text
stringlengths 2.5k
6.39M
| kind
stringclasses 3
values |
|---|---|
# SLU09 - Classification With Logistic Regression: Exercise notebook
```
import pandas as pd
import numpy as np
import hashlib
```
In this notebook you will practice the following:
- What classification is for
- Logistic regression
- Cost function
- Binary classification
You thought that you would get away without implementing your own little Logistic Regression? Hah!
# Exercise 1. Implement the Exponential part of Sigmoid Function
In the first exercise, you will implement **only the piece** of the sigmoid function where you have to use an exponential.
Here's a quick reminder of the formula:
$$\hat{p} = \frac{1}{1 + e^{-z}}$$
In this exercise we only want you to complete the exponential part given the values of b0, b1, x1, b2 and x2:
$$e^{-z}$$
Recall that z has the following formula:
$$z = \beta_0 + \beta_1 x_1 + \beta_2 x_2$$
**Hint: Divide your z into pieces by Betas, I've left the placeholders in there!**
```
def exponential_z_function(beta0, beta1, beta2, x1, x2):
"""
Implementation of the exponential part of
the sigmoid function manually. In this exercise you
have to compute the e raised to the power -z. Z is calculated
according to the following formula: b0+b1x1+b2x2.
You can use the inputs given to generate the z.
Args:
beta0 (np.float64): value of the intercept
beta1 (np.float64): value of first coefficient
beta2 (np.float64): value of second coefficient
x1 (np.float64): value of first variable
x2 (np.float64): value of second variable
Returns:
exp_z (np.float64): the exponential part of
the sigmoid function
"""
# hint: obtain the exponential part
# using np.exp()
# Complete the following
#beta0 = ...
#b1_x1 = ...
#b2_x2 = ...
#exp_z = ...
# YOUR CODE HERE
raise NotImplementedError()
return exp_z
value_arr = [1, 2, 1, 1, 0.5]
exponential = exponential_z_function(
value_arr[0], value_arr[1], value_arr[2], value_arr[3], value_arr[4])
np.testing.assert_almost_equal(np.round(exponential,3), 0.030)
```
Expected output:
Exponential part: 0.03
# Exercise 2: Make a Prediction
The next step is to implement a function that receives an observation and returns the predicted probability with the sigmoid function.
For instance, we can make a prediction given a model with data and coefficients by using the sigmoid:
$$\hat{p} = \frac{1}{1 + e^{-z}}$$
Where Z is the linear equation - you can't use the same function that you used above for the Z part as the input are now two arrays, one with the train data (x1, x2, ..., xn) and another with the coefficients (b0, b1, .., bn).
**Complete here:**
```
def predict_proba(data, coefs):
"""
Implementation of a function that returns
predicted probabilities for an observation.
In the train array you will have
the data values (corresponding to the x1, x2, .. , xn).
In the coefficients array you will have
the coefficients values (corresponding to the b0, b1, .., bn).
In this exercise you should be able to return a float
with the calculated probabilities given an array of size (1, n).
The resulting value should be a float (the predicted probability)
with a value between 0 and 1.
Note: Be mindful that the input is completely different from
the function above - you receive two arrays in this functions while
in the function above you received 5 floats - each corresponding
to the x's and b's.
Args:
data (np.array): a numpy array of shape (n)
- n: number of variables
coefs (np.array): a numpy array of shape (n + 1, 1)
- coefs[0]: intercept
- coefs[1:]: remaining coefficients
Returns:
proba (float): the predicted probability for a data example.
"""
# hint: you have to implement your z in a vectorized
# way aka using vector multiplications - it's different from what you have done above
# hint: don't forget about adding an intercept to the train data!
# YOUR CODE HERE
raise NotImplementedError()
return proba
x = np.array([-1.2, -1.5])
coefficients = np.array([0 ,4, -1])
np.testing.assert_almost_equal(round(predict_proba(x, coefficients),3),0.036)
x_1 = np.array([-1.5, -1, 3, 0])
coefficients_1 = np.array([0 ,2.1, -1, 0.5, 0])
np.testing.assert_almost_equal(round(predict_proba(x_1, coefficients_1),3),0.343)
```
Expected output:
Predicted probabilities for example with 2 variables: 0.036
Predicted probabilities for example with 3 variables: 0.343
# Exercise 3: Compute the Maximum Log-Likelihood Cost Function
As you will implement stochastic gradient descent, you need to calculate the cost function (the Maximum Log-Likelihood) for each prediction, checking how much you will penalize each example according to the difference between the calculated probability and its true value:
$$H_{\hat{p}}(y) = - (y \log(\hat{p}) + (1-y) \log (1-\hat{p}))$$
In the next exercise, you will loop through some examples stored in an array and calculate the cost function for the full dataset. Recall that the formula to generalize the cost function across several examples is:
$$H_{\hat{p}}(y) = - \frac{1}{N}\sum_{i=1}^{N} \left [{ y_i \ \log(\hat{p}_i) + (1-y_i) \ \log (1-\hat{p}_i)} \right ]$$
You will basically simulate what stochastic gradient descent does without updating the coefficients - computing the log for each example, sum each log-loss and then averaging the result across the number of observations in the x dataset/array.
```
import math
def max_log_likelihood_cost_function(var_x, coefs, var_y):
"""
Implementation of a function that returns the Maximum-Log-Likelihood loss
Args:
var_x (np.array): array with x training data of size (m, n) shape
where m is the number of observations and n the number of columns
coefs (float64): an array with the coefficients to apply of size (1, n+1)
where n is the number of columns plus the intercept.
var_y (float64): an array with integers with the real outcome per
example.
Returns:
loss (np.float): a float with the resulting log loss for the
entire data.
"""
# A list of hints that you can follow:
# - you already computed a probability for an example so you might be able to reuse the function
# - Store number of examples that you have to loop through
#Steps to follow:
# 1. Initialize loss
# 2. Loop through every example
# Hint: if you don't use the function from above to predict probas
# don't forget to add the intercept to the X_array!
# 2.1 Calculate probability for each example
# 2.2 Compute log loss
# Hint: maybe separating the log loss will help you avoiding get confused inside all the parenthesis
# 2.3 Sum the computed loss for the example to the total log loss
# 3. Divide log loss by the number of examples (don't forget that the log loss
# has to return a positive number!)
# YOUR CODE HERE
raise NotImplementedError()
return total_loss
x = np.array([[-2, -2], [3.5, 0], [6, 4]])
coefficients = np.array([[0 ,2, -1]])
y = np.array([[1],[1],[0]])
np.testing.assert_almost_equal(round(max_log_likelihood_cost_function(x, coefficients, y),3),3.376)
coefficients_1 = np.array([[3 ,4, -0.6]])
x_1 = np.array([[-4, -4], [6, 0], [3, 2], [4, 0]])
y_1 = np.array([[4],[4],[2],[1.5]])
np.testing.assert_almost_equal(round(max_log_likelihood_cost_function(x_1, coefficients_1, y_1),3),-15.475)
```
Expected output:
Computed log loss for first training set: 3.376
Computed log loss for second training set: -15.475
# Exercise 4: Compute a first pass on Stochastic Gradient Descent
Now that we know how to calculate probabilities and the cost function, let's do an interesting exercise - computing the derivatives and updating our coefficients. Here you will do a full pass on a bunch of examples, computing the gradient descent for each time you see one of them.
In this exercise, you should compute a single iteration of the gradient descent!
You will basically use stochastic gradient descent but you will have to update the coefficients after
you see a new example - so each time your algorithm knows that he saw something way off (for example,
returning a low probability for an example with outcome = 1) he will have a way (the gradient) to
change the coefficients so that he is able to minimize the cost function.
## Quick reminders:
Remember our formulas for the gradient:
$$\beta_{0(t+1)} = \beta_{0(t)} - learning\_rate \frac{\partial H_{\hat{p}}(y)}{\partial \beta_{0(t)}}$$
$$\beta_{t+1} = \beta_t - learning\_rate \frac{\partial H_{\hat{p}}(y)}{\partial \beta_t}$$
which can be simplified to
$$\beta_{0(t+1)} = \beta_{0(t)} + learning\_rate \left [(y - \hat{p}) \ \hat{p} \ (1 - \hat{p})\right]$$
$$\beta_{t+1} = \beta_t + learning\_rate \left [(y - \hat{p}) \ \hat{p} \ (1 - \hat{p}) \ x \right]$$
You will have to initialize the coefficients in some way. If you have a training set $X$, you can initialize them to zero, this way:
```python
coefficients = np.zeros(X.shape[1]+1)
```
where the $+1$ is adding the intercept.
Note: We are doing a stochastic gradient descent so don't forget to go observation by observation and updating the coefficients every time!
**Complete here:**
```
def compute_coefs_sgd(x_train, y_train, learning_rate = 0.1, verbose = False):
"""
Implementation of a function that returns the a first iteration of
stochastic gradient descent.
Args:
x_train (np.array): a numpy array of shape (m, n)
m: number of training observations
n: number of variables
y_train (np.array): a numpy array of shape (m,) with
the real value of the target.
learning_rate (np.float64): a float
Returns:
coefficients (np.array): a numpy array of shape (n+1,)
"""
# A list of hints that might help you:
# 1. Calculate the number of observations
# 2. Initialize the coefficients array with zeros
# hint: use np.zeros()
# 3. Run the stochastic gradient descent and update the coefficients after each observation
# 3.1 Compute the predicted probability - you can use a function we have done previously
# 3.2 Update intercept
# 3.3 Update the rest of the coefficients by looping through each variable
# YOUR CODE HERE
raise NotImplementedError()
return coefficients
#Test 1
x_train = np.array([[1,2,4], [2,4,9], [2,1,4], [9,2,10]])
y_train = np.array([0,2.2,0,2.3])
learning_rate = 0.1
np.testing.assert_almost_equal(round(compute_coefs_sgd(x_train, y_train, learning_rate)[0],3),0.022)
np.testing.assert_almost_equal(round(compute_coefs_sgd(x_train, y_train, learning_rate)[1],3),0.081)
np.testing.assert_almost_equal(round(compute_coefs_sgd(x_train, y_train, learning_rate)[2],3),0.140)
np.testing.assert_almost_equal(round(compute_coefs_sgd(x_train, y_train, learning_rate)[3],3),0.320)
#Test 2
x_train_1 = np.array([[4,4,2,6], [1,5,7,2], [3,1,2,1], [8,2,9,5], [2,2,9,4]])
y_train_1 = np.array([0,1.3,0,1.3,1.2])
np.testing.assert_almost_equal(round(compute_coefs_sgd(x_train_1, y_train_1, learning_rate).max(),3) ,0.277)
np.testing.assert_almost_equal(round(compute_coefs_sgd(x_train_1, y_train_1, learning_rate).min(),3) ,0.015)
np.testing.assert_almost_equal(round(compute_coefs_sgd(x_train_1, y_train_1, learning_rate).mean(),3),0.102)
np.testing.assert_almost_equal(round(compute_coefs_sgd(x_train_1, y_train_1, learning_rate).var(),3) ,0.008)
```
# Exercise 5: Normalize Data
To get this concept in your head, let's do a quick and easy function to normalize the data using a MaxMin approach. It is crucial that your variables are adjusted between $[0;1]$ (normalized) or standardized so that you can correctly analyze some logistic regression coefficients for your possible future employer.
You only have to implement this formula
$$ x_{normalized} = \frac{x - x_{min}}{x_{max} - x_{min}}$$
Don't forget that the `axis` argument is critical when obtaining the maximum, minimum and mean values! As you want to obtain the maximum and minimum values of each individual feature, you have to specify `axis=0`. Thus, if you wanted to obtain the maximum values of each feature of data $X$, you would do the following:
```python
X_max = np.max(X, axis=0)
```
Not an assertable question but can you remember why it is important to normalize data for Logistic Regression?
**Complete here:**
```
def normalize_data_function(data):
"""
Implementation of a function that normalizes your data variables
Args:
data (np.array): a numpy array of shape (m, n)
m: number of observations
n: number of variables
Returns:
normalized_data (np.array): a numpy array of shape (m, n)
"""
# Compute the numerator first
# you can use np.min()
# numerator = ...
# Compute the denominator
# you can use np.max() and np.min()
# denominator = ...
# YOUR CODE HERE
raise NotImplementedError()
return normalized_data
data = np.array([[7,7,3], [2,2,11], [9,5,2], [0,9,5], [10,1,3], [1,5,2]])
normalized_data = normalize_data_function(data)
print('Before normalization:')
print(data)
print('\n-------------------\n')
print('After normalization:')
print(normalized_data)
```
Expected output:
Before normalization:
[[ 7 7 3]
[ 2 2 11]
[ 9 5 2]
[ 0 9 5]
[10 1 3]
[ 1 5 2]]
-------------------
After normalization:
[[0.7 0.75 0.11111111]
[0.2 0.125 1. ]
[0.9 0.5 0. ]
[0. 1. 0.33333333]
[1. 0. 0.11111111]
[0.1 0.5 0. ]]
```
data = np.array([[2,2,11,1], [7,5,1,3], [9,5,2,6]])
normalized_data = normalize_data_function(data)
np.testing.assert_almost_equal(round(normalized_data.max(),3),1.0)
np.testing.assert_almost_equal(round(normalized_data.mean(),3),0.518)
np.testing.assert_almost_equal(round(normalized_data.var(),3),0.205)
data = np.array([[1,3,1,3], [9,5,3,1], [2,2,4,6]])
normalized_data = normalize_data_function(data)
np.testing.assert_almost_equal(round(normalized_data.mean(),3),0.460)
np.testing.assert_almost_equal(round(normalized_data.std(),3),0.427)
```
# Exercise 6: Training a Logistic Regression with Sklearn
In this exercise, we will load a dataset related to direct marketing campaigns (phone calls) of a Portuguese banking institution. The goal is to predict whether the client will subscribe (1/0) to a term deposit (variable y) ([link to dataset](http://archive.ics.uci.edu/ml/datasets/Bank+Marketing))
Prepare to use your sklearn skills!
```
# We will load the dataset for you
bank = pd.read_csv('data/bank.csv', delimiter=";")
bank.head()
```
In this exercise, you need to do the following:
- Select an array/Series with the target variable (y)
- Select an array/dataframe with the X numeric variables (age, balance, day, month, duration, campaign and pdays)
- Scale all the X variables - normalize using Max / Min method.
- Fit a logistic regression for a maximum of 100 epochs and random state = 100.
- Return an array of the predicted probas and return the coefficients
After this, feel free to explore your predictions! As a bonus why don't you construct a decision boundary using two variables eh? :-)
```
from sklearn.linear_model import LogisticRegression
def train_model_sklearn(dataset):
'''
Returns the predicted probas and coefficients
of a trained logistic regression on the Titanic Dataset.
Args:
dataset(pd.DataFrame): dataset to train on.
Returns:
probas (np.array): Array of floats with the probability
of surviving for each passenger
coefficients (np.array): Returned coefficients of the
trained logistic regression.
'''
# leave this np.random seed here
np.random.seed(100)
# List of hints:
# 1. Use the Survived variable as y
# 2. Select the Numerical variables for X
# hint: use pandas .loc or indexing!
# 3. Scale the X dataset - you can use a function we have already
# constructed or resort to the sklearn implementation
# 4. Define logistic regression from sklearn with max iter = 100 also add random_state = 100
# Hint: for epochs look at the max_iter hyper param!
# 5. Fit logistic
# 6. Obtain probability of surviving
# 7. Obtain Coefficients from logistic regression
# Hint: see the sklearn logistic regression documentation if you do not know how to do this
# No need to return the intercept, just the variable coefficients!
# YOUR CODE HERE
raise NotImplementedError()
return probas, coef
probas, coef = train_model_sklearn(bank)
# Testing Probas
max_probas = probas.max()
np.testing.assert_almost_equal(max_probas, 0.997, 2)
min_probas = probas.min()
np.testing.assert_almost_equal(min_probas, 0.008, 2)
mean_probas = probas.mean()
np.testing.assert_almost_equal(mean_probas, 0.115, 2)
std_probas = probas.std()
np.testing.assert_almost_equal(std_probas, 0.115, 2)
sum_probas = probas.sum()
np.testing.assert_almost_equal(sum_probas*0.001, 0.521, 2)
# Testing Coefs
max_coef = coef[0].max()
np.testing.assert_almost_equal(max_coef*0.1, 0.87, 1)
min_coef = coef[0].min()
np.testing.assert_almost_equal(min_coef*0.1, -0.18, 1)
mean_coef = coef[0].mean()
np.testing.assert_almost_equal(mean_coef*0.1, 0.21, 1)
std_coef = coef[0].std()
np.testing.assert_almost_equal(std_coef*0.1, 0.35, 1)
sum_probas = coef[0].sum()
np.testing.assert_almost_equal(sum_probas*0.1, 1.06, 1)
```
|
github_jupyter
|
# Baby boy/girl classifier model preparation
*based on: Francisco Ingham and Jeremy Howard. Inspired by [Adrian Rosebrock](https://www.pyimagesearch.com/2017/12/04/how-to-create-a-deep-learning-dataset-using-google-images/)*
*by: Artyom Vorobyov*
Notebook execution and model training is made in Google Colab
```
from fastai.vision import *
from pathlib import Path
# Check if running in Google Colab and save it to bool variable
try:
import google.colab
IS_COLAB = True
except:
IS_COLAB = False
print("Is Colab:", IS_COLAB)
```
## Get a list of URLs
### How to get a dataset from Google Images
Go to [Google Images](http://images.google.com) and search for the images you are interested in. The more specific you are in your Google Search, the better the results and the less manual pruning you will have to do.
Scroll down until you've seen all the images you want to download, or until you see a button that says 'Show more results'. All the images you scrolled past are now available to download. To get more, click on the button, and continue scrolling. The maximum number of images Google Images shows is 700.
It is a good idea to put things you want to exclude into the search query, for instance if you are searching for the Eurasian wolf, "canis lupus lupus", it might be a good idea to exclude other variants:
"canis lupus lupus" -dog -arctos -familiaris -baileyi -occidentalis
You can also limit your results to show only photos by clicking on Tools and selecting Photos from the Type dropdown.
### How to download image URLs
Now you must run some Javascript code in your browser which will save the URLs of all the images you want for you dataset.
Press <kbd>Ctrl</kbd><kbd>Shift</kbd><kbd>J</kbd> in Windows/Linux and <kbd>Cmd</kbd><kbd>Opt</kbd><kbd>J</kbd> in Mac, and a small window the javascript 'Console' will appear. That is where you will paste the JavaScript commands.
You will need to get the urls of each of the images. Before running the following commands, you may want to disable ad blocking extensions (uBlock, AdBlockPlus etc.) in Chrome. Otherwise the window.open() command doesn't work. Then you can run the following commands:
```javascript
urls = Array.from(document.querySelectorAll('.rg_di .rg_meta')).map(el=>JSON.parse(el.textContent).ou);
window.open('data:text/csv;charset=utf-8,' + escape(urls.join('\n')));
```
### What to do with babies
For this particular application (baby boy/girl classifier) you can just search for "baby boys" and "baby girls". Then run the script mentioned above and save the URLs in "boys_urls.csv" and "girls_urls.csv".
## Download images
fast.ai has a function that allows you to do just that. You just have to specify the urls filename as well as the destination folder and this function will download and save all images that can be opened. If they have some problem in being opened, they will not be saved.
Let's download our images! Notice you can choose a maximum number of images to be downloaded. In this case we will not download all the urls.
```
class_boy = 'boys'
class_girl = 'girls'
classes = [class_boy, class_girl]
path = Path('./data')
path.mkdir(parents=True, exist_ok=True)
def download_dataset(is_colab):
if is_colab:
from google.colab import drive
import shutil
import zipfile
# You'll be asked to sign in Google Account and copy-paste a code here. Do it.
drive.mount('/content/gdrive')
# Copy this model from Google Drive after export and manually put it in the "ai_models" folder in the repository
# If there'll be an error during downloading the model - share it with some other Google account and download
# from this 2nd account - it should work fine.
zip_remote_path = Path('/content/gdrive/My Drive/Colab/boyorgirl/train.zip')
shutil.copy(str(zip_remote_path), str(path))
zip_local_path = path/'train.zip'
with zipfile.ZipFile(zip_local_path, 'r') as zip_ref:
zip_ref.extractall(path)
print("train folder contents:", (path/'train').ls())
else:
data_sources = [
('./boys_urls.csv', path/'train'/class_boy),
('./girls_urls.csv', path/'train'/class_girl)
]
# Download the images listed in URL's files
for urls_path, dest_path in data_sources:
dest = Path(dest_path)
dest.mkdir(parents=True, exist_ok=True)
download_images(urls_path, dest, max_pics=800)
# If you have problems download, try the code below with `max_workers=0` to see exceptions:
# download_images(urls_path, dest, max_pics=20, max_workers=0)
# Then we can remove any images that can't be opened:
for _, dest_path in data_sources:
verify_images(dest_path, delete=True, max_size=800)
# If running from colab - zip your train set (train folder) and put it to "Colab/boyorgirl/train.zip" in your Google Drive
download_dataset(IS_COLAB)
```
## Cleaning the data
Now it's a good moment to review the downloaded images and clean them. There will be some non-relevant images - photos of adults, photos of the baby clothes without the babies etc. Just review the images and remove non-relevant ones. For 2x400 images it'll take just 10-20 minutes in total.
There's also another way to clean the data - use the `fastiai.widgets.ImageCleaner`. It's used after you've trained your model. Even if you plan to use `ImageCleaner` later - it still makes sense to review the dataset briefly by yourself at the beginning.
## Load the data
```
np.random.seed(42)
data = ImageDataBunch.from_folder(path, train='train', valid_pct=0.2,
ds_tfms=get_transforms(), size=224, num_workers=4).normalize(imagenet_stats)
```
Good! Let's take a look at some of our pictures then.
```
# Check if all the classes were correctly read
print(data.classes)
print(data.classes == classes)
data.show_batch(rows=3, figsize=(7,8), ds_type=DatasetType.Train)
data.show_batch(rows=3, figsize=(7,8), ds_type=DatasetType.Valid)
print('Train set size: {}. Validation set size: {}'.format(len(data.train_ds), len(data.valid_ds)))
```
## Train model
```
learn = cnn_learner(data, models.resnet50, metrics=error_rate)
learn.fit_one_cycle(4)
learn.save('stage-1')
learn.unfreeze()
learn.lr_find()
# If the plot is not showing try to give a start and end learning rate
# learn.lr_find(start_lr=1e-5, end_lr=1e-1)
learn.recorder.plot()
learn.fit_one_cycle(2, max_lr=slice(1e-5,1e-3))
learn.save('stage-2')
```
## Interpretation
```
learn.load('stage-2');
interp = ClassificationInterpretation.from_learner(learn)
interp.plot_confusion_matrix()
```
## Putting your model in production
First thing first, let's export the content of our `Learner` object for production. Below are 2 variants of export - for local environment and for colab environment:
```
# Use this cell to export model from local environment within the repository
def get_export_path(is_colab):
if is_colab:
from google.colab import drive
# You'll be asked to sign in Google Account and copy-paste a code here. Do it.
# force_remount=True is needed to write model if it was deleted from Google Drive, but remains in Colab local file system
drive.mount('/content/gdrive', force_remount=True)
# Copy this model from Google Drive after export and manually put it in the "ai_models" folder in the repository
# If there'll be an error during downloading the model - share it with some other Google account and download
# from this 2nd account - it should work fine.
return Path('/content/gdrive/My Drive/Colab/boyorgirl/ai_models/export.pkl')
else:
# Used in case when notebook is run from the repository, but not in the Colab
return Path('../backend/ai_models/export.pkl')
# In case of Colab - model will be exported to 'Colab/boyorgirl/ai_models/export.pkl'. Download and save it in your repository manually
# in the 'ai_models' folder
export_path = get_export_path(IS_COLAB)
# ensure folder exists
export_path.parents[0].mkdir(parents=True, exist_ok=True)
# absolute path is passed as learn object attaches relative path to it's data folder rather than to notebook folder
learn.export(export_path.absolute())
print("Export folder contents:", export_path.parents[0].ls())
```
This will create a file named 'export.pkl' in the given directory. This exported model contains everything we need to deploy our model (the model, the weights but also some metadata like the classes or the transforms/normalization used).
```
```
|
github_jupyter
|
```
import torch
import datasets as nlp
from transformers import LongformerTokenizerFast
tokenizer = LongformerTokenizerFast.from_pretrained('allenai/longformer-base-4096')
def get_correct_alignement(context, answer):
""" Some original examples in SQuAD have indices wrong by 1 or 2 character. We test and fix this here. """
gold_text = answer['text'][0]
start_idx = answer['answer_start'][0]
end_idx = start_idx + len(gold_text)
if context[start_idx:end_idx] == gold_text:
return start_idx, end_idx # When the gold label position is good
elif context[start_idx-1:end_idx-1] == gold_text:
return start_idx-1, end_idx-1 # When the gold label is off by one character
elif context[start_idx-2:end_idx-2] == gold_text:
return start_idx-2, end_idx-2 # When the gold label is off by two character
else:
raise ValueError()
# Tokenize our training dataset
def convert_to_features(example):
# Tokenize contexts and questions (as pairs of inputs)
encodings = tokenizer.encode_plus(example['question'], example['context'], pad_to_max_length=True, max_length=512, truncation=True)
context_encodings = tokenizer.encode_plus(example['context'])
# Compute start and end tokens for labels using Transformers's fast tokenizers alignement methodes.
# this will give us the position of answer span in the context text
start_idx, end_idx = get_correct_alignement(example['context'], example['answers'])
start_positions_context = context_encodings.char_to_token(start_idx)
end_positions_context = context_encodings.char_to_token(end_idx-1)
# here we will compute the start and end position of the answer in the whole example
# as the example is encoded like this <s> question</s></s> context</s>
# and we know the postion of the answer in the context
# we can just find out the index of the sep token and then add that to position + 1 (+1 because there are two sep tokens)
# this will give us the position of the answer span in whole example
sep_idx = encodings['input_ids'].index(tokenizer.sep_token_id)
start_positions = start_positions_context + sep_idx + 1
end_positions = end_positions_context + sep_idx + 1
if end_positions > 512:
start_positions, end_positions = 0, 0
encodings.update({'start_positions': start_positions,
'end_positions': end_positions,
'attention_mask': encodings['attention_mask']})
return encodings
# load train and validation split of squad
train_dataset = nlp.load_dataset('squad', split='train')
valid_dataset = nlp.load_dataset('squad', split='validation')
# Temp. Only for testing quickly
train_dataset = nlp.Dataset.from_dict(train_dataset[:3])
valid_dataset = nlp.Dataset.from_dict(valid_dataset[:3])
train_dataset = train_dataset.map(convert_to_features)
valid_dataset = valid_dataset.map(convert_to_features, load_from_cache_file=False)
# set the tensor type and the columns which the dataset should return
columns = ['input_ids', 'attention_mask', 'start_positions', 'end_positions']
train_dataset.set_format(type='torch', columns=columns)
valid_dataset.set_format(type='torch', columns=columns)
len(train_dataset), len(valid_dataset)
t = torch.load('train_data.pt')
# Write training script
import json
args_dict = {
"n_gpu": 1,
"model_name_or_path": 'allenai/longformer-base-4096',
"max_len": 512 ,
"output_dir": './models',
"overwrite_output_dir": True,
"per_gpu_train_batch_size": 8,
"per_gpu_eval_batch_size": 8,
"gradient_accumulation_steps": 16,
"learning_rate": 1e-4,
"num_train_epochs": 3,
"do_train": True
}
## SQuAD evaluation script. Modifed slightly for this notebook
from __future__ import print_function
from collections import Counter
import string
import re
import argparse
import json
import sys
def normalize_answer(s):
"""Lower text and remove punctuation, articles and extra whitespace."""
def remove_articles(text):
return re.sub(r'\b(a|an|the)\b', ' ', text)
def white_space_fix(text):
return ' '.join(text.split())
def remove_punc(text):
exclude = set(string.punctuation)
return ''.join(ch for ch in text if ch not in exclude)
def lower(text):
return text.lower()
return white_space_fix(remove_articles(remove_punc(lower(s))))
def f1_score(prediction, ground_truth):
prediction_tokens = normalize_answer(prediction).split()
ground_truth_tokens = normalize_answer(ground_truth).split()
common = Counter(prediction_tokens) & Counter(ground_truth_tokens)
num_same = sum(common.values())
if num_same == 0:
return 0
precision = 1.0 * num_same / len(prediction_tokens)
recall = 1.0 * num_same / len(ground_truth_tokens)
f1 = (2 * precision * recall) / (precision + recall)
return f1
def exact_match_score(prediction, ground_truth):
return (normalize_answer(prediction) == normalize_answer(ground_truth))
def metric_max_over_ground_truths(metric_fn, prediction, ground_truths):
scores_for_ground_truths = []
for ground_truth in ground_truths:
score = metric_fn(prediction, ground_truth)
scores_for_ground_truths.append(score)
return max(scores_for_ground_truths)
def evaluate(gold_answers, predictions):
f1 = exact_match = total = 0
for ground_truths, prediction in zip(gold_answers, predictions):
total += 1
exact_match += metric_max_over_ground_truths(
exact_match_score, prediction, ground_truths)
f1 += metric_max_over_ground_truths(f1_score, prediction, ground_truths)
exact_match = 100.0 * exact_match / total
f1 = 100.0 * f1 / total
return {'exact_match': exact_match, 'f1': f1}
import torch
from transformers import LongformerTokenizerFast, LongformerForQuestionAnswering
from tqdm.auto import tqdm
tokenizer = LongformerTokenizerFast.from_pretrained('models')
model = LongformerForQuestionAnswering.from_pretrained('models')
model = model.cuda()
model.eval()
```
|
github_jupyter
|
```
from IPython.display import Markdown as md
### change to reflect your notebook
_nb_loc = "09_deploying/09c_changesig.ipynb"
_nb_title = "Changing signatures of exported model"
### no need to change any of this
_nb_safeloc = _nb_loc.replace('/', '%2F')
md("""
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://console.cloud.google.com/ai-platform/notebooks/deploy-notebook?name={1}&url=https%3A%2F%2Fgithub.com%2FGoogleCloudPlatform%2Fpractical-ml-vision-book%2Fblob%2Fmaster%2F{2}&download_url=https%3A%2F%2Fgithub.com%2FGoogleCloudPlatform%2Fpractical-ml-vision-book%2Fraw%2Fmaster%2F{2}">
<img src="https://raw.githubusercontent.com/GoogleCloudPlatform/practical-ml-vision-book/master/logo-cloud.png"/> Run in AI Platform Notebook</a>
</td>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/GoogleCloudPlatform/practical-ml-vision-book/blob/master/{0}">
<img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/GoogleCloudPlatform/practical-ml-vision-book/blob/master/{0}">
<img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
<td>
<a href="https://raw.githubusercontent.com/GoogleCloudPlatform/practical-ml-vision-book/master/{0}">
<img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table>
""".format(_nb_loc, _nb_title, _nb_safeloc))
```
# Changing signatures of exported model
In this notebook, we start from an already trained and saved model (as in Chapter 7).
For convenience, we have put this model in a public bucket in gs://practical-ml-vision-book/flowers_5_trained
## Enable GPU and set up helper functions
This notebook and pretty much every other notebook in this repository
will run faster if you are using a GPU.
On Colab:
- Navigate to Edit→Notebook Settings
- Select GPU from the Hardware Accelerator drop-down
On Cloud AI Platform Notebooks:
- Navigate to https://console.cloud.google.com/ai-platform/notebooks
- Create an instance with a GPU or select your instance and add a GPU
Next, we'll confirm that we can connect to the GPU with tensorflow:
```
import tensorflow as tf
print('TensorFlow version' + tf.version.VERSION)
print('Built with GPU support? ' + ('Yes!' if tf.test.is_built_with_cuda() else 'Noooo!'))
print('There are {} GPUs'.format(len(tf.config.experimental.list_physical_devices("GPU"))))
device_name = tf.test.gpu_device_name()
if device_name != '/device:GPU:0':
raise SystemError('GPU device not found')
print('Found GPU at: {}'.format(device_name))
```
## Exported model
We start from a trained and saved model from Chapter 7.
<pre>
model.save(...)
</pre>
```
MODEL_LOCATION='gs://practical-ml-vision-book/flowers_5_trained'
!gsutil ls {MODEL_LOCATION}
!saved_model_cli show --tag_set serve --signature_def serving_default --dir {MODEL_LOCATION}
```
## Passing through an input
Note that the signature doesn't tell us the input filename.
Let's add that.
```
import tensorflow as tf
import os, shutil
model = tf.keras.models.load_model(MODEL_LOCATION)
@tf.function(input_signature=[tf.TensorSpec([None,], dtype=tf.string)])
def predict_flower_type(filenames):
old_fn = model.signatures['serving_default']
result = old_fn(filenames) # has flower_type_int etc.
result['filename'] = filenames
return result
shutil.rmtree('export', ignore_errors=True)
os.mkdir('export')
model.save('export/flowers_model',
signatures={
'serving_default': predict_flower_type
})
!saved_model_cli show --tag_set serve --signature_def serving_default --dir export/flowers_model
import tensorflow as tf
serving_fn = tf.keras.models.load_model('export/flowers_model').signatures['serving_default']
filenames = [
'gs://cloud-ml-data/img/flower_photos/dandelion/9818247_e2eac18894.jpg',
'gs://cloud-ml-data/img/flower_photos/dandelion/9853885425_4a82356f1d_m.jpg',
'gs://cloud-ml-data/img/flower_photos/daisy/9299302012_958c70564c_n.jpg',
'gs://cloud-ml-data/img/flower_photos/tulips/8733586143_3139db6e9e_n.jpg',
'gs://cloud-ml-data/img/flower_photos/tulips/8713397358_0505cc0176_n.jpg'
]
pred = serving_fn(tf.convert_to_tensor(filenames))
print(pred)
```
## Multiple signatures
```
import tensorflow as tf
import os, shutil
model = tf.keras.models.load_model(MODEL_LOCATION)
old_fn = model.signatures['serving_default']
@tf.function(input_signature=[tf.TensorSpec([None,], dtype=tf.string)])
def pass_through_input(filenames):
result = old_fn(filenames) # has flower_type_int etc.
result['filename'] = filenames
return result
shutil.rmtree('export', ignore_errors=True)
os.mkdir('export')
model.save('export/flowers_model2',
signatures={
'serving_default': old_fn,
'input_pass_through': pass_through_input
})
!saved_model_cli show --tag_set serve --dir export/flowers_model2
!saved_model_cli show --tag_set serve --dir export/flowers_model2 --signature_def serving_default
!saved_model_cli show --tag_set serve --dir export/flowers_model2 --signature_def input_pass_through
```
## Deploying multi-signature model as REST API
```
!./caip_deploy.sh --version multi --model_location ./export/flowers_model2
%%writefile request.json
{
"instances": [
{
"filenames": "gs://cloud-ml-data/img/flower_photos/dandelion/9818247_e2eac18894.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/dandelion/9853885425_4a82356f1d_m.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/daisy/9299302012_958c70564c_n.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/tulips/8733586143_3139db6e9e_n.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/tulips/8713397358_0505cc0176_n.jpg"
}
]
}
!gcloud ai-platform predict --model=flowers --version=multi --json-request=request.json
%%writefile request.json
{
"signature_name": "input_pass_through",
"instances": [
{
"filenames": "gs://cloud-ml-data/img/flower_photos/dandelion/9818247_e2eac18894.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/dandelion/9853885425_4a82356f1d_m.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/daisy/9299302012_958c70564c_n.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/tulips/8733586143_3139db6e9e_n.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/tulips/8713397358_0505cc0176_n.jpg"
}
]
}
!gcloud ai-platform predict --model=flowers --version=multi --json-request=request.json
```
that's a bug ... filed a bug report; hope it's fixed by the time you are reading the book.
