text
stringlengths 2.5k
6.39M
| kind
stringclasses 3
values |
|---|---|
```
# -*- coding: utf-8 -*-
# เรียกใช้งานโมดูล
file_name="data"
import codecs
from tqdm import tqdm
from pythainlp.tokenize import word_tokenize
#import deepcut
from pythainlp.tag import pos_tag
from nltk.tokenize import RegexpTokenizer
import glob
import nltk
import re
# thai cut
thaicut="newmm"
from sklearn_crfsuite import scorers,metrics
from sklearn.metrics import make_scorer
from sklearn.model_selection import cross_validate,train_test_split
import sklearn_crfsuite
from pythainlp.corpus.common import thai_stopwords
stopwords = list(thai_stopwords())
#จัดการประโยคซ้ำ
data_not=[]
def Unique(p):
text=re.sub("<[^>]*>","",p)
text=re.sub("\[(.*?)\]","",text)
text=re.sub("\[\/(.*?)\]","",text)
if text not in data_not:
data_not.append(text)
return True
else:
return False
# เตรียมตัวตัด tag ด้วย re
pattern = r'\[(.*?)\](.*?)\[\/(.*?)\]'
tokenizer = RegexpTokenizer(pattern) # ใช้ nltk.tokenize.RegexpTokenizer เพื่อตัด [TIME]8.00[/TIME] ให้เป็น ('TIME','ไง','TIME')
# จัดการกับ tag ที่ไม่ได้ tag
def toolner_to_tag(text):
text=text.strip().replace("FACILITY","LOCATION").replace("[AGO]","").replace("[/AGO]","").replace("[T]","").replace("[/T]","")
text=re.sub("<[^>]*>","",text)
text=re.sub("(\[\/(.*?)\])","\\1***",text)#.replace('(\[(.*?)\])','***\\1')# text.replace('>','>***') # ตัดการกับพวกไม่มี tag word
text=re.sub("(\[\w+\])","***\\1",text)
text2=[]
for i in text.split('***'):
if "[" in i:
text2.append(i)
else:
text2.append("[word]"+i+"[/word]")
text="".join(text2)#re.sub("[word][/word]","","".join(text2))
return text.replace("[word][/word]","")
# แปลง text ให้เป็น conll2002
def text2conll2002(text,pos=True):
"""
ใช้แปลงข้อความให้กลายเป็น conll2002
"""
text=toolner_to_tag(text)
text=text.replace("''",'"')
text=text.replace("’",'"').replace("‘",'"')#.replace('"',"")
tag=tokenizer.tokenize(text)
j=0
conll2002=""
for tagopen,text,tagclose in tag:
word_cut=word_tokenize(text,engine=thaicut) # ใช้ตัวตัดคำ newmm
i=0
txt5=""
while i<len(word_cut):
if word_cut[i]=="''" or word_cut[i]=='"':pass
elif i==0 and tagopen!='word':
txt5+=word_cut[i]
txt5+='\t'+'B-'+tagopen
elif tagopen!='word':
txt5+=word_cut[i]
txt5+='\t'+'I-'+tagopen
else:
txt5+=word_cut[i]
txt5+='\t'+'O'
txt5+='\n'
#j+=1
i+=1
conll2002+=txt5
if pos==False:
return conll2002
return postag(conll2002)
# ใช้สำหรับกำกับ pos tag เพื่อใช้กับ NER
# print(text2conll2002(t,pos=False))
def postag(text):
listtxt=[i for i in text.split('\n') if i!='']
list_word=[]
for data in listtxt:
list_word.append(data.split('\t')[0])
#print(text)
list_word=pos_tag(list_word,engine="perceptron", corpus="orchid_ud")
text=""
i=0
for data in listtxt:
text+=data.split('\t')[0]+'\t'+list_word[i][1]+'\t'+data.split('\t')[1]+'\n'
i+=1
return text
# เขียนไฟล์ข้อมูล conll2002
def write_conll2002(file_name,data):
"""
ใช้สำหรับเขียนไฟล์
"""
with codecs.open(file_name, "w", "utf-8-sig") as temp:
temp.write(data)
return True
# อ่านข้อมูลจากไฟล์
def get_data(fileopen):
"""
สำหรับใช้อ่านทั้งหมดทั้งในไฟล์ทีละรรทัดออกมาเป็น list
"""
with codecs.open(fileopen, 'r',encoding='utf-8-sig') as f:
lines = f.read().splitlines()
return [a for a in tqdm(lines) if Unique(a)] # เอาไม่ซ้ำกัน
def alldata(lists):
text=""
for data in lists:
text+=text2conll2002(data)
text+='\n'
return text
def alldata_list(lists):
data_all=[]
for data in lists:
data_num=[]
try:
txt=text2conll2002(data,pos=True).split('\n')
for d in txt:
tt=d.split('\t')
if d!="":
if len(tt)==3:
data_num.append((tt[0],tt[1],tt[2]))
else:
data_num.append((tt[0],tt[1]))
#print(data_num)
data_all.append(data_num)
except:
print(data)
#print(data_all)
return data_all
def alldata_list_str(lists):
string=""
for data in lists:
string1=""
for j in data:
string1+=j[0]+" "+j[1]+" "+j[2]+"\n"
string1+="\n"
string+=string1
return string
def get_data_tag(listd):
list_all=[]
c=[]
for i in listd:
if i !='':
c.append((i.split("\t")[0],i.split("\t")[1],i.split("\t")[2]))
else:
list_all.append(c)
c=[]
return list_all
def getall(lista):
ll=[]
for i in tqdm(lista):
o=True
for j in ll:
if re.sub("\[(.*?)\]","",i)==re.sub("\[(.*?)\]","",j):
o=False
break
if o==True:
ll.append(i)
return ll
data1=getall(get_data(file_name+".txt"))
print(len(data1))
'''
'''
#del datatofile[0]
datatofile=alldata_list(data1)
tt=[]
#datatofile.reverse()
import random
#random.shuffle(datatofile)
print(len(datatofile))
#training_samples = datatofile[:int(len(datatofile) * 0.8)]
#test_samples = datatofile[int(len(datatofile) * 0.8):]
'''training_samples = datatofile[:2822]
test_samples = datatofile[2822:]'''
#print(test_samples[0])
#tag=TrainChunker(training_samples,test_samples) # Train
#run(training_samples,test_samples)
#import dill
#with open('train.data', 'rb') as file:
# datatofile = dill.load(file)
with open(file_name+"-pos.conll","w") as f:
i=0
while i<len(datatofile):
for j in datatofile[i]:
f.write(j[0]+"\t"+j[1]+"\t"+j[2]+"\n")
if i+1<len(datatofile):
f.write("\n")
i+=1
with open(file_name+".conll","w") as f:
i=0
while i<len(datatofile):
for j in datatofile[i]:
f.write(j[0]+"\t"+j[2]+"\n")
if i+1<len(datatofile):
f.write("\n")
i+=1
def isThai(chr):
cVal = ord(chr)
if(cVal >= 3584 and cVal <= 3711):
return True
return False
def isThaiWord(word):
t=True
for i in word:
l=isThai(i)
if l!=True and i!='.':
t=False
break
return t
def is_stopword(word):
return word in stopwords
def is_s(word):
if word == " " or word =="\t" or word=="":
return True
else:
return False
def lennum(word,num):
if len(word)==num:
return True
return False
def doc2features(doc, i):
word = doc[i][0]
postag = doc[i][1]
# Features from current word
features={
'word.word': word,
'word.stopword': is_stopword(word),
'word.isthai':isThaiWord(word),
'word.isspace':word.isspace(),
'postag':postag,
'word.isdigit()': word.isdigit()
}
if word.isdigit() and len(word)==5:
features['word.islen5']=True
if i > 0:
prevword = doc[i-1][0]
postag1 = doc[i-1][1]
features['word.prevword'] = prevword
features['word.previsspace']=prevword.isspace()
features['word.previsthai']=isThaiWord(prevword)
features['word.prevstopword']=is_stopword(prevword)
features['word.prepostag'] = postag1
features['word.prevwordisdigit'] = prevword.isdigit()
else:
features['BOS'] = True # Special "Beginning of Sequence" tag
# Features from next word
if i < len(doc)-1:
nextword = doc[i+1][0]
postag1 = doc[i+1][1]
features['word.nextword'] = nextword
features['word.nextisspace']=nextword.isspace()
features['word.nextpostag'] = postag1
features['word.nextisthai']=isThaiWord(nextword)
features['word.nextstopword']=is_stopword(nextword)
features['word.nextwordisdigit'] = nextword.isdigit()
else:
features['EOS'] = True # Special "End of Sequence" tag
return features
def extract_features(doc):
return [doc2features(doc, i) for i in range(len(doc))]
def get_labels(doc):
return [tag for (token,postag,tag) in doc]
X_data = [extract_features(doc) for doc in tqdm(datatofile)]
y_data = [get_labels(doc) for doc in tqdm(datatofile)]
X, X_test, y, y_test = train_test_split(X_data, y_data, test_size=0.2)
crf = sklearn_crfsuite.CRF(
algorithm='lbfgs',
c1=0.1,
c2=0.1,
max_iterations=500,
all_possible_transitions=True,
model_filename=file_name+"-pos.model0"
)
crf.fit(X, y);
labels = list(crf.classes_)
labels.remove('O')
y_pred = crf.predict(X_test)
e=metrics.flat_f1_score(y_test, y_pred,
average='weighted', labels=labels)
print(e)
sorted_labels = sorted(
labels,
key=lambda name: (name[1:], name[0])
)
print(metrics.flat_classification_report(
y_test, y_pred, labels=sorted_labels, digits=3
))
#del X_data[0]
#del y_data[0]
!export PYTHONIOENCODING=utf-8
import sklearn_crfsuite
crf2 = sklearn_crfsuite.CRF(
algorithm='lbfgs',
c1=0.1,
c2=0.1,
max_iterations=500,
all_possible_transitions=True,
model_filename=file_name+".model"
)
crf2.fit(X_data, y_data);
import dill
with open("train.data", "wb") as dill_file:
dill.dump(datatofile, dill_file)
# cross_validate
"""
import dill
with open("datatrain.data", "wb") as dill_file:
dill.dump(datatofile, dill_file)
f1_scorer = make_scorer(metrics.flat_f1_score, average='macro')
scores = cross_validate(crf, X, y, scoring=f1_scorer, cv=5)
# save data
print(scores)
"""
```
|
github_jupyter
|
```
import cv2
import numpy as np
import matplotlib.pyplot as plt
import glob
import pandas as pd
import os
def imshow(img):
img = cv2.cvtColor(img,cv2.COLOR_BGR2RGB)
plt.imshow(img)
def get_lane_mask(sample,lane_idx):
points_lane = []
h_max = np.max(data['h_samples'][sample])
h_min = np.min(data['h_samples'][sample])
x_idx = data['lanes'][sample][lane_idx]
y_idx = data['h_samples'][sample]
for x,y in zip(x_idx,y_idx):
offset = (y-h_min)/20
# print(offset)
if x>-100:
points_lane.append([x-offset/2,y])
x_idx_=x_idx.copy()
y_idx_=y_idx.copy()
x_idx_.reverse()
y_idx_.reverse()
for x,y in zip(x_idx_,y_idx_):
offset = (y-h_min)/20
# print(offset)
if x>-100:
points_lane.append([x+offset/2,y])
return points_lane
def create_lane_mask(img_raw,sample):
colors = [[255,0,0],[0,255,0],[0,0,255],[0,255,255]]
laneMask = np.zeros(img_raw.shape, dtype=np.uint8)
for lane_idx in range(len(data.lanes[sample])):
points_lane = get_lane_mask(sample,lane_idx)
if len(points_lane)>0:
pts = np.array(points_lane, np.int32)
pts = pts.reshape((-1,1,2))
laneMask = cv2.fillPoly(laneMask,[pts],colors[lane_idx])
colors = [[255,0,0],[0,255,0],[0,0,255],[0,255,255]]
# create grey-scale label image
label = np.zeros((720,1280),dtype = np.uint8)
for i in range(len(colors)):
label[np.where((laneMask == colors[i]).all(axis = 2))] = i+1
else: continue
return(img_raw, label)
data = pd.read_json(os.path.join(data_dir, 'label_data.json'), lines=True)
data.info()
print(len(data.raw_file))
data
print(len(data.raw_file))
for i in range(len(data.raw_file)):
img_path = data.raw_file[i]
img_path = os.path.join(data_dir,img_path)
print('Reading from: ', img_path)
path_list = img_path.split('/')[:-1]
mask_path_dir = os.path.join(*path_list)
img_raw = cv2.imread(img_path)
img_, mask = create_lane_mask(img_raw,i)
"""
fig = plt.figure(figsize=(15,20))
plt.subplot(211)
imshow(img_raw)
plt.subplot(212)
print(mask.shape)
plt.imshow(mask)
"""
mask_path_dir = mask_path_dir.replace('clips', 'masks')
print('Saving to: ', mask_path_dir)
try:
os.makedirs(mask_path_dir)
except:
pass
for i in range(1, 21):
cv2.imwrite(os.path.join( mask_path_dir, f'{i}.tiff'), mask)
# i = i+1
cv2.imwrite('/Users/srinivas/Projects/Lane_Detection/datasets/LaneDetection/train/masks/0313-1/300/1.tiff', mask)
mask_img = cv2.imread('20.tiff', cv2.IMREAD_GRAYSCALE)
mask_img.shape
plt.imshow(mask_img)
print(np.unique(mask_img))
print(np.unique(mask))
```
|
github_jupyter
|
##### Copyright 2018 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# tf.dataを使って画像をロードする
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/tutorials/load_data/images"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/ja/tutorials/load_data/images.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs-l10n/blob/master/site/ja/tutorials/load_data/images.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
</table>
Note: これらのドキュメントは私たちTensorFlowコミュニティが翻訳したものです。コミュニティによる 翻訳は**ベストエフォート**であるため、この翻訳が正確であることや[英語の公式ドキュメント](https://www.tensorflow.org/?hl=en)の 最新の状態を反映したものであることを保証することはできません。 この翻訳の品質を向上させるためのご意見をお持ちの方は、GitHubリポジトリ[tensorflow/docs](https://github.com/tensorflow/docs)にプルリクエストをお送りください。 コミュニティによる翻訳やレビューに参加していただける方は、 [[email protected] メーリングリスト](https://groups.google.com/a/tensorflow.org/forum/#!forum/docs-ja)にご連絡ください。
このチュートリアルでは、'tf.data' を使って画像データセットをロードする簡単な例を示します。
このチュートリアルで使用するデータセットは、クラスごとに別々のディレクトリに別れた形で配布されています。
## 設定
```
from __future__ import absolute_import, division, print_function, unicode_literals
try:
# Colab only
%tensorflow_version 2.x
except Exception:
pass
import tensorflow as tf
AUTOTUNE = tf.data.experimental.AUTOTUNE
```
## データセットのダウンロードと検査
### 画像の取得
訓練を始める前に、ネットワークに認識すべき新しいクラスを教えるために画像のセットが必要です。最初に使うためのクリエイティブ・コモンズでライセンスされた花の画像のアーカイブを作成してあります。
```
import pathlib
data_root_orig = tf.keras.utils.get_file(origin='https://storage.googleapis.com/download.tensorflow.org/example_images/flower_photos.tgz',
fname='flower_photos', untar=True)
data_root = pathlib.Path(data_root_orig)
print(data_root)
```
218MB をダウンロードすると、花の画像のコピーが使えるようになっているはずです。
```
for item in data_root.iterdir():
print(item)
import random
all_image_paths = list(data_root.glob('*/*'))
all_image_paths = [str(path) for path in all_image_paths]
random.shuffle(all_image_paths)
image_count = len(all_image_paths)
image_count
all_image_paths[:10]
```
### 画像の検査
扱っている画像について知るために、画像のいくつかを見てみましょう。
```
import os
attributions = (data_root/"LICENSE.txt").open(encoding='utf-8').readlines()[4:]
attributions = [line.split(' CC-BY') for line in attributions]
attributions = dict(attributions)
import IPython.display as display
def caption_image(image_path):
image_rel = pathlib.Path(image_path).relative_to(data_root)
return "Image (CC BY 2.0) " + ' - '.join(attributions[str(image_rel)].split(' - ')[:-1])
for n in range(3):
image_path = random.choice(all_image_paths)
display.display(display.Image(image_path))
print(caption_image(image_path))
print()
```
### 各画像のラベルの決定
ラベルを一覧してみます。
```
label_names = sorted(item.name for item in data_root.glob('*/') if item.is_dir())
label_names
```
ラベルにインデックスを割り当てます。
```
label_to_index = dict((name, index) for index,name in enumerate(label_names))
label_to_index
```
ファイルとラベルのインデックスの一覧を作成します。
```
all_image_labels = [label_to_index[pathlib.Path(path).parent.name]
for path in all_image_paths]
print("First 10 labels indices: ", all_image_labels[:10])
```
### 画像の読み込みと整形
TensorFlow には画像を読み込んで処理するために必要なツールが備わっています。
```
img_path = all_image_paths[0]
img_path
```
以下は生のデータです。
```
img_raw = tf.io.read_file(img_path)
print(repr(img_raw)[:100]+"...")
```
画像のテンソルにデコードします。
```
img_tensor = tf.image.decode_image(img_raw)
print(img_tensor.shape)
print(img_tensor.dtype)
```
モデルに合わせてリサイズします。
```
img_final = tf.image.resize(img_tensor, [192, 192])
img_final = img_final/255.0
print(img_final.shape)
print(img_final.numpy().min())
print(img_final.numpy().max())
```
このあと使用するために、簡単な関数にまとめます。
```
def preprocess_image(image):
image = tf.image.decode_jpeg(image, channels=3)
image = tf.image.resize(image, [192, 192])
image /= 255.0 # normalize to [0,1] range
return image
def load_and_preprocess_image(path):
image = tf.io.read_file(path)
return preprocess_image(image)
import matplotlib.pyplot as plt
image_path = all_image_paths[0]
label = all_image_labels[0]
plt.imshow(load_and_preprocess_image(img_path))
plt.grid(False)
plt.xlabel(caption_image(img_path))
plt.title(label_names[label].title())
print()
```
## `tf.data.Dataset`の構築
### 画像のデータセット
`tf.data.Dataset` を構築するもっとも簡単な方法は、`from_tensor_slices` メソッドを使うことです。
文字列の配列をスライスすると、文字列のデータセットが出来上がります。
```
path_ds = tf.data.Dataset.from_tensor_slices(all_image_paths)
```
`shapes` と `types` は、データセット中のそれぞれのアイテムの内容を示しています。この場合には、バイナリ文字列のスカラーのセットです。
```
print(path_ds)
```
`preprocess_image` をファイルパスのデータセットにマップすることで、画像を実行時にロードし整形する新しいデータセットを作成します。
```
image_ds = path_ds.map(load_and_preprocess_image, num_parallel_calls=AUTOTUNE)
import matplotlib.pyplot as plt
plt.figure(figsize=(8,8))
for n,image in enumerate(image_ds.take(4)):
plt.subplot(2,2,n+1)
plt.imshow(image)
plt.grid(False)
plt.xticks([])
plt.yticks([])
plt.xlabel(caption_image(all_image_paths[n]))
plt.show()
```
### `(image, label)`のペアのデータセット
おなじ `from_tensor_slices` メソッドを使ってラベルのデータセットを作ることができます。
```
label_ds = tf.data.Dataset.from_tensor_slices(tf.cast(all_image_labels, tf.int64))
for label in label_ds.take(10):
print(label_names[label.numpy()])
```
これらのデータセットはおなじ順番なので、zip することで `(image, label)` というペアのデータセットができます。
```
image_label_ds = tf.data.Dataset.zip((image_ds, label_ds))
```
新しいデータセットの `shapes` と `types` は、それぞれのフィールドを示すシェイプと型のタプルです。
```
print(image_label_ds)
```
注: `all_image_labels` や `all_image_paths` のような配列がある場合、 `tf.data.dataset.Dataset.zip` メソッドの代わりとなるのは、配列のペアをスライスすることです。
```
ds = tf.data.Dataset.from_tensor_slices((all_image_paths, all_image_labels))
# The tuples are unpacked into the positional arguments of the mapped function
# タプルは展開され、マップ関数の位置引数に割り当てられます
def load_and_preprocess_from_path_label(path, label):
return load_and_preprocess_image(path), label
image_label_ds = ds.map(load_and_preprocess_from_path_label)
image_label_ds
```
### 基本的な訓練手法
このデータセットを使ってモデルの訓練を行うには、データが
* よくシャッフルされ
* バッチ化され
* 限りなく繰り返され
* バッチが出来るだけ早く利用できる
ことが必要です。
これらの特性は `tf.data` APIを使えば簡単に付け加えることができます。
```
BATCH_SIZE = 32
# シャッフルバッファのサイズをデータセットとおなじに設定することで、データが完全にシャッフルされる
# ようにできます。
ds = image_label_ds.shuffle(buffer_size=image_count)
ds = ds.repeat()
ds = ds.batch(BATCH_SIZE)
# `prefetch`を使うことで、モデルの訓練中にバックグラウンドでデータセットがバッチを取得できます。
ds = ds.prefetch(buffer_size=AUTOTUNE)
ds
```
注意すべきことがいくつかあります。
1. 順番が重要です。
* `.repeat` の前に `.shuffle` すると、エポックの境界を越えて要素がシャッフルされます。(ほかの要素がすべて出現する前に2回出現する要素があるかもしれません)
* `.batch` の後に `.shuffle` すると、バッチの順番がシャッフルされますが、要素がバッチを越えてシャッフルされることはありません。
1. 完全なシャッフルのため、 `buffer_size` をデータセットとおなじサイズに設定しています。データセットのサイズ未満の場合、値が大きいほど良くランダム化されますが、より多くのメモリーを使用します。
1. シャッフルバッファがいっぱいになってから要素が取り出されます。そのため、大きな `buffer_size` が `Dataset` を使い始める際の遅延の原因になります。
1. シャッフルされたデータセットは、シャッフルバッファが完全に空になるまでデータセットが終わりであることを伝えません。 `.repeat` によって `Dataset` が再起動されると、シャッフルバッファが一杯になるまでもう一つの待ち時間が発生します。
最後の問題は、 `tf.data.Dataset.apply` メソッドを、融合された `tf.data.experimental.shuffle_and_repeat` 関数と組み合わせることで対処できます。
```
ds = image_label_ds.apply(
tf.data.experimental.shuffle_and_repeat(buffer_size=image_count))
ds = ds.batch(BATCH_SIZE)
ds = ds.prefetch(buffer_size=AUTOTUNE)
ds
```
### データセットをモデルにつなぐ
`tf.keras.applications`からMobileNet v2のコピーを取得します。
これを簡単な転移学習のサンプルに使用します。
MobileNetの重みを訓練不可に設定します。
```
mobile_net = tf.keras.applications.MobileNetV2(input_shape=(192, 192, 3), include_top=False)
mobile_net.trainable=False
```
このモデルは、入力が `[-1,1]` の範囲に正規化されていることを想定しています。
```
help(keras_applications.mobilenet_v2.preprocess_input)
```
<pre>
...
This function applies the "Inception" preprocessing which converts
the RGB values from [0, 255] to [-1, 1]
...
</pre>
このため、データをMobileNetモデルに渡す前に、入力を`[0,1]`の範囲から`[-1,1]`の範囲に変換する必要があります。
```
def change_range(image,label):
return 2*image-1, label
keras_ds = ds.map(change_range)
```
MobileNetは画像ごとに `6x6` の特徴量の空間を返します。
バッチを1つ渡してみましょう。
```
# シャッフルバッファがいっぱいになるまで、データセットは何秒かかかります。
image_batch, label_batch = next(iter(keras_ds))
feature_map_batch = mobile_net(image_batch)
print(feature_map_batch.shape)
```
MobileNet をラップしたモデルを作り、出力層である `tf.keras.layers.Dense` の前に、`tf.keras.layers.GlobalAveragePooling2D` で空間の軸にそって平均値を求めます。
```
model = tf.keras.Sequential([
mobile_net,
tf.keras.layers.GlobalAveragePooling2D(),
tf.keras.layers.Dense(len(label_names))])
```
期待したとおりの形状の出力が得られます。
```
logit_batch = model(image_batch).numpy()
print("min logit:", logit_batch.min())
print("max logit:", logit_batch.max())
print()
print("Shape:", logit_batch.shape)
```
訓練手法を記述するためにモデルをコンパイルします。
```
model.compile(optimizer=tf.keras.optimizers.Adam(),
loss='sparse_categorical_crossentropy',
metrics=["accuracy"])
```
訓練可能な変数は2つ、全結合層の `weights` と `bias` です。
```
len(model.trainable_variables)
model.summary()
```
モデルを訓練します。
普通は、エポックごとの本当のステップ数を指定しますが、ここではデモの目的なので3ステップだけとします。
```
steps_per_epoch=tf.math.ceil(len(all_image_paths)/BATCH_SIZE).numpy()
steps_per_epoch
model.fit(ds, epochs=1, steps_per_epoch=3)
```
## 性能
注:このセクションでは性能の向上に役立ちそうな簡単なトリックをいくつか紹介します。詳しくは、[Input Pipeline Performance](https://www.tensorflow.org/guide/performance/datasets) を参照してください。
上記の単純なパイプラインは、エポックごとにそれぞれのファイルを一つずつ読み込みます。これは、CPU を使ったローカルでの訓練では問題になりませんが、GPU を使った訓練では十分ではなく、いかなる分散訓練でも使うべきではありません。
調査のため、まず、データセットの性能をチェックする簡単な関数を定義します。
```
import time
default_timeit_steps = 2*steps_per_epoch+1
def timeit(ds, steps=default_timeit_steps):
overall_start = time.time()
# Fetch a single batch to prime the pipeline (fill the shuffle buffer),
# before starting the timer
it = iter(ds.take(steps+1))
next(it)
start = time.time()
for i,(images,labels) in enumerate(it):
if i%10 == 0:
print('.',end='')
print()
end = time.time()
duration = end-start
print("{} batches: {} s".format(steps, duration))
print("{:0.5f} Images/s".format(BATCH_SIZE*steps/duration))
print("Total time: {}s".format(end-overall_start))
```
現在のデータセットの性能は次のとおりです。
```
ds = image_label_ds.apply(
tf.data.experimental.shuffle_and_repeat(buffer_size=image_count))
ds = ds.batch(BATCH_SIZE).prefetch(buffer_size=AUTOTUNE)
ds
timeit(ds)
```
### キャッシュ
`tf.data.Dataset.cache` を使うと、エポックを越えて計算結果を簡単にキャッシュできます。特に、データがメモリに収まるときには効果的です。
ここでは、画像が前処理(デコードとリサイズ)された後でキャッシュされます。
```
ds = image_label_ds.cache()
ds = ds.apply(
tf.data.experimental.shuffle_and_repeat(buffer_size=image_count))
ds = ds.batch(BATCH_SIZE).prefetch(buffer_size=AUTOTUNE)
ds
timeit(ds)
```
メモリキャッシュを使う際の欠点のひとつは、実行の都度キャッシュを再構築しなければならないことです。このため、データセットがスタートするたびにおなじだけ起動のための遅延が発生します。
```
timeit(ds)
```
データがメモリに収まらない場合には、キャッシュファイルを使用します。
```
ds = image_label_ds.cache(filename='./cache.tf-data')
ds = ds.apply(
tf.data.experimental.shuffle_and_repeat(buffer_size=image_count))
ds = ds.batch(BATCH_SIZE).prefetch(1)
ds
timeit(ds)
```
キャッシュファイルには、キャッシュを再構築することなくデータセットを再起動できるという利点もあります。2回めがどれほど早いか見てみましょう。
```
timeit(ds)
```
### TFRecord ファイル
#### 生の画像データ
TFRecord ファイルは、バイナリの大きなオブジェクトのシーケンスを保存するための単純なフォーマットです。複数のサンプルをおなじファイルに詰め込むことで、TensorFlow は複数のサンプルを一度に読み込むことができます。これは、特に GCS のようなリモートストレージサービスを使用する際の性能にとって重要です。
最初に、生の画像データから TFRecord ファイルを構築します。
```
image_ds = tf.data.Dataset.from_tensor_slices(all_image_paths).map(tf.io.read_file)
tfrec = tf.data.experimental.TFRecordWriter('images.tfrec')
tfrec.write(image_ds)
```
次に、TFRecord ファイルを読み込み、以前定義した `preprocess_image` 関数を使って画像のデコード/リフォーマットを行うデータセットを構築します。
```
image_ds = tf.data.TFRecordDataset('images.tfrec').map(preprocess_image)
```
これを、前に定義済みのラベルデータセットと zip し、期待どおりの `(image,label)` のペアを得ます。
```
ds = tf.data.Dataset.zip((image_ds, label_ds))
ds = ds.apply(
tf.data.experimental.shuffle_and_repeat(buffer_size=image_count))
ds=ds.batch(BATCH_SIZE).prefetch(AUTOTUNE)
ds
timeit(ds)
```
これは、`cache` バージョンよりも低速です。前処理をキャッシュしていないからです。
#### シリアライズしたテンソル
前処理を TFRecord ファイルに保存するには、前やったように前処理した画像のデータセットを作ります。
```
paths_ds = tf.data.Dataset.from_tensor_slices(all_image_paths)
image_ds = paths_ds.map(load_and_preprocess_image)
image_ds
```
`.jpeg` 文字列のデータセットではなく、これはテンソルのデータセットです。
これを TFRecord ファイルにシリアライズするには、まず、テンソルのデータセットを文字列のデータセットに変換します。
```
ds = image_ds.map(tf.io.serialize_tensor)
ds
tfrec = tf.data.experimental.TFRecordWriter('images.tfrec')
tfrec.write(ds)
```
前処理をキャッシュしたことにより、データは TFRecord ファイルから非常に効率的にロードできます。テンソルを使用する前にデシリアライズすることを忘れないでください。
```
ds = tf.data.TFRecordDataset('images.tfrec')
def parse(x):
result = tf.io.parse_tensor(x, out_type=tf.float32)
result = tf.reshape(result, [192, 192, 3])
return result
ds = ds.map(parse, num_parallel_calls=AUTOTUNE)
ds
```
次にラベルを追加し、以前とおなじような標準的な処理を適用します。
```
ds = tf.data.Dataset.zip((ds, label_ds))
ds = ds.apply(
tf.data.experimental.shuffle_and_repeat(buffer_size=image_count))
ds=ds.batch(BATCH_SIZE).prefetch(AUTOTUNE)
ds
timeit(ds)
```
|
github_jupyter
|
## Moodle Database: Educational Data Log Analysis
The Moodle LMS is a free and open-source learning management system written in PHP and distributed under the GNU General Public License. It is used for blended learning, distance education, flipped classroom and other e-learning projects in schools, universities, workplaces and other sectors. With customizable management features, it is used to create private websites with online courses for educators and trainers to achieve learning goals. Moodle allows for extending and tailoring learning environments using community-sourced plugins .
In this notebokk we are going to explore the 10 Academy Moodle logs stored in the database together with many other relevant tables.
# Table of content
1. Installing the required libraries
2. Importing the required libraries
3. Moodle database understanding
4. Data Extraction Transformation and Loading (ETL)
### Installing the necessary libraries
```
#!pip install ipython-sql
#!pip install sqlalchemy
#!pip install psycopg2
```
### Importing necessary libraries
```
import pandas as pd
import numpy as np
from sqlalchemy import create_engine
import psycopg2
import logging
from IPython.display import display
#allowing connection to the database
%load_ext sql
#ipython-sql
%sql postgresql://bessy:Streetdance53@localhost/moodle
#sqlalchemy
engine = create_engine('postgresql://bessy:Streetdance53@localhost/moodle')
```
### Moodle database Understanding.
Now, let's have a glance of how some of the tables look like.We will consider the following tables;
`mdl_logstore_standard_log`,
`mdl_context`,
`mdl_user`,
`mdl_course`,
`mdl_modules `,
`mdl_course_modules`,
`mdl_course_modules_completion`,
`mdl_grade_items`,
`mdl_grade_grades`,
`mdl_grade_categories`,
`mdl_grade_items_history`,
`mdl_grade_grades_history`,
`mdl_grade_categories_history`,
`mdl_forum`,
`mdl_forum_discussions`,
`mdl_forum_posts`.
`Table:mdl_logstore_standard_log`
```
%%sql
SELECT *FROM mdl_logstore_standard_log LIMIT 3;
```
`Table: mdl_context`
```
%%sql
SELECT * FROM mdl_context LIMIT 3;
```
`mdl_course`
```
%%sql
SELECT * FROM mdl_course LIMIT 3;
```
`mdl_user`
```
%%sql
SELECT * FROM mdl_user LIMIT 3;
```
`mdl_modules`
```
%%sql
SELECT * FROM mdl_modules LIMIT 3;
```
`mdl_course_modules`
```
%%sql
SELECT * FROM mdl_course_modules LIMIT 3;
```
`mdl_course_modules_completion`
```
%%sql
SELECT * FROM mdl_course_modules_completion LIMIT 3
```
`mdl_grade_grades`
```
%%sql
SELECT * FROM mdl_grade_grades LIMIT 3
```
### Number of tables in the database;
```
%%sql
SELECT COUNT(*) FROM information_schema.tables
```
### Number of records in the following tables;
```
mit = ['mdl_logstore_standard_log', 'mdl_context', 'mdl_user', 'mdl_course', 'mdl_modules' , 'mdl_course_modules', 'mdl_course_modules_completion',
'mdl_grade_items', 'mdl_grade_grades', 'mdl_grade_categories', 'mdl_grade_items_history', 'mdl_grade_grades_history',
'mdl_grade_categories_history', 'mdl_forum', 'mdl_forum_discussions', 'mdl_forum_posts']
# fetches and returns number of records of a given table in a moodle database
def table_count(table):
count = %sql SELECT COUNT(*) as {table}_count from {table}
return count
for table in mit:
display(table_count(table))
```
### Number of quiz submission by time
```
%%sql
select date_part('hour', timestamp with time zone 'epoch' + timefinish * interval '1 second') as hour, count(1)
from mdl_quiz_attempts qa
where qa.preview = 0 and qa.timefinish <> 0
group by date_part('hour', timestamp with time zone 'epoch' + timefinish * interval '1 second')
order by hour
%%sql
SELECT COUNT(id), EXTRACT(HOUR FROM to_timestamp(timecreated)) FROM mdl_logstore_standard_log WHERE action ='submitted' AND component='mod_quiz'
group by EXTRACT(HOUR FROM to_timestamp(timecreated));
```
## Monthly usage time of learners who have confirmed and are not deleted
```
%%sql
select extract(month from to_timestamp(mdl_stats_user_monthly.timeend)) as calendar_month,
count(distinct mdl_stats_user_monthly.userid) as total_users
from mdl_stats_user_monthly
inner join mdl_role_assignments on mdl_stats_user_monthly.userid = mdl_role_assignments.userid
inner join mdl_context on mdl_role_assignments.contextid = mdl_context.id
where mdl_stats_user_monthly.stattype = 'activity'
and mdl_stats_user_monthly.courseid <>1
group by extract(month from to_timestamp(mdl_stats_user_monthly.timeend))
order by extract(month from to_timestamp(mdl_stats_user_monthly.timeend))
%%sql
SELECT COUNT(lastaccess - firstaccess) AS usagetime, EXTRACT (MONTH FROM to_timestamp(firstaccess)) AS month
FROM mdl_user WHERE confirmed = 1 AND deleted = 0 GROUP BY EXTRACT (MONTH FROM to_timestamp(firstaccess))
```
## Count of log events per user
```
actions = ['loggedin', 'viewed', 'started', 'submitted', 'uploaded', 'updated', 'searched',
'answered', 'attempted', 'abandoned']
# fetch and return count of log events of an action per user
def event_count(action):
count = %sql SELECT userid, COUNT(action) AS {action}_count FROM mdl_logstore_standard_log WHERE action='{action}' GROUP BY userid limit 5
return count
for action in actions:
display(event_count(action))
```
### python class to pull
* Overall grade of learners
* Number of forum posts
```
class PullGrade():
def __init__(self):
pass
def open_db(self, **kwargs):
# extract args, if they are not provided assign a default value
user = kwargs.get('user', 'briodev')
password = kwargs.get('password', '14ConnectPsq')
db = kwargs.get('db', 'moodle')
# make a connection to PostgreSQL
# use exception to show error message if failed to connect
try:
params = dict(user=user,
password=password,
host="127.0.0.1",
port = "5432",
database = db)
proot = 'postgresql://{user}@{host}:5432/{database}'.format(**params)
logging.info('Connecting to the PostgreSQL database... using sqlalchemy engine')
engine = create_engine(proot)
except (Exception, psycopg2.Error) as error:
logging.error(r"Error while connecting to PostgreSQL {error}")
return engine
# fetch and return number of forum posts
def forum_posts(self):
count = %sql SELECT COUNT(*) from mdl_forum_posts
return count
# fetch and return overall grade of learners
def overall_grade(self):
overall = %sql SELECT userid, round(SUM(finalgrade)/count(*), 2) as overall_grade from mdl_grade_grades WHERE finalgrade is not null group by userid LIMIT 10
return overall
db = PullGrade()
db.open_db()
#Forum_posts
db.forum_posts()
#Overall grade.
db.overall_grade()
```
### Data Extraction Transformation and Loading (ETL)
```
#reading the mdl_logstore_standard_log
log_df = pd.read_sql("select * from mdl_logstore_standard_log", engine)
def top_x(df, percent):
total_len = df.shape[0]
top = int((total_len * percent)/100)
return df.iloc[:top,]
```
### Login count
```
log_df_logged_in = log_df[log_df.action == 'loggedin'][['userid', 'action']]
login_by_user = log_df_logged_in.groupby('userid').count().sort_values('action', ascending=False)
login_by_user.columns = ["login_count"]
top_x(login_by_user, 1)
```
### Activity count
```
activity_log = log_df[['userid', 'action']]
activity_log_by_user = activity_log.groupby('userid').count().sort_values('action', ascending=False)
activity_log_by_user.columns = ['activity_count']
top_x(activity_log_by_user, 1)
log_in_out = log_df[(log_df.action == "loggedin") | (log_df.action == "loggedout")]
user_id = log_df.userid.unique()
d_times = {}
for user in user_id:
log_user = log_df[log_df.userid == user].sort_values('timecreated')
d_time = 0
isLoggedIn = 0
loggedIn_timecreated = 0
for i in range(len(log_user)):
row = log_user.iloc[i,]
row_next = log_user.iloc[i+1,] if i+1 < len(log_user) else row
if(row.action == "loggedin"):
isLoggedIn = 1
loggedIn_timecreated = row.timecreated
if( (i+1 == len(log_user)) | ( (row_next.action == "loggedin") & (isLoggedIn == 1) ) ):
d_time += row.timecreated - loggedIn_timecreated
isLoggedIn = 0
d_times[user] = d_time
dedication_time_df = pd.DataFrame({'userid':list(d_times.keys()),
'dedication_time':list(d_times.values())})
dedication_time_df
top_x(dedication_time_df.sort_values('dedication_time', ascending=False), 35)
```
### References
* https://docs.moodle.org/39/en/Custom_SQL_queries_report
* https://docs.moodle.org/39/en/ad-hoc_contributed_reports
* https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.331.667&rep=rep1&type=pdf
* http://informatics.ue-varna.bg/conference19/Conf.proceedings_Informatics-50.years%20177-187.pdf
|
github_jupyter
|
# Format DataFrame
```
import pandas as pd
from sklearn.datasets import make_regression
data = make_regression(n_samples=600, n_features=50, noise=0.1, random_state=42)
train_df = pd.DataFrame(data[0], columns=["x_{}".format(_) for _ in range(data[0].shape[1])])
train_df["target"] = data[1]
print(train_df.shape)
train_df.head()
```
# Set Up Environment
```
from hyperparameter_hunter import Environment, CVExperiment
from sklearn.metrics import explained_variance_score
env = Environment(
train_dataset=train_df,
results_path="HyperparameterHunterAssets",
metrics=dict(evs=explained_variance_score),
cv_type="KFold",
cv_params=dict(n_splits=3, shuffle=True, random_state=1337),
runs=2,
)
```
Now that HyperparameterHunter has an active `Environment`, we can do two things:
# 1. Perform Experiments
*Note: If this is your first HyperparameterHunter example, the CatBoost classification example may be a better starting point.*
In this Experiment, we're also going to use `model_extra_params` to provide arguments to `CatBoostRegressor`'s `fit` method, just like we would if we weren't using HyperparameterHunter.
We'll be using the `verbose` argument to print evaluations of our `CatBoostRegressor` every 50 iterations, and we'll also be using the dataset sentinels offered by `Environment`. You can read more about the exciting thing you can do with the `Environment` sentinels in the documentation and in the example dedicated to them. For now, though, we'll be using them to provide each fold's `env.validation_input`, and `env.validation_target` to `CatBoostRegressor.fit` via its `eval_set` argument.
You could also easily add `CatBoostRegressor.fit`'s `early_stopping_rounds` argument to `model_extra_params["fit"]` to use early stopping, but doing so here with only `iterations=100` doesn't make much sense.
```
from catboost import CatBoostRegressor
experiment = CVExperiment(
model_initializer=CatBoostRegressor,
model_init_params=dict(
iterations=100,
learning_rate=0.05,
depth=5,
bootstrap_type="Bayesian",
save_snapshot=False,
allow_writing_files=False,
),
model_extra_params=dict(
fit=dict(
verbose=50,
eval_set=[(env.validation_input, env.validation_target)],
),
),
)
```
Notice above that CatBoost printed scores for our `eval_set` every 50 iterations just like we said in `model_extra_params["fit"]`; although, it made our results rather difficult to read, so we'll switch back to `verbose=False` during optimization.
# 2. Hyperparameter Optimization
Notice below that `optimizer` still recognizes the results of `experiment` as valid learning material even though their `verbose` values differ. This is because it knows that `verbose` has no effect on actual results.
```
from hyperparameter_hunter import DummyOptPro, Real, Integer, Categorical
optimizer = DummyOptPro(iterations=10, random_state=777)
optimizer.forge_experiment(
model_initializer=CatBoostRegressor,
model_init_params=dict(
iterations=100,
learning_rate=Real(0.001, 0.2),
depth=Integer(3, 7),
bootstrap_type=Categorical(["Bayesian", "Bernoulli"]),
save_snapshot=False,
allow_writing_files=False,
),
model_extra_params=dict(
fit=dict(
verbose=False,
eval_set=[(env.validation_input, env.validation_target)],
),
),
)
optimizer.go()
```
|
github_jupyter
|
[source](../../api/alibi_detect.od.isolationforest.rst)
# Isolation Forest
## Overview
[Isolation forests](https://cs.nju.edu.cn/zhouzh/zhouzh.files/publication/icdm08b.pdf) (IF) are tree based models specifically used for outlier detection. The IF isolates observations by randomly selecting a feature and then randomly selecting a split value between the maximum and minimum values of the selected feature. The number of splittings required to isolate a sample is equivalent to the path length from the root node to the terminating node. This path length, averaged over a forest of random trees, is a measure of normality and is used to define an anomaly score. Outliers can typically be isolated quicker, leading to shorter paths. The algorithm is suitable for low to medium dimensional tabular data.
## Usage
### Initialize
Parameters:
* `threshold`: threshold value for the outlier score above which the instance is flagged as an outlier.
* `n_estimators`: number of base estimators in the ensemble. Defaults to 100.
* `max_samples`: number of samples to draw from the training data to train each base estimator. If *int*, draw `max_samples` samples. If *float*, draw `max_samples` *times number of features* samples. If *'auto'*, `max_samples` = min(256, number of samples).
* `max_features`: number of features to draw from the training data to train each base estimator. If *int*, draw `max_features` features. If float, draw `max_features` *times number of features* features.
* `bootstrap`: whether to fit individual trees on random subsets of the training data, sampled with replacement.
* `n_jobs`: number of jobs to run in parallel for `fit` and `predict`.
* `data_type`: can specify data type added to metadata. E.g. *'tabular'* or *'image'*.
Initialized outlier detector example:
```python
from alibi_detect.od import IForest
od = IForest(
threshold=0.,
n_estimators=100
)
```
### Fit
We then need to train the outlier detector. The following parameters can be specified:
* `X`: training batch as a numpy array.
* `sample_weight`: array with shape *(batch size,)* used to assign different weights to each instance during training. Defaults to *None*.
```python
od.fit(
X_train
)
```
It is often hard to find a good threshold value. If we have a batch of normal and outlier data and we know approximately the percentage of normal data in the batch, we can infer a suitable threshold:
```python
od.infer_threshold(
X,
threshold_perc=95
)
```
### Detect
We detect outliers by simply calling `predict` on a batch of instances `X` to compute the instance level outlier scores. We can also return the instance level outlier score by setting `return_instance_score` to True.
The prediction takes the form of a dictionary with `meta` and `data` keys. `meta` contains the detector's metadata while `data` is also a dictionary which contains the actual predictions stored in the following keys:
* `is_outlier`: boolean whether instances are above the threshold and therefore outlier instances. The array is of shape *(batch size,)*.
* `instance_score`: contains instance level scores if `return_instance_score` equals True.
```python
preds = od.predict(
X,
return_instance_score=True
)
```
## Examples
### Tabular
[Outlier detection on KDD Cup 99](../../examples/od_if_kddcup.nblink)
|
github_jupyter
|
# Classification models using python and scikit-learn
There are many users of online trading platforms and these companies would like to run analytics on and predict churn based on user activity on the platform. Keeping customers happy so they do not move their investments elsewhere is key to maintaining profitability.
In this notebook, we'll use scikit-learn to predict classes. scikit-learn provides implementations of many classification algorithms. In here, we have chosen the random forest classification algorithm to walk through all the different steps.
<a id="top"></a>
## Table of Contents
1. [Load libraries](#load_libraries)
2. [Data exploration](#explore_data)
3. [Prepare data for building classification model](#prepare_data)
4. [Split data into train and test sets](#split_data)
5. [Helper methods for graph generation](#helper_methods)
6. [Prepare Random Forest classification model](#prepare_model)
7. [Train Random Forest classification model](#train_model)
8. [Test Random Forest classification model](#test_model)
9. [Evaluate Random Forest classification model](#evaluate_model)
10.[Build K-Nearest classification model](#model_knn)
11. [Comparative study of both classification algorithms](#compare_classification)
### Quick set of instructions to work through the notebook
If you are new to Notebooks, here's a quick overview of how to work in this environment.
1. The notebook has 2 types of cells - markdown (text) such as this and code such as the one below.
2. Each cell with code can be executed independently or together (see options under the Cell menu). When working in this notebook, we will be running one cell at a time.
3. To run the cell, position cursor in the code cell and click the Run (arrow) icon. The cell is running when you see the * next to it. Some cells have printable output.
4. Work through this notebook by reading the instructions and executing code cell by cell. Some cells will require modifications before you run them.
<a id="load_libraries"></a>
## 1. Load libraries
[Top](#top)
Install python modules
NOTE! Some pip installs require a kernel restart.
The shell command pip install is used to install Python modules. Some installs require a kernel restart to complete. To avoid confusing errors, run the following cell once and then use the Kernel menu to restart the kernel before proceeding.
```
!pip install pandas==0.24.2
!pip install --user pandas_ml==0.6.1
#downgrade matplotlib to bypass issue with confusion matrix being chopped out
!pip install matplotlib==3.1.0
!pip install --user scikit-learn==0.21.3
!pip install -q scikit-plot
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import StandardScaler
from sklearn.compose import ColumnTransformer, make_column_transformer
from sklearn.pipeline import Pipeline
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, classification_report
import pandas as pd, numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import matplotlib.colors as mcolors
import matplotlib.patches as mpatches
import scikitplot as skplt
```
<a id="explore_data"></a>
## 2. Data exploration
[Top](#top)
In this tutorial, we use a data set that contains information about customers of an online trading platform to classify whether a given customer’s probability of churn will be high, medium, or low. This provides a good example to learn how a classification model is built from start to end.
```
df_churn_pd = pd.read_csv("https://raw.githubusercontent.com/IBM/ml-learning-path-assets/master/data/mergedcustomers_missing_values_GENDER.csv")
df_churn_pd.head()
```
We use numpy and matplotlib to get some statistics and visualize data.
print("The dataset contains columns of the following data types : \n" +str(df_churn_pd.dtypes))
Notice below that Gender has three missing values. This will be handled in one of the preprocessing steps that is to follow.
```
print("The dataset contains following number of records for each of the columns : \n" +str(df_churn_pd.count()))
```
If we are not satisfied with the representational data, now is the time to get more data to be used for training and testing.
```
print( "Each category within the churnrisk column has the following count : ")
print(df_churn_pd.groupby(['CHURNRISK']).size())
#bar chart to show split of data
index = ['High','Medium','Low']
churn_plot = df_churn_pd['CHURNRISK'].value_counts(sort=True, ascending=False).plot(kind='bar',
figsize=(4,4),title="Total number for occurences of churn risk "
+ str(df_churn_pd['CHURNRISK'].count()), color=['#BB6B5A','#8CCB9B','#E5E88B'])
churn_plot.set_xlabel("Churn Risk")
churn_plot.set_ylabel("Frequency")
```
<a id="prepare_data"></a>
## 3. Data preparation
[Top](#top)
Data preparation is a very important step in machine learning model building. This is because the model can perform well only when the data it is trained on is good and well prepared. Hence, this step consumes the bulk of a data scientist's time spent building models.
During this process, we identify categorical columns in the dataset. Categories need to be indexed, which means the string labels are converted to label indices. These label indices are encoded using One-hot encoding to a binary vector with at most a single value indicating the presence of a specific feature value from among the set of all feature values. This encoding allows algorithms which expect continuous features to use categorical features.
```
#remove columns that are not required
df_churn_pd = df_churn_pd.drop(['ID'], axis=1)
df_churn_pd.head()
```
### [Preprocessing Data](https://scikit-learn.org/stable/modules/preprocessing.html)
Scikit-learn provides a method to fill empty values with something that would be applicable in its context. We used the <i><b> SimpleImputer <b></i> class that is provided by Sklearn and filled the missing values with the most frequent value in the column.
### [One Hot Encoder](https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.OneHotEncoder.html)
```
# Defining the categorical columns
categoricalColumns = ['GENDER', 'STATUS', 'HOMEOWNER']
print("Categorical columns : " )
print(categoricalColumns)
impute_categorical = SimpleImputer(strategy="most_frequent")
onehot_categorical = OneHotEncoder(handle_unknown='ignore')
categorical_transformer = Pipeline(steps=[('impute',impute_categorical),('onehot',onehot_categorical)])
```
The numerical columns from the data set are identified, and StandardScaler is applied to each of the columns. This way, each value is subtracted with the mean of its column and divided by its standard deviation.<br>
### [Standard Scaler](https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.StandardScaler.html)
```
# Defining the numerical columns
numericalColumns = df_churn_pd.select_dtypes(include=[np.float,np.int]).columns
print("Numerical columns : " )
print(numericalColumns)
scaler_numerical = StandardScaler()
numerical_transformer = Pipeline(steps=[('scale',scaler_numerical)])
```
The preprocessing techniques that are applied must be customized for each of the columns. Sklearn provides a library called the [ColumnTransformer](https://scikit-learn.org/stable/modules/generated/sklearn.compose.ColumnTransformer.html?highlight=columntransformer#sklearn.compose.ColumnTransformer), which allows a sequence of these techniques to be applied to selective columns using a pipeline.
Only the specified columns in transformers are transformed and combined in the output, and the non-specified columns are dropped. By specifying remainder='passthrough', all remaining columns that were not specified in transformers will be automatically passed through
```
preprocessorForCategoricalColumns = ColumnTransformer(transformers=[('cat', categorical_transformer,
categoricalColumns)],
remainder="passthrough")
preprocessorForAllColumns = ColumnTransformer(transformers=[('cat', categorical_transformer, categoricalColumns),
('num',numerical_transformer,numericalColumns)],
remainder="passthrough")
```
Machine learning algorithms cannot use simple text. We must convert the data from text to a number. Therefore, for each string that is a class we assign a label that is a number. For example, in the customer churn data set, the CHURNRISK output label is classified as high, medium, or low and is assigned labels 0, 1, or 2. We use the [LabelEncoder](https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.LabelEncoder.html?highlight=labelencoder#sklearn.preprocessing.LabelEncoder) class provided by Sklearn for this.
```
# prepare data frame for splitting data into train and test datasets
features = []
features = df_churn_pd.drop(['CHURNRISK'], axis=1)
label_churn = pd.DataFrame(df_churn_pd, columns = ['CHURNRISK'])
label_encoder = LabelEncoder()
label = df_churn_pd['CHURNRISK']
label = label_encoder.fit_transform(label)
print("Encoded value of Churnrisk after applying label encoder : " + str(label))
```
### These are some of the popular preprocessing steps that are applied on the data sets. Look at [Data Processing in detail](https://developer.ibm.com/articles/data-preprocessing-in-detail/) for more information
```
area = 75
x = df_churn_pd['ESTINCOME']
y = df_churn_pd['DAYSSINCELASTTRADE']
z = df_churn_pd['TOTALDOLLARVALUETRADED']
pop_a = mpatches.Patch(color='#BB6B5A', label='High')
pop_b = mpatches.Patch(color='#E5E88B', label='Medium')
pop_c = mpatches.Patch(color='#8CCB9B', label='Low')
def colormap(risk_list):
cols=[]
for l in risk_list:
if l==0:
cols.append('#BB6B5A')
elif l==2:
cols.append('#E5E88B')
elif l==1:
cols.append('#8CCB9B')
return cols
fig = plt.figure(figsize=(12,6))
fig.suptitle('2D and 3D view of churnrisk data')
# First subplot
ax = fig.add_subplot(1, 2,1)
ax.scatter(x, y, alpha=0.8, c=colormap(label), s= area)
ax.set_ylabel('DAYS SINCE LAST TRADE')
ax.set_xlabel('ESTIMATED INCOME')
plt.legend(handles=[pop_a,pop_b,pop_c])
# Second subplot
ax = fig.add_subplot(1,2,2, projection='3d')
ax.scatter(z, x, y, c=colormap(label), marker='o')
ax.set_xlabel('TOTAL DOLLAR VALUE TRADED')
ax.set_ylabel('ESTIMATED INCOME')
ax.set_zlabel('DAYS SINCE LAST TRADE')
plt.legend(handles=[pop_a,pop_b,pop_c])
plt.show()
```
<a id="split_data"></a>
## 4. Split data into test and train
[Top](#top)
Scikit-learn provides in built API to split the original dataset into train and test datasets. random_state is set to a number to be able to reproduce the same data split combination through multiple runs.
[Split arrays or matrices into random train and test subsets](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html)
```
X_train, X_test, y_train, y_test = train_test_split(features,label , random_state=0)
print("Dimensions of datasets that will be used for training : Input features"+str(X_train.shape)+
" Output label" + str(y_train.shape))
print("Dimensions of datasets that will be used for testing : Input features"+str(X_test.shape)+
" Output label" + str(y_test.shape))
```
<a id="helper_methods"></a>
## 5. Helper methods for graph generation
[Top](#top)
```
def colormap(risk_list):
cols=[]
for l in risk_list:
if l==0:
cols.append('#BB6B5A')
elif l==2:
cols.append('#E5E88B')
elif l==1:
cols.append('#8CCB9B')
return cols
def two_d_compare(y_test,y_pred,model_name):
#y_pred = label_encoder.fit_transform(y_pred)
#y_test = label_encoder.fit_transform(y_test)
area = (12 * np.random.rand(40))**2
plt.subplots(ncols=2, figsize=(10,4))
plt.suptitle('Actual vs Predicted data : ' +model_name + '. Accuracy : %.2f' % accuracy_score(y_test, y_pred))
plt.subplot(121)
plt.scatter(X_test['ESTINCOME'], X_test['DAYSSINCELASTTRADE'], alpha=0.8, c=colormap(y_test))
plt.title('Actual')
plt.legend(handles=[pop_a,pop_b,pop_c])
plt.subplot(122)
plt.scatter(X_test['ESTINCOME'], X_test['DAYSSINCELASTTRADE'],alpha=0.8, c=colormap(y_pred))
plt.title('Predicted')
plt.legend(handles=[pop_a,pop_b,pop_c])
plt.show()
x = X_test['TOTALDOLLARVALUETRADED']
y = X_test['ESTINCOME']
z = X_test['DAYSSINCELASTTRADE']
pop_a = mpatches.Patch(color='#BB6B5A', label='High')
pop_b = mpatches.Patch(color='#E5E88B', label='Medium')
pop_c = mpatches.Patch(color='#8CCB9B', label='Low')
def three_d_compare(y_test,y_pred,model_name):
fig = plt.figure(figsize=(12,10))
fig.suptitle('Actual vs Predicted (3D) data : ' +model_name + '. Accuracy : %.2f' % accuracy_score(y_test, y_pred))
ax = fig.add_subplot(121, projection='3d')
ax.scatter(x, y, z, c=colormap(y_test), marker='o')
ax.set_xlabel('TOTAL DOLLAR VALUE TRADED')
ax.set_ylabel('ESTIMATED INCOME')
ax.set_zlabel('DAYS SINCE LAST TRADE')
plt.legend(handles=[pop_a,pop_b,pop_c])
plt.title('Actual')
ax = fig.add_subplot(122, projection='3d')
ax.scatter(x, y, z, c=colormap(y_pred), marker='o')
ax.set_xlabel('TOTAL DOLLAR VALUE TRADED')
ax.set_ylabel('ESTIMATED INCOME')
ax.set_zlabel('DAYS SINCE LAST TRADE')
plt.legend(handles=[pop_a,pop_b,pop_c])
plt.title('Predicted')
plt.show()
def model_metrics(y_test,y_pred):
print("Decoded values of Churnrisk after applying inverse of label encoder : " + str(np.unique(y_pred)))
skplt.metrics.plot_confusion_matrix(y_test,y_pred,text_fontsize="small",cmap='Greens',figsize=(6,4))
plt.show()
print("The classification report for the model : \n\n"+ classification_report(y_test, y_pred))
```
<a id="prepare_model"></a>
## 6. Prepare Random Forest classification model
[Top](#top)
We instantiate a decision-tree based classification algorithm, namely, RandomForestClassifier. Next we define a pipeline to chain together the various transformers and estimators defined during the data preparation step before.
Scikit-learn provides APIs that make it easier to combine multiple algorithms into a single pipeline.
We fit the pipeline to training data and apply the trained model to transform test data and generate churn risk class prediction.
[Understanding Random Forest Classifier](https://towardsdatascience.com/understanding-random-forest-58381e0602d2)
```
from sklearn.ensemble import RandomForestClassifier
model_name = "Random Forest Classifier"
randomForestClassifier = RandomForestClassifier(n_estimators=100, max_depth=2,random_state=0)
```
Pipelines are a convenient way of designing your data processing in a machine learning flow. The following code example shows how pipelines are set up using sklearn.
Read more [Here](https://scikit-learn.org/stable/modules/classes.html?highlight=pipeline#module-sklearn.pipeline)
```
rfc_model = Pipeline(steps=[('preprocessorAll',preprocessorForAllColumns),('classifier', randomForestClassifier)])
```
<a id="train_model"></a>
## 7. Train Random Forest classification model
[Top](#top)
```
# Build models
rfc_model.fit(X_train,y_train)
```
<a id="test_model"></a>
## 8. Test Random Forest classification model
[Top](#top)
```
y_pred_rfc = rfc_model.predict(X_test)
```
<a id="evaluate_model"></a>
## 9. Evaluate Random Forest classification model
[Top](#top)
### Model results
In a supervised classification problem such as churn risk classification, we have a true output and a model-generated predicted output for each data point. For this reason, the results for each data point can be assigned to one of four categories:
1. True Positive (TP) - label is positive and prediction is also positive
2. True Negative (TN) - label is negative and prediction is also negative
3. False Positive (FP) - label is negative but prediction is positive
4. False Negative (FN) - label is positive but prediction is negative
These four numbers are the building blocks for most classifier evaluation metrics. A fundamental point when considering classifier evaluation is that pure accuracy (i.e. was the prediction correct or incorrect) is not generally a good metric. The reason for this is because a dataset may be highly unbalanced. For example, if a model is designed to predict fraud from a dataset where 95% of the data points are not fraud and 5% of the data points are fraud, then a naive classifier that predicts not fraud, regardless of input, will be 95% accurate. For this reason, metrics like precision and recall are typically used because they take into account the type of error. In most applications there is some desired balance between precision and recall, which can be captured by combining the two into a single metric, called the F-measure.
```
two_d_compare(y_test,y_pred_rfc,model_name)
#three_d_compare(y_test,y_pred_rfc,model_name)
```
### Confusion matrix
In the graph below we have printed a confusion matrix and a self-explanotary classification report.
The confusion matrix shows that, 42 mediums were wrongly predicted as high, 2 mediums were wrongly predicted as low and 52 mediums were accurately predicted as mediums.
```
y_test = label_encoder.inverse_transform(y_test)
y_pred_rfc = label_encoder.inverse_transform(y_pred_rfc)
model_metrics(y_test,y_pred_rfc)
```
[Precision Recall Fscore support](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.precision_recall_fscore_support.html)
[Understanding the Confusion Matrix](https://towardsdatascience.com/confusion-matrix-for-your-multi-class-machine-learning-model-ff9aa3bf7826)
### Comparative study
In the bar chart below, we have compared the random forest classification algorithm output classes against the actual values.
```
uniqueValues, occurCount = np.unique(y_test, return_counts=True)
frequency_actual = (occurCount[0],occurCount[2],occurCount[1])
uniqueValues, occurCount = np.unique(y_pred_rfc, return_counts=True)
frequency_predicted_rfc = (occurCount[0],occurCount[2],occurCount[1])
n_groups = 3
fig, ax = plt.subplots(figsize=(10,5))
index = np.arange(n_groups)
bar_width = 0.1
opacity = 0.8
rects1 = plt.bar(index, frequency_actual, bar_width,
alpha=opacity,
color='g',
label='Actual')
rects6 = plt.bar(index + bar_width, frequency_predicted_rfc, bar_width,
alpha=opacity,
color='purple',
label='Random Forest - Predicted')
plt.xlabel('Churn Risk')
plt.ylabel('Frequency')
plt.title('Actual vs Predicted frequency.')
plt.xticks(index + bar_width, ('High', 'Medium', 'Low'))
plt.legend()
plt.tight_layout()
plt.show()
```
<a id="model_knn"></a>
## 10. Build K-Nearest classification model
[Top](#top)
K number of nearest points around the data point to be predicted are taken into consideration. These K points at this time, already belong to a class. The data point under consideration, is said to belong to the class with which most number of points from these k points belong to.
```
from sklearn.neighbors import KNeighborsClassifier
model_name = "K-Nearest Neighbor Classifier"
knnClassifier = KNeighborsClassifier(n_neighbors = 5, metric='minkowski', p=2)
knn_model = Pipeline(steps=[('preprocessorAll',preprocessorForAllColumns),('classifier', knnClassifier)])
knn_model.fit(X_train,y_train)
y_pred_knn = knn_model.predict(X_test)
y_test = label_encoder.transform(y_test)
two_d_compare(y_test,y_pred_knn,model_name)
y_test = label_encoder.inverse_transform(y_test)
y_pred_knn = label_encoder.inverse_transform(y_pred_knn)
model_metrics(y_test,y_pred_knn)
```
<a id="compare_classification"></a>
## 11. Comparative study of both classification algorithms.
[Top](#top)
```
uniqueValues, occurCount = np.unique(y_test, return_counts=True)
frequency_actual = (occurCount[0],occurCount[2],occurCount[1])
uniqueValues, occurCount = np.unique(y_pred_rfc, return_counts=True)
frequency_predicted_rfc = (occurCount[0],occurCount[2],occurCount[1])
uniqueValues, occurCount = np.unique(y_pred_knn, return_counts=True)
frequency_predicted_knn = (occurCount[0],occurCount[2],occurCount[1])
n_groups = 3
fig, ax = plt.subplots(figsize=(10,5))
index = np.arange(n_groups)
bar_width = 0.1
opacity = 0.8
rects1 = plt.bar(index, frequency_actual, bar_width,
alpha=opacity,
color='g',
label='Actual')
rects6 = plt.bar(index + bar_width*2, frequency_predicted_rfc, bar_width,
alpha=opacity,
color='purple',
label='Random Forest - Predicted')
rects4 = plt.bar(index + bar_width*4, frequency_predicted_knn, bar_width,
alpha=opacity,
color='b',
label='K-Nearest Neighbor - Predicted')
plt.xlabel('Churn Risk')
plt.ylabel('Frequency')
plt.title('Actual vs Predicted frequency.')
plt.xticks(index + bar_width, ('High', 'Medium', 'Low'))
plt.legend()
plt.tight_layout()
plt.show()
```
Until evaluation provides satisfactory scores, you would repeat the data preprocessing through evaluating steps by tuning what are called the hyperparameters.
[Choosing the right estimator](https://scikit-learn.org/stable/tutorial/machine_learning_map/index.html)
### For a comparative study of some of the current most popular algorithms Please refer to this [tutorial](https://developer.ibm.com/tutorials/learn-classification-algorithms-using-python-and-scikit-learn/)
<p><font size=-1 color=gray>
© Copyright 2019 IBM Corp. All Rights Reserved.
<p>
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file
except in compliance with the License. You may obtain a copy of the License at
https://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the
License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either
express or implied. See the License for the specific language governing permissions and
limitations under the License.
</font></p>
|
github_jupyter
|
# Fitting distribution with R
```
x.norm <- rnorm(n=200,m=10,sd=2)
hist(x.norm,main="Histogram of observed data")
plot(density(x.norm),main="Density estimate of data")
plot(ecdf(x.norm),main="Empirical cumulative distribution function")
z.norm <- (x.norm-mean(x.norm))/sd(x.norm) # standardize data
qqnorm(z.norm) ## drawing the QQplot
abline(0,1) ## drawing a 45-degree reference line
```
If data differ from a normal distribution (i.e. data belonging from a Weibull pdf) we cna use `qqplot()` in this way:
```
x.wei <- rweibull(n=200,shape=2.1,scale=1.1) ## sampling from a Weibull
x.teo <- rweibull(n=200,shape=2,scale=1) ## theoretical quantiles from a Weibull population
qqplot(x.teo,x.wei,main="QQ-plot distr. Weibull")
abline(0,1)
```
# Model choice
Dealing with discrete data we can refer to Poisson's distribution with probability mass function:
$$ f(x,\lambda)=e^{-\lambda\dfrac{\lambda^x}{x!}} \quad \text{where } x=0,1,2,\ldots$$
```
x.poi <- rpois(n=200, lambda=2.5)
hist(x.poi, main="Poisson distribution")
```
As concern continuous data we have the normal (gaussian) dsitrubition:
$$ f(x,\lambda,\sigma)=\dfrac{1}{\sqrt{2\pi}\sigma} e^{\dfrac{1(x-\mu)^2}{2\sigma^2}} $$
with $x \in \mathbb{R}$.
```
curve(dnorm(x,m=10,sd=2),from=0,to=20,main="Normal distribution")
```
Gamma distribution:
$$ f(x,\alpha,\lambda)=\dfrac{\lambda^\alpha}{\gamma(\alpha)}x^{\alpha-1}e^{-\lambda x} $$
with $x \in \mathbb{R}^+$.
```
curve(dgamma(x, scale=1.5, shape=2), from=0, to=15, main="Gamma distribution")
```
Weibull distribition:
$$ f(x,\alpha,\beta)=\alpha\beta^{-\alpha}x^{\alpha-1}e^{-\left[\left(\dfrac{x}{\beta}\right)^\alpha\right]} $$
```
curve(dweibull(x, scale=2.5, shape=1.5), from=0, to=15, main="Weibull distribution")
h<-hist(x.norm,breaks=15)
xhist<-c(min(h$breaks),h$breaks)
yhist<-c(0,h$density,0)
xfit<-seq(min(x.norm),max(x.norm),length=40)
yfit<-dnorm(xfit,mean=mean(x.norm),sd=sd(x.norm))
plot(xhist,yhist,type="s",ylim=c(0,max(yhist,yfit)), main="Normal pdf and histogram")
lines(xfit,yfit, col="red")
yfit
yhist
ks.test(yfit,yhist)
```
# StackOverflow example
The following is from this StackOverflow example: https://stats.stackexchange.com/questions/132652/how-to-determine-which-distribution-fits-my-data-best
This requires you to install the following packages with the R package manager: `fitdistrplus` and `logspline`.
```
library(fitdistrplus)
library(logspline)
x <- c(37.50,46.79,48.30,46.04,43.40,39.25,38.49,49.51,40.38,36.98,40.00,
38.49,37.74,47.92,44.53,44.91,44.91,40.00,41.51,47.92,36.98,43.40,
42.26,41.89,38.87,43.02,39.25,40.38,42.64,36.98,44.15,44.91,43.40,
49.81,38.87,40.00,52.45,53.13,47.92,52.45,44.91,29.54,27.13,35.60,
45.34,43.37,54.15,42.77,42.88,44.26,27.14,39.31,24.80,16.62,30.30,
36.39,28.60,28.53,35.84,31.10,34.55,52.65,48.81,43.42,52.49,38.00,
38.65,34.54,37.70,38.11,43.05,29.95,32.48,24.63,35.33,41.34)
descdist(x, discrete = FALSE)
fit.weibull <- fitdist(x, "weibull")
fit.norm <- fitdist(x, "norm")
plot(fit.norm)
plot(fit.weibull)
fit.weibull$aic
fit.norm$aic
```
## Kolmogorov-Smirnov test simulation
```
n.sims <- 5e4
stats <- replicate(n.sims, {
r <- rweibull(n = length(x)
, shape= fit.weibull$estimate["shape"]
, scale = fit.weibull$estimate["scale"]
)
as.numeric(ks.test(r
, "pweibull"
, shape= fit.weibull$estimate["shape"]
, scale = fit.weibull$estimate["scale"])$statistic
)
})
plot(ecdf(stats), las = 1, main = "KS-test statistic simulation (CDF)", col = "darkorange", lwd = 1.7)
grid()
fit <- logspline(stats)
1 - plogspline(ks.test(x
, "pweibull"
, shape= fit.weibull$estimate["shape"]
, scale = fit.weibull$estimate["scale"])$statistic
, fit
)
xs <- seq(10, 65, len=500)
true.weibull <- rweibull(1e6, shape= fit.weibull$estimate["shape"]
, scale = fit.weibull$estimate["scale"])
boot.pdf <- sapply(1:1000, function(i) {
xi <- sample(x, size=length(x), replace=TRUE)
MLE.est <- suppressWarnings(fitdist(xi, distr="weibull"))
dweibull(xs, shape=MLE.est$estimate["shape"], scale = MLE.est$estimate["scale"])
}
)
boot.cdf <- sapply(1:1000, function(i) {
xi <- sample(x, size=length(x), replace=TRUE)
MLE.est <- suppressWarnings(fitdist(xi, distr="weibull"))
pweibull(xs, shape= MLE.est$estimate["shape"], scale = MLE.est$estimate["scale"])
}
)
#-----------------------------------------------------------------------------
# Plot PDF
#-----------------------------------------------------------------------------
par(bg="white", las=1, cex=1.2)
plot(xs, boot.pdf[, 1], type="l", col=rgb(.6, .6, .6, .1), ylim=range(boot.pdf),
xlab="x", ylab="Probability density")
for(i in 2:ncol(boot.pdf)) lines(xs, boot.pdf[, i], col=rgb(.6, .6, .6, .1))
# Add pointwise confidence bands
quants <- apply(boot.pdf, 1, quantile, c(0.025, 0.5, 0.975))
min.point <- apply(boot.pdf, 1, min, na.rm=TRUE)
max.point <- apply(boot.pdf, 1, max, na.rm=TRUE)
lines(xs, quants[1, ], col="red", lwd=1.5, lty=2)
lines(xs, quants[3, ], col="red", lwd=1.5, lty=2)
lines(xs, quants[2, ], col="darkred", lwd=2)
#-----------------------------------------------------------------------------
# Plot CDF
#-----------------------------------------------------------------------------
par(bg="white", las=1, cex=1.2)
plot(xs, boot.cdf[, 1], type="l", col=rgb(.6, .6, .6, .1), ylim=range(boot.cdf),
xlab="x", ylab="F(x)")
for(i in 2:ncol(boot.cdf)) lines(xs, boot.cdf[, i], col=rgb(.6, .6, .6, .1))
# Add pointwise confidence bands
quants <- apply(boot.cdf, 1, quantile, c(0.025, 0.5, 0.975))
min.point <- apply(boot.cdf, 1, min, na.rm=TRUE)
max.point <- apply(boot.cdf, 1, max, na.rm=TRUE)
lines(xs, quants[1, ], col="red", lwd=1.5, lty=2)
lines(xs, quants[3, ], col="red", lwd=1.5, lty=2)
lines(xs, quants[2, ], col="darkred", lwd=2)
#lines(xs, min.point, col="purple")
#lines(xs, max.point, col="purple")
```
|
github_jupyter
|
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
data = pd.read_csv('dataset-of-10s.csv')
data.head()
```
# checking basic integrity
```
data.shape
data.info()
```
# no. of rows = non null values for each column -> no null value
```
data.head()
```
# checking unique records using uri
```
# extracting exact id
def extract(x):
splited_list = x.split(':') # spliting text at colons
return splited_list[2] # returning third element
data['uri'] = data['uri'].apply(extract)
data.head() #successfully extracted the id
```
# checking for duplicate rows
```
data['uri'].nunique(),
data['uri'].value_counts()
data['uri'].value_counts().unique()
dupe_mask = data['uri'].value_counts()==2
dupe_ids = dupe_mask[dupe_mask]
dupe_ids.value_counts, dupe_ids.shape
#converting duplicate ids into a list
dupe_ids = dupe_ids.index
dupe_ids = dupe_ids.tolist()
dupe_ids
duplicate_index = data.loc[data['uri'].isin(dupe_ids),:].index # all the duplicted records
duplicate_index = duplicate_index.tolist()
```
# We will be removing all the duplication as they are few compared to data
```
data.drop(duplicate_index,axis=0,inplace=True)
data.shape
data.info()
print("shape of data",data.shape )
print("no. of unique rows",data['uri'].nunique()) # no duplicates
data.head()
```
# now we will be dropping all the unnecessary columns which contain string which cant be eficiently converted into numerics
```
data.drop(['track','artist','uri'],axis=1,inplace=True)
data.head()
```
# Univariate analysis
```
#analysing class imbalance
sns.countplot(data=data,x='target')
data.columns
# checking appropriate data type
data[['danceability', 'energy', 'key', 'loudness']].info() # every feature have appropriate datatype
# checking range of first 4 features
data[['danceability', 'energy', 'key', 'loudness']].describe()
plt.figure(figsize=(10,10))
plt.subplot(2,2,1)
data['danceability'].plot()
plt.subplot(2,2,2)
plt.plot(data['energy'],color='red')
plt.subplot(2,2,3)
plt.plot(data[['key','loudness']])
```
# danceabilty is well inside the range(0,1)
# energy is well inside the range(0,1)
# there's no -1 for keys-> every track has been assigned respective keys
# loudness values are out of range(0,-60)db
```
loudness_error_idnex = data[data['loudness']>0].index
loudness_error_idnex
# removing rows with out of range values in loudness column
data.drop(loudness_error_idnex,axis=0, inplace=True)
data.shape # record is removed
# checking appropriate datatype for next 5 columns
data[['mode', 'speechiness',
'acousticness', 'instrumentalness', 'liveness',]].info() # datatypes are in acoordance with provided info
data[['mode', 'speechiness',
'acousticness', 'instrumentalness', 'liveness',]].describe() # every feautre is within range
sns.countplot(x=data['mode']) # have only two possible values 0 and 1, no noise in the feature
data[['valence', 'tempo',
'duration_ms', 'time_signature', 'chorus_hit', 'sections']].info() # data type is in accordance with provided info
data[['valence', 'tempo',
'duration_ms', 'time_signature', 'chorus_hit', 'sections']].describe() # all the data are in specified range
```
# Performing F-test to know the relation between every feature and target
```
data.head()
x = data.iloc[:,:-1].values
y = data.iloc[:,-1].values
x.shape,y.shape
from sklearn.feature_selection import f_classif
f_stat,p_value = f_classif(x,y)
feat_list = data.iloc[:,:-1].columns.tolist()
# making a dataframe
dict = {'Features':feat_list,'f_statistics':f_stat,'p_value':p_value}
relation = pd.DataFrame(dict)
relation.sort_values(by='p_value')
```
# Multivariate analysis
```
correlation = data.corr()
plt.figure(figsize=(15,12))
sns.heatmap(correlation, annot=True)
plt.tight_layout
```
# strong features(accordance with f-test) -->
danceability, loudness, acousticness, instrumentalness, valence
# less imortant feature(accordance with f-test)-->
duration, section, mode, time_signature, chorus hit
# least imortant-->
energy,key,speecheness,liveliness,tempo
|
github_jupyter
|
# PART 3 - Metadata Knowledge Graph creation in Amazon Neptune.
Amazon Neptune is a fast, reliable, fully managed graph database service that makes it easy to build and run applications that work with highly connected datasets. The core of Neptune is a purpose-built, high-performance graph database engine. This engine is optimized for storing billions of relationships and querying the graph with milliseconds latency. Neptune supports the popular graph query languages Apache TinkerPop Gremlin and W3C’s SPARQL, enabling you to build queries that efficiently navigate highly connected datasets.
https://docs.aws.amazon.com/neptune/latest/userguide/feature-overview.html
In that section we're going to use TinkerPop Gremlin as the language to create and query our graph.
### Important
We need to downgrade the tornado library for the gremlin libraries to work in our notebook.
Without doing this, you'll most likely run into the following error when executing some gremlin queries:
"RuntimeError: Cannot run the event loop while another loop is running"
```
!pip install --upgrade tornado==4.5.3
```
### Restart your kernel
Because the notebook itself has some dependencies with the tornado library, we need to restart the kernel before proceeding.
To do so, go to the top menu > Kernel > Restart Kernel.. > Restart
Then proceed and execute the following cells.
```
!pip install pandas
!pip install jsonlines
!pip install gremlinpython
!pip install networkx
!pip install matplotlib
import os
import jsonlines
import networkx as nx
import matplotlib.pyplot as plt
import pandas as pd
#load stored variable from previous notebooks
%store -r
```
Loading the Gremlin libraries and connecting to our Neptune instance
```
from gremlin_python import statics
from gremlin_python.process.anonymous_traversal import traversal
from gremlin_python.process.graph_traversal import __
from gremlin_python.process.strategies import *
from gremlin_python.driver.driver_remote_connection import DriverRemoteConnection
from gremlin_python.process.traversal import T
from gremlin_python.process.traversal import Order
from gremlin_python.process.traversal import Cardinality
from gremlin_python.process.traversal import Column
from gremlin_python.process.traversal import Direction
from gremlin_python.process.traversal import Operator
from gremlin_python.process.traversal import P
from gremlin_python.process.traversal import Pop
from gremlin_python.process.traversal import Scope
from gremlin_python.process.traversal import Barrier
from gremlin_python.process.traversal import Bindings
from gremlin_python.process.traversal import WithOptions
from gremlin_python.structure.graph import Graph
graph = Graph()
def start_remote_connection_neptune():
remoteConn = DriverRemoteConnection(your_neptune_endpoint_url,'g')
g = graph.traversal().withRemote(remoteConn)
return g
# g is the traversal source to use to query the graph
g = start_remote_connection_neptune()
```
<b>IMPORTANT:</b>
- Note that the remote connection will time out after few minutes if unused so if you're encountering exceptions after having paused the notebook execution for a while, please re-run the above cell.
- <b>Make sure your Neptune DB is created for the sole purpose of this labs as we'll be cleaning it before starting.</b>
```
#CAREFUL - the below line of code empties your graph. Again, make sure you're using a dedicated instance for this workshop
g.V().drop().iterate()
```
## A note on Gremlin
Gremlin is a functional, data-flow language that enables users to succinctly express complex traversals on (or queries of) their application's property graph. Every Gremlin traversal is composed of a sequence of (potentially nested) steps. A step performs an atomic operation on the data stream. Every step is either a map-step (transforming the objects in the stream), a filter-step (removing objects from the stream), or a sideEffect-step (computing statistics about the stream).
More info here: https://tinkerpop.apache.org/gremlin.html
The image below is an extract from:
https://tinkerpop.apache.org/docs/3.5.1/tutorials/getting-started/#_the_next_fifteen_minutes
I highly recommend you to be familiar with the concepts of Vertex and Edges at the very minimum before proceeding with the notebook.

## Vertices and Edges names
See below the variables containing the labels for our vertices and edges that we'll create across the notebook.
```
#Vertex representing a Video
V_VIDEO = "video"
#Vertex representing a "scene" e.g. SHOT, TECHNICAL_CUE
V_VIDEO_SCENE = "video_scene"
#Vertex representing a Video segment. we arbitrary split our video into 1min segments and attach metadata to the segments itselves
V_VIDEO_SEGMENT = 'video_segment'
#Edge between VIDEO and SEGMENT
E_HAS_SEGMENT = 'contains_segment'
#Edge between VIDEO and SCENE
E_HAS_SCENE = 'contains_scene'
#Edge between Scene and Segment
E_BELONG_TO_SEGMENT = 'belong_to_segment'
#Vertex representing a label extracted by Rekognition from the video
V_LABEL = 'label'
#Edge between SEGMENT and LABEL
E_HAS_LABEL = 'has_label'
#Edge between parent LABEL and child LABEL e.g. construction -> bulldozer
E_HAS_CHILD_LABEL = 'has_child_label'
#Vertex representing the NER
V_ENTITY = 'entities'
#Vertex representing the type of NER
V_ENTITY_TYPE = 'entity_type'
#Edge between ENTITY and ENTITY_TYPE
E_IS_OF_ENTITY_TYPE = 'is_of_entity_type'
#Edge between SEGMENT and ENTITY
E_HAS_ENTITY = 'has_entity'
#Vertex representing a TOPIC
V_TOPIC = 'topic'
#Vertex representing a TOPIC_TERM
V_TOPIC_TERM = 'topic_term'
#Edge between a VIDEO_SEGMENT and a TOPIC
E_HAS_TOPIC = 'has_topic'
#Edge between a TOPIC and a TOPIC_TERM
E_HAS_TERM = 'has_term'
#Vertex representing a TERM
V_TERM = 'term'
```
## We start by adding our video to the Graph
Note how I start with g, our traversal graph, then call the addV (V for Vertex) method and then attach properties to the new vertex. I end the line with ".next()" which will return the newly created node (similar to how an iterator would work). all method are "chained" together in one expression.
```
sample_video_vertex = g.addV(V_VIDEO).property("name", video_name).property("filename", video_file) .property('description', 'description of the video').next()
```
[QUERY] We're listing all the vertices in the graph with their metadata. At this stage, we only have one.
Explanation: g.V() gets us all vertices in the graph, the .hasLabel() filters the vertices based on the vertex label(=type), the .valueMap() returns all properties for all vertices and the .toList() returns the full list. Note that you can use .next() instead of toList() to just return the next element in the list.
```
g.V().hasLabel(V_VIDEO).valueMap().toList()
```
[QUERY] Below is a different way to precisely return a vertex based on its name.
Explanation: g.V() gives us all the vertices, .has() allows us to filter based on the name of the vertex and .next() returns the first (and only) item from the iterator. note that we haven't used .valueMap() so what is returned is the ID of the vertex.
```
g.V().has('name', video_name).next()
```
## Creating 1min segments vertices in Neptune
As mentioned in the previous notebook, we are creating metadata segments that we'll use to store labels and other information related to those 1min video segments.
This will give us a more fine grained view of the video's topics and metadata.
```
print(segment_size_ms)
#get the video duration by looking at the end of the last segment.
def get_video_duration_in_ms(segment_detection_output):
return segment_detection_output['Segments'][-1]['EndTimestampMillis']
#create a new segment vertex and connect it to the video
def add_segment_vertex(video_name, start, end, g):
#retrieving the video vertex
video_vertex = g.V().has(V_VIDEO, 'name', video_name).next()
#generating a segment ID
segment_id = video_name + '-' + str(start) + '-' + str(end)
#creating a new vertex for the segment
new_segment_vert = g.addV(V_VIDEO_SEGMENT).property("name", segment_id).property('StartTimestampMillis', start).property('EndTimestampMillis', end).next()
#connecting the video vertex to the segment vertex
g.V(video_vertex).addE(E_HAS_SEGMENT).to(new_segment_vert).iterate()
#generate segment vertices of a specific duration (default 60s) for a specific video
def generate_segment_vertices(video_name, g, duration_in_millisecs, segment_size_in_millisecs=60000):
#retrieve the mod
modulo = duration_in_millisecs % segment_size_in_millisecs
#counter that we'll increment by segment_size_in_millisecs steps
counter = 0
while ((counter + segment_size_in_millisecs) < duration_in_millisecs) :
start = counter
end = counter + segment_size_in_millisecs
add_segment_vertex(video_name, start, end, g)
counter += segment_size_in_millisecs
#adding the segment vertex to the video vertex
add_segment_vertex(video_name, duration_in_millisecs - modulo, duration_in_millisecs, g)
#add a vertex if it doesn't already exist
def add_vertex(vertex_label, vertex_name, g):
g.V().has(vertex_label,'name', vertex_name).fold().coalesce(__.unfold(), __.addV(vertex_label).property('name',vertex_name)).iterate()
#add an edge between 2 vertices
def add_edge(vertex_label_from, vertex_label_to, vertex_name_from, vertex_name_to, edge_name, g, weight=None):
if weight == None:
g.V().has(vertex_label_to, 'name', vertex_name_to).as_('v1').V().has(vertex_label_from, 'name', vertex_name_from).coalesce(__.outE(edge_name).where(__.inV().as_('v1')), __.addE(edge_name).to('v1')).iterate()
else:
g.V().has(vertex_label_to, 'name', vertex_name_to).as_('v1').V().has(vertex_label_from, 'name', vertex_name_from).coalesce(__.outE(edge_name).where(__.inV().as_('v1')), __.addE(edge_name).property('weight', weight).to('v1')).iterate()
```
Note: remember, the SegmentDetectionOutput object contains the output of the Amazon Rekognition segment (=scene) detection job
```
duration = get_video_duration_in_ms(SegmentDetectionOutput)
generate_segment_vertices(video_name, g, duration, segment_size_ms)
```
[QUERY] Let's retrieve the segments that are connected to the video vertex via an edge, ordered by StartTimestampMillis. In that case we limit the result set to 5 items.
Explanation: g.V() get us all vertices, .has(V_VIDEO, 'name', video_name) filters on the video vertices with name=video_name, .out() gives us all vertices connected to this vertex by an outgoing edge, .hasLabel(V_VIDEO_SEGMENT) filters the vertices to video segments only, .order().by() orders the vertices by StartTimestampMillis, .valueMap() gives us all properties for those vertices, .limit(5) reduces the results to 5 items, .toList() gives us the list of items.
```
list_of_segments = g.V().has(V_VIDEO, 'name', video_name).out().hasLabel(V_VIDEO_SEGMENT) \
.order().by('StartTimestampMillis', Order.asc).valueMap().limit(5).toList()
list_of_segments
```
## Graph Visualisation
The networkx library alongside with matplotlib allows us to draw visually the graph.
Let's draw our vertex video and the 1min segments we just created.
```
#Function printing the graph from a start vertex and a list of edges that will be traversed/displayed.
def print_graph(start_vertex_label, start_vertex_name, list_edges, displayLabels=True, node_size=2000, node_limit=200):
#getting the paths between vertices
paths = g.V().has(start_vertex_label, 'name', start_vertex_name)
#adding the edges that we want to traverse
for edge in list_edges:
paths = paths.out(edge)
paths = paths.path().toList()
#creating graph object
G=nx.DiGraph()
#counters to limit the number of nodes being displayed.
limit_nodes_counter = 0
#creating the graph by iterating over the paths
for p in paths:
#depth of the graph
depth = len(p)
#we build our graph
for i in range(0, depth -1):
label1 = g.V(p[i]).valueMap().next()['name'][0]
label2 = g.V(p[i+1]).valueMap().next()['name'][0]
if limit_nodes_counter < node_limit:
G.add_edge(label1, label2)
limit_nodes_counter += 1
plt.figure(figsize=(12,7))
nx.draw(G, node_size=node_size, with_labels=displayLabels)
plt.show()
#please note that we limit the number of nodes being displayed
print_graph(V_VIDEO, video_name, [E_HAS_SEGMENT], node_limit=15)
```
# Add the scenes into our graph
In the below steps we're connecting the scenes to the video itself and not the segments as we want to be able to search and list the different types of scenes at the video level. However, note that we're not going to attach any specific metadata at the scene level, only at the segment level.
```
def store_video_segment(original_video_name, json_segment_detection_output, orig_video_vertex):
shot_counter = 0
tech_cue_counter = 0
for technicalCue in json_segment_detection_output['Segments']:
#start
frameStartValue = technicalCue['StartTimestampMillis'] / 1000
#end
frameEndValue = technicalCue['EndTimestampMillis'] / 1000
#SHOT or TECHNICAL_CUE
segment_type = technicalCue['Type']
counter = -1
if (segment_type == 'SHOT'):
shot_counter += 1
counter = shot_counter
elif (segment_type == 'TECHNICAL_CUE'):
tech_cue_counter += 1
counter = tech_cue_counter
segment_id = original_video_name + '-' + segment_type + '-' + str(counter)
#creating the vertex for the video segment with all the metadata extracted from the segment generation job
new_vert = g.addV(V_VIDEO_SCENE).property("name", segment_id).property("type", segment_type) \
.property('StartTimestampMillis', technicalCue['StartTimestampMillis']).property('EndTimestampMillis', technicalCue['EndTimestampMillis']) \
.property('StartFrameNumber', technicalCue['StartFrameNumber']).property('EndFrameNumber', technicalCue['EndFrameNumber']) \
.property('DurationFrames', technicalCue['DurationFrames']).next()
#creating the edge between the original video vertex and the segment vertex with the type as a property of the relationship
g.V(orig_video_vertex).addE(E_HAS_SCENE).to(new_vert).properties("type", segment_type).iterate()
store_video_segment(video_name, SegmentDetectionOutput, sample_video_vertex)
```
[QUERY] We're retrieving the list of edges/branches created between the video and the scenes.
Explanation: g.V() returns all vertices, .has(V_VIDEO, 'name', video_name) returns the V_VIDEO vertex with name=video_name, .out(E_HAS_SCENE) returns the list of vertices that are connected to the V_VIDEO vertex by a E_HAS_SCENE edge, toList() returns the list of items.
```
list_of_edges = g.V().has(V_VIDEO, 'name', video_name).out(E_HAS_SCENE).toList()
print(f"the sample video vertex has now {len(list_of_edges)} edges connecting to the scenes vertices")
```
[QUERY] Let's search for the technical cues (black and fix screens) at the end of the video.
Explanation: g.V() returns all vertices, .has(V_VIDEO, 'name', video_name) returns the V_VIDEO vertex with name=video_name, .out(E_HAS_SCENE) returns the list of vertices that are connected to the V_VIDEO vertex by a E_HAS_SCENE edge, .has('type', 'TECHNICAL_CUE') filters the list on type=TECHNICAL_CUE, the rest was seen above already.
```
g.V().has(V_VIDEO, 'name', video_name).out(E_HAS_SCENE) \
.has('type', 'TECHNICAL_CUE') \
.order().by('EndTimestampMillis', Order.desc) \
.limit(5).valueMap().toList()
```
</br>
Let's print the graph for those newly created SCENE vertices
```
#please note that we limit the number of nodes being displayed
print_graph(V_VIDEO, video_name, [E_HAS_SCENE], node_limit=15)
```
## Create the labels vertices and link them to the segments
We're now going to create vertices to represent the labels in our graph and connect them to the 1min segments
```
def create_label_vertices(LabelDetectionOutput, video_name, g, confidence_threshold=80):
labels = LabelDetectionOutput['Labels']
for instance in labels:
#keeping only the labels with high confidence
label_details_obj = instance['Label']
confidence = label_details_obj['Confidence']
if confidence > confidence_threshold:
#adding then main label name to the list
label_name = str(label_details_obj['Name']).lower()
#adding the label vertex
add_vertex(V_LABEL, label_name, g)
#adding the link between video and label
add_edge(V_VIDEO, V_LABEL, video_name, label_name, E_HAS_LABEL, g, weight=None)
#adding parent labels too
parents = label_details_obj['Parents']
if len(parents) > 0:
for parent in parents:
#create parent vertex if it doesn't exist
parent_label_name = str(parent['Name']).lower()
add_vertex(V_LABEL, parent_label_name, g)
#create the relationship between parent and children if it doesn't already exist
add_edge(V_LABEL, V_LABEL, parent_label_name, label_name, E_HAS_CHILD_LABEL, g, weight=None)
create_label_vertices(LabelDetectionOutput, video_name, g, 80)
```
[QUERY] Let's list the labels vertices to see what was created above.
Explanation: g.V() returns all vertices, .hasLabel(V_LABEL) returns only the vertices of label/type V_LABEL, .valueMap().limit(20).toList() gives us the list with properties for the first 20 items.
```
#retrieving a list of the first 20 labels
label_list = g.V().hasLabel(V_LABEL).valueMap().limit(20).toList()
label_list
```
Let's display a graph with our video's labels and the child labels relationships in between labels.
```
print_graph(V_VIDEO, video_name, [E_HAS_LABEL, E_HAS_CHILD_LABEL], node_limit=15)
```
[QUERY] A typical query would be to search for videos who have a specific label.
Explanation: g.V().has(V_LABEL, 'name', ..) returns the first label vertex from the previous computed list, .in_(E_HAS_LABEL) returns all vertices who have an incoming edge (inE) pointing to this label vertex, .valueMap().toList() returns the list with properties.
note that in_(E_HAS_LABEL) is equivalent to .inE(E_HAS_LABEL).outV() where .inE(E_HAS_LABEL) returns all incoming edges with the specified label and .outV() will traverse to the vertices attached to that edge.
Obviously we only have the one result as we've only processed one video so far.
```
g.V().has(V_LABEL, 'name', label_list[0]['name'][0]).in_(E_HAS_LABEL).valueMap().toList()
```
## Create the topics and associated topic terms vertices
We are going to re-arrange a bit the raw results from the topic modeling job to make it more readable
```
comprehend_topics_df.head()
```
We extract the segment id/number from the docname column in a separate column, cast it to numeric values, drop the docname column and sort by segment_id
```
comprehend_topics_df['segment_id'] = comprehend_topics_df['docname'].apply(lambda x: x.split(':')[-1])
comprehend_topics_df['segment_id'] = pd.to_numeric(comprehend_topics_df['segment_id'], errors='coerce')
comprehend_topics_df = comprehend_topics_df.drop('docname', axis=1)
comprehend_topics_df = comprehend_topics_df.sort_values(by='segment_id')
comprehend_topics_df.head(5)
```
Looks better!
Note that:
- a segment_id can belong to several topics
- proportion = the proportion of the document that is concerned with the topic
Let's now create our topic vertices
```
def create_topic_vertices(topics_df, terms_df, video_name, g):
#retrieve all segments for the video
segments_vertex_list = g.V().has(V_VIDEO, 'name', video_name).out(E_HAS_SEGMENT).order().by('StartTimestampMillis', Order.asc).valueMap().toList()
for index, row in topics_df.iterrows():
topic = row['topic']
segment_id = int(row['segment_id'])
#string formating to use as name for our vertices
topic_str = str(int(row['topic']))
#adding terms vertices that are associated with that topic and create the topic -> term edge
list_of_terms = terms_df[comprehend_terms_df['topic'] == topic]
#getting the segment name
segment_name = segments_vertex_list[segment_id]['name'][0]
#adding the topic vertex
add_vertex(V_TOPIC, topic_str, g)
#adding the link between entity and entity_type
add_edge(V_VIDEO_SEGMENT, V_TOPIC, segment_name, topic_str, E_HAS_TOPIC, g, weight=None)
#looping across all
for index2, row2 in list_of_terms.iterrows():
term = row2['term']
weight = row2['weight']
add_vertex(V_TERM, term, g)
add_edge(V_TOPIC, V_TERM, topic_str, term, E_HAS_TERM, g, weight=weight)
create_topic_vertices(comprehend_topics_df, comprehend_terms_df, video_name, g)
```
Let's display our video, few segments and their associated topics
```
#please note that we limit the number of nodes being displayed
print_graph(V_VIDEO, video_name, [E_HAS_SEGMENT, E_HAS_TOPIC], node_limit=10)
```
Let's display a partial graph showing relationships between the video -> segment -> topic -> term
```
print_graph(V_VIDEO, video_name, [E_HAS_SEGMENT, E_HAS_TOPIC, E_HAS_TERM], node_limit=20)
```
[QUERY] We're now listing all the segments that are in topic 2 (try different topic numbers if you want)
Explanation: g.V().has(V_TOPIC, 'name', '2') returns the topic vertex with name=2, .in_(E_HAS_TOPIC) returns all vertices that have a edge pointing into that topic vertex, .valueMap().toList() returns the list of items with their properties
```
g.V().has(V_TOPIC, 'name', '2').in_(E_HAS_TOPIC).valueMap().toList()
```
## Create the NER vertices and link them to the segments
```
#create the entity and entity_type vertices including the related edges
def create_ner_vertices(ner_job_data, video_name, g, score_threshold=0.8):
#retrieve all segments for the video
segments_vertex_list = g.V().has(V_VIDEO, 'name', video_name).out(E_HAS_SEGMENT).order().by('StartTimestampMillis', Order.asc).valueMap().toList()
counter_vertex = 0
for doc in ner_job_data:
#each jsonline from the ner job is already segmented by 1min chunks, so we're just matching them to our ordered segments list.
segment_vertex_name = segments_vertex_list[counter_vertex]['name'][0]
for entity in doc:
text = entity['Text']
type_ = entity['Type']
score = entity['Score']
if score > score_threshold:
#adding the entity type vertex
entity_type_vertex = g.V().has(V_ENTITY_TYPE,'name', type_).fold().coalesce(__.unfold(), __.addV(V_ENTITY_TYPE).property('name',type_)).iterate()
#adding the entity type vertex
entity_vertex = g.V().has(V_ENTITY,'name', text).fold().coalesce(__.unfold(), __.addV(V_ENTITY).property('name',text)).iterate()
#adding the link between entity and entity_type
entity_entity_type_edge = g.V().has(V_ENTITY_TYPE, 'name', type_).as_('v1').V().has(V_ENTITY, 'name', text).coalesce(__.outE(E_IS_OF_ENTITY_TYPE).where(__.inV().as_('v1')), __.addE(E_IS_OF_ENTITY_TYPE).to('v1')).iterate()
#adding the edge between entity and segment
segment_entity_edge = g.V().has(V_ENTITY,'name', text).as_('v1').V().has(V_VIDEO_SEGMENT, 'name', segment_vertex_name).coalesce(__.outE(E_HAS_ENTITY).where(__.inV().as_('v1')), __.addE(E_HAS_ENTITY).to('v1')).iterate()
#print(f"attaching entity: {text} to segment: {segment_vertex_name}")
counter_vertex += 1
create_ner_vertices(ner_job_data, video_name, g, 0.8)
```
[QUERY] Let's get a list of the first 20 entities
Explanation: g.V().hasLabel(V_ENTITY) returns all vertices of label/type V_ENTITY, .valueMap().limit(20).toList() returns the list of the first 20 items with their properties (just name in that case).
```
entities_list = g.V().hasLabel(V_ENTITY).valueMap().limit(20).toList()
entities_list
```
[QUERY] Let's now look up the first entity of the previous entities_list and check its type
Explanation: g.V().has(V_ENTITY, 'name', ...) return the first V_ENTITY vertex of the entities_list list, .out(E_IS_OF_ENTITY_TYPE) returns vertices connected to this V_ENTITY vertex by a E_IS_OF_ENTITY_TYPE edge.
```
g.V().has(V_ENTITY, 'name', entities_list[0]['name'][0]).out(E_IS_OF_ENTITY_TYPE).valueMap().toList()
```
[QUERY] Let's see now which video segments contains that entity
Explanation: g.V().has(V_ENTITY, 'name', ...) return the first V_ENTITY vertex of the entities_list list, .in_(E_HAS_ENTITY) returns all vertices that have an incoming edge into that V_ENTITY vertex and .valueMap().toList() returns the list with properties.
```
g.V().has(V_ENTITY, 'name', entities_list[0]['name'][0]).in_(E_HAS_ENTITY).valueMap().toList()
```
[QUERY] Similar query but this time we traverse further the graph and only return the list of videos which have this specific entity.
Explanation: g.V().has(V_ENTITY, 'name', ...) return the first V_ENTITY vertex of the entities_list list, .in_(E_HAS_ENTITY) returns the V_VIDEO_SEGMENT vertices that have an incoming edge into that V_ENTITY vertex, .in_(E_HAS_SEGMENT) returns the V_VIDEO vertices that have an incoming edge into those V_VIDEO_SEGMENT vertices and .valueMap().toList() returns the list with properties.
Note how by chaining the .in_() methods we are able to traverse the graph from one type of vertex to the other.
```
g.V().has(V_ENTITY, 'name', entities_list[0]['name'][0]).in_(E_HAS_ENTITY).in_(E_HAS_SEGMENT).dedup().valueMap().toList()
```
</br>
Let's now display a graph showing the relationship between Video -> Segment -> Entity
```
print_graph(V_VIDEO, video_name, [E_HAS_SEGMENT, E_HAS_ENTITY], node_size=800, node_limit=30)
```
# Summary
This notebook only touched the surface of what you can do with Graph databases but it should give you an idea of how powerful they are at modeling highly dimensional relationships between entities. This specific architecture allows them to be especially scalable and performing even with billions of vertices and edges.
Gremlin is the most widely used query language for graph DB and provides quite an intuitive way to traverse/query those graphs by chaining those instructions but if you want a more traditional SQL language, you can also look into SPARQL as an alternative.
https://graphdb.ontotext.com/documentation/free/devhub/sparql.html#using-sparql-in-graphdb
|
github_jupyter
|
```
import pandas as pd
import numpy as np
df = pd.DataFrame({'Map': [0,0,0,1,1,2,2], 'Values': [1,2,3,5,4,2,5]})
df['S'] = df.groupby('Map')['Values'].transform(np.sum)
df['M'] = df.groupby('Map')['Values'].transform(np.mean)
df['V'] = df.groupby('Map')['Values'].transform(np.var)
print (df)
import numpy as np
import pandas as pd
df = pd.DataFrame({'A': [2,3,1], 'B': [1,2,3], 'C': [5,3,4]})
df = df.drop(df.index[[1]])
print (df)
df = df.drop('B', 1)
print (df)
import pandas as pd
df = pd.DataFrame({'A': [0,0,0,0,0,1,1], 'B': [1,2,3,5,4,2,5],
'C': [5,3,4,1,1,2,3]})
a_group_desc = df.groupby('A').describe()
print (a_group_desc)
unstacked = a_group_desc.unstack()
print (unstacked)
import pandas as pd
import numpy as np
s = pd.Series([1, 2, 3, np.NaN, 5, 6, None])
print (s.isnull())
print (s[s.isnull()])
import pandas as pd
import numpy as np
s = pd.Series([1, 2, 3, np.NaN, 5, 6, None])
print (s.fillna(int(s.mean())))
print (s.dropna())
x = np.array([[[1, 2, 3], [4, 5, 6], [7, 8, 9],], [[11,12,13], [14,15,16], [17,18,19],],
[[21,22,23], [24,25,26], [27,28,29]]])
print(x[[[0]]])
values = [1, 5, 8, 9, 2, 0, 3, 10, 4, 7]
import matplotlib.pyplot as plt
plt.plot(range(1,11), values)
plt.savefig('Image.jpeg', format='jpeg')
values = [1, 5, 8, 9, 2, 0, 3, 10, 4, 7]
import matplotlib.pyplot as plt
plt.plot(range(1,11), values)
plt.savefig('MySamplePlot.png', format='png')
values = [1, 5, 8, 9, 2, 0, 3, 10, 4, 7]
import matplotlib.pyplot as plt
plt.plot(range(1,11), values)
plt.savefig('plt.pdf', format='pdf')
import numpy as np
import matplotlib.pyplot as plt
x1 = 50 * np.random.rand(40)
x2 = 25 * np.random.rand(40) + 25
x = np.concatenate((x1, x2))
y1 = 25 * np.random.rand(40)
y2 = 50 * np.random.rand(40) + 25
y = np.concatenate((y1, y2))
plt.scatter(x, y, s=[100], marker='^', c='m')
plt.show()
pip install matplotlib
pip install --upgrade matplotlib
pip install mpl_toolkits
pip install basemap
from mpl_toolkits.basemap import Basemap
import matplotlib.pyplot as plt
m = Basemap(projection='mill')
m.drawcoastlines()
plt.show()
conda install basemap
pip install mpltoolkits.basemap
per = (11438/500)*100
x = 'result = {r:3.2f}%'.format(r=per)
x
coords = {'lat':'37.25N','long':'-115.45W'}
'Coords : {long}, {lat}'.format(**coords)
l = list(x for x in range(1,20))
l
x = [2,4,8,6,3,1,7,9]
x.sort()
x.reverse()
x
l = [(1,2,3), (4,5,6), (7,8,9)]
for x in l:
for y in x:
print(y)
import numpy as np
import matplotlib.pyplot as plt
x = 20 * np.random.randint(1,10,10000)
plt.hist(x, 25,histtype='stepfilled', align='mid', color='g',label='TestData')
plt.legend()
plt.title('Step Filled Histogram')
plt.show()
import numpy as np
import matplotlib.pyplot as plt
data = 50 * np.random.rand(100) - 25
plt.boxplot(data)
plt.show()
import numpy as np
import matplotlib.pyplot as plt
x1 = 5 * np.random.rand(40)
x2 = 5 * np.random.rand(40) + 25
x3 = 25 * np.random.rand(20)
x = np.concatenate((x1, x2, x3))
y1 = 5 * np.random.rand(40)
y2 = 5 * np.random.rand(40) + 25
y3 = 25 * np.random.rand(20)
y = np.concatenate((y1, y2, y3))
plt.scatter(x, y, s=[100], marker='^', c='m')
plt.show()
import numpy as np
import matplotlib.pyplot as plt
x1 = 5 * np.random.rand(50)
x2 = 5 * np.random.rand(50) + 25
x3 = 30 * np.random.rand(25)
x = np.concatenate((x1, x2, x3))
y1 = 5 * np.random.rand(50)
y2 = 5 * np.random.rand(50) + 25
y3 = 30 * np.random.rand(25)
y = np.concatenate((y1, y2, y3))
color_array = ['b'] * 50 + ['g'] * 50 + ['r'] * 25
plt.scatter(x, y, s=[50], marker='D', c=color_array)
plt.show()
import networkx as nx
g = nx.Graph()
g.add_node(1)
g.add_nodes_from([2,7])
g.add_edge(1,2)
g.add_edges_from([(2,3),(4,5),(6,7),(3,7),(2,5),(4,6)])
nx.draw_networkx(g)
nx.info(g)
import pandas as pd
df = pd.DataFrame({'A': [0,0,0,0,0,1,1], 'B': [1,2,3,5,4,2,5],
'C': [5,3,4,1,1,2,3]})
a_group_desc = df.groupby('A').describe()
print (a_group_desc)
unstacked = a_group_desc.unstack()
print (unstacked)
import nltk
nltk.download()
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
example_sent = "This is a sample sentence, showing off the stop words filtration."
stop_words = set(stopwords.words('english'))
word_tokens = word_tokenize(example_sent)
filtered_sentence = [w for w in word_tokens if not w in stop_words]
print(word_tokens)
print(filtered_sentence)
import networkx as nx
G = nx.cycle_graph(10)
A = nx.adjacency_matrix(G)
print(A.todense())
import numpy as np
import pandas as pd
c = pd.Series(["a", "b", "d", "a", "d"], dtype ="category")
print ("\nCategorical without pandas.Categorical() : \n", c)
c1 = pd.Categorical([1, 2, 3, 1, 2, 3])
print ("\n\nc1 : ", c1)
c2 = pd.Categorical(['e', 'm', 'f', 'i', 'f', 'e', 'h', 'm' ])
print ("\nc2 : ", c2)
import sys
sys.getdefaultencoding( )
from scipy.sparse import csc_matrix
print (csc_matrix([1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0]))
sklearn_hashing_trick = txt.HashingVectorizer( n_features=20, binary=True,norm=None)
text_vector = sklearn_hashing_trick.transform( ['Python for data science','Python for machine learning'])
text_vector
from sklearn.feature_extraction.text import CountVectorizer
document = ["One Geek helps Two Geeks",
"Two Geeks help Four Geeks",
"Each Geek helps many other Geeks at GeeksforGeeks"]
# Create a Vectorizer Object
vectorizer = CountVectorizer()
vectorizer.fit(document)
# Printing the identified Unique words along with their indices
print("Vocabulary: ", vectorizer.vocabulary_)
# Encode the Document
vector = vectorizer.transform(document)
# Summarizing the Encoded Texts
print("Encoded Document is:")
print(vector.toarray())
from sklearn.feature_extraction.text import HashingVectorizer
document = ["One Geek helps Two Geeks",
"Two Geeks help Four Geeks",
"Each Geek helps many other Geeks at GeeksforGeeks"]
# Create a Vectorizer Object
vectorizer = HashingVectorizer()
vectorizer.fit(document)
# Encode the Document
vector = vectorizer.transform(document)
# Summarizing the Encoded Texts
print("Encoded Document is:")
print(vector.toarray())
from sklearn.datasets import load_digits
digits = load_digits()
X, y = digits.data,digits.target
from sklearn.svm import SVC
from sklearn.model_selection import cross_val_score
%timeit single_core_learning = cross_val_score(SVC(), X,y, cv=20, n_jobs=1)
%timeit multi_core_learning = cross_val_score(SVC(), X, y, cv=20, n_jobs=-1)from sklearn.datasets import load_iris
iris = load_iris()
from sklearn.datasets import load_iris
iris = load_iris()
import pandas as pd
import numpy as np
iris_nparray = iris.data
iris_dataframe = pd.DataFrame(iris.data, columns=iris.feature_names)
iris_dataframe['group'] = pd.Series([iris.target_names[k] for k in iris.target],dtype="category")
print (iris_dataframe.mean(numeric_only=True))
print (iris_dataframe.median(numeric_only=True))
print (iris_dataframe.std())
print (iris_dataframe.max(numeric_only=True)-iris_dataframe.min(numeric_only=True) )
print (iris_dataframe.quantile(np.array([0,.25,.50,.75,1])))
pip install scipy
from scipy.stats import kurtosis, kurtosistest
k = kurtosis(iris_dataframe['petal length (cm)'])
zscore, pvalue = kurtosistest(iris_dataframe['petal length (cm)'])
print ('Kurtosis %0.3f\nz-score %0.3f\np-value %0.3f' % (k, zscore, pvalue) )
from scipy.stats import skew, skewtest
s = skew(iris_dataframe['petal length (cm)'])
zscore, pvalue = skewtest(iris_dataframe['petal length (cm)'])
print ('Skewness %0.3f\nz-score %0.3f\np-value %0.3f' % (s, zscore, pvalue))
iris_binned = pd.concat([
pd.qcut(iris_dataframe.iloc[:,0], [0, .25, .5, .75, 1]),
pd.qcut(iris_dataframe.iloc[:,1], [0, .25, .5, .75, 1]),
pd.qcut(iris_dataframe.iloc[:,2], [0, .25, .5, .75, 1]),
pd.qcut(iris_dataframe.iloc[:,3], [0, .25, .5, .75, 1]),
], join='outer', axis = 1)
print(iris_dataframe['group'].value_counts())
print(iris_binned['petal length (cm)'].value_counts())
print(iris_binned.describe())
print (pd.crosstab(iris_dataframe['group'], iris_binned['petal length (cm)']) )
boxplots = iris_dataframe.boxplot(return_type='axes')
from scipy.stats import ttest_ind
group0 = iris_dataframe['group'] == 'setosa'
group1 = iris_dataframe['group'] == 'versicolor'
group2 = iris_dataframe['group'] == 'virginica'
print('var1 %0.3f var2 %03f' % (iris_dataframe['petal length (cm)'][group1].var(),iris_dataframe['petal length (cm)'][group2].var()))
t, pvalue = ttest_ind(iris_dataframe['sepal width (cm)'][group1], iris_dataframe['sepal width (cm)'][group2], axis=0, equal_var=False)
print('t statistic %0.3f p-value %0.3f' % (t, pvalue))
from scipy.stats import f_oneway
f, pvalue = f_oneway(iris_dataframe['sepal width (cm)'][group0],iris_dataframe['sepal width (cm)'][group1],iris_dataframe['sepal width (cm)'][group2])
print("One-way ANOVA F-value %0.3f p-value %0.3f" % (f,pvalue))
from pandas.plotting import parallel_coordinates
iris_dataframe['labels'] = [iris.target_names[k] for k in iris_dataframe['group']]
pll = parallel_coordinates(iris_dataframe,'labels')
densityplot = iris_dataframe[iris_dataframe.columns[:4]].plot(kind='density’)
single_distribution = iris_dataframe['petal length (cm)'].plot(kind='hist')
simple_scatterplot = iris_dataframe.plot(kind='scatter', x='petal length (cm)', y='petal width (cm)')
from pandas import scatter_matrix
matrix_of_scatterplots = scatter_matrix(iris_dataframe, figsize=(6, 6),diagonal='kde')
from sklearn.datasets import load_iris
iris = load_iris()
import pandas as pd
import numpy as np
iris_nparray = iris.data
iris_dataframe = pd.DataFrame(iris.data, columns=iris.feature_names)
iris_dataframe['group'] = pd.Series([iris.target_names[k] for k in iris.target],
dtype="category")
print(iris_dataframe['group'])
from scipy.stats import spearmanr
from scipy.stats.stats import pearsonr
spearmanr_coef, spearmanr_p = spearmanr(iris_dataframe['sepal length (cm)'],iris_dataframe['sepal width (cm)'])
pearsonr_coef, pearsonr_p = pearsonr(iris_dataframe['sepal length (cm)'],iris_dataframe['sepal width (cm)'])
print ('Pearson correlation %0.3f | Spearman correlation %0.3f' % (pearsonr_coef,spearmanr_coef))
from scipy.stats import chi2_contingency
table = pd.crosstab(iris_dataframe['group'], iris_binned['petal length (cm)'])
chi2, p, dof, expected = chi2_contingency(table.values)
print('Chi-square %0.2f p-value %0.3f' % (chi2, p))
from sklearn.preprocessing import scale
stand_sepal_width = scale(iris_dataframe['sepal width (cm)'])
import matplotlib.pyplot as plt
values = [5, 8, 9, 10, 4, 7]
colors = ['b', 'g', 'r', 'c', 'm', 'y']
labels = ['A', 'B', 'C', 'D', 'E', 'F']
explode = (0, 0.2, 0, 0, 0, 0)
plt.pie(values, colors=colors, labels=labels, explode=explode, shadow=True, autopct='%1.2f%%')
plt.title('Values')
plt.show()
import matplotlib.pyplot as plt
values = [5, 8, 9, 10, 4, 7]
widths = [0.7, 0.8, 0.7, 0.7, 0.7, 0.7]
colors = ['b', 'r', 'b', 'b', 'b', 'b']
plt.bar(range(0, 6), values, width=widths,color=colors, align='center')
plt.show()
import numpy as np
import matplotlib.pyplot as plt
x = 100 * np.random.randn(10000)
plt.hist(x, histtype='stepfilled', color='g',label='TestData')
plt.legend()
plt.title('Step Filled Histogram')
plt.show()
import numpy as np
import matplotlib.pyplot as plt
x = 100 * np.random.randn(1000)
plt.boxplot(x)
plt.title('Step Filled Histogram')
plt.show()
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.pylab as plb
x1 = 15 * np.random.rand(50)
x2 = 15 * np.random.rand(50) + 15
x3 = 30 * np.random.rand(30)
x = np.concatenate((x1, x2, x3))
y1 = 15 * np.random.rand(50)
y2 = 15 * np.random.rand(50) + 15
y3 = 30 * np.random.rand(30)
y = np.concatenate((y1, y2, y3))
color_array = ['b'] * 50 + ['g'] * 50 + ['r'] * 30
plt.scatter(x, y, s=[90], marker='*', c=color_array)
z = np.polyfit(x, y, 1)
p = np.poly1d(z)
plb.plot(x, p(x), 'm-')
plt.show()
import matplotlib.pyplot as plt
import datetime
import numpy as np
x = np.array([datetime.datetime(2021, 1, 1, i, 0) for i in range(24)])
y = np.random.randint(100, size=x.shape)
plt.plot(x,y)
plt.show()
from sklearn.datasets import fetch_20newsgroups
import sklearn.feature_extraction.text as ext
categories = ['sci.space']
twenty_train = fetch_20newsgroups(subset='train',categories=categories,remove=('headers', 'footers', 'quotes’), shuffle=True,random_state=42)
import pandas as pd
import numpy as np
df = pd.DataFrame({'A': [2,1,2,3,3,5,4], 'B': [1,2,3,5,4,2,5], 'C': [5,3,4,1,1,2,3]})
df = df.sort_index(by=['A', 'B'], ascending=[True, True])
df = df.reset_index(drop=True)
df
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/allanstar-byte/ESTRELLA/blob/master/SQL_WORLD_SUICIDE_ANALYTICS.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# **SQL DATA CLEANING, OUTLIERS AND ANALYTICS**
# **1. Connecting to our Database**
```
#loading the sql extension into our environment
%load_ext sql
# Then connect to our in memory sqlite database
%sql sqlite://
```
# **2. Importing Data from CSV files**
The dataset we will use contains suicide cases from different countries in the world with different generations, age groups and other factors as outlined below.
