code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
```
import os
from dotenv import load_dotenv, find_dotenv
# find .env automagically by walking up directories until it's found
dotenv_path = find_dotenv()
# load up the entries as environment variables
load_dotenv(dotenv_path)
```
### Dealing with ZIP files
The ZIP files contain a CSV file and a fixed width file. We only want the CSV file. We will store those in the RAW directory.
Lets get those variable for the EXTERNAL and the RAW directories.
```
# Get the project folders that we are interested in
PROJECT_DIR = os.path.dirname(dotenv_path)
EXTERNAL_DATA_DIR = PROJECT_DIR + os.environ.get("EXTERNAL_DATA_DIR")
RAW_DATA_DIR = PROJECT_DIR + os.environ.get("RAW_DATA_DIR")
# Get the list of filenames
files=os.environ.get("FILES").split()
print("Project directory is : {0}".format(PROJECT_DIR))
print("External directory is : {0}".format(EXTERNAL_DATA_DIR))
print("Raw data directory is : {0}".format(RAW_DATA_DIR))
print("Base names of files : {0}".format(" ".join(files)))
```
### zipfile package
While some python packages that read files can handle compressed files, the zipfile package can deal with more complex zip files. The files we downloaded from have 2 files as their content. We just want the CSV files.
<br/>
File objects are a bit more complex than other data structures. Opening, reading from, writing to them can all raise exceptions due to the permissions you may or may not have.
<br/>Access to the file is done via a file handler and not directly. You need to properly close them once you are done, otherwise your program keeps that file open as far as the operating system is concerned, potentially blocking other programs from accessing it.
<br/>
To deal with that, you want to use the <b><code>with zipfile.ZipFile() as zfile</code></b> construction. Once the program leaves that scope, Python will nicely close any handlers to the object reference created. This also works great for database connections and other constructions that have these characteristics.
```
import zipfile
print ("Extracting files to: {}".format(RAW_DATA_DIR))
for file in files:
# format the full zip filename in the EXTERNAL DATA DIR
fn=EXTERNAL_DATA_DIR+'/'+file+'.zip'
# and format the csv member name in that zip file
member=file + '.csv'
print("{0} extract {1}.".format(fn, member))
# To make it easier to deal with files, use the with <> as <>: construction.
# It will deal with opening and closing handlers for you.
with zipfile.ZipFile(fn) as zfile:
zfile.extract(member, path=RAW_DATA_DIR)
```
[Back to Agenda](http://localhost:8000/notebooks/Lunch_And_Learn_Session_2_Index.slides.html)
|
github_jupyter
|
import os
from dotenv import load_dotenv, find_dotenv
# find .env automagically by walking up directories until it's found
dotenv_path = find_dotenv()
# load up the entries as environment variables
load_dotenv(dotenv_path)
# Get the project folders that we are interested in
PROJECT_DIR = os.path.dirname(dotenv_path)
EXTERNAL_DATA_DIR = PROJECT_DIR + os.environ.get("EXTERNAL_DATA_DIR")
RAW_DATA_DIR = PROJECT_DIR + os.environ.get("RAW_DATA_DIR")
# Get the list of filenames
files=os.environ.get("FILES").split()
print("Project directory is : {0}".format(PROJECT_DIR))
print("External directory is : {0}".format(EXTERNAL_DATA_DIR))
print("Raw data directory is : {0}".format(RAW_DATA_DIR))
print("Base names of files : {0}".format(" ".join(files)))
import zipfile
print ("Extracting files to: {}".format(RAW_DATA_DIR))
for file in files:
# format the full zip filename in the EXTERNAL DATA DIR
fn=EXTERNAL_DATA_DIR+'/'+file+'.zip'
# and format the csv member name in that zip file
member=file + '.csv'
print("{0} extract {1}.".format(fn, member))
# To make it easier to deal with files, use the with <> as <>: construction.
# It will deal with opening and closing handlers for you.
with zipfile.ZipFile(fn) as zfile:
zfile.extract(member, path=RAW_DATA_DIR)
| 0.155976 | 0.499451 |
```
import plotly.tools as tls
tls.set_credentials_file(username='blue.black83', api_key='kiqXVDGOu8KRTznVJJpy')
import plotly.plotly as py
import plotly.graph_objs as go
data = [go.Contour(z=[[10, 10.625, 12.5, 15.625, 20],
[5.625, 6.25, 8.125, 11.25, 15.625],
[2.5, 3.125, 5., 8.125, 12.5],
[0.625, 1.25, 3.125, 6.25, 10.625],
[0, 0.625, 2.5, 5.625, 10]])]
data
type(data)
data[:]
py.iplot(data)
data = [go.Contour(z=[[10, 10.625, 12.5, 15.625, 20],
[5.625, 6.25, 8.125, 11.25, 15.625],
[2.5, 3.125, 5., 8.125, 12.5],
[0.625, 1.25, 3.125, 6.25, 10.625],
[0, 0.625, 2.5, 5.625, 10]],
x=[-9, -6, -5 , -3, -1],
y=[0, 1, 4, 5, 7])]
py.iplot(data)
data = [go.Contour(z=[[10, 10.625, 12.5, 15.625, 20],
[5.625, 6.25, 8.125, 11.25, 15.625],
[2.5, 3.125, 5., 8.125, 12.5],
[0.625, 1.25, 3.125, 6.25, 10.625],
[0, 0.625, 2.5, 5.625, 10]],
x=[-9, -6, -5 , -3, -1],
y=[0, 1, 4, 5, 7],
colorscale= 'Jet')]
py.iplot(data)
data = [go.Contour(z=[[10, 10.625, 12.5, 15.625, 20],
[5.625, 6.25, 8.125, 11.25, 15.625],
[2.5, 3.125, 5., 8.125, 12.5],
[0.625, 1.25, 3.125, 6.25, 10.625],
[0, 0.625, 2.5, 5.625, 10]],
x=[-9, -6, -5 , -3, -1],
y=[0, 1, 4, 5, 7],
colorscale= 'Jet',
autocontour=False,
contours=dict(
start=0,
end=8,
size=2,),)]
py.iplot(data)
data = [
{
'z': [[10, 10.625, 12.5, 15.625, 20],
[5.625, 6.25, 8.125, 11.25, 15.625],
[2.5, 3.125, 5., 8.125, 12.5],
[0.625, 1.25, 3.125, 6.25, 10.625],
[0, 0.625, 2.5, 5.625, 10]],
'colorscale':'Jet',
'type': u'contour',
'dx': 10,
'x0': 5,
'dy': 10,
'y0':10,
}
]
py.iplot(data)
from plotly import tools
import plotly.plotly as py
import plotly.graph_objs as go
trace0 = go.Contour(
z=[[2, 4, 7, 12, 13, 14, 15, 16],
[3, 1, 6, 11, 12, 13, 16, 17],
[4, 2, 7, 7, 11, 14, 17, 18],
[5, 3, 8, 8, 13, 15, 18, 19],
[7, 4, 10, 9, 16, 18, 20, 19],
[9, 10, 5, 27, 23, 21, 21, 21],
[11, 14, 17, 26, 25, 24, 23, 22]],
line=dict(smoothing=0),
)
trace1 = go.Contour(
z=[[2, 4, 7, 12, 13, 14, 15, 16],
[3, 1, 6, 11, 12, 13, 16, 17],
[4, 2, 7, 7, 11, 14, 17, 18],
[5, 3, 8, 8, 13, 15, 18, 19],
[7, 4, 10, 9, 16, 18, 20, 19],
[9, 10, 5, 27, 23, 21, 21, 21],
[11, 14, 17, 26, 25, 24, 23, 22]],
line=dict(smoothing=0.85),
)
fig = tools.make_subplots(rows=1, cols=2,
subplot_titles=('Without Smoothing', 'With Smoothing'))
fig.append_trace(trace0, 1, 1)
fig.append_trace(trace1, 1, 2)
py.iplot(fig)
import plotly.graph_objs as go
import pandas as pd
z_data = pd.read_csv('https://raw.githubusercontent.com/plotly/datasets/master/api_docs/mt_bruno_elevation.csv')
data = [
go.Surface(
z=z_data.as_matrix()
)
]
data
import plotly.plotly as py
layout = go.Layout(autosize=False,
width=500,
height=500,
margin=dict(
l=65,
r=50,
b=65,
t=90
)
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='elevations-3d-surface')
```
|
github_jupyter
|
import plotly.tools as tls
tls.set_credentials_file(username='blue.black83', api_key='kiqXVDGOu8KRTznVJJpy')
import plotly.plotly as py
import plotly.graph_objs as go
data = [go.Contour(z=[[10, 10.625, 12.5, 15.625, 20],
[5.625, 6.25, 8.125, 11.25, 15.625],
[2.5, 3.125, 5., 8.125, 12.5],
[0.625, 1.25, 3.125, 6.25, 10.625],
[0, 0.625, 2.5, 5.625, 10]])]
data
type(data)
data[:]
py.iplot(data)
data = [go.Contour(z=[[10, 10.625, 12.5, 15.625, 20],
[5.625, 6.25, 8.125, 11.25, 15.625],
[2.5, 3.125, 5., 8.125, 12.5],
[0.625, 1.25, 3.125, 6.25, 10.625],
[0, 0.625, 2.5, 5.625, 10]],
x=[-9, -6, -5 , -3, -1],
y=[0, 1, 4, 5, 7])]
py.iplot(data)
data = [go.Contour(z=[[10, 10.625, 12.5, 15.625, 20],
[5.625, 6.25, 8.125, 11.25, 15.625],
[2.5, 3.125, 5., 8.125, 12.5],
[0.625, 1.25, 3.125, 6.25, 10.625],
[0, 0.625, 2.5, 5.625, 10]],
x=[-9, -6, -5 , -3, -1],
y=[0, 1, 4, 5, 7],
colorscale= 'Jet')]
py.iplot(data)
data = [go.Contour(z=[[10, 10.625, 12.5, 15.625, 20],
[5.625, 6.25, 8.125, 11.25, 15.625],
[2.5, 3.125, 5., 8.125, 12.5],
[0.625, 1.25, 3.125, 6.25, 10.625],
[0, 0.625, 2.5, 5.625, 10]],
x=[-9, -6, -5 , -3, -1],
y=[0, 1, 4, 5, 7],
colorscale= 'Jet',
autocontour=False,
contours=dict(
start=0,
end=8,
size=2,),)]
py.iplot(data)
data = [
{
'z': [[10, 10.625, 12.5, 15.625, 20],
[5.625, 6.25, 8.125, 11.25, 15.625],
[2.5, 3.125, 5., 8.125, 12.5],
[0.625, 1.25, 3.125, 6.25, 10.625],
[0, 0.625, 2.5, 5.625, 10]],
'colorscale':'Jet',
'type': u'contour',
'dx': 10,
'x0': 5,
'dy': 10,
'y0':10,
}
]
py.iplot(data)
from plotly import tools
import plotly.plotly as py
import plotly.graph_objs as go
trace0 = go.Contour(
z=[[2, 4, 7, 12, 13, 14, 15, 16],
[3, 1, 6, 11, 12, 13, 16, 17],
[4, 2, 7, 7, 11, 14, 17, 18],
[5, 3, 8, 8, 13, 15, 18, 19],
[7, 4, 10, 9, 16, 18, 20, 19],
[9, 10, 5, 27, 23, 21, 21, 21],
[11, 14, 17, 26, 25, 24, 23, 22]],
line=dict(smoothing=0),
)
trace1 = go.Contour(
z=[[2, 4, 7, 12, 13, 14, 15, 16],
[3, 1, 6, 11, 12, 13, 16, 17],
[4, 2, 7, 7, 11, 14, 17, 18],
[5, 3, 8, 8, 13, 15, 18, 19],
[7, 4, 10, 9, 16, 18, 20, 19],
[9, 10, 5, 27, 23, 21, 21, 21],
[11, 14, 17, 26, 25, 24, 23, 22]],
line=dict(smoothing=0.85),
)
fig = tools.make_subplots(rows=1, cols=2,
subplot_titles=('Without Smoothing', 'With Smoothing'))
fig.append_trace(trace0, 1, 1)
fig.append_trace(trace1, 1, 2)
py.iplot(fig)
import plotly.graph_objs as go
import pandas as pd
z_data = pd.read_csv('https://raw.githubusercontent.com/plotly/datasets/master/api_docs/mt_bruno_elevation.csv')
data = [
go.Surface(
z=z_data.as_matrix()
)
]
data
import plotly.plotly as py
layout = go.Layout(autosize=False,
width=500,
height=500,
margin=dict(
l=65,
r=50,
b=65,
t=90
)
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='elevations-3d-surface')
| 0.444324 | 0.59408 |
# Image Classification Inference for High Resolution Images - ONNX Runtime
In this example notebook, we describe how to use a pre-trained Classification model, using high resolution images, for inference.
- The user can choose the model (see section titled *Choosing a Pre-Compiled Model*)
- The models used in this example were trained on the ***ImageNet*** dataset because it is a widely used dataset developed for training and benchmarking image classification AI models.
- We perform inference on a few sample images.
- We also describe the input preprocessing and output postprocessing steps, demonstrate how to collect various benchmarking statistics and how to visualize the data.
## Choosing a Pre-Compiled Model
We provide a set of precompiled artifacts to use with this notebook that will appear as a drop-down list once the first code cell is executed.
<img src=docs/images/drop_down.PNG width="400">
## Image classification
Image classification is a popular computer vision algorithm used in applications such as, object recongnition, traffic sign recongnition and traffic light recongnition. Image classification models are also used as feature extractors for other tasks such as object detection and semantic segmentation.
- The image below shows classification results on few sample images.
- Note: in this example, we used models trained with ***ImageNet*** because it is a widely used dataset developed for training and benchmarking image classifcation AI models
<img src=docs/images/CLS.PNG width="500">
## ONNX Runtime based Work flow
The diagram below describes the steps for ONNX Runtime based workflow.
Note:
- The user needs to compile models(sub-graph creation and quantization) on a PC to generate model artifacts.
- For this notebook we use pre-compiled models artifacts
- The generated artifacts can then be used to run inference on the target.
- Users can run this notebook as-is, only action required is to select a model.
<img src=docs/images/onnx_work_flow_2.png width="400">
```
import os
import cv2
import numpy as np
import ipywidgets as widgets
from scripts.utils import get_eval_configs
last_artifacts_id = selected_model_id.value if "selected_model_id" in locals() else None
prebuilt_configs, selected_model_id = get_eval_configs('classification','onnxrt', num_quant_bits = 8, last_artifacts_id = last_artifacts_id, model_selection='high_resolution')
display(selected_model_id)
print(f'Selected Model: {selected_model_id.label}')
config = prebuilt_configs[selected_model_id.value]
config['session'].set_param('model_id', selected_model_id.value)
config['session'].start()
```
## Define utility function to preprocess input images
Below, we define a utility function to preprocess images for the model. This function takes a path as input, loads the image and preprocesses the images as required by the model. The steps below are shown as a reference (no user action required):
1. Load image
2. Convert BGR image to RGB
3. Scale image
4. Apply per-channel pixel scaling and mean subtraction
5. Convert RGB Image to BGR.
6. Convert the image to NCHW format
- The input arguments of this utility function is selected automatically by this notebook based on the model selected in the drop-down
```
def preprocess(image_path, size, mean, scale, layout, reverse_channels):
# Step 1
img = cv2.imread(image_path)
# Step 2
img = img[:,:,::-1]
# Step 3
img = cv2.resize(img, (size, size), interpolation=cv2.INTER_CUBIC)
# Step 4
img = img.astype('float32')
for mean, scale, ch in zip(mean, scale, range(img.shape[2])):
img[:,:,ch] = ((img.astype('float32')[:,:,ch] - mean) * scale)
# Step 5
if reverse_channels:
img = img[:,:,::-1]
# Step 6
if layout == 'NCHW':
img = np.expand_dims(np.transpose(img, (2,0,1)),axis=0)
else:
img = np.expand_dims(img,axis=0)
return img
```
## Create the model using the stored artifacts
<div class="alert alert-block alert-warning">
<b>Warning:</b> It is recommended to use the ONNX Runtime APIs in the cells below without any modifications.
</div>
```
import onnxruntime as rt
onnx_model_path = config['session'].get_param('model_file')
delegate_options = {}
so = rt.SessionOptions()
delegate_options['artifacts_folder'] = config['session'].get_param('artifacts_folder')
EP_list = ['TIDLExecutionProvider','CPUExecutionProvider']
sess = rt.InferenceSession(onnx_model_path ,providers=EP_list, provider_options=[delegate_options, {}], sess_options=so)
input_details = sess.get_inputs()
output_details = sess.get_outputs()
```
## Run the model for inference
### Preprocessing and Inference
- You can use a portion of images provided in `/sample-images` directory to evaluate the classification inferences. In the cell below, we use a loop to preprocess the selected images, and provide them as the input to the network.
### Postprocessing and Visualization
- Once the inference results are available, we postpocess the results and visualize the inferred classes for each of the input images.
- Classification models return the results as a list of `numpy.ndarray`, containing one element which is an array with `shape` = `(1,1000)` and `dtype` = `'float32'`, where each element represents the activation for a particular ***ImageNet*** class. The results from the these inferences above are postprocessed using `argsort()` to get the `TOP-5` class IDs and the corresponding names using `imagenet_class_to_name()`.
- Then, in this notebook, we use *matplotlib* to plot the original images and the corresponding results.
```
from scripts.utils import get_preproc_props
# use results from the past inferences
images = [
('sample-images/elephant.bmp', 221),
('sample-images/laptop.bmp', 222),
('sample-images/bus.bmp', 223),
('sample-images/zebra.bmp', 224),
]
size, mean, scale, layout, reverse_channels = get_preproc_props(config)
print(f'Image size: {size}')
import tqdm
import matplotlib.pyplot as plt
from scripts.utils import imagenet_class_to_name
plt.figure(figsize=(20,10))
for num in tqdm.trange(len(images)):
image_file, grid = images[num]
img = cv2.imread(image_file)[:,:,::-1]
ax = plt.subplot(grid)
img_in = preprocess(image_file , size, mean, scale, layout, reverse_channels)
if not input_details[0].type == 'tensor(float)':
img_in = np.uint8(img_in)
res = list(sess.run(None, {input_details[0].name: img_in}))[0]
# get the TOP-5 class IDs by argsort()
# and use utility function to get names
output = res.squeeze()
classes = output.argsort()[-5:][::-1]
names = [imagenet_class_to_name(x)[0] for x in classes]
# plot the TOP-5 class names
ax.text(20, 0 * img.shape[0] / 15, names[0], {'color': 'red', 'fontsize': 18, 'ha': 'left', 'va': 'top'})
ax.text(20, 1 * img.shape[0] / 15, names[1], {'color': 'blue', 'fontsize': 14, 'ha': 'left', 'va': 'top'})
ax.text(20, 2 * img.shape[0] / 15, names[2], {'color': 'blue', 'fontsize': 14, 'ha': 'left', 'va': 'top'})
ax.text(20, 3 * img.shape[0] / 15, names[3], {'color': 'blue', 'fontsize': 14, 'ha': 'left', 'va': 'top'})
ax.text(20, 4 * img.shape[0] / 15, names[4], {'color': 'blue', 'fontsize': 14, 'ha': 'left', 'va': 'top'})
# Show the original image
ax.imshow(img)
plt.show()
```
## Plot Inference benchmarking statistics
- During the model execution several benchmarking statistics such as timestamps at different checkpoints, DDR bandwidth are collected and stored. `get_TI_benchmark_data()` can be used to collect these statistics. This function returns a dictionary of `annotations` and the corresponding markers.
- We provide the utility function plot_TI_benchmark_data to visualize these benchmark KPIs
<div class="alert alert-block alert-info">
<b>Note:</b> The values represented by <i>Inferences Per Second</i> and <i>Inference Time Per Image</i> uses the total time taken by the inference except the time taken for copying inputs and outputs. In a performance oriented system, these operations can be bypassed by writing the data directly into shared memory and performing on-the-fly input / output normalization.
</div>
```
from scripts.utils import plot_TI_performance_data, plot_TI_DDRBW_data, get_benchmark_output
stats = sess.get_TI_benchmark_data()
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(10,5))
plot_TI_performance_data(stats, axis=ax)
plt.show()
tt, st, rb, wb = get_benchmark_output(stats)
print(f'SoC: J721E/DRA829/TDA4VM')
print(f' OPP:')
print(f' Cortex-A72 @2GHZ')
print(f' DSP C7x-MMA @1GHZ')
print(f' DDR @4266 MT/s\n')
print(f'{selected_model_id.label} :')
print(f' Inferences Per Second : {1000.0/tt :7.2f} fps')
print(f' Inference Time Per Image : {tt :7.2f} ms')
print(f' DDR usage Per Image : {rb+ wb : 7.2f} MB')
```
|
github_jupyter
|
import os
import cv2
import numpy as np
import ipywidgets as widgets
from scripts.utils import get_eval_configs
last_artifacts_id = selected_model_id.value if "selected_model_id" in locals() else None
prebuilt_configs, selected_model_id = get_eval_configs('classification','onnxrt', num_quant_bits = 8, last_artifacts_id = last_artifacts_id, model_selection='high_resolution')
display(selected_model_id)
print(f'Selected Model: {selected_model_id.label}')
config = prebuilt_configs[selected_model_id.value]
config['session'].set_param('model_id', selected_model_id.value)
config['session'].start()
def preprocess(image_path, size, mean, scale, layout, reverse_channels):
# Step 1
img = cv2.imread(image_path)
# Step 2
img = img[:,:,::-1]
# Step 3
img = cv2.resize(img, (size, size), interpolation=cv2.INTER_CUBIC)
# Step 4
img = img.astype('float32')
for mean, scale, ch in zip(mean, scale, range(img.shape[2])):
img[:,:,ch] = ((img.astype('float32')[:,:,ch] - mean) * scale)
# Step 5
if reverse_channels:
img = img[:,:,::-1]
# Step 6
if layout == 'NCHW':
img = np.expand_dims(np.transpose(img, (2,0,1)),axis=0)
else:
img = np.expand_dims(img,axis=0)
return img
import onnxruntime as rt
onnx_model_path = config['session'].get_param('model_file')
delegate_options = {}
so = rt.SessionOptions()
delegate_options['artifacts_folder'] = config['session'].get_param('artifacts_folder')
EP_list = ['TIDLExecutionProvider','CPUExecutionProvider']
sess = rt.InferenceSession(onnx_model_path ,providers=EP_list, provider_options=[delegate_options, {}], sess_options=so)
input_details = sess.get_inputs()
output_details = sess.get_outputs()
from scripts.utils import get_preproc_props
# use results from the past inferences
images = [
('sample-images/elephant.bmp', 221),
('sample-images/laptop.bmp', 222),
('sample-images/bus.bmp', 223),
('sample-images/zebra.bmp', 224),
]
size, mean, scale, layout, reverse_channels = get_preproc_props(config)
print(f'Image size: {size}')
import tqdm
import matplotlib.pyplot as plt
from scripts.utils import imagenet_class_to_name
plt.figure(figsize=(20,10))
for num in tqdm.trange(len(images)):
image_file, grid = images[num]
img = cv2.imread(image_file)[:,:,::-1]
ax = plt.subplot(grid)
img_in = preprocess(image_file , size, mean, scale, layout, reverse_channels)
if not input_details[0].type == 'tensor(float)':
img_in = np.uint8(img_in)
res = list(sess.run(None, {input_details[0].name: img_in}))[0]
# get the TOP-5 class IDs by argsort()
# and use utility function to get names
output = res.squeeze()
classes = output.argsort()[-5:][::-1]
names = [imagenet_class_to_name(x)[0] for x in classes]
# plot the TOP-5 class names
ax.text(20, 0 * img.shape[0] / 15, names[0], {'color': 'red', 'fontsize': 18, 'ha': 'left', 'va': 'top'})
ax.text(20, 1 * img.shape[0] / 15, names[1], {'color': 'blue', 'fontsize': 14, 'ha': 'left', 'va': 'top'})
ax.text(20, 2 * img.shape[0] / 15, names[2], {'color': 'blue', 'fontsize': 14, 'ha': 'left', 'va': 'top'})
ax.text(20, 3 * img.shape[0] / 15, names[3], {'color': 'blue', 'fontsize': 14, 'ha': 'left', 'va': 'top'})
ax.text(20, 4 * img.shape[0] / 15, names[4], {'color': 'blue', 'fontsize': 14, 'ha': 'left', 'va': 'top'})
# Show the original image
ax.imshow(img)
plt.show()
from scripts.utils import plot_TI_performance_data, plot_TI_DDRBW_data, get_benchmark_output
stats = sess.get_TI_benchmark_data()
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(10,5))
plot_TI_performance_data(stats, axis=ax)
plt.show()
tt, st, rb, wb = get_benchmark_output(stats)
print(f'SoC: J721E/DRA829/TDA4VM')
print(f' OPP:')
print(f' Cortex-A72 @2GHZ')
print(f' DSP C7x-MMA @1GHZ')
print(f' DDR @4266 MT/s\n')
print(f'{selected_model_id.label} :')
print(f' Inferences Per Second : {1000.0/tt :7.2f} fps')
print(f' Inference Time Per Image : {tt :7.2f} ms')
print(f' DDR usage Per Image : {rb+ wb : 7.2f} MB')
| 0.422266 | 0.978219 |
# Section IV. DYNAMICS AND CONTROL
# Chapter 17. Optimal Control
In previous chapters we have seen the use of myopic controllers like PID
or operational space control, as well as some predictive controllers
like trajectory generation. Particularly for complex and nonlinear
systems like robots, predictive control allows the controller to make
better decisions at the current time to account for future
possibilities. However, our previous predictive methods were largely
restricted to one class of systems. Optimal control addresses these
shortcomings in a highly general framework.
Optimal control asks to compute a control function (either open loop or
closed loop) that optimizes some performance metric regarding the
control and the predicted state. For example, a driver of a car would
like to reach a desired location while achieving several other goals:
e.g., avoiding obstacles, not driving erratically, maintaining a
comfortable level of accelerations for human passengers. A driving style
can be composed of some balanced combination of these goals. Optimal
control allows a control designer to specify the *dynamic model* and the
*desired outcomes*, and the algorithm will compute an optimized control.
This relieves some burden by letting the designer reason at the level of
*what* the robot should do, rather than designing *how* it should do it.
In this chapter, we will discuss how to specify optimal control problems
and how to implement and use optimal control techniques. We will
consider only the open loop problem; however we will mention how an open
loop optimizer can be adapted to closed loop control via the use of
*model predictive control*.
Optimal control problem
-----------------------
An optimal control problem is defined by the dynamics function $f$ and a
*cost functional* over the entire trajectory $x$ and $u$:
$$J(x,u) = \int_0^\infty L(x(t),u(t),t) dt.$$ The term *functional*
indicates that this is a function mapping a function to a real number.
The term $L(x,u,t)$ is known as the *instantaneous cost* (or *running cost*) which is
accumulated over time, and should be chosen to be nonnegative and to
penalize certain undesirable states, velocities, or controls. Its units
are cost units per second.
The goal of optimal control is to find state and control trajectories
$x$ and $u$ such that $J(x,u)$ is minimized: $$\begin{gathered}
x^\star, u^\star = \arg \min_{x,u} J(x,u) \text{ such that} \\
\dot{x}(t) = f(x(t),u(t)) \text{ for all }t
\end{gathered}
\label{eq:OptimalControl}$$
(For somewhat technical reasons, there are problems for which no optimal
trajectory exists, but rather only a sequence of trajectories
approaching an optimal cost. Hence, if we prefer to be pedantic, it is
often necessary to prove existence of an optimal solution first, or to
relax the problem to determine only an approximate optimum.)
### Cost functionals
A variety of behaviors can be specified in this framework by modifying
the instantaneous cost. For example:
- Trajectory tracking for a trajectory $x_D(t)$ can be implemented by penalizing squared error $L(x,u,t) = \|x - x_D(t)\|^2$.
- Minimizing effort can be defined in terms of a control penalty $\|u\|^2$.
- Minimum time to hit a target $x_{tgt}$ could be implemented as an indicator function $I[x\neq x_{tgt}]$ where $I[z]$ is 1 if $z$ is true, and 0 otherwise.
- Obstacle avoidance and other feasibility constraints can be implemented as indicator functions as well, $\infty \cdot I[x \notin \mathcal{F}]$ where $\mathcal{F}$ is the free space.
- Smoothed obstacle avoidance can be implemented by a repulsive barrier that decreases to 0 when the distance to the closest obstacle $d$ exceeds some minimum buffer distance $d_{min}$ and incrases to infinity as the distance shrinks to 0. One common form of this barrier is $L(x,u,t) = 1/d^2 - 1/d_{min}^2$ when $d < d_{min}$ and $L(x,u,t) = 0$ otherwise.
It is common to mix and match different types of costs functionals using a _weighted cost functional_ $$J(x,u) = \sum_{i=1}^N w_i J_i(x,u)$$
where each $J_i(x,u)$ is some primitive cost functional and $w_i$ scales its contribution to the final cost. By tuning these weights, a designer can encourage the optimized trajectories to emphasize some aspects of the trajectory over others.
### Finite horizon optimal control and discounting
As stated, this problem is somewhat ill-behaved because it
involves an infinite integral, which could achieve infinite cost even
for relatively well-behaved trajectories. For example, if the cost were
simply squared error, a trajectory that achieves 0.01 steady-state error
would be rated as having the same cost as a trajectory that had an error
of 1: namely, infinitely bad.
There are two general ways to make the cost functional better behaved.
The first method to truncate the problem at some maximum time $T$,
leading to a *finite-horizon optimal control* cost functional
$$J(x,u) = \int_0^T L(x(t),u(t),t) dt + \Phi(x(T))$$ where $\Phi(x)$ is
a nonnegative *terminal cost* that penalizes the state attained at the
terminal time.
The second method is to modify the instantaneous cost functional by
including a *discount factor* that decays to 0 relatively quickly as
$t \rightarrow \infty$. This is usually expressed as the product of a
time-independent term and a time-dependent discount factor term:
$$L(x,u,t) = L(x,u,0) \gamma(t)$$ with $\gamma(t)$ a decaying function,
such as $O(1/t^\alpha)$ or $O(\beta^t)$. It is important to choose a
discount factor that drops relatively rapidly toward 0 to ensure that
the cost is integrable. Discount factors of the form $O(1/t^\alpha)$
must have $\alpha > 1$ to ensure that the cost functional is finite for
all bounded trajectories, and those of the form $O(\beta^t)$ must have
$\beta < 1$.
### State and Control Constraints
Usually, optimal control solvers require that the cost functional is
smooth, and so non-differentiable constraints like minimum time and
obstacle avoidance must be reformulated as hard constraints, external to
the cost functional. As a result the reformulation becomes essentially
an infinite-dimensional constrained optimization problem. Solvers may
differ about whether they can handle constraints on state or constraints
on control.
### Analytical vs Numerical Solvers
Minimization over a space of functions is considerably more difficult to
solve than typical optimization problems: the space of functions is
*uncountably infinite-dimensional*! There are two general ways to tackle
these problems: analytical or numerical. Analytical techniques use the
mathematical conditions of optimality so that the optimal control can be
determined directly through calculus and algebraic manipulation.
Successfully applying analysis typically requires a relatively simple
dynamics and cost. Numerical techniques approximate the problem by
discretizing either the state, time, and/or control space and attempt to cast
the problem as a finite dimensional optimization. These are more
general-purpose and can be applied to complex problems, but are more
computationally expensive and usually require some parameter tuning to
obtain high-quality solutions.
LQR control
-----------
The simplest class of optimal control problems is LTI systems with costs
that are quadratic in $x$ and $u$. Through the calculus of variations,
which is beyond the scope of this book, the optimal control for this
problem class can be determined analytically as a closed-form function
of $x$.
LTI systems with quadratic cost are specified as $\dot{x} = Ax + Bu$ and
$$L(x,u,t) = x^T Q x + u^T R u$$ where $Q$ and $R$ are symmetric
matrices of size $n\times n$ and $m\times m$, respectively. The
magnitude of entries of $Q$ penalize error from the equilibrium point,
and the magnitude of entries of $R$ penalize control effort. The overall
control functional is therefore
$$J(x,u) = \int_0^\infty x(t)^T Q x(t) + u(t)^T R u(t).$$
Here, the optimal control can be shown to be a linear function of $x$:
$$u = -Kx$$ for the gain $K = R^{-1}B^T P$ defined as a function of an
unknown matrix $P$. $P$ is a symmetric $n \times n$ matrix that solves
the following *Riccati equation* :
$$A^TP + PA - PBR^{-1}B^TP + Q = 0.$$ Numerical methods are available
for solving the Riccati equation for $P$. This method is known as the
Linear Quadratic Regulator (LQR) approach.
As we showed in the [section on LTI stability](Control.ipynb#Stability-in-Linear-Time-Invariant-Systems),
traditional pole placement methods can be
used to derive a stable controller for LTI systems. However, the
significance of the LQR controller compared to traditional pole
stability analysis is that the performance metric is made explicit
rather than implicit. Moreover, it gives a closed form solution for any
dynamic model specified by $A,B$, so if more information is gathered
that yields a better estimate for $A$ and $B$, the LQR method can be
applied directly to obtain the optimal gains.
Pointryagin's Minimum Principle
-------------------------------
The "magic" of the LQR solution is obtained through a more generic
principle of optimal controllers called the *Pointryagin's minimum
principle*. It defines a *first order*, *necessary* condition for a
particular state/control trajectory to be an optimum. It is derived from
($\ref{eq:OptimalControl}$) via a combination of calculus of
variations and the method of Lagrange multipliers. This will briefly be
described here.
In (equality) constrained optimization problems, the method of Lagrange
multipliers defines an auxiliary Lagrange multiplier variable
$\lambda_i$ for each equality constraint. However, in optimal control
problems, there are an infinite number of equality constraints
$\dot{x}(t) = f(x(t),u(t))$ defined for each point in time. As a result,
the Lagrange multipliers for this problem are not single variables, but
rather *trajectories* defined over time. This trajectory of multipliers
is known as a *costate trajectory*
$\lambda(t):[0,\infty)\rightarrow \mathbb{R}^n$.
An auxiliary function, called the *Hamiltonian*, is defined over the
system and costate at particular points in time:
$$H(\lambda,x,u,t)=\lambda^T f(x,u) + L(x,u,t).$$ It is also possible to
maintain control constraints $u\in \mathcal{U}\subseteq \mathbb{R}^m$.
Pointryagin's minimum principle is then stated as follows. An optimal
state/ control/ costate trajectory $(x^\star,u^\star,\lambda^\star)$
satisfies for all $t \geq 0$:
1. $H(\lambda^\star(t),x^\star(t),u^\star(t),t) \leq H(\lambda^\star(t),x^\star(t),u,t)$
for all $u \in \mathcal{U}$
2. $\dot{x}^\star(t) = f(x^\star(t),u^\star(t))$
3. $\dot{\lambda}^\star(t) = -\frac{\partial}{\partial x}H(\lambda^\star(t),x^\star(t),u^\star(t),t)$.
The derivation of these equations is outside the scope of this book. But
the conditions can be applied in certain cases to obtain optimal
controls, or at least limit the range of controls possibly optimal.
### Derivation of LQR from PMP
As a result, consider the LQR setting. The Hamiltonian is
$$H(\lambda,x,u,t) = \lambda^T(Ax + Bu) + x^T Q x + u^T R u$$ and the
Pointryagin's minimum principle gives:
1. Subtracting off terms that do not contain $u$,
$\lambda^{\star T}Bu^{\star} + u^{\star T} R u^\star \leq \lambda^{\star T}Bu + u^T R u$
for all $u$.
2. $\dot{x}^\star = Ax^\star + Bu^\star$
3. $\dot{\lambda}^\star = - A^T \lambda^{\star} - 2 Qx$.
Expanding 1, we see that for $u^\star$ to be a minimizer of the
Hamiltonian, given $\lambda^\star$, $x^\star$, and $t$ fixed, we must
have that $B^T \lambda^\star + 2 R u^\star = 0$ so that
$u^\star = \frac{1}{2}R^{-1} B^T \lambda^\star$.
Now replacing this into 2 and 3, we have a system of ODEs:
$$\begin{split}
\dot{x}^\star &= A x^\star + \frac{1}{2} B R^{-1} B^T \lambda^\star \\
\dot{\lambda}^\star &= - 2 Qx^\star -A^T \lambda^\star
\end{split}$$
Hypothesizing that $\lambda^\star = 2 P x^\star$ and
multiplying the first equation by $P$, we obtain the system of equations
$$\begin{split}
P\dot{x}^\star &= (PA + P B R^{-1} B^T P )x^\star \\
2P\dot{x}^\star &= (-2Q -2A^T P)x^\star.
\end{split}$$
After dividing the second equation by 2 and equating the
left hand sides, we have an equation that must be satisfied for all $x$.
Since the equation must hold for all $x$, the matrices must also be
equal, which produces the Riccati equations.
### Bang-bang control
Another result from Pointryagin's minimum principle condition (1) is
that the Hamiltonian must be minimized by the control, keeping the state
and costate fixed. As a result, there are two possibilities: (1a) the
derivative of the Hamiltonian is 0 at $u^\star$, or (1b) the control is
at the boundary of the control set $u^\star \in \partial U$.
This leads to many systems having the characteristic of *bang-bang
control*, which means that the optimal control will jump discontinuously
between extremes of the control set. As an example, consider a race car
driver attempting to minimize time. The optimal control at all points in
time will either maximize acceleration, maximize braking, or
maximize/minimize angular acceleration; otherwise, time could be saved
by making the control more extreme.
If it can be determined that there are a finite number of possible
controls satisfying condition (1), then the optimal control problem
becomes one of simply finding *switching times* between optimal
controls.
Taking the [Dubins car model](WhatAreDynamicsAndControl.ipynb#Dubins-car) as an example,
we have the state variable
$(x,y,\theta)\in SO(2)$ and control variable $(v,\phi)$ denoting
velocity and steering angle: $$\begin{split}
\dot{x} &= v \cos \theta \\
\dot{y} &= v \sin \theta \\
\dot{\theta} &= v/L \tan \phi .
\end{split}$$
Here $(x,y)$ are the coordinates of a point in the middle
of the rear axis, and $L$ is the length between the rear and front axle.
The velocity and steering angle are bounded, $v \in [-1,1]$ and
$\phi \in [-\phi_{min},\phi_{max}]$, and the cost only measures time to
reach a target state. Hence, the Hamiltonian is
$$H(\lambda,x,u,t) = \lambda_1 v \cos \theta + \lambda_2 v \sin \theta + \lambda_3 v/L \tan \phi + I[x \neq x_{tgt}]$$
The latter term does not contribute to the choice of $u$, so we can
ignore it. For $(v,\phi)$ to be a minimum of the Hamiltonian, with
$\lambda$ and $x$ fixed, either $\lambda = 0$ and the control is
irrelevant, or $\lambda \neq 0$ and
$v = -sign(\lambda_1 \cos \theta + \lambda_2 \sin \theta + \lambda_3 /L \tan \phi)$.
Then, since $\tan$ is a monotonic function, we have
$\phi = -sign(\lambda_3 v)\phi_{max}$. As a result, the only options are
the minimum, maximum, and 0 controls on each axis.
The trajectories corresponding to these extrema are straight
forward/backward, moving forward while turning left/right, and moving
backward while turning left/right. The curves traced out by these
trajectories are then either straight line segments or arcs of turning
rate $\pm \tan \phi_{max}/L$. To find all minimum-time paths between two
points, it is then a matter of enumerating all possible arcs and
straight line segments. The solutions are known as Reeds-Shepp curves.
Trajectory Optimization
-----------------------
It is not always possible (or easy) to derive elegant expressions of
optimal controls using Pointryagin's minimum principle for general
nonlinear systems. For example, if any modification is made to an LQR
system, such as a non-quadratic term in the cost functional, or if state
constraints or control constraints are added, the analytical solution no
longer applies. As a result, numerical methods can solve a wider variety
of optimal control problems. We have already seen
a form of trajectory optimization under the discussion of kinematic path
planning, and here we extend this type of formulation to dynamic systems.
Since trajectories are infinite-dimensional, the main challenge of
trajectory optimization is to suitably discretize the space of
trajectories. A second challenge is that the discretized optimization
problem is usually also fairly large, depending on the granularity of
the discretization, and hence optimization may be computationally
inefficient.
In any case, the general method for performing trajectory optimization
follows the following procedure 1) define a set of basis functions for the control
trajectory, 2) defining a state evolution technique, 3) reformulating
($\ref{eq:OptimalControl}$) as a finite-dimensional optimization
problem over the coefficients of the basis functions, and 4) optimizing
using a gradient-based technique.
### Piecewise-constant control and state trajectories
The main choice in trajectory optimization is how to represent a
trajectory of controls? The most typical assumption is that the
trajectory consists of piecewise-constant controls at a fixed time step.
If we define the time step $\Delta t = T/N$ with $N$ an integer, then we
have the *computational grid*
$0, \Delta t, 2\Delta t, \ldots, N\Delta t$. Let these grid points be
denoted $t_0,t_1,\ldots,t_N$ respectively with $t_0=0$ and $t_N=T$.
Then, the entire control trajectory is specified by a control sequence
$u_1,\ldots,u_N$, with each $u_i$ active on the time range
$[t_{i-1},t_i)$. In other words $u(t) = u_i$ with
$i = \lfloor t/\Delta t \rfloor + 1$.
Suppose now we define a *simulation function*, which is a method for
integrating the state trajectory over time. Specifically, given an
initial state $x(0)$, a constant control $u$, and a fixed duration $h$,
the simulation function $g$ computes an approximation
$$x(h) \approx g(x(0),u,h)$$ If the timestep is small enough, the Euler
approximation is a reasonable simulation function:
$$x(h) \approx x(0) +h f(x(0),u).$$ If accuracy of this method is too
low, then Euler integration could be performed at a finer time sub-step
up to time $h$, and/or a more accurate integration technique could be
used.
In any case, given a piecewise-constant control trajectory defined by a
control sequence $u_1,\ldots,u_N$, we can derive corresponding points on
the state trajectory as follows.
1. Set $x_0 \gets x(0)$.
2. For $i=1,\ldots,N$, set $x_i = g(x_{i-1},u_i,\Delta t)$ to arrive at a state sequence $x_0=x(0),x_1,\ldots,x_N$.
With this definition, each $x_i$ is a function of $u_1,\ldots,u_i$. Hence, we can
approximate the cost functional as:
$$J(x,u) \approx \tilde{J}(u_1,\ldots,u_n) = \delta t \sum_{i=0}^{N-1} L(x_i,u_{i+1},t_i) + \Phi(x_N).$$
Using this definition we can express the approximated optimal control
function as a minimization problem:
$$\arg \min_{u_1,\ldots,u_N} \tilde{J}(u_1,\ldots,u_N).
\label{eq:DirectTranscription}$$
With control space $\mathbb{R}^m$, this is an
optimization problem over $mN$ variables. This approach is also known
as _direct transcription_.
There is a tradeoff in determining the resolution $N$. With higher
values of $N$, the control trajectory can obtain lower costs, but the
optimization problem will have more variables, and hence become more
computationally complex. Moreover, it will be more susceptible to local
minimum problems.
### Descent approaches
Standard gradient-based techniques can be used to solve the problem
($\ref{eq:DirectTranscription}$). One difficulty is that to take the gradient
of $\tilde{J}$ with respect to a control variable $u_i$, observe that
this choice of control affects *every* state variable $x_k$ with
$i \leq k \leq N$. Hence,
$$\frac{\partial J}{\partial u_i} = \Delta t \frac{\partial L}{\partial u_i} (x_{i-1},u_i,t_i) + \Delta t \sum_{k=i}^{N-1} \frac{\partial L}{\partial x}(x_k,u_{k+1},t_k)\frac{\partial x_k}{\partial u_i} + \frac{\partial \Phi}{\partial x_N}\frac{\partial x_N}{\partial u_i}
\label{eq:JacobianUi}$$ The expressions for
$\frac{\partial x_k}{\partial u_i}$ are relatively complex because $x_k$
is defined recursively assuming that $x_{i-1}$ is known. $$\begin{split}
x_i &= g(x_{i-1},u_i,\Delta t) \\
x_{i+1} &= g(x_i,u_{i+1},\Delta t) \\
x_{i+2} &= g(x_{i+1},u_{i+2},\Delta t) \\
&\vdots
\end{split}$$
In this list, the only equation directly affected by $u_i$
is the first. The effects on the remaining states are due to cascading
effects of previous states. Hence, we see that
$$\frac{\partial x_i}{\partial u_i} = \frac{\partial g}{\partial u}(x_{i-1},u_i,\Delta t)$$
$$\frac{\partial x_{i+1}}{\partial u_i} = \frac{\partial x_{i+1}}{\partial x_i} \frac{\partial x_i}{\partial u_i} = \frac{\partial g}{\partial x}(x_i,u_{i+1},\Delta t) \frac{\partial x_i}{\partial u_i}$$
And in general, for $k > i$,
$$\frac{\partial x_k}{\partial u_i} = \frac{\partial g}{\partial x}(x_{k-1},u_k,\Delta t) \frac{\partial x_{k-1}}{\partial u_i}.$$
This appears to be extremely computationally expensive, since each
evaluation of ($\ref{eq:JacobianUi}$) requires calculating $O(N)$ derivatives,
leading to an overall $O(N^2)$ algorithm for calculating the gradient
with respect to the entire control sequence.
However, with a clever forward/backward formulation, the gradient can be calculated with $O(N)$ operations.
Note that all expressions of the form
$\frac{\partial x_k}{\partial u_i}$ are equivalent to
$\frac{\partial x_k}{\partial x_i}\frac{\partial x_i}{\partial u_i}$.
So, we observe that
($\ref{eq:JacobianUi}$) is equal to
$$\Delta t \frac{\partial L}{\partial u_i}(x_{i-1},u_i,t) + \frac{\partial J}{\partial x_i} \frac{\partial x_i}{\partial u_i}.$$
Then, we can express:
$$\frac{\partial J}{\partial x_i} = \Delta t \sum_{k=i}^{N-1} \frac{\partial L}{\partial x}(x_k,u_{k+1},t_k) \frac{\partial x_k}{\partial x_i} + \frac{\partial \Phi }{\partial x_N}\frac{\partial x_N}{\partial x_i}.$$
This entire vector can be computed in a single backward pass starting
from $i=N$ back to $i=1$. Starting with $i=N$, see that
$$\frac{\partial J}{\partial x_N} = \frac{\partial \Phi }{\partial x_N}$$
Then, proceeding to $i=N-1$, observe $$\begin{split}
\frac{\partial J}{\partial x_{N-1}} &= \Delta t \frac{\partial L}{\partial x}(x_{N-1},u_N,t_{N-1}) + \frac{\partial \Phi }{\partial x_N}\frac{\partial x_N}{\partial x_{N-1}} \\
&= \Delta t \frac{\partial L}{\partial x}(x_{N-1},u_N,t_{N-1}) + \frac{\partial J}{\partial x_N} \frac{\partial x_N}{\partial x_{N-1}}.
\end{split}$$ In general, with $i<N$, we have the recursive expression
$$\frac{\partial J}{\partial x_i} = \Delta t \frac{\partial L}{\partial x}(x_i,u_{i+1},t_i) + \frac{\partial J}{\partial x_{i+1}} \frac{\partial x_{i+1}}{\partial x_{i}}.$$
The entire set of values can be computed in $O(N)$ time for all
$x_1,\ldots,x_N$.
However, problems of this sort are usually poorly scaled, and hence
standard gradient descent converges slowly. The most commonly used
higher-order technique is known as _Differential Dynamic Programming_
(DDP), which is an efficient recursive method for performing Newton's
method. Given a current control trajectory, it approximates the cost
function as a quadratic function of the controls, and solves for the
minimum of the function. A similar approach is the _Iterative LQR_ (iLQR) algorithm, which is very closely related to DDP but drops the 2nd derivative of the dynamics function. The exact steps for implementing DDP and iLQR are beyond
the scope of this course but are readily available from other sources.
### Pseudospectral / collocation methods
As an alternative to piecewise constant controls, it is also possible to
use other discretizations, such as polynomials or splines. In any case,
the control is specified by a linear combination of *basis functions*
$$u(t) = \sum_{i=1}^k c_i \beta_i(t)$$ where the $c_i \in \mathbb{R}^m$
are control coefficients, which are to be chosen by the optimization,
and the basis functions $\beta_i(t)$ are constant. For example, a set of
polynomial basis functions could be $1$, $t$, $t^2$, \..., $t^{k-1}$.
The difficulty with such parameterizations is that the state trajectory
depends on every control coefficient, so evaluating the gradient is
computationally expensive.
To address this problem, it is typical to include a *state trajectory parameterization* in
which the state trajectory $x(t)$ is also represented as an optimization variable that is
parameterized explicitly along with the control trajectory. Specifically, we suppose that $$x(t) = \sum_{i=1}^k d_i \gamma_i(t)$$ where the $d_i \in \mathbb{R}^n$ are state coefficients to be optimized, and the basis functions $\gamma_i$ are constant. The main challenge is then to enforce dynamic consistency between the $x$ trajectory and the $u$ trajectory over the time domain. Because it is impossible to do this exactly in a continuous infinity of points, the dynamic consistance must then be enforced at a finite number of points
in time, which are known as *collocation points*. The result is an
equality-constrained, finite-dimensional optimization problem.
Specifically, given $N$ points in the time domain $t_1,\ldots,t_N$, dynamic consistency is enforced at the $j$'th time point with a constraint
$$
x^\prime(t_j) = f(x(t_j),u(t_j) )
$$
which can be rewritten in terms of the cofficients
$$
\sum_{i=1}^k d_i \gamma_i^\prime(t_j) = f\left(\sum_{i=1}^k d_i \gamma_i(t_j), \sum_{i=1}^k c_i \beta_i(t_j) \right).
$$
### Handling infinite-horizon problems
There are some challenges when applying trajectory optimization to
infinite-horizon optimal control problems. Specifically, it is not
possible to define a computational grid over the infinite domain
$[0,\infty)$ for the purposes of computing the integral in $J(x,u)$. To
do so, there are two general techniques available. The first is to
simply truncate the problem at some maximum time $T$, leading to a
finite-horizon optimal control problem.
The second method is to reparameterize time so that the range
$[0,\infty)$ is transformed into a finite range, say $[0,1]$. If we let
$s=1-e^t$ then $s$ is in the range $[0,1]$. The cost functional then
becomes:
$$J(x,u) = \int_0^1 L(x(-\ln(1-s)),u(-\ln(1-s)),-\ln(1-s)) / (1-s) ds.$$
This leads to a finite-horizon optimal control problem over the $s$
domain, with $T=1$. Hence, if a uniform grid is defined over
$s \in [0,1]$, then the grid spacing in the time domain becomes
progressively large as $t$ increases.
In the reformulated problem it is necessary to express the derivative of
$x$ with respect to $s$ in the new dynamics:
$$\frac{d}{d s} x(t(s)) = \dot{x}(t(s)) t^\prime(s) = f(x(t(s)),u(t(s))) / (1-s)$$
Care must also be taken as $s$ approaches 1, since the $1/(1-s)$ term
approaches infinity, and if instantaneous cost does not also approach 0,
then cost will become infinite. It is therefore customary to use a
discount factor. With an appropriately defined discount term, the $s=1$
contribution to cost will be dropped.
### Local minima
A major issue with any descent-based trajectory optimization approach is
local minima. Only in a few cases can we prove that the problem is
convex, such as in LTI problems with convex costs and linear control
constraints. As we have seen before, random restarts are one of the most
effective ways to handle local minima, but in the high dimensional spaces of
trajectory optimization, a prohibitive number of restarts is needed to
have a high chance of finding a global optimum.
Hamilton-Jacobi-Bellman Equation
--------------------------------
An alternative method to solve optimal control problems is to find the
solution in *state space* rather than the time domain. In the
Hamilton-Jacobi-Bellman (HJB) equation, so named as an extension of the
Bellman equation for discrete optimal planning problems, a partial
differential equation (PDE) across state space is formulated to
determine the optimal control everywhere. (Contrast this with the
Pointryagin's minimum principle, which is an optimality condition only
along a single trajectory.)
### Derivation
We start by formulating the HJB equation in discrete time. Consider a
finite-horizon optimal control problem, and define the *value function*
as a function
$V(x,t) : \mathbb{R}^n \times \mathbb{R} \rightarrow \mathbb{R}$ that defines the
*minimum possible accumulated cost* that could be obtained by any trajectory
starting from initial state $x$ and time $t$. In other words, let us define the
truncated cost functional
$$J_t(x,u) = \int_t^T L(x,u,s) ds + \Phi(x(T))$$
which truncates the lower point in the integral term of $J(x,u)$ to start from time
$t$. (Obviously, $J(x,u)=J_0(x,u)$.) Then, the value function is the minimizer of
the truncated cost over all possible future controls: $V(x,t) = \min_u(J_t(x,u))$.
The value is also known as the *cost to come*, measuring the remaining cost to reach
a goal. (This stands in contrast to the *cost to go* which is the cost that would be
accumulated to reach a state $x$ from the start.)
It is apparent that at time $T$, the only term that remains is the
terminal cost, so one boundary term is given: $$V(x,T) = \Phi(x).$$ Now
we examine the value function going backwards in time. Suppose we know
$V(x,t+\Delta t)$ for all $x$, and now we are considering time $t$. Let
us also assume that at a state $x$ with control $u$, the resulting state
at time $T$ is approximated by Euler integration, and the incremental
cost is approximately constant over the interval $[t,t+\Delta t)$. Then,
we have the approximation
$$V(x,t) \approx \min_{u\in U} [ \Delta t L(x,u,t) + V(x + \Delta t f(x,u),t+\Delta t)]
\label{eq:DiscreteTimeHJB}$$ The minimization is taken over controls to
find the optimal control for the next time step. The first term of the
minimized term includes the incremental cost from the current state,
time, and chosen control. The second term includes the cost contribution
from the next state under the chosen control, incremented forward in
time.
Note that the first order approximation of $V(x,t)$ is given by:
$$V(x+\Delta x,t+\Delta t) \approx V(x,t) + \frac{\partial V}{\partial x}(x,t) \Delta x + \dot{V}(x,t)\Delta t$$
If we take the limit of
($\ref{eq:DiscreteTimeHJB}$) as the time step $\Delta t$ approaches
0, subtract $V(x,t)$ from both sides, and divide by $\Delta t$, then we
obtain the Hamilton-Jacobi-Bellman PDE :
$$0 = \dot{V}(x,t) + \min_{u \in U} [ L(x,u,t) + \frac{\partial V}{\partial x}(x,t) f(x,u)].
\label{eq:HJB}$$
If these equations were to be solved either in discrete or continuous
time across the $\mathbb{R}^n \times \mathbb{R}$ state space, then we
have a complete description of optimal cost starting from any state. It
is also possible to enforce state constraints simply by setting the
value function at inadmissible states to $\infty$. Moreover, it is a
relatively straightforward process to determine the optimal control
given a value function:
$$u^\star(x,t) = \arg \min_{u \in U} [ \Delta t L(x,u,t) + V(x + \Delta t f(x,u),t + \Delta t)]$$
for the discrete case and
$$u^\star(x,t) = \arg \min_{u \in U} [ L(x,u,t) + \frac{\partial V}{\partial x}(x,t) f(x,u)]$$
for the continuous case. The main challenge here is to represent and
calculate a function over an $n+1$ dimensional grid, which is
prohibitively expensive for high-D state spaces. It is also potentially
difficult to perform the minimization over the control
in ($\ref{eq:DiscreteTimeHJB}$)
and ($\ref{eq:HJB}$),
since it must be performed at each point in time and space.
### Reducing dimension by 1 using time-independence
It is often useful to reduce the dimensionality down to an $n$-D grid if
the incremental cost is time-independent and the problem has an infinite
horizon. With these assumptions, the optimal control is *stationary*,
that is, it is dependent only on state and not time. Then, we can set up
a set of recursive equations on a time-independent value function:
$$V(x) = \min_{u \in U} [ \Delta L(x,u) + V(x+\Delta t f(x,u)) ]
\label{eq:DiscreteHJBStationary}$$
in the discrete time case, or taking
the limit as $\Delta t \rightarrow 0$, we get the continuous PDE
$$0 = \min_{u \in U} [ L(x,u) + \frac{\partial V}{\partial x}(x) f(x,u) ].$$
### Solution methods
It can be rather challenging to solve either the time-dependent or the
stationary equations exactly due the dimensionality of the grids used,
and the recursive nature of the stationary equations. Also, some
discretization of the control set $U$ is usually needed, and a
finer discretization will help the method
compute better estimates. Three general methods exist for solving HJB
equations:
1. Value iteration uses a guess of $V(x)$ and then iteratively improves
it by optimizing
($\ref{eq:DiscreteHJBStationary}$) on each $x$ in the grid. This
is also known as recursive dynamic programming and is a continuous-space variant of the
value iteration algorithm discussed in [Chapter 11](PlanningWithDynamicsAndUncertainty.ipynb#Value-iteration).
2. Policy iteration assigns guesses for the policy $u(x)$, and
iteratively alternates between a) solving for the $V(x)$ induced by
those controls, and b) improving the assigned controls using the
induced $V(x)$. This is a continuous-space variant of the
policy iteration algorithm discussed in [Chapter 11](PlanningWithDynamicsAndUncertainty.ipynb#Policy-iteration).
3. Linear programming uses a set of sample points $x_1,\ldots,x_N$ on a
state space grid and points $u_1,\ldots,u_M$ on a control space
grid, and then sets up a large linear programming problem with
constraints of the form
($\ref{eq:DiscreteHJBStationary}$).
4. The Fast Marching Method can be thought of as a one-pass value iteration, which is
applicable to problems with known terminal sets and positive costs. The principle is similar to
the "brush fire" method for calculating navigation functions as discussed in
[Chapter 11](PlanningWithDynamicsAndUncertainty.ipynb#Navigation-functions-and-the-Dynamic-Window-Approach).
Observe that once a value is defined for the goal states, their value no
longer needs to be updated; they are called *closed*. We can try to
determine all states for which the value is below some threshold $v$, and these
states will be found in a small neighborhood of the closed states, known as the *frontier*.
Once these states' values are determined, they are added to the closed states, and a new set of
frontier states is determined. Next, we increase $v$ and repeat the process, until all
states are visited.
```
#Code for a pendulum HJB problem
from rsbook_code.control.examples.pendulum import Pendulum
from rsbook_code.control.optimalcontrol import OptimalControlProblem,ControlSampler,rollout_policy,LookaheadPolicy
from rsbook_code.control.objective import ObjectiveFunction
from rsbook_code.control.hjb import HJBSolver,OptimalControlTreeSolver
from rsbook_code.control.dynamics import simulate
import numpy as np
import math
#this is needed to sample from the control space
class PendulumControlSampler(ControlSampler):
def __init__(self,umin,umax):
self.umin = umin
self.umax = umax
def sample(self,state):
return [[self.umin],[0],[self.umax]]
class TimeObjectiveFunction(ObjectiveFunction):
def __init__(self,dt):
self.dt = dt
def incremental(self,state,control):
return abs(self.dt)
class EffortObjectiveFunction(ObjectiveFunction):
def __init__(self,dt):
self.dt = dt
def incremental(self,state,control):
return np.linalg.norm(control)**2*self.dt
#create the dynamics function, terminal conditions, and control bounds
dynamics = Pendulum()
umin = -2.5
umax = 2.5
down = np.array([math.pi*3/2,0])
right = np.array([0,0])
up = np.array([math.pi/2,0])
start = down
goal = up
bounds = [(0,math.pi*2),(-6,6)]
controlSampler = PendulumControlSampler(umin,umax)
#need to set dt large enough to have a chance to jump cells
dt = 0.1
objective = TimeObjectiveFunction(dt)
problem = OptimalControlProblem(start,dynamics,objective,goal=goal,controlSampler=controlSampler,dt=dt)
grid_resolution = (50,60)
hjb = HJBSolver(problem,bounds,grid_resolution)
scell = hjb.stateToCell(problem.x0)
print("Start state",start,"goal state",goal)
#print("Start cell",scell)
#print("Start cell center state",hjb.cellToCenterState(scell))
#print("Goal cell",hjb.stateToCell(problem.goal))
from rsbook_code.control.hjb import GridCostFunctionDisplay
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
from IPython.display import display,Markdown
import matplotlib.pyplot as plt
%matplotlib notebook
display(Markdown("#### HJB Solver"))
hjbdisplay = GridCostFunctionDisplay(hjb,hjb.value,hjb.policy,policyDims=1,figsize=(9.5,4))
hjbdisplay.show()
def do_value_iteration(i):
print("Running",i,"value iteration steps")
hjb.valueIteration(iters=i)
hjbdisplay.refresh(hjb.value,hjb.policy)
if hjb.getPolicy(start) is not None:
#show the HJB policy
xs,us = rollout_policy(dynamics,start,(lambda x:hjb.getPolicy(x)),dt*0.5,200)
hjbdisplay.plotTrajectory(xs,color='r',zorder=3)
la_policy = LookaheadPolicy(problem,hjb.interpolateValue,goal=(lambda x:False))
xs,us = rollout_policy(dynamics,start,la_policy,dt,200)
hjbdisplay.plotTrajectory(xs,color='y',zorder=4)
hjbdisplay.plotFlow(lambda x:hjb.getPolicy(x),color='k',linewidth=0.5)
interact_manual(do_value_iteration,i=widgets.IntSlider(min=1, max=101, step=10, value=11));
#this does forward search
tree = OptimalControlTreeSolver(problem,
bounds,[50,60])
tree.maxVisitedPerCell = 5
display(Markdown("# Forward Solver"))
treedisplay = GridCostFunctionDisplay(tree,tree.costToCome(),tree.reversePolicy(),policyDims=1,figsize=(9.5,4))
treedisplay.show()
def do_fw_search(N):
for i in range(N):
tree.search_step()
treedisplay.refresh(tree.costToCome(),tree.reversePolicy())
path = tree.result_path()
if tree.goal is not None:
assert len(path) > 0
if len(path) > 0:
if len(path[0].state)==0:
path = path[1:]
if path[-1].state == None:
path = path[:-1]
xs = np.array([n.state for n in path])
treedisplay.plotTrajectory(xs,color='r',zorder=3)
interact_manual(do_fw_search,N=widgets.IntSlider(min=1, max=10001, step=100, value=1001));
```
Model Predictive Control
------------------------
The method of model predictive control (MPC) is a process for building a
closed-loop controller when given a method that computes open loop
trajectories. Generally speaking, it simply replans a new trajectory
starting from the sensed state at each step. It executes some small
portion of that trajectory, senses the new state, and then replans
again. By repeating this process, MPC is able to cope with unexpected
disturbances by dynamically calculating paths to return to desirable
states.
There are, however, several caveats involved in successful application
of MPC. Let us define the steps more specifically. For a control loop
that operates at rate $\Delta t$, perform the following steps:
1. Sense the current state $x_c$
2. Compute a finite-time optimal trajectory $x,u$ starting at
$x(0) = x_c$.
3. Execute the control $u(t)$ for $t \in [0,\Delta t)$
4. Repeat from step 1.
There are several variables of note when creating an MPC approach.
First, the time step $\Delta t$ must be long enough for step 2 to find
an optimal trajectory. Second, the time horizon used in step 2 is an
important variable, because it should be long enough for MPC to benefit
from predictive lookahead, but not too long to make computational time
exceed $\Delta t$. Third, when recasting the problem as a finite-time
optimal control problem, the terminal cost should be somewhat
approximate the problem's value function, or else the system may not
converge properly. Finally, the optimization method used ought to be
extremely reliable. Failures in step 2 can be tolerated to some extent
simply by using the previous trajectory segment, but to achieve good
performance Step 2 should succeed regularly.
Due to all of these variables, MPC is difficult to analyze, and as
employed in practice it usually does not satisfy many theoretical
stability / convergence properties. However, with careful tuning, MPC
can be an extremely high performing and practical nonlinear optimal control
technique.
Summary
-------
Key takeaways:
- Optimal control functions are specified by a dynamics function, a
cost functional, and optional constraints. By changing the cost
functional, different performance objectives can be specified.
- Analytical optimal control techniques can be challenging to apply to
a given problem, but for some problems can yield computationally
efficient solutions by greatly reducing the search space.
- Numerical optimal control techniques are more general, but require
more computation.
- In trajectory optimization, time is discretized to produce a
finite-dimensional optimization problem. There is a tradeoff between
optimality and speed in the choice of computational grid resolution,
and are also susceptible to local minima.
- In Hamilton-Jacobi-Bellman (HJB) techniques, both time and space are
discretized. There are no local minima, but the technique suffers
from the curse of dimensionality.
- Model predictive control (MPC) turns open-loop trajectory
optimization into a closed-loop controller by means of repeated
replanning.
The [following table](#tab:OptControlSummary) lists an overview of the approaches
covered in this chapter.
********************************************************************
<div class="figcaption"><a name="tab:OptControlSummary">Summary of optimal control approaches</a></div>
| **Approach** | **Type** | **Characteristics** |
:------------------:|:------------:|:-----------------------------------------------------------------------:
| LQR |Analytical |Applies to LTI systems with quadratic costs |
| PMP |Analytical |Defines necessary conditions for optimality |
| Trajectory opt. |Numerical |Optimize a time-discretized control or trajectory space. Local minima |
| HJB |Numerical |Discretize and solve over state space |
| MPC |Numerical |Closed-loop control by repeated optimization |
********************************************************************
Exercises
---------
1. Consider devising an optimal control formulation that describes how
your arm should reach for a cup. What is the state $x$? The control
$u$? The dynamics $f(x,u)$? The objective functional? Is an
infinite-horizon or finite-horizon more appropriate for this
problem? Do the same for the task of balancing on one foot.
2. Recall the point mass double-integrator system:
$$\dot{x} \equiv \begin{bmatrix}{\dot{p}}\\{\dot{v}}\end{bmatrix} = f(x,u) = \begin{bmatrix}{v}\\{u/M}\end{bmatrix}.$$
Express this as an LTI system, and solve for the LQR gain matrix $K$
with the cost terms $Q=\begin{bmatrix}{10}&{0}\\{0}&{1}\end{bmatrix}$ and $R=5$.
3. Let $V^*(x)$ denote the infinite-horizon value function (i.e., the cost incurred by the optimal infinite-horizon control starting at $x$). Now define a finite-horizon optimal control problem with horizon $T$, the same incremental cost $L$, and terminal cost $\Phi(x) = V^*(x)$. Prove that the solution to this finite-horizon optimal control problem is identical to the infinite horizon optimal control problem for all $x$.
4. Let $V^*_T(x)$ denote the $T$-horizon value function for a terminal cost function $\Phi$. Suppose that $0 \leq T_1 < T_2$. Is it always true that $V^*_{T2}(x) \geq V^*_{T1}(x)$? If so, give a proof. If not, define a condition on $\Phi$ that would make this condition true.
5. TBD\...
|
github_jupyter
|
#Code for a pendulum HJB problem
from rsbook_code.control.examples.pendulum import Pendulum
from rsbook_code.control.optimalcontrol import OptimalControlProblem,ControlSampler,rollout_policy,LookaheadPolicy
from rsbook_code.control.objective import ObjectiveFunction
from rsbook_code.control.hjb import HJBSolver,OptimalControlTreeSolver
from rsbook_code.control.dynamics import simulate
import numpy as np
import math
#this is needed to sample from the control space
class PendulumControlSampler(ControlSampler):
def __init__(self,umin,umax):
self.umin = umin
self.umax = umax
def sample(self,state):
return [[self.umin],[0],[self.umax]]
class TimeObjectiveFunction(ObjectiveFunction):
def __init__(self,dt):
self.dt = dt
def incremental(self,state,control):
return abs(self.dt)
class EffortObjectiveFunction(ObjectiveFunction):
def __init__(self,dt):
self.dt = dt
def incremental(self,state,control):
return np.linalg.norm(control)**2*self.dt
#create the dynamics function, terminal conditions, and control bounds
dynamics = Pendulum()
umin = -2.5
umax = 2.5
down = np.array([math.pi*3/2,0])
right = np.array([0,0])
up = np.array([math.pi/2,0])
start = down
goal = up
bounds = [(0,math.pi*2),(-6,6)]
controlSampler = PendulumControlSampler(umin,umax)
#need to set dt large enough to have a chance to jump cells
dt = 0.1
objective = TimeObjectiveFunction(dt)
problem = OptimalControlProblem(start,dynamics,objective,goal=goal,controlSampler=controlSampler,dt=dt)
grid_resolution = (50,60)
hjb = HJBSolver(problem,bounds,grid_resolution)
scell = hjb.stateToCell(problem.x0)
print("Start state",start,"goal state",goal)
#print("Start cell",scell)
#print("Start cell center state",hjb.cellToCenterState(scell))
#print("Goal cell",hjb.stateToCell(problem.goal))
from rsbook_code.control.hjb import GridCostFunctionDisplay
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
from IPython.display import display,Markdown
import matplotlib.pyplot as plt
%matplotlib notebook
display(Markdown("#### HJB Solver"))
hjbdisplay = GridCostFunctionDisplay(hjb,hjb.value,hjb.policy,policyDims=1,figsize=(9.5,4))
hjbdisplay.show()
def do_value_iteration(i):
print("Running",i,"value iteration steps")
hjb.valueIteration(iters=i)
hjbdisplay.refresh(hjb.value,hjb.policy)
if hjb.getPolicy(start) is not None:
#show the HJB policy
xs,us = rollout_policy(dynamics,start,(lambda x:hjb.getPolicy(x)),dt*0.5,200)
hjbdisplay.plotTrajectory(xs,color='r',zorder=3)
la_policy = LookaheadPolicy(problem,hjb.interpolateValue,goal=(lambda x:False))
xs,us = rollout_policy(dynamics,start,la_policy,dt,200)
hjbdisplay.plotTrajectory(xs,color='y',zorder=4)
hjbdisplay.plotFlow(lambda x:hjb.getPolicy(x),color='k',linewidth=0.5)
interact_manual(do_value_iteration,i=widgets.IntSlider(min=1, max=101, step=10, value=11));
#this does forward search
tree = OptimalControlTreeSolver(problem,
bounds,[50,60])
tree.maxVisitedPerCell = 5
display(Markdown("# Forward Solver"))
treedisplay = GridCostFunctionDisplay(tree,tree.costToCome(),tree.reversePolicy(),policyDims=1,figsize=(9.5,4))
treedisplay.show()
def do_fw_search(N):
for i in range(N):
tree.search_step()
treedisplay.refresh(tree.costToCome(),tree.reversePolicy())
path = tree.result_path()
if tree.goal is not None:
assert len(path) > 0
if len(path) > 0:
if len(path[0].state)==0:
path = path[1:]
if path[-1].state == None:
path = path[:-1]
xs = np.array([n.state for n in path])
treedisplay.plotTrajectory(xs,color='r',zorder=3)
interact_manual(do_fw_search,N=widgets.IntSlider(min=1, max=10001, step=100, value=1001));
| 0.571049 | 0.985215 |
```
from google.colab import drive
drive.mount("/content/gdrive")
```
# **Importing Libraries & getting Authentication from Google Cloud**
```
!pip3 install google-cloud-speech
from google.colab import files
uploaded = files.upload()
for fn in uploaded.keys():
print('User uploaded file "{name}" with length {length} bytes'.format(
name=fn, length=len(uploaded[fn])))
!pwd
!ls -l ./gc_creds_STT.json
import os
os.environ['GOOGLE_APPLICATION_CREDENTIALS'] = '/content/gc_creds_STT.json'
!ls -l $GOOGLE_APPLICATION_CREDENTIALS
!pip install pydub
from pydub import AudioSegment
import io
import os
from google.cloud import speech_v1p1beta1 as speech
from google.cloud.speech_v1p1beta1 import enums
from google.cloud.speech_v1p1beta1 import types
import wave
from google.cloud import storage
```
# **Generating Transcripts**
###***For single Audio File***
```
!ffmpeg -i /content/Shubham_Q.No-12.mp4 /content/Shubham_Q.No-12.wav
filepath = '/content/Transcript/'
output_filepath = '/content/Transcript/'
def frame_rate_channel(audio_file_name):
with wave.open(audio_file_name, "rb") as wave_file:
frame_rate = wave_file.getframerate()
channels = wave_file.getnchannels()
return frame_rate,channels
def stereo_to_mono(audio_file_name):
sound = AudioSegment.from_wav(audio_file_name)
sound = sound.set_channels(1)
sound.export(audio_file_name, format="wav")
def upload_blob(bucket_name, source_file_name, destination_blob_name):
"""Uploads a file to the bucket."""
storage_client = storage.Client()
bucket = storage_client.get_bucket(bucket_name)
blob = bucket.blob(destination_blob_name)
blob.upload_from_filename(source_file_name)
def delete_blob(bucket_name, blob_name):
"""Deletes a blob from the bucket."""
storage_client = storage.Client()
bucket = storage_client.get_bucket(bucket_name)
blob = bucket.blob(blob_name)
blob.delete()
def google_transcribe(audio_file_name):
file_name = filepath + audio_file_name
#mp4_to_wav(file_name)
# The name of the audio file to transcribe
frame_rate, channels = frame_rate_channel(file_name)
if channels > 1:
stereo_to_mono(file_name)
bucket_name = 'calls_audio_file'
source_file_name = filepath + audio_file_name
destination_blob_name = audio_file_name
upload_blob(bucket_name, source_file_name, destination_blob_name)
gcs_uri = 'gs://calls_audio_file/' + audio_file_name
transcript = ''
client = speech.SpeechClient()
audio = types.RecognitionAudio(uri=gcs_uri)
config = types.RecognitionConfig(
encoding=enums.RecognitionConfig.AudioEncoding.LINEAR16,
sample_rate_hertz=frame_rate,
language_code='en-IN',
enable_automatic_punctuation=True,
enable_word_confidence=True,
)
# Detects speech in the audio file
operation = client.long_running_recognize(config, audio)
response = operation.result(timeout=10000)
for result in response.results:
transcript += result.alternatives[0].transcript
# The first result includes confidence levels per word
result = response.results[0]
# First alternative is the most probable result
alternative = result.alternatives[0]
transcript_filename = audio_file_name.split('.')[0] + '.txt'
f= write_transcripts(transcript_filename,transcript)
#appending Word Confidence for each word
for word in alternative.words:
f.write("\n\n Word: {} ; Confidence: {}".format(word.word,word.confidence))
f.seek(0) #to move the file reading pointer to the starting position
print(f.read())
f.close()
delete_blob(bucket_name, destination_blob_name)
return transcript
def write_transcripts(transcript_filename,transcript):
#writing transcript
f= open(output_filepath + transcript_filename,"w+")
f.write(transcript)
return f
if __name__ == "__main__":
for audio_file_name in os.listdir(filepath):
exists = os.path.isfile(output_filepath + audio_file_name.split('.')[0] + '.txt')
if exists:
pass
else:
transcript = google_transcribe(audio_file_name)
```
###***For Multiple Audio Files at a time***
```
filepath = '/content/drive/My Drive/EvueMe New Dataset/50 Can_be_Considered Candidates/Audio (50 Can_be_Considered)/'
output_filepath = '/content/drive/My Drive/EvueMe New Dataset/50 Can_be_Considered Candidates/Transcript/'
def frame_rate_channel(audio_file_name):
with wave.open(audio_file_name, "rb") as wave_file:
frame_rate = wave_file.getframerate()
channels = wave_file.getnchannels()
return frame_rate,channels
def stereo_to_mono(audio_file_name):
sound = AudioSegment.from_wav(audio_file_name)
sound = sound.set_channels(1)
sound.export(audio_file_name, format="wav")
def upload_blob(bucket_name, source_file_name, destination_blob_name):
"""Uploads a file to the bucket."""
storage_client = storage.Client()
bucket = storage_client.get_bucket(bucket_name)
blob = bucket.blob(destination_blob_name)
blob.upload_from_filename(source_file_name)
def delete_blob(bucket_name, blob_name):
"""Deletes a blob from the bucket."""
storage_client = storage.Client()
bucket = storage_client.get_bucket(bucket_name)
blob = bucket.blob(blob_name)
blob.delete()
def google_transcribe(audio_file_name):
file_name = filepath + audio_file_name
#mp4_to_wav(file_name)
# The name of the audio file to transcribe
frame_rate, channels = frame_rate_channel(file_name)
if channels > 1:
stereo_to_mono(file_name)
bucket_name = 'calls_audio_file'
source_file_name = filepath + audio_file_name
destination_blob_name = audio_file_name
upload_blob(bucket_name, source_file_name, destination_blob_name)
gcs_uri = 'gs://calls_audio_file/' + audio_file_name
transcript = ''
client = speech.SpeechClient()
audio = types.RecognitionAudio(uri=gcs_uri)
config = types.RecognitionConfig(
encoding=enums.RecognitionConfig.AudioEncoding.LINEAR16,
sample_rate_hertz=frame_rate,
language_code='en-IN',
enable_automatic_punctuation=True,
enable_word_confidence=True,
)
# Detects speech in the audio file
operation = client.long_running_recognize(config, audio)
response = operation.result(timeout=10000)
for result in response.results:
transcript += result.alternatives[0].transcript
# The first result includes confidence levels per word
result = response.results[0]
# First alternative is the most probable result
alternative = result.alternatives[0]
transcript_filename = audio_file_name.split('.')[0] + '.txt'
f= write_transcripts(transcript_filename,transcript)
#appending Word Confidence for each word
for word in alternative.words:
f.write("\n\n Word: {} ; Confidence: {}".format(word.word,word.confidence))
f.seek(0) #to move the file reading pointer to the starting position
print(f.read())
f.close()
delete_blob(bucket_name, destination_blob_name)
return transcript
def write_transcripts(transcript_filename,transcript):
#writing transcript
f= open(output_filepath + transcript_filename,"w+")
f.write(transcript)
return f
if __name__ == "__main__":
for audio_file_name in os.listdir(filepath):
exists = os.path.isfile(output_filepath + audio_file_name.split('.')[0] + '.txt')
if exists:
pass
else:
transcript = google_transcribe(audio_file_name)
```
|
github_jupyter
|
from google.colab import drive
drive.mount("/content/gdrive")
!pip3 install google-cloud-speech
from google.colab import files
uploaded = files.upload()
for fn in uploaded.keys():
print('User uploaded file "{name}" with length {length} bytes'.format(
name=fn, length=len(uploaded[fn])))
!pwd
!ls -l ./gc_creds_STT.json
import os
os.environ['GOOGLE_APPLICATION_CREDENTIALS'] = '/content/gc_creds_STT.json'
!ls -l $GOOGLE_APPLICATION_CREDENTIALS
!pip install pydub
from pydub import AudioSegment
import io
import os
from google.cloud import speech_v1p1beta1 as speech
from google.cloud.speech_v1p1beta1 import enums
from google.cloud.speech_v1p1beta1 import types
import wave
from google.cloud import storage
!ffmpeg -i /content/Shubham_Q.No-12.mp4 /content/Shubham_Q.No-12.wav
filepath = '/content/Transcript/'
output_filepath = '/content/Transcript/'
def frame_rate_channel(audio_file_name):
with wave.open(audio_file_name, "rb") as wave_file:
frame_rate = wave_file.getframerate()
channels = wave_file.getnchannels()
return frame_rate,channels
def stereo_to_mono(audio_file_name):
sound = AudioSegment.from_wav(audio_file_name)
sound = sound.set_channels(1)
sound.export(audio_file_name, format="wav")
def upload_blob(bucket_name, source_file_name, destination_blob_name):
"""Uploads a file to the bucket."""
storage_client = storage.Client()
bucket = storage_client.get_bucket(bucket_name)
blob = bucket.blob(destination_blob_name)
blob.upload_from_filename(source_file_name)
def delete_blob(bucket_name, blob_name):
"""Deletes a blob from the bucket."""
storage_client = storage.Client()
bucket = storage_client.get_bucket(bucket_name)
blob = bucket.blob(blob_name)
blob.delete()
def google_transcribe(audio_file_name):
file_name = filepath + audio_file_name
#mp4_to_wav(file_name)
# The name of the audio file to transcribe
frame_rate, channels = frame_rate_channel(file_name)
if channels > 1:
stereo_to_mono(file_name)
bucket_name = 'calls_audio_file'
source_file_name = filepath + audio_file_name
destination_blob_name = audio_file_name
upload_blob(bucket_name, source_file_name, destination_blob_name)
gcs_uri = 'gs://calls_audio_file/' + audio_file_name
transcript = ''
client = speech.SpeechClient()
audio = types.RecognitionAudio(uri=gcs_uri)
config = types.RecognitionConfig(
encoding=enums.RecognitionConfig.AudioEncoding.LINEAR16,
sample_rate_hertz=frame_rate,
language_code='en-IN',
enable_automatic_punctuation=True,
enable_word_confidence=True,
)
# Detects speech in the audio file
operation = client.long_running_recognize(config, audio)
response = operation.result(timeout=10000)
for result in response.results:
transcript += result.alternatives[0].transcript
# The first result includes confidence levels per word
result = response.results[0]
# First alternative is the most probable result
alternative = result.alternatives[0]
transcript_filename = audio_file_name.split('.')[0] + '.txt'
f= write_transcripts(transcript_filename,transcript)
#appending Word Confidence for each word
for word in alternative.words:
f.write("\n\n Word: {} ; Confidence: {}".format(word.word,word.confidence))
f.seek(0) #to move the file reading pointer to the starting position
print(f.read())
f.close()
delete_blob(bucket_name, destination_blob_name)
return transcript
def write_transcripts(transcript_filename,transcript):
#writing transcript
f= open(output_filepath + transcript_filename,"w+")
f.write(transcript)
return f
if __name__ == "__main__":
for audio_file_name in os.listdir(filepath):
exists = os.path.isfile(output_filepath + audio_file_name.split('.')[0] + '.txt')
if exists:
pass
else:
transcript = google_transcribe(audio_file_name)
filepath = '/content/drive/My Drive/EvueMe New Dataset/50 Can_be_Considered Candidates/Audio (50 Can_be_Considered)/'
output_filepath = '/content/drive/My Drive/EvueMe New Dataset/50 Can_be_Considered Candidates/Transcript/'
def frame_rate_channel(audio_file_name):
with wave.open(audio_file_name, "rb") as wave_file:
frame_rate = wave_file.getframerate()
channels = wave_file.getnchannels()
return frame_rate,channels
def stereo_to_mono(audio_file_name):
sound = AudioSegment.from_wav(audio_file_name)
sound = sound.set_channels(1)
sound.export(audio_file_name, format="wav")
def upload_blob(bucket_name, source_file_name, destination_blob_name):
"""Uploads a file to the bucket."""
storage_client = storage.Client()
bucket = storage_client.get_bucket(bucket_name)
blob = bucket.blob(destination_blob_name)
blob.upload_from_filename(source_file_name)
def delete_blob(bucket_name, blob_name):
"""Deletes a blob from the bucket."""
storage_client = storage.Client()
bucket = storage_client.get_bucket(bucket_name)
blob = bucket.blob(blob_name)
blob.delete()
def google_transcribe(audio_file_name):
file_name = filepath + audio_file_name
#mp4_to_wav(file_name)
# The name of the audio file to transcribe
frame_rate, channels = frame_rate_channel(file_name)
if channels > 1:
stereo_to_mono(file_name)
bucket_name = 'calls_audio_file'
source_file_name = filepath + audio_file_name
destination_blob_name = audio_file_name
upload_blob(bucket_name, source_file_name, destination_blob_name)
gcs_uri = 'gs://calls_audio_file/' + audio_file_name
transcript = ''
client = speech.SpeechClient()
audio = types.RecognitionAudio(uri=gcs_uri)
config = types.RecognitionConfig(
encoding=enums.RecognitionConfig.AudioEncoding.LINEAR16,
sample_rate_hertz=frame_rate,
language_code='en-IN',
enable_automatic_punctuation=True,
enable_word_confidence=True,
)
# Detects speech in the audio file
operation = client.long_running_recognize(config, audio)
response = operation.result(timeout=10000)
for result in response.results:
transcript += result.alternatives[0].transcript
# The first result includes confidence levels per word
result = response.results[0]
# First alternative is the most probable result
alternative = result.alternatives[0]
transcript_filename = audio_file_name.split('.')[0] + '.txt'
f= write_transcripts(transcript_filename,transcript)
#appending Word Confidence for each word
for word in alternative.words:
f.write("\n\n Word: {} ; Confidence: {}".format(word.word,word.confidence))
f.seek(0) #to move the file reading pointer to the starting position
print(f.read())
f.close()
delete_blob(bucket_name, destination_blob_name)
return transcript
def write_transcripts(transcript_filename,transcript):
#writing transcript
f= open(output_filepath + transcript_filename,"w+")
f.write(transcript)
return f
if __name__ == "__main__":
for audio_file_name in os.listdir(filepath):
exists = os.path.isfile(output_filepath + audio_file_name.split('.')[0] + '.txt')
if exists:
pass
else:
transcript = google_transcribe(audio_file_name)
| 0.484136 | 0.32748 |
# packages used
```
%matplotlib inline
# misc
from IPython.display import display, HTML
import numpy as np
# DATA - prep
#kaggle
import pandas as pd
import sklearn.model_selection
# ML - models
import sklearn.linear_model
import sklearn.tree
import sklearn.ensemble
import xgboost.sklearn
# ML - accuracy
import sklearn.metrics
# Plot and visualize
import matplotlib.pyplot as plt
import shap
```
# Get data
Setup:
- follow "API credential step" listed here: https://github.com/Kaggle/kaggle-api
- go to https://www.kaggle.com/ (login)
- go to my_profile (download kaggle.json)
- put it in ~/.kaggle/kaggle.json
- `cp ~/Downloads/kaggle.json ~/.kaggle/kaggle.json`
- `chmod 600 ~/.kaggle/kaggle.json`
- Go to kaggle and join competition:
- https://www.kaggle.com/c/titanic
- install kaggle
- download data
- profit!!!
```
!pip install kaggle -q
# -q is just for quite, so we don't spam the notebook
metadata = {
'basepath' : '../data/',
'dataset':'titanic',
'train' : 'train.csv',
'test' : 'test.csv'}
# make folder
# download .zip
# unzip
# remove the .zip
# (data is placed ../data/titanic)
!mkdir -p {metadata['basepath']}
!kaggle competitions download -c dataset {metadata['dataset']} -p {metadata['basepath']}
!unzip -o {metadata['basepath']}{metadata['dataset']}.zip -d {metadata['basepath']}{metadata['dataset']}/
!rm {metadata['basepath']}{metadata['dataset']}.zip
```
# Load and explore
```
# load
train = pd.read_csv("{basepath}/{dataset}/{train}".format(**metadata))
test = pd.read_csv("{basepath}/{dataset}/{test}".format(**metadata))
# Train
display(HTML("<h1>train</h1>"))
# example data
display(train.head(3))
# summary stats
display(train.describe())
# list missing values
display(pd.DataFrame(train.isna().mean() ,columns=["is na fraction"]))
# list types of column
display(train.dtypes)
# list dimenstion
display(train.shape)
# TODO check test
display(HTML("<h1>test</h1>"))
display(pd.DataFrame(test.isna().mean() ,columns=["is na fraction"]))
```
# Simple imputation + cleaning
```
def clean(df):
dfc = df.copy()
# Simple map
dfc['Sex'] = dfc['Sex'].map({"female":0,"male":1}).astype(int)
# simple Impute
dfc['Age'] = dfc["Age"].fillna(-1)
dfc['Fare'] = dfc["Fare"].fillna(-1)
# Simple feature engineering (combining two variables)
dfc['FamilySize'] = dfc['SibSp'] + dfc['Parch'] + 1
# Simple feature engineering (converting to boolean)
dfc['Has_Cabin'] = dfc["Cabin"].apply(lambda x: 0 if type(x) == float else 1)
dfc = dfc.drop(["Cabin"],axis=1)
# "Stupid feature engineering - apply length
dfc['Name_length'] = dfc['Name'].apply(len)
dfc = dfc.drop(["Name"],axis=1)
dfc['Ticket_length'] = dfc['Ticket'].apply(len)
dfc = dfc.drop(["Ticket"],axis=1)
# 1-hot encoding - different options are encoded as booleans
# ie. 1 categorical - become 3: 0-1 features.
dfc['Embarked_Q'] = dfc['Embarked'].apply(lambda x: 1 if x=="Q" else 0)
dfc['Embarked_S'] = dfc['Embarked'].apply(lambda x: 1 if x=="S" else 0)
dfc['Embarked_C'] = dfc['Embarked'].apply(lambda x: 1 if x=="C" else 0)
dfc = dfc.drop(["Embarked"],axis=1)
return dfc
clean_train = clean(train)
clean_test = clean(test)
display(pd.DataFrame(clean_test.isna().mean() ,columns=["is na fraction"]))
# clean data / build feature
# to_expand
target = "Survived"
# keep numeric features without missing vals
#keep_features = ["Pclass","SibSp","Parch"]
y = clean_train[target]
X = clean_train.drop([target],axis=1)
# Split data in train and validation
target = "Survived"
seed = 42
test_size = 0.7
X_train, X_val, y_train, y_val = sklearn.model_selection.train_test_split(
X,
y,
random_state = seed,
test_size = test_size)
```
# ML
```
# default models
# Logistic regression
model_logreg = sklearn.linear_model.LogisticRegression()
model_logreg.fit(X_train, y_train);
# decision tree
model_decision_tree = sklearn.tree.DecisionTreeClassifier()
model_decision_tree.fit(X_train, y_train);
# randomForest
model_random_forest = sklearn.ensemble.RandomForestClassifier()
model_random_forest.fit(X_train, y_train);
# xgboost
model_xgboost = xgboost.sklearn.XGBClassifier()
model_xgboost.fit(X_train, y_train);
```
# Eval ML
```
# naive model
class naive_model():
# everyone dies
def predict(self, df):
return np.zeros(df.shape[0])
model_naive = naive_model()
models = {
"model_naive" : model_naive,
"model_logreg" : model_logreg,
"model_decision_tree": model_decision_tree,
"model_random_forest": model_random_forest,
"model_xgboost" :model_xgboost
}
for name,model in zip(models.keys(),models.values()):
acc = sklearn.metrics.accuracy_score(
y_true = y_val,
y_pred = model.predict(X_val)
)
print(name,round(acc,4))
```
# Visualize feature importance
```
def plot_feature_graphs(model, X, plot_all=True):
#xgb.plot_importance(model)
plt.show()
explainer = shap.TreeExplainer(model)
shap_values = explainer.shap_values(X)
shap.summary_plot(shap_values, X)
shap.summary_plot(shap_values, X, plot_type='bar')
if plot_all:
vals = list(np.abs(shap_values).mean(0))
labels = list(X.columns.values)
col_order = [x for _,x in sorted(zip(vals,labels),reverse=True)]
for c in col_order:
shap.dependence_plot(c,shap_values,X)
plot_feature_graphs(model_xgboost, X_train)
```
# Output
```
# passengerid
id = "PassengerId"
out = pd.DataFrame(data = test[id], columns = [id])
# target
out_target = model_logreg.predict(test[keep_features])
out[target] = pd.DataFrame(out_target
,columns = [target]
,dtype=np.int32
)
# put them out
outfile = metadata["basepath"] + "output_logreg.csv"
out.to_csv(path_or_buf = outfile,
index = False)
# Submit
#!kaggle competitions submit {metadata['dataset']} -f {outfile} -m "minimal model"
# See submission
!kaggle competitions submissions "{metadata['dataset']}"
```
|
github_jupyter
|
%matplotlib inline
# misc
from IPython.display import display, HTML
import numpy as np
# DATA - prep
#kaggle
import pandas as pd
import sklearn.model_selection
# ML - models
import sklearn.linear_model
import sklearn.tree
import sklearn.ensemble
import xgboost.sklearn
# ML - accuracy
import sklearn.metrics
# Plot and visualize
import matplotlib.pyplot as plt
import shap
!pip install kaggle -q
# -q is just for quite, so we don't spam the notebook
metadata = {
'basepath' : '../data/',
'dataset':'titanic',
'train' : 'train.csv',
'test' : 'test.csv'}
# make folder
# download .zip
# unzip
# remove the .zip
# (data is placed ../data/titanic)
!mkdir -p {metadata['basepath']}
!kaggle competitions download -c dataset {metadata['dataset']} -p {metadata['basepath']}
!unzip -o {metadata['basepath']}{metadata['dataset']}.zip -d {metadata['basepath']}{metadata['dataset']}/
!rm {metadata['basepath']}{metadata['dataset']}.zip
# load
train = pd.read_csv("{basepath}/{dataset}/{train}".format(**metadata))
test = pd.read_csv("{basepath}/{dataset}/{test}".format(**metadata))
# Train
display(HTML("<h1>train</h1>"))
# example data
display(train.head(3))
# summary stats
display(train.describe())
# list missing values
display(pd.DataFrame(train.isna().mean() ,columns=["is na fraction"]))
# list types of column
display(train.dtypes)
# list dimenstion
display(train.shape)
# TODO check test
display(HTML("<h1>test</h1>"))
display(pd.DataFrame(test.isna().mean() ,columns=["is na fraction"]))
def clean(df):
dfc = df.copy()
# Simple map
dfc['Sex'] = dfc['Sex'].map({"female":0,"male":1}).astype(int)
# simple Impute
dfc['Age'] = dfc["Age"].fillna(-1)
dfc['Fare'] = dfc["Fare"].fillna(-1)
# Simple feature engineering (combining two variables)
dfc['FamilySize'] = dfc['SibSp'] + dfc['Parch'] + 1
# Simple feature engineering (converting to boolean)
dfc['Has_Cabin'] = dfc["Cabin"].apply(lambda x: 0 if type(x) == float else 1)
dfc = dfc.drop(["Cabin"],axis=1)
# "Stupid feature engineering - apply length
dfc['Name_length'] = dfc['Name'].apply(len)
dfc = dfc.drop(["Name"],axis=1)
dfc['Ticket_length'] = dfc['Ticket'].apply(len)
dfc = dfc.drop(["Ticket"],axis=1)
# 1-hot encoding - different options are encoded as booleans
# ie. 1 categorical - become 3: 0-1 features.
dfc['Embarked_Q'] = dfc['Embarked'].apply(lambda x: 1 if x=="Q" else 0)
dfc['Embarked_S'] = dfc['Embarked'].apply(lambda x: 1 if x=="S" else 0)
dfc['Embarked_C'] = dfc['Embarked'].apply(lambda x: 1 if x=="C" else 0)
dfc = dfc.drop(["Embarked"],axis=1)
return dfc
clean_train = clean(train)
clean_test = clean(test)
display(pd.DataFrame(clean_test.isna().mean() ,columns=["is na fraction"]))
# clean data / build feature
# to_expand
target = "Survived"
# keep numeric features without missing vals
#keep_features = ["Pclass","SibSp","Parch"]
y = clean_train[target]
X = clean_train.drop([target],axis=1)
# Split data in train and validation
target = "Survived"
seed = 42
test_size = 0.7
X_train, X_val, y_train, y_val = sklearn.model_selection.train_test_split(
X,
y,
random_state = seed,
test_size = test_size)
# default models
# Logistic regression
model_logreg = sklearn.linear_model.LogisticRegression()
model_logreg.fit(X_train, y_train);
# decision tree
model_decision_tree = sklearn.tree.DecisionTreeClassifier()
model_decision_tree.fit(X_train, y_train);
# randomForest
model_random_forest = sklearn.ensemble.RandomForestClassifier()
model_random_forest.fit(X_train, y_train);
# xgboost
model_xgboost = xgboost.sklearn.XGBClassifier()
model_xgboost.fit(X_train, y_train);
# naive model
class naive_model():
# everyone dies
def predict(self, df):
return np.zeros(df.shape[0])
model_naive = naive_model()
models = {
"model_naive" : model_naive,
"model_logreg" : model_logreg,
"model_decision_tree": model_decision_tree,
"model_random_forest": model_random_forest,
"model_xgboost" :model_xgboost
}
for name,model in zip(models.keys(),models.values()):
acc = sklearn.metrics.accuracy_score(
y_true = y_val,
y_pred = model.predict(X_val)
)
print(name,round(acc,4))
def plot_feature_graphs(model, X, plot_all=True):
#xgb.plot_importance(model)
plt.show()
explainer = shap.TreeExplainer(model)
shap_values = explainer.shap_values(X)
shap.summary_plot(shap_values, X)
shap.summary_plot(shap_values, X, plot_type='bar')
if plot_all:
vals = list(np.abs(shap_values).mean(0))
labels = list(X.columns.values)
col_order = [x for _,x in sorted(zip(vals,labels),reverse=True)]
for c in col_order:
shap.dependence_plot(c,shap_values,X)
plot_feature_graphs(model_xgboost, X_train)
# passengerid
id = "PassengerId"
out = pd.DataFrame(data = test[id], columns = [id])
# target
out_target = model_logreg.predict(test[keep_features])
out[target] = pd.DataFrame(out_target
,columns = [target]
,dtype=np.int32
)
# put them out
outfile = metadata["basepath"] + "output_logreg.csv"
out.to_csv(path_or_buf = outfile,
index = False)
# Submit
#!kaggle competitions submit {metadata['dataset']} -f {outfile} -m "minimal model"
# See submission
!kaggle competitions submissions "{metadata['dataset']}"
| 0.356671 | 0.648508 |
```
#export
from local.torch_basics import *
from local.test import *
from local.core import *
from local.layers import *
from local.data.all import *
from local.text.core import *
from local.text.models.awdlstm import *
from local.notebook.showdoc import *
#default_exp text.models.core
#default_cls_lvl 3
```
# Core text modules
> Contain the modules common between different architectures and the generic functions to get models
```
#export
_model_meta = {AWD_LSTM: {'hid_name':'emb_sz', 'url':URLs.WT103_FWD, 'url_bwd':URLs.WT103_BWD,
'config_lm':awd_lstm_lm_config, 'split_lm': awd_lstm_lm_split,
'config_clas':awd_lstm_clas_config, 'split_clas': awd_lstm_clas_split},
AWD_QRNN: {'hid_name':'emb_sz',
'config_lm':awd_qrnn_lm_config, 'split_lm': awd_lstm_lm_split,
'config_clas':awd_qrnn_clas_config, 'split_clas': awd_lstm_clas_split},}
# Transformer: {'hid_name':'d_model', 'url':URLs.OPENAI_TRANSFORMER,
# 'config_lm':tfmer_lm_config, 'split_lm': tfmer_lm_split,
# 'config_clas':tfmer_clas_config, 'split_clas': tfmer_clas_split},
# TransformerXL: {'hid_name':'d_model',
# 'config_lm':tfmerXL_lm_config, 'split_lm': tfmerXL_lm_split,
# 'config_clas':tfmerXL_clas_config, 'split_clas': tfmerXL_clas_split}}
```
## Language models
```
#export
class LinearDecoder(Module):
"To go on top of a RNNCore module and create a Language Model."
initrange=0.1
def __init__(self, n_out, n_hid, output_p=0.1, tie_encoder=None, bias=True):
self.decoder = nn.Linear(n_hid, n_out, bias=bias)
self.decoder.weight.data.uniform_(-self.initrange, self.initrange)
self.output_dp = RNNDropout(output_p)
if bias: self.decoder.bias.data.zero_()
if tie_encoder: self.decoder.weight = tie_encoder.weight
def forward(self, input):
raw_outputs, outputs = input
decoded = self.decoder(self.output_dp(outputs[-1]))
return decoded, raw_outputs, outputs
from local.text.models.awdlstm import *
enc = AWD_LSTM(100, 20, 10, 2)
x = torch.randint(0, 100, (10,5))
r = enc(x)
tst = LinearDecoder(100, 20, 0.1)
y = tst(r)
test_eq(y[1], r[0])
test_eq(y[2], r[1])
test_eq(y[0].shape, [10, 5, 100])
tst = LinearDecoder(100, 20, 0.1, tie_encoder=enc.encoder)
test_eq(tst.decoder.weight, enc.encoder.weight)
#export
class SequentialRNN(nn.Sequential):
"A sequential module that passes the reset call to its children."
def reset(self):
for c in self.children(): getattr(c, 'reset', noop)()
class _TstMod(Module):
def reset(self): print('reset')
tst = SequentialRNN(_TstMod(), _TstMod())
test_stdout(tst.reset, 'reset\nreset')
#export
def get_language_model(arch, vocab_sz, config=None, drop_mult=1.):
"Create a language model from `arch` and its `config`."
meta = _model_meta[arch]
config = ifnone(config, meta['config_lm']).copy()
for k in config.keys():
if k.endswith('_p'): config[k] *= drop_mult
tie_weights,output_p,out_bias = map(config.pop, ['tie_weights', 'output_p', 'out_bias'])
init = config.pop('init') if 'init' in config else None
encoder = arch(vocab_sz, **config)
enc = encoder.encoder if tie_weights else None
decoder = LinearDecoder(vocab_sz, config[meta['hid_name']], output_p, tie_encoder=enc, bias=out_bias)
model = SequentialRNN(encoder, decoder)
return model if init is None else model.apply(init)
```
The default `config` used can be found in `_model_meta[arch]['config_lm']`. `drop_mult` is applied to all the probabilities of dropout in that config.
```
config = awd_lstm_lm_config.copy()
config.update({'n_hid':10, 'emb_sz':20})
tst = get_language_model(AWD_LSTM, 100, config=config)
x = torch.randint(0, 100, (10,5))
y = tst(x)
test_eq(y[0].shape, [10, 5, 100])
test_eq(tst[1].decoder.weight, tst[0].encoder.weight)
for i in range(1,3): test_eq([h_.shape for h_ in y[1]], [[10, 5, 10], [10, 5, 10], [10, 5, 20]])
#test drop_mult
tst = get_language_model(AWD_LSTM, 100, config=config, drop_mult=0.5)
test_eq(tst[1].output_dp.p, config['output_p']*0.5)
for rnn in tst[0].rnns: test_eq(rnn.weight_p, config['weight_p']*0.5)
for dp in tst[0].hidden_dps: test_eq(dp.p, config['hidden_p']*0.5)
test_eq(tst[0].encoder_dp.embed_p, config['embed_p']*0.5)
test_eq(tst[0].input_dp.p, config['input_p']*0.5)
```
## Classification models
```
#export
def _pad_tensor(t, bs, val=0.):
if t.size(0) < bs: return torch.cat([t, val + t.new_zeros(bs-t.size(0), *t.shape[1:])])
return t
#export
class SentenceEncoder(Module):
"Create an encoder over `module` that can process a full sentence."
def __init__(self, bptt, module, pad_idx=1): store_attr(self, 'bptt,module,pad_idx')
def _concat(self, arrs, bs):
return [torch.cat([_pad_tensor(l[si],bs) for l in arrs], dim=1) for si in range(len(arrs[0]))]
def reset(self): getattr(self.module, 'reset', noop)()
def forward(self, input):
bs,sl = input.size()
self.reset()
raw_outputs,outputs,masks = [],[],[]
for i in range(0, sl, self.bptt):
r,o = self.module(input[:,i: min(i+self.bptt, sl)])
masks.append(input[:,i: min(i+self.bptt, sl)] == self.pad_idx)
raw_outputs.append(r)
outputs.append(o)
return self._concat(raw_outputs, bs),self._concat(outputs, bs),torch.cat(masks,dim=1)
class DoubleEmbedding(nn.Embedding):
def forward(self, x):
y = super().forward(x)
return ([y],[y+1])
mod = DoubleEmbedding(5, 10,)
tst = SentenceEncoder(5, mod, pad_idx=0)
x = torch.randint(1, 5, (3, 15))
x[2,10:]=0
raw,out,mask = tst(x)
test_eq(raw[0], mod(x)[0][0])
test_eq(out[0], mod(x)[0][0]+1)
test_eq(mask, x==0)
class PoolingLinearClassifier(nn.Module):
"Create a linear classifier with pooling."
def __init__(self, layers, drops):
super().__init__()
mod_layers = []
activs = [nn.ReLU(inplace=True)] * (len(layers) - 2) + [None]
for n_in, n_out, p, actn in zip(layers[:-1], layers[1:], drops, activs):
mod_layers += bn_drop_lin(n_in, n_out, p=p, actn=actn)
self.layers = nn.Sequential(*mod_layers)
def forward(self, input):
raw_outputs,outputs,mask = input
output = outputs[-1]
lengths = output.size(1) - mask.long().sum(dim=1)
avg_pool = output.masked_fill(mask[:,:,None], 0).sum(dim=1)
avg_pool.div_(lengths.type(avg_pool.dtype)[:,None])
max_pool = output.masked_fill(mask[:,:,None], -float('inf')).max(dim=1)[0]
x = torch.cat([output[torch.arange(0, output.size(0)),lengths-1], max_pool, avg_pool], 1) #Concat pooling.
x = self.layers(x)
return x
#export
def masked_concat_pool(outputs, mask):
"Pool `MultiBatchEncoder` outputs into one vector [last_hidden, max_pool, avg_pool]"
output = outputs[-1]
lens = output.size(1) - mask.long().sum(dim=1)
avg_pool = output.masked_fill(mask[:, :, None], 0).sum(dim=1)
avg_pool.div_(lens.type(avg_pool.dtype)[:,None])
max_pool = output.masked_fill(mask[:,:,None], -float('inf')).max(dim=1)[0]
x = torch.cat([output[torch.arange(0, output.size(0)),lens-1], max_pool, avg_pool], 1) #Concat pooling.
return x
out = torch.randn(2,3,5)
mask = tensor([[False,False,True], [False,False,False]])
x = masked_concat_pool([out], mask)
test_close(x[0,:5], out[0,-2])
test_close(x[1,:5], out[1,-1])
test_close(x[0,5:10], out[0,:2].max(dim=0)[0])
test_close(x[1,5:10], out[1].max(dim=0)[0])
test_close(x[0,10:], out[0,:2].mean(dim=0))
test_close(x[1,10:], out[1].mean(dim=0))
#Test the result is independent of padding
out1 = torch.randn(2,4,5)
out1[:,:-1] = out.clone()
mask1 = tensor([[False,False,True,True], [False,False,False,True]])
x1 = masked_concat_pool([out1], mask1)
test_eq(x, x1)
#export
class PoolingLinearClassifier(Module):
"Create a linear classifier with pooling"
def __init__(self, dims, ps):
mod_layers = []
if len(ps) != len(dims)-1: raise ValueError("Number of layers and dropout values do not match.")
acts = [nn.ReLU(inplace=True)] * (len(dims) - 2) + [None]
layers = [BnDropLin(i, o, p=p, act=a) for i,o,p,a in zip(dims[:-1], dims[1:], ps, acts)]
self.layers = nn.Sequential(*layers)
def forward(self, input):
raw,out,mask = input
x = masked_concat_pool(out, mask)
x = self.layers(x)
return x, raw, out
mod = DoubleEmbedding(5, 10)
tst = nn.Sequential(SentenceEncoder(5, mod, pad_idx=0), PoolingLinearClassifier([10*3,4], [0.]))
x = torch.randint(1, 5, (3, 14))
x[2,10:] = 0
res,raw,out = tst(x)
test_eq(raw[0], mod(x)[0][0])
test_eq(out[0], mod(x)[0][0]+1)
test_eq(res.shape, [3,4])
x1 = torch.cat([x, tensor([0,0,0])[:,None]], dim=1)
res1,raw1,out1 = tst(x1)
test_eq(res, res1)
#export
def get_text_classifier(arch, vocab_sz, n_class, bptt=72, config=None, drop_mult=1., lin_ftrs=None,
ps=None, pad_idx=1):
"Create a text classifier from `arch` and its `config`, maybe `pretrained`"
meta = _model_meta[arch]
config = ifnone(config, meta['config_clas']).copy()
for k in config.keys():
if k.endswith('_p'): config[k] *= drop_mult
if lin_ftrs is None: lin_ftrs = [50]
if ps is None: ps = [0.1]*len(lin_ftrs)
layers = [config[meta['hid_name']] * 3] + lin_ftrs + [n_class]
ps = [config.pop('output_p')] + ps
init = config.pop('init') if 'init' in config else None
encoder = SentenceEncoder(bptt, arch(vocab_sz, **config), pad_idx=pad_idx)
model = SequentialRNN(encoder, PoolingLinearClassifier(layers, ps))
return model if init is None else model.apply(init)
config = awd_lstm_clas_config.copy()
config.update({'n_hid':10, 'emb_sz':20})
tst = get_text_classifier(AWD_LSTM, 100, 3, config=config)
x = torch.randint(2, 100, (10,5))
y = tst(x)
test_eq(y[0].shape, [10, 3])
for i in range(1,3): test_eq([h_.shape for h_ in y[1]], [[10, 5, 10], [10, 5, 10], [10, 5, 20]])
#test padding gives same results
tst.eval()
y = tst(x)
x1 = torch.cat([x, tensor([2,1,1,1,1,1,1,1,1,1])[:,None]], dim=1)
y1 = tst(x1)
test_eq(y[0][1:],y1[0][1:])
#test drop_mult
tst = get_text_classifier(AWD_LSTM, 100, 3, config=config, drop_mult=0.5)
test_eq(tst[1].layers[1][1].p, 0.1)
test_eq(tst[1].layers[0][1].p, config['output_p']*0.5)
for rnn in tst[0].module.rnns: test_eq(rnn.weight_p, config['weight_p']*0.5)
for dp in tst[0].module.hidden_dps: test_eq(dp.p, config['hidden_p']*0.5)
test_eq(tst[0].module.encoder_dp.embed_p, config['embed_p']*0.5)
test_eq(tst[0].module.input_dp.p, config['input_p']*0.5)
```
## Export -
```
#hide
from local.notebook.export import notebook2script
notebook2script(all_fs=True)
```
|
github_jupyter
|
#export
from local.torch_basics import *
from local.test import *
from local.core import *
from local.layers import *
from local.data.all import *
from local.text.core import *
from local.text.models.awdlstm import *
from local.notebook.showdoc import *
#default_exp text.models.core
#default_cls_lvl 3
#export
_model_meta = {AWD_LSTM: {'hid_name':'emb_sz', 'url':URLs.WT103_FWD, 'url_bwd':URLs.WT103_BWD,
'config_lm':awd_lstm_lm_config, 'split_lm': awd_lstm_lm_split,
'config_clas':awd_lstm_clas_config, 'split_clas': awd_lstm_clas_split},
AWD_QRNN: {'hid_name':'emb_sz',
'config_lm':awd_qrnn_lm_config, 'split_lm': awd_lstm_lm_split,
'config_clas':awd_qrnn_clas_config, 'split_clas': awd_lstm_clas_split},}
# Transformer: {'hid_name':'d_model', 'url':URLs.OPENAI_TRANSFORMER,
# 'config_lm':tfmer_lm_config, 'split_lm': tfmer_lm_split,
# 'config_clas':tfmer_clas_config, 'split_clas': tfmer_clas_split},
# TransformerXL: {'hid_name':'d_model',
# 'config_lm':tfmerXL_lm_config, 'split_lm': tfmerXL_lm_split,
# 'config_clas':tfmerXL_clas_config, 'split_clas': tfmerXL_clas_split}}
#export
class LinearDecoder(Module):
"To go on top of a RNNCore module and create a Language Model."
initrange=0.1
def __init__(self, n_out, n_hid, output_p=0.1, tie_encoder=None, bias=True):
self.decoder = nn.Linear(n_hid, n_out, bias=bias)
self.decoder.weight.data.uniform_(-self.initrange, self.initrange)
self.output_dp = RNNDropout(output_p)
if bias: self.decoder.bias.data.zero_()
if tie_encoder: self.decoder.weight = tie_encoder.weight
def forward(self, input):
raw_outputs, outputs = input
decoded = self.decoder(self.output_dp(outputs[-1]))
return decoded, raw_outputs, outputs
from local.text.models.awdlstm import *
enc = AWD_LSTM(100, 20, 10, 2)
x = torch.randint(0, 100, (10,5))
r = enc(x)
tst = LinearDecoder(100, 20, 0.1)
y = tst(r)
test_eq(y[1], r[0])
test_eq(y[2], r[1])
test_eq(y[0].shape, [10, 5, 100])
tst = LinearDecoder(100, 20, 0.1, tie_encoder=enc.encoder)
test_eq(tst.decoder.weight, enc.encoder.weight)
#export
class SequentialRNN(nn.Sequential):
"A sequential module that passes the reset call to its children."
def reset(self):
for c in self.children(): getattr(c, 'reset', noop)()
class _TstMod(Module):
def reset(self): print('reset')
tst = SequentialRNN(_TstMod(), _TstMod())
test_stdout(tst.reset, 'reset\nreset')
#export
def get_language_model(arch, vocab_sz, config=None, drop_mult=1.):
"Create a language model from `arch` and its `config`."
meta = _model_meta[arch]
config = ifnone(config, meta['config_lm']).copy()
for k in config.keys():
if k.endswith('_p'): config[k] *= drop_mult
tie_weights,output_p,out_bias = map(config.pop, ['tie_weights', 'output_p', 'out_bias'])
init = config.pop('init') if 'init' in config else None
encoder = arch(vocab_sz, **config)
enc = encoder.encoder if tie_weights else None
decoder = LinearDecoder(vocab_sz, config[meta['hid_name']], output_p, tie_encoder=enc, bias=out_bias)
model = SequentialRNN(encoder, decoder)
return model if init is None else model.apply(init)
config = awd_lstm_lm_config.copy()
config.update({'n_hid':10, 'emb_sz':20})
tst = get_language_model(AWD_LSTM, 100, config=config)
x = torch.randint(0, 100, (10,5))
y = tst(x)
test_eq(y[0].shape, [10, 5, 100])
test_eq(tst[1].decoder.weight, tst[0].encoder.weight)
for i in range(1,3): test_eq([h_.shape for h_ in y[1]], [[10, 5, 10], [10, 5, 10], [10, 5, 20]])
#test drop_mult
tst = get_language_model(AWD_LSTM, 100, config=config, drop_mult=0.5)
test_eq(tst[1].output_dp.p, config['output_p']*0.5)
for rnn in tst[0].rnns: test_eq(rnn.weight_p, config['weight_p']*0.5)
for dp in tst[0].hidden_dps: test_eq(dp.p, config['hidden_p']*0.5)
test_eq(tst[0].encoder_dp.embed_p, config['embed_p']*0.5)
test_eq(tst[0].input_dp.p, config['input_p']*0.5)
#export
def _pad_tensor(t, bs, val=0.):
if t.size(0) < bs: return torch.cat([t, val + t.new_zeros(bs-t.size(0), *t.shape[1:])])
return t
#export
class SentenceEncoder(Module):
"Create an encoder over `module` that can process a full sentence."
def __init__(self, bptt, module, pad_idx=1): store_attr(self, 'bptt,module,pad_idx')
def _concat(self, arrs, bs):
return [torch.cat([_pad_tensor(l[si],bs) for l in arrs], dim=1) for si in range(len(arrs[0]))]
def reset(self): getattr(self.module, 'reset', noop)()
def forward(self, input):
bs,sl = input.size()
self.reset()
raw_outputs,outputs,masks = [],[],[]
for i in range(0, sl, self.bptt):
r,o = self.module(input[:,i: min(i+self.bptt, sl)])
masks.append(input[:,i: min(i+self.bptt, sl)] == self.pad_idx)
raw_outputs.append(r)
outputs.append(o)
return self._concat(raw_outputs, bs),self._concat(outputs, bs),torch.cat(masks,dim=1)
class DoubleEmbedding(nn.Embedding):
def forward(self, x):
y = super().forward(x)
return ([y],[y+1])
mod = DoubleEmbedding(5, 10,)
tst = SentenceEncoder(5, mod, pad_idx=0)
x = torch.randint(1, 5, (3, 15))
x[2,10:]=0
raw,out,mask = tst(x)
test_eq(raw[0], mod(x)[0][0])
test_eq(out[0], mod(x)[0][0]+1)
test_eq(mask, x==0)
class PoolingLinearClassifier(nn.Module):
"Create a linear classifier with pooling."
def __init__(self, layers, drops):
super().__init__()
mod_layers = []
activs = [nn.ReLU(inplace=True)] * (len(layers) - 2) + [None]
for n_in, n_out, p, actn in zip(layers[:-1], layers[1:], drops, activs):
mod_layers += bn_drop_lin(n_in, n_out, p=p, actn=actn)
self.layers = nn.Sequential(*mod_layers)
def forward(self, input):
raw_outputs,outputs,mask = input
output = outputs[-1]
lengths = output.size(1) - mask.long().sum(dim=1)
avg_pool = output.masked_fill(mask[:,:,None], 0).sum(dim=1)
avg_pool.div_(lengths.type(avg_pool.dtype)[:,None])
max_pool = output.masked_fill(mask[:,:,None], -float('inf')).max(dim=1)[0]
x = torch.cat([output[torch.arange(0, output.size(0)),lengths-1], max_pool, avg_pool], 1) #Concat pooling.
x = self.layers(x)
return x
#export
def masked_concat_pool(outputs, mask):
"Pool `MultiBatchEncoder` outputs into one vector [last_hidden, max_pool, avg_pool]"
output = outputs[-1]
lens = output.size(1) - mask.long().sum(dim=1)
avg_pool = output.masked_fill(mask[:, :, None], 0).sum(dim=1)
avg_pool.div_(lens.type(avg_pool.dtype)[:,None])
max_pool = output.masked_fill(mask[:,:,None], -float('inf')).max(dim=1)[0]
x = torch.cat([output[torch.arange(0, output.size(0)),lens-1], max_pool, avg_pool], 1) #Concat pooling.
return x
out = torch.randn(2,3,5)
mask = tensor([[False,False,True], [False,False,False]])
x = masked_concat_pool([out], mask)
test_close(x[0,:5], out[0,-2])
test_close(x[1,:5], out[1,-1])
test_close(x[0,5:10], out[0,:2].max(dim=0)[0])
test_close(x[1,5:10], out[1].max(dim=0)[0])
test_close(x[0,10:], out[0,:2].mean(dim=0))
test_close(x[1,10:], out[1].mean(dim=0))
#Test the result is independent of padding
out1 = torch.randn(2,4,5)
out1[:,:-1] = out.clone()
mask1 = tensor([[False,False,True,True], [False,False,False,True]])
x1 = masked_concat_pool([out1], mask1)
test_eq(x, x1)
#export
class PoolingLinearClassifier(Module):
"Create a linear classifier with pooling"
def __init__(self, dims, ps):
mod_layers = []
if len(ps) != len(dims)-1: raise ValueError("Number of layers and dropout values do not match.")
acts = [nn.ReLU(inplace=True)] * (len(dims) - 2) + [None]
layers = [BnDropLin(i, o, p=p, act=a) for i,o,p,a in zip(dims[:-1], dims[1:], ps, acts)]
self.layers = nn.Sequential(*layers)
def forward(self, input):
raw,out,mask = input
x = masked_concat_pool(out, mask)
x = self.layers(x)
return x, raw, out
mod = DoubleEmbedding(5, 10)
tst = nn.Sequential(SentenceEncoder(5, mod, pad_idx=0), PoolingLinearClassifier([10*3,4], [0.]))
x = torch.randint(1, 5, (3, 14))
x[2,10:] = 0
res,raw,out = tst(x)
test_eq(raw[0], mod(x)[0][0])
test_eq(out[0], mod(x)[0][0]+1)
test_eq(res.shape, [3,4])
x1 = torch.cat([x, tensor([0,0,0])[:,None]], dim=1)
res1,raw1,out1 = tst(x1)
test_eq(res, res1)
#export
def get_text_classifier(arch, vocab_sz, n_class, bptt=72, config=None, drop_mult=1., lin_ftrs=None,
ps=None, pad_idx=1):
"Create a text classifier from `arch` and its `config`, maybe `pretrained`"
meta = _model_meta[arch]
config = ifnone(config, meta['config_clas']).copy()
for k in config.keys():
if k.endswith('_p'): config[k] *= drop_mult
if lin_ftrs is None: lin_ftrs = [50]
if ps is None: ps = [0.1]*len(lin_ftrs)
layers = [config[meta['hid_name']] * 3] + lin_ftrs + [n_class]
ps = [config.pop('output_p')] + ps
init = config.pop('init') if 'init' in config else None
encoder = SentenceEncoder(bptt, arch(vocab_sz, **config), pad_idx=pad_idx)
model = SequentialRNN(encoder, PoolingLinearClassifier(layers, ps))
return model if init is None else model.apply(init)
config = awd_lstm_clas_config.copy()
config.update({'n_hid':10, 'emb_sz':20})
tst = get_text_classifier(AWD_LSTM, 100, 3, config=config)
x = torch.randint(2, 100, (10,5))
y = tst(x)
test_eq(y[0].shape, [10, 3])
for i in range(1,3): test_eq([h_.shape for h_ in y[1]], [[10, 5, 10], [10, 5, 10], [10, 5, 20]])
#test padding gives same results
tst.eval()
y = tst(x)
x1 = torch.cat([x, tensor([2,1,1,1,1,1,1,1,1,1])[:,None]], dim=1)
y1 = tst(x1)
test_eq(y[0][1:],y1[0][1:])
#test drop_mult
tst = get_text_classifier(AWD_LSTM, 100, 3, config=config, drop_mult=0.5)
test_eq(tst[1].layers[1][1].p, 0.1)
test_eq(tst[1].layers[0][1].p, config['output_p']*0.5)
for rnn in tst[0].module.rnns: test_eq(rnn.weight_p, config['weight_p']*0.5)
for dp in tst[0].module.hidden_dps: test_eq(dp.p, config['hidden_p']*0.5)
test_eq(tst[0].module.encoder_dp.embed_p, config['embed_p']*0.5)
test_eq(tst[0].module.input_dp.p, config['input_p']*0.5)
#hide
from local.notebook.export import notebook2script
notebook2script(all_fs=True)
| 0.749271 | 0.59131 |
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#Aerospike-Hello-World!" data-toc-modified-id="Aerospike-Hello-World!-1"><span class="toc-item-num">1 </span>Aerospike Hello World!</a></span><ul class="toc-item"><li><span><a href="#Ensure-database-is-running" data-toc-modified-id="Ensure-database-is-running-1.1"><span class="toc-item-num">1.1 </span>Ensure database is running</a></span></li><li><span><a href="#Import-the-module" data-toc-modified-id="Import-the-module-1.2"><span class="toc-item-num">1.2 </span>Import the module</a></span></li><li><span><a href="#Configure-the-client" data-toc-modified-id="Configure-the-client-1.3"><span class="toc-item-num">1.3 </span>Configure the client</a></span></li><li><span><a href="#Create-client-object-and-connect-to-the-cluster" data-toc-modified-id="Create-client-object-and-connect-to-the-cluster-1.4"><span class="toc-item-num">1.4 </span>Create client object and connect to the cluster</a></span></li><li><span><a href="#Understand-records-are-addressable-via-a-tuple-of-(namespace,-set,-userkey)" data-toc-modified-id="Understand-records-are-addressable-via-a-tuple-of-(namespace,-set,-userkey)-1.5"><span class="toc-item-num">1.5 </span>Understand records are addressable via a tuple of (namespace, set, userkey)</a></span></li><li><span><a href="#Write-a-record" data-toc-modified-id="Write-a-record-1.6"><span class="toc-item-num">1.6 </span>Write a record</a></span></li><li><span><a href="#Read-a-record" data-toc-modified-id="Read-a-record-1.7"><span class="toc-item-num">1.7 </span>Read a record</a></span></li><li><span><a href="#Display-result" data-toc-modified-id="Display-result-1.8"><span class="toc-item-num">1.8 </span>Display result</a></span></li><li><span><a href="#Clean-up" data-toc-modified-id="Clean-up-1.9"><span class="toc-item-num">1.9 </span>Clean up</a></span></li><li><span><a href="#Next-steps" data-toc-modified-id="Next-steps-1.10"><span class="toc-item-num">1.10 </span>Next steps</a></span></li></ul></li></ul></div>
# Aerospike Hello World!
Hello, World! in Python with Aerospike.
<br>
This notebook requires Aerospike datbase running on localhost and that python and the Aerospike python client have been installed (`pip install aerospike`). Visit [Aerospike notebooks repo](https://github.com/aerospike-examples/interactive-notebooks) for additional details and the docker container.
## Ensure database is running
This notebook requires that Aerospike datbase is running.
```
!asd >& /dev/null
!pgrep -x asd >/dev/null && echo "Aerospike database is running!" || echo "**Aerospike database is not running!**"
```
## Import the module
Import the client library.
```
import aerospike
print("Client module imported")
```
## Configure the client
The configuration is for Aerospike database running on port 3000 of localhost (IP 127.0.0.1) which is the default. Modify config if your environment is different (Aerospike database running on a different host or different port).
```
config = {
'hosts': [ ('127.0.0.1', 3000) ]
}
print("Configuring with seed host:", config['hosts'])
```
## Create client object and connect to the cluster
```
try:
client = aerospike.client(config).connect()
except:
import sys
print("Failed to connect to the cluster with", config['hosts'])
sys.exit(1)
print("Connected to the cluster")
```
## Understand records are addressable via a tuple of (namespace, set, userkey)
The three components namespace, set, and userkey (with set being optional) form the Primary Key (PK) or simply key, of the record. The key serves as a handle to the record, and using it, a record can be read or written. For a detailed description of the data model see the [Data Model overview](https://www.aerospike.com/docs/architecture/data-model.html)
```
key = ('test', 'demo', 'foo')
print('Working with record key ', key)
```
## Write a record
Aerospike is schema-less and records may be written without any other setup. Here the bins or fields: name, age and greeting, are being written to a record with the key as defined above.
```
try:
# Write a record
client.put(key, {
'name': 'John Doe',
'age': 32,
'greeting': 'Hello, World!'
})
except Exception as e:
import sys
print("error: {0}".format(e), file=sys.stderr)
sys.exit(1)
print('Successfully written the record')
```
## Read a record
The record may be retrieved using the same key.
```
(key, metadata, record) = client.get(key)
print('Read back the record')
```
## Display result
Print the record that was just retrieved. We are also printing:
1. The components of the key which are: namespace, set, and userkey. By default userkey is not stored on server, only a hash (appearing as bytearray in the output below) which is the internal representation of the key is stored.
1. The metadata with the time-to-live and the record's generation or version.
1. The actual value of the record's bins.
```
print("Record contents are", record)
print("Key's components are", key)
print("Metadata is", metadata)
```
## Clean up
Finally close the client we created at the beginning.
```
# Close the connection to the Aerospike cluster
client.close()
print('Connection closed.')
```
## Next steps
Visit [Aerospike notebooks repo](https://github.com/aerospike-examples/interactive-notebooks) to run additional Aerospike notebooks. To run a different notebook, download the notebook from the repo to your local machine, and then click on File->Open, and select Upload.
|
github_jupyter
|
!asd >& /dev/null
!pgrep -x asd >/dev/null && echo "Aerospike database is running!" || echo "**Aerospike database is not running!**"
import aerospike
print("Client module imported")
config = {
'hosts': [ ('127.0.0.1', 3000) ]
}
print("Configuring with seed host:", config['hosts'])
try:
client = aerospike.client(config).connect()
except:
import sys
print("Failed to connect to the cluster with", config['hosts'])
sys.exit(1)
print("Connected to the cluster")
key = ('test', 'demo', 'foo')
print('Working with record key ', key)
try:
# Write a record
client.put(key, {
'name': 'John Doe',
'age': 32,
'greeting': 'Hello, World!'
})
except Exception as e:
import sys
print("error: {0}".format(e), file=sys.stderr)
sys.exit(1)
print('Successfully written the record')
(key, metadata, record) = client.get(key)
print('Read back the record')
print("Record contents are", record)
print("Key's components are", key)
print("Metadata is", metadata)
# Close the connection to the Aerospike cluster
client.close()
print('Connection closed.')
| 0.153994 | 0.926703 |
# Developing an AI application
Going forward, AI algorithms will be incorporated into more and more everyday applications. For example, you might want to include an image classifier in a smart phone app. To do this, you'd use a deep learning model trained on hundreds of thousands of images as part of the overall application architecture. A large part of software development in the future will be using these types of models as common parts of applications.
In this project, you'll train an image classifier to recognize different species of flowers. You can imagine using something like this in a phone app that tells you the name of the flower your camera is looking at. In practice you'd train this classifier, then export it for use in your application. We'll be using [this dataset](http://www.robots.ox.ac.uk/~vgg/data/flowers/102/index.html) of 102 flower categories, you can see a few examples below.
<img src='assets/Flowers.png' width=500px>
The project is broken down into multiple steps:
* Load and preprocess the image dataset
* Train the image classifier on your dataset
* Use the trained classifier to predict image content
We'll lead you through each part which you'll implement in Python.
When you've completed this project, you'll have an application that can be trained on any set of labeled images. Here your network will be learning about flowers and end up as a command line application. But, what you do with your new skills depends on your imagination and effort in building a dataset. For example, imagine an app where you take a picture of a car, it tells you what the make and model is, then looks up information about it. Go build your own dataset and make something new.
First up is importing the packages you'll need. It's good practice to keep all the imports at the beginning of your code. As you work through this notebook and find you need to import a package, make sure to add the import up here.
```
# Imports here
from pathlib import Path
import json
from collections import OrderedDict
from PIL import Image
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import torch
from torch import nn, optim
import torchvision as tv
```
## Load the data
Here you'll use `torchvision` to load the data ([documentation](http://pytorch.org/docs/0.3.0/torchvision/index.html)). The data should be included alongside this notebook, otherwise you can [download it here](https://s3.amazonaws.com/content.udacity-data.com/nd089/flower_data.tar.gz). The dataset is split into three parts, training, validation, and testing. For the training, you'll want to apply transformations such as random scaling, cropping, and flipping. This will help the network generalize leading to better performance. You'll also need to make sure the input data is resized to 224x224 pixels as required by the pre-trained networks.
The validation and testing sets are used to measure the model's performance on data it hasn't seen yet. For this you don't want any scaling or rotation transformations, but you'll need to resize then crop the images to the appropriate size.
The pre-trained networks you'll use were trained on the ImageNet dataset where each color channel was normalized separately. For all three sets you'll need to normalize the means and standard deviations of the images to what the network expects. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`, calculated from the ImageNet images. These values will shift each color channel to be centered at 0 and range from -1 to 1.
```
data_dir = 'flowers'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
mean =[0.485, 0.456, 0.406]
std = [0.229, 0.224, 0.225]
batchSize = 64
nThreads = 0
# TODO: Define your transforms for the training, validation, and testing sets
train_transforms = tv.transforms.Compose([
tv.transforms.RandomRotation(30),
tv.transforms.RandomResizedCrop(224),
tv.transforms.RandomHorizontalFlip(),
tv.transforms.ToTensor(),
tv.transforms.Normalize(mean=mean, std=std),
])
test_transforms = tv.transforms.Compose([
tv.transforms.Resize(256),
tv.transforms.CenterCrop(224),
tv.transforms.ToTensor(),
tv.transforms.Normalize(mean=mean, std=std),
])
# TODO: Load the datasets with ImageFolder
train_data = tv.datasets.ImageFolder(train_dir, transform=train_transforms)
valid_data = tv.datasets.ImageFolder(valid_dir, transform=test_transforms)
test_data = tv.datasets.ImageFolder(test_dir, transform=test_transforms)
# TODO: Using the image datasets and the trainforms, define the dataloaders
train_loader = torch.utils.data.DataLoader(train_data, batch_size=batchSize,
shuffle=True, num_workers=nThreads)
valid_loader = torch.utils.data.DataLoader(valid_data, batch_size=batchSize,
shuffle=True, num_workers=nThreads)
test_loader = torch.utils.data.DataLoader(test_data, batch_size=batchSize,
shuffle=True, num_workers=nThreads)
```
### Label mapping
You'll also need to load in a mapping from category label to category name. You can find this in the file `cat_to_name.json`. It's a JSON object which you can read in with the [`json` module](https://docs.python.org/2/library/json.html). This will give you a dictionary mapping the integer encoded categories to the actual names of the flowers.
```
with open('cat_to_name.json', 'r') as f:
cat_to_name = json.load(f)
print(len(cat_to_name))
print(min(cat_to_name, key=lambda x: int(x)))
print(max(cat_to_name, key=lambda x: int(x)))
```
# Building and training the classifier
Now that the data is ready, it's time to build and train the classifier. As usual, you should use one of the pretrained models from `torchvision.models` to get the image features. Build and train a new feed-forward classifier using those features.
We're going to leave this part up to you. Refer to [the rubric](https://review.udacity.com/#!/rubrics/1663/view) for guidance on successfully completing this section. Things you'll need to do:
* Load a [pre-trained network](http://pytorch.org/docs/master/torchvision/models.html) (If you need a starting point, the VGG networks work great and are straightforward to use)
* Define a new, untrained feed-forward network as a classifier, using ReLU activations and dropout
* Train the classifier layers using backpropagation using the pre-trained network to get the features
* Track the loss and accuracy on the validation set to determine the best hyperparameters
We've left a cell open for you below, but use as many as you need. Our advice is to break the problem up into smaller parts you can run separately. Check that each part is doing what you expect, then move on to the next. You'll likely find that as you work through each part, you'll need to go back and modify your previous code. This is totally normal!
When training make sure you're updating only the weights of the feed-forward network. You should be able to get the validation accuracy above 70% if you build everything right. Make sure to try different hyperparameters (learning rate, units in the classifier, epochs, etc) to find the best model. Save those hyperparameters to use as default values in the next part of the project.
One last important tip if you're using the workspace to run your code: To avoid having your workspace disconnect during the long-running tasks in this notebook, please read in the earlier page in this lesson called Intro to
GPU Workspaces about Keeping Your Session Active. You'll want to include code from the workspace_utils.py module.
**Note for Workspace users:** If your network is over 1 GB when saved as a checkpoint, there might be issues with saving backups in your workspace. Typically this happens with wide dense layers after the convolutional layers. If your saved checkpoint is larger than 1 GB (you can open a terminal and check with `ls -lh`), you should reduce the size of your hidden layers and train again.
```
# TODO: Build and train your network
model = tv.models.vgg16(pretrained=True)
model
# TODO: Build and train your network
architecture = 'vgg16'
dropout = 0.2
hidden_units = [512, 128]
output_size = len(cat_to_name)
def create_model(architecture, hidden_units, dropout, output_size):
model = getattr(tv.models, architecture)(pretrained=True)
# Freeze parameters so we don't backprop through them
for param in model.parameters():
param.requires_grad = False
# Get number of input features for classifier
classifier = model.classifier
if hasattr(classifier, '__getitem__'):
i = 0
for i in range(len(classifier)):
if hasattr(classifier[i], 'in_features'):
break
classifier = classifier[i]
in_features = classifier.in_features
classifier = nn.Sequential(OrderedDict([
('dropout', nn.Dropout(dropout)),
('fc1', nn.Linear(in_features, hidden_units[0])),
('relu', nn.ReLU()),
('fc2', nn.Linear(hidden_units[0], hidden_units[1])),
('relu', nn.ReLU()),
('fc3', nn.Linear(hidden_units[1], output_size)),
('output', nn.LogSoftmax(dim=1))
]))
model.classifier = classifier
return model
model = create_model(architecture, hidden_units, dropout, output_size)
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.classifier.parameters(), lr=0.003)
# Use GPU if it's available
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device);
epochs = 6
steps = 0
running_loss = 0
print_every = 10
model.train()
for epoch in range(epochs):
for inputs, labels in train_loader:
steps += 1
# Move input and label tensors to the default device
inputs, labels = inputs.to(device), labels.to(device)
optimizer.zero_grad()
logps = model.forward(inputs)
loss = criterion(logps, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
if steps % print_every == 0:
valid_loss = 0
accuracy = 0
model.eval()
with torch.no_grad():
for inputs, labels in valid_loader:
inputs, labels = inputs.to(device), labels.to(device)
logps = model.forward(inputs)
batch_loss = criterion(logps, labels)
valid_loss += batch_loss.item()
# Calculate accuracy
ps = torch.exp(logps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Epoch {epoch+1}/{epochs}.. "
f"Train loss: {running_loss/print_every:.3f}.. "
f"Validation loss: {valid_loss/len(valid_loader):.3f}.. "
f"Validation accuracy: {accuracy/len(valid_loader):.3f}")
running_loss = 0
model.train()
```
## Testing your network
It's good practice to test your trained network on test data, images the network has never seen either in training or validation. This will give you a good estimate for the model's performance on completely new images. Run the test images through the network and measure the accuracy, the same way you did validation. You should be able to reach around 70% accuracy on the test set if the model has been trained well.
```
# TODO: Do validation on the test set
model.eval()
test_loss = 0
accuracy = 0
with torch.no_grad():
for inputs, labels in test_loader:
inputs, labels = inputs.to(device), labels.to(device)
logps = model.forward(inputs)
batch_loss = criterion(logps, labels)
test_loss += batch_loss.item()
# Calculate accuracy
ps = torch.exp(logps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Test loss: {test_loss/len(test_loader):.3f}.. "
f"Test accuracy: {accuracy/len(test_loader):.3f}")
```
## Save the checkpoint
Now that your network is trained, save the model so you can load it later for making predictions. You probably want to save other things such as the mapping of classes to indices which you get from one of the image datasets: `image_datasets['train'].class_to_idx`. You can attach this to the model as an attribute which makes inference easier later on.
```model.class_to_idx = image_datasets['train'].class_to_idx```
Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
```
# TODO: Save the checkpoint
model.class_to_idx = train_data.class_to_idx
checkpoint = {'architecture' : architecture,
'input_size': 224,
'dropout': dropout,
'hidden_units': hidden_units,
'output_size': output_size,
'class_to_idx': model.class_to_idx,
'state_dict': model.state_dict()}
torch.save(checkpoint, 'checkpoint.pth')
```
## Loading the checkpoint
At this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
```
# TODO: Write a function that loads a checkpoint and rebuilds the model
def load_checkpoint(filepath):
checkpoint = torch.load(filepath, map_location='cpu')
architecture = checkpoint['architecture']
dropout = checkpoint['dropout']
hidden_units = checkpoint['hidden_units']
output_size = checkpoint['output_size']
model = create_model(architecture, hidden_units, dropout, output_size)
model.load_state_dict(checkpoint['state_dict'])
model.class_to_idx = checkpoint['class_to_idx']
return model
model = load_checkpoint('checkpoint.pth')
model
```
# Inference for classification
Now you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like
```python
probs, classes = predict(image_path, model)
print(probs)
print(classes)
> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
> ['70', '3', '45', '62', '55']
```
First you'll need to handle processing the input image such that it can be used in your network.
## Image Preprocessing
You'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training.
First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.
Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.
As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation.
And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
```
def process_image(image):
''' Scales, crops, and normalizes a PIL image for a PyTorch model,
returns an Numpy array
'''
# TODO: Process a PIL image for use in a PyTorch model
image = test_transforms(image).float()
return image
img_path = test_dir + "/1/image_06743.jpg"
image = Image.open(img_path)
img_tensor = process_image(image)
img_tensor.max(), img_tensor.min()
```
To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
```
def imshow(image, ax=None, title=None):
"""Imshow for Tensor."""
if ax is None:
fig, ax = plt.subplots()
if title is not None:
ax.set_title(title)
# PyTorch tensors assume the color channel is the first dimension
# but matplotlib assumes is the third dimension
image = image.numpy().transpose((1, 2, 0))
# Undo preprocessing
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
image = std * image + mean
# Image needs to be clipped between 0 and 1 or it looks like noise when displayed
image = np.clip(image, 0, 1)
ax.imshow(image)
return ax
img_path = test_dir + "/1/image_06743.jpg"
image = Image.open(img_path)
img_tensor = process_image(image)
imshow(img_tensor, title=cat_to_name['1'])
```
## Class Prediction
Once you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.
To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.
Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.
```python
probs, classes = predict(image_path, model)
print(probs)
print(classes)
> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
> ['70', '3', '45', '62', '55']
```
```
def predict(image_path, model, topk=5):
''' Predict the class (or classes) of an image using a trained deep learning model.
'''
# TODO: Implement the code to predict the class from an image file
model.eval()
image = Image.open(image_path)
img_tensor = process_image(image)
# Calculate the class probabilities (softmax) for img
with torch.no_grad():
output = model.forward(img_tensor.reshape((1,3,224,224)))
return torch.exp(output).topk(topk), img_tensor
img_path = test_dir + "/1/image_06743.jpg"
top_k_pred, img_tensor = predict(img_path, model)
top_k_pred
```
## Sanity Checking
Now that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:
<img src='assets/inference_example.png' width=300px>
You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
```
import matplotlib.gridspec as gridspec
idx_to_class = {v: k for k,v in train_data.class_to_idx.items()}
# TODO: Display an image along with the top 5 classes
def display_top_k_predictions(top_k_pred, img_tensor, title):
probs, class_idxs = top_k_pred
probs = probs.data.numpy().squeeze()
class_idxs = class_idxs.data.numpy().squeeze()
class_names = [cat_to_name[idx_to_class[c]] for c in class_idxs]
plt.figure(figsize=(10,7))
G = gridspec.GridSpec(1, 5)
ax1 = plt.subplot(G[0, :2])
#fig, (ax1, ax2) = plt.subplots(figsize=(6,20), ncols=2)
imshow(img_tensor, ax=ax1, title=title)
ax1.axis('off')
ax2 = plt.subplot(G[0,2:])
ax2.barh(np.arange(len(probs)), probs)
ax2.set_aspect(0.1)
ax2.set_yticks(np.arange(len(probs)))
ax2.set_yticklabels(class_names, size='small');
ax2.set_title('Class Probability')
ax2.set_xlim(0, 1.)
ax2.invert_yaxis()
plt.tight_layout()
cat = '1'
img_path = sorted(Path(test_dir + "/" + cat).glob("*.jpg"))[0]
top_k_pred, img_tensor = predict(img_path, model)
display_top_k_predictions(top_k_pred, img_tensor, title=cat_to_name[cat])
```
|
github_jupyter
|
# Imports here
from pathlib import Path
import json
from collections import OrderedDict
from PIL import Image
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import torch
from torch import nn, optim
import torchvision as tv
data_dir = 'flowers'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
mean =[0.485, 0.456, 0.406]
std = [0.229, 0.224, 0.225]
batchSize = 64
nThreads = 0
# TODO: Define your transforms for the training, validation, and testing sets
train_transforms = tv.transforms.Compose([
tv.transforms.RandomRotation(30),
tv.transforms.RandomResizedCrop(224),
tv.transforms.RandomHorizontalFlip(),
tv.transforms.ToTensor(),
tv.transforms.Normalize(mean=mean, std=std),
])
test_transforms = tv.transforms.Compose([
tv.transforms.Resize(256),
tv.transforms.CenterCrop(224),
tv.transforms.ToTensor(),
tv.transforms.Normalize(mean=mean, std=std),
])
# TODO: Load the datasets with ImageFolder
train_data = tv.datasets.ImageFolder(train_dir, transform=train_transforms)
valid_data = tv.datasets.ImageFolder(valid_dir, transform=test_transforms)
test_data = tv.datasets.ImageFolder(test_dir, transform=test_transforms)
# TODO: Using the image datasets and the trainforms, define the dataloaders
train_loader = torch.utils.data.DataLoader(train_data, batch_size=batchSize,
shuffle=True, num_workers=nThreads)
valid_loader = torch.utils.data.DataLoader(valid_data, batch_size=batchSize,
shuffle=True, num_workers=nThreads)
test_loader = torch.utils.data.DataLoader(test_data, batch_size=batchSize,
shuffle=True, num_workers=nThreads)
with open('cat_to_name.json', 'r') as f:
cat_to_name = json.load(f)
print(len(cat_to_name))
print(min(cat_to_name, key=lambda x: int(x)))
print(max(cat_to_name, key=lambda x: int(x)))
# TODO: Build and train your network
model = tv.models.vgg16(pretrained=True)
model
# TODO: Build and train your network
architecture = 'vgg16'
dropout = 0.2
hidden_units = [512, 128]
output_size = len(cat_to_name)
def create_model(architecture, hidden_units, dropout, output_size):
model = getattr(tv.models, architecture)(pretrained=True)
# Freeze parameters so we don't backprop through them
for param in model.parameters():
param.requires_grad = False
# Get number of input features for classifier
classifier = model.classifier
if hasattr(classifier, '__getitem__'):
i = 0
for i in range(len(classifier)):
if hasattr(classifier[i], 'in_features'):
break
classifier = classifier[i]
in_features = classifier.in_features
classifier = nn.Sequential(OrderedDict([
('dropout', nn.Dropout(dropout)),
('fc1', nn.Linear(in_features, hidden_units[0])),
('relu', nn.ReLU()),
('fc2', nn.Linear(hidden_units[0], hidden_units[1])),
('relu', nn.ReLU()),
('fc3', nn.Linear(hidden_units[1], output_size)),
('output', nn.LogSoftmax(dim=1))
]))
model.classifier = classifier
return model
model = create_model(architecture, hidden_units, dropout, output_size)
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.classifier.parameters(), lr=0.003)
# Use GPU if it's available
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device);
epochs = 6
steps = 0
running_loss = 0
print_every = 10
model.train()
for epoch in range(epochs):
for inputs, labels in train_loader:
steps += 1
# Move input and label tensors to the default device
inputs, labels = inputs.to(device), labels.to(device)
optimizer.zero_grad()
logps = model.forward(inputs)
loss = criterion(logps, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
if steps % print_every == 0:
valid_loss = 0
accuracy = 0
model.eval()
with torch.no_grad():
for inputs, labels in valid_loader:
inputs, labels = inputs.to(device), labels.to(device)
logps = model.forward(inputs)
batch_loss = criterion(logps, labels)
valid_loss += batch_loss.item()
# Calculate accuracy
ps = torch.exp(logps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Epoch {epoch+1}/{epochs}.. "
f"Train loss: {running_loss/print_every:.3f}.. "
f"Validation loss: {valid_loss/len(valid_loader):.3f}.. "
f"Validation accuracy: {accuracy/len(valid_loader):.3f}")
running_loss = 0
model.train()
# TODO: Do validation on the test set
model.eval()
test_loss = 0
accuracy = 0
with torch.no_grad():
for inputs, labels in test_loader:
inputs, labels = inputs.to(device), labels.to(device)
logps = model.forward(inputs)
batch_loss = criterion(logps, labels)
test_loss += batch_loss.item()
# Calculate accuracy
ps = torch.exp(logps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Test loss: {test_loss/len(test_loader):.3f}.. "
f"Test accuracy: {accuracy/len(test_loader):.3f}")
Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
## Loading the checkpoint
At this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
# Inference for classification
Now you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like
First you'll need to handle processing the input image such that it can be used in your network.
## Image Preprocessing
You'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training.
First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.
Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.
As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation.
And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
## Class Prediction
Once you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.
To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.
Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.
## Sanity Checking
Now that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:
<img src='assets/inference_example.png' width=300px>
You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
| 0.592549 | 0.982774 |
# T81-558: Applications of Deep Neural Networks
**Class 4: Training a Neural Network**
* Instructor: [Jeff Heaton](https://sites.wustl.edu/jeffheaton/), School of Engineering and Applied Science, [Washington University in St. Louis](https://engineering.wustl.edu/Programs/Pages/default.aspx)
* For more information visit the [class website](https://sites.wustl.edu/jeffheaton/t81-558/).
# Module Video Material
Main video lecture:
* [Part 4.1: Early Stopping and Feature Vector Encoding](https://www.youtube.com/watch?v=ATuyK_HWZgc&index=12&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN)
* [Part 4.2: Evaluating Classification and Regression Networks](https://www.youtube.com/watch?v=hXkZqGi5mB4&index=13&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN)
* [Part 4.3: Cross-Validation for Neural Networks](https://www.youtube.com/watch?v=SIyMm5DFwQ8&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN)
Weekly video update:
* *Will be posted week of this class*
# Helpful Functions
You will see these at the top of every module. These are simply a set of reusable functions that we will make use of. Each of them will be explained as the semester progresses. They are explained in greater detail as the course progresses. Class 4 contains a complete overview of these functions.
```
from sklearn import preprocessing
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import shutil
import os
import requests
import base64
# Encode text values to dummy variables(i.e. [1,0,0],[0,1,0],[0,0,1] for red,green,blue)
def encode_text_dummy(df, name):
dummies = pd.get_dummies(df[name])
for x in dummies.columns:
dummy_name = "{}-{}".format(name, x)
df[dummy_name] = dummies[x]
df.drop(name, axis=1, inplace=True)
# Encode text values to a single dummy variable. The new columns (which do not replace the old) will have a 1
# at every location where the original column (name) matches each of the target_values. One column is added for
# each target value.
def encode_text_single_dummy(df, name, target_values):
for tv in target_values:
l = list(df[name].astype(str))
l = [1 if str(x) == str(tv) else 0 for x in l]
name2 = "{}-{}".format(name, tv)
df[name2] = l
# Encode text values to indexes(i.e. [1],[2],[3] for red,green,blue).
def encode_text_index(df, name):
le = preprocessing.LabelEncoder()
df[name] = le.fit_transform(df[name])
return le.classes_
# Encode a numeric column as zscores
def encode_numeric_zscore(df, name, mean=None, sd=None):
if mean is None:
mean = df[name].mean()
if sd is None:
sd = df[name].std()
df[name] = (df[name] - mean) / sd
# Convert all missing values in the specified column to the median
def missing_median(df, name):
med = df[name].median()
df[name] = df[name].fillna(med)
# Convert all missing values in the specified column to the default
def missing_default(df, name, default_value):
df[name] = df[name].fillna(default_value)
# Convert a Pandas dataframe to the x,y inputs that TensorFlow needs
def to_xy(df, target):
result = []
for x in df.columns:
if x != target:
result.append(x)
# find out the type of the target column. Is it really this hard? :(
target_type = df[target].dtypes
target_type = target_type[0] if hasattr(target_type, '__iter__') else target_type
# Encode to int for classification, float otherwise. TensorFlow likes 32 bits.
if target_type in (np.int64, np.int32):
# Classification
dummies = pd.get_dummies(df[target])
return df.as_matrix(result).astype(np.float32), dummies.as_matrix().astype(np.float32)
else:
# Regression
return df.as_matrix(result).astype(np.float32), df.as_matrix([target]).astype(np.float32)
# Nicely formatted time string
def hms_string(sec_elapsed):
h = int(sec_elapsed / (60 * 60))
m = int((sec_elapsed % (60 * 60)) / 60)
s = sec_elapsed % 60
return "{}:{:>02}:{:>05.2f}".format(h, m, s)
# Regression chart.
def chart_regression(pred,y,sort=True):
t = pd.DataFrame({'pred' : pred, 'y' : y.flatten()})
if sort:
t.sort_values(by=['y'],inplace=True)
a = plt.plot(t['y'].tolist(),label='expected')
b = plt.plot(t['pred'].tolist(),label='prediction')
plt.ylabel('output')
plt.legend()
plt.show()
# Remove all rows where the specified column is +/- sd standard deviations
def remove_outliers(df, name, sd):
drop_rows = df.index[(np.abs(df[name] - df[name].mean()) >= (sd * df[name].std()))]
df.drop(drop_rows, axis=0, inplace=True)
# Encode a column to a range between normalized_low and normalized_high.
def encode_numeric_range(df, name, normalized_low=-1, normalized_high=1,
data_low=None, data_high=None):
if data_low is None:
data_low = min(df[name])
data_high = max(df[name])
df[name] = ((df[name] - data_low) / (data_high - data_low)) \
* (normalized_high - normalized_low) + normalized_low
# This function submits an assignment. You can submit an assignment as much as you like, only the final
# submission counts. The paramaters are as follows:
# data - Pandas dataframe output.
# key - Your student key that was emailed to you.
# no - The assignment class number, should be 1 through 1.
# source_file - The full path to your Python or IPYNB file. This must have "_class1" as part of its name.
# . The number must match your assignment number. For example "_class2" for class assignment #2.
def submit(data,key,no,source_file=None):
if source_file is None and '__file__' not in globals(): raise Exception('Must specify a filename when a Jupyter notebook.')
if source_file is None: source_file = __file__
suffix = '_class{}'.format(no)
if suffix not in source_file: raise Exception('{} must be part of the filename.'.format(suffix))
with open(source_file, "rb") as image_file:
encoded_python = base64.b64encode(image_file.read()).decode('ascii')
ext = os.path.splitext(source_file)[-1].lower()
if ext not in ['.ipynb','.py']: raise Exception("Source file is {} must be .py or .ipynb".format(ext))
r = requests.post("https://api.heatonresearch.com/assignment-submit",
headers={'x-api-key':key}, json={'csv':base64.b64encode(data.to_csv(index=False).encode('ascii')).decode("ascii"),
'assignment': no, 'ext':ext, 'py':encoded_python})
if r.status_code == 200:
print("Success: {}".format(r.text))
else: print("Failure: {}".format(r.text))
```
# Building the Feature Vector
Neural networks require their input to be a fixed number of columns. This is very similar to spreadsheet data. This input must be completely numeric.
It is important to represent the data in a way that the neural network can train from it. In class 6, we will see even more ways to preprocess data. For now, we will look at several of the most basic ways to transform data for a neural network.
Before we look at specific ways to preprocess data, it is important to consider four basic types of data, as defined by [Stanley Smith Stevens](https://en.wikipedia.org/wiki/Stanley_Smith_Stevens). These are commonly referred to as the [levels of measure](https://en.wikipedia.org/wiki/Level_of_measurement):
* Character Data (strings)
* **Nominal** - Individual discrete items, no order. For example: color, zip code, shape.
* **Ordinal** - Individual discrete items that can be ordered. For example: grade level, job title, Starbucks(tm) coffee size (tall, vente, grande)
* Numeric Data
* **Interval** - Numeric values, no defined start. For example, temperature. You would never say "yesterday was twice as hot as today".
* **Ratio** - Numeric values, clearly defined start. For example, speed. You would say that "The first car is going twice as fast as the second."
The following code contains several useful functions to encode the feature vector for various types of data. Encoding data:
* **encode_text_dummy** - Encode text fields, such as the iris species as a single field for each class. Three classes would become "0,0,1" "0,1,0" and "1,0,0". Encode non-target predictors this way. Good for nominal.
* **encode_text_index** - Encode text fields, such as the iris species as a single numeric field as "0" "1" and "2". Encode the target field for a classification this way. Good for nominal.
* **encode_numeric_zscore** - Encode numeric values as a z-score. Neural networks deal well with "centered" fields, zscore is usually a good starting point for interval/ratio.
*Ordinal values can be encoded as dummy or index. Later we will see a more advanced means of encoding*
Dealing with missing data:
* **missing_median** - Fill all missing values with the median value.
Creating the final feature vector:
* **to_xy** - Once all fields are numeric, this function can provide the x and y matrixes that are used to fit the neural network.
Other utility functions:
* **hms_string** - Print out an elapsed time string.
* **chart_regression** - Display a chart to show how well a regression performs.
# Dealing with Outliers
```
import tensorflow.contrib.learn as skflow
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
# create feature vector
missing_median(df, 'horsepower')
df.drop('name',1,inplace=True)
#encode_numeric_binary(df,'mpg',20)
#df['origin'] = df['origin'].astype(str)
#encode_text_tfidf(df, 'origin')
# Drop outliers in horsepower
print("Length before MPG outliers dropped: {}".format(len(df)))
remove_outliers(df,'mpg',2)
print("Length after MPG outliers dropped: {}".format(len(df)))
print(df)
```
# Other Examples: Dealing with Addresses
Addresses can be difficult to encode into a neural network. There are many different approaches, and you must consider how you can transform the address into something more meaningful. Map coordinates can be a good approach. [Latitude and longitude](https://en.wikipedia.org/wiki/Geographic_coordinate_system) can be a useful encoding. Thanks to the power of the Internet, it is relatively easy to transform an address into its latitude and longitude values. The following code determines the coordinates of [Washington University](https://wustl.edu/):
```
import requests
address = "1 Brookings Dr, St. Louis, MO 63130"
response = requests.get('https://maps.googleapis.com/maps/api/geocode/json?address='+address)
resp_json_payload = response.json()
print(resp_json_payload['results'][0]['geometry']['location'])
```
If latitude and longitude are simply fed into the neural network as two features, they might not be overly helpful. These two values would allow your neural network to cluster locations on a map. Sometimes cluster locations on a map can be useful. Consider the percentage of the population that smokes in the USA by state:

The above map shows that certian behaviors, like smoking, can be clustered by global region.
However, often you will want to transform the coordinates into distances. It is reasonably easy to estimate the distance between any two points on Earth by using the [great circle distance](https://en.wikipedia.org/wiki/Great-circle_distance) between any two points on a sphere:
The following code implements this formula:
$\Delta\sigma=\arccos\bigl(\sin\phi_1\cdot\sin\phi_2+\cos\phi_1\cdot\cos\phi_2\cdot\cos(\Delta\lambda)\bigr)$
$d = r \, \Delta\sigma$
```
from math import sin, cos, sqrt, atan2, radians
# Distance function
def distance_lat_lng(lat1,lng1,lat2,lng2):
# approximate radius of earth in km
R = 6373.0
# degrees to radians (lat/lon are in degrees)
lat1 = radians(lat1)
lng1 = radians(lng1)
lat2 = radians(lat2)
lng2 = radians(lng2)
dlng = lng2 - lng1
dlat = lat2 - lat1
a = sin(dlat / 2)**2 + cos(lat1) * cos(lat2) * sin(dlng / 2)**2
c = 2 * atan2(sqrt(a), sqrt(1 - a))
return R * c
# Find lat lon for address
def lookup_lat_lng(address):
response = requests.get('https://maps.googleapis.com/maps/api/geocode/json?address='+address)
json = response.json()
if len(json['results']) == 0:
print("Can't find: {}".format(address))
return 0,0
map = json['results'][0]['geometry']['location']
return map['lat'],map['lng']
# Distance between two locations
import requests
address1 = "1 Brookings Dr, St. Louis, MO 63130"
address2 = "3301 College Ave, Fort Lauderdale, FL 33314"
lat1, lng1 = lookup_lat_lng(address1)
lat2, lng2 = lookup_lat_lng(address2)
print("Distance, St. Louis, MO to Ft. Lauderdale, FL: {} km".format(
distance_lat_lng(lat1,lng1,lat2,lng2)))
```
Distances can be useful to encode addresses as. You must consider what distance might be useful for your dataset. Consider:
* Distance to major metropolitan area
* Distance to competitor
* Distance to distribution center
* Distance to retail outlet
The following code calculates the distance between 10 universities and washu:
```
# Encoding other universities by their distance to Washington University
schools = [
["Princeton University, Princeton, NJ 08544", 'Princeton'],
["Massachusetts Hall, Cambridge, MA 02138", 'Harvard'],
["5801 S Ellis Ave, Chicago, IL 60637", 'University of Chicago'],
["Yale, New Haven, CT 06520", 'Yale'],
["116th St & Broadway, New York, NY 10027", 'Columbia University'],
["450 Serra Mall, Stanford, CA 94305", 'Stanford'],
["77 Massachusetts Ave, Cambridge, MA 02139", 'MIT'],
["Duke University, Durham, NC 27708", 'Duke University'],
["University of Pennsylvania, Philadelphia, PA 19104", 'University of Pennsylvania'],
["Johns Hopkins University, Baltimore, MD 21218", 'Johns Hopkins']
]
lat1, lng1 = lookup_lat_lng("1 Brookings Dr, St. Louis, MO 63130")
for address, name in schools:
lat2,lng2 = lookup_lat_lng(address)
dist = distance_lat_lng(lat1,lng1,lat2,lng2)
print("School '{}', distance to wustl is: {}".format(name,dist))
```
# Training with a Validation Set and Early Stopping
**Overfitting** occurs when a neural network is trained to the point that it begins to memorize rather than generalize.

It is important to segment the original dataset into several datasets:
* **Training Set**
* **Validation Set**
* **Holdout Set**
There are several different ways that these sets can be constructed. The following programs demonstrate some of these.
The first method is a training and validation set. The training data are used to train the neural network until the validation set no longer improves. This attempts to stop at a near optimal training point. This method will only give accurate "out of sample" predictions for the validation set, this is usually 20% or so of the data. The predictions for the training data will be overly optimistic, as these were the data that the neural network was trained on.

```
import pandas as pd
import io
import requests
import numpy as np
import os
from sklearn.model_selection import train_test_split
from sklearn import metrics
from keras.models import Sequential
from keras.layers.core import Dense, Activation
from keras.callbacks import EarlyStopping
path = "./data/"
filename = os.path.join(path,"iris.csv")
df = pd.read_csv(filename,na_values=['NA','?'])
species = encode_text_index(df,"species")
x,y = to_xy(df,"species")
# Split into train/test
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.25, random_state=42)
model = Sequential()
model.add(Dense(10, input_dim=x.shape[1], activation='relu'))
model.add(Dense(5,activation='relu'))
model.add(Dense(y.shape[1],activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
model.fit(x,y,validation_data=(x_test,y_test),callbacks=[monitor],verbose=2,epochs=1000)
```
Now that the neural network is trained, we can make predictions about the test set. The following code predicts the type of iris for test set and displays the first five irises.
```
from sklearn import metrics
import tensorflow as tf
pred = model.predict(x_test)
print(pred[0:5]) # print first five predictions
```
These numbers are in scientific notation. Each line provides the probability that the iris is one of the 3 types of iris in the data set. For the first line, the second type of iris has a 91% probability of being the species of iris.
# Early Stopping and the Best Weights
In the previous section we used early stopping so that the training would halt once the validation set no longer saw score improvements for a number of steps. This number of steps that early stopping will tolerate no improvement is called *patience*. If the patience value is large, the neural network's error may continue to worsen while early stopping is patiently waiting. At some point earlier in the training the optimal set of weights was obtained for the neural network. However, at the end of training we will have the weights for the neural network that finally exhausted the patience of early stopping. The weights of this neural network might not be bad, but it would be better to have the most optimal weights during the entire training operation.
The code presented below does this. An additional monitor is used and saves a copy of the neural network to **best_weights.hdf5** each time the validation score of the neural network improves. Once training is done, we just reload this file and we have the optimal training weights that were found.
This technique is slight overkill for many of the examples for this class. It can also introduce an occasional issue (as described in the next section). Because of this, most of the examples in this course will not use this code to obtain the absolute best weights. However, for the larger, more complex datasets in this course, we will save the absolute best weights as demonstrated here.
```
import pandas as pd
import io
import requests
import numpy as np
import os
from sklearn.model_selection import train_test_split
from sklearn import metrics
from keras.models import Sequential
from keras.layers.core import Dense, Activation
from keras.callbacks import EarlyStopping
from keras.callbacks import ModelCheckpoint
path = "./data/"
filename = os.path.join(path,"iris.csv")
df = pd.read_csv(filename,na_values=['NA','?'])
species = encode_text_index(df,"species")
x,y = to_xy(df,"species")
# Split into train/test
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.25, random_state=42)
model = Sequential()
model.add(Dense(10, input_dim=x.shape[1], activation='relu'))
model.add(Dense(5,activation='relu'))
model.add(Dense(y.shape[1],activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
checkpointer = ModelCheckpoint(filepath="best_weights.hdf5", verbose=0, save_best_only=True) # save best model
model.fit(x,y,validation_data=(x_test,y_test),callbacks=[monitor,checkpointer],verbose=2,epochs=1000)
model.load_weights('best_weights.hdf5') # load weights from best model
```
# Potential Keras Issue on Small Networks and Early Stopping
You might occasionally see this error:
```
OSError: Unable to create file (Unable to open file: name = 'best_weights.hdf5', errno = 22, error message = 'invalid argument', flags = 13, o_flags = 302)
```
Usually you can just run rerun the code and it goes away. This is an unfortnuate result of saving a file each time the validation score improves (as described in the previous section). If the errors improve two rapidly, you might try to save the file twice and get an error from these two saves overlapping. For larger neural networks this will not be a problem because each training step will take longer, allowing for plenty of time for the previous save to complete. Because of this potential issue, this code is not used with every neural network in this course.
# Calculate Classification Accuracy
Accuracy is the number of rows where the neural network correctly predicted the target class. Accuracy is only used for classification, not regression.
$ accuracy = \frac{\textit{#} \ correct}{N} $
Where $N$ is the size of the evaluted set (training or validation). Higher accuracy numbers are desired.
As we just saw, by default, Keras will return the percent probability for each class. We can change these prediction probabilities into the actual iris predicted with **argmax**.
```
pred = np.argmax(pred,axis=1) # raw probabilities to chosen class (highest probability)
print(pred)
```
Now that we have the actual iris flower predicted, we can calculate the percent accuracy (how many were correctly classified).
```
y_compare = np.argmax(y_test,axis=1)
score = metrics.accuracy_score(y_compare, pred)
print("Accuracy score: {}".format(score))
```
# Calculate Classification Log Loss
Accuracy is like a final exam with no partial credit. However, neural networks can predict a probability of each of the target classes. Neural networks will give high probabilities to predictions that are more likely. Log loss is an error metric that penalizes confidence in wrong answers. Lower log loss values are desired.
The following code shows the output of predict_proba:
```
from IPython.display import display
# Don't display numpy in scientific notation
np.set_printoptions(precision=4)
np.set_printoptions(suppress=True)
# Generate predictions
pred = model.predict(x_test)
print("Numpy array of predictions")
print(pred[0]*100)
print("As percent probability")
display(pred[0:5])
score = metrics.log_loss(y_test, pred)
print("Log loss score: {}".format(score))
```
[Log loss](https://www.kaggle.com/wiki/LogarithmicLoss) is calculated as follows:
$ \text{log loss} = -\frac{1}{N}\sum_{i=1}^N {( {y}_i\log(\hat{y}_i) + (1 - {y}_i)\log(1 - \hat{y}_i))} $
The log function is useful to penalizing wrong answers. The following code demonstrates the utility of the log function:
```
%matplotlib inline
from matplotlib.pyplot import figure, show
from numpy import arange, sin, pi
#t = arange(1e-5, 5.0, 0.00001)
#t = arange(1.0, 5.0, 0.00001) # computer scientists
t = arange(0.0, 1.0, 0.00001) # data scientists
fig = figure(1,figsize=(12, 10))
ax1 = fig.add_subplot(211)
ax1.plot(t, np.log(t))
ax1.grid(True)
ax1.set_ylim((-8, 1.5))
ax1.set_xlim((-0.1, 2))
ax1.set_xlabel('x')
ax1.set_ylabel('y')
ax1.set_title('log(x)')
show()
```
# Evaluating Regression Results
Regression results are evaluated differently than classification. Consider the following code that trains a neural network for the [MPG dataset](https://github.com/jeffheaton/t81_558_deep_learning/blob/master/datasets_mpg.ipynb).
```
from sklearn.model_selection import train_test_split
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
cars = df['name']
df.drop('name',1,inplace=True)
missing_median(df, 'horsepower')
x,y = to_xy(df,"mpg")
# Split into train/test
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.25, random_state=45)
model = Sequential()
model.add(Dense(10, input_dim=x.shape[1], activation='relu'))
model.add(Dense(10))
model.add(Dense(10))
model.add(Dense(10))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
model.fit(x,y,validation_data=(x_test,y_test),callbacks=[monitor],verbose=2,epochs=1000)
```
### Mean Square Error
The mean square error is the sum of the squared differences between the prediction ($\hat{y}$) and the expected ($y$). MSE values are not of a particular unit. If an MSE value has decreased for a model, that is good. However, beyond this, there is not much more you can determine. Low MSE values are desired.
$ \text{MSE} = \frac{1}{n} \sum_{i=1}^n \left(\hat{y}_i - y_i\right)^2 $
```
# Predict
pred = model.predict(x_test)
# Measure MSE error.
score = metrics.mean_squared_error(pred,y_test)
print("Final score (MSE): {}".format(score))
```
### Root Mean Square Error
The root mean square (RMSE) is essentially the square root of the MSE. Because of this, the RMSE error is in the same units as the training data outcome. Low RMSE values are desired.
$ \text{RMSE} = \sqrt{\frac{1}{n} \sum_{i=1}^n \left(\hat{y}_i - y_i\right)^2} $
```
# Measure RMSE error. RMSE is common for regression.
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print("Final score (RMSE): {}".format(score))
```
# Training with Cross-Validation
Cross-Validation uses a number of folds, and multiple models, to generate out of sample predictions on the entire dataset. It is important to note that there will be one model (neural network) for each fold. Each model contributes part of the final out-of-sample prediction.

For new data, which is data not present in the training set, predictions from the fold models can be handled in several ways.
* Choose the model that had the highest validation score as the final model.
* Preset new data to the 5 models and average the result (this is an [enesmble](https://en.wikipedia.org/wiki/Ensemble_learning)).
* Retrain a new model (using the same settings as the cross-validation) on the entire dataset. Train for as many steps, and with the same hidden layer structure.
## Regression with Cross-Validation
The following code trains the MPG dataset using a 5-fold cross-validation. The expected performance of a neural network, of the type trained here, would be the score for the generated out-of-sample predictions.
```
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
from sklearn.model_selection import KFold
from keras.models import Sequential
from keras.layers.core import Dense, Activation
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
filename_write = os.path.join(path,"auto-mpg-out-of-sample.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
# Shuffle
np.random.seed(42)
df = df.reindex(np.random.permutation(df.index))
df.reset_index(inplace=True, drop=True)
# Preprocess
cars = df['name']
df.drop('name',1,inplace=True)
missing_median(df, 'horsepower')
# Encode to a 2D matrix for training
x,y = to_xy(df,'mpg')
# Cross-Validate
kf = KFold(5)
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x):
fold+=1
print("Fold #{}".format(fold))
x_train = x[train]
y_train = y[train]
x_test = x[test]
y_test = y[test]
model = Sequential()
model.add(Dense(20, input_dim=x.shape[1], activation='relu'))
model.add(Dense(10, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
model.fit(x_train,y_train,validation_data=(x_test,y_test),callbacks=[monitor],verbose=0,epochs=1000)
pred = model.predict(x_test)
oos_y.append(y_test)
oos_pred.append(pred)
# Measure this fold's RMSE
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print("Fold score (RMSE): {}".format(score))
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
score = np.sqrt(metrics.mean_squared_error(oos_pred,oos_y))
print("Final, out of sample score (RMSE): {}".format(score))
# Write the cross-validated prediction
oos_y = pd.DataFrame(oos_y)
oos_pred = pd.DataFrame(oos_pred)
oosDF = pd.concat( [df, oos_y, oos_pred],axis=1 )
oosDF.to_csv(filename_write,index=False)
```
## Classification with Cross-Validation
The following code trains and fits the iris dataset with Cross-Validation. It also writes out the out of sample (predictions on the test set) results.
```
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
from sklearn.model_selection import KFold
from keras.models import Sequential
from keras.layers.core import Dense, Activation
path = "./data/"
filename_read = os.path.join(path,"iris.csv")
filename_write = os.path.join(path,"iris-out-of-sample.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
# Shuffle
np.random.seed(42)
df = df.reindex(np.random.permutation(df.index))
df.reset_index(inplace=True, drop=True)
# Encode to a 2D matrix for training
species = encode_text_index(df,"species")
x,y = to_xy(df,"species")
# Cross-validate
kf = KFold(5)
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x):
fold+=1
print("Fold #{}".format(fold))
x_train = x[train]
y_train = y[train]
x_test = x[test]
y_test = y[test]
model = Sequential()
model.add(Dense(50, input_dim=x.shape[1], activation='relu')) # Hidden 1
model.add(Dense(25, activation='relu')) # Hidden 2
model.add(Dense(y.shape[1],activation='softmax')) # Output
model.compile(loss='categorical_crossentropy', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=25, verbose=1, mode='auto')
model.fit(x,y,validation_data=(x_test,y_test),callbacks=[monitor],verbose=0,epochs=1000)
pred = model.predict(x_test)
oos_y.append(y_test)
pred = np.argmax(pred,axis=1) # raw probabilities to chosen class (highest probability)
oos_pred.append(pred)
# Measure this fold's accuracy
y_compare = np.argmax(y_test,axis=1) # For accuracy calculation
score = metrics.accuracy_score(y_compare, pred)
print("Fold score (accuracy): {}".format(score))
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
oos_y_compare = np.argmax(oos_y,axis=1) # For accuracy calculation
score = metrics.accuracy_score(oos_y_compare, oos_pred)
print("Final score (accuracy): {}".format(score))
# Write the cross-validated prediction
oos_y = pd.DataFrame(oos_y)
oos_pred = pd.DataFrame(oos_pred)
oosDF = pd.concat( [df, oos_y, oos_pred],axis=1 )
oosDF.to_csv(filename_write,index=False)
```
# Training with both a Cross-Validation and a Holdout Set
If you have a considerable amount of data, it is always valuable to set aside a holdout set before you cross-validate. This hold out set will be the final evaluation before you make use of your model for its real-world use.

The following program makes use of a holdout set, and then still cross-validates.
```
from sklearn.model_selection import train_test_split
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
from sklearn.model_selection import KFold
from keras.callbacks import EarlyStopping
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
filename_write = os.path.join(path,"auto-mpg-holdout.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
# create feature vector
missing_median(df, 'horsepower')
df.drop('name',1,inplace=True)
encode_text_dummy(df, 'origin')
# Shuffle
np.random.seed(42)
df = df.reindex(np.random.permutation(df.index))
df.reset_index(inplace=True, drop=True)
# Encode to a 2D matrix for training
x,y = to_xy(df,'mpg')
# Keep a 10% holdout
x_main, x_holdout, y_main, y_holdout = train_test_split(
x, y, test_size=0.10)
# Cross-validate
kf = KFold(5)
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x_main):
fold+=1
print("Fold #{}".format(fold))
x_train = x_main[train]
y_train = y_main[train]
x_test = x_main[test]
y_test = y_main[test]
model = Sequential()
model.add(Dense(20, input_dim=x.shape[1], activation='relu'))
model.add(Dense(5, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
model.fit(x_train,y_train,validation_data=(x_test,y_test),callbacks=[monitor],verbose=0,epochs=1000)
pred = model.predict(x_test)
oos_y.append(y_test)
oos_pred.append(pred)
# Measure accuracy
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print("Fold score (RMSE): {}".format(score))
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
score = np.sqrt(metrics.mean_squared_error(oos_pred,oos_y))
print()
print("Cross-validated score (RMSE): {}".format(score))
# Write the cross-validated prediction (from the last neural network)
holdout_pred = model.predict(x_holdout)
score = np.sqrt(metrics.mean_squared_error(holdout_pred,y_holdout))
print("Holdout score (RMSE): {}".format(score))
```
# Scikit-Learn Versions: model_selection vs cross_validation
Scikit-Learn changed a bit in how cross-validation is handled. Both versions still work, but you should use the **sklearn.model_selection** import, rather than **sklearn.cross_validation**. The following shows both the new and old forms of cross-validation. All examples from this class will use the newer form.
The following two sections show both forms:
```
# Older scikit-learn syntax for splits/cross-validation
# Still valid, but going away. Do not use.
# (Note the red box warning below)
from sklearn.cross_validation import train_test_split
from sklearn.cross_validation import KFold
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
kf = KFold(len(df), n_folds=5)
fold = 0
for train, test in kf:
fold+=1
print("Fold #{}: train={}, test={}".format(fold,len(train),len(test)))
# Newer scikit-learn syntax for splits/cross-validation
# Use this method (as shown above)
from sklearn.model_selection import train_test_split
from sklearn.model_selection import KFold
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
kf = KFold(5)
fold = 0
for train, test in kf.split(df):
fold+=1
print("Fold #{}: train={}, test={}".format(fold,len(train),len(test)))
```
# How Kaggle Competitions are Scored
[Kaggle](https://www.kaggle.com/) is a platform for competitive data science. Competitions are posted onto Kaggle by companies seeking the best model for their data. Competing in a Kaggle competition is quite a bit of work, I've [competed in one Kaggle competition](https://www.kaggle.com/jeffheaton).
Kaggle awards "tiers", such as:
* Kaggle Grandmaster
* Kaggle Master
* Kaggle Expert
Your [tier](https://www.kaggle.com/progression) is based on your performance in past competitions.
To compete in Kaggle you simply provide predictions for a dataset that they post. You do not need to submit any code. Your prediction output will place you onto the [leaderboard of a competition](https://www.kaggle.com/c/otto-group-product-classification-challenge/leaderboard/public).

An original dataset is sent to Kaggle by the company. From this dataset, Kaggle posts public data that includes "train" and "test. For the "train" data, the outcomes (y) are provided. For the test data, no outcomes are provided. Your submission file contains your predictions for the "test data". When you submit your results, Kaggle will calculate a score on part of your prediction data. They do not publish want part of the submission data are used for the public and private leaderboard scores (this is a secret to prevent overfitting). While the competition is still running, Kaggle publishes the public leaderboard ranks. Once the competition ends, the private leaderboard is revealed to designate the true winners. Due to overfitting, there is sometimes an upset in positions when the final private leaderboard is revealed.
# Managing Hyperparameters
There are many different settings that you can use for a neural network. These can affect performance. The following code changes some of these, beyond their default values:
* **activation:** relu, sigmoid, tanh
* Layers/Neuron Counts
* **optimizer:** adam, sgd, rmsprop, and [others](https://keras.io/optimizers/)
```
%matplotlib inline
from matplotlib.pyplot import figure, show
from sklearn.model_selection import train_test_split
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
import tensorflow as tf
path = "./data/"
preprocess = False
filename_read = os.path.join(path,"auto-mpg.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
# create feature vector
missing_median(df, 'horsepower')
encode_text_dummy(df, 'origin')
df.drop('name',1,inplace=True)
if preprocess:
encode_numeric_zscore(df, 'horsepower')
encode_numeric_zscore(df, 'weight')
encode_numeric_zscore(df, 'cylinders')
encode_numeric_zscore(df, 'displacement')
encode_numeric_zscore(df, 'acceleration')
# Encode to a 2D matrix for training
x,y = to_xy(df,'mpg')
# Split into train/test
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.20, random_state=42)
model = Sequential()
model.add(Dense(100, input_dim=x.shape[1], activation='relu'))
model.add(Dense(50, activation='relu'))
model.add(Dense(25, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
model.fit(x,y,validation_data=(x_test,y_test),callbacks=[monitor],verbose=0,epochs=1000)
# Predict and measure RMSE
pred = model.predict(x_test)
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print("Score (RMSE): {}".format(score))
```
# Module 4 Assignment
You can find the first assignmeht here: [assignment 4](https://github.com/jeffheaton/t81_558_deep_learning/blob/master/assignments/assignment_yourname_class1.ipynb)
|
github_jupyter
|
from sklearn import preprocessing
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import shutil
import os
import requests
import base64
# Encode text values to dummy variables(i.e. [1,0,0],[0,1,0],[0,0,1] for red,green,blue)
def encode_text_dummy(df, name):
dummies = pd.get_dummies(df[name])
for x in dummies.columns:
dummy_name = "{}-{}".format(name, x)
df[dummy_name] = dummies[x]
df.drop(name, axis=1, inplace=True)
# Encode text values to a single dummy variable. The new columns (which do not replace the old) will have a 1
# at every location where the original column (name) matches each of the target_values. One column is added for
# each target value.
def encode_text_single_dummy(df, name, target_values):
for tv in target_values:
l = list(df[name].astype(str))
l = [1 if str(x) == str(tv) else 0 for x in l]
name2 = "{}-{}".format(name, tv)
df[name2] = l
# Encode text values to indexes(i.e. [1],[2],[3] for red,green,blue).
def encode_text_index(df, name):
le = preprocessing.LabelEncoder()
df[name] = le.fit_transform(df[name])
return le.classes_
# Encode a numeric column as zscores
def encode_numeric_zscore(df, name, mean=None, sd=None):
if mean is None:
mean = df[name].mean()
if sd is None:
sd = df[name].std()
df[name] = (df[name] - mean) / sd
# Convert all missing values in the specified column to the median
def missing_median(df, name):
med = df[name].median()
df[name] = df[name].fillna(med)
# Convert all missing values in the specified column to the default
def missing_default(df, name, default_value):
df[name] = df[name].fillna(default_value)
# Convert a Pandas dataframe to the x,y inputs that TensorFlow needs
def to_xy(df, target):
result = []
for x in df.columns:
if x != target:
result.append(x)
# find out the type of the target column. Is it really this hard? :(
target_type = df[target].dtypes
target_type = target_type[0] if hasattr(target_type, '__iter__') else target_type
# Encode to int for classification, float otherwise. TensorFlow likes 32 bits.
if target_type in (np.int64, np.int32):
# Classification
dummies = pd.get_dummies(df[target])
return df.as_matrix(result).astype(np.float32), dummies.as_matrix().astype(np.float32)
else:
# Regression
return df.as_matrix(result).astype(np.float32), df.as_matrix([target]).astype(np.float32)
# Nicely formatted time string
def hms_string(sec_elapsed):
h = int(sec_elapsed / (60 * 60))
m = int((sec_elapsed % (60 * 60)) / 60)
s = sec_elapsed % 60
return "{}:{:>02}:{:>05.2f}".format(h, m, s)
# Regression chart.
def chart_regression(pred,y,sort=True):
t = pd.DataFrame({'pred' : pred, 'y' : y.flatten()})
if sort:
t.sort_values(by=['y'],inplace=True)
a = plt.plot(t['y'].tolist(),label='expected')
b = plt.plot(t['pred'].tolist(),label='prediction')
plt.ylabel('output')
plt.legend()
plt.show()
# Remove all rows where the specified column is +/- sd standard deviations
def remove_outliers(df, name, sd):
drop_rows = df.index[(np.abs(df[name] - df[name].mean()) >= (sd * df[name].std()))]
df.drop(drop_rows, axis=0, inplace=True)
# Encode a column to a range between normalized_low and normalized_high.
def encode_numeric_range(df, name, normalized_low=-1, normalized_high=1,
data_low=None, data_high=None):
if data_low is None:
data_low = min(df[name])
data_high = max(df[name])
df[name] = ((df[name] - data_low) / (data_high - data_low)) \
* (normalized_high - normalized_low) + normalized_low
# This function submits an assignment. You can submit an assignment as much as you like, only the final
# submission counts. The paramaters are as follows:
# data - Pandas dataframe output.
# key - Your student key that was emailed to you.
# no - The assignment class number, should be 1 through 1.
# source_file - The full path to your Python or IPYNB file. This must have "_class1" as part of its name.
# . The number must match your assignment number. For example "_class2" for class assignment #2.
def submit(data,key,no,source_file=None):
if source_file is None and '__file__' not in globals(): raise Exception('Must specify a filename when a Jupyter notebook.')
if source_file is None: source_file = __file__
suffix = '_class{}'.format(no)
if suffix not in source_file: raise Exception('{} must be part of the filename.'.format(suffix))
with open(source_file, "rb") as image_file:
encoded_python = base64.b64encode(image_file.read()).decode('ascii')
ext = os.path.splitext(source_file)[-1].lower()
if ext not in ['.ipynb','.py']: raise Exception("Source file is {} must be .py or .ipynb".format(ext))
r = requests.post("https://api.heatonresearch.com/assignment-submit",
headers={'x-api-key':key}, json={'csv':base64.b64encode(data.to_csv(index=False).encode('ascii')).decode("ascii"),
'assignment': no, 'ext':ext, 'py':encoded_python})
if r.status_code == 200:
print("Success: {}".format(r.text))
else: print("Failure: {}".format(r.text))
import tensorflow.contrib.learn as skflow
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
# create feature vector
missing_median(df, 'horsepower')
df.drop('name',1,inplace=True)
#encode_numeric_binary(df,'mpg',20)
#df['origin'] = df['origin'].astype(str)
#encode_text_tfidf(df, 'origin')
# Drop outliers in horsepower
print("Length before MPG outliers dropped: {}".format(len(df)))
remove_outliers(df,'mpg',2)
print("Length after MPG outliers dropped: {}".format(len(df)))
print(df)
import requests
address = "1 Brookings Dr, St. Louis, MO 63130"
response = requests.get('https://maps.googleapis.com/maps/api/geocode/json?address='+address)
resp_json_payload = response.json()
print(resp_json_payload['results'][0]['geometry']['location'])
from math import sin, cos, sqrt, atan2, radians
# Distance function
def distance_lat_lng(lat1,lng1,lat2,lng2):
# approximate radius of earth in km
R = 6373.0
# degrees to radians (lat/lon are in degrees)
lat1 = radians(lat1)
lng1 = radians(lng1)
lat2 = radians(lat2)
lng2 = radians(lng2)
dlng = lng2 - lng1
dlat = lat2 - lat1
a = sin(dlat / 2)**2 + cos(lat1) * cos(lat2) * sin(dlng / 2)**2
c = 2 * atan2(sqrt(a), sqrt(1 - a))
return R * c
# Find lat lon for address
def lookup_lat_lng(address):
response = requests.get('https://maps.googleapis.com/maps/api/geocode/json?address='+address)
json = response.json()
if len(json['results']) == 0:
print("Can't find: {}".format(address))
return 0,0
map = json['results'][0]['geometry']['location']
return map['lat'],map['lng']
# Distance between two locations
import requests
address1 = "1 Brookings Dr, St. Louis, MO 63130"
address2 = "3301 College Ave, Fort Lauderdale, FL 33314"
lat1, lng1 = lookup_lat_lng(address1)
lat2, lng2 = lookup_lat_lng(address2)
print("Distance, St. Louis, MO to Ft. Lauderdale, FL: {} km".format(
distance_lat_lng(lat1,lng1,lat2,lng2)))
# Encoding other universities by their distance to Washington University
schools = [
["Princeton University, Princeton, NJ 08544", 'Princeton'],
["Massachusetts Hall, Cambridge, MA 02138", 'Harvard'],
["5801 S Ellis Ave, Chicago, IL 60637", 'University of Chicago'],
["Yale, New Haven, CT 06520", 'Yale'],
["116th St & Broadway, New York, NY 10027", 'Columbia University'],
["450 Serra Mall, Stanford, CA 94305", 'Stanford'],
["77 Massachusetts Ave, Cambridge, MA 02139", 'MIT'],
["Duke University, Durham, NC 27708", 'Duke University'],
["University of Pennsylvania, Philadelphia, PA 19104", 'University of Pennsylvania'],
["Johns Hopkins University, Baltimore, MD 21218", 'Johns Hopkins']
]
lat1, lng1 = lookup_lat_lng("1 Brookings Dr, St. Louis, MO 63130")
for address, name in schools:
lat2,lng2 = lookup_lat_lng(address)
dist = distance_lat_lng(lat1,lng1,lat2,lng2)
print("School '{}', distance to wustl is: {}".format(name,dist))
import pandas as pd
import io
import requests
import numpy as np
import os
from sklearn.model_selection import train_test_split
from sklearn import metrics
from keras.models import Sequential
from keras.layers.core import Dense, Activation
from keras.callbacks import EarlyStopping
path = "./data/"
filename = os.path.join(path,"iris.csv")
df = pd.read_csv(filename,na_values=['NA','?'])
species = encode_text_index(df,"species")
x,y = to_xy(df,"species")
# Split into train/test
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.25, random_state=42)
model = Sequential()
model.add(Dense(10, input_dim=x.shape[1], activation='relu'))
model.add(Dense(5,activation='relu'))
model.add(Dense(y.shape[1],activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
model.fit(x,y,validation_data=(x_test,y_test),callbacks=[monitor],verbose=2,epochs=1000)
from sklearn import metrics
import tensorflow as tf
pred = model.predict(x_test)
print(pred[0:5]) # print first five predictions
import pandas as pd
import io
import requests
import numpy as np
import os
from sklearn.model_selection import train_test_split
from sklearn import metrics
from keras.models import Sequential
from keras.layers.core import Dense, Activation
from keras.callbacks import EarlyStopping
from keras.callbacks import ModelCheckpoint
path = "./data/"
filename = os.path.join(path,"iris.csv")
df = pd.read_csv(filename,na_values=['NA','?'])
species = encode_text_index(df,"species")
x,y = to_xy(df,"species")
# Split into train/test
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.25, random_state=42)
model = Sequential()
model.add(Dense(10, input_dim=x.shape[1], activation='relu'))
model.add(Dense(5,activation='relu'))
model.add(Dense(y.shape[1],activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
checkpointer = ModelCheckpoint(filepath="best_weights.hdf5", verbose=0, save_best_only=True) # save best model
model.fit(x,y,validation_data=(x_test,y_test),callbacks=[monitor,checkpointer],verbose=2,epochs=1000)
model.load_weights('best_weights.hdf5') # load weights from best model
OSError: Unable to create file (Unable to open file: name = 'best_weights.hdf5', errno = 22, error message = 'invalid argument', flags = 13, o_flags = 302)
pred = np.argmax(pred,axis=1) # raw probabilities to chosen class (highest probability)
print(pred)
y_compare = np.argmax(y_test,axis=1)
score = metrics.accuracy_score(y_compare, pred)
print("Accuracy score: {}".format(score))
from IPython.display import display
# Don't display numpy in scientific notation
np.set_printoptions(precision=4)
np.set_printoptions(suppress=True)
# Generate predictions
pred = model.predict(x_test)
print("Numpy array of predictions")
print(pred[0]*100)
print("As percent probability")
display(pred[0:5])
score = metrics.log_loss(y_test, pred)
print("Log loss score: {}".format(score))
%matplotlib inline
from matplotlib.pyplot import figure, show
from numpy import arange, sin, pi
#t = arange(1e-5, 5.0, 0.00001)
#t = arange(1.0, 5.0, 0.00001) # computer scientists
t = arange(0.0, 1.0, 0.00001) # data scientists
fig = figure(1,figsize=(12, 10))
ax1 = fig.add_subplot(211)
ax1.plot(t, np.log(t))
ax1.grid(True)
ax1.set_ylim((-8, 1.5))
ax1.set_xlim((-0.1, 2))
ax1.set_xlabel('x')
ax1.set_ylabel('y')
ax1.set_title('log(x)')
show()
from sklearn.model_selection import train_test_split
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
cars = df['name']
df.drop('name',1,inplace=True)
missing_median(df, 'horsepower')
x,y = to_xy(df,"mpg")
# Split into train/test
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.25, random_state=45)
model = Sequential()
model.add(Dense(10, input_dim=x.shape[1], activation='relu'))
model.add(Dense(10))
model.add(Dense(10))
model.add(Dense(10))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
model.fit(x,y,validation_data=(x_test,y_test),callbacks=[monitor],verbose=2,epochs=1000)
# Predict
pred = model.predict(x_test)
# Measure MSE error.
score = metrics.mean_squared_error(pred,y_test)
print("Final score (MSE): {}".format(score))
# Measure RMSE error. RMSE is common for regression.
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print("Final score (RMSE): {}".format(score))
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
from sklearn.model_selection import KFold
from keras.models import Sequential
from keras.layers.core import Dense, Activation
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
filename_write = os.path.join(path,"auto-mpg-out-of-sample.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
# Shuffle
np.random.seed(42)
df = df.reindex(np.random.permutation(df.index))
df.reset_index(inplace=True, drop=True)
# Preprocess
cars = df['name']
df.drop('name',1,inplace=True)
missing_median(df, 'horsepower')
# Encode to a 2D matrix for training
x,y = to_xy(df,'mpg')
# Cross-Validate
kf = KFold(5)
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x):
fold+=1
print("Fold #{}".format(fold))
x_train = x[train]
y_train = y[train]
x_test = x[test]
y_test = y[test]
model = Sequential()
model.add(Dense(20, input_dim=x.shape[1], activation='relu'))
model.add(Dense(10, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
model.fit(x_train,y_train,validation_data=(x_test,y_test),callbacks=[monitor],verbose=0,epochs=1000)
pred = model.predict(x_test)
oos_y.append(y_test)
oos_pred.append(pred)
# Measure this fold's RMSE
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print("Fold score (RMSE): {}".format(score))
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
score = np.sqrt(metrics.mean_squared_error(oos_pred,oos_y))
print("Final, out of sample score (RMSE): {}".format(score))
# Write the cross-validated prediction
oos_y = pd.DataFrame(oos_y)
oos_pred = pd.DataFrame(oos_pred)
oosDF = pd.concat( [df, oos_y, oos_pred],axis=1 )
oosDF.to_csv(filename_write,index=False)
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
from sklearn.model_selection import KFold
from keras.models import Sequential
from keras.layers.core import Dense, Activation
path = "./data/"
filename_read = os.path.join(path,"iris.csv")
filename_write = os.path.join(path,"iris-out-of-sample.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
# Shuffle
np.random.seed(42)
df = df.reindex(np.random.permutation(df.index))
df.reset_index(inplace=True, drop=True)
# Encode to a 2D matrix for training
species = encode_text_index(df,"species")
x,y = to_xy(df,"species")
# Cross-validate
kf = KFold(5)
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x):
fold+=1
print("Fold #{}".format(fold))
x_train = x[train]
y_train = y[train]
x_test = x[test]
y_test = y[test]
model = Sequential()
model.add(Dense(50, input_dim=x.shape[1], activation='relu')) # Hidden 1
model.add(Dense(25, activation='relu')) # Hidden 2
model.add(Dense(y.shape[1],activation='softmax')) # Output
model.compile(loss='categorical_crossentropy', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=25, verbose=1, mode='auto')
model.fit(x,y,validation_data=(x_test,y_test),callbacks=[monitor],verbose=0,epochs=1000)
pred = model.predict(x_test)
oos_y.append(y_test)
pred = np.argmax(pred,axis=1) # raw probabilities to chosen class (highest probability)
oos_pred.append(pred)
# Measure this fold's accuracy
y_compare = np.argmax(y_test,axis=1) # For accuracy calculation
score = metrics.accuracy_score(y_compare, pred)
print("Fold score (accuracy): {}".format(score))
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
oos_y_compare = np.argmax(oos_y,axis=1) # For accuracy calculation
score = metrics.accuracy_score(oos_y_compare, oos_pred)
print("Final score (accuracy): {}".format(score))
# Write the cross-validated prediction
oos_y = pd.DataFrame(oos_y)
oos_pred = pd.DataFrame(oos_pred)
oosDF = pd.concat( [df, oos_y, oos_pred],axis=1 )
oosDF.to_csv(filename_write,index=False)
from sklearn.model_selection import train_test_split
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
from sklearn.model_selection import KFold
from keras.callbacks import EarlyStopping
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
filename_write = os.path.join(path,"auto-mpg-holdout.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
# create feature vector
missing_median(df, 'horsepower')
df.drop('name',1,inplace=True)
encode_text_dummy(df, 'origin')
# Shuffle
np.random.seed(42)
df = df.reindex(np.random.permutation(df.index))
df.reset_index(inplace=True, drop=True)
# Encode to a 2D matrix for training
x,y = to_xy(df,'mpg')
# Keep a 10% holdout
x_main, x_holdout, y_main, y_holdout = train_test_split(
x, y, test_size=0.10)
# Cross-validate
kf = KFold(5)
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x_main):
fold+=1
print("Fold #{}".format(fold))
x_train = x_main[train]
y_train = y_main[train]
x_test = x_main[test]
y_test = y_main[test]
model = Sequential()
model.add(Dense(20, input_dim=x.shape[1], activation='relu'))
model.add(Dense(5, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
model.fit(x_train,y_train,validation_data=(x_test,y_test),callbacks=[monitor],verbose=0,epochs=1000)
pred = model.predict(x_test)
oos_y.append(y_test)
oos_pred.append(pred)
# Measure accuracy
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print("Fold score (RMSE): {}".format(score))
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
score = np.sqrt(metrics.mean_squared_error(oos_pred,oos_y))
print()
print("Cross-validated score (RMSE): {}".format(score))
# Write the cross-validated prediction (from the last neural network)
holdout_pred = model.predict(x_holdout)
score = np.sqrt(metrics.mean_squared_error(holdout_pred,y_holdout))
print("Holdout score (RMSE): {}".format(score))
# Older scikit-learn syntax for splits/cross-validation
# Still valid, but going away. Do not use.
# (Note the red box warning below)
from sklearn.cross_validation import train_test_split
from sklearn.cross_validation import KFold
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
kf = KFold(len(df), n_folds=5)
fold = 0
for train, test in kf:
fold+=1
print("Fold #{}: train={}, test={}".format(fold,len(train),len(test)))
# Newer scikit-learn syntax for splits/cross-validation
# Use this method (as shown above)
from sklearn.model_selection import train_test_split
from sklearn.model_selection import KFold
path = "./data/"
filename_read = os.path.join(path,"auto-mpg.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
kf = KFold(5)
fold = 0
for train, test in kf.split(df):
fold+=1
print("Fold #{}: train={}, test={}".format(fold,len(train),len(test)))
%matplotlib inline
from matplotlib.pyplot import figure, show
from sklearn.model_selection import train_test_split
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
import tensorflow as tf
path = "./data/"
preprocess = False
filename_read = os.path.join(path,"auto-mpg.csv")
df = pd.read_csv(filename_read,na_values=['NA','?'])
