common words found in the corpus the vectors were trained on. **In these set of GloVe vectors, every single word is lower-case only.**
`glove.vectors` is the actual tensor containing the values of the embeddings.
```
glove.vectors.shape
```
We can see what word is associated with each row by checking the `itos` (int to string) list.
Below implies that row 0 is the vector associated with the word 'the', row 1 for ',' (comma), row 2 for '.' (period), etc.
```
glove.itos[:10]
```
We can also use the `stoi` (string to int) dictionary, in which we input a word and receive the associated integer/index. If you try get the index of a word that is not in the vocabulary, you receive an error.
```
glove.stoi['the']
```
We can get the vector of a word by first getting the integer associated with it and then indexing into the word embedding tensor with that index.
```
glove.vectors[glove.stoi['the']].shape
```
We'll be doing this a lot, so we'll create a function that takes in word embeddings and a word then returns the associated vector. It'll also throw an error if the word doesn't exist in the vocabulary.
```
def get_vector(embeddings, word):
assert word in embeddings.stoi, f'*{word}* is not in the vocab!'
return embeddings.vectors[embeddings.stoi[word]]
```
As before, we use a word to get the associated vector.
```
get_vector(glove, 'the').shape
```
## Similar Contexts
Now to start looking at the context of different words.
If we want to find the words similar to a certain input word, we first find the vector of this input word, then we scan through our vocabulary calculating the distance between the vector of each word and our input word vector. We then sort these from closest to furthest away.
The function below returns the closest 10 words to an input word vector:
```
import torch
def closest_words(embeddings, vector, n = 10):
distances = [(word, torch.dist(vector, get_vector(embeddings, word)).item())
for word in embeddings.itos]
return sorted(distances, key = lambda w: w[1])[:n]
```
Let's try it out with 'korea'. The closest word is the word 'korea' itself (not very interesting), however all of the words are related in some way. Pyongyang is the capital of North Korea, DPRK is the official name of North Korea, etc.
Interestingly, we also get 'Japan' and 'China', implies that Korea, Japan and China are frequently talked about together in similar contexts. This makes sense as they are geographically situated near each other.
```
word_vector = get_vector(glove, 'korea')
closest_words(glove, word_vector)
```
Looking at another country, India, we also get nearby countries: Thailand, Malaysia and Sri Lanka (as two separate words). Australia is relatively close to India (geographically), but Thailand and Malaysia are closer. So why is Australia closer to India in vector space? This is most probably due to India and Australia appearing in the context of [cricket](https://en.wikipedia.org/wiki/Cricket) matches together.
```
word_vector = get_vector(glove, 'india')
closest_words(glove, word_vector)
```
We'll also create another function that will nicely print out the tuples returned by our `closest_words` function.
```
def print_tuples(tuples):
for w, d in tuples:
print(f'({d:02.04f}) {w}')
```
A final word to look at, 'sports'. As we can see, the closest words are most of the sports themselves.
```
word_vector = get_vector(glove, 'sports')
print_tuples(closest_words(glove, word_vector))
```
## Analogies
Another property of word embeddings is that they can be operated on just as any standard vector and give interesting results.
We'll show an example of this first, and then explain it:
```
def analogy(embeddings, word1, word2, word3, n=5):
#get vectors for each word
word1_vector = get_vector(embeddings, word1)
word2_vector = get_vector(embeddings, word2)
word3_vector = get_vector(embeddings, word3)
#calculate analogy vector
analogy_vector = word2_vector - word1_vector + word3_vector
#find closest words to analogy vector
candidate_words = closest_words(embeddings, analogy_vector, n+3)
#filter out words already in analogy
candidate_words = [(word, dist) for (word, dist) in candidate_words
if word not in [word1, word2, word3]][:n]
print(f'{word1} is to {word2} as {word3} is to...')
return candidate_words
print_tuples(analogy(glove, 'man', 'king', 'woman'))
```
This is the canonical example which shows off this property of word embeddings. So why does it work? Why does the vector of 'woman' added to the vector of 'king' minus the vector of 'man' give us 'queen'?
If we think about it, the vector calculated from 'king' minus 'man' gives us a "royalty vector". This is the vector associated with traveling from a man to his royal counterpart, a king. If we add this "royality vector" to 'woman', this should travel to her royal equivalent, which is a queen!
We can do this with other analogies too. For example, this gets an "acting career vector":
```
print_tuples(analogy(glove, 'man', 'actor', 'woman'))
```
For a "baby animal vector":
```
print_tuples(analogy(glove, 'cat', 'kitten', 'dog'))
```
A "capital city vector":
```
print_tuples(analogy(glove, 'france', 'paris', 'england'))
```
A "musician's genre vector":
```
print_tuples(analogy(glove, 'elvis', 'rock', 'eminem'))
```
And an "ingredient vector":
```
print_tuples(analogy(glove, 'beer', 'barley', 'wine'))
```
## Correcting Spelling Mistakes
Another interesting property of word embeddings is that they can actually be used to correct spelling mistakes!
We'll put their findings into code and briefly explain them, but to read more about this, check out the [original thread](http://forums.fast.ai/t/nlp-any-libraries-dictionaries-out-there-for-fixing-common-spelling-errors/16411) and the associated [write-up](https://blog.usejournal.com/a-simple-spell-checker-built-from-word-vectors-9f28452b6f26).
First, we need to load up the much larger vocabulary GloVe vectors, this is due to the spelling mistakes not appearing in the smaller vocabulary.
**Note**: these vectors are very large (~2GB), so watch out if you have a limited internet connection.
```
glove = torchtext.vocab.GloVe(name = '840B', dim = 300)
```
Checking the vocabulary size of these embeddings, we can see we now have over 2 million unique words in our vocabulary!
```
glove.vectors.shape
```
As the vectors were trained with a much larger vocabulary on a larger corpus of text, the words that appear are a little different. Notice how the words 'north', 'south', 'pyongyang' and 'dprk' no longer appear in the most closest words to 'korea'.
```
word_vector = get_vector(glove, 'korea')
print_tuples(closest_words(glove, word_vector))
```
Our first step to correcting spelling mistakes is looking at the vector for a misspelling of the word 'reliable'.
```
word_vector = get_vector(glove, 'relieable')
print_tuples(closest_words(glove, word_vector))
```
Notice how the correct spelling, "reliable", does not appear in the top 10 closest words. Surely the misspellings of a word should appear next to the correct spelling of the word as they appear in the same context, right?
The hypothesis is that misspellings of words are all equally shifted away from their correct spelling. This is because articles of text that contain spelling mistakes are usually written in an informal manner where correct spelling doesn't matter as much (such as tweets/blog posts), thus spelling errors will appear together as they appear in context of informal articles.
Similar to how we created analogies before, we can create a "correct spelling" vector. This time, instead of using a single example to create our vector, we'll use the average of multiple examples. This will hopefully give better accuracy!
We first create a vector for the correct spelling, 'reliable', then calculate the difference between the "reliable vector" and each of the 8 misspellings of 'reliable'. As we are going to concatenate these 8 misspelling tensors together we need to unsqueeze a "batch" dimension to them.
```
reliable_vector = get_vector(glove, 'reliable')
reliable_misspellings = ['relieable', 'relyable', 'realible', 'realiable',
'relable', 'relaible', 'reliabe', 'relaiable']
diff_reliable = [(reliable_vector - get_vector(glove, s)).unsqueeze(0)
for s in reliable_misspellings]
```
We take the average of these 8 'difference from reliable' vectors to get our "misspelling vector".
```
misspelling_vector = torch.cat(diff_reliable, dim = 0).mean(dim = 0)
```
We can now correct other spelling mistakes using this "misspelling vector" by finding the closest words to the sum of the vector of a misspelled word and the "misspelling vector".
For a misspelling of "because":
```
word_vector = get_vector(glove, 'becuase')
print_tuples(closest_words(glove, word_vector + misspelling_vector))
```
For a misspelling of "definitely":
```
word_vector = get_vector(glove, 'defintiely')
print_tuples(closest_words(glove, word_vector + misspelling_vector))
```
For a misspelling of "consistent":
```
word_vector = get_vector(glove, 'consistant')
print_tuples(closest_words(glove, word_vector + misspelling_vector))
```
For a misspelling of "package":
```
word_vector = get_vector(glove, 'pakage')
print_tuples(closest_words(glove, word_vector + misspelling_vector))
```
For a more in-depth look at this, check out the [write-up](https://blog.usejournal.com/a-simple-spell-checker-built-from-word-vectors-9f28452b6f26).
| github_jupyter |
### This notebook explores the calendar of Munich listings to answer the question:
## What is the most expensive and the cheapest time to visit Munich?
```
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
import seaborn as sns
sns.set()
LOCATION = 'munich'
df_list = pd.read_csv(LOCATION + '/listings.csv.gz')
df_reviews = pd.read_csv(LOCATION + '/reviews.csv.gz')
df_cal = pd.read_csv(LOCATION + '/calendar.csv.gz')
pd.options.display.max_rows=10
pd.options.display.max_columns=None
pd.options.display.max_colwidth=30
```
___
### Calendar
#### First look into to data and types for each column:
```
df_cal
df_cal.dtypes
```
___
#### Some data types are wrong. In order to be able to work with the data, we need to change some datatypes.
First convert **date** to *datetime* type:
```
df_cal['date'] = pd.to_datetime(df_cal['date'])
```
**Price** needs to converted to *float* in order to be able to work with it.
```
df_cal['price']=df_cal['price'].replace(to_replace='[\$,]', value='', regex=True).astype(float)
df_cal['adjusted_price']=df_cal['adjusted_price'].replace(to_replace='[\$,]', value='', regex=True).astype(float)
```
This is how it looks now:
```
df_cal.head()
```
And this are the corrected data types:
```
df_cal.dtypes
```
___
### First question to be answered is, what is the price distribution over the year:
Let's calculate the mean price over all listings for each day of the year:
First check if we have *NULL* values in the data frame.
```
df_cal.isnull().sum()
```
*NULL* values have impact to the average (even if very small due to the small number of missing values), let's drop all rows with *NULL* **price**.
```
df_cal.dropna(subset=['price'], inplace=True)
```
Now let's group all listings by **date** and calculate the average **price** of all listings for each day:
```
mean_price = df_cal[['date', 'price']].groupby(by='date').mean().reset_index()
mean_price
```
And plot the result:
```
## use the plot method and scale the size based on ploted values
scale_from = mean_price['price'][1:-2].min()*0.95
scale_to = mean_price['price'][1:-2].max()*1.02
mean_price.set_index('date')[1:-2].plot(kind='line', y='price', figsize=(20,10), grid=True).set_ylim(scale_from, scale_to);
```
### HERE WE ARE! There are two interesting observations:
#### 1. There is a peak in the second half of September: **"Welcome to the Octoberfest!**"
#### 2. The price apparently depends on the day of week. Let's have a closer look at it.
___
### Second question: What is the price distribution within a week?
To be able to have a close look at prices, let's introduce the **day_of_week** column.
```
df_cal['day_of_week'] = df_cal['date'].dt.dayofweek
```
Let's group the prices for each day of week and get the average price:
```
mean_price_dow = df_cal[['day_of_week', 'price']].groupby(by='day_of_week').mean().reset_index()
mean_price_dow
```
It's difficult to interpret index-based day of week. Let's convert it to strings from Monday to Sunday:
```
def convert_day_of_week(day_idx):
'''
This function convert index based day of week to string
0 - Monday
6 - Sunday
if the day_idx oís out of this range, this index will be returned
'''
if(day_idx>6 or day_idx<0):
return day_idx
lst = ['Monday', 'Tuesday', 'Wednesday', 'Thursday', 'Friday', 'Saturday', 'Sunday']
return lst[day_idx]
mean_price_dow['day_of_week'] = mean_price_dow['day_of_week'].apply(convert_day_of_week)
mean_price_dow
```
#### Now we can plot the result:
```
scale_from = mean_price_dow['price'].min()*0.95
scale_to = mean_price_dow['price'].max()*1.02
sns.set(rc={'figure.figsize':(15,5)})
fig = sns.barplot(data=mean_price_dow, x='day_of_week', y='price', color='#0080bb');
fig.set_ylim(scale_from, scale_to);
fig.set_title('Prices for day of the week');
fig.set_xlabel('Day of week');
fig.set_ylabel('Price');
```
#### No surprise, the most expensive days are Friday and Saturday. The weekend in Munich can start!
___
## What is the most expensive and the cheapest time to visit Munich?
#### If you want to save your money, don't visit Munich during the Octoberfest, which is end of September, beginning of October. Do it better in April, it's much cheaper and you can enjoy green Munich.
#### If you plan a short trip and you are not constrained by day of week, in order to save the money, Sunday to Thursday is the best choice for you.
| github_jupyter |
```
# second notebook for Yelp1 Labs 18 Project
# data cleanup
# imports
# dataframe
import pandas as pd
import json
# NLP
import gensim
from gensim.utils import simple_preprocess
from gensim.parsing.preprocessing import STOPWORDS
from gensim import corpora
# import review.json file from https://www.yelp.com/dataset
with open('/Users/ianforrest/Desktop/coding/repos/yelp/yelp_dataset/review.json') as f:
review = json.loads("[" +
f.read().replace("}\n{", "},\n{") +
"]")
# convert review.json files to pandas DataFrame 'df_review'
df_review = pd.DataFrame(review)
# check df_review to make sure it was created correctly
df_review.head()
# check column names of df_review
df_review.columns
# check value counts of 'stars' column
df_review['stars'].value_counts()
# check value counts of useful column
df_review['useful'].value_counts()
# check value counts of funny column
df_review['funny'].value_counts()
# check value counts of cool column
df_review['cool'].value_counts()
# check text of random reviews in dataset as part of initial exploration
df_review.iloc[3244,7]
# check text of random reviews in dataset as part of initial exploration
df_review.iloc[2342553,7]
# check text of random reviews in dataset as part of initial exploration
df_review.iloc[3,7]
# export df_review to .csv
#df_review.to_csv(r'/Users/ianforrest/Desktop/coding/repos/yelp/yelp_dataset/df_review.csv')
# create copy of dataframe to manipulate for model
df = df_review.copy()
df.head()
# add 'total_votes' column to dataframe; total of 'useful', 'funny', 'cool' columns
df['total_votes'] = df['useful'] + df['funny'] + df['cool']
df.head()
# drop unused columns from dataframe
df = df.drop(columns=['user_id', 'business_id', 'review_id', 'useful', 'funny', 'cool'])
df.head()
# convert 'date' column to datetime format
df['date'] = pd.to_datetime(df['date'])
df.dtypes
# check value counts of 'total_votes' column
df['total_votes'].value_counts()
# limit dataframe to reviews with 0 or more total votes
df = df.loc[df['total_votes'] >= 0]
# check value counts of 'total_votes' column
df['total_votes'].value_counts()
# remove html code from text column
df['text'] = df['text'].str.replace('(\d{1,2}[/. ](?:\d{1,2}|January|Jan)[/. ]\d{2}(?:\d{2})?)', '')
df['text'] = df['text'].str.replace('\n\n', '')
df['text'] = df['text'].str.replace('\\n', '')
df['text'] = df['text'].str.replace('\n', '')
# check text of random reviews in dataset to make sure HTML code is removed correctly
# backslashes before apostrophes are for display purposes only to indicate apostrophes are not quotation marks
df.iloc[2342553,1]
# initiate STOPWORDS for NLP Processing
STOPWORDS = set(STOPWORDS).union(set(['I', 'We', 'i', 'we', 'it', "it's",
'it', 'the', 'this', 'they', 'They',
'he', 'He', 'she', 'She', '\n', '\n\n']))
# create tokenize function to tokenize review text
def tokenize(text):
return [token for token in simple_preprocess(text, deacc=True, min_len=4, max_len=40) if token not in STOPWORDS]
# add tokens column to dataframe
df['tokens'] = df['text'].apply(tokenize)
# check to make sure tokens were added to dataframe correctly
df.head()
# export cleaned dataframe with tokenized text to .csv file
df.to_csv(r'/Users/ianforrest/Desktop/coding/repos/yelp/yelp_dataset/df.csv')
df.sort_values(['total_votes'], ascending=False)
df.iloc[1292098,1]
df.dtypes
```
| github_jupyter |
##### Copyright 2019 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Deep Convolutional Generative Adversarial Network
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/tutorials/generative/dcgan">
<img src="https://www.tensorflow.org/images/tf_logo_32px.png" />
View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/tutorials/generative/dcgan.ipynb">
<img src="https://www.tensorflow.org/images/colab_logo_32px.png" />
Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/tutorials/generative/dcgan.ipynb">
<img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />
View source on GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/docs/site/en/tutorials/generative/dcgan.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table>
This tutorial demonstrates how to generate images of handwritten digits using a [Deep Convolutional Generative Adversarial Network](https://arxiv.org/pdf/1511.06434.pdf) (DCGAN). The code is written using the [Keras Sequential API](https://www.tensorflow.org/guide/keras) with a `tf.GradientTape` training loop.
## What are GANs?
[Generative Adversarial Networks](https://arxiv.org/abs/1406.2661) (GANs) are one of the most interesting ideas in computer science today. Two models are trained simultaneously by an adversarial process. A *generator* ("the artist") learns to create images that look real, while a *discriminator* ("the art critic") learns to tell real images apart from fakes.

During training, the *generator* progressively becomes better at creating images that look real, while the *discriminator* becomes better at telling them apart. The process reaches equilibrium when the *discriminator* can no longer distinguish real images from fakes.

This notebook demonstrates this process on the MNIST dataset. The following animation shows a series of images produced by the *generator* as it was trained for 50 epochs. The images begin as random noise, and increasingly resemble hand written digits over time.

To learn more about GANs, we recommend MIT's [Intro to Deep Learning](http://introtodeeplearning.com/) course.
### Import TensorFlow and other libraries
```
from __future__ import absolute_import, division, print_function, unicode_literals
try:
# %tensorflow_version only exists in Colab.
%tensorflow_version 2.x
except Exception:
pass
import tensorflow as tf
tf.__version__
# To generate GIFs
!pip install imageio
import glob
import imageio
import matplotlib.pyplot as plt
import numpy as np
import os
import PIL
from tensorflow.keras import layers
import time
from IPython import display
```
### Load and prepare the dataset
You will use the MNIST dataset to train the generator and the discriminator. The generator will generate handwritten digits resembling the MNIST data.
```
(train_images, train_labels), (_, _) = tf.keras.datasets.mnist.load_data()
train_images = train_images.reshape(train_images.shape[0], 28, 28, 1).astype('float32')
train_images = (train_images - 127.5) / 127.5 # Normalize the images to [-1, 1]
BUFFER_SIZE = 60000
BATCH_SIZE = 256
# Batch and shuffle the data
train_dataset = tf.data.Dataset.from_tensor_slices(train_images).shuffle(BUFFER_SIZE).batch(BATCH_SIZE)
```
## Create the models
Both the generator and discriminator are defined using the [Keras Sequential API](https://www.tensorflow.org/guide/keras#sequential_model).
### The Generator
The generator uses `tf.keras.layers.Conv2DTranspose` (upsampling) layers to produce an image from a seed (random noise). Start with a `Dense` layer that takes this seed as input, then upsample several times until you reach the desired image size of 28x28x1. Notice the `tf.keras.layers.LeakyReLU` activation for each layer, except the output layer which uses tanh.
```
def make_generator_model():
model = tf.keras.Sequential()
model.add(layers.Dense(7*7*256, use_bias=False, input_shape=(100,)))
model.add(layers.BatchNormalization())
model.add(layers.LeakyReLU())
model.add(layers.Reshape((7, 7, 256)))
assert model.output_shape == (None, 7, 7, 256) # Note: None is the batch size
model.add(layers.Conv2DTranspose(128, (5, 5), strides=(1, 1), padding='same', use_bias=False))
assert model.output_shape == (None, 7, 7, 128)
model.add(layers.BatchNormalization())
model.add(layers.LeakyReLU())
model.add(layers.Conv2DTranspose(64, (5, 5), strides=(2, 2), padding='same', use_bias=False))
assert model.output_shape == (None, 14, 14, 64)
model.add(layers.BatchNormalization())
model.add(layers.LeakyReLU())
model.add(layers.Conv2DTranspose(1, (5, 5), strides=(2, 2), padding='same', use_bias=False, activation='tanh'))
assert model.output_shape == (None, 28, 28, 1)
return model
```
Use the (as yet untrained) generator to create an image.
```
generator = make_generator_model()
noise = tf.random.normal([1, 100])
generated_image = generator(noise, training=False)
plt.imshow(generated_image[0, :, :, 0], cmap='gray')
```
### The Discriminator
The discriminator is a CNN-based image classifier.
```
def make_discriminator_model():
model = tf.keras.Sequential()
model.add(layers.Conv2D(64, (5, 5), strides=(2, 2), padding='same',
input_shape=[28, 28, 1]))
model.add(layers.LeakyReLU())
model.add(layers.Dropout(0.3))
model.add(layers.Conv2D(128, (5, 5), strides=(2, 2), padding='same'))
model.add(layers.LeakyReLU())
model.add(layers.Dropout(0.3))
model.add(layers.Flatten())
model.add(layers.Dense(1))
return model
```
Use the (as yet untrained) discriminator to classify the generated images as real or fake. The model will be trained to output positive values for real images, and negative values for fake images.
```
discriminator = make_discriminator_model()
decision = discriminator(generated_image)
print (decision)
```
## Define the loss and optimizers
Define loss functions and optimizers for both models.
```
# This method returns a helper function to compute cross entropy loss
cross_entropy = tf.keras.losses.BinaryCrossentropy(from_logits=True)
```
### Discriminator loss
This method quantifies how well the discriminator is able to distinguish real images from fakes. It compares the discriminator's predictions on real images to an array of 1s, and the discriminator's predictions on fake (generated) images to an array of 0s.
```
def discriminator_loss(real_output, fake_output):
real_loss = cross_entropy(tf.ones_like(real_output), real_output)
fake_loss = cross_entropy(tf.zeros_like(fake_output), fake_output)
total_loss = real_loss + fake_loss
return total_loss
```
### Generator loss
The generator's loss quantifies how well it was able to trick the discriminator. Intuitively, if the generator is performing well, the discriminator will classify the fake images as real (or 1). Here, we will compare the discriminators decisions on the generated images to an array of 1s.
```
def generator_loss(fake_output):
return cross_entropy(tf.ones_like(fake_output), fake_output)
```
The discriminator and the generator optimizers are different since we will train two networks separately.
```
generator_optimizer = tf.keras.optimizers.Adam(1e-4)
discriminator_optimizer = tf.keras.optimizers.Adam(1e-4)
```
### Save checkpoints
This notebook also demonstrates how to save and restore models, which can be helpful in case a long running training task is interrupted.
```
checkpoint_dir = './training_checkpoints'
checkpoint_prefix = os.path.join(checkpoint_dir, "ckpt")
checkpoint = tf.train.Checkpoint(generator_optimizer=generator_optimizer,
discriminator_optimizer=discriminator_optimizer,
generator=generator,
discriminator=discriminator)
```
## Define the training loop
```
EPOCHS = 50
noise_dim = 100
num_examples_to_generate = 16
# We will reuse this seed overtime (so it's easier)
# to visualize progress in the animated GIF)
seed = tf.random.normal([num_examples_to_generate, noise_dim])
```
The training loop begins with generator receiving a random seed as input. That seed is used to produce an image. The discriminator is then used to classify real images (drawn from the training set) and fakes images (produced by the generator). The loss is calculated for each of these models, and the gradients are used to update the generator and discriminator.
```
# Notice the use of `tf.function`
# This annotation causes the function to be "compiled".
@tf.function
def train_step(images):
noise = tf.random.normal([BATCH_SIZE, noise_dim])
with tf.GradientTape() as gen_tape, tf.GradientTape() as disc_tape:
generated_images = generator(noise, training=True)
real_output = discriminator(images, training=True)
fake_output = discriminator(generated_images, training=True)
gen_loss = generator_loss(fake_output)
disc_loss = discriminator_loss(real_output, fake_output)
gradients_of_generator = gen_tape.gradient(gen_loss, generator.trainable_variables)
gradients_of_discriminator = disc_tape.gradient(disc_loss, discriminator.trainable_variables)
generator_optimizer.apply_gradients(zip(gradients_of_generator, generator.trainable_variables))
discriminator_optimizer.apply_gradients(zip(gradients_of_discriminator, discriminator.trainable_variables))
def train(dataset, epochs):
for epoch in range(epochs):
start = time.time()
for image_batch in dataset:
train_step(image_batch)
# Produce images for the GIF as we go
display.clear_output(wait=True)
generate_and_save_images(generator,
epoch + 1,
seed)
# Save the model every 15 epochs
if (epoch + 1) % 15 == 0:
checkpoint.save(file_prefix = checkpoint_prefix)
print ('Time for epoch {} is {} sec'.format(epoch + 1, time.time()-start))
# Generate after the final epoch
display.clear_output(wait=True)
generate_and_save_images(generator,
epochs,
seed)
```
**Generate and save images**
```
def generate_and_save_images(model, epoch, test_input):
# Notice `training` is set to False.
# This is so all layers run in inference mode (batchnorm).
predictions = model(test_input, training=False)
fig = plt.figure(figsize=(4,4))
for i in range(predictions.shape[0]):
plt.subplot(4, 4, i+1)
plt.imshow(predictions[i, :, :, 0] * 127.5 + 127.5, cmap='gray')
plt.axis('off')
plt.savefig('image_at_epoch_{:04d}.png'.format(epoch))
plt.show()
```
## Train the model
Call the `train()` method defined above to train the generator and discriminator simultaneously. Note, training GANs can be tricky. It's important that the generator and discriminator do not overpower each other (e.g., that they train at a similar rate).
At the beginning of the training, the generated images look like random noise. As training progresses, the generated digits will look increasingly real. After about 50 epochs, they resemble MNIST digits. This may take about one minute / epoch with the default settings on Colab.
```
train(train_dataset, EPOCHS)
```
Restore the latest checkpoint.
```
checkpoint.restore(tf.train.latest_checkpoint(checkpoint_dir))
```
## Create a GIF
```
# Display a single image using the epoch number
def display_image(epoch_no):
return PIL.Image.open('image_at_epoch_{:04d}.png'.format(epoch_no))
display_image(EPOCHS)
```
Use `imageio` to create an animated gif using the images saved during training.
```
anim_file = 'dcgan.gif'
with imageio.get_writer(anim_file, mode='I') as writer:
filenames = glob.glob('image*.png')
filenames = sorted(filenames)
last = -1
for i,filename in enumerate(filenames):
frame = 2*(i**0.5)
if round(frame) > round(last):
last = frame
else:
continue
image = imageio.imread(filename)
writer.append_data(image)
image = imageio.imread(filename)
writer.append_data(image)
import IPython
if IPython.version_info > (6,2,0,''):
display.Image(filename=anim_file)
```
If you're working in Colab you can download the animation with the code below:
```
try:
from google.colab import files
except ImportError:
pass
else:
files.download(anim_file)
```
## Next steps
This tutorial has shown the complete code necessary to write and train a GAN. As a next step, you might like to experiment with a different dataset, for example the Large-scale Celeb Faces Attributes (CelebA) dataset [available on Kaggle](https://www.kaggle.com/jessicali9530/celeba-dataset). To learn more about GANs we recommend the [NIPS 2016 Tutorial: Generative Adversarial Networks](https://arxiv.org/abs/1701.00160).
| github_jupyter |
```
import pickle
from misc import *
import SYCLOP_env as syc
from RL_brain_b import DeepQNetwork
import cv2
import time
from mnist import MNIST
mnist = MNIST('/home/bnapp/datasets/mnist/')
images, labels = mnist.load_training()
# some_mnistSM =[ cv2.resize(1.+np.reshape(uu,[28,28]), dsize=(256, 256)) for uu in images[:2]]#[:4096]]
some_samples_for_setup= prep_mnist_padded_images(2)
# run_dir = 'saved_runs/run_syclop_generic1.py_noname_1576060868_0/' #padded mnist beta 0.1 speed penalty 5
# result_type = 'nwk2.nwk'
# run_dir = 'saved_runs/run_syclop_generic1.py_noname_1576147784_0/' #padded mnist beta 0.1 speed penalty 20
run_dir = 'saved_runs/run_syclop_generic1.py_noname_1576403573_0/' #padded mnist beta 0.1 speed penalty 0
result_type = 'tempX_1.nwk'
hp = HP()
hp.mem_depth=1
hp.logmode=False
batch_size=256
action_space_size=9
# images = some_mnistSM
number_of_images = len(images)
reward = syc.Rewards()
observation_size = 256*4
RL = DeepQNetwork(action_space_size, observation_size*hp.mem_depth,#sensor.frame_size+2,
reward_decay=0.99,
e_greedy=1-1e-9,
e_greedy0=1-1e-9,
replace_target_iter=10,
memory_size=100000,
e_greedy_increment=0.0001,
learning_rate=0.0025,
double_q=False,
dqn_mode=True,
state_table=np.zeros([1,observation_size*hp.mem_depth]),
soft_q_type='boltzmann',
beta=0.1
)
def local_observer(sensor,agent):
if hp.logmode:
normfactor=1.0
else:
normfactor = 1.0/256.0
return normfactor*np.concatenate([relu_up_and_down(sensor.central_dvs_view),
relu_up_and_down(cv2.resize(1.0*sensor.dvs_view, dsize=(16, 16), interpolation=cv2.INTER_AREA))])
observation = np.random.uniform(0,1,size=[hp.mem_depth, observation_size])
scene_bb = [None]*batch_size
sensor_bb =[None]*batch_size
agent_bb = [None]*batch_size
action_bb = [None]*batch_size
action_list_bb = [None]*batch_size
q_list_bb = [None]*batch_size
observation_bb = [None]*batch_size
with open(run_dir+'/hp.pkl','rb') as f:
this_hp = pickle.load(f)
for bb in range(batch_size):
scene_bb[bb] = syc.Scene(frame_list=some_samples_for_setup[0:1])
sensor_bb[bb] = syc.Sensor()
agent_bb[bb] = syc.Agent(max_q = [scene_bb[bb].maxx-sensor_bb[bb].hp.winx,scene_bb[bb].maxy-sensor_bb[bb].hp.winy])
agent_bb[bb].hp.action_space = this_hp.agent.action_space
RL.dqn.load_nwk_param(run_dir+'/'+ result_type)
with open(run_dir+'/hp.pkl','rb') as f:
this_hp = pickle.load(f)
hp.fading_mem = this_hp.fading_mem +0.0 #to avoid assignment by address
size=(28,28)
offset=(0,0)
action_records=[]
q_records=[]
observation_feeder=np.zeros([batch_size,1024])
for image_num,image in enumerate(images):
step = 0
episode = 0
for batch_num in range(len(images)//batch_size):
for bb in range(batch_size):
action_list_bb[bb] = []
# q_list_bb[bb] = []
observation_bb[bb] = np.random.uniform(0,1,size=[hp.mem_depth, observation_size])
observation_bb[bb] = np.random.uniform(0,1,size=[hp.mem_depth, observation_size])
# scene_bb[bb].current_frame = image_num[bb]
#### sizing story:
image_resized=cv2.resize(0.0+np.reshape(images[batch_num*batch_size+bb],[28,28]), dsize=size)
scene_bb[bb].image = build_mnist_padded([image_resized],y_size=size[1],x_size=size[0],offset=offset)
# scene_bb[bb].image = build_mnist_padded([images[batch_num*batch_size+bb]])
agent_bb[bb].reset()
agent_bb[bb].q_ana[1]=128./2.-32
agent_bb[bb].q_ana[0]=128./2-32
agent_bb[bb].q = np.int32(np.floor(agent_bb[bb].q_ana))
sensor_bb[bb].reset()
sensor_bb[bb].update(scene_bb[bb], agent_bb[bb])
sensor_bb[bb].update(scene_bb[bb], agent_bb[bb])
time1=time.time()
for step_prime in range(1000):
deep_time1=time.time()
# action = RL.choose_action(observation.reshape([-1]))
for bb in range(batch_size):
observation_feeder[bb,:]=observation_bb[bb].reshape([1,-1])
oo = RL.dqn.eval_eval(observation_feeder)
boltzmann_measure = np.exp(RL.beta * (oo-np.max(oo,axis=1).reshape([-1,1]))) #todo here substracted max to avoid exponent exploding. need to be taken into a separate function!
boltzmann_measure = boltzmann_measure / np.sum(boltzmann_measure, axis=1).reshape([-1,1])
for bb in range(batch_size):
action_bb[bb] = np.random.choice(list(range(RL.n_actions)),1, p=boltzmann_measure[bb,:].reshape([-1]))[0]
# action_bb= [a for a in np.argmax(oo,axis=1)]
deep_time2=time.time()
shallow_time1=time.time()
for bb in range(batch_size):
agent_bb[bb].act(action_bb[bb])
action_list_bb[bb].append(action_bb[bb])
# q_list_bb[bb].append(agent_bb[bb].q_ana)
sensor_bb[bb].update(scene_bb[bb],agent_bb[bb])
observation_bb[bb] *= hp.fading_mem
observation_bb[bb] += local_observer(sensor_bb[bb], agent_bb[bb]) # todo: generalize
shallow_time2=time.time()
# print('deep:',deep_time2-deep_time1,'shallow:',shallow_time2-shallow_time1)
time2=time.time()
print('batch num:',batch_num,'wall time consumed:',time2-time1)
for bb in range(batch_size):
action_records.append(action_list_bb[bb])
# q_records.append(q_list_bb[bb])
len(action_records)
with open('mnist_padded_b0p1_v0_X28_Tx0y0_act_full1.pkl','wb') as f:
pickle.dump([action_records[:30000],labels[:30000]],f)
with open('mnist_padded_b0p1_v0_X28_Tx0y0_act_full2.pkl','wb') as f:
pickle.dump([action_records[30000:],labels[30000:]],f)
np.shape(sensor_bb[0].frame_view)
agent_bb[0].q_ana
```
| github_jupyter |
```
import lifelines
import pymc as pm
from pyBMA.CoxPHFitter import CoxPHFitter
import matplotlib.pyplot as plt
import numpy as np
from numpy import log
from datetime import datetime
import pandas as pd
%matplotlib inline
```
The first step in any data analysis is acquiring and munging the data
Our starting data set can be found here:
http://jakecoltman.com in the pyData post
It is designed to be roughly similar to the output from DCM's path to conversion
Download the file and transform it into something with the columns:
id,lifetime,age,male,event,search,brand
where lifetime is the total time that we observed someone not convert for and event should be 1 if we see a conversion and 0 if we don't. Note that all values should be converted into ints
It is useful to note that end_date = datetime.datetime(2016, 5, 3, 20, 36, 8, 92165)
```
running_id = 0
output = [[0]]
with open("E:/output.txt") as file_open:
for row in file_open.read().split("\n"):
cols = row.split(",")
if cols[0] == output[-1][0]:
output[-1].append(cols[1])
output[-1].append(True)
else:
output.append(cols)
output = output[1:]
for row in output:
if len(row) == 6:
row += [datetime(2016, 5, 3, 20, 36, 8, 92165), False]
output = output[1:-1]
def convert_to_days(dt):
day_diff = dt / np.timedelta64(1, 'D')
if day_diff == 0:
return 23.0
else:
return day_diff
df = pd.DataFrame(output, columns=["id", "advert_time", "male","age","search","brand","conversion_time","event"])
df["lifetime"] = pd.to_datetime(df["conversion_time"]) - pd.to_datetime(df["advert_time"])
df["lifetime"] = df["lifetime"].apply(convert_to_days)
df["male"] = df["male"].astype(int)
df["search"] = df["search"].astype(int)
df["brand"] = df["brand"].astype(int)
df["age"] = df["age"].astype(int)
df["event"] = df["event"].astype(int)
df = df.drop('advert_time', 1)
df = df.drop('conversion_time', 1)
df = df.set_index("id")
df = df.dropna(thresh=2)
df.median()
###Parametric Bayes
#Shout out to Cam Davidson-Pilon
## Example fully worked model using toy data
## Adapted from http://blog.yhat.com/posts/estimating-user-lifetimes-with-pymc.html
## Note that we've made some corrections
N = 2500
##Generate some random data
lifetime = pm.rweibull( 2, 5, size = N )
birth = pm.runiform(0, 10, N)
censor = ((birth + lifetime) >= 10)
lifetime_ = lifetime.copy()
lifetime_[censor] = 10 - birth[censor]
alpha = pm.Uniform('alpha', 0, 20)
beta = pm.Uniform('beta', 0, 20)
@pm.observed
def survival(value=lifetime_, alpha = alpha, beta = beta ):
return sum( (1-censor)*(log( alpha/beta) + (alpha-1)*log(value/beta)) - (value/beta)**(alpha))
mcmc = pm.MCMC([alpha, beta, survival ] )
mcmc.sample(50000, 30000)
pm.Matplot.plot(mcmc)
mcmc.trace("alpha")[:]
```
Problems:
1 - Try to fit your data from section 1
2 - Use the results to plot the distribution of the median
Note that the media of a Weibull distribution is:
$$β(log 2)^{1/α}$$
```
censor = np.array(df["event"].apply(lambda x: 0 if x else 1).tolist())
alpha = pm.Uniform("alpha", 0,50)
beta = pm.Uniform("beta", 0,50)
@pm.observed
def survival(value=df["lifetime"], alpha = alpha, beta = beta ):
return sum( (1-censor)*(np.log( alpha/beta) + (alpha-1)*np.log(value/beta)) - (value/beta)**(alpha))
mcmc = pm.MCMC([alpha, beta, survival ] )
mcmc.sample(10000)
def weibull_median(alpha, beta):
return beta * ((log(2)) ** ( 1 / alpha))
plt.hist([weibull_median(x[0], x[1]) for x in zip(mcmc.trace("alpha"), mcmc.trace("beta"))])
```
Problems:
4 - Try adjusting the number of samples for burning and thinnning
5 - Try adjusting the prior and see how it affects the estimate
```
#### Adjust burn and thin, both paramters of the mcmc sample function
#### Narrow and broaden prior
```
Problems:
7 - Try testing whether the median is greater than a different values
```
#### Hypothesis testing
```
If we want to look at covariates, we need a new approach.
We'll use Cox proprtional hazards, a very popular regression model.
To fit in python we use the module lifelines:
http://lifelines.readthedocs.io/en/latest/
```
### Fit a cox proprtional hazards model
```
Once we've fit the data, we need to do something useful with it. Try to do the following things:
1 - Plot the baseline survival function
2 - Predict the functions for a particular set of features
3 - Plot the survival function for two different set of features
4 - For your results in part 3 caculate how much more likely a death event is for one than the other for a given period of time
```
#### Plot baseline hazard function
#### Predict
#### Plot survival functions for different covariates
#### Plot some odds
```
Model selection
Difficult to do with classic tools (here)
Problem:
1 - Calculate the BMA coefficient values
2 - Try running with different priors
```
#### BMA Coefficient values
#### Different priors
```
| github_jupyter |
# Probability Distributions
# Some typical stuff we'll likely use
```
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%config InlineBackend.figure_format = 'retina'
```
# [SciPy](https://scipy.org)
### [scipy.stats](https://docs.scipy.org/doc/scipy-0.14.0/reference/stats.html)
```
import scipy as sp
import scipy.stats as st
```
# Binomial Distribution
### <font color=darkred> **Example**: A couple, who are both carriers for a recessive disease, wish to have 5 children. They want to know the probability that they will have four healthy kids.</font>
In this case the random variable is the number of healthy kids.
```
# number of trials (kids)
n = 5
# probability of success on each trial
# i.e. probability that each child will be healthy = 1 - 0.5 * 0.5 = 0.75
p = 0.75
# a binomial distribution object
dist = st.binom(n, p)
# probability of four healthy kids
dist.pmf(4)
print(f"The probability of having four healthy kids is {dist.pmf(4):.3f}")
```
### <font color=darkred>Probability to have each of 0-5 healthy kids.</font>
```
# all possible # of successes out of n trials
# i.e. all possible outcomes of the random variable
# i.e. all possible number of healthy kids = 0-5
numHealthyKids = np.arange(n+1)
numHealthyKids
# probability of obtaining each possible number of successes
# i.e. probability of having each possible number of healthy children
pmf = dist.pmf(numHealthyKids)
pmf
```
### <font color=darkred>Visualize the probability to have each of 0-5 healthy kids.</font>
```
plt.bar(numHealthyKids, pmf)
plt.xlabel('# healthy children', fontsize=18)
plt.ylabel('probability', fontsize=18);
```
### <font color=darkred>Probability to have at least 4 healthy kids.</font>
```
# sum of probabilities of 4 and 5 healthy kids
pmf[-2:].sum()
# remaining probability after subtracting CDF for 3 kids
1 - dist.cdf(3)
# survival function for 3 kids
dist.sf(3)
```
### <font color=darkred>What is the expected number of healthy kids?</font>
```
print(f"The expected number of healthy kids is {dist.mean()}")
```
### <font color=darkred>How sure are we about the above estimate?</font>
```
print(f"The expected number of healthy kids is {dist.mean()} ± {dist.std():.2f}")
```
# <font color=red> Exercise</font>
Should the couple consider having six children?
1. Plot the *pmf* for the probability of each possible number of healthy children.
2. What's the probability that they will all be healthy?
# Poisson Distribution
### <font color=darkred> **Example**: Assume that the rate of deleterious mutations is ~1.2 per diploid genome. What is the probability that an individual has 8 or more spontaneous deleterious mutations?</font>
In this case the random variable is the number of deleterious mutations within an individuals genome.
```
# the rate of deleterious mutations is 1.2 per diploid genome
rate = 1.2
# poisson distribution describing the predicted number of spontaneous mutations
dist = st.poisson(rate)
# let's look at the probability for 0-10 mutations
numMutations = np.arange(11)
plt.bar(numMutations, dist.pmf(numMutations))
plt.xlabel('# mutations', fontsize=18)
plt.ylabel('probability', fontsize=18);
print(f"Probability of less than 8 mutations = {dist.cdf(7)}")
print(f"Probability of 8 or more mutations = {dist.sf(7)}")
dist.cdf(7) + dist.sf(7)
```
# <font color=red> Exercise</font>
For the above example, what is the probability that an individual has three or fewer mutations?
# Exponential Distribution
### <font color=darkred> **Example**: Assume that a neuron spikes 1.5 times per second on average. Plot the probability density function of interspike intervals from zero to five seconds with a resolution of 0.01 seconds.</font>
In this case the random variable is the interspike interval time.
```
# spike rate per second
rate = 1.5
# exponential distribution describing the neuron's predicted interspike intervals
dist = st.expon(loc=0, scale=1/rate)
# plot interspike intervals from 0-5 seconds at 0.01 sec resolution
intervalsSec = np.linspace(0, 5, 501)
# probability density for each interval
pdf = dist.pdf(intervalsSec)
plt.plot(intervalsSec, pdf)
plt.xlabel('interspike interval (sec)', fontsize=18)
plt.ylabel('pdf', fontsize=18);
```
### <font color=darkred>What is the average interval?</font>
```
print(f"Average interspike interval = {dist.mean():.2f} seconds.")
```
### <font color=darkred>time constant = 1 / rate = mean</font>
```
tau = 1 / rate
tau
```
### <font color=darkred> What is the probability that an interval will be between 1 and 2 seconds?</font>
```
prob1to2 = dist.cdf(2) - dist.cdf(1);
print(f"Probability of an interspike interval being between 1 and 2 seconds is {prob1to2:.2f}")
```
### <font color=darkred> For what time *T* is the probability that an interval is shorter than *T* equal to 25%?</font>
```
timeAtFirst25PercentOfDist = dist.ppf(0.25) # percent point function
print(f"There is a 25% chance that an interval is shorter than {timeAtFirst25PercentOfDist:.2f} seconds.")
```
# <font color=red> Exercise</font>
For the above example, what is the probability that 3 seconds will pass without any spikes?
# Normal Distribution
### <font color=darkred> **Example**: Under basal conditions the resting membrane voltage of a neuron fluctuates around -70 mV with a variance of 10 mV.</font>
In this case the random variable is the neuron's resting membrane voltage.
```
# mean resting membrane voltage (mV)
mu = -70
# standard deviation about the mean
sd = np.sqrt(10)
# normal distribution describing the neuron's predicted resting membrane voltage
dist = st.norm(mu, sd)
# membrane voltages from -85 to -55 mV
mV = np.linspace(-85, -55, 301)
# probability density for each membrane voltage in mV
pdf = dist.pdf(mV)
plt.plot(mV, pdf)
plt.xlabel('membrane voltage (mV)', fontsize=18)
plt.ylabel('pdf', fontsize=18);
```
### <font color=darkred> What range of membrane voltages (centered on the mean) account for 95% of the probability.</font>
```
low = dist.ppf(0.025) # first 2.5% of distribution
high = dist.ppf(0.975) # first 97.5% of distribution
print(f"95% of membrane voltages are expected to fall within {low :.1f} and {high :.1f} mV.")
```
# <font color=red> Exercise</font>
In a resting neuron, what's the probability that you would measure a membrane voltage greater than -65 mV?
If you meaassure -65 mV, is the neuron at rest?
# <font color=red> Exercise</font>
What probability distribution might best describe the number of synapses per millimeter of dendrite?
A) Binomial
B) Poisson
C) Exponential
D) Normal
# <font color=red> Exercise</font>
What probability distribution might best describe the time a protein spends in its active conformation?
A) Binomial
B) Poisson
C) Exponential
D) Normal
# <font color=red> Exercise</font>
What probability distribution might best describe the weights of adult mice in a colony?
A) Binomial
B) Poisson
C) Exponential
D) Normal
# <font color=red> Exercise</font>
What probability distribution might best describe the number of times a subject is able to identify the correct target in a series of trials?
A) Binomial
B) Poisson
C) Exponential
D) Normal
| github_jupyter |
### Dr. Ignaz Semmelweis
```
import pandas as pd
import matplotlib.pyplot as plt
from IPython.display import display
# Read datasets/yearly_deaths_by_clinic.csv into yearly
yearly = pd.read_csv('datasets/yearly_deaths_by_clinic.csv')
# Print out yearly
display(yearly)
```
### The alarming number of deaths
```
# Calculate proportion of deaths per no. births
yearly['proportion_deaths'] = yearly.deaths / yearly.births
# Extract Clinic 1 data into clinic_1 and Clinic 2 data into clinic_2
clinic_1 = yearly[yearly.clinic == 'clinic 1']
clinic_2 = yearly[yearly.clinic == 'clinic 2']
# Print out clinic_1
display(clinic_2)
```
### Death at the clinics
```
# Plot yearly proportion of deaths at the two clinics
ax = clinic_1.plot(x='year', y='proportion_deaths', label='Clinic 1')
clinic_2.plot(x='year', y='proportion_deaths', label='Clinic 2', ax=ax)
plt.ylabel("Proportion deaths")
plt.show()
```
### The handwashing
```
# Read datasets/monthly_deaths.csv into monthly
monthly = pd.read_csv('datasets/monthly_deaths.csv', parse_dates=['date'])
# Calculate proportion of deaths per no. births
monthly["proportion_deaths"] = monthly.deaths/monthly.births
# Print out the first rows in monthly
display(monthly.head())
```
### The effect of handwashing
```
# Date when handwashing was made mandatory
handwashing_start = pd.to_datetime('1847-06-01')
# Split monthly into before and after handwashing_start
before_washing = monthly[monthly.date < handwashing_start]
after_washing = monthly[monthly.date >= handwashing_start]
# Plot monthly proportion of deaths before and after handwashing
ax = before_washing.plot(x='date',
y='proportion_deaths', label='Before Washing')
after_washing.plot(x='date',y='proportion_deaths', label='After Washing', ax=ax)
plt.ylabel("Proportion deaths")
plt.show()
```
### More handwashing, fewer deaths?
```
# Difference in mean monthly proportion of deaths due to handwashing
before_proportion = before_washing.proportion_deaths
after_proportion = after_washing.proportion_deaths
mean_diff = after_proportion.mean() - before_proportion.mean()
print(mean_diff)
```
### Bootstrap analysis
```
# A bootstrap analysis of the reduction of deaths due to handwashing
boot_mean_diff = []
for i in range(3000):
boot_before = before_proportion.sample(replace=True,n=len(before_proportion))
boot_after = after_proportion.sample(replace=True,n=len(after_proportion))
boot_mean_diff.append(boot_after.mean()-boot_before.mean())
# Calculating a 95% confidence interval from boot_mean_diff
confidence_interval = pd.Series(boot_mean_diff).quantile([0.025, 0.975] )
print(confidence_interval)
```
### Conclusion
```
# The data Semmelweis collected points to that:
doctors_should_wash_their_hands = True
print(doctors_should_wash_their_hands)
```
| github_jupyter |
```
# Import the necessary libraries
import numpy as np
import pandas as pd
import os
import time
import warnings
import gc
gc.collect()
import os
from six.moves import urllib
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
import datetime
warnings.filterwarnings('ignore')
%matplotlib inline
plt.style.use('seaborn')
from scipy import stats
from scipy.stats import norm, skew
from sklearn.preprocessing import StandardScaler
#Add All the Models Libraries
# preprocessing
from sklearn.preprocessing import LabelEncoder
label_enc = LabelEncoder()
# Scalers
from sklearn.utils import shuffle
from sklearn.pipeline import Pipeline
from sklearn.pipeline import FeatureUnion
# Models
from sklearn.linear_model import Lasso
from sklearn.metrics import mean_squared_log_error,mean_squared_error, r2_score,mean_absolute_error
from sklearn.model_selection import train_test_split #training and testing data split
from sklearn import metrics #accuracy measure
from sklearn.metrics import confusion_matrix #for confusion matrix
from scipy.stats import reciprocal, uniform
from sklearn.model_selection import StratifiedKFold, RepeatedKFold
# Cross-validation
from sklearn.model_selection import KFold #for K-fold cross validation
from sklearn.model_selection import cross_val_score #score evaluation
from sklearn.model_selection import cross_val_predict #prediction
from sklearn.model_selection import cross_validate
# GridSearchCV
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import RandomizedSearchCV
#Common data processors
from sklearn.preprocessing import OneHotEncoder, LabelEncoder
from sklearn import feature_selection
from sklearn import model_selection
from sklearn import metrics
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.utils import check_array
from scipy import sparse
# to make this notebook's output stable across runs
np.random.seed(123)
gc.collect()
# To plot pretty figures
%matplotlib inline
plt.rcParams['axes.labelsize'] = 14
plt.rcParams['xtick.labelsize'] = 12
plt.rcParams['ytick.labelsize'] = 12
#Reduce the memory usage - by Panchajanya Banerjee
def reduce_mem_usage(df, verbose=True):
numerics = ['int16', 'int32', 'int64', 'float16', 'float32', 'float64']
start_mem = df.memory_usage().sum() / 1024**2
for col in df.columns:
col_type = df[col].dtypes
if col_type in numerics:
c_min = df[col].min()
c_max = df[col].max()
if str(col_type)[:3] == 'int':
if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:
df[col] = df[col].astype(np.int8)
elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:
df[col] = df[col].astype(np.int16)
elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:
df[col] = df[col].astype(np.int32)
elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:
df[col] = df[col].astype(np.int64)
else:
if c_min > np.finfo(np.float16).min and c_max < np.finfo(np.float16).max:
df[col] = df[col].astype(np.float16)
elif c_min > np.finfo(np.float32).min and c_max < np.finfo(np.float32).max:
df[col] = df[col].astype(np.float32)
else:
df[col] = df[col].astype(np.float64)
end_mem = df.memory_usage().sum() / 1024**2
if verbose: print('Mem. usage decreased to {:5.2f} Mb ({:.1f}% reduction)'.format(end_mem, 100 * (start_mem - end_mem) / start_mem))
return df
train = reduce_mem_usage(pd.read_csv('train.csv',parse_dates=["first_active_month"]))
test = reduce_mem_usage(pd.read_csv('test.csv', parse_dates=["first_active_month"]))
test.first_active_month = test.first_active_month.fillna(pd.to_datetime('2017-09-01'))
test.isnull().sum()
# Now extract the month, year, day, weekday
train["month"] = train["first_active_month"].dt.month
train["year"] = train["first_active_month"].dt.year
train['week'] = train["first_active_month"].dt.weekofyear
train['dayofweek'] = train['first_active_month'].dt.dayofweek
train['days'] = (datetime.date(2018, 2, 1) - train['first_active_month'].dt.date).dt.days
train['quarter'] = train['first_active_month'].dt.quarter
test["month"] = test["first_active_month"].dt.month
test["year"] = test["first_active_month"].dt.year
test['week'] = test["first_active_month"].dt.weekofyear
test['dayofweek'] = test['first_active_month'].dt.dayofweek
test['days'] = (datetime.date(2018, 2, 1) - test['first_active_month'].dt.date).dt.days
test['quarter'] = test['first_active_month'].dt.quarter
# Taking Reference from Other Kernels
def aggregate_transaction_hist(trans, prefix):
agg_func = {
'purchase_date' : ['max','min'],
'month_diff' : ['mean', 'min', 'max', 'var'],
'month_diff_lag' : ['mean', 'min', 'max', 'var'],
'weekend' : ['sum', 'mean'],
'authorized_flag': ['sum', 'mean'],
'category_1': ['sum','mean', 'max','min'],
'purchase_amount': ['sum', 'mean', 'max', 'min', 'std'],
'installments': ['sum', 'mean', 'max', 'min', 'std'],
'month_lag': ['max','min','mean','var'],
'card_id' : ['size'],
'month': ['nunique'],
'hour': ['nunique'],
'weekofyear': ['nunique'],
'dayofweek': ['nunique'],
'year': ['nunique'],
'subsector_id': ['nunique'],
'merchant_category_id' : ['nunique', lambda x:stats.mode(x)[0]],
'merchant_id' : ['nunique', lambda x:stats.mode(x)[0]],
'state_id' : ['nunique', lambda x:stats.mode(x)[0]],
}
agg_trans = trans.groupby(['card_id']).agg(agg_func)
agg_trans.columns = [prefix + '_'.join(col).strip() for col in agg_trans.columns.values]
agg_trans.reset_index(inplace=True)
df = (trans.groupby('card_id').size().reset_index(name='{}transactions_count'.format(prefix)))
agg_trans = pd.merge(df, agg_trans, on='card_id', how='left')
return agg_trans
transactions = reduce_mem_usage(pd.read_csv('historical_transactions_clean_outlier.csv'))
transactions = transactions.loc[transactions.purchase_amount < 50,]
transactions['authorized_flag'] = transactions['authorized_flag'].map({'Y': 1, 'N': 0})
transactions['category_1'] = transactions['category_1'].map({'Y': 0, 'N': 1})
#Feature Engineering - Adding new features
transactions['purchase_date'] = pd.to_datetime(transactions['purchase_date'])
transactions['year'] = transactions['purchase_date'].dt.year
transactions['weekofyear'] = transactions['purchase_date'].dt.weekofyear
transactions['month'] = transactions['purchase_date'].dt.month
transactions['dayofweek'] = transactions['purchase_date'].dt.dayofweek
transactions['weekend'] = (transactions.purchase_date.dt.weekday >=5).astype(int)
transactions['hour'] = transactions['purchase_date'].dt.hour
transactions['quarter'] = transactions['purchase_date'].dt.quarter
transactions['month_diff'] = ((pd.to_datetime('01/03/2018') - transactions['purchase_date']).dt.days)//30
transactions['month_diff_lag'] = transactions['month_diff'] + transactions['month_lag']
gc.collect()
def aggregate_bymonth(trans, prefix):
agg_func = {
'purchase_amount': ['sum', 'mean'],
'card_id' : ['size'],
'merchant_category_id' : ['nunique', lambda x:stats.mode(x)[0]],
# 'merchant_id' : ['nunique', lambda x:stats.mode(x)[0]],
}
agg_trans = trans.groupby(['card_id','month','year']).agg(agg_func)
agg_trans.columns = [prefix + '_'.join(col).strip() for col in agg_trans.columns.values]
agg_trans.reset_index(inplace=True)
df = (trans.groupby('card_id').size().reset_index(name='{}transactions_count'.format(prefix)))
agg_trans = pd.merge(df, agg_trans, on='card_id', how='left')
return agg_trans
merge = aggregate_bymonth(transactions, prefix='hist_')
merge = merge.drop(['hist_transactions_count'], axis = 1)
merge['Date'] = pd.to_datetime(merge[['year', 'month']].assign(Day=1))
df1 = merge.groupby(['card_id', 'hist_merchant_category_id_<lambda>']).size().reset_index(name='Count')
df1 = df1.loc[df1.Count > 1]
df1 = df1.groupby(['card_id']).agg({'Count':['sum']})
df1.columns = ['category_repeated_month']
train = pd.merge(train, df1, on='card_id',how='left')
test = pd.merge(test, df1, on='card_id',how='left')
df1
gc.collect()
## Second last month
amerge = merge.sort_values('Date').groupby('card_id',
as_index=False).apply(lambda x: x.iloc[-2])[['card_id','hist_card_id_size','hist_purchase_amount_sum','hist_purchase_amount_mean']]
new_names = [(i,i+'_last2') for i in amerge.iloc[:, 1:].columns.values]
amerge.rename(columns = dict(new_names), inplace=True)
train = pd.merge(train, amerge, on='card_id',how='left')
test = pd.merge(test, amerge, on='card_id',how='left')
gc.collect()
# last month and first month
merge1 = merge.loc[merge.groupby('card_id').Date.idxmax(),:][[ 'card_id','hist_card_id_size',
'hist_purchase_amount_sum','hist_purchase_amount_mean']]
new_names = [(i,i+'_last') for i in merge1.iloc[:, 1:].columns.values]
merge1.rename(columns = dict(new_names), inplace=True)
merge2 = merge.loc[merge.groupby('card_id').Date.idxmin(),:][['card_id','hist_card_id_size',
'hist_purchase_amount_sum','hist_purchase_amount_mean']]
new_names = [(i,i+'_first') for i in merge2.iloc[:, 1:].columns.values]
merge2.rename(columns = dict(new_names), inplace=True)
comb = pd.merge(merge1, merge2, on='card_id',how='left')
train = pd.merge(train, comb, on='card_id',how='left')
test = pd.merge(test, comb, on='card_id',how='left')
gc.collect()
## Same merchant purchase
df = (transactions.groupby(['card_id','merchant_id','purchase_amount']).size().reset_index(name='count_hist'))
df['purchase_amount_hist'] = df.groupby(['card_id','merchant_id'])['purchase_amount'].transform('sum')
df['count_hist'] = df.groupby(['card_id','merchant_id'])['count_hist'].transform('sum')
df = df.drop_duplicates()
df = df.loc[df['count_hist'] >= 2]
agg_func = {
'count_hist' : ['count'],
'purchase_amount_hist':['sum','mean'],
'purchase_amount':['sum','mean'],
}
df = df.groupby(['card_id']).agg(agg_func)
df.columns = [''.join(col).strip() for col in df.columns.values]
new_names = [(i,i+'_merhist') for i in df.iloc[:, 3:].columns.values]
df.rename(columns = dict(new_names), inplace=True)
train = pd.merge(train, df, on='card_id',how='left')
test = pd.merge(test, df, on='card_id',how='left')
# Same category purchase
df = (transactions.groupby(['card_id','merchant_category_id','purchase_amount']).size().reset_index(name='hist_count'))
df['hist_purchase_amount'] = df.groupby(['card_id','merchant_category_id'])['purchase_amount'].transform('sum')
df['hist_count'] = df.groupby(['card_id','merchant_category_id'])['hist_count'].transform('sum')
df = df.drop_duplicates()
df = df.loc[df['hist_count'] >= 2]
df['hist_count_4'] = 0
df.loc[df['hist_count'] >= 4, 'hist_count_4'] = 1
df['hist_mean4'] = 0
df.loc[df['hist_count'] >= 4, 'hist_mean4'] = df['hist_purchase_amount']/df['hist_count']
agg_fun = {
'hist_count' : ['count'],
'hist_count_4' : ['sum'],
'hist_purchase_amount':['sum','mean'],
'hist_mean4' : ['sum','mean'],
'purchase_amount':['sum','mean'],
}
df = df.groupby(['card_id']).agg(agg_fun)
df.columns = [''.join(col).strip() for col in df.columns.values]
new_names = [(i,'hist'+i) for i in df.iloc[:, 6:].columns.values]
df.rename(columns = dict(new_names), inplace=True)
train = pd.merge(train, df, on='card_id',how='left')
test = pd.merge(test, df, on='card_id',how='left')
# agg_func = {'mean': ['mean'],}
# for col in ['category_2','category_3']:
# transactions[col+'_mean'] = transactions['purchase_amount'].groupby(transactions[col]).agg('mean')
# transactions[col+'_max'] = transactions['purchase_amount'].groupby(transactions[col]).agg('max')
# transactions[col+'_min'] = transactions['purchase_amount'].groupby(transactions[col]).agg('min')
# transactions[col+'_var'] = transactions['purchase_amount'].groupby(transactions[col]).agg('var')
# agg_func[col+'_mean'] = ['mean']
# gc.collect()
merchants = reduce_mem_usage(pd.read_csv('merchants_clean.csv'))
merchants = merchants.drop(['Unnamed: 0', 'merchant_group_id', 'merchant_category_id',
'subsector_id', 'numerical_1', 'numerical_2',
'active_months_lag3','active_months_lag6',
'city_id', 'state_id'
], axis = 1)
d = dict(zip(merchants.columns[1:], ['histchant_{}'.format(x) for x in (merchants.columns[1:])]))
d.update({"merchant_id": "hist_merchant_id_<lambda>"})
merchants = merchants.rename(index=str, columns= d)
## convert the month in business to categorical
merchants.histchant_active_months_lag12 = pd.cut(merchants.histchant_active_months_lag12, 4)
merge_trans = aggregate_transaction_hist(transactions, prefix='hist_')
merge_trans = merge_trans.merge(merchants, on = 'hist_merchant_id_<lambda>', how = 'left')
## hist transaction frequency
merge_trans['hist_freq'] = merge_trans.hist_transactions_count/(((merge_trans.hist_purchase_date_max -
merge_trans.hist_purchase_date_min).dt.total_seconds())/86400)
merge_trans['hist_freq_amount'] = merge_trans['hist_freq'] * merge_trans['hist_purchase_amount_mean']
merge_trans['hist_freq_install'] = merge_trans['hist_freq'] * merge_trans['hist_installments_mean']
cols = ['histchant_avg_sales_lag3','histchant_avg_purchases_lag3',
'histchant_avg_sales_lag6','histchant_avg_purchases_lag6',
'histchant_avg_sales_lag12','histchant_avg_purchases_lag12','hist_freq']
for col in cols:
merge_trans[col] = pd.qcut(merge_trans[col], 4)
for col in cols:
merge_trans[col].fillna(merge_trans[col].mode()[0], inplace=True)
label_enc.fit(list(merge_trans[col].values))
merge_trans[col] = label_enc.transform(list(merge_trans[col].values))
for col in ['histchant_category_1','histchant_most_recent_sales_range','histchant_most_recent_purchases_range',
'histchant_active_months_lag12','histchant_category_4','histchant_category_2']:
merge_trans[col].fillna(merge_trans[col].mode()[0], inplace=True)
label_enc.fit(list(merge_trans['hist_merchant_id_<lambda>'].values))
merge_trans['hist_merchant_id_<lambda>'] = label_enc.transform(list(merge_trans['hist_merchant_id_<lambda>'].values))
label_enc.fit(list(merge_trans['histchant_active_months_lag12'].values))
merge_trans['histchant_active_months_lag12'] = label_enc.transform(list(merge_trans['histchant_active_months_lag12'].values))
#del transactions
gc.collect()
train = pd.merge(train, merge_trans, on='card_id',how='left')
test = pd.merge(test, merge_trans, on='card_id',how='left')
#del merge_trans
gc.collect()
#Feature Engineering - Adding new features
train['hist_purchase_date_max'] = pd.to_datetime(train['hist_purchase_date_max'])
train['hist_purchase_date_min'] = pd.to_datetime(train['hist_purchase_date_min'])
train['hist_purchase_date_diff'] = (train['hist_purchase_date_max'] - train['hist_purchase_date_min']).dt.days
train['hist_purchase_date_average'] = train['hist_purchase_date_diff']/train['hist_card_id_size']
train['hist_purchase_date_uptonow'] = (pd.to_datetime('01/03/2018') - train['hist_purchase_date_max']).dt.days
train['hist_purchase_date_uptomin'] = (pd.to_datetime('01/03/2018') - train['hist_purchase_date_min']).dt.days
train['hist_first_buy'] = (train['hist_purchase_date_min'] - train['first_active_month']).dt.days
for feature in ['hist_purchase_date_max','hist_purchase_date_min']:
train[feature] = train[feature].astype(np.int64) * 1e-9
gc.collect()
#Feature Engineering - Adding new features
test['hist_purchase_date_max'] = pd.to_datetime(test['hist_purchase_date_max'])
test['hist_purchase_date_min'] = pd.to_datetime(test['hist_purchase_date_min'])
test['hist_purchase_date_diff'] = (test['hist_purchase_date_max'] - test['hist_purchase_date_min']).dt.days
test['hist_purchase_date_average'] = test['hist_purchase_date_diff']/test['hist_card_id_size']
test['hist_purchase_date_uptonow'] = (pd.to_datetime('01/03/2018') - test['hist_purchase_date_max']).dt.days
test['hist_purchase_date_uptomin'] = (pd.to_datetime('01/03/2018') - test['hist_purchase_date_min']).dt.days
test['hist_first_buy'] = (test['hist_purchase_date_min'] - test['first_active_month']).dt.days
for feature in ['hist_purchase_date_max','hist_purchase_date_min']:
test[feature] = test[feature].astype(np.int64) * 1e-9
gc.collect()
# Taking Reference from Other Kernels
def aggregate_transaction_new(trans, prefix):
agg_func = {
'purchase_date' : ['max','min'],
'month_diff' : ['mean', 'min', 'max'],
'month_diff_lag' : ['mean', 'min', 'max'],
'weekend' : ['sum', 'mean'],
'authorized_flag': ['sum'],
'category_1': ['sum','mean', 'max','min'],
'purchase_amount': ['sum', 'mean', 'max', 'min'],
'installments': ['sum', 'mean', 'max', 'min'],
'month_lag': ['max','min','mean'],
'card_id' : ['size'],
'month': ['nunique'],
'hour': ['nunique'],
'weekofyear': ['nunique'],
'dayofweek': ['nunique'],
'year': ['nunique'],
'subsector_id': ['nunique'],
'merchant_category_id' : ['nunique', lambda x:stats.mode(x)[0]],
'merchant_id' : ['nunique', lambda x:stats.mode(x)[0]],
'state_id' : ['nunique', lambda x:stats.mode(x)[0]],
}
agg_trans = trans.groupby(['card_id']).agg(agg_func)
agg_trans.columns = [prefix + '_'.join(col).strip() for col in agg_trans.columns.values]
agg_trans.reset_index(inplace=True)
df = (trans.groupby('card_id').size().reset_index(name='{}transactions_count'.format(prefix)))
agg_trans = pd.merge(df, agg_trans, on='card_id', how='left')
return agg_trans
# Now extract the data from the new transactions
new_transactions = reduce_mem_usage(pd.read_csv('new_merchant_transactions_clean_outlier.csv'))
new_transactions = new_transactions.loc[new_transactions.purchase_amount < 50,]
new_transactions['authorized_flag'] = new_transactions['authorized_flag'].map({'Y': 1, 'N': 0})
new_transactions['category_1'] = new_transactions['category_1'].map({'Y': 0, 'N': 1})
#Feature Engineering - Adding new features inspired by Chau's first kernel
new_transactions['purchase_date'] = pd.to_datetime(new_transactions['purchase_date'])
new_transactions['year'] = new_transactions['purchase_date'].dt.year
new_transactions['weekofyear'] = new_transactions['purchase_date'].dt.weekofyear
new_transactions['month'] = new_transactions['purchase_date'].dt.month
new_transactions['dayofweek'] = new_transactions['purchase_date'].dt.dayofweek
new_transactions['weekend'] = (new_transactions.purchase_date.dt.weekday >=5).astype(int)
new_transactions['hour'] = new_transactions['purchase_date'].dt.hour
new_transactions['quarter'] = new_transactions['purchase_date'].dt.quarter
new_transactions['is_month_start'] = new_transactions['purchase_date'].dt.is_month_start
new_transactions['month_diff'] = ((pd.to_datetime('01/03/2018') - new_transactions['purchase_date']).dt.days)//30
new_transactions['month_diff_lag'] = new_transactions['month_diff'] + new_transactions['month_lag']
gc.collect()
# new_transactions['Christmas_Day_2017'] = (pd.to_datetime('2017-12-25') -
# new_transactions['purchase_date']).dt.days.apply(lambda x: x if x > 0 and x <= 15 else 0)
# new_transactions['Valentine_Day_2017'] = (pd.to_datetime('2017-06-13') -
# new_transactions['purchase_date']).dt.days.apply(lambda x: x if x > 0 and x <= 7 else 0)
# #Black Friday : 24th November 2017
# new_transactions['Black_Friday_2017'] = (pd.to_datetime('2017-11-27') -
# new_transactions['purchase_date']).dt.days.apply(lambda x: x if x > 0 and x <= 7 else 0)
# aggs = {'mean': ['mean'],}
# for col in ['category_2','category_3']:
# new_transactions[col+'_mean'] = new_transactions['purchase_amount'].groupby(new_transactions[col]).agg('mean')
# new_transactions[col+'_max'] = new_transactions['purchase_amount'].groupby(new_transactions[col]).agg('max')
# new_transactions[col+'_min'] = new_transactions['purchase_amount'].groupby(new_transactions[col]).agg('min')
# new_transactions[col+'_var'] = new_transactions['purchase_amount'].groupby(new_transactions[col]).agg('var')
# aggs[col+'_mean'] = ['mean']
new_merge = aggregate_bymonth(new_transactions, prefix='new_')
new_merge = new_merge.drop(['new_transactions_count'], axis = 1)
new_merge['Date'] = pd.to_datetime(new_merge[['year', 'month']].assign(Day=1))
gc.collect()
merge1 = new_merge.loc[new_merge.groupby('card_id').Date.idxmax(),:][[ 'card_id','new_card_id_size',
'new_purchase_amount_sum','new_purchase_amount_mean']]
new_names = [(i,i+'_last') for i in merge1.iloc[:, 1:].columns.values]
merge1.rename(columns = dict(new_names), inplace=True)
# merge2 = merge.loc[merge.groupby('card_id').Date.idxmin(),:][['card_id','new_card_id_size',
# 'new_purchase_amount_sum','new_purchase_amount_mean']]
# new_names = [(i,i+'_first') for i in merge2.iloc[:, 1:].columns.values]
# merge2.rename(columns = dict(new_names), inplace=True)
# comb = pd.merge(merge1, merge2, on='card_id',how='left')
train = pd.merge(train, merge1, on='card_id',how='left')
test = pd.merge(test, merge1, on='card_id',how='left')
gc.collect()
## Same merchant purchase
df = (new_transactions.groupby(['card_id','merchant_id','purchase_amount']).size().reset_index(name='count_new'))
df['purchase_amount_new'] = df.groupby(['card_id','merchant_id'])['purchase_amount'].transform('sum')
df['count_new'] = df.groupby(['card_id','merchant_id'])['count_new'].transform('sum')
df = df.drop_duplicates()
df = df.loc[df['count_new'] >= 2]
agg_func = {
'count_new' : ['count'],
'purchase_amount_new':['sum','mean'],
'purchase_amount':['sum','mean'],
}
df = df.groupby(['card_id']).agg(agg_func)
df.columns = [''.join(col).strip() for col in df.columns.values]
new_names = [(i,'new'+i) for i in df.iloc[:, 3:].columns.values]
df.rename(columns = dict(new_names), inplace=True)
train = pd.merge(train, df, on='card_id',how='left')
test = pd.merge(test, df, on='card_id',how='left')
# Same category purchase
df = (new_transactions.groupby(['card_id','merchant_category_id']).size().reset_index(name='new_count'))
df['new_count'] = df.groupby(['card_id','merchant_category_id'])['new_count'].transform('sum')
df = df.drop_duplicates()
df = df.loc[df['new_count'] >= 2]
df['new_count_4'] = 0
df.loc[df['new_count'] >= 4, 'new_count_4'] = 1
agg_fun = {
'new_count' : ['count'],
'new_count_4' : ['sum'],
}
df = df.groupby(['card_id']).agg(agg_fun)
df.columns = [''.join(col).strip() for col in df.columns.values]
train = pd.merge(train, df, on='card_id',how='left')
test = pd.merge(test, df, on='card_id',how='left')
merchants = reduce_mem_usage(pd.read_csv('merchants_clean.csv'))
merchants = merchants.drop(['Unnamed: 0', 'merchant_group_id', 'merchant_category_id',
'subsector_id', 'numerical_1', 'numerical_2',
'active_months_lag3','active_months_lag6',
'city_id', 'state_id',
], axis = 1)
d = dict(zip(merchants.columns[1:], ['newchant_{}'.format(x) for x in (merchants.columns[1:])]))
d.update({"merchant_id": "new_merchant_id_<lambda>"})
merchants = merchants.rename(index=str, columns= d)
## convert the month in business to categorical
merchants.newchant_active_months_lag12 = pd.cut(merchants.newchant_active_months_lag12, 4)
merge_new = aggregate_transaction_new(new_transactions, prefix='new_')
merge_new = merge_new.merge(merchants, on = 'new_merchant_id_<lambda>', how = 'left')
## new transaction frequency
merge_new['new_freq'] = merge_new.new_transactions_count/(((merge_new.new_purchase_date_max -
merge_new.new_purchase_date_min).dt.total_seconds())/86400)
merge_new['new_freq_amount'] = merge_new['new_freq'] * merge_new['new_purchase_amount_mean']
merge_new['new_freq_install'] = merge_new['new_freq'] * merge_new['new_installments_mean']
cols = ['newchant_avg_sales_lag3','newchant_avg_purchases_lag3',
'newchant_avg_sales_lag6','newchant_avg_purchases_lag6',
'newchant_avg_sales_lag12','newchant_avg_purchases_lag12','new_freq']
for col in cols:
merge_new[col] = pd.qcut(merge_new[col], 4)
for col in cols:
merge_new[col].fillna(merge_new[col].mode()[0], inplace=True)
label_enc.fit(list(merge_new[col].values))
merge_new[col] = label_enc.transform(list(merge_new[col].values))
for col in ['newchant_category_1','newchant_most_recent_sales_range','newchant_most_recent_purchases_range',
'newchant_active_months_lag12','newchant_category_4','newchant_category_2']:
merge_new[col].fillna(merge_new[col].mode()[0], inplace=True)
label_enc.fit(list(merge_new['new_merchant_id_<lambda>'].values))
merge_new['new_merchant_id_<lambda>'] = label_enc.transform(list(merge_new['new_merchant_id_<lambda>'].values))
label_enc.fit(list(merge_new['newchant_active_months_lag12'].values))
merge_new['newchant_active_months_lag12'] = label_enc.transform(list(merge_new['newchant_active_months_lag12'].values))
#del new_transactions
gc.collect()
train = pd.merge(train, merge_new, on='card_id',how='left')
test = pd.merge(test, merge_new, on='card_id',how='left')
#del merge_new
gc.collect()
train_na = train.isnull().sum()
train_na = train_na.drop(train_na[train_na == 0].index).sort_values(ascending=False)
missing_data = pd.DataFrame({'Missing Value' :train_na})
missing_data.head(5)
for col in ['new_freq','new_purchase_amount_min','new_purchase_amount_max','newchant_category_4','new_weekend_mean',
'new_purchase_amount_mean','newchant_active_months_lag12','new_weekend_sum','newchant_avg_purchases_lag12',
'newchant_avg_sales_lag12','newchant_avg_purchases_lag6','newchant_avg_sales_lag6','new_category_1_sum',
'newchant_avg_purchases_lag3','newchant_avg_sales_lag3','new_category_1_mean','new_category_1_max',
'new_category_1_min','newchant_most_recent_purchases_range','newchant_most_recent_sales_range',
'newchant_category_1'] : # -1
train[col] = train[col].fillna(-1)
test[col] = test[col].fillna(-1)
for col in ['new_installments_min','new_installments_max','new_installments_mean','new_installments_sum',
'new_purchase_amount_sum','new_state_id_<lambda>' ]: # -2
train[col] = train[col].fillna(-2)
test[col] = test[col].fillna(-2)
for col in ['newchant_category_2','new_authorized_flag_sum','new_month_lag_min','new_month_lag_max','new_card_id_size',
'new_month_lag_mean','new_weekofyear_nunique','new_year_nunique','new_state_id_nunique',
'new_merchant_id_<lambda>','new_merchant_id_nunique','new_merchant_category_id_nunique',
'new_subsector_id_nunique','new_dayofweek_nunique','new_hour_nunique','new_month_nunique',
'new_transactions_count','new_count_4sum','new_countcount','hist_count_4sum','hist_countcount',
'hist_purchase_amountmean','hist_purchase_amountsum','purchase_amount_newmean','purchase_amount_newsum',
'count_newcount','purchase_amount_histmean','purchase_amount_histsum','count_histcount','hist_mean4mean',
'hist_mean4sum','newpurchase_amountmean','newpurchase_amountsum','purchase_amountmean_merhist',
'purchase_amountsum_merhist','histpurchase_amountmean','histpurchase_amountsum',
'new_merchant_category_id_<lambda>','category_repeated_month','new_purchase_amount_mean_last',
'new_purchase_amount_sum_last','new_card_id_size_last']: # 0
train[col] = train[col].fillna(0)
test[col] = test[col].fillna(0)
train.new_month_diff_mean = train.new_month_diff_mean.fillna(23)
train.new_month_diff_min = train.new_month_diff_min.fillna(23)
train.new_month_diff_max = train.new_month_diff_max.fillna(24)
train.new_month_diff_lag_mean = train.new_month_diff_lag_mean.fillna(24)
train.new_month_diff_lag_min = train.new_month_diff_lag_min.fillna(24)
train.new_month_diff_lag_max = train.new_month_diff_lag_max.fillna(24)
test.new_month_diff_mean = test.new_month_diff_mean.fillna(23)
test.new_month_diff_min = test.new_month_diff_min.fillna(23)
test.new_month_diff_max = test.new_month_diff_max.fillna(24)
test.new_month_diff_lag_mean = test.new_month_diff_lag_mean.fillna(24)
test.new_month_diff_lag_min = test.new_month_diff_lag_min.fillna(24)
test.new_month_diff_lag_max = test.new_month_diff_lag_max.fillna(24)
for col in ['new_purchase_date_min','new_purchase_date_max']:
train[col] = train[col].fillna(pd.to_datetime(1/9/2017))
test[col] = test[col].fillna(pd.to_datetime(1/9/2017))
#Feature Engineering - Adding new features inspired by Chau's first kernel
train['total_count_merid'] = train['count_newcount'] + train['count_histcount']
train['total_count'] = train['new_countcount'] + train['hist_countcount']
train['new_purchase_date_max'] = pd.to_datetime(train['new_purchase_date_max'])
train['new_purchase_date_min'] = pd.to_datetime(train['new_purchase_date_min'])
train['new_purchase_date_diff'] = (train['new_purchase_date_max'] - train['new_purchase_date_min']).dt.days
train['new_purchase_date_average'] = train['new_purchase_date_diff']/train['new_card_id_size']
train['new_purchase_date_uptonow'] = (pd.to_datetime('01/03/2018') - train['new_purchase_date_max']).dt.days
train['new_purchase_date_uptomin'] = (pd.to_datetime('01/03/2018') - train['new_purchase_date_min']).dt.days
train['new_first_buy'] = (train['new_purchase_date_min'] - train['first_active_month']).dt.days
for feature in ['new_purchase_date_max','new_purchase_date_min']:
train[feature] = train[feature].astype(np.int64) * 1e-9
#Feature Engineering - Adding new features inspired by Chau's first kernel
test['total_count_merid'] = test['count_newcount'] + test['count_histcount']
test['total_count'] = test['new_countcount'] + test['hist_countcount']
test['new_purchase_date_max'] = pd.to_datetime(test['new_purchase_date_max'])
test['new_purchase_date_min'] = pd.to_datetime(test['new_purchase_date_min'])
test['new_purchase_date_diff'] = (test['new_purchase_date_max'] - test['new_purchase_date_min']).dt.days
test['new_purchase_date_average'] = test['new_purchase_date_diff']/test['new_card_id_size']
test['new_purchase_date_uptonow'] = (pd.to_datetime('01/03/2018') - test['new_purchase_date_max']).dt.days
test['new_purchase_date_uptomin'] = (pd.to_datetime('01/03/2018') - test['new_purchase_date_min']).dt.days
test['new_first_buy'] = (test['new_purchase_date_min'] - test['first_active_month']).dt.days
for feature in ['new_purchase_date_max','new_purchase_date_min']:
test[feature] = test[feature].astype(np.int64) * 1e-9
#added new feature - Interactive
train['card_id_total'] = train['new_card_id_size'] + train['hist_card_id_size']
train['purchase_amount_total'] = train['new_purchase_amount_sum'] + train['hist_purchase_amount_sum']
test['card_id_total'] = test['new_card_id_size'] + test['hist_card_id_size']
test['purchase_amount_total'] = test['new_purchase_amount_sum'] + test['hist_purchase_amount_sum']
gc.collect()
cols = ['new_freq_amount',]
for col in cols:
train[col] = train[col].fillna(0)
train[col] = pd.qcut(train[col], 5)
label_enc.fit(list(train[col].values))
train[col] = label_enc.transform(list(train[col].values))
test[col] = test[col].fillna(0)
test[col] = pd.qcut(test[col], 5)
label_enc.fit(list(test[col].values))
test[col] = label_enc.transform(list(test[col].values))
train = train.drop(['new_freq_install'], axis = 1)
test = test.drop(['new_freq_install'], axis = 1)
train.new_purchase_date_average = train.new_purchase_date_average.fillna(-1.0)
test.new_purchase_date_average = test.new_purchase_date_average.fillna(-1.0)
# last month of new over hist
train['amountmean_ratiolastnew'] = train.new_purchase_amount_mean_last/train.hist_purchase_amount_mean
train['amountsum_ratiolastnew'] = train.new_purchase_amount_sum_last/(train.hist_purchase_amount_sum/(train.hist_purchase_date_diff//30))
train['transcount_ratiolastnew'] = train.new_card_id_size_last/(train.hist_transactions_count/(train.hist_purchase_date_diff//30))
test['amountmean_ratiolastnew'] = test.new_purchase_amount_mean_last/test.hist_purchase_amount_mean
test['amountsum_ratiolastnew'] = test.new_purchase_amount_sum_last/(test.hist_purchase_amount_sum/(test.hist_purchase_date_diff//30))
test['transcount_ratiolastnew'] = test.new_card_id_size_last/(test.hist_transactions_count/(test.hist_purchase_date_diff//30))
# last month of hist over hist
train['amountmean_ratiolast'] = train.hist_purchase_amount_mean_last/train.hist_purchase_amount_mean
train['amountsum_ratiolast'] = train.hist_purchase_amount_sum_last/(train.hist_purchase_amount_sum/(train.hist_purchase_date_diff//30))
train['transcount_ratiolast'] = train.hist_card_id_size_last/(train.hist_transactions_count/(train.hist_purchase_date_diff//30))
test['amountmean_ratiolast'] = test.hist_purchase_amount_mean_last/test.hist_purchase_amount_mean
test['amountsum_ratiolast'] = test.hist_purchase_amount_sum_last/(test.hist_purchase_amount_sum/(test.hist_purchase_date_diff//30))
test['transcount_ratiolast'] = test.hist_card_id_size_last/(test.hist_transactions_count/(test.hist_purchase_date_diff//30))
# last 2 month of hist ratio
train['amountmean_lastlast2'] = train.hist_purchase_amount_mean_last/train.hist_purchase_amount_mean_last2
train['amountsum_lastlast2'] = train.hist_purchase_amount_sum_last/train.hist_purchase_amount_sum_last2
train['transcount_lastlast2'] = train.hist_card_id_size_last/train.hist_card_id_size_last2
test['amountmean_lastlast2'] = test.hist_purchase_amount_mean_last/test.hist_purchase_amount_mean_last2
test['amountsum_lastlast2'] = test.hist_purchase_amount_sum_last/test.hist_purchase_amount_sum_last2
test['transcount_lastlast2'] = test.hist_card_id_size_last/test.hist_card_id_size_last2
# train['amountmean_ratiofirst'] = train.hist_purchase_amount_mean_first/train.hist_purchase_amount_mean
# train['amountsum_ratiofirst'] = train.hist_purchase_amount_sum_first/train.hist_purchase_amount_sum
# train['transcount_ratiofirst'] = train.hist_card_id_size_first/(train.hist_transactions_count/(train.hist_purchase_date_diff//30))
# test['amountmean_ratiofirst'] = test.hist_purchase_amount_mean_first/test.hist_purchase_amount_mean
# test['amountsum_ratiofirst'] = test.hist_purchase_amount_sum_first/test.hist_purchase_amount_sum
# test['transcount_ratiofirst'] = test.hist_card_id_size_first/(test.hist_transactions_count/(test.hist_purchase_date_diff//30))
# train['amountmean_lastfirst'] = train.hist_purchase_amount_mean_last/train.hist_purchase_amount_mean_first
# train['amountsum_lastfirst'] = train.hist_purchase_amount_sum_last/train.hist_purchase_amount_sum_first
# train['transcount_lastfirst'] = train.hist_card_id_size_last/train.hist_card_id_size_first
# test['amountmean_lastfirst'] = test.hist_purchase_amount_mean_last/test.hist_purchase_amount_mean_first
# test['amountsum_lastfirst'] = test.hist_purchase_amount_sum_last/test.hist_purchase_amount_sum_first
# test['transcount_lastfirst'] = test.hist_card_id_size_last/test.hist_card_id_size_first
train = train.drop(['hist_purchase_amount_mean_last2','hist_purchase_amount_sum_last2','hist_card_id_size_last2'], axis = 1)
test = test.drop(['hist_purchase_amount_mean_last2','hist_purchase_amount_sum_last2','hist_card_id_size_last2'], axis = 1)
train = train.drop(['hist_card_id_size','new_card_id_size','card_id', 'first_active_month'], axis = 1)
test = test.drop(['hist_card_id_size','new_card_id_size','card_id', 'first_active_month'], axis = 1)
train.shape
# Remove the Outliers if any
train['outliers'] = 0
train.loc[train['target'] < -30, 'outliers'] = 1
train['outliers'].value_counts()
for features in ['feature_1','feature_2','feature_3']:
order_label = train.groupby([features])['outliers'].mean()
train[features] = train[features].map(order_label)
test[features] = test[features].map(order_label)
# Get the X and Y
df_train_columns = [c for c in train.columns if c not in ['target','outliers']]
cat_features = [c for c in df_train_columns if 'feature_' in c]
#df_train_columns
target = train['target']
del train['target']
import lightgbm as lgb
param = {'num_leaves': 31,
'min_data_in_leaf': 30,
'objective':'regression',
'max_depth': -1,
'learning_rate': 0.01,
"min_child_samples": 20,
"boosting": "gbdt",
"feature_fraction": 0.9,
"bagging_freq": 1,
"bagging_fraction": 0.9 ,
"bagging_seed": 11,
"metric": 'rmse',
"lambda_l1": 0.1,
"verbosity": -1,
"nthread": 4,
"random_state": 4590}
folds = StratifiedKFold(n_splits=6, shuffle=True, random_state=4590)
oof = np.zeros(len(train))
predictions = np.zeros(len(test))
feature_importance_df = pd.DataFrame()
for fold_, (trn_idx, val_idx) in enumerate(folds.split(train,train['outliers'].values)):
print("fold {}".format(fold_))
trn_data = lgb.Dataset(train.iloc[trn_idx][df_train_columns], label=target.iloc[trn_idx])
val_data = lgb.Dataset(train.iloc[val_idx][df_train_columns], label=target.iloc[val_idx])
num_round = 10000
clf = lgb.train(param, trn_data, num_round, valid_sets = [trn_data, val_data], verbose_eval=-1, early_stopping_rounds = 200)
oof[val_idx] = clf.predict(train.iloc[val_idx][df_train_columns], num_iteration=clf.best_iteration)
fold_importance_df = pd.DataFrame()
fold_importance_df["Feature"] = df_train_columns
fold_importance_df["importance"] = clf.feature_importance()
fold_importance_df["fold"] = fold_ + 1
feature_importance_df = pd.concat([feature_importance_df, fold_importance_df], axis=0)
predictions += clf.predict(test[df_train_columns], num_iteration=clf.best_iteration) / folds.n_splits
np.sqrt(mean_squared_error(oof, target))
cols = (feature_importance_df[["Feature", "importance"]]
.groupby("Feature")
.mean()
.sort_values(by="importance", ascending=False)[:1000].index)
best_features = feature_importance_df.loc[feature_importance_df.Feature.isin(cols)]
plt.figure(figsize=(14,25))
sns.barplot(x="importance",
y="Feature",
data=best_features.sort_values(by="importance",
ascending=False))
plt.title('LightGBM Features (avg over folds)')
plt.tight_layout()
plt.savefig('lgbm_importances.png')
features = [c for c in train.columns if c not in ['card_id', 'first_active_month','target','outliers']]
cat_features = [c for c in features if 'feature_' in c]
param = {'num_leaves': 31,
'min_data_in_leaf': 30,
'objective':'regression',
'max_depth': -1,
'learning_rate': 0.01,
"min_child_samples": 20,
"boosting": "gbdt",
"feature_fraction": 0.9,
"bagging_freq": 1,
"bagging_fraction": 0.9 ,
"bagging_seed": 11,
"metric": 'rmse',
"lambda_l1": 0.1,
"verbosity": -1,
"nthread": 4,
"random_state": 4590}
folds = RepeatedKFold(n_splits=6, n_repeats=2, random_state=4590)
oof_2 = np.zeros(len(train))
predictions_2 = np.zeros(len(test))
feature_importance_df_2 = pd.DataFrame()
for fold_, (trn_idx, val_idx) in enumerate(folds.split(train.values, target.values)):
print("fold {}".format(fold_))
trn_data = lgb.Dataset(train.iloc[trn_idx][features], label=target.iloc[trn_idx], categorical_feature=cat_features)
val_data = lgb.Dataset(train.iloc[val_idx][features], label=target.iloc[val_idx], categorical_feature=cat_features)
num_round = 10000
clf_r = lgb.train(param, trn_data, num_round, valid_sets = [trn_data, val_data], verbose_eval=-1, early_stopping_rounds = 200)
oof_2[val_idx] = clf_r.predict(train.iloc[val_idx][features], num_iteration=clf_r.best_iteration)
fold_importance_df = pd.DataFrame()
fold_importance_df["Feature"] = features
fold_importance_df["importance"] = clf_r.feature_importance()
fold_importance_df["fold"] = fold_ + 1
feature_importance_df_2 = pd.concat([feature_importance_df_2, fold_importance_df], axis=0)
predictions_2 += clf_r.predict(test[features], num_iteration=clf_r.best_iteration) / (5 * 2)
print("CV score: {:<8.5f}".format(mean_squared_error(oof_2, target)**0.5))
cols = (feature_importance_df_2[["Feature", "importance"]]
.groupby("Feature")
.mean()
.sort_values(by="importance", ascending=False)[:1000].index)
best_features = feature_importance_df_2.loc[feature_importance_df_2.Feature.isin(cols)]
plt.figure(figsize=(14,25))
sns.barplot(x="importance",
y="Feature",
data=best_features.sort_values(by="importance",
ascending=False))
plt.title('LightGBM Features (avg over folds)')
plt.tight_layout()
plt.savefig('lgbm_importances.png')
from sklearn.linear_model import BayesianRidge
train_stack = np.vstack([oof,oof_2]).transpose()
test_stack = np.vstack([predictions, predictions_2]).transpose()
folds_stack = RepeatedKFold(n_splits=6, n_repeats=1, random_state=4590)
oof_stack = np.zeros(train_stack.shape[0])
predictions_3 = np.zeros(test_stack.shape[0])
for fold_, (trn_idx, val_idx) in enumerate(folds_stack.split(train_stack,target)):
print("fold {}".format(fold_))
trn_data, trn_y = train_stack[trn_idx], target.iloc[trn_idx].values
val_data, val_y = train_stack[val_idx], target.iloc[val_idx].values
clf_3 = BayesianRidge()
clf_3.fit(trn_data, trn_y)
oof_stack[val_idx] = clf_3.predict(val_data)
predictions_3 += clf_3.predict(test_stack) / 6
np.sqrt(mean_squared_error(target.values, oof_stack))
sample_submission = pd.read_csv('sample_submission.csv')
sample_submission['target'] = predictions_3
# combine = pd.read_csv('combining_submission.csv')
# sample_submission['target'] = predictions_3*0.7 + combine['target']*0.3
q = sample_submission['target'].quantile(0.002)
# #sample_submission['target'] = sample_submission['target'].apply(lambda x: x if x > q else x*1.04)
# sample_submission.loc[sample_submission.target < -19.3, 'target'] = -33.218750
# for i in [2726,17430,28039,42686]:
# sample_submission['target'][i] = -33.21875
sample_submission.to_csv('submission.csv', index=False)
((sample_submission.target <= -30) & (sample_submission.target > -35)).sum()
sample_submission.iloc[108111]
q
sample_submission.loc[sample_submission.target < -19.5]
sample_submission.head(5)
my = pd.read_csv('submission (1).csv')
my['target'][96354] = -33.218750
my.to_csv('submission96354.csv', index=False)
```
## Classification
```
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, ExtraTreesClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.base import BaseEstimator, TransformerMixin, RegressorMixin, clone
from sklearn.model_selection import KFold, cross_val_score, train_test_split, GridSearchCV, StratifiedKFold
from sklearn.metrics import mean_squared_error, accuracy_score
from sklearn.preprocessing import LabelEncoder
y_train = train['outliers']
del train['outliers']
train['target'] = target
test['target'] = predictions_3
models = [RandomForestClassifier(),ExtraTreesClassifier()]
names = ["RF", "Xtree"]
dict_score = {}
for name, model in zip(names, models):
model.fit(train, y_train)
model_train_pred = model.predict(train)
accy = round(accuracy_score(y_train, model_train_pred), 6)
dict_score[name] = accy
import operator
dict_score = sorted(dict_score.items(), key = operator.itemgetter(1), reverse = True)
dict_score
Xtree = ExtraTreesClassifier()
XtreeMd = Xtree.fit(train, y_train)
y_pred = XtreeMd.predict(test)
sample_submission['outliers'] = y_pred
sample_submission.loc[sample_submission['outliers'] == 1, 'target'] = -33.218750
sample_submission = sample_submission.drop(['outliers'], axis = 1)
sample_submission.to_csv('submission.csv', index=False)
sample_submission.loc[sample_submission['target'] == -33.21875][:40]
```
| github_jupyter |
# What's this PyTorch business?
You've written a lot of code in this assignment to provide a whole host of neural network functionality. Dropout, Batch Norm, and 2D convolutions are some of the workhorses of deep learning in computer vision. You've also worked hard to make your code efficient and vectorized.
For the last part of this assignment, though, we're going to leave behind your beautiful codebase and instead migrate to one of two popular deep learning frameworks: in this instance, PyTorch (or TensorFlow, if you choose to use that notebook).
### What is PyTorch?
PyTorch is a system for executing dynamic computational graphs over Tensor objects that behave similarly as numpy ndarray. It comes with a powerful automatic differentiation engine that removes the need for manual back-propagation.
### Why?
* Our code will now run on GPUs! Much faster training. When using a framework like PyTorch or TensorFlow you can harness the power of the GPU for your own custom neural network architectures without having to write CUDA code directly (which is beyond the scope of this class).
* We want you to be ready to use one of these frameworks for your project so you can experiment more efficiently than if you were writing every feature you want to use by hand.
* We want you to stand on the shoulders of giants! TensorFlow and PyTorch are both excellent frameworks that will make your lives a lot easier, and now that you understand their guts, you are free to use them :)
* We want you to be exposed to the sort of deep learning code you might run into in academia or industry.
### PyTorch versions
This notebook assumes that you are using **PyTorch version 1.0**. In some of the previous versions (e.g. before 0.4), Tensors had to be wrapped in Variable objects to be used in autograd; however Variables have now been deprecated. In addition 1.0 also separates a Tensor's datatype from its device, and uses numpy-style factories for constructing Tensors rather than directly invoking Tensor constructors.
## How will I learn PyTorch?
Justin Johnson has made an excellent [tutorial](https://github.com/jcjohnson/pytorch-examples) for PyTorch.
You can also find the detailed [API doc](http://pytorch.org/docs/stable/index.html) here. If you have other questions that are not addressed by the API docs, the [PyTorch forum](https://discuss.pytorch.org/) is a much better place to ask than StackOverflow.
# Table of Contents
This assignment has 5 parts. You will learn PyTorch on **three different levels of abstraction**, which will help you understand it better and prepare you for the final project.
1. Part I, Preparation: we will use CIFAR-10 dataset.
2. Part II, Barebones PyTorch: **Abstraction level 1**, we will work directly with the lowest-level PyTorch Tensors.
3. Part III, PyTorch Module API: **Abstraction level 2**, we will use `nn.Module` to define arbitrary neural network architecture.
4. Part IV, PyTorch Sequential API: **Abstraction level 3**, we will use `nn.Sequential` to define a linear feed-forward network very conveniently.
5. Part V, CIFAR-10 open-ended challenge: please implement your own network to get as high accuracy as possible on CIFAR-10. You can experiment with any layer, optimizer, hyperparameters or other advanced features.
Here is a table of comparison:
| API | Flexibility | Convenience |
|---------------|-------------|-------------|
| Barebone | High | Low |
| `nn.Module` | High | Medium |
| `nn.Sequential` | Low | High |
# Part I. Preparation
First, we load the CIFAR-10 dataset. This might take a couple minutes the first time you do it, but the files should stay cached after that.
In previous parts of the assignment we had to write our own code to download the CIFAR-10 dataset, preprocess it, and iterate through it in minibatches; PyTorch provides convenient tools to automate this process for us.
```
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import DataLoader
from torch.utils.data import sampler
import torchvision.datasets as dset
import torchvision.transforms as T
import numpy as np
NUM_TRAIN = 49000
# The torchvision.transforms package provides tools for preprocessing data
# and for performing data augmentation; here we set up a transform to
# preprocess the data by subtracting the mean RGB value and dividing by the
# standard deviation of each RGB value; we've hardcoded the mean and std.
transform = T.Compose([
T.ToTensor(),
T.Normalize((0.4914, 0.4822, 0.4465), (0.2023, 0.1994, 0.2010))
])
# We set up a Dataset object for each split (train / val / test); Datasets load
# training examples one at a time, so we wrap each Dataset in a DataLoader which
# iterates through the Dataset and forms minibatches. We divide the CIFAR-10
# training set into train and val sets by passing a Sampler object to the
# DataLoader telling how it should sample from the underlying Dataset.
cifar10_train = dset.CIFAR10('./cs231n/datasets', train=True, download=True,
transform=transform)
loader_train = DataLoader(cifar10_train, batch_size=64,
sampler=sampler.SubsetRandomSampler(range(NUM_TRAIN)))
cifar10_val = dset.CIFAR10('./cs231n/datasets', train=True, download=True,
transform=transform)
loader_val = DataLoader(cifar10_val, batch_size=64,
sampler=sampler.SubsetRandomSampler(range(NUM_TRAIN, 50000)))
cifar10_test = dset.CIFAR10('./cs231n/datasets', train=False, download=True,
transform=transform)
loader_test = DataLoader(cifar10_test, batch_size=64)
```
You have an option to **use GPU by setting the flag to True below**. It is not necessary to use GPU for this assignment. Note that if your computer does not have CUDA enabled, `torch.cuda.is_available()` will return False and this notebook will fallback to CPU mode.
The global variables `dtype` and `device` will control the data types throughout this assignment.
```
USE_GPU = True
dtype = torch.float32 # we will be using float throughout this tutorial
if USE_GPU and torch.cuda.is_available():
device = torch.device('cuda')
else:
device = torch.device('cpu')
# Constant to control how frequently we print train loss
print_every = 100
print('using device:', device)
```
# Part II. Barebones PyTorch
PyTorch ships with high-level APIs to help us define model architectures conveniently, which we will cover in Part II of this tutorial. In this section, we will start with the barebone PyTorch elements to understand the autograd engine better. After this exercise, you will come to appreciate the high-level model API more.
We will start with a simple fully-connected ReLU network with two hidden layers and no biases for CIFAR classification.
This implementation computes the forward pass using operations on PyTorch Tensors, and uses PyTorch autograd to compute gradients. It is important that you understand every line, because you will write a harder version after the example.
When we create a PyTorch Tensor with `requires_grad=True`, then operations involving that Tensor will not just compute values; they will also build up a computational graph in the background, allowing us to easily backpropagate through the graph to compute gradients of some Tensors with respect to a downstream loss. Concretely if x is a Tensor with `x.requires_grad == True` then after backpropagation `x.grad` will be another Tensor holding the gradient of x with respect to the scalar loss at the end.
### PyTorch Tensors: Flatten Function
A PyTorch Tensor is conceptionally similar to a numpy array: it is an n-dimensional grid of numbers, and like numpy PyTorch provides many functions to efficiently operate on Tensors. As a simple example, we provide a `flatten` function below which reshapes image data for use in a fully-connected neural network.
Recall that image data is typically stored in a Tensor of shape N x C x H x W, where:
* N is the number of datapoints
* C is the number of channels
* H is the height of the intermediate feature map in pixels
* W is the height of the intermediate feature map in pixels
This is the right way to represent the data when we are doing something like a 2D convolution, that needs spatial understanding of where the intermediate features are relative to each other. When we use fully connected affine layers to process the image, however, we want each datapoint to be represented by a single vector -- it's no longer useful to segregate the different channels, rows, and columns of the data. So, we use a "flatten" operation to collapse the `C x H x W` values per representation into a single long vector. The flatten function below first reads in the N, C, H, and W values from a given batch of data, and then returns a "view" of that data. "View" is analogous to numpy's "reshape" method: it reshapes x's dimensions to be N x ??, where ?? is allowed to be anything (in this case, it will be C x H x W, but we don't need to specify that explicitly).
```
def flatten(x):
N = x.shape[0] # read in N, C, H, W
return x.view(N, -1) # "flatten" the C * H * W values into a single vector per image
def test_flatten():
x = torch.arange(12).view(2, 1, 3, 2)
print('Before flattening: ', x)
print('After flattening: ', flatten(x))
test_flatten()
```
### Barebones PyTorch: Two-Layer Network
Here we define a function `two_layer_fc` which performs the forward pass of a two-layer fully-connected ReLU network on a batch of image data. After defining the forward pass we check that it doesn't crash and that it produces outputs of the right shape by running zeros through the network.
You don't have to write any code here, but it's important that you read and understand the implementation.
```
import torch.nn.functional as F # useful stateless functions
def two_layer_fc(x, params):
"""
A fully-connected neural networks; the architecture is:
NN is fully connected -> ReLU -> fully connected layer.
Note that this function only defines the forward pass;
PyTorch will take care of the backward pass for us.
The input to the network will be a minibatch of data, of shape
(N, d1, ..., dM) where d1 * ... * dM = D. The hidden layer will have H units,
and the output layer will produce scores for C classes.
Inputs:
- x: A PyTorch Tensor of shape (N, d1, ..., dM) giving a minibatch of
input data.
- params: A list [w1, w2] of PyTorch Tensors giving weights for the network;
w1 has shape (D, H) and w2 has shape (H, C).
Returns:
- scores: A PyTorch Tensor of shape (N, C) giving classification scores for
the input data x.
"""
# first we flatten the image
x = flatten(x) # shape: [batch_size, C x H x W]
w1, w2 = params
# Forward pass: compute predicted y using operations on Tensors. Since w1 and
# w2 have requires_grad=True, operations involving these Tensors will cause
# PyTorch to build a computational graph, allowing automatic computation of
# gradients. Since we are no longer implementing the backward pass by hand we
# don't need to keep references to intermediate values.
# you can also use `.clamp(min=0)`, equivalent to F.relu()
x = F.relu(x.mm(w1))
x = x.mm(w2)
return x
def two_layer_fc_test():
hidden_layer_size = 42
x = torch.zeros((64, 50), dtype=dtype) # minibatch size 64, feature dimension 50
w1 = torch.zeros((50, hidden_layer_size), dtype=dtype)
w2 = torch.zeros((hidden_layer_size, 10), dtype=dtype)
scores = two_layer_fc(x, [w1, w2])
print(scores.size()) # you should see [64, 10]
two_layer_fc_test()
```
### Barebones PyTorch: Three-Layer ConvNet
Here you will complete the implementation of the function `three_layer_convnet`, which will perform the forward pass of a three-layer convolutional network. Like above, we can immediately test our implementation by passing zeros through the network. The network should have the following architecture:
1. A convolutional layer (with bias) with `channel_1` filters, each with shape `KW1 x KH1`, and zero-padding of two
2. ReLU nonlinearity
3. A convolutional layer (with bias) with `channel_2` filters, each with shape `KW2 x KH2`, and zero-padding of one
4. ReLU nonlinearity
5. Fully-connected layer with bias, producing scores for C classes.
Note that we have **no softmax activation** here after our fully-connected layer: this is because PyTorch's cross entropy loss performs a softmax activation for you, and by bundling that step in makes computation more efficient.
**HINT**: For convolutions: http://pytorch.org/docs/stable/nn.html#torch.nn.functional.conv2d; pay attention to the shapes of convolutional filters!
```
def three_layer_convnet(x, params):
"""
Performs the forward pass of a three-layer convolutional network with the
architecture defined above.
Inputs:
- x: A PyTorch Tensor of shape (N, 3, H, W) giving a minibatch of images
- params: A list of PyTorch Tensors giving the weights and biases for the
network; should contain the following:
- conv_w1: PyTorch Tensor of shape (channel_1, 3, KH1, KW1) giving weights
for the first convolutional layer
- conv_b1: PyTorch Tensor of shape (channel_1,) giving biases for the first
convolutional layer
- conv_w2: PyTorch Tensor of shape (channel_2, channel_1, KH2, KW2) giving
weights for the second convolutional layer
- conv_b2: PyTorch Tensor of shape (channel_2,) giving biases for the second
convolutional layer
- fc_w: PyTorch Tensor giving weights for the fully-connected layer. Can you
figure out what the shape should be?
- fc_b: PyTorch Tensor giving biases for the fully-connected layer. Can you
figure out what the shape should be?
Returns:
- scores: PyTorch Tensor of shape (N, C) giving classification scores for x
"""
conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b = params
scores = None
################################################################################
# TODO: Implement the forward pass for the three-layer ConvNet. #
################################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
scores=F.relu_(F.conv2d(x,conv_w1,conv_b1,padding=2))
scores=F.relu_(F.conv2d(scores,conv_w2,conv_b2,padding=1))
scores=F.linear(flatten(scores),fc_w.T,fc_b)
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
################################################################################
# END OF YOUR CODE #
################################################################################
return scores
```
After defining the forward pass of the ConvNet above, run the following cell to test your implementation.
When you run this function, scores should have shape (64, 10).
```
def three_layer_convnet_test():
x = torch.zeros((64, 3, 32, 32), dtype=dtype) # minibatch size 64, image size [3, 32, 32]
conv_w1 = torch.zeros((6, 3, 5, 5), dtype=dtype) # [out_channel, in_channel, kernel_H, kernel_W]
conv_b1 = torch.zeros((6,)) # out_channel
conv_w2 = torch.zeros((9, 6, 3, 3), dtype=dtype) # [out_channel, in_channel, kernel_H, kernel_W]
conv_b2 = torch.zeros((9,)) # out_channel
# you must calculate the shape of the tensor after two conv layers, before the fully-connected layer
fc_w = torch.zeros((9 * 32 * 32, 10))
fc_b = torch.zeros(10)
scores = three_layer_convnet(x, [conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b])
print(scores.size()) # you should see [64, 10]
three_layer_convnet_test()
```
### Barebones PyTorch: Initialization
Let's write a couple utility methods to initialize the weight matrices for our models.
- `random_weight(shape)` initializes a weight tensor with the Kaiming normalization method.
- `zero_weight(shape)` initializes a weight tensor with all zeros. Useful for instantiating bias parameters.
The `random_weight` function uses the Kaiming normal initialization method, described in:
He et al, *Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification*, ICCV 2015, https://arxiv.org/abs/1502.01852
```
def random_weight(shape):
"""
Create random Tensors for weights; setting requires_grad=True means that we
want to compute gradients for these Tensors during the backward pass.
We use Kaiming normalization: sqrt(2 / fan_in)
"""
if len(shape) == 2: # FC weight
fan_in = shape[0]
else:
fan_in = np.prod(shape[1:]) # conv weight [out_channel, in_channel, kH, kW]
# randn is standard normal distribution generator.
w = torch.randn(shape, device=device, dtype=dtype) * np.sqrt(2. / fan_in)
w.requires_grad = True
return w
def zero_weight(shape):
return torch.zeros(shape, device=device, dtype=dtype, requires_grad=True)
# create a weight of shape [3 x 5]
# you should see the type `torch.cuda.FloatTensor` if you use GPU.
# Otherwise it should be `torch.FloatTensor`
random_weight((3, 5))
```
### Barebones PyTorch: Check Accuracy
When training the model we will use the following function to check the accuracy of our model on the training or validation sets.
When checking accuracy we don't need to compute any gradients; as a result we don't need PyTorch to build a computational graph for us when we compute scores. To prevent a graph from being built we scope our computation under a `torch.no_grad()` context manager.
```
def check_accuracy_part2(loader, model_fn, params):
"""
Check the accuracy of a classification model.
Inputs:
- loader: A DataLoader for the data split we want to check
- model_fn: A function that performs the forward pass of the model,
with the signature scores = model_fn(x, params)
- params: List of PyTorch Tensors giving parameters of the model
Returns: Nothing, but prints the accuracy of the model
"""
split = 'val' if loader.dataset.train else 'test'
print('Checking accuracy on the %s set' % split)
num_correct, num_samples = 0, 0
with torch.no_grad():
for x, y in loader:
x = x.to(device=device, dtype=dtype) # move to device, e.g. GPU
y = y.to(device=device, dtype=torch.int64)
scores = model_fn(x, params)
_, preds = scores.max(1)
num_correct += (preds == y).sum()
num_samples += preds.size(0)
acc = float(num_correct) / num_samples
print('Got %d / %d correct (%.2f%%)' % (num_correct, num_samples, 100 * acc))
```
### BareBones PyTorch: Training Loop
We can now set up a basic training loop to train our network. We will train the model using stochastic gradient descent without momentum. We will use `torch.functional.cross_entropy` to compute the loss; you can [read about it here](http://pytorch.org/docs/stable/nn.html#cross-entropy).
The training loop takes as input the neural network function, a list of initialized parameters (`[w1, w2]` in our example), and learning rate.
```
def train_part2(model_fn, params, learning_rate):
"""
Train a model on CIFAR-10.
Inputs:
- model_fn: A Python function that performs the forward pass of the model.
It should have the signature scores = model_fn(x, params) where x is a
PyTorch Tensor of image data, params is a list of PyTorch Tensors giving
model weights, and scores is a PyTorch Tensor of shape (N, C) giving
scores for the elements in x.
- params: List of PyTorch Tensors giving weights for the model
- learning_rate: Python scalar giving the learning rate to use for SGD
Returns: Nothing
"""
for t, (x, y) in enumerate(loader_train):
# Move the data to the proper device (GPU or CPU)
x = x.to(device=device, dtype=dtype)
y = y.to(device=device, dtype=torch.long)
# Forward pass: compute scores and loss
scores = model_fn(x, params)
loss = F.cross_entropy(scores, y)
# Backward pass: PyTorch figures out which Tensors in the computational
# graph has requires_grad=True and uses backpropagation to compute the
# gradient of the loss with respect to these Tensors, and stores the
# gradients in the .grad attribute of each Tensor.
loss.backward()
# Update parameters. We don't want to backpropagate through the
# parameter updates, so we scope the updates under a torch.no_grad()
# context manager to prevent a computational graph from being built.
with torch.no_grad():
for w in params:
w -= learning_rate * w.grad
# Manually zero the gradients after running the backward pass
w.grad.zero_()
if t % print_every == 0:
print('Iteration %d, loss = %.4f' % (t, loss.item()))
check_accuracy_part2(loader_val, model_fn, params)
print()
```
### BareBones PyTorch: Train a Two-Layer Network
Now we are ready to run the training loop. We need to explicitly allocate tensors for the fully connected weights, `w1` and `w2`.
Each minibatch of CIFAR has 64 examples, so the tensor shape is `[64, 3, 32, 32]`.
After flattening, `x` shape should be `[64, 3 * 32 * 32]`. This will be the size of the first dimension of `w1`.
The second dimension of `w1` is the hidden layer size, which will also be the first dimension of `w2`.
Finally, the output of the network is a 10-dimensional vector that represents the probability distribution over 10 classes.
You don't need to tune any hyperparameters but you should see accuracies above 40% after training for one epoch.
```
hidden_layer_size = 4000
learning_rate = 1e-2
w1 = random_weight((3 * 32 * 32, hidden_layer_size))
w2 = random_weight((hidden_layer_size, 10))
train_part2(two_layer_fc, [w1, w2], learning_rate)
```
### BareBones PyTorch: Training a ConvNet
In the below you should use the functions defined above to train a three-layer convolutional network on CIFAR. The network should have the following architecture:
1. Convolutional layer (with bias) with 32 5x5 filters, with zero-padding of 2
2. ReLU
3. Convolutional layer (with bias) with 16 3x3 filters, with zero-padding of 1
4. ReLU
5. Fully-connected layer (with bias) to compute scores for 10 classes
You should initialize your weight matrices using the `random_weight` function defined above, and you should initialize your bias vectors using the `zero_weight` function above.
You don't need to tune any hyperparameters, but if everything works correctly you should achieve an accuracy above 42% after one epoch.
```
learning_rate = 3e-3
channel_1 = 32
channel_2 = 16
conv_w1 = None
conv_b1 = None
conv_w2 = None
conv_b2 = None
fc_w = None
fc_b = None
################################################################################
# TODO: Initialize the parameters of a three-layer ConvNet. #
################################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
conv_w1=random_weight((channel_1,3,5,5))
conv_b1=zero_weight(channel_1)
conv_w2=random_weight((channel_2,channel_1,3,3))
conv_b2=zero_weight(channel_2)
fc_w=random_weight((16*32*32,10))
fc_b=zero_weight(10)
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
################################################################################
# END OF YOUR CODE #
################################################################################
params = [conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b]
train_part2(three_layer_convnet, params, learning_rate)
```
# Part III. PyTorch Module API
Barebone PyTorch requires that we track all the parameter tensors by hand. This is fine for small networks with a few tensors, but it would be extremely inconvenient and error-prone to track tens or hundreds of tensors in larger networks.
PyTorch provides the `nn.Module` API for you to define arbitrary network architectures, while tracking every learnable parameters for you. In Part II, we implemented SGD ourselves. PyTorch also provides the `torch.optim` package that implements all the common optimizers, such as RMSProp, Adagrad, and Adam. It even supports approximate second-order methods like L-BFGS! You can refer to the [doc](http://pytorch.org/docs/master/optim.html) for the exact specifications of each optimizer.
To use the Module API, follow the steps below:
1. Subclass `nn.Module`. Give your network class an intuitive name like `TwoLayerFC`.
2. In the constructor `__init__()`, define all the layers you need as class attributes. Layer objects like `nn.Linear` and `nn.Conv2d` are themselves `nn.Module` subclasses and contain learnable parameters, so that you don't have to instantiate the raw tensors yourself. `nn.Module` will track these internal parameters for you. Refer to the [doc](http://pytorch.org/docs/master/nn.html) to learn more about the dozens of builtin layers. **Warning**: don't forget to call the `super().__init__()` first!
3. In the `forward()` method, define the *connectivity* of your network. You should use the attributes defined in `__init__` as function calls that take tensor as input and output the "transformed" tensor. Do *not* create any new layers with learnable parameters in `forward()`! All of them must be declared upfront in `__init__`.
After you define your Module subclass, you can instantiate it as an object and call it just like the NN forward function in part II.
### Module API: Two-Layer Network
Here is a concrete example of a 2-layer fully connected network:
```
class TwoLayerFC(nn.Module):
def __init__(self, input_size, hidden_size, num_classes):
super().__init__()
# assign layer objects to class attributes
self.fc1 = nn.Linear(input_size, hidden_size)
# nn.init package contains convenient initialization methods
# http://pytorch.org/docs/master/nn.html#torch-nn-init
nn.init.kaiming_normal_(self.fc1.weight)
self.fc2 = nn.Linear(hidden_size, num_classes)
nn.init.kaiming_normal_(self.fc2.weight)
def forward(self, x):
# forward always defines connectivity
x = flatten(x)
scores = self.fc2(F.relu(self.fc1(x)))
return scores
def test_TwoLayerFC():
input_size = 50
x = torch.zeros((64, input_size), dtype=dtype) # minibatch size 64, feature dimension 50
model = TwoLayerFC(input_size, 42, 10)
scores = model(x)
print(scores.size()) # you should see [64, 10]
test_TwoLayerFC()
```
### Module API: Three-Layer ConvNet
It's your turn to implement a 3-layer ConvNet followed by a fully connected layer. The network architecture should be the same as in Part II:
1. Convolutional layer with `channel_1` 5x5 filters with zero-padding of 2
2. ReLU
3. Convolutional layer with `channel_2` 3x3 filters with zero-padding of 1
4. ReLU
5. Fully-connected layer to `num_classes` classes
You should initialize the weight matrices of the model using the Kaiming normal initialization method.
**HINT**: http://pytorch.org/docs/stable/nn.html#conv2d
After you implement the three-layer ConvNet, the `test_ThreeLayerConvNet` function will run your implementation; it should print `(64, 10)` for the shape of the output scores.
```
class ThreeLayerConvNet(nn.Module):
def __init__(self, in_channel, channel_1, channel_2, num_classes):
super().__init__()
########################################################################
# TODO: Set up the layers you need for a three-layer ConvNet with the #
# architecture defined above. #
########################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
self.conv1=nn.Conv2d(in_channel,channel_1,5,padding=2)
nn.init.kaiming_normal_(self.conv1.weight)
self.conv2=nn.Conv2d(channel_1,channel_2,3,padding=1)
nn.init.kaiming_normal_(self.conv2.weight)
self.fc=nn.Linear(channel_2*32*32,num_classes)
nn.init.kaiming_normal_(self.fc.weight)
self.relu=nn.ReLU(inplace=True)
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
########################################################################
# END OF YOUR CODE #
########################################################################
def forward(self, x):
scores = None
########################################################################
# TODO: Implement the forward function for a 3-layer ConvNet. you #
# should use the layers you defined in __init__ and specify the #
# connectivity of those layers in forward() #
########################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
scores=self.relu(self.conv1(x))
scores=self.relu(self.conv2(scores))
scores=self.fc(flatten(scores))
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
########################################################################
# END OF YOUR CODE #
########################################################################
return scores
def test_ThreeLayerConvNet():
x = torch.zeros((64, 3, 32, 32), dtype=dtype) # minibatch size 64, image size [3, 32, 32]
model = ThreeLayerConvNet(in_channel=3, channel_1=12, channel_2=8, num_classes=10)
scores = model(x)
print(scores.size()) # you should see [64, 10]
test_ThreeLayerConvNet()
```
### Module API: Check Accuracy
Given the validation or test set, we can check the classification accuracy of a neural network.
This version is slightly different from the one in part II. You don't manually pass in the parameters anymore.
```
def check_accuracy_part34(loader, model):
if loader.dataset.train:
print('Checking accuracy on validation set')
else:
print('Checking accuracy on test set')
num_correct = 0
num_samples = 0
model.eval() # set model to evaluation mode
with torch.no_grad():
for x, y in loader:
x = x.to(device=device, dtype=dtype) # move to device, e.g. GPU
y = y.to(device=device, dtype=torch.long)
scores = model(x)
_, preds = scores.max(1)
num_correct += (preds == y).sum()
num_samples += preds.size(0)
acc = float(num_correct) / num_samples
print('Got %d / %d correct (%.2f)' % (num_correct, num_samples, 100 * acc))
```
### Module API: Training Loop
We also use a slightly different training loop. Rather than updating the values of the weights ourselves, we use an Optimizer object from the `torch.optim` package, which abstract the notion of an optimization algorithm and provides implementations of most of the algorithms commonly used to optimize neural networks.
```
def train_part34(model, optimizer, epochs=1):
"""
Train a model on CIFAR-10 using the PyTorch Module API.
Inputs:
- model: A PyTorch Module giving the model to train.
- optimizer: An Optimizer object we will use to train the model
- epochs: (Optional) A Python integer giving the number of epochs to train for
Returns: Nothing, but prints model accuracies during training.
"""
model = model.to(device=device) # move the model parameters to CPU/GPU
for e in range(epochs):
for t, (x, y) in enumerate(loader_train):
model.train() # put model to training mode
x = x.to(device=device, dtype=dtype) # move to device, e.g. GPU
y = y.to(device=device, dtype=torch.long)
scores = model(x)
loss = F.cross_entropy(scores, y)
# Zero out all of the gradients for the variables which the optimizer
# will update.
optimizer.zero_grad()
# This is the backwards pass: compute the gradient of the loss with
# respect to each parameter of the model.
loss.backward()
# Actually update the parameters of the model using the gradients
# computed by the backwards pass.
optimizer.step()
if t % print_every == 0:
print('Iteration %d, loss = %.4f' % (t, loss.item()))
check_accuracy_part34(loader_val, model)
print()
```
### Module API: Train a Two-Layer Network
Now we are ready to run the training loop. In contrast to part II, we don't explicitly allocate parameter tensors anymore.
Simply pass the input size, hidden layer size, and number of classes (i.e. output size) to the constructor of `TwoLayerFC`.
You also need to define an optimizer that tracks all the learnable parameters inside `TwoLayerFC`.
You don't need to tune any hyperparameters, but you should see model accuracies above 40% after training for one epoch.
```
hidden_layer_size = 4000
learning_rate = 1e-2
model = TwoLayerFC(3 * 32 * 32, hidden_layer_size, 10)
optimizer = optim.SGD(model.parameters(), lr=learning_rate)
train_part34(model, optimizer)
```
### Module API: Train a Three-Layer ConvNet
You should now use the Module API to train a three-layer ConvNet on CIFAR. This should look very similar to training the two-layer network! You don't need to tune any hyperparameters, but you should achieve above above 45% after training for one epoch.
You should train the model using stochastic gradient descent without momentum.
```
learning_rate = 3e-3
channel_1 = 32
channel_2 = 16
model = None
optimizer = None
################################################################################
# TODO: Instantiate your ThreeLayerConvNet model and a corresponding optimizer #
################################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
model=ThreeLayerConvNet(3,channel_1,channel_2,10)
optimizer=optim.SGD(model.parameters(),lr=learning_rate)
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
################################################################################
# END OF YOUR CODE
################################################################################
train_part34(model, optimizer)
```
# Part IV. PyTorch Sequential API
Part III introduced the PyTorch Module API, which allows you to define arbitrary learnable layers and their connectivity.
For simple models like a stack of feed forward layers, you still need to go through 3 steps: subclass `nn.Module`, assign layers to class attributes in `__init__`, and call each layer one by one in `forward()`. Is there a more convenient way?
Fortunately, PyTorch provides a container Module called `nn.Sequential`, which merges the above steps into one. It is not as flexible as `nn.Module`, because you cannot specify more complex topology than a feed-forward stack, but it's good enough for many use cases.
### Sequential API: Two-Layer Network
Let's see how to rewrite our two-layer fully connected network example with `nn.Sequential`, and train it using the training loop defined above.
Again, you don't need to tune any hyperparameters here, but you shoud achieve above 40% accuracy after one epoch of training.
```
# We need to wrap `flatten` function in a module in order to stack it
# in nn.Sequential
class Flatten(nn.Module):
def forward(self, x):
return flatten(x)
hidden_layer_size = 4000
learning_rate = 1e-2
model = nn.Sequential(
Flatten(),
nn.Linear(3 * 32 * 32, hidden_layer_size),
nn.ReLU(),
nn.Linear(hidden_layer_size, 10),
)
# you can use Nesterov momentum in optim.SGD
optimizer = optim.SGD(model.parameters(), lr=learning_rate,
momentum=0.9, nesterov=True)
train_part34(model, optimizer)
```
### Sequential API: Three-Layer ConvNet
Here you should use `nn.Sequential` to define and train a three-layer ConvNet with the same architecture we used in Part III:
1. Convolutional layer (with bias) with 32 5x5 filters, with zero-padding of 2
2. ReLU
3. Convolutional layer (with bias) with 16 3x3 filters, with zero-padding of 1
4. ReLU
5. Fully-connected layer (with bias) to compute scores for 10 classes
You should initialize your weight matrices using the `random_weight` function defined above, and you should initialize your bias vectors using the `zero_weight` function above.
You should optimize your model using stochastic gradient descent with Nesterov momentum 0.9.
Again, you don't need to tune any hyperparameters but you should see accuracy above 55% after one epoch of training.
```
channel_1 = 32
channel_2 = 16
learning_rate = 1e-2
model = None
optimizer = None
################################################################################
# TODO: Rewrite the 2-layer ConvNet with bias from Part III with the #
# Sequential API. #
################################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
model=nn.Sequential(
nn.Conv2d(3,channel_1,5,padding=2),
nn.ReLU(inplace=True),
nn.Conv2d(channel_1,channel_2,3,padding=1),
nn.ReLU(inplace=True),
Flatten(),
nn.Linear(channel_2*32*32,10)
)
# for i in (0,2,5):
# w_shape=model[i].weight.data.shape
# b_shape=model[i].bias.data.shape
# model[i].weight.data=random_weight(w_shape)
# model[i].bias.data=zero_weight(b_shape)
optimizer=optim.SGD(model.parameters(),nesterov=True,lr=learning_rate, momentum=0.9)
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
################################################################################
# END OF YOUR CODE
################################################################################
train_part34(model, optimizer)
```
# Part V. CIFAR-10 open-ended challenge
In this section, you can experiment with whatever ConvNet architecture you'd like on CIFAR-10.
Now it's your job to experiment with architectures, hyperparameters, loss functions, and optimizers to train a model that achieves **at least 70%** accuracy on the CIFAR-10 **validation** set within 10 epochs. You can use the check_accuracy and train functions from above. You can use either `nn.Module` or `nn.Sequential` API.
Describe what you did at the end of this notebook.
Here are the official API documentation for each component. One note: what we call in the class "spatial batch norm" is called "BatchNorm2D" in PyTorch.
* Layers in torch.nn package: http://pytorch.org/docs/stable/nn.html
* Activations: http://pytorch.org/docs/stable/nn.html#non-linear-activations
* Loss functions: http://pytorch.org/docs/stable/nn.html#loss-functions
* Optimizers: http://pytorch.org/docs/stable/optim.html
### Things you might try:
- **Filter size**: Above we used 5x5; would smaller filters be more efficient?
- **Number of filters**: Above we used 32 filters. Do more or fewer do better?
- **Pooling vs Strided Convolution**: Do you use max pooling or just stride convolutions?
- **Batch normalization**: Try adding spatial batch normalization after convolution layers and vanilla batch normalization after affine layers. Do your networks train faster?
- **Network architecture**: The network above has two layers of trainable parameters. Can you do better with a deep network? Good architectures to try include:
- [conv-relu-pool]xN -> [affine]xM -> [softmax or SVM]
- [conv-relu-conv-relu-pool]xN -> [affine]xM -> [softmax or SVM]
- [batchnorm-relu-conv]xN -> [affine]xM -> [softmax or SVM]
- **Global Average Pooling**: Instead of flattening and then having multiple affine layers, perform convolutions until your image gets small (7x7 or so) and then perform an average pooling operation to get to a 1x1 image picture (1, 1 , Filter#), which is then reshaped into a (Filter#) vector. This is used in [Google's Inception Network](https://arxiv.org/abs/1512.00567) (See Table 1 for their architecture).
- **Regularization**: Add l2 weight regularization, or perhaps use Dropout.
### Tips for training
For each network architecture that you try, you should tune the learning rate and other hyperparameters. When doing this there are a couple important things to keep in mind:
- If the parameters are working well, you should see improvement within a few hundred iterations
- Remember the coarse-to-fine approach for hyperparameter tuning: start by testing a large range of hyperparameters for just a few training iterations to find the combinations of parameters that are working at all.
- Once you have found some sets of parameters that seem to work, search more finely around these parameters. You may need to train for more epochs.
- You should use the validation set for hyperparameter search, and save your test set for evaluating your architecture on the best parameters as selected by the validation set.
### Going above and beyond
If you are feeling adventurous there are many other features you can implement to try and improve your performance. You are **not required** to implement any of these, but don't miss the fun if you have time!
- Alternative optimizers: you can try Adam, Adagrad, RMSprop, etc.
- Alternative activation functions such as leaky ReLU, parametric ReLU, ELU, or MaxOut.
- Model ensembles
- Data augmentation
- New Architectures
- [ResNets](https://arxiv.org/abs/1512.03385) where the input from the previous layer is added to the output.
- [DenseNets](https://arxiv.org/abs/1608.06993) where inputs into previous layers are concatenated together.
- [This blog has an in-depth overview](https://chatbotslife.com/resnets-highwaynets-and-densenets-oh-my-9bb15918ee32)
### Have fun and happy training!
```
################################################################################
# TODO: #
# Experiment with any architectures, optimizers, and hyperparameters. #
# Achieve AT LEAST 70% accuracy on the *validation set* within 10 epochs. #
# #
# Note that you can use the check_accuracy function to evaluate on either #
# the test set or the validation set, by passing either loader_test or #
# loader_val as the second argument to check_accuracy. You should not touch #
# the test set until you have finished your architecture and hyperparameter #
# tuning, and only run the test set once at the end to report a final value. #
################################################################################
model = None
optimizer = None
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
class AlexNet(nn.Module):
def __init__(self, num_classes=10):
super(AlexNet, self).__init__()
self.relu=nn.ReLU(inplace=True)
self.features = nn.Sequential(
nn.Conv2d(3, 64, kernel_size=3, padding=1),
self.relu,
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(64, 192, kernel_size=3, padding=1),
self.relu,
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(192, 384, kernel_size=3, padding=1),
self.relu,
nn.Conv2d(384, 256, kernel_size=3, padding=1),
self.relu,
# nn.Conv2d(256, 256, kernel_size=3, padding=1),
# nn.ReLU(inplace=True),
# nn.MaxPool2d(kernel_size=2),
)
self.avgpool = nn.AdaptiveAvgPool2d((7, 7))
self.classifier = nn.Sequential(
nn.Dropout(),
nn.Linear(256 * 7 * 7, 4096),
nn.ReLU(inplace=True),
nn.Dropout(),
nn.Linear(4096, 4096),
nn.ReLU(inplace=True),
nn.Linear(4096, num_classes)
)
def forward(self, x):
x = self.features(x)
x: Tensor = self.avgpool(x)
x = x.view(-1, 7 * 7 * 256)
x = self.classifier(x)
return x
model=AlexNet()
optimizer=optim.Adam(model.parameters())
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
################################################################################
# END OF YOUR CODE
################################################################################
# You should get at least 70% accuracy
train_part34(model, optimizer, epochs=10)
```
## Describe what you did
In the cell below you should write an explanation of what you did, any additional features that you implemented, and/or any graphs that you made in the process of training and evaluating your network.
TODO: Describe what you did
## Test set -- run this only once
Now that we've gotten a result we're happy with, we test our final model on the test set (which you should store in best_model). Think about how this compares to your validation set accuracy.
```
best_model = model
check_accuracy_part34(loader_test, best_model)
```
| github_jupyter |
<a id=top></a>
# Analysis of Engineered Features
## Table of Contents
**Note:** In this notebook, the engineered features are referred to as "covariates".
----
1. [Preparations](#prep)
2. [Analysis of Covariates](#covar_analysis)
1. [Boxplots](#covar_analysis_boxplots)
2. [Forward Mapping (onto Shape Space)](#covar_analysis_fwdmap)
3. [Back Mapping (Tissue Consensus Map)](#covar_analysis_backmap)
4. [Covariate Correlations](#covar_analysis_correlations)
3. [Covariate-Shape Relationships](#covar_fspace)
1. [Covariate-Shape Correlations](#covar_fspace_correlations)
2. [Covariate Relation Graph](#covar_fspace_graph)
<a id=prep></a>
## 1. Preparations
----
```
### Import modules
# External, general
from __future__ import division
import os, sys
import numpy as np
np.random.seed(42)
import matplotlib.pyplot as plt
%matplotlib inline
# External, specific
import pandas as pd
import ipywidgets as widgets
from IPython.display import display, HTML
from scipy.stats import linregress, pearsonr, gaussian_kde
from scipy.spatial import cKDTree
import seaborn as sns
sns.set_style('white')
import networkx as nx
# Internal
import katachi.utilities.loading as ld
import katachi.utilities.plotting as kp
### Load data
# Prep loader
loader = ld.DataLoaderIDR()
loader.find_imports(r"data/experimentA/extracted_measurements/", recurse=True, verbose=True)
# Import embedded feature space
dataset_suffix = "shape_TFOR_pca_measured.tsv"
#dataset_suffix = "shape_CFOR_pca_measured.tsv"
#dataset_suffix = "tagRFPtUtrCH_TFOR_pca_measured.tsv"
#dataset_suffix = "mKate2GM130_TFOR_pca_measured.tsv"
fspace_pca, prim_IDs, fspace_idx = loader.load_dataset(dataset_suffix)
print "Imported feature space of shape:", fspace_pca.shape
# Import TFOR centroid locations
centroids = loader.load_dataset("_other_measurements.tsv", IDs=prim_IDs)[0][:,3:6][:,::-1]
print "Imported TFOR centroids of shape:", centroids.shape
# Import engineered features
covar_df, _, _ = loader.load_dataset("_other_measurements.tsv", IDs=prim_IDs, force_df=True)
del covar_df['Centroids RAW X']; del covar_df['Centroids RAW Y']; del covar_df['Centroids RAW Z']
covar_names = list(covar_df.columns)
print "Imported covariates of shape:", covar_df.shape
### Report
print "\ncovar_df.head()"
display(covar_df.head())
print "\ncovar_df.describe()"
display(covar_df.describe())
### Z-standardize the covariates
covar_df_z = (covar_df - covar_df.mean()) / covar_df.std()
```
<a id=covar_analysis></a>
## 2. Analysis of Covariates
----
### Boxplots <a id=covar_analysis_boxplots></a>
```
### General boxplot of Covariates
# Interactive selection of covariates
wid = widgets.SelectMultiple(
options=covar_names,
value=covar_names,
description='Covars',
)
# Interactive plot
@widgets.interact(selected=wid, standardized=True)
def covariate_boxplot(selected=covar_names,
standardized=True):
# Select data
if standardized:
covar_df_plot = covar_df_z[list(selected)]
else:
covar_df_plot = covar_df[list(selected)]
# Plot
fig = plt.figure(figsize=(12,3))
covar_df_plot.boxplot(grid=False)
plt.tick_params(axis='both', which='major', labelsize=6)
fig.autofmt_xdate()
if standardized: plt.title("Boxplot of Covariates [standardized]")
if not standardized: plt.title("Boxplot of Covariates [raw]")
plt.show()
```
### Forward Mapping (onto Shape Space) <a id=covar_analysis_fwdmap></a>
```
### Interactive mapping of covariates onto PCA-transformed shape space
# Set interactions
@widgets.interact(covariate=covar_names,
prim_ID=prim_IDs,
PCx=(1, fspace_pca.shape[1], 1),
PCy=(1, fspace_pca.shape[1], 1),
standardized=False,
show_all_prims=True)
# Show
def show_PCs(covariate=covar_names[0], prim_ID=prim_IDs[0],
PCx=1, PCy=2, standardized=False, show_all_prims=True):
# Select covariate data
if standardized:
covar_df_plot = covar_df_z[covariate]
else:
covar_df_plot = covar_df[covariate]
# Prep
plt.figure(figsize=(9,7))
# If all should be shown...
if show_all_prims:
# Plot
plt.scatter(fspace_pca[:,PCx-1], fspace_pca[:,PCy-1],
c=covar_df_plot, cmap=plt.cm.plasma,
s=10, edgecolor='', alpha=0.75)
# Cosmetics
cbar = plt.colorbar()
if standardized:
cbar.set_label(covariate+" [standardized]", rotation=270, labelpad=15)
else:
cbar.set_label(covariate+" [raw]", rotation=270, labelpad=15)
plt.xlabel("PC "+str(PCx))
plt.ylabel("PC "+str(PCy))
plt.title("PCA-Transformed Shape Space [All Prims]")
plt.show()
# If individual prims should be shown...
else:
# Plot
plt.scatter(fspace_pca[fspace_idx==prim_IDs.index(prim_ID), PCx-1],
fspace_pca[fspace_idx==prim_IDs.index(prim_ID), PCy-1],
c=covar_df_plot[fspace_idx==prim_IDs.index(prim_ID)],
cmap=plt.cm.plasma, s=10, edgecolor='',
vmin=covar_df_plot.min(), vmax=covar_df_plot.max())
# Cosmetics
cbar = plt.colorbar()
if standardized:
cbar.set_label(covariate+" [standardized]", rotation=270, labelpad=15)
else:
cbar.set_label(covariate+" [raw]", rotation=270, labelpad=15)
plt.xlabel("PC "+str(PCx))
plt.ylabel("PC "+str(PCy))
plt.title("PCA-Transformed Shape Space [prim "+prim_ID+"]")
plt.show()
```
### Back Mapping (Tissue Consensus Map) <a id=covar_analysis_backmap></a>
```
### Interactive mapping of covariates onto centroids in TFOR
# Axis range
xlim = (-175, 15)
ylim = (- 25, 25)
# Set interactions
@widgets.interact(covariate=covar_names,
standardized=['no','z'])
# Plot
def centroid_backmap(covariate=covar_names[0],
standardized='no'):
# Select covariate data
if standardized=='no':
covar_df_plot = covar_df[covariate]
elif standardized=='z':
covar_df_plot = covar_df_z[covariate]
# Init
fig,ax = plt.subplots(1, figsize=(12,5))
# Back-mapping plot
#zord = np.argsort(covar_df_plot)
zord = np.arange(len(covar_df_plot)); np.random.shuffle(zord) # Random is better!
scat = ax.scatter(centroids[zord,2], centroids[zord,1],
color=covar_df_plot[zord], cmap=plt.cm.plasma,
edgecolor='', s=15, alpha=0.75)
# Cosmetics
ax.set_xlim(xlim)
ax.set_ylim(ylim)
ax.invert_yaxis() # To match images
ax.set_xlabel('TFOR x')
ax.set_ylabel('TFOR y')
cbar = plt.colorbar(scat,ax=ax)
if standardized:
ax.set_title('Centroid Back-Mapping of '+covariate+' [standardized]')
cbar.set_label(covariate+' [standardized]', rotation=270, labelpad=10)
else:
ax.set_title('Centroid Back-Mapping of '+covariate+' [raw]')
cbar.set_label(covariate+' [raw]', rotation=270, labelpad=20)
# Done
plt.tight_layout()
plt.show()
### Contour plot backmapping plot for publication
# Set interactions
@widgets.interact(covariate=covar_names,
standardized=['no','z'])
# Plot
def contour_backmap(covariate=covar_names[0],
standardized='no'):
# Settings
xlim = (-130, 8)
ylim = ( -19, 19)
# Select covariate data
if standardized=='no':
covar_df_plot = covar_df[covariate]
elif standardized=='z':
covar_df_plot = covar_df_z[covariate]
# Tools for smoothing on scatter
from katachi.utilities.pcl_helpers import pcl_gaussian_smooth
from scipy.spatial.distance import pdist, squareform
# Cut off at prim contour outline
kernel_prim = gaussian_kde(centroids[:,1:].T)
f_prim = kernel_prim(centroids[:,1:].T)
f_prim_mask = f_prim > f_prim.min() + (f_prim.max()-f_prim.min())*0.1
plot_values = covar_df_plot[f_prim_mask]
plot_centroids = centroids[f_prim_mask]
# Smoothen
pdists = squareform(pdist(plot_centroids[:,1:]))
plot_values = pcl_gaussian_smooth(pdists, plot_values[:,np.newaxis], sg_percentile=0.5)[:,0]
# Initialize figure
fig, ax = plt.subplots(1, figsize=(8, 3.25))
# Contourf plot
cfset = ax.tricontourf(plot_centroids[:,2], plot_centroids[:,1], plot_values, 20,
cmap='plasma')
# Illustrative centroids from a single prim
plt.scatter(centroids[fspace_idx==prim_IDs.index(prim_IDs[0]), 2],
centroids[fspace_idx==prim_IDs.index(prim_IDs[0]), 1],
c='', alpha=0.5)
# Cosmetics
ax.set_xlabel('TFOR x', fontsize=16)
ax.set_ylabel('TFOR y', fontsize=16)
plt.tick_params(axis='both', which='major', labelsize=13)
plt.xlim(xlim); plt.ylim(ylim)
ax.invert_yaxis() # To match images
# Colorbar
cbar = plt.colorbar(cfset, ax=ax, pad=0.01)
cbar.set_label(covariate, rotation=270, labelpad=10, fontsize=16)
cbar.ax.tick_params(labelsize=13)
# Done
plt.tight_layout()
plt.show()
```
### Covariate Correlations <a id=covar_analysis_correlations></a>
```
### Interactive linear fitting plot
# Set interaction
@widgets.interact(covar_x=covar_names,
covar_y=covar_names)
# Plotting function
def corr_plot_covar(covar_x=covar_names[0],
covar_y=covar_names[1]):
# Prep
plt.figure(figsize=(5,3))
# Scatterplot
plt.scatter(covar_df[covar_x], covar_df[covar_y],
facecolor='darkblue', edgecolor='',
s=5, alpha=0.5)
plt.xlabel(covar_x)
plt.ylabel(covar_y)
# Linear regression and pearson
fitted = linregress(covar_df[covar_x], covar_df[covar_y])
pearson = pearsonr(covar_df[covar_x], covar_df[covar_y])
# Report
print "Linear regression:"
for param,value in zip(["slope","intercept","rvalue","pvalue","stderr"], fitted):
print " {}:\t{:.2e}".format(param,value)
print "Pearson:"
print " r:\t{:.2e}".format(pearson[0])
print " p:\t{:.2e}".format(pearson[1])
# Add fit to plot
xmin,xmax = (covar_df[covar_x].min(), covar_df[covar_x].max())
ymin,ymax = (covar_df[covar_y].min(), covar_df[covar_y].max())
ybot,ytop = (xmin*fitted[0]+fitted[1], xmax*fitted[0]+fitted[1])
plt.plot([xmin,xmax], [ybot,ytop], c='blue', lw=2, alpha=0.5)
# Cosmetics and show
plt.xlim([xmin,xmax])
plt.ylim([ymin,ymax])
plt.show()
### Full pairwise correlation plot
# Create the plot
mclust = sns.clustermap(covar_df_z.corr(method='pearson'),
figsize=(10, 10),
cmap='RdBu')
# Fix the y axis orientation
mclust.ax_heatmap.set_yticklabels(mclust.ax_heatmap.get_yticklabels(),
rotation=0)
# Other cosmetics
mclust.ax_heatmap.set_title("Pairwise Correlations Cluster Plot", y=1.275)
plt.ylabel("Pearson\nCorr. Coef.")
plt.show()
```
<a id=covar_fspace></a>
## 3. Covariate-Shape Relationships
----
### Covariate-Shape Correlations <a id=covar_fspace_correlations></a>
```
### Interactive linear fitting plot
# Set interaction
@widgets.interact(covar_x=covar_names,
PC_y=range(1,fspace_pca.shape[1]+1))
# Plotting function
def corr_plot_covar(covar_x=covar_names[0],
PC_y=1):
# Prep
PC_y = int(PC_y)
plt.figure(figsize=(5,3))
# Scatterplot
plt.scatter(covar_df[covar_x], fspace_pca[:, PC_y-1],
facecolor='darkred', edgecolor='',
s=5, alpha=0.5)
plt.xlabel(covar_x)
plt.ylabel("PC "+str(PC_y))
# Linear regression and pearson
fitted = linregress(covar_df[covar_x], fspace_pca[:, PC_y-1])
pearson = pearsonr(covar_df[covar_x], fspace_pca[:, PC_y-1])
# Report
print "Linear regression:"
for param,value in zip(["slope","intercept","rvalue","pvalue","stderr"], fitted):
print " {}:\t{:.2e}".format(param,value)
print "Pearson:"
print " r:\t{:.2e}".format(pearson[0])
print " p:\t{:.2e}".format(pearson[1])
# Add fit to plot
xmin,xmax = (covar_df[covar_x].min(), covar_df[covar_x].max())
ymin,ymax = (fspace_pca[:, PC_y-1].min(), fspace_pca[:, PC_y-1].max())
ybot,ytop = (xmin*fitted[0]+fitted[1], xmax*fitted[0]+fitted[1])
plt.plot([xmin,xmax], [ybot,ytop], c='red', lw=2, alpha=0.5)
# Cosmetics and show
plt.xlim([xmin,xmax])
plt.ylim([ymin,ymax])
plt.show()
### Selected linear fits
# Settings for TFOR PC 3
if 'TFOR' in dataset_suffix:
covar_x = 'Z Axis Length'
PC_y = 3
x_reduc = 0
lbl_x = 'TFOR PC 3'
lbl_y = 'Z Axis Length\n(Cell Height)'
# Settings for CFOR PC 1
if 'CFOR' in dataset_suffix:
covar_x = 'Sphericity'
PC_y = 1
x_reduc = 2
lbl_x = 'CFOR PC 1'
lbl_y = 'Sphericity'
# Prep
plt.figure(figsize=(6,4))
# Scatterplot
plt.scatter(fspace_pca[:, PC_y-1], covar_df[covar_x],
facecolor='darkblue', edgecolor='',
s=5, alpha=0.25)
plt.xlabel(covar_x)
plt.ylabel("PC "+str(PC_y))
# Linear regression and pearson
fitted = linregress(fspace_pca[:, PC_y-1], covar_df[covar_x])
pearson = pearsonr(fspace_pca[:, PC_y-1], covar_df[covar_x])
# Report
print "Linear regression:"
for param,value in zip(["slope","intercept","rvalue","pvalue","stderr"], fitted):
print " {}:\t{:.2e}".format(param,value)
print "Pearson:"
print " r:\t{:.2e}".format(pearson[0])
print " p:\t{:.2e}".format(pearson[1])
# Add fit to plot
ymin,ymax = (covar_df[covar_x].min(), covar_df[covar_x].max())
xmin,xmax = (fspace_pca[:, PC_y-1].min()-x_reduc, fspace_pca[:, PC_y-1].max())
ybot,ytop = (xmin*fitted[0]+fitted[1], xmax*fitted[0]+fitted[1])
plt.plot([xmin,xmax], [ybot,ytop], c='black', lw=1, alpha=0.5)
# Cosmetics
plt.tick_params(axis='both', which='major', labelsize=16)
plt.xlabel(lbl_x, fontsize=18)
plt.ylabel(lbl_y, fontsize=18)
plt.xlim([xmin,xmax])
plt.ylim([ymin,ymax+0.05])
plt.tight_layout()
# Done
plt.show()
### Full pairwise correlation plot
# Prepare the pairwise correlation
fspace_pca_z = (fspace_pca - fspace_pca.mean(axis=0)) / fspace_pca.std(axis=0)
fspace_pca_z_df = pd.DataFrame(fspace_pca_z[:,:25])
pairwise_corr = covar_df_z.expanding(axis=1).corr(fspace_pca_z_df, pairwise=True).iloc[-1, :, :] # Ouf, pandas...
# Create the plot
mclust = sns.clustermap(pairwise_corr,
figsize=(10, 10),
col_cluster=False,
cmap='RdBu')
# Fix the y axis orientation
mclust.ax_heatmap.set_yticklabels(mclust.ax_heatmap.get_yticklabels(),
rotation=0)
# Other cosmetics
mclust.ax_heatmap.set_title("Pairwise Correlations Cluster Plot", y=1.275)
mclust.ax_heatmap.set_xticklabels(range(1,fspace_pca_z_df.shape[1]+1))
plt.ylabel("Pearson\nCorr. Coef.")
# Done
plt.show()
```
### Covariate Relation Graph <a id=covar_fspace_graph></a>
```
# Parameters
num_PCs = 8 # Number of PCs to include
corr_measure = 'pearsonr' # Correlation measure to use
threshold = 0.30 # Threshold to include a correlation as relevant
# Get relevant data
if corr_measure == 'pearsonr':
covar_fspace_dists = pairwise_corr.get_values()[:, :num_PCs] # Retrieved from above!
else:
raise NotImplementedError()
# Generate the plot
kp.covar_pc_bigraph(covar_fspace_dists, threshold, covar_names,
height=0.6, verbose=True, show=False)
# Done
plt.show()
```
----
[back to top](#top)
| github_jupyter |
**Due Date: Monday, October 19th, 11:59pm**
- Fill out the missing parts.
- Answer the questions (if any) in a separate document or by adding a new `Text` block inside the Colab.
- Save the notebook by going to the menu and clicking `File` > `Download .ipynb`.
- Make sure the saved version is showing your solutions.
- Send the saved notebook by email to your TA.
```
import numpy as np
np.random.seed(0)
```
Simulate a dataset of 100 coin flips for a coin with $p$ = P(head) = 0.6.
```
n = 100 # number of coin flips
p = 0.6 # probability of getting a head (not a fair coin)
# A coin toss experiment can be modeled with a binomial distribution
# if we set n=1 (one trial), which is equivalent to a Bernoulli distribution
y = np.random.binomial(n=1, p=p, size=n)
y
```
## Point Estimation
Estimate the value of $p$ using the data.
```
def estimator(y):
return np.mean(y)
p_hat = estimator(y)
p_hat
```
## Bootstrap
Estimate the standard error of $\hat{p}$ using bootstrap.
```
def bootstrap_se_est(y, stat_function, B=1000):
# 1. Generate bootstrap samples from the observed/simulated data (i.e. y)
# 2. Compute the statistic (using stat_function passed) on the bootstrap
# samples
# 3. Compute the standard error -> std dev
t_boot_list = [stat_function(np.random.choice(y, len(y), replace=True))
for _ in range(B)]
return np.std(t_boot_list)
print("Standard error of p_hat computed by bootstrap:")
print(bootstrap_se_est(y, estimator))
```
Validate the estimated standard error by computing it analytically.
```
def estimator_analytical_se(p, n):
return np.sqrt(p * (1-p) / n)
print("Analytical standard error for the estimator: ", estimator_analytical_se(p, n))
```
Estimate the 95% confidence interval for $p$.
```
def confidence_interval_95_for_p(y):
ci_lower = estimator(y) - 1.96*bootstrap_se_est(y, estimator)
ci_higher = estimator(y) + 1.96*bootstrap_se_est(y, estimator)
return (ci_lower, ci_higher)
lower, higher = confidence_interval_95_for_p(y)
print("95% confidence interval for p: ({},{})".format(lower, higher))
```
Validate the 95% confidence interval for $p$.
```
ci_contains_p_flags = []
for sim in range(1000):
y = np.random.binomial(n=1, p=p, size=n)
ci_lower, ci_higher = confidence_interval_95_for_p(y)
if ci_lower < p and p < ci_higher:
ci_contains_p_flags.append(1)
else:
ci_contains_p_flags.append(0)
coverage = np.mean(ci_contains_p_flags)
print("Coverage of 95% confidence interval for p: ", coverage)
```
## Bayesian Inference
**[Optional]**
Estimate $p$ using Bayesian inference. As the prior for $p$ use Normal(0.5, 0.1).
```
!pip install pystan
import pystan
model_code = """
data {
int<lower=0> n;
int<lower=0,upper=1> y[n];
}
parameters {
real<lower=0,upper=1> p;
}
model {
p ~ normal(0.5, 0.1);
for (i in 1:n)
y[i] ~ bernoulli(p);
}
"""
model = pystan.StanModel(model_code=model_code)
fit = model.sampling(data={"n": n, "y": y}, iter=2000, chains=4, n_jobs=4)
print(fit.stansummary())
```
Compute the Bayesian inference results if our data contains 20 coin tosses instead.
```
n = 20
p = 0.6
y = np.random.binomial(1, p, n)
model = pystan.StanModel(model_code=model_code)
fit = model.sampling(data={"n": n, "y": y}, iter=2000, chains=4, n_jobs=4)
print(fit.stansummary())
```
| github_jupyter |
```
import torch
import torch.nn as nn
import torch.nn.functional as F
import numpy
from fastai.script import *
from fastai.vision import *
from fastai.callbacks import *
from fastai.distributed import *
from fastprogress import fastprogress
from torchvision.models import *
from fastai.vision.models.xresnet import *
from fastai.vision.models.xresnet2 import *
from fastai.vision.models.presnet import *
torch.backends.cudnn.benchmark = True
```
# XResNet baseline
```
#https://github.com/fastai/fastai_docs/blob/master/dev_course/dl2/11_train_imagenette.ipynb
def noop(x): return x
class Flatten(nn.Module):
def forward(self, x): return x.view(x.size(0), -1)
def conv(ni, nf, ks=3, stride=1, bias=False):
return nn.Conv2d(ni, nf, kernel_size=ks, stride=stride, padding=ks//2, bias=bias)
act_fn = nn.ReLU(inplace=True)
def init_cnn(m):
if getattr(m, 'bias', None) is not None: nn.init.constant_(m.bias, 0)
if isinstance(m, (nn.Conv2d,nn.Linear)): nn.init.kaiming_normal_(m.weight)
for l in m.children(): init_cnn(l)
def conv_layer(ni, nf, ks=3, stride=1, zero_bn=False, act=True):
bn = nn.BatchNorm2d(nf)
nn.init.constant_(bn.weight, 0. if zero_bn else 1.)
layers = [conv(ni, nf, ks, stride=stride), bn]
if act: layers.append(act_fn)
return nn.Sequential(*layers)
class ResBlock(nn.Module):
def __init__(self, expansion, ni, nh, stride=1):
super().__init__()
nf,ni = nh*expansion,ni*expansion
layers = [conv_layer(ni, nh, 3, stride=stride),
conv_layer(nh, nf, 3, zero_bn=True, act=False)
] if expansion == 1 else [
conv_layer(ni, nh, 1),
conv_layer(nh, nh, 3, stride=stride),
conv_layer(nh, nf, 1, zero_bn=True, act=False)
]
self.convs = nn.Sequential(*layers)
self.idconv = noop if ni==nf else conv_layer(ni, nf, 1, act=False)
self.pool = noop if stride==1 else nn.AvgPool2d(2, ceil_mode=True)
def forward(self, x): return act_fn(self.convs(x) + self.idconv(self.pool(x)))
class XResNet(nn.Sequential):
@classmethod
def create(cls, expansion, layers, c_in=3, c_out=1000):
nfs = [c_in, (c_in+1)*8, 64, 64]
stem = [conv_layer(nfs[i], nfs[i+1], stride=2 if i==0 else 1)
for i in range(3)]
nfs = [64//expansion,64,128,256,512]
res_layers = [cls._make_layer(expansion, nfs[i], nfs[i+1],
n_blocks=l, stride=1 if i==0 else 2)
for i,l in enumerate(layers)]
res = cls(
*stem,
nn.MaxPool2d(kernel_size=3, stride=2, padding=1),
*res_layers,
nn.AdaptiveAvgPool2d(1), Flatten(),
nn.Linear(nfs[-1]*expansion, c_out),
)
init_cnn(res)
return res
@staticmethod
def _make_layer(expansion, ni, nf, n_blocks, stride):
return nn.Sequential(
*[ResBlock(expansion, ni if i==0 else nf, nf, stride if i==0 else 1)
for i in range(n_blocks)])
def xresnet18 (**kwargs): return XResNet.create(1, [2, 2, 2, 2], **kwargs)
def xresnet34 (**kwargs): return XResNet.create(1, [3, 4, 6, 3], **kwargs)
def xresnet50 (**kwargs): return XResNet.create(4, [3, 4, 6, 3], **kwargs)
def xresnet101(**kwargs): return XResNet.create(4, [3, 4, 23, 3], **kwargs)
def xresnet152(**kwargs): return XResNet.create(4, [3, 8, 36, 3], **kwargs)
```
# XResNet with Self Attention
```
#Unmodified from https://github.com/fastai/fastai/blob/5c51f9eabf76853a89a9bc5741804d2ed4407e49/fastai/layers.py
def conv1d(ni:int, no:int, ks:int=1, stride:int=1, padding:int=0, bias:bool=False):
"Create and initialize a `nn.Conv1d` layer with spectral normalization."
conv = nn.Conv1d(ni, no, ks, stride=stride, padding=padding, bias=bias)
nn.init.kaiming_normal_(conv.weight)
if bias: conv.bias.data.zero_()
return spectral_norm(conv)
# Adapted from SelfAttention layer at https://github.com/fastai/fastai/blob/5c51f9eabf76853a89a9bc5741804d2ed4407e49/fastai/layers.py
# Inspired by https://arxiv.org/pdf/1805.08318.pdf
class SimpleSelfAttention(nn.Module):
def __init__(self, n_in:int, ks=1):#, n_out:int):
super().__init__()
self.conv = conv1d(n_in, n_in, ks, padding=ks//2, bias=False)
self.gamma = nn.Parameter(tensor([0.]))
def forward(self,x):
size = x.size()
x = x.view(*size[:2],-1)
o = torch.bmm(x.permute(0,2,1).contiguous(),self.conv(x))
o = self.gamma * torch.bmm(x,o) + x
return o.view(*size).contiguous()
#unmodified from https://github.com/fastai/fastai/blob/9b9014b8967186dc70c65ca7dcddca1a1232d99d/fastai/vision/models/xresnet.py
def conv(ni, nf, ks=3, stride=1, bias=False):
return nn.Conv2d(ni, nf, kernel_size=ks, stride=stride, padding=ks//2, bias=bias)
def noop(x): return x
def conv_layer(ni, nf, ks=3, stride=1, zero_bn=False, act=True):
bn = nn.BatchNorm2d(nf)
nn.init.constant_(bn.weight, 0. if zero_bn else 1.)
layers = [conv(ni, nf, ks, stride=stride), bn]
if act: layers.append(act_fn)
return nn.Sequential(*layers)
# Modified from https://github.com/fastai/fastai/blob/9b9014b8967186dc70c65ca7dcddca1a1232d99d/fastai/vision/models/xresnet.py
# Added self attention
class ResBlock(nn.Module):
def __init__(self, expansion, ni, nh, stride=1,sa=False):
super().__init__()
nf,ni = nh*expansion,ni*expansion
layers = [conv_layer(ni, nh, 3, stride=stride),
conv_layer(nh, nf, 3, zero_bn=True, act=False)
] if expansion == 1 else [
conv_layer(ni, nh, 1),
conv_layer(nh, nh, 3, stride=stride),
conv_layer(nh, nf, 1, zero_bn=True, act=False)
]
self.sa = SimpleSelfAttention(nf,ks=1) if sa else noop
self.convs = nn.Sequential(*layers)
self.idconv = noop if ni==nf else conv_layer(ni, nf, 1, act=False)
self.pool = noop if stride==1 else nn.AvgPool2d(2, ceil_mode=True)
def forward(self, x):
return act_fn(self.sa(self.convs(x)) + self.idconv(self.pool(x)))
# Modified from https://github.com/fastai/fastai/blob/9b9014b8967186dc70c65ca7dcddca1a1232d99d/fastai/vision/models/xresnet.py
# Added self attention
class XResNet_sa(nn.Sequential):
@classmethod
def create(cls, expansion, layers, c_in=3, c_out=1000):
nfs = [c_in, (c_in+1)*8, 64, 64]
stem = [conv_layer(nfs[i], nfs[i+1], stride=2 if i==0 else 1)
for i in range(3)]
nfs = [64//expansion,64,128,256,512]
res_layers = [cls._make_layer(expansion, nfs[i], nfs[i+1],
n_blocks=l, stride=1 if i==0 else 2, sa = True if i in[len(layers)-4] else False)
for i,l in enumerate(layers)]
res = cls(
*stem,
nn.MaxPool2d(kernel_size=3, stride=2, padding=1),
*res_layers,
nn.AdaptiveAvgPool2d(1), Flatten(),
nn.Linear(nfs[-1]*expansion, c_out),
)
init_cnn(res)
return res
@staticmethod
def _make_layer(expansion, ni, nf, n_blocks, stride, sa = False):
return nn.Sequential(
*[ResBlock(expansion, ni if i==0 else nf, nf, stride if i==0 else 1, sa if i in [n_blocks -1] else False)
for i in range(n_blocks)])
def xresnet50_sa (**kwargs): return XResNet_sa.create(4, [3, 4, 6, 3], **kwargs)
```
# Data loading
```
#https://github.com/fastai/fastai/blob/master/examples/train_imagenette.py
def get_data(size, woof, bs, workers=None):
if size<=128: path = URLs.IMAGEWOOF_160 if woof else URLs.IMAGENETTE_160
elif size<=224: path = URLs.IMAGEWOOF_320 if woof else URLs.IMAGENETTE_320
else : path = URLs.IMAGEWOOF if woof else URLs.IMAGENETTE
path = untar_data(path)
n_gpus = num_distrib() or 1
if workers is None: workers = min(8, num_cpus()//n_gpus)
return (ImageList.from_folder(path).split_by_folder(valid='val')
.label_from_folder().transform(([flip_lr(p=0.5)], []), size=size)
.databunch(bs=bs, num_workers=workers)
.presize(size, scale=(0.35,1))
.normalize(imagenet_stats))
```
# Train
```
opt_func = partial(optim.Adam, betas=(0.9,0.99), eps=1e-6)
```
## Imagewoof
### Image size = 256
```
image_size = 256
data = get_data(image_size,woof =True,bs=64)
```
#### Epochs = 5
```
# we use the same parameters for baseline and new model
epochs = 5
lr = 3e-3
bs = 64
mixup = 0
```
##### Baseline
```
m = xresnet50(c_out=10)
learn = (Learner(data, m, wd=1e-2, opt_func=opt_func,
metrics=[accuracy,top_k_accuracy],
bn_wd=False, true_wd=True,
loss_func = LabelSmoothingCrossEntropy())
)
if mixup: learn = learn.mixup(alpha=mixup)
learn = learn.to_fp16(dynamic=True)
learn.fit_one_cycle(epochs, lr, div_factor=10, pct_start=0.3)
learn.fit_one_cycle(epochs, lr, div_factor=10, pct_start=0.3)
learn.fit_one_cycle(epochs, lr, div_factor=10, pct_start=0.3)
learn.fit_one_cycle(epochs, lr, div_factor=10, pct_start=0.3)
learn.fit_one_cycle(epochs, lr, div_factor=10, pct_start=0.3)
results = [61.8,64.8,57.4,62.4,63,61.8, 57.6,63,62.6, 64.8] #included some from previous notebook iteration
np.mean(results), np.std(results), np.min(results), np.max(results)
```
##### New model
```
m = xresnet50_sa(c_out=10)
learn = None
gc.collect()
learn = (Learner(data, m, wd=1e-2, opt_func=opt_func,
metrics=[accuracy,top_k_accuracy],
bn_wd=False, true_wd=True,
loss_func = LabelSmoothingCrossEntropy())
)
if mixup: learn = learn.mixup(alpha=mixup)
learn = learn.to_fp16(dynamic=True)
learn.fit_one_cycle(5, lr, div_factor=10, pct_start=0.3)
learn.fit_one_cycle(5, lr, div_factor=10, pct_start=0.3)
learn.fit_one_cycle(5, lr, div_factor=10, pct_start=0.3)
learn.fit_one_cycle(5, lr, div_factor=10, pct_start=0.3)
results = [67.4,65.8,70.6,65.8,67.8,69,65.6,66.4, 67.8,70.2]
np.mean(results), np.std(results), np.min(results), np.max(results)
```
| github_jupyter |
##Functions
Let's say that we have some code that does some task, but the code is 25 lines long, we need to run it over 1000 items and it doesn't work in a loop. How in the world will we handle this situation? That is where functions come in really handy. Functions are a generalized block of code that allow you to run code over and over while changing its parameters if you so choose. Functions may take **(arguments)** that you are allowed to change when you call the function. It may also **return** a value.
A function must be defined before you can call it. To define a function, we use the following syntax:
def <function name>(arg0, arg1, arg3,...):
#code here must be indented.
#you can use arg0,...,argn within the function
#you can also return things
return 1
#This code returns 1 no matter what you tell the function
Functions can take as many arguments as you wish, but they may only return 1 thing. A simple example of a familiar function is any mathematical function. Take sin(x), it is a function that takes one argument x and returns one value based on the input. Let's get familiar with functions.
```
def add1(x):
return x+1
print(add1(1))
def xsq(x):
return x**2
print(xsq(5))
for i in range(0,10):
print(xsq(i))
```
The true power of functions is being able to call it as many times as we would like. In the previous example, we called the square function, xsq in a loop 10 times. Let's check out some more complicated examples.
```
def removefs(data):
newdata=''
for d in data:
if(d=="f" or d=="F"):
pass
else:
newdata+=(d)
return newdata
print(removefs('ffffffFFFFFg'))
intro='''##Functions
Let's say that we have some code that does some task, but the code is 25 lines long, we need to run it over 1000 items and it doesn't work in a loop. How in the world will we handle this situation? That is where functions come in really handy. Functions are a generalized block of code that allow you to run code over and over while changing its parameters if you so choose. Functions may take **(arguments)** that you are allowed to change when you call the function. It may also **return** a value.
A function must be defined before you can call it. To define a function, we use the following syntax:
def <function name>(arg0, arg1, arg3,...):
#code here must be indented.
#you can use arg0,...,argn within the function
#you can also return things
return 1
#This code returns 1 no matter what you tell the function
Functions can take as many arguments as you wish, but they may only return 1 thing. A simple example of a familiar function is any mathematical function. Take sin(x), it is a function that takes one argument x and returns one value based on the input. Let's get familiar with functions."'''
print(removefs(intro))
def removevowels(data):
newdata = ''
for d in data:
if(d=='a' or d=='e' or d=='i' or d=='o' or d=='u' or d=='y'):
pass
else:
newdata+=d
return newdata
print(removevowels(intro))
```
So clearly we can do some powerful things. Now let's see why these functions have significant power over loops.
```
def fib(n):
a,b = 1,1
for i in range(n-1):
a,b = b,a+b
return a
def printfib(n):
for i in range(0,n):
print(fib(i))
printfib(15)
```
Here, using loops within functions allows to generate the fibonacci sequence. We then write a function to print out the first n numbers.
##Exercises
1. Write a function that takes two arguments and returns a value that uses the arguments.
2. Write a power function. It should take two arguments and returns the first argument to the power of the second argument.
3. is a semi-guided exercise. If you are stumped ask for help.
3a. Write a function that takes the cost of a dinner as an argument and returns the cost after a .075% sales tax is added.
3b. Write a function that takes the cost of a dinner and tax and adds a 20% tip to the total, then returns the total.
3c. Write a function that takes a list of food names(choose them yourself) as an argument and returns the cost of purchasing all those items.
3d. Write a function that takes a list of food names as an argument and returns the total cost of having a meal including tax and tip.
4 . In the next cell is a 1000-digit number, write a function to solve Project Euler #8 https://projecteuler.net/problem=8
```
thoudigits = 7316717653133062491922511967442657474235534919493496983520312774506326239578318016984801869478851843858615607891129494954595017379583319528532088055111254069874715852386305071569329096329522744304355766896648950445244523161731856403098711121722383113622298934233803081353362766142828064444866452387493035890729629049156044077239071381051585930796086670172427121883998797908792274921901699720888093776657273330010533678812202354218097512545405947522435258490771167055601360483958644670632441572215539753697817977846174064955149290862569321978468622482839722413756570560574902614079729686524145351004748216637048440319989000889524345065854122758866688116427171479924442928230863465674813919123162824586178664583591245665294765456828489128831426076900422421902267105562632111110937054421750694165896040807198403850962455444362981230987879927244284909188845801561660979191338754992005240636899125607176060588611646710940507754100225698315520005593572972571636269561882670428252483600823257530420752963450
```
##Lambda
Next we will look at a special type of function called a lambda. A lambda is a single line, single expression function. It is perfect for evaluating mathematical expressions like x^2 and e^sin(x^cos(x)). To write a lambda function, we use the following syntax:
func = lambda <args>:<expression>
for example:
xsq = lambda x:x**2
xsq(4) #returns 16
Lambdas will return the result of the expression. Let's check it out.
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
#^^^Some junk we will learn later^^^
func = lambda x:np.exp(np.sin(x**np.cos(x)))
#^^^The important part^^^
plt.plot(np.linspace(0,10,1000), func(np.linspace(0,10,1000)))
#^^^We will learn this next^^^
```
##Exercises
1. Write a lambda for x^n where x and n are arguments.
2. Write a function that removes all instances of the letters "p", "h", "y", "s", "i", "c", "s" from any string. Then prints the new string out.
3. Write a function that does the same thing as **in**, that is, write a function that takes two arguments, a variable and a list and check if the variable is in the list. If it is, return True, otherwise, return False.
4. The factorial function takes a number n and returns the product n\*(n-1)\*(n-2)... Write this function.
5. If you want to retrieve the 4th digit of a number, first convert it to a string using the str() command, then take the value at index [3]. Using this information and your factorial function from 4. solve Project Euler #20 https://projecteuler.net/problem=20.
| github_jupyter |
# Exploring colour channels
In this session, we'll be looking at how to explore the different colour channels that compris an image.
```
# We need to include the home directory in our path, so we can read in our own module.
import os
# image processing tools
import cv2
import numpy as np
# utility functions for this course
import sys
sys.path.append(os.path.join("..", "..", "CDS-VIS"))
from utils.imutils import jimshow
from utils.imutils import jimshow_channel
# plotting tool
import matplotlib.pyplot as plt
```
## Rotation
```
filename = os.path.join("..", "..", "CDS-VIS", "img", "terasse.jpeg")
image = cv2.imread(filename)
image.shape
jimshow(image)
```
## Splitting channels
```
(B, G, R) = cv2.split(image)
jimshow_channel(R, "Red")
```
__Empty numpy array__
```
zeros = np.zeros(image.shape[:2], dtype = "uint8")
jimshow(cv2.merge([zeros, zeros, R]))
jimshow(cv2.merge([zeros, G, zeros]))
jimshow(cv2.merge([B, zeros, zeros]))
```
## Histograms
```
jimshow_channel(cv2.cvtColor(image, cv2.COLOR_BGR2GRAY), "Greyscale")
```
__A note on ```COLOR_BRG2GRAY```__
```
greyed_image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
```
```greyed_image.flatten() != image.flatten()```
## A quick greyscale histogram using matplotlib
```
# Create figure
plt.figure()
# Add histogram
plt.hist(image.flatten(), 256, [0,256])
# Plot title
plt.title("Greyscale histogram")
plt.xlabel("Bins")
plt.ylabel("# of Pixels")
plt.show()
```
## Plotting color histograms
```cv2.calcHist(images, channels, mask, histSize, ranges[, hist[, accumulate]])```
- images : it is the source image of type uint8 or float32 represented as “[img]”.
- channels : it is the index of channel for which we calculate histogram.
- For grayscale image, its value is [0] and
- color image, you can pass [0], [1] or [2] to calculate histogram of blue, green or red channel respectively.
- mask : mask image. To find histogram of full image, it is given as “None”.
- histSize : this represents our BIN count. For full scale, we pass [256].
- ranges : this is our RANGE. Normally, it is [0,256].
```
# split channels
channels = cv2.split(image)
# names of colours
colors = ("b", "g", "r")
# create plot
plt.figure()
# add title
plt.title("Histogram")
# Add xlabel
plt.xlabel("Bins")
# Add ylabel
plt.ylabel("# of Pixels")
# for every tuple of channel, colour
for (channel, color) in zip(channels, colors):
# Create a histogram
hist = cv2.calcHist([channel], [0], None, [256], [0, 256])
# Plot histogram
plt.plot(hist, color=color)
# Set limits of x-axis
plt.xlim([0, 256])
# Show plot
plt.show()
```
| github_jupyter |
# [모듈 2.1] SageMaker 클러스터에서 훈련 (No VPC에서 실행)
이 노트북은 아래의 작업을 실행 합니다.
- SageMaker Hosting Cluster 에서 훈련을 실행
- 훈련한 Job 이름을 저장
- 다음 노트북에서 모델 배포 및 추론시에 사용 합니다.
---
SageMaker의 세션을 얻고, role 정보를 가져옵니다.
- 위의 두 정보를 통해서 SageMaker Hosting Cluster에 연결합니다.
```
import os
import sagemaker
from sagemaker import get_execution_role
sagemaker_session = sagemaker.Session()
role = get_execution_role()
```
## 로컬의 데이터 S3 업로딩
로컬의 데이터를 S3에 업로딩하여 훈련시에 Input으로 사용 합니다.
```
# dataset_location = sagemaker_session.upload_data(path='data', key_prefix='data/DEMO-cifar10')
# display(dataset_location)
dataset_location = 's3://sagemaker-ap-northeast-2-057716757052/data/DEMO-cifar10'
dataset_location
# efs_dir = '/home/ec2-user/efs/data'
# ! ls {efs_dir} -al
# ! aws s3 cp {dataset_location} {efs_dir} --recursive
from sagemaker.inputs import FileSystemInput
# Specify EFS ile system id.
file_system_id = 'fs-38dc1558' # 'fs-xxxxxxxx'
print(f"EFS file-system-id: {file_system_id}")
# Specify directory path for input data on the file system.
# You need to provide normalized and absolute path below.
train_file_system_directory_path = '/data/train'
eval_file_system_directory_path = '/data/eval'
validation_file_system_directory_path = '/data/validation'
print(f'EFS file-system data input path: {train_file_system_directory_path}')
print(f'EFS file-system data input path: {eval_file_system_directory_path}')
print(f'EFS file-system data input path: {validation_file_system_directory_path}')
# Specify the access mode of the mount of the directory associated with the file system.
# Directory must be mounted 'ro'(read-only).
file_system_access_mode = 'ro'
# Specify your file system type
file_system_type = 'EFS'
train = FileSystemInput(file_system_id=file_system_id,
file_system_type=file_system_type,
directory_path=train_file_system_directory_path,
file_system_access_mode=file_system_access_mode)
eval = FileSystemInput(file_system_id=file_system_id,
file_system_type=file_system_type,
directory_path=eval_file_system_directory_path,
file_system_access_mode=file_system_access_mode)
validation = FileSystemInput(file_system_id=file_system_id,
file_system_type=file_system_type,
directory_path=validation_file_system_directory_path,
file_system_access_mode=file_system_access_mode)
aws_region = 'ap-northeast-2'# aws-region-code e.g. us-east-1
s3_bucket = 'sagemaker-ap-northeast-2-057716757052'# your-s3-bucket-name
prefix = "cifar10/efs" #prefix in your bucket
s3_output_location = f's3://{s3_bucket}/{prefix}/output'
print(f'S3 model output location: {s3_output_location}')
security_group_ids = ['sg-0192524ef63ec6138'] # ['sg-xxxxxxxx']
# subnets = ['subnet-0a84bcfa36d3981e6','subnet-0304abaaefc2b1c34','subnet-0a2204b79f378b178'] # [ 'subnet-xxxxxxx', 'subnet-xxxxxxx', 'subnet-xxxxxxx']
subnets = ['subnet-0a84bcfa36d3981e6'] # [ 'subnet-xxxxxxx', 'subnet-xxxxxxx', 'subnet-xxxxxxx']
from sagemaker.tensorflow import TensorFlow
estimator = TensorFlow(base_job_name='cifar10',
entry_point='cifar10_keras_sm_tf2.py',
source_dir='training_script',
role=role,
framework_version='2.0.0',
py_version='py3',
script_mode=True,
hyperparameters={'epochs' : 1},
train_instance_count=1,
train_instance_type='ml.p3.2xlarge',
output_path=s3_output_location,
subnets=subnets,
security_group_ids=security_group_ids,
session = sagemaker.Session()
)
estimator.fit({'train': train,
'validation': validation,
'eval': eval,
})
# estimator.fit({'train': 'file://data/train',
# 'validation': 'file://data/validation',
# 'eval': 'file://data/eval'})
```
# VPC_Mode를 True, False 선택
#### **[중요] VPC_Mode에서 실행시에 True로 변경해주세요**
```
VPC_Mode = False
from sagemaker.tensorflow import TensorFlow
def retrieve_estimator(VPC_Mode):
if VPC_Mode:
# VPC 모드 경우에 subnets, security_group을 기술 합니다.
estimator = TensorFlow(base_job_name='cifar10',
entry_point='cifar10_keras_sm_tf2.py',
source_dir='training_script',
role=role,
framework_version='2.0.0',
py_version='py3',
script_mode=True,
hyperparameters={'epochs': 2},
train_instance_count=1,
train_instance_type='ml.p3.8xlarge',
subnets = ['subnet-090c1fad32165b0fa','subnet-0bd7cff3909c55018'],
security_group_ids = ['sg-0f45d634d80aef27e']
)
else:
estimator = TensorFlow(base_job_name='cifar10',
entry_point='cifar10_keras_sm_tf2.py',
source_dir='training_script',
role=role,
framework_version='2.0.0',
py_version='py3',
script_mode=True,
hyperparameters={'epochs': 2},
train_instance_count=1,
train_instance_type='ml.p3.8xlarge')
return estimator
estimator = retrieve_estimator(VPC_Mode)
```
학습을 수행합니다. 이번에는 각각의 채널(`train, validation, eval`)에 S3의 데이터 저장 위치를 지정합니다.<br>
학습 완료 후 Billable seconds도 확인해 보세요. Billable seconds는 실제로 학습 수행 시 과금되는 시간입니다.
```
Billable seconds: <time>
```
참고로, `ml.p2.xlarge` 인스턴스로 5 epoch 학습 시 전체 6분-7분이 소요되고, 실제 학습에 소요되는 시간은 3분-4분이 소요됩니다.
```
%%time
estimator.fit({'train':'{}/train'.format(dataset_location),
'validation':'{}/validation'.format(dataset_location),
'eval':'{}/eval'.format(dataset_location)})
```
## training_job_name 저장
현재의 training_job_name을 저장 합니다.
- training_job_name을 에는 훈련에 관련 내용 및 훈련 결과인 **Model Artifact** 파일의 S3 경로를 제공 합니다.
```
train_job_name = estimator._current_job_name
%store train_job_name
```
| github_jupyter |
<a href="https://colab.research.google.com/github/iotanalytics/IoTTutorial/blob/main/code/preprocessing_and_decomposition/Matrix_Profile.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## Matrix Profile
## Introduction
The matrix profile (MP) is a data structure and associated algorithms that helps solve the dual problem of anomaly detection and motif discovery. It is robust, scalable and largely parameter-free.
MP can be combined with other algorithms to accomplish:
* Motif discovery
* Time series chains
* Anomaly discovery
* Joins
* Semantic segmentation
matrixprofile-ts offers 3 different algorithms to compute Matrix Profile:
* STAMP (Scalable Time Series Anytime Matrix Profile) - Each distance profile is independent of other distance profiles, the order in which they are computed can be random. It is an anytime algorithm.
* STOMP (Scalable Time Series Ordered Matrix Profile) - This algorithm is an exact ordered algorithm. It is significantly faster than STAMP.
* SCRIMP++ (Scalable Column Independent Matrix Profile) - This algorithm combines the anytime component of STAMP with the speed of STOMP.
See: https://towardsdatascience.com/introduction-to-matrix-profiles-5568f3375d90
## Code Example
```
!pip install matrixprofile-ts
import pandas as pd
## example data importing
data = pd.read_csv('https://raw.githubusercontent.com/iotanalytics/IoTTutorial/main/data/SCG_data.csv').drop('Unnamed: 0',1).to_numpy()[0:20,:1000]
import operator
import numpy as np
import matplotlib.pyplot as plt
from matrixprofile import *
import numpy as np
from datetime import datetime
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
from sklearn import neighbors, datasets
# Pull a portion of the data
pattern = data[10,:] + max(abs(data[10,:]))
# Compute Matrix Profile
m = 10
mp = matrixProfile.stomp(pattern,m)
#Append np.nan to Matrix profile to enable plotting against raw data
mp_adj = np.append(mp[0],np.zeros(m-1)+np.nan)
#Plot the signal data
fig, (ax1, ax2) = plt.subplots(2,1,sharex=True,figsize=(20,10))
ax1.plot(np.arange(len(pattern)),pattern)
ax1.set_ylabel('Signal', size=22)
#Plot the Matrix Profile
ax2.plot(np.arange(len(mp_adj)),mp_adj, label="Matrix Profile", color='red')
ax2.set_ylabel('Matrix Profile', size=22)
ax2.set_xlabel('Time', size=22);
```
## Discussion
Pros:
* It is exact: For motif discovery, discord discovery, time series joins etc., the Matrix Profile based methods provide no false positives or false dismissals.
* It is simple and parameter-free: In contrast, the more general algorithms in this space
that typically require building and tuning spatial access methods and/or hash functions.
* It is space efficient: Matrix Profile construction algorithms requires an inconsequential
space overhead, just linear in the time series length with a small constant factor, allowing
massive datasets to be processed in main memory (for most data mining, disk is death).
* It allows anytime algorithms: While exact MP algorithms are extremely scalable, for
extremely large datasets we can compute the Matrix Profile in an anytime fashion, allowing
ultra-fast approximate solutions and real-time data interaction.
* It is incrementally maintainable: Having computed the Matrix Profile for a dataset,
we can incrementally update it very efficiently. In many domains this means we can effectively
maintain exact joins, motifs, discords on streaming data forever.
* It can leverage hardware: Matrix Profile construction is embarrassingly parallelizable,
both on multicore processors, GPUs, distributed systems etc.
* It is free of the curse of dimensionality: That is to say, It has time complexity that is
constant in subsequence length: This is a very unusual and desirable property; virtually all
existing algorithms in the time series scale poorly as the subsequence length grows.
* It can be constructed in deterministic time: Almost all algorithms for time series
data mining can take radically different times to finish on two (even slightly) different datasets.
In contrast, given only the length of the time series, we can precisely predict in advance how
long it will take to compute the Matrix Profile. (this allows resource planning)
* It can handle missing data: Even in the presence of missing data, we can provide
answers which are guaranteed to have no false negatives.
* Finally, and subjectively: Simplicity and Intuitiveness: Seeing the world through
the MP lens often invites/suggests simple and elegant solutions.
Cons:
* Larger datasets can take a long time to compute. Scalability needs to be addressed.
* Cannot be used with Dynamic time Warping as of now.
* DTW is used for one-to-all matching whereas MP is used for all-to-all matching.
* DTW is used for smaller datasets rather than large.
* Need to adjust window size manually for different datasets.
*How to read MP* :
* Where you see relatively low values, you know that the subsequence in the original time
series must have (at least one) relatively similar subsequence elsewhere in the data (such
regions are “motifs” or reoccurring patterns)
* Where you see relatively high values, you know that the subsequence in the original time
series must be unique in its shape (such areas are “discords” or anomalies). In fact, the highest point is exactly the definition of Time
Series Discord, perhaps the best anomaly detector for time series.
##References:
https://www.cs.ucr.edu/~eamonn/MatrixProfile.html (powerpoints on this site - a lot of examples)
https://towardsdatascience.com/introduction-to-matrix-profiles-5568f3375d90
Python implementation: https://github.com/TDAmeritrade/stumpy
| github_jupyter |
## Python Modules
```
%%writefile weather.py
def prognosis():
print("It will rain today")
import weather
weather.prognosis()
```
## How does Python know from where to import packages/modules from?
```
# Python imports work by searching the directories listed in sys.path.
import sys
sys.path
## "__main__" usage
# A module can discover whether or not it is running in the main scope by checking its own __name__,
# which allows a common idiom for conditionally executing code in a module when it is run as a script or with python -m
# but not when it is imported:
%%writefile hw.py
#!/usr/bin/env python
def hw():
print("Running Main")
def hw2():
print("Hello 2")
if __name__ == "__main__":
# execute only if run as a script
print("Running as script")
hw()
hw2()
import main
import hw
main.main()
hw.hw2()
# Running on all 3 OSes from command line:
python main.py
```
## Make main.py self running on Linux (also should work on MacOS):
Add
#!/usr/bin/env python to first line of script
mark it executable using
### need to change permissions too!
$ chmod +x main.py
## Making Standalone .EXEs for Python in Windows
* http://www.py2exe.org/ used to be for Python 2 , now supposedly Python 3 as well
* http://www.pyinstaller.org/
Tutorial: https://medium.com/dreamcatcher-its-blog/making-an-stand-alone-executable-from-a-python-script-using-pyinstaller-d1df9170e263
Need to create exe on a similar system as target system!
```
# Exercise Write a function which returns a list of fibonacci numbers up to starting with 1, 1, 2, 3, 5 up to the nth.
So Fib(4) would return [1,1,2,3]
```