```
# Importing the pandas library
# We will use a function read_csv from pandas to read our datasets as shown
#
import pandas as pd
# Loading our table from the respective CSV files
with open('/content/Suicide.csv','r') as f:
Suicide = pd.read_csv(f, index_col=0, encoding='utf-8')
%sql DROP TABLE if EXISTS Suicide
%sql PERSIST Suicide;
%sql SELECT * FROM Suicide LIMIT 5;
```
# **3. Analytics**
```
#1. identifying top 5 countries with the highest suicide cases in the world
%%sql
SELECT Country,
SUM (Suicides_no)
FROM Suicide
GROUP BY Country
ORDER BY SUM (Suicides_no) DESC
limit 5;
#2. identifying top 5 countries with the lowest suicide cases in the world
%%sql
SELECT Country,
SUM (Suicides_no)
FROM Suicide
GROUP BY Country
ORDER BY SUM (Suicides_no) ASC
limit 5;
#3. identifying the generation with the highest suicide cases
%%sql
SELECT Generation,
SUM (Suicide_rate)
FROM Suicide
GROUP BY Generation
ORDER BY SUM (Suicide_rate) DESC
limit 5;
#4. identifying the generations with the lowest suicide cases
%%sql
SELECT Generation,
SUM (Suicide_rate)
FROM Suicide
GROUP BY Generation
ORDER BY SUM (Suicide_rate) ASC
limit 5;
#5 Investigating which gender has more suicide rates compared to the other one
%%sql
SELECT Sex,
SUM (Suicides_no)
FROM Suicide
GROUP BY Sex
ORDER BY SUM (Suicides_no) DESC
limit 5;
#6. Knowing the age group which most people commit suicide
%%sql
SELECT Age,
SUM (Suicides_no)
FROM Suicide
GROUP BY Age
ORDER BY SUM (Suicide_rate) DESC
limit 5;
#7. Finding out the year where people committed suicide the most
%%sql
SELECT Year,
SUM (Suicides_no)
FROM Suicide
GROUP BY Year
ORDER BY SUM (Suicides_no) DESC
limit 5;
#8. Finding which countries has the most suicides comited at every 100,000
%%sql
SELECT Country,
SUM (Suicides_per_hundred_thousand_pop)
FROM Suicide
GROUP BY Country
ORDER BY SUM (Suicides_per_hundred_thousand_pop) DESC
limit 5;
#9. Finding which countries has the leas suicides comited at every 100,000
%%sql
SELECT Country,
SUM (Suicides_per_hundred_thousand_pop)
FROM Suicide
GROUP BY Country
ORDER BY SUM (Suicides_per_hundred_thousand_pop) ASC
limit 7;
#10. Finding which Age groups has the most suicides commited at every 100,000
%%sql
SELECT Age,
SUM (Suicides_per_hundred_thousand_pop)
FROM Suicide
GROUP BY Age
ORDER BY SUM (Suicides_per_hundred_thousand_pop) DESC
limit 5;
```
|
github_jupyter
|
```
import json
import joblib
import pickle
import pandas as pd
from lightgbm import LGBMClassifier
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline, Pipeline
from sklearn.preprocessing import OneHotEncoder
from sklearn.compose import ColumnTransformer
from sklearn.impute import SimpleImputer
from sklearn.metrics import precision_score, recall_score
import numpy as np
from sklearn.metrics import precision_recall_curve
import matplotlib.pyplot as plt
df = pd.read_csv("data/train_searched.csv")
df.head()
# lowercaes departments and location names
df['Department Name'] = df['Department Name'].apply(lambda x: str(x).lower())
df['InterventionLocationName'] = df['InterventionLocationName'].apply(lambda x: str(x).lower())
train_features = df.columns.drop(['VehicleSearchedIndicator', 'ContrabandIndicator'])
categorical_features = train_features.drop(['InterventionDateTime', 'SubjectAge'])
numerical_features = ['SubjectAge']
target = 'ContrabandIndicator'
# show the most common feature values for all the categorical features
for feature in categorical_features:
display(df[feature].value_counts())
# I'm going to remove less common features.
# Let's create a dictionary with the minimum required number of appearences
min_frequency = {
"Department Name": 50,
"InterventionLocationName": 50,
"ReportingOfficerIdentificationID": 30,
"StatuteReason": 10
}
def filter_values(df: pd.DataFrame, column_name: str, threshold: int):
value_counts = df[column_name].value_counts()
to_keep = value_counts[value_counts > threshold].index
filtered = df[df[column_name].isin(to_keep)]
return filtered
df.shape
for feature, threshold in min_frequency.items():
df = filter_values(df, feature, threshold)
df.shape
X = df[train_features]
y = df[target]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=42)
categorical_transformer = Pipeline(steps=[
('imputer', SimpleImputer(strategy='constant', fill_value='missing')),
('onehot', OneHotEncoder(handle_unknown='ignore'))])
preprocessor = ColumnTransformer(
transformers=[('cat', categorical_transformer, categorical_features)])
pipeline = make_pipeline(
preprocessor,
LGBMClassifier(n_jobs=-1, random_state=42),
)
pipeline.fit(X_train, y_train)
preds = pipeline.predict(X_test)
def verify_success_rate_above(y_true, y_pred, min_success_rate=0.5):
"""
Verifies the success rate on a test set is above a provided minimum
"""
precision = precision_score(y_true, y_pred, pos_label=True)
is_satisfied = (precision >= min_success_rate)
return is_satisfied, precision
def verify_amount_found(y_true, y_pred):
"""
Verifies the amout of contraband found in the test dataset - a.k.a the recall in our test set
"""
recall = recall_score(y_true, y_pred)
return recall
verify_success_rate_above(y_test, preds)
verify_amount_found(y_test, preds)
```
Now let's find the best threshold for our requirements.
Precision needs to be at least 0.5, and recall has to be as max as possible.
It's usually true that the bigger is precision, the lower is the recall.
So we need to find the threshold that coresponds to precision = 0.5
```
proba = pipeline.predict_proba(X_test)
precision, recall, thresholds = precision_recall_curve(y_test, proba[:, 1])
print(len(precision), len(recall), len(thresholds))
# according to documentation, precision and recall
# have 1 and 0 at the end, so we should remove them before plotting.
precision = precision[:-1]
recall = recall[:-1]
fig=plt.figure()
ax1 = plt.subplot(211)
ax2 = plt.subplot(212)
ax1.hlines(y=0.5,xmin=0, xmax=1, colors='red')
ax1.plot(thresholds,precision)
ax2.plot(thresholds,recall)
ax1.get_shared_x_axes().join(ax1, ax2)
ax1.set_xticklabels([])
ax1.set_title('Precision')
ax2.set_title('Recall')
plt.xlabel('Threshold')
plt.show()
```
Red line shows the point where precision is equal 0.5.
It looks like the biggest recall for precision >= 0.5 is around 0.2
Let's find the exact value.
```
min_index = [i for i, prec in enumerate(precision) if prec >= 0.5][0]
print(min_index)
precision[min_index]
recall[min_index]
thresholds[min_index]
best_preds = [1 if pred > thresholds[min_index] else 0 for pred in proba[:, 1]]
verify_success_rate_above(y_test, best_preds)
verify_amount_found(y_test, best_preds)
with open('columns.json', 'w') as fh:
json.dump(X_train.columns.tolist(), fh)
with open('dtypes.pickle', 'wb') as fh:
pickle.dump(X_train.dtypes, fh)
joblib.dump(pipeline, 'pipeline.pickle');
```
|
github_jupyter
|
# Project: Investigate Children Out of School
## Table of Contents
<ul>
<li><a href="#intro">Introduction</a></li>
<li><a href="#wrangling">Data Wrangling</a></li>
<li><a href="#eda">Exploratory Data Analysis</a></li>
<li><a href="#conclusions">Conclusions</a></li>
</ul>
<a id='intro'></a>
## Introduction
> **Key notes**: "Gapminder has collected a lot of information about how people live their lives in different countries, tracked across the years, and on a number of different indicators.
> **Questions to explore**:
><ul>
><li><a href="#q1"> 1. Research Question 1: What is the total numbers of children out of primary school over years, indicate the male and female numbers as well?</a></li>
><li><a href="#q2"> 2. Research Question 2: What is distribution of female children who was out of primary school from 1980 to 1995?</a></li>
><li><a href="#q3"> 3. Research Question 3: What are numbers of children out of school in total, by male and female in China, 1985?</a></li>
><li><a href="#q4"> 4. What are relationship of children out of school of female in China in russian and usa over time? Which has a better trend?</a></li>
><li><a href="#q5"> 5. Research Question 5: What is the overall trend for children out of primary school over the years?</a></li>
```
# Set up import statements for all of the packages that are planed to use;
# Include a 'magic word' so that visualizations are plotted;
# call on dataframe to display the first 5 rows.
import pandas as pd
import numpy as np
import datetime
from statistics import mode
% matplotlib inline
import matplotlib.pyplot as plt
%config InlineBackend.figure_format = 'retina'
import seaborn as sns
sns.set_style('darkgrid')
# Reading an Excel file in python using pandas
# call on dataframe to display the first 5 rows
xl = pd.ExcelFile('Child out of school primary.xlsx')
xl.sheet_names
[u'Data']
df_tot = xl.parse("Data")
df_tot.head()
x2 = pd.ExcelFile('Child out of school primiary female.xlsx')
x2.sheet_names
[u'Data']
df_f = x2.parse("Data")
df_f.head()
x3 = pd.ExcelFile('Child out of school primiary male.xlsx')
x3.sheet_names
[u'Data']
df_m = x3.parse("Data")
df_m.head()
# Check if the three dataframe have the same shape
df_tot.shape, df_m.shape, df_f.shape
# Check if the first columns from the 3 dataframe are exactly the same
assert (df_tot['Children out of school, primary'].tolist() == df_m['Children out of school, primary, male'].tolist()\
== df_f['Children out of school, primary, female'].tolist())
# Merge the 3 dataframe
df1 = df_tot.merge(df_f, how='outer', left_index = True, right_index = True)
df1 = df1.merge(df_m, how='outer', left_index = True, right_index = True)
# Confirm changes
df1.shape
```
<a id='wrangling'></a>
## Data Wrangling
> **Key notes**: In this section of the report, the following work will be done: load the data; check for cleanliness; trim and clean dataset for analysis.
### General Properties
```
# return the datatypes of the columns.
df1.dtypes
# check for duplicates in the data.
sum(df1.duplicated())
# check if any value is NaN in DataFrame and in how many columns
df1.isnull().any().any(), sum(df1.isnull().any())
# Generates descriptive statistics, excluding NaN values.
df1.describe()
```
### Data Cleaning
```
# Locate the columns whose NaN values needs to be treated
col = df1.drop(['Children out of school, primary', 'Children out of school, primary, female'\
, 'Children out of school, primary, male'], axis=1)
# Replace NaN with mean
for c in col:
c_mean = df1[c].mean()
df1[c].fillna(c_mean, inplace = True)
# Confirm changes
df1.isnull().any().any()
# Rename column for simplification
df1.rename(columns = {'Children out of school, primary':'country'}, inplace = True)
# check the new dataframe
df1.head()
```
<a id='eda'></a>
## Exploratory Data Analysis
<a id='q1'></a>
### Research Question 1: What is the total numbers of children out of primary school over years, indicate the male and female numbers as well?
```
# Get the sum for each group
sum_tot = df1.iloc[:, 1:43]
m_tot = df1.iloc[:, 44:86]
f_tot = df1.iloc[:, 87:]
tot = []
for t in sum_tot.columns:
tot.append(sum_tot[t].sum())
m = []
for ma in m_tot.columns:
m.append(m_tot[ma].sum())
f = []
for fa in f_tot.columns:
f.append(f_tot[fa].sum())
# Plot
x = ['total number', 'male number', 'female number']
y = [sum(tot), sum(m), sum(f)]
plt.subplots(figsize=(10,6))
sns.barplot(x,y, alpha = 0.8);
```
<a id='q2'></a>
### Research Question 2: What is distribution of female children who was out of primary school from 1980 to 1995?
```
# Target the year and plot
sum_tot1 = sum_tot.iloc[:, 10:26]
new_col = []
for ele in sum_tot1.columns:
new_col.append(ele.split('_x')[0])
sum_tot1.columns = new_col
plt.figure(figsize=(20,15))
sns.boxplot(data = sum_tot1);
```
<a id='q3'></a>
### Research Question 3: What are numbers of children out of school in total, by male and female in China, 1985?
```
china = df1.copy()
china = china.set_index('country')
tot_chi = china.loc['China', '1985_x']
f_chi = china.loc['China', '1985_y']
m_chi = china.loc['China', '1985']
print('The numbers of children out of school in total, by male and female in China were \
{0:.0f}, {1:.0f} and {2:.0f} in 1985, respectively.'.format(tot_chi, f_chi, m_chi))
```
<a id='q4'></a>
### Research Question 4: What are relationship of children out of school of female in China in russian and usa over time? Which has a better trend?
```
rus_us = df1.iloc[:, 0:42].copy()
new_col1 = []
for ele in rus_us:
new_col1.append(ele.split('_x')[0])
rus_us.columns = new_col1
rus_us = rus_us.set_index('country')
rus_us_df = pd.DataFrame(columns=['USA','Russia'])
rus_us_df['USA'] = rus_us.loc['United States'].values
rus_us_df['Russia'] = rus_us.loc['Russia'].values
sns.lmplot(x = 'USA', y = 'Russia', data = rus_us_df);
sns.boxplot(data=rus_us_df);
rus_us_df['year'] = rus_us.columns
rus_us_df.index = rus_us_df.year
rus_us_df.plot();
plt.ylabel('Numers')
plt.xlabel('Country')
plt.title('Numbers of children out of primary school from 1970 to 2011');
```
> There is a positive correlation between children droped out of primary school in Russia and USA. The estimated linear regression is shown as the blue line, the estimates varies in the light blue shade with 95% confident level. The trend of children out of school in USA is much higher than that of Russia over that past 40 years.
<a id='q5'></a>
### Research Question 5: What is the overall trend for children out of primary school over the years?
```
overall_df = pd.DataFrame(columns=['year','numbers'])
overall_df['year'] = rus_us.columns
n_list =[]
for n in rus_us.columns:
n_list.append(rus_us[n].mean())
overall_df['numbers'] = np.array(n_list)
overall_df.index = overall_df.year
overall_df.plot();
plt.ylabel('Numers')
plt.xlabel('Year')
plt.title('Numbers of children out of primary school from 1970 to 2011');
```
> From the analysis we can conclude that the overall trend of children out of primary school had been descreasing starting between 1970 and 1975 at which point of time the numbers fell down dramatically
<a id='conclusions'></a>
## Conclusions
> In current study, a good amount of profound analysis has been carried out. Prior to each step, deailed instructions was given and interpretions was also provided afterwards. The dataset across 41 years from 1970 to 2011.
> The limitations of current study was that the structure is only 275*42 in shape, thus the analysis would not be much reliable due to small scale samples.
> In addition, the parameters in the dataset is very simple, it only focus on the number of children out of school.
```
from subprocess import call
call(['python', '-m', 'nbconvert', 'Investigate_Children_Out_of_School_20180108.ipynb'])
```
|
github_jupyter
|
# Apache Kafka Integration + Preprocessing / Interactive Analysis with KSQL
This notebook uses the combination of Python, Apache Kafka, KSQL for Machine Learning infrastructures.
It includes code examples using ksql-python and other widespread components from Python’s machine learning ecosystem, like Numpy, pandas, TensorFlow and Keras.
The use case is fraud detection for credit card payments. We use a test data set from Kaggle as foundation to train an unsupervised autoencoder to detect anomalies and potential fraud in payments. Focus of this example is not just model training, but the whole Machine Learning infrastructure including data ingestion, data preprocessing, model training, model deployment and monitoring. All of this needs to be scalable, reliable and performant.
If you want to learn more about the relation between the Apache Kafka open source ecosystem and Machine Learning, please check out these two blog posts:
- [How to Build and Deploy Scalable Machine Learning in Production with Apache Kafka](https://www.confluent.io/blog/build-deploy-scalable-machine-learning-production-apache-kafka/)
- [Using Apache Kafka to Drive Cutting-Edge Machine Learning](https://www.confluent.io/blog/using-apache-kafka-drive-cutting-edge-machine-learning)
##### This notebook is not meant to be perfect using all coding and ML best practices, but just a simple guide how to build your own notebooks where you can combine Python APIs with Kafka and KSQL
### Start Backend Services (Zookeeper, Kafka, KSQL)
The only server requirement is a local KSQL server running (with Kafka broker ZK node). If you don't have it running, just use Confluent CLI:
```
# Shows correct startup but does not work 100% yet. Better run this command from outside Jupyter if you have any problems (e.g. from Terminal)!
! confluent start ksql-server
```
## Data Integration and Preprocessing with Python and KSQL
First of all, create the Kafka Topic 'creditcardfraud_source' if it does not exist already:
```
! kafka-topics --zookeeper localhost:2181 --create --topic creditcardfraud_source --partitions 3 --replication-factor 1
```
Then load KSQL library and initiate connection to KSQL server:
```
from ksql import KSQLAPI
client = KSQLAPI('http://localhost:8088')
```
Consume source data from Kafka Topic "creditcardfraud_source":
```
client.create_stream(table_name='creditcardfraud_source',
columns_type=['Id bigint', 'Timestamp varchar', 'User varchar', 'Time int', 'V1 double', 'V2 double', 'V3 double', 'V4 double', 'V5 double', 'V6 double', 'V7 double', 'V8 double', 'V9 double', 'V10 double', 'V11 double', 'V12 double', 'V13 double', 'V14 double', 'V15 double', 'V16 double', 'V17 double', 'V18 double', 'V19 double', 'V20 double', 'V21 double', 'V22 double', 'V23 double', 'V24 double', 'V25 double', 'V26 double', 'V27 double', 'V28 double', 'Amount double', 'Class string'],
topic='creditcardfraud_source',
value_format='DELIMITED')
```
Preprocessing:
- Filter columns which are not needed
- Filter messages where column 'class' is empty
- Change data format to Avro for more convenient further processing
```
client.create_stream_as(table_name='creditcardfraud_preprocessed_avro',
select_columns=['Time', 'V1', 'V2', 'V3', 'V4', 'V5', 'V6', 'V7', 'V8', 'V9', 'V10', 'V11', 'V12', 'V13', 'V14', 'V15', 'V16', 'V17', 'V18', 'V19', 'V20', 'V21', 'V22', 'V23', 'V24', 'V25', 'V26', 'V27', 'V28', 'Amount', 'Class'],
src_table='creditcardfraud_source',
conditions='Class IS NOT NULL',
kafka_topic='creditcardfraud_preprocessed_avro',
value_format='AVRO')
```
Take a look at the creates KSQL Streams:
```
client.ksql('show streams')
```
Take a look at the metadata of the KSQL Stream:
```
client.ksql('describe CREDITCARDFRAUD_PREPROCESSED_AVRO')
```
Interactive query statement:
```
query = client.query('SELECT * FROM CREDITCARDFRAUD_PREPROCESSED_AVRO LIMIT 1')
for item in query:
print(item)
```
Produce single test data manually (if you did not connect to a real data stream which produces data continuously), e.g. from terminal:
confluent produce creditcardfraud_source
1,"2018-12- 18T12:00:00Z","Hans",0,-1.3598071336738,-0.0727811733098497,2.53634673796914,1.37815522427443,-0.338320769942518,0.462387777762292,0.239598554061257,0.0986979012610507,0.363786969611213,0.0907941719789316,-0.551599533260813,-0.617800855762348,-0.991389847235408,-0.311169353699879,1.46817697209427,-0.470400525259478,0.207971241929242,0.0257905801985591,0.403992960255733,0.251412098239705,-0.018306777944153,0.277837575558899,-0.110473910188767,0.0669280749146731,0.128539358273528,-0.189114843888824,0.133558376740387,-0.0210530534538215,149.62,"0"
*BE AWARE: The KSQL Python API does a REST call. This only waits a few seconds by default and then throws a timeout exception. You need to get data into the query before the timeout (e.g. by using above command).*
```
# TODO How to embed ' ' in Python ???
# See https://github.com/bryanyang0528/ksql-python/issues/54
# client.ksql('SET \'auto.offset.reset\'=\'earliest\'');
```
### Additional (optional) analysis and preprocessing examples
Some more examples for possible data wrangling and preprocessing with KSQL:
- Anonymization
- Augmentation
- Merge / Join data frames
```
query = client.query('SELECT Id, MASK_LEFT(User, 2) FROM creditcardfraud_source LIMIT 1')
for item in query:
print(item)
query = client.query('SELECT Id, IFNULL(Class, \'-1\') FROM creditcardfraud_source LIMIT 1')
for item in query:
print(item)
```
#### Stream-Table-Join
For the STREAM-TABLE-JOIN, you first need to create a Kafka Topic 'Users' (for the corresponding KSQL TABLE 'Users):
```
! kafka-topics --zookeeper localhost:2181 --create --topic users --partitions 3 --replication-factor 1
```
Then create the KSQL Table:
```
client.create_table(table_name='users',
columns_type=['userid varchar', 'gender varchar', 'regionid varchar'],
topic='users',
key='userid',
value_format='AVRO')
client.ksql("CREATE STREAM creditcardfraud_per_user WITH (VALUE_FORMAT='AVRO', KAFKA_TOPIC='creditcardfraud_per_user') AS SELECT Time, Amount, Class FROM creditcardfraud_source c INNER JOIN USERS u on c.user = u.userid WHERE u.USERID = 1")
```
# Mapping from KSQL to NumPy / pandas for Machine Learning tasks
```
import numpy as np
import pandas as pd
import json
```
The query below command returns a Python generator. It can be printed e.g. by reading its values via next(query) or a for loop.
Due to a current [bug in ksql-python library](https://github.com/bryanyang0528/ksql-python/issues/57), we need to to an additional line of Python code to strip out unnecessary info and change to 2D array
```
query = client.query('SELECT * FROM CREDITCARDFRAUD_PREPROCESSED_AVRO LIMIT 8') # Returns a Python generator object
#items = [item for item in query][:-1] # -1 to remove last record that is a dummy msg for "Limit Reached"
#one_record = json.loads(''.join(items)) # Join two records as one as ksql-python is splitting it into two?
#data = [one_record['row']['columns'][2:-1]] # Strip out unnecessary info and change to 2D array
#df = pd.DataFrame(data=data)
records = [json.loads(r) for r in ''.join(query).strip().replace('\n\n\n\n', '').split('\n')]
data = [r['row']['columns'][2:] for r in records[:-1]]
#data = r['row']['columns'][2] for r in records
df = pd.DataFrame(data=data, columns=['Time', 'V1' , 'V2' , 'V3' , 'V4' , 'V5' , 'V6' , 'V7' , 'V8' , 'V9' , 'V10' , 'V11' , 'V12' , 'V13' , 'V14' , 'V15' , 'V16' , 'V17' , 'V18' , 'V19' , 'V20' , 'V21' , 'V22' , 'V23' , 'V24' , 'V25' , 'V26' , 'V27' , 'V28' , 'Amount' , 'Class'])
df
```
### Generate some test data
As discussed in the step-by-step guide, you have various options. Here we - ironically - read messages from a CSV file. This is for simple demo purposes so that you don't have to set up a real continuous Kafka stream.
In real world or more advanced examples, you should connect to a real Kafka data stream (for instance using the Kafka data generator or Kafka Connect).
Here we just consume a few messages for demo purposes so that they get mapped into a pandas dataframe:
cat /Users/kai.waehner/git-projects/python-jupyter-apache-kafka-ksql-tensorflow-keras/data/creditcard_extended.csv | kafka-console-producer --broker-list localhost:9092 --topic creditcardfraud_source
You need to do this from command line because Jupyter cannot execute this in parallel to above KSQL query.
# Preprocessing with Pandas + Model Training with TensorFlow / Keras
#### BE AWARE: You need enough messages in the pandas data frame to train the model in the below cells (if you just play around with ksql-python and just add a few Kafka events, it is not a sufficient number of rows to continue. You can simply change to df = pd.read_csv("data/creditcard.csv") as shown below in this case to get a bigger data set...
This part only includes the steps required for model training of the Autoencoder with Keras and TensorFlow.
If you want to get a better understanding of the model, take a look at the other notebook [Python Tensorflow Keras Fraud Detection Autoencoder.ipynb](http://localhost:8888/notebooks/Python%20Tensorflow%20Keras%20Fraud%20Detection%20Autoencoder.ipynb) which includes many more details, plots and explanations.
[Kudos to David Ellison](https://www.datascience.com/blog/fraud-detection-with-tensorflow).
[The credit card fraud data set is available at Kaggle](https://www.kaggle.com/mlg-ulb/creditcardfraud/data).
```
# import packages
# matplotlib inline
#import pandas as pd
#import numpy as np
from scipy import stats
import tensorflow as tf
import matplotlib.pyplot as plt
import seaborn as sns
import pickle
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix, precision_recall_curve
from sklearn.metrics import recall_score, classification_report, auc, roc_curve
from sklearn.metrics import precision_recall_fscore_support, f1_score
from sklearn.preprocessing import StandardScaler
from pylab import rcParams
from keras.models import Model, load_model
from keras.layers import Input, Dense
from keras.callbacks import ModelCheckpoint, TensorBoard
from keras import regularizers
# Use the dataframe from above (imported and preprocessed with KSQL)
# As alternative directly import from a CSV file ("the normal approach without Kafka and streaming data")
# "data/creditcard_small.csv" is a very small data set (just for quick demo purpose to get a model binary)
# => replace with "data/creditcard.csv" to use a real data set to train a model with good accuracy
#df = pd.read_csv("data/creditcard.csv")
df.head(n=5) #just to check you imported the dataset properly
#set random seed and percentage of test data
RANDOM_SEED = 314 #used to help randomly select the data points
TEST_PCT = 0.2 # 20% of the data
#set up graphic style in this case I am using the color scheme from xkcd.com
rcParams['figure.figsize'] = 14, 8.7 # Golden Mean
LABELS = ["Normal","Fraud"]
#col_list = ["cerulean","scarlet"]# https://xkcd.com/color/rgb/
#sns.set(style='white', font_scale=1.75, palette=sns.xkcd_palette(col_list))
normal_df = [df.Class == 0] #save normal_df observations into a separate df
fraud_df = [df.Class == 1] #do the same for frauds
#data = df.drop(['Time'], axis=1) #if you think the var is unimportant
df_norm = df
df_norm['Time'] = StandardScaler().fit_transform(df_norm['Time'].values.reshape(-1, 1))
df_norm['Amount'] = StandardScaler().fit_transform(df_norm['Amount'].values.reshape(-1, 1))
train_x, test_x = train_test_split(df_norm, test_size=TEST_PCT, random_state=RANDOM_SEED)
train_x = train_x[train_x.Class == 0] #where normal transactions
train_x = train_x.drop(['Class'], axis=1) #drop the class column
test_y = test_x['Class'] #save the class column for the test set
test_x = test_x.drop(['Class'], axis=1) #drop the class column
train_x = train_x.values #transform to ndarray
test_x = test_x.values
```
### My Jupyter Notebook crashed sometimes in the next step 'model training' (probably memory issues):
```
# Reduce number of epochs and batch_size if your Jupyter crashes (due to memory issues)
# nb_epoch = 100
# batch_size = 128
nb_epoch = 5
batch_size = 32
input_dim = train_x.shape[1] #num of columns, 30
encoding_dim = 14
hidden_dim = int(encoding_dim / 2) #i.e. 7
learning_rate = 1e-7
input_layer = Input(shape=(input_dim, ))
encoder = Dense(encoding_dim, activation="tanh", activity_regularizer=regularizers.l1(learning_rate))(input_layer)
encoder = Dense(hidden_dim, activation="relu")(encoder)
decoder = Dense(hidden_dim, activation='tanh')(encoder)
decoder = Dense(input_dim, activation='relu')(decoder)
autoencoder = Model(inputs=input_layer, outputs=decoder)
autoencoder.compile(metrics=['accuracy'],
loss='mean_squared_error',
optimizer='adam')
cp = ModelCheckpoint(filepath="models/autoencoder_fraud.h5",
save_best_only=True,
verbose=0)
tb = TensorBoard(log_dir='./logs',
histogram_freq=0,
write_graph=True,
write_images=True)
history = autoencoder.fit(train_x, train_x,
epochs=nb_epoch,
batch_size=batch_size,
shuffle=True,
validation_data=(test_x, test_x),
verbose=1,
callbacks=[cp, tb]).history
autoencoder = load_model('models/autoencoder_fraud.h5')
test_x_predictions = autoencoder.predict(test_x)
mse = np.mean(np.power(test_x - test_x_predictions, 2), axis=1)
error_df = pd.DataFrame({'Reconstruction_error': mse,
'True_class': test_y})
error_df.describe()
```
The binary 'models/autoencoder_fraud.h5' is the trained model which can then be deployed anywhere to do prediction on new incoming events in real time.
# Model Deployment
This demo focuses on the combination of Python and KSQL for data preprocessing and model training. If you want to understand the relation between Apache Kafka, KSQL and Python-related Machine Learning tools like TensorFlow for model deployment and monitoring, please check out my other Github projects:
Some examples of model deployment in Kafka environments:
- [Analytic models (TensorFlow, Keras, H2O and Deeplearning4j) embedded in Kafka Streams microservices](https://github.com/kaiwaehner/kafka-streams-machine-learning-examples)
- [Anomaly detection of IoT sensor data with a model embedded into a KSQL UDF](https://github.com/kaiwaehner/ksql-udf-deep-learning-mqtt-iot)
- [RPC communication between Kafka Streams application and model server (TensorFlow Serving)](https://github.com/kaiwaehner/tensorflow-serving-java-grpc-kafka-streams)
# Appendix: Pandas analysis with above Fraud Detection Data
```
df = pd.read_csv("data/creditcard.csv")
df.head()
df.shape
df.index
df.columns
df.values
df.describe()
df['Amount']
df[0:3]
df.iloc[1,1]
# Takes a minute or two (big CSV file)...
#df.plot()
```
|
github_jupyter
|
# Multi-Layer Perceptron, MNIST
---
In this notebook, we will train an MLP to classify images from the [MNIST database](http://yann.lecun.com/exdb/mnist/) hand-written digit database.
The process will be broken down into the following steps:
>1. Load and visualize the data
2. Define a neural network
3. Train the model
4. Evaluate the performance of our trained model on a test dataset!
Before we begin, we have to import the necessary libraries for working with data and PyTorch.
```
# import libraries
import torch
import numpy as np
```
---
## Load and Visualize the [Data](http://pytorch.org/docs/stable/torchvision/datasets.html)
Downloading may take a few moments, and you should see your progress as the data is loading. You may also choose to change the `batch_size` if you want to load more data at a time.
This cell will create DataLoaders for each of our datasets.
```
from torchvision import datasets
import torchvision.transforms as transforms
from torch.utils.data.sampler import SubsetRandomSampler
# number of subprocesses to use for data loading
num_workers = 0
# how many samples per batch to load
batch_size = 20
# percentage of training set to use as validation
valid_size = 0.2
# convert data to torch.FloatTensor
transform = transforms.ToTensor()
# choose the training and test datasets
train_data = datasets.MNIST(root='data', train=True,
download=True, transform=transform)
test_data = datasets.MNIST(root='data', train=False,
download=True, transform=transform)
# obtain training indices that will be used for validation
num_train = len(train_data)
indices = list(range(num_train))
np.random.shuffle(indices)
split = int(np.floor(valid_size * num_train))
train_idx, valid_idx = indices[split:], indices[:split]
# define samplers for obtaining training and validation batches
train_sampler = SubsetRandomSampler(train_idx)
valid_sampler = SubsetRandomSampler(valid_idx)
# prepare data loaders
train_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=train_sampler, num_workers=num_workers)
valid_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=valid_sampler, num_workers=num_workers)
test_loader = torch.utils.data.DataLoader(test_data, batch_size=batch_size,
num_workers=num_workers)
```
### Visualize a Batch of Training Data
The first step in a classification task is to take a look at the data, make sure it is loaded in correctly, then make any initial observations about patterns in that data.
```
import matplotlib.pyplot as plt
%matplotlib inline
# obtain one batch of training images
dataiter = iter(train_loader)
images, labels = dataiter.next()
images = images.numpy()
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(25, 4))
for idx in np.arange(20):
ax = fig.add_subplot(2, 20/2, idx+1, xticks=[], yticks=[])
ax.imshow(np.squeeze(images[idx]), cmap='gray')
# print out the correct label for each image
# .item() gets the value contained in a Tensor
ax.set_title(str(labels[idx].item()))
```
### View an Image in More Detail
```
img = np.squeeze(images[1])
fig = plt.figure(figsize = (12,12))
ax = fig.add_subplot(111)
ax.imshow(img, cmap='gray')
width, height = img.shape
thresh = img.max()/2.5
for x in range(width):
for y in range(height):
val = round(img[x][y],2) if img[x][y] !=0 else 0
ax.annotate(str(val), xy=(y,x),
horizontalalignment='center',
verticalalignment='center',
color='white' if img[x][y]<thresh else 'black')
```
---
## Define the Network [Architecture](http://pytorch.org/docs/stable/nn.html)
The architecture will be responsible for seeing as input a 784-dim Tensor of pixel values for each image, and producing a Tensor of length 10 (our number of classes) that indicates the class scores for an input image. This particular example uses two hidden layers and dropout to avoid overfitting.
```
import torch.nn as nn
import torch.nn.functional as F
# define the NN architecture
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
# number of hidden nodes in each layer (512)
hidden_1 = 512
hidden_2 = 512
# linear layer (784 -> hidden_1)
self.fc1 = nn.Linear(28 * 28, hidden_1)
# linear layer (n_hidden -> hidden_2)
self.fc2 = nn.Linear(hidden_1, hidden_2)
# linear layer (n_hidden -> 10)
self.fc3 = nn.Linear(hidden_2, 10)
# dropout layer (p=0.2)
# dropout prevents overfitting of data
self.dropout = nn.Dropout(0.2)
def forward(self, x):
# flatten image input
x = x.view(-1, 28 * 28)
# add hidden layer, with relu activation function
x = F.relu(self.fc1(x))
# add dropout layer
x = self.dropout(x)
# add hidden layer, with relu activation function
x = F.relu(self.fc2(x))
# add dropout layer
x = self.dropout(x)
# add output layer
x = self.fc3(x)
return x
# initialize the NN
model = Net()
print(model)
```
### Specify [Loss Function](http://pytorch.org/docs/stable/nn.html#loss-functions) and [Optimizer](http://pytorch.org/docs/stable/optim.html)
It's recommended that you use cross-entropy loss for classification. If you look at the documentation (linked above), you can see that PyTorch's cross entropy function applies a softmax funtion to the output layer *and* then calculates the log loss.
```
# specify loss function (categorical cross-entropy)
criterion = nn.CrossEntropyLoss()
# specify optimizer (stochastic gradient descent) and learning rate = 0.01
optimizer = torch.optim.SGD(model.parameters(), lr=0.01)
```
---
## Train the Network
The steps for training/learning from a batch of data are described in the comments below:
1. Clear the gradients of all optimized variables
2. Forward pass: compute predicted outputs by passing inputs to the model
3. Calculate the loss
4. Backward pass: compute gradient of the loss with respect to model parameters
5. Perform a single optimization step (parameter update)
6. Update average training loss
The following loop trains for 50 epochs; take a look at how the values for the training loss decrease over time. We want it to decrease while also avoiding overfitting the training data.
```
model.to('cuda')
# number of epochs to train the model
n_epochs = 50
# initialize tracker for minimum validation loss
valid_loss_min = np.Inf # set initial "min" to infinity
for epoch in range(n_epochs):
# monitor training loss
train_loss = 0.0
valid_loss = 0.0
###################
# train the model #
###################
model.train() # prep model for training
for data, target in train_loader:
data, target = data.to('cuda'), target.to('cuda')
# clear the gradients of all optimized variables
optimizer.zero_grad()
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the loss
loss = criterion(output, target)
# backward pass: compute gradient of the loss with respect to model parameters
loss.backward()
# perform a single optimization step (parameter update)
optimizer.step()
# update running training loss
train_loss += loss.item()*data.size(0)
######################
# validate the model #
######################
model.eval() # prep model for evaluation
for data, target in valid_loader:
data, target = data.to('cuda'), target.to('cuda')
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the loss
loss = criterion(output, target)
# update running validation loss
valid_loss += loss.item()*data.size(0)
# print training/validation statistics
# calculate average loss over an epoch
train_loss = train_loss/len(train_loader.sampler)
valid_loss = valid_loss/len(valid_loader.sampler)
print('Epoch: {} \tTraining Loss: {:.6f} \tValidation Loss: {:.6f}'.format(
epoch+1,
train_loss,
valid_loss
))
# save model if validation loss has decreased
if valid_loss <= valid_loss_min:
print('Validation loss decreased ({:.6f} --> {:.6f}). Saving model ...'.format(
valid_loss_min,
valid_loss))
torch.save(model.state_dict(), 'model.pt')
valid_loss_min = valid_loss
```
### Load the Model with the Lowest Validation Loss
```
model.load_state_dict(torch.load('model.pt'))
```
---
## Test the Trained Network
Finally, we test our best model on previously unseen **test data** and evaluate it's performance. Testing on unseen data is a good way to check that our model generalizes well. It may also be useful to be granular in this analysis and take a look at how this model performs on each class as well as looking at its overall loss and accuracy.
```
model.to('cpu')
# initialize lists to monitor test loss and accuracy
test_loss = 0.0
class_correct = list(0. for i in range(10))
class_total = list(0. for i in range(10))
model.eval() # prep model for evaluation
for data, target in test_loader:
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the loss
loss = criterion(output, target)
# update test loss
test_loss += loss.item()*data.size(0)
# convert output probabilities to predicted class
_, pred = torch.max(output, 1)
# compare predictions to true label
correct = np.squeeze(pred.eq(target.data.view_as(pred)))
# calculate test accuracy for each object class
for i in range(len(target)):
label = target.data[i]
class_correct[label] += correct[i].item()
class_total[label] += 1
# calculate and print avg test loss
test_loss = test_loss/len(test_loader.sampler)
print('Test Loss: {:.6f}\n'.format(test_loss))
for i in range(10):
if class_total[i] > 0:
print('Test Accuracy of %5s: %2d%% (%2d/%2d)' % (
str(i), 100 * class_correct[i] / class_total[i],
np.sum(class_correct[i]), np.sum(class_total[i])))
else:
print('Test Accuracy of %5s: N/A (no training examples)' % (classes[i]))
print('\nTest Accuracy (Overall): %2d%% (%2d/%2d)' % (
100. * np.sum(class_correct) / np.sum(class_total),
np.sum(class_correct), np.sum(class_total)))
```
### Visualize Sample Test Results
This cell displays test images and their labels in this format: `predicted (ground-truth)`. The text will be green for accurately classified examples and red for incorrect predictions.
```
# obtain one batch of test images
dataiter = iter(test_loader)
images, labels = dataiter.next()
# get sample outputs
output = model(images)
# convert output probabilities to predicted class
_, preds = torch.max(output, 1)
# prep images for display
images = images.numpy()
# plot the images in the batch, along with predicted and true labels
fig = plt.figure(figsize=(25, 4))
for idx in np.arange(20):
ax = fig.add_subplot(2, 20/2, idx+1, xticks=[], yticks=[])
ax.imshow(np.squeeze(images[idx]), cmap='gray')
ax.set_title("{} ({})".format(str(preds[idx].item()), str(labels[idx].item())),
color=("green" if preds[idx]==labels[idx] else "red"))
```
|
github_jupyter
|
# US Production Data for RBC Modeling
```
import pandas as pd
import numpy as np
import fredpy as fp
import matplotlib.pyplot as plt
plt.style.use('classic')
%matplotlib inline
pd.plotting.register_matplotlib_converters()
# Load API key
fp.api_key = fp.load_api_key('fred_api_key.txt')
# Download nominal GDP, nominal personal consumption expenditures, nominal
# gross private domestic investment, the GDP deflator, and an index of hours
# worked in the nonfarm business sector produced by the BLS. All data are
# from FRED and are quarterly.
gdp = fp.series('GDP')
cons = fp.series('PCEC')
invest = fp.series('GPDI')
hours = fp.series('HOANBS')
defl = fp.series('GDPDEF')
pcec = fp.series('PCEC')
m2 = fp.series('M2SL')
tb3mo = fp.series('TB3MS')
unemp = fp.series('UNRATE')
# Convert monthly M2, 3-mo T-Bill, and unemployment to quarterly
m2 = m2.as_frequency('Q')
tb3mo = tb3mo.as_frequency('Q')
unemp = unemp.as_frequency('Q')
# Convert unemployment and t-bill data to decimals instead of percents
unemp.data = unemp.data/100
tb3mo.data = tb3mo.data/100
# pcec inflation as pecent change over past year
pcec = pcec.apc()
pcec.data = pcec.data/100
# Make sure that all of the downloaded series have the same data ranges
gdp,cons,invest,hours,defl,pcec,m2,tb3mo,unemp = fp.window_equalize([gdp,cons,invest,hours,defl,pcec,m2,tb3mo,unemp])
# Compute real GDP, real consumption, real investment
gdp.data = gdp.data/defl.data*100
cons.data = cons.data/defl.data*100
invest.data = invest.data/defl.data*100
m2.data = m2.data/defl.data*100
# Print units
print('Hours units: ',hours.units)
print('Deflator units:',defl.units)
```
Next, compute the quarterly capital stock series for the US using the perpetual inventory method. The discrete-time Solow growth model is given by:
\begin{align}
Y_t & = A_tK_t^{\alpha}L_t^{1-\alpha} \tag{1}\\
C_t & = (1-s)Y_t \tag{2}\\
Y_t & = C_t + I_t \tag{3}\\
K_{t+1} & = I_t + (1-\delta)K_t \tag{4}\\
A_{t+1} & = (1+g)A_t \tag{5}\\
L_{t+1} & = (1+n)L_t \tag{6}.
\end{align}
Here the model is assumed to be quarterly so $n$ is the *quarterly* growth rate of labor hours, $g$ is the *quarterly* growth rate of TFP, and $\delta$ is the *quarterly* rate of depreciation of the capital stock. Given a value of the quarterly depreciation rate $\delta$, an investment series $I_t$, and an initial capital stock $K_0$, the law of motion for the capital stock, Equation (4), can be used to compute an implied capital series. But we don't know $K_0$ or $\delta$ so we'll have to *calibrate* these values using statistics computed from the data that we've already obtained.
Let lowercase letters denote a variable that's been divided by $A_t^{1/(1-\alpha)}L_t$. E.g.,
\begin{align}
y_t = \frac{Y_t}{A_t^{1/(1-\alpha)}L_t}\tag{7}
\end{align}
Then (after substituting consumption from the model), the scaled version of the model can be written as:
\begin{align}
y_t & = k_t^{\alpha} \tag{8}\\
i_t & = sy_t \tag{9}\\
k_{t+1} & = i_t + (1-\delta-n-g')k_t,\tag{10}
\end{align}
where $g' = g/(1-\alpha)$ is the growth rate of $A_t^{1/(1-\alpha)}$. In the steady state:
\begin{align}
k & = \left(\frac{s}{\delta+n+g'}\right)^{\frac{1}{1-\alpha}} \tag{11}
\end{align}
which means that the ratio of capital to output is constant:
\begin{align}
\frac{k}{y} & = \frac{s}{\delta+n+g'} \tag{12}
\end{align}
and therefore the steady state ratio of depreciation to output is:
\begin{align}
\overline{\delta K/ Y} & = \frac{\delta s}{\delta + n + g'} \tag{13}
\end{align}
where $\overline{\delta K/ Y}$ is the long-run average ratio of depreciation to output. We can use Equation (13) to calibrate $\delta$ given $\overline{\delta K/ Y}$, $s$, $n$, and $g'$.
Furthermore, in the steady state, the growth rate of output is constant:
\begin{align}
\frac{\Delta Y}{Y} & = n + g' \tag{14}
\end{align}
1. Assume $\alpha = 0.35$.
2. Calibrate $s$ as the average of ratio of investment to GDP.
3. Calibrate $n$ as the average quarterly growth rate of labor hours.
4. Calibrate $g'$ as the average quarterly growth rate of real GDP minus n.
5. Calculate the average ratio of depreciation to GDP $\overline{\delta K/ Y}$ and use the result to calibrate $\delta$. That is, find the average ratio of Current-Cost Depreciation of Fixed Assets (FRED series ID: M1TTOTL1ES000) to GDP (FRED series ID: GDPA). Then calibrate $\delta$ from the following steady state relationship:
\begin{align}
\delta & = \frac{\left( \overline{\delta K/ Y} \right)\left(n + g' \right)}{s - \left( \overline{\delta K/ Y} \right)} \tag{15}
\end{align}
6. Calibrate $K_0$ by asusming that the capital stock is initially equal to its steady state value:
\begin{align}
K_0 & = \left(\frac{s}{\delta + n + g'}\right) Y_0 \tag{16}
\end{align}
Then, armed with calibrated values for $K_0$ and $\delta$, compute $K_1, K_2, \ldots$ recursively. See Timothy Kehoe's notes for more information on the perpetual inventory method:
http://users.econ.umn.edu/~tkehoe/classes/GrowthAccountingNotes.pdf
```
# Set the capital share of income
alpha = 0.35
# Average saving rate
s = np.mean(invest.data/gdp.data)
# Average quarterly labor hours growth rate
n = (hours.data[-1]/hours.data[0])**(1/(len(hours.data)-1)) - 1
# Average quarterly real GDP growth rate
g = ((gdp.data[-1]/gdp.data[0])**(1/(len(gdp.data)-1)) - 1) - n
# Compute annual depreciation rate
depA = fp.series('M1TTOTL1ES000')
gdpA = fp.series('gdpa')
gdpA = gdpA.window([gdp.data.index[0],gdp.data.index[-1]])
gdpA,depA = fp.window_equalize([gdpA,depA])
deltaKY = np.mean(depA.data/gdpA.data)
delta = (n+g)*deltaKY/(s-deltaKY)
# print calibrated values:
print('Avg saving rate: ',round(s,5))
print('Avg annual labor growth:',round(4*n,5))
print('Avg annual gdp growth: ',round(4*g,5))
print('Avg annual dep rate: ',round(4*delta,5))
# Construct the capital series. Note that the GPD and investment data are reported on an annualized basis
# so divide by 4 to get quarterly data.
capital = np.zeros(len(gdp.data))
capital[0] = gdp.data[0]/4*s/(n+g+delta)
for t in range(len(gdp.data)-1):
capital[t+1] = invest.data[t]/4 + (1-delta)*capital[t]
# Save in a fredpy series
capital = fp.to_fred_series(data = capital,dates =gdp.data.index,units = gdp.units,title='Capital stock of the US',frequency='Quarterly')
# plot the computed capital series
plt.plot(capital.data.index,capital.data,'-',lw=3,alpha = 0.7)
plt.ylabel(capital.units)
plt.title(capital.title)
plt.grid()
# Compute TFP
tfp = gdp.data/capital.data**alpha/hours.data**(1-alpha)
tfp = fp.to_fred_series(data = tfp,dates =gdp.data.index,units = gdp.units,title='TFP of the US',frequency='Quarterly')
# Plot the computed capital series
plt.plot(tfp.data.index,tfp.data,'-',lw=3,alpha = 0.7)
plt.ylabel(tfp.units)
plt.title(tfp.title)
plt.grid()
# Convert each series into per capita using civilian pop 16 and over
gdp = gdp.per_capita(civ_pop=True)
cons = cons.per_capita(civ_pop=True)
invest = invest.per_capita(civ_pop=True)
hours = hours.per_capita(civ_pop=True)
capital = capital.per_capita(civ_pop=True)
m2 = m2.per_capita(civ_pop=True)
# Put GDP, consumption, investment, and M2 in units of thousands of dollars per person
gdp.data = gdp.data*1000
cons.data = cons.data*1000
invest.data = invest.data*1000
capital.data = capital.data*1000
m2.data = m2.data/1000
# Scale hours per person to equal 100 on October (Quarter III) of 2012
hours.data = hours.data/hours.data.loc['2012-10-01']*100
# Compute and plot log real GDP, log consumption, log investment, log hours
gdp_log = gdp.log()
cons_log = cons.log()
invest_log = invest.log()
hours_log = hours.log()
capital_log = capital.log()
tfp_log = tfp.log()
m2_log = m2.log()
m2_log = m2.log()
# HP filter to isolate trend and cyclical components
gdp_log_cycle,gdp_log_trend = gdp_log.hp_filter()
cons_log_cycle,cons_log_trend = cons_log.hp_filter()
invest_log_cycle,invest_log_trend = invest_log.hp_filter()
hours_log_cycle,hours_log_trend = hours_log.hp_filter()
capital_log_cycle,capital_log_trend = capital_log.hp_filter()
tfp_log_cycle,tfp_log_trend = tfp_log.hp_filter()
m2_log_cycle,m2_log_trend = m2_log.hp_filter()
tb3mo_cycle,tb3mo_trend = tb3mo.hp_filter()
unemp_cycle,unemp_trend = unemp.hp_filter()
pcec_cycle,pcec_trend = pcec.hp_filter()
# Create a DataFrame with actual and trend data
data = pd.DataFrame({
'gdp':gdp.data,
'gdp_trend':np.exp(gdp_log_trend.data),
'gdp_cycle':gdp_log_cycle.data,
'consumption':cons.data,
'consumption_trend':np.exp(cons_log_trend.data),
'consumption_cycle':cons_log_cycle.data,
'investment':invest.data,
'investment_trend':np.exp(invest_log_trend.data),
'investment_cycle':invest_log_cycle.data,
'hours':hours.data,
'hours_trend':np.exp(hours_log_trend.data),
'hours_cycle':hours_log_cycle.data,
'capital':capital.data,
'capital_trend':np.exp(capital_log_trend.data),
'capital_cycle':capital_log_cycle.data,
'tfp':tfp.data,
'tfp_trend':np.exp(tfp_log_trend.data),
'tfp_cycle':tfp_log_cycle.data,
'real_m2':m2.data,
'real_m2_trend':np.exp(m2_log_trend.data),
'real_m2_cycle':m2_log_cycle.data,
't_bill_3mo':tb3mo.data,
't_bill_3mo_trend':tb3mo_trend.data,
't_bill_3mo_cycle':tb3mo_cycle.data,
'pce_inflation':pcec.data,
'pce_inflation_trend':pcec_trend.data,
'pce_inflation_cycle':pcec_cycle.data,
'unemployment':unemp.data,
'unemployment_trend':unemp_trend.data,
'unemployment_cycle':unemp_cycle.data,
},index = gdp.data.index)
# # RBC Data
# columns_ordered =[]
# names = ['gdp','consumption','investment','hours','capital','tfp']
# for name in names:
# columns_ordered.append(name)
# columns_ordered.append(name+'_trend')
# data[columns_ordered].to_csv('../Csv/rbc_data_actual_trend.csv')
# # Create a DataFrame with actual, trend, and cycle data
# columns_ordered =[]
# names = ['gdp','consumption','investment','hours','capital','tfp']
# for name in names:
# columns_ordered.append(name)
# columns_ordered.append(name+'_trend')
# columns_ordered.append(name+'_cycle')
# data[columns_ordered].to_csv('../Csv/rbc_data_actual_trend_cycle.csv')
# Business Cycle Data
columns_ordered =[]
names = ['gdp','consumption','investment','hours','capital','tfp','real_m2','t_bill_3mo','pce_inflation','unemployment']
for name in names:
columns_ordered.append(name)
columns_ordered.append(name+'_trend')
data[columns_ordered].to_csv('../Csv/business_cycle_data_actual_trend.csv')
# Create a DataFrame with actual, trend, and cycle data
columns_ordered =[]
names = ['gdp','consumption','investment','hours','capital','tfp','real_m2','t_bill_3mo','pce_inflation','unemployment']
for name in names:
columns_ordered.append(name)
columns_ordered.append(name+'_trend')
columns_ordered.append(name+'_cycle')
data[columns_ordered].to_csv('../Csv/business_cycle_data_actual_trend_cycle.csv')
```
|
github_jupyter
|
```
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
```
# Reading QoS analysis raw info
Temporarily, this info is saved in a CSV file but it will be in the database
**qos_analysis_13112018.csv**
- columns = ['url','protocol','code','start','end','duration','runid']
- First try of qos analysis.
- It was obtained from 50 repetitions of each of 3921 eepsites gathered.
- Just one i2router (UCA desktop host)
- Time gap between each eepsite request 3921*0.3sec=1176sec/60sec ~ 19 mins
- Total experiment elapsed time 50rep X 19 mins ~ 16 hours
**qos_analysis_29112018_local.csv**
- columns = ['url','code','duration','runid']
- 100 repetions of the first 10 eepsite from the list. Just for testing.
- local i2prouter from my laptop
- Time gap between each eepsite 10*5sec=50sec ~ 60s
- Total experiment elapsed time 100rep x 1min ~ 100 mins
```
# File for processing it
qos_file = 'qos_analysis_13112018.csv'
path_to_file = 'data/' + qos_file
columns = ['url','protocol','code','start','end','duration','runid']
df_qos = pd.read_csv(path_to_file,names=columns,delimiter="|")
# File for processing it - local router
qos_file = 'qos_analysis_29112018_local.csv'
path_to_file = 'data/' + qos_file
columns = ['url','code','duration','runid']
df_qos_local = pd.read_csv(path_to_file,names=columns,delimiter="|")
# File for processing it - local router
qos_file = 'qos_analysis_29112018_remote.csv'
path_to_file = 'data/' + qos_file
columns = ['url','code','duration','runid']
df_qos_remote = pd.read_csv(path_to_file,names=columns,delimiter="|")
# File for testing - to be removed
qos_file = 'analitica.csv'
path_to_file = 'data/' + qos_file
columns = ['url','code','duration','runid','intervals']
df_qos_testing = pd.read_csv(path_to_file,names=columns,delimiter="|")
# DF to analize
df_qos = df_qos_testing.copy()
# Removing not valid rounds
df_qos['runid'] = pd.to_numeric(df_qos['runid'], errors='coerce').dropna()
df_qos.head()
# Duration distribution by http response
fig, ax1 = plt.subplots(figsize=(10, 6))
# http code
code = 200
df_to_plot = df_qos[(df_qos['code']==code)]['duration']
#df_qos[(df_qos['code']==500)]['duration'].hist(bins=100)
df_to_plot.plot(kind='hist',bins=100, ax=ax1, color={'r','g'}, alpha=0.7)
ax1.set_ylabel('Frequency')
ax1.set_xlabel('Duration (seconds)')
ax1.set_title('HTTP ' + str(code))
plt.sca(ax1)# matplotlib only acts over the current axis
plt.xticks(rotation=75)
df_qos['code'].hist(bins=100)
df_qos['code'].unique()
df_qos.code.value_counts()
df_qos.describe()
# Average duration by error code
df = pd.DataFrame({
'code': df_qos['code'],
'duration': df_qos['duration'],
})
df = df.sort_values(by='code')
fig, ax1 = plt.subplots(figsize=(12, 8))
to_drop = []
df = df[~df['code'].isin(to_drop)]
means = df.groupby('code').mean()
std = df.groupby('code').std()
means.plot(kind='bar',yerr=std, ax=ax1, color={'r','g'}, alpha=0.7)
ax1.set_ylabel('Duration average (seconds)')
plt.sca(ax1)# matplotlib only acts over the current axis
plt.xticks(rotation=75)
df.groupby('code').describe()
# Duration by error code
df = pd.DataFrame({
'code': df_qos['code'],
'duration': df_qos['duration'],
})
to_drop = [504]
df = df[~df['code'].isin(to_drop)]
fig, ax1 = plt.subplots(figsize=(12, 8))
ax = sns.boxplot(x="code", y="duration", data=df, ax=ax1)
ax1.set_ylabel('Duration (seconds)')
ax1.set_xticklabels(set(df.code))
plt.sca(ax1)# matplotlib
# Average duration by eepsite
df = pd.DataFrame({
'url': df_qos['url'],
'duration': df_qos['duration'],
})
fig, ax1 = plt.subplots(figsize=(15, 8),)
df = df.sort_values(by='url')
means = df.groupby('url').mean()
std = df.groupby('url').std()
means = means[0:50]
std = std[0:50]
means.plot(kind='bar',yerr=std, ax=ax1, color={'r'}, alpha=0.7)
ax1.set_ylabel('Duration average (seconds)')
plt.sca(ax1)# matplotlib only acts over the current axis
plt.xticks(rotation=90)
# Average duration by eepsite
df = pd.DataFrame({
'url': df_qos['url'],
'duration': df_qos['duration'],
'code': df_qos['code']
})
fig, ax1 = plt.subplots(figsize=(15, 8),)
df = df.sort_values(by='duration',ascending=False)
eepsites = list(df[0:10000].groupby('url').groups.keys())[0:20]
df = df[df['url'].isin(eepsites)]
ax = sns.boxplot(x="url", y="duration", data=df, hue='code', ax=ax1)
ax1.set_ylabel('Duration (seconds)')
#ax1.set_ylim((0,3))
plt.sca(ax1)# matplotlib only acts over the current axis
plt.xticks(rotation=90)
```
# Availability study
```
HTTP_RESPONSE_CODES = {200:'OK',
301:'Moved Permanently',
302:'Found (Previously "Moved temporarily")',
400:'Bad Request',
401:'Unauthorized',
403:'Forbidden',
429:'Too Many Requests',
500:'Internal Server Error',
502:'Bad Gateway',
503:'Service Unavailable',
504:'Gateway Timeout'}
df_qos
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/neurorishika/PSST/blob/master/Tutorial/Day%205%20Optimal%20Mind%20Control/Day%205.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a> <a href="https://kaggle.com/kernels/welcome?src=https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/Day%205.ipynb" target="_parent"><img src="https://kaggle.com/static/images/open-in-kaggle.svg" alt="Open in Kaggle"/></a>
## Day 5: Optimal Mind Control
Welcome to Day 5! Now that we can simulate a model network of conductance-based neurons, we discuss the limitations of our approach and attempts to work around these issues.
### Memory Management
Using Python and TensorFlow allowed us to write code that is readable, parallizable and scalable across a variety of computational devices. However, our implementation is very memory intensive. The iterators in TensorFlow do not follow the normal process of memory allocation and garbage collection. Since, TensorFlow is designed to work on diverse hardware like GPUs, TPUs and distributed platforms, memory allocation is done adaptively during the TensorFlow session and not cleared until the Python kernel has stopped execution. The memory used increases linearly with time as the state matrix is computed recursively by the tf.scan function. The maximum memory used by the computational graph is 2 times the total state matrix size at the point when the computation finishes and copies the final data into the memory. The larger the network and longer the simulation, the larger the solution matrix. Each run is limited by the total available memory. For a system with a limited memory of K bytes, The length of a given simulation (L timesteps) of a given network (N differential equations) with 64-bit floating-point precision will follow:
$$2\times64\times L\times N=K$$
That is, for any given network, our maximum simulation length is limited. One way to improve our maximum length is to divide the simulation into smaller batches. There will be a small queuing time between batches, which will slow down our code by a small amount but we will be able to simulate longer times. Thus, if we split the simulation into K sequential batches, the maximum memory for the simulation becomes $(1+\frac{1}{K})$ times the total matrix size. Thus the memory relation becomes:
$$\Big(1+\frac{1}{K}\Big)\times64\times L\times N=K$$
This way, we can maximize the length of out simulation that we can run in a single python kernel.
Let us implement this batch system for our 3 neuron feed-forward model.
### Implementing the Model
To improve the readability of our code we separate the integrator into a independent import module. The integrator code was placed in a file called tf integrator.py. The file must be present in the same directory as the implementation of the model.
Note: If you are using Jupyter Notebook, remember to remove the %matplotlib inline command as it is specific to jupyter.
#### Importing tf_integrator and other requirements
Once the Integrator is saved in tf_integrator.py in the same directory as the Notebook, we can start importing the essentials including the integrator.
**WARNING: If you are running this notebook using Kaggle, make sure you have logged in to your verified Kaggle account and enabled Internet Access for the kernel. For instructions on enabling Internet on Kaggle Kernels, visit: https://www.kaggle.com/product-feedback/63544**
```
#@markdown Import required files and code from previous tutorials
!wget --no-check-certificate \
"https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/tf_integrator.py" \
-O "tf_integrator.py"
!wget --no-check-certificate \
"https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/call.py" \
-O "call.py"
!wget --no-check-certificate \
"https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/run.py" \
-O "run.py"
import numpy as np
import tf_integrator as tf_int
import matplotlib.pyplot as plt
import seaborn as sns
import tensorflow.compat.v1 as tf
tf.disable_eager_execution()
```
### Recall the Model
For implementing a Batch system, we do not need to change how we construct our model only how we execute it.
#### Step 1: Initialize Parameters and Dynamical Equations; Define Input
```
n_n = 3 # Number of simultaneous neurons to simulate
sim_res = 0.01 # Time Resolution of the Simulation
sim_time = 700 # Length of the Simulation
t = np.arange(0,sim_time,sim_res) # Time points at which to simulate the network
# Acetylcholine
ach_mat = np.zeros((n_n,n_n)) # Ach Synapse Connectivity Matrix
ach_mat[1,0]=1
## PARAMETERS FOR ACETYLCHLOLINE SYNAPSES ##
n_ach = int(np.sum(ach_mat)) # Number of Acetylcholine (Ach) Synapses
alp_ach = [10.0]*n_ach # Alpha for Ach Synapse
bet_ach = [0.2]*n_ach # Beta for Ach Synapse
t_max = 0.3 # Maximum Time for Synapse
t_delay = 0 # Axonal Transmission Delay
A = [0.5]*n_n # Synaptic Response Strength
g_ach = [0.35]*n_n # Ach Conductance
E_ach = [0.0]*n_n # Ach Potential
# GABAa
gaba_mat = np.zeros((n_n,n_n)) # GABAa Synapse Connectivity Matrix
gaba_mat[2,1] = 1
## PARAMETERS FOR GABAa SYNAPSES ##
n_gaba = int(np.sum(gaba_mat)) # Number of GABAa Synapses
alp_gaba = [10.0]*n_gaba # Alpha for GABAa Synapse
bet_gaba = [0.16]*n_gaba # Beta for GABAa Synapse
V0 = [-20.0]*n_n # Decay Potential
sigma = [1.5]*n_n # Decay Time Constant
g_gaba = [0.8]*n_n # fGABA Conductance
E_gaba = [-70.0]*n_n # fGABA Potential
## Storing Firing Thresholds ##
F_b = [0.0]*n_n # Fire threshold
def I_inj_t(t):
"""
This function returns the external current to be injected into the network at any time step from the current_input matrix.
Parameters:
-----------
t: float
The time at which the current injection is being performed.
"""
# Turn indices to integer and extract from matrix
index = tf.cast(t/sim_res,tf.int32)
return tf.constant(current_input.T,dtype=tf.float64)[index]
## Acetylcholine Synaptic Current ##
def I_ach(o,V):
"""
This function returns the synaptic current for the Acetylcholine (Ach) synapses for each neuron.
Parameters:
-----------
o: float
The fraction of open acetylcholine channels for each synapse.
V: float
The membrane potential of the postsynaptic neuron.
"""
o_ = tf.constant([0.0]*n_n**2,dtype=tf.float64) # Initialize the flattened matrix to store the synaptic open fractions
ind = tf.boolean_mask(tf.range(n_n**2),ach_mat.reshape(-1) == 1) # Get the indices of the synapses that exist
o_ = tf.tensor_scatter_nd_update(o_,tf.reshape(ind,[-1,1]),o) # Update the flattened open fraction matrix
o_ = tf.transpose(tf.reshape(o_,(n_n,n_n))) # Reshape and Transpose the matrix to be able to multiply it with the conductance matrix
return tf.reduce_sum(tf.transpose((o_*(V-E_ach))*g_ach),1) # Calculate the synaptic current
## GABAa Synaptic Current ##
def I_gaba(o,V):
"""
This function returns the synaptic current for the GABA synapses for each neuron.
Parameters:
-----------
o: float
The fraction of open GABA channels for each synapse.
V: float
The membrane potential of the postsynaptic neuron.
"""
o_ = tf.constant([0.0]*n_n**2,dtype=tf.float64) # Initialize the flattened matrix to store the synaptic open fractions
ind = tf.boolean_mask(tf.range(n_n**2),gaba_mat.reshape(-1) == 1) # Get the indices of the synapses that exist
o_ = tf.tensor_scatter_nd_update(o_,tf.reshape(ind,[-1,1]),o) # Update the flattened open fraction matrix
o_ = tf.transpose(tf.reshape(o_,(n_n,n_n))) # Reshape and Transpose the matrix to be able to multiply it with the conductance matrix
return tf.reduce_sum(tf.transpose((o_*(V-E_gaba))*g_gaba),1) # Calculate the synaptic current
## Other Currents ##
def I_K(V, n):
"""
This function determines the K-channel current.
Parameters:
-----------
V: float
The membrane potential.
n: float
The K-channel gating variable n.
"""
return g_K * n**4 * (V - E_K)
def I_Na(V, m, h):
"""
This function determines the Na-channel current.
Parameters:
-----------
V: float
The membrane potential.
m: float
The Na-channel gating variable m.
h: float
The Na-channel gating variable h.
"""
return g_Na * m**3 * h * (V - E_Na)
def I_L(V):
"""
This function determines the leak current.
Parameters:
-----------
V: float
The membrane potential.
"""
return g_L * (V - E_L)
def dXdt(X, t):
"""
This function determines the derivatives of the membrane voltage and gating variables for n_n neurons.
Parameters:
-----------
X: float
The state vector given by the [V1,V2,...,Vn_n,m1,m2,...,mn_n,h1,h2,...,hn_n,n1,n2,...,nn_n] where
Vx is the membrane potential for neuron x
mx is the Na-channel gating variable for neuron x
hx is the Na-channel gating variable for neuron x
nx is the K-channel gating variable for neuron x.
t: float
The time points at which the derivatives are being evaluated.
"""
V = X[:1*n_n] # First n_n values are Membrane Voltage
m = X[1*n_n:2*n_n] # Next n_n values are Sodium Activation Gating Variables
h = X[2*n_n:3*n_n] # Next n_n values are Sodium Inactivation Gating Variables
n = X[3*n_n:4*n_n] # Next n_n values are Potassium Gating Variables
o_ach = X[4*n_n : 4*n_n + n_ach] # Next n_ach values are Acetylcholine Synapse Open Fractions
o_gaba = X[4*n_n + n_ach : 4*n_n + n_ach + n_gaba] # Next n_gaba values are GABAa Synapse Open Fractions
fire_t = X[-n_n:] # Last n_n values are the last fire times as updated by the modified integrator
dVdt = (I_inj_t(t) - I_Na(V, m, h) - I_K(V, n) - I_L(V) - I_ach(o_ach,V) - I_gaba(o_gaba,V)) / C_m # The derivative of the membrane potential
## Updation for gating variables ##
m0,tm,h0,th = Na_prop(V) # Calculate the dynamics of the Na-channel gating variables for all n_n neurons
n0,tn = K_prop(V) # Calculate the dynamics of the K-channel gating variables for all n_n neurons
dmdt = - (1.0/tm)*(m-m0) # The derivative of the Na-channel gating variable m for all n_n neurons
dhdt = - (1.0/th)*(h-h0) # The derivative of the Na-channel gating variable h for all n_n neurons
dndt = - (1.0/tn)*(n-n0) # The derivative of the K-channel gating variable n for all n_n neurons
## Updation for o_ach ##
A_ = tf.constant(A,dtype=tf.float64) # Get the synaptic response strengths of the pre-synaptic neurons
Z_ = tf.zeros(tf.shape(A_),dtype=tf.float64) # Create a zero matrix of the same size as A_
T_ach = tf.where(tf.logical_and(tf.greater(t,fire_t+t_delay),tf.less(t,fire_t+t_max+t_delay)),A_,Z_) # Find which synapses would have received an presynaptic spike in the past window and assign them the corresponding synaptic response strength
T_ach = tf.multiply(tf.constant(ach_mat,dtype=tf.float64),T_ach) # Find the postsynaptic neurons that would have received an presynaptic spike in the past window
T_ach = tf.boolean_mask(tf.reshape(T_ach,(-1,)),ach_mat.reshape(-1) == 1) # Get the pre-synaptic activation function for only the existing synapses
do_achdt = alp_ach*(1.0-o_ach)*T_ach - bet_ach*o_ach # Calculate the derivative of the open fraction of the acetylcholine synapses
## Updation for o_gaba ##
T_gaba = 1.0/(1.0+tf.exp(-(V-V0)/sigma)) # Calculate the presynaptic activation function for all n_n neurons
T_gaba = tf.multiply(tf.constant(gaba_mat,dtype=tf.float64),T_gaba) # Find the postsynaptic neurons that would have received an presynaptic spike in the past window
T_gaba = tf.boolean_mask(tf.reshape(T_gaba,(-1,)),gaba_mat.reshape(-1) == 1) # Get the pre-synaptic activation function for only the existing synapses
do_gabadt = alp_gaba*(1.0-o_gaba)*T_gaba - bet_gaba*o_gaba # Calculate the derivative of the open fraction of the GABAa synapses
## Updation for fire times ##
dfdt = tf.zeros(tf.shape(fire_t),dtype=fire_t.dtype) # zero change in fire_t as it will be updated by the modified integrator
out = tf.concat([dVdt,dmdt,dhdt,dndt,do_achdt,do_gabadt,dfdt],0) # Concatenate the derivatives of the membrane potential, gating variables, and open fractions
return out
def K_prop(V):
"""
This function determines the K-channel gating dynamics.
Parameters:
-----------
V: float
The membrane potential.
"""
T = 22 # Temperature
phi = 3.0**((T-36.0)/10) # Temperature-correction factor
V_ = V-(-50) # Voltage baseline shift
alpha_n = 0.02*(15.0 - V_)/(tf.exp((15.0 - V_)/5.0) - 1.0) # Alpha for the K-channel gating variable n
beta_n = 0.5*tf.exp((10.0 - V_)/40.0) # Beta for the K-channel gating variable n
t_n = 1.0/((alpha_n+beta_n)*phi) # Time constant for the K-channel gating variable n
n_0 = alpha_n/(alpha_n+beta_n) # Steady-state value for the K-channel gating variable n
return n_0, t_n
def Na_prop(V):
"""
This function determines the Na-channel gating dynamics.
Parameters:
-----------
V: float
The membrane potential.
"""
T = 22 # Temperature
phi = 3.0**((T-36)/10) # Temperature-correction factor
V_ = V-(-50) # Voltage baseline shift
alpha_m = 0.32*(13.0 - V_)/(tf.exp((13.0 - V_)/4.0) - 1.0) # Alpha for the Na-channel gating variable m
beta_m = 0.28*(V_ - 40.0)/(tf.exp((V_ - 40.0)/5.0) - 1.0) # Beta for the Na-channel gating variable m
alpha_h = 0.128*tf.exp((17.0 - V_)/18.0) # Alpha for the Na-channel gating variable h
beta_h = 4.0/(tf.exp((40.0 - V_)/5.0) + 1.0) # Beta for the Na-channel gating variable h
t_m = 1.0/((alpha_m+beta_m)*phi) # Time constant for the Na-channel gating variable m
t_h = 1.0/((alpha_h+beta_h)*phi) # Time constant for the Na-channel gating variable h
m_0 = alpha_m/(alpha_m+beta_m) # Steady-state value for the Na-channel gating variable m
h_0 = alpha_h/(alpha_h+beta_h) # Steady-state value for the Na-channel gating variable h
return m_0, t_m, h_0, t_h
# Initializing the Parameters
C_m = [1.0]*n_n # Membrane capacitances
g_K = [10.0]*n_n # K-channel conductances
E_K = [-95.0]*n_n # K-channel reversal potentials
g_Na = [100]*n_n # Na-channel conductances
E_Na = [50]*n_n # Na-channel reversal potentials
g_L = [0.15]*n_n # Leak conductances
E_L = [-55.0]*n_n # Leak reversal potentials
# Creating the Current Input
current_input= np.zeros((n_n,t.shape[0])) # The current input to the network
current_input[0,int(100/sim_res):int(200/sim_res)] = 2.5
current_input[0,int(300/sim_res):int(400/sim_res)] = 5.0
current_input[0,int(500/sim_res):int(600/sim_res)] = 7.5
```
#### Step 2: Define the Initial Condition of the Network and Add some Noise to the initial conditions
```
# Initializing the State Vector and adding 1% noise
state_vector = [-71]*n_n+[0,0,0]*n_n+[0]*n_ach+[0]*n_gaba+[-9999999]*n_n
state_vector = np.array(state_vector)
state_vector = state_vector + 0.01*state_vector*np.random.normal(size=state_vector.shape)
```
#### Step 3: Splitting Time Series into independent batches and Run Each Batch Sequentially
Since we will be dividing the computation into batches, we have to split the time array such that for each new call, the final state vector of the last batch will be the initial condition for the current batch. The function $np.array\_split()$ splits the array into non-overlapping vectors. Therefore, we append the last time of the previous batch to the beginning of the current time array batch.
```
# Define the Number of Batches
n_batch = 2
# Split t array into batches using numpy
t_batch = np.array_split(t,n_batch)
# Iterate over the batches of time array
for n,i in enumerate(t_batch):
# Inform start of Batch Computation
print("Batch",(n+1),"Running...",end="")
# In np.array_split(), the split edges are present in only one array and since
# our initial vector to successive calls is corresposnding to the last output
# our first element in the later time array should be the last element of the
# previous output series, Thus, we append the last time to the beginning of
# the current time array batch.
if n>0:
i = np.append(i[0]-sim_res,i)
# Set state_vector as the initial condition
init_state = tf.constant(state_vector, dtype=tf.float64)
# Create the Integrator computation graph over the current batch of t array
tensor_state = tf_int.odeint(dXdt, init_state, i, n_n, F_b)
# Initialize variables and run session
with tf.Session() as sess:
tf.global_variables_initializer().run()
state = sess.run(tensor_state)
sess.close()
# Reset state_vector as the last element of output
state_vector = state[-1,:]
# Save the output of the simulation to a binary file
np.save("part_"+str(n+1),state)
# Clear output
state=None
print("Finished")
```
#### Putting the Output Together
The output from our batch implementation is a set of binary files that store parts of our total simulation. To get the overall output we have to stitch them back together.
```
overall_state = []
# Iterate over the generated output files
for n,i in enumerate(["part_"+str(n+1)+".npy" for n in range(n_batch)]):
# Since the first element in the series was the last output, we remove them
if n>0:
overall_state.append(np.load(i)[1:,:])
else:
overall_state.append(np.load(i))
# Concatenate all the matrix to get a single state matrix
overall_state = np.concatenate(overall_state)
```
#### Visualizing the Overall Data
Finally, we plot the same voltage traces of the 3 neurons from Day 4 as a Voltage vs Time heatmap. While this visualization may seem unnecessary for just 3 neurons, it becomes an useful tool when on visualizes the dynamics of a large network of neurons as illustrated in the Example Implementation of the Locust Antennal Lobe.
```
# Plot the voltage traces of the three neurons
plt.figure(figsize=(12,6))
sns.heatmap(overall_state[::100,:3].T,xticklabels=100,yticklabels=5,cmap='RdBu_r')
plt.xlabel("Time (in ms)")
plt.ylabel("Neuron Number")
plt.title("Voltage vs Time Heatmap for Projection Neurons (PNs)")
plt.tight_layout()
plt.show()
```
By this method, we have maximized the usage of our available memory but we can go further and develop a method to allow indefinitely long simulation. The issue behind this entire algorithm is that the memory is not cleared until the python kernel finishes. One way to overcome this is to save the parameters of the model (such as connectivity matrix) and the state vector in a file, and start a new python kernel from a python script to compute successive batches. This way after each large batch, the memory gets cleaned. By combining the previous batch implementation and this system, we can maximize our computability.
### Implementing a Runner and a Caller
Firstly, we have to create an implementation of the model that takes in previous input as current parameters. Thus, we create a file, which we call "run.py" that takes an argument ie. the current batch number. The implementation for "run.py" is mostly same as the above model but there is a small difference.
When the batch number is 0, we initialize all variable parameters and save them, but otherwise we use the saved values. The parameters we save include: Acetylcholine Matrix, GABAa Matrix and Final/Initial State Vector. It will also save the files with both batch number and sub-batch number listed.
The time series will be created and split initially by the caller, which we call "call.py", and stored in a file. Each execution of the Runner will extract its relevant time series and compute on it.
#### Implementing the Runner code
"run.py" is essentially identical to the batch-implemented model we developed above with the changes described below:
```
# Additional Imports #
import sys
# Duration of Simulation #
# t = np.arange(0,sim_time,sim_res)
t = np.load("time.npy",allow_pickle=True)[int(sys.argv[1])] # get first argument to run.py
# Connectivity Matrix Definitions #
if sys.argv[1] == '0':
ach_mat = np.zeros((n_n,n_n)) # Ach Synapse Connectivity Matrix
ach_mat[1,0]=1 # If connectivity is random, once initialized it will be the same.
np.save("ach_mat",ach_mat)
else:
ach_mat = np.load("ach_mat.npy")
if sys.argv[1] == '0':
gaba_mat = np.zeros((n_n,n_n)) # GABAa Synapse Connectivity Matrix
gaba_mat[2,1] = 1 # If connectivity is random, once initialized it will be the same.
np.save("gaba_mat",gaba_mat)
else:
gaba_mat = np.load("gaba_mat.npy")
# Current Input Definition #
if sys.argv[1] == '0':
current_input= np.zeros((n_n,int(sim_time/sim_res)))
current_input[0,int(100/sim_res):int(200/sim_res)] = 2.5
current_input[0,int(300/sim_res):int(400/sim_res)] = 5.0
current_input[0,int(500/sim_res):int(600/sim_res)] = 7.5
np.save("current_input",current_input)
else:
current_input = np.load("current_input.npy")
# State Vector Definition #
if sys.argv[1] == '0':
state_vector = [-71]*n_n+[0,0,0]*n_n+[0]*n_ach+[0]*n_gaba+[-9999999]*n_n
state_vector = np.array(state_vector)
state_vector = state_vector + 0.01*state_vector*np.random.normal(size=state_vector.shape)
np.save("state_vector",state_vector)
else:
state_vector = np.load("state_vector.npy")
# Saving of Output #
# np.save("part_"+str(n+1),state)
np.save("batch"+str(int(sys.argv[1])+1)+"_part_"+str(n+1),state)
```
#### Implementing the Caller code
The caller will create the time series, split it and use python subprocess module to call "run.py" with appropriate arguments. The code for "call.py" is given below.
```
from subprocess import call
import numpy as np
total_time = 700
n_splits = 2
time = np.split(np.arange(0,total_time,0.01),n_splits)
# Append the last time point to the beginning of the next batch
for n,i in enumerate(time):
if n>0:
time[n] = np.append(i[0]-0.01,i)
np.save("time",time)
# call successive batches with a new python subprocess and pass the batch number
for i in range(n_splits):
call(['python','run.py',str(i)])
print("Simulation Completed.")
```
#### Using call.py
```
!python call.py
```
#### Combining all Data
Just like we merged all the batches, we merge all the sub-batches and batches.
```
n_splits = 2
n_batch = 2
overall_state = []
# Iterate over the generated output files
for n,i in enumerate(["batch"+str(x+1) for x in range(n_splits)]):
for m,j in enumerate(["_part_"+str(x+1)+".npy" for x in range(n_batch)]):
# Since the first element in the series was the last output, we remove them
if n>0 and m>0:
overall_state.append(np.load(i+j)[1:,:])
else:
overall_state.append(np.load(i+j))
# Concatenate all the matrix to get a single state matrix
overall_state = np.concatenate(overall_state)
# Plot the simulation results
plt.figure(figsize=(12,6))
sns.heatmap(overall_state[::100,:3].T,xticklabels=100,yticklabels=5,cmap='RdBu_r')
plt.xlabel("Time (in ms)")
plt.ylabel("Neuron Number")
plt.title("Voltage vs Time Heatmap for Projection Neurons (PNs)")
plt.tight_layout()
plt.show()
```
|
github_jupyter
|
# 📝 Exercise M6.03
This exercise aims at verifying if AdaBoost can over-fit.
We will make a grid-search and check the scores by varying the
number of estimators.
We will first load the California housing dataset and split it into a
training and a testing set.
```
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import train_test_split
data, target = fetch_california_housing(return_X_y=True, as_frame=True)
target *= 100 # rescale the target in k$
data_train, data_test, target_train, target_test = train_test_split(
data, target, random_state=0, test_size=0.5)
```
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">If you want a deeper overview regarding this dataset, you can refer to the
Appendix - Datasets description section at the end of this MOOC.</p>
</div>
Then, create an `AdaBoostRegressor`. Use the function
`sklearn.model_selection.validation_curve` to get training and test scores
by varying the number of estimators. Use the mean absolute error as a metric
by passing `scoring="neg_mean_absolute_error"`.
*Hint: vary the number of estimators between 1 and 60.*
```
# Write your code here.
from sklearn.ensemble import AdaBoostRegressor
from sklearn.model_selection import validation_curve
import pandas as pd
import numpy as np
n_estimators = np.arange(1, 100, 2)
n_estimators
model = AdaBoostRegressor()
model.get_params()
train_scores, test_scores = validation_curve(
model, data, target, param_name="n_estimators", param_range=n_estimators,
cv=5, scoring="neg_mean_absolute_error", n_jobs=2)
train_errors, test_errors = -train_scores, -test_scores
```
Plot both the mean training and test errors. You can also plot the
standard deviation of the errors.
*Hint: you can use `plt.errorbar`.*
```
# Write your code here.
from matplotlib import pyplot as plt
plt.errorbar(n_estimators, train_errors.mean(axis=1),
yerr=train_errors.std(axis=1), label="Training error")
plt.errorbar(n_estimators, test_errors.mean(axis=1),
yerr=train_errors.std(axis=1), label="Testing error")
```
Plotting the validation curve, we can see that AdaBoost is not immune against
overfitting. Indeed, there is an optimal number of estimators to be found.
Adding too many estimators is detrimental for the statistical performance of
the model.
Repeat the experiment using a random forest instead of an AdaBoost regressor.
```
# Write your code here.
from sklearn.ensemble import RandomForestRegressor
model = RandomForestRegressor()
train_scores, test_scores = validation_curve(
model, data, target, param_name="n_estimators", param_range=n_estimators,
cv=5, scoring="neg_mean_absolute_error", n_jobs=2)
# PLOT
plt.errorbar(n_estimators, train_errors.mean(axis=1),
yerr=train_errors.std(axis=1), label="Training error")
plt.errorbar(n_estimators, test_errors.mean(axis=1),
yerr=train_errors.std(axis=1), label="Testing error")
```
|
github_jupyter
|
# SmallPebble
[](https://github.com/sradc/smallpebble/commits/)
**Project status: unstable.**
<br><p align="center"><img src="https://raw.githubusercontent.com/sradc/SmallPebble/master/pebbles.jpg"/></p><br>
SmallPebble is a minimal automatic differentiation and deep learning library written from scratch in [Python](https://www.python.org/), using [NumPy](https://numpy.org/)/[CuPy](https://cupy.dev/).
The implementation is relatively small, and mainly in the file: [smallpebble.py](https://github.com/sradc/SmallPebble/blob/master/smallpebble/smallpebble.py). To help understand it, check out [this](https://sidsite.com/posts/autodiff/) introduction to autodiff, which presents an autodiff framework that works in the same way as SmallPebble (except using scalars instead of NumPy arrays).
SmallPebble's *raison d'etre* is to be a simplified deep learning implementation,
for those who want to learn what’s under the hood of deep learning frameworks.
However, because it is written in terms of vectorised NumPy/CuPy operations,
it performs well enough for non-trivial models to be trained using it.
**Highlights**
- Relatively simple implementation.
- Can run on GPU, using CuPy.
- Various operations, such as matmul, conv2d, maxpool2d.
- Array broadcasting support.
- Eager or lazy execution.
- Powerful API for creating models.
- It's easy to add new SmallPebble functions.
**Notes**
Graphs are built implicitly via Python objects referencing Python objects.
When `get_gradients` is called, autodiff is carried out on the whole sub-graph. The default array library is NumPy.
---
**Read on to see:**
- Example models created and trained using SmallPebble.
- A brief guide to using SmallPebble.
```
import matplotlib.pyplot as plt
import numpy as np
from tqdm.notebook import tqdm
import smallpebble as sp
from smallpebble.misc import load_data
```
## Training a neural network to classify handwritten digits (MNIST)
```
"Load the dataset, and create a validation set."
X_train, y_train, _, _ = load_data('mnist') # load / download from openml.org
X_train = X_train/255 # normalize
# Seperate out data for validation.
X = X_train[:50_000, ...]
y = y_train[:50_000]
X_eval = X_train[50_000:60_000, ...]
y_eval = y_train[50_000:60_000]
"Plot, to check we have the right data."
plt.figure(figsize=(5,5))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(X_train[i,:].reshape(28,28), cmap='gray', vmin=0, vmax=1)
plt.show()
"Create a model, with two fully connected hidden layers."
X_in = sp.Placeholder()
y_true = sp.Placeholder()
h = sp.linearlayer(28*28, 100)(X_in)
h = sp.Lazy(sp.leaky_relu)(h)
h = sp.linearlayer(100, 100)(h)
h = sp.Lazy(sp.leaky_relu)(h)
h = sp.linearlayer(100, 10)(h)
y_pred = sp.Lazy(sp.softmax)(h)
loss = sp.Lazy(sp.cross_entropy)(y_pred, y_true)
learnables = sp.get_learnables(y_pred)
loss_vals = []
validation_acc = []
"Train model, while measuring performance on the validation dataset."
NUM_ITERS = 300
BATCH_SIZE = 200
eval_batch = sp.batch(X_eval, y_eval, BATCH_SIZE)
adam = sp.Adam() # Adam optimization
for i, (xbatch, ybatch) in tqdm(enumerate(sp.batch(X, y, BATCH_SIZE)), total=NUM_ITERS):
if i >= NUM_ITERS: break
X_in.assign_value(sp.Variable(xbatch))
y_true.assign_value(ybatch)
loss_val = loss.run() # run the graph
if np.isnan(loss_val.array):
print("loss is nan, aborting.")
break
loss_vals.append(loss_val.array)
# Compute gradients, and use to carry out learning step:
gradients = sp.get_gradients(loss_val)
adam.training_step(learnables, gradients)
# Compute validation accuracy:
x_eval_batch, y_eval_batch = next(eval_batch)
X_in.assign_value(sp.Variable(x_eval_batch))
predictions = y_pred.run()
predictions = np.argmax(predictions.array, axis=1)
accuracy = (y_eval_batch == predictions).mean()
validation_acc.append(accuracy)
# Plot results:
print(f'Final validation accuracy: {np.mean(validation_acc[-10:])}')
plt.figure(figsize=(14, 4))
plt.subplot(1, 2, 1)
plt.ylabel('Loss')
plt.xlabel('Iteration')
plt.plot(loss_vals)
plt.subplot(1, 2, 2)
plt.ylabel('Validation accuracy')
plt.xlabel('Iteration')
plt.suptitle('Neural network trained on MNIST, using SmallPebble.')
plt.ylim([0, 1])
plt.plot(validation_acc)
plt.show()
```
## Training a convolutional neural network on CIFAR-10, using CuPy
This was run on [Google Colab](https://colab.research.google.com/), with a GPU.
```
"Load the CIFAR dataset."
X_train, y_train, _, _ = load_data('cifar') # load/download from openml.org
X_train = X_train/255 # normalize
"""Plot, to check it's the right data.
(This cell's code is from: https://www.tensorflow.org/tutorials/images/cnn#verify_the_data)
"""
class_names = ['airplane', 'automobile', 'bird', 'cat', 'deer',
'dog', 'frog', 'horse', 'ship', 'truck']
plt.figure(figsize=(8,8))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(X_train[i,:].reshape(32,32,3))
plt.xlabel(class_names[y_train[i]])
plt.show()
"Switch array library to CuPy, so can use GPU."
import cupy
sp.use(cupy)
print(sp.array_library.library.__name__) # should be 'cupy'
"Convert data to CuPy arrays"
X_train = cupy.array(X_train)
y_train = cupy.array(y_train)
# Seperate out data for validation as before.
X = X_train[:45_000, ...]
y = y_train[:45_000]
X_eval = X_train[45_000:50_000, ...]
y_eval = y_train[45_000:50_000]
"""Define a model."""
X_in = sp.Placeholder()
y_true = sp.Placeholder()
h = sp.convlayer(height=3, width=3, depth=3, n_kernels=32)(X_in)
h = sp.Lazy(sp.leaky_relu)(h)
h = sp.Lazy(lambda a: sp.maxpool2d(a, 2, 2, strides=[2, 2]))(h)
h = sp.convlayer(3, 3, 32, 128, padding='VALID')(h)
h = sp.Lazy(sp.leaky_relu)(h)
h = sp.Lazy(lambda a: sp.maxpool2d(a, 2, 2, strides=[2, 2]))(h)
h = sp.convlayer(3, 3, 128, 128, padding='VALID')(h)
h = sp.Lazy(sp.leaky_relu)(h)
h = sp.Lazy(lambda a: sp.maxpool2d(a, 2, 2, strides=[2, 2]))(h)
h = sp.Lazy(lambda x: sp.reshape(x, [-1, 3*3*128]))(h)
h = sp.linearlayer(3*3*128, 10)(h)
h = sp.Lazy(sp.softmax)(h)
y_pred = h
loss = sp.Lazy(sp.cross_entropy)(y_pred, y_true)
learnables = sp.get_learnables(y_pred)
loss_vals = []
validation_acc = []
# Check we get the expected dimensions
X_in.assign_value(sp.Variable(X[0:3, :].reshape([-1, 32, 32, 3])))
h.run().shape
```
Train the model.
```
NUM_ITERS = 3000
BATCH_SIZE = 128
eval_batch = sp.batch(X_eval, y_eval, BATCH_SIZE)
adam = sp.Adam()
for i, (xbatch, ybatch) in tqdm(enumerate(sp.batch(X, y, BATCH_SIZE)), total=NUM_ITERS):
if i >= NUM_ITERS: break
xbatch_images = xbatch.reshape([-1, 32, 32, 3])
X_in.assign_value(sp.Variable(xbatch_images))
y_true.assign_value(ybatch)
loss_val = loss.run()
if np.isnan(loss_val.array):
print("Aborting, loss is nan.")
break
loss_vals.append(loss_val.array)
# Compute gradients, and carry out learning step.
gradients = sp.get_gradients(loss_val)
adam.training_step(learnables, gradients)
# Compute validation accuracy:
x_eval_batch, y_eval_batch = next(eval_batch)
X_in.assign_value(sp.Variable(x_eval_batch.reshape([-1, 32, 32, 3])))
predictions = y_pred.run()
predictions = np.argmax(predictions.array, axis=1)
accuracy = (y_eval_batch == predictions).mean()
validation_acc.append(accuracy)
print(f'Final validation accuracy: {np.mean(validation_acc[-10:])}')
plt.figure(figsize=(14, 4))
plt.subplot(1, 2, 1)
plt.ylabel('Loss')
plt.xlabel('Iteration')
plt.plot(loss_vals)
plt.subplot(1, 2, 2)
plt.ylabel('Validation accuracy')
plt.xlabel('Iteration')
plt.suptitle('CNN trained on CIFAR-10, using SmallPebble.')
plt.ylim([0, 1])
plt.plot(validation_acc)
plt.show()
```
It looks like we could improve our results by training for longer (and we could improve our model architecture).
---
# Brief guide to using SmallPebble
SmallPebble provides the following building blocks to make models with:
- `sp.Variable`
- Operations, such as `sp.add`, `sp.mul`, etc.
- `sp.get_gradients`
- `sp.Lazy`
- `sp.Placeholder` (this is really just `sp.Lazy` on the identity function)
- `sp.learnable`
- `sp.get_learnables`
The following examples show how these are used.
## Switching between NumPy and CuPy
We can dynamically switch between NumPy and CuPy. (Assuming you have a CuPy compatible GPU and CuPy set up. Note, CuPy is available on Google Colab, if you change the runtime to GPU.)
```
import cupy
import numpy
import smallpebble as sp
# Switch to CuPy
sp.use(cupy)
print(sp.array_library.library.__name__) # should be 'cupy'
# Switch back to NumPy:
sp.use(numpy)
print(sp.array_library.library.__name__) # should be 'numpy'
```
## sp.Variable & sp.get_gradients
With SmallPebble, you can:
- Wrap NumPy arrays in `sp.Variable`
- Apply SmallPebble operations (e.g. `sp.matmul`, `sp.add`, etc.)
- Compute gradients with `sp.get_gradients`
```
a = sp.Variable(np.random.random([2, 2]))
b = sp.Variable(np.random.random([2, 2]))
c = sp.Variable(np.random.random([2]))
y = sp.mul(a, b) + c
print('y.array:\n', y.array)
gradients = sp.get_gradients(y)
grad_a = gradients[a]
grad_b = gradients[b]
grad_c = gradients[c]
print('grad_a:\n', grad_a)
print('grad_b:\n', grad_b)
print('grad_c:\n', grad_c)
```
Note that `y` is computed straight away, i.e. the (forward) computation happens immediately.
Also note that `y` is a sp.Variable and we could continue to carry out SmallPebble operations on it.
## sp.Lazy & sp.Placeholder
Lazy graphs are constructed using `sp.Lazy` and `sp.Placeholder`.
```
lazy_node = sp.Lazy(lambda a, b: a + b)(1, 2)
print(lazy_node)
print(lazy_node.run())
a = sp.Lazy(lambda a: a)(2)
y = sp.Lazy(lambda a, b, c: a * b + c)(a, 3, 4)
print(y)
print(y.run())
```
Forward computation does not happen immediately - only when .run() is called.
```
a = sp.Placeholder()
b = sp.Variable(np.random.random([2, 2]))
y = sp.Lazy(sp.matmul)(a, b)
a.assign_value(sp.Variable(np.array([[1,2], [3,4]])))
result = y.run()
print('result.array:\n', result.array)
```
You can use .run() as many times as you like.
Let's change the placeholder value and re-run the graph:
```
a.assign_value(sp.Variable(np.array([[10,20], [30,40]])))
result = y.run()
print('result.array:\n', result.array)
```
Finally, let's compute gradients:
```
gradients = sp.get_gradients(result)
```
Note that `sp.get_gradients` is called on `result`,
which is a `sp.Variable`,
not on `y`, which is a `sp.Lazy` instance.
## sp.learnable & sp.get_learnables
Use `sp.learnable` to flag parameters as learnable,
allowing them to be extracted from a lazy graph with `sp.get_learnables`.
This enables a workflow of: building a model, while flagging parameters as learnable, and then extracting all the parameters in one go at the end.
```
a = sp.Placeholder()
b = sp.learnable(sp.Variable(np.random.random([2, 1])))
y = sp.Lazy(sp.matmul)(a, b)
y = sp.Lazy(sp.add)(y, sp.learnable(sp.Variable(np.array([5]))))
learnables = sp.get_learnables(y)
for learnable in learnables:
print(learnable)
```
|
github_jupyter
|
```
import rioxarray as rio
import xarray as xr
import glob
import os
import numpy as np
import requests
import geopandas as gpd
from pathlib import Path
from datetime import datetime
from rasterio.enums import Resampling
import matplotlib.pyplot as plt
%matplotlib inline
site = "BRC"
# Change site name
chirps_seas_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/ee_season_precip_data_brc')
eeflux_seas_int_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/ee_growing_season_integrated_brc')
chirps_wy_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/wy_total_chirps_brc')
eeflux_seas_mean_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/ee_season_mean_brc')
all_scenes_f_precip = Path('/scratch/waves/rhone-ecostress/rasters/chirps-clipped')
all_scenes_f_et = Path('/home/serdp/rhone/rhone-ecostress/rasters/eeflux/BRC') # Change file path based on site
all_precip_paths = list(all_scenes_f_precip.glob("*"))
all_et_paths = list(all_scenes_f_et.glob("*.tif")) # Variable name agnostic to site?
# for some reason the fll value is not correct. this is the correct bad value to mask by
testf = all_precip_paths[0]
x = rio.open_rasterio(testf)
badvalue = np.unique(x.where(x != x._FillValue).sel(band=1))[0]
def chirps_path_date(path):
_, _, year, month, day, _ = path.name.split(".")
day = day.split("-")[0]
return datetime(int(year), int(month), int(day))
def open_chirps(path):
data_array = rio.open_rasterio(path) #chunks makes i lazyily executed
data_array = data_array.sel(band=1).drop("band") # gets rid of old coordinate dimension since we need bands to have unique coord ids
data_array["date"] = chirps_path_date(path) # makes a new coordinate
return data_array.expand_dims({"date":1}) # makes this coordinate a dimension
### data is not tiled so not a good idea to use chunking
#https://github.com/pydata/xarray/issues/2314
import rasterio
with rasterio.open(testf) as src:
print(src.profile)
len(all_precip_paths) * 41.7 / 10e3 # convert from in to mm
%timeit open_chirps(testf)
all_daily_precip_path = "/home/serdp/ravery/rhone-ecostress/netcdfs/all_chirps_daily_i.nc"
if Path(all_daily_precip_path).exists():
all_chirps_arr = xr.open_dataarray(all_daily_precip_path)
all_chirps_arr = all_chirps_arr.sortby("date")
else:
daily_chirps_arrs = []
for path in all_precip_paths:
daily_chirps_arrs.append(open_chirps(path))
all_chirps_arr = xr.concat(daily_chirps_arrs, dim="date")
all_chirps_arr = all_chirps_arr.sortby("date")
all_chirps_arr.to_netcdf(all_daily_precip_path)
def eeflux_path_date(path):
year, month, day, _, _ = path.name.split("-") # Change this line accordingly based on format of eeflux dates
return datetime(int(year), int(month), int(day))
def open_eeflux(path, da_for_match):
data_array = rio.open_rasterio(path) #chunks makes i lazyily executed
data_array.rio.reproject_match(da_for_match)
data_array = data_array.sel(band=1).drop("band") # gets rid of old coordinate dimension since we need bands to have unique coord ids
data_array["date"] = eeflux_path_date(path) # makes a new coordinate
return data_array.expand_dims({"date":1}) # makes this coordinate a dimension
# The following lines seem to write the lists of rasters to netcdf files? Do we need to replicate for chirps?
da_for_match = rio.open_rasterio(all_et_paths[0])
daily_eeflux_arrs = [open_eeflux(path, da_for_match) for path in all_et_paths]
all_eeflux_arr = xr.concat(daily_eeflux_arrs, dim="date")
all_daily_eeflux_path = "/home/serdp/ravery/rhone-ecostress/netcdfs/all_eeflux_daily_i.nc"
all_eeflux_arr.to_netcdf(all_daily_eeflux_path)
all_eeflux_arr[-3,:,:].plot.imshow()
all_eeflux_arr = all_eeflux_arr.sortby("date")
ey = max(all_eeflux_arr['date.year'].values)
ey
sy = min(all_eeflux_arr['date.year'].values)
sy
all_eeflux_arr['date.dayofyear'].values
# THIS IS IMPORTANT
def years_list(all_arr):
ey = max(all_arr['date.year'].values)
sy = min(all_arr['date.year'].values)
start_years = range(sy, ey)
end_years = range(sy+1, ey+1) # Change to sy+1, ey+1 for across-calendar-year (e.g. winter) calculations
return list(zip(start_years, end_years))
def group_by_custom_doy(all_arr, doy_start, doy_end):
start_end_years = years_list(all_arr)
water_year_arrs = []
for water_year in start_end_years:
start_mask = ((all_arr['date.dayofyear'].values > doy_start) & (all_arr['date.year'].values == water_year[0]))
end_mask = ((all_arr['date.dayofyear'].values < doy_end) & (all_arr['date.year'].values == water_year[1]))
water_year_arrs.append(all_arr[start_mask | end_mask]) # | = or, & = and
return water_year_arrs
def group_by_season(all_arr, doy_start, doy_end):
yrs = np.unique(all_arr['date.year'])
season_arrs = []
for yr in yrs:
start_mask = ((all_arr['date.dayofyear'].values >= doy_start) & (all_arr['date.year'].values == yr))
end_mask = ((all_arr['date.dayofyear'].values <= doy_end) & (all_arr['date.year'].values == yr))
season_arrs.append(all_arr[start_mask & end_mask])
return season_arrs
# THIS IS IMPORTANT
doystart = 125 # Edit these variables to change doy length of year
doyend = 275
# eeflux_water_year_arrs = group_by_custom_doy(all_eeflux_arr, doystart, doyend) # Replaced by eeflux_seas_arrs below
chirps_water_year_arrs = group_by_custom_doy(all_chirps_arr, doyend, doystart)
eeflux_seas_arrs = group_by_season(all_eeflux_arr, doystart, doyend)
eeflux_seas_arrs
chirps_water_year_arrs[-1]
fig = plt.figure()
plt.plot(wy_list,[arr.mean() for arr in chirps_wy_sums],'.')
plt.ylabel('WY Precipitation (mm)')
# Creates figure of ET availability
group_counts = list(map(lambda x: len(x['date']), water_year_arrs))
year_tuples = years_list(all_eeflux_arr)
indexes = np.arange(len(year_tuples))
plt.bar(indexes, group_counts)
degrees = 80
plt.xticks(indexes, year_tuples, rotation=degrees, ha="center")
plt.title("Availability of EEFLUX between DOY 125 and 275")
plt.savefig("eeflux_availability.png")
# Figure below shows empty years in 85, 88, 92, 93, 96; no winter precip rasters generated for these years b/c no ET data w/in winter window
def sum_seasonal_precip(precip_arr, eeflux_group_arr):
return precip_arr.sel(date=slice(eeflux_group_arr.date.min(), eeflux_group_arr.date.max())).sum(dim="date")
# This is matching up precip w/ available ET window for each year
for index, eeflux_group in enumerate(eeflux_seas_arrs):
if len(eeflux_group['date']) > 0:
seasonal_precip = sum_seasonal_precip(all_chirps_arr, eeflux_group) # Variable/array name matters here
seasonal_et = eeflux_group.integrate(coord="date", datetime_unit="D")
year = eeflux_group['date.year'].values[0]
et_doystart = eeflux_group['date.dayofyear'].values[0]
et_doyend = eeflux_group['date.dayofyear'].values[-1]
pname = os.path.join(chirps_seas_out_dir,f"seas_chirps_{site}_{year}_{et_doystart}_{et_doyend}.tif") #Edit output raster labels
eename = os.path.join(eeflux_seas_int_out_dir, f"seasonal_eeflux_integrated_{site}_{year}_{et_doystart}_{et_doyend}.tif")
seasonal_precip.rio.to_raster(pname)
seasonal_et.rio.to_raster(eename)
# This chunk actually outputs the rasters
## Elmera Additions for winter precip:
for index, (eeflux_group,chirps_group) in enumerate(zip(eeflux_seas_arrs,chirps_water_year_arrs[3:])): #changed eeflux_group to eeflux_seas_arrs & changed from water_year_arrs to season_arrs
if len(eeflux_group['date']) > 0: # eeflux_group to eeflux_seas_arrs
mean_seas_et = eeflux_group.mean(dim='date',skipna=False)
chirps_wy_sum = chirps_group.sum(dim='date',skipna=False)
# seasonal_precip = sum_seasonal_precip(chirps_water_year_arrs, eeflux_seas_arr) # Here's where above fxn is applied to rasters, need to replace eeflux_group
year = eeflux_group['date.year'].values[0]
pname = os.path.join(chirps_wy_out_dir,f"wy_total_chirps_{site}_{year}.tif") #Edit output raster labels
eename = os.path.join(eeflux_seas_mean_out_dir,f"mean_daily_seas_et_{site}_{year}.tif")
chirps_wy_sum.rio.to_raster(pname)
mean_seas_et.rio.to_raster(eename)
# This chunk actually outputs the rasters, ET lines removed - including seasonal_precip line?
[arr['date.year'] for arr in chirps_water_year_arrs]
seasonal_precip # This just shows the array - corner cells have empty values b/c of projection mismatch @ edge of raster
water_year_arrs[0][0].plot.imshow()
water_year_arrs[0].integrate(dim="date", datetime_unit="D").plot.imshow()
# This chunk does the actual integration
all_eeflux_arr.integrate(dim="date", datetime_unit="D")
import pandas as pd
import numpy as np
labels = ['<=2', '3-9', '>=10']
bins = [0,2,9, np.inf]
pd.cut(all_eeflux_arr, bins, labels=labels)
all_eeflux_arr
import pandas as pd
all_scene_ids = [str(i) for i in list(all_scenes_f.glob("L*"))]
df = pd.DataFrame({"scene_id":all_scene_ids}).reindex()
split_vals_series = df.scene_id.str.split("/")
dff = pd.DataFrame(split_vals_series.to_list(), columns=['_', '__', '___', '____', '_____', '______', 'fname'])
df['date'] = dff['fname'].str.slice(10,18)
df['pathrow'] = dff['fname'].str.slice(4,10)
df['sensor'] = dff['fname'].str.slice(0,4)
df['datetime'] = pd.to_datetime(df['date'])
df = df.set_index("datetime").sort_index()
marc_df = df['2014-01-01':'2019-12-31']
marc_df = marc_df[marc_df['sensor']=="LC08"]
x.where(x != badvalue).sel(band=1).plot.imshow()
# Evan additions
year_tuples = years_list(all_eeflux_arr)
year_tuples
# Winter precip calculations
year_tuples_p = years_list(all_chirps_arr)
year_tuples_p
def group_p_by_custom_doy(all_chirps_arr, doy_start, doy_end):
start_end_years = years_list(all_chirps_arr)
water_year_arrs = []
for water_year in start_end_years:
start_mask = ((all_chirps_arr['date.dayofyear'].values > doy_start) & (all_chirps_arr['date.year'].values == water_year[0]))
end_mask = ((all_chirps_arr['date.dayofyear'].values < doy_end) & (all_chirps_arr['date.year'].values == water_year[0]))
water_year_arrs.append(all_chirps_arr[start_mask | end_mask])
return water_year_arrs
doystart = 275 # Edit these variables to change doy length of year
doyend = 125
water_year_arrs = group_p_by_custom_doy(all_chirps_arr, doystart, doyend)
water_year_arrs
def sum_seasonal_precip(precip_arr, eeflux_group_arr):
return precip_arr.sel(date=slice(eeflux_group_arr.date.min(), eeflux_group_arr.date.max())).sum(dim="date")
# This is matching up precip w/ available ET window for each year, need to figure out what to feed in for 2nd variable
for index, eeflux_group in enumerate(water_year_arrs):
if len(eeflux_group['date']) > 0:
seasonal_precip = sum_seasonal_precip(all_chirps_arr, eeflux_group) # Here's where above fxn is applied to rasters, need to replace eeflux_group
year_range = year_tuples_p[index]
pname = f"winter_chirps_{year_range[0]}_{year_range[1]}_{doystart}_{doyend}.tif" #Edit output raster labels
seasonal_precip.rio.to_raster(pname)
# This chunk actually outputs the rasters, ET lines removed
```
|
github_jupyter
|
# ChainerRL Quickstart Guide
This is a quickstart guide for users who just want to try ChainerRL for the first time.