# create feature vector
missing_median(df, 'horsepower')
encode_text_dummy(df, 'origin')
df.drop('name',1,inplace=True)
if preprocess:
encode_numeric_zscore(df, 'horsepower')
encode_numeric_zscore(df, 'weight')
encode_numeric_zscore(df, 'cylinders')
encode_numeric_zscore(df, 'displacement')
encode_numeric_zscore(df, 'acceleration')
# Encode to a 2D matrix for training
x,y = to_xy(df,'mpg')
# Split into train/test
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.20, random_state=42)
model = Sequential()
model.add(Dense(100, input_dim=x.shape[1], activation='relu'))
model.add(Dense(50, activation='relu'))
model.add(Dense(25, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3, patience=5, verbose=1, mode='auto')
model.fit(x,y,validation_data=(x_test,y_test),callbacks=[monitor],verbose=0,epochs=1000)
# Predict and measure RMSE
pred = model.predict(x_test)
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print("Score (RMSE): {}".format(score))
| 0.524395 | 0.982237 |
# Read SEG-Y with segyio
This is a relatively new library from Statoil. It is very easy to use... in most cases.
```
import segyio
help(segyio)
```
## Basics
If you don't have the file yet, **[get the large dataset from Agile's S3 bucket](https://s3.amazonaws.com/agilegeo/Penobscot_0-1000ms.sgy.zip)**. It's 140MB.
```
with segyio.open('../data/Penobscot_0-1000ms.sgy') as s:
print("Binary header")
print(s.bin)
print()
print("Text header")
print(s.text[0])
```
This garbled text header is a bug. `segyio` currently (Jan 2019) assumes the header is EBCDIC encoded, but in this file it's ASCII encoded. It has been filed [as an issue](https://github.com/equinor/segyio/issues/317).
## Access the data
```
with segyio.open('../data/Penobscot_0-1000ms.sgy') as s:
c = segyio.cube(s)
```
`c` is just an `ndarray`.
```
type(c)
c.shape
import matplotlib.pyplot as plt
%matplotlib inline
plt.imshow(c[100].T, cmap='Greys')
```
## 2D data
https://github.com/agile-geoscience/geocomputing/blob/master/data/HUN00-ALT-01_STK.sgy
This file does not open with the default `strict=True`:
```
# This should produce an error.
with segyio.open('../data/HUN00-ALT-01_STK.sgy') as s:
c = segyio.cube(s)
```
It's OK if not strict... but then you can't use `cube`
```
with segyio.open('../data/HUN00-ALT-01_STK.sgy', strict=False) as s:
c = segyio.cube(s)
```
So we'll unpack the traces manually...
```
import numpy as np
with segyio.open('../data/HUN00-ALT-01_STK.sgy', strict=False) as s:
data = np.stack(t.astype(np.float) for t in s.trace)
plt.figure(figsize=(15,5))
plt.imshow(data.T)
```
With a bit more work we can also read the file header and the trace headers.
```
import numpy as np
def chunks(s, n):
"""Produce `n`-character chunks from string `s`."""
for start in range(0, len(s), n):
yield s[start:start + n]
with segyio.open('../data/HUN00-ALT-01_STK.sgy', strict=False) as s:
# Read the data.
data = np.stack(t.astype(np.float) for t in s.trace)
# Get the (x, y) locations.
x = [t[segyio.TraceField.GroupX] for t in s.header]
y = [t[segyio.TraceField.GroupY] for t in s.header]
# Get the trace numbers.
cdp = np.array([t[segyio.TraceField.CDP] for t in s.header])
# Get the first textual header.
header = s.text[0].decode('ascii')
formatted = '\n'.join(chunk for chunk in chunks(header, 80))
# Get data from the binary header.
# Get the sample interval in ms (convert from microsec).
sample_interval = s.bin[segyio.BinField.Interval] / 1000
print(formatted)
```
Getting a sub-set of traces using CDP (or trace number or similar) is a little fiddly:
```
cdp
selection = np.where((cdp>500) & (cdp<800))[0]
subset = data[selection]
plt.imshow(subset.T, aspect='auto')
```
## Try another
```
with segyio.open('../data/31_81_PR.sgy') as s:
data = segyio.cube(s)
```
Nope.
```
with segyio.open('../data/31_81_PR.sgy', strict=False) as s:
data = np.stack(t.astype(np.float) for t in s.trace)
data.shape
```
OK, I guess this isn't quite the flow for a 2D file...
```
plt.figure(figsize=(16, 8))
plt.imshow(np.squeeze(data).T, cmap='Greys', aspect=0.2)
```
## Another, known to be 'weird'
```
with segyio.open('../data/31_81_PR.sgy') as s:
data = segyio.cube(s)
```
Nope again.
```
with segyio.open('../data/marmousi/velocity.segy', strict=False) as s:
print("weird dt: ", segyio.dt(s))
data = np.stack(t.astype(np.float) for t in s.trace)
```
This file is improperly organized (time first, so we don't need to transpose it) and the dt header is wrong.
```
plt.figure(figsize=(10, 6))
plt.imshow(data, cmap='viridis', aspect='auto')
plt.show()
```
|
github_jupyter
|
import segyio
help(segyio)
with segyio.open('../data/Penobscot_0-1000ms.sgy') as s:
print("Binary header")
print(s.bin)
print()
print("Text header")
print(s.text[0])
with segyio.open('../data/Penobscot_0-1000ms.sgy') as s:
c = segyio.cube(s)
type(c)
c.shape
import matplotlib.pyplot as plt
%matplotlib inline
plt.imshow(c[100].T, cmap='Greys')
# This should produce an error.
with segyio.open('../data/HUN00-ALT-01_STK.sgy') as s:
c = segyio.cube(s)
with segyio.open('../data/HUN00-ALT-01_STK.sgy', strict=False) as s:
c = segyio.cube(s)
import numpy as np
with segyio.open('../data/HUN00-ALT-01_STK.sgy', strict=False) as s:
data = np.stack(t.astype(np.float) for t in s.trace)
plt.figure(figsize=(15,5))
plt.imshow(data.T)
import numpy as np
def chunks(s, n):
"""Produce `n`-character chunks from string `s`."""
for start in range(0, len(s), n):
yield s[start:start + n]
with segyio.open('../data/HUN00-ALT-01_STK.sgy', strict=False) as s:
# Read the data.
data = np.stack(t.astype(np.float) for t in s.trace)
# Get the (x, y) locations.
x = [t[segyio.TraceField.GroupX] for t in s.header]
y = [t[segyio.TraceField.GroupY] for t in s.header]
# Get the trace numbers.
cdp = np.array([t[segyio.TraceField.CDP] for t in s.header])
# Get the first textual header.
header = s.text[0].decode('ascii')
formatted = '\n'.join(chunk for chunk in chunks(header, 80))
# Get data from the binary header.
# Get the sample interval in ms (convert from microsec).
sample_interval = s.bin[segyio.BinField.Interval] / 1000
print(formatted)
cdp
selection = np.where((cdp>500) & (cdp<800))[0]
subset = data[selection]
plt.imshow(subset.T, aspect='auto')
with segyio.open('../data/31_81_PR.sgy') as s:
data = segyio.cube(s)
with segyio.open('../data/31_81_PR.sgy', strict=False) as s:
data = np.stack(t.astype(np.float) for t in s.trace)
data.shape
plt.figure(figsize=(16, 8))
plt.imshow(np.squeeze(data).T, cmap='Greys', aspect=0.2)
with segyio.open('../data/31_81_PR.sgy') as s:
data = segyio.cube(s)
with segyio.open('../data/marmousi/velocity.segy', strict=False) as s:
print("weird dt: ", segyio.dt(s))
data = np.stack(t.astype(np.float) for t in s.trace)
plt.figure(figsize=(10, 6))
plt.imshow(data, cmap='viridis', aspect='auto')
plt.show()
| 0.553505 | 0.888614 |
# Load and Visualize FashionMNIST
---
In this notebook, we load and look at images from the [Fashion-MNIST database](https://github.com/zalandoresearch/fashion-mnist).
The first step in any classification problem is to look at the dataset you are working with. This will give you some details about the format of images and labels, as well as some insight into how you might approach defining a network to recognize patterns in such an image set.
PyTorch has some built-in datasets that you can use, and FashionMNIST is one of them; it has already been dowloaded into the `data/` directory in this notebook, so all we have to do is load these images using the FashionMNIST dataset class *and* load the data in batches with a `DataLoader`.
### Load the [data](http://pytorch.org/docs/master/torchvision/datasets.html)
#### Dataset class and Tensors
``torch.utils.data.Dataset`` is an abstract class representing a
dataset. The FashionMNIST class is an extension of this Dataset class and it allows us to 1. load batches of image/label data, and 2. uniformly apply transformations to our data, such as turning all our images into Tensor's for training a neural network. *Tensors are similar to numpy arrays, but can also be used on a GPU to accelerate computing.*
Let's see how to construct a training dataset.
```
# our basic libraries
import torch
import torchvision
# data loading and transforming
from torchvision.datasets import FashionMNIST
from torch.utils.data import DataLoader
from torchvision import transforms
# The output of torchvision datasets are PILImage images of range [0, 1].
# We transform them to Tensors for input into a CNN
## Define a transform to read the data in as a tensor
data_transform = transforms.ToTensor()
# choose the training and test datasets
train_data = FashionMNIST(root='./data', train=True,
download=False, transform=data_transform)
# Print out some stats about the training data
print('Train data, number of images: ', len(train_data))
```
#### Data iteration and batching
Next, we'll use ``torch.utils.data.DataLoader`` , which is an iterator that allows us to batch and shuffle the data.
In the next cell, we shuffle the data and load in image/label data in batches of size 20.
```
# prepare data loaders, set the batch_size
## TODO: you can try changing the batch_size to be larger or smaller
## when you get to training your network, see how batch_size affects the loss
batch_size = 20
train_loader = DataLoader(train_data, batch_size=batch_size, shuffle=True)
# specify the image classes
classes = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',
'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']
```
### Visualize some training data
This cell iterates over the training dataset, loading a random batch of image/label data, using `dataiter.next()`. It then plots the batch of images and labels in a `2 x batch_size/2` grid.
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# obtain one batch of training images
dataiter = iter(train_loader)
images, labels = dataiter.next()
images = images.numpy()
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(25, 4))
for idx in np.arange(batch_size):
ax = fig.add_subplot(2, batch_size/2, idx+1, xticks=[], yticks=[])
ax.imshow(np.squeeze(images[idx]), cmap='gray')
ax.set_title(classes[labels[idx]])
```
### View an image in more detail
Each image in this dataset is a `28x28` pixel, normalized, grayscale image.
#### A note on normalization
Normalization ensures that, as we go through a feedforward and then backpropagation step in training our CNN, that each image feature will fall within a similar range of values and not overly activate any particular layer in our network. During the feedfoward step, a network takes in an input image and multiplies each input pixel by some convolutional filter weights (and adds biases!), then it applies some activation and pooling functions. Without normalization, it's much more likely that the calculated gradients in the backpropagaton step will be quite large and cause our loss to increase instead of converge.
```
# select an image by index
idx = 2
img = np.squeeze(images[idx])
# display the pixel values in that image
fig = plt.figure(figsize = (12,12))
ax = fig.add_subplot(111)
ax.imshow(img, cmap='gray')
width, height = img.shape
thresh = img.max()/2.5
for x in range(width):
for y in range(height):
val = round(img[x][y],2) if img[x][y] !=0 else 0
ax.annotate(str(val), xy=(y,x),
horizontalalignment='center',
verticalalignment='center',
color='white' if img[x][y]<thresh else 'black')
```
|
github_jupyter
|
# our basic libraries
import torch
import torchvision
# data loading and transforming
from torchvision.datasets import FashionMNIST
from torch.utils.data import DataLoader
from torchvision import transforms
# The output of torchvision datasets are PILImage images of range [0, 1].
# We transform them to Tensors for input into a CNN
## Define a transform to read the data in as a tensor
data_transform = transforms.ToTensor()
# choose the training and test datasets
train_data = FashionMNIST(root='./data', train=True,
download=False, transform=data_transform)
# Print out some stats about the training data
print('Train data, number of images: ', len(train_data))
# prepare data loaders, set the batch_size
## TODO: you can try changing the batch_size to be larger or smaller
## when you get to training your network, see how batch_size affects the loss
batch_size = 20
train_loader = DataLoader(train_data, batch_size=batch_size, shuffle=True)
# specify the image classes
classes = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',
'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# obtain one batch of training images
dataiter = iter(train_loader)
images, labels = dataiter.next()
images = images.numpy()
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(25, 4))
for idx in np.arange(batch_size):
ax = fig.add_subplot(2, batch_size/2, idx+1, xticks=[], yticks=[])
ax.imshow(np.squeeze(images[idx]), cmap='gray')
ax.set_title(classes[labels[idx]])
# select an image by index
idx = 2
img = np.squeeze(images[idx])
# display the pixel values in that image
fig = plt.figure(figsize = (12,12))
ax = fig.add_subplot(111)
ax.imshow(img, cmap='gray')
width, height = img.shape
thresh = img.max()/2.5
for x in range(width):
for y in range(height):
val = round(img[x][y],2) if img[x][y] !=0 else 0
ax.annotate(str(val), xy=(y,x),
horizontalalignment='center',
verticalalignment='center',
color='white' if img[x][y]<thresh else 'black')
| 0.550366 | 0.994402 |
Author: Arend-Jan Quist\
Date of creation: 6 May 2020\
Last modified: 17 June 2020
Create a artificially created dataset of a moire lattice with Gaussian peaks.\
Apply drizzle and shift-and-add to this dataset. \
Apply various types of data processing to these images.
```
import numpy as np
from scipy import ndimage
from skimage import data, io, filters
import matplotlib.pyplot as plt
from Drizzle import *
save_folder = r"C:\Users\Arend-Jan\Documents\Universiteit\Bachelor Research Project\Thesis\Drizzle_images_moire_May\Artificial datasets"
```
```
def create_gaussian(sigma,size,pos):
"""Create a gaussian with standarddeviation sigma at an array of size with peak
at a given position.
"""
x, y = np.meshgrid(np.arange(0,size[0]) - pos[0], np.arange(0,size[1]) - pos[1])
d = np.sqrt(x*x+y*y)
g = np.exp(-( (d)**2 / ( 2.0 * sigma**2 ) ) )
#plt.imshow(g)
#plt.show()
return (g.T)
def create_lattice_images(mean_shifts=0,sigma_shifts = 3,seed_input=2020,size = [120,120],n_input=50,
sigma = 5.,d=20,h=None):
"""
Create artificial dataset of shifted images of Gaussian bulbs on a lattice.
Parameters:
"sigma_shifts" is the standard deviation of the shifts of the images.
"seed_input" is the seed used for dataset creation.
"size" is image size.
"n_input" is number of input images.
"sigma" is the width of the bulbs in the images.
"d" and "h" are geometrical parameters for the lattice, see sketch above.
"""
np.random.seed(seed_input) #set seed
if h == None:
h = d*np.sqrt(3)/2 #create triangular lattice
# Create shifts of input images
shifts = np.random.normal(mean_shifts,sigma_shifts,[n_input,2])
# Define point of lattice
lattice_points = []
brd = np.ceil(4*sigma/h)*h
for x in np.arange(-brd, size[0] + d + brd, d):
for y in np.arange(-brd, size[1] + 2*h + brd, 2*h):
lattice_points.append(np.array([x,y]))
lattice_points.append(np.array([x+d/2,y+h]))
# Create input images
ims = []
for i,shift in enumerate(shifts):
im = np.zeros(size)
for lp in lattice_points:
im += create_gaussian(sigma,size,lp+shift)
ims.append(im*10000)
return (ims,shifts)
def add_noise(ims,shifts,seed_input=2020,sigm_shift = .3,
sigm_noi_before_blur = 1000.,sigm_blur = 2.5,sigm_noi_after_blur = 1000.):
"""Add noise to given input images and shifts
Parameters:
"seed_input" is the seed used for dataset creation.
sigm_shift is sigma to bias shift.
sigm_noi_before_blur is sigma for gaussian noise.
sigm_blur is sigma for gaussian blurring.
sigm_noi_after_blur is sigma for gaussian noise.
"""
np.random.seed(seed_input) #set seed
# Add noise to the images
noise_ims = []
for im in ims:
noise_im = im + np.random.normal(0,sigm_noi_after_blur,np.shape(im))
noise_im = ndimage.filters.gaussian_filter(noise_im,sigm_blur)
noise_im += np.random.normal(0,sigm_noi_after_blur,np.shape(im))
noise_ims.append(noise_im)
#Add noise to the shifts
shifts_bias = shifts + np.random.normal(0,sigm_shift,np.shape(shifts))
return(noise_ims,shifts_bias)
def correlate(drizzled,mean):
size = [len(drizzled),len(drizzled[0])]
corr = ndimage.correlate(mean,drizzled-mean)
amax = np.argmax(corr)
peak = [amax%size[0],amax//size[0]]
return(peak)
def meansh(x):
"""Calculate the mean of shifts modulo 1"""
av_shift = [np.angle(np.mean(np.exp(1j*2*np.pi*x[:,0])))/2/np.pi%1,np.angle(np.mean(np.exp(1j*2*np.pi*x[:,1])))/2/np.pi%1]
av_shift = np.round(av_shift,2)
return(av_shift)
def center_shifts(shift):
"""Center the input shifts around 0."""
av_shift = meansh(shift)
shift[:,0] = shift[:,0]+1-av_shift[0]
shift[:,1] = shift[:,1]+1-av_shift[1]
return(shift)
```
# Single input image and output image
```
# drizzle parameters
p = 0.5
n = 1
ims,shifts = create_lattice_images(seed_input=2020,size=[120,120],sigma=5,d=20)
#ims = -np.array(ims)+50000
ims,shifts = add_noise(ims,shifts,seed_input=2020,
sigm_shift=0.5,sigm_noi_before_blur = 1000.,sigm_noi_after_blur = 1000.,sigm_blur = 2.5)
drizzled_im = drizzle(ims,shifts,p,n)
mean_im = drizzle(ims,shifts,1,n)
#cut off borders
mean = mean_im[10:-10,10:-10]
drizzled = drizzled_im[10:-10,10:-10]
fig,axs = plt.subplots(1,2,figsize=[10,4])
img = drizzled - mean
mima = max(-np.min(img),np.max(img))
im=axs[0].imshow(img,cmap='seismic',vmin=-mima,vmax=mima,interpolation='none')
plt.colorbar(im,ax=axs[0])
axs[0].set_title("Drizzle minus shift-and-add \n for artificial dataset")
im=axs[0].imshow(mean,cmap='gray',alpha = 0.7,interpolation='none')
plt.colorbar(im,ax=axs[0])
axs[0].set_xlabel("x pixels")
axs[0].set_ylabel("y pixels")
im=axs[1].imshow(ims[0][10:-10,10:-10])
plt.colorbar(im,ax=axs[1])
axs[1].set_title("Single input image \n of artificial dataset")
axs[1].set_xlabel("x pixels")
axs[1].set_ylabel("y pixels")
plt.tight_layout()
#plt.savefig(save_folder+'/Artificial_input_image+Shifted_drizzle_minus_shift_and_add_inverted.pdf', interpolation='none')
plt.show()
```
# Average shift versus peak of cross correlation
```
n=1
p=0.5
N = 50 #number of runs
#argmaxs = []
#av_shifts = []
for i in range(N):
ims,shifts = create_lattice_images(seed_input=i,size=[50,50])
drizzled_im = drizzle(ims,shifts,p,n)
mean_im = drizzle(ims,shifts,1,n)
#cut borders to prevent from border effects
mean = mean_im[10:-10,10:-10]
drizzled = drizzled_im[10:-10,10:-10]
argmax = correlate(drizzled,mean)
av_shift = meansh(shifts)
argmaxs.append(argmax)
av_shifts.append(av_shift)
for i,av in enumerate(av_shifts):
plt.plot(av[0],argmaxs[i][1]-15,"o",color="blue")
plt.title("Average shift vs cross correlation peak position (x-direction)")
plt.xlabel("Average shift (mod 1)")
plt.ylabel("Position of peak of cross correlation")
#plt.savefig(save_folder+'/av shift vs cross corr x-dir -- new average.pdf', interpolation='none')
plt.show()
for i,av in enumerate(av_shifts):
plt.plot(av[1],argmaxs[i][0]-15,"o",color="blue")
plt.title("Average shift vs cross correlation peak position (y-direction)")
plt.xlabel("Average shift (mod 1)")
plt.ylabel("Position of peak of cross correlation")
#plt.savefig(save_folder+'/av shift vs cross corr y-dir -- new average.pdf', interpolation='none')
plt.show()
```
# Center value of cross correlation for a range of pixfracs
```
n = 1 #scale factor 1/n
p_s = [0.1,0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,1] #pixfracs
N = 10 #number of input images to calculate
maxs = [] #Maximum values of the cross correlations
imss = []
shiftss = []
for i in range(N):
print("Image "+str(i))
maxs.append([])
ims,shifts = create_lattice_images(seed_input=i,sigma=5,d=20,size=[120,120],sigma_shifts=3)
ims,shifts = add_noise(ims,shifts,seed_input=i)
shifts = center_shifts(shifts)
imss.append(ims)
shiftss.append(shifts)
ims = imss[i]
shifts = shiftss[i]
mean_im = drizzle(ims,shifts,1,n)
mean = mean_im[10:-10,10:-10] #cutt off borders
for p in p_s:
print("Pixfrac "+str(p))
drizzled_im = drizzle(ims,shifts,p,n)
drizzled = drizzled_im[10:-10,10:-10]
corr = ndimage.correlate(mean,drizzled-mean)
maxs[i].append(corr[50][50])
maxs
for mx in maxs[:]:
plt.plot(p_s,mx/mx[0],"o",alpha=0.5)
plt.title("Center value of cross correlation")
plt.xlabel("Pixfrac")
plt.ylabel("Normalised cross correlation value")
plt.savefig(save_folder+'/Artificial_moire_images_center_of_crosscorrelation_normalised.pdf', interpolation='none')
plt.show()
for mx in maxs[:]:
plt.plot(p_s,mx,"o",alpha=0.5)
plt.title("Center value of cross correlation")
plt.xlabel("Pixfrac")
plt.ylabel("Cross correlation value")
plt.savefig(save_folder+'/Artificial_moire_images_center_of_crosscorrelation.pdf', interpolation='none')
plt.show()
```
|
github_jupyter
|
import numpy as np
from scipy import ndimage
from skimage import data, io, filters
import matplotlib.pyplot as plt
from Drizzle import *
save_folder = r"C:\Users\Arend-Jan\Documents\Universiteit\Bachelor Research Project\Thesis\Drizzle_images_moire_May\Artificial datasets"
def create_gaussian(sigma,size,pos):
"""Create a gaussian with standarddeviation sigma at an array of size with peak
at a given position.
"""
x, y = np.meshgrid(np.arange(0,size[0]) - pos[0], np.arange(0,size[1]) - pos[1])
d = np.sqrt(x*x+y*y)
g = np.exp(-( (d)**2 / ( 2.0 * sigma**2 ) ) )
#plt.imshow(g)
#plt.show()
return (g.T)
def create_lattice_images(mean_shifts=0,sigma_shifts = 3,seed_input=2020,size = [120,120],n_input=50,
sigma = 5.,d=20,h=None):
"""
Create artificial dataset of shifted images of Gaussian bulbs on a lattice.
Parameters:
"sigma_shifts" is the standard deviation of the shifts of the images.
"seed_input" is the seed used for dataset creation.
"size" is image size.
"n_input" is number of input images.
"sigma" is the width of the bulbs in the images.
"d" and "h" are geometrical parameters for the lattice, see sketch above.
"""
np.random.seed(seed_input) #set seed
if h == None:
h = d*np.sqrt(3)/2 #create triangular lattice
# Create shifts of input images
shifts = np.random.normal(mean_shifts,sigma_shifts,[n_input,2])
# Define point of lattice
lattice_points = []
brd = np.ceil(4*sigma/h)*h
for x in np.arange(-brd, size[0] + d + brd, d):
for y in np.arange(-brd, size[1] + 2*h + brd, 2*h):
lattice_points.append(np.array([x,y]))
lattice_points.append(np.array([x+d/2,y+h]))
# Create input images
ims = []
for i,shift in enumerate(shifts):
im = np.zeros(size)
for lp in lattice_points:
im += create_gaussian(sigma,size,lp+shift)
ims.append(im*10000)
return (ims,shifts)
def add_noise(ims,shifts,seed_input=2020,sigm_shift = .3,
sigm_noi_before_blur = 1000.,sigm_blur = 2.5,sigm_noi_after_blur = 1000.):
"""Add noise to given input images and shifts
Parameters:
"seed_input" is the seed used for dataset creation.
sigm_shift is sigma to bias shift.
sigm_noi_before_blur is sigma for gaussian noise.
sigm_blur is sigma for gaussian blurring.
sigm_noi_after_blur is sigma for gaussian noise.
"""
np.random.seed(seed_input) #set seed
# Add noise to the images
noise_ims = []
for im in ims:
noise_im = im + np.random.normal(0,sigm_noi_after_blur,np.shape(im))
noise_im = ndimage.filters.gaussian_filter(noise_im,sigm_blur)
noise_im += np.random.normal(0,sigm_noi_after_blur,np.shape(im))
noise_ims.append(noise_im)
#Add noise to the shifts
shifts_bias = shifts + np.random.normal(0,sigm_shift,np.shape(shifts))
return(noise_ims,shifts_bias)
def correlate(drizzled,mean):
size = [len(drizzled),len(drizzled[0])]
corr = ndimage.correlate(mean,drizzled-mean)
amax = np.argmax(corr)
peak = [amax%size[0],amax//size[0]]
return(peak)
def meansh(x):
"""Calculate the mean of shifts modulo 1"""
av_shift = [np.angle(np.mean(np.exp(1j*2*np.pi*x[:,0])))/2/np.pi%1,np.angle(np.mean(np.exp(1j*2*np.pi*x[:,1])))/2/np.pi%1]
av_shift = np.round(av_shift,2)
return(av_shift)
def center_shifts(shift):
"""Center the input shifts around 0."""
av_shift = meansh(shift)
shift[:,0] = shift[:,0]+1-av_shift[0]
shift[:,1] = shift[:,1]+1-av_shift[1]
return(shift)
# drizzle parameters
p = 0.5
n = 1
ims,shifts = create_lattice_images(seed_input=2020,size=[120,120],sigma=5,d=20)
#ims = -np.array(ims)+50000
ims,shifts = add_noise(ims,shifts,seed_input=2020,
sigm_shift=0.5,sigm_noi_before_blur = 1000.,sigm_noi_after_blur = 1000.,sigm_blur = 2.5)
drizzled_im = drizzle(ims,shifts,p,n)
mean_im = drizzle(ims,shifts,1,n)
#cut off borders
mean = mean_im[10:-10,10:-10]
drizzled = drizzled_im[10:-10,10:-10]
fig,axs = plt.subplots(1,2,figsize=[10,4])
img = drizzled - mean
mima = max(-np.min(img),np.max(img))
im=axs[0].imshow(img,cmap='seismic',vmin=-mima,vmax=mima,interpolation='none')
plt.colorbar(im,ax=axs[0])
axs[0].set_title("Drizzle minus shift-and-add \n for artificial dataset")
im=axs[0].imshow(mean,cmap='gray',alpha = 0.7,interpolation='none')
plt.colorbar(im,ax=axs[0])
axs[0].set_xlabel("x pixels")
axs[0].set_ylabel("y pixels")
im=axs[1].imshow(ims[0][10:-10,10:-10])
plt.colorbar(im,ax=axs[1])
axs[1].set_title("Single input image \n of artificial dataset")
axs[1].set_xlabel("x pixels")
axs[1].set_ylabel("y pixels")
plt.tight_layout()
#plt.savefig(save_folder+'/Artificial_input_image+Shifted_drizzle_minus_shift_and_add_inverted.pdf', interpolation='none')
plt.show()
n=1
p=0.5
N = 50 #number of runs
#argmaxs = []
#av_shifts = []
for i in range(N):
ims,shifts = create_lattice_images(seed_input=i,size=[50,50])
drizzled_im = drizzle(ims,shifts,p,n)
mean_im = drizzle(ims,shifts,1,n)
#cut borders to prevent from border effects
mean = mean_im[10:-10,10:-10]
drizzled = drizzled_im[10:-10,10:-10]
argmax = correlate(drizzled,mean)
av_shift = meansh(shifts)
argmaxs.append(argmax)
av_shifts.append(av_shift)
for i,av in enumerate(av_shifts):
plt.plot(av[0],argmaxs[i][1]-15,"o",color="blue")
plt.title("Average shift vs cross correlation peak position (x-direction)")
plt.xlabel("Average shift (mod 1)")
plt.ylabel("Position of peak of cross correlation")
#plt.savefig(save_folder+'/av shift vs cross corr x-dir -- new average.pdf', interpolation='none')
plt.show()
for i,av in enumerate(av_shifts):
plt.plot(av[1],argmaxs[i][0]-15,"o",color="blue")
plt.title("Average shift vs cross correlation peak position (y-direction)")
plt.xlabel("Average shift (mod 1)")
plt.ylabel("Position of peak of cross correlation")
#plt.savefig(save_folder+'/av shift vs cross corr y-dir -- new average.pdf', interpolation='none')
plt.show()
n = 1 #scale factor 1/n
p_s = [0.1,0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,1] #pixfracs
N = 10 #number of input images to calculate
maxs = [] #Maximum values of the cross correlations
imss = []
shiftss = []
for i in range(N):
print("Image "+str(i))
maxs.append([])
ims,shifts = create_lattice_images(seed_input=i,sigma=5,d=20,size=[120,120],sigma_shifts=3)
ims,shifts = add_noise(ims,shifts,seed_input=i)
shifts = center_shifts(shifts)
imss.append(ims)
shiftss.append(shifts)
ims = imss[i]
shifts = shiftss[i]
mean_im = drizzle(ims,shifts,1,n)
mean = mean_im[10:-10,10:-10] #cutt off borders
for p in p_s:
print("Pixfrac "+str(p))
drizzled_im = drizzle(ims,shifts,p,n)
drizzled = drizzled_im[10:-10,10:-10]
corr = ndimage.correlate(mean,drizzled-mean)
maxs[i].append(corr[50][50])
maxs
for mx in maxs[:]:
plt.plot(p_s,mx/mx[0],"o",alpha=0.5)
plt.title("Center value of cross correlation")
plt.xlabel("Pixfrac")
plt.ylabel("Normalised cross correlation value")
plt.savefig(save_folder+'/Artificial_moire_images_center_of_crosscorrelation_normalised.pdf', interpolation='none')
plt.show()
for mx in maxs[:]:
plt.plot(p_s,mx,"o",alpha=0.5)
plt.title("Center value of cross correlation")
plt.xlabel("Pixfrac")
plt.ylabel("Cross correlation value")
plt.savefig(save_folder+'/Artificial_moire_images_center_of_crosscorrelation.pdf', interpolation='none')
plt.show()
| 0.678327 | 0.97859 |
# Change of basis for vectors

This work by Jephian Lin is licensed under a [Creative Commons Attribution 4.0 International License](http://creativecommons.org/licenses/by/4.0/).
```
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits import mplot3d
from PIL import Image
```
## Main idea
Suppose $\beta = \{{\bf u}_1, \ldots, {\bf u}_n\}$ is an orthonormal basis of $\mathbb{R}^n$.
Then every vector ${\bf v}\in\mathbb{R}^n$ can be written as
$${\bf v} = c_1{\bf u}_1 + \cdots + c_n{\bf u}_n,$$
where $c_i = \langle {\bf v}, {\bf u}_i \rangle$.
We call
$$[{\bf v}]_\beta = \begin{bmatrix} c_1 \\ \vdots \\ c_n \end{bmatrix}$$
the **representation** of ${\bf v}$ with respect to the basis $\beta$.
Let $${\bf c} = [{\bf v}]_\beta \text{ and }
Q = \begin{bmatrix}
| & ~ & | \\
{\bf u}_1 & \cdots & {\bf u}_n \\
| & ~ & | \\
\end{bmatrix}.$$
Then $Q^\top {\bf v} = {\bf c}$ and $Q{\bf c} = {\bf v}$.
## Side stories
- new basis = new coordinates
- change of basis (general basis)
## Experiments
###### Exercise 1
This exercise asks you to draw a new coordinates $\mathbb{R}^2$ by the following steps.
Let
```python
u0 = np.array([1,1]) / np.sqrt(2)
u1 = np.array([-1,1]) / np.sqrt(2)
```
and $\beta = \{{\bf u}_0, {\bf u}_1\}$.
###### 1(a)
Draw the grid using ${\bf u}_0$ and ${\bf u}_1$.
Draw a red vector for $3{\bf u}_0$.
Draw a blue vector for $3{\bf u}_1$.
```
### your answer here
```
###### 1(b)
Draw a green vector for
```python
v = np.array([1,3]) / np.sqrt(2)
```
According to the graph, can you tell what is $[{\bf v}]_\beta$?
```
### your answer here
```
###### 1(c)
Find $[{\bf v}]_\beta$ by matrix multiplication.
```
### your answer here
```
###### 1(d)
Draw a vector for
```python
w = np.array([2,-1])
```
and find $[{\bf w}]_\beta$.
###### Exercise 2
Let
```python
theta = np.pi / 4
Q = np.array([[np.cos(theta), -np.sin(theta)],
[np.sin(theta), np.cos(theta)]])
mu = np.array([0,0])
cov = np.array([[4.1,2],
[2,1.1]])
vs = np.random.multivariate_normal(mu, cov, (100,))
```
Plot the data points (rows) of `vs` .
Draw the coordinates using the columns of $Q$.
Try and find an appropriate `theta` such that the data looks simple on the coordinates.
```
### your answer here
```
## Exercises
###### Exercise 3
Let
```python
x = np.linspace(0, np.pi, 20)
y = x**2 * np.sin(x)
z = np.zeros_like(x)
Q = np.array([[1,1,1],
[-1,1,1],
[0,-2,1]])
Q = Q / np.sqrt((Q**2).sum(axis=0))
```
Use the columns of $Q$ as the coordinates to plot `x`, `y`, `z` .
```
### your answer here
```
###### Exercise 4
This exercise is similar to Exercise 1 but the basis is no more orthonormal.
Let
```python
u0 = np.array([2,1])
u1 = np.array([1,2])
```
and $\beta = \{{\bf u}_0, {\bf u}_1\}$.
###### 4(a)
Draw the grid using ${\bf u}_0$ and ${\bf u}_1$.
Draw a red vector for $3{\bf u}_0$.
Draw a blue vector for $3{\bf u}_1$.
```
### your answer here
```
###### 4(b)
Draw a green vector for
```python
v = np.array([7,5])
```
According to the graph, can you tell what is $[{\bf v}]_\beta$?
```
### your answer here
```
###### 4(c)
Suppose your previous answer is $[{\bf v}]_\beta = (c_0, c_1)^\top$.
Let $Q$ be the matrix whose columns are vectors in $\beta$.
Then $Q[{\bf v}]_\beta = c_0{\bf u}_0 + c_1{\bf u}_1 = {\bf v}$.
Plot $Q[{\bf v}]_\beta$ and double check if your answer is correct.
```
### your answer here
```
###### 4(d)
If $\beta$ is orthogonal, then $Q^{-1} = Q^\top$ and $Q^\top{\bf v} = Q^{-1}{\bf v} = [{\bf v}]_\beta$, but not it is not the case.
However, the formula $Q^{-1}{\bf v} = [{\bf v}]_\beta$ is still valid.
Use this formula to find $[{\bf v}]_\beta$ and compare your answer with 4(b).
```
### your answer here
```
###### 4(e)
Draw a vector for
```python
w = np.array([2,-1])
```
and find $[{\bf w}]_\beta$.
```
### your answer here
```
##### Exercise 5
This exercise ask you to put an image on the plane using the given coordinates.
Let
```python
img = Image.open('incrediville-side.jpg')
width,height = 200,150
img = img.resize((width,height)).convert('L')
arr = np.array(img)
```
###### 5(a)
Let
```python
unit = 0.1
xx,yy = np.meshgrid(unit*np.arange(width), unit*np.arange(height))
xx = xx.ravel()
yy = -yy.ravel()
```
Make a scatter plot of `xx` and `yy` using the colors `arr.ravel()` .
Hint: You need to set `cmap='Greys_r` to make it looks good.
```
### your answer here
```
###### 5(b)
Let
```python
Q = np.array([[1,1],
[-1,1],
[0,-2]])
Q = Q / np.sqrt((Q**2).sum(axis=0))
vs = np.vstack([xx,yy])
new_vs = Q.dot(vs)
```
Make a scatter plot of points (columns) of `new_vs` using the same color setting.
```
### your answer here
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits import mplot3d
from PIL import Image
u0 = np.array([1,1]) / np.sqrt(2)
u1 = np.array([-1,1]) / np.sqrt(2)
### your answer here
v = np.array([1,3]) / np.sqrt(2)
### your answer here
### your answer here
w = np.array([2,-1])
theta = np.pi / 4
Q = np.array([[np.cos(theta), -np.sin(theta)],
[np.sin(theta), np.cos(theta)]])
mu = np.array([0,0])
cov = np.array([[4.1,2],
[2,1.1]])
vs = np.random.multivariate_normal(mu, cov, (100,))
### your answer here
x = np.linspace(0, np.pi, 20)
y = x**2 * np.sin(x)
z = np.zeros_like(x)
Q = np.array([[1,1,1],
[-1,1,1],
[0,-2,1]])
Q = Q / np.sqrt((Q**2).sum(axis=0))
### your answer here
u0 = np.array([2,1])
u1 = np.array([1,2])
### your answer here
v = np.array([7,5])
### your answer here
### your answer here
### your answer here
w = np.array([2,-1])
### your answer here
img = Image.open('incrediville-side.jpg')
width,height = 200,150
img = img.resize((width,height)).convert('L')
arr = np.array(img)
unit = 0.1
xx,yy = np.meshgrid(unit*np.arange(width), unit*np.arange(height))
xx = xx.ravel()
yy = -yy.ravel()
### your answer here
Q = np.array([[1,1],
[-1,1],
[0,-2]])
Q = Q / np.sqrt((Q**2).sum(axis=0))
vs = np.vstack([xx,yy])
new_vs = Q.dot(vs)
### your answer here
| 0.264074 | 0.981239 |
<a href="https://colab.research.google.com/github/brianharnish/cs419/blob/master/labs/brian_regression.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Regression
So far, in our exploration of machine learning, we have build sysems that predict discrete values: mountain bike or not mountain bike, democrat or republican. And not just binary choices, but, for example, deciding whether an image is of a particular digit:
or which of 1,000 categories does a picture represent.
This lab looks at how we can build classifiers that predict **continuous** values and such classifiers are called regression classifiers.
First, let's take a small detour into correlation.
## Correlation
A correlation is the degree of association between two variables. One of my favorite books on this topic is:
<img src="http://zacharski.org/files/courses/cs419/mg_statistics_big.png" width="250" />
and they illustrate it by looking at
## Ladies expenditures on clothes and makeup

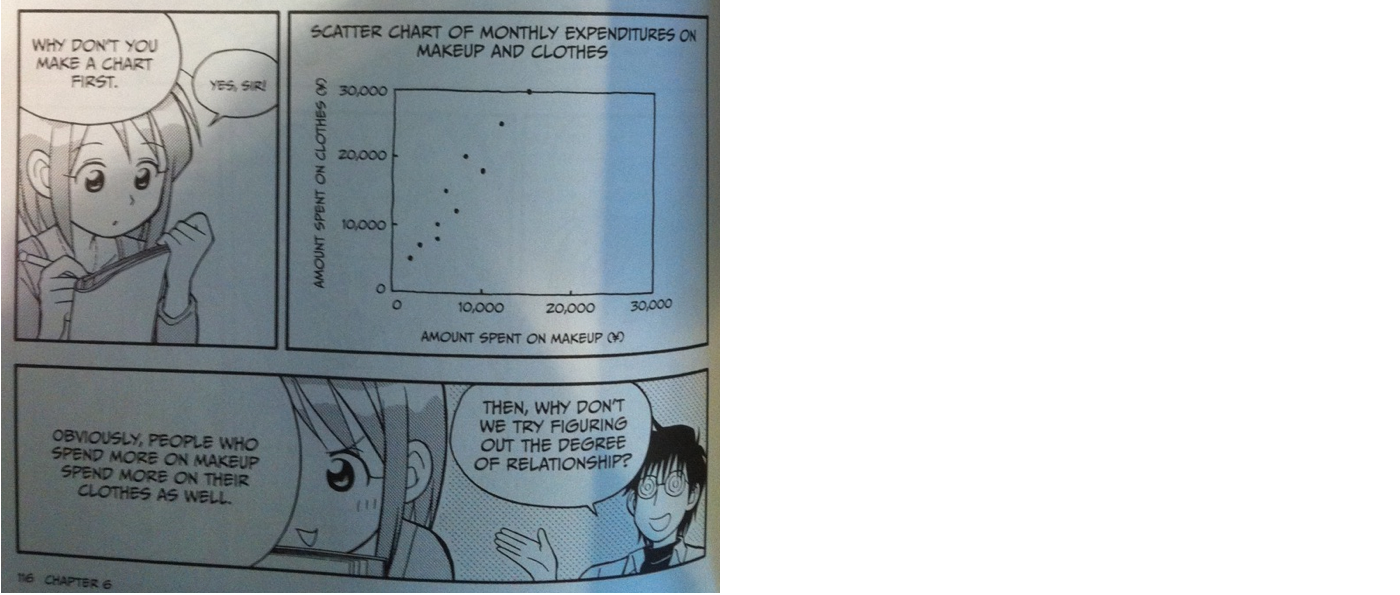
So let's go ahead and create that data in Pandas and show the table:
```
import pandas as pd
from pandas import DataFrame
makeup = [3000, 5000, 12000, 2000, 7000, 15000, 5000, 6000, 8000, 10000]
clothes = [7000, 8000, 25000, 5000, 12000, 30000, 10000, 15000, 20000, 18000]
ladies = ['Ms A','Ms B','Ms C','Ms D','Ms E','Ms F','Ms G','Ms H','Ms I','Ms J',]
monthly = DataFrame({'makeup': makeup, 'clothes': clothes}, index= ladies)
monthly
```
and let's show the scatterplot
```
from bokeh.plotting import figure, output_file, show
from bokeh.io import push_notebook, show, output_notebook
output_notebook()
x = figure(title="Montly Expenditures on Makeup and Clothes", x_axis_label="Money spent on makeup", y_axis_label="Money spent on clothes")
x.circle(monthly['makeup'], monthly['clothes'], size=10, color="navy", alpha=0.5)
output_file("stuff.html")
show(x)
```
When the data points are close to a straight line going up, we say that there is a positive correlation between the two variables. So in the case of the plot above, it visually looks like a postive correlation. Let's look at a few more examples:
## Weight and calories consumed in 1-3 yr/old children
This small but real dataset examines whether young children who weigh more, consume more calories
```
weight = [7.7, 7.8, 8.6, 8.5, 8.6, 9, 10.1, 11.5, 11, 10.2, 11.9, 10.4, 9.3, 9.1, 8.5, 11]
calories = [360, 400, 500, 370, 525, 800, 900, 1200, 1000, 1400, 1600, 850, 575, 425, 950, 800]
kids = DataFrame({'weight': weight, 'calories': calories})
kids
p = figure(title="Weight and calories in 1-3 yr.old children",
x_axis_label="weight (kg)", y_axis_label='weekly calories')
p.circle(kids['weight'], kids['calories'], size=10, color='navy', alpha=0.5)
show(p)
```
And again, there appears to be a positive correlation.
## The stronger the correlation the closer to a straight line
The closer the data points are to a straight line, the higher the correlation. A rising straight line (rising going left to right) would be perfect positive correlation. Here we are comparing the heights in inches of some NHL players with their heights in cm. Obviously, those are perfectly correlated.
```
inches =[68, 73, 69,72,71,77]
cm = [173, 185, 175, 183, 180, 196]
nhlHeights = DataFrame({'heightInches': inches, 'heightCM': cm})
nhlHeights
p = figure(title="Comparison of Height in Inches and Height in CM",
x_axis_label="Height in Inches",
y_axis_label="Height in centimeters")
p.circle(nhlHeights['heightInches'], nhlHeights['heightCM'],
size=10, color='navy', alpha=0.5)
show(p)
```
## No correlation = far from straight line
On the opposite extreme, if the datapoints are scattered and no line is discernable, there is no correlation.
Here we are comparing length of the player's hometown name to his height in inches. We are checking whether a player whose hometown name has more letters, tends to be taller. For example, maybe someone from Medicine Hat is taller than someone from Ledue. Obviously there should be no correlation.
(Again, a small but real dataset)
```
medicineHat = pd.read_csv('https://raw.githubusercontent.com/zacharski/machine-learning-notebooks/master/data/medicineHatTigers.csv')
medicineHat['hometownLength'] = medicineHat['Hometown'].str.len()
medicineHat
p = figure(title="Correlation of the number of Letters in the Hometown to Height",
x_axis_label="Player's Height", y_axis_label="Hometown Name Length")
p.circle(medicineHat['Height'], medicineHat['hometownLength'], size=10, color='navy', alpha=0.5)
show(p)
```
And that does not look at all like a straight line.
## negative correlation has a line going downhill
When the slope goes up, we say there is a positive correlation and when it goes down there is a negative correlation.
#### the relationship of hair length to a person's height
```
height =[62, 64, 65, 68, 69, 70, 67, 65, 72, 73, 74]
hairLength = [7, 10, 6, 4, 5, 4, 5, 8, 1, 1, 3]
cm = [173, 185, 175, 183, 180, 196]
people = DataFrame({'height': height, 'hairLength': hairLength})
p = figure(title="Correlation of hair length to a person's height",
x_axis_label="Person's Height", y_axis_label="Hair Length")
p.circle(people['height'], people['hairLength'], size=10, color='navy', alpha=0.5)
show(p)
```
There is a strong negative correlation between the length of someone's hair and how tall they are. That makes some sense. I am bald and 6'0" and my friend Sara is 5'8" and has long hair.
# Numeric Represenstation of the Strength of the Correlation
So far, we've seen a visual representation of the correlation, but we can also represent the degree of correlation numerically.
## Pearson Correlation Coefficient
This ranges from -1 to 1.
1 is perfect positive correlation, -1 is perfect negative.
$$r=\frac{\sum_{i=1}^n(x_i - \bar{x})(y_i-\bar{y})}{\sqrt{\sum_{i=1}^n(x_i - \bar{x})} \sqrt{\sum_{i=1}^n(y_i - \bar{y})}}$$
In Pandas it is very easy to compute.
### Japanese ladies expensives on makeup and clothes
Let's go back to our first example.
First here is the data:
```
monthly
p = figure(title="Montly Expenditures on Makeup and Clothes",
x_axis_label="Money spent on makeup", y_axis_label="Money spent on clothes")
p.circle(monthly['makeup'], monthly['clothes'], size=10, color='navy', alpha=0.5)
show(p)
```
So that looks like a pretty strong positive correlation. To compute Pearson on this data we do:
```
monthly.corr()
```
There is no surprise that makeup is perfectly correlated with makeup and clothes with clothes (those are the 1.000 on the diagonal). The interesting bit is that the Pearson for makeup to clothes is 0.968. That is a pretty strong correlation.
If you are interesting you can compute the Pearson values for the datasets above, but let's now move to ...
#### Regression
Let's say we know a young lady who spends about ¥22,500 per month on clothes (that's about $200/month). What do you think she spends on makeup, based on the chart below?
```
show(p)
```
I'm guessing you would predict she spends somewhere around ¥10,000 a month on makeup (almost $100/month). And how we do this is when looking at the graph we mentally draw an imaginary straight line through the datapoints and use that line for predictions. We are performing human linear regression. And as humans, we have the training set--the dots representing data points on the graph. and we **fit** our human classifier by mentally drawing that straight line. That straight line is our model. Once we have it, we can throw away the data points. When we want to make a prediction, we see where the money spent on clothes falls on that line.
We just predicted a continuous value (money spent on makeup) from one factor (money spent on clothes).
What happens when we want to predict a continuous value from 2 factors? Suppose we want to predict MPG based on the weight of a car and its horsepower.
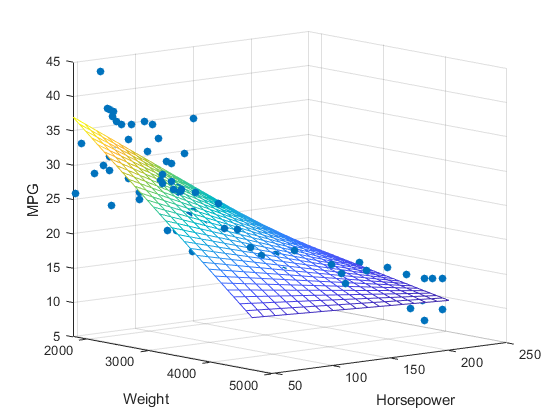
from [Mathworks](https://www.mathworks.com/help/stats/regress.html)
Now instead of a line representing the relationship we have a plane.
Let's create a linear regression classifier and try this out!
First, let's get the data.
```
columnNames = ['mpg', 'cylinders', 'displacement', 'HP', 'weight', 'acceleration', 'year', 'origin', 'model']
cars = pd.read_csv('https://raw.githubusercontent.com/zacharski/ml-class/master/data/auto-mpg.csv',
na_values=['?'], names=columnNames)
cars = cars.set_index('model')
cars = cars.dropna()
cars
```
Now divide the dataset into training and testing. And let's only use the horsepower and weight columns as features.
```
from sklearn.model_selection import train_test_split
cars_train, cars_test = train_test_split(cars, test_size = 0.2)
cars_train_features = cars_train[['HP', 'weight']]
# cars_train_features['HP'] = cars_train_features.HP.astype(float)
cars_train_labels = cars_train['mpg']
cars_test_features = cars_test[['HP', 'weight']]
# cars_test_features['HP'] = cars_test_features.HP.astype(float)
cars_test_labels = cars_test['mpg']
cars_test_features
```
### SKLearn Linear Regression
Now let's create a Linear Regression classifier and fit it.
```
from sklearn.linear_model import LinearRegression
linclf = LinearRegression()
linclf.fit(cars_train_features, cars_train_labels)
```
and finally use the trained classifier to make predictions on our test data
```
predictions = linclf.predict(cars_test_features)
```
Let's take an informal look at how we did:
```
results = cars_test_labels.to_frame()
results['Predicted']= predictions
results
```
Here is what my output looked like:
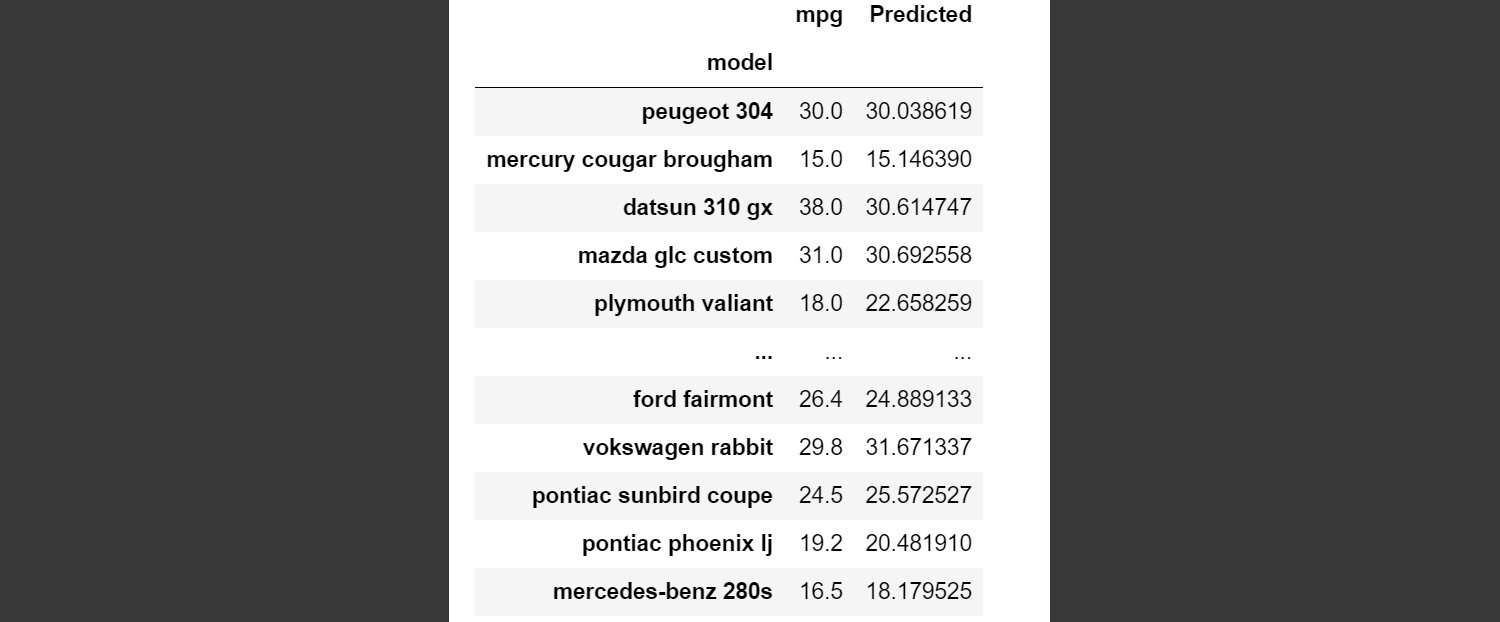
as you can see the first two predictions were pretty close as were a few others.
### Determining how well the classifier performed
With categorical classifiers we used sklearn's accuracy_score:
```
from sklearn.metrics import accuracy_score
```
Consider a task of predicting whether an image is of a dog or a cat. We have 10 instances in our test set. After our classifier makes predictions, for each image we have the actual (true) value, and the value our classifier predicted:
actual | predicted
:-- | :---
dog | dog
**dog** | **cat**
cat | cat
dog | dog
cat | cat
**cat** | **dog**
dog | dog
cat | cat
cat | cat
dog | dog
sklearn's accuracy score just counts how many predicted values matched the actual values and then divides by the total number of test instances. In this case the accuracy score would be .8000. The classifier was correct 80% of the time.
We can't use this method with a regression classifier. In the image above, the actual MPG of the Peugeot 304 was 30 and our classifier predicted 30.038619. Does that count as a match or not? The actual mpg of a Pontiac Sunbird Coupe was 24.5 and we predicted 25.57. Does that count as a match? Instead of accuracy_score, there are different evaluation metrics we can use.
#### Mean Squared Error and Root Mean Square Error
A common metric is Mean Squared Error or MSE. MSE is a measure of the quality of a regression classifier. The closer MSE is to zero, the better the classifier. Let's look at some made up data to see how this works:
vehicle | Actual MPG | Predicted MPG
:---: | ---: | ---:
Ram Promaster 3500 | 18.0 | 20.0
Ford F150 | 20 | 19
Fiat 128 | 33 | 33
First we compute the error (the difference between the predicted and actual values)
vehicle | Actual MPG | Predicted MPG | Error
:---: | ---: | ---: | --:
Ram Promaster 3500 | 18.0 | 20.0 | -2
Ford F150 | 20 | 19 | 1
Fiat 128 | 33 | 33 | 0
Next we square the error and compute the average:
vehicle | Actual MPG | Predicted MPG | Error | Error^2
:---: | ---: | ---: | --: | ---:
Ram Promaster 3500 | 18.0 | 20.0 | -2 | 4
Ford F150 | 20 | 19 | 1 | 1
Fiat 128 | 33 | 33 | 0 | 0
MSE | - | - | - | 1.667
**Root Mean Squared Error** is simply the square root of MSE. The advantage of RMSE is that it has the same units as what we are trying to predict. Let's take a look ...
```
from sklearn.metrics import mean_squared_error
MSE = mean_squared_error(cars_test_labels, predictions)
RMSE = mean_squared_error(cars_test_labels, predictions, squared=False)
print("MSE: %5.3f. RMSE: %5.3f" %(MSE, RMSE))
```
That RMSE tells us on average how many mpg we were off.
---
## So what kind of model does a linear regression classifier build?
You probably know this if you reflect on grade school math classes you took.
Let's go back and look at the young ladies expenditures on clothes and makeup.
```
p = figure(title="Montly Expenditures on Makeup and Clothes",
x_axis_label="Money spent on makeup", y_axis_label="Money spent on clothes")
p.circle(monthly['makeup'], monthly['clothes'], size=10, color='navy', alpha=0.5)
show(p)
```
When we talked about this example above, I mentioned that when we do this, we imagine a line. Let's see if we can use sklearns linear regression classifier to draw that line:
```
regr = LinearRegression()
regr.fit(monthly[['clothes']], monthly['makeup'])
pred = regr.predict(monthly[['clothes']])
import matplotlib.pyplot as plt
# Plot outputs
plt.scatter(monthly['clothes'], monthly['makeup'], color='black')
plt.plot(monthly['clothes'], pred, color='blue', linewidth=3)
plt.xticks(())
plt.yticks(())
plt.show()
```
Hopefully that matches your imaginary line!
The formula for the line is
$$makeup=w_0clothes + y.intercept$$
We can query our classifier for those values ($w_0$, and $i.intercept$):
```
print('w0 = %5.3f' % regr.coef_)
print('y intercept = %5.3f' % regr.intercept_)
```
So the formula for this particular example is
$$ makeup = 0.479 * clothes + 121.782$$
So if a young lady spent ¥22,500 on clothes we would predict she spent the following on makeup:
```
makeup = regr.coef_[0] * 22500 + regr.intercept_
makeup
```
The formula for regression in general is
$$\hat{y}=\theta_0 + \theta_1x_1 + \theta_2x_2 + ... \theta_nx_n$$
where $\theta_0$ is the y intercept. When you fit your classifier it is learning all those $\theta$'s. That is the model your classifier learns.
It is important to understand this as it applies to other classifiers as well!
## Overfitting
Consider two models for our makeup predictor. One is the straight line:
```
plt.scatter(monthly['clothes'], monthly['makeup'], color='black')
plt.plot(monthly['clothes'], pred, color='blue', linewidth=3)
plt.xticks(())
plt.yticks(())
plt.show()
```
And the other looks like:
```
monthly2 = monthly.sort_values(by='clothes')
plt.scatter(monthly2['clothes'], monthly2['makeup'], color='black')
plt.plot(monthly2['clothes'], monthly2['makeup'], color='blue', linewidth=3)
plt.xticks(())
plt.yticks(())
plt.show()
```
The second model fits the training data perfectly. Is it better than the first? Here is what could happen.
Let's say we have been tuning our model using our validation data set. Our error rates look like

As you can see our training error rate keeps going down, but at the very end our validation error increases. This is called **overfitting** the data. The model is highly tuned to the nuances of the training data. So much so, that it hurts the performance on new data--in this case, the validation data. This, obviously, is not a good thing.
---
#### An aside
Imagine preparing for a job interview for a position you really, really, want. Since we are working on machine learning, let's say it is a machine learning job. In their job ad they list a number of things they want the candidate to know:
* Convolutional Neural Networks
* Long Short Term Memory models
* Recurrent Neural Networks
* Generative Deep Learning
And you spend all your waking hours laser focused on these topics. You barely get any sleep and you read articles on these topics while you eat. You know the tiniest intricacies of these topics. You are more than 100% ready.
The day of the interview arrives. After of easy morning of chatting with various people, you are now in a conference room for the technical interview, standing at a whiteboard, ready to hopefully wow them with your wisdom. The first question they ask is for you to write the solution to the fizz buzz problem:
> Write a program that prints the numbers from 1 to 100. But for multiples of three print “Fizz” instead of the number and for the multiples of five print “Buzz”. For numbers which are multiples of both three and five print “FizzBuzz”
And you freeze. This is a simple request and a very common interview question. In fact, to prepare you for this job interview possibility, write it now:
```
def fizzbuzz():
i=0
while i <100:
fizz = False
Buzz = False
if i%3 == 0:
fizz = True
if i%5 == 0:
Buzz = True
if fizz == True and Buzz == True:
print("FizzBuzz")
elif fizz == True and Buzz == False:
print("Fizz")
elif fizz == False and Buzz == True:
print("Buzz")
else:
print(i)
i=i+1
fizzbuzz()
```
Back to the job candidate freezing, this is an example of overfitting. You overfitting to the skills mentioned in the job posting.
At dissertation defenses often faculty will ask the candidate questions outside of the candidate's dissertation. I heard of one case in a physics PhD defense where a faculty member asked "Why is the sky blue?" and the candidate couldn't answer.
Anyway, back to machine learning.
---
There are a number of ways to reduce the likelihood of overfitting including
* We can reduce the complexity of the model. Instead of going with the model of the jagged line immediately above we can go with the simpler straight line model. We have seen this in decision trees where we limit the depth of the tree.
* Another method is to increase the amount of training data.
Let's examine the first. The process of reducing the complexity of a model is called regularization.
The linear regression model we have just used tends to overfit the data and there are some variants that are better and these are called regularized linear models. These include
* Ridge Regression
* Lasso Regression
* Elastic Net - a combination of Ridge and Lasso
Let's explore Elastic Net. And let's use all the columns of the car mpg dataset:
```
newTrain_features = cars_train.drop('mpg', axis=1)
newTrain_labels = cars_train['mpg']
newTest_features = cars_test.drop('mpg', axis=1)
newTest_labels = cars_test['mpg']
newTrain_features
```
First, let's try with our standard Linear Regression classifier:
```
linclf = LinearRegression()
linclf.fit(newTrain_features, newTrain_labels)
predictions = linclf.predict(newTest_features)
MSE = mean_squared_error(newTest_labels, predictions)
RMSE = mean_squared_error(newTest_labels, predictions, squared=False)
print("MSE: %5.3f. RMSE: %5.3f" %(MSE, RMSE))
```
Now let's try with [ElasticNet](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.ElasticNet.html)
```
from sklearn.linear_model import ElasticNet
elastic_net = ElasticNet(alpha=0.1, l1_ratio=0.5)
elastic_net.fit(newTrain_features, newTrain_labels)
ePredictions = elastic_net.predict(newTest_features)
MSE = mean_squared_error(newTest_labels, ePredictions)
RMSE = mean_squared_error(newTest_labels, ePredictions, squared=False)
print("MSE: %5.3f. RMSE: %5.3f" %(MSE, RMSE))
```
I've run this a number of times. Sometimes linear regression is slightly better and sometimes ElasticNet is. Here are the results of one run:
##### RMSE
Linear Regression | Elastic Net
:---: | :---:
2.864 | 2.812
So this is not the most convincing example.
However, in general, it is always better to have some regularization, so (mostly) you should avoid the generic linear regression classifier.
## Happiness
What better way to explore regression then to look at happiness.
From a Zen perspective, happiness is being fully present in the current moment.
But, ignoring that advice, let's see if we can predict happiness, or life satisfaction.

We are going to be investigating the [Better Life Index](https://stats.oecd.org/index.aspx?DataSetCode=BLI). You can download a csv file of that data from that site.
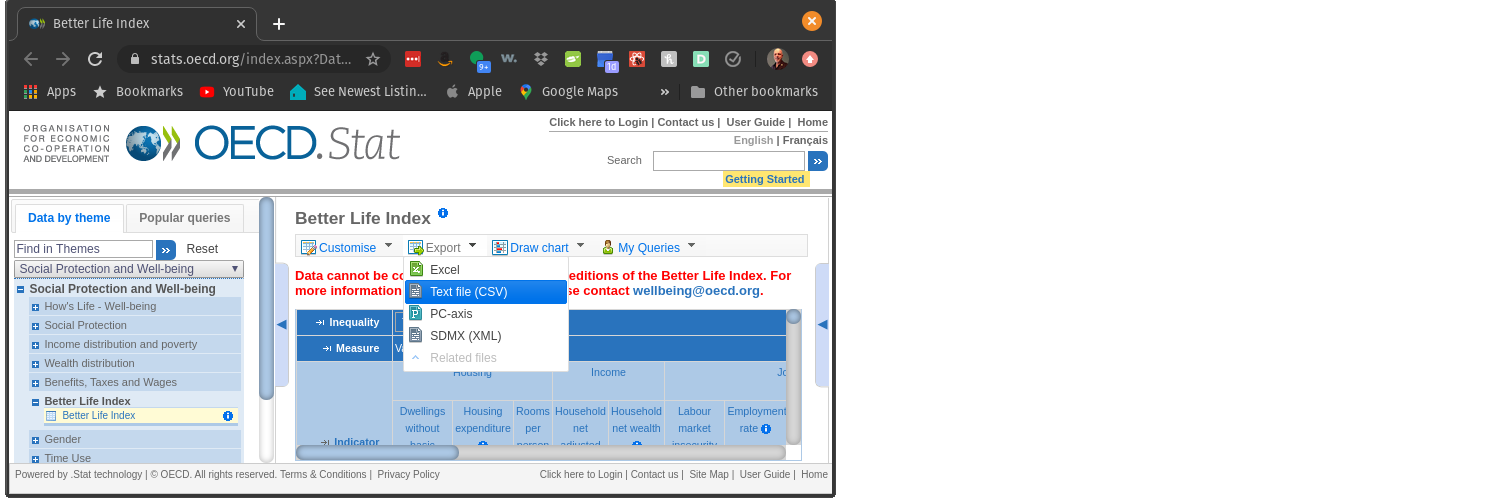
Now that you have the CSV data file on your laptop, you can upload it to Colab.
In Colab, you will see a list of icons on the left.

Select the file folder icon.
Next, select the upload icon (the page with an arrow icon). And upload the file.
Next, let's execute the Linux command `ls`:
```
!ls
```
### Load that file into a Pandas DataFrame
We will load the file into Pandas Dataframe called `bli` for better life index:
```
# TO DO
bli = pd.read_csv('BLI.csv')
bli
```
When examining the DataFrame we can see it has an interesting structure. So the first row we can parse:
* The country is Australia
* The feature is Labour market insecurity
* The Inequality column tells us it is the **total** Labour market insecurity value.
* The unit column tells the us the number is a percentage.
* And the value is 5.40
So, in English, the column is The total labor market insecurity for Australia is 5.40%.
I am curious as to what values other than Total are in the Inequality column:
```
bli.Inequality.unique()
```
Cool. So in addition to the total for each feature, we can get values for just men, just women, and the high and low.
Let's get just the totals and then pivot the DataFrame so it is in a more usable format.
In addition, there are a lot of NaN values in the data, let's replace them with the mean value of the column.
We are just learning about regression and this is a very small dataset, so let's divide training and testing by hand ...
```
bli = bli[bli["INEQUALITY"]=="TOT"]
bli = bli.pivot(index="Country", columns="Indicator", values="Value")
bli.fillna(bli.mean(), inplace=True)
bliTest = bli.loc['Greece':'Italy', :]
bliTrain = pd.concat([bli.loc[:'Germany' , :], bli.loc['Japan':, :]])
bliTrain
```
Now we need to divide both the training and test sets into features and labels.
```
# TO DO
bliTrain_features = bliTrain.drop('Life satisfaction', axis=1)
bliTrain_labels = bliTrain['Life satisfaction']
bliTest_features = bliTest.drop('Life satisfaction', axis=1)
bliTest_labels = bliTest['Life satisfaction']
bliTest_labels
```
### Create and Train an elastic net model
```
# TO DO
from sklearn.linear_model import ElasticNet
elastic_net = ElasticNet(alpha=0.1, l1_ratio=0.5)
elastic_net.fit(bliTrain_features, bliTrain_labels)
```
### Use the trained model to make predictions on our tiny test set
```
# TO DO
predictions = elastic_net.predict(bliTest_features)
predictions
Now let's visually compare the differences between the predictions and the actual values
results = pd.DataFrame(bliTest_labels)
results['Predicted']= predictions
results
```
How did you do? For me Hungary was a lot less happy than what was predicted.
# Prediction Housing Prices
## But first some wonkiness
When doing one hot encoding, sometimes the original datafile has the same type of data in multiple columns. For example...
Title | Genre 1 | Genre 2
:--: | :---: | :---:
Mission: Impossible - Fallout | Action | Drama
Mama Mia: Here We Go Again | Comedy | Musical
Ant-Man and The Wasp | Action | Comedy
BlacKkKlansman | Drama | Comedy
When we one-hot encode this we get something like
Title | Genre1 Action | Genre1 Comedy | Genre1 Drama | Genre2 Drama | Genre2 Musical | Genre2 Comedy
:--: | :--: | :--: | :--: | :--: | :--: | :--:
Mission: Impossible - Fallout | 1 | 0 | 0 | 1 | 0 | 0
Mama Mia: Here We Go Again | 0 | 1 | 0 | 0 | 1 | 0
Ant-Man and The Wasp | 1 | 0 | 0 | 0 | 0 | 1
BlacKkKlansman | 0 | 0 | 1 | 0 | 0 | 1
But this isn't what we probably want. Instead this would be a better representation:
Title | Action | Comedy | Drama | Musical
:---: | :---: | :---: | :---: | :---: |
Mission: Impossible - Fallout | 1 | 0 | 1 | 0
Mama Mia: Here We Go Again | 0 | 1 | 0 | 1
Ant-Man and The Wasp | 1 | 1 | 0 | 0
BlacKkKlansman | 0 | 1 | 1 | 0
Let's see how we might do this in code
```
df = pd.DataFrame({'Title': ['Mission: Impossible - Fallout', 'Mama Mia: Here We Go Again',
'Ant-Man and The Wasp', 'BlacKkKlansman' ],
'Genre1': ['Action', 'Comedy', 'Action', 'Drama'],
'Genre2': ['Drama', 'Musical', 'Comedy', 'Comedy']})
df
one_hot_1 = pd.get_dummies(df['Genre1'])
one_hot_2 = pd.get_dummies(df['Genre2'])
# now get the intersection of the column names
s1 = set(one_hot_1.columns.values)
s2 = set(one_hot_2.columns.values)
intersect = s1 & s2
only_s1 = s1 - intersect
only_s2 = s2 - intersect
# now logically or the intersect
logical_or = one_hot_1[list(intersect)] | one_hot_2[list(intersect)]
# then combine everything
combined = pd.concat([one_hot_1[list(only_s1)], logical_or, one_hot_2[list(only_s2)]], axis=1)
combined
### Now drop the two original columns and add the one hot encoded columns
df= df.drop('Genre1', axis=1)
df= df.drop('Genre2', axis=1)
df = df.join(combined)
df
```
That looks more like it!!!
## The task: Predict Housing Prices
Your task is to create a regession classifier that predicts house prices. The data and a description of
* [The description of the data](https://raw.githubusercontent.com/zacharski/ml-class/master/data/housePrices/data_description.txt)
* [The CSV file](https://raw.githubusercontent.com/zacharski/ml-class/master/data/housePrices/data.csv)
Minimally, your classifier should be trained on the following columns:
```
numericColumns = ['LotFrontage', 'LotArea', 'OverallQual', 'OverallCond', '1stFlrSF', '2ndFlrSF', 'GrLivArea',
'FullBath', 'HalfBath', 'Bedroom', 'Kitchen']
categoryColumns = ['MSZoning', 'Street', 'LotShape', 'LandContour',
'Utilities', 'LotConfig', 'LandSlope', 'Neighborhood', 'BldgType',
'HouseStyle', 'RoofStyle', 'RoofMatl']
# Using multicolumns is optional
multicolumns = [['Condition1', 'Condition2'], ['Exterior1st', 'Exterior2nd']]
```
You are free to use more columns than these. Also, you may need to process some of the columns.
Here are the requirements:
### 1. Drop any data rows that contain Nan in a column.
Once you do this you should have around 1200 rows.
### 2. Use the following train_test_split parameters
```
train_test_split( originalData, test_size=0.20, random_state=42)
```
### 3. You are to compare Linear Regression and Elastic Net
### 4. You should use 10 fold cross validation (it is fine to use grid search)
### 5. When finished tuning your model, determine the accuracy on the test data using RMSE.
# Performance Bonus
You are free to adjust any hyperparameters but do so before you evaluate the test data. You may get up to 15xp bonus for improved accuracy.
Good luck!
```
house = pd.read_csv('https://raw.githubusercontent.com/zacharski/ml-class/master/data/housePrices/data.csv')
house = house.set_index('Id')
house = house.drop('MiscFeature', axis=1)
house = house.drop('PoolQC', axis=1)
house = house.drop('Fence', axis=1)
house = house.drop('Alley', axis=1)
for o in categoryColumns:
one_hot = pd.get_dummies(house[o])
house = house.drop(o, axis=1)
house = house.join(one_hot)
house
import numpy as np
house = pd.read_csv('https://raw.githubusercontent.com/zacharski/ml-class/master/data/housePrices/data.csv')
house = house.set_index('Id')
house = house.drop('PoolQC', axis=1)
house = house.drop('MiscFeature', axis=1)
house = house.drop('Fence', axis=1)
house = house.drop('Alley', axis=1)
house = house.drop('Condition1', axis=1)
house = house.drop('Condition2', axis=1)
house = house.drop('Exterior1st', axis=1)
house = house.drop('Exterior2nd', axis=1)
house = house.drop('MasVnrType', axis=1)
house = house.drop('MasVnrArea', axis=1)
house = house.drop('ExterQual', axis=1)
house = house.drop('ExterCond', axis=1)
house = house.drop('Foundation', axis=1)
house = house.drop('BsmtQual', axis=1)
house = house.drop('BsmtCond', axis=1)
house = house.drop('BsmtExposure', axis=1)
house = house.drop('BsmtFinType1', axis=1)
house = house.drop('Heating', axis=1)
house = house.drop('BsmtFinType2', axis=1)
house = house.drop('HeatingQC', axis=1)
house = house.drop('CentralAir', axis=1)
house = house.drop('Electrical', axis=1)
house = house.drop('KitchenQual', axis=1)
house = house.drop('GarageType', axis=1)
house = house.drop('FireplaceQu', axis=1)
house = house.drop('GarageFinish', axis=1)
house = house.drop('GarageQual', axis=1)
house = house.drop('GarageCond', axis=1)
house = house.drop('PavedDrive', axis=1)
house = house.drop('Functional', axis=1)
house = house.drop('SaleType', axis=1)
house = house.drop('SaleCondition', axis=1)
house = house.drop('GarageYrBlt', axis=1)
house.fillna(house.mean(), inplace=True)
house
#house = house.drop('Alley', axis=1)
#one_hot = pd.get_dummies(house['MSZoning', 'Street', 'Alley', 'LotShape', 'LandContour',
# 'Utilities', 'LotConfig', 'LandSlope', 'Neighborhood', 'BldgType',
# 'HouseStyle', 'RoofStyle', 'RoofMatl', ''])
one_hot = pd.get_dummies(house['MSZoning'])
house = house.drop('MSZoning', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['Street'])
house = house.drop('Street', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['LotShape'])
house = house.drop('LotShape', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['LandContour'])
house = house.drop('LandContour', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['Utilities'])
house = house.drop('Utilities', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['LotConfig'])
house = house.drop('LotConfig', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['LandSlope'])
house = house.drop('LandSlope', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['Neighborhood'])
house = house.drop('Neighborhood', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['BldgType'])
house = house.drop('BldgType', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['HouseStyle'])
house = house.drop('HouseStyle', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['RoofStyle'])
house = house.drop('RoofStyle', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['RoofMatl'])
house = house.drop('RoofMatl', axis=1)
house = house.join(one_hot)
house
#one_hot = pd.get_dummies(house['Street'])
#house = house.drop('Street', axis=1)
#house = house.join(one_hot)
houseTrain, houseTest = train_test_split( house, test_size=0.20, random_state=42)
houseTrain
from sklearn.model_selection import cross_val_score
houseTrain_features = houseTrain.drop('SalePrice', axis=1)
houseTrain_labels = houseTrain['SalePrice']
houseTest_features = houseTest.drop('SalePrice', axis=1)
houseTest_labels = houseTest['SalePrice']
from sklearn.linear_model import ElasticNet
elastic_net = ElasticNet(alpha=0.1, l1_ratio=0.5)
elastic_net.fit(houseTrain_features, houseTrain_labels)
scores = cross_val_score(elastic_net, houseTrain_features, houseTrain_labels, cv=10)
print("elastic score is {}".format(scores.mean()))
predictions = elastic_net.predict(houseTest_features)
results = pd.DataFrame(houseTest_labels)
results['Predicted']= predictions
linclf = LinearRegression()
linclf.fit(houseTrain_features, houseTrain_labels)
predictions = linclf.predict(houseTest_features)
second_score = cross_val_score(linclf, houseTrain_features, houseTrain_labels, cv=10)
print("linear regression score {}".format(second_score.mean()))
```
|
github_jupyter
|
import pandas as pd
from pandas import DataFrame
makeup = [3000, 5000, 12000, 2000, 7000, 15000, 5000, 6000, 8000, 10000]
clothes = [7000, 8000, 25000, 5000, 12000, 30000, 10000, 15000, 20000, 18000]
ladies = ['Ms A','Ms B','Ms C','Ms D','Ms E','Ms F','Ms G','Ms H','Ms I','Ms J',]
monthly = DataFrame({'makeup': makeup, 'clothes': clothes}, index= ladies)
monthly
from bokeh.plotting import figure, output_file, show
from bokeh.io import push_notebook, show, output_notebook
output_notebook()
x = figure(title="Montly Expenditures on Makeup and Clothes", x_axis_label="Money spent on makeup", y_axis_label="Money spent on clothes")
x.circle(monthly['makeup'], monthly['clothes'], size=10, color="navy", alpha=0.5)
output_file("stuff.html")
show(x)
weight = [7.7, 7.8, 8.6, 8.5, 8.6, 9, 10.1, 11.5, 11, 10.2, 11.9, 10.4, 9.3, 9.1, 8.5, 11]
calories = [360, 400, 500, 370, 525, 800, 900, 1200, 1000, 1400, 1600, 850, 575, 425, 950, 800]
kids = DataFrame({'weight': weight, 'calories': calories})
kids
p = figure(title="Weight and calories in 1-3 yr.old children",
x_axis_label="weight (kg)", y_axis_label='weekly calories')
p.circle(kids['weight'], kids['calories'], size=10, color='navy', alpha=0.5)
show(p)
inches =[68, 73, 69,72,71,77]
cm = [173, 185, 175, 183, 180, 196]
nhlHeights = DataFrame({'heightInches': inches, 'heightCM': cm})
nhlHeights
p = figure(title="Comparison of Height in Inches and Height in CM",
x_axis_label="Height in Inches",
y_axis_label="Height in centimeters")
p.circle(nhlHeights['heightInches'], nhlHeights['heightCM'],
size=10, color='navy', alpha=0.5)
show(p)
medicineHat = pd.read_csv('https://raw.githubusercontent.com/zacharski/machine-learning-notebooks/master/data/medicineHatTigers.csv')
medicineHat['hometownLength'] = medicineHat['Hometown'].str.len()
medicineHat
p = figure(title="Correlation of the number of Letters in the Hometown to Height",
x_axis_label="Player's Height", y_axis_label="Hometown Name Length")
p.circle(medicineHat['Height'], medicineHat['hometownLength'], size=10, color='navy', alpha=0.5)
show(p)
height =[62, 64, 65, 68, 69, 70, 67, 65, 72, 73, 74]
hairLength = [7, 10, 6, 4, 5, 4, 5, 8, 1, 1, 3]
cm = [173, 185, 175, 183, 180, 196]
people = DataFrame({'height': height, 'hairLength': hairLength})
p = figure(title="Correlation of hair length to a person's height",
x_axis_label="Person's Height", y_axis_label="Hair Length")
p.circle(people['height'], people['hairLength'], size=10, color='navy', alpha=0.5)
show(p)
monthly
p = figure(title="Montly Expenditures on Makeup and Clothes",
x_axis_label="Money spent on makeup", y_axis_label="Money spent on clothes")
p.circle(monthly['makeup'], monthly['clothes'], size=10, color='navy', alpha=0.5)
show(p)
monthly.corr()
show(p)
columnNames = ['mpg', 'cylinders', 'displacement', 'HP', 'weight', 'acceleration', 'year', 'origin', 'model']
cars = pd.read_csv('https://raw.githubusercontent.com/zacharski/ml-class/master/data/auto-mpg.csv',
na_values=['?'], names=columnNames)
cars = cars.set_index('model')
cars = cars.dropna()
cars
from sklearn.model_selection import train_test_split
cars_train, cars_test = train_test_split(cars, test_size = 0.2)
cars_train_features = cars_train[['HP', 'weight']]
# cars_train_features['HP'] = cars_train_features.HP.astype(float)
cars_train_labels = cars_train['mpg']
cars_test_features = cars_test[['HP', 'weight']]
# cars_test_features['HP'] = cars_test_features.HP.astype(float)
cars_test_labels = cars_test['mpg']
cars_test_features
from sklearn.linear_model import LinearRegression
linclf = LinearRegression()
linclf.fit(cars_train_features, cars_train_labels)
predictions = linclf.predict(cars_test_features)
results = cars_test_labels.to_frame()
results['Predicted']= predictions
results
from sklearn.metrics import accuracy_score
from sklearn.metrics import mean_squared_error
MSE = mean_squared_error(cars_test_labels, predictions)
RMSE = mean_squared_error(cars_test_labels, predictions, squared=False)
print("MSE: %5.3f. RMSE: %5.3f" %(MSE, RMSE))
p = figure(title="Montly Expenditures on Makeup and Clothes",
x_axis_label="Money spent on makeup", y_axis_label="Money spent on clothes")
p.circle(monthly['makeup'], monthly['clothes'], size=10, color='navy', alpha=0.5)
show(p)
regr = LinearRegression()
regr.fit(monthly[['clothes']], monthly['makeup'])
pred = regr.predict(monthly[['clothes']])
import matplotlib.pyplot as plt
# Plot outputs
plt.scatter(monthly['clothes'], monthly['makeup'], color='black')
plt.plot(monthly['clothes'], pred, color='blue', linewidth=3)
plt.xticks(())
plt.yticks(())
plt.show()
print('w0 = %5.3f' % regr.coef_)
print('y intercept = %5.3f' % regr.intercept_)
makeup = regr.coef_[0] * 22500 + regr.intercept_
makeup
plt.scatter(monthly['clothes'], monthly['makeup'], color='black')
plt.plot(monthly['clothes'], pred, color='blue', linewidth=3)
plt.xticks(())
plt.yticks(())
plt.show()
monthly2 = monthly.sort_values(by='clothes')
plt.scatter(monthly2['clothes'], monthly2['makeup'], color='black')
plt.plot(monthly2['clothes'], monthly2['makeup'], color='blue', linewidth=3)
plt.xticks(())
plt.yticks(())
plt.show()
def fizzbuzz():
i=0
while i <100:
fizz = False
Buzz = False
if i%3 == 0:
fizz = True
if i%5 == 0:
Buzz = True
if fizz == True and Buzz == True:
print("FizzBuzz")
elif fizz == True and Buzz == False:
print("Fizz")
elif fizz == False and Buzz == True:
print("Buzz")
else:
print(i)
i=i+1
fizzbuzz()
newTrain_features = cars_train.drop('mpg', axis=1)
newTrain_labels = cars_train['mpg']
newTest_features = cars_test.drop('mpg', axis=1)
newTest_labels = cars_test['mpg']
newTrain_features
linclf = LinearRegression()
linclf.fit(newTrain_features, newTrain_labels)
predictions = linclf.predict(newTest_features)
MSE = mean_squared_error(newTest_labels, predictions)
RMSE = mean_squared_error(newTest_labels, predictions, squared=False)
print("MSE: %5.3f. RMSE: %5.3f" %(MSE, RMSE))
from sklearn.linear_model import ElasticNet
elastic_net = ElasticNet(alpha=0.1, l1_ratio=0.5)
elastic_net.fit(newTrain_features, newTrain_labels)
ePredictions = elastic_net.predict(newTest_features)
MSE = mean_squared_error(newTest_labels, ePredictions)
RMSE = mean_squared_error(newTest_labels, ePredictions, squared=False)
print("MSE: %5.3f. RMSE: %5.3f" %(MSE, RMSE))
!ls
# TO DO
bli = pd.read_csv('BLI.csv')
bli
bli.Inequality.unique()
bli = bli[bli["INEQUALITY"]=="TOT"]
bli = bli.pivot(index="Country", columns="Indicator", values="Value")
bli.fillna(bli.mean(), inplace=True)
bliTest = bli.loc['Greece':'Italy', :]
bliTrain = pd.concat([bli.loc[:'Germany' , :], bli.loc['Japan':, :]])
bliTrain
# TO DO
bliTrain_features = bliTrain.drop('Life satisfaction', axis=1)
bliTrain_labels = bliTrain['Life satisfaction']
bliTest_features = bliTest.drop('Life satisfaction', axis=1)
bliTest_labels = bliTest['Life satisfaction']
bliTest_labels
# TO DO
from sklearn.linear_model import ElasticNet
elastic_net = ElasticNet(alpha=0.1, l1_ratio=0.5)
elastic_net.fit(bliTrain_features, bliTrain_labels)
# TO DO
predictions = elastic_net.predict(bliTest_features)
predictions
Now let's visually compare the differences between the predictions and the actual values
results = pd.DataFrame(bliTest_labels)
results['Predicted']= predictions
results
df = pd.DataFrame({'Title': ['Mission: Impossible - Fallout', 'Mama Mia: Here We Go Again',
'Ant-Man and The Wasp', 'BlacKkKlansman' ],
'Genre1': ['Action', 'Comedy', 'Action', 'Drama'],
'Genre2': ['Drama', 'Musical', 'Comedy', 'Comedy']})
df
one_hot_1 = pd.get_dummies(df['Genre1'])
one_hot_2 = pd.get_dummies(df['Genre2'])
# now get the intersection of the column names
s1 = set(one_hot_1.columns.values)
s2 = set(one_hot_2.columns.values)
intersect = s1 & s2
only_s1 = s1 - intersect
only_s2 = s2 - intersect
# now logically or the intersect
logical_or = one_hot_1[list(intersect)] | one_hot_2[list(intersect)]
# then combine everything
combined = pd.concat([one_hot_1[list(only_s1)], logical_or, one_hot_2[list(only_s2)]], axis=1)
combined
### Now drop the two original columns and add the one hot encoded columns
df= df.drop('Genre1', axis=1)
df= df.drop('Genre2', axis=1)
df = df.join(combined)
df
numericColumns = ['LotFrontage', 'LotArea', 'OverallQual', 'OverallCond', '1stFlrSF', '2ndFlrSF', 'GrLivArea',
'FullBath', 'HalfBath', 'Bedroom', 'Kitchen']
categoryColumns = ['MSZoning', 'Street', 'LotShape', 'LandContour',
'Utilities', 'LotConfig', 'LandSlope', 'Neighborhood', 'BldgType',
'HouseStyle', 'RoofStyle', 'RoofMatl']
# Using multicolumns is optional
multicolumns = [['Condition1', 'Condition2'], ['Exterior1st', 'Exterior2nd']]
train_test_split( originalData, test_size=0.20, random_state=42)
house = pd.read_csv('https://raw.githubusercontent.com/zacharski/ml-class/master/data/housePrices/data.csv')
house = house.set_index('Id')
house = house.drop('MiscFeature', axis=1)
house = house.drop('PoolQC', axis=1)
house = house.drop('Fence', axis=1)
house = house.drop('Alley', axis=1)
for o in categoryColumns:
one_hot = pd.get_dummies(house[o])
house = house.drop(o, axis=1)
house = house.join(one_hot)
house
import numpy as np
house = pd.read_csv('https://raw.githubusercontent.com/zacharski/ml-class/master/data/housePrices/data.csv')
house = house.set_index('Id')
house = house.drop('PoolQC', axis=1)
house = house.drop('MiscFeature', axis=1)
house = house.drop('Fence', axis=1)
house = house.drop('Alley', axis=1)
house = house.drop('Condition1', axis=1)
house = house.drop('Condition2', axis=1)
house = house.drop('Exterior1st', axis=1)
house = house.drop('Exterior2nd', axis=1)
house = house.drop('MasVnrType', axis=1)
house = house.drop('MasVnrArea', axis=1)
house = house.drop('ExterQual', axis=1)
house = house.drop('ExterCond', axis=1)
house = house.drop('Foundation', axis=1)
house = house.drop('BsmtQual', axis=1)
house = house.drop('BsmtCond', axis=1)
house = house.drop('BsmtExposure', axis=1)
house = house.drop('BsmtFinType1', axis=1)
house = house.drop('Heating', axis=1)
house = house.drop('BsmtFinType2', axis=1)
house = house.drop('HeatingQC', axis=1)
house = house.drop('CentralAir', axis=1)
house = house.drop('Electrical', axis=1)
house = house.drop('KitchenQual', axis=1)
house = house.drop('GarageType', axis=1)
house = house.drop('FireplaceQu', axis=1)
house = house.drop('GarageFinish', axis=1)
house = house.drop('GarageQual', axis=1)
house = house.drop('GarageCond', axis=1)
house = house.drop('PavedDrive', axis=1)
house = house.drop('Functional', axis=1)
house = house.drop('SaleType', axis=1)
house = house.drop('SaleCondition', axis=1)
house = house.drop('GarageYrBlt', axis=1)
house.fillna(house.mean(), inplace=True)
house
#house = house.drop('Alley', axis=1)
#one_hot = pd.get_dummies(house['MSZoning', 'Street', 'Alley', 'LotShape', 'LandContour',
# 'Utilities', 'LotConfig', 'LandSlope', 'Neighborhood', 'BldgType',
# 'HouseStyle', 'RoofStyle', 'RoofMatl', ''])
one_hot = pd.get_dummies(house['MSZoning'])
house = house.drop('MSZoning', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['Street'])
house = house.drop('Street', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['LotShape'])
house = house.drop('LotShape', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['LandContour'])
house = house.drop('LandContour', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['Utilities'])
house = house.drop('Utilities', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['LotConfig'])
house = house.drop('LotConfig', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['LandSlope'])
house = house.drop('LandSlope', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['Neighborhood'])
house = house.drop('Neighborhood', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['BldgType'])
house = house.drop('BldgType', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['HouseStyle'])
house = house.drop('HouseStyle', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['RoofStyle'])
house = house.drop('RoofStyle', axis=1)
house = house.join(one_hot)
one_hot = pd.get_dummies(house['RoofMatl'])
house = house.drop('RoofMatl', axis=1)
house = house.join(one_hot)
house
#one_hot = pd.get_dummies(house['Street'])
#house = house.drop('Street', axis=1)
#house = house.join(one_hot)
houseTrain, houseTest = train_test_split( house, test_size=0.20, random_state=42)
houseTrain
from sklearn.model_selection import cross_val_score
houseTrain_features = houseTrain.drop('SalePrice', axis=1)
houseTrain_labels = houseTrain['SalePrice']
houseTest_features = houseTest.drop('SalePrice', axis=1)
houseTest_labels = houseTest['SalePrice']
from sklearn.linear_model import ElasticNet
elastic_net = ElasticNet(alpha=0.1, l1_ratio=0.5)
elastic_net.fit(houseTrain_features, houseTrain_labels)
scores = cross_val_score(elastic_net, houseTrain_features, houseTrain_labels, cv=10)
print("elastic score is {}".format(scores.mean()))
predictions = elastic_net.predict(houseTest_features)
results = pd.DataFrame(houseTest_labels)
results['Predicted']= predictions
linclf = LinearRegression()
linclf.fit(houseTrain_features, houseTrain_labels)
predictions = linclf.predict(houseTest_features)
second_score = cross_val_score(linclf, houseTrain_features, houseTrain_labels, cv=10)
print("linear regression score {}".format(second_score.mean()))
| 0.550607 | 0.983423 |
# Difference in difference analysis to estimate impact of CICIG on Guatemala's homicide rates
This is a replication of the quantitative analysis made by CrisisGroup in the article ["Saving Guatemala’s Fight Against Crime and Impunity"](https://www.crisisgroup.org/latin-america-caribbean/central-america/guatemala/70-saving-guatemalas-fight-against-crime-and-impunity). The analysis uses synthetics controls estimated with an [entropy balancing method](https://web.stanford.edu/~jhain/Paper/PA2012.pdf) to estimate a counterfactual from nearby latinamerican countries.
```
import pandas as pd
import matplotlib as mlp
from matplotlib import pyplot as plt
import numpy as np
import seaborn as sbs
from statsmodels import api as stm
sbs.set(style="whitegrid")
%matplotlib inline
def tryFunc(functionToTry, onError = np.NaN, showErrors = False):
def wrapper(inputValue):
try:
return functionToTry(inputValue)
except:
if showErrors:
print("Bad value: ", inputValue)
return onError
return wrapper
toFloat = lambda x: float(x)
gt1 = pd.read_excel("./guatemala1.xlsx")
gt1.head(3)
gtpop = pd.read_excel("./gtm_pop.xlsx")
gtpop.head(3)
# Ignore Cuba and Haiti
gt1 = gt1[gt1["Country Code"].isin(["CUB", "HTI"]) == False]
gt1.head()
# The available indicators
gt1["Series Name"].value_counts()
cols = gt1.columns[gt1.columns.map(lambda x: x.startswith("YR")) == True]
gt2 = gt1.set_index(["Country Code", "Series Name"])[cols].stack().map(tryFunc(toFloat)).unstack(1).rename(index=lambda x: int(x[2:6]) if str.startswith(x, "YR") else x)
gt2.head().reset_index()
# This gives the same that stata
del gt2["CPIA_accountability_corruption"]
del gt2["CPIA_public_sector"]
gt2[gt2.index.get_level_values(1) < 2007].describe()
cols = ["gdp_per_capita_ppp_2011", "homicide", "household_consumption", "poverty_headcount_320"]
gt2["intercept"] = 1
def trends(df):
results = []
for col in cols:
results.append(np.linalg.lstsq(df[df[col].isna() == False].reset_index()[["intercept", "level_1"]], df[df[col].isna() == False][col])[0][1])
return pd.Series(index = cols, data = results)
trends = gt2[(gt2.index.get_level_values(1) < 2007 )].groupby("Country Code").apply(trends)
# The trends for each country for each indicator
# This is showing the same results as the original file.
trends
trends.columns = trends.columns.map(lambda x: "trend_" + x)
trends = pd.concat([trends, gt2[gt2.index.get_level_values(1) < 2007].groupby(level=[0]).mean()], axis = 1)
trends.to_csv("trends.csv")
gt2.to_csv("data_time_series.csv")
```
Since the entropy balancing method is not available for python, I'll have to defer the synthetic control estimation to R, where the package "ebal", from the same author will be used to estimate the synth control weights.
# Comparing Guatemala with the Synthetic Control
Now reading the weights generated by ebalance package in R and the weights generated by Stata (the same CrisisGroup used)
```
ebal_st = pd.read_csv("./ebalanced_stata.csv", index_col=0)
ebal_st.loc["GTM"] = 1
ebal_st.columns = ["ebal_st"]
ebal_R = pd.read_csv("./ebalanced.csv", index_col=0)
ebal_R.loc["GTM"] = 1
ebal_R.columns = ["ebal_R"]
gt3 = gt2.reset_index()
gt3 = gt2.merge(ebal_st, left_on="Country Code", right_index=True).reset_index()
gt3 = gt3.merge(ebal_R, left_on="level_0", right_index=True)
gt3["logHom"] = np.log10(gt3.homicide)
gt3["Treatment"] = gt3.level_0 == "GTM"
def wmean(df, m, w):
return df[m].multiply(df[w]).sum() / \
df[w][df[m].notna()].sum()
# TEST
# wmean(gt3[gt3.level_0 == "GTM"], "homicide", "ebal_st" ), gt3[gt3.level_0 == "GTM"].homicide.mean()
avgCols = ['logHom','adult_literacy_rate', 'gdp_per_capita_ppp_2011', 'homicide',
'household_consumption', 'poverty_headcount_320',
'under5_mortality_rate', 'youth_literacy_rate']
gt4_st = gt3.groupby(["Treatment", "level_1"]).apply(
lambda x: pd.Series([ wmean(x, col, "ebal_st") for col in avgCols ], avgCols) )
gt4_R = gt3.groupby(["Treatment", "level_1"]).apply(
lambda x: pd.Series([ wmean(x, col, "ebal_R") for col in avgCols ], avgCols) )
gt4_unw = gt3.groupby(["Treatment", "level_1"]).apply(
lambda x: pd.Series([ x[col].mean() for col in avgCols ], avgCols) )
```
# Unweighted means (no entropy balancing & synth control)
```
gt4_unw["prepost"] = gt4_unw.index.get_level_values(1).map(lambda x: ("pre" if x<2007 else "post"), 1)
sbs.lmplot("level_1", "logHom", gt4_unw[gt4_unw.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
# Finally, getting the same graph that CrisisGroup obtained in stata, of course, using their ebalance results.
plt.suptitle("Compare Pre-Post CICIG scenarios for homicides\nwithout weights (simple means)")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "under5_mortality_rate", gt4_unw[gt4_unw.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for under 5 mortality rate\nwithout weights (simple means)")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "household_consumption", gt4_unw[gt4_unw.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for under 5 mortality rate\nwithout weights (simple means)")
plt.tight_layout(4.5)
```
# Using weights estimated with Stata ebalance package
```
gt4_st["prepost"] = gt4_st.index.get_level_values(1).map(lambda x: ("pre" if x<2007 else "post"), 1)
sbs.lmplot("level_1", "homicide", gt4_st[gt4_st.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
# Finally, getting the same graph that CrisisGroup obtained in stata, of course, using their ebalance results.
plt.suptitle("Compare Pre-Post CICIG scenarios for homicides\nwith Stata ebalance weights")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "logHom", gt4_st[gt4_st.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for log(homicides)\nwith Stata ebalance weights")
plt.tight_layout(4.5)
axs.set_yticklabels(np.power(10, axs.axes[0,0].get_yticks()).round())
axs = sbs.lmplot("level_1", "under5_mortality_rate", gt4_st[gt4_st.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for under 5 mortality rate\nwith Stata ebalance weights")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "household_consumption", gt4_st[gt4_st.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for household consumption\nwith Stata ebalance weights")
plt.tight_layout(4.5)
```
# Using weights estimated with R ebal
```
gt4_R["prepost"] = gt4_R.index.get_level_values(1).map(lambda x: ("pre" if x<2007 else "post"), 1)
axs = sbs.lmplot("level_1", "logHom", gt4_R[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
axs.set_yticklabels(np.power(10, axs.axes[0,0].get_yticks()).round())
plt.suptitle("Compare Pre-Post CICIG scenarios for log(homicides)\nwith R ebalance weights")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "under5_mortality_rate", gt4_R[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for under 5 mortality rate\nwith R ebalance weights")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "household_consumption", gt4_R[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for household consumption\nwith R ebalance weights")
plt.tight_layout(4.5)
```
# Estimation of impact with a simple regression
```
gt4_R["Tx"] = gt4_R.index.get_level_values(0).astype(int)
gt4_R["After"] = (gt4_R["prepost"] == "post").astype(int)
reg = stm.GLM.from_formula("homicide ~ After + Tx*After - Tx+ C(level_1)", gt4_R[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))].reset_index())
fit = reg.fit()
print(fit.summary())
print("Net avoided homicides (using R ebalance and Python GLM)")
ci = fit.conf_int().loc["Tx:After"]
print((gtpop.GTM_pop100k * fit.params["Tx:After"]).sum().round(), "(C.I. 95% ", (gtpop.GTM_pop100k * ci[0]).sum().round(),
", ", (gtpop.GTM_pop100k * ci[1]).sum().round() ," )")
gt4_st["Tx"] = gt4_st.index.get_level_values(0).astype(int)
gt4_st["After"] = (gt4_st["prepost"] == "post").astype(int)
reg = stm.GLM.from_formula("homicide ~ After + Tx*After - Tx + C(level_1)", gt4_st[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))].reset_index())
fit = reg.fit()
print(fit.summary())
print("Net avoided homicides (using Stata ebalance and Python GLM)")
ci = fit.conf_int().loc["Tx:After"]
print((gtpop.GTM_pop100k * fit.params["Tx:After"]).sum().round(), "(C.I. 95% ", (gtpop.GTM_pop100k * ci[0]).sum().round(),
", ", (gtpop.GTM_pop100k * ci[1]).sum().round() ," )")
# Crisis Group result in using stata GLM with robust variance is 4658
# Comparing under 5 mort. rate. Compare model with different intercepts and with same ints.
reg = stm.GLM.from_formula("under5_mortality_rate ~ After + Tx*After - Tx + C(level_1)", gt4_st[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))].reset_index())
fit = reg.fit()
print(fit.summary())
reg = stm.GLM.from_formula("under5_mortality_rate ~ After + Tx*After + C(level_1)", gt4_st[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))].reset_index())
fit = reg.fit()
print(fit.summary())
# Estimation of impact over homicides using different intercepts
reg = stm.GLM.from_formula("homicide ~ After + Tx*After + C(level_1)", gt4_st[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))].reset_index())
fit = reg.fit()
print(fit.summary())
print("Net avoided homicides (using Stata ebalance and Python GLM with different intercepts)")
ci = fit.conf_int().loc["Tx:After"]
print((gtpop.GTM_pop100k * fit.params["Tx:After"]).sum().round(), "(C.I. 95% ", (gtpop.GTM_pop100k * ci[0]).sum().round(),
", ", (gtpop.GTM_pop100k * ci[1]).sum().round() ," )")
# Estimation of impact with the simplest DID possible:
a = gt4_st.groupby(["Tx", "After"]).homicide.mean()
print(a.unstack(1))
did = (a[0][0] - a[1][0]) - (a[0][1] - a[1][1])
print("Simplest DID ", (gtpop.GTM_pop100k * did).sum())
```
# Conclusion
I have obtained a similar result to the one CrisisGroup obtained. DID results are not significant, and confidence intervals are very large even though they suggest an effect may be present. This can be due to the very small sample size used to estimate the weights (few years, covariates and countries to estimate the weights)
I think covariates are not very good and I want to attempt to do a similar analysis using microsynth package and other covariates. That package uses time series instead of just the means (in this analysis only mean moments are used to fit ebalance weights). Despite this, if you look at the graphs for **under 5 mortality rate** and **household consumption**, the trends are quite comparable and so, the parallel trends assumption of the diff&diff methodology is satisfied. I am not sure whether or not the GLM regression is correct. I think the synthetic control and treatment group should be allowed to have different intercepts. By looking at the graph of homicides it is evident that both groups have different intercepts. DID models usually include a term to account for such difference. I have also included the same graphics with unweighted data (no synth control) to show how the entropy balancing is succesfully generating a homicide synthetic control with almost parallel trends.
Beyond the statistical analysis and its many shortcomings, it is a very well known fact that homicide rate in Guatemala has been declining in the last years. Attributing that only to CICIG would be a big mistake, however I do believe corruption and organized crime in Guatemala are related and therefore I am biased to think that the fight against corruption has had an impact on this. Unfortunately, now that CICIG is officially gone, many corruption cases are falling apart and we will be able to see the effect of CICIG abscence with actual human lifes, instead of a synthetic counterfactual.
|
github_jupyter
|
import pandas as pd
import matplotlib as mlp
from matplotlib import pyplot as plt
import numpy as np
import seaborn as sbs
from statsmodels import api as stm
sbs.set(style="whitegrid")
%matplotlib inline
def tryFunc(functionToTry, onError = np.NaN, showErrors = False):
def wrapper(inputValue):
try:
return functionToTry(inputValue)
except:
if showErrors:
print("Bad value: ", inputValue)
return onError
return wrapper
toFloat = lambda x: float(x)
gt1 = pd.read_excel("./guatemala1.xlsx")
gt1.head(3)
gtpop = pd.read_excel("./gtm_pop.xlsx")
gtpop.head(3)
# Ignore Cuba and Haiti
gt1 = gt1[gt1["Country Code"].isin(["CUB", "HTI"]) == False]
gt1.head()
# The available indicators
gt1["Series Name"].value_counts()
cols = gt1.columns[gt1.columns.map(lambda x: x.startswith("YR")) == True]
gt2 = gt1.set_index(["Country Code", "Series Name"])[cols].stack().map(tryFunc(toFloat)).unstack(1).rename(index=lambda x: int(x[2:6]) if str.startswith(x, "YR") else x)
gt2.head().reset_index()
# This gives the same that stata
del gt2["CPIA_accountability_corruption"]
del gt2["CPIA_public_sector"]
gt2[gt2.index.get_level_values(1) < 2007].describe()
cols = ["gdp_per_capita_ppp_2011", "homicide", "household_consumption", "poverty_headcount_320"]
gt2["intercept"] = 1
def trends(df):
results = []
for col in cols:
results.append(np.linalg.lstsq(df[df[col].isna() == False].reset_index()[["intercept", "level_1"]], df[df[col].isna() == False][col])[0][1])
return pd.Series(index = cols, data = results)
trends = gt2[(gt2.index.get_level_values(1) < 2007 )].groupby("Country Code").apply(trends)
# The trends for each country for each indicator
# This is showing the same results as the original file.
trends
trends.columns = trends.columns.map(lambda x: "trend_" + x)
trends = pd.concat([trends, gt2[gt2.index.get_level_values(1) < 2007].groupby(level=[0]).mean()], axis = 1)
trends.to_csv("trends.csv")
gt2.to_csv("data_time_series.csv")
ebal_st = pd.read_csv("./ebalanced_stata.csv", index_col=0)
ebal_st.loc["GTM"] = 1
ebal_st.columns = ["ebal_st"]
ebal_R = pd.read_csv("./ebalanced.csv", index_col=0)
ebal_R.loc["GTM"] = 1
ebal_R.columns = ["ebal_R"]
gt3 = gt2.reset_index()
gt3 = gt2.merge(ebal_st, left_on="Country Code", right_index=True).reset_index()
gt3 = gt3.merge(ebal_R, left_on="level_0", right_index=True)
gt3["logHom"] = np.log10(gt3.homicide)
gt3["Treatment"] = gt3.level_0 == "GTM"
def wmean(df, m, w):
return df[m].multiply(df[w]).sum() / \
df[w][df[m].notna()].sum()
# TEST
# wmean(gt3[gt3.level_0 == "GTM"], "homicide", "ebal_st" ), gt3[gt3.level_0 == "GTM"].homicide.mean()
avgCols = ['logHom','adult_literacy_rate', 'gdp_per_capita_ppp_2011', 'homicide',
'household_consumption', 'poverty_headcount_320',
'under5_mortality_rate', 'youth_literacy_rate']
gt4_st = gt3.groupby(["Treatment", "level_1"]).apply(
lambda x: pd.Series([ wmean(x, col, "ebal_st") for col in avgCols ], avgCols) )
gt4_R = gt3.groupby(["Treatment", "level_1"]).apply(
lambda x: pd.Series([ wmean(x, col, "ebal_R") for col in avgCols ], avgCols) )
gt4_unw = gt3.groupby(["Treatment", "level_1"]).apply(
lambda x: pd.Series([ x[col].mean() for col in avgCols ], avgCols) )
gt4_unw["prepost"] = gt4_unw.index.get_level_values(1).map(lambda x: ("pre" if x<2007 else "post"), 1)
sbs.lmplot("level_1", "logHom", gt4_unw[gt4_unw.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
# Finally, getting the same graph that CrisisGroup obtained in stata, of course, using their ebalance results.
plt.suptitle("Compare Pre-Post CICIG scenarios for homicides\nwithout weights (simple means)")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "under5_mortality_rate", gt4_unw[gt4_unw.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for under 5 mortality rate\nwithout weights (simple means)")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "household_consumption", gt4_unw[gt4_unw.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for under 5 mortality rate\nwithout weights (simple means)")
plt.tight_layout(4.5)
gt4_st["prepost"] = gt4_st.index.get_level_values(1).map(lambda x: ("pre" if x<2007 else "post"), 1)
sbs.lmplot("level_1", "homicide", gt4_st[gt4_st.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
# Finally, getting the same graph that CrisisGroup obtained in stata, of course, using their ebalance results.
plt.suptitle("Compare Pre-Post CICIG scenarios for homicides\nwith Stata ebalance weights")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "logHom", gt4_st[gt4_st.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for log(homicides)\nwith Stata ebalance weights")
plt.tight_layout(4.5)
axs.set_yticklabels(np.power(10, axs.axes[0,0].get_yticks()).round())
axs = sbs.lmplot("level_1", "under5_mortality_rate", gt4_st[gt4_st.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for under 5 mortality rate\nwith Stata ebalance weights")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "household_consumption", gt4_st[gt4_st.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for household consumption\nwith Stata ebalance weights")
plt.tight_layout(4.5)
gt4_R["prepost"] = gt4_R.index.get_level_values(1).map(lambda x: ("pre" if x<2007 else "post"), 1)
axs = sbs.lmplot("level_1", "logHom", gt4_R[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
axs.set_yticklabels(np.power(10, axs.axes[0,0].get_yticks()).round())
plt.suptitle("Compare Pre-Post CICIG scenarios for log(homicides)\nwith R ebalance weights")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "under5_mortality_rate", gt4_R[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for under 5 mortality rate\nwith R ebalance weights")
plt.tight_layout(4.5)
axs = sbs.lmplot("level_1", "household_consumption", gt4_R[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))]\
.reset_index(), "Treatment", col = "prepost", sharex=False, legend_out=False)
plt.suptitle("Compare Pre-Post CICIG scenarios for household consumption\nwith R ebalance weights")
plt.tight_layout(4.5)
gt4_R["Tx"] = gt4_R.index.get_level_values(0).astype(int)
gt4_R["After"] = (gt4_R["prepost"] == "post").astype(int)
reg = stm.GLM.from_formula("homicide ~ After + Tx*After - Tx+ C(level_1)", gt4_R[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))].reset_index())
fit = reg.fit()
print(fit.summary())
print("Net avoided homicides (using R ebalance and Python GLM)")
ci = fit.conf_int().loc["Tx:After"]
print((gtpop.GTM_pop100k * fit.params["Tx:After"]).sum().round(), "(C.I. 95% ", (gtpop.GTM_pop100k * ci[0]).sum().round(),
", ", (gtpop.GTM_pop100k * ci[1]).sum().round() ," )")
gt4_st["Tx"] = gt4_st.index.get_level_values(0).astype(int)
gt4_st["After"] = (gt4_st["prepost"] == "post").astype(int)
reg = stm.GLM.from_formula("homicide ~ After + Tx*After - Tx + C(level_1)", gt4_st[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))].reset_index())
fit = reg.fit()
print(fit.summary())
print("Net avoided homicides (using Stata ebalance and Python GLM)")
ci = fit.conf_int().loc["Tx:After"]
print((gtpop.GTM_pop100k * fit.params["Tx:After"]).sum().round(), "(C.I. 95% ", (gtpop.GTM_pop100k * ci[0]).sum().round(),
", ", (gtpop.GTM_pop100k * ci[1]).sum().round() ," )")
# Crisis Group result in using stata GLM with robust variance is 4658
# Comparing under 5 mort. rate. Compare model with different intercepts and with same ints.
reg = stm.GLM.from_formula("under5_mortality_rate ~ After + Tx*After - Tx + C(level_1)", gt4_st[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))].reset_index())
fit = reg.fit()
print(fit.summary())
reg = stm.GLM.from_formula("under5_mortality_rate ~ After + Tx*After + C(level_1)", gt4_st[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))].reset_index())
fit = reg.fit()
print(fit.summary())
# Estimation of impact over homicides using different intercepts
reg = stm.GLM.from_formula("homicide ~ After + Tx*After + C(level_1)", gt4_st[gt4_R.index.get_level_values(1).isin(list(range(2000, 2015)))].reset_index())
fit = reg.fit()
print(fit.summary())
print("Net avoided homicides (using Stata ebalance and Python GLM with different intercepts)")
ci = fit.conf_int().loc["Tx:After"]
print((gtpop.GTM_pop100k * fit.params["Tx:After"]).sum().round(), "(C.I. 95% ", (gtpop.GTM_pop100k * ci[0]).sum().round(),
", ", (gtpop.GTM_pop100k * ci[1]).sum().round() ," )")
# Estimation of impact with the simplest DID possible:
a = gt4_st.groupby(["Tx", "After"]).homicide.mean()
print(a.unstack(1))
did = (a[0][0] - a[1][0]) - (a[0][1] - a[1][1])
print("Simplest DID ", (gtpop.GTM_pop100k * did).sum())
| 0.436622 | 0.879147 |
please use python 3
```
%time
%load_ext autotime
%load_ext autoreload
%autoreload 2
import torch
from torch import nn
from torch.autograd import Variable
from data_loader import DataLoader
from model import UniSkip
from config import *
from datetime import datetime, timedelta
```
### train a new model
```
d = DataLoader('../dir_HugeFiles/instructions/skip_instruction.csv')
mod = UniSkip()
if USE_CUDA:
mod.cuda(CUDA_DEVICE)
lr = 3e-4
optimizer = torch.optim.Adam(params=mod.parameters(), lr=lr)
loss_trail = []
last_best_loss = None
current_time = datetime.utcnow()
def debug(i, loss, prev, nex, prev_pred, next_pred):
global loss_trail
global last_best_loss
global current_time
this_loss = loss.item()
loss_trail.append(this_loss)
loss_trail = loss_trail[-20:]
new_current_time = datetime.utcnow()
time_elapsed = str(new_current_time - current_time)
current_time = new_current_time
print("Iteration {}: time = {} last_best_loss = {}, this_loss = {}".format(
i, time_elapsed, last_best_loss, this_loss))
'''
print("prev = {}\nnext = {}\npred_prev = {}\npred_next = {}".format(
d.convert_indices_to_sentences(prev),
d.convert_indices_to_sentences(nex),
d.convert_indices_to_sentences(prev_pred),
d.convert_indices_to_sentences(next_pred),
))
'''
try:
trail_loss = sum(loss_trail)/len(loss_trail)
if last_best_loss is None or last_best_loss > trail_loss:
print("Loss improved from {} to {}".format(last_best_loss, trail_loss))
save_loc = "./saved_models/skip-best".format(lr, VOCAB_SIZE)
print("saving model at {}".format(save_loc))
torch.save(mod.state_dict(), save_loc)
last_best_loss = trail_loss
except Exception as e:
print("Couldn't save model because {}".format(e))
print("Starting training...")
print('current GPU is on %d ' %(CUDA_DEVICE)) # should change at the config.py
# a million iterations
for i in range(0, 1000000):
sentences, lengths = d.fetch_batch(32 * 8) # remember to change cuda device
loss, prev, nex, prev_pred, next_pred = mod(sentences, lengths)
if i % 100 == 0:
debug(i, loss, prev, nex, prev_pred, next_pred)
optimizer.zero_grad()
loss.backward()
optimizer.step()
```
### resume the training
```
# model to resume
loc = "./prev_model/skip-best"
d = DataLoader('../dir_HugeFiles/instructions/skip_instruction.csv')
mod = UniSkip()
mod.load_state_dict(torch.load(loc, map_location=lambda storage, loc: storage))
if USE_CUDA:
mod.cuda(CUDA_DEVICE)
lr = 3e-4
optimizer = torch.optim.Adam(params=mod.parameters(), lr=lr)
loss_trail = []
last_best_loss = None
current_time = datetime.utcnow()
def debug(i, loss, prev, nex, prev_pred, next_pred):
global loss_trail
global last_best_loss
global current_time
this_loss = loss.item()
loss_trail.append(this_loss)
loss_trail = loss_trail[-20:]
new_current_time = datetime.utcnow()
time_elapsed = str(new_current_time - current_time)
current_time = new_current_time
print("Iteration {}: time = {} last_best_loss = {}, this_loss = {}".format(
i, time_elapsed, last_best_loss, this_loss))
'''
print("prev = {}\nnext = {}\npred_prev = {}\npred_next = {}".format(
d.convert_indices_to_sentences(prev),
d.convert_indices_to_sentences(nex),
d.convert_indices_to_sentences(prev_pred),
d.convert_indices_to_sentences(next_pred),
))
'''
try:
trail_loss = sum(loss_trail)/len(loss_trail)
if last_best_loss is None or last_best_loss > trail_loss:
print("Loss improved from {} to {}".format(last_best_loss, trail_loss))
save_loc = "./prev_model/" + "skip-best-loss{0:.3f}".format(trail_loss)
print("saving model at {}".format(save_loc))
torch.save(mod.state_dict(), save_loc)
last_best_loss = trail_loss
except Exception as e:
print("Couldn't save model because {}".format(e))
print("Starting training...")
print('current GPU is on %d ' %(CUDA_DEVICE)) # should change at the config.py
# a million iterations
for i in range(0, 1000000):
sentences, lengths = d.fetch_batch(32 * 8) # remember to change cuda device
loss, prev, nex, prev_pred, next_pred = mod(sentences, lengths)
if i % 100 == 0:
debug(i, loss, prev, nex, prev_pred, next_pred)
optimizer.zero_grad()
loss.backward()
optimizer.step()
```
|
github_jupyter
|
%time
%load_ext autotime
%load_ext autoreload
%autoreload 2
import torch
from torch import nn
from torch.autograd import Variable
from data_loader import DataLoader
from model import UniSkip
from config import *
from datetime import datetime, timedelta
d = DataLoader('../dir_HugeFiles/instructions/skip_instruction.csv')
mod = UniSkip()
if USE_CUDA:
mod.cuda(CUDA_DEVICE)
lr = 3e-4
optimizer = torch.optim.Adam(params=mod.parameters(), lr=lr)
loss_trail = []
last_best_loss = None
current_time = datetime.utcnow()
def debug(i, loss, prev, nex, prev_pred, next_pred):
global loss_trail
global last_best_loss
global current_time
this_loss = loss.item()
loss_trail.append(this_loss)
loss_trail = loss_trail[-20:]
new_current_time = datetime.utcnow()
time_elapsed = str(new_current_time - current_time)
current_time = new_current_time
print("Iteration {}: time = {} last_best_loss = {}, this_loss = {}".format(
i, time_elapsed, last_best_loss, this_loss))
'''
print("prev = {}\nnext = {}\npred_prev = {}\npred_next = {}".format(
d.convert_indices_to_sentences(prev),
d.convert_indices_to_sentences(nex),
d.convert_indices_to_sentences(prev_pred),
d.convert_indices_to_sentences(next_pred),
))
'''
try:
trail_loss = sum(loss_trail)/len(loss_trail)
if last_best_loss is None or last_best_loss > trail_loss:
print("Loss improved from {} to {}".format(last_best_loss, trail_loss))
save_loc = "./saved_models/skip-best".format(lr, VOCAB_SIZE)
print("saving model at {}".format(save_loc))
torch.save(mod.state_dict(), save_loc)
last_best_loss = trail_loss
except Exception as e:
print("Couldn't save model because {}".format(e))
print("Starting training...")
print('current GPU is on %d ' %(CUDA_DEVICE)) # should change at the config.py
# a million iterations
for i in range(0, 1000000):
sentences, lengths = d.fetch_batch(32 * 8) # remember to change cuda device
loss, prev, nex, prev_pred, next_pred = mod(sentences, lengths)
if i % 100 == 0:
debug(i, loss, prev, nex, prev_pred, next_pred)
optimizer.zero_grad()
loss.backward()
optimizer.step()
# model to resume
loc = "./prev_model/skip-best"
d = DataLoader('../dir_HugeFiles/instructions/skip_instruction.csv')
mod = UniSkip()
mod.load_state_dict(torch.load(loc, map_location=lambda storage, loc: storage))
if USE_CUDA:
mod.cuda(CUDA_DEVICE)
lr = 3e-4
optimizer = torch.optim.Adam(params=mod.parameters(), lr=lr)
loss_trail = []
last_best_loss = None
current_time = datetime.utcnow()
def debug(i, loss, prev, nex, prev_pred, next_pred):
global loss_trail
global last_best_loss
global current_time
this_loss = loss.item()
loss_trail.append(this_loss)
loss_trail = loss_trail[-20:]
new_current_time = datetime.utcnow()
time_elapsed = str(new_current_time - current_time)
current_time = new_current_time
print("Iteration {}: time = {} last_best_loss = {}, this_loss = {}".format(
i, time_elapsed, last_best_loss, this_loss))
'''
print("prev = {}\nnext = {}\npred_prev = {}\npred_next = {}".format(
d.convert_indices_to_sentences(prev),
d.convert_indices_to_sentences(nex),
d.convert_indices_to_sentences(prev_pred),
d.convert_indices_to_sentences(next_pred),
))
'''
try:
trail_loss = sum(loss_trail)/len(loss_trail)
if last_best_loss is None or last_best_loss > trail_loss:
print("Loss improved from {} to {}".format(last_best_loss, trail_loss))
save_loc = "./prev_model/" + "skip-best-loss{0:.3f}".format(trail_loss)
print("saving model at {}".format(save_loc))
torch.save(mod.state_dict(), save_loc)
last_best_loss = trail_loss
except Exception as e:
print("Couldn't save model because {}".format(e))
print("Starting training...")
print('current GPU is on %d ' %(CUDA_DEVICE)) # should change at the config.py
# a million iterations
for i in range(0, 1000000):
sentences, lengths = d.fetch_batch(32 * 8) # remember to change cuda device
loss, prev, nex, prev_pred, next_pred = mod(sentences, lengths)
if i % 100 == 0:
debug(i, loss, prev, nex, prev_pred, next_pred)
optimizer.zero_grad()
loss.backward()
optimizer.step()
| 0.543833 | 0.643007 |
# Calculate Well Trajectory from Different Data Sources
This notebook shows examples of how to calculate the well trajectory from different data sources.
Currently accepting data from CSV, dataframe, dictionary, and json.
The data from csv and dataframe must be in a certain format commonly seen in directional surveys.
The dictionary and json must be in a format specific to the library.
```
from welltrajconvert.wellbore_trajectory import *
from welltrajconvert.data_source import *
```
## Get Wellbore Trajectory object
```
path = './'
```
`get_files` is a great function for accessing your data in a folder. You can access all data in a specific folder or data with a specific extension. It returns said data in a list of items.
```
# use get_files to get all the files in the data directory
file_paths = get_files(path, folders='data')
#gives a path lib path to all your files in the folder of choice
file_paths.items
```
## From Dict
```
json_path = get_files(path, folders='data', extensions='.json')
json_path.items[0]
with open(json_path.items[0]) as json_file:
json_data = json.load(json_file)
json_file.close()
#show the dict keys present in the dict.
json_data.keys()
# call DataSource.from_dictionary and input the dict data
my_data = DataSource.from_dictionary(json_data)
#view the dict data dataclass object
my_data.data
#create a wellboreTrajectory object
dev_obj = WellboreTrajectory(my_data.data)
#view the object
dev_obj.deviation_survey_obj
#calculate the survey points along the wellbore for the object
dev_obj.calculate_survey_points()
#serialize the data
json_ds = dev_obj.serialize()
# view the json in a df
json_ds_obj = json.loads(json_ds)
df_min_curve = pd.DataFrame(json_ds_obj)
df_min_curve.head()
```
## From DF
If you have a df and want to calculate its metadata use the following workflow. Just enter in the column names and calculate its survey points.
```
# FROM DF
csv_path = get_files(path, folders='data', extensions='.csv')
csv_path.items[0]
df = pd.read_csv(csv_path.items[0],sep=',')
df.head()
# call the from_df method, fill in the parameters with the column names
my_data = DataSource.from_df(df, wellId_name='wellId',md_name='md',inc_name='inc',azim_name='azim',
surface_latitude_name='surface_latitude',surface_longitude_name='surface_longitude')
#view the dict data dataclass object
my_data.data
#create a wellboreTrajectory object
dev_obj = WellboreTrajectory(my_data.data)
#view the object
dev_obj.deviation_survey_obj
#calculate the survey points along the wellbore for the object
dev_obj.calculate_survey_points()
#serialize the data
json_ds = dev_obj.serialize()
# view the json in a df
json_ds_obj = json.loads(json_ds)
df_min_curve = pd.DataFrame(json_ds_obj)
df_min_curve.head()
```
# From CSV
If you have a csv and want to calculate its metadata use the following workflow. Just enter in the column names and calculate its survey points.
```
csv_path = get_files(path, folders='data', extensions='.csv')
my_data = DataSource.from_csv(csv_path.items[0], wellId_name='wellId',md_name='md',inc_name='inc',azim_name='azim',
surface_latitude_name='surface_latitude',surface_longitude_name='surface_longitude')
#view the dict data dataclass object
my_data.data
#create a wellboreTrajectory object
dev_obj = WellboreTrajectory(my_data.data)
#view the object
dev_obj.deviation_survey_obj
#calculate the survey points along the wellbore for the object
dev_obj.calculate_survey_points()
#serialize the data
json_ds = dev_obj.serialize()
# view the json in a df
json_ds_obj = json.loads(json_ds)
df_min_curve = pd.DataFrame(json_ds_obj)
df_min_curve.head()
```
## From CSV with multiple wells
If you have a file with multiple wells appened one after another you can use this simple function to break them up by wellid, run it through welltrajconvert and calculate each wells metadata. You can then convert the list of dicts into a dataframe.
```
def from_multiple_wells_to_dict(df: DataFrame, wellId_name: Optional[str] = None, md_name: Optional[str] = None,
inc_name: Optional[str] = None, azim_name: Optional[str] = None,
surface_latitude_name: Optional[str] = None,
surface_longitude_name: Optional[str] = None,
surface_x_name: Optional[str] = None,
surface_y_name: Optional[str] = None):
"""
takes a df of mulitple well deviation surveys in a typical columnar fashion and calculates their survey points.
Then appends them to a list of dicts.
:parameter: df
:returns: list of dict
"""
# group by wellId, ensures this will work with single well or mulitple.
grouped = df.groupby(wellId_name)
# initialize empty dict and list
#appended_df = pd.DataFrame()
dict_list = []
# loop through groups converting them to the proper dict format
for name, group in grouped:
group.reset_index(inplace=True, drop=True)
if surface_latitude_name is not None and surface_longitude_name is not None:
well_obj = DataSource.from_df(group, wellId_name=wellId_name, md_name=md_name,
inc_name=inc_name, azim_name=azim_name,
surface_latitude_name=surface_latitude_name,
surface_longitude_name=surface_longitude_name)
if surface_x_name is not None and surface_y_name is not None:
well_obj = DataSource.from_df(group, wellId_name=wellId_name, md_name=md_name,
inc_name=inc_name, azim_name=azim_name,
surface_x_name=surface_x_name,
surface_y_name=surface_y_name)
well_obj = WellboreTrajectory(well_obj.data)
well_obj.calculate_survey_points()
json_ds = well_obj.serialize()
dict_list.append(json_ds)
res = dict_list
return res
# FROM df with mulitple wells
csv_path = get_files(path, folders='data', extensions='.csv')
csv_path.items[1]
df = pd.read_csv(csv_path.items[1])
# well id unique
print(df['wellId'].unique())
df.head()
# call function, fill in column name params
dicts = from_multiple_wells_to_dict(df,wellId_name='wellId',md_name='md',inc_name='inc',azim_name='azim',
surface_latitude_name='surface_latitude',surface_longitude_name='surface_longitude')
#dicts[:]
def list_of_dicts_to_df(dict_list):
"""takes a list of dicts and converts to a appended df"""
appended_df = pd.DataFrame()
for i in dict_list:
json_ds_obj = json.loads(i)
df_well_obj = pd.DataFrame(json_ds_obj)
appended_df = appended_df.append(df_well_obj)
return appended_df
# convert a list of dicts into a df
df = list_of_dicts_to_df(dicts)
df.head()
```
|
github_jupyter
|
from welltrajconvert.wellbore_trajectory import *
from welltrajconvert.data_source import *
path = './'
# use get_files to get all the files in the data directory
file_paths = get_files(path, folders='data')
#gives a path lib path to all your files in the folder of choice
file_paths.items
json_path = get_files(path, folders='data', extensions='.json')
json_path.items[0]
with open(json_path.items[0]) as json_file:
json_data = json.load(json_file)
json_file.close()
#show the dict keys present in the dict.
json_data.keys()
# call DataSource.from_dictionary and input the dict data
my_data = DataSource.from_dictionary(json_data)
#view the dict data dataclass object
my_data.data
#create a wellboreTrajectory object
dev_obj = WellboreTrajectory(my_data.data)
#view the object
dev_obj.deviation_survey_obj
#calculate the survey points along the wellbore for the object
dev_obj.calculate_survey_points()
#serialize the data
json_ds = dev_obj.serialize()
# view the json in a df
json_ds_obj = json.loads(json_ds)
df_min_curve = pd.DataFrame(json_ds_obj)
df_min_curve.head()
# FROM DF
csv_path = get_files(path, folders='data', extensions='.csv')
csv_path.items[0]
df = pd.read_csv(csv_path.items[0],sep=',')
df.head()
# call the from_df method, fill in the parameters with the column names
my_data = DataSource.from_df(df, wellId_name='wellId',md_name='md',inc_name='inc',azim_name='azim',
surface_latitude_name='surface_latitude',surface_longitude_name='surface_longitude')
#view the dict data dataclass object
my_data.data
#create a wellboreTrajectory object
dev_obj = WellboreTrajectory(my_data.data)
#view the object
dev_obj.deviation_survey_obj
#calculate the survey points along the wellbore for the object
dev_obj.calculate_survey_points()
#serialize the data
json_ds = dev_obj.serialize()
# view the json in a df
json_ds_obj = json.loads(json_ds)
df_min_curve = pd.DataFrame(json_ds_obj)
df_min_curve.head()
csv_path = get_files(path, folders='data', extensions='.csv')
my_data = DataSource.from_csv(csv_path.items[0], wellId_name='wellId',md_name='md',inc_name='inc',azim_name='azim',
surface_latitude_name='surface_latitude',surface_longitude_name='surface_longitude')
#view the dict data dataclass object
my_data.data
#create a wellboreTrajectory object
dev_obj = WellboreTrajectory(my_data.data)
#view the object
dev_obj.deviation_survey_obj
#calculate the survey points along the wellbore for the object
dev_obj.calculate_survey_points()
#serialize the data
json_ds = dev_obj.serialize()
# view the json in a df
json_ds_obj = json.loads(json_ds)
df_min_curve = pd.DataFrame(json_ds_obj)
df_min_curve.head()
def from_multiple_wells_to_dict(df: DataFrame, wellId_name: Optional[str] = None, md_name: Optional[str] = None,
inc_name: Optional[str] = None, azim_name: Optional[str] = None,
surface_latitude_name: Optional[str] = None,
surface_longitude_name: Optional[str] = None,
surface_x_name: Optional[str] = None,
surface_y_name: Optional[str] = None):
"""
takes a df of mulitple well deviation surveys in a typical columnar fashion and calculates their survey points.
Then appends them to a list of dicts.
:parameter: df
:returns: list of dict
"""
# group by wellId, ensures this will work with single well or mulitple.
grouped = df.groupby(wellId_name)
# initialize empty dict and list
#appended_df = pd.DataFrame()
dict_list = []
# loop through groups converting them to the proper dict format
for name, group in grouped:
group.reset_index(inplace=True, drop=True)
if surface_latitude_name is not None and surface_longitude_name is not None:
well_obj = DataSource.from_df(group, wellId_name=wellId_name, md_name=md_name,
inc_name=inc_name, azim_name=azim_name,
surface_latitude_name=surface_latitude_name,
surface_longitude_name=surface_longitude_name)
if surface_x_name is not None and surface_y_name is not None:
well_obj = DataSource.from_df(group, wellId_name=wellId_name, md_name=md_name,
inc_name=inc_name, azim_name=azim_name,
surface_x_name=surface_x_name,
surface_y_name=surface_y_name)
well_obj = WellboreTrajectory(well_obj.data)
well_obj.calculate_survey_points()
json_ds = well_obj.serialize()
dict_list.append(json_ds)
res = dict_list
return res
# FROM df with mulitple wells
csv_path = get_files(path, folders='data', extensions='.csv')
csv_path.items[1]
df = pd.read_csv(csv_path.items[1])
# well id unique
print(df['wellId'].unique())
df.head()
# call function, fill in column name params
dicts = from_multiple_wells_to_dict(df,wellId_name='wellId',md_name='md',inc_name='inc',azim_name='azim',
surface_latitude_name='surface_latitude',surface_longitude_name='surface_longitude')
#dicts[:]
def list_of_dicts_to_df(dict_list):
"""takes a list of dicts and converts to a appended df"""
appended_df = pd.DataFrame()
for i in dict_list:
json_ds_obj = json.loads(i)
df_well_obj = pd.DataFrame(json_ds_obj)
appended_df = appended_df.append(df_well_obj)
return appended_df
# convert a list of dicts into a df
df = list_of_dicts_to_df(dicts)
df.head()
| 0.482673 | 0.920932 |
```
import numpy as np;
import math;
import csv;
import Simplex;
class IntegerSimplex(Simplex.Simplex):
integerSolution = [];
def solve(self):
super().solve();
print("Now will perform integer solution\n\n");
self.integerSolve();
@staticmethod
def divideArraysZero(array1, array2): #divides array1 by array2.
temp = np.zeros(len(array1));
i = 0;
for x in array2:
if( x == 0 ):
temp[i] = -math.inf;
else:
temp[i] = array1[i]/array2[i];
i += 1;
return temp;
@staticmethod
def minNonZero(array):
minVal = math.inf;
minIndex = -1;
i = 0;
for val in array:
if(val > 0 and val < minVal):
minVal = val;
minIndex = i;
i += 1;
#print("MinVal = " + str(minVal));
return minIndex;
def performCheck(self, matrix):
#print("All integers? Let's see...");
#print(matrix[:, -1 ]);
areIntegers = True;
for val in matrix[:, -1 ]:
areIntegers = areIntegers and ( val%1 == 0 or val == math.inf or val == -math.inf);
return areIntegers;
def integerSolve(self):
self.integerSolution = np.copy(self.solutionMatrix);
while(not self.performCheck(self.roundSolution(np.copy(self.integerSolution),4))):
print("Initial matrix for integer: ");
self.printArray(self.integerSolution);
#find the row -> search for maximum "answer" from original solution
row = np.argmax(self.integerSolution[0:len(self.solutionMatrix),-1]%1);
#print("Row: " + str(row) + " with value: " + str( np.max(self.integerSolution[0:-2,-1]%1) ));
#print("From row: ");
#print(self.integerSolution[0:2,-1]%1);
t_matrix = np.copy(self.integerSolution);
t_matrix = np.insert(t_matrix,
[
t_matrix.shape[1]-1
],
np.zeros(1, dtype=np.float32),
axis = 1);
#print("t_matrix is: ");
#self.printArray(t_matrix);
insertRow = -(t_matrix[row,]%1);
insertRow[-2] = 1; #last but one element
#print("Insert row: ");
#print(insertRow);
t_matrix = np.insert(t_matrix, [-1], insertRow, axis=0);
#print("t_matrix is: ");
#self.printArray(t_matrix);
temp = IntegerSimplex.divideArraysZero(t_matrix[t_matrix.shape[0]-1, :-1],t_matrix[t_matrix.shape[0]-2, :-1]);
#print(temp[0:len(self.solutionMatrix[0])-1]);
#print(temp);
#print(abs(temp[0:len(self.solutionMatrix[0])-1]));
indexMin = IntegerSimplex.minNonZero(abs(temp[0:len(self.solutionMatrix[0])-1]));
if(indexMin == -1):
#print("No values except INF or 0");
indexMin = np.argmin(abs(temp[0:len(self.solutionMatrix[0])-1]));
if(temp[indexMin] == -math.inf):
#print("Min is INF. Take the whole row: ");
#print(temp);
indexMin = np.argmin(abs(temp));
elif(temp[indexMin] == 0):
pass;
#print("Min is 0");
#print("Max is " + str(temp[indexMin]) + " and its pos is: " + str(indexMin));
print("Perform elimination at point " + str(t_matrix.shape[0]-2) + " : " + str(indexMin));
self.integerSolution = self.performElimiation(t_matrix, t_matrix.shape[0]-2, indexMin); #at last row and index of greatest negative number from 'temp'
self.printArray(self.integerSolution);
self.roundSolution(self.integerSolution,4);
example1 = IntegerSimplex("example1.csv");
example1.solve();
example1.showResultValues(example1.integerSolution);
print("\n\n\n" + ("=" * 80) + "\n" + ("="*80) + "\n\n\n");
example2 = IntegerSimplex("example2.csv");
example2.solve();
example2.showResultValues(example2.integerSolution);
#13 is in infinite loop!
i = 17;
while (i < 16):
s_min = IntegerSimplex("Var/"+ str(i) + "/" + str(i) + "min.txt");
s_max = IntegerSimplex("Var/"+ str(i) + "/" + str(i) + "max.txt");
s_min.solve();
s_min.showResultValues(s_min.integerSolution);
print("\n\n\n" + ("_" * 40) + "\n" + ("_"*40) + "\n\n\n");
s_max.solve();
s_max.showResultValues(s_max.integerSolution);
print("\n\n\n" + ("_" * 80) + "\n" + ("_"*80) + "\n\n\n");
print("\n\n\n" + ("=" * 80) + "\n" + ("="*80) + "\n\n\n");
i+= 1;
example1 = IntegerSimplex("example1.csv");
example1.solve();
example1.showResultValues(example1.integerSolution);
print("\n\n\n" + ("=" * 80) + "\n" + ("="*80) + "\n\n\n");
example2 = IntegerSimplex("example2.csv");
example2.solve();
example2.showResultValues(example2.integerSolution);
```
|
github_jupyter
|
import numpy as np;
import math;
import csv;
import Simplex;
class IntegerSimplex(Simplex.Simplex):
integerSolution = [];
def solve(self):
super().solve();
print("Now will perform integer solution\n\n");
self.integerSolve();
@staticmethod
def divideArraysZero(array1, array2): #divides array1 by array2.
temp = np.zeros(len(array1));
i = 0;
for x in array2:
if( x == 0 ):
temp[i] = -math.inf;
else:
temp[i] = array1[i]/array2[i];
i += 1;
return temp;
@staticmethod
def minNonZero(array):
minVal = math.inf;
minIndex = -1;
i = 0;
for val in array:
if(val > 0 and val < minVal):
minVal = val;
minIndex = i;
i += 1;
#print("MinVal = " + str(minVal));
return minIndex;
def performCheck(self, matrix):
#print("All integers? Let's see...");
#print(matrix[:, -1 ]);
areIntegers = True;
for val in matrix[:, -1 ]:
areIntegers = areIntegers and ( val%1 == 0 or val == math.inf or val == -math.inf);
return areIntegers;
def integerSolve(self):
self.integerSolution = np.copy(self.solutionMatrix);
while(not self.performCheck(self.roundSolution(np.copy(self.integerSolution),4))):
print("Initial matrix for integer: ");
self.printArray(self.integerSolution);
#find the row -> search for maximum "answer" from original solution
row = np.argmax(self.integerSolution[0:len(self.solutionMatrix),-1]%1);
#print("Row: " + str(row) + " with value: " + str( np.max(self.integerSolution[0:-2,-1]%1) ));
#print("From row: ");
#print(self.integerSolution[0:2,-1]%1);
t_matrix = np.copy(self.integerSolution);
t_matrix = np.insert(t_matrix,
[
t_matrix.shape[1]-1
],
np.zeros(1, dtype=np.float32),
axis = 1);
#print("t_matrix is: ");
#self.printArray(t_matrix);
insertRow = -(t_matrix[row,]%1);
insertRow[-2] = 1; #last but one element
#print("Insert row: ");
#print(insertRow);
t_matrix = np.insert(t_matrix, [-1], insertRow, axis=0);
#print("t_matrix is: ");
#self.printArray(t_matrix);
temp = IntegerSimplex.divideArraysZero(t_matrix[t_matrix.shape[0]-1, :-1],t_matrix[t_matrix.shape[0]-2, :-1]);
#print(temp[0:len(self.solutionMatrix[0])-1]);
#print(temp);
#print(abs(temp[0:len(self.solutionMatrix[0])-1]));
indexMin = IntegerSimplex.minNonZero(abs(temp[0:len(self.solutionMatrix[0])-1]));
if(indexMin == -1):
#print("No values except INF or 0");
indexMin = np.argmin(abs(temp[0:len(self.solutionMatrix[0])-1]));
if(temp[indexMin] == -math.inf):
#print("Min is INF. Take the whole row: ");
#print(temp);
indexMin = np.argmin(abs(temp));
elif(temp[indexMin] == 0):
pass;
#print("Min is 0");
#print("Max is " + str(temp[indexMin]) + " and its pos is: " + str(indexMin));
print("Perform elimination at point " + str(t_matrix.shape[0]-2) + " : " + str(indexMin));
self.integerSolution = self.performElimiation(t_matrix, t_matrix.shape[0]-2, indexMin); #at last row and index of greatest negative number from 'temp'
self.printArray(self.integerSolution);
self.roundSolution(self.integerSolution,4);
example1 = IntegerSimplex("example1.csv");
example1.solve();
example1.showResultValues(example1.integerSolution);
print("\n\n\n" + ("=" * 80) + "\n" + ("="*80) + "\n\n\n");
example2 = IntegerSimplex("example2.csv");
example2.solve();
example2.showResultValues(example2.integerSolution);
#13 is in infinite loop!
i = 17;
while (i < 16):
s_min = IntegerSimplex("Var/"+ str(i) + "/" + str(i) + "min.txt");
s_max = IntegerSimplex("Var/"+ str(i) + "/" + str(i) + "max.txt");
s_min.solve();
s_min.showResultValues(s_min.integerSolution);
print("\n\n\n" + ("_" * 40) + "\n" + ("_"*40) + "\n\n\n");
s_max.solve();
s_max.showResultValues(s_max.integerSolution);
print("\n\n\n" + ("_" * 80) + "\n" + ("_"*80) + "\n\n\n");
print("\n\n\n" + ("=" * 80) + "\n" + ("="*80) + "\n\n\n");
i+= 1;
example1 = IntegerSimplex("example1.csv");
example1.solve();
example1.showResultValues(example1.integerSolution);
print("\n\n\n" + ("=" * 80) + "\n" + ("="*80) + "\n\n\n");
example2 = IntegerSimplex("example2.csv");
example2.solve();
example2.showResultValues(example2.integerSolution);
| 0.180251 | 0.359701 |
# 📃 Solution for Exercise M5.02
The aim of this exercise is to find out whether a decision tree
model is able to extrapolate.
By extrapolation, we refer to values predicted by a model outside of the
range of feature values seen during the training.
We will first load the regression data.
```
import pandas as pd
penguins = pd.read_csv("../datasets/penguins_regression.csv")
data_columns = ["Flipper Length (mm)"]
target_column = "Body Mass (g)"
data_train, target_train = penguins[data_columns], penguins[target_column]
```
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">If you want a deeper overview regarding this dataset, you can refer to the
Appendix - Datasets description section at the end of this MOOC.</p>
</div>
First, create two models, a linear regression model and a decision tree
regression model, and fit them on the training data. Limit the depth at
3 levels for the decision tree.
```
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
linear_regression = LinearRegression()
tree = DecisionTreeRegressor(max_depth=3)
linear_regression.fit(data_train, target_train)
tree.fit(data_train, target_train)
```
Create a testing dataset, ranging from the minimum to the maximum of the
flipper length of the training dataset. Get the predictions of each model
using this test dataset.
```
import numpy as np
data_test = pd.DataFrame(np.arange(data_train[data_columns[0]].min(),
data_train[data_columns[0]].max()),
columns=data_columns)
target_predicted_linear_regression = linear_regression.predict(data_test)
target_predicted_tree = tree.predict(data_test)
```
Create a scatter plot containing the training samples and superimpose the
predictions of both model on the top.
```
import matplotlib.pyplot as plt
import seaborn as sns
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted_linear_regression,
label="Linear regression")
plt.plot(data_test, target_predicted_tree, label="Decision tree")
plt.legend()
_ = plt.title("Prediction of linear model and a decision tree")
```
The predictions that we got were within the range of feature values seen
during training. In some sense, we observe the capabilities of our model to
interpolate.
Now, we will check the extrapolation capabilities of each model. Create a
dataset containing the value of your previous dataset. Besides, add values
below and above the minimum and the maximum of the flipper length seen
during training.
```
offset = 30
data_test = pd.DataFrame(np.arange(data_train[data_columns[0]].min() - offset,
data_train[data_columns[0]].max() + offset),
columns=data_columns)
```
Finally, make predictions with both models on this new testing set. Repeat
the plotting of the previous exercise.
```
target_predicted_linear_regression = linear_regression.predict(data_test)
target_predicted_tree = tree.predict(data_test)
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted_linear_regression,
label="Linear regression")
plt.plot(data_test, target_predicted_tree, label="Decision tree")
plt.legend()
_ = plt.title("Prediction of linear model and a decision tree")
```
The linear model will extrapolate using the fitted model for flipper lengths
< 175 mm and > 235 mm. In fact, we are using the model parametrization to
make this predictions.
As mentioned, decision trees are non-parametric models and we observe that
they cannot extrapolate. For flipper lengths below the minimum, the mass of
the penguin in the training data with the shortest flipper length will always
be predicted. Similarly, for flipper lengths above the maximum, the mass of
the penguin in the training data with the longest flipper will always be
predicted.
|
github_jupyter
|
import pandas as pd
penguins = pd.read_csv("../datasets/penguins_regression.csv")
data_columns = ["Flipper Length (mm)"]
target_column = "Body Mass (g)"
data_train, target_train = penguins[data_columns], penguins[target_column]
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
linear_regression = LinearRegression()
tree = DecisionTreeRegressor(max_depth=3)
linear_regression.fit(data_train, target_train)
tree.fit(data_train, target_train)
import numpy as np
data_test = pd.DataFrame(np.arange(data_train[data_columns[0]].min(),
data_train[data_columns[0]].max()),
columns=data_columns)
target_predicted_linear_regression = linear_regression.predict(data_test)
target_predicted_tree = tree.predict(data_test)
import matplotlib.pyplot as plt
import seaborn as sns
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted_linear_regression,
label="Linear regression")
plt.plot(data_test, target_predicted_tree, label="Decision tree")
plt.legend()
_ = plt.title("Prediction of linear model and a decision tree")
offset = 30
data_test = pd.DataFrame(np.arange(data_train[data_columns[0]].min() - offset,
data_train[data_columns[0]].max() + offset),
columns=data_columns)
target_predicted_linear_regression = linear_regression.predict(data_test)
target_predicted_tree = tree.predict(data_test)
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted_linear_regression,
label="Linear regression")
plt.plot(data_test, target_predicted_tree, label="Decision tree")
plt.legend()
_ = plt.title("Prediction of linear model and a decision tree")
| 0.641759 | 0.994375 |
```
# Import required packages
from __future__ import division, print_function # Imports from __future__ since we're running Python 2
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
random_state=0
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
rng = np.random.RandomState(seed=random_state)
%matplotlib inline
path_data = os.path.join(os.getcwd(),'train_data_ongoing.csv')
flights_train = pd.read_csv(path_data, delimiter = ',')
path_data = os.path.join(os.getcwd(),'test_data_ongoing.csv')
flights_test = pd.read_csv(path_data, delimiter = ',')
flights_train.loc[flights_train['ARR_DELAY_GROUP'] > 0, 'ARR_DELAY_GROUP'] = 1
flights_test.loc[flights_test['ARR_DELAY_GROUP'] > 0, 'ARR_DELAY_GROUP'] = 1
X_train_full = flights_train.drop('ARR_DELAY_GROUP', axis=1).values.astype(np.float) # Training features
y_train_full = flights_train['ARR_DELAY_GROUP'].values # Training labels
X_test = flights_test.drop('ARR_DELAY_GROUP', axis=1).values.astype(np.float) # Training features
y_test = flights_test['ARR_DELAY_GROUP'].values # Training labels
X_train, X_val, y_train, y_val = train_test_split(X_train_full, y_train_full,
test_size=0.2, random_state=random_state)
sc = StandardScaler().fit(X_train)
X_train_sc = sc.transform(X_train)
X_val_sc = sc.transform(X_val)
X_test_sc = sc.transform(X_test)
from sklearn.linear_model import LogisticRegression
from sklearn.neural_network import MLPClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.gaussian_process import GaussianProcessClassifier
from sklearn.gaussian_process.kernels import RBF
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis, QuadraticDiscriminantAnalysis
from sklearn.metrics import log_loss
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score
from scipy.stats import mode # Computes the mode of a signal
from sklearn.metrics import accuracy_score
from sklearn.metrics import cohen_kappa_score, make_scorer
kappa_scorer = make_scorer(cohen_kappa_score)
names = ["Logistic Regression", "Nearest Neighbors",
"Decision Tree", "Random Forest",
"Naive Bayes", "LDA", "QDA","Neural Net (Multi-layer perceptron)"]
classifiers = [
LogisticRegression(solver='lbfgs', multi_class='multinomial'),
KNeighborsClassifier(n_neighbors=10),
DecisionTreeClassifier(max_depth=10),
RandomForestClassifier(max_depth=10, n_estimators=50,random_state=random_state),
GaussianNB(),
LinearDiscriminantAnalysis(),
QuadraticDiscriminantAnalysis(),
MLPClassifier(random_state=random_state)]
ca_score = {} # Classification accuracy
ce_score = {} # Cross-entropy
kc_score = {} #kappa score
print('Classification performance on validation set:')
for name, clf in zip(names, classifiers):
clf.fit(X_train_sc, y_train)
ca_score[name] = clf.score(X_val_sc, y_val)
ce_score[name] = log_loss(y_val, clf.predict_proba(X_val_sc))
kc_score[name] = cohen_kappa_score(y_val,clf.predict(X_val_sc))
print ("{}, accuracy: {:.3f}, log-loss: {:.3f}, kappa score: {:.3f}".format(name, ca_score[name], ce_score[name], kc_score[name]))
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import KFold
cv = KFold(n_splits=3, shuffle=True, random_state=random_state)
svc = LogisticRegression(penalty='l2'
,fit_intercept=True
,solver='newton-cg'
,multi_class='multinomial'
,max_iter=4000)
parameters = {'C': np.logspace(-3,3,7)}
svc_clf = GridSearchCV(estimator=svc, cv=cv, param_grid=parameters, scoring='accuracy')
svc_clf.fit(X_train_sc, y_train)
print("Best setting of C parameter for Logistic Regression: {}".format(svc_clf.best_params_["C"]))
print("Best cross-validated score: {:.3f}".
format(svc_clf.best_score_))
print("Classification accuracy on validation set: {:.3f}".format(svc_clf.score(X_val_sc,y_val)))
from sklearn import metrics
from sklearn.metrics import confusion_matrix
kappa_scorer = make_scorer(cohen_kappa_score)
names = ["Logistic Regression"]
classifiers = [LogisticRegression(penalty='l2'
,C=0.1
,fit_intercept=True
,solver='newton-cg'
,multi_class='multinomial'
,max_iter=4000)]
ca_score = {} # Classification accuracy
ce_score = {} # Cross-entropy
kc_score = {} #kappa score
print('Classification performance on validation set:')
for name, clf in zip(names, classifiers):
clf.fit(X_train_sc, y_train)
ca_score[name] = clf.score(X_val_sc, y_val)
ce_score[name] = log_loss(y_val, clf.predict_proba(X_val_sc))
kc_score[name] = cohen_kappa_score(y_val,clf.predict(X_val_sc))
precision = metrics.precision_score(y_val, clf.predict(X_val_sc), average='macro')
recall = metrics.recall_score(y_val, clf.predict(X_val_sc), average='macro')
f1 = metrics.f1_score(y_val, clf.predict(X_val_sc), average='weighted')
print ("Validation {}, accuracy: {:.3f}, log-loss: {:.3f}, kappa score: {:.3f}".format(name, ca_score[name], ce_score[name], kc_score[name]))
print ("Validation {},precision: {:.3f}, recall: {:.3f}, f1: {:.3f}".format(name, precision,recall,f1))
test1 = clf.score(X_test_sc, y_test)
test2 = log_loss(y_test, clf.predict_proba(X_test_sc))
test3 = cohen_kappa_score(y_test,clf.predict(X_test_sc))
print ("Test {}, accuracy: {:.3f}, log-loss: {:.3f}, kappa score: {:.3f}".format(name, test1, test2, test3))
precision = metrics.precision_score(y_test,clf.predict(X_test_sc), average='macro')
recall = metrics.recall_score(y_test,clf.predict(X_test_sc), average='macro')
f1 = metrics.f1_score(y_test,clf.predict(X_test_sc), average='weighted')
print ("Test {},precision: {:.3f}, recall: {:.3f}, f1: {:.3f}".format(name, precision,recall,f1))
```
|
github_jupyter
|
# Import required packages
from __future__ import division, print_function # Imports from __future__ since we're running Python 2
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
random_state=0
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
rng = np.random.RandomState(seed=random_state)
%matplotlib inline
path_data = os.path.join(os.getcwd(),'train_data_ongoing.csv')
flights_train = pd.read_csv(path_data, delimiter = ',')
path_data = os.path.join(os.getcwd(),'test_data_ongoing.csv')
flights_test = pd.read_csv(path_data, delimiter = ',')
flights_train.loc[flights_train['ARR_DELAY_GROUP'] > 0, 'ARR_DELAY_GROUP'] = 1
flights_test.loc[flights_test['ARR_DELAY_GROUP'] > 0, 'ARR_DELAY_GROUP'] = 1
X_train_full = flights_train.drop('ARR_DELAY_GROUP', axis=1).values.astype(np.float) # Training features
y_train_full = flights_train['ARR_DELAY_GROUP'].values # Training labels
X_test = flights_test.drop('ARR_DELAY_GROUP', axis=1).values.astype(np.float) # Training features
y_test = flights_test['ARR_DELAY_GROUP'].values # Training labels
X_train, X_val, y_train, y_val = train_test_split(X_train_full, y_train_full,
test_size=0.2, random_state=random_state)
sc = StandardScaler().fit(X_train)
X_train_sc = sc.transform(X_train)
X_val_sc = sc.transform(X_val)
X_test_sc = sc.transform(X_test)
from sklearn.linear_model import LogisticRegression
from sklearn.neural_network import MLPClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.gaussian_process import GaussianProcessClassifier
from sklearn.gaussian_process.kernels import RBF
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis, QuadraticDiscriminantAnalysis
from sklearn.metrics import log_loss
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score
from scipy.stats import mode # Computes the mode of a signal
from sklearn.metrics import accuracy_score
from sklearn.metrics import cohen_kappa_score, make_scorer
kappa_scorer = make_scorer(cohen_kappa_score)
names = ["Logistic Regression", "Nearest Neighbors",
"Decision Tree", "Random Forest",
"Naive Bayes", "LDA", "QDA","Neural Net (Multi-layer perceptron)"]
classifiers = [
LogisticRegression(solver='lbfgs', multi_class='multinomial'),
KNeighborsClassifier(n_neighbors=10),
DecisionTreeClassifier(max_depth=10),
RandomForestClassifier(max_depth=10, n_estimators=50,random_state=random_state),
GaussianNB(),
LinearDiscriminantAnalysis(),
QuadraticDiscriminantAnalysis(),
MLPClassifier(random_state=random_state)]
ca_score = {} # Classification accuracy
ce_score = {} # Cross-entropy
kc_score = {} #kappa score
print('Classification performance on validation set:')
for name, clf in zip(names, classifiers):
clf.fit(X_train_sc, y_train)
ca_score[name] = clf.score(X_val_sc, y_val)
ce_score[name] = log_loss(y_val, clf.predict_proba(X_val_sc))
kc_score[name] = cohen_kappa_score(y_val,clf.predict(X_val_sc))
print ("{}, accuracy: {:.3f}, log-loss: {:.3f}, kappa score: {:.3f}".format(name, ca_score[name], ce_score[name], kc_score[name]))
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import KFold
cv = KFold(n_splits=3, shuffle=True, random_state=random_state)
svc = LogisticRegression(penalty='l2'
,fit_intercept=True
,solver='newton-cg'
,multi_class='multinomial'
,max_iter=4000)
parameters = {'C': np.logspace(-3,3,7)}
svc_clf = GridSearchCV(estimator=svc, cv=cv, param_grid=parameters, scoring='accuracy')
svc_clf.fit(X_train_sc, y_train)
print("Best setting of C parameter for Logistic Regression: {}".format(svc_clf.best_params_["C"]))
print("Best cross-validated score: {:.3f}".
format(svc_clf.best_score_))
print("Classification accuracy on validation set: {:.3f}".format(svc_clf.score(X_val_sc,y_val)))
from sklearn import metrics
from sklearn.metrics import confusion_matrix
kappa_scorer = make_scorer(cohen_kappa_score)
names = ["Logistic Regression"]
classifiers = [LogisticRegression(penalty='l2'
,C=0.1
,fit_intercept=True
,solver='newton-cg'
,multi_class='multinomial'
,max_iter=4000)]
ca_score = {} # Classification accuracy
ce_score = {} # Cross-entropy
kc_score = {} #kappa score
print('Classification performance on validation set:')
for name, clf in zip(names, classifiers):
clf.fit(X_train_sc, y_train)
ca_score[name] = clf.score(X_val_sc, y_val)
ce_score[name] = log_loss(y_val, clf.predict_proba(X_val_sc))
kc_score[name] = cohen_kappa_score(y_val,clf.predict(X_val_sc))
precision = metrics.precision_score(y_val, clf.predict(X_val_sc), average='macro')
recall = metrics.recall_score(y_val, clf.predict(X_val_sc), average='macro')
f1 = metrics.f1_score(y_val, clf.predict(X_val_sc), average='weighted')
print ("Validation {}, accuracy: {:.3f}, log-loss: {:.3f}, kappa score: {:.3f}".format(name, ca_score[name], ce_score[name], kc_score[name]))
print ("Validation {},precision: {:.3f}, recall: {:.3f}, f1: {:.3f}".format(name, precision,recall,f1))
test1 = clf.score(X_test_sc, y_test)
test2 = log_loss(y_test, clf.predict_proba(X_test_sc))
test3 = cohen_kappa_score(y_test,clf.predict(X_test_sc))
print ("Test {}, accuracy: {:.3f}, log-loss: {:.3f}, kappa score: {:.3f}".format(name, test1, test2, test3))
precision = metrics.precision_score(y_test,clf.predict(X_test_sc), average='macro')
recall = metrics.recall_score(y_test,clf.predict(X_test_sc), average='macro')
f1 = metrics.f1_score(y_test,clf.predict(X_test_sc), average='weighted')
print ("Test {},precision: {:.3f}, recall: {:.3f}, f1: {:.3f}".format(name, precision,recall,f1))
| 0.687 | 0.39065 |
# Flax seq2seq Example
<a href="https://colab.research.google.com/github/google/flax/blob/main/examples/seq2seq/seq2seq.ipynb" ><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Demonstration notebook for
https://github.com/google/flax/tree/main/examples/seq2seq
The **Flax Notebook Workflow**:
1. Run the entire notebook end-to-end and check out the outputs.
- This will open Python files in the right-hand editor!
- You'll be able to interactively explore metrics in TensorBoard.
2. Change some of the hyperparameters in the command-line flags in `train.py` for different hyperparameters. Check out the updated TensorBoard plots.
3. Update the code in `train.py`, `models.py`, and `input_pipeline.py`.
Thanks to `%autoreload`, any changes you make in the file will
automatically appear in the notebook. Some ideas to get you started:
- Change the model.
- Log some per-batch metrics during training.
- Add new hyperparameters to `models.py` and use them in `train.py`.
- Train on a different vocabulary by initializing `CharacterTable` with a
different character set.
4. At any time, feel free to paste code from the source code into the notebook
and modify it directly there!
## Setup
```
# Install CLU & Flax.
!pip install -q clu flax
example_directory = 'examples/seq2seq'
editor_relpaths = ('train.py', 'input_pipeline.py', 'models.py')
repo, branch = 'https://github.com/google/flax', 'main'
# (If you run this code in Jupyter[lab], then you're already in the
# example directory and nothing needs to be done.)
#@markdown **Fetch newest Flax, copy example code**
#@markdown
#@markdown **If you select no** below, then the files will be stored on the
#@markdown *ephemeral* Colab VM. **After some time of inactivity, this VM will
#@markdown be restarted an any changes are lost**.
#@markdown
#@markdown **If you select yes** below, then you will be asked for your
#@markdown credentials to mount your personal Google Drive. In this case, all
#@markdown changes you make will be *persisted*, and even if you re-run the
#@markdown Colab later on, the files will still be the same (you can of course
#@markdown remove directories inside your Drive's `flax/` root if you want to
#@markdown manually revert these files).
if 'google.colab' in str(get_ipython()):
import os
os.chdir('/content')
# Download Flax repo from Github.
if not os.path.isdir('flaxrepo'):
!git clone --depth=1 -b $branch $repo flaxrepo
# Copy example files & change directory.
mount_gdrive = 'no' #@param ['yes', 'no']
if mount_gdrive == 'yes':
DISCLAIMER = 'Note : Editing in your Google Drive, changes will persist.'
from google.colab import drive
drive.mount('/content/gdrive')
example_root_path = f'/content/gdrive/My Drive/flax/{example_directory}'
else:
DISCLAIMER = 'WARNING : Editing in VM - changes lost after reboot!!'
example_root_path = f'/content/{example_directory}'
from IPython import display
display.display(display.HTML(
f'<h1 style="color:red;" class="blink">{DISCLAIMER}</h1>'))
if not os.path.isdir(example_root_path):
os.makedirs(example_root_path)
!cp -r flaxrepo/$example_directory/* "$example_root_path"
os.chdir(example_root_path)
from google.colab import files
for relpath in editor_relpaths:
s = open(f'{example_root_path}/{relpath}').read()
open(f'{example_root_path}/{relpath}', 'w').write(
f'## {DISCLAIMER}\n' + '#' * (len(DISCLAIMER) + 3) + '\n\n' + s)
files.view(f'{example_root_path}/{relpath}')
# Note : In Colab, above cell changed the working directory.
!pwd
```
## Imports
```
from absl import app
app.parse_flags_with_usage(['seq2seq'])
from absl import logging
logging.set_verbosity(logging.INFO)
import jax
# Local imports from current directory - auto reload.
# Any changes you make to the three imported files will appear automatically.
%load_ext autoreload
%autoreload 2
import input_pipeline
import models
import train
```
## Dataset
```
# Examples are generated on the fly.
ctable = input_pipeline.CharacterTable('0123456789+= ')
list(ctable.generate_examples(5))
batch = ctable.get_batch(5)
# A single query (/answer) is one-hot encoded.
batch['query'][0]
# Note how CTABLE encodes PAD=0, EOS=1, '0'=2, '1'=3, ...
ctable.decode_onehot(batch['query'][:1])
```
## Training
```
# Get a live update during training - use the "refresh" button!
# (In Jupyter[lab] start "tensorboard" in the local directory instead.)
if 'google.colab' in str(get_ipython()):
%load_ext tensorboard
%tensorboard --logdir=./workdirs
import time
workdir = f'./workdirs/{int(time.time())}'
# Train 2k steps & log 20 times.
app.parse_flags_with_usage([
'seq2seq',
'--num_train_steps=2000',
'--decode_frequency=100',
])
state = train.train_and_evaluate(workdir=workdir)
if 'google.colab' in str(get_ipython()):
#@markdown You can upload the training results directly to https://tensorboard.dev
#@markdown
#@markdown Note that everbody with the link will be able to see the data.
upload_data = 'yes' #@param ['yes', 'no']
if upload_data == 'yes':
!tensorboard dev upload --one_shot --logdir ./workdirs --name 'Flax examples/seq2seq (Colab)'
```
## Inference
```
inputs = ctable.encode_onehot(['2+40'])
# batch, max_length, vocab_size
inputs.shape
# Using different random seeds generates different samples.
preds = train.decode(state.params, inputs, jax.random.PRNGKey(0), ctable)
ctable.decode_onehot(preds)
```
|
github_jupyter
|
# Install CLU & Flax.
!pip install -q clu flax
example_directory = 'examples/seq2seq'
editor_relpaths = ('train.py', 'input_pipeline.py', 'models.py')
repo, branch = 'https://github.com/google/flax', 'main'
# (If you run this code in Jupyter[lab], then you're already in the
# example directory and nothing needs to be done.)
#@markdown **Fetch newest Flax, copy example code**
#@markdown
#@markdown **If you select no** below, then the files will be stored on the
#@markdown *ephemeral* Colab VM. **After some time of inactivity, this VM will
#@markdown be restarted an any changes are lost**.
#@markdown
#@markdown **If you select yes** below, then you will be asked for your
#@markdown credentials to mount your personal Google Drive. In this case, all
#@markdown changes you make will be *persisted*, and even if you re-run the
#@markdown Colab later on, the files will still be the same (you can of course
#@markdown remove directories inside your Drive's `flax/` root if you want to
#@markdown manually revert these files).
if 'google.colab' in str(get_ipython()):
import os
os.chdir('/content')
# Download Flax repo from Github.
if not os.path.isdir('flaxrepo'):
!git clone --depth=1 -b $branch $repo flaxrepo
# Copy example files & change directory.
mount_gdrive = 'no' #@param ['yes', 'no']
if mount_gdrive == 'yes':
DISCLAIMER = 'Note : Editing in your Google Drive, changes will persist.'
from google.colab import drive
drive.mount('/content/gdrive')
example_root_path = f'/content/gdrive/My Drive/flax/{example_directory}'
else:
DISCLAIMER = 'WARNING : Editing in VM - changes lost after reboot!!'
example_root_path = f'/content/{example_directory}'
from IPython import display
display.display(display.HTML(
f'<h1 style="color:red;" class="blink">{DISCLAIMER}</h1>'))
if not os.path.isdir(example_root_path):
os.makedirs(example_root_path)
!cp -r flaxrepo/$example_directory/* "$example_root_path"
os.chdir(example_root_path)
from google.colab import files
for relpath in editor_relpaths:
s = open(f'{example_root_path}/{relpath}').read()
open(f'{example_root_path}/{relpath}', 'w').write(
f'## {DISCLAIMER}\n' + '#' * (len(DISCLAIMER) + 3) + '\n\n' + s)
files.view(f'{example_root_path}/{relpath}')
# Note : In Colab, above cell changed the working directory.
!pwd
from absl import app
app.parse_flags_with_usage(['seq2seq'])
from absl import logging
logging.set_verbosity(logging.INFO)
import jax
# Local imports from current directory - auto reload.
# Any changes you make to the three imported files will appear automatically.
%load_ext autoreload
%autoreload 2
import input_pipeline
import models
import train
# Examples are generated on the fly.
ctable = input_pipeline.CharacterTable('0123456789+= ')
list(ctable.generate_examples(5))
batch = ctable.get_batch(5)
# A single query (/answer) is one-hot encoded.
batch['query'][0]
# Note how CTABLE encodes PAD=0, EOS=1, '0'=2, '1'=3, ...
ctable.decode_onehot(batch['query'][:1])
# Get a live update during training - use the "refresh" button!
# (In Jupyter[lab] start "tensorboard" in the local directory instead.)
if 'google.colab' in str(get_ipython()):
%load_ext tensorboard
%tensorboard --logdir=./workdirs
import time
workdir = f'./workdirs/{int(time.time())}'
# Train 2k steps & log 20 times.
app.parse_flags_with_usage([
'seq2seq',
'--num_train_steps=2000',
'--decode_frequency=100',
])
state = train.train_and_evaluate(workdir=workdir)
if 'google.colab' in str(get_ipython()):
#@markdown You can upload the training results directly to https://tensorboard.dev
#@markdown
#@markdown Note that everbody with the link will be able to see the data.
upload_data = 'yes' #@param ['yes', 'no']
if upload_data == 'yes':
!tensorboard dev upload --one_shot --logdir ./workdirs --name 'Flax examples/seq2seq (Colab)'
inputs = ctable.encode_onehot(['2+40'])
# batch, max_length, vocab_size
inputs.shape
# Using different random seeds generates different samples.
preds = train.decode(state.params, inputs, jax.random.PRNGKey(0), ctable)
ctable.decode_onehot(preds)
| 0.595022 | 0.891717 |
# VOOF (Visualizing 1/f)
```
%pylab inline
import matplotlib.pyplot as plt
from foof import syn
from foof.fit import FOOF
import numpy as np
import os
import scipy.io
import scipy.signal
import pandas as pd
from foof import syn
import seaborn as sns
import matplotlib.patches as patches
```
### Making some fake data with foof.syn
```
# Data settings
N = 20 # Number of PSDs
A = 0.1 # Peak height (0.005 give small but still clear; 0.2 gives huge peaks)
A2 = 0.02 # Peak height (0.005 give small but still clear; 0.2 gives huge peaks)
f = 8 # Frequency (3-100, Hz)
f2 = 25 # Frequency (3-100, Hz)
f_sig = 2 # Peak bandwidth (Hz)
f_sig2 = 2 # Peak bandwidth (Hz)
chi = 2 # Slope of the PSD (1-3)
noi = 0.05 # Noise to add to the PSD (0-0.4)
res = 0.5 # Spectral resolution (1-0.1, Hz)
fs, X = syn.synthesize(N, k=A, f=f, f_sig=f_sig, chi=chi,
f0=3, fmax=50, res=res, noi=noi)
fs,X2 = syn.synthesize(N, k=A2, f=f2, f_sig=f_sig2, chi=chi,
f0=3, fmax=50, res=res, noi=noi)
two_peak_spec= mean([X,X2],0)
print two_peak_spec.shape
# And plot
plt.plot(fs, np.mean(two_peak_spec,1))
plt.xlabel("F (Hz)")
plt.ylabel("PSD")
```
### Foof-ing the fake data
```
# Foof settings
max_components = 10 # Max number of Gaussians to consider
min_p = 0.3 # Min total probablity a peak must contain to be included
flatten_thresh = 0.05 # Useful for accounting large peaks and 'wiggly' PSDs, which often occur in real data (0-1)
foof = FOOF(min_p=min_p,
res=res,
fmin=fs.min(),
fmax=fs.max())
foof.model(fs, two_peak_spec) # Fit PSD; Run time: 500-800 ms
print foof.centers_
print foof.stdevs_
print foof.powers_
print foof.chi_
```
### Some helpful functions
```
def find_visual_wavelength_of_frequency_center(cf,freq_lo,freq_hi,vis_lo,vis_hi,res):
"""Finds the visual wavelength of a center frequency"""
spec = np.linspace(freq_lo,freq_hi,(freq_hi+1-freq_lo)/res);
closest_ind = min(range(len(spec)), key=lambda i: abs(spec[i]-cf))
visual_x_frequency_spectrum = np.linspace(visual_spectrum[0],visual_spectrum[1],num = (frequency_spectrum[1]-frequency_spectrum[0])/res)
flipped_ind = len(visual_x_frequency_spectrum) - closest_ind
wavelength = visual_x_frequency_spectrum[flipped_ind]
return wavelength
def chi_to_wavelength(chi,slope_range,visual_spectrum):
"""Takes a slope and returns the 'visual wavelength associated with it'"""
chi_to_wl = np.linspace(0,4,num = (visual_spectrum[1]-visual_spectrum[0]))
closest_wl_ind = min(range(len(chi_to_wl)), key=lambda i: abs(chi_to_wl[i]-chi))
vis = np.linspace(visual_spectrum[0],visual_spectrum[1],num=visual_spectrum[1]-visual_spectrum[0]+1)
return vis[closest_wl_ind]
def wav2RGB(wavelength):
"""very helpful code I stole from http://codingmess.blogspot.com/2009/05/conversion-of-wavelength-in-nanometers.html"""
w = int(wavelength)
# colour
if w >= 380 and w < 440:
R = -(w - 440.) / (440. - 350.)
G = 0.0
B = 1.0
elif w >= 440 and w < 490:
R = 0.0
G = (w - 440.) / (490. - 440.)
B = 1.0
elif w >= 490 and w < 510:
R = 0.0
G = 1.0
B = -(w - 510.) / (510. - 490.)
elif w >= 510 and w < 580:
R = (w - 510.) / (580. - 510.)
G = 1.0
B = 0.0
elif w >= 580 and w < 645:
R = 1.0
G = -(w - 645.) / (645. - 580.)
B = 0.0
elif w >= 645 and w <= 780:
R = 1.0
G = 0.0
B = 0.0
else:
R = 0.0
G = 0.0
B = 0.0
# intensity correction
if w >= 380 and w < 420:
SSS = 0.3 + 0.7*(w - 350) / (420 - 350)
elif w >= 420 and w <= 700:
SSS = 1.0
elif w > 700 and w <= 780:
SSS = 0.3 + 0.7*(780 - w) / (780 - 700)
else:
SSS = 0.0
SSS *= 255
return [int(SSS*R), int(SSS*G), int(SSS*B)]
def rgb_to_hex(rgb):
"""Helpful code stolen from http://stackoverflow.com/questions/214359/converting-hex-color-to-rgb-and-vice-versa"""
return '#%02x%02x%02x' % rgb
```
### defining fx, visual, and slope ranges
```
visual_spectrum = [380, 750];
frequency_spectrum = [1,50];
slope_range = [0,4];
```
# Voofing
```
bandwidth_fracts = [i/sum(foof.stdevs_) for i in foof.stdevs_]
power_fracts = [i/sum(foof.powers_) for i in foof.powers_]
visual_wavelengths = [find_visual_wavelength_of_frequency_center(i,frequency_spectrum[0],frequency_spectrum[1],visual_spectrum[0],visual_spectrum[1],res) for i in foof.centers_]
fig1 = plt.figure(figsize=(8,8))
ax1 = fig1.add_subplot(111, aspect='equal')
i=0
for bw in xrange(len(foof.stdevs_)):
ax1.add_patch(
patches.Rectangle(
(i, 0), # (x,y)
bandwidth_fracts[bw], # width
1, # height
facecolor= rgb_to_hex(tuple(wav2RGB(visual_wavelengths[bw]))),
alpha = power_fracts[bw],
label = 'peak at ' +str(foof.centers_[bw])+'hz. bw='+ str(foof.stdevs_[bw])
)
)
i=i+bandwidth_fracts[bw]
#print bw
ax1.set_xlim([0,1])
bump = 1/foof.chi_
polygon_params = [[0,(foof.chi_/10)+bump],[0,0],[0-bump/(-foof.chi_/10),0]]; #some y=mx+b stuff
ax1.add_patch(patches.Polygon(polygon_params, True,label='slope= -'+str(foof.chi_),
facecolor = rgb_to_hex(tuple(wav2RGB(chi_to_wavelength(foof.chi_,slope_range,visual_spectrum))))))
plt.title('VOOF Demo')
plt.legend()
```
|
github_jupyter
|
%pylab inline
import matplotlib.pyplot as plt
from foof import syn
from foof.fit import FOOF
import numpy as np
import os
import scipy.io
import scipy.signal
import pandas as pd
from foof import syn
import seaborn as sns
import matplotlib.patches as patches
# Data settings
N = 20 # Number of PSDs
A = 0.1 # Peak height (0.005 give small but still clear; 0.2 gives huge peaks)
A2 = 0.02 # Peak height (0.005 give small but still clear; 0.2 gives huge peaks)
f = 8 # Frequency (3-100, Hz)
f2 = 25 # Frequency (3-100, Hz)
f_sig = 2 # Peak bandwidth (Hz)
f_sig2 = 2 # Peak bandwidth (Hz)
chi = 2 # Slope of the PSD (1-3)
noi = 0.05 # Noise to add to the PSD (0-0.4)
res = 0.5 # Spectral resolution (1-0.1, Hz)
fs, X = syn.synthesize(N, k=A, f=f, f_sig=f_sig, chi=chi,
f0=3, fmax=50, res=res, noi=noi)
fs,X2 = syn.synthesize(N, k=A2, f=f2, f_sig=f_sig2, chi=chi,
f0=3, fmax=50, res=res, noi=noi)
two_peak_spec= mean([X,X2],0)
print two_peak_spec.shape
# And plot
plt.plot(fs, np.mean(two_peak_spec,1))
plt.xlabel("F (Hz)")
plt.ylabel("PSD")
# Foof settings
max_components = 10 # Max number of Gaussians to consider
min_p = 0.3 # Min total probablity a peak must contain to be included
flatten_thresh = 0.05 # Useful for accounting large peaks and 'wiggly' PSDs, which often occur in real data (0-1)
foof = FOOF(min_p=min_p,
res=res,
fmin=fs.min(),
fmax=fs.max())
foof.model(fs, two_peak_spec) # Fit PSD; Run time: 500-800 ms
print foof.centers_
print foof.stdevs_
print foof.powers_
print foof.chi_
def find_visual_wavelength_of_frequency_center(cf,freq_lo,freq_hi,vis_lo,vis_hi,res):
"""Finds the visual wavelength of a center frequency"""
spec = np.linspace(freq_lo,freq_hi,(freq_hi+1-freq_lo)/res);
closest_ind = min(range(len(spec)), key=lambda i: abs(spec[i]-cf))
visual_x_frequency_spectrum = np.linspace(visual_spectrum[0],visual_spectrum[1],num = (frequency_spectrum[1]-frequency_spectrum[0])/res)
flipped_ind = len(visual_x_frequency_spectrum) - closest_ind
wavelength = visual_x_frequency_spectrum[flipped_ind]
return wavelength
def chi_to_wavelength(chi,slope_range,visual_spectrum):
"""Takes a slope and returns the 'visual wavelength associated with it'"""
chi_to_wl = np.linspace(0,4,num = (visual_spectrum[1]-visual_spectrum[0]))
closest_wl_ind = min(range(len(chi_to_wl)), key=lambda i: abs(chi_to_wl[i]-chi))
vis = np.linspace(visual_spectrum[0],visual_spectrum[1],num=visual_spectrum[1]-visual_spectrum[0]+1)
return vis[closest_wl_ind]
def wav2RGB(wavelength):
"""very helpful code I stole from http://codingmess.blogspot.com/2009/05/conversion-of-wavelength-in-nanometers.html"""
w = int(wavelength)
# colour
if w >= 380 and w < 440:
R = -(w - 440.) / (440. - 350.)
G = 0.0
B = 1.0
elif w >= 440 and w < 490:
R = 0.0
G = (w - 440.) / (490. - 440.)
B = 1.0
elif w >= 490 and w < 510:
R = 0.0
G = 1.0
B = -(w - 510.) / (510. - 490.)
elif w >= 510 and w < 580:
R = (w - 510.) / (580. - 510.)
G = 1.0
B = 0.0
elif w >= 580 and w < 645:
R = 1.0
G = -(w - 645.) / (645. - 580.)
B = 0.0
elif w >= 645 and w <= 780:
R = 1.0
G = 0.0
B = 0.0
else:
R = 0.0
G = 0.0
B = 0.0
# intensity correction
if w >= 380 and w < 420:
SSS = 0.3 + 0.7*(w - 350) / (420 - 350)
elif w >= 420 and w <= 700:
SSS = 1.0
elif w > 700 and w <= 780:
SSS = 0.3 + 0.7*(780 - w) / (780 - 700)
else:
SSS = 0.0
SSS *= 255
return [int(SSS*R), int(SSS*G), int(SSS*B)]
def rgb_to_hex(rgb):
"""Helpful code stolen from http://stackoverflow.com/questions/214359/converting-hex-color-to-rgb-and-vice-versa"""
return '#%02x%02x%02x' % rgb
visual_spectrum = [380, 750];
frequency_spectrum = [1,50];
slope_range = [0,4];
bandwidth_fracts = [i/sum(foof.stdevs_) for i in foof.stdevs_]
power_fracts = [i/sum(foof.powers_) for i in foof.powers_]
visual_wavelengths = [find_visual_wavelength_of_frequency_center(i,frequency_spectrum[0],frequency_spectrum[1],visual_spectrum[0],visual_spectrum[1],res) for i in foof.centers_]
fig1 = plt.figure(figsize=(8,8))
ax1 = fig1.add_subplot(111, aspect='equal')
i=0
for bw in xrange(len(foof.stdevs_)):
ax1.add_patch(
patches.Rectangle(
(i, 0), # (x,y)
bandwidth_fracts[bw], # width
1, # height
facecolor= rgb_to_hex(tuple(wav2RGB(visual_wavelengths[bw]))),
alpha = power_fracts[bw],
label = 'peak at ' +str(foof.centers_[bw])+'hz. bw='+ str(foof.stdevs_[bw])
)
)
i=i+bandwidth_fracts[bw]
#print bw
ax1.set_xlim([0,1])
bump = 1/foof.chi_
polygon_params = [[0,(foof.chi_/10)+bump],[0,0],[0-bump/(-foof.chi_/10),0]]; #some y=mx+b stuff
ax1.add_patch(patches.Polygon(polygon_params, True,label='slope= -'+str(foof.chi_),
facecolor = rgb_to_hex(tuple(wav2RGB(chi_to_wavelength(foof.chi_,slope_range,visual_spectrum))))))
plt.title('VOOF Demo')
plt.legend()
| 0.47171 | 0.832169 |
## Interest Rate
---
Suppose $W(t)$ is the wealth at time $t$ ($t\geqq 0$) and $r$ is an interest rate.
+ **Simple interest rate**
$$
W(t) = (1+rt)W(0).
$$
+ **One-year compound interest rate**
$$
W(t) = (1+r)^t W(0).
$$
+ **$\frac1{M}$-year compound interest rate**
$$
W(t) = \left(1+\frac{r}{M}\right)^{Mt} W(0).
$$
+ **Continuous compoud interest rate**
$$
W(t) = e^{rt}W(0).
$$
## Python Code: Simple Rate vs. Compounding Rate
---
The following command enables plotting within cells.
```
%matplotlib inline
```
`import` literally imports a package named NumPy in Python. NumPy enable us to use vectors and matrices in Python. It also comes with numerous functions for mathematical computation. `as np` means that we use `np` as a abbreviation of `numpy`.
```
import numpy as np
```
This line imports PyPlot, a collection of functions for 2D/3D graphics.
```
import matplotlib.pyplot as plt
```
`r` is the interest rate. `Maturity` is the number of years until the end of the investment period.
```
r = 0.2
Maturity = 10
```
`Simple_Rate` contains the amount of wealth at each year when we invest funds with simple interest rate.
```
Simple_Rate = 1.0 + r * np.linspace(0, Maturity, Maturity + 1)
```
`linspace(0, Maturity, Maturity + 1)` creates a vector of grid points. The first number in `linspace(0, Maturity, Maturity + 1)` is the starting point, the second is the end point, and the third is the number of grid points. `print` show the content of the object.
```
print(np.linspace(0, Maturity, Maturity + 1))
print(Simple_Rate)
```
`Compound_1year` contains the amount of wealth at each year when we invest funds with one-year compound interest rate.
```
Compound_1year = np.hstack((1.0, np.cumprod(np.tile(1.0 + r, Maturity))))
```
`tile` creates a larger vector/matrix by tiling the same vector/matrix.
```
print(np.tile(1.0 + r, Maturity))
```
`cumprod` computes *cumulative products*, i.e.,
```
print(np.cumprod(np.tile(1.0 + r, Maturity)))
```
`hstack` creates a larger vector/matrix by putting vectors/matrices together horizontally.
```
print(Compound_1year)
```
`Comound_6month` contains the amount of wealth at each year when we invest funds with six-month compound interest rate.
```
Compound_6month = np.hstack((1.0, np.cumprod(np.tile((1.0 + r/2.0)**2, Maturity))))
```
`Continuous_Rate` contains the amount of wealth at each year when we invest funds with continuous compound interest rate.
```
Continuous_Rate = np.exp(r*np.linspace(0, Maturity, Maturity + 1))
```
The following cell creates a figure.
```
fig1 = plt.figure(num=1, facecolor='w')
plt.plot(Simple_Rate, 'b-')
plt.plot(Compound_1year, 'r--')
plt.plot(Compound_6month, 'g-.')
plt.plot(Continuous_Rate, 'm:')
plt.legend(['simple', '1-year compound', '6-month compound', 'continuous'],
loc='upper left', frameon=False)
plt.xlabel('t')
plt.ylabel('W(t)/W(0)')
plt.show()
```
|
github_jupyter
|
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
r = 0.2
Maturity = 10
Simple_Rate = 1.0 + r * np.linspace(0, Maturity, Maturity + 1)
print(np.linspace(0, Maturity, Maturity + 1))
print(Simple_Rate)
Compound_1year = np.hstack((1.0, np.cumprod(np.tile(1.0 + r, Maturity))))
print(np.tile(1.0 + r, Maturity))
print(np.cumprod(np.tile(1.0 + r, Maturity)))
print(Compound_1year)
Compound_6month = np.hstack((1.0, np.cumprod(np.tile((1.0 + r/2.0)**2, Maturity))))
Continuous_Rate = np.exp(r*np.linspace(0, Maturity, Maturity + 1))
fig1 = plt.figure(num=1, facecolor='w')
plt.plot(Simple_Rate, 'b-')
plt.plot(Compound_1year, 'r--')
plt.plot(Compound_6month, 'g-.')
plt.plot(Continuous_Rate, 'm:')
plt.legend(['simple', '1-year compound', '6-month compound', 'continuous'],
loc='upper left', frameon=False)
plt.xlabel('t')
plt.ylabel('W(t)/W(0)')
plt.show()
| 0.379723 | 0.99493 |
```
import pandas as pd, matplotlib.pyplot as plt, numpy as np
from matplotlib.colors import BoundaryNorm
from matplotlib.ticker import MaxNLocator
import matplotlib.patches as patches
%matplotlib inline
```
Fossil fuels
```
fop=np.linspace(10,40,100)
fcap=np.linspace(20,150,100)
eroei_el=30
CR=[50,90]
CF=[50,90]
R=[2,5]
eroei_ccs=np.zeros([len(CR),len(CF),len(fop),len(fcap)])
for i in range(len(R)):
for j in range(len(CF)):
for k in range(len(fop)):
for l in range(len(fcap)):
eroei_ccs[i][j][k,l]=(1-fop[k]/100.0)*((R[i]+1)/(R[i]+1+fcap[l]/100.0))*eroei_el*CF[j]/100.0
fig,axes=plt.subplots(2,2,figsize=(11,9))
plt.subplots_adjust(hspace=0.35)
#levels = MaxNLocator(nbins=15).tick_values(4000, 15000)
for i in range(len(axes)):
for j in range(len(axes[i])):
ax=axes[i][j]
z = eroei_ccs[i][j][:-1, :-1]
#levels = MaxNLocator(nbins=15).tick_values(z.min(), z.max())
levels = MaxNLocator(nbins=30).tick_values(8, 24)
cmap = plt.get_cmap('viridis')
norm = BoundaryNorm(levels, ncolors=cmap.N, clip=True)
im = ax.pcolormesh(fop, fcap, z, cmap=cmap, norm=norm)
fig.colorbar(im, ax=ax)
ax.set_xlim((fop.min(),fop.max()))
ax.set_ylim((fcap.min(),fcap.max()))
t=[12,20,80,140]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'CCGT\nGas')
t=[25,35,75,100]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'Pulverized\nCoal')
t=[15,30,30,50]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'IGCC\nCoal')
ax.set_xlabel('fop')
ax.set_ylabel('fcap')
ax.set_title(u'$EROEI_{CCS}$\nCR='+str(CR[i])+', CF='+str(CF[j])+', R='+str(R[i]))
plt.suptitle(r'$EROEI_{el}='+str(eroei_el)+'$',fontsize=16)
plt.show()
#change CF - CF
#EROEI-el by techn - create
#equation R- EROI-l
```
RE
```
eroei_el=np.linspace(5,40,100)
phi=np.linspace(.10,.60,100)
ESOI=[16,300]
eta=0.78
eroei_disp=np.zeros([len(ESOI),len(phi),len(eroei_el)])
for i in range(len(ESOI)):
for j in range(len(phi)):
for k in range(len(eroei_el)):
eroei_disp[i][j,k]=((1-phi[j])+(eta*phi[j]))/((1/eroei_el[k])+(eta*phi[j]/ESOI[i]))
fig,axes=plt.subplots(2,2,figsize=(11,9))
plt.subplots_adjust(hspace=0.35)
#levels = MaxNLocator(nbins=15).tick_values(4000, 15000)
for i in range(len(axes)):
for j in range(len(axes[i])):
ax=axes[i][j]
z = eroei_disp[i][:-1, :-1]
#levels = MaxNLocator(nbins=15).tick_values(z.min(), z.max())
levels = MaxNLocator(nbins=30).tick_values(2, 40)
cmap = plt.get_cmap('viridis')
norm = BoundaryNorm(levels, ncolors=cmap.N, clip=True)
im = ax.pcolormesh(phi, eroei_el, z, cmap=cmap, norm=norm)
fig.colorbar(im, ax=ax)
ax.set_xlim((phi.min(),phi.max()))
ax.set_ylim((eroei_el.min(),eroei_el.max()))
'''
t=[12,20,80,140]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'CCGT\nGas')
t=[25,35,75,100]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'Pulverized\nCoal')
t=[15,30,30,50]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'IGCC\nCoal')
'''
ax.set_xlabel('phi')
ax.set_ylabel('eroei_el')
ax.set_title(u'$EROEI_{disp}$\nESOI='+str(ESOI[i])+', eta='+str(eta))
plt.show()
#transalte technology CCS PV-EROEI
```
|
github_jupyter
|
import pandas as pd, matplotlib.pyplot as plt, numpy as np
from matplotlib.colors import BoundaryNorm
from matplotlib.ticker import MaxNLocator
import matplotlib.patches as patches
%matplotlib inline
fop=np.linspace(10,40,100)
fcap=np.linspace(20,150,100)
eroei_el=30
CR=[50,90]
CF=[50,90]
R=[2,5]
eroei_ccs=np.zeros([len(CR),len(CF),len(fop),len(fcap)])
for i in range(len(R)):
for j in range(len(CF)):
for k in range(len(fop)):
for l in range(len(fcap)):
eroei_ccs[i][j][k,l]=(1-fop[k]/100.0)*((R[i]+1)/(R[i]+1+fcap[l]/100.0))*eroei_el*CF[j]/100.0
fig,axes=plt.subplots(2,2,figsize=(11,9))
plt.subplots_adjust(hspace=0.35)
#levels = MaxNLocator(nbins=15).tick_values(4000, 15000)
for i in range(len(axes)):
for j in range(len(axes[i])):
ax=axes[i][j]
z = eroei_ccs[i][j][:-1, :-1]
#levels = MaxNLocator(nbins=15).tick_values(z.min(), z.max())
levels = MaxNLocator(nbins=30).tick_values(8, 24)
cmap = plt.get_cmap('viridis')
norm = BoundaryNorm(levels, ncolors=cmap.N, clip=True)
im = ax.pcolormesh(fop, fcap, z, cmap=cmap, norm=norm)
fig.colorbar(im, ax=ax)
ax.set_xlim((fop.min(),fop.max()))
ax.set_ylim((fcap.min(),fcap.max()))
t=[12,20,80,140]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'CCGT\nGas')
t=[25,35,75,100]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'Pulverized\nCoal')
t=[15,30,30,50]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'IGCC\nCoal')
ax.set_xlabel('fop')
ax.set_ylabel('fcap')
ax.set_title(u'$EROEI_{CCS}$\nCR='+str(CR[i])+', CF='+str(CF[j])+', R='+str(R[i]))
plt.suptitle(r'$EROEI_{el}='+str(eroei_el)+'$',fontsize=16)
plt.show()
#change CF - CF
#EROEI-el by techn - create
#equation R- EROI-l
eroei_el=np.linspace(5,40,100)
phi=np.linspace(.10,.60,100)
ESOI=[16,300]
eta=0.78
eroei_disp=np.zeros([len(ESOI),len(phi),len(eroei_el)])
for i in range(len(ESOI)):
for j in range(len(phi)):
for k in range(len(eroei_el)):
eroei_disp[i][j,k]=((1-phi[j])+(eta*phi[j]))/((1/eroei_el[k])+(eta*phi[j]/ESOI[i]))
fig,axes=plt.subplots(2,2,figsize=(11,9))
plt.subplots_adjust(hspace=0.35)
#levels = MaxNLocator(nbins=15).tick_values(4000, 15000)
for i in range(len(axes)):
for j in range(len(axes[i])):
ax=axes[i][j]
z = eroei_disp[i][:-1, :-1]
#levels = MaxNLocator(nbins=15).tick_values(z.min(), z.max())
levels = MaxNLocator(nbins=30).tick_values(2, 40)
cmap = plt.get_cmap('viridis')
norm = BoundaryNorm(levels, ncolors=cmap.N, clip=True)
im = ax.pcolormesh(phi, eroei_el, z, cmap=cmap, norm=norm)
fig.colorbar(im, ax=ax)
ax.set_xlim((phi.min(),phi.max()))
ax.set_ylim((eroei_el.min(),eroei_el.max()))
'''
t=[12,20,80,140]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'CCGT\nGas')
t=[25,35,75,100]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'Pulverized\nCoal')
t=[15,30,30,50]
rect = patches.Rectangle((t[0],t[2]),t[1]-t[0],t[3]-t[2],linewidth=1,edgecolor='k',facecolor='none')
ax.add_patch(rect)
ax.text(t[0]+0.5,t[2]+5,'IGCC\nCoal')
'''
ax.set_xlabel('phi')
ax.set_ylabel('eroei_el')
ax.set_title(u'$EROEI_{disp}$\nESOI='+str(ESOI[i])+', eta='+str(eta))
plt.show()
#transalte technology CCS PV-EROEI
| 0.130812 | 0.765287 |
# Spark SQL Quiz
This quiz uses the same dataset and questions from the Spark Data Frames Programming Quiz. For this quiz, however, use Spark SQL instead of Spark Data Frames.