```
%%writefile fibo.py
# Fibonacci numbers module
def fib(n): # write Fibonacci series up to n
a, b = 1 1
while b < n:
print(b, end=' ')
a, b = b, a+b
print()
def fib2(n): # return Fibonacci series up to n
result = []
a, b = 1, 1
while b < n:
result.append(b)
a, b = b, a+b
return result
import fibo
fibo.fib(100)
fibo.fib2(100)
fib=fibo.fib
```
If you intend to use a function often you can assign it to a local name:
```
fib(300)
```
#### There is a variant of the import statement that imports names from a module directly into the importing module’s symbol table.
```
from fibo import fib, fib2 # we overwrote fib=fibo.fib
fib(100)
fib2(200)
```
This does not introduce the module name from which the imports are taken in the local symbol table (so in the example, fibo is not defined).
There is even a variant to import all names that a module defines: **NOT RECOMMENDED**
```
## DO not do this Namespace collission possible!!
from fibo import *
fib(400)
```
### If the module name is followed by as, then the name following as is bound directly to the imported module.
```
import fibo as fib
dir(fib)
fib.fib(50)
### It can also be used when utilising from with similar effects:
from fibo import fib as fibonacci
fibonacci(200)
```
### Executing modules as scripts¶
When you run a Python module with
python fibo.py <arguments>
the code in the module will be executed, just as if you imported it, but with the \_\_name\_\_ set to "\_\_main\_\_". That means that by adding this code at the end of your module:
```
%%writefile fibbo.py
# Fibonacci numbers module
def fib(n): # write Fibonacci series up to n
a, b = 0, 1
while b < n:
print(b, end=' ')
a, b = b, a+b
print()
def fib2(n): # return Fibonacci series up to n
result = []
a, b = 0, 1
while b < n:
result.append(b)
a, b = b, a+b
return result
if __name__ == "__main__":
import sys
fib(int(sys.argv[1], 10))
import fibbo as fi
fi.fib(200)
```
#### This is often used either to provide a convenient user interface to a module, or for testing purposes (running the module as a script executes a test suite).
### The Module Search Path
When a module named spam is imported, the interpreter first searches for a built-in module with that name. If not found, it then searches for a file named spam.py in a list of directories given by the variable sys.path. sys.path is initialized from these locations:
* The directory containing the input script (or the current directory when no file is specified).
* PYTHONPATH (a list of directory names, with the same syntax as the shell variable PATH).
* The installation-dependent default.
Packages are a way of structuring Python’s module namespace by using “dotted module names”. For example, the module name A.B designates a submodule named B in a package named A. Just like the use of modules saves the authors of different modules from having to worry about each other’s global variable names, the use of dotted module names saves the authors of multi-module packages like NumPy or Pillow from having to worry about each other’s module names.
```
sound/ Top-level package
__init__.py Initialize the sound package
formats/ Subpackage for file format conversions
__init__.py
wavread.py
wavwrite.py
aiffread.py
aiffwrite.py
auread.py
auwrite.py
...
effects/ Subpackage for sound effects
__init__.py
echo.py
surround.py
reverse.py
...
filters/ Subpackage for filters
__init__.py
equalizer.py
vocoder.py
karaoke.py
...
```
The \_\_init\_\_.py files are required to make Python treat the directories as containing packages; this is done to prevent directories with a common name, such as string, from unintentionally hiding valid modules that occur later on the module search path. In the simplest case, \_\_init\_\_.py can just be an empty file
| github_jupyter |
```
# General imports
import numpy as np
import torch
# DeepMoD stuff
from multitaskpinn import DeepMoD
from multitaskpinn.model.func_approx import NN
from multitaskpinn.model.library import Library1D
from multitaskpinn.model.constraint import LeastSquares
from multitaskpinn.model.sparse_estimators import Threshold
from multitaskpinn.training import train, train_multitask
from multitaskpinn.training.sparsity_scheduler import TrainTestPeriodic
from phimal_utilities.data import Dataset
from phimal_utilities.data.burgers import BurgersDelta
if torch.cuda.is_available():
device ='cuda'
else:
device = 'cpu'
device = 'cpu'
# Settings for reproducibility
np.random.seed(0)
torch.manual_seed(0)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
%load_ext autoreload
%autoreload 2
device
# Making dataset
v = 0.1
A = 1.0
x = np.linspace(-3, 4, 100)
t = np.linspace(0.5, 5.0, 50)
x_grid, t_grid = np.meshgrid(x, t, indexing='ij')
dataset = Dataset(BurgersDelta, v=v, A=A)
X, y = dataset.create_dataset(x_grid.reshape(-1, 1), t_grid.reshape(-1, 1), n_samples=1000, noise=0.2, random=True, normalize=False)
X, y = X.to(device), y.to(device)
network = NN(2, [30, 30, 30, 30, 30], 1)
library = Library1D(poly_order=2, diff_order=3) # Library function
estimator = Threshold(0.1) # Sparse estimator
constraint = LeastSquares() # How to constrain
model = DeepMoD(network, library, estimator, constraint).to(device) # Putting it all in the model
sparsity_scheduler = TrainTestPeriodic(patience=8, delta=1e-5, periodicity=50)
optimizer = torch.optim.Adam(model.parameters(), betas=(0.99, 0.999), amsgrad=True, lr=2e-3) # Defining optimizer
train_multitask(model, X, y, optimizer, sparsity_scheduler, write_iterations=25, log_dir='runs/testing_multitask_unnormalized/', max_iterations=15000, delta=1e-3, patience=8) # Running
network = NN(2, [30, 30, 30, 30, 30], 1)
library = Library1D(poly_order=2, diff_order=3) # Library function
estimator = Threshold(0.1) # Sparse estimator
constraint = LeastSquares() # How to constrain
model = DeepMoD(network, library, estimator, constraint).to(device) # Putting it all in the model
sparsity_scheduler = TrainTestPeriodic(patience=8, delta=1e-5, periodicity=50)
optimizer = torch.optim.Adam(model.parameters(), betas=(0.99, 0.999), amsgrad=True, lr=2e-3) # Defining optimizer
train(model, X, y, optimizer, sparsity_scheduler, write_iterations=25, log_dir='runs/testing_normal_unnormalized/', max_iterations=15000, delta=1e-3, patience=8) # Running
```
# Quick analysis
```
from phimal_utilities.analysis import Results
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(context='notebook', style='white')
%config InlineBackend.figure_format = 'svg'
data_mt = Results('runs/testing_multitask_unnormalized//')
data_bl = Results('runs/testing_normal_unnormalized//')
keys = data_mt.keys
fig, axes = plt.subplots(figsize=(10, 3), constrained_layout=True, ncols=2)
ax = axes[0]
ax.semilogy(data_bl.df.index, data_bl.df[keys['mse']], label='Baseline')
ax.semilogy(data_mt.df.index, data_mt.df[keys['mse']], label='Multitask')
ax.set_title('MSE')
ax.set_xlabel('Epoch', weight='bold')
ax.set_ylabel('Cost', weight='bold')
ax.legend()
#ax.set_xlim([0, 8000])
ax = axes[1]
ax.semilogy(data_bl.df.index, data_bl.df[keys['reg']], label='Baseline')
ax.semilogy(data_mt.df.index, data_mt.df[keys['reg']], label='Multitask')
ax.set_title('Regression')
ax.set_xlabel('Epoch', weight='bold')
ax.set_ylabel('Cost', weight='bold')
ax.legend()
#ax.set_xlim([0, 8000])
fig.show()
fig, axes = plt.subplots(ncols=3, constrained_layout=True, figsize=(15, 4))
ax = axes[0]
ax.plot(data_bl.df.index, data_bl.df[keys['coeffs']])
ax.plot(data_bl.df.index, data_bl.df[keys['coeffs'][2]], lw=3)
ax.plot(data_bl.df.index, data_bl.df[keys['coeffs'][5]], lw=3)
ax.set_ylim([-2, 2])
ax.set_title('Coefficients baseline')
ax.set_xlabel('Epoch', weight='bold')
ax.set_ylabel('Cost', weight='bold')
#ax.set_xlim([0, 8000])
ax = axes[1]
ax.plot(data_mt.df.index, data_mt.df[keys['coeffs']])
ax.plot(data_mt.df.index, data_mt.df[keys['coeffs'][2]], lw=3)
ax.plot(data_mt.df.index, data_mt.df[keys['coeffs'][5]], lw=3)
ax.set_ylim([-2, 2])
ax.set_title('Coefficients Multitask')
ax.set_xlabel('Epoch', weight='bold')
ax.set_ylabel('Cost', weight='bold')
#ax.set_xlim([0, 8000])
ax = axes[2]
true_coeffs = np.zeros(len(keys['unscaled_coeffs']))
true_coeffs[2] = 0.1
true_coeffs[5] = -1
ax.semilogy(data_bl.df.index, np.mean(np.abs(data_bl.df[keys['unscaled_coeffs']] - true_coeffs), axis=1), label='Baseline')
ax.semilogy(data_mt.df.index, np.mean(np.abs(data_mt.df[keys['unscaled_coeffs']] - true_coeffs), axis=1), label='Baseline')
ax.set_ylim([-5, 2])
ax.legend()
fig.show()
```
| github_jupyter |
```
%matplotlib inline
```
What is `torch.nn` *really*?
============================
by Jeremy Howard, `fast.ai <https://www.fast.ai>`_. Thanks to Rachel Thomas and Francisco Ingham.
We recommend running this tutorial as a notebook, not a script. To download the notebook (.ipynb) file,
click `here <https://pytorch.org/tutorials/beginner/nn_tutorial.html#sphx-glr-download-beginner-nn-tutorial-py>`_ .
PyTorch provides the elegantly designed modules and classes `torch.nn <https://pytorch.org/docs/stable/nn.html>`_ ,
`torch.optim <https://pytorch.org/docs/stable/optim.html>`_ ,
`Dataset <https://pytorch.org/docs/stable/data.html?highlight=dataset#torch.utils.data.Dataset>`_ ,
and `DataLoader <https://pytorch.org/docs/stable/data.html?highlight=dataloader#torch.utils.data.DataLoader>`_
to help you create and train neural networks.
In order to fully utilize their power and customize
them for your problem, you need to really understand exactly what they're
doing. To develop this understanding, we will first train basic neural net
on the MNIST data set without using any features from these models; we will
initially only use the most basic PyTorch tensor functionality. Then, we will
incrementally add one feature from ``torch.nn``, ``torch.optim``, ``Dataset``, or
``DataLoader`` at a time, showing exactly what each piece does, and how it
works to make the code either more concise, or more flexible.
**This tutorial assumes you already have PyTorch installed, and are familiar
with the basics of tensor operations.** (If you're familiar with Numpy array
operations, you'll find the PyTorch tensor operations used here nearly identical).
MNIST data setup
----------------
We will use the classic `MNIST <http://deeplearning.net/data/mnist/>`_ dataset,
which consists of black-and-white images of hand-drawn digits (between 0 and 9).
We will use `pathlib <https://docs.python.org/3/library/pathlib.html>`_
for dealing with paths (part of the Python 3 standard library), and will
download the dataset using
`requests <http://docs.python-requests.org/en/master/>`_. We will only
import modules when we use them, so you can see exactly what's being
used at each point.
```
from pathlib import Path
import requests
DATA_PATH = Path("data")
PATH = DATA_PATH / "mnist"
PATH.mkdir(parents=True, exist_ok=True)
URL = "http://deeplearning.net/data/mnist/"
FILENAME = "mnist.pkl.gz"
if not (PATH / FILENAME).exists():
content = requests.get(URL + FILENAME).content
(PATH / FILENAME).open("wb").write(content)
```
This dataset is in numpy array format, and has been stored using pickle,
a python-specific format for serializing data.
```
import pickle
import gzip
with gzip.open((PATH / FILENAME).as_posix(), "rb") as f:
((x_train, y_train), (x_valid, y_valid), _) = pickle.load(f, encoding="latin-1")
```
Each image is 28 x 28, and is being stored as a flattened row of length
784 (=28x28). Let's take a look at one; we need to reshape it to 2d
first.
```
from matplotlib import pyplot
import numpy as np
pyplot.imshow(x_train[0].reshape((28, 28)), cmap="gray")
print(x_train.shape)
```
PyTorch uses ``torch.tensor``, rather than numpy arrays, so we need to
convert our data.
```
import torch
x_train, y_train, x_valid, y_valid = map(
torch.tensor, (x_train, y_train, x_valid, y_valid)
)
n, c = x_train.shape
x_train, x_train.shape, y_train.min(), y_train.max()
print(x_train, y_train)
print(x_train.shape)
print(y_train.min(), y_train.max())
```
Neural net from scratch (no torch.nn)
---------------------------------------------
Let's first create a model using nothing but PyTorch tensor operations. We're assuming
you're already familiar with the basics of neural networks. (If you're not, you can
learn them at `course.fast.ai <https://course.fast.ai>`_).
PyTorch provides methods to create random or zero-filled tensors, which we will
use to create our weights and bias for a simple linear model. These are just regular
tensors, with one very special addition: we tell PyTorch that they require a
gradient. This causes PyTorch to record all of the operations done on the tensor,
so that it can calculate the gradient during back-propagation *automatically*!
For the weights, we set ``requires_grad`` **after** the initialization, since we
don't want that step included in the gradient. (Note that a trailling ``_`` in
PyTorch signifies that the operation is performed in-place.)
<div class="alert alert-info"><h4>Note</h4><p>We are initializing the weights here with
`Xavier initialisation <http://proceedings.mlr.press/v9/glorot10a/glorot10a.pdf>`_
(by multiplying with 1/sqrt(n)).</p></div>
```
import math
weights = torch.randn(784, 10) / math.sqrt(784)
weights.requires_grad_()
bias = torch.zeros(10, requires_grad=True)
```
Thanks to PyTorch's ability to calculate gradients automatically, we can
use any standard Python function (or callable object) as a model! So
let's just write a plain matrix multiplication and broadcasted addition
to create a simple linear model. We also need an activation function, so
we'll write `log_softmax` and use it. Remember: although PyTorch
provides lots of pre-written loss functions, activation functions, and
so forth, you can easily write your own using plain python. PyTorch will
even create fast GPU or vectorized CPU code for your function
automatically.
```
def log_softmax(x):
return x - x.exp().sum(-1).log().unsqueeze(-1)
def model(xb):
return log_softmax(xb @ weights + bias)
```
In the above, the ``@`` stands for the dot product operation. We will call
our function on one batch of data (in this case, 64 images). This is
one *forward pass*. Note that our predictions won't be any better than
random at this stage, since we start with random weights.
```
bs = 64 # batch size
xb = x_train[0:bs] # a mini-batch from x
preds = model(xb) # predictions
preds[0], preds.shape
print(preds[0], preds.shape)
```
As you see, the ``preds`` tensor contains not only the tensor values, but also a
gradient function. We'll use this later to do backprop.
Let's implement negative log-likelihood to use as the loss function
(again, we can just use standard Python):
```
def nll(input, target):
return -input[range(target.shape[0]), target].mean()
loss_func = nll
```
Let's check our loss with our random model, so we can see if we improve
after a backprop pass later.
```
yb = y_train[0:bs]
print(loss_func(preds, yb))
```
Let's also implement a function to calculate the accuracy of our model.
For each prediction, if the index with the largest value matches the
target value, then the prediction was correct.
```
def accuracy(out, yb):
preds = torch.argmax(out, dim=1)
return (preds == yb).float().mean()
```
Let's check the accuracy of our random model, so we can see if our
accuracy improves as our loss improves.
```
print(accuracy(preds, yb))
```
We can now run a training loop. For each iteration, we will:
- select a mini-batch of data (of size ``bs``)
- use the model to make predictions
- calculate the loss
- ``loss.backward()`` updates the gradients of the model, in this case, ``weights``
and ``bias``.
We now use these gradients to update the weights and bias. We do this
within the ``torch.no_grad()`` context manager, because we do not want these
actions to be recorded for our next calculation of the gradient. You can read
more about how PyTorch's Autograd records operations
`here <https://pytorch.org/docs/stable/notes/autograd.html>`_.
We then set the
gradients to zero, so that we are ready for the next loop.
Otherwise, our gradients would record a running tally of all the operations
that had happened (i.e. ``loss.backward()`` *adds* the gradients to whatever is
already stored, rather than replacing them).
.. tip:: You can use the standard python debugger to step through PyTorch
code, allowing you to check the various variable values at each step.
Uncomment ``set_trace()`` below to try it out.
```
from IPython.core.debugger import set_trace
lr = 0.5 # learning rate
epochs = 2 # how many epochs to train for
for epoch in range(epochs):
for i in range((n - 1) // bs + 1):
# set_trace()
start_i = i * bs
end_i = start_i + bs
xb = x_train[start_i:end_i]
yb = y_train[start_i:end_i]
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
with torch.no_grad():
weights -= weights.grad * lr
bias -= bias.grad * lr
weights.grad.zero_()
bias.grad.zero_()
```
That's it: we've created and trained a minimal neural network (in this case, a
logistic regression, since we have no hidden layers) entirely from scratch!
Let's check the loss and accuracy and compare those to what we got
earlier. We expect that the loss will have decreased and accuracy to
have increased, and they have.
```
print(loss_func(model(xb), yb), accuracy(model(xb), yb))
```
Using torch.nn.functional
------------------------------
We will now refactor our code, so that it does the same thing as before, only
we'll start taking advantage of PyTorch's ``nn`` classes to make it more concise
and flexible. At each step from here, we should be making our code one or more
of: shorter, more understandable, and/or more flexible.
The first and easiest step is to make our code shorter by replacing our
hand-written activation and loss functions with those from ``torch.nn.functional``
(which is generally imported into the namespace ``F`` by convention). This module
contains all the functions in the ``torch.nn`` library (whereas other parts of the
library contain classes). As well as a wide range of loss and activation
functions, you'll also find here some convenient functions for creating neural
nets, such as pooling functions. (There are also functions for doing convolutions,
linear layers, etc, but as we'll see, these are usually better handled using
other parts of the library.)
If you're using negative log likelihood loss and log softmax activation,
then Pytorch provides a single function ``F.cross_entropy`` that combines
the two. So we can even remove the activation function from our model.
```
import torch.nn.functional as F
loss_func = F.cross_entropy
def model(xb):
return xb @ weights + bias
```
Note that we no longer call ``log_softmax`` in the ``model`` function. Let's
confirm that our loss and accuracy are the same as before:
```
print(loss_func(model(xb), yb), accuracy(model(xb), yb))
```
Refactor using nn.Module
-----------------------------
Next up, we'll use ``nn.Module`` and ``nn.Parameter``, for a clearer and more
concise training loop. We subclass ``nn.Module`` (which itself is a class and
able to keep track of state). In this case, we want to create a class that
holds our weights, bias, and method for the forward step. ``nn.Module`` has a
number of attributes and methods (such as ``.parameters()`` and ``.zero_grad()``)
which we will be using.
<div class="alert alert-info"><h4>Note</h4><p>``nn.Module`` (uppercase M) is a PyTorch specific concept, and is a
class we'll be using a lot. ``nn.Module`` is not to be confused with the Python
concept of a (lowercase ``m``) `module <https://docs.python.org/3/tutorial/modules.html>`_,
which is a file of Python code that can be imported.</p></div>
```
from torch import nn
class Mnist_Logistic(nn.Module):
def __init__(self):
super().__init__()
self.weights = nn.Parameter(torch.randn(784, 10) / math.sqrt(784))
self.bias = nn.Parameter(torch.zeros(10))
def forward(self, xb):
return xb @ self.weights + self.bias
```
Since we're now using an object instead of just using a function, we
first have to instantiate our model:
```
model = Mnist_Logistic()
```
Now we can calculate the loss in the same way as before. Note that
``nn.Module`` objects are used as if they are functions (i.e they are
*callable*), but behind the scenes Pytorch will call our ``forward``
method automatically.
```
print(loss_func(model(xb), yb))
```
Previously for our training loop we had to update the values for each parameter
by name, and manually zero out the grads for each parameter separately, like this:
::
with torch.no_grad():
weights -= weights.grad * lr
bias -= bias.grad * lr
weights.grad.zero_()
bias.grad.zero_()
Now we can take advantage of model.parameters() and model.zero_grad() (which
are both defined by PyTorch for ``nn.Module``) to make those steps more concise
and less prone to the error of forgetting some of our parameters, particularly
if we had a more complicated model:
::
with torch.no_grad():
for p in model.parameters(): p -= p.grad * lr
model.zero_grad()
We'll wrap our little training loop in a ``fit`` function so we can run it
again later.
```
def fit():
for epoch in range(epochs):
for i in range((n - 1) // bs + 1):
start_i = i * bs
end_i = start_i + bs
xb = x_train[start_i:end_i]
yb = y_train[start_i:end_i]
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
with torch.no_grad():
for p in model.parameters():
p -= p.grad * lr
model.zero_grad()
fit()
```
Let's double-check that our loss has gone down:
```
print(loss_func(model(xb), yb))
```
Refactor using nn.Linear
-------------------------
We continue to refactor our code. Instead of manually defining and
initializing ``self.weights`` and ``self.bias``, and calculating ``xb @
self.weights + self.bias``, we will instead use the Pytorch class
`nn.Linear <https://pytorch.org/docs/stable/nn.html#linear-layers>`_ for a
linear layer, which does all that for us. Pytorch has many types of
predefined layers that can greatly simplify our code, and often makes it
faster too.
```
class Mnist_Logistic(nn.Module):
def __init__(self):
super().__init__()
self.lin = nn.Linear(784, 10)
def forward(self, xb):
return self.lin(xb)
```
We instantiate our model and calculate the loss in the same way as before:
```
model = Mnist_Logistic()
print(loss_func(model(xb), yb))
```
We are still able to use our same ``fit`` method as before.
```
fit()
print(loss_func(model(xb), yb))
```
Refactor using optim
------------------------------
Pytorch also has a package with various optimization algorithms, ``torch.optim``.
We can use the ``step`` method from our optimizer to take a forward step, instead
of manually updating each parameter.
This will let us replace our previous manually coded optimization step:
::
with torch.no_grad():
for p in model.parameters(): p -= p.grad * lr
model.zero_grad()
and instead use just:
::
opt.step()
opt.zero_grad()
(``optim.zero_grad()`` resets the gradient to 0 and we need to call it before
computing the gradient for the next minibatch.)
```
from torch import optim
```
We'll define a little function to create our model and optimizer so we
can reuse it in the future.
```
def get_model():
model = Mnist_Logistic()
return model, optim.SGD(model.parameters(), lr=lr)
model, opt = get_model()
print(loss_func(model(xb), yb))
for epoch in range(epochs):
for i in range((n - 1) // bs + 1):
start_i = i * bs
end_i = start_i + bs
xb = x_train[start_i:end_i]
yb = y_train[start_i:end_i]
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
print(loss_func(model(xb), yb))
```
Refactor using Dataset
------------------------------
PyTorch has an abstract Dataset class. A Dataset can be anything that has
a ``__len__`` function (called by Python's standard ``len`` function) and
a ``__getitem__`` function as a way of indexing into it.
`This tutorial <https://pytorch.org/tutorials/beginner/data_loading_tutorial.html>`_
walks through a nice example of creating a custom ``FacialLandmarkDataset`` class
as a subclass of ``Dataset``.
PyTorch's `TensorDataset <https://pytorch.org/docs/stable/_modules/torch/utils/data/dataset.html#TensorDataset>`_
is a Dataset wrapping tensors. By defining a length and way of indexing,
this also gives us a way to iterate, index, and slice along the first
dimension of a tensor. This will make it easier to access both the
independent and dependent variables in the same line as we train.
```
from torch.utils.data import TensorDataset
```
Both ``x_train`` and ``y_train`` can be combined in a single ``TensorDataset``,
which will be easier to iterate over and slice.
```
train_ds = TensorDataset(x_train, y_train)
```
Previously, we had to iterate through minibatches of x and y values separately:
::
xb = x_train[start_i:end_i]
yb = y_train[start_i:end_i]
Now, we can do these two steps together:
::
xb,yb = train_ds[i*bs : i*bs+bs]
```
model, opt = get_model()
for epoch in range(epochs):
for i in range((n - 1) // bs + 1):
xb, yb = train_ds[i * bs: i * bs + bs]
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
print(loss_func(model(xb), yb))
```
Refactor using DataLoader
------------------------------
Pytorch's ``DataLoader`` is responsible for managing batches. You can
create a ``DataLoader`` from any ``Dataset``. ``DataLoader`` makes it easier
to iterate over batches. Rather than having to use ``train_ds[i*bs : i*bs+bs]``,
the DataLoader gives us each minibatch automatically.
```
from torch.utils.data import DataLoader
train_ds = TensorDataset(x_train, y_train)
train_dl = DataLoader(train_ds, batch_size=bs)
```
Previously, our loop iterated over batches (xb, yb) like this:
::
for i in range((n-1)//bs + 1):
xb,yb = train_ds[i*bs : i*bs+bs]
pred = model(xb)
Now, our loop is much cleaner, as (xb, yb) are loaded automatically from the data loader:
::
for xb,yb in train_dl:
pred = model(xb)
```
model, opt = get_model()
for epoch in range(epochs):
for xb, yb in train_dl:
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
print(loss_func(model(xb), yb))
```
Thanks to Pytorch's ``nn.Module``, ``nn.Parameter``, ``Dataset``, and ``DataLoader``,
our training loop is now dramatically smaller and easier to understand. Let's
now try to add the basic features necessary to create effecive models in practice.
Add validation
-----------------------
In section 1, we were just trying to get a reasonable training loop set up for
use on our training data. In reality, you **always** should also have
a `validation set <https://www.fast.ai/2017/11/13/validation-sets/>`_, in order
to identify if you are overfitting.
Shuffling the training data is
`important <https://www.quora.com/Does-the-order-of-training-data-matter-when-training-neural-networks>`_
to prevent correlation between batches and overfitting. On the other hand, the
validation loss will be identical whether we shuffle the validation set or not.
Since shuffling takes extra time, it makes no sense to shuffle the validation data.
We'll use a batch size for the validation set that is twice as large as
that for the training set. This is because the validation set does not
need backpropagation and thus takes less memory (it doesn't need to
store the gradients). We take advantage of this to use a larger batch
size and compute the loss more quickly.
```
train_ds = TensorDataset(x_train, y_train)
train_dl = DataLoader(train_ds, batch_size=bs, shuffle=True)
valid_ds = TensorDataset(x_valid, y_valid)
valid_dl = DataLoader(valid_ds, batch_size=bs * 2)
```
We will calculate and print the validation loss at the end of each epoch.
(Note that we always call ``model.train()`` before training, and ``model.eval()``
before inference, because these are used by layers such as ``nn.BatchNorm2d``
and ``nn.Dropout`` to ensure appropriate behaviour for these different phases.)
```
model, opt = get_model()
for epoch in range(epochs):
model.train()
for xb, yb in train_dl:
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
model.eval()
with torch.no_grad():
valid_loss = sum(loss_func(model(xb), yb) for xb, yb in valid_dl)
print(epoch, valid_loss / len(valid_dl))
```
Create fit() and get_data()
----------------------------------
We'll now do a little refactoring of our own. Since we go through a similar
process twice of calculating the loss for both the training set and the
validation set, let's make that into its own function, ``loss_batch``, which
computes the loss for one batch.
We pass an optimizer in for the training set, and use it to perform
backprop. For the validation set, we don't pass an optimizer, so the
method doesn't perform backprop.
```
def loss_batch(model, loss_func, xb, yb, opt=None):
loss = loss_func(model(xb), yb)
if opt is not None:
loss.backward()
opt.step()
opt.zero_grad()
return loss.item(), len(xb)
```
``fit`` runs the necessary operations to train our model and compute the
training and validation losses for each epoch.
```
import numpy as np
def fit(epochs, model, loss_func, opt, train_dl, valid_dl):
for epoch in range(epochs):
model.train()
for xb, yb in train_dl:
loss_batch(model, loss_func, xb, yb, opt)
model.eval()
with torch.no_grad():
losses, nums = zip(
*[loss_batch(model, loss_func, xb, yb) for xb, yb in valid_dl]
)
val_loss = np.sum(np.multiply(losses, nums)) / np.sum(nums)
print(epoch, val_loss)
```
``get_data`` returns dataloaders for the training and validation sets.
```
def get_data(train_ds, valid_ds, bs):
return (
DataLoader(train_ds, batch_size=bs, shuffle=True),
DataLoader(valid_ds, batch_size=bs * 2),
)
```
Now, our whole process of obtaining the data loaders and fitting the
model can be run in 3 lines of code:
```
train_dl, valid_dl = get_data(train_ds, valid_ds, bs)
model, opt = get_model()
fit(epochs, model, loss_func, opt, train_dl, valid_dl)
```
You can use these basic 3 lines of code to train a wide variety of models.
Let's see if we can use them to train a convolutional neural network (CNN)!
Switch to CNN
-------------
We are now going to build our neural network with three convolutional layers.
Because none of the functions in the previous section assume anything about
the model form, we'll be able to use them to train a CNN without any modification.
We will use Pytorch's predefined
`Conv2d <https://pytorch.org/docs/stable/nn.html#torch.nn.Conv2d>`_ class
as our convolutional layer. We define a CNN with 3 convolutional layers.
Each convolution is followed by a ReLU. At the end, we perform an
average pooling. (Note that ``view`` is PyTorch's version of numpy's
``reshape``)
```
class Mnist_CNN(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1, 16, kernel_size=3, stride=2, padding=1)
self.conv2 = nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1)
self.conv3 = nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1)
def forward(self, xb):
xb = xb.view(-1, 1, 28, 28)
xb = F.relu(self.conv1(xb))
xb = F.relu(self.conv2(xb))
xb = F.relu(self.conv3(xb))
xb = F.avg_pool2d(xb, 4)
return xb.view(-1, xb.size(1))
lr = 0.1
```
`Momentum <https://cs231n.github.io/neural-networks-3/#sgd>`_ is a variation on
stochastic gradient descent that takes previous updates into account as well
and generally leads to faster training.
```
model = Mnist_CNN()
opt = optim.SGD(model.parameters(), lr=lr, momentum=0.9)
fit(epochs, model, loss_func, opt, train_dl, valid_dl)
```
nn.Sequential
------------------------
``torch.nn`` has another handy class we can use to simply our code:
`Sequential <https://pytorch.org/docs/stable/nn.html#torch.nn.Sequential>`_ .
A ``Sequential`` object runs each of the modules contained within it, in a
sequential manner. This is a simpler way of writing our neural network.
To take advantage of this, we need to be able to easily define a
**custom layer** from a given function. For instance, PyTorch doesn't
have a `view` layer, and we need to create one for our network. ``Lambda``
will create a layer that we can then use when defining a network with
``Sequential``.
```
class Lambda(nn.Module):
def __init__(self, func):
super().__init__()
self.func = func
def forward(self, x):
return self.func(x)
def preprocess(x):
return x.view(-1, 1, 28, 28)
```
The model created with ``Sequential`` is simply:
```
model = nn.Sequential(
Lambda(preprocess),
nn.Conv2d(1, 16, kernel_size=3, stride=2, padding=1),
nn.ReLU(),
nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1),
nn.ReLU(),
nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1),
nn.ReLU(),
nn.AvgPool2d(4),
Lambda(lambda x: x.view(x.size(0), -1)),
)
opt = optim.SGD(model.parameters(), lr=lr, momentum=0.9)
fit(epochs, model, loss_func, opt, train_dl, valid_dl)
```
Wrapping DataLoader
-----------------------------
Our CNN is fairly concise, but it only works with MNIST, because:
- It assumes the input is a 28\*28 long vector
- It assumes that the final CNN grid size is 4\*4 (since that's the average
pooling kernel size we used)
Let's get rid of these two assumptions, so our model works with any 2d
single channel image. First, we can remove the initial Lambda layer but
moving the data preprocessing into a generator:
```
def preprocess(x, y):
return x.view(-1, 1, 28, 28), y
class WrappedDataLoader:
def __init__(self, dl, func):
self.dl = dl
self.func = func
def __len__(self):
return len(self.dl)
def __iter__(self):
batches = iter(self.dl)
for b in batches:
yield (self.func(*b))
train_dl, valid_dl = get_data(train_ds, valid_ds, bs)
train_dl = WrappedDataLoader(train_dl, preprocess)
valid_dl = WrappedDataLoader(valid_dl, preprocess)
```
Next, we can replace ``nn.AvgPool2d`` with ``nn.AdaptiveAvgPool2d``, which
allows us to define the size of the *output* tensor we want, rather than
the *input* tensor we have. As a result, our model will work with any
size input.
```
model = nn.Sequential(
nn.Conv2d(1, 16, kernel_size=3, stride=2, padding=1),
nn.ReLU(),
nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1),
nn.ReLU(),
nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1),
nn.ReLU(),
nn.AdaptiveAvgPool2d(1),
Lambda(lambda x: x.view(x.size(0), -1)),
)
opt = optim.SGD(model.parameters(), lr=lr, momentum=0.9)
```
Let's try it out:
```
fit(epochs, model, loss_func, opt, train_dl, valid_dl)
```
Using your GPU
---------------
If you're lucky enough to have access to a CUDA-capable GPU (you can
rent one for about $0.50/hour from most cloud providers) you can
use it to speed up your code. First check that your GPU is working in
Pytorch:
```
print(torch.cuda.is_available())
```
And then create a device object for it:
```
dev = torch.device(
"cuda") if torch.cuda.is_available() else torch.device("cpu")
```
Let's update ``preprocess`` to move batches to the GPU:
```
def preprocess(x, y):
return x.view(-1, 1, 28, 28).to(dev), y.to(dev)
train_dl, valid_dl = get_data(train_ds, valid_ds, bs)
train_dl = WrappedDataLoader(train_dl, preprocess)
valid_dl = WrappedDataLoader(valid_dl, preprocess)
```
Finally, we can move our model to the GPU.
```
model.to(dev)
opt = optim.SGD(model.parameters(), lr=lr, momentum=0.9)
```
You should find it runs faster now:
```
fit(epochs, model, loss_func, opt, train_dl, valid_dl)
```
Closing thoughts
-----------------
We now have a general data pipeline and training loop which you can use for
training many types of models using Pytorch. To see how simple training a model
can now be, take a look at the `mnist_sample` sample notebook.
Of course, there are many things you'll want to add, such as data augmentation,
hyperparameter tuning, monitoring training, transfer learning, and so forth.
These features are available in the fastai library, which has been developed
using the same design approach shown in this tutorial, providing a natural
next step for practitioners looking to take their models further.
We promised at the start of this tutorial we'd explain through example each of
``torch.nn``, ``torch.optim``, ``Dataset``, and ``DataLoader``. So let's summarize
what we've seen:
- **torch.nn**
+ ``Module``: creates a callable which behaves like a function, but can also
contain state(such as neural net layer weights). It knows what ``Parameter`` (s) it
contains and can zero all their gradients, loop through them for weight updates, etc.
+ ``Parameter``: a wrapper for a tensor that tells a ``Module`` that it has weights
that need updating during backprop. Only tensors with the `requires_grad` attribute set are updated
+ ``functional``: a module(usually imported into the ``F`` namespace by convention)
which contains activation functions, loss functions, etc, as well as non-stateful
versions of layers such as convolutional and linear layers.
- ``torch.optim``: Contains optimizers such as ``SGD``, which update the weights
of ``Parameter`` during the backward step
- ``Dataset``: An abstract interface of objects with a ``__len__`` and a ``__getitem__``,
including classes provided with Pytorch such as ``TensorDataset``
- ``DataLoader``: Takes any ``Dataset`` and creates an iterator which returns batches of data.
| github_jupyter |
```
import pandas as pd
#Google colab does not have pickle
try:
import pickle5 as pickle
except:
!pip install pickle5
import pickle5 as pickle
import os
import seaborn as sns
import sys
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.layers import Dense, Input, GlobalMaxPooling1D,Flatten
from keras.layers import Conv1D, MaxPooling1D, Embedding, Concatenate, Lambda
from keras.models import Model
from sklearn.metrics import roc_auc_score,confusion_matrix,roc_curve, auc
from numpy import random
from keras.layers import LSTM, Bidirectional, GlobalMaxPool1D, Dropout
from keras.optimizers import Adam
from keras.utils.vis_utils import plot_model
import sys
sys.path.insert(0,'/content/drive/MyDrive/ML_Data/')
import functions as f
def load_data(D=1,randomize=False):
try:
with open('/content/drive/MyDrive/ML_Data/df_train_'+str(D)+'D.pickle', 'rb') as handle:
df_train = pickle.load(handle)
except:
df_train = pd.read_pickle("C:/Users/nik00/py/proj/hyppi-train.pkl")
try:
with open('/content/drive/MyDrive/ML_Data/df_test_'+str(D)+'D.pickle', 'rb') as handle:
df_test = pickle.load(handle)
except:
df_test = pd.read_pickle("C:/Users/nik00/py/proj/hyppi-independent.pkl")
if randomize:
return shuff_together(df_train,df_test)
else:
return df_train,df_test
df_train,df_test = load_data(5)
print('The data used will be:')
df_train[['Human','Yersinia']]
lengths = sorted(len(s) for s in df_train['Human'])
print("Median length of Human sequence is",lengths[len(lengths)//2])
_ = sns.displot(lengths)
_=plt.title("Most Human sequences seem to be less than 2000 in length")
lengths = sorted(len(s) for s in df_train['Yersinia'])
print("Median length of Yersinia sequence is",lengths[len(lengths)//2])
_ = sns.displot(lengths)
_=plt.title("Most Yersinia sequences seem to be less than 1000 in length")
data1_5D_doubleip_pre,data2_5D_doubleip_pre,data1_test_5D_doubleip_pre,data2_test_5D_doubleip_pre,num_words_5D,MAX_SEQUENCE_LENGTH_5D,MAX_VOCAB_SIZE_5D = f.get_seq_data_doubleip(500000,1000,df_train,df_test,pad = 'pre',show = True)
EMBEDDING_DIM_5D = 15
VALIDATION_SPLIT = 0.2
BATCH_SIZE = 128
EPOCHS = 5
DROP=0.7
x1 = f.conv_model(MAX_SEQUENCE_LENGTH_5D,EMBEDDING_DIM_5D,num_words_5D,DROP)
x2 = f.conv_model(MAX_SEQUENCE_LENGTH_5D,EMBEDDING_DIM_5D,num_words_5D,DROP)
concatenator = Concatenate(axis=1)
x = concatenator([x1.output, x2.output])
x = Dense(128)(x)
x = Dropout(DROP)(x)
output = Dense(1, activation="sigmoid",name="Final")(x)
model5D_CNN_doubleip = Model(inputs=[x1.input, x2.input], outputs=output)
model5D_CNN_doubleip.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
#plot_model(model5D_CNN_doubleip, to_file='model_plot.png', show_shapes=True, show_layer_names=False)
trains = [data1_5D_doubleip_pre,data2_5D_doubleip_pre]
tests = [data1_test_5D_doubleip_pre,data2_test_5D_doubleip_pre]
model5D_CNN_doubleip.fit(trains, df_train['label'].values, epochs=EPOCHS, batch_size=BATCH_SIZE,validation_data=(tests, df_test['label'].values))
print(roc_auc_score(df_test['label'].values, model5D_CNN_doubleip.predict(tests)))
#asd
```
| github_jupyter |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split # for spliting the data into train and test
from sklearn.tree import DecisionTreeClassifier # For creating a decision a tree
from sklearn import tree # for displaying the tree
from sklearn.metrics import classification_report # for calculating accuracy
from sklearn import preprocessing # As we have applied encoding technique we have used this preprocessing library
iris = pd.read_csv("iris.csv", index_col = 0) # In order to set the index to 0 we have mentioned that index_col = 0
iris.head()
# Converting the species column to numbers so we will use encoding technique called as label encoder
label_encoder = preprocessing.LabelEncoder() # This is called function calling
iris['Species'] = label_encoder.fit_transform(iris['Species'])
iris.head()
# Splitting the data in x and y for classification purpose, for performing any classification we are required to split the data first in input and output
x = iris.iloc[:,0:4]
y = iris['Species']
x
y
iris['Species'].unique() # for determining unique values
iris.Species.value_counts()
# Splitting the data into training and test dataset
x_train, x_test, y_train, y_test = train_test_split(x,y,
test_size=0.2,
random_state=40)
```
### Building decision tree classifier using entropy criteria (c5.o)
```
model = DecisionTreeClassifier(criterion = 'entropy',max_depth = 3)
model.fit(x_train,y_train)
```
### Plotting the decision tree
```
tree.plot_tree(model);
model.get_n_leaves()
## As this tree is not visible so we will display it with some another technique
# we will extract the feature names, class names and we will define the figure size so that our tree will be visible in a better way
fn = ['SepalLengthCm', 'SepalWidthCm', 'PetalLengthCm', 'PetalWidthCm']
cn = ['Iris-setosa', 'Iris-versicolar', 'Iris-virginica']
fig,axes = plt.subplots(nrows = 1, ncols =1, figsize =(4,4), dpi = 300) #dpi is the no. of pixels
tree.plot_tree(model, feature_names = fn, class_names = cn, filled = True); # filled = true will fill the values inside the boxes
# Predicting the builded model on our x-test data
preds = model.predict(x_test)
pd.Series(preds).value_counts()
preds
# In order to check whether the predictions are correct or wrong we will create a cross tab on y_test data
crosstable = pd.crosstab(y_test,preds)
crosstable
# Final step we will calculate the accuracy of our model
np.mean(preds==y_test) # We are comparing the predicted values with the actual values and calculating mean for the matches
print(classification_report(preds,y_test))
```
## Building a decision tree using CART method (Classifier model)
```
model_1 = DecisionTreeClassifier(criterion = 'gini',max_depth = 3)
model_1.fit(x_train,y_train)
tree.plot_tree(model_1);
# predicting the values on xtest data
preds = model_1.predict(x_test)
preds
pd.Series(preds).value_counts()
# calculating accuracy of the model using the actual values
np.mean(preds==y_test)
```
## Decision tree Regressor using CART
```
from sklearn.tree import DecisionTreeRegressor
# Just converting the iris data into the following way as I want my Y to be numeric
X = iris.iloc[:,0:3]
Y = iris.iloc[:,3]
X_train,X_test,Y_train,Y_test = train_test_split(X,Y, test_size = 0.33, random_state = 1)
model_reg = DecisionTreeRegressor()
model_reg.fit(X_train,Y_train)
preds1 = model_reg.predict(X_test)
preds1
# Will see the correct and wrong matches
pd.crosstab(Y_test,preds1)
## We will calculate the accuracy by using score method,this is an either way to calculate the accuracy of the model
model_reg.score(X_test,Y_test) # THis model.score function will first calculate the predicted values using the X_test data and then internaly only it will compare those values with the y_test data which is our actual data
```
model_reg.score calculates r squared value and Aic value in background
| github_jupyter |
# Homework
```
import matplotlib.pyplot as plt
%matplotlib inline
import random
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from plotting import overfittingDemo, plot_multiple_linear_regression, overlay_simple_linear_model,plot_simple_residuals
from scipy.optimize import curve_fit
```
**Exercise 1:** What are the two "specialities" of machine learning? Pick one and in your own words, explain what it means. `
Your Answer Here
**Exercise 2:** What is the difference between a regression task and a classification task?
Your Answer Here
**Exercise 3:**
1. What is parametric fitting in your understanding?
2. Given the data $x = 1,2,3,4,5, y_1 = 2,4,6,8,10, y_2 = 2,4,8,16,32,$ what function $f_1, f_2$ will you use to fit $y_1, y_2$? Why do you choose those?
3. Why is parametric fitting somehow not machine learning?
Your Answer Here
**Exercise 4:** Take a look at the following residual plots. Residuals can be helpful in assessing if our model is overpredicting or underpredicting certain values. Assign the variable bestplot to the letter corresponding to which residual plot indicates a good fit for a linear model.
<img src='residplots.png' width="600" height="600">
```
bestplot = 'Put your letter answer between these quotes'
```
**Exercise 5:** Observe the following graphs. Assign each graph variable to one of the following strings: 'overfitting', 'underfitting', or 'bestfit'.
<img src='overfit-underfit.png' width="800" height="800">
```
graph1 = "Put answer here"
graph2 = "Put answer here"
graph3 = "Put answer here"
```
**Exercise 6:** What are the 3 sets we split our initial data set into?
Your Answer Here
**Exercise 7:** Refer to the graphs below when answering the following questions (Exercise 6 and 7).
<img src='training_vs_test_error.png' width="800" height="800">
As we increase the degree of our model, what happens to the training error and what happens to the test error?
Your Answer Here
**Exercise 8:** What is the issue with just increasing the degree of our model to get the lowest training error possible?
Your Answer Here
**Exercise 9:** Find the gradient for ridge loss, most concretely, when $L(\theta, \textbf{y}, \alpha)
= (\frac{1}{n} \sum_{i = 1}^{n}(y_i - \theta)^2) + \frac{\alpha }{2}\sum_{i = 1}^{n}\theta ^2$
find $\frac{\partial}{\partial \hat{\theta}} L(\theta, \textbf{y},\alpha)$, you can have a look at the class example, they are really similar.
Your Answer Here
**Exercise 10:** Following the last part of the exercise, you've already fitted your model, now let's test the performance. Make sure you check the code for the previous example we went through in class.
1. copy what you had from the exercise here.
```
import pandas as pd
mpg = pd.read_csv("./mpg_category.csv", index_col="name")
#exercise part 1
mpg['Old?'] = ...
#exercise part 2
mpg_train, mpg_test = ..., ...
#exercise part 3
from sklearn.linear_model import LogisticRegression
softmax_reg = LogisticRegression(multi_class="multinomial",solver="lbfgs", C=10)
X = ...
Y = ...
softmax_reg.fit(X, Y)
```
2. create the test data set and make the prediction on test dataset
```
X_test = ...
Y_test = ...
pred = softmax_reg.predict(...)
```
3. Make the confusion matrix and tell me how you interpret each of the cell in the confusion matrix. What does different depth of blue means. You can just run the cell below, assumed what you did above is correct. You just have to answer your understanding.
```
from sklearn.metrics import confusion_matrix
confusion_matrix = confusion_matrix(Y_test, pred)
X_label = ['old', 'new']
def plot_confusion_matrix(cm, title='Confusion matrix', cmap=plt.cm.Blues):
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(X_label))
plt.xticks(tick_marks, X_label, rotation=45)
plt.yticks(tick_marks, X_label,)
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
plot_confusion_matrix(confusion_matrix)
# confusion_matrix
```
Your Answer Here
```
# be sure to hit save (File > Save and Checkpoint) or Ctrl/Command-S before you run the cell!
from submit import create_and_submit
create_and_submit(['Intro to Machine Learning Homework.ipynb'], verbose=True)
```
| github_jupyter |
# CS231n_CNN for Visual Recognition
> Stanford University CS231n
- toc: true
- badges: true
- comments: true
- categories: [CNN]
- image: images/
---
- http://cs231n.stanford.edu/
---
# Image Classification
- **Image Classification:** We are given a **Training Set** of labeled images, asked to predict labels on **Test Set.** Common to report the **Accuracy** of predictions(fraction of correctly predicted images)
- We introduced the **k-Nearest Neighbor Classifier**, which predicts the labels based on nearest images in the training set
- We saw that the choice of distance and the value of k are **hyperparameters** that are tuned using a **validation set**, or through **cross-validation** if the size of the data is small.
- Once the best set of hyperparameters is chosen, the classifier is evaluated once on the test set, and reported as the performance of kNN on that data.
- Nearest Neighbor 분류기는 CIFAR-10 데이터셋에서 약 40% 정도의 정확도를 보이는 것을 확인하였다. 이 방법은 구현이 매우 간단하지만, 학습 데이터셋 전체를 메모리에 저장해야 하고, 새로운 테스트 이미지를 분류하고 평가할 때 계산량이 매우 많다.
- 단순히 픽셀 값들의 L1이나 L2 거리는 이미지의 클래스보다 배경이나 이미지의 전체적인 색깔 분포 등에 더 큰 영향을 받기 때문에 이미지 분류 문제에 있어서 충분하지 못하다는 점을 보았다.
---
# Linear Classification
- We defined a **score function** from image pixels to class scores (in this section, a linear function that depends on weights **W** and biases **b**).
- Unlike kNN classifier, the advantage of this **parametric approach** is that once we learn the parameters we can discard the training data. Additionally, the prediction for a new test image is fast since it requires a single matrix multiplication with **W**, not an exhaustive comparison to every single training example.
- We introduced the **bias trick**, which allows us to fold the bias vector into the weight matrix for convenience of only having to keep track of one parameter matrix.
하나의 매개변수 행렬만 추적해야 하는 편의를 위해 편향 벡터를 가중치 행렬로 접을 수 있는 편향 트릭을 도입했습니다 .
- We defined a **loss function** (we introduced two commonly used losses for linear classifiers: the **SVM** and the **Softmax**) that measures how compatible a given set of parameters is with respect to the ground truth labels in the training dataset. We also saw that the loss function was defined in such way that making good predictions on the training data is equivalent to having a small loss.
---
# Optimization
- We developed the intuition of the loss function as a **high-dimensional optimization landscape** in which we are trying to reach the bottom. The working analogy we developed was that of a blindfolded hiker who wishes to reach the bottom. In particular, we saw that the SVM cost function is piece-wise linear and bowl-shaped.
- We motivated the idea of optimizing the loss function with **iterative refinement**, where we start with a random set of weights and refine them step by step until the loss is minimized.
- We saw that the **gradient** of a function gives the steepest ascent direction and we discussed a simple but inefficient way of computing it numerically using the finite difference approximation (the finite difference being the value of h used in computing the numerical gradient).
- We saw that the parameter update requires a tricky setting of the **step size** (or the **learning rate**) that must be set just right: if it is too low the progress is steady but slow. If it is too high the progress can be faster, but more risky. We will explore this tradeoff in much more detail in future sections.
- We discussed the tradeoffs between computing the **numerical** and **analytic** gradient. The numerical gradient is simple but it is approximate and expensive to compute. The analytic gradient is exact, fast to compute but more error-prone since it requires the derivation of the gradient with math. Hence, in practice we always use the analytic gradient and then perform a **gradient check**, in which its implementation is compared to the numerical gradient.
- We introduced the **Gradient Descent** algorithm which iteratively computes the gradient and performs a parameter update in loop.
---
# Backprop
- We developed intuition for what the gradients mean, how they flow backwards in the circuit, and how they communicate which part of the circuit should increase or decrease and with what force to make the final output higher.
- We discussed the importance of **staged computation** for practical implementations of backpropagation. You always want to break up your function into modules for which you can easily derive local gradients, and then chain them with chain rule. Crucially, you almost never want to write out these expressions on paper and differentiate them symbolically in full, because you never need an explicit mathematical equation for the gradient of the input variables. Hence, decompose your expressions into stages such that you can differentiate every stage independently (the stages will be matrix vector multiplies, or max operations, or sum operations, etc.) and then backprop through the variables one step at a time.
---
# Neural Network - 1
- We introduced a very coarse model of a biological **neuron**
- 실제 사용되는 몇몇 **활성화 함수** 에 대해 논의하였고, ReLU가 가장 일반적인 선택이다.
- 활성화 함수 쓰는 이유 : 데이터를 비선형으로 바꾸기 위해서. 선형이면 은닉층이 1개밖에 안나옴
- We introduced **Neural Networks** where neurons are connected with **Fully-Connected layers** where neurons in adjacent layers have full pair-wise connections, but neurons within a layer are not connected.
- 우리는 layered architecture를 통해 활성화 함수의 기능 적용과 결합된 행렬 곱을 기반으로 신경망을 매우 효율적으로 평가할 수 있음을 보았다.
- 우리는 Neural Networks가 **universal function approximators**(NN으로 어떠한 함수든 근사시킬 수 있다)임을 보았지만, 우리는 또한 이 특성이 그들의 편재적인 사용과 거의 관련이 없다는 사실에 대해 논의하였다. They are used because they make certain “right” assumptions about the functional forms of functions that come up in practice.
- 우리는 큰 network가 작은 network보다 항상 더 잘 작동하지만, 더 높은 model capacity는 더 강력한 정규화(높은 가중치 감소같은)로 적절히 해결되어야 하며, 그렇지 않으면 오버핏될 수 있다는 사실에 대해 논의하였다. 이후 섹션에서 더 많은 형태의 정규화(특히 dropout)를 볼 수 있을 것이다.
---
# Neural Network - 2
- 권장되는 전처리는 데이터의 중앙에 평균이 0이 되도록 하고 (zero centered), 스케일을 [-1, 1]로 정규화 하는 것 입니다.
- 올바른 전처리 방법 : 예를들어 평균차감 기법을 쓸 때 학습, 검증, 테스트를 위한 데이터를 먼저 나눈 후 학습 데이터를 대상으로 평균값을 구한 후에 평균차감 전처리를 모든 데이터군(학습, 검증, 테스트)에 적용하는 것이다.
- ReLU를 사용하고 초기화는 $\sqrt{2/n}$ 의 표준 편차를 갖는 정규 분포에서 가중치를 가져와 초기화합니다. 여기서 $n$은 뉴런에 대한 입력 수입니다. E.g. in numpy: `w = np.random.randn(n) * sqrt(2.0/n)`.
- L2 regularization과 드랍아웃을 사용 (the inverted version)
- Batch normalization 사용 (이걸쓰면 드랍아웃은 잘 안씀)
- 실제로 수행할 수 있는 다양한 작업과 각 작업에 대한 가장 일반적인 손실 함수에 대해 논의했다.
---
# Neural Network - 3
신경망(neural network)를 훈련하기 위하여:
- 코드를 짜는 중간중간에 작은 배치로 그라디언트를 체크하고, 뜻하지 않게 튀어나올 위험을 인지하고 있어야 한다.
- 코드가 제대로 돌아가는지 확인하는 방법으로, 손실함수값의 초기값이 합리적인지 그리고 데이터의 일부분으로 100%의 훈련 정확도를 달성할 수 있는지 확인해야한다.
- 훈련 동안, 손실함수와 train/validation 정확도를 계속 살펴보고, (이게 좀 더 멋져 보이면) 현재 파라미터 값 대비 업데이트 값 또한 살펴보라 (대충 ~ 1e-3 정도 되어야 한다). 만약 ConvNet을 다루고 있다면, 첫 층의 웨이트값도 살펴보라.
- 업데이트 방법으로 추천하는 건 SGD+Nesterov Momentum 혹은 Adam이다.
- 학습 속도를 훈련 동안 계속 하강시켜라. 예를 들면, 정해진 에폭 수 뒤에 (혹은 검증 정확도가 상승하다가 하강세로 꺾이면) 학습 속도를 반으로 깎아라.
- Hyper parameter 검색은 grid search가 아닌 random search으로 수행하라. 처음에는 성긴 규모에서 탐색하다가 (넓은 hyper parameter 범위, 1-5 epoch 정도만 학습), 점점 촘촘하게 검색하라. (좁은 범위, 더 많은 에폭에서 학습)
- 추가적인 개선을 위하여 모형 앙상블을 구축하라.
---
# CNN
- ConvNet 아키텍쳐는 여러 레이어를 통해 입력 이미지 볼륨을 출력 볼륨 (클래스 점수)으로 변환시켜 준다.
- ConvNet은 몇 가지 종류의 레이어로 구성되어 있다. CONV/FC/RELU/POOL 레이어가 현재 가장 많이 쓰인다.
- 각 레이어는 3차원의 입력 볼륨을 미분 가능한 함수를 통해 3차원 출력 볼륨으로 변환시킨다.
- parameter가 있는 레이어도 있고 그렇지 않은 레이어도 있다 (FC/CONV는 parameter를 갖고 있고, RELU/POOL 등은 parameter가 없음).
- hyperparameter가 있는 레이어도 있고 그렇지 않은 레이어도 있다 (CONV/FC/POOL 레이어는 hyperparameter를 가지며 ReLU는 가지지 않음).
- stride, zero-padding ...
---
# Spatial Localization and Detection
<img src='img/cs231/detect.png' width="500" height="500">
- Classification : 사진에 대한 라벨이 아웃풋
- Localization : 사진에 대한 상자가 아웃풋 (x, y, w, h)
- Detection : 사진에 대한 여러개의 상자가 아웃풋 DOG(x, y, w, h), CAT(x, y, w, h), ...
- Segmentation : 상자가 아니라 객체의 이미지 형상을 그대로.
- Localization method : localization as Regression, Sliding Window : Overfeat
- Region Proposals : 비슷한 색깔, 텍스쳐를 기준으로 박스를 생성
- Detection :
- R-CNN : Region-based CNN. Region -> CNN
- 문제점 : Region proposal 마다 CNN을 돌려서 시간이 매우 많이든다.
- Fast R-CNN : CNN -> Region
- 문제점 : Region Proposal 과정에서 시간이 든다.
- Faster R-CNN : Region Proposals도 CNN을 이용해서 해보자.
- YOLO(You Only Look Once) : Detection as Regression
- 성능은 Faster R-CNN보다 떨어지지만, 속도가 매우 빠르다.
---
# CNNs in practice
- Data Augmentation
- Change the pixels without changing the label
- Train on transformed data
- VERY widely used
.....
1. Horizontal flips
2. Random crops/scales
3. Color jitter
- Transfer learning
이미지넷의 클래스와 관련있는 데이터라면 사전학습시 성능이 좋아지는게 이해가되는데 관련없는 이미지 (e.g. mri같은 의료이미지)의 경우도 성능이 좋아지는데 그 이유는 무엇인가?
-> 앞단에선 엣지, 컬러같은 low level의 feature를 인식, 뒤로갈수록 상위레벨을 인식. lowlevel을 미리 학습해놓는다는 것은 그 어떤 이미지를 분석할 때도 도움이된다!
- How to stack convolutions:
- Replace large convolutions (5x5, 7x7) with stacks of 3x3 convolutions
- 1x1 "bottleneck" convolutions are very efficient
- Can factor NxN convolutions into 1xN and Nx1
- All of the above give fewer parameters, less compute, more nonlinearity
더 적은 파라미터, 더 적은 컴퓨팅연산, 더 많은 nonlinearity(필터 사이사이 ReLU등이 들어가기에)
- Computing Convolutions:
- im2col : Easy to implement, but big memory overhead.
- FFT : Big speedups for small kernels
- "Fast Algorithms" : seem promising, not widely used yet
---
# Segmentaion
- Semantic Segmentation
- Classify all pixels
- Fully convolutional models, downsample then upsample
- Learnable upsampling: fractionally strided convolution
- Skip connections can help
...
- Instance Segmentation
- Detect instance, generate mask
- Similar pipelines to object detection
| github_jupyter |
##### Copyright 2020 Google LLC.
Licensed under the Apache License, Version 2.0 (the "License");
```
#@title License header
# Copyright 2020 Google LLC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# ResNet
[ResNet](https://arxiv.org/abs/1512.03385) is a deep neural network architecture for image recognition.
This notebook
* Constructs a [ResNet50](https://www.tensorflow.org/api_docs/python/tf/keras/applications/ResNet50) model using `tf.keras`, with weights pretrained using the[ImageNet](http://www.image-net.org/) dataset
* Compiles that model with IREE
* Tests TensorFlow and IREE execution of the model on a sample image
```
#@title Imports and common setup
from pyiree import rt as ireert
from pyiree.tf import compiler as ireec
from pyiree.tf.support import tf_utils
import tensorflow as tf
from matplotlib import pyplot as plt
#@title Construct a pretrained ResNet model with ImageNet weights
tf.keras.backend.set_learning_phase(False)
# Static shape, including batch size (1).
# Can be dynamic once dynamic shape support is ready.
INPUT_SHAPE = [1, 224, 224, 3]
tf_model = tf.keras.applications.resnet50.ResNet50(
weights="imagenet", include_top=True, input_shape=tuple(INPUT_SHAPE[1:]))
# Wrap the model in a tf.Module to compile it with IREE.
class ResNetModule(tf.Module):
def __init__(self):
super(ResNetModule, self).__init__()
self.m = tf_model
self.predict = tf.function(
input_signature=[tf.TensorSpec(INPUT_SHAPE, tf.float32)])(tf_model.call)
#@markdown ### Backend Configuration
backend_choice = "iree_vmla (CPU)" #@param [ "iree_vmla (CPU)", "iree_llvmjit (CPU)", "iree_vulkan (GPU/SwiftShader)" ]
backend_choice = backend_choice.split(" ")[0]
backend = tf_utils.BackendInfo(backend_choice)
#@title Compile ResNet with IREE
# This may take a few minutes.
iree_module = backend.compile(ResNetModule, ["predict"])
#@title Load a test image of a [labrador](https://commons.wikimedia.org/wiki/File:YellowLabradorLooking_new.jpg)
def load_image(path_to_image):
image = tf.io.read_file(path_to_image)
image = tf.image.decode_image(image, channels=3)
image = tf.image.resize(image, (224, 224))
image = image[tf.newaxis, :]
return image
content_path = tf.keras.utils.get_file(
'YellowLabradorLooking_new.jpg',
'https://storage.googleapis.com/download.tensorflow.org/example_images/YellowLabradorLooking_new.jpg')
content_image = load_image(content_path)
print("Test image:")
plt.imshow(content_image.numpy().reshape(224, 224, 3) / 255.0)
plt.axis("off")
plt.tight_layout()
#@title Model pre- and post-processing
input_data = tf.keras.applications.resnet50.preprocess_input(content_image)
def decode_result(result):
return tf.keras.applications.resnet50.decode_predictions(result, top=3)[0]
#@title Run TF model
print("TF prediction:")
tf_result = tf_model.predict(input_data)
print(decode_result(tf_result))
#@title Run the model compiled with IREE
print("IREE prediction:")
iree_result = iree_module.predict(input_data)
print(decode_result(iree_result))
```
| github_jupyter |
# ART for TensorFlow v2 - Keras API
This notebook demonstrate applying ART with the new TensorFlow v2 using the Keras API. The code follows and extends the examples on www.tensorflow.org.
```
import warnings
warnings.filterwarnings('ignore')
import tensorflow as tf
tf.compat.v1.disable_eager_execution()
import numpy as np
from matplotlib import pyplot as plt
from art.estimators.classification import KerasClassifier
from art.attacks.evasion import FastGradientMethod, CarliniLInfMethod
if tf.__version__[0] != '2':
raise ImportError('This notebook requires TensorFlow v2.')
```
# Load MNIST dataset
```
(x_train, y_train), (x_test, y_test) = tf.keras.datasets.mnist.load_data()
x_train, x_test = x_train / 255.0, x_test / 255.0
x_test = x_test[0:100]
y_test = y_test[0:100]
```
# TensorFlow with Keras API
Create a model using Keras API. Here we use the Keras Sequential model and add a sequence of layers. Afterwards the model is compiles with optimizer, loss function and metrics.
```
model = tf.keras.models.Sequential([
tf.keras.layers.InputLayer(input_shape=(28, 28)),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(128, activation='relu'),
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Dense(10, activation='softmax')
])
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy']);
```
Fit the model on training data.
```
model.fit(x_train, y_train, epochs=3);
```
Evaluate model accuracy on test data.
```
loss_test, accuracy_test = model.evaluate(x_test, y_test)
print('Accuracy on test data: {:4.2f}%'.format(accuracy_test * 100))
```
Create a ART Keras classifier for the TensorFlow Keras model.
```
classifier = KerasClassifier(model=model, clip_values=(0, 1))
```
## Fast Gradient Sign Method attack
Create a ART Fast Gradient Sign Method attack.
```
attack_fgsm = FastGradientMethod(estimator=classifier, eps=0.3)
```
Generate adversarial test data.
```
x_test_adv = attack_fgsm.generate(x_test)
```
Evaluate accuracy on adversarial test data and calculate average perturbation.
```
loss_test, accuracy_test = model.evaluate(x_test_adv, y_test)
perturbation = np.mean(np.abs((x_test_adv - x_test)))
print('Accuracy on adversarial test data: {:4.2f}%'.format(accuracy_test * 100))
print('Average perturbation: {:4.2f}'.format(perturbation))
```
Visualise the first adversarial test sample.
```
plt.matshow(x_test_adv[0])
plt.show()
```
## Carlini&Wagner Infinity-norm attack
Create a ART Carlini&Wagner Infinity-norm attack.
```
attack_cw = CarliniLInfMethod(classifier=classifier, eps=0.3, max_iter=100, learning_rate=0.01)
```
Generate adversarial test data.
```
x_test_adv = attack_cw.generate(x_test)
```
Evaluate accuracy on adversarial test data and calculate average perturbation.
```
loss_test, accuracy_test = model.evaluate(x_test_adv, y_test)
perturbation = np.mean(np.abs((x_test_adv - x_test)))
print('Accuracy on adversarial test data: {:4.2f}%'.format(accuracy_test * 100))
print('Average perturbation: {:4.2f}'.format(perturbation))
```
Visualise the first adversarial test sample.
```
plt.matshow(x_test_adv[0, :, :])
plt.show()
```
| github_jupyter |
# Prophet
Time serie forecasting using Prophet
Official documentation: https://facebook.github.io/prophet/docs/quick_start.html
Procedure for forecasting time series data based on an additive model where non-linear trends are fit with yearly, weekly, and daily seasonality, plus holiday effects. It is released by Facebook’s Core Data Science team.
Additive model is a model like:
$Data = seasonal\space effect + trend + residual$
and, multiplicative model:
$Data = seasonal\space effect * trend * residual$
The algorithm provides useful statistics that help visualize the tuning process, e.g. trend, week trend, year trend and their max and min errors.
### Data
The data on which the algorithms will be trained and tested upon comes from Kaggle Hourly Energy Consumption database. It is collected by PJM Interconnection, a company coordinating the continuous buying, selling, and delivery of wholesale electricity through the Energy Market from suppliers to customers in the reagon of South Carolina, USA. All .csv files contains rows with a timestamp and a value. The name of the value column corresponds to the name of the contractor. the timestamp represents a single hour and the value represents the total energy, cunsumed during that hour.
The data we will be using is hourly power consumption data from PJM. Energy consumtion has some unique charachteristics. It will be interesting to see how prophet picks them up.
https://www.kaggle.com/robikscube/hourly-energy-consumption
Pulling the PJM East which has data from 2002-2018 for the entire east region.
```
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from fbprophet import Prophet
from sklearn.metrics import mean_squared_error, mean_absolute_error
plt.style.use('fivethirtyeight') # For plots
dataset_path = './data/hourly-energy-consumption/PJME_hourly.csv'
df = pd.read_csv(dataset_path, index_col=[0], parse_dates=[0])
print("Dataset path:",df.shape)
df.head(10)
# VISUALIZE DATA
# Color pallete for plotting
color_pal = ["#F8766D", "#D39200", "#93AA00",
"#00BA38", "#00C19F", "#00B9E3",
"#619CFF", "#DB72FB"]
df.plot(style='.', figsize=(20,10), color=color_pal[0], title='PJM East Dataset TS')
plt.show()
#Decompose the seasonal data
def create_features(df, label=None):
"""
Creates time series features from datetime index.
"""
df = df.copy()
df['date'] = df.index
df['hour'] = df['date'].dt.hour
df['dayofweek'] = df['date'].dt.dayofweek
df['quarter'] = df['date'].dt.quarter
df['month'] = df['date'].dt.month
df['year'] = df['date'].dt.year
df['dayofyear'] = df['date'].dt.dayofyear
df['dayofmonth'] = df['date'].dt.day
df['weekofyear'] = df['date'].dt.weekofyear
X = df[['hour','dayofweek','quarter','month','year',
'dayofyear','dayofmonth','weekofyear']]
if label:
y = df[label]
return X, y
return X
df.columns
X, y = create_features(df, label='PJME_MW')
features_and_target = pd.concat([X, y], axis=1)
print("Shape",features_and_target.shape)
features_and_target.head(10)
sns.pairplot(features_and_target.dropna(),
hue='hour',
x_vars=['hour','dayofweek',
'year','weekofyear'],
y_vars='PJME_MW',
height=5,
plot_kws={'alpha':0.15, 'linewidth':0}
)
plt.suptitle('Power Use MW by Hour, Day of Week, Year and Week of Year')
plt.show()
```
## Train and Test Split
We use a temporal split, keeping old data and use only new period to do the prediction
```
split_date = '01-Jan-2015'
pjme_train = df.loc[df.index <= split_date].copy()
pjme_test = df.loc[df.index > split_date].copy()
# Plot train and test so you can see where we have split
pjme_test \
.rename(columns={'PJME_MW': 'TEST SET'}) \
.join(pjme_train.rename(columns={'PJME_MW': 'TRAINING SET'}),
how='outer') \
.plot(figsize=(15,5), title='PJM East', style='.')
plt.show()
```
To use prophet you need to correctly rename features and label to correctly pass the input to the engine.
```
# Format data for prophet model using ds and y
pjme_train.reset_index() \
.rename(columns={'Datetime':'ds',
'PJME_MW':'y'})
print(pjme_train.columns)
pjme_train.head(5)
```
### Create and train the model
```
# Setup and train model and fit
model = Prophet()
model.fit(pjme_train.reset_index() \
.rename(columns={'Datetime':'ds',
'PJME_MW':'y'}))
# Predict on training set with model
pjme_test_fcst = model.predict(df=pjme_test.reset_index() \
.rename(columns={'Datetime':'ds'}))
pjme_test_fcst.head()
```
### Plot the results and forecast
```
# Plot the forecast
f, ax = plt.subplots(1)
f.set_figheight(5)
f.set_figwidth(15)
fig = model.plot(pjme_test_fcst,
ax=ax)
plt.show()
# Plot the components of the model
fig = model.plot_components(pjme_test_fcst)
```
| github_jupyter |
```
from PyEIS import *
```
## Frequency range
The first first step needed to simulate an electrochemical impedance spectra is to generate a frequency domain, to do so, use to build-in freq_gen() function, as follows
```
f_range = freq_gen(f_start=10**10, f_stop=0.1, pts_decade=7)
# print(f_range[0]) #First 5 points in the freq. array
print()
# print(f_range[1]) #First 5 points in the angular freq.array
```
Note that all functions included are described, to access these descriptions stay within () and press shift+tab. The freq_gen(), returns both the frequency, which is log seperated based on points/decade between f_start to f_stop, and the angular frequency. This function is quite useful and will be used through this tutorial
## The Equivalent Circuits
There exist a number of equivalent circuits that can be simulated and fitted, these functions are made as definations and can be called at any time. To find these, write: "cir_" and hit tab. All functions are outline in the next cell and can also be viewed in the equivalent circuit overview:
```
cir_RC
cir_RQ
cir_RsRQ
cir_RsRQRQ
cir_Randles
cir_Randles_simplified
cir_C_RC_C
cir_Q_RQ_Q
cir_RCRCZD
cir_RsTLsQ
cir_RsRQTLsQ
cir_RsTLs
cir_RsRQTLs
cir_RsTLQ
cir_RsRQTLQ
cir_RsTL
cir_RsRQTL
cir_RsTL_1Dsolid
cir_RsRQTL_1Dsolid
```
## Simulation of -(RC)-
<img src='https://raw.githubusercontent.com/kbknudsen/PyEIS/master/pyEIS_images/RC_circuit.png' width="300" />
#### Input Parameters:
- w = Angular frequency [1/s]
- R = Resistance [Ohm]
- C = Capacitance [F]
- fs = summit frequency of RC circuit [Hz]
```
RC_example = EIS_sim(frange=f_range[0], circuit=cir_RC(w=f_range[1], R=70, C=10**-6), legend='on')
```
## Simulation of -Rs-(RQ)-
<img src='https://raw.githubusercontent.com/kbknudsen/PyEIS/master/pyEIS_images/RsRQ_circuit.png' width="500" />
#### Input parameters:
- w = Angular frequency [1/s]
- Rs = Series resistance [Ohm]
- R = Resistance [Ohm]
- Q = Constant phase element [s^n/ohm]
- n = Constant phase elelment exponent [-]
- fs = summit frequency of RQ circuit [Hz]
```
RsRQ_example = EIS_sim(frange=f_range[0], circuit=cir_RsRQ(w=f_range[1], Rs=70, R=200, n=.8, Q=10**-5), legend='on')
RsRC_example = EIS_sim(frange=f_range[0], circuit=cir_RsRC(w=f_range[1], Rs=80, R=100, C=10**-5), legend='on')
```
## Simulation of -Rs-(RQ)-(RQ)-
<img src='https://raw.githubusercontent.com/kbknudsen/PyEIS/master/pyEIS_images/RsRQRQ_circuit.png' width="500" />
#### Input parameters:
- w = Angular frequency [1/s]
- Rs = Series Resistance [Ohm]
- R = Resistance [Ohm]
- Q = Constant phase element [s^n/ohm]
- n = Constant phase element exponent [-]
- fs = summit frequency of RQ circuit [Hz]
- R2 = Resistance [Ohm]
- Q2 = Constant phase element [s^n/ohm]
- n2 = Constant phase element exponent [-]
- fs2 = summit frequency of RQ circuit [Hz]
```
RsRQRQ_example = EIS_sim(frange=f_range[0], circuit=cir_RsRQRQ(w=f_range[1], Rs=200, R=150, n=.872, Q=10**-4, R2=50, n2=.853, Q2=10**-6), legend='on')
```
## Simulation of -Rs-(Q(RW))- (Randles-circuit)
This circuit is often used for an experimental setup with a macrodisk working electrode with an outer-sphere heterogeneous charge transfer. This, classical, warburg element is controlled by semi-infinite linear diffusion, which is given by the geometry of the working electrode. Two Randles functions are avaliable for simulations: cir_Randles_simplified() and cir_Randles(). The former contains the Warburg constant (sigma), which summs up all mass transport constants (Dox/Dred, Cred/Cox, number of electrons (n_electron), Faradays constant (F), T, and E0) into a single constant sigma, while the latter contains all of these constants. Only cir_Randles_simplified() is avaliable for fitting, as either D$_{ox}$ or D$_{red}$ and C$_{red}$ or C$_{ox}$ are needed.
<img src='https://raw.githubusercontent.com/kbknudsen/PyEIS/master/pyEIS_images/Randles_circuit.png' width="500" />
#### Input parameters:
- Rs = Series resistance [ohm]
- Rct = charge-transfer resistance [ohm]
- Q = Constant phase element used to model the double-layer capacitance [F]
- n = expononent of the CPE [-]
- sigma = Warburg Constant [ohm/s^1/2]
```
Randles = cir_Randles_simplified(w=f_range[1], Rs=100, R=1000, n=1, sigma=300, Q=10**-5)
Randles_example = EIS_sim(frange=f_range[0], circuit=Randles, legend='off')
Randles_example = EIS_sim(frange=f_range[0], circuit=cir_Randles_simplified(w=f_range[1], Rs=100, R=1000, n=1, sigma=300, Q='none', fs=10**3.3), legend='off')
```
In the following, the Randles circuit with the Warburg constant (sigma) defined is simulated where:
- D$_{red}$/D$_{ox}$ = 10$^{-6}$ cm$^2$/s
- C$_{red}$/C$_{ox}$ = 10 mM
- n_electron = 1
- T = 25 $^o$C
This function is a great tool to simulate expected impedance responses prior to starting experiments as it allows for evaluation of concentrations, diffusion constants, number of electrons, and Temp. to evaluate the feasability of obtaining information on either kinetics, mass-transport, or both.
```
Randles_example = EIS_sim(frange=f_range[0], circuit=cir_Randles(w=f_range[1], Rs=100, Rct=1000, Q=10**-7, n=1, T=298.15, D_ox=10**-9, D_red=10**-9, C_ox=10**-5, C_red=10**-5, n_electron=1, E=0, A=1), legend='off')
```
| github_jupyter |
```
#hide
#default_exp examples.complex_dummy_experiment_manager
from nbdev.showdoc import *
from block_types.utils.nbdev_utils import nbdev_setup, TestRunner
nbdev_setup ()
tst = TestRunner (targets=['dummy'])
```
# Complex Dummy Experiment Manager
> Dummy experiment manager with features that allow additional functionality
```
#export
from hpsearch.examples.dummy_experiment_manager import DummyExperimentManager, FakeModel
import hpsearch
import os
import shutil
import os
import hpsearch.examples.dummy_experiment_manager as dummy_em
from hpsearch.visualization import plot_utils
#for tests
import pytest
from block_types.utils.nbdev_utils import md
```
## ComplexDummyExperimentManager
```
#export
class ComplexDummyExperimentManager (DummyExperimentManager):
def __init__ (self, model_file_name='model_weights.pk', **kwargs):
super().__init__ (model_file_name=model_file_name, **kwargs)
self.raise_error_if_run = False
def run_experiment (self, parameters={}, path_results='./results'):
# useful for testing: in some cases the experiment manager should not call run_experiment
if self.raise_error_if_run:
raise RuntimeError ('run_experiment should not be called')
# extract hyper-parameters used by our model. All the parameters have default values if they are not passed.
offset = parameters.get('offset', 0.5) # default value: 0.5
rate = parameters.get('rate', 0.01) # default value: 0.01
epochs = parameters.get('epochs', 10) # default value: 10
noise = parameters.get('noise', 0.0)
if parameters.get('actual_epochs') is not None:
epochs = parameters.get('actual_epochs')
# other parameters that do not form part of our experiment definition
# changing the values of these other parameters, does not make the ID of the experiment change
verbose = parameters.get('verbose', True)
# build model with given hyper-parameters
model = FakeModel (offset=offset, rate=rate, epochs=epochs, noise = noise, verbose=verbose)
# load training, validation and test data (fake step)
model.load_data()
# start from previous experiment if indicated by parameters
path_results_previous_experiment = parameters.get('prev_path_results')
if path_results_previous_experiment is not None:
model.load_model_and_history (path_results_previous_experiment)
# fit model with training data
model.fit ()
# save model weights and evolution of accuracy metric across epochs
model.save_model_and_history(path_results)
# simulate ctrl-c
if parameters.get ('halt', False):
raise KeyboardInterrupt ('stopped')
# evaluate model with validation and test data
validation_accuracy, test_accuracy = model.score()
# store model
self.model = model
# the function returns a dictionary with keys corresponding to the names of each metric.
# We return result on validation and test set in this example
dict_results = dict (validation_accuracy = validation_accuracy,
test_accuracy = test_accuracy)
return dict_results
```
### Usage
```
#exports tests.examples.test_complex_dummy_experiment_manager
def test_complex_dummy_experiment_manager ():
#em = generate_data ('complex_dummy_experiment_manager')
md (
'''
Extend previous experiment by using a larger number of epochs
We see how to create a experiment that is the same as a previous experiment,
only increasing the number of epochs.
1.a. For test purposes, we first run the full number of epochs, 30, take note of the accuracy,
and remove the experiment
'''
)
em = ComplexDummyExperimentManager (path_experiments='test_complex_dummy_experiment_manager',
verbose=0)
em.create_experiment_and_run (parameters = {'epochs': 30});
reference_accuracy = em.model.accuracy
reference_weight = em.model.weight
from hpsearch.config.hpconfig import get_path_experiments
import os
import pandas as pd
path_experiments = get_path_experiments ()
print (f'experiments folders: {os.listdir(f"{path_experiments}/experiments")}\n')
experiments_data = pd.read_pickle (f'{path_experiments}/experiments_data.pk')
print ('csv data')
display (experiments_data)
md ('we plot the history')
from hpsearch.visualization.experiment_visualization import plot_multiple_histories
plot_multiple_histories ([0], run_number=0, op='max', backend='matplotlib', metrics='validation_accuracy')
md ('1.b. Now we run two experiments: ')
md ('We run the first experiment with 20 epochs:')
# a.- remove previous experiment
em.remove_previous_experiments()
# b.- create first experiment with epochs=20
em.create_experiment_and_run (parameters = {'epochs': 20});
print (f'experiments folders: {os.listdir(f"{path_experiments}/experiments")}\n')
experiments_data = pd.read_pickle (f'{path_experiments}/experiments_data.pk')
print ('csv data')
display(experiments_data)
print (f'weight: {em.model.weight}, accuracy: {em.model.accuracy}')
md ('We run the second experiment resumes from the previous one and increases the epochs to 30')
# 4.- create second experiment with epochs=10
em.create_experiment_and_run (parameters = {'epochs': 30},
other_parameters={'prev_epoch': True,
'name_epoch': 'epochs',
'previous_model_file_name': 'model_weights.pk'});
experiments_data = pd.read_pickle (f'{path_experiments}/experiments_data.pk')
print ('csv data')
display(experiments_data)
new_accuracy = em.model.accuracy
new_weight = em.model.weight
assert new_weight==reference_weight
assert new_accuracy==reference_accuracy
print (f'weight: {new_weight}, accuracy: {new_accuracy}')
md ('We plot the history')
plot_multiple_histories ([1], run_number=0, op='max', backend='matplotlib', metrics='validation_accuracy')
em.remove_previous_experiments()
tst.run (test_complex_dummy_experiment_manager, tag='dummy')
```
## Running experiments and removing experiments
```
# export
def run_multiple_experiments (**kwargs):
dummy_em.run_multiple_experiments (EM=ComplexDummyExperimentManager, **kwargs)
def remove_previous_experiments ():
dummy_em.remove_previous_experiments (EM=ComplexDummyExperimentManager)
#export
def generate_data (name_folder):
em = ComplexDummyExperimentManager (path_experiments=f'test_{name_folder}', verbose=0)
em.remove_previous_experiments ()
run_multiple_experiments (em=em, nruns=5, noise=0.1, verbose=False)
return em
```
| github_jupyter |
# Workshop 13
## _Object-oriented programming._
#### Classes and Objects
```
class MyClass:
pass
obj1 = MyClass()
obj2 = MyClass()
print(obj1)
print(type(obj1))
print(obj2)
print(type(obj2))
```
##### Constructor and destructor
```
class Employee:
def __init__(self):
print('Employee created.')
def __del__(self):
print('Destructor called, Employee deleted.')
obj = Employee()
del obj
```
##### Attributes and methods
```
class Student:
def __init__(self, name, grade):
self.name = name
self.grade = grade
def __str__(self):
return '{' + self.name + ': ' + str(self.grade) + '}'
def learn(self):
print('My name is %s. I am learning Python! My grade is %d.' % (self.name, self.grade))
students = [Student('Steve', 9), Student('Oleg', 10)]
for student in students:
print()
print('student.name = ' + student.name)
print('student.grade = ' + str(student.grade))
print('student = ' + str(student))
student.learn()
```
##### Class and instance attributes
```
class Person:
# class variable shared by all instances
status = 'student'
def __init__(self, name):
# instance variable unique to each instance
self.name = name
a = Person('Steve')
b = Person('Mark')
print('')
print(a.name + ' : ' + a.status)
print(b.name + ' : ' + b.status)
Person.status = 'graduate'
print('')
print(a.name + ' : ' + a.status)
print(b.name + ' : ' + b.status)
Person.status = 'student'
print('')
print(a.name + ' : ' + a.status)
print(b.name + ' : ' + b.status)
```
##### Class and static methods
```
class Env:
os = 'Windows'
@classmethod
def print_os(self):
print(self.os)
@staticmethod
def print_user():
print('guest')
Env.print_os()
Env.print_user()
```
##### Encapsulation
```
class Person:
def __init__(self, name):
self.name = name
def __str__(self):
return 'My name is ' + self.name
person = Person('Steve')
print(person.name)
person.name = 'Said'
print(person.name)
class Identity:
def __init__(self, name):
self.__name = name
def __str__(self):
return 'My name is ' + self.__name
person = Identity('Steve')
print(person.__name)
person.__name = 'Said'
print(person)
```
##### Operator overloading
```
class Number:
def __init__(self, value):
self.__value = value
def __del__(self):
pass
def __str__(self):
return str(self.__value)
def __int__(self):
return self.__value
def __eq__(self, other):
return self.__value == other.__value
def __ne__(self, other):
return self.__value != other.__value
def __lt__(self, other):
return self.__value < other.__value
def __gt__(self, other):
return self.__value > other.__value
def __add__(self, other):
return Number(self.__value + other.__value)
def __mul__(self, other):
return Number(self.__value * other.__value)
def __neg__(self):
return Number(-self.__value)
a = Number(10)
b = Number(20)
c = Number(5)
# Overloaded operators
x = -a + b * c
print(x)
print(a < b)
print(b > c)
# Unsupported operators
print(a <= b)
print(b >= c)
print(a // c)
```
#### Inheritance and polymorphism
```
class Creature:
def say(self):
pass
class Dog(Creature):
def say(self):
print('Woof!')
class Cat(Creature):
def say(self):
print("Meow!")
class Lion(Creature):
def say(self):
print("Roar!")
animals = [Creature(), Dog(), Cat(), Lion()]
for animal in animals:
print(type(animal))
animal.say()
```
##### Multiple inheritance
```
class Person:
def __init__(self, name):
self.name = name
class Student(Person):
def __init__(self, name, grade):
super().__init__(name)
self.grade = grade
class Employee:
def __init__(self, salary):
self.salary = salary
class Teacher(Person, Employee):
def __init__(self, name, salary):
Person.__init__(self, name)
Employee.__init__(self, salary)
class TA(Student, Employee):
def __init__(self, name, grage, salary):
Student.__init__(self, name, grage)
Employee.__init__(self, salary)
x = Student('Oleg', 9)
y = TA('Sergei', 10, 1000)
z = Teacher('Andrei', 2000)
for person in [x, y, z]:
print(person.name)
if isinstance(person, Employee):
print(person.salary)
if isinstance(person, Student):
print(person.grade)
```
##### Function _isinstance_
```
x = 10
print('')
print(isinstance(x, int))
print(isinstance(x, float))
print(isinstance(x, str))
y = 3.14
print('')
print(isinstance(y, int))
print(isinstance(y, float))
print(isinstance(y, str))
z = 'Hello world'
print('')
print(isinstance(z, int))
print(isinstance(z, float))
print(isinstance(z, str))
class A:
pass
class B:
pass
class C(A):
pass
class D(A, B):
pass
a = A()
b = B()
c = C()
d = D()
print('')
print(isinstance(a, object))
print(isinstance(a, A))
print(isinstance(b, B))
print('')
print(isinstance(b, object))
print(isinstance(b, A))
print(isinstance(b, B))
print(isinstance(b, C))
print('')
print(isinstance(c, object))
print(isinstance(c, A))
print(isinstance(c, B))
print(isinstance(c, D))
print('')
print(isinstance(d, object))
print(isinstance(d, A))
print(isinstance(d, B))
print(isinstance(d, C))
print(isinstance(d, D))
```
##### Composition
```
class Teacher:
pass
class Student:
pass
class ClassRoom:
def __init__(self, teacher, students):
self.teacher = teacher
self.students = students
cl = ClassRoom(Teacher(), [Student(), Student(), Student()])
class Set:
def __init__(self, values=None):
self.dict = {}
if values is not None:
for value in values:
self.add(value)
def __repr__(self):
return "Set: " + str(self.dict.keys())
def add(self, value):
self.dict[value] = True
def contains(self, value):
return value in self.dict
def remove(self, value):
del self.dict[value]
s = Set([1,2,3])
s.add(4)
print(s.contains(4))
s.remove(3)
print(s.contains(3))
```
| github_jupyter |
# Scalable GP Classification in 1D (w/ KISS-GP)
This example shows how to use grid interpolation based variational classification with an `ApproximateGP` using a `GridInterpolationVariationalStrategy` module. This classification module is designed for when the inputs of the function you're modeling are one-dimensional.
The use of inducing points allows for scaling up the training data by making computational complexity linear instead of cubic.
In this example, we’re modeling a function that is periodically labeled cycling every 1/8 (think of a square wave with period 1/4)
This notebook doesn't use cuda, in general we recommend GPU use if possible and most of our notebooks utilize cuda as well.
Kernel interpolation for scalable structured Gaussian processes (KISS-GP) was introduced in this paper:
http://proceedings.mlr.press/v37/wilson15.pdf
KISS-GP with SVI for classification was introduced in this paper:
https://papers.nips.cc/paper/6426-stochastic-variational-deep-kernel-learning.pdf
```
import math
import torch
import gpytorch
from matplotlib import pyplot as plt
from math import exp
%matplotlib inline
%load_ext autoreload
%autoreload 2
train_x = torch.linspace(0, 1, 26)
train_y = torch.sign(torch.cos(train_x * (2 * math.pi))).add(1).div(2)
from gpytorch.models import ApproximateGP
from gpytorch.variational import CholeskyVariationalDistribution
from gpytorch.variational import GridInterpolationVariationalStrategy
class GPClassificationModel(ApproximateGP):
def __init__(self, grid_size=128, grid_bounds=[(0, 1)]):
variational_distribution = CholeskyVariationalDistribution(grid_size)
variational_strategy = GridInterpolationVariationalStrategy(self, grid_size, grid_bounds, variational_distribution)
super(GPClassificationModel, self).__init__(variational_strategy)
self.mean_module = gpytorch.means.ConstantMean()
self.covar_module = gpytorch.kernels.ScaleKernel(gpytorch.kernels.RBFKernel())
def forward(self,x):
mean_x = self.mean_module(x)
covar_x = self.covar_module(x)
latent_pred = gpytorch.distributions.MultivariateNormal(mean_x, covar_x)
return latent_pred
model = GPClassificationModel()
likelihood = gpytorch.likelihoods.BernoulliLikelihood()
from gpytorch.mlls.variational_elbo import VariationalELBO
# Find optimal model hyperparameters
model.train()
likelihood.train()
# Use the adam optimizer
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
# "Loss" for GPs - the marginal log likelihood
# n_data refers to the number of training datapoints
mll = VariationalELBO(likelihood, model, num_data=train_y.numel())
def train():
num_iter = 100
for i in range(num_iter):
optimizer.zero_grad()
output = model(train_x)
# Calc loss and backprop gradients
loss = -mll(output, train_y)
loss.backward()
print('Iter %d/%d - Loss: %.3f' % (i + 1, num_iter, loss.item()))
optimizer.step()
# Get clock time
%time train()
# Set model and likelihood into eval mode
model.eval()
likelihood.eval()
# Initialize axes
f, ax = plt.subplots(1, 1, figsize=(4, 3))
with torch.no_grad():
test_x = torch.linspace(0, 1, 101)
predictions = likelihood(model(test_x))
ax.plot(train_x.numpy(), train_y.numpy(), 'k*')
pred_labels = predictions.mean.ge(0.5).float()
ax.plot(test_x.data.numpy(), pred_labels.numpy(), 'b')
ax.set_ylim([-1, 2])
ax.legend(['Observed Data', 'Mean', 'Confidence'])
```
| github_jupyter |
```
import numpy as np
import astropy
from itertools import izip
from pearce.mocks import compute_prim_haloprop_bins, cat_dict
from pearce.mocks.customHODModels import *
from halotools.utils.table_utils import compute_conditional_percentiles
from halotools.mock_observables import hod_from_mock, wp, tpcf, tpcf_one_two_halo_decomp
from math import ceil
from matplotlib import pyplot as plt
%matplotlib inline
import seaborn as sns
sns.set()
shuffle_type = ''#'sh_shuffled'
mag_type = 'vpeak'
mag_cut = -21
min_ptcl = 200
mag_key = 'halo_%s%s_mag'%(shuffle_type, mag_type)
upid_key = 'halo_%supid'%(shuffle_type)
PMASS = 591421440.0000001 #chinchilla 400/ 2048
catalog = astropy.table.Table.read('abmatched_halos.hdf5', format = 'hdf5')
cosmo_params = {'simname':'chinchilla', 'Lbox':400.0, 'scale_factors':[0.658, 1.0]}
cat = cat_dict[cosmo_params['simname']](**cosmo_params)#construct the specified catalog!
cat.load_catalog(1.0)
#cat.h = 1.0
halo_catalog = catalog[catalog['halo_mvir'] > min_ptcl*cat.pmass] #mass cut
galaxy_catalog = halo_catalog[ halo_catalog[mag_key] < mag_cut ] # mag cut
def compute_mass_bins(prim_haloprop, dlog10_prim_haloprop=0.05):
lg10_min_prim_haloprop = np.log10(np.min(prim_haloprop))-0.001
lg10_max_prim_haloprop = np.log10(np.max(prim_haloprop))+0.001
num_prim_haloprop_bins = (lg10_max_prim_haloprop-lg10_min_prim_haloprop)/dlog10_prim_haloprop
return np.logspace(
lg10_min_prim_haloprop, lg10_max_prim_haloprop,
num=int(ceil(num_prim_haloprop_bins)))
mass_bins = compute_mass_bins(halo_catalog['halo_mvir'], 0.2)
mass_bin_centers = (mass_bins[1:]+mass_bins[:-1])/2.0
cen_mask = galaxy_catalog['halo_upid']==-1
cen_hod_sham, _ = hod_from_mock(galaxy_catalog[cen_mask]['halo_mvir_host_halo'],\
halo_catalog['halo_mvir'],\
mass_bins)
sat_hod_sham, _ = hod_from_mock(galaxy_catalog[~cen_mask]['halo_mvir_host_halo'],\
halo_catalog['halo_mvir'],\
mass_bins)
cat.load_model(1.0, HOD=(FSAssembiasTabulatedCens, FSAssembiasTabulatedSats), hod_kwargs = {'prim_haloprop_vals': mass_bin_centers,
#'sec_haloprop_key': 'halo_%s'%(mag_type),
'cen_hod_vals':cen_hod_sham,
'sat_hod_vals':sat_hod_sham,
'split':0.5})
print cat.model.param_dict
#rp_bins = np.logspace(-1,1.5,20)
#rp_bins = np.logspace(-1.1,1.8, 25)
#rp_bins = np.loadtxt('/nfs/slac/g/ki/ki18/des/swmclau2/AB_tests/rp_bins.npy')
rp_bins = np.array([7.943282000000000120e-02,
1.122018500000000057e-01,
1.584893199999999891e-01,
2.238721100000000130e-01,
3.162277700000000191e-01,
4.466835900000000192e-01,
6.309573400000000332e-01,
8.912509400000000470e-01,
1.258925410000000022e+00,
1.778279409999999894e+00,
2.511886430000000114e+00,
3.548133889999999901e+00,
5.011872340000000037e+00,
7.079457839999999891e+00,
1.000000000000000000e+01,
1.412537544999999994e+01,
1.995262315000000086e+01,
2.818382931000000013e+01,
3.981071706000000177e+01])
bin_centers = (rp_bins[:1]+rp_bins[:-1])/2
min_logmass, max_logmass = 9.0, 17.0
names = ['mean_occupation_centrals_assembias_param1','mean_occupation_satellites_assembias_param1',\
'mean_occupation_centrals_assembias_split1','mean_occupation_satellites_assembias_split1']
#mock_wp = cat.calc_wp(rp_bins, RSD= False)
MAP = np.array([ 0.85, -0.3,0.85,0.5])
params = dict(zip(names, MAP))
#print params.keys()
mock_wps = []
mock_wps_1h, mock_wps_2h = [],[]
#mock_nds = []
split = np.linspace(0.1, 0.9, 4)
#split_abcissa = [10**9, 10**13, 10**16]
#cat.model._input_model_dictionary['centrals_occupation']._split_abscissa = split_abcissa
#cat.model._input_model_dictionary['satellites_occupation']._split_abscissa = split_abcissa
for p in split:
#params['mean_occupation_centrals_assembias_split1'] = p
params['mean_occupation_satellites_assembias_split1'] = p
#print params.keys()
#print cat.model.param_dict
cat.populate(params)
#print cat.model.param_dict
#cut_idx = cat.model.mock.galaxy_table['gal_type'] == 'centrals'
mass_cut = np.logical_and(np.log10(cat.model.mock.galaxy_table['halo_mvir'] ) > min_logmass,\
np.log10(cat.model.mock.galaxy_table['halo_mvir'] ) <= max_logmass)
#mass_cut = np.logical_and(mass_cut, cut_idx)
#mock_nds.append(len(cut_idx)/cat.Lbox**3)
mock_pos = np.c_[cat.model.mock.galaxy_table['x'],\
cat.model.mock.galaxy_table['y'],\
cat.model.mock.galaxy_table['z']]
mock_wps.append(wp(mock_pos*cat.h, rp_bins ,40.0*cat.h, period=cat.Lbox*cat.h, num_threads=1))
#oneh, twoh = tpcf_one_two_halo_decomp(mock_pos,cat.model.mock.galaxy_table[mass_cut]['halo_hostid'],\
# rp_bins , period=cat.Lbox, num_threads=1)
#mock_wps_1h.append(oneh)
#mock_wps_2h.append(twoh)
mock_wps = np.array(mock_wps)
wp_errs = np.std(mock_wps, axis = 0)
#mock_wps_1h = np.array(mock_wps_1h)
#mock_wp_no_ab_1h = np.mean(mock_wps_1h, axis = 0)
#mock_wps_2h = np.array(mock_wps_2h)
#mock_wp_no_ab_2h = np.mean(mock_wps_2h, axis = 0)
#mock_nds = np.array(mock_nds)
#mock_nd = np.mean(mock_nds)
#nd_err = np.std(mock_nds)
params
params = dict(zip(names, [0,0,0.5,0.5]))
cat.populate(params)
mass_cut = np.logical_and(np.log10(cat.model.mock.galaxy_table['halo_mvir'] ) > min_logmass,\
np.log10(cat.model.mock.galaxy_table['halo_mvir'] ) <= max_logmass)
print cat.model.param_dict
mock_pos = np.c_[cat.model.mock.galaxy_table['x'],\
cat.model.mock.galaxy_table['y'],\
cat.model.mock.galaxy_table['z']]
noab_wp = wp(mock_pos*cat.h, rp_bins ,40.0*cat.h, period=cat.Lbox*cat.h, num_threads=1)
print np.log10(noab_wp)
from halotools.mock_observables import return_xyz_formatted_array
sham_pos = np.c_[galaxy_catalog['halo_x'],\
galaxy_catalog['halo_y'],\
galaxy_catalog['halo_z']]
distortion_dim = 'z'
v_distortion_dim = galaxy_catalog['halo_v%s' % distortion_dim]
# apply redshift space distortions
#sham_pos = return_xyz_formatted_array(sham_pos[:,0],sham_pos[:,1],sham_pos[:,2], velocity=v_distortion_dim, \
# velocity_distortion_dimension=distortion_dim, period=cat.Lbox)
#sham_wp = wp(sham_pos*cat.h, rp_bins, 40.0*cat.h, period=cat.Lbox*cat.h, num_threads=1)
sham_wp = wp(sham_pos*cat.h, rp_bins, 40.0*cat.h, period=cat.Lbox*cat.h, num_threads=1)
#sham_wp = tpcf(sham_pos, rp_bins , period=cat.Lbox, num_threads=1)
sham_wp
len(galaxy_catalog)/((cat.Lbox*cat.h)**3)
```
```
plt.figure(figsize=(10,8))
for p, mock_wp in zip(split, mock_wps):
plt.plot(bin_centers, mock_wp, label = p)
#plt.plot(bin_centers, sham_wp, ls='--', label = 'SHAM')
plt.plot(bin_centers, noab_wp, ls=':', label = 'No AB')
plt.loglog()
plt.legend(loc='best',fontsize = 15)
plt.xlim([1e-1, 30e0]);
#plt.ylim([1,15000])
plt.xlabel(r'$r$',fontsize = 15)
plt.ylabel(r'$\xi(r)$',fontsize = 15)
plt.show()
np.log10(mock_wps[-1])
plt.figure(figsize=(10,8))
for p, mock_wp in zip(split, mock_wps):
plt.plot(bin_centers, mock_wp/sham_wp, label = p)
#plt.plot(bin_centers, sham_wp, ls='--', label = 'SHAM')
#plt.plot(bin_centers, noab_wp, ls=':', label = 'No AB')
#plt.loglog()
plt.xscale('log')
plt.legend(loc='best',fontsize = 15)
plt.xlim([1e-1, 15e0]);
plt.ylim([0.8,1.2])
plt.xlabel(r'$r$',fontsize = 15)
plt.ylabel(r'$\xi(r)$',fontsize = 15)
plt.show()
plt.figure(figsize=(10,8))
#for p, mock_wp in zip(split, mock_wps):
# plt.plot(bin_centers, mock_wp/sham_wp, label = p)
#plt.plot(bin_centers, sham_wp, ls='--', label = 'SHAM')
plt.plot(bin_centers, noab_wp/sham_wp, label = 'No AB')
#plt.loglog()
plt.xscale('log')
plt.legend(loc='best',fontsize = 15)
plt.xlim([1e-1, 15e0]);
#plt.ylim([1,15000])
plt.xlabel(r'$r$',fontsize = 15)
plt.ylabel(r'$\xi(r)$',fontsize = 15)
plt.show()
plt.figure(figsize=(10,8))
for p, mock_wp in zip(split, mock_wps_1h):
plt.plot(bin_centers, mock_wp, label = p)
#plt.plot(bin_centers, sham_wp, ls='--', label = 'SHAM')
#plt.plot(bin_centers, noab_wp, ls=':', label = 'No AB')
plt.loglog()
plt.legend(loc='best',fontsize = 15)
plt.xlim([1e-1, 30e0]);
#plt.ylim([1,15000])
plt.xlabel(r'$r$',fontsize = 15)
plt.ylabel(r'$\xi(r)$',fontsize = 15)
plt.show()
plt.figure(figsize=(10,8))
for p, mock_wp in zip(split, mock_wps_2h):
plt.plot(bin_centers, mock_wp, label = p)
#plt.plot(bin_centers, sham_wp, ls='--', label = 'SHAM')
plt.plot(bin_centers, noab_wp, ls=':', label = 'No AB')
plt.loglog()
plt.legend(loc='best',fontsize = 15)
plt.xlim([1e-1, 30e0]);
#plt.ylim([1,15000])
plt.xlabel(r'$r$',fontsize = 15)
plt.ylabel(r'$\xi(r)$',fontsize = 15)
plt.show()
plt.figure(figsize=(10,8))
for p, mock_wp in zip(split, mock_wps_2h):
plt.plot(bin_centers, mock_wp/noab_wp, label = p)
#plt.plot(bin_centers, sham_wp, ls='--', label = 'SHAM')
#plt.plot(bin_centers, noab_wp, ls=':', label = 'No AB')
plt.loglog()
plt.legend(loc='best',fontsize = 15)
plt.xlim([1e-1, 30e0]);
#plt.ylim([1,15000])
plt.xlabel(r'$r$',fontsize = 15)
plt.ylabel(r'$\xi(r)$',fontsize = 15)
plt.show()
plt.plot(bin_centers, mock_wps[0, :])
plt.plot(bin_centers, mock_wps_1h[0, :])
plt.plot(bin_centers, mock_wps_2h[0, :])
plt.loglog()
plt.legend(loc='best',fontsize = 15)
plt.xlim([1e-1, 30e0]);
#plt.ylim([1,15000])
plt.xlabel(r'$r$',fontsize = 15)
plt.ylabel(r'$\xi(r)$',fontsize = 15)
plt.show()
plt.figure(figsize=(10,8))
#avg = mock_wps.mean(axis = 0)
for p, mock_wp in zip(split, mock_wps):
plt.plot(bin_centers, mock_wp/sham_wp, label = 'p = %.2f'%p)
plt.plot(bin_centers, noab_wp/sham_wp, label = 'No AB', ls = ':')
#plt.loglog()
plt.xscale('log')
plt.legend(loc='best',fontsize = 15)
plt.xlim([1e-1, 5e0]);
plt.ylim([0.75,1.25]);
plt.xlabel(r'$r$',fontsize = 15)
plt.ylabel(r'$\xi(r)/\xi_{SHAM}(r)$',fontsize = 15)
plt.show()
sats_occ = cat.model._input_model_dictionary['satellites_occupation']
sats_occ._split_ordinates = [0.99]
```
```
print sats_occ
baseline_lower_bound, baseline_upper_bound = 0,np.inf
prim_haloprop = cat.model.mock.halo_table['halo_mvir']
sec_haloprop = cat.model.mock.halo_table['halo_nfw_conc']
from halotools.utils.table_utils import compute_conditional_percentile_values
split = sats_occ.percentile_splitting_function(prim_haloprop)
# Compute the baseline, undecorated result
result = sats_occ.baseline_mean_occupation(prim_haloprop=prim_haloprop)
# We will only decorate values that are not edge cases,
# so first compute the mask for non-edge cases
no_edge_mask = (
(split > 0) & (split < 1) &
(result > baseline_lower_bound) & (result < baseline_upper_bound)
)
# Now create convenient references to the non-edge-case sub-arrays
no_edge_result = result[no_edge_mask]
no_edge_split = split[no_edge_mask]
```
```
from halotools.utils.table_utils import compute_conditional_averages
strength = sats_occ.assembias_strength(prim_haloprop[no_edge_mask])
slope = sats_occ.assembias_slope(prim_haloprop[no_edge_mask])
# the average displacement acts as a normalization we need.
max_displacement = sats_occ._disp_func(sec_haloprop=pv_sub_sec_haloprop/np.max(np.abs(pv_sub_sec_haloprop)), slope=slope)
disp_average = compute_conditional_averages(vals=max_displacement,prim_haloprop=prim_haloprop[no_edge_mask])
#disp_average = np.ones((prim_haloprop.shape[0], ))*0.5
perturbation2 = np.zeros(len(prim_haloprop[no_edge_mask]))
greater_than_half_avg_idx = disp_average > 0.5
less_than_half_avg_idx = disp_average <= 0.5
if len(max_displacement[greater_than_half_avg_idx]) > 0:
base_pos = result[no_edge_mask][greater_than_half_avg_idx]
strength_pos = strength[greater_than_half_avg_idx]
avg_pos = disp_average[greater_than_half_avg_idx]
upper_bound1 = (base_pos - baseline_lower_bound)/avg_pos
upper_bound2 = (baseline_upper_bound - base_pos)/(1-avg_pos)
upper_bound = np.minimum(upper_bound1, upper_bound2)
print upper_bound1, upper_bound2
perturbation2[greater_than_half_avg_idx] = strength_pos*upper_bound*(max_displacement[greater_than_half_avg_idx]-avg_pos)
if len(max_displacement[less_than_half_avg_idx]) > 0:
base_neg = result[no_edge_mask][less_than_half_avg_idx]
strength_neg = strength[less_than_half_avg_idx]
avg_neg = disp_average[less_than_half_avg_idx]
lower_bound1 = (base_neg-baseline_lower_bound)/avg_neg#/(1- avg_neg)
lower_bound2 = (baseline_upper_bound - base_neg)/(1-avg_neg)#(avg_neg)
lower_bound = np.minimum(lower_bound1, lower_bound2)
perturbation2[less_than_half_avg_idx] = strength_neg*lower_bound*(max_displacement[less_than_half_avg_idx]-avg_neg)
print np.unique(max_displacement[indices_of_mb])
print np.unique(disp_average[indices_of_mb])
perturbation
mass_bins = compute_mass_bins(prim_haloprop)
mass_bin_idxs = compute_prim_haloprop_bins(prim_haloprop_bin_boundaries=mass_bins, prim_haloprop = prim_haloprop[no_edge_mask])
mb = 87
indices_of_mb = np.where(mass_bin_idxs == mb)[0]
plt.hist(perturbation[indices_of_mb], bins =100);
plt.yscale('log');
#plt.loglog();
print max(perturbation)
print min(perturbation)
print max(perturbation[indices_of_mb])
print min(perturbation[indices_of_mb])
idxs = np.argsort(perturbation)
print mass_bin_idxs[idxs[-10:]]
plt.hist(perturbation2[indices_of_mb], bins =100);
plt.yscale('log');
#plt.loglog();
print perturbation2
```
| github_jupyter |
# Showing uncertainty
> Uncertainty occurs everywhere in data science, but it's frequently left out of visualizations where it should be included. Here, we review what a confidence interval is and how to visualize them for both single estimates and continuous functions. Additionally, we discuss the bootstrap resampling technique for assessing uncertainty and how to visualize it properly. This is the Summary of lecture "Improving Your Data Visualizations in Python", via datacamp.
- toc: true
- badges: true
- comments: true
- author: Chanseok Kang
- categories: [Python, Datacamp, Visualization]
- image: images/so2_compare.png
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
plt.rcParams['figure.figsize'] = (10, 5)
```
### Point estimate intervals
- When is uncertainty important?
- Estimates from sample
- Average of a subset
- Linear model coefficients
- Why is uncertainty important?
- Helps inform confidence in estimate
- Neccessary for decision making
- Acknowledges limitations of data
### Basic confidence intervals
You are a data scientist for a fireworks manufacturer in Des Moines, Iowa. You need to make a case to the city that your company's large fireworks show has not caused any harm to the city's air. To do this, you look at the average levels for pollutants in the week after the fourth of July and how they compare to readings taken after your last show. By showing confidence intervals around the averages, you can make a case that the recent readings were well within the normal range.
```
average_ests = pd.read_csv('./dataset/average_ests.csv', index_col=0)
average_ests
# Construct CI bounds for averages
average_ests['lower'] = average_ests['mean'] - 1.96 * average_ests['std_err']
average_ests['upper'] = average_ests['mean'] + 1.96 * average_ests['std_err']
# Setup a grid of plots, with non-shared x axes limits
g = sns.FacetGrid(average_ests, row='pollutant', sharex=False, aspect=2);
# Plot CI for average estimate
g.map(plt.hlines, 'y', 'lower', 'upper');
# Plot observed values for comparison and remove axes labels
g.map(plt.scatter, 'seen', 'y', color='orangered').set_ylabels('').set_xlabels('');
```
This simple visualization shows that all the observed values fall well within the confidence intervals for all the pollutants except for $O_3$.
### Annotating confidence intervals
Your data science work with pollution data is legendary, and you are now weighing job offers in both Cincinnati, Ohio and Indianapolis, Indiana. You want to see if the SO2 levels are significantly different in the two cities, and more specifically, which city has lower levels. To test this, you decide to look at the differences in the cities' SO2 values (Indianapolis' - Cincinnati's) over multiple years.
Instead of just displaying a p-value for a significant difference between the cities, you decide to look at the 95% confidence intervals (columns `lower` and `upper`) of the differences. This allows you to see the magnitude of the differences along with any trends over the years.
```
diffs_by_year = pd.read_csv('./dataset/diffs_by_year.csv', index_col=0)
diffs_by_year
# Set start and ends according to intervals
# Make intervals thicker
plt.hlines(y='year', xmin='lower', xmax='upper',
linewidth=5, color='steelblue', alpha=0.7,
data=diffs_by_year);
# Point estimates
plt.plot('mean', 'year', 'k|', data=diffs_by_year);
# Add a 'null' reference line at 0 and color orangered
plt.axvline(x=0, color='orangered', linestyle='--');
# Set descriptive axis labels and title
plt.xlabel('95% CI');
plt.title('Avg SO2 differences between Cincinnati and Indianapolis');
```
By looking at the confidence intervals you can see that the difference flipped from generally positive (more pollution in Cincinnati) in 2013 to negative (more pollution in Indianapolis) in 2014 and 2015. Given that every year's confidence interval contains the null value of zero, no P-Value would be significant, and a plot that only showed significance would have been entirely hidden this trend.
## Confidence bands
### Making a confidence band
Vandenberg Air Force Base is often used as a location to launch rockets into space. You have a theory that a recent increase in the pace of rocket launches could be harming the air quality in the surrounding region. To explore this, you plotted a 25-day rolling average line of the measurements of atmospheric $NO_2$. To help decide if any pattern observed is random-noise or not, you decide to add a 99% confidence band around your rolling mean. Adding a confidence band to a trend line can help shed light on the stability of the trend seen. This can either increase or decrease the confidence in the discovered trend.
```
vandenberg_NO2 = pd.read_csv('./dataset/vandenberg_NO2.csv', index_col=0)
vandenberg_NO2.head()
# Draw 99% interval bands for average NO2
vandenberg_NO2['lower'] = vandenberg_NO2['mean'] - 2.58 * vandenberg_NO2['std_err']
vandenberg_NO2['upper'] = vandenberg_NO2['mean'] + 2.58 * vandenberg_NO2['std_err']
# Plot mean estimate as a white semi-transparent line
plt.plot('day', 'mean', data=vandenberg_NO2, color='white', alpha=0.4);
# Fill between the upper and lower confidence band values
plt.fill_between(x='day', y1='lower', y2='upper', data=vandenberg_NO2);
```
This plot shows that the middle of the year's $NO_2$ values are not only lower than the beginning and end of the year but also are less noisy. If just the moving average line were plotted, then this potentially interesting observation would be completely missed. (Can you think of what may cause reduced variance at the lower values of the pollutant?)
### Separating a lot of bands
It is relatively simple to plot a bunch of trend lines on top of each other for rapid and precise comparisons. Unfortunately, if you need to add uncertainty bands around those lines, the plot becomes very difficult to read. Figuring out whether a line corresponds to the top of one class' band or the bottom of another's can be hard due to band overlap. Luckily in Seaborn, it's not difficult to break up the overlapping bands into separate faceted plots.
To see this, explore trends in SO2 levels for a few cities in the eastern half of the US. If you plot the trends and their confidence bands on a single plot - it's a mess. To fix, use Seaborn's `FacetGrid()` function to spread out the confidence intervals to multiple panes to ease your inspection.
```
eastern_SO2 = pd.read_csv('./dataset/eastern_SO2.csv', index_col=0)
eastern_SO2.head()
# setup a grid of plots with columns divided by location
g = sns.FacetGrid(eastern_SO2, col='city', col_wrap=2);
# Map interval plots to each cities data with coral colored ribbons
g.map(plt.fill_between, 'day', 'lower', 'upper', color='coral');
# Map overlaid mean plots with white line
g.map(plt.plot, 'day', 'mean', color='white');
```
By separating each band into its own plot you can investigate each city with ease. Here, you see that Des Moines and Houston on average have lower SO2 values for the entire year than the two cities in the Midwest. Cincinnati has a high and variable peak near the beginning of the year but is generally more stable and lower than Indianapolis.
### Cleaning up bands for overlaps
You are working for the city of Denver, Colorado and want to run an ad campaign about how much cleaner Denver's air is than Long Beach, California's air. To investigate this claim, you will compare the SO2 levels of both cities for the year 2014. Since you are solely interested in how the cities compare, you want to keep the bands on the same plot. To make the bands easier to compare, decrease the opacity of the confidence bands and set a clear legend.
```
SO2_compare = pd.read_csv('./dataset/SO2_compare.csv', index_col=0)
SO2_compare.head()
for city, color in [('Denver', '#66c2a5'), ('Long Beach', '#fc8d62')]:
# Filter data to desired city
city_data = SO2_compare[SO2_compare.city == city]
# Set city interval color to desired and lower opacity
plt.fill_between(x='day', y1='lower', y2='upper', data=city_data, color=color, alpha=0.4);
# Draw a faint mean line for reference and give a label for legend
plt.plot('day', 'mean', data=city_data, label=city, color=color, alpha=0.25);
plt.legend();
```
From these two curves you can see that during the first half of the year Long Beach generally has a higher average SO2 value than Denver, in the middle of the year they are very close, and at the end of the year Denver seems to have higher averages. However, by showing the confidence intervals, you can see however that almost none of the year shows a statistically meaningful difference in average values between the two cities.
## Beyond 95%
### 90, 95, and 99% intervals
You are a data scientist for an outdoor adventure company in Fairbanks, Alaska. Recently, customers have been having issues with SO2 pollution, leading to costly cancellations. The company has sensors for CO, NO2, and O3 but not SO2 levels.
You've built a model that predicts SO2 values based on the values of pollutants with sensors (loaded as `pollution_model`, a `statsmodels` object). You want to investigate which pollutant's value has the largest effect on your model's SO2 prediction. This will help you know which pollutant's values to pay most attention to when planning outdoor tours. To maximize the amount of information in your report, show multiple levels of uncertainty for the model estimates.
```
from statsmodels.formula.api import ols
pollution = pd.read_csv('./dataset/pollution_wide.csv')
pollution = pollution.query("city == 'Fairbanks' & year == 2014 & month == 11")
pollution_model = ols(formula='SO2 ~ CO + NO2 + O3 + day', data=pollution)
res = pollution_model.fit()
# Add interval percent widths
alphas = [ 0.01, 0.05, 0.1]
widths = [ '99% CI', '95%', '90%']
colors = ['#fee08b','#fc8d59','#d53e4f']
for alpha, color, width in zip(alphas, colors, widths):
# Grab confidence interval
conf_ints = res.conf_int(alpha)
# Pass current interval color and legend label to plot
plt.hlines(y = conf_ints.index, xmin = conf_ints[0], xmax = conf_ints[1],
colors = color, label = width, linewidth = 10)
# Draw point estimates
plt.plot(res.params, res.params.index, 'wo', label = 'Point Estimate')
plt.legend(loc = 'upper right')
```
### 90 and 95% bands
You are looking at a 40-day rolling average of the $NO_2$ pollution levels for the city of Cincinnati in 2013. To provide as detailed a picture of the uncertainty in the trend you want to look at both the 90 and 99% intervals around this rolling estimate.
To do this, set up your two interval sizes and an orange ordinal color palette. Additionally, to enable precise readings of the bands, make them semi-transparent, so the Seaborn background grids show through.
```
cinci_13_no2 = pd.read_csv('./dataset/cinci_13_no2.csv', index_col=0);
cinci_13_no2.head()
int_widths = ['90%', '99%']
z_scores = [1.67, 2.58]
colors = ['#fc8d59', '#fee08b']
for percent, Z, color in zip(int_widths, z_scores, colors):
# Pass lower and upper confidence bounds and lower opacity
plt.fill_between(
x = cinci_13_no2.day, alpha = 0.4, color = color,
y1 = cinci_13_no2['mean'] - Z * cinci_13_no2['std_err'],
y2 = cinci_13_no2['mean'] + Z * cinci_13_no2['std_err'],
label = percent);
plt.legend();
```
This plot shows us that throughout 2013, the average NO2 values in Cincinnati followed a cyclical pattern with the seasons. However, the uncertainty bands show that for most of the year you can't be sure this pattern is not noise at both a 90 and 99% confidence level.
### Using band thickness instead of coloring
You are a researcher investigating the elevation a rocket reaches before visual is lost and pollutant levels at Vandenberg Air Force Base. You've built a model to predict this relationship, and since you are working independently, you don't have the money to pay for color figures in your journal article. You need to make your model results plot work in black and white. To do this, you will plot the 90, 95, and 99% intervals of the effect of each pollutant as successively smaller bars.
```
rocket_model = pd.read_csv('./dataset/rocket_model.csv', index_col=0)
rocket_model
# Decrase interval thickness as interval widens
sizes = [ 15, 10, 5]
int_widths = ['90% CI', '95%', '99%']
z_scores = [ 1.67, 1.96, 2.58]
for percent, Z, size in zip(int_widths, z_scores, sizes):
plt.hlines(y = rocket_model.pollutant,
xmin = rocket_model['est'] - Z * rocket_model['std_err'],
xmax = rocket_model['est'] + Z * rocket_model['std_err'],
label = percent,
# Resize lines and color them gray
linewidth = size,
color = 'gray');
# Add point estimate
plt.plot('est', 'pollutant', 'wo', data = rocket_model, label = 'Point Estimate');
plt.legend(loc = 'center left', bbox_to_anchor = (1, 0.5));
```
While less elegant than using color to differentiate interval sizes, this plot still clearly allows the reader to access the effect each pollutant has on rocket visibility. You can see that of all the pollutants, O3 has the largest effect and also the tightest confidence bounds
## Visualizing the bootstrap
### The bootstrap histogram
You are considering a vacation to Cincinnati in May, but you have a severe sensitivity to NO2. You pull a few years of pollution data from Cincinnati in May and look at a bootstrap estimate of the average $NO_2$ levels. You only have one estimate to look at the best way to visualize the results of your bootstrap estimates is with a histogram.
While you like the intuition of the bootstrap histogram by itself, your partner who will be going on the vacation with you, likes seeing percent intervals. To accommodate them, you decide to highlight the 95% interval by shading the region.
```
# Perform bootstrapped mean on a vector
def bootstrap(data, n_boots):
return [np.mean(np.random.choice(data,len(data))) for _ in range(n_boots) ]
pollution = pd.read_csv('./dataset/pollution_wide.csv')
cinci_may_NO2 = pollution.query("city == 'Cincinnati' & month == 5").NO2
# Generate bootstrap samples
boot_means = bootstrap(cinci_may_NO2, 1000)
# Get lower and upper 95% interval bounds
lower, upper = np.percentile(boot_means, [2.5, 97.5])
# Plot shaded area for interval
plt.axvspan(lower, upper, color = 'gray', alpha = 0.2);
# Draw histogram of bootstrap samples
sns.distplot(boot_means, bins = 100, kde = False);
```
Your bootstrap histogram looks stable and uniform. You're now confident that the average NO2 levels in Cincinnati during your vacation should be in the range of 16 to 23.
### Bootstrapped regressions
While working for the Long Beach parks and recreation department investigating the relationship between $NO_2$ and $SO_2$ you noticed a cluster of potential outliers that you suspect might be throwing off the correlations.
Investigate the uncertainty of your correlations through bootstrap resampling to see how stable your fits are. For convenience, the bootstrap sampling is complete and is provided as `no2_so2_boot` along with `no2_so2` for the non-resampled data.
```
no2_so2 = pd.read_csv('./dataset/no2_so2.csv', index_col=0)
no2_so2_boot = pd.read_csv('./dataset/no2_so2_boot.csv', index_col=0)
sns.lmplot('NO2', 'SO2', data = no2_so2_boot,
# Tell seaborn to a regression line for each sample
hue = 'sample',
# Make lines blue and transparent
line_kws = {'color': 'steelblue', 'alpha': 0.2},
# Disable built-in confidence intervals
ci = None, legend = False, scatter = False);
# Draw scatter of all points
plt.scatter('NO2', 'SO2', data = no2_so2);
```
The outliers appear to drag down the regression lines as evidenced by the cluster of lines with more severe slopes than average. In a single plot, you have not only gotten a good idea of the variability of your correlation estimate but also the potential effects of outliers.
### Lots of bootstraps with beeswarms
As a current resident of Cincinnati, you're curious to see how the average NO2 values compare to Des Moines, Indianapolis, and Houston: a few other cities you've lived in.
To look at this, you decide to use bootstrap estimation to look at the mean NO2 values for each city. Because the comparisons are of primary interest, you will use a swarm plot to compare the estimates.
```
pollution_may = pollution.query("month == 5")
pollution_may
# Initialize a holder DataFrame for bootstrap results
city_boots = pd.DataFrame()
for city in ['Cincinnati', 'Des Moines', 'Indianapolis', 'Houston']:
# Filter to city
city_NO2 = pollution_may[pollution_may.city == city].NO2
# Bootstrap city data & put in DataFrame
cur_boot = pd.DataFrame({'NO2_avg': bootstrap(city_NO2, 100), 'city': city})
# Append to other city's bootstraps
city_boots = pd.concat([city_boots,cur_boot])
# Beeswarm plot of averages with citys on y axis
sns.swarmplot(y = "city", x = "NO2_avg", data = city_boots, color = 'coral');
```
The beeswarm plots show that Indianapolis and Houston both have the highest average NO2 values, with Cincinnati falling roughly in the middle. Interestingly, you can rather confidently say that Des Moines has the lowest as nearly all its sample estimates fall below those of the other cities.
| github_jupyter |
<a href="https://colab.research.google.com/github/mariokart345/DS-Unit-2-Applied-Modeling/blob/master/module3-permutation-boosting/LS_DS_233.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Lambda School Data Science
*Unit 2, Sprint 3, Module 3*
---
# Permutation & Boosting
- Get **permutation importances** for model interpretation and feature selection
- Use xgboost for **gradient boosting**
### Setup
Run the code cell below. You can work locally (follow the [local setup instructions](https://lambdaschool.github.io/ds/unit2/local/)) or on Colab.
Libraries:
- category_encoders
- [**eli5**](https://eli5.readthedocs.io/en/latest/)
- matplotlib
- numpy
- pandas
- scikit-learn
- [**xgboost**](https://xgboost.readthedocs.io/en/latest/)
```
%%capture
import sys
# If you're on Colab:
if 'google.colab' in sys.modules:
DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Applied-Modeling/master/data/'
!pip install category_encoders==2.*
!pip install eli5
# If you're working locally:
else:
DATA_PATH = '../data/'
```
We'll go back to Tanzania Waterpumps for this lesson.
```
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
# Merge train_features.csv & train_labels.csv
train = pd.merge(pd.read_csv(DATA_PATH+'waterpumps/train_features.csv'),
pd.read_csv(DATA_PATH+'waterpumps/train_labels.csv'))
# Read test_features.csv & sample_submission.csv
test = pd.read_csv(DATA_PATH+'waterpumps/test_features.csv')
sample_submission = pd.read_csv(DATA_PATH+'waterpumps/sample_submission.csv')
# Split train into train & val
train, val = train_test_split(train, train_size=0.80, test_size=0.20,
stratify=train['status_group'], random_state=42)
def wrangle(X):
"""Wrangle train, validate, and test sets in the same way"""
# Prevent SettingWithCopyWarning
X = X.copy()
# About 3% of the time, latitude has small values near zero,
# outside Tanzania, so we'll treat these values like zero.
X['latitude'] = X['latitude'].replace(-2e-08, 0)
# When columns have zeros and shouldn't, they are like null values.
# So we will replace the zeros with nulls, and impute missing values later.
# Also create a "missing indicator" column, because the fact that
# values are missing may be a predictive signal.
cols_with_zeros = ['longitude', 'latitude', 'construction_year',
'gps_height', 'population']
for col in cols_with_zeros:
X[col] = X[col].replace(0, np.nan)
X[col+'_MISSING'] = X[col].isnull()
# Drop duplicate columns
duplicates = ['quantity_group', 'payment_type']
X = X.drop(columns=duplicates)
# Drop recorded_by (never varies) and id (always varies, random)
unusable_variance = ['recorded_by', 'id']
X = X.drop(columns=unusable_variance)
# Convert date_recorded to datetime
X['date_recorded'] = pd.to_datetime(X['date_recorded'], infer_datetime_format=True)
# Extract components from date_recorded, then drop the original column
X['year_recorded'] = X['date_recorded'].dt.year
X['month_recorded'] = X['date_recorded'].dt.month
X['day_recorded'] = X['date_recorded'].dt.day
X = X.drop(columns='date_recorded')
# Engineer feature: how many years from construction_year to date_recorded
X['years'] = X['year_recorded'] - X['construction_year']
X['years_MISSING'] = X['years'].isnull()
# return the wrangled dataframe
return X
train = wrangle(train)
val = wrangle(val)
test = wrangle(test)
# Arrange data into X features matrix and y target vector
target = 'status_group'
X_train = train.drop(columns=target)
y_train = train[target]
X_val = val.drop(columns=target)
y_val = val[target]
X_test = test
import category_encoders as ce
from sklearn.impute import SimpleImputer
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='median'),
RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)
)
# Fit on train, score on val
pipeline.fit(X_train, y_train)
print('Validation Accuracy', pipeline.score(X_val, y_val))
```
# Get permutation importances for model interpretation and feature selection
## Overview
Default Feature Importances are fast, but Permutation Importances may be more accurate.
These links go deeper with explanations and examples:
- Permutation Importances
- [Kaggle / Dan Becker: Machine Learning Explainability](https://www.kaggle.com/dansbecker/permutation-importance)
- [Christoph Molnar: Interpretable Machine Learning](https://christophm.github.io/interpretable-ml-book/feature-importance.html)
- (Default) Feature Importances
- [Ando Saabas: Selecting good features, Part 3, Random Forests](https://blog.datadive.net/selecting-good-features-part-iii-random-forests/)
- [Terence Parr, et al: Beware Default Random Forest Importances](https://explained.ai/rf-importance/index.html)
There are three types of feature importances:
### 1. (Default) Feature Importances
Fastest, good for first estimates, but be aware:
>**When the dataset has two (or more) correlated features, then from the point of view of the model, any of these correlated features can be used as the predictor, with no concrete preference of one over the others.** But once one of them is used, the importance of others is significantly reduced since effectively the impurity they can remove is already removed by the first feature. As a consequence, they will have a lower reported importance. This is not an issue when we want to use feature selection to reduce overfitting, since it makes sense to remove features that are mostly duplicated by other features. But when interpreting the data, it can lead to the incorrect conclusion that one of the variables is a strong predictor while the others in the same group are unimportant, while actually they are very close in terms of their relationship with the response variable. — [Selecting good features – Part III: random forests](https://blog.datadive.net/selecting-good-features-part-iii-random-forests/)
> **The scikit-learn Random Forest feature importance ... tends to inflate the importance of continuous or high-cardinality categorical variables.** ... Breiman and Cutler, the inventors of Random Forests, indicate that this method of “adding up the gini decreases for each individual variable over all trees in the forest gives a **fast** variable importance that is often very consistent with the permutation importance measure.” — [Beware Default Random Forest Importances](https://explained.ai/rf-importance/index.html)
```
# Get feature importances
rf = pipeline.named_steps['randomforestclassifier']
importances = pd.Series(rf.feature_importances_, X_train.columns)
# Plot feature importances
%matplotlib inline
import matplotlib.pyplot as plt
n = 20
plt.figure(figsize=(10,n/2))
plt.title(f'Top {n} features')
importances.sort_values()[-n:].plot.barh(color='grey');
```
### 2. Drop-Column Importance
The best in theory, but too slow in practice
```
column = 'wpt_name'
# Fit without column
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='median'),
RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)
)
pipeline.fit(X_train.drop(columns=column), y_train)
score_without = pipeline.score(X_val.drop(columns=column), y_val)
print(f'Validation Accuracy without {column}: {score_without}')
# Fit with column
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='median'),
RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)
)
pipeline.fit(X_train, y_train)
score_with = pipeline.score(X_val, y_val)
print(f'Validation Accuracy with {column}: {score_with}')
# Compare the error with & without column
print(f'Drop-Column Importance for {column}: {score_with - score_without}')
```
### 3. Permutation Importance
Permutation Importance is a good compromise between Feature Importance based on impurity reduction (which is the fastest) and Drop Column Importance (which is the "best.")
[The ELI5 library documentation explains,](https://eli5.readthedocs.io/en/latest/blackbox/permutation_importance.html)
> Importance can be measured by looking at how much the score (accuracy, F1, R^2, etc. - any score we’re interested in) decreases when a feature is not available.
>
> To do that one can remove feature from the dataset, re-train the estimator and check the score. But it requires re-training an estimator for each feature, which can be computationally intensive. ...
>
>To avoid re-training the estimator we can remove a feature only from the test part of the dataset, and compute score without using this feature. It doesn’t work as-is, because estimators expect feature to be present. So instead of removing a feature we can replace it with random noise - feature column is still there, but it no longer contains useful information. This method works if noise is drawn from the same distribution as original feature values (as otherwise estimator may fail). The simplest way to get such noise is to shuffle values for a feature, i.e. use other examples’ feature values - this is how permutation importance is computed.
>
>The method is most suitable for computing feature importances when a number of columns (features) is not huge; it can be resource-intensive otherwise.
### Do-It-Yourself way, for intuition
```
#lets see how permutation works first
nevi_array = [1,2,3,4,5]
nevi_permuted = np.random.permutation(nevi_array)
nevi_permuted
#BEFORE : sequence of the feature to be permuted
feature = 'quantity'
X_val[feature].head()
#BEFORE: distribution
X_val[feature].value_counts()
#PERMUTE
X_val_permuted = X_val.copy()
X_val_permuted[feature] =np.random.permutation(X_val[feature])
#AFTER : sequence of the feature to be permuted
feature = 'quantity'
X_val_permuted[feature].head()
#AFTER: distribution
X_val_permuted[feature].value_counts()
#get the permutation importance
X_val_permuted[feature] =np.random.permutation(X_val[feature])
score_permuted = pipeline.score(X_val_permuted, y_val)
print(f'Validation Accuracy with {feature}: {score_with}')
print(f'Validation Accuracy with {feature} permuted: {score_permuted}')
print(f'Permutation Importance: {score_with - score_permuted}')
feature = 'wpt_name'
X_val_permuted=X_val.copy()
X_val_permuted[feature] = np.random.permutation(X_val[feature])
score_permuted = pipeline.score(X_val_permuted, y_val)
print(f'Validation Accuracy with {feature}: {score_with}')
print(f'Validation Accuracy with {feature} permuted: {score_permuted}')
print(f'Permutation Importance: {score_with - score_permuted}')
X_val[feature]
```
### With eli5 library
For more documentation on using this library, see:
- [eli5.sklearn.PermutationImportance](https://eli5.readthedocs.io/en/latest/autodocs/sklearn.html#eli5.sklearn.permutation_importance.PermutationImportance)
- [eli5.show_weights](https://eli5.readthedocs.io/en/latest/autodocs/eli5.html#eli5.show_weights)
- [scikit-learn user guide, `scoring` parameter](https://scikit-learn.org/stable/modules/model_evaluation.html#the-scoring-parameter-defining-model-evaluation-rules)
eli5 doesn't work with pipelines.
```
# Ignore warnings
transformers = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='median')
)
X_train_transformed = transformers.fit_transform(X_train)
X_val_transformed = transformers.transform(X_val)
model = RandomForestClassifier(n_estimators=50, random_state=42, n_jobs=-1)
model.fit(X_train_transformed, y_train)
import eli5
from eli5.sklearn import PermutationImportance
permuter = PermutationImportance(
model,
scoring='accuracy',
n_iter=5,
random_state=42
)
permuter.fit(X_val_transformed,y_val)
feature_names = X_val.columns.to_list()
pd.Series(permuter.feature_importances_, feature_names).sort_values(ascending=False)
eli5.show_weights(
permuter,
top=None,
feature_names=feature_names
)
```
### We can use importances for feature selection
For example, we can remove features with zero importance. The model trains faster and the score does not decrease.
```
print('Shape before removing feature ', X_train.shape)
#remove features with feature importance <0
minimum_importance = 0
mask=permuter.feature_importances_ > minimum_importance
features = X_train.columns[mask]
X_train=X_train[features]
print('Shape AFTER removing feature ', X_train.shape)
X_val=X_val[features]
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='mean'),
RandomForestClassifier(n_estimators=50, random_state=42, n_jobs=-1)
)
#fit on train, score on val
pipeline.fit(X_train, y_train)
print('Validation accuracy', pipeline.score(X_val, y_val))
```
# Use xgboost for gradient boosting
## Overview
In the Random Forest lesson, you learned this advice:
#### Try Tree Ensembles when you do machine learning with labeled, tabular data
- "Tree Ensembles" means Random Forest or **Gradient Boosting** models.
- [Tree Ensembles often have the best predictive accuracy](https://arxiv.org/abs/1708.05070) with labeled, tabular data.
- Why? Because trees can fit non-linear, non-[monotonic](https://en.wikipedia.org/wiki/Monotonic_function) relationships, and [interactions](https://christophm.github.io/interpretable-ml-book/interaction.html) between features.
- A single decision tree, grown to unlimited depth, will [overfit](http://www.r2d3.us/visual-intro-to-machine-learning-part-1/). We solve this problem by ensembling trees, with bagging (Random Forest) or **[boosting](https://www.youtube.com/watch?v=GM3CDQfQ4sw)** (Gradient Boosting).
- Random Forest's advantage: may be less sensitive to hyperparameters. **Gradient Boosting's advantage:** may get better predictive accuracy.
Like Random Forest, Gradient Boosting uses ensembles of trees. But the details of the ensembling technique are different:
### Understand the difference between boosting & bagging
Boosting (used by Gradient Boosting) is different than Bagging (used by Random Forests).
Here's an excerpt from [_An Introduction to Statistical Learning_](http://www-bcf.usc.edu/~gareth/ISL/ISLR%20Seventh%20Printing.pdf) Chapter 8.2.3, Boosting:
>Recall that bagging involves creating multiple copies of the original training data set using the bootstrap, fitting a separate decision tree to each copy, and then combining all of the trees in order to create a single predictive model.
>
>**Boosting works in a similar way, except that the trees are grown _sequentially_: each tree is grown using information from previously grown trees.**
>
>Unlike fitting a single large decision tree to the data, which amounts to _fitting the data hard_ and potentially overfitting, the boosting approach instead _learns slowly._ Given the current model, we fit a decision tree to the residuals from the model.
>
>We then add this new decision tree into the fitted function in order to update the residuals. Each of these trees can be rather small, with just a few terminal nodes. **By fitting small trees to the residuals, we slowly improve fˆ in areas where it does not perform well.**
>
>Note that in boosting, unlike in bagging, the construction of each tree depends strongly on the trees that have already been grown.
This high-level overview is all you need to know for now. If you want to go deeper, we recommend you watch the StatQuest videos on gradient boosting!
Let's write some code. We have lots of options for which libraries to use:
#### Python libraries for Gradient Boosting
- [scikit-learn Gradient Tree Boosting](https://scikit-learn.org/stable/modules/ensemble.html#gradient-boosting) — slower than other libraries, but [the new version may be better](https://twitter.com/amuellerml/status/1129443826945396737)
- Anaconda: already installed
- Google Colab: already installed
- [xgboost](https://xgboost.readthedocs.io/en/latest/) — can accept missing values and enforce [monotonic constraints](https://xiaoxiaowang87.github.io/monotonicity_constraint/)
- Anaconda, Mac/Linux: `conda install -c conda-forge xgboost`
- Windows: `conda install -c anaconda py-xgboost`
- Google Colab: already installed
- [LightGBM](https://lightgbm.readthedocs.io/en/latest/) — can accept missing values and enforce [monotonic constraints](https://blog.datadive.net/monotonicity-constraints-in-machine-learning/)
- Anaconda: `conda install -c conda-forge lightgbm`
- Google Colab: already installed
- [CatBoost](https://catboost.ai/) — can accept missing values and use [categorical features](https://catboost.ai/docs/concepts/algorithm-main-stages_cat-to-numberic.html) without preprocessing
- Anaconda: `conda install -c conda-forge catboost`
- Google Colab: `pip install catboost`
In this lesson, you'll use a new library, xgboost — But it has an API that's almost the same as scikit-learn, so it won't be a hard adjustment!
#### [XGBoost Python API Reference: Scikit-Learn API](https://xgboost.readthedocs.io/en/latest/python/python_api.html#module-xgboost.sklearn)
```
from xgboost import XGBClassifier
pipeline = make_pipeline(
ce.OrdinalEncoder(),
XGBClassifier(n_estimators=100, random_state=42, n_jobs=-1)
)
pipeline.fit(X_train, y_train)
from sklearn.metrics import accuracy_score
y_pred=pipeline.predict(X_val)
print('Validation score', accuracy_score(y_val, y_pred))
```
#### [Avoid Overfitting By Early Stopping With XGBoost In Python](https://machinelearningmastery.com/avoid-overfitting-by-early-stopping-with-xgboost-in-python/)
Why is early stopping better than a For loop, or GridSearchCV, to optimize `n_estimators`?
With early stopping, if `n_iterations` is our number of iterations, then we fit `n_iterations` decision trees.
With a for loop, or GridSearchCV, we'd fit `sum(range(1,n_rounds+1))` trees.
But it doesn't work well with pipelines. You may need to re-run multiple times with different values of other parameters such as `max_depth` and `learning_rate`.
#### XGBoost parameters
- [Notes on parameter tuning](https://xgboost.readthedocs.io/en/latest/tutorials/param_tuning.html)
- [Parameters documentation](https://xgboost.readthedocs.io/en/latest/parameter.html)
```
encoder = ce.OrdinalEncoder()
X_train_encoded = encoder.fit_transform(X_train)
X_val_encoded = encoder.transform(X_val)
model = XGBClassifier(
n_estimators=1000, # <= 1000 trees, depend on early stopping
max_depth=7, # try deeper trees because of high cardinality categoricals
learning_rate=0.5, # try higher learning rate
n_jobs=-1
)
eval_set = [(X_train_encoded, y_train),
(X_val_encoded, y_val)]
model.fit(X_train_encoded, y_train,
eval_set=eval_set,
eval_metric='merror',
early_stopping_rounds=50) # Stop if the score hasn't improved in 50 rounds
results = model.evals_result()
train_error = results['validation_0']['merror']
val_error = results['validation_1']['merror']
epoch = list(range(1, len(train_error)+1))
plt.plot(epoch, train_error, label='Train')
plt.plot(epoch, val_error, label='Validation')
plt.ylabel('Classification Error')
plt.xlabel('Model Complexity (n_estimators)')
plt.title('Validation Curve for this XGBoost model')
plt.ylim((0.10, 0.25)) # Zoom in
plt.legend();
```
### Try adjusting these hyperparameters
#### Random Forest
- class_weight (for imbalanced classes)
- max_depth (usually high, can try decreasing)
- n_estimators (too low underfits, too high wastes time)
- min_samples_leaf (increase if overfitting)
- max_features (decrease for more diverse trees)
#### Xgboost
- scale_pos_weight (for imbalanced classes)
- max_depth (usually low, can try increasing)
- n_estimators (too low underfits, too high wastes time/overfits) — Use Early Stopping!
- learning_rate (too low underfits, too high overfits)
For more ideas, see [Notes on Parameter Tuning](https://xgboost.readthedocs.io/en/latest/tutorials/param_tuning.html) and [DART booster](https://xgboost.readthedocs.io/en/latest/tutorials/dart.html).
## Challenge
You will use your portfolio project dataset for all assignments this sprint. Complete these tasks for your project, and document your work.
- Continue to clean and explore your data. Make exploratory visualizations.
- Fit a model. Does it beat your baseline?
- Try xgboost.
- Get your model's permutation importances.
You should try to complete an initial model today, because the rest of the week, we're making model interpretation visualizations.
But, if you aren't ready to try xgboost and permutation importances with your dataset today, you can practice with another dataset instead. You may choose any dataset you've worked with previously.
| github_jupyter |
<h1><center>Introductory Data Analysis Workflow</center></h1>