If you have not yet installed ChainerRL, run the command below to install it:
```
%%bash
pip install chainerrl
```
If you have already installed ChainerRL, let's begin!
First, you need to import necessary modules. The module name of ChainerRL is `chainerrl`. Let's import `gym` and `numpy` as well since they are used later.
```
import chainer
import chainer.functions as F
import chainer.links as L
import chainerrl
import gym
import numpy as np
```
ChainerRL can be used for any problems if they are modeled as "environments". [OpenAI Gym](https://github.com/openai/gym) provides various kinds of benchmark environments and defines the common interface among them. ChainerRL uses a subset of the interface. Specifically, an environment must define its observation space and action space and have at least two methods: `reset` and `step`.
- `env.reset` will reset the environment to the initial state and return the initial observation.
- `env.step` will execute a given action, move to the next state and return four values:
- a next observation
- a scalar reward
- a boolean value indicating whether the current state is terminal or not
- additional information
- `env.render` will render the current state.
Let's try 'CartPole-v0', which is a classic control problem. You can see below that its observation space consists of four real numbers while its action space consists of two discrete actions.
```
env = gym.make('CartPole-v0')
print('observation space:', env.observation_space)
print('action space:', env.action_space)
obs = env.reset()
env.render(close=True)
print('initial observation:', obs)
action = env.action_space.sample()
obs, r, done, info = env.step(action)
print('next observation:', obs)
print('reward:', r)
print('done:', done)
print('info:', info)
```
Now you have defined your environment. Next, you need to define an agent, which will learn through interactions with the environment.
ChainerRL provides various agents, each of which implements a deep reinforcement learning algorithm.
To use [DQN (Deep Q-Network)](http://dx.doi.org/10.1038/nature14236), you need to define a Q-function that receives an observation and returns an expected future return for each action the agent can take. In ChainerRL, you can define your Q-function as `chainer.Link` as below. Note that the outputs are wrapped by `chainerrl.action_value.DiscreteActionValue`, which implements `chainerrl.action_value.ActionValue`. By wrapping the outputs of Q-functions, ChainerRL can treat discrete-action Q-functions like this and [NAFs (Normalized Advantage Functions)](https://arxiv.org/abs/1603.00748) in the same way.
```
class QFunction(chainer.Chain):
def __init__(self, obs_size, n_actions, n_hidden_channels=50):
super().__init__(
l0=L.Linear(obs_size, n_hidden_channels),
l1=L.Linear(n_hidden_channels, n_hidden_channels),
l2=L.Linear(n_hidden_channels, n_actions))
def __call__(self, x, test=False):
"""
Args:
x (ndarray or chainer.Variable): An observation
test (bool): a flag indicating whether it is in test mode
"""
h = F.tanh(self.l0(x))
h = F.tanh(self.l1(h))
return chainerrl.action_value.DiscreteActionValue(self.l2(h))
obs_size = env.observation_space.shape[0]
n_actions = env.action_space.n
q_func = QFunction(obs_size, n_actions)
```
If you want to use CUDA for computation, as usual as in Chainer, call `to_gpu`.
```
# Uncomment to use CUDA
# q_func.to_gpu(0)
```
You can also use ChainerRL's predefined Q-functions.
```
_q_func = chainerrl.q_functions.FCStateQFunctionWithDiscreteAction(
obs_size, n_actions,
n_hidden_layers=2, n_hidden_channels=50)
```
As in Chainer, `chainer.Optimizer` is used to update models.
```
# Use Adam to optimize q_func. eps=1e-2 is for stability.
optimizer = chainer.optimizers.Adam(eps=1e-2)
optimizer.setup(q_func)
```
A Q-function and its optimizer are used by a DQN agent. To create a DQN agent, you need to specify a bit more parameters and configurations.
```
# Set the discount factor that discounts future rewards.
gamma = 0.95
# Use epsilon-greedy for exploration
explorer = chainerrl.explorers.ConstantEpsilonGreedy(
epsilon=0.3, random_action_func=env.action_space.sample)
# DQN uses Experience Replay.
# Specify a replay buffer and its capacity.
replay_buffer = chainerrl.replay_buffer.ReplayBuffer(capacity=10 ** 6)
# Since observations from CartPole-v0 is numpy.float64 while
# Chainer only accepts numpy.float32 by default, specify
# a converter as a feature extractor function phi.
phi = lambda x: x.astype(np.float32, copy=False)
# Now create an agent that will interact with the environment.
agent = chainerrl.agents.DoubleDQN(
q_func, optimizer, replay_buffer, gamma, explorer,
replay_start_size=500, update_interval=1,
target_update_interval=100, phi=phi)
```
Now you have an agent and an environment. It's time to start reinforcement learning!
In training, use `agent.act_and_train` to select exploratory actions. `agent.stop_episode_and_train` must be called after finishing an episode. You can get training statistics of the agent via `agent.get_statistics`.
```
n_episodes = 200
max_episode_len = 200
for i in range(1, n_episodes + 1):
obs = env.reset()
reward = 0
done = False
R = 0 # return (sum of rewards)
t = 0 # time step
while not done and t < max_episode_len:
# Uncomment to watch the behaviour
# env.render()
action = agent.act_and_train(obs, reward)
obs, reward, done, _ = env.step(action)
R += reward
t += 1
if i % 10 == 0:
print('episode:', i,
'R:', R,
'statistics:', agent.get_statistics())
agent.stop_episode_and_train(obs, reward, done)
print('Finished.')
```
Now you finished training the agent. How good is the agent now? You can test it by using `agent.act` and `agent.stop_episode` instead. Exploration such as epsilon-greedy is not used anymore.
```
for i in range(10):
obs = env.reset()
done = False
R = 0
t = 0
while not done and t < 200:
env.render(close=True)
action = agent.act(obs)
obs, r, done, _ = env.step(action)
R += r
t += 1
print('test episode:', i, 'R:', R)
agent.stop_episode()
```
If test scores are good enough, the only remaining task is to save the agent so that you can reuse it. What you need to do is to simply call `agent.save` to save the agent, then `agent.load` to load the saved agent.
```
# Save an agent to the 'agent' directory
agent.save('agent')
# Uncomment to load an agent from the 'agent' directory
# agent.load('agent')
```
RL completed!
But writing code like this every time you use RL might be boring. So, ChainerRL has utility functions that do these things.
```
# Set up the logger to print info messages for understandability.
import logging
import sys
gym.undo_logger_setup() # Turn off gym's default logger settings
logging.basicConfig(level=logging.INFO, stream=sys.stdout, format='')
chainerrl.experiments.train_agent_with_evaluation(
agent, env,
steps=2000, # Train the agent for 2000 steps
eval_n_runs=10, # 10 episodes are sampled for each evaluation
max_episode_len=200, # Maximum length of each episodes
eval_interval=1000, # Evaluate the agent after every 1000 steps
outdir='result') # Save everything to 'result' directory
```
That's all of the ChainerRL quickstart guide. To know more about ChainerRL, please look into the `examples` directory and read and run the examples. Thank you!
|
github_jupyter
|
# Advanced Usage Exampes for Seldon Client
## Istio Gateway Request with token over HTTPS - no SSL verification
Test against a current kubeflow cluster with Dex token authentication.
1. Install kubeflow with Dex authentication
```
INGRESS_HOST=!kubectl -n istio-system get service istio-ingressgateway -o jsonpath='{.status.loadBalancer.ingress[0].ip}'
ISTIO_GATEWAY=INGRESS_HOST[0]
ISTIO_GATEWAY
```
Get a token from the Dex gateway. At present as Dex does not support curl password credentials you will need to get it from your browser logged into the cluster. Open up a browser console and run `document.cookie`
```
TOKEN="eyJhbGciOiJIUzUxMiIsInR5cCI6IkpXVCJ9.eyJleHAiOjE1NjM2MjA0ODYsImlhdCI6MTU2MzUzNDA4NiwiaXNzIjoiMzQuNjUuNzMuMjU1IiwianRpIjoiYjllNDQxOGQtZjNmNC00NTIyLTg5ODEtNDcxOTY0ODNmODg3IiwidWlmIjoiZXlKcGMzTWlPaUpvZEhSd2N6b3ZMek0wTGpZMUxqY3pMakkxTlRvMU5UVTJMMlJsZUNJc0luTjFZaUk2SWtOcFVYZFBSMFUwVG1wbk1GbHBNV3RaYW1jMFRGUlNhVTU2VFhSUFZFSm9UMU13ZWxreVVYaE9hbGw0V21wVk1FNXFXVk5DVjNoMldUSkdjeUlzSW1GMVpDSTZJbXQxWW1WbWJHOTNMV0YxZEdoelpYSjJhV05sTFc5cFpHTWlMQ0psZUhBaU9qRTFOak0yTWpBME9EWXNJbWxoZENJNk1UVTJNelV6TkRBNE5pd2lZWFJmYUdGemFDSTZJbE5OWlZWRGJUQmFOVkZoUTNCdVNHTndRMWgwTVZFaUxDSmxiV0ZwYkNJNkltRmtiV2x1UUhObGJHUnZiaTVwYnlJc0ltVnRZV2xzWDNabGNtbG1hV1ZrSWpwMGNuVmxMQ0p1WVcxbElqb2lZV1J0YVc0aWZRPT0ifQ.7CQIz4A1s9m6lJeWTqpz_JKGArGX4e_zpRCOXXjVRJgguB3z48rSfei_KL7niMCWpruhU11c8UIw9E79PwHNNw"
```
## Start Seldon Core
Use the setup notebook to [Install Seldon Core](seldon_core_setup.ipynb#Install-Seldon-Core) with [Istio Ingress](seldon_core_setup.ipynb#Istio). Instructions [also online](./seldon_core_setup.html).
**Note** When running helm install for this example you will need to set the istio.gateway flag to kubeflow-gateway (```--set istio.gateway=kubeflow-gateway```).
```
deployment_name="test1"
namespace="default"
from seldon_core.seldon_client import SeldonClient, SeldonChannelCredentials, SeldonCallCredentials
sc = SeldonClient(deployment_name=deployment_name,namespace=namespace,gateway_endpoint=ISTIO_GATEWAY,debug=True,
channel_credentials=SeldonChannelCredentials(verify=False),
call_credentials=SeldonCallCredentials(token=TOKEN))
r = sc.predict(gateway="istio",transport="rest",shape=(1,4))
print(r)
```
Its not presently possible to use gRPC without getting access to the certificates. We will update this once its clear how to obtain them from a Kubeflow cluser setup.
## Istio - SSL Endpoint - Client Side Verification - No Authentication
1. First run through the [Istio Secure Gateway SDS example](https://istio.io/docs/tasks/traffic-management/ingress/secure-ingress-sds/) and make sure this works for you.
* This will create certificates for `httpbin.example.com` and test them out.
1. Update your `/etc/hosts` file to include an entry for the ingress gateway for `httpbin.example.com` e.g. add a line like: `10.107.247.132 httpbin.example.com` replacing the ip address with your ingress gateway ip address.
```
# Set to folder where the httpbin certificates are
ISTIO_HTTPBIN_CERT_FOLDER='/home/clive/work/istio/httpbin.example.com'
```
## Start Seldon Core
Use the setup notebook to [Install Seldon Core](seldon_core_setup.ipynb#Install-Seldon-Core) with [Istio Ingress](seldon_core_setup.ipynb#Istio). Instructions [also online](./seldon_core_setup.html).
**Note** When running ```helm install``` for this example you will need to set the ```istio.gateway``` flag to ```mygateway``` (```--set istio.gateway=mygateway```) used in the example.
```
deployment_name="mymodel"
namespace="default"
from seldon_core.seldon_client import SeldonClient, SeldonChannelCredentials, SeldonCallCredentials
sc = SeldonClient(deployment_name=deployment_name,namespace=namespace,gateway_endpoint="httpbin.example.com",debug=True,
channel_credentials=SeldonChannelCredentials(certificate_chain_file=ISTIO_HTTPBIN_CERT_FOLDER+'/2_intermediate/certs/ca-chain.cert.pem',
root_certificates_file=ISTIO_HTTPBIN_CERT_FOLDER+'/4_client/certs/httpbin.example.com.cert.pem',
private_key_file=ISTIO_HTTPBIN_CERT_FOLDER+'/4_client/private/httpbin.example.com.key.pem'
))
r = sc.predict(gateway="istio",transport="rest",shape=(1,4))
print(r)
r = sc.predict(gateway="istio",transport="grpc",shape=(1,4))
print(r)
```
|
github_jupyter
|
<table width="100%">
<tr style="border-bottom:solid 2pt #009EE3">
<td style="text-align:left" width="10%">
<a href="prepare_anaconda.dwipynb" download><img src="../../images/icons/download.png"></a>
</td>
<td style="text-align:left" width="10%">
<a href="https://mybinder.org/v2/gh/biosignalsnotebooks/biosignalsnotebooks/biosignalsnotebooks_binder?filepath=biosignalsnotebooks_environment%2Fcategories%2FInstall%2Fprepare_anaconda.dwipynb" target="_blank"><img src="../../images/icons/program.png" title="Be creative and test your solutions !"></a>
</td>
<td></td>
<td style="text-align:left" width="5%">
<a href="../MainFiles/biosignalsnotebooks.ipynb"><img src="../../images/icons/home.png"></a>
</td>
<td style="text-align:left" width="5%">
<a href="../MainFiles/contacts.ipynb"><img src="../../images/icons/contacts.png"></a>
</td>
<td style="text-align:left" width="5%">
<a href="https://github.com/biosignalsnotebooks/biosignalsnotebooks" target="_blank"><img src="../../images/icons/github.png"></a>
</td>
<td style="border-left:solid 2pt #009EE3" width="15%">
<img src="../../images/ost_logo.png">
</td>
</tr>
</table>
<link rel="stylesheet" href="../../styles/theme_style.css">
<!--link rel="stylesheet" href="../../styles/header_style.css"-->
<link rel="stylesheet" href="https://cdnjs.cloudflare.com/ajax/libs/font-awesome/4.7.0/css/font-awesome.min.css">
<table width="100%">
<tr>
<td id="image_td" width="15%" class="header_image_color_13"><div id="image_img"
class="header_image_13"></div></td>
<td class="header_text"> Download, Install and Execute Anaconda </td>
</tr>
</table>
<div id="flex-container">
<div id="diff_level" class="flex-item">
<strong>Difficulty Level:</strong> <span class="fa fa-star checked"></span>
<span class="fa fa-star"></span>
<span class="fa fa-star"></span>
<span class="fa fa-star"></span>
<span class="fa fa-star"></span>
</div>
<div id="tag" class="flex-item-tag">
<span id="tag_list">
<table id="tag_list_table">
<tr>
<td class="shield_left">Tags</td>
<td class="shield_right" id="tags">install☁jupyter☁notebook☁anaconda☁download</td>
</tr>
</table>
</span>
<!-- [OR] Visit https://img.shields.io in order to create a tag badge-->
</div>
</div>
In every journey we always need to prepare our toolbox with the needed resources !
With <strong><span class="color1">biosignalsnotebooks</span></strong> happens the same, being <strong><span class="color4">Jupyter Notebook</span></strong> environment the most relevant application (that supports <strong><span class="color1">biosignalsnotebooks</span></strong>) to take the maximum advantage during your learning process.
In the following sequence of instruction it will be presented the operations that should be completed in order to have <strong><span class="color4">Jupyter Notebook</span></strong> ready to use and to open our <strong>ipynb</strong> files on local server.
<table width="100%">
<tr>
<td style="text-align:left;font-size:12pt;border-top:dotted 2px #62C3EE">
<span class="color1">☌</span> The current <span class="color4"><strong>Jupyter Notebook</strong></span> is focused on a complete Python toolbox called <a href="https://www.anaconda.com/distribution/"><span class="color4"><strong>Anaconda <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></span></a>.
However, there is an alternative approach to get all things ready for starting our journey, which is described on <a href="../Install/prepare_jupyter.ipynb"><span class="color1"><strong>"Download, Install and Execute Jypyter Notebook Environment" <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></span></a>
</td>
</tr>
</table>
<hr>
<hr>
<p class="steps">1 - Access the <strong><span class="color4">Anaconda</span></strong> official page at <a href="https://www.anaconda.com/distribution/">https://www.anaconda.com/distribution/</a></p>
<img src="../../images/other/anaconda_page.png">
<p class="steps">2 - Click on "Download" button, giving a first but strong step into our final objective</p>
<img src="../../images/other/anaconda_download.gif">
<p class="steps">3 - Specify the operating system of your local machine</p>
<img src="../../images/other/anaconda_download_os.gif">
<p class="steps">4 - Select the version of <span class="color1">Python</span> compiler to be included on <span class="color4">Anaconda</span></p>
It is strongly advisable that you chose version <strong>3.-</strong> to ensure that all functionalities of packages like <strong><span class="color1">biosignalsnotebooks</span></strong> are fully operational.
<img src="../../images/other/anaconda_download_version.gif">
<p class="steps">5 - After defining the directory where the downloaded file will be stored, please, wait a few minutes for the end of transfer</p>
<span class="color13" style="font-size:30px">⚠</span>
The waiting time will depend on the quality of the Internet connection !
<p class="steps">6 - When download is finished navigate through your directory tree until reaching the folder where the downloaded file is located</p>
In our case the destination folder was <img src="../../images/other/anaconda_download_location.png" style="display:inline;margin-top:0px">
<p class="steps">7 - Execute <span class="color4">Anaconda</span> installer file with a double-click</p>
<img src="../../images/other/anaconda_download_installer.gif">
<p class="steps">8 - Follow the sequential instructions presented on the <span class="color4">Anaconda</span> installer</p>
<img src="../../images/other/anaconda_download_install_steps.gif">
<p class="steps">9 - <span class="color4">Jupyter Notebook</span> environment is included on the previous installation. For starting your first Notebook execute <span class="color4">Jupyter Notebook</span></p>
Launch from "Anaconda Navigator" or through a command window, like described on the following steps.
<p class="steps">9.1 - For executing <span class="color4">Jupyter Notebook</span> environment you should open a <strong>console</strong> (in your operating system).</p>
<i>If you are a Microsoft Windows native, just type click on Windows logo (bottom-left corner of the screen) and type "cmd". Then press "Enter".</i>
<p class="steps">9.2 - Type <strong>"jupyter notebook"</strong> inside the opened console. A local <span class="color4"><strong>Jupyter Notebook</strong></span> server will be launched.</p>
<img src="../../images/other/open_jupyter.gif">
<p class="steps">10 - Create a blank Notebook</p>
<p class="steps">10.1 - Now, you should navigate through your directories until reaching the folder where you want to create or open a Notebook (as demonstrated in the following video)</p>
<span class="color13" style="font-size:30px">⚠</span>
<p style="margin-top:0px">You should note that your folder hierarchy is unique, so, the steps followed in the next image, will depend on your folder organisation, being merely illustrative </p>
<img src="../../images/other/create_notebook_part1.gif">
<p class="steps">10.2 - For creating a new Notebook, "New" button (top-right zone of Jupyter Notebook interface) should be pressed and <span class="color1"><strong>Python 3</strong></span> option selected.</p>
<i>A blank Notebook will arise and now you just need to be creative and expand your thoughts to others persons!!!</i>
<img src="../../images/other/create_notebook_part2.gif">
This can be the start of something great. Now you have all the software conditions to create and develop interactive tutorials, combining Python with HTML !
<span class="color4"><strong>Anaconda</strong></span> contains lots of additional functionalities, namely <a href="https://anaconda.org/anaconda/spyder"><span class="color7"><strong>Spyder <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></span></a>, which is an intuitive Python editor for creating and testing your own scripts.
<strong><span class="color7">We hope that you have enjoyed this guide. </span><span class="color2">biosignalsnotebooks</span><span class="color4"> is an environment in continuous expansion, so don't stop your journey and learn more with the remaining <a href="../MainFiles/biosignalsnotebooks.ipynb">Notebooks <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></a></span></strong> !
<hr>
<table width="100%">
<tr>
<td style="border-right:solid 3px #009EE3" width="20%">
<img src="../../images/ost_logo.png">
</td>
<td width="40%" style="text-align:left">
<a href="../MainFiles/aux_files/biosignalsnotebooks_presentation.pdf" target="_blank">☌ Project Presentation</a>
<br>
<a href="https://github.com/biosignalsnotebooks/biosignalsnotebooks" target="_blank">☌ GitHub Repository</a>
<br>
<a href="https://pypi.org/project/biosignalsnotebooks/" target="_blank">☌ How to install biosignalsnotebooks Python package ?</a>
<br>
<a href="../MainFiles/signal_samples.ipynb">☌ Signal Library</a>
</td>
<td width="40%" style="text-align:left">
<a href="../MainFiles/biosignalsnotebooks.ipynb">☌ Notebook Categories</a>
<br>
<a href="../MainFiles/by_diff.ipynb">☌ Notebooks by Difficulty</a>
<br>
<a href="../MainFiles/by_signal_type.ipynb">☌ Notebooks by Signal Type</a>
<br>
<a href="../MainFiles/by_tag.ipynb">☌ Notebooks by Tag</a>
</td>
</tr>
</table>
```
from biosignalsnotebooks.__notebook_support__ import css_style_apply
css_style_apply()
%%html
<script>
// AUTORUN ALL CELLS ON NOTEBOOK-LOAD!
require(
['base/js/namespace', 'jquery'],
function(jupyter, $) {
$(jupyter.events).on("kernel_ready.Kernel", function () {
console.log("Auto-running all cells-below...");
jupyter.actions.call('jupyter-notebook:run-all-cells-below');
jupyter.actions.call('jupyter-notebook:save-notebook');
});
}
);
</script>
```
|
github_jupyter
|
# Testing cosmogan
April 19, 2021
Borrowing pieces of code from :
- https://github.com/pytorch/tutorials/blob/11569e0db3599ac214b03e01956c2971b02c64ce/beginner_source/dcgan_faces_tutorial.py
- https://github.com/exalearn/epiCorvid/tree/master/cGAN
```
import os
import random
import logging
import sys
import torch
import torch.nn as nn
import torch.nn.parallel
import torch.backends.cudnn as cudnn
import torch.optim as optim
import torch.utils.data
from torchsummary import summary
from torch.utils.data import DataLoader, TensorDataset
import torch.distributed as dist
from torch.nn.parallel import DistributedDataParallel
# import torch.fft
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.animation as animation
# from IPython.display import HTML
import argparse
import time
from datetime import datetime
import glob
import pickle
import yaml
import collections
import socket
import shutil
# # Import modules from other files
# from utils import *
# from spec_loss import *
%matplotlib widget
```
## Modules
```
### Transformation functions for image pixel values
def f_transform(x):
return 2.*x/(x + 4.) - 1.
def f_invtransform(s):
return 4.*(1. + s)/(1. - s)
# Generator Code
class View(nn.Module):
def __init__(self, shape):
super(View, self).__init__()
self.shape = shape
def forward(self, x):
return x.view(*self.shape)
def f_get_model(gdict):
''' Module to define Generator and Discriminator'''
if gdict['image_size']==64:
class Generator(nn.Module):
def __init__(self, gdict):
super(Generator, self).__init__()
## Define new variables from dict
keys=['ngpu','nz','nc','ngf','kernel_size','stride','g_padding']
ngpu, nz,nc,ngf,kernel_size,stride,g_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
self.main = nn.Sequential(
# nn.ConvTranspose3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
nn.Linear(nz,nc*ngf*8**3),# 262144
nn.BatchNorm3d(nc,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
View(shape=[-1,ngf*8,4,4,4]),
nn.ConvTranspose3d(ngf * 8, ngf * 4, kernel_size, stride, g_padding, output_padding=1, bias=False),
nn.BatchNorm3d(ngf*4,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf*4) x 8 x 8
nn.ConvTranspose3d( ngf * 4, ngf * 2, kernel_size, stride, g_padding, 1, bias=False),
nn.BatchNorm3d(ngf*2,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf*2) x 16 x 16
nn.ConvTranspose3d( ngf * 2, ngf, kernel_size, stride, g_padding, 1, bias=False),
nn.BatchNorm3d(ngf,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf) x 32 x 32
nn.ConvTranspose3d( ngf, nc, kernel_size, stride,g_padding, 1, bias=False),
nn.Tanh()
)
def forward(self, ip):
return self.main(ip)
class Discriminator(nn.Module):
def __init__(self, gdict):
super(Discriminator, self).__init__()
## Define new variables from dict
keys=['ngpu','nz','nc','ndf','kernel_size','stride','d_padding']
ngpu, nz,nc,ndf,kernel_size,stride,d_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
self.main = nn.Sequential(
# input is (nc) x 64 x 64 x 64
# nn.Conv3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
nn.Conv3d(nc, ndf,kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf) x 32 x 32
nn.Conv3d(ndf, ndf * 2, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 2,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*2) x 16 x 16
nn.Conv3d(ndf * 2, ndf * 4, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 4,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*4) x 8 x 8
nn.Conv3d(ndf * 4, ndf * 8, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 8,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*8) x 4 x 4
nn.Flatten(),
nn.Linear(nc*ndf*8*8*8, 1)
# nn.Sigmoid()
)
def forward(self, ip):
# print(ip.shape)
results=[ip]
lst_idx=[]
for i,submodel in enumerate(self.main.children()):
mid_output=submodel(results[-1])
results.append(mid_output)
## Select indices in list corresponding to output of Conv layers
if submodel.__class__.__name__.startswith('Conv'):
# print(submodel.__class__.__name__)
# print(mid_output.shape)
lst_idx.append(i)
FMloss=True
if FMloss:
ans=[results[1:][i] for i in lst_idx + [-1]]
else :
ans=results[-1]
return ans
elif gdict['image_size']==128:
class Generator(nn.Module):
def __init__(self, gdict):
super(Generator, self).__init__()
## Define new variables from dict
keys=['ngpu','nz','nc','ngf','kernel_size','stride','g_padding']
ngpu, nz,nc,ngf,kernel_size,stride,g_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
self.main = nn.Sequential(
# nn.ConvTranspose3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
nn.Linear(nz,nc*ngf*8**3*8),# 262144
nn.BatchNorm3d(nc,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
View(shape=[-1,ngf*8,8,8,8]),
nn.ConvTranspose3d(ngf * 8, ngf * 4, kernel_size, stride, g_padding, output_padding=1, bias=False),
nn.BatchNorm3d(ngf*4,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf*4) x 8 x 8
nn.ConvTranspose3d( ngf * 4, ngf * 2, kernel_size, stride, g_padding, 1, bias=False),
nn.BatchNorm3d(ngf*2,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf*2) x 16 x 16
nn.ConvTranspose3d( ngf * 2, ngf, kernel_size, stride, g_padding, 1, bias=False),
nn.BatchNorm3d(ngf,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf) x 32 x 32
nn.ConvTranspose3d( ngf, nc, kernel_size, stride,g_padding, 1, bias=False),
nn.Tanh()
)
def forward(self, ip):
return self.main(ip)
class Discriminator(nn.Module):
def __init__(self, gdict):
super(Discriminator, self).__init__()
## Define new variables from dict
keys=['ngpu','nz','nc','ndf','kernel_size','stride','d_padding']
ngpu, nz,nc,ndf,kernel_size,stride,d_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
self.main = nn.Sequential(
# input is (nc) x 64 x 64 x 64
# nn.Conv3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
nn.Conv3d(nc, ndf,kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf) x 32 x 32
nn.Conv3d(ndf, ndf * 2, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 2,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*2) x 16 x 16
nn.Conv3d(ndf * 2, ndf * 4, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 4,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*4) x 8 x 8
nn.Conv3d(ndf * 4, ndf * 8, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 8,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*8) x 4 x 4
nn.Flatten(),
nn.Linear(nc*ndf*8*8*8*8, 1)
# nn.Sigmoid()
)
def forward(self, ip):
results=[ip]
lst_idx=[]
for i,submodel in enumerate(self.main.children()):
mid_output=submodel(results[-1])
results.append(mid_output)
## Select indices in list corresponding to output of Conv layers
if submodel.__class__.__name__.startswith('Conv'):
# print(submodel.__class__.__name__)
# print(mid_output.shape)
lst_idx.append(i)
FMloss=True
if FMloss:
ans=[results[1:][i] for i in lst_idx + [-1]]
else :
ans=results[-1]
return ans
return Generator, Discriminator
def f_gen_images(gdict,netG,optimizerG,ip_fname,op_loc,op_strg='inf_img_',op_size=500):
'''Generate images for best saved models
Arguments: gdict, netG, optimizerG,
ip_fname: name of input file
op_strg: [string name for output file]
op_size: Number of images to generate
'''
nz,device=gdict['nz'],gdict['device']
try:# handling cpu vs gpu
if torch.cuda.is_available(): checkpoint=torch.load(ip_fname)
else: checkpoint=torch.load(ip_fname,map_location=torch.device('cpu'))
except Exception as e:
print(e)
print("skipping generation of images for ",ip_fname)
return
## Load checkpoint
if gdict['multi-gpu']:
netG.module.load_state_dict(checkpoint['G_state'])
else:
netG.load_state_dict(checkpoint['G_state'])
## Load other stuff
iters=checkpoint['iters']
epoch=checkpoint['epoch']
optimizerG.load_state_dict(checkpoint['optimizerG_state_dict'])
# Generate batch of latent vectors
noise = torch.randn(op_size, 1, 1, 1, nz, device=device)
# Generate fake image batch with G
netG.eval() ## This is required before running inference
with torch.no_grad(): ## This is important. fails without it for multi-gpu
gen = netG(noise)
gen_images=gen.detach().cpu().numpy()
print(gen_images.shape)
op_fname='%s_epoch-%s_step-%s.npy'%(op_strg,epoch,iters)
np.save(op_loc+op_fname,gen_images)
print("Image saved in ",op_fname)
def f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,netG,netD,optimizerG,optimizerD,save_loc):
''' Checkpoint model '''
if gdict['multi-gpu']: ## Dataparallel
torch.save({'epoch':epoch,'iters':iters,'best_chi1':best_chi1,'best_chi2':best_chi2,
'G_state':netG.module.state_dict(),'D_state':netD.module.state_dict(),'optimizerG_state_dict':optimizerG.state_dict(),
'optimizerD_state_dict':optimizerD.state_dict()}, save_loc)
else :
torch.save({'epoch':epoch,'iters':iters,'best_chi1':best_chi1,'best_chi2':best_chi2,
'G_state':netG.state_dict(),'D_state':netD.state_dict(),'optimizerG_state_dict':optimizerG.state_dict(),
'optimizerD_state_dict':optimizerD.state_dict()}, save_loc)
def f_load_checkpoint(ip_fname,netG,netD,optimizerG,optimizerD,gdict):
''' Load saved checkpoint
Also loads step, epoch, best_chi1, best_chi2'''
print("torch device",torch.device('cuda',torch.cuda.current_device()))
try:
checkpoint=torch.load(ip_fname,map_location=torch.device('cuda',torch.cuda.current_device()))
except Exception as e:
print("Error loading saved checkpoint",ip_fname)
print(e)
raise SystemError
## Load checkpoint
if gdict['multi-gpu']:
netG.module.load_state_dict(checkpoint['G_state'])
netD.module.load_state_dict(checkpoint['D_state'])
else:
netG.load_state_dict(checkpoint['G_state'])
netD.load_state_dict(checkpoint['D_state'])
optimizerD.load_state_dict(checkpoint['optimizerD_state_dict'])
optimizerG.load_state_dict(checkpoint['optimizerG_state_dict'])
iters=checkpoint['iters']
epoch=checkpoint['epoch']
best_chi1=checkpoint['best_chi1']
best_chi2=checkpoint['best_chi2']
netG.train()
netD.train()
return iters,epoch,best_chi1,best_chi2,netD,optimizerD,netG,optimizerG
####################
### Pytorch code ###
####################
def f_get_rad(img):
''' Get the radial tensor for use in f_torch_get_azimuthalAverage '''
height,width,depth=img.shape[-3:]
# Create a grid of points with x and y and z coordinates
z,y,x = np.indices([height,width,depth])
center=[]
if not center:
center = np.array([(x.max()-x.min())/2.0, (y.max()-y.min())/2.0, (z.max()-z.min())/2.0])
# Get the radial coordinate for every grid point. Array has the shape of image
r= torch.tensor(np.sqrt((x-center[0])**2 + (y-center[1])**2 + (z-center[2])**2))
# Get sorted radii
ind = torch.argsort(torch.reshape(r, (-1,)))
return r.detach(),ind.detach()
def f_torch_get_azimuthalAverage(image,r,ind):
"""
Calculate the azimuthally averaged radial profile.
image - The 2D image
center - The [x,y] pixel coordinates used as the center. The default is
None, which then uses the center of the image (including
fracitonal pixels).
source: https://www.astrobetter.com/blog/2010/03/03/fourier-transforms-of-images-in-python/
"""
# height, width = image.shape
# # Create a grid of points with x and y coordinates
# y, x = np.indices([height,width])
# if not center:
# center = np.array([(x.max()-x.min())/2.0, (y.max()-y.min())/2.0])
# # Get the radial coordinate for every grid point. Array has the shape of image
# r = torch.tensor(np.hypot(x - center[0], y - center[1]))
# # Get sorted radii
# ind = torch.argsort(torch.reshape(r, (-1,)))
r_sorted = torch.gather(torch.reshape(r, ( -1,)),0, ind)
i_sorted = torch.gather(torch.reshape(image, ( -1,)),0, ind)
# Get the integer part of the radii (bin size = 1)
r_int=r_sorted.to(torch.int32)
# Find all pixels that fall within each radial bin.
deltar = r_int[1:] - r_int[:-1] # Assumes all radii represented
rind = torch.reshape(torch.where(deltar)[0], (-1,)) # location of changes in radius
nr = (rind[1:] - rind[:-1]).type(torch.float) # number of radius bin
# Cumulative sum to figure out sums for each radius bin
csum = torch.cumsum(i_sorted, axis=-1)
tbin = torch.gather(csum, 0, rind[1:]) - torch.gather(csum, 0, rind[:-1])
radial_prof = tbin / nr
return radial_prof
def f_torch_fftshift(real, imag):
for dim in range(0, len(real.size())):
real = torch.roll(real, dims=dim, shifts=real.size(dim)//2)
imag = torch.roll(imag, dims=dim, shifts=imag.size(dim)//2)
return real, imag
def f_torch_compute_spectrum(arr,r,ind):
GLOBAL_MEAN=1.0
arr=(arr-GLOBAL_MEAN)/(GLOBAL_MEAN)
y1=torch.rfft(arr,signal_ndim=3,onesided=False)
real,imag=f_torch_fftshift(y1[:,:,:,0],y1[:,:,:,1]) ## last index is real/imag part ## Mod for 3D
# # For pytorch 1.8
# y1=torch.fft.fftn(arr,dim=(-3,-2,-1))
# real,imag=f_torch_fftshift(y1.real,y1.imag)
y2=real**2+imag**2 ## Absolute value of each complex number
z1=f_torch_get_azimuthalAverage(y2,r,ind) ## Compute radial profile
return z1
def f_torch_compute_batch_spectrum(arr,r,ind):
batch_pk=torch.stack([f_torch_compute_spectrum(i,r,ind) for i in arr])
return batch_pk
def f_torch_image_spectrum(x,num_channels,r,ind):
'''
Data has to be in the form (batch,channel,x,y)
'''
mean=[[] for i in range(num_channels)]
var=[[] for i in range(num_channels)]
for i in range(num_channels):
arr=x[:,i,:,:,:] # Mod for 3D
batch_pk=f_torch_compute_batch_spectrum(arr,r,ind)
mean[i]=torch.mean(batch_pk,axis=0)
# var[i]=torch.std(batch_pk,axis=0)/np.sqrt(batch_pk.shape[0])
# var[i]=torch.std(batch_pk,axis=0)
var[i]=torch.var(batch_pk,axis=0)
mean=torch.stack(mean)
var=torch.stack(var)
if (torch.isnan(mean).any() or torch.isnan(var).any()):
print("Nans in spectrum",mean,var)
if torch.isnan(x).any():
print("Nans in Input image")
return mean,var
def f_compute_hist(data,bins):
try:
hist_data=torch.histc(data,bins=bins)
## A kind of normalization of histograms: divide by total sum
hist_data=(hist_data*bins)/torch.sum(hist_data)
except Exception as e:
print(e)
hist_data=torch.zeros(bins)
return hist_data
### Losses
def loss_spectrum(spec_mean,spec_mean_ref,spec_var,spec_var_ref,image_size,lambda_spec_mean,lambda_spec_var):
''' Loss function for the spectrum : mean + variance
Log(sum( batch value - expect value) ^ 2 )) '''
if (torch.isnan(spec_mean).any() or torch.isnan(spec_var).any()):
ans=torch.tensor(float("inf"))
return ans
idx=int(image_size/2) ### For the spectrum, use only N/2 indices for loss calc.
### Warning: the first index is the channel number.For multiple channels, you are averaging over them, which is fine.
loss_mean=torch.log(torch.mean(torch.pow(spec_mean[:,:idx]-spec_mean_ref[:,:idx],2)))
loss_var=torch.log(torch.mean(torch.pow(spec_var[:,:idx]-spec_var_ref[:,:idx],2)))
ans=lambda_spec_mean*loss_mean+lambda_spec_var*loss_var
if (torch.isnan(ans).any()) :
print("loss spec mean %s, loss spec var %s"%(loss_mean,loss_var))
print("spec mean %s, ref %s"%(spec_mean, spec_mean_ref))
print("spec var %s, ref %s"%(spec_var, spec_var_ref))
# raise SystemExit
return ans
def loss_hist(hist_sample,hist_ref):
lambda1=1.0
return lambda1*torch.log(torch.mean(torch.pow(hist_sample-hist_ref,2)))
def f_FM_loss(real_output,fake_output,lambda_fm,gdict):
'''
Module to implement Feature-Matching loss. Reads all but last elements of Discriminator ouput
'''
FM=torch.Tensor([0.0]).to(gdict['device'])
for i,j in zip(real_output[:-1],fake_output[:-1]):
# print(i.shape,j.shape)
real_mean=torch.mean(i)
fake_mean=torch.mean(j)
# print(real_mean,fake_mean)
FM=FM.clone()+torch.sum(torch.square(real_mean-fake_mean))
return lambda_fm*FM
def f_gp_loss(grads,l=1.0):
'''
Module to implement gradient penalty loss.
'''
loss=torch.mean(torch.sum(torch.square(grads),dim=[1,2,3]))
return l*loss
```
## Train loop
```
### Train code ###
def f_train_loop(gan_model,Dset,metrics_df,gdict,fixed_noise):
''' Train epochs '''
## Define new variables from dict
keys=['image_size','start_epoch','epochs','iters','best_chi1','best_chi2','save_dir','device','flip_prob','nz','batch_size','bns']
image_size,start_epoch,epochs,iters,best_chi1,best_chi2,save_dir,device,flip_prob,nz,batchsize,bns=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
for epoch in range(start_epoch,epochs):
t_epoch_start=time.time()
for count, data in enumerate(Dset.train_dataloader):
####### Train GAN ########
gan_model.netG.train(); gan_model.netD.train(); ### Need to add these after inference and before training
tme1=time.time()
### Update D network: maximize log(D(x)) + log(1 - D(G(z)))
gan_model.netD.zero_grad()
real_cpu = data[0].to(device)
real_cpu.requires_grad=True
b_size = real_cpu.size(0)
real_label = torch.full((b_size,), 1, device=device,dtype=float)
fake_label = torch.full((b_size,), 0, device=device,dtype=float)
g_label = torch.full((b_size,), 1, device=device,dtype=float) ## No flipping for Generator labels
# Flip labels with probability flip_prob
for idx in np.random.choice(np.arange(b_size),size=int(np.ceil(b_size*flip_prob))):
real_label[idx]=0; fake_label[idx]=1
# Generate fake image batch with G
noise = torch.randn(b_size, 1, 1, 1, nz, device=device) ### Mod for 3D
fake = gan_model.netG(noise)
# Forward pass real batch through D
real_output = gan_model.netD(real_cpu)
errD_real = gan_model.criterion(real_output[-1].view(-1), real_label.float())
errD_real.backward(retain_graph=True)
D_x = real_output[-1].mean().item()
# Forward pass fake batch through D
fake_output = gan_model.netD(fake.detach()) # The detach is important
errD_fake = gan_model.criterion(fake_output[-1].view(-1), fake_label.float())
errD_fake.backward(retain_graph=True)
D_G_z1 = fake_output[-1].mean().item()
errD = errD_real + errD_fake
if gdict['lambda_gp']: ## Add gradient - penalty loss
grads=torch.autograd.grad(outputs=real_output[-1],inputs=real_cpu,grad_outputs=torch.ones_like(real_output[-1]),allow_unused=False,create_graph=True)[0]
gp_loss=f_gp_loss(grads,gdict['lambda_gp'])
gp_loss.backward(retain_graph=True)
errD = errD + gp_loss
else:
gp_loss=torch.Tensor([np.nan])
if gdict['grad_clip']:
nn.utils.clip_grad_norm_(gan_model.netD.parameters(),gdict['grad_clip'])
gan_model.optimizerD.step()
lr_d=gan_model.optimizerD.param_groups[0]['lr']
gan_model.schedulerD.step()
# dict_keys(['train_data_loader', 'r', 'ind', 'train_spec_mean', 'train_spec_var', 'train_hist', 'val_spec_mean', 'val_spec_var', 'val_hist'])
###Update G network: maximize log(D(G(z)))
gan_model.netG.zero_grad()
output = gan_model.netD(fake)
errG_adv = gan_model.criterion(output[-1].view(-1), g_label.float())
# errG_adv.backward(retain_graph=True)
# Histogram pixel intensity loss
hist_gen=f_compute_hist(fake,bins=bns)
hist_loss=loss_hist(hist_gen,Dset.train_hist.to(device))
# Add spectral loss
mean,var=f_torch_image_spectrum(f_invtransform(fake),1,Dset.r.to(device),Dset.ind.to(device))
spec_loss=loss_spectrum(mean,Dset.train_spec_mean.to(device),var,Dset.train_spec_var.to(device),image_size,gdict['lambda_spec_mean'],gdict['lambda_spec_var'])
errG=errG_adv
if gdict['lambda_spec_mean']:
# spec_loss.backward(retain_graph=True)
errG = errG+ spec_loss
if gdict['lambda_fm']:## Add feature matching loss
fm_loss=f_FM_loss([i.detach() for i in real_output],output,gdict['lambda_fm'],gdict)
# fm_loss.backward(retain_graph=True)
errG= errG+ fm_loss
else:
fm_loss=torch.Tensor([np.nan])
if torch.isnan(errG).any():
logging.info(errG)
raise SystemError
# Calculate gradients for G
errG.backward()
D_G_z2 = output[-1].mean().item()
### Implement Gradient clipping
if gdict['grad_clip']:
nn.utils.clip_grad_norm_(gan_model.netG.parameters(),gdict['grad_clip'])
gan_model.optimizerG.step()
lr_g=gan_model.optimizerG.param_groups[0]['lr']
gan_model.schedulerG.step()
tme2=time.time()
####### Store metrics ########
# Output training stats
if gdict['world_rank']==0:
if ((count % gdict['checkpoint_size'] == 0)):
logging.info('[%d/%d][%d/%d]\tLoss_D: %.4f\tLoss_adv: %.4f\tLoss_G: %.4f\tD(x): %.4f\tD(G(z)): %.4f / %.4f'
% (epoch, epochs, count, len(Dset.train_dataloader), errD.item(), errG_adv.item(),errG.item(), D_x, D_G_z1, D_G_z2)),
logging.info("Spec loss: %s,\t hist loss: %s"%(spec_loss.item(),hist_loss.item())),
logging.info("Training time for step %s : %s"%(iters, tme2-tme1))
# Save metrics
cols=['step','epoch','Dreal','Dfake','Dfull','G_adv','G_full','spec_loss','hist_loss','fm_loss','gp_loss','D(x)','D_G_z1','D_G_z2','lr_d','lr_g','time']
vals=[iters,epoch,errD_real.item(),errD_fake.item(),errD.item(),errG_adv.item(),errG.item(),spec_loss.item(),hist_loss.item(),fm_loss.item(),gp_loss.item(),D_x,D_G_z1,D_G_z2,lr_d,lr_g,tme2-tme1]
for col,val in zip(cols,vals): metrics_df.loc[iters,col]=val
### Checkpoint the best model
checkpoint=True
iters += 1 ### Model has been updated, so update iters before saving metrics and model.
### Compute validation metrics for updated model
gan_model.netG.eval()
with torch.no_grad():
fake = gan_model.netG(fixed_noise)
hist_gen=f_compute_hist(fake,bins=bns)
hist_chi=loss_hist(hist_gen,Dset.val_hist.to(device))
mean,var=f_torch_image_spectrum(f_invtransform(fake),1,Dset.r.to(device),Dset.ind.to(device))
spec_chi=loss_spectrum(mean,Dset.val_spec_mean.to(device),var,Dset.val_spec_var.to(device),image_size,gdict['lambda_spec_mean'],gdict['lambda_spec_var'])
# Storing chi for next step
for col,val in zip(['spec_chi','hist_chi'],[spec_chi.item(),hist_chi.item()]): metrics_df.loc[iters,col]=val
# Checkpoint model for continuing run
if count == len(Dset.train_dataloader)-1: ## Check point at last step of epoch
f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_last.tar')
if (checkpoint and (epoch > 1)): # Choose best models by metric
if hist_chi< best_chi1:
f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_best_hist.tar')
best_chi1=hist_chi.item()
logging.info("Saving best hist model at epoch %s, step %s."%(epoch,iters))
if spec_chi< best_chi2:
f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_best_spec.tar')
best_chi2=spec_chi.item()
logging.info("Saving best spec model at epoch %s, step %s"%(epoch,iters))
# if (iters in gdict['save_steps_list']) :
if ((gdict['save_steps_list']=='all') and (iters % gdict['checkpoint_size'] == 0)):
f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_{0}.tar'.format(iters))
logging.info("Saving given-step at epoch %s, step %s."%(epoch,iters))
# Save G's output on fixed_noise
if ((iters % gdict['checkpoint_size'] == 0) or ((epoch == epochs-1) and (count == len(Dset.train_dataloader)-1))):
gan_model.netG.eval()
with torch.no_grad():
fake = gan_model.netG(fixed_noise).detach().cpu()
img_arr=np.array(fake)
fname='gen_img_epoch-%s_step-%s'%(epoch,iters)
np.save(save_dir+'/images/'+fname,img_arr)
t_epoch_end=time.time()
if gdict['world_rank']==0:
logging.info("Time taken for epoch %s, count %s: %s for rank %s"%(epoch,count,t_epoch_end-t_epoch_start,gdict['world_rank']))
# Save Metrics to file after each epoch
metrics_df.to_pickle(save_dir+'/df_metrics.pkle')
logging.info("best chis: {0}, {1}".format(best_chi1,best_chi2))
```
## Start
```
### Setup modules ###
def f_manual_add_argparse():
''' use only in jpt notebook'''
args=argparse.Namespace()
args.config='config_3dgan_128_cori.yaml'
args.mode='fresh'
args.local_rank=0
args.facility='cori'
args.distributed=False
# args.mode='continue'
return args
def f_parse_args():
"""Parse command line arguments.Only for .py file"""
parser = argparse.ArgumentParser(description="Run script to train GAN using pytorch", formatter_class=argparse.ArgumentDefaultsHelpFormatter)
add_arg = parser.add_argument
add_arg('--config','-cfile', type=str, default='config_3d_Cgan.yaml', help='Name of config file')
add_arg('--mode','-m', type=str, choices=['fresh','continue','fresh_load'],default='fresh', help='Whether to start fresh run or continue previous run or fresh run loading a config file.')
add_arg("--local_rank", default=0, type=int,help='Local rank of GPU on node. Using for pytorch DDP. ')
add_arg("--facility", default='cori', choices=['cori','summit'],type=str,help='Facility: cori or summit ')
add_arg("--ddp", dest='distributed' ,default=False,action='store_true',help='use Distributed DataParallel for Pytorch or DataParallel')
return parser.parse_args()
def try_barrier(rank):
"""
Used in Distributed data parallel
Attempt a barrier but ignore any exceptions
"""
print('BAR %d'%rank)
try:
dist.barrier()
except:
pass
def f_init_gdict(args,gdict):
''' Create global dictionary gdict from args and config file'''
## read config file
config_file=args.config
with open(config_file) as f:
config_dict= yaml.load(f, Loader=yaml.SafeLoader)
gdict=config_dict['parameters']
args_dict=vars(args)
## Add args variables to gdict
for key in args_dict.keys():
gdict[key]=args_dict[key]
if gdict['distributed']:
assert not gdict['lambda_gp'],"GP couplings is %s. Cannot use Gradient penalty loss in pytorch DDP"%(gdict['lambda_gp'])
else : print("Not using DDP")
return gdict
def f_get_img_samples(ip_arr,rank=0,num_ranks=1):
'''
Module to get part of the numpy image file
'''
data_size=ip_arr.shape[0]
size=data_size//num_ranks
if gdict['batch_size']>size:
print("Caution: batchsize %s is greater than samples per GPU %s"%(gdict['batch_size'],size))
raise SystemExit
### Get a set of random indices from numpy array
random=False
if random:
idxs=np.arange(ip_arr.shape[0])
np.random.shuffle(idxs)
rnd_idxs=idxs[rank*(size):(rank+1)*size]
arr=ip_arr[rnd_idxs].copy()
else: arr=ip_arr[rank*(size):(rank+1)*size].copy()
return arr
def f_setup(gdict,metrics_df,log):
'''
Set up directories, Initialize random seeds, add GPU info, add logging info.
'''
torch.backends.cudnn.benchmark=True
# torch.autograd.set_detect_anomaly(True)
## New additions. Code taken from Jan B.
os.environ['MASTER_PORT'] = "8885"
if gdict['facility']=='summit':
get_master = "echo $(cat {} | sort | uniq | grep -v batch | grep -v login | head -1)".format(os.environ['LSB_DJOB_HOSTFILE'])
os.environ['MASTER_ADDR'] = str(subprocess.check_output(get_master, shell=True))[2:-3]
os.environ['WORLD_SIZE'] = os.environ['OMPI_COMM_WORLD_SIZE']
os.environ['RANK'] = os.environ['OMPI_COMM_WORLD_RANK']
gdict['local_rank'] = int(os.environ['OMPI_COMM_WORLD_LOCAL_RANK'])
else:
if gdict['distributed']:
os.environ['WORLD_SIZE'] = os.environ['SLURM_NTASKS']
os.environ['RANK'] = os.environ['SLURM_PROCID']
gdict['local_rank'] = int(os.environ['SLURM_LOCALID'])
## Special declarations
gdict['ngpu']=torch.cuda.device_count()
gdict['device']=torch.device("cuda" if (torch.cuda.is_available()) else "cpu")
gdict['multi-gpu']=True if (gdict['device'].type == 'cuda') and (gdict['ngpu'] > 1) else False
########################
###### Set up Distributed Data parallel ######
if gdict['distributed']:
# gdict['local_rank']=args.local_rank ## This is needed when using pytorch -m torch.distributed.launch
gdict['world_size']=int(os.environ['WORLD_SIZE'])
torch.cuda.set_device(gdict['local_rank']) ## Very important
dist.init_process_group(backend='nccl', init_method="env://")
gdict['world_rank']= dist.get_rank()
device = torch.cuda.current_device()
logging.info("World size %s, world rank %s, local rank %s device %s, hostname %s, GPUs on node %s\n"%(gdict['world_size'],gdict['world_rank'],gdict['local_rank'],device,socket.gethostname(),gdict['ngpu']))
# Divide batch size by number of GPUs
# gdict['batch_size']=gdict['batch_size']//gdict['world_size']
else:
gdict['world_size'],gdict['world_rank'],gdict['local_rank']=1,0,0
########################
###### Set up directories #######
### sync up so that time is the same for each GPU for DDP
if gdict['mode'] in ['fresh','fresh_load']:
### Create prefix for foldername
if gdict['world_rank']==0: ### For rank=0, create directory name string and make directories
dt_strg=datetime.now().strftime('%Y%m%d_%H%M%S') ## time format
dt_lst=[int(i) for i in dt_strg.split('_')] # List storing day and time
dt_tnsr=torch.LongTensor(dt_lst).to(gdict['device']) ## Create list to pass to other GPUs
else: dt_tnsr=torch.Tensor([0,0]).long().to(gdict['device'])
### Pass directory name to other ranks
if gdict['distributed']: dist.broadcast(dt_tnsr, src=0)
gdict['save_dir']=gdict['op_loc']+str(int(dt_tnsr[0]))+'_'+str(int(dt_tnsr[1]))+'_'+gdict['run_suffix']
if gdict['world_rank']==0: # Create directories for rank 0
### Create directories
if not os.path.exists(gdict['save_dir']):
os.makedirs(gdict['save_dir']+'/models')
os.makedirs(gdict['save_dir']+'/images')
shutil.copy(gdict['config'],gdict['save_dir'])
elif gdict['mode']=='continue': ## For checkpointed runs
gdict['save_dir']=gdict['ip_fldr']
### Read loss data
metrics_df=pd.read_pickle(gdict['save_dir']+'/df_metrics.pkle').astype(np.float64)
########################
### Initialize random seed
manualSeed = np.random.randint(1, 10000) if gdict['seed']=='random' else int(gdict['seed'])
# print("Seed",manualSeed,gdict['world_rank'])
random.seed(manualSeed)
np.random.seed(manualSeed)
torch.manual_seed(manualSeed)
torch.cuda.manual_seed_all(manualSeed)
if gdict['deterministic']:
logging.info("Running with deterministic sequence. Performance will be slower")
torch.backends.cudnn.deterministic=True
# torch.backends.cudnn.enabled = False
torch.backends.cudnn.benchmark = False
########################
if log:
### Write all logging.info statements to stdout and log file
logfile=gdict['save_dir']+'/log.log'
if gdict['world_rank']==0:
logging.basicConfig(level=logging.DEBUG, filename=logfile, filemode="a+", format="%(asctime)-15s %(levelname)-8s %(message)s")
Lg = logging.getLogger()
Lg.setLevel(logging.DEBUG)
lg_handler_file = logging.FileHandler(logfile)
lg_handler_stdout = logging.StreamHandler(sys.stdout)
Lg.addHandler(lg_handler_file)
Lg.addHandler(lg_handler_stdout)
logging.info('Args: {0}'.format(args))
logging.info('Start: %s'%(datetime.now().strftime('%Y-%m-%d %H:%M:%S')))
if gdict['distributed']: try_barrier(gdict['world_rank'])
if gdict['world_rank']!=0:
logging.basicConfig(level=logging.DEBUG, filename=logfile, filemode="a+", format="%(asctime)-15s %(levelname)-8s %(message)s")
return metrics_df
class Dataset:
def __init__(self,gdict):
'''
Load training dataset and compute spectrum and histogram for a small batch of training and validation dataset.
'''
## Load training dataset
t0a=time.time()
img=np.load(gdict['ip_fname'],mmap_mode='r')[:gdict['num_imgs']]
# print("Shape of input file",img.shape)
img=f_get_img_samples(img,gdict['world_rank'],gdict['world_size'])
t_img=torch.from_numpy(img)
dataset=TensorDataset(t_img)
self.train_dataloader=DataLoader(dataset,batch_size=gdict['batch_size'],shuffle=True,num_workers=0,drop_last=True)
logging.info("Size of dataset for GPU %s : %s"%(gdict['world_rank'],len(self.train_dataloader.dataset)))
t0b=time.time()
logging.info("Time for creating dataloader",t0b-t0a,gdict['world_rank'])
# Precompute spectrum and histogram for small training and validation data for computing losses
with torch.no_grad():
val_img=np.load(gdict['ip_fname'],mmap_mode='r')[-100:].copy()
t_val_img=torch.from_numpy(val_img).to(gdict['device'])
# Precompute radial coordinates
r,ind=f_get_rad(val_img)
self.r,self.ind=r.to(gdict['device']),ind.to(gdict['device'])
# Compute
self.train_spec_mean,self.train_spec_var=f_torch_image_spectrum(f_invtransform(t_val_img),1,self.r,self.ind)
self.train_hist=f_compute_hist(t_val_img,bins=gdict['bns'])
# Repeat for validation dataset
val_img=np.load(gdict['ip_fname'],mmap_mode='r')[-200:-100].copy()
t_val_img=torch.from_numpy(val_img).to(gdict['device'])
# Compute
self.val_spec_mean,self.val_spec_var=f_torch_image_spectrum(f_invtransform(t_val_img),1,self.r,self.ind)
self.val_hist=f_compute_hist(t_val_img,bins=gdict['bns'])
del val_img; del t_val_img; del img; del t_img;
class GAN_model():
def __init__(self,gdict,print_model=False):
def weights_init(m):
'''custom weights initialization called on netG and netD '''
classname = m.__class__.__name__
if classname.find('Conv') != -1:
nn.init.normal_(m.weight.data, 0.0, 0.02)
elif classname.find('BatchNorm') != -1:
nn.init.normal_(m.weight.data, 1.0, 0.02)
nn.init.constant_(m.bias.data, 0)
## Choose model
Generator, Discriminator=f_get_model(gdict) ## Mod for cGAN
# Create Generator
self.netG = Generator(gdict).to(gdict['device'])
self.netG.apply(weights_init)
# Create Discriminator
self.netD = Discriminator(gdict).to(gdict['device'])
self.netD.apply(weights_init)
if print_model:
if gdict['world_rank']==0:
print(self.netG)
# summary(netG,(1,1,64))
print(self.netD)
# summary(netD,(1,128,128))
print("Number of GPUs used %s"%(gdict['ngpu']))
if (gdict['multi-gpu']):
if not gdict['distributed']:
self.netG = nn.DataParallel(self.netG, list(range(gdict['ngpu'])))
self.netD = nn.DataParallel(self.netD, list(range(gdict['ngpu'])))
else:
self.netG=DistributedDataParallel(self.netG,device_ids=[gdict['local_rank']],output_device=[gdict['local_rank']])
self.netD=DistributedDataParallel(self.netD,device_ids=[gdict['local_rank']],output_device=[gdict['local_rank']])
#### Initialize networks ####
# self.criterion = nn.BCELoss()
self.criterion = nn.BCEWithLogitsLoss()
self.optimizerD = optim.Adam(self.netD.parameters(), lr=gdict['learn_rate_d'], betas=(gdict['beta1'], 0.999),eps=1e-7)
self.optimizerG = optim.Adam(self.netG.parameters(), lr=gdict['learn_rate_g'], betas=(gdict['beta1'], 0.999),eps=1e-7)
if gdict['distributed']: try_barrier(gdict['world_rank'])
if gdict['mode']=='fresh':
iters,start_epoch,best_chi1,best_chi2=0,0,1e10,1e10
elif gdict['mode']=='continue':
iters,start_epoch,best_chi1,best_chi2,self.netD,self.optimizerD,self.netG,self.optimizerG=f_load_checkpoint(gdict['save_dir']+'/models/checkpoint_last.tar',\
self.netG,self.netD,self.optimizerG,self.optimizerD,gdict)
if gdict['world_rank']==0: logging.info("\nContinuing existing run. Loading checkpoint with epoch {0} and step {1}\n".format(start_epoch,iters))
if gdict['distributed']: try_barrier(gdict['world_rank'])
start_epoch+=1 ## Start with the next epoch
elif gdict['mode']=='fresh_load':
iters,start_epoch,best_chi1,best_chi2,self.netD,self.optimizerD,self.netG,self.optimizerG=f_load_checkpoint(gdict['chkpt_file'],\
self.netG,self.netD,self.optimizerG,self.optimizerD,gdict)
if gdict['world_rank']==0: logging.info("Fresh run loading checkpoint file {0}".format(gdict['chkpt_file']))
# if gdict['distributed']: try_barrier(gdict['world_rank'])
iters,start_epoch,best_chi1,best_chi2=0,0,1e10,1e10
## Add to gdict
for key,val in zip(['best_chi1','best_chi2','iters','start_epoch'],[best_chi1,best_chi2,iters,start_epoch]): gdict[key]=val
## Set up learn rate scheduler
lr_stepsize=int((gdict['num_imgs'])/(gdict['batch_size']*gdict['world_size'])) # convert epoch number to step
lr_d_epochs=[i*lr_stepsize for i in gdict['lr_d_epochs']]
lr_g_epochs=[i*lr_stepsize for i in gdict['lr_g_epochs']]
self.schedulerD = optim.lr_scheduler.MultiStepLR(self.optimizerD, milestones=lr_d_epochs,gamma=gdict['lr_d_gamma'])
self.schedulerG = optim.lr_scheduler.MultiStepLR(self.optimizerG, milestones=lr_g_epochs,gamma=gdict['lr_g_gamma'])
```
## Main
```
#########################
### Main code #######
#########################
if __name__=="__main__":
jpt=False
jpt=True ##(different for jupyter notebook)
t0=time.time()
t0=time.time()
args=f_parse_args() if not jpt else f_manual_add_argparse()
#################################
### Set up global dictionary###
gdict={}
gdict=f_init_gdict(args,gdict)
# gdict['num_imgs']=200
if jpt: ## override for jpt nbks
gdict['num_imgs']=400
gdict['run_suffix']='nb_test'
### Set up metrics dataframe
cols=['step','epoch','Dreal','Dfake','Dfull','G_adv','G_full','spec_loss','hist_loss','spec_chi','hist_chi','gp_loss','fm_loss','D(x)','D_G_z1','D_G_z2','time']
metrics_df=pd.DataFrame(columns=cols)
# Setup
metrics_df=f_setup(gdict,metrics_df,log=(not jpt))
## Build GAN
gan_model=GAN_model(gdict,False)
fixed_noise = torch.randn(gdict['op_size'], 1, 1, 1, gdict['nz'], device=gdict['device']) #Latent vectors to view G progress # Mod for 3D
if gdict['distributed']: try_barrier(gdict['world_rank'])
## Load data and precompute
Dset=Dataset(gdict)
#################################
########## Train loop and save metrics and images ######
if gdict['distributed']: try_barrier(gdict['world_rank'])
if gdict['world_rank']==0:
logging.info(gdict)
logging.info("Starting Training Loop...")
f_train_loop(gan_model,Dset,metrics_df,gdict,fixed_noise)
if gdict['world_rank']==0: ## Generate images for best saved models ######
op_loc=gdict['save_dir']+'/images/'
ip_fname=gdict['save_dir']+'/models/checkpoint_best_spec.tar'
f_gen_images(gdict,gan_model.netG,gan_model.optimizerG,ip_fname,op_loc,op_strg='best_spec',op_size=32)
ip_fname=gdict['save_dir']+'/models/checkpoint_best_hist.tar'
f_gen_images(gdict,gan_model.netG,gan_model.optimizerG,ip_fname,op_loc,op_strg='best_hist',op_size=32)
tf=time.time()
logging.info("Total time %s"%(tf-t0))
logging.info('End: %s'%(datetime.now().strftime('%Y-%m-%d %H:%M:%S')))
# metrics_df.plot('step','time')
metrics_df
gan_model.optimizerG.param_groups[0]['lr']
# metrics_df['lr_d']
# summary(gan_model.netG,(1,1,64))
summary(gan_model.netD,(1,128,128,128))
# gdict
```
### Debug
```
# class Generator(nn.Module):
# def __init__(self, gdict):
# super(Generator, self).__init__()
# ## Define new variables from dict
# keys=['ngpu','nz','nc','ngf','kernel_size','stride','g_padding']
# ngpu, nz,nc,ngf,kernel_size,stride,g_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
# self.main = nn.Sequential(
# # nn.ConvTranspose2d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
# nn.Linear(nz,nc*ngf*8*8*8),# 32768
# nn.BatchNorm2d(nc,eps=1e-05, momentum=0.9, affine=True),
# nn.ReLU(inplace=True),
# View(shape=[-1,ngf*8,8,8]),
# nn.ConvTranspose2d(ngf * 8, ngf * 4, kernel_size, stride, g_padding, output_padding=1, bias=False),
# nn.BatchNorm2d(ngf*4,eps=1e-05, momentum=0.9, affine=True),
# nn.ReLU(inplace=True),
# # state size. (ngf*4) x 8 x 8
# nn.ConvTranspose2d( ngf * 4, ngf * 2, kernel_size, stride, g_padding, 1, bias=False),
# nn.BatchNorm2d(ngf*2,eps=1e-05, momentum=0.9, affine=True),
# nn.ReLU(inplace=True),
# # state size. (ngf*2) x 16 x 16
# nn.ConvTranspose2d( ngf * 2, ngf, kernel_size, stride, g_padding, 1, bias=False),
# nn.BatchNorm2d(ngf,eps=1e-05, momentum=0.9, affine=True),
# nn.ReLU(inplace=True),
# # state size. (ngf) x 32 x 32
# nn.ConvTranspose2d( ngf, nc, kernel_size, stride,g_padding, 1, bias=False),
# nn.Tanh()
# )
# def forward(self, ip):
# return self.main(ip)
# class Discriminator(nn.Module):
# def __init__(self, gdict):
# super(Discriminator, self).__init__()
# ## Define new variables from dict
# keys=['ngpu','nz','nc','ndf','kernel_size','stride','d_padding']
# ngpu, nz,nc,ndf,kernel_size,stride,d_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
# self.main = nn.Sequential(
# # input is (nc) x 64 x 64
# # nn.Conv2d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
# nn.Conv2d(nc, ndf,kernel_size, stride, d_padding, bias=True),
# nn.BatchNorm2d(ndf,eps=1e-05, momentum=0.9, affine=True),
# nn.LeakyReLU(0.2, inplace=True),
# # state size. (ndf) x 32 x 32
# nn.Conv2d(ndf, ndf * 2, kernel_size, stride, d_padding, bias=True),
# nn.BatchNorm2d(ndf * 2,eps=1e-05, momentum=0.9, affine=True),
# nn.LeakyReLU(0.2, inplace=True),
# # state size. (ndf*2) x 16 x 16
# nn.Conv2d(ndf * 2, ndf * 4, kernel_size, stride, d_padding, bias=True),
# nn.BatchNorm2d(ndf * 4,eps=1e-05, momentum=0.9, affine=True),
# nn.LeakyReLU(0.2, inplace=True),
# # state size. (ndf*4) x 8 x 8
# nn.Conv2d(ndf * 4, ndf * 8, kernel_size, stride, d_padding, bias=True),
# nn.BatchNorm2d(ndf * 8,eps=1e-05, momentum=0.9, affine=True),
# nn.LeakyReLU(0.2, inplace=True),
# # state size. (ndf*8) x 4 x 4
# nn.Flatten(),
# nn.Linear(nc*ndf*8*8*8, 1)
# # nn.Sigmoid()
# )
# def forward(self, ip):
# # print(ip.shape)
# results=[ip]
# lst_idx=[]
# for i,submodel in enumerate(self.main.children()):
# mid_output=submodel(results[-1])
# results.append(mid_output)
# ## Select indices in list corresponding to output of Conv layers
# if submodel.__class__.__name__.startswith('Conv'):
# # print(submodel.__class__.__name__)
# # print(mid_output.shape)
# lst_idx.append(i)
# FMloss=True
# if FMloss:
# ans=[results[1:][i] for i in lst_idx + [-1]]
# else :
# ans=results[-1]
# return ans
# netG = Generator(gdict).to(gdict['device'])
# netG.apply(weights_init)
# # # # print(netG)
# # summary(netG,(1,1,64))
# # Create Discriminator
# netD = Discriminator(gdict).to(gdict['device'])
# netD.apply(weights_init)
# # print(netD)
# summary(netD,(1,128,128))
# noise = torch.randn(gdict['batchsize'], 1, 1, gdict['nz'], device=gdict['device'])
# fake = netG(noise)
# # Forward pass real batch through D
# output = netD(fake)
# print([i.shape for i in output])
0.5**10
70000/(8*6*8)
gdict.keys()
for key in ['batch_size','num_imgs','ngpu']:
print(key,gdict[key])
gdict['world_size']
```
|
github_jupyter
|
```
%matplotlib inline
```
# Decoding in time-frequency space using Common Spatial Patterns (CSP)
The time-frequency decomposition is estimated by iterating over raw data that
has been band-passed at different frequencies. This is used to compute a
covariance matrix over each epoch or a rolling time-window and extract the CSP
filtered signals. A linear discriminant classifier is then applied to these
signals.
```
# Authors: Laura Gwilliams <[email protected]>
# Jean-Remi King <[email protected]>
# Alex Barachant <[email protected]>
# Alexandre Gramfort <[email protected]>
#
# License: BSD (3-clause)
import numpy as np
import matplotlib.pyplot as plt
from mne import Epochs, create_info, events_from_annotations
from mne.io import concatenate_raws, read_raw_edf
from mne.datasets import eegbci
from mne.decoding import CSP
from mne.time_frequency import AverageTFR
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import StratifiedKFold, cross_val_score
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import LabelEncoder
```
Set parameters and read data
```
event_id = dict(hands=2, feet=3) # motor imagery: hands vs feet
subject = 1
runs = [6, 10, 14]
raw_fnames = eegbci.load_data(subject, runs)
raw = concatenate_raws([read_raw_edf(f) for f in raw_fnames])
# Extract information from the raw file
sfreq = raw.info['sfreq']
events, _ = events_from_annotations(raw, event_id=dict(T1=2, T2=3))
raw.pick_types(meg=False, eeg=True, stim=False, eog=False, exclude='bads')
raw.load_data()
# Assemble the classifier using scikit-learn pipeline
clf = make_pipeline(CSP(n_components=4, reg=None, log=True, norm_trace=False),
LinearDiscriminantAnalysis())
n_splits = 5 # how many folds to use for cross-validation
cv = StratifiedKFold(n_splits=n_splits, shuffle=True, random_state=42)
# Classification & time-frequency parameters
tmin, tmax = -.200, 2.000
n_cycles = 10. # how many complete cycles: used to define window size
min_freq = 5.
max_freq = 25.
n_freqs = 8 # how many frequency bins to use
# Assemble list of frequency range tuples
freqs = np.linspace(min_freq, max_freq, n_freqs) # assemble frequencies
freq_ranges = list(zip(freqs[:-1], freqs[1:])) # make freqs list of tuples
# Infer window spacing from the max freq and number of cycles to avoid gaps
window_spacing = (n_cycles / np.max(freqs) / 2.)
centered_w_times = np.arange(tmin, tmax, window_spacing)[1:]
n_windows = len(centered_w_times)
# Instantiate label encoder
le = LabelEncoder()
```
Loop through frequencies, apply classifier and save scores
```
# init scores
freq_scores = np.zeros((n_freqs - 1,))
# Loop through each frequency range of interest
for freq, (fmin, fmax) in enumerate(freq_ranges):
# Infer window size based on the frequency being used
w_size = n_cycles / ((fmax + fmin) / 2.) # in seconds
# Apply band-pass filter to isolate the specified frequencies
raw_filter = raw.copy().filter(fmin, fmax, n_jobs=1, fir_design='firwin',
skip_by_annotation='edge')
# Extract epochs from filtered data, padded by window size
epochs = Epochs(raw_filter, events, event_id, tmin - w_size, tmax + w_size,
proj=False, baseline=None, preload=True)
epochs.drop_bad()
y = le.fit_transform(epochs.events[:, 2])
X = epochs.get_data()
# Save mean scores over folds for each frequency and time window
freq_scores[freq] = np.mean(cross_val_score(estimator=clf, X=X, y=y,
scoring='roc_auc', cv=cv,
n_jobs=1), axis=0)
```
Plot frequency results
```
plt.bar(freqs[:-1], freq_scores, width=np.diff(freqs)[0],
align='edge', edgecolor='black')
plt.xticks(freqs)
plt.ylim([0, 1])
plt.axhline(len(epochs['feet']) / len(epochs), color='k', linestyle='--',
label='chance level')
plt.legend()
plt.xlabel('Frequency (Hz)')
plt.ylabel('Decoding Scores')
plt.title('Frequency Decoding Scores')
```
Loop through frequencies and time, apply classifier and save scores
```
# init scores
tf_scores = np.zeros((n_freqs - 1, n_windows))
# Loop through each frequency range of interest
for freq, (fmin, fmax) in enumerate(freq_ranges):
# Infer window size based on the frequency being used
w_size = n_cycles / ((fmax + fmin) / 2.) # in seconds
# Apply band-pass filter to isolate the specified frequencies
raw_filter = raw.copy().filter(fmin, fmax, n_jobs=1, fir_design='firwin',
skip_by_annotation='edge')
# Extract epochs from filtered data, padded by window size
epochs = Epochs(raw_filter, events, event_id, tmin - w_size, tmax + w_size,
proj=False, baseline=None, preload=True)
epochs.drop_bad()
y = le.fit_transform(epochs.events[:, 2])
# Roll covariance, csp and lda over time
for t, w_time in enumerate(centered_w_times):
# Center the min and max of the window
w_tmin = w_time - w_size / 2.
w_tmax = w_time + w_size / 2.
# Crop data into time-window of interest
X = epochs.copy().crop(w_tmin, w_tmax).get_data()
# Save mean scores over folds for each frequency and time window
tf_scores[freq, t] = np.mean(cross_val_score(estimator=clf, X=X, y=y,
scoring='roc_auc', cv=cv,
n_jobs=1), axis=0)
```
Plot time-frequency results
```
# Set up time frequency object
av_tfr = AverageTFR(create_info(['freq'], sfreq), tf_scores[np.newaxis, :],
centered_w_times, freqs[1:], 1)
chance = np.mean(y) # set chance level to white in the plot
av_tfr.plot([0], vmin=chance, title="Time-Frequency Decoding Scores",
cmap=plt.cm.Reds)
```
|
github_jupyter
|
## week03: Логистическая регрессия и анализ изображений
В этом ноутбуке предлагается построить классификатор изображений на основе логистической регрессии.
*Забегая вперед, мы попробуем решить задачу классификации изображений используя лишь простые методы. В третьей части нашего курса мы вернемся к этой задаче.*
```
import numpy as np
import matplotlib.pyplot as plt
import h5py
%matplotlib inline
```
## 1. Постановка задачи ##
**Задача**: Есть датасет [прямая ссылка](https://drive.google.com/file/d/15tOimf2QYWsMtPJXTUCwgZaOTF8Nxcsm/view?usp=sharing) ("catvnoncat.h5") состоящий из:
- обучающей выборки из m_train изображений, помеченных "cat" (y=1) или "non-cat" (y=0)
- тестовой выборки m_test изображений, помеченных "cat" или "non-cat"
- каждое цветное изображение имеет размер (src_size, src_size, 3), где 3 - число каналов (RGB).
Таким образом, каждый слой - квадрат размера src_size x src_size$.
Давайте построим простой алгоритм классификации изображений на классы "cat"/"non-cat".
Автоматическая загрузка доступна ниже.
<img src="img/LogReg_kiank.png" style="width:650px;height:400px;">
**Recap**:
Для каждого примера $x^{(i)}$:
$$z^{(i)} = w^T x^{(i)} + b \tag{1}$$
$$\hat{y}^{(i)} = a^{(i)} = sigmoid(z^{(i)})\tag{2}$$
$$ \mathcal{L}(a^{(i)}, y^{(i)}) = - y^{(i)} \log(a^{(i)}) - (1-y^{(i)} ) \log(1-a^{(i)})\tag{3}$$
Функция потерь:
$$ J = \frac{1}{m} \sum_{i=1}^m \mathcal{L}(a^{(i)}, y^{(i)})\tag{6}$$
```
# Uncomment this cell to download the data
# !wget "https://downloader.disk.yandex.ru/disk/7ef1d1e30e23740a4a30799a825319154815ddc85bf689542add0a3d11ccb91c/5d7fdcb0/3dcxK38Q0fG3ui0g2gMZgKkLls8ULwVpoYNkWpBm9d24EceJ6mIoH5l3_wKkFv3PfZ0WMGYjfJULynuJkuGaug%3D%3D?uid=76549735&filename=data.zip&disposition=attachment&hash=&limit=0&content_type=application%2Fzip&owner_uid=76549735&fsize=2815580&hid=084389255415f71a92d0f1024ab741d4&media_type=compressed&tknv=v2&etag=2b348ac8eca72d223108e36b2a671210" -O data.zip
# !unzip data.zip
```
### 1.1 Загрузка данных и визуализация ###
```
def load_dataset():
train_data = h5py.File("data/train_catvnoncat.h5", "r")
train_set_x_orig = np.array(train_data["train_set_x"][:]) # признаки
train_set_y_orig = np.array(train_data["train_set_y"][:]) # метки классов
test_data = h5py.File("data/test_catvnoncat.h5", "r")
test_set_x_orig = np.array(test_data["test_set_x"][:]) # признаки
test_set_y_orig = np.array(test_data["test_set_y"][:]) # метки классов
classes = np.array(test_data["list_classes"][:]) # the list of classes
classes = np.array(list(map(lambda x: x.decode('utf-8'), classes)))
train_set_y = train_set_y_orig.reshape(train_set_y_orig.shape[0])
test_set_y = test_set_y_orig.reshape(test_set_y_orig.shape[0])
return train_set_x_orig, train_set_y, test_set_x_orig, test_set_y, classes
train_set_x_orig, train_set_y, test_set_x_orig, test_set_y, classes = load_dataset()
```
Цветные изображения в формате RGB представлены в виде трёхмерных numpy.array.
Порядок измерений $H \times W \times C$: $H$ - высота, $W$ - ширина и $C$ - число каналов.
Значение каждого пиксела находится в интервале $[0;255]$.
```
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
def show_image_interact(i=0):
f, ax = plt.subplots(1,4, figsize=(15,20), sharey=True)
ax[0].imshow(train_set_x_orig[i])
ax[0].set_title('RGB image')
ax[1].imshow(train_set_x_orig[i][:,:,0], cmap='gray')
ax[1].set_title('R channel')
ax[2].imshow(train_set_x_orig[i][:,:,1], cmap='gray')
ax[2].set_title('G channel')
ax[3].imshow(train_set_x_orig[i][:,:,2], cmap='gray')
ax[3].set_title('B channel')
print("y = {} belongs to '{}' class.".format(str(train_set_y[i]),classes[np.squeeze(train_set_y[i])]))
interact(show_image_interact,
i=widgets.IntSlider(min=0, max=len(train_set_y)-1, step=1))
```
При работе с данными полезно будет сохранить размерности входных изображений для дальнейшей обработки.
```
m_train = train_set_x_orig.shape[0]
m_test = test_set_x_orig.shape[0]
src_size = train_set_x_orig.shape[1]
print ("Размер обучающей выборки: m_train = " + str(m_train))
print ("Размер тестовой выборки: m_test = " + str(m_test))
print ("Ширина/Высота каждого изображения: src_size = " + str(src_size))
print ("Размерны трёхмерной матрицы для каждого изображения: (" + str(src_size) + ", " + str(src_size) + ", 3)")
print ("Размерность train_set_x: " + str(train_set_x_orig.shape))
print ("Размерность train_set_y: " + str(train_set_y.shape))
print ("Размерность test_set_x: " + str(test_set_x_orig.shape))
print ("Размерность test_set_y: " + str(test_set_y.shape))
```
## 2. Предварительная обработка
Преобразуем входные изображения размера (num_px, num_px, 3) в вектор признаков размера (num_px $*$ num_px $*$ 3, 1), чтобы сформировать матрицы объект-признак в виде numpy-array для обучающей и тестовой выборок.
Каждой строке матрицы объект-признак соответствует входное развёрнутое в вектор-строку изображение.
Помимо этого, для предварительной обработки (препроцессинга) изображений применяют центрирование значений: из значения каждого пиксела вычитается среднее и делят полученное значение на среднеквадратичное отклонение значений пикселей всего изображения.
Однако, на практике обычно просто делят значения пикселей на 255 (максимальное значение пикселя).
Оформим эти шаги в функцию предварительной обработки
```
def image_preprocessing_simple(data):
assert type(data) == np.ndarray
assert data.ndim == 4
n,h,w,c = data.shape
data_vectorized = <ваш код>
data_normalized = <ваш код>
return data_normalized
# Изменить размеры входных данных
train_set_x_vectorized = image_preprocessing_simple(train_set_x_orig)
test_set_x_vectorized = image_preprocessing_simple(test_set_x_orig)
print('Train set:')
print("Размеры train_set_x_vectorized: {}".format(str(train_set_x_vectorized.shape)))
print("Размеры train_set_y: {}".format(str(train_set_y.shape)))
print("Размеры классов 'cat'/'non-cat': {} / {}".format(sum(train_set_y==1), sum(train_set_y==0)))
print('Test set:')
print("Размеры test_set_x_vectorized: {}".format(str(test_set_x_vectorized.shape)))
print("Размеры test_set_y: {}".format(str(test_set_y.shape)))
print("Размеры классов 'cat'/'non-cat': {} / {}".format(sum(test_set_y==1), sum(test_set_y==0)))
```
## 3. Классификация
```
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV
import warnings
warnings.filterwarnings('ignore')
```
**Вопрос**: Какую метрику качества стоит использовать?
### 3.1 Построение модели
Построим модель с параметрами по умолчанию и посмотрим, как хорошо она справится с задачей.
```
clf = # <ваш код>
score = <ваш код>
print('Точность для простой модели с параметрами по умолчанию: {:.4f}'.format(score))
# from sklearn.metrics import f1_score
y_predicted = clf.predict(test_set_x_vectorized)
correct_score = <ваш код>
print('<Имя метрики> для простой модели: {:.4f}'.format(correct_score))
```
Попробуем подобрать параметры регуляризации в надежде, что это повысит точность предсказаний.
```
<ваш код>
print('Оптимальные параметры: {}'.format(<ваш код>))
print('Наилучшее значение метрики качества: {}'.format(<ваш код>))
```
Обучим модель с оптимальными параметрами на всей обучающей выборке и посмотрим на метрики качества:
```
best_clf = <ваш код>
best_clf.fit(train_set_x_vectorized, train_set_y)
y_predicted = best_clf.predict(test_set_x_vectorized)
metric_score = <ваш код>(y_predicted, test_set_y)
print('Optimal model hyperparameters accuracy score: {:.4f}'.format(metric_score))
```
### 3.2 Анализ ошибок
```
is_outlier = (y_predicted != test_set_y)
test_outliers_x, test_outliers_y, predicted_y = test_set_x_orig[is_outlier], test_set_y[is_outlier], y_predicted[is_outlier]
def show_image_outliers(i=0):
f = plt.figure(figsize=(5,5))
plt.imshow(test_outliers_x[i])
plt.title('RGB image')
fmt_string = "Sample belongs to '{}' class, but '{}' is predicted'"
print(fmt_string.format(classes[test_outliers_y[i]], classes[predicted_y[i]]))
interact(show_image_outliers,
i=widgets.IntSlider(min=0, max=len(test_outliers_y)-1, step=1))
```
**Вопрос**: Как по-вашему можно повысить точность? Каким недостатком обладает данный подход к классификации?
### 3.3 Модель с аугментациями
Как можно увеличить количество данных для обучения?
Сформировать новые примеры из уже имеющихся!
Например, можно пополнить class 'cat' обучающей выборки [зеркально отображёнными](https://docs.scipy.org/doc/numpy/reference/generated/numpy.fliplr.html) изображениями котов.
```
def augment_sample(src, label):
<ваш код>
def image_preprocessing_augment(data, labels):
assert type(data) == np.ndarray
assert data.ndim == 4
## ВАШ КОД ##
data_augmented =
labels_augmented =
## ВАШ КОД ЗАКАНЧИВАЕТСЯ ЗДЕСЬ ##
n,h,w,c = data_augmented.shape
data_vectorized = data_augmented.reshape(n, -1) # <ваш код>
data_normalized = data_vectorized / 255
return data_normalized, labels_augmented
train_set_x_augmented, train_set_y_augmented = image_preprocessing_augment(train_set_x_orig, train_set_y)
clf = LogisticRegression(solver='liblinear')
clf.fit(train_set_x_augmented, train_set_y_augmented)
y_pred = clf.predict(test_set_x_vectorized)
print('F-мера для модели с аугментациями: {:.4f}'.format(f1_score(y_pred, test_set_y)))
```
## 4. Проверьте работу классификатора на своей картинке
Библиотека [OpenCV](https://opencv.org) для работы с изображениями для [python](https://pypi.org/project/opencv-python/):
`pip install opencv-python`
Вместе с contrib-модулями:
`pip install opencv-contrib-python`
```
import cv2
# Путь к картинке на вашем ПК
fname = "cat-non-cat.jpg"
# Считываем картинку через scipy
src = cv2.cvtColor(cv2.imread((fname)), cv2.COLOR_BGR2RGB)
src_resized = cv2.resize(src, (src_size,src_size), interpolation=cv2.INTER_LINEAR).reshape(1, src_size*src_size*3)
my_image_predict = clf.predict(src_resized)[0]
plt.imshow(src)
print("Алгоритм говорит, что это '{}': {}".format(my_image_predict, classes[my_image_predict]))
```
|
github_jupyter
|
```
import numpy as np
import matplotlib.pyplot as plt
from scipy.io import loadmat
from scipy.interpolate import RectBivariateSpline as rbs
from scipy.integrate import romb
import scipy.sparse as sp
import os
import pywt
wvt = 'db12'
%matplotlib inline
import matplotlib as mpl
norm = mpl.colors.Normalize(vmin=0.0,vmax=1.5)
nx = ny = 32
t = np.linspace(0,320,nx+1)
s = np.linspace(0,320,17)
x = y = (t[:-1]+t[1:]) / 2
x = y = (t[:-1]+t[1:]) / 2
xst = yst = (s[:-1]+s[1:]) / 2
xs, ys = np.meshgrid(xst,yst)
xs = xs.flatten()
ys = ys.flatten()
from scipy.signal import butter, lfilter
def butter_bandpass(lowcut, highcut, fs, order=5):
nyq = 0.5 * fs
low = lowcut / nyq
high = highcut / nyq
b, a = butter(order, [low, high], btype='band')
return b, a
def butter_bandpass_filter(data, lowcut, highcut, fs, order=5, axis=0):
b, a = butter_bandpass(lowcut, highcut, fs, order=order)
y = lfilter(b, a, data, axis=axis)
return y
def butter_lowpass(lowcut, fs, order=5):
nyq = 0.5 * fs
low = lowcut / nyq
b, a = butter(order, [low], btype='low')
return b, a
def butter_low_filter(data, lowcut, fs, order=5, axis=0):
b, a = butter_lowpass(lowcut, fs, order=order)
y = lfilter(b, a, data, axis=axis)
return y
shot = np.reshape(np.fromfile("Testing/TestData/shot1.dat", dtype=np.float32), (4001,64,64))
t = np.linspace(0, 0.5, 4001)
shotf = butter_low_filter(shot, 10, 8000)
tf = t[::20]
shotf = shotf[::20,:,:]
tf_freq = 1/(tf[1]-tf[0])
xc = np.linspace(0,320,65)
xc = (xc[:-1]+xc[1:])/2
yc = xc
shotf_itps = [rbs(xc, yc, s) for s in shotf[:-1]]
def reconstruction(w, wvt_lens, wvt):
starts = np.hstack([0,np.cumsum(wvt_lens)])
wcoef = [w[starts[i]:starts[i+1]] for i in range(len(wvt_lens))]
return pywt.waverec(wcoef, wvt)
```
# ZigZag
```
das_template_x = np.array([2.5*np.sqrt(2)*i for i in range(24)])
das_template_y = np.array([2.5*np.sqrt(2)*i for i in range(24)])
das_template_x2 = np.hstack([das_template_x,das_template_x[::-1],das_template_x,das_template_x[::-1]])
das_template_y2 = np.hstack([das_template_y,das_template_y+das_template_y[-1],das_template_y+2*das_template_y[-1],das_template_y+3*das_template_y[-1]])
das_x = np.hstack([das_template_x2+i*das_template_x[-1] for i in range(4)])
das_y = np.hstack([das_template_y2 for i in range(4)])
offset = (320-np.max(das_x))/2
das_x += offset
das_y += offset
azimuth_template_1 = np.array([[[45 for i in range(24)], [-45 for i in range(24)]] for i in range(2)]).flatten()
azimuth_template_2 = np.array([[[135 for i in range(24)], [215 for i in range(24)]] for i in range(2)]).flatten()
das_az = np.hstack([azimuth_template_1, azimuth_template_2,
azimuth_template_1, azimuth_template_2])
raz = np.deg2rad(das_az)
cscale = 2
generate_kernels = True
L = 10 #gauge length
ll = np.linspace(-L/2, L/2, 2**5+1)
dl = ll[1]-ll[0]
p1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]
p2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]
if generate_kernels:
os.makedirs("Kernels", exist_ok=True)
crv = loadmat(f"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat")
G_mat = np.reshape(crv["G_mat"].T, (crv["G_mat"].shape[1], nx, ny))
crvscales = crv["scales"].flatten()
cvtscaler = 2.0**(cscale*crvscales)
G1 = np.zeros((len(raz), G_mat.shape[0]))
G2 = np.zeros((len(raz), G_mat.shape[0]))
G3 = np.zeros((len(xs), G_mat.shape[0]))
for j in range(G_mat.shape[0]):
frame = rbs(x,y,G_mat[j])
#average derivatives of frame along gauge length
fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L
fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L
G1[:,j] = (np.sin(raz)**2*fd1 +
np.sin(2*raz)*fd2/2) / cvtscaler[j]
G2[:,j] = (np.cos(raz)**2*fd2 +
np.sin(2*raz)*fd1/2) / cvtscaler[j]
G3[:,j] = frame.ev(xs, ys) / cvtscaler[j]
G = np.hstack([G1, G2])
Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))
G = G / Gn
# Gn=1
G_zigzag = G
np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))
plt.plot(np.sort(np.diag(G @ np.linalg.solve(G.T@G + 1e-10*np.eye(G.shape[1]), G.T))))
exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])
eyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])
exyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])
edasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr)
das_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])
cuxr = np.array([s.ev(xs, ys, dx=1) for s in shotf_itps])
cuyr = np.array([s.ev(xs, ys, dy=1) for s in shotf_itps])
np.save("Testing/zigzag.npy", das_wvt_data)
wvt_tmp = pywt.wavedec(edasr.T[0], wvt)
wvt_lens = [len(wc) for wc in wvt_tmp]
resi = np.load(f"Testing/zigzag_res.npy")
Gs = np.std(G)
resxi = resi[:G3.shape[1], :]
resyi = resi[G3.shape[1]:, :]
xpredi = (G3/Gn/Gs) @ resxi
ypredi = (G3/Gn/Gs) @ resyi
txpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))
typredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))
res = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))
plt.plot(np.std(resi, axis=1))
cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)
plt.xlim(0,320)
plt.ylim(0,320)
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
plt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))
, norm=norm)
plt.colorbar()
res
plt.plot(cuxr.T[100])
plt.plot(txpredi[100])
cax = plt.scatter(das_x, das_y,c=das_az)
plt.xlim(0,320)
plt.ylim(0,320)
plt.colorbar(cax, label="Cable Azimuth")
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
np.sqrt(np.square(das_x[1:]-das_x[:-1])+np.square(das_y[1:]-das_y[:-1]))
```
# Spiral
```
das_theta2 = np.linspace(0,(360*4)**2, 192*2)
das_theta = np.deg2rad(np.sqrt(das_theta2))
a = 0
b = 1
das_r = b*das_theta
das_x = das_r * np.cos(das_theta)
das_y = das_r * np.sin(das_theta)
raz = np.pi/2-np.arctan2(b*np.tan(das_theta)+(a+b*das_theta), b-(a+b*das_theta)*np.tan(das_theta))
das_az = np.rad2deg(raz)
xwidth = np.max(das_x)-np.min(das_x)
das_x = das_x / xwidth * 320
das_y = das_y / xwidth * 320
x_offset = 320 - np.max(das_x)
das_x = das_x + x_offset
y_offset = np.min(das_y)
das_y = das_y - y_offset
L = 10 #gauge length
ll = np.linspace(-L/2, L/2, 2**5+1)
dl = ll[1]-ll[0]
p1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]
p2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]
if generate_kernels:
os.makedirs("Kernels", exist_ok=True)
crv = loadmat(f"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat")
G_mat = np.reshape(crv["G_mat"].T, (crv["G_mat"].shape[1], nx, ny))
crvscales = crv["scales"].flatten()
cvtscaler = 2.0**(cscale*crvscales)
G1 = np.zeros((len(raz), G_mat.shape[0]))
G2 = np.zeros((len(raz), G_mat.shape[0]))
for j in range(G_mat.shape[0]):
frame = rbs(x,y,G_mat[j])
#average derivatives of frame along gauge length
fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L
fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L
G1[:,j] = (np.sin(raz)**2*fd1 +
np.sin(2*raz)*fd2/2) / cvtscaler[j]
G2[:,j] = (np.cos(raz)**2*fd2 +
np.sin(2*raz)*fd1/2) / cvtscaler[j]
G = np.hstack([G1, G2])
Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))
G = G / Gn
G_spiral = G
np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))
exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])
eyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])
exyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])
edasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr)
das_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])
np.save("Testing/spiral.npy", das_wvt_data)
resi = np.load(f"Testing/spiral_res.npy")
Gs = np.std(G)
resxi = resi[:G3.shape[1], :]
resyi = resi[G3.shape[1]:, :]
xpredi = (G3/Gn/Gs) @ resxi
ypredi = (G3/Gn/Gs) @ resyi
txpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))
typredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))
res = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))
res
plt.plot(cuxr.T[100])
plt.plot(txpredi[100])
cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)
plt.xlim(0,320)
plt.ylim(0,320)
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
plt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))
, norm=norm)
plt.colorbar()
np.hstack([cuxr, cuyr]).shape
cax = plt.scatter(das_x, das_y,c=das_az)
plt.xlim(0,320)
plt.ylim(0,320)
plt.colorbar(cax, label="Cable Azimuth")
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
np.sqrt(np.square(das_x[1:]-das_x[:-1])+np.square(das_y[1:]-das_y[:-1]))
```
# Crossing
```
template = np.linspace(0,320, 65)
template = (template[1:]+template[:-1])/2
das_x = np.hstack([template, template, template,[80 for i in range(len(template))], [160 for i in range(len(template))], [240 for i in range(len(template))]])
das_y = np.hstack([[80 for i in range(len(template))], [160 for i in range(len(template))], [240 for i in range(len(template))],template,template,template])
das_az = np.hstack([[90 for i in range(len(template))], [270 for i in range(len(template))], [90 for i in range(len(template))],[0 for i in range(len(template))], [180 for i in range(len(template))], [0 for i in range(len(template))]])
raz = np.deg2rad(das_az)
L = 10 #gauge length
ll = np.linspace(-L/2, L/2, 2**5+1)
dl = ll[1]-ll[0]
p1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]
p2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]
if generate_kernels:
os.makedirs("Kernels", exist_ok=True)
crv = loadmat(f"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat")
G_mat = np.reshape(crv["G_mat"].T, (crv["G_mat"].shape[1], nx, ny))
crvscales = crv["scales"].flatten()
cvtscaler = 2.0**(cscale*crvscales)
G1 = np.zeros((len(raz), G_mat.shape[0]))
G2 = np.zeros((len(raz), G_mat.shape[0]))
for j in range(G_mat.shape[0]):
frame = rbs(x,y,G_mat[j])
#average derivatives of frame along gauge length
fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L
fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L
G1[:,j] = (np.sin(raz)**2*fd1 +
np.sin(2*raz)*fd2/2) / cvtscaler[j]
G2[:,j] = (np.cos(raz)**2*fd2 +
np.sin(2*raz)*fd1/2) / cvtscaler[j]
G = np.hstack([G1, G2])
Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))
G = G / Gn
G_cross = G
np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))
exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])
eyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])
exyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])
edasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr)
das_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])
np.save("Testing/crossing.npy", das_wvt_data )
resi = np.load(f"Testing/crossing_res.npy")
Gs = np.std(G)
resxi = resi[:G3.shape[1], :]
resyi = resi[G3.shape[1]:, :]
xpredi = (G3/Gn/Gs) @ resxi
ypredi = (G3/Gn/Gs) @ resyi
txpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))
typredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))
res = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))
res
plt.plot(cuxr.T[150])
plt.plot(txpredi[150])
cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)
plt.xlim(0,320)
plt.ylim(0,320)
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
plt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))
, norm=norm)
plt.scatter(xs[150], ys[150], color='r')
plt.colorbar()
cax = plt.scatter(das_x, das_y,c=das_az)
plt.xlim(0,320)
plt.ylim(0,320)
plt.colorbar(cax, label="Cable Azimuth")
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
```
# Random
```
template = np.linspace(0,320, 65)
template = (template[1:]+template[:-1])/2
np.random.seed(94899109)
das_x = np.random.uniform(5,315,384)
das_y = np.random.uniform(5,315,384)
das_az = np.random.uniform(0,360,384)
raz = np.deg2rad(das_az)
L = 10 #gauge length
ll = np.linspace(-L/2, L/2, 2**5+1)
dl = ll[1]-ll[0]
p1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]
p2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]
if generate_kernels:
os.makedirs("Kernels", exist_ok=True)
crv = loadmat(f"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat")
G_mat = np.reshape(crv["G_mat"].T, (crv["G_mat"].shape[1], nx, ny))
crvscales = crv["scales"].flatten()
cvtscaler = 2.0**(cscale*crvscales)
G1 = np.zeros((len(raz), G_mat.shape[0]))
G2 = np.zeros((len(raz), G_mat.shape[0]))
for j in range(G_mat.shape[0]):
frame = rbs(x,y,G_mat[j])
#average derivatives of frame along gauge length
fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L
fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L
G1[:,j] = (np.sin(raz)**2*fd1 +
np.sin(2*raz)*fd2/2) / cvtscaler[j]
G2[:,j] = (np.cos(raz)**2*fd2 +
np.sin(2*raz)*fd1/2) / cvtscaler[j]
G = np.hstack([G1, G2])
Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))
G = G / Gn
G_random = G
np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))
exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])
eyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])
exyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])
edasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr)
das_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])
np.save("Testing/random.npy", das_wvt_data)
resi = np.load(f"Testing/random_res.npy")
Gs = np.std(G)
resxi = resi[:G3.shape[1], :]
resyi = resi[G3.shape[1]:, :]
xpredi = (G3/Gn/Gs) @ resxi
ypredi = (G3/Gn/Gs) @ resyi
txpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))
typredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))
res = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))
plt.plot(np.std(resi, axis=1))
res
plt.plot(cuxr.T[100])
plt.plot(txpredi[100])
cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)
plt.xlim(0,320)
plt.ylim(0,320)
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
plt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))
, norm=norm)
plt.colorbar()
cax = plt.scatter(das_x, das_y,c=das_az)
plt.xlim(0,320)
plt.ylim(0,320)
plt.colorbar(cax, label="Cable Azimuth")
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
np.save("Kernels/G_zigzag.npy", G_zigzag)
np.save("Kernels/G_spiral.npy", G_spiral)
np.save("Kernels/G_cross.npy", G_cross)
np.save("Kernels/G_random.npy", G_random)
```
# Eigenvalue Spectrum
```
G_full = np.vstack([np.hstack([G3, np.zeros(G3.shape)]), np.hstack([np.zeros(G3.shape), G3])])
idet = 1e-10*np.eye(G_zigzag.shape[1])
ezig = np.sort(np.linalg.eigvals(G_zigzag.T@G_zigzag+idet))[::-1]
espi = np.sort(np.linalg.eigvals(G_spiral.T@G_spiral+idet))[::-1]
ecro = np.sort(np.linalg.eigvals(G_cross.T@G_cross+idet))[::-1]
eran = np.sort(np.linalg.eigvals(G_random.T@G_random+idet))[::-1]
# efull= np.sort(np.linalg.eigvals(G_full.T@G_full+idet))[::-1]
ezign = ezig / ezig[0]
espin = espi / espi[0]
ecron = ecro / ecro[0]
erann = eran / eran[0]
# efulln = efull / efull[0]
plt.plot(np.sort(np.diag(G_zigzag @ np.linalg.solve(G_zigzag.T@G_zigzag + 1e-0*np.eye(G_zigzag.shape[1]), G_zigzag.T))), label="ZigZag")
plt.plot(np.sort(np.diag(G_spiral @ np.linalg.solve(G_spiral.T@G_spiral + 1e-0*np.eye(G_spiral.shape[1]), G_spiral.T))), label="Spiral")
plt.plot(np.sort(np.diag(G_cross @ np.linalg.solve(G_cross.T@G_cross + 1e-0*np.eye(G_cross.shape[1]), G_cross.T))), label="Crossing")
plt.plot(np.sort(np.diag(G_random @ np.linalg.solve(G_random.T@G_random + 1e-0*np.eye(G_random.shape[1]), G_random.T))), label="Random")
plt.xlim(0,384)
plt.ylabel("Coherence")
plt.xlabel("Sorted Diagonal")
plt.legend(loc="lower right")
plt.plot(np.log10(np.real(ezig)), label="ZigZag")
plt.plot(np.log10(np.real(espi)), label="Spiral")
plt.plot(np.log10(np.real(ecro)), label="Crossing")
plt.plot(np.log10(np.real(eran)), label="Random")
plt.xlim(0,384)
plt.ylabel("Log10 Normalized Eigenvalues")
plt.xlabel("Eigenvalue Index")
plt.legend(loc="upper right")
```
|
github_jupyter
|
## Reinforcement Learning for seq2seq
This time we'll solve a problem of transribing hebrew words in english, also known as g2p (grapheme2phoneme)
* word (sequence of letters in source language) -> translation (sequence of letters in target language)
Unlike what most deep learning practicioners do, we won't only train it to maximize likelihood of correct translation, but also employ reinforcement learning to actually teach it to translate with as few errors as possible.
### About the task
One notable property of Hebrew is that it's consonant language. That is, there are no wovels in the written language. One could represent wovels with diacritics above consonants, but you don't expect people to do that in everyay life.
Therefore, some hebrew characters will correspond to several english letters and others - to none, so we should use encoder-decoder architecture to figure that out.
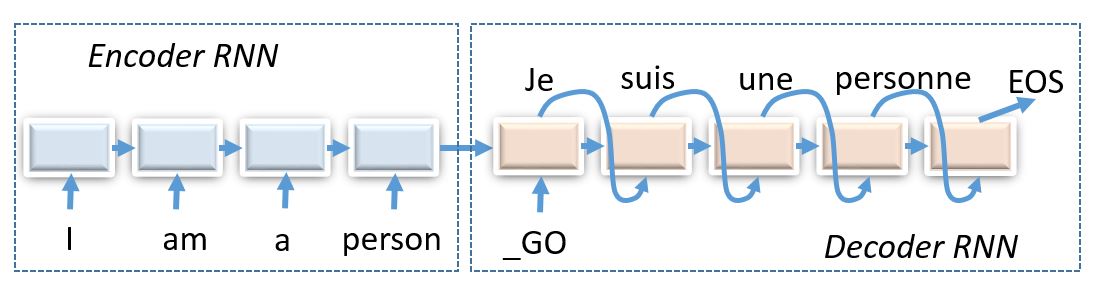
_(img: esciencegroup.files.wordpress.com)_
Encoder-decoder architectures are about converting anything to anything, including
* Machine translation and spoken dialogue systems
* [Image captioning](http://mscoco.org/dataset/#captions-challenge2015) and [image2latex](https://htmlpreview.github.io/?https://github.com/openai/requests-for-research/blob/master/_requests_for_research/im2latex.html) (convolutional encoder, recurrent decoder)
* Generating [images by captions](https://arxiv.org/abs/1511.02793) (recurrent encoder, convolutional decoder)
* Grapheme2phoneme - convert words to transcripts
We chose simplified __Hebrew->English__ machine translation for words and short phrases (character-level), as it is relatively quick to train even without a gpu cluster.
```
import sys
if 'google.colab' in sys.modules:
!wget https://raw.githubusercontent.com/yandexdataschool/Practical_RL/spring20/week07_seq2seq/basic_model_torch.py -O basic_model_torch.py
!wget https://raw.githubusercontent.com/yandexdataschool/Practical_RL/spring20/week07_seq2seq/main_dataset.txt -O main_dataset.txt
!wget https://raw.githubusercontent.com/yandexdataschool/Practical_RL/spring20/week07_seq2seq/voc.py -O voc.py
!pip3 install torch==1.0.0 nltk editdistance
# If True, only translates phrases shorter than 20 characters (way easier).
EASY_MODE = True
# Useful for initial coding.
# If false, works with all phrases (please switch to this mode for homework assignment)
# way we translate. Either "he-to-en" or "en-to-he"
MODE = "he-to-en"
# maximal length of _generated_ output, does not affect training
MAX_OUTPUT_LENGTH = 50 if not EASY_MODE else 20
REPORT_FREQ = 100 # how often to evaluate validation score
```
### Step 1: preprocessing
We shall store dataset as a dictionary
`{ word1:[translation1,translation2,...], word2:[...],...}`.
This is mostly due to the fact that many words have several correct translations.
We have implemented this thing for you so that you can focus on more interesting parts.
__Attention python2 users!__ You may want to cast everything to unicode later during homework phase, just make sure you do it _everywhere_.
```
import numpy as np
from collections import defaultdict
word_to_translation = defaultdict(list) # our dictionary
bos = '_'
eos = ';'
with open("main_dataset.txt", encoding="utf-8") as fin:
for line in fin:
en, he = line[:-1].lower().replace(bos, ' ').replace(eos,
' ').split('\t')
word, trans = (he, en) if MODE == 'he-to-en' else (en, he)
if len(word) < 3:
continue
if EASY_MODE:
if max(len(word), len(trans)) > 20:
continue
word_to_translation[word].append(trans)
print("size = ", len(word_to_translation))
# get all unique lines in source language
all_words = np.array(list(word_to_translation.keys()))
# get all unique lines in translation language
all_translations = np.array(list(set(
[ts for all_ts in word_to_translation.values() for ts in all_ts])))
```
### split the dataset
We hold out 10% of all words to be used for validation.
```
from sklearn.model_selection import train_test_split
train_words, test_words = train_test_split(
all_words, test_size=0.1, random_state=42)
```
### Building vocabularies
We now need to build vocabularies that map strings to token ids and vice versa. We're gonna need these fellas when we feed training data into model or convert output matrices into english words.
```
from voc import Vocab
inp_voc = Vocab.from_lines(''.join(all_words), bos=bos, eos=eos, sep='')
out_voc = Vocab.from_lines(''.join(all_translations), bos=bos, eos=eos, sep='')
# Here's how you cast lines into ids and backwards.
batch_lines = all_words[:5]
batch_ids = inp_voc.to_matrix(batch_lines)
batch_lines_restored = inp_voc.to_lines(batch_ids)
print("lines")
print(batch_lines)
print("\nwords to ids (0 = bos, 1 = eos):")
print(batch_ids)
print("\nback to words")
print(batch_lines_restored)
```
Draw word/translation length distributions to estimate the scope of the task.
```
import matplotlib.pyplot as plt
%matplotlib inline
plt.figure(figsize=[8, 4])
plt.subplot(1, 2, 1)
plt.title("words")
plt.hist(list(map(len, all_words)), bins=20)
plt.subplot(1, 2, 2)
plt.title('translations')
plt.hist(list(map(len, all_translations)), bins=20)
```
### Step 3: deploy encoder-decoder (1 point)
__assignment starts here__
Our architecture consists of two main blocks:
* Encoder reads words character by character and outputs code vector (usually a function of last RNN state)
* Decoder takes that code vector and produces translations character by character
Than it gets fed into a model that follows this simple interface:
* __`model(inp, out, **flags) -> logp`__ - takes symbolic int32 matrices of hebrew words and their english translations. Computes the log-probabilities of all possible english characters given english prefices and hebrew word.
* __`model.translate(inp, **flags) -> out, logp`__ - takes symbolic int32 matrix of hebrew words, produces output tokens sampled from the model and output log-probabilities for all possible tokens at each tick.
* if given flag __`greedy=True`__, takes most likely next token at each iteration. Otherwise samples with next token probabilities predicted by model.
That's all! It's as hard as it gets. With those two methods alone you can implement all kinds of prediction and training.
```
import torch
import torch.nn as nn
import torch.nn.functional as F
from basic_model_torch import BasicTranslationModel
model = BasicTranslationModel(inp_voc, out_voc,
emb_size=64, hid_size=256)
# Play around with symbolic_translate and symbolic_score
inp = torch.tensor(np.random.randint(0, 10, [3, 5]), dtype=torch.int64)
out = torch.tensor(np.random.randint(0, 10, [3, 5]), dtype=torch.int64)
# translate inp (with untrained model)
sampled_out, logp = model.translate(inp, greedy=False)
print("Sample translations:\n", sampled_out)
print("Log-probabilities at each step:\n", logp)
# score logp(out | inp) with untrained input
logp = model(inp, out)
print("Symbolic_score output:\n", logp)
print("Log-probabilities of output tokens:\n",
torch.gather(logp, dim=2, index=out[:, :, None]))
def translate(lines, max_len=MAX_OUTPUT_LENGTH):
"""
You are given a list of input lines.
Make your neural network translate them.
:return: a list of output lines
"""
# Convert lines to a matrix of indices
lines_ix = inp_voc.to_matrix(lines)
lines_ix = torch.tensor(lines_ix, dtype=torch.int64)
# Compute translations in form of indices
trans_ix = <YOUR CODE>
# Convert translations back into strings
return out_voc.to_lines(trans_ix.data.numpy())
print("Sample inputs:", all_words[:3])
print("Dummy translations:", translate(all_words[:3]))
trans = translate(all_words[:3])
assert translate(all_words[:3]) == translate(
all_words[:3]), "make sure translation is deterministic (use greedy=True and disable any noise layers)"
assert type(translate(all_words[:3])) is list and (type(translate(all_words[:1])[0]) is str or type(
translate(all_words[:1])[0]) is unicode), "translate(lines) must return a sequence of strings!"
# note: if translation freezes, make sure you used max_len parameter
print("Tests passed!")
```
### Scoring function
LogLikelihood is a poor estimator of model performance.
* If we predict zero probability once, it shouldn't ruin entire model.
* It is enough to learn just one translation if there are several correct ones.
* What matters is how many mistakes model's gonna make when it translates!
Therefore, we will use minimal Levenshtein distance. It measures how many characters do we need to add/remove/replace from model translation to make it perfect. Alternatively, one could use character-level BLEU/RougeL or other similar metrics.