```
from pyspark.sql import SparkSession
from pyspark.sql import functions as func
from pyspark.sql import types
# TODOS:
# 1) import any other libraries you might need
# 2) instantiate a Spark session
# 3) read in the data set located at the path "data/sparkify_log_small.json"
# 4) create a view to use with your SQL queries
# 5) write code to answer the quiz questions
spark = SparkSession\
.builder\
.appName("My Spark SQL Quiz")\
.getOrCreate()
log_data = spark.read.json("data/sparkify_log_small.json")
log_data.count()
```
# Question 1
Which page did user id ""(empty string) NOT visit?
```
log_data.createOrReplaceTempView("log_data_table")
spark.sql("""
SELECT DISTINCT page
FROM log_data_table
WHERE page NOT IN (SELECT DISTINCT page
FROM log_data_table
WHERE userId == '')
""").show()
spark.sql("""
SELECT DISTINCT page
FROM log_data_table
WHERE page NOT IN (SELECT DISTINCT page
FROM log_data_table
WHERE userId == '')
""").explain()
```
# Question 2 - Reflect
Why might you prefer to use SQL over data frames? Why might you prefer data frames over SQL?
Team may already be familiar or even skilled in SQL than Data Frames / Pandas. Some operations are just easier to do with SQL.. especially operations requiring JOINing two data sets
Data Frames - Offer more flexibility and allows for imperative programming. Shareable and increase in adaptation by Data Science community
# Question 3
How many female users do we have in the data set?
```
spark.sql("""
SELECT COUNT(DISTINCT userId)
FROM log_data_table
WHERE gender == 'F'
""").show()
```
# Question 4
How many songs were played from the most played artist?
```
spark.sql("""
SELECT artist, count(*) as num_songs
FROM log_data_table
WHERE page == 'NextSong'
GROUP BY artist
ORDER BY num_songs DESC
""").show(1)
```
# Question 5 (challenge)
How many songs do users listen to on average between visiting our home page? Please round your answer to the closest integer.
```
songs_by_user_phase = spark.sql("""
SELECT userId, ts, song_play,
SUM(homepage_visit) OVER (PARTITION BY userId ORDER BY int(ts) DESC RANGE UNBOUNDED PRECEDING) as phase
FROM (
SELECT userId, ts,
CASE WHEN page == 'Home' THEN 1 ELSE 0 END as homepage_visit,
CASE WHEN page == 'NextSong' THEN 1 ELSE 0 END as song_play
FROM log_data_table
WHERE page == 'NextSong' OR page == 'Home'
) sub
""")
songs_by_user_phase.createOrReplaceTempView("songs_by_user_phase_table")
spark.sql("""
SELECT AVG(num_songs) AS avg1, AVG(CASE WHEN num_songs == 0 THEN NULL ELSE num_songs END) AS avg2
FROM (
SELECT userId, phase, SUM(song_play) AS num_songs
FROM songs_by_user_phase_table
GROUP BY userId, phase
) sub
""").show()
```
|
github_jupyter
|
from pyspark.sql import SparkSession
from pyspark.sql import functions as func
from pyspark.sql import types
# TODOS:
# 1) import any other libraries you might need
# 2) instantiate a Spark session
# 3) read in the data set located at the path "data/sparkify_log_small.json"
# 4) create a view to use with your SQL queries
# 5) write code to answer the quiz questions
spark = SparkSession\
.builder\
.appName("My Spark SQL Quiz")\
.getOrCreate()
log_data = spark.read.json("data/sparkify_log_small.json")
log_data.count()
log_data.createOrReplaceTempView("log_data_table")
spark.sql("""
SELECT DISTINCT page
FROM log_data_table
WHERE page NOT IN (SELECT DISTINCT page
FROM log_data_table
WHERE userId == '')
""").show()
spark.sql("""
SELECT DISTINCT page
FROM log_data_table
WHERE page NOT IN (SELECT DISTINCT page
FROM log_data_table
WHERE userId == '')
""").explain()
spark.sql("""
SELECT COUNT(DISTINCT userId)
FROM log_data_table
WHERE gender == 'F'
""").show()
spark.sql("""
SELECT artist, count(*) as num_songs
FROM log_data_table
WHERE page == 'NextSong'
GROUP BY artist
ORDER BY num_songs DESC
""").show(1)
songs_by_user_phase = spark.sql("""
SELECT userId, ts, song_play,
SUM(homepage_visit) OVER (PARTITION BY userId ORDER BY int(ts) DESC RANGE UNBOUNDED PRECEDING) as phase
FROM (
SELECT userId, ts,
CASE WHEN page == 'Home' THEN 1 ELSE 0 END as homepage_visit,
CASE WHEN page == 'NextSong' THEN 1 ELSE 0 END as song_play
FROM log_data_table
WHERE page == 'NextSong' OR page == 'Home'
) sub
""")
songs_by_user_phase.createOrReplaceTempView("songs_by_user_phase_table")
spark.sql("""
SELECT AVG(num_songs) AS avg1, AVG(CASE WHEN num_songs == 0 THEN NULL ELSE num_songs END) AS avg2
FROM (
SELECT userId, phase, SUM(song_play) AS num_songs
FROM songs_by_user_phase_table
GROUP BY userId, phase
) sub
""").show()
| 0.394201 | 0.914939 |
```
!pip install pytorch-adapt[lightning,ignite]
```
### Load a toy dataset
```
import torch
from tqdm import tqdm
from pytorch_adapt.datasets import get_mnist_mnistm
# mnist is the source domain
# mnistm is the target domain
datasets = get_mnist_mnistm(["mnist"], ["mnistm"], ".", download=True)
dataloader = torch.utils.data.DataLoader(
datasets["train"], batch_size=32, num_workers=2
)
```
### Load toy models
```
from pytorch_adapt.models import Discriminator, mnistC, mnistG
device = torch.device("cuda")
def get_models():
G = mnistG(pretrained=True).to(device)
C = mnistC(pretrained=True).to(device)
D = Discriminator(in_size=1200, h=256).to(device)
return {"G": G, "C": C, "D": D}
def get_optimizers(models):
G_opt = torch.optim.Adam(models["G"].parameters(), lr=0.0001)
C_opt = torch.optim.Adam(models["C"].parameters(), lr=0.0001)
D_opt = torch.optim.Adam(models["D"].parameters(), lr=0.0001)
return [G_opt, C_opt, D_opt]
```
### Use in vanilla PyTorch
```
from pytorch_adapt.hooks import DANNHook
from pytorch_adapt.utils.common_functions import batch_to_device
models = get_models()
optimizers = get_optimizers(models)
# Assuming that models, optimizers, and dataloader are already created.
hook = DANNHook(optimizers)
for data in tqdm(dataloader):
data = batch_to_device(data, device)
# Optimization is done inside the hook.
# The returned loss is for logging.
loss, _ = hook({}, {**models, **data})
```
### Build complex algorithms
```
from pytorch_adapt.hooks import MCCHook, VATHook
models = get_models()
optimizers = get_optimizers(models)
# G and C are the Generator and Classifier models
G, C = models["G"], models["C"]
misc = {"combined_model": torch.nn.Sequential(G, C)}
hook = DANNHook(optimizers, post_g=[MCCHook(), VATHook()])
for data in tqdm(dataloader):
data = batch_to_device(data, device)
loss, _ = hook({}, {**models, **data, **misc})
```
### Wrap with your favorite PyTorch framework
```
from pytorch_adapt.adapters import DANN
from pytorch_adapt.containers import Models
from pytorch_adapt.datasets import DataloaderCreator
models = get_models()
models_cont = Models(models)
adapter = DANN(models=models_cont)
dc = DataloaderCreator(num_workers=2)
dataloaders = dc(**datasets)
```
#### Lightning
```
import pytorch_lightning as pl
from pytorch_adapt.frameworks.lightning import Lightning
L_adapter = Lightning(adapter)
trainer = pl.Trainer(gpus=1, max_epochs=1)
trainer.fit(L_adapter, dataloaders["train"])
```
#### Ignite
```
from pytorch_adapt.frameworks.ignite import Ignite
models = get_models()
models_cont = Models(models)
adapter = DANN(models=models_cont)
trainer = Ignite(adapter)
trainer.run(datasets, dataloader_creator=dc)
```
### Check your model's performance
```
from pytorch_adapt.validators import SNDValidator
# Random predictions as placeholder
preds = torch.randn(1000, 100)
# Assuming predictions have been collected
target_train = {"preds": preds}
validator = SNDValidator()
score = validator.score(target_train=target_train)
```
#### Lightning
```
from pytorch_adapt.frameworks.utils import filter_datasets
models = get_models()
models_cont = Models(models)
adapter = DANN(models=models_cont)
validator = SNDValidator()
dataloaders = dc(**filter_datasets(datasets, validator))
train_loader = dataloaders.pop("train")
L_adapter = Lightning(adapter, validator=validator)
trainer = pl.Trainer(gpus=1, max_epochs=1)
trainer.fit(L_adapter, train_loader, list(dataloaders.values()))
```
#### Ignite
```
from pytorch_adapt.validators import ScoreHistory
models = get_models()
models_cont = Models(models)
adapter = DANN(models=models_cont)
validator = ScoreHistory(SNDValidator())
trainer = Ignite(adapter, validator=validator)
trainer.run(datasets, dataloader_creator=dc)
```
|
github_jupyter
|
!pip install pytorch-adapt[lightning,ignite]
import torch
from tqdm import tqdm
from pytorch_adapt.datasets import get_mnist_mnistm
# mnist is the source domain
# mnistm is the target domain
datasets = get_mnist_mnistm(["mnist"], ["mnistm"], ".", download=True)
dataloader = torch.utils.data.DataLoader(
datasets["train"], batch_size=32, num_workers=2
)
from pytorch_adapt.models import Discriminator, mnistC, mnistG
device = torch.device("cuda")
def get_models():
G = mnistG(pretrained=True).to(device)
C = mnistC(pretrained=True).to(device)
D = Discriminator(in_size=1200, h=256).to(device)
return {"G": G, "C": C, "D": D}
def get_optimizers(models):
G_opt = torch.optim.Adam(models["G"].parameters(), lr=0.0001)
C_opt = torch.optim.Adam(models["C"].parameters(), lr=0.0001)
D_opt = torch.optim.Adam(models["D"].parameters(), lr=0.0001)
return [G_opt, C_opt, D_opt]
from pytorch_adapt.hooks import DANNHook
from pytorch_adapt.utils.common_functions import batch_to_device
models = get_models()
optimizers = get_optimizers(models)
# Assuming that models, optimizers, and dataloader are already created.
hook = DANNHook(optimizers)
for data in tqdm(dataloader):
data = batch_to_device(data, device)
# Optimization is done inside the hook.
# The returned loss is for logging.
loss, _ = hook({}, {**models, **data})
from pytorch_adapt.hooks import MCCHook, VATHook
models = get_models()
optimizers = get_optimizers(models)
# G and C are the Generator and Classifier models
G, C = models["G"], models["C"]
misc = {"combined_model": torch.nn.Sequential(G, C)}
hook = DANNHook(optimizers, post_g=[MCCHook(), VATHook()])
for data in tqdm(dataloader):
data = batch_to_device(data, device)
loss, _ = hook({}, {**models, **data, **misc})
from pytorch_adapt.adapters import DANN
from pytorch_adapt.containers import Models
from pytorch_adapt.datasets import DataloaderCreator
models = get_models()
models_cont = Models(models)
adapter = DANN(models=models_cont)
dc = DataloaderCreator(num_workers=2)
dataloaders = dc(**datasets)
import pytorch_lightning as pl
from pytorch_adapt.frameworks.lightning import Lightning
L_adapter = Lightning(adapter)
trainer = pl.Trainer(gpus=1, max_epochs=1)
trainer.fit(L_adapter, dataloaders["train"])
from pytorch_adapt.frameworks.ignite import Ignite
models = get_models()
models_cont = Models(models)
adapter = DANN(models=models_cont)
trainer = Ignite(adapter)
trainer.run(datasets, dataloader_creator=dc)
from pytorch_adapt.validators import SNDValidator
# Random predictions as placeholder
preds = torch.randn(1000, 100)
# Assuming predictions have been collected
target_train = {"preds": preds}
validator = SNDValidator()
score = validator.score(target_train=target_train)
from pytorch_adapt.frameworks.utils import filter_datasets
models = get_models()
models_cont = Models(models)
adapter = DANN(models=models_cont)
validator = SNDValidator()
dataloaders = dc(**filter_datasets(datasets, validator))
train_loader = dataloaders.pop("train")
L_adapter = Lightning(adapter, validator=validator)
trainer = pl.Trainer(gpus=1, max_epochs=1)
trainer.fit(L_adapter, train_loader, list(dataloaders.values()))
from pytorch_adapt.validators import ScoreHistory
models = get_models()
models_cont = Models(models)
adapter = DANN(models=models_cont)
validator = ScoreHistory(SNDValidator())
trainer = Ignite(adapter, validator=validator)
trainer.run(datasets, dataloader_creator=dc)
| 0.901621 | 0.930205 |
# 11 - Jointly Learning to Align and Translate
Prepared by Jan Christian Blaise Cruz
DLSU Machine Learning Group
In this notebook, we'll implement the model in the paper Neural Machine Translation by Jointly Learning to Align and Translate (Bahadanau et al., 2014). We'll use attention to improve our translations performance-wise and interpretability-wise.
# Preliminaries
First, let's make sure we have a GPU.
```
!nvidia-smi
```
Download the data and the tokenizers as with the previous notebook. Don't forget to restart the runtime.
```
!wget https://s3.us-east-2.amazonaws.com/blaisecruz.com/datasets/translation/multi30k.zip
!unzip multi30k.zip && rm multi30k.zip
!python -m spacy download de_core_news_sm
!python -m spacy download en_core_web_sm
```
Then it's our usual imports.
```
import torch
import torch.nn as nn
import torch.optim as optim
import torch.utils.data as datautils
import spacy
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
from nltk.translate.bleu_score import corpus_bleu
from nltk.translate.bleu_score import SmoothingFunction
import random
from collections import Counter
from tqdm import tqdm
np.random.seed(42)
torch.manual_seed(42)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
%matplotlib inline
```
# Data Processing
We'll do the same thing here as with the last notebook, so we'll breeze through this. Load the data.
```
with open('multi30k/train.en', 'r') as f:
train_en = [line.strip() for line in f]
with open('multi30k/train.de', 'r') as f:
train_de = [line.strip() for line in f]
with open('multi30k/val.en', 'r') as f:
valid_en = [line.strip() for line in f]
with open('multi30k/val.de', 'r') as f:
valid_de = [line.strip() for line in f]
```
Load the tokenizers then tokenize the dataset and add the delimiter tokens.
```
spacy_de = spacy.load('de_core_news_sm')
spacy_en = spacy.load('en_core_web_sm')
def tokenize_de(text):
return ['<sos>'] + [tok.text.lower() for tok in spacy_de.tokenizer(text)] + ['<eos>']
def tokenize_en(text):
return ['<sos>'] + [tok.text.lower() for tok in spacy_en.tokenizer(text)] + ['<eos>']
# Tokenize the text
train_en = [tokenize_en(text) for text in tqdm(train_en)]
train_de = [tokenize_de(text) for text in tqdm(train_de)]
valid_en = [tokenize_en(text) for text in tqdm(valid_en)]
valid_de = [tokenize_de(text) for text in tqdm(valid_de)]
```
Process each sequence to the maximum sequence length of the dataset.
```
def process(dataset):
max_len = max([len(text) for text in dataset])
temp = []
for text in dataset:
if len(text) < max_len:
text += ['<pad>' for _ in range(max_len - len(text))]
temp.append(text)
return temp
# Pad to maximum length of the dataset
train_en_proc, valid_en_proc = process(train_en), process(valid_en)
train_de_proc, valid_de_proc = process(train_de), process(valid_de)
```
Produce vocabularies for the source and target languages.
```
def get_vocab(dataset, min_freq=2):
# Add all tokens to the list
special_tokens = ['<unk>', '<pad>', '<sos>', '<eos>']
vocab = []
for line in dataset: vocab.extend(line)
# Remove words that are below the minimum frequency, the enforce set
counts = Counter(vocab)
vocab = special_tokens + [word for word in counts.keys() if counts[word] > min_freq]
vocab_set = set(vocab)
# Push all special tokens to the front
idx2word = list(vocab_set)
for token in special_tokens[::-1]:
idx2word.insert(0, idx2word.pop(idx2word.index(token)))
# Produce word2idx then return
word2idx = {idx2word[i]: i for i in range(len(idx2word))}
return vocab_set, idx2word, word2idx
# Get vocabulary and references
vocab_set_en, idx2word_en, word2idx_en = get_vocab(train_en_proc, min_freq=2)
vocab_set_de, idx2word_de, word2idx_de = get_vocab(train_de_proc, min_freq=2)
# Convert unknown tokens
train_en_proc = [[token if token in vocab_set_en else '<unk>' for token in line] for line in train_en_proc]
train_de_proc = [[token if token in vocab_set_de else '<unk>' for token in line] for line in train_de_proc]
valid_en_proc = [[token if token in vocab_set_en else '<unk>' for token in line] for line in valid_en_proc]
valid_de_proc = [[token if token in vocab_set_de else '<unk>' for token in line] for line in valid_de_proc]
```
Finally, convert the sequences of tokens into their corresponding indices.
```
def serialize(dataset, word2idx):
temp = []
for line in dataset: temp.append([word2idx[token] for token in line])
return torch.LongTensor(temp)
# Convert to idx
y_train = serialize(train_en_proc, word2idx_en)
X_train = serialize(train_de_proc, word2idx_de)
y_valid = serialize(valid_en_proc, word2idx_en)
X_valid = serialize(valid_de_proc, word2idx_de)
```
We'll wrap up by making dataloaders.
```
bs = 128
train_dataset = datautils.TensorDataset(X_train, y_train)
valid_dataset = datautils.TensorDataset(X_valid, y_valid)
train_sampler = datautils.RandomSampler(train_dataset)
train_loader = datautils.DataLoader(train_dataset, batch_size=bs, sampler=train_sampler)
valid_loader = datautils.DataLoader(valid_dataset, batch_size=bs, shuffle=False)
```
# Modeling
Let's start with the encoder. The biggest change here is that we're using a bidirectional GRU instead of a vanilla LSTM. Since the RNN is bidirectional, we'll introduce a linear layer to **pool** the forward and backward hidden states.
Encoding the source sequence remains the same. We'll embed and apply dropout. Afterwhich, we'll pack the sequences to disregard padding. Pass that result to the GRU, then unpack the result. We pass the concatenated hidden states to the linear pooling layer, then apply a hyperbolic tangent activation function. This tells us which "candidate information" to keep.
To prepare for attention, we'll output both the RNN output and the pooled hidden states.
```
class Encoder(nn.Module):
def __init__(self, vocab_sz, embedding_dim, enc_hidden_dim, dec_hidden_dim, dropout=0.5):
super(Encoder, self).__init__()
self.embedding = nn.Embedding(vocab_sz, embedding_dim)
self.rnn = nn.GRU(embedding_dim, enc_hidden_dim, bidirectional=True)
self.fc1 = nn.Linear(enc_hidden_dim * 2, dec_hidden_dim)
self.dropout = nn.Dropout(dropout)
self.vocab_sz = vocab_sz
def init_hidden(self, bs):
weight = next(self.parameters())
hidden_dim = self.rnn.hidden_size
h = weight.new_zeros(2, bs, hidden_dim)
return h
def forward(self, x, pad_idx):
msl, bs = x.shape
# Embed then pack the sequence
out = self.embedding(x)
out = self.dropout(out)
# Get the length of the sequences and pack them
lens = ((x.rot90() == pad_idx) == False).int().sum(dim=1)
out = nn.utils.rnn.pack_padded_sequence(out, lens, enforce_sorted=False)
hidden = self.init_hidden(bs)
out, hidden = self.rnn(out, hidden)
out, _ = nn.utils.rnn.pad_packed_sequence(out, total_length=msl)
# Pool the forward and backward states
hidden = torch.cat((hidden[-2, :, :], hidden[-1, :, :]), axis=1)
hidden = torch.tanh(self.fc1(hidden))
return out, hidden
```
For the rest of the modeling part, we'll run through attention and decoding manually as things can get rather confusing.
First, let's get a sample source and target sequence batch.
```
x, y = next(iter(train_loader))
x, y = x.rot90(k=3), y.rot90(k=3)
print(x.shape, y.shape)
```
Here's our hyperparameters.
```
vocab_sz = len(vocab_set_de)
embedding_dim = 128
enc_hidden_dim = 256
dec_hidden_dim = 256
pad_idx = word2idx_de['<pad>']
```
Instantiate an encoder.
```
encoder = Encoder(vocab_sz, embedding_dim, enc_hidden_dim, dec_hidden_dim)
```
Then pass the source sentence.
We get what we expect.
```
out, hidden = encoder(x, pad_idx=pad_idx)
print(out.shape, hidden.shape)
```
Time to compute the attention weights!
Attention weights can be thought of as a form of "feature importance." We'll use the RNN outputs of the encoder and the pooled hidden states to output a matrix of size [bs $\times$ sequence length]. This can be thought of as the "feature importance" of each token in the sequence given the current information (the hidden states).
At every decoding timestep, we will produce this attention matrix (or attention vector per sequence) to tell our model which tokens are important at each step. This gives the model the ability to "look back" and "focus on important context tokens."
Anyhow, we'll implement an attention layer using two linear layers.
```
attn = nn.Linear((enc_hidden_dim * 2) + dec_hidden_dim, dec_hidden_dim)
v = nn.Linear(dec_hidden_dim, 1, bias=False)
```
We'll take the length of the source sentence and the batch size.
```
src_len, bs, _ = out.shape
```
Here's what we're going to do:
1. We need to match the hidden state to each output token. If, for example, the sequence has 50 tokens, we need to 50 copies of the hidden state to "match" to each output token.
2. We'll compute the "energy" between the hidden state and the output. Think of this as "how much does a specific token match the information in the hidden state. If it is more relevant, energy is higher."
3. Once we have our energy, we need to turn them into weights. These are the weights of each token at a certain timestep. In other words, the "importance."
Let's do #1 first. Remember the hidden state's shape:
```
hidden.shape
```
Let's introduce a blank dimension.
```
hidden.unsqueeze(1).shape
```
Then repeat the hidden state src_len times. This makes a number of clones of the hidden state that we can match to every entry in the RNN outputs.
```
hidden_repeated = hidden.unsqueeze(1).repeat(1, src_len, 1)
hidden_repeated.shape
```
As a sanity check:
```
hidden_repeated[:, 0, :] == hidden_repeated[:, 1, :]
```
Now here's our output shape:
```
out.shape
```
We'll permute the dimensions to match our hidden state clones:
```
out_permuted = out.permute(1, 0, 2)
out_permuted.shape
```
Since their dimensions are now matched, we can concatenate them on the last dimension. This should now be of shape [batch size $\times$ src_len $\times$ encoder hidden dim * 2 + decoder hidden dim]
Remember that we're doing encoder hidden dim * 2 because our encoder uses a bidirectional RNN.
```
torch.cat((out_permuted, hidden_repeated), dim=2).shape
```
Pass the concatenated hidden and outputs to a linear layer to compute new features, then apply a hyperbolic tangent activation function.
```
energy = attn(torch.cat((out_permuted, hidden_repeated), dim=2))
energy = torch.tanh(energy)
energy.shape
```
We still have a 3D tensor. Let's project this down to 2 dimensions by applying a final linear transform. This turns our energy into a "weight matrix."
```
a = v(energy)
a.shape
```
Squeeze the final dimension since there's nothing there.
```
a = a.squeeze(2)
a.shape
```
We're not yet done. Lastly, we have to mask out the attention weights on the padding tokens. We can do this by using an attention mask.
```
mask = (x != pad_idx).permute(1, 0)
mask.shape
```
It looks like this.
```
mask
```
To mask the padding tokens, we'll do it like so:
```
a = a.masked_fill(mask == 0, -1e10)
```
The full attention layer looks like this:
```
class Attention(nn.Module):
def __init__(self, enc_hidden_dim, dec_hidden_dim):
super(Attention, self).__init__()
self.attention = nn.Linear((enc_hidden_dim * 2) + dec_hidden_dim, dec_hidden_dim)
self.v = nn.Linear(dec_hidden_dim, 1, bias=False)
def forward(self, out, hidden, mask):
src_len, bs, _ = out.shape
hidden = hidden.unsqueeze(1).repeat(1, src_len, 1)
out = out.permute(1, 0, 2)
# Compute the weighted energy (match between hidden states)
energy = torch.tanh(self.attention(torch.cat((out, hidden), dim=2)))
attn = self.v(energy).squeeze(2)
attn = attn.masked_fill(mask == 0, -1e10)
return torch.softmax(attn, dim=1)
```
Instantiate an attention layer for testing.
```
attention = Attention(enc_hidden_dim, dec_hidden_dim)
```
Using it gives us the outputs we expect.
```
mask = (x != pad_idx).permute(1, 0)
a = attention(out, hidden, mask)
a.shape
```
Lastly, we'll implement the decoder.
We have a few differences from the previous decoder.
1. Our RNN is again a GRU, which accepts encoder hidden dim * 2 + embedding dim shaped inputs. This is because we're passing it the encoded source sentence (remember the encoder is bidirectional) along with the embedded target token.
2. We use attention inside the decoder. At every decoding step, we generate a new attention matrix, which we batch matrix multiply to the outputs of the encoder. This lets the decoder know which tokens in the source sentence it should "pay attention to."
```
class Decoder(nn.Module):
def __init__(self, vocab_sz, embedding_dim, enc_hidden_dim, dec_hidden_dim, dropout=0.5):
super(Decoder, self).__init__()
self.attention = Attention(enc_hidden_dim, dec_hidden_dim)
self.embedding = nn.Embedding(vocab_sz, embedding_dim)
self.rnn = nn.GRU((enc_hidden_dim * 2) + embedding_dim, dec_hidden_dim)
self.fc1 = nn.Linear((enc_hidden_dim * 2) + dec_hidden_dim + embedding_dim, vocab_sz)
self.dropout = nn.Dropout(dropout)
self.vocab_sz = vocab_sz
def forward(self, y, out, hidden, mask):
embedded = self.embedding(y.unsqueeze(0))
embedded = self.dropout(embedded)
# Batch multiply to get weighted outputs
a = self.attention(out, hidden, mask).unsqueeze(1)
out = out.permute(1, 0, 2)
weighted = torch.bmm(a, out).permute(1, 0, 2)
# Prepare RNN Inputs
rnn_inputs = torch.cat((embedded, weighted), dim=2)
hidden = hidden.unsqueeze(0)
out, hidden = self.rnn(rnn_inputs)
# Pool the embeddings, weighted outputs, and new outputs
out = torch.cat((embedded.squeeze(0), out.squeeze(0), weighted.squeeze(0)), dim=1)
out = self.fc1(out)
return out, hidden.squeeze(0), a.squeeze(1)
```
Let's set the first input token for example.
```
token = y[0,:]
print(token.shape)
print(token)
```
Instantiate a decoder.
```
decoder = Decoder(vocab_sz, embedding_dim, enc_hidden_dim, dec_hidden_dim)
```
Produce a mask and pass in the output, hidden states, and the mask.
```
mask = (x != pad_idx).permute(1, 0)
out, hidden, a = decoder(token, out, hidden, mask)
print(out.shape, hidden.shape, a.shape)
```
Getting the maximum of the logits gives us the predicted next output token.
```
out.argmax(1)
```
Let's wrap the encoder and decoder in a seq2seq module. This is largely the same, except we include a helper function to generate an attention mask.
```
class Seq2Seq(nn.Module):
def __init__(self, encoder, decoder, pad_idx, initrange=0.01):
super(Seq2Seq, self).__init__()
self.encoder = encoder
self.decoder = decoder
self.init_weights(initrange)
self.pad_idx = pad_idx
def create_mask(self, x):
with torch.no_grad():
mask = (x != self.pad_idx).permute(1, 0)
return mask
def init_weights(self, initrange=0.01):
for name, param in self.named_parameters():
if 'weight' in name: nn.init.normal_(param.data, mean=0, std=initrange)
else: nn.init.constant_(param.data, 0)
def forward(self, x, y, teacher_forcing=0.5):
src_len, bs = x.shape
trg_len, bs = y.shape
# Make container for outputs
weight = next(self.encoder.parameters())
outputs = weight.new_zeros(trg_len, bs, self.decoder.vocab_sz)
# Encode source then remember context
encoder_out, hidden = self.encoder(x, self.pad_idx)
input_ids = y[0,:]
mask = self.create_mask(x)
# Decode per input token
for i in range(1, trg_len):
out, hidden, _ = self.decoder(input_ids, encoder_out, hidden, mask)
outputs[i] = out
teacher_force = random.random() < teacher_forcing
input_ids = y[i] if teacher_force else out.argmax(1)
return outputs
```
Test out the wrapper and instantiate a loss function.
```
model = Seq2Seq(encoder, decoder, pad_idx)
criterion = nn.CrossEntropyLoss(ignore_index=pad_idx)
```
We get the expected output.
```
out = model(x, y)
print(out.shape)
```
Here's the initial loss.
```
loss = criterion(out[1:].flatten(0, 1), y[1:].flatten(0))
print(loss)
```
# Training
We'll instantiate a training setup with the following hyperparameters:
```
encoder = Encoder(vocab_sz=len(vocab_set_de), embedding_dim=256, enc_hidden_dim=512, dec_hidden_dim=512, dropout=0.5)
decoder = Decoder(vocab_sz=len(vocab_set_en), embedding_dim=256, enc_hidden_dim=512, dec_hidden_dim=512, dropout=0.5)
model = Seq2Seq(encoder, decoder, pad_idx=word2idx_de['<pad>']).to(device)
optimizer = optim.Adam(model.parameters(), lr=3e-3)
criterion = nn.CrossEntropyLoss(ignore_index=word2idx_en['<pad>'])
epochs = 20
iters = epochs * len(train_loader)
scheduler = optim.lr_scheduler.CosineAnnealingLR(optimizer, T_max=iters, eta_min=0)
```
Count the parameters of the model.
```
def count_parameters(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
print("The model has {:,} trainable parameters".format(count_parameters(model)))
```
For transparency, here's the final architecture of our model:
```
model
```
Finally, we'll train it.
```
clip = 1.0
for e in range(1, epochs + 1):
train_loss = 0
model.train()
for x, y in tqdm(train_loader):
x, y = x.rot90(k=3).to(device), y.rot90(k=3).to(device)
out = model(x, y)
loss = criterion(out[1:].flatten(0, 1), y[1:].flatten(0))
optimizer.zero_grad()
loss.backward()
nn.utils.clip_grad_norm_(model.parameters(), clip)
optimizer.step()
scheduler.step()
train_loss += loss.item()
train_loss /= len(train_loader)
valid_loss = 0
model.eval()
with torch.no_grad():
for x, y in tqdm(valid_loader):
x, y = x.rot90(k=3).to(device), y.rot90(k=3).to(device)
out = model(x, y)
loss = criterion(out[1:].flatten(0, 1), y[1:].flatten(0))
valid_loss += loss.item()
valid_loss /= len(valid_loader)
print("\nEpoch {:3} | Train Loss {:.4f} | Train Ppl {:.4f} | Valid Loss {:.4f} | Valid Ppl {:.4f}".format(e, train_loss, np.exp(train_loss), valid_loss, np.exp(valid_loss)))
```
If you train your own model, make sure to save the resulting weights.
```
torch.save(model.state_dict(), 'seq2seq_attention.pt')
```
# Sampling
Let's load the pretrained weights.
```
encoder = Encoder(vocab_sz=len(vocab_set_de), embedding_dim=256, enc_hidden_dim=512, dec_hidden_dim=512, dropout=0.5)
decoder = Decoder(vocab_sz=len(vocab_set_en), embedding_dim=256, enc_hidden_dim=512, dec_hidden_dim=512, dropout=0.5)
model = Seq2Seq(encoder, decoder, pad_idx=word2idx_de['<pad>']).to(device)
criterion = nn.CrossEntropyLoss(ignore_index=word2idx_en['<pad>'])
model.load_state_dict(torch.load('seq2seq_attention.pt'))
model.eval();
```
Evaluate them on the validation set.
```
model.eval()
valid_loss = 0
with torch.no_grad():
for x, y in tqdm(valid_loader):
x, y = x.rot90(k=3).to(device), y.rot90(k=3).to(device)
out = model(x, y)
loss = criterion(out[1:].flatten(0, 1), y[1:].flatten(0))
valid_loss += loss.item()
valid_loss /= len(valid_loader)
print("\nValid Loss {:.4f} | Valid Ppl {:.4f}".format(valid_loss, np.exp(valid_loss)))
```
Let's write a modified translation function. This is largely akin to the seq2seq decoding method, but we're using multinomial sampling and we're not using any teacher forcing.
We'll start with a tokenized sequence of tokens. We encode it with the encoder, then start decoding. We'll save each token as well as the attention weights per timestep so we can plot them later.
```
def translate(src_sentence, model, max_words=20, temperature=1.0, seed=42):
s = [token if token in vocab_set_de else '<unk>' for token in src_sentence]
sample = torch.LongTensor([word2idx_de[token] for token in s]).unsqueeze(1)
predictions = []
torch.manual_seed(seed)
with torch.no_grad():
# Produce an encoding of the source text and a storage for attention
encoder_out, hidden = model.encoder(sample, pad_idx=word2idx_de['<pad>'])
token = sample[0,:]
attentions = torch.zeros(max_words, 1, sample.shape[0])
# Decoding. Sample succeding tokens using attention and encodings
for i in range(max_words):
mask = (sample != pad_idx).permute(1, 0)
out, hidden, attn = model.decoder(token, encoder_out, hidden, mask)
weights = torch.softmax(out / temperature, dim=-1)
token = torch.multinomial(weights, 1).squeeze(0)
predictions.append(token.item())
attentions[i] = attn
if token.item() == word2idx_en['<eos>']:
break
# Convert predictions from indices to text. Cut the attentions to translation length
predictions = [idx2word_en[ix] for ix in predictions]
attentions = attentions.squeeze(1)[:len(predictions),:]
return predictions, attentions
```
The nice thing about attention is that it's interpretable. We can write a function to plot the attention weights in a heatmap for each timestep of decoding.
```
def plot_attention(src_sentence, predictions, attentions):
fig = plt.figure(figsize=(7,7))
ax = fig.add_subplot(111)
attention = attentions.numpy()
cax = ax.matshow(attention, cmap='bone')
ax.tick_params(labelsize=12)
ax.set_xticklabels([''] + [t for t in src_sentence], rotation=45)
ax.set_yticklabels([''] + predictions)
ax.xaxis.set_major_locator(ticker.MultipleLocator(1))
ax.yaxis.set_major_locator(ticker.MultipleLocator(1))
```
Let's put the model in the CPU.
```
model = model.cpu()
```
Then test out some translations.
```
ix = 0
src_sentence, trg_sentence = valid_de[ix], valid_en[ix]
src_sentence = src_sentence[:src_sentence.index('<eos>') + 1]
trg_sentence = trg_sentence[:trg_sentence.index('<eos>') + 1]
print('Source:', ' '.join(src_sentence))
print('Target:', ' '.join(trg_sentence))
predictions, attentions = translate(src_sentence, model, temperature=0.9)
print('Prediction:', ' '.join(predictions))
plot_attention(src_sentence, predictions, attentions)
ix = 6
src_sentence, trg_sentence = valid_de[ix], valid_en[ix]
src_sentence = src_sentence[:src_sentence.index('<eos>') + 1]
trg_sentence = trg_sentence[:trg_sentence.index('<eos>') + 1]
print('Source:', ' '.join(src_sentence))
print('Target:', ' '.join(trg_sentence))
predictions, attentions = translate(src_sentence, model, temperature=0.9)
print('Prediction:', ' '.join(predictions))
plot_attention(src_sentence, predictions, attentions)
ix = 10
src_sentence, trg_sentence = valid_de[ix], valid_en[ix]
src_sentence = src_sentence[:src_sentence.index('<eos>') + 1]
trg_sentence = trg_sentence[:trg_sentence.index('<eos>') + 1]
print('Source:', ' '.join(src_sentence))
print('Target:', ' '.join(trg_sentence))
predictions, attentions = translate(src_sentence, model, temperature=0.9)
print('Prediction:', ' '.join(predictions))
plot_attention(src_sentence, predictions, attentions)
ix = 64
src_sentence, trg_sentence = valid_de[ix], valid_en[ix]
src_sentence = src_sentence[:src_sentence.index('<eos>') + 1]
trg_sentence = trg_sentence[:trg_sentence.index('<eos>') + 1]
print('Source:', ' '.join(src_sentence))
print('Target:', ' '.join(trg_sentence))
predictions, attentions = translate(src_sentence, model, temperature=0.9)
print('Prediction:', ' '.join(predictions))
plot_attention(src_sentence, predictions, attentions)
```
|
github_jupyter
|
!nvidia-smi
!wget https://s3.us-east-2.amazonaws.com/blaisecruz.com/datasets/translation/multi30k.zip
!unzip multi30k.zip && rm multi30k.zip
!python -m spacy download de_core_news_sm
!python -m spacy download en_core_web_sm
import torch
import torch.nn as nn
import torch.optim as optim
import torch.utils.data as datautils
import spacy
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
from nltk.translate.bleu_score import corpus_bleu
from nltk.translate.bleu_score import SmoothingFunction
import random
from collections import Counter
from tqdm import tqdm
np.random.seed(42)
torch.manual_seed(42)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
%matplotlib inline
with open('multi30k/train.en', 'r') as f:
train_en = [line.strip() for line in f]
with open('multi30k/train.de', 'r') as f:
train_de = [line.strip() for line in f]
with open('multi30k/val.en', 'r') as f:
valid_en = [line.strip() for line in f]
with open('multi30k/val.de', 'r') as f:
valid_de = [line.strip() for line in f]
spacy_de = spacy.load('de_core_news_sm')
spacy_en = spacy.load('en_core_web_sm')
def tokenize_de(text):
return ['<sos>'] + [tok.text.lower() for tok in spacy_de.tokenizer(text)] + ['<eos>']
def tokenize_en(text):
return ['<sos>'] + [tok.text.lower() for tok in spacy_en.tokenizer(text)] + ['<eos>']
# Tokenize the text
train_en = [tokenize_en(text) for text in tqdm(train_en)]
train_de = [tokenize_de(text) for text in tqdm(train_de)]
valid_en = [tokenize_en(text) for text in tqdm(valid_en)]
valid_de = [tokenize_de(text) for text in tqdm(valid_de)]
def process(dataset):
max_len = max([len(text) for text in dataset])
temp = []
for text in dataset:
if len(text) < max_len:
text += ['<pad>' for _ in range(max_len - len(text))]
temp.append(text)
return temp
# Pad to maximum length of the dataset
train_en_proc, valid_en_proc = process(train_en), process(valid_en)
train_de_proc, valid_de_proc = process(train_de), process(valid_de)
def get_vocab(dataset, min_freq=2):
# Add all tokens to the list
special_tokens = ['<unk>', '<pad>', '<sos>', '<eos>']
vocab = []
for line in dataset: vocab.extend(line)
# Remove words that are below the minimum frequency, the enforce set
counts = Counter(vocab)
vocab = special_tokens + [word for word in counts.keys() if counts[word] > min_freq]
vocab_set = set(vocab)
# Push all special tokens to the front
idx2word = list(vocab_set)
for token in special_tokens[::-1]:
idx2word.insert(0, idx2word.pop(idx2word.index(token)))
# Produce word2idx then return
word2idx = {idx2word[i]: i for i in range(len(idx2word))}
return vocab_set, idx2word, word2idx
# Get vocabulary and references
vocab_set_en, idx2word_en, word2idx_en = get_vocab(train_en_proc, min_freq=2)
vocab_set_de, idx2word_de, word2idx_de = get_vocab(train_de_proc, min_freq=2)
# Convert unknown tokens
train_en_proc = [[token if token in vocab_set_en else '<unk>' for token in line] for line in train_en_proc]
train_de_proc = [[token if token in vocab_set_de else '<unk>' for token in line] for line in train_de_proc]
valid_en_proc = [[token if token in vocab_set_en else '<unk>' for token in line] for line in valid_en_proc]
valid_de_proc = [[token if token in vocab_set_de else '<unk>' for token in line] for line in valid_de_proc]
def serialize(dataset, word2idx):
temp = []
for line in dataset: temp.append([word2idx[token] for token in line])
return torch.LongTensor(temp)
# Convert to idx
y_train = serialize(train_en_proc, word2idx_en)
X_train = serialize(train_de_proc, word2idx_de)
y_valid = serialize(valid_en_proc, word2idx_en)
X_valid = serialize(valid_de_proc, word2idx_de)
bs = 128
train_dataset = datautils.TensorDataset(X_train, y_train)
valid_dataset = datautils.TensorDataset(X_valid, y_valid)
train_sampler = datautils.RandomSampler(train_dataset)
train_loader = datautils.DataLoader(train_dataset, batch_size=bs, sampler=train_sampler)
valid_loader = datautils.DataLoader(valid_dataset, batch_size=bs, shuffle=False)
class Encoder(nn.Module):
def __init__(self, vocab_sz, embedding_dim, enc_hidden_dim, dec_hidden_dim, dropout=0.5):
super(Encoder, self).__init__()
self.embedding = nn.Embedding(vocab_sz, embedding_dim)
self.rnn = nn.GRU(embedding_dim, enc_hidden_dim, bidirectional=True)
self.fc1 = nn.Linear(enc_hidden_dim * 2, dec_hidden_dim)
self.dropout = nn.Dropout(dropout)
self.vocab_sz = vocab_sz
def init_hidden(self, bs):
weight = next(self.parameters())
hidden_dim = self.rnn.hidden_size
h = weight.new_zeros(2, bs, hidden_dim)
return h
def forward(self, x, pad_idx):
msl, bs = x.shape
# Embed then pack the sequence
out = self.embedding(x)
out = self.dropout(out)
# Get the length of the sequences and pack them
lens = ((x.rot90() == pad_idx) == False).int().sum(dim=1)
out = nn.utils.rnn.pack_padded_sequence(out, lens, enforce_sorted=False)
hidden = self.init_hidden(bs)
out, hidden = self.rnn(out, hidden)
out, _ = nn.utils.rnn.pad_packed_sequence(out, total_length=msl)
# Pool the forward and backward states
hidden = torch.cat((hidden[-2, :, :], hidden[-1, :, :]), axis=1)
hidden = torch.tanh(self.fc1(hidden))
return out, hidden
x, y = next(iter(train_loader))
x, y = x.rot90(k=3), y.rot90(k=3)
print(x.shape, y.shape)
vocab_sz = len(vocab_set_de)
embedding_dim = 128
enc_hidden_dim = 256
dec_hidden_dim = 256
pad_idx = word2idx_de['<pad>']
encoder = Encoder(vocab_sz, embedding_dim, enc_hidden_dim, dec_hidden_dim)
out, hidden = encoder(x, pad_idx=pad_idx)
print(out.shape, hidden.shape)
attn = nn.Linear((enc_hidden_dim * 2) + dec_hidden_dim, dec_hidden_dim)
v = nn.Linear(dec_hidden_dim, 1, bias=False)
src_len, bs, _ = out.shape
hidden.shape
hidden.unsqueeze(1).shape
hidden_repeated = hidden.unsqueeze(1).repeat(1, src_len, 1)
hidden_repeated.shape
hidden_repeated[:, 0, :] == hidden_repeated[:, 1, :]
out.shape
out_permuted = out.permute(1, 0, 2)
out_permuted.shape
torch.cat((out_permuted, hidden_repeated), dim=2).shape
energy = attn(torch.cat((out_permuted, hidden_repeated), dim=2))
energy = torch.tanh(energy)
energy.shape
a = v(energy)
a.shape
a = a.squeeze(2)
a.shape
mask = (x != pad_idx).permute(1, 0)
mask.shape
mask
a = a.masked_fill(mask == 0, -1e10)
class Attention(nn.Module):
def __init__(self, enc_hidden_dim, dec_hidden_dim):
super(Attention, self).__init__()
self.attention = nn.Linear((enc_hidden_dim * 2) + dec_hidden_dim, dec_hidden_dim)
self.v = nn.Linear(dec_hidden_dim, 1, bias=False)
def forward(self, out, hidden, mask):
src_len, bs, _ = out.shape
hidden = hidden.unsqueeze(1).repeat(1, src_len, 1)
out = out.permute(1, 0, 2)
# Compute the weighted energy (match between hidden states)
energy = torch.tanh(self.attention(torch.cat((out, hidden), dim=2)))
attn = self.v(energy).squeeze(2)
attn = attn.masked_fill(mask == 0, -1e10)
return torch.softmax(attn, dim=1)
attention = Attention(enc_hidden_dim, dec_hidden_dim)
mask = (x != pad_idx).permute(1, 0)
a = attention(out, hidden, mask)
a.shape
class Decoder(nn.Module):
def __init__(self, vocab_sz, embedding_dim, enc_hidden_dim, dec_hidden_dim, dropout=0.5):
super(Decoder, self).__init__()
self.attention = Attention(enc_hidden_dim, dec_hidden_dim)
self.embedding = nn.Embedding(vocab_sz, embedding_dim)
self.rnn = nn.GRU((enc_hidden_dim * 2) + embedding_dim, dec_hidden_dim)
self.fc1 = nn.Linear((enc_hidden_dim * 2) + dec_hidden_dim + embedding_dim, vocab_sz)
self.dropout = nn.Dropout(dropout)
self.vocab_sz = vocab_sz
def forward(self, y, out, hidden, mask):
embedded = self.embedding(y.unsqueeze(0))
embedded = self.dropout(embedded)
# Batch multiply to get weighted outputs
a = self.attention(out, hidden, mask).unsqueeze(1)
out = out.permute(1, 0, 2)
weighted = torch.bmm(a, out).permute(1, 0, 2)
# Prepare RNN Inputs
rnn_inputs = torch.cat((embedded, weighted), dim=2)
hidden = hidden.unsqueeze(0)
out, hidden = self.rnn(rnn_inputs)
# Pool the embeddings, weighted outputs, and new outputs
out = torch.cat((embedded.squeeze(0), out.squeeze(0), weighted.squeeze(0)), dim=1)
out = self.fc1(out)
return out, hidden.squeeze(0), a.squeeze(1)
token = y[0,:]
print(token.shape)
print(token)
decoder = Decoder(vocab_sz, embedding_dim, enc_hidden_dim, dec_hidden_dim)
mask = (x != pad_idx).permute(1, 0)
out, hidden, a = decoder(token, out, hidden, mask)
print(out.shape, hidden.shape, a.shape)
out.argmax(1)
class Seq2Seq(nn.Module):
def __init__(self, encoder, decoder, pad_idx, initrange=0.01):
super(Seq2Seq, self).__init__()
self.encoder = encoder
self.decoder = decoder
self.init_weights(initrange)
self.pad_idx = pad_idx
def create_mask(self, x):
with torch.no_grad():
mask = (x != self.pad_idx).permute(1, 0)
return mask
def init_weights(self, initrange=0.01):
for name, param in self.named_parameters():
if 'weight' in name: nn.init.normal_(param.data, mean=0, std=initrange)
else: nn.init.constant_(param.data, 0)
def forward(self, x, y, teacher_forcing=0.5):
src_len, bs = x.shape
trg_len, bs = y.shape
# Make container for outputs
weight = next(self.encoder.parameters())
outputs = weight.new_zeros(trg_len, bs, self.decoder.vocab_sz)
# Encode source then remember context
encoder_out, hidden = self.encoder(x, self.pad_idx)
input_ids = y[0,:]
mask = self.create_mask(x)
# Decode per input token
for i in range(1, trg_len):
out, hidden, _ = self.decoder(input_ids, encoder_out, hidden, mask)
outputs[i] = out
teacher_force = random.random() < teacher_forcing
input_ids = y[i] if teacher_force else out.argmax(1)
return outputs
model = Seq2Seq(encoder, decoder, pad_idx)
criterion = nn.CrossEntropyLoss(ignore_index=pad_idx)
out = model(x, y)
print(out.shape)
loss = criterion(out[1:].flatten(0, 1), y[1:].flatten(0))
print(loss)
encoder = Encoder(vocab_sz=len(vocab_set_de), embedding_dim=256, enc_hidden_dim=512, dec_hidden_dim=512, dropout=0.5)
decoder = Decoder(vocab_sz=len(vocab_set_en), embedding_dim=256, enc_hidden_dim=512, dec_hidden_dim=512, dropout=0.5)
model = Seq2Seq(encoder, decoder, pad_idx=word2idx_de['<pad>']).to(device)
optimizer = optim.Adam(model.parameters(), lr=3e-3)
criterion = nn.CrossEntropyLoss(ignore_index=word2idx_en['<pad>'])
epochs = 20
iters = epochs * len(train_loader)
scheduler = optim.lr_scheduler.CosineAnnealingLR(optimizer, T_max=iters, eta_min=0)
def count_parameters(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
print("The model has {:,} trainable parameters".format(count_parameters(model)))
model
clip = 1.0
for e in range(1, epochs + 1):
train_loss = 0
model.train()
for x, y in tqdm(train_loader):
x, y = x.rot90(k=3).to(device), y.rot90(k=3).to(device)
out = model(x, y)
loss = criterion(out[1:].flatten(0, 1), y[1:].flatten(0))
optimizer.zero_grad()
loss.backward()
nn.utils.clip_grad_norm_(model.parameters(), clip)
optimizer.step()
scheduler.step()
train_loss += loss.item()
train_loss /= len(train_loader)
valid_loss = 0
model.eval()
with torch.no_grad():
for x, y in tqdm(valid_loader):
x, y = x.rot90(k=3).to(device), y.rot90(k=3).to(device)
out = model(x, y)
loss = criterion(out[1:].flatten(0, 1), y[1:].flatten(0))
valid_loss += loss.item()
valid_loss /= len(valid_loader)
print("\nEpoch {:3} | Train Loss {:.4f} | Train Ppl {:.4f} | Valid Loss {:.4f} | Valid Ppl {:.4f}".format(e, train_loss, np.exp(train_loss), valid_loss, np.exp(valid_loss)))
torch.save(model.state_dict(), 'seq2seq_attention.pt')
encoder = Encoder(vocab_sz=len(vocab_set_de), embedding_dim=256, enc_hidden_dim=512, dec_hidden_dim=512, dropout=0.5)
decoder = Decoder(vocab_sz=len(vocab_set_en), embedding_dim=256, enc_hidden_dim=512, dec_hidden_dim=512, dropout=0.5)
model = Seq2Seq(encoder, decoder, pad_idx=word2idx_de['<pad>']).to(device)
criterion = nn.CrossEntropyLoss(ignore_index=word2idx_en['<pad>'])
model.load_state_dict(torch.load('seq2seq_attention.pt'))
model.eval();
model.eval()
valid_loss = 0
with torch.no_grad():
for x, y in tqdm(valid_loader):
x, y = x.rot90(k=3).to(device), y.rot90(k=3).to(device)
out = model(x, y)
loss = criterion(out[1:].flatten(0, 1), y[1:].flatten(0))
valid_loss += loss.item()
valid_loss /= len(valid_loader)
print("\nValid Loss {:.4f} | Valid Ppl {:.4f}".format(valid_loss, np.exp(valid_loss)))
def translate(src_sentence, model, max_words=20, temperature=1.0, seed=42):
s = [token if token in vocab_set_de else '<unk>' for token in src_sentence]
sample = torch.LongTensor([word2idx_de[token] for token in s]).unsqueeze(1)
predictions = []
torch.manual_seed(seed)
with torch.no_grad():
# Produce an encoding of the source text and a storage for attention
encoder_out, hidden = model.encoder(sample, pad_idx=word2idx_de['<pad>'])
token = sample[0,:]
attentions = torch.zeros(max_words, 1, sample.shape[0])
# Decoding. Sample succeding tokens using attention and encodings
for i in range(max_words):
mask = (sample != pad_idx).permute(1, 0)
out, hidden, attn = model.decoder(token, encoder_out, hidden, mask)
weights = torch.softmax(out / temperature, dim=-1)
token = torch.multinomial(weights, 1).squeeze(0)
predictions.append(token.item())
attentions[i] = attn
if token.item() == word2idx_en['<eos>']:
break
# Convert predictions from indices to text. Cut the attentions to translation length
predictions = [idx2word_en[ix] for ix in predictions]
attentions = attentions.squeeze(1)[:len(predictions),:]
return predictions, attentions
def plot_attention(src_sentence, predictions, attentions):
fig = plt.figure(figsize=(7,7))
ax = fig.add_subplot(111)
attention = attentions.numpy()
cax = ax.matshow(attention, cmap='bone')
ax.tick_params(labelsize=12)
ax.set_xticklabels([''] + [t for t in src_sentence], rotation=45)
ax.set_yticklabels([''] + predictions)
ax.xaxis.set_major_locator(ticker.MultipleLocator(1))
ax.yaxis.set_major_locator(ticker.MultipleLocator(1))
model = model.cpu()
ix = 0
src_sentence, trg_sentence = valid_de[ix], valid_en[ix]
src_sentence = src_sentence[:src_sentence.index('<eos>') + 1]
trg_sentence = trg_sentence[:trg_sentence.index('<eos>') + 1]
print('Source:', ' '.join(src_sentence))
print('Target:', ' '.join(trg_sentence))
predictions, attentions = translate(src_sentence, model, temperature=0.9)
print('Prediction:', ' '.join(predictions))
plot_attention(src_sentence, predictions, attentions)
ix = 6
src_sentence, trg_sentence = valid_de[ix], valid_en[ix]
src_sentence = src_sentence[:src_sentence.index('<eos>') + 1]
trg_sentence = trg_sentence[:trg_sentence.index('<eos>') + 1]
print('Source:', ' '.join(src_sentence))
print('Target:', ' '.join(trg_sentence))
predictions, attentions = translate(src_sentence, model, temperature=0.9)
print('Prediction:', ' '.join(predictions))
plot_attention(src_sentence, predictions, attentions)
ix = 10
src_sentence, trg_sentence = valid_de[ix], valid_en[ix]
src_sentence = src_sentence[:src_sentence.index('<eos>') + 1]
trg_sentence = trg_sentence[:trg_sentence.index('<eos>') + 1]
print('Source:', ' '.join(src_sentence))
print('Target:', ' '.join(trg_sentence))
predictions, attentions = translate(src_sentence, model, temperature=0.9)
print('Prediction:', ' '.join(predictions))
plot_attention(src_sentence, predictions, attentions)
ix = 64
src_sentence, trg_sentence = valid_de[ix], valid_en[ix]
src_sentence = src_sentence[:src_sentence.index('<eos>') + 1]
trg_sentence = trg_sentence[:trg_sentence.index('<eos>') + 1]
print('Source:', ' '.join(src_sentence))
print('Target:', ' '.join(trg_sentence))
predictions, attentions = translate(src_sentence, model, temperature=0.9)
print('Prediction:', ' '.join(predictions))
plot_attention(src_sentence, predictions, attentions)
| 0.778313 | 0.920576 |
```
import pandas as pd
import warnings
warnings.filterwarnings("ignore")
file_to_load = ("Resources/purchase_data.csv")
purchase_data = pd.read_csv(file_to_load)
purchase_data.head()
```
### Player Count
Display Total Number of Players
```
Player_count = purchase_data["SN"].nunique()
Player_count
```
### Purchasing Analysis(Total)
```
unique_items = purchase_data["Item ID"].nunique()
unique_items
average_price = purchase_data["Price"].mean()
average_price
Total_purchase_number = purchase_data["Price"].count()
Total_purchase_number
Total_revenue = average_price*Total_purchase_number
Total_revenue
data = {'Number of Unique Items':[unique_items],'Average Purchase Price':[average_price],'Total Number of Purchases':[Total_purchase_number],'Total Revenue':[Total_revenue]}
purchasing_analysis_df = pd.DataFrame(data)
purchasing_analysis_df['Average Purchase Price'] = purchasing_analysis_df['Average Purchase Price'].map("${:.2f}".format)
purchasing_analysis_df['Total Revenue'] = purchasing_analysis_df['Total Revenue'].map("${:.2f}".format)
purchasing_analysis_df
```
### Gender Demographics
```
male_df = purchase_data.loc[purchase_data["Gender"] == "Male",:]
male_df.head()
male_count = male_df["SN"].nunique()
male_count
female_df = purchase_data.loc[purchase_data["Gender"] == "Female",:]
female_df.head()
female_count = female_df["SN"].nunique()
female_count
other_df = purchase_data.loc[purchase_data["Gender"] == "Other / Non-Disclosed",:]
other_df.head()
other_count = other_df["SN"].nunique()
other_count
male_percent = (male_count/(male_count+female_count+other_count))*100
female_percent = (female_count/(male_count+female_count+other_count))*100
other_percent = (other_count/(male_count+female_count+other_count))*100
gender_data = {"":["Male","Female","Other / Non-Disclosed"],"Total Count":[male_count,female_count,other_count],"Percentage of Players":[male_percent,female_percent,other_percent]}
gender_df = pd.DataFrame(gender_data)
gender_df
gender_df = gender_df.set_index("")
gender_df["Percentage of Players"] = gender_df["Percentage of Players"].astype(float).map("{:.2f}%".format)
gender_df
```
### Purchasing Analysis(Gender)
```
gender_group = purchase_data.groupby(['Gender'])
gender_count = gender_group["Purchase ID"].count()
average_purchase_price = gender_group['Price'].mean()
Total_purchase_value = gender_group['Price'].sum()
average_purchase_price_per_person_male = Total_purchase_value['Male']/male_count
average_purchase_price_per_person_female = Total_purchase_value['Female']/female_count
average_purchase_price_per_person_other = Total_purchase_value['Other / Non-Disclosed']/other_count
purchase_analysis_gender_df = pd.DataFrame({
"Purchase Count": gender_count,"Average Purchase Price": average_purchase_price,"Total Purchase Value": Total_purchase_value,"Avg Total Purchase per Person": [average_purchase_price_per_person_female,average_purchase_price_per_person_male,average_purchase_price_per_person_other]
})
purchase_analysis_gender_df["Average Purchase Price"] = purchase_analysis_gender_df["Average Purchase Price"].astype(float).map("${:.2f}".format)
purchase_analysis_gender_df["Total Purchase Value"] = purchase_analysis_gender_df["Total Purchase Value"].astype(float).map("${:,.2f}".format)
purchase_analysis_gender_df["Avg Total Purchase per Person"] = purchase_analysis_gender_df["Avg Total Purchase per Person"].astype(float).map("${:.2f}".format)
purchase_analysis_gender_df
```
### Age Demographics
```
purchase_data.head()
purchase_unique = purchase_data.drop_duplicates('SN',keep='first')
purchase_unique
bins = [0,9,14,19,24,29,34,39,47]
group_labels = ["<10","10-14","15-19","20-24","25-29","30-34","35-39","40+"]
purchase_unique["Age group"] = pd.cut(purchase_unique["Age"],bins,labels = group_labels)
purchase_unique.head()
age_group = purchase_unique.groupby("Age group")["Age"].count().reset_index()
age_group = age_group.rename(columns = {"Age":"Total count"})
age_group["Percentage of Players"] = 100*age_group["Total count"]/age_group["Total count"].sum()
age_group["Percentage of Players"] = age_group["Percentage of Players"].astype(float).map("{:.2f}%".format)
age_group.set_index("Age group")
```
### Purchasing Analysis (Age)
```
purchase_data.head()
bins = [0,9,14,19,24,29,34,39,47]
group_labels = ["<10","10-14","15-19","20-24","25-29","30-34","35-39","40+"]
purchase_data["Age group"] = pd.cut(purchase_data["Age"],bins,labels = group_labels)
purchase_data.head()
pdc = purchase_data.groupby("Age group")["SN"].count()
pdc = pdc.reset_index()
pdc = pdc.rename(columns = {"SN":"Purchase Count"})
pdc
app = purchase_data.groupby("Age group")["Price"].mean()
app = app.reset_index()
# app["Price"] = app["Price"].map("${:.2f}".format)
app = app.rename(columns={"Price":"Average Purchase Price"})
app
tpv = purchase_data.groupby("Age group")["Price"].sum()
tpv = tpv.reset_index()
# tpv["Price"] = tpv["Price"].map("${:.2f}".format).astype(float)
tpv = tpv.rename(columns = {"Price":"Total Purchase Value"})
tpv
tpv["Avg Total Purchase per Person"] = tpv["Total Purchase Value"]/age_group["Total count"]
tpv
combine = pdc.merge(app,on='Age group')
combine = combine.merge(tpv,on='Age group')
combine["Average Purchase Price"] = combine["Average Purchase Price"].map("${:.2f}".format)
combine["Total Purchase Value"] = combine["Total Purchase Value"].map("${:.2f}".format)
combine["Avg Total Purchase per Person"] = combine["Avg Total Purchase per Person"].map("${:.2f}".format)
combine = combine.rename(columns={'Age group':'Age Ranges'})
combine.set_index("Age Ranges")
```
## Top Spenders
```
purchase_data['Total Purchase Value'] = purchase_data.groupby(['SN'])['Price'].transform('sum')
purchase_data['Purchase Count'] = purchase_data.groupby(['SN'])['Price'].transform('count')
purchase_data['Average Purchase Price'] = purchase_data['Total Purchase Value']/purchase_data['Purchase Count']
purchase_data
purchase_unique = purchase_data.drop_duplicates(subset=['SN'])
purchase_unique
top_5 = purchase_unique.nlargest(5,'Total Purchase Value')
top_5
top_spenders = top_5[['SN','Purchase Count','Average Purchase Price','Total Purchase Value']]
top_spenders['Average Purchase Price'] = top_spenders['Average Purchase Price'].map("${:.2f}".format)
top_spenders['Total Purchase Value'] = top_spenders['Total Purchase Value'].map("${:.2f}".format)
top_spenders.set_index('SN')
top_spenders = top_5.loc[:,['SN','Purchase Count','Average Purchase Price','Total Purchase Value']]
top_spenders['Average Purchase Price'] = top_spenders['Average Purchase Price'].map("${:.2f}".format)
top_spenders['Total Purchase Value'] = top_spenders['Total Purchase Value'].map("${:.2f}".format)
top_spenders.set_index('SN')
```
### Most Popular Items
```
purchase_data['Purchase Count'] = purchase_data.groupby(['Item ID'])['Price'].transform('count')
purchase_data
purchase_data['Total Purchase Value'] = purchase_data.groupby(['Item ID'])['Price'].transform('sum')
purchase_data
Item_count = purchase_data.drop_duplicates(subset=['Item ID'])
Item_count
sort_item = Item_count.sort_values('Purchase Count',ascending=False)
sort_item.head(6)
popular = Item_count.nlargest(5,'Purchase Count')
popular['Price'] = popular['Price'].map("${:.2f}".format)
popular['Total Purchase Value'] = popular['Total Purchase Value'].map("${:.2f}".format)
popular = popular.rename(columns={'Price':'Item Price'})
popular
Most_Popular_Items = popular[['Item ID','Item Name','Purchase Count','Item Price','Total Purchase Value']]
Most_Popular_Items.set_index('Item ID','Item Name')
```
### Most Profitable Items
```
most_profitable = Item_count.nlargest(5,'Total Purchase Value')
most_profitable
Most_Profitable_Items = most_profitable[['Item ID','Item Name','Purchase Count','Price','Total Purchase Value']]
Most_Profitable_Items = Most_Profitable_Items.rename(columns={'Price':'Item Price'})
Most_Profitable_Items['Item Price'] = Most_Profitable_Items['Item Price'].map("${:.2f}".format)
Most_Profitable_Items['Total Purchase Value'] = Most_Profitable_Items['Total Purchase Value'].map("${:.2f}".format)
Most_Profitable_Items.set_index('Item ID')
```
### Observable Trends
|
github_jupyter
|
import pandas as pd
import warnings
warnings.filterwarnings("ignore")
file_to_load = ("Resources/purchase_data.csv")
purchase_data = pd.read_csv(file_to_load)
purchase_data.head()
Player_count = purchase_data["SN"].nunique()
Player_count
unique_items = purchase_data["Item ID"].nunique()
unique_items
average_price = purchase_data["Price"].mean()
average_price
Total_purchase_number = purchase_data["Price"].count()
Total_purchase_number
Total_revenue = average_price*Total_purchase_number
Total_revenue
data = {'Number of Unique Items':[unique_items],'Average Purchase Price':[average_price],'Total Number of Purchases':[Total_purchase_number],'Total Revenue':[Total_revenue]}
purchasing_analysis_df = pd.DataFrame(data)
purchasing_analysis_df['Average Purchase Price'] = purchasing_analysis_df['Average Purchase Price'].map("${:.2f}".format)
purchasing_analysis_df['Total Revenue'] = purchasing_analysis_df['Total Revenue'].map("${:.2f}".format)
purchasing_analysis_df
male_df = purchase_data.loc[purchase_data["Gender"] == "Male",:]
male_df.head()
male_count = male_df["SN"].nunique()
male_count
female_df = purchase_data.loc[purchase_data["Gender"] == "Female",:]
female_df.head()
female_count = female_df["SN"].nunique()
female_count
other_df = purchase_data.loc[purchase_data["Gender"] == "Other / Non-Disclosed",:]
other_df.head()
other_count = other_df["SN"].nunique()
other_count
male_percent = (male_count/(male_count+female_count+other_count))*100
female_percent = (female_count/(male_count+female_count+other_count))*100
other_percent = (other_count/(male_count+female_count+other_count))*100
gender_data = {"":["Male","Female","Other / Non-Disclosed"],"Total Count":[male_count,female_count,other_count],"Percentage of Players":[male_percent,female_percent,other_percent]}
gender_df = pd.DataFrame(gender_data)
gender_df
gender_df = gender_df.set_index("")
gender_df["Percentage of Players"] = gender_df["Percentage of Players"].astype(float).map("{:.2f}%".format)
gender_df
gender_group = purchase_data.groupby(['Gender'])
gender_count = gender_group["Purchase ID"].count()
average_purchase_price = gender_group['Price'].mean()
Total_purchase_value = gender_group['Price'].sum()
average_purchase_price_per_person_male = Total_purchase_value['Male']/male_count
average_purchase_price_per_person_female = Total_purchase_value['Female']/female_count
average_purchase_price_per_person_other = Total_purchase_value['Other / Non-Disclosed']/other_count
purchase_analysis_gender_df = pd.DataFrame({
"Purchase Count": gender_count,"Average Purchase Price": average_purchase_price,"Total Purchase Value": Total_purchase_value,"Avg Total Purchase per Person": [average_purchase_price_per_person_female,average_purchase_price_per_person_male,average_purchase_price_per_person_other]
})
purchase_analysis_gender_df["Average Purchase Price"] = purchase_analysis_gender_df["Average Purchase Price"].astype(float).map("${:.2f}".format)
purchase_analysis_gender_df["Total Purchase Value"] = purchase_analysis_gender_df["Total Purchase Value"].astype(float).map("${:,.2f}".format)
purchase_analysis_gender_df["Avg Total Purchase per Person"] = purchase_analysis_gender_df["Avg Total Purchase per Person"].astype(float).map("${:.2f}".format)
purchase_analysis_gender_df
purchase_data.head()
purchase_unique = purchase_data.drop_duplicates('SN',keep='first')
purchase_unique
bins = [0,9,14,19,24,29,34,39,47]
group_labels = ["<10","10-14","15-19","20-24","25-29","30-34","35-39","40+"]
purchase_unique["Age group"] = pd.cut(purchase_unique["Age"],bins,labels = group_labels)
purchase_unique.head()
age_group = purchase_unique.groupby("Age group")["Age"].count().reset_index()
age_group = age_group.rename(columns = {"Age":"Total count"})
age_group["Percentage of Players"] = 100*age_group["Total count"]/age_group["Total count"].sum()
age_group["Percentage of Players"] = age_group["Percentage of Players"].astype(float).map("{:.2f}%".format)
age_group.set_index("Age group")
purchase_data.head()
bins = [0,9,14,19,24,29,34,39,47]
group_labels = ["<10","10-14","15-19","20-24","25-29","30-34","35-39","40+"]
purchase_data["Age group"] = pd.cut(purchase_data["Age"],bins,labels = group_labels)
purchase_data.head()
pdc = purchase_data.groupby("Age group")["SN"].count()
pdc = pdc.reset_index()
pdc = pdc.rename(columns = {"SN":"Purchase Count"})
pdc
app = purchase_data.groupby("Age group")["Price"].mean()
app = app.reset_index()
# app["Price"] = app["Price"].map("${:.2f}".format)
app = app.rename(columns={"Price":"Average Purchase Price"})
app
tpv = purchase_data.groupby("Age group")["Price"].sum()
tpv = tpv.reset_index()
# tpv["Price"] = tpv["Price"].map("${:.2f}".format).astype(float)
tpv = tpv.rename(columns = {"Price":"Total Purchase Value"})
tpv
tpv["Avg Total Purchase per Person"] = tpv["Total Purchase Value"]/age_group["Total count"]
tpv
combine = pdc.merge(app,on='Age group')
combine = combine.merge(tpv,on='Age group')
combine["Average Purchase Price"] = combine["Average Purchase Price"].map("${:.2f}".format)
combine["Total Purchase Value"] = combine["Total Purchase Value"].map("${:.2f}".format)
combine["Avg Total Purchase per Person"] = combine["Avg Total Purchase per Person"].map("${:.2f}".format)
combine = combine.rename(columns={'Age group':'Age Ranges'})
combine.set_index("Age Ranges")
purchase_data['Total Purchase Value'] = purchase_data.groupby(['SN'])['Price'].transform('sum')
purchase_data['Purchase Count'] = purchase_data.groupby(['SN'])['Price'].transform('count')
purchase_data['Average Purchase Price'] = purchase_data['Total Purchase Value']/purchase_data['Purchase Count']
purchase_data
purchase_unique = purchase_data.drop_duplicates(subset=['SN'])
purchase_unique
top_5 = purchase_unique.nlargest(5,'Total Purchase Value')
top_5
top_spenders = top_5[['SN','Purchase Count','Average Purchase Price','Total Purchase Value']]
top_spenders['Average Purchase Price'] = top_spenders['Average Purchase Price'].map("${:.2f}".format)
top_spenders['Total Purchase Value'] = top_spenders['Total Purchase Value'].map("${:.2f}".format)
top_spenders.set_index('SN')
top_spenders = top_5.loc[:,['SN','Purchase Count','Average Purchase Price','Total Purchase Value']]
top_spenders['Average Purchase Price'] = top_spenders['Average Purchase Price'].map("${:.2f}".format)
top_spenders['Total Purchase Value'] = top_spenders['Total Purchase Value'].map("${:.2f}".format)
top_spenders.set_index('SN')
purchase_data['Purchase Count'] = purchase_data.groupby(['Item ID'])['Price'].transform('count')
purchase_data
purchase_data['Total Purchase Value'] = purchase_data.groupby(['Item ID'])['Price'].transform('sum')
purchase_data
Item_count = purchase_data.drop_duplicates(subset=['Item ID'])
Item_count
sort_item = Item_count.sort_values('Purchase Count',ascending=False)
sort_item.head(6)
popular = Item_count.nlargest(5,'Purchase Count')
popular['Price'] = popular['Price'].map("${:.2f}".format)
popular['Total Purchase Value'] = popular['Total Purchase Value'].map("${:.2f}".format)
popular = popular.rename(columns={'Price':'Item Price'})
popular
Most_Popular_Items = popular[['Item ID','Item Name','Purchase Count','Item Price','Total Purchase Value']]
Most_Popular_Items.set_index('Item ID','Item Name')
most_profitable = Item_count.nlargest(5,'Total Purchase Value')
most_profitable
Most_Profitable_Items = most_profitable[['Item ID','Item Name','Purchase Count','Price','Total Purchase Value']]
Most_Profitable_Items = Most_Profitable_Items.rename(columns={'Price':'Item Price'})
Most_Profitable_Items['Item Price'] = Most_Profitable_Items['Item Price'].map("${:.2f}".format)
Most_Profitable_Items['Total Purchase Value'] = Most_Profitable_Items['Total Purchase Value'].map("${:.2f}".format)
Most_Profitable_Items.set_index('Item ID')
| 0.298185 | 0.613729 |
```
import spotipy
import pprint
import json
from spotipy.oauth2 import SpotifyClientCredentials
import pandas as pd
import numpy as np
keys = {}
i=0
def getAudioFeature(id):
feature = sp.audio_features(id)
if feature[0]:
s = pd.Series([feature[(0)]['danceability'], feature[(0)]['energy'], feature[(0)]['key'], feature[(0)]['loudness'],
feature[(0)]['mode'], feature[(0)]['speechiness'], feature[(0)]['acousticness'], feature[(0)]['instrumentalness'],
feature[(0)]['liveness'], feature[(0)]['valence'], feature[(0)]['tempo'], feature[(0)]['duration_ms'],
feature[(0)]['time_signature'] ],
index=['Danceability', 'Energy', 'Key', 'Loudness', 'Mode', 'Speechiness',
'Acousticness', 'Instrumentalness', 'Liveness', 'Valence', 'Tempo',
'Duration_ms', 'Time_signature'])
return s
else:
return pd.Series()
with open('keys.txt') as fp:
for line in fp:
keys[i] = line
i = i+1
print(keys[0])
print(keys[1])
client_credentials_manager = SpotifyClientCredentials(client_id= keys[0],client_secret= keys[1])
sp = spotipy.Spotify(client_credentials_manager=client_credentials_manager)
songTest = pd.DataFrame(columns=['Song Name','Artists Name', 'Danceability', 'Energy',
'Key', 'Loudness', 'Mode', 'Speechiness', 'Acousticness', 'Instrumentalness',
'Liveness', 'Valence', 'Tempo', 'Duration_ms', 'Time_signature'])
results = sp.search(q='year:2017', market='US', type='track', limit = 50, offset = 0)
offset = 0
while(results['tracks']['items']):
for element in results['tracks']['items']:
artistList = []
for artist in element['artists']:
artistList.append(artist['name'])
artistString = ",".join(artistList)
s = pd.Series(element['name'], index=['Song Name'])
s = s.append(pd.Series(artistString, index=['Artists Name']))
audioFeature = getAudioFeature(element['id'])
if not audioFeature.empty:
s = s.append(audioFeature)
songTest = songTest.append(s,ignore_index=True)
if not results['tracks']['next']:
results['tracks']['items'] = False
else:
offset = offset + 50
print(offset)
results = sp.search(q='year:2017', market='US', type='track', limit = 50, offset = offset)
songTest.to_csv('SongReleased2017WithFeatures.csv')
```
|
github_jupyter
|
import spotipy
import pprint
import json
from spotipy.oauth2 import SpotifyClientCredentials
import pandas as pd
import numpy as np
keys = {}
i=0
def getAudioFeature(id):
feature = sp.audio_features(id)
if feature[0]:
s = pd.Series([feature[(0)]['danceability'], feature[(0)]['energy'], feature[(0)]['key'], feature[(0)]['loudness'],
feature[(0)]['mode'], feature[(0)]['speechiness'], feature[(0)]['acousticness'], feature[(0)]['instrumentalness'],
feature[(0)]['liveness'], feature[(0)]['valence'], feature[(0)]['tempo'], feature[(0)]['duration_ms'],
feature[(0)]['time_signature'] ],
index=['Danceability', 'Energy', 'Key', 'Loudness', 'Mode', 'Speechiness',
'Acousticness', 'Instrumentalness', 'Liveness', 'Valence', 'Tempo',
'Duration_ms', 'Time_signature'])
return s
else:
return pd.Series()
with open('keys.txt') as fp:
for line in fp:
keys[i] = line
i = i+1
print(keys[0])
print(keys[1])
client_credentials_manager = SpotifyClientCredentials(client_id= keys[0],client_secret= keys[1])
sp = spotipy.Spotify(client_credentials_manager=client_credentials_manager)
songTest = pd.DataFrame(columns=['Song Name','Artists Name', 'Danceability', 'Energy',
'Key', 'Loudness', 'Mode', 'Speechiness', 'Acousticness', 'Instrumentalness',
'Liveness', 'Valence', 'Tempo', 'Duration_ms', 'Time_signature'])
results = sp.search(q='year:2017', market='US', type='track', limit = 50, offset = 0)
offset = 0
while(results['tracks']['items']):
for element in results['tracks']['items']:
artistList = []
for artist in element['artists']:
artistList.append(artist['name'])
artistString = ",".join(artistList)
s = pd.Series(element['name'], index=['Song Name'])
s = s.append(pd.Series(artistString, index=['Artists Name']))
audioFeature = getAudioFeature(element['id'])
if not audioFeature.empty:
s = s.append(audioFeature)
songTest = songTest.append(s,ignore_index=True)
if not results['tracks']['next']:
results['tracks']['items'] = False
else:
offset = offset + 50
print(offset)
results = sp.search(q='year:2017', market='US', type='track', limit = 50, offset = offset)
songTest.to_csv('SongReleased2017WithFeatures.csv')
| 0.19031 | 0.139133 |
```
import numpy as np
import pandas as pd
import seaborn as sns
from matplotlib import pyplot as plt
```
## Data
Data used here are in the current repository.
Full and additional datasets you can find here:
[Olympic Games, 1986-2021](https://www.kaggle.com/piterfm/olympic-games-medals-19862018)
[Tokyo 2020 Olympics](https://www.kaggle.com/piterfm/tokyo-2020-olympics)
[Tokyo 2020 Paralympics](https://www.kaggle.com/piterfm/tokyo-2020-paralympics). [Data Visualization](https://share.streamlit.io/petroivaniuk/paralympics-2020/main/paralympics-app.py)
```
path_data = 'data/medals.pkl'
df = pd.read_pickle(path_data)
df.shape
df.head()
```
## Step 1. Filtering
```
season = 'Summer'
df[df['game_season']==season].reset_index(drop=True).head(10)
```
## Step 2. Aggregation
```
discipline = 'Archery'
df_agg = df.groupby(['discipline_title', 'game_year'])['participant_type'].count().reset_index()\
.rename(columns={'participant_type':'#medals'})
df_agg[df_agg['discipline_title']==discipline]
```
## Step 3. Pivoting
```
def highlight_zero(s, props=''):
return np.where(s==0, props, '')
df_pivot = df_agg.pivot('discipline_title', 'game_year', '#medals').fillna(0).astype(int)
# The next line of code just show styled first 10 lines of table df_pivot
df_pivot.head(10).style.apply(highlight_zero, props='color:white;background-color:pink', axis=None)
```
## Step 2 + Step 3
```
df.pivot_table(values=['participant_type'],
index=['discipline_title'],
columns=['game_year'],
fill_value=0,
aggfunc=np.size)
```
## Step 4. Visualization
```
# simple heatmap without settings, let's go forward through the notebook
plt.figure(figsize=(10, 10))
sns.heatmap(df_pivot, annot=False, cbar=False,
linewidths=0.8, linecolor='black',
square=True, cmap='Spectral')
plt.show()
```
## All Steps together
```
def get_disciplines_game(df, season):
'''
'''
df = df[df['game_season']==season].reset_index(drop=True).copy()
df_disciplines_year = df.groupby(['discipline_title', 'game_year'])['participant_type']\
.count().reset_index()
df_heatmap = df_disciplines_year.pivot('discipline_title', 'game_year', 'participant_type')
df_heatmap[df_heatmap > 0] = 1
column_list = list(df_heatmap.columns)
column_last = column_list[-1]
disciplines_current = df_heatmap[df_heatmap[column_last]==1].sort_values(column_list)
disciplines_current_not = df_heatmap[df_heatmap[column_last]!=1].sort_values(column_list)
df_heatmap = pd.concat([disciplines_current, disciplines_current_not])
df_heatmap.columns = [str(col)[:-2]+'\n'+str(col)[-2:] for col in column_list]
df_heatmap.index = [idx.replace(' ', '\n', 1) for idx in df_heatmap.index]
return df_heatmap
def plot_disciplines(data, title, size=(16,16)):
'''
Return plot
'''
plt.figure(figsize=size)
ax = sns.heatmap(data, annot=False, cbar=False,
linewidths=0.8, linecolor='black',
square=True, cmap='Spectral')
ax.tick_params(axis='x', labelsize=14)
ax.tick_params(axis='y', labelsize=16)
ax.set_ylabel('')
ax.set_xlabel('')
ax.xaxis.tick_top()
ax.xaxis.set_ticks_position('none')
ax.yaxis.set_ticks_position('none')
ax.spines[['bottom', 'right']].set_visible(True)
ax.set_title('{} Games'.format(title), size=22)
plt.tight_layout()
plt.savefig('images/heatmap_{}_games.png'.format(title.lower()), dpi=200)
plt.show()
```
## Timeline of disciplines contested at the Olympic Summer Games, 1986-2020
```
season = 'Summer'
size = (20, 38)
disciplines = get_disciplines_game(df, season)
plot_disciplines(disciplines, title=season, size=size)
```
## Timeline of disciplines contested at the Olympic Winter Games, 1986-2020
```
season = 'Winter'
size = (16,12)
disciplines = get_disciplines_game(df, season)
plot_disciplines(disciplines, title=season, size=size)
```
## Bonus
```
season = 'Summer'
df_summer = df[df['game_season']==season].reset_index(drop=True)
df_summer.drop(['game_season'], axis=1, inplace=True)
df_summer.shape
def sort_games_name(game_name_list):
'''
Input: ['Sydney 2000', 'Atlanta 1996', 'Beijing 2008', 'Athens 2004']
Output: ['Atlanta 1996', 'Sydney 2000', 'Athens 2004', 'Beijing 2008'
'''
game_name_tuple_split = [(' '.join(i.split(' ')[:-1]), i.split(' ')[-1]) for i in game_name_list]
game_name_tuple_sorted = sorted(game_name_tuple_split, key=lambda x: x[1])
game_name_list_sorted = [' '.join(i) for i in game_name_tuple_sorted]
return game_name_list_sorted
def get_country_medal(data, country):
data_country = data[data['country_name']==country]
data_medal = data_country.groupby(['game_name', 'discipline_title'])['participant_type']\
.count().reset_index()
data_medal = data_medal.pivot('discipline_title', 'game_name', 'participant_type')
data_medal = data_medal[sort_games_name(list(data_medal.columns))]
if len(list(data_medal.columns))<10:
data_medal.columns = [col.replace(' ', '\n') for col in data_medal.columns]
data_medal.index = [idx.replace(' ', '\n', 1) for idx in data_medal.index]
else:
data_medal.columns = [col.split(' ')[-1] for col in data_medal.columns]
data_medal['Total'] = data_medal.sum(axis=1)
data_medal.loc["Total"] = data_medal.sum()
return data_medal
def plot_country_medal(data, title):
plt.figure(figsize=(20, 20))
ax = sns.heatmap(data, annot=True, annot_kws={"fontsize":16},
cbar=False, linewidths=.8, fmt='g', cmap='coolwarm')
ax.tick_params(axis='x', which='major', labelsize=16)
ax.tick_params(axis='y', which='major', labelsize=18)
ax.xaxis.tick_top()
ax.set_ylabel('')
ax.set_xlabel('')
plt.tight_layout()
plt.savefig('images/medals_{}.png'.format(title.lower()), dpi=200)
plt.show()
```
### Medals, Ukraine
**Here could be your country!!!**
```
country = "Ukraine"
country_medal_ua = get_country_medal(df_summer, country)
plot_country_medal(country_medal_ua, title=country)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import seaborn as sns
from matplotlib import pyplot as plt
path_data = 'data/medals.pkl'
df = pd.read_pickle(path_data)
df.shape
df.head()
season = 'Summer'
df[df['game_season']==season].reset_index(drop=True).head(10)
discipline = 'Archery'
df_agg = df.groupby(['discipline_title', 'game_year'])['participant_type'].count().reset_index()\
.rename(columns={'participant_type':'#medals'})
df_agg[df_agg['discipline_title']==discipline]
def highlight_zero(s, props=''):
return np.where(s==0, props, '')
df_pivot = df_agg.pivot('discipline_title', 'game_year', '#medals').fillna(0).astype(int)
# The next line of code just show styled first 10 lines of table df_pivot
df_pivot.head(10).style.apply(highlight_zero, props='color:white;background-color:pink', axis=None)
df.pivot_table(values=['participant_type'],
index=['discipline_title'],
columns=['game_year'],
fill_value=0,
aggfunc=np.size)
# simple heatmap without settings, let's go forward through the notebook
plt.figure(figsize=(10, 10))
sns.heatmap(df_pivot, annot=False, cbar=False,
linewidths=0.8, linecolor='black',
square=True, cmap='Spectral')
plt.show()
def get_disciplines_game(df, season):
'''
'''
df = df[df['game_season']==season].reset_index(drop=True).copy()
df_disciplines_year = df.groupby(['discipline_title', 'game_year'])['participant_type']\
.count().reset_index()
df_heatmap = df_disciplines_year.pivot('discipline_title', 'game_year', 'participant_type')
df_heatmap[df_heatmap > 0] = 1
column_list = list(df_heatmap.columns)
column_last = column_list[-1]
disciplines_current = df_heatmap[df_heatmap[column_last]==1].sort_values(column_list)
disciplines_current_not = df_heatmap[df_heatmap[column_last]!=1].sort_values(column_list)
df_heatmap = pd.concat([disciplines_current, disciplines_current_not])
df_heatmap.columns = [str(col)[:-2]+'\n'+str(col)[-2:] for col in column_list]
df_heatmap.index = [idx.replace(' ', '\n', 1) for idx in df_heatmap.index]
return df_heatmap
def plot_disciplines(data, title, size=(16,16)):
'''
Return plot
'''
plt.figure(figsize=size)
ax = sns.heatmap(data, annot=False, cbar=False,
linewidths=0.8, linecolor='black',
square=True, cmap='Spectral')
ax.tick_params(axis='x', labelsize=14)
ax.tick_params(axis='y', labelsize=16)
ax.set_ylabel('')
ax.set_xlabel('')
ax.xaxis.tick_top()
ax.xaxis.set_ticks_position('none')
ax.yaxis.set_ticks_position('none')
ax.spines[['bottom', 'right']].set_visible(True)
ax.set_title('{} Games'.format(title), size=22)
plt.tight_layout()
plt.savefig('images/heatmap_{}_games.png'.format(title.lower()), dpi=200)
plt.show()
season = 'Summer'
size = (20, 38)
disciplines = get_disciplines_game(df, season)
plot_disciplines(disciplines, title=season, size=size)
season = 'Winter'
size = (16,12)
disciplines = get_disciplines_game(df, season)
plot_disciplines(disciplines, title=season, size=size)
season = 'Summer'
df_summer = df[df['game_season']==season].reset_index(drop=True)
df_summer.drop(['game_season'], axis=1, inplace=True)
df_summer.shape
def sort_games_name(game_name_list):
'''
Input: ['Sydney 2000', 'Atlanta 1996', 'Beijing 2008', 'Athens 2004']
Output: ['Atlanta 1996', 'Sydney 2000', 'Athens 2004', 'Beijing 2008'
'''
game_name_tuple_split = [(' '.join(i.split(' ')[:-1]), i.split(' ')[-1]) for i in game_name_list]
game_name_tuple_sorted = sorted(game_name_tuple_split, key=lambda x: x[1])
game_name_list_sorted = [' '.join(i) for i in game_name_tuple_sorted]
return game_name_list_sorted
def get_country_medal(data, country):
data_country = data[data['country_name']==country]
data_medal = data_country.groupby(['game_name', 'discipline_title'])['participant_type']\
.count().reset_index()
data_medal = data_medal.pivot('discipline_title', 'game_name', 'participant_type')
data_medal = data_medal[sort_games_name(list(data_medal.columns))]
if len(list(data_medal.columns))<10:
data_medal.columns = [col.replace(' ', '\n') for col in data_medal.columns]
data_medal.index = [idx.replace(' ', '\n', 1) for idx in data_medal.index]
else:
data_medal.columns = [col.split(' ')[-1] for col in data_medal.columns]
data_medal['Total'] = data_medal.sum(axis=1)
data_medal.loc["Total"] = data_medal.sum()
return data_medal
def plot_country_medal(data, title):
plt.figure(figsize=(20, 20))
ax = sns.heatmap(data, annot=True, annot_kws={"fontsize":16},
cbar=False, linewidths=.8, fmt='g', cmap='coolwarm')
ax.tick_params(axis='x', which='major', labelsize=16)
ax.tick_params(axis='y', which='major', labelsize=18)
ax.xaxis.tick_top()
ax.set_ylabel('')
ax.set_xlabel('')
plt.tight_layout()
plt.savefig('images/medals_{}.png'.format(title.lower()), dpi=200)
plt.show()
country = "Ukraine"
country_medal_ua = get_country_medal(df_summer, country)
plot_country_medal(country_medal_ua, title=country)
| 0.392803 | 0.858125 |
## Calculate Evaluation Metrics on Mask R-CNN Keypoint Model
```
USE_GPU = False
MODEL_NAME = "run1"
%matplotlib inline
import importlib
import os
if not USE_GPU:
os.environ["CUDA_VISIBLE_DEVICES"] = "-1"
import sys
import random
import math
import re
import time
import numpy as np
import cv2
import matplotlib
import matplotlib.pyplot as plt
import imgaug
import json
# Root directory of the project
ROOT_DIR = os.path.abspath(".")
# Import Mask RCNN
sys.path.append(ROOT_DIR) # To find local version of the library
from mrcnn.config import Config
from mrcnn import utils
import mrcnn.model as modellib
from mrcnn import visualize
from mrcnn.model import log
# Directory to save logs and trained model
MODEL_DIR = os.path.join(ROOT_DIR, "logs", MODEL_NAME)
from coco import coco_keypoints
# COCO dataset dir
COCO_DATA_DIR = "A:/Programming/DeepLearningDatasets/coco" if os.path.isdir("A:/Programming/DeepLearningDatasets/coco") else os.path.join(ROOT_DIR, "data/coco")
# Prepare dataset
dataset_val = coco_keypoints.CocoDataset()
dataset_val.load_coco(COCO_DATA_DIR, subset="val", year="2017", auto_download=True)
dataset_val.prepare()
```
## Initialize Model From Weights
```
class InferenceConfig(coco_keypoints.CocoConfig):
GPU_COUNT = 1
IMAGES_PER_GPU = 1
BACKBONE = "resnet50"
NUM_CLASSES = dataset_val.num_classes
NUM_KEYPOINTS = dataset_val.num_kp_classes
RPN_NMS_THRESHOLD = 0.5
config = InferenceConfig()
# Recreate the model in inference mode
model = modellib.MaskRCNN(mode="inference",
config=config,
model_dir=MODEL_DIR)
# Get path to saved weights
model_path = model.find_last()[1]
# Load trained weights (fill in path to trained weights here)
assert model_path != "", "Provide path to trained weights"
print("Loading weights from ", model_path)
model.load_weights(model_path, by_name=True)
```
## Generate Detections
Detections are written to ```MODEL_DIR/detections.json```
```
def unmold_detections(result, dataset, image_id, config):
# Get scale and padding for image
image_info = dataset.image_info[image_id]
w, h = image_info["width"], image_info["height"]
_, window, scale, padding, crop =\
utils.resize_image(np.zeros((h, w, 3)),
min_dim=config.IMAGE_MIN_DIM,
max_dim=config.IMAGE_MAX_DIM,
min_scale=config.IMAGE_MIN_SCALE,
mode=config.IMAGE_RESIZE_MODE)
pad_top, pad_bot, pad_left, pad_right = padding[0][0], padding[0][1], padding[1][0], padding[1][1]
# Generate detections
detections = []
for instance_masks, score in zip(result["kp_masks"], result["scores"]):
keypoints = []
for kp_mask in instance_masks:
kpy, kpx = np.unravel_index(np.argmax(kp_mask, axis=None), kp_mask.shape)
kpx = (kpx - pad_left) / scale
kpy = (kpy - pad_top) / scale
keypoints.append(int(kpx))
keypoints.append(int(kpy))
keypoints.append(1)
detections.append({
"image_id": int(image_info["id"]),
"category_id": 1,
"keypoints": keypoints,
"score": float(score)
})
return detections
detections = []
N = len(dataset_val.image_ids)
for image_id, image_info in zip(dataset_val.image_ids, dataset_val.image_info):
print("image_id",image_id)
if image_id % 10 == 0: print("Processing {}/{}...".format(image_id+1, N))
# Load an image from the validation set
image = modellib.load_image_gt(dataset_val, config, image_id)[0]
# Detect keypoints
detection_start_time = time.time()
result = model.detect([image])[0]
print("Detection time {}s".format(time.time() - detection_start_time))
detections.extend(unmold_detections(result, dataset_val, image_id, config))
# Save data to .json
with open(os.path.join(MODEL_DIR, "detections.json"), "w") as f:
json.dump(detections, f)
```
## Coco-analyze
```
## general imports
import json
import numpy as np
## COCO imports
from analyze.pycocotools.coco import COCO
from analyze.pycocotools.cocoeval import COCOeval
from analyze.pycocotools.cocoanalyze import COCOanalyze
## plotting imports
%matplotlib inline
import matplotlib.pyplot as plt
import skimage.io as io
## set paths
dataDir = COCO_DATA_DIR
dataType = "val2017"
annType = "person_keypoints"
teamName = "pose_rcnn"
annFile = os.path.join(COCO_DATA_DIR, "annotations/{}_{}.json".format(annType, dataType))
resFile = os.path.join(MODEL_DIR, "detections.json")
print("{:10}[{}]".format("annFile:",annFile))
print("{:10}[{}]".format("resFile:",resFile))
gt_data = json.load(open(annFile,"rb"))
imgs_info = {i["id"]:{"id":i["id"] ,
"width":i["width"],
"height":i["height"]}
for i in gt_data["images"]}
team_dts = json.load(open(resFile,"rb"))
team_dts = [d for d in team_dts if d["image_id"] in imgs_info]
team_img_ids = set([d["image_id"] for d in team_dts])
print("Loaded [{}] instances in [{}] images.".format(len(team_dts),len(imgs_info)))
## load ground truth annotations
coco_gt = COCO( annFile )
## initialize COCO detections api
coco_dt = coco_gt.loadRes( team_dts )
## initialize COCO analyze api
coco_analyze = COCOanalyze(coco_gt, coco_dt, "keypoints")
import skimage.io as io
img_id = list(team_img_ids)[0]
print(img_id)
# Load image
img = coco_gt.loadImgs(img_id)[0]
I = io.imread(img['coco_url'])
plt.imshow(I); plt.axis('off')
anns_gt = coco_gt.loadAnns(coco_gt.getAnnIds(imgIds=img_id))
coco_gt.showAnns(anns_gt)
plt.show()
plt.imshow(I); plt.axis('off')
anns_dt = coco_dt.loadAnns(coco_dt.getAnnIds(imgIds=img_id))
coco_dt.showAnns(anns_dt)
#coco_dt.showAnns(coco_dt.loadAnns(coco_dt.getAnnIds(imgIds=img_id)))
plt.show()
kpx_dt, kpy_dt = np.array(anns_dt[0]["keypoints"][0::3]), np.array(anns_dt[0]["keypoints"][1::3])
kpx_gt, kpy_gt = np.array(anns_gt[0]["keypoints"][0::3]), np.array(anns_gt[0]["keypoints"][1::3])
print(np.sum(np.sqrt((kpx_dt - kpx_gt) ** 2 + (kpy_dt - kpy_gt) ** 2)))
# use evaluate() method for standard coco evaluation
# input arguments:
# - verbose : verbose outputs (default: False)
# - makeplots : plots eval results (default: False)
# - savedir : path to savedir (default: None)
# - team_name : team name string (default: None)
coco_analyze.evaluate(verbose=True, makeplots=True)
## NOTE: the values below are all default
# set OKS threshold of the extended error analysis
coco_analyze.params.oksThrs = [.05,.50,.95]
# set OKS threshold required to match a detection to a ground truth
coco_analyze.params.oksLocThrs = .1
# set KS threshold limits defining jitter errors
coco_analyze.params.jitterKsThrs = [.5,.85]
# set the localization errors to analyze and in what order
# note: different order will show different progressive improvement
# to study impact of single error type, study in isolation
coco_analyze.params.err_types = ["miss","swap","inversion","jitter"]
# area ranges for evaluation
# "all" range is union of medium and large
coco_analyze.params.areaRng = [[32 ** 2, 1e5 ** 2]] #[96 ** 2, 1e5 ** 2],[32 ** 2, 96 ** 2]
coco_analyze.params.areaRngLbl = ["all"] # "large","medium"
coco_analyze.params.maxDets = [20]
coco_analyze.evaluate(verbose=True, makeplots=True)
# use analyze() method for advanced error analysis
# input arguments:
# - check_kpts : analyze keypoint localization errors for detections with a match (default: True)
# : default errors types are ["jitter","inversion","swap","miss"]
# - check_scores : analyze optimal score (maximizing oks over all matches) for every detection (default: True)
# - check_bkgd : analyze background false positives and false negatives (default: True)
coco_analyze.analyze(check_kpts=True, check_scores=True, check_bckgd=True)
# use summarize() method to get the results after progressive correction of errors
# input arguments:
# - makeplots : plots eval results (default: False)
# - savedir : path to savedir (default: None)
# - team_name : team name string (default: None)
coco_analyze.summarize(makeplots=True)
## print the performance summary
for stat in coco_analyze.stats: print(stat)
## after analyze() has been called the following variables are available
# list of the corrected detections
corrected_dts = coco_analyze.corrected_dts["all"]
i = 17
# info on keypoint detection localization error
print "good: %s"%corrected_dts[i]["good"]
print "miss: %s"%corrected_dts[i]["miss"]
print "swap: %s"%corrected_dts[i]["swap"]
print "inv.: %s"%corrected_dts[i]["inversion"]
print "jit.: %s\n"%corrected_dts[i]["jitter"]
# corrected keypoint locations
print "predicted keypoints:\n %s"%corrected_dts[i]["keypoints"]
print "corrected keypoints:\n %s\n"%corrected_dts[i]["opt_keypoints"]
# optimal detection score
print "original score: %s"%corrected_dts[i]["score"]
print "optimal score: %s\n"%corrected_dts[i]["opt_score"]
## after summarize() has been called the following variables are available
# list of the false positive detections and missed ground-truth annotations
false_pos_dts = coco_analyze.false_pos_dts
false_neg_gts = coco_analyze.false_neg_gts
for oks in coco_analyze.params.oksThrs:
print "Oks:[%.2f] - Num.FP:[%d] - Num.FN:[%d]"%(oks,len(false_pos_dts["all",str(oks)]),len(false_neg_gts["all",str(oks)]))
```
|
github_jupyter
|
USE_GPU = False
MODEL_NAME = "run1"
%matplotlib inline
import importlib
import os
if not USE_GPU:
os.environ["CUDA_VISIBLE_DEVICES"] = "-1"
import sys
import random
import math
import re
import time
import numpy as np
import cv2
import matplotlib
import matplotlib.pyplot as plt
import imgaug
import json
# Root directory of the project
ROOT_DIR = os.path.abspath(".")
# Import Mask RCNN
sys.path.append(ROOT_DIR) # To find local version of the library
from mrcnn.config import Config
from mrcnn import utils
import mrcnn.model as modellib
from mrcnn import visualize
from mrcnn.model import log
# Directory to save logs and trained model
MODEL_DIR = os.path.join(ROOT_DIR, "logs", MODEL_NAME)
from coco import coco_keypoints
# COCO dataset dir
COCO_DATA_DIR = "A:/Programming/DeepLearningDatasets/coco" if os.path.isdir("A:/Programming/DeepLearningDatasets/coco") else os.path.join(ROOT_DIR, "data/coco")
# Prepare dataset
dataset_val = coco_keypoints.CocoDataset()
dataset_val.load_coco(COCO_DATA_DIR, subset="val", year="2017", auto_download=True)
dataset_val.prepare()
class InferenceConfig(coco_keypoints.CocoConfig):
GPU_COUNT = 1
IMAGES_PER_GPU = 1
BACKBONE = "resnet50"
NUM_CLASSES = dataset_val.num_classes
NUM_KEYPOINTS = dataset_val.num_kp_classes
RPN_NMS_THRESHOLD = 0.5
config = InferenceConfig()
# Recreate the model in inference mode
model = modellib.MaskRCNN(mode="inference",
config=config,
model_dir=MODEL_DIR)
# Get path to saved weights
model_path = model.find_last()[1]
# Load trained weights (fill in path to trained weights here)
assert model_path != "", "Provide path to trained weights"
print("Loading weights from ", model_path)
model.load_weights(model_path, by_name=True)
## Coco-analyze
| 0.355327 | 0.55652 |
# RadarCOVID-Report
## Data Extraction
```
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import pycountry
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
```
### Constants
```
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
```
### Parameters
```
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
environment_enable_multi_backend_download = \
os.environ.get("RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD")
if environment_enable_multi_backend_download:
report_backend_identifiers = None
else:
report_backend_identifiers = [report_backend_identifier]
report_backend_identifiers
environment_invalid_shared_diagnoses_dates = \
os.environ.get("RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES")
if environment_invalid_shared_diagnoses_dates:
invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(",")
else:
invalid_shared_diagnoses_dates = []
invalid_shared_diagnoses_dates
```
### COVID-19 Cases
```
report_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=report_backend_identifier)
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe():
return pd.read_csv(
"https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv")
confirmed_df_ = download_cases_dataframe()
confirmed_df = confirmed_df_.copy()
confirmed_df = confirmed_df[["date", "new_cases", "iso_code"]]
confirmed_df.rename(
columns={
"date": "sample_date",
"iso_code": "country_code",
},
inplace=True)
def convert_iso_alpha_3_to_alpha_2(x):
try:
return pycountry.countries.get(alpha_3=x).alpha_2
except Exception as e:
logging.info(f"Error converting country ISO Alpha 3 code '{x}': {repr(e)}")
return None
confirmed_df["country_code"] = confirmed_df.country_code.apply(convert_iso_alpha_3_to_alpha_2)
confirmed_df.dropna(inplace=True)
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_days_df["sample_date_string"] = \
confirmed_days_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_days_df.tail()
def sort_source_regions_for_display(source_regions: list) -> list:
if report_backend_identifier in source_regions:
source_regions = [report_backend_identifier] + \
list(sorted(set(source_regions).difference([report_backend_identifier])))
else:
source_regions = list(sorted(source_regions))
return source_regions
report_source_regions = report_backend_client.source_regions_for_date(
date=extraction_datetime.date())
report_source_regions = sort_source_regions_for_display(
source_regions=report_source_regions)
report_source_regions
def get_cases_dataframe(source_regions_for_date_function, columns_suffix=None):
source_regions_at_date_df = confirmed_days_df.copy()
source_regions_at_date_df["source_regions_at_date"] = \
source_regions_at_date_df.sample_date.apply(
lambda x: source_regions_for_date_function(date=x))
source_regions_at_date_df.sort_values("sample_date", inplace=True)
source_regions_at_date_df["_source_regions_group"] = source_regions_at_date_df. \
source_regions_at_date.apply(lambda x: ",".join(sort_source_regions_for_display(x)))
source_regions_at_date_df.tail()
#%%
source_regions_for_summary_df_ = \
source_regions_at_date_df[["sample_date", "_source_regions_group"]].copy()
source_regions_for_summary_df_.rename(columns={"_source_regions_group": "source_regions"}, inplace=True)
source_regions_for_summary_df_.tail()
#%%
confirmed_output_columns = ["sample_date", "new_cases", "covid_cases"]
confirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)
for source_regions_group, source_regions_group_series in \
source_regions_at_date_df.groupby("_source_regions_group"):
source_regions_set = set(source_regions_group.split(","))
confirmed_source_regions_set_df = \
confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()
confirmed_source_regions_group_df = \
confirmed_source_regions_set_df.groupby("sample_date").new_cases.sum() \
.reset_index().sort_values("sample_date")
confirmed_source_regions_group_df["covid_cases"] = \
confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[confirmed_output_columns]
confirmed_source_regions_group_df.fillna(method="ffill", inplace=True)
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[
confirmed_source_regions_group_df.sample_date.isin(
source_regions_group_series.sample_date_string)]
confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)
result_df = confirmed_output_df.copy()
result_df.tail()
#%%
result_df.rename(columns={"sample_date": "sample_date_string"}, inplace=True)
result_df = confirmed_days_df[["sample_date_string"]].merge(result_df, how="left")
result_df.sort_values("sample_date_string", inplace=True)
result_df.fillna(method="ffill", inplace=True)
result_df.tail()
#%%
result_df[["new_cases", "covid_cases"]].plot()
if columns_suffix:
result_df.rename(
columns={
"new_cases": "new_cases_" + columns_suffix,
"covid_cases": "covid_cases_" + columns_suffix},
inplace=True)
return result_df, source_regions_for_summary_df_
confirmed_eu_df, source_regions_for_summary_df = get_cases_dataframe(
report_backend_client.source_regions_for_date)
confirmed_es_df, _ = get_cases_dataframe(
lambda date: [spain_region_country_code],
columns_suffix=spain_region_country_code.lower())
```
### Extract API TEKs
```
raw_zip_path_prefix = "Data/TEKs/Raw/"
fail_on_error_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=fail_on_error_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
```
### Dump API TEKs
```
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_df.head()
```
### Load TEK Dumps
```
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
```
### Daily New TEKs
```
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \
.groupby(["upload_date"]).shared_teks.max().reset_index() \
.sort_values(["upload_date"], ascending=False) \
.rename(columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
invalid_shared_diagnoses_dates_mask = \
estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)
estimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0
estimated_shared_diagnoses_df.head()
```
### Hourly New TEKs
```
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
```
### Official Statistics
```
import requests
import pandas.io.json
official_stats_response = requests.get("https://radarcovidpre.covid19.gob.es/kpi/statistics/basics")
official_stats_response.raise_for_status()
official_stats_df_ = pandas.io.json.json_normalize(official_stats_response.json())
official_stats_df = official_stats_df_.copy()
official_stats_df = official_stats_df.append(pd.DataFrame({
"date": ["06/12/2020"],
"applicationsDownloads.totalAcummulated": [5653519],
"communicatedContagions.totalAcummulated": [21925],
}), sort=False)
official_stats_df["date"] = pd.to_datetime(official_stats_df["date"], dayfirst=True)
official_stats_df.head()
official_stats_column_map = {
"date": "sample_date",
"applicationsDownloads.totalAcummulated": "app_downloads_es_accumulated",
"communicatedContagions.totalAcummulated": "shared_diagnoses_es_accumulated",
}
accumulated_suffix = "_accumulated"
accumulated_values_columns = \
list(filter(lambda x: x.endswith(accumulated_suffix), official_stats_column_map.values()))
interpolated_values_columns = \
list(map(lambda x: x[:-len(accumulated_suffix)], accumulated_values_columns))
official_stats_df = \
official_stats_df[official_stats_column_map.keys()] \
.rename(columns=official_stats_column_map)
official_stats_df.head()
official_stats_df = confirmed_days_df.merge(official_stats_df, how="left")
official_stats_df.sort_values("sample_date", ascending=False, inplace=True)
official_stats_df.head()
official_stats_df[accumulated_values_columns] = \
official_stats_df[accumulated_values_columns] \
.astype(float).interpolate(limit_area="inside")
official_stats_df[interpolated_values_columns] = \
official_stats_df[accumulated_values_columns].diff(periods=-1)
official_stats_df.drop(columns="sample_date", inplace=True)
official_stats_df.head()
```
### Data Merge
```
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
official_stats_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_eu_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df = confirmed_es_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df = result_summary_df.merge(source_regions_for_summary_df, how="left")
result_summary_df.set_index(["sample_date", "source_regions"], inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(result_summary_df.shared_diagnoses_es / result_summary_df.covid_cases_es).fillna(0)
result_summary_df.head(daily_plot_days)
weekly_result_summary_df = result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(7).agg({
"covid_cases": "sum",
"covid_cases_es": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum",
"shared_diagnoses_es": "sum",
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
weekly_result_summary_df = weekly_result_summary_df.fillna(0).astype(int)
weekly_result_summary_df["teks_per_shared_diagnosis"] = \
(weekly_result_summary_df.shared_teks_by_upload_date / weekly_result_summary_df.shared_diagnoses).fillna(0)
weekly_result_summary_df["shared_diagnoses_per_covid_case"] = \
(weekly_result_summary_df.shared_diagnoses / weekly_result_summary_df.covid_cases).fillna(0)
weekly_result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(weekly_result_summary_df.shared_diagnoses_es / weekly_result_summary_df.covid_cases_es).fillna(0)
weekly_result_summary_df.head()
last_7_days_summary = weekly_result_summary_df.to_dict(orient="records")[1]
last_7_days_summary
```
## Report Results
```
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"source_regions": "Source Countries",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases (Source Countries)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date (Source Countries)",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date (Source Countries)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date (Source Countries)",
"shared_diagnoses": "Shared Diagnoses (Source Countries – Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis (Source Countries)",
"shared_diagnoses_per_covid_case": "Usage Ratio (Source Countries)",
"covid_cases_es": "COVID-19 Cases (Spain)",
"app_downloads_es": "App Downloads (Spain – Official)",
"shared_diagnoses_es": "Shared Diagnoses (Spain – Official)",
"shared_diagnoses_per_covid_case_es": "Usage Ratio (Spain)",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
"covid_cases_es",
"app_downloads_es",
"shared_diagnoses_es",
"shared_diagnoses_per_covid_case_es",
]
summary_percentage_columns= [
"shared_diagnoses_per_covid_case_es",
"shared_diagnoses_per_covid_case",
]
```
### Daily Summary Table
```
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
```
### Daily Summary Plots
```
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.droplevel(level=["source_regions"]) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 30), legend=False)
ax_ = summary_ax_list[0]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
for percentage_column in summary_percentage_columns:
percentage_column_index = summary_columns.index(percentage_column)
summary_ax_list[percentage_column_index].yaxis \
.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
```
### Daily Generation to Upload Period Table
```
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
```
### Hourly Summary Plots
```
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
```
### Publish Results
```
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case_es"]: lambda x: f"{x:.2%}" if x != 0 else "",
}
general_columns = \
list(filter(lambda x: x not in display_formatters, display_column_name_mapping.values()))
general_formatter = lambda x: f"{x}" if x != 0 else ""
display_formatters.update(dict(map(lambda x: (x, general_formatter), general_columns)))
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index.get_level_values("sample_date") == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.item()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.item()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.item()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.item()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.item()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
if len(report_source_regions) == 1:
display_brief_source_regions = report_source_regions[0]
else:
display_brief_source_regions = f"{len(report_source_regions)} 🇪🇺"
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
df = df.copy()
df_styler = df.style.format(display_formatters)
media_path = get_temporary_image_path()
dfi.export(df_styler, media_path)
return media_path
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
```
### Save Results
```
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
```
### Publish Results as JSON
```
def generate_summary_api_results(df: pd.DataFrame) -> list:
api_df = df.reset_index().copy()
api_df["sample_date_string"] = \
api_df["sample_date"].dt.strftime("%Y-%m-%d")
api_df["source_regions"] = \
api_df["source_regions"].apply(lambda x: x.split(","))
return api_df.to_dict(orient="records")
summary_api_results = \
generate_summary_api_results(df=result_summary_df)
today_summary_api_results = \
generate_summary_api_results(df=extraction_date_result_summary_df)[0]
summary_results = dict(
backend_identifier=report_backend_identifier,
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=today_summary_api_results,
last_7_days=last_7_days_summary,
daily_results=summary_api_results)
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
```
### Publish on README
```
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
```
### Publish on Twitter
```
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
Source Countries: {display_brief_source_regions}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: ≤{shared_diagnoses_per_covid_case:.2%}
Last 7 Days:
- Shared Diagnoses: ≤{last_7_days_summary["shared_diagnoses"]:.0f}
- Usage Ratio: ≤{last_7_days_summary["shared_diagnoses_per_covid_case"]:.2%}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
```
|
github_jupyter
|
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import pycountry
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
environment_enable_multi_backend_download = \
os.environ.get("RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD")
if environment_enable_multi_backend_download:
report_backend_identifiers = None
else:
report_backend_identifiers = [report_backend_identifier]
report_backend_identifiers
environment_invalid_shared_diagnoses_dates = \
os.environ.get("RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES")
if environment_invalid_shared_diagnoses_dates:
invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(",")
else:
invalid_shared_diagnoses_dates = []
invalid_shared_diagnoses_dates
report_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=report_backend_identifier)
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe():
return pd.read_csv(
"https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv")
confirmed_df_ = download_cases_dataframe()
confirmed_df = confirmed_df_.copy()
confirmed_df = confirmed_df[["date", "new_cases", "iso_code"]]
confirmed_df.rename(
columns={
"date": "sample_date",
"iso_code": "country_code",
},
inplace=True)
def convert_iso_alpha_3_to_alpha_2(x):
try:
return pycountry.countries.get(alpha_3=x).alpha_2
except Exception as e:
logging.info(f"Error converting country ISO Alpha 3 code '{x}': {repr(e)}")
return None
confirmed_df["country_code"] = confirmed_df.country_code.apply(convert_iso_alpha_3_to_alpha_2)
confirmed_df.dropna(inplace=True)
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_days_df["sample_date_string"] = \
confirmed_days_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_days_df.tail()
def sort_source_regions_for_display(source_regions: list) -> list:
if report_backend_identifier in source_regions:
source_regions = [report_backend_identifier] + \
list(sorted(set(source_regions).difference([report_backend_identifier])))
else:
source_regions = list(sorted(source_regions))
return source_regions
report_source_regions = report_backend_client.source_regions_for_date(
date=extraction_datetime.date())
report_source_regions = sort_source_regions_for_display(
source_regions=report_source_regions)
report_source_regions
def get_cases_dataframe(source_regions_for_date_function, columns_suffix=None):
source_regions_at_date_df = confirmed_days_df.copy()
source_regions_at_date_df["source_regions_at_date"] = \
source_regions_at_date_df.sample_date.apply(
lambda x: source_regions_for_date_function(date=x))
source_regions_at_date_df.sort_values("sample_date", inplace=True)
source_regions_at_date_df["_source_regions_group"] = source_regions_at_date_df. \
source_regions_at_date.apply(lambda x: ",".join(sort_source_regions_for_display(x)))
source_regions_at_date_df.tail()
#%%
source_regions_for_summary_df_ = \
source_regions_at_date_df[["sample_date", "_source_regions_group"]].copy()
source_regions_for_summary_df_.rename(columns={"_source_regions_group": "source_regions"}, inplace=True)
source_regions_for_summary_df_.tail()
#%%
confirmed_output_columns = ["sample_date", "new_cases", "covid_cases"]
confirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)
for source_regions_group, source_regions_group_series in \
source_regions_at_date_df.groupby("_source_regions_group"):
source_regions_set = set(source_regions_group.split(","))
confirmed_source_regions_set_df = \
confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()
confirmed_source_regions_group_df = \
confirmed_source_regions_set_df.groupby("sample_date").new_cases.sum() \
.reset_index().sort_values("sample_date")
confirmed_source_regions_group_df["covid_cases"] = \
confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[confirmed_output_columns]
confirmed_source_regions_group_df.fillna(method="ffill", inplace=True)
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[
confirmed_source_regions_group_df.sample_date.isin(
source_regions_group_series.sample_date_string)]
confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)
result_df = confirmed_output_df.copy()
result_df.tail()
#%%
result_df.rename(columns={"sample_date": "sample_date_string"}, inplace=True)
result_df = confirmed_days_df[["sample_date_string"]].merge(result_df, how="left")
result_df.sort_values("sample_date_string", inplace=True)
result_df.fillna(method="ffill", inplace=True)
result_df.tail()
#%%
result_df[["new_cases", "covid_cases"]].plot()
if columns_suffix:
result_df.rename(
columns={
"new_cases": "new_cases_" + columns_suffix,
"covid_cases": "covid_cases_" + columns_suffix},
inplace=True)
return result_df, source_regions_for_summary_df_
confirmed_eu_df, source_regions_for_summary_df = get_cases_dataframe(
report_backend_client.source_regions_for_date)
confirmed_es_df, _ = get_cases_dataframe(
lambda date: [spain_region_country_code],
columns_suffix=spain_region_country_code.lower())
raw_zip_path_prefix = "Data/TEKs/Raw/"
fail_on_error_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=fail_on_error_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_df.head()
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \
.groupby(["upload_date"]).shared_teks.max().reset_index() \
.sort_values(["upload_date"], ascending=False) \
.rename(columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
invalid_shared_diagnoses_dates_mask = \
estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)
estimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0
estimated_shared_diagnoses_df.head()
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
import requests
import pandas.io.json
official_stats_response = requests.get("https://radarcovidpre.covid19.gob.es/kpi/statistics/basics")
official_stats_response.raise_for_status()
official_stats_df_ = pandas.io.json.json_normalize(official_stats_response.json())
official_stats_df = official_stats_df_.copy()
official_stats_df = official_stats_df.append(pd.DataFrame({
"date": ["06/12/2020"],
"applicationsDownloads.totalAcummulated": [5653519],
"communicatedContagions.totalAcummulated": [21925],
}), sort=False)
official_stats_df["date"] = pd.to_datetime(official_stats_df["date"], dayfirst=True)
official_stats_df.head()
official_stats_column_map = {
"date": "sample_date",
"applicationsDownloads.totalAcummulated": "app_downloads_es_accumulated",
"communicatedContagions.totalAcummulated": "shared_diagnoses_es_accumulated",
}
accumulated_suffix = "_accumulated"
accumulated_values_columns = \
list(filter(lambda x: x.endswith(accumulated_suffix), official_stats_column_map.values()))
interpolated_values_columns = \
list(map(lambda x: x[:-len(accumulated_suffix)], accumulated_values_columns))
official_stats_df = \
official_stats_df[official_stats_column_map.keys()] \
.rename(columns=official_stats_column_map)
official_stats_df.head()
official_stats_df = confirmed_days_df.merge(official_stats_df, how="left")
official_stats_df.sort_values("sample_date", ascending=False, inplace=True)
official_stats_df.head()
official_stats_df[accumulated_values_columns] = \
official_stats_df[accumulated_values_columns] \
.astype(float).interpolate(limit_area="inside")
official_stats_df[interpolated_values_columns] = \
official_stats_df[accumulated_values_columns].diff(periods=-1)
official_stats_df.drop(columns="sample_date", inplace=True)
official_stats_df.head()
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
official_stats_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_eu_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df = confirmed_es_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df = result_summary_df.merge(source_regions_for_summary_df, how="left")
result_summary_df.set_index(["sample_date", "source_regions"], inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(result_summary_df.shared_diagnoses_es / result_summary_df.covid_cases_es).fillna(0)
result_summary_df.head(daily_plot_days)
weekly_result_summary_df = result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(7).agg({
"covid_cases": "sum",
"covid_cases_es": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum",
"shared_diagnoses_es": "sum",
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
weekly_result_summary_df = weekly_result_summary_df.fillna(0).astype(int)
weekly_result_summary_df["teks_per_shared_diagnosis"] = \
(weekly_result_summary_df.shared_teks_by_upload_date / weekly_result_summary_df.shared_diagnoses).fillna(0)
weekly_result_summary_df["shared_diagnoses_per_covid_case"] = \
(weekly_result_summary_df.shared_diagnoses / weekly_result_summary_df.covid_cases).fillna(0)
weekly_result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(weekly_result_summary_df.shared_diagnoses_es / weekly_result_summary_df.covid_cases_es).fillna(0)
weekly_result_summary_df.head()
last_7_days_summary = weekly_result_summary_df.to_dict(orient="records")[1]
last_7_days_summary
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"source_regions": "Source Countries",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases (Source Countries)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date (Source Countries)",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date (Source Countries)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date (Source Countries)",
"shared_diagnoses": "Shared Diagnoses (Source Countries – Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis (Source Countries)",
"shared_diagnoses_per_covid_case": "Usage Ratio (Source Countries)",
"covid_cases_es": "COVID-19 Cases (Spain)",
"app_downloads_es": "App Downloads (Spain – Official)",
"shared_diagnoses_es": "Shared Diagnoses (Spain – Official)",
"shared_diagnoses_per_covid_case_es": "Usage Ratio (Spain)",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
"covid_cases_es",
"app_downloads_es",
"shared_diagnoses_es",
"shared_diagnoses_per_covid_case_es",
]
summary_percentage_columns= [
"shared_diagnoses_per_covid_case_es",
"shared_diagnoses_per_covid_case",
]
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.droplevel(level=["source_regions"]) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 30), legend=False)
ax_ = summary_ax_list[0]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
for percentage_column in summary_percentage_columns:
percentage_column_index = summary_columns.index(percentage_column)
summary_ax_list[percentage_column_index].yaxis \
.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case_es"]: lambda x: f"{x:.2%}" if x != 0 else "",
}
general_columns = \
list(filter(lambda x: x not in display_formatters, display_column_name_mapping.values()))
general_formatter = lambda x: f"{x}" if x != 0 else ""
display_formatters.update(dict(map(lambda x: (x, general_formatter), general_columns)))
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index.get_level_values("sample_date") == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.item()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.item()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.item()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.item()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.item()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
if len(report_source_regions) == 1:
display_brief_source_regions = report_source_regions[0]
else:
display_brief_source_regions = f"{len(report_source_regions)} 🇪🇺"
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
df = df.copy()
df_styler = df.style.format(display_formatters)
media_path = get_temporary_image_path()
dfi.export(df_styler, media_path)
return media_path
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
def generate_summary_api_results(df: pd.DataFrame) -> list:
api_df = df.reset_index().copy()
api_df["sample_date_string"] = \
api_df["sample_date"].dt.strftime("%Y-%m-%d")
api_df["source_regions"] = \
api_df["source_regions"].apply(lambda x: x.split(","))
return api_df.to_dict(orient="records")
summary_api_results = \
generate_summary_api_results(df=result_summary_df)
today_summary_api_results = \
generate_summary_api_results(df=extraction_date_result_summary_df)[0]
summary_results = dict(
backend_identifier=report_backend_identifier,
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=today_summary_api_results,
last_7_days=last_7_days_summary,
daily_results=summary_api_results)
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
Source Countries: {display_brief_source_regions}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: ≤{shared_diagnoses_per_covid_case:.2%}
Last 7 Days:
- Shared Diagnoses: ≤{last_7_days_summary["shared_diagnoses"]:.0f}
- Usage Ratio: ≤{last_7_days_summary["shared_diagnoses_per_covid_case"]:.2%}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
| 0.269037 | 0.187449 |
## Example "dumb" trading agent
```
import os
import sys
import warnings
def warn(*args, **kwargs):
pass
warnings.warn = warn
warnings.simplefilter(action='ignore', category=FutureWarning)
sys.path.append(os.path.dirname(os.path.abspath('')))
from tensortrade.environments import TradingEnvironment
from tensortrade.exchanges.simulated import FBMExchange
from tensortrade.actions import DiscreteActionStrategy
from tensortrade.rewards import SimpleProfitStrategy
exchange = FBMExchange()
action_strategy = DiscreteActionStrategy()
reward_strategy = SimpleProfitStrategy()
env = TradingEnvironment(exchange=exchange,
action_strategy=action_strategy,
reward_strategy=reward_strategy)
obs = env.reset()
sell_price = 1e9
stop_price = -1
print('Initial portfolio: ', exchange.portfolio)
for i in range(1000):
action = 0 if obs['close'] < sell_price else 18
action = 19 if obs['close'] < stop_price else action
if i == 0 or portfolio['BTC'] == 0:
action = 16
sell_price = obs['close'] + (obs['close'] / 50)
stop_price = obs['close'] - (obs['close'] / 50)
obs, reward, done, info = env.step(action)
executed_trade = info['executed_trade']
filled_trade = info['filled_trade']
portfolio = exchange.portfolio
print('Obs: ', obs)
print('Reward: ', reward)
print('Portfolio: ', portfolio)
print('Trade executed: ', executed_trade.trade_type, executed_trade.price, executed_trade.amount)
print('Trade filled: ', filled_trade.trade_type, filled_trade.price, filled_trade.amount)
import sys
import os
import warnings
def warn(*args, **kwargs):
pass
warnings.warn = warn
warnings.simplefilter(action='ignore', category=FutureWarning)
import gym
import numpy as np
from tensorforce.agents import Agent
from tensorforce.execution import Runner
from tensorforce.contrib.openai_gym import OpenAIGym
sys.path.append(os.path.dirname(os.path.abspath('')))
from tensortrade.environments import TradingEnvironment
from tensortrade.exchanges.simulated import FBMExchange
from tensortrade.actions import DiscreteActionStrategy
from tensortrade.rewards import SimpleProfitStrategy
exchange = FBMExchange(times_to_generate=100000)
action_strategy = DiscreteActionStrategy()
reward_strategy = SimpleProfitStrategy()
env = TradingEnvironment(exchange=exchange,
action_strategy=action_strategy,
reward_strategy=reward_strategy,
feature_pipeline=None)
agent_config = {
"type": "dqn_agent",
"update_mode": {
"unit": "timesteps",
"batch_size": 64,
"frequency": 4
},
"memory": {
"type": "replay",
"capacity": 10000,
"include_next_states": True
},
"optimizer": {
"type": "clipped_step",
"clipping_value": 0.1,
"optimizer": {
"type": "adam",
"learning_rate": 1e-3
}
},
"discount": 0.999,
"entropy_regularization": None,
"double_q_model": True,
"target_sync_frequency": 1000,
"target_update_weight": 1.0,
"actions_exploration": {
"type": "epsilon_anneal",
"initial_epsilon": 0.5,
"final_epsilon": 0.,
"timesteps": 1000000000
},
"saver": {
"directory": None,
"seconds": 600
},
"summarizer": {
"directory": None,
"labels": ["graph", "total-loss"]
},
"execution": {
"type": "single",
"session_config": None,
"distributed_spec": None
}
}
network_spec = [
dict(type='dense', size=64),
dict(type='dense', size=32)
]
agent = Agent.from_spec(
spec=agent_config,
kwargs=dict(
states=env.states,
actions=env.actions,
network=network_spec,
)
)
# Create the runner
runner = Runner(agent=agent, environment=env)
# Callback function printing episode statistics
def episode_finished(r):
print("Finished episode {ep} after {ts} timesteps (reward: {reward})".format(ep=r.episode, ts=r.episode_timestep,
reward=r.episode_rewards[-1]))
return True
# Start learning
runner.run(episodes=300, max_episode_timesteps=10000, episode_finished=episode_finished)
runner.close()
# Print statistics
print("Learning finished. Total episodes: {ep}. Average reward of last 100 episodes: {ar}.".format(
ep=runner.episode,
ar=np.mean(runner.episode_rewards))
)
```
|
github_jupyter
|
import os
import sys
import warnings
def warn(*args, **kwargs):
pass
warnings.warn = warn
warnings.simplefilter(action='ignore', category=FutureWarning)
sys.path.append(os.path.dirname(os.path.abspath('')))
from tensortrade.environments import TradingEnvironment
from tensortrade.exchanges.simulated import FBMExchange
from tensortrade.actions import DiscreteActionStrategy
from tensortrade.rewards import SimpleProfitStrategy
exchange = FBMExchange()
action_strategy = DiscreteActionStrategy()
reward_strategy = SimpleProfitStrategy()
env = TradingEnvironment(exchange=exchange,
action_strategy=action_strategy,
reward_strategy=reward_strategy)
obs = env.reset()
sell_price = 1e9
stop_price = -1
print('Initial portfolio: ', exchange.portfolio)
for i in range(1000):
action = 0 if obs['close'] < sell_price else 18
action = 19 if obs['close'] < stop_price else action
if i == 0 or portfolio['BTC'] == 0:
action = 16
sell_price = obs['close'] + (obs['close'] / 50)
stop_price = obs['close'] - (obs['close'] / 50)
obs, reward, done, info = env.step(action)
executed_trade = info['executed_trade']
filled_trade = info['filled_trade']
portfolio = exchange.portfolio
print('Obs: ', obs)
print('Reward: ', reward)
print('Portfolio: ', portfolio)
print('Trade executed: ', executed_trade.trade_type, executed_trade.price, executed_trade.amount)
print('Trade filled: ', filled_trade.trade_type, filled_trade.price, filled_trade.amount)
import sys
import os
import warnings
def warn(*args, **kwargs):
pass
warnings.warn = warn
warnings.simplefilter(action='ignore', category=FutureWarning)
import gym
import numpy as np
from tensorforce.agents import Agent
from tensorforce.execution import Runner
from tensorforce.contrib.openai_gym import OpenAIGym
sys.path.append(os.path.dirname(os.path.abspath('')))
from tensortrade.environments import TradingEnvironment
from tensortrade.exchanges.simulated import FBMExchange
from tensortrade.actions import DiscreteActionStrategy
from tensortrade.rewards import SimpleProfitStrategy
exchange = FBMExchange(times_to_generate=100000)
action_strategy = DiscreteActionStrategy()
reward_strategy = SimpleProfitStrategy()
env = TradingEnvironment(exchange=exchange,
action_strategy=action_strategy,
reward_strategy=reward_strategy,
feature_pipeline=None)
agent_config = {
"type": "dqn_agent",
"update_mode": {
"unit": "timesteps",
"batch_size": 64,
"frequency": 4
},
"memory": {
"type": "replay",
"capacity": 10000,
"include_next_states": True
},
"optimizer": {
"type": "clipped_step",
"clipping_value": 0.1,
"optimizer": {
"type": "adam",
"learning_rate": 1e-3
}
},
"discount": 0.999,
"entropy_regularization": None,
"double_q_model": True,
"target_sync_frequency": 1000,
"target_update_weight": 1.0,
"actions_exploration": {
"type": "epsilon_anneal",
"initial_epsilon": 0.5,
"final_epsilon": 0.,
"timesteps": 1000000000
},
"saver": {
"directory": None,
"seconds": 600
},
"summarizer": {
"directory": None,
"labels": ["graph", "total-loss"]
},
"execution": {
"type": "single",
"session_config": None,
"distributed_spec": None
}
}
network_spec = [
dict(type='dense', size=64),
dict(type='dense', size=32)
]
agent = Agent.from_spec(
spec=agent_config,
kwargs=dict(
states=env.states,
actions=env.actions,
network=network_spec,
)
)
# Create the runner
runner = Runner(agent=agent, environment=env)
# Callback function printing episode statistics
def episode_finished(r):
print("Finished episode {ep} after {ts} timesteps (reward: {reward})".format(ep=r.episode, ts=r.episode_timestep,
reward=r.episode_rewards[-1]))
return True
# Start learning
runner.run(episodes=300, max_episode_timesteps=10000, episode_finished=episode_finished)
runner.close()
# Print statistics
print("Learning finished. Total episodes: {ep}. Average reward of last 100 episodes: {ar}.".format(
ep=runner.episode,
ar=np.mean(runner.episode_rewards))
)
| 0.466116 | 0.628507 |
# Task 02: K- Means Clustering
## Author: Rupali Shekhawat
#### KMeans Clustering
Kmeans clustering is a method of vector quantization, originally from signal processing, that aims to partition n observations into k clusters in which each observation belongs to the cluster with the nearest mean (cluster centers or cluster centroid), serving as a prototype of the cluster. This results in a partitioning of the data space into Voronoi cells. It is popular for cluster analysis in data mining. k-means clustering minimizes within-cluster variances (squared Euclidean distances), but not regular Euclidean distances, which would be the more difficult Weber problem: the mean optimizes squared errors, whereas only the geometric median minimizes Euclidean distances. For instance, better Euclidean solutions can be found using k-medians and k-medoids.
#### Advantages and Disadvantages
Here is a list of the main advantages and disadvantages of this algorithm.
Advantages:
1. Means is simple and computationally efficient.
2. It is very intuitive and their results are easy to visualize.
Disadvantages:
1. K-Means is highly scale dependent and is not suitable for data of varying shapes and densities.
2. Evaluating results is more subjective. It requires much more human evaluation than trusted metrics.
### Problem Statement: Predicting the optimum number of clusters and representing it visually. We are using ‘Iris’ dataset and it is taken from: https://bit.ly/3kXTdox
### Importing required libraries
```
# Importing the libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn import datasets
```
### Loading the IRIS dataset
```
# Load the iris dataset
iris_df = pd.read_csv('Iris.csv')
iris_df.head(5)
```
### Dropping redundant data
```
# Id is dropped as it is creating confusion with the S.No.
iris_df.drop('Id', axis =1, inplace = True)
iris_df.head()
```
### Checking for missing data
```
iris_df.isnull().sum()
```
### Finding the optimum number of clusters for K Means & determining the value of K
```
# Finding the optimum number of clusters for k-means classification
x = iris_df.iloc[:, [0, 1, 2, 3]].values
from sklearn.cluster import KMeans
wcss = []
for i in range(1, 11):
kmeans = KMeans(n_clusters = i, init = 'k-means++', max_iter = 300, n_init = 10, random_state = 0)
kmeans.fit(x)
wcss.append(kmeans.inertia_)
# Plotting the results onto a line graph, allows us to observe 'The elbow'
plt.plot(range(1, 11), wcss)
plt.title('The elbow method')
plt.xlabel('Number of clusters')
plt.ylabel('WCSS') # Within cluster sum of squares
plt.show()
```
We can clearly see why it is called 'The elbow method' from the above graph, the optimum clusters is where the elbow occurs. This is when the within cluster sum of squares (WCSS) doesn't decrease significantly with every iteration.
From this we choose the number of clusters as '3'.
```
# Applying kmeans to the dataset / Creating the k-means classifier
kmeans = KMeans(n_clusters = 3, init = 'k-means++', max_iter = 300, n_init = 10, random_state = 0)
y_kmeans = kmeans.fit_predict(x)
# Visualising the clusters - On the first two columns
plt.scatter(x[y_kmeans == 0, 0], x[y_kmeans == 0, 1],
s = 100, c = 'red', label = 'Iris-setosa')
plt.scatter(x[y_kmeans == 1, 0], x[y_kmeans == 1, 1],
s = 100, c = 'blue', label = 'Iris-versicolour')
plt.scatter(x[y_kmeans == 2, 0], x[y_kmeans == 2, 1],
s = 100, c = 'green', label = 'Iris-virginica')
# Plotting the centroids of the clusters
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:,1],
s = 100, c = 'yellow', label = 'Centroids')
plt.legend()
```
|
github_jupyter
|
# Importing the libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn import datasets
# Load the iris dataset
iris_df = pd.read_csv('Iris.csv')
iris_df.head(5)
# Id is dropped as it is creating confusion with the S.No.
iris_df.drop('Id', axis =1, inplace = True)
iris_df.head()
iris_df.isnull().sum()
# Finding the optimum number of clusters for k-means classification
x = iris_df.iloc[:, [0, 1, 2, 3]].values
from sklearn.cluster import KMeans
wcss = []
for i in range(1, 11):
kmeans = KMeans(n_clusters = i, init = 'k-means++', max_iter = 300, n_init = 10, random_state = 0)
kmeans.fit(x)
wcss.append(kmeans.inertia_)
# Plotting the results onto a line graph, allows us to observe 'The elbow'
plt.plot(range(1, 11), wcss)
plt.title('The elbow method')
plt.xlabel('Number of clusters')
plt.ylabel('WCSS') # Within cluster sum of squares
plt.show()
# Applying kmeans to the dataset / Creating the k-means classifier
kmeans = KMeans(n_clusters = 3, init = 'k-means++', max_iter = 300, n_init = 10, random_state = 0)
y_kmeans = kmeans.fit_predict(x)
# Visualising the clusters - On the first two columns
plt.scatter(x[y_kmeans == 0, 0], x[y_kmeans == 0, 1],
s = 100, c = 'red', label = 'Iris-setosa')
plt.scatter(x[y_kmeans == 1, 0], x[y_kmeans == 1, 1],
s = 100, c = 'blue', label = 'Iris-versicolour')
plt.scatter(x[y_kmeans == 2, 0], x[y_kmeans == 2, 1],
s = 100, c = 'green', label = 'Iris-virginica')
# Plotting the centroids of the clusters
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:,1],
s = 100, c = 'yellow', label = 'Centroids')
plt.legend()
| 0.858303 | 0.993417 |
## Product Review Aspect Detection: Screen Guard
### This is a Natural Language Processing based solution which can detect up to 7 aspects from online product reviews for screen guard.
This sample notebook shows you how to deploy Product Review Aspect Detection: Screen Guard using Amazon SageMaker.
> **Note**: This is a reference notebook and it cannot run unless you make changes suggested in the notebook.
#### Pre-requisites:
1. **Note**: This notebook contains elements which render correctly in Jupyter interface. Open this notebook from an Amazon SageMaker Notebook Instance or Amazon SageMaker Studio.
1. Ensure that IAM role used has **AmazonSageMakerFullAccess**
1. To deploy this ML model successfully, ensure that:
1. Either your IAM role has these three permissions and you have authority to make AWS Marketplace subscriptions in the AWS account used:
1. **aws-marketplace:ViewSubscriptions**
1. **aws-marketplace:Unsubscribe**
1. **aws-marketplace:Subscribe**
2. or your AWS account has a subscription to Product Review Aspect Detection: Screen Guard. If so, skip step: [Subscribe to the model package](#1.-Subscribe-to-the-model-package)
#### Contents:
1. [Subscribe to the model package](#1.-Subscribe-to-the-model-package)
2. [Create an endpoint and perform real-time inference](#2.-Create-an-endpoint-and-perform-real-time-inference)
1. [Create an endpoint](#A.-Create-an-endpoint)
2. [Create input payload](#B.-Create-input-payload)
3. [Perform real-time inference](#C.-Perform-real-time-inference)
4. [Visualize output](#D.-Visualize-output)
5. [Delete the endpoint](#E.-Delete-the-endpoint)
3. [Perform batch inference](#3.-Perform-batch-inference)
4. [Clean-up](#4.-Clean-up)
1. [Delete the model](#A.-Delete-the-model)
2. [Unsubscribe to the listing (optional)](#B.-Unsubscribe-to-the-listing-(optional))
#### Usage instructions
You can run this notebook one cell at a time (By using Shift+Enter for running a cell).
### 1. Subscribe to the model package
To subscribe to the model package:
1. Open the model package listing page Product Review Aspect Detection: Screen Guard
1. On the AWS Marketplace listing, click on the **Continue to subscribe** button.
1. On the **Subscribe to this software** page, review and click on **"Accept Offer"** if you and your organization agrees with EULA, pricing, and support terms.
1. Once you click on **Continue to configuration button** and then choose a **region**, you will see a **Product Arn** displayed. This is the model package ARN that you need to specify while creating a deployable model using Boto3. Copy the ARN corresponding to your region and specify the same in the following cell.
```
model_package_arn='arn:aws:sagemaker:us-east-2:786796469737:model-package/screen-guard-aspect'
import base64
import json
import uuid
from sagemaker import ModelPackage
import sagemaker as sage
from sagemaker import get_execution_role
from sagemaker import ModelPackage
from urllib.parse import urlparse
import boto3
from IPython.display import Image
from PIL import Image as ImageEdit
import urllib.request
import numpy as np
role = get_execution_role()
sagemaker_session = sage.Session()
bucket=sagemaker_session.default_bucket()
bucket
```
### 2. Create an endpoint and perform real-time inference
If you want to understand how real-time inference with Amazon SageMaker works, see [Documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/how-it-works-hosting.html).
```
model_name='screen-guard'
content_type='text/plain'
real_time_inference_instance_type='ml.m5.large'
batch_transform_inference_instance_type='ml.m5.large'
```
#### A. Create an endpoint
```
def predict_wrapper(endpoint, session):
return sage.predictor.Predictor(endpoint, session,content_type)
#create a deployable model from the model package.
model = ModelPackage(role=role,
model_package_arn=model_package_arn,
sagemaker_session=sagemaker_session,
predictor_cls=predict_wrapper)
#Deploy the model
predictor = model.deploy(1, real_time_inference_instance_type, endpoint_name=model_name)
```
Once endpoint has been created, you would be able to perform real-time inference.
#### B. Create input payload
```
file_name = 'input/review.txt'
```
<Add code snippet that shows the payload contents>
#### C. Perform real-time inference
```
!aws sagemaker-runtime invoke-endpoint \
--endpoint-name $model_name \
--body fileb://$file_name \
--content-type $content_type \
--region $sagemaker_session.boto_region_name \
output.txt
```
#### D. Visualize output
```
import json
with open('output.txt', 'r') as f:
output = json.load(f)
print(json.dumps(output, indent = 1))
```
#### E. Delete the endpoint
Now that you have successfully performed a real-time inference, you do not need the endpoint any more. You can terminate the endpoint to avoid being charged.
```
predictor=sage.predictor.Predictor(model_name, sagemaker_session,content_type)
predictor.delete_endpoint(delete_endpoint_config=True)
```
### 3. Perform batch inference
In this section, you will perform batch inference using multiple input payloads together. If you are not familiar with batch transform, and want to learn more, see these links:
1. [How it works](https://docs.aws.amazon.com/sagemaker/latest/dg/ex1-batch-transform.html)
2. [How to run a batch transform job](https://docs.aws.amazon.com/sagemaker/latest/dg/how-it-works-batch.html)
```
#upload the batch-transform job input files to S3
transform_input_folder = "input"
transform_input = sagemaker_session.upload_data(transform_input_folder, key_prefix=model_name)
print("Transform input uploaded to " + transform_input)
#Run the batch-transform job
transformer = model.transformer(1, batch_transform_inference_instance_type)
transformer.transform(transform_input, content_type=content_type)
transformer.wait()
import os
s3_conn = boto3.client("s3")
with open('output2.txt', 'wb') as f:
s3_conn.download_fileobj(bucket, os.path.basename(transformer.output_path)+'/review.txt.out', f)
print("Output file loaded from bucket")
with open('output2.txt', 'r') as f:
output = json.load(f)
print(json.dumps(output, indent = 1))
```
### 4. Clean-up
#### A. Delete the model
```
model.delete_model()
```
#### B. Unsubscribe to the listing (optional)
If you would like to unsubscribe to the model package, follow these steps. Before you cancel the subscription, ensure that you do not have any [deployable model](https://console.aws.amazon.com/sagemaker/home#/models) created from the model package or using the algorithm. Note - You can find this information by looking at the container name associated with the model.
**Steps to unsubscribe to product from AWS Marketplace**:
1. Navigate to __Machine Learning__ tab on [__Your Software subscriptions page__](https://aws.amazon.com/marketplace/ai/library?productType=ml&ref_=mlmp_gitdemo_indust)
2. Locate the listing that you want to cancel the subscription for, and then choose __Cancel Subscription__ to cancel the subscription.
|
github_jupyter
|
model_package_arn='arn:aws:sagemaker:us-east-2:786796469737:model-package/screen-guard-aspect'
import base64
import json
import uuid
from sagemaker import ModelPackage
import sagemaker as sage
from sagemaker import get_execution_role
from sagemaker import ModelPackage
from urllib.parse import urlparse
import boto3
from IPython.display import Image
from PIL import Image as ImageEdit
import urllib.request
import numpy as np
role = get_execution_role()
sagemaker_session = sage.Session()
bucket=sagemaker_session.default_bucket()
bucket
model_name='screen-guard'
content_type='text/plain'
real_time_inference_instance_type='ml.m5.large'
batch_transform_inference_instance_type='ml.m5.large'
def predict_wrapper(endpoint, session):
return sage.predictor.Predictor(endpoint, session,content_type)
#create a deployable model from the model package.
model = ModelPackage(role=role,
model_package_arn=model_package_arn,
sagemaker_session=sagemaker_session,
predictor_cls=predict_wrapper)
#Deploy the model
predictor = model.deploy(1, real_time_inference_instance_type, endpoint_name=model_name)
file_name = 'input/review.txt'
!aws sagemaker-runtime invoke-endpoint \
--endpoint-name $model_name \
--body fileb://$file_name \
--content-type $content_type \
--region $sagemaker_session.boto_region_name \
output.txt
import json
with open('output.txt', 'r') as f:
output = json.load(f)
print(json.dumps(output, indent = 1))
predictor=sage.predictor.Predictor(model_name, sagemaker_session,content_type)
predictor.delete_endpoint(delete_endpoint_config=True)
#upload the batch-transform job input files to S3
transform_input_folder = "input"
transform_input = sagemaker_session.upload_data(transform_input_folder, key_prefix=model_name)
print("Transform input uploaded to " + transform_input)
#Run the batch-transform job
transformer = model.transformer(1, batch_transform_inference_instance_type)
transformer.transform(transform_input, content_type=content_type)
transformer.wait()
import os
s3_conn = boto3.client("s3")
with open('output2.txt', 'wb') as f:
s3_conn.download_fileobj(bucket, os.path.basename(transformer.output_path)+'/review.txt.out', f)
print("Output file loaded from bucket")
with open('output2.txt', 'r') as f:
output = json.load(f)
print(json.dumps(output, indent = 1))
model.delete_model()
| 0.402275 | 0.887205 |
# Palkkajakaumien tarkastelua
Verrataan elinkaarimallin palkkajakaumia eri ryhmille havaittuihin
```
import random as rd
import numpy as np
import matplotlib.pyplot as plt
from lifecycle_rl import Lifecycle
import gym
%matplotlib inline
%pylab inline
# varoitukset piiloon (Stable baseline ei ole vielä Tensorflow 2.0-yhteensopiva, ja Tensorflow 1.5 valittaa paljon)
import warnings
warnings.filterwarnings('ignore')
pop_size=1_000
ben=gym.make('unemployment-v2',kwargs={})
n=1000
m=0
palkat_ika_miehet=12.5*np.array([2339.01,2489.09,2571.40,2632.58,2718.03,2774.21,2884.89,2987.55,3072.40,3198.48,3283.81,3336.51,3437.30,3483.45,3576.67,3623.00,3731.27,3809.58,3853.66,3995.90,4006.16,4028.60,4104.72,4181.51,4134.13,4157.54,4217.15,4165.21,4141.23,4172.14,4121.26,4127.43,4134.00,4093.10,4065.53,4063.17,4085.31,4071.25,4026.50,4031.17,4047.32,4026.96,4028.39,4163.14,4266.42,4488.40,4201.40,4252.15,4443.96,3316.92,3536.03,3536.03])
palkat_ika_naiset=12.5*np.array([2223.96,2257.10,2284.57,2365.57,2443.64,2548.35,2648.06,2712.89,2768.83,2831.99,2896.76,2946.37,2963.84,2993.79,3040.83,3090.43,3142.91,3159.91,3226.95,3272.29,3270.97,3297.32,3333.42,3362.99,3381.84,3342.78,3345.25,3360.21,3324.67,3322.28,3326.72,3326.06,3314.82,3303.73,3302.65,3246.03,3244.65,3248.04,3223.94,3211.96,3167.00,3156.29,3175.23,3228.67,3388.39,3457.17,3400.23,3293.52,2967.68,2702.05,2528.84,2528.84])
g_r=[0.77,1.0,1.23]
data_range=np.arange(20,72)
sal20=np.zeros((n,1))
sal25=np.zeros((n,1))
sal30=np.zeros((n,1))
sal50=np.zeros((n,1))
sal=np.zeros((n,72))
p=np.arange(700,17500,100)*12.5
palkka20=np.array([10.3,5.6,4.5,14.2,7.1,9.1,22.8,22.1,68.9,160.3,421.6,445.9,501.5,592.2,564.5,531.9,534.4,431.2,373.8,320.3,214.3,151.4,82.3,138.0,55.6,61.5,45.2,19.4,32.9,13.1,9.6,7.4,12.3,12.5,11.5,5.3,2.4,1.6,1.2,1.2,14.1,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka25=np.array([12.4,11.3,30.2,4.3,28.5,20.3,22.5,23.7,83.3,193.0,407.9,535.0,926.5,1177.1,1540.9,1526.4,1670.2,1898.3,1538.8,1431.5,1267.9,1194.8,1096.3,872.6,701.3,619.0,557.2,465.8,284.3,291.4,197.1,194.4,145.0,116.7,88.7,114.0,56.9,57.3,55.0,25.2,24.4,20.1,25.2,37.3,41.4,22.6,14.1,9.4,6.3,7.5,8.1,9.0,4.0,3.4,5.4,4.1,5.2,1.0,2.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka30=np.array([1.0,2.0,3.0,8.5,12.1,22.9,15.8,21.8,52.3,98.2,295.3,392.8,646.7,951.4,1240.5,1364.5,1486.1,1965.2,1908.9,1729.5,1584.8,1460.6,1391.6,1551.9,1287.6,1379.0,1205.6,1003.6,1051.6,769.9,680.5,601.2,552.0,548.3,404.5,371.0,332.7,250.0,278.2,202.2,204.4,149.8,176.7,149.0,119.6,76.8,71.4,56.3,75.9,76.8,58.2,50.2,46.8,48.9,30.1,32.2,28.8,31.1,45.5,41.2,36.5,18.1,11.6,8.5,10.2,4.3,13.5,12.3,4.9,13.9,5.4,5.9,7.4,14.1,9.6,8.4,11.5,0.0,3.3,9.0,5.2,5.0,3.1,7.4,2.0,4.0,4.1,14.0,2.0,3.0,1.0,0.0,6.2,2.0,1.2,2.0,1.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka50=np.array([2.0,3.1,2.4,3.9,1.0,1.0,11.4,30.1,29.3,34.3,231.9,341.9,514.4,724.0,1076.8,1345.2,1703.0,1545.8,1704.0,1856.1,1805.4,1608.1,1450.0,1391.4,1338.5,1173.2,1186.3,1024.8,1105.6,963.0,953.0,893.7,899.8,879.5,857.0,681.5,650.5,579.2,676.8,498.0,477.5,444.3,409.1,429.0,340.5,297.2,243.1,322.5,297.5,254.1,213.1,249.3,212.1,212.8,164.4,149.3,158.6,157.4,154.1,112.7,93.4,108.4,87.3,86.7,82.0,115.9,66.9,84.2,61.4,43.7,58.1,40.9,73.9,50.0,51.6,25.7,43.2,48.2,43.0,32.6,21.6,22.4,36.3,28.3,19.4,21.1,21.9,21.5,19.2,15.8,22.6,9.3,14.0,22.4,14.0,13.0,11.9,18.7,7.3,21.6,9.5,11.2,12.0,18.2,12.9,2.2,10.7,6.1,11.7,7.6,1.0,4.7,8.5,6.4,3.3,4.6,1.2,3.7,5.8,1.0,1.0,1.0,1.0,3.2,1.2,3.1,2.2,2.3,2.1,1.1,2.0,2.1,2.2,4.6,2.2,1.0,1.0,1.0,0.0,3.0,1.2,0.0,8.2,3.0,1.0,1.0,2.1,1.2,3.2,1.0,5.2,1.1,5.2,1.0,1.2,2.3,1.0,3.1,1.0,1.0,1.1,1.6,1.1,1.1,1.0,1.0,1.0,1.0])
for k in range(n):
g=rd.choices(np.array([0,1,2],dtype=int),weights=[0.3,0.5,0.2])[0]
gender=rd.choices(np.array([0,1],dtype=int),weights=[0.5,0.5])[0]
group=int(g+gender*3)
ben.compute_salary_TK(group=group)
sal20[m]=ben.salary[20]
sal25[m]=ben.salary[25]
sal30[m]=ben.salary[30]
sal50[m]=ben.salary[50]
sal[m,:]=ben.salary
m=m+1
def kuva(sal,ika,p,palkka):
plt.hist(sal,bins=50,density=True)
ave=np.mean(sal)/12
palave=np.sum(palkka*p)/12/np.sum(palkka)
plt.title('{}: ave {} vs {}'.format(ika,ave,palave))
plt.plot(p,palkka/sum(palkka)/2000)
plt.show()
kuva(sal20,20,p,palkka20)
kuva(sal25,25,p,palkka25)
kuva(sal30,30,p,palkka30)
kuva(sal50,50,p,palkka50)
data_range=np.arange(20,72)
plt.plot(np.mean(sal,axis=0),label='arvio')
plt.plot(data_range,0.5*palkat_ika_miehet+0.5*palkat_ika_naiset,label='data')
plt.legend()
plt.show()
ben=gym.make('unemployment-v2',kwargs={})
n=300
x0 = np.linspace(20, 72, num=206, endpoint=True)
for g in range(6):
print('Group {}:'.format(g))
sal=np.zeros((n,206))
for k in range(0,n):
ben.compute_salary_TK_v3(group=g)
sal[k,:]=ben.salary
plt.plot(x0,ben.salary)
plt.show()
plt.plot(x0,np.mean(sal,axis=0),label='arvio')
if g<3:
plt.plot(data_range,palkat_ika_miehet*g_r[g],label='data')
else:
plt.plot(data_range,palkat_ika_naiset*g_r[g-3],label='data')
plt.legend()
plt.show()
import random
random.choices([0,1,2],weights=[0.3,0.3,0.4])
# test benefits
import fin_benefits
import numpy as np
#opiskelija
ben=fin_benefits.Benefits()
p,selite=fin_benefits.perheparametrit(perhetyyppi=50,tulosta=False)
tulot,q=ben.laske_tulot(p)
tulot=tulot-440
print(np.exp(4))
ben.check_p(p)
ben=gym.make('unemployment-v0',kwargs={})
n=1000
m=0
data_range=np.arange(20,72)
sal20=np.zeros((n,1))
sal25=np.zeros((n,1))
sal30=np.zeros((n,1))
sal50=np.zeros((n,1))
sal=np.zeros((n,76))
p=np.arange(700,17500,100)*12.5
palkka20=np.array([10.3,5.6,4.5,14.2,7.1,9.1,22.8,22.1,68.9,160.3,421.6,445.9,501.5,592.2,564.5,531.9,534.4,431.2,373.8,320.3,214.3,151.4,82.3,138.0,55.6,61.5,45.2,19.4,32.9,13.1,9.6,7.4,12.3,12.5,11.5,5.3,2.4,1.6,1.2,1.2,14.1,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka25=np.array([12.4,11.3,30.2,4.3,28.5,20.3,22.5,23.7,83.3,193.0,407.9,535.0,926.5,1177.1,1540.9,1526.4,1670.2,1898.3,1538.8,1431.5,1267.9,1194.8,1096.3,872.6,701.3,619.0,557.2,465.8,284.3,291.4,197.1,194.4,145.0,116.7,88.7,114.0,56.9,57.3,55.0,25.2,24.4,20.1,25.2,37.3,41.4,22.6,14.1,9.4,6.3,7.5,8.1,9.0,4.0,3.4,5.4,4.1,5.2,1.0,2.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka30=np.array([1.0,2.0,3.0,8.5,12.1,22.9,15.8,21.8,52.3,98.2,295.3,392.8,646.7,951.4,1240.5,1364.5,1486.1,1965.2,1908.9,1729.5,1584.8,1460.6,1391.6,1551.9,1287.6,1379.0,1205.6,1003.6,1051.6,769.9,680.5,601.2,552.0,548.3,404.5,371.0,332.7,250.0,278.2,202.2,204.4,149.8,176.7,149.0,119.6,76.8,71.4,56.3,75.9,76.8,58.2,50.2,46.8,48.9,30.1,32.2,28.8,31.1,45.5,41.2,36.5,18.1,11.6,8.5,10.2,4.3,13.5,12.3,4.9,13.9,5.4,5.9,7.4,14.1,9.6,8.4,11.5,0.0,3.3,9.0,5.2,5.0,3.1,7.4,2.0,4.0,4.1,14.0,2.0,3.0,1.0,0.0,6.2,2.0,1.2,2.0,1.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka50=np.array([2.0,3.1,2.4,3.9,1.0,1.0,11.4,30.1,29.3,34.3,231.9,341.9,514.4,724.0,1076.8,1345.2,1703.0,1545.8,1704.0,1856.1,1805.4,1608.1,1450.0,1391.4,1338.5,1173.2,1186.3,1024.8,1105.6,963.0,953.0,893.7,899.8,879.5,857.0,681.5,650.5,579.2,676.8,498.0,477.5,444.3,409.1,429.0,340.5,297.2,243.1,322.5,297.5,254.1,213.1,249.3,212.1,212.8,164.4,149.3,158.6,157.4,154.1,112.7,93.4,108.4,87.3,86.7,82.0,115.9,66.9,84.2,61.4,43.7,58.1,40.9,73.9,50.0,51.6,25.7,43.2,48.2,43.0,32.6,21.6,22.4,36.3,28.3,19.4,21.1,21.9,21.5,19.2,15.8,22.6,9.3,14.0,22.4,14.0,13.0,11.9,18.7,7.3,21.6,9.5,11.2,12.0,18.2,12.9,2.2,10.7,6.1,11.7,7.6,1.0,4.7,8.5,6.4,3.3,4.6,1.2,3.7,5.8,1.0,1.0,1.0,1.0,3.2,1.2,3.1,2.2,2.3,2.1,1.1,2.0,2.1,2.2,4.6,2.2,1.0,1.0,1.0,0.0,3.0,1.2,0.0,8.2,3.0,1.0,1.0,2.1,1.2,3.2,1.0,5.2,1.1,5.2,1.0,1.2,2.3,1.0,3.1,1.0,1.0,1.1,1.6,1.1,1.1,1.0,1.0,1.0,1.0])
for k in range(n):
ben.compute_salary()
sal20[m]=ben.salary[20]
sal25[m]=ben.salary[25]
sal30[m]=ben.salary[30]
sal50[m]=ben.salary[50]
sal[m,:]=ben.salary
m=m+1
def kuva(sal,ika,p,palkka):
plt.hist(sal,bins=50,density=True)
ave=np.mean(sal)/12
palave=np.sum(palkka*p)/12/np.sum(palkka)
plt.title('{}: ave {} vs {}'.format(ika,ave,palave))
plt.plot(p,palkka/sum(palkka)/2000)
plt.show()
kuva(sal20,20,p,palkka20)
kuva(sal25,25,p,palkka25)
kuva(sal30,30,p,palkka30)
kuva(sal50,50,p,palkka50)
data_range=np.arange(20,72)
plt.plot(np.mean(sal,axis=0),label='arvio')
plt.plot(data_range,ben.palkat_ika_miehet,label='data')
plt.legend()
plt.show()
```
|
github_jupyter
|
import random as rd
import numpy as np
import matplotlib.pyplot as plt
from lifecycle_rl import Lifecycle
import gym
%matplotlib inline
%pylab inline
# varoitukset piiloon (Stable baseline ei ole vielä Tensorflow 2.0-yhteensopiva, ja Tensorflow 1.5 valittaa paljon)
import warnings
warnings.filterwarnings('ignore')
pop_size=1_000
ben=gym.make('unemployment-v2',kwargs={})
n=1000
m=0
palkat_ika_miehet=12.5*np.array([2339.01,2489.09,2571.40,2632.58,2718.03,2774.21,2884.89,2987.55,3072.40,3198.48,3283.81,3336.51,3437.30,3483.45,3576.67,3623.00,3731.27,3809.58,3853.66,3995.90,4006.16,4028.60,4104.72,4181.51,4134.13,4157.54,4217.15,4165.21,4141.23,4172.14,4121.26,4127.43,4134.00,4093.10,4065.53,4063.17,4085.31,4071.25,4026.50,4031.17,4047.32,4026.96,4028.39,4163.14,4266.42,4488.40,4201.40,4252.15,4443.96,3316.92,3536.03,3536.03])
palkat_ika_naiset=12.5*np.array([2223.96,2257.10,2284.57,2365.57,2443.64,2548.35,2648.06,2712.89,2768.83,2831.99,2896.76,2946.37,2963.84,2993.79,3040.83,3090.43,3142.91,3159.91,3226.95,3272.29,3270.97,3297.32,3333.42,3362.99,3381.84,3342.78,3345.25,3360.21,3324.67,3322.28,3326.72,3326.06,3314.82,3303.73,3302.65,3246.03,3244.65,3248.04,3223.94,3211.96,3167.00,3156.29,3175.23,3228.67,3388.39,3457.17,3400.23,3293.52,2967.68,2702.05,2528.84,2528.84])
g_r=[0.77,1.0,1.23]
data_range=np.arange(20,72)
sal20=np.zeros((n,1))
sal25=np.zeros((n,1))
sal30=np.zeros((n,1))
sal50=np.zeros((n,1))
sal=np.zeros((n,72))
p=np.arange(700,17500,100)*12.5
palkka20=np.array([10.3,5.6,4.5,14.2,7.1,9.1,22.8,22.1,68.9,160.3,421.6,445.9,501.5,592.2,564.5,531.9,534.4,431.2,373.8,320.3,214.3,151.4,82.3,138.0,55.6,61.5,45.2,19.4,32.9,13.1,9.6,7.4,12.3,12.5,11.5,5.3,2.4,1.6,1.2,1.2,14.1,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka25=np.array([12.4,11.3,30.2,4.3,28.5,20.3,22.5,23.7,83.3,193.0,407.9,535.0,926.5,1177.1,1540.9,1526.4,1670.2,1898.3,1538.8,1431.5,1267.9,1194.8,1096.3,872.6,701.3,619.0,557.2,465.8,284.3,291.4,197.1,194.4,145.0,116.7,88.7,114.0,56.9,57.3,55.0,25.2,24.4,20.1,25.2,37.3,41.4,22.6,14.1,9.4,6.3,7.5,8.1,9.0,4.0,3.4,5.4,4.1,5.2,1.0,2.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka30=np.array([1.0,2.0,3.0,8.5,12.1,22.9,15.8,21.8,52.3,98.2,295.3,392.8,646.7,951.4,1240.5,1364.5,1486.1,1965.2,1908.9,1729.5,1584.8,1460.6,1391.6,1551.9,1287.6,1379.0,1205.6,1003.6,1051.6,769.9,680.5,601.2,552.0,548.3,404.5,371.0,332.7,250.0,278.2,202.2,204.4,149.8,176.7,149.0,119.6,76.8,71.4,56.3,75.9,76.8,58.2,50.2,46.8,48.9,30.1,32.2,28.8,31.1,45.5,41.2,36.5,18.1,11.6,8.5,10.2,4.3,13.5,12.3,4.9,13.9,5.4,5.9,7.4,14.1,9.6,8.4,11.5,0.0,3.3,9.0,5.2,5.0,3.1,7.4,2.0,4.0,4.1,14.0,2.0,3.0,1.0,0.0,6.2,2.0,1.2,2.0,1.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka50=np.array([2.0,3.1,2.4,3.9,1.0,1.0,11.4,30.1,29.3,34.3,231.9,341.9,514.4,724.0,1076.8,1345.2,1703.0,1545.8,1704.0,1856.1,1805.4,1608.1,1450.0,1391.4,1338.5,1173.2,1186.3,1024.8,1105.6,963.0,953.0,893.7,899.8,879.5,857.0,681.5,650.5,579.2,676.8,498.0,477.5,444.3,409.1,429.0,340.5,297.2,243.1,322.5,297.5,254.1,213.1,249.3,212.1,212.8,164.4,149.3,158.6,157.4,154.1,112.7,93.4,108.4,87.3,86.7,82.0,115.9,66.9,84.2,61.4,43.7,58.1,40.9,73.9,50.0,51.6,25.7,43.2,48.2,43.0,32.6,21.6,22.4,36.3,28.3,19.4,21.1,21.9,21.5,19.2,15.8,22.6,9.3,14.0,22.4,14.0,13.0,11.9,18.7,7.3,21.6,9.5,11.2,12.0,18.2,12.9,2.2,10.7,6.1,11.7,7.6,1.0,4.7,8.5,6.4,3.3,4.6,1.2,3.7,5.8,1.0,1.0,1.0,1.0,3.2,1.2,3.1,2.2,2.3,2.1,1.1,2.0,2.1,2.2,4.6,2.2,1.0,1.0,1.0,0.0,3.0,1.2,0.0,8.2,3.0,1.0,1.0,2.1,1.2,3.2,1.0,5.2,1.1,5.2,1.0,1.2,2.3,1.0,3.1,1.0,1.0,1.1,1.6,1.1,1.1,1.0,1.0,1.0,1.0])
for k in range(n):
g=rd.choices(np.array([0,1,2],dtype=int),weights=[0.3,0.5,0.2])[0]
gender=rd.choices(np.array([0,1],dtype=int),weights=[0.5,0.5])[0]
group=int(g+gender*3)
ben.compute_salary_TK(group=group)
sal20[m]=ben.salary[20]
sal25[m]=ben.salary[25]
sal30[m]=ben.salary[30]
sal50[m]=ben.salary[50]
sal[m,:]=ben.salary
m=m+1
def kuva(sal,ika,p,palkka):
plt.hist(sal,bins=50,density=True)
ave=np.mean(sal)/12
palave=np.sum(palkka*p)/12/np.sum(palkka)
plt.title('{}: ave {} vs {}'.format(ika,ave,palave))
plt.plot(p,palkka/sum(palkka)/2000)
plt.show()
kuva(sal20,20,p,palkka20)
kuva(sal25,25,p,palkka25)
kuva(sal30,30,p,palkka30)
kuva(sal50,50,p,palkka50)
data_range=np.arange(20,72)
plt.plot(np.mean(sal,axis=0),label='arvio')
plt.plot(data_range,0.5*palkat_ika_miehet+0.5*palkat_ika_naiset,label='data')
plt.legend()
plt.show()
ben=gym.make('unemployment-v2',kwargs={})
n=300
x0 = np.linspace(20, 72, num=206, endpoint=True)
for g in range(6):
print('Group {}:'.format(g))
sal=np.zeros((n,206))
for k in range(0,n):
ben.compute_salary_TK_v3(group=g)
sal[k,:]=ben.salary
plt.plot(x0,ben.salary)
plt.show()
plt.plot(x0,np.mean(sal,axis=0),label='arvio')
if g<3:
plt.plot(data_range,palkat_ika_miehet*g_r[g],label='data')
else:
plt.plot(data_range,palkat_ika_naiset*g_r[g-3],label='data')
plt.legend()
plt.show()
import random
random.choices([0,1,2],weights=[0.3,0.3,0.4])
# test benefits
import fin_benefits
import numpy as np
#opiskelija
ben=fin_benefits.Benefits()
p,selite=fin_benefits.perheparametrit(perhetyyppi=50,tulosta=False)
tulot,q=ben.laske_tulot(p)
tulot=tulot-440
print(np.exp(4))
ben.check_p(p)
ben=gym.make('unemployment-v0',kwargs={})
n=1000
m=0
data_range=np.arange(20,72)
sal20=np.zeros((n,1))
sal25=np.zeros((n,1))
sal30=np.zeros((n,1))
sal50=np.zeros((n,1))
sal=np.zeros((n,76))
p=np.arange(700,17500,100)*12.5
palkka20=np.array([10.3,5.6,4.5,14.2,7.1,9.1,22.8,22.1,68.9,160.3,421.6,445.9,501.5,592.2,564.5,531.9,534.4,431.2,373.8,320.3,214.3,151.4,82.3,138.0,55.6,61.5,45.2,19.4,32.9,13.1,9.6,7.4,12.3,12.5,11.5,5.3,2.4,1.6,1.2,1.2,14.1,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka25=np.array([12.4,11.3,30.2,4.3,28.5,20.3,22.5,23.7,83.3,193.0,407.9,535.0,926.5,1177.1,1540.9,1526.4,1670.2,1898.3,1538.8,1431.5,1267.9,1194.8,1096.3,872.6,701.3,619.0,557.2,465.8,284.3,291.4,197.1,194.4,145.0,116.7,88.7,114.0,56.9,57.3,55.0,25.2,24.4,20.1,25.2,37.3,41.4,22.6,14.1,9.4,6.3,7.5,8.1,9.0,4.0,3.4,5.4,4.1,5.2,1.0,2.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka30=np.array([1.0,2.0,3.0,8.5,12.1,22.9,15.8,21.8,52.3,98.2,295.3,392.8,646.7,951.4,1240.5,1364.5,1486.1,1965.2,1908.9,1729.5,1584.8,1460.6,1391.6,1551.9,1287.6,1379.0,1205.6,1003.6,1051.6,769.9,680.5,601.2,552.0,548.3,404.5,371.0,332.7,250.0,278.2,202.2,204.4,149.8,176.7,149.0,119.6,76.8,71.4,56.3,75.9,76.8,58.2,50.2,46.8,48.9,30.1,32.2,28.8,31.1,45.5,41.2,36.5,18.1,11.6,8.5,10.2,4.3,13.5,12.3,4.9,13.9,5.4,5.9,7.4,14.1,9.6,8.4,11.5,0.0,3.3,9.0,5.2,5.0,3.1,7.4,2.0,4.0,4.1,14.0,2.0,3.0,1.0,0.0,6.2,2.0,1.2,2.0,1.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0])
palkka50=np.array([2.0,3.1,2.4,3.9,1.0,1.0,11.4,30.1,29.3,34.3,231.9,341.9,514.4,724.0,1076.8,1345.2,1703.0,1545.8,1704.0,1856.1,1805.4,1608.1,1450.0,1391.4,1338.5,1173.2,1186.3,1024.8,1105.6,963.0,953.0,893.7,899.8,879.5,857.0,681.5,650.5,579.2,676.8,498.0,477.5,444.3,409.1,429.0,340.5,297.2,243.1,322.5,297.5,254.1,213.1,249.3,212.1,212.8,164.4,149.3,158.6,157.4,154.1,112.7,93.4,108.4,87.3,86.7,82.0,115.9,66.9,84.2,61.4,43.7,58.1,40.9,73.9,50.0,51.6,25.7,43.2,48.2,43.0,32.6,21.6,22.4,36.3,28.3,19.4,21.1,21.9,21.5,19.2,15.8,22.6,9.3,14.0,22.4,14.0,13.0,11.9,18.7,7.3,21.6,9.5,11.2,12.0,18.2,12.9,2.2,10.7,6.1,11.7,7.6,1.0,4.7,8.5,6.4,3.3,4.6,1.2,3.7,5.8,1.0,1.0,1.0,1.0,3.2,1.2,3.1,2.2,2.3,2.1,1.1,2.0,2.1,2.2,4.6,2.2,1.0,1.0,1.0,0.0,3.0,1.2,0.0,8.2,3.0,1.0,1.0,2.1,1.2,3.2,1.0,5.2,1.1,5.2,1.0,1.2,2.3,1.0,3.1,1.0,1.0,1.1,1.6,1.1,1.1,1.0,1.0,1.0,1.0])
for k in range(n):
ben.compute_salary()
sal20[m]=ben.salary[20]
sal25[m]=ben.salary[25]
sal30[m]=ben.salary[30]
sal50[m]=ben.salary[50]
sal[m,:]=ben.salary
m=m+1
def kuva(sal,ika,p,palkka):
plt.hist(sal,bins=50,density=True)
ave=np.mean(sal)/12
palave=np.sum(palkka*p)/12/np.sum(palkka)
plt.title('{}: ave {} vs {}'.format(ika,ave,palave))
plt.plot(p,palkka/sum(palkka)/2000)
plt.show()
kuva(sal20,20,p,palkka20)
kuva(sal25,25,p,palkka25)
kuva(sal30,30,p,palkka30)
kuva(sal50,50,p,palkka50)
data_range=np.arange(20,72)
plt.plot(np.mean(sal,axis=0),label='arvio')
plt.plot(data_range,ben.palkat_ika_miehet,label='data')
plt.legend()
plt.show()
| 0.189296 | 0.487734 |
```
# Dependencies and Setup
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
# Hide warning messages in notebook
import warnings
warnings.filterwarnings('ignore')
# File to Load (Remember to Change These)
mouse_drug_data_to_load = "mouse_drug_data.csv"
clinical_trial_data_to_load = "clinicaltrial_data.csv"
# Read the Mouse and Drug Data and the Clinical Trial Data
mouse_drug_data = pd.read_csv(mouse_drug_data_to_load)
clinical_trial_data = pd.read_csv(clinical_trial_data_to_load)
# Combine the data into a single dataset
clinical_merge_mouse = pd.merge(clinical_trial_data, mouse_drug_data, on="Mouse ID", how="left")
# Display the data table for preview
clinical_merge_mouse.head()
# Store the Mean Tumor Volume Data Grouped by Drug and Timepoint
grouped_clinical_merge_mouse = clinical_merge_mouse.groupby(["Drug", "Timepoint"])
mean_tumor_volume_data = grouped_clinical_merge_mouse["Tumor Volume (mm3)"].mean()
# Create data frame with reseted index to hold the results (headers at the same level)
grouped_clinical_merge_mouse_df = pd.DataFrame(mean_tumor_volume_data).reset_index()
# Display the data frame for preview
grouped_clinical_merge_mouse_df.head()
# Store the Standard Error of Tumor Volumes Grouped by Drug and Timepoint
std_error_tumor_volume_data = grouped_clinical_merge_mouse["Tumor Volume (mm3)"].sem()
# Create data frame with reseted index to hold the results (headers at the same level)
std_error_clinical_merge_mouse_df = pd.DataFrame(std_error_tumor_volume_data).reset_index()
# Display the data frame for preview
std_error_clinical_merge_mouse_df.head()
# Minor Data Munging to Re-Format the Data Frames
new_tumor_v_reformated_df = grouped_clinical_merge_mouse_df.pivot(index='Timepoint', columns='Drug', values='Tumor Volume (mm3)')
new_tumor_v_reformated_df.head(10)
# Find Errors by treatments (Capomulin, Infubinol, Ketapril, and Placebo)
capomulin_std_error = std_error_clinical_merge_mouse_df[std_error_clinical_merge_mouse_df.Drug == 'Capomulin']['Tumor Volume (mm3)']
Infubinol_std_error = std_error_clinical_merge_mouse_df[std_error_clinical_merge_mouse_df.Drug == 'Infubinol']['Tumor Volume (mm3)']
Infubinol_std_error = std_error_clinical_merge_mouse_df[std_error_clinical_merge_mouse_df.Drug == 'Ketapril']['Tumor Volume (mm3)']
Infubinol_std_error = std_error_clinical_merge_mouse_df[std_error_clinical_merge_mouse_df.Drug == 'Placebo']['Tumor Volume (mm3)']
# Plot values
plt.errorbar(new_tumor_v_reformated_df.index.values.tolist(), new_tumor_v_reformated_df["Capomulin"] , yerr = capomulin_std_error, label= "Capomulin", marker= "o", color="red")
plt.errorbar(new_tumor_v_reformated_df.index.values.tolist(), new_tumor_v_reformated_df["Infubinol"] , yerr = capomulin_std_error, label= "Infubinol", marker= "o", color="green")
plt.errorbar(new_tumor_v_reformated_df.index.values.tolist(), new_tumor_v_reformated_df["Ketapril"] , yerr = capomulin_std_error, label= "Ketapril", marker= "o", color="blue")
plt.errorbar(new_tumor_v_reformated_df.index.values.tolist(), new_tumor_v_reformated_df["Placebo"] , yerr = capomulin_std_error, label= "Placebo", marker= "o", color="darkorange")
plt.legend()
plt.title("Tumor Volume Changes Over Time by Treatment")
plt.xlabel("Time (Days)")
plt.ylabel("Tumor Volume (mm3)")
plt.grid()
# Save the Figure
plt.savefig("Tumor_Volume_Changes.png")
# Show the Figure
fig.show()
# Store the Mean Met. Site Data Grouped by Drug and Timepoint
mean_metastatic_data = grouped_clinical_merge_mouse["Metastatic Sites"].mean()
# Create data frame with reseted index to hold the results (headers at the same level)
grouped_metastatic_df = pd.DataFrame(mean_metastatic_data).reset_index()
# Show results
grouped_metastatic_df.head(10)
# Store the Standard Error of Metastatic Site Grouped by Drug and Timepoint
std_error_metastatic_data = grouped_clinical_merge_mouse["Metastatic Sites"].sem()
# Create data frame with reseted index to hold the results (headers at the same level)
std_error_metastatic_df = pd.DataFrame(std_error_metastatic_data).reset_index()
# Display the data frame for preview
std_error_metastatic_df.head()
# Minor Data Munging to Re-Format the Data Frames
new_reformated_met_df = std_error_metastatic_df.pivot(index='Timepoint', columns='Drug', values='Metastatic Sites')
# Show results
new_reformated_met_df.head()
# Find Errors by treatments (Capomulin, Infubinol, Ketapril, and Placebo)
capomulin_met_std_error = std_error_metastatic_df[std_error_metastatic_df.Drug == 'Capomulin']['Metastatic Sites']
Infubinol_met_std_error = std_error_metastatic_df[std_error_metastatic_df.Drug == 'Infubinol']['Metastatic Sites']
Infubinol_met_std_error = std_error_metastatic_df[std_error_metastatic_df.Drug == 'Ketapril']['Metastatic Sites']
Infubinol_met_std_error = std_error_metastatic_df[std_error_metastatic_df.Drug == 'Placebo']['Metastatic Sites']
# Plot values
plt.errorbar(new_reformated_met_df.index.values.tolist(), new_reformated_met_df["Capomulin"] , yerr = capomulin_met_std_error, label= "Capomulin", marker= "o", color="red")
plt.errorbar(new_reformated_met_df.index.values.tolist(), new_reformated_met_df["Infubinol"] , yerr = capomulin_met_std_error, label= "Infubinol", marker= "o", color="green")
plt.errorbar(new_reformated_met_df.index.values.tolist(), new_reformated_met_df["Ketapril"] , yerr = capomulin_met_std_error, label= "Ketapril", marker= "o", color="blue")
plt.errorbar(new_reformated_met_df.index.values.tolist(), new_reformated_met_df["Placebo"] , yerr = capomulin_met_std_error, label= "Placebo", marker= "o", color="darkorange")
plt.legend()
plt.title("Metastatic Sites Changes Over Time by Treatment")
plt.xlabel("Time (Days)")
plt.ylabel("Metastatic Sites")
plt.grid()
# Save the Figure
plt.savefig("Metastatic_Sites_Changes.png")
# Show the Figure
fig.show()
# Store the Count of Mice Grouped by Drug and Timepoint (W can pass any metric)
count_mice_data = grouped_clinical_merge_mouse["Mouse ID"].nunique()
# Create data frame with reseted index to hold the results (headers at the same level)
grouped_count_mice_df = pd.DataFrame(count_mice_data).reset_index()
# Show results
grouped_count_mice_df.head()
# Minor Data Munging to Re-Format the Data Frames
new_reformated_mice_df = grouped_count_mice_df.pivot(index='Timepoint', columns='Drug', values='Mouse ID')
# Show results
new_reformated_mice_df.head(10)
# Set the total amount of mice that started the treatment at Timepoint = 0
total_mice_started_treatment = 25
# Find mice survival rate per treatment
capomulin_surv_rate = new_reformated_mice_df["Capomulin"]/total_mice_started_treatment * 100
infubinol_surv_rate = new_reformated_mice_df["Infubinol"]/total_mice_started_treatment * 100
ketapril_surv_rate = new_reformated_mice_df["Ketapril"]/total_mice_started_treatment * 100
placebo_surv_rate = new_reformated_mice_df["Placebo"]/total_mice_started_treatment * 100
# Plot Values
plt.errorbar(new_reformated_mice_df.index.values.tolist(), capomulin_surv_rate , label= "Capomulin", marker= "o", color="red")
plt.errorbar(new_reformated_mice_df.index.values.tolist(), infubinol_surv_rate , label= "Infubinol", marker= "o", color="green")
plt.errorbar(new_reformated_mice_df.index.values.tolist(), ketapril_surv_rate , label= "Ketapril", marker= "o", color="blue")
plt.errorbar(new_reformated_mice_df.index.values.tolist(), placebo_surv_rate , label= "Placebo", marker= "o", color="darkorange")
plt.legend()
plt.title("Mice Survival Rate Over Time by Treatment")
plt.xlabel("Time (Days)")
plt.ylabel("Survival Rate (%)")
plt.grid()
# Save the Figure
plt.savefig("Mice_Survival_Rate.png")
# Show the Figure
fig.show()
# Calculate the percent changes for each drug
percent_change_tumor_volume_data = new_tumor_v_reformated_df.iloc[[0,9]].pct_change()*100
percent_change = percent_change_tumor_volume_data.iloc[1]
# Show results
percent_change
# Store all Relevant Percent Changes into a Tuple
tup_percent_change = list(tuple(zip(percent_change.index, percent_change)))
# Show results
tup_percent_change
# Splice the data between passing and failing drugs
# Initialize an empty list to collect passing or failing status
drug_status = []
# Create a new data frame to show the results
status_df = pd.DataFrame(tup_percent_change, columns=["Drug", "Perecent Change"]).sort_values(by=["Perecent Change"]).set_index(['Drug'])
# Create a function to return the second element
def takeSecond(elem):
return elem[1]
# For loop to iterate through the list of tupples and collect the drug status depending on the second elemt's value
# If second element is negative this imply a reduction in the tumor volume and consequently the Drug is considered as "Passed".
# If second element is positive this imply an increment in the tumor volume and consequently the Drug is considered as "Failed".
# The list of tuples is being sorted by second element during iteration to be aligned with the sorted data frame
for drug, value in sorted(tup_percent_change, key=takeSecond):
if value < 0:
drug_status.append("Passed")
else:
drug_status.append("Failed")
status_df["Passing / Failing"] = drug_status
# Show results
status_df
# Orient widths. Add labels, tick marks, etc.
fig, ax = plt.subplots()
y_value1 = [percent_change["Capomulin"]]
y_value2 = [percent_change["Infubinol"], percent_change["Ketapril"], percent_change["Placebo"]]
x_axis1 = np.arange(1)
x_axis2 = np.arange(1,4)
x_labels = ["Capomulin", "Infubinol", "Ketapril", "Placebo"]
rect_1 = ax.bar(x_axis1, y_value1, color='limegreen', alpha=0.9, align="center", width = 0.55)
rect_2 = ax.bar(x_axis2, y_value2 , color='darkorange', alpha=0.9, align="center", width = 0.55)
plt.setp(ax, xticks=[0, 1, 2, 3], xticklabels=["Capomulin", "Infubinol", "Ketapril", "Placebo"],
yticks=[-20, 0, 20, 40, 60])
ax.set_ylabel('Tumor Volume Change (%)', fontsize=12,)
ax.set_xlabel('Drugs', fontsize=12)
ax.set_title('Tumor Change Over 45 Day Treatment',fontsize=14)
# Use functions to label the percentages of changes
def autolabel(rects):
for rect in rects:
height = rect.get_height()
ax.text(rect.get_x() + rect.get_width()/2, .1*height, "%d" %int(height)+ "%",
ha='center', va='top', color="white")
# Call functions to implement the function calls
autolabel(rect_1)
autolabel(rect_2)
fig.tight_layout()
# Save the Figure
plt.savefig("Percentage_Tumor_Volume_Change.png")
# Show the Figure
fig.show()
```
|
github_jupyter
|
# Dependencies and Setup
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
# Hide warning messages in notebook
import warnings
warnings.filterwarnings('ignore')
# File to Load (Remember to Change These)
mouse_drug_data_to_load = "mouse_drug_data.csv"
clinical_trial_data_to_load = "clinicaltrial_data.csv"
# Read the Mouse and Drug Data and the Clinical Trial Data
mouse_drug_data = pd.read_csv(mouse_drug_data_to_load)
clinical_trial_data = pd.read_csv(clinical_trial_data_to_load)
# Combine the data into a single dataset
clinical_merge_mouse = pd.merge(clinical_trial_data, mouse_drug_data, on="Mouse ID", how="left")
# Display the data table for preview
clinical_merge_mouse.head()
# Store the Mean Tumor Volume Data Grouped by Drug and Timepoint
grouped_clinical_merge_mouse = clinical_merge_mouse.groupby(["Drug", "Timepoint"])
mean_tumor_volume_data = grouped_clinical_merge_mouse["Tumor Volume (mm3)"].mean()
# Create data frame with reseted index to hold the results (headers at the same level)
grouped_clinical_merge_mouse_df = pd.DataFrame(mean_tumor_volume_data).reset_index()
# Display the data frame for preview
grouped_clinical_merge_mouse_df.head()
# Store the Standard Error of Tumor Volumes Grouped by Drug and Timepoint
std_error_tumor_volume_data = grouped_clinical_merge_mouse["Tumor Volume (mm3)"].sem()
# Create data frame with reseted index to hold the results (headers at the same level)
std_error_clinical_merge_mouse_df = pd.DataFrame(std_error_tumor_volume_data).reset_index()
# Display the data frame for preview
std_error_clinical_merge_mouse_df.head()
# Minor Data Munging to Re-Format the Data Frames
new_tumor_v_reformated_df = grouped_clinical_merge_mouse_df.pivot(index='Timepoint', columns='Drug', values='Tumor Volume (mm3)')
new_tumor_v_reformated_df.head(10)
# Find Errors by treatments (Capomulin, Infubinol, Ketapril, and Placebo)
capomulin_std_error = std_error_clinical_merge_mouse_df[std_error_clinical_merge_mouse_df.Drug == 'Capomulin']['Tumor Volume (mm3)']
Infubinol_std_error = std_error_clinical_merge_mouse_df[std_error_clinical_merge_mouse_df.Drug == 'Infubinol']['Tumor Volume (mm3)']
Infubinol_std_error = std_error_clinical_merge_mouse_df[std_error_clinical_merge_mouse_df.Drug == 'Ketapril']['Tumor Volume (mm3)']
Infubinol_std_error = std_error_clinical_merge_mouse_df[std_error_clinical_merge_mouse_df.Drug == 'Placebo']['Tumor Volume (mm3)']
# Plot values
plt.errorbar(new_tumor_v_reformated_df.index.values.tolist(), new_tumor_v_reformated_df["Capomulin"] , yerr = capomulin_std_error, label= "Capomulin", marker= "o", color="red")
plt.errorbar(new_tumor_v_reformated_df.index.values.tolist(), new_tumor_v_reformated_df["Infubinol"] , yerr = capomulin_std_error, label= "Infubinol", marker= "o", color="green")
plt.errorbar(new_tumor_v_reformated_df.index.values.tolist(), new_tumor_v_reformated_df["Ketapril"] , yerr = capomulin_std_error, label= "Ketapril", marker= "o", color="blue")
plt.errorbar(new_tumor_v_reformated_df.index.values.tolist(), new_tumor_v_reformated_df["Placebo"] , yerr = capomulin_std_error, label= "Placebo", marker= "o", color="darkorange")
plt.legend()
plt.title("Tumor Volume Changes Over Time by Treatment")
plt.xlabel("Time (Days)")
plt.ylabel("Tumor Volume (mm3)")
plt.grid()
# Save the Figure
plt.savefig("Tumor_Volume_Changes.png")
# Show the Figure
fig.show()
# Store the Mean Met. Site Data Grouped by Drug and Timepoint
mean_metastatic_data = grouped_clinical_merge_mouse["Metastatic Sites"].mean()
# Create data frame with reseted index to hold the results (headers at the same level)
grouped_metastatic_df = pd.DataFrame(mean_metastatic_data).reset_index()
# Show results
grouped_metastatic_df.head(10)
# Store the Standard Error of Metastatic Site Grouped by Drug and Timepoint
std_error_metastatic_data = grouped_clinical_merge_mouse["Metastatic Sites"].sem()
# Create data frame with reseted index to hold the results (headers at the same level)
std_error_metastatic_df = pd.DataFrame(std_error_metastatic_data).reset_index()
# Display the data frame for preview
std_error_metastatic_df.head()
# Minor Data Munging to Re-Format the Data Frames
new_reformated_met_df = std_error_metastatic_df.pivot(index='Timepoint', columns='Drug', values='Metastatic Sites')
# Show results
new_reformated_met_df.head()
# Find Errors by treatments (Capomulin, Infubinol, Ketapril, and Placebo)
capomulin_met_std_error = std_error_metastatic_df[std_error_metastatic_df.Drug == 'Capomulin']['Metastatic Sites']
Infubinol_met_std_error = std_error_metastatic_df[std_error_metastatic_df.Drug == 'Infubinol']['Metastatic Sites']
Infubinol_met_std_error = std_error_metastatic_df[std_error_metastatic_df.Drug == 'Ketapril']['Metastatic Sites']
Infubinol_met_std_error = std_error_metastatic_df[std_error_metastatic_df.Drug == 'Placebo']['Metastatic Sites']
# Plot values
plt.errorbar(new_reformated_met_df.index.values.tolist(), new_reformated_met_df["Capomulin"] , yerr = capomulin_met_std_error, label= "Capomulin", marker= "o", color="red")
plt.errorbar(new_reformated_met_df.index.values.tolist(), new_reformated_met_df["Infubinol"] , yerr = capomulin_met_std_error, label= "Infubinol", marker= "o", color="green")
plt.errorbar(new_reformated_met_df.index.values.tolist(), new_reformated_met_df["Ketapril"] , yerr = capomulin_met_std_error, label= "Ketapril", marker= "o", color="blue")
plt.errorbar(new_reformated_met_df.index.values.tolist(), new_reformated_met_df["Placebo"] , yerr = capomulin_met_std_error, label= "Placebo", marker= "o", color="darkorange")
plt.legend()
plt.title("Metastatic Sites Changes Over Time by Treatment")
plt.xlabel("Time (Days)")
plt.ylabel("Metastatic Sites")
plt.grid()
# Save the Figure
plt.savefig("Metastatic_Sites_Changes.png")
# Show the Figure
fig.show()
# Store the Count of Mice Grouped by Drug and Timepoint (W can pass any metric)
count_mice_data = grouped_clinical_merge_mouse["Mouse ID"].nunique()
# Create data frame with reseted index to hold the results (headers at the same level)
grouped_count_mice_df = pd.DataFrame(count_mice_data).reset_index()
# Show results
grouped_count_mice_df.head()
# Minor Data Munging to Re-Format the Data Frames
new_reformated_mice_df = grouped_count_mice_df.pivot(index='Timepoint', columns='Drug', values='Mouse ID')
# Show results
new_reformated_mice_df.head(10)
# Set the total amount of mice that started the treatment at Timepoint = 0
total_mice_started_treatment = 25
# Find mice survival rate per treatment
capomulin_surv_rate = new_reformated_mice_df["Capomulin"]/total_mice_started_treatment * 100
infubinol_surv_rate = new_reformated_mice_df["Infubinol"]/total_mice_started_treatment * 100
ketapril_surv_rate = new_reformated_mice_df["Ketapril"]/total_mice_started_treatment * 100
placebo_surv_rate = new_reformated_mice_df["Placebo"]/total_mice_started_treatment * 100
# Plot Values
plt.errorbar(new_reformated_mice_df.index.values.tolist(), capomulin_surv_rate , label= "Capomulin", marker= "o", color="red")
plt.errorbar(new_reformated_mice_df.index.values.tolist(), infubinol_surv_rate , label= "Infubinol", marker= "o", color="green")
plt.errorbar(new_reformated_mice_df.index.values.tolist(), ketapril_surv_rate , label= "Ketapril", marker= "o", color="blue")
plt.errorbar(new_reformated_mice_df.index.values.tolist(), placebo_surv_rate , label= "Placebo", marker= "o", color="darkorange")
plt.legend()
plt.title("Mice Survival Rate Over Time by Treatment")
plt.xlabel("Time (Days)")
plt.ylabel("Survival Rate (%)")
plt.grid()
# Save the Figure
plt.savefig("Mice_Survival_Rate.png")
# Show the Figure
fig.show()
# Calculate the percent changes for each drug
percent_change_tumor_volume_data = new_tumor_v_reformated_df.iloc[[0,9]].pct_change()*100
percent_change = percent_change_tumor_volume_data.iloc[1]
# Show results
percent_change
# Store all Relevant Percent Changes into a Tuple
tup_percent_change = list(tuple(zip(percent_change.index, percent_change)))
# Show results
tup_percent_change
# Splice the data between passing and failing drugs
# Initialize an empty list to collect passing or failing status
drug_status = []
# Create a new data frame to show the results
status_df = pd.DataFrame(tup_percent_change, columns=["Drug", "Perecent Change"]).sort_values(by=["Perecent Change"]).set_index(['Drug'])
# Create a function to return the second element
def takeSecond(elem):
return elem[1]
# For loop to iterate through the list of tupples and collect the drug status depending on the second elemt's value
# If second element is negative this imply a reduction in the tumor volume and consequently the Drug is considered as "Passed".
# If second element is positive this imply an increment in the tumor volume and consequently the Drug is considered as "Failed".
# The list of tuples is being sorted by second element during iteration to be aligned with the sorted data frame
for drug, value in sorted(tup_percent_change, key=takeSecond):
if value < 0:
drug_status.append("Passed")
else:
drug_status.append("Failed")
status_df["Passing / Failing"] = drug_status
# Show results
status_df
# Orient widths. Add labels, tick marks, etc.
fig, ax = plt.subplots()
y_value1 = [percent_change["Capomulin"]]
y_value2 = [percent_change["Infubinol"], percent_change["Ketapril"], percent_change["Placebo"]]
x_axis1 = np.arange(1)
x_axis2 = np.arange(1,4)
x_labels = ["Capomulin", "Infubinol", "Ketapril", "Placebo"]
rect_1 = ax.bar(x_axis1, y_value1, color='limegreen', alpha=0.9, align="center", width = 0.55)
rect_2 = ax.bar(x_axis2, y_value2 , color='darkorange', alpha=0.9, align="center", width = 0.55)
plt.setp(ax, xticks=[0, 1, 2, 3], xticklabels=["Capomulin", "Infubinol", "Ketapril", "Placebo"],
yticks=[-20, 0, 20, 40, 60])
ax.set_ylabel('Tumor Volume Change (%)', fontsize=12,)
ax.set_xlabel('Drugs', fontsize=12)
ax.set_title('Tumor Change Over 45 Day Treatment',fontsize=14)
# Use functions to label the percentages of changes
def autolabel(rects):
for rect in rects:
height = rect.get_height()
ax.text(rect.get_x() + rect.get_width()/2, .1*height, "%d" %int(height)+ "%",
ha='center', va='top', color="white")
# Call functions to implement the function calls
autolabel(rect_1)
autolabel(rect_2)
fig.tight_layout()
# Save the Figure
plt.savefig("Percentage_Tumor_Volume_Change.png")
# Show the Figure
fig.show()
| 0.663887 | 0.687014 |
```
# -*- coding: utf-8 -*-
"""The Copy Task performed by LSTM cells using the tensorflow API"""
__author__ = "Aly Shmahell"
__copyright__ = "Copyright © 2019, Aly Shmahell"
__license__ = "All Rights Reserved"
__version__ = "0.1.2"
__maintainer__ = "Aly Shmahell"
__email__ = "[email protected]"
__status__ = "Alpha"
from __future__ import division
from __future__ import print_function
from __future__ import generator_stop
from __future__ import unicode_literals
from __future__ import absolute_import
import re
import os
import sys
import string
import itertools
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
from dataclasses import dataclass
from tensorflow.nn import dynamic_rnn
from tensorflow.nn.rnn_cell import LSTMCell, MultiRNNCell
tf.reset_default_graph()
class Pretty:
def __new__(self, x):
return re.sub(r"\n\s*", "\n", x)
class Oneliner:
def __new__(self, x):
return re.sub(r"\n\s*", " ", x)
@dataclass()
class Data(object):
corpus: np.array
symbols_ordered: dict
symbols_reversed: dict
corpus_size: int
symbols_size: int
def __init__(self):
_text = Oneliner(
"""long ago , the mice had a general council to consider
what measures they could take to outwit their common enemy ,
the cat .
some said this , and some said that but at last a young
mouse got up and said he had a proposal to make , which he
thought would meet the case .
you will all agree , said he , that our chief danger
consists in the sly and treacherous manner
in which the enemy approaches us .
now , if we could receive some signal of her approach ,
we could easily escape from her .