https://xkcd.com/2054
# An example machine learning notebook
* Original Notebook by [Randal S. Olson](http://www.randalolson.com/)
* Supported by [Jason H. Moore](http://www.epistasis.org/)
* [University of Pennsylvania Institute for Bioinformatics](http://upibi.org/)
* Adapted for LU Py-Sem 2018 by [Valdis Saulespurens](valdis.s.coding@gmail.com)
**You can also [execute the code in this notebook on Binder](https://mybinder.org/v2/gh/ValRCS/RigaComm_DataAnalysis/master) - no local installation required.**
```
# text 17.04.2019
import datetime
print(datetime.datetime.now())
print('hello')
```
## Table of contents
1. [Introduction](#Introduction)
2. [License](#License)
3. [Required libraries](#Required-libraries)
4. [The problem domain](#The-problem-domain)
5. [Step 1: Answering the question](#Step-1:-Answering-the-question)
6. [Step 2: Checking the data](#Step-2:-Checking-the-data)
7. [Step 3: Tidying the data](#Step-3:-Tidying-the-data)
- [Bonus: Testing our data](#Bonus:-Testing-our-data)
8. [Step 4: Exploratory analysis](#Step-4:-Exploratory-analysis)
9. [Step 5: Classification](#Step-5:-Classification)
- [Cross-validation](#Cross-validation)
- [Parameter tuning](#Parameter-tuning)
10. [Step 6: Reproducibility](#Step-6:-Reproducibility)
11. [Conclusions](#Conclusions)
12. [Further reading](#Further-reading)
13. [Acknowledgements](#Acknowledgements)
## Introduction
[[ go back to the top ]](#Table-of-contents)
In the time it took you to read this sentence, terabytes of data have been collectively generated across the world — more data than any of us could ever hope to process, much less make sense of, on the machines we're using to read this notebook.
In response to this massive influx of data, the field of Data Science has come to the forefront in the past decade. Cobbled together by people from a diverse array of fields — statistics, physics, computer science, design, and many more — the field of Data Science represents our collective desire to understand and harness the abundance of data around us to build a better world.
In this notebook, I'm going to go over a basic Python data analysis pipeline from start to finish to show you what a typical data science workflow looks like.
In addition to providing code examples, I also hope to imbue in you a sense of good practices so you can be a more effective — and more collaborative — data scientist.
I will be following along with the data analysis checklist from [The Elements of Data Analytic Style](https://leanpub.com/datastyle), which I strongly recommend reading as a free and quick guidebook to performing outstanding data analysis.
**This notebook is intended to be a public resource. As such, if you see any glaring inaccuracies or if a critical topic is missing, please feel free to point it out or (preferably) submit a pull request to improve the notebook.**
## License
[[ go back to the top ]](#Table-of-contents)
Please see the [repository README file](https://github.com/rhiever/Data-Analysis-and-Machine-Learning-Projects#license) for the licenses and usage terms for the instructional material and code in this notebook. In general, I have licensed this material so that it is as widely usable and shareable as possible.
## Required libraries
[[ go back to the top ]](#Table-of-contents)
If you don't have Python on your computer, you can use the [Anaconda Python distribution](http://continuum.io/downloads) to install most of the Python packages you need. Anaconda provides a simple double-click installer for your convenience.
This notebook uses several Python packages that come standard with the Anaconda Python distribution. The primary libraries that we'll be using are:
* **NumPy**: Provides a fast numerical array structure and helper functions.
* **pandas**: Provides a DataFrame structure to store data in memory and work with it easily and efficiently.
* **scikit-learn**: The essential Machine Learning package in Python.
* **matplotlib**: Basic plotting library in Python; most other Python plotting libraries are built on top of it.
* **Seaborn**: Advanced statistical plotting library.
* **watermark**: A Jupyter Notebook extension for printing timestamps, version numbers, and hardware information.
**Note:** I will not be providing support for people trying to run this notebook outside of the Anaconda Python distribution.
## The problem domain
[[ go back to the top ]](#Table-of-contents)
For the purposes of this exercise, let's pretend we're working for a startup that just got funded to create a smartphone app that automatically identifies species of flowers from pictures taken on the smartphone. We're working with a moderately-sized team of data scientists and will be building part of the data analysis pipeline for this app.
We've been tasked by our company's Head of Data Science to create a demo machine learning model that takes four measurements from the flowers (sepal length, sepal width, petal length, and petal width) and identifies the species based on those measurements alone.
<img src="img/petal_sepal.jpg" />
We've been given a [data set](https://github.com/ValRCS/RCS_Data_Analysis_Python/blob/master/data/iris-data.csv) from our field researchers to develop the demo, which only includes measurements for three types of *Iris* flowers:
### *Iris setosa*
<img src="img/iris_setosa.jpg" />
### *Iris versicolor*
<img src="img/iris_versicolor.jpg" />
### *Iris virginica*
<img src="img/iris_virginica.jpg" />
The four measurements we're using currently come from hand-measurements by the field researchers, but they will be automatically measured by an image processing model in the future.
**Note:** The data set we're working with is the famous [*Iris* data set](https://archive.ics.uci.edu/ml/datasets/Iris) — included with this notebook — which I have modified slightly for demonstration purposes.
## Step 1: Answering the question
[[ go back to the top ]](#Table-of-contents)
The first step to any data analysis project is to define the question or problem we're looking to solve, and to define a measure (or set of measures) for our success at solving that task. The data analysis checklist has us answer a handful of questions to accomplish that, so let's work through those questions.
>Did you specify the type of data analytic question (e.g. exploration, association causality) before touching the data?
We're trying to classify the species (i.e., class) of the flower based on four measurements that we're provided: sepal length, sepal width, petal length, and petal width.
Petal - ziedlapiņa, sepal - arī ziedlapiņa