The catch here is that Levenshtein distance is not differentiable: it isn't even continuous. We can't train our neural network to maximize it by gradient descent.
```
import editdistance # !pip install editdistance
def get_distance(word, trans):
"""
A function that takes word and predicted translation
and evaluates (Levenshtein's) edit distance to closest correct translation
"""
references = word_to_translation[word]
assert len(references) != 0, "wrong/unknown word"
return min(editdistance.eval(trans, ref) for ref in references)
def score(words, bsize=100):
"""a function that computes levenshtein distance for bsize random samples"""
assert isinstance(words, np.ndarray)
batch_words = np.random.choice(words, size=bsize, replace=False)
batch_trans = translate(batch_words)
distances = list(map(get_distance, batch_words, batch_trans))
return np.array(distances, dtype='float32')
# should be around 5-50 and decrease rapidly after training :)
[score(test_words, 10).mean() for _ in range(5)]
```
## Step 2: Supervised pre-training (2 points)
Here we define a function that trains our model through maximizing log-likelihood a.k.a. minimizing crossentropy.
```
import random
def sample_batch(words, word_to_translation, batch_size):
"""
sample random batch of words and random correct translation for each word
example usage:
batch_x,batch_y = sample_batch(train_words, word_to_translations,10)
"""
# choose words
batch_words = np.random.choice(words, size=batch_size)
# choose translations
batch_trans_candidates = list(map(word_to_translation.get, batch_words))
batch_trans = list(map(random.choice, batch_trans_candidates))
return batch_words, batch_trans
bx, by = sample_batch(train_words, word_to_translation, batch_size=3)
print("Source:")
print(bx)
print("Target:")
print(by)
from basic_model_torch import infer_length, infer_mask, to_one_hot
def compute_loss_on_batch(input_sequence, reference_answers):
""" Compute crossentropy loss given a batch of sources and translations """
input_sequence = torch.tensor(inp_voc.to_matrix(input_sequence), dtype=torch.int64)
reference_answers = torch.tensor(out_voc.to_matrix(reference_answers), dtype=torch.int64)
# Compute log-probabilities of all possible tokens at each step. Use model interface.
logprobs_seq = <YOUR CODE>
# compute elementwise crossentropy as negative log-probabilities of reference_answers.
crossentropy = - \
torch.sum(logprobs_seq *
to_one_hot(reference_answers, len(out_voc)), dim=-1)
assert crossentropy.dim(
) == 2, "please return elementwise crossentropy, don't compute mean just yet"
# average with mask
mask = infer_mask(reference_answers, out_voc.eos_ix)
loss = torch.sum(crossentropy * mask) / torch.sum(mask)
return loss
# test it
loss = compute_loss_on_batch(*sample_batch(train_words, word_to_translation, 3))
print('loss = ', loss)
assert loss.item() > 0.0
loss.backward()
for w in model.parameters():
assert w.grad is not None and torch.max(torch.abs(w.grad)).item() != 0, \
"Loss is not differentiable w.r.t. a weight with shape %s. Check comput_loss_on_batch." % (
w.size(),)
```
##### Actually train the model
Minibatches and stuff...
```
from IPython.display import clear_output
from tqdm import tqdm, trange # or use tqdm_notebook,tnrange
loss_history = []
editdist_history = []
entropy_history = []
opt = torch.optim.Adam(model.parameters())
for i in trange(25000):
loss = compute_loss_on_batch(*sample_batch(train_words, word_to_translation, 32))
# train with backprop
loss.backward()
opt.step()
opt.zero_grad()
loss_history.append(loss.item())
if (i+1) % REPORT_FREQ == 0:
clear_output(True)
current_scores = score(test_words)
editdist_history.append(current_scores.mean())
print("llh=%.3f, mean score=%.3f" %
(np.mean(loss_history[-10:]), np.mean(editdist_history[-10:])))
plt.figure(figsize=(12, 4))
plt.subplot(131)
plt.title('train loss / traning time')
plt.plot(loss_history)
plt.grid()
plt.subplot(132)
plt.title('val score distribution')
plt.hist(current_scores, bins=20)
plt.subplot(133)
plt.title('val score / traning time (lower is better)')
plt.plot(editdist_history)
plt.grid()
plt.show()
```
__How to interpret the plots:__
* __Train loss__ - that's your model's crossentropy over minibatches. It should go down steadily. Most importantly, it shouldn't be NaN :)
* __Val score distribution__ - distribution of translation edit distance (score) within batch. It should move to the left over time.
* __Val score / training time__ - it's your current mean edit distance. This plot is much whimsier than loss, but make sure it goes below 8 by 2500 steps.
If it doesn't, first try to re-create both model and opt. You may have changed it's weight too much while debugging. If that doesn't help, it's debugging time.
```
for word in train_words[:10]:
print("%s -> %s" % (word, translate([word])[0]))
test_scores = []
for start_i in trange(0, len(test_words), 32):
batch_words = test_words[start_i:start_i+32]
batch_trans = translate(batch_words)
distances = list(map(get_distance, batch_words, batch_trans))
test_scores.extend(distances)
print("Supervised test score:", np.mean(test_scores))
```
## Self-critical policy gradient (2 points)
In this section you'll implement algorithm called self-critical sequence training (here's an [article](https://arxiv.org/abs/1612.00563)).
The algorithm is a vanilla policy gradient with a special baseline.
$$ \nabla J = E_{x \sim p(s)} E_{y \sim \pi(y|x)} \nabla log \pi(y|x) \cdot (R(x,y) - b(x)) $$
Here reward R(x,y) is a __negative levenshtein distance__ (since we minimize it). The baseline __b(x)__ represents how well model fares on word __x__.
In practice, this means that we compute baseline as a score of greedy translation, $b(x) = R(x,y_{greedy}(x)) $.

Luckily, we already obtained the required outputs: `model.greedy_translations, model.greedy_mask` and we only need to compute levenshtein using `compute_levenshtein` function.
```
def compute_reward(input_sequence, translations):
""" computes sample-wise reward given token ids for inputs and translations """
distances = list(map(get_distance,
inp_voc.to_lines(input_sequence.data.numpy()),
out_voc.to_lines(translations.data.numpy())))
# use negative levenshtein distance so that larger reward means better policy
return - torch.tensor(distances, dtype=torch.int64)
def scst_objective_on_batch(input_sequence, max_len=MAX_OUTPUT_LENGTH):
""" Compute pseudo-loss for policy gradient given a batch of sources """
input_sequence = torch.tensor(inp_voc.to_matrix(input_sequence), dtype=torch.int64)
# use model to __sample__ symbolic translations given input_sequence
sample_translations, sample_logp = <YOUR CODE>
# use model to __greedy__ symbolic translations given input_sequence
greedy_translations, greedy_logp = <YOUR CODE>
# compute rewards and advantage
rewards = compute_reward(input_sequence, sample_translations)
baseline = <YOUR CODE: compute __negative__ levenshtein for greedy mode>
# compute advantage using rewards and baseline
advantage = <YOUR CODE>
# compute log_pi(a_t|s_t), shape = [batch, seq_length]
logp_sample = <YOUR CODE>
# ^-- hint: look at how crossentropy is implemented in supervised learning loss above
# mind the sign - this one should not be multiplied by -1 :)
# policy gradient pseudo-loss. Gradient of J is exactly policy gradient.
J = logp_sample * advantage[:, None]
assert J.dim() == 2, "please return elementwise objective, don't compute mean just yet"
# average with mask
mask = infer_mask(sample_translations, out_voc.eos_ix)
loss = - torch.sum(J * mask) / torch.sum(mask)
# regularize with negative entropy. Don't forget the sign!
# note: for entropy you need probabilities for all tokens (sample_logp), not just logp_sample
entropy = <YOUR CODE: compute entropy matrix of shape[batch, seq_length], H = -sum(p*log_p), don't forget the sign!>
# hint: you can get sample probabilities from sample_logp using math :)
assert entropy.dim(
) == 2, "please make sure elementwise entropy is of shape [batch,time]"
reg = - 0.01 * torch.sum(entropy * mask) / torch.sum(mask)
return loss + reg, torch.sum(entropy * mask) / torch.sum(mask)
```
# Policy gradient training
```
entropy_history = [np.nan] * len(loss_history)
opt = torch.optim.Adam(model.parameters(), lr=1e-5)
for i in trange(100000):
loss, ent = scst_objective_on_batch(
sample_batch(train_words, word_to_translation, 32)[0]) # [0] = only source sentence
# train with backprop
loss.backward()
opt.step()
opt.zero_grad()
loss_history.append(loss.item())
entropy_history.append(ent.item())
if (i+1) % REPORT_FREQ == 0:
clear_output(True)
current_scores = score(test_words)
editdist_history.append(current_scores.mean())
plt.figure(figsize=(12, 4))
plt.subplot(131)
plt.title('val score distribution')
plt.hist(current_scores, bins=20)
plt.subplot(132)
plt.title('val score / traning time')
plt.plot(editdist_history)
plt.grid()
plt.subplot(133)
plt.title('policy entropy / traning time')
plt.plot(entropy_history)
plt.grid()
plt.show()
print("J=%.3f, mean score=%.3f" %
(np.mean(loss_history[-10:]), np.mean(editdist_history[-10:])))
```
__Debugging tips:__
<img src=https://github.com/yandexdataschool/Practical_RL/raw/master/yet_another_week/_resource/do_something_scst.png width=400>
* As usual, don't expect improvements right away, but in general the model should be able to show some positive changes by 5k steps.
* Entropy is a good indicator of many problems.
* If it reaches zero, you may need greater entropy regularizer.
* If it has rapid changes time to time, you may need gradient clipping.
* If it oscillates up and down in an erratic manner... it's perfectly okay for entropy to do so. But it should decrease at the end.
* We don't show loss_history cuz it's uninformative for pseudo-losses in policy gradient. However, if something goes wrong you can check it to see if everything isn't a constant zero.
### Results
```
for word in train_words[:10]:
print("%s -> %s" % (word, translate([word])[0]))
test_scores = []
for start_i in trange(0, len(test_words), 32):
batch_words = test_words[start_i:start_i+32]
batch_trans = translate(batch_words)
distances = list(map(get_distance, batch_words, batch_trans))
test_scores.extend(distances)
print("Supervised test score:", np.mean(test_scores))
# ^^ If you get Out Of MemoryError, please replace this with batched computation
```
## Step 6: Make it actually work (5++ pts)
In this section we want you to finally __restart with EASY_MODE=False__ and experiment to find a good model/curriculum for that task.
We recommend you to start with the following architecture
```
encoder---decoder
P(y|h)
^
LSTM -> LSTM
^ ^
biLSTM -> LSTM
^ ^
input y_prev
```
__Note:__ you can fit all 4 state tensors of both LSTMs into a in a single state - just assume that it contains, for example, [h0, c0, h1, c1] - pack it in encode and update in decode.
Here are some cool ideas on what you can do then.
__General tips & tricks:__
* You will likely need to adjust pre-training time for such a network.
* Supervised pre-training may benefit from clipping gradients somehow.
* SCST may indulge a higher learning rate in some cases and changing entropy regularizer over time.
* It's often useful to save pre-trained model parameters to not re-train it every time you want new policy gradient parameters.
* When leaving training for nighttime, try setting REPORT_FREQ to a larger value (e.g. 500) not to waste time on it.
__Formal criteria:__
To get 5 points we want you to build an architecture that:
* _doesn't consist of single GRU_
* _works better_ than single GRU baseline.
* We also want you to provide either learning curve or trained model, preferably both
* ... and write a brief report or experiment log describing what you did and how it fared.
### Attention
There's more than one way to connect decoder to encoder
* __Vanilla:__ layer_i of encoder last state goes to layer_i of decoder initial state
* __Every tick:__ feed encoder last state _on every iteration_ of decoder.
* __Attention:__ allow decoder to "peek" at one (or several) positions of encoded sequence on every tick.
The most effective (and cool) of those is, of course, attention.
You can read more about attention [in this nice blog post](https://distill.pub/2016/augmented-rnns/). The easiest way to begin is to use "soft" attention with "additive" or "dot-product" intermediate layers.
__Tips__
* Model usually generalizes better if you no longer allow decoder to see final encoder state
* Once your model made it through several epochs, it is a good idea to visualize attention maps to understand what your model has actually learned
* There's more stuff [here](https://github.com/yandexdataschool/Practical_RL/blob/master/week8_scst/bonus.ipynb)
* If you opted for hard attention, we recommend [gumbel-softmax](https://blog.evjang.com/2016/11/tutorial-categorical-variational.html) instead of sampling. Also please make sure soft attention works fine before you switch to hard.
### UREX
* This is a way to improve exploration in policy-based settings. The main idea is that you find and upweight under-appreciated actions.
* Here's [video](https://www.youtube.com/watch?v=fZNyHoXgV7M&feature=youtu.be&t=3444)
and an [article](https://arxiv.org/abs/1611.09321).
* You may want to reduce batch size 'cuz UREX requires you to sample multiple times per source sentence.
* Once you got it working, try using experience replay with importance sampling instead of (in addition to) basic UREX.
### Some additional ideas:
* (advanced deep learning) It may be a good idea to first train on small phrases and then adapt to larger ones (a.k.a. training curriculum).
* (advanced nlp) You may want to switch from raw utf8 to something like unicode or even syllables to make task easier.
* (advanced nlp) Since hebrew words are written __with vowels omitted__, you may want to use a small Hebrew vowel markup dataset at `he-pron-wiktionary.txt`.
```
assert not EASY_MODE, "make sure you set EASY_MODE = False at the top of the notebook."
```
`[your report/log here or anywhere you please]`
__Contributions:__ This notebook is brought to you by
* Yandex [MT team](https://tech.yandex.com/translate/)
* Denis Mazur ([DeniskaMazur](https://github.com/DeniskaMazur)), Oleg Vasilev ([Omrigan](https://github.com/Omrigan/)), Dmitry Emelyanenko ([TixFeniks](https://github.com/tixfeniks)) and Fedor Ratnikov ([justheuristic](https://github.com/justheuristic/))
* Dataset is parsed from [Wiktionary](https://en.wiktionary.org), which is under CC-BY-SA and GFDL licenses.
|
github_jupyter
|
# DEAP
DEAP is a novel evolutionary computation framework for rapid prototyping and testing of ideas. It seeks to make algorithms explicit and data structures transparent. It works in perfect harmony with parallelisation mechanism such as multiprocessing and SCOOP. The following documentation presents the key concepts and many features to build your own evolutions.
Library documentation: <a>http://deap.readthedocs.org/en/master/</a>
## One Max Problem (GA)
This problem is very simple, we search for a 1 filled list individual. This problem is widely used in the evolutionary computation community since it is very simple and it illustrates well the potential of evolutionary algorithms.
```
import random
from deap import base
from deap import creator
from deap import tools
# creator is a class factory that can build new classes at run-time
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
creator.create("Individual", list, fitness=creator.FitnessMax)
# a toolbox stores functions and their arguments
toolbox = base.Toolbox()
# attribute generator
toolbox.register("attr_bool", random.randint, 0, 1)
# structure initializers
toolbox.register("individual", tools.initRepeat, creator.Individual, toolbox.attr_bool, 100)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
# evaluation function
def evalOneMax(individual):
return sum(individual),
# register the required genetic operators
toolbox.register("evaluate", evalOneMax)
toolbox.register("mate", tools.cxTwoPoint)
toolbox.register("mutate", tools.mutFlipBit, indpb=0.05)
toolbox.register("select", tools.selTournament, tournsize=3)
random.seed(64)
# instantiate a population
pop = toolbox.population(n=300)
CXPB, MUTPB, NGEN = 0.5, 0.2, 40
# evaluate the entire population
fitnesses = list(map(toolbox.evaluate, pop))
for ind, fit in zip(pop, fitnesses):
ind.fitness.values = fit
print(" Evaluated %i individuals" % len(pop))
# begin the evolution
for g in range(NGEN):
print("-- Generation %i --" % g)
# select the next generation individuals
offspring = toolbox.select(pop, len(pop))
# clone the selected individuals
offspring = list(map(toolbox.clone, offspring))
# apply crossover and mutation on the offspring
for child1, child2 in zip(offspring[::2], offspring[1::2]):
if random.random() < CXPB:
toolbox.mate(child1, child2)
del child1.fitness.values
del child2.fitness.values
for mutant in offspring:
if random.random() < MUTPB:
toolbox.mutate(mutant)
del mutant.fitness.values
# evaluate the individuals with an invalid fitness
invalid_ind = [ind for ind in offspring if not ind.fitness.valid]
fitnesses = map(toolbox.evaluate, invalid_ind)
for ind, fit in zip(invalid_ind, fitnesses):
ind.fitness.values = fit
print(" Evaluated %i individuals" % len(invalid_ind))
# the population is entirely replaced by the offspring
pop[:] = offspring
# gather all the fitnesses in one list and print the stats
fits = [ind.fitness.values[0] for ind in pop]
length = len(pop)
mean = sum(fits) / length
sum2 = sum(x*x for x in fits)
std = abs(sum2 / length - mean**2)**0.5
print(" Min %s" % min(fits))
print(" Max %s" % max(fits))
print(" Avg %s" % mean)
print(" Std %s" % std)
best_ind = tools.selBest(pop, 1)[0]
print("Best individual is %s, %s" % (best_ind, best_ind.fitness.values))
```
## Symbolic Regression (GP)
Symbolic regression is one of the best known problems in GP. It is commonly used as a tuning problem for new algorithms, but is also widely used with real-life distributions, where other regression methods may not work.
All symbolic regression problems use an arbitrary data distribution, and try to fit the most accurately the data with a symbolic formula. Usually, a measure like the RMSE (Root Mean Square Error) is used to measure an individual’s fitness.
In this example, we use a classical distribution, the quartic polynomial (x^4 + x^3 + x^2 + x), a one-dimension distribution. 20 equidistant points are generated in the range [-1, 1], and are used to evaluate the fitness.
```
import operator
import math
import random
import numpy
from deap import algorithms
from deap import base
from deap import creator
from deap import tools
from deap import gp
# define a new function for divison that guards against divide by 0
def protectedDiv(left, right):
try:
return left / right
except ZeroDivisionError:
return 1
# add aritmetic primitives
pset = gp.PrimitiveSet("MAIN", 1)
pset.addPrimitive(operator.add, 2)
pset.addPrimitive(operator.sub, 2)
pset.addPrimitive(operator.mul, 2)
pset.addPrimitive(protectedDiv, 2)
pset.addPrimitive(operator.neg, 1)
pset.addPrimitive(math.cos, 1)
pset.addPrimitive(math.sin, 1)
# constant terminal
pset.addEphemeralConstant("rand101", lambda: random.randint(-1,1))
# define number of inputs
pset.renameArguments(ARG0='x')
# create fitness and individual objects
creator.create("FitnessMin", base.Fitness, weights=(-1.0,))
creator.create("Individual", gp.PrimitiveTree, fitness=creator.FitnessMin)
# register evolution process parameters through the toolbox
toolbox = base.Toolbox()
toolbox.register("expr", gp.genHalfAndHalf, pset=pset, min_=1, max_=2)
toolbox.register("individual", tools.initIterate, creator.Individual, toolbox.expr)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("compile", gp.compile, pset=pset)
# evaluation function
def evalSymbReg(individual, points):
# transform the tree expression in a callable function
func = toolbox.compile(expr=individual)
# evaluate the mean squared error between the expression
# and the real function : x**4 + x**3 + x**2 + x
sqerrors = ((func(x) - x**4 - x**3 - x**2 - x)**2 for x in points)
return math.fsum(sqerrors) / len(points),
toolbox.register("evaluate", evalSymbReg, points=[x/10. for x in range(-10,10)])
toolbox.register("select", tools.selTournament, tournsize=3)
toolbox.register("mate", gp.cxOnePoint)
toolbox.register("expr_mut", gp.genFull, min_=0, max_=2)
toolbox.register("mutate", gp.mutUniform, expr=toolbox.expr_mut, pset=pset)
# prevent functions from getting too deep/complex
toolbox.decorate("mate", gp.staticLimit(key=operator.attrgetter("height"), max_value=17))
toolbox.decorate("mutate", gp.staticLimit(key=operator.attrgetter("height"), max_value=17))
# compute some statistics about the population
stats_fit = tools.Statistics(lambda ind: ind.fitness.values)
stats_size = tools.Statistics(len)
mstats = tools.MultiStatistics(fitness=stats_fit, size=stats_size)
mstats.register("avg", numpy.mean)
mstats.register("std", numpy.std)
mstats.register("min", numpy.min)
mstats.register("max", numpy.max)
random.seed(318)
pop = toolbox.population(n=300)
hof = tools.HallOfFame(1)
# run the algorithm
pop, log = algorithms.eaSimple(pop, toolbox, 0.5, 0.1, 40, stats=mstats,
halloffame=hof, verbose=True)
```
|
github_jupyter
|
```
# !wget https://malaya-dataset.s3-ap-southeast-1.amazonaws.com/crawler/academia/academia-pdf.json
import json
import cleaning
from tqdm import tqdm
with open('../academia/academia-pdf.json') as fopen:
pdf = json.load(fopen)
len(pdf)
import os
os.path.split(pdf[0]['file'])
import malaya
fast_text = malaya.language_detection.fasttext()
fast_text.predict(['Prosiding_Kolokium_Siswazah_JUF_2017.pdf'])
from unidecode import unidecode
def clean(string):
string = [cleaning.cleaning(s) for s in string]
string = [s.strip() for s in string if 'tarikh' not in s.lower() and 'soalan no' not in s.lower()]
string = [s for s in string if not ''.join(s.split()[:1]).isdigit() and '.soalan' not in s.lower() and 'jum ' not in s.lower()]
string = [s for s in string if not s[:3].isdigit() and not s[-3:].isdigit()]
return string
outer = []
for k in tqdm(range(len(pdf))):
c = clean(pdf[k]['content']['content'].split('\n'))
t, last = [], 0
i = 0
while i < len(c):
text = c[i]
if len(text) > 5:
if len(text.split()) > 1:
t.append(text)
last = i
else:
if len(t) and (i - last) > 2:
t.append('')
outer.extend(t)
t = []
last = i
elif not len(t):
last = i
i += 1
if len(t):
t.append('')
outer.extend(t)
len(outer)
%%time
temp_vocab = list(set(cleaning.multiprocessing(outer, cleaning.unique_words)))
%%time
# important
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.duplicate_dots_marks_exclamations, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
# important
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.remove_underscore, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
# important
temp_dict = cleaning.multiprocessing(outer, cleaning.isolate_spamchars, list_mode = False)
print(len(temp_dict))
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.break_short_words, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.break_long_words, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.remove_ending_underscore, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.remove_starting_underscore, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.end_punct, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.start_punct, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.join_dashes, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
results, result = [], []
for i in tqdm(outer):
if not len(i) and len(result):
results.append(result)
result = []
else:
result.append(i)
if len(result):
results.append(result)
import re
alphabets = '([A-Za-z])'
prefixes = (
'(Mr|St|Mrs|Ms|Dr|Prof|Capt|Cpt|Lt|Mt|Puan|puan|Tuan|tuan|sir|Sir)[.]'
)
suffixes = '(Inc|Ltd|Jr|Sr|Co|Mo)'
starters = '(Mr|Mrs|Ms|Dr|He\s|She\s|It\s|They\s|Their\s|Our\s|We\s|But\s|However\s|That\s|This\s|Wherever|Dia|Mereka|Tetapi|Kita|Itu|Ini|Dan|Kami|Beliau|Seri|Datuk|Dato|Datin|Tuan|Puan)'
acronyms = '([A-Z][.][A-Z][.](?:[A-Z][.])?)'
websites = '[.](com|net|org|io|gov|me|edu|my)'
another_websites = '(www|http|https)[.]'
digits = '([0-9])'
before_digits = '([Nn]o|[Nn]ombor|[Nn]umber|[Kk]e|=|al)'
month = '([Jj]an(?:uari)?|[Ff]eb(?:ruari)?|[Mm]a(?:c)?|[Aa]pr(?:il)?|Mei|[Jj]u(?:n)?|[Jj]ula(?:i)?|[Aa]ug(?:ust)?|[Ss]ept?(?:ember)?|[Oo]kt(?:ober)?|[Nn]ov(?:ember)?|[Dd]is(?:ember)?)'
def split_into_sentences(text, minimum_length = 5):
text = text.replace('\x97', '\n')
text = '. '.join([s for s in text.split('\n') if len(s)])
text = text + '.'
text = unidecode(text)
text = ' ' + text + ' '
text = text.replace('\n', ' ')
text = re.sub(prefixes, '\\1<prd>', text)
text = re.sub(websites, '<prd>\\1', text)
text = re.sub(another_websites, '\\1<prd>', text)
text = re.sub('[,][.]+', '<prd>', text)
if '...' in text:
text = text.replace('...', '<prd><prd><prd>')
if 'Ph.D' in text:
text = text.replace('Ph.D.', 'Ph<prd>D<prd>')
text = re.sub('[.]\s*[,]', '<prd>,', text)
text = re.sub(before_digits + '\s*[.]\s*' + digits, '\\1<prd>\\2', text)
text = re.sub(month + '[.]\s*' + digits, '\\1<prd>\\2', text)
text = re.sub('\s' + alphabets + '[.][ ]+', ' \\1<prd> ', text)
text = re.sub(acronyms + ' ' + starters, '\\1<stop> \\2', text)
text = re.sub(
alphabets + '[.]' + alphabets + '[.]' + alphabets + '[.]',
'\\1<prd>\\2<prd>\\3<prd>',
text,
)
text = re.sub(
alphabets + '[.]' + alphabets + '[.]', '\\1<prd>\\2<prd>', text
)
text = re.sub(' ' + suffixes + '[.][ ]+' + starters, ' \\1<stop> \\2', text)
text = re.sub(' ' + suffixes + '[.]', ' \\1<prd>', text)
text = re.sub(' ' + alphabets + '[.]', ' \\1<prd>', text)
text = re.sub(digits + '[.]' + digits, '\\1<prd>\\2', text)
if '”' in text:
text = text.replace('.”', '”.')
if '"' in text:
text = text.replace('."', '".')
if '!' in text:
text = text.replace('!"', '"!')
if '?' in text:
text = text.replace('?"', '"?')
text = text.replace('.', '.<stop>')
text = text.replace('?', '?<stop>')
text = text.replace('!', '!<stop>')
text = text.replace('<prd>', '.')
sentences = text.split('<stop>')
sentences = sentences[:-1]
sentences = [s.strip() for s in sentences if len(s) > minimum_length]
return sentences
split_into_sentences('733 ke . 633 , berlaku penurunan akibat kesan program PMI .')
import malaya
import re
def strip(string):
string = ' '.join(string)
string = re.sub(r'[ ]+', ' ', string.replace('\n', ' ').replace('\t', ' ')).strip()
return split_into_sentences(string)
output = []
for r in tqdm(results):
output.extend(strip(r) + [''])
len(output)
output[10000:11000]
with open('dumping-academia.txt', 'w') as fopen:
fopen.write('\n'.join(output))
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/csy99/dna-nn-theory/blob/master/supervised_UCI_adam256_save_embedding.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
from itertools import product
import re
import time
from sklearn.model_selection import train_test_split
from sklearn.manifold import TSNE
import tensorflow as tf
from tensorflow import keras
```
# Read Data
```
!pip install PyDrive
from google.colab import drive
drive.mount('/content/gdrive')
def convert_label(row):
if row["Classes"] == 'EI':
return 0
if row["Classes"] == 'IE':
return 1
if row["Classes"] == 'N':
return 2
data_path = '/content/gdrive/My Drive/Colab Notebooks/UCI/'
splice_df = pd.read_csv(data_path + 'splice.data', header=None)
splice_df.columns = ['Classes', 'Name', 'Seq']
splice_df["Seq"] = splice_df["Seq"].str.replace(' ', '').str.replace('N', 'A').str.replace('D', 'T').str.replace('S', 'C').str.replace('R', 'G')
splice_df["Label"] = splice_df.apply(lambda row: convert_label(row), axis=1)
print('The shape of the datasize is', splice_df.shape)
splice_df.head()
seq_num = 0
for seq in splice_df["Seq"]:
char_num = 0
for char in seq:
if char != 'A' and char != 'C' and char != 'T' and char != 'G':
print("seq", seq_num, 'char', char_num, 'is', char)
char_num += 1
seq_num += 1
# check if the length of the sequence is the same
seq_len = len(splice_df.Seq[0])
print("The length of the sequence is", seq_len)
for seq in splice_df.Seq[:200]:
assert len(seq) == seq_len
xtrain_full, xtest, ytrain_full, ytest = train_test_split(splice_df, splice_df.Label, test_size=0.2, random_state=100, stratify=splice_df.Label)
xtrain, xval, ytrain, yval = train_test_split(xtrain_full, ytrain_full, test_size=0.2, random_state=100, stratify=ytrain_full)
print("shape of training, validation, test set\n", xtrain.shape, xval.shape, xtest.shape, ytrain.shape, yval.shape, ytest.shape)
word_size = 1
vocab = [''.join(p) for p in product('ACGT', repeat=word_size)]
word_to_idx = {word: i for i, word in enumerate(vocab)}
vocab_size = len(word_to_idx)
print('vocab_size:', vocab_size)
create1gram = keras.layers.experimental.preprocessing.TextVectorization(
standardize=lambda x: tf.strings.regex_replace(x, '(.)', '\\1 '), ngrams=1
)
create1gram.adapt(vocab)
def ds_preprocess(x, y):
x_index = tf.subtract(create1gram(x), 2)
return x_index, y
# not sure the correct way to get mapping from word to its index
create1gram('A C G T') - 2
BATCH_SIZE = 256
xtrain_ds = tf.data.Dataset.from_tensor_slices((xtrain['Seq'], ytrain)).map(ds_preprocess).batch(BATCH_SIZE)
xval_ds = tf.data.Dataset.from_tensor_slices((xval['Seq'], yval)).map(ds_preprocess).batch(BATCH_SIZE)
xtest_ds = tf.data.Dataset.from_tensor_slices((xtest['Seq'], ytest)).map(ds_preprocess).batch(BATCH_SIZE)
latent_size = 30
model = keras.Sequential([
keras.Input(shape=(seq_len,)),
keras.layers.Embedding(seq_len, latent_size),
keras.layers.LSTM(latent_size, return_sequences=False),
keras.layers.Dense(128, activation="relu", input_shape=[latent_size]),
keras.layers.Dropout(0.2),
keras.layers.Dense(64, activation="relu"),
keras.layers.Dropout(0.2),
keras.layers.Dense(32, activation="relu"),
keras.layers.Dropout(0.2),
keras.layers.Dense(16, activation="relu"),
keras.layers.Dropout(0.2),
keras.layers.Dense(3, activation="softmax")
])
model.summary()
es_cb = keras.callbacks.EarlyStopping(patience=100, restore_best_weights=True)
model.compile(keras.optimizers.Adam(), loss=keras.losses.SparseCategoricalCrossentropy(), metrics=['accuracy'])
hist = model.fit(xtrain_ds, validation_data=xval_ds, epochs=4000, callbacks=[es_cb])
def save_hist():
filename = data_path + "baseline_uci_adam256_history.csv"
hist_df = pd.DataFrame(hist.history)
with open(filename, mode='w') as f:
hist_df.to_csv(f)
save_hist()
fig, axes = plt.subplots(1, 2, figsize=(10, 5))
for i in range(1):
ax1 = axes[0]
ax2 = axes[1]
ax1.plot(hist.history['loss'], label='training')
ax1.plot(hist.history['val_loss'], label='validation')
ax1.set_ylim((0.2, 1.2))
ax1.set_title('lstm autoencoder loss')
ax1.set_xlabel('epoch')
ax1.set_ylabel('loss')
ax1.legend(['train', 'validation'], loc='upper left')
ax2.plot(hist.history['accuracy'], label='training')
ax2.plot(hist.history['val_accuracy'], label='validation')
ax2.set_ylim((0.5, 1.0))
ax2.set_title('lstm autoencoder accuracy')
ax2.set_xlabel('epoch')
ax2.set_ylabel('accuracy')
ax2.legend(['train', 'validation'], loc='upper left')
fig.tight_layout()
def eval_model(model, ds, ds_name="Training"):
loss, acc = model.evaluate(ds, verbose=0)
print("{} Dataset: loss = {} and acccuracy = {}%".format(ds_name, np.round(loss, 3), np.round(acc*100, 2)))
eval_model(model, xtrain_ds, "Training")
eval_model(model, xval_ds, "Validation")
eval_model(model, xtest_ds, "Test")
```
|
github_jupyter
|
```
import pandas as pd
import numpy as np
import os
import math
import graphlab
import graphlab as gl
import graphlab.aggregate as agg
from graphlab import SArray
'''钢炮'''
path = '/home/zongyi/bimbo_data/'
train = gl.SFrame.read_csv(path + 'train_lag5.csv', verbose=False)
town = gl.SFrame.read_csv(path + 'towns.csv', verbose=False)
train = train.join(town, on=['Agencia_ID','Producto_ID'], how='left')
train = train.fillna('t_c',1)
train = train.fillna('tcc',0)
train = train.fillna('tp_sum',0)
del train['Town']
del train['id']
del train['Venta_uni_hoy']
del train['Venta_hoy']
del train['Dev_uni_proxima']
del train['Dev_proxima']
del train['Demanda_uni_equil']
# relag_train = gl.SFrame.read_csv(path + 're_lag_train.csv', verbose=False)
# train = train.join(relag_train, on=['Cliente_ID','Producto_ID','Semana'], how='left')
# train = train.fillna('re_lag1',0)
# train = train.fillna('re_lag2',0)
# train = train.fillna('re_lag3',0)
# train = train.fillna('re_lag4',0)
# train = train.fillna('re_lag5',0)
# del relag_train
# pd = gl.SFrame.read_csv(path + 'products.csv', verbose=False)
# train = train.join(pd, on=['Producto_ID'], how='left')
# train = train.fillna('prom',0)
# train = train.fillna('weight',0)
# train = train.fillna('pieces',1)
# train = train.fillna('w_per_piece',0)
# train = train.fillna('healthy',0)
# train = train.fillna('drink',0)
# del train['brand']
# del train['NombreProducto']
# del pd
# client = gl.SFrame.read_csv(path + 'clients.csv', verbose=False)
# train = train.join(client, on=['Cliente_ID'], how='left')
# del client
# cluster = gl.SFrame.read_csv(path + 'prod_cluster.csv', verbose=False)
# cluster = cluster[['Producto_ID','cluster']]
# train = train.join(cluster, on=['Producto_ID'], how='left')
train
# Make a train-test split
train_data, test_data = train.random_split(0.999)
# Create a model.
model = gl.boosted_trees_regression.create(train_data, target='Demada_log',
step_size=0.1,
max_iterations=500,
max_depth = 10,
metric='rmse',
random_seed=395,
column_subsample=0.7,
row_subsample=0.85,
validation_set=test_data,
model_checkpoint_path=path,
model_checkpoint_interval=500)
model1 = gl.boosted_trees_regression.create(train, target='Demada_log',
step_size=0.1,
max_iterations=4,
max_depth = 10,
metric='rmse',
random_seed=395,
column_subsample=0.7,
row_subsample=0.85,
validation_set=None,
resume_from_checkpoint=path+'model_checkpoint_4',
model_checkpoint_path=path,
model_checkpoint_interval=2)
model
w = model.get_feature_importance()
w = w.add_row_number()
w
from IPython.core.pylabtools import figsize
import numpy as np
from matplotlib import pyplot as plt
import seaborn as sns
sns.set_style('darkgrid', {'grid.color': '.8','grid.linestyle': u'--'})
%matplotlib inline
figsize(12, 6)
plt.bar(w['id'], w['count'], tick_label=w['name'])
plt.xticks(rotation=45)
# Save predictions to an SArray
predictions = model.predict(train)
# Evaluate the model and save the results into a dictionary
results = model.evaluate(train)
print results
model.summary()
test = gl.SFrame.read_csv(path + 'test_lag5.csv', verbose=False)
test = test.join(town, on=['Agencia_ID','Producto_ID'], how='left')
del test['Town']
test = test.fillna('t_c',1)
test = test.fillna('tcc',0)
test = test.fillna('tp_sum',0)
test
ids = test['id']
del test['id']
demand_log = model.predict(test)
sub = gl.SFrame({'id':ids,'Demanda_uni_equil':demand_log})
import math
sub['Demanda_uni_equil'] = sub['Demanda_uni_equil'].apply(lambda x: math.expm1(max(0, x)))
sub
sub.save(path+'gbrt_sub3.csv',format='csv')
math.expm1(math.log1p(2))
```
|
github_jupyter
|
## Multi-label classification
```
%reload_ext autoreload
%autoreload 2
%matplotlib inline
from fastai.conv_learner import *
PATH = 'data/planet/'
# Data preparation steps if you are using Crestle:
os.makedirs('data/planet/models', exist_ok=True)
os.makedirs('/cache/planet/tmp', exist_ok=True)
!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/train-jpg {PATH}
!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/test-jpg {PATH}
!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/train_v2.csv {PATH}
!ln -s /cache/planet/tmp {PATH}
ls {PATH}
```
## Multi-label versus single-label classification
```
from fastai.plots import *
def get_1st(path): return glob(f'{path}/*.*')[0]
dc_path = "data/dogscats/valid/"
list_paths = [get_1st(f"{dc_path}cats"), get_1st(f"{dc_path}dogs")]
plots_from_files(list_paths, titles=["cat", "dog"], maintitle="Single-label classification")
```
In single-label classification each sample belongs to one class. In the previous example, each image is either a *dog* or a *cat*.
```
list_paths = [f"{PATH}train-jpg/train_0.jpg", f"{PATH}train-jpg/train_1.jpg"]
titles=["haze primary", "agriculture clear primary water"]
plots_from_files(list_paths, titles=titles, maintitle="Multi-label classification")
```
In multi-label classification each sample can belong to one or more clases. In the previous example, the first images belongs to two clases: *haze* and *primary*. The second image belongs to four clases: *agriculture*, *clear*, *primary* and *water*.
## Multi-label models for Planet dataset
```
from planet import f2
metrics=[f2]
f_model = resnet34
label_csv = f'{PATH}train_v2.csv'
n = len(list(open(label_csv)))-1
val_idxs = get_cv_idxs(n)
```
We use a different set of data augmentations for this dataset - we also allow vertical flips, since we don't expect vertical orientation of satellite images to change our classifications.
```
def get_data(sz):
tfms = tfms_from_model(f_model, sz, aug_tfms=transforms_top_down, max_zoom=1.05)
return ImageClassifierData.from_csv(PATH, 'train-jpg', label_csv, tfms=tfms,
suffix='.jpg', val_idxs=val_idxs, test_name='test-jpg')
data = get_data(256)
x,y = next(iter(data.val_dl))
y
list(zip(data.classes, y[0]))
plt.imshow(data.val_ds.denorm(to_np(x))[0]*1.4);
sz=64
data = get_data(sz)
data = data.resize(int(sz*1.3), 'tmp')
learn = ConvLearner.pretrained(f_model, data, metrics=metrics)
lrf=learn.lr_find()
learn.sched.plot()
lr = 0.05
learn.fit(lr, 3, cycle_len=1, cycle_mult=2)
lrs = np.array([lr/9,lr/3,lr])
learn.unfreeze()
learn.fit(lrs, 3, cycle_len=1, cycle_mult=2)
learn.save(f'{sz}')
learn.sched.plot_loss()
sz=128
learn.set_data(get_data(sz))
learn.freeze()
learn.fit(lr, 3, cycle_len=1, cycle_mult=2)
learn.unfreeze()
learn.fit(lrs, 3, cycle_len=1, cycle_mult=2)
learn.save(f'{sz}')
sz=256
learn.set_data(get_data(sz))
learn.freeze()
learn.fit(lr, 3, cycle_len=1, cycle_mult=2)
learn.unfreeze()
learn.fit(lrs, 3, cycle_len=1, cycle_mult=2)
learn.save(f'{sz}')
multi_preds, y = learn.TTA()
preds = np.mean(multi_preds, 0)
files = !ls {PATH}test/
```
### End
|
github_jupyter
|
```
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.layers import Embedding, LSTM, Dense, Dropout, Bidirectional
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.models import Sequential
from tensorflow.keras.optimizers import Adam
from tensorflow.keras import regularizers
import tensorflow.keras.utils as ku
import numpy as np
tokenizer = Tokenizer()
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/sonnets.txt \
-O /tmp/sonnets.txt
data = open('/tmp/sonnets.txt').read()
corpus = data.lower().split("\n")
tokenizer.fit_on_texts(corpus)
total_words = len(tokenizer.word_index) + 1
# create input sequences using list of tokens
input_sequences = []
for line in corpus:
token_list = tokenizer.texts_to_sequences([line])[0]
for i in range(1, len(token_list)):
n_gram_sequence = token_list[:i+1]
input_sequences.append(n_gram_sequence)
# pad sequences
max_sequence_len = max([len(x) for x in input_sequences])
input_sequences = np.array(pad_sequences(input_sequences, maxlen=max_sequence_len, padding='pre'))
# create predictors and label
predictors, label = input_sequences[:,:-1],input_sequences[:,-1]
label = ku.to_categorical(label, num_classes=total_words)
model = Sequential()
model.add(Embedding(total_words, 100, input_length=max_sequence_len-1))
model.add(Bidirectional(LSTM(150, return_sequences = True)))
model.add(Dropout(0.2))
model.add(LSTM(100))
model.add(Dense(total_words/2, activation='relu', kernel_regularizer=regularizers.l2(0.01)))
model.add(Dense(total_words, activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
print(model.summary())
history = model.fit(predictors, label, epochs=100, verbose=1)
import matplotlib.pyplot as plt
acc = history.history['accuracy']
loss = history.history['loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'b', label='Training accuracy')
plt.title('Training accuracy')
plt.figure()
plt.plot(epochs, loss, 'b', label='Training Loss')
plt.title('Training loss')
plt.legend()
plt.show()
seed_text = "I Love you"
next_words = 100
for _ in range(next_words):
token_list = tokenizer.texts_to_sequences([seed_text])[0]
token_list = pad_sequences([token_list], maxlen=max_sequence_len-1, padding='pre')
predicted = model.predict_classes(token_list, verbose=0)
output_word = ""
for word, index in tokenizer.word_index.items():
if index == predicted:
output_word = word
break
seed_text += " " + output_word
print(seed_text)
```
|
github_jupyter
|
# sentinelRequest
sentinelRequest can be used to colocate a geodataframe (ie areas, trajectories, buoys, etc ...) with sentinel (1, but also 2 , 3 : all known by scihub)
## Install
```
conda install -c conda-forge lxml numpy geopandas shapely requests fiona matplotlib jupyter descartes
pip install --upgrade git+https://github.com/oarcher/sentinelrequest.git
```
## CLI usage
```
!sentinelrequest --help
```
### "One shot" from command line:
`
% sentinelrequest --user=xxxx --password=xxxxx --date='2018-09-23 00:00' --date='2018-09-23 12:00' --filename='S1?_?W_GRD*.SAFE' --cachedir=/home1/scratch/oarcher/scihub_cache/ --wkt='POLYGON ((-10 75, -10 86, 12 86, 12 84, -10 75))'
`
```
INFO:sentinelRequest:from 2018-09-23 00:00:00 to 2018-09-23 12:00:00 : 11 SAFES
INFO:sentinelRequest:Total : 11 SAFES
filename
S1B_EW_GRDM_1SDH_20180923T071854_20180923T071954_012839_017B47_17F2.SAFE
S1B_EW_GRDM_1SDH_20180923T071954_20180923T072054_012839_017B47_1E6F.SAFE
S1B_EW_GRDM_1SDH_20180923T072054_20180923T072154_012839_017B47_CD41.SAFE
S1B_EW_GRDM_1SDH_20180923T072154_20180923T072254_012839_017B47_3682.SAFE
S1A_EW_GRDM_1SDH_20180923T081003_20180923T081107_023823_02997B_049A.SAFE
S1A_EW_GRDM_1SDH_20180923T081107_20180923T081207_023823_02997B_6EA6.SAFE
S1B_EW_GRDM_1SDH_20180923T085656_20180923T085756_012840_017B4E_B07B.SAFE
S1B_EW_GRDM_1SDH_20180923T085756_20180923T085856_012840_017B4E_6CAD.SAFE
S1B_EW_GRDM_1SDH_20180923T085856_20180923T085956_012840_017B4E_1CCD.SAFE
S1B_EW_GRDM_1SDH_20180923T103504_20180923T103604_012841_017B54_DBBC.SAFE
S1B_EW_GRDM_1SDH_20180923T103604_20180923T103704_012841_017B54_B267.SAFE
```
### From csv file
`
% cat test.csv
`
```
index;startdate;stopdate;geometry
area1;2018-10-02 00:00;2018-10-02 21:00;POLYGON ((-12 35, -5 35, -5 45, -12 45, -12 35))
area2;2018-10-13 06:00;2018-10-13 21:00;POLYGON ((-10 32, -3 32, -3 42, -10 42, -10 32))
area3;2018-10-13 00:00;2018-10-13 18:00;POLYGON ((12 35, 5 35, 5 45, 12 45, 12 35))
```
`
% sentinelRequest --user=xxxx --password=xxxx --infile=test.csv --filename='S1?_?W_GRD*.SAFE' --cachedir=/home1/scratch/oarcher/scihub_cache/ --cols=index,filename
`
```
INFO:sentinelRequest:req 1/2 from 2018-10-02 00:00:00 to 2018-10-02 21:00:00 : 9/21 SAFES
INFO:sentinelRequest:req 2/2 from 2018-10-13 00:00:00 to 2018-10-13 21:00:00 : 30/35 SAFES
INFO:sentinelRequest:Total : 39 SAFES
index;filename
area1;S1A_IW_GRDH_1SDV_20181002T061827_20181002T061852_023953_029DA0_C61E.SAFE
area1;S1B_IW_GRDH_1SDV_20181002T181105_20181002T181130_012977_017F7D_FE88.SAFE
area1;S1B_IW_GRDH_1SDV_20181002T181130_20181002T181155_012977_017F7D_93FF.SAFE
area1;S1B_IW_GRDH_1SDV_20181002T181155_20181002T181222_012977_017F7D_CD9A.SAFE
area3;S1A_IW_GRDH_1SDV_20181013T053545_20181013T053610_024113_02A2DB_D121.SAFE
area3;S1A_IW_GRDH_1SDV_20181013T053815_20181013T053840_024113_02A2DB_7D53.SAFE
area2;S1B_IW_GRDH_1SDV_20181013T062502_20181013T062527_013130_018428_1E77.SAFE
area2;S1B_IW_GRDH_1SDV_20181013T062527_20181013T062552_013130_018428_82AB.SAFE
area2;S1B_IW_GRDH_1SDV_20181013T062642_20181013T062707_013130_018428_AB0E.SAFE
area2;S1B_IW_GRDH_1SDV_20181013T062707_20181013T062732_013130_018428_8210.SAFE
```
If `--date` is specified 2 times with `--infile`, it will superseeds ones founds in infile :
`
sentinelRequest --user oarcher --password nliqt6u3 --infile=test.csv --date=last-monday-7days --date=now --filename='S1?_?W_GRD*.SAFE' --cachedir=/home1/scratch/oarcher/scihub_cache/ --cols=index,filename
`
## API usage
```
%matplotlib inline
import geopandas as gpd
import datetime
import matplotlib.pyplot as plt
import shapely.wkt as wkt
# get your own credential from https://scihub.copernicus.eu/dhus
import pickle
user,password = pickle.load(open("credential.pkl","rb"))
import sentinelrequest as sr
# set default values, so we don't have to pass them at every requests
sr.default_user = user
sr.default_password = password
sr.default_cachedir='/tmp/scihub_cache'
sr.default_filename='S1?_?W_GRD*.SAFE'
# optional : debug messages
#import logging
#sr.logger.setLevel(logging.DEBUG)
help(sr.scihubQuery)
```
### Simplest api usage
Just a startdate and a stopdate are given, with no geometry
```
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
startdate=datetime.datetime(2018,10,2),
stopdate=datetime.datetime(2018,10,3),
fig=fig)
```
The result is a geodataframe with most information from scihub:
```
safes.iloc[0]
```
Most fields are converted from str to python type (geometry, datetime, int ...)
```
safes.iloc[1:4]['footprint'].plot()
print('safe was ingested %s after aquisition' % (safes.iloc[0]['ingestiondate']-safes.iloc[0]['endposition']))
```
### Using a geodataframe with geometries
As an example, two areas are defined. Note that the index is named with the area name
```
gdf = gpd.GeoDataFrame({
"beginposition" : [ datetime.datetime(2018,10,2,0) , datetime.datetime(2018,10,13,0) ],
"endposition" : [ datetime.datetime(2018,10,2,21) ,datetime.datetime(2018,10,13,18) ],
"geometry" : [ wkt.loads("POINT (-7.5 53)").buffer(4), wkt.loads("POLYGON ((-12 35, -5 35, -5 45, -12 45, -12 35))")]
},index=["Irland","Portugal"])
gdf
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf,
min_sea_percent=20,
fig=fig)
```
User requested area are in green, and found safes are in blue.
Index from original request are preserved, so it's easy to know the area that belong to a safe. (See end of example 2 for advanced index handling).
```
safes.loc['Portugal']
```
### Working with projection
SentinelRequest works with projections, by defining crs in gdf.
The colocalisation is done using this crs.
get safes around 1000km, at 84° (North pole included)
```
import pyproj
gdf = gpd.GeoDataFrame({
"beginposition" : [ datetime.datetime(2019,12,1,0) ],
"endposition" : [ datetime.datetime(2019,12,4,0)],
"geometry" : [ wkt.loads("POINT (0 84)")]
},index=["Artic"], crs=pyproj.CRS('epsg:4326'))
# to polar projection (units in meters)
gdf.to_crs(pyproj.CRS('epsg:3408'), inplace=True)
gdf.loc["Artic","geometry"]=gdf.loc["Artic"].geometry.buffer(1000 * 1000)
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf,
min_sea_percent=20,
fig=fig)
```
### Cyclone track colocalization
```
import pandas as pd
#ibtracs=gpd.read_file('tmp/IBTrACS.NA.list.v04r00.points.shp')
#gdf_track=ibtracs[ibtracs['SID'] == '2019235N10324']
#gdf_track=gdf_track[['ISO_TIME','USA_WIND','geometry']]
#gdf_track['ISO_TIME']=pd.to_datetime(gdf_track['ISO_TIME'],format="%Y-%m-%d %H:%M:%S")
#gdf_track.reset_index(inplace = True,drop=True)
#gdf_track.to_file("track.gpkg", driver="GPKG")
gdf_track = gpd.read_file('track.gpkg')
gdf_track['ISO_TIME']=pd.to_datetime(gdf_track['ISO_TIME'],format="%Y-%m-%d %H:%M:%S")
gdf_track
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf_track,
date='ISO_TIME', # no startdate/stopdate, but a date ans a dtime
dtime=datetime.timedelta(hours=1.5),
datatake=1, # take adjacents safes, up to one.
fig=fig)
```
#### datatake
Here, `datatake=1` is specified to retrieve adjacents safes from colocated ones (in cyan). When specified, the result contain a `datatake_index` column. 0 means the colocated one, and other values are the range of the adjacent safe (up to -n..n with `datatake=n`)
Positive `datatake_index` are for safes *after* the colocated one, and negative index are fo safes *before* the colocated one.
```
safes[['filename','datatake_index']]
```
#### Time slicing with timedelta_slice
One can see on previous figure that 3 requests are done. gdf rows are grouped to reduce the amount of scihub requests with the `timedelta_slice` parameter (default to `datetime.timedelta(weeks=1)` )
If we reduce `timedelta_slice`, we can see that more scihub request are done, with less uncolocated safes (ie yellow). (be warned with a big `timedelta_slice` : this can produce scihub timeouts).
(with `timedelta_slice=None`, this feature is *disabled* : a scihub request is done for *every* geometry).
```
# same request as above, but with reduced timedelta_slice
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf_track,
date='ISO_TIME',
dtime=datetime.timedelta(hours=1.5),
timedelta_slice=datetime.timedelta(days=1),
datatake=1,
full_fig = True, # to show internals requests and colocs
fig=fig)
```
#### Merging source and result with shared index
As seen before, the result (safes) share the same index as the source. So we can merge the two geodataframe, to associate a wind speed from the cyclone track with the safe, and compute distance from the eye to the safe.
```
# here, we merge the result with the source request, to associate wind speed to each safe.
merged=safes[['filename','datatake_index','footprint']].merge(
gdf_track[['USA_WIND','geometry']],left_index=True,right_index=True)
merged['eye_dist'] = merged.set_geometry('geometry').distance(merged.set_geometry('footprint').exterior)
# negative dist if safe contains eye
merged['eye_dist']=merged['eye_dist']*(((~merged.set_geometry('footprint').contains(merged.set_geometry('geometry'))+1)*2)-3)
merged[['filename','datatake_index','USA_WIND','eye_dist']]
```
## Annexes
### Antimeridian handling: small geometry vs large one
Given 2 points on the earth, there is two possible paths: one short, and one long that wrap around the earth.
Note: only longitude is wrapped, as if earth was a cylinder (epgs 4326 used for computation)
By default, geometry are the smallest ones. To preserve a large geometry, GeometryCollection must be used.
```
from shapely.geometry import GeometryCollection
# the polygon is more than 180 deg wide. It will be wrapped, and will cross antimeridian
large_poly = wkt.loads("POLYGON ((-140 -14, 140 -14, 140 -20, -140 -20, -140 -14))")
gdf = gpd.GeoDataFrame({
"beginposition" : [ datetime.datetime(2018,10,1)],
"endposition" : [ datetime.datetime(2018,10,31) ],
"geometry" : [ large_poly ]
},index=[0])
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf,
fig=fig)
plt.show()
# same polygon, but encapsulated in a GeometryCollection : it will not be wrapped
gdf = gpd.GeoDataFrame({
"beginposition" : [ datetime.datetime(2018,10,1)],
"endposition" : [ datetime.datetime(2018,10,31) ],
"geometry" : [ GeometryCollection([large_poly]) ]
},index=[0])
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf,
fig=fig)
plt.show()
gdf
import shapely.ops
len(shapely.ops.unary_union(gdf_track.geometry).buffer(2).simplify(1.9).wkt)
```
|
github_jupyter
|
```
library(data.table)
library(dplyr)
library(Matrix)
library(BuenColors)
library(stringr)
library(cowplot)
library(SummarizedExperiment)
library(chromVAR)
library(BSgenome.Hsapiens.UCSC.hg19)
library(JASPAR2016)
library(motifmatchr)
library(GenomicRanges)
library(irlba)
library(cicero)
library(umap)
library(cisTopic)
library(prabclus)
library(BrockmanR)
library(jackstraw)
library(RColorBrewer)
```
#### define functions
```
read_FM <- function(filename){
df_FM = data.frame(readRDS(filename),stringsAsFactors=FALSE,check.names=FALSE)
rownames(df_FM) <- make.names(rownames(df_FM), unique=TRUE)
df_FM[is.na(df_FM)] <- 0
return(df_FM)
}
run_pca <- function(mat,num_pcs=50,remove_first_PC=FALSE,scale=FALSE,center=FALSE){
set.seed(2019)
mat = as.matrix(mat)
SVD = irlba(mat, num_pcs, num_pcs,scale=scale,center=center)
sk_diag = matrix(0, nrow=num_pcs, ncol=num_pcs)
diag(sk_diag) = SVD$d
if(remove_first_PC){
sk_diag[1,1] = 0
SVD_vd = (sk_diag %*% t(SVD$v))[2:num_pcs,]
}else{
SVD_vd = sk_diag %*% t(SVD$v)
}
return(SVD_vd)
}
elbow_plot <- function(mat,num_pcs=50,scale=FALSE,center=FALSE,title='',width=3,height=3){
set.seed(2019)
mat = data.matrix(mat)
SVD = irlba(mat, num_pcs, num_pcs,scale=scale,center=center)
options(repr.plot.width=width, repr.plot.height=height)
df_plot = data.frame(PC=1:num_pcs, SD=SVD$d);
# print(SVD$d[1:num_pcs])
p <- ggplot(df_plot, aes(x = PC, y = SD)) +
geom_point(col="#cd5c5c",size = 1) +
ggtitle(title)
return(p)
}
run_umap <- function(fm_mat){
umap_object = umap(t(fm_mat),random_state = 2019)
df_umap = umap_object$layout
return(df_umap)
}
plot_umap <- function(df_umap,labels,title='UMAP',colormap=colormap){
set.seed(2019)
df_umap = data.frame(cbind(df_umap,labels),stringsAsFactors = FALSE)
colnames(df_umap) = c('umap1','umap2','celltype')
df_umap$umap1 = as.numeric(df_umap$umap1)
df_umap$umap2 = as.numeric(df_umap$umap2)
options(repr.plot.width=4, repr.plot.height=4)
p <- ggplot(shuf(df_umap), aes(x = umap1, y = umap2, color = celltype)) +
geom_point(size = 1) + scale_color_manual(values = colormap) +
ggtitle(title)
return(p)
}
```
### Input
```
workdir = '../output/'
path_umap = paste0(workdir,'umap_rds/')
system(paste0('mkdir -p ',path_umap))
path_fm = paste0(workdir,'feature_matrices/')
metadata <- read.table('../input/metadata.tsv',
header = TRUE,
stringsAsFactors=FALSE,quote="",row.names=1)
list.files(path_fm,pattern="^FM*")
# read in feature matrices and double check if cell names of feature matrices are consistent with metadata
flag_identical = c()
for (filename in list.files(path_fm,pattern="^FM*")){
filename_split = unlist(strsplit(sub('\\.rds$', '', filename),'_'))
method_i = filename_split[2]
if(method_i == 'chromVAR'){
method_i = paste(filename_split[2],filename_split[4],sep='_')
}
print(paste0('Read in ','fm_',method_i))
assign(paste0('fm_',method_i),read_FM(paste0(path_fm,filename)))
#check if column names are the same
flag_identical[[method_i]] = identical(colnames(eval(as.name(paste0('fm_',method_i)))),
rownames(metadata))
}
flag_identical
all(flag_identical)
labels = metadata$label
num_colors = length(unique(labels))
colormap = colorRampPalette(brewer.pal(8, "Dark2"))(num_colors)
names(colormap) = unique(metadata$label)
head(labels)
```
### SnapATAC
```
df_umap_SnapATAC <- run_umap(fm_SnapATAC)
head(df_umap_SnapATAC)
p_SnapATAC <- plot_umap(df_umap_SnapATAC,labels = labels,colormap = colormap,title='SnapATAC')
p_SnapATAC
```
### SCRAT
```
df_umap_SCRAT <- run_umap(fm_SCRAT)
p_SCRAT <- plot_umap(df_umap_SCRAT,labels = labels,colormap = colormap,title='SCRAT')
p_SCRAT
```
#### Save feature matrices and UMAP coordinates
```
dataset = 'cusanovich2018subset_no_blacklist_filtering'
saveRDS(df_umap_SnapATAC,paste0(path_umap,'df_umap_SnapATAC.rds'))
saveRDS(df_umap_SCRAT,paste0(path_umap,'df_umap_SCRAT.rds'))
save.image(file = 'run_umap_cusanovich2018subset_no_blacklist_filtering.RData')
fig_width = 8
fig_height = 4
options(repr.plot.width=fig_width, repr.plot.height=fig_height)
combined_fig = cowplot::plot_grid(p_SnapATAC+theme(legend.position = "none"),
p_SCRAT+theme(legend.position = "none"),
labels = "",nrow = 1)
combined_fig
cowplot::ggsave(combined_fig,filename = "Cusanovich_2018_ssubset_no_blacklist_filtering.pdf", width = fig_width, height = fig_height)
cowplot::ggsave(p_SCRAT ,filename = "cusanovich_legend.pdf", width = fig_width, height = fig_height)
```
|
github_jupyter
|
```
# default_exp learner
```
# Learner
> This contains fastai Learner extensions.
```
#export
from tsai.imports import *
from tsai.data.core import *
from tsai.data.validation import *
from tsai.models.all import *
from tsai.models.InceptionTimePlus import *
from fastai.learner import *
from fastai.vision.models.all import *
from fastai.data.transforms import *
#export
@patch
def show_batch(self:Learner, **kwargs):
self.dls.show_batch(**kwargs)
# export
@patch
def remove_all_cbs(self:Learner, max_iters=10):
i = 0
while len(self.cbs) > 0 and i < max_iters:
self.remove_cbs(self.cbs)
i += 1
if len(self.cbs) > 0: print(f'Learner still has {len(self.cbs)} callbacks: {self.cbs}')
#export
@patch
def one_batch(self:Learner, i, b): # this fixes a bug that will be managed in the next release of fastai
self.iter = i
# b_on_device = tuple( e.to(device=self.dls.device) for e in b if hasattr(e, "to")) if self.dls.device is not None else b
b_on_device = to_device(b, device=self.dls.device) if self.dls.device is not None else b
self._split(b_on_device)
self._with_events(self._do_one_batch, 'batch', CancelBatchException)
#export
@patch
def save_all(self:Learner, path='export', dls_fname='dls', model_fname='model', learner_fname='learner', verbose=False):
path = Path(path)
if not os.path.exists(path): os.makedirs(path)
self.dls_type = self.dls.__class__.__name__
if self.dls_type == "MixedDataLoaders":
self.n_loaders = (len(self.dls.loaders), len(self.dls.loaders[0].loaders))
dls_fnames = []
for i,dl in enumerate(self.dls.loaders):
for j,l in enumerate(dl.loaders):
l = l.new(num_workers=1)
torch.save(l, path/f'{dls_fname}_{i}_{j}.pth')
dls_fnames.append(f'{dls_fname}_{i}_{j}.pth')
else:
dls_fnames = []
self.n_loaders = len(self.dls.loaders)
for i,dl in enumerate(self.dls):
dl = dl.new(num_workers=1)
torch.save(dl, path/f'{dls_fname}_{i}.pth')
dls_fnames.append(f'{dls_fname}_{i}.pth')
# Saves the model along with optimizer
self.model_dir = path
self.save(f'{model_fname}', with_opt=True)
# Export learn without the items and the optimizer state for inference
self.export(path/f'{learner_fname}.pkl')
pv(f'Learner saved:', verbose)
pv(f"path = '{path}'", verbose)
pv(f"dls_fname = '{dls_fnames}'", verbose)
pv(f"model_fname = '{model_fname}.pth'", verbose)
pv(f"learner_fname = '{learner_fname}.pkl'", verbose)
def load_all(path='export', dls_fname='dls', model_fname='model', learner_fname='learner', device=None, pickle_module=pickle, verbose=False):
if isinstance(device, int): device = torch.device('cuda', device)
elif device is None: device = default_device()
if device == 'cpu': cpu = True
else: cpu = None
path = Path(path)
learn = load_learner(path/f'{learner_fname}.pkl', cpu=cpu, pickle_module=pickle_module)
learn.load(f'{model_fname}', with_opt=True, device=device)
if learn.dls_type == "MixedDataLoaders":
dls_fnames = []
_dls = []
for i in range(learn.n_loaders[0]):
_dl = []
for j in range(learn.n_loaders[1]):
l = torch.load(path/f'{dls_fname}_{i}_{j}.pth', map_location=device, pickle_module=pickle_module)
l = l.new(num_workers=0)
l.to(device)
dls_fnames.append(f'{dls_fname}_{i}_{j}.pth')
_dl.append(l)
_dls.append(MixedDataLoader(*_dl, path=learn.dls.path, device=device, shuffle=l.shuffle))
learn.dls = MixedDataLoaders(*_dls, path=learn.dls.path, device=device)
else:
loaders = []
dls_fnames = []
for i in range(learn.n_loaders):
dl = torch.load(path/f'{dls_fname}_{i}.pth', map_location=device, pickle_module=pickle_module)
dl = dl.new(num_workers=0)
dl.to(device)
first(dl)
loaders.append(dl)
dls_fnames.append(f'{dls_fname}_{i}.pth')
learn.dls = type(learn.dls)(*loaders, path=learn.dls.path, device=device)
pv(f'Learner loaded:', verbose)
pv(f"path = '{path}'", verbose)
pv(f"dls_fname = '{dls_fnames}'", verbose)
pv(f"model_fname = '{model_fname}.pth'", verbose)
pv(f"learner_fname = '{learner_fname}.pkl'", verbose)
return learn
load_learner_all = load_all
#export
@patch
@delegates(subplots)
def plot_metrics(self: Recorder, nrows=None, ncols=None, figsize=None, final_losses=True, perc=.5, **kwargs):
n_values = len(self.recorder.values)
if n_values < 2:
print('not enough values to plot a chart')
return
metrics = np.stack(self.values)
n_metrics = metrics.shape[1]
names = self.metric_names[1:n_metrics+1]
if final_losses:
sel_idxs = int(round(n_values * perc))
if sel_idxs >= 2:
metrics = np.concatenate((metrics[:,:2], metrics), -1)
names = names[:2] + names
else:
final_losses = False
n = len(names) - 1 - final_losses
if nrows is None and ncols is None:
nrows = int(math.sqrt(n))
ncols = int(np.ceil(n / nrows))
elif nrows is None: nrows = int(np.ceil(n / ncols))
elif ncols is None: ncols = int(np.ceil(n / nrows))
figsize = figsize or (ncols * 6, nrows * 4)
fig, axs = subplots(nrows, ncols, figsize=figsize, **kwargs)
axs = [ax if i < n else ax.set_axis_off() for i, ax in enumerate(axs.flatten())][:n]
axs = ([axs[0]]*2 + [axs[1]]*2 + axs[2:]) if final_losses else ([axs[0]]*2 + axs[1:])
for i, (name, ax) in enumerate(zip(names, axs)):
if i in [0, 1]:
ax.plot(metrics[:, i], color='#1f77b4' if i == 0 else '#ff7f0e', label='valid' if i == 1 else 'train')
ax.set_title('losses')
ax.set_xlim(0, len(metrics)-1)
elif i in [2, 3] and final_losses:
ax.plot(np.arange(len(metrics) - sel_idxs, len(metrics)), metrics[-sel_idxs:, i],
color='#1f77b4' if i == 2 else '#ff7f0e', label='valid' if i == 3 else 'train')
ax.set_title('final losses')
ax.set_xlim(len(metrics) - sel_idxs, len(metrics)-1)
# ax.set_xticks(np.arange(len(metrics) - sel_idxs, len(metrics)))
else:
ax.plot(metrics[:, i], color='#1f77b4' if i == 0 else '#ff7f0e', label='valid' if i > 0 else 'train')
ax.set_title(name if i >= 2 * (1 + final_losses) else 'losses')
ax.set_xlim(0, len(metrics)-1)
ax.legend(loc='best')
ax.grid(color='gainsboro', linewidth=.5)
plt.show()
@patch
@delegates(subplots)
def plot_metrics(self: Learner, **kwargs):
self.recorder.plot_metrics(**kwargs)
#export
@patch
@delegates(subplots)
def show_probas(self:Learner, figsize=(6,6), ds_idx=1, dl=None, one_batch=False, max_n=None, **kwargs):
recorder = copy(self.recorder) # This is to avoid loss of recorded values while generating preds
if one_batch: dl = self.dls.one_batch()
probas, targets = self.get_preds(ds_idx=ds_idx, dl=[dl] if dl is not None else None)
if probas.ndim == 2 and probas.min() < 0 or probas.max() > 1: probas = nn.Softmax(-1)(probas)
if not isinstance(targets[0].item(), Integral): return
targets = targets.flatten()
if max_n is not None:
idxs = np.random.choice(len(probas), max_n, False)
probas, targets = probas[idxs], targets[idxs]
fig = plt.figure(figsize=figsize, **kwargs)
classes = np.unique(targets)
nclasses = len(classes)
vals = np.linspace(.5, .5 + nclasses - 1, nclasses)[::-1]
plt.vlines(.5, min(vals) - 1, max(vals), color='black', linewidth=.5)
cm = plt.get_cmap('gist_rainbow')
color = [cm(1.* c/nclasses) for c in range(1, nclasses + 1)][::-1]
class_probas = np.array([probas[i,t] for i,t in enumerate(targets)])
for i, c in enumerate(classes):
plt.scatter(class_probas[targets == c] if nclasses > 2 or i > 0 else 1 - class_probas[targets == c],
targets[targets == c] + .5 * (np.random.rand((targets == c).sum()) - .5), color=color[i], edgecolor='black', alpha=.2, s=100)
if nclasses > 2: plt.vlines((targets == c).float().mean(), i - .5, i + .5, color='r', linewidth=.5)
plt.hlines(vals, 0, 1)
plt.ylim(min(vals) - 1, max(vals))
plt.xlim(0,1)
plt.xticks(np.linspace(0,1,11), fontsize=12)
plt.yticks(classes, [self.dls.vocab[x] for x in classes], fontsize=12)
plt.title('Predicted proba per true class' if nclasses > 2 else 'Predicted class 1 proba per true class', fontsize=14)
plt.xlabel('Probability', fontsize=12)
plt.ylabel('True class', fontsize=12)
plt.grid(axis='x', color='gainsboro', linewidth=.2)
plt.show()
self.recorder = recorder
#export
all_archs = [FCN, FCNPlus, InceptionTime, InceptionTimePlus, InCoordTime, XCoordTime,
InceptionTimePlus17x17, InceptionTimePlus32x32, InceptionTimePlus47x47, InceptionTimePlus62x62,
InceptionTimeXLPlus, MultiInceptionTimePlus, MiniRocketClassifier, MiniRocketRegressor,
MiniRocketVotingClassifier, MiniRocketVotingRegressor, MiniRocketFeaturesPlus, MiniRocketPlus, MiniRocketHead,
InceptionRocketFeaturesPlus, InceptionRocketPlus, MLP, MultiInputNet, OmniScaleCNN, RNN, LSTM, GRU, RNNPlus, LSTMPlus, GRUPlus,
RNN_FCN, LSTM_FCN, GRU_FCN, MRNN_FCN, MLSTM_FCN, MGRU_FCN, ROCKET, RocketClassifier, RocketRegressor, ResCNNBlock, ResCNN,
ResNet, ResNetPlus, TCN, TSPerceiver, TST, TSTPlus, MultiTSTPlus, TSiTPlus, TSiT, InceptionTSiTPlus, InceptionTSiT,
TabFusionTransformer, TSTabFusionTransformer, TabModel, TabTransformer, TransformerModel, XCM, XCMPlus,
xresnet1d18, xresnet1d34, xresnet1d50, xresnet1d101, xresnet1d152, xresnet1d18_deep,
xresnet1d34_deep, xresnet1d50_deep, xresnet1d18_deeper, xresnet1d34_deeper, xresnet1d50_deeper,
XResNet1dPlus, xresnet1d18plus, xresnet1d34plus, xresnet1d50plus, xresnet1d101plus,
xresnet1d152plus, xresnet1d18_deepplus, xresnet1d34_deepplus, xresnet1d50_deepplus,
xresnet1d18_deeperplus, xresnet1d34_deeperplus, xresnet1d50_deeperplus, XceptionTime, XceptionTimePlus
]
all_archs_names = [arch.__name__ for arch in all_archs]
def get_arch(arch_name):
assert arch_name in all_archs_names, "confirm the name of the architecture"
idx = all_archs_names.index(arch_name)
return all_archs[idx]
arch_name = 'InceptionTimePlus'
test_eq(get_arch('InceptionTimePlus').__name__, arch_name)
#export
@delegates(build_ts_model)
def ts_learner(dls, arch=None, c_in=None, c_out=None, seq_len=None, d=None, splitter=trainable_params,
# learner args
loss_func=None, opt_func=Adam, lr=defaults.lr, cbs=None, metrics=None, path=None,
model_dir='models', wd=None, wd_bn_bias=False, train_bn=True, moms=(0.95,0.85,0.95),
# other model args
**kwargs):
if arch is None: arch = InceptionTimePlus
elif isinstance(arch, str): arch = get_arch(arch)
model = build_ts_model(arch, dls=dls, c_in=c_in, c_out=c_out, seq_len=seq_len, d=d, **kwargs)
try:
model[0], model[1]
subscriptable = True
except:
subscriptable = False
if subscriptable: splitter = ts_splitter
if loss_func is None:
if hasattr(dls, 'loss_func'): loss_func = dls.loss_func
elif hasattr(dls, 'train_ds') and hasattr(dls.train_ds, 'loss_func'): loss_func = dls.train_ds.loss_func
elif hasattr(dls, 'cat') and not dls.cat: loss_func = MSELossFlat()
learn = Learner(dls=dls, model=model,
loss_func=loss_func, opt_func=opt_func, lr=lr, cbs=cbs, metrics=metrics, path=path, splitter=splitter,
model_dir=model_dir, wd=wd, wd_bn_bias=wd_bn_bias, train_bn=train_bn, moms=moms, )
# keep track of args for loggers
store_attr('arch', self=learn)
return learn
#export
@delegates(build_tsimage_model)
def tsimage_learner(dls, arch=None, pretrained=False,
# learner args
loss_func=None, opt_func=Adam, lr=defaults.lr, cbs=None, metrics=None, path=None,
model_dir='models', wd=None, wd_bn_bias=False, train_bn=True, moms=(0.95,0.85,0.95),
# other model args
**kwargs):
if arch is None: arch = xresnet34
elif isinstance(arch, str): arch = get_arch(arch)
model = build_tsimage_model(arch, dls=dls, pretrained=pretrained, **kwargs)
learn = Learner(dls=dls, model=model,
loss_func=loss_func, opt_func=opt_func, lr=lr, cbs=cbs, metrics=metrics, path=path,
model_dir=model_dir, wd=wd, wd_bn_bias=wd_bn_bias, train_bn=train_bn, moms=moms)
# keep track of args for loggers
store_attr('arch', self=learn)
return learn
#export
@patch
def decoder(self:Learner, o): return L([self.dls.decodes(oi) for oi in o])
# export
@patch
@delegates(GatherPredsCallback.__init__)
def get_X_preds(self: Learner, X, y=None, bs=64, with_input=False, with_decoded=True, with_loss=False, **kwargs):
if with_loss and y is None:
print('cannot find loss as y=None')
with_loss = False
dl = self.dls.valid.new_dl(X, y=y)
dl.bs = ifnone(bs, self.dls.bs)
output = list(self.get_preds(dl=dl, with_input=with_input, with_decoded=with_decoded, with_loss=with_loss, reorder=False))
if with_decoded and hasattr(self.dls, 'vocab'):
output[2 + with_input] = L([self.dls.vocab[p] for p in output[2 + with_input]])
return tuple(output)
from tsai.data.all import *
from tsai.data.core import *
from tsai.models.FCNPlus import *
dsid = 'OliveOil'
X, y, splits = get_UCR_data(dsid, verbose=True, split_data=False)
tfms = [None, [Categorize()]]
dls = get_ts_dls(X, y, splits=splits, tfms=tfms)
learn = ts_learner(dls, FCNPlus)
for p in learn.model.parameters():
p.requires_grad=False
test_eq(count_parameters(learn.model), 0)
learn.freeze()
test_eq(count_parameters(learn.model), 1540)
learn.unfreeze()
test_eq(count_parameters(learn.model), 264580)
learn = ts_learner(dls, 'FCNPlus')
for p in learn.model.parameters():
p.requires_grad=False
test_eq(count_parameters(learn.model), 0)
learn.freeze()
test_eq(count_parameters(learn.model), 1540)
learn.unfreeze()
test_eq(count_parameters(learn.model), 264580)
learn.show_batch();
learn.fit_one_cycle(2, lr_max=1e-3)
dsid = 'OliveOil'
X, y, splits = get_UCR_data(dsid, split_data=False)
tfms = [None, [Categorize()]]
dls = get_ts_dls(X, y, tfms=tfms, splits=splits)
learn = ts_learner(dls, FCNPlus, metrics=accuracy)
learn.fit_one_cycle(2)
learn.plot_metrics()
learn.show_probas()
learn.save_all()
del learn
learn = load_all()
test_probas, test_targets, test_preds = learn.get_X_preds(X[0:10], with_decoded=True)
test_probas, test_targets, test_preds
test_probas2, test_targets2, test_preds2 = learn.get_X_preds(X[0:10], y[0:10], with_decoded=True)
test_probas2, test_targets2, test_preds2
test_eq(test_probas, test_probas2)
test_eq(test_preds, test_preds2)
learn.fit_one_cycle(1, lr_max=1e-3)
#hide
from tsai.imports import create_scripts
from tsai.export import get_nb_name
nb_name = get_nb_name()
create_scripts(nb_name);
```
|
github_jupyter
|
# Experiment with variables of given high correlation structure
This notebook is meant to address to a shared concern from two referees. The [motivating example](motivating_example.html) in the manuscript was designed to be a simple toy for illustrating the novel type of inference SuSiE offers. Here are some slightly more complicated examples, based on the motivating example, but with variables in high (rather than perfect) correlations with each other.
## $x_1$ and $x_2$ are highly correlated
Following a reviewer's
suggestion, we simulated two variables, $x_1$ and $x_2$, with high but
not perfect correlation ($0.9$). Specifically, we simulated $n = 600$
samples stored as an $X_{600 \times 2}$ matrix, in which each row was
drawn *i.i.d.* from a normal distribution with mean zero and
$\mathrm{cor}(x_1, x_2) = 0.9$.
We then simulated $y_i = x_{i1} \beta_1 + x_{i2} \beta_2 + \varepsilon_i$,
with $\beta_1 = 1, \beta_2 = 0$,
and $\varepsilon_i$ *i.i.d.* normal with zero mean and standard
deviation of 3. We performed 1,000 replicates of this simulation
(generated with different random number seeds).
In this simulation, the correlation between $x_1$ and $x_2$ is still
sufficiently high (0.9) to make distinguishing between the two
variables somewhat possible, but not non entirely straightforward. For
example, when we run lasso (using `cv.glmnet` from the `glmnet`
R package) on these data it wrongly selected $x_2$ as having
non-zero coefficient in about 10% of the simulations (95 out of
1,000), and correctly selected $x_1$ in about 96% of simulations (956
out of 1,000). Note that the lasso does not assess uncertainty in
variable selection, so these results are not directly comparable
with SuSiE CSs below; however, the lasso results demonstrate that
distinguishing the correct variable here is possible, but not so easy
that the example is uninteresting.
Ideally, then, SuSiE should identify variable $x_1$ as an effect
variable and drop $x_2$ as often as possible. However, due to the high
correlation between the variables, it is inevitable that some
95% SuSiE credible sets (CS) will also contain $x_2$. Most
important is that we should avoid, as much as possible, reporting a CS
that contains *only* $x_2$, since the goal is that 95% of CSs
should contain at least one effect variable. The SuSiE results (SuSiE version 0.9.1 on R 3.5.2)
are summarized below. The code used for the simulation [can be found here](https://github.com/stephenslab/susie-paper/blob/master/src/ref_3_question.R).
| CSs | count |
| :---- | ----: |
| (1) | 829 |
| (1,2) | 169 |
| **(2)** | 2 |
Highlighted in **bold** are CSs that do *not* contain
the true effect variable --- there are 2 of them out of 1,000 CSs
detected. In summary, SuSiE precisely identifies the effect
variable in a single CS in the majority (83%) of the simulations, and
provides a "valid" CS (*i.e.*, one containing an effect
variable) in almost all simulations (998 out of 1,000). Further, even
when SuSiE reports a CS including both variables, it consistently
assigns higher posterior inclusion probability (PIP) to the correct
variable, $x_1$: among the 169 CSs that contain both variables, the
median PIPs for $x_1$ and $x_2$ were 0.86 and 0.14, respectively.
## When an additional non-effect variable highly correlated with both variable groups
Another referee suggested the following:
> Suppose
we have another predictor $x_5$, which is both correlated with $(x_1,
x_2)$ and $(x_3, x_4)$. Say $\mathrm{cor}(x_1, x_5) = 0.9$,
$\mathrm{cor}(x_2, x_5) = 0.7$, and $\mathrm{cor}(x_5, x_3)
= \mathrm{cor}(x_5, x_4) = 0.8$. Does the current method assign $x_5$
to the $(x_1, x_2)$ group or the $(x_3, x_4)$ group?
Following the suggestion, we simulated $x_1, \ldots, x_5$ from a
multivariate normal with zero mean and the covariance matrix
approximately as given in Table below. (Since this matrix is
not quite positive definite, in our R code we used `nearPD` from
the `Matrix` package to generate the nearest positive definite
matrix --- the entries of the resulting covariance matrix differ only
very slightly from those in Table below, with a maximum
absolute difference of 0.0025 between corresponding elements in the
two matrices).
| | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ |
| ------: | ------: | ------: | ------: | ------: | ------: |
| $x_1$ | 1.00 | 0.92 | 0.70 | 0.70 | 0.90 |
| $x_2$ | 0.92 | 1.00 | 0.70 | 0.70 | 0.70 |
| $x_3$ | 0.70 | 0.70 | 1.00 | 0.92 | 0.80 |
| $x_4$ | 0.70 | 0.70 | 0.92 | 1.00 | 0.80 |
| $x_5$ | 0.90 | 0.70 | 0.80 | 0.80 | 1.00 |
We simulated $n = 600$ samples from this
multivariate normal distribution, then we simulated $n = 600$
responses $y_i$ from the regression model $y_i = x_{i1} \beta_1 + \cdots x_{i5} \beta_5 + \varepsilon_i$,
with $\beta = (0, 1, 1, 0, 0)^T$, and $\varepsilon_i$ *i.i.d.* normal with zero mean and
standard deviation of 3. We repeated this simulation procedure 1,000
times with different random seeds, and each time we fit a SuSiE
model to the simulated data by running the IBSS algorithm. To
simplify the example, we ran the IBSS algorithm with $L = 2$, and
fixed the $\sigma_0^2 = 1$. Similar results were obtained when we used
larger values of $L$, and when $\sigma_0^2$ was estimated. For more
details on how the data were simulated and how the SuSiE models
were fitted to the data sets, [see this script](https://github.com/stephenslab/susie-paper/blob/master/src/ref_4_question.R).
Like the toy motivating example given in the paper, in this simulation
the first two predictors are strongly correlated with each other, so
it may be difficult to distinguish among them, and likewise for the
third and fourth predictors. The fifth predictor, which has no effect
on $y$, potentially complicates matters because it is also strongly
correlated with the other predictors. Despite this complication, our
basic goal remains the same: the Credible Sets inferred by SuSiE
should capture the true effects most of the time, while also
minimizing "false positive" CSs that do not contain any true
effects. (Further, each CS should, ideally, be as small as possible.)
Table below summarizes the results of these simulations:
the left-hand column gives a unique result (a combination of CSs), and
the right-hand column gives the number of times this unique result
occurred among the 1,000 simulations. The CS combinations are ordered
by the frequency of their occurrence in the simulations. We highlight
in **bold** CSs that do not contain a true effect.
| CSs | count |
| :------------- | ----: |
| (2), (3) | 551 |
| (2), (3,4) | 212 |
| (1,2), (3) | 176 |
| (1,2), (3,4) | 38 |
| (2), (3,4,5) | 9 |
| **(1)**, (3,4) | 3 |
| (2), **(4)** | 3 |
| (1,2), (3,4,5) | 2 |
| **(1)**, (3) | 1 |
| (1,2), **(4)** | 1 |
| (2), (3,5) | 1 |
| (3), (1,2,5) | 1 |
| (3), (1,2,3) | 1 |
| (3,4), (1,2,4) | 1 |
In the majority (551) of the simulations, SuSiE precisely identiied
the true effect variables, and no others. In most other cases,
SuSiE identified two CSs, each containing a correct effect variable, and
with one or more other variables included due to high correlation with
the true-effect variable. The referee asks specifically about how the
additional variable $x_5$ behaves in this example. In practice, $x_5$
was rarely included in a CS. In the few cases where $x_5$ *was*
included in a CS, the results were consistent with the simulation
setting; $x_5$ was included more frequently with $x_3$ and/or $x_4$
(12 times) rather than $x_2$ and/or $x_1$ (only once). In no
simulations did SuSiE form a large group that contains all five
predictors.
This example actually highlights the benefits of SuSiE compared to
alternative approaches (e.g., hierinf) that *first* cluster the
variables into groups based on the correlation structure, then test
the groups. As we pointed out in the manuscript, this alternative
approach (first cluster variables into groups, then test groups) would
work well in the toy example in the paper, but in general it requires
*ad hoc* decisions about how to cluster variables. In this more
complex example raised by the referee, it is far from clear how to
cluster the variables. SuSiE avoids this problem because there is
no pre-clustering of variables; instead, the SuSiE CSs are computed
directly from an (approximate) posterior distribution (which takes
into account how the variables $x$ are correlated with each other, as
well as their relationship with $y$).
|
github_jupyter
|
## Analyzing Hamlet
```
%load_ext autoreload
%autoreload 2
import src.data
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
from collections import OrderedDict
from IPython.display import display
pd.options.display.max_rows = 999
pd.options.display.max_columns = 999
pd.set_option("display.max_columns", None)
import itertools
file_names = {
'aurora':'2020-01-01-till-2021-02-24-aurora.csv',
'hamlet':'2020-01-01-till-2021-02-24-hamlet.csv',
'mercandia':'2020-01-01-till-2021-02-24-mercandia-iv.csv',
'tycho-brahe':'2020-01-01-till-2021-02-24-tycho-brahe.csv',
}
dfs = OrderedDict()
for ship_name, file_name in file_names.items():
file_path = os.path.join(src.data.path_ships,file_name)
reader = pd.read_csv(file_path, chunksize=1000, iterator=True) # Loading a small part of the data
dfs[ship_name] = next(reader)
for ship_name, df in dfs.items():
display(df.describe())
file_path = os.path.join(src.data.path_ships,file_names['aurora'])
reader = pd.read_csv(file_path, chunksize=1000000, iterator=True) # Loading a small part of the data
df_raw = next(reader)
df_raw.set_index('Tidpunkt [UTC]', inplace=True)
df_raw.index = pd.to_datetime(df_raw.index)
mask = df_raw['Fart över grund (kts)']>1
df = df_raw.loc[mask].copy()
df.hist(column='Kurs över grund (deg)', bins=1000)
mask = df_raw['Kurs över grund (deg)'] < 150
df_direction_1 = df.loc[mask]
df_direction_1.describe()
df_direction_1.plot(x='Longitud (deg)', y = 'Latitud (deg)', style='.', alpha=0.005)
deltas = []
for i in range(1,5):
sin_key = 'Sin EM%i ()' % i
cos_key = 'Cos EM%i ()' % i
delta_key = 'delta_%i' % i
deltas.append(delta_key)
df_direction_1[delta_key] = np.arctan2(df_direction_1[sin_key],df_direction_1[cos_key])
df_plot = df_direction_1.loc['2020-01-01 01:00':'2020-01-01 02:00']
df_plot.plot(y=['Kurs över grund (deg)','Stävad kurs (deg)'],style='.')
df_plot.plot(y='Fart över grund (kts)',style='.')
df_plot.plot(y=deltas,style='.')
df_direction_1.head()
(df_direction_1['Sin EM1 ()']**2 + df_direction_1['Cos EM1 ()']**2).hist()
df_direction_1.columns
descriptions = pd.Series(index = df_direction_1.columns.copy())
descriptions['Latitud (deg)'] = 'Latitud (deg) (WGS84?)'
descriptions['Longitud (deg)'] = 'Longitud (deg) (WGS84?)'
descriptions['Effekt DG Total (kW)'] = '?'
descriptions['Effekt EM Thruster Total (kW)'] = ''
descriptions['Sin EM1 ()'] = ''
descriptions['Sin EM2 ()'] = ''
descriptions['Sin EM3 ()'] = ''
descriptions['Sin EM4 ()'] = ''
descriptions['Cos EM1 ()'] = ''
descriptions['Cos EM2 ()'] = ''
descriptions['Cos EM3 ()'] = ''
descriptions['Cos EM4 ()'] = ''
descriptions['Fart över grund (kts)'] = 'GPS fart'
descriptions['Stävad kurs (deg)'] = 'Kompas kurs'
descriptions['Kurs över grund (deg)'] = 'GPS kurs'
descriptions['Effekt hotell Total (kW)'] = ''
descriptions['Effekt Consumption Total (kW)'] = ''
descriptions['Förbrukning GEN alla (kg/h)'] = '?'
descriptions['delta_1'] = 'Thruster angle 1'
descriptions['delta_2'] = 'Thruster angle 2'
descriptions['delta_3'] = 'Thruster angle 3'
descriptions['delta_4'] = 'Thruster angle 4'
df_numenclature = pd.DataFrame(descriptions, columns=['Description'])
df_numenclature
```
|
github_jupyter
|
```
import sys
sys.path.append(r'C:\Users\moallemie\EMAworkbench-master')
sys.path.append(r'C:\Users\moallemie\EM_analysis')
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from ema_workbench import load_results, ema_logging
from ema_workbench.em_framework.salib_samplers import get_SALib_problem
from SALib.analyze import morris
# Set up number of scenarios, outcome of interest, and number of parallel processors
sc = 500 # Specify the number of scenarios where the convergence in the SA indices occures
t = 2100
outcome_var = 'GWP per Capita Indicator' # Specify the outcome of interest for SA ranking verification
nprocess = 100
```
## Loading the model, uncertainities, and outcomes and generate experiments
```
# Here we only generate experiments for loading the necessary components.
#The actual results will be loaded in the next cell.
# Open Excel input data from the notebook directory before runnign the code in multi-processing.
# Close the folder where the results will be saved in multi-processing.
# This line must be at the beginning for multi processing.
if __name__ == '__main__':
ema_logging.log_to_stderr(ema_logging.INFO)
#The model must be imoorted as .py file in parallel processing.
from Model_init import vensimModel
from ema_workbench import (TimeSeriesOutcome,
perform_experiments,
RealParameter,
CategoricalParameter,
ema_logging,
save_results,
load_results)
directory = 'C:/Users/moallemie/EM_analysis/Model/'
df_unc = pd.read_excel(directory+'ScenarioFramework.xlsx', sheet_name='Uncertainties')
# 0.5/1.5 multiplication is added to previous Min/Max cells for parameters with Reference values 0
#or min/max manually set in the spreadsheet
df_unc['Min'] = df_unc['Min'] + df_unc['Reference'] * 0.75
df_unc['Max'] = df_unc['Max'] + df_unc['Reference'] * 1.25
# From the Scenario Framework (all uncertainties), filter only those top 20 sensitive uncertainties under each outcome
sa_dir='C:/Users/moallemie/EM_analysis/Data/'
mu_df = pd.read_csv(sa_dir+"MorrisIndices_{}_sc5000_t{}.csv".format(outcome_var, t))
mu_df.rename(columns={'Unnamed: 0': 'Uncertainty'}, inplace=True)
mu_df.sort_values(by=['mu_star'], ascending=False, inplace=True)
mu_df = mu_df.head(20)
mu_unc = mu_df['Uncertainty']
mu_unc_df = mu_unc.to_frame()
# Remove the rest of insensitive uncertainties from the Scenario Framework and update df_unc
keys = list(mu_unc_df.columns.values)
i1 = df_unc.set_index(keys).index
i2 = mu_unc_df.set_index(keys).index
df_unc2 = df_unc[i1.isin(i2)]
vensimModel.uncertainties = [RealParameter(row['Uncertainty'], row['Min'], row['Max']) for index, row in df_unc2.iterrows()]
df_out = pd.read_excel(directory+'ScenarioFramework.xlsx', sheet_name='Outcomes')
vensimModel.outcomes = [TimeSeriesOutcome(out) for out in df_out['Outcome']]
from ema_workbench import MultiprocessingEvaluator
from ema_workbench.em_framework.evaluators import (MC, LHS, FAST, FF, PFF, SOBOL, MORRIS)
import time
start = time.time()
with MultiprocessingEvaluator(vensimModel, n_processes=nprocess) as evaluator:
results = evaluator.perform_experiments(scenarios=sc, uncertainty_sampling=MORRIS)
end = time.time()
print("took {} seconds".format(end-start))
experiments, outcomes = results
r_dir = 'D:/moallemie/EM_analysis/Data/'
save_results(results, r_dir+"SDG_experiments_ranking_verification_{}_sc{}.tar.gz".format(outcome_var, sc))
```
## Calculating SA (Morris) metrics
```
# Morris mu_star index calculation as a function of number of scenarios and time
def make_morris_df(scores, problem, outcome_var, sc, t):
scores_filtered = {k:scores[k] for k in ['mu_star','mu_star_conf','mu','sigma']}
Si_df = pd.DataFrame(scores_filtered, index=problem['names'])
indices = Si_df[['mu_star','mu']]
errors = Si_df[['mu_star_conf','sigma']]
return indices, errors
problem = get_SALib_problem(vensimModel.uncertainties)
X = experiments.iloc[:, :-3].values
Y = outcomes[outcome_var][:,-1]
scores = morris.analyze(problem, X, Y, print_to_console=False)
inds, errs = make_morris_df(scores, problem, outcome_var, sc, t)
```
## Where to draw the line between important and not important?
```
'''
Modifed from Waterprogramming blog by Antonia Hadgimichael: https://github.com/antonia-had/SA_verification
The idea is that we create 2 additiopnal Sets (current SA samples are Set 1).
We can create a Set 2, using only the T most important factors from our Set 1 sample,
and fixing all other factors to their default values.
We can also create a Set 3, now fixing the T most important factors to defaults
and using the sampled values of all other factors from Set 1.
If we classified our important and unimportant factors correctly,
then the correlation coefficient between the model outputs of Set 2 and Set 1 should approximate 1
(since we’re fixing all factors that don’t matter),
and the correlation coefficient between outputs from Set 3 and Set 1 should approximate 0
(since the factors we sampled are inconsequential to the output).
'''
# Sort factors by importance
inds_mu = inds['mu_star'].reindex(df_unc2['Uncertainty']).values
factors_sorted = np.argsort(inds_mu)[::-1]
# Set up DataFrame of default values to use for experiment
nsamples = len(experiments.index)
defaultvalues = df_unc2['Reference'].values
X_defaults = np.tile(defaultvalues,(nsamples, 1))
# Create Set 1 from experiments
exp_T = experiments.drop(['scenario', 'policy', 'model'], axis=1).T.reindex(df_unc2['Uncertainty'])
exp_ordered = exp_T.T
X_Set1 = exp_ordered.values
# Create initial Sets 2 and 3
X_Set2 = np.copy(X_defaults)
X_Set3 = np.copy(X_Set1)
# Define a function to convert your Set 2 and Set 3 into experiments structure in the EMA Workbench
def SA_experiments_to_scenarios(experiments, model=None):
'''
"Slighlty modifed from the EMA Workbench"
This function transform a structured experiments array into a list
of Scenarios.
If model is provided, the uncertainties of the model are used.
Otherwise, it is assumed that all non-default columns are
uncertainties.
Parameters
----------
experiments : numpy structured array
a structured array containing experiments
model : ModelInstance, optional
Returns
-------
a list of Scenarios
'''
from ema_workbench import Scenario
# get the names of the uncertainties
uncertainties = [u.name for u in model.uncertainties]
# make list of of tuples of tuples
cases = []
cache = set()
for i in range(experiments.shape[0]):
case = {}
case_tuple = []
for uncertainty in uncertainties:
entry = experiments[uncertainty][i]
case[uncertainty] = entry
case_tuple.append(entry)
case_tuple = tuple(case_tuple)
cases.append(case)
cache.add((case_tuple))
scenarios = [Scenario(**entry) for entry in cases]
return scenarios
# Run the models for the top n factors in Set 2 and Set 3 and generate correlation figures
if __name__ == '__main__':
ema_logging.log_to_stderr(ema_logging.INFO)
#The model must be imoorted as .py file in parallel processing.
from Model_init import vensimModel
from ema_workbench import (TimeSeriesOutcome,
perform_experiments,
RealParameter,
CategoricalParameter,
ema_logging,
save_results,
load_results)
vensimModel.outcomes = [TimeSeriesOutcome(outcome_var)]
from ema_workbench import MultiprocessingEvaluator
coefficient_S1_S3 = 0.99
for f in range(1, len(factors_sorted)+1):
ntopfactors = f
if coefficient_S1_S3 >= 0.1:
for i in range(ntopfactors): #Loop through all important factors
X_Set2[:,factors_sorted[i]] = X_Set1[:,factors_sorted[i]] #Fix use samples for important
X_Set3[:,factors_sorted[i]] = X_defaults[:,factors_sorted[i]] #Fix important to defaults
X_Set2_exp = pd.DataFrame(data=X_Set2, columns=df_unc2['Uncertainty'].tolist())
X_Set3_exp = pd.DataFrame(data=X_Set3, columns=df_unc2['Uncertainty'].tolist())
scenarios_Set2 = SA_experiments_to_scenarios(X_Set2_exp, model=vensimModel)
scenarios_Set3 = SA_experiments_to_scenarios(X_Set3_exp, model=vensimModel)
#experiments_Set2, outcomes_Set2 = perform_experiments(vensimModel, scenarios_Set2)
#experiments_Set3, outcomes_Set3 = perform_experiments(vensimModel, scenarios_Set3)
with MultiprocessingEvaluator(vensimModel, n_processes=nprocess) as evaluator:
experiments_Set2, outcomes_Set2 = evaluator.perform_experiments(scenarios=scenarios_Set2)
experiments_Set3, outcomes_Set3 = evaluator.perform_experiments(scenarios=scenarios_Set3)
# Calculate coefficients of correlation
data_Set1 = Y
data_Set2 = outcomes_Set2[outcome_var][:,-1]
data_Set3 = outcomes_Set3[outcome_var][:,-1]
coefficient_S1_S2 = np.corrcoef(data_Set1,data_Set2)[0][1]
coefficient_S1_S3 = np.corrcoef(data_Set1,data_Set3)[0][1]
# Plot outputs and correlation
fig = plt.figure(figsize=(14,7))
ax1 = fig.add_subplot(1,2,1)
ax1.plot(data_Set1,data_Set1, color='#39566E')
ax1.scatter(data_Set1,data_Set2, color='#8DCCFC')
ax1.set_xlabel("Set 1",fontsize=14)
ax1.set_ylabel("Set 2",fontsize=14)
ax1.tick_params(axis='both', which='major', labelsize=10)
ax1.set_title('Set 1 vs Set 2 - ' + str(f) + ' top factors',fontsize=15)
ax1.text(0.05,0.95,'R= '+"{0:.3f}".format(coefficient_S1_S2),transform = ax1.transAxes,fontsize=16)
ax2 = fig.add_subplot(1,2,2)
ax2.plot(data_Set1,data_Set1, color='#39566E')
ax2.scatter(data_Set1,data_Set3, color='#FFE0D5')
ax2.set_xlabel("Set 1",fontsize=14)
ax2.set_ylabel("Set 3",fontsize=14)
ax2.tick_params(axis='both', which='major', labelsize=10)
ax2.set_title('Set 1 vs Set 3 - ' + str(f) + ' top factors',fontsize=15)
ax2.text(0.05,0.95,'R= '+"{0:.3f}".format(coefficient_S1_S3),transform = ax2.transAxes,fontsize=16)
plt.savefig('{}/{}_{}_topfactors.png'.format(r'C:/Users/moallemie/EM_analysis/Fig/sa_verification', outcome_var, str(f)))
plt.close()
```
|
github_jupyter
|
# Neural Transfer
## Input images
```
%matplotlib inline
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from PIL import Image
import matplotlib.pyplot as plt
import torchvision.transforms as transforms
import torchvision.models as models
import copy
np.random.seed(37)
torch.manual_seed(37)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
def get_device():
return torch.device('cuda' if torch.cuda.is_available() else 'cpu')
def get_image_size():
imsize = 512 if torch.cuda.is_available() else 128
return imsize
def get_loader():
image_size = get_image_size()
loader = transforms.Compose([
transforms.Resize((image_size, image_size)),
transforms.ToTensor()])
return loader
def get_unloader():
unloader = transforms.ToPILImage()
return unloader
def image_loader(image_name):
device = get_device()
image = Image.open(image_name)
# fake batch dimension required to fit network's input dimensions
loader = get_loader()
image = loader(image).unsqueeze(0)
return image.to(device, torch.float)
def imshow(tensor, title=None):
image = tensor.cpu().clone() # we clone the tensor to not do changes on it
image = image.squeeze(0) # remove the fake batch dimension
unloader = get_unloader()
image = unloader(image)
plt.imshow(image)
if title is not None:
plt.title(title)
plt.pause(0.001)
style_img = image_loader("./styles/picasso-01.jpg")
content_img = image_loader("./styles/dancing.jpg")
input_img = content_img.clone()
assert style_img.size() == content_img.size(), \
f'size mismatch, style {style_img.size()}, content {content_img.size()}'
plt.ion()
plt.figure()
imshow(input_img, title='Input Image')
plt.figure()
imshow(style_img, title='Style Image')
plt.figure()
imshow(content_img, title='Content Image')
```
## Loss functions
### Content loss
```
class ContentLoss(nn.Module):
def __init__(self, target,):
super(ContentLoss, self).__init__()
# we 'detach' the target content from the tree used
# to dynamically compute the gradient: this is a stated value,
# not a variable. Otherwise the forward method of the criterion
# will throw an error.
self.target = target.detach()
def forward(self, input):
self.loss = F.mse_loss(input, self.target)
return input
```
### Style loss
```
def gram_matrix(input):
a, b, c, d = input.size() # a=batch size(=1)
# b=number of feature maps
# (c,d)=dimensions of a f. map (N=c*d)
features = input.view(a * b, c * d) # resise F_XL into \hat F_XL
G = torch.mm(features, features.t()) # compute the gram product
# we 'normalize' the values of the gram matrix
# by dividing by the number of element in each feature maps.
return G.div(a * b * c * d)
class StyleLoss(nn.Module):
def __init__(self, target_feature):
super(StyleLoss, self).__init__()
self.target = gram_matrix(target_feature).detach()
def forward(self, input):
G = gram_matrix(input)
self.loss = F.mse_loss(G, self.target)
return input
```
## Model
```
device = get_device()
cnn = models.vgg19(pretrained=True).features.to(device).eval()
```
## Normalization
```
class Normalization(nn.Module):
def __init__(self, mean, std):
super(Normalization, self).__init__()
# .view the mean and std to make them [C x 1 x 1] so that they can
# directly work with image Tensor of shape [B x C x H x W].
# B is batch size. C is number of channels. H is height and W is width.
self.mean = torch.tensor(mean).view(-1, 1, 1)
self.std = torch.tensor(std).view(-1, 1, 1)
def forward(self, img):
# normalize img
return (img - self.mean) / self.std
cnn_normalization_mean = torch.tensor([0.485, 0.456, 0.406]).to(device)
cnn_normalization_std = torch.tensor([0.229, 0.224, 0.225]).to(device)
```
## Loss
```
content_layers_default = ['conv_4']
style_layers_default = ['conv_1', 'conv_2', 'conv_3', 'conv_4', 'conv_5']
def get_style_model_and_losses(cnn, normalization_mean, normalization_std,
style_img, content_img,
content_layers=content_layers_default,
style_layers=style_layers_default):
cnn = copy.deepcopy(cnn)
# normalization module
normalization = Normalization(normalization_mean, normalization_std).to(device)
# just in order to have an iterable access to or list of content/syle
# losses
content_losses = []
style_losses = []
# assuming that cnn is a nn.Sequential, so we make a new nn.Sequential
# to put in modules that are supposed to be activated sequentially
model = nn.Sequential(normalization)
i = 0 # increment every time we see a conv
for layer in cnn.children():
if isinstance(layer, nn.Conv2d):
i += 1
name = 'conv_{}'.format(i)
elif isinstance(layer, nn.ReLU):
name = 'relu_{}'.format(i)
# The in-place version doesn't play very nicely with the ContentLoss
# and StyleLoss we insert below. So we replace with out-of-place
# ones here.
layer = nn.ReLU(inplace=False)
elif isinstance(layer, nn.MaxPool2d):
name = 'pool_{}'.format(i)
elif isinstance(layer, nn.BatchNorm2d):
name = 'bn_{}'.format(i)
else:
raise RuntimeError('Unrecognized layer: {}'.format(layer.__class__.__name__))
model.add_module(name, layer)
if name in content_layers:
# add content loss:
target = model(content_img).detach()
content_loss = ContentLoss(target)
model.add_module("content_loss_{}".format(i), content_loss)
content_losses.append(content_loss)
if name in style_layers:
# add style loss:
target_feature = model(style_img).detach()
style_loss = StyleLoss(target_feature)
model.add_module("style_loss_{}".format(i), style_loss)
style_losses.append(style_loss)
# now we trim off the layers after the last content and style losses
for i in range(len(model) - 1, -1, -1):
if isinstance(model[i], ContentLoss) or isinstance(model[i], StyleLoss):
break
model = model[:(i + 1)]
return model, style_losses, content_losses
```
## Optimizer
```
def get_input_optimizer(input_img):
# this line to show that input is a parameter that requires a gradient
optimizer = optim.LBFGS([input_img.requires_grad_()])
return optimizer
```
## Transfer
```
import warnings
from collections import namedtuple
RESULTS = namedtuple('RESULTS', 'run style content')
results = []
def run_style_transfer(cnn, normalization_mean, normalization_std,
content_img, style_img, input_img, num_steps=600,
style_weight=1000000, content_weight=1):
model, style_losses, content_losses = get_style_model_and_losses(cnn,
normalization_mean, normalization_std, style_img, content_img)
optimizer = get_input_optimizer(input_img)
run = [0]
while run[0] <= num_steps:
def closure():
# correct the values of updated input image
input_img.data.clamp_(0, 1)
optimizer.zero_grad()
model(input_img)
style_score = 0
content_score = 0
for sl in style_losses:
style_score += sl.loss
for cl in content_losses:
content_score += cl.loss
style_score *= style_weight
content_score *= content_weight
loss = style_score + content_score
loss.backward()
run[0] += 1
results.append(RESULTS(run[0], style_score.item(), content_score.item()))
if run[0] % 10 == 0:
s_score = style_score.item()
c_score = content_score.item()
print(f'[{run[0]}/{num_steps}] Style Loss {s_score:.4f}, Content Loss {c_score}')
return style_score + content_score
optimizer.step(closure)
# a last correction...
input_img.data.clamp_(0, 1)
return input_img
with warnings.catch_warnings():
warnings.simplefilter('ignore')
output = run_style_transfer(cnn, cnn_normalization_mean, cnn_normalization_std,
content_img, style_img, input_img)
```
## Results
```
x = [r.run for r in results]
y1 = [r.style for r in results]
y2 = [r.content for r in results]
fig, ax1 = plt.subplots(figsize=(10, 5))
color = 'tab:red'
ax1.plot(x, y1, color=color)
ax1.set_ylabel('Style Loss', color=color)
ax1.tick_params(axis='y', labelcolor=color)
color = 'tab:blue'
ax2 = ax1.twinx()
ax2.plot(x, y2, color=color)
ax2.set_ylabel('Content Loss', color=color)
ax2.tick_params(axis='y', labelcolor=color)
```
## Visualize
```
plt.figure()
imshow(output, title='Output Image')
# sphinx_gallery_thumbnail_number = 4
plt.ioff()
plt.show()
```
|
github_jupyter
|
# Lab: Working with a real world data-set using SQL and Python
## Introduction
This notebook shows how to work with a real world dataset using SQL and Python. In this lab you will:
1. Understand the dataset for Chicago Public School level performance
1. Store the dataset in an Db2 database on IBM Cloud instance
1. Retrieve metadata about tables and columns and query data from mixed case columns
1. Solve example problems to practice your SQL skills including using built-in database functions
## Chicago Public Schools - Progress Report Cards (2011-2012)
The city of Chicago released a dataset showing all school level performance data used to create School Report Cards for the 2011-2012 school year. The dataset is available from the Chicago Data Portal: https://data.cityofchicago.org/Education/Chicago-Public-Schools-Progress-Report-Cards-2011-/9xs2-f89t
This dataset includes a large number of metrics. Start by familiarizing yourself with the types of metrics in the database: https://data.cityofchicago.org/api/assets/AAD41A13-BE8A-4E67-B1F5-86E711E09D5F?download=true
__NOTE__: Do not download the dataset directly from City of Chicago portal. Instead download a more database friendly version from the link below.
Now download a static copy of this database and review some of its contents:
https://ibm.box.com/shared/static/0g7kbanvn5l2gt2qu38ukooatnjqyuys.csv
### Store the dataset in a Table
In many cases the dataset to be analyzed is available as a .CSV (comma separated values) file, perhaps on the internet. To analyze the data using SQL, it first needs to be stored in the database.
While it is easier to read the dataset into a Pandas dataframe and then PERSIST it into the database as we saw in the previous lab, it results in mapping to default datatypes which may not be optimal for SQL querying. For example a long textual field may map to a CLOB instead of a VARCHAR.
Therefore, __it is highly recommended to manually load the table using the database console LOAD tool, as indicated in Week 2 Lab 1 Part II__. The only difference with that lab is that in Step 5 of the instructions you will need to click on create "(+) New Table" and specify the name of the table you want to create and then click "Next".
##### Now open the Db2 console, open the LOAD tool, Select / Drag the .CSV file for the CHICAGO PUBLIC SCHOOLS dataset and load the dataset into a new table called __SCHOOLS__.
<a href="https://cognitiveclass.ai"><img src = "https://ibm.box.com/shared/static/uc4xjh1uxcc78ks1i18v668simioz4es.jpg"></a>
### Connect to the database
Let us now load the ipython-sql extension and establish a connection with the database
```
%load_ext sql
# Enter the connection string for your Db2 on Cloud database instance below
# %sql ibm_db_sa://my-username:my-password@my-hostname:my-port/my-db-name
%sql ibm_db_sa://
```
### Query the database system catalog to retrieve table metadata
#### You can verify that the table creation was successful by retrieving the list of all tables in your schema and checking whether the SCHOOLS table was created
```
# type in your query to retrieve list of all tables in the database for your db2 schema (username)
#In Db2 the system catalog table called SYSCAT.TABLES contains the table metadata
%sql SELECT * from SYSCAT.TABLES where TABNAME = 'SCHOOLS'
#OR
%sql select TABSCHEMA, TABNAME, CREATE_TIME from SYSCAT.TABLES where TABSCHEMA='YOUR-DB2-USERNAME'
#OR
%sql select TABSCHEMA, TABNAME, CREATE_TIME from SYSCAT.TABLES \
where TABSCHEMA not in ('SYSIBM', 'SYSCAT', 'SYSSTAT', 'SYSIBMADM', 'SYSTOOLS', 'SYSPUBLIC')
```
### Query the database system catalog to retrieve column metadata
#### The SCHOOLS table contains a large number of columns. How many columns does this table have?
```
#In Db2 the system catalog table called SYSCAT.COLUMNS contains the column metadata
%sql SELECT COUNT(*) FROM SYSCAT.COLUMNS WHERE TABNAME = 'SCHOOLS'
#Correct answer: 78
```
Now retrieve the the list of columns in SCHOOLS table and their column type (datatype) and length.
```
%sql SELECT COLNAME, TYPENAME, LENGTH FROM SYSCAT.COLUMNS WHERE TABNAME = 'SCHOOLS'
#OR
%sql SELECT DISTINCT(NAME), COLTYPE, LENGTH FROM SYSIBM.SYSCOLUMNS WHERE TABNAME = 'SCHOOLS'
```
### Questions
1. Is the column name for the "SCHOOL ID" attribute in upper or mixed case?
1. What is the name of "Community Area Name" column in your table? Does it have spaces?
1. Are there any columns in whose names the spaces and paranthesis (round brackets) have been replaced by the underscore character "_"?