I venture , therefore , to propose that a small bell
be procured , and attached by a ribbon round the neck
of the cat .
by this means we should always know when she was about ,
and could easily retire while she was in the neighbourhood .
this proposal met with general applause , until an old
mouse got up and said that is all very well ,
but who is to bell the cat ? the mice looked
at one another and nobody spoke .
then the old mouse said it is easy to
propose impossible remedies ."""
)
self.corpus = [
word
for i in range(
len(
[
x.strip() for x in _text
]
)
)
for word in [
x.strip()
for x in _text
][i].split()
]
_counter = itertools.count(0)
self.symbols_ordered = {
chr(i):next(_counter)
for i in range(128)
if chr(i) in string.ascii_letters
or chr(i) in string.digits
or chr(i) in string.punctuation
}
self.symbols_reversed = dict(
zip(
self.symbols_ordered.values(),
self.symbols_ordered.keys()
)
)
self.corpus_size = len(self.corpus)
self.symbols_size = len(self.symbols_ordered)
data = Data()
def get_random_chunck(chunk_len):
if chunk_len > data.corpus_size:
sys.exit("chunk length exceeds the corpus length")
_offset = np.random.randint(0, data.corpus_size-chunk_len-1)
return data.corpus[_offset: _offset+chunk_len]
def cast_chunk_to_ints(chunk):
return [
data.symbols_ordered[c]
for c in chunk
]
def create_batch(num_samples):
raw = [
f"{i:>08b}"
for i in cast_chunk_to_ints(
get_random_chunck(num_samples)
)
]
np.random.shuffle(raw)
raw = [
list(map(int,i))
for i in raw
]
raw = np.array(raw)
source = np.copy(raw)
source = source.reshape(num_samples, 8, 1)
target = np.copy(raw)
target = target.reshape(num_samples, 8)
return source, target
def stringify(result):
return "".join(
[
data.symbols_reversed[x]
for x in [
int(
"".join(
[
str(y)
for y in result[i]
]
),
2
)
for i in range(len(result))
]
]
)
class Architecture(object):
def __init__(self,
units,
tf_source_shape,
tf_source_dtype,
tf_target_shape,
tf_target_dtype):
self.tf_source = tf.placeholder(
shape=tf_source_shape,
dtype=tf_source_dtype
)
self.tf_target = tf.placeholder(
shape=tf_target_shape,
dtype=tf_target_dtype
)
multi_rnn_cells = MultiRNNCell(
[
LSTMCell(
units,
num_proj=len(tf_target_shape)
),
LSTMCell(
units,
num_proj=len(tf_target_shape)
)
]
)
self.output, _ = tf.nn.dynamic_rnn(
multi_rnn_cells,
self.tf_source,
dtype=tf_source_dtype
)
structure = Architecture(
units= 64,
tf_source_shape = [None, None, 1],
tf_source_dtype = tf.float32,
tf_target_shape = [None, None],
tf_target_dtype = tf.int64
)
loss_function = tf.reduce_mean(
tf.nn
.sparse_softmax_cross_entropy_with_logits(
labels=structure.tf_target,
logits=structure.output
)
)
optimizer = tf.train.AdamOptimizer(learning_rate=0.1).minimize(loss_function)
prediction = tf.argmax(structure.output, axis=2)
validity = tf.equal(structure.tf_target, prediction)
precision = tf.reduce_mean(tf.cast(validity, tf.float32))
errors = []
with tf.Session() as session:
session.run(tf.initialize_all_variables())
for epoch in range(100):
source, target = create_batch(200)
_, error, accuracy = session.run(
[
optimizer,
loss_function,
precision
],
feed_dict={
structure.tf_source: source,
structure.tf_target: target
}
)
print(f"Epoch: {epoch}, Error: {error}, Accuracy: {accuracy*100}")
errors.append(error)
source = [
[
[0],[1],[0],[0],[0],[1],[1],[1]
],
[
[0],[1],[0],[0],[0],[1],[0],[0]
],
[
[0],[1],[0],[1],[1],[0],[0],[0]
]
]
print(
Oneliner(
f"""Source:
{
stringify(
np.reshape(
np.array(source),
[3, 8]
)
)
}
"""
)
)
print(
Oneliner(
f"""Predicted:
{
stringify(
session.run(
prediction,
feed_dict={structure.tf_source: source}
)
)
}
"""
)
)
plt.plot(errors,label="Loss Function")
plt.xlabel("Epochs")
plt.ylabel("Errors")
plt.legend()
plt.show()
```
|
github_jupyter
|
# -*- coding: utf-8 -*-
"""The Copy Task performed by LSTM cells using the tensorflow API"""
__author__ = "Aly Shmahell"
__copyright__ = "Copyright © 2019, Aly Shmahell"
__license__ = "All Rights Reserved"
__version__ = "0.1.2"
__maintainer__ = "Aly Shmahell"
__email__ = "[email protected]"
__status__ = "Alpha"
from __future__ import division
from __future__ import print_function
from __future__ import generator_stop
from __future__ import unicode_literals
from __future__ import absolute_import
import re
import os
import sys
import string
import itertools
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
from dataclasses import dataclass
from tensorflow.nn import dynamic_rnn
from tensorflow.nn.rnn_cell import LSTMCell, MultiRNNCell
tf.reset_default_graph()
class Pretty:
def __new__(self, x):
return re.sub(r"\n\s*", "\n", x)
class Oneliner:
def __new__(self, x):
return re.sub(r"\n\s*", " ", x)
@dataclass()
class Data(object):
corpus: np.array
symbols_ordered: dict
symbols_reversed: dict
corpus_size: int
symbols_size: int
def __init__(self):
_text = Oneliner(
"""long ago , the mice had a general council to consider
what measures they could take to outwit their common enemy ,
the cat .
some said this , and some said that but at last a young
mouse got up and said he had a proposal to make , which he
thought would meet the case .
you will all agree , said he , that our chief danger
consists in the sly and treacherous manner
in which the enemy approaches us .
now , if we could receive some signal of her approach ,
we could easily escape from her .
I venture , therefore , to propose that a small bell
be procured , and attached by a ribbon round the neck
of the cat .
by this means we should always know when she was about ,
and could easily retire while she was in the neighbourhood .
this proposal met with general applause , until an old
mouse got up and said that is all very well ,
but who is to bell the cat ? the mice looked
at one another and nobody spoke .
then the old mouse said it is easy to
propose impossible remedies ."""
)
self.corpus = [
word
for i in range(
len(
[
x.strip() for x in _text
]
)
)
for word in [
x.strip()
for x in _text
][i].split()
]
_counter = itertools.count(0)
self.symbols_ordered = {
chr(i):next(_counter)
for i in range(128)
if chr(i) in string.ascii_letters
or chr(i) in string.digits
or chr(i) in string.punctuation
}
self.symbols_reversed = dict(
zip(
self.symbols_ordered.values(),
self.symbols_ordered.keys()
)
)
self.corpus_size = len(self.corpus)
self.symbols_size = len(self.symbols_ordered)
data = Data()
def get_random_chunck(chunk_len):
if chunk_len > data.corpus_size:
sys.exit("chunk length exceeds the corpus length")
_offset = np.random.randint(0, data.corpus_size-chunk_len-1)
return data.corpus[_offset: _offset+chunk_len]
def cast_chunk_to_ints(chunk):
return [
data.symbols_ordered[c]
for c in chunk
]
def create_batch(num_samples):
raw = [
f"{i:>08b}"
for i in cast_chunk_to_ints(
get_random_chunck(num_samples)
)
]
np.random.shuffle(raw)
raw = [
list(map(int,i))
for i in raw
]
raw = np.array(raw)
source = np.copy(raw)
source = source.reshape(num_samples, 8, 1)
target = np.copy(raw)
target = target.reshape(num_samples, 8)
return source, target
def stringify(result):
return "".join(
[
data.symbols_reversed[x]
for x in [
int(
"".join(
[
str(y)
for y in result[i]
]
),
2
)
for i in range(len(result))
]
]
)
class Architecture(object):
def __init__(self,
units,
tf_source_shape,
tf_source_dtype,
tf_target_shape,
tf_target_dtype):
self.tf_source = tf.placeholder(
shape=tf_source_shape,
dtype=tf_source_dtype
)
self.tf_target = tf.placeholder(
shape=tf_target_shape,
dtype=tf_target_dtype
)
multi_rnn_cells = MultiRNNCell(
[
LSTMCell(
units,
num_proj=len(tf_target_shape)
),
LSTMCell(
units,
num_proj=len(tf_target_shape)
)
]
)
self.output, _ = tf.nn.dynamic_rnn(
multi_rnn_cells,
self.tf_source,
dtype=tf_source_dtype
)
structure = Architecture(
units= 64,
tf_source_shape = [None, None, 1],
tf_source_dtype = tf.float32,
tf_target_shape = [None, None],
tf_target_dtype = tf.int64
)
loss_function = tf.reduce_mean(
tf.nn
.sparse_softmax_cross_entropy_with_logits(
labels=structure.tf_target,
logits=structure.output
)
)
optimizer = tf.train.AdamOptimizer(learning_rate=0.1).minimize(loss_function)
prediction = tf.argmax(structure.output, axis=2)
validity = tf.equal(structure.tf_target, prediction)
precision = tf.reduce_mean(tf.cast(validity, tf.float32))
errors = []
with tf.Session() as session:
session.run(tf.initialize_all_variables())
for epoch in range(100):
source, target = create_batch(200)
_, error, accuracy = session.run(
[
optimizer,
loss_function,
precision
],
feed_dict={
structure.tf_source: source,
structure.tf_target: target
}
)
print(f"Epoch: {epoch}, Error: {error}, Accuracy: {accuracy*100}")
errors.append(error)
source = [
[
[0],[1],[0],[0],[0],[1],[1],[1]
],
[
[0],[1],[0],[0],[0],[1],[0],[0]
],
[
[0],[1],[0],[1],[1],[0],[0],[0]
]
]
print(
Oneliner(
f"""Source:
{
stringify(
np.reshape(
np.array(source),
[3, 8]
)
)
}
"""
)
)
print(
Oneliner(
f"""Predicted:
{
stringify(
session.run(
prediction,
feed_dict={structure.tf_source: source}
)
)
}
"""
)
)
plt.plot(errors,label="Loss Function")
plt.xlabel("Epochs")
plt.ylabel("Errors")
plt.legend()
plt.show()
| 0.433981 | 0.210219 |
# AI4M Course 1 week 3 lecture notebook
# Outline
Click on these links to jump to that section of the notebook!
- [Explore the data](#data)
- [Get a subsection](#subsection)
- [U-net model](#unet)
<a name="data"></a>
# Explore the data
```
import numpy as np
import nibabel as nib
```
#### Image of brain
```
image_path = "./BraTS-Data/imagesTr/BRATS_001.nii.gz"
image_obj = nib.load(image_path)
type(image_obj)
image_data = image_obj.get_fdata()
type(image_data)
print("height, width, depth, channels")
image_data.shape
import matplotlib.pyplot as plt
# start at this depth to see more of the middle of the brain
i=65
# run this cell a few times
# to see some slices of the brain
print(f"depth {i}")
plt.imshow(image_data[:,:,i,0]);
i +=1
```
#### Label of diseases
```
label_path = "./BraTS-Data/labelsTr/BRATS_003.nii.gz"
#label = np.array(nib.load(label_nifty_file).get_fdata())
label_obj = nib.load(label_path)
type(label_obj)
label = label_obj.get_fdata()
type(label)
print("height, width, depth")
label.shape
# See that all label values are either 0, 1, 2 or 3
print("""categories:
0: normal
1: edema
2: non-enhancing tumor
3: enhancing tumor""")
np.unique(label)
```
#### Label for edema
```
label_edema = (label == 1.)
# start at this index to see more of the middle of the brain image
i=65
# run this cell a few times
# to see the edema labels at various slices
print(f"depth {i}")
plt.imshow(label_edema[:,:,i])
i +=1
```
### This is the end of this practice section.
Please continue on with the lecture videos!
---
<a name="subsection"></a>
# Get a sub-section
Show how to do this for 1D arrays. the assignment will be 3D.
```
import numpy as np
image = np.array([10,11,12,13,14,15])
image
image_length = image.shape[0]
image_length
patch_length = 3
start_i = 0
# run this a few times to see some valid sub-sections
# and see when it's no longer valid
print(f"start index {start_i}")
end_i = start_i + patch_length
print(f"end index {end_i}")
sub_section = image[start_i: end_i]
print(sub_section)
start_i +=1
# This is a valid patch
start_i = 3
print(f"start index {start_i}")
end_i = start_i + patch_length
print(f"end index {end_i}")
sub_section = image[start_i: end_i]
print(sub_section)
print(f"The largest start index for which "
f"a sub section is still valid is "
f"{image_length - patch_length}")
print(f"The range of valid start indices is:")
valid_start_i = [i for i in range(image_length - patch_length + 1)]
print(valid_start_i)
```
Randomly select a valid integer for the start index
```
start_i = np.random.randint(image_length - patch_length + 1)
print(f"randomly selected start index {start_i}")
for _ in range(10):
start_i = np.random.randint(image_length - patch_length + 1)
print(f"randomly selected start index {start_i}")
```
### This is the end of this practice section.
Please continue on with the lecture videos!
---
<a name="unet"></a>
## U-Net model
```
import keras
from keras import backend as K
from keras.engine import Input, Model
from keras.layers import Conv3D, MaxPooling3D, UpSampling3D, Activation, BatchNormalization, PReLU, Deconvolution3D
from keras.optimizers import Adam
from keras.layers.merge import concatenate
K.set_image_data_format("channels_first")
```
### Choose depth
We'll choose to make a smaller U-Net which has a full depth of 2.
```
u_net_depth = 2
```
#### Input layer
The shape of the input is (num_channels, height, width, depth).
For the assignment, the values will be:
- num_channels: 4
- height: 160
- width: 160
- depth: 16
```
input_layer = Input(shape=(4, 160, 160, 16))
input_layer
```
Notice that the tensor shape has a '?' as the very first dimension. This is the batch size.
(batch_size, num_channels, height, width, depth)
## Contracting path (downward)
We'll start with the downward path on the left side of the U-Net. The (height,width,depth) of the 'image' gets smaller, and the number of channels increases.
### Depth 0
When we say 'depth' here, we're referring to the depth of the U-net and not the depth related to the height, width, depth of an image.
The number of filters is specified for each depth and for each layer within that depth.
At depth 0, for layer 0, we'll use 32 filters.
The formula we're using is:
$$filters_{i} = 32 \times (2^{i})$$
Where $i$ is the current depth.
So at depth $i=0$:
$$filters_{0} = 32 \times (2^{0}) = 32$$
#### Layer 0
There are two convolutional layers for each depth
##### 3d convolution
```
down_depth_0_layer_0 = Conv3D(filters=32,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(input_layer)
```
You can ignore the warning messages.
```
down_depth_0_layer_0
```
##### Add a relu activation
```
down_depth_0_layer_0 = Activation('relu')(down_depth_0_layer_0)
down_depth_0_layer_0
```
### Depth 0
#### Layer 1
For layer 1 of depth 0, we'll choose 64 filters.
The formula we're using is:
$$filters_{i} = 32 \times (2^{i}) \times 2$$
Where $i$ is the current depth.
- Notice the '$\times 2$' at the end of this expression, which isn't there for layer 0.
So at depth $i=0$:
$$filters_{0} = 32 \times (2^{0}) \times 2 = 64$$
```
down_depth_0_layer_1 = Conv3D(filters=64,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(down_depth_0_layer_0)
down_depth_0_layer_1 = Activation('relu')(down_depth_0_layer_1)
down_depth_0_layer_1
```
#### Max pooling
There is max pooling for the earlier depths (when the inputs to the layers are larger).
- To decide if we include max pooling, check if the current depth of 0 is less than the U-Net's full depth of 2 minus 1.
- The current layer depth of 0 is less than 2 minus 1.
- In other words, 0 < 1.
- So we'll include a max pooling layer.
```
down_depth_0_layer_pool = MaxPooling3D(pool_size=(2,2,2))(down_depth_0_layer_1)
down_depth_0_layer_pool
```
### Depth 1
At depth 1, layer 0, we'll choose 64 filters
The formula we're using is:
$$filters_{i} = 32 \times (2^{i})$$
Where $i$ is the current depth.
So at depth $i=1$:
$$filters_{1} = 32 \times (2^{1}) = 64$$
#### layer 0
```
down_depth_1_layer_0 = Conv3D(filters=64,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(down_depth_0_layer_pool)
down_depth_1_layer_0 = Activation('relu')(down_depth_1_layer_0)
down_depth_1_layer_0
```
#### Layer 1
For layer 1 of depth 1, we'll choose 128 filters.
The formula we're using is:
$$filters_{i} = 32 \times (2^{i}) \times 2$$
Where $i$ is the current depth.
- Notice the '$\times 2$' at the end of this expression, which isn't there for layer 0.
So at depth $i=1$:
$$filters_{0} = 32 \times (2^{1}) \times 2 = 128$$
```
down_depth_1_layer_1 = Conv3D(filters=128,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(down_depth_1_layer_0)
down_depth_1_layer_1 = Activation('relu')(down_depth_1_layer_1)
down_depth_1_layer_1
```
##### No max pooling at depth 1
When we get to the "bottom" of the U-net, a the last depth, we don't need to apply max pooling.
- To decide if we include max pooling, check if the current depth of 1 is less than the U-Net's full depth of 2 minus 1.
- The current layer depth of 1 is not less than 2 minus 1.
- In other words, 1 is not less than 1.
- So we won't include a max pooling layer.
## Expanding Path (upward)
Now we'll work on the expanding path of the U-Net, (going up on the right side, when viewing the diagram). The image's (height,width,depth) gets larger in the expanding path.
### Depth 0
#### Up sampling layer 0
We'll use a pool size of (2,2,2) for up sampling.
- This is the default value for [tf.keras.layers.UpSampling3D](https://www.tensorflow.org/api_docs/python/tf/keras/layers/UpSampling3D)
- We'll still set this parameter explicitly for learning purposes.
- As input to the up sampling at depth 1, we'll use the last layer of the down sampling. In this case, it's the depth 1 layer 1.
- Note that we're not adding any activation to this upsampling layer.
```
up_depth_0_layer_0 = UpSampling3D(size=(2,2,2))(down_depth_1_layer_1)
up_depth_0_layer_0
```
#### Concatenate the up sampled layer with the down sampled layer
Use the layers that are both at the same depth of 0.
- up_depth_0_layer_0: shape is ?, 128, 160, 160, 16
- depth_0_layer_1: shape is (?, 64, 160, 160, 16)
- Double check that both of these layers have the same shape.
- If they're the same shape, then they can be concatenated at axis 1 (the channel axis).
- The height, width, depth (depth of image, not of network) is 160, 160, 16 for both.
```
print(up_depth_0_layer_0)
print()
print(down_depth_0_layer_1)
up_depth_1_concat = concatenate([up_depth_0_layer_0,
down_depth_0_layer_1],
axis=1)
up_depth_1_concat
```
The up sampling layer has 128 channels, and the down convolution layer has 64 channels.
- When concatenated, they have 128 + 64 = 192 channels.
#### up convolution layer 1
The number of filters for this layer will be set to the number of channels in the down convolution's layer 1 at the same depth of 0 (down_depth_0_layer_1).
- The shape of down_depth_0_layer_1 is (?, 64, 160, 160, 16)
```
down_depth_0_layer_1
```
- The number of channels for depth_0_layer_1 is 64
```
print(f"number of filters: {down_depth_0_layer_1._keras_shape[1]}")
up_depth_1_layer_1 = Conv3D(filters=64,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(up_depth_1_concat)
up_depth_1_layer_1 = Activation('relu')(up_depth_1_layer_1)
up_depth_1_layer_1
```
#### up convolution layer 2
The number of filters will also be set to 64.
- Again, since we're at depth 0, we look for the number of channels in the downward convolution layer at depth 0.
- The shape of down_depth_0_layer_1 is (?, 4, 160, 160, 64)
```
down_depth_0_layer_1
```
- The number of channels in down_depth_0_layer_1 is 64.
```
print(f"number of filters: {down_depth_0_layer_1._keras_shape[1]}")
up_depth_1_layer_2 = Conv3D(filters=64,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(up_depth_1_layer_1)
up_depth_1_layer_2 = Activation('relu')(up_depth_1_layer_2)
up_depth_1_layer_2
```
### Final Convolution
The number of filters is set to the number of labels.
- The number of labels is the number of categories.
In this case, there are 3 labels
- 1: edema
- 2: non-enhancing tumor
- 3: enhancing tumor
```
final_conv = Conv3D(filters=3, #3 categories
kernel_size=(1,1,1),
padding='valid',
strides=(1,1,1)
)(up_depth_1_layer_2)
final_conv
```
#### Activation for final convolution
```
final_activation = Activation('sigmoid')(final_conv)
final_activation
```
### Create and compile the model
In this example, we're setting the loss and metrics to options that are pre-built in Keras. However, in the assignment, you will implement better loss funtions and metrics for evaluating the model's performance.
- The soft dice loss
- dice coefficient.
```
model = Model(inputs=input_layer, outputs=final_activation)
model.compile(optimizer=Adam(lr=0.00001),
loss='categorical_crossentropy',
metrics=['categorical_accuracy']
)
model.summary()
```
### Double check your model
Use a function that we've provided to create the same model, and check that the layers and the layer dimensions match!
```
import util
model_2 = util.unet_model_3d(depth=2,
loss_function='categorical_crossentropy',
metrics=['categorical_accuracy'])
model_2.summary()
```
### This is the end of this practice section.
Please continue on with the lecture videos!
---
|
github_jupyter
|
import numpy as np
import nibabel as nib
image_path = "./BraTS-Data/imagesTr/BRATS_001.nii.gz"
image_obj = nib.load(image_path)
type(image_obj)
image_data = image_obj.get_fdata()
type(image_data)
print("height, width, depth, channels")
image_data.shape
import matplotlib.pyplot as plt
# start at this depth to see more of the middle of the brain
i=65
# run this cell a few times
# to see some slices of the brain
print(f"depth {i}")
plt.imshow(image_data[:,:,i,0]);
i +=1
label_path = "./BraTS-Data/labelsTr/BRATS_003.nii.gz"
#label = np.array(nib.load(label_nifty_file).get_fdata())
label_obj = nib.load(label_path)
type(label_obj)
label = label_obj.get_fdata()
type(label)
print("height, width, depth")
label.shape
# See that all label values are either 0, 1, 2 or 3
print("""categories:
0: normal
1: edema
2: non-enhancing tumor
3: enhancing tumor""")
np.unique(label)
label_edema = (label == 1.)
# start at this index to see more of the middle of the brain image
i=65
# run this cell a few times
# to see the edema labels at various slices
print(f"depth {i}")
plt.imshow(label_edema[:,:,i])
i +=1
import numpy as np
image = np.array([10,11,12,13,14,15])
image
image_length = image.shape[0]
image_length
patch_length = 3
start_i = 0
# run this a few times to see some valid sub-sections
# and see when it's no longer valid
print(f"start index {start_i}")
end_i = start_i + patch_length
print(f"end index {end_i}")
sub_section = image[start_i: end_i]
print(sub_section)
start_i +=1
# This is a valid patch
start_i = 3
print(f"start index {start_i}")
end_i = start_i + patch_length
print(f"end index {end_i}")
sub_section = image[start_i: end_i]
print(sub_section)
print(f"The largest start index for which "
f"a sub section is still valid is "
f"{image_length - patch_length}")
print(f"The range of valid start indices is:")
valid_start_i = [i for i in range(image_length - patch_length + 1)]
print(valid_start_i)
start_i = np.random.randint(image_length - patch_length + 1)
print(f"randomly selected start index {start_i}")
for _ in range(10):
start_i = np.random.randint(image_length - patch_length + 1)
print(f"randomly selected start index {start_i}")
import keras
from keras import backend as K
from keras.engine import Input, Model
from keras.layers import Conv3D, MaxPooling3D, UpSampling3D, Activation, BatchNormalization, PReLU, Deconvolution3D
from keras.optimizers import Adam
from keras.layers.merge import concatenate
K.set_image_data_format("channels_first")
u_net_depth = 2
input_layer = Input(shape=(4, 160, 160, 16))
input_layer
down_depth_0_layer_0 = Conv3D(filters=32,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(input_layer)
down_depth_0_layer_0
down_depth_0_layer_0 = Activation('relu')(down_depth_0_layer_0)
down_depth_0_layer_0
down_depth_0_layer_1 = Conv3D(filters=64,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(down_depth_0_layer_0)
down_depth_0_layer_1 = Activation('relu')(down_depth_0_layer_1)
down_depth_0_layer_1
down_depth_0_layer_pool = MaxPooling3D(pool_size=(2,2,2))(down_depth_0_layer_1)
down_depth_0_layer_pool
down_depth_1_layer_0 = Conv3D(filters=64,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(down_depth_0_layer_pool)
down_depth_1_layer_0 = Activation('relu')(down_depth_1_layer_0)
down_depth_1_layer_0
down_depth_1_layer_1 = Conv3D(filters=128,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(down_depth_1_layer_0)
down_depth_1_layer_1 = Activation('relu')(down_depth_1_layer_1)
down_depth_1_layer_1
up_depth_0_layer_0 = UpSampling3D(size=(2,2,2))(down_depth_1_layer_1)
up_depth_0_layer_0
print(up_depth_0_layer_0)
print()
print(down_depth_0_layer_1)
up_depth_1_concat = concatenate([up_depth_0_layer_0,
down_depth_0_layer_1],
axis=1)
up_depth_1_concat
down_depth_0_layer_1
print(f"number of filters: {down_depth_0_layer_1._keras_shape[1]}")
up_depth_1_layer_1 = Conv3D(filters=64,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(up_depth_1_concat)
up_depth_1_layer_1 = Activation('relu')(up_depth_1_layer_1)
up_depth_1_layer_1
down_depth_0_layer_1
print(f"number of filters: {down_depth_0_layer_1._keras_shape[1]}")
up_depth_1_layer_2 = Conv3D(filters=64,
kernel_size=(3,3,3),
padding='same',
strides=(1,1,1)
)(up_depth_1_layer_1)
up_depth_1_layer_2 = Activation('relu')(up_depth_1_layer_2)
up_depth_1_layer_2
final_conv = Conv3D(filters=3, #3 categories
kernel_size=(1,1,1),
padding='valid',
strides=(1,1,1)
)(up_depth_1_layer_2)
final_conv
final_activation = Activation('sigmoid')(final_conv)
final_activation
model = Model(inputs=input_layer, outputs=final_activation)
model.compile(optimizer=Adam(lr=0.00001),
loss='categorical_crossentropy',
metrics=['categorical_accuracy']
)
model.summary()
import util
model_2 = util.unet_model_3d(depth=2,
loss_function='categorical_crossentropy',
metrics=['categorical_accuracy'])
model_2.summary()
| 0.620507 | 0.935935 |
#### Helper Code
This code runs out of the box on Google Colab. It is also the recommended way to run this notebook if you do not already have an environment with Python 3 and TensorFlow already set up.
```
import numpy as np
import os
matplotlib_style = "ggplot"
import matplotlib.pyplot as plt; plt.style.use(matplotlib_style)
import matplotlib.axes as axes;
from matplotlib.patches import Ellipse
%matplotlib inline
import seaborn as sns
sns.set_context('notebook')
from IPython.core.pylabtools import figsize
notebook_screen_res = "retina"
%config InlineBackend.figure_format = notebook_screen_res
from IPython.display import clear_output
import tensorflow as tf
try:
tf.enable_eager_execution()
print("Enabled eager execution")
except Exception as e:
print(e)
print("Eager execution:", tf.executing_eagerly() )
import tensorflow_probability as tfp
tfd = tfp.distributions
tfb = tfp.bijectors
print("TensorFlow\t\t", ":", tf.__version__)
print("TensorFlow Probability\t", ":", tfp.__version__)
from tensorflow.python.client import device_lib
local_device_protos = device_lib.list_local_devices()
print("Devices:")
print([x.name for x in local_device_protos])
def evaluate(tensors):
"""Evaluates Tensor or EagerTensor to Numpy `ndarray`s.
Args:
tensors: Object of `Tensor` or EagerTensor`s; can be `list`, `tuple`,
`namedtuple` or combinations thereof.
Returns:
ndarrays: Object with same structure as `tensors` except with `Tensor` or
`EagerTensor`s replaced by Numpy `ndarray`s.
"""
return tf.contrib.framework.nest.pack_sequence_as(tensors,
[t.numpy() if tf.contrib.framework.is_tensor(t) else t
for t in tf.contrib.framework.nest.flatten(tensors)])
from sklearn.metrics import mean_squared_error
```
# 04. Sampling from Distributions
## Sampling
**Conjugate priors**
Family of conjugate priors: If we choose a prior from $\Psi$, then sampling from the prior will result in a posterior will also in $\Psi$. In other words, $\Psi$ is closed under sampling from this given class of distributions.
| Sampling From | Posterior Distribution |
| ------------------------- | ----------------------------------- |
| Bernoulli | beta |
| binomial | beta |
| geometric | beta |
| poisson | gamma |
| exponential | gamma |
| normal | normal |
## Sampling from Exponential Distribution
A lighting company has designed a new light bulb model. They are interested in finding out how long each light bulb lasts.
Consider a statistical model consisting of observable exponential RV $X_1 ... X_{10}$ that are conditionally iid given the parameter $\theta$.
Each $X_i$ represents the lifespan (in hours) of the $i$-th light bulb. Suppose that $\theta$ is a gamma RV with prior hyperparameters 10 and 4500.
```
alpha, beta = 4500, 10
theta = tfd.Gamma(concentration=alpha,
rate=beta)
```
Suppose that the observed values are represented by X:
```
X_sample = theta.sample(10).numpy()
X_sample = X_sample.tolist()
print(X_sample)
```
From the given information, we can determine the posterior distribution of $\theta$ is a gamma distribution with hyperparameters $\alpha'$ and $\beta'$. We can also see how a biased (offset) sample can affect the posterior distribution.
```
#@title Add offset to sample { run: "auto" }
offset = 40 #@param {type:"slider", min:-100, max:100, step:1}
X = theta.sample(10).numpy()
X += offset
X = X.tolist()
alpha_p = alpha + sum(X)
beta_p = beta + len(X)
print("𝛼′ =", alpha_p, "𝛽′ =", beta_p)
theta_p = tfd.Gamma(concentration=alpha_p,
rate=beta_p)
n = 50
start = tf.constant(400., dtype="float32")
space = tf.linspace(start=start, stop=500., num=n, name="linspace")
theta_values = theta.prob(space[:, tf.newaxis])
theta_values = tf.transpose(theta_values)
theta_values = theta_values.numpy().reshape((n))
theta_p_values = theta_p.prob(space[:, tf.newaxis])
theta_p_values = tf.transpose(theta_p_values)
theta_p_values = theta_p_values.numpy().reshape((n))
plt.figure(figsize(8, 4))
plt.plot(space,
theta_values,
label="Prior")
plt.fill_between(space, 0, theta_values, alpha=0.4)
plt.plot(space,
theta_p_values,
label="Posterior")
plt.fill_between(space, 0, theta_p_values, alpha=0.4)
plt.legend()
plt.tight_layout()
plt.show()
```
## References
1. Lecture 15, 16, Introduction to Probability and Statistics, 50.034 (2018), Singapore University of Technology and Design
```
```
|
github_jupyter
|
import numpy as np
import os
matplotlib_style = "ggplot"
import matplotlib.pyplot as plt; plt.style.use(matplotlib_style)
import matplotlib.axes as axes;
from matplotlib.patches import Ellipse
%matplotlib inline
import seaborn as sns
sns.set_context('notebook')
from IPython.core.pylabtools import figsize
notebook_screen_res = "retina"
%config InlineBackend.figure_format = notebook_screen_res
from IPython.display import clear_output
import tensorflow as tf
try:
tf.enable_eager_execution()
print("Enabled eager execution")
except Exception as e:
print(e)
print("Eager execution:", tf.executing_eagerly() )
import tensorflow_probability as tfp
tfd = tfp.distributions
tfb = tfp.bijectors
print("TensorFlow\t\t", ":", tf.__version__)
print("TensorFlow Probability\t", ":", tfp.__version__)
from tensorflow.python.client import device_lib
local_device_protos = device_lib.list_local_devices()
print("Devices:")
print([x.name for x in local_device_protos])
def evaluate(tensors):
"""Evaluates Tensor or EagerTensor to Numpy `ndarray`s.
Args:
tensors: Object of `Tensor` or EagerTensor`s; can be `list`, `tuple`,
`namedtuple` or combinations thereof.
Returns:
ndarrays: Object with same structure as `tensors` except with `Tensor` or
`EagerTensor`s replaced by Numpy `ndarray`s.
"""
return tf.contrib.framework.nest.pack_sequence_as(tensors,
[t.numpy() if tf.contrib.framework.is_tensor(t) else t
for t in tf.contrib.framework.nest.flatten(tensors)])
from sklearn.metrics import mean_squared_error
alpha, beta = 4500, 10
theta = tfd.Gamma(concentration=alpha,
rate=beta)
X_sample = theta.sample(10).numpy()
X_sample = X_sample.tolist()
print(X_sample)
#@title Add offset to sample { run: "auto" }
offset = 40 #@param {type:"slider", min:-100, max:100, step:1}
X = theta.sample(10).numpy()
X += offset
X = X.tolist()
alpha_p = alpha + sum(X)
beta_p = beta + len(X)
print("𝛼′ =", alpha_p, "𝛽′ =", beta_p)
theta_p = tfd.Gamma(concentration=alpha_p,
rate=beta_p)
n = 50
start = tf.constant(400., dtype="float32")
space = tf.linspace(start=start, stop=500., num=n, name="linspace")
theta_values = theta.prob(space[:, tf.newaxis])
theta_values = tf.transpose(theta_values)
theta_values = theta_values.numpy().reshape((n))
theta_p_values = theta_p.prob(space[:, tf.newaxis])
theta_p_values = tf.transpose(theta_p_values)
theta_p_values = theta_p_values.numpy().reshape((n))
plt.figure(figsize(8, 4))
plt.plot(space,
theta_values,
label="Prior")
plt.fill_between(space, 0, theta_values, alpha=0.4)
plt.plot(space,
theta_p_values,
label="Posterior")
plt.fill_between(space, 0, theta_p_values, alpha=0.4)
plt.legend()
plt.tight_layout()
plt.show()
| 0.791338 | 0.948728 |
<a href="https://colab.research.google.com/github/Funmilayo-Aina/-JPMorgan-Chase-Software-Engineering-Virtual-Experience/blob/main/Welcome_To_Colaboratory.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
<h1>Welcome to Colab!</h1>
If you're already familiar with Colab, check out this video to learn about interactive tables, the executed code history view, and the command palette.
<center>
<a href="https://www.youtube.com/watch?v=rNgswRZ2C1Y" target="_blank">
<img alt='Thumbnail for a video showing 3 cool Google Colab features' src="data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAVIAAAC+CAYAAABnJIeiAAAMa2lDQ1BJQ0MgUHJvZmlsZQAASImVlwdYk1cXgO83kpCQsAJhyAh7CbKnjBBWBAGZgouQBBJGjAlBxI2WKli3iOBEqyKKVisgoiJqnUVxW0dRi0qlFrUoisp/M0Br//H853nud9+ce+45557cb1wAtAe4Ekk+qgNAgbhQmhgVxpyYnsEkdQNNYA7IgAQMuTyZhJWQEAugDPd/lzc3AaLor7kofP1z/L+KHl8g4wGATIacxZfxCiC3AYBv5EmkhQAQFXrrmYUSBS+ArC+FCUJeq+AcFe9WcJaKW5U2yYlsyFcA0KByudIcALTuQT2ziJcD/Wh9gOwm5ovEAGiPhhzME3L5kBW5jy4omK7gKsgO0F4CGeYD/LK+8JnzN/9ZI/653JwRVq1LKRrhIpkknzvr/yzN/5aCfPlwDDvYqEJpdKJi/bCGt/OmxyiYCrlXnBUXr6g15AERX1V3AFCKUB6dorJHTXkyNqwfYEB243PDYyCbQo4U58fFqvVZ2aJIDmS4W9BiUSEnGbIR5CUCWUSS2mardHqiOhbamC1ls9T6c1ypMq4i1gN5XgpL7f+VUMBR+8e0SoTJaZApkG2KRKlxkLUgu8rykmLUNmNLhOy4YRupPFGRvw3kRIE4KkzlHyvKlkYmqu3LC2TD68W2CkWcODUfLBQmR6vqg53mcZX5w7VgVwRiVsqwH4FsYuzwWviC8AjV2rFnAnFKktrPgKQwLFE1F6dI8hPU9riVID9KobeC7CUrSlLPxVML4eZU+cezJYUJyao88ZJc7rgEVT74ShAL2CAcMIEctiwwHeQCUUdvUy/8pRqJBFwgBTlAAFzUmuEZacoRMbwmgRLwByQBkI3MC1OOCkAR1H8c0aquLiBbOVqknJEHnkAuADEgH/6WK2eJR6Klgt+gRvSP6FzYeDDffNgU4/9eP6z9rGFBTaxaIx+OyNQetiRGEMOJ0cRIoiNuggfjgXgsvIbC5oH74f7D6/hsT3hC6CQ8ItwgdBHuTBOVSr/Kcjzogv4j1bXI+rIWuB306Y2H4UHQO/SMM3AT4IJ7wTgsPARG9oZatjpvRVWYX/n+2wq++DfUdmQ3Mko2JIeSHb6eqeWk5T3iRVHrL+ujyjVrpN7skZGv47O/qD4f9jFfW2JLsEPYWewkdh5rxZoAEzuBNWOXsGMKHtldvyl313C0RGU+edCP6B/xuOqYikrK3Ordetw+qMYKBcWFihuPPV0ySyrKERYyWfDtIGByxDzX0UwPNw93ABTvGtXj6zVD+Q5BGBc+60qPABDEGRoaav2sizkKwKFl8Pa/9VnnkKV6jp+r5smlRSodrrgQ4FNCG95pxvBdZg0c4Ho8gA8IBKEgAowD8SAZpIOpsMpCuM+lYCaYAxaCMlABVoJ1oBpsAdvBbrAPHARNoBWcBD+Bi+AKuAHuwt3TDZ6DPvAGDCIIQkJoCB0xRiwQW8QZ8UD8kGAkAolFEpF0JBPJQcSIHJmDLEIqkNVINbINqUN+QI4gJ5HzSCdyB3mI9CCvkPcohlJRfdQMtUPHoH4oC41Bk9EpaA46Ay1BF6PL0Sq0Ft2LNqIn0YvoDbQLfY72YwDTxBiYJeaC+WFsLB7LwLIxKTYPK8cqsVqsAWuB//M1rAvrxd7hRJyOM3EXuIOj8RSch8/A5+HL8Gp8N96In8av4Q/xPvwTgUYwJTgTAggcwkRCDmEmoYxQSdhJOEw4A++lbsIbIpHIINoTfeG9mE7MJc4mLiNuIu4nthE7iY+J/SQSyZjkTAoixZO4pEJSGWkDaS/pBOkqqZs0oKGpYaHhoRGpkaEh1ijVqNTYo3Fc46rGU41Bsg7ZlhxAjifzybPIK8g7yC3ky+Ru8iBFl2JPCaIkU3IpCylVlAbKGco9ymtNTU0rTX/NCZoizQWaVZoHNM9pPtR8R9WjOlHZ1MlUOXU5dRe1jXqH+ppGo9nRQmkZtELaclod7RTtAW1Ai67lqsXR4mvN16rRatS6qvVCm6xtq83Snqpdol2pfUj7snavDlnHToetw9WZp1Ojc0Tnlk6/Ll3XXTdet0B3me4e3fO6z/RIenZ6EXp8vcV62/VO6T2mY3RrOpvOoy+i76CfoXfrE/Xt9Tn6ufoV+vv0O/T7DPQMvAxSDYoNagyOGXQxMIYdg8PIZ6xgHGTcZLw3NDNkGQoMlxo2GF41fGs0yijUSGBUbrTf6IbRe2OmcYRxnvEq4ybj+ya4iZPJBJOZJptNzpj0jtIfFTiKN6p81MFRv5iipk6miaazTbebXjLtNzM3izKTmG0wO2XWa84wDzXPNV9rfty8x4JuEWwhslhrccLid6YBk8XMZ1YxTzP7LE0toy3lltssOywHreytUqxKrfZb3bemWPtZZ1uvtW637rOxsBlvM8em3uYXW7Ktn63Qdr3tWdu3dvZ2aXbf2jXZPbM3sufYl9jX299zoDmEOMxwqHW47kh09HPMc9zkeMUJdfJ2EjrVOF12Rp19nEXOm5w7RxNG+48Wj64dfcuF6sJyKXKpd3noynCNdS11bXJ9McZmTMaYVWPOjvnk5u2W77bD7a67nvs491L3FvdXHk4ePI8aj+ueNM9Iz/mezZ4vvZy9BF6bvW57073He3/r3e790cfXR+rT4NPja+Ob6bvR95afvl+C3zK/c/4E/zD/+f6t/u8CfAIKAw4G/BnoEpgXuCfw2Vj7sYKxO8Y+DrIK4gZtC+oKZgZnBm8N7gqxDOGG1IY8CrUO5YfuDH3KcmTlsvayXoS5hUnDDoe9ZQew57LbwrHwqPDy8I4IvYiUiOqIB5FWkTmR9ZF9Ud5Rs6PaognRMdGrom9xzDg8Th2nb5zvuLnjTsdQY5JiqmMexTrFSmNbxqPjx41fM/5enG2cOK4pHsRz4tfE30+wT5iRcHQCcULChJoJTxLdE+cknk2iJ01L2pP0JjkseUXy3RSHFHlKe6p26uTUutS3aeFpq9O6Jo6ZOHfixXSTdFF6cwYpIzVjZ0b/pIhJ6yZ1T/aeXDb55hT7KcVTzk81mZo/9dg07WncaYcyCZlpmXsyP3DjubXc/ixO1sasPh6bt573nB/KX8vvEQQJVgueZgdlr85+lhOUsyanRxgirBT2itiiatHL3OjcLblv8+LzduUN5afl7y/QKMgsOCLWE+eJT083n148vVPiLCmTdM0ImLFuRp80RrpThsimyJoL9eFH/SW5g/wb+cOi4KKaooGZqTMPFesWi4svzXKatXTW05LIku9n47N5s9vnWM5ZOOfhXNbcbfOQeVnz2udbz188v3tB1ILdCykL8xb+XOpWurr0r0Vpi1oWmy1esPjxN1Hf1JdplUnLbn0b+O2WJfgS0ZKOpZ5LNyz9VM4vv1DhVlFZ8WEZb9mF79y/q/puaHn28o4VPis2rySuFK+8uSpk1e7VuqtLVj9eM35N41rm2vK1f62btu58pVfllvWU9fL1XVWxVc0bbDas3PChWlh9oyasZv9G041LN77dxN90dXPo5oYtZlsqtrzfKtp6e1vUtsZau9rK7cTtRduf7EjdcfZ7v+/rdprsrNj5cZd4V9fuxN2n63zr6vaY7llRj9bL63v2Tt57ZV/4vuYGl4Zt+xn7Kw6AA/IDv/+Q+cPNgzEH2w/5HWr40fbHjYfph8sbkcZZjX1Nwqau5vTmziPjjrS3BLYcPup6dFerZWvNMYNjK45Tji8+PnSi5ER/m6St92TOycft09rvnpp46vrpCac7zsScOfdT5E+nzrLOnjgXdK71fMD5Ixf8LjRd9LnYeMn70uGfvX8+3OHT0XjZ93LzFf8rLZ1jO49fDbl68lr4tZ+uc65fvBF3o/Nmys3btybf6rrNv/3sTv6dl78U/TJ4d8E9wr3y+zr3Kx+YPqj91fHX/V0+Xccehj+89Cjp0d3HvMfPf5P99qF78RPak8qnFk/rnnk8a+2J7Lny+6Tfu59Lng/2lv2h+8fGFw4vfvwz9M9LfRP7ul9KXw69Wvba+PWuv7z+au9P6H/wpuDN4NvyAeOB3e/83p19n/b+6eDMD6QPVR8dP7Z8ivl0b6hgaEjClXKVnwIYbGh2NgCvdgFASweADs9tlEmqs6BSENX5VUngP7HqvKgUHwAaYKf4jGe3AXAANrs25VEFKD7hk0MB6uk50tQiy/b0UPmiwpMQYWBo6LUZAKQWAD5Kh4YGNw0NfdwBk70DQNsM1RlUIUR4ZtjqpaCrjOIF4CtRnU+/WOPXPVBkoJz+t/5fpM6PWp0rMUkAAAA4ZVhJZk1NACoAAAAIAAGHaQAEAAAAAQAAABoAAAAAAAKgAgAEAAAAAQAAAVKgAwAEAAAAAQAAAL4AAAAAvqIx7QAAQABJREFUeAHsvWeUXcd171m3c05oNBoZIAIBEswEJYpRFCmJFBUt07L8LMu2xmFsz3jeevNmzZq1Zr7Nh/kw857X8rOXbMt+tpIlipIoiZSYRZFizgkEkTPQOefu+f92nX3vubdvNxpgA6Sk3sDtc06dOnXq1Dn1r51qV2b7FTd1zWSmWzIzmZGwREstoBaYmpysblveEtavaQ8TE5OhvLws9PSPhP0HDoSQKf2V/054vtKyspFV7a3VlZXVYf/Bw/ZM45MT1R/csXJk7erl1UODI7/yz3muPubJycls0WVlZbafTsue/DXYmZienvcpSmZmqktLS0fUCjPV5JzJxO28Vy2d/I1ogakwExoaGg1EJyYnAr+66vJQWVkVBoaHq8vLy/lifoXaIhNKVNvxsTGr8/LWZaG5sba6f3AkHDx0lLTqyqoqDSAzoaSkJPaHkoxtS/SYmZKMXbeQPzMzuoCmSV0yM51rq5mkrEzSQTMlqhmXzNBhuSiXt9j9KD+TSRWeZLL7UoLO+X6x60+X5lUt9sicm5mZsmpmMqVW1JSeg7w8B+dI59jLSd/Pz7OlTbxducbKVWbfT58v3E+Xmd6fVpklujnlpts8nYd9zk9NRoAkf5oow2lK76Q0w5dTnDiv1gjT05kQh5Pi+ZZSf0NbwDr59Lg40WoDUZoBrhQaHR3TvjqL/k2fptPbBe/pnwigYxMTYVpcVEtzU1jR1hbGxsRdHzoWhkdGQ011lTpWaZgCoKxbxApPq5OUqBN5Z/fHmAukHNwMyArapaRUIKMOmgNLAZKAJ5PqtLF8OjEdO9eZ/b5s57q354l1mPt6zzfftgBX8rLauQRA01Wfmo4A6uDKRYXAyDkHSc6n29Xz+nUOgun8dg2AGybtWs9DupODYrFznoftQkCUfA6iXm4aZP08YAotAak1w9IfbwF1b9s9crwj1NXWCmiMMQtHT3SEpsY6pVWHUx3dYXJ6IlRUVgpMoeId3069R394jhl18JGx8VBTUxPa16wMpaVl4djxE6Gvvz9UVFSGuro6ASjDQXyKd1NVQM7B1MuJHVrppZF7iyDpZ+M21+nzOaP8XPGosPx0nhINBgA1v4wGgAja6RyLu++AC6AWAuhU6nNIg+SZ1MC4SrhfkYNuuiw7n0bzJF+uPe3Son8ARkCxEBiLZlbifPkAW76eJSCdq/V+U9MFCADk8PBIeH3X3tDYUB/GxdF19/SGqsoKcXStYcP6NaG3ry90dHaHEunIqioqjKN7fwBq5MhGpOKkvuvXrjLQpL7d3b32Vqs1OACfzk1EgEv1fuWCGy1GaTBLg2cxbtE5RD9nnT9VKOcd+PIZURfP8+uUulS7+ZxnvIfEWpUp/tBUGfn5T39k1ybg5EBZ7Ko0fjm4pcG0+DWqUyb3POn8XoZflwZJB9H0Od9/N1sHU8pwjpP9+UCT84XEN8SzLAFpYcssHSOXGZgiEp841RFKJYYCPlNT0zLMHDFwbV/RFurrag1M+/oHQlVVdSgrE1cknZlURuednJMGQKlve9vyUF9fpwFhWFzoEVNRVEsPqq/eQPRcV9BAUmAdwVIAojZxvjdPbTAtjiZpL8CLPAbhOcwpUtW5GjheZPzvu9SV+k0BzTSocgzwpYHQ8863zUgk12e1YEqD6UIvWgg3mi4rDaDp9LPZXwLSs2m1X+NrvFPDXZpRyQxLdHBE1BITh0dHx8M7e/ebznHNyuXaNoaTpzoNtOACSzRCk/98EeLVxHg0iqEHXdbSHMbHx8LhI0fDqER76lQmfW+s05nVy7lJniULism+HSfsWZpzIt2J6x1APc10rxpwTE8KSpFHnI1kWMti+Q29BPqki0pnYpledrpeliFp7ynVpzSNfPFkXt2TJNukgZIy05e67tPzcwyVSo0AGaiKFwMkz4QA4UIu1K/3cwbUJdEo5efe7RZusxh4prnQ3PevAeQ0n0pOolkS7d/tu/k1v77wS0IcnhEoldmvr68/8GtfsTysXbPagPTEyQ61yWQok3ogUmEZi91kGbsvelBUDtCJk6fCwOBQKC8rN/2occlnCOyliMh61jQBNA5knp4GUNLSAJfNW1CO5cPYpOKnsd4LveZSJWTTDVCjyJ8t1yuR2gKirh/1fNQpXa9UdgM02aazAJoGVsDMgRW9a01NlTw5ZLibioBqnKkKK5PuGZYbC74T3GFh2/i5+baUybUYlRZCZ8K5FgPRhdyjMA8AyuDtxiieeokjLWylpePTt0ACDNHYNB0OHz0eagVk+J5esHF96OruMZ0qIjZ5jHRNerQ//U1OnwNxHlHerfGdXZ2mauC+ACsAyu9sKA2i6RLcbJQu0/MCvsWouLa1WM5cmov/uZSo/4wgWfw+nhcAnQ88PR9bwHJ6SmI3AAyKQgJGvDQyUuWMyU2oulIDp3Thhw8fDmPDvaGsoipUlJVoW5dkL1V710n9I9WJCJ9SJIK0ddwBz7YF7k920Xv4B4B1rhQu9HTfKQDqYOp5l4D0PXyBv6q39o8H8R+NHtbvycmpcPDwMelPGwSoy2ThrzdQgzPEQl4qtUDMvzhPDYhOarIAelpcmpgsgBgfDUlycQHs58ebBVUkDaJc4KBZeHExEDUukCZK14NjiDR+fkzaHBR9TmMhcIZQBEv2ihfgYEqONHl6mvMEJMelGgFQmqSmgZA0IM1YCN1dXeHIvrdDX3dHaABg65vtnP8ZHJ0MdVUC3+raUN/YEOobmqVOqQpVCbDivzvOxI7Ee8HFdr8+vfVzDuoOwOk8vs+5c0FzifTO0Tro5vrBEkd6Lt7Db1iZUdwHKAGxwcFBcy9a3toikb9N3OJYOH6yyzjHSnUsAHCx9KfT0tmhn4UTBUSdC80DrgW/jdgpp2T8gWZzhAsrCKBK0yyROv909IlMX5DsZ0V6HRf6nObKLA6iufOzC/ZzYBA6z/LyCgPlEqFCmRDk4L7d4YR0y4dO9YeWhprQTPsePRB6+obC+tZGA1EAdWQ4Tm7wO3QKKKfH4rBTUlnChIfQunpDWLV2fVi2rDXLpXr+ubYu2vt5B1Q/9i0g6ty0p7GdD3jT+XzfQdGP59um88KVpgfZJY50vpZbOrfgFgAcGcld3MfVqKu3P6wUd7pJ4n5PT4841C4rr0LuUuoFQogIBOmRfcE3TDJqep64qUmz1J/ptQvJP9tpPnKDgHUhaBaW56CVTdfjprkoO680Nw6lud1CIDfRWwX5jCjtZYst3HGuk/TCOk4ixosdZSZXnTjIiYmpcOrkyXBSng3vvPlG6OqZCpMqukpANT1UEjqVDkACjgc7+8L00Z4wOS6vjvrSUCXudFQAWkhDw9NhaHggHDn+mpUJqF546eWhbeX6MK7ZY+iFMUimQbNE6oT0cWGZhcc8g+tR0+A5F/D69biGQbq7J+VtC7nOvJNFDmb08SIkLAFpkcZZSjq7FoiAyAcq7sZ0o9Ph6PGToRtAXdGa1Z92dnWHSon7GK0cTM/ujkH+rAOmSsDJHst9eYWmrxpAF+8op7tPaUmaz8jlLgSk3JnT7BXgXV5HLziXLinNkabT4U5d9IzjUPHnTIOpX49FH3Lx/e23Xg9HDh8PU8M9Ar0BO7e6rTJU11QaxwlIjg9MhYlSVVTgmCUdTwCWYTyblN6pliIZ4IUA4a5TA+HpXzwZtlzUG7Zuu9h0kGMa/DRBzsiAMHG+jynn5q+DKKWzD5g6cBbekfQ0B1p4nuOof48PUbp85fr/Q2UyeXqJllpgkVsgEyrkPoX/KeDJFpG/uakhjGiqKYYiZuTYfPMzvDMWb7jRAakS6uswdLWGvoFBTWEdFUCXmwqhOMTMdSOpKKTnXb2iRXre6oAPLWDEfWyLLAyxKQKAaaA1sOTmRfI5l2r5k7KAKK8rnCjnotdA3NfpPDLtga5V7ZTuV+Zl0VgS02lf8gCiGINqaqrNVe2hhx4Nzzy3K5RNDYbqqoqwpqU2VNaUS+8sjlS/4cHx0D+sMsoyAWAUIynxXHgKR6mtmir7oybCdzs/PiE+USNqVaV+Giin1IAZlQF1d3SEnt6+0CqddpN0qVZ12lgsnYFS8UeJXPwc56zg+c5ZhvhnWm2Sfk8R6lMZtAt4eh6aEEBNmjI/o47gibER6BEml4B0VvMsJSxmC/CpAZQA6rCMDr2aIVUhvVzrshYDQBzmxwSqnAcYFtgnFGRHH7K+YLjarq4eu37VynZxumWhX4aSKbnoYEShTMAwccOc99EA0rXtzQakk+Juuc4JYKIs/p+WyFMsn9KoMz3P6q9scWZMLjMA6kclyT7H6V/+/a2FC26oMrhWv0ndiDGgvr4mjCq2wOuvvBQeeuTJMDEwEFobIjfVvmp16B8atGIdREcElPV1vDe5l0mU79dvRAAJUzsjBJpSmYCrcETow6XUMAIPW8T7CVn8uR6uFDBle+JkXzgpnXZDbak433q9J3G9yme2SC4sQgZsqXdRJMuCkmhPquu/YhcBmmkgnQtE7Vo1rIoUZZaAtFhjLqUtfgvQD+AgmVLap+maWIXhjtpXtMsPscSmnMKVcN475cJqAZiWWnmDgwPidhvlkN+iDj9jaYjIJbrvQvphGkhRE7iFPFePBEyTBJvemQBIej+XP9mLGGMHhfXwTuvXZME7dY2fK7bl+sIyvP3w66xQ29Q1tYdDB94J9979w7DnwMmwvD4TGuA+dR795djIsBXd2LI8DEpF0i8UXdZYYeePdStegThQAKWqQtcJGCsl2vOrEjiWANQJt86rgzNVsZYGdyrG3kT5kZHo/sa5rt6hcOzgIaHxcKhvaQ01VYrZwIk5yJ6vsOHmyFssOc1lFjvvaYj7GT2ot+e8IKqLcoPzEpB6Gy5tz2MLwH3S+bpkkIIjxfLeumyZgd+gxHMs+4AusLUQojMzC4u+eKqj0yzEiPpwvUNSH8Rpo6VmkJqvPxYC6envna5fMUBLSkhlm+/+5IbDQWdYCJB+7J3c8ib54iiRLjk6xyPKV9ctD8//8oFw/89+KYNIRgBZatwicnnLsroAePaKo9/YtiJ09HQqOI3ULTWanTYzGQBRqEp1am8pDctqpQHUdRmxopUAarU4Vt0c3TgA6ioAB1O4V36oAXjfnl6h/QFxuCe6esPMxEho0ky0xqYmYEz59RLTj0IFCo9JOwNKtxmXpYGVfQCTHyAK+fF8Yr3ly77XJSC1hlv6c35bwD5XIUaVDFL4guLADzG1E/9TAqYMCWArNHcfjjJ+3nPXEc6APKgQKJOIT6gQEO3bV6wwcX9oaFgAO2FpcwF0GkjHVQaghghLuXgl2H2SY06SZvpdQz9mQqmTqh78hBU5Su2zy8/ETK4DadgmRLpb89MAkN73vPlb1S+pJ4BQ31BrvrsP/fRH4Zcv7A7NQsPlzRVhXOwlQLh8ZUNY07wsHDp61IxLYWoivH2wT6CnmWt6gq4hZu+E0CBd54plMgxKasD4VE1c2mr5jFYoUIfS0IEivk+gG9WjAKblcKX2SJFb5REdSNNgOqML+rr7w9jA8ZBBtSPLPfEa7Fl1zbkigHLavitUSfFroH7cl6HD37NOGc0FqJZPo7cklyXR/ly9rKVyT98CfMTMQoKbHBbQdQpQAT+4SeKEDirw8phmyGA8ivCTA5y5SrcyTYUQxf1+cbiNApVl4k4R9wd0TP+o0H3y+6o6kawpriPFQZ2ORWdh6z/K55+dS+1TH2XL/dIVLFZtbq6f38OzA6Skm042hcbkK0bRC0A1AkTFzVXLG6K6utK8JR555Odh954joVlgWFtTYqI6uspSWdQxMJ1EIpBRiaIPnZINXmAIwUGiPa1prBQXGvWopNfXV5shqksupAN94zLIyaJv4rtAVOK/G5oiiAqsNKIYMAuREfvJA5DC2JKH+wCynUPj4fiBw2Gk95i8PSpU/3qpYwTcGA65cJHIuU+K4x1aQ+svLevtyyAYVTq54RbgLUZ8G7CvS0BarHWW0s5rCxhQ6UPV0h+aGy/9qSJJDQwMhUbFPl0hbhKuZ1CGEeZ3lwkg+eDRI9o3zJ95CBUCgITHANMWAWi43hEZXVApEHwDII+dan4g9duQt5T789O13gH9vG8N96yHesrsLXWzoM+pU/ZI+lNYbuRyZz8w4rABgm5IOEPosYcfDg88+qym6Q5KB6opuuLyJsSJwjliSUc3Otw/Frrl2gSgjYxNaUKDHkiED6mwLixrFscpF6WJ8anQ0FRjU0ZxkTrRNymPi6lQKZAHFLluCIDU5WUCTICTY1i7CulUywTcEaBj3QFX7gmwA1CAacYAV1b9Xk3e0GSA1Wtadc9lGlyZFVeRJ4pTx7MluFA1bPK+U6XYN4BaIbZBGmA9FwOVX2ucaHyc5D0tifbeTkvb97AF7MPU/dnCfcpXO3QoePSIOBJcpTAeTajH9QtQ3XhEdck/H9EtALtKdUY4zA5NdQRc25c3h9raGgNsON4KBTfBmj4pUXNte6NZ7SNHqs6uewCe/o/7USZ6XMDSRXS2DvCkz0tWMfplzOjdl2sKL7WBQ/nIw4/7AKpJEdqL5eClUCP/T0D0xTcOmoN4vUBsWNM3pwV8jYmFHtEeKzxiOKAGPsApwhkCohCcKMBLnsYGGZ3EeR48OhwGlQ99aVut9KNiFAdHpQYQeNYkgElZPAGgSlm4SQGk/HCbKlPdq9VI0wJhOFmorE7qHQG506jq1C/pZIX8juvqGmTRlzpGg+likPOYqGFpejhPfwex/Fgn50jt3Stf/M6iFBJbvrA2S0Ba2CJLx++LFojg5/pT/E+Zvw+osjxIvv+pvvTTEN0D4xUqBAI892iCAB4Dy1UmU1vhgslDB3MgZdE/QCEN1lFfywwu71Rc452PHpdU5HRV0nkHSK7wy9jnUj+X7uSeByD1fb8NkxAA0RefezY8LX0osKOYIkIycYUCOjhDuFEIEIUb5BhDEfpSfEXhDLmE/IAt4IflvksiPFwoFvpl1fo1VxqH2t0/bnlwkSpTG1AM3CnNAbga2tsdI+dKuYBpXCpJg4HqZkCuaarci/v7jKqTsuqXzYyFzVu22CSLdDtQZNpYZLco8iedx0V6BkNr20Qfmr4sFzHLQdtbOS9X+iC1/z4HUkY2+7BU5fQHnXqC981uuq6/KnV+3zRekYrA3eG2BPgBnDhy43+KQ3+13GUwHhFhCF0n7X2674PyIMrDeIDHAOI+7lJ4DBBYo0cuWResbTWOdBSHfLFYdD4nOiHH3tXS6ZbNX7xOCCcsiaudi2SbJuvUKWDknBVRkA8VgNfCt+S1svUsWOYP7Hk1PPjwswZoFUkFa6QbhUvEADQjkRsQhXB9gsvDzQk1BRwid6ZdILboMTkPOArnQqNAtKYuqg4GNc9+YFD6WKXXKg3DU01VqQBb3Kws+Tjjs0VSzgKm6gGYIvZPqHHoLwC8ccMAvvbhYJkrxf2OneoLy1uqQvvK1QLT/BlUhcBqlS74w709H/sQ30guLb4hP46tmTtPejyXXByLmOPvOQZSxB9GcdU/+yHMUZNZyRbrTy0wrkZEbIHdNh3ZWZQ1q/AFJHD/hdbb8qrh4aDonLw4QMA5mAXc7rxnoc6Ru1nIh3Leq2cfPTXjh2+p+58ODo0I6KLxKMtNqrdG/9O560knsh9Z9Ozl6vX4irrHAPrTcukSV7TIWCOgntQ3l4OvWK4fUw5fR66zJfdVunfMdKv6vgFpcpA35z65nA3fnBH5/Efdk+T0BjAiRuiRQ/vC93/wkJ2qE1ghQvNDNwm3h5sSQAr32STZHJ3oiOR0QBRiC8Pqv2oBIfPpSWemJ79aAZ1f16sZT8018gKQ5Z9we/wQ/+FkuQ+WffSwcL1TOo4ifwLoajwgDMke7rRKeRxsHUx1OwNU3vWWTRt0vQxpZDoDog/6L3uZV8QSlCG79f3c+0tOZt9xGlRzKgG/7pwAqV4AAQnUXL29A2FQriyIY1jfZltKvbq5LR18WqJcV0+f9Dtj2VESl5a+PvkY8mLVCfwRclcuzh4dhA7T0aHgDBI76iUCpoNJpO9CXalHr0TFfhlIqBsvfEyA2t8/aJ2xUu44Md+5qnG6RqffTz8fsizuQryrVBc+fSHvUQ43HrF+FNGe2rSsMsajMQ22GI9y1v2FVdDFfSz5TBBo0NIkq1fUm7sU+lJTes5RFO80jxIQRdSHg1QPzJ4GA2xwVUoaQMlAB01/GXaVJ3CQFONJXANhoWchwl71k3/91r0mUjdKtzkqGXlC4AWQYnDH33NiRAONAHZ5nTh8bWuUVlelgZS+BJCJA+U8YMkx7kvmJ6p9tlUNuJRFPSb5qpUGdwp4dp8cDkMyXI2Jex2SbG4We9Whe2A6DIibnVQ9MrrUDE4Jp8vyxfiS8tmhE81oNlqJ2o12ghtGPUDdBgZGQp0Au619lXlUxCdf2F/aO02Uq6FE78Gn3RZkSDLb+8jmyZXgon9M4dr09ZnJxfMtsIKlexEnRgzKoZGxsPWCdWH5ssbA+uH75N7QIbBZvqzZPir+WKPn6mqA0yt91ZiWstixfVO47OKtYf2adn18pdZR3nx7X3jmhdc0V7grtCqIMKCwWCHZYjUiV8m0w499+IOhQz5uLKlRKXeS7Bed1JfnxN+xt38oXLBhdbjm8u1h3ZpVCaczHvYeOBKef+Utxeg8LmdjdU6JlPpOROkXkBR2HjfMIce6y/OdkEHn4KGjScSm97ZeC2kC89uUSEq4vlGBZ3a5k9WrLboU60sRqm+hRHlwdb4e1QHFU924qi60NNWGEakNIF/iI5aZaiMHuKTHCi4SrinmYZKAk2GGDrNz8DlBNtIKerx9I0qnXvF78VLilimklEPQl/blK8Jjj79gZSDOT8qVCeGbMUB8S6iHixSI1UsGb5W+s0FtV1EXV4UdV5/sF8NCGsT+sDjKYiT/fLVRzMf54XHl1b0gyuYecLAYkkYEpj0CUP23ZyyD9RT4ojc1zwBxu06mSxXoT4wRvQujlJ9RHtJ1ePidty0UX0N9o7lDlUr5CycbATHWqXDeGjrRNLbAQcZhQE2eDIBs88Ex3tvTfOs1Ot02W/XTZTz9+RhoARAd14j0u5/9mNjy9dnL8BO8+8cPhwMClgimtHSOGOEB0TI16Bd/986wft0aOwmngPW2Rbqsa6+5PFx12fbwk4eeNGCOAEe23MvJ7Xv5nCvc9/yk+3l0X3rZektVGq251zt7D4aXX9+VACn3iURdfbD4zO03hssvvchO8Ixd3d2hQYGOL9EgwO/lV98Mjz/1sgXBQD+XT14PT6XrFKb5ufTW65xO831/pnQ58fl5vkkBULmix/N8u97eExicCKmmryopwMv26xdaJ7//ud5G/pm2LNWA0NUzYNzk1k3rLL4m4fti5Cl/nvnrEzucuBQBSqWCES+UAEAX47km17G93fJLAhizXKr27XredfHsWSBwiRYODQJE4Wora5aFp597Prz+2stmoRcOCkDFiSagVS4govBlAsB2cY/1YmgqhVTlkkCMxMlvlOkdFYfTwMCwQE3cpdRTY+L4K+XT6cQx5ADs6b51EAZkRzSLAWAd0WACOFKXMoE69QF0SUM3CqFPBWBH9Vx1Oo8hjHn6cKq02cGOsdC++81wyVXXSn0jSVTLgKcbjXbXpcbJUh4gCjlgsu+gmE4jfTFpUYEULgEQ/YPP3xZWrV5rIPLqW3vD6vbWcOOHdoYv3fWJ8NV/vcespLVaM12PaM8CMCGaMZPly1/4pDlPP/fim+G5l1+XUQFH4SnpW0rD2tUrw+c+8eHwsZs/GP7x6yckTowZyHE9wR9GBGSTakjAuFruLTGdsci/VmZrlOolSok+OpLKq1kgKsOXpfBwY8Ua2sBWzzmgEf33Pn+7DRYA7qNPPGfP5XVt1pS3Oz96Xbj8km3iTN+WsaTXDB2xzKj+oL1Gpb6AyvWRVFVrap729Wnor9eZAV3OyXPmjW1IGVxD3iEZAyYm44dfJ9Bktgj6QHzhMiVMpYzjM0ElIDo1z8XR6Mi4XUsbwvVgEZ6yEGe5+thF7/mfGFC6VtbqMQ20Bw6fCGv0ffRJQoi83Lmvbz43mdwvfdv0q0naK4qO8YSDMYABF+lgmW7awjRAFIf7R+Tm9NwLL5n+MmEq4ypHgJbAdIWmgtZoBhIc58p1q0JLS5NcjuI3Ua04pJWSuhTrLn2r0NYW0XpcfQP1FEQ+9pkVxvUYfkbFQQ6qXxYSYOtA68DaJbWCupsRRi8IkDWLUjy0+sKBGseqr9AmD+jbHKIvK+srr+wLTcuXhws2b5fXxajUEDIw0l/V522AUjlmmaex1D4EfM57DSaqJyNRck82XkYqqSgA+/li+f3cIgKpnlhUK+6msqo2vPbG7vCtex6QVa8mvPTKm5rD2xm+9DufDJds3xwefPxZ+YjpUfWAEDpIgOm37rzFQPS+B34eHnzs6dCmRdUAgmr5rU0LTN/YtUcvUHNz5aw9oReLSMaHCSeLu8oq8ktvRJ6THV1q8JLQpHXZo44zgmi3dErQiuXLLC8zXzo05xdupKGuSi9g1M7P9Yf79Un3e8MHLzMQBfDvvveB0NBQZzo26orC/ZgWgfv2PT+zZTiIGs9yHIB9BCzpYDW/ubqyXIPMcg0S5XKe7tPMnl59/DX6kCqVVxZVfRBsOVelvOtWtZu7DgYS/CybZHSBm2TNHT4uwLJbz1NfXxtWtrUbh3b0BIvR6bvVB1aW4j7Sz4cFl5VBe3VtS4umDja1avCa0DvrUHDfYXM7iuDOxxjfc/r693Kfd4vTdjRKTpp+k1iXarp3R3qHegEFZXDM86fTpRNN0M48A07TPCbeU4K+I6MkP5bqwqaFW0YXar6znk/AVlLeFI4dO2SXE4AZbg4CdKANTZELZd9BlH0H0AotnT0XleshymoaQoVxfjGXlmgyAmAh4hcQTcqt6QArVCfmaEIMTKv24WoHuvr0PedUBnCrENypeKAs+ewntn3yTZWvlfxLdVoO+rhEjWmUeeb518OatReo7zeGIfUngDNpwahXTdqAQgkQvRByTjWdNxc4W+X4DZIMRfMboKvN0oW82330gPj8fe2bP9TDlYhDaDcLKOWyXC9Up4ZOE67NxJFcv3alicIHDx0JP//lSxYEGE4K8IHQW65VeXB2HQISABK9Cnq+ZdJBfkGqBKYBOnVpNsu3fviAARaqBPUK22+XbvV3PvNRzZxRPMSEWHXyu/c+YoBaIyCej+AMAUU4bNQOP3nwcfk4NtvaQeOy9BpXqA7IfYbEbVLfRtV1EtlLr56/3dK9XrR1Y/j07R82i6ffD1H7ez95zMQXrgEQCMKx88pLw+0f+VBeXlQG9z7wRKiV7Me6ReQlgDIA/5GbrvUiFWKuO3z7+z8Td3yD6tIffnD/4zZgeAYGG+rJbKJPffT6rJqC87gEfffeh8NerWVPG/q78Gvf+2380pmXv6KN7iv9nfTWZyLaL8YzONgBfETUz4LkAgp3rvR0WZEiwIq6mhjJ6eTJLuNGDYRSF2NN37IehkKgpvdngCdOEhCt1yBdLkvSTCZfxTSZAs0JoUe5bOayJ4k5iX3Pi6+Q1GSkLjKmSQwsUwKXGj0gIrdbhae+qFzgiiqhQgNx0DJQnVqqRLyoPqqkTDHDPbL+m57UrlAfV79BohyU8aoOw5fqAESzPdk5GJ578a1w9RXbktxRnDd9aAKiAN18XKOfzxbwLne4l9OiAinKe1xS4HzKJYpPjk3I2BIB9LO332z3fFtisE3LEycB8dHBTa5b3WbHT2nkgYslrmQEJktmmDGRPKoEYhqcJFzoV/7Dp1VQSXjq2ZeNi4LLu/aay8Jf/tEXwj/+2/cNeLlig8Aarpj7Pf7k8+GUuMKY9/Lwp1/6bPinb95rhqz6VSuSm+Zv4CaHxe1edOEFBmrPv/yGZWCtoHFxhXCQqCjgrgEonhMC7NknyhGc6NWXXhju+OhNBsRPPveG9FEjYfOGNTaQ/LGczr9x9/0GbnC+N193lQEjgIihbVicI0Y49LK0xbcEkiwuhw/kLddfHW687mpbjvjZl3ZZrMcbP3h5+Is//h1rn5GRPZZmlUr+0BZ4Sdz1yVvCtgs3m174FUkTrAp56w1Xht/77TvCN757n0TnY1b/9wuY8i4wnE2L8wFEW7Uu0AGFZnPuMP2MZ7U/ixv1UtSr56A8PegceUjOVwnkZywEVrhRCKxokATSeeJA+NZ3fmggWgIbK2MOXBvcKEC0tkWxXlsaQ01tXOGzTICHeA4XWia1Tux1uXsCoqQ7ZWYiIHIM6BYS4JoFVJ2kfMR+B9V0fkAVnSxqAC33FMZLo4ELsZ8AJUqRZT9e4Vwpz8HzRDE/PpeXuWfvgbBt6xqTRKkZIGpW/gRIyVeMa0ynLzaYUja0qECq3iqOixBb5cZlXnbx9nDdzottDnWNONHv/+SR8MrrssKtXGHiKEohdy3ClQVCLEfkjRycJdmfnBUuWlr9PHpIQPTv/vm76khHTcTmHi+99nb4HwUgn73jpvDVr//ARNvP3/kRtfR0+LuvfUeA1i2jUpVGuTfCG2/vD1/5/c+GOz5ybfhvX7tbaoMoquTuHvcAfWI8LhPYQSdO9Ui0lgLcBoWom1yjZ8NTQWNIHgGWu/YcMM4OEIUL/vt/ucfy4If39HOvhpu0rDHnbrx2p6kLLpbnAtwlXPo/f+vH2bzU+RO3XW+gefO1V4orfkIf2EY7Ju8/6nnhEgDIF6VWYfDA8Oc60XTFesTd3yadMyDKQPSdH/xM3HqjzUd/6dU3wn/+qy+Lm71eZX7fVAdICbl3kS7p/OyjA0aVgTscA1j7mpUavMvCgUOHTbzH3Yxv8N0S7jhpjoPyiHSUpSKiv4YkE8PJA5fqJGHcd1Nb6XgFGAClD07sx7on99FCfIzFGJx8qePHn3jSyjAxHou4yPa1xWreqviiTs2ydAtd7RCwBCSN41S+mUzkLqvE8Ewl+nQyEvMAIoDLfJQGU8/noMpxPrBKBaVyWZe0QUxGkO6WIKczqtqIdjEs+TNwbXqfY0gaM0lUPaaCYjE9ZqjZt5AC0fm40VjK7L8WA1ffz1wAzBWsy6QWEczofaQ+AUT/eJ3UZrOLfjcpubvQiYWH0bk50TY3yPXCHaER1fHXVFWK3jD3Gc4+zQMRxHfD2lW2UiXcJSC6Yf1qA7Wy0qawd/8R4zrh0FADVMkCCZijfwVEURMAxnCJGIvQde688iIrA65yPgL4IFZhhEyUE5iiC922+co88dgy6A9iMqCGmxR0/8NP2XaVDHEYt/BhfOr518IHrrokXK16oDLYvmWT5fnJg08ah4u6gLyoHx554vlw2Y6t4erLL9IA9XDYpGeHHvz5cwYsnhf96kPSSQOk5TZv0LJl/8DNXrwt3uexp140vXSDys9kms116/mXXjcLPzOKDh89ae3luu1sIedlR76Ouo8ZJVXn9Xr31J0O1dHZrWcT56WB0QyGfPfnmubgWAE9wG8+ovNZ/1cZacAtdo2Xh4THFNB9++ROJ04UvSj6UQgAhTa2R+MROsvlrepr4ijBWuc4DSQFkIAoAWKgyYnct+4gSjr7Dqa+Dwg7jafA19NimMIcd+vpvoX9SIPpsNRkZtFXHc1disEpRWlOm+RS6Ur37t5lcWZZ2hmO9HTE9wBeFCMbKHUqj0tNDYB+TUbo6clgqoMp18XyFeDFMy/OFusvo2qwDvfy67vDL55+xcRJF1FrZCDC2GR6S4FP5DCmTX9HHSok0o+IncMY5dxqrJs3GiKzxDqJuM3NcX3tw8dOGSdKPkZ0ALKpuUEGn6hWWL6sSVPY4gh8srNXoFVragIaBfsehqIjx0+EneEi1aspvJ0YpOJ9c3/hPBHRma4Iofjed+CohlmOZuQc3mQWejwV0BWj5sAd57PyNFizehWZzNeUcxh28C919QWcHoPM7r2HwrXS9cLVNom7QPRGB9gsnS7qAwiOHzp05ISpA5rlytLW2qQqSLEvHWG6XEAXKz7nSgU2hUR7u976P//ll63O2Tx8PeKSIPSwcOPvDdGB5U6jiR0AOmL88PBg2Lf/oNWpWgAq5LePerG5ZVbvTK8rP9fzu55Un9MsKibKg4HTAAdrKhH+SGSgyjoeBcT7ZrXPQ8c6BIoalMmiSxCH65MejJX+6ku3hqamNqmKFDXJDEoz0nXqHuJEnQMFFDOZeBGSV3m5vFsSQGaQnlHejICWrQMrx2XSh3LVZMaBN+caRXXdys8+BKhCcKZpcjCNGm3OSJWg6PkoQwFTdZk8jhQwRW8K1WkAYXnoETElFTAzBUAauUPLan8cRAFM3mH625iVl29dVOT1xfQEjB1ELTH1J3kNqZR3sYuOEFcUnNTpzIhecA1TatTHnnwhbN+8xnSXT7/0hnXumip0NnrZapQDR46HG3XvSyXOviEgAmijhZsH5GMmX7kZVboEQhAcr1Ouk0eRDI7YCXcnJ8CNFzNtVlM+HBYGA/RiU6Tz+jW+BaSxnu8/dMySLrvoAnGyrxooA4QzmlM3INAb7WYOuCKRyzSJZZO1hHpldBqUjy1uXNQBAGcSYiGVJQr96dToT32ZZVVudcxdM4vD5IPRC8+ovXnh8RlpvdzzF96PY2ad4QP72JPPmZEinWdwRB1ielz+sX1aZ6fa2it9/nzsgxuA6GqpTRoaGk2Mxy2uWvXBYSwrDs/VC866kvpO6H1Wbq7d5youb+nmeeriKoPI1Or7K6ImoN96p3Vu9NShE8aFTokbK9EAi06UzwW3onWKWgWIVthE+5x4T10jIOb6AyAJValv6oPU92KHBtJRjOU4crdpcCXVwdWQjwSjCJbmUpWksI91312uSHZDVKUAHFcpV0OMTI2HevUFuind1jwYknK81nwDffiIiTCkjYhhmIvSIj77OOxPqUFLnBEQaKa6V14xrmO3QS6x/uMR48R1gDL58M6Jy0jDQC4SAXpwPljD8a9skqhqLj6at8xa56w9PoQyRPkqGHYSIgWxFkd99HsYUdANIqobVydwhmulwhiX8P/ctnmDgS8uVdAF61fJJSKOlIA5xBRNZkVBXHfsZLft49Pa29NvdTBuWKlcu3FtzNspbhOwnE30DInVGhw65NaB1ZxJA9defUk4cvSEPTsiPrOGUCUw99v0jzd90OqKoQg6KH9HqEkDTbcMT8yQoh54AwC6WzetMS6UoBqdOs8g0yhuEMNTzCuDlnw9MWYxkwrPAe5zSpw2ZNMGNZAB7LQz1ni4SThLBrRC4r30qgzUHnhLoG998vlX7ffgz5+W3vQFUzmgtiBikpC0sIhzfkwMg8aGBgPR3e/ssZgG0egYdfLntAIJt1jsHm4IKnZuvjTnhkwlNEdG+jxG85mSKg0iY8aNmkFGfbpU3CnU2FBiUzW3rWs2ldTQUK/et74TgalxoqmyJ6fEQOgHlZUq4n21jFEC0ZIZcajqW9H4Ey8APCen5Hqkn4MuKgB+iPsu8pO70LIfS4h/ayV9pjlSd5NCX9qQeO8ApkwawCUKVYVDg1vzYRQTZlGAqPuNDImTnzIjU/pexfaNqVBDTicceLE86TQf4AxEkxOuFvA0QHRKaMqW6bJOOUTzlHexHRsbNZ0o+rhbrr/KSjp4+Gg4fuKU9I8Xm54OsIRjxfk8stpygVCnR2T+2aNPG4+GceS6D16pefrDBsZMCQWU69X4n7njFonKt4RLLtpq/qmUxywdjC2AL/nYAsakY9QxX1GBDaCD8WbTxjXS952wvIfFXV5x6TYTkdGVHj503MBydjPkAAT3pwd//oy5FmEcwvDDjAvu7T+s+3d95mNZA9BLr79j+sdX5eIEffrjNxl4UQ+e78TxDrPQMxBh9IED27XnoOX93J3RTcrzdsgl6rYbr7Hrn3zmJeP6d+87bHlvl8EMIi8/2vXOWz9gacWMTZx48bXddv4PNaMMUR91AqoTVA1/8cd3hb/+0y/aeXxNzz8JLMWN4zt88tQpuz369fcC0AuffT79JiDrv7SRClVBjLbEI+S+qcKyOS6TWF4Soll7ZXNNmJZPEtZsANWXO14mZnB526qwqn2tvDiaJKXluCfKcA4SQ1NZ6bSArU5+qNKBKx8gWoxsuqbA1kT6ZEs5/it2TToNMd9/GKBwvcKLAI4UlywnnzkFNw2Yout1QxPPmCYGlWoZqZatXCfPlVHxBTkQS+djn4EKUDTOVN8/y6g4EBbm9WPUAD7AkYZeNM2JApqUAYhCbDn2chdtOWYeGyv4IXGWPONlmtGzU9M50Udeu/OycIOsy3TQe37yqOk4WDkQsR5iywiFm9Ae6Qi3bV4fdly0OVx1yYVynG8Rx7lW1v8d4WMfuc70kDj7v/jKG9bRMYBcdvEW+ZddbL6bdXJm/9DOS5X3ersfrjt0RCLv7JEB6ioZe666/OLQoo5ZL6vmLdddEW6+4RoD2X+/9yHjgtFHXqm6A1hv7N5v7hbUEm6auqJvRWx/c9fesOWCtWH7hZvCNVdeEla1NYtLXB2u3LHZ/DY3rF9jQP6te34qUUJBI6TLPNXZYw72zHj6oAxLtYres6p9WfjYLdeGKy672Ljy+x75pfSAzeG4dLzDw0OWfrXAnqhEa1Ytt7w7NJDQDg89/pxE3uWWd0ofzKUXbwtXXLLVAHGDZrR85o4Pi/ttMXVCT09PeO2tPRY6jufj+J0DR1Snbj1Ttd7ZhfJv3WAc+eYNa+XnepMp9vGC2KPBicHPlhO2t3a+/vDBKkq79Ni8Aqbm4oAPkCbf9CJVRD6Mki09HumoGWUKiqZzpztwcgxg8l0glTmhmvQf9eTsjBL4B6XzGqBmy82VAZcDYPD877zxshanUxAfpcGtEQe0VNLEhtUrwroNGxVIRMoidX4J/aY7nBaAlJRI8ZHoB/DrhhO1WT/az0wjdcGtYXwS8HCdpkZn4FLlSZop1RRc06mL+zLlCWs0AaYYIqtMIuJ+GT0TRh9WG3DjKxM8MlOl0ulruqmYqyznrUczFZjugJ4T4pjobo2aDIP3AAGfUUnh1oQkz1gDl8r+cvlr77z6SrvnlGY+4e2QDk0YdczJRdaMam31/djmse15D/yP3CfnYXl1DcRW1+n2yXklJac9S8yY/1f5F3PNpmg4IpoQ87cBgC2bN0r8XWXgB2cIqJ1Up22TQQcdZrojMOODqOXMSHpO1m1m4qxZ1WauUgBFk8CNMn78wC9Ml8eaPi3NTTYbiI6+csWysHXLRnG9G2TJX26A9K///iMFTBk2dQMgBFC/KLeoVW0t4mA3GQi2yngBJ/qNu+8z1woWDePlrBVgHTpyMhw9qUAYJurbm7EWxAhGVCiiWj0ja/+EAlzgz7pSOjzqSpnogB5/6nlzgCc8G+DMbA9mauFRcEiuTljaN4t7v0CgxfPBid5z32Pm/sXzUee33tkvp/zucKEGl1zeenkkvBB++uhToUEDQpU+ftxjXhdIYqW/UqB7wYZ1Vi5W7a/+97vDDlnm+WBffHWXDW6rV7ba8+HBgAj26pu7dX5CFvyt4tjX2Xujb9/7s8fDk8++IiU/ZgJ9ge8B8dHDoa9sb7eOSbQmiwTFF79olA+k+KjOIvobjeKkfUDUOdO4lG/shJG74QI6cFQ3paNJ0aG9NQ1IY1Zd7KmxtQFSBrxDe96RlBCt/c1NfE+1QXFINAivCs2Kp1qSSeqrfmQksKxIKRsBUUR2A0Wli681ACUv3GmmTLP6JAKbISoxas4kW9AEIIlhAOH2/FigOq1nkwVsWoAKiMflW7AXEO1N2nndEz3pmFRyDAhQmfKXlOheKrRM9aRNHUwry1W+wG+IxxGI8Qi0CIdb1rdpmuhmqcFAt3wQ1ekEMNkTUWF+s0ggb68wOZfkMTVA3vcEyM66OJsAh8q74XJdNpnZfsWNwxop55/Ok7389Dvo+xjZcRCv1ZxgdFtYB+ngPmXTRgEVlbaiJQOnKhf9MYe11jbXo5NE1MBi3dcbjUyN8uMkCo5Po0Q3S36mN1aVK5TYxJjlRdfJ9X4/ymY6KSBOtHU64wizj9ARatYHU0SjZVzLxUq3yPx3n66p5ip4eAaOOG8ffSb+pD7biufH0m46Y+lC0ZtSV8qw51Tdmc0FcQ2uKBipTJ8pDt7zUz6g3acZSRipmO1UopEffWW/VAdMBSWviyUYLZjdRF3aNQV2QmCAeI+I/n/+pz8xDhZgpP0MjATw3j7UGU8CDEqoLuDiiRxP7IBce8/zZdnTnJs/+vTNfYx6YbgDWDo6u+xm6N8hdW3bnt0frpWeWrrnD12+TpNDmqVj10yclA5srnKzQAp3Sn5Z3c24kf5e0npWnU86Xy6f2hrCwEgfSRNh744c3h+ef+xJc17Xp2KR6xHp0S+2bdgWVjVWS6JQDFVmKOk9GundoidFHAdE0XdCxpXimK9yDEDFXUIzM+IcJ8SlzkRAdhBFZQUYumWfvBxDMxOxT0wKxCfH86dWj9O5p+M9fa7+8NCgzdO3i5M/Lup7oBSSmQXVqcj8p9xBQGmDYkl3bGwI19z0MQMwONJCcn/cmJ4MXoWZ5jG8uj40Z3CbdXFegov7jPPFlSR52c/sAMDI6OVjdceAwhRJxIrGpjoDHgOUPOSP5WdBVddXKTBsbU28HtCAYhkOoLD1vEzAdFogGAN+0BFGBDCel0/SGtcxUHkBLq4l6jqWX4J4tMptCYquSPFDZmodFF+OF2BJyZ94b2GdZtYss8EDH0fikXL/KonKdHzu5XXlQtcLx3pMiXOP1zAbLE7DJD8fSSyfa/B1BegANiiXd8rKHh1V3IHK0vDnf3RXOCY3LiYVYFBz+tTHb7FdZpU5AcLqCrpeHU/vo0RcSfqdkY+gJXV1MWiJ64b8+vO5BSSZ9sm3sP/AAZNSLti4Xpx6l9qk33xIqT/Pc07IQbJI4c6NOoiSZVKAyCfugJm9TCA6H5UI+DTK62OPIMD1eFRUKXZFiaIr12u4IA5otdRX6IkrqzWDSXrHaKmfXfLpQNSu0OJ2gOeMRHxuDzmIugFqZorYunwmGrQFnpOJmV/DmwbrHKABnlN5evTYb5iliO4WfWm5jEVEmYIAUWY+OZhaov6krflEi4LKdBsW39s+0C+GSb7Xkzlda8yR/jsXiOby5EAzV//IcJ3+Wi/F9aMcLzqQAgAQYIDFORc6LgcQlmHOP+nrmc+btqA7KOVf7NwIOjyZIpOT0rXkZ7OjCFICCYmzTn59PPbOGOvheebeOqBqbnCqTDq136vYtQ6W6Wvmyk86Yd6q5fbh5NdzzIc6rIFh7z5Nobtwc/hf/vyL0iG/ZVlRdWD8Yx7/2zJe4ZZG3SKlnzG2Le2da3N/hnS+5NLzvlFwEnU6LPjEtkWtwzpOyzQd8oSMdQxIhFWMXDxvXoOD/sZvwJ/3LCsNpzkPmBaW6nyCbzlPtPkEEwxkSRPLwWYWwdECxhArLiBRsPrnqMRxQNQJKcopPV+eNLjRtHUdThRy+9KM2hE8NBG/AAUAUHSnDqwYqJDytV6FuFgNauJc4UKzlHC/uBmMV+i+40nd4VTlKUJ+J6z49foEPeCJgyiWfA/VVyI9LFNKof0nxiwcHxb9LgU0GewnFnGb9S2k3+JES58eEF1SLSzDrfFpoCzMU3hc0ISFpxfj+N18xGd67ZnkP5O8C22HsylzodfMlS86/sOlflvLTdxyfa95Ctzx0bZspdG9MgkCsZ2Pby7AjhfMdZ9sce/ZDgMenCfcDR4Y/JZrajH+pQDpCemztYCwGUnS0x4B4PzB8iweATB1EmdZYqoaT4jbrFSVnywEUpsXpMFtQsa5ItYklFG5RLgvkToAUb+sTBybJLtRAR960QY9O9RQ1yin+zrNDFKYyXGpsFy01zlmNMGNOpA6iNqFihDmRBWQyNLWe+dGzT/SdaTKJ2wVJUyK9lm2BD3wTAJm8XFGbR7+uEDPxHrVC1YNMHV/UgdQSnO/UsAU0R4rPmA6PSmpUmCq/6GiXm9U8Ul12ri+5xRDorV9nVRdisWhNkmDqe8XSk8YxaAosse2VtVFuXbnyMmea86X6bnyt/Gt5KctHf1KtQDmA32UAlI579rU0Wdf3qXVHhUdS1wsM6twN2uRXvT9G1v0TBo8dgrTjQqNiIqPjnqVjHxbt2w21Qazu4g8xlIivX0x8PNZR4SCE3VKwBSOMZXqZ89o66K/A6pf7BMxfEs6wJIG0WzeSSIqyU9bfqO4Nk1KTPdAI4Coi/bkR0eKaJ7WdQKiOfFdxicAMikcQI1BlGOC60U5yon3kSN0/WhaL0q+UunqS6VPLZuJHKlHinLRnjwOpuyzDLNTeemUpuRXGieuocKSiQI1NDQaBqTOWdG2woDU87MFQEudjU6dyHGexYEzlZVS9Nz5KXMBbjrXeQJSfz3cmk/QP8N0erpav2r7PIc/03tR9yiCM5UQfS266U5Z4yE4VdKwDNsHlZY334uqLtY9EwTCWEYQk4OKTsU+U3zdIDUyOmjiPwNKl4xwUWVxhu8pzYlSdwErDjUwXacjqlisuQvBs7AcdKUz4lIdAMZlWBlhmm8iyddIP1jG5IhQa76jMfAI0z1zoi4gCrmRiX2ATejDrjnjMy3TOVCSEtUnu0YALZQG1LiIc46rtQz6w9x7RPrShGnFwFRm6olcQ7lzvov2OOgPaJBHT8rPZzxRJpxpdUV1qNa7tWj/Mt+jp13VUCpVlsL3JQNcvnSFcTZ3P6/b+dieMyB1KzwPEd0E0FnyETNqIF4WCjsLf1ysuPim8aEtnAPnvrgrqA76Lfw66f6t2vGec9XSA/Zy/kzKnqu8M0+PnCnPxpTCcjldY4kG4LMf23vzjZ35oyzgimwbwz7IuAeIIt4PJMtjHDtx0koh4DOR8/stojusxhk0QqFxCOu760tPU0e41tytEIP1zfEdJYYkLkeMhwDNNHEMAOOjiY4Ujg/daGPL8tDX3WFZu8V1s/CfifQqhqdKi/NkctG+srLWRHhuL5k46kZZMYFjEYBp3KiAFQI4HUQ5Zl8eSwmgavqx1K2ZaTnXI28nZNGghCbpYCZmzU9m07lo7/nZYtDFfJqOtu/ivecjyn+ZouwzmQv/2S5Z8zEU12uqcI5TdCzxrV/NlrbNb9/02dn7Z5I3d/U5A1JugWWMjxvndWbY4NqB6xFz23OR4Gfx0bnazdqLnYAAxRDWbIBDf+x4rj8AN1bvjsQli06nr2Ku7LPS/Tk8zuisDEpgSiyis3MRxfKcXVocALJgOG8h3g4OFn4870W/FifxjGBaLi5tvGsI7wn0XVNZXZ+3y+kf2cAslY3PrBh3mcoy564N3gVnHURJZt/BlG/NviFtvbZ8r6XyYnEinmeDxsna5hapXytDyVT05uC8c6J4sMChAqLQpDjGUo0+gKKL86Sb8Ul4WMiNOheaBlTyu4iPeC9Tcr7RSecBVMAUPTaEWD86Nay6RFYVFygIbpQfon1VZaP8wfuMCwVIfcaTgadmRVWXCjwTjGQZElbGWAhFDxpynh04LuQenuccAGn0r0T86ZQ1leUwrtt5ic0Lr1cYPVj2PfsPh9fe3GNTI+NqoFhXF9LpI/gxPZTOcVjRj0pMnPFPzh8rf4uoiw7x6ssvMR9EdGoLd5ch0lSMOrT9wlZVtMg0Sc0gwe0I3dxC6pNfu7mOYjty785fKx3n7OclhgDxFghIwtwHYrk2CymY3ZUmYg/09I9k8/T399n03+i2Fjlx3i0zyljfi6nBa1avlpqjU8YO9IeIqgv5ztJ3Pbt9t8gbZ5oqwsZ9agHXWYQjNbBOwNSBu7KmUcsny8CWcKNwaS0S7av1TWfkq0lkp3RAZjhRNzhx65x4r9mE9Hg1Q2ZCAWjKxV3yOQtn8kR8OFR8UkW+LQRUrPdGAmFXKeAKBYh6rKvVMCsAAEAASURBVFLnTqtKa7KA29DYLFBXIOgKBXWWG5SL9gQigcAHgHRCM6NcT5qRN061ApsQpqSS6Fbqz7XZwdEuK/qHgfR8gCg3X2QgFVegf3AEfNDXXLHdAhVnnxIuUNzhegX7uEFz6X/66DPh7Xf2WhSfmMcB0bek+ocfI+mTwtx2AOZvv/bvmtWm2RXZAWf2dYjlLDBHMOY7br3W4o7ef/CJxGeT0iC/Rzwq/IuxhjihzN2fiwhazaws/GX1kAXZ5is/nTedLwYyYcbT+nWrLeYmc91LJLa/O/L7pe9FiXOlF97N85FeWEZh3vmPmRZMKMJa+Q0TgIUZXi61NMjft9HEt1wZfQCnfGk9D25Qh44ck8QzYXFJCQouZY8G2KPSC7dayD3m52PdRwd3tvV18MP6bvtqgqxqIVc9NWFOyplRUGaAEEDNaS7TmVWb3IebPYHEBlfqz+gqo8HRnBhNZkBGU0/kQ6u1kxLDkhcCV4phCcoamaQfxQHfCRA1YtkR+ZGmuVTP41sHUz/GYAWwwuVCADmc50xFTnfqIFoqdyh0AZPT0f2qTLMAQR18SiF4aXeBcjAlneeDqmVIQ63hyz/XzDBRJvYBDEtzSWpMosLYuNg0rQ8gf1qqGB6941zLLsodCWkWbJ0hjz9KeLaHfvG8IiQdN4DlI2E65x2aN/+pj98Yvi3RnzXgmWnEtegx4SBZQgJRLe3T6FUERNP6GdeZ+nWoEbDSxg84dnYiSTlxHh0tq5CybwGB1UB0whygeG7UWrmu8PDPn7LI+DWaI58mggsTVIX1zNEJe138GSghV36sEz6Pno/XkVtFFG40o4XshsJVOy40d6a7733Q/EBRZ0T9slxclKf4h+Rgx31y/pTUgamPU3JdYdJD+hyioEqzNo8zq3LtBQAxQFL/MU3VjHXVvGvVv/j9VXQRYmFElpTZvHGtGYQu3LJZ02nF5SeECIzYRpR7ZmUhmqUJDoPvYUzuMujJrv3ANdnTnZ2dGpT3mGSAxLN73xEbbMnwbtyfDDizd5m94+cBzULyc6Qnk4EKs8x7bA76ypEXR1bfBLZt/C4LF7Fz8ATonAv1mUwZWFFwT8DpIMqceaCeNIUK074IB31RHsab1Z33IbWAztlrSV4NABq50+hf6h5SZsG3knJ/phRwZUQNVTaaAzjcnPgS0ZOaC5Sei22acPmK89hialVN5F4Lvz3/XvhOCkEUAExTIRimz53N/qICqXV+TQW9eNtmi7LE3Piv/tsPzA+uqTGGlptSoAXmxjNllHB4bAEgAh2wCBvggb6xSnohrLG90ocy1ZNQcKwkisogTTiqAw5Mr5zrunR+9gmx52XSYYnYtFCd7Zu7D1rYPAuikSqYZ6jSqM8idExhxGeTOlM+z8AUT5/phM4WAO3WLA1mGKEDAxzTU0EJTk14vN5kFkhv32Do1LTIOG222mY5EQC7RQ7pDCSoRtgyYPRofj35uD9cbJ8GM56XpbIBs3IFsKQtqW//4KiAadREa3wWGaRoj6zOVx8gc92ZWkv8A54BHd6QyvTpo3GQoDHy3w0p3I8gNKxzdcVll0p8bzCwpBz057SPf/TMtx7V/fmdjjA2pq9buXJl2LRpk81+AmT75SLz+htvBtafIorW0ALK9HuiBnAqBEg/TvdLE9HzkEdAZIiTlJMCWb+e8meVQZreoelMjc+Bo9Q70/eBaH+icyygHUS0r1SkJ6zazlA4iFKu6zEtTXp7sXBi+4ArkQBTLRf3k2M208QmFVm0KcAUcE0oUybH+iITibDg4+uJlI2fKU+r2PthTLOzLEKXjsen5QequfYj+t514zCpeAGTk6NaJVTB1UcxGHn4x9osmCa3nbUBX8fLmsyHtNgU0SjKc1nq+WaVsvAEwLYQgLnaudL0uUUFUkR6DEk3X3el1ZbliCFf1oNBDDVVQ127dWCWtrD58erQTH8kgPHtt1xvS37YhfpDQJGfPPC4Onev6VsBgDTBSZVJfi9cAZPwet9XpClzeyH6bUIdnR020+cPv/hpdcTIaRJF6YHHnjKwQAXg8+39mvQWf0xiojLv3T9YP++rcbKO0o1atM6+HP3lGe796WM2aLAMNPPqJ/VhFT4rdSYoyzHFWV2jBfj+9A9+y8RTyr/jlqvDbTft1JLUey0+KBw/C/c9/vTLNgDBqbGcCTFDf1crqiLyPvncawq+siLcfut14dEnnrNlWZACIJZcIf5ou/wv7/r4bdYmpAPETz79YuDdMBhQHm2/VlGGCP3nq6+yGN9PHnrS4sjGKbb5ILpu1cpw07WXG3iuWLHCwBNOEgI8+wT2TgDo2VD6unSZxCrlt2HDhvCRWz4cTp48qZCLr4afP/Wy4noeP6NbuY6T71rjiBEDi+b9xP1Ez8nUzrSoLl5dg5uAKbnGt6RRputQKSQaKGERYnc0H1KpwQBdJKa0AZMljfF8qq2OBWNsQk8KoBKzwbeAqPuMjgxJf6wuMMV0zmRKZ6lsC+UC1YmEAxX8aVpxr2ZEZQTcxCoVGCXn0vrTab27En1rpYoOlVHMOz/nKktAFHIA7RIjMChf3lL5sU4ls6sqpQKZVNxUlvzGOKiJeRrk5dZUwI2m9aRWqP5MKrAOIFoqNwJ7Hj9h2zMH0DQYpotyjnU+ME3nX0QglTg7Omwdt31FmwU+7hK3yUJ3hcCEcQAxlh9cLFwRIPqHX7jTOjuAgngGUDDl8S++8ru2+Jpzr/4AcGS1imP4+3d90pZiLrzuT770W1o07ocCgj5zUNcXGa77wBUCg3pFhzpq90DMvESrcrJ20r98+0cWpi1yiHzys4loSp0qj/pC6LQqK6vsWQi+/Ft3Klaqyjt29HB4a8+R7DP8+Zd/O3z1X79nelSW//j9uz6RfVaCPaPa4Lo/+8PfCX/zj98yzmple5txgNynqWmZQK3KAkMTaR+dIlM/n3j2NQNlfEhH5NIC0JFOEGcCVhNBiuO21maBf51FxUKZ369FyIgY9af/4bMW15TB5JQ4XtZvYp0r6vP9+x8zjwvWxmI1UcIgsj4WU39ZyZS4sQAysVajbrgkXLBurQH+zquuNO6TugNyBBzOhlMj8RwS94NwfWKu+po1a+x30w3Xh+deeNHWtdp36NCcNcB9TFOJ7DyAB/AZoKHjFwGqyL4AncnACXjYydQfB89Uku1yfRp0owivuyRjkQRzA9WYRy5QCUoxu8lv1ZTojytKNBMq+VRjEObIHJjPqO7WL3chA7VerXKQSDdVpZJohgfChBiCcluSRAyOARTLiyjmg1RvtZXSXy9viWAq8R11I9HBAFEjmyIa4QNQhYYnoiQxpAHz8IGD4qC7w0nNrR/uynkVmOGIaa6SICG46/r6Fg3aqBkoJ1+sNzAVo0WsgVGZ7onLaiBaJGiJc4qU61QsjXNzAWj6Ot8v3DrIpstYNCAFEBH1EOGhYyc6NOKx9gtfBy83n2PhGEMQqQSj+MztNxqw3PvTx235Dpb+wDKPAQLu8a5PfST8/X+/RyNuTqFNh4b2HTwh7uv18OjjT9kxf6647CLr6NdceZmtyGknxEkANOg5H5ChCyBhe9OHrjCj2EdvvjZ878ePaM42L5WaJeyEXRz/rF2z2oCTSPQWwV9cLSMrazdhXAMMWUjv7nsfsPIJ/0bg6C987uPh44o5+p0fPWIF7T94TCH99oYHH3k8WzpxTf/kDz4fbr1+p4Wv+7//v6+Fm67/gBnJ7v7xw8bZtsiS3appkU5wIC7SIHq7LtiDOPsxz41BjNVKUUsAigRyJjI+4Q1fFZAS1Jn2+PynPmpSAUFO4E7v+vRtdrv/8vf/Jh3kKYF7rcU1ZalnAJmBZLUGz88o4PaHPvCBLPeJiO3g6Vuv9/nYck+4Vn58h3CpH7vt1nDj9deFXz7zTPiB2oMwiYWUFu2dIyXPtIvvfBbpz7mIb6nlddTzG3gwkqQcOE1zdwKUnbO1ouN3TZrmJ2ngksAsNyAIQIHQj44rhB1ACsW59hHQLF1KgKGhgXBIuuNOxcDFF3VC6o2RlIzer28gDWhty1rtXjLrhgFFUSMC1joNohipmJtvgJmaYw83CsYDskRnIl4JU0BPnTweQTQJxD0m7nF8IJE6ZJc42RWNVBbIWT6ia0O36lVrngjm/zxdfBkR8SwBo5sb4OzBZ/2J95ku9AGele/sE/ieeG9pWjQgpVBG2srEqjwmUGUUtw6UjOTpG/s+nY2ZNywxgghMx127bqUkCwlH4vp2KbAyXBBc0iat/b7rnQN+qensCPrhoHXjdTvNOs969W/qOgxda1e3GkD4RXCtrB/FFEKCfaDTRMSFEwMEf/H0Sxb7kgXV0pZZgAdy0djLY0u9X1IMVYIlI17f+9NHQm1djek/cfl65vlXwlU6B3fd+sQLpkz/geKOAlxeZ1QQr2ixQERmuGN0mhMCdG2MiPPIwMTgYRxRTF7wX4KWsIopK60ycDEDCO8Jlkx5/qXXrD1YiZP4pA889ksD0p0Kgs376FVYM+gDCl794qtvqdNmLDwfS0QzSPxPX/ntcM3OnVLTtJgRyLnP9wI852oQ6gKn6lzqR26+WXW/NPzyqafDfQ8/qY7fk3epidN0lmQsTYviZIQbdQ4Sa77n80LsnANvNtF3Zm+N+yzST7iP1Z0lNsambX2msgRUKQXQhIP1lUJJy4irPHXquJYZ3xu6O44poM1kQCUA1cgKnt63xNQf9JnSsqmteiTBjIZ1m9ZGrtTzmOEpHkxIAoVGZMiaFIdYJs70RNcxTUs+JQkkzqzjXuMsbidiOZHJcTjseGx8qgB4Upb8+mrdt6JPWNBmKqWQAlOp9G2qqGA7DEiVRxyA6cQ9ywrO6kRlMDZpIR/W0pxjzP/u/jqIZlUuwqr8O76L8hnxAc7hJIwWukbAJ3KkxQqWWl3gMCrx/IINkcN6Z+8BAyCMMYj/cKysBuoL47EG0xu79uQVxlxyljH57Cc+opFR1wnIKqRghyMDdI6KM04TYjSARGRu7sEWzhRVAsDSJnHmHe3X0ImMS4g9yblft9rXyyAGxwzn3CWj0KqV+gBkQMHCyvIcvtwtouWIOAHOQehW4f4AoM9pdVEsyl7nOz96o9UZsC8kni3WIenZhRlOc8yaTjw3ejMkhzXSjUIb5XP5v/3Pf5itL2luwMD1Cu73sSefsQGJwYwfYP/Grn1Wnz//ypekdmgykPKQh+8nAOV50kTd4CioKxzqHbd/PHzo2g+Gf/iXb4cnnnvJsiLa01kAMRftOQE4kub7ce/M/mY7X3JZ4bFxqAKD2PlzrlPu/uSO6Vw+oQqh00RP6lSpPnWy42B46tXd4cSJHkVRqggNMkgSlhsi2AkEd+pgZwn6MyLQw93IZyERFKVXQcVpj5pyBX5Wf4LgThHzhxP1VhgftnFkeEwxbXtOBWZdAd6I7YjqrY1RfK9uiOEqR/qjgYmycE870MsUWM10Ess5LN/wjern6Oc155Qs5owfo15JF6x7Y4SGosO9+mmaFrg+U/qS0+3zjtLg6fvp6xYNSLEaI+Id09pD6CK3bl4nXdTTavAYnSjncB9n6lAJPmYaJs3FpisHR0hwZRdP0+fY75eeZf3a1RJFbzPu85sybhGfkjXsN27YYCIx1vLTEe5NwnMjLMHoPSMlvSY5YlNotXfucJkChgBUrBa6e88hWcFzIc64DlEewr923ZoVJurz7ETxx3WKOqM2gONFh+muHHZR8idXr3Rq3PcaOwgW5gCEGejg8q3OmkQAsZ5WjzwCqqWHShP1JTQfuk8cp//pG/eGK3Zskb51g+l9v/D5T1mQEOqJ9f3dEG3ubm5Y7NknzVUWXjb3AgThLFnWBmKftLMhruWHJ8H/+td/Fq6XuP/kL35hRRnApcB0oeUDtMaNLvSCgnwOrOjh0J2WlmrxOy2vDBGLtF8r1EKlZQq+rRs5mJJWXaOg3/K1fWvXbuXrMxDdtuWiUCMdJDpvPEucJmQYHhXTwSzBsZFBieiaK5/4hXoetqwq0Y6uNJGM0udYmK9U8QwG5fI3Kc+P7t7IhRJspNo4yypJZU2yH8gzRQzDZDK7qUxcr9OmjZvCmuNHwytvHzBuFS4VcK0WmPpsKL+1QyZeOrairoHm7D7qZZ/NthA0vQzS6TswIv6O/BzbRQNSCsN4dFIdk1BXO6+8SKLuRRLLnwsE4bW1WfTi0aVi3ScqPctzTE73ZFf4XKPo5+jwCDCMrMQqmCf7OiTSr6L40NHdbxygHegPK2BedvGFdgiIImIjsiMmH3riGfmqfkANHsUIv4b13z2UF+5CEEaZtSsjV8yyw4CvNVaRd9SomVqjctvAfccJVyYi1lMuQPb9nzwsXWyjPoTYvAAmVmqWpkacv+PW6+3Sb91zvy09TZ175ZK1RzreW2+4Oo87RNyGGEzS/qyWqD+0pVrV2qrP1Cmx0rlpkZ4zt6VeuGhB+w4eDQ8+9rQtzEc9AVvADIMWulAGqrGyMS39MBmefektA/lPf+JjoV3LfqCWYS74mXKg7rYEYOIzStu4FZ97n+zokOtXtzpmX7YtaNeWpsbQLPXBiuXLs0DKe6Ct8QgAFKEzBVaeA8nhumtZGHFj+MWTv5Q64x0riz/vBhizhSQ7HpDE04t1yuw5dKTj/aFZFWA991MHDpl4zHnEd3mye9ZQIUv7yPBg2PXmW+HQib6wheVi1l4QmlvVVuYzrHaRVQoARQzH+g0t1zIlIyNV+oZiWfh6jksPWibucnSeACCTssBnxgayIDqs9geIscK3Num9Khg1hitoVGqJoYEerVuvmAhJnwBcSwS4/NatXB3aJNUcPcFCkKfsGiJeiTPRO2Wqq7hqXceXzbuCWNMJiRXy912MU4w5FvbX3wXbYmW5F0Sx0hYVSBkxGhubwpPPvGDLCrPaJ9zm86++bVweHZjOCl22Y6utBoph5pt3/8isyYDvS6+9aWsaIdKj42TxvE/IfQcgeWfvflv6wwpI/vjsCgAKYuVRgPEzd9ysrw3fVOfV7LTpKbFiv/HWXlMbEEmeVUXRjyJSH5PxgfWVWIMGMC9GWOxRWTiXjcUcP1Z8Fpn9dNvNHzS9a62m9uFZwHP/9Z/9nj6CUXkR/DjbBnx0EHXGt/XOj91oxh/0lIWEQeuU6gaHDrneEmDnWcbG6qyMT6kMKGlm2/c/zj2zbtSBw8eyq6DuPXjcJlHw7vr6eg08f1vGvUeeeN70tl/4zK1Zw9kff/n3jVs8Gz0oAArwIYnQId5+++3wzAuvCLjHwn6pXFCRLNTfE/9UgnawjHaVPBM+cNVlYd26dWboAli5h3cwf/65ti7u80xMEPjUnZ8I9/30Z+HIobdngWgaVOE+oXRaej+ezf+bttbnnyl+BAtRXzkTXu2OKip0jHB8aTJQVULv4ICForviog2hbfVGDdq46I1a6LnOnu7Qp5l3ZYqiVFndZBwiZQCAAB0gCPDVKfxiBXpQqcfKBKjsY/VHlHffUET8Sn1g2OjhRPvU1uPiTAFKl//GRgWew6ey6gLAkO99TG5QfMsDA1G8b2puU/8oN65109pVAuF6fQsHg5wMwgqcZlNEc09qgGdBuxJ5HkzL8AznDug5CHr29DFrTEEzqp8DpJ/3Y7+ObbE0T/fr0vnZX7RVRClM8CIDTrmJ3Lv3HggXCrAuEce448KNAqd66dKalbZWq2B+MOy8YofpBp996XUTL0509ITLNYvnmisu0mjDKDNjjtx3ffqjauTqcM+PHwpHjp0SsNbbdE2+3qdkxBnV3NyrLr9IDt+bZIAcMnefO2+7znSQKLRwmQLgmsXNXKGVO9XiYcf2rWbMwVn96sulq7zzNrvn3fc+Yo7yLDzHswCk6GsHBczbtlxgC9tR1oCOEcWd19WrNMMX4LRpw2pb9bNJ4uKgdEWsnfS5T9wsTnml6n8i7BbXDHdFnXds3ywn5GEp6Utl0f+QuPhLbaQdHuyTVXyf3b9ORqvtWmq6Ts6DOGuv0EqKrPoJB8bzbN+ywe5DaDFWDN2qBQcZrTvE1b0ugMWQd6lWWT2itZvgPmtVDiIza1WxEOFlavNLtl2g9pbjtLgVViAFjNEfMnGCNZ+2b90Qrrri0vD5z37GuD7ufSZcKGI4K38Cbj//xRPhh+LYv/uD+8LDjz8ddksffVD3YA2rudQSfFuFRF6u4dq9B4/YwoG/eOq58M6e/dZJ28S11utbAbzhmhdCPBNcLddcJkPUwNBY6JSxZiEEqPJzypSIqxFnA7DyPorRXJ2SvIjtTfpWy/t6QnW9BiBxbIODveFoz2jYKAPqujWrxZFJbcZNBSyoKzuOHQulivq1ZsMme4aJCUlJw93h6KH9EvmHw/oLLpAxdpn5f9o6Z+L+eVa+K4B0Qpb9Eb2jWklOWORL1AfNT1RWd8CuAtEc/1JJivjRMrVzRIPP4FC/1EDDYUZlMFjzGx7qCaPyDW1e1hraxE1XVMX1xmiJaQ0E1QJjPHBOdXUJfCUJCcR5p5yXYBXKtTgexmDqBwD3i9no6FbUJwUlXbVWfslaAXg6cYFyNVjeN5k3ogG2qrfSyJNu9+xxOj/76WNeCFQkDXFfi/kt5iqi3EmV1D9WpeyRE+7Lr++yGTYXSMzAKZ+li1lV1NxnBEis3HlcgU3Wrmo3/8q3ZFnGMn+xgA6QQB9H9J5vfO9+rUy615zTaQTAeEhgs+fAMdPvoWMESAEmgGFZS1345vfkfpTo/RxId2jGlc2JVzDgD9/wAbOy4141Ku7o63ffL9HilPxNfQpm/PgZ8QCdNbKkY1DCso74CTBkgVQ76DWJcvX6m++YIecieQHwDNQJV6n7Hnxc1vBnwgr5hgLuBFzZJj3yju1bLN+KtpbwnR88YC+Z0fsNrcQKmB08clxL1VarTbYYIE6qo+/ac8AGqyF99Nv03Dw792J22Hc1lRSw7ekblgHtoK3+yT0OHDoaDh87aZwlU+t4R0e0lPV+pbPC6EXKg9cB7QG4o7sFpLZv2Rg+efuHw/Uf+pABIUCT98Hy2gsIbh1RGbGdvMfUwb9zz73hW9/7cfi5vABo52G1aVzLquDiszykLMqk7BfkWfDs8y+Ho0ePyn+2xUR/e1/KQ6ebr/6cg5MFfLdv2yYAqbb6T4srm490mfUztvx0I8tu+xqMrfPq/pRvaZzV8VzEGfFcITPeJ1CSZ8uWC0Of/Jf3HDkVVjZWhY0b1kdwSFyshgRkZfoG62TMZOFCfpMjPXJ9itLN5m07NMX4QPjFa6+EQU2weGnvXg3WB8Oew4fD2JC4XRmTYBjKMopWPyoH+ooqfU8usDJVm7WjpJfWNwaVCjQxFPcK6Ps0Qw8R3O/L1ubiC4h7JNIfkw70xd2HNGgeDwdPdcqftTcckdTVUFcf6irLQu/JYzKY0YDUG+u/JiSLi6QcgJQfQDqogDWb1rWECnxg9X3V1CkAit6VvU/aMmlfq2CqbZ1j9a2d9z+pfJ50Jlsul5S9+KuIeiXg5Cw+pPwMiQDFkhA4lBPdhSUhCASCaxDO73xk6FDRbSL6b1i7MrRKtMdHcb+4PIwsKMtpMDqp60m4FmJKI1MiAQGIYBaAGtM+mfUUo6Mjhqjz6l6Ew4NTW792jcThPpvrj/4xLkgXO4AVlP2jqWKqFyMonKiPbu4e5boanoF11ymfmUmrVrSYFwNBh0kj0hV6X0Yx6lwt3yas5uhO9wn00LNiraSOuF/x4ifkZM/qnrQJUz5Znwi9KFb8ThmuUEOsl0g0LhcSvA2gCtUDXScxBLBwwrWja40R8nm+OHee9gTUefaNKqNB76NTqo7DAlhP+49/8UcC660GorS9fbR2l7n/0OEA0gMHDoT7fvZQePblt0zknPuKc3eGwCjXyI3rjo/dGjZs2GAAyUB4OvJnRRWx6+3d4dFHfqZrT3+dl5vnF5qJAq9zQi46cgznyWBdSKQ3z0yGFQrPNDohYKtqDDi6f+cH95sR6ZMfv8P0pHCBGJxkbJDFQyCn7dCYjH8TWrAu5XP91Auvh8Pd4+GOT340HBMXOCI3oh69+2Oa8tzQUhmuufQyDXxlYffuXWGdnN5bNIGjLBNt/SUC1Qb14aYG9bckyAmc6kDPsMo6KXXYiawIX5vMg+/U5JRR9T+8DTqPHwlTlU1iWFSnwfGwSpNgljeUh92vSmJqLDWXJ7wFXPwnoAl1H9KS7p52WP6weCF8SNIZ1CuutnHThVI3iJOVrcQp3baedjZbNyyd7lryCfxHFlW0T98UDSIPWS/RdExTzZj/zfrsiGOsnd2k9dgRFwiEgU4bLqlGHRCDFbEJj5/UCpHKiy4M8RZ9ZPwHDOh9Cij4sY/YC0cCyAAuWM8b5Z5kKzzqIy038UX+bBME66BOtbZU8hFNF+QeXI/IG4OBzP6oMUpxH0Y8oi8hyjuIWrIu8Wdg5c1qlYW+7aimesKZUx9AmjJ4Tp4DPSWQhpiOTpQ6EvFoUp2L5ypJ6sxoXCVVA89F+xFbAFGHa6k3oMDkB+5DmaglWD+cZZsR4c1eoJeN750NRFQ4eR6XHhj0Tkk/SZujjqB9UEn8p7/6Y7PMs9qqXVWkw9sJ/XEulKAjGI6++rV/C9/+/n3hrb2HLBaC5zvfWyy8+w4fl+j/Ujh48GDYtnWTifxwPvNxpz5g4Hfa3r4itLW1h3379umaXKed71nANjU6H2o2m5XJCf/pjN/HM6HH4/3g+FQl56u6jFz0ZNgp1TUNy1vDmCSxk1KDXbhtqwZTcYxwbXovDKz1mntfJV/NaUWegsEr1bZ/sCc8/9puK/6OOz8bNly6M7z6/NNhj9Q2tVJ3rZSagBU5K2XEu+bGW0ONQL+nU+oMfTgtUk9VaKAuQ8TWc1RKvyp5WueYLz8ubnBYNoABAXJ3mFac0Uat7lkuUJvsGQhVUq1c/dHfEXfbH3a9sT+sXbtcagfFolC+1uVN4YZbPqLQeNKT75d0Ih3tsnoFcBZXzPfOt8u7oR0gGAsGkWnehVQcY2LCMnrOsmaVKdDN82ZReywGpQc3wJJXlk6L9eIl27e/2KK9lZv8iY0QAVUvQWIeRgFE4DLN86UKsRrkix8bHZvGIw+gWgH3h2ojSU8Kto+GBtdbtSTO8yFxHT/2PS3uk00NLxBkyzn2vT6AE/X0F0fuNFmt9GWWiNOL16fPpvdj2aTA/cGZpeuTzun18zpTtqfFZ4ttEdsI17JKi4YeAdZTeQ4tfKZnrtJ5mpEyiDjFktgxV6y3rVKQrkCyT36en3pQ32orqyL8xz//Utix42IZKiKIFrk0mwSIIjojDj/x1FPhb//h6+GNd/a/pwCarVyyA6CiqkDkr64qk5pGkz5UZ8T4QjArvBZ1Rpvceco12+fw4X2Fp7PHeeI7oel0xjmkXKb4VrLHqZ3InZIQ+8649KzVAq7qKvn+ygG9tLElLGtoDQcO7A4EaanSe7bc0i9ifspUEg+AQMkyFklMBlK65f1QJb3mmlVrBYjVoblWjIUMsIcO7pf0NxKmBIJSRoYrN28LJVIVDUsdICdT039WKuAyVv9yffsYnjThSWBIH0BzIdWBxHlUCqi+EPkR5wdH+5Smb6ZGa4QJxKdlhBrvPRGGNI+fmKk1VaVhvaTNUt2jX66CVSXyOZWHAH25UutO1ddKPytnIoCUfgCIQgAp8QuW1TWEKakUyuReWNLUakwZ31+O2NdvnkE/l3f2noMmRfrPc/mxb0nHkj89PXMugdRvb5+THyx4m2ua+a6PH9yCC50342KWNe+NzuJkrFuuTfKLgOuc61x+ztMfwQt95fc/F2684Xrjqk9/BUtG1Btn/F//2z+GH/7s58bVLuS69yIPHPcLL74hD41DYedVl1vd4TpPR+TZuHGjgmuEcPzYoVnZrd/qj4PyDC8FjrSAslxpQTqHvEPOs8SI9mxobywTx6l7jgv8SjVolq/ZGE4cPCBD0Lhmki0TGEr6UP4KxZyAZqRbhJvNzMDZzSg4dquMvC1hWoMA/u3dp7rN5WnlCs3sk96zXQFltq5ZL51oZeg6dQJfQOmVl2t24bowI79wIZkxJqgqplU2KiJil07jkC9RHV/UCRltGajK5GrYsmyFLO+tYlonNM+/K9RV14UN69bLDrIyrJfLHD9AbqRPUpiqXC0GAG60WRIqqjEGN7hM9j0Ikhub6lSHuhp5IuAJI+ktNDTrGTV9FWRzshehY9tS/eKqE8/ONg2e6fSF7HPrqqry8wGkC6nOUp73SwvcKVezuz7/uaxOdK56uSiPQQwd4v/zN/8QdslT4/1Ops9WJ2MiwrNyvbpAU2ZXqZPDmc4n6vNccNxbNm+Wr6PCG/ZEd6Q5nzdhWxxYyecca/oa8ysV9JlrjgAwSkYRgGekI0U8b6oCLDVpQcA8I/DIjPTKYHRcRkUB1uSwcUVlkoBYVhkQNYu7QKqsihl+AGyZDJa1MrrVi4uVH7fAhWA2iO7L4TgBLrUJ+ns8SKalShiX2xPjwAxLI4sbtfuLA0bsRrSfkliuTeRIhxWKUUBKG5aihqhmcorUDPIVRX2HM35ZbZ2VgbqvSgBekZwHoCsVIAUpARCFctH4ZSOQrnRULlYZxQ2o0oBQJeAESEul7psS14u1H3coB85YQI4p4jWkRfJiwJrGYbv+DP7AkWqa+RKQnkGb/dpnJYj0X/7ZV8yYB2ikQSD98A6iqBseeuTR8F8VzKRLetH3Pwmw1MemxMVFnJsJL8tvuULgAkDyXPOBKefcmr9HPs0YT+aiyFnGDg2AZo8Leq0UK0kRcQsQOhFsD1fKGsRofDrF3Y10HNW0zc4wKdBsbq6VRBz13wj5M+gwxUFmSUWVyCgEIsK5sgRJhXTY5dK7jksvCiZOykkfZ/ypMU29lPsTTvilUifA41UoSj1LPUOI1SWaS29AqmPASYoEcbiEw1PgGgE5YDouQ1eFuF93xqfNWBpENxdARlUEwIer1eS4VjRIRPdSjXDT0gpHsV7lC6CkF1A9eSdqB7k7rcDPeUzLBKk4gHS6rikBUj1zwoFaZZM/gCaUBtL43tVWOuW/JPtZbWIZ50W0P6v6LV10nlugWeL5//W//7VZ2xFjTweiGLIefuSR8M/f/OGsiObnueoLuh0GQ4wSM/oRY5U1otC/dctL4YWXXw9NdfL62LjRnnsuMKVNOIdHwqZNm8Mrrzy/oHtneyy9LkXoRHPtnAAporlEdWCFuKe4QFWVsKSIuEGl9nR0yLCj8JHyJkBfyfLDGYFQqUDKdPjCPRVhazMZh1oum4O4Np5VurwwI+5xRFxgiRzjR4ZxkRoUgAo4AVxRdYPWyhJAN9bL8IPRU5wwA40NBtOyb4iTjXpfhc2UG1KHOHOMm2VTmtYsHSdcqWqjVT6btLyL9Jh6lqjzV50EiAaiAlzfh3OtNS4YbjeCp3hm40rHNPkAbpRZTg0N8njRvacEvjOoOQSkJS1taqNkFqLuqoNs6zqIkuCAWdD82byn28lKEnnvK3eVBuDJOETk0pb2fgNbAIvsX/0PX7TgI8QKyHXu/MZwThRx7Uc/uS/82/d+qgz54JB/xfvhiM4lTlIcGQbABhkrILw14KAIyEHH/+o3fmiua5/99Kfs/FwcOW2D+x2BWm697dPhwQd+aPkNaAQ8Ttb5/GCOrXg/6+RznLbkIXSh4vBYc2pSXCPUKi+CMonpPQpOgp4Tj1MnCywiWxNPzZuRmci2NdKB2tIiAqYalTOmNZTKygWo45WKji+xWopYgp8Q9KREqlEANiPVAusx5RF6U0jTUX1dJg4zNQLssQgnYyN9oUMzPZfLE4AQ+oj2aWLW4GS97m9GpnimfKbfdibGywSe4o6ZIqr2ccIdamZcUZYE2EEzyKe1OioRrwBuoS4oHX9+gbVA9uBd7zDBIlLuHacLXQLSdGv8hu5ff81l4aqrrjK96FwgStO4k/199/9UIHr/+7y1IpRgcS8TODRp6iGhCIfk2M2MNrgqpvYi2DrkfPXrP1CA8OZw84032bNZJy3ylLQRs7Quvmh7ePHll0Pnif2WayHgWVgc4jsEqEJwqWmaLNXyxlOKWi//zRJZuBVySVZ2Apho0ovUtGPyb66QQcrXSuLauBZTupTUvoBQUKWI9UqTNV/TmJKTeMhotpFE80n5mKIGgMrEZUIsqMcAWkgspzwzHOtsIOeIojB4/b0nbUmUMCmPHbk2lUvXC5VKGqgTFz2G14Tm9qNSwF80TqOX3J6iCKh6JrlajQKaojGtHVUhg5a1FQAKkJ5D4lvhNpGyO/YN+Ts/tzXwey9t37ctgEj/R1/6XZtyChc2H+Ej+vCjj4Wv/fuP58v2np5DhDcxXgDDsjSsPcXyMMRqIPIWIIpFGLc42EHyMpVzRG5e+Po+/+xz4SlFgTrd2um0FVNlP3nH7eLsYjea5epUpCXIAwA4YAKgxpkKUB1U05cRxm8w0fV5es/wSdtlzXpzutfzzEesx+RrMs3M5CYVYFnHH9RXCZ2WrjNG2ReAlhOJPqIiy5bgyzqLCN8nkHSCK4VzZskQfuwDhDPTcsQXx+kTBFj9FKoUmErLkgVY0jIluWBAgCc0rsks/Zpt56uNliq8X4Vc9QzEnBu1nOf3j4Mod10C0vPb9u+7u31RS4i4+9Jc3Cgivc/w+aev36NnyOea3j8PhS5PhhQBHAv7LWuRMUKAyXpgzA4rEeCVSBRmMoVxouIsyUtcB2aYEex6XP6Nu15/xjwReGaevRjRVkyGoO2u2nlzsSxF01Q9A05OFgPO9EUz0mVCiPeTWhakRNNVW9Yt05RRLYR4TDFzpTNMk+lEbWXQdOrs/elMHDDdOm7uPwJVCMB00CwE0PmWbGbNJScAlB/cKj8mpMBxMmOJVUezpLWmpjSwFRIri0Jwnk74OUO9MpLNKFjNwmh+xqCwDIBxrl86L3pyN5J5+hKQekv8Bm5ZoI6lQXzKbbEmAEjgzpip9f/+7dcsGHaxfO9tGmI8c57lBC/xvUUAysqbiPEE/iZeQxRLo7iPTnhG4MlzswTL5k0bxS1N2XLXbNGbPvSgdKZ6Zp59LjDlnpRx6Y4dAos41Zm0+cjBsxgn6uI912NpZsYnxhSIReUwCJVrquimtdu0xIhiCwz3GzhpipHlmetPiaYP83MCcJ1w3+EHqPoPwCYNcqONg64+AFVGS3InwJuR3hlOtJCqShWDVC5P/NIg6uvcW344WlEeuOq4olRxYjXAeZT8CqkxfM17K6uCWYtqnEQvisGtOOWes9j5QtAslqdYGoZWfmnKP0qfWdr/NW+BTPjyFz5lQDGXLpAGQC8KQP3N3/1T6FR80HdLgBjz38+OohiOyM4PMCwmxhOnNivGi5OZ4cPXgGBivxhMYkBgTSY2A6H4jii4yeGjx+XPSIyA6CqEhf+H92opcXSsiYhbrM60HWB77bU3FDs9Ky0NloUnHWRJR/THFQgal350WIwb4DWt+KQV0uPWrdti6zYhLrOGPEuOwGmmRXe7WH/Soj360xkxd/8/e+8BZcd13nnezgndjZwTEZgDmEkxiVkiFRw0HllylMf2euz18Yz3eHzGnrOzZ7zn7J7ZPWfOzK7HM2N7ZcuWaUuWrERJlKhAEiIJggRJEAAJgCAAIqMbHdA57v/3VX3v3Vf93utuoBHZ3+nXVXXr1q2qW3X/9eXr3CWgyU+T3Xv1CctqWf1zpIgtiDnucY1qbpbuNuUgAT//Uae+SlOU6+ecaM248vGqLT4CTj5Jn2+PK7fE0GhNGNdxTHxH2/PmL7N2me8+EeubjHP0Y6r0LCEA1X++L7uMwTO7b7LtmAt1rtTLZoF0st67TPdfowxaNypVHEaTcoR4+8JPXgyvKjZ7JghO5w9/7zfDA3ferOZKcRLFziSxXeAGNStHLcYj0qwhmuPO5GI8U0f3KXoJQESMd0I3OSK9KVzqImWEumLtWkugQ45b6hNYkKVTijnHmEQflCP6kL6c25r/QExFX+ptxn6OXgbg4qTPPj4E7RIn4Q4BMDhCqGXhiqS6RORiVAxUqWfGKIFzBQBIEpIMmcEpbRIQBXStLhZ7fjqGifCwmsNN1smrAAL8EOkBvNNK39evOaYASnS5gKjRqI7XDzCNAXV8JPUGUCU3MLGEA21S/gjaTGYUFbcMcOatP0m7Rf8XivaA6FTJgdLrO2B6+SxH6j3zAV9+4olHTNwtJbZSTuw9eU3/9svfmrHeYp4s5oL67d/8XPgtzZi6VKA2NUr0V8yVhd6TZNCI72QVI6l3t7JuIcbDaXoCYo02NZ2ANVxolfIPbFi3RudvDfsPHAyHj5JWsMHmzfIhhmgf07ZXX7A+oC/K9RWqgxs35bnSeNBmDUwx58m54D6zYEodfm6Uol7vqBInp1Mfu6he3TDPLPfsh5hPyamY9T7HnWKxj0kO/5aIhGxSAi2mXcYPNAe2qh8fi5uVi+TV1cqML88Bfg0Nc0NLq5KrqI1+RSOhA60ViFYLUPnlKAXUyn4ZqBxkczv11FJDkyWDZiaAyoYwoulxRhTZVVqUjxpIV0txoP6hc4DMHulA6cCZ3Z89bpYjzfbQB2B73cqlpteDkyplYKIckfbzf/PUjKfA69Q0M4jMjz78UPiTP/79cO9tN0gvlhnYmeeA3QXOEwIwsb53ptFUXcoGRLpB8nHCvWFIwqDkYn+/LL5LFy8yLrRPDulwoQx0QNTq6n8pQsR/+tvfsr4o11f05VVXXVXAlXqbMah6WXaZZH3Klibb6EkR7y0ngADPnOfRVYqwfCPeV4grHVA+UeZsylFax0GXctOXqp5ztUJehYGS2BkQVrJm9QvO/9Wa7nlM0UTmIsWBasvbcQ8AwLahrtnS7SGCE5kEtWpmiKaKRrkr9RpXOlLEoGQV9W+sQfNOKQcqhIsUOlfagrt1gjOtq9TzVTo/DFexKspF+Sy4JgBaGt78mThg5s6VGpKyQOn7Sy1Ln6nUEbPll3wPfFjTViOulnN3ggPbv3+/5mnaeU7ul8GAMYeJ5/717/7L8K/+5S8HAL4U8eLDiWYnQhyVEYZMV6Q3zBNcaOI4D3d61UZS580xLvSYknozhTccZGn4zLfEGiI+fUGflCL6kj5dufb6UlUKyrNcKTvL6U/Z3yOfSwDP9JuprhLLPWJxMq+9XPMVKZSjLNeZ7shxrQ6iSkAypg8bBJdpOtH0WANNrXPMuGYMqBQI84tpQJ4OIwrlRARvbEpmCq0id6loTB+uUjQkAHWrPT6mtAPRltPQYOKo79uWC9g3Jizhxv03YWeuYLogmTuwzMoskJbpnMtxF4ae2269xXwgS3FY3DdA9/m//fI5tdJzfqzecHN33n57+A//7g/C4w/cZTlos32P61KPEl8j6mFpByAR0TEsYXgChvjBhcLtksQbLnTt6lUC7B7jQhHb4UITkZ/Bmh+w2ihLL2x+voATylbmXvArvfbqaxSTPnFYuegOgDpgxmDKerydbV/xoGFI3FjHgO5V3CkGI3SgtdXSSwqEhtJ56xHHzbLuDaRcqW+yrFCqulg36iAKhwlZtKj0sAPH3g8nX3srvP3mdsuW1SegM0AFVPXrUcLwU+KABxSvz1xQEM71UAyG3QrrzHKlbMdi/fBA/kPobVlDEulxocIValz5U42DLKEfHVeCF//ZsfpXDDTdP5Z92Z8fN91lXhs/3SNn61+SPXCNuDNyazLoSxEzD+zes9emNClVZ6bKHcwBU7jEX//VXwwP3Ht3+MJT/2hJofPnAZzGTJyHuyRxN3kw3bBk9aRrxJiE7pQ5spiH6l3NPMDsAXXilgBaRPmpUFZv2XHqWDh69KhNsFdq+mkAnL6dO2+pcbF+nljPSVkpwHS/Ueq42xPrEOBLlFO7fErHO9o1D5JmvRWnWF03YFwkGZsI8SSMVMnjJbonHKYDpovl1lgKroj0wwNEejG9dWJAIlc0me/7205LhdIVOjVbQoc8BY4ps/0mTW9y1c03hRqpOzimUyDar2Tj/eQ0FY0qi77zqsBqpbjSSs2KAQGmBOfGelJEevSjg3wVUid96kJwoviOzlF6wO5eBfHKCb9ViZz1fS9CAnaBaDHKiu7UcbG+WP3JynClcyIXglN+zUtml5d1D9xyE/MQpTHKZe6UKUIwDJ1PAogAqQ3r14U//oPfC5968sHIVYrkF8nr2qlpLE5pWhoDUXGmGA6w3vfLQX6VAHTtKk3JrYz/TDnDoPEpaabDgWbvm3bgSssRXDx9e8W6DQVGIj8GMIx/Xu5LwDMGULPYSzkMwAK+Y0raPCjAPDxeH44or2hHe5/ALIEukpBAQ/KjBSBzhAog5kodRMVRMjMogAjBjVZU6yMj0R5QJRS1X5x8ncDxiis2hrvXX2n+ou3t3Tp3m3Gi7d2d4WgkepO53n+06dZ31qEe3QWcaMydUuZcLK5P/CAs9Pxw9K+UuqButSb0k0O/WGHbf6b/Yg40BlkAMgZJ3+fLYueLj5nlSIv10GVaxrQtt968ycCq1C0CBCc0t/i2t/aUqnLOyxH3MXT94md+XgEDdwRUDG9q8kO4yUpxAebWKVFaXuMCylEZNIZCq3StizU9Clzo3vcOWx7LRIxPOZAIW870BuBK6Zt58+aZ+qBYO3wIrpHR6bWtL0zgfEpxotl2HExHo2lNAFMHWmFlOK6DxnTf9QJQpSMJLWkIZy2hU+kHB0A1cPQTRCDqnCigNmeu5qCX6kA5SsT6JkC1YPWy0KCJIBHdoZqlDaFVZ2L6ZXSZvZpaHCPXXLG/wwvl3iQVS688Jprky9k7joeEQnH7FD9fdTrURS5WaDwb6pIPIu5RzonS5hBpq1Lql6qidsWKULVwmQVZYIxjqiCpw2W114dFHGg+kYgflV+yjzoAZykwZJ9TzF1S5vtYxgDr9cf1bDwtIGWzHKn3zAdgefWGNZY811+SYrcM9/biy1umPL98sTbOtgxxH+6OuZ+Yq/6P/83vhV/82Y8q87qm4NVA4gfBhTJZIpP/LV+WcKF79x0wDhWDEsBrvxkAUc4HV7pj55sRh0tpIdG3JCieN3+pcaVZsb6wdvEt16dm9xpnKhCBmIOLFHSjmru+srrJ0uJRDiACnlj24TiNIq4U3SYg2tOJRf20idpwo9TlN9yfHIP+kwxTC5avCk0LFuuempUcWi5M8iPFut4oL4sWrTcqOKCuXg7y8qhgsslhuaJBGMYgLPeD8poY1DW44QkA5Qd4+s85UY5Bv1qjj37L3MVKDq25zvg46Hj8R0cVlRWDKOv8smR6XhUCojwT/2XrFdt2TtOXxerEIMr+iVdQ7KjZssuiB65Ys9xAAJAqRuSNhKPa+vqOYrsvSBlzRiHyk97u//j3fxDu3HSDXQflWMmv3LjBttHpdsmtCsd65qDSyDvr6wUE/eeNHT74jvWR59j0cl/St3yMFi5c7EUTlrEudMJOFXBOr+Nc6JiiuPhBvrSNzD/nQLHso+/0PKMmEguMANEOTfOBzhJnecR4wk/5YXQaTrlZwmQHBYYjmpWURNA1NeIGlcgEMGVW0Yam5rBi2Uqz0tfLEEgiZ35EMeHzCcGZ9vSPhW6lmupTvwCm+JYCnhCADDmI1trEfwBp+gHQ+zjIjKgRAablONGo6hmtFuM+yzUEZwrNivbleuky2oef5vp1V5QUc7hVxPo2TXt78PCJi+bOnTvFGIWr1O//7m+El195JXzvB5s1SIc1Z/oxJSVR2KQyHBHNxCytM03OIQJwXacHNGtmh4BSs2+W+CDBBTEX0tu78leSBcb8nsI1r+elvl2JTJsS6wmYKmRSxf0SievEpps/glgxD/9kCZASOkqekj6J9sOKjhpRX82XIadRyZEBULLnQ8AXXmR9JIAOmhJZHGo1AQAK+q9BlRIRYFol63+LONBExwl3mVjv8azgNyRghljaujhWo/7RHIgmBYX//TjaGGWa6ZRyvqLe73qnJ6Ny0pcfWw48syK8H+NL50xnOVLvkct8iSV7zapVZa31AMC77713QcX6co8B3SncKa5Sv/tbnwvLF7fqfgbMpQlf0kIQhXs7O67UATS+JsDzwPvvl/0g4RGxVJO8ERrp5Jylb5/NkrR/TjlwSQuifCRexUCUOH2c9qGW5oUmtvcIZIfhfsnkDFgKRdmGKjTNB1RXV6FZSJWARNyjkuIlZRVD0nsOmfV+jgB5fkurASOcKeGiiPxk30LUBwwhlnCazm06V8o+nPAhdKTOlVYsWKCPhPyGpcoYldtayoybekX6C6s/2b8YREvpSSdrw4EyWw+A9R/7ZoE020OX6TYuQ+WMJNw2HOnLr2y7aHvAuVMA1R35/93/8lth7col0TXjR4pvqfRi9oPbKuSmospTWs2K9+/ue8/6qtTBgD19bQmXi1SCy3RO03cXK/N95ZZcW1UqIgOC8pSdUJ3QT2L0cY8aEgBUabZPxPZhRTLJk1+i8pDsU4oqIqGIgLBeM3Xya5bPcYPADIAkJJQfgIrLkod1esgqJ3VRHWAEUBH1AVTnTi15iUR/wBTuFa7Wj3FDE0sxrIkKCunCuU+1X+C2VARMXQ8aA6h3RrGyKqksynGjfixLZWqwXwye8f5Z0T7ujct0HbF+2eKFZe/O9aNHTkQhhmWPuHA7AVTAlOWmTZvCNddcY6GsL7z8upzve0OzuO85TQmg4CZFspLCyKepXTsgVYxG+o/n9KSlxHuOq2tcEE73Hi7WREGZgypcq697hVgfytxMEFnjbF4nifNwzdJ2imPTLKOaa6ulSVn0K3Tv0keawSlR4Zm+NCiDFIawXqWSGlZdwi2HdVx9ms5uVJn4a6QeATiNPDJKKgGMVBij0KPGvqCnlREMy74bkvAIHUoYTGtnYJhIqeS6fUI82h4VuDvhAeBO+IT+QhZyCucdAeYE3ahAVnKIN6Pl1DhVP4A+k8bZNycsAc80T07BWagIpwqoOs0CqffEZbycLzeWdWtkRY5Ewuztun50cFAD8RIgQBQizBRwwJH/8YcfMFepztN9GvTJS7544TxNn9xmOkXE/6kSgAawFSMFWE2qJ6WvFy1eFk7L13JwsLdYMznQ9POUA1FBjwauN1N4XYjA0PDwkCa0U6o6qSKr0IuqDIOToleNmI9pWNZ4pnkm8mvotJK8iCscF4fZVFdn3GZFCqIkkh7U5HijCguFsPD3ylEfwsm+RR4DHR0C0VNyyE99WBtSFyyrlP4b074R6VER9+MQEAdVZi2FQ62S5wE0NNxrMfW1ivU3az2FEVfK5pkS3GfWzYntmCstB57xeWMQFafcMAukce9cpusYYhYsKJ9lCR0SyUROtiVRKpdKVwCoiNL8Vq5cGf7Nv/7t8NwLm8P3fvyKuMYeMTTi1KSysHmaQJUp6k0d3Ir1A8BIdvzJ9G7MO39cDKkwKweaxdrLAmixOvmyLIjmueZ+cZlNMhxVSWzGRgR5AuZkK/+f/C8KVlK2/c4w0hlCuzjY0DesOZZSg5CqYmWHYj9QtuE+O/XDrcnJHPFD4QeDiKhusXQt8gQYWrTUdKbUBzibmueFKulfh3vE4WLpTwmxvkJ+rfQteRRyIApnCqD60g+YwjIGysmqOwearRfrSgFRpoj2qamHBnr6Z4E022OX4TbcGXPQ8ysX3tglEe18RzPNZHe7Iz9Zpa6SW9RXvv50OHikzYCUKCjM0sTo41t6tjQgzg7wLkX0NQT3CsViu4O0A2g80MfdSTY5LPpfCKC+AwMb3O9wVaLK6GVeefnQVkikTvxIZT3XoMdqz3ekRgmg6QcE6LlypG9Uflj8SDt7OsLJvUcKwLEzDf0EEOfVtliUE4AJzZ03P8T7rbDIP0B0KU71DTqP4vH5oOF32txSbxPfCYTsKEGqAFZ6W6ZvaZxr9zSitIEmFACgzpX6ssi5ShWvycGKAABAAElEQVRluU7qeZ9nOdRSbcQcKHUcRL3+LJB6T1zGyxZN1dBkyTpK3ySDEd/MS5ngTtFZ8rGAO/293/mfwjPf+1741vc3m8GDOHwAlVDDs/U15aNTTlVCP9bLao2qFo50MsKQ4jkyJ6tbbj9caY18NcXyabQTLproHHFzqvN5lZjDRMVMwVyppCcA7MKFy8OC1sUBnSfRTsPSQc/XdMo1em9YhxDjG+bV5UR5wBRa0rSyKACzjxBTpiJh6hG+XyPq+/qGRIzvkkubE+5WXd0nQkOLpokhM5kBpq7zDICTNgHKGCQdTB1A/bwsfZ+tS/cJZYGTspgL9W2W0CyQJv1wWf9v1GBgUJcb+IhS7e2Xllhf7qE5d/rkE0+Ee++5J/z3v/xrJWE5aAlNOk4n+VBx9xHa6Dc1DtX1phh4OgQqJn5OMtAra0grV/iBck40e/2JVbo45xnXxQjGNZgxLFWcci1Y3sFsdKVDSoac9f3MtZH6ZmLFN22HdozbPPJJnH1PavABRFsWMYEgyUKGDVS9DTc4edz8sHSgvXJFO37okFcxrlWK19Ct8srqRMHrRqXT3QoJTblR9KUYnwgLnbtmkfXriHIAVFeKEy1DWWDLVo1B00E1BkgzGKX958fG+71sKstZIJ1KL13idRYqjK9e/n7lCF1iT6TzKlf3Utjn3ClhpuQR/e3f/LWw9bXXwle/8YyAodIyR2Hhx2qLI38sUZfK0+oiOQBG5in6rJR4z0eLPrfwyTSsM+63eJDH5aXXI1BBnC+C/b26CWKF4EpJEdLExHOmQ06OVRCo1BxKBCId3whJnAVUowTup1Rdp+k/mGhO2wNyWwIku092mpXeAdPr+rZzq4BoZ3eH7YYLNcOTQNR8Q5U53wnxvVIqEYBzUJFMdWl8/THp5uFGG1sWCmBlxU/sZ37YhOWIPTBZztUGbkxQxXjCfbPu811hPIL43lnQFkxuarQDNF33OVUABbxjQsQfU+OzQBr3yuz6ZdkDzp3iyH+L3KW++fS3w3d+8JLN8wQgEjWV1Xmdj44o8IucwgnLSRQAhTAzRybi1whYUlGVHSOyiA+NyaqvMjn2WF0T+VMwJQUffqE19Up/VyNFanNj6JMHhINmrvF0JQZRDE+AZ8OKifNbIdbLk9V0oBxqIDrQmzgrSdQ/Le8BqGnVFWWlJqukfwmYJdDFB2lEln8HTK+TA8YIkB1Avc50lwaaGSB1YJ0F0un25mz9y6IHmOsenWTLHHFPyjLPnE+9A9ItimzWUIHKuSAGPgal1GNpyqcoB6LmJhQ1OMh0yuK2u8WVtogrBTghcpZCcKRmKE/ZvhhMicX3BM/UJRafGKcsmAKi/WJmseDjDtWQ5h1NkpDk/LR0pOLlBfBkgvWoJl9irWe9u1cJqlesUISaptCWfneyj9p4BVmt8+fIgijXDSXc5uTPMQe6yWEpUKcbU1zMAukUO2q22qXbA6TTwwhFjP7ffumbATES9m1coYdM28xcUPOU6ciplGiPbtNF8lJ1vA1f+lTOsdVePFmMA141XRaKjuUAlANyvpY44Evc7xfINEjEbcRDo6dTYKqQUIEpBKACojERDUU0U09qUCLkE2B1IjbfmTpyhzph/IcAUXxRibfHJ5SIpWLkGfOJoyfSyV2eevt6LdPTPFn2+/vz7lTehnN8bDvA+jOwslR0Zx2O08R3rSfLvEM9+4vRTIAo7c4CabHevczK2to7Au465QigmRP5EJareynsY8ZPQIwMUZ2dneE//9e/CK/v2BPF46eJouX+gyWfX41CG9GXlpotNL7vZnGy5aKaqEuf9/YkkWKAcBKlVBxoqA9oYjRy8GQ93nYmLGI+dZTrOPPgR1v4aDboVP0Dco2qlSVf0xpDgGnfoHR6yubkhGXf4+xJDM28SJSRzMTUAUokrchyA8uhKoV16pSAJlRJOn6delD5SD1ayZ3tx0Z6dVTeRxQQxRDm8fd9Ak4zMG1cZZ4URFs5WPq1xUsHVQf2eJ+vn4n47u16G2eynAXSM+m1S+yYPnEbA3ppGZSliME7mdN+qWMvxnK4UO73a1//Rvj6d58LbXLryRL+pPQIvqVMqoeudGRkoZhVcatFDETx8fPkGuSAF5f7uvf1YF+7FwmglCCkNI5avbjNeJ2dhQAacaO5M6B/FKql2MpUzZoHRNFVUncyz0eKaYj1ABoEsOINm3gwqE0BaEyDUkMMS4fpSUaGBZhkk4LzBDgdNP0Y305ANTlh4iOagCiAHov0/SODoTnU5vpyKqDmYOncZ7zt6349M7XEoFRJMEAJmgXSEh1zORV39/TLMTz1DC9xYwx8cnleyuRcKJFMh+SG45n1y92TGX+lv8OvtKIyzz3F4qO7zsTttCrjEX1Wjivl48VUGXFbcRtTWXfgzYJoqWOrxFEryN52M1Vzvzjnng4SOBf/iBJr35hx6CCUFPEecJXLfuK8JfUxYIrLUp/0mi6iA5y52HxNo+zk3ClLrPOjafCCZ4FCpI8pp6KIC0usZwHUq00FRGOgLmY88rayy3IgSt1ZIM322GW4XSVnQVK7lZvwDvABHEhwcqlGN8GF4o70ne8+E778zWdlxJioc5vq48WiThb6mLKASJ+Vcn/yviYDVJ/aKkVwnc69Zus4iGbL2S63z+vDTTYoD0GY12xgytK2vYKWJGzWDB4FPqeuIwVQ6+qqgulN5zTrXnUvUgsw3QiE+gCqSKcgMT1pmrTZdugfxiRAFA7Y1QuAKCI91CDVgN441my71D9AshSAljrGywHPYqDpoFpsnx9bagmHGlPhmxLvmV2/bHqAWTQnc7ZnQM/VNMeLFs4Ph4+fvGTuHS6UpCWA2sGDB8P/89//Krx38Ijp+c7uJkpzkg0Nc62vsqJ39nzHjh8XkCTGHTwEpuvuFLeX59jQYbInP5AR56vVB3Xqgzr5zI72SpaXftIJ8BxRNBGcaQymOfFe+8cqpOvU9M4Yn9yRf2ikz2YoxYoPqKIaaCWIQVnyAVUMV7QBsDqIDioQwA1Lfn4/D9twsg6idZX6uMsdq1kRVUgGpqeGo85QzGnG65lqUg8kfVJMz+r7ssewXW5fXD8LnuyDU531I4176TJeP6UsPfsOHCsrisJZkUOzrq48Z3CxdBMAii5z7ty5ptv8q7/5oia9ez/0pVE5+I7WNeDuA+rkDStTuf7E2MSgzIvDgKCHcBL2OVluV7jMtrYTOZVKFkRjEI7X/frgOAHMiZxnYvF38MTpH06PYKXTnafC69v3irfrDvddd5U3ZWCHFT8oeqn/tGKfiB5VtiWMQcwX34xOuIr7Sw4hLym60lqFj5I2zzlUzwQ1ls7LRG3OPSppxy32o3K4d/cmA011o3NrJCQhlh5qaVIUVr9mfpWO9KVXtoebbrrWkmG3t53UxIUJB+kcqB0wjX8AYzEwLdXEZPrP+DgHTi9zcPV79PLZ5WXYA4jqR5VKrhyZ1V4WbrLO7ztYruaF3wfQIcaTGOQnL70UvvTVp8OuPfuVS7NGM4nOD6tWrhB30xeOnsTQoxhzcVRnm6jEQZS7J+yTeZmImipHpNArB6DZY7OgGW9jSU/iz3WUnicfEITq4yeOh4OH2pVA5Fjolm9s56mecPWaFnHpN4Qhi3BKwI7g+uoKOcbLP7ZPv1TzaWCq2VNCMyFRykeaUJIcYFjJbmrQHSuzPhGkDqBwohip6sVs96TWeyqgL0UnCoBCAGf1mDjWShmYtHQQbahNEJt56xvEir69852w793d4Y477wnXXb06nO45bdxpzmqWXFQuIindLFhkucrsdkHlzIaDY1YP6iAZV3fQze6bBdK4ly7j9dOao5y5hspxUnCld95+c3hh6/aLsicAUFyaEOO7NU/TF/7uy+HFrW+aThdgVeBkOHz0uIVmLlEi6/VrVoX3Dx222UarLRvT9DjTUp2w8cqrxeUlYFGsDtdHX4+OJtxXsTrZMgfNQi40P8WGGEYD0FqJ7nMEoodlTHtbU1SfOCn1gQCwWlnvIWY4rmpUdn75khba3223/Etxf0rBNF0HVAHTenHREAb9xib0vwJWC78UTKQGKUCU8FIio8Z1hlFl2h/SFM7D6RwnGJYA0FIEJ5qlOcoK1TU0Hn78/AvhwP5F4UP33GUeJKXUUTG3WQowp6v3dBB1oIyvMQZa9mdBlLqzQBr32GW8TuYj5hpiLqFSIICIuf6KK0KTBmtvKoJdTF0CWEIFjvVWQtwMVGmcKq5M7x04FJYvXWLc6b73DmgfQAPHdXZgigWfua+KieNq3AhO+dixY2FQUUJO5ep7nfwyQSKfp4hyRPlmGQOhnzz/XHhzxwG7k7lzUV8oM1OXcpHKwdOT2pOQRLBq+yrQfwr00JUSOgqYdihpiJNzqAMS95v17KG+3hSGdQyEjydEQmimZIZQC8jFX1ytpjGpHQk9XcwQmnf4hwOtFvcJN4peFBB1H9KhQUIFEhpF7ypdMjml979/Mhz/yjfDffc/GNavX27nG04NO6MyXHEdLvrHIIrBalRo78t4n58nu3RAdBD1/V6e3c6W+36Ws0Aa98ZlvI54z1xDt996a8m7dD3p6hWLw653Lw753rlQQJQ0bv/1z/86vPrWbt1DDIjRulZJSoIr15Fjx8MVdTVKBzc3tGvKETjFMyUX0VsVAVWOq6d9QPPE8aT/pgeg6ESLsHMmys8LPbJ2P/fcZsWm615qK+QEPx46OgftI7Jk4ZwwX2ntdu9JzusT3SGCy2IkFywMRHkwrROYDoozZQkBrHV1eRCkDA41D6CKXEJUVzITBw3296hsTHpOHPKdqhXp1C+1RkzMU58D0RLcvHBUYJ/c1/ee/WE4fPhKawI1SmNjU5jfyrNcYGXdmhkBYHMQjEE0Pi/rDoBeNy7LrrMNOReabE3+3/tk8pqzNS75HnjvwBHTHZK1qJj/o+tJb9t03UUDpAAiYPTVr309fPvZnxR1rI8fTOIXiqO9EllooA8OJiGJ8cybcf1i6w6avozrrF1/bVn9KH2Lfvbw8e4c18p1cA+I727QoU0X5+P2Yy7Uy1vl/H/yxMnw3PObrahZ3vWdnSc1dxOBA+MBEF2xZEE4cPiY2fLn1FdL1NfJhgWEzOWkjw6eDXCmAGOtjpHHkhFg6gRHCc1pSjjfAV2zE37+gCjg6eXE3wOip+ESNTWJc6MjChfFiIRrE/rRBjGcTZmP2PCorkvz3WO5r5K6Riyp6UAB00ZN3DEkYN6x6x0/vS1JDdDcMi/ceMN1YcMVSy2kFKOiAyRgOhVyYC1Xdyp14uO57ln6gPTA23sPmAGilN8i3QAI3H3nHSbeX6huARwApNbWVnNp+pP/8z+FL/zjtycF0fz1uqjPbJTijpThaVwAcLaEX+l1195ofVSqLfoWI09nO+qE0hSDqE05LCd6ljEx7VRzS3NoE4j+4Ic/UgSW5qMX19nWJv2rKtbI4X6BRHvKTnX3hS5xp0vFdG9ctiR0DiT3a36i6DqpL59RQkMRzd2f1DlS2695kgDT9lPtSqmYJHgGLD1hCUtAtF+p72x9tL8ARA1AxYki0gOicKGQc6K2kW5j7QdMoSWN+Xj+YXHZgGiV5teCO+VXp1sxqBWud3R2mC71h89tNeNWs96RqYIe9fg58I6nU6nYRehfse1smdfNLmc50myPXMbb6D1f3fZ6+Mhjj5a0OCPeL168ONx8/cYLZnRCjOc6vvDFvwvP/OjlaTnWw4miIx0Qh3TFmpX2NLs0F1XNDBibFi1ebX3Dx6YUIYaS93QEi01KLt7H3GiyCzic6DdZrY8IngZkij9y9ISBaH3DHMtUdUrTf/T1J/rEWrkhefIPxH3842+94YqwQOnvEOPdAd7BdHx80LjS6hqlDhysMV2pX2PMmTKhYEc7+s360NycT+ZidaNbz3KhbpWnHiDK8U3KeerX6G5Rfk5fLhAj2ddUEY71SgWhQmGdCC5a+UYHmAZZorYAtVLsOnlYcfXat3+/fKOPhcceeUB+qK3htDwoHCAnA9Z4P0BZUZGofHzpZb7N1UxGs0A6WQ9dZvtfe+Pt8PCDHzaOr5h477f7xOOPhC3bdp7XKCd0mPx279kbvvDUP05bvcBsmoBclbjCDevW6B6rA4amGsXSM0CnQ4BfIedepUz795VtwhM9v7dvb0G9mPsETONtwUauLtxntWBjWC5EPfKyOCC3pt3vbDcgqpFcC4h2a84k+MtGGW5GZaBxA1wQt7i8uSIsXb5MIn06hUfNfOOOmSFUE9ELXFN5XsfXVcrrIDI+cRFjStxyOjIWoeeEy4wJcMyWsd9B1LnQOWnGKVIe15nonm+lGKCuml8bTivJy2n5odalelLF12oSP0kXKlPSUbMVKseMcacoHbrkavDNbz0TPvnxx8Kc5jnWZ/mzJGvOUdamKQOdC2avA6XXiY8tVlaqPuWXLZDCmUB5LQ9DKSnjC1dY7sMs2c9xlyvt2iOXmRMnNEfPwqJ6Uu4bMLpi7Zpw9Ya14U252JxLQozHsZ4sTei7/ux//GV48dUd0/Qa4LklSUfmyTVo+bKlApwuuUK9L6yoM0f6hNcpfyfMuDkSAUlce9HC1rBs2bKyYj0fAfr25ImD8aEF63kQTQAUazx6YLjP9w6cDNu3v2WcVq9ydKKiXL1qkYElfTOQ6jMXSieKmN8VJWKpEbfWuKDFuNERJSqxhCU6sxmbtLRoJYEp7Jyk5xy3Snhn2+kO6U1lXVcI6bASOxunqVz7gGaWANFiYEp2e6heH62YPAN+XFZqvVk43yt1Bh8kHKyGpfCuNTcEieQAaUQ8cXj5ft3Md5/dIjB9UMayWkki+tCkHCbVWQcUh3TfcTn7IPYBsuyfjIqBqx9TeNdeekkupWeRczCO06ManMmLpzjfSGHutwXHUiuXiwaFqFRpmkIGM5R0ZeED82MulyXx51tffS188hMftz4CxIoRL/OvfPZT4Q//t//rnHGl9DscFb6hm1980aYB2XfoWLHLKVomFaE97wHpJDEmIco3NiIOH5MurdPmZ9LoYbT4N3RCO4kqQNE4AlHAvF7RUCNSThZyoyHc/+GP2wAv5TrGveD2tHPXThPr84Dpp8xzni7OA6LzpN/sVXKT7373WQUVFAIwRqQG6S37paawfqoZCzVzFS+vMrjTmIalU2xJQWxYGemzA9vzjo5XSOUg0ITMT1RuQwaiSmpCtFOrpgcZkCW/RqANoEJuRGLdwdWXlGXBs0bX58SYqo3eMU9mwn6MTXwbmGmUaZsxSqXh+2aUG0/nU+FdJNTWhqkaZITGoxS96fe+/+PwqMR8/Ef7+rpzAMp5nEoB4VRA1Nsotcz2d6l6F305gEjkB47QACWJeldI6b5oQWtoaW5V1IteHD00kseeUH7OEydPSTTo1cs0EuZKLKhnjho9zCQCpji4XPSdMMUL/NHmV8LDDz1oAFZKvGfe9rVr14Y7br72nOhKGRwA18mTJ8Pn/+apaasRTOLQY4J7bpA+buWKZRJjB00tgGGJtpmq2IZc0ccpnZtGJgMUw8bi+fPMKDUicKuSpRkrMoSIv2zFBusL0uyVIj4GRDq98/aOVHSPgTM5Km+RT/bNX7AgHD16NDz97WeNs6JWksJPBiFhnYvtDqaNDYlVekQT0ZUiy/hUZCfJm2NiojzCQselL21J/VMxJGGZV059We7VAxiXZFCKQTUGybi9Yuv1AtABQ79kr6fYKybaA6bt3J9CWOswvMlJf0TcJu+ncah6hu4ZNqZ2nVjjI3b4aLuB6QP33mmW/dPp/FHObWaXfvx0lmN8YDOJbPz4SxpIGUy8eHAkbZ2nTRd2wzUbwvVXrw/r1ipZrB5OSdIgO/D+kbDj7XfDjnf2hVPyM2TwNTUmL1wyCEsefUnvgOt78623wofuusuSHhfjSnk5AZlf+YVPhzd3vTstg89knUNyFMTgZ559Nvzj17+XZKyf7KCC/Rpk+gAOCjhxusev87hE6jbNglovboiUeOWfn9yG9NEkhrxZxqE5kkx6NAE9XOzIsiSbvBuIqmVdfuKjT6YcUaKGKLgUbdBXvDuvvvqqjB4TQ3HzAJoeKXAARHfvPRye+f4PrNCfQVXFuO6hWuJ+IlYDmtXMoxSJzGxnCdHeSOGcBCZALtbbRvqPOHrgWJegD6km6RjlfR8Shzco16Y6s+rjJjVQATT0FIAqTQCsTrGBysuy4Ml2McKSP2Tfg7xRbrw/v84x8ehVtxgXWqw17zvA9OvffCY8/tiHc2Dq3GZ2WeyaplPmoMqS6b2rFi1b80fCo4lPZjqtnue6AGglhgV1fFtbu4l3gOfPfuzBcIuSHyyQA3YlWulypAfMgN64fq0s1FepftBgbAsnNQ0FqgHEFc7BA7Sv3iTNlTvVxbjvVHt7eODeuw0gil0fLydAytzmjUqltvWNncWqTbsMg9AtN1yl+Phvhi8p1V2PDBxTJz13PY0B6QurJMJtWLfW9GIHDr6v+Oxe+R9KpORBFgh+SeumBtA98TzxpQTIF8xFUlGUjwAUDlxOV2GZxOnGRk0OB9KI7rrn0XDtNddoLiF0b8VfAsrhmv7pn76scFR8MRNQA9uSNWsq+ZeCKCGe3/rOD60M7nNeq7IgzanXPdTK7UuWcklYYybaKhuTBqqFPKb6AtaH9SHok5qGbE91MhqNSJxtluiPFFZDWGeVAFNzUeklNpUVhjenSoFkhVyYxqUK4z4rKyU2j2GQU9SSEpVAI3pOVfqI8BuT8atSWZn41UpvWjkmAB7XvE+6cP/V6Fqr0/6BlwdA6WP/IFmjtDvUH4alt6yE6xQHjOMTMQOjMix1yBvhtLCUKZtJ7UfnMY5Rs1A3tTfBtntzBUuew5ASTu9/7z3plpeHufMXarqVLt1nPuFMwQFT2AAo/Xhfchjr/KDKypqRSwpIjQPVQOKlZvqMBk3MdfNNN4SfffLDYdMNV2sA5HUzdodT/McUE3Cwm667UmqAJhuUJ091CaiJ9lC+RA2SCQNiim1frNXaOrrD1etWhtWrV5tlt9R1AqarFRJ5YP9+JT4hCcjZEfOxv7LtrbBHIZzTo5SLVMq2+fPm6ppWWtan/QcPaVxVmnsTDupFoMtOwyDDLWpMvxZZx5n0jufbKYMNCUEY9IQeAqStyuAxrPsmXv+RR56w/oHrLEVwo2+88UbYteOVgirZI3Brwsg3qBR03/rO8wbewH6tLNMkUU48BRJ9ICGOiJFVAhKAFBrLDVwlXhY7hx2gQe+8AelQT6jWva1duThU1iUO9caR5hgKwBhOVKCvFfoMMK0SivHN4B2vSifH41zVgl//jdfIjUmgWWlgJhck9VO1kmD7D1BVQwrLT+4YH9WaukaBcyKWJ20L/GWooqxaXzPulXLYlFE573eIKWrvU75TbdfoY0KaPr4AzF4A8ez8w1Tqg0Y99g2r4oH9B8T1LwtrlszXNraS7NOg9kRy4IyBkloOmtly9l0CQJrcvBmEdMHdcgnp7u6xgXTfXTeHjz12f7j26nUShRBPqFv8S6UdUyIAddWKpeHWm66WyLhIA2g4HBOX2i1dKjoWBlsyVM/+XFO6oHNciUinB+65w774pYCCF56Pya2bbggv/GSL0tQhOp45YT0HpKZDfEDJKDSuEb965TJxbnPC+4ePyuDSafpR/AdLW+WTd2JUAxPxeJ6kEB5ip3xLeb41GvRgDXo39KPOkVaIo/vMZ3/D7h0H+1KDFzDgY/O1rz0lq7oc/3Vj/vN7FFMjUX2OgKNGYbqHw+bNL4QTknwwSMNl1Zs/KK5bmrpD+lo4Tggw5Z0rBqTsA/iJWAJIq5R9qbtvJKxQ9qt5mld+VNxmLQCcAilcKEDKTzBmP85RLbCG46tQPHytpVBUFFYK2OyHAFS4eHSo7pyf7En+A7DY2R0wbR4n7YrDXTHSsc298eOp8G7BaQKkfYpAO654UzhRn2uej16Vxh2RTzjpg4WlnkN8PXkwfS+QMmDpkkXicmslLfSb9BrXZT0Gz+y+qWzr1am5KDlSF8MQrZkwrFPRFiR9XSFwe+T+O8JHHr5XHMkym6yMG+XFldA2lXsuU4dWILWjnkE9cL30rVeKa+NdbFdOz3ZxqSN6Gfji8vKVHrxJSxf7/y59mJYtnhuu3LjRuK5i12svpQYBjtkb160Oz//kFVN9FKs7k2X+DiCK9xMpIxF2/RVrxakMhQMHD5sPZb3KoCQs1FYL/gHAo9KlYv1tlqTR2KBkLDJOdfWI79G7hahaIU5zjJOorgPpHGU+evyjnworli8vK9JzMlyXtmzdGvbt2W7vCe+Kv0nCRJ1/KLS0zrUPwbefflq66Xekj9X0w8JKmFzAoVbZOmAGEHMHZOCJwRQRn23ADd2og2y/6lXquufM0Rz0AqYxRRINKHFzS2tTWLNigcJHldJOxxi4ybAEAaIxAa4QHOp4up6oxNQ3KeBx3nIgyvEAn4GkrsFBMC53T4cYWFmvxpdV5x0a7EuAtFtcKOoDOEg9HwdR/GUR2dWduR+3wnYihXC2QuK9pW9PSvW3X9zp6lVL9MHR/FR6lz0logMonD9lznUWtjT5VmVVXf9FAqS8enSLHraWzOSIzsqAS524fu3K8NiH7woPP3CnxK0FeshJXTtA/wq3vHS6S1qZ2NIciYAb168Nt9wo1YEGYqc4Yiz+hB1iqa3Vj6O4g0uR9u49GB66/y6zENtLphcwS7yUcGXL5eyN2Lv1jV3ZKjO+zTkTg9KQ6f2WKNoKt6ajx09YmrxqcWrwVZ4Os/ACUAPIWCYQa9Azmz+vVYNECT6kzhjU+2SShb1D8fF5IL3/wcfDLZs2meqA6yhGtIdVHTXTN//pi+KsBAIp0TQ/3JvmaIoOwPCfvvZMaDvVbSGPer0NRHlnGOxCIbWlOHa1B4cWgymc56i+FIj+DqKcBtEeAOADAwF4XEtLQ0VYv1pZkxRjDwGkDphZIIUz5YMC1crlCWYEvSw/jFwG0GqXSKAR3a/rSRHtISz8vt4gbl/Mo3HYtlP/iMOH4DwhB2fnTMd0X+wb0bM6qb5p6x0L9fqIAaajKoPId5DjRq0EjOC+1K3q4wRe0x2ZBc/O3l2B8JDyDlyrZNfoaAfl1gWIOqAWE9czTZXd1JTOFwtHmhgR3IB04mSb3r0qs75/4iP3hbtuv8k4RO6G7i3+ape917PeiXiB2H/7pmvC0kXzJMYN62vXYWI/OrY6qQW4roRLvRBXeGa3iMdDR3tbuE85IBFRS4n4tI4rysYNGzSHT2XYtmP3mZ1wSkclBiVCC9etXW0fTiKU4EwJwbTBr4FdCkT5IPAuLZRLU4M4vQ4kGlnl4ez4+CXPKHshCZB+9p9/Un1xr33Iy/UF7XAtf/8PTymp8vFcY4x/B1Gm/4Bj/c53npGKqN1A1Cvq8nNAwNuCkQmHcu4Z3SfYQ4gkBGDyZrmITxlASrSTlxnoaYqRKhmGNqxZKtcm1F15IK1K85VSlhibkKjyQJrz2RTa1gjIOL+7WQHGRCe5jtQBlQQmY9KN2nb6IYnB1DlQB1LODXk5S/YN9neHo6f6Q5ceKBwtgA+Q8r7xDCThm34UAKWvAFHj0lVeDkg5l1NiUByxPKdLUH3woVb7Ts6dOrh6eXYZA3C874JypIhviMgYINyANEfuR3feen345EcSAxIcYQKfXPbZC/DxzZ/Ruq51oeY1uvG6jTmxv+1Uh1n7x/TQG6RPTKz9cpXRCYoP9jM68zk76NCRExp8y8LatWsNQEpxYbz0vNhXXXllkIeOuUXN5EUhjTCC+6XGWbxoQVgp0Zo4efShxI2jDzQuNNOvrgbg2BEBTLPAC2MSelW4UHRtlRgt9Oz4EBenivCrn35SsduPGPfNR6VUP9AHLS0tYcfOXWHryz8wzhMgMjDS+4wul9G+bOliBRq8EXbKxY7kGxAAocPzlIJEtQAJ0K9X2r8RgWF//7CBTCU3x1WLq4U71S0Y90kZOl8AFMCDI62Vsz160lWLFoWFcq8akSUe0HH9qJ+UD9EYBiKRrwOqMbACpjAHjE8+GmpFIKc4KK07qHI8lnp+AMyInlGlrhEw5ed5RLNAyjU7V0qO0ROn+8LJrgEbL5XSeZCoGk7UP2R6FPbs6EH7qW2kUlWZEpD6czQx/733Qs9QlTx2GhVNJu8eeTjgfwwlHyy1GXGrtiP6FwOt1+O48wikuW6wy8INBuqQ/ybGHCyxGJCefPS+cNVGNyBZFf3zY3374li62O/W/t6+gXBcRoRCa7+PmovzHuhJOLQ3lePzw3JmRlwtJ+L7oECvuqClPmzfuWcGdKYyVOh9GJKxhmdtEUoNjZpC44j0412ae0m+oRo4pThJBgpqADgn3iNcmrrlDsXHGTG+vDFKnJ8Gwq995hMGonBBk4EofYRz/lNf/EsNeo3yGBm1zpTILUqk8fobezUVyhbThzLwXVylzxM9qO5Jr4cwWNYKMlXJ66RC96pXZUQfAa4FsIjBFAMTYr77SAOgEPpT15O2NlYqQGGF+kuW9dTnFC7PCcB0itddBcA+zpO4XyU1WXfRnHPGiT+oUak+d0Blm9ylrjs1Pa2ejb877AdIiXIaV16AdhkNT3XJmJT6cGOx594hQkP1+bB1/nMfuEdB9A0SgAOlFZb4Rx1+WPQB1L3KtwuAtrY0KunJQumJyTolY2HanzFgepMAp6sB2O91KDtPQMqN8/D0kvE1FIeA8QbAWblssRmQnnz0XmUzzxuQ/OIv5iUQyZ25tf8WWfuXLJhrYv9xzRfEB6JSogpf9uSF5Yjojb6Ibg4R/33Nwnm/RHw4AV76Yi8oZb4PznRuS0N4a+deM8Kdye3AhdI3+ES2istbs3qVRSuZW5P24TFQHEDpSTghcaHShSJCt7bMkcg7ZNZ8ngwg6lRKMqgXF/W5z/50ePThhw1AJwNRuDOA4Ut//+fhtOL5eaKGhn4iLWsU37/5+RfD1m3bLes7Az8GUT/E8RcgFbKYjlTSPRsJmCr+OwumLuY7kFIbAtwMTEd7wqCMiCtXLddHEWkuoRhIXbSHG41/1IRTRWfpNEJsvvrXwFPnGGTG0BTkvE68BFCNO02bcDClTgyogDJAOjDYK+ajJ3TCdeoDlOVG6RtEe947Rg734TYS40i1v9h7yvmy5Bwu5bTbJuPxu/sO5QC1qXmeLkFgq48Y4OhA6e04yLLt+5wrPS9AyguPuDskruGE3IkGlOVlwxWrwicev1dc0B1mQLI3x6/4Elkm38X4YvNi/3VXrrXB4WI/g8Z8UvVCaPWiJPxE++XAfJfykZbTlzqY8mICptcpGGLPuwcUcls6hJIb5j2AjRjT4BSDJi5AE0OICx3U+7BKIZ74WGJQOi4deb3yiKInLwmi6kQMYIjEC6QLdcd6vDuqhUa86BwLgJYC0dUynv3+73xO81TdZiqNciDK9QOi+Ix++StfDYcO7qUoeZb24ZFIK+DGIHdS7/iPN79m+xFxHTApSB0ECjhScItfS3OjGY+4Dn4GsDomBlMirfAt5WPmYGoAqoEPOM2p1UepZzAsFEe8ZPES1RPzEulHuQY+XABoTFXyHEA1YaK+n1gVspyog2GpJSALmFYyt1P6ortO1M9XpfmkquTDOjrSYz7bxzTJU79UF5D5j2ppH3M1MKyHZwCqMr0O1m/Ug0wY0HiaKvHexj+O41YB1H0CVAC0GIfqoBkDKeu+zf5zCqQOoB6BhN7lxuuvCZ/86H3hQ3dsssiipBPo8al3SHLMxf0fMW3j+rU5J3+s/ceOnVA467D8BmVUMJCAS7+47nuvXqi5sswTzTNoonbxfuaF5GV3a/69d90aDh44WNJpn3cB0AQA8N0cVj/gmYHBZuMVK8V1jIf39iNuaU4hlaVjsMjJdV6hDvq3uTjWN88xyQb/YgCLtksfm2/uVkWz/dvf/20DPsR07oV7KkfoRZ974YWw7dXnTA9qOlEdh1GJ5CMYL/a+ezS8tm2bAKLfRHoGatwsZ7BfJOqzn3q11fLZlZ6UaTvQhxI1RDnkYKp5kqWqUEhiBKYu3gOkg7LWVwBUOn758sVmOEpaKPwPYDp4ssfuJa2CO5SL9YkuMrkI2vdzOXj70lt3bjXLlVbVCr4dWeXMDzMVFM7adbIjHJM+eDhNpOKce4XmeLLoprRhA1P1EyoeQJznBWPMCIIme3ZJrcL/MajyIQNQd797WJFzPfJOaQhzWhYYhxqL/IUtJFvnAEilR+ELYtxA4kDf1dWj6AsZkG65Lnz88QfCTddfqa+6ixw8IH+1il3ipVyWvHwMbLf2L9d0EOieTigmnH5BJWDuU3opxp1VucC3zFUjqq+T391kUU9+qQAuhqDbb90UliuS5P33j+ZCP90QBIjCza1dszqsXCqdlLgWBvMyOUuT5+DQkWM5tyYQMctFAsRwmCSZIU57fupYf0o5FgBlImAqxZ1iMilHS2Uo/OVPfyL87Cc/ZqI/FvKpDEKufZsA8sc//KYAATlUwCfXI7L4Mwhf2/a2HO1flGFpTw5EdRv2dlM3B6bp684+J/bRBkEDtVJlQAApBJiiI0WMLQemABoE0BGCOTLWHxbJ7au5KZOY2WolXCkOuDGYprts4UDKRhZMAdSkPFk6uFIWA+lY1bCyOVWZ4WlUHKh6TJGrCaCSuAS3pnZ5v3T1DRqQuljPfdIfdJ73E91WrQgwKAFRvQ1WJymbyjO0g0v8c1Cl7VMdPWHPe3qHT3eJ6amxkF3URHCsxQjOdAY5UvSf+mLpTEQg9UrntVj6wg/dfqMs8A8Yd5ZEIMWXknRCXHL5rHNv0f1ptGDtx8l/4xUrTLQ9daozdfJPoofQHwMWF5oIOsD4dNWGNZp1dEnJF8ivk5eQlx/Rd8P69ZI2bgldysKFIRF1DoYgfIOvvVIzlOqjClc6V/HkNYrb3iO3pl4ZhurFhYrdsPsvBqK0AdfUKg4Uf178eDv1nuG07SqAcj3X0tQY7rrl+vCvfvtfhGuuusquoZRRze/Ll3DI7+zeE7799D8Y1wkjMEcBCj2KRHpz++7wwgublbH9kHGSONoLz41b4vHXSD/qYEB7pb6XlMOBjcsCT6ABulD/AXuAKfpGjFCAKro8LPlwpoj4MbihFqhXZvoGuUAtWrBY5+caYHD8jpIlIn6paIYYSKmd3Y5bijlVgJSYfWZN0dXlqrGuRyW3I1Q7xOUrhZ6kj24FSJzQtChDunkc7yHADE6c+4QYRYj1AHqOG2Wf6nBP1D1bIOU8UAyonZq+5b2Dx813vEk+s+RAKAaoM2K1d/GdrymcRa+iRjAgfeTDd4bHH/qQGZCSya2SC539r8nFNBA3rl8bbrx2o2UeIuHGEc3Hnoj9ysBjYv+F7SmMT29s3xWuXL/GxF9Ebl7wUi8s5ewHnDD8fOiu28Mmic84QB89cUrier0G9TzNVgkHyCiosHBT5i7HKm+jYsItM4TUpsRAOLWFijbjHJ3i5hMuFC4sqTPh0LQAAL3vjhvDr//yz2uKlYdzXGi5e+FQ3w8nCoh+59tfUfIW3JPqZPVt03Qiu8KLP9msLPZHcwBqxmRdji7RfnCaDPaYKOKX/Iv2qBBASKKcEid73+sGJsGQgSmuQXDmDqZZZ32AdFCuT/Ua+HMkKSxQwuZxkE2UNTT5OTAyxXpTB06MTb7udYstHUwTjlRgmpwuV7VKiUarqqX3FohWoT/VDyA9JPe2Tn2QxgB73RcfZMj7kHXwH24UEPX9w3KRopx+nEkgpUnIAZV11DTlABWOtOKam+/vk1g57WwfJrLpc8CA60pT2BGBdPdt18vyupLzz1LZHmCE2ZDSWzMW3t69L2x5fZfm9T5qlui5chqu1qC1l4V29GaVCoUse5qz3AkQ/dbnPm0p99AlMkhLgamfChCCO+XrzYtPzs3vPvtjc0citLFP4jQ5Yg8cOmypDB1IeaeMDIFwAUo4FMJ14bpwrB+UXtXSlk0ixiPC36QkNI8//IBltmcQAvLTuX5AlNR4L7/4fXX/uAxqR2zqj4Oae51PAc+GudizHDT3YECgZcI9JkaSnP5P9+nH4Abl5QQMwWEtkQoIB33AEhoWmI0MD+ra86Ll4GACOKTbwx2LnKWeco8ldom5cxeGq69cFW5eslAeARJNNVZjFydrPP3nfqXsR/3kBJBOhSrGBsRRk5gkSck3Opg8TACULPxOcKvs6+vtDAcVIXioo10/DI55lyfqMgmenxkOP56kD5UAl8iUJMMCZi5xsnfSz3+mS56/07y588It16+Sc38yKwA5cacNpHCgXDQHg9SIaldtXK8EwFcrOcBiP9fs8gx74JhCIF9TCOZu+bl1an4e+rdJoiUK7fI5Ns/whFM4DBehz/zM4+FjTz5hBii40+m8uCaq6vo7la7u9TffVKz8obD3vffDOwpPJb7dnOXFuhmQwtlKtQCIYLDj3nGsPy1XMsjrZi+7VkCJ4Wm9IqE23XCl9LW3CEjmCsgIuSycdyh7bLzNgEHfizj/vWefDXt2bbW8oTt2vGHJg6mLiIrIjojqgOjjjG+Agyh1AVJImg19DJIPpwNnvO2AOiSAsJlBlb4Qoh8cSK1A/wBU9ImIwkh79QrvJKMVelKPRuK4SmXKX33lTeGq+Q1hlXK1AKajCpdEL8oyS4Ap2aEgDMNZQEVfCwGuvu7bAOkQnKZmLAUsY/Bk5lIvp36/3p+O7oEwoqmVX9rymlRAp+1enNukfxzL+bDQ1zEBnuI99JwqxNXyDAr3x3Vner0YoLbMXdA/ZSB1B/rObriCIQvZvOnaDWY8akXxbwRqn7+bSk96mSz8i5f0H36VbygM8/Ude8JJpQzEkto6VxmEpBe7EIDK8ydhDIme4e6maqSJHw7HweXB2XL8QfmtbnvzLenUpYt6/5g533dqUAEqzQJF3KNwrDcxHiTSgHFjUpP0iLg9XbFqqSX8uPPWm8w4Bnfm5/CBGV9DuXUGCcdz3NPf+W544bkfGge6XxwoZPrPlIM0XafKgKPCJ0fNtEw7fR9lun27NwdSyqAYUAFSiGlGmGYZYMyCKUBKKClLMkah9kBdRDSXc6WDii0fVG7U+nkbpGK5MixUghDAtLlVKQLTk2TB1IEUdykH1ORqkv/OnU4A0UpZ4AWuDqTUBjyLEYC6/9DBsPbux8MRRax9/e+/GKrn1Np9eH2AstgHiP18vAbVr2C6utPWKZ/Oh536Z0sxoG5ct6w0R+rcARfIwEV854Eul8X15huuta9+nLWbbps+hCadLc2blPC8kjSiHorXk9IPxP+kDwt7kq298tF8VVzquzJo8Aya5zQkLkIa+A4s56uDcBv6tV/6tIxQSw0MpyIqZ68NQIUQ/fGtZYoOwBWCezx+8qTlCW0/dUrcCOGJiLdKMq1EzPPEqS1RCKRnfgI0sZzjOeBW1TMBUFQRgOixY8fCM9/5UvjiU18tyYECos6J2sXpX8yRUobezojXOn2lKcpypg6i1AVgaduBYuWKReojMhblRXzq4Z4DAaZMiIdHRGNjrT4+LSbis495nhYtXmmSY8vidcapjw2eCFdqTqZ5zYmoPZByp9R3UHUwpSxLMScar4+KG3Ug5Rgy7JNcGlGfKaChet3CgB7lEemUF950a1i87Mrw//6X/yhJo9tAFICsTDnPIaYY0TG8JXCdMbEPamio1vsnDlrr5xtE7QLSfymgOkcaCnSkzGWDAhVfr95+vaBixXHhuf+uTdK5rEvALm7tDNfpEiJTht76VqjZcGeo7jgcxpSgtmLdfdZiYRee4Ukuo8MQ+7fIzeadPe8qHdygif0NMuxUpR+7hP85972G3vTXf+lT4e4775y26FzscQCsgCoEkLJOGW50MSGmA5IApgMp69MFzrhN1l318OLLL4evfPkfwvPiRJmdEv2nhyM6yAGggCbfer75LlWWBNL0ZM4b+LmdO/Xt7JLzE5+/RPrNGEypBzfvXCnbrKNjRF/aNGdhqFOmfICU4+66+6GwZft7Yd2Gq6VTVJLysZ5wReNAWCXOFBY7y526rpR2nYpxp77PlwAr4j00rrR9vu77AdX9Rw6H5qXLwtW3PaR+/lJ465XN4kabQucp+fGqX/2jAliCDfFHh3Z4BjDTcwSiqDSYjvlCgqjfm8C0v6p53uI/QZwiKW2/Qjb5MX8NFuQmfaFXr1waHnvgDrPAL1y4wN4cbvJshyttjJ/Q9BVv/KdQvfudMH7N/fKbOhrCT76sh3sqhNalobLW/U39kj+gSxuFybxAuCQxNQrzDLUrIQcZqHAFatDsl0kCbOvZGXhCpft6UOC15dXtYb8c6K+/ZqM4ocRXEaA7E+KrDmfLD6Id1gHJ+EeZn8PrxyLWdM8NYAOicMT/5c8+H57+/vNh/943FPetCfUEYgaY6s4cSOoEvPf2E4gazKcDwQHVlzwFk/t9vwoAXgwntGeWZlXxcEeqx1QjFQKAQv5V3AYBRQxudv/4ZkpvPaSM82zDmdIekzdaFFNFvelND2pOsjtvvyGsW7M8vCk10QJcoZSntEvO7r19+hhpgr8G4vZ14nGdzyObsN4j67iFH7WSO+mjO/V1rpdtvDDGFASAT5/mUrXbqJC+tbZKHKPcuE6P1YaOwZrQLS711ns+EmAIvvfdr2l81ypRe699lKpSbhSw9L6pSf1GadBBFJG+Tq5ITFdtaOsdbme9MP/0iEeqP/Xxh9PEIX25qyBUDRcWEik3ivtIiFeDH1a46VFyVP7/aO+JMLbtG6Gm/c2kLcYhrhF6WYLWaw9vCRWntoShFR8JNdc8rAekkD9O6Z9167zpXsX0rvmiqs0IjIhncrciw267+Xqz9r8pB/r9GjSIgK2arpfIKQDmXIr9qHtefn17OPAnh8MTD98TPnT3XTa/U8ItTW7Zj27nvK7SLy7Gn5Lq4CcvvhSefnazTcCHUa2vX8CUuSLXh1LsnFNsaIqrx6Cbd7lQjegRwmjzKmNQIXF5TUZ89fYQX7FQY4yZp+fqRPo8DFCuJzXuVGho27LuQ3U10pnK8+OV13aGX/ilX5G/ckc4eHR/uO6qdaGte044rjptnT1hYfdwWNY4Jr9YuXaJBR/XOzSoi3XrvnOobAOaTvE6+/BnrUSMlxM+E+yNj1QpO31VODlYF4aaWsOWN34U7pZLHLRz+xthRLpvuFEA0gGFPuVbzMhOtT9Wn39pfIJCOBX5JXWGbv+i4Eb9AquvWLu6f83YWIFo7zsLl2cOXByJHnR0sDuMbv+GASWPRIlujMaZ/2wkb1mtSLwKQs3e74SxjudD5ZqPh/FVd+sA+QzwBvK2XgRfouTqL9x/uJQb5OLDz639u/bsD8c18Phqm8FG/XQujVPHlO3qL//+G+G7P3op/NSTD4UPSdyHQ0VnOV3r/rnsSQDUpuWQThawf/ZHPwr/9K0fhMPHE0MSb6gTfHElA1wUg6gV6F88+L3Ml95KqdHiQAuwVupV1p9ZnrNiLO0Bog3yv8U63ybJo1U6YneLwh0KAjwhwNTJXKVGa81Y9cbr2+QHvCI89MijoeMbX7MZc/Fs6BmU5b4yBVTppxeKQ22uVyJqAWKD5rh3y/64EosAjE4KpzCgZZt1p+qKkdCfcqOdip0/PVAX2saS2U9/8vwrobL/VLh+UwKkRyTi9+nQKrjKIsSHxsV8duMKxTNBpId6pBu92KjqySef/CPpQ/U0ErjLX2CpVyFfo+gaQGcgl3+lWBvZ+Wyo3vJnoVKdWCmgVBRbntQv4+vuV2d3hHD0rSDJxIhl5YBekPffCmNtz0uZtUy9uTj9EtEqokRaN11+UBfu5E9KP7JNdZ4+HdoUx8y8RHXitM51bP9peRm8sm2H3Jt2yRl7MCxauNCs53B/xh2L1Tjf+iznPjFoIcZj0HpOUUh//oWvhGd+9JPANcfE/EXdpxQYga9qykFOGAUqKPcdp74fk7yh8Rny61aHf6rEAnHWuS4X91nCfTUpazziPaK83NmN+3M1iLfokU3Vmr/es+o3NLYaZ/faq2+EG268Ptx62+1h947XQ58s+nX1ebUZ4v6poUrjHjtGKkKHooz6dCYCIRD88ZFHB8pvQFyjr0vuUFTSaOiW0epob3U4Ji70/b7qcKhbQRMatq1yydqz89Ww661t4TOf+UxYuGiJXe5LL/4kHDnepvsgYiuvQqEfEL6AD8pNzBec6JSmammWpNzTI7cpXc/5fpe8n0ssR1IgLTYdM7d1JsTrw9uBnkWdJD3o0A/+W6jZvy3ImGcgOqHVEkBq9fQRckCtOPZqGOpw/Wky/zh1Eos/5zzTa55wRZdsAfH7a1YvD3dsutYy+TPd8ZGjJ6VTkvFB+4jtJ5E2cxVlLc8zcdMd0je+KjB9VSnk+nu75GGgcEpZ1lEVAWwYkVzPORPnK9aGG6vwBQVwjh49Gn78/Avh//viV5SVaWs41d2tw+J3hXdWCYx1bVkgpRo16Sv4L2rGR2qzJJWqRxtOvLLxDz4E4AJU4coAlN7eEeU3bZYevF6O7KflH0qOVcT+REdKfzqQ5sNKpSKT3pGJ8U4oGUfnqXYB6R1hw8arwn4ll6mqUKz7qLhSnYwPiP8GANGR6tDZr58MWEcHKkLbiNoYqp7wOzZQJcCsDkf6G0LXaI0d19ur2UzrmsPCuQqrfWtLeOXVbVJLzAuf+MRPmYEIt77nnvu++aCjB4b4MHGvfEwg1gFRluxjltX5C+bb/XarL6YCohjMaT3+lfI3LVY3Ps4uqjy2OJCOp/zhhMOtjen8s4vVSdGDjr7wF6Hy9e+FKk1uVSlf/QIuNG60HJB6PdQAkjCqu8TRnng+DMk/e7R1tT0c3kTjTcvfrLf0wViqL4jtv0mGqSuvWKKXUgOhrUsZ47v05iqbugZY2mtpf/DsZ45OC7jfemdv+OFzLylF2Xuhu1NhojJeYonn3GRSAgD4uYUeoJ0OxccBnqgUAE6CRdrb28MPf/xc+PrT3w9PfeXb8lfdZR4oHi1FFqFCUqb3CEjN9qEqLtrbyGA78ytsoxBoi72O3KG1lTmQy3EPAdAa0HYwBVAwgFUpfn6+3L9wlEd8Bzz9h2jPOtxoElLqQCwXJFnT6Q+CKgnV3bn9tXBg395wk9wYh8RF9vd26HwJqHJZWPcBWOXpsl+l5roHYIfHZOUv8otvBU/fJjFMb23bLOnkLdtF1BXjc/MLz2u21S/bDAhwo+hE0XXaU9c/7hN1FYl+GuplxGLaEfULnCj31a7pSKz/inVsfBFap4+zVApIi9XNHmsPfkJhrmCk4k//9E/79DJPQUeaO6jkCjfpetCat7dYvQrpyF0XSgH60Hjby0Yf+aNQdepdpdF5KriO1BqI/8XqALnSjTY2hYrlHw0Va+9Up+f1OPEhH9R1e+EyNw83sPX1neHtvfvDMYXnYX0lGoiEIqMWHM0rxY+jz4yIfAMEsLYTAz5Pvo0QjvUQs7KSi3b5sqWKiNtgOUhth/4BpPhyIopzPCAbEyAMYKJ/Rc8ZcyZtin9/Z89ey2dK1NTBwydCr7wZaKdJYjET4AG8fZrFoF3JYogUq9DATYj71UyeavuQrPZdnR250E8HUurFHLxzTVnsj/ud8V5uf9zLWVznOG6fK3RjFFE8zUpxuFhzhmFYxAcT31EMTzEBsoj4OOhDHu20b99R87vE8s0h111zVbj5jg+bvnRQ2eGdxoYTlUdljRuafc/EpdcFyEk319g4R324LezY9U4ugAHAxDcW4pk1CSSHpbJwv1GeC+5MqKdMv6u20P3iI0vUFtQuFymuGe52bFTzO03i+gSXmSWm3C5GxerG9WipFAhbPbk/ZQ2U8fFntD72ypdCzX5Z4/NGRgNPGgNAsyA65ZPEIMpBkuyrhjU49345jGPtxxg1S7keNogEJwAALulJREFUKPbKYO2//57bwj133RL27M3H9nMQ0xXnrf25Zqa84gEco3LBYR4e5p7HRQvXrDi8c5dCX/lB+KPOlTN9k0IXV69cHtatXSVuSi4+aoOQx9aW1oLzt0s8JXGz19m3X4Cp6Uh6lc+SKUm6MzpP8pbWCRwXiTOH4LIwwDHjp0VQwcEV6yjVddBksJtxyVqY+C87NouMXzsoBtWJQzwB3Lgt1oX19klzMZdtAGRo8Jj5lxp3KkAFfPgAwc0BsDXVyTonrtR6rX5jmvfeQyoBbaK0ALvOzrZw131PhAXoH2WAMhJHWgxYk52l/+Plc/TgHmsXYzIfG4i+5Hx8lMg7C9XL57VaEVRuLLPC9B8+sV3dgwaiDfJWOHLkpIHo7bfeHB55/Inw9a9+OXTp2rNUDhA5a7n92bamuz2jQGrdhsxdhABQs85r37TBlI9lFkg5R1qGy0byyCicpcl6AH/Eq6/aYD+s/S9ufSvs3XcgHFEoaosAFX0mnMPUrf3qff0hNjN45uNvLDolNQKWe0R5RKOsOxbAx484+ZPtXUqpd0TAJyu2DBXzNIXJQmWLiqlN10fCE4g63Zrqg5SNQygVixDpAJlQDrUG3BI0qrTqhJ5avk+57SRQZbty/5y/s4GfWu/ZCUg62GXBlboAhr+HXo8B7GW5E0Qrvs/F+rhd2mA/58VBP+bGCFv1MFIAtFuh23SNAyqnsHBRLQFTyFQhZv+WxEAuUsH04aPt4QfPfDPccd/jYdWyBXKNSu4e8T5H8XqusHBlTt1YTidKr8JFx/eiR4HZxKQVzw/gLcB5Jin09JGQpwKEmxMgC4jihP+Ln/2sdLyb/JBpL72fp33gFA+YUSDlpQklnOjHTqgjxaUON68PtSMS4adIo0q8MFq9NNT2pMfkbUy5FvA/neyFzVWeXSnoARLN/LTcllzsf+vtd8NRZfLHUb11botFTZHerBTnhigPFzokLhSjUpYLJQ6coWncQIm3GSBs6+yyX8HFneVGrT4YzDEEiAIicKSAKh+SOt1fYSapwpPFIr3vKfeOOQdLXQdRW/eDM0vvCtrMivWZqrn2brzhuvAzn/pnYeuWLeG117Ya54dj+ipx88Tln5IhFnHfCSBKRHwJcMr2XlV9QjAqoBIwPfbIg+Hw4cPWRodUGc999+/DDXc8ptSOG0ykhjt1rrQAVL3xdEmdBc01ORD1fATsTjhQ3Z+4UX5Wpn/9Uq+MyqDF8yCunr6I57Vqlk8rKgv8ZwHRdWuVcvKmm/ShUCKbnF877RviWLvn8p89qzLn4ipmFEizNwPnCYBCI8vXh8p7fkXGokNh/MV3p8aVwkSgprnnN8NY17thfPffhCp0bRkwneVI6eGzIxf7cfQnpd+2t97RNMjHTVQs5eSPOA9IAZbz5yZcKI7fQwJWdJmQc6GlgPjsrrr80RUavXDJJ9s7wzxZvdEFc32oG0gQPSrDy2k4WulcjWtOm0PANe4yHadw2SPpgC93Rh9rXt/rxsDqZdmli+8cG3NybBuJM967d5d9tG67447A751d28Of/tmfywr/vmYeWCX3ohXKB3zK9MfoLAHZujplipLonyQzyQPPhg3rrFnEe6K40L9ueeG7oe3QO9Kb3iefzUaVZa+ycNtBlHSIWOezU06nioJCQFWh3xORWx5Lz4eusVERWwJ/rv243KM8rn6BJJNazcNVUyMxJCJ7TtH2uVxNn0LRU7DvnAIpIDrUqgQTNz8ZalbebF+eYSV+nvJJU+lC30ZZ/a8Nowv+JAy9/d1QJSf9qr48oFpEVNFbvMQLZV3HONIvv0bciACsc03lnPxJ6QfXiVEIgILTJPMX4jNhxfhpcjx6SQfQc329ZdvXdTIFCdmjAFOMS2NKQA5hcEIvTAYp0vR1KVSRxChQdtDEwAZYlgJGLzfQVSPuzoMdD7E22zawVuxc8TkcYOH0EMP37t4RrrrmBmuLJXpDQGyP9M7kMSUHaZMYjSEFvyDuA0r8bF0p8gA7AOqv/+rzUuHUm1GNc5BXgOe5b/9+WfiPhTvv/FBYvGJjzhCV5UoB0dYGpjY+GbY+/y2Z6gqJvnDY5qMkISAp0Drnn9NMkphg1+CivYNot/SjZPkzY6I6Y948VDzyMUkNyuh7IW/fNi7wv+z9z9jljJ8WF3qzQjyf+LdhPAVRGq/OfFXKnjBRa1kVOg2RrOa6j4bx2/8wDM6/Q3oCFaoOHGlCF1PXppd0FotjJ9pkDGi3Fo4qAgfx+3wSYv8Tjz0QfutX/ll46N7bzC3G0igKeLB6L1403+ZPgstLZu+s0cs/+bxJ5/MeUDCSwzSx0CfrNr+TkIOpSrh2aJGS9DaJQ4X8LYrFddsxjX+AoRMgyqb9QCsRQJMV6R2w2ZdVLTgX98orryYNpFd59dXX2jYWbYD20KEDZlhqVmJoxP36hjkmJmO8g0NF16rmpWsWp65pkB38aQQwAFAxaD3z/R+YuI7uExB1MZ96zol2SB2z+QdftzZJssz15+4hvX/uHQd6+tKt9HzKEO/r6hJ5H8s8elPi7js78yDq7nBLlizjtDkiReDFRlNmDqd64WMdR40LrX7sd0NNXUv6xaVXeXzTJDFgVaO9YVBhpeGGj4cqtQexrLrts1Ib3Gzi/qjSil1uzk9dmoIBWr5ilS3hSNvaTphujw/K+SQX+7H4v/7mzvDtH75kM3iS1q9WLk4Yp3Cuxjh1IUT4Un2Rv5Z0VNs7SA4CSM7q4pxJIs3cWXD7ixfOk6+rMvF3lmoxKXeO0cHNwaPUUX529gMsMXhlQTNWKRRr78gRzbSKKiKVTq5Yv8G4TACSwQwI9vYm3OmS+XNCg37sQ7RfvXqtNXno/f05DtLvhR12bi3hfgE/ON12GfgQ9VsVnjk83COn+xrTicKJPve9r1rbLtLTBh8A+pd7ZsRzf3gbSB1qkgzbwCfGLjhQyABewA7A8mbHbm3svxRoRoGUCKPqh34jVDUoBpREI9o2it+c6faK+rpOCUzCj/NJTIzFp22J+2Otfxgqc3H6ZwDW072e81Qfx3LPxckpaxWVVFktR2mJaVVV517Ez94mBqXX3nhbOVEPhk997GHpRJvDDze/Gt47cCickE9qYY7U9Lmfyccze+JzuS0UgYMGlLg/vAxIWsJAByQR0QtAMLoWM6SIA3Mw9V0FnKgKefUpi4HF69KGc70OoM6JernXZeni/YFDxzTn1xrbtUARP8uWrzGRHEs5OQIAQbjTU5oPi4gg4vNrxXwQ6dXT3a53qVo5TfV+qb4THwO/F/w+4TC5f0R9OM877nlQhkSpQmRY6u/vCy8993QBiPrHxMGYdmmPcjhvQNL7AGBF7wEHGk8VwtVQJ0vNmhq5GJWqX6zuuS6bUSDlc1vdpBAm6w51CZ/fsyXEd4xLWtYe/k4YlX50/Obfs/MQLVENl8oPYJ2J853t9c7Q8Q2yKiPWDw+3mN4RXSlTu1Du3MgMnWpiM7kPYPL83laE0gtbtsvnc054VCkVGbzQpz7xqFLQdYc3NOPojt3vheMn2u1aPZN/OWv/xJOe/5KYY0X09zBLvxL7/us19sHNksFr4JhWcgDxY7JL15Nmy+NtB01fxvvidQM6AdPRQ/tyQMr+VatWGuCxDhC7vhNOFEDFAb++7qQkxOYw0NeZgJu+Fg6eAGf8zXMRnKdPW1j1Ac677n+CU5i7FJyvc6LeTtwXfDzYZkl/yZaUrGvbP7OmZtBO62drufi/+qa8G9z42JA8MaQ3FKmp80Kcxx3y8RTgHcjSzAJprvVip8rtnP4KYIoUoB9W+7HeNmmpBdj+lGjxMgJRbgdxGuPHyRNHjROlDIfnbhl0enu6FBG0+NwBatqX+Jg++9xWM8Y8+sDtRSc1xNjkTv47396bs/aPymWq0Mmf4TPD7wWdMmM09WEZv3bx6SkvSbr9cYFS3AMAp3NtJY8rsuPggQMFpatXr9H2CwZcfgLOAwfLNWG4IWPSNSubw+Kl12kK6R0GjuyH/DpYdw6R9ZgAzi2bf2hFACuZmHBfgpyTtY3MP0Ay9zHRdQypAN0n4jvXOF0aJheApIaLjc4RkJ6j2wRQY6n2MgPPbK8BUohjzFCA3yO6UQAWoxPGpzq5hJBlaaZ1prT//ee3yq3mULj79k3h9lsSg4aGXHqJE4cA1xCn9MPJn1DUU6mTP9Z+KAlFZW1iG5ReCmS9oH/u+1gKDL237J4iEHXx3e+V412NQK9kOTTn+Lw+etJYAlu+IjHG4HMJOHr7ACRtwTnSxq49B8O7+w4asIGh4CD5UCH2A4uEjS5cuCicOHZQH/C8SN128nA43tZh3G0MohwHAdi2xgfDSvL/2Ab6XPfpy3yN8mtDqadF+VrJXvrPzx+vT+XYs6lz8QMpnGgyRU3ClQKmIjrLEpVcwgPSbmSSf7gT8YsJMF0jCzMuPIcOHwkrVyyfMTB9TtmRENOJif+tz/1cdO6kx+PrKLXuTv4u9r+xc+8EsX/U2Dd/5Uu1dD7LGXYTyUAtvUwW8RU7iPjSATAHKjSXMk84pFMOd8aZHIQLjo0b51hRltsDALuks+TZu4oF6YSoJzhFJ06Lny9gyjlop04XgP6U86On1F9OR2tcow7iw/3TP/NJZcw6LlexzrBy7fVS35wO//d//N+t6VE1iH6V++EY8gFA6SLZKPJ/OuDpoZwAe5bwDS5nECzShdkmprxdKja/WAOFI7RYjQtcNjqmRAbzr0sMTrhDpRxp8df+Al/s+Ty9uHEfSCeVsONsp8JGD/rcS69bco+f+8TDubbztzT9Hnex/67bbtDUywdM7N///lFr0sX+qYeh5q/k3Kzlh6DfKZwdQOQq45ypu8wF5MRYryO04ngPkYxdnmLO0b4rOgYw9vN6EywdDNF3Iqa3nTice0bozJcvW2xA6twx91Ap8ET/yT2YHlRlHO9EuZNf12uy1D/08MNh2bIlmhCPXAcV4e++8D8E1M1KhfezcpQ/aq5RsK/eFPrDqClvcsaWGFjPhPJPdOpH+31M91jvi6mf6XzWBDhF1Tf/fBi554/CUPX6MB67pvjbl1T7QP7HLcooN9on6wbQIc8/oAf92y89HV58dUd4/MG7zICUAPR0X6XS52WgI/b/0j//ePjln3si3HDNBg3sEeNSB5RwhKmek+m+Z+6cpa+m1B4fQuoeVQG4DICmeElZzjE+Cxoob8brOSfqYOb7Oc6NPXEbvu77Dh8+5kW2XLFihS0BTAdoB0/f9gM4p5/Xy1iiFoAL/MGzz8rkkDgU/vv/9d8rU9jp8MSTn7Sqt95xt4VsskGP5XvNdp/zf0yzUoxm6jp4DvGzKHauYmUXPUfKRY+PKFmsjEvjD/1uGN/3fLH7+MCWEe5oxGidEumVEweBHpTpQY4eOxluv1kRMjk9qDcyU6+mt5cs16xeaUYrzk9Kv9dl8T8iMZLYflL61Wk0M4nb+Y2MYujk+w8wQYTOcZf5XXYT9EyWc3SOcRwLeIaoaw1q4fUcqOEF7LsWncNBNm7GAdjLmK4jpmbld4UASAfOeD2uO9n6W9tfN1/VF55/2bjcB+67NzS3zFWI6j75NS8TJzzPvAToJ0R2RHFu0e/c12PRuEp1fH+p83MclK03zMwZRtqj97xO3geKTUvLLo7FJQGkdJV3LlM1W4dHXNXF0ZXn/yoAo065Hy1bsmjKJ8dfcvNLr4XXlOj4yvWrw7/4hZ86d9b/MlflTv7E9u/YtVti/55wIBX75ytZCtn8se4mgFpqiJU5wVnsijCtoBVeObuSIhVcpM69p9GRgCXEguNjMKXcMFxtZ6367HJQzQIpjvIxzZNrGsS5OMd0wNSv3RrQv/rGueGU9LBbt75oRVte3hyuu+7qcO/9D5rHyO7du6zc9Z5+z35/vu26Tip7mR1Y4l9cB76gSn0CWJMHwMhujkCKvETlTcXHetmZLum/qbbndS8ZIOWC7fbSzjSWwMuS19u2Pgj/iGwhyglapjDOxK/UH33SU8X6IfYH/dXPfNLi5JNXhmNLH1esrbMt8zNiSNt047X2O3DwUHht+25L6XdSzuRzbXoN5dLMmTJAML/Ps72C+PjCe2cLcDHROB2zubOmIGrgGB3mgOet+v35dnZJffSmVk9tAhzGuWYrRtsOwBQRD8+HlA8StGCRpi/XUt9JhcPql16wL61SiX9+7XZPqjNPk+x1nTou17uE68Pf8++e+sdA9qkD+9+x6Ck4dgyGnNO7wZd+mng713++s8jS63tdX+aqph0UexPk9s3gip83zi7lfqSlTnPJAGnuBqYswuaOuLRXjA3idYU7GZOTfqfSv/UqMUVrCoTsyT16NlLKl6EHfeZHL9vxE/1B/fX1487PsthZXex3a//2XXtCm6zTgG1+RlS/r3N3nQVnoOszDJDvB3jcSONX4/fldSh3gPR9qAx4rNShedrIcoUcBzkn6mCXlOpdkI6ZZDYOpExmRzSSObnHJ/cDSixp312v4ntBB4s7FdeHU363PAKY94ptU1Pq+gHTcjSNy0iaUeP0kQmbfBDSxvv6PcdxUoJnAUT7XscKZvgfbfPLPP6Cs/g9XmJAei67raB/Lp6N9MMBuOCIz4BZsnSFBhivtMiBVsvtO/doXqZ2m8oDUHI96GEB6Z2bboz0oOirzq2lNbm4M/vv1n4y+buTP9Z+MueXSul3ZmcqflQ8cGxQR9Wsu9lOuz/aVbBa6k11EPUmnC8AzCAHXdvQvwkAmvptJrk8PVkP7lTVJpn0a0K8LLlhKQbrnP5X7dk5HRF0cEND3lmb++B4IpMcOO0eVI4Llbs/WT2V+f1oNU/q0Kj5fHlmjb6lnrdhfaiytpMKwDGyvTa9tGZXTMsujsUlBqQXR6edz6swPWiaAapoNFP61j39vedDW0d32Kj5kH784rYwd+c+m35jxdKF4RMf+bk88KYXfyn44MZO/i724+TfpYS/OPgTQ46e7lyFotpATvtrSkAQVYpWNT11IZCwz/w40xM4qMXnK/eOAWhwnoQML12S1CSVYWUVDvSaslkNuUjvIEotQNnF9+y58Gt1Kz8cH/H0EAAakx0vcLN4ee2gHe4nd7/aZxR/aLRu50uBMq1RdkF9/6j0kTLzPJPdk1/AFM593oHUUt4pVHaq042Qd3SEpCTE03+AiLl32k+d0ss9qByTC3IiXLEu2L5jt0JH+829iP03ydXov/3NV8PHHrm3ICY7f2x2GOX3XKxrWbEfa38+k3+zuU/NpJN/jhMEEBwcSnSOc3o+7hysvHoOZNICQKjUE+BYb8ePZ+nnSNYBRV3WUD4bPomP3VHdQTQ+nna5jmLnpT4gaq5VAmhmYW3UxJIQIO8iP/WsjRgkrVb+n+3Pbxaucd+TgKldn+rQ5ROvNSlpSefyok4G5wvPdx63zjuQji25KlTJe6HYbKIF9y2vHuoML7gx1JBN6oNCkm+IWsnrQUkCU546OrvDmpVLc5XQm61XxnRS3F1KhA6Y5NBkviI5i+v/4ntwsR8n/53vvGvW/vePHLcqeSd/hnPxoRi3VW7dwTAr2rvYGR+bA13OyqmddAnxJsUcDzAZsGV2sungkQXU+BzefOFS1uwybkEGzpnzFQNs2uzvT13q2EjRykCUi4va4Hi75hRYs33F4RMIMFVhqbpR87lDT544lByQdv6c1iRpTq7CRbCSdsH5u5LqxoVh9CP/IQytuMOA0ifEC9VJUl2uBKd7ploOt3w61Nz7a3r5Lrdso8X7Gz3ogfePWLIS9KCAxlTouqvXhW3b3wkYlaAXt7wuMO4Ia1YtTw8v9nomg8DeaL3VVsPe7uJ104bO3ULnPn7ssIEo+j6s9qg1ShGeClj78T5wJ/8R5SQgA9VgOkEeTv6ESfKbLpU8BGDxX6ZRB1EDwzIgSj3a91/cjJXxT+Rgnl23nfo3pPmoipE77bOvGGDHx8DpIv7zKwbWfj3csoP7BADWTu7ZxX07Ju2jUoBpfRRdSDkuvev0iLLq56ObWubkdbhRExd09TxzpMkbQuo7EjOPXvNoGHv5qVB95F1lkpVuR9NVBIn9gzc8EqqufixUarBkO/yC9tY5OnmsB827M039ZEQi3XfXzeHbz76ovKCN0p31hU88fl/C/iQQWbQx05NqVLjhaTz94l+IPocLx5DmYa/o/HDxaiRrfXpdRW9ChS72048vbn0jvL1nfzh5MrH2z21pku5Qr7mhUopQCU9UqrmkHCAowmbELXgDsdgNyHKoU64vnROjgRJt+zEss2CV3abOybY4zI+S4sQpDQSL7y5aSkKcmGjDRfy4PLvO/VLXl+wHTK0f6E/vHPrD16lDxUyZ9b/qnO7uML9WD4M2j5XmenPD8nNx+IWk8wyk6a2qZzESTIhWmi/x9LHfDPVK2Jx7AS9k75zjc7sedFCzjC1aUF4POtmlEIK5fu1K4+TySUzKvWbJPv4PvPGNUNX+XhhnpknNRGA5Xic74TnYDyfq5Ims4UQSP1nfU2yZ3AuqgIcfuDvc/6Hbzcl/6xvvhEMS+yvl4eBif+Lk7yO49FvmPRcDQnxm9oMLgChkOK1mKXeyPYCDCgBC41hVh+8C23ZMus+PMcCzA71k4tLOKcORG4S8xphmkyhHfk9xnWJcKPt5J6H4Urj+eNv2614Aypz1Xtv6S/rB79W3o6W1Q8UsRWV+LqZRyRvWxhUBVy9jY5MBafbwC7Wdf3PPyxWoa3KfVh5rwgsRrVTJ07Ck0H4h/kp6d3r5ZbDUvcKB9YiDmqPBv3TxQvVL9Aad4S0CJGv0S8j7r3hj9Dw0tPVvQ5Wypldf/0gYPrgtjG3+fAgP/c/aw37aOPf9T4ABfVFTpUnRZI3HWs98VdDkIEqt+Bo1kVsJJ/8jigbCyR+rdAKoHFu6nxwcqJUlzsirHANR3JJdkR4pXJyJ8tppx2TU1rnhkJ4gu02xg6vrKeNzpodZgu02pbmDbL9dQHKs16GIa+SaaIuN2Krv9VhOsJRreEJEXzlxXQzbLFFG81Buf3qcv+ZebpcZtZkclfynDXZxit7e5CMxPsZ8YbU2yR9Jq9PbTA44z/9zIbDqiPMMpPGdZrrAezhXJbM/V35pr7gYz7Qhy5ctjdLUzfR9Td5/4yd2hgqBaO2Dv6ORpRhmSQJDL/xFGN75g1Bz7cMzfUET2sO4ROYqPBPgyCG2nci1On0qvG8X+9ulc31dU0zv3H0gSunn1n6ZwM9ySDowcL1cQZw2j7KsWGzcKTsccVhPKQZTwKoUxZPCHTtywLJC+YAudRzXZiBaqlGVw9n29aV6yBTkuB/IwDNZnfJ/biEW4+MDc/tKgCm6Uz5op5TtKoRrc4e6U36u4AKveL9f4Mu4/E8P10X88lTcmWa+N/KjMZEB/H9ypqxudHx8yMa3hcjFo3oGLwzDGnkC4MgXRBw5CazHNHKyOVjP+NQgnD4S6F5jsZ/Y/iNK2JLP5F/IKgI4+V4rf/YsaMETxIDjIGqcKexVEfJzcax/ChxsfZvDAEF9f4ziuYx27HjbyhzwkhqF//1Rci7nbl01QU04WXoBzrZDvrqObdaFhd1D9QKarL/8/goOSjfseortUJnfe1aN4e5ZJQ4778WzQHqOu9y5LnROc2WFb22d3J1ppi8pB5uMbo0m2z60LYzsfl7TtnSbjrTm6gfC2N7NgVlgK2//heQSfOTN8AXhXUB/FOPIEetzUVszcV6QKaKs2P/K67ssVypO/gvmT93NjsHvQOfGIwa9D3z2O4AyYyaEOiGLRw4iDqB0uYMcR7HfqZQYzv42zeoJEb8POUfMOu3QbmwUy7YFiPJ6wAGS77QyjIRqXazdo8psFlDapcHCLqUkRzFnniucwkrJ49JzHT6cZLuqqKyx1nx6FevntP24r6ZwyhmtMgukM9qdhY0hTs60HrTwDFPbAjoNPDWaRntPmKcEAFp758+E8QVXh5FtfxdGnvnPobKpJZkFVt4SUALAUzvHVGuh2oArX7NK+TP///auNTau4gof73ptL7YTHOKAQxOSlMRJQ4AmEBQDeRS1UhtSWt5CLeJHf1CkVuofWrWoqH8qtVJVqUUI9U9/lCJVtFWrFkF5BBJCCiSA84I4UmOSyE6ah+MYEyf7sPt9595zPXv32t6179op2kmu773zOHNmdua7Z87MnAmBXKk04opnw35Kx7s7DwBQj0JK9mbCufkyDHrhfAmUKnn6nZ2AZc5AVKVQRoJTa0n+s8Xj3cDAwI5+pgd1adKfIMvdwZyA4RI3c+5Q16qV9JQ4b0hnNA1QFSQBoHRaDj6ALsttO5pYDsbz8Vm/GQYaxhspWJ5hQHRYIPVJu9G1rV6utpaUwjnrI5zvpDMqIWFUmaxOSkhejVJqDbh6UJq4K23CpFTq5cbTZq771DP7X5Cao3sksWSN1K/8qhIiBiSwFM2OzuYw32uqXgdDFyk3w3Hj80A/6oeDnjdu7OkJ5HpdG/bverdNdrz5hp6+aeDBOgrXgr0TrAwbFbgQl+8EH5usYvqw9SB3zSVpWVoDO3ipM/qWh/6afub79+2XjlvXa7yWFu+kTQ3309rNaJMGny0PklEwtYi8g1nTZxK0LxKp4O7acqfMX7BYtr/+MlZEdKkRaPr7wR5e+y+kSz7UsfAWyfcq6+anDVu7ap07R8+i8k4iBUXkMxGYBjz5DJDPAl5LYIy/o7ULN3oVSN3aiOHZdH9TXc4UAys+CV0tKvltT6mkkfjK953lTV7TUvMlvkjhxbYmFh8XRimFJU6fQiLVVm9ijAXO8J3DfgITL5oc3P3ODvngA6hA/M5MTGDNqMOLSXZaW34AgYkd2jobpVA1fAwUs45sIEo6Jrl6RJEWkQh2Bpzm794N/Lq6uqS35xiMLS8Qd+KJ+dOUnuuMV/Mbkz7SGp82/L/n7m/I+vUbwFNOlrc/Js889Ss9SE8P2vMBzAUxS695+XVn+ZZ7N1o8xZTHk3u73UZUDTMHem/O3FudTUTb/ZiZiTzS509n+UxEY6xwto2qi6kGOKFEC01cxxm1vTGmbMomkz3woqZJ4YSBJDZDeI3GmpCPAAHV8HsQEMsD64UL77mDi/V1qbrl7dfKtx5+RH78xJNCC/E8OZOYQIBkzREAA8kOz9RNqh7RBw4CqA7l/fi4qf6RQ2YCpV0EOF50BATSNjul+uz7KbhqrFHw1fWVAxMvyidd49UnofmQfpHzEYFpaEaPh+otW75SDn70nvQc7cJHIiM3rb29KBm/iVHfxcg8ilJHe7BWSNO+Cb09xzUil0AxYP78hf67V2fRVCb2DfOo+U6crCBGVSItqI6pvXwy+KmCRKyTJVNjyUt9tldqFnXoM4fwHL5XQv9ZKqucQefiex4prbP2nOSJ6oWlEqxgPPJ69733yZ1bvi579+yRrVtfUimIIENnRwhRp2rASX+TePhMCTQsedLflRINROmvw2vc2ME9iAVQ+L2dN8Y1N5Lwjkw+evSIeZV0J12HTFEa4ikBjCeTUqfddnW7Cglcw8kD8AJnHw54kJ7xG6bNn9eVWoP0EQ/Gm9IDfaM5MHBOY+ezFzCqqhNXnREmYzTC/hO9B2tDJ4oYCmd9VV1MNUBDG9QBXnKuZb6M9O7x2EKL1nn7Ult1hQpDvSQldzqeP0SVyKXsqOe+ae1aefxHP5XHHv2Onv9OfikV2tA/zD87MydCXBCl3pEXHaXEQBr1vBQ0DDh8r6KbG55OezYqeA69OZJ3wdb8g7z8/M3fvbvNwj4Sz/7h92pEmiqEfZ275NVXt2qSqDyiZvTJr8uzm1/Uc5g9W9Jl51QlU16ZTZ3hf9MKSIVpFARW4KUqkcZYqRy2DsB6EZf3xLVbKQ72UivukOwbz+guprrV90H4wz5qbdlsbuU08Ti4GaVByZ0SX+Zio66xpVpkIpOBo6mn+mRdrfzyt69YJbz4O7/37r9l51vbdckQlzeZo4RCSVSH5L43n8MuMOgcDhjn3aVje+J50qc5ZqOASCbgGN+Vfuk3yinfPMc0mhavBk7Ug1IP+dtf/wJGtWdj7W2fYiX9neIaicg783IBOjLSOJ6Wj51TZUugWq8yozzRia0sDOWz6/juhrthk3n2q3oySatpompAARQBl46UhUE8gDO18VFl9+ILv5Ts4d3oSCqX+i08qltFla4yfpT2aJCCIMoJBQIU7RBUznnLwdzuNZkaIM+bt9wlT/7s5/LA/ffI1W3e7izyTXou4NkzJ3BsEsfi8T6WUxAKBRqw0JujIErzHIK7LqwpofTLdKYiKAAWAKjNrLv1wDiMT2nrPNaWElB5GqhJqlYmN99SngvyLiGBgfBZbGjhihj73bgum7rrKMcheh4X7/ZMdYtd5mfhdrdwVzUTRT/sF81FOFb1vfQaQAtmB+NECq0XUcqKtGxfOsWpx2RLBJjWY5nTMLaFZt75q4x8vFMStzwoSdg3KLdhT52haAqU6GntiXYIeo+fqJD+1ANRrqfNf/SKMpKEFTLWw2QdPwTubL8tEXL1qFF1TGAzfwMwex+LF4YbgM2G8Q6qSDjcpnOlIotDf6oSFMD5TI+Qs7wLvAmuJOjfqS/lRboKxqSJ9wJnaQo843lh3vxY0Lyi2ZOYDbsJlJIHh7w998aP3aNytjoyiTscR+sXnqQxHp1wuuTmzZt/giGJt10gHFp9n3QNJDGZ0gxL3slkSvr6TsunMJZbjw43/RNRkDzR+il/qkjSNE9S7etleGhQ8u//HdYLz0ntvMXoIGyq5qwJRXU7i1OhO3i97LI0zAE2weLPoAycw2QHUKABZvXicd6mhNzW38nInGtgqrER9fBPqfnccnxrPKvwU8lnLtY3rrnpZvni6jUAnhE5c6pXLmSHceImqOLicJ8Aao7AxIuunNommF2/apVcf8ON8srLr+iHx0CCdAokUni4oK2Z2R+CX4RTXpRQRCC8WASN45clKJNTtuiU5flqHkjCemOZFy5okwULr/GJ1Ej34W4tu8ULqFulBh6F9esuhXKiBL+BFiOChhvXec5VgdSpjUo88nz25uZm3RI5cK5PMtk8joTggvSin74S2StNBVE+aZ66alSSrUtEFt0oNd27JLf3XzKSukySmJQKXNDzpo/PIG88JDBDQ1Np9fUYuvb3ySefDOgzP1BTcewgeRhmqVlwndSv2oyPyFKsIEhJ7sDLklx8y1RIF6Ql7yu+sFJuv30jzL414hTOU3IOx8EEgIpq5c9B3aVWNZ4VCH1/8onHyAu4rO7Bh76t4X987jl9D4DUHrxokX85SAl+YieG5sn0YID86bCazPgX02imloYJGBdptHkxPMJZWcJB9A9/XIrikB/wy2LPnXOF1qvFOYbVCv853K11R1p0lpe9e4wxwBMqxgJRpmWYhmth6FOSy5VQ5SURqkYarwbQyjip0jqvTWf1qT/1dD3jJYozbLRpBY0LrZ92R+twAgG3iuYPvCSZrb+RHIa82hTZkLTXxMlH+bQ4bOZsMfWnVJVMVX9q5U/N8vbVs98PN89WmwNcIzkGDpTPuJ+C/K/fsEke/+ET8t3Hvqez/eyoBEMO/am7tFl8V3dqGVp/trv5L1rQqgatd2x/Xb3K7chWTrsbXb6zjlI42tl0kxYWdbf6VJQj0pXpmF+Yh0gSfgGPfNxVEHzttUv0vSQaBSnjfZna5z1eXj7z1LhzhvpT20LKiZWZ0p9yssm2hSZgPq9hC0zoYeE+h7zZhTdI+vrN6FFsvZ5OMegwM/Qrqf4UOlTaLwj0p/g4leYMHgCa0BHzRAYeXJG6eg3EoTrJ7X9VRlBmPludFI6NS8tl3FioSy7y58UybHv9NXlv99s62890VKxwEodgqjpIgJJy7WtcbJG+5bGsfZU+BsY8LGCcO8HYaDPaaK2MJqIfL34AMhFHO4/G9J4MbJlmPDdeuNEYL70CNSJ8fOyU8ERZ2kmgW7xkmW4aoP50JttodWivP8f0/tHhPvSn1P2dhrHhCxcu4CzxtA5np4sTr9Gheauo470lMcy14X5274s63E+0wLgInAcw7A7+FRaRNFbl/1B/2gjDzIMw9EujHXUY6rM+ix34hERN6Y//KGnnMKTPdXdKzdd+AFFwRLLv/Enyh7ZjvDhf6lff63fEwjoppjt1H5bBhv1zWmZhm+cJOY82wGE/L647tZ8Fr+rcwQH9OtZ1SFtbK3YcHdQPC/342eO3z9LbT8Qw/sK8G2iZH7yKHNe+trTMhgV+HP3j80JadrkJXL5c/1KfyddETtfegiduAOOHoLmxQZa1L0dZMlKbqpe+MwTXo1rGMD2+82Ld6JB9oswmF17VkU6u3uJJxQmUy2c1qf70HKTTeCdVSuDReqsTlQZFEgtXQ186F1ah/iEjPZ0imIxK1DX5oISWrJKqk2iaHzlhRx0kQZRSfbT+1NN15S8OSLbzzzK87zUtV91tj0gSk0oJ6Ih1gmnxGkktWqedTYsRUSeVKh71vZw42bhpoyzBMTHn8XE4dZrtwJcccSeo0bmABW9ZsWKZLFy4CPrdnBw+fEw/xhRew2CnxWF6XCbV8dEFHAUb5KOz+whLpWrk8pZ5MoDtpzS8YjQK0nEYr0TpO3nn8jEWFY2DvMgH6+b0qeOyes0t0pD2ToPgyoW339qpxfOrq4gUaVSBtKhaPkMeaB2UUDipwi2mnJDiTH+0lDUd5fbW2glm92sxuz/iz+5n/dl9mG7yOiFFG23h+mc6GCvKw5XsOSHFiTyujOBEFUWv7IcvSX7336SmuUVqOx6W2rYVAQ1K2DX4ONhM/cyVwmNpbuuVBbP9/WdPyoVMTiVUApYBBPkkhumky8qV0oIlQP04jpsSGSdtWHRzBF/89xwSBc/m59+D7yLTMg0uftSzHNrDzyEZPDMO/d2wENn4X5EZT4QdQr2kIJ6aVDoLxzMfOdKtHyGrJzdzJPHK7n4R3AhTf65KpFOvw3goUDqhlEUQpZQ1c8ulUB40OOskKrk5s/uC2f0EZ/cZR3vTtHalyMqmZM+VEVzEn8GRzMnefZLv/ItI9qIkOx6SOkiclLStTMqxU8aZL8FosWy2/1ZYoOKwv7/vJHbLDSl4EhAooVIqa2pMA3jXwsAJz7sakH37D6q/SbCkyG8df6IxERRBXl0wtv+M+KRPEGVSrTP8IQbxIk175p3gPR590o3LWb7krw9rjdeuXQtdrrdd9Fx/v3QdOhT8xpYny6CSKBNXzlWBtHJ1OznKJmXlYUqIw7w89u5P73IpNjj/Qi9UM3AAoSSG+4mI4f7kShl/KnaW2jzWxO56VoZ7DsjJK9dJ8+o7JdXQjMwMEuLPt1IUbdh/620bMKGyVIawseMM2wOLAke1cEfHOuFhn4MYgu/duzcAUk5MqZSJn1GB1EtS9NfiBAEASfxXy/hNs1qwv/5ioLc1/S3DCWR20Z9+xNOJljEhyqSdwqD+AQnkOQSdcvvSxUJJnm7OFa3S+UGn+ptUGkSvLIgy+1x11p7VcAk66n14oiYPg6PJOe+YklkepyoWWHOZDua9mXub3c9++Nro7P4NWwKBxBrudHDEPIgpzJOTDpn3n5eR491St7RDEsu/JFdBOj3T18dowkP0zGCHevyf/bHZfi792vnmNtn25g4ZhBpoCNs2aSQnA2tI5bpgOM+EREI41icdQTIh56WxIQkJPydp7DajpDwLV6I2jXw984f1qWEA1xC2Q5+SHMG7gg1AeQOf5Nuy6cEkHW0e8PfH/BMEjgR2P/llqCQzXhYFfz0gRcdMJGsxRVd1l1INJPCJ57lGON8oTVDgVxjv+J2mC0Sj86m/7suS+fzN6eFdz+vaU241rWsmX9Pvcif2p7nltaZtsdRvflyStQ3KB415sO7Onu1P//dEj67hhd+M8BhXrbA8997/gGzYdEf60MHOIbQHtaGXyWSH8j4YqlV+IA3vY0mjijGGmmQO8S0uQYrWrK5bdXN6y13fxITTgO7nb2puGrPusBwt/czTT3ML55C/WiuuIhfTAd/GPw7ES6OPsOzpk6exD39IK0H5dA3IGPAWE4vBp6aGv8HQ/wBHmhsC2gZn9AAAAABJRU5ErkJggg==" height="188" width="336">
</a>
</center>
<h1>What is Colab?</h1>
Colab, or "Colaboratory", allows you to write and execute Python in your browser, with
- Zero configuration required
- Free access to GPUs
- Easy sharing
Whether you're a **student**, a **data scientist** or an **AI researcher**, Colab can make your work easier. Watch [Introduction to Colab](https://www.youtube.com/watch?v=inN8seMm7UI) to learn more, or just get started below!