>Did you define the metric for success before beginning?
Let's do that now. Since we're performing classification, we can use [accuracy](https://en.wikipedia.org/wiki/Accuracy_and_precision) — the fraction of correctly classified flowers — to quantify how well our model is performing. Our company's Head of Data has told us that we should achieve at least 90% accuracy.
>Did you understand the context for the question and the scientific or business application?
We're building part of a data analysis pipeline for a smartphone app that will be able to classify the species of flowers from pictures taken on the smartphone. In the future, this pipeline will be connected to another pipeline that automatically measures from pictures the traits we're using to perform this classification.
>Did you record the experimental design?
Our company's Head of Data has told us that the field researchers are hand-measuring 50 randomly-sampled flowers of each species using a standardized methodology. The field researchers take pictures of each flower they sample from pre-defined angles so the measurements and species can be confirmed by the other field researchers at a later point. At the end of each day, the data is compiled and stored on a private company GitHub repository.
>Did you consider whether the question could be answered with the available data?
The data set we currently have is only for three types of *Iris* flowers. The model built off of this data set will only work for those *Iris* flowers, so we will need more data to create a general flower classifier.
<hr />
Notice that we've spent a fair amount of time working on the problem without writing a line of code or even looking at the data.
**Thinking about and documenting the problem we're working on is an important step to performing effective data analysis that often goes overlooked.** Don't skip it.
## Step 2: Checking the data
[[ go back to the top ]](#Table-of-contents)
The next step is to look at the data we're working with. Even curated data sets from the government can have errors in them, and it's vital that we spot these errors before investing too much time in our analysis.
Generally, we're looking to answer the following questions:
* Is there anything wrong with the data?
* Are there any quirks with the data?
* Do I need to fix or remove any of the data?
Let's start by reading the data into a pandas DataFrame.
```
import pandas as pd
iris_data = pd.read_csv('../data/iris-data.csv')
# Resources for loading data from nonlocal sources
# Pandas Can generally handle most common formats
# https://pandas.pydata.org/pandas-docs/stable/io.html
# SQL https://stackoverflow.com/questions/39149243/how-do-i-connect-to-a-sql-server-database-with-python
# NoSQL MongoDB https://realpython.com/introduction-to-mongodb-and-python/
# Apache Hadoop: https://dzone.com/articles/how-to-get-hadoop-data-into-a-python-model
# Apache Spark: https://www.datacamp.com/community/tutorials/apache-spark-python
# Data Scraping / Crawling libraries : https://elitedatascience.com/python-web-scraping-libraries Big Topic in itself
# Most data resources have some form of Python API / Library
iris_data.head()
```
We're in luck! The data seems to be in a usable format.
The first row in the data file defines the column headers, and the headers are descriptive enough for us to understand what each column represents. The headers even give us the units that the measurements were recorded in, just in case we needed to know at a later point in the project.
Each row following the first row represents an entry for a flower: four measurements and one class, which tells us the species of the flower.
**One of the first things we should look for is missing data.** Thankfully, the field researchers already told us that they put a 'NA' into the spreadsheet when they were missing a measurement.
We can tell pandas to automatically identify missing values if it knows our missing value marker.
```
iris_data.shape
iris_data.info()
iris_data.describe()
iris_data = pd.read_csv('../data/iris-data.csv', na_values=['NA', 'N/A'])
```
Voilà! Now pandas knows to treat rows with 'NA' as missing values.
Next, it's always a good idea to look at the distribution of our data — especially the outliers.
Let's start by printing out some summary statistics about the data set.
```
iris_data.describe()
```
We can see several useful values from this table. For example, we see that five `petal_width_cm` entries are missing.
If you ask me, though, tables like this are rarely useful unless we know that our data should fall in a particular range. It's usually better to visualize the data in some way. Visualization makes outliers and errors immediately stand out, whereas they might go unnoticed in a large table of numbers.
Since we know we're going to be plotting in this section, let's set up the notebook so we can plot inside of it.
```
# This line tells the notebook to show plots inside of the notebook
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sb
```
Next, let's create a **scatterplot matrix**. Scatterplot matrices plot the distribution of each column along the diagonal, and then plot a scatterplot matrix for the combination of each variable. They make for an efficient tool to look for errors in our data.
We can even have the plotting package color each entry by its class to look for trends within the classes.
```
# We have to temporarily drop the rows with 'NA' values
# because the Seaborn plotting function does not know
# what to do with them
sb.pairplot(iris_data.dropna(), hue='class')
```
From the scatterplot matrix, we can already see some issues with the data set:
1. There are five classes when there should only be three, meaning there were some coding errors.
2. There are some clear outliers in the measurements that may be erroneous: one `sepal_width_cm` entry for `Iris-setosa` falls well outside its normal range, and several `sepal_length_cm` entries for `Iris-versicolor` are near-zero for some reason.
3. We had to drop those rows with missing values.
In all of these cases, we need to figure out what to do with the erroneous data. Which takes us to the next step...
## Step 3: Tidying the data
### GIGO principle
[[ go back to the top ]](#Table-of-contents)
Now that we've identified several errors in the data set, we need to fix them before we proceed with the analysis.
Let's walk through the issues one-by-one.
>There are five classes when there should only be three, meaning there were some coding errors.
After talking with the field researchers, it sounds like one of them forgot to add `Iris-` before their `Iris-versicolor` entries. The other extraneous class, `Iris-setossa`, was simply a typo that they forgot to fix.
Let's use the DataFrame to fix these errors.
```
iris_data['class'].unique()
# Copy and Replace
iris_data.loc[iris_data['class'] == 'versicolor', 'class'] = 'Iris-versicolor'
iris_data['class'].unique()
# So we take a row where a specific column('class' here) matches our bad values
# and change them to good values
iris_data.loc[iris_data['class'] == 'Iris-setossa', 'class'] = 'Iris-setosa'
iris_data['class'].unique()
iris_data.tail()
iris_data[98:103]
```
Much better! Now we only have three class types. Imagine how embarrassing it would've been to create a model that used the wrong classes.
>There are some clear outliers in the measurements that may be erroneous: one `sepal_width_cm` entry for `Iris-setosa` falls well outside its normal range, and several `sepal_length_cm` entries for `Iris-versicolor` are near-zero for some reason.
Fixing outliers can be tricky business. It's rarely clear whether the outlier was caused by measurement error, recording the data in improper units, or if the outlier is a real anomaly. For that reason, we should be judicious when working with outliers: if we decide to exclude any data, we need to make sure to document what data we excluded and provide solid reasoning for excluding that data. (i.e., "This data didn't fit my hypothesis" will not stand peer review.)
In the case of the one anomalous entry for `Iris-setosa`, let's say our field researchers know that it's impossible for `Iris-setosa` to have a sepal width below 2.5 cm. Clearly this entry was made in error, and we're better off just scrapping the entry than spending hours finding out what happened.
```
smallpetals = iris_data.loc[(iris_data['sepal_width_cm'] < 2.5) & (iris_data['class'] == 'Iris-setosa')]
smallpetals
iris_data.loc[iris_data['class'] == 'Iris-setosa', 'sepal_width_cm'].hist()
# This line drops any 'Iris-setosa' rows with a separal width less than 2.5 cm
# Let's go over this command in class
iris_data = iris_data.loc[(iris_data['class'] != 'Iris-setosa') | (iris_data['sepal_width_cm'] >= 2.5)]
iris_data.loc[iris_data['class'] == 'Iris-setosa', 'sepal_width_cm'].hist()
```
Excellent! Now all of our `Iris-setosa` rows have a sepal width greater than 2.5.
The next data issue to address is the several near-zero sepal lengths for the `Iris-versicolor` rows. Let's take a look at those rows.
```
iris_data.loc[(iris_data['class'] == 'Iris-versicolor') &
(iris_data['sepal_length_cm'] < 1.0)]
```
How about that? All of these near-zero `sepal_length_cm` entries seem to be off by two orders of magnitude, as if they had been recorded in meters instead of centimeters.
After some brief correspondence with the field researchers, we find that one of them forgot to convert those measurements to centimeters. Let's do that for them.
```
iris_data.loc[iris_data['class'] == 'Iris-versicolor', 'sepal_length_cm'].hist()
iris_data['sepal_length_cm'].hist()
# Here we fix the wrong units
iris_data.loc[(iris_data['class'] == 'Iris-versicolor') &
(iris_data['sepal_length_cm'] < 1.0),
'sepal_length_cm'] *= 100.0
iris_data.loc[iris_data['class'] == 'Iris-versicolor', 'sepal_length_cm'].hist()
;
iris_data['sepal_length_cm'].hist()
```
Phew! Good thing we fixed those outliers. They could've really thrown our analysis off.
>We had to drop those rows with missing values.
Let's take a look at the rows with missing values:
```
iris_data.loc[(iris_data['sepal_length_cm'].isnull()) |
(iris_data['sepal_width_cm'].isnull()) |
(iris_data['petal_length_cm'].isnull()) |
(iris_data['petal_width_cm'].isnull())]
```
It's not ideal that we had to drop those rows, especially considering they're all `Iris-setosa` entries. Since it seems like the missing data is systematic — all of the missing values are in the same column for the same *Iris* type — this error could potentially bias our analysis.
One way to deal with missing data is **mean imputation**: If we know that the values for a measurement fall in a certain range, we can fill in empty values with the average of that measurement.
Let's see if we can do that here.
```
iris_data.loc[iris_data['class'] == 'Iris-setosa', 'petal_width_cm'].hist()
```
Most of the petal widths for `Iris-setosa` fall within the 0.2-0.3 range, so let's fill in these entries with the average measured petal width.
```
iris_data.loc[iris_data['class'] == 'Iris-setosa', 'petal_width_cm'].mean()
average_petal_width = iris_data.loc[iris_data['class'] == 'Iris-setosa', 'petal_width_cm'].mean()
print(average_petal_width)
iris_data.loc[(iris_data['class'] == 'Iris-setosa') &
(iris_data['petal_width_cm'].isnull()),
'petal_width_cm'] = average_petal_width
iris_data.loc[(iris_data['class'] == 'Iris-setosa') &
(iris_data['petal_width_cm'] == average_petal_width)]
iris_data.loc[(iris_data['sepal_length_cm'].isnull()) |
(iris_data['sepal_width_cm'].isnull()) |
(iris_data['petal_length_cm'].isnull()) |
(iris_data['petal_width_cm'].isnull())]
```
Great! Now we've recovered those rows and no longer have missing data in our data set.
**Note:** If you don't feel comfortable imputing your data, you can drop all rows with missing data with the `dropna()` call:
iris_data.dropna(inplace=True)
After all this hard work, we don't want to repeat this process every time we work with the data set. Let's save the tidied data file *as a separate file* and work directly with that data file from now on.
```
iris_data.to_json('../data/iris-clean.json')
iris_data.to_csv('../data/iris-data-clean.csv', index=False)
cleanedframe = iris_data.dropna()
iris_data_clean = pd.read_csv('../data/iris-data-clean.csv')
```
Now, let's take a look at the scatterplot matrix now that we've tidied the data.
```
myplot = sb.pairplot(iris_data_clean, hue='class')
myplot.savefig('irises.png')
import scipy.stats as stats
iris_data = pd.read_csv('../data/iris-data.csv')
iris_data.columns.unique()
stats.entropy(iris_data_clean['sepal_length_cm'])
iris_data.columns[:-1]
# we go through list of column names except last one and get entropy
# for data (without missing values) in each column
for col in iris_data.columns[:-1]:
print("Entropy for: ", col, stats.entropy(iris_data[col].dropna()))
```
Of course, I purposely inserted numerous errors into this data set to demonstrate some of the many possible scenarios you may face while tidying your data.
The general takeaways here should be:
* Make sure your data is encoded properly
* Make sure your data falls within the expected range, and use domain knowledge whenever possible to define that expected range
* Deal with missing data in one way or another: replace it if you can or drop it
* Never tidy your data manually because that is not easily reproducible
* Use code as a record of how you tidied your data
* Plot everything you can about the data at this stage of the analysis so you can *visually* confirm everything looks correct
## Bonus: Testing our data
[[ go back to the top ]](#Table-of-contents)
At SciPy 2015, I was exposed to a great idea: We should test our data. Just how we use unit tests to verify our expectations from code, we can similarly set up unit tests to verify our expectations about a data set.
We can quickly test our data using `assert` statements: We assert that something must be true, and if it is, then nothing happens and the notebook continues running. However, if our assertion is wrong, then the notebook stops running and brings it to our attention. For example,
```Python
assert 1 == 2
```
will raise an `AssertionError` and stop execution of the notebook because the assertion failed.
Let's test a few things that we know about our data set now.
```
# We know that we should only have three classes
assert len(iris_data_clean['class'].unique()) == 3
# We know that sepal lengths for 'Iris-versicolor' should never be below 2.5 cm
assert iris_data_clean.loc[iris_data_clean['class'] == 'Iris-versicolor', 'sepal_length_cm'].min() >= 2.5
# We know that our data set should have no missing measurements
assert len(iris_data_clean.loc[(iris_data_clean['sepal_length_cm'].isnull()) |
(iris_data_clean['sepal_width_cm'].isnull()) |
(iris_data_clean['petal_length_cm'].isnull()) |
(iris_data_clean['petal_width_cm'].isnull())]) == 0
# We know that our data set should have no missing measurements
assert len(iris_data.loc[(iris_data['sepal_length_cm'].isnull()) |
(iris_data['sepal_width_cm'].isnull()) |
(iris_data['petal_length_cm'].isnull()) |
(iris_data['petal_width_cm'].isnull())]) == 0
```
And so on. If any of these expectations are violated, then our analysis immediately stops and we have to return to the tidying stage.
### Data Cleanup & Wrangling > 80% time spent in Data Science
## Step 4: Exploratory analysis
[[ go back to the top ]](#Table-of-contents)
Now after spending entirely too much time tidying our data, we can start analyzing it!
Exploratory analysis is the step where we start delving deeper into the data set beyond the outliers and errors. We'll be looking to answer questions such as:
* How is my data distributed?
* Are there any correlations in my data?
* Are there any confounding factors that explain these correlations?
This is the stage where we plot all the data in as many ways as possible. Create many charts, but don't bother making them pretty — these charts are for internal use.
Let's return to that scatterplot matrix that we used earlier.
```
sb.pairplot(iris_data_clean)
;
```
Our data is normally distributed for the most part, which is great news if we plan on using any modeling methods that assume the data is normally distributed.
There's something strange going on with the petal measurements. Maybe it's something to do with the different `Iris` types. Let's color code the data by the class again to see if that clears things up.
```
sb.pairplot(iris_data_clean, hue='class')
;
```
Sure enough, the strange distribution of the petal measurements exist because of the different species. This is actually great news for our classification task since it means that the petal measurements will make it easy to distinguish between `Iris-setosa` and the other `Iris` types.
Distinguishing `Iris-versicolor` and `Iris-virginica` will prove more difficult given how much their measurements overlap.
There are also correlations between petal length and petal width, as well as sepal length and sepal width. The field biologists assure us that this is to be expected: Longer flower petals also tend to be wider, and the same applies for sepals.
We can also make [**violin plots**](https://en.wikipedia.org/wiki/Violin_plot) of the data to compare the measurement distributions of the classes. Violin plots contain the same information as [box plots](https://en.wikipedia.org/wiki/Box_plot), but also scales the box according to the density of the data.
```
plt.figure(figsize=(10, 10))
for column_index, column in enumerate(iris_data_clean.columns):
if column == 'class':
continue
plt.subplot(2, 2, column_index + 1)
sb.violinplot(x='class', y=column, data=iris_data_clean)
```
Enough flirting with the data. Let's get to modeling.
## Step 5: Classification
[[ go back to the top ]](#Table-of-contents)
Wow, all this work and we *still* haven't modeled the data!
As tiresome as it can be, tidying and exploring our data is a vital component to any data analysis. If we had jumped straight to the modeling step, we would have created a faulty classification model.
Remember: **Bad data leads to bad models.** Always check your data first.
<hr />
Assured that our data is now as clean as we can make it — and armed with some cursory knowledge of the distributions and relationships in our data set — it's time to make the next big step in our analysis: Splitting the data into training and testing sets.
A **training set** is a random subset of the data that we use to train our models.
A **testing set** is a random subset of the data (mutually exclusive from the training set) that we use to validate our models on unforseen data.
Especially in sparse data sets like ours, it's easy for models to **overfit** the data: The model will learn the training set so well that it won't be able to handle most of the cases it's never seen before. This is why it's important for us to build the model with the training set, but score it with the testing set.
Note that once we split the data into a training and testing set, we should treat the testing set like it no longer exists: We cannot use any information from the testing set to build our model or else we're cheating.
Let's set up our data first.
```
iris_data_clean = pd.read_csv('../data/iris-data-clean.csv')
# We're using all four measurements as inputs
# Note that scikit-learn expects each entry to be a list of values, e.g.,
# [ [val1, val2, val3],
# [val1, val2, val3],
# ... ]
# such that our input data set is represented as a list of lists
# We can extract the data in this format from pandas like this:
all_inputs = iris_data_clean[['sepal_length_cm', 'sepal_width_cm',
'petal_length_cm', 'petal_width_cm']].values
# Similarly, we can extract the class labels
all_labels = iris_data_clean['class'].values
# Make sure that you don't mix up the order of the entries
# all_inputs[5] inputs should correspond to the class in all_labels[5]
# Here's what a subset of our inputs looks like:
all_inputs[:5]
all_labels[:5]
type(all_inputs)
all_labels[:5]
type(all_labels)
```
Now our data is ready to be split.
```
from sklearn.model_selection import train_test_split
all_inputs[:3]
iris_data_clean.head(3)
all_labels[:3]
# Here we split our data into training and testing data
(training_inputs,
testing_inputs,
training_classes,
testing_classes) = train_test_split(all_inputs, all_labels, test_size=0.25, random_state=1)
training_inputs[:5]
testing_inputs[:5]
testing_classes[:5]
training_classes[:5]
```
With our data split, we can start fitting models to our data. Our company's Head of Data is all about decision tree classifiers, so let's start with one of those.
Decision tree classifiers are incredibly simple in theory. In their simplest form, decision tree classifiers ask a series of Yes/No questions about the data — each time getting closer to finding out the class of each entry — until they either classify the data set perfectly or simply can't differentiate a set of entries. Think of it like a game of [Twenty Questions](https://en.wikipedia.org/wiki/Twenty_Questions), except the computer is *much*, *much* better at it.
Here's an example decision tree classifier:
<img src="img/iris_dtc.png" />
Notice how the classifier asks Yes/No questions about the data — whether a certain feature is <= 1.75, for example — so it can differentiate the records. This is the essence of every decision tree.
The nice part about decision tree classifiers is that they are **scale-invariant**, i.e., the scale of the features does not affect their performance, unlike many Machine Learning models. In other words, it doesn't matter if our features range from 0 to 1 or 0 to 1,000; decision tree classifiers will work with them just the same.
There are several [parameters](http://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeClassifier.html) that we can tune for decision tree classifiers, but for now let's use a basic decision tree classifier.
```
from sklearn.tree import DecisionTreeClassifier
# Create the classifier
decision_tree_classifier = DecisionTreeClassifier()
# Train the classifier on the training set
decision_tree_classifier.fit(training_inputs, training_classes)
# Validate the classifier on the testing set using classification accuracy
decision_tree_classifier.score(testing_inputs, testing_classes)
150*0.25
len(testing_inputs)
37/38
from sklearn import svm
svm_classifier = svm.SVC(gamma = 'scale')
svm_classifier.fit(training_inputs, training_classes)
svm_classifier.score(testing_inputs, testing_classes)
svm_classifier = svm.SVC(gamma = 'scale')
svm_classifier.fit(training_inputs, training_classes)
svm_classifier.score(testing_inputs, testing_classes)
```
Heck yeah! Our model achieves 97% classification accuracy without much effort.
However, there's a catch: Depending on how our training and testing set was sampled, our model can achieve anywhere from 80% to 100% accuracy:
```
import matplotlib.pyplot as plt
# here we randomly split data 1000 times in differrent training and test sets
model_accuracies = []
for repetition in range(1000):
(training_inputs,
testing_inputs,
training_classes,
testing_classes) = train_test_split(all_inputs, all_labels, test_size=0.25)
decision_tree_classifier = DecisionTreeClassifier()
decision_tree_classifier.fit(training_inputs, training_classes)
classifier_accuracy = decision_tree_classifier.score(testing_inputs, testing_classes)
model_accuracies.append(classifier_accuracy)
plt.hist(model_accuracies)
;
100/38
```
It's obviously a problem that our model performs quite differently depending on the subset of the data it's trained on. This phenomenon is known as **overfitting**: The model is learning to classify the training set so well that it doesn't generalize and perform well on data it hasn't seen before.
### Cross-validation
[[ go back to the top ]](#Table-of-contents)
This problem is the main reason that most data scientists perform ***k*-fold cross-validation** on their models: Split the original data set into *k* subsets, use one of the subsets as the testing set, and the rest of the subsets are used as the training set. This process is then repeated *k* times such that each subset is used as the testing set exactly once.
10-fold cross-validation is the most common choice, so let's use that here. Performing 10-fold cross-validation on our data set looks something like this:
(each square is an entry in our data set)
```
# new text
import numpy as np
from sklearn.model_selection import StratifiedKFold
def plot_cv(cv, features, labels):
masks = []
for train, test in cv.split(features, labels):
mask = np.zeros(len(labels), dtype=bool)
mask[test] = 1
masks.append(mask)
plt.figure(figsize=(15, 15))
plt.imshow(masks, interpolation='none', cmap='gray_r')
plt.ylabel('Fold')
plt.xlabel('Row #')
plot_cv(StratifiedKFold(n_splits=10), all_inputs, all_labels)
```
You'll notice that we used **Stratified *k*-fold cross-validation** in the code above. Stratified *k*-fold keeps the class proportions the same across all of the folds, which is vital for maintaining a representative subset of our data set. (e.g., so we don't have 100% `Iris setosa` entries in one of the folds.)
We can perform 10-fold cross-validation on our model with the following code:
```
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import cross_val_score
decision_tree_classifier = DecisionTreeClassifier()
# cross_val_score returns a list of the scores, which we can visualize
# to get a reasonable estimate of our classifier's performance
cv_scores = cross_val_score(decision_tree_classifier, all_inputs, all_labels, cv=10)
plt.hist(cv_scores)
plt.title('Average score: {}'.format(np.mean(cv_scores)))
;
len(all_inputs.T[1])
print("Entropy for: ", stats.entropy(all_inputs.T[1]))
# we go through list of column names except last one and get entropy
# for data (without missing values) in each column
def printEntropy(npdata):
for i, col in enumerate(npdata.T):
print("Entropy for column:", i, stats.entropy(col))
printEntropy(all_inputs)
```
Now we have a much more consistent rating of our classifier's general classification accuracy.
### Parameter tuning
[[ go back to the top ]](#Table-of-contents)
Every Machine Learning model comes with a variety of parameters to tune, and these parameters can be vitally important to the performance of our classifier. For example, if we severely limit the depth of our decision tree classifier:
```
decision_tree_classifier = DecisionTreeClassifier(max_depth=1)
cv_scores = cross_val_score(decision_tree_classifier, all_inputs, all_labels, cv=10)
plt.hist(cv_scores)
plt.title('Average score: {}'.format(np.mean(cv_scores)))
;
```
the classification accuracy falls tremendously.
Therefore, we need to find a systematic method to discover the best parameters for our model and data set.
The most common method for model parameter tuning is **Grid Search**. The idea behind Grid Search is simple: explore a range of parameters and find the best-performing parameter combination. Focus your search on the best range of parameters, then repeat this process several times until the best parameters are discovered.
Let's tune our decision tree classifier. We'll stick to only two parameters for now, but it's possible to simultaneously explore dozens of parameters if we want.
```
from sklearn.model_selection import GridSearchCV
decision_tree_classifier = DecisionTreeClassifier()
parameter_grid = {'max_depth': [1, 2, 3, 4, 5],
'max_features': [1, 2, 3, 4]}
cross_validation = StratifiedKFold(n_splits=10)
grid_search = GridSearchCV(decision_tree_classifier,
param_grid=parameter_grid,
cv=cross_validation)
grid_search.fit(all_inputs, all_labels)
print('Best score: {}'.format(grid_search.best_score_))
print('Best parameters: {}'.format(grid_search.best_params_))
```
Now let's visualize the grid search to see how the parameters interact.
```
grid_search.cv_results_['mean_test_score']
grid_visualization = grid_search.cv_results_['mean_test_score']
grid_visualization.shape = (5, 4)
sb.heatmap(grid_visualization, cmap='Reds', annot=True)
plt.xticks(np.arange(4) + 0.5, grid_search.param_grid['max_features'])
plt.yticks(np.arange(5) + 0.5, grid_search.param_grid['max_depth'])
plt.xlabel('max_features')
plt.ylabel('max_depth')
;
```
Now we have a better sense of the parameter space: We know that we need a `max_depth` of at least 2 to allow the decision tree to make more than a one-off decision.
`max_features` doesn't really seem to make a big difference here as long as we have 2 of them, which makes sense since our data set has only 4 features and is relatively easy to classify. (Remember, one of our data set's classes was easily separable from the rest based on a single feature.)
Let's go ahead and use a broad grid search to find the best settings for a handful of parameters.
```
decision_tree_classifier = DecisionTreeClassifier()
parameter_grid = {'criterion': ['gini', 'entropy'],
'splitter': ['best', 'random'],
'max_depth': [1, 2, 3, 4, 5],
'max_features': [1, 2, 3, 4]}
cross_validation = StratifiedKFold(n_splits=10)
grid_search = GridSearchCV(decision_tree_classifier,
param_grid=parameter_grid,
cv=cross_validation)
grid_search.fit(all_inputs, all_labels)
print('Best score: {}'.format(grid_search.best_score_))
print('Best parameters: {}'.format(grid_search.best_params_))
```
Now we can take the best classifier from the Grid Search and use that:
```
decision_tree_classifier = grid_search.best_estimator_
decision_tree_classifier
```
We can even visualize the decision tree with [GraphViz](http://www.graphviz.org/) to see how it's making the classifications:
```
import sklearn.tree as tree
from sklearn.externals.six import StringIO
with open('iris_dtc.dot', 'w') as out_file:
out_file = tree.export_graphviz(decision_tree_classifier, out_file=out_file)
```
<img src="img/iris_dtc.png" />
(This classifier may look familiar from earlier in the notebook.)
Alright! We finally have our demo classifier. Let's create some visuals of its performance so we have something to show our company's Head of Data.
```
dt_scores = cross_val_score(decision_tree_classifier, all_inputs, all_labels, cv=10)
sb.boxplot(dt_scores)
sb.stripplot(dt_scores, jitter=True, color='black')
;
```
Hmmm... that's a little boring by itself though. How about we compare another classifier to see how they perform?
We already know from previous projects that Random Forest classifiers usually work better than individual decision trees. A common problem that decision trees face is that they're prone to overfitting: They complexify to the point that they classify the training set near-perfectly, but fail to generalize to data they have not seen before.
**Random Forest classifiers** work around that limitation by creating a whole bunch of decision trees (hence "forest") — each trained on random subsets of training samples (drawn with replacement) and features (drawn without replacement) — and have the decision trees work together to make a more accurate classification.
Let that be a lesson for us: **Even in Machine Learning, we get better results when we work together!**
Let's see if a Random Forest classifier works better here.
The great part about scikit-learn is that the training, testing, parameter tuning, etc. process is the same for all models, so we only need to plug in the new classifier.
```
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import RandomForestClassifier
random_forest_classifier = RandomForestClassifier()
parameter_grid = {'n_estimators': [10, 25, 50, 100],
'criterion': ['gini', 'entropy'],
'max_features': [1, 2, 3, 4]}
cross_validation = StratifiedKFold(n_splits=10)
grid_search = GridSearchCV(random_forest_classifier,
param_grid=parameter_grid,
cv=cross_validation)
grid_search.fit(all_inputs, all_labels)
print('Best score: {}'.format(grid_search.best_score_))
print('Best parameters: {}'.format(grid_search.best_params_))
grid_search.best_estimator_
```
Now we can compare their performance:
```
random_forest_classifier = grid_search.best_estimator_
rf_df = pd.DataFrame({'accuracy': cross_val_score(random_forest_classifier, all_inputs, all_labels, cv=10),
'classifier': ['Random Forest'] * 10})
dt_df = pd.DataFrame({'accuracy': cross_val_score(decision_tree_classifier, all_inputs, all_labels, cv=10),
'classifier': ['Decision Tree'] * 10})
both_df = rf_df.append(dt_df)
sb.boxplot(x='classifier', y='accuracy', data=both_df)
sb.stripplot(x='classifier', y='accuracy', data=both_df, jitter=True, color='black')
;
```
How about that? They both seem to perform about the same on this data set. This is probably because of the limitations of our data set: We have only 4 features to make the classification, and Random Forest classifiers excel when there's hundreds of possible features to look at. In other words, there wasn't much room for improvement with this data set.
## Step 6: Reproducibility
[[ go back to the top ]](#Table-of-contents)
Ensuring that our work is reproducible is the last and — arguably — most important step in any analysis. **As a rule, we shouldn't place much weight on a discovery that can't be reproduced**. As such, if our analysis isn't reproducible, we might as well not have done it.
Notebooks like this one go a long way toward making our work reproducible. Since we documented every step as we moved along, we have a written record of what we did and why we did it — both in text and code.
Beyond recording what we did, we should also document what software and hardware we used to perform our analysis. This typically goes at the top of our notebooks so our readers know what tools to use.
[Sebastian Raschka](http://sebastianraschka.com/) created a handy [notebook tool](https://github.com/rasbt/watermark) for this:
```
!pip install watermark
%load_ext watermark
pd.show_versions()
%watermark -a 'RCS_April_2019' -nmv --packages numpy,pandas,sklearn,matplotlib,seaborn
```
Finally, let's extract the core of our work from Steps 1-5 and turn it into a single pipeline.
```
%matplotlib inline
import pandas as pd
import seaborn as sb
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split, cross_val_score
# We can jump directly to working with the clean data because we saved our cleaned data set
iris_data_clean = pd.read_csv('../data/iris-data-clean.csv')
# Testing our data: Our analysis will stop here if any of these assertions are wrong
# We know that we should only have three classes
assert len(iris_data_clean['class'].unique()) == 3
# We know that sepal lengths for 'Iris-versicolor' should never be below 2.5 cm
assert iris_data_clean.loc[iris_data_clean['class'] == 'Iris-versicolor', 'sepal_length_cm'].min() >= 2.5
# We know that our data set should have no missing measurements
assert len(iris_data_clean.loc[(iris_data_clean['sepal_length_cm'].isnull()) |
(iris_data_clean['sepal_width_cm'].isnull()) |
(iris_data_clean['petal_length_cm'].isnull()) |
(iris_data_clean['petal_width_cm'].isnull())]) == 0
all_inputs = iris_data_clean[['sepal_length_cm', 'sepal_width_cm',
'petal_length_cm', 'petal_width_cm']].values
all_labels = iris_data_clean['class'].values
# This is the classifier that came out of Grid Search
random_forest_classifier = RandomForestClassifier(criterion='gini', max_features=3, n_estimators=50)
# All that's left to do now is plot the cross-validation scores
rf_classifier_scores = cross_val_score(random_forest_classifier, all_inputs, all_labels, cv=10)
sb.boxplot(rf_classifier_scores)
sb.stripplot(rf_classifier_scores, jitter=True, color='black')
# ...and show some of the predictions from the classifier
(training_inputs,
testing_inputs,
training_classes,
testing_classes) = train_test_split(all_inputs, all_labels, test_size=0.25)
random_forest_classifier.fit(training_inputs, training_classes)
for input_features, prediction, actual in zip(testing_inputs[:10],
random_forest_classifier.predict(testing_inputs[:10]),
testing_classes[:10]):
print('{}\t-->\t{}\t(Actual: {})'.format(input_features, prediction, actual))
%matplotlib inline
import pandas as pd
import seaborn as sb
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split, cross_val_score
def processData(filename):
# We can jump directly to working with the clean data because we saved our cleaned data set
iris_data_clean = pd.read_csv(filename)
# Testing our data: Our analysis will stop here if any of these assertions are wrong
# We know that we should only have three classes
assert len(iris_data_clean['class'].unique()) == 3
# We know that sepal lengths for 'Iris-versicolor' should never be below 2.5 cm
assert iris_data_clean.loc[iris_data_clean['class'] == 'Iris-versicolor', 'sepal_length_cm'].min() >= 2.5
# We know that our data set should have no missing measurements
assert len(iris_data_clean.loc[(iris_data_clean['sepal_length_cm'].isnull()) |
(iris_data_clean['sepal_width_cm'].isnull()) |
(iris_data_clean['petal_length_cm'].isnull()) |
(iris_data_clean['petal_width_cm'].isnull())]) == 0
all_inputs = iris_data_clean[['sepal_length_cm', 'sepal_width_cm',
'petal_length_cm', 'petal_width_cm']].values
all_labels = iris_data_clean['class'].values
# This is the classifier that came out of Grid Search
random_forest_classifier = RandomForestClassifier(criterion='gini', max_features=3, n_estimators=50)
# All that's left to do now is plot the cross-validation scores
rf_classifier_scores = cross_val_score(random_forest_classifier, all_inputs, all_labels, cv=10)
sb.boxplot(rf_classifier_scores)
sb.stripplot(rf_classifier_scores, jitter=True, color='black')
# ...and show some of the predictions from the classifier
(training_inputs,
testing_inputs,
training_classes,
testing_classes) = train_test_split(all_inputs, all_labels, test_size=0.25)
random_forest_classifier.fit(training_inputs, training_classes)
for input_features, prediction, actual in zip(testing_inputs[:10],
random_forest_classifier.predict(testing_inputs[:10]),
testing_classes[:10]):
print('{}\t-->\t{}\t(Actual: {})'.format(input_features, prediction, actual))
return rf_classifier_scores
myscores = processData('../data/iris-data-clean.csv')
myscores
```
There we have it: We have a complete and reproducible Machine Learning pipeline to demo to our company's Head of Data. We've met the success criteria that we set from the beginning (>90% accuracy), and our pipeline is flexible enough to handle new inputs or flowers when that data set is ready. Not bad for our first week on the job!
## Conclusions
[[ go back to the top ]](#Table-of-contents)
I hope you found this example notebook useful for your own work and learned at least one new trick by reading through it.
* [Submit an issue](https://github.com/ValRCS/LU-pysem/issues) on GitHub
* Fork the [notebook repository](https://github.com/ValRCS/LU-pysem), make the fix/addition yourself, then send over a pull request
## Further reading
[[ go back to the top ]](#Table-of-contents)
This notebook covers a broad variety of topics but skips over many of the specifics. If you're looking to dive deeper into a particular topic, here's some recommended reading.
**Data Science**: William Chen compiled a [list of free books](http://www.wzchen.com/data-science-books/) for newcomers to Data Science, ranging from the basics of R & Python to Machine Learning to interviews and advice from prominent data scientists.
**Machine Learning**: /r/MachineLearning has a useful [Wiki page](https://www.reddit.com/r/MachineLearning/wiki/index) containing links to online courses, books, data sets, etc. for Machine Learning. There's also a [curated list](https://github.com/josephmisiti/awesome-machine-learning) of Machine Learning frameworks, libraries, and software sorted by language.
**Unit testing**: Dive Into Python 3 has a [great walkthrough](http://www.diveintopython3.net/unit-testing.html) of unit testing in Python, how it works, and how it should be used
**pandas** has [several tutorials](http://pandas.pydata.org/pandas-docs/stable/tutorials.html) covering its myriad features.
**scikit-learn** has a [bunch of tutorials](http://scikit-learn.org/stable/tutorial/index.html) for those looking to learn Machine Learning in Python. Andreas Mueller's [scikit-learn workshop materials](https://github.com/amueller/scipy_2015_sklearn_tutorial) are top-notch and freely available.
**matplotlib** has many [books, videos, and tutorials](http://matplotlib.org/resources/index.html) to teach plotting in Python.
**Seaborn** has a [basic tutorial](http://stanford.edu/~mwaskom/software/seaborn/tutorial.html) covering most of the statistical plotting features.
## Acknowledgements
[[ go back to the top ]](#Table-of-contents)
Many thanks to [Andreas Mueller](http://amueller.github.io/) for some of his [examples](https://github.com/amueller/scipy_2015_sklearn_tutorial) in the Machine Learning section. I drew inspiration from several of his excellent examples.
The photo of a flower with annotations of the petal and sepal was taken by [Eric Guinther](https://commons.wikimedia.org/wiki/File:Petal-sepal.jpg).
The photos of the various *Iris* flower types were taken by [Ken Walker](http://www.signa.org/index.pl?Display+Iris-setosa+2) and [Barry Glick](http://www.signa.org/index.pl?Display+Iris-virginica+3).
## Further questions?
Feel free to contact [Valdis Saulespurens]
(email:valdis.s.coding@gmail.com)
| github_jupyter |
## Example 2: Sensitivity analysis on a NetLogo model with SALib
This notebook provides a more advanced example of interaction between NetLogo and a Python environment, using the SALib library (Herman & Usher, 2017; available through the pip package manager) to sample and analyze a suitable experimental design for a Sobol global sensitivity analysis. All files used in the example are available from the pyNetLogo repository at https://github.com/quaquel/pyNetLogo.
```
#Ensuring compliance of code with both python2 and python3
from __future__ import division, print_function
try:
from itertools import izip as zip
except ImportError: # will be 3.x series
pass
%matplotlib inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import pyNetLogo
#Import the sampling and analysis modules for a Sobol variance-based sensitivity analysis
from SALib.sample import saltelli
from SALib.analyze import sobol
```
SALib relies on a problem definition dictionary which contains the number of input parameters to sample, their names (which should here correspond to a NetLogo global variable), and the sampling bounds. Documentation for SALib can be found at https://salib.readthedocs.io/en/latest/.
```
problem = {
'num_vars': 6,
'names': ['random-seed',
'grass-regrowth-time',
'sheep-gain-from-food',
'wolf-gain-from-food',
'sheep-reproduce',
'wolf-reproduce'],
'bounds': [[1, 100000],
[20., 40.],
[2., 8.],
[16., 32.],
[2., 8.],
[2., 8.]]
}
```
We start by instantiating the wolf-sheep predation example model, specifying the _gui=False_ flag to run in headless mode.
```
netlogo = pyNetLogo.NetLogoLink(gui=False)
netlogo.load_model(r'Wolf Sheep Predation_v6.nlogo')
```
The SALib sampler will automatically generate an appropriate number of samples for Sobol analysis. To calculate first-order, second-order and total sensitivity indices, this gives a sample size of _n*(2p+2)_, where _p_ is the number of input parameters, and _n_ is a baseline sample size which should be large enough to stabilize the estimation of the indices. For this example, we use _n_ = 1000, for a total of 14000 experiments.
For more complex analyses, parallelizing the experiments can significantly improve performance. An additional notebook in the pyNetLogo repository demonstrates the use of the ipyparallel library; parallel processing for NetLogo models is also supported by the Exploratory Modeling Workbench (Kwakkel, 2017).
```
n = 1000
param_values = saltelli.sample(problem, n, calc_second_order=True)
```
The sampler generates an input array of shape (_n*(2p+2)_, _p_) with rows for each experiment and columns for each input parameter.
```
param_values.shape
```
Assuming we are interested in the mean number of sheep and wolf agents over a timeframe of 100 ticks, we first create an empty dataframe to store the results.
```
results = pd.DataFrame(columns=['Avg. sheep', 'Avg. wolves'])
```
We then simulate the model over the 14000 experiments, reading input parameters from the param_values array generated by SALib. The repeat_report command is used to track the outcomes of interest over time.
To later compare performance with the ipyparallel implementation of the analysis, we also keep track of the elapsed runtime.
```
import time
t0=time.time()
for run in range(param_values.shape[0]):
#Set the input parameters
for i, name in enumerate(problem['names']):
if name == 'random-seed':
#The NetLogo random seed requires a different syntax
netlogo.command('random-seed {}'.format(param_values[run,i]))
else:
#Otherwise, assume the input parameters are global variables
netlogo.command('set {0} {1}'.format(name, param_values[run,i]))
netlogo.command('setup')
#Run for 100 ticks and return the number of sheep and wolf agents at each time step
counts = netlogo.repeat_report(['count sheep','count wolves'], 100)
#For each run, save the mean value of the agent counts over time
results.loc[run, 'Avg. sheep'] = counts['count sheep'].values.mean()
results.loc[run, 'Avg. wolves'] = counts['count wolves'].values.mean()
elapsed=time.time()-t0 #Elapsed runtime in seconds
elapsed
```
The "to_csv" dataframe method provides a simple way of saving the results to disk.
Pandas supports several more advanced storage options, such as serialization with msgpack, or hierarchical HDF5 storage.
```
results.to_csv('Sobol_sequential.csv')
results = pd.read_csv('Sobol_sequential.csv', header=0, index_col=0)
results.head(5)
```
We can then proceed with the analysis, first using a histogram to visualize output distributions for each outcome:
```
sns.set_style('white')
sns.set_context('talk')
fig, ax = plt.subplots(1,len(results.columns), sharey=True)
for i, n in enumerate(results.columns):
ax[i].hist(results[n], 20)
ax[i].set_xlabel(n)
ax[0].set_ylabel('Counts')
fig.set_size_inches(10,4)
fig.subplots_adjust(wspace=0.1)
#plt.savefig('JASSS figures/SA - Output distribution.pdf', bbox_inches='tight')
#plt.savefig('JASSS figures/SA - Output distribution.png', dpi=300, bbox_inches='tight')
plt.show()
```
Bivariate scatter plots can be useful to visualize relationships between each input parameter and the outputs. Taking the outcome for the average sheep count as an example, we obtain the following, using the scipy library to calculate the Pearson correlation coefficient (r) for each parameter:
```
%matplotlib
import scipy
nrow=2
ncol=3
fig, ax = plt.subplots(nrow, ncol, sharey=True)
sns.set_context('talk')
y = results['Avg. sheep']
for i, a in enumerate(ax.flatten()):
x = param_values[:,i]
sns.regplot(x, y, ax=a, ci=None, color='k',scatter_kws={'alpha':0.2, 's':4, 'color':'gray'})
pearson = scipy.stats.pearsonr(x, y)
a.annotate("r: {:6.3f}".format(pearson[0]), xy=(0.15, 0.85), xycoords='axes fraction',fontsize=13)
if divmod(i,ncol)[1]>0:
a.get_yaxis().set_visible(False)
a.set_xlabel(problem['names'][i])
a.set_ylim([0,1.1*np.max(y)])
fig.set_size_inches(9,9,forward=True)
fig.subplots_adjust(wspace=0.2, hspace=0.3)
#plt.savefig('JASSS figures/SA - Scatter.pdf', bbox_inches='tight')
#plt.savefig('JASSS figures/SA - Scatter.png', dpi=300, bbox_inches='tight')
plt.show()
```
This indicates a positive relationship between the "sheep-gain-from-food" parameter and the mean sheep count, and negative relationships for the "wolf-gain-from-food" and "wolf-reproduce" parameters.
We can then use SALib to calculate first-order (S1), second-order (S2) and total (ST) Sobol indices, to estimate each input's contribution to output variance. By default, 95% confidence intervals are estimated for each index.
```
Si = sobol.analyze(problem, results['Avg. sheep'].values, calc_second_order=True, print_to_console=False)
```
As a simple example, we first select and visualize the first-order and total indices for each input, converting the dictionary returned by SALib to a dataframe.
```
Si_filter = {k:Si[k] for k in ['ST','ST_conf','S1','S1_conf']}
Si_df = pd.DataFrame(Si_filter, index=problem['names'])
Si_df
sns.set_style('white')
fig, ax = plt.subplots(1)
indices = Si_df[['S1','ST']]
err = Si_df[['S1_conf','ST_conf']]
indices.plot.bar(yerr=err.values.T,ax=ax)
fig.set_size_inches(8,4)
#plt.savefig('JASSS figures/SA - Indices.pdf', bbox_inches='tight')
#plt.savefig('JASSS figures/SA - Indices.png', dpi=300, bbox_inches='tight')
plt.show()
```
The "sheep-gain-from-food" parameter has the highest ST index, indicating that it contributes over 50% of output variance when accounting for interactions with other parameters. However, it can be noted that the confidence bounds are overly broad due to the small _n_ value used for sampling, so that a larger sample would be required for reliable results. For instance, the S1 index is estimated to be larger than ST for the "random-seed" parameter, which is an artifact of the small sample size.
We can use a more sophisticated visualization to include the second-order interactions between inputs.
```
import itertools
from math import pi
def normalize(x, xmin, xmax):
return (x-xmin)/(xmax-xmin)
def plot_circles(ax, locs, names, max_s, stats, smax, smin, fc, ec, lw,
zorder):
s = np.asarray([stats[name] for name in names])
s = 0.01 + max_s * np.sqrt(normalize(s, smin, smax))
fill = True
for loc, name, si in zip(locs, names, s):
if fc=='w':
fill=False
else:
ec='none'
x = np.cos(loc)
y = np.sin(loc)
circle = plt.Circle((x,y), radius=si, ec=ec, fc=fc, transform=ax.transData._b,
zorder=zorder, lw=lw, fill=True)
ax.add_artist(circle)
def filter(sobol_indices, names, locs, criterion, threshold):
if criterion in ['ST', 'S1', 'S2']:
data = sobol_indices[criterion]
data = np.abs(data)
data = data.flatten() # flatten in case of S2
# TODO:: remove nans
filtered = ([(name, locs[i]) for i, name in enumerate(names) if
data[i]>threshold])
filtered_names, filtered_locs = zip(*filtered)
elif criterion in ['ST_conf', 'S1_conf', 'S2_conf']:
raise NotImplementedError
else:
raise ValueError('unknown value for criterion')
return filtered_names, filtered_locs
def plot_sobol_indices(sobol_indices, criterion='ST', threshold=0.01):
'''plot sobol indices on a radial plot
Parameters
----------
sobol_indices : dict
the return from SAlib
criterion : {'ST', 'S1', 'S2', 'ST_conf', 'S1_conf', 'S2_conf'}, optional
threshold : float
only visualize variables with criterion larger than cutoff
'''
max_linewidth_s2 = 15#25*1.8
max_s_radius = 0.3
# prepare data
# use the absolute values of all the indices
#sobol_indices = {key:np.abs(stats) for key, stats in sobol_indices.items()}
# dataframe with ST and S1
sobol_stats = {key:sobol_indices[key] for key in ['ST', 'S1']}
sobol_stats = pd.DataFrame(sobol_stats, index=problem['names'])
smax = sobol_stats.max().max()
smin = sobol_stats.min().min()
# dataframe with s2
s2 = pd.DataFrame(sobol_indices['S2'], index=problem['names'],
columns=problem['names'])
s2[s2<0.0]=0. #Set negative values to 0 (artifact from small sample sizes)
s2max = s2.max().max()
s2min = s2.min().min()
names = problem['names']
n = len(names)
ticklocs = np.linspace(0, 2*pi, n+1)
locs = ticklocs[0:-1]
filtered_names, filtered_locs = filter(sobol_indices, names, locs,
criterion, threshold)
# setup figure
fig = plt.figure()
ax = fig.add_subplot(111, polar=True)
ax.grid(False)
ax.spines['polar'].set_visible(False)
ax.set_xticks(ticklocs)
ax.set_xticklabels(names)
ax.set_yticklabels([])
ax.set_ylim(ymax=1.4)
legend(ax)
# plot ST
plot_circles(ax, filtered_locs, filtered_names, max_s_radius,
sobol_stats['ST'], smax, smin, 'w', 'k', 1, 9)
# plot S1
plot_circles(ax, filtered_locs, filtered_names, max_s_radius,
sobol_stats['S1'], smax, smin, 'k', 'k', 1, 10)
# plot S2
for name1, name2 in itertools.combinations(zip(filtered_names, filtered_locs), 2):
name1, loc1 = name1
name2, loc2 = name2
weight = s2.ix[name1, name2]
lw = 0.5+max_linewidth_s2*normalize(weight, s2min, s2max)
ax.plot([loc1, loc2], [1,1], c='darkgray', lw=lw, zorder=1)
return fig
from matplotlib.legend_handler import HandlerPatch
class HandlerCircle(HandlerPatch):
def create_artists(self, legend, orig_handle,
xdescent, ydescent, width, height, fontsize, trans):
center = 0.5 * width - 0.5 * xdescent, 0.5 * height - 0.5 * ydescent
p = plt.Circle(xy=center, radius=orig_handle.radius)
self.update_prop(p, orig_handle, legend)
p.set_transform(trans)
return [p]
def legend(ax):
some_identifiers = [plt.Circle((0,0), radius=5, color='k', fill=False, lw=1),
plt.Circle((0,0), radius=5, color='k', fill=True),
plt.Line2D([0,0.5], [0,0.5], lw=8, color='darkgray')]
ax.legend(some_identifiers, ['ST', 'S1', 'S2'],
loc=(1,0.75), borderaxespad=0.1, mode='expand',
handler_map={plt.Circle: HandlerCircle()})
sns.set_style('whitegrid')
fig = plot_sobol_indices(Si, criterion='ST', threshold=0.005)
fig.set_size_inches(7,7)
#plt.savefig('JASSS figures/Figure 8 - Interactions.pdf', bbox_inches='tight')
#plt.savefig('JASSS figures/Figure 8 - Interactions.png', dpi=300, bbox_inches='tight')
plt.show()
```
In this case, the sheep-gain-from-food variable has strong interactions with the wolf-gain-from-food and sheep-reproduce inputs in particular. The size of the ST and S1 circles correspond to the normalized variable importances.
Finally, the kill_workspace() function shuts down the NetLogo instance.
```
netlogo.kill_workspace()
```
| github_jupyter |
```
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
players_time = pd.read_csv("players_time.csv")
events_time = pd.read_csv("events_time.csv")
serve_time = pd.read_csv("serve_times.csv")
players_time
events_time
pd.options.display.max_rows = None
events_time
serve_time
```
## 1. Visualize The 10 Most Slow Players
```
most_slow_Players = players_time[players_time["seconds_added_per_point"] > 0].sort_values(by="seconds_added_per_point", ascending=False).head(10)
most_slow_Players
sns.set(style="darkgrid")
plt.figure(figsize = (10,5))
ax= sns.barplot(x="seconds_added_per_point", y="player", data=most_slow_Players)
ax.set_title("TOP 10 MOST SLOW PLAYERS", fontsize=17)
plt.xlabel("Seconds", fontsize=17)
plt.ylabel("Players", fontsize=17)
plt.yticks(size=17)
plt.xticks(size=17)
plt.show()
```
## 2. Visualize The 10 Most Fast Players
```
most_fast_Players = players_time[players_time["seconds_added_per_point"] < 0].sort_values(by="seconds_added_per_point").head(10)
most_fast_Players
sns.set(style="darkgrid")
plt.figure(figsize = (10,5))
ax= sns.barplot(x="seconds_added_per_point", y="player", data=most_fast_Players)
ax.set_title("TOP 10 MOST FAST PLAYERS", fontsize=17)
plt.xlabel("Seconds", fontsize=17)
plt.ylabel("Players", fontsize=17)
plt.yticks(size=17)
plt.xticks(size=17)
plt.show()
```
## 3. Visualize The Time Of The Big 3
```
big_three_time = players_time[(players_time["player"] == "Novak Djokovic") | (players_time["player"] == "Roger Federer") | (players_time["player"] == "Rafael Nadal")]
big_three_time
sns.set(style="darkgrid")
plt.figure(figsize = (10,5))
ax= sns.barplot(x="seconds_added_per_point", y="player", data=big_three_time)
ax.set_title("TIME OF THE BIG THREE", fontsize=17)
plt.xlabel("Seconds", fontsize=17)
plt.ylabel("Players", fontsize=17)
plt.yticks(size=17)
plt.xticks(size=17)
plt.show()
```
## 4. Figure Out The Top 10 Surfaces That Take The Longest Time
```
longest_time_surfaces = events_time[events_time["seconds_added_per_point"] > 0].sort_values(by="seconds_added_per_point", ascending=False).head(10)
longest_time_surfaces
sns.set(style="darkgrid")
plt.figure(figsize = (10,5))
ax= sns.barplot(x="seconds_added_per_point", y="tournament", hue="surface", data=longest_time_surfaces)
ax.set_title("TOP 10 SURFACES THAT TAKE THE LONGEST TIME", fontsize=17)
plt.xlabel("Seconds", fontsize=17)
plt.ylabel("Tournament", fontsize=17)
plt.yticks(size=17)
plt.xticks(size=17)
plt.show()
```
## 5. Figure Out The Top 10 Surfaces That Take The Shortest Time
```
shortest_time_surfaces = events_time[events_time["seconds_added_per_point"] < 0].sort_values(by="seconds_added_per_point").head(10)
shortest_time_surfaces
sns.set(style="darkgrid")
plt.figure(figsize = (10,5))
ax = sns.barplot(x="seconds_added_per_point", y="tournament", hue="surface", data=shortest_time_surfaces)
ax.set_title("TOP 10 SURFACES THAT TAKE THE SHORTEST TIME", fontsize=17)
plt.xlabel("Seconds", fontsize=17)
plt.ylabel("Tournament", fontsize=17)
plt.yticks(size=17)
plt.xticks(size=17)
plt.show()
```
## 6. Figure Out How The Time For The Clay Surface Has Progressed Throughout The Years
```
years = events_time[~events_time["years"].str.contains("-")]
sorted_years_clay = years[years["surface"] == "Clay"].sort_values(by="years")
sorted_years_clay
sns.set(style="darkgrid")
plt.figure(figsize = (10,5))
ax= sns.lineplot(x="years", y="seconds_added_per_point", hue="surface", data=sorted_years_clay)
ax.set_title("PROGRESSION OF TIME FOR THE CLAY SURFACE THROUGHOUT THE YEARS", fontsize=17)
plt.xlabel("Years", fontsize=17)
plt.ylabel("Seconds", fontsize=17)
plt.yticks(size=17)
plt.xticks(size=17)
plt.show()
```
## 7. Figure Out How The Time For The Hard Surface Has Progressed Throughout The Years
```
sorted_years_hard = years[years["surface"] == "Hard"].sort_values(by="years")
sns.set(style="darkgrid")
plt.figure(figsize = (10,5))
ax= sns.lineplot(x="years", y="seconds_added_per_point", hue="surface", data=sorted_years_hard)
ax.set_title("PROGRESSION OF TIME FOR THE HARD SURFACE THROUGHOUT THE YEARS", fontsize=17)
plt.xlabel("Years", fontsize=17)
plt.ylabel("Seconds", fontsize=17)
plt.yticks(size=17)
plt.xticks(size=17)
plt.show()
```
## 8. Figure Out How The Time For The Carpet Surface Has Progressed Throughout The Years
```
sorted_years_carpet = years[years["surface"] == "Carpet"].sort_values(by="years")
sns.set(style="darkgrid")
plt.figure(figsize = (10,5))
ax= sns.lineplot(x="years", y="seconds_added_per_point", hue="surface", data=sorted_years_carpet)
ax.set_title("PROGRESSION OF TIME FOR THE CARPET SURFACE THROUGHOUT THE YEARS", fontsize=17)
plt.xlabel("Years", fontsize=17)
plt.ylabel("Seconds", fontsize=17)
plt.yticks(size=17)
plt.xticks(size=17)
plt.show()
```
## 9. Figure Out How The Time For The Grass Surface Has Progressed Throughout The Years
```
sorted_years_grass = events_time[events_time["surface"] == "Grass"].sort_values(by="years").head(5)
sns.set(style="darkgrid")
plt.figure(figsize = (10,5))
ax= sns.lineplot(x="years", y="seconds_added_per_point", hue="surface", data=sorted_years_grass)
ax.set_title("PROGRESSION OF TIME FOR THE GRASS SURFACE THROUGHOUT THE YEARS", fontsize=17)
plt.xlabel("Years", fontsize=17)
plt.ylabel("Seconds", fontsize=17)
plt.yticks(size=17)
plt.xticks(size=17)
plt.show()
```
## 10. Figure Out The Person Who Took The Most Time Serving In 2015
```
serve_time
serve_time_visualization = serve_time.groupby("server")["seconds_before_next_point"].agg("sum")
serve_time_visualization
serve_time_visual_data = serve_time_visualization.reset_index()
serve_time_visual_data
serve_time_visual_sorted = serve_time_visual_data.sort_values(by="seconds_before_next_point", ascending = False)
sns.set(style="darkgrid")
plt.figure(figsize = (10,5))
ax = sns.barplot(x="seconds_before_next_point", y="server", data=serve_time_visual_sorted)
ax.set_title("PLAYERS TOTAL SERVING TIME(2015) ", fontsize=17)
plt.xlabel("Seconds", fontsize=17)
plt.ylabel("Player", fontsize=17)
plt.yticks(size=17)
plt.xticks(size=17)
plt.show()
```
### BIG THREE TOTAL SERVING TIME IN 2015
```
big_three_total_serving_time = serve_time_visual_sorted[(serve_time_visual_sorted["server"] == "Roger Federer") | (serve_time_visual_sorted["server"] == "Rafael Nadal") | (serve_time_visual_sorted["server"] == "Novak Djokovic")]
big_three_total_serving_time
sns.set(style="darkgrid")
plt.figure(figsize = (10,5))
ax = sns.barplot(x="seconds_before_next_point", y="server", data=big_three_total_serving_time)
ax.set_title("BIG THREE TOTAL SERVING TIME(2015) ", fontsize=17)
plt.xlabel("Seconds", fontsize=17)
plt.ylabel("Player", fontsize=17)
plt.yticks(size=17)
plt.xticks(size=17)
plt.show()
```
## Conclusion
### Matches are short when they are played on a Grass, Carpet or Hard Surface. Grass however has proved to let matches be way more short compared to the other 2.
### Clay Surfaces have proved to make matches last so long.
### In 2015, among the Big Three, Novak Djokovic took the shortest time serving followed by Rafael Nadal. Roger Federer took the longest time serving. Overall however, Roger Federer proved to have the shortest time serving over the past years, followed by Novak Djokovic. Rafael Nadal has proved to have the longest time serving over the past years, making the matches that he is involved in last longer.
| github_jupyter |
```
%matplotlib inline
%reload_ext autoreload
%autoreload 2
from fastai.conv_learner import *
from fastai.dataset import *
from fastai.models.resnet import vgg_resnet50
import json
#torch.cuda.set_device(2)
torch.backends.cudnn.benchmark=True
```
## Data
```
PATH = Path('/home/giles/Downloads/fastai_data/salt/')
MASKS_FN = 'train_masks.csv'
META_FN = 'metadata.csv'
masks_csv = pd.read_csv(PATH/MASKS_FN)
meta_csv = pd.read_csv(PATH/META_FN)
def show_img(im, figsize=None, ax=None, alpha=None):
if not ax: fig,ax = plt.subplots(figsize=figsize)
ax.imshow(im, alpha=alpha)
ax.set_axis_off()
return ax
(PATH/'train_masks-128').mkdir(exist_ok=True)
def resize_img(fn):
Image.open(fn).resize((128,128)).save((fn.parent.parent)/'train_masks-128'/fn.name)
files = list((PATH/'train_masks').iterdir())
with ThreadPoolExecutor(8) as e: e.map(resize_img, files)
(PATH/'train-128').mkdir(exist_ok=True)
def resize_img(fn):
Image.open(fn).resize((128,128)).save((fn.parent.parent)/'train-128'/fn.name)
files = list((PATH/'train').iterdir())
with ThreadPoolExecutor(8) as e: e.map(resize_img, files)
TRAIN_DN = 'train-128'
MASKS_DN = 'train_masks-128'
sz = 32
bs = 64
nw = 16
```
TRAIN_DN = 'train'
MASKS_DN = 'train_masks_png'
sz = 128
bs = 64
nw = 16
```
class MatchedFilesDataset(FilesDataset):
def __init__(self, fnames, y, transform, path):
self.y=y
assert(len(fnames)==len(y))
super().__init__(fnames, transform, path)
def get_y(self, i): return open_image(os.path.join(self.path, self.y[i]))
def get_c(self): return 0
x_names = np.array(glob(f'{PATH}/{TRAIN_DN}/*'))
y_names = np.array(glob(f'{PATH}/{MASKS_DN}/*'))
val_idxs = list(range(800))
((val_x,trn_x),(val_y,trn_y)) = split_by_idx(val_idxs, x_names, y_names)
aug_tfms = [RandomFlip(tfm_y=TfmType.CLASS)]
tfms = tfms_from_model(resnet34, sz, crop_type=CropType.NO, tfm_y=TfmType.CLASS, aug_tfms=aug_tfms)
datasets = ImageData.get_ds(MatchedFilesDataset, (trn_x,trn_y), (val_x,val_y), tfms, path=PATH)
md = ImageData(PATH, datasets, bs, num_workers=16, classes=None)
denorm = md.trn_ds.denorm
x,y = next(iter(md.trn_dl))
x.shape,y.shape
denorm = md.val_ds.denorm
def show_aug_img(ims, idx, figsize=(5,5), normed=True, ax=None, nchannels=3):
if ax is None: fig,ax = plt.subplots(figsize=figsize)
if normed: ims = denorm(ims)
else: ims = np.rollaxis(to_np(ims),1,nchannels+1)
ax.imshow(np.clip(ims,0,1)[idx])
ax.axis('off')
batches = [next(iter(md.aug_dl)) for i in range(9)]
fig, axes = plt.subplots(3, 6, figsize=(18, 9))
for i,(x,y) in enumerate(batches):
show_aug_img(x,1, ax=axes.flat[i*2])
show_aug_img(y,1, ax=axes.flat[i*2+1], nchannels=1, normed=False)
```
## Simple upsample
```
f = resnet34
cut,lr_cut = model_meta[f]
def get_base():
layers = cut_model(f(True), cut)
return nn.Sequential(*layers)
def dice(pred, targs):
pred = (pred>0.5).float()
return 2. * (pred*targs).sum() / (pred+targs).sum()
```
## U-net (ish)
```
class SaveFeatures():
features=None
def __init__(self, m): self.hook = m.register_forward_hook(self.hook_fn)
def hook_fn(self, module, input, output): self.features = output
def remove(self): self.hook.remove()
class UnetBlock(nn.Module):
def __init__(self, up_in, x_in, n_out):
super().__init__()
up_out = x_out = n_out//2
self.x_conv = nn.Conv2d(x_in, x_out, 1)
self.tr_conv = nn.ConvTranspose2d(up_in, up_out, 2, stride=2)
self.bn = nn.BatchNorm2d(n_out)
def forward(self, up_p, x_p):
up_p = self.tr_conv(up_p)
x_p = self.x_conv(x_p)
cat_p = torch.cat([up_p,x_p], dim=1)
return self.bn(F.relu(cat_p))
class Unet34(nn.Module):
def __init__(self, rn):
super().__init__()
self.rn = rn
self.sfs = [SaveFeatures(rn[i]) for i in [2,4,5,6]]
self.up1 = UnetBlock(512,256,256)
self.up2 = UnetBlock(256,128,256)
self.up3 = UnetBlock(256,64,256)
self.up4 = UnetBlock(256,64,256)
self.up5 = UnetBlock(256,3,16)
self.up6 = nn.ConvTranspose2d(16, 1, 1)
def forward(self,x):
inp = x
x = F.relu(self.rn(x))
x = self.up1(x, self.sfs[3].features)
x = self.up2(x, self.sfs[2].features)
x = self.up3(x, self.sfs[1].features)
x = self.up4(x, self.sfs[0].features)
x = self.up5(x, inp)
x = self.up6(x)
return x[:,0]
def close(self):
for sf in self.sfs: sf.remove()
class UnetModel():
def __init__(self,model,name='unet'):
self.model,self.name = model,name
def get_layer_groups(self, precompute):
lgs = list(split_by_idxs(children(self.model.rn), [lr_cut]))
return lgs + [children(self.model)[1:]]
m_base = get_base()
m = to_gpu(Unet34(m_base))
models = UnetModel(m)
learn = ConvLearner(md, models)
learn.opt_fn=optim.Adam
learn.crit=nn.BCEWithLogitsLoss()
learn.metrics=[accuracy_thresh(0.5),dice]
learn.summary()
[o.features.size() for o in m.sfs]
learn.freeze_to(1)
learn.lr_find()
learn.sched.plot()
lr=1e-2
wd=1e-7
lrs = np.array([lr/9,lr/3,lr])
learn.fit(lr,1,wds=wd,cycle_len=10,use_clr=(5,8))
learn.save('32urn-tmp')
learn.load('32urn-tmp')
learn.unfreeze()
learn.bn_freeze(True)
learn.fit(lrs/4, 1, wds=wd, cycle_len=20,use_clr=(20,10))
learn.sched.plot_lr()
learn.save('32urn-0')
learn.load('32urn-0')
x,y = next(iter(md.val_dl))
py = to_np(learn.model(V(x)))
show_img(py[0]>0.5);
show_img(y[0]);
show_img(x[0][0]);
m.close()
```
## 64x64
```
sz=64
bs=64
tfms = tfms_from_model(resnet34, sz, crop_type=CropType.NO, tfm_y=TfmType.CLASS, aug_tfms=aug_tfms)
datasets = ImageData.get_ds(MatchedFilesDataset, (trn_x,trn_y), (val_x,val_y), tfms, path=PATH)
md = ImageData(PATH, datasets, bs, num_workers=16, classes=None)
denorm = md.trn_ds.denorm
m_base = get_base()
m = to_gpu(Unet34(m_base))
models = UnetModel(m)
learn = ConvLearner(md, models)
learn.opt_fn=optim.Adam
learn.crit=nn.BCEWithLogitsLoss()
learn.metrics=[accuracy_thresh(0.5),dice]
learn.freeze_to(1)
learn.load('32urn-0')
learn.fit(lr/2,1,wds=wd, cycle_len=10,use_clr=(10,10))
learn.sched.plot_lr()
learn.save('64urn-tmp')
learn.unfreeze()
learn.bn_freeze(True)
learn.load('64urn-tmp')
learn.fit(lrs/4,1,wds=wd, cycle_len=8,use_clr=(20,8))
learn.sched.plot_lr()
learn.save('64urn')
learn.load('64urn')
x,y = next(iter(md.val_dl))
py = to_np(learn.model(V(x)))
show_img(py[0]>0.5);
show_img(y[0]);
show_img(x[0][0]);
m.close()
```
## 128x128
```
sz=128
bs=64
tfms = tfms_from_model(resnet34, sz, crop_type=CropType.NO, tfm_y=TfmType.CLASS, aug_tfms=aug_tfms)
datasets = ImageData.get_ds(MatchedFilesDataset, (trn_x,trn_y), (val_x,val_y), tfms, path=PATH)
md = ImageData(PATH, datasets, bs, num_workers=16, classes=None)
denorm = md.trn_ds.denorm
m_base = get_base()
m = to_gpu(Unet34(m_base))
models = UnetModel(m)
learn = ConvLearner(md, models)
learn.opt_fn=optim.Adam
learn.crit=nn.BCEWithLogitsLoss()
learn.metrics=[accuracy_thresh(0.5),dice]
learn.load('64urn')
learn.fit(lr/2,1, wds=wd, cycle_len=6,use_clr=(6,4))
learn.save('128urn-tmp')
learn.load('128urn-tmp')
learn.unfreeze()
learn.bn_freeze(True)
#lrs = np.array([lr/200,lr/30,lr])
learn.fit(lrs/5,1, wds=wd,cycle_len=8,use_clr=(20,8))
learn.sched.plot_lr()
learn.sched.plot_loss()
learn.save('128urn')
learn.load('128urn')
x,y = next(iter(md.val_dl))
py = to_np(learn.model(V(x)))
show_img(py[0]>0.5);
show_img(y[0]);
show_img(x[0][0]);
y.shape
batches = [next(iter(md.aug_dl)) for i in range(9)]
fig, axes = plt.subplots(3, 6, figsize=(18, 9))
for i,(x,y) in enumerate(batches):
show_aug_img(x,1, ax=axes.flat[i*2])
show_aug_img(y,1, ax=axes.flat[i*2+1], nchannels=1, normed=False)
```
# Test on original validation
```
x_names_orig = np.array(glob(f'{PATH}/train/*'))
y_names_orig = np.array(glob(f'{PATH}/train_masks/*'))
val_idxs_orig = list(range(800))
((val_x_orig,trn_x_orig),(val_y_orig,trn_y_orig)) = split_by_idx(val_idxs_orig, x_names_orig, y_names_orig)
sz=128
bs=64
tfms = tfms_from_model(resnet34, sz, crop_type=CropType.NO, tfm_y=TfmType.CLASS, aug_tfms=aug_tfms)
datasets = ImageData.get_ds(MatchedFilesDataset, (trn_x,trn_y), (val_x,val_y), tfms, path=PATH)
md = ImageData(PATH, datasets, bs, num_workers=16, classes=None)
denorm = md.trn_ds.denorm
m_base = get_base()
m = to_gpu(Unet34(m_base))
models = UnetModel(m)
learn = ConvLearner(md, models)
learn.opt_fn=optim.Adam
learn.crit=nn.BCEWithLogitsLoss()
learn.metrics=[accuracy_thresh(0.5),dice]
learn.load('128urn')
probs = learn.predict()
probs.shape
_, y = learn.TTA(n_aug=1)
y.shape
idx=0
show_img(probs[idx]>0.5);
show_img(probs[idx]);
show_img(y[idx]);
show_img(x[idx][0]);
```
# Optimise threshold
```
# src: https://www.kaggle.com/aglotero/another-iou-metric
def iou_metric(y_true_in, y_pred_in, print_table=False):
labels = y_true_in
y_pred = y_pred_in
true_objects = 2
pred_objects = 2
intersection = np.histogram2d(labels.flatten(), y_pred.flatten(), bins=(true_objects, pred_objects))[0]
# Compute areas (needed for finding the union between all objects)
area_true = np.histogram(labels, bins = true_objects)[0]
area_pred = np.histogram(y_pred, bins = pred_objects)[0]
area_true = np.expand_dims(area_true, -1)
area_pred = np.expand_dims(area_pred, 0)
# Compute union
union = area_true + area_pred - intersection
# Exclude background from the analysis
intersection = intersection[1:,1:]
union = union[1:,1:]
union[union == 0] = 1e-9
# Compute the intersection over union
iou = intersection / union
# Precision helper function
def precision_at(threshold, iou):
matches = iou > threshold
true_positives = np.sum(matches, axis=1) == 1 # Correct objects
false_positives = np.sum(matches, axis=0) == 0 # Missed objects
false_negatives = np.sum(matches, axis=1) == 0 # Extra objects
tp, fp, fn = np.sum(true_positives), np.sum(false_positives), np.sum(false_negatives)
return tp, fp, fn
# Loop over IoU thresholds
prec = []
if print_table:
print("Thresh\tTP\tFP\tFN\tPrec.")
for t in np.arange(0.5, 1.0, 0.05):
tp, fp, fn = precision_at(t, iou)
if (tp + fp + fn) > 0:
p = tp / (tp + fp + fn)
else:
p = 0
if print_table:
print("{:1.3f}\t{}\t{}\t{}\t{:1.3f}".format(t, tp, fp, fn, p))
prec.append(p)
if print_table:
print("AP\t-\t-\t-\t{:1.3f}".format(np.mean(prec)))
return np.mean(prec)
def iou_metric_batch(y_true_in, y_pred_in):
batch_size = y_true_in.shape[0]
metric = []
for batch in range(batch_size):
value = iou_metric(y_true_in[batch], y_pred_in[batch])
metric.append(value)
return np.mean(metric)
thres = np.linspace(-1, 1, 10)
thres_ioc = [iou_metric_batch(y, np.int32(probs > t)) for t in tqdm_notebook(thres)]
plt.plot(thres, thres_ioc);
best_thres = thres[np.argmax(thres_ioc)]
best_thres, max(thres_ioc)
thres = np.linspace(-0.5, 0.5, 50)
thres_ioc = [iou_metric_batch(y, np.int32(probs > t)) for t in tqdm_notebook(thres)]
plt.plot(thres, thres_ioc);
best_thres = thres[np.argmax(thres_ioc)]
best_thres, max(thres_ioc)
show_img(probs[0]>best_thres);
```
# Run on test
```
(PATH/'test-128').mkdir(exist_ok=True)
def resize_img(fn):
Image.open(fn).resize((128,128)).save((fn.parent.parent)/'test-128'/fn.name)
files = list((PATH/'test').iterdir())
with ThreadPoolExecutor(8) as e: e.map(resize_img, files)
testData = np.array(glob(f'{PATH}/test-128/*'))
class TestFilesDataset(FilesDataset):
def __init__(self, fnames, y, transform, path):
self.y=y
assert(len(fnames)==len(y))
super().__init__(fnames, transform, path)
def get_y(self, i): return open_image(os.path.join(self.path, self.fnames[i]))
def get_c(self): return 0
tfms_from_model(resnet34, sz, crop_type=CropType.NO, tfm_y=TfmType.CLASS, aug_tfms=aug_tfms)
datasets = ImageData.get_ds(TestFilesDataset, (trn_x,trn_y), (val_x,val_y), tfms, test=testData, path=PATH)
md = ImageData(PATH, datasets, bs, num_workers=16, classes=None)
denorm = md.trn_ds.denorm
m_base = get_base()
m = to_gpu(Unet34(m_base))
models = UnetModel(m)
learn = ConvLearner(md, models)
learn.opt_fn=optim.Adam
learn.crit=nn.BCEWithLogitsLoss()
learn.metrics=[accuracy_thresh(0.5),dice]
learn.load('128urn')
x,y = next(iter(md.test_dl))
py = to_np(learn.model(V(x)))
show_img(py[6]>best_thres);
show_img(py[6]);
show_img(y[6]);
probs = learn.predict(is_test=True)
show_img(probs[12]>best_thres);
show_img(probs[12]);
show_img(y[12]);
show_img(x[12][0]);
with open(f'{PATH}/probs.pkl', 'wb') as fout: #Save results
pickle.dump(probs, fout)
probs.shape
def resize_img(fn):
return np.array(Image.fromarray(fn).resize((101,101)))
resizePreds = np.array([resize_img(x) for x in probs])
resizePreds.shape
show_img(resizePreds[12]);
testData
f'{PATH}/test'
test_ids = next(os.walk(f'{PATH}/test'))[2]
def RLenc(img, order='F', format=True):
"""
img is binary mask image, shape (r,c)
order is down-then-right, i.e. Fortran
format determines if the order needs to be preformatted (according to submission rules) or not
returns run length as an array or string (if format is True)
"""
bytes = img.reshape(img.shape[0] * img.shape[1], order=order)
runs = [] ## list of run lengths
r = 0 ## the current run length
pos = 1 ## count starts from 1 per WK
for c in bytes:
if (c == 0):
if r != 0:
runs.append((pos, r))
pos += r
r = 0
pos += 1
else:
r += 1
# if last run is unsaved (i.e. data ends with 1)
if r != 0:
runs.append((pos, r))
pos += r
r = 0
if format:
z = ''
for rr in runs:
z += '{} {} '.format(rr[0], rr[1])
return z[:-1]
else:
return runs
pred_dict = {id_[:-4]:RLenc(np.round(resizePreds[i] > best_thres)) for i,id_ in tqdm_notebook(enumerate(test_ids))}
sub = pd.DataFrame.from_dict(pred_dict,orient='index')
sub.index.names = ['id']
sub.columns = ['rle_mask']
sub.to_csv('submission.csv')
sub
```
| github_jupyter |
```
# feature extractoring and preprocessing data
# 음원 데이터를 분석
import librosa
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# notebook을 실행한 브라우저에서 바로 그림을 볼 수 있게 해주는 것
%matplotlib inline
# 운영체제와의 상호작용을 돕는 다양한 기능을 제공
# 1. 현재 디렉토리 확인하기
# 2. 디렉토리 변경
# 3. 현재 디렉토리의 파일 목록 확인하기
# 4. csv 파일 호출
import os
# 파이썬에서의 이미지 처리
from PIL import Image
import pathlib
import csv
# Preprocessing
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder, StandardScaler
from sklearn.metrics import mean_squared_error
#Keras
import keras
# 경고 메시지를 무시하고 숨기거나 -> warnings.filterwarnings(action='ignore')
# 일치하는 경고를 인쇄하지 않습니다 = ('ignore')
import warnings
warnings.filterwarnings('ignore')
# 원하는 종류의 색깔만 넘겨주는 것
cmap = plt.get_cmap('inferno')
plt.figure(figsize=(10,10))
genres = 'blues classical country disco hiphop jazz metal pop reggae rock'.split()
for g in genres:
pathlib.Path(f'img_data/{g}').mkdir(parents=True, exist_ok=True)
for filename in os.listdir(f'./MIR/genres/{g}'):
songname = f'./MIR/genres/{g}/{filename}'
y, sr = librosa.load(songname, mono=True, duration=5)
plt.specgram(y, NFFT=2048, Fs=2, Fc=0, noverlap=128, cmap=cmap, sides='default', mode='default', scale='dB');
plt.axis('off');
plt.savefig(f'img_data/{g}/{filename[:-3].replace(".", "")}.png')
plt.clf()
header = 'filename chroma_stft rmse spectral_centroid spectral_bandwidth rolloff zero_crossing_rate'
for i in range(1, 21):
header += f' mfcc{i}'
header += ' label'
header = header.split()
file = open('data.csv', 'w', newline='')
with file:
writer = csv.writer(file)
writer.writerow(header)
genres = 'blues classical country disco hiphop jazz metal pop reggae rock'.split()
for g in genres:
for filename in os.listdir(f'./MIR/genres/{g}'):
songname = f'./MIR/genres/{g}/{filename}'
y, sr = librosa.load(songname, mono=True, duration=30)
chroma_stft = librosa.feature.chroma_stft(y=y, sr=sr)
spec_cent = librosa.feature.spectral_centroid(y=y, sr=sr)
spec_bw = librosa.feature.spectral_bandwidth(y=y, sr=sr)
rolloff = librosa.feature.spectral_rolloff(y=y, sr=sr)
zcr = librosa.feature.zero_crossing_rate(y)
mfcc = librosa.feature.mfcc(y=y, sr=sr)
#rmse = mean_squared_error(y, y_pred=sr)**0.5
rmse = librosa.feature.rms(y=y)
to_append = f'{filename} {np.mean(chroma_stft)} {np.mean(rmse)} {np.mean(spec_cent)} {np.mean(spec_bw)} {np.mean(rolloff)} {np.mean(zcr)}'
for e in mfcc:
to_append += f' {np.mean(e)}'
to_append += f' {g}'
file = open('data.csv', 'a', newline='')
with file:
writer = csv.writer(file)
writer.writerow(to_append.split())
# mfcc = 오디오 신호에서 추출할 수 있는 feature로, 소리의 고유한 특징을 나타내는 수치
# = 등록된 음성과 현재 입력된 음성의 유사도를 판별하는 근거의 일부로 쓰입니다.
# = MFCC(Mel-Frequency Cepstral Coefficient)는
# Mel Spectrum(멜 스펙트럼)에서 Cepstral(켑스트럴) 분석을 통해 추출된 값
#
# 이해하기 위해 먼저
# - Spectrum(스펙트럼)
# - Cepstrum(켑스트럼)
# - Mel Spectrum(멜 스펙트럼) 들을 알아야 한다.
data = pd.read_csv('data.csv')
data.head()
# chroma_stft = 채도_? , 크로마 표준
# spectral_centroid = 스펙트럼 중심
# spectral_bandwidth = 스펙트럼 대역폭
# rolloff = 롤 오프
# zero_crossing_rate = 제로 크로싱 비율
#
# mfcc[n] =
data.shape
# Dropping unneccesary columns
data = data.drop(['filename'],axis=1)
genre_list = data.iloc[:, -1]
encoder = LabelEncoder()
y = encoder.fit_transform(genre_list)
scaler = StandardScaler()
X = scaler.fit_transform(np.array(data.iloc[:, :-1], dtype = float))
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
len(y_train)
len(y_test)
X_train[10]
from keras import models
from keras import layers
model = models.Sequential()
model.add(layers.Dense(256, activation='relu', input_shape=(X_train.shape[1],)))
model.add(layers.Dense(128, activation='relu'))
model.add(layers.Dense(64, activation='relu'))
model.add(layers.Dense(10, activation='softmax'))
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
history = model.fit(X_train,
y_train,
epochs=20,
batch_size=128)
test_loss, test_acc = model.evaluate(X_test,y_test)
print('test_acc: ',test_acc)
x_val = X_train[:200]
partial_x_train = X_train[200:]
y_val = y_train[:200]
partial_y_train = y_train[200:]
model = models.Sequential()
model.add(layers.Dense(512, activation='relu', input_shape=(X_train.shape[1],)))
model.add(layers.Dense(256, activation='relu'))
model.add(layers.Dense(128, activation='relu'))
model.add(layers.Dense(64, activation='relu'))
model.add(layers.Dense(10, activation='softmax'))
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
model.fit(partial_x_train,
partial_y_train,
epochs=30,
batch_size=512,
validation_data=(x_val, y_val))
results = model.evaluate(X_test, y_test)
results
predictions = model.predict(X_test)
predictions[0].shape
np.sum(predictions[0])
np.argmax(predictions[0])
```
| github_jupyter |
# AWS Elastic Kubernetes Service (EKS) Deep MNIST
In this example we will deploy a tensorflow MNIST model in Amazon Web Services' Elastic Kubernetes Service (EKS).
This tutorial will break down in the following sections:
1) Train a tensorflow model to predict mnist locally
2) Containerise the tensorflow model with our docker utility
3) Send some data to the docker model to test it
4) Install and configure AWS tools to interact with AWS
5) Use the AWS tools to create and setup EKS cluster with Seldon
6) Push and run docker image through the AWS Container Registry
7) Test our Elastic Kubernetes deployment by sending some data
Let's get started! 🚀🔥
## Dependencies:
* Helm v3.0.0+
* A Kubernetes cluster running v1.13 or above (minkube / docker-for-windows work well if enough RAM)
* kubectl v1.14+
* EKS CLI v0.1.32
* AWS Cli v1.16.163
* Python 3.6+
* Python DEV requirements
## 1) Train a tensorflow model to predict mnist locally
We will load the mnist images, together with their labels, and then train a tensorflow model to predict the right labels
```
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("MNIST_data/", one_hot = True)
import tensorflow as tf
if __name__ == '__main__':
x = tf.placeholder(tf.float32, [None,784], name="x")
W = tf.Variable(tf.zeros([784,10]))
b = tf.Variable(tf.zeros([10]))
y = tf.nn.softmax(tf.matmul(x,W) + b, name="y")
y_ = tf.placeholder(tf.float32, [None, 10])
cross_entropy = tf.reduce_mean(-tf.reduce_sum(y_ * tf.log(y), reduction_indices=[1]))
train_step = tf.train.GradientDescentOptimizer(0.5).minimize(cross_entropy)
init = tf.initialize_all_variables()
sess = tf.Session()
sess.run(init)
for i in range(1000):
batch_xs, batch_ys = mnist.train.next_batch(100)
sess.run(train_step, feed_dict={x: batch_xs, y_: batch_ys})
correct_prediction = tf.equal(tf.argmax(y,1), tf.argmax(y_,1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
print(sess.run(accuracy, feed_dict = {x: mnist.test.images, y_:mnist.test.labels}))
saver = tf.train.Saver()
saver.save(sess, "model/deep_mnist_model")
```
## 2) Containerise the tensorflow model with our docker utility
First you need to make sure that you have added the .s2i/environment configuration file in this folder with the following content:
```
!cat .s2i/environment
```
Now we can build a docker image named "deep-mnist" with the tag 0.1
```
!s2i build . seldonio/seldon-core-s2i-python36:1.5.0-dev deep-mnist:0.1
```
## 3) Send some data to the docker model to test it
We first run the docker image we just created as a container called "mnist_predictor"
```
!docker run --name "mnist_predictor" -d --rm -p 5000:5000 deep-mnist:0.1
```
Send some random features that conform to the contract
```
import matplotlib.pyplot as plt
# This is the variable that was initialised at the beginning of the file
i = [0]
x = mnist.test.images[i]
y = mnist.test.labels[i]
plt.imshow(x.reshape((28, 28)), cmap='gray')
plt.show()
print("Expected label: ", np.sum(range(0,10) * y), ". One hot encoding: ", y)
from seldon_core.seldon_client import SeldonClient
import math
import numpy as np
# We now test the REST endpoint expecting the same result
endpoint = "0.0.0.0:5000"
batch = x
payload_type = "ndarray"
sc = SeldonClient(microservice_endpoint=endpoint)
# We use the microservice, instead of the "predict" function
client_prediction = sc.microservice(
data=batch,
method="predict",
payload_type=payload_type,
names=["tfidf"])
for proba, label in zip(client_prediction.response.data.ndarray.values[0].list_value.ListFields()[0][1], range(0,10)):
print(f"LABEL {label}:\t {proba.number_value*100:6.4f} %")
!docker rm mnist_predictor --force
```
## 4) Install and configure AWS tools to interact with AWS
First we install the awscli
```
!pip install awscli --upgrade --user
```
### Configure aws so it can talk to your server
(if you are getting issues, make sure you have the permmissions to create clusters)
```
%%bash
# You must make sure that the access key and secret are changed
aws configure << END_OF_INPUTS
YOUR_ACCESS_KEY
YOUR_ACCESS_SECRET
us-west-2
json
END_OF_INPUTS
```
### Install EKCTL
*IMPORTANT*: These instructions are for linux
Please follow the official installation of ekctl at: https://docs.aws.amazon.com/eks/latest/userguide/getting-started-eksctl.html
```
!curl --silent --location "https://github.com/weaveworks/eksctl/releases/download/latest_release/eksctl_$(uname -s)_amd64.tar.gz" | tar xz
!chmod 755 ./eksctl
!./eksctl version
```
## 5) Use the AWS tools to create and setup EKS cluster with Seldon
In this example we will create a cluster with 2 nodes, with a minimum of 1 and a max of 3. You can tweak this accordingly.
If you want to check the status of the deployment you can go to AWS CloudFormation or to the EKS dashboard.
It will take 10-15 minutes (so feel free to go grab a ☕).
*IMPORTANT*: If you get errors in this step it is most probably IAM role access requirements, which requires you to discuss with your administrator.
```
%%bash
./eksctl create cluster \
--name demo-eks-cluster \
--region us-west-2 \
--nodes 2
```
### Configure local kubectl
We want to now configure our local Kubectl so we can actually reach the cluster we've just created
```
!aws eks --region us-west-2 update-kubeconfig --name demo-eks-cluster
```
And we can check if the context has been added to kubectl config (contexts are basically the different k8s cluster connections)
You should be able to see the context as "...aws:eks:eu-west-1:27...".
If it's not activated you can activate that context with kubectlt config set-context <CONTEXT_NAME>
```
!kubectl config get-contexts
```
## Setup Seldon Core
Use the setup notebook to [Setup Cluster](https://docs.seldon.io/projects/seldon-core/en/latest/examples/seldon_core_setup.html#Setup-Cluster) with [Ambassador Ingress](https://docs.seldon.io/projects/seldon-core/en/latest/examples/seldon_core_setup.html#Ambassador) and [Install Seldon Core](https://docs.seldon.io/projects/seldon-core/en/latest/examples/seldon_core_setup.html#Install-Seldon-Core). Instructions [also online](https://docs.seldon.io/projects/seldon-core/en/latest/examples/seldon_core_setup.html).
## Push docker image
In order for the EKS seldon deployment to access the image we just built, we need to push it to the Elastic Container Registry (ECR).
If you have any issues please follow the official AWS documentation: https://docs.aws.amazon.com/AmazonECR/latest/userguide/what-is-ecr.html
### First we create a registry
You can run the following command, and then see the result at https://us-west-2.console.aws.amazon.com/ecr/repositories?#
```
!aws ecr create-repository --repository-name seldon-repository --region us-west-2
```
### Now prepare docker image
We need to first tag the docker image before we can push it
```
%%bash
export AWS_ACCOUNT_ID=""
export AWS_REGION="us-west-2"
if [ -z "$AWS_ACCOUNT_ID" ]; then
echo "ERROR: Please provide a value for the AWS variables"
exit 1
fi
docker tag deep-mnist:0.1 "$AWS_ACCOUNT_ID.dkr.ecr.$AWS_REGION.amazonaws.com/seldon-repository"
```
### We now login to aws through docker so we can access the repository
```
!`aws ecr get-login --no-include-email --region us-west-2`
```
### And push the image
Make sure you add your AWS Account ID
```
%%bash
export AWS_ACCOUNT_ID=""
export AWS_REGION="us-west-2"
if [ -z "$AWS_ACCOUNT_ID" ]; then
echo "ERROR: Please provide a value for the AWS variables"
exit 1
fi
docker push "$AWS_ACCOUNT_ID.dkr.ecr.$AWS_REGION.amazonaws.com/seldon-repository"
```
## Running the Model
We will now run the model.
Let's first have a look at the file we'll be using to trigger the model:
```
!cat deep_mnist.json
```
Now let's trigger seldon to run the model.
We basically have a yaml file, where we want to replace the value "REPLACE_FOR_IMAGE_AND_TAG" for the image you pushed
```
%%bash
export AWS_ACCOUNT_ID=""
export AWS_REGION="us-west-2"
if [ -z "$AWS_ACCOUNT_ID" ]; then
echo "ERROR: Please provide a value for the AWS variables"
exit 1
fi
sed 's|REPLACE_FOR_IMAGE_AND_TAG|'"$AWS_ACCOUNT_ID"'.dkr.ecr.'"$AWS_REGION"'.amazonaws.com/seldon-repository|g' deep_mnist.json | kubectl apply -f -
```
And let's check that it's been created.
You should see an image called "deep-mnist-single-model...".
We'll wait until STATUS changes from "ContainerCreating" to "Running"
```
!kubectl get pods
```
## Test the model
Now we can test the model, let's first find out what is the URL that we'll have to use:
```
!kubectl get svc ambassador -o jsonpath='{.status.loadBalancer.ingress[0].hostname}'
```
We'll use a random example from our dataset
```
import matplotlib.pyplot as plt
# This is the variable that was initialised at the beginning of the file
i = [0]
x = mnist.test.images[i]
y = mnist.test.labels[i]
plt.imshow(x.reshape((28, 28)), cmap='gray')
plt.show()
print("Expected label: ", np.sum(range(0,10) * y), ". One hot encoding: ", y)
```
We can now add the URL above to send our request:
```
from seldon_core.seldon_client import SeldonClient
import math
import numpy as np
host = "a68bbac487ca611e988060247f81f4c1-707754258.us-west-2.elb.amazonaws.com"
port = "80" # Make sure you use the port above
batch = x
payload_type = "ndarray"
sc = SeldonClient(
gateway="ambassador",
ambassador_endpoint=host + ":" + port,
namespace="default",
oauth_key="oauth-key",
oauth_secret="oauth-secret")
client_prediction = sc.predict(
data=batch,
deployment_name="deep-mnist",
names=["text"],
payload_type=payload_type)
print(client_prediction)
```
### Let's visualise the probability for each label
It seems that it correctly predicted the number 7
```
for proba, label in zip(client_prediction.response.data.ndarray.values[0].list_value.ListFields()[0][1], range(0,10)):
print(f"LABEL {label}:\t {proba.number_value*100:6.4f} %")
```
| github_jupyter |
# **Introduction to TinyAutoML**
---
TinyAutoML is a Machine Learning Python3.9 library thought as an extension of Scikit-Learn. It builds an adaptable and auto-tuned pipeline to handle binary classification tasks.
In a few words, your data goes through 2 main preprocessing steps. The first one is scaling and NonStationnarity correction, which is followed by Lasso Feature selection.
Finally, one of the three MetaModels is fitted on the transformed data.
Let's import the library !
```
%pip install TinyAutoML==0.2.3.3
from TinyAutoML.Models import *
from TinyAutoML import MetaPipeline
```
## MetaModels
MetaModels inherit from the MetaModel Abstract Class. They all implement ensemble methods and therefore are based on EstimatorPools.
When training EstimatorPools, you are faced with a choice : doing parameterTuning on entire pipelines with the estimators on the top or training the estimators using the same pipeline and only training the top. The first case refers to what we will be calling **comprehensiveSearch**.
Moreover, as we will see in details later, those EstimatorPools can be shared across MetaModels.
They are all initialised with those minimum arguments :
```python
MetaModel(comprehensiveSearch: bool = True, parameterTuning: bool = True, metrics: str = 'accuracy', nSplits: int=10)
```
- nSplits corresponds to the number of split of the cross validation
- The other parameters are equivoque
**They need to be put in the MetaPipeline wrapper to work**
**There are 3 MetaModels**
1- BestModel : selects the best performing model of the pool
```
best_model = MetaPipeline(BestModel(comprehensiveSearch = False, parameterTuning = False))
```
2- OneRulerForAll : implements Stacking using a RandomForestClassifier by default. The user is free to use another classifier using the ruler arguments
```
orfa_model = MetaPipeline(OneRulerForAll(comprehensiveSearch=False, parameterTuning=False))
```
3- DemocraticModel : implements Soft and Hard voting models through the voting argument
```
democratic_model = MetaPipeline(DemocraticModel(comprehensiveSearch=False, parameterTuning=False, voting='soft'))
```
As of release v0.2.3.2 (13/04/2022) there are 5 models on which these MetaModels rely in the EstimatorPool:
- Random Forest Classifier
- Logistic Regression
- Gaussian Naive Bayes
- Linear Discriminant Analysis
- XGBoost
***
We'll use the breast_cancer dataset from sklearn as an example:
```
import pandas as pd
from sklearn.datasets import load_breast_cancer
cancer = load_breast_cancer()
X = pd.DataFrame(data=cancer.data, columns=cancer.feature_names)
y = cancer.target
cut = int(len(y) * 0.8)
X_train, X_test = X[:cut], X[cut:]
y_train, y_test = y[:cut], y[cut:]
```
Let's train a BestModel first and reuse its Pool for the other MetaModels
```
best_model.fit(X_train,y_train)
```
We can now extract the pool
```
pool = best_model.get_pool()
```
And use it when fitting the other MetaModels to skip the fitting of the underlying models:
```
orfa_model.fit(X_train,y_train,pool=pool)
democratic_model.fit(X_train,y_train,pool=pool)
```
Great ! Let's look at the results with the sk_learn classification report :
```
orfa_model.classification_report(X_test,y_test)
```
Looking good! What about the ROC Curve ?
```
democratic_model.roc_curve(X_test,y_test)
```
Let's see how the estimators of the pool are doing individually:
```
best_model.get_scores(X_test,y_test)
```
## What's next ?
You can do the same steps with comprehensiveSearch set to True if you have the time and if you want to improve your results. You can also try new rulers and so on.
Maria, Thomas and Lucas.
| github_jupyter |
# Computer Vision Nanodegree
## Project: Image Captioning
---
In this notebook, you will train your CNN-RNN model.
You are welcome and encouraged to try out many different architectures and hyperparameters when searching for a good model.
This does have the potential to make the project quite messy! Before submitting your project, make sure that you clean up:
- the code you write in this notebook. The notebook should describe how to train a single CNN-RNN architecture, corresponding to your final choice of hyperparameters. You should structure the notebook so that the reviewer can replicate your results by running the code in this notebook.
- the output of the code cell in **Step 2**. The output should show the output obtained when training the model from scratch.
This notebook **will be graded**.
Feel free to use the links below to navigate the notebook:
- [Step 1](#step1): Training Setup
- [Step 2](#step2): Train your Model
- [Step 3](#step3): (Optional) Validate your Model
<a id='step1'></a>
## Step 1: Training Setup
In this step of the notebook, you will customize the training of your CNN-RNN model by specifying hyperparameters and setting other options that are important to the training procedure. The values you set now will be used when training your model in **Step 2** below.
You should only amend blocks of code that are preceded by a `TODO` statement. **Any code blocks that are not preceded by a `TODO` statement should not be modified**.
### Task #1
Begin by setting the following variables:
- `batch_size` - the batch size of each training batch. It is the number of image-caption pairs used to amend the model weights in each training step.
- `vocab_threshold` - the minimum word count threshold. Note that a larger threshold will result in a smaller vocabulary, whereas a smaller threshold will include rarer words and result in a larger vocabulary.
- `vocab_from_file` - a Boolean that decides whether to load the vocabulary from file.
- `embed_size` - the dimensionality of the image and word embeddings.
- `hidden_size` - the number of features in the hidden state of the RNN decoder.
- `num_epochs` - the number of epochs to train the model. We recommend that you set `num_epochs=3`, but feel free to increase or decrease this number as you wish. [This paper](https://arxiv.org/pdf/1502.03044.pdf) trained a captioning model on a single state-of-the-art GPU for 3 days, but you'll soon see that you can get reasonable results in a matter of a few hours! (_But of course, if you want your model to compete with current research, you will have to train for much longer._)
- `save_every` - determines how often to save the model weights. We recommend that you set `save_every=1`, to save the model weights after each epoch. This way, after the `i`th epoch, the encoder and decoder weights will be saved in the `models/` folder as `encoder-i.pkl` and `decoder-i.pkl`, respectively.
- `print_every` - determines how often to print the batch loss to the Jupyter notebook while training. Note that you **will not** observe a monotonic decrease in the loss function while training - this is perfectly fine and completely expected! You are encouraged to keep this at its default value of `100` to avoid clogging the notebook, but feel free to change it.
- `log_file` - the name of the text file containing - for every step - how the loss and perplexity evolved during training.
If you're not sure where to begin to set some of the values above, you can peruse [this paper](https://arxiv.org/pdf/1502.03044.pdf) and [this paper](https://arxiv.org/pdf/1411.4555.pdf) for useful guidance! **To avoid spending too long on this notebook**, you are encouraged to consult these suggested research papers to obtain a strong initial guess for which hyperparameters are likely to work best. Then, train a single model, and proceed to the next notebook (**3_Inference.ipynb**). If you are unhappy with your performance, you can return to this notebook to tweak the hyperparameters (and/or the architecture in **model.py**) and re-train your model.
### Question 1
**Question:** Describe your CNN-RNN architecture in detail. With this architecture in mind, how did you select the values of the variables in Task 1? If you consulted a research paper detailing a successful implementation of an image captioning model, please provide the reference.
**Answer:** I used a pretrained Resnet152 network to extract features (deep CNN network). In the literature other architectures like VGG16 are also used, but Resnet152 is claimed to diminish the vanishing gradient problem.I'm using 2 layers of LSTM currently (as it is taking a lot of time), in the future I will explore with more layers.
vocab_threshold is 6, I tried with 9 (meaning lesser elements in vocab) but the training seemed to converge faster in the case of 6. Many paper suggest taking batch_size of 64 or 128, I went with 64. embed_size and hidden_size are both 512. I consulted several blogs and famous papers like "Show, attend and tell - Xu et al" although i did not use attention currently.
### (Optional) Task #2
Note that we have provided a recommended image transform `transform_train` for pre-processing the training images, but you are welcome (and encouraged!) to modify it as you wish. When modifying this transform, keep in mind that:
- the images in the dataset have varying heights and widths, and
- if using a pre-trained model, you must perform the corresponding appropriate normalization.
### Question 2
**Question:** How did you select the transform in `transform_train`? If you left the transform at its provided value, why do you think that it is a good choice for your CNN architecture?
**Answer:** The transform value is the same. Empirically, these parameters values worked well in my past projects.
### Task #3
Next, you will specify a Python list containing the learnable parameters of the model. For instance, if you decide to make all weights in the decoder trainable, but only want to train the weights in the embedding layer of the encoder, then you should set `params` to something like:
```
params = list(decoder.parameters()) + list(encoder.embed.parameters())
```
### Question 3
**Question:** How did you select the trainable parameters of your architecture? Why do you think this is a good choice?
**Answer:** Since resnet was pretrained, i trained only the embedding layer and all layers of the decoder. The resnet is already fitting for feature extraction as it is pretrained, hence only the other parts of the architecture should be trained.
### Task #4
Finally, you will select an [optimizer](http://pytorch.org/docs/master/optim.html#torch.optim.Optimizer).
### Question 4
**Question:** How did you select the optimizer used to train your model?
**Answer:** I used the Adam optimizer, since in my past similar projects it gave me better performance than SGD. I have found Adam to perform better than vanilla SGD almost in all cases, aligning with intuition.
```
import nltk
nltk.download('punkt')
import torch
import torch.nn as nn
from torchvision import transforms
import sys
sys.path.append('/opt/cocoapi/PythonAPI')
from pycocotools.coco import COCO
from data_loader import get_loader
from model import EncoderCNN, DecoderRNN
import math
## TODO #1: Select appropriate values for the Python variables below.
batch_size = 64 # batch size
vocab_threshold = 6 # minimum word count threshold
vocab_from_file = True # if True, load existing vocab file
embed_size = 512 # dimensionality of image and word embeddings
hidden_size = 512 # number of features in hidden state of the RNN decoder
num_epochs = 3 # number of training epochs
save_every = 1 # determines frequency of saving model weights
print_every = 100 # determines window for printing average loss
log_file = 'training_log.txt' # name of file with saved training loss and perplexity
# (Optional) TODO #2: Amend the image transform below.
transform_train = transforms.Compose([
transforms.Resize(256), # smaller edge of image resized to 256
transforms.RandomCrop(224), # get 224x224 crop from random location
transforms.RandomHorizontalFlip(), # horizontally flip image with probability=0.5
transforms.ToTensor(), # convert the PIL Image to a tensor
transforms.Normalize((0.485, 0.456, 0.406), # normalize image for pre-trained model
(0.229, 0.224, 0.225))])
# Build data loader.
data_loader = get_loader(transform=transform_train,
mode='train',
batch_size=batch_size,
vocab_threshold=vocab_threshold,
vocab_from_file=vocab_from_file)
# The size of the vocabulary.
vocab_size = len(data_loader.dataset.vocab)
# Initialize the encoder and decoder.
encoder = EncoderCNN(embed_size)
decoder = DecoderRNN(embed_size, hidden_size, vocab_size)
# Move models to GPU if CUDA is available.
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
encoder.to(device)
decoder.to(device)
# Define the loss function.
criterion = nn.CrossEntropyLoss().cuda() if torch.cuda.is_available() else nn.CrossEntropyLoss()
# TODO #3: Specify the learnable parameters of the model.
params = list(decoder.parameters()) + list(encoder.embed.parameters())
# TODO #4: Define the optimizer.
optimizer = torch.optim.Adam(params, lr=0.001, betas=(0.9,0.999), eps=1e-8)
# Set the total number of training steps per epoch.
total_step = math.ceil(len(data_loader.dataset.caption_lengths) / data_loader.batch_sampler.batch_size)
```
<a id='step2'></a>
## Step 2: Train your Model
Once you have executed the code cell in **Step 1**, the training procedure below should run without issue.
It is completely fine to leave the code cell below as-is without modifications to train your model. However, if you would like to modify the code used to train the model below, you must ensure that your changes are easily parsed by your reviewer. In other words, make sure to provide appropriate comments to describe how your code works!
You may find it useful to load saved weights to resume training. In that case, note the names of the files containing the encoder and decoder weights that you'd like to load (`encoder_file` and `decoder_file`). Then you can load the weights by using the lines below:
```python
# Load pre-trained weights before resuming training.
encoder.load_state_dict(torch.load(os.path.join('./models', encoder_file)))
decoder.load_state_dict(torch.load(os.path.join('./models', decoder_file)))
```
While trying out parameters, make sure to take extensive notes and record the settings that you used in your various training runs. In particular, you don't want to encounter a situation where you've trained a model for several hours but can't remember what settings you used :).
### A Note on Tuning Hyperparameters
To figure out how well your model is doing, you can look at how the training loss and perplexity evolve during training - and for the purposes of this project, you are encouraged to amend the hyperparameters based on this information.
However, this will not tell you if your model is overfitting to the training data, and, unfortunately, overfitting is a problem that is commonly encountered when training image captioning models.
For this project, you need not worry about overfitting. **This project does not have strict requirements regarding the performance of your model**, and you just need to demonstrate that your model has learned **_something_** when you generate captions on the test data. For now, we strongly encourage you to train your model for the suggested 3 epochs without worrying about performance; then, you should immediately transition to the next notebook in the sequence (**3_Inference.ipynb**) to see how your model performs on the test data. If your model needs to be changed, you can come back to this notebook, amend hyperparameters (if necessary), and re-train the model.
That said, if you would like to go above and beyond in this project, you can read about some approaches to minimizing overfitting in section 4.3.1 of [this paper](http://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=7505636). In the next (optional) step of this notebook, we provide some guidance for assessing the performance on the validation dataset.
```
import torch.utils.data as data
import numpy as np
import os
import requests
import time
# Open the training log file.
f = open(log_file, 'w')
old_time = time.time()
response = requests.request("GET",
"http://metadata.google.internal/computeMetadata/v1/instance/attributes/keep_alive_token",
headers={"Metadata-Flavor":"Google"})
for epoch in range(1, num_epochs+1):
for i_step in range(1, total_step+1):
if time.time() - old_time > 60:
old_time = time.time()
requests.request("POST",
"https://nebula.udacity.com/api/v1/remote/keep-alive",
headers={'Authorization': "STAR " + response.text})
# Randomly sample a caption length, and sample indices with that length.
indices = data_loader.dataset.get_train_indices()
# Create and assign a batch sampler to retrieve a batch with the sampled indices.
new_sampler = data.sampler.SubsetRandomSampler(indices=indices)
data_loader.batch_sampler.sampler = new_sampler
# Obtain the batch.
images, captions = next(iter(data_loader))
# Move batch of images and captions to GPU if CUDA is available.
images = images.to(device)
captions = captions.to(device)
# Zero the gradients.
decoder.zero_grad()
encoder.zero_grad()
# Pass the inputs through the CNN-RNN model.
features = encoder(images)
outputs = decoder(features, captions)
# Calculate the batch loss.
loss = criterion(outputs.view(-1, vocab_size), captions.view(-1))
# Backward pass.
loss.backward()
# Update the parameters in the optimizer.
optimizer.step()
# Get training statistics.
stats = 'Epoch [%d/%d], Step [%d/%d], Loss: %.4f, Perplexity: %5.4f' % (epoch, num_epochs, i_step, total_step, loss.item(), np.exp(loss.item()))
# Print training statistics (on same line).
print('\r' + stats, end="")
sys.stdout.flush()
# Print training statistics to file.
f.write(stats + '\n')
f.flush()
# Print training statistics (on different line).
if i_step % print_every == 0:
print('\r' + stats)
# Save the weights.
if epoch % save_every == 0:
torch.save(decoder.state_dict(), os.path.join('./models', 'decoder-%d.pkl' % epoch))
torch.save(encoder.state_dict(), os.path.join('./models', 'encoder-%d.pkl' % epoch))
# Close the training log file.
f.close()
```
<a id='step3'></a>
## Step 3: (Optional) Validate your Model
To assess potential overfitting, one approach is to assess performance on a validation set. If you decide to do this **optional** task, you are required to first complete all of the steps in the next notebook in the sequence (**3_Inference.ipynb**); as part of that notebook, you will write and test code (specifically, the `sample` method in the `DecoderRNN` class) that uses your RNN decoder to generate captions. That code will prove incredibly useful here.
If you decide to validate your model, please do not edit the data loader in **data_loader.py**. Instead, create a new file named **data_loader_val.py** containing the code for obtaining the data loader for the validation data. You can access:
- the validation images at filepath `'/opt/cocoapi/images/train2014/'`, and
- the validation image caption annotation file at filepath `'/opt/cocoapi/annotations/captions_val2014.json'`.
The suggested approach to validating your model involves creating a json file such as [this one](https://github.com/cocodataset/cocoapi/blob/master/results/captions_val2014_fakecap_results.json) containing your model's predicted captions for the validation images. Then, you can write your own script or use one that you [find online](https://github.com/tylin/coco-caption) to calculate the BLEU score of your model. You can read more about the BLEU score, along with other evaluation metrics (such as TEOR and Cider) in section 4.1 of [this paper](https://arxiv.org/pdf/1411.4555.pdf). For more information about how to use the annotation file, check out the [website](http://cocodataset.org/#download) for the COCO dataset.
```
# (Optional) TODO: Validate your model.
```
| github_jupyter |
# Mount google drive to colab
```
from google.colab import drive
drive.mount("/content/drive")
```
# Import libraries
```
import os
import random
import numpy as np
import shutil
import time
from PIL import Image, ImageOps
import cv2
import pandas as pd
import math
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style('darkgrid')
import tensorflow as tf
from keras import models
from keras import layers
from keras import optimizers
from keras.callbacks import EarlyStopping
from keras.callbacks import ModelCheckpoint
from keras.callbacks import LearningRateScheduler
from keras.utils import np_utils
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.preprocessing import LabelBinarizer
from sklearn.preprocessing import MinMaxScaler
from keras.preprocessing.image import ImageDataGenerator
from keras import models, layers, optimizers
from keras.callbacks import ModelCheckpoint
from keras import losses
```
# Initialize basic working directories
```
directory = "drive/MyDrive/Datasets/Sign digits/Dataset"
trainDir = "train"
testDir = "test"
os.chdir(directory)
```
# Augmented dataframes
```
augDir = "augmented/"
classNames_train = os.listdir(augDir+'train/')
classNames_test = os.listdir(augDir+'test/')
classes_train = []
data_train = []
paths_train = []
classes_test = []
data_test = []
paths_test = []
classes_val = []
data_val = []
paths_val = []
for className in range(0,10):
temp_train = os.listdir(augDir+'train/'+str(className))
temp_test = os.listdir(augDir+'test/'+str(className))
for dataFile in temp_train:
path_train = augDir+'train/'+str(className)+'/'+dataFile
paths_train.append(path_train)
classes_train .append(str(className))
testSize = [i for i in range(math.floor(len(temp_test)/2),len(temp_test))]
valSize = [i for i in range(0,math.floor(len(temp_test)/2))]
for dataFile in testSize:
path_test = augDir+'test/'+str(className)+'/'+temp_test[dataFile]
paths_test.append(path_test)
classes_test .append(str(className))
for dataFile in valSize:
path_val = augDir+'test/'+str(className)+'/'+temp_test[dataFile]
paths_val.append(path_val)
classes_val .append(str(className))
augTrain_df = pd.DataFrame({'fileNames': paths_train, 'labels': classes_train})
augTest_df = pd.DataFrame({'fileNames': paths_test, 'labels': classes_test})
augVal_df = pd.DataFrame({'fileNames': paths_val, 'labels': classes_val})
augTrain_df.head(10)
augTrain_df['labels'].hist(figsize=(10,5))
augTest_df['labels'].hist(figsize=(10,5))
augTest_df['labels'].hist(figsize=(10,5))
augVal_df['labels'].hist(figsize=(10,5))
augTrainX=[]
augTrainY=[]
augTestX=[]
augTestY=[]
augValX=[]
augValY=[]
iter = -1
#read images from train set
for path in augTrain_df['fileNames']:
iter = iter + 1
#image = np.array((Image.open(path)))
image = cv2.imread(path)
augTrainX.append(image)
label = augTrain_df['labels'][iter]
augTrainY.append(label)
iter = -1
for path in augTest_df['fileNames']:
iter = iter + 1
#image = np.array((Image.open(path)))
image = cv2.imread(path)
augTestX.append(image)
augTestY.append(augTest_df['labels'][iter])
iter = -1
for path in augVal_df['fileNames']:
iter = iter + 1
#image = np.array((Image.open(path)))
image = cv2.imread(path)
augValX.append(image)
augValY.append(augVal_df['labels'][iter])
augTrainX = np.array(augTrainX)
augTestX = np.array(augTestX)
augValX = np.array(augValX)
augTrainX = augTrainX / 255
augTestX = augTestX / 255
augValX = augValX / 255
# OneHot Encode the Output
augTrainY = np_utils.to_categorical(augTrainY, 10)
augTestY = np_utils.to_categorical(augTestY, 10)
augValY = np_utils.to_categorical(augValY, 10)
train_datagen = ImageDataGenerator(rescale=1./255)
validation_datagen = ImageDataGenerator(rescale=1./255)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_dataframe(dataframe=augTrain_df,
x_col="fileNames",
y_col="labels",
batch_size=16,
class_mode="categorical",
color_mode="grayscale",
target_size=(100,100),
shuffle=True)
validation_generator = validation_datagen.flow_from_dataframe(dataframe=augVal_df,
x_col="fileNames",
y_col="labels",
batch_size=16,
class_mode="categorical",
color_mode="grayscale",
target_size=(100,100),
shuffle=True)
test_generator = test_datagen.flow_from_dataframe(dataframe=augTest_df,
x_col="fileNames",
y_col="labels",
batch_size=16,
class_mode="categorical",
color_mode="grayscale",
target_size=(100,100),
shuffle=True)
model_best = models.Sequential()
model_best.add(layers.Conv2D(64, (3,3), input_shape=(100, 100,1), padding='same', activation='relu'))
model_best.add(layers.BatchNormalization(momentum=0.1))
model_best.add(layers.MaxPooling2D(pool_size=(2,2)))
model_best.add(layers.Conv2D(32, (3,3), padding='same', activation='relu'))
model_best.add(layers.BatchNormalization(momentum=0.1))
model_best.add(layers.MaxPooling2D(pool_size=(2,2)))
model_best.add(layers.Conv2D(16, (3,3), padding='same', activation='relu'))
model_best.add(layers.BatchNormalization(momentum=0.1))
model_best.add(layers.MaxPooling2D(pool_size=(2,2)))
model_best.add(layers.Flatten())
model_best.add(layers.Dense(128, activation='relu'))
model_best.add(layers.Dropout(0.2))
model_best.add(layers.Dense(10, activation='softmax'))
model_best.summary()
print("[INFO] Model is training...")
time1 = time.time() # to measure time taken
# Compile the model
model_best.compile(loss='categorical_crossentropy',
optimizer=optimizers.Adam(learning_rate=1e-3),
metrics=['acc'])
history_best = model_best.fit(
train_generator,
steps_per_epoch=train_generator.samples/train_generator.batch_size ,
epochs=20,
validation_data=validation_generator,
validation_steps=validation_generator.samples/validation_generator.batch_size,
)
print('Time taken: {:.1f} seconds'.format(time.time() - time1)) # to measure time taken
print("[INFO] Model is trained.")
score = model_best.evaluate(test_generator)
print('===Testing loss and accuracy===')
print('Test loss: ', score[0])
print('Test accuracy: ', score[1])
import matplotlib.pyplot as plot
plot.plot(history_best.history['acc'])
plot.plot(history_best.history['val_acc'])
plot.title('Model accuracy')
plot.ylabel('Accuracy')
plot.xlabel('Epoch')
plot.legend(['Train', 'Vall'], loc='upper left')
plot.show()
plot.plot(history_best.history['loss'])
plot.plot(history_best.history['val_loss'])
plot.title('Model loss')
plot.ylabel('Loss')
plot.xlabel('Epoch')
plot.legend(['Train', 'Vall'], loc='upper left')
plot.show()
```
| github_jupyter |
```
import numpy as np
import scipy.sparse as sp
from sklearn.datasets import load_svmlight_file
from oracle import Oracle, make_oracle
import scipy as sc
from methods import OptimizeLassoProximal, OptimizeGD, NesterovLineSearch
import matplotlib.pyplot as plt
from sklearn import linear_model
```
Решаем задачу логистической регрессии и l1-регуляризацией:
$$F(w) = - \frac{1}{N}\sum\limits_{i=1}^Ny_i\ln(\sigma_w(x_i)) + (1 - y_i)\ln(1 - \sigma_w(x_i)) + \lambda\|w\|_1,$$
где $\lambda$ -- параметр регуляризации.
Задачу решаем проксимальным градиентным методом. Убедимся сначала, что при $\lambda = 0$ наше решение совпадает с решением метода градиентного спуска с оценкой длины шага методом Нестерова.
```
orac = make_oracle('a1a.txt', penalty='l1', reg=0)
orac1 = make_oracle('a1a.txt')
x, y = load_svmlight_file('a1a.txt', zero_based=False)
m = x[0].shape[1] + 1
w0 = np.zeros((m, 1))
optimizer = OptimizeLassoProximal()
optimizer1 = OptimizeGD()
point = optimizer(orac, w0)
point1 = optimizer1(orac1, w0, NesterovLineSearch())
np.allclose(point, point1)
```
Изучим скорость сходимости метода на датасете a1a.txt ($\lambda = 0.001$)
```
def convergence_plot(xs, ys, xlabel, title=None):
plt.figure(figsize = (12, 3))
plt.xlabel(xlabel)
plt.ylabel('F(w_{k+1} - F(w_k)')
plt.plot(xs, ys)
plt.yscale('log')
if title:
plt.title(title)
plt.tight_layout()
plt.show()
orac = make_oracle('a1a.txt', penalty='l1', reg=0.001)
point = optimizer(orac, w0)
errs = optimizer.errs
title = 'lambda = 0.001'
convergence_plot(optimizer.times, errs, 'вермя работы, с', title)
convergence_plot(optimizer.orac_calls, errs, 'кол-во вызовов оракула', title)
convergence_plot(list(range(1, optimizer.n_iter + 1)), errs, 'кол-во итераций', title)
```
Заметим, что было использовано условие остановки $F(w_{k+1}) - F(w_k) \leq tol = 10^{-16}$. Из математических соображений кажется, что это ок, так как в вещественных числах сходимость последовательности равносильна её фундаментальности. Я также пытался использовать в качестве условия остановки $\|\nabla_w f(w_k)\|_2^2 / \|\nabla_w f(w_0)\|_2^2 <= tol$, где $f$ -- лосс логистической регрессии без регуляризации ($F = f + reg$), но, вообще говоря, не очень понятно, можно ли так делать, потому что оно учитывает только часть функции.
Из графиков видно, что метод обладает линейной скоростью сходимости
Изучим теперь зависимость скорости сходимости и количества ненулевых компонент в решении от параметра регуляризации $\lambda$
```
def plot(x, ys, ylabel, legend=False):
plt.figure(figsize = (12, 3))
plt.xlabel("lambda")
plt.ylabel(ylabel)
plt.plot(x, ys, 'o')
plt.xscale('log')
if legend:
plt.legend()
plt.tight_layout()
plt.show()
lambdas = [10**(-i) for i in range(8, 0, -1)]
non_zeros = []
for reg in lambdas:
orac = make_oracle('a1a.txt', penalty='l1', reg=reg)
point = optimizer(orac, w0)
convergence_plot(list(range(1, optimizer.n_iter + 1)), optimizer.errs, 'кол-во итераций',
f"lambda = {reg}")
non_zeros.append(len(np.nonzero(point)[0]))
plot(lambdas, non_zeros, '# nonzero components')
```
Видно, что параметр регуляризации практически не влияет на скорость сходимости (она всегда линейная), но количество итераций метода падает с увеличением параметра регуляризации. Так же из последнего графика делаем ожидаемый вывод, что число ненулевых компонент в решении уменьшается с ростом параметра регуляризации
Построим еще графики зависимости значения оптимизируемой функции и критерия остновки (ещё разок) в зависимости от итерации ($\lambda = 0.001$)
```
def value_plot(xs, ys, xlabel, title=None):
plt.figure(figsize = (12, 3))
plt.xlabel(xlabel)
plt.ylabel('F(w_k)')
plt.plot(xs, ys)
# plt.yscale('log')
if title:
plt.title(title)
plt.tight_layout()
plt.show()
orac = make_oracle('a1a.txt', penalty='l1', reg=0.001)
point = optimizer(orac, w0)
title = 'lambda = 0.001'
value_plot(list(range(1, optimizer.n_iter + 1)), optimizer.values, 'кол-во итераций', title)
convergence_plot(list(range(1, optimizer.n_iter + 1)), optimizer.errs, 'кол-во итераций', title)
```
Для подтверждения сделаных выводов проверим их ещё на breast-cancer_scale датасете.
Проверка равносильности GD + Nesterov и Proximal + $\lambda = 0$:
```
orac = make_oracle('breast-cancer_scale.txt', penalty='l1', reg=0)
orac1 = make_oracle('breast-cancer_scale.txt')
x, y = load_svmlight_file('breast-cancer_scale.txt', zero_based=False)
m = x[0].shape[1] + 1
w0 = np.zeros((m, 1))
optimizer = OptimizeLassoProximal()
optimizer1 = OptimizeGD()
point = optimizer(orac, w0)
point1 = optimizer1(orac1, w0, NesterovLineSearch())
np.allclose(point, point1)
print(abs(orac.value(point) - orac1.value(point1)))
```
Сами вектора весов не совпали, но значения оптимизируемой функции близки, так что будем считать, что все ок.
Изучаем скорость сходимости для $\lambda = 0.001$:
```
orac = make_oracle('breast-cancer_scale.txt', penalty='l1', reg=0.001)
point = optimizer(orac, w0)
errs = optimizer.errs
title = 'lambda = 0.001'
convergence_plot(optimizer.times, errs, 'вермя работы, с', title)
convergence_plot(optimizer.orac_calls, errs, 'кол-во вызовов оракула', title)
convergence_plot(list(range(1, optimizer.n_iter + 1)), errs, 'кол-во итераций', title)
```
Кажется, что скорость сходимости опять линейная
Изучаем зависимость скорости сходимости и количества ненулевых компонент в решении от $\lambda$
```
lambdas = [10**(-i) for i in range(8, 0, -1)]
non_zeros = []
for reg in lambdas:
orac = make_oracle('breast-cancer_scale.txt', penalty='l1', reg=reg)
point = optimizer(orac, w0)
convergence_plot(list(range(1, optimizer.n_iter + 1)), optimizer.errs, 'кол-во итераций',
f"lambda = {reg}")
non_zeros.append(len(np.nonzero(point)[0]))
plot(lambdas, non_zeros, '# nonzero components')
```
Делаем те же выводы
Построим напоследок грфики для значений оптимизируемой функции и критерия остановки (ещё разок) в зависимости от итерации ($\lambda = 0.001$)
```
orac = make_oracle('breast-cancer_scale.txt', penalty='l1', reg=0.001)
point = optimizer(orac, w0)
title = 'lambda = 0.001'
value_plot(list(range(1, optimizer.n_iter + 1)), optimizer.values, 'кол-во итераций', title)
convergence_plot(list(range(1, optimizer.n_iter + 1)), optimizer.errs, 'кол-во итераций', title)
```
Конец.
| github_jupyter |
## Implementing BERT with SNGP
```
!pip install tensorflow_text==2.7.3
!pip install -U tf-models-official==2.7.0
import matplotlib.pyplot as plt
import matplotlib.colors as colors
import sklearn.metrics
import sklearn.calibration
import tensorflow_hub as hub
import tensorflow_datasets as tfds
import numpy as np
import tensorflow as tf
import pandas as pd
import json
import official.nlp.modeling.layers as layers
import official.nlp.optimization as optimization
```
### Implement a standard BERT classifier following which classifies text
```
gpus = tf.config.list_physical_devices('GPU')
gpus
# Standard BERT model
PREPROCESS_HANDLE = 'https://tfhub.dev/tensorflow/bert_en_uncased_preprocess/3'
MODEL_HANDLE = 'https://tfhub.dev/tensorflow/bert_en_uncased_L-12_H-768_A-12/3'
class BertClassifier(tf.keras.Model):
def __init__(self,
num_classes=150, inner_dim=768, dropout_rate=0.1,
**classifier_kwargs):
super().__init__()
self.classifier_kwargs = classifier_kwargs
# Initiate the BERT encoder components.
self.bert_preprocessor = hub.KerasLayer(PREPROCESS_HANDLE, name='preprocessing')
self.bert_hidden_layer = hub.KerasLayer(MODEL_HANDLE, trainable=True, name='bert_encoder')
# Defines the encoder and classification layers.
self.bert_encoder = self.make_bert_encoder()
self.classifier = self.make_classification_head(num_classes, inner_dim, dropout_rate)
def make_bert_encoder(self):
text_inputs = tf.keras.layers.Input(shape=(), dtype=tf.string, name='text')
encoder_inputs = self.bert_preprocessor(text_inputs)
encoder_outputs = self.bert_hidden_layer(encoder_inputs)
return tf.keras.Model(text_inputs, encoder_outputs)
def make_classification_head(self, num_classes, inner_dim, dropout_rate):
return layers.ClassificationHead(
num_classes=num_classes,
inner_dim=inner_dim,
dropout_rate=dropout_rate,
**self.classifier_kwargs)
def call(self, inputs, **kwargs):
encoder_outputs = self.bert_encoder(inputs)
classifier_inputs = encoder_outputs['sequence_output']
return self.classifier(classifier_inputs, **kwargs)
```
### Build SNGP model
To implement a BERT-SNGP model designed by Google researchers
```
class ResetCovarianceCallback(tf.keras.callbacks.Callback):
def on_epoch_begin(self, epoch, logs=None):
"""Resets covariance matrix at the begining of the epoch."""
if epoch > 0:
self.model.classifier.reset_covariance_matrix()
class SNGPBertClassifier(BertClassifier):
def make_classification_head(self, num_classes, inner_dim, dropout_rate):
return layers.GaussianProcessClassificationHead(
num_classes=num_classes,
inner_dim=inner_dim,
dropout_rate=dropout_rate,
gp_cov_momentum=-1,
temperature=30.,
**self.classifier_kwargs)
def fit(self, *args, **kwargs):
"""Adds ResetCovarianceCallback to model callbacks."""
kwargs['callbacks'] = list(kwargs.get('callbacks', []))
kwargs['callbacks'].append(ResetCovarianceCallback())
return super().fit(*args, **kwargs)
```
### Load train and test datasets
```
is_train = pd.read_json('is_train.json')
is_train.columns = ['question','intent']
is_test = pd.read_json('is_test.json')
is_test.columns = ['question','intent']
oos_test = pd.read_json('oos_test.json')
oos_test.columns = ['question','intent']
is_test.shape
```
Make the train and test data.
```
#Generate codes
is_data = is_train.append(is_test)
is_data.intent = pd.Categorical(is_data.intent)
is_data['code'] = is_data.intent.cat.codes
#in-scope evaluation data
is_test = is_data[15000:19500]
is_test_queries = is_test.question
is_test_labels = is_test.intent
is_test_codes = is_test.code
is_eval_data = (tf.convert_to_tensor(is_test_queries), tf.convert_to_tensor(is_test_codes))
is_train = is_data[0:15000]
is_train_queries = is_train.question
is_train_labels = is_train.intent
is_train_codes = is_train.code
training_ds_queries = tf.convert_to_tensor(is_train_queries)
training_ds_labels = tf.convert_to_tensor(is_train_codes)
is_test.shape
```
Create a OOD evaluation dataset. For this, combine the in-scope test data 'is_test' and out-of-scope 'oos_test' data. Assign label 0 for in-scope and label 1 for out-of-scope data
```
train_size = len(is_train)
test_size = len(is_test)
oos_size = len(oos_test)
# Combines the in-domain and out-of-domain test examples.
oos_queries= tf.concat([is_test['question'], oos_test['question']], axis=0)
oos_labels = tf.constant([0] * test_size + [1] * oos_size)
# Converts into a TF dataset.
oos_eval_dataset = tf.data.Dataset.from_tensor_slices(
{"text": oos_queries, "label": oos_labels})
```
### Train and evaluate
```
TRAIN_EPOCHS = 4
TRAIN_BATCH_SIZE = 16
EVAL_BATCH_SIZE = 256
#@title
def bert_optimizer(learning_rate,
batch_size=TRAIN_BATCH_SIZE, epochs=TRAIN_EPOCHS,
warmup_rate=0.1):
"""Creates an AdamWeightDecay optimizer with learning rate schedule."""
train_data_size = train_size
steps_per_epoch = int(train_data_size / batch_size)
num_train_steps = steps_per_epoch * epochs
num_warmup_steps = int(warmup_rate * num_train_steps)
# Creates learning schedule.
lr_schedule = tf.keras.optimizers.schedules.PolynomialDecay(
initial_learning_rate=learning_rate,
decay_steps=num_train_steps,
end_learning_rate=0.0)
return optimization.AdamWeightDecay(
learning_rate=lr_schedule,
weight_decay_rate=0.01,
epsilon=1e-6,
exclude_from_weight_decay=['LayerNorm', 'layer_norm', 'bias'])
optimizer = bert_optimizer(learning_rate=1e-4)
loss = tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True)
metrics = tf.metrics.SparseCategoricalAccuracy()
fit_configs = dict(batch_size=TRAIN_BATCH_SIZE,
epochs=TRAIN_EPOCHS,
validation_batch_size=EVAL_BATCH_SIZE,
validation_data=is_eval_data)
```
### Model 1 - Batch size of 32 & 3 epochs
```
sngp_model = SNGPBertClassifier()
sngp_model.compile(optimizer=optimizer, loss=loss, metrics=metrics)
sngp_model.fit(training_ds_queries, training_ds_labels, **fit_configs)
```
### Model 2 - Batch size of 16 & 2 epochs
```
sngp_model2 = SNGPBertClassifier()
sngp_model2.compile(optimizer=optimizer, loss=loss, metrics=metrics)
sngp_model2.fit(training_ds_queries, training_ds_labels, **fit_configs)
```
### Model 3 - Batch size of 16 & 4 epochs
```
sngp_model3 = SNGPBertClassifier()
sngp_model3.compile(optimizer=optimizer, loss=loss, metrics=metrics)
sngp_model3.fit(training_ds_queries, training_ds_labels, **fit_configs)
```
### Evaluate OOD performance
Evaluate how well the model can detect the unfamiliar out-of-domain queries.
```
def oos_predict(model, ood_eval_dataset, **model_kwargs):
oos_labels = []
oos_probs = []
ood_eval_dataset = ood_eval_dataset.batch(EVAL_BATCH_SIZE)
for oos_batch in ood_eval_dataset:
oos_text_batch = oos_batch["text"]
oos_label_batch = oos_batch["label"]
pred_logits = model(oos_text_batch, **model_kwargs)
pred_probs_all = tf.nn.softmax(pred_logits, axis=-1)
pred_probs = tf.reduce_max(pred_probs_all, axis=-1)
oos_labels.append(oos_label_batch)
oos_probs.append(pred_probs)
oos_probs = tf.concat(oos_probs, axis=0)
oos_labels = tf.concat(oos_labels, axis=0)
return oos_probs, oos_labels
```
Computes the OOD probabilities as $1 - p(x)$, where $p(x)=softmax(logit(x))$ is the predictive probability.
```
sngp_probs, ood_labels = oos_predict(sngp_model, oos_eval_dataset)
sngp_probs2, ood_labels2 = oos_predict(sngp_model2, oos_eval_dataset)
sngp_probs3, ood_labels3 = oos_predict(sngp_model3, oos_eval_dataset)
ood_probs = 1 - sngp_probs
ood_probs2 = 1 - sngp_probs2
ood_probs3 = 1 - sngp_probs3
plt.rcParams['figure.dpi'] = 140
DEFAULT_X_RANGE = (-3.5, 3.5)
DEFAULT_Y_RANGE = (-2.5, 2.5)
DEFAULT_CMAP = colors.ListedColormap(["#377eb8", "#ff7f00"])
DEFAULT_NORM = colors.Normalize(vmin=0, vmax=1,)
DEFAULT_N_GRID = 100
ood_uncertainty = ood_probs * (1 - ood_probs)
ood_uncertainty2 = ood_probs2 * (1 - ood_probs2)
ood_uncertainty3 = ood_probs3 * (1 - ood_probs3)
s1 = np.array(sngp_probs.numpy())
print(s1[3000])
s2 = np.array(sngp_probs2.numpy())
print(s2[2000])
s3 = np.array(sngp_probs3.numpy())
print(s3[1000])
```
### Compute the Area under precision-recall curve (AUPRC) for OOD probability v.s. OOD detection accuracy.
```
precision, recall, _ = sklearn.metrics.precision_recall_curve(ood_labels, ood_probs)
precision2, recall2, _ = sklearn.metrics.precision_recall_curve(ood_labels2, ood_probs2)
precision3, recall3, _ = sklearn.metrics.precision_recall_curve(ood_labels3, ood_probs3)
print((precision3)
print(recall3)
```
[0.23380874 0.23362956 0.23368421 ... 1. 1. 1. ]
[1. 0.999 0.999 ... 0.002 0.001 0. ]
```
sklearn.metrics.recall_score(oos_labels, ood_labels3, average='weighted')
sklearn.metrics.precision_score(oos_labels, ood_labels3, average='weighted')
auprc = sklearn.metrics.auc(recall, precision)
print(f'SNGP AUPRC: {auprc:.4f}')
auprc2 = sklearn.metrics.auc(recall2, precision2)
print(f'SNGP AUPRC 2: {auprc2:.4f}')
auprc3 = sklearn.metrics.auc(recall3, precision3)
print(f'SNGP AUPRC 3: {auprc3:.4f}')
prob_true, prob_pred = sklearn.calibration.calibration_curve(
ood_labels, ood_probs, n_bins=10, strategy='quantile')
prob_true2, prob_pred2 = sklearn.calibration.calibration_curve(
ood_labels2, ood_probs2, n_bins=10, strategy='quantile')
prob_true3, prob_pred3 = sklearn.calibration.calibration_curve(
ood_labels3, ood_probs3, n_bins=10, strategy='quantile')
plt.plot(prob_pred, prob_true)
plt.plot([0., 1.], [0., 1.], c='k', linestyle="--")
plt.xlabel('Predictive Probability')
plt.ylabel('Predictive Accuracy')
plt.title('Calibration Plots, SNGP')
plt.show()
plt.plot(prob_pred2, prob_true2)
plt.plot([0., 1.], [0., 1.], c='k', linestyle="--")
plt.xlabel('Predictive Probability')
plt.ylabel('Predictive Accuracy')
plt.title('Calibration Plots, SNGP')
plt.show()
plt.plot(prob_pred3, prob_true3)
plt.plot([0., 1.], [0., 1.], c='k', linestyle="--")
plt.xlabel('Predictive Probability')
plt.ylabel('Predictive Accuracy')
plt.title('Calibration Plots, SNGP')
plt.show()
# calculate scores
auc1 = roc_auc_score(oos_labels, ood_probs)
auc2 = roc_auc_score(oos_labels, ood_probs2)
auc3 = roc_auc_score(oos_labels, ood_probs3)
# summarize scores
print('SNGP Model 1: ROC AUC=%.3f' % (auc1))
print('SNGP Model 2: ROC AUC=%.3f' % (auc2))
print('SNGP Model 3: ROC AUC=%.3f' % (auc3))
# calculate roc curves
fpr1, tpr1, _ = roc_curve(oos_labels, ood_probs)
fpr2, tpr2, _ = roc_curve(oos_labels, ood_probs2)
fpr3, tpr3, _ = roc_curve(oos_labels, ood_probs3)
# plot the roc curve for the model
pyplot.plot(fpr1, tpr1, marker='.', label='SNGP Model 1')
pyplot.plot(fpr2, tpr2, marker='*', label='SNGP Model 2')
pyplot.plot(fpr3, tpr3, marker='+', label='SNGP Model 3')
# axis labels
pyplot.xlabel('False Positive Rate (Precision)')
pyplot.ylabel('True Positive Rate (Recall)')
# show the legend
pyplot.legend()
# show the plot
pyplot.show()
```
| github_jupyter |
<a href="https://colab.research.google.com/github/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_02_4_pandas_functional.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# T81-558: Applications of Deep Neural Networks
**Module 2: Python for Machine Learning**
* Instructor: [Jeff Heaton](https://sites.wustl.edu/jeffheaton/), McKelvey School of Engineering, [Washington University in St. Louis](https://engineering.wustl.edu/Programs/Pages/default.aspx)
* For more information visit the [class website](https://sites.wustl.edu/jeffheaton/t81-558/).
# Module 2 Material
Main video lecture:
* Part 2.1: Introduction to Pandas [[Video]](https://www.youtube.com/watch?v=bN4UuCBdpZc&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_02_1_python_pandas.ipynb)
* Part 2.2: Categorical Values [[Video]](https://www.youtube.com/watch?v=4a1odDpG0Ho&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_02_2_pandas_cat.ipynb)
* Part 2.3: Grouping, Sorting, and Shuffling in Python Pandas [[Video]](https://www.youtube.com/watch?v=YS4wm5gD8DM&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_02_3_pandas_grouping.ipynb)
* **Part 2.4: Using Apply and Map in Pandas for Keras** [[Video]](https://www.youtube.com/watch?v=XNCEZ4WaPBY&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_02_4_pandas_functional.ipynb)
* Part 2.5: Feature Engineering in Pandas for Deep Learning in Keras [[Video]](https://www.youtube.com/watch?v=BWPTj4_Mi9E&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_02_5_pandas_features.ipynb)
# Google CoLab Instructions
The following code ensures that Google CoLab is running the correct version of TensorFlow.
```
try:
%tensorflow_version 2.x
COLAB = True
print("Note: using Google CoLab")
except:
print("Note: not using Google CoLab")
COLAB = False
```
# Part 2.4: Apply and Map
If you've ever worked with Big Data or functional programming languages before, you've likely heard of map/reduce. Map and reduce are two functions that apply a task that you create to a data frame. Pandas supports functional programming techniques that allow you to use functions across en entire data frame. In addition to functions that you write, Pandas also provides several standard functions for use with data frames.
### Using Map with Dataframes
The map function allows you to transform a column by mapping certain values in that column to other values. Consider the Auto MPG data set that contains a field **origin_name** that holds a value between one and three that indicates the geographic origin of each car. We can see how to use the map function to transform this numeric origin into the textual name of each origin.
We will begin by loading the Auto MPG data set.
```
import os
import pandas as pd
import numpy as np
df = pd.read_csv(
"https://data.heatonresearch.com/data/t81-558/auto-mpg.csv",
na_values=['NA', '?'])
pd.set_option('display.max_columns', 7)
pd.set_option('display.max_rows', 5)
display(df)
```
The **map** method in Pandas operates on a single column. You provide **map** with a dictionary of values to transform the target column. The map keys specify what values in the target column should be turned into values specified by those keys. The following code shows how the map function can transform the numeric values of 1, 2, and 3 into the string values of North America, Europe and Asia.
```
# Apply the map
df['origin_name'] = df['origin'].map(
{1: 'North America', 2: 'Europe', 3: 'Asia'})
# Shuffle the data, so that we hopefully see
# more regions.
df = df.reindex(np.random.permutation(df.index))
# Display
pd.set_option('display.max_columns', 7)
pd.set_option('display.max_rows', 10)
display(df)
```
### Using Apply with Dataframes
The **apply** function of the data frame can run a function over the entire data frame. You can use either be a traditional named function or a lambda function. Python will execute the provided function against each of the rows or columns in the data frame. The **axis** parameter specifies of the function is run across rows or columns. For axis = 1, rows are used. The following code calculates a series called **efficiency** that is the **displacement** divided by **horsepower**.
```
efficiency = df.apply(lambda x: x['displacement']/x['horsepower'], axis=1)
display(efficiency[0:10])
```
You can now insert this series into the data frame, either as a new column or to replace an existing column. The following code inserts this new series into the data frame.
```
df['efficiency'] = efficiency
```
### Feature Engineering with Apply and Map
In this section, we will see how to calculate a complex feature using map, apply, and grouping. The data set is the following CSV:
* https://www.irs.gov/pub/irs-soi/16zpallagi.csv
This URL contains US Government public data for "SOI Tax Stats - Individual Income Tax Statistics." The entry point to the website is here:
* https://www.irs.gov/statistics/soi-tax-stats-individual-income-tax-statistics-2016-zip-code-data-soi
Documentation describing this data is at the above link.
For this feature, we will attempt to estimate the adjusted gross income (AGI) for each of the zip codes. The data file contains many columns; however, you will only use the following:
* STATE - The state (e.g., MO)
* zipcode - The zipcode (e.g. 63017)
* agi_stub - Six different brackets of annual income (1 through 6)
* N1 - The number of tax returns for each of the agi_stubs
Note, the file will have six rows for each zip code, for each of the agi_stub brackets. You can skip zip codes with 0 or 99999.
We will create an output CSV with these columns; however, only one row per zip code. Calculate a weighted average of the income brackets. For example, the following six rows are present for 63017:
|zipcode |agi_stub | N1 |
|--|--|-- |
|63017 |1 | 4710 |
|63017 |2 | 2780 |
|63017 |3 | 2130 |
|63017 |4 | 2010 |
|63017 |5 | 5240 |
|63017 |6 | 3510 |
We must combine these six rows into one. For privacy reasons, AGI's are broken out into 6 buckets. We need to combine the buckets and estimate the actual AGI of a zipcode. To do this, consider the values for N1:
* 1 = 1 to 25,000
* 2 = 25,000 to 50,000
* 3 = 50,000 to 75,000
* 4 = 75,000 to 100,000
* 5 = 100,000 to 200,000
* 6 = 200,000 or more
The median of each of these ranges is approximately:
* 1 = 12,500
* 2 = 37,500
* 3 = 62,500
* 4 = 87,500
* 5 = 112,500
* 6 = 212,500
Using this you can estimate 63017's average AGI as:
```
>>> totalCount = 4710 + 2780 + 2130 + 2010 + 5240 + 3510
>>> totalAGI = 4710 * 12500 + 2780 * 37500 + 2130 * 62500
+ 2010 * 87500 + 5240 * 112500 + 3510 * 212500
>>> print(totalAGI / totalCount)
88689.89205103042
```
We begin by reading in the government data.
```
import pandas as pd
df=pd.read_csv('https://www.irs.gov/pub/irs-soi/16zpallagi.csv')
```
First, we trim all zip codes that are either 0 or 99999. We also select the three fields that we need.
```
df=df.loc[(df['zipcode']!=0) & (df['zipcode']!=99999),
['STATE','zipcode','agi_stub','N1']]
pd.set_option('display.max_columns', 0)
pd.set_option('display.max_rows', 10)
display(df)
```
We replace all of the **agi_stub** values with the correct median values with the **map** function.
```
medians = {1:12500,2:37500,3:62500,4:87500,5:112500,6:212500}
df['agi_stub']=df.agi_stub.map(medians)
pd.set_option('display.max_columns', 0)
pd.set_option('display.max_rows', 10)
display(df)
```
Next, we group the data frame by zip code.
```
groups = df.groupby(by='zipcode')
```
The program applies a lambda is applied across the groups, and then calculates the AGI estimate.
```
df = pd.DataFrame(groups.apply(
lambda x:sum(x['N1']*x['agi_stub'])/sum(x['N1']))) \
.reset_index()
pd.set_option('display.max_columns', 0)
pd.set_option('display.max_rows', 10)
display(df)
```
We can now rename the new agi_estimate column.
```
df.columns = ['zipcode','agi_estimate']
pd.set_option('display.max_columns', 0)
pd.set_option('display.max_rows', 10)
display(df)
```
Finally, we check to see that our zip code of 63017 got the correct value.
```
df[ df['zipcode']==63017 ]
```
| github_jupyter |
# Charting a path into the data science field
This project attempts to shed light on the path or paths to becoming a data science professional in the United States.
Data science is a rapidly growing field, and the demand for data scientists is outpacing supply. In the past, most Data Scientist positions went to people with PhDs in Computer Science. I wanted to know if that is changing in light of both the increased job openings and the expanding definition of data science that has come with more companies realizing the wealth of raw data they have available for analysis, and how that can help to grow and refine their businesses.
## Business Questions
1. Do you need a a formal degree?
2. What programming language(s) do data science professionals need to know?
3. What are the preferred online learning platforms to gain data science knowledge and skills?
## Data
Since 2017, Kaggle ('The world's largest data science community') has annually surveyed its users on demographics, practices, and preferences. This notebook explores the data from Kaggle's 2020 Machine Learning and Data Science survey. A caveat: Kaggle is heavy on Machine Learning and competitions, and while it claims over 8 million users the group may not be representative of the overall data science community. Additionally,survey respondents are self-selected, so we can't extrapolate any findings to the data science community as a whole, but the trends and demographics amongst Kaggle survey takers may still offer insights about data science professionals.
The first step is importing the necessary libraries and data.
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import textwrap
%matplotlib inline
from matplotlib.ticker import PercentFormatter
import warnings
warnings.filterwarnings('ignore')
df = pd.read_csv('./kaggle_survey_2020_responses.csv')
low_memory = False
```
### Initial data exploration and cleaning
Let's take a look at the survey data.
```
# Let's look at the first 5 rows of the dataset
df.head()
```
One thing we can see from this: some questions are tied to a single column, with a number of answers possible; these questions only allowed survey respondents to choose one answer from among the options. Other questions take up multiple columns, with each column tied to a specific answer; these were questions that allowed users to choose more than one option as the answer ('select all that apply'). The two types of questions will require different approaches to data preparation.
But first, we'll do some cleaning. The top row of data contains the question titles. We'll remove that, as well as the first column of survey completion time values.
```
# Removing the first column and the first row
df.drop(['Time from Start to Finish (seconds)'], axis=1, inplace=True)