```
from oauth2client.client import GoogleCredentials
import requests
import json
PROJECT = 'ai-analytics-solutions' # CHANGE
MODEL_NAME = 'flowers'
MODEL_VERSION = 'multi'
token = GoogleCredentials.get_application_default().get_access_token().access_token
api = 'https://ml.googleapis.com/v1/projects/{}/models/{}/versions/{}:predict' \
.format(PROJECT, MODEL_NAME, MODEL_VERSION)
headers = {'Authorization': 'Bearer ' + token }
data = {
"signature_name": "input_pass_through",
"instances": [
{
"filenames": "gs://cloud-ml-data/img/flower_photos/dandelion/9818247_e2eac18894.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/dandelion/9853885425_4a82356f1d_m.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/daisy/9299302012_958c70564c_n.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/tulips/8733586143_3139db6e9e_n.jpg"
},
{
"filenames": "gs://cloud-ml-data/img/flower_photos/tulips/8713397358_0505cc0176_n.jpg"
}
]
}
response = requests.post(api, json=data, headers=headers)
print(response.content.decode('utf-8'))
```
## License
Copyright 2020 Google Inc. Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
|
github_jupyter
|
<a href="https://colab.research.google.com/github/Miseq/naive_imdb_reviews_model/blob/master/naive_imdb_model.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
from keras.datasets import imdb
from keras import optimizers
from keras import losses
from keras import metrics
from keras import models
from keras import layers
import matplotlib.pyplot as plt
import numpy as np
def vectorize_data(data, dimension=10000):
result = np.zeros((len(data), dimension))
for i, seq in enumerate(data):
result[i, seq] = 1.
return result
(train_data, train_labels), (test_data, test_labels) = imdb.load_data(num_words=10000)
x_train = vectorize_data(train_data)
x_test = vectorize_data(test_data)
y_train = np.asarray(train_labels).astype('float32')
y_test = np.asarray(test_labels).astype('float32')
model = models.Sequential()
model.add(layers.Dense(16, activation='relu', input_shape=(10000,)))
model.add(layers.Dense(1, activation='sigmoid'))
model.compile(optimizer=optimizers.RMSprop(lr=0.001), loss=losses.binary_crossentropy, metrics=['accuracy'])
x_val = x_train[:20000]
partial_x_train = x_train[20000:]
y_val = y_train[:20000]
partial_y_train = y_train[20000:]
history = model.fit(partial_x_train, partial_y_train, epochs=20, batch_size=512, validation_data=(x_val, y_val))
history_dict = history.history
history_dict.keys()
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(1, len(acc)+1)
# Tworzenie wykresu start tenowania i walidacji
plt.plot(epochs, loss, 'bo', label='Strata trenowania')
plt.plot(epochs, val_loss, 'b', label='Strata walidacji')
plt.title('Strata renowania i walidacji')
plt.xlabel('Epoki')
plt.ylabel('Strata')
plt.legend()
plt.show()
# Tworzenie wykresu dokładności trenowania i walidacji
plt.clf() # Czyszczenie rysunku(wazne)
acc_values = history_dict['acc']
val_acc_vales = history_dict['val_acc']
plt.plot(epochs, acc, 'bo', label='Dokladnosc trenowania')
plt.plot(epochs, val_acc, 'b', label='Dokladnosc walidacji')
plt.title('Dokladnosc trenowania i walidacji')
plt.xlabel('Epoki')
plt.ylabel('Strata')
plt.legend()
plt.show()
min_loss_val = min(val_loss)
max_acc_val = max(val_acc)
min_loss_ix = val_loss.index(min_loss_val)
max_acc_ix = val_acc.index(max_acc_val)
print(f'{min_loss_ix} --- {max_acc_ix}')
```
Po 7 epoce model zaczyna być przetrenowany
```
model.fit(x_train, y_train, epochs=7, batch_size=512, validation_data=(x_val, y_val))
results = model.evaluate(x_test, y_test)
results
```
## Wiecej warstw ukrytych
```
model = models.Sequential()
model.add(layers.Dense(16, activation='relu', input_shape=(10000,)))
model.add(layers.Dense(8, activation='relu'))
model.add(layers.Dense(4, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
model.compile(optimizer=optimizers.RMSprop(lr=0.001), loss=losses.binary_crossentropy, metrics=['accuracy'])
model.fit(x_train, y_train, epochs=10, batch_size=512, validation_data=(x_val, y_val))
results = model.evaluate(x_test, y_test)
results
```
## Wieksza ilosc jednostek ukrytych
```
model = models.Sequential()
model.add(layers.Dense(64, activation='relu', input_shape=(10000,)))
model.add(layers.Dense(32, activation='relu'))
model.add(layers.Dense(16, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
model.compile(optimizer=optimizers.RMSprop(lr=0.001), loss=losses.binary_crossentropy, metrics=['accuracy'])
model.fit(x_train, y_train, epochs=10, batch_size=512, validation_data=(x_val, y_val))
results = model.evaluate(x_test, y_test)
results
```
## Funkcja straty mse
```
model = models.Sequential()
model.add(layers.Dense(64, activation='relu', input_shape=(10000,)))
model.add(layers.Dense(32, activation='relu'))
model.add(layers.Dense(16, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
model.compile(optimizer=optimizers.RMSprop(lr=0.001), loss=losses.mse, metrics=['accuracy'])
model.fit(x_train, y_train, epochs=10, batch_size=512, validation_data=(x_val, y_val))
results = model.evaluate(x_test, y_test)
results
```
|
github_jupyter
|
# Convolutional Neural Networks: Application
Welcome to Course 4's second assignment! In this notebook, you will:
- Implement helper functions that you will use when implementing a TensorFlow model
- Implement a fully functioning ConvNet using TensorFlow
**After this assignment you will be able to:**
- Build and train a ConvNet in TensorFlow for a classification problem
We assume here that you are already familiar with TensorFlow. If you are not, please refer the *TensorFlow Tutorial* of the third week of Course 2 ("*Improving deep neural networks*").
### <font color='darkblue'> Updates to Assignment <font>
#### If you were working on a previous version
* The current notebook filename is version "1a".
* You can find your work in the file directory as version "1".
* To view the file directory, go to the menu "File->Open", and this will open a new tab that shows the file directory.
#### List of Updates
* `initialize_parameters`: added details about tf.get_variable, `eval`. Clarified test case.
* Added explanations for the kernel (filter) stride values, max pooling, and flatten functions.
* Added details about softmax cross entropy with logits.
* Added instructions for creating the Adam Optimizer.
* Added explanation of how to evaluate tensors (optimizer and cost).
* `forward_propagation`: clarified instructions, use "F" to store "flatten" layer.
* Updated print statements and 'expected output' for easier visual comparisons.
* Many thanks to Kevin P. Brown (mentor for the deep learning specialization) for his suggestions on the assignments in this course!
## 1.0 - TensorFlow model
In the previous assignment, you built helper functions using numpy to understand the mechanics behind convolutional neural networks. Most practical applications of deep learning today are built using programming frameworks, which have many built-in functions you can simply call.
As usual, we will start by loading in the packages.
```
import math
import numpy as np
import h5py
import matplotlib.pyplot as plt
import scipy
from PIL import Image
from scipy import ndimage
import tensorflow as tf
from tensorflow.python.framework import ops
from cnn_utils import *
%matplotlib inline
np.random.seed(1)
```
Run the next cell to load the "SIGNS" dataset you are going to use.
```
# Loading the data (signs)
X_train_orig, Y_train_orig, X_test_orig, Y_test_orig, classes = load_dataset()
```
As a reminder, the SIGNS dataset is a collection of 6 signs representing numbers from 0 to 5.
<img src="images/SIGNS.png" style="width:800px;height:300px;">
The next cell will show you an example of a labelled image in the dataset. Feel free to change the value of `index` below and re-run to see different examples.
```
# Example of a picture
index = 6
plt.imshow(X_train_orig[index])
print ("y = " + str(np.squeeze(Y_train_orig[:, index])))
```
In Course 2, you had built a fully-connected network for this dataset. But since this is an image dataset, it is more natural to apply a ConvNet to it.
To get started, let's examine the shapes of your data.
```
X_train = X_train_orig/255.
X_test = X_test_orig/255.
Y_train = convert_to_one_hot(Y_train_orig, 6).T
Y_test = convert_to_one_hot(Y_test_orig, 6).T
print ("number of training examples = " + str(X_train.shape[0]))
print ("number of test examples = " + str(X_test.shape[0]))
print ("X_train shape: " + str(X_train.shape))
print ("Y_train shape: " + str(Y_train.shape))
print ("X_test shape: " + str(X_test.shape))
print ("Y_test shape: " + str(Y_test.shape))
conv_layers = {}
```
### 1.1 - Create placeholders
TensorFlow requires that you create placeholders for the input data that will be fed into the model when running the session.
**Exercise**: Implement the function below to create placeholders for the input image X and the output Y. You should not define the number of training examples for the moment. To do so, you could use "None" as the batch size, it will give you the flexibility to choose it later. Hence X should be of dimension **[None, n_H0, n_W0, n_C0]** and Y should be of dimension **[None, n_y]**. [Hint: search for the tf.placeholder documentation"](https://www.tensorflow.org/api_docs/python/tf/placeholder).
```
# GRADED FUNCTION: create_placeholders
def create_placeholders(n_H0, n_W0, n_C0, n_y):
"""
Creates the placeholders for the tensorflow session.
Arguments:
n_H0 -- scalar, height of an input image
n_W0 -- scalar, width of an input image
n_C0 -- scalar, number of channels of the input
n_y -- scalar, number of classes
Returns:
X -- placeholder for the data input, of shape [None, n_H0, n_W0, n_C0] and dtype "float"
Y -- placeholder for the input labels, of shape [None, n_y] and dtype "float"
"""
### START CODE HERE ### (≈2 lines)
X = tf.placeholder(tf.float32, [None, n_H0, n_W0, n_C0])
Y = tf.placeholder(tf.float32, [None, n_y])
### END CODE HERE ###
return X, Y
X, Y = create_placeholders(64, 64, 3, 6)
print ("X = " + str(X))
print ("Y = " + str(Y))
```
**Expected Output**
<table>
<tr>
<td>
X = Tensor("Placeholder:0", shape=(?, 64, 64, 3), dtype=float32)
</td>
</tr>
<tr>
<td>
Y = Tensor("Placeholder_1:0", shape=(?, 6), dtype=float32)
</td>
</tr>
</table>
### 1.2 - Initialize parameters
You will initialize weights/filters $W1$ and $W2$ using `tf.contrib.layers.xavier_initializer(seed = 0)`. You don't need to worry about bias variables as you will soon see that TensorFlow functions take care of the bias. Note also that you will only initialize the weights/filters for the conv2d functions. TensorFlow initializes the layers for the fully connected part automatically. We will talk more about that later in this assignment.
**Exercise:** Implement initialize_parameters(). The dimensions for each group of filters are provided below. Reminder - to initialize a parameter $W$ of shape [1,2,3,4] in Tensorflow, use:
```python
W = tf.get_variable("W", [1,2,3,4], initializer = ...)
```
#### tf.get_variable()
[Search for the tf.get_variable documentation](https://www.tensorflow.org/api_docs/python/tf/get_variable). Notice that the documentation says:
```
Gets an existing variable with these parameters or create a new one.
```
So we can use this function to create a tensorflow variable with the specified name, but if the variables already exist, it will get the existing variable with that same name.
```
# GRADED FUNCTION: initialize_parameters
def initialize_parameters():
"""
Initializes weight parameters to build a neural network with tensorflow. The shapes are:
W1 : [4, 4, 3, 8]
W2 : [2, 2, 8, 16]
Note that we will hard code the shape values in the function to make the grading simpler.
Normally, functions should take values as inputs rather than hard coding.
Returns:
parameters -- a dictionary of tensors containing W1, W2
"""
tf.set_random_seed(1) # so that your "random" numbers match ours
### START CODE HERE ### (approx. 2 lines of code)
W1 = tf.get_variable("W1", [4, 4, 3, 8], initializer=tf.contrib.layers.xavier_initializer(seed=0))
W2 = tf.get_variable("W2", [2, 2, 8, 16], initializer=tf.contrib.layers.xavier_initializer(seed=0))
### END CODE HERE ###
parameters = {"W1": W1,
"W2": W2}
return parameters
tf.reset_default_graph()
with tf.Session() as sess_test:
parameters = initialize_parameters()
init = tf.global_variables_initializer()
sess_test.run(init)
print("W1[1,1,1] = \n" + str(parameters["W1"].eval()[1,1,1]))
print("W1.shape: " + str(parameters["W1"].shape))
print("\n")
print("W2[1,1,1] = \n" + str(parameters["W2"].eval()[1,1,1]))
print("W2.shape: " + str(parameters["W2"].shape))
```
** Expected Output:**
```
W1[1,1,1] =
[ 0.00131723 0.14176141 -0.04434952 0.09197326 0.14984085 -0.03514394
-0.06847463 0.05245192]
W1.shape: (4, 4, 3, 8)
W2[1,1,1] =
[-0.08566415 0.17750949 0.11974221 0.16773748 -0.0830943 -0.08058
-0.00577033 -0.14643836 0.24162132 -0.05857408 -0.19055021 0.1345228
-0.22779644 -0.1601823 -0.16117483 -0.10286498]
W2.shape: (2, 2, 8, 16)
```
### 1.3 - Forward propagation
In TensorFlow, there are built-in functions that implement the convolution steps for you.
- **tf.nn.conv2d(X,W, strides = [1,s,s,1], padding = 'SAME'):** given an input $X$ and a group of filters $W$, this function convolves $W$'s filters on X. The third parameter ([1,s,s,1]) represents the strides for each dimension of the input (m, n_H_prev, n_W_prev, n_C_prev). Normally, you'll choose a stride of 1 for the number of examples (the first value) and for the channels (the fourth value), which is why we wrote the value as `[1,s,s,1]`. You can read the full documentation on [conv2d](https://www.tensorflow.org/api_docs/python/tf/nn/conv2d).
- **tf.nn.max_pool(A, ksize = [1,f,f,1], strides = [1,s,s,1], padding = 'SAME'):** given an input A, this function uses a window of size (f, f) and strides of size (s, s) to carry out max pooling over each window. For max pooling, we usually operate on a single example at a time and a single channel at a time. So the first and fourth value in `[1,f,f,1]` are both 1. You can read the full documentation on [max_pool](https://www.tensorflow.org/api_docs/python/tf/nn/max_pool).
- **tf.nn.relu(Z):** computes the elementwise ReLU of Z (which can be any shape). You can read the full documentation on [relu](https://www.tensorflow.org/api_docs/python/tf/nn/relu).
- **tf.contrib.layers.flatten(P)**: given a tensor "P", this function takes each training (or test) example in the batch and flattens it into a 1D vector.
* If a tensor P has the shape (m,h,w,c), where m is the number of examples (the batch size), it returns a flattened tensor with shape (batch_size, k), where $k=h \times w \times c$. "k" equals the product of all the dimension sizes other than the first dimension.
* For example, given a tensor with dimensions [100,2,3,4], it flattens the tensor to be of shape [100, 24], where 24 = 2 * 3 * 4. You can read the full documentation on [flatten](https://www.tensorflow.org/api_docs/python/tf/contrib/layers/flatten).
- **tf.contrib.layers.fully_connected(F, num_outputs):** given the flattened input F, it returns the output computed using a fully connected layer. You can read the full documentation on [full_connected](https://www.tensorflow.org/api_docs/python/tf/contrib/layers/fully_connected).
In the last function above (`tf.contrib.layers.fully_connected`), the fully connected layer automatically initializes weights in the graph and keeps on training them as you train the model. Hence, you did not need to initialize those weights when initializing the parameters.
#### Window, kernel, filter
The words "window", "kernel", and "filter" are used to refer to the same thing. This is why the parameter `ksize` refers to "kernel size", and we use `(f,f)` to refer to the filter size. Both "kernel" and "filter" refer to the "window."
**Exercise**
Implement the `forward_propagation` function below to build the following model: `CONV2D -> RELU -> MAXPOOL -> CONV2D -> RELU -> MAXPOOL -> FLATTEN -> FULLYCONNECTED`. You should use the functions above.
In detail, we will use the following parameters for all the steps:
- Conv2D: stride 1, padding is "SAME"
- ReLU
- Max pool: Use an 8 by 8 filter size and an 8 by 8 stride, padding is "SAME"
- Conv2D: stride 1, padding is "SAME"
- ReLU
- Max pool: Use a 4 by 4 filter size and a 4 by 4 stride, padding is "SAME"
- Flatten the previous output.
- FULLYCONNECTED (FC) layer: Apply a fully connected layer without an non-linear activation function. Do not call the softmax here. This will result in 6 neurons in the output layer, which then get passed later to a softmax. In TensorFlow, the softmax and cost function are lumped together into a single function, which you'll call in a different function when computing the cost.
```
# GRADED FUNCTION: forward_propagation
def forward_propagation(X, parameters):
"""
Implements the forward propagation for the model:
CONV2D -> RELU -> MAXPOOL -> CONV2D -> RELU -> MAXPOOL -> FLATTEN -> FULLYCONNECTED
Note that for simplicity and grading purposes, we'll hard-code some values
such as the stride and kernel (filter) sizes.
Normally, functions should take these values as function parameters.
Arguments:
X -- input dataset placeholder, of shape (input size, number of examples)
parameters -- python dictionary containing your parameters "W1", "W2"
the shapes are given in initialize_parameters
Returns:
Z3 -- the output of the last LINEAR unit
"""
# Retrieve the parameters from the dictionary "parameters"
W1 = parameters['W1']
W2 = parameters['W2']
### START CODE HERE ###
# CONV2D: stride of 1, padding 'SAME'
Z1 = tf.nn.conv2d(X, W1, strides=[1, 1, 1, 1], padding='SAME')
# RELU
A1 = tf.nn.relu(Z1)
# MAXPOOL: window 8x8, stride 8, padding 'SAME'
P1 = tf.nn.max_pool(A1, ksize=[1, 8, 8, 1], strides=[1, 8, 8, 1], padding='SAME')
# CONV2D: filters W2, stride 1, padding 'SAME'
Z2 = tf.nn.conv2d(P1, W2, strides=[1, 1, 1, 1], padding='SAME')
# RELU
A2 = tf.nn.relu(Z2)
# MAXPOOL: window 4x4, stride 4, padding 'SAME'
P2 = tf.nn.max_pool(A2, ksize=[1, 4, 4, 1], strides=[1, 4, 4, 1], padding='SAME')
# FLATTEN
F = tf.contrib.layers.flatten(P2)
# FULLY-CONNECTED without non-linear activation function (not not call softmax).
# 6 neurons in output layer. Hint: one of the arguments should be "activation_fn=None"
Z3 = tf.contrib.layers.fully_connected(F, 6, activation_fn=None)
### END CODE HERE ###
return Z3
tf.reset_default_graph()
with tf.Session() as sess:
np.random.seed(1)
X, Y = create_placeholders(64, 64, 3, 6)
parameters = initialize_parameters()
Z3 = forward_propagation(X, parameters)
init = tf.global_variables_initializer()
sess.run(init)
a = sess.run(Z3, {X: np.random.randn(2,64,64,3), Y: np.random.randn(2,6)})
print("Z3 = \n" + str(a))
```
**Expected Output**:
```
Z3 =
[[-0.44670227 -1.57208765 -1.53049231 -2.31013036 -1.29104376 0.46852064]
[-0.17601591 -1.57972014 -1.4737016 -2.61672091 -1.00810647 0.5747785 ]]
```
### 1.4 - Compute cost
Implement the compute cost function below. Remember that the cost function helps the neural network see how much the model's predictions differ from the correct labels. By adjusting the weights of the network to reduce the cost, the neural network can improve its predictions.
You might find these two functions helpful:
- **tf.nn.softmax_cross_entropy_with_logits(logits = Z, labels = Y):** computes the softmax entropy loss. This function both computes the softmax activation function as well as the resulting loss. You can check the full documentation [softmax_cross_entropy_with_logits](https://www.tensorflow.org/api_docs/python/tf/nn/softmax_cross_entropy_with_logits).
- **tf.reduce_mean:** computes the mean of elements across dimensions of a tensor. Use this to calculate the sum of the losses over all the examples to get the overall cost. You can check the full documentation [reduce_mean](https://www.tensorflow.org/api_docs/python/tf/reduce_mean).
#### Details on softmax_cross_entropy_with_logits (optional reading)
* Softmax is used to format outputs so that they can be used for classification. It assigns a value between 0 and 1 for each category, where the sum of all prediction values (across all possible categories) equals 1.
* Cross Entropy is compares the model's predicted classifications with the actual labels and results in a numerical value representing the "loss" of the model's predictions.
* "Logits" are the result of multiplying the weights and adding the biases. Logits are passed through an activation function (such as a relu), and the result is called the "activation."
* The function is named `softmax_cross_entropy_with_logits` takes logits as input (and not activations); then uses the model to predict using softmax, and then compares the predictions with the true labels using cross entropy. These are done with a single function to optimize the calculations.
** Exercise**: Compute the cost below using the function above.
```
# GRADED FUNCTION: compute_cost
def compute_cost(Z3, Y):
"""
Computes the cost
Arguments:
Z3 -- output of forward propagation (output of the last LINEAR unit), of shape (number of examples, 6)
Y -- "true" labels vector placeholder, same shape as Z3
Returns:
cost - Tensor of the cost function
"""
### START CODE HERE ### (1 line of code)
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=Z3, labels=Y))
### END CODE HERE ###
return cost
tf.reset_default_graph()
with tf.Session() as sess:
np.random.seed(1)
X, Y = create_placeholders(64, 64, 3, 6)
parameters = initialize_parameters()
Z3 = forward_propagation(X, parameters)
cost = compute_cost(Z3, Y)
init = tf.global_variables_initializer()
sess.run(init)
a = sess.run(cost, {X: np.random.randn(4,64,64,3), Y: np.random.randn(4,6)})
print("cost = " + str(a))
```
**Expected Output**:
```
cost = 2.91034
```
## 1.5 Model
Finally you will merge the helper functions you implemented above to build a model. You will train it on the SIGNS dataset.
**Exercise**: Complete the function below.
The model below should:
- create placeholders
- initialize parameters
- forward propagate
- compute the cost
- create an optimizer
Finally you will create a session and run a for loop for num_epochs, get the mini-batches, and then for each mini-batch you will optimize the function. [Hint for initializing the variables](https://www.tensorflow.org/api_docs/python/tf/global_variables_initializer)
#### Adam Optimizer
You can use `tf.train.AdamOptimizer(learning_rate = ...)` to create the optimizer. The optimizer has a `minimize(loss=...)` function that you'll call to set the cost function that the optimizer will minimize.
For details, check out the documentation for [Adam Optimizer](https://www.tensorflow.org/api_docs/python/tf/train/AdamOptimizer)
#### Random mini batches
If you took course 2 of the deep learning specialization, you implemented `random_mini_batches()` in the "Optimization" programming assignment. This function returns a list of mini-batches. It is already implemented in the `cnn_utils.py` file and imported here, so you can call it like this:
```Python
minibatches = random_mini_batches(X, Y, mini_batch_size = 64, seed = 0)
```
(You will want to choose the correct variable names when you use it in your code).
#### Evaluating the optimizer and cost
Within a loop, for each mini-batch, you'll use the `tf.Session` object (named `sess`) to feed a mini-batch of inputs and labels into the neural network and evaluate the tensors for the optimizer as well as the cost. Remember that we built a graph data structure and need to feed it inputs and labels and use `sess.run()` in order to get values for the optimizer and cost.
You'll use this kind of syntax:
```
output_for_var1, output_for_var2 = sess.run(
fetches=[var1, var2],
feed_dict={var_inputs: the_batch_of_inputs,
var_labels: the_batch_of_labels}
)
```
* Notice that `sess.run` takes its first argument `fetches` as a list of objects that you want it to evaluate (in this case, we want to evaluate the optimizer and the cost).
* It also takes a dictionary for the `feed_dict` parameter.
* The keys are the `tf.placeholder` variables that we created in the `create_placeholders` function above.
* The values are the variables holding the actual numpy arrays for each mini-batch.
* The sess.run outputs a tuple of the evaluated tensors, in the same order as the list given to `fetches`.
For more information on how to use sess.run, see the documentation [tf.Sesssion#run](https://www.tensorflow.org/api_docs/python/tf/Session#run) documentation.
```
# GRADED FUNCTION: model
def model(X_train, Y_train, X_test, Y_test, learning_rate = 0.009,
num_epochs = 100, minibatch_size = 64, print_cost = True):
"""
Implements a three-layer ConvNet in Tensorflow:
CONV2D -> RELU -> MAXPOOL -> CONV2D -> RELU -> MAXPOOL -> FLATTEN -> FULLYCONNECTED
Arguments:
X_train -- training set, of shape (None, 64, 64, 3)
Y_train -- test set, of shape (None, n_y = 6)
X_test -- training set, of shape (None, 64, 64, 3)
Y_test -- test set, of shape (None, n_y = 6)
learning_rate -- learning rate of the optimization
num_epochs -- number of epochs of the optimization loop
minibatch_size -- size of a minibatch
print_cost -- True to print the cost every 100 epochs
Returns:
train_accuracy -- real number, accuracy on the train set (X_train)
test_accuracy -- real number, testing accuracy on the test set (X_test)
parameters -- parameters learnt by the model. They can then be used to predict.
"""
ops.reset_default_graph() # to be able to rerun the model without overwriting tf variables
tf.set_random_seed(1) # to keep results consistent (tensorflow seed)
seed = 3 # to keep results consistent (numpy seed)
(m, n_H0, n_W0, n_C0) = X_train.shape
n_y = Y_train.shape[1]
costs = [] # To keep track of the cost
# Create Placeholders of the correct shape
### START CODE HERE ### (1 line)
X, Y = create_placeholders(n_H0, n_W0, n_C0, n_y)
### END CODE HERE ###
# Initialize parameters
### START CODE HERE ### (1 line)
parameters = initialize_parameters()
### END CODE HERE ###
# Forward propagation: Build the forward propagation in the tensorflow graph
### START CODE HERE ### (1 line)
Z3 = forward_propagation(X, parameters)
### END CODE HERE ###
# Cost function: Add cost function to tensorflow graph
### START CODE HERE ### (1 line)
cost = compute_cost(Z3, Y)
### END CODE HERE ###
# Backpropagation: Define the tensorflow optimizer. Use an AdamOptimizer that minimizes the cost.
### START CODE HERE ### (1 line)
optimizer = tf.train.AdamOptimizer(learning_rate=learning_rate).minimize(cost)
### END CODE HERE ###
# Initialize all the variables globally
init = tf.global_variables_initializer()
# Start the session to compute the tensorflow graph
with tf.Session() as sess:
# Run the initialization
sess.run(init)
# Do the training loop
for epoch in range(num_epochs):
minibatch_cost = 0.
num_minibatches = int(m / minibatch_size) # number of minibatches of size minibatch_size in the train set
seed = seed + 1
minibatches = random_mini_batches(X_train, Y_train, minibatch_size, seed)
for minibatch in minibatches:
# Select a minibatch
(minibatch_X, minibatch_Y) = minibatch
"""
# IMPORTANT: The line that runs the graph on a minibatch.
# Run the session to execute the optimizer and the cost.
# The feedict should contain a minibatch for (X,Y).
"""
### START CODE HERE ### (1 line)
_ , temp_cost = sess.run([optimizer, cost], feed_dict={X:minibatch_X, Y:minibatch_Y})
### END CODE HERE ###
minibatch_cost += temp_cost / num_minibatches
# Print the cost every epoch
if print_cost == True and epoch % 5 == 0:
print ("Cost after epoch %i: %f" % (epoch, minibatch_cost))
if print_cost == True and epoch % 1 == 0:
costs.append(minibatch_cost)
# plot the cost
plt.plot(np.squeeze(costs))
plt.ylabel('cost')
plt.xlabel('iterations (per tens)')
plt.title("Learning rate =" + str(learning_rate))
plt.show()
# Calculate the correct predictions
predict_op = tf.argmax(Z3, 1)
correct_prediction = tf.equal(predict_op, tf.argmax(Y, 1))
# Calculate accuracy on the test set
accuracy = tf.reduce_mean(tf.cast(correct_prediction, "float"))
print(accuracy)
train_accuracy = accuracy.eval({X: X_train, Y: Y_train})
test_accuracy = accuracy.eval({X: X_test, Y: Y_test})
print("Train Accuracy:", train_accuracy)
print("Test Accuracy:", test_accuracy)
return train_accuracy, test_accuracy, parameters
```
Run the following cell to train your model for 100 epochs. Check if your cost after epoch 0 and 5 matches our output. If not, stop the cell and go back to your code!
```
_, _, parameters = model(X_train, Y_train, X_test, Y_test)
```
**Expected output**: although it may not match perfectly, your expected output should be close to ours and your cost value should decrease.
<table>
<tr>
<td>
**Cost after epoch 0 =**
</td>
<td>
1.917929
</td>
</tr>
<tr>
<td>
**Cost after epoch 5 =**
</td>
<td>
1.506757
</td>
</tr>
<tr>
<td>
**Train Accuracy =**
</td>
<td>
0.940741
</td>
</tr>
<tr>
<td>
**Test Accuracy =**
</td>
<td>
0.783333
</td>
</tr>
</table>
Congratulations! You have finished the assignment and built a model that recognizes SIGN language with almost 80% accuracy on the test set. If you wish, feel free to play around with this dataset further. You can actually improve its accuracy by spending more time tuning the hyperparameters, or using regularization (as this model clearly has a high variance).
Once again, here's a thumbs up for your work!
```
fname = "images/thumbs_up.jpg"
image = np.array(ndimage.imread(fname, flatten=False))
my_image = scipy.misc.imresize(image, size=(64,64))
plt.imshow(my_image)
```
|
github_jupyter
|
### Deep Kung-Fu with advantage actor-critic
In this notebook you'll build a deep reinforcement learning agent for atari [KungFuMaster](https://gym.openai.com/envs/KungFuMaster-v0/) and train it with advantage actor-critic.

```
from __future__ import print_function, division
from IPython.core import display
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
#If you are running on a server, launch xvfb to record game videos
#Please make sure you have xvfb installed
import os
if os.environ.get("DISPLAY") is str and len(os.environ.get("DISPLAY"))!=0:
!bash ../xvfb start
%env DISPLAY=:1
```
For starters, let's take a look at the game itself:
* Image resized to 42x42 and grayscale to run faster
* Rewards divided by 100 'cuz they are all divisible by 100
* Agent sees last 4 frames of game to account for object velocity
```
import gym
from atari_util import PreprocessAtari
# We scale rewards to avoid exploding gradients during optimization.
reward_scale = 0.01
def make_env():
env = gym.make("KungFuMasterDeterministic-v0")
env = PreprocessAtari(
env, height=42, width=42,
crop=lambda img: img[60:-30, 5:],
dim_order='tensorflow',
color=False, n_frames=4,
reward_scale=reward_scale)
return env
env = make_env()
obs_shape = env.observation_space.shape
n_actions = env.action_space.n
print("Observation shape:", obs_shape)
print("Num actions:", n_actions)
print("Action names:", env.env.env.get_action_meanings())
s = env.reset()
for _ in range(100):
s, _, _, _ = env.step(env.action_space.sample())
plt.title('Game image')
plt.imshow(env.render('rgb_array'))
plt.show()
plt.title('Agent observation (4-frame buffer)')
plt.imshow(s.transpose([0,2,1]).reshape([42,-1]))
plt.show()
```
### Build an agent
We now have to build an agent for actor-critic training - a convolutional neural network that converts states into action probabilities $\pi$ and state values $V$.
Your assignment here is to build and apply a neural network - with any framework you want.
For starters, we want you to implement this architecture:

After your agent gets mean reward above 50, we encourage you to experiment with model architecture to score even better.
```
import tensorflow as tf
tf.reset_default_graph()
sess = tf.InteractiveSession()
from keras.layers import Conv2D, Dense, Flatten
class Agent:
def __init__(self, name, state_shape, n_actions, reuse=False):
"""A simple actor-critic agent"""
with tf.variable_scope(name, reuse=reuse):
# Prepare neural network architecture
### Your code here: prepare any necessary layers, variables, etc.
# prepare a graph for agent step
self.state_t = tf.placeholder('float32', [None,] + list(state_shape))
self.agent_outputs = self.symbolic_step(self.state_t)
def symbolic_step(self, state_t):
"""Takes agent's previous step and observation, returns next state and whatever it needs to learn (tf tensors)"""
# Apply neural network
### Your code here: apply agent's neural network to get policy logits and state values.
logits = <logits go here>
state_value = <state values go here>
assert tf.is_numeric_tensor(state_value) and state_value.shape.ndims == 1, \
"please return 1D tf tensor of state values [you got %s]" % repr(state_value)
assert tf.is_numeric_tensor(logits) and logits.shape.ndims == 2, \
"please return 2d tf tensor of logits [you got %s]" % repr(logits)
# hint: if you triggered state_values assert with your shape being [None, 1],
# just select [:, 0]-th element of state values as new state values
return (logits, state_value)
def step(self, state_t):
"""Same as symbolic step except it operates on numpy arrays"""
sess = tf.get_default_session()
return sess.run(self.agent_outputs, {self.state_t: state_t})
def sample_actions(self, agent_outputs):
"""pick actions given numeric agent outputs (np arrays)"""
logits, state_values = agent_outputs
policy = np.exp(logits) / np.sum(np.exp(logits), axis=-1, keepdims=True)
return np.array([np.random.choice(len(p), p=p) for p in policy])
agent = Agent("agent", obs_shape, n_actions)
sess.run(tf.global_variables_initializer())
state = [env.reset()]
logits, value = agent.step(state)
print("action logits:\n", logits)
print("state values:\n", value)
```
### Let's play!
Let's build a function that measures agent's average reward.
```
def evaluate(agent, env, n_games=1):
"""Plays an a game from start till done, returns per-game rewards """
game_rewards = []
for _ in range(n_games):
state = env.reset()
total_reward = 0
while True:
action = agent.sample_actions(agent.step([state]))[0]
state, reward, done, info = env.step(action)
total_reward += reward
if done: break
# We rescale the reward back to ensure compatibility
# with other evaluations.
game_rewards.append(total_reward / reward_scale)
return game_rewards
env_monitor = gym.wrappers.Monitor(env, directory="kungfu_videos", force=True)
rw = evaluate(agent, env_monitor, n_games=3,)
env_monitor.close()
print (rw)
#show video
import os
from IPython.display import HTML
video_names = [s for s in os.listdir("./kungfu_videos/") if s.endswith(".mp4")]
HTML("""
<video width="640" height="480" controls>
<source src="{}" type="video/mp4">
</video>
""".format("./kungfu_videos/" + video_names[-1])) #this may or may not be _last_ video. Try other indices
```
### Training on parallel games

To make actor-critic training more stable, we shall play several games in parallel. This means ya'll have to initialize several parallel gym envs, send agent's actions there and .reset() each env if it becomes terminated. To minimize learner brain damage, we've taken care of them for ya - just make sure you read it before you use it.
```
class EnvBatch:
def __init__(self, n_envs = 10):
""" Creates n_envs environments and babysits them for ya' """
self.envs = [make_env() for _ in range(n_envs)]
def reset(self):
""" Reset all games and return [n_envs, *obs_shape] observations """
return np.array([env.reset() for env in self.envs])
def step(self, actions):
"""
Send a vector[batch_size] of actions into respective environments
:returns: observations[n_envs, *obs_shape], rewards[n_envs], done[n_envs,], info[n_envs]
"""
results = [env.step(a) for env, a in zip(self.envs, actions)]
new_obs, rewards, done, infos = map(np.array, zip(*results))
# reset environments automatically
for i in range(len(self.envs)):
if done[i]:
new_obs[i] = self.envs[i].reset()
return new_obs, rewards, done, infos
```
__Let's try it out:__
```
env_batch = EnvBatch(10)
batch_states = env_batch.reset()
batch_actions = agent.sample_actions(agent.step(batch_states))
batch_next_states, batch_rewards, batch_done, _ = env_batch.step(batch_actions)
print("State shape:", batch_states.shape)
print("Actions:", batch_actions[:3])
print("Rewards:", batch_rewards[:3])
print("Done:", batch_done[:3])
```
# Actor-critic
Here we define a loss functions and learning algorithms as usual.