## **Getting started**
The document you are reading is not a static web page, but an interactive environment called a **Colab notebook** that lets you write and execute code.
For example, here is a **code cell** with a short Python script that computes a value, stores it in a variable, and prints the result:
```
seconds_in_a_day = 24 * 60 * 60
seconds_in_a_day
```
To execute the code in the above cell, select it with a click and then either press the play button to the left of the code, or use the keyboard shortcut "Command/Ctrl+Enter". To edit the code, just click the cell and start editing.
Variables that you define in one cell can later be used in other cells:
```
seconds_in_a_week = 7 * seconds_in_a_day
seconds_in_a_week
```
Colab notebooks allow you to combine **executable code** and **rich text** in a single document, along with **images**, **HTML**, **LaTeX** and more. When you create your own Colab notebooks, they are stored in your Google Drive account. You can easily share your Colab notebooks with co-workers or friends, allowing them to comment on your notebooks or even edit them. To learn more, see [Overview of Colab](/notebooks/basic_features_overview.ipynb). To create a new Colab notebook you can use the File menu above, or use the following link: [create a new Colab notebook](http://colab.research.google.com#create=true).
Colab notebooks are Jupyter notebooks that are hosted by Colab. To learn more about the Jupyter project, see [jupyter.org](https://www.jupyter.org).
## Data science
With Colab you can harness the full power of popular Python libraries to analyze and visualize data. The code cell below uses **numpy** to generate some random data, and uses **matplotlib** to visualize it. To edit the code, just click the cell and start editing.
```
import numpy as np
from matplotlib import pyplot as plt
ys = 200 + np.random.randn(100)
x = [x for x in range(len(ys))]
plt.plot(x, ys, '-')
plt.fill_between(x, ys, 195, where=(ys > 195), facecolor='g', alpha=0.6)
plt.title("Sample Visualization")
plt.show()
```
You can import your own data into Colab notebooks from your Google Drive account, including from spreadsheets, as well as from Github and many other sources. To learn more about importing data, and how Colab can be used for data science, see the links below under [Working with Data](#working-with-data).
## Machine learning
With Colab you can import an image dataset, train an image classifier on it, and evaluate the model, all in just [a few lines of code](https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/tutorials/quickstart/beginner.ipynb). Colab notebooks execute code on Google's cloud servers, meaning you can leverage the power of Google hardware, including [GPUs and TPUs](#using-accelerated-hardware), regardless of the power of your machine. All you need is a browser.
Colab is used extensively in the machine learning community with applications including:
- Getting started with TensorFlow
- Developing and training neural networks
- Experimenting with TPUs
- Disseminating AI research
- Creating tutorials
To see sample Colab notebooks that demonstrate machine learning applications, see the [machine learning examples](#machine-learning-examples) below.
## More Resources
### Working with Notebooks in Colab
- [Overview of Colaboratory](/notebooks/basic_features_overview.ipynb)
- [Guide to Markdown](/notebooks/markdown_guide.ipynb)
- [Importing libraries and installing dependencies](/notebooks/snippets/importing_libraries.ipynb)
- [Saving and loading notebooks in GitHub](https://colab.research.google.com/github/googlecolab/colabtools/blob/main/notebooks/colab-github-demo.ipynb)
- [Interactive forms](/notebooks/forms.ipynb)
- [Interactive widgets](/notebooks/widgets.ipynb)
- <img src="/img/new.png" height="20px" align="left" hspace="4px" alt="New"></img>
[TensorFlow 2 in Colab](/notebooks/tensorflow_version.ipynb)
<a name="working-with-data"></a>
### Working with Data
- [Loading data: Drive, Sheets, and Google Cloud Storage](/notebooks/io.ipynb)
- [Charts: visualizing data](/notebooks/charts.ipynb)
- [Getting started with BigQuery](/notebooks/bigquery.ipynb)
### Machine Learning Crash Course
These are a few of the notebooks from Google's online Machine Learning course. See the [full course website](https://developers.google.com/machine-learning/crash-course/) for more.
- [Intro to Pandas DataFrame](https://colab.research.google.com/github/google/eng-edu/blob/main/ml/cc/exercises/pandas_dataframe_ultraquick_tutorial.ipynb)
- [Linear regression with tf.keras using synthetic data](https://colab.research.google.com/github/google/eng-edu/blob/main/ml/cc/exercises/linear_regression_with_synthetic_data.ipynb)
<a name="using-accelerated-hardware"></a>
### Using Accelerated Hardware
- [TensorFlow with GPUs](/notebooks/gpu.ipynb)
- [TensorFlow with TPUs](/notebooks/tpu.ipynb)
<a name="machine-learning-examples"></a>
### Featured examples
- [NeMo Voice Swap](https://colab.research.google.com/github/NVIDIA/NeMo/blob/stable/tutorials/VoiceSwapSample.ipynb): Use Nvidia's NeMo conversational AI Toolkit to swap a voice in an audio fragment with a computer generated one.
- [Retraining an Image Classifier](https://tensorflow.org/hub/tutorials/tf2_image_retraining): Build a Keras model on top of a pre-trained image classifier to distinguish flowers.
- [Text Classification](https://tensorflow.org/hub/tutorials/tf2_text_classification): Classify IMDB movie reviews as either *positive* or *negative*.
- [Style Transfer](https://tensorflow.org/hub/tutorials/tf2_arbitrary_image_stylization): Use deep learning to transfer style between images.
- [Multilingual Universal Sentence Encoder Q&A](https://tensorflow.org/hub/tutorials/retrieval_with_tf_hub_universal_encoder_qa): Use a machine learning model to answer questions from the SQuAD dataset.
- [Video Interpolation](https://tensorflow.org/hub/tutorials/tweening_conv3d): Predict what happened in a video between the first and the last frame.
|
github_jupyter
|
seconds_in_a_day = 24 * 60 * 60
seconds_in_a_day
seconds_in_a_week = 7 * seconds_in_a_day
seconds_in_a_week
import numpy as np
from matplotlib import pyplot as plt
ys = 200 + np.random.randn(100)
x = [x for x in range(len(ys))]
plt.plot(x, ys, '-')
plt.fill_between(x, ys, 195, where=(ys > 195), facecolor='g', alpha=0.6)
plt.title("Sample Visualization")
plt.show()
| 0.45302 | 0.562237 |
# "Wide multiple baseline stereo in simple terms"
> "Or how to explain it to everyone"
- toc: false
- image: images/doll-theater.jpg
- branch: master
- badges: true
- comments: true
- hide: false
- search_exclude: false
## What is wide multiple baseline stereo?

Imagine you have a nice photo you took in autumn and would like to take one in summer, from the same spot. How would you achieve that? You go to the place and you start to compare what you see on the camera screen and on the printed photo. Specifically, you would probably try to locate the same objects, e.g., "that high lamppost" or "this wall clock". Then one would estimate how differently they are arranged on the old photo and camera screen. For example, by checking whether the lamppost is occluding the clock on the tower or not. That would give an idea of how you should move your camera.
Now, what if you are not allowed to take that photo with you, because it is a museum photo and taking pictures is prohibited there. Instead you can create a description of it. In that case, it is likely that you would try to make a list of features and objects in the photo together with the descriptions, which are sufficient to distinguish the objects. For example, "a long staircase on the left side", "The nice building with a dark roof and two towers" or "the top of the lampost". It would be useful to also describe where these objects and features are pictured in the photo, "The lamp posts are on the left, the closest to the viewer is in front of the left tower with a clock. The clock tower is not the part of the building and stands on its own". Then when arriving, you would try to find those objects, match them to the description you have, and try to estimate where you should go. You repeat the procedure until the camera screen shows a picture which is fitting the description you have and the image you have in your memory.
Congratulations! You just have successfully registered the two images, which have a significant difference in viewpoint, appearance, and illumination. In the process of doing so, you were solving multiple times the wide multiple-baseline stereo problem (WxBS) -- estimating the relative camera pose from a pair of images, different in many aspects, yet depicting the same scene.
Let us write down the steps, which we took.
1. Identify salient objects and features in the images -- "trees", "statues", “tip of the tower”, etc in images.
2. Describe the objects and features, taking into account their neighborhood: “statue with a blue left ear".
3. Establish potential correspondences between features in the different images, based on their descriptors.
4. Estimate, which direction one should move the camera to align the objects and features.
That is it! detailed explanation of the each of the steps is [in this post](https://ducha-aiki.github.io/wide-baseline-stereo-blog/2021/02/11/WxBS-step-by-step.html). If you are interested in the formal definition, [check here](https://ducha-aiki.github.io/wide-baseline-stereo-blog/2020/07/09/wxbs.html), and the [history of the WxBS is here](https://ducha-aiki.github.io/wide-baseline-stereo-blog/2020/03/27/intro.html).
|
github_jupyter
|
# "Wide multiple baseline stereo in simple terms"
> "Or how to explain it to everyone"
- toc: false
- image: images/doll-theater.jpg
- branch: master
- badges: true
- comments: true
- hide: false
- search_exclude: false
## What is wide multiple baseline stereo?

Imagine you have a nice photo you took in autumn and would like to take one in summer, from the same spot. How would you achieve that? You go to the place and you start to compare what you see on the camera screen and on the printed photo. Specifically, you would probably try to locate the same objects, e.g., "that high lamppost" or "this wall clock". Then one would estimate how differently they are arranged on the old photo and camera screen. For example, by checking whether the lamppost is occluding the clock on the tower or not. That would give an idea of how you should move your camera.
Now, what if you are not allowed to take that photo with you, because it is a museum photo and taking pictures is prohibited there. Instead you can create a description of it. In that case, it is likely that you would try to make a list of features and objects in the photo together with the descriptions, which are sufficient to distinguish the objects. For example, "a long staircase on the left side", "The nice building with a dark roof and two towers" or "the top of the lampost". It would be useful to also describe where these objects and features are pictured in the photo, "The lamp posts are on the left, the closest to the viewer is in front of the left tower with a clock. The clock tower is not the part of the building and stands on its own". Then when arriving, you would try to find those objects, match them to the description you have, and try to estimate where you should go. You repeat the procedure until the camera screen shows a picture which is fitting the description you have and the image you have in your memory.
Congratulations! You just have successfully registered the two images, which have a significant difference in viewpoint, appearance, and illumination. In the process of doing so, you were solving multiple times the wide multiple-baseline stereo problem (WxBS) -- estimating the relative camera pose from a pair of images, different in many aspects, yet depicting the same scene.
Let us write down the steps, which we took.
1. Identify salient objects and features in the images -- "trees", "statues", “tip of the tower”, etc in images.
2. Describe the objects and features, taking into account their neighborhood: “statue with a blue left ear".
3. Establish potential correspondences between features in the different images, based on their descriptors.
4. Estimate, which direction one should move the camera to align the objects and features.
That is it! detailed explanation of the each of the steps is [in this post](https://ducha-aiki.github.io/wide-baseline-stereo-blog/2021/02/11/WxBS-step-by-step.html). If you are interested in the formal definition, [check here](https://ducha-aiki.github.io/wide-baseline-stereo-blog/2020/07/09/wxbs.html), and the [history of the WxBS is here](https://ducha-aiki.github.io/wide-baseline-stereo-blog/2020/03/27/intro.html).
| 0.654453 | 0.706646 |
# File formats converter
.cooler to .hic
Note that currently .hic doesn't include interchromosomal interatcions.
TODO: include interchromosomal interactions to output .hic.
```
import os
import glob
import cooler
java_path = "java"
juicer_path = "./juicer_tools.1.8.9_jcuda.0.8.jar"
import basic_utils
genome_file = {}
genome_file["hg19"] = "../data/genomes/hg19.reduced.chrom.sizes"
OUT_COOL = "../data/cool/"
OUT_HIC = "../data/hic/"
OUT_COMP = "../data/eigenvectors/"
if not os.path.isdir(OUT_HIC):
os.mkdir(OUT_HIC)
if not os.path.isdir(OUT_COMP):
os.mkdir(OUT_COMP)
files = sorted(glob.glob( OUT_COOL + "*.*" ))
for file in files:
resolution = int(file.split(".")[-2])
if resolution!=100000:
continue
experiment_id = file.split("/")[-1].split(".")[0]
cool_file = os.path.join(OUT_COOL, "{}.{}.cool".format(experiment_id, resolution))
hic_file = os.path.join(OUT_HIC, "{}.{}.hic".format(experiment_id, resolution))
c = cooler.Cooler(cool_file)
genome = genome_file[ c.info["genome-assembly"] ]
resolutions = [resolution, resolution*2, resolution*5, resolution*10, resolution*100, resolution*1000]
# basic_utils.cooler2hic(cool_file, hic_file,
# genome = genome,
# resolutions = resolutions,
# remove_intermediary_files = True,
# juicer_path = juicer_path,
# java_path = java_path)
```
# Compartments calling
With juicer tools, per chromosome calling.
TODO: implement calling with cooltools, include interchromosomal interactions.
```
chromnames = cooler.Cooler("../data/cool/A549_NA_NA.100000.cool").chromnames
for file in files:
resolution = int(file.split(".")[-2])
if resolution!=100000:
continue
experiment_id = file.split("/")[-1].split('.')[0]
cool_file = os.path.join(OUT_COOL, "{}.{}.cool".format(experiment_id, resolution))
hic_file = os.path.join(OUT_HIC, "{}.{}.hic".format(experiment_id, resolution))
for ch in chromnames:
if ch=="chrM" or ch=="chrY":
continue
comp_file = os.path.join(OUT_COMP, "{}.{}.{}.comp.bed".format(experiment_id, resolution, ch))
if os.path.isfile(comp_file):
continue
command = "{} -Xmx2g -jar {} eigenvector -p KR {} {} BP {} {}".format(java_path, juicer_path, hic_file, ch, resolution, comp_file+".txt")
basic_utils.call_and_check_errors(command)
command = "awk -v OFS='\t' '{{print \"{0}\", {1}*(NR-1), {1}*NR, $1}}' {3} > {2}".format(ch, resolution, comp_file, comp_file+'.txt')
basic_utils.call_and_check_errors(command)
```
# Compartments calling of public datasets from AidenLab
For full datasets see: https://aidenlab.org/data.html
```
data_dict = {"K562": "https://hicfiles.s3.amazonaws.com/hiseq/k562/in-situ/combined.hic",
"HeLa": "https://hicfiles.s3.amazonaws.com/hiseq/hela/in-situ/combined.hic",
"HMEC": "https://hicfiles.s3.amazonaws.com/hiseq/hmec/in-situ/combined.hic",
"NHEK": "https://hicfiles.s3.amazonaws.com/hiseq/nhek/in-situ/combined.hic",
"HUVEC": "https://hicfiles.s3.amazonaws.com/hiseq/huvec/in-situ/combined.hic",
"IMR90": "https://hicfiles.s3.amazonaws.com/hiseq/imr90/in-situ/combined.hic",
"GM12878": "https://hicfiles.s3.amazonaws.com/hiseq/gm12878/in-situ/combined.hic"}
resolution = 100000
for k in data_dict.keys():
experiment_id = k
hic_file = data_dict[k]
for ch in chromnames:
if ch=="chrM" or ch=="chrY":
continue
comp_file = os.path.join(OUT_COMP, "{}.{}.{}.comp.bed".format(experiment_id, resolution, ch))
command = "{} -Xmx2g -jar {} eigenvector -p KR {} {} BP {} {}".format(java_path, juicer_path, hic_file, ch, resolution, comp_file+".txt")
basic_utils.call_and_check_errors(command)
command = "awk -v OFS='\t' '{{print \"{0}\", {1}*(NR-1), {1}*NR, $1}}' {3} > {2}".format(ch, resolution, comp_file, comp_file+'.txt')
basic_utils.call_and_check_errors(command)
```
|
github_jupyter
|
import os
import glob
import cooler
java_path = "java"
juicer_path = "./juicer_tools.1.8.9_jcuda.0.8.jar"
import basic_utils
genome_file = {}
genome_file["hg19"] = "../data/genomes/hg19.reduced.chrom.sizes"
OUT_COOL = "../data/cool/"
OUT_HIC = "../data/hic/"
OUT_COMP = "../data/eigenvectors/"
if not os.path.isdir(OUT_HIC):
os.mkdir(OUT_HIC)
if not os.path.isdir(OUT_COMP):
os.mkdir(OUT_COMP)
files = sorted(glob.glob( OUT_COOL + "*.*" ))
for file in files:
resolution = int(file.split(".")[-2])
if resolution!=100000:
continue
experiment_id = file.split("/")[-1].split(".")[0]
cool_file = os.path.join(OUT_COOL, "{}.{}.cool".format(experiment_id, resolution))
hic_file = os.path.join(OUT_HIC, "{}.{}.hic".format(experiment_id, resolution))
c = cooler.Cooler(cool_file)
genome = genome_file[ c.info["genome-assembly"] ]
resolutions = [resolution, resolution*2, resolution*5, resolution*10, resolution*100, resolution*1000]
# basic_utils.cooler2hic(cool_file, hic_file,
# genome = genome,
# resolutions = resolutions,
# remove_intermediary_files = True,
# juicer_path = juicer_path,
# java_path = java_path)
chromnames = cooler.Cooler("../data/cool/A549_NA_NA.100000.cool").chromnames
for file in files:
resolution = int(file.split(".")[-2])
if resolution!=100000:
continue
experiment_id = file.split("/")[-1].split('.')[0]
cool_file = os.path.join(OUT_COOL, "{}.{}.cool".format(experiment_id, resolution))
hic_file = os.path.join(OUT_HIC, "{}.{}.hic".format(experiment_id, resolution))
for ch in chromnames:
if ch=="chrM" or ch=="chrY":
continue
comp_file = os.path.join(OUT_COMP, "{}.{}.{}.comp.bed".format(experiment_id, resolution, ch))
if os.path.isfile(comp_file):
continue
command = "{} -Xmx2g -jar {} eigenvector -p KR {} {} BP {} {}".format(java_path, juicer_path, hic_file, ch, resolution, comp_file+".txt")
basic_utils.call_and_check_errors(command)
command = "awk -v OFS='\t' '{{print \"{0}\", {1}*(NR-1), {1}*NR, $1}}' {3} > {2}".format(ch, resolution, comp_file, comp_file+'.txt')
basic_utils.call_and_check_errors(command)
data_dict = {"K562": "https://hicfiles.s3.amazonaws.com/hiseq/k562/in-situ/combined.hic",
"HeLa": "https://hicfiles.s3.amazonaws.com/hiseq/hela/in-situ/combined.hic",
"HMEC": "https://hicfiles.s3.amazonaws.com/hiseq/hmec/in-situ/combined.hic",
"NHEK": "https://hicfiles.s3.amazonaws.com/hiseq/nhek/in-situ/combined.hic",
"HUVEC": "https://hicfiles.s3.amazonaws.com/hiseq/huvec/in-situ/combined.hic",
"IMR90": "https://hicfiles.s3.amazonaws.com/hiseq/imr90/in-situ/combined.hic",
"GM12878": "https://hicfiles.s3.amazonaws.com/hiseq/gm12878/in-situ/combined.hic"}
resolution = 100000
for k in data_dict.keys():
experiment_id = k
hic_file = data_dict[k]
for ch in chromnames:
if ch=="chrM" or ch=="chrY":
continue
comp_file = os.path.join(OUT_COMP, "{}.{}.{}.comp.bed".format(experiment_id, resolution, ch))
command = "{} -Xmx2g -jar {} eigenvector -p KR {} {} BP {} {}".format(java_path, juicer_path, hic_file, ch, resolution, comp_file+".txt")
basic_utils.call_and_check_errors(command)
command = "awk -v OFS='\t' '{{print \"{0}\", {1}*(NR-1), {1}*NR, $1}}' {3} > {2}".format(ch, resolution, comp_file, comp_file+'.txt')
basic_utils.call_and_check_errors(command)
| 0.193909 | 0.332107 |
```
%tensorflow_version 2.x
import tensorflow as tf
import tensorflow_hub as hub
import cv2
import numpy as np
import matplotlib.pyplot as plt
# grab the model from Tensorflow hub and append a softmax activation
model = tf.keras.Sequential([
hub.KerasLayer('https://tfhub.dev/google/tf2-preview/inception_v3/classification/4'),
tf.keras.layers.Activation('softmax')
])
# build the model based on a specified batch input shape
model.build([None, 300, 300, 3])
!wget -O image.jpg https://cdn.pixabay.com/photo/2018/02/27/14/11/the-pacific-ocean-3185553_960_720.jpg
# If you want to try the cat, uncomment this line
# !wget -O image.jpg https://cdn.pixabay.com/photo/2018/02/27/14/11/the-pacific-ocean-3185553_960_720.jpg
# read the image
img = cv2.imread('image.jpg')
# format it to be in the RGB colorspace
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
# resize to 300x300 and normalize pixel values to be in the range [0, 1]
img = cv2.resize(img, (300, 300)) / 255.0
# add a batch dimension in front
image = np.expand_dims(img, axis=0)
plt.figure(figsize=(8, 8))
plt.imshow(img)
plt.axis('off')
plt.show()
# Siberian Husky's class ID in ImageNet
class_index = 251
# If you downloaded the cat, use this line instead
#class_index = 282 # Tabby Cat in ImageNet
# number of classes in the model's training data
num_classes = 1001
# convert to one hot representation to match our softmax activation in the model definition
expected_output = tf.one_hot([class_index] * image.shape[0], num_classes)
with tf.GradientTape() as tape:
# cast image to float
inputs = tf.cast(image, tf.float32)
# watch the input pixels
tape.watch(inputs)
# generate the predictions
predictions = model(inputs)
# get the loss
loss = tf.keras.losses.categorical_crossentropy(
expected_output, predictions
)
# get the gradient with respect to the inputs
gradients = tape.gradient(loss, inputs)
# reduce the RGB image to grayscale
grayscale_tensor = tf.reduce_sum(tf.abs(gradients), axis=-1)
# normalize the pixel values to be in the range [0, 255].
# the max value in the grayscale tensor will be pushed to 255.
# the min value will be pushed to 0.
normalized_tensor = tf.cast(
255
* (grayscale_tensor - tf.reduce_min(grayscale_tensor))
/ (tf.reduce_max(grayscale_tensor) - tf.reduce_min(grayscale_tensor)),
tf.uint8,
)
# remove the channel dimension to make the tensor a 2d tensor
normalized_tensor = tf.squeeze(normalized_tensor)
# max and min value in the grayscale tensor
print(np.max(grayscale_tensor[0]))
print(np.min(grayscale_tensor[0]))
print()
# coordinates of the first pixel where the max and min values are located
max_pixel = np.unravel_index(np.argmax(grayscale_tensor[0]), grayscale_tensor[0].shape)
min_pixel = np.unravel_index(np.argmin(grayscale_tensor[0]), grayscale_tensor[0].shape)
print(max_pixel)
print(min_pixel)
print()
# these coordinates should have the max (255) and min (0) value in the normalized tensor
print(normalized_tensor[max_pixel])
print(normalized_tensor[min_pixel])
plt.figure(figsize=(8, 8))
plt.axis('off')
plt.imshow(normalized_tensor, cmap='gray')
plt.show()
gradient_color = cv2.applyColorMap(normalized_tensor.numpy(), cv2.COLORMAP_HOT)
gradient_color = gradient_color / 255.0
super_imposed = cv2.addWeighted(img, 0.5, gradient_color, 0.5, 0.0)
plt.figure(figsize=(8, 8))
plt.imshow(super_imposed)
plt.axis('off')
plt.show()
```
|
github_jupyter
|
%tensorflow_version 2.x
import tensorflow as tf
import tensorflow_hub as hub
import cv2
import numpy as np
import matplotlib.pyplot as plt
# grab the model from Tensorflow hub and append a softmax activation
model = tf.keras.Sequential([
hub.KerasLayer('https://tfhub.dev/google/tf2-preview/inception_v3/classification/4'),
tf.keras.layers.Activation('softmax')
])
# build the model based on a specified batch input shape
model.build([None, 300, 300, 3])
!wget -O image.jpg https://cdn.pixabay.com/photo/2018/02/27/14/11/the-pacific-ocean-3185553_960_720.jpg
# If you want to try the cat, uncomment this line
# !wget -O image.jpg https://cdn.pixabay.com/photo/2018/02/27/14/11/the-pacific-ocean-3185553_960_720.jpg
# read the image
img = cv2.imread('image.jpg')
# format it to be in the RGB colorspace
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
# resize to 300x300 and normalize pixel values to be in the range [0, 1]
img = cv2.resize(img, (300, 300)) / 255.0
# add a batch dimension in front
image = np.expand_dims(img, axis=0)
plt.figure(figsize=(8, 8))
plt.imshow(img)
plt.axis('off')
plt.show()
# Siberian Husky's class ID in ImageNet
class_index = 251
# If you downloaded the cat, use this line instead
#class_index = 282 # Tabby Cat in ImageNet
# number of classes in the model's training data
num_classes = 1001
# convert to one hot representation to match our softmax activation in the model definition
expected_output = tf.one_hot([class_index] * image.shape[0], num_classes)
with tf.GradientTape() as tape:
# cast image to float
inputs = tf.cast(image, tf.float32)
# watch the input pixels
tape.watch(inputs)
# generate the predictions
predictions = model(inputs)
# get the loss
loss = tf.keras.losses.categorical_crossentropy(
expected_output, predictions
)
# get the gradient with respect to the inputs
gradients = tape.gradient(loss, inputs)
# reduce the RGB image to grayscale
grayscale_tensor = tf.reduce_sum(tf.abs(gradients), axis=-1)
# normalize the pixel values to be in the range [0, 255].
# the max value in the grayscale tensor will be pushed to 255.
# the min value will be pushed to 0.
normalized_tensor = tf.cast(
255
* (grayscale_tensor - tf.reduce_min(grayscale_tensor))
/ (tf.reduce_max(grayscale_tensor) - tf.reduce_min(grayscale_tensor)),
tf.uint8,
)
# remove the channel dimension to make the tensor a 2d tensor
normalized_tensor = tf.squeeze(normalized_tensor)
# max and min value in the grayscale tensor
print(np.max(grayscale_tensor[0]))
print(np.min(grayscale_tensor[0]))
print()
# coordinates of the first pixel where the max and min values are located
max_pixel = np.unravel_index(np.argmax(grayscale_tensor[0]), grayscale_tensor[0].shape)
min_pixel = np.unravel_index(np.argmin(grayscale_tensor[0]), grayscale_tensor[0].shape)
print(max_pixel)
print(min_pixel)
print()
# these coordinates should have the max (255) and min (0) value in the normalized tensor
print(normalized_tensor[max_pixel])
print(normalized_tensor[min_pixel])
plt.figure(figsize=(8, 8))
plt.axis('off')
plt.imshow(normalized_tensor, cmap='gray')
plt.show()
gradient_color = cv2.applyColorMap(normalized_tensor.numpy(), cv2.COLORMAP_HOT)
gradient_color = gradient_color / 255.0
super_imposed = cv2.addWeighted(img, 0.5, gradient_color, 0.5, 0.0)
plt.figure(figsize=(8, 8))
plt.imshow(super_imposed)
plt.axis('off')
plt.show()
| 0.871516 | 0.788705 |
# Decision tree for regression
In this notebook, we present how decision trees are working in regression
problems. We show differences with the decision trees previously presented in
a classification setting.
First, we load the penguins dataset specifically for solving a regression
problem.
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">If you want a deeper overview regarding this dataset, you can refer to the
Appendix - Datasets description section at the end of this MOOC.</p>
</div>
```
import pandas as pd
penguins = pd.read_csv("../datasets/penguins_regression.csv")
data_columns = ["Flipper Length (mm)"]
target_column = "Body Mass (g)"
data_train, target_train = penguins[data_columns], penguins[target_column]
```
To illustrate how decision trees are predicting in a regression setting, we
will create a synthetic dataset containing all possible flipper length from
the minimum to the maximum of the original data.
```
import numpy as np
data_test = pd.DataFrame(np.arange(data_train[data_columns[0]].min(),
data_train[data_columns[0]].max()),
columns=data_columns)
data_test.head()
import matplotlib.pyplot as plt
import seaborn as sns
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
_ = plt.title("Illustration of the regression dataset used")
```
We will first illustrate the difference between a linear model and a decision
tree.
```
from sklearn.linear_model import LinearRegression
linear_model = LinearRegression()
linear_model.fit(data_train, target_train)
target_predicted = linear_model.predict(data_test)
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted, label="Linear regression")
plt.legend()
_ = plt.title("Prediction function using a LinearRegression")
```
On the plot above, we see that a non-regularized `LinearRegression` is able
to fit the data. A feature of this model is that all new predictions
will be on the line.
```
ax = sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted, label="Linear regression",
linestyle="--")
plt.scatter(data_test[::3], target_predicted[::3], label="Test predictions",
color="tab:orange")
plt.legend()
_ = plt.title("Prediction function using a LinearRegression")
```
Contrary to linear models, decision trees are non-parametric models:
they do not make assumptions about the way data is distributed.
This will affect the prediction scheme. Repeating the above experiment
will highlight the differences.
```
from sklearn.tree import DecisionTreeRegressor
tree = DecisionTreeRegressor(max_depth=1)
tree.fit(data_train, target_train)
target_predicted = tree.predict(data_test)
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted, label="Decision tree")
plt.legend()
_ = plt.title("Prediction function using a DecisionTreeRegressor")
```
We see that the decision tree model does not have an *a priori* distribution
for the data and we do not end-up with a straight line to regress flipper
length and body mass.
Instead, we observe that the predictions of the tree are piecewise constant.
Indeed, our feature space was split into two partitions. Let's check the
tree structure to see what was the threshold found during the training.
```
from sklearn.tree import plot_tree
_, ax = plt.subplots(figsize=(8, 6))
_ = plot_tree(tree, feature_names=data_columns, ax=ax)
```
The threshold for our feature (flipper length) is 206.5 mm. The predicted
values on each side of the split are two constants: 3683.50 g and 5023.62 g.
These values corresponds to the mean values of the training samples in each
partition.
In classification, we saw that increasing the depth of the tree allowed us to
get more complex decision boundaries.
Let's check the effect of increasing the depth in a regression setting:
```
tree = DecisionTreeRegressor(max_depth=3)
tree.fit(data_train, target_train)
target_predicted = tree.predict(data_test)
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted, label="Decision tree")
plt.legend()
_ = plt.title("Prediction function using a DecisionTreeRegressor")
```
Increasing the depth of the tree will increase the number of partition and
thus the number of constant values that the tree is capable of predicting.
In this notebook, we highlighted the differences in behavior of a decision
tree used in a classification problem in contrast to a regression problem.
|
github_jupyter
|
import pandas as pd
penguins = pd.read_csv("../datasets/penguins_regression.csv")
data_columns = ["Flipper Length (mm)"]
target_column = "Body Mass (g)"
data_train, target_train = penguins[data_columns], penguins[target_column]
import numpy as np
data_test = pd.DataFrame(np.arange(data_train[data_columns[0]].min(),
data_train[data_columns[0]].max()),
columns=data_columns)
data_test.head()
import matplotlib.pyplot as plt
import seaborn as sns
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
_ = plt.title("Illustration of the regression dataset used")
from sklearn.linear_model import LinearRegression
linear_model = LinearRegression()
linear_model.fit(data_train, target_train)
target_predicted = linear_model.predict(data_test)
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted, label="Linear regression")
plt.legend()
_ = plt.title("Prediction function using a LinearRegression")
ax = sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted, label="Linear regression",
linestyle="--")
plt.scatter(data_test[::3], target_predicted[::3], label="Test predictions",
color="tab:orange")
plt.legend()
_ = plt.title("Prediction function using a LinearRegression")
from sklearn.tree import DecisionTreeRegressor
tree = DecisionTreeRegressor(max_depth=1)
tree.fit(data_train, target_train)
target_predicted = tree.predict(data_test)
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted, label="Decision tree")
plt.legend()
_ = plt.title("Prediction function using a DecisionTreeRegressor")
from sklearn.tree import plot_tree
_, ax = plt.subplots(figsize=(8, 6))
_ = plot_tree(tree, feature_names=data_columns, ax=ax)
tree = DecisionTreeRegressor(max_depth=3)
tree.fit(data_train, target_train)
target_predicted = tree.predict(data_test)
sns.scatterplot(data=penguins, x="Flipper Length (mm)", y="Body Mass (g)",
color="black", alpha=0.5)
plt.plot(data_test, target_predicted, label="Decision tree")
plt.legend()
_ = plt.title("Prediction function using a DecisionTreeRegressor")
| 0.752377 | 0.991985 |
# Dynamic SSSP SC'18 Figures
```
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns # you can install this with "pip install seaborn"
import sys
import pprint, math
import glob
from pylab import MaxNLocator
%matplotlib inline
#print(plt.rcParams.keys())
params = {'font.size': 18, 'figure.titlesize':'medium','legend.fontsize': 'medium','legend.handlelength':1,
'lines.linewidth':2, 'lines.markersize':12, 'legend.handletextpad':0.2,'axes.formatter.useoffset':False,}
plt.rcParams.update(params)
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
sns.set_style("whitegrid")
#print('Baseline power',baseline_power)
```
## Settings
Edit the variables below to indicate which plots to generate, the algs list is a list of subdirectories with log files containing time, power, and energy measurements.
```
whats = {'Energy':'Total CPU Energy (J)','Power':'Average CPU Power (W)', 'Memory':'Total Memory (GB)','Time':'Time (seconds)'}
whats = {'Memory':'Total Memory (GB)','Time':'Time (seconds)'}
resultsdir = 'output-8epv-new/'
epvs = ['8']
nws = ['ER','G']
insperc = ['100','75']
speedup = False
printData = True
## Misc stuff
whats = {
'Total Time for Creating tree' : 'SSSP Create Tree',
'Total Time for Initial Update' : 'SSSP Initial Update',
'Total Time for Complete Update':'SSSP Complete Update',
'TotalTime' : 'Galois'}
seq_time = {}
```
## Functions
A couple of functions that extract the data and generate the plots.
```
def processGaloisLine(line):
path,contents = line.strip().split(':')
d,fname = path.split('/')
experiment = fname.split('.')[0]
parts = experiment.split('-')[-1].split('_')
network = '_'.join(parts[:-1])
threads = parts[-1].strip('t')
network = '_'.join(experiment.split('-')[-1].split('_')[:-1])
return experiment,network,threads,contents
def processSSSPLine(path,line):
contents = line
d,fname = path.split('/')
experiment = fname.split('.')[0]
parts = experiment.split('-')[-1].split('_')
network = '_'.join(parts[:-1])
threads = parts[-1].strip('t')
network = '_'.join(experiment.split('-')[-1].split('_')[:-1])
parts2 = contents.split()
time = float(parts2[-1])
memory = 0
what = whats[' '.join(parts2[:-1])]
return experiment,network,threads,what,time,memory
def parseTimeData():
global seq_time
# Galois: times-galois.txt (divide time by 1000 to get seconds)
# output-8epv/galois-248_G_75i_1000000_4t.log:STAT,(NULL),TotalTime,4,18838,18838,0,0,0
# SSSP: times-sssp.txt
# output-8epv/sssp-248_G_75i_1000000_32t.log:Total Time for Complete Update 34.470351
galois_time = {}; sssp_time = {}
lines = open(resultsdir+'times-galois.txt').readlines()
for l in lines:
experiment,network,threads,contents = processGaloisLine(l)
parts = contents.split(',')
time = float(parts[4])/1000 # seconds
exp = network+'-'+threads
if not exp in galois_time.keys(): galois_time[exp] = []
galois_time[exp].append(time)
files = glob.glob(resultsdir+'sssp*.log')
seq_time = {}
for f in files:
lines = open(f,'r').readlines()
tmptotals = {}; totals = {}
skip = True; localtimes = []
for l in lines:
if l.startswith('==Update Starts ======='):
skip = False; counter = 0
if not skip and l.find('Time')>=0:
experiment,network,threads,what,time,memory = processSSSPLine(f,l)
exp = network+'-'+threads
if not exp in sssp_time.keys(): sssp_time[exp] = []
if not l.startswith('Total Time for Creating tree'):
sssp_time[exp].append((what,time))
localtimes.append(time)
if len(localtimes) == 2:
total = sum(localtimes)
sssp_time[exp].append(('SSSP Total',total))
if not network in seq_time: seq_time[network] = []
if threads == '1': seq_time[network].append(total)
if l.find('Exit status:') >=0:
skip = True; localtimes = []
return galois_time,sssp_time
```
## Galois comparison plots
All the data structures are populated by the functions above
```
galois_time, sssp_time = parseTimeData()
columns=['Experiment','Threads','Time (sec.)','Algorithm']
df = pd.DataFrame(columns=columns)
nets = []
for k in sorted(galois_time.keys()):
net,th = k.split('-')
for i in range(0,len(galois_time[k])):
tmpdf = pd.DataFrame([[net,int(th),galois_time[k][i],'Galois']],columns=columns)
df = df.append(tmpdf, ignore_index=True).fillna(0)
for i in range(0,len(sssp_time[k])):
tmpdf = pd.DataFrame([[net,int(th),sssp_time[k][i][1],sssp_time[k][i][0]]],columns=columns)
df = df.append(tmpdf, ignore_index=True).fillna(0)
if not net in nets: nets.append(net)
#df.loc[df['Experiment']=='248_ER_100i_1000000']
#Improvement over Galois:
def s(x): return '%s' % 'RMAT24_'+'_'.join(x.split('_')[1:-1])
def f(x): return '%1.1f' % x
def i(x): return '%d' % x
#fig, ax = plt.subplots(); fig.set_size_inches(8,6)
for net in nets:
print(net)
#print("Seq time",net,seq_time[net])
df2 = df.loc[df['Experiment']==net]
dfg = df2.loc[df2['Algorithm']=='Galois'].drop(['Algorithm'],axis=1).rename(columns={'Time (sec.)':'Galois'})
dfs = df2.loc[df2['Algorithm']=='SSSP Total'].drop(['Algorithm'],axis=1).rename(columns={'Time (sec.)':'SSSP Update'})
df3 = pd.merge(dfg,dfs, on=['Experiment', 'Threads'])
df3['Improvement'] = df3['Galois'] / df3['SSSP Update']
groupedvalues=df3.groupby(['Experiment','Threads']).mean().reset_index()
print(groupedvalues.to_latex(columns=['Experiment','Threads','Galois','SSSP Update','Improvement'],
index=False, formatters=[s,i,f,f,f]))
#dfs['Speedup'] = dfg['Time (sec.)']/dfs['Time (sec.)'])
#avgseqtime = sum(seq_time[net])/len(seq_time[net])
#df2['Ideal'] = avgseqtime / df2.loc[df2['Algorithm']=='SSSP Total']['Time (sec.)']
#df2['Speedup'] = df2.loc[df2['Algorithm']=='SSSP Total']['Time (sec.)'] / avgseqtime
#print(df)
# Process data for plotting
colors = ['#af0000','#019cad','#7f8cff','#2f7c00','#5566fc']
sns.set(font_scale=2)
for net in nets:
df2 = df.loc[df['Experiment']==net].copy(deep=True)
df2['Threads'] = df2['Threads'].astype('int', copy=False)
fig, ax = plt.subplots(); fig.set_size_inches(8,5)
sns.set_palette(sns.color_palette(colors))
g1 = sns.pointplot(x='Threads',y='Time (sec.)',data=df2,hue='Algorithm', errwidth=1, capsize=0.1,
markers=['^', '1','*','o','.'], #,'+','2','p','d','s','.'],
linestyles=["-", "--", "-.",':','-'])
g1.set_ylim(-5)
g1.set(axis_bgcolor='white');
legend = plt.legend(frameon=True); frame = legend.get_frame(); frame.set_facecolor('white'); frame.set_edgecolor('#cbcfd6')
ax.grid(color='#e1e3e8', linestyle='-', linewidth=1)
parts = net.split('_')
titlestr = 'Graph RMAT24' + '-' + parts[1] + " %s%% Insertions" % parts[2].strip('i')
plt.title(titlestr)
plt.tight_layout()
plt.savefig(net + "_Time.pdf")
#g2 = sns.pointplot(x='Threads',y='Ideal',data=df3,color='black',markers=['.'],linestyles=['--'],legend=None)
sns.set_style("whitegrid")
```
## Scaling
The data for these plots is in the RMAT.csv and Real.csv files, which must be in the same directory as this notebook.
```
df3 = pd.read_csv('RMAT.csv',skip_blank_lines=True).dropna()
df4 = pd.melt(df3, id_vars=["Graph", "Insertion %"], var_name='Threads',
value_vars = ['1','2','4','8','16','32','64'],value_name="Time (sec.)")
df4['Threads'] = df4['Threads'].astype('int', copy=False)
df4['Insertion %'] = df4['Insertion %'].astype('int', copy=False)
df4.head()
for ins in [50,75,100]:
fig, ax = plt.subplots(); fig.set_size_inches(6,5)
g1 = sns.pointplot(x='Threads',y='Time (sec.)',data=df4.loc[df4['Insertion %']==ins],
hue='Graph', errwidth=1, capsize=0.1,
markers=['^', '1','*','o','.'], #,'+','2','p','d','s','.'],
linestyles=["-", "--", "-.",':','-'])
g1.text(.3, .98, '%d%% Insertions'%ins, horizontalalignment='center',verticalalignment='top',transform=g1.transAxes)
#g1 = sns.factorplot(x="Threads", y="Time (sec.)", hue="Graph", data=df4.loc[df4['Insertion %']==ins],
# size=6, kind="bar", palette="bright", legend_out=False)
#plt.suptitle('%d%% Insertions'%ins, x=.45, y=0.92)
plt.tight_layout()
plt.savefig("Scaling-RMAT-%d.pdf" % ins)
df5 = pd.read_csv('Real.csv',skip_blank_lines=True).dropna()
df6 = pd.melt(df5, id_vars=["Graph", "Insertion %"], var_name='Threads',
value_vars = ['1','2','4','8','16','32','64'],value_name="Time (sec.)")
df6['Threads'] = df6['Threads'].astype('int', copy=False)
print(df6.head())
for ins in [50,75,100]:
fig, ax = plt.subplots(); fig.set_size_inches(6,5)
g1 = sns.pointplot(x='Threads',y='Time (sec.)',data=df6.loc[df4['Insertion %']==ins],
hue='Graph', errwidth=1, capsize=0.1,
markers=['^', '1','*','o','.'], #,'+','2','p','d','s','.'],
linestyles=["-", "--", "-.",':','-'])
g1.set_ylim(bottom=-5)
g1.text(.3, .98, '%d%% Insertions'%ins,horizontalalignment='center',verticalalignment='top',transform=g1.transAxes)
#g1 = sns.factorplot(x="Threads", y="Time (sec.)", hue="Graph", data=df6.loc[df4['Insertion %']==ins],
# size=6, kind="bar", palette="bright", legend_out=False)
#plt.suptitle('%d%% Insertions'%ins, x=.45, y=0.92)
plt.tight_layout()
plt.savefig("Scaling-Real-%d.pdf" % ins)
ddf = pd.read_csv('DifferentSource.csv',skip_blank_lines=True).dropna()
dd = pd.melt(ddf, id_vars=["Graph", "Insertion %"], var_name='Threads',
value_vars = ['1','2','4','8','16','32','64'],value_name="Time (sec.)")
dd['Threads'] = dd['Threads'].astype('int', copy=False)
print(dd.head())
fig, ax = plt.subplots(); fig.set_size_inches(8,4)
g = sns.pointplot(x='Threads',y='Time (sec.)',data=dd, hue='Graph', errwidth=1, capsize=0.1,
markers=['^', '1','*','o','.'], #,'+','2','p','d','s','.'],
linestyles=["-", "--", "-.",':','-'])
g.set_ylim(-5)
plt.tight_layout()
plt.savefig("DiffSource.pdf")
```
## Distributed-memory SSSPInc algorithm results
The data for these results is in the SSSPInc.csv file in the same directory as this notebook.
```
df7 = pd.read_csv('SSSPInc.csv',skip_blank_lines=True).dropna()
print(df7.head())
df7['Update Size'] = df7['Update Size'].astype('int', copy=False)
df7_time = df7.loc[df7['What']=='Time (sec.)'].rename(columns={'Value':'Time (sec.)'})
print(df7_time.head())
df7_msg = df7.loc[df7['What']=='Max Messages'].rename(columns={'Value':'Max Messages'})
df7_steps = df7.loc[df7['What']=='Supersteps'].rename(columns={'Value':'Supersteps'})
for net in ['Friendster','Twitter MPI','Twitter']:
## Time
fig, ax = plt.subplots(); fig.set_size_inches(6,4)
g1 = sns.barplot(x="Update Size", y="Time (sec.)", hue="Algorithm", data=df7_time[df7_time['Graph']==net],
ci=None, palette=sns.color_palette(['#96ff4c','#4286f4']))
#g1.set_xticklabels(rotation=30)
plt.suptitle('Graph: %s'% net, x=.45, y=.98)
plt.legend(loc='upper right', ncol = 1, labelspacing=0.5)
g1.set_xticklabels(['1M','10M','20M','50M'])
g1.set_xlabel('Update size (number of edges)')
plt.tight_layout();
plt.savefig("SSSPInc-Time-%s.pdf" % net)
# Maximum number of messages
fig, ax = plt.subplots(); fig.set_size_inches(6,4)
g2 = sns.barplot(x="Update Size", y="Max Messages", hue="Algorithm", data=df7_msg[df7_msg['Graph']==net],
ci=None, palette=sns.color_palette(['#96ff4c','#4286f4']))
plt.suptitle('Graph: %s'% net, x=.45, y=.98)
g2.set_xticklabels(['1M','10M','20M','50M'])
g2.set_ylabel('Maximum number of messages')
g2.set_xlabel('Update size (number of edges)')
plt.legend(loc='upper left', ncol = 2, labelspacing=0.5, bbox_to_anchor=(0.1, 1.05))
plt.tight_layout();
plt.savefig("SSSPInc-MaxMsgs-%s.pdf" % net)
# Supersteps
fig, ax = plt.subplots(); fig.set_size_inches(6,4)
g3 = sns.barplot(x="Update Size", y="Supersteps", hue="Algorithm", data=df7_steps[df7_steps['Graph']==net],
ci=None, palette=sns.color_palette(['#96ff4c','#4286f4']))
plt.suptitle('Graph: %s'% net, x=.45, y=.98)
plt.legend(loc='upper right', ncol = 1, labelspacing=0.5, bbox_to_anchor=(1, 1.05))
g3.set_xticklabels(['1M','10M','20M','50M'])
g3.set_xlabel('Update size (number of edges)')
plt.tight_layout();
plt.savefig("SSSPInc-Supersteps-%s.pdf" % net)
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns # you can install this with "pip install seaborn"
import sys
import pprint, math
import glob
from pylab import MaxNLocator
%matplotlib inline
#print(plt.rcParams.keys())
params = {'font.size': 18, 'figure.titlesize':'medium','legend.fontsize': 'medium','legend.handlelength':1,
'lines.linewidth':2, 'lines.markersize':12, 'legend.handletextpad':0.2,'axes.formatter.useoffset':False,}
plt.rcParams.update(params)
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
sns.set_style("whitegrid")
#print('Baseline power',baseline_power)
whats = {'Energy':'Total CPU Energy (J)','Power':'Average CPU Power (W)', 'Memory':'Total Memory (GB)','Time':'Time (seconds)'}
whats = {'Memory':'Total Memory (GB)','Time':'Time (seconds)'}
resultsdir = 'output-8epv-new/'
epvs = ['8']
nws = ['ER','G']
insperc = ['100','75']
speedup = False
printData = True
## Misc stuff
whats = {
'Total Time for Creating tree' : 'SSSP Create Tree',
'Total Time for Initial Update' : 'SSSP Initial Update',
'Total Time for Complete Update':'SSSP Complete Update',
'TotalTime' : 'Galois'}
seq_time = {}
def processGaloisLine(line):
path,contents = line.strip().split(':')
d,fname = path.split('/')
experiment = fname.split('.')[0]
parts = experiment.split('-')[-1].split('_')
network = '_'.join(parts[:-1])
threads = parts[-1].strip('t')
network = '_'.join(experiment.split('-')[-1].split('_')[:-1])
return experiment,network,threads,contents
def processSSSPLine(path,line):
contents = line
d,fname = path.split('/')
experiment = fname.split('.')[0]
parts = experiment.split('-')[-1].split('_')
network = '_'.join(parts[:-1])
threads = parts[-1].strip('t')
network = '_'.join(experiment.split('-')[-1].split('_')[:-1])
parts2 = contents.split()
time = float(parts2[-1])
memory = 0
what = whats[' '.join(parts2[:-1])]
return experiment,network,threads,what,time,memory
def parseTimeData():
global seq_time
# Galois: times-galois.txt (divide time by 1000 to get seconds)
# output-8epv/galois-248_G_75i_1000000_4t.log:STAT,(NULL),TotalTime,4,18838,18838,0,0,0
# SSSP: times-sssp.txt
# output-8epv/sssp-248_G_75i_1000000_32t.log:Total Time for Complete Update 34.470351
galois_time = {}; sssp_time = {}
lines = open(resultsdir+'times-galois.txt').readlines()
for l in lines:
experiment,network,threads,contents = processGaloisLine(l)
parts = contents.split(',')
time = float(parts[4])/1000 # seconds
exp = network+'-'+threads
if not exp in galois_time.keys(): galois_time[exp] = []
galois_time[exp].append(time)
files = glob.glob(resultsdir+'sssp*.log')
seq_time = {}
for f in files:
lines = open(f,'r').readlines()
tmptotals = {}; totals = {}
skip = True; localtimes = []
for l in lines:
if l.startswith('==Update Starts ======='):
skip = False; counter = 0
if not skip and l.find('Time')>=0:
experiment,network,threads,what,time,memory = processSSSPLine(f,l)
exp = network+'-'+threads
if not exp in sssp_time.keys(): sssp_time[exp] = []
if not l.startswith('Total Time for Creating tree'):
sssp_time[exp].append((what,time))
localtimes.append(time)
if len(localtimes) == 2:
total = sum(localtimes)
sssp_time[exp].append(('SSSP Total',total))
if not network in seq_time: seq_time[network] = []
if threads == '1': seq_time[network].append(total)
if l.find('Exit status:') >=0:
skip = True; localtimes = []
return galois_time,sssp_time
galois_time, sssp_time = parseTimeData()
columns=['Experiment','Threads','Time (sec.)','Algorithm']
df = pd.DataFrame(columns=columns)
nets = []
for k in sorted(galois_time.keys()):
net,th = k.split('-')
for i in range(0,len(galois_time[k])):
tmpdf = pd.DataFrame([[net,int(th),galois_time[k][i],'Galois']],columns=columns)
df = df.append(tmpdf, ignore_index=True).fillna(0)
for i in range(0,len(sssp_time[k])):
tmpdf = pd.DataFrame([[net,int(th),sssp_time[k][i][1],sssp_time[k][i][0]]],columns=columns)
df = df.append(tmpdf, ignore_index=True).fillna(0)
if not net in nets: nets.append(net)
#df.loc[df['Experiment']=='248_ER_100i_1000000']
#Improvement over Galois:
def s(x): return '%s' % 'RMAT24_'+'_'.join(x.split('_')[1:-1])
def f(x): return '%1.1f' % x
def i(x): return '%d' % x
#fig, ax = plt.subplots(); fig.set_size_inches(8,6)
for net in nets:
print(net)
#print("Seq time",net,seq_time[net])
df2 = df.loc[df['Experiment']==net]
dfg = df2.loc[df2['Algorithm']=='Galois'].drop(['Algorithm'],axis=1).rename(columns={'Time (sec.)':'Galois'})
dfs = df2.loc[df2['Algorithm']=='SSSP Total'].drop(['Algorithm'],axis=1).rename(columns={'Time (sec.)':'SSSP Update'})
df3 = pd.merge(dfg,dfs, on=['Experiment', 'Threads'])
df3['Improvement'] = df3['Galois'] / df3['SSSP Update']
groupedvalues=df3.groupby(['Experiment','Threads']).mean().reset_index()
print(groupedvalues.to_latex(columns=['Experiment','Threads','Galois','SSSP Update','Improvement'],
index=False, formatters=[s,i,f,f,f]))
#dfs['Speedup'] = dfg['Time (sec.)']/dfs['Time (sec.)'])
#avgseqtime = sum(seq_time[net])/len(seq_time[net])
#df2['Ideal'] = avgseqtime / df2.loc[df2['Algorithm']=='SSSP Total']['Time (sec.)']
#df2['Speedup'] = df2.loc[df2['Algorithm']=='SSSP Total']['Time (sec.)'] / avgseqtime
#print(df)
# Process data for plotting
colors = ['#af0000','#019cad','#7f8cff','#2f7c00','#5566fc']
sns.set(font_scale=2)
for net in nets:
df2 = df.loc[df['Experiment']==net].copy(deep=True)
df2['Threads'] = df2['Threads'].astype('int', copy=False)
fig, ax = plt.subplots(); fig.set_size_inches(8,5)
sns.set_palette(sns.color_palette(colors))
g1 = sns.pointplot(x='Threads',y='Time (sec.)',data=df2,hue='Algorithm', errwidth=1, capsize=0.1,
markers=['^', '1','*','o','.'], #,'+','2','p','d','s','.'],
linestyles=["-", "--", "-.",':','-'])
g1.set_ylim(-5)
g1.set(axis_bgcolor='white');
legend = plt.legend(frameon=True); frame = legend.get_frame(); frame.set_facecolor('white'); frame.set_edgecolor('#cbcfd6')
ax.grid(color='#e1e3e8', linestyle='-', linewidth=1)
parts = net.split('_')
titlestr = 'Graph RMAT24' + '-' + parts[1] + " %s%% Insertions" % parts[2].strip('i')
plt.title(titlestr)
plt.tight_layout()
plt.savefig(net + "_Time.pdf")
#g2 = sns.pointplot(x='Threads',y='Ideal',data=df3,color='black',markers=['.'],linestyles=['--'],legend=None)
sns.set_style("whitegrid")
df3 = pd.read_csv('RMAT.csv',skip_blank_lines=True).dropna()
df4 = pd.melt(df3, id_vars=["Graph", "Insertion %"], var_name='Threads',
value_vars = ['1','2','4','8','16','32','64'],value_name="Time (sec.)")
df4['Threads'] = df4['Threads'].astype('int', copy=False)
df4['Insertion %'] = df4['Insertion %'].astype('int', copy=False)
df4.head()
for ins in [50,75,100]:
fig, ax = plt.subplots(); fig.set_size_inches(6,5)
g1 = sns.pointplot(x='Threads',y='Time (sec.)',data=df4.loc[df4['Insertion %']==ins],
hue='Graph', errwidth=1, capsize=0.1,
markers=['^', '1','*','o','.'], #,'+','2','p','d','s','.'],
linestyles=["-", "--", "-.",':','-'])
g1.text(.3, .98, '%d%% Insertions'%ins, horizontalalignment='center',verticalalignment='top',transform=g1.transAxes)
#g1 = sns.factorplot(x="Threads", y="Time (sec.)", hue="Graph", data=df4.loc[df4['Insertion %']==ins],
# size=6, kind="bar", palette="bright", legend_out=False)
#plt.suptitle('%d%% Insertions'%ins, x=.45, y=0.92)
plt.tight_layout()
plt.savefig("Scaling-RMAT-%d.pdf" % ins)
df5 = pd.read_csv('Real.csv',skip_blank_lines=True).dropna()
df6 = pd.melt(df5, id_vars=["Graph", "Insertion %"], var_name='Threads',
value_vars = ['1','2','4','8','16','32','64'],value_name="Time (sec.)")
df6['Threads'] = df6['Threads'].astype('int', copy=False)
print(df6.head())
for ins in [50,75,100]:
fig, ax = plt.subplots(); fig.set_size_inches(6,5)
g1 = sns.pointplot(x='Threads',y='Time (sec.)',data=df6.loc[df4['Insertion %']==ins],
hue='Graph', errwidth=1, capsize=0.1,
markers=['^', '1','*','o','.'], #,'+','2','p','d','s','.'],
linestyles=["-", "--", "-.",':','-'])
g1.set_ylim(bottom=-5)
g1.text(.3, .98, '%d%% Insertions'%ins,horizontalalignment='center',verticalalignment='top',transform=g1.transAxes)
#g1 = sns.factorplot(x="Threads", y="Time (sec.)", hue="Graph", data=df6.loc[df4['Insertion %']==ins],
# size=6, kind="bar", palette="bright", legend_out=False)
#plt.suptitle('%d%% Insertions'%ins, x=.45, y=0.92)
plt.tight_layout()
plt.savefig("Scaling-Real-%d.pdf" % ins)
ddf = pd.read_csv('DifferentSource.csv',skip_blank_lines=True).dropna()
dd = pd.melt(ddf, id_vars=["Graph", "Insertion %"], var_name='Threads',
value_vars = ['1','2','4','8','16','32','64'],value_name="Time (sec.)")
dd['Threads'] = dd['Threads'].astype('int', copy=False)
print(dd.head())
fig, ax = plt.subplots(); fig.set_size_inches(8,4)
g = sns.pointplot(x='Threads',y='Time (sec.)',data=dd, hue='Graph', errwidth=1, capsize=0.1,
markers=['^', '1','*','o','.'], #,'+','2','p','d','s','.'],
linestyles=["-", "--", "-.",':','-'])
g.set_ylim(-5)
plt.tight_layout()
plt.savefig("DiffSource.pdf")
df7 = pd.read_csv('SSSPInc.csv',skip_blank_lines=True).dropna()
print(df7.head())
df7['Update Size'] = df7['Update Size'].astype('int', copy=False)
df7_time = df7.loc[df7['What']=='Time (sec.)'].rename(columns={'Value':'Time (sec.)'})
print(df7_time.head())
df7_msg = df7.loc[df7['What']=='Max Messages'].rename(columns={'Value':'Max Messages'})
df7_steps = df7.loc[df7['What']=='Supersteps'].rename(columns={'Value':'Supersteps'})
for net in ['Friendster','Twitter MPI','Twitter']:
## Time
fig, ax = plt.subplots(); fig.set_size_inches(6,4)
g1 = sns.barplot(x="Update Size", y="Time (sec.)", hue="Algorithm", data=df7_time[df7_time['Graph']==net],
ci=None, palette=sns.color_palette(['#96ff4c','#4286f4']))
#g1.set_xticklabels(rotation=30)
plt.suptitle('Graph: %s'% net, x=.45, y=.98)
plt.legend(loc='upper right', ncol = 1, labelspacing=0.5)
g1.set_xticklabels(['1M','10M','20M','50M'])
g1.set_xlabel('Update size (number of edges)')
plt.tight_layout();
plt.savefig("SSSPInc-Time-%s.pdf" % net)
# Maximum number of messages
fig, ax = plt.subplots(); fig.set_size_inches(6,4)
g2 = sns.barplot(x="Update Size", y="Max Messages", hue="Algorithm", data=df7_msg[df7_msg['Graph']==net],
ci=None, palette=sns.color_palette(['#96ff4c','#4286f4']))
plt.suptitle('Graph: %s'% net, x=.45, y=.98)
g2.set_xticklabels(['1M','10M','20M','50M'])
g2.set_ylabel('Maximum number of messages')
g2.set_xlabel('Update size (number of edges)')
plt.legend(loc='upper left', ncol = 2, labelspacing=0.5, bbox_to_anchor=(0.1, 1.05))
plt.tight_layout();
plt.savefig("SSSPInc-MaxMsgs-%s.pdf" % net)
# Supersteps
fig, ax = plt.subplots(); fig.set_size_inches(6,4)
g3 = sns.barplot(x="Update Size", y="Supersteps", hue="Algorithm", data=df7_steps[df7_steps['Graph']==net],
ci=None, palette=sns.color_palette(['#96ff4c','#4286f4']))
plt.suptitle('Graph: %s'% net, x=.45, y=.98)
plt.legend(loc='upper right', ncol = 1, labelspacing=0.5, bbox_to_anchor=(1, 1.05))
g3.set_xticklabels(['1M','10M','20M','50M'])
g3.set_xlabel('Update size (number of edges)')
plt.tight_layout();
plt.savefig("SSSPInc-Supersteps-%s.pdf" % net)
| 0.114827 | 0.685654 |
***
# <font size=06>Base de Dados do IBGE</font>
***
# I. INTRODUÇÃO
## Contexto
Este notebook é resultado da minha prática com dados para treinar estatística com Python.
## Fonte dos Dados
- https://ww2.ibge.gov.br/home/estatistica/populacao/trabalhoerendimento/pnad2015/microdados.shtm
- Disponível em: https://www.kaggle.com/upadorprofzs/testes
- **Observação:** Dados já tratados.
A **Pesquisa Nacional por Amostra de Domicílios - PNAD** investiga anualmente, de forma permanente, características gerais da população, de educação, trabalho, rendimento e habitação e outras, com periodicidade variável, de acordo com as necessidades de informação para o país, como as características sobre migração, fecundidade, nupcialidade, saúde, segurança alimentar, entre outros temas. O levantamento dessas estatísticas constitui, ao longo dos 49 anos de realização da pesquisa, um importante instrumento para formulação, validação e avaliação de políticas orientadas para o desenvolvimento socioeconômico e a melhoria das condições de vida no Brasil.
## Variáveis utilizadas
**Renda**
Rendimento mensal do trabalho principal para pessoas de 10 anos ou mais de idade.
**Idade**
Idade do morador na data de referência em anos.
**Altura**
Altura do morador em metros.
**UF**
|Código|Descrição|
|---|---|
|11|Rondônia|
|12|Acre|
|13|Amazonas|
|14|Roraima|
|15|Pará|
|16|Amapá|
|17|Tocantins|
|21|Maranhão|
|22|Piauí|
|23|Ceará|
|24|Rio Grande do Norte|
|25|Paraíba|
|26|Pernambuco|
|27|Alagoas|
|28|Sergipe|
|29|Bahia|
|31|Minas Gerais|
|32|Espírito Santo|
|33|Rio de Janeiro|
|35|São Paulo|
|41|Paraná|
|42|Santa Catarina|
|43|Rio Grande do Sul|
|50|Mato Grosso do Sul|
|51|Mato Grosso|
|52|Goiás|
|53|Distrito Federal|
**Sexo**
|Código|Descrição|
|---|---|
|0|Masculino|
|1|Feminino|
**Anos de Estudo**
|Código|Descrição|
|---|---|
|1|Sem instrução e menos de 1 ano|
|2|1 ano|
|3|2 anos|
|4|3 anos|
|5|4 anos|
|6|5 anos|
|7|6 anos|
|8|7 anos|
|9|8 anos|
|10|9 anos|
|11|10 anos|
|12|11 anos|
|13|12 anos|
|14|13 anos|
|15|14 anos|
|16|15 anos ou mais|
|17|Não determinados|
||Não aplicável|
**Cor**
|Código|Descrição|
|---|---|
|0|Indígena|
|2|Branca|
|4|Preta|
|6|Amarela|
|8|Parda|
|9|Sem declaração|
## Objetivos
**Objetivo principal:** treino, prática e aprendizado de componentes de estatística.
# II. Desenvolvimento
### Bibliotecas importadas
```
import pandas as pd
import numpy as np
import seaborn as sns
from scipy.special import comb
from scipy.stats import binom
```
### Extração de dados
```
dados = pd.read_csv('dataset.csv')
```
### Breve visualização dos dados
```
# Visualizando o início
dados.head(3)
# Informações gerais
dados.info()
```
## Parte I: Classificação
### 1.1. Classificação de variáveis
**Variáveis qualitativas ordinais:** Variáveis que podem ser ordenadas ou hierarquizadas.
```
#Exemplo: anos de estudo, idade
print('Anos de estudo:' , sorted(dados['Anos de Estudo'].unique()))
print('Idade:' , sorted(dados['Idade'].unique()))
```
**Variáveis qualitativas nominais:** Variáveis que não podem ser ordenadas ou hierarquizardas.
```
# Exemplo:
print('Sexo: ', sorted(dados['Sexo'].unique()))
print('UF: ', dados['UF'].unique())
print('Cor: ', dados['Cor'].unique())
```
**Variáveis quantitativas discretas:** Variáveis que representam uma contagem onde os valores possíveis formam um conjunto finito ou enumerável.
```
#mostra a idade minima e maxima
print(f"Variável 'Idade' vai de {dados.Idade.min()} até {dados.Idade.max()} anos.")
```
**Variáveis quantitativas contínuas:** Variáveis que representam uma contagem ou mensuração que assumem valores em uma escala contínua (números reais).
```
print("A variável 'Altura' vai de %s até %s metros." % (dados.Altura.min(), dados.Altura.max()))
```
## Parte II: Frequências
### 2.1. Distribuição de frequências para variáveis qualitativas
**Uma Variável**
```
# Quantidade total:
frequencia_sexo = dados['Sexo'].value_counts()
frequencia_sexo
# Proporção:
porcentual_sexo = dados['Sexo'].value_counts(normalize = True) * 100
porcentual_sexo
# Montando um dataframe, renomeando variáveis e colunas:
dist_freq_qualitativas = pd.DataFrame({'Frequência': frequencia_sexo, 'Porcentagem (%)': porcentual_sexo})
dist_freq_qualitativas.rename(index = {0: 'Masculino', 1: 'Feminino'}, inplace = True)
dist_freq_qualitativas.rename_axis('Sexo', axis = 'columns', inplace = True)
dist_freq_qualitativas
```
**Duas variáveis**
```
sexo = {0: 'Masculino', 1: 'Feminino'}
cor = {0: 'Indígena',
2: 'Branca',
4: 'Preta',
6: 'Amarela',
8: 'Parda',
9: 'Sem declaração'}
# Tabela com frequências de pessoas com cada cor
percentualSC = pd.crosstab(dados.Sexo,
dados.Cor,
normalize = True) * 100
percentualSC.rename(index = sexo, inplace = True)
percentualSC.rename(columns = cor, inplace = True)
percentualSC
```
---------------------
### 2.2. Distribuição de frequências para variáveis quantitativas (classes personalizadas)
**Variável Salário**
**Passo 1** Especificar os limites de cada classe, classificando pelo Salário Mínimo (SM - R$ 788,00 em 2015)
**Classe**|SM|Valor
:---:|:---:|:---:
**A**|Acima de 20 SM|Acima de 15.760
**B**|De 10 a 20 SM|De 7.880 a 15.760
**C**|De 4 a 10 SM|De 3.152 a 7.880
**D**|De 2 a 4 SM|De 1.576 a 3.152
**E**|Até 2 SM|Até 1.576
**Passo 2** Definir extremos e categorias
```
# Extremos dos dados:
print(f"Renda máxima: {dados.Renda.max()}\t Renda mínima: {dados.Renda.min()}.")
# Variáveis para dicionário:
classes = [0, 1576, 3152, 7880, 15760, 200000]
labels = ['E', 'D', 'C', 'B', 'A']
```
**Passo 3** Criando uma series para o dataframe
```
# Criará uma series com um índice
pd.cut(x = dados.Renda,
bins = classes,
labels = labels,
include_lowest = True)
frequencia_renda = pd.value_counts(
pd.cut(x = dados.Renda,
bins = classes,
labels = labels,
include_lowest = True)
)
frequencia_renda
percentual_renda = pd.value_counts(
pd.cut(x = dados.Renda,
bins = classes,
labels = labels,
include_lowest = True),
normalize = True
)
percentual_renda
```
**Passo 5** Criando o Dataframe
```
# Criando o dataframe e ordenando
dist_freq_quantitativas_renda = pd.DataFrame(
{'Frequência': frequencia_renda, 'Porcentagem (%)': percentual_renda}
)
dist_freq_quantitativas_renda.sort_index(ascending = False, inplace=True)
dist_freq_quantitativas_renda
```
**Variável Altura**
```
# Definindo categorias
classes_altura = [dados.Altura.min(), 1.65, 1.75, dados.Altura.max()]
labels_altura = ['1 - Baixa', '2 - Média', '3 - Alta']
frequencia_altura = pd.value_counts(
pd.cut(
x = dados.Altura,
bins = classes_altura,
labels = labels_altura,
include_lowest = True
)
)
percentual_altura = pd.value_counts(
pd.cut(
x = dados.Altura,
bins = classes_altura,
labels = labels_altura,
include_lowest = True
), normalize = True
) * 100
# Criando Dataframe e renomando
dist_freq_altura = pd.DataFrame(
{'Frequência': frequencia_altura, 'Porcentagem (%)': percentual_altura}
)
dist_freq_altura.rename_axis('Estaturas', axis= 'columns', inplace = True)
dist_freq_altura.sort_index(ascending = True, inplace = True)
dist_freq_altura
```
---------------------
### 2.3. Distribuição de frequências para variáveis quantitativas (classes de amplitude fixa)
**Variável Renda**
*Utilizando a Regra de Sturges*
**PS:** Regra de Sturges: otimiza a escolha da quantidade de classes que teremos nas tabelas de distribuições, considerando somente o número de observações que temos na variável. Fórmula:
$$k = 1 + \frac {10}{3}\log_{10}n$$
```
# Quantidade de observações: n
n = dados.shape[0]
# número de classes de amplitude fixa por meio da regra de Sturges
k = 1 + (10 /3) * np.log10(n)
k = int(k.round(0)) # arredonda e tranforma em numero inteiro
k
# Criando series
frequencia_renda_rs = pd.value_counts(
pd.cut(
x = dados.Renda,
bins = 17, # valor de k
include_lowest = True # para incluir o item mais baixo
),
sort = False # ordenado pela primeira coluna
)
percentual_renda_rs = pd.value_counts(
pd.cut(
x = dados.Renda,
bins = 17,
include_lowest = True
),
sort = False,
normalize = True
) * 100
# Criando o Dataframe
dist_freq_quantitativas_amplitude_fixa = pd.DataFrame(
{'Frequência': frequencia_renda_rs, 'Porcentagem (%)': percentual_renda_rs})
dist_freq_quantitativas_amplitude_fixa
```
------------------
### 2.4. Representação gráfica das frequências
**Histograma da Altura**
```
# Histograma das alturas
ax_altura = sns.displot(data=dados.Altura, kde=True, height=6)
ax_altura
# Histograma das classificações das alturas
dist_freq_altura['Frequência'].plot.bar(width= 0.7, alpha = 0.2, figsize= (6, 5))
```
**Distribuição de Frequências Acumulada** - Idade
```
ax = sns.distplot(dados.Idade, hist_kws = {'cumulative': True}, kde_kws = {'cumulative': True})
ax.figure.set_size_inches(10, 6)
ax.set_title('Distribuição de Frequências Acumulada', fontsize=18)
ax.set_ylabel('Acumulado', fontsize=14)
ax.set_xlabel('Anos', fontsize=14)
ax
ax = sns.displot(data=dados.Idade, kind="ecdf")
ax
```
## Parte III: Medidas de Tendência Central
**Notebook 2**
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
from scipy.special import comb
from scipy.stats import binom
dados = pd.read_csv('dataset.csv')
# Visualizando o início
dados.head(3)
# Informações gerais
dados.info()
#Exemplo: anos de estudo, idade
print('Anos de estudo:' , sorted(dados['Anos de Estudo'].unique()))
print('Idade:' , sorted(dados['Idade'].unique()))
# Exemplo:
print('Sexo: ', sorted(dados['Sexo'].unique()))
print('UF: ', dados['UF'].unique())
print('Cor: ', dados['Cor'].unique())
#mostra a idade minima e maxima
print(f"Variável 'Idade' vai de {dados.Idade.min()} até {dados.Idade.max()} anos.")
print("A variável 'Altura' vai de %s até %s metros." % (dados.Altura.min(), dados.Altura.max()))
# Quantidade total:
frequencia_sexo = dados['Sexo'].value_counts()
frequencia_sexo
# Proporção:
porcentual_sexo = dados['Sexo'].value_counts(normalize = True) * 100
porcentual_sexo
# Montando um dataframe, renomeando variáveis e colunas:
dist_freq_qualitativas = pd.DataFrame({'Frequência': frequencia_sexo, 'Porcentagem (%)': porcentual_sexo})
dist_freq_qualitativas.rename(index = {0: 'Masculino', 1: 'Feminino'}, inplace = True)
dist_freq_qualitativas.rename_axis('Sexo', axis = 'columns', inplace = True)
dist_freq_qualitativas
sexo = {0: 'Masculino', 1: 'Feminino'}
cor = {0: 'Indígena',
2: 'Branca',
4: 'Preta',
6: 'Amarela',
8: 'Parda',
9: 'Sem declaração'}
# Tabela com frequências de pessoas com cada cor
percentualSC = pd.crosstab(dados.Sexo,
dados.Cor,
normalize = True) * 100
percentualSC.rename(index = sexo, inplace = True)
percentualSC.rename(columns = cor, inplace = True)
percentualSC
# Extremos dos dados:
print(f"Renda máxima: {dados.Renda.max()}\t Renda mínima: {dados.Renda.min()}.")
# Variáveis para dicionário:
classes = [0, 1576, 3152, 7880, 15760, 200000]
labels = ['E', 'D', 'C', 'B', 'A']
# Criará uma series com um índice
pd.cut(x = dados.Renda,
bins = classes,
labels = labels,
include_lowest = True)
frequencia_renda = pd.value_counts(
pd.cut(x = dados.Renda,
bins = classes,
labels = labels,
include_lowest = True)
)
frequencia_renda
percentual_renda = pd.value_counts(
pd.cut(x = dados.Renda,
bins = classes,
labels = labels,
include_lowest = True),
normalize = True
)
percentual_renda
# Criando o dataframe e ordenando
dist_freq_quantitativas_renda = pd.DataFrame(
{'Frequência': frequencia_renda, 'Porcentagem (%)': percentual_renda}
)
dist_freq_quantitativas_renda.sort_index(ascending = False, inplace=True)
dist_freq_quantitativas_renda
# Definindo categorias
classes_altura = [dados.Altura.min(), 1.65, 1.75, dados.Altura.max()]
labels_altura = ['1 - Baixa', '2 - Média', '3 - Alta']
frequencia_altura = pd.value_counts(
pd.cut(
x = dados.Altura,
bins = classes_altura,
labels = labels_altura,
include_lowest = True
)
)
percentual_altura = pd.value_counts(
pd.cut(
x = dados.Altura,
bins = classes_altura,
labels = labels_altura,
include_lowest = True
), normalize = True
) * 100
# Criando Dataframe e renomando
dist_freq_altura = pd.DataFrame(
{'Frequência': frequencia_altura, 'Porcentagem (%)': percentual_altura}
)
dist_freq_altura.rename_axis('Estaturas', axis= 'columns', inplace = True)
dist_freq_altura.sort_index(ascending = True, inplace = True)
dist_freq_altura
# Quantidade de observações: n
n = dados.shape[0]
# número de classes de amplitude fixa por meio da regra de Sturges
k = 1 + (10 /3) * np.log10(n)
k = int(k.round(0)) # arredonda e tranforma em numero inteiro
k
# Criando series
frequencia_renda_rs = pd.value_counts(
pd.cut(
x = dados.Renda,
bins = 17, # valor de k
include_lowest = True # para incluir o item mais baixo
),
sort = False # ordenado pela primeira coluna
)
percentual_renda_rs = pd.value_counts(
pd.cut(
x = dados.Renda,
bins = 17,
include_lowest = True
),
sort = False,
normalize = True
) * 100
# Criando o Dataframe
dist_freq_quantitativas_amplitude_fixa = pd.DataFrame(
{'Frequência': frequencia_renda_rs, 'Porcentagem (%)': percentual_renda_rs})
dist_freq_quantitativas_amplitude_fixa
# Histograma das alturas
ax_altura = sns.displot(data=dados.Altura, kde=True, height=6)
ax_altura
# Histograma das classificações das alturas
dist_freq_altura['Frequência'].plot.bar(width= 0.7, alpha = 0.2, figsize= (6, 5))
ax = sns.distplot(dados.Idade, hist_kws = {'cumulative': True}, kde_kws = {'cumulative': True})
ax.figure.set_size_inches(10, 6)
ax.set_title('Distribuição de Frequências Acumulada', fontsize=18)
ax.set_ylabel('Acumulado', fontsize=14)
ax.set_xlabel('Anos', fontsize=14)
ax
ax = sns.displot(data=dados.Idade, kind="ecdf")
ax
| 0.271928 | 0.906239 |
### Start dask cluster
```
from dask.distributed import Client
client = Client("tcp://10.32.5.46:44639")
client
```
### Import required packages
```
import intake
import xarray as xr
from matplotlib import pyplot as plt
import numpy as np
import xgcm
import dask
import pprint
import gsw
import xesmf as xe
from xhistogram.xarray import histogram
from datetime import datetime
# local file for CMIP6 preprocessing
import preprocessing
```
### Locate UKESM1 ozone data
```
col_url='https://raw.githubusercontent.com/NCAR/intake-esm-datastore/master/catalogs/pangeo-cmip6.json'
col = intake.open_esm_datastore(col_url)
cat = col.search(institution_id='MOHC',
source_id='UKESM1-0-LL',
table_id='Amon',
experiment_id=['historical','ssp126','ssp585'],
variable_id=['o3'],
member_id=['r1i1p1f2'],
grid_label='gn')
cat.df
dset_dict = cat.to_dataset_dict(zarr_kwargs={'consolidated': True, 'decode_times': True},
cdf_kwargs={'chunks': {}, 'decode_times': True})
dset_dict.keys()
```
### Preprocess the historical, ssp126, and ssp585 datasets
```
historical = dset_dict['CMIP.MOHC.UKESM1-0-LL.historical.Amon.gn']
ssp126 = dset_dict['ScenarioMIP.MOHC.UKESM1-0-LL.ssp126.Amon.gn']
ssp585 = dset_dict['ScenarioMIP.MOHC.UKESM1-0-LL.ssp585.Amon.gn']
historical = preprocessing.combined_preprocessing(historical)
ssp126 = preprocessing.combined_preprocessing(ssp126)
ssp585 = preprocessing.combined_preprocessing(ssp585)
```
### Drop the coordinates that are not needed
```
historical = historical.drop(('lon_bnds','time_bounds','lat_bnds','member_id','bnds'))
ssp126 = ssp126.drop(('lon_bnds','time_bounds','lat_bnds','member_id','bnds'))
ssp585 = ssp585.drop(('lon_bnds','time_bounds','lat_bnds','member_id','bnds'))
```
### Calculate annual mean profiles
```
historical = historical.groupby('time.year').mean('time')
ssp126 = ssp126.groupby('time.year').mean('time')
ssp585 = ssp585.groupby('time.year').mean('time')
```
### Slice one decade from each experiment
```
historical = historical.sel(year=slice(2004,2014))
ssp126 = ssp126.sel(year=slice(2090,2100))
ssp585 = ssp585.sel(year=slice(2090,2100))
historical = historical.squeeze()
ssp126 = ssp126.squeeze()
ssp585 = ssp585.squeeze()
# scale to get units in ppmv ()
historical['o3'] = historical.o3*1e6
ssp126['o3'] = ssp126.o3*1e6
ssp585['o3'] = ssp585.o3*1e6
```
### Prepare attributes for combined NetCDF file
```
historical.attrs['Prepared by'] = 'D. Jones'
historical.attrs['Institute'] = 'British Antarctic Survey'
historical.attrs['Units'] = 'ppmv'
historical.attrs['Model Info'] = 'UK Earth System Model 1'
historical.attrs['Description'] = 'Annual mean ozone profiles from historical experiment'
historical.attrs['Years covered'] = '2004-2014'
ssp126.attrs['Prepared by'] = 'D. Jones'
ssp126.attrs['Institute'] = 'British Antarctic Survey'
ssp126.attrs['Units'] = 'ppmv'
ssp126.attrs['Model Info'] = 'UK Earth System Model 1'
ssp126.attrs['Description'] = 'Annual mean ozone profiles from ssp126 experiment (strong emissions reductions)'
ssp126.attrs['Years covered'] = '2090-2100'
ssp585.attrs['Prepared by'] = 'D. Jones'
ssp585.attrs['Institute'] = 'British Antarctic Survey'
ssp585.attrs['Units'] = 'ppmv'
ssp585.attrs['Model Info'] = 'UK Earth System Model 1'
ssp585.attrs['Description'] = 'Annual mean ozone profiles from ssp585 experiment (strong emissions reductions)'
ssp585.attrs['Years covered'] = '2090-2100'
```
### Save results to NetCDF files
```
historical.load()
historical.to_netcdf(path='./data_out/UKESM_O3_historical_v2.nc')
ssp126.load()
ssp126.to_netcdf(path='./data_out/UKESM_O3_ssp126_v2.nc')
ssp585.load()
ssp585.to_netcdf(path='./data_out/UKESM_O3_ssp585_v2.nc')
```
### Create merged dataset, save to single NetCDF file
```
merged = xr.concat([historical,ssp126,ssp585],dim='year')
merged.to_netcdf(path='./data_out/UKESM_O3_merged.nc')
```
|
github_jupyter
|
from dask.distributed import Client
client = Client("tcp://10.32.5.46:44639")
client
import intake
import xarray as xr
from matplotlib import pyplot as plt
import numpy as np
import xgcm
import dask
import pprint
import gsw
import xesmf as xe
from xhistogram.xarray import histogram
from datetime import datetime
# local file for CMIP6 preprocessing
import preprocessing
col_url='https://raw.githubusercontent.com/NCAR/intake-esm-datastore/master/catalogs/pangeo-cmip6.json'
col = intake.open_esm_datastore(col_url)
cat = col.search(institution_id='MOHC',
source_id='UKESM1-0-LL',
table_id='Amon',
experiment_id=['historical','ssp126','ssp585'],
variable_id=['o3'],
member_id=['r1i1p1f2'],
grid_label='gn')
cat.df
dset_dict = cat.to_dataset_dict(zarr_kwargs={'consolidated': True, 'decode_times': True},
cdf_kwargs={'chunks': {}, 'decode_times': True})
dset_dict.keys()
historical = dset_dict['CMIP.MOHC.UKESM1-0-LL.historical.Amon.gn']
ssp126 = dset_dict['ScenarioMIP.MOHC.UKESM1-0-LL.ssp126.Amon.gn']
ssp585 = dset_dict['ScenarioMIP.MOHC.UKESM1-0-LL.ssp585.Amon.gn']
historical = preprocessing.combined_preprocessing(historical)
ssp126 = preprocessing.combined_preprocessing(ssp126)
ssp585 = preprocessing.combined_preprocessing(ssp585)
historical = historical.drop(('lon_bnds','time_bounds','lat_bnds','member_id','bnds'))
ssp126 = ssp126.drop(('lon_bnds','time_bounds','lat_bnds','member_id','bnds'))
ssp585 = ssp585.drop(('lon_bnds','time_bounds','lat_bnds','member_id','bnds'))
historical = historical.groupby('time.year').mean('time')
ssp126 = ssp126.groupby('time.year').mean('time')
ssp585 = ssp585.groupby('time.year').mean('time')
historical = historical.sel(year=slice(2004,2014))
ssp126 = ssp126.sel(year=slice(2090,2100))
ssp585 = ssp585.sel(year=slice(2090,2100))
historical = historical.squeeze()
ssp126 = ssp126.squeeze()
ssp585 = ssp585.squeeze()
# scale to get units in ppmv ()
historical['o3'] = historical.o3*1e6
ssp126['o3'] = ssp126.o3*1e6
ssp585['o3'] = ssp585.o3*1e6
historical.attrs['Prepared by'] = 'D. Jones'
historical.attrs['Institute'] = 'British Antarctic Survey'
historical.attrs['Units'] = 'ppmv'
historical.attrs['Model Info'] = 'UK Earth System Model 1'
historical.attrs['Description'] = 'Annual mean ozone profiles from historical experiment'
historical.attrs['Years covered'] = '2004-2014'
ssp126.attrs['Prepared by'] = 'D. Jones'
ssp126.attrs['Institute'] = 'British Antarctic Survey'
ssp126.attrs['Units'] = 'ppmv'
ssp126.attrs['Model Info'] = 'UK Earth System Model 1'
ssp126.attrs['Description'] = 'Annual mean ozone profiles from ssp126 experiment (strong emissions reductions)'
ssp126.attrs['Years covered'] = '2090-2100'
ssp585.attrs['Prepared by'] = 'D. Jones'
ssp585.attrs['Institute'] = 'British Antarctic Survey'
ssp585.attrs['Units'] = 'ppmv'
ssp585.attrs['Model Info'] = 'UK Earth System Model 1'
ssp585.attrs['Description'] = 'Annual mean ozone profiles from ssp585 experiment (strong emissions reductions)'
ssp585.attrs['Years covered'] = '2090-2100'
historical.load()
historical.to_netcdf(path='./data_out/UKESM_O3_historical_v2.nc')
ssp126.load()
ssp126.to_netcdf(path='./data_out/UKESM_O3_ssp126_v2.nc')
ssp585.load()
ssp585.to_netcdf(path='./data_out/UKESM_O3_ssp585_v2.nc')
merged = xr.concat([historical,ssp126,ssp585],dim='year')
merged.to_netcdf(path='./data_out/UKESM_O3_merged.nc')
| 0.327991 | 0.738598 |
# Maximum likelihood
In Bayesian modelling, the **likelihood**, $L$, is the name given to the measure of the goodness of fit between the model, with some given variables, and the data.
When the likelihood is maximised, $\hat{L}$, the most probable statistical model has been found for the given data.
In this tutorial we will see how `uravu` can be used to maximize the likelihood of a model for some dataset.
In `uravu`, when the sample is normally distributed the likelihood is calculated as follows,
$$ \ln L = -0.5 \sum_{i=1}^n \bigg[ \frac{(y_i - m_i) ^2}{\delta y_i^2} + \ln(2 \pi \delta y_i^2) \bigg], $$
where, $y$ is the data ordinate, $m$ is the model ordinate, and $\delta y_i$ is uncertainty in $y$.
`uravu` is able to maximize this function using the [scipy.optimize.minimize()](https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.minimize.html) function (we minimize the negative of the likelihood).
Before we maximise the likelihood, is it necessary to create some *synthetic* data to analyse.
```
import numpy as np
import matplotlib.pyplot as plt
from uravu import plotting
from uravu.relationship import Relationship
np.random.seed(1)
x = np.linspace(0, 10, 20)
y = np.exp(0.5 * x) * 4
y += y * np.random.randn(20) * 0.1
dy = y * 0.2
plt.errorbar(x, y, dy, marker='o', ls='')
plt.yscale('log')
plt.show()
```
The data plotted above (note the logarthimic $y$-axis) may be modelled with the following relationship,
$$ y = a\exp(bx), $$
where $a$ and $b$ are the variables of interest in the modelling process.
We want to find the values for these variables, which maximises the likelihood.
First, we must write a function to describe the model (more about the function specification can be found in teh [Input functions](./input_functions.html) tutorial).
```
def my_model(x, a, b):
"""
A function to describe the model under investgation.
Args:
x (array_like): Abscissa data.
a (float): The pre-exponential factor.
b (float): The x-multiplicative factor.
Returns
y (array_like): Ordinate data.
"""
return a * np.exp(b * x)
```
With our model defined, we can construct a `Relationship` object.
```
modeller = Relationship(my_model, x, y, ordinate_error=dy)
```
The `Relationship` object gives us access to a few powerful Bayesian modelling methods.
However, this tutorial is focused on maximising the likelihood, this is achieved with the `max_likelihood()` class method, where the keyword `'mini'` indicates the standard [scipy.optimize.minimize()](https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.minimize.html) function should be used.
```
modeller.max_likelihood('mini')
print(modeller.variable_modes)
```
We can see that the variables are close to the values used in the data synthesis.
Note, that here `variable_modes` are in fact the variable values that maximise the likelihood.
Let's inspect the model visually.
This can be achieved easily with the `plotting` module in `uravu`.
```
ax = plotting.plot_relationship(modeller)
plt.yscale('log')
plt.show()
```
Above, we can see that the orange line of maximum likelihood agrees well with the data.
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from uravu import plotting
from uravu.relationship import Relationship
np.random.seed(1)
x = np.linspace(0, 10, 20)
y = np.exp(0.5 * x) * 4
y += y * np.random.randn(20) * 0.1
dy = y * 0.2
plt.errorbar(x, y, dy, marker='o', ls='')
plt.yscale('log')
plt.show()
def my_model(x, a, b):
"""
A function to describe the model under investgation.
Args:
x (array_like): Abscissa data.
a (float): The pre-exponential factor.
b (float): The x-multiplicative factor.
Returns
y (array_like): Ordinate data.
"""
return a * np.exp(b * x)
modeller = Relationship(my_model, x, y, ordinate_error=dy)
modeller.max_likelihood('mini')
print(modeller.variable_modes)
ax = plotting.plot_relationship(modeller)
plt.yscale('log')
plt.show()
| 0.733833 | 0.994563 |
<a href="https://colab.research.google.com/github/mandarup/deeplearning/blob/master/models/keras_nmt_with_attention_for_chat.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
##### Copyright 2018 The TensorFlow Authors.
Licensed under the Apache License, Version 2.0 (the "License").
# Neural Machine Translation with Attention
<table class="tfo-notebook-buttons" align="left"><td>
<a target="_blank" href="https://colab.sandbox.google.com/github/tensorflow/tensorflow/blob/master/tensorflow/contrib/eager/python/examples/nmt_with_attention/nmt_with_attention.ipynb">
<img src="https://www.tensorflow.org/images/colab_logo_32px.png" /><span>Run in Google Colab</span></a>
</td><td>
<a target="_blank" href="https://github.com/tensorflow/tensorflow/tree/master/tensorflow/contrib/eager/python/examples/nmt_with_attention/nmt_with_attention.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /><span>View source on GitHub</span></a></td></table>
This notebook trains a sequence to sequence (seq2seq) model for Spanish to English translation using [tf.keras](https://www.tensorflow.org/programmers_guide/keras) and [eager execution](https://www.tensorflow.org/programmers_guide/eager). This is an advanced example that assumes some knowledge of sequence to sequence models.
After training the model in this notebook, you will be able to input a Spanish sentence, such as *"¿todavia estan en casa?"*, and return the English translation: *"are you still at home?"*
The translation quality is reasonable for a toy example, but the generated attention plot is perhaps more interesting. This shows which parts of the input sentence has the model's attention while translating:
<img src="https://tensorflow.org/images/spanish-english.png" alt="spanish-english attention plot">
Note: This example takes approximately 10 mintues to run on a single P100 GPU.
```
from __future__ import absolute_import, division, print_function
# Import TensorFlow >= 1.9 and enable eager execution
import tensorflow as tf
tf.enable_eager_execution()
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
import unicodedata
import re
import numpy as np
import os
import time
print(tf.__version__)
from google.colab import drive
drive.mount('/content/drive/')
!ls "/content/drive/"
datadir = "/content/drive/My Drive/Colab Notebooks/data"
checkpoints_dir = "/content/drive/My Drive/Colab Notebooks/checkpoints/curiousbot/seq2seq/v1"
```
## Download and prepare the dataset
We'll use a language dataset provided by http://www.manythings.org/anki/. This dataset contains language translation pairs in the format:
```
May I borrow this book? ¿Puedo tomar prestado este libro?
```
There are a variety of languages available, but we'll use the English-Spanish dataset. For convenience, we've hosted a copy of this dataset on Google Cloud, but you can also download your own copy. After downloading the dataset, here are the steps we'll take to prepare the data:
1. Add a *start* and *end* token to each sentence.
2. Clean the sentences by removing special characters.
3. Create a word index and reverse word index (dictionaries mapping from word → id and id → word).
4. Pad each sentence to a maximum length.
```
# # Download the file
# path_to_zip = tf.keras.utils.get_file(
# 'spa-eng.zip', origin='http://download.tensorflow.org/data/spa-eng.zip',
# extract=True)
# path_to_file = os.path.dirname(path_to_zip)+"/spa-eng/spa.txt"
corpus_name = "cornell-movie-dialogs-corpus"
corpus = os.path.join(datadir, corpus_name)
path_to_file = os.path.join(corpus, "formatted_movie_lines.txt")
# Converts the unicode file to ascii
def unicode_to_ascii(s):
return ''.join(c for c in unicodedata.normalize('NFD', s)
if unicodedata.category(c) != 'Mn')
def preprocess_sentence(w):
w = unicode_to_ascii(w.lower().strip())
# creating a space between a word and the punctuation following it
# eg: "he is a boy." => "he is a boy ."
# Reference:- https://stackoverflow.com/questions/3645931/python-padding-punctuation-with-white-spaces-keeping-punctuation
w = re.sub(r"([?.!,¿])", r" \1 ", w)
w = re.sub(r'[" "]+', " ", w)
# replacing everything with space except (a-z, A-Z, ".", "?", "!", ",")
w = re.sub(r"[^a-zA-Z?.!,¿]+", " ", w)
w = w.rstrip().strip()
# adding a start and an end token to the sentence
# so that the model know when to start and stop predicting.
w = '<start> ' + w + ' <end>'
return w
# 1. Remove the accents
# 2. Clean the sentences
# 3. Return word pairs in the format: [ENGLISH, SPANISH]
def create_dataset(path, num_examples=None):
lines = open(path, encoding='UTF-8').read().strip().split('\n')
# load all examples if not none
if num_examples is None:
num_examples = -1
word_pairs = [[preprocess_sentence(w) for w in l.split('\t')] for l in lines[:num_examples]]
return word_pairs
# Default word tokens
PAD_token = 0 # Used for padding short sentences
SOS_token = 1 # Start-of-sentence token
EOS_token = 2 # End-of-sentence token
class Voc:
def __init__(self, name):
self.name = name
self.trimmed = False
self.word2index = {}
self.word2count = {}
self.index2word = {PAD_token: "PAD", SOS_token: "SOS", EOS_token: "EOS"}
self.num_words = 3 # Count SOS, EOS, PAD
def addSentence(self, sentence):
for word in sentence.split(' '):
self.addWord(word)
def addWord(self, word):
if word not in self.word2index:
self.word2index[word] = self.num_words
self.word2count[word] = 1
self.index2word[self.num_words] = word
self.num_words += 1
else:
self.word2count[word] += 1
# Remove words below a certain count threshold
def trim(self, min_count):
if self.trimmed:
return
self.trimmed = True
keep_words = []
for k, v in self.word2count.items():
if v >= min_count:
keep_words.append(k)
print('keep_words {} / {} = {:.4f}'.format(
len(keep_words), len(self.word2index), len(keep_words) / len(self.word2index)
))
# Reinitialize dictionaries
self.word2index = {}
self.word2count = {}
self.index2word = {PAD_token: "PAD", SOS_token: "SOS", EOS_token: "EOS"}
self.num_words = 3 # Count default tokens
for word in keep_words:
self.addWord(word)
# This class creates a word -> index mapping (e.g,. "dad" -> 5) and vice-versa
# (e.g., 5 -> "dad") for each language,
class LanguageIndex():
def __init__(self, lang):
self.lang = lang
self.word2idx = {}
self.idx2word = {}
self.vocab = set()
self.create_index()
def create_index(self):
for phrase in self.lang:
self.vocab.update(phrase.split(' '))
self.vocab = sorted(self.vocab)
self.word2idx['<pad>'] = 0
for index, word in enumerate(self.vocab):
self.word2idx[word] = index + 1
for word, index in self.word2idx.items():
self.idx2word[index] = word
def max_length(tensor):
return max(len(t) for t in tensor)
def load_dataset(path, num_examples):
# creating cleaned input, output pairs
pairs = create_dataset(path, num_examples)
# index language using the class defined above
inp_lang = LanguageIndex(sp for en, sp in pairs)
targ_lang = LanguageIndex(en for en, sp in pairs)
# Vectorize the input and target languages
# Spanish sentences
input_tensor = [[inp_lang.word2idx[s] for s in sp.split(' ')] for en, sp in pairs]
# English sentences
target_tensor = [[targ_lang.word2idx[s] for s in en.split(' ')] for en, sp in pairs]
# Calculate max_length of input and output tensor
# Here, we'll set those to the longest sentence in the dataset
max_length_inp, max_length_tar = max_length(input_tensor), max_length(target_tensor)
# Padding the input and output tensor to the maximum length
input_tensor = tf.keras.preprocessing.sequence.pad_sequences(input_tensor,
maxlen=max_length_inp,
padding='post')
target_tensor = tf.keras.preprocessing.sequence.pad_sequences(target_tensor,
maxlen=max_length_tar,
padding='post')
return input_tensor, target_tensor, inp_lang, targ_lang, max_length_inp, max_length_tar
```
### Limit the size of the dataset to experiment faster (optional)
Training on the complete dataset of >100,000 sentences will take a long time. To train faster, we can limit the size of the dataset to 30,000 sentences (of course, translation quality degrades with less data):
```
pairs = create_dataset(path_to_file, num_examples=None)
len(pairs)
del(pairs)
# pairs[-3:]
# min([len(conv) for conv in pairs])
# inp_lang = LanguageIndex(sp for en, sp in pairs)
# inp_lang.word2idx
# input_tensor = [[inp_lang.word2idx[s] for s in sp.split(' ')] for en, sp in pairs]
# tar_lang = LanguageIndex(en for en, sp in pairs)
# tar_tensor = [[tar_lang.word2idx[e] for e in en.split(' ')] for en, sp in pairs]
# targ_lang.word2idx
# Try experimenting with the size of that dataset
num_examples = 10000
input_tensor, target_tensor, inp_lang, targ_lang, max_length_inp, max_length_targ = load_dataset(path_to_file, num_examples)
# Creating training and validation sets using an 80-20 split
input_tensor_train, input_tensor_val, target_tensor_train, target_tensor_val = train_test_split(input_tensor, target_tensor, test_size=0.2)
# Show length
len(input_tensor_train), len(target_tensor_train), len(input_tensor_val), len(target_tensor_val)
```
### Create a tf.data dataset
```
BUFFER_SIZE = len(input_tensor_train)
BATCH_SIZE = 8
embedding_dim = 256
units = 256
vocab_inp_size = len(inp_lang.word2idx)
vocab_tar_size = len(targ_lang.word2idx)
dataset = tf.data.Dataset.from_tensor_slices((input_tensor_train, target_tensor_train)).shuffle(BUFFER_SIZE)
dataset = dataset.apply(tf.contrib.data.batch_and_drop_remainder(BATCH_SIZE))
```
## Write the encoder and decoder model
Here, we'll implement an encoder-decoder model with attention which you can read about in the TensorFlow [Neural Machine Translation (seq2seq) tutorial](https://www.tensorflow.org/tutorials/seq2seq). This example uses a more recent set of APIs. This notebook implements the [attention equations](https://www.tensorflow.org/tutorials/seq2seq#background_on_the_attention_mechanism) from the seq2seq tutorial. The following diagram shows that each input words is assigned a weight by the attention mechanism which is then used by the decoder to predict the next word in the sentence.
<img src="https://www.tensorflow.org/images/seq2seq/attention_mechanism.jpg" width="500" alt="attention mechanism">
The input is put through an encoder model which gives us the encoder output of shape *(batch_size, max_length, hidden_size)* and the encoder hidden state of shape *(batch_size, hidden_size)*.
Here are the equations that are implemented:
<img src="https://www.tensorflow.org/images/seq2seq/attention_equation_0.jpg" alt="attention equation 0" width="800">
<img src="https://www.tensorflow.org/images/seq2seq/attention_equation_1.jpg" alt="attention equation 1" width="800">
We're using *Bahdanau attention*. Lets decide on notation before writing the simplified form:
* FC = Fully connected (dense) layer
* EO = Encoder output
* H = hidden state
* X = input to the decoder
And the pseudo-code:
* `score = FC(tanh(FC(EO) + FC(H)))`
* `attention weights = softmax(score, axis = 1)`. Softmax by default is applied on the last axis but here we want to apply it on the *1st axis*, since the shape of score is *(batch_size, max_length, hidden_size)*. `Max_length` is the length of our input. Since we are trying to assign a weight to each input, softmax should be applied on that axis.
* `context vector = sum(attention weights * EO, axis = 1)`. Same reason as above for choosing axis as 1.
* `embedding output` = The input to the decoder X is passed through an embedding layer.
* `merged vector = concat(embedding output, context vector)`
* This merged vector is then given to the GRU
The shapes of all the vectors at each step have been specified in the comments in the code:
```
def gru(units):
# If you have a GPU, we recommend using CuDNNGRU(provides a 3x speedup than GRU)
# the code automatically does that.
# if tf.test.is_gpu_available():
# return tf.keras.layers.CuDNNGRU(units,
# return_sequences=True,
# return_state=True,
# recurrent_initializer='glorot_uniform')
# else:
# return tf.keras.layers.GRU(units,
# return_sequences=True,
# return_state=True,
# recurrent_activation='sigmoid',
# recurrent_initializer='glorot_uniform')
return tf.keras.layers.GRU(units,
return_sequences=True,
return_state=True,
recurrent_activation='sigmoid',
recurrent_initializer='glorot_uniform')
class Encoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, enc_units, batch_sz):
super(Encoder, self).__init__()
self.batch_sz = batch_sz
self.enc_units = enc_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = gru(self.enc_units)
def call(self, x, hidden):
x = self.embedding(x)
output, state = self.gru(x, initial_state = hidden)
return output, state
def initialize_hidden_state(self):
return tf.zeros((self.batch_sz, self.enc_units))
class Decoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, dec_units, batch_sz):
super(Decoder, self).__init__()
self.batch_sz = batch_sz
self.dec_units = dec_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = gru(self.dec_units)
self.fc = tf.keras.layers.Dense(vocab_size)
# used for attention
self.W1 = tf.keras.layers.Dense(self.dec_units)
self.W2 = tf.keras.layers.Dense(self.dec_units)
self.V = tf.keras.layers.Dense(1)
def call(self, x, hidden, enc_output):
# enc_output shape == (batch_size, max_length, hidden_size)
# hidden shape == (batch_size, hidden size)
# hidden_with_time_axis shape == (batch_size, 1, hidden size)
# we are doing this to perform addition to calculate the score
hidden_with_time_axis = tf.expand_dims(hidden, 1)
# score shape == (batch_size, max_length, hidden_size)
score = tf.nn.tanh(self.W1(enc_output) + self.W2(hidden_with_time_axis))
# attention_weights shape == (batch_size, max_length, 1)
# we get 1 at the last axis because we are applying score to self.V
attention_weights = tf.nn.softmax(self.V(score), axis=1)
# context_vector shape after sum == (batch_size, hidden_size)
context_vector = attention_weights * enc_output
context_vector = tf.reduce_sum(context_vector, axis=1)
# x shape after passing through embedding == (batch_size, 1, embedding_dim)
x = self.embedding(x)
# x shape after concatenation == (batch_size, 1, embedding_dim + hidden_size)
x = tf.concat([tf.expand_dims(context_vector, 1), x], axis=-1)
# passing the concatenated vector to the GRU
output, state = self.gru(x)
# output shape == (batch_size * max_length, hidden_size)
output = tf.reshape(output, (-1, output.shape[2]))
# output shape == (batch_size * max_length, vocab)
x = self.fc(output)
return x, state, attention_weights
def initialize_hidden_state(self):
return tf.zeros((self.batch_sz, self.dec_units))
encoder = Encoder(vocab_inp_size, embedding_dim, units, BATCH_SIZE)
decoder = Decoder(vocab_tar_size, embedding_dim, units, BATCH_SIZE)
```
## Define the optimizer and the loss function
```
optimizer = tf.train.AdamOptimizer()
def loss_function(real, pred):
mask = 1 - np.equal(real, 0)
loss_ = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=real, logits=pred) * mask
return tf.reduce_mean(loss_)
```
## Training
1. Pass the *input* through the *encoder* which return *encoder output* and the *encoder hidden state*.
2. The encoder output, encoder hidden state and the decoder input (which is the *start token*) is passed to the decoder.
3. The decoder returns the *predictions* and the *decoder hidden state*.
4. The decoder hidden state is then passed back into the model and the predictions are used to calculate the loss.
5. Use *teacher forcing* to decide the next input to the decoder.
6. *Teacher forcing* is the technique where the *target word* is passed as the *next input* to the decoder.
7. The final step is to calculate the gradients and apply it to the optimizer and backpropagate.
```
EPOCHS = 10
for epoch in range(EPOCHS):
start = time.time()
hidden = encoder.initialize_hidden_state()
total_loss = 0
for (batch, (inp, targ)) in enumerate(dataset):
loss = 0
with tf.GradientTape() as tape:
enc_output, enc_hidden = encoder(inp, hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word2idx['<start>']] * BATCH_SIZE, 1)
# Teacher forcing - feeding the target as the next input
for t in range(1, targ.shape[1]):
# passing enc_output to the decoder
predictions, dec_hidden, _ = decoder(dec_input, dec_hidden, enc_output)
loss += loss_function(targ[:, t], predictions)
# using teacher forcing
dec_input = tf.expand_dims(targ[:, t], 1)
total_loss += (loss / int(targ.shape[1]))
variables = encoder.variables + decoder.variables
gradients = tape.gradient(loss, variables)
optimizer.apply_gradients(zip(gradients, variables), tf.train.get_or_create_global_step())
if batch % 100 == 0:
print('Epoch {} Batch {} Loss {:.4f}'.format(epoch + 1,
batch,
loss.numpy() / int(targ.shape[1])))
print('Epoch {} Loss {:.4f}'.format(epoch + 1,
total_loss/len(input_tensor)))
print('Time taken for 1 epoch {} sec\n'.format(time.time() - start))
```
## Translate
* The evaluate function is similar to the training loop, except we don't use *teacher forcing* here. The input to the decoder at each time step is its previous predictions along with the hidden state and the encoder output.
* Stop predicting when the model predicts the *end token*.
* And store the *attention weights for every time step*.
Note: The encoder output is calculated only once for one input.
```
def evaluate(sentence, encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ):
attention_plot = np.zeros((max_length_targ, max_length_inp))
sentence = preprocess_sentence(sentence)
try:
inputs = [inp_lang.word2idx[i] for i in sentence.split(' ')]
except KeyError as e:
return 'unknown word', sentence, None
inputs = tf.keras.preprocessing.sequence.pad_sequences([inputs], maxlen=max_length_inp, padding='post')
inputs = tf.convert_to_tensor(inputs)
result = ''
hidden = [tf.zeros((1, units))]
enc_out, enc_hidden = encoder(inputs, hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word2idx['<start>']], 0)
for t in range(max_length_targ):
predictions, dec_hidden, attention_weights = decoder(dec_input, dec_hidden, enc_out)
# storing the attention weigths to plot later on
attention_weights = tf.reshape(attention_weights, (-1, ))
attention_plot[t] = attention_weights.numpy()
predicted_id = tf.multinomial(tf.exp(predictions), num_samples=1)[0][0].numpy()
result += targ_lang.idx2word[predicted_id] + ' '
if targ_lang.idx2word[predicted_id] == '<end>':
return result, sentence, attention_plot
# the predicted ID is fed back into the model
dec_input = tf.expand_dims([predicted_id], 0)
return result, sentence, attention_plot
# function for plotting the attention weights
def plot_attention(attention, sentence, predicted_sentence):
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot(1, 1, 1)
ax.matshow(attention, cmap='viridis')
fontdict = {'fontsize': 14}
ax.set_xticklabels([''] + sentence, fontdict=fontdict, rotation=90)
ax.set_yticklabels([''] + predicted_sentence, fontdict=fontdict)
plt.show()
def translate(sentence, encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ):
result, sentence, attention_plot = evaluate(sentence, encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
print('Input: {}'.format(sentence))
print('Predicted translation: {}'.format(result))
attention_plot = attention_plot[:len(result.split(' ')), :len(sentence.split(' '))]
plot_attention(attention_plot, sentence.split(' '), result.split(' '))
translate('now ?', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
translate('esta es mi vida.', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
translate('¿todavia estan en casa?', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
# wrong translation
translate('trata de averiguarlo.', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
```
## Next steps
* [Download a different dataset](http://www.manythings.org/anki/) to experiment with translations, for example, English to German, or English to French.
* Experiment with training on a larger dataset, or using more epochs
```
```
|
github_jupyter
|
from __future__ import absolute_import, division, print_function
# Import TensorFlow >= 1.9 and enable eager execution
import tensorflow as tf
tf.enable_eager_execution()
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
import unicodedata
import re
import numpy as np
import os
import time
print(tf.__version__)
from google.colab import drive
drive.mount('/content/drive/')
!ls "/content/drive/"
datadir = "/content/drive/My Drive/Colab Notebooks/data"
checkpoints_dir = "/content/drive/My Drive/Colab Notebooks/checkpoints/curiousbot/seq2seq/v1"
May I borrow this book? ¿Puedo tomar prestado este libro?
# # Download the file
# path_to_zip = tf.keras.utils.get_file(
# 'spa-eng.zip', origin='http://download.tensorflow.org/data/spa-eng.zip',
# extract=True)
# path_to_file = os.path.dirname(path_to_zip)+"/spa-eng/spa.txt"
corpus_name = "cornell-movie-dialogs-corpus"
corpus = os.path.join(datadir, corpus_name)
path_to_file = os.path.join(corpus, "formatted_movie_lines.txt")
# Converts the unicode file to ascii
def unicode_to_ascii(s):
return ''.join(c for c in unicodedata.normalize('NFD', s)
if unicodedata.category(c) != 'Mn')
def preprocess_sentence(w):
w = unicode_to_ascii(w.lower().strip())
# creating a space between a word and the punctuation following it
# eg: "he is a boy." => "he is a boy ."
# Reference:- https://stackoverflow.com/questions/3645931/python-padding-punctuation-with-white-spaces-keeping-punctuation
w = re.sub(r"([?.!,¿])", r" \1 ", w)
w = re.sub(r'[" "]+', " ", w)
# replacing everything with space except (a-z, A-Z, ".", "?", "!", ",")
w = re.sub(r"[^a-zA-Z?.!,¿]+", " ", w)
w = w.rstrip().strip()
# adding a start and an end token to the sentence
# so that the model know when to start and stop predicting.
w = '<start> ' + w + ' <end>'
return w
# 1. Remove the accents
# 2. Clean the sentences
# 3. Return word pairs in the format: [ENGLISH, SPANISH]
def create_dataset(path, num_examples=None):
lines = open(path, encoding='UTF-8').read().strip().split('\n')
# load all examples if not none
if num_examples is None:
num_examples = -1
word_pairs = [[preprocess_sentence(w) for w in l.split('\t')] for l in lines[:num_examples]]
return word_pairs
# Default word tokens
PAD_token = 0 # Used for padding short sentences
SOS_token = 1 # Start-of-sentence token
EOS_token = 2 # End-of-sentence token
class Voc:
def __init__(self, name):
self.name = name
self.trimmed = False
self.word2index = {}
self.word2count = {}
self.index2word = {PAD_token: "PAD", SOS_token: "SOS", EOS_token: "EOS"}
self.num_words = 3 # Count SOS, EOS, PAD
def addSentence(self, sentence):
for word in sentence.split(' '):
self.addWord(word)
def addWord(self, word):
if word not in self.word2index:
self.word2index[word] = self.num_words
self.word2count[word] = 1
self.index2word[self.num_words] = word
self.num_words += 1
else:
self.word2count[word] += 1
# Remove words below a certain count threshold
def trim(self, min_count):
if self.trimmed:
return
self.trimmed = True
keep_words = []
for k, v in self.word2count.items():
if v >= min_count:
keep_words.append(k)
print('keep_words {} / {} = {:.4f}'.format(
len(keep_words), len(self.word2index), len(keep_words) / len(self.word2index)
))
# Reinitialize dictionaries
self.word2index = {}
self.word2count = {}
self.index2word = {PAD_token: "PAD", SOS_token: "SOS", EOS_token: "EOS"}
self.num_words = 3 # Count default tokens
for word in keep_words:
self.addWord(word)
# This class creates a word -> index mapping (e.g,. "dad" -> 5) and vice-versa
# (e.g., 5 -> "dad") for each language,
class LanguageIndex():
def __init__(self, lang):
self.lang = lang
self.word2idx = {}
self.idx2word = {}
self.vocab = set()
self.create_index()
def create_index(self):
for phrase in self.lang:
self.vocab.update(phrase.split(' '))
self.vocab = sorted(self.vocab)
self.word2idx['<pad>'] = 0
for index, word in enumerate(self.vocab):
self.word2idx[word] = index + 1
for word, index in self.word2idx.items():
self.idx2word[index] = word
def max_length(tensor):
return max(len(t) for t in tensor)
def load_dataset(path, num_examples):
# creating cleaned input, output pairs
pairs = create_dataset(path, num_examples)
# index language using the class defined above
inp_lang = LanguageIndex(sp for en, sp in pairs)
targ_lang = LanguageIndex(en for en, sp in pairs)
# Vectorize the input and target languages
# Spanish sentences
input_tensor = [[inp_lang.word2idx[s] for s in sp.split(' ')] for en, sp in pairs]
# English sentences
target_tensor = [[targ_lang.word2idx[s] for s in en.split(' ')] for en, sp in pairs]
# Calculate max_length of input and output tensor
# Here, we'll set those to the longest sentence in the dataset
max_length_inp, max_length_tar = max_length(input_tensor), max_length(target_tensor)
# Padding the input and output tensor to the maximum length
input_tensor = tf.keras.preprocessing.sequence.pad_sequences(input_tensor,
maxlen=max_length_inp,
padding='post')
target_tensor = tf.keras.preprocessing.sequence.pad_sequences(target_tensor,
maxlen=max_length_tar,
padding='post')
return input_tensor, target_tensor, inp_lang, targ_lang, max_length_inp, max_length_tar
pairs = create_dataset(path_to_file, num_examples=None)
len(pairs)
del(pairs)
# pairs[-3:]
# min([len(conv) for conv in pairs])
# inp_lang = LanguageIndex(sp for en, sp in pairs)
# inp_lang.word2idx
# input_tensor = [[inp_lang.word2idx[s] for s in sp.split(' ')] for en, sp in pairs]
# tar_lang = LanguageIndex(en for en, sp in pairs)
# tar_tensor = [[tar_lang.word2idx[e] for e in en.split(' ')] for en, sp in pairs]
# targ_lang.word2idx
# Try experimenting with the size of that dataset
num_examples = 10000
input_tensor, target_tensor, inp_lang, targ_lang, max_length_inp, max_length_targ = load_dataset(path_to_file, num_examples)
# Creating training and validation sets using an 80-20 split
input_tensor_train, input_tensor_val, target_tensor_train, target_tensor_val = train_test_split(input_tensor, target_tensor, test_size=0.2)
# Show length
len(input_tensor_train), len(target_tensor_train), len(input_tensor_val), len(target_tensor_val)
BUFFER_SIZE = len(input_tensor_train)
BATCH_SIZE = 8
embedding_dim = 256
units = 256
vocab_inp_size = len(inp_lang.word2idx)
vocab_tar_size = len(targ_lang.word2idx)
dataset = tf.data.Dataset.from_tensor_slices((input_tensor_train, target_tensor_train)).shuffle(BUFFER_SIZE)
dataset = dataset.apply(tf.contrib.data.batch_and_drop_remainder(BATCH_SIZE))
def gru(units):
# If you have a GPU, we recommend using CuDNNGRU(provides a 3x speedup than GRU)
# the code automatically does that.
# if tf.test.is_gpu_available():
# return tf.keras.layers.CuDNNGRU(units,
# return_sequences=True,
# return_state=True,
# recurrent_initializer='glorot_uniform')
# else:
# return tf.keras.layers.GRU(units,
# return_sequences=True,
# return_state=True,
# recurrent_activation='sigmoid',
# recurrent_initializer='glorot_uniform')
return tf.keras.layers.GRU(units,
return_sequences=True,
return_state=True,
recurrent_activation='sigmoid',
recurrent_initializer='glorot_uniform')
class Encoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, enc_units, batch_sz):
super(Encoder, self).__init__()
self.batch_sz = batch_sz
self.enc_units = enc_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = gru(self.enc_units)
def call(self, x, hidden):
x = self.embedding(x)
output, state = self.gru(x, initial_state = hidden)
return output, state
def initialize_hidden_state(self):
return tf.zeros((self.batch_sz, self.enc_units))
class Decoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, dec_units, batch_sz):
super(Decoder, self).__init__()
self.batch_sz = batch_sz
self.dec_units = dec_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = gru(self.dec_units)
self.fc = tf.keras.layers.Dense(vocab_size)
# used for attention
self.W1 = tf.keras.layers.Dense(self.dec_units)
self.W2 = tf.keras.layers.Dense(self.dec_units)
self.V = tf.keras.layers.Dense(1)
def call(self, x, hidden, enc_output):
# enc_output shape == (batch_size, max_length, hidden_size)
# hidden shape == (batch_size, hidden size)
# hidden_with_time_axis shape == (batch_size, 1, hidden size)
# we are doing this to perform addition to calculate the score
hidden_with_time_axis = tf.expand_dims(hidden, 1)
# score shape == (batch_size, max_length, hidden_size)
score = tf.nn.tanh(self.W1(enc_output) + self.W2(hidden_with_time_axis))
# attention_weights shape == (batch_size, max_length, 1)
# we get 1 at the last axis because we are applying score to self.V
attention_weights = tf.nn.softmax(self.V(score), axis=1)
# context_vector shape after sum == (batch_size, hidden_size)
context_vector = attention_weights * enc_output
context_vector = tf.reduce_sum(context_vector, axis=1)
# x shape after passing through embedding == (batch_size, 1, embedding_dim)
x = self.embedding(x)
# x shape after concatenation == (batch_size, 1, embedding_dim + hidden_size)
x = tf.concat([tf.expand_dims(context_vector, 1), x], axis=-1)
# passing the concatenated vector to the GRU
output, state = self.gru(x)
# output shape == (batch_size * max_length, hidden_size)
output = tf.reshape(output, (-1, output.shape[2]))
# output shape == (batch_size * max_length, vocab)
x = self.fc(output)
return x, state, attention_weights
def initialize_hidden_state(self):
return tf.zeros((self.batch_sz, self.dec_units))
encoder = Encoder(vocab_inp_size, embedding_dim, units, BATCH_SIZE)
decoder = Decoder(vocab_tar_size, embedding_dim, units, BATCH_SIZE)
optimizer = tf.train.AdamOptimizer()
def loss_function(real, pred):
mask = 1 - np.equal(real, 0)
loss_ = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=real, logits=pred) * mask
return tf.reduce_mean(loss_)
EPOCHS = 10
for epoch in range(EPOCHS):
start = time.time()
hidden = encoder.initialize_hidden_state()
total_loss = 0
for (batch, (inp, targ)) in enumerate(dataset):
loss = 0
with tf.GradientTape() as tape:
enc_output, enc_hidden = encoder(inp, hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word2idx['<start>']] * BATCH_SIZE, 1)
# Teacher forcing - feeding the target as the next input
for t in range(1, targ.shape[1]):
# passing enc_output to the decoder
predictions, dec_hidden, _ = decoder(dec_input, dec_hidden, enc_output)
loss += loss_function(targ[:, t], predictions)
# using teacher forcing
dec_input = tf.expand_dims(targ[:, t], 1)
total_loss += (loss / int(targ.shape[1]))
variables = encoder.variables + decoder.variables
gradients = tape.gradient(loss, variables)
optimizer.apply_gradients(zip(gradients, variables), tf.train.get_or_create_global_step())
if batch % 100 == 0:
print('Epoch {} Batch {} Loss {:.4f}'.format(epoch + 1,
batch,
loss.numpy() / int(targ.shape[1])))
print('Epoch {} Loss {:.4f}'.format(epoch + 1,
total_loss/len(input_tensor)))
print('Time taken for 1 epoch {} sec\n'.format(time.time() - start))
def evaluate(sentence, encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ):
attention_plot = np.zeros((max_length_targ, max_length_inp))
sentence = preprocess_sentence(sentence)
try:
inputs = [inp_lang.word2idx[i] for i in sentence.split(' ')]
except KeyError as e:
return 'unknown word', sentence, None
inputs = tf.keras.preprocessing.sequence.pad_sequences([inputs], maxlen=max_length_inp, padding='post')
inputs = tf.convert_to_tensor(inputs)
result = ''
hidden = [tf.zeros((1, units))]
enc_out, enc_hidden = encoder(inputs, hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word2idx['<start>']], 0)
for t in range(max_length_targ):
predictions, dec_hidden, attention_weights = decoder(dec_input, dec_hidden, enc_out)
# storing the attention weigths to plot later on
attention_weights = tf.reshape(attention_weights, (-1, ))
attention_plot[t] = attention_weights.numpy()
predicted_id = tf.multinomial(tf.exp(predictions), num_samples=1)[0][0].numpy()
result += targ_lang.idx2word[predicted_id] + ' '
if targ_lang.idx2word[predicted_id] == '<end>':
return result, sentence, attention_plot
# the predicted ID is fed back into the model
dec_input = tf.expand_dims([predicted_id], 0)
return result, sentence, attention_plot
# function for plotting the attention weights
def plot_attention(attention, sentence, predicted_sentence):
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot(1, 1, 1)
ax.matshow(attention, cmap='viridis')
fontdict = {'fontsize': 14}
ax.set_xticklabels([''] + sentence, fontdict=fontdict, rotation=90)
ax.set_yticklabels([''] + predicted_sentence, fontdict=fontdict)
plt.show()
def translate(sentence, encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ):
result, sentence, attention_plot = evaluate(sentence, encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
print('Input: {}'.format(sentence))
print('Predicted translation: {}'.format(result))
attention_plot = attention_plot[:len(result.split(' ')), :len(sentence.split(' '))]
plot_attention(attention_plot, sentence.split(' '), result.split(' '))
translate('now ?', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
translate('esta es mi vida.', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
translate('¿todavia estan en casa?', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
# wrong translation
translate('trata de averiguarlo.', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
| 0.586404 | 0.983597 |
<a href="https://colab.research.google.com/github/Daniel-Loaiza/CodingSamples/blob/main/Titanic_Machine_Learning_from_Disaster.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
from google.colab import drive
drive.mount('/content/drive')
# We import the required modules and packages
import matplotlib.pyplot as plt
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
import seaborn as sns
from sklearn import tree
%matplotlib inline
os.chdir('/content/drive/MyDrive/Titanic - Machine Learning from Disaster')
os.listdir()
for dirname, _, filenames in os.walk('/content/drive/MyDrive/Titanic - Machine Learning from Disaster'):
for filename in filenames:
print(os.path.join(dirname, filename))
train = pd.read_csv('/content/drive/MyDrive/Titanic - Machine Learning from Disaster/train.csv')
test = pd.read_csv('/content/drive/MyDrive/Titanic - Machine Learning from Disaster/test.csv')
```
**Exploratory Data Analysis**
```
print(train.head())
print(test.head())
train.describe()
train.shape
# Passengers that survived vs passengers that passed away
print(train["Survived"].value_counts())
# As proportions
print(train["Survived"].value_counts(normalize = True))
# Males that survived vs males that passed away
print(train["Survived"][train["Sex"] == 'male'].value_counts())
# Females that survived vs Females that passed away
print(train["Survived"][train["Sex"] == 'female'].value_counts())
# Normalized male survival
print(train["Survived"][train["Sex"] == 'male'].value_counts(normalize = True))
# Normalized female survival
print(train["Survived"][train["Sex"] == 'female'].value_counts(normalize = True))
# Create the column Child and assign to 'NaN'
train["Child"] = float('NaN')
# Assign 1 to passengers under 18, 0 to those 18 or older. Print the new column.
train["Child"][train["Age"] < 18] = 1
train["Child"][train["Age"] >= 18] = 0
# Print normalized Survival Rates for passengers under 18
print(train["Survived"][train["Child"] == 1].value_counts(normalize = True))
# Print normalized Survival Rates for passengers 18 or older
print(train["Survived"][train["Child"] == 0].value_counts(normalize = True))
# Create a copy of test: test_one
test_one=test
# Initialize a Survived column to 0
test_one["Survived"]=0
# Set Survived to 1 if Sex equals "female" and print the `Survived` column from `test_one`
test_one["Survived"][test_one["Sex"] == "female"] = 1
print(test_one.Survived[:10])
```
**Decision Tree**
```
# Convert the male and female groups to integer form
train["Sex"][train["Sex"] == "male"] = 0
train["Sex"][train["Sex"] == "female"] = 1
# Impute the Embarked variable
train["Embarked"] = train["Embarked"].fillna("S")
# Impute the Age variable
train["Age"] = train["Age"].fillna(train["Age"].median())
# Convert the Embarked classes to integer form
train["Embarked"][train["Embarked"] == "S"] = 0
train["Embarked"][train["Embarked"] == "C"] = 1
train["Embarked"][train["Embarked"] == "Q"] = 2
#Print the Sex and Embarked columns
print(train["Sex"])
print(train["Embarked"])
# Print the train data to see the available features
print(train)
# Create the target and features numpy arrays: target, features_one
target = train["Survived"].values
features_one = train[["Pclass", "Sex", "Age", "Fare"]].values
target[:10]
features_one[:10]
# Fit your first decision tree: my_tree_one
my_tree_one = tree.DecisionTreeClassifier()
my_tree_one = my_tree_one.fit(features_one, target)
# Look at the importance and score of the included features
print(my_tree_one.feature_importances_)
print(my_tree_one.score(features_one, target))
my_tree_one.feature_importances_
# Impute the missing value with the median
test.Fare[153] = test["Fare"].median()
test["Sex"][test["Sex"] == "male"] = 0
test["Sex"][test["Sex"] == "female"] = 1
test["Age"] = test["Age"].fillna(train["Age"].median())
# Extract the features from the test set: Pclass, Sex, Age, and Fare.
test_features = test[['Pclass', 'Sex', 'Age', 'Fare']].values
# Make your prediction using the test set
my_prediction = my_tree_one.predict(test_features)
# Create a data frame with two columns: PassengerId & Survived. Survived contains your predictions
PassengerId =np.array(test["PassengerId"]).astype(int)
my_solution = pd.DataFrame(my_prediction, PassengerId, columns = ["Survived"])
print(my_solution)
# Check that your data frame has 418 entries
print(my_solution.shape)
# Write your solution to a csv file with the name my_solution.csv
my_solution.to_csv("/content/drive/MyDrive/Titanic - Machine Learning from Disaster/my_solution_one.csv", index_label = ["PassengerId"])
# Create a new array with the added features: features_two
features_two = train[["Pclass","Age","Sex","Fare", "SibSp", "Parch", "Embarked"]].values
#Control overfitting by setting "max_depth" to 10 and "min_samples_split" to 5 : my_tree_two
max_depth = 10
min_samples_split = 5
my_tree_two = tree.DecisionTreeClassifier(max_depth = max_depth, min_samples_split = min_samples_split, random_state = 1)
my_tree_two = my_tree_two.fit(features_two, target)
# Create train_two with the newly defined feature
train_two = train.copy()
train_two["family_size"] = train_two["SibSp"]+train_two["Parch"]+1
# Create a new feature set and add the new feature
features_three = train_two[["Pclass", "Sex", "Age", "Fare", "SibSp", "Parch", "family_size"]].values
# Define the tree classifier, then fit the model
my_tree_three = tree.DecisionTreeClassifier()
my_tree_three = my_tree_three.fit(features_three, target)
# Print the score of this decision tree
print(my_tree_three.score(features_three, target))
```
**Random Forest Classifier**
```
# Import the `RandomForestClassifier`
from sklearn.ensemble import RandomForestClassifier
# We want the Pclass, Age, Sex, Fare, SibSp, Parch, and Embarked variables
features_forest = train[["Pclass", "Age", "Sex", "Fare", "SibSp", "Parch", "Embarked"]].values
# Building and fitting my_forest
forest = RandomForestClassifier(max_depth = 10, min_samples_split=2, n_estimators = 100, random_state = 1)
my_forest = forest.fit(features_forest, target)
# Print the score of the fitted random forest
print(my_forest.score(features_forest, target))
# Compute predictions on our test set features then print the length of the prediction vector
test["Embarked"][test["Embarked"] == "S"] = 0
test["Embarked"][test["Embarked"] == "C"] = 1
test["Embarked"][test["Embarked"] == "Q"] = 2
test_features = test[["Pclass", "Age", "Sex", "Fare", "SibSp", "Parch", "Embarked"]].values
pred_forest = my_forest.predict(test_features)
print(len(pred_forest))
test[["Pclass", "Age", "Sex", "Fare", "SibSp", "Parch", "Embarked"]].values
test.Embarked.value_counts()
features_two
#Request and print the `.feature_importances_` attribute
print(my_tree_two.feature_importances_)
print(my_forest.feature_importances_)
#Compute and print the mean accuracy score for both models
print(my_tree_two.score(features_two, target))
print(my_forest.score(features_two, target))
```
|
github_jupyter
|
from google.colab import drive
drive.mount('/content/drive')
# We import the required modules and packages
import matplotlib.pyplot as plt
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
import seaborn as sns
from sklearn import tree
%matplotlib inline
os.chdir('/content/drive/MyDrive/Titanic - Machine Learning from Disaster')
os.listdir()
for dirname, _, filenames in os.walk('/content/drive/MyDrive/Titanic - Machine Learning from Disaster'):
for filename in filenames:
print(os.path.join(dirname, filename))
train = pd.read_csv('/content/drive/MyDrive/Titanic - Machine Learning from Disaster/train.csv')
test = pd.read_csv('/content/drive/MyDrive/Titanic - Machine Learning from Disaster/test.csv')
print(train.head())
print(test.head())
train.describe()
train.shape
# Passengers that survived vs passengers that passed away
print(train["Survived"].value_counts())
# As proportions
print(train["Survived"].value_counts(normalize = True))
# Males that survived vs males that passed away
print(train["Survived"][train["Sex"] == 'male'].value_counts())
# Females that survived vs Females that passed away
print(train["Survived"][train["Sex"] == 'female'].value_counts())
# Normalized male survival
print(train["Survived"][train["Sex"] == 'male'].value_counts(normalize = True))
# Normalized female survival
print(train["Survived"][train["Sex"] == 'female'].value_counts(normalize = True))
# Create the column Child and assign to 'NaN'
train["Child"] = float('NaN')
# Assign 1 to passengers under 18, 0 to those 18 or older. Print the new column.
train["Child"][train["Age"] < 18] = 1
train["Child"][train["Age"] >= 18] = 0
# Print normalized Survival Rates for passengers under 18
print(train["Survived"][train["Child"] == 1].value_counts(normalize = True))
# Print normalized Survival Rates for passengers 18 or older
print(train["Survived"][train["Child"] == 0].value_counts(normalize = True))
# Create a copy of test: test_one
test_one=test
# Initialize a Survived column to 0
test_one["Survived"]=0
# Set Survived to 1 if Sex equals "female" and print the `Survived` column from `test_one`
test_one["Survived"][test_one["Sex"] == "female"] = 1
print(test_one.Survived[:10])
# Convert the male and female groups to integer form
train["Sex"][train["Sex"] == "male"] = 0
train["Sex"][train["Sex"] == "female"] = 1
# Impute the Embarked variable
train["Embarked"] = train["Embarked"].fillna("S")
# Impute the Age variable
train["Age"] = train["Age"].fillna(train["Age"].median())
# Convert the Embarked classes to integer form
train["Embarked"][train["Embarked"] == "S"] = 0
train["Embarked"][train["Embarked"] == "C"] = 1
train["Embarked"][train["Embarked"] == "Q"] = 2
#Print the Sex and Embarked columns
print(train["Sex"])
print(train["Embarked"])
# Print the train data to see the available features
print(train)
# Create the target and features numpy arrays: target, features_one
target = train["Survived"].values
features_one = train[["Pclass", "Sex", "Age", "Fare"]].values
target[:10]
features_one[:10]
# Fit your first decision tree: my_tree_one
my_tree_one = tree.DecisionTreeClassifier()
my_tree_one = my_tree_one.fit(features_one, target)
# Look at the importance and score of the included features
print(my_tree_one.feature_importances_)
print(my_tree_one.score(features_one, target))
my_tree_one.feature_importances_
# Impute the missing value with the median
test.Fare[153] = test["Fare"].median()
test["Sex"][test["Sex"] == "male"] = 0
test["Sex"][test["Sex"] == "female"] = 1
test["Age"] = test["Age"].fillna(train["Age"].median())
# Extract the features from the test set: Pclass, Sex, Age, and Fare.
test_features = test[['Pclass', 'Sex', 'Age', 'Fare']].values
# Make your prediction using the test set
my_prediction = my_tree_one.predict(test_features)
# Create a data frame with two columns: PassengerId & Survived. Survived contains your predictions
PassengerId =np.array(test["PassengerId"]).astype(int)
my_solution = pd.DataFrame(my_prediction, PassengerId, columns = ["Survived"])
print(my_solution)
# Check that your data frame has 418 entries
print(my_solution.shape)
# Write your solution to a csv file with the name my_solution.csv
my_solution.to_csv("/content/drive/MyDrive/Titanic - Machine Learning from Disaster/my_solution_one.csv", index_label = ["PassengerId"])
# Create a new array with the added features: features_two
features_two = train[["Pclass","Age","Sex","Fare", "SibSp", "Parch", "Embarked"]].values
#Control overfitting by setting "max_depth" to 10 and "min_samples_split" to 5 : my_tree_two
max_depth = 10
min_samples_split = 5
my_tree_two = tree.DecisionTreeClassifier(max_depth = max_depth, min_samples_split = min_samples_split, random_state = 1)
my_tree_two = my_tree_two.fit(features_two, target)
# Create train_two with the newly defined feature
train_two = train.copy()
train_two["family_size"] = train_two["SibSp"]+train_two["Parch"]+1
# Create a new feature set and add the new feature
features_three = train_two[["Pclass", "Sex", "Age", "Fare", "SibSp", "Parch", "family_size"]].values
# Define the tree classifier, then fit the model
my_tree_three = tree.DecisionTreeClassifier()
my_tree_three = my_tree_three.fit(features_three, target)
# Print the score of this decision tree
print(my_tree_three.score(features_three, target))
# Import the `RandomForestClassifier`
from sklearn.ensemble import RandomForestClassifier
# We want the Pclass, Age, Sex, Fare, SibSp, Parch, and Embarked variables
features_forest = train[["Pclass", "Age", "Sex", "Fare", "SibSp", "Parch", "Embarked"]].values
# Building and fitting my_forest
forest = RandomForestClassifier(max_depth = 10, min_samples_split=2, n_estimators = 100, random_state = 1)
my_forest = forest.fit(features_forest, target)
# Print the score of the fitted random forest
print(my_forest.score(features_forest, target))
# Compute predictions on our test set features then print the length of the prediction vector
test["Embarked"][test["Embarked"] == "S"] = 0
test["Embarked"][test["Embarked"] == "C"] = 1
test["Embarked"][test["Embarked"] == "Q"] = 2
test_features = test[["Pclass", "Age", "Sex", "Fare", "SibSp", "Parch", "Embarked"]].values
pred_forest = my_forest.predict(test_features)
print(len(pred_forest))
test[["Pclass", "Age", "Sex", "Fare", "SibSp", "Parch", "Embarked"]].values
test.Embarked.value_counts()
features_two
#Request and print the `.feature_importances_` attribute
print(my_tree_two.feature_importances_)
print(my_forest.feature_importances_)
#Compute and print the mean accuracy score for both models
print(my_tree_two.score(features_two, target))
print(my_forest.score(features_two, target))
| 0.495117 | 0.833053 |
# Saving model architecture only
In this reading you will learn how to save a model's architecture, but not its weights.
```
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
import json
import numpy as np
```
In previous videos and notebooks you have have learned how to save a model's weights, as well as the entire model - weights and architecture.
### Accessing a model's configuration
A model's *configuration* refers to its architecture. TensorFlow has a convenient way to retrieve a model's architecture as a dictionary. We start by creating a simple fully connected feedforward neural network with 1 hidden layer.
```
# Build the model
model = Sequential([
Dense(units=32, input_shape=(32, 32, 3), activation='relu', name='dense_1'),
Dense(units=10, activation='softmax', name='dense_2')
])
```
A TensorFlow model has an inbuilt method `get_config` which returns the model's architecture as a dictionary:
```
# Get the model config
config_dict = model.get_config()
print(config_dict)
```
### Creating a new model from the config
A new TensorFlow model can be created from this config dictionary. This model will have reinitialized weights, which are not the same as the original model.
```
# Create a model from the config dictionary
model_same_config = tf.keras.Sequential.from_config(config_dict)
```
We can check explicitly that the config of both models is the same, but the weights are not:
```
# Check the new model is the same architecture
print('Same config:',
model.get_config() == model_same_config.get_config())
print('Same value for first weight matrix:',
np.allclose(model.weights[0].numpy(), model_same_config.weights[0].numpy()))
```
For models that are not `Sequential` models, use `tf.keras.Model.from_config` instead of `tf.keras.Sequential.from_config`.
### Other file formats: JSON and YAML
It is also possible to obtain a model's config in JSON or YAML formats. This follows the same pattern:
```
# Convert the model to JSON
json_string = model.to_json()
print(json_string)
```
The JSON format can easily be written out and saved as a file:
```
# Write out JSON config file
with open('config.json', 'w') as f:
json.dump(json_string, f)
del json_string
# Read in JSON config file again
with open('config.json', 'r') as f:
json_string = json.load(f)
# Reinitialize the model
model_same_config = tf.keras.models.model_from_json(json_string)
# Check the new model is the same architecture, but different weights
print('Same config:',
model.get_config() == model_same_config.get_config())
print('Same value for first weight matrix:',
np.allclose(model.weights[0].numpy(), model_same_config.weights[0].numpy()))
```
The YAML format is similar. The details of writing out YAML files, loading them and using them to create a new model are similar as for the JSON files, so we won't show it here.
```
# Convert the model to YAML
yaml_string = model.to_yaml()
print(yaml_string)
```
Writing out, reading in and using YAML files to create models is similar to JSON files.
### Further reading and resources
* https://www.tensorflow.org/guide/keras/save_and_serialize#architecture-only_saving
* https://keras.io/getting-started/faq/#how-can-i-save-a-keras-model
|
github_jupyter
|
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
import json
import numpy as np
# Build the model
model = Sequential([
Dense(units=32, input_shape=(32, 32, 3), activation='relu', name='dense_1'),
Dense(units=10, activation='softmax', name='dense_2')
])
# Get the model config
config_dict = model.get_config()
print(config_dict)
# Create a model from the config dictionary
model_same_config = tf.keras.Sequential.from_config(config_dict)
# Check the new model is the same architecture
print('Same config:',
model.get_config() == model_same_config.get_config())
print('Same value for first weight matrix:',
np.allclose(model.weights[0].numpy(), model_same_config.weights[0].numpy()))
# Convert the model to JSON
json_string = model.to_json()
print(json_string)
# Write out JSON config file
with open('config.json', 'w') as f:
json.dump(json_string, f)
del json_string
# Read in JSON config file again
with open('config.json', 'r') as f:
json_string = json.load(f)
# Reinitialize the model
model_same_config = tf.keras.models.model_from_json(json_string)
# Check the new model is the same architecture, but different weights
print('Same config:',
model.get_config() == model_same_config.get_config())
print('Same value for first weight matrix:',
np.allclose(model.weights[0].numpy(), model_same_config.weights[0].numpy()))
# Convert the model to YAML
yaml_string = model.to_yaml()
print(yaml_string)
| 0.790449 | 0.973919 |
```
import pandas as pd
def supres(low, high, n=28, min_touches=2, stat_likeness_percent=1.5, bounce_percent=5):
"""Support and Resistance Testing
Identifies support and resistance levels of provided price action data.
Args:
n(int): Number of frames to evaluate
low(pandas.Series): A pandas Series of lows from price action data.
high(pandas.Series): A pandas Series of highs from price action data.
min_touches(int): Minimum # of touches for established S&R.
stat_likeness_percent(int/float): Acceptable margin of error for level.
bounce_percent(int/float): Percent of price action for established bounce.
Returns:
sup(float): Established level of support or None (if no level)
res(float): Established level of resistance or None (if no level)
"""
import pandas as pd
import numpy as np
# Collapse into dataframe
df = pd.concat([high, low], keys = ['high', 'low'], axis=1)
df['sup'] = pd.Series(np.zeros(len(low)))
df['res'] = pd.Series(np.zeros(len(low)))
df['sup_break'] = pd.Series(np.zeros(len(low)))
df['sup_break'] = 0
df['res_break'] = pd.Series(np.zeros(len(high)))
df['res_break'] = 0
for x in range((n-1)+n, len(df)):
# Split into defined timeframes for analysis
tempdf = df[x-n:x+1]
# Setting default values for support and resistance to None
sup = None
res = None
# Identifying local high and local low
maxima = tempdf.high.max()
minima = tempdf.low.min()
# Calculating distance between max and min (total price movement)
move_range = maxima - minima
# Calculating bounce distance and allowable margin of error for likeness
move_allowance = move_range * (stat_likeness_percent / 100)
bounce_distance = move_range * (bounce_percent / 100)
# Test resistance by iterating through data to check for touches delimited by bounces
touchdown = 0
awaiting_bounce = False
for y in range(0, len(tempdf)):
if abs(maxima - tempdf.high.iloc[y]) < move_allowance and not awaiting_bounce:
touchdown = touchdown + 1
awaiting_bounce = True
elif abs(maxima - tempdf.high.iloc[y]) > bounce_distance:
awaiting_bounce = False
if touchdown >= min_touches:
res = maxima
# Test support by iterating through data to check for touches delimited by bounces
touchdown = 0
awaiting_bounce = False
for y in range(0, len(tempdf)):
if abs(tempdf.low.iloc[y] - minima) < move_allowance and not awaiting_bounce:
touchdown = touchdown + 1
awaiting_bounce = True
elif abs(tempdf.low.iloc[y] - minima) > bounce_distance:
awaiting_bounce = False
if touchdown >= min_touches:
sup = minima
if sup:
df['sup'].iloc[x] = sup
if res:
df['res'].iloc[x] = res
res_break_indices = list(df[(np.isnan(df['res']) & ~np.isnan(df.shift(1)['res'])) & (df['high'] > df.shift(1)['res'])].index)
for index in res_break_indices:
df['res_break'].at[index] = 1
sup_break_indices = list(df[(np.isnan(df['sup']) & ~np.isnan(df.shift(1)['sup'])) & (df['low'] < df.shift(1)['sup'])].index)
for index in sup_break_indices:
df['sup_break'].at[index] = 1
ret_df = pd.concat([df['sup'], df['res'], df['sup_break'], df['res_break']], keys = ['sup', 'res', 'sup_break', 'res_break'], axis=1)
return ret_df
tic = 'SPY'
start = '2017-01-01'
end = '2019-06-18'
url = f"https://hw3nhrdos1.execute-api.us-east-2.amazonaws.com/api/history/{tic}/{start}/{end}"
df = pd.read_json(url)[['Open','Close','High','Low','Volume']]
df.tail()
%%time
levels = supres(df.Low, df.High)
%%time
for column in levels.columns:
df[column] = levels[column]
df.columns
# Where resistance level is not nan
df[~np.isnan(df['res'])][['Close','res']]
# Where previous support level has been broken
df[df['sup_break'] == 1][['Close','sup_break']]
# Where previous resistance level has been broken
df[df['res_break'] == 1][['Close','res_break']]
df.head()
# Round numbers within 10% of the closing price
len([i for i in df['Close'] if (i % 2 <= 0.1 or i % 3 <= 0.1)])
df['around_round'] = [1 if (i % 2 <= 0.1 or i % 3 <= 0.1) else 0 for i in df['Close']]
df.around_round.value_counts()
df.sample(5)
# Investability index
# trace = go.Candlestick(x=df.index,
# open=df.Open,
# high=df.High,
# low=df.Low,
# close=df.Close)
# data = [trace]
# layout = {
# 'title': 'The Great Recession',
# 'yaxis': {'title': 'AAPL Stock'},
# 'xaxis': {'rangeslider':{'visible': False}},
# 'shapes': [{
# 'x0': '2017-12-09', 'x1': '2017-12-09',
# 'y0': 0, 'y1': 1, 'xref': 'x', 'yref': 'paper',
# 'line': {'color': 'rgb(30,30,30)', 'width': 1}
# }],
# 'annotations': [{
# 'x': '2017-12-09', 'y': 0.05, 'xref': 'x', 'yref': 'paper',
# 'showarrow': False, 'xanchor': 'left',
# 'text': 'Increase Period Begins'
# }]
# }
# fig = dict(data=data, layout=layout)
# py.iplot(fig, filename='aapl-recession-candlestick')
```
|
github_jupyter
|
import pandas as pd
def supres(low, high, n=28, min_touches=2, stat_likeness_percent=1.5, bounce_percent=5):
"""Support and Resistance Testing
Identifies support and resistance levels of provided price action data.
Args:
n(int): Number of frames to evaluate
low(pandas.Series): A pandas Series of lows from price action data.
high(pandas.Series): A pandas Series of highs from price action data.
min_touches(int): Minimum # of touches for established S&R.
stat_likeness_percent(int/float): Acceptable margin of error for level.
bounce_percent(int/float): Percent of price action for established bounce.
Returns:
sup(float): Established level of support or None (if no level)
res(float): Established level of resistance or None (if no level)
"""
import pandas as pd
import numpy as np
# Collapse into dataframe
df = pd.concat([high, low], keys = ['high', 'low'], axis=1)
df['sup'] = pd.Series(np.zeros(len(low)))
df['res'] = pd.Series(np.zeros(len(low)))
df['sup_break'] = pd.Series(np.zeros(len(low)))
df['sup_break'] = 0
df['res_break'] = pd.Series(np.zeros(len(high)))
df['res_break'] = 0
for x in range((n-1)+n, len(df)):
# Split into defined timeframes for analysis
tempdf = df[x-n:x+1]
# Setting default values for support and resistance to None
sup = None
res = None
# Identifying local high and local low
maxima = tempdf.high.max()
minima = tempdf.low.min()
# Calculating distance between max and min (total price movement)
move_range = maxima - minima
# Calculating bounce distance and allowable margin of error for likeness
move_allowance = move_range * (stat_likeness_percent / 100)
bounce_distance = move_range * (bounce_percent / 100)
# Test resistance by iterating through data to check for touches delimited by bounces
touchdown = 0
awaiting_bounce = False
for y in range(0, len(tempdf)):
if abs(maxima - tempdf.high.iloc[y]) < move_allowance and not awaiting_bounce:
touchdown = touchdown + 1
awaiting_bounce = True
elif abs(maxima - tempdf.high.iloc[y]) > bounce_distance:
awaiting_bounce = False
if touchdown >= min_touches:
res = maxima
# Test support by iterating through data to check for touches delimited by bounces
touchdown = 0
awaiting_bounce = False
for y in range(0, len(tempdf)):
if abs(tempdf.low.iloc[y] - minima) < move_allowance and not awaiting_bounce:
touchdown = touchdown + 1
awaiting_bounce = True
elif abs(tempdf.low.iloc[y] - minima) > bounce_distance:
awaiting_bounce = False
if touchdown >= min_touches:
sup = minima
if sup:
df['sup'].iloc[x] = sup
if res:
df['res'].iloc[x] = res
res_break_indices = list(df[(np.isnan(df['res']) & ~np.isnan(df.shift(1)['res'])) & (df['high'] > df.shift(1)['res'])].index)
for index in res_break_indices:
df['res_break'].at[index] = 1
sup_break_indices = list(df[(np.isnan(df['sup']) & ~np.isnan(df.shift(1)['sup'])) & (df['low'] < df.shift(1)['sup'])].index)
for index in sup_break_indices:
df['sup_break'].at[index] = 1
ret_df = pd.concat([df['sup'], df['res'], df['sup_break'], df['res_break']], keys = ['sup', 'res', 'sup_break', 'res_break'], axis=1)
return ret_df
tic = 'SPY'
start = '2017-01-01'
end = '2019-06-18'
url = f"https://hw3nhrdos1.execute-api.us-east-2.amazonaws.com/api/history/{tic}/{start}/{end}"
df = pd.read_json(url)[['Open','Close','High','Low','Volume']]
df.tail()
%%time
levels = supres(df.Low, df.High)
%%time
for column in levels.columns:
df[column] = levels[column]
df.columns
# Where resistance level is not nan
df[~np.isnan(df['res'])][['Close','res']]
# Where previous support level has been broken
df[df['sup_break'] == 1][['Close','sup_break']]
# Where previous resistance level has been broken
df[df['res_break'] == 1][['Close','res_break']]
df.head()
# Round numbers within 10% of the closing price
len([i for i in df['Close'] if (i % 2 <= 0.1 or i % 3 <= 0.1)])
df['around_round'] = [1 if (i % 2 <= 0.1 or i % 3 <= 0.1) else 0 for i in df['Close']]
df.around_round.value_counts()
df.sample(5)
# Investability index
# trace = go.Candlestick(x=df.index,
# open=df.Open,
# high=df.High,
# low=df.Low,
# close=df.Close)
# data = [trace]
# layout = {
# 'title': 'The Great Recession',
# 'yaxis': {'title': 'AAPL Stock'},
# 'xaxis': {'rangeslider':{'visible': False}},
# 'shapes': [{
# 'x0': '2017-12-09', 'x1': '2017-12-09',
# 'y0': 0, 'y1': 1, 'xref': 'x', 'yref': 'paper',
# 'line': {'color': 'rgb(30,30,30)', 'width': 1}
# }],
# 'annotations': [{
# 'x': '2017-12-09', 'y': 0.05, 'xref': 'x', 'yref': 'paper',
# 'showarrow': False, 'xanchor': 'left',
# 'text': 'Increase Period Begins'
# }]
# }
# fig = dict(data=data, layout=layout)
# py.iplot(fig, filename='aapl-recession-candlestick')
| 0.571169 | 0.597549 |
# Publications markdown generator for academicpages
Takes a TSV of publications with metadata and converts them for use with [academicpages.github.io](academicpages.github.io). This is an interactive Jupyter notebook ([see more info here](http://jupyter-notebook-beginner-guide.readthedocs.io/en/latest/what_is_jupyter.html)). The core python code is also in `publications.py`. Run either from the `markdown_generator` folder after replacing `publications.tsv` with one containing your data.
TODO: Make this work with BibTex and other databases of citations, rather than Stuart's non-standard TSV format and citation style.
## Data format
The TSV needs to have the following columns: pub_date, title, venue, excerpt, citation, site_url, and paper_url, with a header at the top.
- `excerpt` and `paper_url` can be blank, but the others must have values.
- `pub_date` must be formatted as YYYY-MM-DD.
- `url_slug` will be the descriptive part of the .md file and the permalink URL for the page about the paper. The .md file will be `YYYY-MM-DD-[url_slug].md` and the permalink will be `https://[yourdomain]/publications/YYYY-MM-DD-[url_slug]`
This is how the raw file looks (it doesn't look pretty, use a spreadsheet or other program to edit and create).
```
!cat publications.tsv
```
## Import pandas
We are using the very handy pandas library for dataframes.
```
import pandas as pd
```
## Import TSV
Pandas makes this easy with the read_csv function. We are using a TSV, so we specify the separator as a tab, or `\t`.
I found it important to put this data in a tab-separated values format, because there are a lot of commas in this kind of data and comma-separated values can get messed up. However, you can modify the import statement, as pandas also has read_excel(), read_json(), and others.
```
publications = pd.read_csv("publications.tsv", sep="\t", header=0)
publications
```
## Escape special characters
YAML is very picky about how it takes a valid string, so we are replacing single and double quotes (and ampersands) with their HTML encoded equivilents. This makes them look not so readable in raw format, but they are parsed and rendered nicely.
```
html_escape_table = {
"&": "&",
'"': """,
"'": "'"
}
def html_escape(text):
"""Produce entities within text."""
return "".join(html_escape_table.get(c,c) for c in text)
```
## Creating the markdown files
This is where the heavy lifting is done. This loops through all the rows in the TSV dataframe, then starts to concatentate a big string (```md```) that contains the markdown for each type. It does the YAML metadata first, then does the description for the individual page.
```
import os
for row, item in publications.iterrows():
md_filename = str(item.pub_date) + "-" + item.url_slug + ".md"
html_filename = str(item.pub_date) + "-" + item.url_slug
year = item.pub_date[:4]
## YAML variables
md = "---\ntitle: \"" + item.title + '"\n'
md += """collection: publications"""
md += """\npermalink: /publication/""" + html_filename
if len(str(item.excerpt)) > 5:
md += "\nexcerpt: '" + html_escape(item.excerpt) + "'"
md += "\ndate: " + str(item.pub_date)
md += "\nvenue: '" + html_escape(item.venue) + "'"
if len(str(item.paper_url)) > 5:
md += "\npaperurl: '" + item.paper_url + "'"
md += "\ncitation: '" + html_escape(item.citation) + "'"
md += "\n---"
## Markdown description for individual page
if len(str(item.excerpt)) > 5:
md += "\n" + html_escape(item.excerpt) + "\n"
if len(str(item.paper_url)) > 5:
md += "\n[Download paper here](" + item.paper_url + ")\n"
md += "\n" + item.citation
md_filename = os.path.basename(md_filename)
with open("../_publications/" + md_filename, 'w') as f:
f.write(md)
```
These files are in the publications directory, one directory below where we're working from.