df = df.loc[1:, :]
df.head()
df.shape
```
There are over 20,000 responses, with 354 answer fields.
#### Data preparation and filtering
To improve readability of visualizations, we'll aggregate some fields, shorten some labels, and re-order categories.
```
# Aggregating the nonbinary answers
df.loc[(df.Q2 == 'Prefer not to say'), 'Q2'] = 'Other Response'
df.loc[(df.Q2 == 'Prefer to self-describe'),'Q2'] = 'Other Response'
df.loc[(df.Q2 == 'Nonbinary'), 'Q2'] = 'Other Response'
# Abbreviating country name
df.loc[(df.Q3 == 'United States of America'),'Q3']='USA'
# Shortening education level descriptions
df.loc[(df.Q4 == 'Doctoral degree'),'Q4']='PhD'
df.loc[(df.Q4 == 'Master’s degree'),'Q4']='Master’s'
df.loc[(df.Q4 == 'Bachelor’s degree'),'Q4']='Bachelor’s'
df.loc[(df.Q4 == "Some college/university study without earning a bachelor’s degree"), 'Q4']='Some college/university'
df.loc[(df.Q4 == 'No formal education past high school'), 'Q4']='High school'
df.loc[(df.Q4 == 'I prefer not to answer'), 'Q4']='Prefer not to answer'
# Ordering education levels by reverse typical chronological completion
q4_order = [
'PhD',
'Master’s',
'Professional degree',
'Bachelor’s',
'Some college/university',
'High school',
'Prefer not to answer']
# Putting coding experience answers in order from shortest time to longest
q6_order = [
'I have never written code',
'< 1 years',
'1-2 years',
'3-5 years',
'5-10 years',
'10-20 years',
'20+ years']
df.loc[(df.Q37_Part_9 == 'Cloud-certification programs (direct from AWS, Azure, GCP, or similar)'), 'Q37_Part_9']='Cloud-certification programs'
df.loc[(df.Q37_Part_10 == 'University Courses (resulting in a university degree)'), 'Q37_Part_10']='University Courses resulting in a degree'
```
We're going to focus on the US answers from currently employed Kagglers.
```
# Filtering for just US responses
us_df = df[df['Q3'] == 'USA']
# Filtering to only include currently employed Kagglers
q5_order = [
'Data Scientist',
'Software Engineer',
'Data Analyst',
'Research Scientist',
'Product/Project Manager',
'Business Analyst',
'Machine Learning Engineer',
'Data Engineer',
'Statistician',
'DBA/Database Engineer',
'Other']
us_df = us_df[us_df['Q5'].isin(q5_order)]
```
We're interested in the demographic questions at the beginning, plus coding experience, coding languages used, and online learning platforms used.
```
# Filtering to only include specific question columns
us_df = us_df.loc[:, ['Q1', 'Q2', 'Q3', 'Q4', 'Q5', 'Q6', 'Q7_Part_1', 'Q7_Part_2','Q7_Part_3','Q7_Part_4','Q7_Part_5',
'Q7_Part_6', 'Q7_Part_7','Q7_Part_8','Q7_Part_9','Q7_Part_10','Q7_Part_11', 'Q7_Part_12', 'Q7_OTHER',
'Q37_Part_1', 'Q37_Part_2', 'Q37_Part_3', 'Q37_Part_4', 'Q37_Part_5', 'Q37_Part_6', 'Q37_Part_7',
'Q37_Part_8', 'Q37_Part_9', 'Q37_Part_10','Q37_Part_11', 'Q37_OTHER']]
us_df.isna().sum()
```
Not much in the way of missing values in the first 6 questions; that changes for the multiple-column questions, as expected, since users only filled in the column when they were choosing that particular option. We'll address that by converting the missing values to zeros in the helper functions.
```
us_df.shape
```
This will be the data for our analysis -- covering 1680 currently employed Kagglers in the US.
## Helper functions
A few functions to help with data visualizations. The first two plot a barchart with a corresponding list of the counts and percentages for the values; one handles single-column questions and the other handles multiple-column questions. The third and fourth are heatmap functions -- one for single-column questions, and one for multiple-column questions.
```
def list_and_bar(qnum, q_order, title):
'''
INPUT:
qnum - the y-axis variable, a single-column question
q_order - the order to display responses on the barchart
title - the title of the barchart
OUTPUT:
1. A list of responses to the selected question, in descending order
2. A horizontal barchart showing the values, in sorted order
'''
# creating a dataframe of values to include both raw counts and percentages
val_list = pd.DataFrame()
val_list['Count'] = us_df[qnum].value_counts()
pct = round(val_list * 100/us_df[qnum].count(),2)
val_list['Pct'] = pct
print(val_list)
fig, ax = plt.subplots(1, 1, figsize=(12,6))
ax = us_df[qnum].value_counts()[q_order].plot(kind='barh')
# reversing the order of y axis --
# the horizontal barchart displays values in the reverse order of a regular barchart (i.e., where the barchart might show
# a - b - c left to right, the corresponding horizontal barchart would show c at the top, and a at the bottom)
ax.invert_yaxis()
plt.title(title, fontsize = 14, fontweight = 'bold')
plt.show()
def list_and_bar_mc(mc_df, title):
'''
INPUT:
mc_df - a dataframe consisting of answers to a specific multiple-column question
title - the title of the barchart
OUTPUT:
1. A list of responses to the selected question, in descending order
2. A horizontal barchart showing the values, also in descending order
'''
print(mc_df)
fig, ax = plt.subplots(1, 1, figsize=(12,6))
mc_df['Count'].sort_values().plot(kind='barh')
plt.title(title, fontsize = 14, fontweight = 'bold')
plt.show()
def heatmap(qnum_a, qnum_b, title, order_rows, columns):
'''
INPUT:
qnum_a - the x-axis variable, a single-column question
qnum_b - the y-axis variable, a single-column question
title - the title of the heatmap, describing the variables in the visualization
order_rows - sorted order for the y-axis
columns - sorted order for the x-axis
OUTPUT:
A heatmap showing the correlation between the two chosen variables
'''
vals = us_df[[qnum_a, qnum_b]].groupby(qnum_b)[qnum_a].value_counts().unstack()
# getting the total number of responses for the columns in order to calculate the % of the total
vals_rowsums = pd.DataFrame([vals.sum(axis=0).tolist()], columns=vals.columns, index=['All'])
vals = pd.concat([vals_rowsums, vals], axis=0)
# convert to %
vals = ((vals.T / (vals.sum(axis=1) + 0.001)).T) * 100
order = order_rows
columns = columns
vals = vals.reindex(order).reindex(columns = columns)
fig, ax = plt.subplots(1, 1, figsize=[12,6])
ax = sns.heatmap(ax = ax, data = vals, cmap = 'GnBu', cbar_kws = {'format': '%.0f%%'})
plt.title(title, fontsize = 14, fontweight = 'bold')
ax.set_xlabel('')
ax.set_ylabel('')
plt.show()
def heatmap_mc(qnum, qnum_mc, title, columns, order_rows):
'''
INPUT:
qnum - the y-axis variable, a single-column question
qnum_mc - the x-axis variable, a question with multiple columns of answers
title - the title of the heatmap, describing the variables in the visualization
order_rows - sorted order for the y-axis
columns - a list of column names, representing the multiple-column answer options, ordered
OUTPUT:
1. A heatmap showing the correlation between the two specified variables
2. avg_num - the average number of answer options chosen for the multiple column question
'''
# creating a dataframe with the single-column question
df_qnum = us_df[qnum]
df_qnum = pd.DataFrame(df_qnum)
# creating a dataframe containing all the columns for a given multiple-column question
cols_mc = [col for col in us_df if col.startswith(qnum_mc)]
df_mc = us_df[cols_mc]
df_mc.columns = columns
# converting column values to binary 0 or 1 values (1 if the user chose that answer, 0 if not)
df_mc = df_mc.notnull().astype(int)
# joining the dataframes together
df_join = df_qnum.join(df_mc)
# aggregating counts for each answer option and re-ordering dataframe
df_agg = df_join.groupby([qnum]).agg('sum')
df_agg = df_agg.reindex(order_rows)
df_agg['users'] = df_join.groupby(qnum)[qnum].count()
df_agg = df_agg.div(df_agg.loc[:, 'users'], axis=0)
df_agg.drop(columns='users', inplace=True)
fig, ax = plt.subplots(1, 1, figsize=(12, 6))
ax = sns.heatmap(ax = ax, data = df_agg, cmap = 'GnBu')
cbar = ax.collections[0].colorbar
cbar.ax.yaxis.set_major_formatter(PercentFormatter(1, 0))
plt.title(title, fontsize = 14, fontweight = 'bold')
ax.set_xlabel('')
ax.set_ylabel('')
plt.show()
# finding the average number of answers chosen for the multiple column options, minus tabulations for 'None'
df_temp = df_join
df_temp.drop('None', axis = 1, inplace = True)
rowsums = df_temp.sum(axis = 1)
avg_num = round(rowsums.mean(), 2)
print('Average number of options chosen by survey respondents: ' + str(avg_num) + '.')
```
## Analysis and visualizations
We'll start by looking at the age and gender distribution, just to get an overview of the response community.
```
plt.figure(figsize=[12,6])
us_ages = us_df['Q1'].value_counts().sort_index()
sns.countplot(data = us_df, x = 'Q1', hue = 'Q2', order = us_ages.index)
plt.title('Age and Gender Distribution')
```
The survey response pool skews heavily male, with most US Kagglers between the ages of 25 and 45.
```
list_and_bar('Q6', q6_order, 'Years of Coding Experience')
```
Around 80 percent of those responding have 3 or more years experience coding.
### 1. Do you need a formal degree to become a data science professional?
Let's look at formal education, and how it correlates with job title.
```
list_and_bar('Q4', q4_order, 'Highest Level of Education Attained')
list_and_bar('Q5', q5_order, 'Current Job Title')
heatmap('Q4', 'Q5', 'Roles by Education Level', q5_order, q4_order)
```
### Question 1 analysis
With almost 49% of the responses, a Master's degree was by far the most common level of education listed, more than double the next most popular answer. Other notable observations:
* Sixty-eight percent of US Kagglers hold a Master's Degree or higher.
* Research scientists and statisticians are most likely to hold PhDs, followed by Data Scientists.
* Relatively few survey respondents (around 5%) indicate they do not have at least a Bachelor's degree.
* Only 23% of those responding hold the title of Data Scientist, but it is nonetheless the title with the highest count.
Arguably anyone who is active on Kaggle and who would complete their survey considers themself to be either in, or
interested in, the data science field, if not actively working as a Data Scientist.
### Question 2. What programming language(s) do Data Scientists need to know?
Now we'll turn to programming languages used. As this is a "Select all that apply" question, with each language option appearing as a separate column, we need to do some processing to get the data into a format for easier graphing and analysis.
```
# creating a dataframe of the language options and the number of times each language was selected
languages = pd.DataFrame()
for col in us_df.columns:
if(col.startswith('Q7_')):
language = us_df[col].value_counts()
languages = languages.append({'Language':language.index[0], 'Count':language[0]}, ignore_index=True)
languages = languages.set_index('Language')
languages = languages.sort_values(by = 'Count', ascending = False)
languages_tot = sum(languages.Count)
languages['Pct'] = round((languages['Count'] * 100 / languages_tot), 2)
list_and_bar_mc(languages, 'Programming Languages Used')
heatmap_mc('Q5', 'Q7', 'Language Use by Role', languages.index, q5_order)
heatmap_mc('Q4', 'Q7','Language Use by Education Level', languages.index, q4_order)
heatmap_mc('Q6', 'Q7', 'Language Use by Years Coding', languages.index, q6_order)
```
### Question 2 analysis
Python was the most widely used language, followed by SQL and R. Python held the top spot across almost all job roles -- only Statisticians listed another language (SQL) higher -- and for all education levels and coding experience. R enjoys widespread popularity across education level and years coding as well; SQL shows a high number of users overall, but they are more concentrated in people holding Master's or PhD degrees, working as Statisticians, Data Scientists and Data Analysts.
Kagglers reported using 2-3 languages on a regular basis.
### 3. What are the preferred online learning platforms to gain data science knowledge and skills?
Regarding online learning, Kaggle's survey asked, "On which platforms have you begun or completed data science courses? (Select all that apply)." We'll handle the answers similarly to the language data.
```
# creating a dataframe of online course providers and the number of times each was selected by users
platforms = pd.DataFrame()
for col in us_df.columns:
if(col.startswith('Q37_')):
platform = us_df[col].value_counts()
platforms = platforms.append({'Platform':platform.index[0], 'Count':platform[0]}, ignore_index=True)
platforms = platforms.set_index('Platform')
platforms = platforms.sort_values(by = 'Count', ascending=False)
platforms_tot = sum(platforms.Count)
platforms['Pct'] = round((platforms['Count'] * 100 / platforms_tot), 2)
list_and_bar_mc(platforms, 'Learning Platforms Used')
heatmap_mc('Q5', 'Q37', 'Learning Platform Use by Role', platforms.index, q5_order)
heatmap_mc('Q4', 'Q37', 'Learning Platform Use by Education Level', platforms.index, q4_order)
```
### Question 3 analysis
Coursera was the most popular response, by a good margin. Kaggle Learn, University Courses towards a degree and Udemy followed, with Datacamp and edX not far behind. Kaggle Learn is a relatively new entrant into this area, offering short, narrowly-focused, skill-based courses for free which offer certificates upon completion. These factors may all contribute to the platform's popularity, as it is easy to try out for the cost of a few hours and no money.
Kagglers reported trying data science courses on two platforms, on average.
Coursera's popularity was high across almost education levels and job titles. Kaggle Learn's usage was fairly uniform across categories. Fast.ai was popular with Research Scientists, Data Scientists, Machine Learnig Engineers, and Statisticians. Other platforms seem to enjoy popularity with some groups more than others, but not in ways that make it easy to extrapolate much.
## Conclusion
The most well-travelled path into the data science field, at least for those responding to the 2020 Kaggle survey:
* Get at least a Bachelor's degree, though a Master's degree may be preferable
* Learn at least 2 coding languages -- Python and R are the top data science languages; depending on the role you want,
you might want to get comfortable with another language, such as SQL or C.
* Take classes on online learning platforms to update your skills and learn new ones. Coursera is the standard, while
Kaggle Learn is a good option for short,targeted learning.
| github_jupyter |
# 基本程序设计
- 一切代码输入,请使用英文输入法
```
print('hello word')
print 'hello'
```
## 编写一个简单的程序
- 圆公式面积: area = radius \* radius \* 3.1415
```
radius = 1.0
area = radius * radius * 3.14 # 将后半部分的结果赋值给变量area
# 变量一定要有初始值!!!
# radius: 变量.area: 变量!
# int 类型
print(area)
```
### 在Python里面不需要定义数据的类型
## 控制台的读取与输入
- input 输入进去的是字符串
- eval
```
radius = input('请输入半径') # input得到的结果是字符串类型
radius = float(radius)
area = radius * radius * 3.14
print('面积为:',area)
```
- 在jupyter用shift + tab 键可以跳出解释文档
## 变量命名的规范
- 由字母、数字、下划线构成
- 不能以数字开头 \*
- 标识符不能是关键词(实际上是可以强制改变的,但是对于代码规范而言是极其不适合)
- 可以是任意长度
- 驼峰式命名
## 变量、赋值语句和赋值表达式
- 变量: 通俗理解为可以变化的量
- x = 2 \* x + 1 在数学中是一个方程,而在语言中它是一个表达式
- test = test + 1 \* 变量在赋值之前必须有值
## 同时赋值
var1, var2,var3... = exp1,exp2,exp3...
## 定义常量
- 常量:表示一种定值标识符,适合于多次使用的场景。比如PI
- 注意:在其他低级语言中如果定义了常量,那么,该常量是不可以被改变的,但是在Python中一切皆对象,常量也是可以被改变的
## 数值数据类型和运算符
- 在Python中有两种数值类型(int 和 float)适用于加减乘除、模、幂次
<img src = "../Photo/01.jpg"></img>
## 运算符 /、//、**
## 运算符 %
## EP:
- 25/4 多少,如果要将其转变为整数该怎么改写
- 输入一个数字判断是奇数还是偶数
- 进阶: 输入一个秒,数,写一个程序将其转换成分和秒:例如500秒等于8分20秒
- 进阶: 如果今天是星期六,那么10天以后是星期几? 提示:每个星期的第0天是星期天
```
day = eval(input('week'))
plus_day = eval(input('plus'))
```
## 计算表达式和运算优先级
<img src = "../Photo/02.png"></img>
<img src = "../Photo/03.png"></img>
## 增强型赋值运算
<img src = "../Photo/04.png"></img>
## 类型转换
- float -> int
- 四舍五入 round
## EP:
- 如果一个年营业税为0.06%,那么对于197.55e+2的年收入,需要交税为多少?(结果保留2为小数)
- 必须使用科学计数法
# Project
- 用Python写一个贷款计算器程序:输入的是月供(monthlyPayment) 输出的是总还款数(totalpayment)

# Homework
- 1
<img src="../Photo/06.png"></img>
```
celsius = input('请输入温度')
celsius = float(celsius)
fahrenheit = (9/5) * celsius + 32
print(celsius,'Celsius is',fahrenheit,'Fahrenheit')
```
- 2
<img src="../Photo/07.png"></img>
```
radius = input('请输入半径')
length = input('请输入高')
radius = float(radius)
length = float(length)
area = radius * radius * 3.14
volume = area * length
print('The area is',area)
print('The volume is',volume)
```
- 3
<img src="../Photo/08.png"></img>
```
feet = input('请输入英尺')
feet = float(feet)
meter = feet * 0.305
print(feet,'feet is',meter,'meters')
```
- 4
<img src="../Photo/10.png"></img>
```
M = input('请输入水量')
initial = input('请输入初始温度')
final = input('请输入最终温度')
M = float(M)
initial = float(initial)
final = float(final)
Q = M * (final - initial) * 4184
print('The energy needed is ',Q)
```
- 5
<img src="../Photo/11.png"></img>
```
cha = input('请输入差额')
rate = input('请输入年利率')
cha = float(cha)
rate = float(rate)
interest = cha * (rate/1200)
print(interest)
```
- 6
<img src="../Photo/12.png"></img>
```
start = input('请输入初始速度')
end = input('请输入末速度')
time = input('请输入时间')
start = float(start)
end =float(end)
time = float(time)
a = (end - start)/time
print(a)
```
- 7 进阶
<img src="../Photo/13.png"></img>
- 8 进阶
<img src="../Photo/14.png"></img>
```
a,b = eval(input('>>'))
print(a,b)
print(type(a),type(b))
a = eval(input('>>'))
print(a)
```
| github_jupyter |
```
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
from __future__ import unicode_literals
import torch
from torch.jit import script, trace
import torch.nn as nn
from torch import optim
import torch.nn.functional as F
import csv
import random
import re
import os
import unicodedata
import codecs
from io import open
import itertools
import math
import gensim
USE_CUDA = torch.cuda.is_available()
device = torch.device("cuda" if USE_CUDA else "cpu")
```
# Load & Preprocess Data
### Cornell Movie Dialogues Corpus
```
corpus_name = "cornell movie-dialogs corpus"
corpus = os.path.join("data", corpus_name)
def printLines(file, n=10):
with open(file, 'rb') as datafile:
lines = datafile.readlines()
for line in lines[:n]:
print(line)
printLines(os.path.join(corpus, "movie_lines.txt"))
# Splits each line of the file into a dictionary of fields
def loadLines(fileName, fields):
lines = {}
with open(fileName, 'r', encoding='iso-8859-1') as f:
for line in f:
values = line.split(" +++$+++ ")
# Extract fields
lineObj = {}
for i, field in enumerate(fields):
lineObj[field] = values[i]
lines[lineObj['lineID']] = lineObj
return lines
# Groups fields of lines from `loadLines` into conversations based on *movie_conversations.txt*
def loadConversations(fileName, lines, fields):
conversations = []
with open(fileName, 'r', encoding='iso-8859-1') as f:
for line in f:
values = line.split(" +++$+++ ")
# Extract fields
convObj = {}
for i, field in enumerate(fields):
convObj[field] = values[i]
# Convert string to list (convObj["utteranceIDs"] == "['L598485', 'L598486', ...]")
utterance_id_pattern = re.compile('L[0-9]+')
lineIds = utterance_id_pattern.findall(convObj["utteranceIDs"])
# Reassemble lines
convObj["lines"] = []
for lineId in lineIds:
convObj["lines"].append(lines[lineId])
conversations.append(convObj)
return conversations
# Extracts pairs of sentences from conversations
def extractSentencePairs(conversations):
qa_pairs = []
for conversation in conversations:
# Iterate over all the lines of the conversation
for i in range(len(conversation["lines"]) - 1): # We ignore the last line (no answer for it)
inputLine = conversation["lines"][i]["text"].strip()
targetLine = conversation["lines"][i+1]["text"].strip()
# Filter wrong samples (if one of the lists is empty)
if inputLine and targetLine:
qa_pairs.append([inputLine, targetLine])
return qa_pairs
# Define path to new file
datafile = os.path.join(corpus, "formatted_movie_lines.txt")
delimiter = '\t'
# Unescape the delimiter
delimiter = str(codecs.decode(delimiter, "unicode_escape"))
# Initialize lines dict, conversations list, and field ids
lines = {}
conversations = []
MOVIE_LINES_FIELDS = ["lineID", "characterID", "movieID", "character", "text"]
MOVIE_CONVERSATIONS_FIELDS = ["character1ID", "character2ID", "movieID", "utteranceIDs"]
# Load lines and process conversations
print("\nProcessing corpus...")
lines = loadLines(os.path.join(corpus, "movie_lines.txt"), MOVIE_LINES_FIELDS)
print("\nLoading conversations...")
conversations = loadConversations(os.path.join(corpus, "movie_conversations.txt"),
lines, MOVIE_CONVERSATIONS_FIELDS)
# Write new csv file
print("\nWriting newly formatted file...")
with open(datafile, 'w', encoding='utf-8') as outputfile:
writer = csv.writer(outputfile, delimiter=delimiter, lineterminator='\n')
for pair in extractSentencePairs(conversations):
writer.writerow(pair)
# Print a sample of lines
print("\nSample lines from file:")
printLines(datafile)
# Default word tokens
PAD_token = 0 # Used for padding short sentences
SOS_token = 1 # Start-of-sentence token
EOS_token = 2 # End-of-sentence token
class Voc:
def __init__(self, name):
self.name = name
self.trimmed = False
self.word2index = {}
self.word2count = {}
self.index2word = {PAD_token: "PAD", SOS_token: "SOS", EOS_token: "EOS"}
self.num_words = 3 # Count SOS, EOS, PAD
def addSentence(self, sentence):
for word in sentence.split(' '):
self.addWord(word)
def addWord(self, word):
if word not in self.word2index:
self.word2index[word] = self.num_words
self.word2count[word] = 1
self.index2word[self.num_words] = word
self.num_words += 1
else:
self.word2count[word] += 1
# Remove words below a certain count threshold
def trim(self, min_count):
if self.trimmed:
return
self.trimmed = True
keep_words = []
for k, v in self.word2count.items():
if v >= min_count:
keep_words.append(k)
print('keep_words {} / {} = {:.4f}'.format(
len(keep_words), len(self.word2index), len(keep_words) / len(self.word2index)
))
# Reinitialize dictionaries
self.word2index = {}
self.word2count = {}
self.index2word = {PAD_token: "PAD", SOS_token: "SOS", EOS_token: "EOS"}
self.num_words = 3 # Count default tokens
for word in keep_words:
self.addWord(word)
MAX_LENGTH = 10 # Maximum sentence length to consider
# Turn a Unicode string to plain ASCII, thanks to
# https://stackoverflow.com/a/518232/2809427
def unicodeToAscii(s):
return ''.join(
c for c in unicodedata.normalize('NFD', s)
if unicodedata.category(c) != 'Mn'
)
# Lowercase, trim, and remove non-letter characters
def normalizeString(s):
s = unicodeToAscii(s.lower().strip())
s = re.sub(r"([.!?])", r" \1", s)
s = re.sub(r"[^a-zA-Z.!?']+", r" ", s)
s = re.sub(r"\s+", r" ", s).strip()
return s
# Read query/response pairs and return a voc object
def readVocs(datafile, corpus_name):
print("Reading lines...")
# Read the file and split into lines
lines = open(datafile, encoding='utf-8').\
read().strip().split('\n')
# Split every line into pairs and normalize
pairs = [[normalizeString(s) for s in l.split('\t')] for l in lines]
voc = Voc(corpus_name)
return voc, pairs
# Returns True iff both sentences in a pair 'p' are under the MAX_LENGTH threshold
def filterPair(p):
# Input sequences need to preserve the last word for EOS token
return len(p[0].split(' ')) < MAX_LENGTH and len(p[1].split(' ')) < MAX_LENGTH
# Filter pairs using filterPair condition
def filterPairs(pairs):
return [pair for pair in pairs if filterPair(pair)]
# Using the functions defined above, return a populated voc object and pairs list
def loadPrepareData(corpus, corpus_name, datafile, save_dir):
print("Start preparing training data ...")
voc, pairs = readVocs(datafile, corpus_name)
print("Read {!s} sentence pairs".format(len(pairs)))
pairs = filterPairs(pairs)
print("Trimmed to {!s} sentence pairs".format(len(pairs)))
print("Counting words...")
for pair in pairs:
voc.addSentence(pair[0])
voc.addSentence(pair[1])
print("Counted words:", voc.num_words)
return voc, pairs
# Load/Assemble voc and pairs
save_dir = os.path.join("data", "save")
voc, pairs = loadPrepareData(corpus, corpus_name, datafile, save_dir)
# Print some pairs to validate
print("\npairs:")
for pair in pairs[:10]:
print(pair)
MIN_COUNT = 3 # Minimum word count threshold for trimming
def trimRareWords(voc, pairs, MIN_COUNT):
# Trim words used under the MIN_COUNT from the voc
voc.trim(MIN_COUNT)
# Filter out pairs with trimmed words
keep_pairs = []
for pair in pairs:
input_sentence = pair[0]
output_sentence = pair[1]
keep_input = True
keep_output = True
# Check input sentence
for word in input_sentence.split(' '):
if word not in voc.word2index:
keep_input = False
break
# Check output sentence
for word in output_sentence.split(' '):
if word not in voc.word2index:
keep_output = False
break
# Only keep pairs that do not contain trimmed word(s) in their input or output sentence
if keep_input and keep_output:
keep_pairs.append(pair)
print("Trimmed from {} pairs to {}, {:.4f} of total".format(len(pairs), len(keep_pairs), len(keep_pairs) / len(pairs)))
return keep_pairs
# Trim voc and pairs
pairs = trimRareWords(voc, pairs, MIN_COUNT)
```
# Prepare Data for Models
```
def indexesFromSentence(voc, sentence):
return [voc.word2index[word] for word in sentence.split(' ')] + [EOS_token]
def zeroPadding(l, fillvalue=PAD_token):
return list(itertools.zip_longest(*l, fillvalue=fillvalue))
def binaryMatrix(l, value=PAD_token):
m = []
for i, seq in enumerate(l):
m.append([])
for token in seq:
if token == PAD_token:
m[i].append(0)
else:
m[i].append(1)
return m
# Returns padded input sequence tensor and lengths
def inputVar(l, voc):
indexes_batch = [indexesFromSentence(voc, sentence) for sentence in l]
lengths = torch.tensor([len(indexes) for indexes in indexes_batch])
padList = zeroPadding(indexes_batch)
padVar = torch.LongTensor(padList)
return padVar, lengths
# Returns padded target sequence tensor, padding mask, and max target length
def outputVar(l, voc):
indexes_batch = [indexesFromSentence(voc, sentence) for sentence in l]
max_target_len = max([len(indexes) for indexes in indexes_batch])
padList = zeroPadding(indexes_batch)
mask = binaryMatrix(padList)
mask = torch.BoolTensor(mask)
padVar = torch.LongTensor(padList)
return padVar, mask, max_target_len
# Returns all items for a given batch of pairs
def batch2TrainData(voc, pair_batch):
pair_batch.sort(key=lambda x: len(x[0].split(" ")), reverse=True)
input_batch, output_batch = [], []
for pair in pair_batch:
input_batch.append(pair[0])
output_batch.append(pair[1])
inp, lengths = inputVar(input_batch, voc)
output, mask, max_target_len = outputVar(output_batch, voc)
return inp, lengths, output, mask, max_target_len
# Example for validation
small_batch_size = 5
batches = batch2TrainData(voc, [random.choice(pairs) for _ in range(small_batch_size)])
input_variable, lengths, target_variable, mask, max_target_len = batches
print("input_variable:", input_variable)
print("lengths:", lengths)
print("target_variable:", target_variable)
print("mask:", mask)
print("max_target_len:", max_target_len)
```
# Encoder
```
class EncoderRNN(nn.Module):
def __init__(self, hidden_size, embedding, n_layers=1, dropout=0):
super(EncoderRNN, self).__init__()
self.n_layers = n_layers
self.hidden_size = hidden_size
self.embedding = embedding
# Initialize GRU; the input_size and hidden_size params are both set to 'hidden_size'
# because our input size is a word embedding with number of features == hidden_size
self.gru = nn.GRU(hidden_size, hidden_size, n_layers,
dropout=(0 if n_layers == 1 else dropout), bidirectional=True)
def forward(self, input_seq, input_lengths, hidden=None):
# Convert word indexes to embeddings
embedded = self.embedding(input_seq)
# Pack padded batch of sequences for RNN module
packed = nn.utils.rnn.pack_padded_sequence(embedded, input_lengths)
# Forward pass through GRU
outputs, hidden = self.gru(packed, hidden)
# Unpack padding
outputs, _ = nn.utils.rnn.pad_packed_sequence(outputs)
# Sum bidirectional GRU outputs
outputs = outputs[:, :, :self.hidden_size] + outputs[:, : ,self.hidden_size:]
# Return output and final hidden state
return outputs, hidden
```
# Decoder
```
# Luong attention layer
class Attn(nn.Module):
def __init__(self, method, hidden_size):
super(Attn, self).__init__()
self.method = method
if self.method not in ['dot', 'general', 'concat']:
raise ValueError(self.method, "is not an appropriate attention method.")
self.hidden_size = hidden_size
if self.method == 'general':
self.attn = nn.Linear(self.hidden_size, hidden_size)
elif self.method == 'concat':
self.attn = nn.Linear(self.hidden_size * 2, hidden_size)
self.v = nn.Parameter(torch.FloatTensor(hidden_size))
def dot_score(self, hidden, encoder_output):
return torch.sum(hidden * encoder_output, dim=2)
def general_score(self, hidden, encoder_output):
energy = self.attn(encoder_output)
return torch.sum(hidden * energy, dim=2)
def concat_score(self, hidden, encoder_output):
energy = self.attn(torch.cat((hidden.expand(encoder_output.size(0), -1, -1), encoder_output), 2)).tanh()
return torch.sum(self.v * energy, dim=2)
def forward(self, hidden, encoder_outputs):
# Calculate the attention weights (energies) based on the given method
if self.method == 'general':
attn_energies = self.general_score(hidden, encoder_outputs)
elif self.method == 'concat':
attn_energies = self.concat_score(hidden, encoder_outputs)
elif self.method == 'dot':
attn_energies = self.dot_score(hidden, encoder_outputs)
# Transpose max_length and batch_size dimensions
attn_energies = attn_energies.t()
# Return the softmax normalized probability scores (with added dimension)
return F.softmax(attn_energies, dim=1).unsqueeze(1)
class LuongAttnDecoderRNN(nn.Module):
def __init__(self, attn_model, embedding, hidden_size, output_size, n_layers=1, dropout=0.1):
super(LuongAttnDecoderRNN, self).__init__()
# Keep for reference
self.attn_model = attn_model
self.hidden_size = hidden_size
self.output_size = output_size
self.n_layers = n_layers
self.dropout = dropout
# Define layers
self.embedding = embedding
self.embedding_dropout = nn.Dropout(dropout)
self.gru = nn.GRU(hidden_size, hidden_size, n_layers, dropout=(0 if n_layers == 1 else dropout))
self.concat = nn.Linear(hidden_size * 2, hidden_size)
self.out = nn.Linear(hidden_size, output_size)
self.attn = Attn(attn_model, hidden_size)
def forward(self, input_step, last_hidden, encoder_outputs):
# Note: we run this one step (word) at a time
# Get embedding of current input word
embedded = self.embedding(input_step)
embedded = self.embedding_dropout(embedded)
# Forward through unidirectional GRU
rnn_output, hidden = self.gru(embedded, last_hidden)
# Calculate attention weights from the current GRU output
attn_weights = self.attn(rnn_output, encoder_outputs)
# Multiply attention weights to encoder outputs to get new "weighted sum" context vector
context = attn_weights.bmm(encoder_outputs.transpose(0, 1))
# Concatenate weighted context vector and GRU output using Luong eq. 5
rnn_output = rnn_output.squeeze(0)
context = context.squeeze(1)
concat_input = torch.cat((rnn_output, context), 1)
concat_output = torch.tanh(self.concat(concat_input))
# Predict next word using Luong eq. 6
output = self.out(concat_output)
output = F.softmax(output, dim=1)
# Return output and final hidden state
return output, hidden
```
# Training Procedure
```
def maskNLLLoss(inp, target, mask):
nTotal = mask.sum()
crossEntropy = -torch.log(torch.gather(inp, 1, target.view(-1, 1)).squeeze(1))
loss = crossEntropy.masked_select(mask).mean()
loss = loss.to(device)
return loss, nTotal.item()
def train(input_variable, lengths, target_variable, mask, max_target_len, encoder, decoder, embedding,
encoder_optimizer, decoder_optimizer, batch_size, clip, max_length=MAX_LENGTH):
# Zero gradients
encoder_optimizer.zero_grad()
decoder_optimizer.zero_grad()
# Set device options
input_variable = input_variable.to(device)
target_variable = target_variable.to(device)
mask = mask.to(device)
# Lengths for rnn packing should always be on the cpu
lengths = lengths.to("cpu")
# Initialize variables
loss = 0
print_losses = []
n_totals = 0
# Forward pass through encoder
encoder_outputs, encoder_hidden = encoder(input_variable, lengths)
# Create initial decoder input (start with SOS tokens for each sentence)
decoder_input = torch.LongTensor([[SOS_token for _ in range(batch_size)]])
decoder_input = decoder_input.to(device)
# Set initial decoder hidden state to the encoder's final hidden state
decoder_hidden = encoder_hidden[:decoder.n_layers]
# Determine if we are using teacher forcing this iteration
use_teacher_forcing = True if random.random() < teacher_forcing_ratio else False
# Forward batch of sequences through decoder one time step at a time
if use_teacher_forcing:
for t in range(max_target_len):
decoder_output, decoder_hidden = decoder(
decoder_input, decoder_hidden, encoder_outputs
)
# Teacher forcing: next input is current target
decoder_input = target_variable[t].view(1, -1)
# Calculate and accumulate loss
mask_loss, nTotal = maskNLLLoss(decoder_output, target_variable[t], mask[t])
loss += mask_loss
print_losses.append(mask_loss.item() * nTotal)
n_totals += nTotal
else:
for t in range(max_target_len):
decoder_output, decoder_hidden = decoder(
decoder_input, decoder_hidden, encoder_outputs
)
# No teacher forcing: next input is decoder's own current output
_, topi = decoder_output.topk(1)
decoder_input = torch.LongTensor([[topi[i][0] for i in range(batch_size)]])
decoder_input = decoder_input.to(device)
# Calculate and accumulate loss
mask_loss, nTotal = maskNLLLoss(decoder_output, target_variable[t], mask[t])
loss += mask_loss
print_losses.append(mask_loss.item() * nTotal)
n_totals += nTotal
# Perform backpropatation
loss.backward()
# Clip gradients: gradients are modified in place
_ = nn.utils.clip_grad_norm_(encoder.parameters(), clip)
_ = nn.utils.clip_grad_norm_(decoder.parameters(), clip)
# Adjust model weights
encoder_optimizer.step()
decoder_optimizer.step()
return sum(print_losses) / n_totals
def trainIters(model_name, voc, pairs, encoder, decoder, encoder_optimizer, decoder_optimizer, embedding, encoder_n_layers, decoder_n_layers, save_dir, n_iteration, batch_size, print_every, save_every, clip, corpus_name, loadFilename):
# Load batches for each iteration
training_batches = [batch2TrainData(voc, [random.choice(pairs) for _ in range(batch_size)])
for _ in range(n_iteration)]
# Initializations
print('Initializing ...')
start_iteration = 1
print_loss = 0
if loadFilename:
start_iteration = checkpoint['iteration'] + 1
# Training loop
print("Training...")
for iteration in range(start_iteration, n_iteration + 1):
training_batch = training_batches[iteration - 1]
# Extract fields from batch
input_variable, lengths, target_variable, mask, max_target_len = training_batch
# Run a training iteration with batch
loss = train(input_variable, lengths, target_variable, mask, max_target_len, encoder,
decoder, embedding, encoder_optimizer, decoder_optimizer, batch_size, clip)
print_loss += loss
# Print progress
if iteration % print_every == 0:
print_loss_avg = print_loss / print_every
print("Iteration: {}; Percent complete: {:.1f}%; Average loss: {:.4f}".format(iteration, iteration / n_iteration * 100, print_loss_avg))
print_loss = 0
# Save checkpoint
if (iteration % save_every == 0):
directory = os.path.join(save_dir, model_name, corpus_name, '{}-{}_{}'.format(encoder_n_layers, decoder_n_layers, hidden_size))
if not os.path.exists(directory):
os.makedirs(directory)
torch.save({
'iteration': iteration,
'en': encoder.state_dict(),
'de': decoder.state_dict(),
'en_opt': encoder_optimizer.state_dict(),
'de_opt': decoder_optimizer.state_dict(),
'loss': loss,
'voc_dict': voc.__dict__,
'embedding': embedding.state_dict()
}, os.path.join(directory, '{}_{}.tar'.format(iteration, 'checkpoint')))
```
# Evaluation
```
class GreedySearchDecoder(nn.Module):
def __init__(self, encoder, decoder, voc):
super(GreedySearchDecoder, self).__init__()
self.encoder = encoder
self.decoder = decoder
self.voc = voc
def forward(self, input_seq, input_length, max_length):
# Forward input through encoder model
encoder_outputs, encoder_hidden = self.encoder(input_seq, input_length)
# Prepare encoder's final hidden layer to be first hidden input to the decoder
decoder_hidden = encoder_hidden[:decoder.n_layers]
# Initialize decoder input with SOS_token
decoder_input = torch.ones(1, 1, device=device, dtype=torch.long) * SOS_token
# Initialize tensors to append decoded words to
all_tokens = torch.zeros([0], device=device, dtype=torch.long)
all_scores = torch.zeros([0], device=device)
# Iteratively decode one word token at a time
for _ in range(max_length):
# Forward pass through decoder
decoder_output, decoder_hidden = self.decoder(decoder_input, decoder_hidden, encoder_outputs)
# Obtain most likely word token and its softmax score
decoder_scores, decoder_input = torch.max(decoder_output, dim=1)
# Print words and scores
# print('all tokens', all_tokens)
print('all tokens words', [voc.index2word[token.item()] for token in all_tokens])
if all_tokens.nelement() > 0 and int(decoder_input[0]) == self.voc.word2index['.']: # and int(all_tokens[-1]) == 2
decoder_scores, decoder_input = torch.kthvalue(decoder_output, 2)
# Record token and score
all_tokens = torch.cat((all_tokens, decoder_input), dim=0)
all_scores = torch.cat((all_scores, decoder_scores), dim=0)
# Prepare current token to be next decoder input (add a dimension)
decoder_input = torch.unsqueeze(decoder_input, 0)
# Return collections of word tokens and scores
return all_tokens, all_scores
def evaluate(encoder, decoder, searcher, voc, sentence, max_length=MAX_LENGTH):
### Format input sentence as a batch
# words -> indexes
indexes_batch = [indexesFromSentence(voc, sentence)]
# Create lengths tensor
lengths = torch.tensor([len(indexes) for indexes in indexes_batch])
# Transpose dimensions of batch to match models' expectations
input_batch = torch.LongTensor(indexes_batch).transpose(0, 1)
# Use appropriate device
input_batch = input_batch.to(device)
lengths = lengths.to("cpu")
# Decode sentence with searcher
tokens, scores = searcher(input_batch, lengths, max_length)
# indexes -> words
decoded_words = [voc.index2word[token.item()] for token in tokens]
return decoded_words
def evaluateInput(encoder, decoder, searcher, voc):
input_sentence = ''
while True:
try:
# Get input sentence
input_sentence = input('> ')
# Check if it is quit case
if input_sentence == 'q' or input_sentence == 'quit': break
# Normalize sentence
input_sentence = normalizeString(input_sentence)
# Evaluate sentence
output_words = evaluate(encoder, decoder, searcher, voc, input_sentence)
# Format and print response sentence
output_words[:] = [x for x in output_words if not (x == 'EOS' or x == 'PAD')] # or x == '.'
print('human:', input_sentence)
print('Bot:', ' '.join(output_words))
except KeyError:
print("Error: Encountered unknown word.")
```
# Embeddings
```
# load pre-trained word2Vec model
import gensim.downloader as api
model = api.load('word2vec-google-news-300')
weights_w2v = torch.FloatTensor(model.vectors)
# load pre-trained Gloves 42B-300d model
# model = gensim.models.KeyedVectors.load_word2vec_format('glove.42B.300d.w2vformat.txt')
corpus = os.path.join("glove", "glove.42B.300d.w2vformat.txt")
model = gensim.models.KeyedVectors.load_word2vec_format(corpus)
weights_42b = torch.FloatTensor(model.vectors)
# load pre-trained Gloves 6B-300d model
corpus = os.path.join("glove", "glove.6B.300d.w2vformat.txt")
model = gensim.models.KeyedVectors.load_word2vec_format(corpus)
weights_6b = torch.FloatTensor(model.vectors)
# Configure models
model_name = 'cb_model'
# attn_model = 'dot'
#attn_model = 'general'
attn_model = 'concat'
hidden_size = 300 # 500 -> 300 to fit Gloves model
encoder_n_layers = 3 # 2 -> 3
decoder_n_layers = 3 # 2 -> 3
dropout = 0.1
batch_size = 64
# Set checkpoint to load from; set to None if starting from scratch
loadFilename = None
checkpoint_iter = 5000
# loadFilename = os.path.join(save_dir, model_name, corpus_name,
# '{}-{}_{}'.format(encoder_n_layers, decoder_n_layers, hidden_size),
# '{}_checkpoint.tar'.format(checkpoint_iter))
# Load model if a loadFilename is provided
if loadFilename:
# If loading on same machine the model was trained on
checkpoint = torch.load(loadFilename)
# If loading a model trained on GPU to CPU
#checkpoint = torch.load(loadFilename, map_location=torch.device('cpu'))
encoder_sd = checkpoint['en']
decoder_sd = checkpoint['de']
encoder_optimizer_sd = checkpoint['en_opt']
decoder_optimizer_sd = checkpoint['de_opt']
embedding_sd = checkpoint['embedding']
voc.__dict__ = checkpoint['voc_dict']
print('Building encoder and decoder ...')
# Initialize word embeddings
# embedding = nn.Embedding(voc.num_words, hidden_size)
embedding = nn.Embedding.from_pretrained(weights_w2v) # Choose embedding model
if loadFilename:
embedding.load_state_dict(embedding_sd)
# Initialize encoder & decoder models
encoder = EncoderRNN(hidden_size, embedding, encoder_n_layers, dropout)
decoder = LuongAttnDecoderRNN(attn_model, embedding, hidden_size, voc.num_words, decoder_n_layers, dropout)
if loadFilename:
encoder.load_state_dict(encoder_sd)
decoder.load_state_dict(decoder_sd)
# Use appropriate device
encoder = encoder.to(device)
decoder = decoder.to(device)
print('Models built and ready to go!')
```
# Run Model
### Training
```
# Configure training/optimization
clip = 50.0
teacher_forcing_ratio = 1.0
learning_rate = 0.0001
decoder_learning_ratio = 6.0 # 5.0 -> 4.0
n_iteration = 5000 # 4000 -> 5000
print_every = 1
save_every = 500
# Ensure dropout layers are in train mode
encoder.train()
decoder.train()
# Initialize optimizers
print('Building optimizers ...')
encoder_optimizer = optim.Adam(encoder.parameters(), lr=learning_rate)
decoder_optimizer = optim.Adam(decoder.parameters(), lr=learning_rate * decoder_learning_ratio)
if loadFilename:
encoder_optimizer.load_state_dict(encoder_optimizer_sd)
decoder_optimizer.load_state_dict(decoder_optimizer_sd)
# If you have cuda, configure cuda to call
for state in encoder_optimizer.state.values():
for k, v in state.items():
if isinstance(v, torch.Tensor):
state[k] = v.cuda()
for state in decoder_optimizer.state.values():
for k, v in state.items():
if isinstance(v, torch.Tensor):
state[k] = v.cuda()
# Run training iterations
print("Starting Training!")
trainIters(model_name, voc, pairs, encoder, decoder, encoder_optimizer, decoder_optimizer,
embedding, encoder_n_layers, decoder_n_layers, save_dir, n_iteration, batch_size,
print_every, save_every, clip, corpus_name, loadFilename)
```
### Evaluation
```
# Set dropout layers to eval mode
encoder.eval()
decoder.eval()
# Initialize search module
searcher = GreedySearchDecoder(encoder, decoder, voc)
evaluateInput(encoder, decoder, searcher, voc)
```
| github_jupyter |
# 0.0. IMPORTS
```
import math
import pandas as pd
import inflection
import numpy as np
import seaborn as sns
import matplotlib as plt
import datetime
from IPython.display import Image
```
## 0.1. Helper Functions
## 0.2. Loading Data
```
# read_csv é um metodo da classe Pandas
# Preciso "unzipar" o arquivo antes?
# low_memory para dizer se ele lê o arquivo todo (False) ou em pedações (True), ele costuma avisar qual o melhor para a situação
df_sales_raw = pd.read_csv("data/train.csv.zip", low_memory=False)
df_store_raw = pd.read_csv("data/store.csv", low_memory=False)
# Merge (arquivo de referencia, arquivo a ser anexado a essa referencia, como quero fazer o merge, coluna que é igual nos 2 datasets para servir de chave )
# Merge também é um método da classe Pandas
df_raw = pd.merge( df_sales_raw, df_store_raw, how="left", on="Store" )
df_sales_raw.head()
df_store_raw.head()
# Plotar uma linha aleatória para ver se deu certo com o método sample
df_raw.sample()
```
# 1.0. STEP 01 - DATA DESCRIPTION
```
df1 = df_raw.copy()
```
## 1.1. Rename Columns
### Para ganhar velocidade no desenvolvimento!
```
df_raw.columns
# Estão até bem organizadas, formato candle (ou camble?) case, mas no mundo real pode ser bem diferente! rs
cols_old = ['Store', 'DayOfWeek', 'Date', 'Sales', 'Customers', 'Open', 'Promo',
'StateHoliday', 'SchoolHoliday', 'StoreType', 'Assortment',
'CompetitionDistance', 'CompetitionOpenSinceMonth',
'CompetitionOpenSinceYear', 'Promo2', 'Promo2SinceWeek',
'Promo2SinceYear', 'PromoInterval']
snakecase = lambda x: inflection.underscore( x )
cols_new = list( map( snakecase, cols_old) )
# Rename
df1.columns = cols_new
df1.columns
```
## 1.2. Data Dimensions
### Saber qual a quantidade de linhas e colunas do dataset
```
# O shape printa linhas e colunas do dataframe em que primeiro elemento são as rows
# Pq ali são as chaves que ele usa? Isso tem a ver com placeholder?
print( "Number of Rows: {}".format( df1.shape[0] ) )
print( "Number of Cols: {}".format( df1.shape[1] ) )
```
## 1.3. Data Types
```
# Atente que não usamos os parênteses aqui. Isso pq estamos vendo uma propriedade e não usando um método?
# O default do pandas é assumir o que não for int como object. Object é o "caracter" dentro do Pandas
# Atente para o date, precisamos mudar de object para datetime!
df1.dtypes
df1["date"] = pd.to_datetime( df1["date"] )
df1.dtypes
```
## 1.4. Check NA
```
# O método isna vai mostrar todas as linhas que tem pelo menos uma coluna com um NA (vazia)
# Mas como eu quero ver a soma disso por coluna, uso o método sum
df1.isna().sum()
# Precisamos tratar esses NAs.
# Existem basicamente 3 maneiras:
# 1. Descartar essas linhas (fácil e rápido; mas jogando dado fora)
# 2. Usando algoritmos de machine learning. Tem alguns metodos de input NA que voce pode, por exemplo, substituir as colunas vazias pelo proprio comportamento da coluna (e.g. mediana, media...)
# 3. Entendendo o negócio para colocar valores nos NAs e recuperar dados.
```
## 1.5. Fillout NA
```
df1["competition_distance"].max()
#competition_distance: distance in meters to the nearest competitor store
# Se pensarmos que não ter o dado nessa coluna significa um competidor estar muito longe geograficamente e, portanto, se assumirmos os valores como muito maiores que a distancia máxima encontrada resolveria o problema?
# Quando uso função lambda, posso usar tudo conforme o nome da variável que defino, no caso x
# Função apply vai aplicar essa logica a todas as linhas do dataset
# Aplica função apply só na coluna competition_distance
# O resultado eu quero sobrescrever na minha coluna original
df1["competition_distance"] = df1["competition_distance"].apply( lambda x: 200000.0 if math.isnan( x ) else x)
#competition_open_since_month - gives the approximate year and month of the time the nearest competitor was opened
# PREMISSA: Podemos assumir que se essa coluna for NA eu vou copiar a data de venda (extrair o mês)
# Pq isso? já pensando na etapa a frente de feature engineering... tem algumas variaveis que derivamos do tempo que são muito importantes pra representar o comportamento, uma delas é: quanto tempo faz desde que o evento aconteceu
# A informação de competição proxima é muito importante pois influencia nas vendas! (entao evitamos ao maximo excluir esses dados)
# Primeiro tenho que ver se é NA, uso a classe math. Se isso for verdade, vou pegar a coluna "date" e extrair o mês dela. Se não for verdade, mantem.
# Vou usar função lambda, então posso colocar como x os df1.
# Vou aplicar (função apply) isso ao longo das colunas (axis=1). Não precisamos fazer isso no "competition_distance" pois lá estavamos avaliando apenas 1 coluna. Preciso explicitar para a função apply quando tenho mais de uma coluna
# O resultado disso eu vou sobrescrever a coluna "competition_open_since_month"
df1["competition_open_since_month"] = df1.apply( lambda x: x["date"].month if math.isnan( x["competition_open_since_month"] ) else x["competition_open_since_month"] , axis=1)
#competition_open_since_year - gives the approximate year and month of the time the nearest competitor was opened
# Mesma lógica da coluna acima, só que em anos
df1["competition_open_since_year"] = df1.apply( lambda x: x["date"].year if math.isnan( x["competition_open_since_year"] ) else x["competition_open_since_year"] , axis=1)
#promo2 - Promo2 is a continuing and consecutive promotion for some stores: 0 = store is not participating, 1 = store is participating
#promo2_since_week - describes the year and calendar week when the store started participating in Promo2
# Dados NA nessa coluna querem dizer que a loja não participa da promoção
# Similar ao de cima
df1["promo2_since_week"] = df1.apply( lambda x: x["date"].week if math.isnan( x["promo2_since_week"] ) else x["promo2_since_week"] , axis=1)
#promo2_since_year
df1["promo2_since_year"] = df1.apply( lambda x: x["date"].year if math.isnan( x["promo2_since_year"] ) else x["promo2_since_year"] , axis=1)
#promo_interval - describes the consecutive intervals Promo2 is started, naming the months the promotion is started anew. E.g. "Feb,May,Aug,Nov" means each round starts in February, May, August, November of any given year for that store (meses que a promoção ficou ativa)
# Vamos fazer um split dessa coluna e criar uma lista: se a minha data estiver dentro dessa lista (promoção ativa) eu vou criar uma coluna falando que a promo2 foi ativa
# Cria coluna auxiliar
month_map = {1: "Jan",2: "Feb",3: "Mar",4: "Apr",5: "May",6: "Jun",7: "Jul",8: "Aug",9: "Sep",10: "Oct",11: "Nov",12: "Dec"}
# Se o valor na coluna promo_interval for NA, substituo por 0 (não há promoção ativa). inplace=True pois não quero que ele retorne nenhum valor (faça a modificação direto na coluna)
df1["promo_interval"].fillna(0, inplace=True)
# ??? Pq aqui usamos o map ao inves do apply?
df1["month_map"] = df1["date"].dt.month.map( month_map )
# Se o mês da coluna month_map estiver na promoção, vamos colocar 1, se não estiver, 0
# Temos aluns zeros na coluna "promo_interval" que são lojas que não aderiram a promo2
# 0 if df1["promo_interval"] == 0 else 1 if df1["month_map"] in df1["promo_interval"].split( "," ) else 0
# Como vou usar mais de uma coluna preciso especificar a direção
# apply(lambda x: 0 if x["promo_interval"] == 0 else 1 if df1["month_map"] in x["promo_interval"].split( "," ) else 0, axis=1 )
# Não vou aplicar no dataset todo, vou filtrar pra ficar mais fácil:
# Vou criar uma nova coluna is_promo que vai ser 1 ou 0
df1["is_promo"] = df1[["promo_interval","month_map"]].apply(lambda x: 0 if x["promo_interval"] == 0 else 1 if x["month_map"] in x["promo_interval"].split( "," ) else 0, axis=1 )
df1.isna().sum()
# Agora a coluna "competition_distance" não tem mais NA e o valor maximo é 200000
df1["competition_distance"].max()
# Pegando linhas aleatorias. T para mostrar a transposta
df1.sample(5).T
```
## 1.6. Change Types
```
# Importante checar se alguma operação feita na etapa anterior alterou algum dado anterior
# Método dtypes
# competition_open_since_month float64
# competition_open_since_year float64
# promo2_since_week float64
# promo2_since_year float64
# Na verdade essas variaveis acima deveriam ser int (mês e ano)
df1.dtypes
# Método astype nesse caso vai aplicar o int sob essa coluna e vai salvar de volta
df1["competition_open_since_month"] = df1["competition_open_since_month"].astype(int)
df1["competition_open_since_year"] = df1["competition_open_since_year"].astype(int)
df1["promo2_since_week"] = df1["promo2_since_week"].astype(int)
df1["promo2_since_year"] = df1["promo2_since_year"].astype(int)
df1.dtypes
```
## 1.7. Descriptive Statistics
### Ganhar conhecimento de negócio e detectar alguns erros
```
# Central Tendency = mean, median
# Dispersion = std, min, max, range, skew, kurtosis
# Precisamos separar nossas variáveis entre numéricas e categóricas.
# A estatística descritiva funciona para os dois tipos de variáveis, mas a forma com que eu construo a estatistica
# descritiva é diferente.
# Vou separar todas as colunas que são numéricas:
# método select_dtypes e vou passar uma lista de todos os tipos de variaveis que quero selecionar
# datetime64(ns) = dado de tempo (date)
# ??? Qual a diferença do int64 e int32?
num_attributes = df1.select_dtypes( include=["int64","int32","float64"] )
cat_attributes = df1.select_dtypes( exclude=["int64", "float64","int32","datetime64[ns]"] )
num_attributes.sample(2)
cat_attributes.sample(2)
```
## 1.7.1 Numerical Attributes
```
# Apply para aplicar uma operação em todas as colunas e transformar num dataframe pra facilitar a visualização
# Transpostas para ter metricas nas colunas e features nas linhas
# central tendency
ct1 = pd.DataFrame( num_attributes.apply ( np.mean) ).T
ct2 = pd.DataFrame( num_attributes.apply ( np.median ) ).T
# dispersion
d1 = pd.DataFrame( num_attributes.apply( np.std )).T
d2 = pd.DataFrame( num_attributes.apply( min )).T
d3 = pd.DataFrame( num_attributes.apply( max )).T
d4 = pd.DataFrame( num_attributes.apply( lambda x: x.max() - x.min() )).T
d5 = pd.DataFrame( num_attributes.apply( lambda x: x.skew() )).T
d6 = pd.DataFrame( num_attributes.apply( lambda x: x.kurtosis() )).T
# Para concatenar todas essas métricas na ordem que quero ver:
# obs: Classe Pandas
# Tem que transpor e resetar o index (Pq???)
m = pd.concat([d2,d3,d4,ct1,ct2,d1,d5,d6]).T.reset_index()
# Vamos nomear as colunas para não aparecer o index padrão
m.columns = ["attributes","min","max","range","mean","median","std","skew","kurtosis"]
m
# Avaliando por exemplo vendas: min 0, max 41k. Media e mediana parecidas, nao tenho um deslocamento da Normal muito grande.
# Skew proxima de 0 - muito proxima de uma normal
# Kurtosis proximo de 1 - nao tem um pico muuuito grande
# Plotando as sales passando as colunas que quero mostrar
# Obs: Você consegue mudar o tamanho do plot usando os parâmetros height e aspect. Um exemplo ficaria assim:
# sns.displot(df1['sales'], height=8, aspect=2)
# Descobri isso procurando a função displot direto na documentação do seaborn: https://seaborn.pydata.org/generated/seaborn.displot.html#seaborn.displot
sns.displot( df1["sales"], height=8, aspect=2)
# Skew alta, alta concentração de valores no começo
# Meus competidores estão muito proximos
sns.displot( df1["competition_distance"])
```
## 1.7.2 Categorical Attributes
### Vai de boxblot!
```
# ??? No do Meigarom só apareceu os: state_holiday, store_type, assortment, promo_interval e month_map
# Tirei os int32 tambem dos categoricos
cat_attributes.apply( lambda x: x.unique().shape[0] )
# Meigarom prefere o seaborn do que o matplotlib
# sns.boxplot( x= y=, data= )
# x = linha que vai ficar como referencia
# y = o que quero medir (no caso, as vendas)
sns.boxplot( x="state_holiday", y="sales", data=df1 )
# Se plotamos da forma acima não da pra ver nada... (variaveis com ranges mt diferentes)
# Vamos filtrar os dados para plotar:
# ??? Pq esse 0 é uma string e nao um numero? df1["state_holiday"] != "0"
aux1 = df1[(df1["state_holiday"] != "0") & (df1["sales"] > 0)]
# plt.subplot = para plotar um do lado do outro
plt.pyplot.subplot( 1, 3, 1)
sns.boxplot( x="state_holiday", y="sales", data=aux1)
plt.pyplot.subplot( 1, 3, 2)
sns.boxplot( x="store_type", y="sales", data=aux1)
plt.pyplot.subplot( 1, 3, 3)
sns.boxplot( x="assortment", y="sales", data=aux1)
# Boxplot:
# Linha do meio é a mediana: chegou na metade dos valores (em termos de posição), aquele valor é sua mediana
# Limite inferior da barra: 25º quartil (quartil 25) e o limite superior é o quartil 75
# Os ultimos tracinhos são em cima o maximo e embaixo o minimo. Todos os pontos acima do tracinho de maximo são considerados outliers (3x o desvio padrão)
# assortment = mix de produtos
```
# 2.0. STEP 02 - FEATURE ENGINEERING
Para quê fazer a Feature Engineering? Para ter as variáveis DISPONÍVEIS para ESTUDO durante a Análise Exploratória dos Dados. Pra não ter bagunça, crie as variáveis ANTES na análise exploratória!!!
Vou usar uma classe Image para colocar a imagem do mapa mental:
```
df2 = df1.copy()
```
## 2.1. Hypothesis Mind Map
```
Image ("img/mind-map-hypothesis.png")
```
## 2.2. Hypothesis Creation
### 2.2.1 Store Hypothesis
1. Stores with greater number of employees should sell more.
2. Stores with greater stock size should sell more.
3. Stores with bigger size should sell more.
4. Stores with smaller size should sell less.
5. Stores with greater assortment should sell more.
6. Stores with more competitors nearby should sell less.
7. Stores with competitors for longer should sell more.
### 2.2.2 Product Hypothesis
1. Stores with more marketing should sell more.
2. Stores that exhibit more products in the showcase sell more.
3. Stores that have lower prices on products should sell more.
4. Stores that have lower prices for longer on products should sell more.
5. Stores with more consecutive sales should sell more.
### 2.2.3Time-based Hypothesis
1. Stores with more days in holidays should sell less.
2. Stores that open in the first 6 months should sell more.
3. Stores that open on weekends should sell more.
## 2.3. Final Hypothesis List
### As hipóteses das quais temos os dados, vão para a lista final de hipóteses.
1. Stores with greater assortment should sell more.
2. Stores with more competitors nearby should sell less.
3. Stores with competitors for longer should sell more.
4. Stores with active sales for longer should sell more.
5. Stores with more days on sale should sell more.
7. Stores with more consecutive sales should sell more.
8. Stores opened during the Christmas holiday should sell more.
9. Stores should sell more over the years.
10. Stores should sell more in the second half of the year.
11. Stores should sell more after the 10th of each month.
12. Stores should sell less on weekends.
13. Stores should sell less during school holidays.
## 2.4. Feature Engineering
```
# year
df2['year'] = df2['date'].dt.year
# month
df2['month'] = df2['date'].dt.month
# day
df2['day'] = df2['date'].dt.day
# week of year
df2['week_of_year'] = df2['date'].dt.isocalendar().week
# year week
# aqui não usaremos nenhum metodo, e sim mudaremos a formatação da data apenas
# ele fala do strftime no bônus
df2['year_week'] = df2['date'].dt.strftime( '%Y-%W' )
# week of year
# ps: <ipython-input-35-d06c5b7375c4>:9: FutureWarning: Series.dt.weekofyear and Series.dt.week have been deprecated. Please use Series.dt.isocalendar().week instead.
# df2["week_of_year"] = df2["date"].dt.weekofyear
df2["week_of_year"] = df2["date"].dt.isocalendar().week
# ??? Não era pra week_of_year ser igual à semana que aparece na coluna "year_week"? é diferente!
df2.sample(10).T
# competition since
# ja temos a coluna "date" para comparar, mas a informação de competition since está quebrada, temos coluna com year
# e outra com month
# Precisamos juntar as duas em uma data e fazer a substração das duas
# método datetime vem de uma classe também chamada datetime
# datetime.datetime( year=, month=, day= )
# datetime.datetime( year= df2["competition_open_since_year"], month= df2["competition_open_since_month"], day= 1 )
# Vamos usar a função acima para todas as linhas do dataframe vamos usar lambda com variavel x e depois usar o apply
# day = 1 pois nao temos informação sobre o dia
# o apply vai precisar do axis pois estou usando duas colunas diferentes
df2["competition_since"] = df2.apply(lambda x: datetime.datetime( year= x["competition_open_since_year"], month= x["competition_open_since_month"], day= 1), axis=1 )
# com esse comando acima geramos a coluna "competition_since" no formato 2008-09-01 00:00:00.
# Agora precisamos ver a diferença dessa data com a date para saber o tempo de "competition since"
# df2['date'] - df2['competition_since'] )/30
# divido por 30 pq quero manter a glanularidade em dias
# o .days vai extrair os dias desse datetime e salva como inteiro em uma nova coluna 'competition_time_month'
df2['competition_time_month'] = ( ( df2['date'] - df2['competition_since'] )/30 ).apply( lambda x: x.days ).astype( int )
df2.head().T
# promo since, mesma estratégia acima
# Mas para as promoçoes temos uma dificuldade a mais pois temos a coluna promo2 e informação de ano e semana
# não temos de mês
# Vamos fazer um join dos caracteres e depois converter na data
# Mas para juntar as variáveis assim precisamos que as 2 sejam strings (astype converte)
# colocamos o "-" pra ficar no formato ano - semana do ano
# df2['promo_since'] = df2['promo2_since_year'].astype( str ) + '-' + df2['promo2_since_week'].astype( str )
# "promo_since" agora e string, nao é datetime
df2['promo_since'] = df2['promo2_since_year'].astype( str ) + '-' + df2['promo2_since_week'].astype( str )
# Deu uma complicada nesse promo, mas bora lá...
# Truque para converter o que geramos aqui em cima que ficou como string para data: datetime.datetime.strptime( x + '-1', '%Y-%W-%w' ). strptime( o que vai
# mostrar, "formato")
# x pq vamos aplicar para todas as linhas do dataframe
# /7 para ter em semanas
df2['promo_since'] = df2['promo_since'].apply( lambda x: datetime.datetime.strptime( x + '-1', '%Y-%W-%w' ) - datetime.timedelta( days=7 ) )
# Agora que temos duas datas só falta subtrair...
df2['promo_time_week'] = ( ( df2['date'] - df2['promo_since'] )/7 ).apply( lambda x: x.days ).astype( int )
#Obs:
# %W Week number of the year (Monday as the first day of the week).
# All days in a new year preceding the first Monday are considered to be in week 0
# %w Weekday as a decimal number.
# assortment (describes an assortment level: a = basic, b = extra, c = extended)
# Mudar as letras para o que isso representa pra ficar mais facil a leitura:
# Pq else e não elif na estrutura dentro do lambda???
# ??? object type é tipo string?
# Nao preciso usar o axis pq só vou usar a coluna "assortment"
# assortment
df2['assortment'] = df2['assortment'].apply( lambda x: 'basic' if x == 'a' else 'extra' if x == 'b' else 'extended' )
# Mesma coisa do assortment no "state holiday"
# state holiday
df2['state_holiday'] = df2['state_holiday'].apply( lambda x: 'public_holiday' if x == 'a' else 'easter_holiday' if x == 'b' else 'christmas' if x == 'c' else 'regular_day' )
df2.head().T
```
# 3.0. STEP 03 - VARIABLES FILTERING
```
# Antes de qualquer coisa, ao começar um novo passo, copia o dataset do passo anterior e passa a trabalhar com um novo
df3 = df2.copy()
df3.head()
```
## 3.1. ROWS FILTERING
```
# "open" != 0 & "sales" > 0
df3 = df3[(df3["open"] != 0) & (df3["sales"] > 0)]
```
## 3.2. COLUMNS SELECTION
```
# Vamos "dropar" as colunas que não queremos
# A "open" está aqui pois após tirarmos as linhas cujos dados da coluna "open" eram 0, só sobraram valores 1, então é uma coluna 'inútil'
cols_drop = ['customers', 'open', 'promo_interval', 'month_map']
# Drop é um metodo da classe Pandas (quais colunas e sentido); axis 0 = linhas, axis 1 = colunas
df3 = df3.drop( cols_drop, axis=1 )
df3.columns
```
| github_jupyter |
## Dependencies
```
import os
import cv2
import shutil
import random
import warnings
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from tensorflow import set_random_seed
from sklearn.utils import class_weight
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix, cohen_kappa_score
from keras import backend as K
from keras.models import Model
from keras.utils import to_categorical
from keras import optimizers, applications
from keras.preprocessing.image import ImageDataGenerator
from keras.callbacks import EarlyStopping, ReduceLROnPlateau, Callback, LearningRateScheduler
from keras.layers import Dense, Dropout, GlobalAveragePooling2D, Input
# Set seeds to make the experiment more reproducible.
def seed_everything(seed=0):
random.seed(seed)
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
set_random_seed(0)
seed = 0
seed_everything(seed)
%matplotlib inline
sns.set(style="whitegrid")
warnings.filterwarnings("ignore")
```
## Load data
```
hold_out_set = pd.read_csv('../input/aptos-data-split/hold-out.csv')
X_train = hold_out_set[hold_out_set['set'] == 'train']
X_val = hold_out_set[hold_out_set['set'] == 'validation']
test = pd.read_csv('../input/aptos2019-blindness-detection/test.csv')
print('Number of train samples: ', X_train.shape[0])
print('Number of validation samples: ', X_val.shape[0])
print('Number of test samples: ', test.shape[0])
# Preprocecss data
X_train["id_code"] = X_train["id_code"].apply(lambda x: x + ".png")
X_val["id_code"] = X_val["id_code"].apply(lambda x: x + ".png")
test["id_code"] = test["id_code"].apply(lambda x: x + ".png")
X_train['diagnosis'] = X_train['diagnosis'].astype('str')
X_val['diagnosis'] = X_val['diagnosis'].astype('str')
display(X_train.head())
```
# Model parameters
```
# Model parameters
N_CLASSES = X_train['diagnosis'].nunique()
BATCH_SIZE = 16
EPOCHS = 40
WARMUP_EPOCHS = 5
LEARNING_RATE = 1e-4
WARMUP_LEARNING_RATE = 1e-3
HEIGHT = 320
WIDTH = 320
CHANNELS = 3
ES_PATIENCE = 5
RLROP_PATIENCE = 3
DECAY_DROP = 0.5
def kappa(y_true, y_pred, n_classes=5):
y_trues = K.cast(K.argmax(y_true), K.floatx())
y_preds = K.cast(K.argmax(y_pred), K.floatx())
n_samples = K.cast(K.shape(y_true)[0], K.floatx())
distance = K.sum(K.abs(y_trues - y_preds))
max_distance = n_classes - 1
kappa_score = 1 - ((distance**2) / (n_samples * (max_distance**2)))
return kappa_score
def step_decay(epoch):
lrate = 30e-5
if epoch > 3:
lrate = 15e-5
if epoch > 7:
lrate = 7.5e-5
if epoch > 11:
lrate = 3e-5
if epoch > 15:
lrate = 1e-5
return lrate
def focal_loss(y_true, y_pred):
gamma = 2.0
epsilon = K.epsilon()
pt = y_pred * y_true + (1-y_pred) * (1-y_true)
pt = K.clip(pt, epsilon, 1-epsilon)
CE = -K.log(pt)
FL = K.pow(1-pt, gamma) * CE
loss = K.sum(FL, axis=1)
return loss
```
# Pre-procecess images
```
train_base_path = '../input/aptos2019-blindness-detection/train_images/'
test_base_path = '../input/aptos2019-blindness-detection/test_images/'
train_dest_path = 'base_dir/train_images/'
validation_dest_path = 'base_dir/validation_images/'
test_dest_path = 'base_dir/test_images/'
# Making sure directories don't exist
if os.path.exists(train_dest_path):
shutil.rmtree(train_dest_path)
if os.path.exists(validation_dest_path):
shutil.rmtree(validation_dest_path)
if os.path.exists(test_dest_path):
shutil.rmtree(test_dest_path)
# Creating train, validation and test directories
os.makedirs(train_dest_path)
os.makedirs(validation_dest_path)
os.makedirs(test_dest_path)
def crop_image(img, tol=7):
if img.ndim ==2:
mask = img>tol
return img[np.ix_(mask.any(1),mask.any(0))]
elif img.ndim==3:
gray_img = cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)
mask = gray_img>tol
check_shape = img[:,:,0][np.ix_(mask.any(1),mask.any(0))].shape[0]
if (check_shape == 0): # image is too dark so that we crop out everything,
return img # return original image
else:
img1=img[:,:,0][np.ix_(mask.any(1),mask.any(0))]
img2=img[:,:,1][np.ix_(mask.any(1),mask.any(0))]
img3=img[:,:,2][np.ix_(mask.any(1),mask.any(0))]
img = np.stack([img1,img2,img3],axis=-1)
return img
def circle_crop(img):
img = crop_image(img)
height, width, depth = img.shape
largest_side = np.max((height, width))
img = cv2.resize(img, (largest_side, largest_side))
height, width, depth = img.shape
x = width//2
y = height//2
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def preprocess_image(base_path, save_path, image_id, HEIGHT, WIDTH, sigmaX=10):
image = cv2.imread(base_path + image_id)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = circle_crop(image)
image = cv2.resize(image, (HEIGHT, WIDTH))
image = cv2.addWeighted(image, 4, cv2.GaussianBlur(image, (0,0), sigmaX), -4 , 128)
cv2.imwrite(save_path + image_id, image)
# Pre-procecss train set
for i, image_id in enumerate(X_train['id_code']):
preprocess_image(train_base_path, train_dest_path, image_id, HEIGHT, WIDTH)
# Pre-procecss validation set
for i, image_id in enumerate(X_val['id_code']):
preprocess_image(train_base_path, validation_dest_path, image_id, HEIGHT, WIDTH)
# Pre-procecss test set
for i, image_id in enumerate(test['id_code']):
preprocess_image(test_base_path, test_dest_path, image_id, HEIGHT, WIDTH)
```
# Data generator
```
train_datagen=ImageDataGenerator(rescale=1./255,
rotation_range=360,
horizontal_flip=True,
vertical_flip=True)
valid_datagen=ImageDataGenerator(rescale=1./255)
train_generator=train_datagen.flow_from_dataframe(
dataframe=X_train,
directory=train_dest_path,
x_col="id_code",
y_col="diagnosis",
class_mode="categorical",
batch_size=BATCH_SIZE,
target_size=(HEIGHT, WIDTH),
seed=seed)
valid_generator=valid_datagen.flow_from_dataframe(
dataframe=X_val,
directory=validation_dest_path,
x_col="id_code",
y_col="diagnosis",
class_mode="categorical",
batch_size=BATCH_SIZE,
target_size=(HEIGHT, WIDTH),
seed=seed)
test_generator=valid_datagen.flow_from_dataframe(
dataframe=test,
directory=test_dest_path,
x_col="id_code",
batch_size=1,
class_mode=None,
shuffle=False,
target_size=(HEIGHT, WIDTH),
seed=seed)
```
# Model
```
def create_model(input_shape, n_out):
input_tensor = Input(shape=input_shape)
base_model = applications.DenseNet169(weights=None,
include_top=False,
input_tensor=input_tensor)
base_model.load_weights('../input/keras-notop/densenet169_weights_tf_dim_ordering_tf_kernels_notop.h5')
x = GlobalAveragePooling2D()(base_model.output)
x = Dropout(0.5)(x)
x = Dense(2048, activation='relu')(x)
x = Dropout(0.5)(x)
final_output = Dense(n_out, activation='softmax', name='final_output')(x)
model = Model(input_tensor, final_output)
return model
```
# Train top layers
```
model = create_model(input_shape=(HEIGHT, WIDTH, CHANNELS), n_out=N_CLASSES)
for layer in model.layers:
layer.trainable = False
for i in range(-5, 0):
model.layers[i].trainable = True
class_weights = class_weight.compute_class_weight('balanced', np.unique(X_train['diagnosis'].astype('int').values), X_train['diagnosis'].astype('int').values)
metric_list = ["accuracy", kappa]
optimizer = optimizers.Adam(lr=WARMUP_LEARNING_RATE)
model.compile(optimizer=optimizer, loss='categorical_crossentropy', metrics=metric_list)
model.summary()
STEP_SIZE_TRAIN = train_generator.n//train_generator.batch_size
STEP_SIZE_VALID = valid_generator.n//valid_generator.batch_size
history_warmup = model.fit_generator(generator=train_generator,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_generator,
validation_steps=STEP_SIZE_VALID,
epochs=WARMUP_EPOCHS,
class_weight=class_weights,
verbose=1).history
```
# Fine-tune the complete model
```
for layer in model.layers:
layer.trainable = True
# lrstep = LearningRateScheduler(step_decay)
es = EarlyStopping(monitor='val_loss', mode='min', patience=ES_PATIENCE, restore_best_weights=True, verbose=1)
rlrop = ReduceLROnPlateau(monitor='val_loss', mode='min', patience=RLROP_PATIENCE, factor=DECAY_DROP, min_lr=1e-6, verbose=1)
callback_list = [es, rlrop]
optimizer = optimizers.Adam(lr=LEARNING_RATE)
model.compile(optimizer=optimizer, loss='categorical_crossentropy', metrics=metric_list)
model.summary()
history = model.fit_generator(generator=train_generator,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_generator,
validation_steps=STEP_SIZE_VALID,
epochs=EPOCHS,
callbacks=callback_list,
class_weight=class_weights,
verbose=1).history
```
# Model loss graph
```
sns.set_style("whitegrid")
fig, (ax1, ax2, ax3) = plt.subplots(3, 1, sharex='col', figsize=(20, 18))
ax1.plot(history['loss'], label='Train loss')
ax1.plot(history['val_loss'], label='Validation loss')
ax1.legend(loc='best')
ax1.set_title('Loss')
ax2.plot(history['acc'], label='Train accuracy')
ax2.plot(history['val_acc'], label='Validation accuracy')
ax2.legend(loc='best')
ax2.set_title('Accuracy')
ax3.plot(history['kappa'], label='Train kappa')
ax3.plot(history['val_kappa'], label='Validation kappa')
ax3.legend(loc='best')
ax3.set_title('Kappa')
plt.xlabel('Epochs')
sns.despine()
plt.show()
# Create empty arays to keep the predictions and labels
lastFullTrainPred = np.empty((0, N_CLASSES))
lastFullTrainLabels = np.empty((0, N_CLASSES))
lastFullValPred = np.empty((0, N_CLASSES))
lastFullValLabels = np.empty((0, N_CLASSES))
# Add train predictions and labels
for i in range(STEP_SIZE_TRAIN+1):
im, lbl = next(train_generator)
scores = model.predict(im, batch_size=train_generator.batch_size)
lastFullTrainPred = np.append(lastFullTrainPred, scores, axis=0)
lastFullTrainLabels = np.append(lastFullTrainLabels, lbl, axis=0)
# Add validation predictions and labels
for i in range(STEP_SIZE_VALID+1):
im, lbl = next(valid_generator)
scores = model.predict(im, batch_size=valid_generator.batch_size)
lastFullValPred = np.append(lastFullValPred, scores, axis=0)
lastFullValLabels = np.append(lastFullValLabels, lbl, axis=0)
lastFullComPred = np.concatenate((lastFullTrainPred, lastFullValPred))
lastFullComLabels = np.concatenate((lastFullTrainLabels, lastFullValLabels))
train_preds = [np.argmax(pred) for pred in lastFullTrainPred]
train_labels = [np.argmax(label) for label in lastFullTrainLabels]
validation_preds = [np.argmax(pred) for pred in lastFullValPred]
validation_labels = [np.argmax(label) for label in lastFullValLabels]
complete_labels = [np.argmax(label) for label in lastFullComLabels]
```
# Model Evaluation
## Confusion Matrix
### Original thresholds
```
labels = ['0 - No DR', '1 - Mild', '2 - Moderate', '3 - Severe', '4 - Proliferative DR']
def plot_confusion_matrix(train, validation, labels=labels):
train_labels, train_preds = train
validation_labels, validation_preds = validation
fig, (ax1, ax2) = plt.subplots(1, 2, sharex='col', figsize=(24, 7))
train_cnf_matrix = confusion_matrix(train_labels, train_preds)
validation_cnf_matrix = confusion_matrix(validation_labels, validation_preds)
train_cnf_matrix_norm = train_cnf_matrix.astype('float') / train_cnf_matrix.sum(axis=1)[:, np.newaxis]
validation_cnf_matrix_norm = validation_cnf_matrix.astype('float') / validation_cnf_matrix.sum(axis=1)[:, np.newaxis]
train_df_cm = pd.DataFrame(train_cnf_matrix_norm, index=labels, columns=labels)
validation_df_cm = pd.DataFrame(validation_cnf_matrix_norm, index=labels, columns=labels)
sns.heatmap(train_df_cm, annot=True, fmt='.2f', cmap="Blues",ax=ax1).set_title('Train')
sns.heatmap(validation_df_cm, annot=True, fmt='.2f', cmap=sns.cubehelix_palette(8),ax=ax2).set_title('Validation')
plt.show()
plot_confusion_matrix((train_labels, train_preds), (validation_labels, validation_preds))
```
## Quadratic Weighted Kappa
```
def evaluate_model(train, validation):
train_labels, train_preds = train
validation_labels, validation_preds = validation
print("Train Cohen Kappa score: %.3f" % cohen_kappa_score(train_preds, train_labels, weights='quadratic'))
print("Validation Cohen Kappa score: %.3f" % cohen_kappa_score(validation_preds, validation_labels, weights='quadratic'))
print("Complete set Cohen Kappa score: %.3f" % cohen_kappa_score(train_preds+validation_preds, train_labels+validation_labels, weights='quadratic'))
evaluate_model((train_preds, train_labels), (validation_preds, validation_labels))
```
## Apply model to test set and output predictions
```
step_size = test_generator.n//test_generator.batch_size
test_generator.reset()
preds = model.predict_generator(test_generator, steps=step_size)
predictions = np.argmax(preds, axis=1)
results = pd.DataFrame({'id_code':test['id_code'], 'diagnosis':predictions})
results['id_code'] = results['id_code'].map(lambda x: str(x)[:-4])
# Cleaning created directories
if os.path.exists(train_dest_path):
shutil.rmtree(train_dest_path)
if os.path.exists(validation_dest_path):
shutil.rmtree(validation_dest_path)
if os.path.exists(test_dest_path):
shutil.rmtree(test_dest_path)
```
# Predictions class distribution
```
fig = plt.subplots(sharex='col', figsize=(24, 8.7))
sns.countplot(x="diagnosis", data=results, palette="GnBu_d").set_title('Test')
sns.despine()
plt.show()
results.to_csv('submission.csv', index=False)
display(results.head())
```
| github_jupyter |
<!--TITLE:Custom Convnets-->
# Introduction #
Now that you've seen the layers a convnet uses to extract features, it's time to put them together and build a network of your own!
# Simple to Refined #
In the last three lessons, we saw how convolutional networks perform **feature extraction** through three operations: **filter**, **detect**, and **condense**. A single round of feature extraction can only extract relatively simple features from an image, things like simple lines or contrasts. These are too simple to solve most classification problems. Instead, convnets will repeat this extraction over and over, so that the features become more complex and refined as they travel deeper into the network.
<figure>
<img src="https://i.imgur.com/VqmC1rm.png" alt="Features extracted from an image of a car, from simple to refined." width=800>
</figure>
# Convolutional Blocks #
It does this by passing them through long chains of **convolutional blocks** which perform this extraction.
<figure>
<img src="https://i.imgur.com/pr8VwCZ.png" width="400" alt="Extraction as a sequence of blocks.">
</figure>
These convolutional blocks are stacks of `Conv2D` and `MaxPool2D` layers, whose role in feature extraction we learned about in the last few lessons.
<figure>
<!-- <img src="./images/2-block-crp.png" width="400" alt="A kind of extraction block: convolution, ReLU, pooling."> -->
<img src="https://i.imgur.com/8D6IhEw.png" width="400" alt="A kind of extraction block: convolution, ReLU, pooling.">
</figure>
Each block represents a round of extraction, and by composing these blocks the convnet can combine and recombine the features produced, growing them and shaping them to better fit the problem at hand. The deep structure of modern convnets is what allows this sophisticated feature engineering and has been largely responsible for their superior performance.
# Example - Design a Convnet #
Let's see how to define a deep convolutional network capable of engineering complex features. In this example, we'll create a Keras `Sequence` model and then train it on our Cars dataset.
## Step 1 - Load Data ##
This hidden cell loads the data.
```
#$HIDE_INPUT$
# Imports
import os, warnings
import matplotlib.pyplot as plt
from matplotlib import gridspec
import numpy as np
import tensorflow as tf
from tensorflow.keras.preprocessing import image_dataset_from_directory
# Reproducability
def set_seed(seed=31415):
np.random.seed(seed)
tf.random.set_seed(seed)
os.environ['PYTHONHASHSEED'] = str(seed)
os.environ['TF_DETERMINISTIC_OPS'] = '1'
set_seed()
# Set Matplotlib defaults
plt.rc('figure', autolayout=True)
plt.rc('axes', labelweight='bold', labelsize='large',
titleweight='bold', titlesize=18, titlepad=10)
plt.rc('image', cmap='magma')
warnings.filterwarnings("ignore") # to clean up output cells
# Load training and validation sets
ds_train_ = image_dataset_from_directory(
'../input/car-or-truck/train',
labels='inferred',
label_mode='binary',
image_size=[128, 128],
interpolation='nearest',
batch_size=64,
shuffle=True,
)
ds_valid_ = image_dataset_from_directory(
'../input/car-or-truck/valid',
labels='inferred',
label_mode='binary',
image_size=[128, 128],
interpolation='nearest',
batch_size=64,
shuffle=False,
)
# Data Pipeline
def convert_to_float(image, label):
image = tf.image.convert_image_dtype(image, dtype=tf.float32)
return image, label
AUTOTUNE = tf.data.experimental.AUTOTUNE
ds_train = (
ds_train_
.map(convert_to_float)
.cache()
.prefetch(buffer_size=AUTOTUNE)
)
ds_valid = (
ds_valid_
.map(convert_to_float)
.cache()
.prefetch(buffer_size=AUTOTUNE)
)
```
## Step 2 - Define Model ##
Here is a diagram of the model we'll use:
<figure>
<!-- <img src="./images/2-convmodel-1.png" width="200" alt="Diagram of a convolutional model."> -->
<img src="https://i.imgur.com/U1VdoDJ.png" width="250" alt="Diagram of a convolutional model.">
</figure>
Now we'll define the model. See how our model consists of three blocks of `Conv2D` and `MaxPool2D` layers (the base) followed by a head of `Dense` layers. We can translate this diagram more or less directly into a Keras `Sequential` model just by filling in the appropriate parameters.
```
import tensorflow.keras as keras
import tensorflow.keras.layers as layers
model = keras.Sequential([
# First Convolutional Block
layers.Conv2D(filters=32, kernel_size=5, activation="relu", padding='same',
# give the input dimensions in the first layer
# [height, width, color channels(RGB)]
input_shape=[128, 128, 3]),
layers.MaxPool2D(),
# Second Convolutional Block
layers.Conv2D(filters=64, kernel_size=3, activation="relu", padding='same'),
layers.MaxPool2D(),
# Third Convolutional Block
layers.Conv2D(filters=128, kernel_size=3, activation="relu", padding='same'),
layers.MaxPool2D(),
# Classifier Head
layers.Flatten(),
layers.Dense(units=6, activation="relu"),
layers.Dense(units=1, activation="sigmoid"),
])
model.summary()
```
Notice in this definition is how the number of filters doubled block-by-block: 64, 128, 256. This is a common pattern. Since the `MaxPool2D` layer is reducing the *size* of the feature maps, we can afford to increase the *quantity* we create.
## Step 3 - Train ##
We can train this model just like the model from Lesson 1: compile it with an optimizer along with a loss and metric appropriate for binary classification.
```
model.compile(
optimizer=tf.keras.optimizers.Adam(epsilon=0.01),
loss='binary_crossentropy',
metrics=['binary_accuracy']
)
history = model.fit(
ds_train,
validation_data=ds_valid,
epochs=40,
)
import pandas as pd
history_frame = pd.DataFrame(history.history)
history_frame.loc[:, ['loss', 'val_loss']].plot()
history_frame.loc[:, ['binary_accuracy', 'val_binary_accuracy']].plot();
```
This model is much smaller than the VGG16 model from Lesson 1 -- only 3 convolutional layers versus the 16 of VGG16. It was nevertheless able to fit this dataset fairly well. We might still be able to improve this simple model by adding more convolutional layers, hoping to create features better adapted to the dataset. This is what we'll try in the exercises.
# Conclusion #
In this tutorial, you saw how to build a custom convnet composed of many **convolutional blocks** and capable of complex feature engineering.
# Your Turn #
In the exercises, you'll create a convnet that performs as well on this problem as VGG16 does -- without pretraining! [**Try it now!**](#$NEXT_NOTEBOOK_URL$)
| github_jupyter |
# Looking up Trig Ratios
There are three ways you could find the value of a trig function at a particular angle.
**1. Use a table** - This is how engineers used to find trig ratios before the days of computers. For example, from the table below I can see that $\sin(60)=0.866$
| angle | sin | cos | tan |
| :---: | :---: | :---: | :---: |
| 0 | 0.000 | 1.000 | 0.000 |
| 10 | 0.174 | 0.985 | 0.176 |
| 20 | 0.342 | 0.940 | 0.364 |
| 30 | 0.500 | 0.866 | 0.577 |
| 40 | 0.643 | 0.766 | 0.839 |
| 50 | 0.766 | 0.643 | 1.192 |
| 60 | 0.866 | 0.500 | 1.732 |
| 70 | 0.940 | 0.342 | 2.747 |
| 80 | 0.985 | 0.174 | 5.671 |
The problem with this technique is that there will always be gaps in a table.
**2. Use a graph** - One way to try to fill these gaps is by consulting a graph of a trigonometric function. For example, the image below shows a plot of $\sin(\theta)$ for $0 \leq \theta \leq 360$
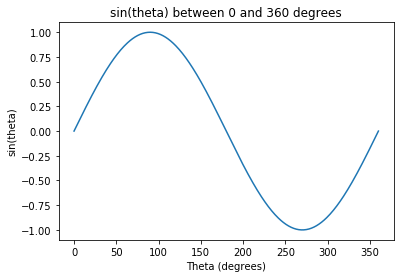
These graphs are nice because they give a good visual sense for how these ratios behave, but they aren't great for getting accurate values. Which leads us to the **best** way to look up trig ratios...
**3. Use a computer!** This probably isn't a surprise, but python has built in functions to calculate sine, cosine, and tangent...
In fact, you can even type "sin(60 degrees)" into **Google** and you'll get the correct answer!
Note how I wrote in "sin(60 degrees)" instead of just "sin(60)". That's because these functions generally expect their input to be in **radians**.
Now let's calculate these ratios with Python.
```
# Python's math module has functions called sin, cos, and tan
# as well as the constant "pi" (which we will find useful shortly)
from math import sin, cos, tan, pi
# Run this cell. What do you expect the output to be?
print(sin(60))
```
Did the output match what you expected?
If not, it's probably because we didn't convert our angle to radians.
### EXERCISE 1 - Write a function that converts degrees to radians
Implement the following math in code:
$$\theta_{\text{radians}} = \theta_{\text{degrees}} \times \frac{\pi}{180}$$
```
from math import pi
def deg2rad(theta):
"""Converts degrees to radians"""
return theta * (pi/180)
# TODO - implement this function (solution
# code at end of notebook)
assert(deg2rad(45.0) == pi / 4)
assert(deg2rad(90.0) == pi / 2)
print("Nice work! Your degrees to radians function works!")
for theta in [0, 30, 45, 60, 90]:
theta_rad = deg2rad(theta)
sin_theta = sin(theta_rad)
print("sin(", theta, "degrees) =", sin_theta)
```
### EXERCISE 2 - Make plots of cosine and tangent
```
import numpy as np
from matplotlib import pyplot as plt
def plot_sine(min_theta, max_theta):
"""
Generates a plot of sin(theta) between min_theta
and max_theta (both of which are specified in degrees).
"""
angles_degrees = np.linspace(min_theta, max_theta)
angles_radians = deg2rad(angles_degrees)
values = np.sin(angles_radians)
X = angles_degrees
Y = values
plt.plot(X,Y)
plt.show()
# EXERCISE 2.1 Implement this! Try not to look at the
# implementation of plot_sine TOO much...
def plot_cosine(min_theta, max_theta):
"""
Generates a plot of sin(theta) between min_theta
and max_theta (both of which are specified in degrees).
"""
angles_degrees = np.linspace(min_theta, max_theta)
angles_radians = deg2rad(angles_degrees)
values = np.cos(angles_radians)
X = angles_degrees
Y = values
plt.plot(X,Y)
plt.show()
plot_sine(0, 360)
plot_cosine(0, 360)
#
#
#
#
# SOLUTION CODE
#
#
#
#
from math import pi
def deg2rad_solution(theta):
"""Converts degrees to radians"""
return theta * pi / 180
assert(deg2rad_solution(45.0) == pi / 4)
assert(deg2rad_solution(90.0) == pi / 2)
import numpy as np
from matplotlib import pyplot as plt
def plot_cosine_solution(min_theta, max_theta):
"""
Generates a plot of sin(theta) between min_theta
and max_theta (both of which are specified in degrees).
"""
angles_degrees = np.linspace(min_theta, max_theta)
angles_radians = deg2rad_solution(angles_degrees)
values = np.cos(angles_radians)
X = angles_degrees
Y = values
plt.plot(X,Y)
plt.show()
plot_cosine_solution(0, 360)
```
| github_jupyter |
```
from gs_quant.data import Dataset
from gs_quant.markets.securities import Asset, AssetIdentifier, SecurityMaster
from gs_quant.timeseries import *
from gs_quant.target.instrument import FXOption, IRSwaption
from gs_quant.markets import PricingContext, HistoricalPricingContext, BackToTheFuturePricingContext
from gs_quant.risk import CarryScenario, MarketDataPattern, MarketDataShock, MarketDataShockBasedScenario, MarketDataShockType, CurveScenario,CarryScenario
from gs_quant.markets.portfolio import Portfolio
from gs_quant.risk import IRAnnualImpliedVol
from gs_quant.timeseries import percentiles
from gs_quant.datetime import business_day_offset
import seaborn as sns
import pandas as pd
import matplotlib.pyplot as plt
from scipy import stats
import warnings
from datetime import date
warnings.filterwarnings('ignore')
sns.set(style="darkgrid", color_codes=True)
from gs_quant.session import GsSession
# external users should substitute their client id and secret; please skip this step if using internal jupyterhub
GsSession.use(client_id=None, client_secret=None, scopes=('run_analytics',))
```
In this notebook, we'll look at entry points for G10 vol, look for crosses with the largest downside sensivity to SPX, indicatively price several structures and analyze their carry profile.
* [1: FX entry point vs richness](#1:-FX-entry-point-vs-richness)
* [2: Downside sensitivity to SPX](#2:-Downside-sensitivity-to-SPX)
* [3: AUDJPY conditional relationship with SPX](#3:-AUDJPY-conditional-relationship-with-SPX)
* [4: Price structures](#4:-Price-structures)
* [5: Analyse rates package](#5:-Analyse-rates-package)
### 1: FX entry point vs richness
Let's pull [GS FX Spot](https://marquee.gs.com/s/developer/datasets/FXSPOT_PREMIUM) and [GS FX Implied Volatility](https://marquee.gs.com/s/developer/datasets/FXIMPLIEDVOL_PREMIUM) and look at implied vs realized vol as well as current implied level as percentile relative to the last 2 years.
```
def format_df(data_dict):
df = pd.concat(data_dict, axis=1)
df.columns = data_dict.keys()
return df.fillna(method='ffill').dropna()
g10 = ['USDJPY', 'EURUSD', 'AUDUSD', 'GBPUSD', 'USDCAD', 'USDNOK', 'NZDUSD', 'USDSEK', 'USDCHF', 'AUDJPY']
start_date = date(2005, 8, 26)
end_date = business_day_offset(date.today(), -1, roll='preceding')
fxspot_dataset, fxvol_dataset = Dataset('FXSPOT_PREMIUM'), Dataset('FXIMPLIEDVOL_PREMIUM')
spot_data, impvol_data, spot_fx = {}, {}, {}
for cross in g10:
spot = fxspot_dataset.get_data(start_date, end_date, bbid=cross)[['spot']].drop_duplicates(keep='last')
spot_fx[cross] = spot['spot']
spot_data[cross] = volatility(spot['spot'], 63) # realized vol
vol = fxvol_dataset.get_data(start_date, end_date, bbid=cross, tenor='3m', deltaStrike='DN', location='NYC')[['impliedVolatility']]
impvol_data[cross] = vol.drop_duplicates(keep='last') * 100
spdata, ivdata = format_df(spot_data), format_df(impvol_data)
diff = ivdata.subtract(spdata).dropna()
_slice = ivdata['2018-09-01': '2020-09-08']
pct_rank = {}
for x in _slice.columns:
pct = percentiles(_slice[x])
pct_rank[x] = pct.iloc[-1]
for fx in pct_rank:
plt.scatter(pct_rank[fx], diff[fx]['2020-09-08'])
plt.legend(pct_rank.keys(),loc='best', bbox_to_anchor=(0.9, -0.13), ncol=3)
plt.xlabel('Percentile of Current Implied Vol')
plt.ylabel('Implied vs Realized Vol')
plt.title('Entry Point vs Richness')
plt.show()
```
### 2: Downside sensitivity to SPX
Let's now look at beta and correlation with SPX across G10.
```
spx_spot = Dataset('TREOD').get_data(start_date, end_date, bbid='SPX')[['closePrice']]
spx_spot = spx_spot.fillna(method='ffill').dropna()
df = pd.DataFrame(spx_spot)
#FX Spot data
fx_spots = format_df(spot_fx)
data = pd.concat([spx_spot, fx_spots], axis=1).dropna()
data.columns = ['SPX'] + g10
beta_spx, corr_spx = {}, {}
#calculate rolling 84d or 4m beta to S&P
for cross in g10:
beta_spx[cross] = beta(data[cross],data['SPX'], 84)
corr_spx[cross] = correlation(data['SPX'], data[cross], 84)
fig, axs = plt.subplots(5, 2, figsize=(18, 20))
for j in range(2):
for i in range(5):
color='tab:blue'
axs[i,j].plot(beta_spx[g10[i + j*5]], color=color)
axs[i,j].set_title(g10[i + j*5])
color='tab:blue'
axs[i,j].set_ylabel('Beta', color=color)
axs[i,j].plot(beta_spx[g10[i + j*5]], color=color)
ax2 = axs[i,j].twinx()
color = 'tab:orange'
ax2.plot(corr_spx[g10[i + j*5]], color=color)
ax2.set_ylabel('Correlation', color=color)
plt.show()
```
### Part 3: AUDJPY conditional relationship with SPX
Let's focus on AUDJPY and look at its relationship with SPX when SPX is significantly up and down.
```
# resample data to weekly from daily & get weekly returns
wk_data = data.resample('W-FRI').last()
rets = returns(wk_data, 1)
sns.set(style='white', color_codes=True)
spx_returns = [-.1, -.05, .05, .1]
r2 = lambda x,y: stats.pearsonr(x,y)[0]**2
betas = pd.DataFrame(index=spx_returns, columns=g10)
for ret in spx_returns:
dns = rets[rets.SPX <= ret].dropna() if ret < 0 else rets[rets.SPX >= ret].dropna()
j = sns.jointplot(x='SPX', y='AUDJPY', data=dns, kind='reg')
j.set_axis_labels('SPX with {}% Returns'.format(ret*100), 'AUDJPY')
j.fig.subplots_adjust(wspace=.02)
plt.show()
```
Let's use the beta for all S&P returns to price a structure
```
sns.jointplot(x='SPX', y='AUDJPY', data=rets, kind='reg', stat_func=r2)
```
### 4: Price structures
##### Let's now look at a few AUDJPY structures as potential hedges
* Buy 4m AUDJPY put using spx beta to size. Max loss limited to premium paid.
* Buy 4m AUDJPY put spread (4.2%/10.6% OTMS). Max loss limited to premium paid.
For more info on this trade, check out our market strats piece [here](https://marquee.gs.com/content/#/article/2020/08/28/gs-marketstrats-audjpy-as-us-election-hedge)
```
#buy 4m AUDJPY put
audjpy_put = FXOption(option_type='Put', pair='AUDJPY', strike_price= 's-4.2%', expiration_date='4m', buy_sell='Buy')
print('cost in bps: {:,.2f}'.format(audjpy_put.premium / audjpy_put.notional_amount * 1e4))
#buy 4m AUDJPY put spread (5.3%/10.6% OTMS)
from gs_quant.markets.portfolio import Portfolio
put1 = FXOption(option_type='Put', pair='AUDJPY', strike_price= 's-4.2%', expiration_date='4m', buy_sell='Buy')
put2 = FXOption(option_type='Put', pair='AUDJPY', strike_price= 's-10.6%', expiration_date='4m', buy_sell='Sell')
fx_package = Portfolio((put1, put2))
cost = put2.premium/put2.notional_amount - put1.premium/put1.notional_amount
print('cost in bps: {:,.2f}'.format(cost * 1e4))
```
##### ...And some rates ideas
* Sell straddle. Max loss unlimited.
* Sell 3m30y straddle, buy 2y30y straddle in a 0 pv package. Max loss unlimited.
```
leg = IRSwaption('Straddle', '30y', notional_currency='USD', expiration_date='3m', buy_sell='Sell')
print('PV in USD: {:,.2f}'.format(leg.dollar_price()))
leg1 = IRSwaption('Straddle', '30y', notional_currency='USD', expiration_date='3m', buy_sell='Sell',name='3m30y ATM Straddle')
leg2 = IRSwaption('Straddle', '30y', notional_currency='USD', expiration_date='2y', notional_amount='{}/pv'.format(leg1.price()), buy_sell='Buy', name = '2y30y ATM Straddle')
rates_package = Portfolio((leg1, leg2))
rates_package.resolve()
print('Package cost in USD: {:,.2f}'.format(rates_package.price().aggregate()))
print('PV Flat notionals ($$m):', round(leg1.notional_amount/1e6, 1),' by ',round(leg2.notional_amount/1e6, 1))
```
### 5: Analyse rates package
```
dates = pd.bdate_range(date(2020, 6, 8), leg1.expiration_date, freq='5B').date.tolist()
with BackToTheFuturePricingContext(dates=dates, roll_to_fwds=True):
future = rates_package.price()
rates_future = future.result().aggregate()
rates_future.plot(figsize=(10, 6), title='Historical PV and carry for rates package')
print('PV breakdown between legs:')
results = future.result().to_frame()
results /= 1e6
results.index=[leg1.name,leg2.name]
results.loc['Total'] = results.sum()
results.round(1)
```
Let's focus on the next 3m and how the calendar carries in different rates shocks.
```
dates = pd.bdate_range(dt.date.today(), leg1.expiration_date, freq='5B').date.tolist()
shocked_pv = pd.DataFrame(columns=['Base', '5bp per week', '50bp instantaneous'], index=dates)
p1, p2, p3 = [], [], []
with PricingContext(is_batch=True):
for t, d in enumerate(dates):
with CarryScenario(date=d, roll_to_fwds=True):
p1.append(rates_package.price())
with MarketDataShockBasedScenario({MarketDataPattern('IR', 'USD'): MarketDataShock(MarketDataShockType.Absolute, t*0.0005)}):
p2.append(rates_package.price())
with MarketDataShockBasedScenario({MarketDataPattern('IR', 'USD'): MarketDataShock(MarketDataShockType.Absolute, 0.005)}):
p3.append(rates_package.price())
shocked_pv.Base = [p.result().aggregate() for p in p1]
shocked_pv['5bp per week'] = [p.result().aggregate() for p in p2]
shocked_pv['50bp instantaneous'] = [p.result().aggregate() for p in p3]
shocked_pv/=1e6
shocked_pv.round(1)
shocked_pv.plot(figsize=(10, 6), title='Carry + scenario analysis')
```
### Disclaimers
Scenarios/predictions: Simulated results are for illustrative purposes only. GS provides no assurance or guarantee that the strategy will operate or would have operated in the past in a manner consistent with the above analysis. Past performance figures are not a reliable indicator of future results.
Indicative Terms/Pricing Levels: This material may contain indicative terms only, including but not limited to pricing levels. There is no representation that any transaction can or could have been effected at such terms or prices. Proposed terms and conditions are for discussion purposes only. Finalized terms and conditions are subject to further discussion and negotiation.
www.goldmansachs.com/disclaimer/sales-and-trading-invest-rec-disclosures.html If you are not accessing this material via Marquee ContentStream, a list of the author's investment recommendations disseminated during the preceding 12 months and the proportion of the author's recommendations that are 'buy', 'hold', 'sell' or other over the previous 12 months is available by logging into Marquee ContentStream using the link below. Alternatively, if you do not have access to Marquee ContentStream, please contact your usual GS representative who will be able to provide this information to you.
Backtesting, Simulated Results, Sensitivity/Scenario Analysis or Spreadsheet Calculator or Model: There may be data presented herein that is solely for illustrative purposes and which may include among other things back testing, simulated results and scenario analyses. The information is based upon certain factors, assumptions and historical information that Goldman Sachs may in its discretion have considered appropriate, however, Goldman Sachs provides no assurance or guarantee that this product will operate or would have operated in the past in a manner consistent with these assumptions. In the event any of the assumptions used do not prove to be true, results are likely to vary materially from the examples shown herein. Additionally, the results may not reflect material economic and market factors, such as liquidity, transaction costs and other expenses which could reduce potential return.
OTC Derivatives Risk Disclosures:
Terms of the Transaction: To understand clearly the terms and conditions of any OTC derivative transaction you may enter into, you should carefully review the Master Agreement, including any related schedules, credit support documents, addenda and exhibits. You should not enter into OTC derivative transactions unless you understand the terms of the transaction you are entering into as well as the nature and extent of your risk exposure. You should also be satisfied that the OTC derivative transaction is appropriate for you in light of your circumstances and financial condition. You may be requested to post margin or collateral to support written OTC derivatives at levels consistent with the internal policies of Goldman Sachs.
Liquidity Risk: There is no public market for OTC derivative transactions and, therefore, it may be difficult or impossible to liquidate an existing position on favorable terms. Transfer Restrictions: OTC derivative transactions entered into with one or more affiliates of The Goldman Sachs Group, Inc. (Goldman Sachs) cannot be assigned or otherwise transferred without its prior written consent and, therefore, it may be impossible for you to transfer any OTC derivative transaction to a third party.
Conflict of Interests: Goldman Sachs may from time to time be an active participant on both sides of the market for the underlying securities, commodities, futures, options or any other derivative or instrument identical or related to those mentioned herein (together, "the Product"). Goldman Sachs at any time may have long or short positions in, or buy and sell Products (on a principal basis or otherwise) identical or related to those mentioned herein. Goldman Sachs hedging and trading activities may affect the value of the Products.
Counterparty Credit Risk: Because Goldman Sachs, may be obligated to make substantial payments to you as a condition of an OTC derivative transaction, you must evaluate the credit risk of doing business with Goldman Sachs or its affiliates.
Pricing and Valuation: The price of each OTC derivative transaction is individually negotiated between Goldman Sachs and each counterparty and Goldman Sachs does not represent or warrant that the prices for which it offers OTC derivative transactions are the best prices available, possibly making it difficult for you to establish what is a fair price for a particular OTC derivative transaction; The value or quoted price of the Product at any time, however, will reflect many factors and cannot be predicted. If Goldman Sachs makes a market in the offered Product, the price quoted by Goldman Sachs would reflect any changes in market conditions and other relevant factors, and the quoted price (and the value of the Product that Goldman Sachs will use for account statements or otherwise) could be higher or lower than the original price, and may be higher or lower than the value of the Product as determined by reference to pricing models used by Goldman Sachs. If at any time a third party dealer quotes a price to purchase the Product or otherwise values the Product, that price may be significantly different (higher or lower) than any price quoted by Goldman Sachs. Furthermore, if you sell the Product, you will likely be charged a commission for secondary market transactions, or the price will likely reflect a dealer discount. Goldman Sachs may conduct market making activities in the Product. To the extent Goldman Sachs makes a market, any price quoted for the OTC derivative transactions, Goldman Sachs may differ significantly from (i) their value determined by reference to Goldman Sachs pricing models and (ii) any price quoted by a third party. The market price of the OTC derivative transaction may be influenced by many unpredictable factors, including economic conditions, the creditworthiness of Goldman Sachs, the value of any underlyers, and certain actions taken by Goldman Sachs.
Market Making, Investing and Lending: Goldman Sachs engages in market making, investing and lending businesses for its own account and the accounts of its affiliates in the same or similar instruments underlying OTC derivative transactions (including such trading as Goldman Sachs deems appropriate in its sole discretion to hedge its market risk in any OTC derivative transaction whether between Goldman Sachs and you or with third parties) and such trading may affect the value of an OTC derivative transaction.
Early Termination Payments: The provisions of an OTC Derivative Transaction may allow for early termination and, in such cases, either you or Goldman Sachs may be required to make a potentially significant termination payment depending upon whether the OTC Derivative Transaction is in-the-money to Goldman Sachs or you at the time of termination. Indexes: Goldman Sachs does not warrant, and takes no responsibility for, the structure, method of computation or publication of any currency exchange rates, interest rates, indexes of such rates, or credit, equity or other indexes, unless Goldman Sachs specifically advises you otherwise.
Risk Disclosure Regarding futures, options, equity swaps, and other derivatives as well as non-investment-grade securities and ADRs: Please ensure that you have read and understood the current options, futures and security futures disclosure document before entering into any such transactions. Current United States listed options, futures and security futures disclosure documents are available from our sales representatives or at http://www.theocc.com/components/docs/riskstoc.pdf, http://www.goldmansachs.com/disclosures/risk-disclosure-for-futures.pdf and https://www.nfa.futures.org/investors/investor-resources/files/security-futures-disclosure.pdf, respectively. Certain transactions - including those involving futures, options, equity swaps, and other derivatives as well as non-investment-grade securities - give rise to substantial risk and are not available to nor suitable for all investors. If you have any questions about whether you are eligible to enter into these transactions with Goldman Sachs, please contact your sales representative. Foreign-currency-denominated securities are subject to fluctuations in exchange rates that could have an adverse effect on the value or price of, or income derived from, the investment. In addition, investors in securities such as ADRs, the values of which are influenced by foreign currencies, effectively assume currency risk.
Options Risk Disclosures: Options may trade at a value other than that which may be inferred from the current levels of interest rates, dividends (if applicable) and the underlier due to other factors including, but not limited to, expectations of future levels of interest rates, future levels of dividends and the volatility of the underlier at any time prior to maturity. Note: Options involve risk and are not suitable for all investors. Please ensure that you have read and understood the current options disclosure document before entering into any standardized options transactions. United States listed options disclosure documents are available from our sales representatives or at http://theocc.com/publications/risks/riskstoc.pdf. A secondary market may not be available for all options. Transaction costs may be a significant factor in option strategies calling for multiple purchases and sales of options, such as spreads. When purchasing long options an investor may lose their entire investment and when selling uncovered options the risk is potentially unlimited. Supporting documentation for any comparisons, recommendations, statistics, technical data, or other similar information will be supplied upon request.
This material is for the private information of the recipient only. This material is not sponsored, endorsed, sold or promoted by any sponsor or provider of an index referred herein (each, an "Index Provider"). GS does not have any affiliation with or control over the Index Providers or any control over the computation, composition or dissemination of the indices. While GS will obtain information from publicly available sources it believes reliable, it will not independently verify this information. Accordingly, GS shall have no liability, contingent or otherwise, to the user or to third parties, for the quality, accuracy, timeliness, continued availability or completeness of the data nor for any special, indirect, incidental or consequential damages which may be incurred or experienced because of the use of the data made available herein, even if GS has been advised of the possibility of such damages.
Standard & Poor's ® and S&P ® are registered trademarks of The McGraw-Hill Companies, Inc. and S&P GSCI™ is a trademark of The McGraw-Hill Companies, Inc. and have been licensed for use by the Issuer. This Product (the "Product") is not sponsored, endorsed, sold or promoted by S&P and S&P makes no representation, warranty or condition regarding the advisability of investing in the Product.
Notice to Brazilian Investors
Marquee is not meant for the general public in Brazil. The services or products provided by or through Marquee, at any time, may not be offered or sold to the general public in Brazil. You have received a password granting access to Marquee exclusively due to your existing relationship with a GS business located in Brazil. The selection and engagement with any of the offered services or products through Marquee, at any time, will be carried out directly by you. Before acting to implement any chosen service or products, provided by or through Marquee you should consider, at your sole discretion, whether it is suitable for your particular circumstances and, if necessary, seek professional advice. Any steps necessary in order to implement the chosen service or product, including but not limited to remittance of funds, shall be carried out at your discretion. Accordingly, such services and products have not been and will not be publicly issued, placed, distributed, offered or negotiated in the Brazilian capital markets and, as a result, they have not been and will not be registered with the Brazilian Securities and Exchange Commission (Comissão de Valores Mobiliários), nor have they been submitted to the foregoing agency for approval. Documents relating to such services or products, as well as the information contained therein, may not be supplied to the general public in Brazil, as the offering of such services or products is not a public offering in Brazil, nor used in connection with any offer for subscription or sale of securities to the general public in Brazil.
The offer of any securities mentioned in this message may not be made to the general public in Brazil. Accordingly, any such securities have not been nor will they be registered with the Brazilian Securities and Exchange Commission (Comissão de Valores Mobiliários) nor has any offer been submitted to the foregoing agency for approval. Documents relating to the offer, as well as the information contained therein, may not be supplied to the public in Brazil, as the offer is not a public offering of securities in Brazil. These terms will apply on every access to Marquee.
Ouvidoria Goldman Sachs Brasil: 0800 727 5764 e/ou ouvidoriagoldmansachs@gs.com
Horário de funcionamento: segunda-feira à sexta-feira (exceto feriados), das 9hs às 18hs.
Ombudsman Goldman Sachs Brazil: 0800 727 5764 and / or ouvidoriagoldmansachs@gs.com
Available Weekdays (except holidays), from 9 am to 6 pm.
| github_jupyter |
# 💡 Solutions
Before trying out these solutions, please start the [gqlalchemy-workshop notebook](../workshop/gqlalchemy-workshop.ipynb) to import all data. Also, this solutions manual is here to help you out, and it is recommended you try solving the exercises first by yourself.
## Exercise 1
**Find out how many genres there are in the database.**
The correct Cypher query is:
```
MATCH (g:Genre)
RETURN count(g) AS num_of_genres;
```
You can try it out in Memgraph Lab at `localhost:3000`.
With GQLAlchemy's query builder, the solution is:
```
from gqlalchemy import match
total_genres = (
match()
.node(labels="Genre", variable="g")
.return_({"count(g)": "num_of_genres"})
.execute()
)
results = list(total_genres)
for result in results:
print(result["num_of_genres"])
```
## Exercise 2
**Find out to how many genres movie 'Matrix, The (1999)' belongs to.**
The correct Cypher query is:
```
MATCH (:Movie {title: 'Matrix, The (1999)'})-[:OF_GENRE]->(g:Genre)
RETURN count(g) AS num_of_genres;
```
You can try it out in Memgraph Lab at `localhost:3000`.
With GQLAlchemy's query builder, the solution is:
```
matrix = (
match()
.node(labels="Movie", variable="m")
.to("OF_GENRE")
.node(labels="Genre", variable="g")
.where("m.title", "=", "Matrix, The (1999)")
.return_({"count(g)": "num_of_genres"})
.execute()
)
results = list(matrix)
for result in results:
print(result["num_of_genres"])
```
## Exercise 3
**Find out the title of the movies that the user with `id` 1 rated.**
The correct Cypher query is:
```
MATCH (:User {id: 1})-[:RATED]->(m:Movie)
RETURN m.title;
```
You can try it out in Memgraph Lab at `localhost:3000`.
With GQLAlchemy's query builder, the solution is:
```
movies = (
match()
.node(labels="User", variable="u")
.to("RATED")
.node(labels="Movie", variable="m")
.where("u.id", "=", 1)
.return_({"m.title": "movie"})
.execute()
)
results = list(movies)
for result in results:
print(result["movie"])
```
## Exercise 4
**List 15 movies of 'Documentary' and 'Comedy' genres and sort them by title descending.**
The correct Cypher query is:
```
MATCH (m:Movie)-[:OF_GENRE]->(:Genre {name: "Documentary"})
MATCH (m)-[:OF_GENRE]->(:Genre {name: "Comedy"})
RETURN m.title
ORDER BY m.title DESC
LIMIT 15;
```
You can try it out in Memgraph Lab at `localhost:3000`.
With GQLAlchemy's query builder, the solution is:
```
movies = (
match()
.node(labels="Movie", variable="m")
.to("OF_GENRE")
.node(labels="Genre", variable="g1")
.where("g1.name", "=", "Documentary")
.match()
.node(labels="Movie", variable="m")
.to("OF_GENRE")
.node(labels="Genre", variable="g2")
.where("g2.name", "=", "Comedy")
.return_({"m.title": "movie"})
.order_by("m.title DESC")
.limit(15)
.execute()
)
results = list(movies)
for result in results:
print(result["movie"])
```
## Exercise 5
**Find out the minimum rating of the 'Star Wars: Episode I - The Phantom Menace (1999)' movie.**
The correct Cypher query is:
```
MATCH (:User)-[r:RATED]->(:Movie {title: 'Star Wars: Episode I - The Phantom Menace (1999)'})
RETURN min(r.rating);
```
You can try it out in Memgraph Lab at `localhost:3000`.
With GQLAlchemy's query builder, the solution is:
```
rating = (
match()
.node(labels="User")
.to("RATED", variable="r")
.node(labels="Movie", variable="m")
.where("m.title", "=", "Star Wars: Episode I - The Phantom Menace (1999)")
.return_({"min(r.rating)": "min_rating"})
.execute()
)
results = list(rating)
for result in results:
print(result["min_rating"])
```
And that's it! If you have any issues with this notebook, feel free to open an issue on the [GitHub repository](https://github.com/pyladiesams/graphdbs-gqlalchemy-beginner-mar2022), or [join the Discord server](https://discord.gg/memgraph) and get your answer instantly. If you are interested in the Cypher query language and want to learn more, sign up for the free [Cypher Email Course](https://memgraph.com/learn-cypher-query-language).
| github_jupyter |
```
# "PGA Tour Wins Classification"
```
Can We Predict If a PGA Tour Player Won a Tournament in a Given Year?
Golf is picking up popularity, so I thought it would be interesting to focus my project here. I set out to find what sets apart the best golfers from the rest.
I decided to explore their statistics and to see if I could predict which golfers would win in a given year. My original dataset was found on Kaggle, and the data was scraped from the PGA Tour website.
From this data, I performed an exploratory data analysis to explore the distribution of players on numerous aspects of the game, discover outliers, and further explore how the game has changed from 2010 to 2018. I also utilized numerous supervised machine learning models to predict a golfer's earnings and wins.
To predict the golfer's win, I used classification methods such as logisitic regression and Random Forest Classification. The best performance came from the Random Forest Classification method.
1. The Data
pgaTourData.csv contains 1674 rows and 18 columns. Each row indicates a golfer's performance for that year.
```
# Player Name: Name of the golfer
# Rounds: The number of games that a player played
# Fairway Percentage: The percentage of time a tee shot lands on the fairway
# Year: The year in which the statistic was collected
# Avg Distance: The average distance of the tee-shot
# gir: (Green in Regulation) is met if any part of the ball is touching the putting surface while the number of strokes taken is at least two fewer than par
# Average Putts: The average number of strokes taken on the green
# Average Scrambling: Scrambling is when a player misses the green in regulation, but still makes par or better on a hole
# Average Score: Average Score is the average of all the scores a player has played in that year
# Points: The number of FedExCup points a player earned in that year
# Wins: The number of competition a player has won in that year
# Top 10: The number of competitions where a player has placed in the Top 10
# Average SG Putts: Strokes gained: putting measures how many strokes a player gains (or loses) on the greens
# Average SG Total: The Off-the-tee + approach-the-green + around-the-green + putting statistics combined
# SG:OTT: Strokes gained: off-the-tee measures player performance off the tee on all par-4s and par-5s
# SG:APR: Strokes gained: approach-the-green measures player performance on approach shots
# SG:ARG: Strokes gained: around-the-green measures player performance on any shot within 30 yards of the edge of the green
# Money: The amount of prize money a player has earned from tournaments
#collapse
# importing packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# Importing the data
df = pd.read_csv('pgaTourData.csv')
# Examining the first 5 data
df.head()
#collapse
df.info()
#collapse
df.shape
```
2. Data Cleaning
After looking at the dataframe, the data needs to be cleaned:
-For the columns Top 10 and Wins, convert the NaNs to 0s
-Change Top 10 and Wins into an int
-Drop NaN values for players who do not have the full statistics
-Change the columns Rounds into int
-Change points to int
-Remove the dollar sign ($) and commas in the column Money
```
# Replace NaN with 0 in Top 10
df['Top 10'].fillna(0, inplace=True)
df['Top 10'] = df['Top 10'].astype(int)
# Replace NaN with 0 in # of wins
df['Wins'].fillna(0, inplace=True)
df['Wins'] = df['Wins'].astype(int)
# Drop NaN values
df.dropna(axis = 0, inplace=True)
# Change Rounds to int
df['Rounds'] = df['Rounds'].astype(int)
# Change Points to int
df['Points'] = df['Points'].apply(lambda x: x.replace(',',''))
df['Points'] = df['Points'].astype(int)
# Remove the $ and commas in money
df['Money'] = df['Money'].apply(lambda x: x.replace('$',''))
df['Money'] = df['Money'].apply(lambda x: x.replace(',',''))
df['Money'] = df['Money'].astype(float)
#collapse
df.info()
#collapse
df.describe()
```
3. Exploratory Data Analysis
```
#collapse_output
# Looking at the distribution of data
f, ax = plt.subplots(nrows = 6, ncols = 3, figsize=(20,20))
distribution = df.loc[:,df.columns!='Player Name'].columns
rows = 0
cols = 0
for i, column in enumerate(distribution):
p = sns.distplot(df[column], ax=ax[rows][cols])
cols += 1
if cols == 3:
cols = 0
rows += 1
```
From the distributions plotted, most of the graphs are normally distributed. However, we can observe that Money, Points, Wins, and Top 10s are all skewed to the right. This could be explained by the separation of the best players and the average PGA Tour player. The best players have multiple placings in the Top 10 with wins that allows them to earn more from tournaments, while the average player will have no wins and only a few Top 10 placings that prevent them from earning as much.
```
#collapse_output
# Looking at the number of players with Wins for each year
win = df.groupby('Year')['Wins'].value_counts()
win = win.unstack()
win.fillna(0, inplace=True)
# Converting win into ints
win = win.astype(int)
print(win)
```
From this table, we can see that most players end the year without a win. It's pretty rare to find a player that has won more than once!
```
# Looking at the percentage of players without a win in that year
players = win.apply(lambda x: np.sum(x), axis=1)
percent_no_win = win[0]/players
percent_no_win = percent_no_win*100
print(percent_no_win)
#collapse_output
# Plotting percentage of players without a win each year
fig, ax = plt.subplots()
bar_width = 0.8
opacity = 0.7
index = np.arange(2010, 2019)
plt.bar(index, percent_no_win, bar_width, alpha = opacity)
plt.xticks(index)
plt.xlabel('Year')
plt.ylabel('%')
plt.title('Percentage of Players without a Win')
```
From the box plot above, we can observe that the percentages of players without a win are around 80%. There was very little variation in the percentage of players without a win in the past 8 years.
```
#collapse_output
# Plotting the number of wins on a bar chart
fig, ax = plt.subplots()
index = np.arange(2010, 2019)
bar_width = 0.2
opacity = 0.7
def plot_bar(index, win, labels):
plt.bar(index, win, bar_width, alpha=opacity, label=labels)
# Plotting the bars
rects = plot_bar(index, win[0], labels = '0 Wins')
rects1 = plot_bar(index + bar_width, win[1], labels = '1 Wins')
rects2 = plot_bar(index + bar_width*2, win[2], labels = '2 Wins')
rects3 = plot_bar(index + bar_width*3, win[3], labels = '3 Wins')
rects4 = plot_bar(index + bar_width*4, win[4], labels = '4 Wins')
rects5 = plot_bar(index + bar_width*5, win[5], labels = '5 Wins')
plt.xticks(index + bar_width, index)
plt.xlabel('Year')
plt.ylabel('Number of Players')
plt.title('Distribution of Wins each Year')
plt.legend()
```
By looking at the distribution of Wins each year, we can see that it is rare for most players to even win a tournament in the PGA Tour. Majority of players do not win, and a very few number of players win more than once a year.
```
# Percentage of people who did not place in the top 10 each year
top10 = df.groupby('Year')['Top 10'].value_counts()
top10 = top10.unstack()
top10.fillna(0, inplace=True)
players = top10.apply(lambda x: np.sum(x), axis=1)
no_top10 = top10[0]/players * 100
print(no_top10)
```
By looking at the percentage of players that did not place in the top 10 by year, We can observe that only approximately 20% of players did not place in the Top 10. In addition, the range for these player that did not place in the Top 10 is only 9.47%. This tells us that this statistic does not vary much on a yearly basis.
```
# Who are some of the longest hitters
distance = df[['Year','Player Name','Avg Distance']].copy()
distance.sort_values(by='Avg Distance', inplace=True, ascending=False)
print(distance.head())
```
Rory McIlroy is one of the longest hitters in the game, setting the average driver distance to be 319.7 yards in 2018. He was also the longest hitter in 2017 with an average of 316.7 yards.
```
# Who made the most money
money_ranking = df[['Year','Player Name','Money']].copy()
money_ranking.sort_values(by='Money', inplace=True, ascending=False)
print(money_ranking.head())
```
We can see that Jordan Spieth has made the most amount of money in a year, earning a total of 12 million dollars in 2015.
```
#collapse_output
# Who made the most money each year
money_rank = money_ranking.groupby('Year')['Money'].max()
money_rank = pd.DataFrame(money_rank)
indexs = np.arange(2010, 2019)
names = []
for i in range(money_rank.shape[0]):
temp = df.loc[df['Money'] == money_rank.iloc[i,0],'Player Name']
names.append(str(temp.values[0]))
money_rank['Player Name'] = names
print(money_rank)
```
With this table, we can examine the earnings of each player by year. Some of the most notable were Jordan Speith's earning of 12 million dollars and Justin Thomas earning the most money in both 2017 and 2018.
```
#collapse_output
# Plot the correlation matrix between variables
corr = df.corr()
sns.heatmap(corr,
xticklabels=corr.columns.values,
yticklabels=corr.columns.values,
cmap='coolwarm')
df.corr()['Wins']
```
From the correlation matrix, we can observe that Money is highly correlated to wins along with the FedExCup Points. We can also observe that the fairway percentage, year, and rounds are not correlated to Wins.
4. Machine Learning Model (Classification)
To predict winners, I used multiple machine learning models to explore which models could accurately classify if a player is going to win in that year.
To measure the models, I used Receiver Operating Characterisitc Area Under the Curve. (ROC AUC) The ROC AUC tells us how capable the model is at distinguishing players with a win. In addition, as the data is skewed with 83% of players having no wins in that year, ROC AUC is a much better metric than the accuracy of the model.
```
#collapse
# Importing the Machine Learning modules
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_curve, roc_auc_score
from sklearn.metrics import confusion_matrix
from sklearn.feature_selection import RFE
from sklearn.metrics import classification_report
from sklearn.preprocessing import PolynomialFeatures
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.preprocessing import MinMaxScaler
```
Preparing the Data for Classification
We know from the calculation above that the data for wins is skewed. Even without machine learning we know that approximately 83% of the players does not lead to a win. Therefore, we will be utilizing ROC AUC as the metric of these models
```
# Adding the Winner column to determine if the player won that year or not
df['Winner'] = df['Wins'].apply(lambda x: 1 if x>0 else 0)
# New DataFrame
ml_df = df.copy()
# Y value for machine learning is the Winner column
target = df['Winner']
# Removing the columns Player Name, Wins, and Winner from the dataframe to avoid leakage
ml_df.drop(['Player Name','Wins','Winner'], axis=1, inplace=True)
print(ml_df.head())
## Logistic Regression Baseline
per_no_win = target.value_counts()[0] / (target.value_counts()[0] + target.value_counts()[1])
per_no_win = per_no_win.round(4)*100
print(str(per_no_win)+str('%'))
#collapse_show
# Function for the logisitic regression
def log_reg(X, y):
X_train, X_test, y_train, y_test = train_test_split(X, y,
random_state = 10)
clf = LogisticRegression().fit(X_train, y_train)
y_pred = clf.predict(X_test)
print('Accuracy of Logistic regression classifier on training set: {:.2f}'
.format(clf.score(X_train, y_train)))
print('Accuracy of Logistic regression classifier on test set: {:.2f}'
.format(clf.score(X_test, y_test)))
cf_mat = confusion_matrix(y_test, y_pred)
confusion = pd.DataFrame(data = cf_mat)
print(confusion)
print(classification_report(y_test, y_pred))
# Returning the 5 important features
#rfe = RFE(clf, 5)
# rfe = rfe.fit(X, y)
# print('Feature Importance')
# print(X.columns[rfe.ranking_ == 1].values)
print('ROC AUC Score: {:.2f}'.format(roc_auc_score(y_test, y_pred)))
#collapse_show
log_reg(ml_df, target)
```
From the logisitic regression, we got an accuracy of 0.9 on the training set and an accuracy of 0.91 on the test set. This was surprisingly accurate for a first run. However, the ROC AUC Score of 0.78 could be improved. Therefore, I decided to add more features as a way of possibly improving the model.
```
## Feature Engineering
# Adding Domain Features
ml_d = ml_df.copy()
# Top 10 / Money might give us a better understanding on how well they placed in the top 10
ml_d['Top10perMoney'] = ml_d['Top 10'] / ml_d['Money']
# Avg Distance / Fairway Percentage to give us a ratio that determines how accurate and far a player hits
ml_d['DistanceperFairway'] = ml_d['Avg Distance'] / ml_d['Fairway Percentage']
# Money / Rounds to see on average how much money they would make playing a round of golf
ml_d['MoneyperRound'] = ml_d['Money'] / ml_d['Rounds']
#collapse_show
log_reg(ml_d, target)
#collapse_show
# Adding Polynomial Features to the ml_df
mldf2 = ml_df.copy()
poly = PolynomialFeatures(2)
poly = poly.fit(mldf2)
poly_feature = poly.transform(mldf2)
print(poly_feature.shape)
# Creating a DataFrame with the polynomial features
poly_feature = pd.DataFrame(poly_feature, columns = poly.get_feature_names(ml_df.columns))
print(poly_feature.head())
#collapse_show
log_reg(poly_feature, target)
```
From feature engineering, there were no improvements in the ROC AUC Score. In fact as I added more features, the accuracy and the ROC AUC Score decreased. This could signal to us that another machine learning algorithm could better predict winners.
```
#collapse_show
## Randon Forest Model
def random_forest(X, y):
X_train, X_test, y_train, y_test = train_test_split(X, y,
random_state = 10)
clf = RandomForestClassifier(n_estimators=200).fit(X_train, y_train)
y_pred = clf.predict(X_test)
print('Accuracy of Random Forest classifier on training set: {:.2f}'
.format(clf.score(X_train, y_train)))
print('Accuracy of Random Forest classifier on test set: {:.2f}'
.format(clf.score(X_test, y_test)))
cf_mat = confusion_matrix(y_test, y_pred)
confusion = pd.DataFrame(data = cf_mat)
print(confusion)
print(classification_report(y_test, y_pred))
# Returning the 5 important features
rfe = RFE(clf, 5)
rfe = rfe.fit(X, y)
print('Feature Importance')
print(X.columns[rfe.ranking_ == 1].values)
print('ROC AUC Score: {:.2f}'.format(roc_auc_score(y_test, y_pred)))
#collapse_show
random_forest(ml_df, target)
#collapse_show
random_forest(ml_d, target)
#collapse_show
random_forest(poly_feature, target)
```
The Random Forest Model scored highly on the ROC AUC Score, obtaining a value of 0.89. With this, we observed that the Random Forest Model could accurately classify players with and without a win.
6. Conclusion
It's been interesting to learn about numerous aspects of the game that differentiate the winner and the average PGA Tour player. For example, we can see that the fairway percentage and greens in regulations do not seem to contribute as much to a player's win. However, all the strokes gained statistics contribute pretty highly to wins for these players. It was interesting to see which aspects of the game that the professionals should put their time into. This also gave me the idea of track my personal golf statistics, so that I could compare it to the pros and find areas of my game that need the most improvement.
Machine Learning Model
I've been able to examine the data of PGA Tour players and classify if a player will win that year or not. With the random forest classification model, I was able to achieve an ROC AUC of 0.89 and an accuracy of 0.95 on the test set. This was a significant improvement from the ROC AUC of 0.78 and accuracy of 0.91. Because the data is skewed with approximately 80% of players not earning a win, the primary measure of the model was the ROC AUC. I was able to improve my model from ROC AUC score of 0.78 to a score of 0.89 by simply trying 3 different models, adding domain features, and polynomial features.
The End!!
| github_jupyter |
# Monte Carlo Methods
In this notebook, you will write your own implementations of many Monte Carlo (MC) algorithms.
While we have provided some starter code, you are welcome to erase these hints and write your code from scratch.
### Part 0: Explore BlackjackEnv
We begin by importing the necessary packages.
```
import sys
import gym
import numpy as np
from collections import defaultdict
from plot_utils import plot_blackjack_values, plot_policy
```
Use the code cell below to create an instance of the [Blackjack](https://github.com/openai/gym/blob/master/gym/envs/toy_text/blackjack.py) environment.
```
env = gym.make('Blackjack-v0')
```
Each state is a 3-tuple of:
- the player's current sum $\in \{0, 1, \ldots, 31\}$,
- the dealer's face up card $\in \{1, \ldots, 10\}$, and
- whether or not the player has a usable ace (`no` $=0$, `yes` $=1$).
The agent has two potential actions:
```
STICK = 0
HIT = 1
```
Verify this by running the code cell below.
```
print(f"Observation space: \t{env.observation_space}")
print(f"Action space: \t\t{env.action_space}")
```
Execute the code cell below to play Blackjack with a random policy.
(_The code currently plays Blackjack three times - feel free to change this number, or to run the cell multiple times. The cell is designed for you to get some experience with the output that is returned as the agent interacts with the environment._)
```
for i_episode in range(3):
state = env.reset()
while True:
print(state)
action = env.action_space.sample()
state, reward, done, info = env.step(action)
if done:
print('End game! Reward: ', reward)
print('You won :)\n') if reward > 0 else print('You lost :(\n')
break
```
### Part 1: MC Prediction
In this section, you will write your own implementation of MC prediction (for estimating the action-value function).
We will begin by investigating a policy where the player _almost_ always sticks if the sum of her cards exceeds 18. In particular, she selects action `STICK` with 80% probability if the sum is greater than 18; and, if the sum is 18 or below, she selects action `HIT` with 80% probability. The function `generate_episode_from_limit_stochastic` samples an episode using this policy.
The function accepts as **input**:
- `bj_env`: This is an instance of OpenAI Gym's Blackjack environment.
It returns as **output**:
- `episode`: This is a list of (state, action, reward) tuples (of tuples) and corresponds to $(S_0, A_0, R_1, \ldots, S_{T-1}, A_{T-1}, R_{T})$, where $T$ is the final time step. In particular, `episode[i]` returns $(S_i, A_i, R_{i+1})$, and `episode[i][0]`, `episode[i][1]`, and `episode[i][2]` return $S_i$, $A_i$, and $R_{i+1}$, respectively.
```
def generate_episode_from_limit_stochastic(bj_env):
episode = []
state = bj_env.reset()
while True:
probs = [0.8, 0.2] if state[0] > 18 else [0.2, 0.8]
action = np.random.choice(np.arange(2), p=probs)
next_state, reward, done, info = bj_env.step(action)
episode.append((state, action, reward))
state = next_state
if done:
break
return episode
```
Execute the code cell below to play Blackjack with the policy.
(*The code currently plays Blackjack three times - feel free to change this number, or to run the cell multiple times. The cell is designed for you to gain some familiarity with the output of the `generate_episode_from_limit_stochastic` function.*)
```
for i in range(5):
print(generate_episode_from_limit_stochastic(env))
```
Now, you are ready to write your own implementation of MC prediction. Feel free to implement either first-visit or every-visit MC prediction; in the case of the Blackjack environment, the techniques are equivalent.
Your algorithm has three arguments:
- `env`: This is an instance of an OpenAI Gym environment.
- `num_episodes`: This is the number of episodes that are generated through agent-environment interaction.
- `generate_episode`: This is a function that returns an episode of interaction.
- `gamma`: This is the discount rate. It must be a value between 0 and 1, inclusive (default value: `1`).
The algorithm returns as output:
- `Q`: This is a dictionary (of one-dimensional arrays) where `Q[s][a]` is the estimated action value corresponding to state `s` and action `a`.
```
def mc_prediction_q(env, num_episodes, generate_episode, gamma=1.0):
# initialize empty dictionaries of arrays
returns_sum = defaultdict(lambda: np.zeros(env.action_space.n))
N = defaultdict(lambda: np.zeros(env.action_space.n))
Q = defaultdict(lambda: np.zeros(env.action_space.n))
R = defaultdict(lambda: np.zeros(env.action_space.n))
# loop over episodes
for i_episode in range(1, num_episodes+1):
# monitor progress
if i_episode % 1000 == 0:
print("\rEpisode {}/{}.".format(i_episode, num_episodes), end="")
sys.stdout.flush()
episode = generate_episode(env)
n = len(episode)
states, actions, rewards = zip(*episode)
discounts = np.array([gamma**i for i in range(n+1)])
for i, state in enumerate(states):
returns_sum[state][actions[i]] += sum(rewards[i:] * discounts[:-(i+1)])
N[state][actions[i]] += 1
# comnpute Q table
for state in returns_sum.keys():
for action in range(env.action_space.n):
Q[state][action] = returns_sum[state][action] / N[state][action]
return Q, returns_sum, N
```
Use the cell below to obtain the action-value function estimate $Q$. We have also plotted the corresponding state-value function.
To check the accuracy of your implementation, compare the plot below to the corresponding plot in the solutions notebook **Monte_Carlo_Solution.ipynb**.
```
# obtain the action-value function
Q, R, N = mc_prediction_q(env, 500000, generate_episode_from_limit_stochastic)
# obtain the corresponding state-value function
V_to_plot = dict((k,(k[0]>18)*(np.dot([0.8, 0.2],v)) + (k[0]<=18)*(np.dot([0.2, 0.8],v))) \
for k, v in Q.items())
# plot the state-value function
plot_blackjack_values(V_to_plot)
```
### Part 2: MC Control
In this section, you will write your own implementation of constant-$\alpha$ MC control.
Your algorithm has four arguments:
- `env`: This is an instance of an OpenAI Gym environment.
- `num_episodes`: This is the number of episodes that are generated through agent-environment interaction.
- `alpha`: This is the step-size parameter for the update step.
- `gamma`: This is the discount rate. It must be a value between 0 and 1, inclusive (default value: `1`).
The algorithm returns as output:
- `Q`: This is a dictionary (of one-dimensional arrays) where `Q[s][a]` is the estimated action value corresponding to state `s` and action `a`.
- `policy`: This is a dictionary where `policy[s]` returns the action that the agent chooses after observing state `s`.
(_Feel free to define additional functions to help you to organize your code._)
```
def generate_episode_from_Q(env, Q, epsilon, n):
""" generates an episode following the epsilon-greedy policy"""
episode = []
state = env.reset()
while True:
if state in Q:
action = np.random.choice(np.arange(n), p=get_props(Q[state], epsilon, n))
else:
action = env.action_space.sample()
next_state, reward, done, _ = env.step(action)
episode.append((state, action, reward))
state = next_state
if done:
break
return episode
def get_props(Q_s, epsilon, n):
policy_s = np.ones(n) * epsilon / n
best_a = np.argmax(Q_s)
policy_s[best_a] = 1 - epsilon + (epsilon / n)
return policy_s
def update_Q(episode, Q, alpha, gamma):
n = len(episode)
states, actions, rewards = zip(*episode)
discounts = np.array([gamma**i for i in range(n+1)])
for i, state in enumerate(states):
R = sum(rewards[i:] * discounts[:-(1+i)])
Q[state][actions[i]] = Q[state][actions[i]] + alpha * (R - Q[state][actions[i]])
return Q
def mc_control(env, num_episodes, alpha, gamma=1.0, eps_start=1.0, eps_decay=.99999, eps_min=0.05):
nA = env.action_space.n
# initialize empty dictionary of arrays
Q = defaultdict(lambda: np.zeros(nA))
epsilon = eps_start
# loop over episodes
for i_episode in range(1, num_episodes+1):
# monitor progress
if i_episode % 1000 == 0:
print("\rEpisode {}/{}.".format(i_episode, num_episodes), end="")
sys.stdout.flush()
epsilon = max(eps_min, epsilon * eps_decay)
episode = generate_episode_from_Q(env, Q, epsilon, nA)
Q = update_Q(episode, Q, alpha, gamma)
policy = dict((s, np.argmax(v)) for s, v in Q.items())
return policy, Q
```
Use the cell below to obtain the estimated optimal policy and action-value function. Note that you should fill in your own values for the `num_episodes` and `alpha` parameters.
```
# obtain the estimated optimal policy and action-value function
policy, Q = mc_control(env, 500000, 0.02)
```
Next, we plot the corresponding state-value function.
```
# obtain the corresponding state-value function
V = dict((k,np.max(v)) for k, v in Q.items())
# plot the state-value function
plot_blackjack_values(V)
```
Finally, we visualize the policy that is estimated to be optimal.
```
# plot the policy
plot_policy(policy)
```
The **true** optimal policy $\pi_*$ can be found in Figure 5.2 of the [textbook](http://go.udacity.com/rl-textbook) (and appears below). Compare your final estimate to the optimal policy - how close are you able to get? If you are not happy with the performance of your algorithm, take the time to tweak the decay rate of $\epsilon$, change the value of $\alpha$, and/or run the algorithm for more episodes to attain better results.

| github_jupyter |
```
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
#%matplotlib inline
from IPython.core.pylabtools import figsize
figsize(8, 6)
sns.set()
```
## Carregando dados dos usuários premium
```
df = pd.read_csv("../data/processed/premium_students.csv",parse_dates=[1,2],index_col=[0])
print(df.shape)
df.head()
```
---
### Novas colunas auxiliares
```
df['diffDate'] = (df.SubscriptionDate - df.RegisteredDate)
df['diffDays'] = [ item.days for item in df['diffDate']]
df['register_time'] = df.RegisteredDate.map( lambda x : int(x.strftime("%H")) )
df['register_time_AM_PM'] = df.register_time.map( lambda x : 1 if x>=12 else 0)
df['register_num_week'] = df.RegisteredDate.map( lambda x : int(x.strftime("%V")) )
df['register_week_day'] = df.RegisteredDate.map( lambda x : int(x.weekday()) )
df['register_month'] = df.RegisteredDate.map( lambda x : int(x.strftime('%m')) )
df['subscription_time'] = df.SubscriptionDate.map( lambda x : int(x.strftime("%H") ))
df['subscription_time_AM_PM'] = df.subscription_time.map( lambda x : 1 if x>=12 else 0)
df['subscription_num_week'] = df.SubscriptionDate.map( lambda x : int(x.strftime("%V")) )
df['subscription_week_day'] = df.SubscriptionDate.map( lambda x : int(x.weekday()) )
df['subscription_month'] = df.SubscriptionDate.map( lambda x : int(x.strftime('%m')) )
df.tail()
```
---
### Verificando distribuições
```
df.register_time.hist()
df.subscription_time.hist()
df.register_time_AM_PM.value_counts()
df.subscription_time_AM_PM.value_counts()
df.subscription_week_day.value_counts()
df.diffDays.hist()
df.diffDays.quantile([.25,.5,.75,.95])
```
Separando os dados em 2 momentos.
```
lt_50 = df.loc[(df.diffDays <50) & (df.diffDays >3)]
lt_50.diffDays.hist()
lt_50.diffDays.value_counts()
lt_50.diffDays.quantile([.25,.5,.75,.95])
range_0_3 = df.loc[(df.diffDays < 3)]
range_3_18 = df.loc[(df.diffDays >= 3)&(df.diffDays < 18)]
range_6_11 = df.loc[(df.diffDays >= 6) & (df.diffDays < 11)]
range_11_18 = df.loc[(df.diffDays >= 11) & (df.diffDays < 18)]
range_18_32 = df.loc[(df.diffDays >= 18 )& (df.diffDays <= 32)]
range_32 = df.loc[(df.diffDays >=32)]
total_subs = df.shape[0]
(
round(range_0_3.shape[0] / total_subs,2),
round(range_3_18.shape[0] / total_subs,2),
round(range_18_32.shape[0] / total_subs,2),
round(range_32.shape[0] / total_subs,2)
)
gte_30 = df.loc[df.diffDays >=32]
gte_30.diffDays.hist()
gte_30.diffDays.value_counts()
gte_30.shape
gte_30.diffDays.quantile([.25,.5,.75,.95])
range_32_140 = df.loc[(df.diffDays > 32)&(df.diffDays <=140)]
range_140_168 = df.loc[(df.diffDays > 140)&(df.diffDays <=168)]
range_168_188 = df.loc[(df.diffDays > 168)&(df.diffDays <=188)]
range_188 = df.loc[(df.diffDays > 188)]
total_subs_gte_32 = gte_30.shape[0]
(
round(range_32_140.shape[0] / total_subs,2),
round(range_140_168.shape[0] / total_subs,2),
round(range_168_188.shape[0] / total_subs,2),
round(range_188.shape[0] / total_subs,2)
)
(
round(range_32_140.shape[0] / total_subs_gte_32,2),
round(range_140_168.shape[0] / total_subs_gte_32,2),
round(range_168_188.shape[0] / total_subs_gte_32,2),
round(range_188.shape[0] / total_subs_gte_32,2)
)
```
----
## Questão 1:
Dentre os usuários cadastrados em Nov/2017 que assinaram o Plano Premium,
qual a probabilidade do usuário virar Premium após o cadastro em ranges de dias? A escolha
dos ranges deve ser feita por você, tendo em vista os insights que podemos tirar para o
negócio.
- De 0 a 3 dias -> 53%
- De 3 a 18 dias -> 12%
- De 18 a 32 -> 3%
- Mais 32 dias -> 33%
Analisando as inscrições feitas depois do primeiro mês (33%)
* De 32 a 140 -> 8%
* De 140 a 168 -> 8%
* De 168 a 188 -> 8%
* De 188 a 216 -> 8%
Um pouco mais da metade das conversões acontecem nos primeiros 3 dias.
A taxa conversão chega a 65% até 18 dias após o registro.
Após 100 dias acontece outro momento relevante que representa 33%.
Possivelmente essa janela coincide com o calendário de provas das instituições.
Insights:
* Maioria das conversões no período da tarde
* Maioria das conversões no começo da semana ( anúncios aos domingos )
* Direcionar anúncios de instagram geolocalizados (instituições) nos períodos que antecede o calendário de provas.
* Tentar converter usuários ativos 100 dias após o registro
* Tentar converter usuários com base no calendário de provas da instituição
| github_jupyter |
# Minimum spanning trees
*Selected Topics in Mathematical Optimization*
**Michiel Stock** ([email](michiel.stock@ugent.be))