```
# These placeholders mean exactly the same as in "Let's try it out" section above
states_ph = tf.placeholder('float32', [None,] + list(obs_shape))
next_states_ph = tf.placeholder('float32', [None,] + list(obs_shape))
actions_ph = tf.placeholder('int32', (None,))
rewards_ph = tf.placeholder('float32', (None,))
is_done_ph = tf.placeholder('float32', (None,))
# logits[n_envs, n_actions] and state_values[n_envs, n_actions]
logits, state_values = agent.symbolic_step(states_ph)
next_logits, next_state_values = agent.symbolic_step(next_states_ph)
next_state_values = next_state_values * (1 - is_done_ph)
# probabilities and log-probabilities for all actions
probs = tf.nn.softmax(logits) # [n_envs, n_actions]
logprobs = tf.nn.log_softmax(logits) # [n_envs, n_actions]
# log-probabilities only for agent's chosen actions
logp_actions = tf.reduce_sum(logprobs * tf.one_hot(actions_ph, n_actions), axis=-1) # [n_envs,]
# compute advantage using rewards_ph, state_values and next_state_values
gamma = 0.99
advantage = <YOUR CODE>
assert advantage.shape.ndims == 1, "please compute advantage for each sample, vector of shape [n_envs,]"
# compute policy entropy given logits_seq. Mind the "-" sign!
entropy = <YOUR CODE>
assert entropy.shape.ndims == 1, "please compute pointwise entropy vector of shape [n_envs,] "
actor_loss = - tf.reduce_mean(logp_actions * tf.stop_gradient(advantage)) - 0.001 * tf.reduce_mean(entropy)
# compute target state values using temporal difference formula. Use rewards_ph and next_step_values
target_state_values = <YOUR CODE>
critic_loss = tf.reduce_mean((state_values - tf.stop_gradient(target_state_values))**2 )
train_step = tf.train.AdamOptimizer(1e-4).minimize(actor_loss + critic_loss)
sess.run(tf.global_variables_initializer())
# Sanity checks to catch some errors. Specific to KungFuMaster in assignment's default setup.
l_act, l_crit, adv, ent = sess.run([actor_loss, critic_loss, advantage, entropy], feed_dict = {
states_ph: batch_states,
actions_ph: batch_actions,
next_states_ph: batch_states,
rewards_ph: batch_rewards,
is_done_ph: batch_done,
})
assert abs(l_act) < 100 and abs(l_crit) < 100, "losses seem abnormally large"
assert 0 <= ent.mean() <= np.log(n_actions), "impossible entropy value, double-check the formula pls"
if ent.mean() < np.log(n_actions) / 2: print("Entropy is too low for untrained agent")
print("You just might be fine!")
```
# Train
Just the usual - play a bit, compute loss, follow the graidents, repeat a few million times.

```
from IPython.display import clear_output
from tqdm import trange
from pandas import DataFrame
ewma = lambda x, span=100: DataFrame({'x':np.asarray(x)}).x.ewm(span=span).mean().values
env_batch = EnvBatch(10)
batch_states = env_batch.reset()
rewards_history = []
entropy_history = []
for i in trange(100000):
batch_actions = agent.sample_actions(agent.step(batch_states))
batch_next_states, batch_rewards, batch_done, _ = env_batch.step(batch_actions)
feed_dict = {
states_ph: batch_states,
actions_ph: batch_actions,
next_states_ph: batch_next_states,
rewards_ph: batch_rewards,
is_done_ph: batch_done,
}
batch_states = batch_next_states
_, ent_t = sess.run([train_step, entropy], feed_dict)
entropy_history.append(np.mean(ent_t))
if i % 500 == 0:
if i % 2500 == 0:
rewards_history.append(np.mean(evaluate(agent, env, n_games=3)))
if rewards_history[-1] >= 50:
print("Your agent has earned the yellow belt" % color)
clear_output(True)
plt.figure(figsize=[8, 4])
plt.subplot(1, 2, 1)
plt.plot(rewards_history, label='rewards')
plt.plot(ewma(np.array(rewards_history), span=10), marker='.', label='rewards ewma@10')
plt.title("Session rewards")
plt.grid()
plt.legend()
plt.subplot(1, 2, 2)
plt.plot(entropy_history, label='entropy')
plt.plot(ewma(np.array(entropy_history), span=1000), label='entropy ewma@1000')
plt.title("Policy entropy")
plt.grid()
plt.legend()
plt.show()
```
Relax and grab some refreshments while your agent is locked in an infinite loop of violence and death.
__How to interpret plots:__
The session reward is the easy thing: it should in general go up over time, but it's okay if it fluctuates ~~like crazy~~. It's also OK if it reward doesn't increase substantially before some 10k initial steps. However, if reward reaches zero and doesn't seem to get up over 2-3 evaluations, there's something wrong happening.
Since we use a policy-based method, we also keep track of __policy entropy__ - the same one you used as a regularizer. The only important thing about it is that your entropy shouldn't drop too low (`< 0.1`) before your agent gets the yellow belt. Or at least it can drop there, but _it shouldn't stay there for long_.
If it does, the culprit is likely:
* Some bug in entropy computation. Remember that it is $ - \sum p(a_i) \cdot log p(a_i) $
* Your agent architecture converges too fast. Increase entropy coefficient in actor loss.
* Gradient explosion - just [clip gradients](https://stackoverflow.com/a/43486487) and maybe use a smaller network
* Us. Or TF developers. Or aliens. Or lizardfolk. Contact us on forums before it's too late!
If you're debugging, just run `logits, values = agent.step(batch_states)` and manually look into logits and values. This will reveal the problem 9 times out of 10: you'll likely see some NaNs or insanely large numbers or zeros. Try to catch the moment when this happens for the first time and investigate from there.
### "Final" evaluation
```
env_monitor = gym.wrappers.Monitor(env, directory="kungfu_videos", force=True)
final_rewards = evaluate(agent, env_monitor, n_games=20)
env_monitor.close()
print("Final mean reward:", np.mean(final_rewards))
video_names = list(filter(lambda s: s.endswith(".mp4"), os.listdir("./kungfu_videos/")))
HTML("""
<video width="640" height="480" controls>
<source src="{}" type="video/mp4">
</video>
""".format("./kungfu_videos/"+video_names[-1]))
HTML("""
<video width="640" height="480" controls>
<source src="{}" type="video/mp4">
</video>
""".format("./kungfu_videos/" + video_names[-2])) # try other indices
# if you don't see videos, just navigate to ./kungfu_videos and download .mp4 files from there.
from submit import submit_kungfu
env = make_env()
submit_kungfu(agent, env, evaluate, <EMAIL>, <TOKEN>)
```
```
```
```
```
```
```
```
```
```
```
```
```
```
```
```
```
### Now what?
Well, 5k reward is [just the beginning](https://www.buzzfeed.com/mattjayyoung/what-the-color-of-your-karate-belt-actually-means-lg3g). Can you get past 200? With recurrent neural network memory, chances are you can even beat 400!
* Try n-step advantage and "lambda"-advantage (aka GAE) - see [this article](https://arxiv.org/abs/1506.02438)
* This change should improve early convergence a lot
* Try recurrent neural network
* RNN memory will slow things down initially, but in will reach better final reward at this game
* Implement asynchronuous version
* Remember [A3C](https://arxiv.org/abs/1602.01783)? The first "A" stands for asynchronuous. It means there are several parallel actor-learners out there.
* You can write custom code for synchronization, but we recommend using [redis](https://redis.io/)
* You can store full parameter set in redis, along with any other metadate
* Here's a _quick_ way to (de)serialize parameters for redis
```
import joblib
from six import BytesIO
```
```
def dumps(data):
"converts whatever to string"
s = BytesIO()
joblib.dump(data,s)
return s.getvalue()
```
```
def loads(string):
"converts string to whatever was dumps'ed in it"
return joblib.load(BytesIO(string))
```
|
github_jupyter
|
# Project 3: Smart Beta Portfolio and Portfolio Optimization
## Overview
Smart beta has a broad meaning, but we can say in practice that when we use the universe of stocks from an index, and then apply some weighting scheme other than market cap weighting, it can be considered a type of smart beta fund. By contrast, a purely alpha fund may create a portfolio of specific stocks, not related to an index, or may choose from the global universe of stocks. The other characteristic that makes a smart beta portfolio "beta" is that it gives its investors a diversified broad exposure to a particular market.
Imagine you're a portfolio manager, and wish to try out some different portfolio weighting methods.
One way to design portfolio is to look at certain accounting measures (fundamentals) that, based on past trends, indicate stocks that produce better results.
For instance, you may start with a hypothesis that dividend-issuing stocks tend to perform better than stocks that do not. This may not always be true of all companies; for instance, Apple does not issue dividends, but has had good historical performance. The hypothesis about dividend-paying stocks may go something like this:
Companies that regularly issue dividends may also be more prudent in allocating their available cash, and may indicate that they are more conscious of prioritizing shareholder interests. For example, a CEO may decide to reinvest cash into pet projects that produce low returns. Or, the CEO may do some analysis, identify that reinvesting within the company produces lower returns compared to a diversified portfolio, and so decide that shareholders would be better served if they were given the cash (in the form of dividends). So according to this hypothesis, dividends may be both a proxy for how the company is doing (in terms of earnings and cash flow), but also a signal that the company acts in the best interest of its shareholders. Of course, it's important to test whether this works in practice.
You may also have another hypothesis, with which you wish to design a portfolio that can then be made into an ETF. You may find that investors may wish to invest in passive beta funds, but wish to have less risk exposure (less volatility) in their investments. The goal of having a low volatility fund that still produces returns similar to an index may be appealing to investors who have a shorter investment time horizon, and so are more risk averse.
So the objective of your proposed portfolio is to design a portfolio that closely tracks an index, while also minimizing the portfolio variance. Also, if this portfolio can match the returns of the index with less volatility, then it has a higher risk-adjusted return (same return, lower volatility).
Smart Beta ETFs can be designed with both of these two general methods (among others): alternative weighting and minimum volatility ETF.
## Instructions
Each problem consists of a function to implement and instructions on how to implement the function. The parts of the function that need to be implemented are marked with a `# TODO` comment. After implementing the function, run the cell to test it against the unit tests we've provided. For each problem, we provide one or more unit tests from our `project_tests` package. These unit tests won't tell you if your answer is correct, but will warn you of any major errors. Your code will be checked for the correct solution when you submit it to Udacity.
## Packages
When you implement the functions, you'll only need to you use the packages you've used in the classroom, like [Pandas](https://pandas.pydata.org/) and [Numpy](http://www.numpy.org/). These packages will be imported for you. We recommend you don't add any import statements, otherwise the grader might not be able to run your code.
The other packages that we're importing are `helper`, `project_helper`, and `project_tests`. These are custom packages built to help you solve the problems. The `helper` and `project_helper` module contains utility functions and graph functions. The `project_tests` contains the unit tests for all the problems.
### Install Packages
```
import sys
!{sys.executable} -m pip install -r requirements.txt
```
### Load Packages
```
import pandas as pd
import numpy as np
import helper
import project_helper
import project_tests
```
## Market Data
### Load Data
For this universe of stocks, we'll be selecting large dollar volume stocks. We're using this universe, since it is highly liquid.
```
df = pd.read_csv('data/eod-quotemedia.csv')
percent_top_dollar = 0.2
high_volume_symbols = project_helper.large_dollar_volume_stocks(df, 'adj_close', 'adj_volume', percent_top_dollar)
df = df[df['ticker'].isin(high_volume_symbols)]
close = df.reset_index().pivot(index='date', columns='ticker', values='adj_close')
volume = df.reset_index().pivot(index='date', columns='ticker', values='adj_volume')
dividends = df.reset_index().pivot(index='date', columns='ticker', values='dividends')
```
### View Data
To see what one of these 2-d matrices looks like, let's take a look at the closing prices matrix.
```
project_helper.print_dataframe(close)
```
# Part 1: Smart Beta Portfolio
In Part 1 of this project, you'll build a portfolio using dividend yield to choose the portfolio weights. A portfolio such as this could be incorporated into a smart beta ETF. You'll compare this portfolio to a market cap weighted index to see how well it performs.
Note that in practice, you'll probably get the index weights from a data vendor (such as companies that create indices, like MSCI, FTSE, Standard and Poor's), but for this exercise we will simulate a market cap weighted index.
## Index Weights
The index we'll be using is based on large dollar volume stocks. Implement `generate_dollar_volume_weights` to generate the weights for this index. For each date, generate the weights based on dollar volume traded for that date. For example, assume the following is close prices and volume data:
```
Prices
A B ...
2013-07-08 2 2 ...
2013-07-09 5 6 ...
2013-07-10 1 2 ...
2013-07-11 6 5 ...
... ... ... ...
Volume
A B ...
2013-07-08 100 340 ...
2013-07-09 240 220 ...
2013-07-10 120 500 ...
2013-07-11 10 100 ...
... ... ... ...
```
The weights created from the function `generate_dollar_volume_weights` should be the following:
```
A B ...
2013-07-08 0.126.. 0.194.. ...
2013-07-09 0.759.. 0.377.. ...
2013-07-10 0.075.. 0.285.. ...
2013-07-11 0.037.. 0.142.. ...
... ... ... ...
```
```
def generate_dollar_volume_weights(close, volume):
"""
Generate dollar volume weights.
Parameters
----------
close : DataFrame
Close price for each ticker and date
volume : str
Volume for each ticker and date
Returns
-------
dollar_volume_weights : DataFrame
The dollar volume weights for each ticker and date
"""
assert close.index.equals(volume.index)
assert close.columns.equals(volume.columns)
#TODO: Implement function
dollar_volume = close * volume
return (dollar_volume.T / dollar_volume.T.sum()).T
project_tests.test_generate_dollar_volume_weights(generate_dollar_volume_weights)
```
### View Data
Let's generate the index weights using `generate_dollar_volume_weights` and view them using a heatmap.
```
index_weights = generate_dollar_volume_weights(close, volume)
project_helper.plot_weights(index_weights, 'Index Weights')
```
## Portfolio Weights
Now that we have the index weights, let's choose the portfolio weights based on dividends.
Implement `calculate_dividend_weights` to returns the weights for each stock based on its total dividend yield over time. This is similar to generating the weight for the index, but it's using dividend data instead.
For example, assume the following is `dividends` data:
```
Prices
A B
2013-07-08 0 0
2013-07-09 0 1
2013-07-10 0.5 0
2013-07-11 0 0
2013-07-12 2 0
... ... ...
```
The weights created from the function `calculate_dividend_weights` should be the following:
```
A B
2013-07-08 NaN NaN
2013-07-09 0 1
2013-07-10 0.333.. 0.666..
2013-07-11 0.333.. 0.666..
2013-07-12 0.714.. 0.285..
... ... ...
```
```
def calculate_dividend_weights(dividends):
"""
Calculate dividend weights.
Parameters
----------
ex_dividend : DataFrame
Ex-dividend for each stock and date
Returns
-------
dividend_weights : DataFrame
Weights for each stock and date
"""
#TODO: Implement function
dividend_cumsum_per_ticker = dividends.cumsum().T
return (dividend_cumsum_per_ticker/dividend_cumsum_per_ticker.sum()).T
project_tests.test_calculate_dividend_weights(calculate_dividend_weights)
```
### View Data
Just like the index weights, let's generate the ETF weights and view them using a heatmap.
```
etf_weights = calculate_dividend_weights(dividends)
project_helper.plot_weights(etf_weights, 'ETF Weights')
```
## Returns
Implement `generate_returns` to generate returns data for all the stocks and dates from price data. You might notice we're implementing returns and not log returns. Since we're not dealing with volatility, we don't have to use log returns.
```
def generate_returns(prices):
"""
Generate returns for ticker and date.
Parameters
----------
prices : DataFrame
Price for each ticker and date
Returns
-------
returns : Dataframe
The returns for each ticker and date
"""
#TODO: Implement function
return prices / prices.shift(1) - 1
project_tests.test_generate_returns(generate_returns)
```
### View Data
Let's generate the closing returns using `generate_returns` and view them using a heatmap.
```
returns = generate_returns(close)
project_helper.plot_returns(returns, 'Close Returns')
```
## Weighted Returns
With the returns of each stock computed, we can use it to compute the returns for an index or ETF. Implement `generate_weighted_returns` to create weighted returns using the returns and weights.
```
def generate_weighted_returns(returns, weights):
"""
Generate weighted returns.
Parameters
----------
returns : DataFrame
Returns for each ticker and date
weights : DataFrame
Weights for each ticker and date
Returns
-------
weighted_returns : DataFrame
Weighted returns for each ticker and date
"""
assert returns.index.equals(weights.index)
assert returns.columns.equals(weights.columns)
#TODO: Implement function
return returns * weights
project_tests.test_generate_weighted_returns(generate_weighted_returns)
```
### View Data
Let's generate the ETF and index returns using `generate_weighted_returns` and view them using a heatmap.
```
index_weighted_returns = generate_weighted_returns(returns, index_weights)
etf_weighted_returns = generate_weighted_returns(returns, etf_weights)
project_helper.plot_returns(index_weighted_returns, 'Index Returns')
project_helper.plot_returns(etf_weighted_returns, 'ETF Returns')
```
## Cumulative Returns
To compare performance between the ETF and Index, we're going to calculate the tracking error. Before we do that, we first need to calculate the index and ETF comulative returns. Implement `calculate_cumulative_returns` to calculate the cumulative returns over time given the returns.
```
def calculate_cumulative_returns(returns):
"""
Calculate cumulative returns.
Parameters
----------
returns : DataFrame
Returns for each ticker and date
Returns
-------
cumulative_returns : Pandas Series
Cumulative returns for each date
"""
#TODO: Implement function
return (returns.T.sum() + 1).cumprod()
project_tests.test_calculate_cumulative_returns(calculate_cumulative_returns)
```
### View Data
Let's generate the ETF and index cumulative returns using `calculate_cumulative_returns` and compare the two.
```
index_weighted_cumulative_returns = calculate_cumulative_returns(index_weighted_returns)
etf_weighted_cumulative_returns = calculate_cumulative_returns(etf_weighted_returns)
project_helper.plot_benchmark_returns(index_weighted_cumulative_returns, etf_weighted_cumulative_returns, 'Smart Beta ETF vs Index')
```
## Tracking Error
In order to check the performance of the smart beta portfolio, we can calculate the annualized tracking error against the index. Implement `tracking_error` to return the tracking error between the ETF and benchmark.
For reference, we'll be using the following annualized tracking error function:
$$ TE = \sqrt{252} * SampleStdev(r_p - r_b) $$
Where $ r_p $ is the portfolio/ETF returns and $ r_b $ is the benchmark returns.
```
def tracking_error(benchmark_returns_by_date, etf_returns_by_date):
"""
Calculate the tracking error.
Parameters
----------
benchmark_returns_by_date : Pandas Series
The benchmark returns for each date
etf_returns_by_date : Pandas Series
The ETF returns for each date
Returns
-------
tracking_error : float
The tracking error
"""
assert benchmark_returns_by_date.index.equals(etf_returns_by_date.index)
#TODO: Implement function
return np.sqrt(252) * (etf_returns_by_date - benchmark_returns_by_date).std()
project_tests.test_tracking_error(tracking_error)
```
### View Data
Let's generate the tracking error using `tracking_error`.
```
smart_beta_tracking_error = tracking_error(np.sum(index_weighted_returns, 1), np.sum(etf_weighted_returns, 1))
print('Smart Beta Tracking Error: {}'.format(smart_beta_tracking_error))
```
# Part 2: Portfolio Optimization
Now, let's create a second portfolio. We'll still reuse the market cap weighted index, but this will be independent of the dividend-weighted portfolio that we created in part 1.
We want to both minimize the portfolio variance and also want to closely track a market cap weighted index. In other words, we're trying to minimize the distance between the weights of our portfolio and the weights of the index.
$Minimize \left [ \sigma^2_p + \lambda \sqrt{\sum_{1}^{m}(weight_i - indexWeight_i)^2} \right ]$ where $m$ is the number of stocks in the portfolio, and $\lambda$ is a scaling factor that you can choose.
Why are we doing this? One way that investors evaluate a fund is by how well it tracks its index. The fund is still expected to deviate from the index within a certain range in order to improve fund performance. A way for a fund to track the performance of its benchmark is by keeping its asset weights similar to the weights of the index. We’d expect that if the fund has the same stocks as the benchmark, and also the same weights for each stock as the benchmark, the fund would yield about the same returns as the benchmark. By minimizing a linear combination of both the portfolio risk and distance between portfolio and benchmark weights, we attempt to balance the desire to minimize portfolio variance with the goal of tracking the index.
## Covariance
Implement `get_covariance_returns` to calculate the covariance of the `returns`. We'll use this to calculate the portfolio variance.
If we have $m$ stock series, the covariance matrix is an $m \times m$ matrix containing the covariance between each pair of stocks. We can use [numpy.cov](https://docs.scipy.org/doc/numpy/reference/generated/numpy.cov.html) to get the covariance. We give it a 2D array in which each row is a stock series, and each column is an observation at the same period of time.
The covariance matrix $\mathbf{P} =
\begin{bmatrix}
\sigma^2_{1,1} & ... & \sigma^2_{1,m} \\
... & ... & ...\\
\sigma_{m,1} & ... & \sigma^2_{m,m} \\
\end{bmatrix}$
```
def get_covariance_returns(returns):
"""
Calculate covariance matrices.
Parameters
----------
returns : DataFrame
Returns for each ticker and date
Returns
-------
returns_covariance : 2 dimensional Ndarray
The covariance of the returns
"""
#TODO: Implement function
return np.cov(returns.T.fillna(0))
project_tests.test_get_covariance_returns(get_covariance_returns)
```
### View Data
Let's look at the covariance generated from `get_covariance_returns`.
```
covariance_returns = get_covariance_returns(returns)
covariance_returns = pd.DataFrame(covariance_returns, returns.columns, returns.columns)
covariance_returns_correlation = np.linalg.inv(np.diag(np.sqrt(np.diag(covariance_returns))))
covariance_returns_correlation = pd.DataFrame(
covariance_returns_correlation.dot(covariance_returns).dot(covariance_returns_correlation),
covariance_returns.index,
covariance_returns.columns)
project_helper.plot_covariance_returns_correlation(
covariance_returns_correlation,
'Covariance Returns Correlation Matrix')
```
### portfolio variance
We can write the portfolio variance $\sigma^2_p = \mathbf{x^T} \mathbf{P} \mathbf{x}$
Recall that the $\mathbf{x^T} \mathbf{P} \mathbf{x}$ is called the quadratic form.
We can use the cvxpy function `quad_form(x,P)` to get the quadratic form.
### Distance from index weights
We want portfolio weights that track the index closely. So we want to minimize the distance between them.
Recall from the Pythagorean theorem that you can get the distance between two points in an x,y plane by adding the square of the x and y distances and taking the square root. Extending this to any number of dimensions is called the L2 norm. So: $\sqrt{\sum_{1}^{n}(weight_i - indexWeight_i)^2}$ Can also be written as $\left \| \mathbf{x} - \mathbf{index} \right \|_2$. There's a cvxpy function called [norm()](https://www.cvxpy.org/api_reference/cvxpy.atoms.other_atoms.html#norm)
`norm(x, p=2, axis=None)`. The default is already set to find an L2 norm, so you would pass in one argument, which is the difference between your portfolio weights and the index weights.
### objective function
We want to minimize both the portfolio variance and the distance of the portfolio weights from the index weights.
We also want to choose a `scale` constant, which is $\lambda$ in the expression.
$\mathbf{x^T} \mathbf{P} \mathbf{x} + \lambda \left \| \mathbf{x} - \mathbf{index} \right \|_2$
This lets us choose how much priority we give to minimizing the difference from the index, relative to minimizing the variance of the portfolio. If you choose a higher value for `scale` ($\lambda$).
We can find the objective function using cvxpy `objective = cvx.Minimize()`. Can you guess what to pass into this function?
### constraints
We can also define our constraints in a list. For example, you'd want the weights to sum to one. So $\sum_{1}^{n}x = 1$. You may also need to go long only, which means no shorting, so no negative weights. So $x_i >0 $ for all $i$. you could save a variable as `[x >= 0, sum(x) == 1]`, where x was created using `cvx.Variable()`.
### optimization
So now that we have our objective function and constraints, we can solve for the values of $\mathbf{x}$.
cvxpy has the constructor `Problem(objective, constraints)`, which returns a `Problem` object.
The `Problem` object has a function solve(), which returns the minimum of the solution. In this case, this is the minimum variance of the portfolio.
It also updates the vector $\mathbf{x}$.
We can check out the values of $x_A$ and $x_B$ that gave the minimum portfolio variance by using `x.value`
```
import cvxpy as cvx
def get_optimal_weights(covariance_returns, index_weights, scale=2.0):
"""
Find the optimal weights.
Parameters
----------
covariance_returns : 2 dimensional Ndarray
The covariance of the returns
index_weights : Pandas Series
Index weights for all tickers at a period in time
scale : int
The penalty factor for weights the deviate from the index
Returns
-------
x : 1 dimensional Ndarray
The solution for x
"""
assert len(covariance_returns.shape) == 2
assert len(index_weights.shape) == 1
assert covariance_returns.shape[0] == covariance_returns.shape[1] == index_weights.shape[0]
#TODO: Implement function
x = cvx.Variable(len(index_weights))
objective = cvx.Minimize(cvx.quad_form(x, covariance_returns) + scale*cvx.norm(x - index_weights, 2))
constraints = [
x >= 0,
sum(x) == 1]
cvx.Problem(objective, constraints).solve()
return x.value
project_tests.test_get_optimal_weights(get_optimal_weights)
```
## Optimized Portfolio
Using the `get_optimal_weights` function, let's generate the optimal ETF weights without rebalanceing. We can do this by feeding in the covariance of the entire history of data. We also need to feed in a set of index weights. We'll go with the average weights of the index over time.
```
raw_optimal_single_rebalance_etf_weights = get_optimal_weights(covariance_returns.values, index_weights.iloc[-1])
optimal_single_rebalance_etf_weights = pd.DataFrame(
np.tile(raw_optimal_single_rebalance_etf_weights, (len(returns.index), 1)),
returns.index,
returns.columns)
```
With our ETF weights built, let's compare it to the index. Run the next cell to calculate the ETF returns and compare it to the index returns.
```
optim_etf_returns = generate_weighted_returns(returns, optimal_single_rebalance_etf_weights)
optim_etf_cumulative_returns = calculate_cumulative_returns(optim_etf_returns)
project_helper.plot_benchmark_returns(index_weighted_cumulative_returns, optim_etf_cumulative_returns, 'Optimized ETF vs Index')
optim_etf_tracking_error = tracking_error(np.sum(index_weighted_returns, 1), np.sum(optim_etf_returns, 1))
print('Optimized ETF Tracking Error: {}'.format(optim_etf_tracking_error))
```
## Rebalance Portfolio Over Time
The single optimized ETF portfolio used the same weights for the entire history. This might not be the optimal weights for the entire period. Let's rebalance the portfolio over the same period instead of using the same weights. Implement `rebalance_portfolio` to rebalance a portfolio.
Reblance the portfolio every n number of days, which is given as `shift_size`. When rebalancing, you should look back a certain number of days of data in the past, denoted as `chunk_size`. Using this data, compute the optoimal weights using `get_optimal_weights` and `get_covariance_returns`.
```
def rebalance_portfolio(returns, index_weights, shift_size, chunk_size):
"""
Get weights for each rebalancing of the portfolio.
Parameters
----------
returns : DataFrame
Returns for each ticker and date
index_weights : DataFrame
Index weight for each ticker and date
shift_size : int
The number of days between each rebalance
chunk_size : int
The number of days to look in the past for rebalancing
Returns
-------
all_rebalance_weights : list of Ndarrays
The ETF weights for each point they are rebalanced
"""
assert returns.index.equals(index_weights.index)
assert returns.columns.equals(index_weights.columns)
assert shift_size > 0
assert chunk_size >= 0
#TODO: Implement function
all_rebalance_weights = []
for shift in range(chunk_size, len(returns), shift_size):
start_idx = shift - chunk_size
covariance_returns = get_covariance_returns(returns.iloc[start_idx:shift])
all_rebalance_weights.append(get_optimal_weights(covariance_returns, index_weights.iloc[shift-1]))
return all_rebalance_weights
project_tests.test_rebalance_portfolio(rebalance_portfolio)
```
Run the following cell to rebalance the portfolio using `rebalance_portfolio`.
```
chunk_size = 250
shift_size = 5
all_rebalance_weights = rebalance_portfolio(returns, index_weights, shift_size, chunk_size)
```
## Portfolio Turnover
With the portfolio rebalanced, we need to use a metric to measure the cost of rebalancing the portfolio. Implement `get_portfolio_turnover` to calculate the annual portfolio turnover. We'll be using the formulas used in the classroom:
$ AnnualizedTurnover =\frac{SumTotalTurnover}{NumberOfRebalanceEvents} * NumberofRebalanceEventsPerYear $
$ SumTotalTurnover =\sum_{t,n}{\left | x_{t,n} - x_{t+1,n} \right |} $ Where $ x_{t,n} $ are the weights at time $ t $ for equity $ n $.
$ SumTotalTurnover $ is just a different way of writing $ \sum \left | x_{t_1,n} - x_{t_2,n} \right | $
```
def get_portfolio_turnover(all_rebalance_weights, shift_size, rebalance_count, n_trading_days_in_year=252):
"""
Calculage portfolio turnover.
Parameters
----------
all_rebalance_weights : list of Ndarrays
The ETF weights for each point they are rebalanced
shift_size : int
The number of days between each rebalance
rebalance_count : int
Number of times the portfolio was rebalanced
n_trading_days_in_year: int
Number of trading days in a year
Returns
-------
portfolio_turnover : float
The portfolio turnover
"""
assert shift_size > 0
assert rebalance_count > 0
#TODO: Implement function
all_rebalance_weights_df = pd.DataFrame(np.array(all_rebalance_weights))
rebalance_total = (all_rebalance_weights_df - all_rebalance_weights_df.shift(-1)).abs().sum().sum()
rebalance_avg = rebalance_total / rebalance_count
rebanaces_per_year = n_trading_days_in_year / shift_size
return rebalance_avg * rebanaces_per_year
project_tests.test_get_portfolio_turnover(get_portfolio_turnover)
```
Run the following cell to get the portfolio turnover from `get_portfolio turnover`.
```
print(get_portfolio_turnover(all_rebalance_weights, shift_size, returns.shape[1]))
```
That's it! You've built a smart beta portfolio in part 1 and did portfolio optimization in part 2. You can now submit your project.
## Submission
Now that you're done with the project, it's time to submit it. Click the submit button in the bottom right. One of our reviewers will give you feedback on your project with a pass or not passed grade. You can continue to the next section while you wait for feedback.
|
github_jupyter
|
# 作業 : (Kaggle)鐵達尼生存預測
https://www.kaggle.com/c/titanic
# [作業目標]
- 試著調整特徵篩選的門檻值, 觀察會有什麼影響效果
# [作業重點]
- 調整相關係數過濾法的篩選門檻, 看看篩選結果的影響 (In[5]~In[8], Out[5]~Out[8])
- 調整L1 嵌入法篩選門檻, 看看篩選結果的影響 (In[9]~In[11], Out[9]~Out[11])
```
# 做完特徵工程前的所有準備 (與前範例相同)
import pandas as pd
import numpy as np
import copy
from sklearn.preprocessing import LabelEncoder, MinMaxScaler
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
data_path = '../../data/'
df = pd.read_csv(data_path + 'titanic_train.csv')
train_Y = df['Survived']
df = df.drop(['PassengerId'] , axis=1)
df.head()
%matplotlib inline
# 計算df整體相關係數, 並繪製成熱圖
import seaborn as sns
import matplotlib.pyplot as plt
corr = df.corr()
sns.heatmap(corr)
plt.show()
# 記得刪除 Survived
df = df.drop(['Survived'] , axis=1)
#只取 int64, float64 兩種數值型欄位, 存於 num_features 中
num_features = []
for dtype, feature in zip(df.dtypes, df.columns):
if dtype == 'float64' or dtype == 'int64':
num_features.append(feature)
print(f'{len(num_features)} Numeric Features : {num_features}\n')
# 削減文字型欄位, 只剩數值型欄位
df = df[num_features]
df = df.fillna(-1)
MMEncoder = MinMaxScaler()
df.head()
```
# 作業1
* 鐵達尼生存率預測中,試著變更兩種以上的相關係數門檻值,觀察預測能力是否提升?
```
# 原始特徵 + 邏輯斯迴歸
train_X = MMEncoder.fit_transform(df.astype(np.float64))
estimator = LogisticRegression(solver='lbfgs')
cross_val_score(estimator, train_X, train_Y, cv=5).mean()
# 篩選相關係數1
high_list = list(corr[(corr['Survived']>0.1) | (corr['Survived']<-0.1)].index)
high_list.remove('Survived')
print(high_list)
# 特徵1 + 邏輯斯迴歸
train_X = MMEncoder.fit_transform(df[high_list].astype(np.float64))
cross_val_score(estimator, train_X, train_Y, cv=5).mean()
# 篩選相關係數2
"""
Your Code Here
"""
high_list = list(corr[(corr['Survived']<0.1) | (corr['Survived']>-0.1)].index)
high_list.remove('Survived')
print(high_list)
# 特徵2 + 邏輯斯迴歸
train_X = MMEncoder.fit_transform(df[high_list].astype(np.float64))
cross_val_score(estimator, train_X, train_Y, cv=5).mean()
```
# 作業2
* 續上題,使用 L1 Embedding 做特徵選擇(自訂門檻),觀察預測能力是否提升?
```
from sklearn.linear_model import Lasso
"""
Your Code Here, select parameter alpha
"""
L1_Reg = Lasso(alpha=0.003)
train_X = MMEncoder.fit_transform(df.astype(np.float64))
L1_Reg.fit(train_X, train_Y)
L1_Reg.coef_
from itertools import compress
L1_mask = list((L1_Reg.coef_>0) | (L1_Reg.coef_<0))
L1_list = list(compress(list(df), list(L1_mask)))
L1_list
# L1_Embedding 特徵 + 線性迴歸
train_X = MMEncoder.fit_transform(df[L1_list].astype(np.float64))
cross_val_score(estimator, train_X, train_Y, cv=5).mean()
```
|
github_jupyter
|
<script async src="https://www.googletagmanager.com/gtag/js?id=UA-59152712-8"></script>
<script>
window.dataLayer = window.dataLayer || [];
function gtag(){dataLayer.push(arguments);}
gtag('js', new Date());
gtag('config', 'UA-59152712-8');
</script>
# Convert LaTeX Sentence to SymPy Expression
## Author: Ken Sible
## The following module will demonstrate a recursive descent parser for LaTeX.
### NRPy+ Source Code for this module:
1. [latex_parser.py](../edit/latex_parser.py); [\[**tutorial**\]](Tutorial-LaTeX_SymPy_Conversion.ipynb) The latex_parser.py script will convert a LaTeX sentence to a SymPy expression using the following function: parse(sentence).