```
!ls ../_publications/
!cat ../_publications/2009-10-01-paper-title-number-1.md
```
|
github_jupyter
|
!cat publications.tsv
import pandas as pd
publications = pd.read_csv("publications.tsv", sep="\t", header=0)
publications
html_escape_table = {
"&": "&",
'"': """,
"'": "'"
}
def html_escape(text):
"""Produce entities within text."""
return "".join(html_escape_table.get(c,c) for c in text)
import os
for row, item in publications.iterrows():
md_filename = str(item.pub_date) + "-" + item.url_slug + ".md"
html_filename = str(item.pub_date) + "-" + item.url_slug
year = item.pub_date[:4]
## YAML variables
md = "---\ntitle: \"" + item.title + '"\n'
md += """collection: publications"""
md += """\npermalink: /publication/""" + html_filename
if len(str(item.excerpt)) > 5:
md += "\nexcerpt: '" + html_escape(item.excerpt) + "'"
md += "\ndate: " + str(item.pub_date)
md += "\nvenue: '" + html_escape(item.venue) + "'"
if len(str(item.paper_url)) > 5:
md += "\npaperurl: '" + item.paper_url + "'"
md += "\ncitation: '" + html_escape(item.citation) + "'"
md += "\n---"
## Markdown description for individual page
if len(str(item.excerpt)) > 5:
md += "\n" + html_escape(item.excerpt) + "\n"
if len(str(item.paper_url)) > 5:
md += "\n[Download paper here](" + item.paper_url + ")\n"
md += "\n" + item.citation
md_filename = os.path.basename(md_filename)
with open("../_publications/" + md_filename, 'w') as f:
f.write(md)
!ls ../_publications/
!cat ../_publications/2009-10-01-paper-title-number-1.md
| 0.380989 | 0.750278 |
```
import gym
env = gym.make('Blackjack-v0')
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
def color_plot(arr, x_bot=None, x_top=None, y_bot=None, y_top=None):
fig = plt.figure()
ax = fig.add_subplot(111)
plt.imshow(arr)
ax.set_aspect('equal')
plt.colorbar(orientation='vertical')
if x_bot and x_top:
plt.xlim(x_bot, x_top)
if y_bot and y_top:
plt.ylim(y_bot, y_top)
```
## First Visit MC
For the purpose of policy evaluation
```
# Only stick if our hand is 20 or 21
policy = np.ones((32, 11, 2))
policy[20:22, :, :] = 0
policy[9:11, :, 1] = 0
V_s = np.zeros((32, 11, 2))
ret = np.zeros((32, 11, 2))
count = np.zeros((32, 11, 2))
DISCOUNT = 1
for _ in range(500000):
hand, show, ace = env.reset()
done = False
episode = []
while not done:
state = (hand, show, int(ace))
(hand, show, ace), reward, done, _ = env.step(int(policy[state]))
episode.append((state, reward))
g = 0
while len(episode) > 0:
state, reward = episode.pop()
g = DISCOUNT * g + reward
if (state, reward) not in episode:
count[state] += 1
V_s[state] += (g - V_s[state])/count[state]
color_plot(V_s[:,:,0], 0.5, 10.5, 11.5, 21.5)
plt.savefig('first_no_ace.png')
color_plot(V_s[:,:,1], 0.5, 10.5, 11.5, 21.5)
plt.savefig('first_ace.png')
```
## Monte Carlo Exploring Starts
For the purpose of policy improvement
```
usable = np.zeros((32, 11, 2, 2))
usable[1:22, 1:12] = 1
q = np.random.random((32, 11, 2, 2)) * usable
policy = np.argmax(q, axis=3)
ret = np.zeros((32, 11, 2, 2))
count = np.zeros((32, 11, 2, 2))
DISCOUNT = 1
for _ in range(10000000):
# Environment already has positive chance for all states
hand, show, ace = env.reset()
state = (hand, show, int(ace))
done = False
episode = []
action = np.random.randint(0, 2)
(hand, show, ace), reward, done, _ = env.step(action)
episode.append((state, action, reward))
while not done:
state = (hand, show, int(ace))
action = int(policy[state])
(hand, show, ace), reward, done, _ = env.step(action)
episode.append((state, action, reward))
g = 0
while len(episode) > 0:
state, action, reward = episode.pop()
g = DISCOUNT * g + reward
if (state, action, reward) not in episode:
count[state + tuple([action])] += 1
q[state + tuple([action])] += (g - q[state + tuple([action])])/count[state + tuple([action])]
policy[state] = np.argmax(q[state])
for i in range(2):
for j in range(2):
color_plot(q[:,:,i,j], 0.5, 10.5, 11.5, 21.5)
str_i = 'ace' if i else 'no_ace'
str_j = 'hit' if j else 'stick'
plt.savefig('es_' + str_i + '_' + str_j + '.png')
```
## On-Policy First-Visit Monte Carlo
$\epsilon$ soft policies
```
usable = np.zeros((32, 11, 2, 2))
usable[1:22, 1:12] = 1
q = np.random.random((32, 11, 2, 2)) * usable
policy = np.argmax(q, axis=3)
ret = np.zeros((32, 11, 2, 2))
count = np.zeros((32, 11, 2, 2))
epsilon = 0.1
DISCOUNT = 1
for _ in range(1000000):
hand, show, ace = env.reset()
done = False
g = 0
episode = []
while not done:
state = (hand, show, int(ace))
action = int(policy[state])
(hand, show, ace), reward, done, _ = env.step(action)
episode.append((state, action, reward))
while len(episode) > 0:
state, action, reward = episode.pop()
g = DISCOUNT * g + reward
if (state, action, reward) not in episode:
count[state + tuple([action])] += 1
q[state + tuple([action])] += (g - q[state + tuple([action])])/count[state + tuple([action])]
g_action = np.argmax(q[state])
if np.random.random() < epsilon:
policy[state] = np.random.randint(0, 2)
else:
policy[state] = g_action
for i in range(2):
for j in range(2):
color_plot(q[:,:,i,j], 0.5, 10.5, 11.5, 21.5)
str_i = 'ace' if i else 'no_ace'
str_j = 'hit' if j else 'stick'
plt.savefig('on_' + str_i + '_' + str_j + '.png')
```
## Off-Policy Monte Carlo Prediction
For policy evaluation
```
pi = (np.random.random((32, 11, 2)) < 0.5).astype(int)
b = np.random.random((32, 11, 2))
b = np.stack((b, 1-b), axis=3)
q = np.zeros((32, 11, 2, 2))
count = np.zeros((32, 11, 2, 2))
DISCOUNT = 1
for _ in range(1000000):
hand, show, ace = env.reset()
done = False
episode = []
while not done:
state = (hand, show, int(ace))
action = 0 if np.random.random() < b[state][0] else 1
(hand, show, ace), reward, done, _ = env.step(action)
episode.append((state, action, reward))
g = 0
w = 1
while len(episode) > 0 and w != 0:
state, action, reward = episode.pop()
g = DISCOUNT * g + reward
sa = state + tuple([action])
count[sa] += w
q[sa] += (w * (g - q[sa])) / count[sa]
# pi[state] = np.argmax(q[state])
w *= pi[state]/b[sa]
for i in range(2):
for j in range(2):
color_plot(q[:,:,i,j], 0.5, 10.5, 11.5, 21.5)
str_i = 'ace' if i else 'no_ace'
str_j = 'hit' if j else 'stick'
plt.savefig('off_eval_' + str_i + '_' + str_j + '.png')
```
## Off Policy Monte Carlo Control
```
b = np.ones((32, 11, 2, 2)) * 0.5
q = np.random.random((32, 11, 2, 2))
count = np.zeros((32, 11, 2, 2))
pi = np.argmax(q, axis=3)
DISCOUNT = 1
for _ in range(10000000):
hand, show, ace = env.reset()
done = False
episode = []
while not done:
state = (hand, show, int(ace))
action = np.random.choice(range(len(b[state])), p=b[state])
(hand, show, ace), reward, done, _ = env.step(action)
episode.append((state, action, reward))
g = 0.0
w = 1.0
while len(episode) > 0:
state, action, reward = episode.pop()
sa = state + tuple([action])
g = DISCOUNT * g + reward
count[sa] += w
q[sa] += (w * (g - q[sa])) / count[sa]
pi[state] = np.argmax(q[state])
if pi[state] != action:
break
w *= 1/b[sa]
for i in range(2):
for j in range(2):
color_plot(q[:,:,i,j], 0.5, 10.5, 11.5, 21.5)
str_i = 'ace' if i else 'no_ace'
str_j = 'hit' if j else 'stick'
plt.savefig('off_' + str_i + '_' + str_j + '.png')
```
|
github_jupyter
|
import gym
env = gym.make('Blackjack-v0')
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
def color_plot(arr, x_bot=None, x_top=None, y_bot=None, y_top=None):
fig = plt.figure()
ax = fig.add_subplot(111)
plt.imshow(arr)
ax.set_aspect('equal')
plt.colorbar(orientation='vertical')
if x_bot and x_top:
plt.xlim(x_bot, x_top)
if y_bot and y_top:
plt.ylim(y_bot, y_top)
# Only stick if our hand is 20 or 21
policy = np.ones((32, 11, 2))
policy[20:22, :, :] = 0
policy[9:11, :, 1] = 0
V_s = np.zeros((32, 11, 2))
ret = np.zeros((32, 11, 2))
count = np.zeros((32, 11, 2))
DISCOUNT = 1
for _ in range(500000):
hand, show, ace = env.reset()
done = False
episode = []
while not done:
state = (hand, show, int(ace))
(hand, show, ace), reward, done, _ = env.step(int(policy[state]))
episode.append((state, reward))
g = 0
while len(episode) > 0:
state, reward = episode.pop()
g = DISCOUNT * g + reward
if (state, reward) not in episode:
count[state] += 1
V_s[state] += (g - V_s[state])/count[state]
color_plot(V_s[:,:,0], 0.5, 10.5, 11.5, 21.5)
plt.savefig('first_no_ace.png')
color_plot(V_s[:,:,1], 0.5, 10.5, 11.5, 21.5)
plt.savefig('first_ace.png')
usable = np.zeros((32, 11, 2, 2))
usable[1:22, 1:12] = 1
q = np.random.random((32, 11, 2, 2)) * usable
policy = np.argmax(q, axis=3)
ret = np.zeros((32, 11, 2, 2))
count = np.zeros((32, 11, 2, 2))
DISCOUNT = 1
for _ in range(10000000):
# Environment already has positive chance for all states
hand, show, ace = env.reset()
state = (hand, show, int(ace))
done = False
episode = []
action = np.random.randint(0, 2)
(hand, show, ace), reward, done, _ = env.step(action)
episode.append((state, action, reward))
while not done:
state = (hand, show, int(ace))
action = int(policy[state])
(hand, show, ace), reward, done, _ = env.step(action)
episode.append((state, action, reward))
g = 0
while len(episode) > 0:
state, action, reward = episode.pop()
g = DISCOUNT * g + reward
if (state, action, reward) not in episode:
count[state + tuple([action])] += 1
q[state + tuple([action])] += (g - q[state + tuple([action])])/count[state + tuple([action])]
policy[state] = np.argmax(q[state])
for i in range(2):
for j in range(2):
color_plot(q[:,:,i,j], 0.5, 10.5, 11.5, 21.5)
str_i = 'ace' if i else 'no_ace'
str_j = 'hit' if j else 'stick'
plt.savefig('es_' + str_i + '_' + str_j + '.png')
usable = np.zeros((32, 11, 2, 2))
usable[1:22, 1:12] = 1
q = np.random.random((32, 11, 2, 2)) * usable
policy = np.argmax(q, axis=3)
ret = np.zeros((32, 11, 2, 2))
count = np.zeros((32, 11, 2, 2))
epsilon = 0.1
DISCOUNT = 1
for _ in range(1000000):
hand, show, ace = env.reset()
done = False
g = 0
episode = []
while not done:
state = (hand, show, int(ace))
action = int(policy[state])
(hand, show, ace), reward, done, _ = env.step(action)
episode.append((state, action, reward))
while len(episode) > 0:
state, action, reward = episode.pop()
g = DISCOUNT * g + reward
if (state, action, reward) not in episode:
count[state + tuple([action])] += 1
q[state + tuple([action])] += (g - q[state + tuple([action])])/count[state + tuple([action])]
g_action = np.argmax(q[state])
if np.random.random() < epsilon:
policy[state] = np.random.randint(0, 2)
else:
policy[state] = g_action
for i in range(2):
for j in range(2):
color_plot(q[:,:,i,j], 0.5, 10.5, 11.5, 21.5)
str_i = 'ace' if i else 'no_ace'
str_j = 'hit' if j else 'stick'
plt.savefig('on_' + str_i + '_' + str_j + '.png')
pi = (np.random.random((32, 11, 2)) < 0.5).astype(int)
b = np.random.random((32, 11, 2))
b = np.stack((b, 1-b), axis=3)
q = np.zeros((32, 11, 2, 2))
count = np.zeros((32, 11, 2, 2))
DISCOUNT = 1
for _ in range(1000000):
hand, show, ace = env.reset()
done = False
episode = []
while not done:
state = (hand, show, int(ace))
action = 0 if np.random.random() < b[state][0] else 1
(hand, show, ace), reward, done, _ = env.step(action)
episode.append((state, action, reward))
g = 0
w = 1
while len(episode) > 0 and w != 0:
state, action, reward = episode.pop()
g = DISCOUNT * g + reward
sa = state + tuple([action])
count[sa] += w
q[sa] += (w * (g - q[sa])) / count[sa]
# pi[state] = np.argmax(q[state])
w *= pi[state]/b[sa]
for i in range(2):
for j in range(2):
color_plot(q[:,:,i,j], 0.5, 10.5, 11.5, 21.5)
str_i = 'ace' if i else 'no_ace'
str_j = 'hit' if j else 'stick'
plt.savefig('off_eval_' + str_i + '_' + str_j + '.png')
b = np.ones((32, 11, 2, 2)) * 0.5
q = np.random.random((32, 11, 2, 2))
count = np.zeros((32, 11, 2, 2))
pi = np.argmax(q, axis=3)
DISCOUNT = 1
for _ in range(10000000):
hand, show, ace = env.reset()
done = False
episode = []
while not done:
state = (hand, show, int(ace))
action = np.random.choice(range(len(b[state])), p=b[state])
(hand, show, ace), reward, done, _ = env.step(action)
episode.append((state, action, reward))
g = 0.0
w = 1.0
while len(episode) > 0:
state, action, reward = episode.pop()
sa = state + tuple([action])
g = DISCOUNT * g + reward
count[sa] += w
q[sa] += (w * (g - q[sa])) / count[sa]
pi[state] = np.argmax(q[state])
if pi[state] != action:
break
w *= 1/b[sa]
for i in range(2):
for j in range(2):
color_plot(q[:,:,i,j], 0.5, 10.5, 11.5, 21.5)
str_i = 'ace' if i else 'no_ace'
str_j = 'hit' if j else 'stick'
plt.savefig('off_' + str_i + '_' + str_j + '.png')
| 0.286568 | 0.849285 |
## Time-lagged conversion rates and cure models
Suppose in our population we have a subpopulation that will never experience the event of interest. Or, for some subjects the event will occur so far in the future that it's essentially at time infinity. The survival function should not asymptically approach zero, but _some positive value_. Models that describe this are sometimes called cure models or time-lagged conversion models.
There's a serious fault in using parametric models for these types of problems that non-parametric models don't have. The most common parametric models like Weibull, Log-Normal, etc. all have strictly increasing cumulative hazard functions, which means the corresponding survival function will always converge to 0.
Let's look at an example of this problem. Below I generated some data that has individuals who _will not experience the event_, not matter how long we wait.
```
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
from matplotlib import pyplot as plt
import autograd.numpy as np
from autograd.scipy.special import expit, logit
import pandas as pd
plt.style.use('bmh')
N = 200
U = np.random.rand(N)
T = -(logit(-np.log(U) / 0.5) - np.random.exponential(2, N) - 6.00) / 0.50
E = ~np.isnan(T)
T[np.isnan(T)] = 50
from lifelines import KaplanMeierFitter
kmf = KaplanMeierFitter().fit(T, E)
kmf.plot(figsize=(8,4))
plt.ylim(0, 1);
```
It should be clear that there is an asymptote at around 0.6. The non-parametric model will always show this. If this is true, then the cumulative hazard function should have a horizontal asymptote as well. Let's use the Nelson-Aalen model to see this.
```
from lifelines import NelsonAalenFitter
naf = NelsonAalenFitter().fit(T, E)
naf.plot(figsize=(8,4))
```
_However_, when we try a parametric model, we will see that it won't extrapolate very well. Let's use the flexible two parameter LogLogisticFitter model.
```
from lifelines import LogLogisticFitter
fig, ax = plt.subplots(nrows=2, ncols=2, figsize=(10, 6))
t = np.linspace(0, 40)
llf = LogLogisticFitter().fit(T, E, timeline=t)
llf.plot_survival_function(ax=ax[0][0])
kmf.plot(ax=ax[0][0])
llf.plot_cumulative_hazard(ax=ax[0][1])
naf.plot(ax=ax[0][1])
t = np.linspace(0, 100)
llf = LogLogisticFitter().fit(T, E, timeline=t)
llf.plot_survival_function(ax=ax[1][0])
kmf.plot(ax=ax[1][0])
llf.plot_cumulative_hazard(ax=ax[1][1])
naf.plot(ax=ax[1][1])
```
The LogLogistic model does a quite terrible job of capturing the not only the asymptotes, but also the intermediate values as well. If we extended the survival function out further, we would see that it eventually nears 0.
### Custom parametric models to handle asymptotes
Focusing on modeling the cumulative hazard function, what we would like is a function that increases up to a limit and then tapers off to an asymptote. We can think long and hard about these (I did), but there's a family of functions that have this property that we are very familiar with: cumulative distribution functions! By their nature, they will asympotically approach 1. And, they are readily available in the SciPy and autograd libraries. So our new model of the cumulative hazard function is:
$$H(t; c, \theta) = c F(t; \theta)$$
where $c$ is the (unknown) horizontal asymptote, and $\theta$ is a vector of (unknown) parameters for the CDF, $F$.
We can create a custom cumulative hazard model using `ParametricUnivariateFitter` (for a tutorial on how to create custom models, see [this here](Piecewise Exponential Models and Creating Custom Models.ipynb)). Let's choose the Normal CDF for our model.
Remember we **must** use the imports from `autograd` for this, i.e. `from autograd.scipy.stats import norm`.
```
from autograd.scipy.stats import norm
from lifelines.fitters import ParametricUnivariateFitter
class UpperAsymptoteFitter(ParametricUnivariateFitter):
_fitted_parameter_names = ["c_", "mu_", "sigma_"]
_bounds = ((0, None), (None, None), (0, None))
def _cumulative_hazard(self, params, times):
c, mu, sigma = params
return c * norm.cdf((times - mu) / sigma, loc=0, scale=1)
uaf = UpperAsymptoteFitter().fit(T, E)
uaf.print_summary(3)
uaf.plot(figsize=(8,4))
```
We get a lovely asympotical cumulative hazard. The summary table suggests that the best value of $c$ is 0.586. This can be translated into the survival function asymptote by $\exp(-0.586) \approx 0.56$.
Let's compare this fit to the non-parametric models.
```
fig, ax = plt.subplots(nrows=2, ncols=2, figsize=(10, 6))
t = np.linspace(0, 40)
uaf = UpperAsymptoteFitter().fit(T, E, timeline=t)
uaf.plot_survival_function(ax=ax[0][0])
kmf.plot(ax=ax[0][0])
uaf.plot_cumulative_hazard(ax=ax[0][1])
naf.plot(ax=ax[0][1])
t = np.linspace(0, 100)
uaf = UpperAsymptoteFitter().fit(T, E, timeline=t)
uaf.plot_survival_function(ax=ax[1][0])
kmf.survival_function_.plot(ax=ax[1][0])
uaf.plot_cumulative_hazard(ax=ax[1][1])
naf.plot(ax=ax[1][1])
```
I wasn't expect this good of a fit. But there it is. This was some artificial data, but let's try this technique on some real life data.
```
from lifelines.datasets import load_leukemia, load_kidney_transplant
T, E = load_leukemia()['t'], load_leukemia()['status']
uaf.fit(T, E)
ax = uaf.plot_survival_function(figsize=(8,4))
uaf.print_summary()
kmf.fit(T, E).plot(ax=ax, ci_show=False)
print("---")
print("Estimated lower bound: {:.2f} ({:.2f}, {:.2f})".format(
np.exp(-uaf.summary.loc['c_', 'coef']),
np.exp(-uaf.summary.loc['c_', 'coef upper 95%']),
np.exp(-uaf.summary.loc['c_', 'coef lower 95%']),
)
)
```
So we might expect that about 20% will not have the even in this population (but make note of the large CI bounds too!)
```
# Another, less obvious, dataset.
T, E = load_kidney_transplant()['time'], load_kidney_transplant()['death']
uaf.fit(T, E)
ax = uaf.plot_survival_function(figsize=(8,4))
uaf.print_summary()
kmf.fit(T, E).plot(ax=ax)
print("---")
print("Estimated lower bound: {:.2f} ({:.2f}, {:.2f})".format(
np.exp(-uaf.summary.loc['c_', 'coef']),
np.exp(-uaf.summary.loc['c_', 'coef upper 95%']),
np.exp(-uaf.summary.loc['c_', 'coef lower 95%']),
)
)
```
#### Using alternative functional forms
An even simplier model might look like $c \left(1 - \frac{1}{\lambda t + 1}\right)$, however this model cannot handle any "inflection points" like our artificial dataset has above. However, it works well for this Lung dataset.
With all cure models, one important feature is the ability to extrapolate to unforseen times.
```
from autograd.scipy.stats import norm
from lifelines.fitters import ParametricUnivariateFitter
class SimpleUpperAsymptoteFitter(ParametricUnivariateFitter):
_fitted_parameter_names = ["c_", "lambda_"]
_bounds = ((0, None), (0, None))
def _cumulative_hazard(self, params, times):
c, lambda_ = params
return c * (1 - 1 /(lambda_ * times + 1))
# Another, less obvious, dataset.
saf = SimpleUpperAsymptoteFitter().fit(T, E, timeline=np.arange(1, 10000))
ax = saf.plot_survival_function(figsize=(8,4))
saf.print_summary(4)
kmf.fit(T, E).plot(ax=ax)
print("---")
print("Estimated lower bound: {:.2f} ({:.2f}, {:.2f})".format(
np.exp(-saf.summary.loc['c_', 'coef']),
np.exp(-saf.summary.loc['c_', 'coef upper 95%']),
np.exp(-saf.summary.loc['c_', 'coef lower 95%']),
)
)
```
|
github_jupyter
|
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
from matplotlib import pyplot as plt
import autograd.numpy as np
from autograd.scipy.special import expit, logit
import pandas as pd
plt.style.use('bmh')
N = 200
U = np.random.rand(N)
T = -(logit(-np.log(U) / 0.5) - np.random.exponential(2, N) - 6.00) / 0.50
E = ~np.isnan(T)
T[np.isnan(T)] = 50
from lifelines import KaplanMeierFitter
kmf = KaplanMeierFitter().fit(T, E)
kmf.plot(figsize=(8,4))
plt.ylim(0, 1);
from lifelines import NelsonAalenFitter
naf = NelsonAalenFitter().fit(T, E)
naf.plot(figsize=(8,4))
from lifelines import LogLogisticFitter
fig, ax = plt.subplots(nrows=2, ncols=2, figsize=(10, 6))
t = np.linspace(0, 40)
llf = LogLogisticFitter().fit(T, E, timeline=t)
llf.plot_survival_function(ax=ax[0][0])
kmf.plot(ax=ax[0][0])
llf.plot_cumulative_hazard(ax=ax[0][1])
naf.plot(ax=ax[0][1])
t = np.linspace(0, 100)
llf = LogLogisticFitter().fit(T, E, timeline=t)
llf.plot_survival_function(ax=ax[1][0])
kmf.plot(ax=ax[1][0])
llf.plot_cumulative_hazard(ax=ax[1][1])
naf.plot(ax=ax[1][1])
from autograd.scipy.stats import norm
from lifelines.fitters import ParametricUnivariateFitter
class UpperAsymptoteFitter(ParametricUnivariateFitter):
_fitted_parameter_names = ["c_", "mu_", "sigma_"]
_bounds = ((0, None), (None, None), (0, None))
def _cumulative_hazard(self, params, times):
c, mu, sigma = params
return c * norm.cdf((times - mu) / sigma, loc=0, scale=1)
uaf = UpperAsymptoteFitter().fit(T, E)
uaf.print_summary(3)
uaf.plot(figsize=(8,4))
fig, ax = plt.subplots(nrows=2, ncols=2, figsize=(10, 6))
t = np.linspace(0, 40)
uaf = UpperAsymptoteFitter().fit(T, E, timeline=t)
uaf.plot_survival_function(ax=ax[0][0])
kmf.plot(ax=ax[0][0])
uaf.plot_cumulative_hazard(ax=ax[0][1])
naf.plot(ax=ax[0][1])
t = np.linspace(0, 100)
uaf = UpperAsymptoteFitter().fit(T, E, timeline=t)
uaf.plot_survival_function(ax=ax[1][0])
kmf.survival_function_.plot(ax=ax[1][0])
uaf.plot_cumulative_hazard(ax=ax[1][1])
naf.plot(ax=ax[1][1])
from lifelines.datasets import load_leukemia, load_kidney_transplant
T, E = load_leukemia()['t'], load_leukemia()['status']
uaf.fit(T, E)
ax = uaf.plot_survival_function(figsize=(8,4))
uaf.print_summary()
kmf.fit(T, E).plot(ax=ax, ci_show=False)
print("---")
print("Estimated lower bound: {:.2f} ({:.2f}, {:.2f})".format(
np.exp(-uaf.summary.loc['c_', 'coef']),
np.exp(-uaf.summary.loc['c_', 'coef upper 95%']),
np.exp(-uaf.summary.loc['c_', 'coef lower 95%']),
)
)
# Another, less obvious, dataset.
T, E = load_kidney_transplant()['time'], load_kidney_transplant()['death']
uaf.fit(T, E)
ax = uaf.plot_survival_function(figsize=(8,4))
uaf.print_summary()
kmf.fit(T, E).plot(ax=ax)
print("---")
print("Estimated lower bound: {:.2f} ({:.2f}, {:.2f})".format(
np.exp(-uaf.summary.loc['c_', 'coef']),
np.exp(-uaf.summary.loc['c_', 'coef upper 95%']),
np.exp(-uaf.summary.loc['c_', 'coef lower 95%']),
)
)
from autograd.scipy.stats import norm
from lifelines.fitters import ParametricUnivariateFitter
class SimpleUpperAsymptoteFitter(ParametricUnivariateFitter):
_fitted_parameter_names = ["c_", "lambda_"]
_bounds = ((0, None), (0, None))
def _cumulative_hazard(self, params, times):
c, lambda_ = params
return c * (1 - 1 /(lambda_ * times + 1))
# Another, less obvious, dataset.
saf = SimpleUpperAsymptoteFitter().fit(T, E, timeline=np.arange(1, 10000))
ax = saf.plot_survival_function(figsize=(8,4))
saf.print_summary(4)
kmf.fit(T, E).plot(ax=ax)
print("---")
print("Estimated lower bound: {:.2f} ({:.2f}, {:.2f})".format(
np.exp(-saf.summary.loc['c_', 'coef']),
np.exp(-saf.summary.loc['c_', 'coef upper 95%']),
np.exp(-saf.summary.loc['c_', 'coef lower 95%']),
)
)
| 0.632389 | 0.986598 |
<a href="https://colab.research.google.com/github/easonwwg/Informer2020/blob/main/Informer.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Informer Demo
## Download code and dataset
```
!git clone https://github.com/zhouhaoyi/Informer2020.git
!git clone https://github.com/zhouhaoyi/ETDataset.git
!ls
import sys
if not 'Informer2020' in sys.path:
sys.path += ['Informer2020']
# !pip install -r ./Informer2020/requirements.txt
```
## Experiments: Train and Test
```
from utils.tools import dotdict
from exp.exp_informer import Exp_Informer
import torch
args = dotdict()
args.model = 'informer' # model of experiment, options: [informer, informerstack, informerlight(TBD)]
args.data = 'ETTh1' # data
args.root_path = './ETDataset/ETT-small/' # root path of data file
args.data_path = 'ETTh1.csv' # data file
args.features = 'M' # forecasting task, options:[M, S, MS]; M:multivariate predict multivariate, S:univariate predict univariate, MS:multivariate predict univariate
args.target = 'OT' # target feature in S or MS task
args.freq = 'h' # freq for time features encoding, options:[s:secondly, t:minutely, h:hourly, d:daily, b:business days, w:weekly, m:monthly], you can also use more detailed freq like 15min or 3h
args.checkpoints = './informer_checkpoints' # location of model checkpoints
args.seq_len = 96 # input sequence length of Informer encoder
args.label_len = 48 # start token length of Informer decoder
args.pred_len = 24 # prediction sequence length
# Informer decoder input: concat[start token series(label_len), zero padding series(pred_len)]
args.enc_in = 7 # encoder input size
args.dec_in = 7 # decoder input size
args.c_out = 7 # output size
args.factor = 5 # probsparse attn factor
args.d_model = 512 # dimension of model
args.n_heads = 8 # num of heads
args.e_layers = 2 # num of encoder layers
args.d_layers = 1 # num of decoder layers
args.d_ff = 2048 # dimension of fcn in model
args.dropout = 0.05 # dropout
args.attn = 'prob' # attention used in encoder, options:[prob, full]
args.embed = 'timeF' # time features encoding, options:[timeF, fixed, learned]
args.activation = 'gelu' # activation
args.distil = True # whether to use distilling in encoder
args.output_attention = False # whether to output attention in ecoder
args.mix = True
args.padding = 0
args.freq = 'h'
args.batch_size = 32
args.learning_rate = 0.0001
args.loss = 'mse'
args.lradj = 'type1'
args.use_amp = False # whether to use automatic mixed precision training
args.num_workers = 0
args.itr = 1
args.train_epochs = 6
args.patience = 3
args.des = 'exp'
args.use_gpu = True if torch.cuda.is_available() else False
args.gpu = 0
args.use_multi_gpu = False
args.devices = '0,1,2,3'
args.use_gpu = True if torch.cuda.is_available() and args.use_gpu else False
if args.use_gpu and args.use_multi_gpu:
args.devices = args.devices.replace(' ','')
device_ids = args.devices.split(',')
args.device_ids = [int(id_) for id_ in device_ids]
args.gpu = args.device_ids[0]
# Set augments by using data name
data_parser = {
'ETTh1':{'data':'ETTh1.csv','T':'OT','M':[7,7,7],'S':[1,1,1],'MS':[7,7,1]},
'ETTh2':{'data':'ETTh2.csv','T':'OT','M':[7,7,7],'S':[1,1,1],'MS':[7,7,1]},
'ETTm1':{'data':'ETTm1.csv','T':'OT','M':[7,7,7],'S':[1,1,1],'MS':[7,7,1]},
'ETTm2':{'data':'ETTm2.csv','T':'OT','M':[7,7,7],'S':[1,1,1],'MS':[7,7,1]},
}
if args.data in data_parser.keys():
data_info = data_parser[args.data]
args.data_path = data_info['data']
args.target = data_info['T']
args.enc_in, args.dec_in, args.c_out = data_info[args.features]
args.detail_freq = args.freq
args.freq = args.freq[-1:]
print('Args in experiment:')
print(args)
Exp = Exp_Informer
for ii in range(args.itr):
# setting record of experiments
setting = '{}_{}_ft{}_sl{}_ll{}_pl{}_dm{}_nh{}_el{}_dl{}_df{}_at{}_fc{}_eb{}_dt{}_mx{}_{}_{}'.format(args.model, args.data, args.features,
args.seq_len, args.label_len, args.pred_len,
args.d_model, args.n_heads, args.e_layers, args.d_layers, args.d_ff, args.attn, args.factor, args.embed, args.distil, args.mix, args.des, ii)
# set experiments
exp = Exp(args)
# train
print('>>>>>>>start training : {}>>>>>>>>>>>>>>>>>>>>>>>>>>'.format(setting))
exp.train(setting)
# test
print('>>>>>>>testing : {}<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<'.format(setting))
exp.test(setting)
torch.cuda.empty_cache()
```
## Prediction
```
import os
# set saved model path
setting = 'informer_ETTh1_ftM_sl96_ll48_pl24_dm512_nh8_el2_dl1_df2048_atprob_fc5_ebtimeF_dtTrue_mxTrue_exp_0'
# path = os.path.join(args.checkpoints,setting,'checkpoint.pth')
# If you already have a trained model, you can set the arguments and model path, then initialize a Experiment and use it to predict
# Prediction is a sequence which is adjacent to the last date of the data, and does not exist in the data
# If you want to get more information about prediction, you can refer to code `exp/exp_informer.py function predict()` and `data/data_loader.py class Dataset_Pred`
exp = Exp(args)
exp.predict(setting, True)
# the prediction will be saved in ./results/{setting}/real_prediction.npy
import numpy as np
prediction = np.load('./results/'+setting+'/real_prediction.npy')
prediction.shape
```
### More details about Prediction - prediction function
```
# here is the detailed code of function predict
def predict(exp, setting, load=False):
pred_data, pred_loader = exp._get_data(flag='pred')
if load:
path = os.path.join(exp.args.checkpoints, setting)
best_model_path = path+'/'+'checkpoint.pth'
exp.model.load_state_dict(torch.load(best_model_path))
exp.model.eval()
preds = []
for i, (batch_x,batch_y,batch_x_mark,batch_y_mark) in enumerate(pred_loader):
batch_x = batch_x.float().to(exp.device)
batch_y = batch_y.float()
batch_x_mark = batch_x_mark.float().to(exp.device)
batch_y_mark = batch_y_mark.float().to(exp.device)
# decoder input
if exp.args.padding==0:
dec_inp = torch.zeros([batch_y.shape[0], exp.args.pred_len, batch_y.shape[-1]]).float()
elif exp.args.padding==1:
dec_inp = torch.ones([batch_y.shape[0], exp.args.pred_len, batch_y.shape[-1]]).float()
else:
dec_inp = torch.zeros([batch_y.shape[0], exp.args.pred_len, batch_y.shape[-1]]).float()
dec_inp = torch.cat([batch_y[:,:exp.args.label_len,:], dec_inp], dim=1).float().to(exp.device)
# encoder - decoder
if exp.args.use_amp:
with torch.cuda.amp.autocast():
if exp.args.output_attention:
outputs = exp.model(batch_x, batch_x_mark, dec_inp, batch_y_mark)[0]
else:
outputs = exp.model(batch_x, batch_x_mark, dec_inp, batch_y_mark)
else:
if exp.args.output_attention:
outputs = exp.model(batch_x, batch_x_mark, dec_inp, batch_y_mark)[0]
else:
outputs = exp.model(batch_x, batch_x_mark, dec_inp, batch_y_mark)
f_dim = -1 if exp.args.features=='MS' else 0
batch_y = batch_y[:,-exp.args.pred_len:,f_dim:].to(exp.device)
pred = outputs.detach().cpu().numpy()#.squeeze()
preds.append(pred)
preds = np.array(preds)
preds = preds.reshape(-1, preds.shape[-2], preds.shape[-1])
# result save
folder_path = './results/' + setting +'/'
if not os.path.exists(folder_path):
os.makedirs(folder_path)
np.save(folder_path+'real_prediction.npy', preds)
return preds
# you can also use this prediction function to get result
prediction = predict(exp, setting, True)
import matplotlib.pyplot as plt
plt.figure()
plt.plot(prediction[0,:,-1])
plt.show()
```
### More details about Prediction - prediction dataset
You can give a `root_path` and `data_path` of the data you want to forecast, and set `seq_len`, `label_len`, `pred_len` and other arguments as other Dataset. The difference is that you can set a more detailed freq such as `15min` or `3h` to generate the timestamp of prediction series.
`Dataset_Pred` only has one sample (including `encoder_input: [1, seq_len, dim]`, `decoder_token: [1, label_len, dim]`, `encoder_input_timestamp: [1, seq_len, date_dim]`, `decoder_input_timstamp: [1, label_len+pred_len, date_dim]`). It will intercept the last sequence of the given data (seq_len data) to forecast the unseen future sequence (pred_len data).
```
from data.data_loader import Dataset_Pred
from torch.utils.data import DataLoader
Data = Dataset_Pred
timeenc = 0 if args.embed!='timeF' else 1
flag = 'pred'; shuffle_flag = False; drop_last = False; batch_size = 1
freq = args.detail_freq
data_set = Data(
root_path=args.root_path,
data_path=args.data_path,
flag=flag,
size=[args.seq_len, args.label_len, args.pred_len],
features=args.features,
target=args.target,
timeenc=timeenc,
freq=freq
)
data_loader = DataLoader(
data_set,
batch_size=batch_size,
shuffle=shuffle_flag,
num_workers=args.num_workers,
drop_last=drop_last)
len(data_set), len(data_loader)
```
## Visualization
```
# When we finished exp.train(setting) and exp.test(setting), we will get a trained model and the results of test experiment
# The results of test experiment will be saved in ./results/{setting}/pred.npy (prediction of test dataset) and ./results/{setting}/true.npy (groundtruth of test dataset)
preds = np.load('./results/'+setting+'/pred.npy')
trues = np.load('./results/'+setting+'/true.npy')
# [samples, pred_len, dimensions]
preds.shape, trues.shape
import matplotlib.pyplot as plt
import seaborn as sns
# draw OT prediction
plt.figure()
plt.plot(trues[0,:,-1], label='GroundTruth')
plt.plot(preds[0,:,-1], label='Prediction')
plt.legend()
plt.show()
# draw HUFL prediction
plt.figure()
plt.plot(trues[0,:,0], label='GroundTruth')
plt.plot(preds[0,:,0], label='Prediction')
plt.legend()
plt.show()
from data.data_loader import Dataset_ETT_hour
from torch.utils.data import DataLoader
Data = Dataset_ETT_hour
timeenc = 0 if args.embed!='timeF' else 1
flag = 'test'; shuffle_flag = False; drop_last = True; batch_size = 1
data_set = Data(
root_path=args.root_path,
data_path=args.data_path,
flag=flag,
size=[args.seq_len, args.label_len, args.pred_len],
features=args.features,
timeenc=timeenc,
freq=args.freq
)
data_loader = DataLoader(
data_set,
batch_size=batch_size,
shuffle=shuffle_flag,
num_workers=args.num_workers,
drop_last=drop_last)
import os
args.output_attention = True
exp = Exp(args)
model = exp.model
setting = 'informer_ETTh1_ftM_sl96_ll48_pl24_dm512_nh8_el2_dl1_df2048_atprob_fc5_ebtimeF_dtTrue_mxTrue_exp_0'
path = os.path.join(args.checkpoints,setting,'checkpoint.pth')
model.load_state_dict(torch.load(path))
# attention visualization
idx = 0
for i, (batch_x,batch_y,batch_x_mark,batch_y_mark) in enumerate(data_loader):
if i!=idx:
continue
batch_x = batch_x.float().to(exp.device)
batch_y = batch_y.float()
batch_x_mark = batch_x_mark.float().to(exp.device)
batch_y_mark = batch_y_mark.float().to(exp.device)
dec_inp = torch.zeros_like(batch_y[:,-args.pred_len:,:]).float()
dec_inp = torch.cat([batch_y[:,:args.label_len,:], dec_inp], dim=1).float().to(exp.device)
outputs,attn = model(batch_x, batch_x_mark, dec_inp, batch_y_mark)
attn[0].shape, attn[1].shape #, attn[2].shape
layer = 0
distil = 'Distil' if args.distil else 'NoDistil'
for h in range(0,8):
plt.figure(figsize=[10,8])
plt.title('Informer, {}, attn:{} layer:{} head:{}'.format(distil, args.attn, layer, h))
A = attn[layer][0,h].detach().cpu().numpy()
ax = sns.heatmap(A, vmin=0, vmax=A.max()+0.01)
plt.show()
layer = 1
distil = 'Distil' if args.distil else 'NoDistil'
for h in range(0,8):
plt.figure(figsize=[10,8])
plt.title('Informer, {}, attn:{} layer:{} head:{}'.format(distil, args.attn, layer, h))
A = attn[layer][0,h].detach().cpu().numpy()
ax = sns.heatmap(A, vmin=0, vmax=A.max()+0.01)
plt.show()
```
## Custom Data
Custom data (xxx.csv) has to include at least 2 features: `date`(format: `YYYY-MM-DD hh:mm:ss`) and `target feature`.
```
from data.data_loader import Dataset_Custom
from torch.utils.data import DataLoader
import pandas as pd
import os
# custom data: xxx.csv
# data features: ['date', ...(other features), target feature]
# we take ETTh2 as an example
args.root_path = './ETDataset/ETT-small/'
args.data_path = 'ETTh2.csv'
df = pd.read_csv(os.path.join(args.root_path, args.data_path))
df.head()
'''
We set 'HULL' as target instead of 'OT'
The following frequencies are supported:
Y - yearly
alias: A
M - monthly
W - weekly
D - daily
B - business days
H - hourly
T - minutely
alias: min
S - secondly
'''
args.target = 'HULL'
args.freq = 'h'
Data = Dataset_Custom
timeenc = 0 if args.embed!='timeF' else 1
flag = 'test'; shuffle_flag = False; drop_last = True; batch_size = 1
data_set = Data(
root_path=args.root_path,
data_path=args.data_path,
flag=flag,
size=[args.seq_len, args.label_len, args.pred_len],
features=args.features,
timeenc=timeenc,
target=args.target, # HULL here
freq=args.freq # 'h': hourly, 't':minutely
)
data_loader = DataLoader(
data_set,
batch_size=batch_size,
shuffle=shuffle_flag,
num_workers=args.num_workers,
drop_last=drop_last)
batch_x,batch_y,batch_x_mark,batch_y_mark = data_set[0]
```
|
github_jupyter
|
!git clone https://github.com/zhouhaoyi/Informer2020.git
!git clone https://github.com/zhouhaoyi/ETDataset.git
!ls
import sys
if not 'Informer2020' in sys.path:
sys.path += ['Informer2020']
# !pip install -r ./Informer2020/requirements.txt
from utils.tools import dotdict
from exp.exp_informer import Exp_Informer
import torch
args = dotdict()
args.model = 'informer' # model of experiment, options: [informer, informerstack, informerlight(TBD)]
args.data = 'ETTh1' # data
args.root_path = './ETDataset/ETT-small/' # root path of data file
args.data_path = 'ETTh1.csv' # data file
args.features = 'M' # forecasting task, options:[M, S, MS]; M:multivariate predict multivariate, S:univariate predict univariate, MS:multivariate predict univariate
args.target = 'OT' # target feature in S or MS task
args.freq = 'h' # freq for time features encoding, options:[s:secondly, t:minutely, h:hourly, d:daily, b:business days, w:weekly, m:monthly], you can also use more detailed freq like 15min or 3h
args.checkpoints = './informer_checkpoints' # location of model checkpoints
args.seq_len = 96 # input sequence length of Informer encoder
args.label_len = 48 # start token length of Informer decoder
args.pred_len = 24 # prediction sequence length
# Informer decoder input: concat[start token series(label_len), zero padding series(pred_len)]
args.enc_in = 7 # encoder input size
args.dec_in = 7 # decoder input size
args.c_out = 7 # output size
args.factor = 5 # probsparse attn factor
args.d_model = 512 # dimension of model
args.n_heads = 8 # num of heads
args.e_layers = 2 # num of encoder layers
args.d_layers = 1 # num of decoder layers
args.d_ff = 2048 # dimension of fcn in model
args.dropout = 0.05 # dropout
args.attn = 'prob' # attention used in encoder, options:[prob, full]
args.embed = 'timeF' # time features encoding, options:[timeF, fixed, learned]
args.activation = 'gelu' # activation
args.distil = True # whether to use distilling in encoder
args.output_attention = False # whether to output attention in ecoder
args.mix = True
args.padding = 0
args.freq = 'h'
args.batch_size = 32
args.learning_rate = 0.0001
args.loss = 'mse'
args.lradj = 'type1'
args.use_amp = False # whether to use automatic mixed precision training
args.num_workers = 0
args.itr = 1
args.train_epochs = 6
args.patience = 3
args.des = 'exp'
args.use_gpu = True if torch.cuda.is_available() else False
args.gpu = 0
args.use_multi_gpu = False
args.devices = '0,1,2,3'
args.use_gpu = True if torch.cuda.is_available() and args.use_gpu else False
if args.use_gpu and args.use_multi_gpu:
args.devices = args.devices.replace(' ','')
device_ids = args.devices.split(',')
args.device_ids = [int(id_) for id_ in device_ids]
args.gpu = args.device_ids[0]
# Set augments by using data name
data_parser = {
'ETTh1':{'data':'ETTh1.csv','T':'OT','M':[7,7,7],'S':[1,1,1],'MS':[7,7,1]},
'ETTh2':{'data':'ETTh2.csv','T':'OT','M':[7,7,7],'S':[1,1,1],'MS':[7,7,1]},
'ETTm1':{'data':'ETTm1.csv','T':'OT','M':[7,7,7],'S':[1,1,1],'MS':[7,7,1]},
'ETTm2':{'data':'ETTm2.csv','T':'OT','M':[7,7,7],'S':[1,1,1],'MS':[7,7,1]},
}
if args.data in data_parser.keys():
data_info = data_parser[args.data]
args.data_path = data_info['data']
args.target = data_info['T']
args.enc_in, args.dec_in, args.c_out = data_info[args.features]
args.detail_freq = args.freq
args.freq = args.freq[-1:]
print('Args in experiment:')
print(args)
Exp = Exp_Informer
for ii in range(args.itr):
# setting record of experiments
setting = '{}_{}_ft{}_sl{}_ll{}_pl{}_dm{}_nh{}_el{}_dl{}_df{}_at{}_fc{}_eb{}_dt{}_mx{}_{}_{}'.format(args.model, args.data, args.features,
args.seq_len, args.label_len, args.pred_len,
args.d_model, args.n_heads, args.e_layers, args.d_layers, args.d_ff, args.attn, args.factor, args.embed, args.distil, args.mix, args.des, ii)
# set experiments
exp = Exp(args)
# train
print('>>>>>>>start training : {}>>>>>>>>>>>>>>>>>>>>>>>>>>'.format(setting))
exp.train(setting)
# test
print('>>>>>>>testing : {}<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<'.format(setting))
exp.test(setting)
torch.cuda.empty_cache()
import os
# set saved model path
setting = 'informer_ETTh1_ftM_sl96_ll48_pl24_dm512_nh8_el2_dl1_df2048_atprob_fc5_ebtimeF_dtTrue_mxTrue_exp_0'
# path = os.path.join(args.checkpoints,setting,'checkpoint.pth')
# If you already have a trained model, you can set the arguments and model path, then initialize a Experiment and use it to predict
# Prediction is a sequence which is adjacent to the last date of the data, and does not exist in the data
# If you want to get more information about prediction, you can refer to code `exp/exp_informer.py function predict()` and `data/data_loader.py class Dataset_Pred`
exp = Exp(args)
exp.predict(setting, True)
# the prediction will be saved in ./results/{setting}/real_prediction.npy
import numpy as np
prediction = np.load('./results/'+setting+'/real_prediction.npy')
prediction.shape
# here is the detailed code of function predict
def predict(exp, setting, load=False):
pred_data, pred_loader = exp._get_data(flag='pred')
if load:
path = os.path.join(exp.args.checkpoints, setting)
best_model_path = path+'/'+'checkpoint.pth'
exp.model.load_state_dict(torch.load(best_model_path))
exp.model.eval()
preds = []
for i, (batch_x,batch_y,batch_x_mark,batch_y_mark) in enumerate(pred_loader):
batch_x = batch_x.float().to(exp.device)
batch_y = batch_y.float()
batch_x_mark = batch_x_mark.float().to(exp.device)
batch_y_mark = batch_y_mark.float().to(exp.device)
# decoder input
if exp.args.padding==0:
dec_inp = torch.zeros([batch_y.shape[0], exp.args.pred_len, batch_y.shape[-1]]).float()
elif exp.args.padding==1:
dec_inp = torch.ones([batch_y.shape[0], exp.args.pred_len, batch_y.shape[-1]]).float()
else:
dec_inp = torch.zeros([batch_y.shape[0], exp.args.pred_len, batch_y.shape[-1]]).float()
dec_inp = torch.cat([batch_y[:,:exp.args.label_len,:], dec_inp], dim=1).float().to(exp.device)
# encoder - decoder
if exp.args.use_amp:
with torch.cuda.amp.autocast():
if exp.args.output_attention:
outputs = exp.model(batch_x, batch_x_mark, dec_inp, batch_y_mark)[0]
else:
outputs = exp.model(batch_x, batch_x_mark, dec_inp, batch_y_mark)
else:
if exp.args.output_attention:
outputs = exp.model(batch_x, batch_x_mark, dec_inp, batch_y_mark)[0]
else:
outputs = exp.model(batch_x, batch_x_mark, dec_inp, batch_y_mark)
f_dim = -1 if exp.args.features=='MS' else 0
batch_y = batch_y[:,-exp.args.pred_len:,f_dim:].to(exp.device)
pred = outputs.detach().cpu().numpy()#.squeeze()
preds.append(pred)
preds = np.array(preds)
preds = preds.reshape(-1, preds.shape[-2], preds.shape[-1])
# result save
folder_path = './results/' + setting +'/'
if not os.path.exists(folder_path):
os.makedirs(folder_path)
np.save(folder_path+'real_prediction.npy', preds)
return preds
# you can also use this prediction function to get result
prediction = predict(exp, setting, True)
import matplotlib.pyplot as plt
plt.figure()
plt.plot(prediction[0,:,-1])
plt.show()
from data.data_loader import Dataset_Pred
from torch.utils.data import DataLoader
Data = Dataset_Pred
timeenc = 0 if args.embed!='timeF' else 1
flag = 'pred'; shuffle_flag = False; drop_last = False; batch_size = 1
freq = args.detail_freq
data_set = Data(
root_path=args.root_path,
data_path=args.data_path,
flag=flag,
size=[args.seq_len, args.label_len, args.pred_len],
features=args.features,
target=args.target,
timeenc=timeenc,
freq=freq
)
data_loader = DataLoader(
data_set,
batch_size=batch_size,
shuffle=shuffle_flag,
num_workers=args.num_workers,
drop_last=drop_last)
len(data_set), len(data_loader)
# When we finished exp.train(setting) and exp.test(setting), we will get a trained model and the results of test experiment
# The results of test experiment will be saved in ./results/{setting}/pred.npy (prediction of test dataset) and ./results/{setting}/true.npy (groundtruth of test dataset)
preds = np.load('./results/'+setting+'/pred.npy')
trues = np.load('./results/'+setting+'/true.npy')
# [samples, pred_len, dimensions]
preds.shape, trues.shape
import matplotlib.pyplot as plt
import seaborn as sns
# draw OT prediction
plt.figure()
plt.plot(trues[0,:,-1], label='GroundTruth')
plt.plot(preds[0,:,-1], label='Prediction')
plt.legend()
plt.show()
# draw HUFL prediction
plt.figure()
plt.plot(trues[0,:,0], label='GroundTruth')
plt.plot(preds[0,:,0], label='Prediction')
plt.legend()
plt.show()
from data.data_loader import Dataset_ETT_hour
from torch.utils.data import DataLoader
Data = Dataset_ETT_hour
timeenc = 0 if args.embed!='timeF' else 1
flag = 'test'; shuffle_flag = False; drop_last = True; batch_size = 1
data_set = Data(
root_path=args.root_path,
data_path=args.data_path,
flag=flag,
size=[args.seq_len, args.label_len, args.pred_len],
features=args.features,
timeenc=timeenc,
freq=args.freq
)
data_loader = DataLoader(
data_set,
batch_size=batch_size,
shuffle=shuffle_flag,
num_workers=args.num_workers,
drop_last=drop_last)
import os
args.output_attention = True
exp = Exp(args)
model = exp.model
setting = 'informer_ETTh1_ftM_sl96_ll48_pl24_dm512_nh8_el2_dl1_df2048_atprob_fc5_ebtimeF_dtTrue_mxTrue_exp_0'
path = os.path.join(args.checkpoints,setting,'checkpoint.pth')
model.load_state_dict(torch.load(path))
# attention visualization
idx = 0
for i, (batch_x,batch_y,batch_x_mark,batch_y_mark) in enumerate(data_loader):
if i!=idx:
continue
batch_x = batch_x.float().to(exp.device)
batch_y = batch_y.float()
batch_x_mark = batch_x_mark.float().to(exp.device)
batch_y_mark = batch_y_mark.float().to(exp.device)
dec_inp = torch.zeros_like(batch_y[:,-args.pred_len:,:]).float()
dec_inp = torch.cat([batch_y[:,:args.label_len,:], dec_inp], dim=1).float().to(exp.device)
outputs,attn = model(batch_x, batch_x_mark, dec_inp, batch_y_mark)
attn[0].shape, attn[1].shape #, attn[2].shape
layer = 0
distil = 'Distil' if args.distil else 'NoDistil'
for h in range(0,8):
plt.figure(figsize=[10,8])
plt.title('Informer, {}, attn:{} layer:{} head:{}'.format(distil, args.attn, layer, h))
A = attn[layer][0,h].detach().cpu().numpy()
ax = sns.heatmap(A, vmin=0, vmax=A.max()+0.01)
plt.show()
layer = 1
distil = 'Distil' if args.distil else 'NoDistil'
for h in range(0,8):
plt.figure(figsize=[10,8])
plt.title('Informer, {}, attn:{} layer:{} head:{}'.format(distil, args.attn, layer, h))
A = attn[layer][0,h].detach().cpu().numpy()
ax = sns.heatmap(A, vmin=0, vmax=A.max()+0.01)
plt.show()
from data.data_loader import Dataset_Custom
from torch.utils.data import DataLoader
import pandas as pd
import os
# custom data: xxx.csv
# data features: ['date', ...(other features), target feature]
# we take ETTh2 as an example
args.root_path = './ETDataset/ETT-small/'
args.data_path = 'ETTh2.csv'
df = pd.read_csv(os.path.join(args.root_path, args.data_path))
df.head()
'''
We set 'HULL' as target instead of 'OT'
The following frequencies are supported:
Y - yearly
alias: A
M - monthly
W - weekly
D - daily
B - business days
H - hourly
T - minutely
alias: min
S - secondly
'''
args.target = 'HULL'
args.freq = 'h'
Data = Dataset_Custom
timeenc = 0 if args.embed!='timeF' else 1
flag = 'test'; shuffle_flag = False; drop_last = True; batch_size = 1
data_set = Data(
root_path=args.root_path,
data_path=args.data_path,
flag=flag,
size=[args.seq_len, args.label_len, args.pred_len],
features=args.features,
timeenc=timeenc,
target=args.target, # HULL here
freq=args.freq # 'h': hourly, 't':minutely
)
data_loader = DataLoader(
data_set,
batch_size=batch_size,
shuffle=shuffle_flag,
num_workers=args.num_workers,
drop_last=drop_last)
batch_x,batch_y,batch_x_mark,batch_y_mark = data_set[0]
| 0.431105 | 0.865167 |
<a href="https://colab.research.google.com/github/x-cloud/deep-learning-v2-pytorch/blob/master/intro-to-pytorch/Part%204%20-%20Fashion-MNIST%20(Exercises).ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Classifying Fashion-MNIST
Now it's your turn to build and train a neural network. You'll be using the [Fashion-MNIST dataset](https://github.com/zalandoresearch/fashion-mnist), a drop-in replacement for the MNIST dataset. MNIST is actually quite trivial with neural networks where you can easily achieve better than 97% accuracy. Fashion-MNIST is a set of 28x28 greyscale images of clothes. It's more complex than MNIST, so it's a better representation of the actual performance of your network, and a better representation of datasets you'll use in the real world.
<img src='https://github.com/x-cloud/deep-learning-v2-pytorch/blob/master/intro-to-pytorch/assets/fashion-mnist-sprite.png?raw=1' width=500px>
In this notebook, you'll build your own neural network. For the most part, you could just copy and paste the code from Part 3, but you wouldn't be learning. It's important for you to write the code yourself and get it to work. Feel free to consult the previous notebooks though as you work through this.
First off, let's load the dataset through torchvision.
```
import torch
from torchvision import datasets, transforms
import helper
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5,), (0.5,))])
# Download and load the training data
trainset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
# Download and load the test data
testset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True)
```
Here we can see one of the images.
```
import matplotlib.pyplot as plt
import numpy as np
def imshow(image, ax=None, title=None, normalize=True):
"""Imshow for Tensor."""
if ax is None:
fig, ax = plt.subplots()
image = image.numpy().transpose((1, 2, 0))
if normalize:
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
image = std * image + mean
image = np.clip(image, 0, 1)
ax.imshow(image)
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.spines['bottom'].set_visible(False)
ax.tick_params(axis='both', length=0)
ax.set_xticklabels('')
ax.set_yticklabels('')
return ax
image, label = next(iter(trainloader))
#helper.imshow(image[0,:]);
imshow(image[0,:]);
```
## Building the network
Here you should define your network. As with MNIST, each image is 28x28 which is a total of 784 pixels, and there are 10 classes. You should include at least one hidden layer. We suggest you use ReLU activations for the layers and to return the logits or log-softmax from the forward pass. It's up to you how many layers you add and the size of those layers.
```
# TODO: Define your network architecture here
import torch.nn as nn
from collections import OrderedDict
model = nn.Sequential(OrderedDict([
('fc1', nn.Linear(784, 256)),
('relu2', nn.ReLU()),
('fc3', nn.Linear(256, 128)),
('relu4', nn.ReLU()),
('fc5', nn.Linear(128, 10)),
('logsoftmax6', nn.LogSoftmax(dim=1))
]))
print(model)
```
# Train the network
Now you should create your network and train it. First you'll want to define [the criterion](http://pytorch.org/docs/master/nn.html#loss-functions) ( something like `nn.CrossEntropyLoss`) and [the optimizer](http://pytorch.org/docs/master/optim.html) (typically `optim.SGD` or `optim.Adam`).
Then write the training code. Remember the training pass is a fairly straightforward process:
* Make a forward pass through the network to get the logits
* Use the logits to calculate the loss
* Perform a backward pass through the network with `loss.backward()` to calculate the gradients
* Take a step with the optimizer to update the weights
By adjusting the hyperparameters (hidden units, learning rate, etc), you should be able to get the training loss below 0.4.
```
# TODO: Create the network, define the criterion and optimizer
from torch import optim
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.parameters(), lr=0.001)
# TODO: Train the network here
epochs = 5
for e in range(epochs):
running_loss = 0
for images, labels in trainloader:
images = images.view(images.shape[0], -1)
optimizer.zero_grad()
output = model(images)
loss = criterion(output, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
else:
print('loss: ', running_loss / len(trainloader))
def view_classify(img, ps, version="MNIST"):
''' Function for viewing an image and it's predicted classes.
'''
ps = ps.data.numpy().squeeze()
fig, (ax1, ax2) = plt.subplots(figsize=(6,9), ncols=2)
ax1.imshow(img.resize_(1, 28, 28).numpy().squeeze())
ax1.axis('off')
ax2.barh(np.arange(10), ps)
ax2.set_aspect(0.1)
ax2.set_yticks(np.arange(10))
if version == "MNIST":
ax2.set_yticklabels(np.arange(10))
elif version == "Fashion":
ax2.set_yticklabels(['T-shirt/top',
'Trouser',
'Pullover',
'Dress',
'Coat',
'Sandal',
'Shirt',
'Sneaker',
'Bag',
'Ankle Boot'], size='small');
ax2.set_title('Class Probability')
ax2.set_xlim(0, 1.1)
plt.tight_layout()
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import helper
# Test out your network!
dataiter = iter(testloader)
images, labels = dataiter.next()
img = images[0]
# Convert 2D image to 1D vector
img = img.resize_(1, 784)
# TODO: Calculate the class probabilities (softmax) for img
with torch.no_grad():
ps = torch.exp(model(img))
# Plot the image and probabilities
# helper.view_classify(img.resize_(1, 28, 28), ps, version='Fashion')
view_classify(img.resize_(1, 28, 28), ps, version='Fashion')
```
|
github_jupyter
|
import torch
from torchvision import datasets, transforms
import helper
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5,), (0.5,))])
# Download and load the training data
trainset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
# Download and load the test data
testset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True)
import matplotlib.pyplot as plt
import numpy as np
def imshow(image, ax=None, title=None, normalize=True):
"""Imshow for Tensor."""
if ax is None:
fig, ax = plt.subplots()
image = image.numpy().transpose((1, 2, 0))
if normalize:
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
image = std * image + mean
image = np.clip(image, 0, 1)
ax.imshow(image)
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.spines['bottom'].set_visible(False)
ax.tick_params(axis='both', length=0)
ax.set_xticklabels('')
ax.set_yticklabels('')
return ax
image, label = next(iter(trainloader))
#helper.imshow(image[0,:]);
imshow(image[0,:]);
# TODO: Define your network architecture here
import torch.nn as nn
from collections import OrderedDict
model = nn.Sequential(OrderedDict([
('fc1', nn.Linear(784, 256)),
('relu2', nn.ReLU()),
('fc3', nn.Linear(256, 128)),
('relu4', nn.ReLU()),
('fc5', nn.Linear(128, 10)),
('logsoftmax6', nn.LogSoftmax(dim=1))
]))
print(model)
# TODO: Create the network, define the criterion and optimizer
from torch import optim
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.parameters(), lr=0.001)
# TODO: Train the network here
epochs = 5
for e in range(epochs):
running_loss = 0
for images, labels in trainloader:
images = images.view(images.shape[0], -1)
optimizer.zero_grad()
output = model(images)
loss = criterion(output, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
else:
print('loss: ', running_loss / len(trainloader))
def view_classify(img, ps, version="MNIST"):
''' Function for viewing an image and it's predicted classes.
'''
ps = ps.data.numpy().squeeze()
fig, (ax1, ax2) = plt.subplots(figsize=(6,9), ncols=2)
ax1.imshow(img.resize_(1, 28, 28).numpy().squeeze())
ax1.axis('off')
ax2.barh(np.arange(10), ps)
ax2.set_aspect(0.1)
ax2.set_yticks(np.arange(10))
if version == "MNIST":
ax2.set_yticklabels(np.arange(10))
elif version == "Fashion":
ax2.set_yticklabels(['T-shirt/top',
'Trouser',
'Pullover',
'Dress',
'Coat',
'Sandal',
'Shirt',
'Sneaker',
'Bag',
'Ankle Boot'], size='small');
ax2.set_title('Class Probability')
ax2.set_xlim(0, 1.1)
plt.tight_layout()
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import helper
# Test out your network!
dataiter = iter(testloader)
images, labels = dataiter.next()
img = images[0]
# Convert 2D image to 1D vector
img = img.resize_(1, 784)
# TODO: Calculate the class probabilities (softmax) for img
with torch.no_grad():
ps = torch.exp(model(img))
# Plot the image and probabilities
# helper.view_classify(img.resize_(1, 28, 28), ps, version='Fashion')
view_classify(img.resize_(1, 28, 28), ps, version='Fashion')
| 0.770724 | 0.991756 |
# Working with model paths
In this example we show how to deal with model paths, which can be tricky when working with a GSFLOW model or individual parts of a GSFLOW model.
Relative paths in the GSFLOW control file and in the MODFLOW NAM file are always set relative to the shell or bat file the model is being run from. This file does not need to be in the same directory as any of the model files. We show how to work with these cases.
```
import os
import gsflow
from gsflow.modflow import Modflow
```
### How pyGSFLOW automatically determines the "base path" for a model
The default method `pyGSFLOW` uses to determine the base path is by the location of the Control file. When a model is loaded using `gsflow.GsflowModel.load_from_file()` the code determines the location of the control file and applies this as the base directory for all relative paths within the control file and the MODFLOW NAM file.
Here are a couple of examples of this case
```
control_file = os.path.join(".", "data", "sagehen", "gsflow", "saghen_new_cont.control")
gsf = gsflow.GsflowModel.load_from_file(control_file)
```
### Loading only the modflow files using the standard method
we first specify the `model_ws` which is the base directory that paths in the NAM file are relative to and then we can pass this to the `Modflow.load()` method.
```
model_ws = os.path.join(".", "data", "sagehen", "gsflow")
name_file = "saghen_new.nam"
ml = Modflow.load(name_file, model_ws=model_ws)
ml
```
## Specifying the "base path"
Specifying the "base path" is useful when paths are not relative to the GSFLOW control file. This is the case when the model run file (.bat, .sh) is located in a different directory.
The `model_ws` parameter in the `GsflowModel.load_from_file()` allows the user to override the default behaviour and set the base path for all model files.
Here is an example where the model run file is located one directory up from the control file:
```
# set the location of model run file
model_ws = os.path.join(".", "data", "sagehen", "gsflow_paths")
# set the location of the gsflow control file
control_file = os.path.join(model_ws, "control", "saghen_paths_cont.control")
# load the model and override the default path creation by passing in model_ws
gsf = gsflow.GsflowModel.load_from_file(control_file, model_ws=model_ws)
```
### Loading only MODFLOW or PRMS with `GsflowModel` after specifying "base path"
```
# set the location of model run file
model_ws = os.path.join(".", "data", "sagehen", "gsflow_paths")
# set the location of the gsflow control file
control_file = os.path.join(model_ws, "control", "saghen_paths_cont.control")
# load only modflow into the GsflowModel object and override the default path creation by passing in model_ws
gsf = gsflow.GsflowModel.load_from_file(control_file, modflow_only=True, model_ws=model_ws)
# get the modflow model
ml = gsf.mf
ml
# set the location of model run file
model_ws = os.path.join(".", "data", "sagehen", "gsflow_paths")
# set the location of the gsflow control file
control_file = os.path.join(model_ws, "control", "saghen_paths_cont.control")
# load only modflow into the GsflowModel object and override the default path creation by passing in model_ws
gsf = gsflow.GsflowModel.load_from_file(control_file, prms_only=True, model_ws=model_ws)
# get the prms model
prms = gsf.prms
```
### Loading MODFLOW with `Modflow` after specifying the base path
```
# specify our base path
model_ws = os.path.join(".", "data", "sagehen", "gsflow_paths")
# specify the relative path to the NAM file
name_file = os.path.join("modflow", "saghen_paths.nam")
ml = Modflow.load(name_file, model_ws=model_ws)
```
|
github_jupyter
|
import os
import gsflow
from gsflow.modflow import Modflow
control_file = os.path.join(".", "data", "sagehen", "gsflow", "saghen_new_cont.control")
gsf = gsflow.GsflowModel.load_from_file(control_file)
model_ws = os.path.join(".", "data", "sagehen", "gsflow")
name_file = "saghen_new.nam"
ml = Modflow.load(name_file, model_ws=model_ws)
ml
# set the location of model run file
model_ws = os.path.join(".", "data", "sagehen", "gsflow_paths")
# set the location of the gsflow control file
control_file = os.path.join(model_ws, "control", "saghen_paths_cont.control")
# load the model and override the default path creation by passing in model_ws
gsf = gsflow.GsflowModel.load_from_file(control_file, model_ws=model_ws)
# set the location of model run file
model_ws = os.path.join(".", "data", "sagehen", "gsflow_paths")
# set the location of the gsflow control file
control_file = os.path.join(model_ws, "control", "saghen_paths_cont.control")
# load only modflow into the GsflowModel object and override the default path creation by passing in model_ws
gsf = gsflow.GsflowModel.load_from_file(control_file, modflow_only=True, model_ws=model_ws)
# get the modflow model
ml = gsf.mf
ml
# set the location of model run file
model_ws = os.path.join(".", "data", "sagehen", "gsflow_paths")
# set the location of the gsflow control file
control_file = os.path.join(model_ws, "control", "saghen_paths_cont.control")
# load only modflow into the GsflowModel object and override the default path creation by passing in model_ws
gsf = gsflow.GsflowModel.load_from_file(control_file, prms_only=True, model_ws=model_ws)
# get the prms model
prms = gsf.prms
# specify our base path
model_ws = os.path.join(".", "data", "sagehen", "gsflow_paths")
# specify the relative path to the NAM file
name_file = os.path.join("modflow", "saghen_paths.nam")
ml = Modflow.load(name_file, model_ws=model_ws)
| 0.366363 | 0.950686 |
<table style="float:left; border:none">
<tr style="border:none">
<td style="border:none">
<a href="https://bokeh.org/">
<img
src="assets/bokeh-transparent.png"
style="width:50px"
>
</a>
</td>
<td style="border:none">
<h1>Bokeh Tutorial</h1>
</td>
</tr>
</table>
<div style="float:right;"><h2>02. Styling and Theming</h2></div>
In this chapter we will learn how to configure various visual aspects of our plots, and how to find out more about what an be configured.
# Imports and Setup
First, let's make the standard imports
```
from bokeh.io import output_notebook, show
from bokeh.plotting import figure
output_notebook()
```
This notebook uses Bokeh sample data. If you haven't downloaded it already, this can be downloaded by running the following:
```
import bokeh.sampledata
bokeh.sampledata.download()
```
Before we get started, it's useful to describe how colors and properties are specified in Bokeh.
# Colors
There are many places where you may need to specify colors. Bokeh can accept colors in a variety of different ways:
* any of the [140 named HTML/CSS colors](https://www.w3schools.com/colors/colors_names.asp), e.g ``'green'``, ``'indigo'``
* an RGB(A) hex value, e.g., ``'#FF0000'``, ``'#44444444'``
* a 3-tuple of integers *(r,g,b)* between 0 and 255
* a 4-tuple of *(r,g,b,a)* where *r*, *g*, *b* are integers between 0 and 255 and *a* is a floating point value between 0 and 1
## Properties
Regardless of how a Bokeh plot is created, styling the visual aspects of the plot can always be accomplished by setting attributes on the Bokeh objects that comprise the resulting plot. Visual properties come in three kinds: line, fill, and text properties. For full information with code and examples see the [Styling Visual Properties](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html) section of the user guide.
----
### Line Properties
Set the visual appearance of lines. The most common are ``line_color``, ``line_alpha``, ``line_width`` and ``line_dash``.
### Fill Properties
Set the visual appearance of filled areas: ``fill_color`` and ``fill_alpha``.
### Text Properties
Set the visual appearance of lines of text. The most common are ``text_font``, ``text_font_size``, ``text_color``, and ``text_alpha``.
----
Sometimes a prefix is used with property names, e.g. to distinguish between different line properties on the same object, or to give a more meaningful name. For example, to set the line width of the plot outline, you would say ``myplot.outline_line_width = 2``.
# Plots
Many top-level attributes of plots (outline, border, etc.) can be configured. See the [Plots](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#plots) section of the styling guide for full information.
Here is an example that tweaks the plot outline:
```
# create a new plot with a title
p = figure(plot_width=400, plot_height=400)
p.outline_line_width = 7
p.outline_line_alpha = 0.3
p.outline_line_color = "navy"
p.circle([1,2,3,4,5], [2,5,8,2,7], size=10)
show(p)
# EXERCISE Create a plot of your own and customize several plot-level properties
```
# Glyphs
It's also possible to style the visual properties of glyphs (see the [Glyphs](https://docs.bokeh.org/en/latest/docs/user_guide/styling.html#glyphs) section of the styling guide for more information). When using `bokeh.plotting` this is often done when calling the glyph methods:
```python
p.circle(line_color="red", fill_alpha=0.2, ...)
```
But it is also possible to set these properties directly on glyph objects. Glyph objects are found on `GlyphRenderer` objects, which are returned by the `Plot.add_glyph` and `bokeh.plotting` glyph methods like `circle`, `rect`, etc. Let's look at an example:
```
p = figure(plot_width=400, plot_height=400)
# keep a reference to the returned GlyphRenderer
r = p.circle([1,2,3,4,5], [2,5,8,2,7])
r.glyph.size = 50
r.glyph.fill_alpha = 0.2
r.glyph.line_color = "firebrick"
r.glyph.line_dash = [5, 1]
r.glyph.line_width = 2
show(p)
```
### Selection and non-selection visuals
You can also control how glyphs look when there are selections involved. The set of "selected" points is displayed according to the optional `.selection_glyph` property of a `GlyphRenderer`:
```python
r.selection_glyph = Circle(fill_alpha=1, fill_color="firebrick", line_color=None)
```
When there is a non-empty selection, the set of "unselected" points is displayed according to the optional `.nonselection_glyph` property of a `GlyphRenderer`:
```python
r.nonselection_glyph = Circle(fill_alpha=0.2, fill_color="grey", line_color=None)
```
When using the `bokeh.plotting` interface, it is easier to pass these visual properties to the glyph methods as shown below. The glyph method will create the selection or nonselection glyphs and attach them to the renderer for you.
```
p = figure(plot_width=400, plot_height=400, tools="tap", title="Select a circle")
renderer = p.circle([1, 2, 3, 4, 5], [2, 5, 8, 2, 7], size=50,
# set visual properties for selected glyphs
selection_color="firebrick",
# set visual properties for non-selected glyphs
nonselection_fill_alpha=0.2,
nonselection_fill_color="grey",
nonselection_line_color="firebrick",
nonselection_line_alpha=1.0)
show(p)
```
It is also possible to specify the visual appearance of glyphs when they are "inspected", e.g. by a hover tool. This is accomplished by setting an optional `hover_glyph` on the glyph renderer:
```python
r.hover_glyph = Circle(fill_alpha=1, fill_color="firebrick", line_color=None)
```
Or if using `bokeh.plotting` glyph methods, by passing `hover_fill_alpha`, etc. to the glyph method. Lets look at an example that works together with a `HoverTool` configured for "hline" hit-testing.
```
from bokeh.models.tools import HoverTool
from bokeh.sampledata.glucose import data
subset = data.loc['2010-10-06']
x, y = subset.index.to_series(), subset['glucose']
# Basic plot setup
p = figure(width=600, height=300, x_axis_type="datetime", title='Hover over points')
p.line(x, y, line_dash="4 4", line_width=1, color='gray')
cr = p.circle(x, y, size=20,
fill_color="grey", hover_fill_color="firebrick",
fill_alpha=0.05, hover_alpha=0.3,
line_color=None, hover_line_color="white")
p.add_tools(HoverTool(tooltips=None, renderers=[cr], mode='hline'))
show(p)
# EXERCISE: experiment with standard, selected, hover glyph visual properties
```
# Axes
Next we will take a look at [stlying of Axes](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#axes).
To style axes, you first must get ahold of `Axis` objects. The simplest way is to use some convenience methods on `Plot`: `axis`, `xaxis`, and `yaxis`. These methods return lists of axis objects:
```
>>> p.xaxis
[<bokeh.models.axes.LinearAxis at 0x106fa2390>]
```
However, you can set properties on all the elements of the list as if it was a single object:
```
p.xaxis.axis_label = "Temperature"
p.axis.major_label_text_color = "orange"
```
These are referred to as "splattable" lists, and tab completion works on them as well.
```
# EXERCISE Try out tab completion. Type p.xaxis.<press tab key> to see a list of attributes that can be set.
```
## Axis properties
Axes objects have many configurable properties that afford control over most visual aspects of an axis. These can be grouped by function according to prefix:
* **axis** [line properties](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#line-properties) e.g `axis_line_width`
* **axis_label** [text properties](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#text-properties) e.g. `axis_label_text_color`, as well as ``axis_label_standoff``
* **major_label** [text properties](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#text-properties) e.g. `major_label_text_font_size`, as well as ``major_label_orientation``
* **major_tick** [line_properties](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#line-properties) e.g. `major_tick_line_dash`, as well as ``major_tick_in`` and ``major_tick_out``
* **minor_tick** [line properties](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#line-properties) e.g. `minor_tick_line_width`, as well as ``minor_tick_in`` and ``minor_tick_out``
As a simple first case, let's change the orientation of the major tick labels on both axes of a plot:
```
from math import pi
p = figure(plot_width=400, plot_height=400)
p.x([1,2,3,4,5], [2,5,8,2,7], size=10, line_width=2)
p.xaxis.major_label_orientation = pi/4
p.yaxis.major_label_orientation = "vertical"
show(p)
```
The next example shows customizations on several of the different Axis properties at once:
```
p = figure(plot_width=400, plot_height=400)
p.asterisk([1,2,3,4,5], [2,5,8,2,7], size=12, color="olive")
# change just some things about the x-axes
p.xaxis.axis_label = "Temp"
p.xaxis.axis_line_width = 3
p.xaxis.axis_line_color = "red"
# change just some things about the y-axes
p.yaxis.axis_label = "Pressure"
p.yaxis.major_label_text_color = "orange"
p.yaxis.major_label_orientation = "vertical"
# change things on all axes
p.axis.minor_tick_in = -3
p.axis.minor_tick_out = 6
show(p)
# EXERCISE Create a plot of your own and customize several axis properties
```
## Configuring tick labels
All Bokeh axes have a `formatter` property, whose value is a `TickFormatter` object that Bokeh uses to format the ticks displayed by that axis. Bokeh will configure default tick formatters for numeric, datetime, or categotical axes. But often we would like to customize the appearance of tick labels. This can be accomplished by changing properties on the default formatter that Bokeh chooses, or by replacing the formatter with a new type entirely.
Let's first look at changing the properties of a default formatter. The default datetime formatter is configured to show *month/day* when the axis is on the scale of days. If would like the also always show the year, we can change the `days` property to a format that includes the year, as done below.
```
from math import pi
from bokeh.sampledata.glucose import data
week = data.loc['2010-10-01':'2010-10-08']
p = figure(x_axis_type="datetime", title="Glocose Range", plot_height=350, plot_width=800)
p.xaxis.formatter.days = '%m/%d/%Y'
p.xaxis.major_label_orientation = pi/3
p.line(week.index, week.glucose)
show(p)
```
See the reference guide entry for [DatetimeTickFormatter](https://bokeh.pydata.org/en/latest/docs/reference/models/formatters.html#bokeh.models.formatters.DatetimeTickFormatter) to see other properties that can be updated.
In addition to the tick formatters that Bokeh will use by default, there are others such as the [`NumeralTickFormatter`](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#numeraltickformatter) that we can configure explicitly. The example below shows how to set a formatter on each axis.
```
from bokeh.models import NumeralTickFormatter
p = figure(plot_height=300, plot_width=800)
p.circle([1,2,3,4,5], [2,5,8,2,7], size=10)
p.xaxis.formatter = NumeralTickFormatter(format="0.0%")
p.yaxis.formatter = NumeralTickFormatter(format="$0.00")
show(p)
```
Try experimenting with the format argument and re-execute the cell above.
There are many other possibilities for controlling tick formatting, including the possibility of supplying a JavaScript snippet to perform arbitrary formatting in the browser. See the [Tick Label Formats](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#tick-label-formats) for more details.
It is also possible to customize *where* ticks will be drawn. See the [Tick Locations](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#tick-locations) section of the User's Guide for more information.
# Grids
It is also possible to control the [styling of Grids](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#grids)
Grids properties in Bokeh have two possible prefixes:
* **grid** properties (which are [line properties](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#line-properties)) control the "grid lines"
* **band** properties (which are [fill properties](https://bokeh.pydata.org/en/latest/docs/user_guide/styling.html#fill-properties)) control shaded bands between grid lines
In this first example we turn off the vertical grid lines (by setting the line color to None) and set the horizontal grid to be light and dashed.
```
p = figure(plot_width=400, plot_height=400)
p.circle([1,2,3,4,5], [2,5,8,2,7], size=10)
# change just some things about the x-grid
p.xgrid.grid_line_color = None
# change just some things about the y-grid
p.ygrid.grid_line_alpha = 0.5
p.ygrid.grid_line_dash = [6, 4]
show(p)
```
The next example shows how the "band" properties of a plot can be specified
```
p = figure(plot_width=400, plot_height=400)
p.circle([1,2,3,4,5], [2,5,8,2,7], size=10)
# change just some things about the x-grid
p.xgrid.grid_line_color = None
# change just some things about the y-grid
p.ygrid.band_fill_alpha = 0.1
p.ygrid.band_fill_color = "navy"
show(p)
# EXERCISE Create a plot of your own and customize several grid properties
```
# Next Section
Click on this link to go to the next notebook: [03 - Data Sources and Transformations](03%20-%20Data%20Sources%20and%20Transformations.ipynb).
To go back to the overview, click [here](00%20-%20Introduction%20and%20Setup.ipynb).
|
github_jupyter
|
from bokeh.io import output_notebook, show
from bokeh.plotting import figure
output_notebook()
import bokeh.sampledata
bokeh.sampledata.download()
# create a new plot with a title
p = figure(plot_width=400, plot_height=400)
p.outline_line_width = 7
p.outline_line_alpha = 0.3
p.outline_line_color = "navy"
p.circle([1,2,3,4,5], [2,5,8,2,7], size=10)
show(p)
# EXERCISE Create a plot of your own and customize several plot-level properties
p.circle(line_color="red", fill_alpha=0.2, ...)
p = figure(plot_width=400, plot_height=400)
# keep a reference to the returned GlyphRenderer
r = p.circle([1,2,3,4,5], [2,5,8,2,7])
r.glyph.size = 50
r.glyph.fill_alpha = 0.2
r.glyph.line_color = "firebrick"
r.glyph.line_dash = [5, 1]
r.glyph.line_width = 2
show(p)
r.selection_glyph = Circle(fill_alpha=1, fill_color="firebrick", line_color=None)
r.nonselection_glyph = Circle(fill_alpha=0.2, fill_color="grey", line_color=None)
p = figure(plot_width=400, plot_height=400, tools="tap", title="Select a circle")
renderer = p.circle([1, 2, 3, 4, 5], [2, 5, 8, 2, 7], size=50,
# set visual properties for selected glyphs
selection_color="firebrick",
# set visual properties for non-selected glyphs
nonselection_fill_alpha=0.2,
nonselection_fill_color="grey",
nonselection_line_color="firebrick",
nonselection_line_alpha=1.0)
show(p)
r.hover_glyph = Circle(fill_alpha=1, fill_color="firebrick", line_color=None)
from bokeh.models.tools import HoverTool
from bokeh.sampledata.glucose import data
subset = data.loc['2010-10-06']
x, y = subset.index.to_series(), subset['glucose']
# Basic plot setup
p = figure(width=600, height=300, x_axis_type="datetime", title='Hover over points')
p.line(x, y, line_dash="4 4", line_width=1, color='gray')
cr = p.circle(x, y, size=20,
fill_color="grey", hover_fill_color="firebrick",
fill_alpha=0.05, hover_alpha=0.3,
line_color=None, hover_line_color="white")
p.add_tools(HoverTool(tooltips=None, renderers=[cr], mode='hline'))
show(p)
# EXERCISE: experiment with standard, selected, hover glyph visual properties
>>> p.xaxis
[<bokeh.models.axes.LinearAxis at 0x106fa2390>]
p.xaxis.axis_label = "Temperature"
p.axis.major_label_text_color = "orange"
# EXERCISE Try out tab completion. Type p.xaxis.<press tab key> to see a list of attributes that can be set.
from math import pi
p = figure(plot_width=400, plot_height=400)
p.x([1,2,3,4,5], [2,5,8,2,7], size=10, line_width=2)
p.xaxis.major_label_orientation = pi/4
p.yaxis.major_label_orientation = "vertical"
show(p)
p = figure(plot_width=400, plot_height=400)
p.asterisk([1,2,3,4,5], [2,5,8,2,7], size=12, color="olive")
# change just some things about the x-axes
p.xaxis.axis_label = "Temp"
p.xaxis.axis_line_width = 3
p.xaxis.axis_line_color = "red"
# change just some things about the y-axes
p.yaxis.axis_label = "Pressure"
p.yaxis.major_label_text_color = "orange"
p.yaxis.major_label_orientation = "vertical"
# change things on all axes
p.axis.minor_tick_in = -3
p.axis.minor_tick_out = 6
show(p)
# EXERCISE Create a plot of your own and customize several axis properties
from math import pi
from bokeh.sampledata.glucose import data
week = data.loc['2010-10-01':'2010-10-08']
p = figure(x_axis_type="datetime", title="Glocose Range", plot_height=350, plot_width=800)
p.xaxis.formatter.days = '%m/%d/%Y'
p.xaxis.major_label_orientation = pi/3
p.line(week.index, week.glucose)
show(p)
from bokeh.models import NumeralTickFormatter
p = figure(plot_height=300, plot_width=800)
p.circle([1,2,3,4,5], [2,5,8,2,7], size=10)
p.xaxis.formatter = NumeralTickFormatter(format="0.0%")
p.yaxis.formatter = NumeralTickFormatter(format="$0.00")
show(p)
p = figure(plot_width=400, plot_height=400)
p.circle([1,2,3,4,5], [2,5,8,2,7], size=10)
# change just some things about the x-grid
p.xgrid.grid_line_color = None
# change just some things about the y-grid
p.ygrid.grid_line_alpha = 0.5
p.ygrid.grid_line_dash = [6, 4]
show(p)
p = figure(plot_width=400, plot_height=400)
p.circle([1,2,3,4,5], [2,5,8,2,7], size=10)
# change just some things about the x-grid
p.xgrid.grid_line_color = None
# change just some things about the y-grid
p.ygrid.band_fill_alpha = 0.1
p.ygrid.band_fill_color = "navy"
show(p)
# EXERCISE Create a plot of your own and customize several grid properties
| 0.760651 | 0.985384 |
# Image analysis in Python with SciPy and scikit-image
<div style="border: solid 1px; background: #abcfef; font-size: 150%; padding: 1em; margin: 1em; width: 75%;">
<p>To participate, please follow the preparation instructions at</p>
<p>https://github.com/scikit-image/skimage-tutorials/</p>
<p>(click on **preparation.md**).</p>
</div>
<hr/>
TL;DR: Install Python 3.6, scikit-image, and the Jupyter notebook. Then clone this repo:
```python
git clone --depth=1 https://github.com/scikit-image/skimage-tutorials
```
<hr/>
scikit-image is a collection of image processing algorithms for the
SciPy ecosystem. It aims to have a Pythonic API (read: does what you'd expect),
is well documented, and provides researchers and practitioners with well-tested,
fundamental building blocks for rapidly constructing sophisticated image
processing pipelines.
In this tutorial, we provide an interactive overview of the library,
where participants have the opportunity to try their hand at various
image processing challenges.
Attendees are expected to have a working knowledge of NumPy, SciPy, and Matplotlib.
Across domains, modalities, and scales of exploration, images form an integral subset of scientific measurements. Despite a deep appeal to human intuition, gaining understanding of image content remains challenging, and often relies on heuristics. Even so, the wealth of knowledge contained inside of images cannot be understated, and <a href="http://scikit-image.org">scikit-image</a>, along with <a href="http://scipy.org">SciPy</a>, provides a strong foundation upon which to build algorithms and applications for exploring this domain.
# Prerequisites
Please see the [preparation instructions](https://github.com/scikit-image/skimage-tutorials/blob/master/preparation.md).
# Schedule
- 1:30–2:20: Introduction & [images as NumPy arrays](../../lectures/00_images_are_arrays.ipynb)
- 2:30–3:20: [Filters](../../lectures/1_image_filters.ipynb)
- 3:30–4:20: [Segmentation](../../lectures/4_segmentation.ipynb)
- 4:30–5:15: [StackOverflow Challenges | BYO problem](../../lectures/stackoverflow_challenges.ipynb)
- 5:15–5:30: Q&A
**Note:** Snacks are available 2:15-4:00; coffee & tea until 5.
# For later
- Check out the other [lectures](../../lectures)
- Check out a [3D segmentation workflow](../../lectures/three_dimensional_image_processing.ipynb)
- Some [real world use cases](http://bit.ly/skimage_real_world)
# After the tutorial
Stay in touch!
- Follow the project's progress [on GitHub](https://github.com/scikit-image/scikit-image).
- Ask the team questions on the [mailing list](https://mail.python.org/mailman/listinfo/scikit-image)
- [Contribute!](http://scikit-image.org/docs/dev/contribute.html)
- Read [our paper](https://peerj.com/articles/453/) (or [this other paper, for skimage in microscopy](https://ascimaging.springeropen.com/articles/10.1186/s40679-016-0031-0))
```
%run ../../check_setup.py
```
|
github_jupyter
|
git clone --depth=1 https://github.com/scikit-image/skimage-tutorials
%run ../../check_setup.py
| 0.44553 | 0.986098 |
# Use Astropy to analyze FITS images
### Based on a tutorial by Lia Corrales
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from astropy.io import fits
```
### Open a FITS file
```
fname = "/Users/labuser/Desktop/HorseHead.fits"
hdu_list = fits.open(fname)
hdu_list.info()
```
### Generally, the image information is located in the PRIMARY block. The blocks are numbered and can be accessed by indexing hdu_list.
```
image_data = hdu_list[0].data
header = hdu_list[0].header
```
### Our data is now stored as a 2-D numpy array. But how do we know the dimensions of the image? We can simply look at the shape of the array.
```
print(header)
print(type(image_data))
print(image_data.shape)
```
### At this point, we can close the FITS file because we've stored everything we wanted to a variable.
```
hdu_list.close()
```
### Shortcut: use "getdata()" to just read in the image data and close the file.
```
image_data = fits.getdata(fname)
print(type(image_data))
print(image_data.shape)
```
### Let's show the data
```
fig = plt.figure(figsize = (7,7))
plt.imshow(image_data, cmap='magma')
plt.colorbar()
```
### Let's get some basic statistics about our image:
```
print('Min:', np.min(image_data))
print('Max:', np.max(image_data))
print('Mean:', np.mean(image_data))
print('Stdev:', np.std(image_data))
```
### Plotting a histogram
To make a histogram with matplotlib.pyplot.hist(), we'll need to cast the data from a 2-D array to something one dimensional. In this case, let's use the ndarray.flatten() to return to a 1-D numpy array./
```
histogram = plt.hist(image_data.flatten(), bins='auto')
```
### Displaying the image with a logarithmis scale
What if we want to use a logarithmic color scale? To do so, we can load the LogNorm object from matplotlib.
```
from matplotlib.colors import LogNorm
plt.imshow(image_data, cmap='gray', norm=LogNorm())
#Choose the tick marks based on the histogram above
cbar = plt.colorbar(ticks=[5.e3,1.e4,2.e4])
cbar.ax.set_yticklabels(['5,000','10,000','20,000'])
```
### Stacking Images
Since the noise in an image results from a random process, we use stacking of separate images to improve the signal to noise ratio of objects we observe. Here we are going to stack 5 images of M13 taken witha 10 inch telescope.
```
#make a list of filenames
image_list = ['/Users/labuser/Desktop/M13_blue_0001.fits','/Users/labuser/Desktop/M13_blue_0002.fits','/Users/labuser/Desktop/M13_blue_0003.fits',\
'/Users/labuser/Desktop/M13_blue_0004.fits','/Users/labuser/Desktop/M13_blue_0005.fits']
print(image_list)
#make an array of images from the list of images
image_concat = [fits.getdata(image) for image in image_list]
#sum the images together
final_image = np.sum(image_concat, axis=0)
#plot a histogram of the image pixel values
image_hist = plt.hist(final_image.flatten(), bins='auto')
```
We'll use the keywords vmin and vmax to set limits on the color scaling for imshow.
```
plt.imshow(final_image, cmap='gray', vmin=2E3, vmax=3E3)
plt.colorbar()
```
### Writing a new FITS file
We can easily do this with the writeto() method.
Warning: you'll receive an error if the file you are trying to write already exists. That's why we've set clobber=True
```
outfile = 'stacked_M13_blue.fits'
hdu = fits.PrimaryHDU(final_image)
hdu.writeto(outfile, overwrite=True)
hdu_list = fits.open(outfile)
header = hdu_list[0].header
data = hdu_list[0].data
plt.imshow(data, cmap='gray', vmin=2.5e3, vmax=3e3)
print(header)
```
|
github_jupyter
|
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from astropy.io import fits
fname = "/Users/labuser/Desktop/HorseHead.fits"
hdu_list = fits.open(fname)
hdu_list.info()
image_data = hdu_list[0].data
header = hdu_list[0].header
print(header)
print(type(image_data))
print(image_data.shape)
hdu_list.close()
image_data = fits.getdata(fname)
print(type(image_data))
print(image_data.shape)
fig = plt.figure(figsize = (7,7))
plt.imshow(image_data, cmap='magma')
plt.colorbar()
print('Min:', np.min(image_data))
print('Max:', np.max(image_data))
print('Mean:', np.mean(image_data))
print('Stdev:', np.std(image_data))
histogram = plt.hist(image_data.flatten(), bins='auto')
from matplotlib.colors import LogNorm
plt.imshow(image_data, cmap='gray', norm=LogNorm())
#Choose the tick marks based on the histogram above
cbar = plt.colorbar(ticks=[5.e3,1.e4,2.e4])
cbar.ax.set_yticklabels(['5,000','10,000','20,000'])
#make a list of filenames
image_list = ['/Users/labuser/Desktop/M13_blue_0001.fits','/Users/labuser/Desktop/M13_blue_0002.fits','/Users/labuser/Desktop/M13_blue_0003.fits',\
'/Users/labuser/Desktop/M13_blue_0004.fits','/Users/labuser/Desktop/M13_blue_0005.fits']
print(image_list)
#make an array of images from the list of images
image_concat = [fits.getdata(image) for image in image_list]
#sum the images together
final_image = np.sum(image_concat, axis=0)
#plot a histogram of the image pixel values
image_hist = plt.hist(final_image.flatten(), bins='auto')
plt.imshow(final_image, cmap='gray', vmin=2E3, vmax=3E3)
plt.colorbar()
outfile = 'stacked_M13_blue.fits'
hdu = fits.PrimaryHDU(final_image)
hdu.writeto(outfile, overwrite=True)
hdu_list = fits.open(outfile)
header = hdu_list[0].header
data = hdu_list[0].data
plt.imshow(data, cmap='gray', vmin=2.5e3, vmax=3e3)
print(header)
| 0.343012 | 0.98847 |
```
import numpy as np
import time
import matplotlib.pyplot as plt
from scipy.interpolate import interp1d, interp2d
from scipy import linalg
def He3_temps(MAT, T_AMP, dat_lin):
%run He_3_after_Tremblay.ipynb
%run new_crank_nicolson_fixed.ipynb
T0 = Temp_fun(MAT, T_AMP, dat_lin, (1/thermnt))
EDT_import = EDT_calculation(MAT, T_AMP, T0)
EDT_trnsfrm = interp1d(x = EDT_import[5], y = EDT_import[3],
kind = 'linear',
fill_value = "extrapolate")
EDT_z = EDT_trnsfrm(x = dat_lin)
return EDT_z
def He3_fxn(grain_conc, EDT_z, ER, dat_lin):
He3P0 = SLHL_He3 * np.exp(-mu * dat_lin)
grain_conc = [MT_He_3(He3P0[i], grain_conc[i], x, 1, dx, EDT_z[i], nx) for i in range(len(dat_lin))]
'''
int_z | return nodes (depths) which remain after erosion;
first_fxn | 100 x 100 mesh, 2d interpolation of grain conc. by depth conc.;
first_data | interpolation function applied over linear nodes;
second_fxn | 100 x 100 mesh, 2d interpolation of grain conc from linear nodes at new depth;
grain_conc | second function applied back onto the original log nodes.
'''
first_fxn = interp2d(x = range(0,nx), y = dat_lin, z = np.stack(grain_conc), kind = 'cubic')
first_data = first_fxn(x = range(0,nx), y = dat_lin)
second_fxn = interp2d(x = range(0,nx), y = (dat_lin - ER), z = np.stack(grain_conc), kind = 'cubic')
grain_conc = second_fxn(x = range(0,nx), y = dat_lin)
return grain_conc
def He3_loop_fxn(total_time, time_ER_shift):
dat_lin = np.arange(0,max_depth[0] + 1,1)
%run He_3_after_Tremblay.ipynb
%run new_crank_nicolson_fixed.ipynb
conc3He = [[0]*nx] * len(dat_lin)
He_surf = np.empty((total_time,))
EDT_z = He3_temps(MAT, T_AMP, dat_lin)
if shift_ER == False:
ER = initial_ER
for i in range(total_time):
conc3He = He3_fxn(conc3He, EDT_z, ER, dat_lin)
He_surf[i] = np.average([np.average(conc3He[i], weights = shell_mass) for i in range(6)])
if shift_ER == True:
for i in range(time_ER_shift):
ER = initial_ER
conc3He = He3_fxn(conc3He, EDT_z, ER, dat_lin)
He_surf[i] = np.average([np.average(conc3He[i], weights = shell_mass) for i in range(6)])
for i in range(time_ER_shift, total_time):
ER = initial_ER * ER_shift_factor
conc3He = He3_fxn(conc3He, EDT_z, ER, dat_lin)
He_surf[i] = np.average([np.average(conc3He[i], weights = shell_mass) for i in range(6)])
if save_output == True:
np.savetxt('CON_He3_ER' + str(ER) + '_shift' + str(shift_ER) + '._surfmat.csv',
He_surf,
delimiter = ',')
return conc3He, He_surf
```
|
github_jupyter
|
import numpy as np
import time
import matplotlib.pyplot as plt
from scipy.interpolate import interp1d, interp2d
from scipy import linalg
def He3_temps(MAT, T_AMP, dat_lin):
%run He_3_after_Tremblay.ipynb
%run new_crank_nicolson_fixed.ipynb
T0 = Temp_fun(MAT, T_AMP, dat_lin, (1/thermnt))
EDT_import = EDT_calculation(MAT, T_AMP, T0)
EDT_trnsfrm = interp1d(x = EDT_import[5], y = EDT_import[3],
kind = 'linear',
fill_value = "extrapolate")
EDT_z = EDT_trnsfrm(x = dat_lin)
return EDT_z
def He3_fxn(grain_conc, EDT_z, ER, dat_lin):
He3P0 = SLHL_He3 * np.exp(-mu * dat_lin)
grain_conc = [MT_He_3(He3P0[i], grain_conc[i], x, 1, dx, EDT_z[i], nx) for i in range(len(dat_lin))]
'''
int_z | return nodes (depths) which remain after erosion;
first_fxn | 100 x 100 mesh, 2d interpolation of grain conc. by depth conc.;
first_data | interpolation function applied over linear nodes;
second_fxn | 100 x 100 mesh, 2d interpolation of grain conc from linear nodes at new depth;
grain_conc | second function applied back onto the original log nodes.
'''
first_fxn = interp2d(x = range(0,nx), y = dat_lin, z = np.stack(grain_conc), kind = 'cubic')
first_data = first_fxn(x = range(0,nx), y = dat_lin)
second_fxn = interp2d(x = range(0,nx), y = (dat_lin - ER), z = np.stack(grain_conc), kind = 'cubic')
grain_conc = second_fxn(x = range(0,nx), y = dat_lin)
return grain_conc
def He3_loop_fxn(total_time, time_ER_shift):
dat_lin = np.arange(0,max_depth[0] + 1,1)
%run He_3_after_Tremblay.ipynb
%run new_crank_nicolson_fixed.ipynb
conc3He = [[0]*nx] * len(dat_lin)
He_surf = np.empty((total_time,))
EDT_z = He3_temps(MAT, T_AMP, dat_lin)
if shift_ER == False:
ER = initial_ER
for i in range(total_time):
conc3He = He3_fxn(conc3He, EDT_z, ER, dat_lin)
He_surf[i] = np.average([np.average(conc3He[i], weights = shell_mass) for i in range(6)])
if shift_ER == True:
for i in range(time_ER_shift):
ER = initial_ER
conc3He = He3_fxn(conc3He, EDT_z, ER, dat_lin)
He_surf[i] = np.average([np.average(conc3He[i], weights = shell_mass) for i in range(6)])
for i in range(time_ER_shift, total_time):
ER = initial_ER * ER_shift_factor
conc3He = He3_fxn(conc3He, EDT_z, ER, dat_lin)
He_surf[i] = np.average([np.average(conc3He[i], weights = shell_mass) for i in range(6)])
if save_output == True:
np.savetxt('CON_He3_ER' + str(ER) + '_shift' + str(shift_ER) + '._surfmat.csv',
He_surf,
delimiter = ',')
return conc3He, He_surf
| 0.483892 | 0.427456 |
```
import dotenv
import pydot
import requests
import numpy as np
import pandas as pd
import ctypes
import shutil
import multiprocessing
import multiprocessing.sharedctypes as sharedctypes
import os.path
import ast
# Number of samples per 30s audio clip.
# TODO: fix dataset to be constant.
NB_AUDIO_SAMPLES = 1321967
SAMPLING_RATE = 44100
# Load the environment from the .env file.
dotenv.load_dotenv(dotenv.find_dotenv())
class FreeMusicArchive:
BASE_URL = 'https://freemusicarchive.org/api/get/'
def __init__(self, api_key):
self.api_key = api_key
def get_recent_tracks(self):
URL = 'https://freemusicarchive.org/recent.json'
r = requests.get(URL)
r.raise_for_status()
tracks = []
artists = []
date_created = []
for track in r.json()['aTracks']:
tracks.append(track['track_id'])
artists.append(track['artist_name'])
date_created.append(track['track_date_created'])
return tracks, artists, date_created
def _get_data(self, dataset, fma_id, fields=None):
url = self.BASE_URL + dataset + 's.json?'
url += dataset + '_id=' + str(fma_id) + '&api_key=' + self.api_key
# print(url)
r = requests.get(url)
r.raise_for_status()
if r.json()['errors']:
raise Exception(r.json()['errors'])
data = r.json()['dataset'][0]
r_id = data[dataset + '_id']
if r_id != str(fma_id):
raise Exception('The received id {} does not correspond to'
'the requested one {}'.format(r_id, fma_id))
if fields is None:
return data
if type(fields) is list:
ret = {}
for field in fields:
ret[field] = data[field]
return ret
else:
return data[fields]
def get_track(self, track_id, fields=None):
return self._get_data('track', track_id, fields)
def get_album(self, album_id, fields=None):
return self._get_data('album', album_id, fields)
def get_artist(self, artist_id, fields=None):
return self._get_data('artist', artist_id, fields)
def get_all(self, dataset, id_range):
index = dataset + '_id'
id_ = 2 if dataset == 'track' else 1
row = self._get_data(dataset, id_)
df = pd.DataFrame(columns=row.keys())
df.set_index(index, inplace=True)
not_found_ids = []
for id_ in id_range:
try:
row = self._get_data(dataset, id_)
except:
not_found_ids.append(id_)
continue
row.pop(index)
df = df.append(pd.Series(row, name=id_))
return df, not_found_ids
def download_track(self, track_file, path):
url = 'https://files.freemusicarchive.org/' + track_file
r = requests.get(url, stream=True)
r.raise_for_status()
with open(path, 'wb') as f:
shutil.copyfileobj(r.raw, f)
def get_track_genres(self, track_id):
genres = self.get_track(track_id, 'track_genres')
genre_ids = []
genre_titles = []
for genre in genres:
genre_ids.append(genre['genre_id'])
genre_titles.append(genre['genre_title'])
return genre_ids, genre_titles
def get_all_genres(self):
df = pd.DataFrame(columns=['genre_parent_id', 'genre_title',
'genre_handle', 'genre_color'])
df.index.rename('genre_id', inplace=True)
page = 1
while True:
url = self.BASE_URL + 'genres.json?limit=50'
url += '&page={}&api_key={}'.format(page, self.api_key)
r = requests.get(url)
for genre in r.json()['dataset']:
genre_id = int(genre.pop(df.index.name))
df.loc[genre_id] = genre
assert (r.json()['page'] == str(page))
page += 1
if page > r.json()['total_pages']:
break
return df
class Genres:
def __init__(self, genres_df):
self.df = genres_df
def create_tree(self, roots, depth=None):
if type(roots) is not list:
roots = [roots]
graph = pydot.Dot(graph_type='digraph', strict=True)
def create_node(genre_id):
title = self.df.at[genre_id, 'title']
ntracks = self.df.at[genre_id, '#tracks']
# name = self.df.at[genre_id, 'title'] + '\n' + str(genre_id)
name = '"{}\n{} / {}"'.format(title, genre_id, ntracks)
return pydot.Node(name)
def create_tree(root_id, node_p, depth):
if depth == 0:
return
children = self.df[self.df['parent'] == root_id]
for child in children.iterrows():
genre_id = child[0]
node_c = create_node(genre_id)
graph.add_edge(pydot.Edge(node_p, node_c))
create_tree(genre_id, node_c,
depth-1 if depth is not None else None)
for root in roots:
node_p = create_node(root)
graph.add_node(node_p)
create_tree(root, node_p, depth)
return graph
def find_roots(self):
roots = []
for gid, row in self.df.iterrows():
parent = row['parent']
title = row['title']
if parent == 0:
roots.append(gid)
elif parent not in self.df.index:
msg = '{} ({}) has parent {} which is missing'.format(
gid, title, parent)
raise RuntimeError(msg)
return roots
def load(filepath):
filename = os.path.basename(filepath)
if 'features' in filename:
return pd.read_csv(filepath, index_col=0, header=[0, 1, 2])
if 'echonest' in filename:
return pd.read_csv(filepath, index_col=0, header=[0, 1, 2])
if 'genres' in filename:
return pd.read_csv(filepath, index_col=0)
if 'tracks' in filename:
tracks = pd.read_csv(filepath, index_col=0, header=[0, 1])
COLUMNS = [('track', 'tags'), ('album', 'tags'), ('artist', 'tags'),
('track', 'genres'), ('track', 'genres_all')]
for column in COLUMNS:
tracks[column] = tracks[column].map(ast.literal_eval)
COLUMNS = [('track', 'date_created'), ('track', 'date_recorded'),
('album', 'date_created'), ('album', 'date_released'),
('artist', 'date_created'), ('artist', 'active_year_begin'),
('artist', 'active_year_end')]
for column in COLUMNS:
tracks[column] = pd.to_datetime(tracks[column])
SUBSETS = ('small', 'medium', 'large')
try:
tracks['set', 'subset'] = tracks['set', 'subset'].astype(
'category', categories=SUBSETS, ordered=True)
except (ValueError, TypeError):
# the categories and ordered arguments were removed in pandas 0.25
tracks['set', 'subset'] = tracks['set', 'subset'].astype(
pd.CategoricalDtype(categories=SUBSETS, ordered=True))
COLUMNS = [('track', 'genre_top'), ('track', 'license'),
('album', 'type'), ('album', 'information'),
('artist', 'bio')]
for column in COLUMNS:
tracks[column] = tracks[column].astype('category')
return tracks
def get_audio_path(audio_dir, track_id):
"""
Return the path to the mp3 given the directory where the audio is stored
and the track ID.
Examples
--------
>>> import utils
>>> AUDIO_DIR = os.environ.get('AUDIO_DIR')
>>> utils.get_audio_path(AUDIO_DIR, 2)
'../data/fma_small/000/000002.mp3'
"""
tid_str = '{:06d}'.format(track_id)
return os.path.join(audio_dir, tid_str[:3], tid_str + '.mp3')
class Loader:
def load(self, filepath):
raise NotImplementedError()
class RawAudioLoader(Loader):
def __init__(self, sampling_rate=SAMPLING_RATE):
self.sampling_rate = sampling_rate
self.shape = (NB_AUDIO_SAMPLES * sampling_rate // SAMPLING_RATE, )
def load(self, filepath):
return self._load(filepath)[:self.shape[0]]
class LibrosaLoader(RawAudioLoader):
def _load(self, filepath):
import librosa
sr = self.sampling_rate if self.sampling_rate != SAMPLING_RATE else None
# kaiser_fast is 3x faster than kaiser_best
# x, sr = librosa.load(filepath, sr=sr, res_type='kaiser_fast')
x, sr = librosa.load(filepath, sr=sr)
return x
class AudioreadLoader(RawAudioLoader):
def _load(self, filepath):
import audioread
a = audioread.audio_open(filepath)
a.read_data()
class PydubLoader(RawAudioLoader):
def _load(self, filepath):
from pydub import AudioSegment
song = AudioSegment.from_file(filepath)
song = song.set_channels(1)
x = song.get_array_of_samples()
# print(filepath) if song.channels != 2 else None
return np.array(x)
class FfmpegLoader(RawAudioLoader):
def _load(self, filepath):
"""Fastest and less CPU intensive loading method."""
import subprocess as sp
command = ['ffmpeg',
'-i', filepath,
'-f', 's16le',
'-acodec', 'pcm_s16le',
'-ac', '1'] # channels: 2 for stereo, 1 for mono
if self.sampling_rate != SAMPLING_RATE:
command.extend(['-ar', str(self.sampling_rate)])
command.append('-')
# 30s at 44.1 kHz ~= 1.3e6
proc = sp.run(command, stdout=sp.PIPE, bufsize=10**7, stderr=sp.DEVNULL, check=True)
return np.fromstring(proc.stdout, dtype="int16")
def build_sample_loader(audio_dir, Y, loader):
class SampleLoader:
def __init__(self, tids, batch_size=4):
self.lock1 = multiprocessing.Lock()
self.lock2 = multiprocessing.Lock()
self.batch_foremost = sharedctypes.RawValue(ctypes.c_int, 0)
self.batch_rearmost = sharedctypes.RawValue(ctypes.c_int, -1)
self.condition = multiprocessing.Condition(lock=self.lock2)
data = sharedctypes.RawArray(ctypes.c_int, tids.data)
self.tids = np.ctypeslib.as_array(data)
self.batch_size = batch_size
self.loader = loader
self.X = np.empty((self.batch_size, *loader.shape))
self.Y = np.empty((self.batch_size, Y.shape[1]), dtype=np.int)
def __iter__(self):
return self
def __next__(self):
with self.lock1:
if self.batch_foremost.value == 0:
np.random.shuffle(self.tids)
batch_current = self.batch_foremost.value
if self.batch_foremost.value + self.batch_size < self.tids.size:
batch_size = self.batch_size
self.batch_foremost.value += self.batch_size
else:
batch_size = self.tids.size - self.batch_foremost.value
self.batch_foremost.value = 0
# print(self.tids, self.batch_foremost.value, batch_current, self.tids[batch_current], batch_size)
# print('queue', self.tids[batch_current], batch_size)
tids = np.array(self.tids[batch_current:batch_current+batch_size])
batch_size = 0
for tid in tids:
try:
audio_path = get_audio_path(audio_dir, tid)
self.X[batch_size] = self.loader.load(audio_path)
self.Y[batch_size] = Y.loc[tid]
batch_size += 1
except Exception as e:
print("\nIgnoring " + audio_path +" (error: " + str(e) +").")
with self.lock2:
while (batch_current - self.batch_rearmost.value) % self.tids.size > self.batch_size:
# print('wait', indices[0], batch_current, self.batch_rearmost.value)
self.condition.wait()
self.condition.notify_all()
# print('yield', indices[0], batch_current, self.batch_rearmost.value)
self.batch_rearmost.value = batch_current
return self.X[:batch_size], self.Y[:batch_size]
return SampleLoader
```
|
github_jupyter
|
import dotenv
import pydot
import requests
import numpy as np
import pandas as pd
import ctypes
import shutil
import multiprocessing
import multiprocessing.sharedctypes as sharedctypes
import os.path
import ast
# Number of samples per 30s audio clip.
# TODO: fix dataset to be constant.
NB_AUDIO_SAMPLES = 1321967
SAMPLING_RATE = 44100
# Load the environment from the .env file.
dotenv.load_dotenv(dotenv.find_dotenv())
class FreeMusicArchive:
BASE_URL = 'https://freemusicarchive.org/api/get/'
def __init__(self, api_key):
self.api_key = api_key
def get_recent_tracks(self):
URL = 'https://freemusicarchive.org/recent.json'
r = requests.get(URL)
r.raise_for_status()
tracks = []
artists = []
date_created = []
for track in r.json()['aTracks']:
tracks.append(track['track_id'])
artists.append(track['artist_name'])
date_created.append(track['track_date_created'])
return tracks, artists, date_created
def _get_data(self, dataset, fma_id, fields=None):
url = self.BASE_URL + dataset + 's.json?'
url += dataset + '_id=' + str(fma_id) + '&api_key=' + self.api_key
# print(url)
r = requests.get(url)
r.raise_for_status()
if r.json()['errors']:
raise Exception(r.json()['errors'])
data = r.json()['dataset'][0]
r_id = data[dataset + '_id']
if r_id != str(fma_id):
raise Exception('The received id {} does not correspond to'
'the requested one {}'.format(r_id, fma_id))
if fields is None:
return data
if type(fields) is list:
ret = {}
for field in fields:
ret[field] = data[field]
return ret
else:
return data[fields]
def get_track(self, track_id, fields=None):
return self._get_data('track', track_id, fields)
def get_album(self, album_id, fields=None):
return self._get_data('album', album_id, fields)
def get_artist(self, artist_id, fields=None):
return self._get_data('artist', artist_id, fields)
def get_all(self, dataset, id_range):
index = dataset + '_id'
id_ = 2 if dataset == 'track' else 1
row = self._get_data(dataset, id_)
df = pd.DataFrame(columns=row.keys())
df.set_index(index, inplace=True)
not_found_ids = []
for id_ in id_range:
try:
row = self._get_data(dataset, id_)
except:
not_found_ids.append(id_)
continue
row.pop(index)
df = df.append(pd.Series(row, name=id_))
return df, not_found_ids
def download_track(self, track_file, path):
url = 'https://files.freemusicarchive.org/' + track_file
r = requests.get(url, stream=True)
r.raise_for_status()
with open(path, 'wb') as f:
shutil.copyfileobj(r.raw, f)
def get_track_genres(self, track_id):
genres = self.get_track(track_id, 'track_genres')
genre_ids = []
genre_titles = []
for genre in genres:
genre_ids.append(genre['genre_id'])
genre_titles.append(genre['genre_title'])
return genre_ids, genre_titles
def get_all_genres(self):
df = pd.DataFrame(columns=['genre_parent_id', 'genre_title',
'genre_handle', 'genre_color'])
df.index.rename('genre_id', inplace=True)
page = 1
while True:
url = self.BASE_URL + 'genres.json?limit=50'
url += '&page={}&api_key={}'.format(page, self.api_key)
r = requests.get(url)
for genre in r.json()['dataset']:
genre_id = int(genre.pop(df.index.name))
df.loc[genre_id] = genre
assert (r.json()['page'] == str(page))
page += 1
if page > r.json()['total_pages']:
break
return df
class Genres:
def __init__(self, genres_df):
self.df = genres_df
def create_tree(self, roots, depth=None):
if type(roots) is not list:
roots = [roots]
graph = pydot.Dot(graph_type='digraph', strict=True)
def create_node(genre_id):
title = self.df.at[genre_id, 'title']
ntracks = self.df.at[genre_id, '#tracks']
# name = self.df.at[genre_id, 'title'] + '\n' + str(genre_id)
name = '"{}\n{} / {}"'.format(title, genre_id, ntracks)
return pydot.Node(name)
def create_tree(root_id, node_p, depth):
if depth == 0:
return
children = self.df[self.df['parent'] == root_id]
for child in children.iterrows():
genre_id = child[0]
node_c = create_node(genre_id)
graph.add_edge(pydot.Edge(node_p, node_c))
create_tree(genre_id, node_c,
depth-1 if depth is not None else None)
for root in roots:
node_p = create_node(root)
graph.add_node(node_p)
create_tree(root, node_p, depth)
return graph
def find_roots(self):
roots = []
for gid, row in self.df.iterrows():
parent = row['parent']
title = row['title']
if parent == 0:
roots.append(gid)
elif parent not in self.df.index:
msg = '{} ({}) has parent {} which is missing'.format(
gid, title, parent)
raise RuntimeError(msg)
return roots
def load(filepath):
filename = os.path.basename(filepath)
if 'features' in filename:
return pd.read_csv(filepath, index_col=0, header=[0, 1, 2])
if 'echonest' in filename:
return pd.read_csv(filepath, index_col=0, header=[0, 1, 2])
if 'genres' in filename:
return pd.read_csv(filepath, index_col=0)
if 'tracks' in filename:
tracks = pd.read_csv(filepath, index_col=0, header=[0, 1])
COLUMNS = [('track', 'tags'), ('album', 'tags'), ('artist', 'tags'),
('track', 'genres'), ('track', 'genres_all')]
for column in COLUMNS:
tracks[column] = tracks[column].map(ast.literal_eval)
COLUMNS = [('track', 'date_created'), ('track', 'date_recorded'),
('album', 'date_created'), ('album', 'date_released'),
('artist', 'date_created'), ('artist', 'active_year_begin'),
('artist', 'active_year_end')]
for column in COLUMNS:
tracks[column] = pd.to_datetime(tracks[column])
SUBSETS = ('small', 'medium', 'large')
try:
tracks['set', 'subset'] = tracks['set', 'subset'].astype(
'category', categories=SUBSETS, ordered=True)
except (ValueError, TypeError):
# the categories and ordered arguments were removed in pandas 0.25
tracks['set', 'subset'] = tracks['set', 'subset'].astype(
pd.CategoricalDtype(categories=SUBSETS, ordered=True))
COLUMNS = [('track', 'genre_top'), ('track', 'license'),
('album', 'type'), ('album', 'information'),
('artist', 'bio')]
for column in COLUMNS:
tracks[column] = tracks[column].astype('category')
return tracks
def get_audio_path(audio_dir, track_id):
"""
Return the path to the mp3 given the directory where the audio is stored
and the track ID.
Examples
--------
>>> import utils
>>> AUDIO_DIR = os.environ.get('AUDIO_DIR')
>>> utils.get_audio_path(AUDIO_DIR, 2)
'../data/fma_small/000/000002.mp3'
"""
tid_str = '{:06d}'.format(track_id)
return os.path.join(audio_dir, tid_str[:3], tid_str + '.mp3')
class Loader:
def load(self, filepath):
raise NotImplementedError()
class RawAudioLoader(Loader):
def __init__(self, sampling_rate=SAMPLING_RATE):
self.sampling_rate = sampling_rate
self.shape = (NB_AUDIO_SAMPLES * sampling_rate // SAMPLING_RATE, )
def load(self, filepath):
return self._load(filepath)[:self.shape[0]]
class LibrosaLoader(RawAudioLoader):
def _load(self, filepath):
import librosa
sr = self.sampling_rate if self.sampling_rate != SAMPLING_RATE else None
# kaiser_fast is 3x faster than kaiser_best
# x, sr = librosa.load(filepath, sr=sr, res_type='kaiser_fast')
x, sr = librosa.load(filepath, sr=sr)
return x
class AudioreadLoader(RawAudioLoader):
def _load(self, filepath):
import audioread
a = audioread.audio_open(filepath)
a.read_data()
class PydubLoader(RawAudioLoader):
def _load(self, filepath):
from pydub import AudioSegment
song = AudioSegment.from_file(filepath)
song = song.set_channels(1)
x = song.get_array_of_samples()
# print(filepath) if song.channels != 2 else None
return np.array(x)
class FfmpegLoader(RawAudioLoader):
def _load(self, filepath):
"""Fastest and less CPU intensive loading method."""
import subprocess as sp
command = ['ffmpeg',
'-i', filepath,
'-f', 's16le',
'-acodec', 'pcm_s16le',
'-ac', '1'] # channels: 2 for stereo, 1 for mono
if self.sampling_rate != SAMPLING_RATE:
command.extend(['-ar', str(self.sampling_rate)])
command.append('-')
# 30s at 44.1 kHz ~= 1.3e6
proc = sp.run(command, stdout=sp.PIPE, bufsize=10**7, stderr=sp.DEVNULL, check=True)
return np.fromstring(proc.stdout, dtype="int16")
def build_sample_loader(audio_dir, Y, loader):
class SampleLoader:
def __init__(self, tids, batch_size=4):
self.lock1 = multiprocessing.Lock()
self.lock2 = multiprocessing.Lock()
self.batch_foremost = sharedctypes.RawValue(ctypes.c_int, 0)
self.batch_rearmost = sharedctypes.RawValue(ctypes.c_int, -1)
self.condition = multiprocessing.Condition(lock=self.lock2)
data = sharedctypes.RawArray(ctypes.c_int, tids.data)
self.tids = np.ctypeslib.as_array(data)
self.batch_size = batch_size
self.loader = loader
self.X = np.empty((self.batch_size, *loader.shape))
self.Y = np.empty((self.batch_size, Y.shape[1]), dtype=np.int)
def __iter__(self):
return self
def __next__(self):
with self.lock1:
if self.batch_foremost.value == 0:
np.random.shuffle(self.tids)
batch_current = self.batch_foremost.value
if self.batch_foremost.value + self.batch_size < self.tids.size:
batch_size = self.batch_size
self.batch_foremost.value += self.batch_size
else:
batch_size = self.tids.size - self.batch_foremost.value
self.batch_foremost.value = 0
# print(self.tids, self.batch_foremost.value, batch_current, self.tids[batch_current], batch_size)
# print('queue', self.tids[batch_current], batch_size)
tids = np.array(self.tids[batch_current:batch_current+batch_size])
batch_size = 0
for tid in tids:
try:
audio_path = get_audio_path(audio_dir, tid)
self.X[batch_size] = self.loader.load(audio_path)
self.Y[batch_size] = Y.loc[tid]
batch_size += 1
except Exception as e:
print("\nIgnoring " + audio_path +" (error: " + str(e) +").")
with self.lock2:
while (batch_current - self.batch_rearmost.value) % self.tids.size > self.batch_size:
# print('wait', indices[0], batch_current, self.batch_rearmost.value)
self.condition.wait()
self.condition.notify_all()
# print('yield', indices[0], batch_current, self.batch_rearmost.value)
self.batch_rearmost.value = batch_current
return self.X[:batch_size], self.Y[:batch_size]
return SampleLoader
| 0.311951 | 0.213152 |
# 3. Importing Data from Databases
**Combine pandas with the powers of SQL to find out just how many problems New Yorkers have with their housing. This chapter features introductory SQL topics like WHERE clauses, aggregate functions, and basic joins.**
## Introduction to databases
In this chapter, you'll learn to build pipelines to relational databases, which underpin the information systems of many organizations.
### Relational Databases
Relational databases organize data about entities in tables, with rows representing instances of entities and columns of attributes. This probably sounds familiar -- data frames, flat files, and many Excel sheets arrange data similarly. Relational databases differ in that tables can be linked, or related, via unique record identifiers, or keys. Databases handle more data and support more simultaneous users than spreadsheets or flat files. They also offer more data quality controls, like enforcing column data types. And we interface with databases via a specific language: Structured Query Language, or SQL.
### Common Relational Databases
Common relational databases include Microsoft SQL Server, Oracle, PostgreSQL, and SQLite, which this course uses. Unlike the others, SQLite databases are stored as regular, self-contained computer files, just as CSVs and Excel files are, making them great for sharing data.
### Connecting to Databases
Reading data from a database is a two-step process. We first make a way to connect to a database then query it with SQL and pandas.
### Creating a Database Engine
To do this, we'll use the `SQLAlchemy` library, which has tools to work with many major relational databases. Specifically, we'll use `SQLAlchemy`'s `create_engine()` function. Create engine takes a string URL to a database and makes an engine object that manages database connections. URLs follow a pattern that varies slightly depending on the database. For SQLite, the pattern is `sqlite:///filename.db`.
### Querying Databases
Once we create the database engine, we can pull data with pandas' read SQL function.
```python
pd.read_sql(query, engine)
```
Read SQL needs two arguments. The first is a string of either a SQL query, or, to load a whole table, just the table name. The second argument is a way to connect to the database. We'll supply the engine we made here.
### SQL Review: SELECT
Let's take a minute to review SQL select statements, which are used to query databases. The basic syntax is `SELECT column_names FROM table_name`. This will get all rows for the specified columns. To get all rows and all columns, use `SELECT * FROM tabel_name` A note about code style: keywords like "select" and "from" are not case sensitive, but it's conventional to type them in **all capital letters**. It's also best practice to mark the end of a SQL statement with a semicolon(;).
### Getting Data from a Database
Now let's put all this together to fetch weather data from a SQLite database containing information about New York City. We import pandas as pd, plus the create engine function from SQLAlchemy. Then we make the engine object, passing the database URL string, sqlite colon slash slash slash data dot db, to the create engine function.
```
# Load pandas and sqlalchemy's create_engine
import pandas as pd
from sqlalchemy import create_engine
# Create database engine to manage connections
engine = create_engine('sqlite:///data.db')
# Load entire weather table by table name
weather = pd.read_sql('weather', engine)
```
Since we want everything in the weather table, we can make the first argument to read SQL the table name as a string, or we can use the SQL statement "select star from weather" as the first argument. We also supply the engine object as the second argument. Then we can check out our new data frame.
```
# Load entire weather table with SQL
weather = pd.read_sql('SELECT * FROM weather', engine)
print(weather.head())
```
## Connect to a database
In order to get data from a database with `pandas`, you first need to be able to connect to one. In this exercise, you'll practice creating a database engine to manage connections to a database, `data.db`. To do this, you'll use `sqlalchemy`'s `create_engine()` function.
`create_engine()` needs a string URL to the database. For SQLite databases, that string consists of `"sqlite:///"`, then the database file name.
- Use `create_engine()` to make a database engine for `data.db`.
- Run the last line of code to show the names of the tables in the database.
```
# Import sqlalchemy's create_engine() function
from sqlalchemy import create_engine
# Create the database engine
engine = create_engine('sqlite:///data.db')
# View the tables in the database
print(engine.table_names())
```
*`sqlalchemy` is a powerful library that can be used with `pandas` to both query databases for analysis and write results back to database tables.*
## Load entire tables
In the last exercise, you saw that `data.db` has two tables. `weather` has historical weather data for New York City. `hpd311calls` is a subset of call records made to the city's 311 help line about housing issues.
In this exercise, you'll use the `read_sql()` function in `pandas` to load both tables. `read_sql()` accepts a string of either a SQL query to run, or a table to load. It also needs a way to connect to the database, like the `engine` in the provided code.
- Use `read_sql()` to load the `hpd311calls` table by name, without any SQL.
```
# Create the database engine
engine = create_engine('sqlite:///data.db')
# Load hpd311calls without any SQL
hpd_calls = pd.read_sql('hpd311calls', engine)
# View the first few rows of data
print(hpd_calls.head())
```
- Use `read_sql()` and a `SELECT * ...` SQL query to load the entire `weather` table.
```
# Create the database engine
engine = create_engine("sqlite:///data.db")
# Create a SQL query to load the entire weather table
query = """
SELECT *
FROM weather;
"""
# Load weather with the SQL query
weather = pd.read_sql(query, engine)
# View the first few rows of data
print(weather.head())
```
*While it's convenient to load tables by name alone, using SQL queries makes it possible to fine-tune imports at the data acquisition phase of an analysis project.*
---
## Refining imports with SQL queries
Now that you know how to connect to a database and query it with pandas, let's focus on refining imports with different SQL queries.
### SELECTing Columns
pandas' read SQL function has fewer arguments than read Excel or read CSV, but SQL lets you customize data imports in even more ways. The last lesson mentioned one such way. Specifying columns in a select statement lets you load only variables you are interested in studying, akin to usecols in read Excel and read CSV.
The statement "select date, t average from weather",
```sql
SELECT date, tavg
FROM weather;
```
for example, gets only the date and average temperatures in the weather table.
### WHERE Clauses
The other common way to selectively get data with SQL is by using a where clause to filter rows. The syntax looks like "select columns from table where" a given condition or conditions are met.
### Filtering by Numbers
To import records based on numeric values, we can use standard operators to compare numbers: equals, greater than and greater than or equal to, less than and less than or equal to, and not equal to. Note that SQL's equality and inequality operators differ from Python's. The weather data contains Fahrenheit temperatures, so the SQL query to get all days with a high temperature above freezing would be "select star from weather where t max is greater than 32."
```sql
SELECT *
FROM weather
WHERE tmax > 32;
```
### Filtering Text
Where clauses can also filter text. To match a string exactly, use the equal sign followed by the text to match in single quotes. Note that string matching is case sensitive. To get call records about incidents in the borough of Brooklyn, for example, the query would be "select star from hpd311calls where borough equals Brooklyn", with Brooklyn in single quotes.
```sql
/* Get records about incidents in Brooklyn */
SELECT *
FROM hpd311calls
WHERE borough = 'BROOKYN';
```
### SQL and pandas
Let's combine SQL and Python to get Brooklyn calls. As before, we import pandas and SQLalchemy's create engine function. We pass create engine the URL to data dot db and assign the result to engine. Since our SQL queries are getting more complicated, we'll write out the query first and assign it to the variable "query." Wrapping the string in triple quotes lets us split it between multiple lines so it's easier to read. Then we pass the query and engine to read SQL. When we check the unique borough values in the resulting data frame, we see there are only Brooklyn calls.
```
# Create database engine
engine = create_engine('sqlite:///data.db')
# Write query to get records from Brooklyn
query = """
SELECT *
FROM hpd311calls
WHERE borough = 'BROOKLYN';
"""
# Query the database
brooklyn_calls = pd.read_sql(query, engine)
print( brooklyn_calls.borough.unique())
```
### Combining Conditions: AND
We can even combine conditions with SQL's and and or operators. And returns only records where all conditions are true. For example, if we create the query "select star from hpd311calls where borough equals Bronx and complaint type equals plumbing", then pass the query and engine to read SQL, we get the 2,016 call records about plumbing from the Bronx.
```
# Write query to get records about plumbing in the Bronx
and_query = """
SELECT *
FROM hpd311calls
WHERE borough = 'BRONX'
AND complaint_type = 'PLUMBING';
"""
# Get calls about plumbing issues in the Bronx
bx_plumbing_calls = pd.read_sql(and_query, engine)
# Check record count
print(bx_plumbing_calls.shape)
```
### Combining Conditions: OR
The or operator returns records with at least one met condition. If we change our query to get records that are about plumbing or water leaks, we get 10,684 records that meet one or both conditions.
```
# Write query to get records about water leaks or plumbing
or_query = """
SELECT *
FROM hpd311calls
WHERE complaint_type = 'WATER LEAK'
OR complaint_type = 'PLUMBING';
"""
# Get calls about plumbing issues in the Bronx
leaks_or_plumbing = pd.read_sql(or_query, engine)
# Check record count
print(leaks_or_plumbing.shape)
```
## Selecting columns with SQL
Datasets can contain columns that are not required for an analysis, like the `weather` table in `data.db` does. Some, such as elevation, are redundant, since all observations occurred at the same place, while others contain variables we are not interested in. After making a database engine, you'll write a query to `SELECT `only the date and temperature columns, and pass both to `read_sql()` to make a data frame of high and low temperature readings.
**Note**: The SQL checker is quite picky about column positions and expects fields to be selected in the specified order.
- Create a database engine for `data.db`.
- Write a SQL query that `SELECT`s the `date`, `tmax`, and `tmin` columns from the `weather` table.
- Make a data frame by passing the query and engine to `read_sql()` and assign the resulting data frame to `temperatures`.
```
# Create database engine for data.db
engine = create_engine('sqlite:///data.db')
# Write query to get date, tmax, and tmin from weather
query = """
SELECT date,
tmax,
tmin
FROM weather;
"""
# Make a data frame by passing query and engine to read_sql()
temperatures = pd.read_sql(query, engine)
# View the resulting data frame
print(temperatures)
```
*Selecting columns is useful when you only want a few columns from a table. If you want most of the columns, it may be easier to load them all and then use `pandas` to drop unwanted columns.*
## Selecting rows
SQL `WHERE` clauses return records whose values meet the given criteria. Passing such a query to `read_sql()` results in a data frame loaded with only records we are interested in, so there is less filtering to do later on.
The `hpd311calls` table in `data.db` has data on calls about various housing issues, from maintenance problems to information requests. In this exercise, you'll use SQL to focus on calls about safety.
- Create a query that selects all columns of records in `hpd311calls` that have `'SAFETY'` as their `complaint_type`.
- Use `read_sql()` to query the database and assign the result to the variable `safety_calls`.
- Run the last section of code to create a graph of safety call counts in each borough.
```
import matplotlib.pyplot as plt
# Create query to get hpd311calls records about safety
query = """
SELECT *
FROM hpd311calls
WHERE complaint_type = 'SAFETY';
"""
# Query the database and assign result to safety_calls
safety_calls = pd.read_sql(query, engine)
# Graph the number of safety calls by borough
call_counts = safety_calls.groupby('borough').unique_key.count()
call_counts.plot.barh()
plt.show()
```
*Filtering data before importing can help you focus on specific records, but it can also be used as a data cleaning technique to exclude records with known errors or missing values.*
## Filtering on multiple conditions
So far, you've selectively imported records that met a single condition, but it's also common to filter datasets on multiple criteria. In this exercise, you'll do just that.
The `weather` table contains daily high and low temperatures and precipitation amounts for New York City. Let's focus on inclement weather, where there was either an inch or more of snow or the high was at or below freezing (32° Fahrenheit). To do this, you'll need to build a query that uses the `OR` operator to look at values in both columns.
- Create a query that selects records in `weather` where `tmax` is less than or equal to 32 degrees `OR` `snow` is greater than or equal to 1 inch.
- Use `read_sql()` to query the database and assign the result to the variable `wintry_days`.
- View summary statistics with the `describe()` method to make sure all records in the data frame meet the given criteria.
```
# Create query for records with max temps <= 32 or snow >= 1
query = """
SELECT *
FROM weather
WHERE tmax <= 32
OR snow >= 1;
"""
# Query database and assign result to wintry_days
wintry_days = pd.read_sql(query, engine)
# View summary stats about the temperatures
print(wintry_days.describe())
```
*`SELECT` statements can use multiple `AND` and `OR` operators to filter data. Like arithmetic, you can control the order of operations with parentheses.*
---
## More complex SQL queries
The SQL we've used so far mimics the functionality of `pandas` keyword arguments like `usecols` and `skiprows`. In this lesson, we'll take advantage of SQL's features to wrangle data in ways that cannot be done at the import stage using pandas alone.
### Getting DISTINCT Values
In an analysis, you might need unique values in a column, or unique combinations of values across several columns. Examples include getting unique values to check data quality and creating crosswalks between values to combine datasets. Alternatively, data might have duplicate records that should be excluded. Pandas has tools to do this wrangling in data frames, but it can be done with SQL during import using select distinct. The syntax is "select distinct column names from table". To remove duplicate records, select distinct on all columns with the query "select distinct star from table". For another example, if we wanted to map buildings with housing complaints, we could use the query "select distinct incident address, borough from hpd311calls" to get unique street addresses and boroughs.
```sql
/* Get unique street addresses and boroughs */
SELECT DISTINCT incident_address, borough
FROM hpd311calls;
```
### Aggregate Functions
Other times, you might not be interested in the details of individual records, particularly when visualizing data. You can query the database directly for descriptive statistics with aggregate functions like sum, average, max, min, and count.
The first four functions all take a single column name in parentheses. For example, the query "select average t max from weather", with tmax in parentheses, returns the average daily high temperature. Count is a little different. While it can accept a single column name, you can do things like get the number of rows that fit a query with count star, or even get the number of unique values in a column with count distinct and the column name.
### GROUP BY
Aggregate functions return a single number on their own. More likely, you want data summarized by categories, such as average high temperatures by month or counts of plumbing complaints by borough. In that case, add a group by clause after the select statement and where clauses. Remember to select the column you're grouping by as well as the aggregate function -- otherwise you'll end up with unlabeled summary figures for each group. The query to get counts of plumbing complaints by borough, for example, would be "select borough, count star from hpd311calls where complaint type equals plumbing, group by borough"
```sql
/* Get counts of plumbing calls by borough */
SELECT borough,
COUNT(*)
FROM hpd311calls
WHERE complaint_type = 'PLUMBING'
GROUP BY borough;
```
### Counting by Groups
Let's see what the results of that query look like in a data frame. With the necessary libraries imported, we create the engine, write out the query, then pass the query and engine to pandas' read SQL function.
When we check the results, we see we have a neat summary data frame ready for plotting.
```
# Create database engine
engine = create_engine('sqlite:///data.db')
# Write query to get plumbing call counts by borough
query = """
SELECT borough, COUNT(*)
FROM hpd311calls
WHERE complaint_type = 'PLUMBING'
GROUP BY borough;
"""
# Query database and create data frame
plumbing_call_counts = pd.read_sql(query, engine)
print(plumbing_call_counts)
```
## Getting distinct values
Sometimes an analysis doesn't need every record, but rather unique values in one or more columns. Duplicate values can be removed after loading data into a data frame, but it can also be done at import with SQL's `DISTINCT` keyword.
Since `hpd311calls` contains data about housing issues, we would expect most records to have a borough listed. Let's test this assumption by querying unique `complaint_type`/`borough` combinations.
**Note**: The SQL checker is quite picky about column positions and expects fields to be selected in the specified order.
- Create a query that gets `DISTINCT` values for `borough` and `complaint_type` (in that order) from `hpd311calls`.
- Use `read_sql()` to load the results of the query to a data frame, `issues_and_boros`.
- Print the data frame to check if the assumption that all issues besides literature requests appear with boroughs listed.
```
# Create query for unique combinations of borough and complaint_type
query = """
SELECT DISTINCT borough, complaint_type
FROM hpd311calls;
"""
# Load results of query to a data frame
issues_and_boros = pd.read_sql(query, engine)
# Check assumption about issues and boroughs
print(issues_and_boros)
```
*Looks like the only issues with no borough are requests for information, rather than housing complaints, which is okay. `SELECT DISTINCT` queries can be an easy way to find data quality issues like misspelled values or combinations of values that violate assumptions.*
## Counting in groups
In previous exercises, you pulled data from tables, then summarized the resulting data frames in `pandas` to create graphs. By using `COUNT` and `GROUP BY` in a SQL query, we can pull those summary figures from the database directly.
The `hpd311calls` table has a column, `complaint_type`, that categorizes call records by issue, such as heating or plumbing. In order to graph call volumes by issue, you'll write a SQL query that `COUNT`s records by complaint type.
- Create a SQL query that gets the `complaint_type` column and counts of all records from `hpd311calls`, grouped by `complaint_type`.
- Create a data frame with `read_sql()` of call counts by issue, `calls_by_issue`.
- Run the last section of code to graph the number of calls for each housing issue.
```
# Create query to get call counts by complaint_type
query = """
SELECT complaint_type,
COUNT(*)
FROM hpd311calls
GROUP BY complaint_type;
"""
# Create data frame of call counts by issue
calls_by_issue = pd.read_sql(query, engine)
# Graph the number of calls for each housing issue
calls_by_issue.plot.barh(x='complaint_type')
plt.show()
```
*Calls about heat and hot water issues vastly outnumber calls about all other issues. The data subset in the table comes from the winter, so this makes sense.*
## Working with aggregate functions
If a table contains data with higher granularity than is needed for an analysis, it can make sense to summarize the data with SQL aggregate functions before importing it. For example, if you have data of flood event counts by month but precipitation data by day, you may decide to `SUM` precipitation by month.
The `weather` table contains daily readings for four months. In this exercise, you'll practice summarizing weather by month with the `MAX`, `MIN`, and `SUM` functions.
- Create a query to pass to `read_sql()` that will get months and the `MAX` value of `tmax` by `month` from `weather`.
```
# Create a query to get month and max tmax by month
query = """
SELECT month,
MAX(tmax)
FROM weather
GROUP BY month;
"""
# Get data frame of monthly weather stats
weather_by_month = pd.read_sql(query, engine)
# View weather stats by month
print(weather_by_month)
```
- Modify the query to also get the `MIN` `tmin` value for each `month`.
```
# Create a query to get month, max tmax, and min tmin by month
query = """
SELECT month,
MAX(tmax),
MIN(tmin)
FROM weather
GROUP BY month;
"""
# Get data frame of monthly weather stats
weather_by_month = pd.read_sql(query, engine)
# View weather stats by month
print(weather_by_month)
```
- Modify the query to also get the total precipitation (`prcp`) for each `month`.
```
# Create query to get temperature and precipitation by month
query = """
SELECT month,
MAX(tmax),
MIN(tmin),
SUM(prcp)
FROM weather
GROUP BY month;
"""
# Get data frame of monthly weather stats
weather_by_month = pd.read_sql(query, engine)
# View weather stats by month
print(weather_by_month)
```
*Aggregate functions can be a useful way to summarize large datasets. Different database management systems even have SQL functions for statistics like standard deviation and percentiles, though these are non-standard and vendor-specific.*
---
## Loading multiple tables with joins
One feature of relational databases is that tables can be linked to one another via unique record identifiers, or keys. This lets users combine tables into custom datasets with SQL joins, which we'll explore in this lesson.
### Keys
As mentioned, records typically have keys that uniquely identify them. At their simplest, keys can be automatically assigned row numbers, like in the 311 call data, but they can also carry meaning, like university course numbers. When records include other tables' keys, you can bring in, or join, data from the referenced table.
For example, given a column of instructor ID numbers and a professor table, we can then join in professors' names. When building data pipelines, this means you're not limited to working with a single table's columns.
### Joining Tables
Let's join weather data to 311 call records to study if certain problems are exacerbated by weather conditions. Both tables contain date columns as text, with unique dates in the weather table, so they'll be the join key.
First, we select star from hpd311calls to get all columns there. Star will also get all columns from weather once we join it.
Then we join weather on hpd311calls dot created date equals weather dot date, which are the key columns. We use dot notation to specify the table and column when querying multiple tables. Two things to note here: join, by default, only returns records with key values that appear in both tables. And key columns must be the same data type or nothing will match.
```sql
SELECT *
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date;
```
### Joining and Filtering
We can incorporate a where clause after the join to refine the dataset. Here, we filter the data to focus on heat and hot water calls, which probably spike in cold weather.
```sql
/* Get only heat/hot water calls and join in weather data */
SELECT *
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date
WHERE hpd311calls.complaint_type = 'HEAT/HOT WATER';
```
It's even possible to summarize data and then join additional columns. Imagine we wanted to compare call counts by borough against census data about population and housing in another table, boro census. Let's build the query in parts. We first get call counts by borough by selecting `hpd311calls.borough` and count star from hpd311calls, grouping by hpd311calls dot borough. Because both tables have borough columns, we have to specify the table here.
```sql
/* Get call counts by borough */
SELECT hpd311calls.borough,
COUNT(*)
FROM hpd311calls
GROUP BY hpd311calls.borough;
```
### Joining and Aggregating
The boro census table has more columns than we need, so we also select its total population and housing units columns. Notice that we don't have to list boro census in the from clause.
Finally, we add the join clause between the from and group by clauses, joining on the borough columns.
```sql
/* Get call counts by borough
and join in population and housing counts */
SELECT hpd311calls.borough,
COUNT(*),
boro_census.total_population,
boro_census.housing_units
FROM hpd311calls
JOIN boro_census
ON hpd311calls.borough = boro_census.borough
GROUP BY hpd311calls.borough;
```
Let's pass this query and the database engine to read SQL and check out the results. Looks like the Bronx is overrepresented in our call data.
```
query = """
SELECT hpd311calls.borough,
COUNT(*),
boro_census.total_population,
boro_census.housing_units
FROM hpd311calls
JOIN boro_census
ON hpd311calls.borough = boro_census.borough
GROUP BY hpd311calls.borough;
"""
call_counts = pd.read_sql(query, engine)
print(call_counts)
```
### Review
As you can tell, SQL queries can get complicated. Let's review the order of keywords. First comes the select statement, including aggregate functions, and the table you're selecting from. Then a join clause if there is one. Then the where clause, if any. Last comes group by.
## Joining tables
Tables in relational databases usually have key columns of unique record identifiers. This lets us build pipelines that combine tables using SQL's `JOIN` operation, instead of having to combine data after importing it.
The records in `hpd311calls` often concern issues, like leaks or heating problems, that are exacerbated by weather conditions. In this exercise, you'll join `weather` data to call records along their common date columns to get everything in one data frame. You can assume these columns have the same data type.
**Note**: The SQL checker is picky about join table order -- it expects specific tables on the left and the right.
- Complete the query to join `weather` to `hpd311calls` by their `date` and `created_date` columns, respectively.
- Query the database and assign the resulting data frame to `calls_with_weather`.
- Print the first few rows of `calls_with_weather` to confirm all columns were joined.
```
# Query to join weather to call records by date columns
query = """
SELECT *
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date;
"""
# Create data frame of joined tables
calls_with_weather = pd.read_sql(query, engine)
# View the data frame to make sure all columns were joined
print(calls_with_weather.head())
```
*The joins you perform in this course only return records whose key values appear in both tables, which is why the resulting data frames have values for all columns. But there are other kinds of joins that can return records that don't have a match.*
## Joining and filtering
Just as you might not always want all the data in a single table, you might not want all columns and rows that result from a `JOIN`. In this exercise, you'll use SQL to refine a data import.
Weather exacerbates some housing problems more than others. Your task is to focus on water leak reports in `hpd311calls` and assemble a dataset that includes the day's precipitation levels from `weather` to see if there is any relationship between the two. The provided SQL gets all columns in `hpd311calls`, but you'll need to modify it to get the necessary `weather` column and filter rows with a `WHERE` clause.
- Complete `query` to get the `prcp` column in `weather` and join `weather` to `hpd311calls` on their `date` and `created_date` columns, respectively.
- Use `read_sql()` to load the results of the query into the `leak_calls` data frame.
```
# Query to get hpd311calls and precipitation values
query = """
SELECT hpd311calls.*, weather.prcp
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date;"""
# Load query results into the leak_calls data frame
leak_calls = pd.read_sql(query, engine)
# View the data frame
print(leak_calls.head())
```
- Modify `query` to get only rows that have `'WATER LEAK'` as their `complaint_type`.
```
# Query to get water leak calls and daily precipitation
query = """
SELECT hpd311calls.*, weather.prcp
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date
WHERE hpd311calls.complaint_type = 'WATER LEAK';"""
# Load query results into the leak_calls data frame
leak_calls = pd.read_sql(query, engine)
# View the data frame
print(leak_calls.head())
```
*How you go about constructing a complicated SQL query can depend on what operators you need and how big the tables you're working with are. If your tables are very big, you may decide to filter or aggregate the data first before attempting a join.*
## oining, filtering, and aggregating
In this exercise, you'll use what you've learned to assemble a dataset to investigate how the number of heating complaints to New York City's 311 line varies with temperature.
In addition to the `hpd311calls` table, `data.db` has a `weather` table with daily high and low temperature readings for NYC. We want to get each day's count of heat/hot water calls with temperatures joined in. This can be done in one query, which we'll build in parts.
In part one, we'll get just the data we want from `hpd311calls`. Then, in part two, we'll modify the query to join in `weather` data.
- Complete the query to get `created_date` and counts of records whose `complaint_type` is `HEAT/HOT WATER` from `hpd311calls` by date.
- Create a data frame, `df`, containing the results of the query.
```
# Query to get heat/hot water call counts by created_date
query = """
SELECT hpd311calls.created_date,
COUNT(*)
FROM hpd311calls
WHERE hpd311calls.complaint_type = 'HEAT/HOT WATER'
GROUP BY hpd311calls.created_date;
"""
# Query database and save results as df
df = pd.read_sql(query, engine)
# View first 5 records
print(df.head())
```
- Modify the query to join `tmax` and `tmin` from the `weather` table. (There is only one record per date in `weather`, so we do not need SQL's `MAX` and `MIN` functions here.) Join the tables on `created_date` in `hpd311calls` and `date` in `weather`.
```
# Modify query to join tmax and tmin from weather by date
query = """
SELECT hpd311calls.created_date,
COUNT(*),
weather.tmax,
weather.tmin
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date
WHERE hpd311calls.complaint_type = 'HEAT/HOT WATER'
GROUP BY hpd311calls.created_date;
"""
# Query database and save results as df
df = pd.read_sql(query, engine)
# View first 5 records
print(df.head())
```
*While SQL joins can only be used in databases, there are analagous `pandas operations to combine datasets.*
|
github_jupyter
|
pd.read_sql(query, engine)
# Load pandas and sqlalchemy's create_engine
import pandas as pd
from sqlalchemy import create_engine
# Create database engine to manage connections
engine = create_engine('sqlite:///data.db')
# Load entire weather table by table name
weather = pd.read_sql('weather', engine)
# Load entire weather table with SQL
weather = pd.read_sql('SELECT * FROM weather', engine)
print(weather.head())
# Import sqlalchemy's create_engine() function
from sqlalchemy import create_engine
# Create the database engine
engine = create_engine('sqlite:///data.db')
# View the tables in the database
print(engine.table_names())
# Create the database engine
engine = create_engine('sqlite:///data.db')
# Load hpd311calls without any SQL
hpd_calls = pd.read_sql('hpd311calls', engine)
# View the first few rows of data
print(hpd_calls.head())
# Create the database engine
engine = create_engine("sqlite:///data.db")
# Create a SQL query to load the entire weather table
query = """
SELECT *
FROM weather;
"""
# Load weather with the SQL query
weather = pd.read_sql(query, engine)
# View the first few rows of data
print(weather.head())
SELECT date, tavg
FROM weather;
SELECT *
FROM weather
WHERE tmax > 32;
/* Get records about incidents in Brooklyn */
SELECT *
FROM hpd311calls
WHERE borough = 'BROOKYN';
# Create database engine
engine = create_engine('sqlite:///data.db')
# Write query to get records from Brooklyn
query = """
SELECT *
FROM hpd311calls
WHERE borough = 'BROOKLYN';
"""
# Query the database
brooklyn_calls = pd.read_sql(query, engine)
print( brooklyn_calls.borough.unique())
# Write query to get records about plumbing in the Bronx
and_query = """
SELECT *
FROM hpd311calls
WHERE borough = 'BRONX'
AND complaint_type = 'PLUMBING';
"""
# Get calls about plumbing issues in the Bronx
bx_plumbing_calls = pd.read_sql(and_query, engine)
# Check record count
print(bx_plumbing_calls.shape)
# Write query to get records about water leaks or plumbing
or_query = """
SELECT *
FROM hpd311calls
WHERE complaint_type = 'WATER LEAK'
OR complaint_type = 'PLUMBING';
"""
# Get calls about plumbing issues in the Bronx
leaks_or_plumbing = pd.read_sql(or_query, engine)
# Check record count
print(leaks_or_plumbing.shape)
# Create database engine for data.db
engine = create_engine('sqlite:///data.db')
# Write query to get date, tmax, and tmin from weather
query = """
SELECT date,
tmax,
tmin
FROM weather;
"""
# Make a data frame by passing query and engine to read_sql()
temperatures = pd.read_sql(query, engine)
# View the resulting data frame
print(temperatures)
import matplotlib.pyplot as plt
# Create query to get hpd311calls records about safety
query = """
SELECT *
FROM hpd311calls
WHERE complaint_type = 'SAFETY';
"""
# Query the database and assign result to safety_calls
safety_calls = pd.read_sql(query, engine)
# Graph the number of safety calls by borough
call_counts = safety_calls.groupby('borough').unique_key.count()
call_counts.plot.barh()
plt.show()
# Create query for records with max temps <= 32 or snow >= 1
query = """
SELECT *
FROM weather
WHERE tmax <= 32
OR snow >= 1;
"""
# Query database and assign result to wintry_days
wintry_days = pd.read_sql(query, engine)
# View summary stats about the temperatures
print(wintry_days.describe())
/* Get unique street addresses and boroughs */
SELECT DISTINCT incident_address, borough
FROM hpd311calls;
/* Get counts of plumbing calls by borough */
SELECT borough,
COUNT(*)
FROM hpd311calls
WHERE complaint_type = 'PLUMBING'
GROUP BY borough;
# Create database engine
engine = create_engine('sqlite:///data.db')
# Write query to get plumbing call counts by borough
query = """
SELECT borough, COUNT(*)
FROM hpd311calls
WHERE complaint_type = 'PLUMBING'
GROUP BY borough;
"""
# Query database and create data frame
plumbing_call_counts = pd.read_sql(query, engine)
print(plumbing_call_counts)
# Create query for unique combinations of borough and complaint_type
query = """
SELECT DISTINCT borough, complaint_type
FROM hpd311calls;
"""
# Load results of query to a data frame
issues_and_boros = pd.read_sql(query, engine)
# Check assumption about issues and boroughs
print(issues_and_boros)
# Create query to get call counts by complaint_type
query = """
SELECT complaint_type,
COUNT(*)
FROM hpd311calls
GROUP BY complaint_type;
"""
# Create data frame of call counts by issue
calls_by_issue = pd.read_sql(query, engine)
# Graph the number of calls for each housing issue
calls_by_issue.plot.barh(x='complaint_type')
plt.show()
# Create a query to get month and max tmax by month
query = """
SELECT month,
MAX(tmax)
FROM weather
GROUP BY month;
"""
# Get data frame of monthly weather stats
weather_by_month = pd.read_sql(query, engine)
# View weather stats by month
print(weather_by_month)
# Create a query to get month, max tmax, and min tmin by month
query = """
SELECT month,
MAX(tmax),
MIN(tmin)
FROM weather
GROUP BY month;
"""
# Get data frame of monthly weather stats
weather_by_month = pd.read_sql(query, engine)
# View weather stats by month
print(weather_by_month)
# Create query to get temperature and precipitation by month
query = """
SELECT month,
MAX(tmax),
MIN(tmin),
SUM(prcp)
FROM weather
GROUP BY month;
"""
# Get data frame of monthly weather stats
weather_by_month = pd.read_sql(query, engine)
# View weather stats by month
print(weather_by_month)
SELECT *
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date;
/* Get only heat/hot water calls and join in weather data */
SELECT *
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date
WHERE hpd311calls.complaint_type = 'HEAT/HOT WATER';
/* Get call counts by borough */
SELECT hpd311calls.borough,
COUNT(*)
FROM hpd311calls
GROUP BY hpd311calls.borough;
/* Get call counts by borough
and join in population and housing counts */
SELECT hpd311calls.borough,
COUNT(*),
boro_census.total_population,
boro_census.housing_units
FROM hpd311calls
JOIN boro_census
ON hpd311calls.borough = boro_census.borough
GROUP BY hpd311calls.borough;
query = """
SELECT hpd311calls.borough,
COUNT(*),
boro_census.total_population,
boro_census.housing_units
FROM hpd311calls
JOIN boro_census
ON hpd311calls.borough = boro_census.borough
GROUP BY hpd311calls.borough;
"""
call_counts = pd.read_sql(query, engine)
print(call_counts)
# Query to join weather to call records by date columns
query = """
SELECT *
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date;
"""
# Create data frame of joined tables
calls_with_weather = pd.read_sql(query, engine)
# View the data frame to make sure all columns were joined
print(calls_with_weather.head())
# Query to get hpd311calls and precipitation values
query = """
SELECT hpd311calls.*, weather.prcp
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date;"""
# Load query results into the leak_calls data frame
leak_calls = pd.read_sql(query, engine)
# View the data frame
print(leak_calls.head())
# Query to get water leak calls and daily precipitation
query = """
SELECT hpd311calls.*, weather.prcp
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date
WHERE hpd311calls.complaint_type = 'WATER LEAK';"""
# Load query results into the leak_calls data frame
leak_calls = pd.read_sql(query, engine)
# View the data frame
print(leak_calls.head())
# Query to get heat/hot water call counts by created_date
query = """
SELECT hpd311calls.created_date,
COUNT(*)
FROM hpd311calls
WHERE hpd311calls.complaint_type = 'HEAT/HOT WATER'
GROUP BY hpd311calls.created_date;
"""
# Query database and save results as df
df = pd.read_sql(query, engine)
# View first 5 records
print(df.head())
# Modify query to join tmax and tmin from weather by date
query = """
SELECT hpd311calls.created_date,
COUNT(*),
weather.tmax,
weather.tmin
FROM hpd311calls
JOIN weather
ON hpd311calls.created_date = weather.date
WHERE hpd311calls.complaint_type = 'HEAT/HOT WATER'
GROUP BY hpd311calls.created_date;
"""
# Query database and save results as df
df = pd.read_sql(query, engine)
# View first 5 records
print(df.head())
| 0.646795 | 0.988369 |
```
! wget https://repo.anaconda.com/miniconda/Miniconda3-py37_4.8.2-Linux-x86_64.sh
! chmod +x Miniconda3-py37_4.8.2-Linux-x86_64.sh
! bash ./Miniconda3-py37_4.8.2-Linux-x86_64.sh -b -f -p /usr/local
! conda install -c rdkit rdkit -y
import sys
sys.path.append('/usr/local/lib/python3.7/site-packages/')
!time conda install -q -y -c openbabel openbabel
import sys
sys.path.append('/usr/local/lib/python3.7/site-packages/')
!pip install py3Dmol # 3D Molecular Visualizer
import os
import numpy as np
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import accuracy_score
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit.Chem import Draw
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem import DataStructs
from rdkit.Chem import RDConfig
from rdkit.Chem import rdBase
import pickle
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
print(rdBase.rdkitVersion)
!wget https://sourceforge.net/projects/smina/files/smina.static/download -O smina.static
!chmod u+x smina.static
!./smina.static
! apt-get install pymol
!wget -O 1OYT.pdb https://files.rcsb.org/download/1oyt.pdb
com_file = open('fetch_and_clean.pml','w')
com_file.write('''
load 1OYT.pdb
remove resn HOH
h_add elem O or elem N
select 1OYT-FSN, resn FSN #Create a selection called 1OYT-FSN from the ligand
select 1OYT-receptor, 1OYT and not 1OYT-FSN #Select all that is not the ligand
save 1OYT-FSN.pdb, 1OYT-FSN
save 1OYT-receptor.pdb, 1OYT-receptor
''')
com_file.close()
!pymol -c fetch_and_clean.pml
!obabel 1OYT-receptor.pdb -xr -O 1OYT-receptor.pdbqt
!obabel 1OYT-FSN.pdb -O 1OYT-FSN.pdbqt
!./smina.static -r 1OYT-receptor.pdbqt -l 1OYT-FSN.pdbqt --autobox_ligand 1OYT-FSN.pdbqt --autobox_add 8 --exhaustiveness 16 -o 1OYT-redock.pdbqt
import py3Dmol
def drawit2(m,confId=-1):
mb = Chem.MolToMolBlock(m,confId=confId)
p = py3Dmol.view(width=400, height=400)
p.addModel(mb,'sdf')
p.setStyle({'stick':{}})
p.setBackgroundColor('0xeeeeee')
p.zoomTo()
return p
def DrawComplex(protein,ligand):
complex_pl = Chem.MolToPDBBlock(Chem.CombineMols(protein,ligand))
#complex_mol=Chem.CombineMols(receptor,mols[-1])
view = py3Dmol.view(width=600,height=600)
view.addModel(complex_pl,'pdb')
#view.addModel(Chem.MolToMolBlock(mols[0]),'sdf')
chA = {'chain':['H','L','I']}
chB = {'resn':'UNL'}
view.setStyle(chA,{'cartoon': {'color':'spectrum'}})
#view.setStyle(chA,{'lines': {}})
view.addSurface(py3Dmol.VDW,{'opacity':0.8}, chB)
view.setStyle(chB,{'stick':{}})
view.zoomTo()
return view
from rdkit import Chem
!obabel 1OYT-redock.pdbqt -O 1OYT-redocked.sdf
mols = [m for m in Chem.SDMolSupplier('1OYT-redocked.sdf') if m is not None]
drawit2(mols[8])
receptor = Chem.MolFromPDBFile('1OYT-receptor.pdb')
DrawComplex(receptor,mols[8])
```
|
github_jupyter
|
! wget https://repo.anaconda.com/miniconda/Miniconda3-py37_4.8.2-Linux-x86_64.sh
! chmod +x Miniconda3-py37_4.8.2-Linux-x86_64.sh
! bash ./Miniconda3-py37_4.8.2-Linux-x86_64.sh -b -f -p /usr/local
! conda install -c rdkit rdkit -y
import sys
sys.path.append('/usr/local/lib/python3.7/site-packages/')
!time conda install -q -y -c openbabel openbabel
import sys
sys.path.append('/usr/local/lib/python3.7/site-packages/')
!pip install py3Dmol # 3D Molecular Visualizer
import os
import numpy as np
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import accuracy_score
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit.Chem import Draw
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem import DataStructs
from rdkit.Chem import RDConfig
from rdkit.Chem import rdBase
import pickle
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
print(rdBase.rdkitVersion)
!wget https://sourceforge.net/projects/smina/files/smina.static/download -O smina.static
!chmod u+x smina.static
!./smina.static
! apt-get install pymol
!wget -O 1OYT.pdb https://files.rcsb.org/download/1oyt.pdb
com_file = open('fetch_and_clean.pml','w')
com_file.write('''
load 1OYT.pdb
remove resn HOH
h_add elem O or elem N
select 1OYT-FSN, resn FSN #Create a selection called 1OYT-FSN from the ligand
select 1OYT-receptor, 1OYT and not 1OYT-FSN #Select all that is not the ligand
save 1OYT-FSN.pdb, 1OYT-FSN
save 1OYT-receptor.pdb, 1OYT-receptor
''')
com_file.close()
!pymol -c fetch_and_clean.pml
!obabel 1OYT-receptor.pdb -xr -O 1OYT-receptor.pdbqt
!obabel 1OYT-FSN.pdb -O 1OYT-FSN.pdbqt
!./smina.static -r 1OYT-receptor.pdbqt -l 1OYT-FSN.pdbqt --autobox_ligand 1OYT-FSN.pdbqt --autobox_add 8 --exhaustiveness 16 -o 1OYT-redock.pdbqt
import py3Dmol
def drawit2(m,confId=-1):
mb = Chem.MolToMolBlock(m,confId=confId)
p = py3Dmol.view(width=400, height=400)
p.addModel(mb,'sdf')
p.setStyle({'stick':{}})
p.setBackgroundColor('0xeeeeee')
p.zoomTo()
return p
def DrawComplex(protein,ligand):
complex_pl = Chem.MolToPDBBlock(Chem.CombineMols(protein,ligand))
#complex_mol=Chem.CombineMols(receptor,mols[-1])
view = py3Dmol.view(width=600,height=600)
view.addModel(complex_pl,'pdb')
#view.addModel(Chem.MolToMolBlock(mols[0]),'sdf')
chA = {'chain':['H','L','I']}
chB = {'resn':'UNL'}
view.setStyle(chA,{'cartoon': {'color':'spectrum'}})
#view.setStyle(chA,{'lines': {}})
view.addSurface(py3Dmol.VDW,{'opacity':0.8}, chB)
view.setStyle(chB,{'stick':{}})
view.zoomTo()
return view
from rdkit import Chem
!obabel 1OYT-redock.pdbqt -O 1OYT-redocked.sdf
mols = [m for m in Chem.SDMolSupplier('1OYT-redocked.sdf') if m is not None]
drawit2(mols[8])
receptor = Chem.MolFromPDBFile('1OYT-receptor.pdb')
DrawComplex(receptor,mols[8])
| 0.433142 | 0.192426 |
# Ownership mechanism and readonly flags
The scipp data structures (variables, data arrays, and datasets) behave mostly like nested Python objects, i.e., sub-objects are shared by default.
Some of the effects are exemplified in the following.
## Shared ownership
### Variables
Slices or other views of variables are also of type `Variable` and all views share ownership of the underlying data.
If a variable refers only to a section of the underlying data buffer this is indicated in the HTML view in the title line as part of the size, here *"16 Bytes out of 96 Bytes"*.
This allows for identification of "small" variables that keep alive potentially large buffers:
```
import scipp as sc
var = sc.arange(dim='x', unit='m', start=0, stop=12)
var['x', 4:6]
```
To create a variable with sole ownership of a buffer, use the `copy()` method:
```
var['x', 4:6].copy()
```
By default, `copy()` returns a deep copy.
Shallow copies can be made by specifying `deep=False`, which preserves shared ownership of underlying buffers:
```
shallow_copy = var['x', 4:6].copy(deep=False)
shallow_copy
```
### Data arrays
As a result of the sharing mechanism, extra care must be taken in some cases, just like when working with any other Python library.
Consider the following example, using the same variable as data and as a coordinate:
```
da = sc.DataArray(data=var, coords={'x': var})
da += 666 * sc.units.m
da
```
The modification unintentionally also affected the coordinate.
However, if we think of data arrays and coordinate dicts as Python-like objects, then the behavior should not be surprising.
Note that the original `var` is also affected:
```
var
```
To avoid this, use `copy()`, e.g.,:
```
da = sc.DataArray(data=var.copy(), coords={'x': var.copy()})
da += 666 * sc.units.m
da
```
Apart from the standard and pythonic behavior, one advantage of this is that creating data arrays from variables is typically cheap, without inflicting copies of potentially large objects.
### Datasets
Just like creating data arrays from variables is cheap (without deep-copies), inserting items into datasets does not inflict potentially expensive deep copies:
```
ds = sc.Dataset()
ds['a'] = da # shallow copy
```
Note that while the buffers are shared, the meta-data dicts such as `coords`, `masks`, or `attrs` are not.
Compare:
```
ds['a'].attrs['attr'] = 1.2 * sc.units.m
'attr' in da.attrs # the attrs *dict* is copied
```
with
```
da.coords['x'] *= -1
ds.coords['x'] # the coords *dict* is copied, but the 'x' coordinate references same buffer
```
## Read-only flags
Consider the following attempt to modify the data via a slice:
```
try:
da['x', 0].data = var['x', 2]
except sc.DataArrayError as e:
print(e)
```
Since `da['x',0]` is itself a data array, assigning to the `data` property would repoint the data to whatever is given on the right-hand side.
However, this would not affect `da`, and the attempt to change the data would silently do nothing, since the temporary `da['x',0]` disappears immediately.
The read-only flag protects us from this.
To actually modify the slice, use `__setitem__` instead:
```
da['x', 0] = var['x', 2]
```
Variables, meta-data dicts (`coords`, `masks`, and `attrs` properties), data arrays, and datasets also have read-only flags.
The flags solve a number of conceptual issues and serve as a safeguard against hidden bugs.
One example is a broadcast of a variable:
```
var = sc.broadcast(sc.scalar(1.0), dims=['x'], shape=[10])
try:
var += 7
except sc.VariableError as e:
print(e)
```
Since `broadcast` returns a view, the readon-only flag is set to avoid multiple additions to the same element.
|
github_jupyter
|
import scipp as sc
var = sc.arange(dim='x', unit='m', start=0, stop=12)
var['x', 4:6]
var['x', 4:6].copy()
shallow_copy = var['x', 4:6].copy(deep=False)
shallow_copy
da = sc.DataArray(data=var, coords={'x': var})
da += 666 * sc.units.m
da
var
da = sc.DataArray(data=var.copy(), coords={'x': var.copy()})
da += 666 * sc.units.m
da
ds = sc.Dataset()
ds['a'] = da # shallow copy
ds['a'].attrs['attr'] = 1.2 * sc.units.m
'attr' in da.attrs # the attrs *dict* is copied
da.coords['x'] *= -1
ds.coords['x'] # the coords *dict* is copied, but the 'x' coordinate references same buffer
try:
da['x', 0].data = var['x', 2]
except sc.DataArrayError as e:
print(e)
da['x', 0] = var['x', 2]
var = sc.broadcast(sc.scalar(1.0), dims=['x'], shape=[10])
try:
var += 7
except sc.VariableError as e:
print(e)
| 0.214445 | 0.969642 |
# AGENT #
An agent, as defined in 2.1 is anything that can perceive its <b>environment</b> through sensors, and act upon that environment through actuators based on its <b>agent program</b>. This can be a dog, robot, or even you. As long as you can perceive the environment and act on it, you are an agent. This notebook will explain how to implement a simple agent, create an environment, and create a program that helps the agent act on the environment based on its percepts.
Before moving on, review the </b>Agent</b> and </b>Environment</b> classes in <b>[agents.py](https://github.com/aimacode/aima-python/blob/master/agents.py)</b>.
Let's begin by importing all the functions from the agents.py module and creating our first agent - a blind dog.
```
from agents import *
class BlindDog(Agent):
def eat(self, thing):
print("Dog: Ate food at {}.".format(self.location))
def drink(self, thing):
print("Dog: Drank water at {}.".format( self.location))
dog = BlindDog()
```
What we have just done is create a dog who can only feel what's in his location (since he's blind), and can eat or drink. Let's see if he's alive...
```
print(dog.alive)
```

This is our dog. How cool is he? Well, he's hungry and needs to go search for food. For him to do this, we need to give him a program. But before that, let's create a park for our dog to play in.
# ENVIRONMENT #
A park is an example of an environment because our dog can perceive and act upon it. The <b>Environment</b> class in agents.py is an abstract class, so we will have to create our own subclass from it before we can use it. The abstract class must contain the following methods:
<li><b>percept(self, agent)</b> - returns what the agent perceives</li>
<li><b>execute_action(self, agent, action)</b> - changes the state of the environment based on what the agent does.</li>
```
class Food(Thing):
pass
class Water(Thing):
pass
class Park(Environment):
def percept(self, agent):
'''prints & return a list of things that are in our agent's location'''
things = self.list_things_at(agent.location)
print(things)
return things
def execute_action(self, agent, action):
'''changes the state of the environment based on what the agent does.'''
if action == "move down":
agent.movedown()
elif action == "eat":
items = self.list_things_at(agent.location, tclass=Food)
if len(items) != 0:
if agent.eat(items[0]): #Have the dog pick eat the first item
self.delete_thing(items[0]) #Delete it from the Park after.
elif action == "drink":
items = self.list_things_at(agent.location, tclass=Water)
if len(items) != 0:
if agent.drink(items[0]): #Have the dog drink the first item
self.delete_thing(items[0]) #Delete it from the Park after.
def is_done(self):
'''By default, we're done when we can't find a live agent,
but to prevent killing our cute dog, we will or it with when there is no more food or water'''
no_edibles = not any(isinstance(thing, Food) or isinstance(thing, Water) for thing in self.things)
dead_agents = not any(agent.is_alive() for agent in self.agents)
return dead_agents or no_edibles
```
## Wumpus Environment
```
from ipythonblocks import BlockGrid
from agents import *
color = {"Breeze": (225, 225, 225),
"Pit": (0,0,0),
"Gold": (253, 208, 23),
"Glitter": (253, 208, 23),
"Wumpus": (43, 27, 23),
"Stench": (128, 128, 128),
"Explorer": (0, 0, 255),
"Wall": (44, 53, 57)
}
def program(percepts):
'''Returns an action based on it's percepts'''
print(percepts)
return input()
w = WumpusEnvironment(program, 7, 7)
grid = BlockGrid(w.width, w.height, fill=(123, 234, 123))
def draw_grid(world):
global grid
grid[:] = (123, 234, 123)
for x in range(0, len(world)):
for y in range(0, len(world[x])):
if len(world[x][y]):
grid[y, x] = color[world[x][y][-1].__class__.__name__]
def step():
global grid, w
draw_grid(w.get_world())
grid.show()
w.step()
step()
```
# PROGRAM #
Now that we have a <b>Park</b> Class, we need to implement a <b>program</b> module for our dog. A program controls how the dog acts upon it's environment. Our program will be very simple, and is shown in the table below.
<table>
<tr>
<td><b>Percept:</b> </td>
<td>Feel Food </td>
<td>Feel Water</td>
<td>Feel Nothing</td>
</tr>
<tr>
<td><b>Action:</b> </td>
<td>eat</td>
<td>drink</td>
<td>move up</td>
</tr>
</table>
```
class BlindDog(Agent):
location = 1
def movedown(self):
self.location += 1
def eat(self, thing):
'''returns True upon success or False otherwise'''
if isinstance(thing, Food):
print("Dog: Ate food at {}.".format(self.location))
return True
return False
def drink(self, thing):
''' returns True upon success or False otherwise'''
if isinstance(thing, Water):
print("Dog: Drank water at {}.".format(self.location))
return True
return False
def program(percepts):
'''Returns an action based on it's percepts'''
for p in percepts:
if isinstance(p, Food):
return 'eat'
elif isinstance(p, Water):
return 'drink'
return 'move down'
park = Park()
dog = BlindDog(program)
dogfood = Food()
water = Water()
park.add_thing(dog, 0)
park.add_thing(dogfood, 5)
park.add_thing(water, 7)
park.run(10)
```
That's how easy it is to implement an agent, its program, and environment. But that was a very simple case. What if our environment was 2-Dimentional instead of 1? And what if we had multiple agents?
To make our Park 2D, we will need to make it a subclass of <b>XYEnvironment</b> instead of Environment. Also, let's add a person to play fetch with the dog.
```
from agents import *
class MAgent(Agent):
def __init__(self):
self.things = []
def symbol(self):
return "a "
def add_percepted_things(self,things):
for thing in things:
add_percepted_thing(thing)
def add_perecpted_thing(self,thing,location):
thing.location = location
self.things.append(thing)
def thing_at(self,location):
for thing in self.things:
if thing.location == location:
return thing.id()
return "? "
def show_state(self):
print(" 0 1 2 3 4 5 ")
for y in range(6):
print(y,end=' ')
for x in range(6):
print(self.thing_at((x,y)),end='')
print()
class Nothing():
pass
class Outside():
pass
class Cosa(Thing):
def symbol(self):
return "c "
class MEnv(Environment):
def symbol(self,thing):
if isinstance(thing,None):
return "o "
elif isinstance(thing,Nothing):
return "- "
else:
self.list_things_at(location)[0].symbol()
def thing_at(self, location):
if location[0] < 0 or location[1] < 0:
return Outside()
elif self.list_things_at(location):
return self.list_things_at(location)[0]
else:
return Nothing()
def percept(self, agent):
location = agent.location
here = self.thing_at(location) # Agent's Location
left = self.thing_at((location[0] - 1,location[1])) # Left
down = self.thing_at((location[0],location[1] + 1)) # Down
right = self.thing_at((location[0] + 1,location[1])) # Right
up = self.thing_at((location[0],location[1] - 1)) # Up
print("Percept: ({},({},{},{},{},{}))".format(location,symbol(here),symbol(left),symbol(down),symbol(up),symbol(right)))
return [here,left,down,up,right]
def draw_grid(self):
print(" 0 1 2 3 4 5 ")
for y in range(6):
print(y,end=' ')
for x in range(6):
print(self.symbol(self.thing_at((x,y))),end='')
print()
e = MEnv()
c = Cosa()
e.add_thing(c,(2,2))
e.add_thing(Cosa(),(4,4))
agent = MAgent()
e.add_thing(agent,(3,4))
e.draw_grid()
print()
#agent.add_perecpted_thing(e.percept(agent))
agent.show_state()
e.run(1)
cha = np.chararray((6,6))
cha[2,2] = "z"
k = []
(1,2)[0]
k = 1
k = "3"
k
```
|
github_jupyter
|
from agents import *
class BlindDog(Agent):
def eat(self, thing):
print("Dog: Ate food at {}.".format(self.location))
def drink(self, thing):
print("Dog: Drank water at {}.".format( self.location))
dog = BlindDog()
print(dog.alive)
class Food(Thing):
pass
class Water(Thing):
pass
class Park(Environment):
def percept(self, agent):
'''prints & return a list of things that are in our agent's location'''
things = self.list_things_at(agent.location)
print(things)
return things
def execute_action(self, agent, action):
'''changes the state of the environment based on what the agent does.'''
if action == "move down":
agent.movedown()
elif action == "eat":
items = self.list_things_at(agent.location, tclass=Food)
if len(items) != 0:
if agent.eat(items[0]): #Have the dog pick eat the first item
self.delete_thing(items[0]) #Delete it from the Park after.
elif action == "drink":
items = self.list_things_at(agent.location, tclass=Water)
if len(items) != 0:
if agent.drink(items[0]): #Have the dog drink the first item
self.delete_thing(items[0]) #Delete it from the Park after.
def is_done(self):
'''By default, we're done when we can't find a live agent,
but to prevent killing our cute dog, we will or it with when there is no more food or water'''
no_edibles = not any(isinstance(thing, Food) or isinstance(thing, Water) for thing in self.things)
dead_agents = not any(agent.is_alive() for agent in self.agents)
return dead_agents or no_edibles
from ipythonblocks import BlockGrid
from agents import *
color = {"Breeze": (225, 225, 225),
"Pit": (0,0,0),
"Gold": (253, 208, 23),
"Glitter": (253, 208, 23),
"Wumpus": (43, 27, 23),
"Stench": (128, 128, 128),
"Explorer": (0, 0, 255),
"Wall": (44, 53, 57)
}
def program(percepts):
'''Returns an action based on it's percepts'''
print(percepts)
return input()
w = WumpusEnvironment(program, 7, 7)
grid = BlockGrid(w.width, w.height, fill=(123, 234, 123))
def draw_grid(world):
global grid
grid[:] = (123, 234, 123)
for x in range(0, len(world)):
for y in range(0, len(world[x])):
if len(world[x][y]):
grid[y, x] = color[world[x][y][-1].__class__.__name__]
def step():
global grid, w
draw_grid(w.get_world())
grid.show()
w.step()
step()
class BlindDog(Agent):
location = 1
def movedown(self):
self.location += 1
def eat(self, thing):
'''returns True upon success or False otherwise'''
if isinstance(thing, Food):
print("Dog: Ate food at {}.".format(self.location))
return True
return False
def drink(self, thing):
''' returns True upon success or False otherwise'''
if isinstance(thing, Water):
print("Dog: Drank water at {}.".format(self.location))
return True
return False
def program(percepts):
'''Returns an action based on it's percepts'''
for p in percepts:
if isinstance(p, Food):
return 'eat'
elif isinstance(p, Water):
return 'drink'
return 'move down'
park = Park()
dog = BlindDog(program)
dogfood = Food()
water = Water()
park.add_thing(dog, 0)
park.add_thing(dogfood, 5)
park.add_thing(water, 7)
park.run(10)
from agents import *
class MAgent(Agent):
def __init__(self):
self.things = []
def symbol(self):
return "a "
def add_percepted_things(self,things):
for thing in things:
add_percepted_thing(thing)
def add_perecpted_thing(self,thing,location):
thing.location = location
self.things.append(thing)
def thing_at(self,location):
for thing in self.things:
if thing.location == location:
return thing.id()
return "? "
def show_state(self):
print(" 0 1 2 3 4 5 ")
for y in range(6):
print(y,end=' ')
for x in range(6):
print(self.thing_at((x,y)),end='')
print()
class Nothing():
pass
class Outside():
pass
class Cosa(Thing):
def symbol(self):
return "c "
class MEnv(Environment):
def symbol(self,thing):
if isinstance(thing,None):
return "o "
elif isinstance(thing,Nothing):
return "- "
else:
self.list_things_at(location)[0].symbol()
def thing_at(self, location):
if location[0] < 0 or location[1] < 0:
return Outside()
elif self.list_things_at(location):
return self.list_things_at(location)[0]
else:
return Nothing()
def percept(self, agent):
location = agent.location
here = self.thing_at(location) # Agent's Location
left = self.thing_at((location[0] - 1,location[1])) # Left
down = self.thing_at((location[0],location[1] + 1)) # Down
right = self.thing_at((location[0] + 1,location[1])) # Right
up = self.thing_at((location[0],location[1] - 1)) # Up
print("Percept: ({},({},{},{},{},{}))".format(location,symbol(here),symbol(left),symbol(down),symbol(up),symbol(right)))
return [here,left,down,up,right]
def draw_grid(self):
print(" 0 1 2 3 4 5 ")
for y in range(6):
print(y,end=' ')
for x in range(6):
print(self.symbol(self.thing_at((x,y))),end='')
print()
e = MEnv()
c = Cosa()
e.add_thing(c,(2,2))
e.add_thing(Cosa(),(4,4))
agent = MAgent()
e.add_thing(agent,(3,4))
e.draw_grid()
print()
#agent.add_perecpted_thing(e.percept(agent))
agent.show_state()
e.run(1)
cha = np.chararray((6,6))
cha[2,2] = "z"
k = []
(1,2)[0]
k = 1
k = "3"
k
| 0.479016 | 0.916372 |
# Simulating Language 1, Why Simulate Language? (lecture)
*This is a first draft of lecture notes for the Simulating Language course. It probably contains lots of typos!*
## Some important questions
Before we get started, it's worth thinking about what it is we're wanting to achieve. A good first step is to ask what the central goal of linguistics as a science actually is. There are many reasonable answers to this question, of course, and they'll differ among the many subdisciplines of linguistics. For example, a descriptive linguist might answer that the goal of linguistics is a careful and detailed account of the structure of individual languages. A historical linguist might be in the business of attempting to reconstruct languages that are no longer spoken. But for many branches of linguistics, I think that a good approximation of the ultimate goal is an answer to a **why** question: "why is language the way it is?".
How might we go about approaching such a question?
## The evolutionary approach
In this course, we will take an evolutionary approach to language, with both language and evolution here taken very broadly indeed. The evolutionary approach attempts to answer the why question by posing instead a **how** question. The idea is that we can only really figure out why language is the way it is if we understand how it came to be that way.
This course will cover work carried out over the past 20 years or so, much of it pioneered here in Edinburgh, that tackles this how question as a way of providing a solid explanatory foundation for the science of language. A recent overview of this work, much of which we will replicate in this course, can be found in [Kirby (2017)](https://doi.org/10.3758/s13423-016-1166-7).
### The *processes* of language evolution
When we hear the term "evolution" we naturally start thinking about genes and natural selection, of ancient prehistory and the survival of the fittest. This is of course part of the story, but one of the exciting things about human behaviour is that it is not determined solely by genes. (Actually, nothing is determined solely by genes. Every organism is the product of development in a particular environment. We'll return to this in a later lecture.) If we are to answer our **how** question as a route to answering the ultimate **why** question, then we need to take a much broader view of what the processes are that are involved in the evolution of language. In other words, what are the different ways the nature of language comes into being?
Broadly speaking there are four different types of process that we will look at in this course, each of which has a role to play in shaping language:
- Language use
- Language learning
- Language change (cultural evolution)
- Language evolution (biological evolution)
You'll notice immediately that terminology is letting us down a little here! I'm using "language evolution" both to refer to the overarching set of processes involved, and more specifically for the kind of evolution that involves changes in gene frequencies. Sorry about that! It is a source of a lot of confusion in the field. Some scholars like to use "language evolution" only in the narrowest sense (we might think of this as the biological evolution of the human faculty for language). Others prefer to cast the net a little wider and include both biological *and* cultural evolution. We'll see in a later lecture what I mean by cultural evolution, but for now you can think of this as the evolution of languages themselves rather that the evolution of the language faculty. Almost no-one takes the approach that I'm going with here by saying that everything from the way we speak from moment to moment (language use) to the way the human genome has evolved can be brought under the rubric "language evolution". However, I think for our purposes right now it's quite useful.
All these processes involve change over time, although the timescales are quite different. Decisions made when choosing how to formulate an utterance happen in milliseconds, whereas biological evolution takes millenia. We can further subdivide these four processes into two groupings. The first two happen at what I will call the *individual level*. Although language use typically involves two or more people, and language learning at the very least requires a language learner and another producer of language, the processes can reasonably be studied by looking at what individuals do. We think about language use and language learning by thinking about the minds of the indivdiuals involved.
Conversely, language change (or cultural evolution, if you prefer) and language evolution (or biological evolution) take place at the *population level*. These aren't really processes that are typically studied by looking at or thinking about individuals. Rather these are phenomenon that arise from the aggregate changes in populations of individuals.
That said, an understanding of the population level in some sense *requires* an understanding of the individual level. After all, what is a population other than a collection of individuals? In fact, an overarching message of this course is that to uncover a truly explanatory model of language you can't rely on a study of any of these processes in isolation. This is because there are deep and important interactions between all these processes, despite the fact that they take place on different timescales.

Learning and use of language rely on properties of our language ready brains (that's what we use to process language in real time, or learn language in the first place). Since our language ready brains are provided by the long process of the biological evolution of our species, then there's clearly an important causal link between biological evolution and learning/use. This is the message of the biolinguistic programme championed by people like Lenneberg and Chomsky in the 1960s.
Equally, the cultural evolution of language arises from the actions of a population of speakers and learners of language across time and space. Child learners and adult speakers shift and change properties of the language that is spoken. Indeed we can think of the dynamics of this cultural process as being ultimately determined by properties of the learning and processing mechanisms of these individuals. The actual universal structural properties of language that are the topic of our ultimate **why** question are the eventual product of this cultural evolutionary process.
But the causal interactions do not stop there. Biological evolution by natural selection is driven by differences in fitness. If we assume (as many do) that possession of language can alter fitness, then we have to assume that the nature of that language, arising from cultural evolution, may have a causal effect on the process of biological evolution.
Here then we have a cycle of causation crossing milliseconds to millenia and back again, and bridging the indivdiual and population levels of description. All to explain why language is the way it is!
This looks hopelessly complicated, and it becomes clear looking at this why researchers have tended to want to simplify the picture and look solely at one process or another.
In this course, I am going to show you that there is another way. We can begin to understand what these interacting processes do in general and on this basis build a modern explanatory framework for language based on evolutionary thinking. The way we're going to do that is to build **models**.
## What is a model?
We tend to think of a model as a miniaturised version of a system we are interested in, whether that's a model of the Eiffel tower that sits on our bookshelf reminding us of a trip to Paris, or a wave tank in a physics laboratory that helps us engineer coastal defences.
The value of having a miniature version of a system in science are, among other things, that: it is simpler than the real thing so that the effects of different sub-parts of the system can be more easily understood; it can be controlled more easily (we can try different coastal defenses against different types of waves without the huge cost of building them and waiting for the right storm); and the behaviour of the system can be more easily understood.
There are problems too, however. It may be difficult to build a model for a system that's being studied. We may not know enough about how that system works, or it may appear so irreducibly complex that any simplification is impossible. If we do simplify, we may not know which parts of the real thing we can ignore, and which are crucial.
Faced with these difficulties, it might appear safer to just study the real thing directly. Why build a model when we can simply observe the phenomenon we want to understand in the first place?
### What is a model for? One answer...
To understand when we might need a model it's worth thinking about the place of models in the scientific process.

One way to think about the actual practice of science is that we test theories about some phenomenon by working out what predictions those theories make and then testing those predictions against observations. The results of those tests may lead us to update our theories and then repeat the cycle again.
In many cases this process is pretty straightforward, but in some cases it turns out that it is not at all obvious what the predictions are that a theory is making. After all, to get from theory to predictions, we need to somehow intuit what would happen in the world if that theory were true. This might be possible for simple phenomena, but it turns out to be extraordinarily hard for what have come to be called *complex systems*. These are systems where there are lots of interacting subcomponents whose aggregate behaviour is somehow "emergent" from local interactions. We'll turn to a simple example from linguistics now.
### A simple example - explaining vowel distributions
Vowels can be thought of as existing in a two dimensional space. This is how they are represented in the IPA for example:

The dimensions of this representation correspond roughly to the position of the highest point of the tongue in the mouth when the vowel is produced. Interestingly, they also map fairly straightforwardly to the first and second formants of the acoustic spectrum of the vowel sound too.
Now, it turns out that if you look at the distribution of vowels in the world's languages only some patterns arise. For example, you never find a language in which the only three vowels are "i", "e", and "y". Specifically, the vowel space tends to be filled symmetrically. As a scientist studying language, we might spot this kind of pattern and look for an explanation. To do so, we first need a theory. A reasonable theory might be something like: **vowels tend to avoid being close to each other in order to maintain perceptual distinctiveness**.
So, how do we tell if this theory is correct? It might not be immediately obvious what predictions this theory makes. And without predictions we can't test the theory against the real data. (Actually, in this case, perhaps you do think it's obvious, but let's go along with this example for now!) If the predictions a theory makes are not immediately obvious, then this is where we need a model - something like the wave tank.
Well it turns out that in the 1970s, [Liljencrants and Lindblom (1972)](https://www.jstor.org/stable/411991) did just that and built a model of the vowel space, and it is similar in many ways to the wave tank model! In their paper they point out that vowels can be modelled using magnets attached to corks floating in water. If the magnets are set up to repel each other than the floating "vowels" will eventually organise themselves in wuch a way to maximise the distances between each other. Now, rather than get their hands wet, they were able to use what is known by physicists about how such repulsion works and predict what this model would do with given numbers of vowels. In this way, they constructed a model based on the theory that vowels maximise distinctiveness and compared this to the real cross-linguistic data.
### What is a model for? An alternative answer
Now, I've given a case for treating models as a bridge between theory and prediction. However, in some ways this sells the scientific process a little short in treating prediction as the only thing we're trying to do when we do science. An alternative or complementary approach sees models as tools for understanding.
*"Predictions are not the pinnacle of science. They are useful, especially for falsifying theories. However, predicting can’t be a model’s only purpose. ... surely the insights offered by a model are at least as important as its predictions: they help in understanding things by playing with them."* ([Sigmund, 1993, p. 4](https://books.google.co.uk/books/about/Games_of_Life.html?id=4G4vDwAAQBAJ&printsec=frontcover&source=kp_read_button&redir_esc=y))
A classic example of this kind of model is the ludicrously simple and abstract model of a kind of artificial life called, appropriately, the Game of Life. It was invented by [John Conway](https://web.archive.org/web/20090603015231/http://ddi.cs.uni-potsdam.de/HyFISCH/Produzieren/lis_projekt/proj_gamelife/ConwayScientificAmerican.htm) in 1970 and there are many implementations of it you can [play with in your web browswer](https://www.bjelic.net/game-of-life/game2.html). In this model, there is a square grid of "cells". Every cell has 8 neighbours that surround it. Cells can either be alive or dead. Each "round" of the game, the cells are updated such that some die and some come alive. The update works according to the following very simple rules:
- Any dead cell with 3 live neighbours becomes alive.
- Any live cell with 2 or 3 live neighbours stays alive.
- Otherwise the cell dies.
A simple sequence in the game of life from a particular starting position is shown here:

This a very short sequence of turns in the Game of Life. However, the remarkable thing about this incredibly simple "game" is that it can lead to enormously complex behaviours depending solely on the initial configuration of cells. In fact, just five living cells in the right organisation can lead to patterns that last thousands of rounds of the game, creating hugely elaborate and evolving structures.
This tells us something very important about the relationship between simplicity and complexity: complex systems can emerge from very simple interacting components!
### Simplicity and complexity in models
This talk of simplicity and complexity (which, you'll note, I have yet to define - more on that later in the course) raises an important methodological point. I have said that models are simplified versions of theories that allow us insight and to generate predictions, but how do we decide how close to reality a model should be? It's not entirely obvious where a model should sit on the spectrum between leaving nothing out and being as close as possible to the real thing, or leaving lots out and being as simple as possible.
In practice, a lot of the art of good modelling is finding the right spot on this spectrum. I tend to think that simpler is better up to a point, because what will we learn by building something as complex as the real thing? Simpler models are easier to build, easier to use, and are in my experience, likely to deliver better insights. But there are limits - you can't miss out important parts of the theory you are testing. Equally you should include as little extra that isn't in your theory that you can, because otherwise you won't know if the predictions you get actually correspond to the theory you're testing.
Anyway, you don't need to take my word for it. Einstein is said to have commented: ["everything should be as simple as possible, but not simpler."](https://quoteinvestigator.com/2011/05/13/einstein-simple/)
## Why use computers for modelling language?
We've seen that you can build physical models, or models based on mathematics borrowed from physics (see, e.g., [Servedio et al, 2014](https://doi.org/10.1371/journal.pbio.1002017), for more discussion of mathematical models), but clearly this course is about using computers to build models. Simulations are basically models that can be run on computers to generate predictions or insights. The Game of Life was originally developed using pieces of paper on the floor of a common room in a university department, but nowadays we explore the insights of that model by simulating it in a computer.
There are several reasons why computers can be good for modelling. Often a physical model simply can't be built that matches the theory you are investigating, or a purely mathematical (pencil and paper) model is too difficult (or indeed impossible) to construct. In these cases, the computer can step in and provide us an alomst unlimited potential for model building.
In particular, we tend to find that mathematical models become difficult to construct in problems that involve dynamic interactions. Notice that these are *precisely* the kinds of things that we're saying are responsible for the evolution of language! For example, if we want to understand how a child's knowledge changes as she responds to hearing thousands of words, or what happens when people interact in groups over thousands of years, or when communicating organisms evolve over millenia - in these cases, computational modelling is the solution.
Computers are absolutely perfect for constructing models with very many simple interacting components. Whereas we may balk at the thought of tracking hundreds of thousands of cells in the game of life over hundreds of thousands of generations, this is really trivial for a computer. The program to run the game of life is not much more complicated than the description of its rules, and - crucially - does not need to get more complicated simply because it is run for longer or for a greater number of cells.
The idea that simulation opens up our understanding of systems with many interacting elements has proved particularly valuable in allowing us to build the fundamentally evolutionary approach to understanding language that we've been aiming at.
In this course, we will be building and playing with models to tackle questions like:
- how do communicating species evolve?
- how are communication systems shaped by cultural evolution?
- where do grammatical generalisations come from?
- what do we mean when we say language is innate?
Despite the apparent magnitude of these questions, the main message I want to get across is how relatively straightforward it is to actually build these models. Even if you have never programmed a computer before, if you work through these lectures and labs carefully, you should be able to replicate some of the cutting edge results in the field of evolutionary linguistics and see how we can answer these kinds of questions without relying on purely rhetorical argument, and instead demonstrate our answers using working simulations.
|
github_jupyter
|
# Simulating Language 1, Why Simulate Language? (lecture)
*This is a first draft of lecture notes for the Simulating Language course. It probably contains lots of typos!*
## Some important questions
Before we get started, it's worth thinking about what it is we're wanting to achieve. A good first step is to ask what the central goal of linguistics as a science actually is. There are many reasonable answers to this question, of course, and they'll differ among the many subdisciplines of linguistics. For example, a descriptive linguist might answer that the goal of linguistics is a careful and detailed account of the structure of individual languages. A historical linguist might be in the business of attempting to reconstruct languages that are no longer spoken. But for many branches of linguistics, I think that a good approximation of the ultimate goal is an answer to a **why** question: "why is language the way it is?".
How might we go about approaching such a question?
## The evolutionary approach
In this course, we will take an evolutionary approach to language, with both language and evolution here taken very broadly indeed. The evolutionary approach attempts to answer the why question by posing instead a **how** question. The idea is that we can only really figure out why language is the way it is if we understand how it came to be that way.
This course will cover work carried out over the past 20 years or so, much of it pioneered here in Edinburgh, that tackles this how question as a way of providing a solid explanatory foundation for the science of language. A recent overview of this work, much of which we will replicate in this course, can be found in [Kirby (2017)](https://doi.org/10.3758/s13423-016-1166-7).
### The *processes* of language evolution
When we hear the term "evolution" we naturally start thinking about genes and natural selection, of ancient prehistory and the survival of the fittest. This is of course part of the story, but one of the exciting things about human behaviour is that it is not determined solely by genes. (Actually, nothing is determined solely by genes. Every organism is the product of development in a particular environment. We'll return to this in a later lecture.) If we are to answer our **how** question as a route to answering the ultimate **why** question, then we need to take a much broader view of what the processes are that are involved in the evolution of language. In other words, what are the different ways the nature of language comes into being?
Broadly speaking there are four different types of process that we will look at in this course, each of which has a role to play in shaping language:
- Language use
- Language learning
- Language change (cultural evolution)
- Language evolution (biological evolution)
You'll notice immediately that terminology is letting us down a little here! I'm using "language evolution" both to refer to the overarching set of processes involved, and more specifically for the kind of evolution that involves changes in gene frequencies. Sorry about that! It is a source of a lot of confusion in the field. Some scholars like to use "language evolution" only in the narrowest sense (we might think of this as the biological evolution of the human faculty for language). Others prefer to cast the net a little wider and include both biological *and* cultural evolution. We'll see in a later lecture what I mean by cultural evolution, but for now you can think of this as the evolution of languages themselves rather that the evolution of the language faculty. Almost no-one takes the approach that I'm going with here by saying that everything from the way we speak from moment to moment (language use) to the way the human genome has evolved can be brought under the rubric "language evolution". However, I think for our purposes right now it's quite useful.
All these processes involve change over time, although the timescales are quite different. Decisions made when choosing how to formulate an utterance happen in milliseconds, whereas biological evolution takes millenia. We can further subdivide these four processes into two groupings. The first two happen at what I will call the *individual level*. Although language use typically involves two or more people, and language learning at the very least requires a language learner and another producer of language, the processes can reasonably be studied by looking at what individuals do. We think about language use and language learning by thinking about the minds of the indivdiuals involved.
Conversely, language change (or cultural evolution, if you prefer) and language evolution (or biological evolution) take place at the *population level*. These aren't really processes that are typically studied by looking at or thinking about individuals. Rather these are phenomenon that arise from the aggregate changes in populations of individuals.
That said, an understanding of the population level in some sense *requires* an understanding of the individual level. After all, what is a population other than a collection of individuals? In fact, an overarching message of this course is that to uncover a truly explanatory model of language you can't rely on a study of any of these processes in isolation. This is because there are deep and important interactions between all these processes, despite the fact that they take place on different timescales.

Learning and use of language rely on properties of our language ready brains (that's what we use to process language in real time, or learn language in the first place). Since our language ready brains are provided by the long process of the biological evolution of our species, then there's clearly an important causal link between biological evolution and learning/use. This is the message of the biolinguistic programme championed by people like Lenneberg and Chomsky in the 1960s.
Equally, the cultural evolution of language arises from the actions of a population of speakers and learners of language across time and space. Child learners and adult speakers shift and change properties of the language that is spoken. Indeed we can think of the dynamics of this cultural process as being ultimately determined by properties of the learning and processing mechanisms of these individuals. The actual universal structural properties of language that are the topic of our ultimate **why** question are the eventual product of this cultural evolutionary process.
But the causal interactions do not stop there. Biological evolution by natural selection is driven by differences in fitness. If we assume (as many do) that possession of language can alter fitness, then we have to assume that the nature of that language, arising from cultural evolution, may have a causal effect on the process of biological evolution.
Here then we have a cycle of causation crossing milliseconds to millenia and back again, and bridging the indivdiual and population levels of description. All to explain why language is the way it is!
This looks hopelessly complicated, and it becomes clear looking at this why researchers have tended to want to simplify the picture and look solely at one process or another.
In this course, I am going to show you that there is another way. We can begin to understand what these interacting processes do in general and on this basis build a modern explanatory framework for language based on evolutionary thinking. The way we're going to do that is to build **models**.
## What is a model?
We tend to think of a model as a miniaturised version of a system we are interested in, whether that's a model of the Eiffel tower that sits on our bookshelf reminding us of a trip to Paris, or a wave tank in a physics laboratory that helps us engineer coastal defences.
The value of having a miniature version of a system in science are, among other things, that: it is simpler than the real thing so that the effects of different sub-parts of the system can be more easily understood; it can be controlled more easily (we can try different coastal defenses against different types of waves without the huge cost of building them and waiting for the right storm); and the behaviour of the system can be more easily understood.
There are problems too, however. It may be difficult to build a model for a system that's being studied. We may not know enough about how that system works, or it may appear so irreducibly complex that any simplification is impossible. If we do simplify, we may not know which parts of the real thing we can ignore, and which are crucial.
Faced with these difficulties, it might appear safer to just study the real thing directly. Why build a model when we can simply observe the phenomenon we want to understand in the first place?