```
import matplotlib.pyplot as plt
%matplotlib inline
from minimumspanningtrees import red, green, blue, orange, yellow
```
## Graphs in python
Consider the following example graph:

This graph can be represented using an *adjacency list*. We do this using a `dict`. Every vertex is a key with the adjacent vertices given as a `set` containing tuples `(weight, neighbor)`. The weight is first because this makes it easy to compare the weights of two edges. Note that for every ingoing edges, there is also an outgoing edge, this is an undirected graph.
```
graph = {
'A' : set([(2, 'B'), (3, 'D')]),
'B' : set([(2, 'A'), (1, 'C'), (2, 'E')]),
'C' : set([(1, 'B'), (2, 'D'), (1, 'E')]),
'D' : set([(2, 'C'), (3, 'A'), (3, 'E')]),
'E' : set([(2, 'B'), (1, 'C'), (3, 'D')])
}
```
Sometimes we will use an *edge list*, i.e. a list of (weighted) edges. This is often a more compact way of storing a graph. The edge list is given below. Note that again every edge is double: an in- and outgoing edge is included.
```
edges = [
(2, 'B', 'A'),
(3, 'D', 'A'),
(2, 'C', 'D'),
(3, 'A', 'D'),
(3, 'E', 'D'),
(2, 'B', 'E'),
(3, 'D', 'E'),
(1, 'C', 'E'),
(2, 'E', 'B'),
(2, 'A', 'B'),
(1, 'C', 'B'),
(1, 'E', 'C'),
(1, 'B', 'C'),
(2, 'D', 'C')]
```
We can easily turn one representation in the other (with a time complexity proportional to the number of edges) using the provided functions `edges_to_adj_list` and `adj_list_to_edges`.
```
from minimumspanningtrees import edges_to_adj_list, adj_list_to_edges
adj_list_to_edges(graph)
edges_to_adj_list(edges)
```
## Disjoint-set data structure
Implementing an algorithm for finding the minimum spanning tree is fairly straightforward. The only bottleneck is that the algorithm requires the a disjoint-set data structure to keep track of a set partitioned in a number of disjoined subsets.
For example, consider the following inital set of eight elements.

We decide to group elements A, B and C together in a subset and F and G in another subset.

The disjoint-set data structure support the following operations:
- **Find**: check which subset an element is in. Is typically used to check whether two objects are in the same subset;
- **Union** merges two subsets into a single subset.
A python implementation of a disjoint-set is available using an union-set forest. A simple example will make everything clear!
```
from union_set_forest import USF
animals = ['mouse', 'bat', 'robin', 'trout', 'seagull', 'hummingbird',
'salmon', 'goldfish', 'hippopotamus', 'whale', 'sparrow']
union_set_forest = USF(animals)
# group mammals together
union_set_forest.union('mouse', 'bat')
union_set_forest.union('mouse', 'hippopotamus')
union_set_forest.union('whale', 'bat')
# group birds together
union_set_forest.union('robin', 'seagull')
union_set_forest.union('seagull', 'sparrow')
union_set_forest.union('seagull', 'hummingbird')
union_set_forest.union('robin', 'hummingbird')
# group fishes together
union_set_forest.union('goldfish', 'salmon')
union_set_forest.union('trout', 'salmon')
# mouse and whale in same subset?
print(union_set_forest.find('mouse') == union_set_forest.find('whale'))
# robin and salmon in the same subset?
print(union_set_forest.find('robin') == union_set_forest.find('salmon'))
```
## Heap queue
Can be used to find the minimum of a changing list without having to sort the list every update.
```
from heapq import heapify, heappop, heappush
heap = [(5, 'A'), (3, 'B'), (2, 'C'), (7, 'D')]
heapify(heap) # turn in a heap
print(heap)
# return item lowest value while retaining heap property
print(heappop(heap))
print(heap)
# add new item and retain heap prop
heappush(heap, (4, 'E'))
print(heap)
```
## Prim's algorithm
Prim's algorithm starts with a single vertex and add $|V|-1$ edges to it, always taking the next edge with minimal weight that connects a vertex on the MST to a vertex not yet in the MST.
```
def prim(vertices, edges, start):
"""
Prim's algorithm for finding a minimum spanning tree.
Inputs :
- vertices : a set of the vertices of the Graph
- edges : a list of weighted edges (e.g. (0.7, 'A', 'B') for an
edge from node A to node B with weigth 0.7)
- start : a vertex to start with
Output:
- edges : a minumum spanning tree represented as a list of edges
- total_cost : total cost of the tree
"""
adj_list = edges_to_adj_list(edges) # easier using an adjacency list
... # to complete
return mst_edges, total_cost
```
## Kruskal's algorithm
Kruskal's algorithm is a very simple algorithm to find the minimum spanning tree. The main idea is to start with an intial 'forest' of the individual nodes of the graph. In each step of the algorithm we add an edge with the smallest possible value that connects two disjoint trees in the forest. This process is continued until we have a single tree, which is a minimum spanning tree, or until all edges are considered. In the latter case, the algoritm returns a minimum spanning forest.
```
from minimumspanningtrees import kruskal
def kruskal(vertices, edges):
"""
Kruskal's algorithm for finding a minimum spanning tree.
Inputs :
- vertices : a set of the vertices of the Graph
- edges : a list of weighted edges (e.g. (0.7, 'A', 'B') for an
edge from node A to node B with weigth 0.7)
Output:
- edges : a minumum spanning tree represented as a list of edges
- total_cost : total cost of the tree
"""
... # to complete
return mst_edges, total_cost
```
```
print(vertices)
print(edges[:5])
# compute the minimum spanning tree of the ticket to ride data set
...
```
## Clustering
Minimum spanning trees on a distance graph can be used to cluster a data set.
```
# import features and distance
from clustering import X, D
fig, ax = plt.subplots()
ax.scatter(X[:,0], X[:,1], color=green)
# cluster the data based on the distance
```
| github_jupyter |
<a href="https://colab.research.google.com/github/yukinaga/bert_nlp/blob/main/section_2/03_simple_bert.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# シンプルなBERTの実装
訓練済みのモデルを使用し、文章の一部の予測、及び2つの文章が連続しているかどうかの判定を行います。
## ライブラリのインストール
PyTorch-Transformers、および必要なライブラリのインストールを行います。
```
!pip install folium==0.2.1
!pip install urllib3==1.25.11
!pip install transformers==4.13.0
```
## 文章の一部の予測
文章における一部の単語をMASKし、それをBERTのモデルを使って予測します。
```
import torch
from transformers import BertForMaskedLM
from transformers import BertTokenizer
text = "[CLS] I played baseball with my friends at school yesterday [SEP]"
tokenizer = BertTokenizer.from_pretrained("bert-base-uncased")
words = tokenizer.tokenize(text)
print(words)
```
文章の一部をMASKします。
```
msk_idx = 3
words[msk_idx] = "[MASK]" # 単語を[MASK]に置き換える
print(words)
```
単語を対応するインデックスに変換します。
```
word_ids = tokenizer.convert_tokens_to_ids(words) # 単語をインデックスに変換
word_tensor = torch.tensor([word_ids]) # テンソルに変換
print(word_tensor)
```
BERTのモデルを使って予測を行います。
```
msk_model = BertForMaskedLM.from_pretrained("bert-base-uncased")
msk_model.cuda() # GPU対応
msk_model.eval()
x = word_tensor.cuda() # GPU対応
y = msk_model(x) # 予測
result = y[0]
print(result.size()) # 結果の形状
_, max_ids = torch.topk(result[0][msk_idx], k=5) # 最も大きい5つの値
result_words = tokenizer.convert_ids_to_tokens(max_ids.tolist()) # インデックスを単語に変換
print(result_words)
```
## 文章が連続しているかどうかの判定
BERTのモデルを使って、2つの文章が連続しているかどうかの判定を行います。
以下の関数`show_continuity`では、2つの文章の連続性を判定し、表示します。
```
from transformers import BertForNextSentencePrediction
def show_continuity(text, seg_ids):
words = tokenizer.tokenize(text)
word_ids = tokenizer.convert_tokens_to_ids(words) # 単語をインデックスに変換
word_tensor = torch.tensor([word_ids]) # テンソルに変換
seg_tensor = torch.tensor([seg_ids])
nsp_model = BertForNextSentencePrediction.from_pretrained('bert-base-uncased')
nsp_model.cuda() # GPU対応
nsp_model.eval()
x = word_tensor.cuda() # GPU対応
s = seg_tensor.cuda() # GPU対応
y = nsp_model(x, token_type_ids=s) # 予測
result = torch.softmax(y[0], dim=1)
print(result) # Softmaxで確率に
print(str(result[0][0].item()*100) + "%の確率で連続しています。")
```
`show_continuity`関数に、自然につながる2つの文章を与えます。
```
text = "[CLS] What is baseball ? [SEP] It is a game of hitting the ball with the bat [SEP]"
seg_ids = [0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1 ,1, 1] # 0:前の文章の単語、1:後の文章の単語
show_continuity(text, seg_ids)
```
`show_continuity`関数に、自然につながらない2つの文章を与えます。
```
text = "[CLS] What is baseball ? [SEP] This food is made with flour and milk [SEP]"
seg_ids = [0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1] # 0:前の文章の単語、1:後の文章の単語
show_continuity(text, seg_ids)
```
| github_jupyter |
# Binary Search or Bust
> Binary search is useful for searching, but its implementation often leaves us searching for edge cases
- toc: true
- badges: true
- comments: true
- categories: [data structures & algorithms, coding interviews, searching]
- image: images/binary_search_gif.gif
# Why should you care?
Binary search is useful for searching through a set of values (which typically are sorted) efficiently. At each step, it reduces the search space by half, thereby running in $O(log(n))$ complexity. While it sounds simple enough to understand, it is deceptively tricky to implement and use in problems. Over the next few sections, let's take a look at binary search and it can be applied to some commonly encountered interview problems.
# A Recipe for Binary Searching
How does binary search reduce the search space by half? It leverages the fact that the input is sorted (_most of the time_) and compares the middle value of the search space at any step with the target value that we're searching for. If the middle value is smaller than the target, then we know that the target can only lie to its right, thus eliminating all the values to the left of the middle value and vice versa. So what information do we need to implement binary search?
1. The left and right ends of the search space
2. The target value we're searching for
3. What to store at each step if any
Here's a nice video which walks through the binary search algorithm:
> youtube: https://youtu.be/P3YID7liBug
Next, let's look at an implementation of vanilla binary search.
```
#hide
from typing import List, Dict, Tuple
def binary_search(nums: List[int], target: int) -> int:
"""Vanilla Binary Search.
Given a sorted list of integers and a target value,
find the index of the target value in the list.
If not present, return -1.
"""
# Left and right boundaries of the search space
left, right = 0, len(nums) - 1
while left <= right:
# Why not (left + right) // 2 ?
# Hint: Doesn't matter for Python
middle = left + (right - left) // 2
# Found the target, return the index
if nums[middle] == target:
return middle
# The middle value is less than the
# target, so look to the right
elif nums[middle] < target:
left = middle + 1
# The middle value is greater than the
# target, so look to the left
else:
right = middle - 1
return -1 # Target not found
```
Here're a few examples of running our binary search implementation on a list and target values
```
#hide_input
nums = [1,4,9,54,100,123]
targets = [4, 100, 92]
for val in targets:
print(f"Result of searching for {val} in {nums} : \
{binary_search(nums, val)}\n")
```
> Tip: Using the approach middle = left + (right - left) // 2 helps avoid overflow. While this isn't a concern in Python, it becomes a tricky issue to debug in other programming languages such as C++. For more on overflow, check out this [article](https://ai.googleblog.com/2006/06/extra-extra-read-all-about-it-nearly.html).
Before we look at some problems that can be solved using binary search, let's run a quick comparison of linear search and binary search on some large input.
```
def linear_search(nums: List[int], target: int) -> int:
"""Linear Search.
Given a list of integers and a target value, return
find the index of the target value in the list.
If not present, return -1.
"""
for idx, elem in enumerate(nums):
# Found the target value
if elem == target:
return idx
return -1 # Target not found
#hide
n = 1000000
large_nums = range((1, n + 1))
target = 99999
```
Let's see the time it takes linear search and binary search to find $99999$ in a sorted list of numbers from $[1, 1000000]$
- Linear Search
```
#hide_input
%timeit linear_search(large_nums, target)
```
- Binary Search
```
#hide_input
%timeit binary_search(large_nums, target)
```
Hopefully, that drives the point home :wink:.
# Naïve Binary Search Problems
Here's a list of problems that can be solved using vanilla binary search (or slightly modifying it). Anytime you see a problem statement which goes something like _"Given a sorted list.."_ or _"Find the position of an element"_, think of using binary search. You can also consider **sorting** the input in case it is an unordered collection of items to reduce it to a binary search problem. Note that this list is by no means exhaustive, but is a good starting point to practice binary search:
- [Search Insert Position](https://leetcode.com/problems/search-insert-position/
)
- [Find the Square Root of x](https://leetcode.com/problems/sqrtx/)
- [Find First and Last Position of Element in Sorted Array](https://leetcode.com/problems/find-first-and-last-position-of-element-in-sorted-array/)
- [Search in a Rotated Sorted Array](https://leetcode.com/problems/search-in-rotated-sorted-array/)
In the problems above, we can either directly apply binary search or adapt it slightly to solve the problem. For example, take the square root problem. We know that the square root of a positive number $n$ has to lie between $[1, n / 2]$. This gives us the bounds for the search space. Applying binary search over this space allows us to find the a good approximation of the square root. See the implementation below for details:
```
def find_square_root(n: int) -> int:
"""Integer square root.
Given a positive integer, return
its square root.
"""
left, right = 1, n // 2 + 1
while left <= right:
middle = left + (right - left) // 2
if middle * middle == n:
return middle # Found an exact match
elif middle * middle < n:
left = middle + 1 # Go right
else:
right = middle - 1 # Go left
return right # This is the closest value to the actual square root
#hide_input
nums = [1,4,8,33,100]
for val in nums:
print(f"Square root of {val} is: {find_square_root(val)}\n")
```
# To Be Continued
- Applying binary search to unordered data
- Problems where using binary search isn't obvious
| github_jupyter |
**INITIALIZATION:**
- I use these three lines of code on top of my each notebooks because it will help to prevent any problems while reloading the same project. And the third line of code helps to make visualization within the notebook.
```
#@ INITIALIZATION:
%reload_ext autoreload
%autoreload 2
%matplotlib inline
```
**LIBRARIES AND DEPENDENCIES:**
- I have downloaded all the libraries and dependencies required for the project in one particular cell.
```
#@ IMPORTING NECESSARY LIBRARIES AND DEPENDENCIES:
from keras.models import Sequential
from keras.layers import BatchNormalization
from keras.layers.convolutional import Conv2D
from keras.layers.convolutional import MaxPooling2D
from keras.layers.core import Activation
from keras.layers.core import Flatten
from keras.layers.core import Dense, Dropout
from keras import backend as K
from tensorflow.keras.optimizers import SGD
from tensorflow.keras.datasets import cifar10
from keras.callbacks import LearningRateScheduler
from sklearn.preprocessing import LabelBinarizer
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report
import matplotlib.pyplot as plt
import numpy as np
```
**VGG ARCHITECTURE:**
- I will define the build method of Mini VGGNet architecture below. It requires four parameters: width of input image, height of input image, depth of image, number of class labels in the classification task. The Sequential class, the building block of sequential networks sequentially stack one layer on top of the other layer initialized below. Batch Normalization operates over the channels, so in order to apply BN, we need to know which axis to normalize over.
```
#@ DEFINING VGGNET ARCHITECTURE:
class MiniVGGNet: # Defining VGG Network.
@staticmethod
def build(width, height, depth, classes): # Defining Build Method.
model = Sequential() # Initializing Sequential Model.
inputShape = (width, height, depth) # Initializing Input Shape.
chanDim = -1 # Index of Channel Dimension.
if K.image_data_format() == "channels_first":
inputShape = (depth, width, height) # Initializing Input Shape.
chanDim = 1 # Index of Channel Dimension.
model.add(Conv2D(32, (3, 3), padding='same',
input_shape=inputShape)) # Adding Convolutional Layer.
model.add(Activation("relu")) # Adding RELU Activation Function.
model.add(BatchNormalization(axis=chanDim)) # Adding Batch Normalization Layer.
model.add(Conv2D(32, (3, 3), padding='same')) # Adding Convolutional Layer.
model.add(Activation("relu")) # Adding RELU Activation Function.
model.add(BatchNormalization(axis=chanDim)) # Adding Batch Normalization Layer.
model.add(MaxPooling2D(pool_size=(2, 2))) # Adding Max Pooling Layer.
model.add(Dropout(0.25)) # Adding Dropout Layer.
model.add(Conv2D(64, (3, 3), padding="same")) # Adding Convolutional Layer.
model.add(Activation("relu")) # Adding RELU Activation Function.
model.add(BatchNormalization(axis=chanDim)) # Adding Batch Normalization Layer.
model.add(Conv2D(64, (3, 3), padding='same')) # Adding Convolutional Layer.
model.add(Activation("relu")) # Adding RELU Activation Function.
model.add(BatchNormalization(axis=chanDim)) # Adding Batch Normalization Layer.
model.add(MaxPooling2D(pool_size=(2, 2))) # Adding Max Pooling Layer.
model.add(Dropout(0.25)) # Adding Dropout Layer.
model.add(Flatten()) # Adding Flatten Layer.
model.add(Dense(512)) # Adding FC Dense Layer.
model.add(Activation("relu")) # Adding Activation Layer.
model.add(BatchNormalization()) # Adding Batch Normalization Layer.
model.add(Dropout(0.5)) # Adding Dropout Layer.
model.add(Dense(classes)) # Adding Dense Output Layer.
model.add(Activation("softmax")) # Adding Softmax Layer.
return model
#@ CUSTOM LEARNING RATE SCHEDULER:
def step_decay(epoch): # Definig step decay function.
initAlpha = 0.01 # Initializing initial LR.
factor = 0.25 # Initializing drop factor.
dropEvery = 5 # Initializing epochs to drop.
alpha = initAlpha*(factor ** np.floor((1 + epoch) / dropEvery))
return float(alpha)
```
**VGGNET ON CIFAR10**
```
#@ GETTING THE DATASET:
((trainX, trainY), (testX, testY)) = cifar10.load_data() # Loading Dataset.
trainX = trainX.astype("float") / 255.0 # Normalizing Dataset.
testX = testX.astype("float") / 255.0 # Normalizing Dataset.
#@ PREPARING THE DATASET:
lb = LabelBinarizer() # Initializing LabelBinarizer.
trainY = lb.fit_transform(trainY) # Converting Labels to Vectors.
testY = lb.transform(testY) # Converting Labels to Vectors.
labelNames = ["airplane", "automobile", "bird", "cat", "deer",
"dog", "frog", "horse", "ship", "truck"] # Initializing LabelNames.
#@ INITIALIZING OPTIMIZER AND MODEL:
callbacks = [LearningRateScheduler(step_decay)] # Initializing Callbacks.
opt = SGD(0.01, nesterov=True, momentum=0.9) # Initializing SGD Optimizer.
model = MiniVGGNet.build(width=32, height=32, depth=3, classes=10) # Initializing VGGNet Architecture.
model.compile(loss="categorical_crossentropy", optimizer=opt,
metrics=["accuracy"]) # Compiling VGGNet Model.
H = model.fit(trainX, trainY,
validation_data=(testX, testY), batch_size=64,
epochs=40, verbose=1, callbacks=callbacks) # Training VGGNet Model.
```
**MODEL EVALUATION:**
```
#@ INITIALIZING MODEL EVALUATION:
predictions = model.predict(testX, batch_size=64) # Getting Model Predictions.
print(classification_report(testY.argmax(axis=1),
predictions.argmax(axis=1),
target_names=labelNames)) # Inspecting Classification Report.
#@ INSPECTING TRAINING LOSS AND ACCURACY:
plt.style.use("ggplot")
plt.figure()
plt.plot(np.arange(0, 40), H.history["loss"], label="train_loss")
plt.plot(np.arange(0, 40), H.history["val_loss"], label="val_loss")
plt.plot(np.arange(0, 40), H.history["accuracy"], label="train_acc")
plt.plot(np.arange(0, 40), H.history["val_accuracy"], label="val_acc")
plt.title("Training Loss and Accuracy")
plt.xlabel("Epoch")
plt.ylabel("Loss/Accuracy")
plt.legend()
plt.show();
```
**Note:**
- Batch Normalization can lead to a faster, more stable convergence with higher accuracy.
- Batch Normalization will require more wall time to train the network even though the network will obtain higher accuracy in less epochs.
| github_jupyter |
```
# Import packages
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# Read in data. If data is zipped, unzip the file and change file path accordingly
yelp = pd.read_csv("../yelp_academic_dataset_business.csv",
dtype={'attributes': str, 'postal_code': str}, low_memory=False)
# Reorder columns
# https://stackoverflow.com/questions/41968732/set-order-of-columns-in-pandas-dataframe
cols_to_order = ['name', 'stars', 'review_count', 'categories', 'city', 'state',
'postal_code', 'latitude', 'longitude', 'address']
new_cols = cols_to_order + (yelp.columns.drop(cols_to_order).tolist())
yelp = yelp[new_cols]
print(yelp.shape)
print(yelp.info())
# Remove entries with null in columns: name, categories, city, postal code
yelp = yelp[(pd.isna(yelp['name'])==False) &
(pd.isna(yelp['city'])==False) &
(pd.isna(yelp['categories'])==False) &
(pd.isna(yelp['postal_code'])==False)]
print(yelp.shape)
# Remove columns with <0.5% non-null values (<894) except BYOB=641 non-null
# and non-relevant columns
yelp = yelp.drop(yelp.columns[[6,9,17,26,31,33,34,37,38]], axis=1)
print(yelp.shape)
# Remove entries with < 1000 businesses in each state
state_counts = yelp['state'].value_counts()
yelp = yelp[~yelp['state'].isin(state_counts[state_counts < 1000].index)]
print(yelp.shape)
# Create new column of grouped star rating
conds = [
((yelp['stars'] == 1) | (yelp['stars'] == 1.5)),
((yelp['stars'] == 2) | (yelp['stars'] == 2.5)),
((yelp['stars'] == 3) | (yelp['stars'] == 3.5)),
((yelp['stars'] == 4) | (yelp['stars'] == 4.5)),
(yelp['stars'] == 5) ]
values = [1, 2, 3, 4, 5]
yelp['star-rating'] = np.select(conds, values)
print(yelp.shape)
# Convert 'hours' columns to total hours open that day for each day column
from datetime import timedelta, time
# Monday ---------------------------------------------------------
yelp[['hours.Monday.start', 'hours.Monday.end']] = yelp['hours.Monday'].str.split('-', 1, expand=True)
# Monday start time
hr_min = []
for row in yelp['hours.Monday.start']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el]) #change elements in list to int
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Monday.start'] = time_obj
# Monday end time
hr_min = []
for row in yelp['hours.Monday.end']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Monday.end'] = time_obj
# Create column of time difference
yelp['Monday.hrs.open'] = yelp['hours.Monday.end'] - yelp['hours.Monday.start']
# Convert seconds to minutes
hour_calc = []
for ob in yelp['Monday.hrs.open']:
hour_calc.append(ob.seconds//3600) #convert seconds to hours for explainability
yelp['Monday.hrs.open'] = hour_calc
# Tuesday -------------------------------------------------------------
yelp[['hours.Tuesday.start', 'hours.Tuesday.end']] = yelp['hours.Tuesday'].str.split('-', 1, expand=True)
hr_min = []
for row in yelp['hours.Tuesday.start']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Tuesday.start'] = time_obj
hr_min = []
for row in yelp['hours.Tuesday.end']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Tuesday.end'] = time_obj
yelp['Tuesday.hrs.open'] = yelp['hours.Tuesday.end'] - yelp['hours.Tuesday.start']
hour_calc = []
for ob in yelp['Tuesday.hrs.open']:
hour_calc.append(ob.seconds//3600)
yelp['Tuesday.hrs.open'] = hour_calc
# Wednesday ---------------------------------------------------------
yelp[['hours.Wednesday.start', 'hours.Wednesday.end']] = yelp['hours.Wednesday'].str.split('-', 1, expand=True)
hr_min = []
for row in yelp['hours.Wednesday.start']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Wednesday.start'] = time_obj
hr_min = []
for row in yelp['hours.Wednesday.end']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Wednesday.end'] = time_obj
yelp['Wednesday.hrs.open'] = yelp['hours.Wednesday.end'] - yelp['hours.Wednesday.start']
hour_calc = []
for ob in yelp['Wednesday.hrs.open']:
hour_calc.append(ob.seconds//3600)
yelp['Wednesday.hrs.open'] = hour_calc
# Thursday --------------------------------------------------------------------
yelp[['hours.Thursday.start', 'hours.Thursday.end']] = yelp['hours.Thursday'].str.split('-', 1, expand=True)
hr_min = []
for row in yelp['hours.Thursday.start']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Thursday.start'] = time_obj
hr_min = []
for row in yelp['hours.Thursday.end']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Thursday.end'] = time_obj
yelp['Thursday.hrs.open'] = yelp['hours.Thursday.end'] - yelp['hours.Thursday.start']
hour_calc = []
for ob in yelp['Thursday.hrs.open']:
hour_calc.append(ob.seconds//3600)
yelp['Thursday.hrs.open'] = hour_calc
# Friday -----------------------------------------------------------------------
yelp[['hours.Friday.start', 'hours.Friday.end']] = yelp['hours.Friday'].str.split('-', 1, expand=True)
hr_min = []
for row in yelp['hours.Friday.start']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Friday.start'] = time_obj
hr_min = []
for row in yelp['hours.Friday.end']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Friday.end'] = time_obj
yelp['Friday.hrs.open'] = yelp['hours.Friday.end'] - yelp['hours.Friday.start']
hour_calc = []
for ob in yelp['Friday.hrs.open']:
hour_calc.append(ob.seconds//3600)
yelp['Friday.hrs.open'] = hour_calc
# Saturday ------------------------------------------------------------------------
yelp[['hours.Saturday.start', 'hours.Saturday.end']] = yelp['hours.Saturday'].str.split('-', 1, expand=True)
hr_min = []
for row in yelp['hours.Saturday.start']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Saturday.start'] = time_obj
hr_min = []
for row in yelp['hours.Saturday.end']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Saturday.end'] = time_obj
yelp['Saturday.hrs.open'] = yelp['hours.Saturday.end'] - yelp['hours.Saturday.start']
hour_calc = []
for ob in yelp['Saturday.hrs.open']:
hour_calc.append(ob.seconds//3600)
yelp['Saturday.hrs.open'] = hour_calc
# Sunday ----------------------------------------------------------------------
yelp[['hours.Sunday.start', 'hours.Sunday.end']] = yelp['hours.Sunday'].str.split('-', 1, expand=True)
hr_min = []
for row in yelp['hours.Sunday.start']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Sunday.start'] = time_obj
hr_min = []
for row in yelp['hours.Sunday.end']:
hr_min.append(str(row).split(':'))
new_el = []
for el in hr_min:
if len(el) == 1:
new_el.append([0,0])
else:
new_el.append([int(i) for i in el])
time_obj = []
for el_split in new_el:
time_obj.append(timedelta(hours=el_split[0], minutes=el_split[1]))
yelp['hours.Sunday.end'] = time_obj
yelp['Sunday.hrs.open'] = yelp['hours.Sunday.end'] - yelp['hours.Sunday.start']
hour_calc = []
for ob in yelp['Sunday.hrs.open']:
hour_calc.append(ob.seconds//3600)
yelp['Sunday.hrs.open'] = hour_calc
# Remove old target variable (stars) and
# unecessary time columns that were created. Only keep 'day.hrs.open' columns
yelp = yelp.drop(yelp.columns[[1,10,11,12,16,18,41,48,52,53,55,56,
58,59,61,62,64,65,67,68,70,71]], axis=1)
print(yelp.shape)
# Delete columns with unworkable form (dict)
del yelp['attributes.BusinessParking']
del yelp['attributes.Music']
del yelp['attributes.Ambience']
del yelp['attributes.GoodForKids']
del yelp['attributes.RestaurantsDelivery']
del yelp['attributes.BestNights']
del yelp['attributes.HairSpecializesIn']
del yelp['attributes.GoodForMeal']
# Look at final DF before saving
print(yelp.info())
# Save as CSV for faster loading -------------------------------------------------
yelp.to_csv('/Data/yelp-clean.csv')
```
| github_jupyter |
# 내가 닮은 연예인은?
사진 모으기
얼굴 영역 자르기
얼굴 영역 Embedding 추출
연예인들의 얼굴과 거리 비교하기
시각화
회고
1. 사진 모으기
2. 얼굴 영역 자르기
이미지에서 얼굴 영역을 자름
image.fromarray를 이용하여 PIL image로 변환한 후, 추후에 시각화에 사용
```
# 필요한 모듈 불러오기
import os
import re
import glob
import glob
import pickle
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.image as img
import face_recognition
%matplotlib inline
from PIL import Image
import numpy as np
import face_recognition
import os
from PIL import Image
dir_path = os.getenv('HOME')+'/aiffel/EXP_07_face_embedding/data'
file_list = os.listdir(dir_path)
print(len(file_list))
# 이미지 파일 불러오기
print('연예인 이미지 파일 갯수:', len(file_list) - 5) # 추가한 내 사진 수를 뺀 나머지 사진 수 세기
# 이미지 파일 리스트 확인
print ("파일 리스트:\n{}".format(file_list))
# 이미지 파일 일부 확인
# Set figsize here
fig, axes = plt.subplots(nrows=2, ncols=3, figsize=(24,10))
# flatten axes for easy iterating
for i, ax in enumerate(axes.flatten()):
image = img.imread(dir_path+'/'+file_list[i])
ax.imshow(image)
plt.show()
fig.tight_layout()
# 이미지 파일 경로를 파라미터로 넘기면 얼굴 영역만 잘라주는 함수
def get_cropped_face(image_file):
image = face_recognition.load_image_file(image_file)
face_locations = face_recognition.face_locations(image)
a, b, c, d = face_locations[0]
cropped_face = image[a:c,d:b,:]
return cropped_face
# 얼굴 영역이 정확히 잘리는 지 확인
image_path = os.getenv('HOME')+'/aiffel/EXP_07_face_embedding/data/이원재_02.jpg'
cropped_face = get_cropped_face(image_path)
plt.imshow(cropped_face)
```
## Step3. 얼굴 영역의 임베딩 추출하기
```
# 얼굴 영역을 가지고 얼굴 임베딩 벡터를 구하는 함수
def get_face_embedding(face):
return face_recognition.face_encodings(face, model='cnn')
# 파일 경로를 넣으면 embedding_dict를 리턴하는 함수
def get_face_embedding_dict(dir_path):
file_list = os.listdir(dir_path)
embedding_dict = {}
for file in file_list:
try:
img_path = os.path.join(dir_path, file)
face = get_cropped_face(img_path)
embedding = get_face_embedding(face)
if len(embedding) > 0:
# 얼굴영역 face가 제대로 detect되지 않으면 len(embedding)==0인 경우가 발생하므로
# os.path.splitext(file)[0]에는 이미지파일명에서 확장자를 제거한 이름이 담깁니다.
embedding_dict[os.path.splitext(file)[0]] = embedding[0]
# embedding_dict[] 이미지 파일의 임베딩을 구해 담음 키=사람이름, 값=임베딩 벡터
# os.path.splitext(file)[0] 파일의 확장자를 제거한 이름만 추출
# embedding[0]은 넣고 싶은 요소값
except:
continue
return embedding_dict
embedding_dict = get_face_embedding_dict(dir_path)
```
## Step4. 모은 연예인들과 비교하기
```
# 이미지 간 거리를 구하는 함수
def get_distance(name1, name2):
return np.linalg.norm(embedding_dict[name1]-embedding_dict[name2], ord=2)
# 본인 사진의 거리를 확인해보자
print('내 사진끼리의 거리는?:', get_distance('이원재_01', '이원재_02'))
# name1과 name2의 거리를 비교하는 함수를 생성하되, name1은 미리 지정하고, name2는 호출시에 인자로 받도록 합니다.
def get_sort_key_func(name1):
def get_distance_from_name1(name2):
return get_distance(name1, name2)
return get_distance_from_name1
# 닮은꼴 순위, 이름, 임베딩 거리를 포함한 Top-5 리스트 출력하는 함수
def get_nearest_face(name, top=5):
sort_key_func = get_sort_key_func(name)
sorted_faces = sorted(embedding_dict.items(), key=lambda x:sort_key_func(x[0]))
rank_cnt = 1 # 순위를 세는 변수
pass_cnt = 1 # 건너뛴 숫자를 세는 변수(본인 사진 카운트)
end = 0 # 닮은 꼴 5번 출력시 종료하기 위해 세는 변수
for i in range(top+15):
rank_cnt += 1
if sorted_faces[i][0].find('이원재_02') == 0: # 본인 사진인 mypicture라는 파일명으로 시작하는 경우 제외합니다.
pass_cnt += 1
continue
if sorted_faces[i]:
print('순위 {} : 이름({}), 거리({})'.format(rank_cnt - pass_cnt, sorted_faces[i][0], sort_key_func(sorted_faces[i][0])))
end += 1
if end == 5: # end가 5가 된 경우 연예인 5명 출력되었기에 종료합니다.
break
# '이원재_01'과 가장 닮은 사람은 누굴까요?
get_nearest_face('이원재_01')
# '이원재_02'와 가장 닮은 사람은 누굴까요?
get_nearest_face('이원재_02')
```
## Step5. 다양한 재미있는 시각화 시도해 보기
```
# 사진 경로 설정
mypicture1 = os.getenv('HOME')+'/aiffel/EXP_07_face_embedding/data/이원재_01.jpg'
mypicture2 = os.getenv('HOME')+'/aiffel/EXP_07_face_embedding/data/이원재_02.jpg'
mc= os.getenv('HOME')+'/aiffel/EXP_07_face_embedding/data/MC몽.jpg'
gahee = os.getenv('HOME')+'/aiffel/EXP_07_face_embedding/data/가희.jpg'
seven = os.getenv('HOME')+'/aiffel/EXP_07_face_embedding/data/SE7EN.jpg'
gam = os.getenv('HOME')+'/aiffel/EXP_07_face_embedding/data/감우성.jpg'
gang = os.getenv('HOME')+'/aiffel/EXP_07_face_embedding/data/강경준.jpg'
gyung = os.getenv('HOME')+'/aiffel/EXP_07_face_embedding/data/강경현.jpg'
gi = os.getenv('HOME')+'/aiffel/EXP_07_face_embedding/data/강기영.jpg'
# 크롭한 얼굴을 저장해 보자
a1 = get_cropped_face(mypicture1)
a2 = get_cropped_face(mypicture2)
b1 = get_cropped_face(mc)
b2 = get_cropped_face(gahee)
b3 = get_cropped_face(gam)
plt.figure(figsize=(10,8))
plt.subplot(231)
plt.imshow(a1)
plt.axis('off')
plt.title('1st')
plt.subplot(232)
plt.imshow(a2)
plt.axis('off')
plt.title('me')
plt.subplot(233)
plt.imshow(b1)
plt.axis('off')
plt.title('2nd')
plt.subplot(234)
print('''mypicture의 순위
순위 1 : 이름(사쿠라), 거리(0.36107689719729225)
순위 2 : 이름(트와이스나연), 거리(0.36906292012955577)
순위 3 : 이름(아이유), 거리(0.3703590842312735)
순위 4 : 이름(유트루), 거리(0.3809516850126146)
순위 5 : 이름(지호), 거리(0.3886670633997685)''')
```
| github_jupyter |
#### loading the libraries
```
import os
import sys
import pyvista as pv
import trimesh as tm
import numpy as np
import topogenesis as tg
import pickle as pk
sys.path.append(os.path.realpath('..\..')) # no idea how or why this is not working without adding this to the path TODO: learn about path etc.
from notebooks.resources import RES as res
```
#### loading the configuration of the test
```
# load base lattice CSV file
lattice_path = os.path.relpath('../../data/macrovoxels.csv')
macro_lattice = tg.lattice_from_csv(lattice_path)
# load random configuration for testing
config_path = os.path.relpath('../../data/random_lattice.csv')
configuration = tg.lattice_from_csv(config_path)
# load environment
environment_path = os.path.relpath("../../data/movedcontext.obj")
environment_mesh = tm.load(environment_path)
# load solar vectors
vectors = pk.load(open("../../data/sunvectors.pk", "rb"))
# load vector intensities
intensity = pk.load(open("../../data/dnival.pk", "rb"))
```
#### during optimization, arrays like these will be passed to the function:
```
variable = [0, 0, 0, 1, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 0, 0, 1, 1, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 1, 1, 0, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 1, 1, 1, 0, 0, 1, 0, 1, 0, 0]
```
#### calling the objective function
```
# input is the decision variables, a referenca lattice, the visibility vectors, their magnitude (i.e. direct normal illuminance for daylight), and a mesh of the environment
# output is the total objective score in 100s of lux on the facade, and 100s of lux per each surface (voxel roofs)
crit, voxcrit = res.crit_2_DL(variable, macro_lattice, vectors, intensity, environment_mesh)
```
#### generating mesh
```
meshes, _, _ = res.construct_vertical_mesh(configuration, configuration.unit)
facademesh = tm.util.concatenate(meshes)
```
#### visualisation
```
p = pv.Plotter(notebook=True)
configuration.fast_vis(p,False,False,opacity=0.1)
# p.add_arrows(ctr_per_ray, -ray_per_ctr, mag=5, show_scalar_bar=False)
# p.add_arrows(ctr_per_ray, nrm_per_ray, mag=5, show_scalar_bar=False)
# p.add_mesh(roof_mesh)
p.add_mesh(environment_mesh)
p.add_mesh(facademesh, cmap='fire', scalars=np.repeat(voxcrit,2))
p.add_points(vectors*-300)
# p.add_points(horizontal_test_points)
p.show(use_ipyvtk=True)
```
| github_jupyter |
<h1>Notebook Content</h1>
1. [Import Packages](#1)
1. [Helper Functions](#2)
1. [Input](#3)
1. [Model](#4)
1. [Prediction](#5)
1. [Complete Figure](#6)
<h1 id="1">1. Import Packages</h1>
Importing all necessary and useful packages in single cell.
```
import numpy as np
import keras
import tensorflow as tf
from numpy import array
from keras.models import Sequential
from keras.layers import LSTM
from keras.layers import Dense
from keras.layers import Flatten
from keras.layers import TimeDistributed
from keras.layers.convolutional import Conv1D
from keras.layers.convolutional import MaxPooling1D
from keras_tqdm import TQDMNotebookCallback
from sklearn.preprocessing import MinMaxScaler
from tqdm import tqdm_notebook
import matplotlib.pyplot as plt
import pandas as pd
import random
from random import randint
```
<h1 id="2">2. Helper Functions</h1>
Defining Some helper functions which we will need later in code
```
# split a univariate sequence into samples
def split_sequence(sequence, n_steps, look_ahead=0):
X, y = list(), list()
for i in range(len(sequence)-look_ahead):
# find the end of this pattern
end_ix = i + n_steps
# check if we are beyond the sequence
if end_ix > len(sequence)-1-look_ahead:
break
# gather input and output parts of the pattern
seq_x, seq_y = sequence[i:end_ix], sequence[end_ix+look_ahead]
X.append(seq_x)
y.append(seq_y)
return array(X), array(y)
def plot_multi_graph(xAxis,yAxes,title='',xAxisLabel='number',yAxisLabel='Y'):
linestyles = ['-', '--', '-.', ':']
plt.figure()
plt.title(title)
plt.xlabel(xAxisLabel)
plt.ylabel(yAxisLabel)
for key, value in yAxes.items():
plt.plot(xAxis, np.array(value), label=key, linestyle=linestyles[randint(0,3)])
plt.legend()
def normalize(values):
values = array(values, dtype="float64").reshape((len(values), 1))
# train the normalization
scaler = MinMaxScaler(feature_range=(0, 1))
scaler = scaler.fit(values)
#print('Min: %f, Max: %f' % (scaler.data_min_, scaler.data_max_))
# normalize the dataset and print the first 5 rows
normalized = scaler.transform(values)
return normalized,scaler
```
<h1 id="3">3. Input</h1>
<h3 id="3-1">3-1. Sequence PreProcessing</h3>
Splitting and Reshaping
```
n_features = 1
n_seq = 20
n_steps = 1
def sequence_preprocessed(values, sliding_window, look_ahead=0):
# Normalization
normalized,scaler = normalize(values)
# Try the following if randomizing the sequence:
# random.seed('sam') # set the seed
# raw_seq = random.sample(raw_seq, 100)
# split into samples
X, y = split_sequence(normalized, sliding_window, look_ahead)
# reshape from [samples, timesteps] into [samples, subsequences, timesteps, features]
X = X.reshape((X.shape[0], n_seq, n_steps, n_features))
return X,y,scaler
```
<h3 id="3-2">3-2. Providing Sequence</h3>
Defining a raw sequence, sliding window of data to consider and look ahead future timesteps
```
# define input sequence
sequence_val = [i for i in range(5000,7000)]
sequence_train = [i for i in range(1000,2000)]
sequence_test = [i for i in range(10000,14000)]
# choose a number of time steps for sliding window
sliding_window = 20
# choose a number of further time steps after end of sliding_window till target start (gap between data and target)
look_ahead = 20
X_train, y_train, scaler_train = sequence_preprocessed(sequence_train, sliding_window, look_ahead)
X_val, y_val ,scaler_val = sequence_preprocessed(sequence_val, sliding_window, look_ahead)
X_test,y_test,scaler_test = sequence_preprocessed(sequence_test, sliding_window, look_ahead)
```
<h1 id="4">4. Model</h1>
<h3 id="4-1">4-1. Defining Layers</h3>
Adding 1D Convolution, Max Pooling, LSTM and finally Dense (MLP) layer
```
# define model
model = Sequential()
model.add(TimeDistributed(Conv1D(filters=64, kernel_size=1, activation='relu'),
input_shape=(None, n_steps, n_features)
))
model.add(TimeDistributed(MaxPooling1D(pool_size=1)))
model.add(TimeDistributed(Flatten()))
model.add(LSTM(50, activation='relu', stateful=False))
model.add(Dense(1))
```
<h3 id="4-2">4-2. Training Model</h3>
Defined early stop, can be used in callbacks param of model fit, not using for now since it's not recommended at first few iterations of experimentation with new data
```
# Defining multiple metrics, leaving it to a choice, some may be useful and few may even surprise on some problems
metrics = ['mean_squared_error',
'mean_absolute_error',
'mean_absolute_percentage_error',
'mean_squared_logarithmic_error',
'logcosh']
# Compiling Model
model.compile(optimizer='adam', loss='mape', metrics=metrics)
# Defining early stop, call it in model fit callback
early_stop = keras.callbacks.EarlyStopping(monitor='val_loss', patience=10)
# Fit model
history = model.fit(X_train, y_train, epochs=100, verbose=3, validation_data=(X_val,y_val))
```
<h3 id="4-3">4-3. Evaluating Model</h3>
Plotting Training and Validation mean square error
```
# Plot Errors
for metric in metrics:
xAxis = history.epoch
yAxes = {}
yAxes["Training"]=history.history[metric]
yAxes["Validation"]=history.history['val_'+metric]
plot_multi_graph(xAxis,yAxes, title=metric,xAxisLabel='Epochs')
```
<h1 id="5">5. Prediction</h1>
<h3 id="5-1">5-1. Single Value Prediction</h3>
Predicting a single value slided 20 (our provided figure for look_ahead above) values ahead
```
# demonstrate prediction
x_input = array([i for i in range(100,120)])
print(x_input)
x_input = x_input.reshape((1, n_seq, n_steps, n_features))
yhat = model.predict(x_input)
print(yhat)
```
<h3 id="5-2">5-2. Sequence Prediction</h3>
Predicting complete sequence (determining closeness to target) based on data <br />
<i>change variable for any other sequence though</i>
```
# Prediction from Training Set
predict_train = model.predict(X_train)
# Prediction from Test Set
predict_test = model.predict(X_test)
"""
df = pd.DataFrame(({"normalized y_train":y_train.flatten(),
"normalized predict_train":predict_train.flatten(),
"actual y_train":scaler_train.inverse_transform(y_train).flatten(),
"actual predict_train":scaler_train.inverse_transform(predict_train).flatten(),
}))
"""
df = pd.DataFrame(({
"normalized y_test":y_test.flatten(),
"normalized predict_test":predict_test.flatten(),
"actual y_test":scaler_test.inverse_transform(y_test).flatten(),
"actual predict_test":scaler_test.inverse_transform(predict_test).flatten()
}))
df
```
<h1 id="6">6. Complete Figure</h1>
Data, Target, Prediction - all in one single graph
```
xAxis = [i for i in range(len(y_train))]
yAxes = {}
yAxes["Data"]=sequence_train[sliding_window:len(sequence_train)-look_ahead]
yAxes["Target"]=scaler_train.inverse_transform(y_train)
yAxes["Prediction"]=scaler_train.inverse_transform(predict_train)
plot_multi_graph(xAxis,yAxes,title='')
xAxis = [i for i in range(len(y_test))]
yAxes = {}
yAxes["Data"]=sequence_test[sliding_window:len(sequence_test)-look_ahead]
yAxes["Target"]=scaler_test.inverse_transform(y_test)
yAxes["Prediction"]=scaler_test.inverse_transform(predict_test)
plot_multi_graph(xAxis,yAxes,title='')
print(metrics)
print(model.evaluate(X_test,y_test))
```
| github_jupyter |
# **Libraries**
```
from google.colab import drive
drive.mount('/content/drive')
# ***********************
# *****| LIBRARIES |*****
# ***********************
%tensorflow_version 2.x
import pandas as pd
import numpy as np
import os
import json
from sklearn.model_selection import train_test_split
import tensorflow as tf
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.layers import Input, Embedding, Activation, Flatten, Dense
from keras.layers import Conv1D, MaxPooling1D, Dropout
from keras.models import Model
from keras.utils import to_categorical
from keras.optimizers import SGD
from keras.wrappers.scikit_learn import KerasClassifier
from sklearn.model_selection import RandomizedSearchCV, GridSearchCV
device_name = tf.test.gpu_device_name()
if device_name != '/device:GPU:0':
print("GPU not found")
else:
print('Found GPU at: {}'.format(device_name))
# ******************************
# *****| GLOBAL VARIABLES |*****
# ******************************
test_size = 0.2
convsize = 256
convsize2 = 1024
embedding_size = 27
input_size = 1000
conv_layers = [
[convsize, 7, 3],
[convsize, 7, 3],
[convsize, 3, -1],
[convsize, 3, -1],
[convsize, 3, -1],
[convsize, 3, 3]
]
fully_connected_layers = [convsize2, convsize2]
num_of_classes= 2
dropout_p = 0.5
optimizer= 'adam'
batch = 128
loss = 'categorical_crossentropy'
```
# **Utility functions**
```
# *****************
# *** GET FILES ***
# *****************
def getFiles( driverPath, directory, basename, extension): # Define a function that will return a list of files
pathList = [] # Declare an empty array
directory = os.path.join( driverPath, directory) #
for root, dirs, files in os.walk( directory): # Iterate through roots, dirs and files recursively
for file in files: # For every file in files
if os.path.basename(root) == basename: # If the parent directory of the current file is equal with the parameter
if file.endswith('.%s' % (extension)): # If the searched file ends in the parameter
path = os.path.join(root, file) # Join together the root path and file name
pathList.append(path) # Append the new path to the list
return pathList
# ****************************************
# *** GET DATA INTO A PANDAS DATAFRAME ***
# ****************************************
def getDataFrame( listFiles, maxFiles, minWords, limit):
counter_real, counter_max, limitReached = 0, 0, 0
text_list, label_list = [], []
print("Word min set to: %i." % ( minWords))
# Iterate through all the files
for file in listFiles:
# Open each file and look into it
with open(file) as f:
if(limitReached):
break
if maxFiles == 0:
break
else:
maxFiles -= 1
objects = json.loads( f.read())['data'] # Get the data from the JSON file
# Look into each object from the file and test for limiters
for object in objects:
if limit > 0 and counter_real >= (limit * 1000):
limitReached = 1
break
if len( object['text'].split()) >= minWords:
text_list.append(object['text'])
label_list.append(object['label'])
counter_real += 1
counter_max += 1
if(counter_real > 0 and counter_max > 0):
ratio = counter_real / counter_max * 100
else:
ratio = 0
# Print the final result
print("Lists created with %i/%i (%.2f%%) data objects." % ( counter_real, counter_max, ratio))
print("Rest ignored due to minimum words limit of %i or the limit of %i data objects maximum." % ( minWords, limit * 1000))
# Return the final Pandas DataFrame
return text_list, label_list, counter_real
```
# **Gather the path to files**
```
# ***********************************
# *** GET THE PATHS FOR THE FILES ***
# ***********************************
# Path to the content of the Google Drive
driverPath = "/content/drive/My Drive"
# Sub-directories in the driver
paths = ["processed/depression/submission",
"processed/depression/comment",
"processed/AskReddit/submission",
"processed/AskReddit/comment"]
files = [None] * len(paths)
for i in range(len(paths)):
files[i] = getFiles( driverPath, paths[i], "text", "json")
print("Gathered %i files from %s." % ( len(files[i]), paths[i]))
```
# **Gather the data from files**
```
# ************************************
# *** GATHER THE DATA AND SPLIT IT ***
# ************************************
# Local variables
rand_state_splitter = 1000
test_size = 0.2
min_files = [ 750, 0, 1300, 0]
max_words = [ 50, 0, 50, 0]
limit_packets = [300, 0, 300, 0]
message = ["Depression submissions", "Depression comments", "AskReddit submissions", "AskReddit comments"]
text, label = [], []
# Get the pandas data frames for each category
print("Build the Pandas DataFrames for each category.")
for i in range(4):
dummy_text, dummy_label, counter = getDataFrame( files[i], min_files[i], max_words[i], limit_packets[i])
if counter > 0:
text += dummy_text
label += dummy_label
dummy_text, dummy_label = None, None
print("Added %i samples to data list: %s.\n" % ( counter ,message[i]) )
# Splitting the data
x_train, x_test, y_train, y_test = train_test_split(text,
label,
test_size = test_size,
shuffle = True,
random_state = rand_state_splitter)
print("Training data: %i samples." % ( len(y_train)) )
print("Testing data: %i samples." % ( len(y_test)) )
# Clear data no longer needed
del rand_state_splitter, min_files, max_words, message, dummy_label, dummy_text
```
# **Process the data at a character-level**
```
# *******************************
# *** CONVERT STRING TO INDEX ***
# *******************************
print("Convert the strings to indexes.")
tk = Tokenizer(num_words = None, char_level = True, oov_token='UNK')
tk.fit_on_texts(x_train)
print("Original:", x_train[0])
# *********************************
# *** CONSTRUCT A NEW VOCABULARY***
# *********************************
print("Construct a new vocabulary")
alphabet = "abcdefghijklmnopqrstuvwxyz"
char_dict = {}
for i, char in enumerate(alphabet):
char_dict[char] = i + 1
print("dictionary")
tk.word_index = char_dict.copy() # Use char_dict to replace the tk.word_index
print(tk.word_index)
tk.word_index[tk.oov_token] = max(char_dict.values()) + 1 # Add 'UNK' to the vocabulary
print(tk.word_index)
# *************************
# *** TEXT TO SEQUENCES ***
# *************************
print("Text to sequence.")
x_train = tk.texts_to_sequences(x_train)
x_test = tk.texts_to_sequences(x_test)
print("After sequences:", x_train[0])
# ***************
# *** PADDING ***
# ***************
print("Padding the sequences.")
x_train = pad_sequences( x_train, maxlen = input_size, padding = 'post')
x_test = pad_sequences( x_test, maxlen= input_size , padding = 'post')
# ************************
# *** CONVERT TO NUMPY ***
# ************************
print("Convert to Numpy arrays")
x_train = np.array( x_train, dtype = 'float32')
x_test = np.array(x_test, dtype = 'float32')
# **************************************
# *** GET CLASSES FOR CLASSIFICATION ***
# **************************************
y_test_copy = y_test
y_train_list = [x-1 for x in y_train]
y_test_list = [x-1 for x in y_test]
y_train = to_categorical( y_train_list, num_of_classes)
y_test = to_categorical( y_test_list, num_of_classes)
```
# **Load embedding words**
```
# ***********************
# *** LOAD EMBEDDINGS ***
# ***********************
embedding_weights = []
vocab_size = len(tk.word_index)
embedding_weights.append(np.zeros(vocab_size))
for char, i in tk.word_index.items():
onehot = np.zeros(vocab_size)
onehot[i-1] = 1
embedding_weights.append(onehot)
embedding_weights = np.array(embedding_weights)
print("Vocabulary size: ",vocab_size)
print("Embedding weights: ", embedding_weights)
```
# **Build the CNN model**
```
def KerasModel():
# ***************************************
# *****| BUILD THE NEURAL NETWORK |******
# ***************************************
embedding_layer = Embedding(vocab_size+1,
embedding_size,
input_length = input_size,
weights = [embedding_weights])
# Input layer
inputs = Input(shape=(input_size,), name='input', dtype='int64')
# Embedding layer
x = embedding_layer(inputs)
# Convolution
for filter_num, filter_size, pooling_size in conv_layers:
x = Conv1D(filter_num, filter_size)(x)
x = Activation('relu')(x)
if pooling_size != -1:
x = MaxPooling1D( pool_size = pooling_size)(x)
x = Flatten()(x)
# Fully Connected layers
for dense_size in fully_connected_layers:
x = Dense( dense_size, activation='relu')(x)
x = Dropout( dropout_p)(x)
# Output Layer
predictions = Dense(num_of_classes, activation = 'softmax')(x)
# BUILD MODEL
model = Model( inputs = inputs, outputs = predictions)
model.compile(optimizer = optimizer, loss = loss, metrics = ['accuracy'])
model.summary()
return model
```
# **Train the CNN**
```
#with tf.device("/gpu:0"):
# history = model.fit(x_train, y_train,
# validation_data = ( x_test, y_test),
# epochs = 10,
# batch_size = batch,
# verbose = True)
with tf.device("/gpu:0"):
grid = KerasClassifier(build_fn = KerasModel, epochs = 15, verbose= True)
param_grid = dict(
epochs = [15]
)
#grid = GridSearchCV(estimator = model,
# param_grid = param_grid,
# cv = 5,
# verbose = 10,
# return_train_score = True)
grid_result = grid.fit(x_train, y_train)
```
# **Test the CNN**
```
#loss, accuracy = model.evaluate( x_train, y_train, verbose = True)
#print("Training Accuracy: {:.4f}".format( accuracy))
#loss, accuracy = model.evaluate( x_test, y_test, verbose = True)
#print("Testing Accuracy: {:.4f}".format( accuracy))
from sklearn.metrics import classification_report, confusion_matrix
y_predict = grid.predict( x_test)
# Build the confusion matrix
y_tested = y_test
print( type(y_test))
print(y_tested)
y_tested = np.argmax( y_tested, axis = 1)
print(y_tested)
confMatrix = confusion_matrix(y_tested, y_predict)
tn, fp, fn, tp = confMatrix.ravel()
# Build a classification report
classification_reports = classification_report( y_tested, y_predict, target_names = ['Non-depressed', 'Depressed'], digits=3)
print(confMatrix)
print(classification_reports)
```
| github_jupyter |
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from scipy import stats
from statsmodels.formula.api import ols
import researchpy as rp
from pingouin import kruskal
from pybedtools import BedTool
RootChomatin_bp_covered = '../../data/promoter_analysis/responsivepromotersRootOpenChrom.bp_covered.txt'
ShootChomatin_bp_covered = '../../data/promoter_analysis/responsivepromotersShootOpenChrom.bp_covered.txt'
RootShootIntersect_bp_covered = '../../data/promoter_analysis/responsivepromotersShootRootIntersectOpenChrom.bp_covered.txt'
def add_chr_linestart(input_location,output_location):
"""this function adds chr to the beginning of the line if it starts with a digit and saves a file"""
output = open(output_location, 'w') #make output file with write capability
#open input file
with open(input_location, 'r') as infile:
#iterate over lines in file
for line in infile:
line = line.strip() # removes hidden characters/spaces
if line[0].isdigit():
line = 'chr' + line #prepend chr to the beginning of line if starts with a digit
output.write(line + '\n') #output to new file
output.close()
def percent_coverage(bp_covered):
"""function to calculate the % coverage from the output file of bedtools coverage"""
coverage_df = pd.read_table(bp_covered, sep='\t', header=None)
col = ['chr','start','stop','gene','dot','strand','source', 'type', 'dot2', 'details', 'no._of_overlaps', 'no._of_bases_covered','promoter_length','fraction_bases_covered']
coverage_df.columns = col
#add % bases covered column
coverage_df['percentage_bases_covered'] = coverage_df.fraction_bases_covered * 100
#remove unnecessary columns
coverage_df_reduced_columns = coverage_df[['chr','start','stop','gene','strand', 'no._of_overlaps', 'no._of_bases_covered','promoter_length','fraction_bases_covered','percentage_bases_covered']]
return coverage_df_reduced_columns
root_coverage = percent_coverage(RootChomatin_bp_covered)
shoot_coverage = percent_coverage(ShootChomatin_bp_covered)
rootshootintersect_coverage = percent_coverage(RootShootIntersect_bp_covered)
sns.set(color_codes=True)
sns.set_style("whitegrid")
#distribution plot
dist_plot = root_coverage['percentage_bases_covered']
#create figure with no transparency
dist_plot_fig = sns.distplot(dist_plot).get_figure()
#save to file
#dist_plot_fig.savefig('../../data/plots/TFBS_coverage/all_genes_bp_covered_dist.pdf', format='pdf')
dist_plot = shoot_coverage['percentage_bases_covered']
#create figure with no transparency
dist_plot_fig = sns.distplot(dist_plot).get_figure()
#save to file
#dist_plot_fig.savefig('../../data/plots/TFBS_coverage/all_genes_bp_covered_dist.pdf', format='pdf')
dist_plot = rootshootintersect_coverage['percentage_bases_covered']
#create figure with no transparency
dist_plot_fig = sns.distplot(dist_plot).get_figure()
#save to file
#dist_plot_fig.savefig('../../data/plots/TFBS_coverage/all_genes_bp_covered_dist.pdf', format='pdf')
```
## constitutive vs variable
```
def add_genetype(coverage):
"""function to add gene type to the df, and remove random genes"""
select_genes_file = '../../data/genomes/ara_housekeeping_list.out'
select_genes = pd.read_table(select_genes_file, sep='\t', header=None)
cols = ['gene','gene_type']
select_genes.columns = cols
merged = pd.merge(coverage, select_genes, on='gene')
merged_renamed = merged.copy()
merged_renamed.gene_type.replace('housekeeping','constitutive', inplace=True)
merged_renamed.gene_type.replace('highVar','variable', inplace=True)
merged_renamed.gene_type.replace('randCont','random', inplace=True)
# no_random = merged_renamed[merged_renamed.gene_type != 'random']
# no_random.reset_index(drop=True, inplace=True)
return merged_renamed
roots_merged = add_genetype(root_coverage)
no_random_roots = roots_merged[roots_merged.gene_type != 'random']
shoots_merged = add_genetype(shoot_coverage)
no_random_shoots = shoots_merged[shoots_merged.gene_type != 'random']
rootsshootsintersect_merged = add_genetype(rootshootintersect_coverage)
no_random_rootsshoots = rootsshootsintersect_merged[rootsshootsintersect_merged.gene_type != 'random']
#how many have open chromatin??
print('root openchromatin present:')
print(len(no_random_roots)-len(no_random_roots[no_random_roots.percentage_bases_covered == 0]))
print('shoot openchromatin present:')
print(len(no_random_shoots)-len(no_random_shoots[no_random_shoots.percentage_bases_covered == 0]))
print('root-shoot intersect openchromatin present:')
print(len(no_random_rootsshoots)-len(no_random_rootsshoots[no_random_rootsshoots.percentage_bases_covered == 0]))
#how many have open chromatin??
print('root openchromatin present variable promoters:')
print(len(no_random_roots[no_random_roots.gene_type=='variable'])-len(no_random_roots[no_random_roots.gene_type=='variable'][no_random_roots[no_random_roots.gene_type=='variable'].percentage_bases_covered == 0]))
print('root openchromatin present constitutive promoters:')
print(len(no_random_roots[no_random_roots.gene_type=='constitutive'])-len(no_random_roots[no_random_roots.gene_type=='constitutive'][no_random_roots[no_random_roots.gene_type=='constitutive'].percentage_bases_covered == 0]))
print('shoot openchromatin present variable promoters:')
print(len(no_random_shoots[no_random_shoots.gene_type=='variable'])-len(no_random_shoots[no_random_shoots.gene_type=='variable'][no_random_shoots[no_random_shoots.gene_type=='variable'].percentage_bases_covered == 0]))
print('shoot openchromatin present constitutive promoters:')
print(len(no_random_shoots[no_random_shoots.gene_type=='constitutive'])-len(no_random_shoots[no_random_shoots.gene_type=='constitutive'][no_random_shoots[no_random_shoots.gene_type=='constitutive'].percentage_bases_covered == 0]))
print('root-shoot intersect openchromatin present variable promoters:')
print(len(no_random_rootsshoots[no_random_rootsshoots.gene_type=='variable'])-len(no_random_rootsshoots[no_random_rootsshoots.gene_type=='variable'][no_random_rootsshoots[no_random_rootsshoots.gene_type=='variable'].percentage_bases_covered == 0]))
print('root-shoot intersect openchromatin present constitutive promoters:')
print(len(no_random_rootsshoots[no_random_rootsshoots.gene_type=='constitutive'])-len(no_random_rootsshoots[no_random_rootsshoots.gene_type=='constitutive'][no_random_rootsshoots[no_random_rootsshoots.gene_type=='constitutive'].percentage_bases_covered == 0]))
sns.catplot(x="gene_type", y="percentage_bases_covered", data=roots_merged) #.savefig('../../data/plots/TFBS_coverage/responsive_bp_covered.pdf', format='pdf')
sns.catplot(x="gene_type", y="percentage_bases_covered", data=shoots_merged) #.savefig('../../data/plots/TFBS_coverage/responsive_bp_covered.pdf', format='pdf')
#roots
plot = sns.catplot(x="gene_type", y="percentage_bases_covered", kind='box', data=no_random_roots)
#plot points
ax = sns.swarmplot(x="gene_type", y="percentage_bases_covered", data=no_random_roots, color=".25")
plt.ylabel('Percentage bases covered')
plt.xlabel('Gene type');
#ax.get_figure() #.savefig('../../data/plots/TFBS_coverage/responsive_bp_covered_boxplot.pdf', format='pdf')
#shoots
plot = sns.catplot(x="gene_type", y="percentage_bases_covered", kind='box', data=no_random_shoots)
#plot points
ax = sns.swarmplot(x="gene_type", y="percentage_bases_covered", data=no_random_shoots, color=".25")
plt.ylabel('Percentage bases covered')
plt.xlabel('Gene type');
#ax.get_figure() #.savefig('../../data/plots/TFBS_coverage/responsive_bp_covered_boxplot.pdf', format='pdf')
#roots-shoots intersect
plot = sns.catplot(x="gene_type", y="percentage_bases_covered", kind='box', data=no_random_rootsshoots)
#plot points
ax = sns.swarmplot(x="gene_type", y="percentage_bases_covered", data=no_random_rootsshoots, color=".25")
plt.ylabel('Percentage bases covered')
plt.xlabel('Gene type');
#ax.get_figure() #.savefig('../../data/plots/TFBS_coverage/responsive_bp_covered_boxplot.pdf', format='pdf')
#Get names of each promoter
def normality(input_proms):
"""function to test normality of data - returns test statistic, p-value"""
#Get names of each promoter
pd.Categorical(input_proms.gene_type)
names = input_proms.gene_type.unique()
# for name in names:
# print(name)
for name in names:
print('{}: {}'.format(name, stats.shapiro(input_proms.percentage_bases_covered[input_proms.gene_type == name])))
def variance(input_proms):
"""function to test variance of data"""
#test variance
constitutive = input_proms[input_proms.gene_type == 'constitutive']
#reset indexes so residuals can be calculated later
constitutive.reset_index(inplace=True)
responsive = input_proms[input_proms.gene_type == 'variable']
responsive.reset_index(inplace=True)
control = input_proms[input_proms.gene_type == 'random']
control.reset_index(inplace=True)
print(stats.levene(constitutive.percentage_bases_covered, responsive.percentage_bases_covered))
normality(no_random_roots)
normality(no_random_shoots)
normality(no_random_rootsshoots)
```
## Not normal
```
variance(no_random_roots)
variance(no_random_shoots)
variance(no_random_rootsshoots)
```
## unequal variance for shoots
```
def kruskal_test(input_data):
"""function to do kruskal-wallis test on data"""
#print('\033[1m' +promoter + '\033[0m')
print(kruskal(data=input_data, dv='percentage_bases_covered', between='gene_type'))
#print('')
no_random_roots
kruskal_test(no_random_roots)
kruskal_test(no_random_shoots)
kruskal_test(no_random_rootsshoots)
```
## try gat enrichment
```
#add Chr to linestart of chromatin bed files
add_chr_linestart('../../data/ATAC-seq/potter2018/Shoots_NaOH_peaks_all.bed','../../data/ATAC-seq/potter2018/Shoots_NaOH_peaks_all_renamed.bed')
add_chr_linestart('../../data/ATAC-seq/potter2018/Roots_NaOH_peaks_all.bed','../../data/ATAC-seq/potter2018/Roots_NaOH_peaks_all_renamed.bed')
add_chr_linestart('../../data/ATAC-seq/potter2018/intersectRootsShoots_PeaksInBoth.bed','../../data/ATAC-seq/potter2018/intersectRootsShoots_PeaksInBoth_renamed.bed')
#create a bed file containing all 100 constitutive/responsive promoters with the fourth column annotating whether it's constitutive or responsive
proms_file = '../../data/genes/constitutive-variable-random_100_each.csv'
promoters = pd.read_csv(proms_file)
promoters
cols2 = ['delete','promoter_AGI', 'gene_type']
promoters_df = promoters[['promoter_AGI','gene_type']]
promoters_no_random = promoters_df.copy()
#drop randCont rows
promoters_no_random = promoters_df[~(promoters_df.gene_type == 'randCont')]
promoters_no_random
#merge promoters with genetype selected
promoterbedfile = '../../data/FIMO/responsivepromoters.bed'
promoters_bed = pd.read_table(promoterbedfile, sep='\t', header=None)
cols = ['chr', 'start', 'stop', 'promoter_AGI', 'score', 'strand', 'source', 'feature_name', 'dot2', 'attributes']
promoters_bed.columns = cols
merged = pd.merge(promoters_bed,promoters_no_random, on='promoter_AGI')
#add gene_type to column3
merged = merged[['chr','start','stop','gene_type','promoter_AGI', 'score', 'strand', 'source', 'feature_name', 'dot2', 'attributes']]
#write to bed file
promoter_file = '../../data/promoter_analysis/old1000bpproms_variable_constitutive_workspace.bed'
with open(promoter_file,'w') as f:
merged.to_csv(f,index=False,sep='\t',header=None)
# new_merged = merged.astype({'start': 'int'})
# new_merged = merged.astype({'stop': 'int'})
# new_merged = merged.astype({'chr': 'int'})
#add Chr to linestart of promoter bed file
add_chr_linestart('../../data/promoter_analysis/old1000bpproms_variable_constitutive_workspace.bed','../../data/promoter_analysis/old1000bpproms_variable_constitutive_workspace_renamed.bed')
#create separate variable and constitutive and gat workspace
promoter_file_renamed = '../../data/promoter_analysis/old1000bpproms_variable_constitutive_workspace_renamed.bed'
promoters = pd.read_table(promoter_file_renamed, sep='\t', header=None)
#make a new gat workspace file with all promoters (first 3 columns)
bed = BedTool.from_dataframe(promoters[[0,1,2]]).saveas('../../data/promoter_analysis/chromatin/variable_constitutive_promoters_1000bp_workspace.bed')
#select only variable promoters
variable_promoters = promoters[promoters[3] == 'highVar']
sorted_variable = variable_promoters.sort_values([0,1])
bed = BedTool.from_dataframe(sorted_variable).saveas('../../data/promoter_analysis/chromatin/variable_promoters_1000bp.bed')
#make a constitutive only file
constitutive_promoters = promoters[promoters[3] == 'housekeeping']
sorted_constitutive = constitutive_promoters.sort_values([0,1])
bed = BedTool.from_dataframe(sorted_constitutive).saveas('../../data/promoter_analysis/chromatin/constitutive_promoters_1000bp.bed')
```
## now I will do the plots with non-overlapping promoters including the 5'UTR
```
#merge promoters with genetype selected
promoter_UTR = '../../data/FIMO/non-overlapping_includingbidirectional_all_genes/promoters_5UTR_renamedChr.bed'
promoters_bed = pd.read_table(promoter_UTR, sep='\t', header=None)
cols = ['chr', 'start', 'stop', 'promoter_AGI', 'score', 'strand', 'source', 'feature_name', 'dot2', 'attributes']
promoters_bed.columns = cols
merged = pd.merge(promoters_bed,promoters_no_random, on='promoter_AGI')
#how many constitutive genes left after removed/shortened overlapping
len(merged[merged.gene_type == 'housekeeping'])
#how many variable genes left after removed/shortened overlapping
len(merged[merged.gene_type == 'highVar'])
merged['length'] = (merged.start - merged.stop).abs()
merged.sort_values('length',ascending=True)
#plot of lengths
dist_plot = merged['length']
#create figure with no transparency
dist_plot_fig = sns.distplot(dist_plot).get_figure()
#remove 2 genes from constitutive group so equal sample size to variable
#random sample of 98, using seed 1
merged[merged.gene_type == 'housekeeping'] = merged[merged.gene_type == 'housekeeping'].sample(98, random_state=1)
#drop rows with at least 2 NaNs
merged = merged.dropna(thresh=2)
merged
#write to bed file so can run OpenChromatin_coverage.py
new_promoter_file = '../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutive.bed'
cols = ['chr', 'start', 'stop', 'promoter_AGI', 'score', 'strand', 'source', 'feature_name', 'dot2', 'attributes']
#remove trailing decimal .0 from start and stop
merged = merged.astype({'start': 'int'})
merged = merged.astype({'stop': 'int'})
merged = merged.astype({'chr': 'int'})
merged_coverage = merged[cols]
with open(new_promoter_file,'w') as f:
merged_coverage.to_csv(f,index=False,sep='\t',header=None)
#write to bed file so can run gat
new_promoter_file_gat = '../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutive_gat.bed'
cols_gat = ['chr', 'start', 'stop', 'gene_type','promoter_AGI', 'score', 'strand', 'source', 'feature_name', 'dot2', 'attributes']
merged_gat = merged[cols_gat]
with open(new_promoter_file_gat,'w') as f:
merged_gat.to_csv(f,index=False,sep='\t',header=None)
#Read in new files
RootChomatin_bp_covered = '../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutiveRootOpenChrom.bp_covered.txt'
ShootChomatin_bp_covered = '../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutiveShootOpenChrom.bp_covered.txt'
RootShootIntersect_bp_covered = '../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutiveShootRootIntersectOpenChrom.bp_covered.txt'
root_coverage = percent_coverage(RootChomatin_bp_covered)
shoot_coverage = percent_coverage(ShootChomatin_bp_covered)
rootshootintersect_coverage = percent_coverage(RootShootIntersect_bp_covered)
#add Chr to linestart of promoter bed file
add_chr_linestart('../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutive_gat.bed','../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutive_gat_renamed.bed')
#create separate variable and constitutive and gat workspace
promoter_file_renamed = '../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutive_gat_renamed.bed'
promoters = pd.read_table(promoter_file_renamed, sep='\t', header=None)
#make a new gat workspace file with all promoters (first 3 columns)
bed = BedTool.from_dataframe(promoters[[0,1,2]]).saveas('../../data/promoter_analysis/chromatin/non-overlapping_includingbidirectional_variable_constitutive_workspace.bed')
#select only variable promoters
variable_promoters = promoters[promoters[3] == 'highVar']
sorted_variable = variable_promoters.sort_values([0,1])
bed = BedTool.from_dataframe(sorted_variable).saveas('../../data/promoter_analysis/chromatin/non-overlapping_includingbidirectional_variable_promoters.bed')
#make a constitutive only file
constitutive_promoters = promoters[promoters[3] == 'housekeeping']
sorted_constitutive = constitutive_promoters.sort_values([0,1])
bed = BedTool.from_dataframe(sorted_constitutive).saveas('../../data/promoter_analysis/chromatin/non-overlapping_includingbidirectional_constitutive_promoters.bed')
#show distribution of the distance from the closest end of the open chromatin peak to the ATG (if overlapping already then distance is 0)
root_peaks_bed = '../../data/ATAC-seq/potter2018/Roots_NaOH_peaks_all_renamed.bed'
shoot_peaks_bed = '../../data/ATAC-seq/potter2018/Shoots_NaOH_peaks_all_renamed.bed'
rootshootintersect_peaks_bed = '../../data/ATAC-seq/potter2018/intersectRootsShoots_PeaksInBoth_renamed.bed'
promoters_bed = '../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutive_renamed.bed'
promoter_openchrom_intersect = '../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutive_chromintersect.bed'
add_chr_linestart('../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutive.bed','../../data/promoter_analysis/non-overlapping_includingbidirectional_variable_constitutive_renamed.bed')
def distr_distance_ATG(peaks_bed, promoter_bed, output_file):
"""function to show the distribution of the distance rom the closest end
of the open chromatin peak to the ATG (if overlapping already then distance is 0)"""
# peaks = pd.read_table(peaks_bed, sep='\t', header=None)
# cols = ['chr','start', 'stop']
# peaks.columns = cols
# promoters = pd.read_table(promoter_bed, sep='\t', header=None)
# cols_proms = ['chr', 'start', 'stop', 'gene_type','promoter_AGI', 'score', 'strand', 'source', 'feature_name', 'dot2', 'attributes']
# promoters.columns = cols_proms
proms = BedTool(promoter_bed) #read in files using BedTools
peaks = BedTool(peaks_bed)
#report chromosome position of overlapping feature, along with the promoter which overlaps it (only reports the overlapping nucleotides, not the whole promoter length. Can use u=True to get whole promoter length)
#f, the minimum overlap as fraction of A. F, nucleotide fraction of B (genes) that need to be overlapping with A (promoters)
#wa, Write the original entry in A for each overlap.
#wo, Write the original A and B entries plus the number of base pairs of overlap between the two features. Only A features with overlap are reported.
#u, write original A entry only once even if more than one overlap
intersect = proms.intersect(peaks, wo=True) #could add u=True which indicates we want to see the promoters that overlap features in the genome
#Write to output_file
with open(output_file, 'w') as output:
#Each line in the file contains bed entry a and bed entry b that it overlaps plus the number of bp in the overlap so 19 columns
output.write(str(intersect))
#read in intersect bed file
overlapping_proms = pd.read_table(output_file, sep='\t', header=None)
cols = ['chrA', 'startA', 'stopA', 'promoter_AGI','dot1','strand','source','type','dot2','attributes','chrB', 'startB','stopB','bp_overlap']
overlapping_proms.columns = cols
#add empty openchrom_distance_from_ATG column
overlapping_proms['openchrom_distance_from_ATG'] = int()
for i, v in overlapping_proms.iterrows():
#if positive strand feature A
if overlapping_proms.loc[i,'strand'] == '+':
#if end of open chromatin is downstream or equal to ATG, distance is 0
if overlapping_proms.loc[i,'stopA'] <= overlapping_proms.loc[i, 'stopB']:
overlapping_proms.loc[i,'openchrom_distance_from_ATG'] = 0
#else if upstream and chromatin stop is after promoter start, add distance from chromatin stop to ATG
elif overlapping_proms.loc[i,'startA'] <= overlapping_proms.loc[i, 'stopB']:
overlapping_proms.loc[i,'openchrom_distance_from_ATG'] = overlapping_proms.loc[i,'stopA'] - overlapping_proms.loc[i, 'stopB']
elif overlapping_proms.loc[i,'strand'] == '-':
#if end of open chromatin is downstream or equal to ATG, distance is 0
if overlapping_proms.loc[i,'startA'] >= overlapping_proms.loc[i, 'startB']:
overlapping_proms.loc[i,'openchrom_distance_from_ATG'] = 0
#else if upstream and chromatin stop is after promoter start, add distance from chromatin stop to ATG
elif overlapping_proms.loc[i,'stopA'] >= overlapping_proms.loc[i, 'startB']:
overlapping_proms.loc[i,'openchrom_distance_from_ATG'] = overlapping_proms.loc[i, 'startB'] - overlapping_proms.loc[i,'startB']
return overlapping_proms
#show length of open chromatin peaks
rootshootintersect = distr_distance_ATG(rootshootintersect_peaks_bed)
rootshootintersect['length'] = (rootshootintersect.start - rootshootintersect.stop).abs()
rootshootintersect.sort_values('length',ascending=True)
rootshootintersect = distr_distance_ATG(rootshootintersect_peaks_bed,promoters_bed,promoter_openchrom_intersect)
rootshootintersect
rootshootintersect.sort_values('openchrom_distance_from_ATG',ascending=True)
#plot of distances of chomatin to ATG
dist_plot = rootshootintersect['openchrom_distance_from_ATG']
#create figure with no transparency
dist_plot_fig = sns.distplot(dist_plot).get_figure()
#now split constitutive and variable
merged_distances = pd.merge(merged, rootshootintersect, on='promoter_AGI')
merged_distances.gene_type
#VARIABLE
#plot of distances of chomatin to ATG
dist_plot = merged_distances[merged_distances.gene_type=='highVar']['openchrom_distance_from_ATG']
#create figure with no transparency
dist_plot_fig = sns.distplot(dist_plot).get_figure()
merged_distances[merged_distances.gene_type=='housekeeping']['openchrom_distance_from_ATG']
#CONSTITUTIVE
#plot of distances of chomatin to ATG
dist_plot = merged_distances[merged_distances.gene_type=='housekeeping']['openchrom_distance_from_ATG']
#create figure with no transparency
dist_plot_fig = sns.distplot(dist_plot).get_figure()
```
| github_jupyter |
```
dataset = 'load' # 'load' or 'generate'
retrain_models = False # False or True or 'save'
import numpy as np
import pandas as pd
import tensorflow as tf
tf.logging.set_verbosity(tf.logging.FATAL)
import gpflow
import library.models.deep_vmgp as deep_vmgp
import library.models.vmgp as vmgp
from doubly_stochastic_dgp.dgp import DGP
import matplotlib as mpl
import matplotlib.pyplot as plt
import cplot
import sklearn.model_selection
import pickle
from pathlib import Path
from types import SimpleNamespace
from library.helper import TrainTestSplit, initial_inducing_points
from library import metrics
%matplotlib inline
random_seed = 19960111
def reset_seed():
np.random.seed(random_seed)
tf.random.set_random_seed(random_seed)
if dataset == 'generate':
s = 0.4
n = 500//2
reset_seed()
rng = np.random.default_rng(random_seed)
m1, m2 = np.array([[-1,1],[2,1]])
X1 = rng.multivariate_normal(m1,s*np.eye(2), size=n)
X2 = rng.multivariate_normal(m2,s*np.eye(2), size=n)
y1 = X1[:,0]**2 + X1[:,0]
y2 = X2[:,1]**2 + X2[:,1]
X = np.concatenate([X1,X2],axis=0)
y = np.concatenate([y1,y2],axis=0)[:,None]
X_all, y_all = X,y
n = X_all.shape[0]
kfold = sklearn.model_selection.KFold(2,shuffle=True,random_state=random_seed)
folds = [
[TrainTestSplit(X_all[train],X_all[test]), TrainTestSplit(y_all[train],y_all[test])]
for train, test in kfold.split(X_all, y_all)
]
X,y = folds[0]
elif dataset == 'load':
with open('./dataset.pkl','rb') as f:
X, y = pickle.load(f)
X_all, y_all = np.concatenate(X,axis=0), np.concatenate(y,axis=0)
scalers = SimpleNamespace(x=sklearn.preprocessing.StandardScaler(),y=sklearn.preprocessing.StandardScaler())
scalers.x.fit(X.train)
X = X.apply(lambda x: scalers.x.transform(x))
scalers.y.fit(y.train)
y = y.apply(lambda y: scalers.y.transform(y))
models = pd.Series(index=pd.Index([],dtype='object'), dtype=object)
parameters = pd.Series({p.stem:p for p in Path('./optimized_parameters/').glob('*.pkl')}, dtype=object).map(read_parameters)
y_pred = pd.DataFrame(dtype=float, index=range(y.test.size), columns=pd.MultiIndex(levels=[[],['mean','var']],codes=[[],[]],names=['model','']))
results = pd.DataFrame(columns=['RMSE','NLPD','MRAE'],dtype=float)
def read_parameters(p):
try:
with p.open('rb') as f:
return pickle.load(f)
except:
return None
def train_model(model_label):
m = models[model_label]
if retrain_models == True or retrain_models == 'save' or model_label not in parameters.index:
print('Training',model_label)
variance_parameter = m.likelihood.variance if not isinstance(m, DGP) else m.likelihood.likelihood.variance
variance_parameter.assign(0.01)
# First round
variance_parameter.trainable = False
opt = gpflow.train.AdamOptimizer(0.01)
opt.minimize(m, maxiter=2000)
# Second round
variance_parameter.trainable = True
opt = gpflow.train.AdamOptimizer(0.01)
opt.minimize(m, maxiter=5000)
if retrain_models == 'save' or model_label not in parameters.index:
with open(f'./optimized_parameters/{model_label}.pkl','wb') as f:
pickle.dump(m.read_trainables(), f)
else:
m.assign(parameters[model_label])
```
# Create, train, and predict with models
```
n,D = X.train.shape
m_v = 25
m_u, Q, = 50, D
Z_v = (m_v,D)
Z_u = (m_u,Q)
sample_size = 200
```
### SGPR
```
models['sgpr'] = gpflow.models.SGPR(X.train, y.train, gpflow.kernels.RBF(D, ARD=True), initial_inducing_points(X.train, m_u))
train_model('sgpr')
y_pred[('sgpr','mean')], y_pred[('sgpr','var')] = models['sgpr'].predict_y(X.test)
```
### Deep Mahalanobis GP
```
reset_seed()
with gpflow.defer_build():
models['dvmgp'] = deep_vmgp.DeepVMGP(
X.train, y.train, Z_u, Z_v,
[gpflow.kernels.RBF(D,ARD=True) for i in range(Q)],
full_qcov=False, diag_qmu=False
)
models['dvmgp'].compile()
train_model('dvmgp')
y_pred[('dvmgp','mean')], y_pred[('dvmgp','var')] = models['dvmgp'].predict_y(X.test)
```
### Show scores
```
for m in models.index:
scaled_y_test = scalers.y.inverse_transform(y.test)
scaled_y_pred = [
scalers.y.inverse_transform(y_pred[m].values[:,[0]]),
scalers.y.var_ * y_pred[m].values[:,[1]]
]
results.at[m,'MRAE'] = metrics.mean_relative_absolute_error(scaled_y_test, scaled_y_pred[0]).squeeze()
results.at[m,'RMSE'] = metrics.root_mean_squared_error(scaled_y_test, scaled_y_pred[0]).squeeze()
results.at[m,'NLPD'] = metrics.negative_log_predictive_density(scaled_y_test, *scaled_y_pred).squeeze()
results
```
# Plot results
```
class MidpointNormalize(mpl.colors.Normalize):
def __init__(self, vmin=None, vmax=None, midpoint=None, clip=False):
self.midpoint = midpoint
mpl.colors.Normalize.__init__(self, vmin, vmax, clip)
def __call__(self, value, clip=None):
x, y = [self.vmin, self.midpoint, self.vmax], [0, 0.5, 1]
return np.ma.masked_array(np.interp(value, x, y), np.isnan(value))
f = plt.figure()
ax = plt.gca()
ax.scatter(scalers.x.transform(X_all)[:,0],scalers.x.transform(X_all)[:,1],edgecolors='white',facecolors='none')
lims = (ax.get_xlim(), ax.get_ylim())
plt.close(f)
n = 50
grid_points = np.dstack(np.meshgrid(np.linspace(*lims[0],n), np.linspace(*lims[1],n))).reshape(-1,2)
grid_y = np.empty((len(models.index),grid_points.shape[0]))
for i,m in enumerate(models.index):
reset_seed()
grid_pred = models[m].predict_y(grid_points, sample_size)[0]
if len(grid_pred.shape) == 3:
grid_y[i] = grid_pred.mean(axis=0)[:,0]
else:
grid_y[i] = grid_pred[:,0]
grid_points = grid_points.reshape(n,n,2)
grid_y = grid_y.reshape(-1,n,n)
f = plt.figure(constrained_layout=True,figsize=(8,7))
gs = f.add_gridspec(ncols=4, nrows=2)
axs = np.empty(3,dtype=object)
axs[0] = f.add_subplot(gs[0,0:2])
axs[1] = f.add_subplot(gs[0,2:4],sharey=axs[0])
axs[2] = f.add_subplot(gs[1,1:3])
axs[1].yaxis.set_visible(False)
axs[2].yaxis.set_visible(False)
axs[0].set_title('SGPR')
axs[1].set_title('DVMGP')
axs[2].set_title('Full Dataset')
ims = np.empty((2,4),dtype=object)
for i,m in enumerate(['sgpr', 'dvmgp']):
ax = axs[i]
ims[0,i] = ax.contourf(grid_points[:,:,0],grid_points[:,:,1],grid_y[i],30)
# Plot features
Z = None
if m == 'dgp':
Z = models[m].layers[0].feature.Z.value
elif m in ['sgpr','vmgp']:
Z = models[m].feature.Z.value
elif m == 'dvmgp':
Z = models[m].Z_v.Z.value
if Z is not None:
ax.scatter(Z[:,0],Z[:,1],marker='^',edgecolors='white',facecolors='none')
# ims[1,i] = ax.scatter(X.test[:,0],X.test[:,1],edgecolors='white',c=y.test)
ims[0,3] = axs[2].scatter(X.test[:,0],X.test[:,1],c=y.test)
ims[1,3] = axs[2].scatter(X.train[:,0],X.train[:,1],c=y.train)
for ax in axs:
ax.set_xlim(lims[0]);
ax.set_ylim(lims[1]);
clim = np.array([i.get_clim() for i in ims.flat if i is not None])
clim = (clim.min(), clim.max())
norm = mpl.colors.Normalize(vmin=clim[0], vmax=clim[1])
# norm = MidpointNormalize(vmin=clim[0], vmax=clim[1], midpoint=0)
for im in ims.flat:
if im is not None:
im.set_norm(norm)
f.colorbar(ims[0,0], ax=axs, orientation='vertical', fraction=1, aspect=50)
for im in ims[0,:3].flat:
if im is not None:
for c in im.collections:
c.set_edgecolor("face")
f.savefig('./figs/outputs.pdf')
n = 50
grid_points = np.dstack(np.meshgrid(np.linspace(*lims[0],n), np.linspace(*lims[1],n))).reshape(-1,2)
grid_y = np.empty((grid_points.shape[0],2))
grid_y = models['dvmgp'].enquire_session().run(tf.matmul(
tf.transpose(models['dvmgp'].compute_qW(grid_points)[0][...,0],[2,0,1]),grid_points[:,:,None]
)[:,:,0])
grid_points = grid_points.reshape(n,n,2)
grid_y = grid_y.reshape(n,n,2)
f = plt.figure(constrained_layout=True,figsize=(8,4))
gs = f.add_gridspec(ncols=2, nrows=1)
axs = np.empty(4,dtype=object)
axs[0] = f.add_subplot(gs[0,0])
axs[1] = f.add_subplot(gs[0,1])
extent = (*lims[0], *lims[1])
colorspace = 'cielab'
alpha = 0.7
axs[0].imshow(
cplot.get_srgb1(grid_points[:,:,0] + grid_points[:,:,1]*1j, colorspace=colorspace, alpha=alpha),
origin='lower',
extent=extent,
aspect='auto',
interpolation='gaussian'
)
axs[0].set_title('Identity map')
axs[1].imshow(
cplot.get_srgb1(grid_y[:,:,0] + grid_y[:,:,1]*1j, colorspace=colorspace, alpha=alpha),
origin='lower',
extent=extent,
aspect='auto',
interpolation='gaussian'
)
axs[1].set_title('DVMGP: $Wx^\intercal$');
f.savefig('./figs/layers.pdf')
dvmgp_var = np.array([k.variance.value for k in models['dvmgp'].w_kerns])
f,ax = plt.subplots(1,1,figsize=(3,3))
ax.bar(np.arange(2), dvmgp_var/dvmgp_var.max(), color='C2')
ax.set_ylabel('1st layer variance\nrelative to largest value')
ax.set_xlabel('Latent dimension')
ax.set_xticks([])
ax.set_title('DVMGP')
f.tight_layout()
f.savefig('./figs/dims.pdf')
```
| github_jupyter |
# Notebook para o PAN - Atribuição Autoral - 2018
```
%matplotlib inline
#python basic libs
import os;
from os.path import join as pathjoin;
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
from sklearn.exceptions import UndefinedMetricWarning
warnings.simplefilter(action='ignore', category=UndefinedMetricWarning)
import re;
import json;
import codecs;
from collections import defaultdict;
from pprint import pprint
from time import time
import logging
#data analysis libs
import numpy as np;
import pandas as pd;
from pandas.plotting import scatter_matrix;
import matplotlib.pyplot as plt;
import random;
#machine learning libs
#feature extraction
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer, TfidfTransformer
#preprocessing and transformation
from sklearn import preprocessing
from sklearn.preprocessing import normalize, MaxAbsScaler, RobustScaler;
from sklearn.decomposition import PCA;
from sklearn.base import BaseEstimator, ClassifierMixin
#classifiers
from sklearn import linear_model;
from sklearn.linear_model import LogisticRegression
from sklearn.svm import LinearSVC, SVC
from sklearn.multiclass import OneVsOneClassifier, OneVsRestClassifier
from sklearn.neural_network import MLPClassifier
#
from sklearn import feature_selection;
from sklearn import ensemble;
from sklearn.model_selection import train_test_split;
from sklearn.model_selection import GridSearchCV
from sklearn.pipeline import Pipeline
#model valuation
from sklearn.metrics import roc_auc_score, f1_score, precision_score, recall_score, accuracy_score;
import seaborn as sns;
sns.set(color_codes=True);
import spacy
get_ipython().config.get('IPKernelApp', {})['parent_appname'] = "" #spacy causes a bug on pandas and this code fix it
import platform;
import sklearn;
import scipy;
print("|%-15s|%-40s|"%("PACK","VERSION"))
print("|%-15s|%-40s|"%('-'*15,'-'*40))
print('\n'.join(
"|%-15s|%-40s|" % (pack, version)
for pack, version in
zip(['SO','NumPy','SciPy','Scikit-Learn','seaborn','spacy'],
[platform.platform(), np.__version__, scipy.__version__, sklearn.__version__, sns.__version__, spacy.__version__])
))
np.set_printoptions(precision=4)
pd.options.display.float_format = '{:,.4f}'.format
#externalizing codes that is used in many notebooks and it is not experiment specific
import pan
#convert a sparse matrix into a dense for being used on PCA
from skleanExtensions import DenseTransformer;
#convert an array of text into an array of tokenized texts each token must contain text, tag_, pos_, dep_
from skleanExtensions import POSTagTransformer
```
### paths configuration
```
baseDir = '/Users/joseeleandrocustodio/Dropbox/mestrado/02 - Pesquisa/code';
inputDir= pathjoin(baseDir,'pan18aa');
outputDir= pathjoin(baseDir,'out',"oficial");
if not os.path.exists(outputDir):
os.mkdir(outputDir);
```
## loading the dataset
```
problems = pan.readCollectionsOfProblems(inputDir);
print(problems[0]['problem'])
print(problems[0].keys())
pd.DataFrame(problems)
def cachingPOSTAG(problem, taggingVersion='TAG'):
import json;
print ("Tagging: %s, language: %s, " %(problem['problem'],problem['language']), end=' ');
if not os.path.exists('POSTAG_cache'):
os.makedirs('POSTAG_cache');
_id = problem['problem']+problem['language'];
filename = os.path.join('POSTAG_cache',taggingVersion+'_'+_id+'.json')
if not os.path.exists(filename):
lang = problem['language'];
if lang == 'sp':
lang = 'es';
elif lang =='pl':
print(lang, ' not supported');
return ;
train_docs, train_labels, _ = zip(*problem['candidates'])
problem['training_docs_size'] = len(train_docs);
test_docs, _, test_filename = zip(*problem['unknown'])
t0 = time()
tagger = POSTagTransformer(language=lang);
train_docs = tagger.fit_transform(train_docs);
test_docs = tagger.fit_transform(test_docs);
print("Annotation time %0.3fs" % (time() - t0))
with open(filename,'w') as f:
json.dump({
'train':train_docs,
'train_labels':train_labels,
'test':test_docs,
'test_filename':test_filename
},f);
else:
with open(filename,'r') as f:
data = json.load(f);
train_docs = data['train'];
train_labels = data['train_labels'];
test_docs = data['test'];
test_filename = data['test_filename'];
print('tagged')
return train_docs, train_labels, test_docs, test_filename;
for problem in problems:
cachingPOSTAG(problem)
train_docs, train_labels, test_docs, test_filename = cachingPOSTAG(problem)
class FilterTagTransformer(BaseEstimator):
def __init__(self,token='POS', parts=None):
self.token = token;
self.parts = parts;
def transform(self, X, y=None):
""" Return An array of tokens
Parameters
----------
X : {array-like}, shape = [n_samples, n_tokens]
Array documents, where each document consists of a list of node
and each node consist of a token and its correspondent tag
[
[('a','TAG1'),('b','TAG2')],
[('a','TAG1')]
]
y : array-like, shape = [n_samples] (default: None)
Returns
---------
X_dense : dense version of the input X array.
"""
if self.token == 'TAG':
X = [' '.join([d[1].split('__')[0] for d in doc]) for doc in X]
elif self.token == 'POS':
if self.parts is None:
X = [' '.join([d[2] for d in doc]) for doc in X];
else:
X = [' '.join([d[0] for d in doc if d[2] in self.parts]) for doc in X]
elif self.token == 'DEP':
X = [' '.join([d[3] for d in doc]) for doc in X]
elif self.token == 'word_POS':
if self.parts is None:
X = [' '.join([d[0]+'/'+d[2] for d in doc]) for doc in X]
elif self.token == 'filter':
if self.parts is None:
X = [' '.join([d[2] for d in doc]) for doc in X];
else:
X = [' '.join([d[0] for d in doc if d[2] in self.parts]) for doc in X]
else:
X = [' '.join([d[0] for d in doc]) for doc in X]
return np.array(X);
def fit(self, X, y=None):
self.is_fitted = True
return self
def fit_transform(self, X, y=None):
return self.transform(X=X, y=y)
```
### analisando os demais parametros
```
def spaceTokenizer(x):
return x.split(" ");
def runML(problem):
print ("\nProblem: %s, language: %s, " %(problem['problem'],problem['language']), end=' ');
lang = problem['language'];
if lang == 'sp':
lang = 'es';
elif lang =='pl':
print(lang, ' not supported');
return None,None,None,None;
train_docs, train_labels, test_docs, test_filename = cachingPOSTAG(problem)
problem['training_docs_size'] = len(train_docs);
t0 = time()
pipeline = Pipeline([
('filter',FilterTagTransformer(token='TAG')),
('vect', CountVectorizer(
tokenizer=spaceTokenizer,
min_df=0.01,
lowercase=False
)),
('tfidf', TfidfTransformer()),
('scaler', MaxAbsScaler()),
('dense', DenseTransformer()),
('transf', PCA(0.999)),
('clf', LogisticRegression(random_state=0,multi_class='multinomial', solver='newton-cg')),
])
# uncommenting more parameters will give better exploring power but will
# increase processing time in a combinatorial way
parameters = {
'vect__ngram_range' :((1,1),(1,2),(1,3),(1,5)),
'tfidf__use_idf' :(True, False),
'tfidf__sublinear_tf':(True, False),
'tfidf__norm':('l1','l2'),
'clf__C':(0.1,1,10),
}
grid_search = GridSearchCV(pipeline,
parameters,
cv=4,
iid=False,
n_jobs=-1,
verbose=False,
scoring='f1_macro')
t0 = time()
grid_search.fit(train_docs, train_labels)
print("Gridsearh %0.3fs" % (time() - t0), end=' ')
print("Best score: %0.3f" % grid_search.best_score_)
print("Best parameters set:")
best_parameters = grid_search.best_estimator_.get_params()
for param_name in sorted(parameters.keys()):
print("\t%s: %r" % (param_name, best_parameters[param_name]))
train_pred=grid_search.predict(train_docs);
test_pred=grid_search.predict(test_docs);
# Writing output file
out_data=[]
for i,v in enumerate(test_pred):
out_data.append({'unknown-text': test_filename[i],'predicted-author': v})
answerFile = pathjoin(outputDir,'answers-'+problem['problem']+'.json');
with open(answerFile, 'w') as f:
json.dump(out_data, f, indent=4)
#calculating the performance using PAN evaluation code
f1,precision,recall,accuracy=pan.evaluate(
pathjoin(inputDir, problem['problem'], 'ground-truth.json'),
answerFile)
return {
'problem-name' : problem['problem'],
"language" : problem['language'],
'AuthorCount' : len(set(train_labels)),
'macro-f1' : round(f1,3),
'macro-precision': round(precision,3),
'macro-recall' : round(recall,3),
'micro-accuracy' : round(accuracy,3),
}, grid_search.cv_results_,best_parameters, grid_search.best_estimator_;
result = [];
cv_result = [];
best_parameters = [];
estimators = [];
for problem in problems:
with warnings.catch_warnings():
warnings.filterwarnings("ignore");
r, c, b, e = runML(problem);
if r is None:
continue;
result.append(r);
cv_result.append(c);
estimators.append(e);
b['problem'] = problem['problem'];
best_parameters.append(b);
df=pd.DataFrame(result)[['problem-name',
"language",
'AuthorCount',
'macro-f1','macro-precision','macro-recall' ,'micro-accuracy']]
df
df[['macro-f1']].mean()
languages={
'en':'inglesa',
'sp':'espanhola',
'it':'italiana',
'pl':'polonesa',
'fr':'francesa'
}
cv_result2 = [];
dfCV = pd.DataFrame();
for i, c in enumerate(cv_result):
temp = pd.DataFrame(c);
temp['language'] = result[i]['AuthorCount']
temp['problem'] = int(re.sub('\D','',result[i]['problem-name']));
temp['language'] = languages[result[i]['language']]
dfCV = dfCV.append(temp);
for p in [
'mean_test_score','std_test_score','mean_train_score',
'split0_test_score',
'split1_test_score',
'split2_test_score']:
dfCV[p]=dfCV[p].astype(np.float32);
dfCV =dfCV[[
'problem',
'language',
'rank_test_score',
'param_vect__ngram_range',
'param_tfidf__sublinear_tf',
'param_tfidf__norm',
'param_clf__C',
'mean_test_score',
'std_test_score',
'split0_test_score',
'split1_test_score',
'split2_test_score',
'mean_score_time',
'mean_fit_time',
'std_fit_time',
'std_score_time',
'std_train_score',
]];
dfCV.rename(columns={
'param_vect__ngram_range':'ngram_range',
'param_tfidf__sublinear_tf':'sublinear_tf',
'param_tfidf__smooth_idf':'smooth_idf',
'param_tfidf__norm':'norm',
'param_clf__C':'regularization',
},inplace=True);
#print('\',\n\''.join(dfCV.columns))
dfCV.head()
```
## Saving the model
```
dfCV.to_csv('PANAA2018_POSTAG.csv', index=False)
dfCV = pd.read_csv('PANAA2018_POSTAG.csv', na_values='')
import pickle;
with open("PANAA2018_POSTAG.pkl","wb") as f:
pickle.dump(estimators,f)
```
## understanding the model with reports
Podemos ver que para um mesmo problema mais de uma configuração é possível
```
print(' | '.join(best_parameters[0]['vect'].get_feature_names()[0:20]))
(dfCV[dfCV.rank_test_score == 1]).drop_duplicates()[
['problem',
'language',
'mean_test_score',
'std_test_score',
'ngram_range',
'sublinear_tf',
'norm']
].sort_values(by=[
'problem',
'mean_test_score',
'std_test_score',
'ngram_range',
'sublinear_tf'
], ascending=[True, False,True,False,False])
dfCV.pivot_table(
index=['problem','language','norm','sublinear_tf'],
columns=[ 'ngram_range','regularization'],
values='mean_test_score'
)
```
O score retornado vem do conjunto de teste da validação cruzada e não do conjunto de testes
```
pd.options.display.precision = 3
print(u"\\begin{table}[h]\n\\centering\n\\caption{Medida F1 para os parâmetros }")
print(re.sub(r'[ ]{2,}',' ',dfCV.pivot_table(
index=['problem','language','sublinear_tf','norm'],
columns=['ngram_range'],
values='mean_test_score'
).to_latex()))
print ("\label{tab:modelocaracter}")
print(r"\end{table}")
d = dfCV.copy()
d = d.rename(columns={'language':u'Língua', 'sublinear_tf':'TF Sublinear'})
d = d [ d.norm.isna() == False]
d['autorNumber'] = d.problem.map(lambda x: 20 if x % 2==0 else 5)
d.problem = d.apply(lambda x: x[u'Língua'] +" "+ str(x[u'problem']), axis=1)
#d.ngram_range = d.apply(lambda x: str(x[u'ngram_range'][0]) +" "+ str(x[u'ngram_range'][1]), axis=1)
d.std_test_score =d.std_test_score / d.std_test_score.quantile(0.95) *500;
d.std_test_score +=1;
d.std_test_score = d.std_test_score.astype(np.int64)
g = sns.FacetGrid(d, col='Língua', hue='TF Sublinear', row="regularization", height=3,palette="Set1")
g.map(plt.scatter, "ngram_range", "mean_test_score",s=d.std_test_score.values).add_legend();
#sns.pairplot(d, hue="TF Sublinear", vars=["autorNumber", "mean_test_score"])
g = sns.FacetGrid(d, row='autorNumber', hue='TF Sublinear', col=u"Língua", height=3,palette="Set1")
g.map(plt.scatter, "ngram_range", "mean_test_score", alpha=0.5, s=d.std_test_score.values).add_legend();
sns.distplot(dfCV.std_test_score, bins=25);
import statsmodels.api as sm
d = dfCV[['mean_test_score','problem', 'language','sublinear_tf','norm','ngram_range']].copy();
d.sublinear_tf=d.sublinear_tf.apply(lambda x: 1 if x else 0)
d.norm=d.norm.apply(lambda x: 1 if x=='l1' else 0)
d['autorNumber'] = d.problem.map(lambda x: 20 if x % 2==0 else 5)
d.norm.fillna(value='None', inplace=True);
_, d['ngram_max'] = zip(*d.ngram_range.str.replace(r'[^\d,]','').str.split(',').values.tolist())
#d.ngram_min = d.ngram_min.astype(np.uint8);
d.ngram_max = d.ngram_max.astype(np.uint8);
d.drop(columns=['ngram_range','problem'], inplace=True)
#d['intercept'] = 1;
d=pd.get_dummies(d, columns=['language'])
d.describe()
mod = sm.OLS( d.iloc[:,0], d.iloc[:,1:])
res = mod.fit()
res.summary()
sns.distplot(res.predict()-d.iloc[:,0].values, bins=25)
sns.jointplot(x='F1',y='F1-estimated',data=pd.DataFrame({'F1':d.iloc[:,0].values, 'F1-estimated':res.predict()}));
```
# tests
```
problem = problems[0]
print ("\nProblem: %s, language: %s, " %(problem['problem'],problem['language']), end=' ');
def d(estimator, n_features=5):
from IPython.display import Markdown, display, HTML
names = np.array(estimator.named_steps['vect'].get_feature_names());
classes_ = estimator.named_steps['clf'].classes_;
weights = estimator.named_steps['clf'].coef_;
def tag(tag, content, attrib=''):
if attrib != '':
attrib = ' style="' + attrib+'"';
return ''.join(['<',tag,attrib,' >',content,'</',tag,'>']);
def color(baseColor, intensity):
r,g,b = baseColor[0:2],baseColor[2:4],baseColor[4:6]
r,g,b = int(r, 16), int(g, 16), int(b, 16)
f= (1-np.abs(intensity))/2;
r = r + int((255-r)*f)
g = g + int((255-g)*f)
b = b + int((255-b)*f)
rgb = '#%02x%x%x' % (r, g, b);
#print(baseColor,rgb,r,g,b,intensity,f)
return rgb
spanStyle ='border-radius: 5px;margin:4px;padding:3px; color:#FFF !important;';
lines = '<table>'+tag('thead',tag('th','Classes')+tag('th','positive')+tag('th','negative'))
lines += '<tbody>'
for i,c in enumerate(weights):
c = np.round(c / np.abs(c).max(),2);
positive = names[np.argsort(-c)][:n_features];
positiveV = c[np.argsort(-c)][:n_features]
negative = names[np.argsort(c)][:n_features];
negativeV = c[np.argsort(c)][:n_features]
lines += tag('tr',
tag('td', re.sub('\D0*','',classes_[i]))
+ tag('td',''.join([tag('span',d.upper()+' '+str(v),spanStyle+'background:'+color('51A3DD',v)) for d,v in zip(positive,positiveV)]))
+ tag('td',''.join([tag('span',d.upper()+' '+str(v),spanStyle+'background:'+color('DD5555',v)) for d,v in zip(negative,negativeV)]))
)
lines+= '</tbody></table>'
display(HTML(lines))
#print(lines)
d(estimators[0])
%%HTML
<table><tbody><tr><th>POS</th><th>Description</th><th>Examples</th></tr><tr >
<td class="c-table__cell u-text"><code>ADJ</code></td><td class="c-table__cell u-text u-text-small">adjective</td><td class="c-table__cell u-text u-text-small"><em>big, old, green, incomprehensible, first</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>ADP</code></td><td class="c-table__cell u-text u-text-small">adposition</td><td class="c-table__cell u-text u-text-small"><em>in, to, during</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>ADV</code></td><td class="c-table__cell u-text u-text-small">adverb</td><td class="c-table__cell u-text u-text-small"><em>very, tomorrow, down, where, there</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>AUX</code></td><td class="c-table__cell u-text u-text-small">auxiliary</td><td class="c-table__cell u-text u-text-small"><em>is, has (done), will (do), should (do)</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>CONJ</code></td><td class="c-table__cell u-text u-text-small">conjunction</td><td class="c-table__cell u-text u-text-small"><em>and, or, but</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>CCONJ</code></td><td class="c-table__cell u-text u-text-small">coordinating conjunction</td><td class="c-table__cell u-text u-text-small"><em>and, or, but</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>DET</code></td><td class="c-table__cell u-text u-text-small">determiner</td><td class="c-table__cell u-text u-text-small"><em>a, an, the</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>INTJ</code></td><td class="c-table__cell u-text u-text-small">interjection</td><td class="c-table__cell u-text u-text-small"><em>psst, ouch, bravo, hello</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>NOUN</code></td><td class="c-table__cell u-text u-text-small">noun</td><td class="c-table__cell u-text u-text-small"><em>girl, cat, tree, air, beauty</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>NUM</code></td><td class="c-table__cell u-text u-text-small">numeral</td><td class="c-table__cell u-text u-text-small"><em>1, 2017, one, seventy-seven, IV, MMXIV</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PART</code></td><td class="c-table__cell u-text u-text-small">particle</td><td class="c-table__cell u-text u-text-small"><em>'s, not, </em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PRON</code></td><td class="c-table__cell u-text u-text-small">pronoun</td><td class="c-table__cell u-text u-text-small"><em>I, you, he, she, myself, themselves, somebody</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PROPN</code></td><td class="c-table__cell u-text u-text-small">proper noun</td><td class="c-table__cell u-text u-text-small"><em>Mary, John, London, NATO, HBO</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text u-text-small">punctuation</td><td class="c-table__cell u-text u-text-small"><em>., (, ), ?</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>SCONJ</code></td><td class="c-table__cell u-text u-text-small">subordinating conjunction</td><td class="c-table__cell u-text u-text-small"><em>if, while, that</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>SYM</code></td><td class="c-table__cell u-text u-text-small">symbol</td><td class="c-table__cell u-text u-text-small"><em>$, %, §, ©, +, −, ×, ÷, =, :), 😝</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text u-text-small">verb</td><td class="c-table__cell u-text u-text-small"><em>run, runs, running, eat, ate, eating</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>X</code></td><td class="c-table__cell u-text u-text-small">other</td><td class="c-table__cell u-text u-text-small"><em>sfpksdpsxmsa</em></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>SPACE</code></td><td class="c-table__cell u-text u-text-small">space</td></tr></tbody></table>
%%HTML
<h1>English</h1>
<table class="c-table o-block"><tbody><tr class="c-table__row c-table__row--head"><th class="c-table__head-cell u-text-label">Tag</th><th class="c-table__head-cell u-text-label">POS</th><th class="c-table__head-cell u-text-label">Morphology</th><th class="c-table__head-cell u-text-label">Description</th></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>-LRB-</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>PunctType=brck</code> <code>PunctSide=ini</code></td><td class="c-table__cell u-text u-text-small">left round bracket</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>-RRB-</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>PunctType=brck</code> <code>PunctSide=fin</code></td><td class="c-table__cell u-text u-text-small">right round bracket</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>,</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>PunctType=comm</code></td><td class="c-table__cell u-text u-text-small">punctuation mark, comma</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>:</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">punctuation mark, colon or ellipsis</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>.</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>PunctType=peri</code></td><td class="c-table__cell u-text u-text-small">punctuation mark, sentence closer</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>''</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>PunctType=quot</code> <code>PunctSide=fin</code></td><td class="c-table__cell u-text u-text-small">closing quotation mark</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>""</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>PunctType=quot</code> <code>PunctSide=fin</code></td><td class="c-table__cell u-text u-text-small">closing quotation mark</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>#</code></td><td class="c-table__cell u-text"><code>SYM</code></td><td class="c-table__cell u-text"> <code>SymType=numbersign</code></td><td class="c-table__cell u-text u-text-small">symbol, number sign</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>``</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>PunctType=quot</code> <code>PunctSide=ini</code></td><td class="c-table__cell u-text u-text-small">opening quotation mark</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>$</code></td><td class="c-table__cell u-text"><code>SYM</code></td><td class="c-table__cell u-text"> <code>SymType=currency</code></td><td class="c-table__cell u-text u-text-small">symbol, currency</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>ADD</code></td><td class="c-table__cell u-text"><code>X</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">email</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>AFX</code></td><td class="c-table__cell u-text"><code>ADJ</code></td><td class="c-table__cell u-text"> <code>Hyph=yes</code></td><td class="c-table__cell u-text u-text-small">affix</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>BES</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">auxiliary "be"</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>CC</code></td><td class="c-table__cell u-text"><code>CONJ</code></td><td class="c-table__cell u-text"> <code>ConjType=coor</code></td><td class="c-table__cell u-text u-text-small">conjunction, coordinating</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>CD</code></td><td class="c-table__cell u-text"><code>NUM</code></td><td class="c-table__cell u-text"> <code>NumType=card</code></td><td class="c-table__cell u-text u-text-small">cardinal number</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>DT</code></td><td class="c-table__cell u-text"><code>DET</code></td><td class="c-table__cell u-text"> <code>determiner</code></td><td class="c-table__cell u-text u-text-small"></td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>EX</code></td><td class="c-table__cell u-text"><code>ADV</code></td><td class="c-table__cell u-text"> <code>AdvType=ex</code></td><td class="c-table__cell u-text u-text-small">existential there</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>FW</code></td><td class="c-table__cell u-text"><code>X</code></td><td class="c-table__cell u-text"> <code>Foreign=yes</code></td><td class="c-table__cell u-text u-text-small">foreign word</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>GW</code></td><td class="c-table__cell u-text"><code>X</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">additional word in multi-word expression</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>HVS</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">forms of "have"</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>HYPH</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>PunctType=dash</code></td><td class="c-table__cell u-text u-text-small">punctuation mark, hyphen</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>IN</code></td><td class="c-table__cell u-text"><code>ADP</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">conjunction, subordinating or preposition</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>JJ</code></td><td class="c-table__cell u-text"><code>ADJ</code></td><td class="c-table__cell u-text"> <code>Degree=pos</code></td><td class="c-table__cell u-text u-text-small">adjective</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>JJR</code></td><td class="c-table__cell u-text"><code>ADJ</code></td><td class="c-table__cell u-text"> <code>Degree=comp</code></td><td class="c-table__cell u-text u-text-small">adjective, comparative</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>JJS</code></td><td class="c-table__cell u-text"><code>ADJ</code></td><td class="c-table__cell u-text"> <code>Degree=sup</code></td><td class="c-table__cell u-text u-text-small">adjective, superlative</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>LS</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>NumType=ord</code></td><td class="c-table__cell u-text u-text-small">list item marker</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>MD</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>VerbType=mod</code></td><td class="c-table__cell u-text u-text-small">verb, modal auxiliary</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>NFP</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">superfluous punctuation</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>NIL</code></td><td class="c-table__cell u-text"><code></code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">missing tag</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>NN</code></td><td class="c-table__cell u-text"><code>NOUN</code></td><td class="c-table__cell u-text"> <code>Number=sing</code></td><td class="c-table__cell u-text u-text-small">noun, singular or mass</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>NNP</code></td><td class="c-table__cell u-text"><code>PROPN</code></td><td class="c-table__cell u-text"> <code>NounType=prop</code> <code>Number=sign</code></td><td class="c-table__cell u-text u-text-small">noun, proper singular</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>NNPS</code></td><td class="c-table__cell u-text"><code>PROPN</code></td><td class="c-table__cell u-text"> <code>NounType=prop</code> <code>Number=plur</code></td><td class="c-table__cell u-text u-text-small">noun, proper plural</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>NNS</code></td><td class="c-table__cell u-text"><code>NOUN</code></td><td class="c-table__cell u-text"> <code>Number=plur</code></td><td class="c-table__cell u-text u-text-small">noun, plural</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PDT</code></td><td class="c-table__cell u-text"><code>ADJ</code></td><td class="c-table__cell u-text"> <code>AdjType=pdt</code> <code>PronType=prn</code></td><td class="c-table__cell u-text u-text-small">predeterminer</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>POS</code></td><td class="c-table__cell u-text"><code>PART</code></td><td class="c-table__cell u-text"> <code>Poss=yes</code></td><td class="c-table__cell u-text u-text-small">possessive ending</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PRP</code></td><td class="c-table__cell u-text"><code>PRON</code></td><td class="c-table__cell u-text"> <code>PronType=prs</code></td><td class="c-table__cell u-text u-text-small">pronoun, personal</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PRP$</code></td><td class="c-table__cell u-text"><code>ADJ</code></td><td class="c-table__cell u-text"> <code>PronType=prs</code> <code>Poss=yes</code></td><td class="c-table__cell u-text u-text-small">pronoun, possessive</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>RB</code></td><td class="c-table__cell u-text"><code>ADV</code></td><td class="c-table__cell u-text"> <code>Degree=pos</code></td><td class="c-table__cell u-text u-text-small">adverb</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>RBR</code></td><td class="c-table__cell u-text"><code>ADV</code></td><td class="c-table__cell u-text"> <code>Degree=comp</code></td><td class="c-table__cell u-text u-text-small">adverb, comparative</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>RBS</code></td><td class="c-table__cell u-text"><code>ADV</code></td><td class="c-table__cell u-text"> <code>Degree=sup</code></td><td class="c-table__cell u-text u-text-small">adverb, superlative</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>RP</code></td><td class="c-table__cell u-text"><code>PART</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">adverb, particle</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>_SP</code></td><td class="c-table__cell u-text"><code>SPACE</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">space</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>SYM</code></td><td class="c-table__cell u-text"><code>SYM</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">symbol</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>TO</code></td><td class="c-table__cell u-text"><code>PART</code></td><td class="c-table__cell u-text"> <code>PartType=inf</code> <code>VerbForm=inf</code></td><td class="c-table__cell u-text u-text-small">infinitival to</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>UH</code></td><td class="c-table__cell u-text"><code>INTJ</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">interjection</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VB</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>VerbForm=inf</code></td><td class="c-table__cell u-text u-text-small">verb, base form</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VBD</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>VerbForm=fin</code> <code>Tense=past</code></td><td class="c-table__cell u-text u-text-small">verb, past tense</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VBG</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>VerbForm=part</code> <code>Tense=pres</code> <code>Aspect=prog</code></td><td class="c-table__cell u-text u-text-small">verb, gerund or present participle</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VBN</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>VerbForm=part</code> <code>Tense=past</code> <code>Aspect=perf</code></td><td class="c-table__cell u-text u-text-small">verb, past participle</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VBP</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>VerbForm=fin</code> <code>Tense=pres</code></td><td class="c-table__cell u-text u-text-small">verb, non-3rd person singular present</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VBZ</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>VerbForm=fin</code> <code>Tense=pres</code> <code>Number=sing</code> <code>Person=3</code></td><td class="c-table__cell u-text u-text-small">verb, 3rd person singular present</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>WDT</code></td><td class="c-table__cell u-text"><code>ADJ</code></td><td class="c-table__cell u-text"> <code>PronType=int</code> <code>rel</code></td><td class="c-table__cell u-text u-text-small">wh-determiner</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>WP</code></td><td class="c-table__cell u-text"><code>NOUN</code></td><td class="c-table__cell u-text"> <code>PronType=int</code> <code>rel</code></td><td class="c-table__cell u-text u-text-small">wh-pronoun, personal</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>WP$</code></td><td class="c-table__cell u-text"><code>ADJ</code></td><td class="c-table__cell u-text"> <code>Poss=yes PronType=int</code> <code>rel</code></td><td class="c-table__cell u-text u-text-small">wh-pronoun, possessive</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>WRB</code></td><td class="c-table__cell u-text"><code>ADV</code></td><td class="c-table__cell u-text"> <code>PronType=int</code> <code>rel</code></td><td class="c-table__cell u-text u-text-small">wh-adverb</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>XX</code></td><td class="c-table__cell u-text"><code>X</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">unknown</td></tr></tbody></table>
%%HTML
<h1>German</h1>
<p> The German part-of-speech tagger uses the <a href="http://www.ims.uni-stuttgart.de/forschung/ressourcen/korpora/TIGERCorpus/annotation/index.html" target="_blank" rel="noopener nofollow">TIGER Treebank</a> annotation scheme. We also map the tags to the simpler Google
Universal POS tag set.</p>
<table class="c-table o-block"><tbody><tr class="c-table__row c-table__row--head"><th class="c-table__head-cell u-text-label">Tag</th><th class="c-table__head-cell u-text-label">POS</th><th class="c-table__head-cell u-text-label">Morphology</th><th class="c-table__head-cell u-text-label">Description</th></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>$(</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>PunctType=brck</code></td><td class="c-table__cell u-text u-text-small">other sentence-internal punctuation mark</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>$,</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>PunctType=comm</code></td><td class="c-table__cell u-text u-text-small">comma</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>$.</code></td><td class="c-table__cell u-text"><code>PUNCT</code></td><td class="c-table__cell u-text"> <code>PunctType=peri</code></td><td class="c-table__cell u-text u-text-small">sentence-final punctuation mark</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>ADJA</code></td><td class="c-table__cell u-text"><code>ADJ</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">adjective, attributive</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>ADJD</code></td><td class="c-table__cell u-text"><code>ADJ</code></td><td class="c-table__cell u-text"> <code>Variant=short</code></td><td class="c-table__cell u-text u-text-small">adjective, adverbial or predicative</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>ADV</code></td><td class="c-table__cell u-text"><code>ADV</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">adverb</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>APPO</code></td><td class="c-table__cell u-text"><code>ADP</code></td><td class="c-table__cell u-text"> <code>AdpType=post</code></td><td class="c-table__cell u-text u-text-small">postposition</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>APPR</code></td><td class="c-table__cell u-text"><code>ADP</code></td><td class="c-table__cell u-text"> <code>AdpType=prep</code></td><td class="c-table__cell u-text u-text-small">preposition; circumposition left</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>APPRART</code></td><td class="c-table__cell u-text"><code>ADP</code></td><td class="c-table__cell u-text"> <code>AdpType=prep</code> <code>PronType=art</code></td><td class="c-table__cell u-text u-text-small">preposition with article</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>APZR</code></td><td class="c-table__cell u-text"><code>ADP</code></td><td class="c-table__cell u-text"> <code>AdpType=circ</code></td><td class="c-table__cell u-text u-text-small">circumposition right</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>ART</code></td><td class="c-table__cell u-text"><code>DET</code></td><td class="c-table__cell u-text"> <code>PronType=art</code></td><td class="c-table__cell u-text u-text-small">definite or indefinite article</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>CARD</code></td><td class="c-table__cell u-text"><code>NUM</code></td><td class="c-table__cell u-text"> <code>NumType=card</code></td><td class="c-table__cell u-text u-text-small">cardinal number</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>FM</code></td><td class="c-table__cell u-text"><code>X</code></td><td class="c-table__cell u-text"> <code>Foreign=yes</code></td><td class="c-table__cell u-text u-text-small">foreign language material</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>ITJ</code></td><td class="c-table__cell u-text"><code>INTJ</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">interjection</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>KOKOM</code></td><td class="c-table__cell u-text"><code>CONJ</code></td><td class="c-table__cell u-text"> <code>ConjType=comp</code></td><td class="c-table__cell u-text u-text-small">comparative conjunction</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>KON</code></td><td class="c-table__cell u-text"><code>CONJ</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">coordinate conjunction</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>KOUI</code></td><td class="c-table__cell u-text"><code>SCONJ</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">subordinate conjunction with "zu" and infinitive</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>KOUS</code></td><td class="c-table__cell u-text"><code>SCONJ</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">subordinate conjunction with sentence</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>NE</code></td><td class="c-table__cell u-text"><code>PROPN</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">proper noun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>NNE</code></td><td class="c-table__cell u-text"><code>PROPN</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">proper noun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>NN</code></td><td class="c-table__cell u-text"><code>NOUN</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">noun, singular or mass</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PAV</code></td><td class="c-table__cell u-text"><code>ADV</code></td><td class="c-table__cell u-text"> <code>PronType=dem</code></td><td class="c-table__cell u-text u-text-small">pronominal adverb</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PROAV</code></td><td class="c-table__cell u-text"><code>ADV</code></td><td class="c-table__cell u-text"> <code>PronType=dem</code></td><td class="c-table__cell u-text u-text-small">pronominal adverb</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PDAT</code></td><td class="c-table__cell u-text"><code>DET</code></td><td class="c-table__cell u-text"> <code>PronType=dem</code></td><td class="c-table__cell u-text u-text-small">attributive demonstrative pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PDS</code></td><td class="c-table__cell u-text"><code>PRON</code></td><td class="c-table__cell u-text"> <code>PronType=dem</code></td><td class="c-table__cell u-text u-text-small">substituting demonstrative pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PIAT</code></td><td class="c-table__cell u-text"><code>DET</code></td><td class="c-table__cell u-text"> <code>PronType=ind</code> <code>neg</code> <code>tot</code></td><td class="c-table__cell u-text u-text-small">attributive indefinite pronoun without determiner</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PIDAT</code></td><td class="c-table__cell u-text"><code>DET</code></td><td class="c-table__cell u-text"> <code>AdjType=pdt PronType=ind</code> <code>neg</code> <code>tot</code></td><td class="c-table__cell u-text u-text-small">attributive indefinite pronoun with determiner</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PIS</code></td><td class="c-table__cell u-text"><code>PRON</code></td><td class="c-table__cell u-text"> <code>PronType=ind</code> <code>neg</code> <code>tot</code></td><td class="c-table__cell u-text u-text-small">substituting indefinite pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PPER</code></td><td class="c-table__cell u-text"><code>PRON</code></td><td class="c-table__cell u-text"> <code>PronType=prs</code></td><td class="c-table__cell u-text u-text-small">non-reflexive personal pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PPOSAT</code></td><td class="c-table__cell u-text"><code>DET</code></td><td class="c-table__cell u-text"> <code>Poss=yes</code> <code>PronType=prs</code></td><td class="c-table__cell u-text u-text-small">attributive possessive pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PPOSS</code></td><td class="c-table__cell u-text"><code>PRON</code></td><td class="c-table__cell u-text"> <code>PronType=rel</code></td><td class="c-table__cell u-text u-text-small">substituting possessive pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PRELAT</code></td><td class="c-table__cell u-text"><code>DET</code></td><td class="c-table__cell u-text"> <code>PronType=rel</code></td><td class="c-table__cell u-text u-text-small">attributive relative pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PRELS</code></td><td class="c-table__cell u-text"><code>PRON</code></td><td class="c-table__cell u-text"> <code>PronType=rel</code></td><td class="c-table__cell u-text u-text-small">substituting relative pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PRF</code></td><td class="c-table__cell u-text"><code>PRON</code></td><td class="c-table__cell u-text"> <code>PronType=prs</code> <code>Reflex=yes</code></td><td class="c-table__cell u-text u-text-small">reflexive personal pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PTKA</code></td><td class="c-table__cell u-text"><code>PART</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">particle with adjective or adverb</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PTKANT</code></td><td class="c-table__cell u-text"><code>PART</code></td><td class="c-table__cell u-text"> <code>PartType=res</code></td><td class="c-table__cell u-text u-text-small">answer particle</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PTKNEG</code></td><td class="c-table__cell u-text"><code>PART</code></td><td class="c-table__cell u-text"> <code>Negative=yes</code></td><td class="c-table__cell u-text u-text-small">negative particle</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PTKVZ</code></td><td class="c-table__cell u-text"><code>PART</code></td><td class="c-table__cell u-text"> <code>PartType=vbp</code></td><td class="c-table__cell u-text u-text-small">separable verbal particle</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PTKZU</code></td><td class="c-table__cell u-text"><code>PART</code></td><td class="c-table__cell u-text"> <code>PartType=inf</code></td><td class="c-table__cell u-text u-text-small">"zu" before infinitive</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PWAT</code></td><td class="c-table__cell u-text"><code>DET</code></td><td class="c-table__cell u-text"> <code>PronType=int</code></td><td class="c-table__cell u-text u-text-small">attributive interrogative pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PWAV</code></td><td class="c-table__cell u-text"><code>ADV</code></td><td class="c-table__cell u-text"> <code>PronType=int</code></td><td class="c-table__cell u-text u-text-small">adverbial interrogative or relative pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>PWS</code></td><td class="c-table__cell u-text"><code>PRON</code></td><td class="c-table__cell u-text"> <code>PronType=int</code></td><td class="c-table__cell u-text u-text-small">substituting interrogative pronoun</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>TRUNC</code></td><td class="c-table__cell u-text"><code>X</code></td><td class="c-table__cell u-text"> <code>Hyph=yes</code></td><td class="c-table__cell u-text u-text-small">word remnant</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VAFIN</code></td><td class="c-table__cell u-text"><code>AUX</code></td><td class="c-table__cell u-text"> <code>Mood=ind</code> <code>VerbForm=fin</code></td><td class="c-table__cell u-text u-text-small">finite verb, auxiliary</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VAIMP</code></td><td class="c-table__cell u-text"><code>AUX</code></td><td class="c-table__cell u-text"> <code>Mood=imp</code> <code>VerbForm=fin</code></td><td class="c-table__cell u-text u-text-small">imperative, auxiliary</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VAINF</code></td><td class="c-table__cell u-text"><code>AUX</code></td><td class="c-table__cell u-text"> <code>VerbForm=inf</code></td><td class="c-table__cell u-text u-text-small">infinitive, auxiliary</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VAPP</code></td><td class="c-table__cell u-text"><code>AUX</code></td><td class="c-table__cell u-text"> <code>Aspect=perf</code> <code>VerbForm=fin</code></td><td class="c-table__cell u-text u-text-small">perfect participle, auxiliary</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VMFIN</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>Mood=ind</code> <code>VerbForm=fin</code> <code>VerbType=mod</code></td><td class="c-table__cell u-text u-text-small">finite verb, modal</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VMINF</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>VerbForm=fin</code> <code>VerbType=mod</code></td><td class="c-table__cell u-text u-text-small">infinitive, modal</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VMPP</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>Aspect=perf</code> <code>VerbForm=part</code> <code>VerbType=mod</code></td><td class="c-table__cell u-text u-text-small">perfect participle, modal</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VVFIN</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>Mood=ind</code> <code>VerbForm=fin</code></td><td class="c-table__cell u-text u-text-small">finite verb, full</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VVIMP</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>Mood=imp</code> <code>VerbForm=fin</code></td><td class="c-table__cell u-text u-text-small">imperative, full</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VVINF</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>VerbForm=inf</code></td><td class="c-table__cell u-text u-text-small">infinitive, full</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VVIZU</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>VerbForm=inf</code></td><td class="c-table__cell u-text u-text-small">infinitive with "zu", full</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>VVPP</code></td><td class="c-table__cell u-text"><code>VERB</code></td><td class="c-table__cell u-text"> <code>Aspect=perf</code> <code>VerbForm=part</code></td><td class="c-table__cell u-text u-text-small">perfect participle, full</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>XY</code></td><td class="c-table__cell u-text"><code>X</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">non-word containing non-letter</td></tr><tr class="c-table__row"><td class="c-table__cell u-text"><code>SP</code></td><td class="c-table__cell u-text"><code>SPACE</code></td><td class="c-table__cell u-text"></td><td class="c-table__cell u-text u-text-small">space</td></tr></tbody></table>
```
| github_jupyter |
# 电影评论文本分类
此笔记本(notebook)使用评论文本将影评分为*积极(positive)*或*消极(nagetive)*两类。这是一个*二元(binary)*或者二分类问题,一种重要且应用广泛的机器学习问题。
我们将使用来源于[网络电影数据库(Internet Movie Database)](https://www.imdb.com/)的 [IMDB 数据集(IMDB dataset)](https://tensorflow.google.cn/api_docs/python/tf/keras/datasets/imdb),其包含 50,000 条影评文本。从该数据集切割出的25,000条评论用作训练,另外 25,000 条用作测试。训练集与测试集是*平衡的(balanced)*,意味着它们包含相等数量的积极和消极评论。
此笔记本(notebook)使用了 [tf.keras](https://tensorflow.google.cn/guide/keras),它是一个 Tensorflow 中用于构建和训练模型的高级API。有关使用 `tf.keras` 进行文本分类的更高级教程,请参阅 [MLCC文本分类指南(MLCC Text Classification Guide)](https://developers.google.com/machine-learning/guides/text-classification/)。
```
from __future__ import absolute_import, division, print_function, unicode_literals
try:
# Colab only
%tensorflow_version 2.x
except Exception:
pass
import tensorflow as tf
from tensorflow import keras
import numpy as np
print(tf.__version__)
```
## 下载 IMDB 数据集
IMDB 数据集已经打包在 Tensorflow 中。该数据集已经经过预处理,评论(单词序列)已经被转换为整数序列,其中每个整数表示字典中的特定单词。
以下代码将下载 IMDB 数据集到您的机器上(如果您已经下载过将从缓存中复制):
```
imdb = keras.datasets.imdb
(train_data, train_labels), (test_data, test_labels) = imdb.load_data(num_words=10000)
```
参数 `num_words=10000` 保留了训练数据中最常出现的 10,000 个单词。为了保持数据规模的可管理性,低频词将被丢弃。
## 探索数据
让我们花一点时间来了解数据格式。该数据集是经过预处理的:每个样本都是一个表示影评中词汇的整数数组。每个标签都是一个值为 0 或 1 的整数值,其中 0 代表消极评论,1 代表积极评论。
```
print("Training entries: {}, labels: {}".format(len(train_data), len(train_labels)))
```
评论文本被转换为整数值,其中每个整数代表词典中的一个单词。首条评论是这样的:
```
print(train_data[0])
```
电影评论可能具有不同的长度。以下代码显示了第一条和第二条评论的中单词数量。由于神经网络的输入必须是统一的长度,我们稍后需要解决这个问题。
```
len(train_data[0]), len(train_data[1])
```
### 将整数转换回单词
了解如何将整数转换回文本对您可能是有帮助的。这里我们将创建一个辅助函数来查询一个包含了整数到字符串映射的字典对象:
```
# 一个映射单词到整数索引的词典
word_index = imdb.get_word_index()
# 保留第一个索引
word_index = {k:(v+3) for k,v in word_index.items()}
word_index["<PAD>"] = 0
word_index["<START>"] = 1
word_index["<UNK>"] = 2 # unknown
word_index["<UNUSED>"] = 3
reverse_word_index = dict([(value, key) for (key, value) in word_index.items()])
def decode_review(text):
return ' '.join([reverse_word_index.get(i, '?') for i in text])
```
现在我们可以使用 `decode_review` 函数来显示首条评论的文本:
```
decode_review(train_data[0])
```
## 准备数据
影评——即整数数组必须在输入神经网络之前转换为张量。这种转换可以通过以下两种方式来完成:
* 将数组转换为表示单词出现与否的由 0 和 1 组成的向量,类似于 one-hot 编码。例如,序列[3, 5]将转换为一个 10,000 维的向量,该向量除了索引为 3 和 5 的位置是 1 以外,其他都为 0。然后,将其作为网络的首层——一个可以处理浮点型向量数据的稠密层。不过,这种方法需要大量的内存,需要一个大小为 `num_words * num_reviews` 的矩阵。
* 或者,我们可以填充数组来保证输入数据具有相同的长度,然后创建一个大小为 `max_length * num_reviews` 的整型张量。我们可以使用能够处理此形状数据的嵌入层作为网络中的第一层。
在本教程中,我们将使用第二种方法。
由于电影评论长度必须相同,我们将使用 [pad_sequences](https://tensorflow.google.cn/api_docs/python/tf/keras/preprocessing/sequence/pad_sequences) 函数来使长度标准化:
```
train_data = keras.preprocessing.sequence.pad_sequences(train_data,
value=word_index["<PAD>"],
padding='post',
maxlen=256)
test_data = keras.preprocessing.sequence.pad_sequences(test_data,
value=word_index["<PAD>"],
padding='post',
maxlen=256)
```
现在让我们看下样本的长度:
```
len(train_data[0]), len(train_data[1])
```
并检查一下首条评论(当前已经填充):
```
print(train_data[0])
```
## 构建模型
神经网络由堆叠的层来构建,这需要从两个主要方面来进行体系结构决策:
* 模型里有多少层?
* 每个层里有多少*隐层单元(hidden units)*?
在此样本中,输入数据包含一个单词索引的数组。要预测的标签为 0 或 1。让我们来为该问题构建一个模型:
```
# 输入形状是用于电影评论的词汇数目(10,000 词)
vocab_size = 10000
model = keras.Sequential()
model.add(keras.layers.Embedding(vocab_size, 16))
model.add(keras.layers.GlobalAveragePooling1D())
model.add(keras.layers.Dense(16, activation='relu'))
model.add(keras.layers.Dense(1, activation='sigmoid'))
model.summary()
```
层按顺序堆叠以构建分类器:
1. 第一层是`嵌入(Embedding)`层。该层采用整数编码的词汇表,并查找每个词索引的嵌入向量(embedding vector)。这些向量是通过模型训练学习到的。向量向输出数组增加了一个维度。得到的维度为:`(batch, sequence, embedding)`。
2. 接下来,`GlobalAveragePooling1D` 将通过对序列维度求平均值来为每个样本返回一个定长输出向量。这允许模型以尽可能最简单的方式处理变长输入。
3. 该定长输出向量通过一个有 16 个隐层单元的全连接(`Dense`)层传输。
4. 最后一层与单个输出结点密集连接。使用 `Sigmoid` 激活函数,其函数值为介于 0 与 1 之间的浮点数,表示概率或置信度。
### 隐层单元
上述模型在输入输出之间有两个中间层或“隐藏层”。输出(单元,结点或神经元)的数量即为层表示空间的维度。换句话说,是学习内部表示时网络所允许的自由度。
如果模型具有更多的隐层单元(更高维度的表示空间)和/或更多层,则可以学习到更复杂的表示。但是,这会使网络的计算成本更高,并且可能导致学习到不需要的模式——一些能够在训练数据上而不是测试数据上改善性能的模式。这被称为*过拟合(overfitting)*,我们稍后会对此进行探究。
### 损失函数与优化器
一个模型需要损失函数和优化器来进行训练。由于这是一个二分类问题且模型输出概率值(一个使用 sigmoid 激活函数的单一单元层),我们将使用 `binary_crossentropy` 损失函数。
这不是损失函数的唯一选择,例如,您可以选择 `mean_squared_error` 。但是,一般来说 `binary_crossentropy` 更适合处理概率——它能够度量概率分布之间的“距离”,或者在我们的示例中,指的是度量 ground-truth 分布与预测值之间的“距离”。
稍后,当我们研究回归问题(例如,预测房价)时,我们将介绍如何使用另一种叫做均方误差的损失函数。
现在,配置模型来使用优化器和损失函数:
```
model.compile(optimizer='adam',
loss='binary_crossentropy',
metrics=['accuracy'])
```
## 创建一个验证集
在训练时,我们想要检查模型在未见过的数据上的准确率(accuracy)。通过从原始训练数据中分离 10,000 个样本来创建一个*验证集*。(为什么现在不使用测试集?我们的目标是只使用训练数据来开发和调整模型,然后只使用一次测试数据来评估准确率(accuracy))。
```
x_val = train_data[:10000]
partial_x_train = train_data[10000:]
y_val = train_labels[:10000]
partial_y_train = train_labels[10000:]
```
## 训练模型
以 512 个样本的 mini-batch 大小迭代 40 个 epoch 来训练模型。这是指对 `x_train` 和 `y_train` 张量中所有样本的的 40 次迭代。在训练过程中,监测来自验证集的 10,000 个样本上的损失值(loss)和准确率(accuracy):
```
history = model.fit(partial_x_train,
partial_y_train,
epochs=40,
batch_size=512,
validation_data=(x_val, y_val),
verbose=1)
```
## 评估模型
我们来看一下模型的性能如何。将返回两个值。损失值(loss)(一个表示误差的数字,值越低越好)与准确率(accuracy)。
```
results = model.evaluate(test_data, test_labels, verbose=2)
print(results)
```
这种十分朴素的方法得到了约 87% 的准确率(accuracy)。若采用更好的方法,模型的准确率应当接近 95%。
## 创建一个准确率(accuracy)和损失值(loss)随时间变化的图表
`model.fit()` 返回一个 `History` 对象,该对象包含一个字典,其中包含训练阶段所发生的一切事件:
```
history_dict = history.history
history_dict.keys()
```
有四个条目:在训练和验证期间,每个条目对应一个监控指标。我们可以使用这些条目来绘制训练与验证过程的损失值(loss)和准确率(accuracy),以便进行比较。
```
import matplotlib.pyplot as plt
acc = history_dict['accuracy']
val_acc = history_dict['val_accuracy']
loss = history_dict['loss']
val_loss = history_dict['val_loss']
epochs = range(1, len(acc) + 1)
# “bo”代表 "蓝点"
plt.plot(epochs, loss, 'bo', label='Training loss')
# b代表“蓝色实线”
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
plt.clf() # 清除数字
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend()
plt.show()
```
在该图中,点代表训练损失值(loss)与准确率(accuracy),实线代表验证损失值(loss)与准确率(accuracy)。
注意训练损失值随每一个 epoch *下降*而训练准确率(accuracy)随每一个 epoch *上升*。这在使用梯度下降优化时是可预期的——理应在每次迭代中最小化期望值。
验证过程的损失值(loss)与准确率(accuracy)的情况却并非如此——它们似乎在 20 个 epoch 后达到峰值。这是过拟合的一个实例:模型在训练数据上的表现比在以前从未见过的数据上的表现要更好。在此之后,模型过度优化并学习*特定*于训练数据的表示,而不能够*泛化*到测试数据。
对于这种特殊情况,我们可以通过在 20 个左右的 epoch 后停止训练来避免过拟合。稍后,您将看到如何通过回调自动执行此操作。
| github_jupyter |
# Just Plot It!
## Introduction
### The System
In this course we will work with a set of "experimental" data to illustrate going from "raw" measurement (or simulation) data through exploratory visualization to an (almost) paper ready figure.
In this scenario, we have fabricated (or simulated) 25 cantilevers. There is some value (suggestively called "control") that varies between the cantilevers and we want to see how the properties of the cantilever are affect by "control".
To see what this will look like physically, take part a "clicky" pen. Hold one end of the spring in your fingers and flick the free end.
Or just watch this cat:
```
from IPython.display import YouTubeVideo
YouTubeVideo('4aTagDSnclk?start=19')
```
Springs, and our cantilevers, are part of a class of systems known as (Damped) Harmonic Oscillators. We are going to measure the natural frequency and damping rate we deflect each cantilever by the same amount and then observe the position as a function of time as the vibrations damp out.
### The Tools
We are going make use of:
- [jupyter](https://jupyter.org)
- [numpy](https://numpy.org)
- [matplotlib](https://matplotlib.org)
- [scipy](https://www.scipy.org/scipylib/index.html)
- [xarray](http://xarray.pydata.org/en/stable/index.html)
- [pandas](https://pandas.pydata.org/docs/)
We are only going to scratch the surface of what any of these libraries can do! For the purposes of this course we assume you know numpy and Matplotlib at least to the level of LINKS TO OTHER COURSES. We will only be using one aspect (least square fitting) from scipy so no prior familiarity is needed. Similarly, we will only be superficially making use of pandas and xarray to provided access to structured data. No prior familiarity is required and if you want to learn more see LINK TO OTHER COURSES.
```
# interactive figures, requires ipypml!
%matplotlib widget
#%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import scipy
import xarray as xa
```
### Philsophy
While this coures uses Matplotlib for the visualization, the high-level lessons of this course are transferable to any plotting tools (in any language).
At its core, programing in the process of taking existing tools (libraries) and building new tools more fit to your purpose. This course will walk through a concrete example, starting with a pile of data and ending with a paper figure, of how to think about and design scientific visualizations tools tuned to exactly *your* data and questions.
## The Data
### Accessing data
As a rule-of-thumb I/O logic should be kept out of the inner loops of analysis or plotting. This will, in the medium term, lead to more re-usable and maintainable code. Remember your most frequent collaborator is yourself in 6 months. Be kind to your (future) self and write re-usable, maintainable, and understandable code now ;)
In this case, we have a data (simulation) function `get_data` that will simulate the experiment and returns to us a [`xarray.DataArray`](http://xarray.pydata.org/en/stable/quick-overview.html#create-a-dataarray). `xarray.DataArray` is (roughly) a N-dimensional numpy array that is enriched by the concept of coordinates and indies on the the axes and meta-data.
`xarray` has much more functionality than we will use in this course!
```
# not sure how else to get the helpers on the path!
import sys
sys.path.append('../scripts')
from data_gen import get_data, fit
```
### First look
Using the function `get_data` we can pull an `xarray.DataArray` into our namespace and the use the html repr from xarray to get a first look at the data
```
d = get_data(25)
d
```
From this we can see that we have a, more-or-less, 2D array with 25 rows, each of which is a measurement that is a 4,112 point time series. Because this is an DataArray it also caries **coordinates** giving the value of **control** for each row and the time for each column.
If we pull out just one row we can see a single experimental measurement.
```
d[6]
```
We can see that the **control** coordinate now gives 1 value, but the **time** coordinate is still a vector. We can access these values via attribute access (which we will use later):
```
d[6].control
d[6].time
```
## The Plotting
### Plot it?
Looking at (truncated) lists of numbers is not intuitive or informative for most people, to get a better sense of what this data looks like lets plot it! We know that `Axes.plot` can plot multiple lines at once so lets try naively throwing `d` at `ax.plot`!
```
fig, ax = plt.subplots()
ax.plot(d);
```
While this does look sort of cool, it is not *useful*. What has happened is that Matplotlib has looked at our `(25, 4_112)` array and said "Clearly, you have a table that is 4k columns wide and 25 rows long. What you want is each column plotted!". Thus, what we are seeing is "The deflection at a fixed time as a function of cantilever ID number". This plot does accurately reflect that data that we passed in, but this is a nearly meaningless plot!
Visualization, just like writing, is a tool for communication and you need to think about the story you want to tell as you make the plots.
### Sidebar: Explicit vs Implicit Matplotlib API
There are two related but distinct APIs to use Matplotlib: the "Explicit" (nee "Object Oriented") and "Implicit" (nee "pyplot/pylab"). The Implicit API is implemented using the Explicit API; anything you can do with the Implicit API you can do with the Explicit API, but there is some functionality of the Explicit API that is not exposed through the Implicit API. It is also possible, but with one exception not suggested, to mix the two APIs.
The core conceptual difference is than in the Implicit API Matplotlib has a notion of the "current figure" and "current axes" that all of the calls re-directed to. For example, the implementation of `plt.plot` (once you scroll past the docstring) is only 1 line:
```
?? plt.plot
```
While the Implicit API reduces the boilerplate required to get some things done and is convenient when working in a terminal, it comes at the cost of Matplotlib maintaining global state of which Axes is currently active! When scripting this can quickly become a headache to manage.
When using Matplotlib with one of the GUI backends, we do need to, at the library level, keep track of some global state so that the plot windows remain responsive. If you are embedding Matplotlib in your own GUI application you are responsible for this, but when working at an IPython prompt,`pyplot` takes care of this for you.
This course is going to, with the exception of creating new figures, always use the Explict API.
### Plot it!
What we really want to see is the transpose of the above (A line per experiment as a function of time):
```
fig, ax = plt.subplots()
ax.plot(d.T);
```
Which is better! If we squint a bit (or zoom in if we are using `ipympl` or a GUI backend) can sort of see each of the individual oscillators ringing-down over time.
### Just one at a time
To make it easier to see lets plot just one of the curves:
```
fig, ax = plt.subplots()
ax.plot(d[6]);
```
### Pass freshman physics
While we do have just one line on the axes and can see what is going on, this plot would, right, be marked as little-to-no credit if turned in as part of a freshman Physics lab! We do not have a meaningful value on the x-axis, no legend, and no axis labels!
```
fig, ax = plt.subplots()
m = d[6]
ax.plot(m.time, m, label=f'control = {float(m.control):.1f}')
ax.set_xlabel('time (ms)')
ax.set_ylabel('displacement (mm)')
ax.legend();
```
At this point we have a minimally acceptable plot! It shows us one curve with axis labels (with units!) and a legend. With
### sidebar: xarray plotting
Because xarray knows more about the structure of your data than a couple of numpy arrays in your local namespace or dictionary, it can make smarter choices about the automatic visualization:
```
fig, ax = plt.subplots()
m.plot(ax=ax)
```
While this is helpful exploritory plotting, `xarray` makes some choices that make it difficult to compose plotting multiple data sets.
| github_jupyter |
## 8. Classification
[Data Science Playlist on YouTube](https://www.youtube.com/watch?v=VLKEj9EN2ew&list=PLLBUgWXdTBDg1Qgmwt4jKtVn9BWh5-zgy)
[](https://www.youtube.com/watch?v=VLKEj9EN2ew&list=PLLBUgWXdTBDg1Qgmwt4jKtVn9BWh5-zgy "Python Data Science")
**Classification** predicts *discrete labels (outcomes)* such as `yes`/`no`, `True`/`False`, or any number of discrete levels such as a letter from text recognition, or a word from speech recognition. There are two main methods for training classifiers: unsupervised and supervised learning. The difference between the two is that unsupervised learning does not use labels while supervised learning uses labels to build the classifier. The goal of unsupervised learning is to cluster input features but without labels to guide the grouping.