<a id='toc'></a>
# Table of Contents
$$\label{toc}$$
1. [Step 1](#intro): Introduction: Lexical Analysis and Syntax Analysis
1. [Step 2](#sandbox): Demonstration and Sandbox (LaTeX Parser)
1. [Step 3](#tensor): Tensor Support with Einstein Notation (WIP)
1. [Step 4](#latex_pdf_output): $\LaTeX$ PDF Output
<a id='intro'></a>
# Step 1: Lexical Analysis and Syntax Analysis \[Back to [top](#toc)\]
$$\label{intro}$$
In the following section, we discuss [lexical analysis](https://en.wikipedia.org/wiki/Lexical_analysis) (lexing) and [syntax analysis](https://en.wikipedia.org/wiki/Parsing) (parsing). In the process of lexical analysis, a lexer will tokenize a character string, called a sentence, using substring pattern matching (or tokenizing). We implemented a regex-based lexer for NRPy+, which does pattern matching using a [regular expression](https://en.wikipedia.org/wiki/Regular_expression) for each token pattern. In the process of syntax analysis, a parser will receive a token iterator from the lexer and build a parse tree containing all syntactic information of the language, as specified by a [formal grammar](https://en.wikipedia.org/wiki/Formal_grammar). We implemented a [recursive descent parser](https://en.wikipedia.org/wiki/Recursive_descent_parser) for NRPy+, which will build a parse tree in [preorder](https://en.wikipedia.org/wiki/Tree_traversal#Pre-order_(NLR)), starting from the root [nonterminal](https://en.wikipedia.org/wiki/Terminal_and_nonterminal_symbols), using a [right recursive](https://en.wikipedia.org/wiki/Left_recursion) grammar. The following right recursive, [context-free grammar](https://en.wikipedia.org/wiki/Context-free_grammar) was written for parsing [LaTeX](https://en.wikipedia.org/wiki/LaTeX), adhering to the canonical (extended) [BNF](https://en.wikipedia.org/wiki/Backus%E2%80%93Naur_form) notation used for describing a context-free grammar:
```
<ROOT> -> <EXPRESSION> | <STRUCTURE> { <LINE_BREAK> <STRUCTURE> }*
<STRUCTURE> -> <CONFIG> | <ENVIROMENT> | <ASSIGNMENT>
<ENVIROMENT> -> <BEGIN_ALIGN> <ASSIGNMENT> { <LINE_BREAK> <ASSIGNMENT> }* <END_ALIGN>
<ASSIGNMENT> -> <VARIABLE> = <EXPRESSION>
<EXPRESSION> -> <TERM> { ( '+' | '-' ) <TERM> }*
<TERM> -> <FACTOR> { [ '/' ] <FACTOR> }*
<FACTOR> -> <BASE> { '^' <EXPONENT> }*
<BASE> -> [ '-' ] ( <ATOM> | '(' <EXPRESSION> ')' | '[' <EXPRESSION> ']' )
<EXPONENT> -> <BASE> | '{' <BASE> '}'
<ATOM> -> <VARIABLE> | <NUMBER> | <COMMAND>
<VARIABLE> -> <ARRAY> | <SYMBOL> [ '_' ( <SYMBOL> | <INTEGER> ) ]
<NUMBER> -> <RATIONAL> | <DECIMAL> | <INTEGER>
<COMMAND> -> <SQRT> | <FRAC>
<SQRT> -> '\\sqrt' [ '[' <INTEGER> ']' ] '{' <EXPRESSION> '}'
<FRAC> -> '\\frac' '{' <EXPRESSION> '}' '{' <EXPRESSION> '}'
<CONFIG> -> '%' <ARRAY> '[' <INTEGER> ']' [ ':' <SYMMETRY> ] { ',' <ARRAY> '[' <INTEGER> ']' [ ':' <SYMMETRY> ] }*
<ARRAY> -> ( <SYMBOL | <TENSOR> )
[ '_' ( <SYMBOL> | '{' { <SYMBOL> }+ '}' ) [ '^' ( <SYMBOL> | '{' { <SYMBOL> }+ '}' ) ]
| '^' ( <SYMBOL> | '{' { <SYMBOL> }+ '}' ) [ '_' ( <SYMBOL> | '{' { <SYMBOL> }+ '}' ) ] ]
```
<small>**Source**: Robert W. Sebesta. Concepts of Programming Languages. Pearson Education Limited, 2016.</small>
```
from latex_parser import * # Import NRPy+ module for lexing and parsing LaTeX
from sympy import srepr # Import SymPy function for expression tree representation
lexer = Lexer(); lexer.initialize(r'\sqrt{5}(x + 2/3)^2')
print(', '.join(token for token in lexer.tokenize()))
expr = parse(r'\sqrt{5}(x + 2/3)^2', expression=True)
print(expr, ':', srepr(expr))
```
#### `Grammar Derivation: (x + 2/3)^2`
```
<EXPRESSION> -> <TERM>
-> <FACTOR>
-> <BASE>^<EXPONENT>
-> (<EXPRESSION>)^<EXPONENT>
-> (<TERM> + <TERM>)^<EXPONENT>
-> (<FACTOR> + <TERM>)^<EXPONENT>
-> (<BASE> + <TERM>)^<EXPONENT>
-> (<ATOM> + <TERM>)^<EXPONENT>
-> (<VARIABLE> + <TERM>)^<EXPONENT>
-> (<SYMBOL> + <TERM>)^<EXPONENT>
-> (x + <TERM>)^<EXPONENT>
-> (x + <FACTOR>)^<EXPONENT>
-> (x + <BASE>)^<EXPONENT>
-> (x + <ATOM>)^<EXPONENT>
-> (x + <NUMBER>)^<EXPONENT>
-> (x + <RATIONAL>)^<EXPONENT>
-> (x + 2/3)^<EXPONENT>
-> (x + 2/3)^<BASE>
-> (x + 2/3)^<ATOM>
-> (x + 2/3)^<NUMBER>
-> (x + 2/3)^<INTEGER>
-> (x + 2/3)^2
```
<a id='sandbox'></a>
# Step 2: Demonstration and Sandbox (LaTeX Parser) \[Back to [top](#toc)\]
$$\label{sandbox}$$
We implemented a wrapper function for the `parse()` method that will accept a LaTeX sentence and return a SymPy expression. Furthermore, the entire parsing module was designed for extendibility. We apply the following procedure for extending parser functionality to include an unsupported LaTeX command: append that command to the grammar dictionary in the Lexer class with the mapping regex:token, write a grammar abstraction (similar to a regular expression) for that command, add the associated nonterminal (the command name) to the command abstraction in the Parser class, and finally implement the straightforward (private) method for parsing the grammar abstraction. We shall demonstrate the extension procedure using the `\sqrt` LaTeX command.
```<SQRT> -> '\\sqrt' [ '[' <INTEGER> ']' ] '{' <EXPRESSION> '}'```
```
def _sqrt(self):
if self.accept('LEFT_BRACKET'):
integer = self.lexer.lexeme
self.expect('INTEGER')
root = Rational(1, integer)
self.expect('RIGHT_BRACKET')
else: root = Rational(1, 2)
self.expect('LEFT_BRACE')
expr = self.__expr()
self.expect('RIGHT_BRACE')
return Pow(expr, root)
```
```
print(parse(r'\sqrt[3]{\alpha_0}', expression=True))
```
In addition to expression parsing, we included support for equation parsing, which will produce a dictionary mapping LHS $\mapsto$ RHS, where LHS must be a symbol, and insert that mapping into the global namespace of the previous stack frame, as demonstrated below.
```
parse(r'x = n\sqrt{2}^n'); print(x)
```
We implemented robust error messaging using the custom `ParseError` exception, which should handle every conceivable case to identify, as detailed as possible, invalid syntax inside of a LaTeX sentence. The following are runnable examples of possible error messages (simply uncomment and run the cell):
```
# parse(r'\sqrt[*]{2}')
# ParseError: \sqrt[*]{2}
# ^
# unexpected '*' at position 6
# parse(r'\sqrt[0.5]{2}')
# ParseError: \sqrt[0.5]{2}
# ^
# expected token INTEGER at position 6
# parse(r'\command{}')
# ParseError: \command{}
# ^
# unsupported command '\command' at position 0
from warnings import filterwarnings # Import Python function for warning suppression
filterwarnings('ignore', category=OverrideWarning); del Parser.namespace['x']
```
In the sandbox code cell below, you can experiment with the LaTeX parser using the wrapper function `parse(sentence)`, where sentence must be a [raw string](https://docs.python.org/3/reference/lexical_analysis.html) to interpret a backslash as a literal character rather than an [escape sequence](https://en.wikipedia.org/wiki/Escape_sequence).
```
# Write Sandbox Code Here
```
<a id='tensor'></a>
# Step 3: Tensor Support with Einstein Notation (WIP) \[Back to [top](#toc)\]
$$\label{tensor}$$
In the following section, we demonstrate the current parser support for tensor notation using the Einstein summation convention. The first example will parse an equation for a tensor contraction, the second will parse an equation for raising an index using the metric tensor, and the third will parse an align enviroment with an equation dependency. In each example, every tensor should appear either on the LHS of an equation or inside of a configuration before appearing on the RHS of an equation. Moreover, the parser will raise an exception upon violation of the Einstein summation convention, i.e. an invalid free or bound index.
**Configuration Syntax** `% <TENSOR> [<DIMENSION>]: <SYMMETRY>, <TENSOR> [<DIMENSION>]: <SYMMETRY>, ... ;`
#### Example 1
LaTeX Source | Rendered LaTeX
:----------- | :-------------
<pre lang="latex"> h = h^\\mu{}_\\mu </pre> | $$ h = h^\mu{}_\mu $$
```
parse(r"""
% h^\mu_\mu [4]: nosym;
h = h^\mu{}_\mu
""")
print('h =', h)
```
#### Example 2
LaTeX Source | Rendered LaTeX
:----------- | :-------------
<pre lang="latex"> v^\\mu = g^{\\mu\\nu}v_\\nu </pre> | $$ v^\mu = g^{\mu\nu}v_\nu $$
```
parse(r"""
% g^{\mu\nu} [3]: metric, v_\nu [3];
v^\mu = g^{\mu\nu}v_\nu
""")
print('vU =', vU)
```
#### Example 3
LaTeX Source | Rendered LaTeX
:----------- | :-------------
<pre lang="latex"> \\begin{align\*}<br>    R &= g_{ab}R^{ab} \\\\ <br>    G^{ab} &= R^{ab} - \\frac{1}{2}g^{ab}R <br> \\end{align\*} </pre> | $$ \begin{align*} R &= g_{ab}R^{ab} \\ G^{ab} &= R^{ab} - \frac{1}{2}g^{ab}R \end{align*} $$
```
parse(r"""
% g_{ab} [2]: metric, R^{ab} [2]: sym01;
\begin{align*}
R &= g_{ab}R^{ab} \\
G^{ab} &= R^{ab} - \frac{1}{2}g^{ab}R
\end{align*}
""")
print('R =', R)
display(GUU)
```
The static variable `namespace` for the `Parser` class will provide access to the global namespace of the parser across each instance of the class.
```
Parser.namespace
```
We extended our robust error messaging using the custom `TensorError` exception, which should handle any inconsistent tensor dimension and any violation of the Einstein summation convention, specifically that a bound index must appear exactly once as a superscript and exactly once as a subscript in any single term and that a free index must appear in every term with the same position and cannot be summed over in any term. The following are runnable examples of possible error messages (simply uncomment and run the cell):
```
# parse(r"""
# % h^{\mu\mu}_{\mu\mu} [4]: nosym;
# h = h^{\mu\mu}_{\mu\mu}
# """)
# TensorError: illegal bound index
# parse(r"""
# % g^\mu_\nu [3]: sym01, v_\nu [3];
# v^\mu = g^\mu_\nu v_\nu
# """)
# TensorError: illegal bound index
# parse(r"""
# % g^{\mu\nu} [3]: sym01, v_\mu [3], w_\nu [3];
# u^\mu = g^{\mu\nu}(v_\mu + w_\nu)
# """)
# TensorError: unbalanced free index
```
<a id='latex_pdf_output'></a>
# Step 4: Output this notebook to $\LaTeX$-formatted PDF file \[Back to [top](#toc)\]
$$\label{latex_pdf_output}$$
The following code cell converts this Jupyter notebook into a proper, clickable $\LaTeX$-formatted PDF file. After the cell is successfully run, the generated PDF may be found in the root NRPy+ tutorial directory, with filename
[Tutorial-LaTeX_SymPy_Conversion.pdf](Tutorial-LaTeX_SymPy_Conversion.pdf) (Note that clicking on this link may not work; you may need to open the PDF file through another means.)
```
import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface
cmd.output_Jupyter_notebook_to_LaTeXed_PDF("Tutorial-LaTeX_SymPy_Conversion")
```
|
github_jupyter
|
# Analyzing data with Pandas
First a little setup. Importing the pandas library as ```pd```
```
import pandas as pd
```
Set some helpful display options. Uncomment the boilerplate in this cell.
```
%matplotlib inline
pd.set_option("max_columns", 150)
pd.set_option('max_colwidth',40)
pd.options.display.float_format = '{:,.2f}'.format
```
open and read in the Master.csv and Salaries.csv tables in the ```data/2017/``` directory
```
master = pd.read_csv('../project3/data/2017/Master.csv') # File with player details
salary = pd.read_csv('../project3/data/2017/Salaries.csv') #File with baseball players' salaries
```
check to see what type each object is with `print(table_name)`. You can also use the ```.info()``` method to explore the data's structure.
```
master.info()
salary.info()
```
print out sample data for each table with `table.head()`<br>
see additional options by pressing `tab` after you type the `head()` method
```
master.head()
salary.head()
```
Now we join the two csv's using `pd.merge`.<br>
We want to keep all the players names in the `master` data set<br>
even if their salary is missing from the `salary` data set.<br>
We can always filter the NaN values out later
```
joined = pd.merge(left=master, right=salary, how="left")
```
see what columns the `joined` table contains
```
joined.info()
```
check if all the players have a salary assigned. The easiest way is to deduct the length of the `joined` table from the `master` table
```
len(master) - len(joined)
```
Something went wrong. There are now more players in the `joined` data set than in the `master` data set.<br>
Some entries probably got duplicated<br>
Let's check if we have duplicate `playerIDs` by using `.value_counts()`
```
joined["playerID"].value_counts()
```
Yep, we do.<br>
Let's filter out an arbitrary player to see why there is duplication
```
joined[joined["playerID"] == "moyerja01"]
```
As we can see, there are now salaries in the dataset for each year of the players carreer.<br>
We only want to have the most recent salary though.<br>
We therefore need to 'deduplicate' the data set.
But first, let's make sure we get the newest year. We can do this by sorting the data on the newest entry
```
joined = joined.sort_values(["playerID","yearID"])
```
Now we deduplicate
```
deduplicated = joined.drop_duplicates("playerID", keep="last")
```
And let's do the check again
```
len(master) - len(deduplicated)
```
Now we van get into the interesting part: analysis!
## What is the average (mean, median, max, min) salary?
```
deduplicated["salary"].describe()
```
## Who makes the most money?
```
max_salary = deduplicated["salary"].max()
deduplicated[deduplicated["salary"] == max_salary]
```
## What are the most common baseball players salaries?
Draw a histogram. <br>
```
deduplicated.hist("salary")
```
We can do the same with the column `yearID` to see how recent our data is.<br>
We have 30 years in our data set, so we need to do some minor tweaking
```
deduplicated.hist("yearID", bins=30)
```
## Who are the top 10% highest-paid players?
calculate the 90 percentile cutoff
```
top_10_p = deduplicated["salary"].quantile(q=0.9)
top_10_p
```
filter out players that make more money than the cutoff
```
best_paid = deduplicated[deduplicated["salary"] >= top_10_p]
best_paid
```
use the `nlargest` to see the top 10 best paid players
```
best_paid_top_10 = best_paid.nlargest(10, "salary")
best_paid_top_10
```
draw a chart
```
best_paid_top_10.plot(kind="barh", x="nameLast", y="salary")
```
save the data
```
best_paid.to_csv('highest-paid.csv', index=False)
```
|
github_jupyter
|
```
library(caret, quiet=TRUE);
library(base64enc)
library(httr, quiet=TRUE)
```
# Build a Model
```
set.seed(1960)
create_model = function() {
model <- train(Species ~ ., data = iris, method = "ctree2")
return(model)
}
# dataset
model = create_model()
# pred <- predict(model, as.matrix(iris[, -5]) , type="prob")
pred_labels <- predict(model, as.matrix(iris[, -5]) , type="raw")
sum(pred_labels != iris$Species)/length(pred_labels)
```
# SQL Code Generation
```
test_ws_sql_gen = function(mod) {
WS_URL = "https://sklearn2sql.herokuapp.com/model"
WS_URL = "http://localhost:1888/model"
model_serialized <- serialize(mod, NULL)
b64_data = base64encode(model_serialized)
data = list(Name = "caret_rpart_test_model", SerializedModel = b64_data , SQLDialect = "postgresql" , Mode="caret")
r = POST(WS_URL, body = data, encode = "json")
# print(r)
content = content(r)
# print(content)
lSQL = content$model$SQLGenrationResult[[1]]$SQL # content["model"]["SQLGenrationResult"][0]["SQL"]
return(lSQL);
}
lModelSQL = test_ws_sql_gen(model)
cat(lModelSQL)
```
# Execute the SQL Code
```
library(RODBC)
conn = odbcConnect("pgsql", uid="db", pwd="db", case="nochange")
odbcSetAutoCommit(conn , autoCommit = TRUE)
dataset = iris[,-5]
df_sql = as.data.frame(dataset)
names(df_sql) = sprintf("Feature_%d",0:(ncol(df_sql)-1))
df_sql$KEY = seq.int(nrow(dataset))
sqlDrop(conn , "INPUT_DATA" , errors = FALSE)
sqlSave(conn, df_sql, tablename = "INPUT_DATA", verbose = FALSE)
head(df_sql)
# colnames(df_sql)
# odbcGetInfo(conn)
# sqlTables(conn)
df_sql_out = sqlQuery(conn, lModelSQL)
head(df_sql_out)
```
# R Caret Rpart Output
```
# pred_proba = predict(model, as.matrix(iris[,-5]), type = "prob")
df_r_out = data.frame(seq.int(nrow(dataset))) # (pred_proba)
names(df_r_out) = c("KEY") # sprintf("Proba_%s",model$levels)
df_r_out$KEY = seq.int(nrow(dataset))
df_r_out$Score_setosa = NA
df_r_out$Score_versicolor = NA
df_r_out$Score_virginica = NA
df_r_out$Proba_setosa = NA
df_r_out$Proba_versicolor = NA
df_r_out$Proba_virginica = NA
df_r_out$LogProba_setosa = log(df_r_out$Proba_setosa)
df_r_out$LogProba_versicolor = log(df_r_out$Proba_versicolor)
df_r_out$LogProba_virginica = log(df_r_out$Proba_virginica)
df_r_out$Decision = predict(model, as.matrix(iris[,-5]), type = "raw")
df_r_out$DecisionProba = NA
head(df_r_out)
```
# Compare R and SQL output
```
df_merge = merge(x = df_r_out, y = df_sql_out, by = "KEY", all = TRUE, , suffixes = c("_1","_2"))
head(df_merge)
diffs_df = df_merge[df_merge$Decision_1 != df_merge$Decision_2,]
head(diffs_df)
stopifnot(nrow(diffs_df) == 0)
summary(df_r_out)
summary(df_sql_out)
```
|
github_jupyter
|
# Predicting NYC Taxi Fares with RAPIDS
Process 380 million rides in NYC from 2015-2017.
RAPIDS is a suite of GPU accelerated data science libraries with APIs that should be familiar to users of Pandas, Dask, and Scikitlearn.
This notebook focuses on showing how to use cuDF with Dask & XGBoost to scale GPU DataFrame ETL-style operations & model training out to multiple GPUs.
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import cupy
import cudf
import dask
import dask_cudf
import xgboost as xgb
from dask.distributed import Client, wait
from dask.utils import parse_bytes
from dask_cuda import LocalCUDACluster
cluster = LocalCUDACluster(rmm_pool_size=parse_bytes("25 GB"),
scheduler_port=9888,
dashboard_address=9787,
)
client = Client(cluster)
client
```
# Inspecting the Data
Now that we have a cluster of GPU workers, we'll use [dask-cudf](https://github.com/rapidsai/dask-cudf/) to load and parse a bunch of CSV files into a distributed DataFrame.
```
base_path = "/raid/vjawa/nyc_taxi/data/"
import dask_cudf
df_2014 = dask_cudf.read_csv(base_path+'2014/yellow_*.csv', chunksize='256 MiB')
df_2014.head()
```
# Data Cleanup
As usual, the data needs to be massaged a bit before we can start adding features that are useful to an ML model.
For example, in the 2014 taxi CSV files, there are `pickup_datetime` and `dropoff_datetime` columns. The 2015 CSVs have `tpep_pickup_datetime` and `tpep_dropoff_datetime`, which are the same columns. One year has `rate_code`, and another `RateCodeID`.
Also, some CSV files have column names with extraneous spaces in them.
Worst of all, starting in the July 2016 CSVs, pickup & dropoff latitude and longitude data were replaced by location IDs, making the second half of the year useless to us.
We'll do a little string manipulation, column renaming, and concatenating of DataFrames to sidestep the problems.
```
#Dictionary of required columns and their datatypes
must_haves = {
'pickup_datetime': 'datetime64[s]',
'dropoff_datetime': 'datetime64[s]',
'passenger_count': 'int32',
'trip_distance': 'float32',
'pickup_longitude': 'float32',
'pickup_latitude': 'float32',
'rate_code': 'int32',
'dropoff_longitude': 'float32',
'dropoff_latitude': 'float32',
'fare_amount': 'float32'
}
def clean(ddf, must_haves):
# replace the extraneous spaces in column names and lower the font type
tmp = {col:col.strip().lower() for col in list(ddf.columns)}
ddf = ddf.rename(columns=tmp)
ddf = ddf.rename(columns={
'tpep_pickup_datetime': 'pickup_datetime',
'tpep_dropoff_datetime': 'dropoff_datetime',
'ratecodeid': 'rate_code'
})
ddf['pickup_datetime'] = ddf['pickup_datetime'].astype('datetime64[ms]')
ddf['dropoff_datetime'] = ddf['dropoff_datetime'].astype('datetime64[ms]')
for col in ddf.columns:
if col not in must_haves:
ddf = ddf.drop(columns=col)
continue
# if column was read as a string, recast as float
if ddf[col].dtype == 'object':
ddf[col] = ddf[col].str.fillna('-1')
ddf[col] = ddf[col].astype('float32')
else:
# downcast from 64bit to 32bit types
# Tesla T4 are faster on 32bit ops
if 'int' in str(ddf[col].dtype):
ddf[col] = ddf[col].astype('int32')
if 'float' in str(ddf[col].dtype):
ddf[col] = ddf[col].astype('float32')
ddf[col] = ddf[col].fillna(-1)
return ddf
```
<b> NOTE: </b>We will realize that some of 2015 data has column name as `RateCodeID` and others have `RatecodeID`. When we rename the columns in the clean function, it internally doesn't pass meta while calling map_partitions(). This leads to the error of column name mismatch in the returned data. For this reason, we will call the clean function with map_partition and pass the meta to it. Here is the link to the bug created for that: https://github.com/rapidsai/cudf/issues/5413
```
df_2014 = df_2014.map_partitions(clean, must_haves, meta=must_haves)
```
We still have 2015 and the first half of 2016's data to read and clean. Let's increase our dataset.
```
df_2015 = dask_cudf.read_csv(base_path+'2015/yellow_*.csv', chunksize='1024 MiB')
df_2015 = df_2015.map_partitions(clean, must_haves, meta=must_haves)
```
# Handling 2016's Mid-Year Schema Change
In 2016, only January - June CSVs have the columns we need. If we try to read base_path+2016/yellow_*.csv, Dask will not appreciate having differing schemas in the same DataFrame.
Instead, we'll need to create a list of the valid months and read them independently.
```
months = [str(x).rjust(2, '0') for x in range(1, 7)]
valid_files = [base_path+'2016/yellow_tripdata_2016-'+month+'.csv' for month in months]
#read & clean 2016 data and concat all DFs
df_2016 = dask_cudf.read_csv(valid_files, chunksize='512 MiB').map_partitions(clean, must_haves, meta=must_haves)
#concatenate multiple DataFrames into one bigger one
taxi_df = dask.dataframe.multi.concat([df_2014, df_2015, df_2016])
```
## Exploratory Data Analysis (EDA)
Here, we are checking out if there are any non-sensical records and outliers, and in such case, we need to remove them from the dataset.
```
# check out if there is any negative total trip time
taxi_df[taxi_df.dropoff_datetime <= taxi_df.pickup_datetime].head()
# check out if there is any abnormal data where trip distance is short, but the fare is very high.
taxi_df[(taxi_df.trip_distance < 10) & (taxi_df.fare_amount > 300)].head()
# check out if there is any abnormal data where trip distance is long, but the fare is very low.
taxi_df[(taxi_df.trip_distance > 50) & (taxi_df.fare_amount < 50)].head()
```
EDA visuals and additional analysis yield the filter logic below.
```
# apply a list of filter conditions to throw out records with missing or outlier values
query_frags = [
'fare_amount > 1 and fare_amount < 500',
'passenger_count > 0 and passenger_count < 6',
'pickup_longitude > -75 and pickup_longitude < -73',
'dropoff_longitude > -75 and dropoff_longitude < -73',
'pickup_latitude > 40 and pickup_latitude < 42',
'dropoff_latitude > 40 and dropoff_latitude < 42',
'trip_distance > 0 and trip_distance < 500',
'not (trip_distance > 50 and fare_amount < 50)',
'not (trip_distance < 10 and fare_amount > 300)',
'not dropoff_datetime <= pickup_datetime'
]
taxi_df = taxi_df.query(' and '.join(query_frags))
# reset_index and drop index column
taxi_df = taxi_df.reset_index(drop=True)
taxi_df.head()
```
# Adding Interesting Features
Dask & cuDF provide standard DataFrame operations, but also let you run "user defined functions" on the underlying data. Here we use [dask.dataframe's map_partitions](https://docs.dask.org/en/latest/dataframe-api.html#dask.dataframe.DataFrame.map_partitions) to apply user defined python function on each DataFrame partition.
We'll use a Haversine Distance calculation to find total trip distance, and extract additional useful variables from the datetime fields.
```
## add features
taxi_df['hour'] = taxi_df['pickup_datetime'].dt.hour
taxi_df['year'] = taxi_df['pickup_datetime'].dt.year
taxi_df['month'] = taxi_df['pickup_datetime'].dt.month
taxi_df['day'] = taxi_df['pickup_datetime'].dt.day
taxi_df['day_of_week'] = taxi_df['pickup_datetime'].dt.weekday
taxi_df['is_weekend'] = (taxi_df['day_of_week']>=5).astype('int32')
#calculate the time difference between dropoff and pickup.
taxi_df['diff'] = taxi_df['dropoff_datetime'].astype('int64') - taxi_df['pickup_datetime'].astype('int64')
taxi_df['diff']=(taxi_df['diff']/1000).astype('int64')
taxi_df['pickup_latitude_r'] = taxi_df['pickup_latitude']//.01*.01
taxi_df['pickup_longitude_r'] = taxi_df['pickup_longitude']//.01*.01
taxi_df['dropoff_latitude_r'] = taxi_df['dropoff_latitude']//.01*.01
taxi_df['dropoff_longitude_r'] = taxi_df['dropoff_longitude']//.01*.01
taxi_df = taxi_df.drop('pickup_datetime', axis=1)
taxi_df = taxi_df.drop('dropoff_datetime', axis=1)
import cupy as cp
def cudf_haversine_distance(lon1, lat1, lon2, lat2):
lon1, lat1, lon2, lat2 = map(cp.radians, [lon1, lat1, lon2, lat2])
newlon = lon2 - lon1
newlat = lat2 - lat1
haver_formula = cp.sin(newlat/2.0)**2 + cp.cos(lat1) * cp.cos(lat2) * cp.sin(newlon/2.0)**2
dist = 2 * cp.arcsin(cp.sqrt(haver_formula ))
km = 6367 * dist #6367 for distance in KM for miles use 3958
return km
def haversine_dist(df):
df['h_distance']= cudf_haversine_distance(
df['pickup_longitude'],
df['pickup_latitude'],
df['dropoff_longitude'],
df['dropoff_latitude']
)
df['h_distance']= df['h_distance'].astype('float32')
return df
taxi_df = taxi_df.map_partitions(haversine_dist)
taxi_df.head()
len(taxi_df)
%%time
taxi_df = taxi_df.persist()
x = wait(taxi_df);
```
# Pick a Training Set
Let's imagine you're making a trip to New York on the 25th and want to build a model to predict what fare prices will be like the last few days of the month based on the first part of the month. We'll use a query expression to identify the `day` of the month to use to divide the data into train and test sets.
The wall-time below represents how long it takes your GPU cluster to load data from the Google Cloud Storage bucket and the ETL portion of the workflow.
```
#since we calculated the h_distance let's drop the trip_distance column, and then do model training with XGB.
taxi_df = taxi_df.drop('trip_distance', axis=1)
# this is the original data partition for train and test sets.
X_train = taxi_df.query('day < 25')
# create a Y_train ddf with just the target variable
Y_train = X_train[['fare_amount']].persist()
# drop the target variable from the training ddf
X_train = X_train[X_train.columns.difference(['fare_amount'])].persist()
# # this wont return until all data is in GPU memory
a = wait([X_train, Y_train]);
```
# Train the XGBoost Regression Model
The wall time output below indicates how long it took your GPU cluster to train an XGBoost model over the training set.
```
dtrain = xgb.dask.DaskDMatrix(client, X_train, Y_train)
%%time
params = {
'learning_rate': 0.3,
'max_depth': 8,
'objective': 'reg:squarederror',
'subsample': 0.6,
'gamma': 1,
'silent': False,
'verbose_eval': True,
'tree_method':'gpu_hist'
}
trained_model = xgb.dask.train(
client,
params,
dtrain,
num_boost_round=12,
evals=[(dtrain, 'train')]
)
ax = xgb.plot_importance(trained_model['booster'], height=0.8, max_num_features=10, importance_type="gain")
ax.grid(False, axis="y")
ax.set_title('Estimated feature importance')
ax.set_xlabel('importance')
plt.show()
```
# How Good is Our Model?
Now that we have a trained model, we need to test it with the 25% of records we held out.
Based on the filtering conditions applied to this dataset, many of the DataFrame partitions will wind up having 0 rows. This is a problem for XGBoost which doesn't know what to do with 0 length arrays. We'll repartition the data.
```
def drop_empty_partitions(df):
lengths = df.map_partitions(len).compute()
nonempty = [length > 0 for length in lengths]
return df.partitions[nonempty]
X_test = taxi_df.query('day >= 25').persist()
X_test = drop_empty_partitions(X_test)
# Create Y_test with just the fare amount
Y_test = X_test[['fare_amount']].persist()
# Drop the fare amount from X_test
X_test = X_test[X_test.columns.difference(['fare_amount'])]
# this wont return until all data is in GPU memory
done = wait([X_test, Y_test])
# display test set size
len(X_test)
```
## Calculate Prediction
```
# generate predictions on the test set
booster = trained_model["booster"] # "Booster" is the trained model
booster.set_param({'predictor': 'gpu_predictor'})
prediction = xgb.dask.inplace_predict(client, booster, X_test).persist()
wait(prediction);
y = Y_test['fare_amount'].reset_index(drop=True)
# Calculate RMSE
squared_error = ((prediction-y)**2)
cupy.sqrt(squared_error.mean().compute())
```
## Save Trained Model for Later Use¶
We often need to store our models on a persistent filesystem for future deployment. Let's save our model.
```
trained_model
import joblib
# Save the booster to file
joblib.dump(trained_model, "xgboost-model")
len(taxi_df)
```
## Reload a Saved Model from Disk
You can also read the saved model back into a normal XGBoost model object.
```
with open("xgboost-model", 'rb') as fh:
loaded_model = joblib.load(fh)
# Generate predictions on the test set again, but this time using the reloaded model
loaded_booster = loaded_model["booster"]
loaded_booster.set_param({'predictor': 'gpu_predictor'})
new_preds = xgb.dask.inplace_predict(client, loaded_booster, X_test).persist()
# Verify that the predictions result in the same RMSE error
squared_error = ((new_preds - y)**2)
cp.sqrt(squared_error.mean().compute())
```
|
github_jupyter
|
```
#importing modules
import os
import codecs
import numpy as np
import string
import pandas as pd
```
# **Data Preprocessing**
```
#downloading and extracting the files on colab server
import urllib.request
urllib.request.urlretrieve ("https://archive.ics.uci.edu/ml/machine-learning-databases/20newsgroups-mld/20_newsgroups.tar.gz", "a.tar.gz")
import tarfile
tar = tarfile.open("a.tar.gz")
tar.extractall()
tar.close()
#making a list of all the file paths and their corresponding class
f_paths=[]
i=-1
path="20_newsgroups"
folderlist=os.listdir(path)
if ".DS_Store" in folderlist:
folderlist.remove('.DS_Store')
for folder in folderlist:
i+=1
filelist=os.listdir(path+'/'+folder)
for file in filelist:
f_paths.append((path+'/'+folder+'/'+file,i))
len(f_paths)
#splitting the list of paths into training and testing data
from sklearn import model_selection
x_train,x_test=model_selection.train_test_split(f_paths)
len(x_train),len(x_test)
#Making the lists X_train and X_test containg only the paths of the files in training and testing data
#First making lists Y_train and Y_test containing the classes of the training and testing data
X_train=[]
X_test=[]
Y_train=[]
Y_test=[]
for i in range(len(x_train)):
X_train.append(x_train[i][0])
Y_train.append(x_train[i][1])
for i in range(len(x_test)):
X_test.append(x_test[i][0])
Y_test.append(x_test[i][1])
#Transforming Y_train and Y_test into 1 dimensional np arrays
Y_train=(np.array([Y_train])).reshape(-1)
Y_test=(np.array([Y_test])).reshape(-1)
#shape of Y_train and Y_test np arrays
Y_train.shape,Y_test.shape
import nltk
nltk.download('stopwords')
from nltk.corpus import stopwords
stop=set(stopwords.words("english"))
#adding all the above lists and including punctuations to stop words
stop_words=list(stop)+list(set(string.punctuation))
len(stop_words)
#making vocabulary from the files in X_train i.e. training data
vocab={}
count =0
for filename in X_train:
count+=1
f = open(filename,'r',errors='ignore')
record=f.read()
words=record.split()
for word in words:
if len(word)>2:
if word.lower() not in stop_words:
if word.lower() in vocab:
vocab[word.lower()]+=1
else:
vocab[word.lower()]=1
f.close()
#length of the vocabulary
len(vocab)
#sorting the vocabulary on the basis of the frequency of the word
#making the sorted vocabulary
import operator
sorted_vocab = sorted(vocab.items(), key= operator.itemgetter(1), reverse= True) # sort the vocab based on frequency
#making the list feature_names containg the words with the frequency of the top 2000 words
feature_names = []
for i in range(len(sorted_vocab)):
if(sorted_vocab[2000][1] <= sorted_vocab[i][1]):
feature_names.append(sorted_vocab[i][0])
#length of the feature_names i.e. number of our features
print(len(feature_names))
#making dataframes df_train and df_test with columns having the feature names i.e. the words
df_train=pd.DataFrame(columns=feature_names)
df_test=pd.DataFrame(columns=feature_names)
count_train,count_test=0,0
#transforming each file in X_train into a row in the dataframe df_train having columns as feature names and values as the frequency of that feature name i.e that word
for filename in X_train:
count_train+=1
#adding a row of zeros for each file
df_train.loc[len(df_train)]=np.zeros(len(feature_names))
f = open(filename,'r',errors='ignore')
record=f.read()
words=record.split()
#parsing through all the words of the file
for word in words:
if word.lower() in df_train.columns:
df_train[word.lower()][len(df_train)-1]+=1 #if the word is in the column names then adding 1 to the frequency of that word in the row
f.close()
#transforming each file in X_test into a row in the dataframe df_test having columns as feature names and values as the frequency of that feature name i.e that word
for filename in X_test:
count_test+=1
#adding a row of zeros for each file
df_test.loc[len(df_test)]=np.zeros(len(feature_names))
f = open(filename,'r',errors='ignore')
record=f.read()
words=record.split()
#parsing through all the words of the file
for word in words:
if word.lower() in df_test.columns:
df_test[word.lower()][len(df_test)-1]+=1 #if the word is in the column names then adding 1 to the frequency of that word in the row
f.close()
#printing the number files tranformed in training and testing data
print(count_train,count_test)
#putting the values of the datafames into X_train and X_test
X_train=df_train.values
X_test=df_test.values
```
# **Using the inbuilt Multinomial Naive Bayes classifier from sklearn**
```
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import classification_report,confusion_matrix
clf=MultinomialNB()
#fitting the classifier on training data
clf.fit(X_train,Y_train)
#prediciting the classes of the testing data
Y_pred=clf.predict(X_test)
#classification report
print(classification_report(Y_test,Y_pred))
#testing score
print("Testing: ",clf.score(X_test,Y_test))
```
# **Self implemented Multinomial Naive Bayes**
```
#makes the nested dictionary required for NB using the training data
def fit(X,Y):
dictionary={}
y_classes=set(Y)
#iterating over each class of y
for y_class in y_classes:
#adding the class as a key to the dictionary
dictionary[y_class]={}
n_features=X.shape[1]
rows=(Y==y_class)
#making the arrays having only those rows where class is y_class
X_y_class=X[rows]
Y_y_class=Y[rows]
#adding the total number of files as total_data
dictionary["total_data"]=X.shape[0]
#iterating over each feature
for i in range(n_features):
#adding the feature as a key which has the count of that word in Y=y_class as its value
dictionary[y_class][i]=X_y_class[:,i].sum()
#adding the total number of files as total_class
dictionary[y_class]["total_class"]=X_y_class.shape[0]
#adding the sum of all the words in Y=y_class i.e. total no. of words in Y=y_class
dictionary[y_class]["total_words"]=X_y_class.sum()
return dictionary
#calculates the probability of the feature vector belonging to a particular class and the probability of the class
#returns the product of the above 2 probabilities
def probability(x,dictionary,y_class):
#output intially has probability of the particular class in log terms
output=np.log(dictionary[y_class]["total_class"])-np.log(dictionary["total_data"])
n_features=len(dictionary[y_class].keys())-2
#calculates probability of x being in a particular class by calulating probability of each word being in that class
for i in range(n_features):
if x[i]>0:
#probability of the ith word being in this class in terms of log
p_i=x[i]*(np.log(dictionary[y_class][i] + 1) - np.log(dictionary[y_class]["total_words"]+n_features))
output+=p_i
return output
#predicts the class to which a single file feature vector belongs to
def predictSinglePoint(x,dictionary):
classes=dictionary.keys()
#contains the class having the max probability
best_class=1
#max probability
best_prob=-1000
first=True
#iterating over all the classes
for y_class in classes:
if y_class=="total_data":
continue
#finding probability of this file feature vector belonging to y_class
p_class=probability(x,dictionary,y_class)
if(first or p_class>best_prob):
best_prob=p_class
best_class=y_class
first=False
return best_class
#predicts the classes to which all the file feature vectors belong in the testing data
def predict(X_test,dictionary):
y_pred=[]
#iterates over all the file feature vectors
for x in X_test:
#predicts the class of a particular file feature vector
x_class=predictSinglePoint(x,dictionary)
y_pred.append(x_class)
return y_pred
dictionary=fit(X_train,Y_train) #makes the required dictionary
y_pred=predict(X_test,dictionary)# predicts the classes
print(classification_report(Y_test,y_pred)) #classification report for testing data
```
# **Comparison of results between inbuilt and self implemented Multinomial NB**
```
print("----------------------------------------------------------------------------")
print("Classification report for inbuilt Multinomial NB on testing data: ")
print("----------------------------------------------------------------------------")
print(classification_report(Y_test,Y_pred))
print("----------------------------------------------------------------------------")
print("Classification report for self implemented Multinomial NB on testing data: ")
print("----------------------------------------------------------------------------")
print(classification_report(Y_test,y_pred))
```
|
github_jupyter
|
```
```
# INTRODUCTION TO UNSUPERVISED LEARNING
Unsupervised learning is the training of a machine using information that is neither classified nor labeled and allowing the algorithm to act on that information without guidance. Here the task of the machine is to group unsorted information according to similarities, patterns, and differences without any prior training of data.