## Problems
### Problem 1
##### How many Elementary Schools are in the dataset?
```
%sql select count(*) from SCHOOLS where "Elementary, Middle, or High School" = 'ES'
#Correct answer: 462
```
### Problem 2
##### What is the highest Safety Score?
```
%sql SELECT MAX("Safety_Score") AS MAX_SAFETY FROM SCHOOLS
#Correct answer: 99
```
### Problem 3
##### Which schools have highest Safety Score?
```
%sql SELECT NAME_OF_SCHOOL FROM SCHOOLS WHERE "Safety_Score" = 99
#OR
%sql SELECT NAME_OF_SCHOOL FROM SCHOOLS WHERE "Safety_Score" = (SELECT MAX("Safety_Score") FROM SCHOOLS)
```
### Problem 4
##### What are the top 10 schools with the highest "Average Student Attendance"?
```
%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS ORDER BY Average_Student_Attendance DESC LIMIT 10
```
### Problem 5
#### Retrieve the list of 5 Schools with the lowest Average Student Attendance sorted in ascending order based on attendance
```
%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS ORDER BY Average_Student_Attendance LIMIT 5
```
### Problem 6
#### Now remove the '%' sign from the above result set for Average Student Attendance column
```
%sql SELECT NAME_OF_SCHOOL, REPLACE(Average_Student_Attendance, '%', '') FROM SCHOOLS ORDER BY Average_Student_Attendance LIMIT 5
```
### Problem 7
#### Which Schools have Average Student Attendance lower than 70%?
```
%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS WHERE CAST(REPLACE(Average_Student_Attendance, '%', '') AS DOUBLE) < 70
#OR
%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS WHERE DECIMAL(REPLACE(Average_Student_Attendance, '%', '')) < 70 ORDER BY Average_Student_Attendance
```
### Problem 8
#### Get the total College Enrollment for each Community Area
```
%sql SELECT COMMUNITY_AREA_NAME, SUM(COLLEGE_ENROLLMENT) AS TOTAL_ENROLLMENT FROM SCHOOLS GROUP BY COMMUNITY_AREA_NAME
```
### Problem 9
##### Get the 5 Community Areas with the least total College Enrollment sorted in ascending order
```
%sql SELECT COMMUNITY_AREA_NAME, SUM(COLLEGE_ENROLLMENT) AS TOTAL_ENROLLMENT FROM SCHOOLS GROUP BY COMMUNITY_AREA_NAME ORDER BY TOTAL_ENROLLMENT LIMIT 5
```
## Summary
#### In this lab you learned how to work with a real word dataset using SQL and Python. You learned how to query columns with spaces or special characters in their names and with mixed case names. You also used built in database functions and practiced how to sort, limit, and order result sets.
Copyright © 2018 [cognitiveclass.ai](cognitiveclass.ai?utm_source=bducopyrightlink&utm_medium=dswb&utm_campaign=bdu). This notebook and its source code are released under the terms of the [MIT License](https://bigdatauniversity.com/mit-license/).
|
github_jupyter
|
<center>
<img src="https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/Logos/organization_logo/organization_logo.png" width="300" alt="cognitiveclass.ai logo" />
</center>
# Access DB2 on Cloud using Python
Estimated time needed: **15** minutes
## Objectives
After completing this lab you will be able to:
- Create a table
- Insert data into the table
- Query data from the table
- Retrieve the result set into a pandas dataframe
- Close the database connection
**Notice:** Please follow the instructions given in the first Lab of this course to Create a database service instance of Db2 on Cloud.
## Task 1: Import the `ibm_db` Python library
The `ibm_db` [API ](https://pypi.python.org/pypi/ibm_db?cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ&cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ&cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ&cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ) provides a variety of useful Python functions for accessing and manipulating data in an IBM® data server database, including functions for connecting to a database, preparing and issuing SQL statements, fetching rows from result sets, calling stored procedures, committing and rolling back transactions, handling errors, and retrieving metadata.
We import the ibm_db library into our Python Application
```
import ibm_db
```
When the command above completes, the `ibm_db` library is loaded in your notebook.
## Task 2: Identify the database connection credentials
Connecting to dashDB or DB2 database requires the following information:
- Driver Name
- Database name
- Host DNS name or IP address
- Host port
- Connection protocol
- User ID
- User Password
**Notice:** To obtain credentials please refer to the instructions given in the first Lab of this course
Now enter your database credentials below
Replace the placeholder values in angular brackets <> below with your actual database credentials
e.g. replace "database" with "BLUDB"
```
#Replace the placeholder values with the actuals for your Db2 Service Credentials
dsn_driver = "{IBM DB2 ODBC DRIVER}"
dsn_database = "database" # e.g. "BLUDB"
dsn_hostname = "hostname" # e.g.: "dashdb-txn-sbox-yp-dal09-04.services.dal.bluemix.net"
dsn_port = "port" # e.g. "50000"
dsn_protocol = "protocol" # i.e. "TCPIP"
dsn_uid = "username" # e.g. "abc12345"
dsn_pwd = "password" # e.g. "7dBZ3wWt9XN6$o0J"
```
## Task 3: Create the database connection
Ibm_db API uses the IBM Data Server Driver for ODBC and CLI APIs to connect to IBM DB2 and Informix.
Create the database connection
```
#Create database connection
#DO NOT MODIFY THIS CELL. Just RUN it with Shift + Enter
dsn = (
"DRIVER={0};"
"DATABASE={1};"
"HOSTNAME={2};"
"PORT={3};"
"PROTOCOL={4};"
"UID={5};"
"PWD={6};").format(dsn_driver, dsn_database, dsn_hostname, dsn_port, dsn_protocol, dsn_uid, dsn_pwd)
try:
conn = ibm_db.connect(dsn, "", "")
print ("Connected to database: ", dsn_database, "as user: ", dsn_uid, "on host: ", dsn_hostname)
except:
print ("Unable to connect: ", ibm_db.conn_errormsg() )
```
## Task 4: Create a table in the database
In this step we will create a table in the database with following details:
<img src = "https://ibm.box.com/shared/static/ztd2cn4xkdoj5erlk4hhng39kbp63s1h.jpg" align="center">
```
#Lets first drop the table INSTRUCTOR in case it exists from a previous attempt
dropQuery = "drop table INSTRUCTOR"
#Now execute the drop statment
dropStmt = ibm_db.exec_immediate(conn, dropQuery)
```
## Dont worry if you get this error:
If you see an exception/error similar to the following, indicating that INSTRUCTOR is an undefined name, that's okay. It just implies that the INSTRUCTOR table does not exist in the table - which would be the case if you had not created it previously.
Exception: [IBM][CLI Driver][DB2/LINUXX8664] SQL0204N "ABC12345.INSTRUCTOR" is an undefined name. SQLSTATE=42704 SQLCODE=-204
```
#Construct the Create Table DDL statement
createQuery = "create table INSTRUCTOR(ID INTEGER PRIMARY KEY NOT NULL, FNAME VARCHAR(20), LNAME VARCHAR(20), CITY VARCHAR(20), CCODE CHAR(2))"
#Execute the statement
createStmt = ibm_db.exec_immediate(conn,createQuery)
```
<details><summary>Click here for the solution</summary>
```python
createQuery = "create table INSTRUCTOR(ID INTEGER PRIMARY KEY NOT NULL, FNAME VARCHAR(20), LNAME VARCHAR(20), CITY VARCHAR(20), CCODE CHAR(2))"
createStmt = ibm_db.exec_immediate(conn,createQuery)
```
</details>
## Task 5: Insert data into the table
In this step we will insert some rows of data into the table.
The INSTRUCTOR table we created in the previous step contains 3 rows of data:
<img src="https://ibm.box.com/shared/static/j5yjassxefrjknivfpekj7698dqe4d8i.jpg" align="center">
We will start by inserting just the first row of data, i.e. for instructor Rav Ahuja
```
#Construct the query
insertQuery = "insert into INSTRUCTOR values (1, 'Rav', 'Ahuja', 'TORONTO', 'CA')"
#execute the insert statement
insertStmt = ibm_db.exec_immediate(conn, insertQuery)
```
<details><summary>Click here for the solution</summary>
```python
insertQuery = "insert into INSTRUCTOR values (1, 'Rav', 'Ahuja', 'TORONTO', 'CA')"
insertStmt = ibm_db.exec_immediate(conn, insertQuery)
```
</details>
Now use a single query to insert the remaining two rows of data
```
#Inerts the remaining two rows of data
insertQuery2 = "insert into INSTRUCTOR values (2, 'Raul', 'Chong', 'Markham', 'CA'), (3, 'Hima', 'Vasudevan', 'Chicago', 'US')"
#execute the statement
insertStmt2 = ibm_db.exec_immediate(conn, insertQuery2)
```
<details><summary>Click here for the solution</summary>
```python
insertQuery2 = "insert into INSTRUCTOR values (2, 'Raul', 'Chong', 'Markham', 'CA'), (3, 'Hima', 'Vasudevan', 'Chicago', 'US')"
insertStmt2 = ibm_db.exec_immediate(conn, insertQuery2)
```
</details>
## Task 6: Query data in the table
In this step we will retrieve data we inserted into the INSTRUCTOR table.
```
#Construct the query that retrieves all rows from the INSTRUCTOR table
selectQuery = "select * from INSTRUCTOR"
#Execute the statement
selectStmt = ibm_db.exec_immediate(conn, selectQuery)
#Fetch the Dictionary (for the first row only)
ibm_db.fetch_both(selectStmt)
```
<details><summary>Click here for the solution</summary>
```python
#Construct the query that retrieves all rows from the INSTRUCTOR table
selectQuery = "select * from INSTRUCTOR"
#Execute the statement
selectStmt = ibm_db.exec_immediate(conn, selectQuery)
#Fetch the Dictionary (for the first row only)
ibm_db.fetch_both(selectStmt)
```
</details>
```
#Fetch the rest of the rows and print the ID and FNAME for those rows
while ibm_db.fetch_row(selectStmt) != False:
print (" ID:", ibm_db.result(selectStmt, 0), " FNAME:", ibm_db.result(selectStmt, "FNAME"))
```
<details><summary>Click here for the solution</summary>
```python
#Fetch the rest of the rows and print the ID and FNAME for those rows
while ibm_db.fetch_row(selectStmt) != False:
print (" ID:", ibm_db.result(selectStmt, 0), " FNAME:", ibm_db.result(selectStmt, "FNAME"))
```
</details>
Bonus: now write and execute an update statement that changes the Rav's CITY to MOOSETOWN
```
updateQuery = "update INSTRUCTOR set CITY='MOOSETOWN' where FNAME='Rav'"
updateStmt = ibm_db.exec_immediate(conn, updateQuery))
```
<details><summary>Click here for the solution</summary>
```python
updateQuery = "update INSTRUCTOR set CITY='MOOSETOWN' where FNAME='Rav'"
updateStmt = ibm_db.exec_immediate(conn, updateQuery))
```
</details>
## Task 7: Retrieve data into Pandas
In this step we will retrieve the contents of the INSTRUCTOR table into a Pandas dataframe
```
import pandas
import ibm_db_dbi
#connection for pandas
pconn = ibm_db_dbi.Connection(conn)
#query statement to retrieve all rows in INSTRUCTOR table
selectQuery = "select * from INSTRUCTOR"
#retrieve the query results into a pandas dataframe
pdf = pandas.read_sql(selectQuery, pconn)
#print just the LNAME for first row in the pandas data frame
pdf.LNAME[0]
#print the entire data frame
pdf
```
Once the data is in a Pandas dataframe, you can do the typical pandas operations on it.
For example you can use the shape method to see how many rows and columns are in the dataframe
```
pdf.shape
```
## Task 8: Close the Connection
We free all resources by closing the connection. Remember that it is always important to close connections so that we can avoid unused connections taking up resources.
```
ibm_db.close(conn)
```
## Summary
In this tutorial you established a connection to a database instance of DB2 Warehouse on Cloud from a Python notebook using ibm_db API. Then created a table and insert a few rows of data into it. Then queried the data. You also retrieved the data into a pandas dataframe.
## Author
<a href="https://www.linkedin.com/in/ravahuja/" target="_blank">Rav Ahuja</a>
## Change Log
| Date (YYYY-MM-DD) | Version | Changed By | Change Description |
| ----------------- | ------- | ---------- | ---------------------------------- |
| 2020-08-28 | 2.0 | Lavanya | Moved lab to course repo in GitLab |
<hr>
## <h3 align="center"> © IBM Corporation 2020. All rights reserved. <h3/>
|
github_jupyter
|
```
import folium
import branca
import geopandas
from folium.plugins import Search
print(folium.__version__)
```
Let's get some JSON data from the web - both a point layer and a polygon GeoJson dataset with some population data.
```
states = geopandas.read_file(
'https://rawcdn.githack.com/PublicaMundi/MappingAPI/master/data/geojson/us-states.json',
driver='GeoJSON'
)
cities = geopandas.read_file(
'https://d2ad6b4ur7yvpq.cloudfront.net/naturalearth-3.3.0/ne_50m_populated_places_simple.geojson',
driver='GeoJSON'
)
```
And take a look at what our data looks like:
```
states.describe()
```
Look how far the minimum and maximum values for the density are from the top and bottom quartile breakpoints! We have some outliers in our data that are well outside the meat of most of the distribution. Let's look into this to find the culprits within the sample.
```
states_sorted = states.sort_values(by='density', ascending=False)
states_sorted.head(5).append(states_sorted.tail(5))[['name','density']]
```
Looks like Washington D.C. and Alaska were the culprits on each end of the range. Washington was more dense than the next most dense state, New Jersey, than the least dense state, Alaska was from Wyoming, however. Washington D.C. has a has a relatively small land area for the amount of people that live there, so it makes sense that it's pretty dense. And Alaska has a lot of land area, but not much of it is habitable for humans.
<br><br>
However, we're looking at all of the states in the US to look at things on a more regional level. That high figure at the top of our range for Washington D.C. will really hinder the ability for us to differentiate between the other states, so let's account for that in the min and max values for our color scale, by getting the quantile values close to the end of the range. Anything higher or lower than those values will just fall into the 'highest' and 'lowest' bins for coloring.
```
min, max = states['density'].quantile([0.05,0.95]).apply(lambda x: round(x, 2))
mean = round(states['density'].mean(),2)
print(f"Min: {min}", f"Max: {max}", f"Mean: {mean}", sep="\n\n")
```
This looks better. Our min and max values for the colorscale are much closer to the mean value now. Let's run with these values, and make a colorscale. I'm just going to use a sequential light-to-dark color palette from the [ColorBrewer](http://colorbrewer2.org/#type=sequential&scheme=Purples&n=5).
```
colormap = branca.colormap.LinearColormap(
colors=['#f2f0f7','#cbc9e2','#9e9ac8','#756bb1','#54278f'],
index=states['density'].quantile([0.2,0.4,0.6,0.8]),
vmin=min,
vmax=max
)
colormap.caption="Population Density in the United States"
colormap
```
Let's narrow down these cities to United states cities, by using GeoPandas' spatial join functionality between two GeoDataFrame objects, using the Point 'within' Polygon functionality.
```
us_cities = geopandas.sjoin(cities, states, how='inner', op='within')
pop_ranked_cities = us_cities.sort_values(
by='pop_max',
ascending=False
)[
[
'nameascii',
'pop_max',
'geometry'
]
].iloc[:20]
```
Ok, now we have a new GeoDataFrame with our top 20 populated cities. Let's see the top 5.
```
pop_ranked_cities.head(5)
```
Alright, let's build a map!
```
m = folium.Map(location=[38,-97], zoom_start=4)
style_function = lambda x: {
'fillColor': colormap(x['properties']['density']),
'color': 'black',
'weight':2,
'fillOpacity':0.5
}
stategeo = folium.GeoJson(
states,
name='US States',
style_function=style_function,
tooltip=folium.GeoJsonTooltip(
fields=['name', 'density'],
aliases=['State', 'Density'],
localize=True
)
).add_to(m)
citygeo = folium.GeoJson(
pop_ranked_cities,
name='US Cities',
tooltip=folium.GeoJsonTooltip(
fields=['nameascii','pop_max'],
aliases=['','Population Max'],
localize=True)
).add_to(m)
statesearch = Search(
layer=stategeo,
geom_type='Polygon',
placeholder='Search for a US State',
collapsed=False,
search_label='name',
weight=3
).add_to(m)
citysearch = Search(
layer=citygeo,
geom_type='Point',
placeholder='Search for a US City',
collapsed=True,
search_label='nameascii'
).add_to(m)
folium.LayerControl().add_to(m)
colormap.add_to(m)
m
```
|
github_jupyter
|
# NumPy Tutorial: Data analysis with Python
[Source](https://www.dataquest.io/blog/numpy-tutorial-python/)
NumPy is a commonly used Python data analysis package. By using NumPy, you can speed up your workflow, and interface with other packages in the Python ecosystem, like scikit-learn, that use NumPy under the hood. NumPy was originally developed in the mid 2000s, and arose from an even older package called Numeric. This longevity means that almost every data analysis or machine learning package for Python leverages NumPy in some way.
In this tutorial, we'll walk through using NumPy to analyze data on wine quality. The data contains information on various attributes of wines, such as pH and fixed acidity, along with a quality score between 0 and 10 for each wine. The quality score is the average of at least 3 human taste testers. As we learn how to work with NumPy, we'll try to figure out more about the perceived quality of wine.
The wines we'll be analyzing are from the Minho region of Portugal.
The data was downloaded from the UCI Machine Learning Repository, and is available [here](https://archive.ics.uci.edu/ml/datasets/Wine+Quality). Here are the first few rows of the winequality-red.csv file, which we'll be using throughout this tutorial:
``` text
"fixed acidity";"volatile acidity";"citric acid";"residual sugar";"chlorides";"free sulfur dioxide";"total sulfur dioxide";"density";"pH";"sulphates";"alcohol";"quality"
7.4;0.7;0;1.9;0.076;11;34;0.9978;3.51;0.56;9.4;5
7.8;0.88;0;2.6;0.098;25;67;0.9968;3.2;0.68;9.8;5
```
The data is in what I'm going to call ssv (semicolon separated values) format -- each record is separated by a semicolon (;), and rows are separated by a new line. There are 1600 rows in the file, including a header row, and 12 columns.
Before we get started, a quick version note -- we'll be using Python 3.5. Our code examples will be done using Jupyter notebook.
If you want to jump right into a specific area, here are the topics:
* Creating an Array
* Reading Text Files
* Array Indexing
* N-Dimensional Arrays
* Data Types
* Array Math
* Array Methods
* Array Comparison and Filtering
* Reshaping and Combining Arrays
Lists Of Lists for CSV Data
Before using NumPy, we'll first try to work with the data using Python and the csv package. We can read in the file using the csv.reader object, which will allow us to read in and split up all the content from the ssv file.
In the below code, we:
* Import the csv library.
* Open the winequality-red.csv file.
* With the file open, create a new csv.reader object.
* Pass in the keyword argument delimiter=";" to make sure that the records are split up on the semicolon character instead of the default comma character.
* Call the list type to get all the rows from the file.
* Assign the result to wines.
```
import csv
with open("winequality-red.csv", 'r') as f:
wines = list(csv.reader(f, delimiter=";"))
# print(wines[:3])
headers = wines[0]
wines_only = wines[1:]
# print the headers
print(headers)
# print the 1st row of data
print(wines_only[0])
# print the 1st three rows of data
print(wines_only[:3])
```
The data has been read into a list of lists. Each inner list is a row from the ssv file. As you may have noticed, each item in the entire list of lists is represented as a string, which will make it harder to do computations.
As you can see from the table above, we've read in three rows, the first of which contains column headers. Each row after the header row represents a wine. The first element of each row is the fixed acidity, the second is the volatile acidity, and so on.
## Calculate Average Wine Quality
We can find the average quality of the wines. The below code will:
* Extract the last element from each row after the header row.
* Convert each extracted element to a float.
* Assign all the extracted elements to the list qualities.
* Divide the sum of all the elements in qualities by the total number of elements in qualities to the get the mean.
```
# calculate average wine quality with a loop
qualities = []
for row in wines[1:]:
qualities.append(float(row[-1]))
sum(qualities) / len(wines[1:])
# calculate average wine quality with a list comprehension
qualities = [float(row[-1]) for row in wines[1:]]
sum(qualities) / len(wines[1:])
```
Although we were able to do the calculation we wanted, the code is fairly complex, and it won't be fun to have to do something similar every time we want to compute a quantity. Luckily, we can use NumPy to make it easier to work with our data.
# Numpy 2-Dimensional Arrays
With NumPy, we work with multidimensional arrays. We'll dive into all of the possible types of multidimensional arrays later on, but for now, we'll focus on 2-dimensional arrays. A 2-dimensional array is also known as a matrix, and is something you should be familiar with. In fact, it's just a different way of thinking about a list of lists. A matrix has rows and columns. By specifying a row number and a column number, we're able to extract an element from a matrix.
If we picked the element at the first row and the second column, we'd get volatile acidity. If we picked the element in the third row and the second column, we'd get 0.88.
In a NumPy array, the number of dimensions is called the **rank**, and each dimension is called an **axis**. So
* the rows are the first axis
* the columns are the second axis
Now that you understand the basics of matrices, let's see how we can get from our list of lists to a NumPy array.
## Creating A NumPy Array
We can create a NumPy array using the numpy.array function. If we pass in a list of lists, it will automatically create a NumPy array with the same number of rows and columns. Because we want all of the elements in the array to be float elements for easy computation, we'll leave off the header row, which contains strings. One of the limitations of NumPy is that all the elements in an array have to be of the same type, so if we include the header row, all the elements in the array will be read in as strings. Because we want to be able to do computations like find the average quality of the wines, we need the elements to all be floats.
In the below code, we:
* Import the ```numpy``` package.
* Pass the ```list``` of lists wines into the array function, which converts it into a NumPy array.
* Exclude the header row with list slicing.
* Specify the keyword argument ```dtype``` to make sure each element is converted to a ```float```. We'll dive more into what the ```dtype``` is later on.
```
import numpy as np
np.set_printoptions(precision=2) # set the output print precision for readability
# create the numpy array skipping the headers
wines = np.array(wines[1:], dtype=np.float)
# If we display wines, we'll now get a NumPy array:
print(type(wines), wines)
# We can check the number of rows and columns in our data using the shape property of NumPy arrays:
wines.shape
```
## Alternative NumPy Array Creation Methods
There are a variety of methods that you can use to create NumPy arrays. It's useful to create an array with all zero elements in cases when you need an array of fixed size, but don't have any values for it yet. To start with, you can create an array where every element is zero. The below code will create an array with 3 rows and 4 columns, where every element is 0, using ```numpy.zeros```:
```
empty_array = np.zeros((3, 4))
empty_array
```
Creating arrays full of random numbers can be useful when you want to quickly test your code with sample arrays. You can also create an array where each element is a random number using ```numpy.random.rand```.
```
np.random.rand(2, 3)
```
### Using NumPy To Read In Files
It's possible to use NumPy to directly read ```csv``` or other files into arrays. We can do this using the ```numpy.genfromtxt``` function. We can use it to read in our initial data on red wines.
In the below code, we:
* Use the ``` genfromtxt ``` function to read in the ``` winequality-red.csv ``` file.
* Specify the keyword argument ``` delimiter=";" ``` so that the fields are parsed properly.
* Specify the keyword argument ``` skip_header=1 ``` so that the header row is skipped.
```
wines = np.genfromtxt("winequality-red.csv", delimiter=";", skip_header=1)
wines
```
Wines will end up looking the same as if we read it into a list then converted it to an array of ```floats```. NumPy will automatically pick a data type for the elements in an array based on their format.
## Indexing NumPy Arrays
We now know how to create arrays, but unless we can retrieve results from them, there isn't a lot we can do with NumPy. We can use array indexing to select individual elements, groups of elements, or entire rows and columns.
One important thing to keep in mind is that just like Python lists, NumPy is **zero-indexed**, meaning that:
* The index of the first row is 0
* The index of the first column is 0
* If we want to work with the fourth row, we'd use index 3
* If we want to work with the second row, we'd use index 1, and so on.
We'll again work with the wines array:
|||||||||||||
|-:|-:|-:|-:|-:|-:|-:|-:|-:|-:|-:|-:|
|7.4 |0.70 |0.00 |1.9 |0.076 |11 |34 |0.9978 |3.51 |0.56 |9.4 |5|
|7.8 |0.88 |0.00 |2.6 |0.098 |25 |67 |0.9968 |3.20 |0.68 |9.8 |5|
|7.8 |0.76 |0.04 |2.3 |0.092 |15 |54 |0.9970 |3.26 |0.65 |9.8 |5|
|11.2|0.28 |0.56 |1.9 |0.075 |17 |60 |0.9980 |3.16 |0.58 |9.8 |6|
|7.4 |0.70 |0.00 |1.9 |0.076 |11 |34 |0.9978 |3.51 |0.56 |9.4 |5|
Let's select the element at **row 3** and **column 4**.
We pass:
* 2 as the row index
* 3 as the column index.
This retrieves the value from the **third row** and **fourth column**
```
wines[2, 3]
wines[2][3]
```
Since we're working with a 2-dimensional array in NumPy we specify 2 indexes to retrieve an element.
* The first index is the row, or **axis 1**, index
* The second index is the column, or **axis 2**, index
Any element in wines can be retrieved using 2 indexes.
```
# rows 1, 2, 3 and column 4
wines[0:3, 3]
# all rows and column 3
wines[:, 2]
```
Just like with ```list``` slicing, it's possible to omit the 0 to just retrieve all the elements from the beginning up to element 3:
```
# rows 1, 2, 3 and column 4
wines[:3, 3]
```
We can select an entire column by specifying that we want all the elements, from the first to the last. We specify this by just using the colon ```:```, with no starting or ending indices. The below code will select the entire fourth column:
```
# all rows and column 4
wines[:, 3]
```
We selected an entire column above, but we can also extract an entire row:
```
# row 4 and all columns
wines[3, :]
```
If we take our indexing to the extreme, we can select the entire array using two colons to select all the rows and columns in wines. This is a great party trick, but doesn't have a lot of good applications:
```
wines[:, :]
```
## Assigning Values To NumPy Arrays
We can also use indexing to assign values to certain elements in arrays. We can do this by assigning directly to the indexed value:
```
# assign the value of 10 to the 2nd row and 6th column
print('Before', wines[1, 4:7])
wines[1, 5] = 10
print('After', wines[1, 4:7])
```
We can do the same for slices. To overwrite an entire column, we can do this:
```
# Overwrites all the values in the eleventh column with 50.
print('Before', wines[:, 9:12])
wines[:, 10] = 50
print('After', wines[:, 9:12])
```
## 1-Dimensional NumPy Arrays
So far, we've worked with 2-dimensional arrays, such as wines. However, NumPy is a package for working with multidimensional arrays.
One of the most common types of multidimensional arrays is the **1-dimensional array**, or **vector**. As you may have noticed above, when we sliced wines, we retrieved a 1-dimensional array.
* A 1-dimensional array only needs a single index to retrieve an element.
* Each row and column in a 2-dimensional array is a 1-dimensional array.
Just like a list of lists is analogous to a 2-dimensional array, a single list is analogous to a 1-dimensional array.
If we slice wines and only retrieve the third row, we get a 1-dimensional array:
```
third_wine = wines[3,:]
third_wine
```
We can retrieve individual elements from ```third_wine``` using a single index.
```
# display the second item in third_wine
third_wine[1]
```
Most NumPy functions that we've worked with, such as ```numpy.random.rand```, can be used with multidimensional arrays. Here's how we'd use ```numpy.random.rand``` to generate a random vector:
```
np.random.rand(3)
```
Previously, when we called ```np.random.rand```, we passed in a shape for a 2-dimensional array, so the result was a 2-dimensional array. This time, we passed in a shape for a single dimensional array. The shape specifies the number of dimensions, and the size of the array in each dimension.
A shape of ```(10,10)``` will be a 2-dimensional array with **10 rows** and **10 columns**. A shape of ```(10,)``` will be a **1-dimensional** array with **10 elements**.
Where NumPy gets more complex is when we start to deal with arrays that have more than 2 dimensions.
## N-Dimensional NumPy Arrays
This doesn't happen extremely often, but there are cases when you'll want to deal with arrays that have greater than 3 dimensions. One way to think of this is as a list of lists of lists. Let's say we want to store the monthly earnings of a store, but we want to be able to quickly lookup the results for a quarter, and for a year. The earnings for one year might look like this:
``` python
[500, 505, 490, 810, 450, 678, 234, 897, 430, 560, 1023, 640]
```
The store earned \$500 in January, \$505 in February, and so on. We can split up these earnings by quarter into a list of lists:
```
year_one = [
[500,505,490], # 1st quarter
[810,450,678], # 2nd quarter
[234,897,430], # 3rd quarter
[560,1023,640] # 4th quarter
]
```
We can retrieve the earnings from January by calling ``` year_one[0][0] ```. If we want the results for a whole quarter, we can call ``` year_one[0] ``` or ``` year_one[1] ```.
We now have a 2-dimensional array, or matrix. But what if we now want to add the results from another year? We have to add a third dimension:
```
earnings = [
[ # year 1
[500,505,490], # year 1, 1st quarter
[810,450,678], # year 1, 2nd quarter
[234,897,430], # year 1, 3rd quarter
[560,1023,640] # year 1, 4th quarter
],
[ # year =2
[600,605,490], # year 2, 1st quarter
[345,900,1000],# year 2, 2nd quarter
[780,730,710], # year 2, 3rd quarter
[670,540,324] # year 2, 4th quarter
]
]
```
We can retrieve the earnings from January of the first year by calling ``` earnings[0][0][0] ```.
We now need three indexes to retrieve a single element. A three-dimensional array in NumPy is much the same. In fact, we can convert earnings to an array and then get the earnings for January of the first year:
```
earnings = np.array(earnings)
# year 1, 1st quarter, 1st month (January)
earnings[0,0,0]
# year 2, 3rd quarter, 1st month (July)
earnings[1,2,0]
# we can also find the shape of the array
earnings.shape
```
Indexing and slicing work the exact same way with a 3-dimensional array, but now we have an extra axis to pass in. If we wanted to get the earnings for **January of all years**, we could do this:
```
# all years, 1st quarter, 1st month (January)
earnings[:,0,0]
```
If we wanted to get first quarter earnings from both years, we could do this:
```
# all years, 1st quarter, all months (January, February, March)
earnings[:,0,:]
```
Adding more dimensions can make it much easier to query your data if it's organized in a certain way. As we go from 3-dimensional arrays to 4-dimensional and larger arrays, the same properties apply, and they can be indexed and sliced in the same ways.
## NumPy Data Types
As we mentioned earlier, each NumPy array can store elements of a single data type. For example, wines contains only float values.
NumPy stores values using its own data types, **which are distinct from Python types** like ```float``` and ```str```.
This is because the core of NumPy is written in a programming language called ```C```, **which stores data differently than the Python data types**. NumPy data types map between Python and C, allowing us to use NumPy arrays without any conversion hitches.
You can find the data type of a NumPy array by accessing the dtype property:
```
wines.dtype
```
NumPy has several different data types, which mostly map to Python data types, like ```float```, and ```str```. You can find a full listing of NumPy data types [here](https://www.dataquest.io/blog/numpy-tutorial-python/), but here are a few important ones:
* ```float``` -- numeric floating point data.
* ```int``` -- integer data.
* ```string``` -- character data.
* ```object``` -- Python objects.
Data types additionally end with a suffix that indicates how many bits of memory they take up. So ```int32``` is a **32 bit integer data type**, and ```float64``` is a **64 bit float data type**.
### Converting Data Types
You can use the numpy.ndarray.astype method to convert an array to a different type. The method will actually **copy the array**, and **return a new array with the specified data type**.
For instance, we can convert wines to the ```int``` data type:
```
# convert wines to the int data type
wines.astype(int)
```
As you can see above, all of the items in the resulting array are integers. Note that we used the Python ```int``` type instead of a NumPy data type when converting wines. This is because several Python data types, including ```float```, ```int```, and ```string```, can be used with NumPy, and are automatically converted to NumPy data types.
We can check the name property of the ```dtype``` of the resulting array to see what data type NumPy mapped the resulting array to:
```
# convert to int
int_wines = wines.astype(int)
# check the data type
int_wines.dtype.name
```
The array has been converted to a **64-bit integer** data type. This allows for very long integer values, **but takes up more space in memory** than storing the values as 32-bit integers.
If you want more control over how the array is stored in memory, you can directly create NumPy dtype objects like ```numpy.int32```
```
np.int32
```
You can use these directly to convert between types:
```
# convert to a 64-bit integer
wines.astype(np.int64)
# convert to a 32-bit integer
wines.astype(np.int32)
# convert to a 16-bit integer
wines.astype(np.int16)
# convert to a 8-bit integer
wines.astype(np.int8)
```
## NumPy Array Operations
NumPy makes it simple to perform mathematical operations on arrays. This is one of the primary advantages of NumPy, and makes it quite easy to do computations.
### Single Array Math
If you do any of the basic mathematical operations ```/```, ```*```, ```-```, ```+```, ```^``` with an array and a value, it will apply the operation to each of the elements in the array.
Let's say we want to add 10 points to each quality score because we're feeling generous. Here's how we'd do that:
```
# add 10 points to the quality score
wines[:,-1] + 10
```
*Note: that the above operation won't change the wines array -- it will return a new 1-dimensional array where 10 has been added to each element in the quality column of wines.*
If we instead did ```+=```, we'd modify the array in place:
```
print('Before', wines[:,11])
# modify the data in place
wines[:,11] += 10
print('After', wines[:,11])
```
All the other operations work the same way. For example, if we want to multiply each of the quality score by 2, we could do it like this:
```
# multiply the quality score by 2
wines[:,11] * 2
```
### Multiple Array Math
It's also possible to do mathematical operations between arrays. This will apply the operation to pairs of elements. For example, if we add the quality column to itself, here's what we get:
```
# add the quality column to itself
wines[:,11] + wines[:,11]
```
Note that this is equivalent to ```wines[:,11] * 2``` -- this is because NumPy adds each pair of elements. The first element in the first array is added to the first element in the second array, the second to the second, and so on.
```
# add the quality column to itself
wines[:,11] * 2
```
We can also use this to multiply arrays. Let's say we want to pick a wine that maximizes alcohol content and quality. We'd multiply alcohol by quality, and select the wine with the highest score:
```
# multiply alcohol content by quality
alcohol_by_quality = wines[:,10] * wines[:,11]
print(alcohol_by_quality)
alcohol_by_quality.sort()
print(alcohol_by_quality, alcohol_by_quality[-1])
```
All of the common operations ```/```, ```*```, ```-```, ```+```, ```^``` will work between arrays.
## NumPy Array Methods
In addition to the common mathematical operations, NumPy also has several methods that you can use for more complex calculations on arrays. An example of this is the ```numpy.ndarray.sum``` method. This finds the sum of all the elements in an array by default:
```
# find the sum of all rows and the quality column
total = 0
for row in wines:
total += row[11]
print(total)
# find the sum of all rows and the quality column
wines[:,11].sum(axis=0)
# find the sum of the rows 1, 2, and 3 across all columns
totals = []
for i in range(3):
total = 0
for col in wines[i,:]:
total += col
totals.append(total)
print(totals)
# find the sum of the rows 1, 2, and 3 across all columns
wines[0:3,:].sum(axis=1)
```
We can pass the ```axis``` keyword argument into the sum method to find sums over an axis.
If we call sum across the wines matrix, and pass in ```axis=0```, we'll find the sums over the first axis of the array. This will give us the **sum of all the values in every column**.
This may seem backwards that the sums over the first axis would give us the sum of each column, but one way to think about this is that **the specified axis is the one "going away"**.
So if we specify ```axis=0```, we want the **rows to go away**, and we want to find **the sums for each of the remaining axes across each row**:
```
# sum each column for all rows
totals = [0] * len(wines[0])
for i, total in enumerate(totals):
for row_val in wines[:,i]:
total += row_val
totals[i] = total
print(totals)
# sum each column for all rows
wines.sum(axis=0)
```
We can verify that we did the sum correctly by checking the shape. The shape should be 12, corresponding to the number of columns:
```
wines.sum(axis=0).shape
```
If we pass in axis=1, we'll find the sums over the second axis of the array. This will give us the sum of each row:
```
# sum each row for all columns
totals = [0] * len(wines)
for i, total in enumerate(totals):
for col_val in wines[i,:]:
total += col_val
totals[i] = total
print(totals[0:3], '...', totals[-3:])
# sum each row for all columns
wines.sum(axis=1)
wines.sum(axis=1).shape
```
There are several other methods that behave like the sum method, including:
* ```numpy.ndarray.mean``` — finds the mean of an array.
* ```numpy.ndarray.std``` — finds the standard deviation of an array.
* ```numpy.ndarray.min``` — finds the minimum value in an array.
* ```numpy.ndarray.max``` — finds the maximum value in an array.
You can find a full list of array methods [here](http://docs.scipy.org/doc/numpy/reference/arrays.ndarray.html).
## NumPy Array Comparisons
NumPy makes it possible to test to see if rows match certain values using mathematical comparison operations like ```<```, ```>```, ```>=```, ```<=```, and ```==```. For example, if we want to see which wines have a quality rating higher than 5, we can do this:
```
# return True for all rows in the Quality column that are greater than 5
wines[:,11] > 5
```
We get a Boolean array that tells us which of the wines have a quality rating greater than 5. We can do something similar with the other operators. For instance, we can see if any wines have a quality rating equal to 10:
```
# return True for all rows that have a Quality rating of 10
wines[:,11] == 10
```
### Subsetting
One of the powerful things we can do with a Boolean array and a NumPy array is select only certain rows or columns in the NumPy array. For example, the below code will only select rows in wines where the quality is over 7:
```
# create a boolean array for wines with quality greater than 15
high_quality = wines[:,11] > 15
print(len(high_quality), high_quality)
# use boolean indexing to find high quality wines
high_quality_wines = wines[high_quality,:]
print(len(high_quality_wines), high_quality_wines)
```
We select only the rows where ```high_quality``` contains a ```True``` value, and all of the columns. This subsetting makes it simple to filter arrays for certain criteria.
For example, we can look for wines with a lot of alcohol and high quality. In order to specify multiple conditions, we have to place each condition in **parentheses** ```(...)```, and separate conditions with an **ampersand** ```&```:
```
# create a boolean array for high alcohol content and high quality
high_alcohol_and_quality = (wines[:,11] > 7) & (wines[:,10] > 10)
print(high_alcohol_and_quality)
# use boolean indexing to select out the wines
wines[high_alcohol_and_quality,:]
```
We can combine subsetting and assignment to overwrite certain values in an array:
```
high_alcohol_and_quality = (wines[:,10] > 10) & (wines[:,11] > 7)
wines[high_alcohol_and_quality,10:] = 20
```
## Reshaping NumPy Arrays
We can change the shape of arrays while still preserving all of their elements. This often can make it easier to access array elements. The simplest reshaping is to flip the axes, so rows become columns, and vice versa. We can accomplish this with the ```numpy.transpose``` function:
```
np.transpose(wines).shape
```
We can use the ```numpy.ravel``` function to turn an array into a one-dimensional representation. It will essentially flatten an array into a long sequence of values:
```
wines.ravel()
```
Here's an example where we can see the ordering of ```numpy.ravel```:
```
array_one = np.array(
[
[1, 2, 3, 4],
[5, 6, 7, 8]
]
)
array_one.ravel()
```
Finally, we can use the numpy.reshape function to reshape an array to a certain shape we specify. The below code will turn the second row of wines into a 2-dimensional array with 2 rows and 6 columns:
```
# print the current shape of the 2nd row and all columns
wines[1,:].shape
# reshape the 2nd row to a 2 by 6 matrix
wines[1,:].reshape((2,6))
```
## Combining NumPy Arrays
With NumPy, it's very common to combine multiple arrays into a single unified array. We can use ```numpy.vstack``` to vertically stack multiple arrays.
Think of it like the second arrays's items being added as new rows to the first array. We can read in the ```winequality-white.csv``` dataset that contains information on the quality of white wines, then combine it with our existing dataset, wines, which contains information on red wines.
In the below code, we:
* Read in ```winequality-white.csv```.
* Display the shape of white_wines.
```
white_wines = np.genfromtxt("winequality-white.csv", delimiter=";", skip_header=1)
white_wines.shape
```
As you can see, we have attributes for 4898 wines. Now that we have the white wines data, we can combine all the wine data.
In the below code, we:
* Use the ```vstack``` function to combine wines and white_wines.
* Display the shape of the result.
```
all_wines = np.vstack((wines, white_wines))
all_wines.shape
```
As you can see, the result has 6497 rows, which is the sum of the number of rows in wines and the number of rows in red_wines.
If we want to combine arrays horizontally, where the number of rows stay constant, but the columns are joined, then we can use the ```numpy.hstack``` function. The arrays we combine need to have the same number of rows for this to work.
Finally, we can use ```numpy.concatenate``` as a general purpose version of ```hstack``` and ```vstack```. If we want to concatenate two arrays, we pass them into concatenate, then specify the axis keyword argument that we want to concatenate along.
* Concatenating along the first axis is similar to ```vstack```
* Concatenating along the second axis is similar to ```hstack```:
```
x = np.concatenate((wines, white_wines), axis=0)
print(x.shape, x)
```
## Broadcasting
Unless the arrays that you're operating on are the exact same size, it's not possible to do elementwise operations. In cases like this, NumPy performs broadcasting to try to match up elements. Essentially, broadcasting involves a few steps:
* The last dimension of each array is compared.
* If the dimension lengths are equal, or one of the dimensions is of length 1, then we keep going.
* If the dimension lengths aren't equal, and none of the dimensions have length 1, then there's an error.
* Continue checking dimensions until the shortest array is out of dimensions.
For example, the following two shapes are compatible:
``` python
A: (50,3)
B (3,)
```
This is because the length of the trailing dimension of array A is 3, and the length of the trailing dimension of array B is 3. They're equal, so that dimension is okay. Array B is then out of elements, so we're okay, and the arrays are compatible for mathematical operations.
The following two shapes are also compatible:
``` python
A: (1,2)
B (50,2)
```
The last dimension matches, and A is of length 1 in the first dimension.
These two arrays don't match:
``` python
A: (50,50)
B: (49,49)
```
The lengths of the dimensions aren't equal, and neither array has either dimension length equal to 1.
There's a detailed explanation of broadcasting [here](http://docs.scipy.org/doc/numpy/user/basics.broadcasting.html), but we'll go through a few examples to illustrate the principle:
```
wines * np.array([1,2])
```
The above example didn't work because the two arrays don't have a matching trailing dimension. Here's an example where the last dimension does match:
```
array_one = np.array(
[
[1,2],
[3,4]
]
)
array_two = np.array([4,5])
array_one + array_two
```
As you can see, array_two has been broadcasted across each row of array_one. Here's an example with our wines data:
```
rand_array = np.random.rand(12)
wines + rand_array
```
|
github_jupyter
|
```
%matplotlib inline
import numpy as np
import yt
```
This notebook shows how to use yt to make plots and examine FITS X-ray images and events files.
## Sloshing, Shocks, and Bubbles in Abell 2052
This example uses data provided by [Scott Randall](http://hea-www.cfa.harvard.edu/~srandall/), presented originally in [Blanton, E.L., Randall, S.W., Clarke, T.E., et al. 2011, ApJ, 737, 99](https://ui.adsabs.harvard.edu/abs/2011ApJ...737...99B). They consist of two files, a "flux map" in counts/s/pixel between 0.3 and 2 keV, and a spectroscopic temperature map in keV.
```
ds = yt.load(
"xray_fits/A2052_merged_0.3-2_match-core_tmap_bgecorr.fits",
auxiliary_files=["xray_fits/A2052_core_tmap_b1_m2000_.fits"],
)
```
Since the flux and projected temperature images are in two different files, we had to use one of them (in this case the "flux" file) as a master file, and pass in the "temperature" file with the `auxiliary_files` keyword to `load`.
Next, let's derive some new fields for the number of counts, the "pseudo-pressure", and the "pseudo-entropy":
```
def _counts(field, data):
exposure_time = data.get_field_parameter("exposure_time")
return data["fits", "flux"] * data["fits", "pixel"] * exposure_time
ds.add_field(
("gas", "counts"),
function=_counts,
sampling_type="cell",
units="counts",
take_log=False,
)
def _pp(field, data):
return np.sqrt(data["gas", "counts"]) * data["fits", "projected_temperature"]
ds.add_field(
("gas", "pseudo_pressure"),
function=_pp,
sampling_type="cell",
units="sqrt(counts)*keV",
take_log=False,
)
def _pe(field, data):
return data["fits", "projected_temperature"] * data["gas", "counts"] ** (-1.0 / 3.0)
ds.add_field(
("gas", "pseudo_entropy"),
function=_pe,
sampling_type="cell",
units="keV*(counts)**(-1/3)",
take_log=False,
)
```
Here, we're deriving a "counts" field from the "flux" field by passing it a `field_parameter` for the exposure time of the time and multiplying by the pixel scale. Second, we use the fact that the surface brightness is strongly dependent on density ($S_X \propto \rho^2$) to use the counts in each pixel as a "stand-in". Next, we'll grab the exposure time from the primary FITS header of the flux file and create a `YTQuantity` from it, to be used as a `field_parameter`:
```
exposure_time = ds.quan(ds.primary_header["exposure"], "s")
```
Now, we can make the `SlicePlot` object of the fields we want, passing in the `exposure_time` as a `field_parameter`. We'll also set the width of the image to 250 pixels.
```
slc = yt.SlicePlot(
ds,
"z",
[
("fits", "flux"),
("fits", "projected_temperature"),
("gas", "pseudo_pressure"),
("gas", "pseudo_entropy"),
],
origin="native",
field_parameters={"exposure_time": exposure_time},
)
slc.set_log(("fits", "flux"), True)
slc.set_log(("gas", "pseudo_pressure"), False)
slc.set_log(("gas", "pseudo_entropy"), False)
slc.set_width(250.0)
slc.show()
```
To add the celestial coordinates to the image, we can use `PlotWindowWCS`, if you have a recent version of AstroPy (>= 1.3) installed:
```
from yt.frontends.fits.misc import PlotWindowWCS
wcs_slc = PlotWindowWCS(slc)
wcs_slc.show()
```
We can make use of yt's facilities for profile plotting as well.
```
v, c = ds.find_max(("fits", "flux")) # Find the maximum flux and its center
my_sphere = ds.sphere(c, (100.0, "code_length")) # Radius of 150 pixels
my_sphere.set_field_parameter("exposure_time", exposure_time)
```
Such as a radial profile plot:
```
radial_profile = yt.ProfilePlot(
my_sphere,
"radius",
["counts", "pseudo_pressure", "pseudo_entropy"],
n_bins=30,
weight_field="ones",
)
radial_profile.set_log("counts", True)
radial_profile.set_log("pseudo_pressure", True)
radial_profile.set_log("pseudo_entropy", True)
radial_profile.set_xlim(3, 100.0)
radial_profile.show()
```
Or a phase plot:
```
phase_plot = yt.PhasePlot(
my_sphere, "pseudo_pressure", "pseudo_entropy", ["counts"], weight_field=None
)
phase_plot.show()
```
Finally, we can also take an existing [ds9](http://ds9.si.edu/site/Home.html) region and use it to create a "cut region", using `ds9_region` (the [pyregion](https://pyregion.readthedocs.io) package needs to be installed for this):
```
from yt.frontends.fits.misc import ds9_region
reg_file = [
"# Region file format: DS9 version 4.1\n",
"global color=green dashlist=8 3 width=3 include=1 source=1 fk5\n",
'circle(15:16:44.817,+7:01:19.62,34.6256")',
]
f = open("circle.reg", "w")
f.writelines(reg_file)
f.close()
circle_reg = ds9_region(
ds, "circle.reg", field_parameters={"exposure_time": exposure_time}
)
```
This region may now be used to compute derived quantities:
```
print(
circle_reg.quantities.weighted_average_quantity("projected_temperature", "counts")
)
```
Or used in projections:
```
prj = yt.ProjectionPlot(
ds,
"z",
[
("fits", "flux"),
("fits", "projected_temperature"),
("gas", "pseudo_pressure"),
("gas", "pseudo_entropy"),
],
origin="native",
field_parameters={"exposure_time": exposure_time},
data_source=circle_reg,
method="sum",
)
prj.set_log(("fits", "flux"), True)
prj.set_log(("gas", "pseudo_pressure"), False)
prj.set_log(("gas", "pseudo_entropy"), False)
prj.set_width(250.0)
prj.show()
```
## The Bullet Cluster
This example uses an events table file from a ~100 ks exposure of the "Bullet Cluster" from the [Chandra Data Archive](http://cxc.harvard.edu/cda/). In this case, the individual photon events are treated as particle fields in yt. However, you can make images of the object in different energy bands using the `setup_counts_fields` function.
```
from yt.frontends.fits.api import setup_counts_fields
```
`load` will handle the events file as FITS image files, and will set up a grid using the WCS information in the file. Optionally, the events may be reblocked to a new resolution. by setting the `"reblock"` parameter in the `parameters` dictionary in `load`. `"reblock"` must be a power of 2.
```
ds2 = yt.load("xray_fits/acisf05356N003_evt2.fits.gz", parameters={"reblock": 2})
```
`setup_counts_fields` will take a list of energy bounds (emin, emax) in keV and create a new field from each where the photons in that energy range will be deposited onto the image grid.
```
ebounds = [(0.1, 2.0), (2.0, 5.0)]
setup_counts_fields(ds2, ebounds)
```
The "x", "y", "energy", and "time" fields in the events table are loaded as particle fields. Each one has a name given by "event\_" plus the name of the field:
```
dd = ds2.all_data()
print(dd["io", "event_x"])
print(dd["io", "event_y"])
```
Now, we'll make a plot of the two counts fields we made, and pan and zoom to the bullet:
```
slc = yt.SlicePlot(
ds2, "z", [("gas", "counts_0.1-2.0"), ("gas", "counts_2.0-5.0")], origin="native"
)
slc.pan((100.0, 100.0))
slc.set_width(500.0)
slc.show()
```
The counts fields can take the field parameter `"sigma"` and use [AstroPy's convolution routines](https://astropy.readthedocs.io/en/latest/convolution/) to smooth the data with a Gaussian:
```
slc = yt.SlicePlot(
ds2,
"z",
[("gas", "counts_0.1-2.0"), ("gas", "counts_2.0-5.0")],
origin="native",
field_parameters={"sigma": 2.0},
) # This value is in pixel scale
slc.pan((100.0, 100.0))
slc.set_width(500.0)
slc.set_zlim(("gas", "counts_0.1-2.0"), 0.01, 100.0)
slc.set_zlim(("gas", "counts_2.0-5.0"), 0.01, 50.0)
slc.show()
```
|
github_jupyter
|
tobac example: Tracking deep convection based on OLR from geostationary satellite retrievals
==
This example notebook demonstrates the use of tobac to track isolated deep convective clouds based on outgoing longwave radiation (OLR) calculated based on a combination of two different channels of the GOES-13 imaging instrument.
The data used in this example is downloaded from "zenodo link" automatically as part of the notebooks (This only has to be done once for all the tobac example notebooks).
```
# Import libraries:
import iris
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
import iris.plot as iplt
import iris.quickplot as qplt
import zipfile
from six.moves import urllib
from glob import glob
%matplotlib inline
# Import tobac itself:
import tobac
# Disable a few warnings:
import warnings
warnings.filterwarnings('ignore', category=UserWarning, append=True)
warnings.filterwarnings('ignore', category=RuntimeWarning, append=True)
warnings.filterwarnings('ignore', category=FutureWarning, append=True)
warnings.filterwarnings('ignore',category=pd.io.pytables.PerformanceWarning)
```
**Download example data:**
This has to be done only once for all tobac examples.
```
data_out='../'
# # Download the data: This only has to be done once for all tobac examples and can take a while
# file_path='https://zenodo.org/record/3195910/files/climate-processes/tobac_example_data-v1.0.1.zip'
# tempfile='temp.zip'
# print('start downloading data')
# request=urllib.request.urlretrieve(file_path,tempfile)
# print('start extracting data')
# zf = zipfile.ZipFile(tempfile)
# zf.extractall(data_out)
# print('example data saved in')
```
**Load data:**
```
data_file=os.path.join(data_out,'*','data','Example_input_OLR_satellite.nc')
data_file = glob(data_file)[0]
print(data_file)
# Load Data from downloaded file:
OLR=iris.load_cube(data_file,'OLR')
# Display information about the input data cube:
display(OLR)
#Set up directory to save output and plots:
savedir='Save'
if not os.path.exists(savedir):
os.makedirs(savedir)
plot_dir="Plot"
if not os.path.exists(plot_dir):
os.makedirs(plot_dir)
```
**Feature identification:**
Identify features based on OLR field and a set of threshold values
```
# Determine temporal and spatial sampling of the input data:
dxy,dt=tobac.get_spacings(OLR,grid_spacing=4000)
# Keyword arguments for the feature detection step
parameters_features={}
parameters_features['position_threshold']='weighted_diff'
parameters_features['sigma_threshold']=0.5
parameters_features['min_num']=4
parameters_features['target']='minimum'
parameters_features['threshold']=[250,225,200,175,150]
# Feature detection and save results to file:
print('starting feature detection')
Features=tobac.feature_detection_multithreshold(OLR,dxy,**parameters_features)
Features.to_hdf(os.path.join(savedir,'Features.h5'),'table')
print('feature detection performed and saved')
```
**Segmentation:**
Segmentation is performed based on the OLR field and a threshold value to determine the cloud areas.
```
# Keyword arguments for the segmentation step:
parameters_segmentation={}
parameters_segmentation['target']='minimum'
parameters_segmentation['method']='watershed'
parameters_segmentation['threshold']=250
# Perform segmentation and save results to files:
Mask_OLR,Features_OLR=tobac.segmentation_2D(Features,OLR,dxy,**parameters_segmentation)
print('segmentation OLR performed, start saving results to files')
iris.save([Mask_OLR],os.path.join(savedir,'Mask_Segmentation_OLR.nc'),zlib=True,complevel=4)
Features_OLR.to_hdf(os.path.join(savedir,'Features_OLR.h5'),'table')
print('segmentation OLR performed and saved')
```
**Trajectory linking:**
The detected features are linked into cloud trajectories using the trackpy library (http://soft-matter.github.io/trackpy). This takes the feature positions determined in the feature detection step into account but does not include information on the shape of the identified objects.
```
# keyword arguments for linking step
parameters_linking={}
parameters_linking['v_max']=20
parameters_linking['stubs']=2
parameters_linking['order']=1
parameters_linking['extrapolate']=1
parameters_linking['memory']=0
parameters_linking['adaptive_stop']=0.2
parameters_linking['adaptive_step']=0.95
parameters_linking['subnetwork_size']=100
parameters_linking['method_linking']= 'predict'
# Perform linking and save results to file:
Track=tobac.linking_trackpy(Features,OLR,dt=dt,dxy=dxy,**parameters_linking)
Track.to_hdf(os.path.join(savedir,'Track.h5'),'table')
```
**Visualisation:**
```
# Set extent of maps created in the following cells:
axis_extent=[-95,-89,28,32]
# Plot map with all individual tracks:
import cartopy.crs as ccrs
fig_map,ax_map=plt.subplots(figsize=(10,10),subplot_kw={'projection': ccrs.PlateCarree()})
ax_map=tobac.map_tracks(Track,axis_extent=axis_extent,axes=ax_map)
# Create animation of tracked clouds and outlines with OLR as a background field
animation_test_tobac=tobac.animation_mask_field(Track,Features,OLR,Mask_OLR,
axis_extent=axis_extent,#figsize=figsize,orientation_colorbar='horizontal',pad_colorbar=0.2,
vmin=80,vmax=330,cmap='Blues_r',
plot_outline=True,plot_marker=True,marker_track='x',plot_number=True,plot_features=True)
# Display animation:
from IPython.display import HTML, Image, display
HTML(animation_test_tobac.to_html5_video())
# # Save animation to file:
# savefile_animation=os.path.join(plot_dir,'Animation.mp4')
# animation_test_tobac.save(savefile_animation,dpi=200)
# print(f'animation saved to {savefile_animation}')
# Lifetimes of tracked clouds:
fig_lifetime,ax_lifetime=plt.subplots()
tobac.plot_lifetime_histogram_bar(Track,axes=ax_lifetime,bin_edges=np.arange(0,200,20),density=False,width_bar=10)
ax_lifetime.set_xlabel('lifetime (min)')
ax_lifetime.set_ylabel('counts')
```
|
github_jupyter
|
# Building a Fraud Prediction Model with EvalML
In this demo, we will build an optimized fraud prediction model using EvalML. To optimize the pipeline, we will set up an objective function to minimize the percentage of total transaction value lost to fraud. At the end of this demo, we also show you how introducing the right objective during the training results in a much better than using a generic machine learning metric like AUC.
```
import evalml
from evalml import AutoMLSearch
from evalml.objectives import FraudCost
```
## Configure "Cost of Fraud"
To optimize the pipelines toward the specific business needs of this model, we can set our own assumptions for the cost of fraud. These parameters are
* `retry_percentage` - what percentage of customers will retry a transaction if it is declined?
* `interchange_fee` - how much of each successful transaction do you collect?
* `fraud_payout_percentage` - the percentage of fraud will you be unable to collect
* `amount_col` - the column in the data the represents the transaction amount
Using these parameters, EvalML determines attempt to build a pipeline that will minimize the financial loss due to fraud.
```
fraud_objective = FraudCost(retry_percentage=.5,
interchange_fee=.02,
fraud_payout_percentage=.75,
amount_col='amount')
```
## Search for best pipeline
In order to validate the results of the pipeline creation and optimization process, we will save some of our data as the holdout set.
```
X, y = evalml.demos.load_fraud(n_rows=5000)
```
EvalML natively supports one-hot encoding. Here we keep 1 out of the 6 categorical columns to decrease computation time.
```
cols_to_drop = ['datetime', 'expiration_date', 'country', 'region', 'provider']
for col in cols_to_drop:
X.ww.pop(col)
X_train, X_holdout, y_train, y_holdout = evalml.preprocessing.split_data(X, y, problem_type='binary', test_size=0.2, random_seed=0)
X.ww
```
Because the fraud labels are binary, we will use `AutoMLSearch(X_train=X_train, y_train=y_train, problem_type='binary')`. When we call `.search()`, the search for the best pipeline will begin.
```
automl = AutoMLSearch(X_train=X_train, y_train=y_train,
problem_type='binary',
objective=fraud_objective,
additional_objectives=['auc', 'f1', 'precision'],
allowed_model_families=["random_forest", "linear_model"],
max_batches=1,
optimize_thresholds=True,
verbose=True)
automl.search()
```
### View rankings and select pipelines
Once the fitting process is done, we can see all of the pipelines that were searched, ranked by their score on the fraud detection objective we defined.
```
automl.rankings
```
To select the best pipeline we can call `automl.best_pipeline`.
```
best_pipeline = automl.best_pipeline
```
### Describe pipelines
We can get more details about any pipeline created during the search process, including how it performed on other objective functions, by calling the `describe_pipeline` method and passing the `id` of the pipeline of interest.
```
automl.describe_pipeline(automl.rankings.iloc[1]["id"])
```
## Evaluate on holdout data
Finally, since the best pipeline is already trained, we evaluate it on the holdout data.
Now, we can score the pipeline on the holdout data using both our fraud cost objective and the AUC (Area under the ROC Curve) objective.
```
best_pipeline.score(X_holdout, y_holdout, objectives=["auc", fraud_objective])
```
## Why optimize for a problem-specific objective?
To demonstrate the importance of optimizing for the right objective, let's search for another pipeline using AUC, a common machine learning metric. After that, we will score the holdout data using the fraud cost objective to see how the best pipelines compare.
```
automl_auc = AutoMLSearch(X_train=X_train, y_train=y_train,
problem_type='binary',
objective='auc',
additional_objectives=['f1', 'precision'],
max_batches=1,
allowed_model_families=["random_forest", "linear_model"],
optimize_thresholds=True,
verbose=True)
automl_auc.search()
```
Like before, we can look at the rankings of all of the pipelines searched and pick the best pipeline.
```
automl_auc.rankings
best_pipeline_auc = automl_auc.best_pipeline
# get the fraud score on holdout data
best_pipeline_auc.score(X_holdout, y_holdout, objectives=["auc", fraud_objective])
# fraud score on fraud optimized again
best_pipeline.score(X_holdout, y_holdout, objectives=["auc", fraud_objective])
```
When we optimize for AUC, we can see that the AUC score from this pipeline performs better compared to the AUC score from the pipeline optimized for fraud cost; however, the losses due to fraud are a much larger percentage of the total transaction amount when optimized for AUC and much smaller when optimized for fraud cost. As a result, we lose a noticable percentage of the total transaction amount by not optimizing for fraud cost specifically.
Optimizing for AUC does not take into account the user-specified `retry_percentage`, `interchange_fee`, `fraud_payout_percentage` values, which could explain the decrease in fraud performance. Thus, the best pipelines may produce the highest AUC but may not actually reduce the amount loss due to your specific type fraud.
This example highlights how performance in the real world can diverge greatly from machine learning metrics.
|
github_jupyter
|
##### Copyright 2019 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/guide/keras/custom_callback"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/guide/keras/custom_callback.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/guide/keras/custom_callback.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/docs/site/en/guide/keras/custom_callback.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table>
# Keras custom callbacks
A custom callback is a powerful tool to customize the behavior of a Keras model during training, evaluation, or inference, including reading/changing the Keras model. Examples include `tf.keras.callbacks.TensorBoard` where the training progress and results can be exported and visualized with TensorBoard, or `tf.keras.callbacks.ModelCheckpoint` where the model is automatically saved during training, and more. In this guide, you will learn what Keras callback is, when it will be called, what it can do, and how you can build your own. Towards the end of this guide, there will be demos of creating a couple of simple callback applications to get you started on your custom callback.
## Setup
```
from __future__ import absolute_import, division, print_function, unicode_literals
try:
# %tensorflow_version only exists in Colab.
%tensorflow_version 2.x
except Exception:
pass
import tensorflow as tf
```
## Introduction to Keras callbacks
In Keras, `Callback` is a python class meant to be subclassed to provide specific functionality, with a set of methods called at various stages of training (including batch/epoch start and ends), testing, and predicting. Callbacks are useful to get a view on internal states and statistics of the model during training. You can pass a list of callbacks (as the keyword argument `callbacks`) to any of `tf.keras.Model.fit()`, `tf.keras.Model.evaluate()`, and `tf.keras.Model.predict()` methods. The methods of the callbacks will then be called at different stages of training/evaluating/inference.
To get started, let's import tensorflow and define a simple Sequential Keras model:
```
# Define the Keras model to add callbacks to
def get_model():
model = tf.keras.Sequential()
model.add(tf.keras.layers.Dense(1, activation = 'linear', input_dim = 784))
model.compile(optimizer=tf.keras.optimizers.RMSprop(lr=0.1), loss='mean_squared_error', metrics=['mae'])
return model
```
Then, load the MNIST data for training and testing from Keras datasets API:
```
# Load example MNIST data and pre-process it
(x_train, y_train), (x_test, y_test) = tf.keras.datasets.mnist.load_data()
x_train = x_train.reshape(60000, 784).astype('float32') / 255
x_test = x_test.reshape(10000, 784).astype('float32') / 255
```
Now, define a simple custom callback to track the start and end of every batch of data. During those calls, it prints the index of the current batch.
```
import datetime
class MyCustomCallback(tf.keras.callbacks.Callback):
def on_train_batch_begin(self, batch, logs=None):
print('Training: batch {} begins at {}'.format(batch, datetime.datetime.now().time()))
def on_train_batch_end(self, batch, logs=None):
print('Training: batch {} ends at {}'.format(batch, datetime.datetime.now().time()))
def on_test_batch_begin(self, batch, logs=None):
print('Evaluating: batch {} begins at {}'.format(batch, datetime.datetime.now().time()))
def on_test_batch_end(self, batch, logs=None):
print('Evaluating: batch {} ends at {}'.format(batch, datetime.datetime.now().time()))
```
Providing a callback to model methods such as `tf.keras.Model.fit()` ensures the methods are called at those stages:
```
model = get_model()
_ = model.fit(x_train, y_train,
batch_size=64,
epochs=1,
steps_per_epoch=5,
verbose=0,
callbacks=[MyCustomCallback()])
```
## Model methods that take callbacks
Users can supply a list of callbacks to the following `tf.keras.Model` methods:
#### [`fit()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#fit), [`fit_generator()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#fit_generator)
Trains the model for a fixed number of epochs (iterations over a dataset, or data yielded batch-by-batch by a Python generator).
#### [`evaluate()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#evaluate), [`evaluate_generator()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#evaluate_generator)
Evaluates the model for given data or data generator. Outputs the loss and metric values from the evaluation.
#### [`predict()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#predict), [`predict_generator()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#predict_generator)
Generates output predictions for the input data or data generator.
```
_ = model.evaluate(x_test, y_test, batch_size=128, verbose=0, steps=5,
callbacks=[MyCustomCallback()])
```
## An overview of callback methods
### Common methods for training/testing/predicting
For training, testing, and predicting, following methods are provided to be overridden.
#### `on_(train|test|predict)_begin(self, logs=None)`
Called at the beginning of `fit`/`evaluate`/`predict`.
#### `on_(train|test|predict)_end(self, logs=None)`
Called at the end of `fit`/`evaluate`/`predict`.
#### `on_(train|test|predict)_batch_begin(self, batch, logs=None)`
Called right before processing a batch during training/testing/predicting. Within this method, `logs` is a dict with `batch` and `size` available keys, representing the current batch number and the size of the batch.
#### `on_(train|test|predict)_batch_end(self, batch, logs=None)`
Called at the end of training/testing/predicting a batch. Within this method, `logs` is a dict containing the stateful metrics result.
### Training specific methods
In addition, for training, following are provided.
#### on_epoch_begin(self, epoch, logs=None)
Called at the beginning of an epoch during training.
#### on_epoch_end(self, epoch, logs=None)
Called at the end of an epoch during training.
### Usage of `logs` dict
The `logs` dict contains the loss value, and all the metrics at the end of a batch or epoch. Example includes the loss and mean absolute error.
```
class LossAndErrorPrintingCallback(tf.keras.callbacks.Callback):
def on_train_batch_end(self, batch, logs=None):
print('For batch {}, loss is {:7.2f}.'.format(batch, logs['loss']))
def on_test_batch_end(self, batch, logs=None):
print('For batch {}, loss is {:7.2f}.'.format(batch, logs['loss']))
def on_epoch_end(self, epoch, logs=None):
print('The average loss for epoch {} is {:7.2f} and mean absolute error is {:7.2f}.'.format(epoch, logs['loss'], logs['mae']))
model = get_model()
_ = model.fit(x_train, y_train,
batch_size=64,
steps_per_epoch=5,
epochs=3,
verbose=0,
callbacks=[LossAndErrorPrintingCallback()])
```
Similarly, one can provide callbacks in `evaluate()` calls.
```
_ = model.evaluate(x_test, y_test, batch_size=128, verbose=0, steps=20,
callbacks=[LossAndErrorPrintingCallback()])
```
## Examples of Keras callback applications
The following section will guide you through creating simple Callback applications.
### Early stopping at minimum loss
First example showcases the creation of a `Callback` that stops the Keras training when the minimum of loss has been reached by mutating the attribute `model.stop_training` (boolean). Optionally, the user can provide an argument `patience` to specify how many epochs the training should wait before it eventually stops.
`tf.keras.callbacks.EarlyStopping` provides a more complete and general implementation.
```
import numpy as np
class EarlyStoppingAtMinLoss(tf.keras.callbacks.Callback):
"""Stop training when the loss is at its min, i.e. the loss stops decreasing.
Arguments:
patience: Number of epochs to wait after min has been hit. After this
number of no improvement, training stops.
"""
def __init__(self, patience=0):
super(EarlyStoppingAtMinLoss, self).__init__()
self.patience = patience
# best_weights to store the weights at which the minimum loss occurs.
self.best_weights = None
def on_train_begin(self, logs=None):
# The number of epoch it has waited when loss is no longer minimum.
self.wait = 0
# The epoch the training stops at.
self.stopped_epoch = 0
# Initialize the best as infinity.
self.best = np.Inf
def on_epoch_end(self, epoch, logs=None):
current = logs.get('loss')
if np.less(current, self.best):
self.best = current
self.wait = 0
# Record the best weights if current results is better (less).
self.best_weights = self.model.get_weights()
else:
self.wait += 1
if self.wait >= self.patience:
self.stopped_epoch = epoch
self.model.stop_training = True
print('Restoring model weights from the end of the best epoch.')
self.model.set_weights(self.best_weights)
def on_train_end(self, logs=None):
if self.stopped_epoch > 0:
print('Epoch %05d: early stopping' % (self.stopped_epoch + 1))
model = get_model()
_ = model.fit(x_train, y_train,
batch_size=64,
steps_per_epoch=5,
epochs=30,
verbose=0,
callbacks=[LossAndErrorPrintingCallback(), EarlyStoppingAtMinLoss()])
```
### Learning rate scheduling
One thing that is commonly done in model training is changing the learning rate as more epochs have passed. Keras backend exposes get_value API which can be used to set the variables. In this example, we're showing how a custom Callback can be used to dynamically change the learning rate.
Note: this is just an example implementation see `callbacks.LearningRateScheduler` and `keras.optimizers.schedules` for more general implementations.
```
class LearningRateScheduler(tf.keras.callbacks.Callback):
"""Learning rate scheduler which sets the learning rate according to schedule.
Arguments:
schedule: a function that takes an epoch index
(integer, indexed from 0) and current learning rate
as inputs and returns a new learning rate as output (float).
"""
def __init__(self, schedule):
super(LearningRateScheduler, self).__init__()
self.schedule = schedule
def on_epoch_begin(self, epoch, logs=None):
if not hasattr(self.model.optimizer, 'lr'):
raise ValueError('Optimizer must have a "lr" attribute.')
# Get the current learning rate from model's optimizer.
lr = float(tf.keras.backend.get_value(self.model.optimizer.lr))
# Call schedule function to get the scheduled learning rate.
scheduled_lr = self.schedule(epoch, lr)
# Set the value back to the optimizer before this epoch starts
tf.keras.backend.set_value(self.model.optimizer.lr, scheduled_lr)
print('\nEpoch %05d: Learning rate is %6.4f.' % (epoch, scheduled_lr))
LR_SCHEDULE = [
# (epoch to start, learning rate) tuples
(3, 0.05), (6, 0.01), (9, 0.005), (12, 0.001)
]
def lr_schedule(epoch, lr):
"""Helper function to retrieve the scheduled learning rate based on epoch."""
if epoch < LR_SCHEDULE[0][0] or epoch > LR_SCHEDULE[-1][0]:
return lr
for i in range(len(LR_SCHEDULE)):
if epoch == LR_SCHEDULE[i][0]:
return LR_SCHEDULE[i][1]
return lr
model = get_model()
_ = model.fit(x_train, y_train,
batch_size=64,
steps_per_epoch=5,
epochs=15,
verbose=0,
callbacks=[LossAndErrorPrintingCallback(), LearningRateScheduler(lr_schedule)])
```
### Standard Keras callbacks
Be sure to check out the existing Keras callbacks by [visiting the API doc](https://www.tensorflow.org/api_docs/python/tf/keras/callbacks). Applications include logging to CSV, saving the model, visualizing on TensorBoard and a lot more.
|
github_jupyter
|
# Imports
```
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Dense, Dropout, Flatten, Input, Concatenate
from tensorflow.keras.optimizers import Adam, RMSprop
import numpy as np
import matplotlib.pyplot as plt
import copy
```
# Global Variables
```
epochs = 500
batch_size = 16
number_of_particles = epochs * 2 * batch_size
dt = 0.1
```
# Classes
```
class Particle:
def __str__(self):
return "Position: %s, Velocity: %s, Accleration: %s" % (self.position, self.velocity, self.acceleration)
def __repr__(self):
return "Position: %s, Velocity: %s, Accleration: %s" % (self.position, self.velocity, self.acceleration)
def __init__(self):
self.position = np.array([np.random.sample()*2-1,np.random.sample()*2-1]) # Position X, Y
self.velocity = np.array([np.random.sample()*2-1,np.random.sample()*2-1]) # Velocity X, Y
self.acceleration = np.array([np.random.sample()*2-1,np.random.sample()*2-1]) # Acceleration X, Y
def apply_physics(self,dt):
nextParticle = copy.deepcopy(self) # Copy to retain initial values
nextParticle.position += self.velocity * dt
nextParticle.velocity += self.acceleration * dt
return nextParticle
def get_list(self):
return [self.position[0],self.position[1],self.velocity[0], self.velocity[1], self.acceleration[0], self.acceleration[1]]
def get_list_physics(self,dt):
n = self.apply_physics(dt)
return [self.position[0],self.position[1],self.velocity[0], self.velocity[1], self.acceleration[0],
self.acceleration[1], n.position[0], n.position[1], n.velocity[0], n.velocity[1]]
class GAN:
def __init__(self,input_size,output_size,dropout=0.4):
self.input_size = input_size
self.output_size = output_size
self.dropout = dropout
self.generator = self.generator_network()
self.discriminator = self.discriminator_network()
self.adverserial = self.adverserial_network()
def discriminator_trainable(self, val):
self.discriminator.trainable = val
for l in self.discriminator.layers:
l.trainable = val
def generator_network(self): # Generator : Object(6) - Dense - Object(4)
self.g_input = Input(shape=(self.input_size,), name="Generator_Input")
g = Dense(128, activation='relu')(self.g_input)
g = Dropout(self.dropout)(g)
g = Dense(256, activation='relu')(g)
g = Dropout(self.dropout)(g)
g = Dense(512, activation='relu')(g)
g = Dropout(self.dropout)(g)
g = Dense(256, activation='relu')(g)
g = Dropout(self.dropout)(g)
g = Dense(128, activation='relu')(self.g_input)
g = Dropout(self.dropout)(g)
self.g_output = Dense(self.output_size, activation='tanh', name="Generator_Output")(g)
m = Model(self.g_input, self.g_output, name="Generator")
return m
def discriminator_network(self): # Discriminator : Object(10) - Dense - Probability
d_opt = RMSprop(lr=0.000125,decay=6e-8)
d_input = Input(shape=(self.input_size+self.output_size,), name="Discriminator_Input")
d = Dense(128, activation='relu')(d_input)
d = Dense(256, activation='relu')(d)
d = Dense(512, activation='relu')(d)
d = Dense(256, activation='relu')(d)
d = Dense(128, activation='relu')(d)
d_output = Dense(1, activation='sigmoid', name="Discriminator_Output")(d)
m = Model(d_input, d_output, name="Discriminator")
m.compile(loss='binary_crossentropy', optimizer=d_opt)
return m
def adverserial_network(self): # Adverserial : Object(6) - Generator - Discriminator - Probability
a_opt = RMSprop(lr=0.0001,decay=3e-8)
d_input = Concatenate(name="Generator_Input_Output")([self.g_input,self.g_output])
m=Model(self.g_input, self.discriminator(d_input))
m.compile(loss='binary_crossentropy', optimizer=a_opt)
return m
def train_discriminator(self,val):
self.discriminator.trainable = val
for l in self.discriminator.layers:
l.trainable = val
def train(self, adverserial_set, discriminator_set, epochs, batch_size):
losses = {"d":[], "g":[]}
for i in range(epochs):
batch = discriminator_set[int(i/2*batch_size/2):int((i/2+1)*batch_size/2)] # Gets a batch of real data
for j in adverserial_set[int(i/2*batch_size/2):int((i/2+1)*batch_size/2)]: # Gets a batch of generated data
n = copy.deepcopy(j)
p = self.predict(j)
for e in p:
n.append(e)
batch.append(n)
#self.train_discriminator(True) # Turns on discriminator weights
output = np.zeros(batch_size) # Sets output weight 0 for real and 1 for fakes
output[int(batch_size/2):] = 1
losses["d"].append(self.discriminator.train_on_batch(np.array(batch), np.array(output))) # Train discriminator
batch = adverserial_set[(i*batch_size):((i+1)*batch_size)] # Gets real data to train generator
output = np.zeros(batch_size)
#self.train_discriminator(False) # Turns off discriminator weights
losses["g"].append(self.adverserial.train_on_batch(np.array(batch), np.array(output))) # Train generator
print('Epoch %s - Adverserial Loss : %s, Discriminator Loss : %s' % (i+1, losses["g"][-1], losses["d"][-1]))
self.generator.save("Generator.h5")
self.discriminator.save("Discriminator.h5")
return losses
def predict(self, pred):
return self.generator.predict(np.array(pred).reshape(-1,6))[0]
```
# Training Data
```
training_set = []
actual_set = []
for i in range(number_of_particles):
p = Particle()
if(i%2==0):
training_set.append(p.get_list())
else:
actual_set.append(p.get_list_physics(dt))
```
# Training
```
network = GAN(input_size=6,output_size=4,dropout=0)
loss = network.train(adverserial_set=training_set,discriminator_set=actual_set,epochs=epochs,batch_size=batch_size)
fig = plt.figure(figsize=(13,7))
plt.title("Loss Function over Epochs")
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.plot(loss["g"], label="Adversarial Loss")
plt.plot(loss["d"], label="Discriminative Loss")
plt.legend()
plt.show()
network.predict([0.1,0.2,0.1,0.1,0.1,0.1])
network.generator.summary()
network.discriminator.summary()
network.adverserial.summary()
```
|
github_jupyter
|
# Field operations
There are several convenience methods that can be used to analyse the field. Let us first define the mesh we are going to work with.
```
import discretisedfield as df
p1 = (-50, -50, -50)
p2 = (50, 50, 50)
n = (2, 2, 2)
mesh = df.Mesh(p1=p1, p2=p2, n=n)
```
We are going to initialise the vector field (`dim=3`), with
$$\mathbf{f}(x, y, z) = (xy, 2xy, xyz)$$
For that, we are going to use the following Python function.
```
def value_function(pos):
x, y, z = pos
return x*y, 2*x*y, x*y*z
```
Finally, our field is
```
field = df.Field(mesh, dim=3, value=value_function)
```
## 1. Sampling the field
As we have shown previously, a field can be sampled by calling it. The argument must be a 3-length iterable and it contains the coordinates of the point.
```
point = (0, 0, 0)
field(point)
```
However if the point is outside the mesh, an exception is raised.
```
point = (100, 100, 100)
try:
field(point)
except ValueError:
print('Exception raised.')
```
## 2. Extracting the component of a vector field
A three-dimensional vector field can be understood as three separate scalar fields, where each scalar field is a component of a vector field value. A scalar field of a component can be extracted by accessing `x`, `y`, or `z` attribute of the field.
```
x_component = field.x
x_component((0, 0, 0))
```
Default names `x`, `y`, and (for dim 3) `z` are only available for fields with dimensionality 2 or 3.
```
field.components
```
It is possible to change the component names:
```
field.components = ['mx', 'my', 'mz']
field.mx((0, 0, 0))
```
This overrides the component labels and the old `x`, `y` and `z` cannot be used anymore:
```
try:
field.x
except AttributeError as e:
print(e)
```
We change the component labels back to `x`, `y`, and `z` for the rest of this notebook.
```
field.components = ['x', 'y', 'z']
```
Custom component names can optionally also be specified during field creation. If not specified, the default values are used for fields with dimensions 2 or 3. Higher-dimensional fields have no defaults and custom labes have to be specified in order to access individual field components:
```
field_4d = df.Field(mesh, dim=4, value=[1, 1, 1, 1], components=['c1', 'c2', 'c3', 'c4'])
field_4d
field_4d.c1((0, 0, 0))
```
## 3. Computing the average
The average of the field can be obtained by calling `discretisedfield.Field.average` property.
```
field.average
```
Average always return a tuple, independent of the dimension of the field's value.
```
field.x.average
```
## 4. Iterating through the field
The field object itself is an iterable. That means that it can be iterated through. As a result it returns a tuple, where the first element is the coordinate of the mesh point, whereas the second one is its value.
```
for coordinate, value in field:
print(coordinate, value)
```
## 5. Sampling the field along the line
To sample the points of the field which are on a certain line, `discretisedfield.Field.line` method is used. It takes two points `p1` and `p2` that define the line and an integer `n` which defines how many mesh coordinates on that line are required. The default value of `n` is 100.
```
line = field.line(p1=(-10, 0, 0), p2=(10, 0, 0), n=5)
```
## 6. Intersecting the field with a plane
If we intersect the field with a plane, `discretisedfield.Field.plane` will return a new field object which contains only discretisation cells that belong to that plane. The planes allowed are the planes perpendicular to the axes of the Cartesian coordinate system. For instance, a plane parallel to the $yz$-plane (perpendicular to the $x$-axis) which intesects the $x$-axis at 1, can be written as
$$x = 1$$
```
field.plane(x=1)
```
If we want to cut through the middle of the mesh, we do not need to provide a particular value for a coordinate.
```
field.plane('x')
```
## 7. Cascading the operations
Let us say we want to compute the average of an $x$ component of the field on the plane $y=10$. In order to do that, we can cascade several operation in a single line.
```
field.plane(y=10).x.average
```
This gives the same result as for instance
```
field.x.plane(y=10).average
```
## 8. Complex fields
`discretisedfield` supports complex-valued fields.
```
cfield = df.Field(mesh, dim=3, value=(1+1.5j, 2, 3j))
```
We can extract `real` and `imaginary` part.
```
cfield.real((0, 0, 0))
cfield.imag((0, 0, 0))
```
Similarly we get `real` and `imaginary` parts of individual components.
```
cfield.x.real((0, 0, 0))
cfield.x.imag((0, 0, 0))
```
Complex conjugate.
```
cfield.conjugate((0, 0, 0))
```
Phase in the complex plane.
```
cfield.phase((0, 0, 0))
```
## 9. Applying `numpys` universal functions
All numpy universal functions can be applied to `discretisedfield.Field` objects. Below we show a different examples. For available functions please refer to the `numpy` [documentation](https://numpy.org/doc/stable/reference/ufuncs.html#available-ufuncs).
```
import numpy as np
f1 = df.Field(mesh, dim=1, value=1)
f2 = df.Field(mesh, dim=1, value=np.pi)
f3 = df.Field(mesh, dim=1, value=2)
np.sin(f1)
np.sin(f2)((0, 0, 0))
np.sum((f1, f2, f3))((0, 0, 0))
np.exp(f1)((0, 0, 0))
np.power(f3, 2)((0, 0, 0))
```
## Other
Full description of all existing functionality can be found in the [API Reference](https://discretisedfield.readthedocs.io/en/latest/_autosummary/discretisedfield.Field.html).
|
github_jupyter
|
# [NTDS'19] tutorial 5: machine learning with scikit-learn
[ntds'19]: https://github.com/mdeff/ntds_2019
[Nicolas Aspert](https://people.epfl.ch/nicolas.aspert), [EPFL LTS2](https://lts2.epfl.ch).
* Dataset: [digits](https://archive.ics.uci.edu/ml/datasets/Pen-Based+Recognition+of+Handwritten+Digits)
* Tools: [scikit-learn](https://scikit-learn.org/stable/), [numpy](http://www.numpy.org), [scipy](https://www.scipy.org), [matplotlib](https://matplotlib.org)
*scikit-learn* is a machine learning python library. Most commonly used algorithms for classification, clustering and regression are implemented as part of the library, e.g.
* [Logistic regression](https://en.wikipedia.org/wiki/Logistic_regression)
* [k-means clustering](https://en.wikipedia.org/wiki/K-means_clustering)
* [Support vector machines](https://en.wikipedia.org/wiki/Support-vector_machine)
* ...
The aim of this tutorial is to show basic usage of some simple machine learning techniques.
Check the official [documentation](https://scikit-learn.org/stable/documentation.html) for more information, especially the [tutorials](https://scikit-learn.org/stable/tutorial/index.html) section.
```
%matplotlib inline
import numpy as np
from matplotlib import pyplot as plt
import sklearn
```
## Data loading
We will use a dataset named *digits*.
It is made of 1797 handwritten digits images (of size 8x8 pixels each) acquired from 44 different writers.
Each image is labelled according to the digit present in the image.
You can find more information about this dataset [here](https://archive.ics.uci.edu/ml/datasets/Pen-Based+Recognition+of+Handwritten+Digits).
Load the dataset.
```
from sklearn.datasets import load_digits
digits = load_digits()
```
The `digits` variable contains several fields.
In `images` you have all samples as 2-dimensional arrays.
```
print(digits.images.shape)
print(digits.images[0])
plt.imshow(digits.images[0], cmap=plt.cm.gray);
```
In `data`, the same samples are represented as 1-d vectors of length 64.
```
print(digits.data.shape)
print(digits.data[0])
```
In `target` you have the label corresponding to each image.
```
print(digits.target.shape)
print(digits.target)
```
Let us visualize the 20 first entries of the dataset (image display kept small on purpose)
```
fig = plt.figure(figsize=(15, 0.5))
for index, (image, label) in enumerate(zip(digits.images[0:20], digits.target[0:20])):
ax = fig.add_subplot(1, 20, index+1)
ax.imshow(image, cmap=plt.cm.gray)
ax.set_title(label)
ax.axis('off')
```
### Training/Test set
Before training our model, the [`train_test_split`](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html) function will separate our dataset into a training set and a test set. The samples from the test set are never used during the training phase. This allows for a fair evaluation of the model's performance.
```
from sklearn.model_selection import train_test_split
train_img, test_img, train_lbl, test_lbl = train_test_split(
digits.data, digits.target, test_size=1/6) # keep ~300 images as test set
```
We can check that all classes are well balanced in the training and test sets.
```
np.histogram(train_lbl, bins=10)
np.histogram(test_lbl, bins=10)
```
## Supervised learning: logistic regression
### Linear regression reminder
Linear regression is used to predict an dependent value $y$ from an n-dimensional vector $x$.
The assumption made here is that the output depends linearly on the input components, i.e. $y = mx + b$.
Given a set of input and output values, the goal is to compute $m$ and $b$ minimizing the [mean squared error (MSE)](https://en.wikipedia.org/wiki/Mean_squared_error) between the predicted and actual outputs.
In scikit-learn this method is available through [`LinearRegression`](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LinearRegression.html).
### Logistic regression
Logistic regression is used to predict categorical data (e.g. yes/no, member/non-member, ham/spam, benign/malignant, ...).
It uses the output of a linear predictor, and maps it to a probability using a [sigmoid function](https://en.wikipedia.org/wiki/Sigmoid_function), such as the logistic function $s(z) = \frac{1}{1+e^{-z}}$.
The output is a probability score between 0 and 1, and using a simple thresholding the class output will be positive if the probability is greater than 0.5, negative if not.
A [log-loss cost function](http://wiki.fast.ai/index.php/Logistic_Regression#Cost_Function) (not just the MSE as for linear regression) is used to train logistic regression (using gradient descent for instance).
[Multinomial logistic regression](https://en.wikipedia.org/wiki/Multinomial_logistic_regression) is an extension of the binary classification problem to a $n$-classes problem.
We can now create a logistic regression object and fit the parameters using the training data.
NB: as the dataset is quite simple, default parameters will give good results. Check the [documentation](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html) for fine-tuning possibilities.
```
from sklearn.linear_model import LogisticRegression
# All unspecified parameters are left to their default values.
logisticRegr = LogisticRegression(verbose=1, solver='liblinear', multi_class='auto') # set solver and multi_class to silence warnings
logisticRegr.fit(train_img, train_lbl)
```
## Model performance evaluation
For a binary classification problem, let us denote by $TP$, $TN$, $FP$, and $FN$ the number of true positives, true negatives, false positives and false negatives.
### Accuracy
The *accuracy* is defined by $a = \frac{TP}{TP + TN + FP + FN}$
NB: in scikit-learn, models may have different definitions of the `score` method. For multi-class logistic regression, the value is the mean accuracy for each class.
```
score = logisticRegr.score(test_img, test_lbl)
print(f'accuracy = {score:.4f}')
```
### F1 score
Accuracy only provides partial information about the performance of a model. Many other [metrics](https://scikit-learn.org/stable/modules/model_evaluation.html#classification-metrics) are part of scikit-learn.
A metric that provides a more complete overview of the classification performance is the [F1 score](https://en.wikipedia.org/wiki/F1_score). It takes into account not only the valid predictions but also the incorrect ones, by combining precision and recall.
*Precision* is the number of positive predictions divided by the total number of positive class values predicted, i.e. $p=\frac{TP}{TP+FP}$. A low precision indicates a high number of false positives.
*Recall* is the number of positive predictions divided by the number of positive class values in the test data, i.e. $r=\frac{TP}{TP+FN}$. A low recall indicates a high number of false negatives.
Finally the F1 score is the harmonic mean between precision and recall, i.e. $F1=2\frac{p.r}{p+r}$
Let us compute the predicted labels in the test set:
```
pred_lbl = logisticRegr.predict(test_img)
from sklearn.metrics import f1_score, classification_report
from sklearn.utils.multiclass import unique_labels
```
The [`f1_score`](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.f1_score.html#sklearn.metrics.f1_score) function computes the F1 score. The `average` parameter controls whether the result is computed globally over all classes (`average='micro'`) or if the F1 score is computed for each class then averaged (`average='macro'`).
```
f1_score(test_lbl, pred_lbl, average='micro')
f1_score(test_lbl, pred_lbl, average='macro')
```
`classification_report` provides a synthetic overview of all results for each class, as well as globally.
```
print(classification_report(test_lbl, pred_lbl))
```
### Confusion matrix
In the case of a multi-class problem, the *confusion matrix* is often used to present the results.
```
from sklearn.metrics import confusion_matrix
def plot_confusion_matrix(y_true, y_pred, classes,
normalize=False,
title=None,
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if not title:
if normalize:
title = 'Normalized confusion matrix'
else:
title = 'Confusion matrix, without normalization'
# Compute confusion matrix
cm = confusion_matrix(y_true, y_pred)
# Only use the labels that appear in the data
classes = classes[unique_labels(y_true, y_pred)]
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
fig, ax = plt.subplots()
im = ax.imshow(cm, interpolation='nearest', cmap=cmap)
ax.figure.colorbar(im, ax=ax)
# We want to show all ticks...
ax.set(xticks=np.arange(cm.shape[1]),
yticks=np.arange(cm.shape[0]),
# ... and label them with the respective list entries
xticklabels=classes, yticklabels=classes,
title=title,
ylabel='True label',
xlabel='Predicted label')
# Rotate the tick labels and set their alignment.
plt.setp(ax.get_xticklabels(), rotation=45, ha="right",
rotation_mode="anchor")
# Loop over data dimensions and create text annotations.
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i in range(cm.shape[0]):
for j in range(cm.shape[1]):
ax.text(j, i, format(cm[i, j], fmt),
ha="center", va="center",
color="white" if cm[i, j] > thresh else "black")
fig.tight_layout()
return ax
plot_confusion_matrix(test_lbl, pred_lbl, np.array(list(map(lambda x: str(x), range(10)))), normalize=False)
```
## Supervised learning: support-vector machines
[Support-vector machines (SVM)](https://en.wikipedia.org/wiki/Support-vector_machine) are also used for classification tasks.
For a binary classification task of $n$-dimensional feature vectors, a linear SVM try to return the ($n-1$)-dimensional hyperplane that separate the two classes with the largest possible margin.
Nonlinear SVMs fit the maximum-margin hyperplane in a transformed feature space.
Although the classifier is a hyperplane in the transformed feature space, it may be nonlinear in the original input space.
The goal here is to show that a method (e.g. the previously used logistic regression) can be substituted transparently for another one.
```
from sklearn import svm
```
Default parameters perform well on this dataset.
It might be needed to adjust $C$ and $\gamma$ (e.g. via a grid search) for optimal performance (cf. [SVC documentation](https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html#sklearn.svm.SVC)).
```
clf = svm.SVC(gamma='scale') # default kernel is RBF
clf.fit(train_img, train_lbl)
```
The classification accuracy improves with respect to logistic regression (here `score` also computes mean accuracy, as in logistic regression).
```
clf.score(test_img, test_lbl)
```
The F1 score is also improved.
```
pred_lbl_svm = clf.predict(test_img)
print(classification_report(test_lbl, pred_lbl_svm))
```
## Unsupervised learning: $k$-means
[$k$-means](https://en.wikipedia.org/wiki/K-means_clustering) aims at partitioning a samples into $k$ clusters, s.t. each sample belongs to the cluster having the closest mean. Its implementation is iterative, and relies on a prior knowledge of the number of clusters present.
One important step in $k$-means clustering is the initialization, i.e. the choice of initial clusters to be refined.
This choice can have a significant impact on results.
```
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters=10)
kmeans.fit(digits.data)
km_labels = kmeans.predict(digits.data)
digits.target
km_labels
```
Since we have ground truth information of classes, we can check if the $k$-means results make sense.
However as you can see, the labels produced by $k$-means and the ground truth ones do not match.
An agreement score based on [mutual information](https://scikit-learn.org/stable/modules/clustering.html#clustering-evaluation), insensitive to labels permutation can be used to evaluate the results.
```
from sklearn.metrics import adjusted_mutual_info_score
adjusted_mutual_info_score(digits.target, kmeans.labels_)
```
## Unsupervized learning: dimensionality reduction
You can also try to visualize the clusters as in this [scikit-learn demo](https://scikit-learn.org/stable/auto_examples/cluster/plot_kmeans_digits.html). Mapping the input features to lower dimensional embeddings (2D or 3D), e.g. using PCA otr tSNE is required for visualization. [This demo](https://scikit-learn.org/stable/auto_examples/manifold/plot_lle_digits.html) provides an overview of the possibilities.
```
from matplotlib import offsetbox
def plot_embedding(X, y, title=None):
"""Scale and visualize the embedding vectors."""
x_min, x_max = np.min(X, 0), np.max(X, 0)
X = (X - x_min) / (x_max - x_min)
plt.figure()
ax = plt.subplot(111)
for i in range(X.shape[0]):
plt.text(X[i, 0], X[i, 1], str(y[i]),
color=plt.cm.Set1(y[i] / 10.),
fontdict={'weight': 'bold', 'size': 9})
if hasattr(offsetbox, 'AnnotationBbox'):
# only print thumbnails with matplotlib > 1.0
shown_images = np.array([[1., 1.]]) # just something big
for i in range(X.shape[0]):
dist = np.sum((X[i] - shown_images) ** 2, 1)
if np.min(dist) < 4e-3:
# don't show points that are too close
continue
shown_images = np.r_[shown_images, [X[i]]]
imagebox = offsetbox.AnnotationBbox(
offsetbox.OffsetImage(digits.images[i], cmap=plt.cm.gray_r),
X[i])
ax.add_artist(imagebox)
plt.xticks([]), plt.yticks([])
if title is not None:
plt.title(title)
from sklearn import manifold
tsne = manifold.TSNE(n_components=2, init='pca', random_state=0)
X_tsne = tsne.fit_transform(digits.data)
plot_embedding(X_tsne, digits.target,
"t-SNE embedding of the digits (ground truth labels)")
plot_embedding(X_tsne, km_labels,
"t-SNE embedding of the digits (kmeans labels)")
```
|
github_jupyter
|
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
from scipy.stats import kurtosis
from sklearn.decomposition import PCA
import seaborn as sns
from scipy.stats import pearsonr
%matplotlib
gov_pop_area_data = pd.read_excel('/Users/Rohil/Documents/iGEM/yemen/gov_area_pop_data.xlsx')
gov_pop_area_data = gov_pop_area_data[gov_pop_area_data.iso != 'YE-HD']
gov_pop_area_data.head()
cholera_case_crosstab = pd.read_csv(r'C:\Users\Rohil\Documents\iGEM\yemen\cholera_epi_data\yemen_cholera_case_data_differenced.csv', dayfirst = True)
cholera_case_crosstab.tail()
norm_cholera_case_crosstab = cholera_case_crosstab
for index, row in gov_pop_area_data[['iso', 'population']].iterrows():
norm_cholera_case_crosstab[row.iso] = (norm_cholera_case_crosstab[row.iso] * 10000) / row.population
norm_cholera_case_crosstab.tail()
cholera_death_crosstab = pd.read_csv(r'C:\Users\Rohil\Documents\iGEM\yemen\cholera_epi_data\yemen_cholera_death_data_differenced.csv', dayfirst = True)
cholera_death_crosstab.head()
norm_cholera_death_crosstab = cholera_death_crosstab
for index, row in gov_pop_area_data[['iso', 'population']].iterrows():
norm_cholera_death_crosstab[row.iso] = (norm_cholera_death_crosstab[row.iso] * 10000) / row.population
norm_cholera_death_crosstab.head()
mean_rainfall_crosstab = pd.read_csv(r'C:\Users\Rohil\Documents\iGEM\yemen\rainfall\yemen_daily_mean_rainfall_crosstab.csv', dayfirst = True)
max_rainfall_crosstab = pd.read_csv(r'C:\Users\Rohil\Documents\iGEM\yemen\rainfall\yemen_daily_max_rainfall_crosstab.csv', dayfirst = True)
mean_rainfall_crosstab.head()
max_rainfall_crosstab.head()
cases_unstacked = norm_cholera_case_crosstab.set_index('date').unstack().reset_index()
cases_unstacked.columns = ['gov_iso', 'date', 'new_cases']
deaths_unstacked = norm_cholera_death_crosstab.set_index('date').unstack().reset_index()
deaths_unstacked.columns = ['gov_iso', 'date', 'new_deaths']
max_rainfall_unstacked = max_rainfall_crosstab.set_index('date').unstack().reset_index()
max_rainfall_unstacked.columns = ['gov_iso', 'date', 'max_rainfall']
mean_rainfall_unstacked = mean_rainfall_crosstab.set_index('date').unstack().reset_index()
mean_rainfall_unstacked.columns = ['gov_iso', 'date', 'mean_rainfall']
cases_unstacked.shape
cases_unstacked.head()
deaths_unstacked.shape
deaths_unstacked.head()
mean_rainfall_unstacked.shape
## date formatting has been fixed
mean_rainfall_unstacked.head()
mean_rainfall_unstacked.date.tail()
cases_unstacked.date.tail()
deaths_unstacked.date.tail()
case_death_rainfall_data = cases_unstacked.merge(deaths_unstacked, on =['date', 'gov_iso']).merge(mean_rainfall_unstacked, on =['date', 'gov_iso'], how = 'left')
case_death_rainfall_data.date = pd.to_datetime(case_death_rainfall_data.date, dayfirst = True)
case_death_rainfall_data.sort_values(by = 'date')
# YE-HD-AL refers to Al Mukulla
neighboring_gov_dict = {"YE-SA" : ["YE-SN"],
"YE-AB" : ["YE-LA", "YE-SH", "YE-BA"],
"YE-AD" : ["YE-LA"],
"YE-DA" : ["YE-LA", "YE-TA", "YE-IB", "YE-BA"],
"YE-BA" : ["YE-DH", "YE-IB", "YE-DA", "YE-AB", "YE-SH", "YE-MA", "YE-SN"],
"YE-HU" : ["YE-HJ", "YE-MW", "YE-SN", "YE-RA", "YE-DH", "YE-TA"],
"YE-JA" : ["YE-MA", "YE-SN", "YE-AM", "YE-SD"],
"YE-MR" : ["YE-HD-AL"],
"YE-MW" : ["YE-HU", "YE-HJ", "YE-AM", "YE-SN"],
"YE-AM" : ["YE-HJ", "YE-SD", "YE-JA", "YE-SN", "YE-MW"],
"YE-DH" : ["YE-IB", "YE-RA", "YE-SN", "YE-BA"],
"YE-HD-AL" : ["YE-SH", "YE-MR"],
"YE-HJ" : ["YE-MW", "YE-HU", "YE-MR"],
"YE-IB" : ["YE-TA", "YE-HU", "YE-DH", "YE-BA", "YE-DA"],
"YE-LA" : ["YE-AD", "YE-TA", "YE-DA", "YE-BA", "YE-AB"],
"YE-MA" : ["YE-BA", "YE-SN", "YE-JA", "YE-SH"],
"YE-RA" : ["YE-DH", "YE-HU", "YE-SN"],
"YE-SD" : ["YE-HJ", "YE-AM", "YE-JA"],
"YE-SN" : ["YE-BA", "YE-DH", "YE-RA", "YE-MW", "YE-AM", "YE-JA", "YE-MA"],
"YE-SH" : ["YE-AB", "YE-BA", "YE-MA", "YE-HD-AL"],
"YE-TA" : ["YE-LA", "YE-DA", "YE-IB", "YE-HU"]}
def get_past_days_features(row, var, daysback):
stock_data = full_data[full_data.stock_id == row.stock_id].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_stock_data = stock_data.loc[(stock_data.index >= x_days_date) & (stock_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_stock_data[var]), np.max(relevant_stock_data[var]), kurtosis(relevant_stock_data[var])]))
def get_past_days_features(row, var, daysback):
other_stock_data = full_data[full_data.stock_id.isin(neighboring_stocks[row.stock_id])].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_other_stock_data = other_stock_data.loc[(other_stock_data.index >= x_days_date) & (other_stock_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_other_stock_data[var]), np.max(relevant_other_stock_data[var]), kurtosis(relevant_other_stock_data[var])]))
def get_past_days_features(row, var, daysback):
if 'rainfall' in var:
rainfall_df = mean_rainfall_unstacked
rainfall_df.date = pd.to_datetime(rainfall_df.date, dayfirst = True)
gov_data = rainfall_df[rainfall_df.gov_iso == row.gov_iso].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_gov_data = gov_data.loc[(gov_data.index >= x_days_date) & (gov_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_gov_data[var]), np.max(relevant_gov_data[var]), kurtosis(relevant_gov_data[var])]))
else:
gov_data = case_death_rainfall_data[case_death_rainfall_data.gov_iso == row.gov_iso].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_gov_data = gov_data.loc[(gov_data.index >= x_days_date) & (gov_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_gov_data[var]), np.max(relevant_gov_data[var]), kurtosis(relevant_gov_data[var])]))
def get_neighbor_past_days_features(row, var, daysback):
if 'rainfall' in var:
rainfall_df = mean_rainfall_unstacked
rainfall_df.date = pd.to_datetime(rainfall_df.date, dayfirst = True)
other_gov_data = rainfall_df[rainfall_df.gov_iso.isin(neighboring_gov_dict[row.gov_iso])].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_other_gov_data = other_gov_data.loc[(other_gov_data.index >= x_days_date) & (other_gov_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_other_gov_data[var]), np.max(relevant_other_gov_data[var]), kurtosis(relevant_other_gov_data[var])]))
else:
other_gov_data = case_death_rainfall_data[case_death_rainfall_data.gov_iso.isin(neighboring_gov_dict[row.gov_iso])].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_other_gov_data = other_gov_data.loc[(other_gov_data.index >= x_days_date) & (other_gov_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_other_gov_data[var]), np.max(relevant_other_gov_data[var]), kurtosis(relevant_other_gov_data[var])]))
past_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 7), axis = 1)
past_week_cases.columns = ['mean_past_week_cases', 'max_past_week_cases', 'kurtosis_past_week_cases']
neighbor_past_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 7), axis = 1)
neighbor_past_week_cases.columns = ['neighbor_mean_past_week_cases', 'neighbor_max_past_week_cases', 'neighbor_kurtosis_past_week_cases']
past_2_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 14), axis = 1)
past_2_week_cases.columns = ['mean_past_2_week_cases', 'max_past_2_week_cases', 'kurtosis_past_2_week_cases']
neighbor_past_2_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 14), axis = 1)
neighbor_past_2_week_cases.columns = ['neighbor_mean_past_2_week_cases', 'neighbor_max_past_2_week_cases', 'neighbor_kurtosis_past_2_week_cases']
past_3_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 21), axis = 1)
past_3_week_cases.columns = ['mean_past_3_week_cases', 'max_past_3_week_cases', 'kurtosis_past_3_week_cases']
neighbor_past_3_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 21), axis = 1)
neighbor_past_3_week_cases.columns = ['neighbor_mean_past_3_week_cases', 'neighbor_max_past_3_week_cases', 'neighbor_kurtosis_past_3_week_cases']
past_month_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 30), axis = 1)
past_month_cases.columns = ['mean_past_month_cases', 'max_past_month_cases', 'kurtosis_past_month_cases']
neighbor_past_month_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 30), axis = 1)
neighbor_past_month_cases.columns = ['neighbor_mean_past_month_cases', 'neighbor_max_past_month_cases', 'neighbor_kurtosis_past_month_cases']
past_6_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 42), axis = 1)
past_6_week_cases.columns = ['mean_past_6_week_cases', 'max_past_6_week_cases', 'kurtosis_past_6_week_cases']
neighbor_past_6_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 42), axis = 1)
neighbor_past_6_week_cases.columns = ['neighbor_mean_past_6_week_cases', 'neighbor_max_past_6_week_cases', 'neighbor_kurtosis_past_6_week_cases']
past_week_deaths = case_death_rainfall_data.apply(get_past_days_features, args = ('new_deaths', 7), axis = 1)
past_week_deaths.columns = ['mean_past_week_deaths', 'max_past_week_deaths', 'kurtosis_past_week_deaths']
neighbor_past_week_deaths = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_deaths', 7), axis = 1)
neighbor_past_week_deaths.columns = ['neighbor_mean_past_week_deaths', 'neighbor_max_past_week_deaths', 'neighbor_kurtosis_past_week_deaths']
past_2_week_deaths = case_death_rainfall_data.apply(get_past_days_features, args = ('new_deaths', 14), axis = 1)
past_2_week_deaths.columns = ['mean_past_2_week_deaths', 'max_past_2_week_deaths', 'kurtosis_past_2_week_deaths']
neighbor_past_2_week_deaths = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_deaths', 14), axis = 1)
neighbor_past_2_week_deaths.columns = ['neighbor_mean_past_2_week_deaths', 'neighbor_max_past_2_week_deaths', 'neighbor_kurtosis_past_2_week_deaths']
past_month_deaths = case_death_rainfall_data.apply(get_past_days_features, args = ('new_deaths', 30), axis = 1)
past_month_deaths.columns = ['mean_past_month_deaths', 'max_past_month_deaths', 'kurtosis_past_month_deaths']
neighbor_past_month_deaths = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_deaths', 30), axis = 1)
neighbor_past_month_deaths.columns = ['neighbor_mean_past_month_deaths', 'neighbor_max_past_month_deaths', 'neighbor_kurtosis_past_month_deaths']
past_week_rainfall = case_death_rainfall_data.apply(get_past_days_features, args = ('mean_rainfall', 7), axis = 1)
past_week_rainfall.columns = ['mean_past_week_rainfall', 'max_past_week_rainfall', 'kurtosis_past_week_rainfall']
neighbor_past_week_rainfall = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('mean_rainfall', 7), axis = 1)
neighbor_past_week_rainfall.columns = ['neighbor_mean_past_week_rainfall', 'neighbor_max_past_week_rainfall', 'neighbor_kurtosis_past_week_rainfall']
past_2_week_rainfall = case_death_rainfall_data.apply(get_past_days_features, args = ('mean_rainfall', 14), axis = 1)
past_2_week_rainfall.columns = ['mean_past_2_week_rainfall', 'max_past_2_week_rainfall', 'kurtosis_past_2_week_rainfall']
neighbor_past_2_week_rainfall = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('mean_rainfall', 14), axis = 1)
neighbor_past_2_week_rainfall.columns = ['neighbor_mean_past_2_week_rainfall', 'neighbor_max_past_2_week_rainfall', 'neighbor_kurtosis_past_2_week_rainfall']
past_month_rainfall = case_death_rainfall_data.apply(get_past_days_features, args = ('mean_rainfall', 30), axis = 1)
past_month_rainfall.columns = ['mean_past_month_rainfall', 'max_past_month_rainfall', 'kurtosis_past_month_rainfall']
neighbor_past_month_rainfall = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('mean_rainfall', 30), axis = 1)
neighbor_past_month_rainfall.columns = ['neighbor_mean_past_month_rainfall', 'neighbor_max_past_month_rainfall', 'neighbor_kurtosis_past_month_rainfall']
training_data = pd.concat([case_death_rainfall_data[['gov_iso', 'date', 'weekly_cases']],
past_week_cases, past_2_week_cases, past_month_cases, neighbor_past_week_cases, neighbor_past_2_week_cases, neighbor_past_month_cases,
past_week_deaths, past_2_week_deaths, past_month_deaths, neighbor_past_week_deaths, neighbor_past_2_week_deaths, neighbor_past_month_deaths,
past_week_rainfall, past_2_week_rainfall, past_month_rainfall, neighbor_past_week_rainfall, neighbor_past_2_week_rainfall, neighbor_past_month_rainfall], axis = 1)
training_data.to_csv('/Users/Rohil/Documents/iGEM/yemen/full_feature_data.csv', index = False)
col_list = []
for col in training_data.columns:
if ('max' not in col) and ('kurtosis' not in col) & ('deaths' not in col):
col_list.append(col)
# want to have at least 7 days of data for most of these examples
trunc_training_data = training_data[col_list]
trunc_training_data = trunc_training_data[(trunc_training_data['date'] > '2017-05-30')].sort_values('date')
features = trunc_training_data.iloc[:,3:].columns.tolist()
target = trunc_training_data.iloc[:,2].name
correlations = {}
for f in features:
data_temp = trunc_training_data[[f,target]]
x1 = data_temp[f].values
x2 = data_temp[target].values
key = f + ' vs ' + target
correlations[key] = pearsonr(x1,x2)[0]
data_correlations = pd.DataFrame(correlations, index=['Value']).T
data_correlations.loc[data_correlations['Value'].abs().sort_values(ascending=False).index]
trunc_training_data = pd.concat([trunc_training_data, pd.get_dummies(trunc_training_data.gov_iso).sort_index()], axis=1)
trunc_training_data.to_csv('/Users/Rohil/Documents/iGEM/yemen/prelim_training_data.csv', index = False)
trunc_training_data[trunc_training_data.isnull().any(axis=1)]
trunc_training_data.shape
trunc_training_data.head()
whole_standard_scaler = StandardScaler()
trunc_training_features = trunc_training_data.iloc[:,3:]
trunc_training_features.shape
norm_features = whole_standard_scaler.fit_transform(trunc_training_features)
pca = PCA(n_components = 33)
pca.fit(norm_features)
pca.explained_variance_ratio_
pd.DataFrame(pca.components_, columns = trunc_training_features.columns)
pca.components_
sns.heatmap(np.log(pca.inverse_transform(np.eye(12))))
# plots of normalized cases x days back vs today
for column in trunc_training_features.columns:
fig, ax = plt.subplots(1,1)
ax.scatter(trunc_training_features[column], trunc_training_data['weekly_cases'])
ax.set_ylabel('weekly cholera cases')
ax.set_xlabel(column)
fig.savefig('/Users/Rohil/Documents/iGEM/yemen/feature_engineering/old/' + column + '_vs_cases.png')
plt.close()
norm_features = pd.DataFrame(data=norm_features, columns = trunc_training_features.columns)
norm_features
```
|
github_jupyter
|
```
import torch
from torch import optim
import torch.nn as nn
import torch.nn.functional as F
import torch.autograd as autograd
from torch.autograd import Variable
from sklearn.preprocessing import OneHotEncoder
import os, math, glob, argparse
from utils.torch_utils import *
from utils.utils import *
from mpradragonn_predictor_pytorch import *
import matplotlib.pyplot as plt
import utils.language_helpers
#plt.switch_backend('agg')
import numpy as np
from models import *
from wgan_gp_mpradragonn_analyzer_quantile_cutoff import *
use_cuda = torch.cuda.is_available()
device = torch.device('cuda:0' if use_cuda else 'cpu')
from torch.distributions import Normal as torch_normal
class IdentityEncoder :
def __init__(self, seq_len, channel_map) :
self.seq_len = seq_len
self.n_channels = len(channel_map)
self.encode_map = channel_map
self.decode_map = {
nt: ix for ix, nt in self.encode_map.items()
}
def encode(self, seq) :
encoding = np.zeros((self.seq_len, self.n_channels))
for i in range(len(seq)) :
if seq[i] in self.encode_map :
channel_ix = self.encode_map[seq[i]]
encoding[i, channel_ix] = 1.
return encoding
def encode_inplace(self, seq, encoding) :
for i in range(len(seq)) :
if seq[i] in self.encode_map :
channel_ix = self.encode_map[seq[i]]
encoding[i, channel_ix] = 1.
def encode_inplace_sparse(self, seq, encoding_mat, row_index) :
raise NotImplementError()
def decode(self, encoding) :
seq = ''
for pos in range(0, encoding.shape[0]) :
argmax_nt = np.argmax(encoding[pos, :])
max_nt = np.max(encoding[pos, :])
seq += self.decode_map[argmax_nt]
return seq
def decode_sparse(self, encoding_mat, row_index) :
raise NotImplementError()
class ActivationMaximizer(nn.Module) :
def __init__(self, generator_dir, batch_size=1, seq_len=145, latent_size=128, sequence_template=None):
super(ActivationMaximizer, self).__init__()
self.generator = Generator_lang(4, seq_len, batch_size, 512)
self.predictor = DragoNNClassifier(batch_size=batch_size).cnn
self.load_generator(generator_dir)
self.use_cuda = torch.cuda.is_available()
self.x_mask = None
self.x_template = None
if sequence_template is not None :
onehot_mask = np.zeros((seq_len, 4))
onehot_template = np.zeros((seq_len, 4))
for j in range(len(sequence_template)) :
if sequence_template[j] == 'N' :
onehot_mask[j, :] = 1.
elif sequence_template[j] == 'A' :
onehot_template[j, 0] = 1.
elif sequence_template[j] == 'C' :
onehot_template[j, 1] = 1.
elif sequence_template[j] == 'G' :
onehot_template[j, 2] = 1.
elif sequence_template[j] == 'T' :
onehot_template[j, 3] = 1.
self.x_mask = Variable(torch.FloatTensor(onehot_mask).unsqueeze(0))
self.x_template = Variable(torch.FloatTensor(onehot_template).unsqueeze(0))
if self.use_cuda :
self.x_mask = self.x_mask.to(device)
self.x_template = self.x_template.to(device)
self.predictor.eval()
if self.use_cuda :
self.generator.cuda()
self.predictor.cuda()
self.cuda()
def load_generator(self, directory, iteration=None) :
list_generator = glob.glob(directory + "G*.pth")
generator_file = max(list_generator, key=os.path.getctime)
self.generator.load_state_dict(torch.load(generator_file))
def forward(self, z) :
x = self.generator.forward(z)
if self.x_mask is not None :
x = x * self.x_mask + self.x_template
return self.predictor.forward(x.unsqueeze(2).transpose(1, 3))
def get_pattern(self, z) :
x = self.generator.forward(z)
if self.x_mask is not None :
x = x * self.x_mask + self.x_template
return x
#Sequence length
seq_len = 145
batch_size = 64
#Sequence decoder
acgt_encoder = IdentityEncoder(seq_len, {'A':0, 'C':1, 'G':2, 'T':3})
#Sequence template
sequence_template = 'N' * 145
#Activation maximization model (pytorch)
act_maximizer = ActivationMaximizer(batch_size=batch_size, seq_len=seq_len, generator_dir='./checkpoint/' + 'mpradragonn_sample' + '/', sequence_template=sequence_template)
#Function for optimizing n sequences for a target predictor
def optimize_sequences(act_maximizer, n_seqs, batch_size=1, latent_size=128, n_iters=100, eps1=0., eps2=0.1, noise_std=1e-6, use_adam=True, run_name='default', store_intermediate_n_seqs=None, store_every_iter=100) :
z = Variable(torch.randn(batch_size, latent_size, device="cuda"), requires_grad=True)
norm_var = torch_normal(0, 1)
optimizer = None
if use_adam :
optimizer = optim.Adam([z], lr=eps2)
else :
optimizer = optim.SGD([z], lr=1)
z.register_hook(lambda grad, batch_size=batch_size, latent_size=latent_size, noise_std=noise_std: grad + noise_std * torch.randn(batch_size, latent_size, device="cuda"))
seqs = []
fitness_histo = []
n_batches = n_seqs // batch_size
for batch_i in range(n_batches) :
if batch_i % 4 == 0 :
print("Optimizing sequence batch " + str(batch_i))
#Re-initialize latent GAN seed
z.data = torch.randn(batch_size, latent_size, device="cuda")
fitness_scores_batch = [act_maximizer(z)[:, 0].data.cpu().numpy().reshape(-1, 1)]
for curr_iter in range(n_iters) :
fitness_score = act_maximizer(z)[:, 0]
fitness_loss = -torch.sum(fitness_score)
z_prior = -torch.sum(norm_var.log_prob(z))
loss = None
if use_adam :
loss = fitness_loss
else :
loss = eps1 * z_prior + eps2 * fitness_loss
optimizer.zero_grad()
loss.backward()
optimizer.step()
fitness_scores_batch.append(fitness_score.data.cpu().numpy().reshape(-1, 1))
if store_intermediate_n_seqs is not None and batch_i * batch_size < store_intermediate_n_seqs and curr_iter % store_every_iter == 0 :
onehot_batch = act_maximizer.get_pattern(z).data.cpu().numpy()
seq_batch = [
acgt_encoder.decode(onehot_batch[k]) for k in range(onehot_batch.shape[0])
]
with open(run_name + "_curr_iter_" + str(curr_iter) + ".txt", "a+") as f :
for i in range(len(seq_batch)) :
seq = seq_batch[i]
f.write(seq + "\n")
onehot_batch = act_maximizer.get_pattern(z).data.cpu().numpy()
seq_batch = [
acgt_encoder.decode(onehot_batch[k]) for k in range(onehot_batch.shape[0])
]
seqs.extend(seq_batch)
fitness_histo.append(np.concatenate(fitness_scores_batch, axis=1))
fitness_histo = np.concatenate(fitness_histo, axis=0)
return seqs, fitness_histo
n_seqs = 4096#960
n_iters = 1000
run_name = 'killoran_mpradragonn_' + str(n_seqs) + "_sequences" + "_" + str(n_iters) + "_iters_sample_wgan"
seqs, fitness_scores = optimize_sequences(
act_maximizer,
n_seqs,
batch_size=64,
latent_size=128,
n_iters=n_iters,
eps1=0.,
eps2=0.1,
noise_std=1e-6,
use_adam=True,
run_name="samples/killoran_mpradragonn/" + run_name,
store_intermediate_n_seqs=None,#960,
store_every_iter=100
)
#Plot fitness statistics of optimization runs
#Plot k trajectories
plot_n_traj = 100
f = plt.figure(figsize=(8, 6))
for i in range(min(plot_n_traj, n_seqs)) :
plt.plot(fitness_scores[i, :], linewidth=2, alpha=0.75)
plt.xlabel("Training iteration", fontsize=14)
plt.ylabel("Fitness score", fontsize=14)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlim(0, n_iters)
plt.ylim(-3, 3)
plt.tight_layout()
plt.show()
#Plot mean trajectory
f = plt.figure(figsize=(8, 6))
plt.plot(np.mean(fitness_scores, axis=0), linewidth=2, alpha=0.75)
plt.xlabel("Training iteration", fontsize=14)
plt.ylabel("Fitness score", fontsize=14)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlim(0, n_iters)
plt.ylim(-3, 3)
plt.tight_layout()
plt.show()
#Save sequences to file
with open(run_name + ".txt", "wt") as f :
for i in range(len(seqs)) :
seq = seqs[i]
f.write(seq + "\n")
```
|
github_jupyter
|
```
import os
import sys
sys.path.append('../')
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from pprint import pprint
from scipy.optimize import curve_fit
import src.io as sio
import src.preprocessing as spp
import src.fitting as sft
AFM_FOLDER = sio.get_folderpath("20200818_Akiyama_AFM")
AFM_FOLDER1 = sio.get_folderpath("20200721_Akiyama_AFM")
AFM_FOLDER2 = sio.get_folderpath("20200824_Akiyama_AFM")
AFM_FOLDER3 = sio.get_folderpath("20200826_TFSC_Preamp_AFM/11613_Tip_5/Akiyama_Tip_Stage")
AFM_FOLDER4 = sio.get_folderpath("20200826_TFSC_Preamp_AFM/11613_Tip_5/Custom_Tip_Stage")
AFM_FOLDER5 = sio.get_folderpath("20200828_Tip_Approach1")
AFM_FOLDER6 = sio.get_folderpath("20200901_Tip_Approach_2/Actual_tip_approach")
```
# Approach
```
params, data = sio.read_dat(AFM_FOLDER6 + "HistoryData001.dat")
amplitude = data["Amplitude (m)"].values
fig, ax = plt.subplots()
ax.plot(amplitude*1e9)
ax.set_ylabel("Amplitude (nm)")
ax.set_xlabel("Time (a.u.)")