### Supervised Learning to Classify Numbers
A dataset that is included with sklearn is a set of 1797 images of numbers that are 64 pixels (8x8) each. There are labels with each to indicate the correct answer. A Support Vector Classifier is trained on the first half of the images.
```
from sklearn import datasets, svm
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
# train classifier
digits = datasets.load_digits()
n_samples = len(digits.images)
data = digits.images.reshape((n_samples, -1))
svc = svm.SVC(gamma=0.001)
X_train, X_test, y_train, y_test = train_test_split(
data, digits.target, test_size=0.5, shuffle=False)
svc.fit(X_train, y_train)
print('SVC Trained')
```

### Test Number Classifier
The image classification is trained on 10 randomly selected images from the other half of the data set to evaluate the training. Run the classifier test until you observe a misclassified number.
```
plt.figure(figsize=(10,4))
for i in range(10):
n = np.random.randint(int(n_samples/2),n_samples)
predict = svc.predict(digits.data[n:n+1])[0]
plt.subplot(2,5,i+1)
plt.imshow(digits.images[n], cmap=plt.cm.gray_r, interpolation='nearest')
plt.text(0,7,'Actual: ' + str(digits.target[n]),color='r')
plt.text(0,1,'Predict: ' + str(predict),color='b')
if predict==digits.target[n]:
plt.text(0,4,'Correct',color='g')
else:
plt.text(0,4,'Incorrect',color='orange')
plt.show()
```

### Classification with Supervised Learning
Select data set option with `moons`, `cirlces`, or `blobs`. Run the following cell to generate the data that will be used to test the classifiers.
```
option = 'moons' # moons, circles, or blobs
n = 2000 # number of data points
X = np.random.random((n,2))
mixing = 0.0 # add random mixing element to data
xplot = np.linspace(0,1,100)
if option=='moons':
X, y = datasets.make_moons(n_samples=n,noise=0.1)
yplot = xplot*0.0
elif option=='circles':
X, y = datasets.make_circles(n_samples=n,noise=0.1,factor=0.5)
yplot = xplot*0.0
elif option=='blobs':
X, y = datasets.make_blobs(n_samples=n,centers=[[-5,3],[5,-3]],cluster_std=2.0)
yplot = xplot*0.0
# Split into train and test subsets (50% each)
XA, XB, yA, yB = train_test_split(X, y, test_size=0.5, shuffle=False)
# Plot regression results
def assess(P):
plt.figure()
plt.scatter(XB[P==1,0],XB[P==1,1],marker='^',color='blue',label='True')
plt.scatter(XB[P==0,0],XB[P==0,1],marker='x',color='red',label='False')
plt.scatter(XB[P!=yB,0],XB[P!=yB,1],marker='s',color='orange',\
alpha=0.5,label='Incorrect')
plt.legend()
```

### S.1 Logistic Regression
**Definition:** Logistic regression is a machine learning algorithm for classification. In this algorithm, the probabilities describing the possible outcomes of a single trial are modelled using a logistic function.
**Advantages:** Logistic regression is designed for this purpose (classification), and is most useful for understanding the influence of several independent variables on a single outcome variable.
**Disadvantages:** Works only when the predicted variable is binary, assumes all predictors are independent of each other, and assumes data is free of missing values.
```
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(solver='lbfgs')
lr.fit(XA,yA)
yP = lr.predict(XB)
assess(yP)
```

### S.2 Naïve Bayes
**Definition:** Naive Bayes algorithm based on Bayes’ theorem with the assumption of independence between every pair of features. Naive Bayes classifiers work well in many real-world situations such as document classification and spam filtering.
**Advantages:** This algorithm requires a small amount of training data to estimate the necessary parameters. Naive Bayes classifiers are extremely fast compared to more sophisticated methods.
**Disadvantages:** Naive Bayes is known to be a bad estimator.
```
from sklearn.naive_bayes import GaussianNB
nb = GaussianNB()
nb.fit(XA,yA)
yP = nb.predict(XB)
assess(yP)
```

### S.3 Stochastic Gradient Descent
**Definition:** Stochastic gradient descent is a simple and very efficient approach to fit linear models. It is particularly useful when the number of samples is very large. It supports different loss functions and penalties for classification.
**Advantages:** Efficiency and ease of implementation.
**Disadvantages:** Requires a number of hyper-parameters and it is sensitive to feature scaling.
```
from sklearn.linear_model import SGDClassifier
sgd = SGDClassifier(loss='modified_huber', shuffle=True,random_state=101)
sgd.fit(XA,yA)
yP = sgd.predict(XB)
assess(yP)
```

### S.4 K-Nearest Neighbours
**Definition:** Neighbours based classification is a type of lazy learning as it does not attempt to construct a general internal model, but simply stores instances of the training data. Classification is computed from a simple majority vote of the k nearest neighbours of each point.
**Advantages:** This algorithm is simple to implement, robust to noisy training data, and effective if training data is large.
**Disadvantages:** Need to determine the value of `K` and the computation cost is high as it needs to computer the distance of each instance to all the training samples. One possible solution to determine `K` is to add a feedback loop to determine the number of neighbors.
```
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(XA,yA)
yP = knn.predict(XB)
assess(yP)
```

### S.5 Decision Tree
**Definition:** Given a data of attributes together with its classes, a decision tree produces a sequence of rules that can be used to classify the data.
**Advantages:** Decision Tree is simple to understand and visualise, requires little data preparation, and can handle both numerical and categorical data.
**Disadvantages:** Decision tree can create complex trees that do not generalise well, and decision trees can be unstable because small variations in the data might result in a completely different tree being generated.
```
from sklearn.tree import DecisionTreeClassifier
dtree = DecisionTreeClassifier(max_depth=10,random_state=101,\
max_features=None,min_samples_leaf=5)
dtree.fit(XA,yA)
yP = dtree.predict(XB)
assess(yP)
```

### S.6 Random Forest
**Definition:** Random forest classifier is a meta-estimator that fits a number of decision trees on various sub-samples of datasets and uses average to improve the predictive accuracy of the model and controls over-fitting. The sub-sample size is always the same as the original input sample size but the samples are drawn with replacement.
**Advantages:** Reduction in over-fitting and random forest classifier is more accurate than decision trees in most cases.
**Disadvantages:** Slow real time prediction, difficult to implement, and complex algorithm.
```
from sklearn.ensemble import RandomForestClassifier
rfm = RandomForestClassifier(n_estimators=70,oob_score=True,\
n_jobs=1,random_state=101,max_features=None,\
min_samples_leaf=3) #change min_samples_leaf from 30 to 3
rfm.fit(XA,yA)
yP = rfm.predict(XB)
assess(yP)
```

### S.7 Support Vector Classifier
**Definition:** Support vector machine is a representation of the training data as points in space separated into categories by a clear gap that is as wide as possible. New examples are then mapped into that same space and predicted to belong to a category based on which side of the gap they fall.
**Advantages:** Effective in high dimensional spaces and uses a subset of training points in the decision function so it is also memory efficient.
**Disadvantages:** The algorithm does not directly provide probability estimates, these are calculated using an expensive five-fold cross-validation.
```
from sklearn.svm import SVC
svm = SVC(gamma='scale', C=1.0, random_state=101)
svm.fit(XA,yA)
yP = svm.predict(XB)
assess(yP)
```

### S.8 Neural Network
The `MLPClassifier` implements a multi-layer perceptron (MLP) algorithm that trains using Backpropagation.
**Definition:** A neural network is a set of neurons (activation functions) in layers that are processed sequentially to relate an input to an output.
**Advantages:** Effective in nonlinear spaces where the structure of the relationship is not linear. No prior knowledge or specialized equation structure is defined although there are different network architectures that may lead to a better result.
**Disadvantages:** Neural networks do not extrapolate well outside of the training domain. They may also require longer to train by adjusting the parameter weights to minimize a loss (objective) function. It is also more challenging to explain the outcome of the training and changes in initialization or number of epochs (iterations) may lead to different results. Too many epochs may lead to overfitting, especially if there are excess parameters beyond the minimum needed to capture the input to output relationship.

MLP trains on two arrays: array X of size (n_samples, n_features), which holds the training samples represented as floating point feature vectors; and array y of size (n_samples,), which holds the target values (class labels) for the training samples.
MLP can fit a non-linear model to the training data. clf.coefs_ contains the weight matrices that constitute the model parameters. Currently, MLPClassifier supports only the Cross-Entropy loss function, which allows probability estimates by running the predict_proba method. MLP trains using Backpropagation. More precisely, it trains using some form of gradient descent and the gradients are calculated using Backpropagation. For classification, it minimizes the Cross-Entropy loss function, giving a vector of probability estimates. MLPClassifier supports multi-class classification by applying Softmax as the output function. Further, the model supports multi-label classification in which a sample can belong to more than one class. For each class, the raw output passes through the logistic function. Values larger or equal to 0.5 are rounded to 1, otherwise to 0. For a predicted output of a sample, the indices where the value is 1 represents the assigned classes of that sample.
```
from sklearn.neural_network import MLPClassifier
clf = MLPClassifier(solver='lbfgs',alpha=1e-5,max_iter=200,activation='relu',\
hidden_layer_sizes=(10,30,10), random_state=1, shuffle=True)
clf.fit(XA,yA)
yP = clf.predict(XB)
assess(yP)
```
### Unsupervised Classification
Additional examples show the potential for unsupervised learning to classify the groups. Unsupervised learning does not use the labels (`True`/`False`) so the results may need to be switched to align with the test set with `if len(XB[yP!=yB]) > n/4: yP = 1 - yP
`

### U.1 K-Means Clustering
**Definition:** Specify how many possible clusters (or K) there are in the dataset. The algorithm then iteratively moves the K-centers and selects the datapoints that are closest to that centroid in the cluster.
**Advantages:** The most common and simplest clustering algorithm.
**Disadvantages:** Must specify the number of clusters although this can typically be determined by increasing the number of clusters until the objective function does not change significantly.
```
from sklearn.cluster import KMeans
km = KMeans(n_clusters=2)
km.fit(XA)
yP = km.predict(XB)
if len(XB[yP!=yB]) > n/4: yP = 1 - yP
assess(yP)
```

### U.2 Gaussian Mixture Model
**Definition:** Data points that exist at the boundary of clusters may simply have similar probabilities of being on either clusters. A mixture model predicts a probability instead of a hard classification such as K-Means clustering.
**Advantages:** Incorporates uncertainty into the solution.
**Disadvantages:** Uncertainty may not be desirable for some applications. This method is not as common as the K-Means method for clustering.
```
from sklearn.mixture import GaussianMixture
gmm = GaussianMixture(n_components=2)
gmm.fit(XA)
yP = gmm.predict_proba(XB) # produces probabilities
if len(XB[np.round(yP[:,0])!=yB]) > n/4: yP = 1 - yP
assess(np.round(yP[:,0]))
```

### U.3 Spectral Clustering
**Definition:** Spectral clustering is known as segmentation-based object categorization. It is a technique with roots in graph theory, where identify communities of nodes in a graph are based on the edges connecting them. The method is flexible and allows clustering of non graph data as well.
It uses information from the eigenvalues of special matrices built from the graph or the data set.
**Advantages:** Flexible approach for finding clusters when data doesn’t meet the requirements of other common algorithms.
**Disadvantages:** For large-sized graphs, the second eigenvalue of the (normalized) graph Laplacian matrix is often ill-conditioned, leading to slow convergence of iterative eigenvalue solvers. Spectral clustering is computationally expensive unless the graph is sparse and the similarity matrix can be efficiently constructed.
```
from sklearn.cluster import SpectralClustering
sc = SpectralClustering(n_clusters=2,eigen_solver='arpack',\
affinity='nearest_neighbors')
yP = sc.fit_predict(XB) # No separation between fit and predict calls
# need to fit and predict on same dataset
if len(XB[yP!=yB]) > n/4: yP = 1 - yP
assess(yP)
```