Unlike supervised learning, no teacher is provided that means no training will be given to the machine. Therefore the machine is restricted to find the hidden structure in unlabeled data by itself.
#Example of Unsupervised Machine Learning
For instance, suppose a image having both dogs and cats which it has never seen.
Thus the machine has no idea about the features of dogs and cats so we can’t categorize it as ‘dogs and cats ‘. But it can categorize them according to their similarities, patterns, and differences, i.e., we can easily categorize the picture into two parts. The first may contain all pics having dogs in it and the second part may contain all pics having cats in it. Here you didn’t learn anything before, which means no training data or examples.
It allows the model to work on its own to discover patterns and information that was previously undetected. It mainly deals with unlabelled data.
#Why Unsupervised Learning?
Here, are prime reasons for using Unsupervised Learning in Machine Learning:
>Unsupervised machine learning finds all kind of unknown patterns in data.
>Unsupervised methods help you to find features which can be useful for categorization.
>It is taken place in real time, so all the input data to be analyzed and labeled in the presence of learners.
>It is easier to get unlabeled data from a computer than labeled data, which needs manual intervention.
#Unsupervised Learning Algorithms
Unsupervised Learning Algorithms allow users to perform more complex processing tasks compared to supervised learning. Although, unsupervised learning can be more unpredictable compared with other natural learning methods. Unsupervised learning algorithms include clustering, anomaly detection, neural networks, etc.
#Unsupervised learning is classified into two categories of algorithms:
>Clustering: A clustering problem is where you want to discover the inherent groupings in the data, such as grouping customers by purchasing behavior.
>Association: An association rule learning problem is where you want to discover rules that describe large portions of your data, such as people that buy X also tend to buy Y.
#a)Clustering
Clustering is an important concept when it comes to unsupervised learning.
It mainly deals with finding a structure or pattern in a collection of uncategorized data.
Unsupervised Learning Clustering algorithms will process your data and find natural clusters(groups) if they exist in the data.
You can also modify how many clusters your algorithms should identify. It allows you to adjust the granularity of these groups.
#There are different types of clustering you can utilize:
#1.Exclusive (partitioning)
In this clustering method, Data are grouped in such a way that one data can belong to one cluster only.
Example: K-means
#2.Agglomerative
In this clustering technique, every data is a cluster. The iterative unions between the two nearest clusters reduce the number of clusters.
Example: Hierarchical clustering
#3.Overlapping
In this technique, fuzzy sets is used to cluster data. Each point may belong to two or more clusters with separate degrees of membership.
Here, data will be associated with an appropriate membership value.
Example: Fuzzy C-Means
#4.Probabilistic
This technique uses probability distribution to create the clusters
Example: Following keywords
“man’s shoe.”
“women’s shoe.”
“women’s glove.”
“man’s glove.”
can be clustered into two categories “shoe” and “glove” or “man” and “women.”
#Clustering Types
Following are the clustering types of Machine Learning:
Hierarchical clustering
K-means clustering
K-NN (k nearest neighbors)
Principal Component Analysis
Singular value decomposition
Independent Component Analysis
#1.Hierarchical Clustering
>Hierarchical clustering is an algorithm which builds a hierarchy of clusters. It begins with all the data which is assigned to a cluster of their own. Here, two close cluster are going to be in the same cluster. This algorithm ends when there is only one cluster left.
#2.K-means Clustering
>K-means it is an iterative clustering algorithm which helps you to find the highest value for every iteration. Initially, the desired number of clusters are selected. In this clustering method, you need to cluster the data points into k groups. A larger k means smaller groups with more granularity in the same way. A lower k means larger groups with less granularity.
>The output of the algorithm is a group of “labels.” It assigns data point to one of the k groups. In k-means clustering, each group is defined by creating a centroid for each group. The centroids are like the heart of the cluster, which captures the points closest to them and adds them to the cluster.
K-mean clustering further defines two subgroups:
Agglomerative clustering
Dendrogram
#Agglomerative clustering
>This type of K-means clustering starts with a fixed number of clusters. It allocates all data into the exact number of clusters. This clustering method does not require the number of clusters K as an input. Agglomeration process starts by forming each data as a single cluster.
>This method uses some distance measure, reduces the number of clusters (one in each iteration) by merging process. Lastly, we have one big cluster that contains all the objects.
#Dendrogram
>In the Dendrogram clustering method, each level will represent a possible cluster. The height of dendrogram shows the level of similarity between two join clusters. The closer to the bottom of the process they are more similar cluster which is finding of the group from dendrogram which is not natural and mostly subjective.
#K- Nearest neighbors
>K- nearest neighbour is the simplest of all machine learning classifiers. It differs from other machine learning techniques, in that it doesn’t produce a model. It is a simple algorithm which stores all available cases and classifies new instances based on a similarity measure.
It works very well when there is a distance between examples. The learning speed is slow when the training set is large, and the distance calculation is nontrivial.
#4.Principal Components Analysis
>In case you want a higher-dimensional space. You need to select a basis for that space and only the 200 most important scores of that basis. This base is known as a principal component. The subset you select constitute is a new space which is small in size compared to original space. It maintains as much of the complexity of data as possible.
#5.Singular value decomposition
>The singular value decomposition of a matrix is usually referred to as the SVD.
This is the final and best factorization of a matrix:
A = UΣV^T
where U is orthogonal, Σ is diagonal, and V is orthogonal.
In the decomoposition A = UΣV^T, A can be any matrix. We know that if A
is symmetric positive definite its eigenvectors are orthogonal and we can write
A = QΛQ^T. This is a special case of a SVD, with U = V = Q. For more general
A, the SVD requires two different matrices U and V.
We’ve also learned how to write A = SΛS^−1, where S is the matrix of n
distinct eigenvectors of A. However, S may not be orthogonal; the matrices U
and V in the SVD will be.
#6.Independent Component Analysis
>Independent Component Analysis (ICA) is a machine learning technique to separate independent sources from a mixed signal. Unlike principal component analysis which focuses on maximizing the variance of the data points, the independent component analysis focuses on independence, i.e. independent components.
#b)Association
>Association rules allow you to establish associations amongst data objects inside large databases. This unsupervised technique is about discovering interesting relationships between variables in large databases. For example, people that buy a new home most likely to buy new furniture.
>Other Examples:
>A subgroup of cancer patients grouped by their gene expression measurements
>Groups of shopper based on their browsing and purchasing histories
>Movie group by the rating given by movies viewers
#Applications of Unsupervised Machine Learning
Some application of Unsupervised Learning Techniques are:
1.Clustering automatically split the dataset into groups base on their similarities
2.Anomaly detection can discover unusual data points in your dataset. It is useful for finding fraudulent transactions
3.Association mining identifies sets of items which often occur together in your dataset
4.Latent variable models are widely used for data preprocessing. Like reducing the number of features in a dataset or decomposing the dataset into multiple components.
#Real-life Applications Of Unsupervised Learning
Machines are not that quick, unlike humans. It takes a lot of resources to train a model based on patterns in data. Below are a few of the wonderful real-life simulations of unsupervised learning.
1.Anomaly detection –The advent of technology and the internet has given birth to enormous anomalies in the past and is still growing. Unsupervised learning has huge scope when it comes to anomaly detection.
2.Segmentation – Unsupervised learning can be used to segment the customers based on certain patterns. Each cluster of customers is different whereas customers within a cluster share common properties. Customer segmentation is a widely opted approach used in devising marketing plans.
#Advantages of unsupervised learning
1.It can see what human minds cannot visualize.
2.It is used to dig hidden patterns which hold utmost importance in the industry and has widespread applications in real-time.
3.The outcome of an unsupervised task can yield an entirely new business vertical or venture.
4.There is lesser complexity compared to the supervised learning task. Here, no one is required to interpret the associated labels and hence it holds lesser complexities.
5.It is reasonably easier to obtain unlabeled data.
#Disadvantages of Unsupervised Learning
1.You cannot get precise information regarding data sorting, and the output as data used in unsupervised learning is labeled and not known.
2.Less accuracy of the results is because the input data is not known and not labeled by people in advance. This means that the machine requires to do this itself.
3.The spectral classes do not always correspond to informational classes.
The user needs to spend time interpreting and label the classes which follow that classification.
4.Spectral properties of classes can also change over time so you can’t have the same class information while moving from one image to another.
#How to use Unsupervised learning to find patterns in data
#CODE:
from sklearn import datasets
import matplotlib.pyplot as plt
iris_df = datasets.load_iris()
print(dir(iris_df)
print(iris_df.feature_names)
print(iris_df.target)
print(iris_df.target_names)
label = {0: 'red', 1: 'blue', 2: 'green'}
x_axis = iris_df.data[:, 0]
y_axis = iris_df.data[:, 2]
plt.scatter(x_axis, y_axis, c=iris_df.target)
plt.show()
#Explanation:
As the above code shows, we have used the Iris dataset to make predictions.The dataset contains a records under four attributes-petal length, petal width, sepal length, sepal width.And also it contains three iris classes:setosa, virginica and versicolor .We'll feed the four features of our flowers to the unsupervised algorithm and it will predict which class the iris belongs.For that we have used scikit-learn library in python to load the Iris dataset and matplotlib for data visualisation.
#OUTPUT:
As we can see here the violet colour represents setosa,green colour represents versicolor and yellow
colour represents virginica.

```
```
|
github_jupyter
|
```
%load_ext rpy2.ipython
%matplotlib inline
from fbprophet import Prophet
import pandas as pd
from matplotlib import pyplot as plt
import logging
logging.getLogger('fbprophet').setLevel(logging.ERROR)
import warnings
warnings.filterwarnings("ignore")
df = pd.read_csv('../examples/example_wp_log_peyton_manning.csv')
m = Prophet()
m.fit(df)
future = m.make_future_dataframe(periods=366)
%%R
library(prophet)
df <- read.csv('../examples/example_wp_log_peyton_manning.csv')
m <- prophet(df)
future <- make_future_dataframe(m, periods=366)
```
### Modeling Holidays and Special Events
If you have holidays or other recurring events that you'd like to model, you must create a dataframe for them. It has two columns (`holiday` and `ds`) and a row for each occurrence of the holiday. It must include all occurrences of the holiday, both in the past (back as far as the historical data go) and in the future (out as far as the forecast is being made). If they won't repeat in the future, Prophet will model them and then not include them in the forecast.
You can also include columns `lower_window` and `upper_window` which extend the holiday out to `[lower_window, upper_window]` days around the date. For instance, if you wanted to include Christmas Eve in addition to Christmas you'd include `lower_window=-1,upper_window=0`. If you wanted to use Black Friday in addition to Thanksgiving, you'd include `lower_window=0,upper_window=1`. You can also include a column `prior_scale` to set the prior scale separately for each holiday, as described below.
Here we create a dataframe that includes the dates of all of Peyton Manning's playoff appearances:
```
%%R
library(dplyr)
playoffs <- data_frame(
holiday = 'playoff',
ds = as.Date(c('2008-01-13', '2009-01-03', '2010-01-16',
'2010-01-24', '2010-02-07', '2011-01-08',
'2013-01-12', '2014-01-12', '2014-01-19',
'2014-02-02', '2015-01-11', '2016-01-17',
'2016-01-24', '2016-02-07')),
lower_window = 0,
upper_window = 1
)
superbowls <- data_frame(
holiday = 'superbowl',
ds = as.Date(c('2010-02-07', '2014-02-02', '2016-02-07')),
lower_window = 0,
upper_window = 1
)
holidays <- bind_rows(playoffs, superbowls)
playoffs = pd.DataFrame({
'holiday': 'playoff',
'ds': pd.to_datetime(['2008-01-13', '2009-01-03', '2010-01-16',
'2010-01-24', '2010-02-07', '2011-01-08',
'2013-01-12', '2014-01-12', '2014-01-19',
'2014-02-02', '2015-01-11', '2016-01-17',
'2016-01-24', '2016-02-07']),
'lower_window': 0,
'upper_window': 1,
})
superbowls = pd.DataFrame({
'holiday': 'superbowl',
'ds': pd.to_datetime(['2010-02-07', '2014-02-02', '2016-02-07']),
'lower_window': 0,
'upper_window': 1,
})
holidays = pd.concat((playoffs, superbowls))
```
Above we have included the superbowl days as both playoff games and superbowl games. This means that the superbowl effect will be an additional additive bonus on top of the playoff effect.
Once the table is created, holiday effects are included in the forecast by passing them in with the `holidays` argument. Here we do it with the Peyton Manning data from the [Quickstart](https://facebook.github.io/prophet/docs/quick_start.html):
```
%%R
m <- prophet(df, holidays = holidays)
forecast <- predict(m, future)
m = Prophet(holidays=holidays)
forecast = m.fit(df).predict(future)
```
The holiday effect can be seen in the `forecast` dataframe:
```
%%R
forecast %>%
select(ds, playoff, superbowl) %>%
filter(abs(playoff + superbowl) > 0) %>%
tail(10)
forecast[(forecast['playoff'] + forecast['superbowl']).abs() > 0][
['ds', 'playoff', 'superbowl']][-10:]
```
The holiday effects will also show up in the components plot, where we see that there is a spike on the days around playoff appearances, with an especially large spike for the superbowl:
```
%%R -w 9 -h 12 -u in
prophet_plot_components(m, forecast)
fig = m.plot_components(forecast)
```
Individual holidays can be plotted using the `plot_forecast_component` function (imported from `fbprophet.plot` in Python) like `plot_forecast_component(m, forecast, 'superbowl')` to plot just the superbowl holiday component.
### Built-in Country Holidays
You can use a built-in collection of country-specific holidays using the `add_country_holidays` method (Python) or function (R). The name of the country is specified, and then major holidays for that country will be included in addition to any holidays that are specified via the `holidays` argument described above:
```
%%R
m <- prophet(holidays = holidays)
m <- add_country_holidays(m, country_name = 'US')
m <- fit.prophet(m, df)
m = Prophet(holidays=holidays)
m.add_country_holidays(country_name='US')
m.fit(df)
```
You can see which holidays were included by looking at the `train_holiday_names` (Python) or `train.holiday.names` (R) attribute of the model:
```
%%R
m$train.holiday.names
m.train_holiday_names
```
The holidays for each country are provided by the `holidays` package in Python. A list of available countries, and the country name to use, is available on their page: https://github.com/dr-prodigy/python-holidays. In addition to those countries, Prophet includes holidays for these countries: Brazil (BR), Indonesia (ID), India (IN), Malaysia (MY), Vietnam (VN), Thailand (TH), Philippines (PH), Turkey (TU), Pakistan (PK), Bangladesh (BD), Egypt (EG), China (CN), and Russian (RU), Korea (KR), Belarus (BY), and United Arab Emirates (AE).
In Python, most holidays are computed deterministically and so are available for any date range; a warning will be raised if dates fall outside the range supported by that country. In R, holiday dates are computed for 1995 through 2044 and stored in the package as `data-raw/generated_holidays.csv`. If a wider date range is needed, this script can be used to replace that file with a different date range: https://github.com/facebook/prophet/blob/master/python/scripts/generate_holidays_file.py.
As above, the country-level holidays will then show up in the components plot:
```
%%R -w 9 -h 12 -u in
forecast <- predict(m, future)
prophet_plot_components(m, forecast)
forecast = m.predict(future)
fig = m.plot_components(forecast)
```
### Fourier Order for Seasonalities
Seasonalities are estimated using a partial Fourier sum. See [the paper](https://peerj.com/preprints/3190/) for complete details, and [this figure on Wikipedia](https://en.wikipedia.org/wiki/Fourier_series#/media/File:Fourier_Series.svg) for an illustration of how a partial Fourier sum can approximate an aribtrary periodic signal. The number of terms in the partial sum (the order) is a parameter that determines how quickly the seasonality can change. To illustrate this, consider the Peyton Manning data from the [Quickstart](https://facebook.github.io/prophet/docs/quick_start.html). The default Fourier order for yearly seasonality is 10, which produces this fit:
```
%%R -w 9 -h 3 -u in
m <- prophet(df)
prophet:::plot_yearly(m)
from fbprophet.plot import plot_yearly
m = Prophet().fit(df)
a = plot_yearly(m)
```
The default values are often appropriate, but they can be increased when the seasonality needs to fit higher-frequency changes, and generally be less smooth. The Fourier order can be specified for each built-in seasonality when instantiating the model, here it is increased to 20:
```
%%R -w 9 -h 3 -u in
m <- prophet(df, yearly.seasonality = 20)
prophet:::plot_yearly(m)
from fbprophet.plot import plot_yearly
m = Prophet(yearly_seasonality=20).fit(df)
a = plot_yearly(m)
```
Increasing the number of Fourier terms allows the seasonality to fit faster changing cycles, but can also lead to overfitting: N Fourier terms corresponds to 2N variables used for modeling the cycle
### Specifying Custom Seasonalities
Prophet will by default fit weekly and yearly seasonalities, if the time series is more than two cycles long. It will also fit daily seasonality for a sub-daily time series. You can add other seasonalities (monthly, quarterly, hourly) using the `add_seasonality` method (Python) or function (R).
The inputs to this function are a name, the period of the seasonality in days, and the Fourier order for the seasonality. For reference, by default Prophet uses a Fourier order of 3 for weekly seasonality and 10 for yearly seasonality. An optional input to `add_seasonality` is the prior scale for that seasonal component - this is discussed below.
As an example, here we fit the Peyton Manning data from the [Quickstart](https://facebook.github.io/prophet/docs/quick_start.html), but replace the weekly seasonality with monthly seasonality. The monthly seasonality then will appear in the components plot:
```
%%R -w 9 -h 9 -u in
m <- prophet(weekly.seasonality=FALSE)
m <- add_seasonality(m, name='monthly', period=30.5, fourier.order=5)
m <- fit.prophet(m, df)
forecast <- predict(m, future)
prophet_plot_components(m, forecast)
m = Prophet(weekly_seasonality=False)
m.add_seasonality(name='monthly', period=30.5, fourier_order=5)
forecast = m.fit(df).predict(future)
fig = m.plot_components(forecast)
```
### Seasonalities that depend on other factors
In some instances the seasonality may depend on other factors, such as a weekly seasonal pattern that is different during the summer than it is during the rest of the year, or a daily seasonal pattern that is different on weekends vs. on weekdays. These types of seasonalities can be modeled using conditional seasonalities.
Consider the Peyton Manning example from the [Quickstart](https://facebook.github.io/prophet/docs/quick_start.html). The default weekly seasonality assumes that the pattern of weekly seasonality is the same throughout the year, but we'd expect the pattern of weekly seasonality to be different during the on-season (when there are games every Sunday) and the off-season. We can use conditional seasonalities to construct separate on-season and off-season weekly seasonalities.
First we add a boolean column to the dataframe that indicates whether each date is during the on-season or the off-season:
```
%%R
is_nfl_season <- function(ds) {
dates <- as.Date(ds)
month <- as.numeric(format(dates, '%m'))
return(month > 8 | month < 2)
}
df$on_season <- is_nfl_season(df$ds)
df$off_season <- !is_nfl_season(df$ds)
def is_nfl_season(ds):
date = pd.to_datetime(ds)
return (date.month > 8 or date.month < 2)
df['on_season'] = df['ds'].apply(is_nfl_season)
df['off_season'] = ~df['ds'].apply(is_nfl_season)
```
Then we disable the built-in weekly seasonality, and replace it with two weekly seasonalities that have these columns specified as a condition. This means that the seasonality will only be applied to dates where the `condition_name` column is `True`. We must also add the column to the `future` dataframe for which we are making predictions.
```
%%R -w 9 -h 12 -u in
m <- prophet(weekly.seasonality=FALSE)
m <- add_seasonality(m, name='weekly_on_season', period=7, fourier.order=3, condition.name='on_season')
m <- add_seasonality(m, name='weekly_off_season', period=7, fourier.order=3, condition.name='off_season')
m <- fit.prophet(m, df)
future$on_season <- is_nfl_season(future$ds)
future$off_season <- !is_nfl_season(future$ds)
forecast <- predict(m, future)
prophet_plot_components(m, forecast)
m = Prophet(weekly_seasonality=False)
m.add_seasonality(name='weekly_on_season', period=7, fourier_order=3, condition_name='on_season')
m.add_seasonality(name='weekly_off_season', period=7, fourier_order=3, condition_name='off_season')
future['on_season'] = future['ds'].apply(is_nfl_season)
future['off_season'] = ~future['ds'].apply(is_nfl_season)
forecast = m.fit(df).predict(future)
fig = m.plot_components(forecast)
```
Both of the seasonalities now show up in the components plots above. We can see that during the on-season when games are played every Sunday, there are large increases on Sunday and Monday that are completely absent during the off-season.
### Prior scale for holidays and seasonality
If you find that the holidays are overfitting, you can adjust their prior scale to smooth them using the parameter `holidays_prior_scale`. By default this parameter is 10, which provides very little regularization. Reducing this parameter dampens holiday effects:
```
%%R
m <- prophet(df, holidays = holidays, holidays.prior.scale = 0.05)
forecast <- predict(m, future)
forecast %>%
select(ds, playoff, superbowl) %>%
filter(abs(playoff + superbowl) > 0) %>%
tail(10)
m = Prophet(holidays=holidays, holidays_prior_scale=0.05).fit(df)
forecast = m.predict(future)
forecast[(forecast['playoff'] + forecast['superbowl']).abs() > 0][
['ds', 'playoff', 'superbowl']][-10:]
```
The magnitude of the holiday effect has been reduced compared to before, especially for superbowls, which had the fewest observations. There is a parameter `seasonality_prior_scale` which similarly adjusts the extent to which the seasonality model will fit the data.
Prior scales can be set separately for individual holidays by including a column `prior_scale` in the holidays dataframe. Prior scales for individual seasonalities can be passed as an argument to `add_seasonality`. For instance, the prior scale for just weekly seasonality can be set using:
```
%%R
m <- prophet()
m <- add_seasonality(
m, name='weekly', period=7, fourier.order=3, prior.scale=0.1)
m = Prophet()
m.add_seasonality(
name='weekly', period=7, fourier_order=3, prior_scale=0.1)
```
### Additional regressors
Additional regressors can be added to the linear part of the model using the `add_regressor` method or function. A column with the regressor value will need to be present in both the fitting and prediction dataframes. For example, we can add an additional effect on Sundays during the NFL season. On the components plot, this effect will show up in the 'extra_regressors' plot:
```
%%R -w 9 -h 12 -u in
nfl_sunday <- function(ds) {
dates <- as.Date(ds)
month <- as.numeric(format(dates, '%m'))
as.numeric((weekdays(dates) == "Sunday") & (month > 8 | month < 2))
}
df$nfl_sunday <- nfl_sunday(df$ds)
m <- prophet()
m <- add_regressor(m, 'nfl_sunday')
m <- fit.prophet(m, df)
future$nfl_sunday <- nfl_sunday(future$ds)
forecast <- predict(m, future)
prophet_plot_components(m, forecast)
def nfl_sunday(ds):
date = pd.to_datetime(ds)
if date.weekday() == 6 and (date.month > 8 or date.month < 2):
return 1
else:
return 0
df['nfl_sunday'] = df['ds'].apply(nfl_sunday)
m = Prophet()
m.add_regressor('nfl_sunday')
m.fit(df)
future['nfl_sunday'] = future['ds'].apply(nfl_sunday)
forecast = m.predict(future)
fig = m.plot_components(forecast)
```
NFL Sundays could also have been handled using the "holidays" interface described above, by creating a list of past and future NFL Sundays. The `add_regressor` function provides a more general interface for defining extra linear regressors, and in particular does not require that the regressor be a binary indicator. Another time series could be used as a regressor, although its future values would have to be known.
[This notebook](https://nbviewer.jupyter.org/github/nicolasfauchereau/Auckland_Cycling/blob/master/notebooks/Auckland_cycling_and_weather.ipynb) shows an example of using weather factors as extra regressors in a forecast of bicycle usage, and provides an excellent illustration of how other time series can be included as extra regressors.
The `add_regressor` function has optional arguments for specifying the prior scale (holiday prior scale is used by default) and whether or not the regressor is standardized - see the docstring with `help(Prophet.add_regressor)` in Python and `?add_regressor` in R. Note that regressors must be added prior to model fitting.
The extra regressor must be known for both the history and for future dates. It thus must either be something that has known future values (such as `nfl_sunday`), or something that has separately been forecasted elsewhere. Prophet will also raise an error if the regressor is constant throughout the history, since there is nothing to fit from it.
Extra regressors are put in the linear component of the model, so the underlying model is that the time series depends on the extra regressor as either an additive or multiplicative factor (see the next section for multiplicativity).
|
github_jupyter
|
### Demonstration of `flopy.utils.get_transmissivities` method
for computing open interval transmissivities (for weighted averages of heads or fluxes)
In practice this method might be used to:
* compute vertically-averaged head target values representative of observation wells of varying open intervals (including variability in saturated thickness due to the position of the water table)
* apportion boundary fluxes (e.g. from an analytic element model) among model layers based on transmissivity.
* any other analysis where a distribution of transmissivity by layer is needed for a specified vertical interval of the model.
```
import os
import sys
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
# run installed version of flopy or add local path
try:
import flopy
except:
fpth = os.path.abspath(os.path.join('..', '..'))
sys.path.append(fpth)
import flopy
print(sys.version)
print('numpy version: {}'.format(np.__version__))
print('matplotlib version: {}'.format(mpl.__version__))
print('flopy version: {}'.format(flopy.__version__))
```
### Make up some open interval tops and bottoms and some heads
* (these could be lists of observation well screen tops and bottoms)
* the heads array contains the simulated head in each model layer,
at the location of each observation well (for example, what you would get back from HYDMOD if you had an entry for each layer at the location of each head target).
* make up a model grid with uniform horizontal k of 2.
```
sctop = [-.25, .5, 1.7, 1.5, 3., 2.5] # screen tops
scbot = [-1., -.5, 1.2, 0.5, 1.5, -.2] # screen bottoms
# head in each layer, for 6 head target locations
heads = np.array([[1., 2.0, 2.05, 3., 4., 2.5],
[1.1, 2.1, 2.2, 2., 3.5, 3.],
[1.2, 2.3, 2.4, 0.6, 3.4, 3.2]
])
nl, nr = heads.shape
nc = nr
botm = np.ones((nl, nr, nc), dtype=float)
top = np.ones((nr, nc), dtype=float) * 2.1
hk = np.ones((nl, nr, nc), dtype=float) * 2.
for i in range(nl):
botm[nl-i-1, :, :] = i
botm
```
### Make a flopy modflow model
```
m = flopy.modflow.Modflow('junk', version='mfnwt', model_ws='data')
dis = flopy.modflow.ModflowDis(m, nlay=nl, nrow=nr, ncol=nc, botm=botm, top=top)
upw = flopy.modflow.ModflowUpw(m, hk=hk)
```
### Get transmissivities along the diagonal cells
* alternatively, if a model's coordinate information has been set up, the real-world x and y coordinates could be supplied with the `x` and `y` arguments
* if `sctop` and `scbot` arguments are given, the transmissivites are computed for the open intervals only
(cells that are partially within the open interval have reduced thickness, cells outside of the open interval have transmissivities of 0). If no `sctop` or `scbot` arguments are supplied, trasmissivites reflect the full saturated thickness in each column of cells (see plot below, which shows different open intervals relative to the model layering)
```
r, c = np.arange(nr), np.arange(nc)
T = flopy.utils.get_transmissivities(heads, m, r=r, c=c, sctop=sctop, scbot=scbot)
np.round(T, 2)
m.dis.botm.array[:, r, c]
```
### Plot the model top and layer bottoms (colors)
open intervals are shown as boxes
* well 0 has zero transmissivities for each layer, as it is below the model bottom
* well 1 has T values of 0 for layers 1 and 2, and 1 for layer 3 (K=2 x 0.5 thickness)
```
fig, ax = plt.subplots()
plt.plot(m.dis.top.array[r, c], label='model top')
for i, l in enumerate(m.dis.botm.array[:, r, c]):
label = 'layer {} bot'.format(i+1)
if i == m.nlay -1:
label = 'model bot'
plt.plot(l, label=label)
plt.plot(heads[0], label='piezometric surface', color='b', linestyle=':')
for iw in range(len(sctop)):
ax.fill_between([iw-.25, iw+.25], scbot[iw], sctop[iw],
facecolor='None', edgecolor='k')
ax.legend(loc=2)
```
### example of transmissivites without `sctop` and `scbot`
* well zero has T=0 in layer 1 because it is dry; T=0.2 in layer 2 because the sat. thickness there is only 0.1
```
T = flopy.utils.get_transmissivities(heads, m, r=r, c=c)
np.round(T, 2)
```
|
github_jupyter
|
```
import pandas as pd
import warnings
warnings.filterwarnings('ignore')
# Dependencies for interaction with database:
from sqlalchemy import create_engine
from sqlalchemy.orm import Session
# Machine Learning dependencies:
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
# Validation libraries
from sklearn import metrics
from sklearn.metrics import accuracy_score, mean_squared_error, precision_recall_curve
from sklearn.model_selection import cross_val_score
from collections import Counter
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
from sklearn.metrics import confusion_matrix
from imblearn.metrics import classification_report_imbalanced
# Create engine and link to local postgres database:
engine = create_engine('postgresql://postgres:[email protected]:5432/postgres')
connect = engine.connect()
# Create session:
session = Session(engine)
# Import clean_dataset_2016 table:
clean_2016_df = pd.read_sql(sql = 'SELECT * FROM "survey_2016"',con=connect)
clean_2016_df.head()
# Check data for insights:
print(clean_2016_df.shape)
print(clean_2016_df.columns.tolist())
print(clean_2016_df.value_counts)
##Test:
#Dataset filtered on tech_company = "yes"
#Target:
#Features: company_size, age, gender, country_live, identified_with_mh, mh_employer, employer_discus_mh, employer_provide_mh_coverage,treatment_mh_from_professional, employers_options_help, protected_anonymity_mh
# Filter tech_or_not columns:
clean_2016_df["tech_company"].head()
tech_df = pd.read_sql('SELECT * FROM "survey_2016" WHERE "tech_company" = 1', connect)
tech_df.shape
ml_df = tech_df[["mh_sought_pro_tx","mh_dx_pro","company_size","mh_discussion_coworkers", "mh_discussion_supervisors","mh_employer_discussion","prev_mh_discussion_coworkers","prev_mh_discussion_supervisors","mh_sharing_friends_family"]]
ml_df
# Encode dataset:
# Create label encoder instance:
le = LabelEncoder()
# Make a copy of desire data:
encoded_df = ml_df.copy()
# Apply encoder:
#encoded_df["age"] = le.fit_transform(encoded_df["age"] )
#encoded_df["company_size"] = le.fit_transform(encoded_df["company_size"])
#encoded_df["gender"] = le.fit_transform(encoded_df["gender"])
#encoded_df["country_live"] = le.fit_transform(encoded_df["country_live"])
#encoded_df["identified_with_mh"] = le.fit_transform(encoded_df["identified_with_mh"])
#encoded_df["dx_mh_disorder"] = le.fit_transform(encoded_df["dx_mh_disorder"])
#encoded_df["employer_discus_mh"] = le.fit_transform(encoded_df["employer_discus_mh"])
#encoded_df["mh_employer"] = le.fit_transform(encoded_df["mh_employer"])
#encoded_df["treatment_mh_from_professional"] = le.fit_transform(encoded_df["treatment_mh_from_professional"])
#encoded_df["employer_provide_mh_coverage"] = le.fit_transform(encoded_df["employer_provide_mh_coverage"])
#encoded_df["employers_options_help"] = le.fit_transform(encoded_df["employers_options_help"])
#encoded_df["protected_anonymity_mh"] = le.fit_transform(encoded_df["protected_anonymity_mh"])
features = encoded_df.columns.tolist()
for feature in features:
encoded_df[feature] = le.fit_transform(encoded_df[feature])
# Check:
encoded_df.head()
from sklearn.preprocessing import OneHotEncoder
encoder = OneHotEncoder(sparse = False)
encoded_df1 = ml_df.copy()
# Apply encoder:
encoded_df1 = encoder.fit_transform(encoded_df1)
encoded_df1
# Create our target:
y = encoded_df["mh_sought_pro_tx"]
# Create our features:
X = encoded_df.drop(columns = "mh_sought_pro_tx", axis =1)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,
y,
random_state=40,
stratify=y)
X_train.shape
from sklearn.linear_model import LogisticRegression
classifier = LogisticRegression(solver='lbfgs',
max_iter=200,
random_state=1)
classifier.fit(X_train, y_train)
y_pred = classifier.predict(X_test)
results = pd.DataFrame({"Prediction": y_pred, "Actual": y_test}).reset_index(drop=True)
results.head(20)
from sklearn.metrics import accuracy_score
print(accuracy_score(y_test, y_pred))
from sklearn.metrics import confusion_matrix, classification_report
matrix = confusion_matrix(y_test,y_pred)
print(matrix)
report = classification_report(y_test, y_pred)
print(report)
```
|
github_jupyter
|
##### Copyright 2018 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Keras
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/guide/keras"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/guide/keras.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/guide/keras.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
</table>
Keras is a high-level API to build and train deep learning models. It's used for
fast prototyping, advanced research, and production, with three key advantages:
- *User friendly*<br>
Keras has a simple, consistent interface optimized for common use cases. It
provides clear and actionable feedback for user errors.