#plt.savefig("snap.jpg", dpi=600)
```
## 20200721_Akiyama_AFM
```
params, data = sio.read_dat(AFM_FOLDER1 + "frq-sweep002.dat")
freq_shift = data["Frequency Shift (Hz)"].values
amplitude = data["Amplitude (m)"].values
phase = data["Phase (deg)"].values
amp_freq_sweep = sft.fit_fano(freq_shift, amplitude, linear_offset=True)
phase_freq_sweep = sft.fit_fano(freq_shift, phase)
%matplotlib inline
fig, (ax1, ax2) = plt.subplots(nrows=2, sharex=True)
ax1.plot(freq_shift, amplitude*1e12)
#ax1.plot(freq_shift, amp_freq_sweep.best_fit)
ax1.set_ylabel("Amplitude (pm)")
ax2.plot(freq_shift, phase)
#ax2.plot(freq_shift, phase_freq_sweep.best_fit)
ax2.set_ylabel(data.columns[3])
ax2.set_xlabel(data.columns[0])
#plt.savefig("second.jpg", dpi=600)
```
Quality factor can be calculated as $ Q = \frac{f_R}{\Delta f} $
```
print(f'Q-factor= {params["f_res (Hz)"] / amp_freq_sweep.params["fwhm"].value}')
```
## 20200818_Akiyama_AFM
```
params, data = sio.read_dat(AFM_FOLDER + "frq-sweep001.dat")
#pprint(params, sort_dicts=False)
freq_shift = data["Frequency Shift (Hz)"]
amplitude = data["Amplitude (m)"]
phase = data["Phase (deg)"]
fano = sft.fit_fano(freq_shift, amplitude)
lorentzian = sft.fit_fano(freq_shift, phase)
params
```
## Equations for calculating Q factor
$$ Q = \frac{f_R}{\Delta f} $$
$$ Q = \frac{A(\omega_0)}{A_{in}} $$
```
f_res = 44379.7064
sigma = 62.2841355
print(f_res/sigma)
A_drive = 50e-3
A_res = 28.3e-6 * 1 / 500e-6
print(A_res/A_drive)
# Calibration
A_drive = 50e-3
osc_amp = 50e-9
print(osc_amp/A_drive)
```
## Plot frequency sweep curves
```
fig, (ax1, ax2) = plt.subplots(nrows=2, sharex=True)
ax1.plot(freq_shift, amplitude)
ax1.plot(freq_shift, fano.best_fit)
ax1.set_ylabel(data.columns[2])
ax2.plot(freq_shift, phase)
ax2.plot(freq_shift, lorentzian.best_fit)
ax2.set_ylabel(data.columns[3])
ax2.set_xlabel(data.columns[1])
```
## Extract fit values
```
print("{} = {:.1f} +- {:.1f}".format(fano.params["sigma"].name, fano.params["sigma"].value, fano.params["sigma"].stderr))
print("{} = {:.2e} +- {:.0e}".format(fano.params["amplitude"].name, fano.params["amplitude"].value, fano.params["amplitude"].stderr))
```
# 20200824_Akiyama_AFM
## Automatically read files from disk
Reads all files stored in **AFM_FOLDER2 = "20200824_Akiyama_AFM/"** and plots the amplitude and phase data.
Optionally, the data can be fit to Fano resonances by setting the variable
```python
fit = True
```
The Q-factor is calculated as:
$$ Q = \frac{f_R}{\Delta f} = \frac{f_R}{2 \sigma} $$
Errors are calculated as (this also gives an estimate of the SNR):
$$ \frac{\Delta Q}{Q} = \sqrt{ \left( \frac{\Delta (\Delta f)}{\Delta f} \right)^2 + \left( \frac{\Delta (\sigma)}{\sigma} \right)^2 } $$
Another estimate of the SNR, is the Chi square or weighted sum of squared deviations (lower is better):
$$ \chi^2 = \sum_{i} {\frac{(O_i - C_i)^2}{\sigma_i^2}} $$
```
%matplotlib inline
fit = False # Setting to True will take slightly longer due to the fitting protocols
files = []
for file in os.listdir(AFM_FOLDER2):
if file.endswith(".dat"):
files.append(file)
fig, ax = plt.subplots(nrows=len(files), ncols=2)
for idx, file in enumerate(files):
params, data = sio.read_dat(AFM_FOLDER2 + file)
freq_shift = data["Frequency Shift (Hz)"]
amplitude = data["Amplitude (m)"]
phase = data["Phase (deg)"]
ax[idx, 0].plot(freq_shift, amplitude)
ax[idx, 0].set_ylabel(data.columns[2])
ax[idx, 0].set_title(file)
ax[idx, 1].plot(freq_shift, phase)
ax[idx, 1].set_ylabel(data.columns[3])
ax[idx, 1].set_title(file)
if fit:
fano1 = sft.fit_fano(freq_shift, amplitude)
q_factor = (params["Center Frequency (Hz)"] + fano1.params["center"].value) / (2 * fano1.params["sigma"].value)
q_factor_err = q_factor * np.sqrt((fano1.params["center"].stderr/fano1.params["center"].value)**2 + (fano1.params["sigma"].stderr/fano1.params["sigma"].value)**2)
ax[idx, 0].plot(freq_shift, fano1.best_fit, label="Q={:.0f}$\pm{:.0f}$".format(q_factor, q_factor_err))
ax[idx, 0].legend()
fano2 = sft.fit_fano(freq_shift, phase, linear_offset=True)
ax[idx, 1].plot(freq_shift, fano2.best_fit)
print("chi-square ({}) = {:.2e}".format(file, fano1.chisqr))
fig.tight_layout()
fig.text(0.5, 0.02, data.columns[1], ha='center', va='center')
```
## 20200826_TFSC_Preamp_AFM
### 11613_Tip_5
```
%matplotlib inline
fit = False # Setting to True will take slightly longer due to the fitting protocols
files = []
for file in os.listdir(AFM_FOLDER4):
if file.endswith(".dat"):
files.append(file)
fig, ax = plt.subplots(nrows=len(files), ncols=2)
for idx, file in enumerate(files):
params, data = sio.read_dat(AFM_FOLDER4 + file)
freq_shift = data["Frequency Shift (Hz)"]
amplitude = data["Amplitude (m)"]
phase = data["Phase (deg)"]
ax[idx, 0].plot(freq_shift, amplitude)
ax[idx, 0].set_ylabel(data.columns[2])
ax[idx, 0].set_title(file)
ax[idx, 1].plot(freq_shift, phase)
ax[idx, 1].set_ylabel(data.columns[3])
ax[idx, 1].set_title(file)
if fit:
fano1 = sft.fit_fano(freq_shift, amplitude)
q_factor = (params["Center Frequency (Hz)"] + fano1.params["center"].value) / (fano1.params["sigma"].value)
q_factor_err = q_factor * np.sqrt((fano1.params["center"].stderr/fano1.params["center"].value)**2 + (fano1.params["sigma"].stderr/fano1.params["sigma"].value)**2)
ax[idx, 0].plot(freq_shift, fano1.best_fit, label="Q={:.0f}$\pm{:.0f}$".format(q_factor, q_factor_err))
ax[idx, 0].legend()
fano2 = sft.fit_fano(freq_shift, phase, linear_offset=True)
ax[idx, 1].plot(freq_shift, fano2.best_fit)
print("chi-square ({}) = {:.2e}".format(file, fano1.chisqr))
fig.tight_layout()
fig.text(0.5, 0.02, data.columns[1], ha='center', va='center')
omega_0 = 1
omega = np.linspace(0, 2, 1000)
Q = 1
ratio = omega / omega_0
phi = np.arctan(-ratio / (Q * (1 - ratio**2)))
fid, ax = plt.subplots()
ax.plot(ratio, phi)
```
# Calibration from Thermal Noise density
From Atomic Force Microscopy, Second Edition by Bert Voigtländer
Section 11.6.5 Experimental Determination of the Sensitivity and Spring Constant in AFM Without Tip-Sample Contact
Eq. 11.28 and 11.26
```
%matplotlib widget
file = "SignalAnalyzer_Spectrum001"
params, data = sio.read_dat(AFM_FOLDER4 + file)
calibration_params = sft.find_afm_calibration_parameters(data, frequency_range=[40000, 48000], Q=1000, f_0_guess=44000)
fig, ax = plt.subplots()
ax.plot(calibration_params["Frequency (Hz)"], calibration_params["PSD squared (V**2/Hz)"])
ax.plot(calibration_params["Frequency (Hz)"], calibration_params["PSD squared fit (V**2/Hz)"])
print("Calibration (m/V) =", calibration_params["Calibration (m/V)"])
%matplotlib inline
fit = False # Setting to True will take slightly longer due to the fitting protocols
files = []
for file in os.listdir("../../Data/" + AFM_FOLDER4):
if file.endswith(".dat"):
files.append(file)
files = ["frq-sweep002.dat"]
fig, ax = plt.subplots(nrows=len(files), ncols=2)
for idx, file in enumerate(files):
params, data = sio.read_dat(AFM_FOLDER4 + file)
freq_shift = data["Frequency Shift (Hz)"]
amplitude = data["Amplitude (m)"]
phase = data["Phase (deg)"]
if len(files) == 1:
ax[0].plot(freq_shift, amplitude)
ax[0].set_ylabel(data.columns[2])
ax[1].plot(freq_shift, phase)
ax[1].set_ylabel(data.columns[3])
else:
ax[idx, 0].plot(freq_shift, amplitude)
ax[idx, 0].set_ylabel(data.columns[2])
ax[idx, 0].set_title(file)
ax[idx, 1].plot(freq_shift, phase)
ax[idx, 1].set_ylabel(data.columns[3])
ax[idx, 1].set_title(file)
if fit:
fano1 = sft.fit_fano(freq_shift, amplitude)
#q_factor = (params["Center Frequency (Hz)"] + fano1.params["center"].value) / (fano1.params["sigma"].value)
#q_factor_err = q_factor * np.sqrt((fano1.params["center"].stderr/fano1.params["center"].value)**2 + (fano1.params["sigma"].stderr/fano1.params["sigma"].value)**2)
ax[idx, 0].plot(freq_shift, fano1.best_fit)
ax[idx, 0].legend()
fano2 = sft.fit_fano(freq_shift, phase, linear_offset=True)
ax[idx, 1].plot(freq_shift, fano2.best_fit)
print("chi-square ({}) = {:.2e}".format(file, fano1.chisqr))
fig.tight_layout()
fig.text(0.5, 0.02, data.columns[1], ha='center', va='center')
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/yarusx/cat-vs-dogo/blob/main/cat_vs_dog_0_0_2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import matplotlib.pyplot as plt
import numpy as np
import os
import tensorflow as tf
from tensorflow.keras.preprocessing import image_dataset_from_directory
_URL = 'https://storage.googleapis.com/mledu-datasets/cats_and_dogs_filtered.zip'
path_to_zip = tf.keras.utils.get_file('cats_and_dogs.zip', origin=_URL, extract=True)
PATH = os.path.join(os.path.dirname(path_to_zip), 'cats_and_dogs_filtered')
train_dir = os.path.join(PATH, 'train')
validation_dir = os.path.join(PATH, 'validation')
BATCH_SIZE = 32
IMG_SIZE = (160, 160)
train_dataset = image_dataset_from_directory(train_dir,
shuffle=True,
batch_size=BATCH_SIZE,
image_size=IMG_SIZE)
validation_dataset = image_dataset_from_directory(validation_dir,
shuffle=True,
batch_size=BATCH_SIZE,
image_size=IMG_SIZE)
class_names = train_dataset.class_names
# plt.figure(figsize=(10, 10))
# for images, labels in train_dataset.take(1):
# for i in range(9):
# ax = plt.subplot(3, 3, i + 1)
# plt.imshow(images[i].numpy().astype("uint8"))
# plt.title(class_names[labels[i]])
# plt.axis("off")
val_batches = tf.data.experimental.cardinality(validation_dataset)
test_dataset = validation_dataset.take(val_batches // 5)
validation_dataset = validation_dataset.skip(val_batches // 5)
AUTOTUNE = tf.data.experimental.AUTOTUNE
train_dataset = train_dataset.prefetch(buffer_size=AUTOTUNE)
validation_dataset = validation_dataset.prefetch(buffer_size=AUTOTUNE)
test_dataset = test_dataset.prefetch(buffer_size=AUTOTUNE)
data_augmentation = tf.keras.Sequential([
tf.keras.layers.experimental.preprocessing.RandomFlip('horizontal'),
tf.keras.layers.experimental.preprocessing.RandomRotation(0.2),
])
# for image, _ in train_dataset.take(1):
# plt.figure(figsize=(10, 10))
# first_image = image[0]
# for i in range(9):
# ax = plt.subplot(3, 3, i + 1)
# augmented_image = data_augmentation(tf.expand_dims(first_image, 0))
# plt.imshow(augmented_image[0] / 255)
# plt.axis('off')
rescale = tf.keras.layers.experimental.preprocessing.Rescaling(1./127.5, offset= -1)
# Create the base model from the pre-trained model MobileNet V2
IMG_SHAPE = IMG_SIZE + (3,)
base_model = tf.keras.applications.MobileNetV2(input_shape=IMG_SHAPE,
include_top=False,
weights='imagenet')
image_batch, label_batch = next(iter(train_dataset))
feature_batch = base_model(image_batch)
print(feature_batch.shape)
base_model.trainable = False
global_average_layer = tf.keras.layers.GlobalAveragePooling2D()
feature_batch_average = global_average_layer(feature_batch)
print(feature_batch_average.shape)
prediction_layer = tf.keras.layers.Dense(1)
prediction_batch = prediction_layer(feature_batch_average)
print(prediction_batch.shape)
inputs = tf.keras.Input(shape=(160, 160, 3))
x = data_augmentation(inputs)
x = rescale(x)
x = base_model(x, training=False)
x = global_average_layer(x)
x = tf.keras.layers.Dropout(0.4)(x)
outputs = prediction_layer(x)
model = tf.keras.Model(inputs, outputs)
base_learning_rate = 0.0001
model.compile(optimizer=tf.keras.optimizers.Adam(lr=base_learning_rate),
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'])
loss0, accuracy0 = model.evaluate(validation_dataset)
print("initial loss: {:.2f}".format(loss0))
print("initial accuracy: {:.2f}".format(accuracy0))
initial_epochs = 1
history = model.fit(train_dataset,
epochs=initial_epochs,
validation_data=validation_dataset)
val_acc = history.history['val_accuracy']
while np.mean(val_acc)*100 < 98.5:
initial_epochs = 3
history = model.fit(train_dataset,
epochs=initial_epochs,
validation_data=validation_dataset)
val_acc = history.history['val_accuracy']
try:
!mkdir -p saved_model
except:
pass
model.save('saved_model/dvc/')
!zip -r dvc.zip saved_model/dvc/
from google.colab import files
files.download("dvc.zip")
from google.colab import drive
drive.mount('/content/drive')
!unzip -q /content/drive/MyDrive/dvc.zip
dvc = tf.keras.models.load_model('/content/saved_model/dvc')
try:
!mkdir -p saved_model
except:
pass
model.save('saved_model/dvc/')
!zip -r dvc.zip saved_model/dvc/
from google.colab import files
files.download("dvc.zip")
from keras.preprocessing.image import load_img, img_to_array
# load and prepare the image
def load_image(filename):
# load the image
img = load_img(filename, target_size=(160, 160))
# convert to array
img = img_to_array(img)
# reshape into a single sample with 3 channels
img = img.reshape(1, 160, 160, 3)
return img
img = load_image('/content/drive/MyDrive/dayana_1.JPG')
categories = ["Cat", "Dog"]
prediction = dvc_model.predict(img)
prediction = tf.nn.sigmoid(prediction)
print(prediction)
plt.figure()
plt.imshow(img[0]/255)
plt.title(categories[int(np.round_(prediction))])
loss0, accuracy0 = model.evaluate(validation_dataset)
# #Retrieve a batch of images from the test set
# image_batch, label_batch = test_dataset.as_numpy_iterator().next()
# predictions = model.predict_on_batch(image_batch).flatten()
# # Apply a sigmoid since our model returns logits
# predictions = tf.nn.sigmoid(predictions)
# predictions = tf.where(predictions < 0.5, 0, 1)
# print('Predictions:\n', predictions.numpy())
# print('Labels:\n', label_batch)
# plt.figure(figsize=(10, 10))
# for i in range(9):
# ax = plt.subplot(3, 3, i + 1)
# plt.imshow(image_batch[i].astype("uint8"))
# plt.title(class_names[predictions[i]])
# plt.axis("off")
```
|
github_jupyter
|
```
# some_file.py
import sys
# insert at 1, 0 is the script path (or '' in REPL)
sys.path.insert(1, "/Users/dhruvbalwada/work_root/sogos/")
import os
from numpy import *
import pandas as pd
import xarray as xr
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
from xgcm import Grid
from xgcm.autogenerate import generate_grid_ds
import sogos.download_product as dlp
import sogos.load_product as ldp
import sogos.time_tools as tt
import sogos.geo_tools as gt
import sogos.download_file as df
import gsw
import cmocean as cmocean
```
# Download Latest Data
```
data_dir = "/Users/dhruvbalwada/work_root/sogos/data/raw/climatology/"
```
FTP ADDRESS: ftp://kakapo.ucsd.edu/pub/gilson/argo_climatology/
Data prior to 2017 (till Dec 2016) is in a single file
```
# download the big climatology files
wget.download(
"ftp://kakapo.ucsd.edu/pub/gilson/argo_climatology/RG_ArgoClim_Salinity_2017.nc.gz",
data_dir,
)
wget.download(
"ftp://kakapo.ucsd.edu/pub/gilson/argo_climatology/RG_ArgoClim_Temperature_2017.nc.gz",
data_dir,
)
from ftplib import FTP
ftp_address = "ftp://kakapo.ucsd.edu/pub/gilson/argo_climatology/RG_ArgoClim_2019"
url_root = "/pub/gilson/argo_climatology/"
ftp_root = "kakapo.ucsd.edu"
ftp = FTP(ftp_root)
ftp.login()
ftp.cwd(url_root)
contents = ftp.nlst("RG_ArgoClim_2017*")
contents = ftp.nlst("RG_ArgoClim_201*")
for i in contents:
print("Downloading" + i)
wget.download("ftp://kakapo.ucsd.edu/pub/gilson/argo_climatology/" + i, data_dir)
```
## Load some data
```
Tclim = xr.open_dataset(data_dir + "RG_ArgoClim_Temperature_2017.nc", decode_times=False)
Sclim = xr.open_dataset(data_dir + "RG_ArgoClim_Salinity_2017.nc", decode_times=False)
Climextra = xr.open_mfdataset(data_dir+ 'RG_ArgoClim_201*', decode_times=False)
RG_clim = xr.merge([Tclim, Sclim, Climextra])
# Calendar type was missing, and giving errors in decoding time
RG_clim.TIME.attrs['calendar'] = '360_day'
RG_clim = xr.decode_cf(RG_clim)
## Add density and other things
SA = xr.apply_ufunc(gsw.SA_from_SP, RG_clim.ARGO_SALINITY_MEAN+RG_clim.ARGO_SALINITY_ANOMALY, RG_clim.PRESSURE ,
RG_clim.LONGITUDE, RG_clim.LATITUDE,
dask='parallelized', output_dtypes=[float,]).rename('SA')
CT = xr.apply_ufunc(gsw.CT_from_t, SA, RG_clim.ARGO_TEMPERATURE_MEAN+RG_clim.ARGO_SALINITY_ANOMALY, RG_clim.PRESSURE,
dask='parallelized', output_dtypes=[float,]).rename('CT')
SIGMA0 = xr.apply_ufunc(gsw.sigma0, SA, CT, dask='parallelized', output_dtypes=[float,]).rename('SIGMA0')
RG_clim = xr.merge([RG_clim, SIGMA0])
T_region = RG_clim.ARGO_TEMPERATURE_ANOMALY.groupby('TIME.season').mean() + RG_clim.ARGO_TEMPERATURE_MEAN
S_region = RG_clim.ARGO_SALINITY_ANOMALY.groupby('TIME.season').mean() + RG_clim.ARGO_SALINITY_MEAN
rho_region = RG_clim.SIGMA0.groupby('TIME.season').mean()
plt.figure(figsize=(18,3))
plt.subplot(141)
T_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='DJF').plot.contourf(levels=11, vmin=-9, vmax=9);
plt.subplot(142)
T_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='MAM').plot.contourf(levels=11, vmin=-9, vmax=9);
plt.subplot(143)
T_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='JJA').plot.contourf(levels=11, vmin=-9, vmax=9);
plt.subplot(144)
T_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='SON').plot.contourf(levels=11, vmin=-9, vmax=9);
plt.figure(figsize=(18,3))
plt.subplot(141)
S_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='DJF').plot.contourf(levels=11, vmin=33.7, vmax=34.2)
plt.subplot(142)
S_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='MAM').plot.contourf(levels=11, vmin=33.7, vmax=34.2)
plt.subplot(143)
S_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='JJA').plot.contourf(levels=11, vmin=33.7, vmax=34.2)
plt.subplot(144)
S_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='SON').plot.contourf(levels=11, vmin=33.7, vmax=34.2)
plt.tight_layoutout()
plt.figure(figsize=(18,3))
plt.subplot(141)
rho_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='DJF').plot.contourf(levels=11)
plt.subplot(142)
rho_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='MAM').plot.contourf(levels=11)
plt.subplot(143)
rho_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='JJA').plot.contourf(levels=11)
plt.subplot(144)
rho_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='SON').plot.contourf(levels=11)
```
### Some Climatological Mean Sections
```
dens_section30 = RG_clim.SIGMA0.sel(LONGITUDE=30, method='nearest').sel(LATITUDE=slice(-70,-40)).load()
dens_section40 = RG_clim.SIGMA0.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).load()
glider = {"start_month": 4.99, "end_month":7.8, "start_lat": -51.5, "end_lat": -53, "max_depth": 1000}
plt.figure(figsize=(15,4))
plt.subplot(121)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=30, method='nearest').sel(LATITUDE=slice(-70,-40)
).plot.contourf(vmin=-10, levels=24)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=30, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(linestyles='-.',levels=[1])
dens_section.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C0',
linestyles='dashed', linewidths=4)
plt.plot([glider['start_lat'], glider['start_lat']], [4, glider['max_depth']], color='C2', alpha=0.5)
plt.plot([glider['end_lat'], glider['end_lat']], [4, glider['max_depth']], color='C2', alpha=0.5)
plt.gca().invert_yaxis()
plt.subplot(122)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contourf(vmin=33.77, vmax=35.21, levels=24, cmap=cmocean.cm.haline)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(levels=[34.2], linestyles='dashdot')
dens_section.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.plot([glider['start_lat'], glider['start_lat']], [4, 1900], color='C3', alpha=0.5)
plt.gca().invert_yaxis()
plt.tight_layout()
#plt.savefig('../figures/clim_TS_30E.png')
plt.figure(figsize=(15,4))
plt.subplot(121)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)
).plot.contourf(vmin=-10,levels=24)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(linestyles='-.',levels=[1])
dens_section40.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section40.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C0',
linestyles='dashed', linewidths=4)
plt.gca().invert_yaxis()
plt.subplot(122)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contourf(vmin=33.77, vmax=35.21,levels=24, cmap=cmocean.cm.haline)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(levels=[34.2], linestyles='dashdot')
dens_section40.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section40.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.gca().invert_yaxis()
plt.tight_layout()
plt.savefig('../figures/clim_TS_40E.png')
plt.figure(figsize=(12,4))
plt.subplot(121)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contourf(levels=24)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(levels=[1])
plt.gca().invert_yaxis()
plt.subplot(122)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contourf(levels=24)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(levels=[34.2])
plt.gca().invert_yaxis()
plt.tight_layout()
# Not much seasonality below 200m
dens_section.groupby('TIME.month').mean().mean('month').sel(PRESSURE=slice(0,1000)).plot.contourf(cmap='Blues')
dens_section.groupby('TIME.month').mean().isel(month=0).sel(PRESSURE=slice(0,1000)).plot.contour(levels=[27.2])
dens_section.groupby('TIME.month').mean().isel(month=3).sel(PRESSURE=slice(0,1000)).plot.contour(levels=[27.2])
dens_section.groupby('TIME.month').mean().isel(month=6).sel(PRESSURE=slice(0,1000)).plot.contour(levels=[27.2])
dens_section.groupby('TIME.month').mean().isel(month=9).sel(PRESSURE=slice(0,1000)).plot.contour(levels=[27.2])
plt.gca().invert_yaxis()
```
## N2
\begin{equation}
N^2 = db/dz
\end{equation}
$b = -\frac{g}{\rho_0} \rho'$
$b = g(\alpha \triangle T - \beta \triangle S)$
```
RG_clim
RG_clim = generate_grid_ds(RG_clim,
{'Z':'PRESSURE', 'X':'LONGITUDE', 'Y':'LATITUDE'})
grid = Grid(RG_clim, periodic='X')
g = 9.81
rho0 = 1000
dens_clim_monthly = RG_clim.SIGMA0.groupby('TIME.month').mean()
dens_clim_monthly
N2_clim_monthly = grid.interp(-g/rho0* grid.diff(dens_clim_monthly, 'Z', boundary='extend') / -(grid.diff(RG_clim.PRESSURE, 'Z', boundary='extend')), 'Z', boundary='extend')
N2_clim_monthly_SO = N2_clim_monthly.sel(LATITUDE=slice(-70, -30)).load()
N2_clim_monthly_SO = N2_clim_monthly_SO.rename('N2')
plt.figure(figsize=(18,12))
plt.subplot(221)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=1).plot(vmin=-5e-5)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=1).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('January')
plt.gca().invert_yaxis()
plt.subplot(222)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=4).plot(vmin=-5e-5)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=4).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('April')
plt.gca().invert_yaxis()
plt.subplot(223)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=7).plot(vmin=-5e-5)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=7).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('July')
plt.gca().invert_yaxis()
plt.subplot(224)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=10).plot(vmin=-5e-5)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=10).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('October')
plt.gca().invert_yaxis()
plt.savefig('../figures/clim_N2_30E.png')
plt.figure(figsize=(8,3))
plt.subplot(121)
#plt.pcolormesh(N2_clim_monthly_SO.LATITUDE.sel(LATITUDE=slice(-70, -40)),
# N2_clim_monthly_SO.LATITUDE.sel(=slice(-70, -40)),
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=4).plot(vmin=-5e-5,
rasterized=True,add_colorbar=False)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=4).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('April')
plt.gca().invert_yaxis()
plt.ylim([1500, 0])
plt.xlabel('Latitude')
plt.ylabel('Depth (m)')
plt.subplot(122)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=9).plot(vmin=-5e-5,
rasterized=True,add_colorbar=False)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=9).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('September')
plt.gca().invert_yaxis()
plt.ylim([1500, 0])
plt.xlabel('Latitude')
plt.ylabel('Depth (m)')
plt.tight_layout()
plt.savefig('N2_climatology.pdf')
N2.sel(LATITUDE=slice(-60, -40)).sel(LONGITUDE=30, method='nearest').isel(TIME=-1).plot()
```
The apply ufunc way, not working yet.
```
CT_clim = CT.groupby('TIME.month').mean()
SA_clim = SA.groupby('TIME.month').mean()
CT_clim_region = CT_clim.sel(LATITUDE=slice(-65,-35), LONGITUDE=slice(20,50)).load()
SA_clim_region = SA_clim.sel(LATITUDE=slice(-65,-35), LONGITUDE=slice(20,50)).load()
(N2, pmid) = xr.apply_ufunc(gsw.Nsquared, SA_clim_region, CT_clim_region, RG_clim.PRESSURE,
dask='parallelized',
input_core_dims=[['PRESSURE'],['PRESSURE'],['PRESSURE']],
output_core_dims=[['PRESSURE'],['PRESSURE']], exclude_dims=set(['PRESSURE']))
```
### Gestrophic Velocities
```
psi = xr.apply_ufunc(gsw.geo_strf_dyn_height, SA, CT , RG_clim.PRESSURE,
dask='parallelized', output_dtypes=[float,]).rename('psi')
psi
vels = xr.apply_ufunc(gsw.geostrophic_velocity, psi, psi.LONGITUDE, psi.LATITUDE,
dask='parallelized', output_core_dims=[4,4], output_dtypes=[float,]).rename('vels')
vels
```
### Mixed Layer Depth
Ended up going with Holte's climatology for MLD work
```
delta_dens = RG_clim.SIGMA0 - RG_clim.SIGMA0.isel(PRESSURE=0)
import nc_time_axis
RG_clim.SIGMA0.sel(LONGITUDE=30, method='nearest').sel( LATITUDE=slice(-63,-45)).isel(TIME=-1).plot.contourf()
plt.gca().invert_yaxis()
RG_clim.SIGMA0.sel(LONGITUDE=30, LATITUDE=-50, method='nearest').isel(TIME=-1).plot()
RG_clim.SIGMA0.sel(LONGITUDE=30, LATITUDE=-50, method='nearest').isel(TIME=-7).plot()
RG_clim.SIGMA0.sel(LONGITUDE=30, LATITUDE=-60, method='nearest').isel(TIME=-1).plot()
RG_clim.SIGMA0.sel(LONGITUDE=30, LATITUDE=-60, method='nearest').isel(TIME=-7).plot()
delta_dens.sel(LONGITUDE=30, LATITUDE=-50, method='nearest').isel(TIME=-1).plot()
temp = delta_dens.where(delta_dens>0.03).sel(LONGITUDE=30, LATITUDE=-50, method='nearest').isel(TIME=-1).plot()
temp = delta_dens.where(delta_dens>0.03)
MLD = temp.PRESSURE.where(temp == temp.min('PRESSURE')).min('PRESSURE')
MLD_clim = temp.PRESSURE.where(temp == temp.min('PRESSURE')).min('PRESSURE').groupby('TIME.month').mean()
MLD_clim.load()
MLD_clim.month
plt.figure(figsize=(12,4))
plt.subplot(121)
MLD_clim.sel(LATITUDE=slice(-75,-25), LONGITUDE=slice(20,90)).sel(month=1).plot(vmin=0, vmax=120)
plt.subplot(122)
MLD_clim.sel(LATITUDE=slice(-75,-25), LONGITUDE=slice(20,90)).sel(month=7).plot(vmin=0, vmax=120)
MLD_clim.max('month').sel(LATITUDE=slice(-75,-25), LONGITUDE=slice(20,380)).plot(vmin=0, vmax=150)
deltaH = MLD_clim.max('month') - MLD_clim.min('month')
deltaH.sel(LATITUDE=slice(-75,-25), LONGITUDE=slice(20,380)).plot(vmin=0, vmax=80)
MLD_clim.sel(LATITUDE=-45, LONGITUDE=35, method='nearest').plot(label='45S')
MLD_clim.sel(LATITUDE=-50, LONGITUDE=35, method='nearest').plot(label='50S')
MLD_clim.sel(LATITUDE=-55, LONGITUDE=35, method='nearest').plot(label='55S')
MLD_clim.sel(LATITUDE=-60, LONGITUDE=35, method='nearest').plot(label='60S')
plt.legend()
MLD_clim.sel(LATITUDE=-45, LONGITUDE=45, method='nearest').plot(label='45S')
MLD_clim.sel(LATITUDE=-50, LONGITUDE=45, method='nearest').plot(label='50S')
MLD_clim.sel(LATITUDE=-55, LONGITUDE=45, method='nearest').plot(label='55S')
MLD_clim.sel(LATITUDE=-60, LONGITUDE=45, method='nearest').plot(label='60S')
plt.legend()
```
|
github_jupyter
|
# User testing for for Scikit-Yellowbrick
### Using data that was recorded from sensors during Data Science Certificate Program at GW
https://github.com/georgetown-analytics/classroom-occupancy
Data consist of temperature, humidity, CO2 levels, light, # of bluetooth devices, noise levels and count of people in the room.
```
import pandas as pd
%matplotlib inline
dataset = pd.read_csv('dataset.csv')
dataset.head(5)
dataset.count_total.describe()
#add a new column to create a binary class for room occupancy
countmed = dataset.count_total.median()
dataset['room_occupancy'] = dataset['count_total'].apply(lambda x: 'occupied' if x > 4 else 'empty')
# map room occupancy to a number
dataset['room_occupancy_num'] = dataset.room_occupancy.map({'empty':0, 'occupied':1})
dataset.head(5)
dataset.room_occupancy.describe()
import os
import sys
# Modify the path
sys.path.append("..")
import pandas as pd
import yellowbrick as yb
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (12, 8)
g = yb.anscombe()
```
## Feature Analysis
Feature analysis visualizers are designed to visualize instances in data space in order to detect features or targets that might impact downstream fitting. Because ML operates on high-dimensional data sets (usually at least 35), the visualizers focus on aggregation, optimization, and other techniques to give overviews of the data. It is our intent that the steering process will allow the data scientist to zoom and filter and explore the relationships between their instances and between dimensions.
At the moment we have three feature analysis visualizers implemented:
Rank2D: rank pairs of features to detect covariance
RadViz: plot data points along axes ordered around a circle to detect separability
Parallel Coordinates: plot instances as lines along vertical axes to detect clusters
Feature analysis visualizers implement the Transformer API from Scikit-Learn, meaning they can be used as intermediate transform steps in a Pipeline (particularly a VisualPipeline). They are instantiated in the same way, and then fit and transform are called on them, which draws the instances correctly. Finally show or show is called which displays the image.
```
from yellowbrick.features.rankd import Rank2D
from yellowbrick.features.radviz import RadViz
from yellowbrick.features.pcoords import ParallelCoordinates
```
### Rank2D
Rank1D and Rank2D evaluate single features or pairs of features using a variety of metrics that score the features on the scale [-1, 1] or [0, 1] allowing them to be ranked. A similar concept to SPLOMs, the scores are visualized on a lower-left triangle heatmap so that patterns between pairs of features can be easily discerned for downstream analysis.
```
# Load the classification data set
data = dataset
# Specify the features of interest
features = ['temperature','humidity','co2','light','noise','bluetooth_devices']
# Extract the numpy arrays from the data frame
X = data[features].as_matrix()
y = data['count_total'].as_matrix()
# Instantiate the visualizer with the Covariance ranking algorithm
visualizer = Rank2D(features=features, algorithm='covariance')
visualizer.fit(X, y) # Fit the data to the visualizer
visualizer.transform(X) # Transform the data
visualizer.show() # Draw/show/show the data
# Instantiate the visualizer with the Pearson ranking algorithm
visualizer = Rank2D(features=features, algorithm='pearson')
visualizer.fit(X, y) # Fit the data to the visualizer
visualizer.transform(X) # Transform the data
visualizer.show() # Draw/show/show the data
```
### RadViz
RadViz is a multivariate data visualization algorithm that plots each feature dimension uniformely around the circumference of a circle then plots points on the interior of the circle such that the point normalizes its values on the axes from the center to each arc. This meachanism allows as many dimensions as will easily fit on a circle, greatly expanding the dimensionality of the visualization.
Data scientists use this method to dect separability between classes. E.g. is there an opportunity to learn from the feature set or is there just too much noise?
```
# Specify the features of interest and the classes of the target
features = ['temperature','humidity','co2','light','noise','bluetooth_devices']
classes = ['empty', 'occupied']
# Extract the numpy arrays from the data frame
X = data[features].as_matrix()
y = data.room_occupancy_num.as_matrix()
# Instantiate the visualizer
visualizer = visualizer = RadViz(classes=classes, features=features)
visualizer.fit(X, y) # Fit the data to the visualizer
visualizer.transform(X) # Transform the data
visualizer.show() # Draw/show/show the data
```
For regression, the RadViz visualizer should use a color sequence to display the target information, as opposed to discrete colors.
## Parallel Coordinates
### !!! On this step notebook crashes and has to be restarted
```
# Specify the features of interest and the classes of the target
#features = ['temperature','humidity','co2','light','noise','bluetooth_devices']
#classes = ['empty', 'occupied']
# Extract the numpy arrays from the data frame
#X = data[features].as_matrix()
#y = data.room_occupancy_num.as_matrix()
# Instantiate the visualizer
#visualizer = visualizer = ParallelCoordinates(classes=classes, features=features)
#visualizer.fit(X, y) # Fit the data to the visualizer
#visualizer.transform(X) # Transform the data
#visualizer.show() # Draw/show/show the data
```
## Regressor Evaluation
Regression models attempt to predict a target in a continuous space. Regressor score visualizers display the instances in model space to better understand how the model is making predictions. We currently have implemented two regressor evaluations:
Residuals Plot: plot the difference between the expected and actual values
Prediction Error: plot expected vs. the actual values in model space
Estimator score visualizers wrap Scikit-Learn estimators and expose the Estimator API such that they have fit(), predict(), and score() methods that call the appropriate estimator methods under the hood. Score visualizers can wrap an estimator and be passed in as the final step in a Pipeline or VisualPipeline.
```
# Regression Evaluation Imports
from sklearn.linear_model import Ridge, Lasso
from sklearn.model_selection import train_test_split
from yellowbrick.regressor import PredictionError, ResidualsPlot
```
### Residuals Plot
A residual plot shows the residuals on the vertical axis and the independent variable on the horizontal axis. If the points are randomly dispersed around the horizontal axis, a linear regression model is appropriate for the data; otherwise, a non-linear model is more appropriate.
```
# Load the data
df = data
feature_names = ['temperature','humidity','co2','light','noise','bluetooth_devices']
target_name = 'count_total'
# Get the X and y data from the DataFrame
X = df[feature_names].as_matrix()
y = df[target_name].as_matrix()
# Create the train and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
# Instantiate the linear model and visualizer
ridge = Ridge()
visualizer = ResidualsPlot(ridge)
visualizer.fit(X_train, y_train) # Fit the training data to the visualizer
visualizer.score(X_test, y_test) # Evaluate the model on the test data
g = visualizer.show() # Draw/show/show the data
```
### Prediction Error Plot
Plots the actual targets from the dataset against the predicted values generated by our model. This allows us to see how much variance is in the model. Data scientists diagnose this plot by comparing against the 45 degree line, where the prediction exactly matches the model.
```
# Load the data
df = data
feature_names = ['temperature','humidity','co2','light','noise','bluetooth_devices']
target_name = 'count_total'
# Get the X and y data from the DataFrame
X = df[feature_names].as_matrix()
y = df[target_name].as_matrix()
# Create the train and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
# Instantiate the linear model and visualizer
lasso = Lasso()
visualizer = PredictionError(lasso)
visualizer.fit(X_train, y_train) # Fit the training data to the visualizer
visualizer.score(X_test, y_test) # Evaluate the model on the test data
g = visualizer.show() # Draw/show/show the data
```
## Classifier Evaluation
Classification models attempt to predict a target in a discrete space, that is assign an instance of dependent variables one or more categories. Classification score visualizers display the differences between classes as well as a number of classifier-specific visual evaluations. We currently have implemented three classifier evaluations:
ClassificationReport: Presents the confusion matrix of the classifier as a heatmap
ROCAUC: Presents the graph of receiver operating characteristics along with area under the curve
ClassBalance: Displays the difference between the class balances and support
Estimator score visualizers wrap Scikit-Learn estimators and expose the Estimator API such that they have fit(), predict(), and score() methods that call the appropriate estimator methods under the hood. Score visualizers can wrap an estimator and be passed in as the final step in a Pipeline or VisualPipeline.
```
# Classifier Evaluation Imports
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from yellowbrick.classifier import ClassificationReport, ROCAUC, ClassBalance
```
### Classification report
The classification report visualizer displays the precision, recall, and F1 scores for the model. Integrates numerical scores as well color-coded heatmap in order for easy interpretation and detection.
```
# Load the classification data set
data = dataset
# Specify the features of interest and the classes of the target
features = ['temperature','humidity','co2','light','noise','bluetooth_devices']
classes = ['empty', 'occupied']
# Extract the numpy arrays from the data frame
X = data[features].as_matrix()
y = data.room_occupancy_num.as_matrix()
# Create the train and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
# Instantiate the classification model and visualizer
bayes = GaussianNB()
visualizer = ClassificationReport(bayes, classes=classes)
visualizer.fit(X_train, y_train) # Fit the training data to the visualizer
visualizer.score(X_test, y_test) # Evaluate the model on the test data
g = visualizer.show() # Draw/show/show the data
```
### ROCAUC
Plot the ROC to visualize the tradeoff between the classifier's sensitivity and specificity.
```
# Instantiate the classification model and visualizer
logistic = LogisticRegression()
visualizer = ROCAUC(logistic)
visualizer.fit(X_train, y_train) # Fit the training data to the visualizer
visualizer.score(X_test, y_test) # Evaluate the model on the test data
g = visualizer.show() # Draw/show/show the data
```
### ClassBalance
Class balance chart that shows the support for each class in the fitted classification model.
```
# Instantiate the classification model and visualizer
forest = RandomForestClassifier()
visualizer = ClassBalance(forest, classes=classes)
visualizer.fit(X_train, y_train) # Fit the training data to the visualizer
visualizer.score(X_test, y_test) # Evaluate the model on the test data
g = visualizer.show() # Draw/show/show the data
```
|
github_jupyter
|
# 5. Putting it all together
**Bring together all of the skills you acquired in the previous chapters to work on a real-life project. From connecting to a database and populating it, to reading and querying it.**
It's time to put all your effort so far to good use on a census case study.
### Census case study
The case study is broken down into three parts.
1. we are going to prepare SQLAlchemy and the database.
2. we will load the data into the database.
3. we solve a few data science type problems with our query knowledge.
### Part 1: preparing SQLAlchemy and the database
For part 1 we are going to focus on preparing SQLAlchemy and the database. You might remember this example from Chapter 1. We import `create_engine` and `Metadata`, then create the engine and initialize the metadata.
```python
from sqlalchemy import create_engine, MetaData
engine = create_engine('sqlite:///census_nyc.sqlite')
metadata = MetaData()
```
### Part 1: preparing SQLAlchemy and the database
Then we will build the census table to hold our data. You might remember the employees table we built in Chapter 4. We begin by importing the `Table` and `Column` objects along with all the types we are going to use in our table. Next we define our Table using the Table object by giving it a name, the metadata object, and then each of the columns we want in our table. Finally we create the table in the database by using the create all method on the metadata with the engine.
```python
from sqlalchemy import Table, Column, String, Integer, Numeric, Boolean
engine = create_engine('sqlite:///')
metadata = MetaData()
employees = Table('employees', metadata,
Column('id', Integer()),
Column('name', String(255)),
Column('salary', Numeric()),
Column('active', Boolean()))
metadata.create_all(engine)
```
## Setup the engine and metadata
In this exercise, your job is to create an engine to the database that will be used in this chapter. Then, you need to initialize its metadata.
Recall how you did this in Chapter 1 by leveraging `create_engine()` and `MetaData()`.
- Import `create_engine` and `MetaData` from `sqlalchemy`.
- Create an `engine` to the chapter 5 database by using `'sqlite:///chapter5.sqlite'` as the connection string.
- Create a MetaData object as `metadata`.
```
# Import create_engine, MetaData
from sqlalchemy import create_engine, MetaData
# Define an engine to connect to chapter5.sqlite: engine
engine = create_engine('sqlite:///chapter5.sqlite')
# Initialize MetaData: metadata
metadata = MetaData()
```
## Create the table to the database
Having setup the engine and initialized the metadata, you will now define the `census` table object and then create it in the database using the `metadata` and `engine` from the previous exercise. To create it in the database, you will have to use the `.create_all()` method on the `metadata` with `engine` as the argument.
- Import `Table`, `Column`, `String`, and `Integer` from `sqlalchemy`.
- Define a `census` table with the following columns:
- `'state'` - String - length of 30
- `'sex'` - String - length of 1
- `'age'` - Integer
- `'pop2000'` - Integer
- `'pop2008'` - Integer
- Create the table in the database using the `metadata` and `engine`.
```
# Import Table, Column, String, and Integer
from sqlalchemy import Table, Column, String, Integer
# Build a census table: census
census = Table('census', metadata,
Column('state', String(30)),
Column('sex', String(1)),
Column('age', Integer),
Column('pop2000', Integer),
Column('pop2008', Integer))
# Create the table in the database
metadata.create_all(engine)
```
---
## Populating the database
With our table in place, we can now load the data into it. The US Census Agency gave us a CSV file full of data that we need to load into the table.
### Part 2: populating the database
We'll start that by building a `values_list` like we did in chapter 4 with this exercise.
```python
values_list = []
for row in csv_reader:
data = {'state': row[0], 'sex': row[1], 'age': row[2],
'pop2000': row[3], 'pop2008': row[4]}
values_list.append(data)
```
We begin by defining an empty list then looping over the rows of the CSV. Then we build a dictionary for each CSV row that has the data for that row matched up with the column we want to store it in. Then we append the dictionary to the values list.
### Part 2: Populating the Database
Now we can insert that `values_list` as we did in Chapter 4 like this example. We we start by importing the `insert` statement. Then we build an insert statement for our table, finally we use the execute method on our connection with the statement and values list to insert the data into the table.
```python
from sqlalchemy import insert
stmt = insert(employees)
result_proxy = connection.execute(stmt, values_list)
print(result_proxy.rowcount)
```
```
2
```
To review how many rows were inserted, we use the `rowcount` method of the `ResultProxy`.
## Reading the data from the CSV
Leverage the Python CSV module from the standard library and load the data into a list of dictionaries.
- Create an empty list called `values_list`.
- Iterate over the rows of `csv_reader` with a for loop, creating a dictionary called `data` for each row and append it to `values_list`.
- Within the for loop, `row` will be a list whose entries are `'state'`, `'sex'`, `'age'`, `'pop2000'` and `'pop2008'` (in that order).
```
import csv
csv_reader = csv.reader(open('census.csv'))
# Create an empty list: values_list
values_list = []
# Iterate over the rows
for row in csv_reader:
# Create a dictionary with the values
data = {'state': row[0], 'sex': row[1], 'age': row[2],
'pop2000': row[3], 'pop2008': row[4]}
# Append the dictionary to the values list
values_list.append(data)
```
## Load data from a list into the Table
Using the multiple insert pattern, in this exercise, you will load the data from `values_list` into the table.
- Import `insert` from `sqlalchemy`.
- Build an insert statement for the `census` table.
- Execute the statement `stmt` along with `values_list`. You will need to pass them both as arguments to `connection.execute()`.
- Print the `rowcount` attribute of `results`.
```
# Import insert
from sqlalchemy import insert
# Build insert statement: stmt
stmt = insert(census)
# Use values_list to insert data: results
results = connection.execute(stmt, values_list)
# Print rowcount
print(results.rowcount)
```
---
## Querying the database
### Part 3: answering data science questions with queries
Here is an example of how we calculated an average in an exercise from Chapter 3. We began by importing the select statement. Next we built a select statement that creates a weighted average. We do this by summing the result of multiplying the age with the population and dividing that by the sum of the total population and labeling that average age. Next we grouped by the sex column to determine the average `age` for each `sex`. Finally, we executed the query and fetched all the results.
```python
from sqlalchemy import select
stmt = select([census.columns.sex,
(func.sum(census.columns.pop2008 *
census.columns.age) /
func.sum(census.columns.pop2008)
). label('avarage_age')])
stmt = stmt.group_by(census.columns.sex)
resutls = connection.execute(stmt).fetchall()
```
### Part 3: answering data science questions with queries
We learned how to calculate a percentage by using the case and cast clauses in Chapter 3. We begin by importing `case`, `cast`, and `Float`. Then we build a select statement that calculates the sum of the `pop2008` column in cases where the state is New York. Then we divided that by the sum of the total population which is cast to a Float so we would get Decimal values. Finally, we multiplied by 100 to get a percentage and labeled it `ny_percent`.
```python
from sqlalchemy import case, cast, Float
stmt = select([
(func.sum(
case([
(census.columns.state == 'New York',
census.columns.pop2008)
], else_=0)) /
cast(func.sum(census.columns.pop2008),
Float) * 100). label('ny_percent')])
```
Also from Chapter 3, we learned how calculate the difference between two columns grouped by another column. We start by building a `select` statement, that selects the column we want to determine the change by, which in this case is `age`. Then we calculate the difference between the population in 2008 and in 2000, and we label that `pop_change`. Remember to wrap the difference calculation in parentheses so you can label it. Next, we order by `pop_change` and finally we limit it to just 5 results.
```python
stmt = select([census.columns.age,
(census.columns.pop2008 -
census.columns.pop2000).label('pop_chage')
])
stmt = stmt.order_by('pop_change')
stmt = stmt.limit(5)
```
## Determine the average age by population
To calculate a weighted average, we first find the total sum of weights multiplied by the values we're averaging, then divide by the sum of all the weights.
For example, if we wanted to find a weighted average of `data = [10, 30, 50]` weighted by `weights = [2,4,6]`, we would compute *(2*10 + 4*30 + 6*50) / (2+4+6)*, or `sum(weights * data) / sum(weights)`.
In this exercise, however, you will make use of **`func.sum()`** together with select to `select` the weighted average of a column from a table. You will still work with the `census` data, and you will compute the average of age weighted by state population in the year 2000, and then group this weighted average by sex.
- Import `select` and `func` from `sqlalchemy`.
- Write a statement to `select` the average of age (`age`) weighted by population in **2000** (`pop2000`) from `census`.
```
# Import select and func
from sqlalchemy import select, func
# Select the average of age weighted by pop2000
stmt = select([func.sum(census.columns.pop2000 *
census.columns.age) /
func.sum(census.columns.pop2000)])
```
- Modify the select statement to alias the new column with weighted average as `'average_age'` using `.label()`.
```
# Import select and func
from sqlalchemy import select, func
# Relabel the new column as average_age
stmt = select([(func.sum(census.columns.pop2000 *
census.columns.age) /
func.sum(census.columns.pop2000)).label('average_age')
])
```
- Modify the select statement to select the `sex` column of `census` in addition to the weighted average, with the `sex` column coming first.
- Group by the `sex` column of `census`.
```
# Import select and func
from sqlalchemy import select, func
# Add the sex column to the select statement
stmt = select([census.columns.sex,
(func.sum(census.columns.pop2000 *
census.columns.age) /
func.sum(census.columns.pop2000)).label('average_age'),
])
# Group by sex
stmt = stmt.group_by(census.columns.sex)
```
- Execute the statement on the `connection` and fetch all the results.
- Loop over the results and print the values in the `sex` and `average_age` columns for each record in the results.
```
# Import select and func
from sqlalchemy import select, func
# Select sex and average age weighted by 2000 population
stmt = select([census.columns.sex,
(func.sum(census.columns.pop2000 *
census.columns.age) /
func.sum(census.columns.pop2000)).label('average_age')
])
# Group by sex
stmt = stmt.group_by(census.columns.sex)
# Execute the query and fetch all the results
connection = engine.connect()
results = connection.execute(stmt).fetchall()
# Print the sex and average age column for each result
for result in results:
print(result.sex, result.average_age)
```
## Determine the percentage of population by gender and state
In this exercise, you will write a query to determine the percentage of the population in 2000 that comprised of women. You will group this query by state.
- Import `case`, `cast` and `Float` from `sqlalchemy`.
- Define a statement to select `state` and the percentage of women in 2000.
- Inside `func.sum()`, use `case()` to select women (using the `sex` column) from `pop2000`. Remember to specify `else_=0` if the `sex` is not `'F'`.
- To get the percentage, divide the number of women in the year 2000 by the overall population in 2000. Cast the divisor - `census.columns.pop2000` - to `Float` before multiplying by 100.
- Group the query by `state`.
- Execute the query and store it as `results`.
- Print `state` and `percent_female` for each record.
```
# import case, cast and Float from sqlalchemy
from sqlalchemy import case, cast, Float, desc
# Build a query to calculate the percentage of women in 2000: stmt
stmt = select([census.columns.state,
(func.sum(
case([
(census.columns.sex == 'F',
census.columns.pop2000)
], else_=0)) /
cast(func.sum(census.columns.pop2000),
Float) * 100).label('percent_female')
])
# Group By state
stmt = stmt.group_by(census.columns.state)
stmt = stmt.order_by(desc('percent_female'))
# Execute the query and store the results: results
results = connection.execute(stmt).fetchall()
# Print the percentage
for result in results:
print(result.state, result.percent_female)
```
*Interestingly, the District of Columbia had the highest percentage of women in 2000, while Alaska had the highest percentage of males.*
## Determine the difference by state from the 2000 and 2008 censuses
In this final exercise, you will write a query to calculate the states that changed the most in population. You will limit your query to display only the top 10 states.
- Build a statement to:
- Select `state`.
- Calculate the difference in population between 2008 (`pop2008`) and 2000 (`pop2000`).
- Group the query by `census.columns.state` using the `.group_by()` method on `stmt`.
- Order by `'pop_change'` in descending order using the `.order_by()` method with the `desc()` function on `'pop_change'`.
- ~Limit the query to the top `10` states using the `.limit()` method.~
- Execute the query and store it as `results`.
- Print the state and the population change for each result.
```
# Build query to return state name and population difference from 2008 to 2000
stmt = select([census.columns.state,
(census.columns.pop2008-
census.columns.pop2000).label('pop_change')
])
# Group by State
stmt = stmt.group_by(census.columns.state)
# Order by Population Change
stmt = stmt.order_by(desc('pop_change'))
# Limit to top 10
##stmt = stmt.limit(10)
# Use connection to execute the statement and fetch all results
results = connection.execute(stmt).fetchall()
# Print the state and population change for each record
for result in results:
print('{}:{}'.format(result.state, result.pop_change))
```
|
github_jupyter
|
# Prepare environment
```
!pip install git+https://github.com/katarinagresova/ensembl_scraper.git@6d3bba8e6be7f5ead58a3bbaed6a4e8cd35e62fd
```
# Create config file
```
import yaml
config = {
"root_dir": "../../datasets/",
"organisms": {
"homo_sapiens": {
"regulatory_feature"
}
}
}
user_config = 'user_config.yaml'
with open(user_config, 'w') as handle:
yaml.dump(config, handle)
```
# Prepare directories
```
from pathlib import Path
BASE_FILE_PATH = Path("../../datasets/human_ensembl_regulatory/")
# copied from https://stackoverflow.com/a/57892171
def rm_tree(pth: Path):
for child in pth.iterdir():
if child.is_file():
child.unlink()
else:
rm_tree(child)
pth.rmdir()
if BASE_FILE_PATH.exists():
rm_tree(BASE_FILE_PATH)
```
# Run tool
```
!python -m scraper.ensembl_scraper -c user_config.yaml
```
# Reformating
```
!mkdir -p ../../datasets/human_ensembl_regulatory/train
!mkdir -p ../../datasets/human_ensembl_regulatory/test
!mv ../../datasets/homo_sapiens_regulatory_feature_open_chromatin_region/train/positive.csv ../../datasets/human_ensembl_regulatory/train/ocr.csv
!mv ../../datasets/homo_sapiens_regulatory_feature_open_chromatin_region/test/positive.csv ../../datasets/human_ensembl_regulatory/test/ocr.csv
!mv ../../datasets/homo_sapiens_regulatory_feature_promoter/train/positive.csv ../../datasets/human_ensembl_regulatory/train/promoter.csv
!mv ../../datasets/homo_sapiens_regulatory_feature_promoter/test/positive.csv ../../datasets/human_ensembl_regulatory/test/promoter.csv
!mv ../../datasets/homo_sapiens_regulatory_feature_enhancer/train/positive.csv ../../datasets/human_ensembl_regulatory/train/enhancer.csv
!mv ../../datasets/homo_sapiens_regulatory_feature_enhancer/test/positive.csv ../../datasets/human_ensembl_regulatory/test/enhancer.csv
def chop_sequnces(file_path, max_len):
df = pd.read_csv(file_path)
df_array = df.values
new_df_array = []
index = 0
for row in df_array:
splits = ((row[3] - row[2]) // max_len) + 1
if splits == 1:
new_df_array.append([index, row[1], row[2], row[3], row[4]])
index += 1
elif splits == 2:
length = (row[3] - row[2]) // 2
new_df_array.append([
index,
row[1],
row[2],
row[2] + length,
row[4]
])
index += 1
new_df_array.append([
index,
row[1],
row[2] + length + 1,
row[3],
row[4]
])
index += 1
else:
length = (row[3] - row[2]) // splits
new_df_array.append([
index,
row[1],
row[2],
row[2] + length,
row[4]
])
index += 1
for i in range(1, splits - 1):
new_df_array.append([
index,
row[1],
row[2] + i*length + 1,
row[2] + (i + 1)*length,
row[4]
])
index += 1
new_df_array.append([
index,
row[1],
row[2] + (splits - 1)*length + 1,
row[3],
row[4]
])
index += 1
new_df = pd.DataFrame(new_df_array, columns=df.columns)
new_df.to_csv(file_path, index=False)
chop_sequnces("../../datasets/human_ensembl_regulatory/train/promoter.csv", 700)
chop_sequnces("../../datasets/human_ensembl_regulatory/test/promoter.csv", 700)
!find ../../datasets/human_ensembl_regulatory/ -type f -name "*.csv" -exec gzip {} \;
!mv ../../datasets/homo_sapiens_regulatory_feature_enhancer/metadata.yaml ../../datasets/human_ensembl_regulatory/metadata.yaml
with open("../../datasets/human_ensembl_regulatory/metadata.yaml", "r") as stream:
try:
config = yaml.safe_load(stream)
except yaml.YAMLError as exc:
print(exc)
config
new_config = {
'classes' : {
'ocr': {
'type': config['classes']['positive']['type'],
'url': config['classes']['positive']['url'],
'extra_processing': 'ENSEMBL_HUMAN_GENOME'
},
'promoter': {
'type': config['classes']['positive']['type'],
'url': config['classes']['positive']['url'],
'extra_processing': 'ENSEMBL_HUMAN_GENOME'
},
'enhancer': {
'type': config['classes']['positive']['type'],
'url': config['classes']['positive']['url'],
'extra_processing': 'ENSEMBL_HUMAN_GENOME'
}
},
'version': config['version']
}
new_config
with open("../../datasets/human_ensembl_regulatory/metadata.yaml", 'w') as handle:
yaml.dump(new_config, handle)
```
# Cleaning
```
!rm user_config.yaml
!rm -rf ../../datasets/tmp/
!rm -rf ../../datasets/homo_sapiens_regulatory_feature_CTCF_binding_site
!rm -rf ../../datasets/homo_sapiens_regulatory_feature_enhancer
!rm -rf ../../datasets/homo_sapiens_regulatory_feature_promoter
!rm -rf ../../datasets/homo_sapiens_regulatory_feature_promoter_flanking_region
!rm -rf ../../datasets/homo_sapiens_regulatory_feature_TF_binding_site
!rm -rf ../../datasets/homo_sapiens_regulatory_feature_open_chromatin_region
```
# Testing
```
from genomic_benchmarks.loc2seq import download_dataset
download_dataset("human_ensembl_regulatory", local_repo=True)
from genomic_benchmarks.data_check import info
info("human_ensembl_regulatory", 0, local_repo=True)
```
|
github_jupyter
|
```
import nltk
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from nltk.stem import WordNetLemmatizer
from nltk.corpus import wordnet
import re, collections
from collections import defaultdict
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.metrics import mean_squared_error, r2_score, cohen_kappa_score
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression, Ridge, Lasso
from sklearn.svm import SVR
from sklearn.ensemble import RandomForestRegressor
from sklearn.ensemble import AdaBoostRegressor
from spellchecker import SpellChecker
from nltk.tokenize import word_tokenize
import string
from sklearn.metrics import classification_report
from sklearn import svm
from sklearn.model_selection import cross_val_score
from sklearn.metrics import classification_report
from sklearn.preprocessing import MinMaxScaler
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib import rcParams
```
## Loading data
```
#dataframe = pd.read_csv('all_essaysets.csv', encoding = 'latin-1')
dataframe = pd.read_csv('training.tsv', encoding = 'latin-1', sep='\t')
dataframe.describe()
dataframe.head()
```
## Methods
```
# selecting which set to be used 1-8
# in order to combine them all assign set number to 9
def select_set(dataframe,setNumber):
if setNumber == 9:
dataframe2 = dataframe[dataframe.essay_set ==1]
texts = dataframe2['essay']
scores = dataframe2['domain1_score']
scores = scores.apply(lambda x: (x*3)/scores.max())
for i in range(1,9):
dataframe2 = dataframe[dataframe.essay_set == i]
texts = texts.append(dataframe2['essay'])
s = dataframe2['domain1_score']
s = s.apply(lambda x: (x*3)/s.max())
scores = scores.append(s)
else:
dataframe2 = dataframe[dataframe.essay_set ==setNumber]
texts = dataframe2['essay']
scores = dataframe2['domain1_score']
scores = scores.apply(lambda x: (x*3)/scores.max())
return texts, scores
# get histogram plot of scores and average score
def get_hist_avg(scores,bin_count):
print(sum(scores)/len(scores))
scores.hist(bins=bin_count)
#average word length for a text
def avg_word_len(text):
clean_essay = re.sub(r'\W', ' ', text)
words = nltk.word_tokenize(clean_essay)
total = 0
for word in words:
total = total + len(word)
average = total / len(words)
return average
# word count in a given text
def word_count(text):
clean_essay = re.sub(r'\W', ' ', text)
return len(nltk.word_tokenize(clean_essay))
# char count in a given text
def char_count(text):
return len(re.sub(r'\s', '', str(text).lower()))
# sentence count in a given text
def sent_count(text):
return len(nltk.sent_tokenize(text))
#tokenization of texts to sentences
def sent_tokenize(text):
stripped_essay = text.strip()
tokenizer = nltk.data.load('tokenizers/punkt/english.pickle')
raw_sentences = tokenizer.tokenize(stripped_essay)
tokenized_sentences = []
for raw_sentence in raw_sentences:
if len(raw_sentence) > 0:
clean_sentence = re.sub("[^a-zA-Z0-9]"," ", raw_sentence)
tokens = nltk.word_tokenize(clean_sentence)
tokenized_sentences.append(tokens)
return tokenized_sentences
# lemma, noun, adjective, verb, adverb count for a given text
def count_lemmas(text):
noun_count = 0
adj_count = 0
verb_count = 0
adv_count = 0
lemmas = []
lemmatizer = WordNetLemmatizer()
tokenized_sentences = sent_tokenize(text)
for sentence in tokenized_sentences:
tagged_tokens = nltk.pos_tag(sentence)
for token_tuple in tagged_tokens:
pos_tag = token_tuple[1]
if pos_tag.startswith('N'):
noun_count += 1
pos = wordnet.NOUN
lemmas.append(lemmatizer.lemmatize(token_tuple[0], pos))
elif pos_tag.startswith('J'):
adj_count += 1
pos = wordnet.ADJ
lemmas.append(lemmatizer.lemmatize(token_tuple[0], pos))
elif pos_tag.startswith('V'):
verb_count += 1
pos = wordnet.VERB
lemmas.append(lemmatizer.lemmatize(token_tuple[0], pos))
elif pos_tag.startswith('R'):
adv_count += 1
pos = wordnet.ADV
lemmas.append(lemmatizer.lemmatize(token_tuple[0], pos))
else:
pos = wordnet.NOUN
lemmas.append(lemmatizer.lemmatize(token_tuple[0], pos))
lemma_count = len(set(lemmas))
return noun_count, adj_count, verb_count, adv_count, lemma_count
def token_word(text):
text = "".join([ch.lower() for ch in text if ch not in string.punctuation])
tokens = nltk.word_tokenize(text)
return tokens
def misspell_count(text):
spell = SpellChecker()
# find those words that may be misspelled
misspelled = spell.unknown(token_word(text))
#print(misspelled)
return len(misspelled)
def create_features(texts):
data = pd.DataFrame(columns=('Average_Word_Length','Sentence_Count','Word_Count',
'Character_Count', 'Noun_Count','Adjective_Count',
'Verb_Count', 'Adverb_Count', 'Lemma_Count' , 'Misspell_Count'
))
data['Average_Word_Length'] = texts.apply(avg_word_len)
data['Sentence_Count'] = texts.apply(sent_count)
data['Word_Count'] = texts.apply(word_count)
data['Character_Count'] = texts.apply(char_count)
temp=texts.apply(count_lemmas)
noun_count,adj_count,verb_count,adverb_count,lemma_count = zip(*temp)
data['Noun_Count'] = noun_count
data['Adjective_Count'] = adj_count
data['Verb_Count'] = verb_count
data['Adverb_Count'] = adverb_count
data['Lemma_Count'] = lemma_count
data['Misspell_Count'] = texts.apply(misspell_count)
return data
def data_prepare(texts,scores):
#create features from the texts and clean non graded essays
data = create_features(texts)
data.describe()
t1=np.where(np.asanyarray(np.isnan(scores)))
scores=scores.drop(scores.index[t1])
data=data.drop(scores.index[t1])
#scaler = MinMaxScaler()
#data = scaler.fit_transform(data)
#train test split
X_train, X_test, y_train, y_test = train_test_split(data, scores, test_size = 0.3)
#checking is there any nan cells
print(np.any(np.isnan(scores)))
print(np.all(np.isfinite(scores)))
return X_train, X_test, y_train, y_test, data
def lin_regression(X_train,y_train,X_test,y_test):
regr = LinearRegression()
regr.fit(X_train, y_train)
y_pred = regr.predict(X_test)
# The mean squared error
mse=mean_squared_error(y_test, y_pred)
mse_per= 100*mse/3
print("Mean squared error: {}".format(mse))
print("Mean squared error in percentage: {}".format(mse_per))
#explained variance score
print('Variance score: {}'.format(regr.score(X_test, y_test)))
def adaBoost_reg(X_train,y_train,X_test,y_test):
#regr = RandomForestRegressor(max_depth=2, n_estimators=300)
#regr = SVR(gamma='scale', C=1, kernel='linear')
regr = AdaBoostRegressor()
regr.fit(X_train, y_train)
y_pred = regr.predict(X_test)
# The mean squared error
mse=mean_squared_error(y_test, y_pred)
mse_per= 100*mse/3
print("Mean squared error: {}".format(mse))
print("Mean squared error in percentage: {}".format(mse_per))
#explained variance score
print('Variance score: {}'.format(regr.score(X_test, y_test)))
feature_importance = regr.feature_importances_
# make importances relative to max importance
feature_importance = 100.0 * (feature_importance / feature_importance.max())
feature_names = list(('Average_Word_Length','Sentence_Count','Word_Count',
'Character_Count', 'Noun_Count','Adjective_Count',
'Verb_Count', 'Adverb_Count', 'Lemma_Count' ,'Misspell_Count'
))
feature_names = np.asarray(feature_names)
sorted_idx = np.argsort(feature_importance)
pos = np.arange(sorted_idx.shape[0]) + .5
plt.subplot(1, 2, 2)
plt.barh(pos, feature_importance[sorted_idx], align='center')
plt.yticks(pos, feature_names[sorted_idx])
plt.xlabel('Relative Importance')
plt.title('Variable Importance')
plt.show()
# convert numerical scores to labels
# (0-1.5) bad (1.5-2.3) average (2.3-3) good
# bad: '0'
# average '1'
# good '2'
def convert_scores(scores):
def mapping(x):
if x < np.percentile(scores,25):
return 0
elif x < np.percentile(scores,75):
return 1
else:
return 2
return scores.apply(mapping)
# selecting which set to be used 1-8
# in order to combine them all assign set number to 9
def select_set_classification(dataframe,setNumber):
if setNumber == 9:
dataframe2 = dataframe[dataframe.essay_set ==1]
texts = dataframe2['essay']
scores = dataframe2['domain1_score']
scores = scores.apply(lambda x: (x*3)/scores.max())
scores = convert_scores(scores)
for i in range(1,9):
dataframe2 = dataframe[dataframe.essay_set == i]
texts = texts.append(dataframe2['essay'])
s = dataframe2['domain1_score']
s = s.apply(lambda x: (x*3)/s.max())
s = convert_scores(s)
scores = scores.append(s)
else:
dataframe2 = dataframe[dataframe.essay_set ==setNumber]
texts = dataframe2['essay']
scores = dataframe2['domain1_score']
scores = scores.apply(lambda x: (x*3)/scores.max())
scores = convert_scores(scores)
return texts, scores
```
## Dataset selection
```
# 1-8
# 9:all sets combined
texts, scores = select_set(dataframe,1)
get_hist_avg(scores,5)
X_train, X_test, y_train, y_test, data = data_prepare(texts,scores)
```
## Regression Analysis
```
print('Testing for Linear Regression \n')
lin_regression(X_train,y_train,X_test,y_test)
print('Testing for Adaboost Regression \n')
adaBoost_reg(X_train,y_train,X_test,y_test)
```
## Dataset selection 2
```
# 1-8
# 9:all sets combined
texts, scores = select_set_classification(dataframe,1)
X_train, X_test, y_train, y_test, data = data_prepare(texts,scores)
```
## Classification analysis
```
a=[0.1,1,10,100,500,1000]
for b in a:
clf = svm.SVC(C=b, gamma=0.00001)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
print (b)
print (clf.score(X_test,y_test))
print (np.mean(cross_val_score(clf, X_train, y_train, cv=3)))
clf = svm.SVC(C=100, gamma=0.00001)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
print('Cohen’s kappa score: {}'.format(cohen_kappa_score(y_test,y_pred)))
print(classification_report(y_test, y_pred))
```
## Data Analysis
```
sns.countplot(scores)
zero = data[(data["Character_Count"] > 0) & (scores == 0)]
one = data[(data["Character_Count"] > 0) & (scores == 1)]
two = data[(data["Character_Count"] > 0) & (scores == 2)]
sns.distplot(zero["Character_Count"], bins=10, color='r')
sns.distplot(one["Character_Count"], bins=10, color='g')
sns.distplot(two["Character_Count"], bins=10, color='b')
plt.title("Score Distribution with respect to Character_Count",fontsize=20)
plt.xlabel("Character_Count",fontsize=15)
plt.ylabel("Distribuition of the scores",fontsize=15)
plt.show()
zero = data[(data["Lemma_Count"] > 0) & (scores == 0)]
one = data[(data["Lemma_Count"] > 0) & (scores == 1)]
two = data[(data["Lemma_Count"] > 0) & (scores == 2)]
sns.distplot(zero["Lemma_Count"], bins=10, color='r')
sns.distplot(one["Lemma_Count"], bins=10, color='g')
sns.distplot(two["Lemma_Count"], bins=10, color='b')
plt.title("Score Distribution with respect to lemma count",fontsize=20)
plt.xlabel("Lemma Count",fontsize=15)
plt.ylabel("Distribuition of the scores",fontsize=15)
plt.show()
zero = data[(data["Sentence_Count"] > 0) & (scores == 0)]
one = data[(data["Sentence_Count"] > 0) & (scores == 1)]
two = data[(data["Sentence_Count"] > 0) & (scores == 2)]
sns.distplot(zero["Sentence_Count"], bins=10, color='r')
sns.distplot(one["Sentence_Count"], bins=10, color='g')
sns.distplot(two["Sentence_Count"], bins=10, color='b')
plt.title("Score Distribution with respect to sentence count",fontsize=20)
plt.xlabel("Sentence Count",fontsize=15)
plt.ylabel("Distribuition of the scores",fontsize=15)
plt.show()
zero = data[(data["Word_Count"] > 0) & (scores == 0)]
one = data[(data["Word_Count"] > 0) & (scores == 1)]
two = data[(data["Word_Count"] > 0) & (scores == 2)]
sns.distplot(zero["Word_Count"], bins=10, color='r')
sns.distplot(one["Word_Count"], bins=10, color='g')
sns.distplot(two["Word_Count"], bins=10, color='b')
plt.title("Score Distribution with respect to word count",fontsize=20)
plt.xlabel("Word_Count",fontsize=15)
plt.ylabel("Distribuition of the scores",fontsize=15)
plt.show()
zero = data[(data["Average_Word_Length"] > 0) & (scores == 0)]
one = data[(data["Average_Word_Length"] > 0) & (scores == 1)]
two = data[(data["Average_Word_Length"] > 0) & (scores == 2)]
sns.distplot(zero["Average_Word_Length"], bins=10, color='r')
sns.distplot(one["Average_Word_Length"], bins=10, color='g')
sns.distplot(two["Average_Word_Length"], bins=10, color='b')
plt.title("Score Distribution with respect to Average_Word_Length",fontsize=20)
plt.xlabel("Average_Word_Length",fontsize=15)
plt.ylabel("Distribuition of the scores",fontsize=15)
plt.show()
```
### Kappa Score Reliability
According to Cohen's original article, values ≤ 0 as indicating no agreement and 0.01–0.20 as none to slight, 0.21–0.40 as fair, 0.41– 0.60 as moderate, 0.61–0.80 as substantial, and 0.81–1.00 as almost perfect agreement. McHugh says that many texts recommend 80% agreement as the minimum acceptable interrater agreement.
|
github_jupyter
|
# Hello Segmentation
A very basic introduction to using segmentation models with OpenVINO.
We use the pre-trained [road-segmentation-adas-0001](https://docs.openvinotoolkit.org/latest/omz_models_model_road_segmentation_adas_0001.html) model from the [Open Model Zoo](https://github.com/openvinotoolkit/open_model_zoo/). ADAS stands for Advanced Driver Assistance Services. The model recognizes four classes: background, road, curb and mark.
## Imports
```
import cv2
import matplotlib.pyplot as plt
import numpy as np
import sys
from openvino.inference_engine import IECore
sys.path.append("../utils")
from notebook_utils import segmentation_map_to_image
```
## Load the Model
```
ie = IECore()
net = ie.read_network(
model="model/road-segmentation-adas-0001.xml")
exec_net = ie.load_network(net, "CPU")
output_layer_ir = next(iter(exec_net.outputs))
input_layer_ir = next(iter(exec_net.input_info))
```
## Load an Image
A sample image from the [Mapillary Vistas](https://www.mapillary.com/dataset/vistas) dataset is provided.
```
# The segmentation network expects images in BGR format
image = cv2.imread("data/empty_road_mapillary.jpg")
rgb_image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image_h, image_w, _ = image.shape
# N,C,H,W = batch size, number of channels, height, width
N, C, H, W = net.input_info[input_layer_ir].tensor_desc.dims
# OpenCV resize expects the destination size as (width, height)
resized_image = cv2.resize(image, (W, H))
# reshape to network input shape
input_image = np.expand_dims(
resized_image.transpose(2, 0, 1), 0
)
plt.imshow(rgb_image)
```
## Do Inference
```
# Run the infernece
result = exec_net.infer(inputs={input_layer_ir: input_image})
result_ir = result[output_layer_ir]
# Prepare data for visualization
segmentation_mask = np.argmax(result_ir, axis=1)
plt.imshow(segmentation_mask[0])
```
## Prepare Data for Visualization
```
# Define colormap, each color represents a class
colormap = np.array([[68, 1, 84], [48, 103, 141], [53, 183, 120], [199, 216, 52]])
# Define the transparency of the segmentation mask on the photo
alpha = 0.3
# Use function from notebook_utils.py to transform mask to an RGB image
mask = segmentation_map_to_image(segmentation_mask, colormap)
resized_mask = cv2.resize(mask, (image_w, image_h))
# Create image with mask put on
image_with_mask = cv2.addWeighted(resized_mask, alpha, rgb_image, 1 - alpha, 0)
```
## Visualize data
```
# Define titles with images
data = {"Base Photo": rgb_image, "Segmentation": mask, "Masked Photo": image_with_mask}
# Create subplot to visualize images
f, axs = plt.subplots(1, len(data.items()), figsize=(15, 10))
# Fill subplot
for ax, (name, image) in zip(axs, data.items()):
ax.axis('off')
ax.set_title(name)
ax.imshow(image)
# Display image
plt.show(f)
```
|
github_jupyter
|
```
from __future__ import print_function
import math
from IPython import display
from matplotlib import cm
from matplotlib import gridspec
from matplotlib import pyplot as plt
import numpy as np
import pandas as pd
from sklearn import metrics
import tensorflow as tf
from tensorflow.python.data import Dataset
tf.logging.set_verbosity(tf.logging.ERROR)
pd.options.display.max_rows = 10
pd.options.display.float_format = '{:.1f}'.format
ls
dataset_all = pd.read_csv('prices.csv')
dataset_all.head()
dataset_all
def my_input_fn(features, targets, batch_size=1, shuffle=True, num_epochs=None):
"""Trains a linear regression model of one feature.
Args:
features: pandas DataFrame of features
targets: pandas DataFrame of targets
batch_size: Size of batches to be passed to the model
shuffle: True or False. Whether to shuffle the data.
num_epochs: Number of epochs for which data should be repeated. None = repeat indefinitely
Returns:
Tuple of (features, labels) for next data batch
"""
# Convert pandas data into a dict of np arrays.
features = {key:np.array(value) for key,value in dict(features).items()}
# Construct a dataset, and configure batching/repeating.
ds = Dataset.from_tensor_slices((features,targets)) # warning: 2GB limit
ds = ds.batch(batch_size).repeat(num_epochs)
# Shuffle the data, if specified.
if shuffle:
ds = ds.shuffle(buffer_size=10000)
# Return the next batch of data.
features, labels = ds.make_one_shot_iterator().get_next()
return features, labels
def train_model(learning_rate, steps, batch_size, input_feature="close"):
"""Trains a linear regression model of one feature.
Args:
learning_rate: A `float`, the learning rate.
steps: A non-zero `int`, the total number of training steps. A training step
consists of a forward and backward pass using a single batch.
batch_size: A non-zero `int`, the batch size.
input_feature: A `string` specifying a column from `california_housing_dataframe`
to use as input feature.
"""
periods = 10
steps_per_period = steps / periods
my_feature = input_feature
my_feature_data = dataset_all[[my_feature]]
my_label = "open"
targets = dataset_all[my_label]
# Create feature columns.
feature_columns = [tf.feature_column.numeric_column(my_feature)]
# Create input functions.
training_input_fn = lambda:my_input_fn(my_feature_data, targets, batch_size=batch_size)
prediction_input_fn = lambda: my_input_fn(my_feature_data, targets, num_epochs=1, shuffle=False)
# Create a linear regressor object.
my_optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)
my_optimizer = tf.contrib.estimator.clip_gradients_by_norm(my_optimizer, 5.0)
linear_regressor = tf.estimator.LinearRegressor(
feature_columns=feature_columns,
optimizer=my_optimizer
)
# Set up to plot the state of our model's line each period.
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Learned Line by Period")
plt.ylabel(my_label)
plt.xlabel(my_feature)
sample = dataset_all.sample(n=300)
plt.scatter(sample[my_feature], sample[my_label])
colors = [cm.coolwarm(x) for x in np.linspace(-1, 1, periods)]
# Train the model, but do so inside a loop so that we can periodically assess
# loss metrics.
print("Training model...")
print("RMSE (on training data):")
root_mean_squared_errors = []
for period in range (0, periods):
# Train the model, starting from the prior state.
linear_regressor.train(
input_fn=training_input_fn,
steps=steps_per_period
)
# Take a break and compute predictions.
predictions = linear_regressor.predict(input_fn=prediction_input_fn)
predictions = np.array([item['predictions'][0] for item in predictions])
# Compute loss.
root_mean_squared_error = math.sqrt(
metrics.mean_squared_error(predictions, targets))
# Occasionally print the current loss.
print(" period %02d : %0.2f" % (period, root_mean_squared_error))
# Add the loss metrics from this period to our list.
root_mean_squared_errors.append(root_mean_squared_error)
# Finally, track the weights and biases over time.
# Apply some math to ensure that the data and line are plotted neatly.
y_extents = np.array([0, sample[my_label].max()])
weight = linear_regressor.get_variable_value('linear/linear_model/%s/weights' % input_feature)[0]
bias = linear_regressor.get_variable_value('linear/linear_model/bias_weights')
x_extents = (y_extents - bias) / weight
x_extents = np.maximum(np.minimum(x_extents,
sample[my_feature].max()),
sample[my_feature].min())
y_extents = weight * x_extents + bias
plt.plot(x_extents, y_extents, color=colors[period])
print("Model training finished.")
# Output a graph of loss metrics over periods.
plt.subplot(1, 2, 2)
plt.ylabel('RMSE')
plt.xlabel('Periods')
plt.title("Root Mean Squared Error vs. Periods")
plt.tight_layout()
plt.plot(root_mean_squared_errors)
# Output a table with calibration data.
calibration_data = pd.DataFrame()
calibration_data["predictions"] = pd.Series(predictions)
calibration_data["targets"] = pd.Series(targets)
display.display(calibration_data.describe())
print("Final RMSE (on training data): %0.2f" % root_mean_squared_error)
train_model(
learning_rate=0.002,
steps=100,
batch_size=10,
input_feature="close"
)
def train_model(learning_rate, steps, batch_size, input_feature="close"):
"""Trains a linear regression model of one feature.
Args:
learning_rate: A `float`, the learning rate.
steps: A non-zero `int`, the total number of training steps. A training step
consists of a forward and backward pass using a single batch.
batch_size: A non-zero `int`, the batch size.
input_feature: A `string` specifying a column from `california_housing_dataframe`
to use as input feature.
"""
periods = 10
steps_per_period = steps / periods
my_feature = input_feature
my_feature_data = dataset_all[[my_feature]]
my_label = "open"
targets = dataset_all[my_label]
# Create feature columns.
feature_columns = [tf.feature_column.numeric_column(my_feature)]
# Create input functions.
training_input_fn = lambda:my_input_fn(my_feature_data, targets, batch_size=batch_size)
prediction_input_fn = lambda: my_input_fn(my_feature_data, targets, num_epochs=1, shuffle=False)
# Create a linear regressor object.
my_optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)
my_optimizer = tf.contrib.estimator.clip_gradients_by_norm(my_optimizer, 5.0)
linear_regressor = tf.estimator.LinearRegressor(
feature_columns=feature_columns,
optimizer=my_optimizer
)
# Set up to plot the state of our model's line each period.
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Learned Line by Period")
plt.ylabel(my_label)
plt.xlabel(my_feature)
sample = dataset_all.sample(n=300)
plt.scatter(sample[my_feature], sample[my_label])
colors = [cm.coolwarm(x) for x in np.linspace(-1, 1, periods)]
# Train the model, but do so inside a loop so that we can periodically assess
# loss metrics.
print("Training model...")
print("RMSE (on training data):")
root_mean_squared_errors = []
for period in range (0, periods):
# Train the model, starting from the prior state.
linear_regressor.train(
input_fn=training_input_fn,
steps=steps_per_period
)
# Take a break and compute predictions.
predictions = linear_regressor.predict(input_fn=prediction_input_fn)
predictions = np.array([item['predictions'][0] for item in predictions])
# Compute loss.
root_mean_squared_error = math.sqrt(
metrics.mean_squared_error(predictions, targets))
# Occasionally print the current loss.
print(" period %02d : %0.2f" % (period, root_mean_squared_error))
# Add the loss metrics from this period to our list.
root_mean_squared_errors.append(root_mean_squared_error)
# Finally, track the weights and biases over time.
# Apply some math to ensure that the data and line are plotted neatly.
y_extents = np.array([0, sample[my_label].max()])
weight = linear_regressor.get_variable_value('linear/linear_model/%s/weights' % input_feature)[0]
bias = linear_regressor.get_variable_value('linear/linear_model/bias_weights')
x_extents = (y_extents - bias) / weight
x_extents = np.maximum(np.minimum(x_extents,
sample[my_feature].max()),
sample[my_feature].min())
y_extents = weight * x_extents + bias
plt.plot(x_extents, y_extents, color=colors[period])
print("Model training finished.")
# Output a graph of loss metrics over periods.
plt.subplot(1, 2, 2)
plt.ylabel('RMSE')
plt.xlabel('Periods')
plt.title("Root Mean Squared Error vs. Periods")
plt.tight_layout()
plt.plot(root_mean_squared_errors)
# Output a table with calibration data.
calibration_data = pd.DataFrame()
calibration_data["predictions"] = pd.Series(predictions)
calibration_data["targets"] = pd.Series(targets)
display.display(calibration_data.describe())
print("Final RMSE (on training data): %0.2f" % root_mean_squared_error)
train_model(
learning_rate=0.002,
steps=100,
batch_size=10,
input_feature="high"
)
```
|
github_jupyter
|
```
import os
import torch
from torch.utils.data import DataLoader, Dataset
from torchvision.transforms import ToTensor, ToPILImage
from torchvision.models.detection import fasterrcnn_resnet50_fpn
from torchvision.models.detection.faster_rcnn import FastRCNNPredictor
from PIL import Image
class PlayerDataset(Dataset):
def __init__(self, root):
self.root = root
self.images = list(sorted(os.listdir(root + '/images')))
self.targets = [target for target in list(sorted(os.listdir(root + '/targets'))) if target != 'classes.txt']
def __len__(self):
return len(self.images)
def __getitem__(self, idx):
image_path = os.path.join(self.root, 'images', self.images[idx])
target_path = os.path.join(self.root, 'targets', self.targets[idx])
image = ToTensor()(Image.open(image_path).convert("RGB"))
f = open(target_path)
target = f.readline().strip().split()
w = 1280
h = 720
center_x = float(target[1]) * w
center_y = float(target[2]) * h
bbox_w = float(target[3]) * w
bbox_h = float(target[4]) * h
x0 = round(center_x - (bbox_w / 2))
x1 = round(center_x + (bbox_w / 2))
y0 = round(center_y - (bbox_h / 2))
y1 = round(center_y + (bbox_h / 2))
print(x1 - x0)
print(y1 - y0)
boxes = torch.as_tensor([x0, y0, x1, y1], dtype=torch.float32)
labels = torch.as_tensor(0, dtype=torch.int64)
target = [{'boxes': boxes, 'labels': labels}]
return image, target
def train_model(model, optimizer, lr_scheduler, data_loader, device, num_epochs):
model.train()
for epoch in range(num_epochs):
running_loss = 0.0
for images, targets in data_loader:
images = list(image.to(device) for image in images)
targets = [{k: v.to(device) for k, v in t.items()} for t in targets]
print(targets)
loss_dict = model(images, targets)
losses = sum(loss for loss in loss_dict.values())
optimizer.zero_grad()
losses.backward()
optimizer.step()
lr_scheduler.step()
running_loss += losses.item()
print('epoch:%d loss: %.3f' % (epoch + 1, running_loss))
def evaluate(model, data_loader, device):
model.eval()
cpu_device = torch.device("cpu")
with torch.no_grad():
for images, targets in data_loader:
images = list(image.to(device) for image in images)
targets = [{k: v.to(device) for k, v in t.items()} for t in targets]
outputs = [{k: v.to(cpu_device) for k, v in t.items()} for t in model(images)]
print(outputs)
model = fasterrcnn_resnet50_fpn(num_classes=1)
device = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
model.to(device)
train_dataset = PlayerDataset('data/train')
test_dataset = PlayerDataset('data/test')
train_data_loader = DataLoader(train_dataset, batch_size=1, shuffle=True, num_workers=4)
test_data_loader = DataLoader(test_dataset, batch_size=1, shuffle=False, num_workers=4)
params = [p for p in model.parameters() if p.requires_grad]
optimizer = torch.optim.SGD(params, lr=0.005, momentum=0.9, weight_decay=0.0005)
lr_scheduler = torch.optim.lr_scheduler.StepLR(optimizer, step_size=3, gamma=0.1)
train_model(model, optimizer, lr_scheduler, train_data_loader, device, 1)
evaluate(model, test_data_loader, device)
```
|
github_jupyter
|
```
!git clone https://github.com/GraphGrailAi/ruGPT3-ZhirV
cd ruGPT3-ZhirV
cd ..
!pip3 install -r requirements.txt
```
Обучение эссе
!python pretrain_transformers.py \
--output_dir=/home/jovyan/ruGPT3-ZhirV/ \
--overwrite_output_dir \
--model_type=gpt2 \
--model_name_or_path=sberbank-ai/rugpt3large_based_on_gpt2 \
--do_train \
--train_data_file=/home/jovyan/ruGPT3-ZhirV/data/all_essays.jsonl \
--do_eval \
--eval_data_file=/home/jovyan/ruGPT3-ZhirV/data/valid_essays.jsonl \
--num_train_epochs 10 \
--overwrite_cache \
--block_size=1024 \
--per_gpu_train_batch_size 1 \
--gradient_accumulation_steps 8
# Обучение Жириновский
```
!python pretrain_transformers.py \
--output_dir=/home/jovyan/ruGPT3-ZhirV/ \
--overwrite_output_dir \
--model_type=gpt2 \
--model_name_or_path=sberbank-ai/rugpt3large_based_on_gpt2 \
--do_train \
--train_data_file=/home/jovyan/ruGPT3-ZhirV/data/girik_all2.jsonl \
--do_eval \
--eval_data_file=/home/jovyan/ruGPT3-ZhirV/data/girik_valid.jsonl \
--num_train_epochs 20 \
--overwrite_cache \
--block_size=1024 \
--per_gpu_train_batch_size 1 \
--gradient_accumulation_steps 8
```
# Генерация Жириновский
```
from transformers import GPT2Tokenizer, GPT2LMHeadModel
tokenizer = GPT2Tokenizer.from_pretrained("checkpoint-1000")
model = GPT2LMHeadModel.from_pretrained("checkpoint-1000")
model.to("cuda")
import copy
bad_word_ids = [
[203], # \n
[225], # weird space 1
[28664], # weird space 2
[13298], # weird space 3
[206], # \r
[49120], # html
[25872], # http
[3886], # amp
[38512], # nbsp
[10], # &
[5436], # & (another)
[5861], # http
[372], # yet another line break
[421, 4395], # МСК
[64], # \
[33077], # https
[1572], # ru
[11101], # Источник
]
def gen_fragment(context, bad_word_ids=bad_word_ids, print_debug_output=False):
input_ids = tokenizer.encode(context, add_special_tokens=False, return_tensors="pt").to("cuda")
input_ids = input_ids[:, -1700:]
input_size = input_ids.size(1)
output_sequences = model.generate(
input_ids=input_ids,
max_length=175 + input_size,
min_length=40 + input_size,
top_p=0.95,
#top_k=0,
do_sample=True,
num_return_sequences=1,
temperature=1.0, # 0.9,
pad_token_id=0,
eos_token_id=2,
bad_words_ids=bad_word_ids,
no_repeat_ngram_size=6
)
if len(output_sequences.shape) > 3:
output_sequences.squeeze_()
generated_sequence = output_sequences[0].tolist()[input_size:]
if print_debug_output:
for idx in generated_sequence:
print(idx, tokenizer.decode([idx], clean_up_tokenization_spaces=True).strip())
text = tokenizer.decode(generated_sequence, clean_up_tokenization_spaces=True)
text = text[: text.find("</s>")]
text = text[: text.rfind(".") + 1]
return context + text
def gen_girik(context, sign, bad_word_ids, print_debug_output=False):
bad_word_ids_girik = copy.copy(bad_word_ids)
bad_word_ids_girik += [tokenizer.encode(bad_word, add_prefix_space=True) for bad_word in signs]
bad_word_ids_girik += [tokenizer.encode("." + bad_word, add_prefix_space=False) for bad_word in signs]
return gen_fragment(context + "\n\n" + sign + "\n", bad_word_ids_girik, print_debug_output=False)
signs = ["Лингвистическому мусору и иностранным словам в русском языке не место!",
"Будет ли Путин президентом после 2024 года?",
"Кто победил: Армения или Азербайджан?",
"И последнее. Когда в России настанет долгожданный мир во всём мире? И чтобы больше таких вопросов не было.",
"Почему Европа постоянно вводит санкции против России?",
"Не надо шутить с войной. Здесь другие ребята.",
"Ночью наши учёные чуть-чуть изменят гравитационное поле Земли, и твоя страна будет под водой.",
"Что было бы, если бы Жириновский стал президентом?",
"Когда Россия станет самой богатой и могущественной страной в мире?",
"Джордж, Джордж! Посмотри ковбойские фильмы!",
"От чего коровы с ума сходят? От британской демократии.",
]
beginning = "Жириновский говорит:."
current_text = beginning
for sign in signs:
current_text = gen_girik(current_text, sign, bad_word_ids)
print(current_text)
```
|
github_jupyter
|
```
# Copyright 2022 Google LLC.
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# 4. Model Training
This notebook demonstrates how to train a Propensity Model using BigQuery ML.
### Requirements
* Input features used for training needs to be stored as a BigQuery table. This can be done using [2. ML Data Preparation Notebook](2.ml_data_preparation.ipynb).
### Install and import required modules
```
# Uncomment to install required python modules
# !sh ../utils/setup.sh
# Add custom utils module to Python environment
import os
import sys
sys.path.append(os.path.abspath(os.pardir))
from gps_building_blocks.cloud.utils import bigquery as bigquery_utils
from utils import model
from utils import helpers
```
### Set paramaters
```
configs = helpers.get_configs('config.yaml')
dest_configs = configs.destination
# GCP project ID
PROJECT_ID = dest_configs.project_id
# Name of the BigQuery dataset
DATASET_NAME = dest_configs.dataset_name
# To distinguish the separate runs of the training pipeline
RUN_ID = 'TRAIN_01'
# BigQuery table name containing model development dataset
FEATURES_DEV_TABLE = f'features_dev_table_{RUN_ID}'
# BigQuery table name containing model testing dataset
FEATURES_TEST_TABLE = f'features_test_table_{RUN_ID}'
# Output model name to save in BigQuery
MODEL_NAME = f'propensity_model_{RUN_ID}'
```
Next, let's configure modeling options.
### Model and features configuration
Model options can be configured in detail based on BigQuery ML specifications
listed in [The CREATE MODEL statement](https://cloud.google.com/bigquery-ml/docs/reference/standard-sql/bigqueryml-syntax-create).
**NOTE**: Propensity modeling supports only following four types of models available in BigQuery ML:
- LOGISTIC_REG
- [AUTOML_CLASSIFIER](https://cloud.google.com/bigquery-ml/docs/reference/standard-sql/bigqueryml-syntax-create-automl)
- [BOOSTED_TREE_CLASSIFIER](https://cloud.google.com/bigquery-ml/docs/reference/standard-sql/bigqueryml-syntax-create-boosted-tree)
- [DNN_CLASSIFIER](https://cloud.google.com/bigquery-ml/docs/reference/standard-sql/bigqueryml-syntax-create-dnn-models)
In order to use specific model options, you can add options to following configuration exactly same as listed in the [The CREATE MODEL statement](https://cloud.google.com/bigquery-ml/docs/reference/standard-sql/bigqueryml-syntax-create). For example, if you want to trian [AUTOML_CLASSIFIER](https://cloud.google.com/bigquery-ml/docs/reference/standard-sql/bigqueryml-syntax-create-automl) with `BUDGET_HOURS=1`, you can specify it as:
```python
params = {
'model_type': 'AUTOML_CLASSIFIER',
'budget_hours': 1
}
```
```
# Read in Features table schema to select feature names for model training
sql = ("SELECT column_name "
f"FROM `{PROJECT_ID}.{DATASET_NAME}`.INFORMATION_SCHEMA.COLUMNS "
f"WHERE table_name='{FEATURES_DEV_TABLE}';")
print(sql)
features_schema = bq_utils.run_query(sql).to_dataframe()
# Columns to remove from the feature list
to_remove = ['window_start_ts', 'window_end_ts', 'snapshot_ts', 'user_id',
'label', 'key', 'data_split']
# Selected features for model training
training_features = [v for v in features_schema['column_name']
if v not in to_remove]
print('Number of training features:', len(training_features))
print(training_features)
# Set parameters for AUTOML_CLASSIFIER model
FEATURE_COLUMNS = training_features
TARGET_COLUMN = 'label'
params = {
'model_path': f'{PROJECT_ID}.{DATASET_NAME}.{MODEL_NAME}',
'features_table_path': f'{PROJECT_ID}.{DATASET_NAME}.{FEATURES_DEV_TABLE}',
'feature_columns': FEATURE_COLUMNS,
'target_column': TARGET_COLUMN,
'MODEL_TYPE': 'AUTOML_CLASSIFIER',
'BUDGET_HOURS': 1.0,
# Enable data_split_col if you want to use custom data split.
# Details on AUTOML data split column:
# https://cloud.google.com/automl-tables/docs/prepare#split
# 'DATA_SPLIT_COL': 'data_split',
'OPTIMIZATION_OBJECTIVE': 'MAXIMIZE_AU_ROC'
}
```
## Train the model
First, we initialize `PropensityModel` with config parameters.
```
bq_utils = bigquery_utils.BigQueryUtils(project_id=PROJECT_ID)
propensity_model = model.PropensityModel(bq_utils=bq_utils,
params=params)
```
Next cell triggers model training job in BigQuery which takes some time to finish depending on dataset size and model complexity. Set `verbose=True`, if you want to verify training query details.
```
propensity_model.train(verbose=False)
```
Following cell allows you to see detailed information about the input features used to train a model. It provides following columns:
- input — The name of the column in the input training data.
- min — The sample minimum. This column is NULL for non-numeric inputs.
- max — The sample maximum. This column is NULL for non-numeric inputs.
- mean — The average. This column is NULL for non-numeric inputs.
- stddev — The standard deviation. This column is NULL for non-numeric inputs.
- category_count — The number of categories. This column is NULL for non-categorical columns.
- null_count — The number of NULLs.
For more details refer to [help page](https://cloud.google.com/bigquery-ml/docs/reference/standard-sql/bigqueryml-syntax-feature).
```
propensity_model.get_feature_info()
```
### Evaluate the model
This section helps to do quick model evaluation to get following model metrics:
* recall
* accuracy
* f1_score
* log_loss
* roc_auc
Two optional parameters can be specified for evaluation:
* eval_table: BigQuery table containing evaluation dataset
* threshold: Custom probability threshold to be used for evaluation (to binarize the predictions). Default value is 0.5.
If neither of these options are specified, the model is evaluated using evaluation dataset split during training with default threshold of 0.5.
**NOTE:** This evaluation provides basic model performance metrics. For thorough evaluation refer to [5. Model evaluation notebook](5.model_evaluation_and_diagnostics.ipynb) notebook.
TODO(): Add sql code to calculate the proportion of positive examples in the evaluation dataset to be used as the *threshold*.
```
# Model performance on the model development dataset on which the final
# model has been trained
EVAL_TABLE_NAME = FEATURES_DEV_TABLE
eval_params = {
'eval_table_path': f'{PROJECT_ID}.{DATASET_NAME}.{EVAL_TABLE_NAME}',
'threshold': 0.5
}
propensity_model.evaluate(eval_params, verbose=False)
# Model performance on the held out test dataset
EVAL_TABLE_NAME = FEATURES_TEST_TABLE
eval_params = {
'eval_table_path': f'{PROJECT_ID}.{DATASET_NAME}.{EVAL_TABLE_NAME}',
'threshold': 0.5
}
propensity_model.evaluate(eval_params, verbose=False)
```
## Next
Use [5. Model evaluation notebook](5.model_evaluation_and_diagnostics.ipynb) to get detailed performance metrics of the model and decide of model actually solves the business problem.
|
github_jupyter
|
```
import torch
import torch.nn as nn
import onmt
import onmt.inputters
import onmt.modules
import onmt.utils
```
We begin by loading in the vocabulary for the model of interest. This will let us check vocab size and to get the special ids for padding.
```
vocab = dict(torch.load("../../data/data.vocab.pt"))
src_padding = vocab["src"].stoi[onmt.inputters.PAD_WORD]
tgt_padding = vocab["tgt"].stoi[onmt.inputters.PAD_WORD]
```
Next we specify the core model itself. Here we will build a small model with an encoder and an attention based input feeding decoder. Both models will be RNNs and the encoder will be bidirectional
```
emb_size = 10
rnn_size = 6
# Specify the core model.
encoder_embeddings = onmt.modules.Embeddings(emb_size, len(vocab["src"]),
word_padding_idx=src_padding)
encoder = onmt.encoders.RNNEncoder(hidden_size=rnn_size, num_layers=1,
rnn_type="LSTM", bidirectional=True,
embeddings=encoder_embeddings)
decoder_embeddings = onmt.modules.Embeddings(emb_size, len(vocab["tgt"]),
word_padding_idx=tgt_padding)
decoder = onmt.decoders.decoder.InputFeedRNNDecoder(hidden_size=rnn_size, num_layers=1,
bidirectional_encoder=True,
rnn_type="LSTM", embeddings=decoder_embeddings)
model = onmt.models.model.NMTModel(encoder, decoder)
# Specify the tgt word generator and loss computation module
model.generator = nn.Sequential(
nn.Linear(rnn_size, len(vocab["tgt"])),
nn.LogSoftmax())
loss = onmt.utils.loss.NMTLossCompute(model.generator, vocab["tgt"])
```
Now we set up the optimizer. This could be a core torch optim class, or our wrapper which handles learning rate updates and gradient normalization automatically.
```
optim = onmt.utils.optimizers.Optimizer(method="sgd", lr=1, max_grad_norm=2)
optim.set_parameters(model.named_parameters())
```
Now we load the data from disk. Currently will need to call a function to load the fields into the data as well.
```
# Load some data
data = torch.load("../../data/data.train.1.pt")
valid_data = torch.load("../../data/data.valid.1.pt")
data.load_fields(vocab)
valid_data.load_fields(vocab)
data.examples = data.examples[:100]
```
To iterate through the data itself we use a torchtext iterator class. We specify one for both the training and test data.
```
train_iter = onmt.inputters.OrderedIterator(
dataset=data, batch_size=10,
device=-1,
repeat=False)
valid_iter = onmt.inputters.OrderedIterator(
dataset=valid_data, batch_size=10,
device=-1,
train=False)
```
Finally we train.
```
trainer = onmt.Trainer(model, loss, loss, optim)
def report_func(*args):
stats = args[-1]
stats.output(args[0], args[1], 10, 0)
return stats
for epoch in range(2):
trainer.train(epoch, report_func)
val_stats = trainer.validate()
print("Validation")
val_stats.output(epoch, 11, 10, 0)
trainer.epoch_step(val_stats.ppl(), epoch)
```
To use the model, we need to load up the translation functions
```
import onmt.translate
translator = onmt.translate.Translator(beam_size=10, fields=data.fields, model=model)
builder = onmt.translate.TranslationBuilder(data=valid_data, fields=data.fields)
valid_data.src_vocabs
for batch in valid_iter:
trans_batch = translator.translate_batch(batch=batch, data=valid_data)
translations = builder.from_batch(trans_batch)
for trans in translations:
print(trans.log(0))
break
```
|
github_jupyter
|
# 决策树
- 非参数学习算法
- 天然解决多分类问题
- 也可以解决回归问题
- 非常好的可解释性
```
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import cross_val_score
from sklearn import datasets
iris = datasets.load_iris()
print(iris.DESCR)
X = iris.data[:, 2:] # 取后两个特征
y = iris.target
plt.scatter(X[y==0, 0], X[y==0, 1])
plt.scatter(X[y==1, 0], X[y==1, 1])
plt.scatter(X[y==2, 0], X[y==2, 1])
```
### 1. scikit-learn 中的决策树
```
from sklearn.tree import DecisionTreeClassifier
# entropy : 熵
dt_clf = DecisionTreeClassifier(max_depth=3, criterion="entropy")
dt_clf.fit(X, y)
def plot_decision_boundary(model, axis):
x0, x1 = np.meshgrid(
np.linspace(axis[0], axis[1], int((axis[1] - axis[0])*100)).reshape(1, -1),
np.linspace(axis[2], axis[3], int((axis[3] - axis[2])*100)).reshape(-1, 1)
)
X_new = np.c_[x0.ravel(), x1.ravel()]
y_predic = model.predict(X_new)
zz = y_predic.reshape(x0.shape)
from matplotlib.colors import ListedColormap
custom_cmap = ListedColormap(['#EF9A9A', '#FFF590', '#90CAF9'])
plt.contourf(x0, x1, zz, linewidth=5, cmap=custom_cmap)
plot_decision_boundary(dt_clf, axis=(0.5, 7.5, 0, 3))
plt.scatter(X[y==0, 0], X[y==0, 1])
plt.scatter(X[y==1, 0], X[y==1, 1])
plt.scatter(X[y==2, 0], X[y==2, 1])
```
### 2. 如何构建决策树
**问题**
- 每个节点在那个维度做划分?