### TCLab Activity
Train a classifier to predict if the heater is on (100%) or off (0%). Generate data with 10 minutes of 1 second data. If you do not have a TCLab, use one of the sample data sets.
- [Sample Data Set 1 (10 min)](http://apmonitor.com/do/uploads/Main/tclab_data5.txt): http://apmonitor.com/do/uploads/Main/tclab_data5.txt
- [Sample Data Set 2 (60 min)](http://apmonitor.com/do/uploads/Main/tclab_data6.txt): http://apmonitor.com/do/uploads/Main/tclab_data6.txt
```
# 10 minute data collection
import tclab, time
import numpy as np
import pandas as pd
with tclab.TCLab() as lab:
n = 600; on=100; t = np.linspace(0,n-1,n)
Q1 = np.zeros(n); T1 = np.zeros(n)
Q2 = np.zeros(n); T2 = np.zeros(n)
Q1[20:41]=on; Q1[60:91]=on; Q1[150:181]=on
Q1[190:206]=on; Q1[220:251]=on; Q1[260:291]=on
Q1[300:316]=on; Q1[340:351]=on; Q1[400:431]=on
Q1[500:521]=on; Q1[540:571]=on; Q1[20:41]=on
Q1[60:91]=on; Q1[150:181]=on; Q1[190:206]=on
Q1[220:251]=on; Q1[260:291]=on
print('Time Q1 Q2 T1 T2')
for i in range(n):
T1[i] = lab.T1; T2[i] = lab.T2
lab.Q1(Q1[i])
if i%5==0:
print(int(t[i]),Q1[i],Q2[i],T1[i],T2[i])
time.sleep(1)
data = np.column_stack((t,Q1,Q2,T1,T2))
data8 = pd.DataFrame(data,columns=['Time','Q1','Q2','T1','T2'])
data8.to_csv('08-tclab.csv',index=False)
```
Use the data file `08-tclab.csv` to train and test the classifier. Select and scale (0-1) the features of the data including `T1`, `T2`, and the 1st and 2nd derivatives of `T1`. Use the measured temperatures, derivatives, and heater value label to create a classifier that predicts when the heater is on or off. Validate the classifier with new data that was not used for training. Starting code is provided below but does not include `T2` as a feature input. **Add `T2` as an input feature to the classifer. Does it improve the classifier performance?**
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import gridspec
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
try:
data = pd.read_csv('08-tclab.csv')
except:
print('Warning: Unable to load 08-tclab.csv, using online data')
url = 'http://apmonitor.com/do/uploads/Main/tclab_data5.txt'
data = pd.read_csv(url)
# Input Features: Temperature and 1st / 2nd Derivatives
# Cubic polynomial fit of temperature using 10 data points
data['dT1'] = np.zeros(len(data))
data['d2T1'] = np.zeros(len(data))
for i in range(len(data)):
if i<len(data)-10:
x = data['Time'][i:i+10]-data['Time'][i]
y = data['T1'][i:i+10]
p = np.polyfit(x,y,3)
# evaluate derivatives at mid-point (5 sec)
t = 5.0
data['dT1'][i] = 3.0*p[0]*t**2 + 2.0*p[1]*t+p[2]
data['d2T1'][i] = 6.0*p[0]*t + 2.0*p[1]
else:
data['dT1'][i] = np.nan
data['d2T1'][i] = np.nan
# Remove last 10 values
X = np.array(data[['T1','dT1','d2T1']][0:-10])
y = np.array(data[['Q1']][0:-10])
# Scale data
# Input features (Temperature and 2nd derivative at 5 sec)
s1 = MinMaxScaler(feature_range=(0,1))
Xs = s1.fit_transform(X)
# Output labels (heater On / Off)
ys = [True if y[i]>50.0 else False for i in range(len(y))]
# Split into train and test subsets (50% each)
XA, XB, yA, yB = train_test_split(Xs, ys, \
test_size=0.5, shuffle=False)
# Supervised Classification
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import SGDClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn.neural_network import MLPClassifier
# Create supervised classification models
lr = LogisticRegression(solver='lbfgs') # Logistic Regression
nb = GaussianNB() # Naïve Bayes
sgd = SGDClassifier(loss='modified_huber', shuffle=True,\
random_state=101) # Stochastic Gradient Descent
knn = KNeighborsClassifier(n_neighbors=5) # K-Nearest Neighbors
dtree = DecisionTreeClassifier(max_depth=10,random_state=101,\
max_features=None,min_samples_leaf=5) # Decision Tree
rfm = RandomForestClassifier(n_estimators=70,oob_score=True,n_jobs=1,\
random_state=101,max_features=None,min_samples_leaf=3) # Random Forest
svm = SVC(gamma='scale', C=1.0, random_state=101) # Support Vector Classifier
clf = MLPClassifier(solver='lbfgs',alpha=1e-5,max_iter=200,\
activation='relu',hidden_layer_sizes=(10,30,10),\
random_state=1, shuffle=True) # Neural Network
models = [lr,nb,sgd,knn,dtree,rfm,svm,clf]
# Supervised learning
yP = [None]*(len(models)+3) # 3 for unsupervised learning
for i,m in enumerate(models):
m.fit(XA,yA)
yP[i] = m.predict(XB)
# Unsupervised learning modules
from sklearn.cluster import KMeans
from sklearn.mixture import GaussianMixture
from sklearn.cluster import SpectralClustering
km = KMeans(n_clusters=2)
gmm = GaussianMixture(n_components=2)
sc = SpectralClustering(n_clusters=2,eigen_solver='arpack',\
affinity='nearest_neighbors')
km.fit(XA)
yP[8] = km.predict(XB)
gmm.fit(XA)
yP[9] = gmm.predict_proba(XB)[:,0]
yP[10] = sc.fit_predict(XB)
plt.figure(figsize=(10,7))
gs = gridspec.GridSpec(3, 1, height_ratios=[1,1,5])
plt.subplot(gs[0])
plt.plot(data['Time']/60,data['T1'],'r-',\
label='Temperature (°C)')
plt.ylabel('T (°C)')
plt.legend()
plt.subplot(gs[1])
plt.plot(data['Time']/60,data['dT1'],'b:',\
label='dT/dt (°C/sec)')
plt.plot(data['Time']/60,data['d2T1'],'k--',\
label=r'$d^2T/dt^2$ ($°C^2/sec^2$)')
plt.ylabel('Derivatives')
plt.legend()
plt.subplot(gs[2])
plt.plot(data['Time']/60,data['Q1']/100,'k-',\
label='Heater (On=1/Off=0)')
t2 = data['Time'][len(yA):-10].values
desc = ['Logistic Regression','Naïve Bayes','Stochastic Gradient Descent',\
'K-Nearest Neighbors','Decision Tree','Random Forest',\
'Support Vector Classifier','Neural Network',\
'K-Means Clustering','Gaussian Mixture Model','Spectral Clustering']
for i in range(11):
plt.plot(t2/60,yP[i]-i-1,label=desc[i])
plt.ylabel('Heater')
plt.legend()
plt.xlabel(r'Time (min)')
plt.legend()
plt.show()
```
| github_jupyter |
## Installing & importing necsessary libs
```
!pip install -q transformers
import numpy as np
import pandas as pd
from sklearn import metrics
import transformers
import torch
from torch.utils.data import Dataset, DataLoader, RandomSampler, SequentialSampler
from transformers import AlbertTokenizer, AlbertModel, AlbertConfig
from tqdm.notebook import tqdm
from transformers import get_linear_schedule_with_warmup
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
n_gpu = torch.cuda.device_count()
torch.cuda.get_device_name(0)
```
## Data Preprocessing
```
df = pd.read_csv("../input/avjantahack/data/train.csv")
df['list'] = df[df.columns[3:]].values.tolist()
new_df = df[['ABSTRACT', 'list']].copy()
new_df.head()
```
## Model configurations
```
# Defining some key variables that will be used later on in the training
MAX_LEN = 512
TRAIN_BATCH_SIZE = 16
VALID_BATCH_SIZE = 8
EPOCHS = 5
LEARNING_RATE = 3e-05
tokenizer = AlbertTokenizer.from_pretrained('albert-base-v2')
```
## Custom Dataset Class
```
class CustomDataset(Dataset):
def __init__(self, dataframe, tokenizer, max_len):
self.tokenizer = tokenizer
self.data = dataframe
self.abstract = dataframe.ABSTRACT
self.targets = self.data.list
self.max_len = max_len
def __len__(self):
return len(self.abstract)
def __getitem__(self, index):
abstract = str(self.abstract[index])
abstract = " ".join(abstract.split())
inputs = self.tokenizer.encode_plus(
abstract,
None,
add_special_tokens = True,
max_length = self.max_len,
pad_to_max_length = True,
return_token_type_ids=True,
truncation = True
)
ids = inputs['input_ids']
mask = inputs['attention_mask']
token_type_ids = inputs['token_type_ids']
return{
'ids': torch.tensor(ids, dtype=torch.long),
'mask': torch.tensor(mask, dtype=torch.long),
'token_type_ids': torch.tensor(token_type_ids, dtype=torch.long),
'targets': torch.tensor(self.targets[index], dtype=torch.float)
}
train_size = 0.8
train_dataset=new_df.sample(frac=train_size,random_state=200)
test_dataset=new_df.drop(train_dataset.index).reset_index(drop=True)
train_dataset = train_dataset.reset_index(drop=True)
print("FULL Dataset: {}".format(new_df.shape))
print("TRAIN Dataset: {}".format(train_dataset.shape))
print("TEST Dataset: {}".format(test_dataset.shape))
training_set = CustomDataset(train_dataset, tokenizer, MAX_LEN)
testing_set = CustomDataset(test_dataset, tokenizer, MAX_LEN)
train_params = {'batch_size': TRAIN_BATCH_SIZE,
'shuffle': True,
'num_workers': 0
}
test_params = {'batch_size': VALID_BATCH_SIZE,
'shuffle': True,
'num_workers': 0
}
training_loader = DataLoader(training_set, **train_params)
testing_loader = DataLoader(testing_set, **test_params)
```
## Albert model
```
class AlbertClass(torch.nn.Module):
def __init__(self):
super(AlbertClass, self).__init__()
self.albert = transformers.AlbertModel.from_pretrained('albert-base-v2')
self.drop = torch.nn.Dropout(0.1)
self.linear = torch.nn.Linear(768, 6)
def forward(self, ids, mask, token_type_ids):
_, output= self.albert(ids, attention_mask = mask)
output = self.drop(output)
output = self.linear(output)
return output
model = AlbertClass()
model.to(device)
```
## Hyperparameters & Loss function
```
def loss_fn(outputs, targets):
return torch.nn.BCEWithLogitsLoss()(outputs, targets)
param_optimizer = list(model.named_parameters())
no_decay = ["bias", "LayerNorm.bias", "LayerNorm.weight"]
optimizer_parameters = [
{
"params": [
p for n, p in param_optimizer if not any(nd in n for nd in no_decay)
],
"weight_decay": 0.001,
},
{
"params": [
p for n, p in param_optimizer if any(nd in n for nd in no_decay)
],
"weight_decay": 0.0,
},
]
optimizer = torch.optim.AdamW(optimizer_parameters, lr=1e-5)
num_training_steps = int(len(train_dataset) / TRAIN_BATCH_SIZE * EPOCHS)
scheduler = get_linear_schedule_with_warmup(
optimizer,
num_warmup_steps = 0,
num_training_steps = num_training_steps
)
```
## Train & Eval Functions
```
def train(epoch):
model.train()
for _,data in tqdm(enumerate(training_loader, 0), total=len(training_loader)):
ids = data['ids'].to(device, dtype = torch.long)
mask = data['mask'].to(device, dtype = torch.long)
token_type_ids = data['token_type_ids'].to(device, dtype = torch.long)
targets = data['targets'].to(device, dtype = torch.float)
outputs = model(ids, mask, token_type_ids)
optimizer.zero_grad()
loss = loss_fn(outputs, targets)
if _%1000==0:
print(f'Epoch: {epoch}, Loss: {loss.item()}')
optimizer.zero_grad()
loss.backward()
optimizer.step()
scheduler.step()
def validation(epoch):
model.eval()
fin_targets=[]
fin_outputs=[]
with torch.no_grad():
for _, data in tqdm(enumerate(testing_loader, 0), total=len(testing_loader)):
ids = data['ids'].to(device, dtype = torch.long)
mask = data['mask'].to(device, dtype = torch.long)
token_type_ids = data['token_type_ids'].to(device, dtype = torch.long)
targets = data['targets'].to(device, dtype = torch.float)
outputs = model(ids, mask, token_type_ids)
fin_targets.extend(targets.cpu().detach().numpy().tolist())
fin_outputs.extend(torch.sigmoid(outputs).cpu().detach().numpy().tolist())
return fin_outputs, fin_targets
```
## Training Model
```
MODEL_PATH = "/kaggle/working/albert-multilabel-model.bin"
best_micro = 0
for epoch in range(EPOCHS):
train(epoch)
outputs, targets = validation(epoch)
outputs = np.array(outputs) >= 0.5
accuracy = metrics.accuracy_score(targets, outputs)
f1_score_micro = metrics.f1_score(targets, outputs, average='micro')
f1_score_macro = metrics.f1_score(targets, outputs, average='macro')
print(f"Accuracy Score = {accuracy}")
print(f"F1 Score (Micro) = {f1_score_micro}")
print(f"F1 Score (Macro) = {f1_score_macro}")
if f1_score_micro > best_micro:
torch.save(model.state_dict(), MODEL_PATH)
best_micro = f1_score_micro
def predict(id, abstract):
MAX_LENGTH = 512
inputs = tokenizer.encode_plus(
abstract,
None,
add_special_tokens=True,
max_length=512,
pad_to_max_length=True,
return_token_type_ids=True,
truncation = True
)
ids = inputs['input_ids']
mask = inputs['attention_mask']
token_type_ids = inputs['token_type_ids']
ids = torch.tensor(ids, dtype=torch.long).unsqueeze(0)
mask = torch.tensor(mask, dtype=torch.long).unsqueeze(0)
token_type_ids = torch.tensor(token_type_ids, dtype=torch.long).unsqueeze(0)
ids = ids.to(device)
mask = mask.to(device)
token_type_ids = token_type_ids.to(device)
with torch.no_grad():
outputs = model(ids, mask, token_type_ids)
outputs = torch.sigmoid(outputs).squeeze()
outputs = np.round(outputs.cpu().numpy())
out = np.insert(outputs, 0, id)
return out
def submit():
test_df = pd.read_csv('../input/avjantahack/data/test.csv')
sample_submission = pd.read_csv('../input/avjantahack/data/sample_submission_UVKGLZE.csv')
y = []
for id, abstract in tqdm(zip(test_df['ID'], test_df['ABSTRACT']),
total=len(test_df)):
out = predict(id, abstract)
y.append(out)
y = np.array(y)
submission = pd.DataFrame(y, columns=sample_submission.columns).astype(int)
return submission
submission = submit()
submission
submission.to_csv('/kaggle/working/alberta-tuned-lr-ws-dr.csv', index=False)
```
| github_jupyter |
# Droplet Evaporation
```
import numpy as np
import matplotlib.pyplot as plt
from scipy import optimize
# Ethyl Acetate
#time_in_sec = np.array([0,5,10,15,20,25,30,35,40,45,50,55,60,65,70,75,80,85,90,95,100,105,110])
#diameter = np.array([2.79,2.697,2.573,2.542,2.573,2.48,2.449,2.449,2.387,2.356,2.263,2.232,2.201,2.139,1.82,1.426,1.178,1.085,0.992,0.496,0.403,0.372,0.11])
# Gasoline
#time_in_min = np.array([0,15,30,45,60,75,90,105,120,135,150,165,180,210,235,250,265])
#diameter = np.array([2,1.85,1.82,1.8,1.77,1.74,1.72,1.68,1.57,1.3,1.166,1.091,0.94,0.81,0.74,0.66,0.59])
```
# Ethyl Acetate
```
time_in_sec = np.array([0,5,10,15,20,25,30,35,40,45,50,55,60,65,70,75,80,85,90,95,100,105,110])
diameter = np.array([2.79,2.697,2.573,2.542,2.573,2.48,2.449,2.449,2.387,2.356,2.263,2.232,2.201,2.139,1.82,1.426,1.178,1.085,0.992,0.496,0.403,0.372,0.11])
x = time_in_sec.tolist()
y = diameter.tolist()
polynomial_coeff_1=np.polyfit(x,y,1)
polynomial_coeff_2=np.polyfit(x,y,2)
polynomial_coeff_3=np.polyfit(x,y,3)
xnew=np.linspace(0,110 ,100)
ynew_1=np.poly1d(polynomial_coeff_1)
ynew_2=np.poly1d(polynomial_coeff_2)
ynew_3=np.poly1d(polynomial_coeff_3)
plt.plot(x,y,'o')
plt.plot(xnew,ynew_1(xnew))
plt.plot(xnew,ynew_2(xnew))
plt.plot(xnew,ynew_3(xnew))
print(ynew_1)
print(ynew_2)
print(ynew_3)
plt.title("Diameter vs Time(s)")
plt.xlabel("Time(s)")
plt.ylabel("Diameter")
plt.show()
# Coeficients
# LINEAR : -0.02386 x + 3.139
# QUADRATIC : -0.0002702 x^2 + 0.005868 x + 2.619
# CUBIC : -4.771e-07 x^3 - 0.0001915 x^2 + 0.002481 x + 2.646
#
# Using Desmos to find the roots of the best fit polynomials
# Root of linear fit = 131.559
# Root of quadratic fit = 109.908
# Root of cubic fit = 109.414
def d_square_law(x, C, n):
y = C/(x**n)
return y
```
# Linear Fit
```
# Calculating time taken for vaporization for different diameters. (LINEAR FIT)
diameter = np.array([2.79,2.697,2.573,2.542,2.573,2.48,2.449,2.449,2.387,2.356,2.263,2.232,2.201,2.139,1.82,1.426,1.178,1.085,0.992,0.496,0.403,0.372,0.11])
time_in_sec = np.array([0,5,10,15,20,25,30,35,40,45,50,55,60,65,70,75,80,85,90,95,100,105,110])
t_vap = time_in_sec
t_vap = t_vap*0
t_vap = t_vap + 131.559
t_vap = t_vap - time_in_sec
print(t_vap.tolist())
# Finding C and n for d-square law
#initial_diameter = np.array([2.79,2.697,2.573,2.542,2.573,2.48,2.449,2.449,2.387,2.356,2.263,2.232,2.201,2.139,1.82,1.426,1.178,1.085,0.992,0.496,0.403,0.372,0.11])
#vap_time = np.array([109.908, 104.908, 99.908, 94.908, 89.908, 84.908, 79.908, 74.908, 69.908, 64.908, 59.908, 54.908, 49.908, 44.908, 39.908, 34.908, 29.908, 24.908, 19.908, 14.908000000000001, 9.908000000000001, 4.908000000000001, -0.09199999999999875])
# Linear
initial_diameter = np.array([2.79,2.697,2.573,2.542,2.573,2.48,2.449,2.449,2.387,2.356,2.263,2.232,2.201,2.139,1.82,1.426,1.178,1.085,0.992,0.496,0.403,0.372,0.11])
vap_time_lin = np.array([131.559, 126.559, 121.559, 116.559, 111.559, 106.559, 101.559, 96.559, 91.559, 86.559, 81.559, 76.559, 71.559, 66.559, 61.559, 56.559, 51.559, 46.559, 41.559, 36.559, 31.558999999999997, 26.558999999999997, 21.558999999999997])
# Linear
parameters_lin = optimize.curve_fit(d_square_law, xdata = initial_diameter, ydata = vap_time_lin)[0]
print("Linear : ",parameters_lin)
#C = parameters_lin[0]
#n = parameters_lin[1]
```
# Quadratic Fit
```
# Calculating time taken for vaporization for different diameters. (QUADRATIC FIT)
diameter = np.array([2.79,2.697,2.573,2.542,2.573,2.48,2.449,2.449,2.387,2.356,2.263,2.232,2.201,2.139,1.82,1.426,1.178,1.085,0.992,0.496,0.403,0.372,0.11])
time_in_sec = np.array([0,5,10,15,20,25,30,35,40,45,50,55,60,65,70,75,80,85,90,95,100,105,110])
t_vap = time_in_sec
t_vap = t_vap*0
t_vap = t_vap + 109.908
t_vap = t_vap - time_in_sec
print(t_vap.tolist())
# Quadratic Fit
initial_diameter = np.array([2.79,2.697,2.573,2.542,2.573,2.48,2.449,2.449,2.387,2.356,2.263,2.232,2.201,2.139,1.82,1.426,1.178,1.085,0.992,0.496,0.403,0.372])
vap_time_quad = np.array([109.908, 104.908, 99.908, 94.908, 89.908, 84.908, 79.908, 74.908, 69.908, 64.908, 59.908, 54.908, 49.908, 44.908, 39.908, 34.908, 29.908, 24.908, 19.908, 14.908000000000001, 9.908000000000001, 4.908000000000001])
# Quadratic
parameters_quad = optimize.curve_fit(d_square_law, xdata = initial_diameter, ydata = vap_time_quad)[0]
print("Linear : ",parameters_quad)
#C = parameters_lin[0]
#n = parameters_lin[1]
```
# Ethyl Acetate - After finding d-square Law
```
# Linear
C = 41.72856231
n = -0.97941652
# Quadratic
# C = 11.6827828
# n = -2.13925924
x = vap_time.tolist()
y = initial_diameter.tolist()
ynew=np.linspace(0,3 ,100)
xnew=[]
for item in ynew:
v1 = C/(item**n)
xnew.append(v1)
plt.plot(x,y,'o')
plt.plot(xnew,ynew)
plt.title("Initial Diameter vs Vaporization Time(s)")
plt.xlabel("Vaporization Time(s)")
plt.ylabel("Initial Diameter")
plt.show()
```
# Gasoline
```
time_in_min = np.array([0,15,30,45,60,75,90,105,120,135,150,165,180,210,235,250,265])
diameter = np.array([2,1.85,1.82,1.8,1.77,1.74,1.72,1.68,1.57,1.3,1.166,1.091,0.94,0.81,0.74,0.66,0.59])
x = time_in_min.tolist()
y = diameter.tolist()
polynomial_coeff_1=np.polyfit(x,y,1)
polynomial_coeff_2=np.polyfit(x,y,2)
polynomial_coeff_3=np.polyfit(x,y,3)
xnew=np.linspace(0,300 ,100)
ynew_1=np.poly1d(polynomial_coeff_1)
ynew_2=np.poly1d(polynomial_coeff_2)
ynew_3=np.poly1d(polynomial_coeff_3)
plt.plot(x,y,'o')
plt.plot(xnew,ynew_1(xnew))
plt.plot(xnew,ynew_2(xnew))
plt.plot(xnew,ynew_3(xnew))
print(ynew_1)
print(ynew_2)
print(ynew_3)
plt.title("Diameter vs Time(min)")
plt.xlabel("Time(min)")
plt.ylabel("Diameter")
plt.show()
# Coeficients
# LINEAR : -0.005637 x + 2.074
# QUADRATIC : -6.67e-06 x^2 - 0.003865 x + 2
# CUBIC : 1.481e-07 x^3 - 6.531e-05 x^2 + 0.00207 x + 1.891
#
# Using Desmos to find the roots of the best fit polynomials
# Root of linear fit = 367.926
# Root of quadratic fit = 329.781
# Root of cubic fit = No Positive Root
```
# Linear Fit
```
# Calculating time taken for vaporization for different diameters. (LINEAR FIT)
time_in_min = np.array([0,15,30,45,60,75,90,105,120,135,150,165,180,210,235,250,265])
diameter = np.array([2,1.85,1.82,1.8,1.77,1.74,1.72,1.68,1.57,1.3,1.166,1.091,0.94,0.81,0.74,0.66,0.59])
t_vap = time_in_min
t_vap = t_vap*0
t_vap = t_vap + 367.926
t_vap = t_vap - time_in_min
print(t_vap.tolist())
initial_diameter_g_lin = np.array([2,1.85,1.82,1.8,1.77,1.74,1.72,1.68,1.57,1.3,1.166,1.091,0.94,0.81,0.74,0.66,0.59])
vap_time_g_lin = np.array([367.926, 352.926, 337.926, 322.926, 307.926, 292.926, 277.926, 262.926, 247.926, 232.926, 217.926, 202.926, 187.926, 157.926, 132.926, 117.92599999999999, 102.92599999999999])
parameters_g_lin = optimize.curve_fit(d_square_law, xdata = initial_diameter_g_lin, ydata = vap_time_g_lin)[0]
print(parameters_g_lin)
C_g = parameters_g_lin[0]
n_g = parameters_g_lin[1]
```
# Quadratic Fit
```
# Calculating time taken for vaporization for different diameters.
time_in_min = np.array([0,15,30,45,60,75,90,105,120,135,150,165,180,210,235,250,265])
diameter = np.array([2,1.85,1.82,1.8,1.77,1.74,1.72,1.68,1.57,1.3,1.166,1.091,0.94,0.81,0.74,0.66,0.59])
t_vap = time_in_min
t_vap = t_vap*0
t_vap = t_vap + 329.781
t_vap = t_vap - time_in_min
print(t_vap.tolist())
initial_diameter_g_quad = np.array([2,1.85,1.82,1.8,1.77,1.74,1.72,1.68,1.57,1.3,1.166,1.091,0.94,0.81,0.74,0.66,0.59])
vap_time_g_quad = np.array([329.781, 314.781, 299.781, 284.781, 269.781, 254.781, 239.781, 224.781, 209.781, 194.781, 179.781, 164.781, 149.781, 119.781, 94.781, 79.781, 64.781])
parameters_g_quad = optimize.curve_fit(d_square_law, xdata = initial_diameter_g_quad, ydata = vap_time_g_quad)[0]
print(parameters_g_quad)
C_g = parameters_g_quad[0]
n_g = parameters_g_quad[1]
```
# Gasoline - After finding Vaporization Time Data
```
#Linear
C_g = 140.10666889
n_g = -1.1686059
# Quadratic
C_g = 140.10666889
n_g = -1.1686059
x_g = vap_time_g.tolist()
y_g = initial_diameter_g.tolist()
ynew_g=np.linspace(0,2.2 ,100)
xnew_g=[]
for item in ynew_g:
v1 = C_g/(item**n_g)
xnew_g.append(v1)
print(ynew_g)
print(xnew_g)
plt.plot(x_g,y_g,'o')
plt.plot(xnew_g,ynew_g)
plt.title("Initial Diameter vs Vaporization Time(min)")
plt.xlabel("Vaporization Time(min)")
plt.ylabel("Initial Diameter")
plt.show()
```
# Optimization Methods (IGNORE)
```
import numpy as np
from scipy import optimize
import matplotlib.pyplot as plt
plt.style.use('seaborn-poster')
time_in_sec = np.array([5,10,15,20,25,30,35,40,45,50,55,60,65,70,75,80,85,90,95,100,105,110])
diameter = np.array([2.697,2.573,2.542,2.573,2.48,2.449,2.449,2.387,2.356,2.263,2.232,2.201,2.139,1.82,1.426,1.178,1.085,0.992,0.496,0.403,0.372,0.11])
def func(x, a, b):
y = a/(x**b)
return y
parameters = optimize.curve_fit(func, xdata = time_in_sec, ydata = diameter)[0]
print(parameters)
C = parameters[0]
n = parameters[1]
plt.plot(time_in_sec,diameter,'o',label='data')
y_new = []
for val in time_in_sec:
v1 = C/(val**n)
y_new.append(v1)
plt.plot(time_in_sec,y_new,'-',label='fit')
log_time = np.log(time_in_min)
log_d = np.log(diameter)
print(log_d)
print(log_time)
x = log_time.tolist()
y = log_d.tolist()
polynomial_coeff=np.polyfit(x,y,1)
xnew=np.linspace(2.5,6,100)
ynew=np.poly1d(polynomial_coeff)
plt.plot(xnew,ynew(xnew),x,y,'o')
print(ynew)
plt.title("log(diameter) vs log(Time(s))")
plt.xlabel("log(Time(s))")
plt.ylabel("log(diameter)")
plt.show()
```
| github_jupyter |
# Enable GPU
```
import torch
device = torch.device('cuda:0' if torch.cuda.is_available else 'cpu')
```
# Actor and Critic Network
```
import torch.nn as nn
import torch.nn.functional as F
from torch.distributions import Categorical
class Actor_Net(nn.Module):
def __init__(self, input_dims, output_dims, num_neurons = 128):
super(Actor_Net, self).__init__()
self.fc1 = nn.Linear(input_dims, num_neurons)
self.actor = nn.Linear(num_neurons, output_dims)
self.log_probs = []
self.entropies = []
def forward(self, state):
x = F.relu(self.fc1(state))
x = F.softmax(self.actor(x), dim = 1)
return x
def get_action(self, state):
with torch.no_grad():
probs = self.forward(state)
dist = Categorical(probs = probs)
action = dist.sample()
return action
def eval_action(self, state):
probs = self.forward(state)
dist = Categorical(probs = probs)
action = dist.sample().to(device)
log_prob = dist.log_prob(action)
entropy = dist.entropy()
self.log_probs.append(log_prob)
self.entropies.append(entropy)
return action
class Critic_Net(nn.Module):
def __init__ (self, input_dims, output_dims, num_neurons = 128):
super(Critic_Net, self).__init__()
self.values = []
self.next_values = []
self.fc1 = nn.Linear(input_dims, num_neurons)
self.critic = nn.Linear(num_neurons, 1)
def forward (self, state):
x = F.relu(self.fc1(state))
x = self.critic(x)
return x
import torch.optim as optim
import numpy as np
import gym
class Actor_Critic_Agent(nn.Module):
def __init__(self, input_dims, output_dims, optimizer = 'RMSprop', num_neurons = 128 , gamma = 0.99, actor_lr=0.001, critic_lr = 0.01):
super(Actor_Critic_Agent, self).__init__()
self.actor_net = Actor_Net(input_dims= input_dims, output_dims= output_dims, num_neurons= num_neurons).to(device)
self.critic_net = Critic_Net(input_dims=input_dims, output_dims= output_dims, num_neurons= num_neurons).to(device)
self.gamma = gamma
if optimizer == 'RMSprop':
self.actor_optimizer = optim.RMSprop(params = self.actor_net.parameters(), lr =actor_lr)
self.critic_optimizer = optim.RMSprop(params = self.critic_net.parameters(), lr = critic_lr)
else:
self.actor_optimizer = optim.Adam(params = self.actor_net.parameters(), lr = actor_lr)
self.critic_optimizer = optim.Adam(params = self.critic_net.parameters(), lr = critic_lr)
def learn_mean(self, rewards, dones):
value_criteration = nn.MSELoss()
value_losses = []
actor_losses = []
self.critic_net.next_values = torch.cat(self.critic_net.next_values, dim = 0).squeeze(0)
self.critic_net.values = torch.cat(self.critic_net.values, dim = 0).squeeze(0)
self.actor_net.log_probs = torch.cat(self.actor_net.log_probs, dim = 0)
self.actor_net.entropies = torch.cat(self.actor_net.entropies, dim = 0)
for reward, entropy, log_prob, v, v_next, done in zip(rewards ,self.actor_net.entropies, self.actor_net.log_probs, self.critic_net.values, self.critic_net.next_values, dones):
td_target = reward + self.gamma * v_next * done
td_error = td_target - v
value_loss = value_criteration(v, td_target.detach())- 0.001 * entropy.detach()
actor_loss = - log_prob * td_error.detach()
value_losses.append(value_loss)
actor_losses.append(actor_loss)
self.critic_optimizer.zero_grad()
value_losses = torch.stack(value_losses).sum()
value_losses.backward()
self.critic_optimizer.step()
self.actor_optimizer.zero_grad()
actor_losses = torch.stack(actor_losses).sum()
actor_losses.backward()
self.actor_optimizer.step()
# clear out memory
self.actor_net.log_probs = []
self.actor_net.entropies = []
self.critic_net.values = []
self.critic_net.next_values = []
```
# Without Wandb
```
import gym
import time
import pdb
env = gym.make('CartPole-v1')
env.seed(543)
torch.manual_seed(543)
state_dims = env.observation_space.shape[0]
action_dims = env.action_space.n
agent = Actor_Critic_Agent(input_dims= state_dims, output_dims = action_dims)
def train():
num_ep = 2000
print_every = 100
running_score = 10
start = time.time()
rewards = []
dones = []
for ep in range(1, num_ep + 1):
state = env.reset()
score = 0
done = False
rewards = []
dones = []
while not done:
state = torch.tensor([state]).float().to(device)
action = agent.actor_net.eval_action(state)
v = agent.critic_net(state)
next_state, reward, done, _ = env.step(action.item())
v_next = agent.critic_net(torch.tensor([next_state]).float().to(device))
agent.critic_net.values.append(v.squeeze(0))
agent.critic_net.next_values.append(v_next.squeeze(0))
rewards.append(reward)
dones.append(1 - done)
# update episode
score += reward
state = next_state
if done:
break
# update agent
#pdb.set_trace()
agent.learn_mean(rewards,dones)
# calculating score and running score
running_score = 0.05 * score + (1 - 0.05) * running_score
if ep % print_every == 0:
print('episode: {}, running score: {}, time elapsed: {}'.format(ep, running_score, time.time() - start))
train() #RMS
```
# Wtih wandb
```
!pip install wandb
!wandb login
import wandb
sweep_config = dict()
sweep_config['method'] = 'grid'
sweep_config['metric'] = {'name': 'running_score', 'goal': 'maximize'}
sweep_config['parameters'] = {'learning': {'value': 'learn_mean'}, 'actor_learning_rate': {'values' : [0.01, 0.001, 0.0001,0.0003,0.00001]}, 'critic_learning_rate' : {'values': [0.01, 0.001, 0.0001, 0.0003, 0.00001]}
, 'num_neurons': {'value': 128 }, 'optimizer': {'values' : ['RMSprop', 'Adam']}}
sweep_id = wandb.sweep(sweep_config, project = 'Advantage_Actor_Critic')
import gym
import torch
import time
import wandb
def train():
wandb.init(config = {'env':'CartPole-v1','algorithm:': 'Actor_Critic','architecture': 'seperate','num_laeyrs':'2'}, project = 'Advantage_Actor_Critic',group = 'Cart_128_neurons_2_layer')
config = wandb.config
env = gym.make('CartPole-v1')
env.seed(543)
torch.manual_seed(543)
state_dim = env.observation_space.shape[0]
action_dim = env.action_space.n
device = torch.device('cuda:0' if torch.cuda.is_available else 'cpu')
agent = Actor_Critic_Agent(input_dims= state_dim, output_dims= action_dim, optimizer = config.optimizer, num_neurons= config.num_neurons, actor_lr = config.actor_learning_rate, critic_lr = config.critic_learning_rate)
num_ep = 3000
print_interval = 100
save_interval = 1000
running_score = 10
start = time.time()
wandb.watch(agent)
for ep in range(1,num_ep+1):
state = env.reset()
score = 0
done = False
rewards = []
dones = []
while not done:
state = torch.tensor([state]).float().to(device)
action = agent.actor_net.eval_action(state)
v = agent.critic_net(state)
next_state, reward, done, _ = env.step(action.item())
v_next = agent.critic_net(torch.tensor([next_state]).float().to(device))
agent.critic_net.values.append(v.squeeze(0))
agent.critic_net.next_values.append(v_next.squeeze(0))
rewards.append(reward)
dones.append(1 - done)
# update episode
score += reward
state = next_state
if done:
break
# update agent
agent.learn_mean(rewards,dones)
# calculating score and running score
running_score = 0.05 * score + (1 - 0.05) * running_score
wandb.log({'episode': ep, 'running_score': running_score})
if ep % print_interval == 0:
print('episode {} average reward {}, ended at {:.01f}'.format(ep, running_score, time.time() - start))
if ep % save_interval == 0:
save_name_actor = 'actor_' + str(ep) + '.pt'
torch.save(agent.actor_net.state_dict(),save_name_actor)
save_name_critic = 'critic_' + str(ep) + '.pt'
torch.save(agent.critic_net.state_dict(),save_name_critic)
wandb.save(save_name_actor)
wandb.save(save_name_critic)
if ep == num_ep:
dummy_input = torch.rand(1,4).to(device)
torch.onnx.export(agent.actor_net,dummy_input,'final_model_actor.onnx')
wandb.save('final_model_actor.onnx')
torch.onnx.export(agent.critic_net, dummy_input, 'final_model_critic.onnx')
wandb.save('final_model_critic.onnx')
wandb.agent(sweep_id, train)
```
# You can see the result here!
[Report Link](https://wandb.ai/ko120/Advantage_Actor_Critic/reports/TD-Actor-Critic-Learning-rate-tune---Vmlldzo4OTIwODg)
| github_jupyter |
## Import Necessary Packages
```
import numpy as np
import pandas as pd
import datetime
import os
np.random.seed(1337) # for reproducibility
from sklearn.model_selection import train_test_split
from sklearn.metrics.classification import accuracy_score
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics.regression import r2_score, mean_squared_error, mean_absolute_error
from dbn.tensorflow import SupervisedDBNRegression
```
## Define Model Settings
```
RBM_EPOCHS = 5
DBN_EPOCHS = 150
RBM_LEARNING_RATE = 0.01
DBN_LEARNING_RATE = 0.01
HIDDEN_LAYER_STRUCT = [20, 50, 100]
ACTIVE_FUNC = 'relu'
BATCH_SIZE = 28
```
## Define Directory, Road, and Year
```
# Read the dataset
ROAD = "Vicente Cruz"
YEAR = "2015"
EXT = ".csv"
DATASET_DIVISION = "seasonWet"
DIR = "../../../datasets/Thesis Datasets/"
OUTPUT_DIR = "PM1/Rolling 3/"
MODEL_DIR = "PM1/Rolling 3/"
'''''''Training dataset'''''''
WP = False
WEEKDAY = False
CONNECTED_ROADS = False
CONNECTED_1 = ["Antipolo"]
trafficDT = "recon_traffic" #orig_traffic recon_traffic
featureEngineering = "Rolling" #Rolling Expanding Rolling and Expanding
timeFE = "today" #today yesterday
timeConnected = "today"
ROLLING_WINDOW = 3
EXPANDING_WINDOW = 3
RECON_SHIFT = 96
# RECON_FE_WINDOW = 48
def addWorkingPeakFeatures(df):
result_df = df.copy()
# Converting the index as date
result_df.index = pd.to_datetime(result_df.index)
# Create column work_day
result_df['work_day'] = ((result_df.index.dayofweek) < 5).astype(int)
# Consider non-working holiday
if DATASET_DIVISION is not "seasonWet":
# Jan
result_df.loc['2015-01-01', 'work_day'] = 0
result_df.loc['2015-01-02', 'work_day'] = 0
# Feb
result_df.loc['2015-02-19', 'work_day'] = 0
result_df.loc['2015-02-25', 'work_day'] = 0
# Apr
result_df.loc['2015-04-02', 'work_day'] = 0
result_df.loc['2015-04-03', 'work_day'] = 0
result_df.loc['2015-04-09', 'work_day'] = 0
# May
result_df.loc['2015-05-01', 'work_day'] = 0
# Jun
result_df.loc['2015-06-12', 'work_day'] = 0
result_df.loc['2015-06-24', 'work_day'] = 0
# Jul
result_df.loc['2015-07-17', 'work_day'] = 0
# Aug
result_df.loc['2015-08-21', 'work_day'] = 0
result_df.loc['2015-08-31', 'work_day'] = 0
# Sep
result_df.loc['2015-08-25', 'work_day'] = 0
if DATASET_DIVISION is not "seasonWet":
# Nov
result_df.loc['2015-11-30', 'work_day'] = 0
# Dec
result_df.loc['2015-12-24', 'work_day'] = 0
result_df.loc['2015-12-25', 'work_day'] = 0
result_df.loc['2015-12-30', 'work_day'] = 0
result_df.loc['2015-12-31', 'work_day'] = 0
# Consider class suspension
if DATASET_DIVISION is not "seasonWet":
# Jan
result_df.loc['2015-01-08', 'work_day'] = 0
result_df.loc['2015-01-09', 'work_day'] = 0
result_df.loc['2015-01-14', 'work_day'] = 0
result_df.loc['2015-01-15', 'work_day'] = 0
result_df.loc['2015-01-16', 'work_day'] = 0
result_df.loc['2015-01-17', 'work_day'] = 0
# Jul
result_df.loc['2015-07-06', 'work_day'] = 0
result_df.loc['2015-07-08', 'work_day'] = 0
result_df.loc['2015-07-09', 'work_day'] = 0
result_df.loc['2015-07-10', 'work_day'] = 0
# Aug
result_df.loc['2015-08-10', 'work_day'] = 0
result_df.loc['2015-08-11', 'work_day'] = 0
# Sep
result_df.loc['2015-09-10', 'work_day'] = 0
# Oct
result_df.loc['2015-10-02', 'work_day'] = 0
result_df.loc['2015-10-19', 'work_day'] = 0
if DATASET_DIVISION is not "seasonWet":
# Nov
result_df.loc['2015-11-16', 'work_day'] = 0
result_df.loc['2015-11-17', 'work_day'] = 0
result_df.loc['2015-11-18', 'work_day'] = 0
result_df.loc['2015-11-19', 'work_day'] = 0
result_df.loc['2015-11-20', 'work_day'] = 0
# Dec
result_df.loc['2015-12-16', 'work_day'] = 0
result_df.loc['2015-12-18', 'work_day'] = 0
result_df['peak_hour'] = 0
# Set morning peak hour
start = datetime.time(7,0,0)
end = datetime.time(10,0,0)
result_df.loc[result_df.between_time(start, end).index, 'peak_hour'] = 1
# Set afternoon peak hour
start = datetime.time(16,0,0)
end = datetime.time(19,0,0)
result_df.loc[result_df.between_time(start, end).index, 'peak_hour'] = 1
result_df
return result_df
def reconstructDT(df, pastTraffic=False, trafficFeatureNeeded=[]):
result_df = df.copy()
# Converting the index as date
result_df.index = pd.to_datetime(result_df.index, format='%d/%m/%Y %H:%M')
result_df['month'] = result_df.index.month
result_df['day'] = result_df.index.day
result_df['hour'] = result_df.index.hour
result_df['min'] = result_df.index.minute
result_df['dayOfWeek'] = result_df.index.dayofweek
if pastTraffic:
for f in trafficFeatureNeeded:
result_df[f + '-' + str(RECON_SHIFT*15) + "mins"] = result_df[f].shift(RECON_SHIFT)
result_df = result_df.iloc[RECON_SHIFT:, :]
for f in range(len(result_df.columns)):
result_df[result_df.columns[f]] = normalize(result_df[result_df.columns[f]])
return result_df
def getNeededFeatures(columns, arrFeaturesNeed, featureEngineering="Original"):
to_remove = []
if len(arrFeaturesNeed) == 0: #all features aren't needed
to_remove += range(0, len(columns))
else:
if featureEngineering == "Original":
compareTo = " "
elif featureEngineering == "Rolling" or featureEngineering == "Expanding":
compareTo = "_"
for f in arrFeaturesNeed:
for c in range(0, len(columns)):
if f not in columns[c].split(compareTo)[0] and columns[c].split(compareTo)[0] not in arrFeaturesNeed:
to_remove.append(c)
if len(columns[c].split(compareTo)) > 1:
if "Esum" in columns[c].split(compareTo)[1]: #Removing all Expanding Sum
to_remove.append(c)
return to_remove
def normalize(data):
y = pd.to_numeric(data)
y = np.array(y.reshape(-1, 1))
scaler = MinMaxScaler()
y = scaler.fit_transform(y)
y = y.reshape(1, -1)[0]
return y
```
<br><br>
### Preparing Traffic Dataset
#### Importing Original Traffic (wo new features)
```
TRAFFIC_DIR = DIR + "mmda/"
TRAFFIC_FILENAME = "mmda_" + ROAD + "_" + YEAR + "_" + DATASET_DIVISION
orig_traffic = pd.read_csv(TRAFFIC_DIR + TRAFFIC_FILENAME + EXT, skipinitialspace=True)
orig_traffic = orig_traffic.fillna(0)
#Converting index to date and time, and removing 'dt' column
orig_traffic.index = pd.to_datetime(orig_traffic.dt, format='%d/%m/%Y %H:%M')
cols_to_remove = [0]
cols_to_remove = getNeededFeatures(orig_traffic.columns, ["statusN"])
orig_traffic.drop(orig_traffic.columns[[cols_to_remove]], axis=1, inplace=True)
orig_traffic.head()
if WEEKDAY:
orig_traffic = orig_traffic[((orig_traffic.index.dayofweek) < 5)]
orig_traffic.head()
TRAFFIC_DIR = DIR + "mmda/Rolling/" + DATASET_DIVISION + "/"
TRAFFIC_FILENAME = "eng_win" + str(ROLLING_WINDOW) + "_mmda_" + ROAD + "_" + YEAR + "_" + DATASET_DIVISION
rolling_traffic = pd.read_csv(TRAFFIC_DIR + TRAFFIC_FILENAME + EXT, skipinitialspace=True)
cols_to_remove = [0, 1, 2]
cols_to_remove += getNeededFeatures(rolling_traffic.columns, ["statusN"], "Rolling")
rolling_traffic.index = pd.to_datetime(rolling_traffic.dt, format='%Y-%m-%d %H:%M')
rolling_traffic.drop(rolling_traffic.columns[[cols_to_remove]], axis=1, inplace=True)
if WEEKDAY:
rolling_traffic = rolling_traffic[((rolling_traffic.index.dayofweek) < 5)]
rolling_traffic.head()
TRAFFIC_DIR = DIR + "mmda/Expanding/" + DATASET_DIVISION + "/"
TRAFFIC_FILENAME = "eng_win" + str(EXPANDING_WINDOW) + "_mmda_" + ROAD + "_" + YEAR + "_" + DATASET_DIVISION
expanding_traffic = pd.read_csv(TRAFFIC_DIR + TRAFFIC_FILENAME + EXT, skipinitialspace=True)
cols_to_remove = [0, 1, 2, 5]
cols_to_remove += getNeededFeatures(expanding_traffic.columns, ["statusN"], "Rolling")
expanding_traffic.index = pd.to_datetime(expanding_traffic.dt, format='%d/%m/%Y %H:%M')
expanding_traffic.drop(expanding_traffic.columns[[cols_to_remove]], axis=1, inplace=True)
if WEEKDAY:
expanding_traffic = expanding_traffic[((expanding_traffic.index.dayofweek) < 5)]
expanding_traffic.head()
recon_traffic = reconstructDT(orig_traffic, pastTraffic=True, trafficFeatureNeeded=['statusN'])
recon_traffic.head()
connected_roads = []
for c in CONNECTED_1:
TRAFFIC_DIR = DIR + "mmda/"
TRAFFIC_FILENAME = "mmda_" + c + "_" + YEAR + "_" + DATASET_DIVISION
temp = pd.read_csv(TRAFFIC_DIR + TRAFFIC_FILENAME + EXT, skipinitialspace=True)
temp = temp.fillna(0)
#Converting index to date and time, and removing 'dt' column
temp.index = pd.to_datetime(temp.dt, format='%d/%m/%Y %H:%M')
cols_to_remove = [0]
cols_to_remove = getNeededFeatures(temp.columns, ["statusN"])
temp.drop(temp.columns[[cols_to_remove]], axis=1, inplace=True)
if WEEKDAY:
temp = temp[((temp.index.dayofweek) < 5)]
for f in range(len(temp.columns)):
temp[temp.columns[f]] = normalize(temp[temp.columns[f]])
temp = temp.rename(columns={temp.columns[f]: temp.columns[f] +"(" + c + ")"})
connected_roads.append(temp)
connected_roads[0].head()
```
### Merging datasets
```
if trafficDT == "orig_traffic":
arrDT = [orig_traffic]
if CONNECTED_ROADS:
for c in connected_roads:
arrDT.append(c)
elif trafficDT == "recon_traffic":
arrDT = [recon_traffic]
if CONNECTED_ROADS:
timeConnected = "today"
print("TimeConnected = " + timeConnected)
for c in connected_roads:
if timeConnected == "today":
startIndex = np.absolute(len(arrDT[0])-len(c))
endIndex = len(c)
elif timeConnected == "yesterday":
startIndex = 0
endIndex = len(rolling_traffic) - RECON_SHIFT
c = c.rename(columns={c.columns[0]: c.columns[0] + "-" + str(RECON_SHIFT*15) + "mins"})
c = c.iloc[startIndex:endIndex, :]
print("Connected Road Start time: " + str(c.index[0]))
c.index = arrDT[0].index
arrDT.append(c)
print(str(startIndex) + " " + str(endIndex))
if featureEngineering != "":
print("Adding Feature Engineering")
print("TimeConnected = " + timeFE)
if timeFE == "today":
startIndex = np.absolute(len(arrDT[0])-len(rolling_traffic))
endIndex = len(rolling_traffic)
elif timeFE == "yesterday":
startIndex = 0
endIndex = len(rolling_traffic) - RECON_SHIFT
if featureEngineering == "Rolling":
temp = rolling_traffic.iloc[startIndex:endIndex, :]
arrDT.append(temp)
elif featureEngineering == "Expanding":
temp = expanding_traffic.iloc[startIndex:endIndex, :]
arrDT.append(temp)
elif featureEngineering == "Rolling and Expanding":
print(str(startIndex) + " " + str(endIndex))
#Rolling
temp = rolling_traffic.iloc[startIndex:endIndex, :]
temp.index = arrDT[0].index
arrDT.append(temp)
#Expanding
temp = expanding_traffic.iloc[startIndex:endIndex, :]
temp.index = arrDT[0].index
arrDT.append(temp)
merged_dataset = pd.concat(arrDT, axis=1)
if "Rolling" in featureEngineering:
merged_dataset = merged_dataset.iloc[ROLLING_WINDOW+1:, :]
if WP:
merged_dataset = addWorkingPeakFeatures(merged_dataset)
print("Adding working / peak days")
merged_dataset
```
### Adding Working / Peak Features
```
if WP:
merged_dataset = addWorkingPeakFeatures(merged_dataset)
print("Adding working / peak days")
```
## Preparing Training dataset
### Merge Original (and Rolling and Expanding)
```
# To-be Predicted variable
Y = merged_dataset.statusN
Y = Y.fillna(0)
# Training Data
X = merged_dataset
X = X.drop(X.columns[[0]], axis=1)
# Splitting data
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.67, shuffle=False)
X_train = np.array(X_train)
X_test = np.array(X_test)
Y_train = np.array(Y_train)
Y_test = np.array(Y_test)
# Data scaling
# min_max_scaler = MinMaxScaler()
# X_train = min_max_scaler.fit_transform(X_train)
#Print training and testing data
pd.concat([X, Y.to_frame()], axis=1).head()
```
<br><br>
## Training Model
```
# Training
regressor = SupervisedDBNRegression(hidden_layers_structure=HIDDEN_LAYER_STRUCT,
learning_rate_rbm=RBM_LEARNING_RATE,
learning_rate=DBN_LEARNING_RATE,
n_epochs_rbm=RBM_EPOCHS,
n_iter_backprop=DBN_EPOCHS,
batch_size=BATCH_SIZE,
activation_function=ACTIVE_FUNC)
regressor.fit(X_train, Y_train)
#To check RBM Loss Errors:
rbm_error = regressor.unsupervised_dbn.rbm_layers[0].rbm_loss_error
#To check DBN Loss Errors
dbn_error = regressor.dbn_loss_error
```
<br><br>
## Testing Model
```
# Test
min_max_scaler = MinMaxScaler()
X_test = min_max_scaler.fit_transform(X_test)
Y_pred = regressor.predict(X_test)
r2score = r2_score(Y_test, Y_pred)
rmse = np.sqrt(mean_squared_error(Y_test, Y_pred))
mae = mean_absolute_error(Y_test, Y_pred)
print('Done.\nR-squared: %.3f\nRMSE: %.3f \nMAE: %.3f' % (r2score, rmse, mae))
print(len(Y_pred))
temp = []
for i in range(len(Y_pred)):
temp.append(Y_pred[i][0])
d = {'Predicted': temp, 'Actual': Y_test}
df = pd.DataFrame(data=d)
df.head()
# Save the model
if MODEL_DIR != "":
directory = "models/" + MODEL_DIR
if not os.path.exists(directory):
print("Making Directory")
os.makedirs(directory)
regressor.save('models/' + MODEL_DIR + 'pm1_' + ROAD + '_' + YEAR + '.pkl')
```
### Results and Analysis below
```
import matplotlib.pyplot as plt
```
##### Printing Predicted and Actual Results
```
startIndex = merged_dataset.shape[0] - Y_pred.shape[0]
dt = merged_dataset.index[startIndex:,]
temp = []
for i in range(len(Y_pred)):
temp.append(Y_pred[i][0])
d = {'Predicted': temp, 'Actual': Y_test, 'dt': dt}
df = pd.DataFrame(data=d)
df.head()
df.tail()
```
#### Visualize Actual and Predicted Traffic
```
print(df.dt[0])
startIndex = 0
endIndex = 96
line1 = df.Actual.rdiv(1)
line2 = df.Predicted.rdiv(1)
x = range(0, RBM_EPOCHS * len(HIDDEN_LAYER_STRUCT))
plt.figure(figsize=(20, 4))
plt.plot(line1[startIndex:endIndex], c='red', label="Actual-Congestion")
plt.plot(line2[startIndex:endIndex], c='blue', label="Predicted-Congestion")
plt.legend()
plt.xlabel("Date")
plt.ylabel("Traffic Congestion")
plt.show()
if OUTPUT_DIR != "":
directory = "output/" + OUTPUT_DIR
if not os.path.exists(directory):
print("Making Directory")
os.makedirs(directory)
df.to_csv("output/" + OUTPUT_DIR + "pm1_" + ROAD + '_' + YEAR + EXT, index=False, encoding='utf-8')
```
#### Visualize trend of loss of RBM and DBN Training
```
line1 = rbm_error
line2 = dbn_error
x = range(0, RBM_EPOCHS * len(HIDDEN_LAYER_STRUCT))
plt.plot(range(0, RBM_EPOCHS * len(HIDDEN_LAYER_STRUCT)), line1, c='red')
plt.xticks(x)
plt.xlabel("Iteration")
plt.ylabel("Error")
plt.show()
plt.plot(range(DBN_EPOCHS), line2, c='blue')
plt.xticks(x)
plt.xlabel("Iteration")
plt.ylabel("Error")
plt.show()
plt.plot(range(0, RBM_EPOCHS * len(HIDDEN_LAYER_STRUCT)), line1, c='red')
plt.plot(range(DBN_EPOCHS), line2, c='blue')
plt.xticks(x)
plt.xlabel("Iteration")
plt.ylabel("Error")
plt.show()
```
| github_jupyter |
# 3D Map
While representing the configuration space in 3 dimensions isn't entirely practical it's fun (and useful) to visualize things in 3D.
In this exercise you'll finish the implementation of `create_grid` such that a 3D grid is returned where cells containing a voxel are set to `True`. We'll then plot the result!
```
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
%matplotlib inline
plt.rcParams['figure.figsize'] = 16, 16
# This is the same obstacle data from the previous lesson.
filename = 'colliders.csv'
data = np.loadtxt(filename, delimiter=',', dtype='Float64', skiprows=2)
print(data)
def create_voxmap(data, voxel_size=5):
"""
Returns a grid representation of a 3D configuration space
based on given obstacle data.
The `voxel_size` argument sets the resolution of the voxel map.
"""
# minimum and maximum north coordinates
north_min = np.floor(np.amin(data[:, 0] - data[:, 3]))
north_max = np.ceil(np.amax(data[:, 0] + data[:, 3]))
# minimum and maximum east coordinates
east_min = np.floor(np.amin(data[:, 1] - data[:, 4]))
east_max = np.ceil(np.amax(data[:, 1] + data[:, 4]))
alt_max = np.ceil(np.amax(data[:, 2] + data[:, 5]))
# given the minimum and maximum coordinates we can
# calculate the size of the grid.
north_size = int(np.ceil((north_max - north_min))) // voxel_size
east_size = int(np.ceil((east_max - east_min))) // voxel_size
alt_size = int(alt_max) // voxel_size
voxmap = np.zeros((north_size, east_size, alt_size), dtype=np.bool)
for datum in data:
x, y, z, dx, dy, dz = datum.astype(np.int32)
obstacle = np.array(((x-dx, x+dx),
(y-dy, y+dy),
(z-dz, z+dz)))
obstacle[0] = (obstacle[0] - north_min) // voxel_size
obstacle[1] = (obstacle[1] - east_min) // voxel_size
obstacle[2] = obstacle[2] // voxel_size
voxmap[obstacle[0][0]:obstacle[0][1], obstacle[1][0]:obstacle[1][1], obstacle[2][0]:obstacle[2][1]] = True
return voxmap
```
Create 3D grid.
```
voxel_size = 10
voxmap = create_voxmap(data, voxel_size)
print(voxmap.shape)
```
Plot the 3D grid.
```
fig = plt.figure()
ax = fig.gca(projection='3d')
ax.voxels(voxmap, edgecolor='k')
ax.set_xlim(voxmap.shape[0], 0)
ax.set_ylim(0, voxmap.shape[1])
# add 100 to the height so the buildings aren't so tall
ax.set_zlim(0, voxmap.shape[2]+100//voxel_size)
plt.xlabel('North')
plt.ylabel('East')
plt.show()
```
Isn't the city pretty?
| github_jupyter |
# Prologue
For this project we will use the logistic regression function to model the growth of confirmed Covid-19 case population growth in Bangladesh. The logistic regression function is commonly used in classification problems, and in this project we will be examining how it fares as a regression tool. Both cumulative case counts over time and logistic regression curves have a sigmoid shape and we shall try to fit a theoretically predicted curve over the actual cumulative case counts over time to reach certain conclusions about the case count growth, such as the time of peak daily new cases and the total cases that may be reached during this outbreak.
# Import the necessary modules
```
import pandas as pd
import numpy as np
from datetime import datetime,timedelta
from sklearn.metrics import mean_squared_error
from scipy.optimize import curve_fit
from scipy.optimize import fsolve
import matplotlib.pyplot as plt
%matplotlib inline
```
# Connect to Google Drive (where the data is kept)
```
from google.colab import drive
drive.mount('/content/drive')
```
# Import data and format as needed
```
df = pd.read_csv('/content/drive/My Drive/Corona-Cases.n-1.csv')
df.tail()
```
As you can see, the format of the date is 'month-day-year'. Let's specify the date column is datetime type. Let's also specify the formatting as %m-%d-%Y. And then, let's find the day when the first confirmed cases of Covid-19 were reported in Bangladesh.
```
FMT = '%m-%d-%Y'
df['Date'] = pd.to_datetime(df['Date'], format=FMT)
```
We have to initialize the first date of confirmed Covid-19 cases as the datetime variable start_date because we would need it later to calculate the peak.
```
# Initialize the start date
start_date = datetime.date(df.loc[0, 'Date'])
print('Start date: ', start_date)
```
Now, for the logistic regression function, we would need a timestep column instead of a date column in the dataframe. So we create a new dataframe called data where we drop the date column and use the index as the timestep column.
```
# drop date column
data = df['Total cases']
# reset index and create a timestep
data = data.reset_index(drop=False)
# rename columns
data.columns = ['Timestep', 'Total Cases']
# check
data.tail()
```
# Defining the logistic regression function
```
def logistic_model(x,a,b,c):
return c/(1+np.exp(-(x-b)/a))
```
In this formula, we have the variable x that is the time and three parameters: a, b, c.
* a is a metric for the speed of infections
* b is the day with the estimated maximum growth rate of confirmed Covid-19 cases
* c is the maximum number the cumulative confirmed cases will reach by the end of the first outbreak here in Bangladesh
The growth of cumulative cases follows a sigmoid shape like the logistic regression curve and hence, this may be a good way to model the growth of the confirmed Covid-19 case population over time. For the first outbreak at least. It makes sense because, for an outbreak, the rise in cumulative case counts is initially exponential. Then there is a point of inflection where the curve nearly becomes linear. We assume that this point of inflection is the time around which the daily new case numbers will peak. After that the curve eventually flattens out.
# Fit the logistic function and extrapolate
```
# Initialize all the timesteps as x
x = list(data.iloc[:,0])
# Initialize all the Total Cases values as y
y = list(data.iloc[:,1])
# Fit the curve using sklearn's curve_fit method we initialize the parameter p0 with arbitrary values
fit = curve_fit(logistic_model,x,y,p0=[2,100,20000])
(a, b, c), cov = fit
# Print outputs
print('Metric for speed of infections: ', a)
print('Days from start when cumulative case counts will peak: ', b)
print('Total cumulative cases that will be reached: ', c)
# Print errors for a, b, c
errors = [np.sqrt(fit[1][i][i]) for i in [0,1,2]]
print('Errors in a, b and c respectively:\n', errors)
# estimated time of peak
print('Estimated time of peak between', start_date + timedelta(days=(b-errors[1])), ' and ', start_date + timedelta(days=(b+errors[1])))
# estimated total number of infections
print('Estimated total number of infections betweeen ', (c - errors[2]), ' and ', (c + errors[2]))
```
To extrapolate the curve to the future, use the fsolve function from scipy.
```
# Extrapolate
sol = int(fsolve(lambda x : logistic_model(x,a,b,c) - int(c),b))
```
# Plot the graph
```
pred_x = list(range(max(x),sol))
plt.rcParams['figure.figsize'] = [7, 7]
plt.rc('font', size=14)
# Real data
plt.scatter(x,y,label="Real data",color="red")
# Predicted logistic curve
plt.plot(x+pred_x, [logistic_model(i,fit[0][0],fit[0][1],fit[0][2]) for i in x+pred_x], label="Logistic model" )
plt.legend()
plt.xlabel("Days since 8th March 2020")
plt.ylabel("Total number of infected people")
plt.ylim((min(y)*0.9,c*1.1))
plt.show()
```
# Evaluate the MSE error
Evaluating the mean squared error (MSE) is not very meaningful on its own until we can compare it with another predictive method. We can compare MSE of our regression with MSE from another method to check if our logistic regression model works better than the other predictive model. The model with the lower MSE performs better.
```
y_pred_logistic = [logistic_model(i,fit[0][0],fit[0][1],fit[0][2])
for i in x]
print('Mean squared error: ', mean_squared_error(y,y_pred_logistic))
```
# Epilogue
We should be mindful of some caveats:
* These predictions will only be meaningful when the peak has actually been crossed definitively.
* Also, the reliability of the reported cases would also influence the dependability of the model. Developing countries, especially the South Asian countries have famously failed to report accurate disaster statisticcs in the past.
* Also, the testing numbers are low overall, especially in cities outside Dhaka where the daily new cases still have not peaked yet.
* Since most of the cases reported were in Dhaka, the findings indicate that the peak in Dhaka may have been reached already.
* If there is a second outbreak before the first outbreak subsides, the curve may not be sigmoid shaped and hence the results may not be as meaningful.
* The total reported case numbers will possibly be greater than 260000, because the daily new cases is still rising in some cities other than Dhaka. It is not unsound to expect that the total reported case count for this first instance of Covid-19 outbreak could very well reach 300000 or more.
* The government recently hiked the prices of tests which may have led to increased unwillingness in suspected candidates to actually test for the disease, and that may have influenced the recent confirmed case counts.
# References
Inspiration for theory and code from the following articles:
* [Covid-19 infection in Italy. Mathematical models and predictions](https://towardsdatascience.com/covid-19-infection-in-italy-mathematical-models-and-predictions-7784b4d7dd8d)
* [Logistic growth modelling of COVID-19 proliferation in China and its international implications](https://www.sciencedirect.com/science/article/pii/S1201971220303039)
* [Logistic Growth Model for COVID-19](https://www.wolframcloud.com/obj/covid-19/Published/Logistic-Growth-Model-for-COVID-19.nb)
| github_jupyter |
# FloPy
## Plotting SWR Process Results
This notebook demonstrates the use of the `SwrObs` and `SwrStage`, `SwrBudget`, `SwrFlow`, and `SwrExchange`, `SwrStructure`, classes to read binary SWR Process observation, stage, budget, reach to reach flows, reach-aquifer exchange, and structure files. It demonstrates these capabilities by loading these binary file types and showing examples of plotting SWR Process data. An example showing how the simulated water surface profile at a selected time along a selection of reaches can be plotted is also presented.
```
%matplotlib inline
from IPython.display import Image
import os
import sys
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
# run installed version of flopy or add local path
try:
import flopy
except:
fpth = os.path.abspath(os.path.join('..', '..'))
sys.path.append(fpth)
import flopy
print(sys.version)
print('numpy version: {}'.format(np.__version__))
print('matplotlib version: {}'.format(mpl.__version__))
print('flopy version: {}'.format(flopy.__version__))
#Set the paths
datapth = os.path.join('..', 'data', 'swr_test')
# SWR Process binary files
files = ('SWR004.obs', 'SWR004.vel', 'SWR004.str', 'SWR004.stg', 'SWR004.flow')
```
### Load SWR Process observations
Create an instance of the `SwrObs` class and load the observation data.
```
sobj = flopy.utils.SwrObs(os.path.join(datapth, files[0]))
ts = sobj.get_data()
```
#### Plot the data from the binary SWR Process observation file
```
fig = plt.figure(figsize=(6, 12))
ax1 = fig.add_subplot(3, 1, 1)
ax1.semilogx(ts['totim']/3600., -ts['OBS1'], label='OBS1')
ax1.semilogx(ts['totim']/3600., -ts['OBS2'], label='OBS2')
ax1.semilogx(ts['totim']/3600., -ts['OBS9'], label='OBS3')
ax1.set_ylabel('Flow, in cubic meters per second')
ax1.legend()
ax = fig.add_subplot(3, 1, 2, sharex=ax1)
ax.semilogx(ts['totim']/3600., -ts['OBS4'], label='OBS4')
ax.semilogx(ts['totim']/3600., -ts['OBS5'], label='OBS5')
ax.set_ylabel('Flow, in cubic meters per second')
ax.legend()
ax = fig.add_subplot(3, 1, 3, sharex=ax1)
ax.semilogx(ts['totim']/3600., ts['OBS6'], label='OBS6')
ax.semilogx(ts['totim']/3600., ts['OBS7'], label='OBS7')
ax.set_xlim(1, 100)
ax.set_ylabel('Stage, in meters')
ax.set_xlabel('Time, in hours')
ax.legend();
```
### Load the same data from the individual binary SWR Process files
Load discharge data from the flow file. The flow file contains the simulated flow between connected reaches for each connection in the model.
```
sobj = flopy.utils.SwrFlow(os.path.join(datapth, files[1]))
times = np.array(sobj.get_times())/3600.
obs1 = sobj.get_ts(irec=1, iconn=0)
obs2 = sobj.get_ts(irec=14, iconn=13)
obs4 = sobj.get_ts(irec=4, iconn=3)
obs5 = sobj.get_ts(irec=5, iconn=4)
```
Load discharge data from the structure file. The structure file contains the simulated structure flow for each reach with a structure.
```
sobj = flopy.utils.SwrStructure(os.path.join(datapth, files[2]))
obs3 = sobj.get_ts(irec=17, istr=0)
```
Load stage data from the stage file. The flow file contains the simulated stage for each reach in the model.
```
sobj = flopy.utils.SwrStage(os.path.join(datapth, files[3]))
obs6 = sobj.get_ts(irec=13)
```
Load budget data from the budget file. The budget file contains the simulated budget for each reach group in the model. The budget file also contains the stage data for each reach group. In this case the number of reach groups equals the number of reaches in the model.
```
sobj = flopy.utils.SwrBudget(os.path.join(datapth, files[4]))
obs7 = sobj.get_ts(irec=17)
```
#### Plot the data loaded from the individual binary SWR Process files.
Note that the plots are identical to the plots generated from the binary SWR observation data.
```
fig = plt.figure(figsize=(6, 12))
ax1 = fig.add_subplot(3, 1, 1)
ax1.semilogx(times, obs1['flow'], label='OBS1')
ax1.semilogx(times, obs2['flow'], label='OBS2')
ax1.semilogx(times, -obs3['strflow'], label='OBS3')
ax1.set_ylabel('Flow, in cubic meters per second')
ax1.legend()
ax = fig.add_subplot(3, 1, 2, sharex=ax1)
ax.semilogx(times, obs4['flow'], label='OBS4')
ax.semilogx(times, obs5['flow'], label='OBS5')
ax.set_ylabel('Flow, in cubic meters per second')
ax.legend()
ax = fig.add_subplot(3, 1, 3, sharex=ax1)
ax.semilogx(times, obs6['stage'], label='OBS6')
ax.semilogx(times, obs7['stage'], label='OBS7')
ax.set_xlim(1, 100)
ax.set_ylabel('Stage, in meters')
ax.set_xlabel('Time, in hours')
ax.legend();
```
### Plot simulated water surface profiles
Simulated water surface profiles can be created using the `ModelCrossSection` class.
Several things that we need in addition to the stage data include reach lengths and bottom elevations. We load these data from an existing file.
```
sd = np.genfromtxt(os.path.join(datapth, 'SWR004.dis.ref'), names=True)
```
The contents of the file are shown in the cell below.
```
fc = open(os.path.join(datapth, 'SWR004.dis.ref')).readlines()
fc
```
Create an instance of the `SwrStage` class for SWR Process stage data.
```
sobj = flopy.utils.SwrStage(os.path.join(datapth, files[3]))
```
Create a selection condition (`iprof`) that can be used to extract data for the reaches of interest (reaches 0, 1, and 8 through 17). Use this selection condition to extract reach lengths (from `sd['RLEN']`) and the bottom elevation (from `sd['BELEV']`) for the reaches of interest. The selection condition will also be used to extract the stage data for reaches of interest.
```
iprof = sd['IRCH'] > 0
iprof[2:8] = False
dx = np.extract(iprof, sd['RLEN'])
belev = np.extract(iprof, sd['BELEV'])
```
Create a fake model instance so that the `ModelCrossSection` class can be used.
```
ml = flopy.modflow.Modflow()
dis = flopy.modflow.ModflowDis(ml, nrow=1, ncol=dx.shape[0], delr=dx, top=4.5, botm=belev.reshape(1,1,12))
```
Create an array with the x position at the downstream end of each reach, which will be used to color the plots below each reach.
```
x = np.cumsum(dx)
```
Plot simulated water surface profiles for 8 times.
```
fig = plt.figure(figsize=(12, 12))
for idx, v in enumerate([19, 29, 34, 39, 44, 49, 54, 59]):
ax = fig.add_subplot(4, 2, idx+1)
s = sobj.get_data(idx=v)
stage = np.extract(iprof, s['stage'])
xs = flopy.plot.ModelCrossSection(model=ml, line={'Row': 0})
xs.plot_fill_between(stage.reshape(1,1,12), colors=['none', 'blue'], ax=ax, edgecolors='none')
linecollection = xs.plot_grid(ax=ax, zorder=10)
ax.fill_between(np.append(0., x), y1=np.append(belev[0], belev), y2=-0.5,
facecolor='0.5', edgecolor='none', step='pre')
ax.set_title('{} hours'.format(times[v]))
ax.set_ylim(-0.5, 4.5)
```
## Summary
This notebook demonstrates flopy functionality for reading binary output generated by the SWR Process. Binary files that can be read include observations, stages, budgets, flow, reach-aquifer exchanges, and structure data. The binary stage data can also be used to create water-surface profiles.
Hope this gets you started!
| github_jupyter |
[제가 미리 만들어놓은 이 링크](https://colab.research.google.com/github/heartcored98/Standalone-DeepLearning/blob/master/Lec4/Lab6_result_report.ipynb)를 통해 Colab에서 바로 작업하실 수 있습니다!
런타임 유형은 python3, GPU 가속 확인하기!
```
!mkdir results
import torch
import torchvision
import torchvision.transforms as transforms
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import argparse
import numpy as np
import time
from copy import deepcopy # Add Deepcopy for args
```
## Data Preparation
```
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
trainset = torchvision.datasets.CIFAR10(root='./data', train=True,
download=True, transform=transform)
trainset, valset = torch.utils.data.random_split(trainset, [40000, 10000])
testset = torchvision.datasets.CIFAR10(root='./data', train=False,
download=True, transform=transform)
partition = {'train': trainset, 'val':valset, 'test':testset}
```
## Model Architecture
```
class MLP(nn.Module):
def __init__(self, in_dim, out_dim, hid_dim, n_layer, act, dropout, use_bn, use_xavier):
super(MLP, self).__init__()
self.in_dim = in_dim
self.out_dim = out_dim
self.hid_dim = hid_dim
self.n_layer = n_layer
self.act = act
self.dropout = dropout
self.use_bn = use_bn
self.use_xavier = use_xavier
# ====== Create Linear Layers ====== #
self.fc1 = nn.Linear(self.in_dim, self.hid_dim)
self.linears = nn.ModuleList()
self.bns = nn.ModuleList()
for i in range(self.n_layer-1):
self.linears.append(nn.Linear(self.hid_dim, self.hid_dim))
if self.use_bn:
self.bns.append(nn.BatchNorm1d(self.hid_dim))
self.fc2 = nn.Linear(self.hid_dim, self.out_dim)
# ====== Create Activation Function ====== #
if self.act == 'relu':
self.act = nn.ReLU()
elif self.act == 'tanh':
self.act == nn.Tanh()
elif self.act == 'sigmoid':
self.act = nn.Sigmoid()
else:
raise ValueError('no valid activation function selected!')
# ====== Create Regularization Layer ======= #
self.dropout = nn.Dropout(self.dropout)
if self.use_xavier:
self.xavier_init()
def forward(self, x):
x = self.act(self.fc1(x))
for i in range(len(self.linears)):
x = self.act(self.linears[i](x))
x = self.bns[i](x)
x = self.dropout(x)
x = self.fc2(x)
return x
def xavier_init(self):
for linear in self.linears:
nn.init.xavier_normal_(linear.weight)
linear.bias.data.fill_(0.01)
net = MLP(3072, 10, 100, 4, 'relu', 0.1, True, True) # Testing Model Construction
```
## Train, Validate, Test and Experiment
```
def train(net, partition, optimizer, criterion, args):
trainloader = torch.utils.data.DataLoader(partition['train'],
batch_size=args.train_batch_size,
shuffle=True, num_workers=2)
net.train()
correct = 0
total = 0
train_loss = 0.0
for i, data in enumerate(trainloader, 0):
optimizer.zero_grad() # [21.01.05 오류 수정] 매 Epoch 마다 .zero_grad()가 실행되는 것을 매 iteration 마다 실행되도록 수정했습니다.
# get the inputs
inputs, labels = data
inputs = inputs.view(-1, 3072)
inputs = inputs.cuda()
labels = labels.cuda()
outputs = net(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
train_loss += loss.item()
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
train_loss = train_loss / len(trainloader)
train_acc = 100 * correct / total
return net, train_loss, train_acc
def validate(net, partition, criterion, args):
valloader = torch.utils.data.DataLoader(partition['val'],
batch_size=args.test_batch_size,
shuffle=False, num_workers=2)
net.eval()
correct = 0
total = 0
val_loss = 0
with torch.no_grad():
for data in valloader:
images, labels = data
images = images.view(-1, 3072)
images = images.cuda()
labels = labels.cuda()
outputs = net(images)
loss = criterion(outputs, labels)
val_loss += loss.item()
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
val_loss = val_loss / len(valloader)
val_acc = 100 * correct / total
return val_loss, val_acc
def test(net, partition, args):
testloader = torch.utils.data.DataLoader(partition['test'],
batch_size=args.test_batch_size,
shuffle=False, num_workers=2)
net.eval()
correct = 0
total = 0
with torch.no_grad():
for data in testloader:
images, labels = data
images = images.view(-1, 3072)
images = images.cuda()
labels = labels.cuda()
outputs = net(images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
test_acc = 100 * correct / total
return test_acc
def experiment(partition, args):
net = MLP(args.in_dim, args.out_dim, args.hid_dim, args.n_layer, args.act, args.dropout, args.use_bn, args.use_xavier)
net.cuda()
criterion = nn.CrossEntropyLoss()
if args.optim == 'SGD':
optimizer = optim.RMSprop(net.parameters(), lr=args.lr, weight_decay=args.l2)
elif args.optim == 'RMSprop':
optimizer = optim.RMSprop(net.parameters(), lr=args.lr, weight_decay=args.l2)
elif args.optim == 'Adam':
optimizer = optim.Adam(net.parameters(), lr=args.lr, weight_decay=args.l2)
else:
raise ValueError('In-valid optimizer choice')
# ===== List for epoch-wise data ====== #
train_losses = []
val_losses = []
train_accs = []
val_accs = []
# ===================================== #
for epoch in range(args.epoch): # loop over the dataset multiple times
ts = time.time()
net, train_loss, train_acc = train(net, partition, optimizer, criterion, args)
val_loss, val_acc = validate(net, partition, criterion, args)
te = time.time()
# ====== Add Epoch Data ====== #
train_losses.append(train_loss)
val_losses.append(val_loss)
train_accs.append(train_acc)
val_accs.append(val_acc)
# ============================ #
print('Epoch {}, Acc(train/val): {:2.2f}/{:2.2f}, Loss(train/val) {:2.2f}/{:2.2f}. Took {:2.2f} sec'.format(epoch, train_acc, val_acc, train_loss, val_loss, te-ts))
test_acc = test(net, partition, args)
# ======= Add Result to Dictionary ======= #
result = {}
result['train_losses'] = train_losses
result['val_losses'] = val_losses
result['train_accs'] = train_accs
result['val_accs'] = val_accs
result['train_acc'] = train_acc
result['val_acc'] = val_acc
result['test_acc'] = test_acc
return vars(args), result
# ===================================== #
```
# Manage Experiment Result
```
import hashlib
import json
from os import listdir
from os.path import isfile, join
import pandas as pd
def save_exp_result(setting, result):
exp_name = setting['exp_name']
del setting['epoch']
del setting['test_batch_size']
hash_key = hashlib.sha1(str(setting).encode()).hexdigest()[:6]
filename = './results/{}-{}.json'.format(exp_name, hash_key)
result.update(setting)
with open(filename, 'w') as f:
json.dump(result, f)
def load_exp_result(exp_name):
dir_path = './results'
filenames = [f for f in listdir(dir_path) if isfile(join(dir_path, f)) if '.json' in f]
list_result = []
for filename in filenames:
if exp_name in filename:
with open(join(dir_path, filename), 'r') as infile:
results = json.load(infile)
list_result.append(results)
df = pd.DataFrame(list_result) # .drop(columns=[])
return df
```
## Experiment
```
# ====== Random Seed Initialization ====== #
seed = 123
np.random.seed(seed)
torch.manual_seed(seed)
parser = argparse.ArgumentParser()
args = parser.parse_args("")
args.exp_name = "exp1_n_layer_hid_dim"
# ====== Model Capacity ====== #
args.in_dim = 3072
args.out_dim = 10
args.hid_dim = 100
args.act = 'relu'
# ====== Regularization ======= #
args.dropout = 0.2
args.use_bn = True
args.l2 = 0.00001
args.use_xavier = True
# ====== Optimizer & Training ====== #
args.optim = 'RMSprop' #'RMSprop' #SGD, RMSprop, ADAM...
args.lr = 0.0015
args.epoch = 10
args.train_batch_size = 256
args.test_batch_size = 1024
# ====== Experiment Variable ====== #
name_var1 = 'n_layer'
name_var2 = 'hid_dim'
list_var1 = [1, 2, 3]
list_var2 = [500, 300]
for var1 in list_var1:
for var2 in list_var2:
setattr(args, name_var1, var1)
setattr(args, name_var2, var2)
print(args)
setting, result = experiment(partition, deepcopy(args))
save_exp_result(setting, result)
import seaborn as sns
import matplotlib.pyplot as plt
df = load_exp_result('exp1')
fig, ax = plt.subplots(1, 3)
fig.set_size_inches(15, 6)
sns.set_style("darkgrid", {"axes.facecolor": ".9"})
sns.barplot(x='n_layer', y='train_acc', hue='hid_dim', data=df, ax=ax[0])
sns.barplot(x='n_layer', y='val_acc', hue='hid_dim', data=df, ax=ax[1])
sns.barplot(x='n_layer', y='test_acc', hue='hid_dim', data=df, ax=ax[2])
var1 = 'n_layer'
var2 = 'hid_dim'
df = load_exp_result('exp1')
list_v1 = df[var1].unique()
list_v2 = df[var2].unique()
list_data = []
for value1 in list_v1:
for value2 in list_v2:
row = df.loc[df[var1]==value1]
row = row.loc[df[var2]==value2]
train_losses = list(row.train_losses)[0]
val_losses = list(row.val_losses)[0]
for epoch, train_loss in enumerate(train_losses):
list_data.append({'type':'train', 'loss':train_loss, 'epoch':epoch, var1:value1, var2:value2})
for epoch, val_loss in enumerate(val_losses):
list_data.append({'type':'val', 'loss':val_loss, 'epoch':epoch, var1:value1, var2:value2})
df = pd.DataFrame(list_data)
g = sns.FacetGrid(df, row=var2, col=var1, hue='type', margin_titles=True, sharey=False)
g = g.map(plt.plot, 'epoch', 'loss', marker='.')
g.add_legend()
g.fig.suptitle('Train loss vs Val loss')
plt.subplots_adjust(top=0.89)
var1 = 'n_layer'
var2 = 'hid_dim'
df = load_exp_result('exp1')
list_v1 = df[var1].unique()
list_v2 = df[var2].unique()
list_data = []
for value1 in list_v1:
for value2 in list_v2:
row = df.loc[df[var1]==value1]
row = row.loc[df[var2]==value2]
train_accs = list(row.train_accs)[0]
val_accs = list(row.val_accs)[0]
test_acc = list(row.test_acc)[0]
for epoch, train_acc in enumerate(train_accs):
list_data.append({'type':'train', 'Acc':train_acc, 'test_acc':test_acc, 'epoch':epoch, var1:value1, var2:value2})
for epoch, val_acc in enumerate(val_accs):
list_data.append({'type':'val', 'Acc':val_acc, 'test_acc':test_acc, 'epoch':epoch, var1:value1, var2:value2})
df = pd.DataFrame(list_data)
g = sns.FacetGrid(df, row=var2, col=var1, hue='type', margin_titles=True, sharey=False)
g = g.map(plt.plot, 'epoch', 'Acc', marker='.')
def show_acc(x, y, metric, **kwargs):
plt.scatter(x, y, alpha=0.3, s=1)
metric = "Test Acc: {:1.3f}".format(list(metric.values)[0])
plt.text(0.05, 0.95, metric, horizontalalignment='left', verticalalignment='center', transform=plt.gca().transAxes, bbox=dict(facecolor='yellow', alpha=0.5, boxstyle="round,pad=0.1"))
g = g.map(show_acc, 'epoch', 'Acc', 'test_acc')
g.add_legend()
g.fig.suptitle('Train Accuracy vs Val Accuracy')
plt.subplots_adjust(top=0.89)
```
| github_jupyter |
```
import pandas as pd
import numpy as np
df_properti = pd.read_csv("https://raw.githubusercontent.com/ardhiraka/PFDS_sources/master/property_data.csv")
df_properti
df_properti.shape
df_properti.columns
df_properti["ST_NAME"]
df_properti["ST_NUM"].isna()
list_missing_values = ["n/a", "--", "na"]
df_properti = pd.read_csv(
"https://raw.githubusercontent.com/ardhiraka/PFDS_sources/master/property_data.csv",
na_values = list_missing_values
)
df_properti
df_properti["OWN_OCCUPIED"].isna()
cnt=0
df_properti_own = df_properti["OWN_OCCUPIED"]
for row in df_properti_own:
try:
int(row)
df_properti[cnt, "OWN_OCCUPIED"]=np.nan
except ValueError:
pass
cnt+=1
df_properti
df_properti["NEW_OWN_OCCUPIEW"] = df_properti["OWN_OCCUPIED"].apply(
lambda val: 1 if val == "Y" else 0
)
df_properti
df_properti.isna().sum()
df_properti.isna().sum().sum()
df_properti
cnt=0
df_properti_num_bat = df_properti["NUM_BATH"]
for row in df_properti_num_bat:
try:
float(row)
df_properti.loc[cnt, "NEW_NUM_BATH"]=row
except ValueError:
df_properti.loc[cnt, "NEW_NUM_BATH"]=np.nan
cnt+=1
df_properti
df_properti["ST_NUM"].fillna(125)
obes = pd.ExcelFile("csv/obes.xls")
obes.sheet_names
obes_age = obes.parse("7.2", skiprows=4, skipfooter=14)
obes_age
obes_age.set_index('Year', inplace=True)
obes_age.plot()
obes_age.drop("Total", axis=1).plot()
from datetime import datetime
datetime.now().date()
opsd_daily = pd.read_csv(
'https://raw.githubusercontent.com/ardhiraka/PFDS_sources/master/opsd_germany_daily.csv',
index_col=0, parse_dates=True
)
opsd_daily.head()
opsd_daily['Year'] = opsd_daily.index.year
opsd_daily['Month'] = opsd_daily.index.month
opsd_daily['Weekday'] = opsd_daily.index.weekday
opsd_daily
opsd_daily["Consumption"].plot(
linewidth=.3,
figsize=(12, 5)
)
df_canada = pd.read_excel(
"https://github.com/ardhiraka/PFDS_sources/blob/master/Canada.xlsx?raw=true",
sheet_name="Canada by Citizenship",
skiprows=range(20),
skipfooter=2
)
df_canada.head()
df_canada.columns
df_canada.drop(
columns=[
"AREA", "REG", "DEV",
"Type", "Coverage"
],
axis=1,
inplace=True
)
df_canada.head()
df_canada.rename(
columns={
"OdName": "Country",
"AreaName": "Continent",
"RegName": "Region"
},
inplace=True
)
df_canada.head()
df_canada_total = df_canada.sum(axis=1)
df_canada["Total"] = df_canada_total
df_canada.head()
df_canada.describe()
df_canada.Country
df_canada[
[
"Country",
2000,
2001,
2002,
2003,
2004,
2005,
2006,
2007,
2008,
2009,
2010,
2011,
2012,
2013,
]
]
df_canada["Continent"] == "Africa"
df_canada[(df_canada["Continent"]=="Asia") & (df_canada["Region"]=="Southern Asia")]
```
| github_jupyter |
## 1、可视化DataGeneratorHomographyNet模块都干了什么
```
import glob
import os
import cv2
import numpy as np
from dataGenerator import DataGeneratorHomographyNet
img_dir = os.path.join(os.path.expanduser("~"), "/home/nvidia/test2017")
img_ext = ".jpg"
img_paths = glob.glob(os.path.join(img_dir, '*' + img_ext))
dg = DataGeneratorHomographyNet(img_paths, input_dim=(240, 240))
data, label = dg.__getitem__(0)
for idx in range(dg.batch_size):
cv2.imshow("orig", data[idx, :, :, 0])
cv2.imshow("transformed", data[idx, :, :, 1])
cv2.waitKey(0)
```
## 2、开始训练
```
import os
import glob
import datetime
import pandas as pd
import matplotlib.pyplot as plt
import keras
from keras.callbacks import ModelCheckpoint
from sklearn.model_selection import train_test_split
import tensorflow as tf
from homographyNet import HomographyNet
import dataGenerator as dg
keras.__version__
batch_size = 2
#取值0,1,2 0-安静模式 1-进度条 2-每一行都有输出
verbose = 1
#Epoch
nb_epo = 150
#计时开始
start_ts = datetime.datetime.now().strftime("%Y%m%d-%H%M%S")
#用于训练的图片目录
data_path = "/home/nvidia/test2017"
#模型保存的目录
model_dir = "/home/nvidia"
img_dir = os.path.join(os.path.expanduser("~"), data_path)
model_dir = os.path.join(os.path.expanduser("~"), model_dir, start_ts)
#以时间为名创建目录
if not os.path.exists(model_dir):
os.makedirs(model_dir)
img_ext = ".jpg"
#获取所有图像目录
img_paths = glob.glob(os.path.join(img_dir, '*' + img_ext))
input_size = (360, 360, 2)
#划分训练集和验证集,验证集搞小一点,不然每个epoch跑完太慢了
train_idx, val_idx = train_test_split(img_paths, test_size=0.01)
#拿到训练数据
train_dg = dg.DataGeneratorHomographyNet(train_idx, input_dim=input_size[0:2], batch_size=batch_size)
#拿到既定事实的标签
val_dg = dg.DataGeneratorHomographyNet(val_idx, input_dim=input_size[0:2], batch_size=batch_size)
#对于神经网络来说这个鬼一样的图就是输入,它自己从这幅图的左边和右边学习出单应性矩阵,神奇吧?
#修正网络输入头
homo_net = HomographyNet(input_size)
#实例化网络结构
model = homo_net.build_model()
#输出模型
model.summary()
#检查点回调,没写tensorboard的回调,真正的大师都是直接看loss输出的
checkpoint = ModelCheckpoint(
os.path.join(model_dir, 'model.h5'),
monitor='val_loss',
verbose=verbose,
save_best_only=True,
save_weights_only=False,
mode='auto'
)
#我嫌弃在上面改太麻烦,直接在这重定义了
#开始训练
#如果不加steps_per_epoch= 32, 就是每次全跑
history = model.fit_generator(train_dg,
validation_data = val_dg,
#steps_per_epoch = 32,
callbacks = [checkpoint],
epochs = 15,
verbose = 1)
```
```
#整个图看看
history_df = pd.DataFrame(history.history)
history_df.to_csv(os.path.join(model_dir, 'history.csv'))
history_df[['loss', 'val_loss']].plot()
history_df[['mean_squared_error', 'val_mean_squared_error']].plot()
plt.show()
```
## 预测&评估
```
TODO
```
| github_jupyter |
# Diamond Prices: Model Tuning and Improving Performance
#### Importing libraries
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
pd.options.mode.chained_assignment = None
%matplotlib inline
```
#### Loading the dataset
```
DATA_DIR = '../data'
FILE_NAME = 'diamonds.csv'
data_path = os.path.join(DATA_DIR, FILE_NAME)
diamonds = pd.read_csv(data_path)
```
#### Preparing the dataset
```
## Preparation done from Chapter 2
diamonds = diamonds.loc[(diamonds['x']>0) | (diamonds['y']>0)]
diamonds.loc[11182, 'x'] = diamonds['x'].median()
diamonds.loc[11182, 'z'] = diamonds['z'].median()
diamonds = diamonds.loc[~((diamonds['y'] > 30) | (diamonds['z'] > 30))]
diamonds = pd.concat([diamonds, pd.get_dummies(diamonds['cut'], prefix='cut', drop_first=True)], axis=1)
diamonds = pd.concat([diamonds, pd.get_dummies(diamonds['color'], prefix='color', drop_first=True)], axis=1)
diamonds = pd.concat([diamonds, pd.get_dummies(diamonds['clarity'], prefix='clarity', drop_first=True)], axis=1)
## Dimensionality reduction
from sklearn.decomposition import PCA
pca = PCA(n_components=1, random_state=123)
diamonds['dim_index'] = pca.fit_transform(diamonds[['x','y','z']])
diamonds.drop(['x','y','z'], axis=1, inplace=True)
diamonds.columns
```
#### Train-test split
```
X = diamonds.drop(['cut','color','clarity','price'], axis=1)
y = diamonds['price']
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1, random_state=7)
```
#### Standarization: centering and scaling
```
numerical_features = ['carat', 'depth', 'table', 'dim_index']
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train[numerical_features])
X_train.loc[:, numerical_features] = scaler.fit_transform(X_train[numerical_features])
X_test.loc[:, numerical_features] = scaler.transform(X_test[numerical_features])
```
## Optimizing a single hyper-parameter
```
X_train, X_val, y_train, y_val = train_test_split(X_train, y_train, test_size=0.1, random_state=13)
from sklearn.neighbors import KNeighborsRegressor
from sklearn.metrics import mean_absolute_error
candidates = np.arange(4,16)
mae_metrics = []
for k in candidates:
model = KNeighborsRegressor(n_neighbors=k, weights='distance', metric='minkowski', leaf_size=50, n_jobs=4)
model.fit(X_train, y_train)
y_pred = model.predict(X_val)
metric = mean_absolute_error(y_true=y_val, y_pred=y_pred)
mae_metrics.append(metric)
fig, ax = plt.subplots(figsize=(8,5))
ax.plot(candidates, mae_metrics, "o-")
ax.set_xlabel('Hyper-parameter K', fontsize=14)
ax.set_ylabel('MAE', fontsize=14)
ax.set_xticks(candidates)
ax.grid();
```
#### Recalculating train-set split
```
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1, random_state=7)
scaler = StandardScaler()
scaler.fit(X_train[numerical_features])
X_train.loc[:, numerical_features] = scaler.fit_transform(X_train[numerical_features])
X_test.loc[:, numerical_features] = scaler.transform(X_test[numerical_features])
```
#### Optimizing with cross-validation
```
from sklearn.model_selection import cross_val_score
candidates = np.arange(4,16)
mean_mae = []
std_mae = []
for k in candidates:
model = KNeighborsRegressor(n_neighbors=k, weights='distance', metric='minkowski', leaf_size=50, n_jobs=4)
cv_results = cross_val_score(model, X_train, y_train, scoring='neg_mean_absolute_error', cv=10)
mean_score, std_score = -1*cv_results.mean(), cv_results.std()
mean_mae.append(mean_score)
std_mae.append(std_score)
fig, ax = plt.subplots(figsize=(8,5))
ax.plot(candidates, mean_mae, "o-")
ax.set_xlabel('Hyper-parameter K', fontsize=14)
ax.set_ylabel('Mean MAE', fontsize=14)
ax.set_xticks(candidates)
ax.grid();
fig, ax = plt.subplots(figsize=(8,5))
ax.plot(candidates, std_mae, "o-")
ax.set_xlabel('Hyper-parameter K', fontsize=14)
ax.set_ylabel('Standard deviation of MAE', fontsize=14)
ax.set_xticks(candidates)
ax.grid();
```
# Improving Performance
## Improving our diamond price predictions
### Fitting a neural network
```
from keras.models import Sequential
from keras.layers import Dense
n_input = X_train.shape[1]
n_hidden1 = 32
n_hidden2 = 16
n_hidden3 = 8
nn_reg = Sequential()
nn_reg.add(Dense(units=n_hidden1, activation='relu', input_shape=(n_input,)))
nn_reg.add(Dense(units=n_hidden2, activation='relu'))
nn_reg.add(Dense(units=n_hidden3, activation='relu'))
# output layer
nn_reg.add(Dense(units=1, activation=None))
batch_size = 32
n_epochs = 40
nn_reg.compile(loss='mean_absolute_error', optimizer='adam')
nn_reg.fit(X_train, y_train, epochs=n_epochs, batch_size=batch_size, validation_split=0.05)
y_pred = nn_reg.predict(X_test).flatten()
mae_neural_net = mean_absolute_error(y_test, y_pred)
print("MAE Neural Network: {:0.2f}".format(mae_neural_net))
```
### Transforming the target
```
diamonds['price'].hist(bins=25, ec='k', figsize=(8,5))
plt.title("Distribution of diamond prices", fontsize=16)
plt.grid(False);
y_train = np.log(y_train)
pd.Series(y_train).hist(bins=25, ec='k', figsize=(8,5))
plt.title("Distribution of log diamond prices", fontsize=16)
plt.grid(False);
nn_reg = Sequential()
nn_reg.add(Dense(units=n_hidden1, activation='relu', input_shape=(n_input,)))
nn_reg.add(Dense(units=n_hidden2, activation='relu'))
nn_reg.add(Dense(units=n_hidden3, activation='relu'))
# output layer
nn_reg.add(Dense(units=1, activation=None))
batch_size = 32
n_epochs = 40
nn_reg.compile(loss='mean_absolute_error', optimizer='adam')
nn_reg.fit(X_train, y_train, epochs=n_epochs, batch_size=batch_size, validation_split=0.05)
y_pred = nn_reg.predict(X_test).flatten()
y_pred = np.exp(y_pred)
mae_neural_net2 = mean_absolute_error(y_test, y_pred)
print("MAE Neural Network (modified target): {:0.2f}".format(mae_neural_net2))
100*(mae_neural_net - mae_neural_net2)/mae_neural_net2
```
#### Analyzing the results
```
fig, ax = plt.subplots(figsize=(8,5))
residuals = y_test - y_pred
ax.scatter(y_test, residuals, s=3)
ax.set_title('Residuals vs. Observed Prices', fontsize=16)
ax.set_xlabel('Observed prices', fontsize=14)
ax.set_ylabel('Residuals', fontsize=14)
ax.grid();
mask_7500 = y_test <=7500
mae_neural_less_7500 = mean_absolute_error(y_test[mask_7500], y_pred[mask_7500])
print("MAE considering price <= 7500: {:0.2f}".format(mae_neural_less_7500))
fig, ax = plt.subplots(figsize=(8,5))
percent_residuals = (y_test - y_pred)/y_test
ax.scatter(y_test, percent_residuals, s=3)
ax.set_title('Pecent residuals vs. Observed Prices', fontsize=16)
ax.set_xlabel('Observed prices', fontsize=14)
ax.set_ylabel('Pecent residuals', fontsize=14)
ax.axhline(y=0.15, color='r'); ax.axhline(y=-0.15, color='r');
ax.grid();
```
| github_jupyter |
# Visualizing COVID-19 Hospital Dataset with Seaborn
**Pre-Work:**
1. Ensure that Jupyter Notebook, Python 3, and seaborn (which will also install dependency libraries if not already installed) are installed. (See resources below for installation instructions.)
### **Instructions:**
1. Using Python, import main visualization library, `seaborn`, and its dependencies: `pandas`, `numpy`, and `matplotlib`.
2. Define dataset and read in data using pandas function, `read_json()`. [Notes: a) we're reading in data as an API endpoint; for more about this, see associated workshop slides or resources at bottom of notebook. b) If, instead, you prefer to use your own data, see comment with alternative for `read_csv()`.]
3. Check data has been read is as expected using `head()` function.
4. Graph two variables with `seaborn`as a lineplot using the `lineplot()` function.
5. Graph these same variables, plus a third, from the source dataset with `seaborn` as a scatterplot using the `relplot()` function.
6. See additional methods, using filtered data and other graphs. Feel free to open a new notebook, and try out your own ideas, using different variables or charts. (Or try out your own data!)
7. When ready, save figure using `matplotlib`'s `savefig`.
**Note:**
*If you're new to Jupyter Notebook, see resources below.*
### **Data source:**
[COVID-19 Reported Patient Impact and Hospital Capacity by State Timeseries](https://healthdata.gov/Hospital/COVID-19-Reported-Patient-Impact-and-Hospital-Capa/g62h-syeh)," created by the U.S. Department of Health & Human Services, on [HealthData.gov](https://healthdata.gov/).
```
# import libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# read JSON data in via healthdata.gov's API endpoint - https://healthdata.gov/resource/g62h-syeh.json?$limit=50000
# because the SODA API defaults to 1,000 rows, we're going to change that with the $limit parameter
# define data as 'covid' and set equal to read function
# if we want filtered data to compare to, define more datasets
covid = pd.read_json("https://healthdata.gov/resource/g62h-syeh.json?$limit=50000")
covid_ct = pd.read_json("https://healthdata.gov/resource/g62h-syeh.json?state=CT")
covid_maytopresent = pd.read_json("https://healthdata.gov/resource/g62h-syeh.json?$limit=50000&$where=date%20between%20%272021-05-01T12:00:00%27%20and%20%272021-08-01T12:00:00%27")
# if you want to read in your own data, see resources below, or if you have a CSV, try: mydata = pd.read_csv('')
# and add data filepath inside ''
# be sure to change covid to mydata in code below
# use head function and call our dataset (covid) to see the first few rows
# the default argument for this function is 5 rows, but you can set this to anything, e.g. covid.head(20)
covid.head()
# example of head with more rows
covid_ct.head(20)
# use seaborn to plot inpatient beds used versus whether a critical staffing shortage is occuring
# we also need to tell seaborn what dataset to use; in this case it's 'covid' as defined above
# variables: inpatient_beds_used_covid; critical_staffing_shortage_today_yes
sns.lineplot(x='inpatient_beds_used_covid', y="critical_staffing_shortage_today_yes", data=covid)
# save and name fig; uncomment below to run
# plt.savefig('covid_lineplot.png')
# use seaborn to plot inpatient beds used versus whether a critical staffing shortage is occuring
# this time, with a bar plot
# variables: inpatient_beds_used_covid; critical_staffing_shortage_today_yes
sns.barplot(x='inpatient_beds_used_covid', y="critical_staffing_shortage_today_yes", data=covid)
# save and name fig; uncomment below to run
# plt.savefig('covid_barplot.png')
# now we're going to try another graph type, a relational graph that will be scatterplot, with the same variables
# and add one more variable, deaths_covid, to color dots based on prevalance of COVID-19 deaths by setting hue
# though feel free to try new variables by browsing them here (scroll down to Columns in this Dataset): https://healthdata.gov/Hospital/COVID-19-Reported-Patient-Impact-and-Hospital-Capa/g62h-syeh
# variables: inpatient_beds_used_covid; critical_staffing_shortage_today_yes; deaths_covid
sns.relplot(x='inpatient_beds_used_covid', y="critical_staffing_shortage_today_yes", hue="deaths_covid", data=covid)
# save and name fig; uncomment below to run
# plt.savefig('covid_scatterplot.png')
# now let's try some graphs with the more limited datasets above, for instance, just the CT data
sns.relplot(x='inpatient_beds_used_covid', y="critical_staffing_shortage_today_yes", hue="deaths_covid", data=covid_ct)
# or just the May - August (present) 2021 date range
sns.relplot(x='inpatient_beds_used_covid', y="critical_staffing_shortage_today_yes", hue="deaths_covid", data=covid_maytopresent)
```
### Final Note:
It's important to remember that we can't necessarily infer any causation or directionality from these charts, but they can be a good place to start for further analysis and exploration, and can point us in the right direction of where to apply more advanced statistical methods, such as linear regression. Even with more advanced methods, though, we still want to stick the principles we're using here: keep charts as simple as possible, using only a few variables, and adding color only where needed. We want our charts to be readable and understandable -- see resources below for more advice and guidance on this.
Ultimately, these quick-start methods are helpful for idea generation and early investigation, and can get that process up and running quickly.
#### Code/Tools Resources:
- Jupyter notebook - about: https://jupyter-notebook.readthedocs.io/en/stable/notebook.html#introduction
- Jupyter notebook - how to use this tool: https://jupyter-notebook.readthedocs.io/en/stable/notebook.html
- Python: https://www.python.org/
- Seaborn: https://seaborn.pydata.org/index.html
- Seaborn tutorial: https://seaborn.pydata.org/tutorial.html
- Seaborn gallery: https://seaborn.pydata.org/examples/index.html
- Seaborn `lineplot()` function: https://seaborn.pydata.org/generated/seaborn.lineplot.html#seaborn.lineplot + https://seaborn.pydata.org/examples/errorband_lineplots.html
- Seaborn `relplot()` function: https://seaborn.pydata.org/generated/seaborn.relplot.html#seaborn.relplot + https://seaborn.pydata.org/examples/faceted_lineplot.html
- Pandas: https://pandas.pydata.org/
- Pandas - how to read / write tabular data: https://pandas.pydata.org/docs/getting_started/intro_tutorials/02_read_write.html
- Pandas `read.json()` function: https://pandas.pydata.org/docs/reference/api/pandas.io.json.read_json.html?highlight=read_json#pandas.io.json.read_json
- Pandas `head()` function: https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.head.html?highlight=head#pandas.DataFrame.head
- Matplotlib: https://matplotlib.org/
- Matplotlib `savefig` function: https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.savefig.html
- Socrata Open Data API (SODA) Docs: https://dev.socrata.com/
- SODA Docs for [Dataset](https://healthdata.gov/Hospital/COVID-19-Reported-Patient-Impact-and-Hospital-Capa/g62h-syeh): https://dev.socrata.com/foundry/healthdata.gov/g62h-syeh
- SODA Docs - what is an endpoint: https://dev.socrata.com/docs/endpoints.html
#### Visualization Resources:
- 10 Simple Rules for Better Figures | *PLOS Comp Bio*: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003833
- How to Choose the Right Data Visualization | *Chartio*: https://chartio.com/learn/charts/how-to-choose-data-visualization/
#### Additional Note:
This notebook was created by Kaitlin Throgmorton for a data analysis workshop, as part of an interview for Yale University.
| github_jupyter |
# Temporal-Difference Methods
In this notebook, you will write your own implementations of many Temporal-Difference (TD) methods.
While we have provided some starter code, you are welcome to erase these hints and write your code from scratch.
---
### Part 0: Explore CliffWalkingEnv
We begin by importing the necessary packages.
```
import sys
import gym
import numpy as np
import random
import math
from collections import defaultdict, deque
import matplotlib.pyplot as plt
%matplotlib inline
import check_test
from plot_utils import plot_values
```
Use the code cell below to create an instance of the [CliffWalking](https://github.com/openai/gym/blob/master/gym/envs/toy_text/cliffwalking.py) environment.
```
env = gym.make('CliffWalking-v0')
```
The agent moves through a $4\times 12$ gridworld, with states numbered as follows:
```
[[ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11],
[12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23],
[24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35],
[36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47]]
```
At the start of any episode, state `36` is the initial state. State `47` is the only terminal state, and the cliff corresponds to states `37` through `46`.
The agent has 4 potential actions:
```
UP = 0
RIGHT = 1
DOWN = 2
LEFT = 3
```
Thus, $\mathcal{S}^+=\{0, 1, \ldots, 47\}$, and $\mathcal{A} =\{0, 1, 2, 3\}$. Verify this by running the code cell below.
```
print(env.action_space)
print(env.observation_space)
```
In this mini-project, we will build towards finding the optimal policy for the CliffWalking environment. The optimal state-value function is visualized below. Please take the time now to make sure that you understand _why_ this is the optimal state-value function.
_**Note**: You can safely ignore the values of the cliff "states" as these are not true states from which the agent can make decisions. For the cliff "states", the state-value function is not well-defined._
```
# define the optimal state-value function
V_opt = np.zeros((4,12))
V_opt[0][0:13] = -np.arange(3, 15)[::-1]
V_opt[1][0:13] = -np.arange(3, 15)[::-1] + 1
V_opt[2][0:13] = -np.arange(3, 15)[::-1] + 2
V_opt[3][0] = -13
plot_values(V_opt)
```
### Part 1: TD Control: Sarsa
In this section, you will write your own implementation of the Sarsa control algorithm.
Your algorithm has four arguments:
- `env`: This is an instance of an OpenAI Gym environment.
- `num_episodes`: This is the number of episodes that are generated through agent-environment interaction.
- `alpha`: This is the step-size parameter for the update step.
- `gamma`: This is the discount rate. It must be a value between 0 and 1, inclusive (default value: `1`).
The algorithm returns as output:
- `Q`: This is a dictionary (of one-dimensional arrays) where `Q[s][a]` is the estimated action value corresponding to state `s` and action `a`.
Please complete the function in the code cell below.
(_Feel free to define additional functions to help you to organize your code._)
```
def update_Q_sarsa(alpha, gamma, Q, state, action, reward, next_state=None, next_action=None):
"""Returns updated Q-value for the most recent experience."""
current = Q[state][action] # estimate in Q-table (for current state, action pair)
# get value of state, action pair at next time step
Qsa_next = Q[next_state][next_action] if next_state is not None else 0
target = reward + (gamma * Qsa_next) # construct TD target
new_value = current + (alpha * (target - current)) # get updated value
return new_value
def epsilon_greedy(Q, state, nA, eps):
"""Selects epsilon-greedy action for supplied state.
Params
======
Q (dictionary): action-value function
state (int): current state
nA (int): number actions in the environment
eps (float): epsilon
"""
if random.random() > eps: # select greedy action with probability epsilon
return np.argmax(Q[state])
else: # otherwise, select an action randomly
return random.choice(np.arange(env.action_space.n))
def sarsa(env, num_episodes, alpha, gamma=1.0, plot_every=100):
nA = env.action_space.n # number of actions
Q = defaultdict(lambda: np.zeros(nA)) # initialize empty dictionary of arrays
# monitor performance
tmp_scores = deque(maxlen=plot_every) # deque for keeping track of scores
avg_scores = deque(maxlen=num_episodes) # average scores over every plot_every episodes
for i_episode in range(1, num_episodes+1):
# monitor progress
if i_episode % 100 == 0:
print("\rEpisode {}/{}".format(i_episode, num_episodes), end="")
sys.stdout.flush()
score = 0 # initialize score
state = env.reset() # start episode
eps = 1.0 / i_episode # set value of epsilon
action = epsilon_greedy(Q, state, nA, eps) # epsilon-greedy action selection
while True:
next_state, reward, done, info = env.step(action) # take action A, observe R, S'
score += reward # add reward to agent's score
if not done:
next_action = epsilon_greedy(Q, next_state, nA, eps) # epsilon-greedy action
Q[state][action] = update_Q_sarsa(alpha, gamma, Q, \
state, action, reward, next_state, next_action)
state = next_state # S <- S'
action = next_action # A <- A'
if done:
Q[state][action] = update_Q_sarsa(alpha, gamma, Q, \
state, action, reward)
tmp_scores.append(score) # append score
break
if (i_episode % plot_every == 0):
avg_scores.append(np.mean(tmp_scores))
# plot performance
plt.plot(np.linspace(0,num_episodes,len(avg_scores),endpoint=False), np.asarray(avg_scores))
plt.xlabel('Episode Number')
plt.ylabel('Average Reward (Over Next %d Episodes)' % plot_every)
plt.show()
# print best 100-episode performance
print(('Best Average Reward over %d Episodes: ' % plot_every), np.max(avg_scores))
return Q
```
Use the next code cell to visualize the **_estimated_** optimal policy and the corresponding state-value function.
If the code cell returns **PASSED**, then you have implemented the function correctly! Feel free to change the `num_episodes` and `alpha` parameters that are supplied to the function. However, if you'd like to ensure the accuracy of the unit test, please do not change the value of `gamma` from the default.
```
# obtain the estimated optimal policy and corresponding action-value function
Q_sarsa = sarsa(env, 50000, .01)
# print the estimated optimal policy
policy_sarsa = np.array([np.argmax(Q_sarsa[key]) if key in Q_sarsa else -1 for key in np.arange(48)]).reshape(4,12)
check_test.run_check('td_control_check', policy_sarsa)
print("\nEstimated Optimal Policy (UP = 0, RIGHT = 1, DOWN = 2, LEFT = 3, N/A = -1):")
print(policy_sarsa)
# plot the estimated optimal state-value function
V_sarsa = ([np.max(Q_sarsa[key]) if key in Q_sarsa else 0 for key in np.arange(48)])
plot_values(V_sarsa)
```
### Part 2: TD Control: Q-learning
In this section, you will write your own implementation of the Q-learning control algorithm.
Your algorithm has four arguments:
- `env`: This is an instance of an OpenAI Gym environment.
- `num_episodes`: This is the number of episodes that are generated through agent-environment interaction.
- `alpha`: This is the step-size parameter for the update step.
- `gamma`: This is the discount rate. It must be a value between 0 and 1, inclusive (default value: `1`).
The algorithm returns as output:
- `Q`: This is a dictionary (of one-dimensional arrays) where `Q[s][a]` is the estimated action value corresponding to state `s` and action `a`.
Please complete the function in the code cell below.
(_Feel free to define additional functions to help you to organize your code._)
```
def update_Q_sarsamax(alpha, gamma, Q, state, action, reward, next_state=None):
"""Returns updated Q-value for the most recent experience."""
current = Q[state][action] # estimate in Q-table (for current state, action pair)
Qsa_next = np.max(Q[next_state]) if next_state is not None else 0 # value of next state
target = reward + (gamma * Qsa_next) # construct TD target
new_value = current + (alpha * (target - current)) # get updated value
return new_value
def q_learning(env, num_episodes, alpha, gamma=1.0, plot_every=100):
"""Q-Learning - TD Control
Params
======
num_episodes (int): number of episodes to run the algorithm
alpha (float): learning rate
gamma (float): discount factor
plot_every (int): number of episodes to use when calculating average score
"""
nA = env.action_space.n # number of actions
Q = defaultdict(lambda: np.zeros(nA)) # initialize empty dictionary of arrays
# monitor performance
tmp_scores = deque(maxlen=plot_every) # deque for keeping track of scores
avg_scores = deque(maxlen=num_episodes) # average scores over every plot_every episodes
for i_episode in range(1, num_episodes+1):
# monitor progress
if i_episode % 100 == 0:
print("\rEpisode {}/{}".format(i_episode, num_episodes), end="")
sys.stdout.flush()
score = 0 # initialize score
state = env.reset() # start episode
eps = 1.0 / i_episode # set value of epsilon
while True:
action = epsilon_greedy(Q, state, nA, eps) # epsilon-greedy action selection
next_state, reward, done, info = env.step(action) # take action A, observe R, S'
score += reward # add reward to agent's score
Q[state][action] = update_Q_sarsamax(alpha, gamma, Q, \
state, action, reward, next_state)
state = next_state # S <- S'
if done:
tmp_scores.append(score) # append score
break
if (i_episode % plot_every == 0):
avg_scores.append(np.mean(tmp_scores))
# plot performance
plt.plot(np.linspace(0,num_episodes,len(avg_scores),endpoint=False), np.asarray(avg_scores))
plt.xlabel('Episode Number')
plt.ylabel('Average Reward (Over Next %d Episodes)' % plot_every)
plt.show()
# print best 100-episode performance
print(('Best Average Reward over %d Episodes: ' % plot_every), np.max(avg_scores))
return Q
```
Use the next code cell to visualize the **_estimated_** optimal policy and the corresponding state-value function.
If the code cell returns **PASSED**, then you have implemented the function correctly! Feel free to change the `num_episodes` and `alpha` parameters that are supplied to the function. However, if you'd like to ensure the accuracy of the unit test, please do not change the value of `gamma` from the default.
```
# obtain the estimated optimal policy and corresponding action-value function
Q_sarsamax = q_learning(env, 5000, .01)
# print the estimated optimal policy
policy_sarsamax = np.array([np.argmax(Q_sarsamax[key]) if key in Q_sarsamax else -1 for key in np.arange(48)]).reshape((4,12))
check_test.run_check('td_control_check', policy_sarsamax)
print("\nEstimated Optimal Policy (UP = 0, RIGHT = 1, DOWN = 2, LEFT = 3, N/A = -1):")
print(policy_sarsamax)
# plot the estimated optimal state-value function
plot_values([np.max(Q_sarsamax[key]) if key in Q_sarsamax else 0 for key in np.arange(48)])
```
### Part 3: TD Control: Expected Sarsa
In this section, you will write your own implementation of the Expected Sarsa control algorithm.
Your algorithm has four arguments:
- `env`: This is an instance of an OpenAI Gym environment.
- `num_episodes`: This is the number of episodes that are generated through agent-environment interaction.
- `alpha`: This is the step-size parameter for the update step.
- `gamma`: This is the discount rate. It must be a value between 0 and 1, inclusive (default value: `1`).
The algorithm returns as output:
- `Q`: This is a dictionary (of one-dimensional arrays) where `Q[s][a]` is the estimated action value corresponding to state `s` and action `a`.
Please complete the function in the code cell below.
(_Feel free to define additional functions to help you to organize your code._)
```
def update_Q_expsarsa(alpha, gamma, nA, eps, Q, state, action, reward, next_state=None):
"""Returns updated Q-value for the most recent experience."""
current = Q[state][action] # estimate in Q-table (for current state, action pair)
policy_s = np.ones(nA) * eps / nA # current policy (for next state S')
policy_s[np.argmax(Q[next_state])] = 1 - eps + (eps / nA) # greedy action
Qsa_next = np.dot(Q[next_state], policy_s) # get value of state at next time step
target = reward + (gamma * Qsa_next) # construct target
new_value = current + (alpha * (target - current)) # get updated value
return new_value
def expected_sarsa(env, num_episodes, alpha, gamma=1.0, plot_every=100):
"""Expected SARSA - TD Control
Params
======
num_episodes (int): number of episodes to run the algorithm
alpha (float): step-size parameters for the update step
gamma (float): discount factor
plot_every (int): number of episodes to use when calculating average score
"""
nA = env.action_space.n # number of actions
Q = defaultdict(lambda: np.zeros(nA)) # initialize empty dictionary of arrays
# monitor performance
tmp_scores = deque(maxlen=plot_every) # deque for keeping track of scores
avg_scores = deque(maxlen=num_episodes) # average scores over every plot_every episodes
for i_episode in range(1, num_episodes+1):
# monitor progress
if i_episode % 100 == 0:
print("\rEpisode {}/{}".format(i_episode, num_episodes), end="")
sys.stdout.flush()
score = 0 # initialize score
state = env.reset() # start episode
eps = 0.005 # set value of epsilon
while True:
action = epsilon_greedy(Q, state, nA, eps) # epsilon-greedy action selection
next_state, reward, done, info = env.step(action) # take action A, observe R, S'
score += reward # add reward to agent's score
# update Q
Q[state][action] = update_Q_expsarsa(alpha, gamma, nA, eps, Q, \
state, action, reward, next_state)
state = next_state # S <- S'
if done:
tmp_scores.append(score) # append score
break
if (i_episode % plot_every == 0):
avg_scores.append(np.mean(tmp_scores))
# plot performance
plt.plot(np.linspace(0,num_episodes,len(avg_scores),endpoint=False), np.asarray(avg_scores))
plt.xlabel('Episode Number')
plt.ylabel('Average Reward (Over Next %d Episodes)' % plot_every)
plt.show()
# print best 100-episode performance
print(('Best Average Reward over %d Episodes: ' % plot_every), np.max(avg_scores))
return Q
```
Use the next code cell to visualize the **_estimated_** optimal policy and the corresponding state-value function.
If the code cell returns **PASSED**, then you have implemented the function correctly! Feel free to change the `num_episodes` and `alpha` parameters that are supplied to the function. However, if you'd like to ensure the accuracy of the unit test, please do not change the value of `gamma` from the default.
```
# obtain the estimated optimal policy and corresponding action-value function
Q_expsarsa = expected_sarsa(env, 50000, 1)
# print the estimated optimal policy
policy_expsarsa = np.array([np.argmax(Q_expsarsa[key]) if key in Q_expsarsa else -1 for key in np.arange(48)]).reshape(4,12)
check_test.run_check('td_control_check', policy_expsarsa)
print("\nEstimated Optimal Policy (UP = 0, RIGHT = 1, DOWN = 2, LEFT = 3, N/A = -1):")
print(policy_expsarsa)
# plot the estimated optimal state-value function
plot_values([np.max(Q_expsarsa[key]) if key in Q_expsarsa else 0 for key in np.arange(48)])
```
| github_jupyter |
```
import re
import pandas as pd
import os
import html
os.chdir('/Users/lindsayduca/Desktop/Downloads')
#file="US20220000001A1-20220106.XML"
#file="USD0864516-20191029.XML"
file = open(file="ipa220106.txt", mode='r') #opening the file in read mode
file_content_raw = file.read()
file.close()
text1=re.compile("<\?xml version=\"1\.0\" encoding\=\"UTF\-8\"\?>")
file_content = text1.split(file_content_raw)
while '' in file_content:
file_content.remove('')
print("No of patents :", len(file_content))
# Writing regular expressions for the desired columns
#grant_id = re.compile('file\=\"([U][S]\w\w\d{6})\-\d{8}\.XML\"')
grant_id = re.compile('file\=\"([U][S]\w{13})\-\w{8}\.[X][M][L]\"')
#patent_title = re.compile("<invention-title id=\"\w{5,6}\">(.*?)</invention-title>")
kind = re.compile("<kind>([A-Z]\d)</kind>")
#number_of_claim = re.compile("\<number\-of\-claims\>(\d{1,4})\<\/number\-of\-claims\>")
first_name=re.compile("<first-name>(.*?)</first-name>")
last_name=re.compile("<last-name>(.*?)</last-name>")
citation_by_examiner = re.compile("\<category\>cited by examiner<\/category\>")
citation_by_applicant = re.compile("\<category>cited by applicant\<\/category\>")
claim_text=re.compile("<claim-text>[\s\S<]*</claim-text>")
abstract=re.compile("\<abstract id\=\"abstract\"\>\n\<p id\=\"p\-0001\" num\=\"0000\"\>(.*?)\<\/p\>\n\<\/abstract\>")
# some extra care with claim-text
cleaner = re.compile('<.*?>')
cleaner2 = re.compile('\n')
cleaner3 = re.compile('\,\,\,')
cleaner4 = re.compile("[\.][\,][\,]")
cleaner5 = re.compile("[\,][\,]")
cleaner6 = re.compile("[\;][\,]")
for line in file_content:
gid=grant_id.findall(line) #to find grant_id
# checking length of gid is not equal to 0 then do append to all the lists
if len(gid)!=0:
gid_list.append(gid[0])
data_frame = pd.DataFrame(
{'grant_id': gid_list})
data_frame
gid_list, title_list, kind_list, no_of_claim_list, name_list, applicant_list, examiners_list, claim_list, abstract_list, = ([] for i in range(9))
for line in file_content:
gid=grant_id.findall(line) #to find grant_id
title=patent_title.findall(line) #to find patent_title
kinds=kind.findall(line) #to find kind
# sclaim=number_of_claim.findall(line) #to find no_of_claims
#to find inventors
inventors=re.findall("<inventor.*?>[\s\S]*</inventor>",line)
for person in inventors:
first=first_name.findall(person)
last=last_name.findall(person)
name = [firstName +" "+ lastName for firstName, lastName in zip(first,last)]
if len(name)==0:
names="NA"
else:
names=name
# here we count citation_by_applicant
if len(citation_by_applicant.findall(line))==0:
citation_by_applicants=0
else:
citation_by_applicants=len(citation_by_applicant.findall(line))
# count for citation_by_examiner
if len(citation_by_examiner.findall(line))==0:
citation_by_examiners=0
else:
citation_by_examiners=len(citation_by_examiner.findall(line))
# For claim_text
if (len(re.findall("<claim-text>[\s\S<]*</claim-text>",line))==0):
claim_text=["NA"]
else:
claim_text=re.findall("<claim-text>[\s\S<]*</claim-text>",line)
# For abstract
abst=abstract.findall(line)
if len(abst)==0:
abstracts=["NA"]
else:
abstracts=abst
# checking length of gid is not equal to 0 then do append to all the lists
if len(gid)!=0:
gid_list.append(gid[0])
# title_list.append(title[0])
kind_list.append(kinds[0])
# no_of_claim_list.append(sclaim[0])
name_list.append(names)
applicant_list.append(citation_by_applicants)
examiners_list.append(citation_by_examiners)
claim_list.append(claim_text[0])
abstract_list.append(abstracts[0])
"""
# cleaning claim text
element=0
for items in claim_list:
claim_list[element]=re.sub(cleaner,'',claim_list[element])
claim_list[element]=re.sub(cleaner2,',',claim_list[element])
claim_list[element]=re.sub(cleaner3,',',claim_list[element])
claim_list[element]=re.sub(cleaner4,'.,',claim_list[element])
claim_list[element]=re.sub(cleaner5,',',claim_list[element])
claim_list[element]=re.sub(cleaner6,'; ',claim_list[element])
element=element+1
"""
# For kind
Kind1 = [w.replace('P2', 'Plant Patent Grant(with a published application) issued on or after January 2, 2001') for w in kind_list]
Kind2 = [w.replace('B2', 'Utility Patent Grant (with a published application) issued on or after January 2, 2001.') for w in Kind1]
Kind3 = [w.replace('S1', 'Design Patent') for w in Kind2]
Kind4 = [w.replace('B1', 'Utility Patent Grant (no published application) issued on or after January 2, 2001.') for w in Kind3]
# Creating data frame
data_frame = pd.DataFrame(
{'grant_id': gid_list,
#'patent_title': title_list,
'kind': Kind4,
#'number_of_claims':no_of_claim_list,
'inventors':name_list,
'citations_applicant_count':applicant_list,
'citations_examiner_count':examiners_list,
'claims_text':claim_list,
'abstract':abstract_list
})
data_frame.isnull().sum()
data_frame
data_frame.to_csv('ParsedPatentGrant.csv')
import requests
from bs4 import BeautifulSoup
import json
r = requests.get("https://developer.uspto.gov/ibd-api/v1/patent/application?patentNumber=9876543&start=0&rows=100")
soup = BeautifulSoup(r.text, "html.parser")
text = soup.get_text()
r_dict = json.loads(str(text))
print(r_dict['response']['docs'][0]['inventor'][0])
```
| github_jupyter |
# Loads pre-trained model and get prediction on validation samples
### 1. Info
Please provide path to the relevant config file
```
config_file_path = "../configs/pretrained/config_model1.json"
```
### 2. Importing required modules
```
import os
import cv2
import sys
import importlib
import torch
import torchvision
import numpy as np
sys.path.insert(0, "../")
# imports for displaying a video an IPython cell
import io
import base64
from IPython.display import HTML
from data_parser import WebmDataset
from data_loader_av import VideoFolder
from models.multi_column import MultiColumn
from transforms_video import *
from utils import load_json_config, remove_module_from_checkpoint_state_dict
from pprint import pprint
```
### 3. Loading configuration file, model definition and its path
```
# Load config file
config = load_json_config(config_file_path)
# set column model
column_cnn_def = importlib.import_module("{}".format(config['conv_model']))
model_name = config["model_name"]
print("=> Name of the model -- {}".format(model_name))
# checkpoint path to a trained model
checkpoint_path = os.path.join("../", config["output_dir"], config["model_name"], "model_best.pth.tar")
print("=> Checkpoint path --> {}".format(checkpoint_path))
```
### 3. Load model
_Note: without cuda() for ease_
```
model = MultiColumn(config['num_classes'], column_cnn_def.Model, int(config["column_units"]))
model.eval();
print("=> loading checkpoint")
checkpoint = torch.load(checkpoint_path)
checkpoint['state_dict'] = remove_module_from_checkpoint_state_dict(
checkpoint['state_dict'])
model.load_state_dict(checkpoint['state_dict'])
print("=> loaded checkpoint '{}' (epoch {})"
.format(checkpoint_path, checkpoint['epoch']))
```
### 4. Load data
```
# Center crop videos during evaluation
transform_eval_pre = ComposeMix([
[Scale(config['input_spatial_size']), "img"],
[torchvision.transforms.ToPILImage(), "img"],
[torchvision.transforms.CenterCrop(config['input_spatial_size']), "img"]
])
transform_post = ComposeMix([
[torchvision.transforms.ToTensor(), "img"],
[torchvision.transforms.Normalize(
mean=[0.485, 0.456, 0.406], # default values for imagenet
std=[0.229, 0.224, 0.225]), "img"]
])
val_data = VideoFolder(root=config['data_folder'],
json_file_input=config['json_data_val'],
json_file_labels=config['json_file_labels'],
clip_size=config['clip_size'],
nclips=config['nclips_val'],
step_size=config['step_size_val'],
is_val=True,
transform_pre=transform_eval_pre,
transform_post=transform_post,
get_item_id=True,
)
dict_two_way = val_data.classes_dict
```
### 5. Get predictions
#### 5.1. Select random sample (or specify the index)
```
selected_indx = np.random.randint(len(val_data))
# selected_indx = 136
```
#### 5.2 Get data in required format
```
input_data, target, item_id = val_data[selected_indx]
input_data = input_data.unsqueeze(0)
print("Id of the video sample = {}".format(item_id))
print("True label --> {} ({})".format(target, dict_two_way[target]))
if config['nclips_val'] > 1:
input_var = list(input_data.split(config['clip_size'], 2))
for idx, inp in enumerate(input_var):
input_var[idx] = torch.autograd.Variable(inp)
else:
input_var = [torch.autograd.Variable(input_data)]
```
#### 5.3 Compute output from the model
```
output = model(input_var).squeeze(0)
output = torch.nn.functional.softmax(output, dim=0)
# compute top5 predictions
pred_prob, pred_top5 = output.data.topk(5)
pred_prob = pred_prob.numpy()
pred_top5 = pred_top5.numpy()
```
#### 5.4 Visualize predictions
```
print("Id of the video sample = {}".format(item_id))
print("True label --> {} ({})".format(target, dict_two_way[target]))
print("\nTop-5 Predictions:")
for i, pred in enumerate(pred_top5):
print("Top {} :== {}. Prob := {:.2f}%".format(i + 1, dict_two_way[pred], pred_prob[i] * 100))
path_to_vid = os.path.join(config["data_folder"], item_id + ".webm")
video = io.open(path_to_vid, 'r+b').read()
encoded = base64.b64encode(video)
HTML(data='''<video alt="test" controls>
<source src="data:video/mp4;base64,{0}" type="video/mp4" />
</video>'''.format(encoded.decode('ascii')))
```
| github_jupyter |
# Bar charts
This is 'abusing' the scatter object to create a 3d bar chart
```
import ipyvolume as ipv
import numpy as np
# set up data similar to animation notebook
u_scale = 10
Nx, Ny = 30, 15
u = np.linspace(-u_scale, u_scale, Nx)
v = np.linspace(-u_scale, u_scale, Ny)
x, y = np.meshgrid(u, v, indexing='ij')
r = np.sqrt(x**2+y**2)
x = x.flatten()
y = y.flatten()
r = r.flatten()
time = np.linspace(0, np.pi*2, 15)
z = np.array([(np.cos(r + t) * np.exp(-r/5)) for t in time])
zz = z
fig = ipv.figure()
s = ipv.scatter(x, 0, y, aux=zz, marker="sphere")
dx = u[1] - u[0]
dy = v[1] - v[0]
# make the x and z lim half a 'box' larger
ipv.xlim(-u_scale-dx/2, u_scale+dx/2)
ipv.zlim(-u_scale-dx/2, u_scale+dx/2)
ipv.ylim(-1.2, 1.2)
ipv.show()
```
We now make boxes, that fit exactly in the volume, by giving them a size of 1, in domain coordinates (so 1 unit as read of by the x-axis etc)
```
# make the size 1, in domain coordinates (so 1 unit as read of by the x-axis etc)
s.geo = 'box'
s.size = 1
s.size_x_scale = fig.scales['x']
s.size_y_scale = fig.scales['y']
s.size_z_scale = fig.scales['z']
s.shader_snippets = {'size':
'size_vector.y = SCALE_SIZE_Y(aux_current); '
}
```
Using a shader snippet (that runs on the GPU), we set the y size equal to the aux value. However, since the box has size 1 around the origin of (0,0,0), we need to translate it up in the y direction by 0.5.
```
s.shader_snippets = {'size':
'size_vector.y = SCALE_SIZE_Y(aux_current) - SCALE_SIZE_Y(0.0) ; '
}
s.geo_matrix = [dx, 0, 0, 0, 0, 1, 0, 0, 0, 0, dy, 0, 0.0, 0.5, 0, 1]
```
Since we see the boxes with negative sizes inside out, we made the material double sided
```
# since we see the boxes with negative sizes inside out, we made the material double sided
s.material.side = "DoubleSide"
# Now also include, color, which containts rgb values
color = np.array([[np.cos(r + t), 1-np.abs(z[i]), 0.1+z[i]*0] for i, t in enumerate(time)])
color = np.transpose(color, (0, 2, 1)) # flip the last axes
s.color = color
ipv.animation_control(s, interval=200)
```
# Spherical bar charts
```
# Create spherical coordinates
u = np.linspace(0, 1, Nx)
v = np.linspace(0, 1, Ny)
u, v = np.meshgrid(u, v, indexing='ij')
phi = u * 2 * np.pi
theta = v * np.pi
radius = 1
xs = radius * np.cos(phi) * np.sin(theta)
ys = radius * np.sin(phi) * np.sin(theta)
zs = radius * np.cos(theta)
xs = xs.flatten()
ys = ys.flatten()
zs = zs.flatten()
fig = ipv.figure()
# we use the coordinates as the normals, and thus direction
s = ipv.scatter(xs, ys, zs, vx=xs, vy=ys, vz=zs, aux=zz, color=color, marker="cylinder_hr")
ipv.xyzlim(2)
ipv.show()
ipv.animation_control(s, interval=200)
import bqplot
# the aux range is from -1 to 1, but if we put 0 as min, negative values will go inside
# the max determines the 'height' of the bars
aux_scale = bqplot.LinearScale(min=0, max=5)
s.aux_scale = aux_scale
s.shader_snippets = {'size':
'''float sc = (SCALE_AUX(aux_current) - SCALE_AUX(0.0)); size_vector.y = sc;
'''}
s.material.side = "DoubleSide"
s.size = 2
s.geo_matrix = [1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0.0, 0.5, 0, 1]
ipv.style.box_off()
ipv.style.axes_off()
```
[screenshot](screenshot/bars.gif)
| github_jupyter |
# ReinforcementLearning: a)UCB, b)ThompsonSampling
**--------------------------------------------------------------------------------------------------------------------------**
**--------------------------------------------------------------------------------------------------------------------------**
**--------------------------------------------------------------------------------------------------------------------------**
**---------------------------------------------------**
**STRUCTURE**
*In this notebook, the use of two models (**Part A**: UCB and **Part B**: Thompson Sampling) for an online advertising (Click-through rate) case study is demonstrated. Both models are part of Reinforcement Learning (RL) which is a machine learning category that is focused on different types of rewards depending on the actions taken at each step of the learning process. RL algorithms are capable of learning based on their interactions with the environment, where a reward is given each time the correct decision has been taken, in contrast to the supervised ML models where the presence of labels is required.*
*For this demonstration, a dataset has been generated to represent 9 web advertisements of a product on its columns(dataset features) and the user selections (dataset rows). This dataset is based on the assumption that every time a user visits this web page, a different advertisement (ADV1 - ADV9) is displayed. The goal is to apply a Reinforcement Learning algorithm that will try to learn as quickly as possible which advertisement is selected the most (click-through rate) so as to be presented when users visit the site. Initially, the models display different advertisements to each user but as the algorithms gain more information with respect to the users selections (clicks), the advertisement that leads to the highest reward is chosen to be diplayed. The difference between the 'Upper Confidence Bound' (UCB) and the 'Thompson Sampling' algorithm lies in the selection process of the next advertisement that is to be displayed. UCB is a deterministic model, whereas Thompson Sampling is based on random variation (probabilistic model). In order to evaluate their ability to choose the advertisement with the highest conversion rate for different number of samples (users), the total reward for each model is provided, together with plots presenting the number of times each advertisement has been displayed at the web page.*
```
# Importing the libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
import random
warnings.filterwarnings('ignore')
# Creating the dataset by generating random values(0 & 1) with different probabilities for each 'Adv'
# Len.Dataset=20000
np.random.seed(0)
dataset={'Adv1':np.random.choice(2, 20000,p=[0.6,0.4]),
'Adv2':np.random.choice(2, 20000,p=[0.65,0.35]),
'Adv3':np.random.choice(2, 20000,p=[0.44,0.56]),
'Adv4':np.random.choice(2, 20000,p=[0.6,0.4]),
'Adv5':np.random.choice(2, 20000,p=[0.50,0.50]),
'Adv6':np.random.choice(2, 20000,p=[0.49,0.51]),
'Adv7':np.random.choice(2, 20000,p=[0.4,0.6]),
'Adv8':np.random.choice(2, 20000,p=[0.52,0.48]),
'Adv9':np.random.choice(2, 20000,p=[0.47,0.53])}
data=pd.DataFrame(data=dataset)
# Dataset-First ten records
data.head(10)
```
## UCB
```
#Upper Confidence Bound Algorithm
def ucb_rewards(Users_Num):
#Total Number of Advertisements
Ad_Num=9
#List of advertisements that are selected by the algorithm based on the user clicks at each step (initially empty)
Ad_to_Display=[]
# Count how many times each advertisement is selected
Ad_Cnt_Selection=[0]*Ad_Num
# For each advertisement compute sum of its rewards (initially empty)
Ad_Rewards=[0]*Ad_Num
# Total number of rewards (initially zero)
Ad_Total_Rewards=0
for x in range(1,Users_Num+1):
Ad=0
UCB_max=0
for j in range(0,Ad_Num):
if Ad_Cnt_Selection[j]>0:
Ad_Avg_Reward=Ad_Rewards[j]/Ad_Cnt_Selection[j]
UCB= Ad_Avg_Reward + np.sqrt(3*np.log(x)/(2*Ad_Cnt_Selection[j]))
else:# The purpose of the else statement is to ensure that all Ads are selected (in order to determine the UCB)
UCB=1e309
if UCB>UCB_max:
UCB_max=UCB
Ad=j
Ad_to_Display.append(Ad)
Ad_Cnt_Selection[Ad]+=1
Ad_Rewards[Ad]+=data.values[x-1,Ad]
Ad_Total_Rewards+=data.values[x-1,Ad]
return Ad_to_Display,Ad_Total_Rewards
# The algorithm is to be executed for different samples, whose number progressively increases, so as to observe how many
# samples were required for the model to be able to identify clearly the Ad with the highest conversion rate
selected_Ad_2000=ucb_rewards(Users_Num=2000)
selected_Ad_5000=ucb_rewards(Users_Num=5000)
selected_Ad_10000=ucb_rewards(Users_Num=10000)
selected_Ad_20000=ucb_rewards(Users_Num=20000)
# Conversion to pandas dataframe
df_selected_Ad_2000=pd.DataFrame(data=selected_Ad_2000[0],columns=['Advertisements - Users:2000'])
df_selected_Ad_5000=pd.DataFrame(data=selected_Ad_5000[0],columns=['Advertisements - Users:5000'])
df_selected_Ad_10000=pd.DataFrame(data=selected_Ad_10000[0],columns=['Advertisements - Users:10000'])
df_selected_Ad_20000=pd.DataFrame(data=selected_Ad_20000[0],columns=['Advertisements - Users:20000'])
# As it can be observed, the model managed to identify clearly the Ad with the highest conversion rate at the first 10000
# samples, with good performance at the first 2000 & 5000 samples as well
fig,axs=plt.subplots(2,2,figsize=(14,8))
sns.countplot(data=df_selected_Ad_2000, x="Advertisements - Users:2000",label='Users:2000',ax=axs[0,0])
sns.countplot(data=df_selected_Ad_5000, x="Advertisements - Users:5000",label='Users:5000',ax=axs[0,1])
sns.countplot(data=df_selected_Ad_10000, x="Advertisements - Users:10000",label='Users:10000',ax=axs[1,0])
sns.countplot(data=df_selected_Ad_20000, x="Advertisements - Users:20000",label='Users:20000',ax=axs[1,1])
for ax in axs.flat:
fig.suptitle("Displayed Advertisements - Upper Confidence Bound", fontweight='bold',fontsize=18)
ax.set_xlabel('Advertisements',fontsize=12,fontweight='bold')
ax.set_ylabel('Count',fontsize=12,fontweight='bold')
ax.legend()
ax.figure.tight_layout(pad=2);
```
## Thompson Sampling
```
#Thompson Sampling Algorithm
def TSampling_rewards(Users_Num):
#Total Number of Advertisements
Ad_Num=9
#List of advertisements that are selected by the algorithm based on the user clicks at each step (initially empty)
Ad_to_Display=[]
# Count each time an advertisement gets reward=1
Ad_Count_Reward_1=[0]*Ad_Num
# Count each time an advertisement gets reward=0
Ad_Count_Reward_0=[0]*Ad_Num
# Total number of rewards (initially zero)
Ad_Total_Rewards=0
for x in range(1,Users_Num+1):
Ad=0
draw_max=0
for j in range(0,Ad_Num):
draw_rndm=random.betavariate(Ad_Count_Reward_1[j]+1,Ad_Count_Reward_0[j]+1)
if draw_rndm>draw_max:
draw_max=draw_rndm
Ad=j
Ad_to_Display.append(Ad)
Tsample_reward = data.values[x-1, Ad]
if Tsample_reward == 1:
Ad_Count_Reward_1[Ad]+= 1
else:
Ad_Count_Reward_0[Ad]+= 1
Ad_Total_Rewards+= Tsample_reward
return Ad_to_Display,Ad_Total_Rewards
# The algorithm is to be executed for different samples, whose number progressively increases, so as to observe how many
# samples were required for the model to be able to identify clearly the Ad with the highest conversion rate
select_Ad_2000=TSampling_rewards(Users_Num=2000)
select_Ad_5000=TSampling_rewards(Users_Num=5000)
select_Ad_10000=TSampling_rewards(Users_Num=10000)
select_Ad_20000=TSampling_rewards(Users_Num=20000)
# Conversion to pandas dataframe
df_select_Ad_2000=pd.DataFrame(data=select_Ad_2000[0],columns=['Advertisements - Users:2000'])
df_select_Ad_5000=pd.DataFrame(data=select_Ad_5000[0],columns=['Advertisements - Users:5000'])
df_select_Ad_10000=pd.DataFrame(data=select_Ad_10000[0],columns=['Advertisements - Users:10000'])
df_select_Ad_20000=pd.DataFrame(data=select_Ad_20000[0],columns=['Advertisements - Users:20000'])
# As it can be observed, the Thompson Sampling algorithm managed to outperform UCB as it has clearly identified the Ad with
# the highest conversion rate at the first 5000 samples, with almost excellent performance at the first 2000 samples as well
fig,axs=plt.subplots(2,2,figsize=(14,8))
sns.countplot(data=df_select_Ad_2000, x="Advertisements - Users:2000",label='Users:2000',ax=axs[0,0])
sns.countplot(data=df_select_Ad_5000, x="Advertisements - Users:5000",label='Users:5000',ax=axs[0,1])
sns.countplot(data=df_select_Ad_10000, x="Advertisements - Users:10000",label='Users:10000',ax=axs[1,0])
sns.countplot(data=df_select_Ad_20000, x="Advertisements - Users:20000",label='Users:20000',ax=axs[1,1])
for ax in axs.flat:
fig.suptitle("Displayed Advertisements - Thompson Sampling", fontweight='bold',fontsize=18)
ax.set_xlabel('Advertisements',fontsize=12,fontweight='bold')
ax.set_ylabel('Count',fontsize=12,fontweight='bold')
ax.legend()
ax.figure.tight_layout(pad=2);
# Total rewards for selected data samples
UCB_total_rewards2000=selected_Ad_2000[1]
UCB_total_rewards5000=selected_Ad_5000[1]
UCB_total_rewards10000=selected_Ad_10000[1]
UCB_total_rewards20000=selected_Ad_20000[1]
print('UCB Total Rewards 2000 samples: {}'.format(UCB_total_rewards2000))
print('UCB Total Rewards 5000 samples: {}'.format(UCB_total_rewards5000))
print('UCB Total Rewards 10000 samples: {}'.format(UCB_total_rewards10000))
print('UCB Total Rewards 20000 samples: {}'.format(UCB_total_rewards20000))
print('\r')
TSampling_total_rewards2000=select_Ad_2000[1]
TSampling_total_rewards5000=select_Ad_5000[1]
TSampling_total_rewards10000=select_Ad_10000[1]
TSampling_total_rewards20000=select_Ad_20000[1]
print('TSampling Total Rewards 2000 samples: {}'.format(TSampling_total_rewards2000))
print('TSampling Total Rewards 5000 samples: {}'.format(TSampling_total_rewards5000))
print('TSampling Total Rewards 10000 samples: {}'.format(TSampling_total_rewards10000))
print('TSampling Total Rewards 20000 samples: {}'.format(TSampling_total_rewards20000))
```
| github_jupyter |
# Emukit tutorials
Emukit tutorials can be added and used through the links below. The goal of each of these tutorials is to explain a particular functionality of the Emukit project. These tutorials are stand-alone notebooks that don't require any extra files and fully sit on Emukit components (apart from the creation of the model).
Some tutorials have been written with the purpose of explaining some scientific concepts and can be used for learning about different topics in emulation and uncertainty quantification. Other tutorials are a small guide to describe some feature of the library.
Another great resource to learn Emukit are the [examples](../emukit/examples) which are more elaborated modules focused either on the implementation of a new method with Emukit components or on the analysis and solution of some specific problem.
### Getting Started
Tutorials in this section will get you up and running with Emukit as quickly as possible.
* [5 minutes introduction to Emukit](Emukit-tutorial-intro.ipynb)
* [Philosophy and Basic use of the library](Emukit-tutorial-basic-use-of-the-library.ipynb)
### Scientific tutorials
Tutorials in this section will teach you about the theoretical foundations of surrogate optimization using Emukit.
* [Introduction to Bayesian optimization](Emukit-tutorial-Bayesian-optimization-introduction.ipynb)
* [Introduction to multi-fidelity Gaussian processes](Emukit-tutorial-multi-fidelity.ipynb)
* [Introduction to sensitivity analysis](Emukit-tutorial-sensitivity-montecarlo.ipynb)
* [Introduction to Bayesian Quadrature](Emukit-tutorial-Bayesian-quadrature-introduction.ipynb)
* [Introduction to Experimental Design](Emukit-tutorial-experimental-design-introduction.ipynb)
### Features tutorials
Tutorials in this section will give you code snippets and explanations of various practical features included in the Emukit project.
* [Bayesian optimization with external evaluation of the objective](Emukit-tutorial-bayesian-optimization-external-objective-evaluation.ipynb)
* [Bayesian optimization with context variables](Emukit-tutorial-bayesian-optimization-context-variables.ipynb)
* [Learn how to to combine an acquisition function (entropy search) with a multi-source (fidelity) Gaussian process](Emukit-tutorial-multi-fidelity-bayesian-optimization.ipynb)
* [How to benchmark several Bayesian optimization methods with Emukit](Emukit-tutorial-bayesian-optimization-benchmark.ipynb)
* [How to perform Bayesian optimization with non-linear constraints](Emukit-tutorial-constrained-optimization.ipynb)
* [Bayesian optimization integrating the hyper-parameters of the model](Emukit-tutorial-bayesian-optimization-integrating-model-hyperparameters.ipynb)
* [How to use custom model](Emukit-tutorial-custom-model.ipynb)
* [How to select neural network hyperparameters: categorical variables in Emukit](Emukit-tutorial-select-neural-net-hyperparameters.ipynb)
* [How to parallelize external objective function evaluations in Bayesian optimization](Emukit-tutorial-parallel-eval-of-obj-fun.ipynb)
## Contribution guide
Community contributions are vital to the success of any open source project. [Tutorials](Emukit-tutorial-how-to-write-a-notebook.ipynb) and [examples](https://github.com/emukit/emukit/tree/main/emukit/examples) are a great way to spread what you have learned about Emukit across the community and an excellent way to showcase new features. If you want to contribute with a new tutorial please follow [these steps](Emukit-tutorial-how-to-write-a-notebook.ipynb).
We also welcome feedback, so if there is any aspect of Emukit that we can improve, please [raise an issue](https://github.com/EmuKit/emukit/issues/new)!
| github_jupyter |
Subsets and Splits