- *Modular and composable*<br>
Keras models are made by connecting configurable building blocks together,
with few restrictions.
- *Easy to extend*<br> Write custom building blocks to express new ideas for
research. Create new layers, loss functions, and develop state-of-the-art
models.
## Import tf.keras
`tf.keras` is TensorFlow's implementation of the
[Keras API specification](https://keras.io). This is a high-level
API to build and train models that includes first-class support for
TensorFlow-specific functionality, such as [eager execution](#eager_execution),
`tf.data` pipelines, and [Estimators](./estimators.md).
`tf.keras` makes TensorFlow easier to use without sacrificing flexibility and
performance.
To get started, import `tf.keras` as part of your TensorFlow program setup:
```
!pip install -q pyyaml # Required to save models in YAML format
from __future__ import absolute_import, division, print_function, unicode_literals
import tensorflow as tf
from tensorflow.keras import layers
print(tf.VERSION)
print(tf.keras.__version__)
```
`tf.keras` can run any Keras-compatible code, but keep in mind:
* The `tf.keras` version in the latest TensorFlow release might not be the same
as the latest `keras` version from PyPI. Check `tf.keras.__version__`.
* When [saving a model's weights](#weights_only), `tf.keras` defaults to the
[checkpoint format](./checkpoints.md). Pass `save_format='h5'` to
use HDF5.
## Build a simple model
### Sequential model
In Keras, you assemble *layers* to build *models*. A model is (usually) a graph
of layers. The most common type of model is a stack of layers: the
`tf.keras.Sequential` model.
To build a simple, fully-connected network (i.e. multi-layer perceptron):
```
model = tf.keras.Sequential()
# Adds a densely-connected layer with 64 units to the model:
model.add(layers.Dense(64, activation='relu'))
# Add another:
model.add(layers.Dense(64, activation='relu'))
# Add a softmax layer with 10 output units:
model.add(layers.Dense(10, activation='softmax'))
```
### Configure the layers
There are many `tf.keras.layers` available with some common constructor
parameters:
* `activation`: Set the activation function for the layer. This parameter is
specified by the name of a built-in function or as a callable object. By
default, no activation is applied.
* `kernel_initializer` and `bias_initializer`: The initialization schemes
that create the layer's weights (kernel and bias). This parameter is a name or
a callable object. This defaults to the `"Glorot uniform"` initializer.
* `kernel_regularizer` and `bias_regularizer`: The regularization schemes
that apply the layer's weights (kernel and bias), such as L1 or L2
regularization. By default, no regularization is applied.
The following instantiates `tf.keras.layers.Dense` layers using constructor
arguments:
```
# Create a sigmoid layer:
layers.Dense(64, activation='sigmoid')
# Or:
layers.Dense(64, activation=tf.sigmoid)
# A linear layer with L1 regularization of factor 0.01 applied to the kernel matrix:
layers.Dense(64, kernel_regularizer=tf.keras.regularizers.l1(0.01))
# A linear layer with L2 regularization of factor 0.01 applied to the bias vector:
layers.Dense(64, bias_regularizer=tf.keras.regularizers.l2(0.01))
# A linear layer with a kernel initialized to a random orthogonal matrix:
layers.Dense(64, kernel_initializer='orthogonal')
# A linear layer with a bias vector initialized to 2.0s:
layers.Dense(64, bias_initializer=tf.keras.initializers.constant(2.0))
```
## Train and evaluate
### Set up training
After the model is constructed, configure its learning process by calling the
`compile` method:
```
model = tf.keras.Sequential([
# Adds a densely-connected layer with 64 units to the model:
layers.Dense(64, activation='relu', input_shape=(32,)),
# Add another:
layers.Dense(64, activation='relu'),
# Add a softmax layer with 10 output units:
layers.Dense(10, activation='softmax')])
model.compile(optimizer=tf.train.AdamOptimizer(0.001),
loss='categorical_crossentropy',
metrics=['accuracy'])
```
`tf.keras.Model.compile` takes three important arguments:
* `optimizer`: This object specifies the training procedure. Pass it optimizer
instances from the `tf.train` module, such as
`tf.train.AdamOptimizer`, `tf.train.RMSPropOptimizer`, or
`tf.train.GradientDescentOptimizer`.
* `loss`: The function to minimize during optimization. Common choices include
mean square error (`mse`), `categorical_crossentropy`, and
`binary_crossentropy`. Loss functions are specified by name or by
passing a callable object from the `tf.keras.losses` module.
* `metrics`: Used to monitor training. These are string names or callables from
the `tf.keras.metrics` module.
The following shows a few examples of configuring a model for training:
```
# Configure a model for mean-squared error regression.
model.compile(optimizer=tf.train.AdamOptimizer(0.01),
loss='mse', # mean squared error
metrics=['mae']) # mean absolute error
# Configure a model for categorical classification.
model.compile(optimizer=tf.train.RMSPropOptimizer(0.01),
loss=tf.keras.losses.categorical_crossentropy,
metrics=[tf.keras.metrics.categorical_accuracy])
```
### Input NumPy data
For small datasets, use in-memory [NumPy](https://www.numpy.org/)
arrays to train and evaluate a model. The model is "fit" to the training data
using the `fit` method:
```
import numpy as np
def random_one_hot_labels(shape):
n, n_class = shape
classes = np.random.randint(0, n_class, n)
labels = np.zeros((n, n_class))
labels[np.arange(n), classes] = 1
return labels
data = np.random.random((1000, 32))
labels = random_one_hot_labels((1000, 10))
model.fit(data, labels, epochs=10, batch_size=32)
```
`tf.keras.Model.fit` takes three important arguments:
* `epochs`: Training is structured into *epochs*. An epoch is one iteration over
the entire input data (this is done in smaller batches).
* `batch_size`: When passed NumPy data, the model slices the data into smaller
batches and iterates over these batches during training. This integer
specifies the size of each batch. Be aware that the last batch may be smaller
if the total number of samples is not divisible by the batch size.
* `validation_data`: When prototyping a model, you want to easily monitor its
performance on some validation data. Passing this argument—a tuple of inputs
and labels—allows the model to display the loss and metrics in inference mode
for the passed data, at the end of each epoch.
Here's an example using `validation_data`:
```
import numpy as np
data = np.random.random((1000, 32))
labels = random_one_hot_labels((1000, 10))
val_data = np.random.random((100, 32))
val_labels = random_one_hot_labels((100, 10))
model.fit(data, labels, epochs=10, batch_size=32,
validation_data=(val_data, val_labels))
```
### Input tf.data datasets
Use the [Datasets API](./datasets.md) to scale to large datasets
or multi-device training. Pass a `tf.data.Dataset` instance to the `fit`
method:
```
# Instantiates a toy dataset instance:
dataset = tf.data.Dataset.from_tensor_slices((data, labels))
dataset = dataset.batch(32)
dataset = dataset.repeat()
# Don't forget to specify `steps_per_epoch` when calling `fit` on a dataset.
model.fit(dataset, epochs=10, steps_per_epoch=30)
```
Here, the `fit` method uses the `steps_per_epoch` argument—this is the number of
training steps the model runs before it moves to the next epoch. Since the
`Dataset` yields batches of data, this snippet does not require a `batch_size`.
Datasets can also be used for validation:
```
dataset = tf.data.Dataset.from_tensor_slices((data, labels))
dataset = dataset.batch(32).repeat()
val_dataset = tf.data.Dataset.from_tensor_slices((val_data, val_labels))
val_dataset = val_dataset.batch(32).repeat()
model.fit(dataset, epochs=10, steps_per_epoch=30,
validation_data=val_dataset,
validation_steps=3)
```
### Evaluate and predict
The `tf.keras.Model.evaluate` and `tf.keras.Model.predict` methods can use NumPy
data and a `tf.data.Dataset`.
To *evaluate* the inference-mode loss and metrics for the data provided:
```
data = np.random.random((1000, 32))
labels = random_one_hot_labels((1000, 10))
model.evaluate(data, labels, batch_size=32)
model.evaluate(dataset, steps=30)
```
And to *predict* the output of the last layer in inference for the data provided,
as a NumPy array:
```
result = model.predict(data, batch_size=32)
print(result.shape)
```
## Build advanced models
### Functional API
The `tf.keras.Sequential` model is a simple stack of layers that cannot
represent arbitrary models. Use the
[Keras functional API](https://keras.io/getting-started/functional-api-guide/)
to build complex model topologies such as:
* Multi-input models,
* Multi-output models,
* Models with shared layers (the same layer called several times),
* Models with non-sequential data flows (e.g. residual connections).
Building a model with the functional API works like this:
1. A layer instance is callable and returns a tensor.
2. Input tensors and output tensors are used to define a `tf.keras.Model`
instance.
3. This model is trained just like the `Sequential` model.
The following example uses the functional API to build a simple, fully-connected
network:
```
inputs = tf.keras.Input(shape=(32,)) # Returns a placeholder tensor
# A layer instance is callable on a tensor, and returns a tensor.
x = layers.Dense(64, activation='relu')(inputs)
x = layers.Dense(64, activation='relu')(x)
predictions = layers.Dense(10, activation='softmax')(x)
```
Instantiate the model given inputs and outputs.
```
model = tf.keras.Model(inputs=inputs, outputs=predictions)
# The compile step specifies the training configuration.
model.compile(optimizer=tf.train.RMSPropOptimizer(0.001),
loss='categorical_crossentropy',
metrics=['accuracy'])
# Trains for 5 epochs
model.fit(data, labels, batch_size=32, epochs=5)
```
### Model subclassing
Build a fully-customizable model by subclassing `tf.keras.Model` and defining
your own forward pass. Create layers in the `__init__` method and set them as
attributes of the class instance. Define the forward pass in the `call` method.
Model subclassing is particularly useful when
[eager execution](./eager.md) is enabled since the forward pass
can be written imperatively.
Key Point: Use the right API for the job. While model subclassing offers
flexibility, it comes at a cost of greater complexity and more opportunities for
user errors. If possible, prefer the functional API.
The following example shows a subclassed `tf.keras.Model` using a custom forward
pass:
```
class MyModel(tf.keras.Model):
def __init__(self, num_classes=10):
super(MyModel, self).__init__(name='my_model')
self.num_classes = num_classes
# Define your layers here.
self.dense_1 = layers.Dense(32, activation='relu')
self.dense_2 = layers.Dense(num_classes, activation='sigmoid')
def call(self, inputs):
# Define your forward pass here,
# using layers you previously defined (in `__init__`).
x = self.dense_1(inputs)
return self.dense_2(x)
def compute_output_shape(self, input_shape):
# You need to override this function if you want to use the subclassed model
# as part of a functional-style model.
# Otherwise, this method is optional.
shape = tf.TensorShape(input_shape).as_list()
shape[-1] = self.num_classes
return tf.TensorShape(shape)
```
Instantiate the new model class:
```
model = MyModel(num_classes=10)
# The compile step specifies the training configuration.
model.compile(optimizer=tf.train.RMSPropOptimizer(0.001),
loss='categorical_crossentropy',
metrics=['accuracy'])
# Trains for 5 epochs.
model.fit(data, labels, batch_size=32, epochs=5)
```
### Custom layers
Create a custom layer by subclassing `tf.keras.layers.Layer` and implementing
the following methods:
* `build`: Create the weights of the layer. Add weights with the `add_weight`
method.
* `call`: Define the forward pass.
* `compute_output_shape`: Specify how to compute the output shape of the layer
given the input shape.
* Optionally, a layer can be serialized by implementing the `get_config` method
and the `from_config` class method.
Here's an example of a custom layer that implements a `matmul` of an input with
a kernel matrix:
```
class MyLayer(layers.Layer):
def __init__(self, output_dim, **kwargs):
self.output_dim = output_dim
super(MyLayer, self).__init__(**kwargs)
def build(self, input_shape):
shape = tf.TensorShape((input_shape[1], self.output_dim))
# Create a trainable weight variable for this layer.
self.kernel = self.add_weight(name='kernel',
shape=shape,
initializer='uniform',
trainable=True)
# Make sure to call the `build` method at the end
super(MyLayer, self).build(input_shape)
def call(self, inputs):
return tf.matmul(inputs, self.kernel)
def compute_output_shape(self, input_shape):
shape = tf.TensorShape(input_shape).as_list()
shape[-1] = self.output_dim
return tf.TensorShape(shape)
def get_config(self):
base_config = super(MyLayer, self).get_config()
base_config['output_dim'] = self.output_dim
return base_config
@classmethod
def from_config(cls, config):
return cls(**config)
```
Create a model using your custom layer:
```
model = tf.keras.Sequential([
MyLayer(10),
layers.Activation('softmax')])
# The compile step specifies the training configuration
model.compile(optimizer=tf.train.RMSPropOptimizer(0.001),
loss='categorical_crossentropy',
metrics=['accuracy'])
# Trains for 5 epochs.
model.fit(data, labels, batch_size=32, epochs=5)
```
## Callbacks
A callback is an object passed to a model to customize and extend its behavior
during training. You can write your own custom callback, or use the built-in
`tf.keras.callbacks` that include:
* `tf.keras.callbacks.ModelCheckpoint`: Save checkpoints of your model at
regular intervals.
* `tf.keras.callbacks.LearningRateScheduler`: Dynamically change the learning
rate.
* `tf.keras.callbacks.EarlyStopping`: Interrupt training when validation
performance has stopped improving.
* `tf.keras.callbacks.TensorBoard`: Monitor the model's behavior using
[TensorBoard](./summaries_and_tensorboard.md).
To use a `tf.keras.callbacks.Callback`, pass it to the model's `fit` method:
```
callbacks = [
# Interrupt training if `val_loss` stops improving for over 2 epochs
tf.keras.callbacks.EarlyStopping(patience=2, monitor='val_loss'),
# Write TensorBoard logs to `./logs` directory
tf.keras.callbacks.TensorBoard(log_dir='./logs')
]
model.fit(data, labels, batch_size=32, epochs=5, callbacks=callbacks,
validation_data=(val_data, val_labels))
```
<a id='weights_only'></a>
## Save and restore
### Weights only
Save and load the weights of a model using `tf.keras.Model.save_weights`:
```
model = tf.keras.Sequential([
layers.Dense(64, activation='relu', input_shape=(32,)),
layers.Dense(10, activation='softmax')])
model.compile(optimizer=tf.train.AdamOptimizer(0.001),
loss='categorical_crossentropy',
metrics=['accuracy'])
# Save weights to a TensorFlow Checkpoint file
model.save_weights('./weights/my_model')
# Restore the model's state,
# this requires a model with the same architecture.
model.load_weights('./weights/my_model')
```
By default, this saves the model's weights in the
[TensorFlow checkpoint](./checkpoints.md) file format. Weights can
also be saved to the Keras HDF5 format (the default for the multi-backend
implementation of Keras):
```
# Save weights to a HDF5 file
model.save_weights('my_model.h5', save_format='h5')
# Restore the model's state
model.load_weights('my_model.h5')
```
### Configuration only
A model's configuration can be saved—this serializes the model architecture
without any weights. A saved configuration can recreate and initialize the same
model, even without the code that defined the original model. Keras supports
JSON and YAML serialization formats:
```
# Serialize a model to JSON format
json_string = model.to_json()
json_string
import json
import pprint
pprint.pprint(json.loads(json_string))
```
Recreate the model (newly initialized) from the JSON:
```
fresh_model = tf.keras.models.model_from_json(json_string)
```
Serializing a model to YAML format requires that you install `pyyaml` *before you import TensorFlow*:
```
yaml_string = model.to_yaml()
print(yaml_string)
```
Recreate the model from the YAML:
```
fresh_model = tf.keras.models.model_from_yaml(yaml_string)
```
Caution: Subclassed models are not serializable because their architecture is
defined by the Python code in the body of the `call` method.
### Entire model
The entire model can be saved to a file that contains the weight values, the
model's configuration, and even the optimizer's configuration. This allows you
to checkpoint a model and resume training later—from the exact same
state—without access to the original code.
```
# Create a trivial model
model = tf.keras.Sequential([
layers.Dense(10, activation='softmax', input_shape=(32,)),
layers.Dense(10, activation='softmax')
])
model.compile(optimizer='rmsprop',
loss='categorical_crossentropy',
metrics=['accuracy'])
model.fit(data, labels, batch_size=32, epochs=5)
# Save entire model to a HDF5 file
model.save('my_model.h5')
# Recreate the exact same model, including weights and optimizer.
model = tf.keras.models.load_model('my_model.h5')
```
## Eager execution
[Eager execution](./eager.md) is an imperative programming
environment that evaluates operations immediately. This is not required for
Keras, but is supported by `tf.keras` and useful for inspecting your program and
debugging.
All of the `tf.keras` model-building APIs are compatible with eager execution.
And while the `Sequential` and functional APIs can be used, eager execution
especially benefits *model subclassing* and building *custom layers*—the APIs
that require you to write the forward pass as code (instead of the APIs that
create models by assembling existing layers).
See the [eager execution guide](./eager.md#build_a_model) for
examples of using Keras models with custom training loops and `tf.GradientTape`.
## Distribution
### Estimators
The [Estimators](./estimators.md) API is used for training models
for distributed environments. This targets industry use cases such as
distributed training on large datasets that can export a model for production.
A `tf.keras.Model` can be trained with the `tf.estimator` API by converting the
model to an `tf.estimator.Estimator` object with
`tf.keras.estimator.model_to_estimator`. See
[Creating Estimators from Keras models](./estimators.md#creating_estimators_from_keras_models).
```
model = tf.keras.Sequential([layers.Dense(10,activation='softmax'),
layers.Dense(10,activation='softmax')])
model.compile(optimizer=tf.train.RMSPropOptimizer(0.001),
loss='categorical_crossentropy',
metrics=['accuracy'])
estimator = tf.keras.estimator.model_to_estimator(model)
```
Note: Enable [eager execution](./eager.md) for debugging
[Estimator input functions](./premade_estimators.md#create_input_functions)
and inspecting data.
### Multiple GPUs
`tf.keras` models can run on multiple GPUs using
`tf.contrib.distribute.DistributionStrategy`. This API provides distributed
training on multiple GPUs with almost no changes to existing code.
Currently, `tf.contrib.distribute.MirroredStrategy` is the only supported
distribution strategy. `MirroredStrategy` does in-graph replication with
synchronous training using all-reduce on a single machine. To use
`DistributionStrategy` with Keras, convert the `tf.keras.Model` to a
`tf.estimator.Estimator` with `tf.keras.estimator.model_to_estimator`, then
train the estimator
The following example distributes a `tf.keras.Model` across multiple GPUs on a
single machine.
First, define a simple model:
```
model = tf.keras.Sequential()
model.add(layers.Dense(16, activation='relu', input_shape=(10,)))
model.add(layers.Dense(1, activation='sigmoid'))
optimizer = tf.train.GradientDescentOptimizer(0.2)
model.compile(loss='binary_crossentropy', optimizer=optimizer)
model.summary()
```
Define an *input pipeline*. The `input_fn` returns a `tf.data.Dataset` object
used to distribute the data across multiple devices—with each device processing
a slice of the input batch.
```
def input_fn():
x = np.random.random((1024, 10))
y = np.random.randint(2, size=(1024, 1))
x = tf.cast(x, tf.float32)
dataset = tf.data.Dataset.from_tensor_slices((x, y))
dataset = dataset.repeat(10)
dataset = dataset.batch(32)
return dataset
```
Next, create a `tf.estimator.RunConfig` and set the `train_distribute` argument
to the `tf.contrib.distribute.MirroredStrategy` instance. When creating
`MirroredStrategy`, you can specify a list of devices or set the `num_gpus`
argument. The default uses all available GPUs, like the following:
```
strategy = tf.contrib.distribute.MirroredStrategy()
config = tf.estimator.RunConfig(train_distribute=strategy)
```
Convert the Keras model to a `tf.estimator.Estimator` instance:
```
keras_estimator = tf.keras.estimator.model_to_estimator(
keras_model=model,
config=config,
model_dir='/tmp/model_dir')
```
Finally, train the `Estimator` instance by providing the `input_fn` and `steps`
arguments:
```
keras_estimator.train(input_fn=input_fn, steps=10)
```
|
github_jupyter
|
```
import math
import numpy as np
import pandas as pd
```
### Initial conditions
```
initial_rating = 400
k = 100
things = ['Malted Milk','Rich Tea','Hobnob','Digestive']
```
### Elo Algos
```
def expected_win(r1, r2):
"""
Expected probability of player 1 beating player 2
if player 1 has rating 1 (r1) and player 2 has rating 2 (r2)
"""
return 1.0 / (1 + math.pow(10, (r2-r1)/400))
def update_rating(R, k, P, d):
"""
d = 1 = WIN
d = 0 = LOSS
"""
return R + k*(d-P)
def elo(Ra, Rb, k, d):
"""
d = 1 when player A wins
d = 0 when player B wins
"""
Pa = expected_win(Ra, Rb)
Pb = expected_win(Rb, Ra)
# update if A wins
if d == 1:
Ra = update_rating(Ra, k, Pa, d)
Rb = update_rating(Rb, k, Pb, d-1)
# update if B wins
elif d == 0:
Ra = update_rating(Ra, k, Pa, d)
Rb = update_rating(Rb, k, Pb, d+1)
return Pa, Pb, Ra, Rb
def elo_sequence(things, initial_rating, k, results):
"""
Initialises score dictionary, and runs through sequence of pairwise results, returning final dictionary of Elo rankings
"""
dic_scores = {i:initial_rating for i in things}
for result in results:
winner, loser = result
Ra, Rb = dic_scores[winner], dic_scores[loser]
_, _, newRa, newRb = elo(Ra, Rb, k, 1)
dic_scores[winner], dic_scores[loser] = newRa, newRb
return dic_scores
```
### Mean Elo
```
def mElo(things, initial_rating, k, results, numEpochs):
"""
Randomises the sequence of the pairwise comparisons, running the Elo sequence in a random
sequence for a number of epochs
Returns the mean Elo ratings over the randomised epoch sequences
"""
lst_outcomes = []
for i in np.arange(numEpochs):
np.random.shuffle(results)
lst_outcomes.append(elo_sequence(things, initial_rating, k, results))
return pd.DataFrame(lst_outcomes).mean().sort_values(ascending=False)
```
### Pairwise Outcomes from Christian's Taste Test
> **Format** (Winner, Loser)
```
results = np.array([('Malted Milk','Rich Tea'),('Malted Milk','Digestive'),('Malted Milk','Hobnob')\
,('Hobnob','Rich Tea'),('Hobnob','Digestive'),('Digestive','Rich Tea')])
jenResults = np.array([('Rich Tea','Malted Milk'),('Digestive','Malted Milk'),('Hobnob','Malted Milk')\
,('Hobnob','Rich Tea'),('Hobnob','Digestive'),('Digestive','Rich Tea')])
mElo(things, initial_rating, k, jenResults, 1000)
```
|
github_jupyter
|
# Benchmark FRESA.CAD BSWIMS final Script
This algorithm implementation uses R code and a Python library (rpy2) to connect with it, in order to run the following it is necesary to have installed both on your computer:
- R (you can download in https://www.r-project.org/) <br>
- install rpy2 by <code> pip install rpy2 </code>
```
import numpy as np
import pandas as pd
import sys
from pathlib import Path
sys.path.append("../tadpole-algorithms")
import tadpole_algorithms
from tadpole_algorithms.models import Benchmark_FRESACAD_R
from tadpole_algorithms.preprocessing.split import split_test_train_tadpole
#rpy2 libs and funcs
import rpy2.robjects.packages as rpackages
from rpy2.robjects.vectors import StrVector
from rpy2.robjects import r, pandas2ri
from rpy2 import robjects
from rpy2.robjects.conversion import localconverter
# Load D1_D2 train and possible test data set
data_path_train_test = Path("data/TADPOLE_D1_D2.csv")
data_df_train_test = pd.read_csv(data_path_train_test)
# Load data Dictionary
data_path_Dictionaty = Path("data/TADPOLE_D1_D2_Dict.csv")
data_Dictionaty = pd.read_csv(data_path_Dictionaty)
# Load D3 possible test set
data_path_test = Path("data/TADPOLE_D3.csv")
data_D3 = pd.read_csv(data_path_test)
# Load D4 evaluation data set
data_path_eval = Path("data/TADPOLE_D4_corr.csv")
data_df_eval = pd.read_csv(data_path_eval)
# Split data in test, train and evaluation data
train_df, test_df, eval_df = split_test_train_tadpole(data_df_train_test, data_df_eval)
#instanciate the model to get the functions
model = Benchmark_FRESACAD_R()
#set the flag to true to use a preprocessed data
USE_PREPROC = False
#preprocess the data
D1Train,D2Test,D3Train,D3Test = model.extractTrainTestDataSets_R("data/TADPOLE_D1_D2.csv","data/TADPOLE_D3.csv")
# AdjustedTrainFrame,testingFrame,Train_Imputed,Test_Imputed = model.preproc_tadpole_D1_D2(data_df_train_test,USE_PREPROC)
AdjustedTrainFrame,testingFrame,Train_Imputed,Test_Imputed = model.preproc_with_R(D1Train,D2Test,data_Dictionaty,usePreProc=USE_PREPROC)
#Train Congitive Models
modelfilename = model.Train_Congitive(AdjustedTrainFrame,usePreProc=USE_PREPROC)
#Train ADAS/Ventricles Models
regresionModelfilename = model.Train_Regression(AdjustedTrainFrame,Train_Imputed,usePreProc=USE_PREPROC)
print(regresionModelfilename)
print(regresionModelfilename)
print(type(regresionModelfilename))
#Predict
Forecast_D2 = model.Forecast_All(modelfilename,
regresionModelfilename,
testingFrame,
Test_Imputed,
submissionTemplateFileName="data/TADPOLE_Simple_Submission_TeamName.xlsx",
usePreProc=USE_PREPROC)
#data_forecast_test = Path("data/_ForecastFRESACAD.csv")
#Forecast_D2 = pd.read_csv(data_forecast_test)
from tadpole_algorithms.evaluation import evaluate_forecast
from tadpole_algorithms.evaluation import print_metrics
# Evaluate the model
dictionary = evaluate_forecast(eval_df, Forecast_D2)
# Print metrics
print_metrics(dictionary)
# AdjustedTrainFrame,testingFrame,Train_Imputed,Test_Imputed = model.preproc_tadpole_D1_D2(data_df_train_test,USE_PREPROC)
D3AdjustedTrainFrame,D3testingFrame,D3Train_Imputed,D3Test_Imputed = model.preproc_with_R(D3Train,
D3Test,
data_Dictionaty,
MinVisit=18,
colImputeThreshold=0.15,
rowImputeThreshold=0.10,
includeID=False,
usePreProc=USE_PREPROC)
#Train Congitive Models
D3modelfilename = model.Train_Congitive(D3AdjustedTrainFrame,usePreProc=USE_PREPROC)
#Train ADAS/Ventricles Models
D3regresionModelfilename = model.Train_Regression(D3AdjustedTrainFrame,D3Train_Imputed,usePreProc=USE_PREPROC)
#Predict
Forecast_D3 = model.Forecast_All(D3modelfilename,
D3regresionModelfilename,
D3testingFrame,
D3Test_Imputed,
submissionTemplateFileName="data/TADPOLE_Simple_Submission_TeamName.xlsx",
usePreProc=USE_PREPROC)
from tadpole_algorithms.evaluation import evaluate_forecast
from tadpole_algorithms.evaluation import print_metrics
# Evaluate the D3 model
dictionary = evaluate_forecast(eval_df,Forecast_D3)
# Print metrics
print_metrics(dictionary)
```
|
github_jupyter
|
This notebook contains an implementation of the third place result in the Rossman Kaggle competition as detailed in Guo/Berkhahn's [Entity Embeddings of Categorical Variables](https://arxiv.org/abs/1604.06737).
The motivation behind exploring this architecture is it's relevance to real-world application. Much of our focus has been computer-vision and NLP tasks, which largely deals with unstructured data.
However, most of the data informing KPI's in industry are structured, time-series data. Here we explore the end-to-end process of using neural networks with practical structured data problems.
```
%matplotlib inline
import math, keras, datetime, pandas as pd, numpy as np, keras.backend as K
import matplotlib.pyplot as plt, xgboost, operator, random, pickle
from utils2 import *
np.set_printoptions(threshold=50, edgeitems=20)
limit_mem()
from isoweek import Week
from pandas_summary import DataFrameSummary
%cd data/rossman/
```
## Create datasets
In addition to the provided data, we will be using external datasets put together by participants in the Kaggle competition. You can download all of them [here](http://files.fast.ai/part2/lesson14/rossmann.tgz).
For completeness, the implementation used to put them together is included below.
```
def concat_csvs(dirname):
os.chdir(dirname)
filenames=glob.glob("*.csv")
wrote_header = False
with open("../"+dirname+".csv","w") as outputfile:
for filename in filenames:
name = filename.split(".")[0]
with open(filename) as f:
line = f.readline()
if not wrote_header:
wrote_header = True
outputfile.write("file,"+line)
for line in f:
outputfile.write(name + "," + line)
outputfile.write("\n")
os.chdir("..")
# concat_csvs('googletrend')
# concat_csvs('weather')
```
Feature Space:
* train: Training set provided by competition
* store: List of stores
* store_states: mapping of store to the German state they are in
* List of German state names
* googletrend: trend of certain google keywords over time, found by users to correlate well w/ given data
* weather: weather
* test: testing set
```
table_names = ['train', 'store', 'store_states', 'state_names',
'googletrend', 'weather', 'test']
```
We'll be using the popular data manipulation framework pandas.
Among other things, pandas allows you to manipulate tables/data frames in python as one would in a database.
We're going to go ahead and load all of our csv's as dataframes into a list `tables`.
```
tables = [pd.read_csv(fname+'.csv', low_memory=False) for fname in table_names]
from IPython.display import HTML
```
We can use `head()` to get a quick look at the contents of each table:
* train: Contains store information on a daily basis, tracks things like sales, customers, whether that day was a holdiay, etc.
* store: general info about the store including competition, etc.
* store_states: maps store to state it is in
* state_names: Maps state abbreviations to names
* googletrend: trend data for particular week/state
* weather: weather conditions for each state
* test: Same as training table, w/o sales and customers
```
for t in tables: display(t.head())
```
This is very representative of a typical industry dataset.
The following returns summarized aggregate information to each table accross each field.
```
for t in tables: display(DataFrameSummary(t).summary())
```
## Data Cleaning / Feature Engineering
As a structured data problem, we necessarily have to go through all the cleaning and feature engineering, even though we're using a neural network.
```
train, store, store_states, state_names, googletrend, weather, test = tables
len(train),len(test)
```
Turn state Holidays to Bool
```
train.StateHoliday = train.StateHoliday!='0'
test.StateHoliday = test.StateHoliday!='0'
```
Define function for joining tables on specific fields.
By default, we'll be doing a left outer join of `right` on the `left` argument using the given fields for each table.
Pandas does joins using the `merge` method. The `suffixes` argument describes the naming convention for duplicate fields. We've elected to leave the duplicate field names on the left untouched, and append a "_y" to those on the right.
```
def join_df(left, right, left_on, right_on=None):
if right_on is None: right_on = left_on
return left.merge(right, how='left', left_on=left_on, right_on=right_on,
suffixes=("", "_y"))
```
Join weather/state names.
```
weather = join_df(weather, state_names, "file", "StateName")
```
In pandas you can add new columns to a dataframe by simply defining it. We'll do this for googletrends by extracting dates and state names from the given data and adding those columns.
We're also going to replace all instances of state name 'NI' with the usage in the rest of the table, 'HB,NI'. This is a good opportunity to highlight pandas indexing. We can use `.ix[rows, cols]` to select a list of rows and a list of columns from the dataframe. In this case, we're selecting rows w/ statename 'NI' by using a boolean list `googletrend.State=='NI'` and selecting "State".
```
googletrend['Date'] = googletrend.week.str.split(' - ', expand=True)[0]
googletrend['State'] = googletrend.file.str.split('_', expand=True)[2]
googletrend.loc[googletrend.State=='NI', "State"] = 'HB,NI'
```
The following extracts particular date fields from a complete datetime for the purpose of constructing categoricals.
You should always consider this feature extraction step when working with date-time. Without expanding your date-time into these additional fields, you can't capture any trend/cyclical behavior as a function of time at any of these granularities.
```
def add_datepart(df):
df.Date = pd.to_datetime(df.Date)
df["Year"] = df.Date.dt.year
df["Month"] = df.Date.dt.month
df["Week"] = df.Date.dt.week
df["Day"] = df.Date.dt.day
```
We'll add to every table w/ a date field.
```
add_datepart(weather)
add_datepart(googletrend)
add_datepart(train)
add_datepart(test)
trend_de = googletrend[googletrend.file == 'Rossmann_DE']
```
Now we can outer join all of our data into a single dataframe.
Recall that in outer joins everytime a value in the joining field on the left table does not have a corresponding value on the right table, the corresponding row in the new table has Null values for all right table fields.
One way to check that all records are consistent and complete is to check for Null values post-join, as we do here.
*Aside*: Why note just do an inner join?
If you are assuming that all records are complete and match on the field you desire, an inner join will do the same thing as an outer join. However, in the event you are wrong or a mistake is made, an outer join followed by a null-check will catch it. (Comparing before/after # of rows for inner join is equivalent, but requires keeping track of before/after row #'s. Outer join is easier.)
```
store = join_df(store, store_states, "Store")
len(store[store.State.isnull()])
joined = join_df(train, store, "Store")
len(joined[joined.StoreType.isnull()])
joined = join_df(joined, googletrend, ["State","Year", "Week"])
len(joined[joined.trend.isnull()])
joined = joined.merge(trend_de, 'left', ["Year", "Week"], suffixes=('', '_DE'))
len(joined[joined.trend_DE.isnull()])
joined = join_df(joined, weather, ["State","Date"])
len(joined[joined.Mean_TemperatureC.isnull()])
joined_test = test.merge(store, how='left', left_on='Store', right_index=True)
len(joined_test[joined_test.StoreType.isnull()])
```
Next we'll fill in missing values to avoid complications w/ na's.
```
joined.CompetitionOpenSinceYear = joined.CompetitionOpenSinceYear.fillna(1900).astype(np.int32)
joined.CompetitionOpenSinceMonth = joined.CompetitionOpenSinceMonth.fillna(1).astype(np.int32)
joined.Promo2SinceYear = joined.Promo2SinceYear.fillna(1900).astype(np.int32)
joined.Promo2SinceWeek = joined.Promo2SinceWeek.fillna(1).astype(np.int32)
```
Next we'll extract features "CompetitionOpenSince" and "CompetitionDaysOpen". Note the use of `apply()` in mapping a function across dataframe values.
```
joined["CompetitionOpenSince"] = pd.to_datetime(joined.apply(lambda x: datetime.datetime(
x.CompetitionOpenSinceYear, x.CompetitionOpenSinceMonth, 15), axis=1).astype(pd.datetime))
joined["CompetitionDaysOpen"] = joined.Date.subtract(joined["CompetitionOpenSince"]).dt.days
```
We'll replace some erroneous / outlying data.