### What is a model for? One answer...
To understand when we might need a model it's worth thinking about the place of models in the scientific process.

One way to think about the actual practice of science is that we test theories about some phenomenon by working out what predictions those theories make and then testing those predictions against observations. The results of those tests may lead us to update our theories and then repeat the cycle again.
In many cases this process is pretty straightforward, but in some cases it turns out that it is not at all obvious what the predictions are that a theory is making. After all, to get from theory to predictions, we need to somehow intuit what would happen in the world if that theory were true. This might be possible for simple phenomena, but it turns out to be extraordinarily hard for what have come to be called *complex systems*. These are systems where there are lots of interacting subcomponents whose aggregate behaviour is somehow "emergent" from local interactions. We'll turn to a simple example from linguistics now.
### A simple example - explaining vowel distributions
Vowels can be thought of as existing in a two dimensional space. This is how they are represented in the IPA for example:

The dimensions of this representation correspond roughly to the position of the highest point of the tongue in the mouth when the vowel is produced. Interestingly, they also map fairly straightforwardly to the first and second formants of the acoustic spectrum of the vowel sound too.
Now, it turns out that if you look at the distribution of vowels in the world's languages only some patterns arise. For example, you never find a language in which the only three vowels are "i", "e", and "y". Specifically, the vowel space tends to be filled symmetrically. As a scientist studying language, we might spot this kind of pattern and look for an explanation. To do so, we first need a theory. A reasonable theory might be something like: **vowels tend to avoid being close to each other in order to maintain perceptual distinctiveness**.
So, how do we tell if this theory is correct? It might not be immediately obvious what predictions this theory makes. And without predictions we can't test the theory against the real data. (Actually, in this case, perhaps you do think it's obvious, but let's go along with this example for now!) If the predictions a theory makes are not immediately obvious, then this is where we need a model - something like the wave tank.
Well it turns out that in the 1970s, [Liljencrants and Lindblom (1972)](https://www.jstor.org/stable/411991) did just that and built a model of the vowel space, and it is similar in many ways to the wave tank model! In their paper they point out that vowels can be modelled using magnets attached to corks floating in water. If the magnets are set up to repel each other than the floating "vowels" will eventually organise themselves in wuch a way to maximise the distances between each other. Now, rather than get their hands wet, they were able to use what is known by physicists about how such repulsion works and predict what this model would do with given numbers of vowels. In this way, they constructed a model based on the theory that vowels maximise distinctiveness and compared this to the real cross-linguistic data.
### What is a model for? An alternative answer
Now, I've given a case for treating models as a bridge between theory and prediction. However, in some ways this sells the scientific process a little short in treating prediction as the only thing we're trying to do when we do science. An alternative or complementary approach sees models as tools for understanding.
*"Predictions are not the pinnacle of science. They are useful, especially for falsifying theories. However, predicting can’t be a model’s only purpose. ... surely the insights offered by a model are at least as important as its predictions: they help in understanding things by playing with them."* ([Sigmund, 1993, p. 4](https://books.google.co.uk/books/about/Games_of_Life.html?id=4G4vDwAAQBAJ&printsec=frontcover&source=kp_read_button&redir_esc=y))
A classic example of this kind of model is the ludicrously simple and abstract model of a kind of artificial life called, appropriately, the Game of Life. It was invented by [John Conway](https://web.archive.org/web/20090603015231/http://ddi.cs.uni-potsdam.de/HyFISCH/Produzieren/lis_projekt/proj_gamelife/ConwayScientificAmerican.htm) in 1970 and there are many implementations of it you can [play with in your web browswer](https://www.bjelic.net/game-of-life/game2.html). In this model, there is a square grid of "cells". Every cell has 8 neighbours that surround it. Cells can either be alive or dead. Each "round" of the game, the cells are updated such that some die and some come alive. The update works according to the following very simple rules:
- Any dead cell with 3 live neighbours becomes alive.
- Any live cell with 2 or 3 live neighbours stays alive.
- Otherwise the cell dies.
A simple sequence in the game of life from a particular starting position is shown here:

This a very short sequence of turns in the Game of Life. However, the remarkable thing about this incredibly simple "game" is that it can lead to enormously complex behaviours depending solely on the initial configuration of cells. In fact, just five living cells in the right organisation can lead to patterns that last thousands of rounds of the game, creating hugely elaborate and evolving structures.
This tells us something very important about the relationship between simplicity and complexity: complex systems can emerge from very simple interacting components!
### Simplicity and complexity in models
This talk of simplicity and complexity (which, you'll note, I have yet to define - more on that later in the course) raises an important methodological point. I have said that models are simplified versions of theories that allow us insight and to generate predictions, but how do we decide how close to reality a model should be? It's not entirely obvious where a model should sit on the spectrum between leaving nothing out and being as close as possible to the real thing, or leaving lots out and being as simple as possible.
In practice, a lot of the art of good modelling is finding the right spot on this spectrum. I tend to think that simpler is better up to a point, because what will we learn by building something as complex as the real thing? Simpler models are easier to build, easier to use, and are in my experience, likely to deliver better insights. But there are limits - you can't miss out important parts of the theory you are testing. Equally you should include as little extra that isn't in your theory that you can, because otherwise you won't know if the predictions you get actually correspond to the theory you're testing.
Anyway, you don't need to take my word for it. Einstein is said to have commented: ["everything should be as simple as possible, but not simpler."](https://quoteinvestigator.com/2011/05/13/einstein-simple/)
## Why use computers for modelling language?
We've seen that you can build physical models, or models based on mathematics borrowed from physics (see, e.g., [Servedio et al, 2014](https://doi.org/10.1371/journal.pbio.1002017), for more discussion of mathematical models), but clearly this course is about using computers to build models. Simulations are basically models that can be run on computers to generate predictions or insights. The Game of Life was originally developed using pieces of paper on the floor of a common room in a university department, but nowadays we explore the insights of that model by simulating it in a computer.
There are several reasons why computers can be good for modelling. Often a physical model simply can't be built that matches the theory you are investigating, or a purely mathematical (pencil and paper) model is too difficult (or indeed impossible) to construct. In these cases, the computer can step in and provide us an alomst unlimited potential for model building.
In particular, we tend to find that mathematical models become difficult to construct in problems that involve dynamic interactions. Notice that these are *precisely* the kinds of things that we're saying are responsible for the evolution of language! For example, if we want to understand how a child's knowledge changes as she responds to hearing thousands of words, or what happens when people interact in groups over thousands of years, or when communicating organisms evolve over millenia - in these cases, computational modelling is the solution.
Computers are absolutely perfect for constructing models with very many simple interacting components. Whereas we may balk at the thought of tracking hundreds of thousands of cells in the game of life over hundreds of thousands of generations, this is really trivial for a computer. The program to run the game of life is not much more complicated than the description of its rules, and - crucially - does not need to get more complicated simply because it is run for longer or for a greater number of cells.
The idea that simulation opens up our understanding of systems with many interacting elements has proved particularly valuable in allowing us to build the fundamentally evolutionary approach to understanding language that we've been aiming at.
In this course, we will be building and playing with models to tackle questions like:
- how do communicating species evolve?
- how are communication systems shaped by cultural evolution?
- where do grammatical generalisations come from?
- what do we mean when we say language is innate?
Despite the apparent magnitude of these questions, the main message I want to get across is how relatively straightforward it is to actually build these models. Even if you have never programmed a computer before, if you work through these lectures and labs carefully, you should be able to replicate some of the cutting edge results in the field of evolutionary linguistics and see how we can answer these kinds of questions without relying on purely rhetorical argument, and instead demonstrate our answers using working simulations.
| 0.797636 | 0.985622 |
<a href="https://colab.research.google.com/github/jpslaga/AI-Crash-Course/blob/master/Front_Door_Detector.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Introduction
This is a rough pass to get a baseline for how effective we can be at searching through the images in a listing and identifying which, if any, are a picture of the front of the house.
Frankly, this is a difficult challenge. The dataset is primarily composed of pictures of the outside of houses, but we are trying to find the ones that are the picture of the front of the house. It's one thing to classify one image as a bicycle and another a bannana, but these images are all very similar. To make things even harder, we only have 700 images of front doors to train on.
I modeled much of this code on this blog post: https://blog.keras.io/building-powerful-image-classification-models-using-very-little-data.html
Another great post: https://blog.keras.io/building-powerful-image-classification-models-using-very-little-data.html
I wish I could remember where I got the data. Note to self, always cite your data source!! I think the original data source may have had photos from inside the house as well which might help the real production version deal with what it sees on Airbnb.
For now a copy of the data I used can be found on the shared drive in the demos folder. You can mount the drive using the controls on the left, or download the folder here: https://drive.google.com/drive/u/0/folders/1lTwGjUy9-MJtfnapxW-1ERT-p0Ius2Dc
The following code is written assuming that you mounted the shared drive.
```
import numpy as np
import os
import shutil
from keras.preprocessing.image import ImageDataGenerator
import matplotlib.pyplot as plt
from keras.models import Sequential
from keras.layers import Dropout, Flatten, Dense
from keras import applications
img_width, img_height = 224, 224
raw_train_data_dir = '/content/drive/Shared drives/BI Share/demos/front_door_data/train_raw'
raw_validation_data_dir = '/content/drive/Shared drives/BI Share/demos/front_door_data/validate_raw'
nbr_train_samples = 4687
nbr_validation_samples = 1172
epochs = 20
batch_size = 128
def plot_history(history):
plt.figure()
plt.xlabel('Epoch')
plt.ylabel('Loss: Binary Crossentropy')
plt.plot(history.epoch, np.array(history.history['loss']), label='Train Loss')
plt.plot(history.epoch, np.array(history.history['val_loss']), label = 'Val loss')
plt.legend()
plt.figure()
plt.xlabel('Epoch')
plt.ylabel('Accuracy')
plt.plot(history.epoch, np.array(history.history['accuracy']), label='Train Accuracy')
plt.plot(history.epoch, np.array(history.history['val_accuracy']), label = 'Val Accuracy')
plt.legend()
_, accuracy = model.evaluate(train_data, train_labels)
print('Train Accuracy: %.2f' % (accuracy*100))
_, accuracy = model.evaluate(validation_data, validation_labels)
print('Validation Accuracy: %.2f' % (accuracy*100))
# This is a data generator. It takes images as input and performs transformations for us.
# In this case we are using it to resize the images down to a much smaller size.
# This allows a smaller DNN to process the image, making the processing run faster.
datagen = ImageDataGenerator(rescale=1. / 255)
train_generator = datagen.flow_from_directory(
raw_train_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode="binary",
shuffle=False)
validation_generator = datagen.flow_from_directory(
raw_validation_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode='binary',
shuffle=False)
```
# Transfer Learning
Transfer learning is a technique where you can take a DNN that has been trained on a great deal of data, and has already figured out a lot of basic tasks, and then use that knowledge to power a slightly different task. In this case, we will be taking some DNNs that were trained to recognize a variety of household objects, and will respecialize them to recognize front doors. They are already good at recognizing shapes, colors, and patters, we just need a new way to interpret the final results of all that image processing.
```
# Load a pretrained model with the 'head' cut off. The output from this network will be an intermediate
# product, the result of the calculations from this DNN on the image, but no conclusion as to what
# to make of the image at the end.
# We are starting with the NASNetMobile DNN, which is fairly lightweight and high performance.
model = applications.NASNetMobile(include_top=False)
# Here we take the training data and run it through the loaded DNN. We save the output of
# this network and the labels to use as the input to a smaller, simpler network
# which sill only have the job of interpreting the results of the image processing
# and making a final decision about what the image represents.
train_data = model.predict(train_generator, nbr_train_samples)
train_labels = train_generator.labels
# Same treatment for the validation set, though one should not use
# data augmentation on validation data. You want the validation data
# to represent the task that will be seen in practice as closely as possible.
validation_data = model.predict(validation_generator, nbr_validation_samples)
validation_labels = validation_generator.labels
# Here we build a small DNN that takes the output of the pretrained model as input, interprets it,
# and renders a final verdict about what to make of the image.
model = Sequential()
model.add(Flatten(input_shape=train_data.shape[1:]))
model.add(Dense(8, activation='relu'))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(train_data, train_labels,
epochs=epochs,
batch_size=batch_size,
validation_data=(validation_data, validation_labels))
plot_history(history)
```
Now let's make two changes that might make this more accurate. Lets use a larger DNN for the image processing, and a larger DNN for interpreting the results.
```
train_generator = datagen.flow_from_directory(
raw_train_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode="binary",
shuffle=False)
validation_generator = datagen.flow_from_directory(
raw_validation_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode='binary',
shuffle=False)
# Here we are trying out the VGG16 DNN which is somewhat larger and slower to process, but offers
# somewhat better accuracy than NASNetMobile on industry standard benchmarking tests.
model = applications.VGG16(include_top=False)
train_data = model.predict(train_generator, nbr_train_samples)
train_labels = train_generator.labels
validation_data = model.predict(validation_generator, nbr_validation_samples)
validation_labels = validation_generator.labels
# Here we build a somewhat larger DNN that takes the output of the pretrained model as input, interprets it,
# and renders a final verdict about what to make of the image.
model = Sequential()
model.add(Flatten(input_shape=train_data.shape[1:]))
model.add(Dense(256, activation='relu'))
model.add(Dense(32, activation='relu'))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(train_data, train_labels,
epochs=epochs,
batch_size=batch_size,
validation_data=(validation_data, validation_labels))
plot_history(history)
```
There are a lot of lessons to be learned from this experience.
Bigger is not always better, and even if it is, suffers from diminishing returns.
The validation loss and accuracy had plateaued before 10 epochs were complete. After that, the validation loss actually gets worse over time. This is overfitting. We are getting better at guessing the training set, but getting worse when exposed to new data. The model has figured out that it can do better in the training exercises if it just memorizes the answers to these specific examples rather than learning generally applicable rules.
Notice that the loss gets worse for the validation set, while the accuracy remains stable. That's because accuracy only takes into account the binary value of whether the prediction was correct (True/False), while the loss measures just how much right or wrong the predictions were. If the actual value is 1, and the model outputs 0.49, it is wrong, but not **as** wrong as if it had output a 0.01. In this case, loss is the metric to watch.
We have already covered some techniques for dealing with overfitting, but we will continue to address the issue as we go.
# Data Augmentation
Now, let's see if we can make more of the data we have. We can use the images we have to generate some similar images that we might see. For instance, maybe we would see images under different lighting conditions, or rotated slightly however the photographer was holding the camera, or with things slightly off-center, or otherwise slightly different than what we have here. Keras has built-in functionality to make this easy.
```
augment_datagen = ImageDataGenerator(
rotation_range=15,
width_shift_range=0.15,
height_shift_range=0.15,
rescale=1./255,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
vertical_flip=False,
fill_mode='nearest',
brightness_range=(0.5, 1.5)
)
train_generator = augment_datagen.flow_from_directory(
raw_train_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode="binary",
shuffle=False)
# Note that we are using the non-augmented data generator for the validation dataset
validation_generator = datagen.flow_from_directory(
raw_validation_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode='binary',
shuffle=False)
# Let's go back to the smaller model for faster runtimes.
model = applications.NASNetMobile(include_top=False)
# Notice that we are looping over the training data 10 times taking different augmentations.
# This increased data volume should help our training efforts.
train_data = model.predict(train_generator, nbr_train_samples*10)
train_labels = train_generator.labels
validation_data = model.predict(validation_generator, nbr_validation_samples)
validation_labels = validation_generator.labels
model = Sequential()
model.add(Flatten(input_shape=train_data.shape[1:]))
model.add(Dense(32, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(8, activation='relu'))
model.add(Dropout(0.3))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(train_data, train_labels,
epochs=epochs,
batch_size=batch_size,
validation_data=(validation_data, validation_labels))
plot_history(history)
```
You might have noticed that our validation set has a lower loss and higher accuracy than our training set. How could we be performing better on the validation set, which the model has not seen, than we are doing on the training set? The answer is that while testing performance Keras uses dropout for the training data, but not for the validation data. This mimics what we would do in production. We use dropout while training the model, but when we deploy the model we want it to use all it's neurons at full power. Because the model is hamstringed on the training data it doesn't perform as well, even though it is the data it has seen before.
# Conclusion
While things will fluctuate from run to run, in my run I got a validation accuracy of 96.50%, not bad for a quick and dirty model on a highly challenging problem! Adding some dropout regularization and data augmentation took that model up to 96.93%. That's almost half a percent improved performance with some extra training-time effort to create a model that processes data just as fast. The larger, slower, VGG16 model performed the best with an accuracy on the validation set of 97.61%.
All of this was done with about two hours of processing time on the free service provided by colab. Everything for free, an afternoon of coding, and two hours processing and a very difficult image processing problem is all but solved!
# Next Steps
There are several ways we could try to make this model better.
For one, you could try out some different image processing DNNs. There are lots available. For a list of the ones available as part of the Keras applications package we were using here see: https://keras.io/api/applications/
.
You could try out different sizes and shapes of DNNs for interpreting the results of the output of the image processing DNN. That includes trying different activation functions, optimizers, learning rates, and regularization. I used a generic dropout here, but you could try different dropout approaches, and or use L2 or other regularization techniques. I got the best results with the VGG16 model, but is that improved performance due to better image processing from the VGG16 model, or from the larger DNN I used to interpret the results?
.
You could try finding more data to train with. I'm not sure I included all the original data in this example (though since I didn't remember to cite my data source I can't be sure). There might also be other data sets we could add to this one, or you could even go to Airbnb and download images yourself.
.
You could try different data augmentation strategies. I tried one setting here, but what happens if we try others?
.
I leave the exploration as an exercise for the student. If you do choose to pursue this, please let us know. We plan to build this as a production system near the beginning of 2021, and would love to know ahead of time what works well and what's a dead end.
|
github_jupyter
|
import numpy as np
import os
import shutil
from keras.preprocessing.image import ImageDataGenerator
import matplotlib.pyplot as plt
from keras.models import Sequential
from keras.layers import Dropout, Flatten, Dense
from keras import applications
img_width, img_height = 224, 224
raw_train_data_dir = '/content/drive/Shared drives/BI Share/demos/front_door_data/train_raw'
raw_validation_data_dir = '/content/drive/Shared drives/BI Share/demos/front_door_data/validate_raw'
nbr_train_samples = 4687
nbr_validation_samples = 1172
epochs = 20
batch_size = 128
def plot_history(history):
plt.figure()
plt.xlabel('Epoch')
plt.ylabel('Loss: Binary Crossentropy')
plt.plot(history.epoch, np.array(history.history['loss']), label='Train Loss')
plt.plot(history.epoch, np.array(history.history['val_loss']), label = 'Val loss')
plt.legend()
plt.figure()
plt.xlabel('Epoch')
plt.ylabel('Accuracy')
plt.plot(history.epoch, np.array(history.history['accuracy']), label='Train Accuracy')
plt.plot(history.epoch, np.array(history.history['val_accuracy']), label = 'Val Accuracy')
plt.legend()
_, accuracy = model.evaluate(train_data, train_labels)
print('Train Accuracy: %.2f' % (accuracy*100))
_, accuracy = model.evaluate(validation_data, validation_labels)
print('Validation Accuracy: %.2f' % (accuracy*100))
# This is a data generator. It takes images as input and performs transformations for us.
# In this case we are using it to resize the images down to a much smaller size.
# This allows a smaller DNN to process the image, making the processing run faster.
datagen = ImageDataGenerator(rescale=1. / 255)
train_generator = datagen.flow_from_directory(
raw_train_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode="binary",
shuffle=False)
validation_generator = datagen.flow_from_directory(
raw_validation_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode='binary',
shuffle=False)
# Load a pretrained model with the 'head' cut off. The output from this network will be an intermediate
# product, the result of the calculations from this DNN on the image, but no conclusion as to what
# to make of the image at the end.
# We are starting with the NASNetMobile DNN, which is fairly lightweight and high performance.
model = applications.NASNetMobile(include_top=False)
# Here we take the training data and run it through the loaded DNN. We save the output of
# this network and the labels to use as the input to a smaller, simpler network
# which sill only have the job of interpreting the results of the image processing
# and making a final decision about what the image represents.
train_data = model.predict(train_generator, nbr_train_samples)
train_labels = train_generator.labels
# Same treatment for the validation set, though one should not use
# data augmentation on validation data. You want the validation data
# to represent the task that will be seen in practice as closely as possible.
validation_data = model.predict(validation_generator, nbr_validation_samples)
validation_labels = validation_generator.labels
# Here we build a small DNN that takes the output of the pretrained model as input, interprets it,
# and renders a final verdict about what to make of the image.
model = Sequential()
model.add(Flatten(input_shape=train_data.shape[1:]))
model.add(Dense(8, activation='relu'))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(train_data, train_labels,
epochs=epochs,
batch_size=batch_size,
validation_data=(validation_data, validation_labels))
plot_history(history)
train_generator = datagen.flow_from_directory(
raw_train_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode="binary",
shuffle=False)
validation_generator = datagen.flow_from_directory(
raw_validation_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode='binary',
shuffle=False)
# Here we are trying out the VGG16 DNN which is somewhat larger and slower to process, but offers
# somewhat better accuracy than NASNetMobile on industry standard benchmarking tests.
model = applications.VGG16(include_top=False)
train_data = model.predict(train_generator, nbr_train_samples)
train_labels = train_generator.labels
validation_data = model.predict(validation_generator, nbr_validation_samples)
validation_labels = validation_generator.labels
# Here we build a somewhat larger DNN that takes the output of the pretrained model as input, interprets it,
# and renders a final verdict about what to make of the image.
model = Sequential()
model.add(Flatten(input_shape=train_data.shape[1:]))
model.add(Dense(256, activation='relu'))
model.add(Dense(32, activation='relu'))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(train_data, train_labels,
epochs=epochs,
batch_size=batch_size,
validation_data=(validation_data, validation_labels))
plot_history(history)
augment_datagen = ImageDataGenerator(
rotation_range=15,
width_shift_range=0.15,
height_shift_range=0.15,
rescale=1./255,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
vertical_flip=False,
fill_mode='nearest',
brightness_range=(0.5, 1.5)
)
train_generator = augment_datagen.flow_from_directory(
raw_train_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode="binary",
shuffle=False)
# Note that we are using the non-augmented data generator for the validation dataset
validation_generator = datagen.flow_from_directory(
raw_validation_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode='binary',
shuffle=False)
# Let's go back to the smaller model for faster runtimes.
model = applications.NASNetMobile(include_top=False)
# Notice that we are looping over the training data 10 times taking different augmentations.
# This increased data volume should help our training efforts.
train_data = model.predict(train_generator, nbr_train_samples*10)
train_labels = train_generator.labels
validation_data = model.predict(validation_generator, nbr_validation_samples)
validation_labels = validation_generator.labels
model = Sequential()
model.add(Flatten(input_shape=train_data.shape[1:]))
model.add(Dense(32, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(8, activation='relu'))
model.add(Dropout(0.3))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(train_data, train_labels,
epochs=epochs,
batch_size=batch_size,
validation_data=(validation_data, validation_labels))
plot_history(history)
| 0.847684 | 0.984381 |
```
import pandas as pd
import numpy as np
from matplotlib.pyplot import figure
import matplotlib.pyplot as plt
#path = "lstm-out/NO-SCALE-1-5-100-120-60-30-0.01-1.csv"
def readFile(path):
df = pd.read_csv("lstm-out/"+path, delimiter=',', index_col=0, parse_dates=True)
return df
def getMean(df, m="MAE"):
#print(df[m])
print(df[m].mean())
def getMeanNaive(df, m="NAIVE_MAPE"):
columns = ['MAE', 'RMSE', 'MAPE', 'MEAN_MAE', 'MEAN_RMSE', 'MEAN_MAPE', 'NAIVE_MAE', 'NAIVE_RMSE', 'NAIVE_MAE']
df = df.set_axis(['MAE', 'RMSE', 'MAPE', 'MEAN_MAE', 'MEAN_RMSE', 'MEAN_MAPE', 'NAIVE_MAE', 'NAIVE_RMSE', 'NAIVE_MAPE'], axis=1, inplace=False)
print(df[m].mean())
def graph(df):
columns = ['MAE', 'RMSE', 'MAPE', 'MEAN_MAE', 'MEAN_RMSE', 'MEAN_MAPE', 'NAIVE_MAE', 'NAIVE_RMSE', 'NAIVE_MAE']
df = df.set_axis(['MAE', 'RMSE', 'MAPE', 'MEAN_MAE', 'MEAN_RMSE', 'MEAN_MAPE', 'NAIVE_MAE', 'NAIVE_RMSE', 'NAIVE_MAPE'], axis=1, inplace=False)
#print(df)
figure(num=None, figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
X = np.arange(50)
X = X[::-1]
states = list(df.index)
#plt.bar(X, df.iloc[:, 0], color='blue', width = 0.25, tick_label=states)
#plt.bar(X+0.25, df.iloc[:, 4], color='red', width = 0.25, tick_label=states)
handle1 = plt.bar(X, df['MAPE'], color='blue', width = 0.25, tick_label=states, label="LSTM")
#plt.bar(X+0.5, df['NAIVE_MAE'], color='green', width = 0.25, tick_label=states)
handle2 = plt.bar(X+0.25, df['MEAN_MAPE'], color='red', width = 0.25, tick_label=states, label="Naive Mean")
plt.legend(handles = [handle1, handle2])
plt.ylabel("Mean Absolute Error")
plt.xlabel("Time Series")
plt.savefig("lstm-out/concur_perf", dpi=200, bbox_inches='tight')
plt.show()
def arima_graph(df):
figure(num=None, figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
X = np.arange(50)
X = X[::-1]
states = list(df.index)
#plt.bar(X, df.iloc[:, 0], color='blue', width = 0.25, tick_label=states)
#plt.bar(X+0.25, df.iloc[:, 4], color='red', width = 0.25, tick_label=states)
handle1 = plt.bar(X, df['3pt MAE'], color='blue', width = 0.25, tick_label=states, label="ARIMA")
#plt.bar(X+0.5, df['NAIVE_MAE'], color='green', width = 0.25, tick_label=states)
handle2 = plt.bar(X+0.25, df['3pt MEAN MAE'], color='red', width = 0.25, tick_label=states, label="Naive Mean")
plt.legend(handles = [handle1, handle2])
plt.ylabel("Mean Absolute Error")
plt.xlabel("Time Series")
plt.savefig("lstm-out/arima_short", dpi=200, bbox_inches='tight')
plt.show()
print("0 dropout")
paths = []
# number of hidden parameters
paths.append("1-2-100-6-3-30-0.01-0.csv")
paths.append("1-5-100-6-3-30-0.01-0.csv")
paths.append("1-10-100-6-3-30-0.01-0.csv")
paths.append("1-20-100-6-3-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("0.5 dropout")
paths = []
# number of hidden parameters
paths.append("1-2-100-6-3-30-0.01-0.5.csv")
paths.append("1-5-100-6-3-30-0.01-0.5.csv")
paths.append("1-10-100-6-3-30-0.01-0.5.csv")
paths.append("1-20-100-6-3-30-0.01-0.5.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("1 dropout")
paths = []
# number of hidden parameters
paths.append("1-2-100-6-3-30-0.01-1.csv")
paths.append("1-5-100-6-3-30-0.01-1.csv")
paths.append("1-10-100-6-3-30-0.01-1.csv")
paths.append("1-20-100-6-3-30-0.01-1.csv")
for path in paths:
df = readFile(path)
getMean(df)
#graph(readFile(paths[0]))
#getMeanNaive(readFile(paths[0]))
print("naive errors")
getMeanNaive(readFile(paths[0]), "NAIVE_MAE")
getMeanNaive(readFile(paths[0]), "MEAN_MAE")
getMeanNaive(readFile(paths[0]), "NAIVE_MAPE")
getMeanNaive(readFile(paths[0]), "MEAN_MAPE")
# performance is best with 5-10 hidden parameters and lower drop out
# Same but 2 layers
print("0 dropout")
paths = []
# number of hidden parameters
paths.append("2-2-100-6-3-30-0.01-0.csv")
paths.append("2-5-100-6-3-30-0.01-0.csv")
paths.append("2-10-100-6-3-30-0.01-0.csv")
paths.append("2-20-100-6-3-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("0.5 dropout")
paths = []
# number of hidden parameters
paths.append("2-2-100-6-3-30-0.01-0.5.csv")
paths.append("2-5-100-6-3-30-0.01-0.5.csv")
paths.append("2-10-100-6-3-30-0.01-0.5.csv")
paths.append("2-20-100-6-3-30-0.01-0.5.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("1 dropout")
paths = []
# number of hidden parameters
paths.append("2-2-100-6-3-30-0.01-1.csv")
paths.append("2-5-100-6-3-30-0.01-1.csv")
paths.append("2-10-100-6-3-30-0.01-1.csv")
paths.append("2-20-100-6-3-30-0.01-1.csv")
for path in paths:
df = readFile(path)
getMean(df)
#graph(readFile(paths[0]))
# performance is best with 5-10 hidden parameters and lower drop out
# Same but 3 layers
print("0 dropout")
paths = []
# number of hidden parameters
paths.append("3-2-100-6-3-30-0.01-0.csv")
paths.append("3-5-100-6-3-30-0.01-0.csv")
paths.append("3-10-100-6-3-30-0.01-0.csv")
paths.append("3-20-100-6-3-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("0.5 dropout")
paths = []
# number of hidden parameters
paths.append("3-2-100-6-3-30-0.01-0.5.csv")
paths.append("3-5-100-6-3-30-0.01-0.5.csv")
paths.append("3-10-100-6-3-30-0.01-0.5.csv")
paths.append("3-20-100-6-3-30-0.01-0.5.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("1 dropout")
paths = []
# number of hidden parameters
paths.append("3-2-100-6-3-30-0.01-1.csv")
paths.append("3-5-100-6-3-30-0.01-1.csv")
paths.append("3-10-100-6-3-30-0.01-1.csv")
paths.append("3-20-100-6-3-30-0.01-1.csv")
for path in paths:
df = readFile(path)
getMean(df)
#graph(readFile(paths[0]))
# performance is best with 5-10 hidden parameters and lower drop out
# Long Term Analysis
print("1 layer")
paths = []
paths.append("1-5-100-432-216-30-0.01-0.csv")
paths.append("1-10-100-432-216-30-0.01-0.csv")
paths.append("1-20-100-432-216-30-0.01-0.csv")
paths.append("1-30-100-432-216-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("2 layers")
paths = []
paths.append("2-5-100-432-216-30-0.01-0.csv")
paths.append("2-10-100-432-216-30-0.01-0.csv")
paths.append("2-20-100-432-216-30-0.01-0.csv")
paths.append("2-30-100-432-216-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("3 layers")
paths = []
paths.append("3-5-100-432-216-30-0.01-0.csv")
paths.append("3-10-100-432-216-30-0.01-0.csv")
paths.append("3-20-100-432-216-30-0.01-0.csv")
paths.append("3-30-100-432-216-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
#graph(readFile(paths[0]))
# performance is best with 5-10 hidden parameters and lower drop out
# no resample
paths = []
paths.append("NO-SCALE-1-5-100-120-60-30-0.01-1.csv")
paths.append("NO-SCALE-1-10-100-120-60-30-0.01-1.csv")
paths.append("NO-SCALE-1-20-100-120-60-30-0.01-1.csv")
paths.append("NO-SCALE-2-5-100-120-60-30-0.01-1.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("naive perf")
for path in paths:
df = readFile(path)
getMeanNaive(df)
#df = readFile("3-20-100-532-216-40-0.01-0.1.csv")
#df = readFile("trash/NO-SCALE-1-10-100-120-60-30-0.01-1.csv")
df = readFile("concur-2-10-100-6-3-50-0.01-0.5.csv")
#df = readFile("4-20-100-6-3-30-0.01-0.csv")
#df = readFile("2-20-100-6-3-30-0.01-0.csv")
#print(df)
#graph(df)
#print(df)
print(getMean(df, m="MAE"))
print(count)
print(df['MAE'].mean())
print(df['MAPE'].mean())
# arima analysis
df = pd.read_csv("arima_results.csv", delimiter=',', index_col=0, parse_dates=True)
#print(df)
print(df['3pt MAE'].mean())
print(df['MAE'].mean())
print(df['3pt MAPE'].mean())
print(df['MAPE'].mean())
#arima_graph(df)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from matplotlib.pyplot import figure
import matplotlib.pyplot as plt
#path = "lstm-out/NO-SCALE-1-5-100-120-60-30-0.01-1.csv"
def readFile(path):
df = pd.read_csv("lstm-out/"+path, delimiter=',', index_col=0, parse_dates=True)
return df
def getMean(df, m="MAE"):
#print(df[m])
print(df[m].mean())
def getMeanNaive(df, m="NAIVE_MAPE"):
columns = ['MAE', 'RMSE', 'MAPE', 'MEAN_MAE', 'MEAN_RMSE', 'MEAN_MAPE', 'NAIVE_MAE', 'NAIVE_RMSE', 'NAIVE_MAE']
df = df.set_axis(['MAE', 'RMSE', 'MAPE', 'MEAN_MAE', 'MEAN_RMSE', 'MEAN_MAPE', 'NAIVE_MAE', 'NAIVE_RMSE', 'NAIVE_MAPE'], axis=1, inplace=False)
print(df[m].mean())
def graph(df):
columns = ['MAE', 'RMSE', 'MAPE', 'MEAN_MAE', 'MEAN_RMSE', 'MEAN_MAPE', 'NAIVE_MAE', 'NAIVE_RMSE', 'NAIVE_MAE']
df = df.set_axis(['MAE', 'RMSE', 'MAPE', 'MEAN_MAE', 'MEAN_RMSE', 'MEAN_MAPE', 'NAIVE_MAE', 'NAIVE_RMSE', 'NAIVE_MAPE'], axis=1, inplace=False)
#print(df)
figure(num=None, figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
X = np.arange(50)
X = X[::-1]
states = list(df.index)
#plt.bar(X, df.iloc[:, 0], color='blue', width = 0.25, tick_label=states)
#plt.bar(X+0.25, df.iloc[:, 4], color='red', width = 0.25, tick_label=states)
handle1 = plt.bar(X, df['MAPE'], color='blue', width = 0.25, tick_label=states, label="LSTM")
#plt.bar(X+0.5, df['NAIVE_MAE'], color='green', width = 0.25, tick_label=states)
handle2 = plt.bar(X+0.25, df['MEAN_MAPE'], color='red', width = 0.25, tick_label=states, label="Naive Mean")
plt.legend(handles = [handle1, handle2])
plt.ylabel("Mean Absolute Error")
plt.xlabel("Time Series")
plt.savefig("lstm-out/concur_perf", dpi=200, bbox_inches='tight')
plt.show()
def arima_graph(df):
figure(num=None, figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
X = np.arange(50)
X = X[::-1]
states = list(df.index)
#plt.bar(X, df.iloc[:, 0], color='blue', width = 0.25, tick_label=states)
#plt.bar(X+0.25, df.iloc[:, 4], color='red', width = 0.25, tick_label=states)
handle1 = plt.bar(X, df['3pt MAE'], color='blue', width = 0.25, tick_label=states, label="ARIMA")
#plt.bar(X+0.5, df['NAIVE_MAE'], color='green', width = 0.25, tick_label=states)
handle2 = plt.bar(X+0.25, df['3pt MEAN MAE'], color='red', width = 0.25, tick_label=states, label="Naive Mean")
plt.legend(handles = [handle1, handle2])
plt.ylabel("Mean Absolute Error")
plt.xlabel("Time Series")
plt.savefig("lstm-out/arima_short", dpi=200, bbox_inches='tight')
plt.show()
print("0 dropout")
paths = []
# number of hidden parameters
paths.append("1-2-100-6-3-30-0.01-0.csv")
paths.append("1-5-100-6-3-30-0.01-0.csv")
paths.append("1-10-100-6-3-30-0.01-0.csv")
paths.append("1-20-100-6-3-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("0.5 dropout")
paths = []
# number of hidden parameters
paths.append("1-2-100-6-3-30-0.01-0.5.csv")
paths.append("1-5-100-6-3-30-0.01-0.5.csv")
paths.append("1-10-100-6-3-30-0.01-0.5.csv")
paths.append("1-20-100-6-3-30-0.01-0.5.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("1 dropout")
paths = []
# number of hidden parameters
paths.append("1-2-100-6-3-30-0.01-1.csv")
paths.append("1-5-100-6-3-30-0.01-1.csv")
paths.append("1-10-100-6-3-30-0.01-1.csv")
paths.append("1-20-100-6-3-30-0.01-1.csv")
for path in paths:
df = readFile(path)
getMean(df)
#graph(readFile(paths[0]))
#getMeanNaive(readFile(paths[0]))
print("naive errors")
getMeanNaive(readFile(paths[0]), "NAIVE_MAE")
getMeanNaive(readFile(paths[0]), "MEAN_MAE")
getMeanNaive(readFile(paths[0]), "NAIVE_MAPE")
getMeanNaive(readFile(paths[0]), "MEAN_MAPE")
# performance is best with 5-10 hidden parameters and lower drop out
# Same but 2 layers
print("0 dropout")
paths = []
# number of hidden parameters
paths.append("2-2-100-6-3-30-0.01-0.csv")
paths.append("2-5-100-6-3-30-0.01-0.csv")
paths.append("2-10-100-6-3-30-0.01-0.csv")
paths.append("2-20-100-6-3-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("0.5 dropout")
paths = []
# number of hidden parameters
paths.append("2-2-100-6-3-30-0.01-0.5.csv")
paths.append("2-5-100-6-3-30-0.01-0.5.csv")
paths.append("2-10-100-6-3-30-0.01-0.5.csv")
paths.append("2-20-100-6-3-30-0.01-0.5.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("1 dropout")
paths = []
# number of hidden parameters
paths.append("2-2-100-6-3-30-0.01-1.csv")
paths.append("2-5-100-6-3-30-0.01-1.csv")
paths.append("2-10-100-6-3-30-0.01-1.csv")
paths.append("2-20-100-6-3-30-0.01-1.csv")
for path in paths:
df = readFile(path)
getMean(df)
#graph(readFile(paths[0]))
# performance is best with 5-10 hidden parameters and lower drop out
# Same but 3 layers
print("0 dropout")
paths = []
# number of hidden parameters
paths.append("3-2-100-6-3-30-0.01-0.csv")
paths.append("3-5-100-6-3-30-0.01-0.csv")
paths.append("3-10-100-6-3-30-0.01-0.csv")
paths.append("3-20-100-6-3-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("0.5 dropout")
paths = []
# number of hidden parameters
paths.append("3-2-100-6-3-30-0.01-0.5.csv")
paths.append("3-5-100-6-3-30-0.01-0.5.csv")
paths.append("3-10-100-6-3-30-0.01-0.5.csv")
paths.append("3-20-100-6-3-30-0.01-0.5.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("1 dropout")
paths = []
# number of hidden parameters
paths.append("3-2-100-6-3-30-0.01-1.csv")
paths.append("3-5-100-6-3-30-0.01-1.csv")
paths.append("3-10-100-6-3-30-0.01-1.csv")
paths.append("3-20-100-6-3-30-0.01-1.csv")
for path in paths:
df = readFile(path)
getMean(df)
#graph(readFile(paths[0]))
# performance is best with 5-10 hidden parameters and lower drop out
# Long Term Analysis
print("1 layer")
paths = []
paths.append("1-5-100-432-216-30-0.01-0.csv")
paths.append("1-10-100-432-216-30-0.01-0.csv")
paths.append("1-20-100-432-216-30-0.01-0.csv")
paths.append("1-30-100-432-216-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("2 layers")
paths = []
paths.append("2-5-100-432-216-30-0.01-0.csv")
paths.append("2-10-100-432-216-30-0.01-0.csv")
paths.append("2-20-100-432-216-30-0.01-0.csv")
paths.append("2-30-100-432-216-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("3 layers")
paths = []
paths.append("3-5-100-432-216-30-0.01-0.csv")
paths.append("3-10-100-432-216-30-0.01-0.csv")
paths.append("3-20-100-432-216-30-0.01-0.csv")
paths.append("3-30-100-432-216-30-0.01-0.csv")
for path in paths:
df = readFile(path)
getMean(df)
#graph(readFile(paths[0]))
# performance is best with 5-10 hidden parameters and lower drop out
# no resample
paths = []
paths.append("NO-SCALE-1-5-100-120-60-30-0.01-1.csv")
paths.append("NO-SCALE-1-10-100-120-60-30-0.01-1.csv")
paths.append("NO-SCALE-1-20-100-120-60-30-0.01-1.csv")
paths.append("NO-SCALE-2-5-100-120-60-30-0.01-1.csv")
for path in paths:
df = readFile(path)
getMean(df)
print("naive perf")
for path in paths:
df = readFile(path)
getMeanNaive(df)
#df = readFile("3-20-100-532-216-40-0.01-0.1.csv")
#df = readFile("trash/NO-SCALE-1-10-100-120-60-30-0.01-1.csv")
df = readFile("concur-2-10-100-6-3-50-0.01-0.5.csv")
#df = readFile("4-20-100-6-3-30-0.01-0.csv")
#df = readFile("2-20-100-6-3-30-0.01-0.csv")
#print(df)
#graph(df)
#print(df)
print(getMean(df, m="MAE"))
print(count)
print(df['MAE'].mean())
print(df['MAPE'].mean())
# arima analysis
df = pd.read_csv("arima_results.csv", delimiter=',', index_col=0, parse_dates=True)
#print(df)
print(df['3pt MAE'].mean())
print(df['MAE'].mean())
print(df['3pt MAPE'].mean())
print(df['MAPE'].mean())
#arima_graph(df)
| 0.281109 | 0.396535 |
---
# MODEL BUILDING PROCESSING
---
```
import pandas as pd
import numpy as np
import random
import seaborn as sn
import matplotlib.pyplot as plt
import pickle
dataset = pd.read_csv(r'./data/coords.csv')
```
## Spliting Data
---
```
X = dataset.drop(columns = ["class"])
y = dataset["class"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.25)
```
## Feature Scaling
```
from sklearn.preprocessing import StandardScaler
sc_X = StandardScaler()
X_train2 = pd.DataFrame(sc_X.fit_transform(X_train))
# reassingn index and columns
X_train2.columns = X_train.columns.values
X_train2.index = X_train.index.values
X_train_scaled = X_train2
X_test2 = pd.DataFrame(sc_X.transform(X_test))
X_test2.columns = X_test.columns.values
X_test2.index = X_test.index.values
X_test_scaled = X_test2
```
## Function to check perfomance
```
from sklearn.metrics import confusion_matrix, accuracy_score, f1_score, precision_score, recall_score
def perfomance_check(name: str):
acc = accuracy_score(y_test, y_pred)
prec = precision_score(y_test, y_pred, pos_label='positive',
average='micro')
rec = recall_score(y_test, y_pred, pos_label='positive',
average='micro')
f1 = f1_score(y_test, y_pred, pos_label='positive',
average='micro')
model_results = pd.DataFrame([[name, acc, prec, rec, f1]],
columns = ['Model', 'Accuracy', 'Precision', 'Recall', 'F1 Score'])
return results.append(model_results, ignore_index = True)
results = pd.DataFrame(columns = ['Model', 'Accuracy', 'Precision', 'Recall', 'F1 Score'])
```
---
# CLASSIFICATIONS
---
## Logical Regression
```
from sklearn.linear_model import LogisticRegression
LR_classifier = LogisticRegression(random_state = 0, penalty = 'l1', solver='saga')
LR_classifier.fit(X_train, y_train.values.ravel())
y_pred = LR_classifier.predict(X_test)
results = perfomance_check('Linear Regression (Lasso)')
```
## KNN (K-Nearest Neighbours)
```
from sklearn.neighbors import KNeighborsClassifier
KNN_classifier = KNeighborsClassifier(n_neighbors = 5, metric = 'minkowski', p = 2)
KNN_classifier.fit(X_train_scaled, y_train.values.ravel())
y_pred = KNN_classifier.predict(X_test_scaled)
results = perfomance_check('K-Nearest Neighbours')
```
## SVM
```
from sklearn.svm import SVC
SVML_classifier = SVC(random_state = 0, kernel = 'linear')
SVML_classifier.fit(X_train, y_train.values.ravel())
y_pred = SVML_classifier.predict(X_test)
results = perfomance_check('SVM (Linear)')
```
## Kernal SVM
```
from sklearn.svm import SVC
K_SVM_classifier = SVC(random_state = 0, kernel = 'rbf')
K_SVM_classifier.fit(X_train, y_train.values.ravel())
y_pred = K_SVM_classifier.predict(X_test)
results = perfomance_check('SVM (RBF)')
```
## Naive Bayes
```
from sklearn.naive_bayes import GaussianNB
NB_classifier = GaussianNB()
NB_classifier.fit(X_train, y_train.values.ravel())
y_pred = NB_classifier.predict(X_test)
results = perfomance_check('Naive Bayes')
```
## Decision Tree Classification
```
from sklearn.tree import DecisionTreeClassifier
DTC_classifier = DecisionTreeClassifier(criterion = 'entropy', random_state = 0)
DTC_classifier.fit(X_train, y_train.values.ravel())
y_pred = DTC_classifier.predict(X_test)
results = perfomance_check('Decision Tree Classification')
```
## Random Forest Classification
```
from sklearn.ensemble import RandomForestClassifier
RF_classifier = RandomForestClassifier(random_state = 0, n_estimators = 100,
criterion = 'entropy')
RF_classifier.fit(X_train, y_train.values.ravel())
y_pred = RF_classifier.predict(X_test)
results = perfomance_check('Random Forest (n=100)')
```
## XGBoost Classifier
```
from xgboost import XGBClassifier
xgb_classifier = XGBClassifier()
xgb_classifier.fit(X_train, y_train.values.ravel())
y_pred = xgb_classifier.predict(X_test)
results = perfomance_check('XGBoost ')
```
## CatBoost Classifier
from catboost import CatBoostClassifier
CB_classifier = CatBoostClassifier()
CB_classifier.fit(X_train, y_train.values.ravel())
y_pred = CB_classifier.predict(X_test)
results = perfomance_check('CatBoost')
```
results
```
## Save Model
```
with open('./saved_model/body_language.pkl', 'wb') as f:
pickle.dump(RF_classifier, f)
with open('./saved_model/body_language.pkl', 'rb') as f:
model = pickle.load(f)
```
---
# MODEL SELECTION
---
### K-fold Cross Validation
---
* Conside which model perform the best
```
from sklearn.model_selection import cross_val_score
model_lst = [LR_classifier, # 0
KNN_classifier, # 1
SVML_classifier, # 2
K_SVM_classifier, # 3
NB_classifier, # 4
DTC_classifier, # 5
RF_classifier, # 6
xgb_classifier] # 7
# CB_classifier] # 8
msg = []
for i in range(len(model_lst)):
accuracies = cross_val_score(estimator =model_lst[i] , X = X_train, y = y_train, cv = 10)
msg.append(f"Model Accuracy {i}: %0.3f (+/- %0.3f)" % (accuracies.mean(), accuracies.std() * 2))
for i in msg:
print(i)
for i in msg:
print(i)
# This is the script to compare result with prediction
comparision = pd.DataFrame(columns = ['Result', 'Prediction'])
comparision.Prediction = pd.Series(y_pred)
comparision.Result = y_test.values
```
## Parameter Tuning
---
Base on the critera we can choose what type of parameter tuning algorithm to use.
#### Grid Search: Entropy
* Meant to maximize the information content (in random forest we maximize the info. at every split)
* pip install joblib
* update joblib if there are some problem in GridSearch
#### Grid Search: Gini
* Meant to minimize the probability of mislabelling
### Input parameters
---
* Input different parameter based on the "Model" that you tring to tune
* Look into documentaion of specific algorithm for avaiable parameters
* Based on the best found parametes, slim down the range of the setting and test again
.
***1st setting***
{"max_depth": [3, None],
"max_features": [1, 5, 10],
'min_samples_split': [2, 5, 10],
'min_samples_leaf': [1, 5, 10],
"bootstrap": [True, False],
"criterion": ["entropy"]}
.
***Slimed down version***
{"max_depth": [None],
"max_features": [3, 5, 7],
'min_samples_split': [8, 10, 12],
'min_samples_leaf': [1, 2, 3],
"bootstrap": [True],
"criterion": ["entropy"]}
```
parameters = {"max_depth": [3, None],
"max_features": [1, 5, 10],
'min_samples_split': [2, 5, 10],
'min_samples_leaf': [1, 5, 10],
"bootstrap": [True, False],
"criterion": ["entropy"]}
```
### Grid Search: Entropy
```
from sklearn.model_selection import GridSearchCV
grid_search_entropy = GridSearchCV(estimator = RF_classifier, # Make sure classifier points to the RF model
param_grid = parameters,
scoring = "accuracy",
cv = 10,
n_jobs = -1)
t0 = time.time() # RECORD THE DURATION ALGORITHM TOOK
grid_search = grid_search.fit(X_train, y_train)
t1 = time.time()
print("Took %0.2f seconds" % (t1 - t0))
rf_best_accuracy = grid_search.best_score_
rf_best_parameters = grid_search.best_params_
rf_best_accuracy, rf_best_parameters
```
### Grid Search: Gini
```
parameters = {"max_depth": [3, None],
"max_features": [1, 5, 10],
'min_samples_split': [2, 5, 10],
'min_samples_leaf': [1, 5, 10],
"bootstrap": [True, False],
"criterion": ["gini"]}
from sklearn.model_selection import GridSearchCV
grid_search_gini = GridSearchCV(estimator = classifier, # Make sure classifier points to the RF model
param_grid = parameters,
scoring = "accuracy",
cv = 10,
n_jobs = -1)
t0 = time.time()
grid_search = grid_search.fit(X_train, y_train)
t1 = time.time()
print("Took %0.2f seconds" % (t1 - t0))
rf_best_accuracy = grid_search.best_score_
rf_best_parameters = grid_search.best_params_
rf_best_accuracy, rf_best_parameters
```
## Save Model
```
with open('body_language.pkl', 'wb') as f:
pickle.dump(grid_search_entropy, f)
with open('body_language.pkl', 'rb') as f:
grid_search_entropy = pickle.load(f)
```
### Predicting Test Set
---
* Base on the best grid result test model, using its specific grid_serach parameter
```
# Use Correct grid_search 'entropy' or 'gini'
y_pred = grid_search_entropy.predict(X_test)
from sklearn.metrics import classification_report, confusion_matrix
print(classification_report(y_test,y_pred))
cm = confusion_matrix(y_test, y_pred)
sn.set(font_scale=2.8)
mask = np.zeros_like(corr, dtype=np.bool)
mask[np.triu_indices_from(mask)] = True
plt.figure(figsize = (50,40))
sn.heatmap(corr, annot=True,mask=mask);
```
### Formatting Final Results
```
final_results = pd.concat([y_test, users], axis = 1).dropna()
final_results['predictions'] = y_pred
final_results = final_results[['entry_id', 'e_signed', 'predictions']]
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import random
import seaborn as sn
import matplotlib.pyplot as plt
import pickle
dataset = pd.read_csv(r'./data/coords.csv')
X = dataset.drop(columns = ["class"])
y = dataset["class"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.25)
from sklearn.preprocessing import StandardScaler
sc_X = StandardScaler()
X_train2 = pd.DataFrame(sc_X.fit_transform(X_train))
# reassingn index and columns
X_train2.columns = X_train.columns.values
X_train2.index = X_train.index.values
X_train_scaled = X_train2
X_test2 = pd.DataFrame(sc_X.transform(X_test))
X_test2.columns = X_test.columns.values
X_test2.index = X_test.index.values
X_test_scaled = X_test2
from sklearn.metrics import confusion_matrix, accuracy_score, f1_score, precision_score, recall_score
def perfomance_check(name: str):
acc = accuracy_score(y_test, y_pred)
prec = precision_score(y_test, y_pred, pos_label='positive',
average='micro')
rec = recall_score(y_test, y_pred, pos_label='positive',
average='micro')
f1 = f1_score(y_test, y_pred, pos_label='positive',
average='micro')
model_results = pd.DataFrame([[name, acc, prec, rec, f1]],
columns = ['Model', 'Accuracy', 'Precision', 'Recall', 'F1 Score'])
return results.append(model_results, ignore_index = True)
results = pd.DataFrame(columns = ['Model', 'Accuracy', 'Precision', 'Recall', 'F1 Score'])
from sklearn.linear_model import LogisticRegression
LR_classifier = LogisticRegression(random_state = 0, penalty = 'l1', solver='saga')
LR_classifier.fit(X_train, y_train.values.ravel())
y_pred = LR_classifier.predict(X_test)
results = perfomance_check('Linear Regression (Lasso)')
from sklearn.neighbors import KNeighborsClassifier
KNN_classifier = KNeighborsClassifier(n_neighbors = 5, metric = 'minkowski', p = 2)
KNN_classifier.fit(X_train_scaled, y_train.values.ravel())
y_pred = KNN_classifier.predict(X_test_scaled)
results = perfomance_check('K-Nearest Neighbours')
from sklearn.svm import SVC
SVML_classifier = SVC(random_state = 0, kernel = 'linear')
SVML_classifier.fit(X_train, y_train.values.ravel())
y_pred = SVML_classifier.predict(X_test)
results = perfomance_check('SVM (Linear)')
from sklearn.svm import SVC
K_SVM_classifier = SVC(random_state = 0, kernel = 'rbf')
K_SVM_classifier.fit(X_train, y_train.values.ravel())
y_pred = K_SVM_classifier.predict(X_test)
results = perfomance_check('SVM (RBF)')
from sklearn.naive_bayes import GaussianNB
NB_classifier = GaussianNB()
NB_classifier.fit(X_train, y_train.values.ravel())
y_pred = NB_classifier.predict(X_test)
results = perfomance_check('Naive Bayes')
from sklearn.tree import DecisionTreeClassifier
DTC_classifier = DecisionTreeClassifier(criterion = 'entropy', random_state = 0)
DTC_classifier.fit(X_train, y_train.values.ravel())
y_pred = DTC_classifier.predict(X_test)
results = perfomance_check('Decision Tree Classification')
from sklearn.ensemble import RandomForestClassifier
RF_classifier = RandomForestClassifier(random_state = 0, n_estimators = 100,
criterion = 'entropy')
RF_classifier.fit(X_train, y_train.values.ravel())
y_pred = RF_classifier.predict(X_test)
results = perfomance_check('Random Forest (n=100)')
from xgboost import XGBClassifier
xgb_classifier = XGBClassifier()
xgb_classifier.fit(X_train, y_train.values.ravel())
y_pred = xgb_classifier.predict(X_test)
results = perfomance_check('XGBoost ')
results
with open('./saved_model/body_language.pkl', 'wb') as f:
pickle.dump(RF_classifier, f)
with open('./saved_model/body_language.pkl', 'rb') as f:
model = pickle.load(f)
from sklearn.model_selection import cross_val_score
model_lst = [LR_classifier, # 0
KNN_classifier, # 1
SVML_classifier, # 2
K_SVM_classifier, # 3
NB_classifier, # 4
DTC_classifier, # 5
RF_classifier, # 6
xgb_classifier] # 7
# CB_classifier] # 8
msg = []
for i in range(len(model_lst)):
accuracies = cross_val_score(estimator =model_lst[i] , X = X_train, y = y_train, cv = 10)
msg.append(f"Model Accuracy {i}: %0.3f (+/- %0.3f)" % (accuracies.mean(), accuracies.std() * 2))
for i in msg:
print(i)
for i in msg:
print(i)
# This is the script to compare result with prediction
comparision = pd.DataFrame(columns = ['Result', 'Prediction'])
comparision.Prediction = pd.Series(y_pred)
comparision.Result = y_test.values
parameters = {"max_depth": [3, None],
"max_features": [1, 5, 10],
'min_samples_split': [2, 5, 10],
'min_samples_leaf': [1, 5, 10],
"bootstrap": [True, False],
"criterion": ["entropy"]}
from sklearn.model_selection import GridSearchCV
grid_search_entropy = GridSearchCV(estimator = RF_classifier, # Make sure classifier points to the RF model
param_grid = parameters,
scoring = "accuracy",
cv = 10,
n_jobs = -1)
t0 = time.time() # RECORD THE DURATION ALGORITHM TOOK
grid_search = grid_search.fit(X_train, y_train)
t1 = time.time()
print("Took %0.2f seconds" % (t1 - t0))
rf_best_accuracy = grid_search.best_score_
rf_best_parameters = grid_search.best_params_
rf_best_accuracy, rf_best_parameters
parameters = {"max_depth": [3, None],
"max_features": [1, 5, 10],
'min_samples_split': [2, 5, 10],
'min_samples_leaf': [1, 5, 10],
"bootstrap": [True, False],
"criterion": ["gini"]}
from sklearn.model_selection import GridSearchCV
grid_search_gini = GridSearchCV(estimator = classifier, # Make sure classifier points to the RF model
param_grid = parameters,
scoring = "accuracy",
cv = 10,
n_jobs = -1)
t0 = time.time()
grid_search = grid_search.fit(X_train, y_train)
t1 = time.time()
print("Took %0.2f seconds" % (t1 - t0))
rf_best_accuracy = grid_search.best_score_
rf_best_parameters = grid_search.best_params_
rf_best_accuracy, rf_best_parameters
with open('body_language.pkl', 'wb') as f:
pickle.dump(grid_search_entropy, f)
with open('body_language.pkl', 'rb') as f:
grid_search_entropy = pickle.load(f)
# Use Correct grid_search 'entropy' or 'gini'
y_pred = grid_search_entropy.predict(X_test)
from sklearn.metrics import classification_report, confusion_matrix
print(classification_report(y_test,y_pred))
cm = confusion_matrix(y_test, y_pred)
sn.set(font_scale=2.8)
mask = np.zeros_like(corr, dtype=np.bool)
mask[np.triu_indices_from(mask)] = True
plt.figure(figsize = (50,40))
sn.heatmap(corr, annot=True,mask=mask);
final_results = pd.concat([y_test, users], axis = 1).dropna()
final_results['predictions'] = y_pred
final_results = final_results[['entry_id', 'e_signed', 'predictions']]
| 0.606615 | 0.821008 |
# Modelling discharge at Mohembo using uppercatchment rainfall
* **Products used:**
ERA5
## Description
The discharge data at Mohembo becomes much more sparse during the era of good quality satellite observations, making it unreliable for comparing discharge with surface water extent. This notebook will try to model the discharge at Mohembo using the upstream rainfall extracted from ERA5 in a previous notebook
## Getting started
To run this analysis, run all the cells in the notebook, starting with the "Load packages" cell.
### Load packages
Import Python packages that are used for the analysis.
```
%matplotlib inline
import warnings
import datetime as dt
import numpy as np
import pandas as pd
import seaborn as sns
from scipy import stats
import matplotlib.pyplot as plt
from sklearn.metrics import mean_absolute_error
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from deafrica_tools.spatial import xr_rasterize
from deafrica_tools.load_era5 import load_era5
warnings.filterwarnings("ignore")
```
## Analysis Parameters
```
upstream_rainfall_1989_2009 = 'results/upstream_rainfall_daily_1989-01_to_2009-12.csv'
upstream_rainfall_2010_2021 = 'results/upstream_rainfall_daily_2010-01_to_2021-05-26.csv'
freq='Q-DEC'
max_lags = 3
sklearn_model = RandomForestRegressor
```
## Retrieve historical rainfall data over all areas of interest
This data has already been retrieved from ERA5 so we can simply pull in the csv on disk
```
upstream_rainfall_1989_2009 = pd.read_csv(upstream_rainfall_1989_2009, index_col='time',parse_dates=True)
upstream_rainfall_2010_2021 = pd.read_csv(upstream_rainfall_2010_2021, index_col='time',parse_dates=True)
rain = pd.concat([upstream_rainfall_1989_2009, upstream_rainfall_2010_2021])
```
#### Code block that loads rainfall from ERA5
No need to re-run this since the data has been saved in the `results/` folder
```
# # Original shapefile from https://data.apps.fao.org/map/catalog/srv/api/records/57bb1c95-2f00-4def-886f
# vector_file = 'data/OB_FWR_Hydrography_Okavango_Subasins_polygon.geojson'
# # define time period of interest
# time_range = '1989-01', '2009-12'
# # load basin polygons
# # Original shapefile from https://data.apps.fao.org/map/catalog/srv/api/records/57bb1c95-2f00-4def-886f-caee3d756da9
# basin = gpd.read_file(vector_file)
# # upstream include Cuito and Cubango subbasins
# upstream = basin[basin.Subbasin.isin(['Cuito', 'Cubango'])]
# print(upstream)
# # get historical rainfall for upstream and delta
# bounds = upstream.total_bounds
# lat = bounds[1], bounds[3]
# lon = bounds[0], bounds[2]
# # download ERA5 rainfall and aggregate to monthly
# var = 'precipitation_amount_1hour_Accumulation'
# precip = load_era5(var, lat, lon, time_range, reduce_func=np.sum, resample='1D').compute()
# # fix inconsistency in axis names
# precip = precip.rename({'lat':'latitude', 'lon':'longitude'})
# upstream_raster = xr_rasterize(upstream, precip, x_dim='longitude', y_dim='latitude')
# upstream_rainfall = precip[var].where(upstream_raster).sum(['latitude','longitude'])
# upstream_rainfall.to_dataframe().drop('spatial_ref',axis=1).rename({'precipitation_amount_1hour_Accumulation':'cumulative daily rainfall (mm)'},axis=1).to_csv(f'results/upstream_rainfall_daily_{time_range[0]}_to_{time_range[1]}.csv')
```
## Import discharge data
```
discharge = 'data/mohembo_daily_water_discharge_data.csv'
dis=pd.read_csv(discharge)
dis['date'] = pd.to_datetime(dis['date'], dayfirst=True)
dis = dis.set_index('date')
```
### Match discharge with rainfall
```
dis = dis.loc[(dis.index >= rain.index[0])]
df = rain.join(dis, how='outer')
df.tail()
```
### Resample seasonal or monthly cumulative totals
By intergrating rainfall over a longer time-period (months to seasons) we can better correlate cumulative upstream rainfall with discharge at Mohembo.
```
#total rainfall per month
df = df.resample(freq).sum()
df.head()
```
Split dataset at 2010. That way we can build a model on the historical, complete dataset, and then predict on the incomplete record from 2010 onwards
```
df_2010 = df.loc[(df.index >= pd.to_datetime('2010-01-01'))]
df_1989 = df.loc[(df.index < pd.to_datetime('2010-01-01'))]
df_1989.tail()
```
## Explore correlations with lags in rainfall
```
def crosscorr(datay, datax, lag=0):
"""
Lag-N cross correlation.
lag : int, default 0
datax, datay : pandas.Series objects of equal length
"""
return datay.corr(datax.shift(lag))
# calculate cross correlation for each lag period
xcov = [crosscorr(df_1989['water_discharge'], df_1989['cumulative daily rainfall (mm)'], lag=i) for i in range(max_lags)]
# Scatter plots of the relationship between rainfall and discharge at each lag
fig, ax = plt.subplots(1,max_lags, figsize=(20,4), sharey=True)
for lag in range(max_lags):
df_1989['rainfall_L'+str(lag)] = df_1989['cumulative daily rainfall (mm)'].shift(lag)
sns.regplot(x='rainfall_L'+str(lag), y='water_discharge', data=df_1989, ax=ax[lag])
r, p = stats.pearsonr(df_1989['rainfall_L'+str(lag)][lag:], df_1989['water_discharge'][lag:])
ax[lag].text(.05, .8, 'r={:.2f}, p={:.2g}'.format(r, p), transform=ax[lag].transAxes)
```
## Regression modelling
Model the relationship between rainfall and discharge using the lag that corresponds with the highest correlation
Conduct a test/train split first to get an idea of the general accuracy of this approach
```
max_value = max(xcov)
best_lag = xcov.index(max_value)
print("Best lag:",best_lag)
#convert data into a format that scikit learn likes
X = df_1989['rainfall_L'+str(best_lag)][best_lag:].values.reshape(-1,1)
y = df_1989['water_discharge'][best_lag:].values.reshape(-1,1)
#conduct a train test split
X_train,X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=1)
#model the relationship using just the training data, then predict on test data
model = sklearn_model()
model.fit(X_train,y_train)
y_pred = model.predict(X_test)
print('MAE: ',mean_absolute_error(y_test,y_pred))
#plot result of test data
sns.regplot(x=y_test, y=y_pred)
plt.xlim(10000, 50000)
plt.ylim(10000, 50000)
plt.plot([10000, 50000], [10000, 50000], 'k-', color = 'r')
plt.title('test prediction using linear model');
```
## Predict Discharge at Mohembo
Grab the rainfall from the period we want to predict
```
fc_rainfall=df['cumulative daily rainfall (mm)'].shift(best_lag)
X_fc = fc_rainfall[best_lag:].values.reshape(-1,1)
```
Make a prediction using rainfall data
```
# model = LinearRegression()
model = sklearn_model()
model.fit(X,y) #train 1989-2010
fc = model.predict(X_fc) # predict on all data
```
## Plot the observed vs predicted discharge as a time series
```
df_predicted = df[best_lag:]
df_predicted['predicted_discharge'] = fc.reshape(-1)
df_predicted.head(2)
```
### Print mean absolute error
```
mean_absolute_error(df_predicted.loc[(df_predicted.index < pd.to_datetime('2010-01-01'))]['water_discharge'],
df_predicted.loc[(df_predicted.index < pd.to_datetime('2010-01-01'))]['predicted_discharge'])
fig, ax = plt.subplots(figsize=(25,7))
ax.plot(df_predicted['water_discharge'], label='Obs discharge', linestyle='dashed', marker='o')
ax.plot(df_predicted['predicted_discharge'], label = 'Predicted discharge', linestyle='dashed', marker='o')
ax.legend()
plt.title('Observed vs Predicted Discharge')
ax.set_ylabel('Cumulative '+freq+' Discharge')
ax.axvline(dt.datetime(2010, 1, 1), c='red',linestyle='dashed')
ax.set_xlabel('')
plt.tight_layout();
```
### Plot rainfall
```
df['cumulative daily rainfall (mm)'].plot(linestyle='dashed', marker='o',figsize=(25,7))
plt.title('Upstream rainfall from ERA5')
plt.ylabel('Cumulative '+freq+' rainfall (mm)')
plt.xlabel('');
data = 'results/okavango_all_datasets.csv'
ok_r=pd.read_csv(data, index_col='time', parse_dates=True)[['okavango_rain']]
```
### Save result to disk
```
df_predicted.rename({'cumulative daily rainfall (mm)':'upstream_rainfall'}, axis=1).to_csv('results/modelled_discharge_'+freq+'.csv')
```
***
## Additional information
**License:** The code in this notebook is licensed under the [Apache License, Version 2.0](https://www.apache.org/licenses/LICENSE-2.0).
Digital Earth Africa data is licensed under the [Creative Commons by Attribution 4.0](https://creativecommons.org/licenses/by/4.0/) license.
**Contact:** If you need assistance, please post a question on the [Open Data Cube Slack channel](http://slack.opendatacube.org/) or on the [GIS Stack Exchange](https://gis.stackexchange.com/questions/ask?tags=open-data-cube) using the `open-data-cube` tag (you can view previously asked questions [here](https://gis.stackexchange.com/questions/tagged/open-data-cube)).
If you would like to report an issue with this notebook, you can file one on [Github](https://github.com/digitalearthafrica/deafrica-sandbox-notebooks).
**Last Tested:**
```
from datetime import datetime
datetime.today().strftime('%Y-%m-%d')
```
|
github_jupyter
|
%matplotlib inline
import warnings
import datetime as dt
import numpy as np
import pandas as pd
import seaborn as sns
from scipy import stats
import matplotlib.pyplot as plt
from sklearn.metrics import mean_absolute_error
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from deafrica_tools.spatial import xr_rasterize
from deafrica_tools.load_era5 import load_era5
warnings.filterwarnings("ignore")
upstream_rainfall_1989_2009 = 'results/upstream_rainfall_daily_1989-01_to_2009-12.csv'
upstream_rainfall_2010_2021 = 'results/upstream_rainfall_daily_2010-01_to_2021-05-26.csv'
freq='Q-DEC'
max_lags = 3
sklearn_model = RandomForestRegressor
upstream_rainfall_1989_2009 = pd.read_csv(upstream_rainfall_1989_2009, index_col='time',parse_dates=True)
upstream_rainfall_2010_2021 = pd.read_csv(upstream_rainfall_2010_2021, index_col='time',parse_dates=True)
rain = pd.concat([upstream_rainfall_1989_2009, upstream_rainfall_2010_2021])
# # Original shapefile from https://data.apps.fao.org/map/catalog/srv/api/records/57bb1c95-2f00-4def-886f
# vector_file = 'data/OB_FWR_Hydrography_Okavango_Subasins_polygon.geojson'
# # define time period of interest
# time_range = '1989-01', '2009-12'
# # load basin polygons
# # Original shapefile from https://data.apps.fao.org/map/catalog/srv/api/records/57bb1c95-2f00-4def-886f-caee3d756da9
# basin = gpd.read_file(vector_file)
# # upstream include Cuito and Cubango subbasins
# upstream = basin[basin.Subbasin.isin(['Cuito', 'Cubango'])]
# print(upstream)
# # get historical rainfall for upstream and delta
# bounds = upstream.total_bounds
# lat = bounds[1], bounds[3]
# lon = bounds[0], bounds[2]
# # download ERA5 rainfall and aggregate to monthly
# var = 'precipitation_amount_1hour_Accumulation'
# precip = load_era5(var, lat, lon, time_range, reduce_func=np.sum, resample='1D').compute()
# # fix inconsistency in axis names
# precip = precip.rename({'lat':'latitude', 'lon':'longitude'})
# upstream_raster = xr_rasterize(upstream, precip, x_dim='longitude', y_dim='latitude')
# upstream_rainfall = precip[var].where(upstream_raster).sum(['latitude','longitude'])
# upstream_rainfall.to_dataframe().drop('spatial_ref',axis=1).rename({'precipitation_amount_1hour_Accumulation':'cumulative daily rainfall (mm)'},axis=1).to_csv(f'results/upstream_rainfall_daily_{time_range[0]}_to_{time_range[1]}.csv')
discharge = 'data/mohembo_daily_water_discharge_data.csv'
dis=pd.read_csv(discharge)
dis['date'] = pd.to_datetime(dis['date'], dayfirst=True)
dis = dis.set_index('date')
dis = dis.loc[(dis.index >= rain.index[0])]
df = rain.join(dis, how='outer')
df.tail()
#total rainfall per month
df = df.resample(freq).sum()
df.head()
df_2010 = df.loc[(df.index >= pd.to_datetime('2010-01-01'))]
df_1989 = df.loc[(df.index < pd.to_datetime('2010-01-01'))]
df_1989.tail()
def crosscorr(datay, datax, lag=0):
"""
Lag-N cross correlation.
lag : int, default 0
datax, datay : pandas.Series objects of equal length
"""
return datay.corr(datax.shift(lag))
# calculate cross correlation for each lag period
xcov = [crosscorr(df_1989['water_discharge'], df_1989['cumulative daily rainfall (mm)'], lag=i) for i in range(max_lags)]
# Scatter plots of the relationship between rainfall and discharge at each lag
fig, ax = plt.subplots(1,max_lags, figsize=(20,4), sharey=True)
for lag in range(max_lags):
df_1989['rainfall_L'+str(lag)] = df_1989['cumulative daily rainfall (mm)'].shift(lag)
sns.regplot(x='rainfall_L'+str(lag), y='water_discharge', data=df_1989, ax=ax[lag])
r, p = stats.pearsonr(df_1989['rainfall_L'+str(lag)][lag:], df_1989['water_discharge'][lag:])
ax[lag].text(.05, .8, 'r={:.2f}, p={:.2g}'.format(r, p), transform=ax[lag].transAxes)
max_value = max(xcov)
best_lag = xcov.index(max_value)
print("Best lag:",best_lag)
#convert data into a format that scikit learn likes
X = df_1989['rainfall_L'+str(best_lag)][best_lag:].values.reshape(-1,1)
y = df_1989['water_discharge'][best_lag:].values.reshape(-1,1)
#conduct a train test split
X_train,X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=1)
#model the relationship using just the training data, then predict on test data
model = sklearn_model()
model.fit(X_train,y_train)
y_pred = model.predict(X_test)
print('MAE: ',mean_absolute_error(y_test,y_pred))
#plot result of test data
sns.regplot(x=y_test, y=y_pred)
plt.xlim(10000, 50000)
plt.ylim(10000, 50000)
plt.plot([10000, 50000], [10000, 50000], 'k-', color = 'r')
plt.title('test prediction using linear model');
fc_rainfall=df['cumulative daily rainfall (mm)'].shift(best_lag)
X_fc = fc_rainfall[best_lag:].values.reshape(-1,1)
# model = LinearRegression()
model = sklearn_model()
model.fit(X,y) #train 1989-2010
fc = model.predict(X_fc) # predict on all data
df_predicted = df[best_lag:]
df_predicted['predicted_discharge'] = fc.reshape(-1)
df_predicted.head(2)
mean_absolute_error(df_predicted.loc[(df_predicted.index < pd.to_datetime('2010-01-01'))]['water_discharge'],
df_predicted.loc[(df_predicted.index < pd.to_datetime('2010-01-01'))]['predicted_discharge'])
fig, ax = plt.subplots(figsize=(25,7))
ax.plot(df_predicted['water_discharge'], label='Obs discharge', linestyle='dashed', marker='o')
ax.plot(df_predicted['predicted_discharge'], label = 'Predicted discharge', linestyle='dashed', marker='o')
ax.legend()
plt.title('Observed vs Predicted Discharge')
ax.set_ylabel('Cumulative '+freq+' Discharge')
ax.axvline(dt.datetime(2010, 1, 1), c='red',linestyle='dashed')
ax.set_xlabel('')
plt.tight_layout();
df['cumulative daily rainfall (mm)'].plot(linestyle='dashed', marker='o',figsize=(25,7))
plt.title('Upstream rainfall from ERA5')
plt.ylabel('Cumulative '+freq+' rainfall (mm)')
plt.xlabel('');
data = 'results/okavango_all_datasets.csv'
ok_r=pd.read_csv(data, index_col='time', parse_dates=True)[['okavango_rain']]
df_predicted.rename({'cumulative daily rainfall (mm)':'upstream_rainfall'}, axis=1).to_csv('results/modelled_discharge_'+freq+'.csv')
from datetime import datetime
datetime.today().strftime('%Y-%m-%d')
| 0.633977 | 0.915394 |
<a href="https://colab.research.google.com/github/harshit54/Group-Project/blob/notebook/bert_ind_rnn.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## This notebook is licensed with The Unlicense
For more information, please refer [here](https://unlicense.org).
# Classifying text with BERT
In this notebook, we will:
- Load the IMDB dataset
- Load a BERT model from TensorFlow Hub
- Build the IndRNN Cell
- Build the model by combining BERT with a classifier and IndRNN
- Training the model for fine-tuning BERT
- Saving the model and classifying some reviews
## About BERT
[BERT](https://arxiv.org/abs/1810.04805) and other Transformer encoder architectures have been wildly successful on a variety of tasks in NLP (natural language processing). They compute word embedding of natural language that are suitable for use in deep learning models. BERT uses the Transformer encoder architecture to process each token of input text in the full context of all tokens before and after, hence the name: Bidirectional Encoder Representations from Transformers.
BERT models are usually pre-trained on a large corpus of text, then fine-tuned for specific tasks.
## Setup
```
# Installing TensorFlow Text Library for using in Preprocessing
!pip install -q tensorflow-text
```
Importing the AdamW optimizer from `tensorflow/models`
```
!pip install -q tf-models-official
import os
import shutil
import tensorflow as tf
import tensorflow_hub as hub
import tensorflow_text as text
from official.nlp import optimization # to create AdamW optmizer
import matplotlib.pyplot as plt
tf.get_logger().setLevel('ERROR')
```
## Sentiment Analysis
In this notebook we will create a sentiment analysis model to classify movie reviews as *positive* or *negative*, based on the text of the review.
We have used the [Large Movie Review Dataset](https://ai.stanford.edu/~amaas/data/sentiment/) that contains the text of 50,000 movie reviews from the [Internet Movie Database](https://www.imdb.com/).
### Downloading the IMDB dataset
```
url = 'https://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz'
dataset = tf.keras.utils.get_file('aclImdb_v1.tar.gz', url,
untar=True, cache_dir='.',
cache_subdir='')
dataset_dir = os.path.join(os.path.dirname(dataset), 'aclImdb')
train_dir = os.path.join(dataset_dir, 'train')
remove_dir = os.path.join(train_dir, 'unsup')
shutil.rmtree(remove_dir)
```
Next, you will use the `text_dataset_from_directory` utility to create a labeled `tf.data.Dataset`.
The IMDB dataset has already been divided into train and test, but it lacks a validation set. Let's create a validation set using an 80:20 split of the training data by using the `validation_split` argument below.
Note: When using the `validation_split` and `subset` arguments, make sure to either specify a random seed, or to pass `shuffle=False`, so that the validation and training splits have no overlap.
```
AUTOTUNE = tf.data.experimental.AUTOTUNE
batch_size = 32
seed = 42
raw_train_ds = tf.keras.preprocessing.text_dataset_from_directory(
'aclImdb/train',
batch_size=batch_size,
validation_split=0.2,
subset='training',
seed=seed)
class_names = raw_train_ds.class_names
train_ds = raw_train_ds.cache().prefetch(buffer_size=AUTOTUNE)
val_ds = tf.keras.preprocessing.text_dataset_from_directory(
'aclImdb/train',
batch_size=batch_size,
validation_split=0.2,
subset='validation',
seed=seed)
val_ds = val_ds.cache().prefetch(buffer_size=AUTOTUNE)
test_ds = tf.keras.preprocessing.text_dataset_from_directory(
'aclImdb/test',
batch_size=batch_size)
test_ds = test_ds.cache().prefetch(buffer_size=AUTOTUNE)
tfhub_handle_encoder = 'https://tfhub.dev/tensorflow/small_bert/bert_en_uncased_L-4_H-512_A-8/1'
tfhub_handle_preprocess = 'https://tfhub.dev/tensorflow/bert_en_uncased_preprocess/1'
print(f'BERT model selected : {tfhub_handle_encoder}')
print(f'Preprocess model auto-selected: {tfhub_handle_preprocess}')
```
## The Preprocessing Model
```
bert_preprocess_model = hub.KerasLayer(tfhub_handle_preprocess)
```
##Trying the Preprocessing Block
```
text_test = ['IIIT Sonepat Is Awesome!']
text_preprocessed = bert_preprocess_model(text_test)
print(f'Keys : {list(text_preprocessed.keys())}')
print(f'Shape : {text_preprocessed["input_word_ids"].shape}')
print(f'Word Ids : {text_preprocessed["input_word_ids"][0, :12]}')
print(f'Input Mask : {text_preprocessed["input_mask"][0, :12]}')
print(f'Type Ids : {text_preprocessed["input_type_ids"][0, :12]}')
```
## Using the BERT model
Before putting BERT into your own model, let's take a look at its outputs. You will load it from TF Hub and see the returned values.
```
bert_model = hub.KerasLayer(tfhub_handle_encoder)
bert_results = bert_model(text_preprocessed)
print(f'Loaded BERT: {tfhub_handle_encoder}')
print(f'Pooled Outputs Shape:{bert_results["pooled_output"].shape}')
print(f'Pooled Outputs Values:{bert_results["pooled_output"][0, :12]}')
print(f'Sequence Outputs Shape:{bert_results["sequence_output"].shape}')
print(f'Sequence Outputs Values:{bert_results["sequence_output"][0, :12]}')
```
The BERT models return a map with 3 important keys: `pooled_output`, `sequence_output`, `encoder_outputs`:
- `pooled_output` to represent each input sequence as a whole. The shape is `[batch_size, H]`. You can think of this as an embedding for the entire movie review.
- `sequence_output` represents each input token in the context. The shape is `[batch_size, seq_length, H]`. You can think of this as a contextual embedding for every token in the movie review.
- `encoder_outputs` are the intermediate activations of the `L` Transformer blocks. `outputs["encoder_outputs"][i]` is a Tensor of shape `[batch_size, seq_length, 1024]` with the outputs of the i-th Transformer block, for `0 <= i < L`. The last value of the list is equal to `sequence_output`.
For the fine-tuning you are going to use the `pooled_output` array.
##Defining The IndRNN Cell
```
from tensorflow.python.ops import math_ops
from tensorflow.python.ops import init_ops
from tensorflow.python.ops import nn_ops
from tensorflow.python.ops import clip_ops
from tensorflow.python.layers import base as base_layer
try:
from tensorflow.python.ops.rnn_cell_impl import LayerRNNCell
except ImportError:
from tensorflow.python.ops.rnn_cell_impl import _LayerRNNCell as LayerRNNCell
class IndRNNCell(LayerRNNCell):
def __init__(self,
num_units,
recurrent_min_abs=0,
recurrent_max_abs=None,
recurrent_kernel_initializer=None,
input_kernel_initializer=None,
activation=None,
reuse=None,
name=None):
super(IndRNNCell, self).__init__(_reuse=reuse, name=name)
self.input_spec = base_layer.InputSpec(ndim=2)
self._num_units = num_units
self._recurrent_min_abs = recurrent_min_abs
self._recurrent_max_abs = recurrent_max_abs
self._recurrent_initializer = recurrent_kernel_initializer
self._input_initializer = input_kernel_initializer
self._activation = activation or nn_ops.relu
@property
def state_size(self):
return self._num_units
@property
def output_size(self):
return self._num_units
def build(self, inputs_shape):
if inputs_shape[1].value is None:
raise ValueError("Expected inputs.shape[-1] to be known, saw shape: %s"
% inputs_shape)
input_depth = inputs_shape[1].value
if self._input_initializer is None:
self._input_initializer = init_ops.random_normal_initializer(mean=0.0,
stddev=0.001)
self._input_kernel = self.add_variable(
"input_kernel",
shape=[input_depth, self._num_units],
initializer=self._input_initializer)
if self._recurrent_initializer is None:
self._recurrent_initializer = init_ops.constant_initializer(1.)
self._recurrent_kernel = self.add_variable(
"recurrent_kernel",
shape=[self._num_units],
initializer=self._recurrent_initializer)
if self._recurrent_min_abs:
abs_kernel = math_ops.abs(self._recurrent_kernel)
min_abs_kernel = math_ops.maximum(abs_kernel, self._recurrent_min_abs)
self._recurrent_kernel = math_ops.multiply(
math_ops.sign(self._recurrent_kernel),
min_abs_kernel
)
if self._recurrent_max_abs:
self._recurrent_kernel = clip_ops.clip_by_value(self._recurrent_kernel,
-self._recurrent_max_abs,
self._recurrent_max_abs)
self._bias = self.add_variable(
"bias",
shape=[self._num_units],
initializer=init_ops.zeros_initializer(dtype=self.dtype))
self.built = True
def call(self, inputs, state):
gate_inputs = math_ops.matmul(inputs, self._input_kernel)
recurrent_update = math_ops.multiply(state, self._recurrent_kernel)
gate_inputs = math_ops.add(gate_inputs, recurrent_update)
gate_inputs = nn_ops.bias_add(gate_inputs, self._bias)
output = self._activation(gate_inputs)
return output, output
```
## Defining The Model
```
def build_classifier_model():
tf.config.run_functions_eagerly(False)
text_input = tf.keras.layers.Input(shape=(), dtype=tf.string, name='text')
preprocessing_layer = hub.KerasLayer(tfhub_handle_preprocess, name='preprocessing')
encoder_inputs = preprocessing_layer(text_input)
encoder = hub.KerasLayer(tfhub_handle_encoder, trainable=True, name='BERT_encoder')
outputs = encoder(encoder_inputs)
net = outputs['pooled_output']
net = tf.keras.layers.Dropout(0.1)(net)
# IndRNN Layer
TIME_STEPS = 0.1
recurrent_max = pow(2, 1 / TIME_STEPS)
net2 = tf.expand_dims(net, axis=1)
net2 = tf.expand_dims(net2, axis=2)
cell = tf.compat.v1.nn.rnn_cell.MultiRNNCell([IndRNNCell(128, recurrent_max_abs=recurrent_max),
IndRNNCell(128, recurrent_max_abs=recurrent_max)])
net = tf.keras.layers.Dense(1, activation=None, name='IndRNN-Layer')(net)
# Single Neuron as Ouput Layer
net = tf.keras.layers.Dense(1, activation=None, name='classifier')(net)
return tf.keras.Model(text_input, net)
```
Let's check that the model runs with the output of the preprocessing model.
```
classifier_model = build_classifier_model()
bert_raw_result = classifier_model(tf.constant(text_test))
print(tf.sigmoid(bert_raw_result))
```
The output is useless right now because the model has not been trained yet.
## Model Diagram
```
tf.keras.utils.plot_model(classifier_model)
```
## Model training
You now have all the pieces to train a model, including the preprocessing module, BERT encoder, data, and classifier.
### Loss function
Since this is a binary classification problem and the model outputs a probability (a single-unit layer), you'll use `losses.BinaryCrossentropy` loss function.
```
loss = tf.keras.losses.BinaryCrossentropy(from_logits=True)
metrics = tf.metrics.BinaryAccuracy()
```
### Optimizer
For fine-tuning, let's use the same optimizer that BERT was originally trained with: the "Adaptive Moments" (Adam). This optimizer minimizes the prediction loss and does regularization by weight decay (not using moments), which is also known as [AdamW](https://arxiv.org/abs/1711.05101).
```
epochs = 5
steps_per_epoch = tf.data.experimental.cardinality(train_ds).numpy()
num_train_steps = steps_per_epoch * epochs
num_warmup_steps = int(0.1*num_train_steps)
init_lr = 3e-5
optimizer = optimization.create_optimizer(init_lr=init_lr,
num_train_steps=num_train_steps,
num_warmup_steps=num_warmup_steps,
optimizer_type='adamw')
```
### Loading the BERT model and training
Using the `classifier_model` you created earlier, you can compile the model with the loss, metric and optimizer.
```
classifier_model.compile(optimizer=optimizer,
loss=loss,
metrics=metrics)
```
## Setting Up Tensorboard Callbacks
```
%load_ext tensorboard
import tensorflow as tf
import datetime
log_dir="./logs"
tensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir=log_dir)
print(f'Training model with {tfhub_handle_encoder}')
history = classifier_model.fit(x=train_ds,
validation_data=val_ds,
epochs=epochs,
callbacks=[tensorboard_callback])
%tensorboard --logdir logs/fit
```
### Evaluate the model
Let's see how the model performs. Two values will be returned. Loss (a number which represents the error, lower values are better), and accuracy.
```
loss, accuracy = classifier_model.evaluate(test_ds)
print(f'Loss: {loss}')
print(f'Accuracy: {accuracy}')
```
### Plotting the accuracy and loss over time
Based on the `History` object returned by `model.fit()`. You can plot the training and validation loss for comparison, as well as the training and validation accuracy:
```
history_dict = history.history
print(history_dict.keys())
acc = history_dict['binary_accuracy']
val_acc = history_dict['val_binary_accuracy']
loss = history_dict['loss']
val_loss = history_dict['val_loss']
epochs = range(1, len(acc) + 1)
fig = plt.figure(figsize=(10, 6))
fig.tight_layout()
plt.subplot(2, 1, 1)
# "bo" is for "blue dot"
plt.plot(epochs, loss, 'r', label='Training loss')
# b is for "solid blue line"
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
# plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.subplot(2, 1, 2)
plt.plot(epochs, acc, 'r', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend(loc='lower right')
```
In this plot, the red lines represents the training loss and accuracy, and the blue lines are the validation loss and accuracy.
## Testing The Model
```
def print_my_examples(inputs, results):
result_for_printing = \
[f'input: {inputs[i]:<30} : score: {results[i][0]:.6f}'
for i in range(len(inputs))]
print(*result_for_printing, sep='\n')
print()
examples = [
'this is such an amazing movie!',
'Best Movie Ever!',
'Totally worth the wait.',
'The movie was okish.',
'The movie was a nightmare'
]
reloaded_results = tf.sigmoid(reloaded_model(tf.constant(examples)))
original_results = tf.sigmoid(classifier_model(tf.constant(examples)))
print('Results from the saved model:')
print_my_examples(examples, reloaded_results)
print('Results from the model in memory:')
print_my_examples(examples, original_results)
```
|
github_jupyter
|
# Installing TensorFlow Text Library for using in Preprocessing
!pip install -q tensorflow-text
!pip install -q tf-models-official
import os
import shutil
import tensorflow as tf
import tensorflow_hub as hub
import tensorflow_text as text
from official.nlp import optimization # to create AdamW optmizer
import matplotlib.pyplot as plt
tf.get_logger().setLevel('ERROR')
url = 'https://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz'
dataset = tf.keras.utils.get_file('aclImdb_v1.tar.gz', url,
untar=True, cache_dir='.',
cache_subdir='')
dataset_dir = os.path.join(os.path.dirname(dataset), 'aclImdb')
train_dir = os.path.join(dataset_dir, 'train')
remove_dir = os.path.join(train_dir, 'unsup')
shutil.rmtree(remove_dir)
AUTOTUNE = tf.data.experimental.AUTOTUNE
batch_size = 32
seed = 42
raw_train_ds = tf.keras.preprocessing.text_dataset_from_directory(
'aclImdb/train',
batch_size=batch_size,
validation_split=0.2,
subset='training',
seed=seed)
class_names = raw_train_ds.class_names
train_ds = raw_train_ds.cache().prefetch(buffer_size=AUTOTUNE)
val_ds = tf.keras.preprocessing.text_dataset_from_directory(
'aclImdb/train',
batch_size=batch_size,
validation_split=0.2,
subset='validation',
seed=seed)
val_ds = val_ds.cache().prefetch(buffer_size=AUTOTUNE)
test_ds = tf.keras.preprocessing.text_dataset_from_directory(
'aclImdb/test',
batch_size=batch_size)
test_ds = test_ds.cache().prefetch(buffer_size=AUTOTUNE)
tfhub_handle_encoder = 'https://tfhub.dev/tensorflow/small_bert/bert_en_uncased_L-4_H-512_A-8/1'
tfhub_handle_preprocess = 'https://tfhub.dev/tensorflow/bert_en_uncased_preprocess/1'
print(f'BERT model selected : {tfhub_handle_encoder}')
print(f'Preprocess model auto-selected: {tfhub_handle_preprocess}')
bert_preprocess_model = hub.KerasLayer(tfhub_handle_preprocess)
text_test = ['IIIT Sonepat Is Awesome!']
text_preprocessed = bert_preprocess_model(text_test)
print(f'Keys : {list(text_preprocessed.keys())}')
print(f'Shape : {text_preprocessed["input_word_ids"].shape}')
print(f'Word Ids : {text_preprocessed["input_word_ids"][0, :12]}')
print(f'Input Mask : {text_preprocessed["input_mask"][0, :12]}')
print(f'Type Ids : {text_preprocessed["input_type_ids"][0, :12]}')
bert_model = hub.KerasLayer(tfhub_handle_encoder)
bert_results = bert_model(text_preprocessed)
print(f'Loaded BERT: {tfhub_handle_encoder}')
print(f'Pooled Outputs Shape:{bert_results["pooled_output"].shape}')
print(f'Pooled Outputs Values:{bert_results["pooled_output"][0, :12]}')
print(f'Sequence Outputs Shape:{bert_results["sequence_output"].shape}')
print(f'Sequence Outputs Values:{bert_results["sequence_output"][0, :12]}')
from tensorflow.python.ops import math_ops
from tensorflow.python.ops import init_ops
from tensorflow.python.ops import nn_ops
from tensorflow.python.ops import clip_ops
from tensorflow.python.layers import base as base_layer
try:
from tensorflow.python.ops.rnn_cell_impl import LayerRNNCell
except ImportError:
from tensorflow.python.ops.rnn_cell_impl import _LayerRNNCell as LayerRNNCell
class IndRNNCell(LayerRNNCell):
def __init__(self,
num_units,
recurrent_min_abs=0,
recurrent_max_abs=None,
recurrent_kernel_initializer=None,
input_kernel_initializer=None,
activation=None,
reuse=None,
name=None):
super(IndRNNCell, self).__init__(_reuse=reuse, name=name)
self.input_spec = base_layer.InputSpec(ndim=2)
self._num_units = num_units
self._recurrent_min_abs = recurrent_min_abs
self._recurrent_max_abs = recurrent_max_abs
self._recurrent_initializer = recurrent_kernel_initializer
self._input_initializer = input_kernel_initializer
self._activation = activation or nn_ops.relu
@property
def state_size(self):
return self._num_units
@property
def output_size(self):
return self._num_units
def build(self, inputs_shape):
if inputs_shape[1].value is None:
raise ValueError("Expected inputs.shape[-1] to be known, saw shape: %s"
% inputs_shape)
input_depth = inputs_shape[1].value
if self._input_initializer is None:
self._input_initializer = init_ops.random_normal_initializer(mean=0.0,
stddev=0.001)
self._input_kernel = self.add_variable(
"input_kernel",
shape=[input_depth, self._num_units],
initializer=self._input_initializer)
if self._recurrent_initializer is None:
self._recurrent_initializer = init_ops.constant_initializer(1.)
self._recurrent_kernel = self.add_variable(
"recurrent_kernel",
shape=[self._num_units],
initializer=self._recurrent_initializer)
if self._recurrent_min_abs:
abs_kernel = math_ops.abs(self._recurrent_kernel)
min_abs_kernel = math_ops.maximum(abs_kernel, self._recurrent_min_abs)
self._recurrent_kernel = math_ops.multiply(
math_ops.sign(self._recurrent_kernel),
min_abs_kernel
)
if self._recurrent_max_abs:
self._recurrent_kernel = clip_ops.clip_by_value(self._recurrent_kernel,
-self._recurrent_max_abs,
self._recurrent_max_abs)
self._bias = self.add_variable(
"bias",
shape=[self._num_units],
initializer=init_ops.zeros_initializer(dtype=self.dtype))
self.built = True
def call(self, inputs, state):
gate_inputs = math_ops.matmul(inputs, self._input_kernel)
recurrent_update = math_ops.multiply(state, self._recurrent_kernel)
gate_inputs = math_ops.add(gate_inputs, recurrent_update)
gate_inputs = nn_ops.bias_add(gate_inputs, self._bias)
output = self._activation(gate_inputs)
return output, output
def build_classifier_model():
tf.config.run_functions_eagerly(False)
text_input = tf.keras.layers.Input(shape=(), dtype=tf.string, name='text')
preprocessing_layer = hub.KerasLayer(tfhub_handle_preprocess, name='preprocessing')
encoder_inputs = preprocessing_layer(text_input)
encoder = hub.KerasLayer(tfhub_handle_encoder, trainable=True, name='BERT_encoder')
outputs = encoder(encoder_inputs)
net = outputs['pooled_output']
net = tf.keras.layers.Dropout(0.1)(net)
# IndRNN Layer
TIME_STEPS = 0.1
recurrent_max = pow(2, 1 / TIME_STEPS)
net2 = tf.expand_dims(net, axis=1)
net2 = tf.expand_dims(net2, axis=2)
cell = tf.compat.v1.nn.rnn_cell.MultiRNNCell([IndRNNCell(128, recurrent_max_abs=recurrent_max),
IndRNNCell(128, recurrent_max_abs=recurrent_max)])
net = tf.keras.layers.Dense(1, activation=None, name='IndRNN-Layer')(net)
# Single Neuron as Ouput Layer
net = tf.keras.layers.Dense(1, activation=None, name='classifier')(net)
return tf.keras.Model(text_input, net)
classifier_model = build_classifier_model()
bert_raw_result = classifier_model(tf.constant(text_test))
print(tf.sigmoid(bert_raw_result))
tf.keras.utils.plot_model(classifier_model)
loss = tf.keras.losses.BinaryCrossentropy(from_logits=True)
metrics = tf.metrics.BinaryAccuracy()
epochs = 5
steps_per_epoch = tf.data.experimental.cardinality(train_ds).numpy()
num_train_steps = steps_per_epoch * epochs
num_warmup_steps = int(0.1*num_train_steps)
init_lr = 3e-5
optimizer = optimization.create_optimizer(init_lr=init_lr,
num_train_steps=num_train_steps,
num_warmup_steps=num_warmup_steps,
optimizer_type='adamw')
classifier_model.compile(optimizer=optimizer,
loss=loss,
metrics=metrics)
%load_ext tensorboard
import tensorflow as tf
import datetime
log_dir="./logs"
tensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir=log_dir)
print(f'Training model with {tfhub_handle_encoder}')
history = classifier_model.fit(x=train_ds,
validation_data=val_ds,
epochs=epochs,
callbacks=[tensorboard_callback])
%tensorboard --logdir logs/fit
loss, accuracy = classifier_model.evaluate(test_ds)
print(f'Loss: {loss}')
print(f'Accuracy: {accuracy}')
history_dict = history.history
print(history_dict.keys())
acc = history_dict['binary_accuracy']
val_acc = history_dict['val_binary_accuracy']
loss = history_dict['loss']
val_loss = history_dict['val_loss']
epochs = range(1, len(acc) + 1)
fig = plt.figure(figsize=(10, 6))
fig.tight_layout()
plt.subplot(2, 1, 1)
# "bo" is for "blue dot"
plt.plot(epochs, loss, 'r', label='Training loss')
# b is for "solid blue line"
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
# plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.subplot(2, 1, 2)
plt.plot(epochs, acc, 'r', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend(loc='lower right')
def print_my_examples(inputs, results):
result_for_printing = \
[f'input: {inputs[i]:<30} : score: {results[i][0]:.6f}'
for i in range(len(inputs))]
print(*result_for_printing, sep='\n')
print()
examples = [
'this is such an amazing movie!',
'Best Movie Ever!',
'Totally worth the wait.',
'The movie was okish.',
'The movie was a nightmare'
]
reloaded_results = tf.sigmoid(reloaded_model(tf.constant(examples)))
original_results = tf.sigmoid(classifier_model(tf.constant(examples)))
print('Results from the saved model:')
print_my_examples(examples, reloaded_results)
print('Results from the model in memory:')
print_my_examples(examples, original_results)
| 0.733833 | 0.965348 |
GP Regression with GPflow
--
*James Hensman, 2015, 2016*
GP regression (with Gaussian noise) is the most straightforward GP model in GPflow. Because of the conjugacy of the latent process and the noise, the marginal likelihood $p(\mathbf y\,|\,\theta)$ can be computed exactly.
This notebook shows how to build a GPR model, estimate the parameters $\theta$ by both maximum likelihood and MCMC.
## Editted by @fujiisoup
In this note, I demonstrate the updated implementation where the model can be reused with different data.
Please see
`m.X = X_new
m.Y = Y_new`
```
from __future__ import print_function
import GPflow
import numpy as np
from matplotlib import pyplot as plt
%matplotlib inline
import time
# build a very simple data set:
N = 12
X = np.random.rand(N,1)
Y = np.sin(12*X) + 0.66*np.cos(25*X) + np.random.randn(N,1)*0.1 + 3
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
```
Maximum Likelihood estimation
--
```
#build the GPR object
k = GPflow.kernels.Matern52(1)
meanf = GPflow.mean_functions.Linear(1,0)
m = GPflow.gpr.GPR(X, Y, k, meanf)
m.likelihood.variance = 0.01
print("Here are the parameters before optimization")
m
t = time.time()
m.optimize()
print("Here are the parameters after optimization")
print(time.time() - t, " (s) for compilation and optimization")
m
#plot!
xx = np.linspace(-0.1, 1.1, 100)[:,None]
mean, var = m.predict_y(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, mean, 'b', lw=2)
plt.plot(xx, mean + 2*np.sqrt(var), 'b--', xx, mean - 2*np.sqrt(var), 'b--', lw=1.2)
```
### For another data
```
N = 12
X = np.random.rand(N,1)
Y = np.sin(12*X) + 0.66*np.cos(25*X) + np.random.randn(N,1)*0.1 + 3
# update data
m.X = X
m.Y = Y
t = time.time()
m.optimize()
print("Here are the parameters after optimization")
print(time.time() - t, " (s) for optimization")
m
#plot!
xx = np.linspace(-0.1, 1.1, 100)[:,None]
mean, var = m.predict_y(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, mean, 'b', lw=2)
plt.plot(xx, mean + 2*np.sqrt(var), 'b--', xx, mean - 2*np.sqrt(var), 'b--', lw=1.2)
```
### For another data with different shape
```
# shape changed!
N = 13
X = np.random.rand(N,1)
Y = np.sin(12*X) + 0.66*np.cos(25*X) + np.random.randn(N,1)*0.1 + 3
# update data
m.X = X
m.Y = Y
t = time.time()
m.optimize()
print("Here are the parameters after optimization")
print(time.time() - t, " (s) for optimization")
m
#plot!
xx = np.linspace(-0.1, 1.1, 100)[:,None]
mean, var = m.predict_y(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, mean, 'b', lw=2)
plt.plot(xx, mean + 2*np.sqrt(var), 'b--', xx, mean - 2*np.sqrt(var), 'b--', lw=1.2)
```
## Fixing the parameters
```
# Fix the lengthscale
m.kern.lengthscales.fixed = True
m.kern.lengthscales = 0.2
t = time.time()
m.optimize()
print("Here are the parameters after optimization")
print(time.time() - t, " (s) for compilation and optimization")
m
#plot!
xx = np.linspace(-0.1, 1.1, 100)[:,None]
mean, var = m.predict_y(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, mean, 'b', lw=2)
plt.plot(xx, mean + 2*np.sqrt(var), 'b--', xx, mean - 2*np.sqrt(var), 'b--', lw=1.2)
```
### With different value for fixed param
```
# Change the lengthscale
m.kern.lengthscales = 0.1
t = time.time()
m.optimize()
print("Here are the parameters after optimization")
print(time.time() - t, " (s) for optimization")
m
#plot!
xx = np.linspace(-0.1, 1.1, 100)[:,None]
mean, var = m.predict_y(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, mean, 'b', lw=2)
plt.plot(xx, mean + 2*np.sqrt(var), 'b--', xx, mean - 2*np.sqrt(var), 'b--', lw=1.2)
```
MCMC
--
First, we'll set come priors on the kernel parameters, then we'll run mcmc and see how much posterior uncertainty there is in the parameters.
```
m.kern.lengthscales.fixed = False
#we'll choose rather arbitrary priors.
m.kern.lengthscales.prior = GPflow.priors.Gamma(1., 1.)
m.kern.variance.prior = GPflow.priors.Gamma(1., 1.)
m.likelihood.variance.prior = GPflow.priors.Gamma(1., 1.)
m.mean_function.A.prior = GPflow.priors.Gaussian(0., 10.)
m.mean_function.b.prior = GPflow.priors.Gaussian(0., 10.)
m
samples = m.sample(500, epsilon = 0.1, verbose=1)
plt.figure()
plt.plot(samples)
#Note. All these labels are wrong (or, most probably wrong). We need some machinery for labelling posterior samples!
f, axs = plt.subplots(1,3, figsize=(12,4), tight_layout=True)
axs[0].plot(samples[:,0], samples[:,1], 'k.', alpha = 0.15)
axs[0].set_xlabel('noise_variance')
axs[0].set_ylabel('signal_variance')
axs[1].plot(samples[:,0], samples[:,2], 'k.', alpha = 0.15)
axs[1].set_xlabel('noise_variance')
axs[1].set_ylabel('lengthscale')
axs[2].plot(samples[:,2], samples[:,1], 'k.', alpha = 0.1)
axs[2].set_xlabel('lengthscale')
axs[2].set_ylabel('signal_variance')
#an attempt to plot the function posterior
#Note that we should really sample the function values here, instead of just using the mean.
#We are under-representing the uncertainty here.
# TODO: get full_covariance of the predictions (predict_f only?)
plt.figure()
for s in samples:
m.set_state(s)
mean, _ = m.predict_y(xx)
plt.plot(xx, mean, 'b', lw=2, alpha = 0.05)
plt.plot(X, Y, 'kx', mew=2)
```
## Variational inference
```
N = 25
X = np.arange(N, dtype=np.float64).reshape(-1,1)
Y = np.random.poisson(np.exp(np.sin(12*X) + 0.66*np.cos(25*X)))*1.0
#build the GP object
k = GPflow.kernels.RBF(1)
lik = GPflow.likelihoods.Poisson()
m = GPflow.vgp.VGP(X, Y, k, likelihood=lik)
t = time.time()
m.optimize()
print(time.time() - t, " (s) for compilation and optimization")
#plot!
xx = np.linspace(-1., N+1., 100)[:,None]
mean, var = m.predict_f(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, np.exp(mean), 'b', lw=2)
plt.plot(xx, np.exp(mean + 2*np.sqrt(var)), 'b--', xx, np.exp(mean - 2*np.sqrt(var)), 'b--', lw=1.2)
```
### For another data with the same shape
```
Y = 30.*np.exp(-((X-np.ones((N,1))*5.)*0.1)**2.) + np.random.rand(N,1)*3.
m.Y = Y
t = time.time()
m.optimize()
print(time.time() - t, " (s) for optimization")
#plot!
xx = np.linspace(-1., N+1., 100)[:,None]
mean, var = m.predict_f(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, np.exp(mean), 'b', lw=2)
plt.plot(xx, np.exp(mean + 2*np.sqrt(var)), 'b--', xx, np.exp(mean - 2*np.sqrt(var)), 'b--', lw=1.2)
```
### For another data with different shape
```
# shape changed
N = 34
X = np.arange(N, dtype=np.float64).reshape(-1,1)
Y = 14.*np.exp(-((X-np.ones((N,1))*5.)*0.1)**2.) + np.random.rand(N,1)*3.
m.X = X
m.Y = Y
t = time.time()
m.optimize()
print(time.time() - t, " (s) for compilation and optimization")
#plot!
xx = np.linspace(-1., N+1., 100)[:,None]
mean, var = m.predict_f(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, np.exp(mean), 'b', lw=2)
plt.plot(xx, np.exp(mean + 2*np.sqrt(var)), 'b--', xx, np.exp(mean - 2*np.sqrt(var)), 'b--', lw=1.2)
```
|
github_jupyter
|
from __future__ import print_function
import GPflow
import numpy as np
from matplotlib import pyplot as plt
%matplotlib inline
import time
# build a very simple data set:
N = 12
X = np.random.rand(N,1)
Y = np.sin(12*X) + 0.66*np.cos(25*X) + np.random.randn(N,1)*0.1 + 3
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
#build the GPR object
k = GPflow.kernels.Matern52(1)
meanf = GPflow.mean_functions.Linear(1,0)
m = GPflow.gpr.GPR(X, Y, k, meanf)
m.likelihood.variance = 0.01
print("Here are the parameters before optimization")
m
t = time.time()
m.optimize()
print("Here are the parameters after optimization")
print(time.time() - t, " (s) for compilation and optimization")
m
#plot!
xx = np.linspace(-0.1, 1.1, 100)[:,None]
mean, var = m.predict_y(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, mean, 'b', lw=2)
plt.plot(xx, mean + 2*np.sqrt(var), 'b--', xx, mean - 2*np.sqrt(var), 'b--', lw=1.2)
N = 12
X = np.random.rand(N,1)
Y = np.sin(12*X) + 0.66*np.cos(25*X) + np.random.randn(N,1)*0.1 + 3
# update data
m.X = X
m.Y = Y
t = time.time()
m.optimize()
print("Here are the parameters after optimization")
print(time.time() - t, " (s) for optimization")
m
#plot!
xx = np.linspace(-0.1, 1.1, 100)[:,None]
mean, var = m.predict_y(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, mean, 'b', lw=2)
plt.plot(xx, mean + 2*np.sqrt(var), 'b--', xx, mean - 2*np.sqrt(var), 'b--', lw=1.2)
# shape changed!
N = 13
X = np.random.rand(N,1)
Y = np.sin(12*X) + 0.66*np.cos(25*X) + np.random.randn(N,1)*0.1 + 3
# update data
m.X = X
m.Y = Y
t = time.time()
m.optimize()
print("Here are the parameters after optimization")
print(time.time() - t, " (s) for optimization")
m
#plot!
xx = np.linspace(-0.1, 1.1, 100)[:,None]
mean, var = m.predict_y(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, mean, 'b', lw=2)
plt.plot(xx, mean + 2*np.sqrt(var), 'b--', xx, mean - 2*np.sqrt(var), 'b--', lw=1.2)
# Fix the lengthscale
m.kern.lengthscales.fixed = True
m.kern.lengthscales = 0.2
t = time.time()
m.optimize()
print("Here are the parameters after optimization")
print(time.time() - t, " (s) for compilation and optimization")
m
#plot!
xx = np.linspace(-0.1, 1.1, 100)[:,None]
mean, var = m.predict_y(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, mean, 'b', lw=2)
plt.plot(xx, mean + 2*np.sqrt(var), 'b--', xx, mean - 2*np.sqrt(var), 'b--', lw=1.2)
# Change the lengthscale
m.kern.lengthscales = 0.1
t = time.time()
m.optimize()
print("Here are the parameters after optimization")
print(time.time() - t, " (s) for optimization")
m
#plot!
xx = np.linspace(-0.1, 1.1, 100)[:,None]
mean, var = m.predict_y(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, mean, 'b', lw=2)
plt.plot(xx, mean + 2*np.sqrt(var), 'b--', xx, mean - 2*np.sqrt(var), 'b--', lw=1.2)
m.kern.lengthscales.fixed = False
#we'll choose rather arbitrary priors.
m.kern.lengthscales.prior = GPflow.priors.Gamma(1., 1.)
m.kern.variance.prior = GPflow.priors.Gamma(1., 1.)
m.likelihood.variance.prior = GPflow.priors.Gamma(1., 1.)
m.mean_function.A.prior = GPflow.priors.Gaussian(0., 10.)
m.mean_function.b.prior = GPflow.priors.Gaussian(0., 10.)
m
samples = m.sample(500, epsilon = 0.1, verbose=1)
plt.figure()
plt.plot(samples)
#Note. All these labels are wrong (or, most probably wrong). We need some machinery for labelling posterior samples!
f, axs = plt.subplots(1,3, figsize=(12,4), tight_layout=True)
axs[0].plot(samples[:,0], samples[:,1], 'k.', alpha = 0.15)
axs[0].set_xlabel('noise_variance')
axs[0].set_ylabel('signal_variance')
axs[1].plot(samples[:,0], samples[:,2], 'k.', alpha = 0.15)
axs[1].set_xlabel('noise_variance')
axs[1].set_ylabel('lengthscale')
axs[2].plot(samples[:,2], samples[:,1], 'k.', alpha = 0.1)
axs[2].set_xlabel('lengthscale')
axs[2].set_ylabel('signal_variance')
#an attempt to plot the function posterior
#Note that we should really sample the function values here, instead of just using the mean.
#We are under-representing the uncertainty here.
# TODO: get full_covariance of the predictions (predict_f only?)
plt.figure()
for s in samples:
m.set_state(s)
mean, _ = m.predict_y(xx)
plt.plot(xx, mean, 'b', lw=2, alpha = 0.05)
plt.plot(X, Y, 'kx', mew=2)
N = 25
X = np.arange(N, dtype=np.float64).reshape(-1,1)
Y = np.random.poisson(np.exp(np.sin(12*X) + 0.66*np.cos(25*X)))*1.0
#build the GP object
k = GPflow.kernels.RBF(1)
lik = GPflow.likelihoods.Poisson()
m = GPflow.vgp.VGP(X, Y, k, likelihood=lik)
t = time.time()
m.optimize()
print(time.time() - t, " (s) for compilation and optimization")
#plot!
xx = np.linspace(-1., N+1., 100)[:,None]
mean, var = m.predict_f(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, np.exp(mean), 'b', lw=2)
plt.plot(xx, np.exp(mean + 2*np.sqrt(var)), 'b--', xx, np.exp(mean - 2*np.sqrt(var)), 'b--', lw=1.2)
Y = 30.*np.exp(-((X-np.ones((N,1))*5.)*0.1)**2.) + np.random.rand(N,1)*3.
m.Y = Y
t = time.time()
m.optimize()
print(time.time() - t, " (s) for optimization")
#plot!
xx = np.linspace(-1., N+1., 100)[:,None]
mean, var = m.predict_f(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, np.exp(mean), 'b', lw=2)
plt.plot(xx, np.exp(mean + 2*np.sqrt(var)), 'b--', xx, np.exp(mean - 2*np.sqrt(var)), 'b--', lw=1.2)
# shape changed
N = 34
X = np.arange(N, dtype=np.float64).reshape(-1,1)
Y = 14.*np.exp(-((X-np.ones((N,1))*5.)*0.1)**2.) + np.random.rand(N,1)*3.
m.X = X
m.Y = Y
t = time.time()
m.optimize()
print(time.time() - t, " (s) for compilation and optimization")
#plot!
xx = np.linspace(-1., N+1., 100)[:,None]
mean, var = m.predict_f(xx)
plt.figure()
plt.plot(X, Y, 'kx', mew=2)
plt.plot(xx, np.exp(mean), 'b', lw=2)
plt.plot(xx, np.exp(mean + 2*np.sqrt(var)), 'b--', xx, np.exp(mean - 2*np.sqrt(var)), 'b--', lw=1.2)
| 0.566378 | 0.974043 |
<script async src="https://www.googletagmanager.com/gtag/js?id=UA-59152712-8"></script>
<script>
window.dataLayer = window.dataLayer || [];
function gtag(){dataLayer.push(arguments);}
gtag('js', new Date());
gtag('config', 'UA-59152712-8');
</script>
# Exact Wald `GiRaFFEfood` Initial Data for `GiRaFFE`
## Author: Zach Etienne & Patrick Nelson
### Formatting improvements courtesy Brandon Clark
[comment]: <> (Abstract: TODO)
### NRPy+ Source Code for this module: [GiRaFFEfood_NRPy/GiRaFFEfood_NRPy_Exact_Wald.py](../../edit/in_progress/GiRaFFEfood_NRPy/GiRaFFEfood_NRPy_Exact_Wald.py)
**Notebook Status:** <font color='green'><b> Validated </b></font>
**Validation Notes:** This tutorial notebook has been confirmed to be self-consistent with its corresponding NRPy+ module, as documented [below](#code_validation1). The initial data has validated against the original `GiRaFFE`, as documented [here](Tutorial-Start_to_Finish_UnitTest-GiRaFFEfood_NRPy.ipynb).
## Introduction:
With the `GiRaFFE` evolution thorn constructed, we now need to "feed" our giraffe with initial data to evolve. There are several different choices of initial data we can use here; here, we will only be implementing the "Exact Wald" initial data, given by Table 3 in [the original paper](https://arxiv.org/pdf/1704.00599.pdf):
\begin{align}
A_{\phi} &= \frac{C_0}{2} r^2 \sin^2 \theta \\
E_{\phi} &= 2 M C_0 \left( 1+ \frac {2M}{r} \right)^{-1/2} \sin^2 \theta \\
\end{align}
(the unspecified components are set to 0). Here, $C_0$ is a constant that we will set to $1$ in our simulations. Now, to use this initial data scheme, we need to transform the above into the quantities actually tracked by `GiRaFFE` and HydroBase: $A_i$, $B^i$, $\tilde{S}_i$, $v^i$, and $\Phi$. Of these quantities, `GiRaFFEfood` will only set $A_i$, $v^i$, and $\Phi=0$, then call a separate function to calculate $\tilde{S}_i$; `GiRaFFE` itself will call a function to set $B^i$ before the time-evolution begins. This can be done with eqs. 16 and 18, here given in that same order:
\begin{align}
v^i &= \alpha \frac{\epsilon^{ijk} E_j B_k}{B^2} -\beta^i \\
B^i &= \frac{[ijk]}{\sqrt{\gamma}} \partial_j A_k \\
\end{align}
In the simulations, $B^i$ will be calculated numerically from $A_i$; however, it will be useful to analytically calculate $B^i$ to use calculating the initial $v^i$.
This module requires the use of the NRPy+ [Shifted Kerr-Schild initial data module](../Tutorial-ADM_Initial_Data-ShiftedKerrSchild.ipynb)
<a id='toc'></a>
# Table of Contents:
$$\label{toc}$$
This notebook is organized as follows
1. [Step 1](#initializenrpy): Import core NRPy+ modules and set NRPy+ parameters
1. [Step 2](#set_aphi_ephi): Set the vectors $A_{\phi}$ and $E_{\phi}$ in Spherical coordinates
1. [Step 3](#jacobian): Use the Jacobian matrix to transform the vectors to Cartesian coordinates
1. [Step 4](#vi): Calculate $v^i$ from $A_i$ and $E_i$
1. [Step 5](#code_validation1): Code Validation against `GiRaFFEfood_NRPy.GiRaFFEfood_NRPy` NRPy+ Module
1. [Step 6](#latex_pdf_output): Output this notebook to $\LaTeX$-formatted PDF file
<a id='initializenrpy'></a>
# Step 1: Import core NRPy+ modules and set NRPy+ parameters \[Back to [top](#toc)\]
$$\label{initializenrpy}$$
Here, we will import the NRPy+ core modules, set the reference metric to Cartesian, and set commonly used NRPy+ parameters. We will also set up a parameter to determine what initial data is set up, although it won't do much yet.
```
# Step 0: Add NRPy's directory to the path
# https://stackoverflow.com/questions/16780014/import-file-from-parent-directory
import os,sys
nrpy_dir_path = os.path.join("..")
if nrpy_dir_path not in sys.path:
sys.path.append(nrpy_dir_path)
# Step 0.a: Import the NRPy+ core modules and set the reference metric to Cartesian
import sympy as sp # SymPy: The Python computer algebra package upon which NRPy+ depends
import NRPy_param_funcs as par # NRPy+: Parameter interface
import indexedexp as ixp # NRPy+: Symbolic indexed expression (e.g., tensors, vectors, etc.) support
import reference_metric as rfm
par.set_parval_from_str("reference_metric::CoordSystem","Cartesian")
rfm.reference_metric()
# Step 1a: Set commonly used parameters.
thismodule = "GiRaFFEfood_NRPy"
# KerrSchild_radial_shift = par.Cparameters("REAL",thismodule,"KerrSchild_radial_shift",0.4) # Default value for ExactWald
KerrSchild_radial_shift = sp.symbols("KerrSchild_radial_shift")
M = sp.symbols("M") # Black hole mass
```
<a id='set_aphi_ephi'></a>
# Step 2: Set the vectors $A_{\phi}$ and $E_{\phi}$ in Spherical coordinates \[Back to [top](#toc)\]
$$\label{set_aphi_ephi}$$
We will first build the fundamental vectors $A_i$ and $E_i$ in spherical coordinates (see [Table 3](https://arxiv.org/pdf/1704.00599.pdf)). Note that we use reference_metric.py to set $r$ and $\theta$ in terms of Cartesian coordinates; this will save us a step later when we convert to Cartesian coordinates. Since $C_0 = 1$,
\begin{align}
A_{\phi} &= \frac{1}{2} r^2 \sin^2 \theta \\
E_{\phi} &= 2 M \left( 1+ \frac {2M}{r} \right)^{-1/2} \sin^2 \theta. \\
\end{align}
While we have $E_i$ set as a variable in NRPy+, note that the final C code won't store these values.
```
# Step 2: Set the vectors A and E in Spherical coordinates
r = rfm.xxSph[0] + KerrSchild_radial_shift # We are setting the data up in Shifted Kerr-Schild coordinates
theta = rfm.xxSph[1]
# Initialize all components of A and E in the *spherical basis* to zero
ASphD = ixp.zerorank1()
ESphD = ixp.zerorank1()
ASphD[2] = (r * r * sp.sin(theta)**2)/2
ESphD[2] = 2 * M * sp.sin(theta)**2 / sp.sqrt(1+2*M/r)
```
<a id='jacobian'></a>
# Step 3: Use the Jacobian matrix to transform the vectors to Cartesian coordinates \[Back to [top](#toc)\]
$$\label{jacobian}$$
Now, we will use the coordinate transformation definitions provided by reference_metric.py to build the Jacobian
$$
\frac{\partial x_{\rm Sph}^j}{\partial x_{\rm Cart}^i},
$$
where $x_{\rm Sph}^j \in \{r,\theta,\phi\}$ and $x_{\rm Cart}^i \in \{x,y,z\}$. We would normally compute its inverse, but since none of the quantities we need to transform have upper indices, it is not necessary. Then, since both $A_i$ and $E_i$ have one lower index, both will need to be multiplied by the Jacobian:
\begin{align}
A_i^{\rm Cart} &= A_j^{\rm Sph} \frac{\partial x_{\rm Sph}^j}{\partial x_{\rm Cart}^i} \\
{\rm and\ }E_i^{\rm Cart} &= E_j^{\rm Sph} \frac{\partial x_{\rm Sph}^j}{\partial x_{\rm Cart}^i}.
\end{align}
```
# Step 3: Use the Jacobian matrix to transform the vectors to Cartesian coordinates.
drrefmetric__dx_0UDmatrix = sp.Matrix([[sp.diff(rfm.xxSph[0],rfm.xx[0]), sp.diff(rfm.xxSph[0],rfm.xx[1]), sp.diff(rfm.xxSph[0],rfm.xx[2])],
[sp.diff(rfm.xxSph[1],rfm.xx[0]), sp.diff(rfm.xxSph[1],rfm.xx[1]), sp.diff(rfm.xxSph[1],rfm.xx[2])],
[sp.diff(rfm.xxSph[2],rfm.xx[0]), sp.diff(rfm.xxSph[2],rfm.xx[1]), sp.diff(rfm.xxSph[2],rfm.xx[2])]])
#dx__drrefmetric_0UDmatrix = drrefmetric__dx_0UDmatrix.inv() # We don't actually need this in this case.
AD = ixp.zerorank1()
ED = ixp.zerorank1()
for i in range(3):
for j in range(3):
AD[i] = drrefmetric__dx_0UDmatrix[(j,i)]*ASphD[j]
ED[i] = drrefmetric__dx_0UDmatrix[(j,i)]*ESphD[j]
#Step 4: Declare the basic spacetime quantities
alpha = sp.symbols("alpha",real=True)
betaU = ixp.declarerank1("betaU",DIM=3)
gammaDD = ixp.declarerank2("gammaDD", "sym01",DIM=3)
import GRHD.equations as GRHD
GRHD.compute_sqrtgammaDET(gammaDD)
```
<a id='vi'></a>
# Step 4: Calculate $v^i$ from $A_i$ and $E_i$ \[Back to [top](#toc)\]
$$\label{vi}$$
We will now find the magnetic field using equation 18 in [the original paper](https://arxiv.org/pdf/1704.00599.pdf) $$B^i = \frac{[ijk]}{\sqrt{\gamma}} \partial_j A_k. $$ We will need the metric quantites: the lapse $\alpha$, the shift $\beta^i$, and the three-metric $\gamma_{ij}$. We will also need the Levi-Civita symbol.
```
# Step 4: Calculate v^i from A_i and E_i
# Step 4a: Calculate the magnetic field B^i
GRHD.compute_sqrtgammaDET(gammaDD)
LeviCivitaTensorUUU = ixp.LeviCivitaTensorUUU_dim3_rank3(GRHD.sqrtgammaDET)
# For the initial data, we can analytically take the derivatives of A_i
ADdD = ixp.zerorank2()
for i in range(3):
for j in range(3):
ADdD[i][j] = sp.simplify(sp.diff(AD[i],rfm.xxCart[j]))
BU = ixp.zerorank1()
for i in range(3):
for j in range(3):
for k in range(3):
BU[i] += LeviCivitaTensorUUU[i][j][k] * ADdD[k][j]
```
We will now build the initial velocity using equation 152 in [this paper,](https://arxiv.org/pdf/1310.3274v2.pdf) cited in the original `GiRaFFE` code: $$ v^i = \alpha \frac{\epsilon^{ijk} E_j B_k}{B^2} -\beta^i. $$
However, our code needs the Valencia 3-velocity while this expression is for the drift velocity. So, we will need to transform it to the Valencia 3-velocity using the rule $\bar{v}^i = \frac{1}{\alpha} \left(v^i +\beta^i \right)$.
Thus, $$\bar{v}^i = \frac{\epsilon^{ijk} E_j B_k}{B^2}$$
```
# Step 4b: Calculate B^2 and B_i
# B^2 is an inner product defined in the usual way:
B2 = sp.sympify(0)
for i in range(3):
for j in range(3):
B2 += gammaDD[i][j] * BU[i] * BU[j]
# Lower the index on B^i
BD = ixp.zerorank1()
for i in range(3):
for j in range(3):
BD[i] += gammaDD[i][j] * BU[j]
# Step 4c: Calculate the Valencia 3-velocity
ValenciavU = ixp.zerorank1()
for i in range(3):
for j in range(3):
for k in range(3):
ValenciavU[i] += LeviCivitaTensorUUU[i][j][k]*ED[j]*BD[k]/B2
```
<a id='code_validation1'></a>
# Step 5: Code Validation against `GiRaFFEfood_NRPy.GiRaFFEfood_NRPy` NRPy+ module \[Back to [top](#toc)\]
$$\label{code_validation1}$$
Here, as a code validation check, we verify agreement in the SymPy expressions for the `GiRaFFE` Exact Wald initial data equations we intend to use between
1. this tutorial and
2. the NRPy+ [GiRaFFEfood_NRPy.GiRaFFEfood_NRPy_Exact_Wald](../edit/GiRaFFEfood_NRPy/GiRaFFEfood_NRPy_Exact_Wald.py) module.
```
import GiRaFFEfood_NRPy.GiRaFFEfood_NRPy_Exact_Wald as gfho
gfho.GiRaFFEfood_NRPy_Exact_Wald(gammaDD,M,KerrSchild_radial_shift)
def consistency_check(quantity1,quantity2,string):
if quantity1-quantity2==0:
print(string+" is in agreement!")
else:
print(string+" does not agree!")
sys.exit(1)
print("Consistency check between GiRaFFEfood_NRPy tutorial and NRPy+ module:")
for i in range(3):
consistency_check(ValenciavU[i],gfho.ValenciavU[i],"ValenciavU"+str(i))
consistency_check(AD[i],gfho.AD[i],"AD"+str(i))
```
<a id='latex_pdf_output'></a>
# Step 6: Output this notebook to $\LaTeX$-formatted PDF file \[Back to [top](#toc)\]
$$\label{latex_pdf_output}$$
The following code cell converts this Jupyter notebook into a proper, clickable $\LaTeX$-formatted PDF file. After the cell is successfully run, the generated PDF may be found in the root NRPy+ tutorial directory, with filename
[Tutorial-GiRaFFEfood_NRPy.pdf](Tutorial-GiRaFFEfood_NRPy.pdf) (Note that clicking on this link may not work; you may need to open the PDF file through another means.)
```
import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface
cmd.output_Jupyter_notebook_to_LaTeXed_PDF("Tutorial-GiRaFFEfood_NRPy_Exact_Wald",location_of_template_file=os.path.join(".."))
```
|
github_jupyter
|
# Step 0: Add NRPy's directory to the path
# https://stackoverflow.com/questions/16780014/import-file-from-parent-directory
import os,sys
nrpy_dir_path = os.path.join("..")
if nrpy_dir_path not in sys.path:
sys.path.append(nrpy_dir_path)
# Step 0.a: Import the NRPy+ core modules and set the reference metric to Cartesian
import sympy as sp # SymPy: The Python computer algebra package upon which NRPy+ depends
import NRPy_param_funcs as par # NRPy+: Parameter interface
import indexedexp as ixp # NRPy+: Symbolic indexed expression (e.g., tensors, vectors, etc.) support
import reference_metric as rfm
par.set_parval_from_str("reference_metric::CoordSystem","Cartesian")
rfm.reference_metric()
# Step 1a: Set commonly used parameters.
thismodule = "GiRaFFEfood_NRPy"
# KerrSchild_radial_shift = par.Cparameters("REAL",thismodule,"KerrSchild_radial_shift",0.4) # Default value for ExactWald
KerrSchild_radial_shift = sp.symbols("KerrSchild_radial_shift")
M = sp.symbols("M") # Black hole mass
# Step 2: Set the vectors A and E in Spherical coordinates
r = rfm.xxSph[0] + KerrSchild_radial_shift # We are setting the data up in Shifted Kerr-Schild coordinates
theta = rfm.xxSph[1]
# Initialize all components of A and E in the *spherical basis* to zero
ASphD = ixp.zerorank1()
ESphD = ixp.zerorank1()
ASphD[2] = (r * r * sp.sin(theta)**2)/2
ESphD[2] = 2 * M * sp.sin(theta)**2 / sp.sqrt(1+2*M/r)
# Step 3: Use the Jacobian matrix to transform the vectors to Cartesian coordinates.
drrefmetric__dx_0UDmatrix = sp.Matrix([[sp.diff(rfm.xxSph[0],rfm.xx[0]), sp.diff(rfm.xxSph[0],rfm.xx[1]), sp.diff(rfm.xxSph[0],rfm.xx[2])],
[sp.diff(rfm.xxSph[1],rfm.xx[0]), sp.diff(rfm.xxSph[1],rfm.xx[1]), sp.diff(rfm.xxSph[1],rfm.xx[2])],
[sp.diff(rfm.xxSph[2],rfm.xx[0]), sp.diff(rfm.xxSph[2],rfm.xx[1]), sp.diff(rfm.xxSph[2],rfm.xx[2])]])
#dx__drrefmetric_0UDmatrix = drrefmetric__dx_0UDmatrix.inv() # We don't actually need this in this case.
AD = ixp.zerorank1()
ED = ixp.zerorank1()
for i in range(3):
for j in range(3):
AD[i] = drrefmetric__dx_0UDmatrix[(j,i)]*ASphD[j]
ED[i] = drrefmetric__dx_0UDmatrix[(j,i)]*ESphD[j]
#Step 4: Declare the basic spacetime quantities
alpha = sp.symbols("alpha",real=True)
betaU = ixp.declarerank1("betaU",DIM=3)
gammaDD = ixp.declarerank2("gammaDD", "sym01",DIM=3)
import GRHD.equations as GRHD
GRHD.compute_sqrtgammaDET(gammaDD)
# Step 4: Calculate v^i from A_i and E_i
# Step 4a: Calculate the magnetic field B^i
GRHD.compute_sqrtgammaDET(gammaDD)
LeviCivitaTensorUUU = ixp.LeviCivitaTensorUUU_dim3_rank3(GRHD.sqrtgammaDET)
# For the initial data, we can analytically take the derivatives of A_i
ADdD = ixp.zerorank2()
for i in range(3):
for j in range(3):
ADdD[i][j] = sp.simplify(sp.diff(AD[i],rfm.xxCart[j]))
BU = ixp.zerorank1()
for i in range(3):
for j in range(3):
for k in range(3):
BU[i] += LeviCivitaTensorUUU[i][j][k] * ADdD[k][j]
# Step 4b: Calculate B^2 and B_i
# B^2 is an inner product defined in the usual way:
B2 = sp.sympify(0)
for i in range(3):
for j in range(3):
B2 += gammaDD[i][j] * BU[i] * BU[j]
# Lower the index on B^i
BD = ixp.zerorank1()
for i in range(3):
for j in range(3):
BD[i] += gammaDD[i][j] * BU[j]
# Step 4c: Calculate the Valencia 3-velocity
ValenciavU = ixp.zerorank1()
for i in range(3):
for j in range(3):
for k in range(3):
ValenciavU[i] += LeviCivitaTensorUUU[i][j][k]*ED[j]*BD[k]/B2
import GiRaFFEfood_NRPy.GiRaFFEfood_NRPy_Exact_Wald as gfho
gfho.GiRaFFEfood_NRPy_Exact_Wald(gammaDD,M,KerrSchild_radial_shift)
def consistency_check(quantity1,quantity2,string):
if quantity1-quantity2==0:
print(string+" is in agreement!")
else:
print(string+" does not agree!")
sys.exit(1)
print("Consistency check between GiRaFFEfood_NRPy tutorial and NRPy+ module:")
for i in range(3):
consistency_check(ValenciavU[i],gfho.ValenciavU[i],"ValenciavU"+str(i))
consistency_check(AD[i],gfho.AD[i],"AD"+str(i))
import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface
cmd.output_Jupyter_notebook_to_LaTeXed_PDF("Tutorial-GiRaFFEfood_NRPy_Exact_Wald",location_of_template_file=os.path.join(".."))
| 0.509764 | 0.932638 |
```
import numpy as np
import medleydb as mdb
from medleydb import download
import librosa
import os
import matplotlib.pyplot as plt
%matplotlib inline
def get_hcqt_params():
bins_per_octave=120
n_octaves = 5
harmonics = [1, 2, 3, 4, 5, 6]
sr = 22050
fmin = 32.7
hop_length = 128
return bins_per_octave, n_octaves, harmonics, sr, fmin, hop_length
def compute_hcqt(audio_fpath):
bins_per_octave, n_octaves, harmonics, sr, f_min, hop_length = get_hcqt_params()
y, fs = librosa.load(audio_fpath, sr=sr)
cqt_list = []
shapes = []
for h in harmonics:
cqt = librosa.cqt(
y, sr=fs, hop_length=hop_length, fmin=f_min*float(h),
n_bins=bins_per_octave*n_octaves,
bins_per_octave=bins_per_octave
)
cqt_list.append(cqt)
shapes.append(cqt.shape)
shapes_equal = [s == shapes[0] for s in shapes]
if not all(shapes_equal):
min_time = np.min([s[1] for s in shapes])
new_cqt_list = []
for i, cqt in enumerate(cqt_list):
new_cqt_list.append(cqt[:, :min_time])
cqt_list.pop(i)
cqt_list = new_cqt_list
log_hcqt = 20.0*np.log10(np.abs(np.array(cqt_list)) + 0.0001)
log_hcqt = log_hcqt - np.min(log_hcqt)
log_hcqt = log_hcqt / np.max(log_hcqt)
return log_hcqt
def get_freq_grid():
bins_per_octave, n_octaves, harmonics, sr, f_min, hop_length = get_hcqt_params()
freq_grid = librosa.cqt_frequencies(
bins_per_octave*n_octaves, f_min, bins_per_octave=bins_per_octave
)
return freq_grid
def get_time_grid(n_time_frames):
bins_per_octave, n_octaves, harmonics, sr, f_min, hop_length = get_hcqt_params()
time_grid = librosa.core.frames_to_time(
range(n_time_frames), sr=sr, hop_length=hop_length)
return time_grid
def grid_to_bins(grid, start_bin_val, end_bin_val):
bin_centers = (grid[1:] + grid[:-1])/2.0
bins = np.concatenate([[start_bin_val], bin_centers, [end_bin_val]])
return bins
def create_annotation_target(freq_grid, time_grid, annotation_times, annotation_freqs):
time_bins = grid_to_bins(time_grid, 0.0, time_grid[-1])
freq_bins = grid_to_bins(freq_grid, 0.0, freq_grid[-1])
annot_time_idx = np.digitize(annotation_times, time_bins) - 1
annot_freq_idx = np.digitize(annotation_freqs, freq_bins) - 1
annotation_target = np.zeros((len(freq_grid), len(time_grid)))
annotation_target[annot_freq_idx, annot_time_idx] = 1
return annotation_target
def get_all_pitch_annotations(mtrack):
annot_times = []
annot_freqs = []
for stem in mtrack.stems.values():
data = stem.pitch_annotation
data2 = stem.pitch_estimate_pyin
if data is not None:
annot = data
elif data2 is not None:
annot = data2
else:
continue
annot = np.array(annot).T
annot_times.append(annot[0])
annot_freqs.append(annot[1])
annot_times = np.concatenate(annot_times)
annot_freqs = np.concatenate(annot_freqs)
return annot_times, annot_freqs
def plot_annot_target(annot_target, hcqt, annot_times, annot_freqs):
plt.figure(figsize=(15,30))
plt.subplot(3, 1, 1)
plt.imshow(hcqt, origin='lower')
plt.axis('auto')
plt.axis('tight')
plt.subplot(3, 1, 2)
plt.imshow(annot_target, origin='lower')
plt.axis('auto')
plt.axis('tight')
plt.subplot(3, 1, 3)
plt.plot(annot_times, annot_freqs, ',')
plt.axis('tight')
plt.show()
def get_input_output_pairs(mtrack):
hcqt = compute_hcqt(mtrack.mix_path)
freq_grid = get_freq_grid()
time_grid = get_time_grid(len(hcqt[0][0]))
annot_times, annot_freqs = get_all_pitch_annotations(mtrack)
annot_target = create_annotation_target(
freq_grid, time_grid, annot_times, annot_freqs
)
plot_annot_target(annot_target, hcqt[0], annot_times, annot_freqs)
return hcqt, annot_target
def get_input_output_pairs_solo_pitch(audio_path, annot_times, annot_freqs, plot=False):
hcqt = compute_hcqt(audio_path)
freq_grid = get_freq_grid()
time_grid = get_time_grid(len(hcqt[0][0]))
annot_target = create_annotation_target(
freq_grid, time_grid, annot_times, annot_freqs
)
if plot:
plot_annot_target(annot_target, hcqt[0], annot_times, annot_freqs)
return hcqt, annot_target, freq_grid, time_grid
save_dir = "../output/training_data/"
failed_tracks = [
'ChrisJacoby_BoothShotLincoln',
'HezekiahJones_BorrowedHeart',
'Handel_TornamiAVagheggiar',
'JoelHelander_Definition',
'JoelHelander_ExcessiveResistancetoChange',
'JoelHelander_IntheAtticBedroom'
]
mtracks = mdb.load_all_multitracks(dataset_version=['V1'])
for mtrack in mtracks:
print(mtrack.track_id)
if mtrack.track_id in failed_tracks:
continue
stem = mtrack.predominant_stem
if stem is None:
continue
data = stem.pitch_annotation
save_path = os.path.join(
save_dir,
"{}_STEM_{}.npz".format(mtrack.track_id, stem.stem_idx)
)
if data is not None:
print(" > Stem {} {}".format(stem.stem_idx, stem.instrument))
annot = np.array(data).T
else:
continue
if os.path.exists(save_path):
one_stem_done = True
continue
if not os.path.exists(stem.audio_path):
print(" >downloading stem...")
download.download_stem(mtrack, stem.stem_idx)
print(" done!")
try:
data_in, data_out, freq, time = get_input_output_pairs_solo_pitch(
stem.audio_path, annot[0], annot[1]
)
np.savez(save_path, data_in=data_in, data_out=data_out, freq=freq, time=time)
except:
print(" > Something failed :(")
mtrack = mdb.MultiTrack("MusicDelta_Beatles")
data_input, data_target = get_input_output_pairs(mtrack)
```
|
github_jupyter
|
import numpy as np
import medleydb as mdb
from medleydb import download
import librosa
import os
import matplotlib.pyplot as plt
%matplotlib inline
def get_hcqt_params():
bins_per_octave=120
n_octaves = 5
harmonics = [1, 2, 3, 4, 5, 6]
sr = 22050
fmin = 32.7
hop_length = 128
return bins_per_octave, n_octaves, harmonics, sr, fmin, hop_length
def compute_hcqt(audio_fpath):
bins_per_octave, n_octaves, harmonics, sr, f_min, hop_length = get_hcqt_params()
y, fs = librosa.load(audio_fpath, sr=sr)
cqt_list = []
shapes = []
for h in harmonics:
cqt = librosa.cqt(
y, sr=fs, hop_length=hop_length, fmin=f_min*float(h),
n_bins=bins_per_octave*n_octaves,
bins_per_octave=bins_per_octave
)
cqt_list.append(cqt)
shapes.append(cqt.shape)
shapes_equal = [s == shapes[0] for s in shapes]
if not all(shapes_equal):
min_time = np.min([s[1] for s in shapes])
new_cqt_list = []
for i, cqt in enumerate(cqt_list):
new_cqt_list.append(cqt[:, :min_time])
cqt_list.pop(i)
cqt_list = new_cqt_list
log_hcqt = 20.0*np.log10(np.abs(np.array(cqt_list)) + 0.0001)
log_hcqt = log_hcqt - np.min(log_hcqt)
log_hcqt = log_hcqt / np.max(log_hcqt)
return log_hcqt
def get_freq_grid():
bins_per_octave, n_octaves, harmonics, sr, f_min, hop_length = get_hcqt_params()
freq_grid = librosa.cqt_frequencies(
bins_per_octave*n_octaves, f_min, bins_per_octave=bins_per_octave
)
return freq_grid
def get_time_grid(n_time_frames):
bins_per_octave, n_octaves, harmonics, sr, f_min, hop_length = get_hcqt_params()
time_grid = librosa.core.frames_to_time(
range(n_time_frames), sr=sr, hop_length=hop_length)
return time_grid
def grid_to_bins(grid, start_bin_val, end_bin_val):
bin_centers = (grid[1:] + grid[:-1])/2.0
bins = np.concatenate([[start_bin_val], bin_centers, [end_bin_val]])
return bins
def create_annotation_target(freq_grid, time_grid, annotation_times, annotation_freqs):
time_bins = grid_to_bins(time_grid, 0.0, time_grid[-1])
freq_bins = grid_to_bins(freq_grid, 0.0, freq_grid[-1])
annot_time_idx = np.digitize(annotation_times, time_bins) - 1
annot_freq_idx = np.digitize(annotation_freqs, freq_bins) - 1
annotation_target = np.zeros((len(freq_grid), len(time_grid)))
annotation_target[annot_freq_idx, annot_time_idx] = 1
return annotation_target
def get_all_pitch_annotations(mtrack):
annot_times = []
annot_freqs = []
for stem in mtrack.stems.values():
data = stem.pitch_annotation
data2 = stem.pitch_estimate_pyin
if data is not None:
annot = data
elif data2 is not None:
annot = data2
else:
continue
annot = np.array(annot).T
annot_times.append(annot[0])
annot_freqs.append(annot[1])
annot_times = np.concatenate(annot_times)
annot_freqs = np.concatenate(annot_freqs)
return annot_times, annot_freqs
def plot_annot_target(annot_target, hcqt, annot_times, annot_freqs):
plt.figure(figsize=(15,30))
plt.subplot(3, 1, 1)
plt.imshow(hcqt, origin='lower')
plt.axis('auto')
plt.axis('tight')
plt.subplot(3, 1, 2)
plt.imshow(annot_target, origin='lower')
plt.axis('auto')
plt.axis('tight')
plt.subplot(3, 1, 3)
plt.plot(annot_times, annot_freqs, ',')
plt.axis('tight')
plt.show()
def get_input_output_pairs(mtrack):
hcqt = compute_hcqt(mtrack.mix_path)
freq_grid = get_freq_grid()
time_grid = get_time_grid(len(hcqt[0][0]))
annot_times, annot_freqs = get_all_pitch_annotations(mtrack)
annot_target = create_annotation_target(
freq_grid, time_grid, annot_times, annot_freqs
)
plot_annot_target(annot_target, hcqt[0], annot_times, annot_freqs)
return hcqt, annot_target
def get_input_output_pairs_solo_pitch(audio_path, annot_times, annot_freqs, plot=False):
hcqt = compute_hcqt(audio_path)
freq_grid = get_freq_grid()
time_grid = get_time_grid(len(hcqt[0][0]))
annot_target = create_annotation_target(
freq_grid, time_grid, annot_times, annot_freqs
)
if plot:
plot_annot_target(annot_target, hcqt[0], annot_times, annot_freqs)
return hcqt, annot_target, freq_grid, time_grid
save_dir = "../output/training_data/"
failed_tracks = [
'ChrisJacoby_BoothShotLincoln',
'HezekiahJones_BorrowedHeart',
'Handel_TornamiAVagheggiar',
'JoelHelander_Definition',
'JoelHelander_ExcessiveResistancetoChange',
'JoelHelander_IntheAtticBedroom'
]
mtracks = mdb.load_all_multitracks(dataset_version=['V1'])
for mtrack in mtracks:
print(mtrack.track_id)
if mtrack.track_id in failed_tracks:
continue
stem = mtrack.predominant_stem
if stem is None:
continue
data = stem.pitch_annotation
save_path = os.path.join(
save_dir,
"{}_STEM_{}.npz".format(mtrack.track_id, stem.stem_idx)
)
if data is not None:
print(" > Stem {} {}".format(stem.stem_idx, stem.instrument))
annot = np.array(data).T
else:
continue
if os.path.exists(save_path):
one_stem_done = True
continue
if not os.path.exists(stem.audio_path):
print(" >downloading stem...")
download.download_stem(mtrack, stem.stem_idx)
print(" done!")
try:
data_in, data_out, freq, time = get_input_output_pairs_solo_pitch(
stem.audio_path, annot[0], annot[1]
)
np.savez(save_path, data_in=data_in, data_out=data_out, freq=freq, time=time)
except:
print(" > Something failed :(")
mtrack = mdb.MultiTrack("MusicDelta_Beatles")
data_input, data_target = get_input_output_pairs(mtrack)
| 0.463444 | 0.328462 |
# 9.5 Working with Files
Now that we've learned about how to open, close, read, and write to/from files, let's examine some common scenarios we'll encounter and how to acheive them in Python. The scenarios we'll cover are:
- File statistics such as file size, number of words in a file, and number of lines.
- Searching within a file.
- Appending data to a file.
- Working with two files at the same time.
## File Statistics
There are lots of statistics that we could try to calculate or find for a file, but the most common that I've run into, espcially text based files, are:
- File Size
- Number of Words
- Number of Lines
### Getting the File Size
Getting the size of a file is actually something that you can do without opening the file. There are two different ways using either the `os` or the `pathlib` module.
```
import os
os.path.getsize('resources/darth_plagueis_tragedy.txt')
import pathlib
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
print(path.stat().st_size)
```
<div class="alert alert-info">
<b>Note:</b> <code>pathlib</code> is only available for Python 3.4+.
</div>
### Getting the number of words of a file
```
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
with open(path) as reader:
num_words = 0
for line in reader:
# remember that .split() will take a line and turn it into
# a list with each delimited string as an entry. The
# default delimeter is a space (' ')
num_words += len(line.split())
print(num_words)
```
### Getting the number of lines in a file
```
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
with open(path) as reader:
# Remember that .readlines() returns a list of all the lines
num_lines = len(reader.readlines())
print(f'There are {num_lines} lines in {path}')
```
## Searching Within a File
```
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
with open(path) as reader:
for line in reader:
if line.find('Jedi') >= 0:
print(line)
```
<div class="alert alert-warning">
<b>Warning:</b> <code>find()</code> is case sensitive which means you'll need to account for that.
</div>
```
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
with open(path) as reader:
for line in reader:
if line.find('jedi') >= 0:
print(line)
```
### Searching for a non-case sensitive string
```
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
with open(path) as reader:
for line in reader:
# we take the search string and make sure all characters
# are lower case and then search for the string
location = line.lower().find('jedi')
if location >= 0:
print(line)
print('-' * location + 'ᐃ')
```
## Appending Data to a File
Appending data to a file is done by passing the `'a'` permission flag into the `open()` function. Let's start with a `test_data.txt` file that contains some numbers in it:
```
import random
path = pathlib.Path('resources/test_data.txt')
with open(path, 'w') as fh:
for _ in range(10):
fh.write(f'{random.randint(0, 100)}|')
with open(path) as reader:
print(reader.read())
```
Next, let's append some more numbers to the file
```
with open(path, 'a') as fh:
for _ in range(5):
fh.write(f'{random.randint(0, 100)}|')
with open(path) as reader:
print(reader.read())
```
## Working with Two Files at the Same Time
`open()` statements can be chained together with the `with` statement to properly open and close two files at the same time.
```
with open('resources/darth_plagueis_tragedy.txt', 'r') as reader, open('resources/darth_stats.txt', 'w') as writer:
for i, line in enumerate(reader):
words = line.lower().strip().split()
num_entries = words.count('the')
writer.write(f'"the" appears {num_entries} times in line {i + 1}\n')
with open('resources/darth_stats.txt') as fh:
print(fh.read())
```
## In-Class Assignments
1. Open the file 'resources/darth_plagueis_tragedy.txt' and find which non-case sensitive word is most often used (Hint: look back at module 1).
1. Do the same thing but ignoring the following words in your count:
- the,
- of,
- a,
- and
```
with open('resources/darth_plagueis_tragedy.txt') as reader:
counts = dict()
for line in reader:
words = line.lower().split()
for word in words:
counts[word] = counts.get(word, 0) + 1
# Get the max value based on the key's value
most_used = max(counts, key=lambda key: counts[key])
print(f"The most used word is '{most_used}' appearing {counts[most_used]} times.")
filtered_words = ['the', 'of', 'a', 'and']
with open('resources/darth_plagueis_tragedy.txt') as reader:
counts = dict()
# iterate through each line
for line in reader:
words = line.lower().split()
# iterate through each word of the line
for word in words:
# make sure that the word we have isn't one
# of the ones that should be filtered
if word not in filtered_words:
counts[word] = counts.get(word, 0) + 1
# Get the max value based on the key's value
most_used = max(counts, key=lambda key: counts[key])
print(f"The most used word is '{most_used}' appearing {counts[most_used]} times.")
```
|
github_jupyter
|
import os
os.path.getsize('resources/darth_plagueis_tragedy.txt')
import pathlib
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
print(path.stat().st_size)
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
with open(path) as reader:
num_words = 0
for line in reader:
# remember that .split() will take a line and turn it into
# a list with each delimited string as an entry. The
# default delimeter is a space (' ')
num_words += len(line.split())
print(num_words)
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
with open(path) as reader:
# Remember that .readlines() returns a list of all the lines
num_lines = len(reader.readlines())
print(f'There are {num_lines} lines in {path}')
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
with open(path) as reader:
for line in reader:
if line.find('Jedi') >= 0:
print(line)
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
with open(path) as reader:
for line in reader:
if line.find('jedi') >= 0:
print(line)
path = pathlib.Path('resources/darth_plagueis_tragedy.txt')
with open(path) as reader:
for line in reader:
# we take the search string and make sure all characters
# are lower case and then search for the string
location = line.lower().find('jedi')
if location >= 0:
print(line)
print('-' * location + 'ᐃ')
import random
path = pathlib.Path('resources/test_data.txt')
with open(path, 'w') as fh:
for _ in range(10):
fh.write(f'{random.randint(0, 100)}|')
with open(path) as reader:
print(reader.read())
with open(path, 'a') as fh:
for _ in range(5):
fh.write(f'{random.randint(0, 100)}|')
with open(path) as reader:
print(reader.read())
with open('resources/darth_plagueis_tragedy.txt', 'r') as reader, open('resources/darth_stats.txt', 'w') as writer:
for i, line in enumerate(reader):
words = line.lower().strip().split()
num_entries = words.count('the')
writer.write(f'"the" appears {num_entries} times in line {i + 1}\n')
with open('resources/darth_stats.txt') as fh:
print(fh.read())
with open('resources/darth_plagueis_tragedy.txt') as reader:
counts = dict()
for line in reader:
words = line.lower().split()
for word in words:
counts[word] = counts.get(word, 0) + 1
# Get the max value based on the key's value
most_used = max(counts, key=lambda key: counts[key])
print(f"The most used word is '{most_used}' appearing {counts[most_used]} times.")
filtered_words = ['the', 'of', 'a', 'and']
with open('resources/darth_plagueis_tragedy.txt') as reader:
counts = dict()
# iterate through each line
for line in reader:
words = line.lower().split()
# iterate through each word of the line
for word in words:
# make sure that the word we have isn't one
# of the ones that should be filtered
if word not in filtered_words:
counts[word] = counts.get(word, 0) + 1
# Get the max value based on the key's value
most_used = max(counts, key=lambda key: counts[key])
print(f"The most used word is '{most_used}' appearing {counts[most_used]} times.")
| 0.24599 | 0.967101 |
# 3. Awkward Array
<br><br><br><br><br>
## What about an array of lists?
```
import awkward as ak
import numpy as np
import uproot
events = uproot.open("data/HZZ.root:events")
events.show()
events["Muon_Px"].array()
events["Muon_Px"].array(entry_stop=20).tolist()
```
This is what Awkward Array was made for. NumPy's equivalent is cumbersome and inefficient.
```
jagged_numpy = events["Muon_Px"].array(entry_stop=20, library="np")
jagged_numpy
```
What if I want the first item in each list as an array?
```
np.array([x[0] for x in jagged_numpy])
```
This violates the rule from [1-python-performance.ipynb](1-python-performance.ipynb): don't iterate in Python.
```
jagged_awkward = events["Muon_Px"].array(entry_stop=20, library="ak")
jagged_awkward
jagged_awkward[:, 0]
```
<br><br><br><br><br>
### Jaggedness in Pandas
It can be done by putting the distinction between events in the DataFrame index.
```
events.arrays(filter_name="Muon_*", library="pd")
```
But if you want multiple particles, they can't be in the same DataFrame.
(A DataFrame has only one index; how would you relate jet subentry #1 with muon subentry #1?)
```
dataframes = events.arrays(filter_name="/(Muon_|Jet_).*/", library="pd")
len(dataframes)
dataframes[0]
dataframes[1]
```
Again, that's why we have Awkward Array.
```
array = events.arrays(filter_name="/(Muon_|Jet_).*/", library="ak", how="zip")
array
array.Jet
array.Jet.Px
array.Muon
array.Muon.Px
ak.num(array.Jet), ak.num(array.Muon)
```
<br><br><br><br><br>
## Awkward Array is a general-purpose library: NumPy-like idioms on JSON-like data
<img src="img/pivarski-one-slide-summary.svg" style="width: 70%">
<br><br><br><br><br>
## Main idea: slicing through structure is computationally inexpensive
Slicing by field name doesn't modify any large buffers and [ak.zip](https://awkward-array.readthedocs.io/en/latest/_auto/ak.zip.html) only scans them to ensure they're compatible (not even that if `depth_limit=1`).
```
array = events.arrays()
array
```
Think of this as zero-cost:
```
array.Muon_Px, array.Muon_Py, array.Muon_Pz
```
Think of this as zero-cost:
```
ak.zip({"px": array.Muon_Px, "py": array.Muon_Py, "pz": array.Muon_Pz})
```
(The above is a manual version of `how="zip"`.)
<br><br><br>
NumPy ufuncs work on these arrays (if they're "[broadcastable](https://awkward-array.readthedocs.io/en/latest/_auto/ak.broadcast_arrays.html)").
```
np.sqrt(array.Muon_Px**2 + array.Muon_Py**2)
```
<br><br><br>
And there are specialized operations that only make sense in a variable-length context.
<table style="margin-left: 0px">
<tr style="background: white"><td style="font-size: 1.75em; font-weight: bold; text-align: center"><a href="https://awkward-array.readthedocs.io/en/latest/_auto/ak.cartesian.html">ak.cartesian</a></td><td style="font-size: 1.75em; font-weight: bold; text-align: center"><a href="https://awkward-array.readthedocs.io/en/latest/_auto/ak.combinations.html">ak.combinations</a></td></tr>
<tr style="background: white"><td><img src="img/cartoon-cartesian.png"></td><td><img src="img/cartoon-combinations.png"></td></tr>
</table>
```
ak.cartesian((array.Muon_Px, array.Jet_Px))
ak.combinations(array.Muon_Px, 2)
```
<br><br><br><br><br>
## Arrays can have custom [behavior](https://awkward-array.readthedocs.io/en/latest/ak.behavior.html)
The following come from the new [Vector](https://github.com/scikit-hep/vector#readme) library.
```
import vector
vector.register_awkward()
muons = ak.zip({"px": array.Muon_Px, "py": array.Muon_Py, "pz": array.Muon_Pz, "E": array.Muon_E}, with_name="Momentum4D")
muons
```
This is an array of lists of vectors, and methods like `pt`, `eta`, `phi` apply through the whole array.
```
muons.pt
muons.eta
muons.phi
```
<br><br><br>
Let's try an example: ΔR(muons, jets)
```
jets = ak.zip({"px": array.Jet_Px, "py": array.Jet_Py, "pz": array.Jet_Pz, "E": array.Jet_E}, with_name="Momentum4D")
jets
ak.num(muons), ak.num(jets)
ms, js = ak.unzip(ak.cartesian((muons, jets)))
ms, js
ak.num(ms), ak.num(js)
ms.deltaR(js)
```
<br><br><br>
And another: muon pairs (all combinations, not just the first two per event).
```
ak.num(muons)
m1, m2 = ak.unzip(ak.combinations(muons, 2))
m1, m2
ak.num(m1), ak.num(m2)
m1 + m2
(m1 + m2).mass
import hist
hist.Hist.new.Reg(120, 0, 120, name="mass").Double().fill(
ak.flatten((m1 + m2).mass)
).plot()
```
<br><br><br>
### It doesn't matter which coordinates were used to construct it
```
array2 = uproot.open(
"root://eospublic.cern.ch//eos/opendata/cms/derived-data/AOD2NanoAODOutreachTool/Run2012BC_DoubleMuParked_Muons.root:Events"
).arrays(["Muon_pt", "Muon_eta", "Muon_phi", "Muon_charge"], entry_stop=100000)
import particle
muons2 = ak.zip({"pt": array2.Muon_pt, "eta": array2.Muon_eta, "phi": array2.Muon_phi, "q": array2.Muon_charge}, with_name="Momentum4D")
muons2["mass"] = particle.Particle.from_string("mu").mass / 1000.0
muons2
```
As long as you use properties (dots, not strings in brackets), you don't need to care what coordinates it's based on.
```
muons2.px
muons2.py
muons2.pz
muons2.E
m1, m2 = ak.unzip(ak.combinations(muons2, 2))
hist.Hist.new.Log(200, 0.1, 120, name="mass").Double().fill(
ak.flatten((m1 + m2).mass)
).plot()
```
<br><br><br>
## Awkward Arrays and Vector in Numba
Remember Numba, the JIT-compiler from [1-python-performance.ipynb](1-python-performance.ipynb)? Awkward Array and Vector have been implemented in Numba's compiler.
```
import numba as nb
@nb.njit
def first_big_dimuon(events):
for event in events:
for i in range(len(event)):
mu1 = event[i]
for j in range(i + 1, len(event)):
mu2 = event[j]
dimuon = mu1 + mu2
if dimuon.mass > 10:
return dimuon
first_big_dimuon(muons2)
```
|
github_jupyter
|
import awkward as ak
import numpy as np
import uproot
events = uproot.open("data/HZZ.root:events")
events.show()
events["Muon_Px"].array()
events["Muon_Px"].array(entry_stop=20).tolist()
jagged_numpy = events["Muon_Px"].array(entry_stop=20, library="np")
jagged_numpy
np.array([x[0] for x in jagged_numpy])
jagged_awkward = events["Muon_Px"].array(entry_stop=20, library="ak")
jagged_awkward
jagged_awkward[:, 0]
events.arrays(filter_name="Muon_*", library="pd")
dataframes = events.arrays(filter_name="/(Muon_|Jet_).*/", library="pd")
len(dataframes)
dataframes[0]
dataframes[1]
array = events.arrays(filter_name="/(Muon_|Jet_).*/", library="ak", how="zip")
array
array.Jet
array.Jet.Px
array.Muon
array.Muon.Px
ak.num(array.Jet), ak.num(array.Muon)
array = events.arrays()
array
array.Muon_Px, array.Muon_Py, array.Muon_Pz
ak.zip({"px": array.Muon_Px, "py": array.Muon_Py, "pz": array.Muon_Pz})
np.sqrt(array.Muon_Px**2 + array.Muon_Py**2)
ak.cartesian((array.Muon_Px, array.Jet_Px))
ak.combinations(array.Muon_Px, 2)
import vector
vector.register_awkward()
muons = ak.zip({"px": array.Muon_Px, "py": array.Muon_Py, "pz": array.Muon_Pz, "E": array.Muon_E}, with_name="Momentum4D")
muons
muons.pt
muons.eta
muons.phi
jets = ak.zip({"px": array.Jet_Px, "py": array.Jet_Py, "pz": array.Jet_Pz, "E": array.Jet_E}, with_name="Momentum4D")
jets
ak.num(muons), ak.num(jets)
ms, js = ak.unzip(ak.cartesian((muons, jets)))
ms, js
ak.num(ms), ak.num(js)
ms.deltaR(js)
ak.num(muons)
m1, m2 = ak.unzip(ak.combinations(muons, 2))
m1, m2
ak.num(m1), ak.num(m2)
m1 + m2
(m1 + m2).mass
import hist
hist.Hist.new.Reg(120, 0, 120, name="mass").Double().fill(
ak.flatten((m1 + m2).mass)
).plot()
array2 = uproot.open(
"root://eospublic.cern.ch//eos/opendata/cms/derived-data/AOD2NanoAODOutreachTool/Run2012BC_DoubleMuParked_Muons.root:Events"
).arrays(["Muon_pt", "Muon_eta", "Muon_phi", "Muon_charge"], entry_stop=100000)
import particle
muons2 = ak.zip({"pt": array2.Muon_pt, "eta": array2.Muon_eta, "phi": array2.Muon_phi, "q": array2.Muon_charge}, with_name="Momentum4D")
muons2["mass"] = particle.Particle.from_string("mu").mass / 1000.0
muons2
muons2.px
muons2.py
muons2.pz
muons2.E
m1, m2 = ak.unzip(ak.combinations(muons2, 2))
hist.Hist.new.Log(200, 0.1, 120, name="mass").Double().fill(
ak.flatten((m1 + m2).mass)
).plot()
import numba as nb
@nb.njit
def first_big_dimuon(events):
for event in events:
for i in range(len(event)):
mu1 = event[i]
for j in range(i + 1, len(event)):
mu2 = event[j]
dimuon = mu1 + mu2
if dimuon.mass > 10:
return dimuon
first_big_dimuon(muons2)
| 0.325092 | 0.917488 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from pandas.io.json import json_normalize
from pymongo import MongoClient
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
%matplotlib inline
course_cluster_uri = "mongodb://agg-student:[email protected]:27017,cluster0-shard-00-01-jxeqq.mongodb.net:27017,cluster0-shard-00-02-jxeqq.mongodb.net:27017/test?ssl=true&replicaSet=Cluster0-shard-0&authSource=admin"
course_client = MongoClient(course_cluster_uri)
# Lichman, M. (2013). UCI Machine Learning Repository [http://archive.ics.uci.edu/ml]. Irvine, CA: University of California, School of Information and Computer Science.
wine = course_client['coursera-agg']['wine']
pipeline = [
{
"$project": {
"_id": 0
}
}
]
cursor = wine.aggregate(pipeline)
docs = list(cursor)
df = json_normalize(docs)
df.head()
X = df.drop(['Alcohol'], axis=1).values.astype('float64')
X = preprocessing.scale(X)
cov_matrix = np.cov(X.T)
eigenvalues, eigenvectors = np.linalg.eig(cov_matrix)
for val in eigenvalues:
print(val)
eigen_map = list(zip(eigenvalues, eigenvectors.T))
eigen_map.sort(key=lambda x: x[0], reverse=True)
sorted_eigenvalues = [pair[0] for pair in eigen_map]
sorted_eigenvectors = [pair[1] for pair in eigen_map]
sorted_eigenvalues
print(pd.DataFrame(sorted_eigenvectors, columns=df.drop(['Alcohol'], axis=1).columns))
eigenvalue_sum = sum(eigenvalues)
var_exp = [(v / eigenvalue_sum)*100 for v in sorted_eigenvalues]
cum_var_exp = np.cumsum(var_exp)
dims = len(df.drop(['Alcohol'], axis=1).columns)
plt.clf()
fig, ax = plt.subplots()
ax.plot(range(dims), cum_var_exp, '-o')
plt.xlabel('Number of Components')
plt.ylabel('Percent of Variance Explained')
plt.show()
ev1 = sorted_eigenvectors[0]
ev2 = sorted_eigenvectors[1]
eigen_matrix = np.hstack((ev1.reshape(dims,1), ev2.reshape(dims,1)))
eigen_matrix
Y = X.dot(eigen_matrix)
plt.clf()
fig, ax = plt.subplots()
ax.scatter(Y.T[0], Y.T[1], alpha=0.2)
plt.xlabel('PC1')
plt.ylabel('PC2')
plt.show()
from sklearn.decomposition import PCA
pca = PCA(n_components=2)
Y_sklearn = pca.fit_transform(X)
plt.clf()
fig, ax = plt.subplots()
ax.scatter(Y_sklearn.T[0], Y_sklearn.T[1], alpha=0.2)
plt.xlabel('PC1')
plt.ylabel('PC2')
plt.show()
y = df['Alcohol'].values
# Let's split the model for training and testing, and use a logistic regression
X_train, X_test, y_train, y_test = train_test_split(df.drop('Alcohol', axis=1), y, test_size=0.25)
classifier = LogisticRegression(random_state=0)
classifier.fit(X_train, y_train)
y_pred = classifier.score(X_test, y_test)
y_pred
# now with PCA applied
X_train, X_test, y_train, y_test = train_test_split(Y_sklearn, y, test_size=0.3)
classifier_with_pca = LogisticRegression(random_state=0)
classifier_with_pca.fit(X_train, y_train)
y_pred = classifier_with_pca.score(X_test, y_test)
y_pred
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from pandas.io.json import json_normalize
from pymongo import MongoClient
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
%matplotlib inline
course_cluster_uri = "mongodb://agg-student:[email protected]:27017,cluster0-shard-00-01-jxeqq.mongodb.net:27017,cluster0-shard-00-02-jxeqq.mongodb.net:27017/test?ssl=true&replicaSet=Cluster0-shard-0&authSource=admin"
course_client = MongoClient(course_cluster_uri)
# Lichman, M. (2013). UCI Machine Learning Repository [http://archive.ics.uci.edu/ml]. Irvine, CA: University of California, School of Information and Computer Science.
wine = course_client['coursera-agg']['wine']
pipeline = [
{
"$project": {
"_id": 0
}
}
]
cursor = wine.aggregate(pipeline)
docs = list(cursor)
df = json_normalize(docs)
df.head()
X = df.drop(['Alcohol'], axis=1).values.astype('float64')
X = preprocessing.scale(X)
cov_matrix = np.cov(X.T)
eigenvalues, eigenvectors = np.linalg.eig(cov_matrix)
for val in eigenvalues:
print(val)
eigen_map = list(zip(eigenvalues, eigenvectors.T))
eigen_map.sort(key=lambda x: x[0], reverse=True)
sorted_eigenvalues = [pair[0] for pair in eigen_map]
sorted_eigenvectors = [pair[1] for pair in eigen_map]
sorted_eigenvalues
print(pd.DataFrame(sorted_eigenvectors, columns=df.drop(['Alcohol'], axis=1).columns))
eigenvalue_sum = sum(eigenvalues)
var_exp = [(v / eigenvalue_sum)*100 for v in sorted_eigenvalues]
cum_var_exp = np.cumsum(var_exp)
dims = len(df.drop(['Alcohol'], axis=1).columns)
plt.clf()
fig, ax = plt.subplots()
ax.plot(range(dims), cum_var_exp, '-o')
plt.xlabel('Number of Components')
plt.ylabel('Percent of Variance Explained')
plt.show()
ev1 = sorted_eigenvectors[0]
ev2 = sorted_eigenvectors[1]
eigen_matrix = np.hstack((ev1.reshape(dims,1), ev2.reshape(dims,1)))
eigen_matrix
Y = X.dot(eigen_matrix)
plt.clf()
fig, ax = plt.subplots()
ax.scatter(Y.T[0], Y.T[1], alpha=0.2)
plt.xlabel('PC1')
plt.ylabel('PC2')
plt.show()
from sklearn.decomposition import PCA
pca = PCA(n_components=2)
Y_sklearn = pca.fit_transform(X)
plt.clf()
fig, ax = plt.subplots()
ax.scatter(Y_sklearn.T[0], Y_sklearn.T[1], alpha=0.2)
plt.xlabel('PC1')
plt.ylabel('PC2')
plt.show()
y = df['Alcohol'].values
# Let's split the model for training and testing, and use a logistic regression
X_train, X_test, y_train, y_test = train_test_split(df.drop('Alcohol', axis=1), y, test_size=0.25)
classifier = LogisticRegression(random_state=0)
classifier.fit(X_train, y_train)
y_pred = classifier.score(X_test, y_test)
y_pred
# now with PCA applied
X_train, X_test, y_train, y_test = train_test_split(Y_sklearn, y, test_size=0.3)
classifier_with_pca = LogisticRegression(random_state=0)
classifier_with_pca.fit(X_train, y_train)
y_pred = classifier_with_pca.score(X_test, y_test)
y_pred
| 0.749362 | 0.689502 |
# PyTorch
## __Tensors__
Одно из основных понятий в PyTorch -- это __Tenosor__.
https://pytorch.org/docs/master/tensors.html
__Tensor__ -- это такой же массив, как и в __numpy.array__, размерность и тип данных которого мы можем задать. Tensor в отличие от numpy.array может вычисляться на __GPU__.
```
import numpy as np
import torch
N = 100
D_in = 50
dtype = torch.float
device = torch.device("cpu")
# device = torch.device("cuda:0") # Uncomment this to run on GPU
x = np.random.randn(N, D_in)
x_torch = torch.randn(N, D_in, device=device, dtype=dtype)
x
x_torch
x_torch = torch.Tensor(np.ones((N, D_in)))
x_torch
x_torch = torch.FloatTensor([1, 2, 3])
x_torch
x1 = torch.IntTensor([1, 2, 3])
x2 = torch.FloatTensor([3, 4, 5])
```
В PyTorch можно найти много операций, которые похожи на то, что есть в numpy :
```
- torch.add (np.add) -> сложение тензоров (поэлементное)
- torch.sub (np.subtract) -> вычитание (поэлементное)
- torch.mul (np.multiply) -> умнажение скаляров / матриц (поэлементное)
- torch.mm (np.matmul) -> перемножение матриц
- torch.ones (np.ones) -> создание тензора из единиц
```
```
# Давайте попробуем вышепересчисленные операции
x1 = torch.FloatTensor([[1, 2, 3], [4, 5, 6]])
x2 = torch.FloatTensor([[7, 8], [9, 1], [2, 3]])
out = torch.mm(x1, x2)
out
```
```
- torch.reshape (np.reshape) -> изменения порядка элементов в тензоре, не путать с транспонированием.
```
## Dynamic Computational Graph
После того, как были реализованы архитектура модели и весь процес обучения и валидация сети, при запуске кода в PyTorch происходят следующие этапы:
1. Строится вычислительный граф (направленный ациклический граф), где каждый узел -- это тензор, а ребро, ведущее к дргуому узлу, это выполнение операции над данным тензором, которое ведет к результату - другому тензору.
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/Graph.png?raw=1" alt="Drawing" style="width: 300px;"/>
Реализуем двухслойную сеть для задачи регрессии. И граф для такой архитектуры бдует выглядить следующим образом:
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/RegGraph.png?raw=1" alt="Drawing" />
```
batch_size = 64
input_size = 3
hidden_size = 2
output_size = 1
# Create random input and output data
x = torch.randn(batch_size, input_size, device=device, dtype=dtype)
y = torch.randn(batch_size, output_size, device=device, dtype=dtype)
# Randomly initialize weights
w1 = torch.randn(input_size, hidden_size, device=device, dtype=dtype)
w2 = torch.randn(hidden_size, output_size, device=device, dtype=dtype)
learning_rate = 1e-6
for t in range(500):
# Forward pass: compute predicted y
#TODO
# Compute and print loss
loss = (y_pred - y).pow(2).sum().item()
```
## Autograd
2. Еще одно фундаментальное понятие и важный элемент при построении графа -- это __Autograd__ -- автоматическое дифференцирование.
Для того чтобы с помощью стохастического градиентного спуска обновить обучаемые параметры сети, нужно посчитать градиенты. И как известно, обновление весов, которые учавтсвуют в нескольких операциях, происходит по `правилу дифференцирования сложной функции` (цепное правило или __chain rule__).
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/RegChainRule.png?raw=1" alt="Drawing" />
То есть (1) вычислительный граф позволяет определить последовательность операций, а (2) автоматическое дифференцирование посчитать нужные градиенты.
Если бы `Autograd` не было, то тогда backprop надо было бы реализовывать самим, и как это бы выглядело?
Рассмотрим на примере, как посчиать градиенты для весов из входного слоя, где входной вектора `X` состоит из 3-х компонент. А входной слой вторую размерность имеет равной 2.
После чего это идет в `ReLU`, но для простоты опустим на время ее, и посмотрим как дальше это идет по сети.
Ниже написано, как это все вычисляется и приводит нас к значению целевой функции для одного наблюдения
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/1.png?raw=1" alt="1" style="width: 600px;"/>
Тогда, чтобы посчитать градиент по первому элементу из обучаемой матрицы на первом слое, необходимо взять производоную у сложной функции. А этот как раз делается по `chain rule`: сначала берем у внешней, потом спускаемся на уровень ниже, и так пока не додйдем до то функции, после которой эта перменная уже нигде не участвует:
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/2.png?raw=1" alt="2" style="width: 400px;"/>
Перепишем это все в матричном виде, то есть сделаем аналог вида матрицы весов из первого слоя, но там уже будут её градиенты, котоыре будут нужны чтобы как раз обновить эти веса:
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/3.jpg?raw=1" alt="3" style="width: 600px;"/>
Как видно, здесь можно вектор X вынести, то есть разделить на две матрицы:
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/4.jpg?raw=1" alt="4" style="width: 500px;"/>
То есть уже видно, что будем траспонировать входной вектор(матрицу). Но надо понимать, что в реальности у нас не одно наблюдение в батче, а несколько, тогда запись немного изменит свой вид:
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/5.jpg?raw=1" alt="5" style="width: 500px;"/>
Теперь мы видим, как на самом деле вычисляется вот те самые частные производные для вектора X, то есть видно, как математически это можно записать, а именно:
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/6.jpg?raw=1" alt="6" style="width: 500px;"/>
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/7.jpg?raw=1" alt="7" style="width: 500px;"/>
Уже можно реализовать. Понятно, что транспонируется, что нет, и что на что умножается.
Но помним про ReLU. Для простоты опустили, но теперь её учесть будет легче.
Так как после первого слоя идет ReLU, а значит, занулились те выходы первого слоя, которые были __меньше__ нуля. Получается, что во второй слой не все дошло, тогда нужно обнулить, что занулил ReLU.
Что занулил ReLU, мы можем выяснить при `forward pass`, а где именно поставить нули, то надо уже смотреть относительно `backward propagation`, на том выходе, где последний раз участвовал выход после ReLU, то есть:
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/8.jpg?raw=1" alt="8" style="width: 600px;"/>
Теперь реализуем эти формулы на PyTorch:
```
#TODO
grad_w2 =
# TODO
grad_w1 =
# Update weights using gradient descent
w1 -= learning_rate * grad_w1
w2 -= learning_rate * grad_w2
```
Благодаря `Autograd` реализацию `chain rule` можно избежать, так как для более сложных нейронных сетей вручную такое реализовать сложно, при этом сделать это эффективным.
Для того чтобы PyTorch понял, за какими переменными надо "следить", то есть указать, что именно "эти" переменные являются обучаемыми, необходимо при создании тензора в качестве аттрибута указать __requires_grad=True__:
```
w1 = torch.randn(input_size, hidden_size, device=device, dtype=dtype, requires_grad=True)
w2 = torch.randn(hidden_size, output_size, device=device, dtype=dtype, requires_grad=True)
learning_rate = 1e-6
for t in range(500):
y_pred = x.mm(w1).clamp(min=0).mm(w2)
loss = (y_pred - y).pow(2).sum()
if t % 100 == 99:
print(t, loss.item())
# Теперь подсчет градиентов для весов происходит при вызове backward
loss.backward()
# Обновляем значение весов, но укзаываем, чтобы PyTorch не считал эту операцию,
# которая бы учавствовала бы при подсчете градиентов в chain rule
with torch.no_grad():
w1 -= learning_rate * w1.grad
w2 -= learning_rate * w2.grad
# Теперь обнуляем значение градиентов, чтобы на следующем шаге
# они не учитывались при подсчете новых градиентов,
# иначе произойдет суммирвоание старых и новых градиентов
w1.grad.zero_()
w2.grad.zero_()
```
Осталось еще не вручную обновлять веса, а использовать адаптивные методы градинетного спсука. Для этого нужно использовать модуль __optim__. А помимо оптимайзера, еще можно использовать готовые целевые функции из модлуя __nn__.
```
import torch.optim as optim
loss_fn = torch.nn.MSELoss(reduction='sum')
learning_rate = 1e-6
optimizer = torch.optim.Adam([w1, w2], lr=learning_rate)
for t in range(500):
optimizer.zero_grad()
y_pred = x.mm(w1).clamp(min=0).mm(w2)
loss = loss_fn(y_pred, y)
if t % 100 == 99:
print(t, loss.item())
loss.backward()
optimizer.step()
```
После того, как мы сделали backward, в этот момент посчитались градиенты и граф уничтожился, то есть стёрлись все пути, которые связывали тензоры между собой. Это значит, что еще раз backward сделать не поулчится, будет ошибка. Но если вдруг нужно считать градиенты еще раз, то нужно при вызове backward задать `retain_graph=True`.
Еще важный аттрибут, который есть у Tensor -- это `grad_fn`. В этом аттрибуте указывается та функция, посредством которой был создан этот тензор. Так PyTorch понимает, как именно считать по нему градиент.
```
y_pred.grad_fn
```
Также можно контролировать, должны ли градиенты течь или нет.
```
x = torch.tensor([1], requires_grad=True)
with torch.no_grad():
with torch.enable_grad():
y = x * 2
y.requires_grad
```
## Почему Backprop надо понимать
1. Backprop позволяет понимать, как те или иные операции, сложные конструкции в сети влияют на обнолвение весов.
Почему лучше сделать конкатенацию тензоров, а не поэлементное сложение. Для этого нужно посмотреть на backprop, как будут обновляться веса.
2. Даже на таком маленьком пример двуслойной MLP можно уже увидеть, когда `ReLU`, как функция активация, не очень хорошо применять. Если разреженные данные, то получить на выходе много нулей вероятнее, чем при использовании `LeakyReLU`, то есть градиенты будут нулевыми и веса никак не будут обновляться => сеть не обучается!
3. В архитектуре могут встречаться недифференцируемые операции, и первое - это нужно понять, потому что при обучении сети это может быть не сразу заметно, просто качество модели будет плохое, и точность хорошую не поулчится достичь.
Например, в одной из статей было предложено в качестве механизма внимания применить распределение Бернулли, которое умножается на выход промежуточного слоя сети. И эта опреация недифференцируема, нужно реализовывать backprop самим, тем самым обеспечить корректное протекание градиентов.
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/Bernoulli.png?raw=1" alt="8" style="width: 600px;"/>
Так же любая статья, которая предлагет новую целевую функцию для той или иной задачи, там всегда будут представлены градиенты, чтобы было понимание, как это влияет на обновление весов. Не просто так !
<img src="https://github.com/RiskModellingResearch/RiskManagementDL_Autumn21/blob/main/week03/images/BernoulliBackProp.png?raw=1" alt="8" style="width: 600px;"/>
## nn.Module
В предыдущем примере архитектуру сети создавали используя последовательной способ объявления слоев сети -- `nn.Sequential`.
Но еще можно это сделать более гибким подходом:
```
class TwoLayerNet(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
"""
TwoLayerNet наследуется от nn.Module и тем самым полчаем возможность
переопределять методы класса.
В конструктуре создаем слои (обучаемые веса) и другие нужные перменные/функции,
которые нужны для модели
"""
super(TwoLayerNet, self).__init__()
self.linear1 = torch.nn.Linear(input_size, hidden_size)
self.linear2 = torch.nn.Linear(hidden_size, output_size)
def forward(self, x):
"""
Метод forward отвечает за прямое распростронение модели,
поэтому данный метод нужно переопределять обязательно,
чтобы задать логику прямого распростронения.
Именно в этот момент начинает строится динамический граф
"""
h_relu = self.linear1(x).clamp(min=0)
y_pred = self.linear2(h_relu)
return y_pred
batch_size = 64
input_size = 1000
hidden_size = 100
output_size = 10
x = torch.randn(batch_size, input_size, device=device, dtype=dtype)
y = torch.randn(batch_size, output_size, device=device, dtype=dtype)
model = TwoLayerNet(Dinput_size_in, hidden_size, output_size)
loss_fn = torch.nn.MSELoss(reduction='sum')
optimizer = torch.optim.SGD(model.parameters(), lr=1e-4)
for t in range(500):
y_pred = model(x)
loss = loss_fn(y_pred, y)
if t % 100 == 99:
print(t, loss.item())
optimizer.zero_grad()
loss.backward()
optimizer.step()
```
|
github_jupyter
|
import numpy as np
import torch
N = 100
D_in = 50
dtype = torch.float
device = torch.device("cpu")
# device = torch.device("cuda:0") # Uncomment this to run on GPU
x = np.random.randn(N, D_in)
x_torch = torch.randn(N, D_in, device=device, dtype=dtype)
x
x_torch
x_torch = torch.Tensor(np.ones((N, D_in)))
x_torch
x_torch = torch.FloatTensor([1, 2, 3])
x_torch
x1 = torch.IntTensor([1, 2, 3])
x2 = torch.FloatTensor([3, 4, 5])
- torch.add (np.add) -> сложение тензоров (поэлементное)
- torch.sub (np.subtract) -> вычитание (поэлементное)
- torch.mul (np.multiply) -> умнажение скаляров / матриц (поэлементное)
- torch.mm (np.matmul) -> перемножение матриц
- torch.ones (np.ones) -> создание тензора из единиц
# Давайте попробуем вышепересчисленные операции
x1 = torch.FloatTensor([[1, 2, 3], [4, 5, 6]])
x2 = torch.FloatTensor([[7, 8], [9, 1], [2, 3]])
out = torch.mm(x1, x2)
out
- torch.reshape (np.reshape) -> изменения порядка элементов в тензоре, не путать с транспонированием.
batch_size = 64
input_size = 3
hidden_size = 2
output_size = 1
# Create random input and output data
x = torch.randn(batch_size, input_size, device=device, dtype=dtype)
y = torch.randn(batch_size, output_size, device=device, dtype=dtype)
# Randomly initialize weights
w1 = torch.randn(input_size, hidden_size, device=device, dtype=dtype)
w2 = torch.randn(hidden_size, output_size, device=device, dtype=dtype)
learning_rate = 1e-6
for t in range(500):
# Forward pass: compute predicted y
#TODO
# Compute and print loss
loss = (y_pred - y).pow(2).sum().item()
#TODO
grad_w2 =
# TODO
grad_w1 =
# Update weights using gradient descent
w1 -= learning_rate * grad_w1
w2 -= learning_rate * grad_w2
w1 = torch.randn(input_size, hidden_size, device=device, dtype=dtype, requires_grad=True)
w2 = torch.randn(hidden_size, output_size, device=device, dtype=dtype, requires_grad=True)
learning_rate = 1e-6
for t in range(500):
y_pred = x.mm(w1).clamp(min=0).mm(w2)
loss = (y_pred - y).pow(2).sum()
if t % 100 == 99:
print(t, loss.item())
# Теперь подсчет градиентов для весов происходит при вызове backward
loss.backward()
# Обновляем значение весов, но укзаываем, чтобы PyTorch не считал эту операцию,
# которая бы учавствовала бы при подсчете градиентов в chain rule
with torch.no_grad():
w1 -= learning_rate * w1.grad
w2 -= learning_rate * w2.grad
# Теперь обнуляем значение градиентов, чтобы на следующем шаге
# они не учитывались при подсчете новых градиентов,
# иначе произойдет суммирвоание старых и новых градиентов
w1.grad.zero_()
w2.grad.zero_()
import torch.optim as optim
loss_fn = torch.nn.MSELoss(reduction='sum')
learning_rate = 1e-6
optimizer = torch.optim.Adam([w1, w2], lr=learning_rate)
for t in range(500):
optimizer.zero_grad()
y_pred = x.mm(w1).clamp(min=0).mm(w2)
loss = loss_fn(y_pred, y)
if t % 100 == 99:
print(t, loss.item())
loss.backward()
optimizer.step()
y_pred.grad_fn
x = torch.tensor([1], requires_grad=True)
with torch.no_grad():
with torch.enable_grad():
y = x * 2
y.requires_grad
class TwoLayerNet(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
"""
TwoLayerNet наследуется от nn.Module и тем самым полчаем возможность
переопределять методы класса.
В конструктуре создаем слои (обучаемые веса) и другие нужные перменные/функции,
которые нужны для модели
"""
super(TwoLayerNet, self).__init__()
self.linear1 = torch.nn.Linear(input_size, hidden_size)
self.linear2 = torch.nn.Linear(hidden_size, output_size)
def forward(self, x):
"""
Метод forward отвечает за прямое распростронение модели,
поэтому данный метод нужно переопределять обязательно,
чтобы задать логику прямого распростронения.
Именно в этот момент начинает строится динамический граф
"""
h_relu = self.linear1(x).clamp(min=0)
y_pred = self.linear2(h_relu)
return y_pred
batch_size = 64
input_size = 1000
hidden_size = 100
output_size = 10
x = torch.randn(batch_size, input_size, device=device, dtype=dtype)
y = torch.randn(batch_size, output_size, device=device, dtype=dtype)
model = TwoLayerNet(Dinput_size_in, hidden_size, output_size)
loss_fn = torch.nn.MSELoss(reduction='sum')
optimizer = torch.optim.SGD(model.parameters(), lr=1e-4)
for t in range(500):
y_pred = model(x)
loss = loss_fn(y_pred, y)
if t % 100 == 99:
print(t, loss.item())
optimizer.zero_grad()
loss.backward()
optimizer.step()
| 0.529263 | 0.962848 |
```
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import datetime, nltk, warnings
import matplotlib.cm as cm
import itertools
from sklearn.preprocessing import StandardScaler
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_samples, silhouette_score
from sklearn import preprocessing, model_selection, metrics, feature_selection
from sklearn.model_selection import GridSearchCV, learning_curve
from sklearn.metrics import confusion_matrix
from sklearn import neighbors, linear_model, svm, tree, ensemble
```
This function analyzes the content of the **Description** column by performing the following operations:
- extract names appearing in the products description
- for each name, extract the root of the word and aggregate the set of names associated with this particular root
- count the number of times each root appears in the dataframe
- when several words are listed in the same root, choose the shortest name.This systematically selects the singular when there are singular/plural variants)
```
is_noun = lambda pos: pos[:2] == 'NN'
def keywords_inventory(dataframe, column = 'Description'):
"""Extract features from text"""
stemmer = nltk.stem.SnowballStemmer("english")
keywords_roots = dict() # collect the words / root
keywords_select = dict() # association: root <-> keyword
category_keys = []
count_keywords = dict()
icount = 0
for s in dataframe[column]:
if pd.isnull(s): continue
lines = s.lower()
tokenized = nltk.word_tokenize(lines)
nouns = [word for (word, pos) in nltk.pos_tag(tokenized) if is_noun(pos)]
for t in nouns:
t = t.lower() ; stm = stemmer.stem(t)
if stm in keywords_roots:
keywords_roots[stm].add(t)
count_keywords[stm] += 1
else:
keywords_roots[stm] = {t}
count_keywords[stm] = 1
for s in keywords_roots.keys():
if len(keywords_roots[s]) > 1:
min_length = 1000
for k in keywords_roots[s]:
if len(k) < min_length:
clef = k ; min_length = len(k)
category_keys.append(clef)
keywords_select[s] = clef
else:
category_keys.append(list(keywords_roots[s])[0])
keywords_select[s] = list(keywords_roots[s])[0]
print("Number of keywords in variable '{}': {}".format(column,len(category_keys)))
return category_keys, keywords_roots, keywords_select, count_keywords
def graph_component_silhouette(n_clusters, lim_x, mat_size, sample_silhouette_values, clusters):
"""Create the silhouette value plot for each group"""
plt.rcParams["patch.force_edgecolor"] = True
plt.style.use('fivethirtyeight')
mpl.rc('patch', edgecolor = 'dimgray', linewidth=1)
fig, ax1 = plt.subplots(1, 1)
fig.set_size_inches(8, 8)
ax1.set_xlim([lim_x[0], lim_x[1]])
ax1.set_ylim([0, mat_size + (n_clusters + 1) * 10])
y_lower = 10
for i in range(n_clusters):
# Aggregate the silhouette scores for samples belonging to cluster i, and sort them
ith_cluster_silhouette_values = sample_silhouette_values[clusters == i]
ith_cluster_silhouette_values.sort()
size_cluster_i = ith_cluster_silhouette_values.shape[0]
y_upper = y_lower + size_cluster_i
cmap = cm.get_cmap("Spectral")
color = cmap(float(i) / n_clusters)
ax1.fill_betweenx(np.arange(y_lower, y_upper), 0, ith_cluster_silhouette_values,
facecolor=color, edgecolor=color, alpha=0.8)
# Label the silhouette plots with their cluster numbers at the middle
ax1.text(-0.03, y_lower + 0.5 * size_cluster_i, str(i), color = 'red', fontweight = 'bold',
bbox=dict(facecolor='white', edgecolor='black', boxstyle='round, pad=0.3'))
# Compute the new y_lower for next plot
y_lower = y_upper + 10
def random_color_func(word=None, font_size=None, position=None,
orientation=None, font_path=None, random_state=None):
h = int(360.0 * tone / 255.0)
s = int(100.0 * 255.0 / 255.0)
l = int(100.0 * float(random_state.randint(70, 120)) / 255.0)
return "hsl({}, {}%, {}%)".format(h, s, l)
def make_wordcloud(liste, increment):
"""Plot word cloud of each group"""
ax1 = fig.add_subplot(5,2,increment)
words = dict()
trunc_occurences = liste[0:150]
for s in trunc_occurences:
words[s[0]] = s[1]
wordcloud = WordCloud(width=1000,height=400, background_color='lightgrey',
max_words=1628,relative_scaling=1,
color_func = random_color_func,
normalize_plurals=False)
wordcloud.generate_from_frequencies(words)
ax1.imshow(wordcloud, interpolation="bilinear")
ax1.axis('off')
plt.title('cluster nº{}'.format(increment-1))
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import datetime, nltk, warnings
import matplotlib.cm as cm
import itertools
from sklearn.preprocessing import StandardScaler
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_samples, silhouette_score
from sklearn import preprocessing, model_selection, metrics, feature_selection
from sklearn.model_selection import GridSearchCV, learning_curve
from sklearn.metrics import confusion_matrix
from sklearn import neighbors, linear_model, svm, tree, ensemble
is_noun = lambda pos: pos[:2] == 'NN'
def keywords_inventory(dataframe, column = 'Description'):
"""Extract features from text"""
stemmer = nltk.stem.SnowballStemmer("english")
keywords_roots = dict() # collect the words / root
keywords_select = dict() # association: root <-> keyword
category_keys = []
count_keywords = dict()
icount = 0
for s in dataframe[column]:
if pd.isnull(s): continue
lines = s.lower()
tokenized = nltk.word_tokenize(lines)
nouns = [word for (word, pos) in nltk.pos_tag(tokenized) if is_noun(pos)]
for t in nouns:
t = t.lower() ; stm = stemmer.stem(t)
if stm in keywords_roots:
keywords_roots[stm].add(t)
count_keywords[stm] += 1
else:
keywords_roots[stm] = {t}
count_keywords[stm] = 1
for s in keywords_roots.keys():
if len(keywords_roots[s]) > 1:
min_length = 1000
for k in keywords_roots[s]:
if len(k) < min_length:
clef = k ; min_length = len(k)
category_keys.append(clef)
keywords_select[s] = clef
else:
category_keys.append(list(keywords_roots[s])[0])
keywords_select[s] = list(keywords_roots[s])[0]
print("Number of keywords in variable '{}': {}".format(column,len(category_keys)))
return category_keys, keywords_roots, keywords_select, count_keywords
def graph_component_silhouette(n_clusters, lim_x, mat_size, sample_silhouette_values, clusters):
"""Create the silhouette value plot for each group"""
plt.rcParams["patch.force_edgecolor"] = True
plt.style.use('fivethirtyeight')
mpl.rc('patch', edgecolor = 'dimgray', linewidth=1)
fig, ax1 = plt.subplots(1, 1)
fig.set_size_inches(8, 8)
ax1.set_xlim([lim_x[0], lim_x[1]])
ax1.set_ylim([0, mat_size + (n_clusters + 1) * 10])
y_lower = 10
for i in range(n_clusters):
# Aggregate the silhouette scores for samples belonging to cluster i, and sort them
ith_cluster_silhouette_values = sample_silhouette_values[clusters == i]
ith_cluster_silhouette_values.sort()
size_cluster_i = ith_cluster_silhouette_values.shape[0]
y_upper = y_lower + size_cluster_i
cmap = cm.get_cmap("Spectral")
color = cmap(float(i) / n_clusters)
ax1.fill_betweenx(np.arange(y_lower, y_upper), 0, ith_cluster_silhouette_values,
facecolor=color, edgecolor=color, alpha=0.8)
# Label the silhouette plots with their cluster numbers at the middle
ax1.text(-0.03, y_lower + 0.5 * size_cluster_i, str(i), color = 'red', fontweight = 'bold',
bbox=dict(facecolor='white', edgecolor='black', boxstyle='round, pad=0.3'))
# Compute the new y_lower for next plot
y_lower = y_upper + 10
def random_color_func(word=None, font_size=None, position=None,
orientation=None, font_path=None, random_state=None):
h = int(360.0 * tone / 255.0)
s = int(100.0 * 255.0 / 255.0)
l = int(100.0 * float(random_state.randint(70, 120)) / 255.0)
return "hsl({}, {}%, {}%)".format(h, s, l)
def make_wordcloud(liste, increment):
"""Plot word cloud of each group"""
ax1 = fig.add_subplot(5,2,increment)
words = dict()
trunc_occurences = liste[0:150]
for s in trunc_occurences:
words[s[0]] = s[1]
wordcloud = WordCloud(width=1000,height=400, background_color='lightgrey',
max_words=1628,relative_scaling=1,
color_func = random_color_func,
normalize_plurals=False)
wordcloud.generate_from_frequencies(words)
ax1.imshow(wordcloud, interpolation="bilinear")
ax1.axis('off')
plt.title('cluster nº{}'.format(increment-1))
| 0.644449 | 0.791217 |
# Determining the Market Value of Debt
If we cannot get access to data on the individual debt instruments issued by the company, then all we can do is use the book value of debt from the balance sheet as an estimate of the market value of debt.
If we still don't have data on the individual debt instruments, but do have an average maturity of the debt, we can do a bit better job with estimating the market value of debt.
If we have data on all the individual debt instruments, the combined with an estimate of the cost of debt, we can get an accurate estimate the market value of debt to be used in the WACC calculation.
This example is divided into three sections, depending on what data are available.
# Some Setup
These inputs will be used throughout.
```
total_book_debt = 1000000
interest_expense = 60000
cost_of_debt = 0.08
```
# MV Debt with No Additional Data
Since we don't have access to an average maturity or the individual debt instruments, all we can do is just use the book value of debt.