- 某个维度在那个值上做划分?
- 划分的标准就是,**划分后使得信息熵降低**
**信息熵**
- 熵在信息论中代表随机变量不确定的度量
- 熵越大,数据的不确定性越高
- 熵越小,数据的不确定性越低
$$H = -\sum_{i=1}^kp_i\log{(p_i)}$$
- 其中 $p_i$ 表示每一类信息在所有信息类别中占的比例

- 对于二分类,香农公式为:
$$H=-x\log(x)-(1-x)\log(1-x)$$
**信息熵函数**
```
def entropy(p):
return -p * np.log(p) - (1-p) * np.log(1-p)
x = np.linspace(0.01, 0.99)
plt.plot(x, entropy(x))
```
- 可以看出,当 x 越接近0.5,熵越高
### 3. 模拟使用信息熵进行划分
```
# 基于维度 d 的 value 值进行划分
def split(X, y, d, value):
index_a = (X[:, d] <= value)
index_b = (X[:, d] > value)
return X[index_a], X[index_b], y[index_a], y[index_b]
from collections import Counter
from math import log
# 计算每一类样本点的熵的和
def entropy(y):
counter = Counter(y)
res = 0.0
for num in counter.values():
p = num / len(y)
res += -p * log(p)
return res
# 寻找要划分的 value 值,寻找最小信息熵及相应的点
def try_split(X, y):
best_entropy = float('inf') # 最小的熵的值
best_d, best_v = -1, -1 # 划分的维度,划分的位置
# 遍历每一个维度
for d in range(X.shape[1]):
# 每两个样本点在 d 这个维度中间的值. 首先把 d 维所有样本排序
sorted_index = np.argsort(X[:, d])
for i in range(1, len(X)):
if X[sorted_index[i-1], d] != X[sorted_index[i], d]:
v = (X[sorted_index[i-1], d] + X[sorted_index[i], d]) / 2
x_l, x_r, y_l, y_r = split(X, y, d, v)
# 计算当前划分后的两部分结果熵是多少
e = entropy(y_l) + entropy(y_r)
if e < best_entropy:
best_entropy, best_d, best_v = e, d, v
return best_entropy, best_d, best_v
best_entropy, best_d, best_v = try_split(X, y)
print("best_entropy = ", best_entropy)
print("best_d", best_d)
print("best_v", best_v)
```
**即在第 0 个维度的 2.45 位置进行划分,可以得到最低的熵,值为 0.693**
```
X1_l, X1_r, y1_l, y1_r = split(X, y, best_d, best_v)
entropy(y1_r)
entropy(y1_l) # 从上图可以看出,粉红色部分只有一类,故熵为 0
best_entropy2, best_d2, best_v2 = try_split(X1_r, y1_r)
print("best_entropy = ", best_entropy2)
print("best_d", best_d2)
print("best_v", best_v2)
X2_l, X2_r, y2_l, y2_r = split(X1_r, y1_r, best_d2, best_v2)
entropy(y2_r)
entropy(y2_l)
```
|
github_jupyter
|
```
import pandas as pd
import numpy as np
from PIL import Image
import os
import sys
!pip install ipython-autotime
%load_ext autotime
%matplotlib inline
```
1. Extract your dataset and split into train_x, train_y, test_x and test_y.
2. Execute the following cells
---
## Hybrid Social Group Optimization
---
```
N = 5 # Number of persons in population
D = len(train_x.columns) # Number of features in dataset
g = 10 # Number of generations
c = 0.6 # Self Introspection factor
r0 = 1
r1 = 0.4
r2 = 0.6
print(r1, r2)
```
**Population Initialization**
```
population = np.random.choice([0,1,2,3,4,5,6,7,8,9], (N,D), p=[0.16, 0.16, 0.16, 0.16, 0.16, 0.04, 0.04, 0.04, 0.04, 0.04]) #Determines no. of features selected by probablity
population = population.astype(float)
print(population.shape)
population
fitness = np.zeros(N)
test_x.shape
def fitter_trait(X_old, X_new):
if X_new > X_old:
return X_old
else:
return X_new
```
**Fitness Function**
```
global classifier
classifier = Svc #Change classifier here
select = train_x.columns
selectno = len(train_x.columns)
classifier.fit(train_x, train_y)
select_acc = classifier.score(test_x, test_y)
def fitness_function(pop): #Fitness Function
for i in range(N):
new_train_x = train_x
new_test_x = test_x
global select
global selectno
global select_acc
new_train_x = new_train_x.drop(train_x.columns[pop[i] < 4], axis = 1)
new_test_x = new_test_x.drop(test_x.columns[pop[i] < 4], axis = 1)
classifier.fit(new_train_x, train_y)
fitness[i] = classifier.score(new_test_x, test_y)
if (fitness[i] > select_acc):
select = new_train_x.columns
# print(select.shape)
selectno = new_train_x.shape[1]
select_acc = fitness[i]
elif fitness[i] == select_acc and new_train_x.shape[1] < selectno:
select = new_train_x.columns
selectno = new_train_x.shape[1]
print("\nPerson "+ str(i+1))
print("No. of Features Used = "+ str(new_train_x.shape[1])+ "/"+str(D)+"\nFitness = " + str(fitness[i]))
print("Feature Used = ", end = " ")
#print(new_train_x.columns)
print(new_train_x.shape[1])
# Initializing Fitness values of population
# fitness_function(population)
# selectno
```
**Gbest : Fittest person in population**
```
###Determining GBest
gbest = 0
gbest_i = 0
def find_gbest():
gbest = max(fitness)#This can be any function
gbest_i = fitness.argmax()
print("Best fitness value for the generation = "+str(gbest) + " Person " + str(gbest_i+1)+"\n")
find_gbest()
#we chose maximum fitness value to be better for simplicity
def cal_fitness(person):
new_train_x = train_x
new_test_x = test_x
new_train_x = new_train_x.drop(train_x.columns[person < 4], axis = 1)
new_test_x = new_test_x.drop(test_x.columns[person < 4], axis = 1)
classifier.fit(new_train_x, train_y)
return classifier.score(new_test_x, test_y)
cal_fitness(population[0])
# new_train_x = train_x
# new_test_x = test_x
# new_train_x = new_train_x.drop(train_x.columns[person < 4], axis = 1)
# new_test_x = new_test_x.drop(test_x.columns[person < 4], axis = 1)
per1 = np.zeros((1,10000))
print(per1.shape)
per1[0][5] = 8
per1[0][89] = 7
per1[0][45] = 6
cal_fitness(per1[0])
```
---
**Mutation Phase**
```
def mutate():
gworst_i = fitness.argmin()
gworst = min(fitness)
mut = np.random.randint(0,2,size=(1,D))[0]
print("Mutating the Generation's Worst....Person "+ str(gworst_i+1))
for i in range(D):
if mut[i] > 0:
mut[i] = population[gbest_i][i]
else:
mut[i] = population[gworst_i][i]
if cal_fitness(mut) > gworst:
population[gworst_i] = mut
print("Person "+str(gworst_i)+" mutated")
else:
print("No Mutations in this generation")
mut = np.random.randint(0,2,size=(1,D))[0]
mut
div = pd.DataFrame(np.random.randint(0,2,size=(1,D))[0])
# div.iloc[:,div > 0] = population[2][div>0]
# div
```
---
**Improving Phase**
```
## Improving Phase
# i = 1
def improve():
print("Improving.......")
for i in range(N):
Xnew = population[i]
print('Persona '+ str(i+1))
for j in range(D):
Xnew[j] = c * population[i][j] + r0 * (population[gbest_i][j] - population[i][j])
try:
if cal_fitness(Xnew) > fitness[i]:
population[i] = Xnew
except:
print("Oops!", sys.exc_info()[0], "occurred.")
print("Next entry.")
```
---
**Acquiring Phase**
```
## Acquiring Phase
def acquire():
random_person = np.random.randint(low=0, high=N)
for i in range(N):
if random_person == i:
random_person = np.random.randint(low=0, high=N)
i = i - 1
continue
X_new = population[i]
if fitness[random_person] > fitness[i]:
for j in range(D):
X_new[j] = population[i][j] + r1*(population[random_person][j]-population[i][j]) + r2*(population[gbest_i][j]-population[i][j])
if cal_fitness(X_new) > fitness[i]:
population[i] = X_new
else:
for j in range(D):
X_new[j] = population[i][j] + r1*(population[i][j]-population[random_person][j]) + r2*(population[gbest_i][j]-population[i][j])
if cal_fitness(X_new) > fitness[i]:
population[i] = X_new
#Run
try:
for k in range(g):
print("Generation "+ str(k+1) + "\n---------------")
fitness_function(population)
find_gbest()
mutate()
improve()
acquire()
except:
print()
print("........................")
print("Optimal Solution Reached")
print("........................")
select.shape
```
|
github_jupyter
|
# ClusterFinder Reference genomes reconstruction
This notebook validates the 10 genomes we obtained from NCBI based on the ClusterFinder supplementary table.
We check that the gene locations from the supplementary table match locations in the GenBank files.
```
from Bio import SeqIO
from Bio.SeqFeature import FeatureLocation
import pandas as pd
from Bio import Entrez
import seaborn as sns
def get_features_of_type(sequence, feature_type):
return [feature for feature in sequence.features if feature.type == feature_type]
def get_reference_gene_location(gene_csv_row):
start = gene_csv_row['gene start'] - 1
end = gene_csv_row['gene stop']
strand = 1 if gene_csv_row['gene strand'] == '+' else (-1 if gene_csv_row['gene strand'] == '-' else None)
return FeatureLocation(start, end, strand)
def feature_locus_matches(feature, reference_locus):
return feature.qualifiers.get('locus_tag',[None])[0] == reference_locus
```
# Loading reference cluster gene locations
```
reference_genes = pd.read_csv('../data/clusterfinder/labelled/CF_labelled_genes_orig.csv', sep=';')
reference_genes.head()
```
# Genes with no sequence
```
no_sequence_genes = reference_genes[reference_genes['NCBI ID'] == '?']
no_sequence_counts = no_sequence_genes.groupby('Genome ID')['gene locus'].count()
print('{} genes don\'t have a sequence!'.format(len(no_sequence_genes)))
pd.DataFrame({'missing genes':no_sequence_counts})
reference_ids = reference_genes[reference_genes['NCBI ID'] != '?']['NCBI ID'].unique()
reference_ids
```
# Validating that reference genes are found in our sequences
```
def validate_genome(record, record_reference_genes):
print('Validating {}'.format(record.id))
record_genes = get_features_of_type(record, 'gene')
record_cds = get_features_of_type(record, 'CDS')
validation = []
record_length = len(record.seq)
min_location = record_length
max_location = -1
prev_gene_index = None
prev_cluster_start = None
for i, reference_gene in record_reference_genes.iterrows():
reference_gene_location = get_reference_gene_location(reference_gene)
reference_gene_locus = reference_gene['gene locus']
reference_cluster_start = reference_gene['NPL start']
gene_matches_locus = [f for f in record_genes if feature_locus_matches(f, reference_gene_locus)]
cds_matches_locus = [f for f in record_cds if feature_locus_matches(f, reference_gene_locus)]
gene_matches_location = [f for f in gene_matches_locus if reference_gene_location == f.location]
cds_matches_location = [f for f in cds_matches_locus if reference_gene_location == f.location]
validation.append({
'gene_locus_not_found':not gene_matches_locus,
'cds_locus_not_found':not cds_matches_locus,
'gene_location_correct': bool(gene_matches_location),
'cds_location_correct': bool(cds_matches_location)
})
if not cds_matches_locus:
print('No CDS found for gene locus {}'.format(reference_gene_locus))
if gene_matches_locus:
gene_match = gene_matches_locus[0]
if not cds_matches_locus:
print(' Gene: ', gene_match.qualifiers)
# Use gene index to check if we have a consecutive sequence of genes (except when going from one cluster to another)
gene_index = [gi for gi,f in enumerate(record_genes) if feature_locus_matches(f, reference_gene_locus)][0]
if reference_cluster_start == prev_cluster_start and gene_index != prev_gene_index + 1:
print('Additional unexpected genes found before {} (index {} -> {}) at cluster start {}'.format(reference_gene_locus, prev_gene_index, gene_index, reference_cluster_start))
# Calculate min and max cluster gene location to see how much of the sequence is covered by the reference genes
min_location = min(gene_match.location.start, min_location)
max_location = max(gene_match.location.end, max_location)
prev_gene_index = gene_index
prev_cluster_start = reference_cluster_start
result = pd.DataFrame(validation).sum().to_dict()
result['location correct'] = min(result['gene_location_correct'], result['cds_location_correct']) / len(validation)
result['ID'] = record.id
result['genome'] = record_reference_genes.iloc[0]['Genome ID']
result['sequence length'] = record_length
result['total genes'] = len(record_genes)
result['reference genes'] = len(record_reference_genes)
result['first location'] = min_location / record_length
result['last location'] = max_location / record_length
result['covered'] = (max_location - min_location) / record_length
return result
validations = []
reference_gene_groups = reference_genes.groupby('NCBI ID')
records = SeqIO.parse('../data/clusterfinder/labelled/CF_labelled_contigs.gbk', 'genbank')
for record in records:
ncbi_id = record.id
print(ncbi_id)
record_reference_genes = reference_gene_groups.get_group(ncbi_id)
validations.append(validate_genome(record, record_reference_genes))
validations = pd.DataFrame(validations)
validations.set_index('ID', inplace=True)
validations
validations['location correct'].mean()
1 - validations['location correct'].mean()
validations[['genome','first location','last location','covered','location correct','reference genes','total genes']]
```
# Cluster genes
```
genes = pd.read_csv('../data/clusterfinder/labelled/CF_labelled_genes.csv', sep=';')
genes.head()
cluster_counts = genes.groupby('contig_id')['cluster_id'].nunique()
cluster_counts.sort_values().plot.barh()
gene_counts = genes.groupby('cluster_id')['locus_tag'].count()
gene_counts.hist(bins=50)
```
|
github_jupyter
|
# Realization of Recursive Filters
*This jupyter notebook is part of a [collection of notebooks](../index.ipynb) on various topics of Digital Signal Processing. Please direct questions and suggestions to [[email protected]](mailto:[email protected]).*
## Cascaded Structures
The realization of recursive filters with a high order may be subject to numerical issues. For instance, when the coefficients span a wide amplitude range, their quantization may require a small quantization step or may impose a large relative error for small coefficients. The basic concept of cascaded structures is to decompose a high order filter into a cascade of lower order filters, typically first and second order recursive filters.
### Decomposition into Second-Order Sections
The rational transfer function $H(z)$ of a linear time-invariant (LTI) recursive system can be [expressed by its zeros and poles](introduction.ipynb#Transfer-Function) as
\begin{equation}
H(z) = \frac{b_M}{a_N} \cdot \frac{\prod_{\mu=1}^{P} (z - z_{0\mu})^{m_\mu}}{\prod_{\nu=1}^{Q} (z - z_{\infty\nu})^{n_\nu}}
\end{equation}
where $z_{0\mu}$ and $z_{\infty\nu}$ denote the $\mu$-th zero and $\nu$-th pole of degree $m_\mu$ and $n_\nu$ of $H(z)$, respectively. The total number of zeros and poles is denoted by $P$ and $Q$.
The poles and zeros of a real-valued filter $h[k] \in \mathbb{R}$ are either single real valued or conjugate complex pairs. This motivates to split the transfer function into
* first order filters constructed from a single pole and zero
* second order filters constructed from a pair of conjugated complex poles and zeros
Decomposing the transfer function into these two types by grouping the poles and zeros into single poles/zeros and conjugate complex pairs of poles/zeros results in
\begin{equation}
H(z) = K \cdot \prod_{\eta=1}^{S_1} \frac{(z - z_{0\eta})}{(z - z_{\infty\eta})}
\cdot \prod_{\eta=1}^{S_2} \frac{(z - z_{0\eta}) (z - z_{0\eta}^*)} {(z - z_{\infty\eta})(z - z_{\infty\eta}^*)}
\end{equation}
where $K$ denotes a constant and $S_1 + 2 S_2 = N$ with $N$ denoting the order of the system. The cascade of two systems results in a multiplication of their transfer functions. Above decomposition represents a cascade of first- and second-order recursive systems. The former can be treated as a special case of second-order recursive systems. The decomposition is therefore known as decomposition into second-order sections (SOSs) or [biquad filters](https://en.wikipedia.org/wiki/Digital_biquad_filter). Using a cascade of SOSs the transfer function of the recursive system can be rewritten as
\begin{equation}
H(z) = \prod_{\mu=1}^{S} \frac{b_{0, \mu} + b_{1, \mu} \, z^{-1} + b_{2, \mu} \, z^{-2}}{1 + a_{1, \mu} \, z^{-1} + a_{2, \mu} \, z^{-2}}
\end{equation}
where $S = \lceil \frac{N}{2} \rceil$ denotes the total number of SOSs. These results state that any real valued system of order $N > 2$ can be decomposed into SOSs. This has a number of benefits
* quantization effects can be reduced by sensible grouping of poles/zeros, e.g. such that the spanned amplitude range of the filter coefficients is limited
* A SOS may be extended by a gain factor to further reduce quantization effects by normalization of the coefficients
* efficient and numerically stable SOSs serve as generic building blocks for higher-order recursive filters
### Example - Cascaded second-order section realization of a lowpass
The following example illustrates the decomposition of a higher-order recursive Butterworth lowpass filter into a cascade of second-order sections.
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.markers import MarkerStyle
from matplotlib.patches import Circle
import scipy.signal as sig
N = 9 # order of recursive filter
def zplane(z, p, title='Poles and Zeros'):
"Plots zero and pole locations in the complex z-plane"
ax = plt.gca()
ax.plot(np.real(z), np.imag(z), 'bo', fillstyle='none', ms = 10)
ax.plot(np.real(p), np.imag(p), 'rx', fillstyle='none', ms = 10)
unit_circle = Circle((0,0), radius=1, fill=False,
color='black', ls='solid', alpha=0.9)
ax.add_patch(unit_circle)
ax.axvline(0, color='0.7')
ax.axhline(0, color='0.7')
plt.title(title)
plt.xlabel(r'Re{$z$}')
plt.ylabel(r'Im{$z$}')
plt.axis('equal')
plt.xlim((-2, 2))
plt.ylim((-2, 2))
plt.grid()
# design filter
b, a = sig.butter(N, 0.2)
# decomposition into SOS
sos = sig.tf2sos(b, a, pairing='nearest')
# print filter coefficients
print('Coefficients of the recursive part \n')
print(['%1.2f'%ai for ai in a])
print('\n')
print('Coefficients of the recursive part of the individual SOS \n')
print('Section \t a1 \t\t a2')
for n in range(sos.shape[0]):
print('%d \t\t %1.5f \t %1.5f'%(n, sos[n, 4], sos[n, 5]))
# plot pole and zero locations
plt.figure(figsize=(5,5))
zplane(np.roots(b), np.roots(a), 'Poles and Zeros - Overall')
plt.figure(figsize=(10, 7))
for n in range(sos.shape[0]):
plt.subplot(231+n)
zplane(np.roots(sos[n, 0:3]), np.roots(sos[n, 3:6]), title='Poles and Zeros - Section %d'%n)
plt.tight_layout()
# compute and plot frequency response of sections
plt.figure(figsize=(10,5))
for n in range(sos.shape[0]):
Om, H = sig.freqz(sos[n, 0:3], sos[n, 3:6])
plt.plot(Om, 20*np.log10(np.abs(H)), label=r'Section %d'%n)
plt.xlabel(r'$\Omega$')
plt.ylabel(r'$|H_n(e^{j \Omega})|$ in dB')
plt.legend()
plt.grid()
```
**Exercise**
* What amplitude range is spanned by the filter coefficients?
* What amplitude range is spanned by the SOS coefficients?
* Change the pole/zero grouping strategy from `pairing='nearest'` to `pairing='keep_odd'`. What changes?
* Increase the order `N` of the filter. What changes?
Solution: Inspecting both the coefficients of the recursive part of the original filter and of the individual SOS reveals that the spanned amplitude range is lower for the latter. The choice of the pole/zero grouping strategy influences the locations of the poles/zeros in the individual SOS, the spanned amplitude range of their coefficients and the transfer functions of the individual sections. The total number of SOS scales with the order of the original filter.
**Copyright**
This notebook is provided as [Open Educational Resource](https://en.wikipedia.org/wiki/Open_educational_resources). Feel free to use the notebook for your own purposes. The text is licensed under [Creative Commons Attribution 4.0](https://creativecommons.org/licenses/by/4.0/), the code of the IPython examples under the [MIT license](https://opensource.org/licenses/MIT). Please attribute the work as follows: *Sascha Spors, Digital Signal Processing - Lecture notes featuring computational examples, 2016-2018*.
|
github_jupyter
|
# Utilizing daal4py in Data Science Workflows
The notebook below has been made to demonstrate daal4py in a data science context. It utilizes a Cycling Dataset for pyworkout-toolkit, and attempts to create a linear regression model from the 5 features collected for telemetry to predict the user's Power output in the absence of a power meter.
```
import pandas as pd
import matplotlib.pyplot as plt
import glob
import sys
%matplotlib inline
sys.version
```
This example will be exploring workout data pulled from Strava, processed into a CSV for Pandas and daal4py usage. Below, we utilize pandas to read in the CSV file, and look at the head of dataframe with .head()
```
workout_data_dd= pd.read_csv('data/batch/cycling_dataset.csv', index_col=0)
workout_data_dd.head()
```
The data above has several key features that would be of great use here.
- Altitude can affect performance, so it might be a useful feature.
- Cadence is the revolutions per minute of the crank, and may have possible influence.
- Heart Rate is a measure of the body's workout strain, and would have a high possibly of influence.
- Distance may have a loose correlation as it is highly route dependent, but might be possible.
- Speed has possible correlations as it ties directly into power.
## Explore and visualize some of the data
In general, we are trying to predict on the 'power' in Watts to see if we can generate a model that can predict one's power output without the usage of a cycling power meter. Below are some basic scatterplots as we explore the data. Scatterplots are great for looking for patterns and correlation in the data itself. Below, we can see that cadence and speed are positively correlated.
```
workout_data_dd.plot.scatter('cadence','power')
plt.show()
workout_data_dd.plot.scatter('hr','power')
plt.show()
workout_data_dd.plot.scatter('cadence','speed')
plt.show()
workout_data_dd.plot.scatter('speed','power')
plt.show()
workout_data_dd.plot.scatter('altitude','power')
plt.show()
workout_data_dd.plot.scatter('distance','power')
plt.show()
```
## Using daal4py for Machine Learning tasks
In the sections below, we will be using daal4py directly. After importing the model, we will arrange it in a separate independent and dependent dataframes, then use the daal4py's training and prediction classes to generate a workable model.
```
import daal4py as d4p
```
It is now the time to split the dataset into train and test sets. This is demonstrated below.
```
print(workout_data_dd.shape)
train_set = workout_data_dd[0:3000]
test_set = workout_data_dd[3000:]
print(train_set.shape, test_set.shape)
# Reduce the dataset, create X. We drop the target, and other non-essential features.
reduced_dataset = train_set.drop(['time','power','latitude','longitude'], axis=1)
# Get the target, create Y
target = train_set.power.values.reshape((-1,1))
# This is essentially doing np.array(dataset.power.values, ndmin=2).T
# as it needs to force a 2 dimensional array as we only have 1 target
```
X is 5 features by 3k rows, Y is 3k rows by 1 column
```
print(reduced_dataset.values.shape, target.shape)
```
## Training the model
Create the Linear Regression Model, and train the model with the data. We utilize daal4py's linear_regression_training class to create the model, then call .compute() with the independent and dependent data as the parameters.
```
d4p_lm = d4p.linear_regression_training(interceptFlag=True)
lm_trained = d4p_lm.compute(reduced_dataset.values, target)
print("Model has this number of features: ", lm_trained.model.NumberOfFeatures)
```
## Prediction (inference) with the trained model
Now that the model is trained, we can test it with the test part of the dataset. We drop the same features to match that of the trained model, and put it into daal4py's linear_regression_prediction class.
```
subset = test_set.drop(['time','power','latitude','longitude'], axis=1)
```
Now we can create the Prediction object and use the reduced dataset for prediction. The class's arguments use the independent data and the trained model from above as the parameters.
```
lm_predictor_component = d4p.linear_regression_prediction()
result = lm_predictor_component.compute(subset.values, lm_trained.model)
plt.plot(result.prediction[0:300])
plt.plot(test_set.power.values[0:300])
plt.show()
```
The graph above shows the Orange (predicted) result over the Blue (original data). This data is notoriously sparse in features leading to a difficult to predict target!
## Model properties
Another aspect of the model is the trained model's properties, which are explored below.
```
print("Betas:",lm_trained.model.Beta)
print("Number of betas:", lm_trained.model.NumberOfBetas)
print("Number of Features:", lm_trained.model.NumberOfFeatures)
```
## Additional metrics
We can generate metrics on the independent data with daal4py's low_order_moments() class.
```
metrics_processor = d4p.low_order_moments()
data = metrics_processor.compute(reduced_dataset.values)
data.standardDeviation
```
## Migrating the trained model for inference on external systems
Occasionally one may need to migrate the trained model to another system for inference only--this use case allows the training on a much more powerful machine with a larger dataset, and placing the trained model for inference-only on a smaller machine.
```
import pickle
with open('trained_model2.pickle', 'wb') as model_pi:
pickle.dump(lm_trained.model, model_pi)
model_pi.close
```
The trained model file above can be moved to an inference-only or embedded system. This is useful if the training is extreamly heavy or computed-limited.
```
with open('trained_model2.pickle', 'rb') as model_import:
lm_import = pickle.load(model_import)
```
The imported model from file is now usable again. We can check the betas from the model to ensure that the trained model is present.
```
lm_import.Beta
```
|
github_jupyter
|
# **JIVE: Joint and Individual Variation Explained**
JIVE (Joint and Individual Variation Explained) is a dimensional reduction algorithm that can be used when there are multiple data matrices (data blocks). The multiple data block setting means there are $K$ different data matrices, with the same number of observations $n$ and (possibly) different numbers of variables ($d_1, \dots, d_k$). JIVE finds modes of variation which are common (joint) to all $K$ data blocks and modes of individual variation which are specific to each block. For a detailed discussion of JIVE see [Angle-Based Joint and Individual Variation Explained](https://arxiv.org/pdf/1704.02060.pdf).[^1]
For a concrete example, consider a two block example from a medical study. Suppose there are $n=500$ patients (observations). For each patient we have $d_1 = 100$ bio-medical variables (e.g. hit, weight, etc). Additionally we have $d_2 = 10,000$ gene expression measurements for each patient.
## **The JIVE decomposition**
Suppose we have $K$ data data matrices (blocks) with the same number of observations, but possibly different numbers of variables; in particular let $X^{(1)}, \dots, X^{(K)}$ where $X^{(k)} \in \mathbb{R}^{n \times d_k}$. JIVE will then decompose each matrix into three components: joint signal, individual signal and noise
\begin{equation}
X^{(k)} = J^{(k)} + I^{(k)} + E^{(k)}
\end{equation}
where $J^{(k)}$ is the joint signal estimate, $I^{(k)}$ is the individual signal estimate and $E^{(k)}$ is the noise estimate (each of these matrices must the same shape as the original data block: $\mathbb{R}^{n \times d_k}$). Note: **we assume each data matrix** $X^{(k)}$ **has been column mean centered**.
The matrices satisfy the following constraints:
1. The joint matrices have a common rank: $rk(J^{(k)}) = r_{joint}$ for $k=1, \dots, K$.
2. The individual matrices have block specific ranks $rk(I^{(k)}) = r_{individual}^{(k)}$.
3. The columns of the joint matrices share a common space called the joint score space (a subspace of $\mathbb{R}^n$); in particular the $\text{col-span}(J^{(1)}) = \dots = \text{col-span}(J^{(K)})$ (hence the name joint).
4. Each individual spaces score subspace (of $\mathbb{R}^n$) is orthogonal to the the joint space; in particular $\text{col-span}(J^{(k)}) \perp \text{col-span}(I^{(k)})$ for $k=1, \dots, K$.
Note that JIVE may be more natural if we think about data matrices subspaces of $\mathbb{R}^n$ (the score space perspective). Typically we think of a data matrix as $n$ points in $\mathbb{R}^d$. The score space perspective views a data matrix as $d$ vectors in $\mathbb{R}^n$ (or rather the span of these vectors). One important consequence of this perspective is that it makes sense to related the data blocks in score space (e.g. as subspaces of $\mathbb{R}^n$) since they share observtions.
## Quantities of interest
There are a number of potential quantities of interest depending on the application. For example the user may be interested in the full matrices $J^{(k)}$ and/or $I^{(k)}$. By construction these matrices are not full rank and we may also be interested in their singular value decomposition which we define as
\begin{align}
& U^{(k)}_{joint}, D^{(k)}_{joint}, V^{(k)}_{joint} = \text{rank } r_{joint} \text{ SVD of } J^{(k)} \\
& U^{(k)}_{individual}, D^{(k)}_{individual}, V^{(k)}_{individual} = \text{rank } r_{individual}^{{k}} \text{ SVD of } I^{(k)}
\end{align}
One additional quantity of interest is $U_{joint} \in \mathbb{R}^{n \times r_{joint}}$ which is an orthogonal basis of $\text{col-span}(J^{(k)})$. This matrix is produced from an intermediate JIVE computation.
## **PCA analogy**
We give a brief discussion of the PCA/SVD decomposition (assuming the reading is already familiar).
#### Basic decomposition
Suppose we have a data matrix $X \in \mathbb{n \times d}$. Assume that $X$ has been column mean centered and consider the SVD decomposition (this is PCA since we have mean centered the data):
\begin{equation}
X = U D V^T.
\end{equation}
where $U \in \mathbb{R}^{n \times m}$, $D \in \mathbb{R}^{m \times m}$ is diagonal, and $V \in \mathbb{R}^{d \times m}$ with $m = min(n, d)$. Note $U^TU = V^TV = I_{m \times m}$.
Suppose we have decided to use a rank $r$ approximation. We can then decompose $X$ into a signal matrix ($A$) and an noise matrix ($E$)
\begin{equation}
X = A + E,
\end{equation}
where $A$ is the rank $r$ SVD approximation of $X$ i.e.
\begin{align}
A := & U_{:, 1:r} D_{1:r, 1:r} V_{:, 1:r}^T \\
= & \widetilde{U}, \widetilde{D} \widetilde{V}^T
\end{align}
The notation $U_{:, 1:r} \in \mathbb{R}^{n \times r}$ means the first $r$ columns of $U$. Similarly we can see the error matrix is $E :=U_{:, r+1:n} D_{r+1:m, r_1:m} V_{:, r+1:d}^T$.
#### Quantities of interest
There are many ways to use a PCA/SVD decomposition. Some common quantities of interest include
- The normalized scores: $\widetilde{U} \in \mathbb{R}^{n \times r}$
- The unnormalized scores: $\widetilde{U}\widetilde{D} \in \mathbb{R}^{n \times r}$
- The loadings: $\widetilde{V} \in \mathbb{R}^{d \times r}$
- The full signal approximation: $A \in \mathbb{R}^{n \times d}$
#### Scores and loadings
For both PCA and JIVE we use the notation $U$ (scores) and $V$ (loadings). These show up in several places.
We refer to all $U \in \mathbb{R}^{n \times r}$ matrices as scores. We can view the $n$ rows of $U$ as representing the $n$ data points with $r$ derived variables (put differently, columns of $U$ are $r$ derived variables). The columns of $U$ are orthonormal: $U^TU = I_{r \times r}$.
Sometimes we may want $UD$ i.e scale the columns of $U$ by $D$ (the columns are still orthogonal). The can be useful when we want to represent the original data by $r$ variables. We refer to $UD$ as unnormalized scores.
We refer to all $V\in \mathbb{R}^{d \times r}$ matrices as loadings[^2]. The j$th$ column of $V$ gives the linear combination of the original $d$ variables which is equal to the j$th$ unnormalized scores (j$th$ column of $UD$). Equivalently, if we project the $n$ data points (rows of $X$) onto the j$th$ column of $V$ we get the j$th$ unnormalized scores.
The typical geometric perspective of PCA is that the scores represent $r$ new derived variables. For example, if $r = 2$ we can look at a scatter plot that gives a two dimensional approximation of the data. In other words, the rows of the scores matrix are $n$ data points living in $\mathbb{R}^r$.
An alternative geometric perspective is the $r$ columns of the scores matrix are vectors living in $\mathbb{R}^n$. The original $d$ variables span a subspace of $\mathbb{R}^n$ given by $\text{col-span}(X)$. The scores then span a lower dimensional subspace of $\mathbb{R}^n$ that approximates $\text{col-span}(X)$.
The first perspective says PCA finds a lower dimensional approximation to a subspace in $\mathbb{R}^d$ (spanned by the $n$ data points). The second perspective says PCA finds a lower dimensional approximation to a subspace in $\mathbb{R}^n$ (spanned by the $d$ data points).
## **JIVE operating in score space**
For a data matrix $X$ let's call the span of the variables (columns) the *score subpace*, $\text{col-span}(X) \subset \mathbb{R}^n$. Typically we think of a data matrix as $n$ points in $\mathbb{R}^d$. The score space perspective reverses this and says a data matrix is $d$ points in $\mathbb{R}^n$. When thinking in the score space it's common to consider about subspaces i.e. the span of the $d$ variables in $\mathbb{R}^n$. In other words, if two data matrices have the same column span then their score subspaces are the same[^3].
JIVE partitions the score space of each data matrix into three subspaces: joint, individual and noise. The joint score subspace for each data block is the same. The individual score subspace, however, is (possibly) different for each of the $K$ blocks. The k$th$ block's individual score subspace is orthogonal to the joint score subspace. Recall that the $K$ data matrices have the same number of observations ($n$) so it makes sense to think about how the data matrices relate to each other in score space.
PCA partitions the score space into two subspaces: signal and noise (see above). For JIVE we might combine the joint and individual score subspaces and call this the signal score subspace.
# Footnotes
[^1]: Note this paper calls the algorithm AJIVE (angle based JIVE) however, we simply use JIVE. Additionally, the paper uses columns as observations in data matrices where as we use rows as observations.
[^2]: For PCA we used tildes (e.g. $\widetilde{U}$) to denote the "partial" SVD approximation however for the final JIVE decomposition we do not use tildes. This is intentional since for JIVE the SVD comes from the $I$ and $J$ matrices which are exactly rank $r$. Therefore we view this SVD as the "full" SVD.
[^3]: This might remind the reader of TODO
|
github_jupyter
|
```
import tensorflow as tf
from tensorflow.keras import layers, Model
from tensorflow.keras.activations import relu
from tensorflow.keras.models import Sequential, load_model
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
from tensorflow.keras.losses import BinaryCrossentropy
from tensorflow.keras.optimizers import RMSprop, Adam
from tensorflow.keras.metrics import binary_accuracy
import tensorflow_datasets as tfds
from tensorflow_addons.layers import InstanceNormalization
import numpy as np
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings('ignore')
print("Tensorflow", tf.__version__)
from packaging.version import parse as parse_version
assert parse_version(tf.__version__) < parse_version("2.4.0"), \
f"Please install TensorFlow version 2.3.1 or older. Your current version is {tf.__version__}."
gpus = tf.config.experimental.list_physical_devices('GPU')
tf.config.experimental.set_visible_devices(gpus[0], 'GPU')
logical_gpus = tf.config.experimental.list_logical_devices('GPU')
print(len(gpus), "Physical GPUs,", len(logical_gpus), "Logical GPU")
ds_train, ds_info = tfds.load('mnist', split='train', shuffle_files=True, with_info=True)
fig = tfds.show_examples(ds_info, ds_train)
batch_size = 400
global_batch_size = batch_size * 1
image_shape = (32, 32, 1)
def preprocess(features):
image = tf.image.resize(features['image'], image_shape[:2])
image = tf.cast(image, tf.float32)
image = (image-127.5)/127.5
label = features['label']
return image, label
ds_train = ds_train.map(preprocess)
ds_train = ds_train.cache() # put dataset into memory
ds_train = ds_train.shuffle(ds_info.splits['train'].num_examples)
ds_train = ds_train.batch(global_batch_size).repeat()
train_num = ds_info.splits['train'].num_examples
train_steps_per_epoch = round(train_num/batch_size)
print(train_steps_per_epoch)
class cDCGAN():
def __init__(self, input_shape):
self.z_dim = 100
self.input_shape = input_shape
self.num_classes = 10
# discriminator
self.n_discriminator = 1
self.discriminator = self.build_discriminator()
self.discriminator.trainable = False
self.optimizer_discriminator = RMSprop(1e-4)
# build generator pipeline with frozen discriminator
self.generator = self.build_generator()
discriminator_output = self.discriminator([self.generator.output, self.generator.input[1]])
self.model = Model(self.generator.input, discriminator_output)
self.model.compile(loss = self.bce_loss,
optimizer = RMSprop(1e-4))
self.discriminator.trainable = True
self.bce = tf.keras.losses.BinaryCrossentropy()
def conv_block(self, channels, kernels, strides=1,
batchnorm=True, activation=True):
model = tf.keras.Sequential()
model.add(layers.Conv2D(channels, kernels, strides=strides, padding='same'))
if batchnorm:
model.add(layers.BatchNormalization())
if activation:
model.add(layers.LeakyReLU(0.2))
return model
def bce_loss(self, y_true, y_pred):
loss = self.bce(y_true, y_pred)
return loss
def build_generator(self):
DIM = 64
input_label = layers.Input(shape=1, dtype=tf.int32, name='ClassLabel')
one_hot_label = tf.one_hot(input_label, self.num_classes)
one_hot_label = layers.Reshape((self.num_classes,))(one_hot_label)
input_z = layers.Input(shape=self.z_dim, name='LatentVector')
x = layers.Concatenate()([input_z, one_hot_label])
x = layers.Dense(4*4*4*DIM, activation=None)(x)
x = layers.Reshape((4,4,4*DIM))(x)
#x = layers.Concatenate()([x, embedding])
x = layers.UpSampling2D((2,2), interpolation="bilinear")(x)
x = self.conv_block(2*DIM, 5)(x)
x = layers.UpSampling2D((2,2), interpolation="bilinear")(x)
x = self.conv_block(DIM, 5)(x)
x = layers.UpSampling2D((2,2), interpolation="bilinear")(x)
output = layers.Conv2D(image_shape[-1], 5, padding='same', activation='tanh')(x)
return Model([input_z, input_label], output)
def build_discriminator(self):
DIM = 64
# label
input_label = layers.Input(shape=[1], dtype =tf.int32, name='ClassLabel')
encoded_label = tf.one_hot(input_label, self.num_classes)
embedding = layers.Dense(32 * 32 * 1, activation=None)(encoded_label)
embedding = layers.Reshape((32, 32, 1))(embedding)
# discriminator
input_image = layers.Input(shape=self.input_shape, name='Image')
x = layers.Concatenate()([input_image, embedding])
x = self.conv_block(DIM, 5, 2, batchnorm=False)(x)
x = self.conv_block(2*DIM, 5, 2)(x)
x = self.conv_block(4*DIM, 5, 2)(x)
x = layers.Flatten()(x)
output = layers.Dense(1, activation='sigmoid')(x)
return Model([input_image, input_label], output)
def train_discriminator(self, real_images, class_labels, batch_size):
real_labels = tf.ones(batch_size)
fake_labels = tf.zeros(batch_size)
g_input = tf.random.normal((batch_size, self.z_dim))
fake_class_labels = tf.random.uniform((batch_size,1), minval=0, maxval=10, dtype=tf.dtypes.int32)
fake_images = self.generator.predict([g_input, fake_class_labels])
with tf.GradientTape() as gradient_tape:
# forward pass
pred_fake = self.discriminator([fake_images, fake_class_labels])
pred_real = self.discriminator([real_images, class_labels])
# calculate losses
loss_fake = self.bce_loss(fake_labels, pred_fake)
loss_real = self.bce_loss(real_labels, pred_real)
# total loss
total_loss = 0.5*(loss_fake + loss_real)
# apply gradients
gradients = gradient_tape.gradient(total_loss, self.discriminator.trainable_variables)
self.optimizer_discriminator.apply_gradients(zip(gradients, self.discriminator.trainable_variables))
return loss_fake, loss_real
def train(self, data_generator, batch_size, steps, interval=100):
val_g_input = tf.random.normal((self.num_classes, self.z_dim))
val_class_labels = np.arange(self.num_classes)
real_labels = tf.ones(batch_size)
for i in range(steps):
real_images, class_labels = next(data_generator)
loss_fake, loss_real = self.train_discriminator(real_images, class_labels, batch_size)
discriminator_loss = 0.5*(loss_fake + loss_real)
# train generator
g_input = tf.random.normal((batch_size, self.z_dim))
fake_class_labels = tf.random.uniform((batch_size, 1),
minval=0, maxval=self.num_classes, dtype=tf.dtypes.int32)
g_loss = self.model.train_on_batch([g_input, fake_class_labels], real_labels)
if i%interval == 0:
msg = "Step {}: discriminator_loss {:.4f} g_loss {:.4f}"\
.format(i, discriminator_loss, g_loss)
print(msg)
fake_images = self.generator.predict([val_g_input,val_class_labels])
self.plot_images(fake_images)
def plot_images(self, images):
grid_row = 1
grid_col = 10
f, axarr = plt.subplots(grid_row, grid_col, figsize=(grid_col*1.5, grid_row*1.5))
for col in range(grid_col):
axarr[col].imshow((images[col,:,:,0]+1)/2, cmap='gray')
axarr[col].axis('off')
plt.show()
def sample_images(self, class_labels):
z = tf.random.normal((len(class_labels), self.z_dim))
images = self.generator.predict([z,class_labels])
self.plot_images(images)
return images
cdcgan = cDCGAN(image_shape)
tf.keras.utils.plot_model(cdcgan.discriminator, show_shapes=True)
tf.keras.utils.plot_model(cdcgan.generator, show_shapes=True)
cdcgan.train(iter(ds_train), batch_size, 2000, 200)
for i in range(5):
images = cdcgan.sample_images(np.array([0,1,2,3,4,5,6,7,8,9]))
```
|
github_jupyter
|
An illustration of the metric and non-metric MDS on generated noisy data.
The reconstructed points using the metric MDS and non metric MDS are slightly shifted to avoid overlapping.
#### New to Plotly?
Plotly's Python library is free and open source! [Get started](https://plot.ly/python/getting-started/) by downloading the client and [reading the primer](https://plot.ly/python/getting-started/).
<br>You can set up Plotly to work in [online](https://plot.ly/python/getting-started/#initialization-for-online-plotting) or [offline](https://plot.ly/python/getting-started/#initialization-for-offline-plotting) mode, or in [jupyter notebooks](https://plot.ly/python/getting-started/#start-plotting-online).
<br>We also have a quick-reference [cheatsheet](https://images.plot.ly/plotly-documentation/images/python_cheat_sheet.pdf) (new!) to help you get started!
### Version
```
import sklearn
sklearn.__version__
```
### Imports
```
print(__doc__)
import plotly.plotly as py
import plotly.graph_objs as go
import numpy as np
from sklearn import manifold
from sklearn.metrics import euclidean_distances
from sklearn.decomposition import PCA
```
### Calculations
```
n_samples = 20
seed = np.random.RandomState(seed=3)
X_true = seed.randint(0, 20, 2 * n_samples).astype(np.float)
X_true = X_true.reshape((n_samples, 2))
# Center the data
X_true -= X_true.mean()
similarities = euclidean_distances(X_true)
# Add noise to the similarities
noise = np.random.rand(n_samples, n_samples)
noise = noise + noise.T
noise[np.arange(noise.shape[0]), np.arange(noise.shape[0])] = 0
similarities += noise
mds = manifold.MDS(n_components=2, max_iter=3000, eps=1e-9, random_state=seed,
dissimilarity="precomputed", n_jobs=1)
pos = mds.fit(similarities).embedding_
nmds = manifold.MDS(n_components=2, metric=False, max_iter=3000, eps=1e-12,
dissimilarity="precomputed", random_state=seed, n_jobs=1,
n_init=1)
npos = nmds.fit_transform(similarities, init=pos)
# Rescale the data
pos *= np.sqrt((X_true ** 2).sum()) / np.sqrt((pos ** 2).sum())
npos *= np.sqrt((X_true ** 2).sum()) / np.sqrt((npos ** 2).sum())
# Rotate the data
clf = PCA(n_components=2)
X_true = clf.fit_transform(X_true)
pos = clf.fit_transform(pos)
npos = clf.fit_transform(npos)
```
### Plot Results
```
data = []
p1 = go.Scatter(x=X_true[:, 0], y=X_true[:, 1],
mode='markers+lines',
marker=dict(color='navy', size=10),
line=dict(width=1),
name='True Position')
data.append(p1)
p2 = go.Scatter(x=pos[:, 0], y=pos[:, 1],
mode='markers+lines',
marker=dict(color='turquoise', size=10),
line=dict(width=1),
name='MDS')
data.append(p2)
p3 = go.Scatter(x=npos[:, 0], y=npos[:, 1],
mode='markers+lines',
marker=dict(color='orange', size=10),
line=dict(width=1),
name='NMDS')
data.append(p3)
similarities = similarities.max() / similarities * 100
similarities[np.isinf(similarities)] = 0
# Plot the edges
start_idx, end_idx = np.where(pos)
# a sequence of (*line0*, *line1*, *line2*), where::
# linen = (x0, y0), (x1, y1), ... (xm, ym)
segments = [[X_true[i, :], X_true[j, :]]
for i in range(len(pos)) for j in range(len(pos))]
values = np.abs(similarities)
for i in range(len(segments)):
p4 = go.Scatter(x=[segments[i][0][0],segments[i][1][0]],
y=[segments[i][0][1],segments[i][1][1]],
mode = 'lines',
showlegend=False,
line = dict(
color = 'lightblue',
width = 0.5))
data.append(p4)
layout = go.Layout(xaxis=dict(zeroline=False, showgrid=False,
ticks='', showticklabels=False),
yaxis=dict(zeroline=False, showgrid=False,
ticks='', showticklabels=False),
height=900, hovermode='closest')
fig = go.Figure(data=data, layout=layout)
py.iplot(fig)
```
### License
Author:
Nelle Varoquaux <[email protected]>
License:
BSD
```
from IPython.display import display, HTML
display(HTML('<link href="//fonts.googleapis.com/css?family=Open+Sans:600,400,300,200|Inconsolata|Ubuntu+Mono:400,700" rel="stylesheet" type="text/css" />'))
display(HTML('<link rel="stylesheet" type="text/css" href="http://help.plot.ly/documentation/all_static/css/ipython-notebook-custom.css">'))
! pip install git+https://github.com/plotly/publisher.git --upgrade
import publisher
publisher.publish(
'Multi-Dimensional Scaling.ipynb', 'scikit-learn/plot-mds/', 'Multi-Dimensional Scaling | plotly',
'',
title = 'Multi-Dimensional Scaling | plotly',
name = 'Multi-Dimensional Scaling',
has_thumbnail='true', thumbnail='thumbnail/mds.jpg',
language='scikit-learn', page_type='example_index',
display_as='manifold_learning', order=2,
ipynb= '~Diksha_Gabha/3320')
```
|
github_jupyter
|
# Python datetime module
We will look at an important standard library, the [datetime library][1] which contains many powerful functions to support date, time and datetime manipulation. Pandas does not rely on this object and instead creates its own, a `Timestamp`, discussed in other notebooks.
The datetime library is part of the standard library, so it comes shipped along with every Python installation. Let's get started by importing it into our namespace.
[1]: https://docs.python.org/3/library/datetime.html
```
import datetime
```
## Create a date, a time and a datetime
The datetime module provides three separate objects for dates, times, and datetimes. Let's use the `date` type to construct a date. It takes three integers, the year, month and day. Here we create the date April 11, 2016.
```
my_date = datetime.date(2016, 4, 11)
my_date
type(my_date)
```
Use the `time` type to construct a time. It takes 4 integers - hours, minutes, seconds, and microseconds (one millionth of a second). Here we create the time 10:54:32.034512
```
my_time = datetime.time(10, 54, 32, 34512)
my_time
type(my_time)
```
Only the hour component is mandatory. For instance, we can create the time 5:44 with this:
```
datetime.time(5, 44)
```
Or you can specify just a particular component of time.
```
datetime.time(second=34)
```
Finally, we can construct a datetime with the `datetime` type, which takes up to 7 parameters - three for the date, and four for the time.
```
my_datetime = datetime.datetime(2016, 4, 11, 10, 54, 32, 34512)
my_datetime
type(my_datetime)
```
### Format changes when printed to the screen
Printing the objects from above to the screen provides a more readable view.
```
print(my_date)
print(my_time)
print(my_datetime)
```
## Attributes of date, time, and datetimes
Each individual component of the date, time, and datetime is available as an attribute.
```
my_date.year
my_date.month
my_date.day
my_time.hour
my_time.minute
my_time.second
my_datetime.day
my_datetime.microsecond
my_date.weekday()
```
## Methods of date, time, and datetimes
Several methods exist for each of these objects. The methods that begin with ISO represent the [International Standards Organization][1] formatting rules for dates, times, and datetimes. The particular standard here is [ISO 8601][2]. Python will return according to this standard.
[1]: https://www.iso.org/home.html
[2]: https://en.wikipedia.org/wiki/ISO_8601
```
my_date.weekday()
my_date.isoformat()
my_date.isocalendar()
my_time.isoformat()
my_datetime.isoformat()
# get the date from a datetime
my_datetime.date()
# get the time from a datetime
my_datetime.time()
```
## Alternate Constructors
You can create dates and datetimes from a single integer which represents the number of seconds since the Unix epoch, January 1, 1970 UTC. UTC is the timezone, [Coordinated Universal Time][1] and is 0 degrees longitude or 5 hours ahead of Easter Standard Time.
Passing the integer 0 to the `fromtimestamp` datetime constructor will return a datetime at the Unix epoch adjusted to your local timezone. If you are located in EST, then you will get returned December 31, 1969 7 p.m.
[1]: https://en.wikipedia.org/wiki/Coordinated_Universal_Time
```
datetime.datetime.fromtimestamp(0)
# 1 billion seconds from the unix epoch
datetime.datetime.fromtimestamp(10 ** 9)
```
The date type also has this constructor, but not time.
```
# also works for date
datetime.date.fromtimestamp(10 ** 9)
```
Can get todays date or datetime:
```
datetime.date.today()
datetime.datetime.now()
```
### Constructing from strings
The `strptime` alternate datetime constructor has the ability to convert a string into a datetime. In addition to the string, you must pass it a specific **format** to alert the constructor which part of the string corresponds to which component of the datetime. There are special character codes called **directives** which must be used to form this correspondence.
## Directives
All the directives can be found in the official [Python documentation for the datetime module][1]. Below are some common ones.
* **%y** - two digit year
* **%Y** - four digit year
* **%m** - Month
* **%d** - Day of the month
* **%H** - Hour (24-hour clock)
* **%I** - Hour (12-hour clock)
* **%M** - Minute
[1]: https://docs.python.org/3/library/datetime.html#strftime-strptime-behavior
### Examples of string parsing to datetimes
The `strptime` alternate constructor stands for string parse time (though it only parses datetimes). You must create a string with the correct directives that represents the format of the date string you are trying to convert to a datetime. For instance, the string '2016-10-22' can use the format '%Y-%m-%d' to parse it correctly.
```
s = '2016-10-22'
fmt = '%Y-%m-%d'
datetime.datetime.strptime(s, fmt)
s = '2016/1/22 5:32:44'
fmt = '%Y/%m/%d %H:%M:%S'
datetime.datetime.strptime(s, fmt)
s = 'January 23, 2019 5:22 PM'
fmt = '%B %d, %Y %H:%M %p'
datetime.datetime.strptime(s, fmt)
s = 'On January the 23rd 2019 at 5:22 PM'
fmt = 'On %B the %drd %Y at %H:%M %p'
datetime.datetime.strptime(s, fmt)
```
### Converting datetimes to string
The **strftime** method converts a date, time, or datetime to a string. It stands for **string format time**. Begin with a date, time, or datetime and use a string with directives to make the conversion.
```
# Convert directly into a string of your choice. Lookup directives online
my_date.strftime("%Y-%m-%d")
# Another more involved directive
my_date.strftime("Remembering back to %A, %B %d, %Y.... What a fantastic day that was.")
```
## Date and Datetime addition
It's possible to add an amount of time to a date or datetime object using the timedelta function. timedelta simply produces some amount of time measured in days, seconds and microseconds. You can then use this object to add to date or datetime objects.
**`timedelta`** objects are constructed with the following definition:
**`timedelta(days=0, seconds=0, microseconds=0, milliseconds=0, minutes=0, hours=0, weeks=0)`**
```
my_timedelta = datetime.timedelta(seconds=5000)
my_timedelta
type(my_timedelta)
# add to datetime
my_datetime + my_timedelta
# original
my_datetime
# add to date
my_date + my_timedelta
# original date. Nothing changed since 5000 seconds wasn't long enough to make an extra day
my_date
# now there is a change
my_date + datetime.timedelta(days = 5)
# add weeks
a = my_datetime + datetime.timedelta(weeks=72, days=4, hours=44)
# the difference between the underlying string representation and the print function
print(a.__repr__())
print(a)
datetime.timedelta(weeks=72, days=4, hours=44)
```
## Third-Party library `dateutil`
For improved datetime handling, you can use [dateutil][1], a more advanced third-party library. Pandas actually uses this library to for its complex date handling. Two of it's most useful features are string parsing and datetime addition.
### Advanced string handling
The `parse` function handles a wide variety of strings. It returns the same datetime type from above. See [many more examples][2] in the documentation.
[1]: https://dateutil.readthedocs.io/en/stable/
[2]: https://dateutil.readthedocs.io/en/stable/examples.html#parse-examples
```
from dateutil.parser import parse
parse('Jan 3, 2003 and 5:22')
```
Pandas uses this under the hood.
```
import pandas as pd
pd.Timestamp('Jan 3, 2003 and 5:22')
```
### Advanced datetime addition
An upgrade to the **`timedelta`** class exists with the **`relativedelta`** class. Check [this stackoverflow][1] post for more detail or see the [documentation for examples][2].
[1]: http://stackoverflow.com/questions/12433233/what-is-the-difference-between-datetime-timedelta-and-dateutil-relativedelta
[2]: https://dateutil.readthedocs.io/en/stable/relativedelta.html#examples
```
from dateutil.relativedelta import relativedelta
```
There are two ways to use it. First, you can pass it two datetimes to find the difference between the two.
```
dt1 = datetime.datetime(2016, 1, 20, 5, 33)
dt2 = datetime.datetime(2018, 3, 20, 6, 22)
relativedelta(dt1, dt2)
```
Second, create an amount of time with the parameters years, months, weeks, days, etc... and then add that to a datetime.
```
rd = relativedelta(months=3)
dt1 + rd
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
import time
import os
from pyspark.ml.clustering import KMeans
from pyspark.ml.evaluation import ClusteringEvaluator
from pyspark.ml.linalg import Vectors
from matplotlib import pyplot as plt
from pyspark.sql import SparkSession
# from pyspark.ml.clustering import KMeans, KMeansModel
import networkx as nx # thư viện để tạo, thao tác, học cấu trúc.... của các mạng phức tạp
import seaborn as sns # thư viện để trực quan hóa dữ liệu
sns.set_style('darkgrid', {'axes.facecolor': '.9'})
sns.set_palette(palette='deep')
sns_c = sns.color_palette(palette='deep')
float_formatter = lambda x: "%.6f" % x
np.set_printoptions(formatter={'float_kind':float_formatter})
def draw_graph(G):
pos = nx.spring_layout(G)
nx.draw_networkx_nodes(G, pos, node_size=10)
# nx.draw_networkx_labels(G, pos)
nx.draw_networkx_edges(G, pos, width=0.1, alpha=0.5)
def draw_graph_cluster(G, labels):
pos = nx.spring_layout(G)
nx.draw(
G,
pos,
node_color=node_colors,
node_size=10,
width=0.1,
alpha=0.5,
with_labels=False,
)
def get_node_color(label):
switcher = {
0: 'red',
1: 'blue',
2: 'orange',
3: 'gray',
4: 'violet',
5: 'pink',
6: 'purple',
7: 'brown',
8: 'yellow',
9: 'lime',
10: 'cyan'
}
return switcher.get(label, 'Invalid label')
spark = SparkSession.builder \
.master("local") \
.appName("CLustering") \
.config("spark.some.config.option", "some-value") \
.getOrCreate()
spark
base_path = os.getcwd()
# file_input = base_path + "/facebook_combined.txt"
file_input = base_path + "/ChG-Miner_miner-chem-gene.tsv"
file_input
pdf = pd.read_table(file_input, sep='\t', names=['src', 'dst'])
pdf.head()
pdf = pdf.to_numpy()
G = nx.Graph()
G.add_edges_from(pdf)
len(G.nodes())
len(G.edges())
# adjacency matrix
W = nx.adjacency_matrix(G)
print(W.todense())
# degree matrix
D = np.diag(np.sum(np.array(W.todense()), axis=1))
print(D)
# Laplacian matrix
L = D - W
print(L)
# eigenvalues, eigenvector
eigenvals, eigenvcts = np.linalg.eigh(L)
eigenvcts
eigenvals
eigenvals_sorted_indices = np.argsort(eigenvals)
eigenvals_sorted = eigenvals[eigenvals_sorted_indices]
eigenvals_sorted_indices
eigenvals_sorted
fig, ax = plt.subplots(figsize=(10, 6))
sns.lineplot(x=range(1, eigenvals_sorted_indices.size + 1), y=eigenvals_sorted, ax=ax)
ax.set(title='Sorted Eigenvalues Graph Laplacian', xlabel='index', ylabel=r'$\lambda$')
index_lim = 250
fig, ax = plt.subplots(figsize=(10, 6))
sns.scatterplot(x=range(1, eigenvals_sorted_indices[: index_lim].size + 1), y=eigenvals_sorted[: index_lim], s=80, ax=ax)
sns.lineplot(x=range(1, eigenvals_sorted_indices[: index_lim].size + 1), y=eigenvals_sorted[: index_lim], alpha=0.5, ax=ax)
ax.axvline(x=1, color=sns_c[3], label='zero eigenvalues', linestyle='--')
ax.legend()
ax.set(title=f'Sorted Eigenvalues Graph Laplacian (First {index_lim})', xlabel='index', ylabel=r'$\lambda$')
zero_eigenvals_index = np.argwhere(abs(eigenvals) < 0.02)
zero_eigenvals_index.squeeze()
proj_df = pd.DataFrame(eigenvcts[:, zero_eigenvals_index.squeeze()[206]])
# proj_df = proj_df.transpose()
proj_df = proj_df.rename(columns={0: 'features'})
proj_df
U = []
for x in proj_df['features']:
U.append(Vectors.dense(x))
pdf_train = pd.DataFrame(U, columns=['features'])
df = spark.createDataFrame(pdf_train)
display(df)
# train k-means model
cost = np.zeros(15)
for i in range(2,15):
kmeans = KMeans(k=i, seed=1)
model = kmeans.fit(df)
cost[i] = model.computeCost(df) # requires Spark 2.0 or later
fig, ax = plt.subplots(1,1, figsize =(8,6))
ax.plot(range(2,15),cost[2:15])
ax.set_xlabel('k')
ax.set_ylabel('cost')
plt.show()
# train
kmeans = KMeans(k=9, seed=1)
model = kmeans.fit(df)
# Make predictions
predictions = model.transform(df)
rows = predictions.select("features","prediction").collect()
# Evaluate clustering by computing Silhouette score
evaluator = ClusteringEvaluator()
silhouette = evaluator.evaluate(predictions)
silhouette
rows
node_colors = []
for label in rows:
node_colors.append(get_node_color(label.prediction))
```
|
github_jupyter
|
# Multi-wavelength maps
New in version `0.2.1` is the ability for users to instantiate wavelength-dependent maps. Nearly all of the computational overhead in `starry` comes from computing rotation matrices and integrals of the Green's basis functions, which makes it **really** fast to compute light curves at different wavelengths if we simply recycle the results of all of these operations.
By "wavelength-dependent map" we mean a map whose spherical harmonic coefficients are a function of wavelength. Specifically, instead of setting the coefficient at $l, m$ to a scalar value, we can set it to a vector, where each element corresponds to the coefficient in a particular wavelength bin. Let's look at some examples.
## Instantiating multi-wavelength maps
The key is to pass the `nwav` keyword when instantiating a `starry` object. For simplicity, let's do `nwav=3`, corresponding to three wavelength bins.
```
%matplotlib inline
from starry import Map
map = Map(lmax=2, nwav=3)
```
Recall that the map coefficients are now *vectors*. Here's what the coefficient *matrix* now looks like:
```
map.y
```
Each row corresponds to a given spherical harmonic, and each column to a given wavelength bin. Let's set the $Y_{1,0}$ coefficient:
```
map[1, 0] = [0.3, 0.4, 0.5]
```
Here's our new map vector:
```
map.y
```
To visualize the map, we can call `map.show()` as usual, but now we actually get an *animation* showing us what the map looks like at each wavelength.
```
map.show()
```
(*Caveat: the* `map.animate()` *routine is disabled for multi-wavelength maps.*)
Let's set a few more coefficients:
```
map[1, -1] = [0, 0.1, -0.1]
map[2, -1] = [-0.1, -0.2, -0.1]
map[2, 2] = [0.3, 0.2, 0.1]
map.show()
```
OK, our map now has some interesting wavelength-dependent features. Let's compute some light curves! First, a simple phase curve:
```
import numpy as np
theta = np.linspace(0, 360, 1000)
map.axis = [0, 1, 0]
phase_curve = map.flux(theta=theta)
```
Let's plot it. The blue line is the first wavelength bin, the orange line is the second bin, and the green line is the third:
```
import matplotlib.pyplot as pl
%matplotlib inline
fig, ax = pl.subplots(1, figsize=(14, 6))
ax.plot(theta, phase_curve);
ax.set_xlabel(r'$\theta$ (degrees)', fontsize=16)
ax.set_ylabel('Flux', fontsize=16);
```
We can also compute an occultation light curve:
```
xo = np.linspace(-1.5, 1.5, 1000)
light_curve = map.flux(xo=xo, yo=0.2, ro=0.1)
```
Let's plot it. This time we normalize the light curve by the baseline for better plotting, since the map has a different total flux at each wavelength:
```
fig, ax = pl.subplots(1, figsize=(14, 6))
ax.plot(theta, light_curve / light_curve[0]);
ax.set_xlabel('Occultor position', fontsize=16)
ax.set_ylabel('Flux', fontsize=16);
```
As we mentioned above, there's not that much overhead to computing light curves in many different wavelength bins. Check it out:
```
import time
np.random.seed(1234)
def runtime(nwav, N=10):
total_time = 0
xo = np.linspace(-1.5, 1.5, 1000)
for n in range(N):
map = Map(lmax=2, nwav=nwav)
map[:, :] = np.random.randn(9, nwav)
tstart = time.time()
map.flux(xo=xo, yo=0.2, ro=0.1)
total_time += time.time() - tstart
return total_time / N
nwav = np.arange(1, 50)
t = [runtime(n) for n in nwav]
fig, ax = pl.subplots(1, figsize=(14, 7))
ax.plot(nwav, t, '.')
ax.plot(nwav, t, '-', color='C0', lw=1, alpha=0.3)
ax.set_xlabel('nwav', fontsize=16)
ax.set_ylabel('time (s)', fontsize=16);
ax.set_ylim(0, 0.003);
```
|
github_jupyter
|
# Fingerprint Generators
## Creating and using a fingerprint generator
Fingerprint generators can be created by using the functions that return the type of generator desired.
```
from rdkit import Chem
from rdkit.Chem import rdFingerprintGenerator
mol = Chem.MolFromSmiles('CC(O)C(O)(O)C')
generator = rdFingerprintGenerator.GetAtomPairGenerator()
fingerprint = generator.GetSparseCountFingerprint(mol)
non_zero = fingerprint.GetNonzeroElements()
print(non_zero)
```
We can set the parameters for the fingerprint while creating the generator for it.
```
generator = rdFingerprintGenerator.GetAtomPairGenerator(minDistance = 1, maxDistance = 2, includeChirality = False)
fingerprint = generator.GetSparseCountFingerprint(mol)
non_zero = fingerprint.GetNonzeroElements()
print(non_zero)
```
We can provide the molecule dependent arguments while creating the fingerprint.
```
fingerprint = generator.GetSparseCountFingerprint(mol, fromAtoms = [1])
non_zero = fingerprint.GetNonzeroElements()
print(non_zero)
fingerprint = generator.GetSparseCountFingerprint(mol, ignoreAtoms = [1, 5])
non_zero = fingerprint.GetNonzeroElements()
print(non_zero)
```
## Types of fingerprint generators
Currently 4 fingerprint types are supported by fingerprint generators
```
generator = rdFingerprintGenerator.GetAtomPairGenerator()
fingerprint = generator.GetSparseCountFingerprint(mol)
non_zero = fingerprint.GetNonzeroElements()
print("Atom pair", non_zero)
generator = rdFingerprintGenerator.GetMorganGenerator(radius = 3)
fingerprint = generator.GetSparseCountFingerprint(mol)
non_zero = fingerprint.GetNonzeroElements()
print("Morgan", non_zero)
generator = rdFingerprintGenerator.GetRDKitFPGenerator()
fingerprint = generator.GetSparseCountFingerprint(mol)
non_zero = fingerprint.GetNonzeroElements()
print("RDKitFingerprint", non_zero)
generator = rdFingerprintGenerator.GetTopologicalTorsionGenerator()
fingerprint = generator.GetSparseCountFingerprint(mol)
non_zero = fingerprint.GetNonzeroElements()
print("TopologicalTorsion", non_zero)
```
## Invariant generators
It is possible to use a custom invariant generators while creating fingerprints. Invariant generators provide values to be used as invariants for each atom or bond in the molecule and these values affect the generated fingerprint.
```
simpleMol = Chem.MolFromSmiles('CCC')
generator = rdFingerprintGenerator.GetRDKitFPGenerator()
fingerprint = generator.GetSparseCountFingerprint(simpleMol)
non_zero = fingerprint.GetNonzeroElements()
print("RDKitFingerprint", non_zero)
atomInvariantsGen = rdFingerprintGenerator.GetAtomPairAtomInvGen()
generator = rdFingerprintGenerator.GetRDKitFPGenerator(atomInvariantsGenerator = atomInvariantsGen)
fingerprint = generator.GetSparseCountFingerprint(simpleMol)
non_zero = fingerprint.GetNonzeroElements()
print("RDKitFingerprint", non_zero)
```
Currently avaliable invariants generators are:
```
atomInvariantsGen = rdFingerprintGenerator.GetAtomPairAtomInvGen()
generator = rdFingerprintGenerator.GetMorganGenerator(radius = 3, atomInvariantsGenerator = atomInvariantsGen)
fingerprint = generator.GetSparseCountFingerprint(mol)
non_zero = fingerprint.GetNonzeroElements()
print("Morgan with AtomPairAtomInvGen", non_zero)
atomInvariantsGen = rdFingerprintGenerator.GetMorganAtomInvGen()
generator = rdFingerprintGenerator.GetMorganGenerator(radius = 3, atomInvariantsGenerator = atomInvariantsGen)
fingerprint = generator.GetSparseCountFingerprint(mol)
non_zero = fingerprint.GetNonzeroElements()
# Default for Morgan FP
print("Morgan with MorganAtomInvGen", non_zero)
atomInvariantsGen = rdFingerprintGenerator.GetMorganFeatureAtomInvGen()
generator = rdFingerprintGenerator.GetMorganGenerator(radius = 3, atomInvariantsGenerator = atomInvariantsGen)
fingerprint = generator.GetSparseCountFingerprint(mol)
non_zero = fingerprint.GetNonzeroElements()
print("Morgan with MorganFeatureAtomInvGen", non_zero)
atomInvariantsGen = rdFingerprintGenerator.GetRDKitAtomInvGen()
generator = rdFingerprintGenerator.GetMorganGenerator(radius = 3, atomInvariantsGenerator = atomInvariantsGen)
fingerprint = generator.GetSparseCountFingerprint(mol)
non_zero = fingerprint.GetNonzeroElements()
print("Morgan with RDKitAtomInvGen", non_zero)
bondInvariantsGen = rdFingerprintGenerator.GetMorganBondInvGen()
generator = rdFingerprintGenerator.GetMorganGenerator(radius = 3, bondInvariantsGenerator = bondInvariantsGen)
fingerprint = generator.GetSparseCountFingerprint(mol)
non_zero = fingerprint.GetNonzeroElements()
# Default for Morgan FP
print("Morgan with MorganBondInvGen", non_zero)
```
## Custom Invariants
It is also possible to provide custom invariants instead of using a invariants generator
```
generator = rdFingerprintGenerator.GetAtomPairGenerator()
fingerprint = generator.GetSparseCountFingerprint(simpleMol)
non_zero = fingerprint.GetNonzeroElements()
print(non_zero)
customAtomInvariants = [1, 1, 1]
fingerprint = generator.GetSparseCountFingerprint(simpleMol, customAtomInvariants = customAtomInvariants)
non_zero = fingerprint.GetNonzeroElements()
print(non_zero)
```
## Convenience functions
## Bulk fingerprint
|
github_jupyter
|
# Part 3: Launch a Grid Network Locally
In this tutorial, you'll learn how to deploy a grid network into a local machine and then interact with it using PySyft.
_WARNING: Grid nodes publish datasets online and are for EXPERIMENTAL use only. Deploy nodes at your own risk. Do not use OpenGrid with any data/models you wish to keep private._
In order to run an grid network locally you will need to run two different apps: a grid gateway and one or more grid workers. In this tutorial we will use the websocket app available [here](https://github.com/OpenMined/PyGrid/tree/dev/app/websocket) to start the grid workers.
## Starting the Grid Gateway
### Step 1: Download the repository
```bash
git clone https://github.com/OpenMined/PyGrid/
```
### Step 2: Download dependencies
You'll need to have the app dependencies installed. We recommend setting up an independent [conda environment](https://docs.conda.io/projects/conda/en/latest/user-guide/concepts/environments.html) to avoid problems with library versions.
You can install the dependencies by running:
```bash
cd PyGrid/gateway/
pip install -r requirements.txt
```
### Step 3: Make grid importable
Install grid as a python package
```bash
cd PyGrid
python setup.py install (or python setup.py develop)
```
### Step 4: Start gateway app
Then to start the app just run the `gateway.py` script. The `--start_local_db` automatically starts a local database so you don't have to configure one yourself.
```bash
python gateway.py --start_local_db --port=<port_number>
```
This will start the app on address: `http://0.0.0.0/<port_number>`.
To check what other arguments you can use when running this app, run:
```bash
python gateway.py --help
```
Let's start a grid gateway on port `5000`
```bash
python gateway.py --port=5000
```
Great, so if your app started successfully the script should still be running.
## Starting the Grid Worker App
### Step 5: Starting the Grid Worker app
This is the same procedure already described at Part 1. But we add a new argument when starting the app called `--gateway_url` this should equal to the address used by the grid network here it's "http://localhost:5000"
Let's start two workers:
* bob on port `3000`
* alice on port `3001`
```bash
python websocket_app.py --db_url=redis:///redis:6379 --id=bob --port=3000 --gateway_url=http://localhost:5000
```
```bash
python websocket_app.py --db_url=redis:///redis:6379 --id=alice --port=3001 --gateway_url=http://localhost:5000
```
We should always start the workers after starting the grid gateway!!
Great, so if your app started successfully the script should still be running.
### Step 6: Start communication with the Grid Gateway and workers
Let's start communication with the Gateway and the workers.
```
# General dependencies
import torch as th
import syft as sy
import grid as gr
hook = sy.TorchHook(th)
gateway = gr.GridNetwork("http://localhost:5000")
# WARNING: We should use the same id and port as the one used to start the app!!!
bob = gr.WebsocketGridClient(hook, id="bob", address="http://localhost:3000")
# If you don't connect to the worker you can't send messages to it
bob.connect()
# WARNING: We should use the same id and port as the one used to start the app!!!
alice = gr.WebsocketGridClient(hook, id="alice", address="http://localhost:3001")
# If you don't connect to the worker you can't send messages to it
alice.connect()
```
### Step 7: Use PySyft Like Normal
Now you can simply use the worker you created like you would any other normal PySyft worker. For more on how PySyft works, please see the PySyft tutorials: https://github.com/OpenMined/PySyft/tree/dev/examples/tutorials
```
x = th.tensor([1,2,3,4]).send(bob)
x
y = x + x
y
y.get()
```
### Step 7: Perform operations on Grid Network
So far we haven't done anything different, but here is the magic: we can interact with the network to query general information about it.
```
x = th.tensor([1, 2, 3, 4, 5]).tag("#tensor").send(bob)
```
We can search for a tensor in the entire network, and get pointers to all tensors.
```
gateway.search("#tensor")
y = th.tensor([1, 2, 3, 4, 5]).tag("#tensor").send(alice)
gateway.search("#tensor")
```
|
github_jupyter
|
#### Reactions processing with AQME - substrates + TS
```
# cell with import, system name and PATHs
import os, glob, subprocess
import shutil
from pathlib import Path
from aqme.csearch import csearch
from aqme.qprep import qprep
from aqme.qcorr import qcorr
from rdkit import Chem
import pandas as pd
```
###### Step 1: Determining the constraints for SN2 TS
```
# Provide the TS smiles to detemine the numbering for constraints
smi = 'C(C)(F)C.[OH-]'
mol = Chem.MolFromSmiles(smi)
mol = Chem.AddHs(mol)
for i,atom in enumerate(mol.GetAtoms()):
atom.SetAtomMapNum(i)
smi_new = Chem.MolToSmiles(mol)
print(smi_new)
mol
# distance and angle to fix are
# constraits_dist = [[0,2,1.8],[0,4,1.8]]
# constraits_angle = [[2,0,4,180]]
```
###### Step 2: Create a CSV as follows
```
data = pd.read_csv('example2.csv')
data
```
###### Step 3: Running CSEARCH on the CSV
```
# run CSEARCH conformational sampling, specifying:
# choose program for conformer sampling
# 1) RDKit ('rdkit'): Fast sampling, only works for systems with one molecule
# 2) CREST ('crest'): Slower sampling, works for noncovalent complexes and
# transition structures (see example of TS in the CSEARCH_CREST_TS.ipynb notebook
# from the CSEARCH_CMIN_conformer_generation folder)
# 3) Program for conformer sampling (program=program)
# 4) SMILES string (smi=smi)
# 5) Name for the output SDF files (name=name)
# 6) Include CREGEN post-analysis for CREST sampling (cregen=True)
csearch(input='example2.csv', program='crest', cregen=True, cregen_keywords='--ethr 0.1 --rthr 0.2 --bthr 0.3 --ewin 1')
```
###### Step 4: Create input files using QPREP
###### a. for TS with TS keywords
###### b. for substrates with substrate keywords
```
# set SDF filenames and directory where the new com files will be created
sdf_rdkit_files = ['CSEARCH/crest/TS_SN2_crest.sdf']
# choose program for input file generation, with the corresponding keywords line, memory and processors:
# 1) Gaussian ('gaussian')
program = 'gaussian'
qm_input = 'B3LYP/6-31G(d) opt=(ts,calcfc,noeigen) freq'
mem='40GB'
nprocs=36
# run QPREP input files generator, with:
# 1) Working directory (w_dir_main=sdf_path)
# 2) PATH to create the new SDF files (destination=com_path)
# 3) Files to convert (files=sdf_rdkit_files)
# 4) QM program for the input (program=program)
# 5) Keyword line for the Gaussian inputs (qm_input=qm_input)
# 6) Memory to use in the calculations (mem='24GB')
# 7) Processors to use in the calcs (nprocs=8)
qprep(files=sdf_rdkit_files,program=program,
qm_input=qm_input,mem=mem,nprocs=nprocs)
# set SDF filenames and directory where the new com files will be created
sdf_rdkit_files = ['CSEARCH/crest/F_crest.sdf', 'CSEARCH/crest/O_anion_crest.sdf']
# choose program for input file generation, with the corresponding keywords line, memory and processors:
# 1) Gaussian ('gaussian')
program = 'gaussian'
qm_input = 'B3LYP/6-31G(d) opt freq'
mem='40GB'
nprocs=36
# run QPREP input files generator, with:
# 1) Working directory (w_dir_main=sdf_path)
# 2) PATH to create the new SDF files (destination=com_path)
# 3) Files to convert (files=sdf_rdkit_files)
# 4) QM program for the input (program=program)
# 5) Keyword line for the Gaussian inputs (qm_input=qm_input)
# 6) Memory to use in the calculations (mem='24GB')
# 7) Processors to use in the calcs (nprocs=8)
qprep(files=sdf_rdkit_files,program=program,
qm_input=qm_input,mem=mem,nprocs=nprocs)
```
###### Step 5: Checking with QPREP for corrections
```
w_dir_main=os.getcwd()+'/QCALC'
# run the QCORR analyzer, with:
# 1) Working directory (w_dir_main=com_path)
# 2) Names of the QM output files (files='*.log')
# 3) Detect and fix calcs that converged during geometry optimization but didn't converge during frequency calcs (freq_conv='opt=(calcfc,maxstep=5)')
# 4) Type of initial input files where the LOG files come from (isom_type='com')
# 5) Folder with the initial input files (isom_inputs=com_path)
qcorr(w_dir_main=w_dir_main,files='*.log',freq_conv='opt=(calcfc,maxstep=5)')
```
###### Step 6: creation of DLPNO input files for ORCA single-point energy calculations
```
# choose output files to get atoms and coordinates to generate inputs for single-point energy calculations
success_dir = os.getcwd()+'/QCALC/successful_QM_outputs'
qm_files = '*.log'
# choose program for input file generation with QPREP, with the corresponding keywords line, memory and processors:
# 1) ORCA ('orca')
program = 'orca'
# a DLPNO example keywords line for ORCA calculations
# qm_input = 'Extrapolate(2/3,cc) def2/J cc-pVTZ/C DLPNO-CCSD(T) NormalPNO TightSCF RIJCOSX\n'
qm_input ='DLPNO-CCSD(T) def2-tzvpp def2-tzvpp/C\n'
qm_input += '%scf maxiter 500\n'
qm_input += 'end\n'
qm_input += '% mdci\n'
qm_input += 'Density None\n'
qm_input += 'end\n'
qm_input += '% elprop\n'
qm_input += 'Dipole False\n'
qm_input += 'end'
mem='4GB'
nprocs=8
# run QPREP input files generator, with:
# 1) Working directory (w_dir_main=sdf_path)
# 2) PATH to create the new SDF files (destination=com_path)
# 3) Files to convert (files=sdf_rdkit_files)
# 4) QM program for the input (program=program)
# 5) Keyword line for the Gaussian inputs (qm_input=qm_input)
# 6) Memory to use in the calculations (mem='24GB')
# 7) Processors to use in the calcs (nprocs=8)
qprep(w_dir_main=success_dir,destination=success_dir,files=qm_files,program=program,
qm_input=qm_input,mem=mem,nprocs=nprocs, suffix='DLPNO')
```
###### Step 7: Analysis with goodvibes
```
# track all the output files from Gaussian and ORCA
opt_files = glob.glob(f'{success_dir}/*.log')
spc_files = glob.glob(f'{success_dir}/*.out')
all_files = opt_files + spc_files
# move all the output files together to a folder called "GoodVibes_analysis" for simplicity
w_dir_main = Path(os.getcwd())
GV_folder = w_dir_main.joinpath('GoodVibes_analysis')
GV_folder.mkdir(exist_ok=True, parents=True)
for file in all_files:
shutil.copy(file, GV_folder)
# this commands runs GoodVibes, including the population % of each conformer
# (final results in the GoodVibes.out file)
os.chdir(GV_folder)
subprocess.run(['python', '-m', 'goodvibes', '--xyz','--pes', '../pes.yaml','--graph','../pes.yaml', '--spc', 'DLPNO', '*.log',])
os.chdir(w_dir_main)
```
|
github_jupyter
|
```
import sys
sys.path.append('../../code/')
import os
import json
from datetime import datetime
import time
from math import *
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import scipy.stats as stats
import igraph as ig
import networkx as nx
from load_data import load_citation_network, case_info
%load_ext autoreload
%autoreload 2
%matplotlib inline
data_dir = '../../data/'
court_name = 'scotus'
case_metadata = pd.read_csv(data_dir + 'clean/case_metadata_master.csv')
edgelist = pd.read_csv(data_dir + 'clean/edgelist_master.csv')
# net_dir = data_dir + 'clean/' + court_name + '/'
# case_metadata = pd.read_csv(net_dir + 'case_metadata.csv')
# edgelist = pd.read_csv(net_dir + 'edgelist.csv')
# edgelist.drop('Unnamed: 0', inplace=True, axis=1)
```
# Compare iterrows vs itertuples
```
start = time.time()
# create graph and add metadata
G = nx.DiGraph()
G.add_nodes_from(case_metadata.index.tolist())
nx.set_node_attributes(G, 'date', case_metadata['date'].to_dict())
for index, edge in edgelist.iterrows():
ing = edge['citing']
ed = edge['cited']
G.add_edge(ing, ed)
end = time.time()
print 'pandas took %d seconds to go though %d edges using iterrows' % (end - start, edgelist.shape[0])
# go through edglist using itertuples
start = time.time()
# create graph and add metadata
G = nx.DiGraph()
G.add_nodes_from(case_metadata.index.tolist())
nx.set_node_attributes(G, 'date', case_metadata['date'].to_dict())
for row in edgelist.itertuples():
ing = row[1]
ed = row[2]
G.add_edge(ing, ed)
end = time.time()
print 'pandas took %d seconds to go though %d edges using itertuples' % (end - start, edgelist.shape[0])
```
# load into igraph
```
# create a dictonary that maps court listener ids to igraph ids
cl_to_ig_id = {}
cl_ids = case_metadata['id'].tolist()
for i in range(case_metadata['id'].size):
cl_to_ig_id[cl_ids[i]] = i
start = time.time()
V = case_metadata.shape[0]
g = ig.Graph(n=V, directed=True)
g.vs['date'] = case_metadata['date'].tolist()
g.vs['name'] = case_metadata['id'].tolist()
ig_edgelist = []
missing_cases = 0
start = time.time()
# i = 1
for row in edgelist.itertuples():
# if log(i, 2) == int(log(i, 2)):
# print 'edge %d' % i
# i += 1
cl_ing = row[1]
cl_ed = row[2]
if (cl_ing in cl_to_ig_id.keys()) and (cl_ed in cl_to_ig_id.keys()):
ing = cl_to_ig_id[cl_ing]
ed = cl_to_ig_id[cl_ed]
else:
missing_cases += 0
ig_edgelist.append((ing, ed))
intermediate = time.time()
g.add_edges(ig_edgelist)
end = time.time()
print 'itertuples took %d seconds to go through %d edges' % (intermediate - start, edgelist.shape[0])
print 'igraph took %d seconds to add %d edges' % (end - start, edgelist.shape[0])
```
# igraph find vs. select
```
start = time.time()
R = 1000
for i in range(R):
g.vs.find(name='92891')
end = time.time()
print 'g.vs.find took %E seconds per lookup' % ((end - start)/R)
start = time.time()
R = 1000
for i in range(R):
g.vs.select(name='92891')
end = time.time()
print 'g.vs.select took %E seconds per lookup' % ((end - start)/R)
start = time.time()
R = 1000
for i in range(R):
cl_to_ig_id[92891]
end = time.time()
print 'pandas df lookup took %E seconds per lookup' % ((end - start)/R)
```
|
github_jupyter
|
## Download and extract zip from web
- Specifies the source link, destination url and file name to download and extract data files
- Currently reading from external folder as github does not support large files
- To rerun function for testing before submission
- To add checks and conditions for the function
- Link to zip download here: "https://s3-ap-southeast-1.amazonaws.com/grab-aiforsea-dataset/safety.zip"
```
import zipfile
import urllib.request
import pandas as pd
import numpy as np
import pickle
from tqdm import tqdm
SOURCE = "https://s3-ap-southeast-1.amazonaws.com/grab-aiforsea-dataset/safety.zip"
OUTPUT_PATH = "../grab-ai-safety-data"
FILE_NAME = ""
class DownloadProgressBar(tqdm):
'''Class for tqdm progress bar.'''
def update_to(self, b=1, bsize=1, tsize=None):
if tsize is not None:
self.total = tsize
self.update(b * bsize - self.n)
def maybe_download(url, output_path, dest_file_name):
'''Function that checks the validity of a desired URL,
downloads and extracts a ZIP file for the purposes of
the Grab AI challenge.
Args:
url (str): Download path of the dataset in question
output_path(str): path of the desired download destination
dest_file_name(str): Desired file name.
To include .zip extension
Returns:
None.
Extracts all relevant data files into a desired folder for
download.
'''
full_path = output_path+'/'+dest_file_name
with DownloadProgressBar(
unit='B',
unit_scale=True,
miniters=1,
desc=url.split("/")[-1]
) as t:
urllib.request.urlretrieve(
url,
filename=full_path,
reporthook=t.update_to
)
with zipfile.ZipFile(full_path, "r") as zip_ref:
zip_ref.extractall(output_path)
# download_url(SOURCE, OUTPUT_PATH, FILE_NAME)
df0 = pd.read_csv("../grab-ai-safety-data/features/part-00000-e6120af0-10c2-4248-97c4-81baf4304e5c-c000.csv")
df1 = pd.read_csv("../grab-ai-safety-data/features/part-00001-e6120af0-10c2-4248-97c4-81baf4304e5c-c000.csv")
df2 = pd.read_csv("../grab-ai-safety-data/features/part-00002-e6120af0-10c2-4248-97c4-81baf4304e5c-c000.csv")
df3 = pd.read_csv("../grab-ai-safety-data/features/part-00003-e6120af0-10c2-4248-97c4-81baf4304e5c-c000.csv")
df4 = pd.read_csv("../grab-ai-safety-data/features/part-00004-e6120af0-10c2-4248-97c4-81baf4304e5c-c000.csv")
df5 = pd.read_csv("../grab-ai-safety-data/features/part-00005-e6120af0-10c2-4248-97c4-81baf4304e5c-c000.csv")
df6 = pd.read_csv("../grab-ai-safety-data/features/part-00006-e6120af0-10c2-4248-97c4-81baf4304e5c-c000.csv")
df7 = pd.read_csv("../grab-ai-safety-data/features/part-00007-e6120af0-10c2-4248-97c4-81baf4304e5c-c000.csv")
df8 = pd.read_csv("../grab-ai-safety-data/features/part-00008-e6120af0-10c2-4248-97c4-81baf4304e5c-c000.csv")
df9 = pd.read_csv("../grab-ai-safety-data/features/part-00009-e6120af0-10c2-4248-97c4-81baf4304e5c-c000.csv")
response = pd.read_csv("../grab-ai-safety-data/labels/part-00000-e9445087-aa0a-433b-a7f6-7f4c19d78ad6-c000.csv")
```
## Merge and drop duplicates
- Join the feautres together with the labels
- Get rid of any obvious duplicates in the features and response
- No data cleaning or formatting to minimize data leakage
```
df_features = pd.concat(
[df1, df2, df3, df4, df5, df6, df7, df8, df9],
axis=0
).drop_duplicates(
keep=False
)
response = response.drop_duplicates(
subset="bookingID",
keep=False
)
df = pd.merge(
df_features,
response,
how="inner",
on="bookingID"
).sort_values(
["bookingID", "second"],
ascending=True
)
with open('../grab-ai-safety-data/df_full.pickle', 'wb') as f:
pickle.dump(df, f)
```
|
github_jupyter
|
```
import numpy as np
from bokeh.plotting import figure, output_file, show
from bokeh.io import output_notebook
from nsopy import SGMDoubleSimpleAveraging as DSA
from nsopy.loggers import EnhancedDualMethodLogger
output_notebook()
%cd ..
from smpspy.oracles import TwoStage_SMPS_InnerProblem
```
# Solving dual model using DSA with Entropy Prox Term
Instantiating inner problem
### Solve battery of problems
```
# Setup
BENCHMARKS_PATH = './smpspy/benchmark_problems/2_caroe_schultz/'
n_S_exp = [10, 50, 100, 500]
N_STEPS = 200
GAMMA = 1.0
# First generate traditional DSA
inner_problems = {}
methods = {}
method_loggers = {}
for n_S in n_S_exp:
ip = TwoStage_SMPS_InnerProblem(BENCHMARKS_PATH+'caroe_schultz_{}'.format(n_S))
dsa = DSA(ip.oracle, ip.projection_function, dimension=ip.dimension, gamma=GAMMA)
logger_dsa = EnhancedDualMethodLogger(dsa)
inner_problems[n_S] = ip
methods[n_S] = dsa
method_loggers[n_S] = logger_dsa
for n_S, method in methods.items():
for step in range(N_STEPS):
if not step % 100:
print('[n_S={}] step: {} of method {}'.format(n_S, str(step), str(method.desc)))
method.dual_step()
inner_problems_entropy = {}
methods_entropy = {}
method_loggers_entropy = {}
for n_S in n_S_exp:
R_a_posteriori = np.linalg.norm(methods[n_S].lambda_k, ord=np.inf)
R_safe = R_a_posteriori*1.1
ip = TwoStage_SMPS_InnerProblem(BENCHMARKS_PATH+'caroe_schultz_{}'.format(n_S), R=R_safe)
dsa_entropy = DSA(ip.oracle, ip.softmax_projection, dimension=ip.dimension, gamma=GAMMA)
logger_dsa_entropy = EnhancedDualMethodLogger(dsa_entropy)
inner_problems_entropy[n_S] = ip
methods_entropy[n_S] = dsa_entropy
method_loggers_entropy[n_S] = logger_dsa_entropy
for n_S, method in methods_entropy.items():
for step in range(N_STEPS):
if not step % 100:
print('[n_S={}] step: {} of method {}'.format(n_S, str(step), str(method.desc)))
method.dual_step()
# find "d*"
d_stars = {}
EPS = 0.01
for n_S in n_S_exp:
d_star_dsa = max(method_loggers[n_S].d_k_iterates)
d_star_dsa_entropy = max(method_loggers_entropy[n_S].d_k_iterates)
d_stars[n_S] = max(d_star_dsa, d_star_dsa_entropy) + EPS
p = figure(title="comparison", x_axis_label='iteration', y_axis_label='d* - d_k', y_axis_type='log', toolbar_location='above')
plot_colors = {
10: 'blue',
50: 'green',
100: 'red',
500: 'orange',
1000: 'purple',
}
for n_S in n_S_exp:
logger = method_loggers[n_S]
p.line(range(len(logger.d_k_iterates)), d_stars[n_S] - np.array(logger.d_k_iterates), legend="DSA, n_scen={}, gamma={}".format(n_S, GAMMA, inner_problems[n_S].R),
color=plot_colors[n_S], line_dash='dashed')
for n_S in n_S_exp:
logger = method_loggers_entropy[n_S]
p.line(range(len(logger.d_k_iterates)), d_stars[n_S] - np.array(logger.d_k_iterates), legend="DSA Entropy, n_scen={}, gamma={}, R={}".format(n_S, GAMMA, inner_problems_entropy[n_S].R),
color=plot_colors[n_S])
p.legend.location = "top_right"
p.legend.visible = True
p.legend.background_fill_alpha = 0.5
show(p)
```
### Single run
```
ip = TwoStage_SMPS_InnerProblem('./smpspy/benchmark_problems/2_caroe_schultz/caroe_schultz_10')
```
First solving it with DSA
```
GAMMA = 1.0
dsa = DSA(ip.oracle, ip.projection_function, dimension=ip.dimension, gamma=GAMMA)
logger_dsa = EnhancedDualMethodLogger(dsa)
for iteration in range(1000):
if not iteration%50:
print('Iteration: {}, d_k={}'.format(iteration, dsa.d_k))
dsa.dual_step()
```
Then get the required parameters (R is derived a posteriori)
```
R_a_posteriori = np.linalg.norm(dsa.lambda_k, ord=np.inf)
R_safe = R_a_posteriori*1.1
ip = TwoStage_SMPS_InnerProblem('./smpspy/benchmark_problems/2_caroe_schultz/caroe_schultz_10', R=R_safe)
print('A-posteriori R={}'.format(R_a_posteriori))
```
Solve it using DSA with Entropy prox function. **Note that the only difference is that we pass in softmax projection function!**
```
dsa_entropy = DSA(ip.oracle, ip.softmax_projection, dimension=ip.dimension, gamma=GAMMA)
logger_dsa_entropy = EnhancedDualMethodLogger(dsa_entropy)
for iteration in range(1000):
if not iteration%50:
print('Iteration: {}, d_k={}'.format(iteration, dsa_entropy.d_k))
dsa_entropy.dual_step()
logger_dsa.lambda_k_iterates[-1]
logger_dsa_entropy.lambda_k_iterates[-1]
p = figure(title="comparison", x_axis_label='iteration', y_axis_label='d_k')
p.line(range(len(logger_dsa.d_k_iterates)), logger_dsa.d_k_iterates, legend="DSA, gamma={}".format(GAMMA, R_safe))
p.line(range(len(logger_dsa_entropy.d_k_iterates)), logger_dsa_entropy.d_k_iterates, legend="DSA Entropy, gamma={}, R={}".format(GAMMA, R_safe), color='red')
p.legend.location = "bottom_right"
show(p)
```
|
github_jupyter
|
## Imperfect Tests and The Effects of False Positives
The US government has been widely criticized for its failure to test as many of its citizens for COVID-19 infections as other countries. But is mass testing really as easy as it seems? This analysis of the false positive and false negative rates of tests, using published sensitivities and specificities for COVID-19 rt-PCR and antigen tests, shows that even tests with slightly less than perfect results can produce very large numbers of false positives.
```
import sys
# Install required packages
#!{sys.executable} -mpip -q install matplotlib seaborn statsmodels pandas publicdata metapack
%matplotlib inline
import pandas as pd
import geopandas as gpd
import numpy as np
import metapack as mp
import rowgenerators as rg
import publicdata as pub
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(color_codes=True)
```
As the world became more aware of the threat posed by COVID-19 in February 2020, US media began to draw attention to the disparity between the extent of testing being done in other countries versus the United States. The CDC released [fairly restrictive guidelines](https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-criteria.html) for what conditions qualified a patient for a lab test for COVID-19 infections, and many media outlets criticized the US CDC for being unprepared to test for the virus.
Criticism intensified when the first version of tests created by the CDC [proved to be unreliable](https://www.forbes.com/sites/rachelsandler/2020/03/02/how-the-cdc-botched-its-initial-coronavirus-response-with-faulty-tests/#5bbf1d50670e). But there are important considerations that these reports have largely ignored, the most important of which is the false positive and false negative rates of the tests, which can produce results that are worse than useless when the prevalence of the condition — the percentage of people who are infected — is very low.
Every test — for nearly any sort of test — has an error rate: false positives and false negatives. False negatives are fairly easy to understand. If a 1,000 women who have breast cancer take a test that has a false positive rate of 1%, the test will report that 999 of them have cancer, and 1 who does not, even though she actually does.
The false positive rate is trickier, because it is multipled not by the number of women who have cancer, but by the number of women who take the test. If the situation is that a large number of women are tested, but few have cancer, the test can report many more false positives than women who actually have cancer.
There is evidence that the tests for the COVID-19 virus have a false positive rate large enough that if a large number of people are tested when the prevalence of COVID-19 infections are small, most of the reported positives are false positives.
# Primer on False Positives and Negatives
Research related to epidemiological tests typically does not report the false positive rate directly; instead it reports two parameters, the Selectivity and Specificity. [Wikipedia has an excellent article](https://en.wikipedia.org/wiki/Sensitivity_and_specificity) describing these parameters and how they related to false positive and false negative rates, and [Health News Review](https://www.healthnewsreview.org/) publishes this [very accessible overview of the most important concepts](https://www.healthnewsreview.org/toolkit/tips-for-understanding-studies/understanding-medical-tests-sensitivity-specificity-and-positive-predictive-value/). The most important part of the Wikipedia article to understand is the table in the [worked example](https://en.wikipedia.org/wiki/Sensitivity_and_specificity#Worked_example). When a test is administered, there are four possible outcomes. The test can return a positive result, which can be a true positive or a false positive, or it can return a negative result, which is a true negative or a false negative. If you organize those posibilities by what is the true condition ( does the patient have the vius or not ):
* Patient has virus
* True Positive ($\mathit{TP}$)
* False negative ($\mathit{FN}$)
* Patient does not have virus
* True Negative ($\mathit{TN}$)
* False Positive. ($\mathit{FP}$)
In the Wikipedia worked example table:
* The number of people who do have the virus is $\mathit{TP}+\mathit{FN}$, the true positives plus the false negatives, which are the cases that should have been reported positive, but were not.
* The number of people who do not have the virus is $\mathit{TN}+\mathit{FP}$, the true negatives and the false positives, which are the cases should have been reported positive, but were not.
The values of Sensitivity and Specificity are defined as:
$$\begin{array}{ll}
Sn = \frac{\mathit{TP}}{\mathit{TP} + \mathit{FN}} & \text{True positives outcomes divided by all positive conditions} \tag{1}\label{eq1}\\
Sp = \frac{\mathit{TN}}{\mathit{FP} + \mathit{TN}} & \text{True negatives outcomes divided by all negative conditions}\\
\end{array}$$
We want to know the number of false positives($\mathit{FP}$) given the number of positive conditions ($\mathit{TP}+\mathit{FN}$) and the total number of tests. To compute these, we need to have some more information about the number of people tested, and how common the disease is:
* Total test population $P$, the number of people being tested, which equals $\mathit{TP}+\mathit{FP}+\mathit{FN}+\mathit{TN}$
* The prevalence $p$, the population rate of positive condition.
We can do a little math to get:
$$\begin{array}{ll}
\mathit{TP} = Pp\mathit{Sn} & \text{}\\
\mathit{FP} = P(1-p)(1-\mathit{Sp}) \text{}\\
\mathit{TN} = P(1-p)\mathit{Sp} & \text{}\\
\mathit{FN} = Pp(1-\mathit{Sn})& \text{}\\
\end{array}$$
You can see examples of these equations worked out in the third line in the red and green cells of the [Worked Example](https://en.wikipedia.org/wiki/Sensitivity_and_specificity#Worked_example) on the Sensitivity and Specificity Wikipedia page.
It is important to note that when these four values are used to calculate $\mathit{Sp}$ and $\mathit{Sn}$, the population value $P$ cancels out, so $\mathit{Sp}$ and $\mathit{Sn}$ do not depend on the number of people tested.
One of the interesting questions when test results are reported is "What percentage of the positive results are true positives?" This is a particularly important question for the COVID-19 pandemic because there are a lot of reports that most people with the virus are asymptomatic. Are they really asymptomatic, or just false positives?
The metric we're interested here is the portion of positive results that are true positives, the positive predictive value, $\mathit{PPV}$:
$$\mathit{PPV} = \frac{\mathit{TP} }{ \mathit{TP} +\mathit{FP} } $$
Which expands to:
$$\mathit{PPV} = \frac{p\mathit{Sn} }{ p\mathit{Sn} + (1-p)(1-\mathit{Sp}) }\tag{2}\label{eq2} $$
It is important to note that $\mathit{PPV}$ is not dependent on $P$, the size of the population being tested. It depends only on the quality parameters of the test, $\mathit{Sn}$ and $\mathit{Sp}$, and the prevalence, $p$. For a given test, only the prevalence will change over time.
# Selctivity and Specificity Values
It has been dificult to find specificity and sensitivity values for COVID-19 tests, or any rt-PCR tests; research papers rarely publish the values. Howver, there are a few reports for the values for serology tests, and a few reports of values for rt-PRC tests for the MERS-CoV virus.
We can get values for an antibidy test for COVID-19 from a a recently published paper, _Development and Clinical Application of A Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis_<sup><a href="#fnote2" rel="noopener" target="_self">2</a></sup>, which reports:
> The overall testing sensitivity was 88.66% and specificity was 90.63%
This test is significantly different from the most common early tests for COVID-19; this test looks for antibodies in the patient's blood, while most COVID-19 tests are rt-PCR assays that look for fragments of RNA from the virus.
The article _MERS-CoV diagnosis: An update._<sup><a href="#fnote4" rel="noopener" target="_self">4</a></sup> reports that for MERS-CoV:
> Song et al. developed a rapid immunochromatographic assay for the detection of MERS-CoV nucleocapsid protein from camel nasal swabs with 93.9% sensitivity and 100% specificity compared to RT-rtPCR
The article _Performance Evaluation of the PowerChek MERS (upE & ORF1a) Real-Time PCR Kit for the Detection of Middle East Respiratory Syndrome Coronavirus RNA_<sup><a href="#fnote5" rel="noopener" target="_self">5</a></sup> reports:
> The diagnostic sensitivity and specificity of the PowerChek MERS assay were both 100% (95% confidence interval, 91.1–100%).
The [Emergency Use Authorization for LabCorp's rt-PCR test](https://www.fda.gov/media/136151/download)<sup><a href="#fnote6" rel="noopener" target="_self">6</a></sup> reports:
~~~
Performance of the COVID-19 RT-PCR test against the expected results [ with NP swabs ] are:
Positive Percent Agreement 40/40 = 100% (95% CI: 91.24%-100%)
Negative Percent Agreement 50/50 = 100% (95% CI: 92.87% -100%)
~~~
Using the lower bound of the 95% CI, values convert to a specificity of .90 and sensitivity of .94.
A recent report characterizes Abbott Labs ID NOW system, used for influenza tests. [Abbott Labs recieved an EUA](https://www.fda.gov/media/136525/download), on 27 March 2020, for a version of the device for use with COVID-19. The study of the the influenza version states:
> The sensitivities of ID NOW 2 for influenza A were 95.9% and 95.7% in NPS and NPA, respectively, and for influenza B were 100% and 98.7% in NPS and NPA, respectively. The specificity was 100% for both influenza A and influenza B in NPS and NPA.
The results section of the paper provides these parameters, when compared to rRT-PCR:
<table>
<tr>
<th>Virus</th>
<th>Parameter</th>
<th>ID NOW 2</th>
<th> ID NOW 2 VTM</th>
</tr>
<tr>
<td>type A</td>
<td>Sensitivity (95% CI)</td>
<td>95.7 (89.2-98.8)</td>
<td>96.7 (90.8-99.3)</td>
</tr>
<tr>
<td></td>
<td>Specificity (95% CI)</td>
<td>100 (89.3-100) </td>
<td>100 (89.3-100)</td>
</tr>
<tr>
<td>Type B</td>
<td>Sensitivity (95% CI)</td>
<td>98.7 (93.0-100)</td>
<td>100 (96.2-100)</td>
</tr>
<tr>
<td></td>
<td>Specificity (95% CI)</td>
<td>100 (98.5-100)</td>
<td>100 (98.5-100)</td>
</tr>
</table>
A recent Medscape article<a href="#fnote7" rel="noopener" target="_self">7</a></sup> on the specificity and sensitivity of Influenza tests reports:
> In a study of the nucleic acid amplification tests ID Now (Abbott), Cobas Influenza A/B Assay (Roche Molecular Diagnostics), and Xpert Xpress Flu (Cepheid), Kanwar et al found the three products to have comparable sensitivities for influenza A (93.2%, 100%, 100%, respectively) and B (97.2%, 94.4%, 91.7%, respectively) detection. In addition, each product had greater than 97% specificity for influenza A and B detection.
> Rapid antigen tests generally have a sensitivity of 50-70% and a specificity of 90-95%. Limited studies have demonstrated very low sensitivity for detection of 2009 H1N1 with some commercial brands.
Based on these values, we'll explore the effects of sensitivity and specificities in the range of .9 to 1.
# PPV For Serology Test
First we'll look at the positive prediction value for the antibody test in reference (<a href="#fnote2" rel="noopener" target="_self">2</a>), which has the lowest published Sp and Sn values at .9063 and .8866. The plot below shows the portion of positive test results that are true positives s a function of the prevalence.
```
def p_vs_tpr(Sp, Sn):
for p in np.power(10,np.linspace(-7,np.log10(.5), num=100)): # range from 1 per 10m to 50%
ppv = (p*Sn) / ( (p*Sn)+(1-p)*(1-Sp))
yield (p, ppv)
def plot_ppv(Sp, Sn):
df = pd.DataFrame(list(p_vs_tpr(Sp, Sn)), columns='p ppv'.split())
df.head()
fig, ax = plt.subplots(figsize=(12,8))
df.plot(ax=ax, x='p',y='ppv', figsize=(10,10))
fig.suptitle(f'Portion of Positives that Are True Vs Prevalence\nFor test with Sp={Sp} and Sn={Sn}', fontsize=20)
ax.set_xlabel('Condition Prevalence in Portion of Tested Population', fontsize=18)
ax.set_ylabel('Portion of Positive Test Results that are True Positives', fontsize=18);
#ax.set_xscale('log')
#ax.set_yscale('log')
plot_ppv(Sp = .9063, Sn = .8866)
```
The important implication of this curve is that using a test with low Sp and Sn values in conditions of low prevalence will result in a very large portion of false positives.
# False Positives for LabCorp's test
Although the published results for the LabCorp test are 100% true positives and true negative rates, the 95% error margin is substantial, because the test was validatd with a relatively small number of samples. This analysis will use the published error margins to produce a distribution of positive prediction values. First, let's look at the distributions of the true positive and true negative rates, accounting for the published confidence intervals. These distributions are generated by converting the published true and false rates, and their CIs into gaussian distributions, and selecting only values that are 1 or lower from those distributions.
```
# Convert CI to standard error. The values are reported for a one-sided 95% CI,
# so we're multiplying by the conversion for a two-sided 90% ci
p_se = (1-.9124) * 1.645
n_se = (1-.9287) * 1.645
def select_v(se):
"""get a distribution value, which must be less than or equal to 1"""
while True:
v = np.random.normal(1, se)
if v <= 1:
return v
# These values are not TP and FP counts; they are normalized to
# prevalence
TP = np.array(list(select_v(p_se) for _ in range(100_000)))
TN = np.array(list(select_v(n_se) for _ in range(100_000)))
fig, ax = plt.subplots(1,2, figsize=(12,8))
sns.distplot( TP, ax=ax[0], kde=False);
ax[0].set_title('Distribution of Posibile True Positives Rates');
sns.distplot( TN, ax=ax[1], kde=False);
ax[1].set_title('Distribution of Posibile True Negative Rates');
fig.suptitle(f'Distribution of True Positive and Negative Rates'
'\nFor published confidence intervals and 4K random samples', fontsize=20);
```
It is important to note that these are not the distributions
From these distributions, we can calculate the distributions for the positive prediction value, the portion of all positive results that are true positives.
With these distributions, we can use ([Eq 2](#MathJax-Span-5239)) to compute the distributions of PPV for a variety of prevalences. In each chart, the 'mean' is the expectation value of the distribution, the weighted mean of the values. It is the most likely PPV valule for the given prevalence.
```
FP = 1-TN
FN = 1-TP
Sn = TP / (TP+FN)
Sp = TN / (TN+FP)
def ppv_dist_ufunc(p, Sp, Sn):
return (p*Sn) / ( (p*Sn)+(1-p)*(1-Sp))
def ppv_dist(p, Sp, Sn):
sp = np.random.choice(Sp, 1_000_000, replace=True)
sn = np.random.choice(Sn, 1_000_000, replace=True)
return ppv_dist_ufunc(p,sp, sn)
fig, axes = plt.subplots( 2,2, figsize=(15,15))
axes = axes.flat
def plot_axis(axn, prevalence):
ppvd = ppv_dist(prevalence, Sp, Sn)
wmean = (ppvd.sum()/len(ppvd)).round(4)
sns.distplot( ppvd, ax=axes[axn], kde=False);
axes[axn].set_title(f' prevalence = {prevalence}, mean={wmean}');
axes[axn].set_xlabel('Positive Prediction Value (PPV)')
axes[axn].set_ylabel('PPV Frequency')
plot_axis(0, .001)
plot_axis(1, .01)
plot_axis(2, .10)
plot_axis(3, .5)
fig.suptitle(f'Distribution of PPV Values for LabCorp Test\nBy condition prevalence', fontsize=20);
```
The implication of these charts is that, even for a test with published true positive and true negative rate of 100%, the uncertainties in the measurements can mean that there still a substantial problem of false positives for low prevalences.
Computing the mean PPV value or a range of prevalence values results in the following relationship.
```
def ppv_vs_p():
for p in np.power(10,np.linspace(-7,np.log10(1), num=100)): # range from 1 per 10m to 50%
ppvd = ppv_dist(p, Sp, Sn)
yield p, ppvd.sum()/len(ppvd)
ppv_v_p = pd.DataFrame(list(ppv_vs_p()), columns='p ppv'.split())
fig, ax = plt.subplots(figsize=(8,8))
sns.lineplot(x='p', y='ppv', data=ppv_v_p, ax=ax)
ax.set_xlabel('Prevalence')
ax.set_ylabel('Positive Predictive Value')
fig.suptitle("Positive Predictive Value vs Prevalence\nFor LabCorp Test", fontsize=18);
```
Compare this curve to the one presented earlier, for the antibody test with published sensitivity of 88.66% and specificity of 90.63%; The relationship between P and PPV for the rt-PCR test isn't much better.
But what if the tests are really, really good: .99 for both sensitivity and specificity? Here is the curve for that case:
```
def ppv_vs_p():
for p in np.power(10,np.linspace(-7,np.log10(1), num=100)): # range from 1 per 10m to 50%
ppvd = ppv_dist_ufunc(p, .99, .99)
yield p, ppvd
ppv_v_p = pd.DataFrame(list(ppv_vs_p()), columns='p ppv'.split())
fig, ax = plt.subplots(figsize=(8,8))
sns.lineplot(x='p', y='ppv', data=ppv_v_p, ax=ax)
ax.set_xlabel('Prevalence')
ax.set_ylabel('Positive Predictive Value')
fig.suptitle("Positive Predictive Value vs Prevalence\nFor Sp=.99, Sn=.99", fontsize=18);
```
This table shows the PPVs and false positive rate for a logrhythimic range of prevalences.