```
joined.loc[joined.CompetitionDaysOpen<0, "CompetitionDaysOpen"] = 0
joined.loc[joined.CompetitionOpenSinceYear<1990, "CompetitionDaysOpen"] = 0
```
Added "CompetitionMonthsOpen" field, limit the maximum to 2 years to limit number of unique embeddings.
```
joined["CompetitionMonthsOpen"] = joined["CompetitionDaysOpen"]//30
joined.loc[joined.CompetitionMonthsOpen>24, "CompetitionMonthsOpen"] = 24
joined.CompetitionMonthsOpen.unique()
```
Same process for Promo dates.
```
joined["Promo2Since"] = pd.to_datetime(joined.apply(lambda x: Week(
x.Promo2SinceYear, x.Promo2SinceWeek).monday(), axis=1).astype(pd.datetime))
joined["Promo2Days"] = joined.Date.subtract(joined["Promo2Since"]).dt.days
joined.loc[joined.Promo2Days<0, "Promo2Days"] = 0
joined.loc[joined.Promo2SinceYear<1990, "Promo2Days"] = 0
joined["Promo2Weeks"] = joined["Promo2Days"]//7
joined.loc[joined.Promo2Weeks<0, "Promo2Weeks"] = 0
joined.loc[joined.Promo2Weeks>25, "Promo2Weeks"] = 25
joined.Promo2Weeks.unique()
```
## Durations
It is common when working with time series data to extract data that explains relationships across rows as opposed to columns, e.g.:
* Running averages
* Time until next event
* Time since last event
This is often difficult to do with most table manipulation frameworks, since they are designed to work with relationships across columns. As such, we've created a class to handle this type of data.
```
columns = ["Date", "Store", "Promo", "StateHoliday", "SchoolHoliday"]
```
We've defined a class `elapsed` for cumulative counting across a sorted dataframe.
Given a particular field `fld` to monitor, this object will start tracking time since the last occurrence of that field. When the field is seen again, the counter is set to zero.
Upon initialization, this will result in datetime na's until the field is encountered. This is reset every time a new store is seen.
We'll see how to use this shortly.
```
class elapsed(object):
def __init__(self, fld):
self.fld = fld
self.last = pd.to_datetime(np.nan)
self.last_store = 0
def get(self, row):
if row.Store != self.last_store:
self.last = pd.to_datetime(np.nan)
self.last_store = row.Store
if (row[self.fld]): self.last = row.Date
return row.Date-self.last
df = train[columns]
```
And a function for applying said class across dataframe rows and adding values to a new column.
```
def add_elapsed(fld, prefix):
sh_el = elapsed(fld)
df[prefix+fld] = df.apply(sh_el.get, axis=1)
```
Let's walk through an example.
Say we're looking at School Holiday. We'll first sort by Store, then Date, and then call `add_elapsed('SchoolHoliday', 'After')`:
This will generate an instance of the `elapsed` class for School Holiday:
* Instance applied to every row of the dataframe in order of store and date
* Will add to the dataframe the days since seeing a School Holiday
* If we sort in the other direction, this will count the days until another promotion.
```
fld = 'SchoolHoliday'
df = df.sort_values(['Store', 'Date'])
add_elapsed(fld, 'After')
df = df.sort_values(['Store', 'Date'], ascending=[True, False])
add_elapsed(fld, 'Before')
```
We'll do this for two more fields.
```
fld = 'StateHoliday'
df = df.sort_values(['Store', 'Date'])
add_elapsed(fld, 'After')
df = df.sort_values(['Store', 'Date'], ascending=[True, False])
add_elapsed(fld, 'Before')
fld = 'Promo'
df = df.sort_values(['Store', 'Date'])
add_elapsed(fld, 'After')
df = df.sort_values(['Store', 'Date'], ascending=[True, False])
add_elapsed(fld, 'Before')
display(df.head())
```
We're going to set the active index to Date.
```
df = df.set_index("Date")
```
Then set null values from elapsed field calculations to 0.
```
columns = ['SchoolHoliday', 'StateHoliday', 'Promo']
for o in ['Before', 'After']:
for p in columns:
a = o+p
df[a] = df[a].fillna(pd.Timedelta(0)).dt.days
```
Next we'll demonstrate window functions in pandas to calculate rolling quantities.
Here we're sorting by date (`sort_index()`) and counting the number of events of interest (`sum()`) defined in `columns` in the following week (`rolling()`), grouped by Store (`groupby()`). We do the same in the opposite direction.
```
bwd = df[['Store']+columns].sort_index().groupby("Store").rolling(7, min_periods=1).sum()
fwd = df[['Store']+columns].sort_index(ascending=False
).groupby("Store").rolling(7, min_periods=1).sum()
```
Next we want to drop the Store indices grouped together in the window function.
Often in pandas, there is an option to do this in place. This is time and memory efficient when working with large datasets.
```
bwd.drop('Store',1,inplace=True)
bwd.reset_index(inplace=True)
fwd.drop('Store',1,inplace=True)
fwd.reset_index(inplace=True)
df.reset_index(inplace=True)
```
Now we'll merge these values onto the df.
```
df = df.merge(bwd, 'left', ['Date', 'Store'], suffixes=['', '_bw'])
df = df.merge(fwd, 'left', ['Date', 'Store'], suffixes=['', '_fw'])
df.drop(columns,1,inplace=True)
df.head()
```
It's usually a good idea to back up large tables of extracted / wrangled features before you join them onto another one, that way you can go back to it easily if you need to make changes to it.
```
df.to_csv('df.csv')
df = pd.read_csv('df.csv', index_col=0)
df["Date"] = pd.to_datetime(df.Date)
df.columns
joined = join_df(joined, df, ['Store', 'Date'])
```
We'll back this up as well.
```
joined.to_csv('joined.csv')
```
We now have our final set of engineered features.
```
joined = pd.read_csv('joined.csv', index_col=0)
joined["Date"] = pd.to_datetime(joined.Date)
joined.columns
```
While these steps were explicitly outlined in the paper, these are all fairly typical feature engineering steps for dealing with time series data and are practical in any similar setting.
## Create features
Now that we've engineered all our features, we need to convert to input compatible with a neural network.
This includes converting categorical variables into contiguous integers or one-hot encodings, normalizing continuous features to standard normal, etc...
```
from sklearn_pandas import DataFrameMapper
from sklearn.preprocessing import LabelEncoder, Imputer, StandardScaler
```
This dictionary maps categories to embedding dimensionality. In generally, categories we might expect to be conceptually more complex have larger dimension.
```
cat_var_dict = {'Store': 50, 'DayOfWeek': 6, 'Year': 2, 'Month': 6,
'Day': 10, 'StateHoliday': 3, 'CompetitionMonthsOpen': 2,
'Promo2Weeks': 1, 'StoreType': 2, 'Assortment': 3, 'PromoInterval': 3,
'CompetitionOpenSinceYear': 4, 'Promo2SinceYear': 4, 'State': 6,
'Week': 2, 'Events': 4, 'Promo_fw': 1,
'Promo_bw': 1, 'StateHoliday_fw': 1,
'StateHoliday_bw': 1, 'SchoolHoliday_fw': 1,
'SchoolHoliday_bw': 1}
```
Name categorical variables
```
cat_vars = [o[0] for o in
sorted(cat_var_dict.items(), key=operator.itemgetter(1), reverse=True)]
"""cat_vars = ['Store', 'DayOfWeek', 'Year', 'Month', 'Day', 'StateHoliday',
'StoreType', 'Assortment', 'Week', 'Events', 'Promo2SinceYear',
'CompetitionOpenSinceYear', 'PromoInterval', 'Promo', 'SchoolHoliday', 'State']"""
```
Likewise for continuous
```
# mean/max wind; min temp; cloud; min/mean humid;
contin_vars = ['CompetitionDistance',
'Max_TemperatureC', 'Mean_TemperatureC', 'Min_TemperatureC',
'Max_Humidity', 'Mean_Humidity', 'Min_Humidity', 'Max_Wind_SpeedKm_h',
'Mean_Wind_SpeedKm_h', 'CloudCover', 'trend', 'trend_DE',
'AfterStateHoliday', 'BeforeStateHoliday', 'Promo', 'SchoolHoliday']
"""contin_vars = ['CompetitionDistance', 'Max_TemperatureC', 'Mean_TemperatureC',
'Max_Humidity', 'trend', 'trend_DE', 'AfterStateHoliday', 'BeforeStateHoliday']"""
```
Replace nulls w/ 0 for continuous, "" for categorical.
```
for v in contin_vars: joined.loc[joined[v].isnull(), v] = 0
for v in cat_vars: joined.loc[joined[v].isnull(), v] = ""
```
Here we create a list of tuples, each containing a variable and an instance of a transformer for that variable.
For categoricals, we use a label encoder that maps categories to continuous integers. For continuous variables, we standardize them.
```
cat_maps = [(o, LabelEncoder()) for o in cat_vars]
contin_maps = [([o], StandardScaler()) for o in contin_vars]
```
The same instances need to be used for the test set as well, so values are mapped/standardized appropriately.
DataFrame mapper will keep track of these variable-instance mappings.
```
cat_mapper = DataFrameMapper(cat_maps)
cat_map_fit = cat_mapper.fit(joined)
cat_cols = len(cat_map_fit.features)
cat_cols
contin_mapper = DataFrameMapper(contin_maps)
contin_map_fit = contin_mapper.fit(joined)
contin_cols = len(contin_map_fit.features)
contin_cols
```
Example of first five rows of zeroth column being transformed appropriately.
```
cat_map_fit.transform(joined)[0,:5], contin_map_fit.transform(joined)[0,:5]
```
We can also pickle these mappings, which is great for portability!
```
pickle.dump(contin_map_fit, open('contin_maps.pickle', 'wb'))
pickle.dump(cat_map_fit, open('cat_maps.pickle', 'wb'))
[len(o[1].classes_) for o in cat_map_fit.features]
```
## Sample data
Next, the authors removed all instances where the store had zero sale / was closed.
```
joined_sales = joined[joined.Sales!=0]
n = len(joined_sales)
```
We speculate that this may have cost them a higher standing in the competition. One reason this may be the case is that a little EDA reveals that there are often periods where stores are closed, typically for refurbishment. Before and after these periods, there are naturally spikes in sales that one might expect. Be ommitting this data from their training, the authors gave up the ability to leverage information about these periods to predict this otherwise volatile behavior.
```
n
```
We're going to run on a sample.
```
samp_size = 100000
np.random.seed(42)
idxs = sorted(np.random.choice(n, samp_size, replace=False))
joined_samp = joined_sales.iloc[idxs].set_index("Date")
samp_size = n
joined_samp = joined_sales.set_index("Date")
```
In time series data, cross-validation is not random. Instead, our holdout data is always the most recent data, as it would be in real application.
We've taken the last 10% as our validation set.
```
train_ratio = 0.9
train_size = int(samp_size * train_ratio)
train_size
joined_valid = joined_samp[train_size:]
joined_train = joined_samp[:train_size]
len(joined_valid), len(joined_train)
```
Here's a preprocessor for our categoricals using our instance mapper.
```
def cat_preproc(dat):
return cat_map_fit.transform(dat).astype(np.int64)
cat_map_train = cat_preproc(joined_train)
cat_map_valid = cat_preproc(joined_valid)
```
Same for continuous.
```
def contin_preproc(dat):
return contin_map_fit.transform(dat).astype(np.float32)
contin_map_train = contin_preproc(joined_train)
contin_map_valid = contin_preproc(joined_valid)
```
Grab our targets.
```
y_train_orig = joined_train.Sales
y_valid_orig = joined_valid.Sales
```
Finally, the authors modified the target values by applying a logarithmic transformation and normalizing to unit scale by dividing by the maximum log value.
Log transformations are used on this type of data frequently to attain a nicer shape.
Further by scaling to the unit interval we can now use a sigmoid output in our neural network. Then we can multiply by the maximum log value to get the original log value and transform back.
```
max_log_y = np.max(np.log(joined_samp.Sales))
y_train = np.log(y_train_orig)/max_log_y
y_valid = np.log(y_valid_orig)/max_log_y
```
Note: Some testing shows this doesn't make a big difference.
```
"""#y_train = np.log(y_train)
ymean=y_train_orig.mean()
ystd=y_train_orig.std()
y_train = (y_train_orig-ymean)/ystd
#y_valid = np.log(y_valid)
y_valid = (y_valid_orig-ymean)/ystd"""
```
Root-mean-squared percent error is the metric Kaggle used for this competition.
```
def rmspe(y_pred, targ = y_valid_orig):
pct_var = (targ - y_pred)/targ
return math.sqrt(np.square(pct_var).mean())
```
These undo the target transformations.
```
def log_max_inv(preds, mx = max_log_y):
return np.exp(preds * mx)
# - This can be used if ymean and ystd are calculated above (they are currently commented out)
def normalize_inv(preds):
return preds * ystd + ymean
```
## Create models
Now we're ready to put together our models.
Much of the following code has commented out portions / alternate implementations.
```
"""
1 97s - loss: 0.0104 - val_loss: 0.0083
2 93s - loss: 0.0076 - val_loss: 0.0076
3 90s - loss: 0.0071 - val_loss: 0.0076
4 90s - loss: 0.0068 - val_loss: 0.0075
5 93s - loss: 0.0066 - val_loss: 0.0075
6 95s - loss: 0.0064 - val_loss: 0.0076
7 98s - loss: 0.0063 - val_loss: 0.0077
8 97s - loss: 0.0062 - val_loss: 0.0075
9 95s - loss: 0.0061 - val_loss: 0.0073
0 101s - loss: 0.0061 - val_loss: 0.0074
"""
def split_cols(arr):
return np.hsplit(arr,arr.shape[1])
# - This gives the correct list length for the model
# - (list of 23 elements: 22 embeddings + 1 array of 16-dim elements)
map_train = split_cols(cat_map_train) + [contin_map_train]
map_valid = split_cols(cat_map_valid) + [contin_map_valid]
len(map_train)
# map_train = split_cols(cat_map_train) + split_cols(contin_map_train)
# map_valid = split_cols(cat_map_valid) + split_cols(contin_map_valid)
```
Helper function for getting categorical name and dim.
```
def cat_map_info(feat): return feat[0], len(feat[1].classes_)
cat_map_info(cat_map_fit.features[1])
# - In Keras 2 the "initializations" module is not available.
# - To keep here the custom initializer the code from Keras 1 "uniform" initializer is exploited
def my_init(scale):
# return lambda shape, name=None: initializations.uniform(shape, scale=scale, name=name)
return K.variable(np.random.uniform(low=-scale, high=scale, size=shape),
name=name)
# - In Keras 2 the "initializations" module is not available.
# - To keep here the custom initializer the code from Keras 1 "uniform" initializer is exploited
def emb_init(shape, name=None):
# return initializations.uniform(shape, scale=2/(shape[1]+1), name=name)
return K.variable(np.random.uniform(low=-2/(shape[1]+1), high=2/(shape[1]+1), size=shape),
name=name)
```
Helper function for constructing embeddings. Notice commented out codes, several different ways to compute embeddings at play.
Also, note we're flattening the embedding. Embeddings in Keras come out as an element of a sequence like we might use in a sequence of words; here we just want to concatenate them so we flatten the 1-vector sequence into a vector.
```
def get_emb(feat):
name, c = cat_map_info(feat)
#c2 = cat_var_dict[name]
c2 = (c+1)//2
if c2>50: c2=50
inp = Input((1,), dtype='int64', name=name+'_in')
# , kernel_regularizer=l2(1e-6) # Keras 2
u = Flatten(name=name+'_flt')(Embedding(c, c2, input_length=1, embeddings_initializer=emb_init)(inp)) # Keras 2
# u = Flatten(name=name+'_flt')(Embedding(c, c2, input_length=1)(inp))
return inp,u
```
Helper function for continuous inputs.
```
def get_contin(feat):
name = feat[0][0]
inp = Input((1,), name=name+'_in')
return inp, Dense(1, name=name+'_d', kernel_initializer=my_init(1.))(inp) # Keras 2
```
Let's build them.
```
contin_inp = Input((contin_cols,), name='contin')
contin_out = Dense(contin_cols*10, activation='relu', name='contin_d')(contin_inp)
#contin_out = BatchNormalization()(contin_out)
```
Now we can put them together. Given the inputs, continuous and categorical embeddings, we're going to concatenate all of them.
Next, we're going to pass through some dropout, then two dense layers w/ ReLU activations, then dropout again, then the sigmoid activation we mentioned earlier.
```
embs = [get_emb(feat) for feat in cat_map_fit.features]
#conts = [get_contin(feat) for feat in contin_map_fit.features]
#contin_d = [d for inp,d in conts]
x = concatenate([emb for inp,emb in embs] + [contin_out]) # Keras 2
#x = concatenate([emb for inp,emb in embs] + contin_d) # Keras 2
x = Dropout(0.02)(x)
x = Dense(1000, activation='relu', kernel_initializer='uniform')(x)
x = Dense(500, activation='relu', kernel_initializer='uniform')(x)
x = Dropout(0.2)(x)
x = Dense(1, activation='sigmoid')(x)
model = Model([inp for inp,emb in embs] + [contin_inp], x)
#model = Model([inp for inp,emb in embs] + [inp for inp,d in conts], x)
model.compile('adam', 'mean_absolute_error')
#model.compile(Adam(), 'mse')
```
### Start training
```
%%time
hist = model.fit(map_train, y_train, batch_size=128, epochs=25,
verbose=1, validation_data=(map_valid, y_valid))
hist.history
plot_train(hist)
preds = np.squeeze(model.predict(map_valid, 1024))
```
Result on validation data: 0.1678 (samp 150k, 0.75 trn)
```
log_max_inv(preds)
# - This will work if ymean and ystd are calculated in the "Data" section above (in this case uncomment)
# normalize_inv(preds)
```
## Using 3rd place data
```
pkl_path = '/data/jhoward/github/entity-embedding-rossmann/'
def load_pickle(fname):
return pickle.load(open(pkl_path+fname + '.pickle', 'rb'))
[x_pkl_orig, y_pkl_orig] = load_pickle('feature_train_data')
max_log_y_pkl = np.max(np.log(y_pkl_orig))
y_pkl = np.log(y_pkl_orig)/max_log_y_pkl
pkl_vars = ['Open', 'Store', 'DayOfWeek', 'Promo', 'Year', 'Month', 'Day',
'StateHoliday', 'SchoolHoliday', 'CompetitionMonthsOpen', 'Promo2Weeks',
'Promo2Weeks_L', 'CompetitionDistance',
'StoreType', 'Assortment', 'PromoInterval', 'CompetitionOpenSinceYear',
'Promo2SinceYear', 'State', 'Week', 'Max_TemperatureC', 'Mean_TemperatureC',
'Min_TemperatureC', 'Max_Humidity', 'Mean_Humidity', 'Min_Humidity', 'Max_Wind_SpeedKm_h',
'Mean_Wind_SpeedKm_h', 'CloudCover','Events', 'Promo_fw', 'Promo_bw',
'StateHoliday_fw', 'StateHoliday_bw', 'AfterStateHoliday', 'BeforeStateHoliday',
'SchoolHoliday_fw', 'SchoolHoliday_bw', 'trend_DE', 'trend']
x_pkl = np.array(x_pkl_orig)
gt_enc = StandardScaler()
gt_enc.fit(x_pkl[:,-2:])
x_pkl[:,-2:] = gt_enc.transform(x_pkl[:,-2:])
x_pkl.shape
x_pkl = x_pkl[idxs]
y_pkl = y_pkl[idxs]
x_pkl_trn, x_pkl_val = x_pkl[:train_size], x_pkl[train_size:]
y_pkl_trn, y_pkl_val = y_pkl[:train_size], y_pkl[train_size:]
x_pkl_trn.shape
xgb_parms = {'learning_rate': 0.1, 'subsample': 0.6,
'colsample_bylevel': 0.6, 'silent': True, 'objective': 'reg:linear'}
xdata_pkl = xgboost.DMatrix(x_pkl_trn, y_pkl_trn, feature_names=pkl_vars)
xdata_val_pkl = xgboost.DMatrix(x_pkl_val, y_pkl_val, feature_names=pkl_vars)
xgb_parms['seed'] = random.randint(0,1e9)
model_pkl = xgboost.train(xgb_parms, xdata_pkl)
model_pkl.eval(xdata_val_pkl)
#0.117473
importance = model_pkl.get_fscore()
importance = sorted(importance.items(), key=operator.itemgetter(1))
df = pd.DataFrame(importance, columns=['feature', 'fscore'])
df['fscore'] = df['fscore'] / df['fscore'].sum()
df.plot(kind='barh', x='feature', y='fscore', legend=False, figsize=(6, 10))
plt.title('XGBoost Feature Importance')
plt.xlabel('relative importance');
```
### Neural net
```
#np.savez_compressed('vars.npz', pkl_cats, pkl_contins)
#np.savez_compressed('deps.npz', y_pkl)
pkl_cats = np.stack([x_pkl[:,pkl_vars.index(f)] for f in cat_vars], 1)
pkl_contins = np.stack([x_pkl[:,pkl_vars.index(f)] for f in contin_vars], 1)
co_enc = StandardScaler().fit(pkl_contins)
pkl_contins = co_enc.transform(pkl_contins)
pkl_contins_trn, pkl_contins_val = pkl_contins[:train_size], pkl_contins[train_size:]
pkl_cats_trn, pkl_cats_val = pkl_cats[:train_size], pkl_cats[train_size:]
y_pkl_trn, y_pkl_val = y_pkl[:train_size], y_pkl[train_size:]
def get_emb_pkl(feat):
name, c = cat_map_info(feat)
c2 = (c+2)//3
if c2>50: c2=50
inp = Input((1,), dtype='int64', name=name+'_in')
u = Flatten(name=name+'_flt')(Embedding(c, c2, input_length=1, init=emb_init)(inp))
return inp,u
n_pkl_contin = pkl_contins_trn.shape[1]
contin_inp = Input((n_pkl_contin,), name='contin')
contin_out = BatchNormalization()(contin_inp)
map_train_pkl = split_cols(pkl_cats_trn) + [pkl_contins_trn]
map_valid_pkl = split_cols(pkl_cats_val) + [pkl_contins_val]
def train_pkl(bs=128, ne=10):
return model_pkl.fit(map_train_pkl, y_pkl_trn, batch_size=bs, nb_epoch=ne,
verbose=0, validation_data=(map_valid_pkl, y_pkl_val))
def get_model_pkl():
conts = [get_contin_pkl(feat) for feat in contin_map_fit.features]
embs = [get_emb_pkl(feat) for feat in cat_map_fit.features]
x = merge([emb for inp,emb in embs] + [contin_out], mode='concat')
x = Dropout(0.02)(x)
x = Dense(1000, activation='relu', init='uniform')(x)
x = Dense(500, activation='relu', init='uniform')(x)
x = Dense(1, activation='sigmoid')(x)
model_pkl = Model([inp for inp,emb in embs] + [contin_inp], x)
model_pkl.compile('adam', 'mean_absolute_error')
#model.compile(Adam(), 'mse')
return model_pkl
model_pkl = get_model_pkl()
train_pkl(128, 10).history['val_loss']
K.set_value(model_pkl.optimizer.lr, 1e-4)
train_pkl(128, 5).history['val_loss']
"""
1 97s - loss: 0.0104 - val_loss: 0.0083
2 93s - loss: 0.0076 - val_loss: 0.0076
3 90s - loss: 0.0071 - val_loss: 0.0076
4 90s - loss: 0.0068 - val_loss: 0.0075
5 93s - loss: 0.0066 - val_loss: 0.0075
6 95s - loss: 0.0064 - val_loss: 0.0076
7 98s - loss: 0.0063 - val_loss: 0.0077
8 97s - loss: 0.0062 - val_loss: 0.0075
9 95s - loss: 0.0061 - val_loss: 0.0073
0 101s - loss: 0.0061 - val_loss: 0.0074
"""
plot_train(hist)
preds = np.squeeze(model_pkl.predict(map_valid_pkl, 1024))
y_orig_pkl_val = log_max_inv(y_pkl_val, max_log_y_pkl)
rmspe(log_max_inv(preds, max_log_y_pkl), y_orig_pkl_val)
```
## XGBoost
Xgboost is extremely quick and easy to use. Aside from being a powerful predictive model, it gives us information about feature importance.
```
X_train = np.concatenate([cat_map_train, contin_map_train], axis=1)
X_valid = np.concatenate([cat_map_valid, contin_map_valid], axis=1)
all_vars = cat_vars + contin_vars
xgb_parms = {'learning_rate': 0.1, 'subsample': 0.6,
'colsample_bylevel': 0.6, 'silent': True, 'objective': 'reg:linear'}
xdata = xgboost.DMatrix(X_train, y_train, feature_names=all_vars)
xdata_val = xgboost.DMatrix(X_valid, y_valid, feature_names=all_vars)
xgb_parms['seed'] = random.randint(0,1e9)
model = xgboost.train(xgb_parms, xdata)
model.eval(xdata_val)
model.eval(xdata_val)
```
Easily, competition distance is the most important, while events are not important at all.
In real applications, putting together a feature importance plot is often a first step. Oftentimes, we can remove hundreds of thousands of features from consideration with importance plots.
```
importance = model.get_fscore()
importance = sorted(importance.items(), key=operator.itemgetter(1))
df = pd.DataFrame(importance, columns=['feature', 'fscore'])
df['fscore'] = df['fscore'] / df['fscore'].sum()
df.plot(kind='barh', x='feature', y='fscore', legend=False, figsize=(6, 10))
plt.title('XGBoost Feature Importance')
plt.xlabel('relative importance');
```
## End
|
github_jupyter
|
<a href="https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/verbose/alphafold_noTemplates_noMD.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
#AlphaFold
```
#################
# WARNING
#################
# - This notebook is intended as a "quick" demo, it disables many aspects of the full alphafold2 pipeline
# (input MSA/templates, amber MD refinement, and number of models). For best results, we recommend using the
# full pipeline: https://github.com/deepmind/alphafold
# - That being said, it was found that input templates and amber-relax did not help much.
# The key input features are the MSA (Multiple Sequence Alignment) of related proteins. Where you see a
# significant drop in predicted accuracy when MSA < 30, but only minor improvements > 100.
# - This notebook does NOT include the alphafold2 MSA generation pipeline, and is designed to work with a
# single sequence, custom MSA input (that you can upload) or MMseqs2 webserver.
# - Single sequence mode is particularly useful for denovo designed proteins (where there are no sequence
# homologs by definition). For natural proteins, an MSA input will make a huge difference.
#################
# EXTRA
#################
# For other related notebooks see: https://github.com/sokrypton/ColabFold
# install/import alphafold (and required libs)
import os, sys
if not os.path.isdir("/content/alphafold"):
%shell git clone -q https://github.com/deepmind/alphafold.git; cd alphafold; git checkout -q 1d43aaff941c84dc56311076b58795797e49107b
%shell mkdir --parents params; curl -fsSL https://storage.googleapis.com/alphafold/alphafold_params_2021-07-14.tar | tar x -C params
%shell pip -q install biopython dm-haiku==0.0.5 ml-collections py3Dmol
if '/content/alphafold' not in sys.path:
sys.path.append('/content/alphafold')
import numpy as np
import py3Dmol
import matplotlib.pyplot as plt
from alphafold.common import protein
from alphafold.data import pipeline
from alphafold.data import templates
from alphafold.model import data
from alphafold.model import config
from alphafold.model import model
# setup which model params to use
# note: for this demo, we only use model 1, for all five models uncomments the others!
model_runners = {}
models = ["model_1"] #,"model_2","model_3","model_4","model_5"]
for model_name in models:
model_config = config.model_config(model_name)
model_config.data.eval.num_ensemble = 1
model_params = data.get_model_haiku_params(model_name=model_name, data_dir=".")
model_runner = model.RunModel(model_config, model_params)
model_runners[model_name] = model_runner
def mk_mock_template(query_sequence):
'''create blank template'''
ln = len(query_sequence)
output_templates_sequence = "-"*ln
templates_all_atom_positions = np.zeros((ln, templates.residue_constants.atom_type_num, 3))
templates_all_atom_masks = np.zeros((ln, templates.residue_constants.atom_type_num))
templates_aatype = templates.residue_constants.sequence_to_onehot(output_templates_sequence,templates.residue_constants.HHBLITS_AA_TO_ID)
template_features = {'template_all_atom_positions': templates_all_atom_positions[None],
'template_all_atom_masks': templates_all_atom_masks[None],
'template_aatype': np.array(templates_aatype)[None],
'template_domain_names': [f'none'.encode()]}
return template_features
def predict_structure(prefix, feature_dict, model_runners, random_seed=0):
"""Predicts structure using AlphaFold for the given sequence."""
# Run the models.
plddts = {}
for model_name, model_runner in model_runners.items():
processed_feature_dict = model_runner.process_features(feature_dict, random_seed=random_seed)
prediction_result = model_runner.predict(processed_feature_dict)
unrelaxed_protein = protein.from_prediction(processed_feature_dict,prediction_result)
unrelaxed_pdb_path = f'{prefix}_unrelaxed_{model_name}.pdb'
plddts[model_name] = prediction_result['plddt']
print(f"{model_name} {plddts[model_name].mean()}")
with open(unrelaxed_pdb_path, 'w') as f:
f.write(protein.to_pdb(unrelaxed_protein))
return plddts
```
# Single sequence input (no MSA)
```
# Change this line to sequence you want to predict
query_sequence = "GWSTELEKHREELKEFLKKEGITNVEIRIDNGRLEVRVEGGTERLKRFLEELRQKLEKKGYTVDIKIE"
%%time
feature_dict = {
**pipeline.make_sequence_features(sequence=query_sequence,
description="none",
num_res=len(query_sequence)),
**pipeline.make_msa_features(msas=[[query_sequence]],
deletion_matrices=[[[0]*len(query_sequence)]]),
**mk_mock_template(query_sequence)
}
plddts = predict_structure("test",feature_dict,model_runners)
# confidence per position
plt.figure(dpi=100)
for model,value in plddts.items():
plt.plot(value,label=model)
plt.legend()
plt.ylim(0,100)
plt.ylabel("predicted LDDT")
plt.xlabel("positions")
plt.show()
p = py3Dmol.view(js='https://3dmol.org/build/3Dmol.js')
p.addModel(open("test_unrelaxed_model_1.pdb",'r').read(),'pdb')
p.setStyle({'cartoon': {'color':'spectrum'}})
p.zoomTo()
p.show()
p = py3Dmol.view(js='https://3dmol.org/build/3Dmol.js')
p.addModel(open("test_unrelaxed_model_1.pdb",'r').read(),'pdb')
p.setStyle({'cartoon': {'color':'spectrum'},'stick':{}})
p.zoomTo()
p.show()
```
# Custom MSA input
```
%%bash
# for this demo we will download a premade MSA input
wget -qnc --no-check-certificate https://gremlin2.bakerlab.org/db/ECOLI/fasta/P0A8I3.fas
msa, deletion_matrix = pipeline.parsers.parse_a3m("".join(open("P0A8I3.fas","r").readlines()))
query_sequence = msa[0]
%%time
feature_dict = {
**pipeline.make_sequence_features(sequence=query_sequence,
description="none",
num_res=len(query_sequence)),
**pipeline.make_msa_features(msas=[msa],deletion_matrices=[deletion_matrix]),
**mk_mock_template(query_sequence)
}
plddts = predict_structure("yaaa",feature_dict,model_runners)
# confidence per position
plt.figure(dpi=100)
for model,value in plddts.items():
plt.plot(value,label=model)
plt.legend()
plt.ylim(0,100)
plt.ylabel("predicted LDDT")
plt.xlabel("positions")
plt.show()
p = py3Dmol.view(js='https://3dmol.org/build/3Dmol.js')
p.addModel(open("yaaa_unrelaxed_model_1.pdb",'r').read(),'pdb')
p.setStyle({'cartoon': {'color':'spectrum'}})
p.zoomTo()
p.show()
```
#MSA from MMseqs2
```
##############################
# Where do I get an MSA?
##############################
# For any "serious" use, I would recommend using the alphafold2 pipeline to make the MSAs,
# since this is what it was trained on.
# That being said, part of the MSA generation pipeline (specifically searching against uniprot database using hhblits)
# can be done here: https://toolkit.tuebingen.mpg.de/tools/hhblits
# Alternatively, using the SUPER FAST MMseqs2 pipeline below
# for a HUMAN FRIENDLY version see:
# https://colab.research.google.com/drive/1LVPSOf4L502F21RWBmYJJYYLDlOU2NTL
%%bash
apt-get -qq -y update 2>&1 1>/dev/null
apt-get -qq -y install jq curl zlib1g gawk 2>&1 1>/dev/null
# save query sequence to file
name = "YAII"
query_sequence = "MTIWVDADACPNVIKEILYRAAERMQMPLVLVANQSLRVPPSRFIRTLRVAAGFDVADNEIVRQCEAGDLVITADIPLAAEAIEKGAAALNPRGERYTPATIRERLTMRDFMDTLRASGIQTGGPDSLSQRDRQAFAAELEKWWLEVQRSRG"
with open(f"{name}.fasta","w") as out: out.write(f">{name}\n{query_sequence}\n")
%%bash -s "$name"
# build msa using the MMseqs2 search server
ID=$(curl -s -F q=@$1.fasta -F mode=all https://a3m.mmseqs.com/ticket/msa | jq -r '.id')
STATUS=$(curl -s https://a3m.mmseqs.com/ticket/${ID} | jq -r '.status')
while [ "${STATUS}" == "RUNNING" ]; do
STATUS=$(curl -s https://a3m.mmseqs.com/ticket/${ID} | jq -r '.status')
sleep 1
done
if [ "${STATUS}" == "COMPLETE" ]; then
curl -s https://a3m.mmseqs.com/result/download/${ID} > $1.tar.gz
tar xzf $1.tar.gz
tr -d '\000' < uniref.a3m > $1.a3m
else
echo "MMseqs2 server did not return a valid result."
exit 1
fi
echo "Found $(grep -c ">" $1.a3m) sequences (after redundacy filtering)"
msa, deletion_matrix = pipeline.parsers.parse_a3m("".join(open(f"{name}.a3m","r").readlines()))
query_sequence = msa[0]
%%time
feature_dict = {
**pipeline.make_sequence_features(sequence=query_sequence,
description="none",
num_res=len(query_sequence)),
**pipeline.make_msa_features(msas=[msa],deletion_matrices=[deletion_matrix]),
**mk_mock_template(query_sequence)
}
plddts = predict_structure(name,feature_dict,model_runners)
plt.figure(dpi=100)
plt.plot((feature_dict["msa"] != 21).sum(0))
plt.plot([0,len(query_sequence)],[30,30])
plt.xlabel("positions")
plt.ylabel("number of sequences")
plt.show()
# confidence per position
plt.figure(dpi=100)
for model,value in plddts.items():
plt.plot(value,label=model)
plt.legend()
plt.ylim(0,100)
plt.ylabel("predicted LDDT")
plt.xlabel("positions")
plt.show()
p = py3Dmol.view(js='https://3dmol.org/build/3Dmol.js')
p.addModel(open(f"{name}_unrelaxed_model_1.pdb",'r').read(),'pdb')
p.setStyle({'cartoon': {'color':'spectrum'}})
p.zoomTo()
p.show()
```
|
github_jupyter
|
# mlrose Tutorial Examples - Genevieve Hayes
## Overview
mlrose is a Python package for applying some of the most common randomized optimization and search algorithms to a range of different optimization problems, over both discrete- and continuous-valued parameter spaces. This notebook contains the examples used in the mlrose tutorial.