```
mv_debt = total_book_debt
mv_debt
```
And we are done. It's not a very good estimate though.
# MV Debt with Average Maturity
Let's just say we also knew that the average maturity of the debt was 5 years. We may be able to find this info when the individual debt instruments are not available. The general approach is then to treat the entire total book debt from the balance sheet as a single coupon bond, and determine the value of that coupon bond.
```
average_maturity = 5
```
## Determine the Coupon Payment on the Hypothetical Bond
The coupon payment on this hypothetical bond can be estimated by taking the interest expense divided by the total book debt.
```
coupon_rate = interest_expense / total_book_debt
coupon_rate
```
Now that we have a coupon rate, we can form the bond cash flows. It should be the coupon rate multiplied by principal for each period up until the final period, which is (1 + coupon rate) multiplied by principal.
## Calculate Hypothetical Bond Cash Flows
```
principal = total_book_debt
coupon_payment = coupon_rate * principal
cash_flows = [coupon_rate * principal for i in range(average_maturity - 1)] + [(1 + coupon_rate) * principal]
cash_flows
```
## Calculate Value of Hypothetical Bond as MV of Debt
Now we can calculate the value of this bond by taking the NPV with the cost of debt. Recall that `numpy`'s NPV function takes the first cash flow as year 0, so we will need to add a 0 at the beginning.
```
import numpy_financial as npf
mv_debt = npf.npv(cost_of_debt, [0] + cash_flows)
mv_debt
```
Therefore we have \\$9.2 million as our estimate for the market value of debt.
## Complete the Preceding Steps with One Forumla
We can wrap all the prior steps into one formula, which calculates the value of an annuity with an additional terminal payment. The formula is: $$V = C (\frac{1 - (1 + r_d)^{-t}}{r_d}) + \frac{P}{(1 + r_d)^t} $$
```
mv_debt = coupon_payment * ((1 - (1 + cost_of_debt)**(-average_maturity))/cost_of_debt) + principal/(1 + cost_of_debt)**average_maturity
mv_debt
```
We can wrap this all up in a function.
```
def mv_debt_by_average_maturity(average_maturity, cost_of_debt, total_book_debt, interest_expense):
"""
Calculate the market value of debt based off financial statement data, cost of debt, and average maturity.
"""
coupon_rate = interest_expense / total_book_debt
principal = total_book_debt
coupon_payment = coupon_rate * principal
return coupon_payment * ((1 - (1 + cost_of_debt)**(-average_maturity))/cost_of_debt) + principal/(1 + cost_of_debt)**average_maturity
mv_debt_by_average_maturity(average_maturity, cost_of_debt, total_book_debt, interest_expense)
```
This formula is especially useful when the average maturity is not a whole number of years, say 5.5 years.
```
mv_debt_by_average_maturity(5.5, cost_of_debt, total_book_debt, interest_expense)
```
# MV Debt with Individual Debt Instruments
The basic process is to value each debt instrument based on its coupon rate and the overall cost of debt, similar to what we did for the hypothetical bond in the last section. Then we sum of the values of all the instruments to find the total market value of debt.
Please note that we must have data on all the debt instruments to use this approach. If any are missing, then it is better to determine the average maturity from the ones we have, and use the approach described in the last section.
Another approach for missing some instruments is to calculate the ratio of the total instrument principal to total balance sheet debt, then after determining the total market value of the instruments, divide that by the ratio to get an estimate of the market value of debt. But we will not explore this option here.
Here we will load in some data which has information on the individual debt instruments for this company.
```
import pandas as pd
df = pd.read_excel('debt data.xlsx')
df
```
## Checking if the Approach will be Valid
Let's see if the total of the principal on the instruments matches up to the firm's total debt.
```
df['Principal'].sum()
total_book_debt
```
We can see these two are the same, so we are fine to go ahead with this approach. For a real company, it will likely never match exactly. If we're close (within a couple percent) then this approach should be fine.
## Calculating the Value of a Debt Instrument
Here we will follow a similar valuation approach to the last section. Since we need to do this for every bond, we will create a function which does this for a single bond and apply it to all the bonds. Let's create this function before working with the `DataFrame`. Here we use the exact same logic from the last section to get the cash flows.
```
principal = 300000
coupon_rate = 0.07
maturity = 2
coupon_payment = coupon_rate * principal
cash_flows = [coupon_rate * principal for i in range(maturity - 1)] + [(1 + coupon_rate) * principal]
cash_flows
```
And get the value in the same way as the last section.
```
mv_debt = npf.npv(cost_of_debt, [0] + cash_flows)
mv_debt
```
Let's wrap this up in one function.
```
def mv_bond(principal, coupon_rate, maturity, cost_of_debt):
"""
Calculate the market value of bond
"""
coupon_payment = coupon_rate * principal
cash_flows = [coupon_payment for i in range(maturity - 1)] + [coupon_payment + principal]
return npf.npv(cost_of_debt, [0] + cash_flows)
mv_bond(principal, coupon_rate, maturity, cost_of_debt)
```
### A Quick Aside to `DataFrame.apply`
Here we have just three securities, it would be pretty easy to manually call this on each of them. But what if there are 50 securities? We want an automated approach to apply this function to all the rows in the `DataFrame`. This is where we bring in `.apply` on the `DataFrame`. When we do `.apply(function, axis=1)` on a `DataFrame`, it will take each row of the `DataFrame` (the row is now a `Series`) and pass it to the function. Then it will take the result of the function call and put it into a new `Series`.
Let's see how this works with a simple example that just prints out what we're getting from the `.apply` call.
```
def understand_apply(series):
"""
For demonstration purposes to understand DataFrame.apply
"""
print('Calling understand_apply')
print(f'Got values:\n{series}\n')
print(f'Values have type {type(series)}')
principal = series['Principal']
print(f'Value of principal in the series: {principal}')
print('\n\n\n') # separate output
df.apply(understand_apply, axis=1)
```
## Calculating the Value of All the Debt Instruments
Now that we understand `.apply`, we can use it to apply this `mv_bond` function to all the `DataFrame` rows. But we'll need to make a new function based off the `mv_bond` function which can take a `Series` rather than the individual values.
```
def mv_bond_for_apply(series, cost_of_debt):
"""
Calculate market value of a bond. This version to be used with DataFrame.apply
"""
principal = series['Principal']
coupon_rate = series['Coupon Rate']
maturity = series['Maturity (years)']
return mv_bond(principal, coupon_rate, maturity, cost_of_debt)
df.apply(mv_bond_for_apply, axis=1, cost_of_debt=cost_of_debt)
```
We can now see we get the value of each bond in a `Series`. The last thing to do is assign that back to the `DataFrame` as a column.
```
df['Value'] = df.apply(mv_bond_for_apply, axis=1, cost_of_debt=cost_of_debt)
df
```
Now we can simply take the sum of those values to get the market value of debt for the firm.
```
mv_debt = df['Value'].sum()
mv_debt
```
## Using Dates Instead of Years to Maturity
Usually with individual debt instruments we will not get a number of years remaining until maturity, but instead a maturity date, and we need to calculate the years to maturity.
When using this approach, the annuity formula should be used to handle non-integer years. Let's first swap that out and make sure it works the same.
```
def mv_bond_annuity_approach(principal, coupon_rate, maturity, cost_of_debt):
"""
Calculate the market value of bond with non-integer maturity
"""
coupon_payment = coupon_rate * principal
return coupon_payment * ((1 - (1 + cost_of_debt)**(-maturity))/cost_of_debt) + principal/(1 + cost_of_debt)**maturity
def mv_bond_annuity_approach_for_apply(series, cost_of_debt):
"""
Calculate market value of a bond with non-integer maturity. This version to be used with DataFrame.apply
"""
principal = series['Principal']
coupon_rate = series['Coupon Rate']
maturity = series['Maturity (years)']
return mv_bond_annuity_approach(principal, coupon_rate, maturity, cost_of_debt)
df.apply(mv_bond_annuity_approach_for_apply, axis=1, cost_of_debt=cost_of_debt)
```
We see we get the same results. Now let's go to calculating the maturities. First, so we're not cheating, let's drop the years to maturity and the value we already calculated.
```
df = df.drop(['Maturity (years)', 'Value'], axis=1) # axis=1 here means column, the command can also be used to drop rows with axis=0
df
```
### Setting the Data Type Correctly
Depending on our data, `pandas` may or may not know where our dates are. We can check this by looking at the data types (`dtypes`) of the columns:
```
df.dtypes
```
We can see here the data type for maturity is listed as `datetime64[ns]`, which means it's a date, which is what we want. If it instead was coming as `object`, then we would have to run the following command:
```
df['Maturity (Date)'] = pd.to_datetime(df['Maturity (Date)'])
df
df.dtypes
```
As we already had it as a date, it had no effect, but it didn't hurt.
### Calculating the Difference Between the Maturity Date and Today
Let's first pull a single date from the `DataFrame` to work with.
```
date = df.loc[0, 'Maturity (Date)'] # get the first row (0-indexed) value from the Maturity (Date) column
date
```
We can see this value shows as a `Timestamp`, which is the way `pandas` represents dates.
Next we need to get the value for today. We can use the built-in `datetime` module for this.
```
import datetime
today = datetime.datetime.today()
today
```
This is a `datetime.datetime` object, which works just fine with `pandas`' `Timestamp`. We can directly do math with them:
```
diff = date - today
diff
```
The difference between two datetime objects is a `Timedelta` object. Now to get a number of years (or any time horizon), we convert it into the total number of seconds, then convert the seconds into years.
```
seconds = diff.total_seconds()
seconds
seconds_per_year = 60 * 60 * 24 * 365
seconds_per_year
years_elapsed = seconds / seconds_per_year
years_elapsed
```
It may be a little bit off due to leap years. We can certainly adjust for this, but this is usually "good enough" and handling leap years is outside the scope of the class.
Let's create a function for this.
```
def years_until_from_date(date):
"""
Calculate the number of years until a date, starting from today.
"""
today = datetime.datetime.today()
diff = date - today
seconds = diff.total_seconds()
seconds_per_year = 60 * 60 * 24 * 365
years_elapsed = seconds / seconds_per_year
return years_elapsed
years_until_from_date(date)
```
We notice it's slightly different, because some seconds have elapsed since running the prior commands! Now let's create the version to use with `DataFrame.apply`.
```
def years_until_from_date_for_apply(series, date_col='Maturity (Date)'):
date = series[date_col]
return years_until_from_date(date)
df.apply(years_until_from_date_for_apply, axis=1)
```
Now to wrap it up we can assign back to the `DataFrame`.
```
df['Maturity (years)'] = df.apply(years_until_from_date_for_apply, axis=1)
df
```
Which now allows us to calculate the value, this time using our calculated maturity.
```
df['Value'] = df.apply(mv_bond_annuity_approach_for_apply, axis=1, cost_of_debt=cost_of_debt)
df
```
And again, we can simply take the sum of those values to get the market value of debt for the firm.
```
df['Value'].sum()
```
|
github_jupyter
|
total_book_debt = 1000000
interest_expense = 60000
cost_of_debt = 0.08
mv_debt = total_book_debt
mv_debt
average_maturity = 5
coupon_rate = interest_expense / total_book_debt
coupon_rate
principal = total_book_debt
coupon_payment = coupon_rate * principal
cash_flows = [coupon_rate * principal for i in range(average_maturity - 1)] + [(1 + coupon_rate) * principal]
cash_flows
import numpy_financial as npf
mv_debt = npf.npv(cost_of_debt, [0] + cash_flows)
mv_debt
mv_debt = coupon_payment * ((1 - (1 + cost_of_debt)**(-average_maturity))/cost_of_debt) + principal/(1 + cost_of_debt)**average_maturity
mv_debt
def mv_debt_by_average_maturity(average_maturity, cost_of_debt, total_book_debt, interest_expense):
"""
Calculate the market value of debt based off financial statement data, cost of debt, and average maturity.
"""
coupon_rate = interest_expense / total_book_debt
principal = total_book_debt
coupon_payment = coupon_rate * principal
return coupon_payment * ((1 - (1 + cost_of_debt)**(-average_maturity))/cost_of_debt) + principal/(1 + cost_of_debt)**average_maturity
mv_debt_by_average_maturity(average_maturity, cost_of_debt, total_book_debt, interest_expense)
mv_debt_by_average_maturity(5.5, cost_of_debt, total_book_debt, interest_expense)
import pandas as pd
df = pd.read_excel('debt data.xlsx')
df
df['Principal'].sum()
total_book_debt
principal = 300000
coupon_rate = 0.07
maturity = 2
coupon_payment = coupon_rate * principal
cash_flows = [coupon_rate * principal for i in range(maturity - 1)] + [(1 + coupon_rate) * principal]
cash_flows
mv_debt = npf.npv(cost_of_debt, [0] + cash_flows)
mv_debt
def mv_bond(principal, coupon_rate, maturity, cost_of_debt):
"""
Calculate the market value of bond
"""
coupon_payment = coupon_rate * principal
cash_flows = [coupon_payment for i in range(maturity - 1)] + [coupon_payment + principal]
return npf.npv(cost_of_debt, [0] + cash_flows)
mv_bond(principal, coupon_rate, maturity, cost_of_debt)
def understand_apply(series):
"""
For demonstration purposes to understand DataFrame.apply
"""
print('Calling understand_apply')
print(f'Got values:\n{series}\n')
print(f'Values have type {type(series)}')
principal = series['Principal']
print(f'Value of principal in the series: {principal}')
print('\n\n\n') # separate output
df.apply(understand_apply, axis=1)
def mv_bond_for_apply(series, cost_of_debt):
"""
Calculate market value of a bond. This version to be used with DataFrame.apply
"""
principal = series['Principal']
coupon_rate = series['Coupon Rate']
maturity = series['Maturity (years)']
return mv_bond(principal, coupon_rate, maturity, cost_of_debt)
df.apply(mv_bond_for_apply, axis=1, cost_of_debt=cost_of_debt)
df['Value'] = df.apply(mv_bond_for_apply, axis=1, cost_of_debt=cost_of_debt)
df
mv_debt = df['Value'].sum()
mv_debt
def mv_bond_annuity_approach(principal, coupon_rate, maturity, cost_of_debt):
"""
Calculate the market value of bond with non-integer maturity
"""
coupon_payment = coupon_rate * principal
return coupon_payment * ((1 - (1 + cost_of_debt)**(-maturity))/cost_of_debt) + principal/(1 + cost_of_debt)**maturity
def mv_bond_annuity_approach_for_apply(series, cost_of_debt):
"""
Calculate market value of a bond with non-integer maturity. This version to be used with DataFrame.apply
"""
principal = series['Principal']
coupon_rate = series['Coupon Rate']
maturity = series['Maturity (years)']
return mv_bond_annuity_approach(principal, coupon_rate, maturity, cost_of_debt)
df.apply(mv_bond_annuity_approach_for_apply, axis=1, cost_of_debt=cost_of_debt)
df = df.drop(['Maturity (years)', 'Value'], axis=1) # axis=1 here means column, the command can also be used to drop rows with axis=0
df
df.dtypes
df['Maturity (Date)'] = pd.to_datetime(df['Maturity (Date)'])
df
df.dtypes
date = df.loc[0, 'Maturity (Date)'] # get the first row (0-indexed) value from the Maturity (Date) column
date
import datetime
today = datetime.datetime.today()
today
diff = date - today
diff
seconds = diff.total_seconds()
seconds
seconds_per_year = 60 * 60 * 24 * 365
seconds_per_year
years_elapsed = seconds / seconds_per_year
years_elapsed
def years_until_from_date(date):
"""
Calculate the number of years until a date, starting from today.
"""
today = datetime.datetime.today()
diff = date - today
seconds = diff.total_seconds()
seconds_per_year = 60 * 60 * 24 * 365
years_elapsed = seconds / seconds_per_year
return years_elapsed
years_until_from_date(date)
def years_until_from_date_for_apply(series, date_col='Maturity (Date)'):
date = series[date_col]
return years_until_from_date(date)
df.apply(years_until_from_date_for_apply, axis=1)
df['Maturity (years)'] = df.apply(years_until_from_date_for_apply, axis=1)
df
df['Value'] = df.apply(mv_bond_annuity_approach_for_apply, axis=1, cost_of_debt=cost_of_debt)
df
df['Value'].sum()
| 0.551574 | 0.990971 |
# Now You Code 4: Guess A Number
Write a program to play the classic "Guess a number" game.
In this game the computer selects a random number between 1 and 10.
It's your job to guess the number. Whenever you guess, the computer will
give you a hint of higher or lower. This process repeats until you guess
the number, after which the computer reports the number of guesses it took you.
For Example:
I'm thinking of a number between 1 and 10...
Your guess: 5
Too low. Guess higher.
Your guess: 7
Too high. Guess lower.
Your guess: 6
You guessed it in 3 tries.
Your loop should continue until your input guess equals the
computer generated random number.
### How do you make Python generate a random number?
```
# Sample code which demostrates how to generate a number between 1 and 10
import random
number = random.randint(1,70)
print(number)
```
Run the cell above a couple of times. Notice how each time you execute the code, it comes up with a different number.
Here's a breakdown of the code
```
line 1 imports the random module
line 2 randomly selects an integer between 1 and 10
line 3 prints the number
```
Now that you understand how to generate a random number, try to design then write code for the program. The first step in your program should be to generate the random number.
## Step 1: Problem Analysis
Inputs:
- a number between one and ten (user input)
- random number
-(generated), random_number
Outputs:
- your number is too high, try again
- your number is too high, try again
- you guessed the number in number of attempts
Algorithm (Steps in Program):
1. Import random
2. Set number of guesses to zero
3. Set random number to be between one and ten
4. Prompt for number between one and ten
5. Compare user guess to random
5. If user guess is low. Output that the number is too low and that they should try a larger number and then prompt for a new number.
6. If user guess is high. Output that the number is too high and that they should try a smaller number and then prompt for a new number.
7. If guess is correct break the loop.
8. Output that they are correct along with the number of tries it took them.
```
# Step 2: write code for program
import random
guesses = 0
computer_number = random.randint(1,10)
try:
while True:
user_number = int(input("I'm thinking of a number between 1 and 10. What is your guess? "))
guesses = guesses + 1
if user_number < computer_number:
print ("Your guess of %d is too low. Please guess a higher number." % user_number)
elif user_number > computer_number:
print ("Your guess of %d is too high. Please guess a lower number." % user_number)
elif user_number == computer_number:
break
print("Your guess of %d is right. You guessed it in %d tries." % (user_number, guesses))
except ValueError:
print("Your input was not an integer. Please try again.")
```
## Step 3: Questions
1. Which loop did you use to solve the problem? Was it a definite or indefinite loop?
- I used a indefinite loop.
2. Modify this program to allow you to guess a number between 1 and 100. How much of your code did you need to change to make this work?
- I only had to change random.randint(1,10) to (1,100). I also changed the prompt to ask for a number between 1 and 100.
3. This program is a good example of a difficult problem to conceptualize which has a simple solution when you look at actual lines of code. I assume you did not write this in a single try, so explain where you got stuck and describe your approach to overcoming it.
- My main issues were getting my indents correct. I fixed this by just going checking the errors and making chnages based off what the error told me.
- The second was getting it to ask for an input again after the first guess. I overcame this by removing the break statements after the if statemnts regarding the guess being too low or high.
- The other issue I had was that I got type errors because I didn't assign the variables as integers. I fixed this by assigning both guesses and user_number as integers.
## Reminder of Evaluation Criteria
1. Was the problem attempted (analysis, code, and answered questions) ?
2. Was the problem analysis thought out? (does the program match the plan?)
3. Does the code execute without syntax error?
4. Does the code solve the intended problem?
5. Is the code well written? (easy to understand, modular, and self-documenting, handles errors)
|
github_jupyter
|
# Sample code which demostrates how to generate a number between 1 and 10
import random
number = random.randint(1,70)
print(number)
line 1 imports the random module
line 2 randomly selects an integer between 1 and 10
line 3 prints the number
# Step 2: write code for program
import random
guesses = 0
computer_number = random.randint(1,10)
try:
while True:
user_number = int(input("I'm thinking of a number between 1 and 10. What is your guess? "))
guesses = guesses + 1
if user_number < computer_number:
print ("Your guess of %d is too low. Please guess a higher number." % user_number)
elif user_number > computer_number:
print ("Your guess of %d is too high. Please guess a lower number." % user_number)
elif user_number == computer_number:
break
print("Your guess of %d is right. You guessed it in %d tries." % (user_number, guesses))
except ValueError:
print("Your input was not an integer. Please try again.")
| 0.412885 | 0.963472 |
<img src="CdeC.png">
```
from IPython.display import YouTubeVideo, HTML
YouTubeVideo('VIxciS1B9eo')
```
## Funciones
Las funciones nos facilitan la programación porque no tenemos que escribir nuevamente todo el codigo de una rutina que vamos a reutilizar
Una función se define en python como:
```python
def mi_funcion(var1,var2):
# el algoritmo
return x
```
```
# ejemplo
def mi_funcion():
return 'Clubes de Ciencia!!'
print (mi_funcion())
#ejemplo
def contar_letras(texto):
n = len(texto)
return n
def contar_palabras(texto):
lista = texto.split(' ')
n = len(lista)
return n
def contar_palabras_letras(texto):
palabras = contar_palabras(texto)
letras = contar_letras(texto)
return [palabras, letras]
print (contar_palabras_letras('contar palabras y letras'))
# ejemplo
def potencia(x,n):
a = 1
for i in range(n): # range(n) genera una lista de numeros de 0 a n-1 de 1 en 1
a = a*x
return a
def factorial(n):
if n == 0:
return 1
if n < 0:
return 'valor negativo'
factorial = 1
for i in range(1,n+1):
factorial = factorial*i
return factorial
print (potencia(3,3))
print (factorial(4))
```
## Reto de Programación
- Construya una función que retorne el nombre de una de sus compañeros de grupo cuando se ingresa el número de
tarjeta de identidad
```python
def encontrar_nombre(numero_identidad):
# codigo
return nombre_completo
```
- La serie de Fibonacci es muy importante en varias areas del conocimiento. Esta se define como:
$$f_{0} = 0 ,$$
$$f_{1} = 1,$$
$$f_{n} = f_{n-1} + f_{n-2}$$
Es decir, el siguiente valor es la suma de los dos anteriores.
$$ f_{2} = 1 + 0,$$
$$f_{3} = 1 + 1,$$
$$f_{4} = 2 + 1$$
Escriba una función que retorne la serie de Fibonacci de algun número $n$.
Por ejemplo para $n=4$, la función debe devolver la lista [0,1,1,2,3]
# Librerias
Las librerias contienen funciones que nos ayudan resolver problemas complejos y nos facilitan la programación.
```python
import pandas # Pandas nos permite leer archivos de excel, filtrar, y hacer estadisticas sobre tabalas
import numpy # Numpy contiene funciones de operaciones matematicas y algebra de matrices
import matplotlib # Matplotlib es una libreria que nos ayuda a graficar datos y funciones matematicas
```
```
# ejemplo, La hora actual del servidor
import datetime
print (datetime.datetime.now())
# ejemplo, Transpuesta de una matriz
import numpy as np
A = np.matrix([[3, 6, -5],
[1, -3, 2],
[5, -1, 4]])
print (A.shape) # las dimensiones de la matriz
print (A.transpose()) # tarnspuesta de la matriz A
%matplotlib notebook
# ejemplo, Gráfica de y = x**2
import matplotlib.pylab as plt
x = list(range(-50,50))
y = [i**2 for i in x]
plt.figure()
plt.scatter(x,y)
plt.title('$y = x^{2}$') # titulo
plt.xlabel('x') # titulo eje x
plt.ylabel('y') # titulo eje y
plt.show()
x = np.linspace(0, 2 * np.pi, 500)
y1 = np.sin(x)
y2 = np.sin(3 * x)
fig, ax = plt.subplots()
ax.fill(x, y1, 'b', x, y2, 'r', alpha=0.3)
plt.show()
# ejemplo, Crear una tabal de datos de sus compañeros
import pandas as pd
nombres = ['Jocelyn', 'Laura','Luis Alejandro']
apellidos = ['Kshi', 'Diaz', 'Mahecha']
pais = ['Estados Unidos', 'Colombia', 'Colombia']
pd.DataFrame({'nombre': nombres, 'apellido': apellidos, 'pais': pais})
```
## Reto de Programación
Cree un dataframe ó tabla que tenga las siguinetes columnas: t, a, v, y:
- t es el tiempo y va de 0 a 100
- a es la aceleración de la gravedad a = 10
- v es la velocidad, y es función de t : $v = 20 - at$
- y es función de t: $y = -5t^{2}$
Grafique y, v, a en función de t
# Pandas y Tablas de datos
```
temperatura_global = pd.read_csv('GlobalTemperatures.csv')
```
# Analisis Temperaturas
https://www.dkrz.de/Nutzerportal-en/doku/vis/sw/python-matplotlib/matplotlib-sourcecode/python-matplotlib-example-contour-filled-plot
https://data.giss.nasa.gov/gistemp/maps/
```
from __future__ import (absolute_import, division, print_function)
from six.moves import (filter, input, map, range, zip) # noqa
import matplotlib.cm as mpl_cm
import matplotlib.pyplot as plt
import iris
import iris.quickplot as qplt
fname = iris.sample_data_path('air_temp.pp')
temperature_cube = iris.load_cube(fname)
# Load a Cynthia Brewer palette.
brewer_cmap = mpl_cm.get_cmap('brewer_OrRd_09')
# Draw the contour with 25 levels.
plt.figure()
qplt.contourf(temperature_cube, 25)
# Add coastlines to the map created by contourf.
plt.gca().coastlines()
plt.show()
# Draw the contours, with n-levels set for the map colours (9).
# NOTE: needed as the map is non-interpolated, but matplotlib does not provide
# any special behaviour for these.
plt.figure()
qplt.contourf(temperature_cube, brewer_cmap.N, cmap=brewer_cmap)
# Add coastlines to the map created by contourf.
plt.gca().coastlines()
plt.show()
```
|
github_jupyter
|
from IPython.display import YouTubeVideo, HTML
YouTubeVideo('VIxciS1B9eo')
def mi_funcion(var1,var2):
# el algoritmo
return x
# ejemplo
def mi_funcion():
return 'Clubes de Ciencia!!'
print (mi_funcion())
#ejemplo
def contar_letras(texto):
n = len(texto)
return n
def contar_palabras(texto):
lista = texto.split(' ')
n = len(lista)
return n
def contar_palabras_letras(texto):
palabras = contar_palabras(texto)
letras = contar_letras(texto)
return [palabras, letras]
print (contar_palabras_letras('contar palabras y letras'))
# ejemplo
def potencia(x,n):
a = 1
for i in range(n): # range(n) genera una lista de numeros de 0 a n-1 de 1 en 1
a = a*x
return a
def factorial(n):
if n == 0:
return 1
if n < 0:
return 'valor negativo'
factorial = 1
for i in range(1,n+1):
factorial = factorial*i
return factorial
print (potencia(3,3))
print (factorial(4))
def encontrar_nombre(numero_identidad):
# codigo
return nombre_completo
import pandas # Pandas nos permite leer archivos de excel, filtrar, y hacer estadisticas sobre tabalas
import numpy # Numpy contiene funciones de operaciones matematicas y algebra de matrices
import matplotlib # Matplotlib es una libreria que nos ayuda a graficar datos y funciones matematicas
# ejemplo, La hora actual del servidor
import datetime
print (datetime.datetime.now())
# ejemplo, Transpuesta de una matriz
import numpy as np
A = np.matrix([[3, 6, -5],
[1, -3, 2],
[5, -1, 4]])
print (A.shape) # las dimensiones de la matriz
print (A.transpose()) # tarnspuesta de la matriz A
%matplotlib notebook
# ejemplo, Gráfica de y = x**2
import matplotlib.pylab as plt
x = list(range(-50,50))
y = [i**2 for i in x]
plt.figure()
plt.scatter(x,y)
plt.title('$y = x^{2}$') # titulo
plt.xlabel('x') # titulo eje x
plt.ylabel('y') # titulo eje y
plt.show()
x = np.linspace(0, 2 * np.pi, 500)
y1 = np.sin(x)
y2 = np.sin(3 * x)
fig, ax = plt.subplots()
ax.fill(x, y1, 'b', x, y2, 'r', alpha=0.3)
plt.show()
# ejemplo, Crear una tabal de datos de sus compañeros
import pandas as pd
nombres = ['Jocelyn', 'Laura','Luis Alejandro']
apellidos = ['Kshi', 'Diaz', 'Mahecha']
pais = ['Estados Unidos', 'Colombia', 'Colombia']
pd.DataFrame({'nombre': nombres, 'apellido': apellidos, 'pais': pais})
temperatura_global = pd.read_csv('GlobalTemperatures.csv')
from __future__ import (absolute_import, division, print_function)
from six.moves import (filter, input, map, range, zip) # noqa
import matplotlib.cm as mpl_cm
import matplotlib.pyplot as plt
import iris
import iris.quickplot as qplt
fname = iris.sample_data_path('air_temp.pp')
temperature_cube = iris.load_cube(fname)
# Load a Cynthia Brewer palette.
brewer_cmap = mpl_cm.get_cmap('brewer_OrRd_09')
# Draw the contour with 25 levels.
plt.figure()
qplt.contourf(temperature_cube, 25)
# Add coastlines to the map created by contourf.
plt.gca().coastlines()
plt.show()
# Draw the contours, with n-levels set for the map colours (9).
# NOTE: needed as the map is non-interpolated, but matplotlib does not provide
# any special behaviour for these.
plt.figure()
qplt.contourf(temperature_cube, brewer_cmap.N, cmap=brewer_cmap)
# Add coastlines to the map created by contourf.
plt.gca().coastlines()
plt.show()
| 0.568416 | 0.882933 |
```
# !pip install git+https://github.com/ClimateImpactLab/xclim@63023d27f89a457c752568ffcec2e9ce9ad7a81a
%matplotlib inline
import xarray as xr
import numpy as np
import matplotlib.pyplot as plt
import os
import gcsfs
from matplotlib import cm
import warnings
from science_validation_manual import *
from xclim import sdba, set_options
from xclim.sdba.utils import equally_spaced_nodes
ref = read_gcs_zarr('gs://scratch-170cd6ec/643c4b73-399d-427a-87be-cc0ab1b401d9/e2e-bcc-csm2-mr-dtr-qxg22-2721721062/rechunked.zarr')
hist = read_gcs_zarr('gs://scratch-170cd6ec/643c4b73-399d-427a-87be-cc0ab1b401d9/e2e-bcc-csm2-mr-dtr-qxg22-1083920341/thresholded.zarr')
sim = read_gcs_zarr('gs://scratch-170cd6ec/643c4b73-399d-427a-87be-cc0ab1b401d9/e2e-bcc-csm2-mr-dtr-qxg22-3490169582/thresholded.zarr')
bc = read_gcs_zarr('gs://biascorrected-492e989a/stage/CMIP/BCC/BCC-CSM2-MR/historical/r1i1p1f1/day/dtr/gn/v20211230093107.zarr')
# define off the rails lat/lon
target_lat = 70.5
target_lon = 41.5
# QDM model specs
quantiles_n = 100
window_n = 31
ref_dtr = ref['dtr'].sel(lon=target_lon, lat=target_lat, method="nearest").load()
hist_dtr = hist['dtr'].sel(lon=target_lon, lat=target_lat, method="nearest").load()
bc_dtr = bc['dtr'].sel(lon=target_lon, lat=target_lat, method="nearest").load()
ref_dtr
sim_slice = sim.sel(lon=target_lon, lat=target_lat, method="nearest").load()
ref_dtr_timeslice = ref_dtr.sel(time=slice("1994-12-17", "2014-12-31"))
hist_dtr_timeslice = hist_dtr.sel(time=slice("1994-12-17", "2014-12-31"))
qdm_extrapolate = sdba.adjustment.QuantileDeltaMapping.train(
ref=ref_dtr_timeslice,
hist=hist_dtr_timeslice,
kind='*',
group=sdba.Grouper("time.dayofyear", window=int(window_n)),
nquantiles=equally_spaced_nodes(int(quantiles_n), eps=None),
)
qdm_NO_extrapolate = sdba.adjustment.QuantileDeltaMapping.train(
ref=ref_dtr_timeslice,
hist=hist_dtr_timeslice,
kind='*',
group=sdba.Grouper("time.dayofyear", window=int(window_n)),
nquantiles=quantiles_n,
)
qdm_NO_extrapolate_timeslice = sdba.adjustment.QuantileDeltaMapping.train(
ref=ref_dtr_timeslice,
hist=hist_dtr_timeslice,
kind='*',
group=sdba.Grouper("time.dayofyear", window=int(window_n)),
nquantiles=equally_spaced_nodes(int(quantiles_n), eps=1e-09),
)
# dtr_sim = sim_slice['dtr'].sel(time=slice("1959-12-17", "1979"))
dtr_sim = sim_slice['dtr'].sel(time=slice("1960-12-17", "1980-12-31"))
print(len(ref_dtr_timeslice))
print(len(hist_dtr_timeslice))
print(len(ref_dtr))
print(len(hist_dtr))
print(len(dtr_sim))
bc_extrapolate_here = qdm_extrapolate.adjust(dtr_sim)
bc_NO_extrapolate_here = qdm_NO_extrapolate.adjust(dtr_sim)
bc_NO_extrapolate_here_timeslice = qdm_NO_extrapolate_timeslice.adjust(dtr_sim)
fig, axes = plt.subplots(1, 5, figsize=(25, 4))
dtr_sim.plot(ax=axes[0])
axes[0].set_title('historical BCC-CSM2-MR')
bc_dtr.sel(time=slice("1960", "1980")).plot(ax=axes[1])
axes[1].set_title('bias corrected, pipeline')
bc_extrapolate_here.plot(ax=axes[2])
axes[2].set_title('bias corrected, notebook, \n pipeline model parameters')
bc_NO_extrapolate_here.plot(ax=axes[3])
axes[3].set_title('bias corrected, notebook, \n pipeline time slicing, \n nquantiles updated')
bc_NO_extrapolate_here_timeslice.plot(ax=axes[4])
axes[4].set_title('bias corrected, notebook, \n time slicing updated, \n nquantiles updated')
print(bc_NO_extrapolate_here_timeslice[4340].values)
print(dtr_sim[4340].values)
fig, axes = plt.subplots(1, 3, figsize=(14, 4))
qdm_NO_extrapolate.ds.af.sel(dayofyear=311).plot(ax=axes[0], linestyle='-')
axes[0].set_ylim([0, 6])
qdm_extrapolate.ds.af.sel(dayofyear=311).plot(ax=axes[1],linestyle='--')
qdm_NO_extrapolate_timeslice.ds.af.sel(dayofyear=311).plot(ax=axes[2],linestyle=':')
axes[1].set_ylim([0, 6])
axes[2].set_ylim([0, 6])
```
|
github_jupyter
|
# !pip install git+https://github.com/ClimateImpactLab/xclim@63023d27f89a457c752568ffcec2e9ce9ad7a81a
%matplotlib inline
import xarray as xr
import numpy as np
import matplotlib.pyplot as plt
import os
import gcsfs
from matplotlib import cm
import warnings
from science_validation_manual import *
from xclim import sdba, set_options
from xclim.sdba.utils import equally_spaced_nodes
ref = read_gcs_zarr('gs://scratch-170cd6ec/643c4b73-399d-427a-87be-cc0ab1b401d9/e2e-bcc-csm2-mr-dtr-qxg22-2721721062/rechunked.zarr')
hist = read_gcs_zarr('gs://scratch-170cd6ec/643c4b73-399d-427a-87be-cc0ab1b401d9/e2e-bcc-csm2-mr-dtr-qxg22-1083920341/thresholded.zarr')
sim = read_gcs_zarr('gs://scratch-170cd6ec/643c4b73-399d-427a-87be-cc0ab1b401d9/e2e-bcc-csm2-mr-dtr-qxg22-3490169582/thresholded.zarr')
bc = read_gcs_zarr('gs://biascorrected-492e989a/stage/CMIP/BCC/BCC-CSM2-MR/historical/r1i1p1f1/day/dtr/gn/v20211230093107.zarr')
# define off the rails lat/lon
target_lat = 70.5
target_lon = 41.5
# QDM model specs
quantiles_n = 100
window_n = 31
ref_dtr = ref['dtr'].sel(lon=target_lon, lat=target_lat, method="nearest").load()
hist_dtr = hist['dtr'].sel(lon=target_lon, lat=target_lat, method="nearest").load()
bc_dtr = bc['dtr'].sel(lon=target_lon, lat=target_lat, method="nearest").load()
ref_dtr
sim_slice = sim.sel(lon=target_lon, lat=target_lat, method="nearest").load()
ref_dtr_timeslice = ref_dtr.sel(time=slice("1994-12-17", "2014-12-31"))
hist_dtr_timeslice = hist_dtr.sel(time=slice("1994-12-17", "2014-12-31"))
qdm_extrapolate = sdba.adjustment.QuantileDeltaMapping.train(
ref=ref_dtr_timeslice,
hist=hist_dtr_timeslice,
kind='*',
group=sdba.Grouper("time.dayofyear", window=int(window_n)),
nquantiles=equally_spaced_nodes(int(quantiles_n), eps=None),
)
qdm_NO_extrapolate = sdba.adjustment.QuantileDeltaMapping.train(
ref=ref_dtr_timeslice,
hist=hist_dtr_timeslice,
kind='*',
group=sdba.Grouper("time.dayofyear", window=int(window_n)),
nquantiles=quantiles_n,
)
qdm_NO_extrapolate_timeslice = sdba.adjustment.QuantileDeltaMapping.train(
ref=ref_dtr_timeslice,
hist=hist_dtr_timeslice,
kind='*',
group=sdba.Grouper("time.dayofyear", window=int(window_n)),
nquantiles=equally_spaced_nodes(int(quantiles_n), eps=1e-09),
)
# dtr_sim = sim_slice['dtr'].sel(time=slice("1959-12-17", "1979"))
dtr_sim = sim_slice['dtr'].sel(time=slice("1960-12-17", "1980-12-31"))
print(len(ref_dtr_timeslice))
print(len(hist_dtr_timeslice))
print(len(ref_dtr))
print(len(hist_dtr))
print(len(dtr_sim))
bc_extrapolate_here = qdm_extrapolate.adjust(dtr_sim)
bc_NO_extrapolate_here = qdm_NO_extrapolate.adjust(dtr_sim)
bc_NO_extrapolate_here_timeslice = qdm_NO_extrapolate_timeslice.adjust(dtr_sim)
fig, axes = plt.subplots(1, 5, figsize=(25, 4))
dtr_sim.plot(ax=axes[0])
axes[0].set_title('historical BCC-CSM2-MR')
bc_dtr.sel(time=slice("1960", "1980")).plot(ax=axes[1])
axes[1].set_title('bias corrected, pipeline')
bc_extrapolate_here.plot(ax=axes[2])
axes[2].set_title('bias corrected, notebook, \n pipeline model parameters')
bc_NO_extrapolate_here.plot(ax=axes[3])
axes[3].set_title('bias corrected, notebook, \n pipeline time slicing, \n nquantiles updated')
bc_NO_extrapolate_here_timeslice.plot(ax=axes[4])
axes[4].set_title('bias corrected, notebook, \n time slicing updated, \n nquantiles updated')
print(bc_NO_extrapolate_here_timeslice[4340].values)
print(dtr_sim[4340].values)
fig, axes = plt.subplots(1, 3, figsize=(14, 4))
qdm_NO_extrapolate.ds.af.sel(dayofyear=311).plot(ax=axes[0], linestyle='-')
axes[0].set_ylim([0, 6])
qdm_extrapolate.ds.af.sel(dayofyear=311).plot(ax=axes[1],linestyle='--')
qdm_NO_extrapolate_timeslice.ds.af.sel(dayofyear=311).plot(ax=axes[2],linestyle=':')
axes[1].set_ylim([0, 6])
axes[2].set_ylim([0, 6])
| 0.37399 | 0.348132 |

## Equilibrium of a system of weights connected by strings/springs
In this notebook we show how to solve the following problem: Find the equlibrium of a system of masses connected by a system of strings, with some masses being assigned fixed coordinates (attached to the wall, say). See the next picture.

Suppose we have $n$ masses with weights $w_1,\ldots,w_n$, and the length of the string between $i$ and $j$ is $\ell_{ij}$ for some set $L$ of pairs of indices $(i,j)$ (we assume $\ell_{ij}$ is not defined if there is no connection). The strings themselves have no mass. We also have a set $F$ of indices such that the $i$-th point is fixed to have coordinates $f_i$ if $i\in F$. The equilibrium of the system is a configuration which minimizes potential energy. With this setup we can write our problem as:
\begin{equation}
\begin{array}{ll}
minimize & g\cdot \sum_i w_ix_i^{(2)} \\
s.t. & \|x_i-x_j\|\leq \ell_{ij},\ ij\in L \\
& x_i = f_i,\ i\in F
\end{array}
\end{equation}
where $x\in (\mathbf{R}^n)^2$, $x_i^{(2)}$ denotes the second (vertical) coordinate of $x_i$ and $g$ is the gravitational constant.
Here is a sample problem description.
```
w = [0.0, 1.1, 2.2, 0.0, 2.1, 2.2, 0.2]
l = {(0,1): 1.0, (1,2): 1.0, (2,3): 1.0, (1,4): 1.0, (4,5): 0.3, (5,2): 1.0, (5,6): 0.5, (1,3): 8.0}
f = {0: (0.0,1.0), 3: (2.0,1.0)}
g = 9.81
```
Now we can formulate the problem using Mosek Fusion:
```
from mosek.fusion import *
# w - masses of points
# l - lengths of strings
# f - coordinates of fixed points
# g - gravitational constant
def stringModel(w, l, f, g):
n, m = len(w), len(l)
starts = [ lKey[0] for lKey in l.keys() ]
ends = [ lKey[1] for lKey in l.keys() ]
M = Model("strings")
# Coordinates of points
x = M.variable("x", [n, 2])
# A is the signed incidence matrix of points and strings
A = Matrix.sparse(m, n, list(range(m))+list(range(m)), starts+ends, [1.0]*m+[-1.0]*m)
# ||x_i-x_j|| <= l_{i,j}
c = M.constraint("c", Expr.hstack(Expr.constTerm(list(l.values())), Expr.mul(A, x)),
Domain.inQCone() )
# x_i = f_i for fixed points
for i in f:
M.constraint(x.slice([i,0], [i+1,2]), Domain.equalsTo(list(f[i])).withShape([1,2]))
# sum (g w_i x_i_2)
M.objective(ObjectiveSense.Minimize,
Expr.mul(g, Expr.dot(w, x.slice([0,1], [n,2]))))
# Solve
M.solve()
if M.getProblemStatus(SolutionType.Interior) == ProblemStatus.PrimalAndDualFeasible:
return x.level().reshape([n,2]), c.dual().reshape([m,3])
else:
return None, None
```
Here is a quick description of how we use vectorization to deal with all the conic constraints in one go. The matrix $A$ is the incidence matrix between the masses and the strings, with coefficients $+1, -1$ for the two endpoints of each string. It is chosen so that the product $Ax$ has rows of the form
$$
(x_i^{(1)} - x_j^{(1)}, x_i^{(2)} - x_j^{(2)})
$$
for all pairs $i,j$ for which $\ell_{ij}$ is bounded. Stacking the values of $\ell$ in the left column produces a matrix with each row of the form
$$
(\ell_{ij}, x_i^{(1)} - x_j^{(1)}, x_i^{(2)} - x_j^{(2)})
$$
and a conic constraint is imposed on all the rows, as required.
The objective and linear constraints show examples of slicing the variable $x$.
The function returns the coordinates of the masses and the values of the dual conic variables. A zero dual value indicates that a particular string is hanging loose, and a nonzero value means it is fully stretched.
All we need now is to define a display function and we can look at some plots.
```
%matplotlib inline
# x - coordinates of the points
# c - dual values of string length constraints
# d - pairs of points to connect
def display(x, c, d):
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
# Plot points
ax.scatter(x[:,0], x[:,1], color="r")
# Plot fully stretched strings (nonzero dual value) as solid lines, else dotted lines
for i in range(len(c)):
col = "b" if c[i][0] > 1e-4 else "b--"
ax.plot([x[d[i][0]][0], x[d[i][1]][0]], [x[d[i][0]][1], x[d[i][1]][1]], col)
ax.axis("equal")
plt.show()
x,c = stringModel(w, l, f, g)
if x is not None:
display(x, c, list(l.keys()))
```
How about we find a discrete approximation to the [catenary](#https://en.wikipedia.org/wiki/Catenary):
```
n = 1000
w = [1.0]*n
l = {(i,i+1): 1.0/n for i in range(n-1)}
f = {0: (0.0,1.0), n-1: (0.7,1.0)}
g = 9.81
x,c = stringModel(w, l, f, g)
if x is not None:
display(x, c, list(l.keys()))
```
We can also have more suspension points and more complicated shapes:
```
n = 20
w = [1.0]*n
l = {(i,i+1): 0.09 for i in range(n-1)}
l.update({(5,14): 0.3})
f = {0: (0.0,1.0), 13: (0.5,0.9), 17: (0.7,1.1)}
g = 9.81
x,c = stringModel(w, l, f, g)
if x is not None:
display(x, c, list(l.keys()))
```
## Duality and feasibility
The dual problem is as follows:
\begin{equation}
\begin{array}{ll}
maximize & -\sum_{ij\in L}\ell_{ij}y_{ij} - \sum_{i\in F}f_i\circ z_i\\
s.t. & y_{ij}\geq \|v_{ij}\|,\ ij\in L \\
& \sum_{j~:~ij\in L} v_{ij}\mathrm{sgn}_{ij} + \left(\begin{array}{c}0\\ gw_i\end{array}\right) +z_i = 0, \ i=1,\ldots,n
\end{array}
\end{equation}
where $\mathrm{sgn}_{ij}=+1$ if $i>j$ and $-1$ otherwise and $\circ$ is the dot product. The variables are $(y_{ij},v_{ij})\in \mathbf{R}\times\mathbf{R}^2$ for $ij\in L$ and $z_i\in\mathbf{R}^2$ for $i\in F$ (we assume $z_i=0$ for $i\not\in F$).
Obviously (!) the linear constraints describe the equilibrium of forces at every mass. The ingredients are: the vectors of forces applied through adjacent strings ($v_{ij}$), gravity, and the attaching force holding a fixed point in its position. By proper use of vectorization this is much easier to express in Fusion than it looks:
```
def dualStringModel(w, l, f, g):
n, m = len(w), len(l)
starts = [ lKey[0] for lKey in l.keys() ]
ends = [ lKey[1] for lKey in l.keys() ]
M = Model("dual strings")
x = M.variable(Domain.inQCone(m,3)) #(y,v)
y = x.slice([0,0],[m,1])
v = x.slice([0,1],[m,3])
z = M.variable([n,2])
# z_i = 0 if i is not fixed
for i in range(n):
if i not in f:
M.constraint(z.slice([i,0], [i+1,2]), Domain.equalsTo(0.0))
B = Matrix.sparse(m, n, list(range(m))+list(range(m)), starts+ends, [1.0]*m+[-1.0]*m).transpose()
w2 = Matrix.sparse(n, 2, range(n), [1]*n, [-wT*g for wT in w])
# sum(v_ij *sgn(ij)) + z_i = -(0, gw_i) for all vertices i
M.constraint(Expr.add( Expr.mul(B, v), z ), Domain.equalsTo(w2))
# Objective -l*y -fM*z
fM = Matrix.sparse(n, 2, list(f.keys())+list(f.keys()), [0]*len(f)+[1]*len(f),
[pt[0] for pt in f.values()] + [pt[1] for pt in f.values()])
M.objective(ObjectiveSense.Maximize, Expr.neg(Expr.add(Expr.dot(list(l.values()), y),Expr.dot(fM, z))))
M.solve()
```
Let us quickly discuss the possible situations regarding feasibility:
* The system has an equilibrium --- the problem is **primal feasible** and **dual feasible**.
* The strings are too short and it is impossible to stretch the required distance between fixed points --- the problem is **primal infeasible**.
* The system has a component that is not connected to any fixed point, hence some masses can keep falling down indefinitely, causing the problem **primal unbounded**. Clearly the forces within such component cannot be balanced, so the problem is **dual infeasible**.
## Springs
We can extend this to consider infinitely strechable springs instead of fixed-length strings connecting the masses. The next model appears in [Applications of SOCP](#http://stanford.edu/~boyd/papers/pdf/socp.pdf) by Lobo, Boyd, Vandenberghe, Lebret. We will now interpret $\ell_{ij}$ as the base length of the spring and assume that the elastic potential energy stored in the spring at length $x$ is
$$
E_{ij}=\left\{\begin{array}{ll}0 & x\leq \ell_{ij}\\ \frac{k}{2}(x-\ell_{ij})^2 & x>\ell_{ij}\end{array}\right.
$$
That leads us to consider the following second order cone program minimizing the total potential energy:
\begin{equation}
\begin{array}{ll}
minimize & g\cdot \sum_i w_ix_i^{(2)} + \frac{k}{2}\sum_{ij\in L} t_{ij}^2 \\
s.t. & \|x_i-x_j\|\leq \ell_{ij}+t_{ij},\ ij\in L \\
& 0\leq t_{ij},\ ij\in L \\
& x_i = f_i,\ i\in F
\end{array}
\end{equation}
If $t$ denotes the vector of $t_{ij}$ then using a rotated quadratic cone for $(1,T,t)$:
$$
2\cdot 1\cdot T\geq \|t\|^2
$$
will place a bound on $\frac12\sum t_{ij}^2$. We now have a simple extension of the first model.
```
# w - masses of points
# l - lengths of strings
# f - coordinates of fixed points
# g - gravitational constant
# k - stiffness coefficient
def elasticModel(w, l, f, g, k):
n, m = len(w), len(l)
starts = [ lKey[0] for lKey in l.keys() ]
ends = [ lKey[1] for lKey in l.keys() ]
M = Model("strings")
x = M.variable("x", [n, 2]) # Coordinates
t = M.variable(m, Domain.greaterThan(0.0)) # Streching
T = M.variable(1) # Upper bound
M.constraint(Expr.vstack(T, Expr.constTerm(1.0), t), Domain.inRotatedQCone())
# A is the signed incidence matrix of points and strings
A = Matrix.sparse(m, n, list(range(m))+list(range(m)), starts+ends, [1.0]*m+[-1.0]*m)
# ||x_i-x_j|| <= l_{i,j} + t_{i,j}
c = M.constraint("c", Expr.hstack(Expr.add(t, Expr.constTerm(list(l.values()))), Expr.mul(A, x)),
Domain.inQCone() )
# x_i = f_i for fixed points
for i in f:
M.constraint(x.slice([i,0], [i+1,2]), Domain.equalsTo(list(f[i])).withShape([1,2]))
# sum (g w_i x_i_2) + k*T
M.objective(ObjectiveSense.Minimize,
Expr.add(Expr.mul(k,T), Expr.mul(g, Expr.dot(w, x.slice([0,1], [n,2])))))
# Solve
M.solve()
if M.getProblemStatus(SolutionType.Interior) == ProblemStatus.PrimalAndDualFeasible:
return x.level().reshape([n,2]), c.dual().reshape([m,3])
else:
return None, None
n = 20
w = [1.0]*n
l = {(i,i+1): 0.09 for i in range(n-1)}
l.update({(5,14): 0.3})
f = {0: (0.0,1.0), 13: (0.5,0.9), 17: (0.7,1.1)}
g = 9.81
k = 800
x, c = elasticModel(w, l, f, g, k)
if x is not None:
display(x, c, list(l.keys()))
```
<a rel="license" href="http://creativecommons.org/licenses/by/4.0/"><img alt="Creative Commons License" style="border-width:0" src="https://i.creativecommons.org/l/by/4.0/80x15.png" /></a><br />This work is licensed under a <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Creative Commons Attribution 4.0 International License</a>. The **MOSEK** logo and name are trademarks of <a href="http://mosek.com">Mosek ApS</a>. The code is provided as-is. Compatibility with future release of **MOSEK** or the `Fusion API` are not guaranteed. For more information contact our [support](mailto:[email protected]).
|
github_jupyter
|
w = [0.0, 1.1, 2.2, 0.0, 2.1, 2.2, 0.2]
l = {(0,1): 1.0, (1,2): 1.0, (2,3): 1.0, (1,4): 1.0, (4,5): 0.3, (5,2): 1.0, (5,6): 0.5, (1,3): 8.0}
f = {0: (0.0,1.0), 3: (2.0,1.0)}
g = 9.81
from mosek.fusion import *
# w - masses of points
# l - lengths of strings
# f - coordinates of fixed points
# g - gravitational constant
def stringModel(w, l, f, g):
n, m = len(w), len(l)
starts = [ lKey[0] for lKey in l.keys() ]
ends = [ lKey[1] for lKey in l.keys() ]
M = Model("strings")
# Coordinates of points
x = M.variable("x", [n, 2])
# A is the signed incidence matrix of points and strings
A = Matrix.sparse(m, n, list(range(m))+list(range(m)), starts+ends, [1.0]*m+[-1.0]*m)
# ||x_i-x_j|| <= l_{i,j}
c = M.constraint("c", Expr.hstack(Expr.constTerm(list(l.values())), Expr.mul(A, x)),
Domain.inQCone() )
# x_i = f_i for fixed points
for i in f:
M.constraint(x.slice([i,0], [i+1,2]), Domain.equalsTo(list(f[i])).withShape([1,2]))
# sum (g w_i x_i_2)
M.objective(ObjectiveSense.Minimize,
Expr.mul(g, Expr.dot(w, x.slice([0,1], [n,2]))))
# Solve
M.solve()
if M.getProblemStatus(SolutionType.Interior) == ProblemStatus.PrimalAndDualFeasible:
return x.level().reshape([n,2]), c.dual().reshape([m,3])
else:
return None, None
%matplotlib inline
# x - coordinates of the points
# c - dual values of string length constraints
# d - pairs of points to connect
def display(x, c, d):
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
# Plot points
ax.scatter(x[:,0], x[:,1], color="r")
# Plot fully stretched strings (nonzero dual value) as solid lines, else dotted lines
for i in range(len(c)):
col = "b" if c[i][0] > 1e-4 else "b--"
ax.plot([x[d[i][0]][0], x[d[i][1]][0]], [x[d[i][0]][1], x[d[i][1]][1]], col)
ax.axis("equal")
plt.show()
x,c = stringModel(w, l, f, g)
if x is not None:
display(x, c, list(l.keys()))
n = 1000
w = [1.0]*n
l = {(i,i+1): 1.0/n for i in range(n-1)}
f = {0: (0.0,1.0), n-1: (0.7,1.0)}
g = 9.81
x,c = stringModel(w, l, f, g)
if x is not None:
display(x, c, list(l.keys()))
n = 20
w = [1.0]*n
l = {(i,i+1): 0.09 for i in range(n-1)}
l.update({(5,14): 0.3})
f = {0: (0.0,1.0), 13: (0.5,0.9), 17: (0.7,1.1)}
g = 9.81
x,c = stringModel(w, l, f, g)
if x is not None:
display(x, c, list(l.keys()))
def dualStringModel(w, l, f, g):
n, m = len(w), len(l)
starts = [ lKey[0] for lKey in l.keys() ]
ends = [ lKey[1] for lKey in l.keys() ]
M = Model("dual strings")
x = M.variable(Domain.inQCone(m,3)) #(y,v)
y = x.slice([0,0],[m,1])
v = x.slice([0,1],[m,3])
z = M.variable([n,2])
# z_i = 0 if i is not fixed
for i in range(n):
if i not in f:
M.constraint(z.slice([i,0], [i+1,2]), Domain.equalsTo(0.0))
B = Matrix.sparse(m, n, list(range(m))+list(range(m)), starts+ends, [1.0]*m+[-1.0]*m).transpose()
w2 = Matrix.sparse(n, 2, range(n), [1]*n, [-wT*g for wT in w])
# sum(v_ij *sgn(ij)) + z_i = -(0, gw_i) for all vertices i
M.constraint(Expr.add( Expr.mul(B, v), z ), Domain.equalsTo(w2))
# Objective -l*y -fM*z
fM = Matrix.sparse(n, 2, list(f.keys())+list(f.keys()), [0]*len(f)+[1]*len(f),
[pt[0] for pt in f.values()] + [pt[1] for pt in f.values()])
M.objective(ObjectiveSense.Maximize, Expr.neg(Expr.add(Expr.dot(list(l.values()), y),Expr.dot(fM, z))))
M.solve()
# w - masses of points
# l - lengths of strings
# f - coordinates of fixed points
# g - gravitational constant
# k - stiffness coefficient
def elasticModel(w, l, f, g, k):
n, m = len(w), len(l)
starts = [ lKey[0] for lKey in l.keys() ]
ends = [ lKey[1] for lKey in l.keys() ]
M = Model("strings")
x = M.variable("x", [n, 2]) # Coordinates
t = M.variable(m, Domain.greaterThan(0.0)) # Streching
T = M.variable(1) # Upper bound
M.constraint(Expr.vstack(T, Expr.constTerm(1.0), t), Domain.inRotatedQCone())
# A is the signed incidence matrix of points and strings
A = Matrix.sparse(m, n, list(range(m))+list(range(m)), starts+ends, [1.0]*m+[-1.0]*m)
# ||x_i-x_j|| <= l_{i,j} + t_{i,j}
c = M.constraint("c", Expr.hstack(Expr.add(t, Expr.constTerm(list(l.values()))), Expr.mul(A, x)),
Domain.inQCone() )
# x_i = f_i for fixed points
for i in f:
M.constraint(x.slice([i,0], [i+1,2]), Domain.equalsTo(list(f[i])).withShape([1,2]))
# sum (g w_i x_i_2) + k*T
M.objective(ObjectiveSense.Minimize,
Expr.add(Expr.mul(k,T), Expr.mul(g, Expr.dot(w, x.slice([0,1], [n,2])))))
# Solve
M.solve()
if M.getProblemStatus(SolutionType.Interior) == ProblemStatus.PrimalAndDualFeasible:
return x.level().reshape([n,2]), c.dual().reshape([m,3])
else:
return None, None
n = 20
w = [1.0]*n
l = {(i,i+1): 0.09 for i in range(n-1)}
l.update({(5,14): 0.3})
f = {0: (0.0,1.0), 13: (0.5,0.9), 17: (0.7,1.1)}
g = 9.81
k = 800
x, c = elasticModel(w, l, f, g, k)
if x is not None:
display(x, c, list(l.keys()))
| 0.558688 | 0.964522 |
## MNIST Training with MXNet and Gluon
MNIST is a widely used dataset for handwritten digit classification. It consists of 70,000 labeled 28x28 pixel grayscale images of hand-written digits. The dataset is split into 60,000 training images and 10,000 test images. There are 10 classes (one for each of the 10 digits). This tutorial will show how to train and test an MNIST model on SageMaker using MXNet and the Gluon API.
```
import os
import boto3
import sagemaker
from sagemaker.mxnet import MXNet
from mxnet import gluon
from sagemaker import get_execution_role
sagemaker_session = sagemaker.Session()
role = get_execution_role()
```
## Download training and test data
```
gluon.data.vision.MNIST('./data/train', train=True)
gluon.data.vision.MNIST('./data/test', train=False)
```
## Uploading the data
We use the `sagemaker.Session.upload_data` function to upload our datasets to an S3 location. The return value `inputs` identifies the location -- we will use this later when we start the training job.
```
inputs = sagemaker_session.upload_data(path='data', key_prefix='data/DEMO-mnist')
```
## Implement the training function
We need to provide a training script that can run on the SageMaker platform. The training scripts are essentially the same as one you would write for local training, except that you need to provide a `train` function. The `train` function will check for the validation accuracy at the end of every epoch and checkpoints the best model so far, along with the optimizer state, in the folder `/opt/ml/checkpoints` if the folder path exists, else it will skip the checkpointing. When SageMaker calls your function, it will pass in arguments that describe the training environment. Check the script below to see how this works.
The script here is an adaptation of the [Gluon MNIST example](https://github.com/apache/incubator-mxnet/blob/master/example/gluon/mnist.py) provided by the [Apache MXNet](https://mxnet.incubator.apache.org/) project.
```
!cat 'mnist.py'
```
## Run the training script on SageMaker
The ```MXNet``` class allows us to run our training function on SageMaker infrastructure. We need to configure it with our training script, an IAM role, the number of training instances, and the training instance type. In this case we will run our training job on a single c4.xlarge instance.
```
m = MXNet("mnist.py",
role=role,
train_instance_count=2,
train_instance_type="ml.p2.xlarge",
framework_version="1.4.1",
py_version="py3",
distributions={'parameter_server': {'enabled': True}},
hyperparameters={'batch-size': 100,
'epochs': 20,
'learning-rate': 0.1,
'momentum': 0.9,
'log-interval': 100})
```
After we've constructed our `MXNet` object, we can fit it using the data we uploaded to S3. SageMaker makes sure our data is available in the local filesystem, so our training script can simply read the data from disk.
```
m.fit(inputs)
```
After training, we use the MXNet object to build and deploy an MXNetPredictor object. This creates a SageMaker endpoint that we can use to perform inference.
This allows us to perform inference on json encoded multi-dimensional arrays.
```
predictor = m.deploy(initial_instance_count=1, instance_type='ml.m4.xlarge')
```
We can now use this predictor to classify hand-written digits. Drawing into the image box loads the pixel data into a 'data' variable in this notebook, which we can then pass to the mxnet predictor.
```
from IPython.display import HTML
HTML(open("input.html").read())
```
The predictor runs inference on our input data and returns the predicted digit (as a float value, so we convert to int for display).
```
response = predictor.predict(data)
print(int(response))
```
## Cleanup
After you have finished with this example, remember to delete the prediction endpoint to release the instance(s) associated with it.
```
predictor.delete_endpoint()
```
|
github_jupyter
|
import os
import boto3
import sagemaker
from sagemaker.mxnet import MXNet
from mxnet import gluon
from sagemaker import get_execution_role
sagemaker_session = sagemaker.Session()
role = get_execution_role()
gluon.data.vision.MNIST('./data/train', train=True)
gluon.data.vision.MNIST('./data/test', train=False)
inputs = sagemaker_session.upload_data(path='data', key_prefix='data/DEMO-mnist')
!cat 'mnist.py'
m = MXNet("mnist.py",
role=role,
train_instance_count=2,
train_instance_type="ml.p2.xlarge",
framework_version="1.4.1",
py_version="py3",
distributions={'parameter_server': {'enabled': True}},
hyperparameters={'batch-size': 100,
'epochs': 20,
'learning-rate': 0.1,
'momentum': 0.9,
'log-interval': 100})
m.fit(inputs)
predictor = m.deploy(initial_instance_count=1, instance_type='ml.m4.xlarge')
from IPython.display import HTML
HTML(open("input.html").read())
response = predictor.predict(data)
print(int(response))
predictor.delete_endpoint()
| 0.250363 | 0.989791 |
```
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import torchvision
import torchvision.transforms as transforms
import torchvision.datasets as datasets
import os
import random
import numpy as np
SEED = 1234
random.seed(SEED)
np.random.seed(SEED)
torch.manual_seed(SEED)
torch.cuda.manual_seed(SEED)
torch.backends.cudnn.deterministic = True
data_transforms = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))
])
train_data = datasets.MNIST('data', train=True, download=True, transform=data_transforms)
test_data = datasets.MNIST('data', train=False, download=True, transform=data_transforms)
n_train_examples = int(len(train_data)*0.9)
n_valid_examples = len(train_data) - n_train_examples
train_data, valid_data = torch.utils.data.random_split(train_data, [n_train_examples, n_valid_examples])
print(f'Number of training examples: {len(train_data)}')
print(f'Number of validation examples: {len(valid_data)}')
print(f'Number of testing examples: {len(test_data)}')
BATCH_SIZE = 64
train_iterator = torch.utils.data.DataLoader(train_data, shuffle=True, batch_size=BATCH_SIZE)
valid_iterator = torch.utils.data.DataLoader(valid_data, batch_size=BATCH_SIZE)
test_iterator = torch.utils.data.DataLoader(test_data, batch_size=BATCH_SIZE)
class MLP(nn.Module):
def __init__(self, hidden_neurons):
super().__init__()
self.hidden_neurons = hidden_neurons
self.input_fc = nn.Linear(28*28, hidden_neurons[0])
self.fcs = nn.ModuleList([nn.Linear(hidden_neurons[i], hidden_neurons[i+1]) for i in range(len(hidden_neurons)-1)])
self.output_fc = nn.Linear(hidden_neurons[-1], 10)
def forward(self, x):
#flatten
x = x.view(x.shape[0], -1)
x = F.relu(self.input_fc(x))
for i in range(len(self.hidden_neurons)-1):
x = F.relu(self.fcs[i](x))
x = self.output_fc(x)
return x
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model = MLP([1000, 500, 250]).to(device)
model
optimizer = optim.Adam(model.parameters())
criterion = nn.CrossEntropyLoss()
def calculate_accuracy(fx, y):
preds = fx.max(1, keepdim=True)[1]
correct = preds.eq(y.view_as(preds)).sum()
acc = correct.float()/preds.shape[0]
return acc
def train(model, device, iterator, optimizer, criterion):
epoch_loss = 0
epoch_acc = 0
model.train()
for (x, y) in iterator:
x = x.to(device)
y = y.to(device)
optimizer.zero_grad()
fx = model(x)
loss = criterion(fx, y)
acc = calculate_accuracy(fx, y)
loss.backward()
optimizer.step()
epoch_loss += loss.item()
epoch_acc += acc.item()
return epoch_loss / len(iterator), epoch_acc / len(iterator)
def evaluate(model, device, iterator, criterion):
epoch_loss = 0
epoch_acc = 0
model.eval()
with torch.no_grad():
for (x, y) in iterator:
x = x.to(device)
y = y.to(device)
fx = model(x)
loss = criterion(fx, y)
acc = calculate_accuracy(fx, y)
epoch_loss += loss.item()
epoch_acc += acc.item()
return epoch_loss / len(iterator), epoch_acc / len(iterator)
EPOCHS = 10
SAVE_DIR = 'models'
MODEL_SAVE_PATH = os.path.join(SAVE_DIR, 'mlp-mnist.pt')
best_valid_loss = float('inf')
if not os.path.isdir(f'{SAVE_DIR}'):
os.makedirs(f'{SAVE_DIR}')
for epoch in range(EPOCHS):
train_loss, train_acc = train(model, device, train_iterator, optimizer, criterion)
valid_loss, valid_acc = evaluate(model, device, valid_iterator, criterion)
if valid_loss < best_valid_loss:
best_valid_loss = valid_loss
torch.save(model.state_dict(), MODEL_SAVE_PATH)
print(f'| Epoch: {epoch+1:02} | Train Loss: {train_loss:.3f} | Train Acc: {train_acc*100:05.2f}% | Val. Loss: {valid_loss:.3f} | Val. Acc: {valid_acc*100:05.2f}% |')
model.load_state_dict(torch.load(MODEL_SAVE_PATH))
test_loss, test_acc = evaluate(model, device, valid_iterator, criterion)
print(f'| Test Loss: {test_loss:.3f} | Test Acc: {test_acc*100:05.2f}% |')
```
|
github_jupyter
|
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import torchvision
import torchvision.transforms as transforms
import torchvision.datasets as datasets
import os
import random
import numpy as np
SEED = 1234
random.seed(SEED)
np.random.seed(SEED)
torch.manual_seed(SEED)
torch.cuda.manual_seed(SEED)
torch.backends.cudnn.deterministic = True
data_transforms = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))
])
train_data = datasets.MNIST('data', train=True, download=True, transform=data_transforms)
test_data = datasets.MNIST('data', train=False, download=True, transform=data_transforms)
n_train_examples = int(len(train_data)*0.9)
n_valid_examples = len(train_data) - n_train_examples
train_data, valid_data = torch.utils.data.random_split(train_data, [n_train_examples, n_valid_examples])
print(f'Number of training examples: {len(train_data)}')
print(f'Number of validation examples: {len(valid_data)}')
print(f'Number of testing examples: {len(test_data)}')
BATCH_SIZE = 64
train_iterator = torch.utils.data.DataLoader(train_data, shuffle=True, batch_size=BATCH_SIZE)
valid_iterator = torch.utils.data.DataLoader(valid_data, batch_size=BATCH_SIZE)
test_iterator = torch.utils.data.DataLoader(test_data, batch_size=BATCH_SIZE)
class MLP(nn.Module):
def __init__(self, hidden_neurons):
super().__init__()
self.hidden_neurons = hidden_neurons
self.input_fc = nn.Linear(28*28, hidden_neurons[0])
self.fcs = nn.ModuleList([nn.Linear(hidden_neurons[i], hidden_neurons[i+1]) for i in range(len(hidden_neurons)-1)])
self.output_fc = nn.Linear(hidden_neurons[-1], 10)
def forward(self, x):
#flatten
x = x.view(x.shape[0], -1)
x = F.relu(self.input_fc(x))
for i in range(len(self.hidden_neurons)-1):
x = F.relu(self.fcs[i](x))
x = self.output_fc(x)
return x
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model = MLP([1000, 500, 250]).to(device)
model
optimizer = optim.Adam(model.parameters())
criterion = nn.CrossEntropyLoss()
def calculate_accuracy(fx, y):
preds = fx.max(1, keepdim=True)[1]
correct = preds.eq(y.view_as(preds)).sum()
acc = correct.float()/preds.shape[0]
return acc
def train(model, device, iterator, optimizer, criterion):
epoch_loss = 0
epoch_acc = 0
model.train()
for (x, y) in iterator:
x = x.to(device)
y = y.to(device)
optimizer.zero_grad()
fx = model(x)
loss = criterion(fx, y)
acc = calculate_accuracy(fx, y)
loss.backward()
optimizer.step()
epoch_loss += loss.item()
epoch_acc += acc.item()
return epoch_loss / len(iterator), epoch_acc / len(iterator)
def evaluate(model, device, iterator, criterion):
epoch_loss = 0
epoch_acc = 0
model.eval()
with torch.no_grad():
for (x, y) in iterator:
x = x.to(device)
y = y.to(device)
fx = model(x)
loss = criterion(fx, y)
acc = calculate_accuracy(fx, y)
epoch_loss += loss.item()
epoch_acc += acc.item()
return epoch_loss / len(iterator), epoch_acc / len(iterator)
EPOCHS = 10
SAVE_DIR = 'models'
MODEL_SAVE_PATH = os.path.join(SAVE_DIR, 'mlp-mnist.pt')
best_valid_loss = float('inf')
if not os.path.isdir(f'{SAVE_DIR}'):
os.makedirs(f'{SAVE_DIR}')
for epoch in range(EPOCHS):
train_loss, train_acc = train(model, device, train_iterator, optimizer, criterion)
valid_loss, valid_acc = evaluate(model, device, valid_iterator, criterion)
if valid_loss < best_valid_loss:
best_valid_loss = valid_loss
torch.save(model.state_dict(), MODEL_SAVE_PATH)
print(f'| Epoch: {epoch+1:02} | Train Loss: {train_loss:.3f} | Train Acc: {train_acc*100:05.2f}% | Val. Loss: {valid_loss:.3f} | Val. Acc: {valid_acc*100:05.2f}% |')
model.load_state_dict(torch.load(MODEL_SAVE_PATH))
test_loss, test_acc = evaluate(model, device, valid_iterator, criterion)
print(f'| Test Loss: {test_loss:.3f} | Test Acc: {test_acc*100:05.2f}% |')
| 0.928026 | 0.71202 |
# ASTR 598 Astrostatistics
## HW2 Part 3
## Hayden Smotherman, Chris Suberlack, Winnie Wang
## To run this Notebook:
The Galfast data must be extracted from the projects/ directory as a .txt file and this notebook must be run in the homeworks/group2/HW_2/ directory.
```
# Imports
%matplotlib inline
from astropy.table import Table
from astropy.coordinates import SkyCoord
from astropy import units as u
from astropy.table import hstack
from astropy.table import vstack
import matplotlib.pyplot as plt
import matplotlib.pyplot as plt
from matplotlib.patches import Circle
import os
import numpy as np
from astropy.io import fits
import pandas as pd
from scipy.stats import binned_statistic_2d as bs2d
from scipy.stats import binned_statistic as bs1d
import seaborn as sns
GalFastData = np.loadtxt('../../../project/Galfast-Stripe82.txt',usecols=(0,1,4,5,12,13,14))
GalFastTable = Table(rows=GalFastData, names=('ra','dec','pmra','pmdec','gmag','rmag','imag'))
# Generate the magnitude mask used in the Hess diagram data analysis
def Hess_r_v_gminusi(aptable,total_mask):
constant = 3 # This is a fudge parameter in determining number of bins
nObjects = np.sum(total_mask)
num_bins = int(constant * nObjects ** (1.0 / 4.0))
# Now calculate the binned proper motions
proper_motion = np.sqrt(aptable['pmra']**2+aptable['pmdec']**2)
total_mask = proper_motion<100*total_mask
Binned_PM = bs2d(aptable['ra'][total_mask],aptable['rmag'][total_mask],proper_motion[total_mask], bins = num_bins)
#cmin = min(np.log10(Binned_PM.statistic.T[Binned_PM.statistic.T > 0]))
#cmax = max(np.log10(Binned_PM.statistic.T[Binned_PM.statistic.T > 0]))
# Define custom colormaps: Set pixels with no sources to white
cmap = plt.cm.viridis
cmap.set_bad('w', 1.)
plt.figure(figsize=[8,8])
plt.imshow(Binned_PM.statistic.T, origin='lower',
extent=[Binned_PM.x_edge[0], Binned_PM.x_edge[-1], Binned_PM.y_edge[0], Binned_PM.y_edge[-1]],
aspect='auto', interpolation='nearest', cmap=cmap)
cb = plt.colorbar(orientation='horizontal')
cb.set_label(r'Proper Motion [mas/yr]',fontsize=16)
#plt.clim(0, 30) # This was set by hand to draw out as much detail as possible
plt.xlabel(r'RA [degree]',fontsize=16)
plt.ylabel(r'r',fontsize=16)
plt.gca().invert_yaxis()
plt.gca().invert_xaxis()
# Make the color masks
r_mask = (GalFastTable['rmag']>20.5) & (GalFastTable['rmag']<21)
gminusi_mask = ((GalFastTable['gmag']-GalFastTable['imag']) > 0.3) & ((GalFastTable['gmag']-GalFastTable['imag']) < 0.4)
mag_mask = r_mask * gminusi_mask
# Make the RA Masks
RA_mask_25to40 = (GalFastTable['ra'] > 25) & (GalFastTable['ra'] < 40)
RA_mask_0to15 = (GalFastTable['ra'] > 0) & (GalFastTable['ra'] < 15)
# Make the net masks
mask_25to40 = mag_mask * RA_mask_25to40
mask_0to15 = mag_mask * RA_mask_0to15
# Make the Hess diagram for 25 < RA < 40
Hess_r_v_gminusi(GalFastTable,mask_25to40)
plt.title(r'Proper Motion for $25^\circ < \mathrm{RA} < 40^\circ$',fontsize=20)
plt.savefig('hw2_3_GalFast_pm_Hess_Diagram_RA25to40') # Save the figure
# Make the Hess diagram for 0 < RA < 15
Hess_r_v_gminusi(GalFastTable,mask_0to15)
plt.title(r'Proper Motion for $0^\circ < \mathrm{RA} < 15^\circ$',fontsize=20)
plt.savefig('hw2_3_GalFast_pm_Hess_Diagram_RA0to15') # Save the figure
```
The Galfast data is consistent with the NSC data in the stripe 82 region. The Galfast data shows a slight proper motion bimoodaility for stars in $25^\circ < \mathrm{RA} < 40^\circ$ while it shows little to no bimodality for stars in $0^\circ < \mathrm{RA} < 15^\circ$. Note that the color bar scale is different for each image. This mirrors what we saw in the NSC data in the same region, although the NSC dataset is too sparse to fully flesh-out the diagram.
```
def ProperMotionHist(aptable,mask_noRAcuts,xmin=-5,xmax=5,normed='True'):
# This function makes two histograms of RA and DEC Proper motions for two different RA cuts
# Calculate the RA Proper Motion
mask_noRAcuts *= aptable['pmra']<50
# Make two masks that have the RA cuts included
RA_mask_0to15 = (aptable['ra'] > 0) & (aptable['ra'] < 15)
RA_mask_25to40 = (aptable['ra'] > 25) & (aptable['ra'] < 40)
# Mask things outside the range of the histogram
mask_pm = (xmin < aptable['pmra'])*(aptable['pmra'] < xmax)
# Combine masks
mask_0to15 = mask_noRAcuts * RA_mask_0to15 * mask_pm
mask_25to40 = mask_noRAcuts * RA_mask_25to40 * mask_pm
#Plot the two distributions with different RA cuts
plt.figure(figsize=[12,8])
plt.hist(aptable['pmra'][mask_0to15],alpha=0.5,bins=30,
normed=normed,linewidth=3,color='r')
plt.hist(aptable['pmra'][mask_25to40],alpha=0.5,bins=30,
normed=normed,linewidth=3,color='b')
plt.legend([r'$0^\circ < \mathrm{RA} < 15^\circ$',
r'$25^\circ < \mathrm{RA} < 40^\circ$'],fontsize=16)
plt.title('Distribution of RA Proper Motions for different RA cuts',fontsize=20)
plt.xlabel('RA Proper Motion [mas/yr]',fontsize=16)
plt.ylabel('Normed number density',fontsize=16)
plt.xlim([xmin,xmax])
#plt.ylim([1,200])
plt.savefig('hw2_3_pm_hist_ra.png', bbox_inches='tight')
# Calculate the DEC Proper Motion
mask_noRAcuts *= aptable['pmdec']<50
# Make two masks that have the RA cuts included
RA_mask_0to15 = (aptable['ra'] > 0) & (aptable['ra'] < 15)
RA_mask_25to40 = (aptable['ra'] > 25) & (aptable['ra'] < 40)
# Mask things outside the range of the histogram
mask_pm = (xmin < aptable['pmdec'])*(aptable['pmdec'] < xmax)
# Combine masks
mask_0to15 = mask_noRAcuts * RA_mask_0to15 * mask_pm
mask_25to40 = mask_noRAcuts * RA_mask_25to40 * mask_pm
#Plot the two distributions with different RA cuts
plt.figure(figsize=[12,8])
plt.hist(aptable['pmdec'][mask_0to15],alpha=0.5,bins=30,
normed=normed,linewidth=3,color='r')
plt.hist(aptable['pmdec'][mask_25to40],alpha=0.5,bins=30,
normed=normed,linewidth=3,color='b')
plt.legend([r'$0^\circ < \mathrm{RA} < 15^\circ$',
r'$25^\circ < \mathrm{RA} < 40^\circ$'],fontsize=16)
plt.title('Distribution of DEC Proper Motions for different RA cuts',fontsize=20)
plt.xlabel('DEC Proper Motion [mas/yr]',fontsize=16)
plt.ylabel('Normed number density',fontsize=16)
plt.xlim([xmin,xmax])
#plt.ylim([1,200])
plt.savefig('hw2_3_pm_hist_dec.png', bbox_inches='tight')
# Make some universal cuts
r_mask = (GalFastTable['rmag']>20.5) & (GalFastTable['rmag']<21)
gminusi_mask = ((GalFastTable['gmag']-GalFastTable['imag']) > 0.3) &\
((GalFastTable['gmag']-GalFastTable['imag']) < 0.4)
mag_mask = r_mask * gminusi_mask
ProperMotionHist(GalFastTable,mag_mask, -5,5)
```
Here, we see a that according ot GALFAST simulation there is no difference in proper motions between the $25^\circ < \mathrm{RA} < 40^\circ$ region and $0^\circ < \mathrm{RA} < 15^\circ$ region. This is *NOT* mirrored in the NSC data, because the proper motion distributions are not consistent consistent. NSC data has most stars with proper motions of $0 \ \mathrm{[mas/yr]} < \mathrm{PM} < 30 \ \mathrm{ [mas/yr]}$, while Galfast data has most stars with proper motions of $0 \ \mathrm{[mas/yr]} < \mathrm{PM} < 6 \ \mathrm{ [mas/yr]}$.
|
github_jupyter
|
# Imports
%matplotlib inline
from astropy.table import Table
from astropy.coordinates import SkyCoord
from astropy import units as u
from astropy.table import hstack
from astropy.table import vstack
import matplotlib.pyplot as plt
import matplotlib.pyplot as plt
from matplotlib.patches import Circle
import os
import numpy as np
from astropy.io import fits
import pandas as pd
from scipy.stats import binned_statistic_2d as bs2d
from scipy.stats import binned_statistic as bs1d
import seaborn as sns
GalFastData = np.loadtxt('../../../project/Galfast-Stripe82.txt',usecols=(0,1,4,5,12,13,14))
GalFastTable = Table(rows=GalFastData, names=('ra','dec','pmra','pmdec','gmag','rmag','imag'))
# Generate the magnitude mask used in the Hess diagram data analysis
def Hess_r_v_gminusi(aptable,total_mask):
constant = 3 # This is a fudge parameter in determining number of bins
nObjects = np.sum(total_mask)
num_bins = int(constant * nObjects ** (1.0 / 4.0))
# Now calculate the binned proper motions
proper_motion = np.sqrt(aptable['pmra']**2+aptable['pmdec']**2)
total_mask = proper_motion<100*total_mask
Binned_PM = bs2d(aptable['ra'][total_mask],aptable['rmag'][total_mask],proper_motion[total_mask], bins = num_bins)
#cmin = min(np.log10(Binned_PM.statistic.T[Binned_PM.statistic.T > 0]))
#cmax = max(np.log10(Binned_PM.statistic.T[Binned_PM.statistic.T > 0]))
# Define custom colormaps: Set pixels with no sources to white
cmap = plt.cm.viridis
cmap.set_bad('w', 1.)
plt.figure(figsize=[8,8])
plt.imshow(Binned_PM.statistic.T, origin='lower',
extent=[Binned_PM.x_edge[0], Binned_PM.x_edge[-1], Binned_PM.y_edge[0], Binned_PM.y_edge[-1]],
aspect='auto', interpolation='nearest', cmap=cmap)
cb = plt.colorbar(orientation='horizontal')
cb.set_label(r'Proper Motion [mas/yr]',fontsize=16)
#plt.clim(0, 30) # This was set by hand to draw out as much detail as possible
plt.xlabel(r'RA [degree]',fontsize=16)
plt.ylabel(r'r',fontsize=16)
plt.gca().invert_yaxis()
plt.gca().invert_xaxis()
# Make the color masks
r_mask = (GalFastTable['rmag']>20.5) & (GalFastTable['rmag']<21)
gminusi_mask = ((GalFastTable['gmag']-GalFastTable['imag']) > 0.3) & ((GalFastTable['gmag']-GalFastTable['imag']) < 0.4)
mag_mask = r_mask * gminusi_mask
# Make the RA Masks
RA_mask_25to40 = (GalFastTable['ra'] > 25) & (GalFastTable['ra'] < 40)
RA_mask_0to15 = (GalFastTable['ra'] > 0) & (GalFastTable['ra'] < 15)
# Make the net masks
mask_25to40 = mag_mask * RA_mask_25to40
mask_0to15 = mag_mask * RA_mask_0to15
# Make the Hess diagram for 25 < RA < 40
Hess_r_v_gminusi(GalFastTable,mask_25to40)
plt.title(r'Proper Motion for $25^\circ < \mathrm{RA} < 40^\circ$',fontsize=20)
plt.savefig('hw2_3_GalFast_pm_Hess_Diagram_RA25to40') # Save the figure
# Make the Hess diagram for 0 < RA < 15
Hess_r_v_gminusi(GalFastTable,mask_0to15)
plt.title(r'Proper Motion for $0^\circ < \mathrm{RA} < 15^\circ$',fontsize=20)
plt.savefig('hw2_3_GalFast_pm_Hess_Diagram_RA0to15') # Save the figure
def ProperMotionHist(aptable,mask_noRAcuts,xmin=-5,xmax=5,normed='True'):
# This function makes two histograms of RA and DEC Proper motions for two different RA cuts
# Calculate the RA Proper Motion
mask_noRAcuts *= aptable['pmra']<50
# Make two masks that have the RA cuts included
RA_mask_0to15 = (aptable['ra'] > 0) & (aptable['ra'] < 15)
RA_mask_25to40 = (aptable['ra'] > 25) & (aptable['ra'] < 40)
# Mask things outside the range of the histogram
mask_pm = (xmin < aptable['pmra'])*(aptable['pmra'] < xmax)
# Combine masks
mask_0to15 = mask_noRAcuts * RA_mask_0to15 * mask_pm
mask_25to40 = mask_noRAcuts * RA_mask_25to40 * mask_pm
#Plot the two distributions with different RA cuts
plt.figure(figsize=[12,8])
plt.hist(aptable['pmra'][mask_0to15],alpha=0.5,bins=30,
normed=normed,linewidth=3,color='r')
plt.hist(aptable['pmra'][mask_25to40],alpha=0.5,bins=30,
normed=normed,linewidth=3,color='b')
plt.legend([r'$0^\circ < \mathrm{RA} < 15^\circ$',
r'$25^\circ < \mathrm{RA} < 40^\circ$'],fontsize=16)
plt.title('Distribution of RA Proper Motions for different RA cuts',fontsize=20)
plt.xlabel('RA Proper Motion [mas/yr]',fontsize=16)
plt.ylabel('Normed number density',fontsize=16)
plt.xlim([xmin,xmax])
#plt.ylim([1,200])
plt.savefig('hw2_3_pm_hist_ra.png', bbox_inches='tight')
# Calculate the DEC Proper Motion
mask_noRAcuts *= aptable['pmdec']<50
# Make two masks that have the RA cuts included
RA_mask_0to15 = (aptable['ra'] > 0) & (aptable['ra'] < 15)
RA_mask_25to40 = (aptable['ra'] > 25) & (aptable['ra'] < 40)
# Mask things outside the range of the histogram
mask_pm = (xmin < aptable['pmdec'])*(aptable['pmdec'] < xmax)
# Combine masks
mask_0to15 = mask_noRAcuts * RA_mask_0to15 * mask_pm
mask_25to40 = mask_noRAcuts * RA_mask_25to40 * mask_pm
#Plot the two distributions with different RA cuts
plt.figure(figsize=[12,8])
plt.hist(aptable['pmdec'][mask_0to15],alpha=0.5,bins=30,
normed=normed,linewidth=3,color='r')
plt.hist(aptable['pmdec'][mask_25to40],alpha=0.5,bins=30,
normed=normed,linewidth=3,color='b')
plt.legend([r'$0^\circ < \mathrm{RA} < 15^\circ$',
r'$25^\circ < \mathrm{RA} < 40^\circ$'],fontsize=16)
plt.title('Distribution of DEC Proper Motions for different RA cuts',fontsize=20)
plt.xlabel('DEC Proper Motion [mas/yr]',fontsize=16)
plt.ylabel('Normed number density',fontsize=16)
plt.xlim([xmin,xmax])
#plt.ylim([1,200])
plt.savefig('hw2_3_pm_hist_dec.png', bbox_inches='tight')
# Make some universal cuts
r_mask = (GalFastTable['rmag']>20.5) & (GalFastTable['rmag']<21)
gminusi_mask = ((GalFastTable['gmag']-GalFastTable['imag']) > 0.3) &\
((GalFastTable['gmag']-GalFastTable['imag']) < 0.4)
mag_mask = r_mask * gminusi_mask
ProperMotionHist(GalFastTable,mag_mask, -5,5)
| 0.641984 | 0.775647 |
[](https://mybinder.org/v2/gh/oddrationale/AdventOfCode2020CSharp/main?urlpath=lab%2Ftree%2FDay11.ipynb)
# --- Day 11: Seating System ---
```
using System.IO;
var initialSeatLayout = File.ReadAllLines(@"input/11.txt").Select(line => line.ToCharArray()).ToArray();
char[][] GenerateNextLayout(char[][] layout)
{
var nextLayout = layout.Select(arr => (char[])arr.Clone()).ToArray();
var maxRow = layout.Length;
var maxCol = layout.First().Length;
ValueTuple<int, int>[] directions = {
(1, 0),
(-1, 0),
(0, -1),
(0, 1),
(-1, 1),
(1, 1),
(1, -1),
(-1, -1),
};
for (int i = 0; i < maxRow; i++)
{
for (int j = 0; j < maxCol; j++)
{
if (layout[i][j] == '.')
{
continue;
}
var adjacents = new List<char>();
foreach (var d in directions)
{
if (0 <= i + d.Item1 && i + d.Item1 < maxRow &&
0 <= j + d.Item2 && j + d.Item2 < maxCol &&
layout[i + d.Item1][j + d.Item2] != '.')
{
adjacents.Add(layout[i + d.Item1][j + d.Item2]);
}
}
if (layout[i][j] == 'L' && !adjacents.Where(seat => seat == '#').Any())
{
nextLayout[i][j] = '#';
}
else if (layout[i][j] == '#' && adjacents.Where(seat => seat == '#').Count() >= 4)
{
nextLayout[i][j] = 'L';
}
}
}
return nextLayout;
}
int CountFinalOccupiedSeats(char[][] layout)
{
char[][] currentLayout;
var nextLayout = layout;
do
{
currentLayout = nextLayout;
nextLayout = GenerateNextLayout(currentLayout);
} while (string.Join("\n", currentLayout.Select(row => string.Join(string.Empty, row)))
!= string.Join("\n", nextLayout.Select(row => string.Join(string.Empty, row))));
return currentLayout.SelectMany(row => row).Where(seat => seat == '#').Count();
}
CountFinalOccupiedSeats(initialSeatLayout)
```
# --- Part Two ---
```
char[][] GenerateNextLayout2(char[][] layout)
{
var nextLayout = layout.Select(arr => (char[])arr.Clone()).ToArray();
var maxRow = layout.Length;
var maxCol = layout.First().Length;
ValueTuple<int, int>[] directions = {
(1, 0),
(-1, 0),
(0, -1),
(0, 1),
(-1, 1),
(1, 1),
(1, -1),
(-1, -1),
};
for (int i = 0; i < maxRow; i++)
{
for (int j = 0; j < maxCol; j++)
{
if (layout[i][j] == '.')
{
continue;
}
var adjacents = new List<char>();
foreach (var d in directions)
{
var steps = 1;
while (0 <= i + steps*d.Item1 && i + steps*d.Item1 < maxRow
&& 0 <= j + steps*d.Item2 && j + steps*d.Item2 < maxCol)
{
if (layout[i + steps*d.Item1][j + steps*d.Item2] != '.')
{
adjacents.Add(layout[i + steps*d.Item1][j + steps*d.Item2]);
break;
}
steps++;
}
}
if (layout[i][j] == 'L' && !adjacents.Where(seat => seat == '#').Any())
{
nextLayout[i][j] = '#';
}
else if (layout[i][j] == '#' && adjacents.Where(seat => seat == '#').Count() >= 5)
{
nextLayout[i][j] = 'L';
}
}
}
return nextLayout;
}
int CountFinalOccupiedSeats2(char[][] layout)
{
char[][] currentLayout;
var nextLayout = layout;
do
{
currentLayout = nextLayout;
nextLayout = GenerateNextLayout2(currentLayout);
} while (string.Join("\n", currentLayout.Select(row => string.Join(string.Empty, row)))
!= string.Join("\n", nextLayout.Select(row => string.Join(string.Empty, row))));
return currentLayout.SelectMany(row => row).Where(seat => seat == '#').Count();
}
CountFinalOccupiedSeats2(initialSeatLayout)
```
|
github_jupyter
|
using System.IO;
var initialSeatLayout = File.ReadAllLines(@"input/11.txt").Select(line => line.ToCharArray()).ToArray();
char[][] GenerateNextLayout(char[][] layout)
{
var nextLayout = layout.Select(arr => (char[])arr.Clone()).ToArray();
var maxRow = layout.Length;
var maxCol = layout.First().Length;
ValueTuple<int, int>[] directions = {
(1, 0),
(-1, 0),
(0, -1),
(0, 1),
(-1, 1),
(1, 1),
(1, -1),
(-1, -1),
};
for (int i = 0; i < maxRow; i++)
{
for (int j = 0; j < maxCol; j++)
{
if (layout[i][j] == '.')
{
continue;
}
var adjacents = new List<char>();
foreach (var d in directions)
{
if (0 <= i + d.Item1 && i + d.Item1 < maxRow &&
0 <= j + d.Item2 && j + d.Item2 < maxCol &&
layout[i + d.Item1][j + d.Item2] != '.')
{
adjacents.Add(layout[i + d.Item1][j + d.Item2]);
}
}
if (layout[i][j] == 'L' && !adjacents.Where(seat => seat == '#').Any())
{
nextLayout[i][j] = '#';
}
else if (layout[i][j] == '#' && adjacents.Where(seat => seat == '#').Count() >= 4)
{
nextLayout[i][j] = 'L';
}
}
}
return nextLayout;
}
int CountFinalOccupiedSeats(char[][] layout)
{
char[][] currentLayout;
var nextLayout = layout;
do
{
currentLayout = nextLayout;
nextLayout = GenerateNextLayout(currentLayout);
} while (string.Join("\n", currentLayout.Select(row => string.Join(string.Empty, row)))
!= string.Join("\n", nextLayout.Select(row => string.Join(string.Empty, row))));
return currentLayout.SelectMany(row => row).Where(seat => seat == '#').Count();
}
CountFinalOccupiedSeats(initialSeatLayout)
char[][] GenerateNextLayout2(char[][] layout)
{
var nextLayout = layout.Select(arr => (char[])arr.Clone()).ToArray();
var maxRow = layout.Length;
var maxCol = layout.First().Length;
ValueTuple<int, int>[] directions = {
(1, 0),
(-1, 0),
(0, -1),
(0, 1),
(-1, 1),
(1, 1),
(1, -1),
(-1, -1),
};
for (int i = 0; i < maxRow; i++)
{
for (int j = 0; j < maxCol; j++)
{
if (layout[i][j] == '.')
{
continue;
}
var adjacents = new List<char>();
foreach (var d in directions)
{
var steps = 1;
while (0 <= i + steps*d.Item1 && i + steps*d.Item1 < maxRow
&& 0 <= j + steps*d.Item2 && j + steps*d.Item2 < maxCol)
{
if (layout[i + steps*d.Item1][j + steps*d.Item2] != '.')
{
adjacents.Add(layout[i + steps*d.Item1][j + steps*d.Item2]);
break;
}
steps++;
}
}
if (layout[i][j] == 'L' && !adjacents.Where(seat => seat == '#').Any())
{
nextLayout[i][j] = '#';
}
else if (layout[i][j] == '#' && adjacents.Where(seat => seat == '#').Count() >= 5)
{
nextLayout[i][j] = 'L';
}
}
}
return nextLayout;
}
int CountFinalOccupiedSeats2(char[][] layout)
{
char[][] currentLayout;
var nextLayout = layout;
do
{
currentLayout = nextLayout;
nextLayout = GenerateNextLayout2(currentLayout);
} while (string.Join("\n", currentLayout.Select(row => string.Join(string.Empty, row)))
!= string.Join("\n", nextLayout.Select(row => string.Join(string.Empty, row))));
return currentLayout.SelectMany(row => row).Where(seat => seat == '#').Count();
}
CountFinalOccupiedSeats2(initialSeatLayout)
| 0.177918 | 0.686767 |
Each function is represented by a **frame**.
* A frame is a box with the name of a function beside it and the parameters and variables of the function inside it
* The frames are arranged in a stack that indicates which function called which, and so on.
* When you create a variable outside of any function, it belongs to __main__. __main__ is a special name for the top-most frame.
* Each parameter refers to the same value as its corresponding argument
* If an error occurs during a function call, Python prints the name of the function, and the name of the function that called it, and the name of the function that called that, all the way back to __main__. This list of functions is called a **traceback**. It tells you what program file the error occurred in, and what line, and what functions were executing at the time. It also shows the line of code that caused the error. The order of the functions in the traceback is the same as the order of the frames in the stack diagram. The function that is currently running is at the bottom.
```
def print_twice(bruce):
"""Print a given parameter twice
"""
print(bruce)
print(bruce)
# Call the function and pass any name to parameter bruce
print_twice('daniel')
def cat_twice(part1, part2):
"""concatenate params part1 and part2
then print it out
"""
cat = part1 + part2
print_twice(cat)
# Call the function with two names using the previous function
print(cat_twice('mary', 'martha'))
```
**Why functions?**
It may not be clear why it is worth the trouble to divide a program into functions. There
are several reasons:
• Creating a new function gives you an opportunity to name a group of statements,
which makes your program easier to read and debug.
• Functions can make a program smaller by eliminating repetitive code. Later, if you
make a change, you only have to make it in one place.
• Dividing a long program into functions allows you to debug the parts one at a time
and then assemble them into a working whole.
• Well-designed functions are often useful for many programs. Once you write and
debug one, you can reuse it.
<h3>Importing modules</h3>
Python provides two ways to import modules
1. `import math`
2. `from math import pi` or `from math import *`
The first one imports a module object named math.This module object has constants like pi and functions like pow(), sin() cosine(). But if we try to access pi directly, we get an error.
```
import math
x = pi
```
Alternatively, we can import an object from a module like so...
* `from math import pi`
* `from math import *`
The advantage of importing everything from the math module is that your code can be
more concise. The disadvantage is that there might be conflicts between names defined in
different modules, or between a name from a module and one of your variables.
It's not a good practice to import everything with the asterisk operator. Rather import the functions you need from the modu;es.
<h3>Glossary</h3>
1. **function:** A named sequence of statements that performs some useful operation. Functions may or may not take arguments and may or may not produce a result.
2. **function definition:** A statement that creates a new function, specifying its name, parameters, and the statements it executes.
3. **function object:** A value created by a function definition. The name of the function is a variable that refers to a function object.
4. **header:** The first line of a function definition.
5. **body:** The sequence of statements inside a function definition.
6. **parameter:** A name used inside a function to refer to the value passed as an argument.
7. **function call:** A statement that executes a function. It consists of the function name followed by an argument list.
8. **argument:** A value provided to a function when the function is called. This value is assigned to the corresponding parameter in the function.
9. **local variable:** A variable defined inside a function. A local variable can only be used inside its function.
10. **return value:** The result of a function. If a function call is used as an expression, the return value is the value of the expression.
11. **fruitful function:** A function that returns a value.
12. **void function:** A function that doesn’t return a value.
13. **module:** A file that contains a collection of related functions and other definitions.
14. **import statement:** A statement that reads a module file and creates a module object.
15. **module object:** A value created by an import statement that provides access to the values defined in a module.
16. **dot notation:** The syntax for calling a function in another module by specifying the module name followed by a dot (period) and the function name.
17. **composition:** Using an expression as part of a larger expression, or a statement as part of a larger statement.
18. **flow of execution:** The order in which statements are executed during a program run.
19. **stack diagram:** A graphical representation of a stack of functions, their variables, and the values they refer to.
20. **frame:** A box in a stack diagram that represents a function call. It contains the local variables and parameters of the function.
21. **traceback:** A list of the functions that are executing, printed when an exception occurs.
<h3>Exercises</h3>
**Exercise 3.3.** Python provides a built-in function called len that returns the length of a string, so
the value of len('allen') is 5.
Write a function named right_justify that takes a string named `s` as a parameter and prints the
string with enough leading spaces so that the last letter of the string is in column 70 of the display.
**Exercise 3.4.** A function object is a value you can assign to a variable or pass as an argument. For
example, `do_twice` is a function that takes a function object as an argument and calls it twice:
`
def do_twice(f):
f()
f()
`<br>
Here’s an example that uses `do_twice` to call a function named `print_spam` twice.
`
def print_spam():
print 'spam'
do_twice(print_spam)
`
1. Type this example into a script and test it.
2. Modify `do_twice` so that it takes two arguments, a function object and a value, and calls the function twice, passing the value as an argument.
3. Write a more general version of `print_spam`, called `print_twice`, that takes a string as a parameter and prints it twice.
4. Use the modified version of `do_twice` to call `print_twice` twice, passing `'spam'` as an argument.
5. Define a new function called `do_four` that takes a function object and a value and calls the function four times, passing the value as a parameter. There should be only two statements in the body of this function, not four.
**Exercise 3.5.** This exercise can be done using only the statements and other features we have learned
so far.
1. Write a function that draws a grid like the following:
|
github_jupyter
|
def print_twice(bruce):
"""Print a given parameter twice
"""
print(bruce)
print(bruce)
# Call the function and pass any name to parameter bruce
print_twice('daniel')
def cat_twice(part1, part2):
"""concatenate params part1 and part2
then print it out
"""
cat = part1 + part2
print_twice(cat)
# Call the function with two names using the previous function
print(cat_twice('mary', 'martha'))
import math
x = pi
| 0.371935 | 0.988154 |
```
#!/usr/bin/python
# -*- coding: utf-8 -*-
"""
This notebook adds the WLE per contributor, per site and per autonomous
community categories to all the files in the WLE log.
"""
import os, sys, inspect
try :
import pywikibot as pb
from pywikibot import textlib
import mwparserfromhell as mwh
except :
current_folder = os.path.realpath(os.path.abspath(os.path.split(inspect.getfile(inspect.currentframe()))[0]))
folder_parts = current_folder.split(os.sep)
pywikibot_folder = os.sep.join(folder_parts[0:-1])
if current_folder not in sys.path:
sys.path.insert(0, current_folder)
if pywikibot_folder not in sys.path:
sys.path.insert(0, pywikibot_folder)
import pywikibot as pb
from pywikibot import textlib
import mwparserfromhell as mwh
import numpy as np
import pandas as pd
from io import StringIO
import re
from mako.template import Template
from datetime import datetime, timedelta
YEAR = 2016
TAG = 'WLE'
TAG_EXT = 'Wiki Loves Earth'
COUNTRY = "Spain"
CODE_TEMPLATE = "lic"
BASE_NAME = "Commons:Wiki Loves in {2}/{1}/{0}".format(YEAR, TAG_EXT, COUNTRY)
LOG_PAGE = BASE_NAME + '/Log'
BASE_SITE_DB_NAME = "Commons:Wiki Loves in {1}/{0}".format(TAG_EXT, COUNTRY)
SITE_DB_PAGE = BASE_SITE_DB_NAME + "/Sites DB"
START_TIME = datetime(YEAR, 5, 1, 0, 0, 0)
END_TIME = datetime(YEAR, 6, 1, 0, 0, 0) + timedelta(hours=1)
commons_site = pb.Site("commons", "commons")
communities = {
'ES-AN': u'Andalusia',
'ES-AR': u'Aragon',
'ES-AS': u'Asturias',
'ES-CB': u'Cantabria',
'ES-CM': u'Castile-La Mancha',
'ES-CL': u'Castile and León',
'ES-CT': u'Catalonia',
'ES-MD': u'Community of Madrid',
'ES-VC': u'Valencian Community',
'ES-EX': u'Extremadura',
'ES-IB': u'Balearic Islands',
'ES-CN': u'Canary Islands',
'ES-GA': u'Galicia',
'ES-RI': u'La Rioja',
'ES-NC': u'Navarre',
'ES-MC': u'Region of Murcia',
'ES-CE': u'Ceuta',
'ES-ML': u'Melilla',
'ES-PV': u'Basque Country',
'ES-MAGRAMA': u'MAGRAMA'
}
def get_community (x) :
try:
return communities[x]
except :
return np.nan
```
### Retrieval of list of sites of community importance
```
pb.output('Retrieving --> WLE SCI list')
site_list_page = pb.Page(commons_site, SITE_DB_PAGE)
site_list_text = StringIO(site_list_page.text[site_list_page.text.find('\n') + 1:site_list_page.text.rfind('\n')])
site_df = pd.read_csv(site_list_text, sep=";",
index_col=False,
names=["name", "lic_id", "magrama_url", "community",
"bio_region", "continent", "min_altitude",
"max_altitude", "avg_altitude", "longitude",
"latitude", "area", "marine_percentage",
"marine_area", "image", "commons_cat", "wikidata_id"])
pb.output('Retrieved --> WLE SCI list')
site_df['community_name'] = site_df['community'].apply(get_community)
codes = site_df["lic_id"].tolist()
site_df.describe(include="all")
```
### Retrieval of the image log
```
pb.output('Retrieving --> {0} {1} images list from cache'.format(TAG, YEAR))
list_page = pb.Page(commons_site, LOG_PAGE)
list_page_text = StringIO(list_page.text[list_page.text.find('\n') + 1:list_page.text.rfind('\n')])
images_df = pd.read_csv(list_page_text,
sep=";",
index_col=False,
names=['image_title', 'lic_id',
'uploader', 'uploader_registration',
'timestamp', 'date', 'size',
'height', 'width', 'qi',
'finalist'])
images_df['timestamp'] = pd.to_datetime(images_df['timestamp'], format="%Y-%m-%d %H:%M:%S")
pb.output('Retrieved --> {0} {1} images list from cache'.format(TAG, YEAR))
images_df = images_df.merge(site_df, on='lic_id', how="left")
site_cat_template = """'''Site of Community Importance''': ${lic_name} ([http://natura2000.eea.europa.eu/Natura2000/SDF.aspx?site=${lic_code} ${lic_code}])
{{hiddencat}}
[[Category:Images from ${tag} in ${country} by site| ${lic_code}]]
"""
site_vars = {
"lic_name": None,
"lic_code": None,
"country": COUNTRY,
"tag": TAG_EXT
}
yearly_site_cat_template = """'''Site of Community Importance''': ${lic_name} ([http://natura2000.eea.europa.eu/Natura2000/SDF.aspx?site=${lic_code} ${lic_code}])
{{hiddencat}}
[[Category:Images from ${tag} ${year} in ${country} by site| ${lic_code}]]
"""
yearly_site_vars = {
"lic_name": None,
"lic_code": None,
"country": COUNTRY,
"tag": TAG_EXT,
"year": YEAR
}
autcom_cat_template = """{{hiddencat}}
[[Category:Images from ${tag} ${year} in ${country} by autonomous community| ${aut_com}]]"""
autcom_vars = {
"aut_com": None,
"country": COUNTRY,
"tag": TAG_EXT,
"year": YEAR}
author_cat_template = """{{hiddencat}}
[[Category:Images from ${tag} ${year} in ${country} by author| ${author_name}]]"""
author_vars = {
"author_name": None,
"country": COUNTRY,
"tag": TAG_EXT,
"year": YEAR}
implicit_cats = ["Sites of Community Importance in Spain with known IDs",
"CC-BY-SA-4.0", "CC-BY-4.0", "GFDL",
"Media with locations",
"License migration redundant",
"Self-published work",
"Pages with maps",
"Images with watermarks",
"All media needing categories as of {1}",
"Images from {0} {1}",
"Images from {0} {1} in {2}"]
implicit_cats = [cat.format(TAG_EXT, YEAR, COUNTRY) for cat in implicit_cats]
c = ["Images from {0} {1} in {2} with a wrong code",
"Images from {0} {1} in {2} without code",
"Images from {0} {1} in {2} without valid template",
"Uncategorized images from {0} {1} in {2}",
"Unqualified images from {0} {1} in {2} (wrong submission time)",
"Unqualified images from {0} {1} in {2} (too small)"]
to_remove_cats = [cat.format(TAG_EXT, YEAR, COUNTRY) for cat in c]
to_remove_cats
autcom_with_article = ['Region', 'Basque', 'Balearic', 'Canary', 'Valencian', 'Community']
lost_cats = []
for image_counter, row in images_df.iterrows():
#print(row["image_title"])
page = pb.FilePage(commons_site, row["image_title"])
text = page.text
if (image_counter != 0) and (image_counter % 50 == 0) :
pb.output ('Reviewing --> %d image pages downloaded' %(image_counter))
nocat_text = textlib.removeCategoryLinks(text)
cats = [cat for cat in page.categories()]
cat_titles = [cat.title(withNamespace=False) for cat in cats]
lost_cats.extend([cat.title(withNamespace=False) for cat in cats if (not cat.isHiddenCategory() and not cat.exists())])
visible_cats = [cat.title(withNamespace=False) for cat in cats if (not cat.isHiddenCategory())]
cleaned_visible_cats = [cat.title(withNamespace=False) for cat in cats if (not cat.isHiddenCategory() and cat.exists())]
hidden_cats = [cat.title(withNamespace=False) for cat in cats if cat.isHiddenCategory()]
cleaned_hidden_cats = [cat.title(withNamespace=False) for cat in cats if cat.isHiddenCategory() and not
(re.match('Images of a site of community importance with code ES\d+ from Wiki Loves Earth 2017 in Spain', cat.title(withNamespace=False)) or
re.match('Images of a site of community importance with code ES\d+ from Wiki Loves Earth in Spain', cat.title(withNamespace=False)) or
re.match('Images of a site of community importance in [a-zA-Z ]+ from Wiki Loves Earth 2017 in Spain', cat.title(withNamespace=False)) or
re.match('Images from Wiki Loves Earth 2017 in Spain by .+', cat.title(withNamespace=False)) or
cat.title(withNamespace=False) in to_remove_cats)]
wikicode = mwh.parse(text)
wle_templates = [template for template in wikicode.filter_templates()
if template.name.lower().strip() == CODE_TEMPLATE]
# authorship classification
if 'flickr' in row['uploader'] :
author = '{0} (flickr)'.format(' '.join(row['uploader'].split(' ')[1:]))
else :
author = row['uploader']
author_category = 'Images from {1} {2} in {3} by {0}'.format(author, TAG_EXT, YEAR, COUNTRY)
cleaned_hidden_cats.append(author_category)
category = pb.Category(commons_site, author_category)
if not category.exists() :
t = Template(author_cat_template)
category.text = t.render(**author_vars)
category.save("{0} {2} {1}: user category creation".format(TAG, YEAR, COUNTRY))
# size classification
if page.latest_file_info["width"] * page.latest_file_info["height"] < 2000000 :
print ("too small ({})".format(row["image_title"]))
cleaned_hidden_cats.append('Unqualified images from {0} {1} in {2} (too small)'.format(TAG_EXT, YEAR, COUNTRY))
try:
cleaned_hidden_cats.remove("Images from {0} {1} in {2} to be evaluated".format(TAG_EXT, YEAR, COUNTRY))
except :
pass
# date classification
if row["timestamp"] > END_TIME or row["timestamp"] < START_TIME :
print ("uploaded too late or too soon ({})".format(row["image_title"]))
cleaned_hidden_cats.append('Unqualified images from {0} {1} in {2} (wrong submission time)'.format(TAG_EXT, YEAR, COUNTRY))
try :
cleaned_hidden_cats.remove("Images from {0} {1} in {2} to be evaluated".format(TAG_EXT, YEAR, COUNTRY))
except:
pass
#uncategorized classification
if '{{Uncategorized|year=' in text :
p = re.compile('\{\{Uncategorized\|year=(\d+)\|month=([a-zA-Z]*)\|day=(\d+)\}\}')
m = p.search(text)
uncategorized_cat = 'Media needing categories as of {0} {1} {2}'.format(m.groups()[2], m.groups()[1], m.groups()[0])
try :
cleaned_hidden_cats.remove(uncategorized_cat)
except :
pass
cleaned_hidden_cats.append('Uncategorized images from {0} {1} in {2}'.format(TAG_EXT, YEAR, COUNTRY))
elif len(cleaned_visible_cats) == 0 or '{{Uncategorized|year=' in text:
cleaned_hidden_cats.append('Uncategorized images from {0} {1} in {2}'.format(TAG_EXT, YEAR, COUNTRY))
# site/autcom classification
if len(wle_templates) == 0:
print ("No template ({})".format(row["image_title"]))
if (("Unqualified images from {0} {1} in {2} (not from a site of community importance)".format(TAG_EXT, YEAR, COUNTRY) not in cleaned_hidden_cats) and
("Unqualified images from {0} {1} in {2} (not from {2})".format(TAG_EXT, YEAR, COUNTRY) not in cleaned_hidden_cats) and
("Unqualified images from {0} {1} in {2} (unidentified locations)".format(TAG_EXT, YEAR, COUNTRY) not in cleaned_hidden_cats)) :
cleaned_hidden_cats.append("Images from {0} {1} in {2} without valid template".format(TAG_EXT, YEAR, COUNTRY))
else :
for template in wle_templates :
#print (template)
code = template.get(1).value.strip()
if code in codes :
is_lic = True
#print ("Valid code")
yearly_site_category = 'Images of a site of community importance with code {0} from {1} {2} in {3}'.format(code, TAG_EXT, YEAR, COUNTRY)
cleaned_hidden_cats.append(yearly_site_category)
site_category = 'Images of a site of community importance with code {0} from {1} in {3}'.format(code, TAG_EXT, YEAR, COUNTRY)
cleaned_hidden_cats.append(site_category)
autcom_name = site_df[site_df['lic_id'] == code]['community_name'].values[0]
if len ([i for i in autcom_with_article if autcom_name.startswith(i)]) != 0 :
autcom_name = 'the ' + autcom_name
if 'Images of a site of community importance in {0} from {1} {2} in {3}'.format(autcom_name, TAG_EXT, YEAR, COUNTRY) not in cleaned_hidden_cats:
community_category = 'Images of a site of community importance in {0} from {1} {2} in {3}'.format(autcom_name, TAG_EXT, YEAR, COUNTRY)
cleaned_hidden_cats.append(community_category)
category = pb.Category(commons_site, community_category)
if not category.exists() :
yearly_site_vars['lic_name'] = row['name']
yearly_site_vars['lic_code'] = code
t = Template(yearly_site_cat_template)
category.text = t.render(**yearly_site_vars)
category.save("{0} {2} {1}: site category creation".format(TAG, YEAR, COUNTRY))
category = pb.Category(commons_site, yearly_site_category)
if not category.exists() :
yearly_site_vars['lic_name'] = row['name']
yearly_site_vars['lic_code'] = code
t = Template(yearly_site_cat_template)
category.text = t.render(**yearly_site_vars)
category.save("{0} {2} {1}: site category creation".format(TAG, YEAR, COUNTRY))
category = pb.Category(commons_site, site_category)
if not category.exists() :
autcom_vars['aut_com'] = row["community_name"]
t = Template(autcom_cat_template)
category.text = t.render(**autcom_vars)
category.save("{0} {2} {1}: autonomous community category creation".format(TAG, YEAR, COUNTRY))
elif code.startswith('ES'):
print ("Code not from a LIC")
cleaned_hidden_cats.append("Images from {0} {1} in {2} with a wrong code".format(TAG_EXT, YEAR, COUNTRY))
else :
print ("Invalid code")
cleaned_hidden_cats.append("Images from {0} {1} in {2} without code".format(TAG_EXT, YEAR, COUNTRY))
if len(set(cat_titles) ^ set(visible_cats + cleaned_hidden_cats)) > 0:
print (set(cat_titles) ^ set(cleaned_visible_cats + cleaned_hidden_cats))
cat_text = '\n'.join(['[[Category:{0}]]'.format(cat) for cat in set(visible_cats+cleaned_hidden_cats) if cat not in implicit_cats])
page_text = nocat_text + '\n' + cat_text
page.text = page_text
#page.save("{1} {0} in {2}: Classification".format(YEAR, TAG, COUNTRY))
for i in set(lost_cats) :
print (i)
```
|
github_jupyter
|
#!/usr/bin/python
# -*- coding: utf-8 -*-
"""
This notebook adds the WLE per contributor, per site and per autonomous
community categories to all the files in the WLE log.
"""
import os, sys, inspect
try :
import pywikibot as pb
from pywikibot import textlib
import mwparserfromhell as mwh
except :
current_folder = os.path.realpath(os.path.abspath(os.path.split(inspect.getfile(inspect.currentframe()))[0]))
folder_parts = current_folder.split(os.sep)
pywikibot_folder = os.sep.join(folder_parts[0:-1])
if current_folder not in sys.path:
sys.path.insert(0, current_folder)
if pywikibot_folder not in sys.path:
sys.path.insert(0, pywikibot_folder)
import pywikibot as pb
from pywikibot import textlib
import mwparserfromhell as mwh
import numpy as np
import pandas as pd
from io import StringIO
import re
from mako.template import Template
from datetime import datetime, timedelta
YEAR = 2016
TAG = 'WLE'
TAG_EXT = 'Wiki Loves Earth'
COUNTRY = "Spain"
CODE_TEMPLATE = "lic"
BASE_NAME = "Commons:Wiki Loves in {2}/{1}/{0}".format(YEAR, TAG_EXT, COUNTRY)
LOG_PAGE = BASE_NAME + '/Log'
BASE_SITE_DB_NAME = "Commons:Wiki Loves in {1}/{0}".format(TAG_EXT, COUNTRY)
SITE_DB_PAGE = BASE_SITE_DB_NAME + "/Sites DB"
START_TIME = datetime(YEAR, 5, 1, 0, 0, 0)
END_TIME = datetime(YEAR, 6, 1, 0, 0, 0) + timedelta(hours=1)
commons_site = pb.Site("commons", "commons")
communities = {
'ES-AN': u'Andalusia',
'ES-AR': u'Aragon',
'ES-AS': u'Asturias',
'ES-CB': u'Cantabria',
'ES-CM': u'Castile-La Mancha',
'ES-CL': u'Castile and León',
'ES-CT': u'Catalonia',
'ES-MD': u'Community of Madrid',
'ES-VC': u'Valencian Community',
'ES-EX': u'Extremadura',
'ES-IB': u'Balearic Islands',
'ES-CN': u'Canary Islands',
'ES-GA': u'Galicia',
'ES-RI': u'La Rioja',
'ES-NC': u'Navarre',
'ES-MC': u'Region of Murcia',
'ES-CE': u'Ceuta',
'ES-ML': u'Melilla',
'ES-PV': u'Basque Country',
'ES-MAGRAMA': u'MAGRAMA'
}
def get_community (x) :
try:
return communities[x]
except :
return np.nan
pb.output('Retrieving --> WLE SCI list')
site_list_page = pb.Page(commons_site, SITE_DB_PAGE)
site_list_text = StringIO(site_list_page.text[site_list_page.text.find('\n') + 1:site_list_page.text.rfind('\n')])
site_df = pd.read_csv(site_list_text, sep=";",
index_col=False,
names=["name", "lic_id", "magrama_url", "community",
"bio_region", "continent", "min_altitude",
"max_altitude", "avg_altitude", "longitude",
"latitude", "area", "marine_percentage",
"marine_area", "image", "commons_cat", "wikidata_id"])
pb.output('Retrieved --> WLE SCI list')
site_df['community_name'] = site_df['community'].apply(get_community)
codes = site_df["lic_id"].tolist()
site_df.describe(include="all")
pb.output('Retrieving --> {0} {1} images list from cache'.format(TAG, YEAR))
list_page = pb.Page(commons_site, LOG_PAGE)
list_page_text = StringIO(list_page.text[list_page.text.find('\n') + 1:list_page.text.rfind('\n')])
images_df = pd.read_csv(list_page_text,
sep=";",
index_col=False,
names=['image_title', 'lic_id',
'uploader', 'uploader_registration',
'timestamp', 'date', 'size',
'height', 'width', 'qi',
'finalist'])
images_df['timestamp'] = pd.to_datetime(images_df['timestamp'], format="%Y-%m-%d %H:%M:%S")
pb.output('Retrieved --> {0} {1} images list from cache'.format(TAG, YEAR))
images_df = images_df.merge(site_df, on='lic_id', how="left")
site_cat_template = """'''Site of Community Importance''': ${lic_name} ([http://natura2000.eea.europa.eu/Natura2000/SDF.aspx?site=${lic_code} ${lic_code}])
{{hiddencat}}
[[Category:Images from ${tag} in ${country} by site| ${lic_code}]]
"""
site_vars = {
"lic_name": None,
"lic_code": None,
"country": COUNTRY,
"tag": TAG_EXT
}
yearly_site_cat_template = """'''Site of Community Importance''': ${lic_name} ([http://natura2000.eea.europa.eu/Natura2000/SDF.aspx?site=${lic_code} ${lic_code}])
{{hiddencat}}
[[Category:Images from ${tag} ${year} in ${country} by site| ${lic_code}]]
"""
yearly_site_vars = {
"lic_name": None,
"lic_code": None,
"country": COUNTRY,
"tag": TAG_EXT,
"year": YEAR
}
autcom_cat_template = """{{hiddencat}}
[[Category:Images from ${tag} ${year} in ${country} by autonomous community| ${aut_com}]]"""
autcom_vars = {
"aut_com": None,
"country": COUNTRY,
"tag": TAG_EXT,
"year": YEAR}
author_cat_template = """{{hiddencat}}
[[Category:Images from ${tag} ${year} in ${country} by author| ${author_name}]]"""
author_vars = {
"author_name": None,
"country": COUNTRY,
"tag": TAG_EXT,
"year": YEAR}
implicit_cats = ["Sites of Community Importance in Spain with known IDs",
"CC-BY-SA-4.0", "CC-BY-4.0", "GFDL",
"Media with locations",
"License migration redundant",
"Self-published work",
"Pages with maps",
"Images with watermarks",
"All media needing categories as of {1}",
"Images from {0} {1}",
"Images from {0} {1} in {2}"]
implicit_cats = [cat.format(TAG_EXT, YEAR, COUNTRY) for cat in implicit_cats]
c = ["Images from {0} {1} in {2} with a wrong code",
"Images from {0} {1} in {2} without code",
"Images from {0} {1} in {2} without valid template",
"Uncategorized images from {0} {1} in {2}",
"Unqualified images from {0} {1} in {2} (wrong submission time)",
"Unqualified images from {0} {1} in {2} (too small)"]
to_remove_cats = [cat.format(TAG_EXT, YEAR, COUNTRY) for cat in c]
to_remove_cats
autcom_with_article = ['Region', 'Basque', 'Balearic', 'Canary', 'Valencian', 'Community']
lost_cats = []
for image_counter, row in images_df.iterrows():
#print(row["image_title"])
page = pb.FilePage(commons_site, row["image_title"])
text = page.text
if (image_counter != 0) and (image_counter % 50 == 0) :
pb.output ('Reviewing --> %d image pages downloaded' %(image_counter))
nocat_text = textlib.removeCategoryLinks(text)
cats = [cat for cat in page.categories()]
cat_titles = [cat.title(withNamespace=False) for cat in cats]
lost_cats.extend([cat.title(withNamespace=False) for cat in cats if (not cat.isHiddenCategory() and not cat.exists())])
visible_cats = [cat.title(withNamespace=False) for cat in cats if (not cat.isHiddenCategory())]
cleaned_visible_cats = [cat.title(withNamespace=False) for cat in cats if (not cat.isHiddenCategory() and cat.exists())]
hidden_cats = [cat.title(withNamespace=False) for cat in cats if cat.isHiddenCategory()]
cleaned_hidden_cats = [cat.title(withNamespace=False) for cat in cats if cat.isHiddenCategory() and not
(re.match('Images of a site of community importance with code ES\d+ from Wiki Loves Earth 2017 in Spain', cat.title(withNamespace=False)) or
re.match('Images of a site of community importance with code ES\d+ from Wiki Loves Earth in Spain', cat.title(withNamespace=False)) or
re.match('Images of a site of community importance in [a-zA-Z ]+ from Wiki Loves Earth 2017 in Spain', cat.title(withNamespace=False)) or
re.match('Images from Wiki Loves Earth 2017 in Spain by .+', cat.title(withNamespace=False)) or
cat.title(withNamespace=False) in to_remove_cats)]
wikicode = mwh.parse(text)
wle_templates = [template for template in wikicode.filter_templates()
if template.name.lower().strip() == CODE_TEMPLATE]
# authorship classification
if 'flickr' in row['uploader'] :
author = '{0} (flickr)'.format(' '.join(row['uploader'].split(' ')[1:]))
else :
author = row['uploader']
author_category = 'Images from {1} {2} in {3} by {0}'.format(author, TAG_EXT, YEAR, COUNTRY)
cleaned_hidden_cats.append(author_category)
category = pb.Category(commons_site, author_category)
if not category.exists() :
t = Template(author_cat_template)
category.text = t.render(**author_vars)
category.save("{0} {2} {1}: user category creation".format(TAG, YEAR, COUNTRY))
# size classification
if page.latest_file_info["width"] * page.latest_file_info["height"] < 2000000 :
print ("too small ({})".format(row["image_title"]))
cleaned_hidden_cats.append('Unqualified images from {0} {1} in {2} (too small)'.format(TAG_EXT, YEAR, COUNTRY))
try:
cleaned_hidden_cats.remove("Images from {0} {1} in {2} to be evaluated".format(TAG_EXT, YEAR, COUNTRY))
except :
pass
# date classification
if row["timestamp"] > END_TIME or row["timestamp"] < START_TIME :
print ("uploaded too late or too soon ({})".format(row["image_title"]))
cleaned_hidden_cats.append('Unqualified images from {0} {1} in {2} (wrong submission time)'.format(TAG_EXT, YEAR, COUNTRY))
try :
cleaned_hidden_cats.remove("Images from {0} {1} in {2} to be evaluated".format(TAG_EXT, YEAR, COUNTRY))
except:
pass
#uncategorized classification
if '{{Uncategorized|year=' in text :
p = re.compile('\{\{Uncategorized\|year=(\d+)\|month=([a-zA-Z]*)\|day=(\d+)\}\}')
m = p.search(text)
uncategorized_cat = 'Media needing categories as of {0} {1} {2}'.format(m.groups()[2], m.groups()[1], m.groups()[0])
try :
cleaned_hidden_cats.remove(uncategorized_cat)
except :
pass
cleaned_hidden_cats.append('Uncategorized images from {0} {1} in {2}'.format(TAG_EXT, YEAR, COUNTRY))
elif len(cleaned_visible_cats) == 0 or '{{Uncategorized|year=' in text:
cleaned_hidden_cats.append('Uncategorized images from {0} {1} in {2}'.format(TAG_EXT, YEAR, COUNTRY))
# site/autcom classification
if len(wle_templates) == 0:
print ("No template ({})".format(row["image_title"]))
if (("Unqualified images from {0} {1} in {2} (not from a site of community importance)".format(TAG_EXT, YEAR, COUNTRY) not in cleaned_hidden_cats) and
("Unqualified images from {0} {1} in {2} (not from {2})".format(TAG_EXT, YEAR, COUNTRY) not in cleaned_hidden_cats) and
("Unqualified images from {0} {1} in {2} (unidentified locations)".format(TAG_EXT, YEAR, COUNTRY) not in cleaned_hidden_cats)) :
cleaned_hidden_cats.append("Images from {0} {1} in {2} without valid template".format(TAG_EXT, YEAR, COUNTRY))
else :
for template in wle_templates :
#print (template)
code = template.get(1).value.strip()
if code in codes :
is_lic = True
#print ("Valid code")
yearly_site_category = 'Images of a site of community importance with code {0} from {1} {2} in {3}'.format(code, TAG_EXT, YEAR, COUNTRY)
cleaned_hidden_cats.append(yearly_site_category)
site_category = 'Images of a site of community importance with code {0} from {1} in {3}'.format(code, TAG_EXT, YEAR, COUNTRY)
cleaned_hidden_cats.append(site_category)
autcom_name = site_df[site_df['lic_id'] == code]['community_name'].values[0]
if len ([i for i in autcom_with_article if autcom_name.startswith(i)]) != 0 :
autcom_name = 'the ' + autcom_name
if 'Images of a site of community importance in {0} from {1} {2} in {3}'.format(autcom_name, TAG_EXT, YEAR, COUNTRY) not in cleaned_hidden_cats:
community_category = 'Images of a site of community importance in {0} from {1} {2} in {3}'.format(autcom_name, TAG_EXT, YEAR, COUNTRY)
cleaned_hidden_cats.append(community_category)
category = pb.Category(commons_site, community_category)
if not category.exists() :
yearly_site_vars['lic_name'] = row['name']
yearly_site_vars['lic_code'] = code
t = Template(yearly_site_cat_template)
category.text = t.render(**yearly_site_vars)
category.save("{0} {2} {1}: site category creation".format(TAG, YEAR, COUNTRY))
category = pb.Category(commons_site, yearly_site_category)
if not category.exists() :
yearly_site_vars['lic_name'] = row['name']
yearly_site_vars['lic_code'] = code
t = Template(yearly_site_cat_template)
category.text = t.render(**yearly_site_vars)
category.save("{0} {2} {1}: site category creation".format(TAG, YEAR, COUNTRY))
category = pb.Category(commons_site, site_category)
if not category.exists() :
autcom_vars['aut_com'] = row["community_name"]
t = Template(autcom_cat_template)
category.text = t.render(**autcom_vars)
category.save("{0} {2} {1}: autonomous community category creation".format(TAG, YEAR, COUNTRY))
elif code.startswith('ES'):
print ("Code not from a LIC")
cleaned_hidden_cats.append("Images from {0} {1} in {2} with a wrong code".format(TAG_EXT, YEAR, COUNTRY))
else :
print ("Invalid code")
cleaned_hidden_cats.append("Images from {0} {1} in {2} without code".format(TAG_EXT, YEAR, COUNTRY))
if len(set(cat_titles) ^ set(visible_cats + cleaned_hidden_cats)) > 0:
print (set(cat_titles) ^ set(cleaned_visible_cats + cleaned_hidden_cats))
cat_text = '\n'.join(['[[Category:{0}]]'.format(cat) for cat in set(visible_cats+cleaned_hidden_cats) if cat not in implicit_cats])
page_text = nocat_text + '\n' + cat_text
page.text = page_text
#page.save("{1} {0} in {2}: Classification".format(YEAR, TAG, COUNTRY))
for i in set(lost_cats) :
print (i)
| 0.319121 | 0.339882 |
# Model Deployment to SageMaker Hosted Inference Endpoint
This notebook deploys iso20022 pacs.008 model trained in previous notebook, [pacs008_automl_model_training.ipynb](./pacs008_automl_model_training.ipynb) to predict if a pacs.008 XML message will be successfuly processed (Success) or fail proccessing (Failure) leading to exception processing. It uses Amazon SageMaker hosting to deploy an Inference Endpoint that can be called as REST api by iso20022 pacs.008 message processors in real time and take actions, if needed, early in the payment processing flow.
The diagram below shows the places in the cross-border payment message flow where a call to ML inference endpoint can be injected to get inference from the ML model. The inference result can be used to take additional actions, including corrective actions before sending the message downstream.

### Amazon SageMaker Inference Endpoint
The SageMaker Inference Endpoint name after the endpoint has been created is stored in a notebook store magic variable to make it easy to test. If the Inference Endoint was created outside of this notebook then you have to obtain the inference endpoint name from SageMaker console or via aws cli.
### Request Payload
To test inference endpoint, you need to know the algorithm-specific format of the model artifacts that were generated by model training. For more information about output formats supported by SageMaker algorithms, see the section corresponding to the algorithm you are using in [Common Data Formats for Training](https://docs.aws.amazon.com/sagemaker/latest/dg/cdf-training.html).
Supervised learning algorithms generally expect input data during inference to be in CSV or JSON format. See [Common Data Formats for Inference](https://docs.aws.amazon.com/sagemaker/latest/dg/cdf-inference.html) documentation for more details on inference request payload formats.
The request payload **must** have values for features in the **same order** as they were during model training. Also note that the input payload **must** not contain target variable as that is what the model will predict based on input data.
To learn about order of features examine the features used during data preparation and pre-processing stage to create training data set. For the prototype example here, the order of payload values must be the full features in the labeled raw dataset which was created from `pacs.008 XML message`.
You can examine the training dataset to confirm that the order of features in it. You can also examine [00_gen_synthetic_dataset.ipynb](../synthetic-data/00_gen_synthetic_dataset.ipynb) notebook to see features in raw labeled dataset that was used in training.
### Further Reading
See SageMaker documentation for information on [Real-time Inference](https://docs.aws.amazon.com/sagemaker/latest/dg/realtime-endpoints.html).
It is strongly recommended that you review [Best Practices for Deploying Models on SageMaker Hosting Services](https://docs.aws.amazon.com/sagemaker/latest/dg/how-it-works-hosting-related-considerations.html) for additional information.
# Deploying Model
```
import boto3
import json
import numpy as np
import pandas as pd
import os
import sagemaker
from sagemaker import get_execution_role
from time import sleep, gmtime, strftime
import time
role = get_execution_role()
sess = sagemaker.Session()
region = boto3.session.Session().region_name
sm = boto3.Session().client('sagemaker')
print ("Executing in region {} with role {}".format (region, role))
# retrieve stored variables from previous notebook
%store -r
training_job_name
sm_client = boto3.client('sagemaker')
training_job_description = sm_client.describe_training_job(TrainingJobName=training_job_name)
model_data = training_job_description['ModelArtifacts']['S3ModelArtifacts']
container_uri = training_job_description['AlgorithmSpecification']['TrainingImage']
sm_client.describe_training_job(TrainingJobName=training_job_name)
# model_name was restored from %store -r
model_name
# Create a endpoint config based on the previous model
endpoint_config = sm_client.create_endpoint_config(
EndpointConfigName="{}-config".format(model_name),
ProductionVariants=[
{
'VariantName': "{}-variant".format(model_name),
'ModelName': model_name,
'InitialInstanceCount': 1,
'InstanceType': 'ml.m5.xlarge',
'InitialVariantWeight': 1.0,
}
]
)
endpoint = sm_client.create_endpoint(
EndpointName="{}-endpoint".format(model_name),
EndpointConfigName="{}-config".format(model_name)
)
from time import sleep
endpoint_name = "{}-endpoint".format(model_name)
status = sm_client.describe_endpoint(EndpointName=endpoint_name)['EndpointStatus']
print(status)
while status == 'Creating':
sleep (60)
status = sm_client.describe_endpoint(EndpointName=endpoint_name)['EndpointStatus']
print (status)
```
#### Store Endpoint Name
```
%store endpoint_name
```
## Test Inference Endpoint
**NOTE**
If inference endpoint has been created, you can test it here without executing any of the previous cells. The endpoint name is stored in the magic store.
See SageMaker documentation on [testing interence endpoints](https://docs.aws.amazon.com/sagemaker/latest/dg/realtime-endpoints-test-endpoints.html).
### Restore Inference Endpoint Name
```
%store -r
print(endpoint_name)
```
### Make Prediction by Calling Inference Endpoint
```
runtime_client = boto3.client('sagemaker-runtime')
endpoint_name = 'pacs008-automl-2021-11-25-17-00-endpoint'
# failure transaction
failed_input = 'urn:iso:std:iso:20022:tech:xsd:head.001.001.02,ZHKQUSQQ,WELUIN4E,F8YrsBmbaTqOsPCj,pacs.008.001.08,swift.cbprplus.01,2021-11-15 03:11:41.584,urn:iso:std:iso:20022:tech:xsd:pacs.008.001.08,F8YrsBmbaTqOsPCj,2021-11-15 03:11:41.584,1.0,INDA,HydxEjO7izKjNEV4,O4MhN69232MeToG2,6df7e54c-bb3b-4535-acb7-97d6d947dee5,HIGH,PENS,USD,7952193915.0,2021-11-14 05:00:00.000,USD,7952193905.0,CRED,USD,8738055655.0,ZHKQUSQQ,HLXSUSDM,ZHKQUSQQ,WELUIN4E,XQIBINGE,Thomas Jefferson University Master Trust - Investment Grade Long Bond Account,Scott Building,19107,Philadelphia,US,KSQUUSDB,FYOAINJS,TEKION INDIA PRIVATE LIMITED,"No 680, Fortuna 1 Building,8th Main Road, 15 Cross J P Nagar, 2nd Phas",560078,Bangalore,IN,6723847BB,INR,TEKION INDIA PRIVATE LIMITED,EMAL,webmaster-services-peter-crazy-but-oh-so-ubber-cool-english-alphabet-loverer-abcdefghijklmnopqrstuvwxyz@please-try-to.send-me-an-email-if-you-can-possibly-begin-to-remember-this-coz.this-is-the-longest-email-address-known-to-man-but-to-be-honest.this-is-such-a-stupidly-long-sub-domain-it-could-go-on-forever.pacraig.com,PHOB,Please call the creditor as soon as funds are credited to the account.The phone number is 4234421443 or 324979347. Leave a message.,/REG/13.P1301,COMC,CRED,Reserve Bank of India,IN,Export Reporting,2021-11-14 00:00:00,IN,13.P1301,USD,7952193905.0'
resp = runtime_client.invoke_endpoint(
EndpointName=endpoint_name,
Body=str.encode(failed_input),
ContentType='text/csv',
Accept='text/csv'
)
print(resp['Body'].read())
# successful transaction
successful_input = 'urn:iso:std:iso:20022:tech:xsd:head.001.001.02,GQTCUS19,OZRHTHO5,P89PDKtOJh0xRbNf,pacs.008.001.08,swift.cbprplus.01,2021-11-15 03:15:58.886,urn:iso:std:iso:20022:tech:xsd:pacs.008.001.08,P89PDKtOJh0xRbNf,2021-11-15 03:15:58.886,1.0,INDA,zQGr1MRgKAa25Vag,pcVAPp0dP3Z002xX,3c29c71e-4711-49a4-a664-e58b9b9d86e3,NORM,PENS,USD,9620483936.0,2021-11-14 05:00:00.000,USD,9620483868.0,CRED,USD,9279977441.0,GQTCUS19,HUMMUSAD,GQTCUS19,OZRHTHO5,DIPNTHGR,"AB BOND FUND, INC. - AB Bond Inflation Strategy",c/o The Corporation Trust Incorporated,21093-2252,Lutherville Timonium,US,WERGUSVB,HKTUTH7P,BARCLAYS CAPITAL (THAILAND) LIMITED,989 SAIM TOWER BUILDING FL.14,10330,PATHUM WAN,TH,6723847BB,THB,BARCLAYS CAPITAL (THAILAND) LIMITED,EMAL,webmaster-services-peter-crazy-but-oh-so-ubber-cool-english-alphabet-loverer-abcdefghijklmnopqrstuvwxyz@please-try-to.send-me-an-email-if-you-can-possibly-begin-to-remember-this-coz.this-is-the-longest-email-address-known-to-man-but-to-be-honest.this-is-such-a-stupidly-long-sub-domain-it-could-go-on-forever.pacraig.com,PHOB,Please call the creditor as soon as funds are credited to the account.The phone number is 4234421443 or 324979347. Leave a message.,,COMC,,,,,,,,,'
resp = runtime_client.invoke_endpoint(
EndpointName=endpoint_name,
Body=str.encode(successful_input),
ContentType='text/csv',
Accept='text/csv'
)
print(resp['Body'].read())
# successful transaction
successful_input = 'urn:iso:std:iso:20022:tech:xsd:head.001.001.02,TVFVGBQO,HKTUTH7P,lXZYU4dlh7BgtV9Y,pacs.008.001.08,swift.cbprplus.01,2021-11-15 03:15:38.791,urn:iso:std:iso:20022:tech:xsd:pacs.008.001.08,lXZYU4dlh7BgtV9Y,2021-11-15 03:15:38.791,1.0,INDA,H04QwlcnIvOnDVWk,geZVvAzWiDQnPqip,9687e8e7-73d3-4594-a889-bbbc9e58643b,HIGH,VATX,GBP,6490589583.0,2021-11-14 05:00:00.000,GBP,6490589528.0,CRED,GBP,6887326681.0,TVFVGBQO,WSTQGBGD,TVFVGBQO,HKTUTH7P,WPVLTHYK,POLLARD FAMILY PENSION SCHEME,C/O DCD TRUSTEES LIMITED,EC2A 4PJ,LONDON,GB,ZXLCGBYY,ITZWTHXB,1YM2,"7th-8th Floor, SCB Park Plaza 1",10900,Bangkok,TH,6723847BB,THB,1YM2,EMAL,webmaster-services-peter-crazy-but-oh-so-ubber-cool-english-alphabet-loverer-abcdefghijklmnopqrstuvwxyz@please-try-to.send-me-an-email-if-you-can-possibly-begin-to-remember-this-coz.this-is-the-longest-email-address-known-to-man-but-to-be-honest.this-is-such-a-stupidly-long-sub-domain-it-could-go-on-forever.pacraig.com,PHOB,Please call the creditor as soon as funds are credited to the account.The phone number is 4234421443 or 324979347. Leave a message.,,COMC,,,,,,,,,'
resp = runtime_client.invoke_endpoint(
EndpointName=endpoint_name,
Body=str.encode(successful_input),
ContentType='text/csv',
Accept='text/csv'
)
print(resp['Body'].read())
# another example with failure prediction
failed_input = "urn:iso:std:iso:20022:tech:xsd:head.001.001.02,VGBBCA66,LXFTGBGS,c49gS37XG5Wtc9Ca,pacs.008.001.08,swift.cbprplus.01,2021-11-15 03:41:23.243,urn:iso:std:iso:20022:tech:xsd:pacs.008.001.08,c49gS37XG5Wtc9Ca,2021-11-15 03:41:23.243,1.0,INDA,aTB6f0l7qCqNYqQh,mkgE0j7JcJCSGNMq,561aee1b-ee4f-4e9f-8315-07a7038f0fa4,NORM,ICCP,CAD,3053772770.0,2021-11-14 05:00:00.000,CAD,3053772726.0,SHAR,,,,AYZLCA7F,VGBBCA66,LXFTGBGS,ZJKCGBZ9,8532630 CANADA INC,1801-200 AVENUE DES SOMMETS,H3E 2B4,VERDUN,CA,JCBACAIB,MRNSGBPD,IBM UNITED KINGDOM LIMITED,P.O. BOX 41,PO6 3AU,PORTSMOUTH,GB,84349274229,GBP,IBM UNITED KINGDOM LIMITED,EMAL,webmaster-services-peter-crazy-but-oh-so-ubber-cool-english-alphabet-loverer-abcdefghijklmnopqrstuvwxyz@please-try-to.send-me-an-email-if-you-can-possibly-begin-to-remember-this-coz.this-is-the-longest-email-address-known-to-man-but-to-be-honest.this-is-such-a-stupidly-long-sub-domain-it-could-go-on-forever.pacraig.com,PHOB,Please call the creditor as soon as funds are credited to the account.The phone number is 4234421443 or 324979347. Leave a message.,/SVC/It is to be delivered in one business day. Two day penalty 7bp;greater than two days penalty add 8bp per day.,COMC,,,,,,,,,"
resp = runtime_client.invoke_endpoint(
EndpointName=endpoint_name,
Body=str.encode(failed_input),
ContentType='text/csv',
Accept='text/csv'
)
print(resp['Body'].read())
```
|
github_jupyter
|
import boto3
import json
import numpy as np
import pandas as pd
import os
import sagemaker
from sagemaker import get_execution_role
from time import sleep, gmtime, strftime
import time
role = get_execution_role()
sess = sagemaker.Session()
region = boto3.session.Session().region_name
sm = boto3.Session().client('sagemaker')
print ("Executing in region {} with role {}".format (region, role))
# retrieve stored variables from previous notebook
%store -r
training_job_name
sm_client = boto3.client('sagemaker')
training_job_description = sm_client.describe_training_job(TrainingJobName=training_job_name)
model_data = training_job_description['ModelArtifacts']['S3ModelArtifacts']
container_uri = training_job_description['AlgorithmSpecification']['TrainingImage']
sm_client.describe_training_job(TrainingJobName=training_job_name)
# model_name was restored from %store -r
model_name
# Create a endpoint config based on the previous model
endpoint_config = sm_client.create_endpoint_config(
EndpointConfigName="{}-config".format(model_name),
ProductionVariants=[
{
'VariantName': "{}-variant".format(model_name),
'ModelName': model_name,
'InitialInstanceCount': 1,
'InstanceType': 'ml.m5.xlarge',
'InitialVariantWeight': 1.0,
}
]
)
endpoint = sm_client.create_endpoint(
EndpointName="{}-endpoint".format(model_name),
EndpointConfigName="{}-config".format(model_name)
)
from time import sleep
endpoint_name = "{}-endpoint".format(model_name)
status = sm_client.describe_endpoint(EndpointName=endpoint_name)['EndpointStatus']
print(status)
while status == 'Creating':
sleep (60)
status = sm_client.describe_endpoint(EndpointName=endpoint_name)['EndpointStatus']
print (status)
%store endpoint_name
%store -r
print(endpoint_name)
runtime_client = boto3.client('sagemaker-runtime')
endpoint_name = 'pacs008-automl-2021-11-25-17-00-endpoint'
# failure transaction
failed_input = 'urn:iso:std:iso:20022:tech:xsd:head.001.001.02,ZHKQUSQQ,WELUIN4E,F8YrsBmbaTqOsPCj,pacs.008.001.08,swift.cbprplus.01,2021-11-15 03:11:41.584,urn:iso:std:iso:20022:tech:xsd:pacs.008.001.08,F8YrsBmbaTqOsPCj,2021-11-15 03:11:41.584,1.0,INDA,HydxEjO7izKjNEV4,O4MhN69232MeToG2,6df7e54c-bb3b-4535-acb7-97d6d947dee5,HIGH,PENS,USD,7952193915.0,2021-11-14 05:00:00.000,USD,7952193905.0,CRED,USD,8738055655.0,ZHKQUSQQ,HLXSUSDM,ZHKQUSQQ,WELUIN4E,XQIBINGE,Thomas Jefferson University Master Trust - Investment Grade Long Bond Account,Scott Building,19107,Philadelphia,US,KSQUUSDB,FYOAINJS,TEKION INDIA PRIVATE LIMITED,"No 680, Fortuna 1 Building,8th Main Road, 15 Cross J P Nagar, 2nd Phas",560078,Bangalore,IN,6723847BB,INR,TEKION INDIA PRIVATE LIMITED,EMAL,webmaster-services-peter-crazy-but-oh-so-ubber-cool-english-alphabet-loverer-abcdefghijklmnopqrstuvwxyz@please-try-to.send-me-an-email-if-you-can-possibly-begin-to-remember-this-coz.this-is-the-longest-email-address-known-to-man-but-to-be-honest.this-is-such-a-stupidly-long-sub-domain-it-could-go-on-forever.pacraig.com,PHOB,Please call the creditor as soon as funds are credited to the account.The phone number is 4234421443 or 324979347. Leave a message.,/REG/13.P1301,COMC,CRED,Reserve Bank of India,IN,Export Reporting,2021-11-14 00:00:00,IN,13.P1301,USD,7952193905.0'
resp = runtime_client.invoke_endpoint(
EndpointName=endpoint_name,
Body=str.encode(failed_input),
ContentType='text/csv',
Accept='text/csv'
)
print(resp['Body'].read())
# successful transaction
successful_input = 'urn:iso:std:iso:20022:tech:xsd:head.001.001.02,GQTCUS19,OZRHTHO5,P89PDKtOJh0xRbNf,pacs.008.001.08,swift.cbprplus.01,2021-11-15 03:15:58.886,urn:iso:std:iso:20022:tech:xsd:pacs.008.001.08,P89PDKtOJh0xRbNf,2021-11-15 03:15:58.886,1.0,INDA,zQGr1MRgKAa25Vag,pcVAPp0dP3Z002xX,3c29c71e-4711-49a4-a664-e58b9b9d86e3,NORM,PENS,USD,9620483936.0,2021-11-14 05:00:00.000,USD,9620483868.0,CRED,USD,9279977441.0,GQTCUS19,HUMMUSAD,GQTCUS19,OZRHTHO5,DIPNTHGR,"AB BOND FUND, INC. - AB Bond Inflation Strategy",c/o The Corporation Trust Incorporated,21093-2252,Lutherville Timonium,US,WERGUSVB,HKTUTH7P,BARCLAYS CAPITAL (THAILAND) LIMITED,989 SAIM TOWER BUILDING FL.14,10330,PATHUM WAN,TH,6723847BB,THB,BARCLAYS CAPITAL (THAILAND) LIMITED,EMAL,webmaster-services-peter-crazy-but-oh-so-ubber-cool-english-alphabet-loverer-abcdefghijklmnopqrstuvwxyz@please-try-to.send-me-an-email-if-you-can-possibly-begin-to-remember-this-coz.this-is-the-longest-email-address-known-to-man-but-to-be-honest.this-is-such-a-stupidly-long-sub-domain-it-could-go-on-forever.pacraig.com,PHOB,Please call the creditor as soon as funds are credited to the account.The phone number is 4234421443 or 324979347. Leave a message.,,COMC,,,,,,,,,'
resp = runtime_client.invoke_endpoint(
EndpointName=endpoint_name,
Body=str.encode(successful_input),
ContentType='text/csv',
Accept='text/csv'
)
print(resp['Body'].read())
# successful transaction
successful_input = 'urn:iso:std:iso:20022:tech:xsd:head.001.001.02,TVFVGBQO,HKTUTH7P,lXZYU4dlh7BgtV9Y,pacs.008.001.08,swift.cbprplus.01,2021-11-15 03:15:38.791,urn:iso:std:iso:20022:tech:xsd:pacs.008.001.08,lXZYU4dlh7BgtV9Y,2021-11-15 03:15:38.791,1.0,INDA,H04QwlcnIvOnDVWk,geZVvAzWiDQnPqip,9687e8e7-73d3-4594-a889-bbbc9e58643b,HIGH,VATX,GBP,6490589583.0,2021-11-14 05:00:00.000,GBP,6490589528.0,CRED,GBP,6887326681.0,TVFVGBQO,WSTQGBGD,TVFVGBQO,HKTUTH7P,WPVLTHYK,POLLARD FAMILY PENSION SCHEME,C/O DCD TRUSTEES LIMITED,EC2A 4PJ,LONDON,GB,ZXLCGBYY,ITZWTHXB,1YM2,"7th-8th Floor, SCB Park Plaza 1",10900,Bangkok,TH,6723847BB,THB,1YM2,EMAL,webmaster-services-peter-crazy-but-oh-so-ubber-cool-english-alphabet-loverer-abcdefghijklmnopqrstuvwxyz@please-try-to.send-me-an-email-if-you-can-possibly-begin-to-remember-this-coz.this-is-the-longest-email-address-known-to-man-but-to-be-honest.this-is-such-a-stupidly-long-sub-domain-it-could-go-on-forever.pacraig.com,PHOB,Please call the creditor as soon as funds are credited to the account.The phone number is 4234421443 or 324979347. Leave a message.,,COMC,,,,,,,,,'
resp = runtime_client.invoke_endpoint(
EndpointName=endpoint_name,
Body=str.encode(successful_input),
ContentType='text/csv',
Accept='text/csv'
)
print(resp['Body'].read())
# another example with failure prediction
failed_input = "urn:iso:std:iso:20022:tech:xsd:head.001.001.02,VGBBCA66,LXFTGBGS,c49gS37XG5Wtc9Ca,pacs.008.001.08,swift.cbprplus.01,2021-11-15 03:41:23.243,urn:iso:std:iso:20022:tech:xsd:pacs.008.001.08,c49gS37XG5Wtc9Ca,2021-11-15 03:41:23.243,1.0,INDA,aTB6f0l7qCqNYqQh,mkgE0j7JcJCSGNMq,561aee1b-ee4f-4e9f-8315-07a7038f0fa4,NORM,ICCP,CAD,3053772770.0,2021-11-14 05:00:00.000,CAD,3053772726.0,SHAR,,,,AYZLCA7F,VGBBCA66,LXFTGBGS,ZJKCGBZ9,8532630 CANADA INC,1801-200 AVENUE DES SOMMETS,H3E 2B4,VERDUN,CA,JCBACAIB,MRNSGBPD,IBM UNITED KINGDOM LIMITED,P.O. BOX 41,PO6 3AU,PORTSMOUTH,GB,84349274229,GBP,IBM UNITED KINGDOM LIMITED,EMAL,webmaster-services-peter-crazy-but-oh-so-ubber-cool-english-alphabet-loverer-abcdefghijklmnopqrstuvwxyz@please-try-to.send-me-an-email-if-you-can-possibly-begin-to-remember-this-coz.this-is-the-longest-email-address-known-to-man-but-to-be-honest.this-is-such-a-stupidly-long-sub-domain-it-could-go-on-forever.pacraig.com,PHOB,Please call the creditor as soon as funds are credited to the account.The phone number is 4234421443 or 324979347. Leave a message.,/SVC/It is to be delivered in one business day. Two day penalty 7bp;greater than two days penalty add 8bp per day.,COMC,,,,,,,,,"
resp = runtime_client.invoke_endpoint(
EndpointName=endpoint_name,
Body=str.encode(failed_input),
ContentType='text/csv',
Accept='text/csv'
)
print(resp['Body'].read())
| 0.267121 | 0.898455 |
<a href="https://colab.research.google.com/github/chopstickexe/pandas_exercises/blob/master/03_Grouping/Regiment/Exercises.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Regiment
### Introduction:
Special thanks to: http://chrisalbon.com/ for sharing the dataset and materials.
### Step 1. Import the necessary libraries
```
import pandas as pd
```
### Step 2. Create the DataFrame with the following values:
```
raw_data = {'regiment': ['Nighthawks', 'Nighthawks', 'Nighthawks', 'Nighthawks', 'Dragoons', 'Dragoons', 'Dragoons', 'Dragoons', 'Scouts', 'Scouts', 'Scouts', 'Scouts'],
'company': ['1st', '1st', '2nd', '2nd', '1st', '1st', '2nd', '2nd','1st', '1st', '2nd', '2nd'],
'name': ['Miller', 'Jacobson', 'Ali', 'Milner', 'Cooze', 'Jacon', 'Ryaner', 'Sone', 'Sloan', 'Piger', 'Riani', 'Ali'],
'preTestScore': [4, 24, 31, 2, 3, 4, 24, 31, 2, 3, 2, 3],
'postTestScore': [25, 94, 57, 62, 70, 25, 94, 57, 62, 70, 62, 70]}
```
### Step 3. Assign it to a variable called regiment.
#### Don't forget to name each column
```
regiment = pd.DataFrame(raw_data, columns=raw_data.keys())
regiment.head()
```
### Step 4. What is the mean preTestScore from the regiment Nighthawks?
```
regiment.groupby("regiment")["preTestScore"].mean()
```
### Step 5. Present general statistics by company
```
regiment.groupby("company").describe()
```
### Step 6. What is the mean of each company's preTestScore?
```
regiment.groupby("company")["preTestScore"].mean()
```
### Step 7. Present the mean preTestScores grouped by regiment and company
```
regiment.groupby(["regiment", "company"])["preTestScore"].mean()
```
### Step 8. Present the mean preTestScores grouped by regiment and company without heirarchical indexing
```
regiment.groupby(["regiment", "company"])["preTestScore"].mean().unstack()
# FYI: index and columns of the unstacked dataframe
result = regiment.groupby(["regiment", "company"])["preTestScore"].mean().unstack()
print(result.index)
print(result.columns)
```
### Step 9. Group the entire dataframe by regiment and company
```
regiment.groupby(["regiment", "company"]).mean()
```
### Step 10. What is the number of observations in each regiment and company
```
regiment.groupby(["regiment", "company"]).size()
```
### Step 11. Iterate over a group and print the name and the whole data from the regiment
```
for name, group in regiment.groupby("regiment"):
print(name)
print(group)
```
|
github_jupyter
|
import pandas as pd
raw_data = {'regiment': ['Nighthawks', 'Nighthawks', 'Nighthawks', 'Nighthawks', 'Dragoons', 'Dragoons', 'Dragoons', 'Dragoons', 'Scouts', 'Scouts', 'Scouts', 'Scouts'],
'company': ['1st', '1st', '2nd', '2nd', '1st', '1st', '2nd', '2nd','1st', '1st', '2nd', '2nd'],
'name': ['Miller', 'Jacobson', 'Ali', 'Milner', 'Cooze', 'Jacon', 'Ryaner', 'Sone', 'Sloan', 'Piger', 'Riani', 'Ali'],
'preTestScore': [4, 24, 31, 2, 3, 4, 24, 31, 2, 3, 2, 3],
'postTestScore': [25, 94, 57, 62, 70, 25, 94, 57, 62, 70, 62, 70]}
regiment = pd.DataFrame(raw_data, columns=raw_data.keys())
regiment.head()
regiment.groupby("regiment")["preTestScore"].mean()
regiment.groupby("company").describe()
regiment.groupby("company")["preTestScore"].mean()
regiment.groupby(["regiment", "company"])["preTestScore"].mean()
regiment.groupby(["regiment", "company"])["preTestScore"].mean().unstack()
# FYI: index and columns of the unstacked dataframe
result = regiment.groupby(["regiment", "company"])["preTestScore"].mean().unstack()
print(result.index)
print(result.columns)
regiment.groupby(["regiment", "company"]).mean()
regiment.groupby(["regiment", "company"]).size()
for name, group in regiment.groupby("regiment"):
print(name)
print(group)
| 0.20347 | 0.982356 |
# COROBOT - Chatbot infos corona
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#imports" data-toc-modified-id="imports-1"><span class="toc-item-num">1 </span>imports</a></span><ul class="toc-item"><li><span><a href="#lib" data-toc-modified-id="lib-1.1"><span class="toc-item-num">1.1 </span>lib</a></span></li><li><span><a href="#data" data-toc-modified-id="data-1.2"><span class="toc-item-num">1.2 </span>data</a></span></li></ul></li><li><span><a href="#Prétraitements" data-toc-modified-id="Prétraitements-2"><span class="toc-item-num">2 </span>Prétraitements</a></span><ul class="toc-item"><li><span><a href="#tokenization" data-toc-modified-id="tokenization-2.1"><span class="toc-item-num">2.1 </span>tokenization</a></span></li></ul></li><li><span><a href="#matrice-TF-IDF" data-toc-modified-id="matrice-TF-IDF-3"><span class="toc-item-num">3 </span>matrice TF-IDF</a></span></li><li><span><a href="#Phrases-génériques" data-toc-modified-id="Phrases-génériques-4"><span class="toc-item-num">4 </span>Phrases génériques</a></span><ul class="toc-item"><li><span><a href="#input" data-toc-modified-id="input-4.1"><span class="toc-item-num">4.1 </span>input</a></span></li><li><span><a href="#output" data-toc-modified-id="output-4.2"><span class="toc-item-num">4.2 </span>output</a></span></li></ul></li><li><span><a href="#Chatbot" data-toc-modified-id="Chatbot-5"><span class="toc-item-num">5 </span>Chatbot</a></span></li></ul></div>
## imports
### lib
```
import nltk
import numpy as np
import random
import string # to process standard python strings
import re
from stop_words import get_stop_words
from sklearn.feature_extraction.text import TfidfVectorizer
```
### data
```
f=open('../data/infos_corona.txt','r',errors = 'ignore', encoding = "utf8")
raw=f.read()
type(raw)
```
## Prétraitements
```
raw=raw.lower()# converts to lowercase
raw = re.sub(r"\ufeff", "", raw)
raw = re.sub(r"\[.{1,2}\]", "", raw)
raw = re.sub(r"n.c.a.", "", raw)
raw = re.sub(r"covid.*", "coronavirus", raw)
raw = re.sub(r"\n", " ", raw)
print(raw[0:1000])
```
### tokenization
Création d'une liste de phrases
```
sent_tokens = nltk.sent_tokenize(raw)
i = 1
for n in range(len(sent_tokens)):
a = len(sent_tokens)- i
if (re.search("\?", sent_tokens[a])):
del sent_tokens[a]
i += 1
sent_tokens
```
## matrice TF-IDF
```
french_stop_words = get_stop_words('french')
TfidfVec = TfidfVectorizer(stop_words = french_stop_words)
tfidf = TfidfVec.fit(sent_tokens)
phrases_tf = tfidf.transform(sent_tokens)
phrases_tf = tfidf.transform(sent_tokens)
phrases_tf, tfidf
```
## Phrases génériques
### input
```
in_bonjour = r"salut.*|bonjour.*|coucou.*|cc.*|hi.*|hello.*"
in_good_by = r"au revoir|quit|ciao|hasta la vista|à \+"
```
### output
```
out_bonjour = ["bonjour", "hello", "salut"]
out_neutre = ["ok"]
out_fin = ["merci pour votre visite"]
out_inconnu = ["posez une autre question"]
out_super = ["c'est cool !", "parfait"]
```
## Chatbot
```
flag = True
print("""Bienvenue sur ce bot \n Écrivez votre question : \n
Dites moi au revoir pour quitter""")
while (flag == True):
text_user = input("> ")
text_user = text_user.lower()
if (re.search(q_good_by, text_user)):
print(random.choice(msg_fin))
flag = False
elif (re.fullmatch(q_bonjour,text_user)):
print(random.choice(msg_bot))
# question inconnue
else:
print(random.choice(msg_inconnu))
```
|
github_jupyter
|
import nltk
import numpy as np
import random
import string # to process standard python strings
import re
from stop_words import get_stop_words
from sklearn.feature_extraction.text import TfidfVectorizer
f=open('../data/infos_corona.txt','r',errors = 'ignore', encoding = "utf8")
raw=f.read()
type(raw)
raw=raw.lower()# converts to lowercase
raw = re.sub(r"\ufeff", "", raw)
raw = re.sub(r"\[.{1,2}\]", "", raw)
raw = re.sub(r"n.c.a.", "", raw)
raw = re.sub(r"covid.*", "coronavirus", raw)
raw = re.sub(r"\n", " ", raw)
print(raw[0:1000])
sent_tokens = nltk.sent_tokenize(raw)
i = 1
for n in range(len(sent_tokens)):
a = len(sent_tokens)- i
if (re.search("\?", sent_tokens[a])):
del sent_tokens[a]
i += 1
sent_tokens
french_stop_words = get_stop_words('french')
TfidfVec = TfidfVectorizer(stop_words = french_stop_words)
tfidf = TfidfVec.fit(sent_tokens)
phrases_tf = tfidf.transform(sent_tokens)
phrases_tf = tfidf.transform(sent_tokens)
phrases_tf, tfidf
in_bonjour = r"salut.*|bonjour.*|coucou.*|cc.*|hi.*|hello.*"
in_good_by = r"au revoir|quit|ciao|hasta la vista|à \+"
out_bonjour = ["bonjour", "hello", "salut"]
out_neutre = ["ok"]
out_fin = ["merci pour votre visite"]
out_inconnu = ["posez une autre question"]
out_super = ["c'est cool !", "parfait"]
flag = True
print("""Bienvenue sur ce bot \n Écrivez votre question : \n
Dites moi au revoir pour quitter""")
while (flag == True):
text_user = input("> ")
text_user = text_user.lower()
if (re.search(q_good_by, text_user)):
print(random.choice(msg_fin))
flag = False
elif (re.fullmatch(q_bonjour,text_user)):
print(random.choice(msg_bot))
# question inconnue
else:
print(random.choice(msg_inconnu))
| 0.085072 | 0.898988 |
# Casual Coded Correspondence: The Project
In this project, you will be working to code and decode various messages between you and your fictional cryptography enthusiast pen pal Vishal. You and Vishal have been exchanging letters for quite some time now and have started to provide a puzzle in each one of your letters. Here is his most recent letter:
Hey there! How have you been? I've been great! I just learned about this really cool type of cipher called a Caesar Cipher. Here's how it works: You take your message, something like "hello" and then you shift all of the letters by a certain offset. For example, if I chose an offset of 3 and a message of "hello", I would code my message by shifting each letter 3 places to the left (with respect to the alphabet). So "h" becomes "e", "e" becomes, "b", "l" becomes "i", and "o" becomes "l". Then I have my coded message,"ebiil"! Now I can send you my message and the offset and you can decode it. The best thing is that Julius Caesar himself used this cipher, that's why it's called the Caesar Cipher! Isn't that so cool! Okay, now I'm going to send you a longer coded message that you have to decode yourself!
xuo jxuhu! jxyi yi qd unqcfbu ev q squiqh syfxuh. muhu oek qrbu je tusetu yj? y xefu ie! iudt cu q cuiiqwu rqsa myjx jxu iqcu evviuj!
This message has an offset of 10. Can you decode it?
#### Step 1: Decode Vishal's Message
In the cell below, use your Python skills to decode Vishal's message and print the result. Hint: you can account for shifts that go past the end of the alphabet using the modulus operator, but I'll let you figure out how!
```
vishals_offset = 10
vishals_message = 'xuo jxuhu! jxyi yi qd unqcfbu ev q squiqh syfxuh. muhu oek qrbu je tusetu yj? y xefu ie! iudt cu q cuiiqwu rqsa myjx jxu iqcu evviuj!'
def decoder(message, offset):
alphabet = 'abcdefghijklmnopqrstuvwxyz'
message_decoded = ''
for char in message:
if char in alphabet:
#add decoded character
pos = (alphabet.find(char) + offset) % 26
message_decoded += alphabet[pos]
else:
#add character if not a letter
message_decoded += char
return message_decoded
print(decoder(vishals_message, vishals_offset))
```
#### Step 2: Send Vishal a Coded Message
Great job! Now send Vishal back a message using the same offset. Your message can be anything you want! Remember, coding happens in opposite direction of decoding.
```
my_message = 'hi vishal! i had a bit of difficulty, but i was able to write a python function to decode your message. it was so much fun that i built a function to encode this one!'
my_offset = 5
def coder(message, offset):
alphabet = 'abcdefghijklmnopqrstuvwxyz'
message_encoded = ''
for char in message:
if char in alphabet:
#add decoded character
pos = alphabet.find(char) - offset
message_encoded += alphabet[pos]
else:
#add character
message_encoded += char
return message_encoded
my_encoded_message = coder(my_message, my_offset)
print(my_encoded_message)
print(decoder(my_encoded_message, my_offset))
```
#### Step 3: Make functions for decoding and coding
Vishal sent over another reply, this time with two coded messages!
You're getting the hang of this! Okay here are two more messages, the first one is coded just like before with an offset of ten, and it contains the hint for decoding the second message!
First message:
jxu evviuj veh jxu iusedt cuiiqwu yi vekhjuud.
Second message:
bqdradyuzs ygxfubxq omqemd oubtqde fa oapq kagd yqeemsqe ue qhqz yadq eqogdq!
Decode both of these messages.
If you haven't already, define two functions `decoder(message, offset)` and `coder(message, offset)` that can be used to quickly decode and code messages given any offset.
```
step3_offset = 10
first_message = 'jxu evviuj veh jxu iusedt cuiiqwu yi vekhjuud.'
print(decoder(first_message, step3_offset))
second_message = 'bqdradyuzs ygxfubxq omqemd oubtqde fa oapq kagd yqeemsqe ue qhqz yadq eqogdq!'
print(decoder(second_message, 14))
```
#### Step 4: Solving a Caesar Cipher without knowing the shift value
Awesome work! While you were working to decode his last two messages, Vishal sent over another letter! He's really been bitten by the crytpo-bug. Read it and see what interesting task he has lined up for you this time.
Hello again friend! I knew you would love the Caesar Cipher, it's a cool simple way to encrypt messages. Did you know that back in Caesar's time, it was considered a very secure way of communication and it took a lot of effort to crack if you were unaware of the value of the shift? That's all changed with computers! Now we can brute force these kinds of ciphers very quickly, as I'm sure you can imagine.
To test your cryptography skills, this next coded message is going to be harder than the last couple to crack. It's still going to be coded with a Caesar Cipher but this time I'm not going to tell you the value of the shift. You'll have to brute force it yourself.
Here's the coded message:
vhfinmxkl atox kxgwxkxw tee hy maxlx hew vbiaxkl tl hulhexmx. px'ee atox mh kxteer lmxi ni hnk ztfx by px ptgm mh dxxi hnk fxlltzxl ltyx.
Good luck!
Decode Vishal's most recent message and see what it says!
```
vishals_message = "vhfinmxkl atox kxgwxkxw tee hy maxlx hew vbiaxkl tl hulhexmx. px'ee atox mh kxteer lmxi ni hnk ztfx by px ptgm mh dxxi hnk fxlltzxl ltyx."
for i in range(0, 25):
print(decoder(vishals_message, i))
```
#### Step 5: The Vigenère Cipher
Great work! While you were working on the brute force cracking of the cipher, Vishal sent over another letter. That guy is a letter machine!
Salutations! As you can see, technology has made brute forcing simple ciphers like the Caesar Cipher extremely easy, and us crypto-enthusiasts have had to get more creative and use more complicated ciphers. This next cipher I'm going to teach you is the Vigenère Cipher, invented by an Italian cryptologist named Giovan Battista Bellaso (cool name eh?) in the 16th century, but named after another cryptologist from the 16th century, Blaise de Vigenère.
The Vigenère Cipher is a polyalphabetic substitution cipher, as opposed to the Caesar Cipher which was a monoalphabetic substitution cipher. What this means is that opposed to having a single shift that is applied to every letter, the Vigenère Cipher has a different shift for each individual letter. The value of the shift for each letter is determined by a given keyword.
Consider the message
barry is the spy
If we want to code this message, first we choose a keyword. For this example, we'll use the keyword
dog
Now we use the repeat the keyword over and over to generate a _keyword phrase_ that is the same length as the message we want to code. So if we want to code the message "barry is the spy" our _keyword phrase_ is "dogdo gd ogd ogd". Now we are ready to start coding our message. We shift the each letter of our message by the place value of the corresponding letter in the keyword phrase, assuming that "a" has a place value of 0, "b" has a place value of 1, and so forth. Remember, we zero-index because this is Python we're talking about!
message: b a r r y i s t h e s p y
keyword phrase: d o g d o g d o g d o g d
resulting place value: 4 14 15 12 16 24 11 21 25 22 22 17 5
So we shift "b", which has an index of 1, by the index of "d", which is 3. This gives us an place value of 4, which is "e". Then continue the trend: we shift "a" by the place value of "o", 14, and get "o" again, we shift "r" by the place value of "g", 15, and get "x", shift the next "r" by 12 places and "u", and so forth. Once we complete all the shifts we end up with our coded message:
eoxum ov hnh gvb
As you can imagine, this is a lot harder to crack without knowing the keyword! So now comes the hard part. I'll give you a message and the keyword, and you'll see if you can figure out how to crack it! Ready? Okay here's my message:
dfc aruw fsti gr vjtwhr wznj? vmph otis! cbx swv jipreneo uhllj kpi rahjib eg fjdkwkedhmp!
and the keyword to decode my message is
friends
Because that's what we are! Good luck friend!
And there it is. Vishal has given you quite the assignment this time! Try to decode his message. It may be helpful to create a function that takes two parameters, the coded message and the keyword and then work towards a solution from there.
**NOTE:** Watch out for spaces and punctuation! When there's a space or punctuation mark in the original message, there should be a space/punctuation mark in the corresponding repeated-keyword string as well!
```
vishals_message = 'dfc aruw fsti gr vjtwhr wznj? vmph otis! cbx swv jipreneo uhllj kpi rahjib eg fjdkwkedhmp!'
vishals_keyword = 'friends'
def vignere_decoder(message, keyword):
alphabet = 'abcdefghijklmnopqrstuvwxyz'
result = ''
counter = 0
keyword_pos = 0
keyword_length = len(keyword)
for char in message:
if char in alphabet:
#get decoded character position
pos = (alphabet.find(char) - alphabet.find(keyword[keyword_pos]))
result += alphabet[pos]
#increment counter and move to next keyword index
counter += 1
keyword_pos = counter % keyword_length
else:
#add character
result += char
return result
print(vignere_decoder(vishals_message, vishals_keyword))
```
#### Step 6: Send a message with the Vigenère Cipher
Great work decoding the message. For your final task, write a function that can encode a message using a given keyword and write out a message to send to Vishal!
*As a bonus, try calling your decoder function on the result of your encryption function. You should get the original message back!*
```
my_message = 'vishal, that was quite a challenge! my brain is tingling!'
my_keyword = 'kittens'
def vignere_coder(message, keyword):
alphabet = 'abcdefghijklmnopqrstuvwxyz'
result = ''
counter = 0
keyword_pos = 0
keyword_length = len(keyword)
for char in message:
if char in alphabet:
#get decoded character position
pos = (alphabet.find(char) + alphabet.find(keyword[keyword_pos])) % 26
result += alphabet[pos]
#increment counter and move to next keyword index
counter += 1
keyword_pos = counter % keyword_length
else:
#add character
result += char
return result
my_message_coded = vignere_coder(my_message, my_keyword)
print(my_message_coded)
print(vignere_decoder(my_message_coded, my_keyword))
```
#### Conclusion
Over the course of this project you've learned about two different cipher methods and have used your Python skills to code and decode messages. There are all types of other facinating ciphers out there to explore, and Python is the perfect language to implement them with, so go exploring!
|
github_jupyter
|
vishals_offset = 10
vishals_message = 'xuo jxuhu! jxyi yi qd unqcfbu ev q squiqh syfxuh. muhu oek qrbu je tusetu yj? y xefu ie! iudt cu q cuiiqwu rqsa myjx jxu iqcu evviuj!'
def decoder(message, offset):
alphabet = 'abcdefghijklmnopqrstuvwxyz'
message_decoded = ''
for char in message:
if char in alphabet:
#add decoded character
pos = (alphabet.find(char) + offset) % 26
message_decoded += alphabet[pos]
else:
#add character if not a letter
message_decoded += char
return message_decoded
print(decoder(vishals_message, vishals_offset))
my_message = 'hi vishal! i had a bit of difficulty, but i was able to write a python function to decode your message. it was so much fun that i built a function to encode this one!'
my_offset = 5
def coder(message, offset):
alphabet = 'abcdefghijklmnopqrstuvwxyz'
message_encoded = ''
for char in message:
if char in alphabet:
#add decoded character
pos = alphabet.find(char) - offset
message_encoded += alphabet[pos]
else:
#add character
message_encoded += char
return message_encoded
my_encoded_message = coder(my_message, my_offset)
print(my_encoded_message)
print(decoder(my_encoded_message, my_offset))
step3_offset = 10
first_message = 'jxu evviuj veh jxu iusedt cuiiqwu yi vekhjuud.'
print(decoder(first_message, step3_offset))
second_message = 'bqdradyuzs ygxfubxq omqemd oubtqde fa oapq kagd yqeemsqe ue qhqz yadq eqogdq!'
print(decoder(second_message, 14))
vishals_message = "vhfinmxkl atox kxgwxkxw tee hy maxlx hew vbiaxkl tl hulhexmx. px'ee atox mh kxteer lmxi ni hnk ztfx by px ptgm mh dxxi hnk fxlltzxl ltyx."
for i in range(0, 25):
print(decoder(vishals_message, i))
vishals_message = 'dfc aruw fsti gr vjtwhr wznj? vmph otis! cbx swv jipreneo uhllj kpi rahjib eg fjdkwkedhmp!'
vishals_keyword = 'friends'
def vignere_decoder(message, keyword):
alphabet = 'abcdefghijklmnopqrstuvwxyz'
result = ''
counter = 0
keyword_pos = 0
keyword_length = len(keyword)
for char in message:
if char in alphabet:
#get decoded character position
pos = (alphabet.find(char) - alphabet.find(keyword[keyword_pos]))
result += alphabet[pos]
#increment counter and move to next keyword index
counter += 1
keyword_pos = counter % keyword_length
else:
#add character
result += char
return result
print(vignere_decoder(vishals_message, vishals_keyword))
my_message = 'vishal, that was quite a challenge! my brain is tingling!'
my_keyword = 'kittens'
def vignere_coder(message, keyword):
alphabet = 'abcdefghijklmnopqrstuvwxyz'
result = ''
counter = 0
keyword_pos = 0
keyword_length = len(keyword)
for char in message:
if char in alphabet:
#get decoded character position
pos = (alphabet.find(char) + alphabet.find(keyword[keyword_pos])) % 26
result += alphabet[pos]
#increment counter and move to next keyword index
counter += 1
keyword_pos = counter % keyword_length
else:
#add character
result += char
return result
my_message_coded = vignere_coder(my_message, my_keyword)
print(my_message_coded)
print(vignere_decoder(my_message_coded, my_keyword))
| 0.264928 | 0.795102 |
Description
---------
This is a test of SimulatedAnnealing.
Basics
------
```
import sys
sys.path.append('../sample/')
from simulated_annealing import Temperature, SimulatedAnnealing
from random import uniform, gauss
import numpy as np
import matplotlib.pyplot as plt
```
### Set Parameters
`State` in MetropolisSampler is `np.array([float])`.
**The smaller the `re_scaling` be, the faster it anneals.**
```
def temperature_of_time(t, re_scaling, max_temperature):
""" int * int -> float
"""
return max_temperature / (1 + np.exp(t / re_scaling))
def initialize_state():
""" None -> [float]
"""
return np.array([uniform(-10, 10) for i in range(dim)])
def markov_process(x, step_length):
""" [float] -> [float]
"""
result = x.copy()
for i, item in enumerate(result):
result[i] = item + gauss(0, 1) * step_length
return result
```
### Create SimulatedAnnealing Object
```
def get_sa(dim, iterations, re_scaling, max_temperature, step_length):
sa = SimulatedAnnealing(
lambda i: temperature_of_time(i, re_scaling, max_temperature),
iterations, initialize_state,
lambda x: markov_process(x, step_length)
)
return sa
```
### Target Function on Arbitrary Dimension
```
def N(mu, sigma):
""" float * float -> ([float] -> float)
"""
return lambda x: np.exp(- np.sum(np.square((x - mu) / sigma)))
## Recall SimulatedAnnealing is searching the argmin, instead of argmax.
def target_function(x):
""" [float] -> float
"""
return -1 * (N(0, 5)(x) + 100 * N(10, 3)(x))
```
Test on 1-Dim
---------
```
dim = 1
## Needs tuning
iterations = int(10 ** 3)
re_scaling = int(iterations / 10)
max_temperature = 1000
step_length = 1
sa = get_sa(dim, iterations, re_scaling, max_temperature, step_length)
```
Get argmin
```
argmin = sa(target_function)
print('argmin = {0}'.format(argmin))
print('target(argmin) = {0}, which shall be about -100'.format(target_function(argmin)))
```
Plot the MCMC
```
def t(x):
return np.log(-1 * target_function(x))
step_list = np.arange(len(sa.chain))
t_lst = [t(_) for _ in sa.chain]
plt.plot(step_list, t_lst)
plt.xlabel('step')
plt.ylabel('log(-1 * value of target function)')
plt.show()
for i in range(dim):
x_lst = [_[i] for _ in sa.chain]
plt.plot(step_list, x_lst)
plt.xlabel('step')
plt.ylabel('x[{0}]'.format(i))
plt.show()
```
### Conclusion
Splendid.
Test on 2-Dim
---------
```
dim = 2
## Needs tuning
iterations = int(10 ** 3)
re_scaling = int(iterations / 10)
max_temperature = 1000
step_length = 1
sa = get_sa(dim, iterations, re_scaling, max_temperature, step_length)
```
Get argmin
```
argmin = sa(target_function)
print('argmin = {0}'.format(argmin))
print('target(argmin) = {0}, which shall be about -100'.format(target_function(argmin)))
```
Plot the MCMC
```
def t(x):
return np.log(-1 * target_function(x))
step_list = np.arange(len(sa.chain))
t_lst = [t(_) for _ in sa.chain]
plt.plot(step_list, t_lst)
plt.xlabel('step')
plt.ylabel('log(-1 * value of target function)')
plt.show()
for i in range(dim):
x_lst = [_[i] for _ in sa.chain]
plt.plot(step_list, x_lst)
plt.xlabel('step')
plt.ylabel('x[{0}]'.format(i))
plt.show()
```
### Conclusion
Splendid.
Test on 4-Dim
---------
```
dim = 4
## Needs tuning
iterations = int(10 ** 6)
re_scaling = int(iterations / 100)
max_temperature = 1000
step_length = 3
sa = get_sa(dim, iterations, re_scaling, max_temperature, step_length)
```
Get argmin
```
argmin = sa(target_function)
print('argmin = {0}'.format(argmin))
print('target(argmin) = {0}, which shall be about -100'.format(target_function(argmin)))
p = np.argmin([target_function(_) for _ in sa.chain])
argmin = sa.chain[p]
print(argmin)
```
Plot the MCMC
```
def t(x):
return np.log(-1 * target_function(x))
step_list = np.arange(len(sa.chain))
t_lst = [t(_) for _ in sa.chain]
plt.plot(step_list, t_lst)
plt.xlabel('step')
plt.ylabel('log(-1 * value of target function)')
plt.show()
for i in range(dim):
x_lst = [_[i] for _ in sa.chain]
plt.plot(step_list, x_lst)
plt.xlabel('step')
plt.ylabel('x[{0}]'.format(i))
plt.show()
for axis_to_plot in range(dim):
x_lst = [_[axis_to_plot] for _ in sa.chain]
plt.plot(x_lst, t_lst)
plt.xlabel('x[{0}]'.format(axis_to_plot))
plt.ylabel('log(-1 * value of target function)')
plt.show()
```
### Conclusion
As the dimension increases, the accept-ratio also increaes. That is, for a random move in the Markov process, the new value of target function is almost always greater than that of the temporal. So, we wonder **why the greater dimenson triggers the greater value of the new value of target function in the random move?**
The reason of so is that a random move has more probability of making `sum([x[i] for i in range(dim)])` invariant as the `dim` increases.
|
github_jupyter
|
import sys
sys.path.append('../sample/')
from simulated_annealing import Temperature, SimulatedAnnealing
from random import uniform, gauss
import numpy as np
import matplotlib.pyplot as plt
def temperature_of_time(t, re_scaling, max_temperature):
""" int * int -> float
"""
return max_temperature / (1 + np.exp(t / re_scaling))
def initialize_state():
""" None -> [float]
"""
return np.array([uniform(-10, 10) for i in range(dim)])
def markov_process(x, step_length):
""" [float] -> [float]
"""
result = x.copy()
for i, item in enumerate(result):
result[i] = item + gauss(0, 1) * step_length
return result
def get_sa(dim, iterations, re_scaling, max_temperature, step_length):
sa = SimulatedAnnealing(
lambda i: temperature_of_time(i, re_scaling, max_temperature),
iterations, initialize_state,
lambda x: markov_process(x, step_length)
)
return sa
def N(mu, sigma):
""" float * float -> ([float] -> float)
"""
return lambda x: np.exp(- np.sum(np.square((x - mu) / sigma)))
## Recall SimulatedAnnealing is searching the argmin, instead of argmax.
def target_function(x):
""" [float] -> float
"""
return -1 * (N(0, 5)(x) + 100 * N(10, 3)(x))
dim = 1
## Needs tuning
iterations = int(10 ** 3)
re_scaling = int(iterations / 10)
max_temperature = 1000
step_length = 1
sa = get_sa(dim, iterations, re_scaling, max_temperature, step_length)
argmin = sa(target_function)
print('argmin = {0}'.format(argmin))
print('target(argmin) = {0}, which shall be about -100'.format(target_function(argmin)))
def t(x):
return np.log(-1 * target_function(x))
step_list = np.arange(len(sa.chain))
t_lst = [t(_) for _ in sa.chain]
plt.plot(step_list, t_lst)
plt.xlabel('step')
plt.ylabel('log(-1 * value of target function)')
plt.show()
for i in range(dim):
x_lst = [_[i] for _ in sa.chain]
plt.plot(step_list, x_lst)
plt.xlabel('step')
plt.ylabel('x[{0}]'.format(i))
plt.show()
dim = 2
## Needs tuning
iterations = int(10 ** 3)
re_scaling = int(iterations / 10)
max_temperature = 1000
step_length = 1
sa = get_sa(dim, iterations, re_scaling, max_temperature, step_length)
argmin = sa(target_function)
print('argmin = {0}'.format(argmin))
print('target(argmin) = {0}, which shall be about -100'.format(target_function(argmin)))
def t(x):
return np.log(-1 * target_function(x))
step_list = np.arange(len(sa.chain))
t_lst = [t(_) for _ in sa.chain]
plt.plot(step_list, t_lst)
plt.xlabel('step')
plt.ylabel('log(-1 * value of target function)')
plt.show()
for i in range(dim):
x_lst = [_[i] for _ in sa.chain]
plt.plot(step_list, x_lst)
plt.xlabel('step')
plt.ylabel('x[{0}]'.format(i))
plt.show()
dim = 4
## Needs tuning
iterations = int(10 ** 6)
re_scaling = int(iterations / 100)
max_temperature = 1000
step_length = 3
sa = get_sa(dim, iterations, re_scaling, max_temperature, step_length)
argmin = sa(target_function)
print('argmin = {0}'.format(argmin))
print('target(argmin) = {0}, which shall be about -100'.format(target_function(argmin)))
p = np.argmin([target_function(_) for _ in sa.chain])
argmin = sa.chain[p]
print(argmin)
def t(x):
return np.log(-1 * target_function(x))
step_list = np.arange(len(sa.chain))
t_lst = [t(_) for _ in sa.chain]
plt.plot(step_list, t_lst)
plt.xlabel('step')
plt.ylabel('log(-1 * value of target function)')
plt.show()
for i in range(dim):
x_lst = [_[i] for _ in sa.chain]
plt.plot(step_list, x_lst)
plt.xlabel('step')
plt.ylabel('x[{0}]'.format(i))
plt.show()
for axis_to_plot in range(dim):
x_lst = [_[axis_to_plot] for _ in sa.chain]
plt.plot(x_lst, t_lst)
plt.xlabel('x[{0}]'.format(axis_to_plot))
plt.ylabel('log(-1 * value of target function)')
plt.show()
| 0.497559 | 0.910744 |
<a href="https://colab.research.google.com/github/jzsyuan/TaipowerTutorial/blob/main/Codis_Tutorial.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# 網路爬蟲
氣溫網址: http://e-service.cwb.gov.tw/HistoryDataQuery/QueryDataController.do?command=viewMain
工具:
1. URLencode and URLdecode: https://tool.chinaz.com/tools/urlencode.aspx
```
import requests # 爬蟲
import pandas as pd # 資料處理
import re # regex 文字正規化
import urllib # 爬蟲及處理url字串轉ASCII
import time # time sleep
import logging # log
FORMAT = '%(asctime)s | %(thread)d |: [%(levelname)s]: %(message)s' # log format time : thread id | Info/success/error | message
logging.basicConfig(level=logging.INFO,format = FORMAT)
```
# 先看如何得到資料
* 台北市鞍部2021-12-01: http://e-service.cwb.gov.tw/HistoryDataQuery/DayDataController.do?command=viewMain&station=466910&stname=%25E9%259E%258D%25E9%2583%25A8&datepicker=2021-12-01
```
url = 'http://e-service.cwb.gov.tw/HistoryDataQuery/QueryDataController.do?command=viewMain'
r = requests.get(url,verify=False)
print(r)
print(r.text)
r.encoding = 'utf-8'
vlistStr = r.text.split("stList = {")[1].split("areaList = {")[0].replace("\r\n","").replace("\t","").replace(" ","").split("};")[0]
print(vlistStr)
# 使用regex 取出 站號,站名,縣市英文,縣市中文,該站的形態
regex = re.compile('"([0-9a-zA-Z]+?)"\:\["(\w+?)","(\w+?)","(\w+?)","(\w+?)"')
match = regex.findall(vlistStr)
match = [x for x in match if x[4]=="1"]
print(match)
code,cname,name,city,status = match[0][0],match[0][1],match[0][2],match[0][3],match[0][4]
datepicker = '2021-12-07'
headers={'User-agent':'Mozilla/5.0 (Windows NT 6.3; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/94.0.4606.61 Safari/537.36'}
url = 'http://e-service.cwb.gov.tw/HistoryDataQuery/DayDataController.do'
f = {'command': 'viewMain', 'station': code, 'stname': urllib.parse.urlencode({"a": cname})[2:],
'datepicker': datepicker}
url = url + "?" + urllib.parse.urlencode(f)
# 萃取id = MyTable
df=pd.read_html(requests.get(url,headers=headers).text,attrs={'id':'MyTable'},encoding='UTF-8')[0]
def Codis(datepicker,Match):
"""
:param datepicker: "%Y-%m-%d"
:param Match: list of tuple [(code,cname,name,city,status),(code,cname,name,city,status),...]
:return dataframe codis
"""
headers={'User-agent':'Mozilla/5.0 (Windows NT 6.3; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/94.0.4606.61 Safari/537.36'}
for index,(code,cname,name,city,status) in enumerate(Match):
url = 'http://e-service.cwb.gov.tw/HistoryDataQuery/DayDataController.do'
f = {
'command': 'viewMain',
'station': code,
'stname': urllib.parse.urlencode({"a": cname})[2:],
'datepicker': datepicker
}
url = url + "?" + urllib.parse.urlencode(f)
resp =requests.get(url,headers=headers,verify=False)
if resp.status_code == 200:
logging.info(f'{resp.status_code}:|{index}/{len(Match)}|{datepicker}-{city}-{cname}')
df=pd.read_html(resp.text,attrs={'id':'MyTable'},encoding='UTF-8')[0]
else:
logging.error(f'{resp.status_code}:|{index}/{len(Match)}|{datepicker}-{city}-{cname}')
continue
# columns name
# 確認columns 是否為tuple
if isinstance(df.columns[0], tuple):
# 一般狀況
df.columns = [i[2].strip().replace(" ", "_") for i in df]
else:
# 曾經的例外狀況
df = df[2:]
df.columns = [i.strip().replace(" ", "_") for i in df.loc[2]]
df = df[1:]
df.index = range(1, len(df) + 1)
# 插入測站代號、測站名、測站中文名、城市、日期
df.insert(loc=0, column='StationCode', value=code)
df.insert(loc=1, column='StationName', value=name)
df.insert(loc=2, column='StationCName', value=cname)
df.insert(loc=3, column='City', value=city)
df.insert(loc=4, column='Date', value=datepicker)
# 看有沒有dataSet 沒有的話去generate dataframe 欄位為
try:
len(dataSet)
except:
dataSet = pd.DataFrame(columns=df.columns)
dataSet = pd.concat([dataSet, df], axis=0, ignore_index=True)
dataSet = dataSet.fillna("NA")
dataSet = dataSet.replace(("-", "", "...", "X"), "NA")
dataSet.to_csv(f'CODIS_{datepicker}.csv', index=False)
return dataSet
Codis('2021-12-08',Match=match[0:10])
```
# Threading 運用子程序爬蟲
```
from threading import Thread
target = pd.date_range('2021-12-01', '2021-12-11').to_pydatetime().tolist()
target_f =[Date.strftime("%Y-%m-%d") for Date in target]
thread_list = []
for index,date in enumerate(target_f):
thread = Thread(target=Codis,args=(date,match[0:12],))
thread_list.append(thread)
thread_list[index].start()
# show logging
for t in thread_list:
t.join()
```
|
github_jupyter
|
import requests # 爬蟲
import pandas as pd # 資料處理
import re # regex 文字正規化
import urllib # 爬蟲及處理url字串轉ASCII
import time # time sleep
import logging # log
FORMAT = '%(asctime)s | %(thread)d |: [%(levelname)s]: %(message)s' # log format time : thread id | Info/success/error | message
logging.basicConfig(level=logging.INFO,format = FORMAT)
url = 'http://e-service.cwb.gov.tw/HistoryDataQuery/QueryDataController.do?command=viewMain'
r = requests.get(url,verify=False)
print(r)
print(r.text)
r.encoding = 'utf-8'
vlistStr = r.text.split("stList = {")[1].split("areaList = {")[0].replace("\r\n","").replace("\t","").replace(" ","").split("};")[0]
print(vlistStr)
# 使用regex 取出 站號,站名,縣市英文,縣市中文,該站的形態
regex = re.compile('"([0-9a-zA-Z]+?)"\:\["(\w+?)","(\w+?)","(\w+?)","(\w+?)"')
match = regex.findall(vlistStr)
match = [x for x in match if x[4]=="1"]
print(match)
code,cname,name,city,status = match[0][0],match[0][1],match[0][2],match[0][3],match[0][4]
datepicker = '2021-12-07'
headers={'User-agent':'Mozilla/5.0 (Windows NT 6.3; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/94.0.4606.61 Safari/537.36'}
url = 'http://e-service.cwb.gov.tw/HistoryDataQuery/DayDataController.do'
f = {'command': 'viewMain', 'station': code, 'stname': urllib.parse.urlencode({"a": cname})[2:],
'datepicker': datepicker}
url = url + "?" + urllib.parse.urlencode(f)
# 萃取id = MyTable
df=pd.read_html(requests.get(url,headers=headers).text,attrs={'id':'MyTable'},encoding='UTF-8')[0]
def Codis(datepicker,Match):
"""
:param datepicker: "%Y-%m-%d"
:param Match: list of tuple [(code,cname,name,city,status),(code,cname,name,city,status),...]
:return dataframe codis
"""
headers={'User-agent':'Mozilla/5.0 (Windows NT 6.3; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/94.0.4606.61 Safari/537.36'}
for index,(code,cname,name,city,status) in enumerate(Match):
url = 'http://e-service.cwb.gov.tw/HistoryDataQuery/DayDataController.do'
f = {
'command': 'viewMain',
'station': code,
'stname': urllib.parse.urlencode({"a": cname})[2:],
'datepicker': datepicker
}
url = url + "?" + urllib.parse.urlencode(f)
resp =requests.get(url,headers=headers,verify=False)
if resp.status_code == 200:
logging.info(f'{resp.status_code}:|{index}/{len(Match)}|{datepicker}-{city}-{cname}')
df=pd.read_html(resp.text,attrs={'id':'MyTable'},encoding='UTF-8')[0]
else:
logging.error(f'{resp.status_code}:|{index}/{len(Match)}|{datepicker}-{city}-{cname}')
continue
# columns name
# 確認columns 是否為tuple
if isinstance(df.columns[0], tuple):
# 一般狀況
df.columns = [i[2].strip().replace(" ", "_") for i in df]
else:
# 曾經的例外狀況
df = df[2:]
df.columns = [i.strip().replace(" ", "_") for i in df.loc[2]]
df = df[1:]
df.index = range(1, len(df) + 1)
# 插入測站代號、測站名、測站中文名、城市、日期
df.insert(loc=0, column='StationCode', value=code)
df.insert(loc=1, column='StationName', value=name)
df.insert(loc=2, column='StationCName', value=cname)
df.insert(loc=3, column='City', value=city)
df.insert(loc=4, column='Date', value=datepicker)
# 看有沒有dataSet 沒有的話去generate dataframe 欄位為
try:
len(dataSet)
except:
dataSet = pd.DataFrame(columns=df.columns)
dataSet = pd.concat([dataSet, df], axis=0, ignore_index=True)
dataSet = dataSet.fillna("NA")
dataSet = dataSet.replace(("-", "", "...", "X"), "NA")
dataSet.to_csv(f'CODIS_{datepicker}.csv', index=False)
return dataSet
Codis('2021-12-08',Match=match[0:10])
from threading import Thread
target = pd.date_range('2021-12-01', '2021-12-11').to_pydatetime().tolist()
target_f =[Date.strftime("%Y-%m-%d") for Date in target]
thread_list = []
for index,date in enumerate(target_f):
thread = Thread(target=Codis,args=(date,match[0:12],))
thread_list.append(thread)
thread_list[index].start()
# show logging
for t in thread_list:
t.join()
| 0.165829 | 0.662585 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.