```
prevs = [1e-6, 1e-5, 1e-4, 1e-3, 1e-2, 1e-1]
names = ["1 per {}".format(round(1/p,0)) for p in prevs]
ppvs = [ppv_v_p.loc[(ppv_v_p.p-p).abs().idxmin()].ppv for p in prevs]
fp = [ str(round((1-ppv)*100,1))+"%" for ppv in ppvs]
df = pd.DataFrame({
'Rate': names,
'Prevalence': prevs,
'PPV': ppvs,
'False Positives Rate': fp
}).set_index('Prevalence')
df
```
This case is much better, across the range of prevalences, but for low prevalence, there are still a lot of false positives, and below 1 per 1000, it is nearly all false positives. Here is the same chart, but for Sp and Sn at 99.99%
```
def ppv_vs_p():
for p in np.power(10,np.linspace(-7,np.log10(1), num=100)): # range from 1 per 10m to 50%
ppvd = ppv_dist_ufunc(p, .9999, .9999)
yield p, ppvd
ppv_v_p = pd.DataFrame(list(ppv_vs_p()), columns='p ppv'.split())
ppvs = [ppv_v_p.loc[(ppv_v_p.p-p).abs().idxmin()].ppv for p in prevs]
fp = [ str(round((1-ppv)*100,1))+"%" for ppv in ppvs]
df = pd.DataFrame({
'Rate': names,
'Prevalence': prevs,
'PPV': ppvs,
'False Positives Rate': fp
}).set_index('Prevalence')
df
```
Even a very accurate test will not be able to distinguish healthy from sick better than a coin flip if the prevalence is less than 1 per 10,000.
# Conclusion
Tests with less than 100% specificity and selectivity, including those with published values of 100% but with a moderate confidence interval, are very sensitive to low condition prevalences. Considering the confidence intervals, to ensure that 50% of positive results are true positives requires a prevalence of about 10%, and 80% PPV requires about a 30% prevalence. This suggests that using rt-PCR tests to test a large population that has a low prevalence is likely to produce a large number of false positive results.
# References
* <a name="fnote1">1</a> Parikh, Rajul et al. “[Understanding and using sensitivity, specificity and predictive values.](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2636062/)” Indian journal of ophthalmology vol. 56,1 (2008): 45-50. doi:10.4103/0301-4738.37595
* <a name="fnote2">2</a> Li, Zhengtu et al. “[Development and Clinical Application of A Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis.](https://pubmed.ncbi.nlm.nih.gov/32104917/)” Journal of medical virology, 10.1002/jmv.25727. 27 Feb. 2020, doi:10.1002/jmv.25727
* <a name="fnote3">3</a> Zhuang, G H et al. “[Potential False-Positive Rate Among the 'Asymptomatic Infected Individuals' in Close Contacts of COVID-19 Patients](https://pubmed.ncbi.nlm.nih.gov/32133832)” Zhonghua liuxingbingxue zazhi, vol. 41,4 485-488. 5 Mar. 2020, doi:10.3760/cma.j.cn112338-20200221-00144
* <a name="fnote4">4</a> Al Johani, Sameera, and Ali H Hajeer. “[MERS-CoV diagnosis: An update.](https://www.sciencedirect.com/science/article/pii/S1876034116300223)” Journal of infection and public health vol. 9,3 (2016): 216-9. doi:10.1016/j.jiph.2016.04.005
* <a name="fnote5">5</a> Huh, Hee Jae et al. “[Performance Evaluation of the PowerChek MERS (upE & ORF1a) Real-Time PCR Kit for the Detection of Middle East Respiratory Syndrome Coronavirus RNA.](http://www.annlabmed.org/journal/view.html?volume=37&number=6&spage=494)” Annals of laboratory medicine vol. 37,6 (2017): 494-498. doi:10.3343/alm.2017.37.6.494
* <a name="fnote7">7</a> [Emergency Use Authorization summary](https://www.fda.gov/media/136151/download) for LabCorp's COVID-19 rt-PCR test.
* Mitamura, Keiko et al. “[Clinical evaluation of ID NOW influenza A & B 2, a rapid influenza virus detection kit using isothermal nucleic acid amplification technology - A comparison with currently available tests.](https://pubmed.ncbi.nlm.nih.gov/31558351/?from_single_result=31558351)” Journal of infection and chemotherapy : official journal of the Japan Society of Chemotherapy vol. 26,2 (2020): 216-221. doi:10.1016/j.jiac.2019.08.015
* <a name="fnote7">8</a> Blanco, E. M. (2020, January 22). [What is the sensitivity and specificity of diagnostic influenza tests?](https://www.medscape.com/answers/2053517-197226/what-is-the-sensitivity-and-specificity-of-diagnostic-influenza-tests) Retrieved March 27, 2020, from https://www.medscape.com/answers/2053517-197226/what-is-the-sensitivity-and-specificity-of-diagnostic-influenza-tests
## Supporting Web Articles
The World Health Organization has a [web page with links to information the COVID-19 tests](https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance) from many countries.
The CDC's page for [Rapid Diagnostic Testing for Influenza: Information for Clinical Laboratory Directors](https://www.cdc.gov/flu/professionals/diagnosis/rapidlab.htm) describes the minimum specificity and sensitivity for rapid influenza diagnostic tests, and shows some examples of PPV and flase positive rates.
Washington Post: [A ‘negative’ coronavirus test result doesn’t always mean you aren’t infected](https://www.washingtonpost.com/science/2020/03/26/negative-coronavirus-test-result-doesnt-always-mean-you-arent-infected/)
Prague Morning: [80% of Rapid COVID-19 Tests the Czech Republic Bought From China are Wrong](https://www.praguemorning.cz/80-of-rapid-covid-19-tests-the-czech-republic-bought-from-china-are-wrong/)
BusinessInsider: [Spain, Europe's worst-hit country after Italy, says coronavirus tests it bought from China are failing to detect positive cases](https://www.businessinsider.com/coronavirus-spain-says-rapid-tests-sent-from-china-missing-cases-2020-3?op=1)
Wikipedia has a good discussion of the false positives problem in the articl about the [Base Rate Falacy](https://en.wikipedia.org/wiki/Base_rate_fallacy#False_positive_paradox).
## Other References
The following references were referenced by Blanco <a href="#fnote6" rel="noopener" target="_self">6</a></sup>, but I haven't evaluated them yet.
Kanwar N, Michael J, Doran K, Montgomery E, Selvarangan R. Comparison of the ID NOWTM Influenza A & B 2, Cobas® Influenza A/B, and Xpert® Xpress Flu Point-of-Care Nucleic Acid Amplification Tests for Influenza A/B Detection in Children. J Clin Microbiol. 2020 Jan 15.
Blyth CC, Iredell JR, Dwyer DE. Rapid-test sensitivity for novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009 Dec 17. 361(25):2493.
Evaluation of rapid influenza diagnostic tests for detection of novel influenza A (H1N1) Virus - United States, 2009. MMWR Morb Mortal Wkly Rep. 2009 Aug 7. 58(30):826-9.
Faix DJ, Sherman SS, Waterman SH. Rapid-test sensitivity for novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009 Aug 13. 361(7):728-9.
Ginocchio CC, Zhang F, Manji R, Arora S, Bornfreund M, Falk L. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. J Clin Virol. 2009 Jul. 45(3):191-5.
Sambol AR, Abdalhamid B, Lyden ER, Aden TA, Noel RK, Hinrichs SH. Use of rapid influenza diagnostic tests under field conditions as a screening tool during an outbreak of the 2009 novel influenza virus: practical considerations. J Clin Virol. 2010 Mar. 47(3):229-33.
# Updates
* 2020-03-25: Changed conversion from CI to SE from 1.96 to 1.645; using the factor for a two sided 90% ci for the 95% one sided CI.
* 2020-03-27: Added parameters for Sp and Sn for the influenza version of Abbott Labs ID NOW device.
|
github_jupyter
|
<h1>Model Deployment</h1>
Once we have built and trained our models for feature engineering (using Amazon SageMaker Processing and SKLearn) and binary classification (using the XGBoost open-source container for Amazon SageMaker), we can choose to deploy them in a pipeline on Amazon SageMaker Hosting, by creating an Inference Pipeline.
https://docs.aws.amazon.com/sagemaker/latest/dg/inference-pipelines.html
This notebook demonstrates how to create a pipeline with the SKLearn model for feature engineering and the XGBoost model for binary classification.
Let's define the variables first.
```
import sagemaker
import sys
import IPython
# Let's make sure we have the required version of the SM PySDK.
required_version = '2.49.2'
def versiontuple(v):
return tuple(map(int, (v.split("."))))
if versiontuple(sagemaker.__version__) < versiontuple(required_version):
!{sys.executable} -m pip install -U sagemaker=={required_version}
IPython.Application.instance().kernel.do_shutdown(True)
import sagemaker
print(sagemaker.__version__)
import boto3
role = sagemaker.get_execution_role()
region = boto3.Session().region_name
sagemaker_session = sagemaker.Session()
bucket_name = sagemaker_session.default_bucket()
prefix = 'endtoendmlsm'
print(region)
print(role)
print(bucket_name)
```
## Retrieve model artifacts
First, we need to create two Amazon SageMaker **Model** objects, which associate the artifacts of training (serialized model artifacts in Amazon S3) to the Docker container used for inference. In order to do that, we need to get the paths to our serialized models in Amazon S3.
<ul>
<li>For the SKLearn model, in Step 02 (data exploration and feature engineering) we defined the path where the artifacts are saved</li>
<li>For the XGBoost model, we need to find the path based on Amazon SageMaker's naming convention. We are going to use a utility function to get the model artifacts of the last training job matching a specific base job name.</li>
</ul>
```
from notebook_utilities import get_latest_training_job_name, get_training_job_s3_model_artifacts
# SKLearn model artifacts path.
sklearn_model_path = 's3://{0}/{1}/output/sklearn/model.tar.gz'.format(bucket_name, prefix)
# XGBoost model artifacts path.
training_base_job_name = 'end-to-end-ml-sm-xgb'
latest_training_job_name = get_latest_training_job_name(training_base_job_name)
xgboost_model_path = get_training_job_s3_model_artifacts(latest_training_job_name)
print('SKLearn model path: ' + sklearn_model_path)
print('XGBoost model path: ' + xgboost_model_path)
```
## SKLearn Featurizer Model
Let's build the SKLearn model. For hosting this model we also provide a custom inference script, that is used to process the inputs and outputs and execute the transform.
The inference script is implemented in the `sklearn_source_dir/inference.py` file. The custom script defines:
- a custom `input_fn` for pre-processing inference requests. Our input function accepts only CSV input, loads the input in a Pandas dataframe and assigns feature column names to the dataframe
- a custom `predict_fn` for running the transform over the inputs
- a custom `output_fn` for returning either JSON or CSV
- a custom `model_fn` for deserializing the model
```
!pygmentize sklearn_source_dir/inference.py
```
Now, let's create the `SKLearnModel` object, by providing the custom script and S3 model artifacts as input.
```
import time
from sagemaker.sklearn import SKLearnModel
code_location = 's3://{0}/{1}/code'.format(bucket_name, prefix)
sklearn_model = SKLearnModel(name='end-to-end-ml-sm-skl-model-{0}'.format(str(int(time.time()))),
model_data=sklearn_model_path,
entry_point='inference.py',
source_dir='sklearn_source_dir/',
code_location=code_location,
role=role,
sagemaker_session=sagemaker_session,
framework_version='0.20.0',
py_version='py3')
```
## XGBoost Model
Similarly to the previous steps, we can create an `XGBoost` model object. Also here, we have to provide a custom inference script.
The inference script is implemented in the `xgboost_source_dir/inference.py` file. The custom script defines:
- a custom `input_fn` for pre-processing inference requests. This input function is able to handle JSON requests, plus all content types supported by the default XGBoost container. For additional information please visit: https://github.com/aws/sagemaker-xgboost-container/blob/master/src/sagemaker_xgboost_container/encoder.py. The reason for adding the JSON content type is that the container-to-container default request content type in an inference pipeline is JSON.
- a custom `model_fn` for deserializing the model
```
!pygmentize xgboost_source_dir/inference.py
```
Now, let's create the `XGBoostModel` object, by providing the custom script and S3 model artifacts as input.
```
import time
from sagemaker.xgboost import XGBoostModel
code_location = 's3://{0}/{1}/code'.format(bucket_name, prefix)
xgboost_model = XGBoostModel(name='end-to-end-ml-sm-xgb-model-{0}'.format(str(int(time.time()))),
model_data=xgboost_model_path,
entry_point='inference.py',
source_dir='xgboost_source_dir/',
code_location=code_location,
framework_version='0.90-2',
py_version='py3',
role=role,
sagemaker_session=sagemaker_session)
```
## Pipeline Model
Once we have models ready, we can deploy them in a pipeline, by building a `PipelineModel` object and calling the `deploy()` method.
```
import sagemaker
import time
from sagemaker.pipeline import PipelineModel
pipeline_model_name = 'end-to-end-ml-sm-xgb-skl-pipeline-{0}'.format(str(int(time.time())))
pipeline_model = PipelineModel(
name=pipeline_model_name,
role=role,
models=[
sklearn_model,
xgboost_model],
sagemaker_session=sagemaker_session)
endpoint_name = 'end-to-end-ml-sm-pipeline-endpoint-{0}'.format(str(int(time.time())))
print(endpoint_name)
pipeline_model.deploy(initial_instance_count=1,
instance_type='ml.m5.xlarge',
endpoint_name=endpoint_name)
```
<span style="color: red; font-weight:bold">Please take note of the endpoint name, since it will be used in the next workshop module.</span>
## Getting inferences
Finally we can try invoking our pipeline of models and get some inferences:
```
from sagemaker.serializers import CSVSerializer
from sagemaker.deserializers import JSONDeserializer
from sagemaker.predictor import Predictor
predictor = Predictor(
endpoint_name=endpoint_name,
sagemaker_session=sagemaker_session,
serializer=CSVSerializer(),
deserializer=JSONDeserializer())
#'Type', 'Air temperature [K]', 'Process temperature [K]', 'Rotational speed [rpm]', 'Torque [Nm]', 'Tool wear [min]'
payload = "L,298.4,308.2,1582,70.7,216"
print(predictor.predict(payload))
payload = "M,298.4,308.2,1582,30.2,214"
print(predictor.predict(payload))
payload = "L,298.4,308.2,30,70.7,216"
print(predictor.predict(payload))
#predictor.delete_endpoint()
```
Once we have tested the endpoint, we can move to the next workshop module. Please access the module <a href="https://github.com/aws-samples/amazon-sagemaker-build-train-deploy/tree/master/05_API_Gateway_and_Lambda" target="_blank">05_API_Gateway_and_Lambda</a> on GitHub to continue.
|
github_jupyter
|
# SimFin Test All Datasets
This Notebook performs automated testing of all the bulk datasets from SimFin. The datasets are first downloaded from the SimFin server and then various tests are performed on the data. An exception is raised if any problems are found.
This Notebook can be run as usual if you have `simfin` installed, by running the following command from the directory where this Notebook is located:
jupyter notebook
This Notebook can also be run using `pytest` which makes automated testing easier. You need to have the Python packages `simfin` and `nbval` installed. Then execute the following command from the directory where this Notebook is located:
pytest --nbval-lax -v test_bulk_data.ipynb
This runs the entire Notebook and outputs error messages for all the cells that raised an exception.
## IMPORTANT!
- When you make changes to this Notebook, remember to clear all cells before pushing it back to github, because that makes it easier to see the difference from the previous version. Select menu-item "Kernel / Restart & Clear Output".
- If you set `refresh_days=0` then it will force a new download of all the datasets.
```
# Set this to 0 to force a new download of all datasets.
refresh_days = 30
```
## Imports
```
import pandas as pd
import numpy as np
import warnings
import sys
import os
from IPython.display import display
import simfin as sf
from simfin.names import *
from simfin.datasets import *
```
## Are We Running Pytest?
```
# Boolean whether this is being run under pytest.
# This is useful when printing examples of errors
# if they take a long time to compute, because it
# is not necessary when running pytest.
running_pytest = ('PYTEST_CURRENT_TEST' in os.environ)
```
## Configure SimFin
```
sf.set_data_dir('~/simfin_data/')
sf.load_api_key(path='~/simfin_api_key.txt', default_key='free')
```
## Load All Datasets
```
%%time
data = AllDatasets(refresh_days=refresh_days)
# Example for annual Income Statements.
data.get(dataset='income', variant='annual', market='us').head()
```
## Lists of Datasets
These are in addition to the lists of datasets from `datasets.py`.
```
# Datasets that have a column named TICKER.
# Some tests are probably only necessary for 'companies'
# but we might as well test all datasets that use tickers.
datasets_tickers = ['companies'] + datasets_fundamental() + datasets_shareprices()
```
## Function for Testing Datasets
```
def test_datasets(test_name, datasets=None, variants=None,
markets=None,
test_func=None,
test_func_rows=None,
test_func_groups=None,
group_index=SIMFIN_ID,
process_df_none=False, raise_exception=True):
"""
Helper-function for running tests on many Pandas DataFrames.
:param test_name:
String with the name of the test.
:param datasets:
By default (datasets=None) all possible datasets
will be tested. Otherwise datasets is a list of
strings with dataset names to be tested.
:param variants:
By default (variants=None) all possible variants
for each dataset will be tested, as defined in
simfin.datasets.valid_variants. Otherwise variants
is a list of strings and only those variants
will be tested.
:param markets:
By default (markets=None) all possible markets
for each dataset will be tested, as defined in
simfin.datasets.valid_markets. Otherwise markets
is a list of strings and only those markets
will be tested.
:param test_func:
Function to be called on the Pandas DataFrame for
each dataset. If there are problems with the DataFrame
then return True, otherwise return False.
This is generally used for testing problems with the
entire DataFrame. For example, if the dataset is empty:
test_func = lambda df: len(df) == 0
If this returns True then there is a problem with df.
:param test_func_rows:
Similar to test_func but for testing individual rows
of a DataFrame. For example, test if SHARES_BASIC is
None, zero or negative:
test_func_rows = lambda df: (df[SHARES_BASIC] is None or
df[SHARES_BASIC] <= 0)
:param test_func_groups:
Similar to test_func but for testing groups of rows
in a DataFrame. For example, test on a per-stock basis
whether SHARES_BASIC is greater than twice its mean:
test_func_groups = lambda df: (df[SHARES_BASIC] >
df[SHARES_BASIC].mean() * 2).any()
:param group_index:
String with the column-name used to create groups when
using test_func_groups e.g. SIMFIN_ID for grouping by companies.
:param process_df_none:
Boolean whether to process (True) or skip (False)
DataFrames that are None, because they could not be loaded.
:param raise_exception:
Boolean. If True then raise an exception if there were
any problems, but wait until all datasets have been
tested, so we can print the list of datasets with problems.
If False then only show a warning if there were problems.
:return:
None
"""
# Convert to test_func.
if test_func_rows is not None:
# Convert test_func_rows to test_func.
test_func = lambda df: test_func_rows(df).any()
elif test_func_groups is not None:
# Convert test_func_groups to test_func.
# NOTE: We must use .any(axis=None) because if the DataFrame
# is empty then the groupby returns an empty DataFrame, and
# .any() then returns an empty Series, but we need a boolean.
# By using .any(axis=None) it is reduced to a boolean value.
test_func = lambda df: df.groupby(group_index, group_keys=False).apply(test_func_groups).any(axis=None)
# Number of problems found.
num_problems = 0
# For all datasets, variants and markets.
for dataset, variant, market, df in data.iter(datasets=datasets,
variants=variants,
markets=markets):
# Also process DataFrames that are None,
# because they could not be loaded?
if df is not None or process_df_none:
try:
# Perform the user-supplied test.
problem_found = test_func(df)
except:
# An exception occurred so we consider
# that to be a problem.
problem_found = True
if problem_found:
# Increase the number of problems found.
num_problems += 1
# Print the test's name. Only done once.
if num_problems==1:
print(test_name, file=sys.stderr)
# Print the dataset details.
msg = "dataset='{}', variant='{}', market='{}'"
msg = msg.format(dataset, variant, market)
print(msg, file=sys.stderr)
# Raise exception or generate warning?
if num_problems>0:
if raise_exception:
raise Exception(test_name)
else:
warnings.warn(test_name)
```
## Function for Getting Rows with Problems
When a test has found problems in a dataset, it does not show which specific rows have the problem. You can get all the problematic rows using this function:
```
def get_problem_rows(df, test_func_rows):
"""
Perform the given test on all rows of the given DataFrame
and return a DataFrame with only the problematic rows.
:param df:
Pandas DataFrame.
:param test_func_rows:
Function used for testing each row. This takes
a Pandas DataFrame as an argument and returns
a Pandas Series of booleans whether each row
in the original DataFrame has the error.
For example:
test_func_rows = lambda df: (df[SHARES_BASIC] is None or
df[SHARES_BASIC] <= 0)
:return:
Pandas DataFrame with only the problematic rows.
"""
# Index of the rows with problems.
idx = test_func_rows(df)
# Extract the rows with problems.
df2 = df[idx]
return df2
```
## Function for Getting Rows with Missing Data
```
def get_missing_data_rows(df, column):
"""
Return the rows of `df` where the data for the given
column is missing i.e. it is either NaN, None, or Null.
:param df:
Pandas DataFrame.
:param column:
Name of the column.
:return:
Pandas Series with the rows where the
column-data is missing.
"""
# Index for the rows where column-data is missing.
idx = df[column].isnull()
# Get those rows from the DataFrame.
df2 = df[idx]
return df2
```
## Function for Getting Problematic Groups
```
def get_problem_groups(df, test_func_groups, group_index):
"""
Perform the given test on the given DataFrame grouped by
the given index, and return a DataFrame with only the
problematic groups.
This is used to perform tests on a DataFrame on a per-group
basis, e.g. per-stock or per-company, and return a new
DataFrame with only the rows for the stocks that had problems.
:param df:
Pandas DataFrame.
:param test_func_groups:
Similar to test_func but for testing groups of rows
in a DataFrame. For example, test on a per-stock basis
whether SHARES_BASIC is greater than twice its mean:
test_func_groups = lambda df: (df[SHARES_BASIC] >
df[SHARES_BASIC].mean() * 2)
:param group_index:
String with the column-name used to create groups when
using test_func_groups e.g. SIMFIN_ID for grouping by companies.
:return:
Pandas DataFrame with only the problematic groups.
"""
return df.groupby(group_index).filter(test_func_groups)
```
## Function for Testing Equality with Tolerance
This function is useful when comparing floating point numbers, or when comparing accounting numbers that are supposed to have a strict relationship (e.g. Assets = Liabilities + Equity) but we might tolerate a small degree of error in the data e.g. 1%.
```
def isclose(x, y, tolerance=0.01):
"""
Compare whether x and y are approximately equal within
the given tolerance, which is a ratio so tolerance=0.01
means that we tolerate max 1% difference between x and y.
This is similar to numpy.isclose() but is a more efficient
implementation for Pandas which apparently does not have
this built-in already (v. 0.25.1)
:param x:
Pandas DataFrame or Series.
:param y:
Pandas DataFrame or Series.
:param tolerance:
Max allowed difference as a ratio e.g. 0.01 = 1%.
:return:
Pandas DataFrame or Series with booleans whether
x and y are approx. equal.
"""
return (x-y).abs() <= tolerance * y.abs()
```
# Tests
## Dataset could not be loaded
```
test_name = "Dataset could not be loaded"
test_func = lambda df: df is None
test_datasets(datasets=datasets_all(),
test_name=test_name, test_func=test_func,
process_df_none=True)
```
## Dataset is empty
```
test_name = "Dataset is empty"
test_func = lambda df: len(df) == 0
# Test for all markets. This only raises a warning,
# because some markets do have some of their datasets empty.
test_datasets(datasets=datasets_all(),
test_name=test_name, test_func=test_func,
raise_exception=False)
# Test only for the 'us' market. This raises an exception.
# It happened once that all the datasets were empty
# because of some bug on the server or whatever, so it
# is important to raise an exception in case this happens again.
test_datasets(datasets=datasets_all(), markets=['us'],
test_name=test_name, test_func=test_func,
raise_exception=True)
data.get(dataset='income-insurance', variant='quarterly', market='de')
```
## Shares Basic is None or <= 0
```
test_name = "SHARES_BASIC is None or <= 0"
test_func_rows = lambda df: (df[SHARES_BASIC] is None or
df[SHARES_BASIC] <= 0)
test_datasets(datasets=datasets_fundamental(),
test_name=test_name, test_func_rows=test_func_rows)
# Show the problematic rows for a dataset.
df = data.get(dataset='income', variant='annual', market='us')
get_problem_rows(df=df, test_func_rows=test_func_rows)
```
## Shares Diluted is None or <= 0
```
test_name = "SHARES_DILUTED is None or <= 0"
test_func_rows = lambda df: (df[SHARES_DILUTED] is None or
df[SHARES_DILUTED] <= 0)
test_datasets(datasets=datasets_fundamental(),
test_name=test_name, test_func_rows=test_func_rows)
# Show the problematic rows for a dataset.
df = data.get(dataset='income', variant='annual', market='us')
get_problem_rows(df=df, test_func_rows=test_func_rows)
```
## Shares Basic or Diluted looks strange
```
# List of SimFin-Id's to ignore in this test.
# Use this list when a company's share-counts look strange,
# but after manual inspection of the financial reports, the
# share-counts are actually correct.
ignore_simfin_ids = \
[ 53151, 61372, 82753, 99062, 148380, 166965, 258731, 378110,
498391, 520475, 543421, 543877, 546550, 592461, 620342, 652016,
652547, 658464, 658467, 659836, 667668, 689587, 698616, 704562,
768206, 778777, 794492, 798464, 826389, 867483, 890308, 896087,
899362, 951586]
# Ensure they are all unique.
ignore_simfin_ids = np.unique(ignore_simfin_ids)
ignore_simfin_ids
def test_func_groups(df_grp):
# Perform various tests on the share-counts.
# Assume `df_grp` only contains data for a single company,
# because this function should be called using:
# df.groupby(SIMFIN_ID).apply(test_func_groups)
# Ignore this company?
if df_grp[SIMFIN_ID].iloc[0] in ignore_simfin_ids:
return False
# Helper-function for calculating absolute ratio between
# a value and its average.
abs_ratio = lambda df: (df / df.mean() - 1).abs()
# Max absolute ratio allowed.
max_abs_ratio = 2
# Test whether Shares Basic is much different from its mean.
test1 = (abs_ratio(df_grp[SHARES_BASIC]) > max_abs_ratio).any()
# Test whether Shares Diluted is much different from its mean.
test2 = (abs_ratio(df_grp[SHARES_DILUTED]) > max_abs_ratio).any()
return (test1 | test2)
%%time
test_name = "Shares Basic or Shares Diluted looks strange"
test_datasets(datasets=datasets_fundamental(),
test_name=test_name,
test_func_groups=test_func_groups,
group_index=SIMFIN_ID)
# Show the problematic groups for a dataset.
if not running_pytest:
# Get the dataset.
df = data.get(dataset='income', variant='annual', market='us')
# Get the problematic groups.
df_problems = get_problem_groups(df=df,
test_func_groups=test_func_groups,
group_index=SIMFIN_ID)
# Print the problematic groups.
for _, df2 in df_problems.groupby(SIMFIN_ID):
display(df2[[SIMFIN_ID, REPORT_DATE, SHARES_BASIC, SHARES_DILUTED]])
```
## Share-Prices are Zero or Negative
```
test_name = "Share-prices are zero"
def test_func_rows(df):
return (df[OPEN] <= 0.0) & (df[LOW] <= 0.0) & \
(df[HIGH] <= 0.0) & (df[CLOSE] <= 0.0) & \
(df[VOLUME] <= 0.0)
test_datasets(datasets=['shareprices'],
test_name=test_name, test_func_rows=test_func_rows)
# Show the problematic rows for a dataset.
df = data.get(dataset='shareprices', variant='daily', market='us')
get_problem_rows(df=df, test_func_rows=test_func_rows)
```
## Revenue is negative
```
test_name = "REVENUE < 0"
test_func_rows = lambda df: (df[REVENUE] < 0)
# It is possible that Revenue is negative for banks and
# insurance companies, so we only test it for "normal" companies
# in the 'income' dataset.
test_datasets(datasets=['income'],
test_name=test_name, test_func_rows=test_func_rows)
# Show the problematic rows for a dataset.
df = data.get(dataset='income-insurance', variant='quarterly', market='us')
get_problem_rows(df=df, test_func_rows=test_func_rows)
```
## Assets != Liabilities + Equity (Exact Comparison)
This only generates a warning, because sometimes there are tiny rounding errors.
```
test_name = "Assets != Liabilities + Equity (Exact Comparison)"
test_func_rows = lambda df: (df[TOTAL_ASSETS] != df[TOTAL_LIABILITIES] + df[TOTAL_EQUITY])
test_datasets(datasets=datasets_balance(),
test_name=test_name, test_func_rows=test_func_rows,
raise_exception=False)
# Get the problematic rows for a dataset.
df = data.get(dataset='balance', variant='quarterly', market='us')
df2 = get_problem_rows(df=df, test_func_rows=test_func_rows)
# Only show the relevant columns.
df2[[TICKER, SIMFIN_ID, REPORT_DATE, TOTAL_ASSETS, TOTAL_LIABILITIES, TOTAL_EQUITY]]
```
## Assets != Liabilities + Equity (1% Tolerance)
The above test used exact comparison. We now allow for 1% error. This raises an exception.
```
def test_func_rows(df):
x = df[TOTAL_ASSETS]
y = df[TOTAL_LIABILITIES] + df[TOTAL_EQUITY]
# Compare x and y within 1% tolerance. Note the resulting
# boolean array is negated because we want to indicate
# which rows are problematic so x and y are not close.
return ~isclose(x=x, y=y, tolerance=0.01)
test_name = "Assets != Liabilities + Equity (1% Tolerance)"
test_datasets(datasets=datasets_balance(),
test_name=test_name, test_func_rows=test_func_rows)
# Get the problematic rows for a dataset.
df = data.get(dataset='balance', variant='annual', market='us')
df2 = get_problem_rows(df=df, test_func_rows=test_func_rows)
# Only show the relevant columns.
df2[[TICKER, SIMFIN_ID, REPORT_DATE, TOTAL_ASSETS, TOTAL_LIABILITIES, TOTAL_EQUITY]]
```
## Dates are invalid (Fundamentals)
```
# Lambda function for converting strings to dates. Format: YYYY-MM-DD
# This will raise an exception if invalid dates are encountered.
date_parser = lambda column: pd.to_datetime(column, yearfirst=True, dayfirst=False)
# Test function for the entire DataFrame.
# This cannot show which individual rows have problems.
def test_func(df):
result1 = date_parser(df[REPORT_DATE])
result2 = date_parser(df[PUBLISH_DATE])
# We only get to this point if date_parser() does not
# raise any exceptions, in which case we assume the
# data did not have any problems.
return False
test_name = "REPORT_DATE or PUBLISH_DATE is invalid"
test_datasets(datasets=datasets_fundamental(),
test_name=test_name, test_func=test_func)
```
## Dates are invalid (Share-Prices)
```
# Test function for the entire DataFrame.
# This cannot show which individual rows have problems.
def test_func(df):
result1 = date_parser(df[DATE])
# We only get to this point if date_parser() does not
# raise any exceptions, in which case we assume the
# data did not have any problems.
return False
test_name = "DATE is invalid"
test_datasets(datasets=datasets_shareprices(),
test_name=test_name, test_func=test_func)
```
## Duplicate Tickers
```
def get_duplicate_tickers(df):
"""
Return the rows of `df` where multiple SIMFIN_ID
have the same TICKER.
:param df: Pandas DataFrame with TICKER column.
:return: Pandas DataFrame.
"""
# Remove duplicate rows of [TICKER, SIMFIN_ID] pairs.
# For the 'companies' dataset this is not necessary,
# but for e.g. the 'income' dataset we have many rows
# for each [TICKER, SIMFIN_ID] pair because there are
# many financial reports for each of these ID pairs.
idx = df[[TICKER, SIMFIN_ID]].duplicated()
df2 = df[~idx]
# Now the DataFrame df2 only contains unique rows of
# [TICKER, SIMFIN_ID] so we need to check if there are
# any duplicate TICKER.
# Index for rows where TICKER is a duplicate.
idx1 = df2[TICKER].duplicated()
# Index for rows where TICKER is not NaN.
# These would otherwise show up as duplicates.
idx2 = df2[TICKER].notna()
# Index for rows where TICKER is a duplicate but not NaN.
idx = idx1 & idx2
# Get those rows from the DataFrame.
df2 = df2[idx]
return df2
# Test-function whether a DataFrame has duplicate tickers.
test_func = lambda df: (len(get_duplicate_tickers(df=df)) > 0)
test_name = "Duplicate Tickers"
test_datasets(datasets=datasets_tickers,
test_name=test_name, test_func=test_func)
# Show duplicate tickers in the 'companies' dataset.
df = data.get(dataset='companies', market='us')
get_duplicate_tickers(df=df)
# Show duplicate tickers in the 'income-annual' dataset.
df = data.get(dataset='income', variant='annual', market='us')
get_duplicate_tickers(df=df)
```
## Missing Tickers
```
# Test-function whether a DataFrame has missing tickers.
test_func = lambda df: (len(get_missing_data_rows(df=df, column=TICKER)) > 0)
test_name = "Missing Tickers"
test_datasets(datasets=datasets_tickers,
test_name=test_name, test_func=test_func)
# Show missing tickers in the 'companies' dataset.
df = data.get(dataset='companies', market='us')
get_missing_data_rows(df=df, column=TICKER)
# Show missing tickers in the 'income-annual' dataset.
df = data.get(dataset='income', variant='annual', market='us')
get_missing_data_rows(df=df, column=TICKER)
# Show missing tickers in the 'shareprices-daily' dataset.
df = data.get(dataset='shareprices', variant='daily', market='us')
get_missing_data_rows(df=df, column=TICKER)
```
## Missing Company Names
```
# Test-function whether a DataFrame has missing company names.
test_func = lambda df: (len(get_missing_data_rows(df=df, column=COMPANY_NAME)) > 0)
test_name = "Missing Company Name"
test_datasets(datasets=['companies'],
test_name=test_name, test_func=test_func)
# Show missing company names in the 'companies' dataset.
df = data.get(dataset='companies', market='us')
get_missing_data_rows(df=df, column=COMPANY_NAME)
```
## Missing Annual Reports
```
def missing_annual_reports(df):
"""
Return a list of the SIMFIN_ID's from the given DataFrame
that have missing annual reports.
:param df:
Pandas DataFrame with a dataset e.g. 'income-annual'.
It must have columns SIMFIN_ID and FISCAL_YEAR.
:return:
List of integers with SIMFIN_ID's that have missing reports.
"""
# The idea is to test for each SIMFIN_ID individually,
# whether the DataFrame has all the expected reports for
# consecutive Fiscal Years between the min/max years.
# Helper-function for processing a DataFrame for one SIMFIN_ID.
def _missing(df):
# Get the Fiscal Years from the DataFrame.
fiscal_years = df[FISCAL_YEAR]
# How many years between min and max fiscal years.
num_years = fiscal_years.max() - fiscal_years.min() + 1
# We expect the Series to have the same length, otherwise
# some reports must be missing between min and max years.
missing = (num_years != len(fiscal_years))
return missing
# Process all companies individually and get a Pandas
# DataFrame with a boolean for each SIMFIN_ID whether
# it has some missing Fiscal Years.
idx = df.groupby(SIMFIN_ID).apply(_missing)
# List of the SIMFIN_ID's that have missing reports.
simfin_ids = list(idx[idx].index.values)
return simfin_ids
test_name = "Missing annual reports"
test_func = lambda df: len(missing_annual_reports(df=df)) > 0
test_datasets(datasets=datasets_fundamental(),
variants=['annual'],
test_name=test_name, test_func=test_func)
# Get list of SIMFIN_ID's that have missing reports for a dataset.
if not running_pytest:
df = data.get(dataset='income', variant='annual', market='de')
display(missing_annual_reports(df=df))
def sort_annual_reports(df, simfin_id):
"""
Get the data for a given SIMFIN_ID and set the index to be
the sorted Fiscal Year so it is easier to see which are missing.
"""
return df.set_index([SIMFIN_ID, FISCAL_YEAR]).sort_index().loc[simfin_id]
# Show all the reports for a given SIMFIN_ID sorted by
# Fiscal Year so it is easier to see which are missing.
if not running_pytest:
display(sort_annual_reports(df=df, simfin_id=936426))
```
## Missing Quarterly Reports
```
def missing_quarterly_reports(df):
"""
Return a list of the SIMFIN_ID's from the given DataFrame
that have missing quarterly or ttm reports.
:param df:
Pandas DataFrame with a dataset e.g. 'income-annual'.
It must have columns SIMFIN_ID, FISCAL_YEAR, FISCAL_PERIOD.
:return:
List of integers with SIMFIN_ID's that have missing reports.
"""
# The idea is to test for each SIMFIN_ID individually,
# whether the DataFrame has all the expected reports for
# consecutive Fiscal Years and Periods between the min/max.
# Helper-function for processing a DataFrame for one SIMFIN_ID.
def _missing(df):
# Get the Fiscal Years and Periods from the DataFrame.
fiscal_years_periods = df[[FISCAL_YEAR, FISCAL_PERIOD]]
# The first Fiscal Year and Period.
min_year = fiscal_years_periods[FISCAL_YEAR].min()
min_idx = (fiscal_years_periods[FISCAL_YEAR] == min_year)
min_period = fiscal_years_periods[min_idx][FISCAL_PERIOD].min()
# The last Fiscal Year and Period.
max_year = fiscal_years_periods[FISCAL_YEAR].max()
max_idx = (fiscal_years_periods[FISCAL_YEAR] == max_year)
max_period = fiscal_years_periods[max_idx][FISCAL_PERIOD].max()
# How many years between min and max fiscal years.
num_years = max_year - min_year + 1
# Total number of Fiscal Periods between first and
# last Fiscal Years - if all Fiscal Periods were included.
num_periods = num_years * 4
# Used to map from Fiscal Period strings to ints.
# This is safer and easier to understand than
# e.g. def map_period(x): int(x[1])
map_period = \
{
'Q1': 1,
'Q2': 2,
'Q3': 3,
'Q4': 4
}
# Number of Fiscal Periods missing in the first year.
adj_min_period = map_period[min_period] - 1
# Number of Fiscal Periods missing in the last year.
adj_max_period = 4 - map_period[max_period]
# Adjust the number of Fiscal Periods between the min/max
# Fiscal Years and Periods by subtracting those periods
# missing in the first and last years.
expected_periods = num_periods - adj_min_period - adj_max_period
# If the expected number of Fiscal Periods between the
# min and max dates, is different from the actual number
# of Fiscal Periods in the DataFrame, then some are missing.
missing = (expected_periods != len(fiscal_years_periods))
return missing
# Process all companies individually and get a Pandas
# DataFrame with a boolean for each SIMFIN_ID whether
# it has some missing Fiscal Years.
idx = df.groupby(SIMFIN_ID).apply(_missing)
# List of the SIMFIN_ID's that have missing reports.
simfin_ids = list(idx[idx].index.values)
return simfin_ids
%%time
test_name = "Missing quarterly reports"
test_func = lambda df: len(missing_quarterly_reports(df=df)) > 0
test_datasets(datasets=datasets_fundamental(),
variants=['quarterly'],
test_name=test_name, test_func=test_func)
# Get list of SIMFIN_ID's that have missing reports for a dataset.
if not running_pytest:
df = data.get(dataset='income', variant='quarterly', market='us')
display(missing_quarterly_reports(df=df))
def sort_quarterly_reports(df, simfin_id):
"""
Get the data for a given SIMFIN_ID and set the index to be
the sorted Fiscal Year and Period so it is easier to see
which ones are missing.
"""
return df.set_index([SIMFIN_ID, FISCAL_YEAR, FISCAL_PERIOD]).sort_index().loc[simfin_id]
# Show all the reports for a given SIMFIN_ID sorted by
# Fiscal Year and Period so it is easier to see which are missing.
if not running_pytest:
display(sort_quarterly_reports(df=df, simfin_id=139560))
```
## Missing TTM Reports
Trailing-Twelve-Months (TTM) data is also quarterly so we can use the same helper-functions from above.
```
test_name = "Missing ttm reports"
test_func = lambda df: len(missing_quarterly_reports(df=df)) > 0
test_datasets(datasets=datasets_fundamental(),
variants=['ttm'],
test_name=test_name, test_func=test_func)
# Get list of SIMFIN_ID's that have missing reports for a dataset.
if not running_pytest:
df = data.get(dataset='income', variant='ttm', market='us')
display(missing_quarterly_reports(df=df))
# Show all the reports for a given SIMFIN_ID sorted by
# Fiscal Year and Period so it is easier to see which are missing.
if not running_pytest:
display(sort_quarterly_reports(df=df, simfin_id=89750))
```
|
github_jupyter
|

[](https://colab.research.google.com/github/JohnSnowLabs/spark-nlp-workshop/blob/master/tutorials/streamlit_notebooks/healthcare/NER_LEGAL_DE.ipynb)
# **Detect legal entities in German**
To run this yourself, you will need to upload your license keys to the notebook. Just Run The Cell Below in order to do that. Also You can open the file explorer on the left side of the screen and upload `license_keys.json` to the folder that opens.
Otherwise, you can look at the example outputs at the bottom of the notebook.
## 1. Colab Setup
Import license keys
```
import os
import json
from google.colab import files
license_keys = files.upload()
with open(list(license_keys.keys())[0]) as f:
license_keys = json.load(f)
sparknlp_version = license_keys["PUBLIC_VERSION"]
jsl_version = license_keys["JSL_VERSION"]
print ('SparkNLP Version:', sparknlp_version)
print ('SparkNLP-JSL Version:', jsl_version)
```
Install dependencies
```
%%capture
for k,v in license_keys.items():
%set_env $k=$v
!wget https://raw.githubusercontent.com/JohnSnowLabs/spark-nlp-workshop/master/jsl_colab_setup.sh
!bash jsl_colab_setup.sh
# Install Spark NLP Display for visualization
!pip install --ignore-installed spark-nlp-display
```
Import dependencies into Python and start the Spark session
```
import pandas as pd
from pyspark.ml import Pipeline
from pyspark.sql import SparkSession
import pyspark.sql.functions as F
import sparknlp
from sparknlp.annotator import *
from sparknlp_jsl.annotator import *
from sparknlp.base import *
import sparknlp_jsl
spark = sparknlp_jsl.start(license_keys['SECRET'])
# manually start session
# params = {"spark.driver.memory" : "16G",
# "spark.kryoserializer.buffer.max" : "2000M",
# "spark.driver.maxResultSize" : "2000M"}
# spark = sparknlp_jsl.start(license_keys['SECRET'],params=params)
```
## 2. Construct the pipeline
For more details: https://github.com/JohnSnowLabs/spark-nlp-models#pretrained-models---spark-nlp-for-healthcare
```
document_assembler = DocumentAssembler() \
.setInputCol('text')\
.setOutputCol('document')
sentence_detector = SentenceDetector() \
.setInputCols(['document'])\
.setOutputCol('sentence')
tokenizer = Tokenizer()\
.setInputCols(['sentence']) \
.setOutputCol('token')
# German word embeddings
word_embeddings = WordEmbeddingsModel.pretrained('w2v_cc_300d','de', 'clinical/models') \
.setInputCols(["sentence", 'token'])\
.setOutputCol("embeddings")
# German NER model
clinical_ner = MedicalNerModel.pretrained('ner_legal','de', 'clinical/models') \
.setInputCols(["sentence", "token", "embeddings"]) \
.setOutputCol("ner")
ner_converter = NerConverter()\
.setInputCols(['sentence', 'token', 'ner']) \
.setOutputCol('ner_chunk')
nlp_pipeline = Pipeline(stages=[
document_assembler,
sentence_detector,
tokenizer,
word_embeddings,
clinical_ner,
ner_converter])
```
## 3. Create example inputs
```
# Enter examples as strings in this array
input_list = [
"""Dementsprechend hat der Bundesgerichtshof mit Beschluss vom 24 August 2017 ( - III ZA 15/17 - ) das bei ihm von der Antragstellerin anhängig gemachte „ Prozesskostenhilfeprüfungsverfahre“ an das Bundesarbeitsgericht abgegeben. 2 Die Antragstellerin hat mit Schriftsatz vom 21 März 2016 und damit mehr als sechs Monate vor der Anbringung des Antrags auf Gewährung von Prozesskostenhilfe für die beabsichtigte Klage auf Entschädigung eine Verzögerungsrüge iSv § 198 Abs 5 Satz 1 GVG erhoben. 3 Nach § 198 Abs 1 Satz 1 GVG wird angemessen entschädigt , wer infolge unangemessener Dauer eines Gerichtsverfahrens als Verfahrensbeteiligter einen Nachteil erleidet. a ) Die Angemessenheit der Verfahrensdauer richtet sich gemäß § 198 Abs 1 Satz 2 GVG nach den Umständen des Einzelfalls , insbesondere nach der Schwierigkeit und Bedeutung des Verfahrens sowie nach dem Verhalten der Verfahrensbeteiligten und Dritter. Hierbei handelt es sich um eine beispielhafte , nicht abschließende Auflistung von Umständen , die für die Beurteilung der Angemessenheit besonders bedeutsam sind ( BT-Drs 17/3802 S 18 ). Weitere gewichtige Beurteilungskriterien sind die Verfahrensführung durch das Gericht sowie die zur Verfahrensbeschleunigung gegenläufigen Rechtsgüter der Gewährleistung der inhaltlichen Richtigkeit von Entscheidungen , der Beachtung der richterlichen Unabhängigkeit und des gesetzlichen Richters.""",
]
```
## 4. Use the pipeline to create outputs
```
empty_df = spark.createDataFrame([['']]).toDF('text')
pipeline_model = nlp_pipeline.fit(empty_df)
df = spark.createDataFrame(pd.DataFrame({'text': input_list}))
result = pipeline_model.transform(df)
```
## 5. Visualize results
Visualize outputs as data frame
```
from sparknlp_display import NerVisualizer
NerVisualizer().display(
result = result.collect()[0],
label_col = 'ner_chunk',
document_col = 'document'
)
```
|
github_jupyter
|
Let's design a LNA using Infineon's BFU520 transistor. First we need to import scikit-rf and a bunch of other utilities:
```
import numpy as np
import skrf
from skrf.media import DistributedCircuit
import skrf.frequency as freq
import skrf.network as net
import skrf.util
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams['figure.figsize'] = [10, 10]
f = freq.Frequency(0.4, 2, 101)
tem = DistributedCircuit(f, z0=50)
# import the scattering parameters/noise data for the transistor
bjt = net.Network('BFU520_05V0_010mA_NF_SP.s2p').interpolate(f)
bjt
```
Let's plot the smith chart for it:
```
bjt.plot_s_smith()
```
Now let's calculate the source and load stablity curves.
I'm slightly misusing the `Network` type to plot the curves; normally the curves you pass in to `Network` should be a function of frequency, but it also works to draw these circles as long as you don't try to use any other functions on them
```
sqabs = lambda x: np.square(np.absolute(x))
delta = bjt.s11.s*bjt.s22.s - bjt.s12.s*bjt.s21.s
rl = np.absolute((bjt.s12.s * bjt.s21.s)/(sqabs(bjt.s22.s) - sqabs(delta)))
cl = np.conj(bjt.s22.s - delta*np.conj(bjt.s11.s))/(sqabs(bjt.s22.s) - sqabs(delta))
rs = np.absolute((bjt.s12.s * bjt.s21.s)/(sqabs(bjt.s11.s) - sqabs(delta)))
cs = np.conj(bjt.s11.s - delta*np.conj(bjt.s22.s))/(sqabs(bjt.s11.s) - sqabs(delta))
def calc_circle(c, r):
theta = np.linspace(0, 2*np.pi, 1000)
return c + r*np.exp(1.0j*theta)
for i, f in enumerate(bjt.f):
# decimate it a little
if i % 100 != 0:
continue
n = net.Network(name=str(f/1.e+9), s=calc_circle(cs[i][0, 0], rs[i][0, 0]))
n.plot_s_smith()
for i, f in enumerate(bjt.f):
# decimate it a little
if i % 100 != 0:
continue
n = net.Network(name=str(f/1.e+9), s=calc_circle(cl[i][0, 0], rl[i][0, 0]))
n.plot_s_smith()
```
So we can see that we need to avoid inductive loads near short circuit in the input matching network and high impedance inductive loads on the output.
Let's draw some constant noise circles. First we grab the noise parameters for our target frequency from the network model:
```
idx_915mhz = skrf.util.find_nearest_index(bjt.f, 915.e+6)
# we need the normalized equivalent noise and optimum source coefficient to calculate the constant noise circles
rn = bjt.rn[idx_915mhz]/50
gamma_opt = bjt.g_opt[idx_915mhz]
fmin = bjt.nfmin[idx_915mhz]
for nf_added in [0, 0.1, 0.2, 0.5]:
nf = 10**(nf_added/10) * fmin
N = (nf - fmin)*abs(1+gamma_opt)**2/(4*rn)
c_n = gamma_opt/(1+N)
r_n = 1/(1-N)*np.sqrt(N**2 + N*(1-abs(gamma_opt)**2))
n = net.Network(name=str(nf_added), s=calc_circle(c_n, r_n))
n.plot_s_smith()
print("the optimum source reflection coefficient is ", gamma_opt)
```
So we can see from the chart that just leaving the input at 50 ohms gets us under 0.1 dB of extra noise, which seems pretty good. I'm actually not sure that these actually correspond to the noise figure level increments I have listed up there, but the circles should at least correspond to increasing noise figures
So let's leave the input at 50 ohms and figure out how to match the output network to maximize gain and stability. Let's see what matching the load impedance with an unmatched input gives us:
```
gamma_s = 0.0
gamma_l = np.conj(bjt.s22.s - bjt.s21.s*gamma_s*bjt.s12.s/(1-bjt.s11.s*gamma_s))
gamma_l = gamma_l[idx_915mhz, 0, 0]
is_gamma_l_stable = np.absolute(gamma_l - cl[idx_915mhz]) > rl[idx_915mhz]
gamma_l, is_gamma_l_stable
```
This looks like it may be kind of close to the load instability circles, so it might make sense to pick a load point with less gain for more stability, or to pick a different source impedance with more noise.
But for now let's just build a matching network for this and see how it performs:
```
def calc_matching_network_vals(z1, z2):
flipped = np.real(z1) < np.real(z2)
if flipped:
z2, z1 = z1, z2
# cancel out the imaginary parts of both input and output impedances
z1_par = 0.0
if abs(np.imag(z1)) > 1e-6:
# parallel something to cancel out the imaginary part of
# z1's impedance
z1_par = 1/(-1j*np.imag(1/z1))
z1 = 1/(1./z1 + 1/z1_par)
z2_ser = 0.0
if abs(np.imag(z2)) > 1e-6:
z2_ser = -1j*np.imag(z2)
z2 = z2 + z2_ser
Q = np.sqrt((np.real(z1) - np.real(z2))/np.real(z2))
x1 = -1.j * np.real(z1)/Q
x2 = 1.j * np.real(z2)*Q
x1_tot = 1/(1/z1_par + 1/x1)
x2_tot = z2_ser + x2
if flipped:
return x2_tot, x1_tot
else:
return x1_tot, x2_tot
z_l = net.s2z(np.array([[[gamma_l]]]))[0,0,0]
# note that we're matching against the conjugate;
# this is because we want to see z_l from the BJT side
# if we plugged in z the matching network would make
# the 50 ohms look like np.conj(z) to match against it, so
# we use np.conj(z_l) so that it'll look like z_l from the BJT's side
z_par, z_ser = calc_matching_network_vals(np.conj(z_l), 50)
z_l, z_par, z_ser
```
Let's calculate what the component values are:
```
c_par = np.real(1/(2j*np.pi*915e+6*z_par))
l_ser = np.real(z_ser/(2j*np.pi*915e+6))
c_par, l_ser
```
The capacitance is kind of low but the inductance seems reasonable. Let's test it out:
```
output_network = tem.shunt_capacitor(c_par) ** tem.inductor(l_ser)
amplifier = bjt ** output_network
amplifier.plot_s_smith()
```
That looks pretty reasonable; let's take a look at the S21 to see what we got:
```
amplifier.s21.plot_s_db()
```
So about 18 dB gain; let's see what our noise figure is:
```
10*np.log10(amplifier.nf(50.)[idx_915mhz])
```
So 0.96 dB NF, which is reasonably close to the BJT tombstone optimal NF of 0.95 dB
|
github_jupyter
|
# Model Selection

## Model Selection
- The process of selecting the model among a collection of candidates machine learning models
### Problem type
- What kind of problem are you looking into?
- **Classification**: *Predict labels on data with predefined classes*
- Supervised Machine Learning
- **Clustering**: *Identify similarieties between objects and group them in clusters*
- Unsupervised Machine Learning
- **Regression**: *Predict continuous values*
- Supervised Machine Learning
- Resource: [Sklearn cheat sheet](https://scikit-learn.org/stable/tutorial/machine_learning_map/index.html)
### What is the "best" model?
- All models have some **predictive error**
- We should seek a model that is *good enough*
### Model Selection Techniques
- **Probabilistic Measures**: Scoring by performance and complexity of model.
- **Resampling Methods**: Splitting in sub-train and sub-test datasets and scoring by mean values of repeated runs.
```
import pandas as pd
data = pd.read_parquet('files/house_sales.parquet')
data.head()
data.describe()
data['SalePrice'].plot.hist(bins=20)
```
### Converting to Categories
- [`cut()`](https://pandas.pydata.org/docs/reference/api/pandas.cut.html) Bin values into discrete intervals.
- Data in bins based on data distribution.
- [`qcut()`](https://pandas.pydata.org/docs/reference/api/pandas.qcut.html) Quantile-based discretization function.
- Data in equal size bins
#### Invstigation
- Figure out why `cut` is not suitable for 3 bins here.
```
data['Target'] = pd.cut(data['SalePrice'], bins=3, labels=[1, 2, 3])
data['Target'].value_counts()/len(data)
data['Target'] = pd.qcut(data['SalePrice'], q=3, labels=[1, 2, 3])
data['Target'].value_counts()/len(data)
from sklearn.model_selection import train_test_split
from sklearn.svm import SVC, LinearSVC
from sklearn.metrics import accuracy_score
X = data.drop(['SalePrice', 'Target'], axis=1).fillna(-1)
y = data['Target']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=.2, random_state=42)
svc = LinearSVC()
svc.fit(X_train, y_train)
y_pred = svc.predict(X_test)
accuracy_score(y_test, y_pred)
from sklearn.neighbors import KNeighborsClassifier
neigh = KNeighborsClassifier()
neigh.fit(X_train, y_train)
y_pred = neigh.predict(X_test)
accuracy_score(y_test, y_pred)
svc = SVC(kernel='rbf')
svc.fit(X_train, y_train)
y_pred = svc.predict(X_test)
accuracy_score(y_test, y_pred)
svc = SVC(kernel='sigmoid')
svc.fit(X_train, y_train)
y_pred = svc.predict(X_test)
accuracy_score(y_test, y_pred)
svc = SVC(kernel='poly', degree=5)
svc.fit(X_train, y_train)
y_pred = svc.predict(X_test)
accuracy_score(y_test, y_pred)
```
|
github_jupyter
|
```
import numpy as np
arr = np.load('MAPS.npy')
print(arr)
print(np.shape(arr))
arr2 = np.empty((20426, 88), dtype = int)
for i in range(arr.shape[0]):
for j in range(arr.shape[1]):
if arr[i,j]==False:
arr2[i,j]=int(0)
int(arr2[i,j])
elif arr[i,j]==True:
arr2[i,j]=int(1)
print(arr2)
!pip install midiutil
from midiutil.MidiFile import MIDIFile
mf = MIDIFile(1)
track = 0
time = 0
delta = 0.000005
mf.addTrackName(track, time, "Output")
mf.addTempo(track, time, 120)
channel = 0
volume = 100
duration = 0.01
for i in range(arr2.shape[0]):
time=time + i*delta
for j in range(arr2.shape[1]):
if arr[i,j] == 1:
pitch = j
mf.addNote(track, channel, pitch, time, duration, volume)
with open("output.mid", 'wb') as outf:
mf.writeFile(outf)
!pip install pretty_midi
import pretty_midi
import pandas as pd
path = "output.mid"
midi_data = pretty_midi.PrettyMIDI(path)
midi_list = []
pretty_midi.pretty_midi.MAX_TICK = 1e10
midi_data.tick_to_time(14325216)
for instrument in midi_data.instruments:
for note in instrument.notes:
start = note.start
end = note.end
pitch = note.pitch
velocity = note.velocity
midi_list.append([start, end, pitch, velocity, instrument.name])
midi_list = sorted(midi_list, key=lambda x: (x[0], x[2]))
df = pd.DataFrame(midi_list, columns=['Start', 'End', 'Pitch', 'Velocity', 'Instrument'])
print(df)
import matplotlib
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
import numpy as np
fig, ax = plt.subplots()
i = 0
while(i<108934) :
start = float(midi_list[i][0])
pitch = float(midi_list[i][2])
duration = float(midi_list[i][1]-midi_list[i][0])
# if my_reader[i][4]=='Right Hand' :
# color1 = 'royalblue'
# else :
# color1 = 'darkorange'
rect = matplotlib.patches.Rectangle((start, pitch),duration, 1, ec='black', linewidth=10)
ax.add_patch(rect)
i+=1
# plt.xlabel("Time (seconds)", fontsize=150)
# plt.ylabel("Pitch", fontsize=150)
plt.xlim([0, 550])
plt.ylim([0, 88])
plt.grid(color='grey',linewidth=1)
plt.show()
```
ACTUAL
```
import pretty_midi
import pandas as pd
path = "MAPS.mid"
midi_data = pretty_midi.PrettyMIDI(path)
midi_list = []
pretty_midi.pretty_midi.MAX_TICK = 1e10
midi_data.tick_to_time(14325216)
for instrument in midi_data.instruments:
for note in instrument.notes:
start = note.start
end = note.end
pitch = note.pitch
velocity = note.velocity
midi_list.append([start, end, pitch, velocity, instrument.name])
midi_list = sorted(midi_list, key=lambda x: (x[0], x[2]))
df = pd.DataFrame(midi_list, columns=['Start', 'End', 'Pitch', 'Velocity', 'Instrument'])
print(df)
import matplotlib
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
import numpy as np
fig, ax = plt.subplots()
i = 0
while(i<2200) :
start = float(midi_list[i][0])
pitch = float(midi_list[i][2])
duration = float(midi_list[i][1]-midi_list[i][0])
# if my_reader[i][4]=='Right Hand' :
# color1 = 'royalblue'
# else :
# color1 = 'darkorange'
rect = matplotlib.patches.Rectangle((start, pitch),duration, 1, ec='black', linewidth=10)
ax.add_patch(rect)
i+=1
# plt.xlabel("Time (seconds)", fontsize=150)
# plt.ylabel("Pitch", fontsize=150)
plt.xlim([0, 240])
plt.ylim([0, 88])
plt.grid(color='grey',linewidth=1)
plt.show()
```
|
github_jupyter
|
```
# Necessary imports
import warnings
warnings.filterwarnings('ignore')
import re
import os
import numpy as np
import scipy as sp
from scipy.sparse import csr_matrix
from sklearn import datasets
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
from sklearn.naive_bayes import MultinomialNB
from active_tester import ActiveTester
from active_tester.estimators.learned import Learned
from active_tester.estimators.naive import Naive
from active_tester.query_strategy.noisy_label_uncertainty import LabelUncertainty
from active_tester.query_strategy.classifier_uncertainty import ClassifierUncertainty
from active_tester.query_strategy.MCM import MCM
from active_tester.query_strategy.random import Random
from sklearn.metrics import accuracy_score
from active_tester.label_estimation.methods import oracle_one_label, no_oracle, oracle_multiple_labels
```
# Active Testing Using Text Data
This is an example of using the ActT library with a text dataset. To walk through this example, download __[a sentiment analysis dataset](https://archive.ics.uci.edu/ml/machine-learning-databases/00331/)__ from the UCI machine learning repository and place the contents in the text_data directory. Additionally, this tutorial follows Scikit Learn's steps on __[Working with Text Data](https://scikit-learn.org/stable/tutorial/text_analytics/working_with_text_data.html)__. Before we employ ActT, using the example dataset, we must preprocess the data to create textfiles for each sentence and to divide the dataset into a train and test set.
## Data Processing
Using the preprocessing scripts below, we will combine all of the files into one file containing all 3000 sentence. Then, we will separate the sentences into a test and training set containing the individual sentences as files, then place them in their respective class folders.
After the dataset is created, set `create_datasets` to `False` to avoid creating duplicate files.
```
create_datasets = False
# get rid of temporary files inserted to preserve directory structure
if create_datasets:
myfile = 'text_data/temp.txt'
if os.path.isfile(myfile):
os.remove(myfile)
myfile = 'train/positive/temp.txt'
if os.path.isfile(myfile):
os.remove(myfile)
myfile = 'train/negative/temp.txt'
if os.path.isfile(myfile):
os.remove(myfile)
myfile = 'test/positive/temp.txt'
if os.path.isfile(myfile):
os.remove(myfile)
myfile = 'test/negative/temp.txt'
if os.path.isfile(myfile):
os.remove(myfile)
if create_datasets:
#Combine all sentence files into one file
try:
sentences = open('sentences.txt', 'a')
#Renamed files with dashes
filenames = ['text_data/imdb_labelled.txt',
'text_data/amazon_cells_labelled.txt',
'text_data/yelp_labelled.txt']
for filename in filenames:
print(filename)
with open(filename) as file:
for line in file:
line = line.rstrip()
sentences.write(line + '\n')
except Exception:
print('File not found')
if create_datasets:
#Separate sentences into a test and training set
#Write each sentence to a file and place that file in its respective class folder
filename = 'sentences.txt'
with open(filename) as file:
count = 1
for line in file:
if count <= 2000:
line = line.rstrip()
if line[-1:] == '0':
input_file = open('train/negative/inputfile-' + str(count) + '.txt', 'a')
line = line[:-1]
line = line.rstrip()
input_file.write(line)
if line[-1:] == '1':
input_file = open('train/positive/inputfile-' + str(count) + '.txt', 'a')
line = line[:-1]
line = line.rstrip()
input_file.write(line)
if count > 2000:
line = line.rstrip()
if line[-1:] == '0':
input_file = open('test/negative/inputfile-' + str(count) + '.txt', 'a')
line = line[:-1]
line = line.rstrip()
input_file.write(line)
if line[-1:] == '1':
input_file = open('test/positive/inputfile-' + str(count) + '.txt', 'a')
line = line[:-1]
line = line.rstrip()
input_file.write(line)
count = count + 1
```
## Loading Data and Training a Model
Below, we load the training data, create term frequency features, and then fit a classifier to the data.
```
#Load training data from files
categories = ['positive', 'negative']
sent_data = datasets.load_files(container_path='train', categories=categories, shuffle=True)
X_train, y_train = sent_data.data, sent_data.target
#Extract features
count_vect = CountVectorizer()
X_train_counts = count_vect.fit_transform(X_train)
#Transform occurance matrix to a frequency matrix
tf_transformer = TfidfTransformer(use_idf=False).fit(X_train_counts)
X_train_tf = tf_transformer.transform(X_train_counts)
#Build a classifier
clf = MultinomialNB().fit(X_train_tf, sent_data.target)
```
Now, we transform the test dataset to use the same features, apply the classifier to the test dataset and compute the classifier's true accuracy.
```
#Load the test data from files
sent_data_test = datasets.load_files(container_path='test', categories=categories, shuffle=False)
X_test, y_test = sent_data_test.data, sent_data_test.target
#Extract features
X_test_counts = count_vect.transform(sent_data_test.data)
#Transform occurance matrix to a frequency matrix
X_test_tf = tf_transformer.transform(X_test_counts)
#Compute the true accuracy of the classifier
label_predictions = clf.predict(X_test_tf)
true_accuracy = accuracy_score(y_test, label_predictions)
```
## Using Active Tester
The following code creates a set of noisy labels, reshapes the true labels, and converts the test features to a dense array.
```
#Initialize key variables: X, Y_noisy, and vetted
Y_noisy = []
noisy_label_accuracy = 0.75
for i in range(len(y_test)):
if np.random.rand() < noisy_label_accuracy:
# noisy label is correct
Y_noisy.append(y_test[i])
else:
# noisy label is incorrect
Y_noisy.append(np.random.choice(np.delete(np.arange(2),y_test[i])))
Y_noisy = np.asarray(Y_noisy, dtype=int)
#Note that if your y_noisy array is shape (L,), you will need to reshape it to be (L,1)
Y_noisy = np.reshape(Y_noisy,(len(Y_noisy),1))
Y_ground_truth = np.reshape(y_test, (len(y_test), 1))
#Note that if using sklearn's transformer, you may recieve an error about a sparse
#matrix. Using scipy's sparse csr_matrix.toarray() method can resolve this issue
X = csr_matrix.toarray(X_test_tf)
```
Now to display the sentences to the vetter in an interactive session, we need to create a list of all the test data files. This will serve as raw input to the `query_vetted` method of `active_tester`.
```
#Create a list with all of the test data files to serve as the raw input to query vetted
file_list = []
sentence_dirs = os.path.join(os.getcwd(),'test')
for root, dirs, files in os.walk(sentence_dirs):
for name in files:
if name.endswith('.txt'):
local_path = os.path.join(root, name)
file_list.append(os.path.join(sentence_dirs, local_path))
```
Now, we are ready to estimate the performance of the classifier by querying the oracle.
```
#Active Tester with a Naive Estimator, Classifier Uncertainty Query Method, and Interactive Query Vetting
budget = 5
active_test = ActiveTester(Naive(metric=accuracy_score),
ClassifierUncertainty())
active_test.standardize_data(X=X,
classes=sent_data.target_names,
Y_noisy=Y_noisy)
active_test.gen_model_predictions(clf)
active_test.query_vetted(True, budget, raw=file_list)
active_test.test()
results = active_test.get_test_results()
# View the result and compare to the true accuracy
print('Test metric with budget of', budget,': ', results['tester_metric'])
print('True accuracy of classifier: ', true_accuracy)
```
## A Comparison of Query Strategies and Estimators
Below, we compare a couple of query strategies and estimators.
```
import matplotlib.pyplot as plt
abs_error_array = []
# Initialize the estimators
learned = Learned(metric=accuracy_score, estimation_method=oracle_multiple_labels)
naive = Naive(metric=accuracy_score)
estimator_list = {'Naive': naive, 'Learned': learned}
# Initialize a few query strategies
rand = Random()
classifier_uncertainty = ClassifierUncertainty()
mcm = MCM(estimation_method=oracle_multiple_labels)
query_strategy_list = {'Random': rand, 'Classifier Uncertainty': classifier_uncertainty,
'Most Common Mistake': mcm}
# Run active testing for each estimator-query pair, for a range of sample sizes
sample_sizes = [100, 200, 300, 400, 500]
for est_k, est_v in estimator_list.items():
for query_k, query_v in query_strategy_list.items():
abs_error_array = []
for i in sample_sizes:
at = ActiveTester(est_v, query_v)
#Set dataset and model values in the active tester object
at.standardize_data(X=X,
classes=sent_data.target_names,
Y_ground_truth=Y_ground_truth,
Y_noisy=Y_noisy)
at.gen_model_predictions(clf)
at.query_vetted(False, i)
at.test()
results = at.get_test_results()
abs_error_array.append(np.abs(results['tester_metric'] - true_accuracy))
plt.ylabel("Absolute Error")
plt.xlabel("Number Vetted")
plt.plot(sample_sizes, abs_error_array, label=est_k + '+' + query_k)
plt.legend(loc='best')
plt.title('Absolute Error vs Number Vetted')
plt.grid(True)
plt.show()
```
As you can see from the graph, the absolute error for the learned estimation method is smaller than for the naive method. There is not a large difference between the different query strategies.
|
github_jupyter
|
# Python course Day 4
## Dictionaries
```
student = {"number": 570, "name":"Simon", "age":23, "height":165}
print(student)
print(student['name'])
print(student['age'])
my_list = {1: 23, 2:56, 3:78, 4:14, 5:67}
my_list[1]
my_list.keys()
my_list.values()
student.keys()
student.values()
student['number'] = 111
print(student)
student.items()
# Iterate over a list
numbers = [23,45,12,67,88,34,11]
for x in numbers:
print(x)
student.keys()
for k in student.keys():
print("student", k, student[k])
# initialize key and value with pairs from student dictionary
for key, value in student.items():
print(key , "--->", value)
print(student)
festival = 4
print(festival)
print(key)
# Iterate over two lists at a time
for x,y in zip(a,b):
print(x , ":", y)
test = zip(a,b)
type(test)
# Iterate over two lists at a time
a = [1,2,3,4,5]
b = [1,4,9,16,25]
c = [1,8,27,64,125]
for x,y,z in zip(a,b,c):
print(x , ":", y , "- ", z)
# items that do not have a corresponding item in the other list
# are ignored
a = [1,2,3,4,5,6,7]
b = [1,4,9,16,25]
for x,y in zip(a,b):
print(x , ":", y)
for x in numbers:
print(x)
for i in range(10):
print(i)
numbers
# Iterate over a list using index
for index in range(len(numbers)):
print(index , ":", numbers[index])
```
## List Comprehension
```
# Pythonic ways to create lists
# 1
numbers = []
for i in range(1,101):
numbers.append(i)
#1
'''
numbers contains all i's
such that
i takes value in range(1,101)
'''
numbers = [i for i in range(1, 101)]
print(numbers)
def square(num):
return num ** 2
square(5)
#2. call a function to make list of squares of numbers from 1-30
squared_numbers = [square(i) for i in range(1,31)]
print(squared_numbers)
#3) even numbers from 1 to 30
even_numbers = [i for i in range(1,31) if i%2 == 0]
print(even_numbers)
my_list = []
for i in range(1,31):
if i%2 == 0:
my_list.append(i)
print(my_list)
#4) squares of even numbers from 1 to 30
squared_even_numbers = [square(i) for i in range(1,31) if i%2 == 0]
print(squared_even_numbers)
#5) list of pairs
numbers_and_letters = [(chr(a),i) for a in range(65,68) for i in range(1,3)]
print(numbers_and_squares)
fun_list = []
for a in range(65,68):
for i in range(1,3):
fun_list.append((chr(a),i))
#print() # prints a new line
print(fun_list)
even_numbers
36 in even_numbers
35 in even_numbers
if 36 in even_numbers:
print("List contains 36")
even_numbers
all(even_numbers)
test = [True, True, False, True]
all(test)
not any(test)
even_numbers
import random
import math
math.factorial(6)
math.log2(16)
math.log10(1000)
math.pi
def fact(x):
if x == 0:
return 1
elif x < 0:
return -1
answer = 1
multiplier = 1
while multiplier <= x:
answer *= multiplier
multiplier += 1
return answer
fact(0)
fact(-5)
fact(4)
fact(6)
def fact_recur(x):
if x == 0:
return 1
if x < 0 :
return -1
return x * fact_recur(x-1)
fact_recur(5)
def fibo(n):
if n == 1:
return 0
if n == 2:
return 1
return fibo(n-1) + fibo(n-2)
fibo(2)
fibo(8)
fibo(9)
def simple_interest(principal, years, rate):
return (principal * years * rate ) /100
print(simple_interest(5000, 5, 2))
# Function with default argument
def simple_interest(principal, years, rate=2):
return (principal * years * rate ) /100
simple_interest(5000,5)
def simple_interest(principal=5000, years, rate=2):
return(principal * years * rate ) /100
def simple_interest(principal=5000, years=5, rate=2):
print("p = ", principal)
print("y = ", years)
print("r = ", rate)
return (principal * years * rate) / 100
simple_interest()
simple_interest(410)
simple_interest(410, 10)
# Call the function with keyword arguments/parameters
simple_interest(principal=7000, rate=10)
simple_interest(rate=3.4)
fun()
def fun():
print("fun")
gun()
def gun():
print("gun")
hun()
def hun():
print("hun")
def good():
print("good")
better()
# good() will cause error because better() function is not defined yet
def better():
print("better")
best()
def best():
print("best")
good()
```
|
github_jupyter
|
## Statistical Analysis
We have learned null hypothesis, and compared two-sample test to check whether two samples are the same or not
To add more to statistical analysis, the follwoing topics should be covered:
1- Approxite the histogram of data with combination of Gaussian (Normal) distribution functions:
Gaussian Mixture Model (GMM)
Kernel Density Estimation (KDE)
2- Correlation among features
## Review
Write a function that computes and plot histogram of a given data
Histogram is one method for estimating density
## What is Gaussian Mixture Model (GMM)?
GMM is a probabilistic model for representing normally distributed subpopulations within an overall population
<img src="Images/gmm_fig.png" width="300">
$p(x) = \sum_{i = 1}^{K} w_i \ \mathcal{N}(x \ | \ \mu_i,\ \sigma_i)$
$\sum_{i=1}^{K} w_i = 1$
https://brilliant.org/wiki/gaussian-mixture-model/
## Activity : Fit a GMM to a given data sample
Task:
1- Generate the concatination of the random variables as follows:
`x_1 = np.random.normal(-5, 1, 3000)
x_2 = np.random.normal(2, 3, 7000)
x = np.concatenate((x_1, x_2))`
2- Plot the histogram of `x`
3- Obtain the weights, mean and variances of each Gassuian
Steps needed:
`from sklearn import mixture
gmm = mixture.GaussianMixture(n_components=2)
gmm.fit(x.reshape(-1,1))`
```
import numpy as np
import matplotlib.pyplot as plt
from sklearn import mixture
# Generate data samples and plot its histogram
x_1 = np.random.normal(-5, 1, 3000)
x_2 = np.random.normal(2, 3, 7000)
x = np.concatenate((x_1, x_2))
plt.hist(x, bins=20, density=1)
plt.show()
# Define a GMM model and obtain its parameters
gmm = mixture.GaussianMixture(n_components=2)
gmm.fit(x.reshape(-1,1))
print(gmm.means_)
print(gmm.covariances_)
print(gmm.weights_)
```
## The GMM has learn the probability density function of our data sample
Lets the model generate sample from it model:
```
z = gmm.sample(10000)
plt.hist(z[0], bins=20, density=1)
plt.show()
```
## Kernel Density Estimation (KDE)
Kernel density estimation (KDE) is a non-parametric way to estimate the probability density function of a random variable. In other words the aim of KDE is to find probability density function (PDF) for a given dataset.
Approximate the pdf of dataset:
$p(x) = \frac{1}{Nh}\sum_{i = 1}^{N} \ K(\frac{x - x_i}{h})$
where $h$ is a bandwidth and $N$ is the number of data points
## Activity: Apply KDE on a given data sample
Task: Apply KDE on previous generated sample data `x`
Hint: use
`kde = KernelDensity(kernel='gaussian', bandwidth=0.6)`
```
from sklearn.neighbors import KernelDensity
kde = KernelDensity(kernel='gaussian', bandwidth=0.6)
kde.fit(x.reshape(-1,1))
s = np.linspace(np.min(x), np.max(x))
log_pdf = kde.score_samples(s.reshape(-1,1))
plt.plot(s, np.exp(log_pdf))
m = kde.sample(10000)
plt.hist(m, bins=20, density=1)
plt.show()
```
## KDE can learn handwitten digits distribution and generate new digits
http://scikit-learn.org/stable/auto_examples/neighbors/plot_digits_kde_sampling.html
## Correlation
Correlation is used to test relationships between quantitative variables
Some examples of data that have a high correlation:
1- Your caloric intake and your weight
2- The amount of time your study and your GPA
Question what is negative correlation?
Correlations are useful because we can find out what relationship variables have, we can make predictions about future behavior.
## Activity: Obtain the correlation among all features of iris dataset
1- Review the iris dataset. What are the features?
2- Eliminate two columns `['Id', 'Species']`
3- Compute the correlation among all features.
Hint: Use `df.corr()`
4- Plot the correlation by heatmap and corr plot in Seaborn -> `sns.heatmap`, `sns.corrplot`
5- Write a function that computes the correlation (Pearson formula)
Hint: https://en.wikipedia.org/wiki/Pearson_correlation_coefficient
6- Compare your answer with `scipy.stats.pearsonr` for any given two features
```
import pandas as pd
import numpy as np
import scipy.stats
import seaborn as sns
import scipy.stats
df = pd.read_csv('Iris.csv')
df = df.drop(columns=['Id', 'Species'])
sns.heatmap(df.corr(), annot=True)
def pearson_corr(x, y):
x_mean = np.mean(x)
y_mean = np.mean(y)
num = [(i - x_mean)*(j - y_mean) for i,j in zip(x,y)]
den_1 = [(i - x_mean)**2 for i in x]
den_2 = [(j - y_mean)**2 for j in y]
correlation_x_y = np.sum(num)/np.sqrt(np.sum(den_1))/np.sqrt(np.sum(den_2))
return correlation_x_y
print(pearson_corr(df['SepalLengthCm'], df['PetalLengthCm']))
print(scipy.stats.pearsonr(df['SepalLengthCm'], df['PetalLengthCm']))
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
import torch
import torchvision
from torch.utils.data import Dataset, DataLoader
from torchvision import transforms, utils
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from matplotlib import pyplot as plt
%matplotlib inline
class MosaicDataset1(Dataset):
"""MosaicDataset dataset."""
def __init__(self, mosaic_list, mosaic_label,fore_idx):
"""
Args:
csv_file (string): Path to the csv file with annotations.
root_dir (string): Directory with all the images.
transform (callable, optional): Optional transform to be applied
on a sample.
"""
self.mosaic = mosaic_list
self.label = mosaic_label
self.fore_idx = fore_idx
def __len__(self):
return len(self.label)
def __getitem__(self, idx):
return self.mosaic[idx] , self.label[idx] , self.fore_idx[idx]
data = np.load("type4_data.npy",allow_pickle=True)
mosaic_list_of_images = data[0]["mosaic_list"]
mosaic_label = data[0]["mosaic_label"]
fore_idx = data[0]["fore_idx"]
batch = 250
msd = MosaicDataset1(mosaic_list_of_images, mosaic_label, fore_idx)
train_loader = DataLoader( msd,batch_size= batch ,shuffle=True)
class Focus_deep(nn.Module):
'''
deep focus network averaged at zeroth layer
input : elemental data
'''
def __init__(self,inputs,output,K,d):
super(Focus_deep,self).__init__()
self.inputs = inputs
self.output = output
self.K = K
self.d = d
self.linear1 = nn.Linear(self.inputs,50) #,self.output)
self.linear2 = nn.Linear(50,50)
self.linear3 = nn.Linear(50,self.output)
def forward(self,z):
batch = z.shape[0]
x = torch.zeros([batch,self.K],dtype=torch.float64)
y = torch.zeros([batch,50], dtype=torch.float64) # number of features of output
features = torch.zeros([batch,self.K,50],dtype=torch.float64)
x,y = x.to("cuda"),y.to("cuda")
features = features.to("cuda")
for i in range(self.K):
alp,ftrs = self.helper(z[:,i] ) # self.d*i:self.d*i+self.d
x[:,i] = alp[:,0]
features[:,i] = ftrs
log_x = F.log_softmax(x,dim=1) #log alpha
x = F.softmax(x,dim=1) # alphas
for i in range(self.K):
x1 = x[:,i]
y = y+torch.mul(x1[:,None],features[:,i]) # self.d*i:self.d*i+self.d
return y , x,log_x
def helper(self,x):
x = self.linear1(x)
x = F.relu(x)
x = self.linear2(x)
x1 = F.tanh(x)
x = F.relu(x)
x = self.linear3(x)
#print(x1.shape)
return x,x1
class Classification_deep(nn.Module):
'''
input : elemental data
deep classification module data averaged at zeroth layer
'''
def __init__(self,inputs,output):
super(Classification_deep,self).__init__()
self.inputs = inputs
self.output = output
self.linear1 = nn.Linear(self.inputs,50)
#self.linear2 = nn.Linear(50,50)
self.linear2 = nn.Linear(50,self.output)
def forward(self,x):
x = F.relu(self.linear1(x))
#x = F.relu(self.linear2(x))
x = self.linear2(x)
return x
criterion = nn.CrossEntropyLoss()
def my_cross_entropy(x, y,alpha,log_alpha,k):
loss = criterion(x,y)
b = -1.0* alpha * log_alpha
b = torch.mean(torch.sum(b,dim=1))
closs = loss
entropy = b
loss = (1-k)*loss + ((k)*b)
return loss,closs,entropy
def calculate_attn_loss(dataloader,what,where,k):
what.eval()
where.eval()
r_loss = 0
cc_loss = 0
cc_entropy = 0
alphas = []
lbls = []
pred = []
fidices = []
with torch.no_grad():
for i, data in enumerate(dataloader, 0):
inputs, labels,fidx = data
lbls.append(labels)
fidices.append(fidx)
inputs = inputs.double()
inputs, labels = inputs.to("cuda"),labels.to("cuda")
avg,alpha,log_alpha = where(inputs)
outputs = what(avg)
_, predicted = torch.max(outputs.data, 1)
pred.append(predicted.cpu().numpy())
alphas.append(alpha.cpu().numpy())
loss,closs,entropy = my_cross_entropy(outputs,labels,alpha,log_alpha,k)
r_loss += loss.item()
cc_loss += closs.item()
cc_entropy += entropy.item()
alphas = np.concatenate(alphas,axis=0)
pred = np.concatenate(pred,axis=0)
lbls = np.concatenate(lbls,axis=0)
fidices = np.concatenate(fidices,axis=0)
#print(alphas.shape,pred.shape,lbls.shape,fidices.shape)
analysis = analyse_data(alphas,lbls,pred,fidices)
return r_loss/i,cc_loss/i,cc_entropy/i,analysis
def analyse_data(alphas,lbls,predicted,f_idx):
'''
analysis data is created here
'''
batch = len(predicted)
amth,alth,ftpt,ffpt,ftpf,ffpf = 0,0,0,0,0,0
for j in range (batch):
focus = np.argmax(alphas[j])
if(alphas[j][focus] >= 0.5):
amth +=1
else:
alth +=1
if(focus == f_idx[j] and predicted[j] == lbls[j]):
ftpt += 1
elif(focus != f_idx[j] and predicted[j] == lbls[j]):
ffpt +=1
elif(focus == f_idx[j] and predicted[j] != lbls[j]):
ftpf +=1
elif(focus != f_idx[j] and predicted[j] != lbls[j]):
ffpf +=1
#print(sum(predicted==lbls),ftpt+ffpt)
return [ftpt,ffpt,ftpf,ffpf,amth,alth]
number_runs = 20
full_analysis = []
FTPT_analysis = pd.DataFrame(columns = ["FTPT","FFPT", "FTPF","FFPF"])
k = 0.005
r_loss = []
r_closs = []
r_centropy = []
for n in range(number_runs):
print("--"*40)
# instantiate focus and classification Model
torch.manual_seed(n)
where = Focus_deep(2,1,9,2).double()
torch.manual_seed(n)
what = Classification_deep(50,3).double()
where = where.to("cuda")
what = what.to("cuda")
# instantiate optimizer
optimizer_where = optim.Adam(where.parameters(),lr =0.001)#,momentum=0.9)
optimizer_what = optim.Adam(what.parameters(), lr=0.001)#,momentum=0.9)
#criterion = nn.CrossEntropyLoss()
acti = []
analysis_data = []
loss_curi = []
cc_loss_curi = []
cc_entropy_curi = []
epochs = 3000
# calculate zeroth epoch loss and FTPT values
running_loss,_,_,anlys_data = calculate_attn_loss(train_loader,what,where,k)
loss_curi.append(running_loss)
analysis_data.append(anlys_data)
print('epoch: [%d ] loss: %.3f' %(0,running_loss))
# training starts
for epoch in range(epochs): # loop over the dataset multiple times
ep_lossi = []
running_loss = 0.0
what.train()
where.train()
for i, data in enumerate(train_loader, 0):
# get the inputs
inputs, labels,_ = data
inputs = inputs.double()
inputs, labels = inputs.to("cuda"),labels.to("cuda")
# zero the parameter gradients
optimizer_where.zero_grad()
optimizer_what.zero_grad()
# forward + backward + optimize
avg, alpha,log_alpha = where(inputs)
outputs = what(avg)
loss,_,_ = my_cross_entropy( outputs,labels,alpha,log_alpha,k)
# print statistics
running_loss += loss.item()
loss.backward()
optimizer_where.step()
optimizer_what.step()
running_loss,ccloss,ccentropy,anls_data = calculate_attn_loss(train_loader,what,where,k)
analysis_data.append(anls_data)
print('epoch: [%d] loss: %.3f celoss: %.3f entropy: %.3f' %(epoch + 1,running_loss,ccloss,ccentropy))
loss_curi.append(running_loss) #loss per epoch
cc_loss_curi.append(ccloss)
cc_entropy_curi.append(ccentropy)
if running_loss<=0.01:
break
print('Finished Training run ' +str(n))
analysis_data = np.array(analysis_data)
FTPT_analysis.loc[n] = analysis_data[-1,:4]/30
full_analysis.append((epoch, analysis_data))
r_loss.append(np.array(loss_curi))
r_closs.append(np.array(cc_loss_curi))
r_centropy.append(np.array(cc_entropy_curi))
correct = 0
total = 0
with torch.no_grad():
for data in train_loader:
images, labels,_ = data
images = images.double()
images, labels = images.to("cuda"), labels.to("cuda")
avg, alpha,_ = where(images)
outputs = what(avg)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 3000 train images: %d %%' % ( 100 * correct / total))
a,b= full_analysis[0]
print(a)
cnt=1
for epoch, analysis_data in full_analysis:
analysis_data = np.array(analysis_data)
# print("="*20+"run ",cnt,"="*20)
plt.figure(figsize=(6,6))
plt.plot(np.arange(0,epoch+2,1),analysis_data[:,0],label="ftpt")
plt.plot(np.arange(0,epoch+2,1),analysis_data[:,1],label="ffpt")
plt.plot(np.arange(0,epoch+2,1),analysis_data[:,2],label="ftpf")
plt.plot(np.arange(0,epoch+2,1),analysis_data[:,3],label="ffpf")
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.title("Training trends for run "+str(cnt))
#plt.savefig("/content/drive/MyDrive/Research/alpha_analysis/100_300/k"+str(k)+"/"+"run"+str(cnt)+name+".png",bbox_inches="tight")
#plt.savefig("/content/drive/MyDrive/Research/alpha_analysis/100_300/k"+str(k)+"/"+"run"+str(cnt)+name+".pdf",bbox_inches="tight")
cnt+=1
# plt.figure(figsize=(6,6))
# plt.plot(np.arange(0,epoch+2,1),analysis_data[:,0],label="ftpt")
# plt.plot(np.arange(0,epoch+2,1),analysis_data[:,1],label="ffpt")
# plt.plot(np.arange(0,epoch+2,1),analysis_data[:,2],label="ftpf")
# plt.plot(np.arange(0,epoch+2,1),analysis_data[:,3],label="ffpf")
# plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.plot(loss_curi)
np.mean(np.array(FTPT_analysis),axis=0)
FTPT_analysis.to_csv("type4_first_k_value_01_lr_001.csv",index=False)
```
```
FTPT_analysis
```
# Entropy
```
entropy_1 = r_centropy[11] # FTPT 100 ,FFPT 0 k value =0.01
loss_1 = r_loss[11]
ce_loss_1 = r_closs[11]
entropy_2 = r_centropy[16] # kvalue = 0 FTPT 99.96, FFPT 0.03
ce_loss_2 = r_closs[16]
# plt.plot(r_closs[1])
plt.plot(entropy_1,label = "entropy k_value=0.01")
plt.plot(loss_1,label = "overall k_value=0.01")
plt.plot(ce_loss_1,label = "ce kvalue = 0.01")
plt.plot(entropy_2,label = "entropy k_value = 0")
plt.plot(ce_loss_2,label = "ce k_value=0")
plt.legend(bbox_to_anchor=(1.05, 1), loc='upper left')
plt.savefig("second_layer.png")
```
|
github_jupyter
|
<a href="http://landlab.github.io"><img style="float: left" src="../../landlab_header.png"></a>
# Using plotting tools associated with the Landlab NetworkSedimentTransporter component
<hr>
<small>For more Landlab tutorials, click here: <a href="https://landlab.readthedocs.io/en/latest/user_guide/tutorials.html">https://landlab.readthedocs.io/en/latest/user_guide/tutorials.html</a></small>
<hr>
This tutorial illustrates how to plot the results of the NetworkSedimentTransporter Landlab component using the `plot_network_and_parcels` tool.
In this example we will:
- create a simple instance of the NetworkSedimentTransporter using a *synthetic river network
- create a simple instance of the NetworkSedimentTransporter using an *input shapefile for the river network
- show options for setting the color and line widths of network links
- show options for setting the color of parcels (marked as dots on the network)
- show options for setting the size of parcels
- show options for plotting a subset of the parcels
- demonstrate changing the timestep plotted
- show an example combining many plotting controls
First, import the necessary libraries:
```
import warnings
warnings.filterwarnings('ignore')
import os
import pathlib
import matplotlib.pyplot as plt
import matplotlib.colors as colors
import numpy as np
from landlab import ExampleData
from landlab.components import FlowDirectorSteepest, NetworkSedimentTransporter
from landlab.data_record import DataRecord
from landlab.grid.network import NetworkModelGrid
from landlab.plot import plot_network_and_parcels
from landlab.io import read_shapefile
from matplotlib.colors import Normalize
```
## 1. Create and run the synthetic example of NST
First, we need to create an implementation of the Landlab NetworkModelGrid to plot. This example creates a synthetic grid, defining the location of each node and link.
```
y_of_node = (0, 100, 200, 200, 300, 400, 400, 125)
x_of_node = (0, 0, 100, -50, -100, 50, -150, -100)
nodes_at_link = ((1, 0), (2, 1), (1, 7), (3, 1), (3, 4), (4, 5), (4, 6))
grid1 = NetworkModelGrid((y_of_node, x_of_node), nodes_at_link)
grid1.at_node["bedrock__elevation"] = [0.0, 0.05, 0.2, 0.1, 0.25, 0.4, 0.8, 0.8]
grid1.at_node["topographic__elevation"] = [0.0, 0.05, 0.2, 0.1, 0.25, 0.4, 0.8, 0.8]
grid1.at_link["flow_depth"] = 2.5 * np.ones(grid1.number_of_links) # m
grid1.at_link["reach_length"] = 200*np.ones(grid1.number_of_links) # m
grid1.at_link["channel_width"] = 1*np.ones(grid1.number_of_links) # m
# element_id is the link on which the parcel begins.
element_id = np.repeat(np.arange(grid1.number_of_links),30)
element_id = np.expand_dims(element_id, axis=1)
volume = 0.1*np.ones(np.shape(element_id)) # (m3)
active_layer = np.ones(np.shape(element_id)) # 1= active, 0 = inactive
density = 2650 * np.ones(np.size(element_id)) # (kg/m3)
abrasion_rate = 0 * np.ones(np.size(element_id)) # (mass loss /m)
# Lognormal GSD
medianD = 0.05 # m
mu = np.log(medianD)
sigma = np.log(2) #assume that D84 = sigma*D50
np.random.seed(0)
D = np.random.lognormal(
mu,
sigma,
np.shape(element_id)
) # (m) the diameter of grains in each parcel
time_arrival_in_link = np.random.rand(np.size(element_id), 1)
location_in_link = np.random.rand(np.size(element_id), 1)
variables = {
"abrasion_rate": (["item_id"], abrasion_rate),
"density": (["item_id"], density),
"time_arrival_in_link": (["item_id", "time"], time_arrival_in_link),
"active_layer": (["item_id", "time"], active_layer),
"location_in_link": (["item_id", "time"], location_in_link),
"D": (["item_id", "time"], D),
"volume": (["item_id", "time"], volume)
}
items = {"grid_element": "link", "element_id": element_id}
parcels1 = DataRecord(
grid1,
items=items,
time=[0.0],
data_vars=variables,
dummy_elements={"link": [NetworkSedimentTransporter.OUT_OF_NETWORK]},
)
fd1 = FlowDirectorSteepest(grid1, "topographic__elevation")
fd1.run_one_step()
nst1 = NetworkSedimentTransporter(
grid1,
parcels1,
fd1,
bed_porosity=0.3,
g=9.81,
fluid_density=1000,
transport_method="WilcockCrowe",
)
timesteps = 10 # total number of timesteps
dt = 60 * 60 * 24 *1 # length of timestep (seconds)
for t in range(0, (timesteps * dt), dt):
nst1.run_one_step(dt)
```
## 2. Create and run an example of NST using a shapefile to define the network
First, we need to create an implementation of the Landlab NetworkModelGrid to plot. This example creates a grid based on a polyline shapefile.
```
datadir = ExampleData("io/shapefile", case="methow").base
shp_file = datadir / "MethowSubBasin.shp"
points_shapefile = datadir / "MethowSubBasin_Nodes_4.shp"
grid2 = read_shapefile(
shp_file,
points_shapefile=points_shapefile,
node_fields=["usarea_km2", "Elev_m"],
link_fields=["usarea_km2", "Length_m"],
link_field_conversion={"usarea_km2": "drainage_area", "Slope":"channel_slope", "Length_m":"reach_length"},
node_field_conversion={
"usarea_km2": "drainage_area",
"Elev_m": "topographic__elevation",
},
threshold=0.01,
)
grid2.at_node["bedrock__elevation"] = grid2.at_node["topographic__elevation"].copy()
grid2.at_link["channel_width"] = 1 * np.ones(grid2.number_of_links)
grid2.at_link["flow_depth"] = 0.9 * np.ones(grid2.number_of_links)
# element_id is the link on which the parcel begins.
element_id = np.repeat(np.arange(grid2.number_of_links), 50)
element_id = np.expand_dims(element_id, axis=1)
volume = 1*np.ones(np.shape(element_id)) # (m3)
active_layer = np.ones(np.shape(element_id)) # 1= active, 0 = inactive
density = 2650 * np.ones(np.size(element_id)) # (kg/m3)
abrasion_rate = 0 * np.ones(np.size(element_id)) # (mass loss /m)
# Lognormal GSD
medianD = 0.15 # m
mu = np.log(medianD)
sigma = np.log(2) #assume that D84 = sigma*D50
np.random.seed(0)
D = np.random.lognormal(
mu,
sigma,
np.shape(element_id)
) # (m) the diameter of grains in each parcel
time_arrival_in_link = np.random.rand(np.size(element_id), 1)
location_in_link = np.random.rand(np.size(element_id), 1)
variables = {
"abrasion_rate": (["item_id"], abrasion_rate),
"density": (["item_id"], density),
"time_arrival_in_link": (["item_id", "time"], time_arrival_in_link),
"active_layer": (["item_id", "time"], active_layer),
"location_in_link": (["item_id", "time"], location_in_link),
"D": (["item_id", "time"], D),
"volume": (["item_id", "time"], volume)
}
items = {"grid_element": "link", "element_id": element_id}
parcels2 = DataRecord(
grid2,
items=items,
time=[0.0],
data_vars=variables,
dummy_elements={"link": [NetworkSedimentTransporter.OUT_OF_NETWORK]},
)
fd2 = FlowDirectorSteepest(grid2, "topographic__elevation")
fd2.run_one_step()
nst2 = NetworkSedimentTransporter(
grid2,
parcels2,
fd2,
bed_porosity=0.3,
g=9.81,
fluid_density=1000,
transport_method="WilcockCrowe",
)
for t in range(0, (timesteps * dt), dt):
nst2.run_one_step(dt)
```
## 3. Options for link color and link line widths
The dictionary below (`link_color_options`) outlines 4 examples of link color and line width choices:
1. The default output of `plot_network_and_parcels`
2. Some simple modifications: the whole network is red, with a line width of 7, and no parcels.
3. Coloring links by an existing grid link attribute, in this case the total volume of sediment on the link (`grid.at_link.["sediment_total_volume"]`, which is created by the `NetworkSedimentTransporter`)
4. Similar to #3 above, but taking advantange of additional flexiblity in plotting
```
network_norm = Normalize(-1, 6) # see matplotlib.colors.Normalize
link_color_options = [
{# empty dictionary = defaults
},
{
"network_color":'r', # specify some simple modifications.
"network_linewidth":7,
"parcel_alpha":0 # make parcels transparent (not visible)
},
{
"link_attribute": "sediment_total_volume", # color links by an existing grid link attribute
"parcel_alpha":0
},
{
"link_attribute": "sediment_total_volume",
"network_norm": network_norm, # and normalize color scheme
"link_attribute_title": "Total Sediment Volume", # title on link color legend
"parcel_alpha":0,
"network_linewidth":3
}
]
```
Below, we implement these 4 plotting options, first for the synthetic network, and then for the shapefile-delineated network:
```
for grid, parcels in zip([grid1, grid2], [parcels1, parcels2]):
for l_opts in link_color_options:
fig = plot_network_and_parcels(
grid, parcels,
parcel_time_index=0, **l_opts)
plt.show()
```
In addition to plotting link coloring using an existing link attribute, we can pass any array of size link. In this example, we color links using an array of random values.
```
random_link = np.random.randn(grid2.size("link"))
l_opts = {
"link_attribute": random_link, # use an array of size link
"network_cmap": "jet", # change colormap
"network_norm": network_norm, # and normalize
"link_attribute_title": "A random number",
"parcel_alpha":0,
"network_linewidth":3
}
fig = plot_network_and_parcels(
grid2, parcels2,
parcel_time_index=0, **l_opts)
plt.show()
```
## 4. Options for parcel color
The dictionary below (`parcel_color_options`) outlines 4 examples of link color and line width choices:
1. The default output of `plot_network_and_parcels`
2. Some simple modifications: all parcels are red, with a parcel size of 10
3. Color parcels by an existing parcel attribute, in this case the sediment diameter of the parcel (`parcels1.dataset['D']`)
4. Color parcels by an existing parcel attribute, but change the colormap.
```
parcel_color_norm = Normalize(0, 1) # Linear normalization
parcel_color_norm2=colors.LogNorm(vmin=0.01, vmax=1)
parcel_color_options = [
{# empty dictionary = defaults
},
{
"parcel_color":'r', # specify some simple modifications.
"parcel_size":10
},
{
"parcel_color_attribute": "D", # existing parcel attribute.
"parcel_color_norm": parcel_color_norm,
"parcel_color_attribute_title":"Diameter [m]",
"parcel_alpha":1.0,
},
{
"parcel_color_attribute": "abrasion_rate", # silly example, does not vary in our example
"parcel_color_cmap": "bone",
},
]
for grid, parcels in zip([grid1, grid2], [parcels1, parcels2]):
for pc_opts in parcel_color_options:
fig = plot_network_and_parcels(
grid, parcels,
parcel_time_index=0, **pc_opts)
plt.show()
```
## 5. Options for parcel size
The dictionary below (`parcel_size_options`) outlines 4 examples of link color and line width choices:
1. The default output of `plot_network_and_parcels`
2. Set a uniform parcel size and color
3. Size parcels by an existing parcel attribute, in this case the sediment diameter (`parcels1.dataset['D']`), and making the parcel markers entirely opaque.
4. Normalize parcel size on a logarithmic scale, and change the default maximum and minimum parcel sizes.
```
parcel_size_norm = Normalize(0, 1)
parcel_size_norm2=colors.LogNorm(vmin=0.01, vmax=1)
parcel_size_options = [
{# empty dictionary = defaults
},
{
"parcel_color":'b', # specify some simple modifications.
"parcel_size":10
},
{
"parcel_size_attribute": "D", # use a parcel attribute.
"parcel_size_norm": parcel_color_norm,
"parcel_size_attribute_title":"Diameter [m]",
"parcel_alpha":1.0, # default parcel_alpha = 0.5
},
{
"parcel_size_attribute": "D",
"parcel_size_norm": parcel_size_norm2,
"parcel_size_min": 10, # default = 5
"parcel_size_max": 100, # default = 40
"parcel_alpha": 0.1
},
]
for grid, parcels in zip([grid1, grid2], [parcels1, parcels2]):
for ps_opts in parcel_size_options:
fig = plot_network_and_parcels(
grid, parcels,
parcel_time_index=0, **ps_opts)
plt.show()
```
## 6. Plotting a subset of the parcels
In some cases, we might want to plot only a subset of the parcels on the network. Below, we plot every 50th parcel in the `DataRecord`.
```
parcel_filter = np.zeros((parcels2.dataset.dims["item_id"]), dtype=bool)
parcel_filter[::50] = True
pc_opts= {
"parcel_color_attribute": "D", # a more complex normalization and a parcel filter.
"parcel_color_norm": parcel_color_norm2,
"parcel_color_attribute_title":"Diameter [m]",
"parcel_alpha": 1.0,
"parcel_size": 40,
"parcel_filter": parcel_filter
}
fig = plot_network_and_parcels(
grid2, parcels2,
parcel_time_index=0, **pc_opts
)
plt.show()
```
## 7. Select the parcel timestep to be plotted
As a default, `plot_network_and_parcels` plots parcel positions for the last timestep of the model run. However, `NetworkSedimentTransporter` tracks the motion of parcels for all timesteps. We can plot the location of parcels on the link at any timestep using `parcel_time_index`.
```
parcel_time_options = [0,4,7]
for grid, parcels in zip([grid1, grid2], [parcels1, parcels2]):
for pt_opts in parcel_time_options:
fig = plot_network_and_parcels(
grid, parcels,
parcel_size = 20,
parcel_alpha = 0.1,
parcel_time_index=pt_opts)
plt.show()
```
## 7. Combining network and parcel plotting options
Nothing will stop us from making all of the choices at once.
```
parcel_color_norm=colors.LogNorm(vmin=0.01, vmax=1)
parcel_filter = np.zeros((parcels2.dataset.dims["item_id"]), dtype=bool)
parcel_filter[::30] = True
fig = plot_network_and_parcels(grid2,
parcels2,
parcel_time_index=0,
parcel_filter=parcel_filter,
link_attribute="sediment_total_volume",
network_norm=network_norm,
network_linewidth=4,
network_cmap='bone_r',
parcel_alpha=1.0,
parcel_color_attribute="D",
parcel_color_norm=parcel_color_norm2,
parcel_size_attribute="D",
parcel_size_min=5,
parcel_size_max=150,
parcel_size_norm=parcel_size_norm,
parcel_size_attribute_title="D")
```
|
github_jupyter
|
- Scipy의 stats 서브 패키지에 있는 binom 클래스는 이항 분포 클래스이다. n 인수와 p 인수를 사용하여 모수를 설정한다
```
N = 10
theta = 0.6
rv = sp.stats.binom(N, theta)
rv
```
- pmf 메서드를 사용하면, 확률 질량 함수 (pmf: probability mass function)를 계산할 수 있다.
```
%matplotlib inline
xx = np.arange(N + 1)
plt.bar(xx, rv.pmf(xx), align='center')
plt.ylabel('p(x)')
plt.title('binomial pmf')
plt.show()
```
- 시뮬레이션을 하려면 rvs 메서드를 사용한다.
```
np.random.seed(0)
x = rv.rvs(100)
x
sns.countplot(x)
plt.title("Binomial Distribution's Simulation")
plt.xlabel('Sample')
plt.show()
```
- 이론적인 확률 분포와 샘플의 확률 분포를 동시에 나타내려면 다음과 같은 코드를 사용한다.
```
y = np.bincount(x, minlength=N+1)/float(len(x))
df = pd.DataFrame({'Theory': rv.pmf(xx), 'simulation': y}).stack()
df = df.reset_index()
df.columns = ['values', 'type', 'ratio']
df.pivot('values', 'type', 'ratio')
df
sns.barplot(x='values', y='ratio', hue='type', data=df)
plt.show()
```
#### 연습 문제 1
- 이항 확률 분포의 모수가 다음과 같을 경우에 각각 샘플을 생성한 후, 기댓값과 분산을 구하고 앞의 예제와 같이 확률 밀도 함수와 비교한 카운트 플롯을 그린다.
- 샘풀의 갯수가 10개인 경우와 1000개인 경우에 대해 각각 위의 계산을 한다.
- 1. Theta = 0.5, N = 5
- 2. Theta = 0.9, N = 20
```
# 연습문제 1 - 1
N = 5
theta = 0.5
rv = sp.stats.binom(N, theta)
xx10 = np.arange(N + 1)
plt.bar(xx, rv.pmf(xx10), align='center')
plt.ylabel('P(x)')
plt.title('Binomail Distribution pmdf')
plt.show()
# sample 갯수 10개 일 때
np.random.seed(0)
x10 = rv.rvs(10)
sns.countplot(x10)
plt.title('binomail distribution Simulation 10')
plt.xlabel('values')
plt.show()
# sample 갯수가 1000개 일 때
x1000 = rv.rvs(1000)
sns.countplot(x1000)
plt.title('binomail distribution Simulation 10')
plt.xlabel('values')
plt.show()
y10 = np.bincount(x10, minlength = N + 1)/float(len(x10))
df = pd.DataFrame({'Theory': rv.pmf(xx10), 'Simulation': y10}).stack()
df = df.reset_index()
df.columns = ['values', 'type', 'ratio']
df.pivot('values', 'type', 'ratio')
sns.barplot(x='values', y='ratio', hue='type', data=df)
plt.show()
df
```
#### 샘플 갯수가 1000개일 경우에 theta = 0.9, N = 20
```
N = 20
theta = 0.9
rv = sp.stats.binom(N, theta)
xx = np.arange(N + 1)
plt.bar(xx, rv.pmf(xx), align = 'center')
plt.ylabel('P(x)')
plt.title('binomial pmf when N=20')
plt.show()
x1000 = rv.rvs(1000) # sample 1000개 생성
sns.countplot(x1000)
plt.title("Binomial Distribution's Simulation")
plt.xlabel('values')
plt.show()
y1000 = np.bincount(x1000, minlength = N + 1)/float(len(x1000))
df = pd.DataFrame({'Theory':rv.pmf(xx), 'Simulation': y1000}).stack()
df = df.reset_index()
df.columns = ['values', 'type', 'ratio']
df.pivot('values', 'type', 'ratio')
df
sns.barplot(x='values', y='ratio', hue='type', data=df)
plt.show()
```
|
github_jupyter
|
<table style="float:left; border:none">
<tr style="border:none; background-color: #ffffff">
<td style="border:none">
<a href="http://bokeh.pydata.org/">
<img
src="assets/bokeh-transparent.png"
style="width:50px"
>
</a>
</td>
<td style="border:none">
<h1>Bokeh Tutorial</h1>
</td>
</tr>
</table>
<div style="float:right;"><h2>07. Exporting and Embedding</h2></div>
So far we have seen how to generate interactive Bokeh output directly inline in Jupyter notbeooks. It also possible to embed interactive Bokeh plots and layouts in other contexts, such as standalone HTML files, or Jinja templates. Additionally, Bokeh can export plots to static (non-interactive) PNG and SVG formats.
We will look at all of these possibilities in this chapter. First we make the usual imports.
```
from bokeh.io import output_notebook, show
output_notebook()
```
And also load some data that will be used throughout this chapter
```
import pandas as pd
from bokeh.plotting import figure
from bokeh.sampledata.stocks import AAPL
df = pd.DataFrame(AAPL)
df['date'] = pd.to_datetime(df['date'])
```
# Embedding Interactive Content
To start we will look differnet ways of embedding live interactive Bokeh output in various situations.
## Displaying in the Notebook
The first way to embed Bokeh output is in the Jupyter Notebooks, as we have already, seen. As a reminder, the cell below will generate a plot inline as output, because we executed `output_notebook` above.
```
p = figure(plot_width=800, plot_height=250, x_axis_type="datetime")
p.line(df['date'], df['close'], color='navy', alpha=0.5)
show(p)
```
## Saving to an HTML File
It is also often useful to generate a standalone HTML script containing Bokeh content. This is accomplished by calling the `output_file(...)` function. It is especially common to do this from standard Python scripts, but here we see that it works in the notebook as well.
```
from bokeh.io import output_file, show
output_file("plot.html")
show(p) # save(p) will save without opening a new browser tab
```
In addition the inline plot above, you should also have seen a new browser tab open with the contents of the newly saved "plot.html" file. It is important to note that `output_file` initiates a *persistent mode of operation*. That is, all subsequent calls to show will generate output to the specified file. We can "reset" where output will go by calling `reset_output`:
```
from bokeh.io import reset_output
reset_output()
```
## Templating in HTML Documents
Another use case is to embed Bokeh content in a Jinja HTML template. We will look at a simple explicit case first, and then see how this technique might be used in a web app framework such as Flask.
The simplest way to embed standalone (i.e. not Bokeh server) content is to use the `components` function. This function takes a Bokeh object, and returns a `<script>` tag and `<div>` tag that can be put in any HTML tempate. The script will eecute and load the Bokeh content into the associated div.
The cells below show a complete example, including loading BokehJS JS and CSS resources in the temlpate.
```
import jinja2
from bokeh.embed import components
# IMPORTANT NOTE!! The version of BokehJS loaded in the template should match
# the version of Bokeh installed locally.
template = jinja2.Template("""
<!DOCTYPE html>
<html lang="en-US">
<link
href="http://cdn.pydata.org/bokeh/dev/bokeh-0.13.0.min.css"
rel="stylesheet" type="text/css"
>
<script
src="http://cdn.pydata.org/bokeh/dev/bokeh-0.13.0.min.js"
></script>
<body>
<h1>Hello Bokeh!</h1>
<p> Below is a simple plot of stock closing prices </p>
{{ script }}
{{ div }}
</body>
</html>
""")
p = figure(plot_width=800, plot_height=250, x_axis_type="datetime")
p.line(df['date'], df['close'], color='navy', alpha=0.5)
script, div = components(p)
from IPython.display import HTML
HTML(template.render(script=script, div=div))
```
Note that it is possible to pass multiple objects to a single call to `components`, in order to template multiple Bokeh objects at once. See the [User's Guide for components](https://bokeh.pydata.org/en/latest/docs/user_guide/embed.html#components) for more information.
Once we have the script and div from `components`, it is straighforward to serve a rendered page containing Bokeh content in a web application, e.g. a Flask app as shown below.
```
from flask import Flask
app = Flask(__name__)
@app.route('/')
def hello_bokeh():
return template.render(script=script, div=div)
# Uncomment to run the Flask Server. Use Kernel -> Interrupt from Notebook menubar to stop
#app.run(port=5050)
# EXERCISE: Create your own template (or modify the one above)
```
# Exporting Static Images
Sometimes it is desirable to produce static images of plots or other Bokeh output, without any interactive capabilities. Bokeh supports exports to PNG and SVG formats.
## PNG Export
Bokeh supports exporting a plot or layout to PNG image format with the `export_png` function. This function is alled with a Bokeh object to export, and a filename to write the PNG output to. Often the Bokeh object passed to `export_png` is a single plot, but it need not be. If a layout is exported, the entire lahyout is saved to one PNG image.
***Important Note:*** *the PNG export capability requires installing some additional optional dependencies. The simplest way to obtain them is via conda:*
conda install selenium phantomjs pillow
```
from bokeh.io import export_png
p = figure(plot_width=800, plot_height=250, x_axis_type="datetime")
p.line(df['date'], df['close'], color='navy', alpha=0.5)
export_png(p, filename="plot.png")
from IPython.display import Image
Image('plot.png')
# EXERCISE: Save a layout of plots (e.g. row or column) as SVG and see what happens
```
## SVG Export
Bokeh can also generate SVG output in the browser, instead of rendering to HTML canvas. This is accomplished by setting `output_backend='svg'` on a figure. This can be be used to generate SVGs in `output_file` HTML files, or in content emebdded with `components`. It can also be used with the `export_svgs` function to save `.svg` files. Note that an SVG is created for *each canvas*. It is not possible to capture entire layouts or widgets in SVG output.
***Important Note:*** *There a currently some known issue with SVG output, it may not work for all use-cases*
```
from bokeh.io import export_svgs
p = figure(plot_width=800, plot_height=250, x_axis_type="datetime", output_backend='svg')
p.line(df['date'], df['close'], color='navy', alpha=0.5)
export_svgs(p, filename="plot.svg")
from IPython.display import SVG
SVG('plot.svg')
# EXERCISE: Save a layout of plots (e.g. row or column) as SVG and see what happens
```
|
github_jupyter
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.