### Import Libraries
```
import mlrose
import numpy as np
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler, OneHotEncoder
from sklearn.metrics import accuracy_score
```
### Example 1: 8-Queens Using Pre-Defined Fitness Function
```
# Initialize fitness function object using pre-defined class
fitness = mlrose.Queens()
# Define optimization problem object
problem = mlrose.DiscreteOpt(length = 8, fitness_fn = fitness, maximize=False, max_val=8)
# Define decay schedule
schedule = mlrose.ExpDecay()
# Solve using simulated annealing - attempt 1
np.random.seed(1)
init_state = np.array([0, 1, 2, 3, 4, 5, 6, 7])
best_state, best_fitness = mlrose.simulated_annealing(problem, schedule = schedule, max_attempts = 10,
max_iters = 1000, init_state = init_state)
print(best_state)
print(best_fitness)
# Solve using simulated annealing - attempt 2
np.random.seed(1)
best_state, best_fitness = mlrose.simulated_annealing(problem, schedule = schedule, max_attempts = 100,
max_iters = 1000, init_state = init_state)
print(best_state)
print(best_fitness)
```
### Example 2: 8-Queens Using Custom Fitness Function
```
# Define alternative N-Queens fitness function for maximization problem
def queens_max(state):
# Initialize counter
fitness = 0
# For all pairs of queens
for i in range(len(state) - 1):
for j in range(i + 1, len(state)):
# Check for horizontal, diagonal-up and diagonal-down attacks
if (state[j] != state[i]) \
and (state[j] != state[i] + (j - i)) \
and (state[j] != state[i] - (j - i)):
# If no attacks, then increment counter
fitness += 1
return fitness
# Check function is working correctly
state = np.array([1, 4, 1, 3, 5, 5, 2, 7])
# The fitness of this state should be 22
queens_max(state)
# Initialize custom fitness function object
fitness_cust = mlrose.CustomFitness(queens_max)
# Define optimization problem object
problem_cust = mlrose.DiscreteOpt(length = 8, fitness_fn = fitness_cust, maximize = True, max_val = 8)
# Solve using simulated annealing - attempt 1
np.random.seed(1)
best_state, best_fitness = mlrose.simulated_annealing(problem_cust, schedule = schedule,
max_attempts = 10, max_iters = 1000,
init_state = init_state)
print(best_state)
print(best_fitness)
# Solve using simulated annealing - attempt 2
np.random.seed(1)
best_state, best_fitness = mlrose.simulated_annealing(problem_cust, schedule = schedule,
max_attempts = 100, max_iters = 1000,
init_state = init_state)
print(best_state)
print(best_fitness)
```
### Example 3: Travelling Salesperson Using Coordinate-Defined Fitness Function
```
# Create list of city coordinates
coords_list = [(1, 1), (4, 2), (5, 2), (6, 4), (4, 4), (3, 6), (1, 5), (2, 3)]
# Initialize fitness function object using coords_list
fitness_coords = mlrose.TravellingSales(coords = coords_list)
# Define optimization problem object
problem_fit = mlrose.TSPOpt(length = 8, fitness_fn = fitness_coords, maximize = False)
# Solve using genetic algorithm - attempt 1
np.random.seed(2)
best_state, best_fitness = mlrose.genetic_alg(problem_fit)
print(best_state)
print(best_fitness)
# Solve using genetic algorithm - attempt 2
np.random.seed(2)
best_state, best_fitness = mlrose.genetic_alg(problem_fit, mutation_prob = 0.2, max_attempts = 100)
print(best_state)
print(best_fitness)
```
### Example 4: Travelling Salesperson Using Distance-Defined Fitness Function
```
# Create list of distances between pairs of cities
dist_list = [(0, 1, 3.1623), (0, 2, 4.1231), (0, 3, 5.8310), (0, 4, 4.2426), (0, 5, 5.3852), \
(0, 6, 4.0000), (0, 7, 2.2361), (1, 2, 1.0000), (1, 3, 2.8284), (1, 4, 2.0000), \
(1, 5, 4.1231), (1, 6, 4.2426), (1, 7, 2.2361), (2, 3, 2.2361), (2, 4, 2.2361), \
(2, 5, 4.4721), (2, 6, 5.0000), (2, 7, 3.1623), (3, 4, 2.0000), (3, 5, 3.6056), \
(3, 6, 5.0990), (3, 7, 4.1231), (4, 5, 2.2361), (4, 6, 3.1623), (4, 7, 2.2361), \
(5, 6, 2.2361), (5, 7, 3.1623), (6, 7, 2.2361)]
# Initialize fitness function object using dist_list
fitness_dists = mlrose.TravellingSales(distances = dist_list)
# Define optimization problem object
problem_fit2 = mlrose.TSPOpt(length = 8, fitness_fn = fitness_dists, maximize = False)
# Solve using genetic algorithm
np.random.seed(2)
best_state, best_fitness = mlrose.genetic_alg(problem_fit2, mutation_prob = 0.2, max_attempts = 100)
print(best_state)
print(best_fitness)
```
### Example 5: Travelling Salesperson Defining Fitness Function as Part of Optimization Problem Definition Step
```
# Create list of city coordinates
coords_list = [(1, 1), (4, 2), (5, 2), (6, 4), (4, 4), (3, 6), (1, 5), (2, 3)]
# Define optimization problem object
problem_no_fit = mlrose.TSPOpt(length = 8, coords = coords_list, maximize = False)
# Solve using genetic algorithm
np.random.seed(2)
best_state, best_fitness = mlrose.genetic_alg(problem_no_fit, mutation_prob = 0.2, max_attempts = 100)
print(best_state)
print(best_fitness)
```
### Example 6: Fitting a Neural Network to the Iris Dataset
```
# Load the Iris dataset
data = load_iris()
# Get feature values of first observation
print(data.data[0])
# Get feature names
print(data.feature_names)
# Get target value of first observation
print(data.target[0])
# Get target name of first observation
print(data.target_names[data.target[0]])
# Get minimum feature values
print(np.min(data.data, axis = 0))
# Get maximum feature values
print(np.max(data.data, axis = 0))
# Get unique target values
print(np.unique(data.target))
# Split data into training and test sets
X_train, X_test, y_train, y_test = train_test_split(data.data, data.target, test_size = 0.2,
random_state = 3)
# Normalize feature data
scaler = MinMaxScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
# One hot encode target values
one_hot = OneHotEncoder()
y_train_hot = one_hot.fit_transform(y_train.reshape(-1, 1)).todense()
y_test_hot = one_hot.transform(y_test.reshape(-1, 1)).todense()
# Initialize neural network object and fit object - attempt 1
np.random.seed(3)
nn_model1 = mlrose.NeuralNetwork(hidden_nodes = [2], activation ='relu',
algorithm ='random_hill_climb',
max_iters = 1000, bias = True, is_classifier = True,
learning_rate = 0.0001, early_stopping = True,
clip_max = 5, max_attempts = 100)
nn_model1.fit(X_train_scaled, y_train_hot)
# Predict labels for train set and assess accuracy
y_train_pred = nn_model1.predict(X_train_scaled)
y_train_accuracy = accuracy_score(y_train_hot, y_train_pred)
print(y_train_accuracy)
# Predict labels for test set and assess accuracy
y_test_pred = nn_model1.predict(X_test_scaled)
y_test_accuracy = accuracy_score(y_test_hot, y_test_pred)
print(y_test_accuracy)
# Initialize neural network object and fit object - attempt 2
np.random.seed(3)
nn_model2 = mlrose.NeuralNetwork(hidden_nodes = [2], activation = 'relu',
algorithm = 'gradient_descent',
max_iters = 1000, bias = True, is_classifier = True,
learning_rate = 0.0001, early_stopping = True,
clip_max = 5, max_attempts = 100)
nn_model2.fit(X_train_scaled, y_train_hot)
# Predict labels for train set and assess accuracy
y_train_pred = nn_model2.predict(X_train_scaled)
y_train_accuracy = accuracy_score(y_train_hot, y_train_pred)
print(y_train_accuracy)
# Predict labels for test set and assess accuracy
y_test_pred = nn_model2.predict(X_test_scaled)
y_test_accuracy = accuracy_score(y_test_hot, y_test_pred)
print(y_test_accuracy)
```
### Example 7: Fitting a Logistic Regression to the Iris Data
```
# Initialize logistic regression object and fit object - attempt 1
np.random.seed(3)
lr_model1 = mlrose.LogisticRegression(algorithm = 'random_hill_climb', max_iters = 1000,
bias = True, learning_rate = 0.0001,
early_stopping = True, clip_max = 5, max_attempts = 100)
lr_model1.fit(X_train_scaled, y_train_hot)
# Predict labels for train set and assess accuracy
y_train_pred = lr_model1.predict(X_train_scaled)
y_train_accuracy = accuracy_score(y_train_hot, y_train_pred)
print(y_train_accuracy)
# Predict labels for test set and assess accuracy
y_test_pred = lr_model1.predict(X_test_scaled)
y_test_accuracy = accuracy_score(y_test_hot, y_test_pred)
print(y_test_accuracy)
# Initialize logistic regression object and fit object - attempt 2
np.random.seed(3)
lr_model2 = mlrose.LogisticRegression(algorithm = 'random_hill_climb', max_iters = 1000,
bias = True, learning_rate = 0.01,
early_stopping = True, clip_max = 5, max_attempts = 100)
lr_model2.fit(X_train_scaled, y_train_hot)
# Predict labels for train set and assess accuracy
y_train_pred = lr_model2.predict(X_train_scaled)
y_train_accuracy = accuracy_score(y_train_hot, y_train_pred)
print(y_train_accuracy)
# Predict labels for test set and assess accuracy
y_test_pred = lr_model2.predict(X_test_scaled)
y_test_accuracy = accuracy_score(y_test_hot, y_test_pred)
print(y_test_accuracy)
```
### Example 8: Fitting a Logistic Regression to the Iris Data using the NeuralNetwork() class
```
# Initialize neural network object and fit object - attempt 1
np.random.seed(3)
lr_nn_model1 = mlrose.NeuralNetwork(hidden_nodes = [], activation = 'sigmoid',
algorithm = 'random_hill_climb',
max_iters = 1000, bias = True, is_classifier = True,
learning_rate = 0.0001, early_stopping = True,
clip_max = 5, max_attempts = 100)
lr_nn_model1.fit(X_train_scaled, y_train_hot)
# Predict labels for train set and assess accuracy
y_train_pred = lr_nn_model1.predict(X_train_scaled)
y_train_accuracy = accuracy_score(y_train_hot, y_train_pred)
print(y_train_accuracy)
# Predict labels for test set and assess accuracy
y_test_pred = lr_nn_model1.predict(X_test_scaled)
y_test_accuracy = accuracy_score(y_test_hot, y_test_pred)
print(y_test_accuracy)
# Initialize neural network object and fit object - attempt 2
np.random.seed(3)
lr_nn_model2 = mlrose.NeuralNetwork(hidden_nodes = [], activation = 'sigmoid',
algorithm = 'random_hill_climb',
max_iters = 1000, bias = True, is_classifier = True,
learning_rate = 0.01, early_stopping = True,
clip_max = 5, max_attempts = 100)
lr_nn_model2.fit(X_train_scaled, y_train_hot)
# Predict labels for train set and assess accuracy
y_train_pred = lr_nn_model2.predict(X_train_scaled)
y_train_accuracy = accuracy_score(y_train_hot, y_train_pred)
print(y_train_accuracy)
# Predict labels for test set and assess accuracy
y_test_pred = lr_nn_model2.predict(X_test_scaled)
y_test_accuracy = accuracy_score(y_test_hot, y_test_pred)
print(y_test_accuracy)
```
|
github_jupyter
|
<!--NAVIGATION-->
_______________
Este documento puede ser utilizado de forma interactiva en las siguientes plataformas:
- [Google Colab](https://colab.research.google.com/github/masdeseiscaracteres/ml_course/blob/master/material/05_random_forests.ipynb)
- [MyBinder](https://mybinder.org/v2/gh/masdeseiscaracteres/ml_course/master)
- [Deepnote](https://deepnote.com/launch?template=python_3.6&url=https%3A%2F%2Fgithub.com%2Fmasdeseiscaracteres%2Fml_course%2Fblob%2Fmaster%2Fmaterial%2F05_random_forests.ipynb)
_______________
# Bagging: Random Forests
Vamos a analizar el funcionamiento de la técnica de *bagging* aplicada a árboles. En concreto veremos los *random forest* para [clasificación](http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html#sklearn.ensemble.RandomForestClassifier) y [regresión](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestRegressor.html) mediante ejemplos ilustrativos.
Este notebook se estructura de la siguiente manera:
1. Ejemplo en clasificación
2. Ejemplo en regresión
## 0. Configuración del entorno
```
# clonar el resto del repositorio si no está disponible
import os
curr_dir = os.getcwd()
if not os.path.exists(os.path.join(curr_dir, '../.ROOT_DIR')):
!git clone https://github.com/masdeseiscaracteres/ml_course.git ml_course
os.chdir(os.path.join(curr_dir, 'ml_course/material'))
```
En primer lugar, preparamos el entorno con las bibliotecas y datos necesarios:
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
%matplotlib inline
cm = plt.cm.RdBu
cm_bright = ListedColormap(['#FF0000', '#0000FF'])
import warnings
warnings.filterwarnings('ignore')
```
## 1. Ejemplo en clasificación
En este primer ejemplo vamos a explorar el conjunto de datos para la detección de cáncer de mama ([Breast Cancer](https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+%28Diagnostic%29)). Este dataset está también incluido en el módulo `sklearn.datasets`.
El objetivo es detectar si un cáncer es beningo o maligno (B/N) a partir de la información de atributos numéricos que caracterizan los núcleos celulares de las imágenes digitalizadas de biopsias realizadas a distintos pacientes.
La variable objetivo es binaria.
```
from sklearn.datasets import load_breast_cancer
bunch = load_breast_cancer()
X = bunch['data']
y = bunch['target']
target_lut = {k:v for k,v in zip([0,1], bunch['target_names'])} # conversión de variable objetivo a string
feature_names = bunch['feature_names']
# verificamos que los tamaños de los datos son consistentes entre sí
print(X.shape)
print(y.shape)
```
En primer lugar vemos cómo se distribuye la variable objetivo:
```
# Calculamos los valores distintos que toma y el número de veces que aparece cada uno
np.unique(y, return_counts=True)
```
Conviene echar un vistazo a los datos. Como todos los datos son numéricos, un histograma puede ser una buena opción:
```
# Pintamos histogramas para cada clase
plt.figure(figsize=(20,20))
idx_0 = (y==0)
idx_1 = (y==1)
for i, feature in enumerate(feature_names):
plt.subplot(6, 5, i+1)
plt.hist(X[idx_0, i], density=1, alpha=0.6, label='y=0')
plt.hist(X[idx_1, i], density=1, facecolor='red', alpha=0.6, label='y=1')
plt.legend()
plt.title(feature)
plt.show()
```
Vamos a reservar una parte de nuestros datos para la evaluación final y entrenamos sobre el resto, como si se tratase de una situación real: disponemos de los datos recogidos para construir nuestro modelo que después será aplicado sobre nuevos pacientes.
```
from sklearn.model_selection import train_test_split
# separamos los datos
X_train, X_test, y_train, y_test = train_test_split(X, y, stratify=y, test_size=0.2, random_state=0)
print('Datos train: ', X_train.shape)
print('Datos test: ', X_test.shape)
```
## 1.1. Árbol de decisión convencional
En primer lugar entrenamos un árbol de decisión convencional para hacernos una idea de las prestaciones que alcanzamos.
```
from sklearn.model_selection import GridSearchCV
from sklearn.tree import DecisionTreeClassifier
from sklearn import tree
max_depth_arr = range(1, 10+1)
param_grid = {'max_depth': max_depth_arr}
n_folds = 10
clf = DecisionTreeClassifier(random_state=0)
grid = GridSearchCV(clf, param_grid=param_grid, cv=n_folds, return_train_score=True)
grid.fit(X_train, y_train)
print("best mean cross-validation score: {:.3f}".format(grid.best_score_))
print("best parameters: {}".format(grid.best_params_))
scores_test = np.array(grid.cv_results_['mean_test_score'])
scores_train = np.array(grid.cv_results_['mean_train_score'])
plt.plot(max_depth_arr, scores_test, '-o', label='Validación')
plt.plot(max_depth_arr, scores_train, '-o', label='Entrenamiento')
plt.xlabel('max_depth', fontsize=16)
plt.ylabel('{}-Fold accuracy'.format(n_folds))
plt.ylim((0.8, 1))
plt.show()
best_max_depth = grid.best_params_['max_depth']
tree_model = DecisionTreeClassifier(max_depth=best_max_depth)
tree_model.fit(X_train, y_train)
print("Train: ", tree_model.score(X_train, y_train))
print("Test: ", tree_model.score(X_test, y_test))
from sklearn.tree import export_graphviz
import graphviz
tree_dot = export_graphviz(tree_model, out_file=None, feature_names=feature_names, class_names=['M', 'B'],
filled=True, rounded=True, special_characters=True)
graph = graphviz.Source(tree_dot)
# Mostrar grafo como SVG
# graph
# Mostrar grafo como PNG
from IPython.display import Image
Image(graph.pipe(format='png'))
importances = tree_model.feature_importances_
importances = importances / np.max(importances)
indices = np.argsort(importances)[::-1]
plt.figure(figsize=(10, 10))
plt.barh(range(X_train.shape[1]), importances[indices])
plt.yticks(range(X_train.shape[1]), feature_names[indices])
plt.show()
```
## 1.2. Random forest
En un *random forest* aparecen nuevos parámetros libres:
- Número de árboles construidos: aquí hemos de asegurarnos que la función de coste es estable para el número de árboles elegido.
- Número máximo de características a seleccionar aleatoriamente para ajustar cada árbol.
Además de los propios de los árboles de decisión:
- Complejidad de los mismos (normalmente `max_depth` o `min_samples_leaf`)
```
from sklearn.ensemble import RandomForestClassifier
# grid search
max_depth_arr = range(1, 15)
params = {'max_depth': max_depth_arr}
n_folds = 10
clf = RandomForestClassifier(random_state=0, n_estimators=200, max_features='sqrt')
grid = GridSearchCV(clf, param_grid=params, cv=n_folds, return_train_score=True)
grid.fit(X_train, y_train)
print("best mean cross-validation score: {:.3f}".format(grid.best_score_))
print("best parameters: {}".format(grid.best_params_))
scores_test = np.array(grid.cv_results_['mean_test_score'])
scores_train = np.array(grid.cv_results_['mean_train_score'])
plt.plot(max_depth_arr, scores_test, '-o', label='Validación')
plt.plot(max_depth_arr, scores_train, '-o', label='Entrenamiento')
plt.xlabel('max_depth', fontsize=16)
plt.ylabel('{}-Fold accuracy'.format(n_folds))
plt.ylim((0.8, 1))
plt.show()
best_max_depth = grid.best_params_['max_depth']
bag_model = RandomForestClassifier(max_depth=best_max_depth, n_estimators=200, max_features='sqrt')
bag_model.fit(X_train, y_train)
print("Train: ", bag_model.score(X_train, y_train))
print("Test: ", bag_model.score(X_test, y_test))
```
## 1.3. Importancia de las variables
Una propiedad muy interesante de los algoritmos basados en árboles es que podemos medir la importancia de las variables
```
importances = bag_model.feature_importances_
importances = importances / np.max(importances)
indices = np.argsort(importances)[::-1]
plt.figure(figsize=(10,10))
plt.barh(range(X_train.shape[1]), importances[indices])
plt.yticks(range(X_train.shape[1]), feature_names[indices])
plt.show()
```
Utilizando este ranking, podemos hacer selección de características:
```
from sklearn.model_selection import KFold
N, N_features = X_train.shape
rf = RandomForestClassifier(max_depth=best_max_depth, n_estimators=200, max_features='sqrt')
n_folds = 10
kf = KFold(n_splits=n_folds, shuffle=True, random_state=1)
cv_error = []
cv_std = []
for nfeatures in range(N_features, 0, -1):
error_i = []
for idxTrain, idxVal in kf.split(X_train):
Xt = X_train[idxTrain,:]
yt = y_train[idxTrain]
Xv = X_train[idxVal,:]
yv = y_train[idxVal]
rf.fit(Xt, yt)
ranking = rf.feature_importances_
indices = np.argsort(ranking)[::-1]
selected = indices[0:(N_features-nfeatures+1)]
Xs = Xt[:, selected]
rf.fit(Xs, yt)
error = (1.0 - rf.score(Xv[:, selected], yv))
error_i.append(error)
cv_error.append(np.mean(error_i))
cv_std.append(np.std(error_i))
print('# features: ' + str(len(selected)) + ', error: ' + str(np.mean(error_i)) + ' +/- ' + str(np.std(error_i)))
plt.plot(range(1, N_features+1,1), cv_error, '-o')
plt.errorbar(range(1, N_features+1,1), cv_error, yerr=cv_std, fmt='o')
plt.xlabel('# features')
plt.ylabel('CV error')
plt.show()
```
Como se puede ver, seleccionando las primeras 7 u 8 características obtenemos unos resultados muy buenos. De esta manera reducimos la complejidad del algoritmo y facilitamos su explicación.
## 2. Ejemplo en regresión
```
# cargamos datos
house_data = pd.read_csv("./data/kc_house_data.csv") # cargamos fichero
# Eliminamos las columnas id y date
house_data = house_data.drop(['id','date'], axis=1)
# convertir las variables en pies al cuadrado en metros al cuadrado
feetFeatures = ['sqft_living', 'sqft_lot', 'sqft_above', 'sqft_basement', 'sqft_living15', 'sqft_lot15']
house_data[feetFeatures] = house_data[feetFeatures].apply(lambda x: x * 0.3048 * 0.3048)
# renombramos
house_data.columns = ['price','bedrooms','bathrooms','sqm_living','sqm_lot','floors','waterfront','view','condition',
'grade','sqm_above','sqm_basement','yr_built','yr_renovated','zip_code','lat','long',
'sqm_living15','sqm_lot15']
# añadimos las nuevas variables
house_data['years'] = pd.Timestamp('today').year - house_data['yr_built']
#house_data['bedrooms_squared'] = house_data['bedrooms'].apply(lambda x: x**2)
#house_data['bed_bath_rooms'] = house_data['bedrooms']*house_data['bathrooms']
house_data['sqm_living'] = house_data['sqm_living'].apply(lambda x: np.log(x))
house_data['price'] = house_data['price'].apply(lambda x: np.log(x))
#house_data['lat_plus_long'] = house_data['lat']*house_data['long']
house_data.head()
# convertimos el DataFrame al formato necesario para scikit-learn
data = house_data.values
y = data[:, 0:1] # nos quedamos con la 1ª columna, price
X = data[:, 1:] # nos quedamos con el resto
feature_names = house_data.columns[1:]
# Dividimos los datos en entrenamiento y test (80 training, 20 test)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=.25, random_state = 2)
print('Datos entrenamiento: ', X_train.shape)
print('Datos test: ', X_test.shape)
```
### 2.1 Árbol de decisión
#### Ejercicio
Entrena un árbol de decisión y devuelve las prestaciones para el conjunto de test.
#### Solución
```
from sklearn.tree import DecisionTreeRegressor
# parámetros para GridSearch
max_depth = range(1, 15)
tuned_parameters = {'max_depth': max_depth}
n_folds = 5
clf = DecisionTreeRegressor(random_state=0)
grid = GridSearchCV(clf, param_grid=tuned_parameters, cv=n_folds, return_train_score=True)
grid.fit(X_train, y_train)
print("best mean cross-validation score: {:.3f}".format(grid.best_score_))
print("best parameters: {}".format(grid.best_params_))
scores_test = np.array(grid.cv_results_['mean_test_score'])
scores_train = np.array(grid.cv_results_['mean_train_score'])
plt.plot(max_depth_arr, scores_test, '-o', label='Validación')
plt.plot(max_depth_arr, scores_train, '-o', label='Entrenamiento')
plt.xlabel('max_depth', fontsize=16)
plt.ylabel('{}-fold R-squared'.format(n_folds))
plt.ylim((0.5, 1))
plt.show()
best_max_depth = grid.best_params_['max_depth']
dt = DecisionTreeRegressor(max_depth=best_max_depth)
dt.fit(X_train, y_train)
print("Train: ", dt.score(X_train, y_train))
print("Test: ", dt.score(X_test, y_test))
```
### 2.2. Random forest
#### Ejercicio
Entrena un algoritmo de random forest y devuelve las prestaciones para el conjunto de test.
#### Solución
```
from sklearn.ensemble import RandomForestRegressor
# parámetros para GridSearch
max_depth_arr = range(1, 20+1)
tuned_parameters = {'max_depth': max_depth_arr}
n_folds = 3 # ponemos este valor algo bajo para que no tarde demasiado
clf = RandomForestRegressor(random_state=0, n_estimators=100, max_features='sqrt')
grid = GridSearchCV(clf, param_grid=tuned_parameters, cv=n_folds, return_train_score=True)
grid.fit(X_train, y_train)
print("best mean cross-validation score: {:.3f}".format(grid.best_score_))
print("best parameters: {}".format(grid.best_params_))
scores_test = np.array(grid.cv_results_['mean_test_score'])
scores_train = np.array(grid.cv_results_['mean_train_score'])
plt.plot(max_depth_arr, scores_test, '-o', label='Validación')
plt.plot(max_depth_arr, scores_train, '-o', label='Entrenamiento')
plt.xlabel('max_depth', fontsize=16)
plt.ylabel('{}-fold R-squared'.format(n_folds))
plt.ylim((0.5, 1))
plt.show()
best_max_depth = grid.best_params_['max_depth']
bag_model = RandomForestRegressor(max_depth=best_max_depth)
bag_model.fit(X_train, y_train)
print("Train: ", bag_model.score(X_train, y_train))
print("Test: ", bag_model.score(X_test, y_test))
```
#### Ejercicio
¿Qué características son las más relevantes?¿Coincide este ranking de características con las variables importantes seleccionadas por el algoritmo Lasso?
#### Solución
```
importances = bag_model.feature_importances_
importances = importances / np.max(importances)
indices = np.argsort(importances)[::-1]
plt.figure(figsize=(10,10))
plt.barh(range(X_train.shape[1]), importances[indices])
plt.yticks(range(X_train.shape[1]), feature_names[indices])
plt.show()
from sklearn.linear_model import Lasso
lasso = Lasso(alpha=1e-4, normalize=True)
lasso.fit(X_train, y_train)
print("Train: ", lasso.score(X_train, y_train))
print("Test: ", lasso.score(X_test, y_test))
importances = lasso.coef_
importances = importances / np.max(importances)
indices = np.argsort(importances)[::-1]
plt.figure(figsize=(10, 10))
plt.barh(range(X_train.shape[1]), importances[indices])
plt.yticks(range(X_train.shape[1]), feature_names[indices])
plt.show()
```
Como se puede ver, algunas variables coinciden y otras no. Es natural que esto ocurra, pues cada modelo explica la relación entre las características y la variable objetivo de una forma diferente. Los modelos de árboles son capaces de encontrar relaciones no lineales, mientras que Lasso solo puede encontrar relaciones lineales.
|
github_jupyter
|
```
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
% matplotlib inline
```
### Loading Training Transactions Data
```
tr_tr = pd.read_csv('data/train_transaction.csv', index_col='TransactionID')
print('Rows :', tr_tr.shape[0],' Columns : ',tr_tr.shape[1] )
tr_tr.tail()
print('Memory Usage : ', (tr_tr.memory_usage(deep=True).sum()/1024).round(0))
tr_tr.tail()
tr_tr.isFraud.describe()
tr_tr.isFraud.value_counts().plot(kind='bar')
tr_tr.isFraud.value_counts()
f, (ax1, ax2) = plt.subplots(2, 1, sharex=True, figsize=(12,4))
ax1.hist(tr_tr.TransactionAmt[tr_tr.isFraud == 1], bins = 10)
ax1.set_title('Fraud Transactions ='+str(tr_tr.isFraud.value_counts()[1]))
ax2.hist(tr_tr.TransactionAmt[tr_tr.isFraud == 0], bins = 10)
ax2.set_title('Normal Transactions ='+str(tr_tr.isFraud.value_counts()[0]))
plt.xlabel('Amount ($)')
plt.ylabel('Number of Transactions')
plt.yscale('log')
plt.show()
```
### Exploratory Analysis of category Items in Training Transactions data
```
for i in tr_tr.columns:
if tr_tr[i].dtypes == str('object'):
print('Column Name :', i)
print('Unique Items :', tr_tr[i].unique())
print('Number of NaNs :', tr_tr[i].isna().sum())
print('Number of Frauds :','\n', tr_tr[tr_tr.isFraud==1][i].value_counts(dropna=False))
print('*'*50)
```
### Exploratory Analysis of Float Items in Training Transactions data
```
for i in tr_tr.columns:
if tr_tr[i].dtypes == 'float64':
print('Column Name :', i)
print('Number of NaNs :', tr_tr[i].isna().sum())
print('*'*50)
```
### Exploratory Analysis of Int Items in Training Transactions data
```
for i in tr_tr.columns:
if tr_tr[i].dtypes == 'int64':
print('Column Name :', i)
print('Number of NaNs :', tr_tr[i].isna().sum())
print('*'*50)
```
### Loading Test Transactions Data
```
te_tr = pd.read_csv('data/test_transaction.csv', index_col='TransactionID')
print(te_tr.shape)
te_tr.tail()
```
### Exploratory Analysis of category Items in Test Transactions data
```
for i in te_tr.columns:
if te_tr[i].dtypes == str('object'):
print('Column Name :', i)
print('Unique Items :', te_tr[i].unique())
print('Number of NaNs :', te_tr[i].isna().sum())
print('*'*50)
```
### Exploratory Analysis of Float Items in Test Transactions data
```
for i in te_tr.columns:
if te_tr[i].dtypes == 'float64':
print('Column Name :', i)
print('Number of NaNs :', te_tr[i].isna().sum())
print('*'*50)
```
### Check for any missing column in Test transaction data for integrity
```
for i in tr_tr.columns:
if i in te_tr.columns:
pass
elif i == str('isFraud'):
print('All columns are present in Test Transactions data')
else:
print(i)
```
### Check for any mismatching category items between Training and Test transaction data
```
for i in te_tr.columns:
if te_tr[i].dtypes == str('object'):
for j in te_tr[i].unique():
if j in tr_tr[i].unique():
pass
else:
print(j,': item is in test but not in training for category : ',i)
```
### Loading Training Identity Data
```
tr_id = pd.read_csv('data/train_identity.csv', index_col='TransactionID')
print(tr_id.shape)
tr_id.tail()
```
### Exploratory Analysis of category Items in Training Identity data
```
for i in tr_id.columns:
if tr_id[i].dtypes == str('object'):
print('Column Name :', i)
print('Unique Items :', tr_id[i].unique())
print('Number of NaNs :', tr_id[i].isna().sum())
print('*'*50)
```
### Exploratory Analysis of Float Items in Training Identity data
```
for i in tr_id.columns:
if tr_id[i].dtypes == 'float64':
print('Column Name :', i)
print('Number of NaNs :', tr_id[i].isna().sum())
print('*'*50)
```
### Combining training transactions and identity data
```
tr = tr_tr.join(tr_id)
print(tr.shape)
tr.head()
print('percent of NaN data : ',tr.isna().any().mean())
print('Top 10 columns with NaN data :','\n',tr.isna().mean().sort_values(ascending=False).head(10))
```
### Fraud Counts by Category Items for Training Data
```
for i in tr.columns:
if tr[i].dtypes == str('object'):
print('Fraud Counts for : ',i)
print('-'*30)
print(tr[tr.isFraud==1][i].value_counts(dropna=False))
```
### Create categories for items with more than 100 counts of Fraud
```
def map_categories(*args):
columns = [col for col in args]
for column in columns:
if column == index:
return 1
else:
return 0
new_tr_categories = []
for i in tr.columns:
if tr[i].dtypes == str('object'):
fraud_count = tr[tr_tr.isFraud==1][i].value_counts(dropna=False)
for index, value in fraud_count.items():
if value>100:
tr[(str(i)+'_'+str(index))]=list(map(map_categories, tr[i]))
new_tr_categories.append((str(i)+'_'+str(index)))
# else:
# tr[(str(i)+'_'+str('other'))]=list(map(map_categories, tr[i]))
# new_tr_categories.append((str(i)+'_'+str('other')))
tr.drop([i], axis=1, inplace=True)
print(new_tr_categories)
```
### Replace NaN with zero for combined training data
```
tr.fillna(0, inplace=True)
tr.head()
```
### Loading Test Transactions Data
```
te_tr = pd.read_csv('data/test_transaction.csv', index_col='TransactionID')
print(te_tr.shape)
te_tr.tail()
```
### Loading Test Identity Data
```
te_id = pd.read_csv('data/test_identity.csv', index_col='TransactionID')
print(te_id.shape)
te_id.tail()
```
### Exploratory Analysis of category Items in Test Identity data
```
for i in te_id.columns:
if te_id[i].dtypes == str('object'):
print('Column Name :', i)
print('Unique Items :', te_id[i].unique())
print('Number of NaNs :', te_id[i].isna().sum())
print('*'*50)
```
### Exploratory Analysis of Float Items in Test Identity data
```
for i in te_id.columns:
if te_id[i].dtypes == 'float64':
print('Column Name :', i)
print('Number of NaNs :', te_id[i].isna().sum())
print('*'*50)
### check for any missing column in Test identity data for integrity
for i in tr_id.columns:
if i in te_id.columns:
pass
else:
print(i)
for i in te_id.columns:
if te_id[i].dtypes == str('object'):
for j in te_id[i].unique():
if j in tr_id[i].unique():
pass
else:
print(j,': item is in test but not in training for category : ',i)
```
### Combining Test transactions and identity data
```
te = te_tr.join(te_id)
print(te.shape)
te.head()
print('percent of NaN data : ',te.isna().any().mean())
print('Top 10 columns with NaN data :','\n',te.isna().mean().sort_values(ascending=False).head(10))
```
|
github_jupyter
|
```
midifile = 'data/chopin-fantaisie.mid'
import time
import copy
import subprocess
from abc import abstractmethod
import numpy as np
import midi # Midi file parser
from midipattern import MidiPattern
from distorter import *
from align import align_frame_to_frame, read_align, write_align
MidiPattern.MIDI_DEVICE = 2
```
Init Pygame and Audio
--------
Midi Pattern
--------
```
pattern = MidiPattern(midi.read_midifile(midifile))
simple = pattern.simplified(bpm=160)
simple.stamp_time('t0')
midi.write_midifile("generated/simple.mid", simple)
print simple.attributes[0][-40:]
pattern[0]
pattern.play(180)
simple.play()
```
Distorter
--------
```
distorter = VelocityNoiseDistorter(sigma=20.)
distorter.randomize()
print distorter
dist_pattern = distorter.distort(simple)
midi.write_midifile('generated/velocity-noise.mid', dist_pattern)
dist_pattern.play(bpm=180)
print dist_pattern.attributes[0][-4:]
distorter = VelocityWalkDistorter(sigma=0.1)
distorter.randomize()
print distorter
dist_pattern = distorter.distort(simple)
midi.write_midifile('generated/velocity-walk.mid', dist_pattern)
dist_pattern.play(bpm=180)
distorter = ProgramDistorter()
distorter.randomize()
# for some reason GM 1- 3 makes no sound in pygame?
print distorter
dist_pattern = distorter.distort(simple)
midi.write_midifile('generated/program.mid', dist_pattern)
dist_pattern.play(bpm=180)
distorter = TempoDistorter(sigma=0, min=0.5, max=2.)
distorter.randomize()
print distorter
dist_pattern = distorter.distort(simple)
print 'time warp', dist_pattern.attributes[0][-4:]
midi.write_midifile('generated/tempo.mid', dist_pattern)
dist_pattern.play(bpm=180)
distorter = TimeNoiseDistorter()
distorter.randomize()
print distorter
dist_pattern = distorter.distort(simple)
print 'time warp', dist_pattern.attributes[0][-4:]
midi.write_midifile('generated/time.mid', dist_pattern)
dist_pattern.play(bpm=180)
```
Individual Note Times to Global Alignment
-------
```
stride = 1.
align = align_frame_to_frame(dist_pattern, stride)
align
write_align('generated/align.txt', align, stride)
align2, stride2 = read_align('generated/align.txt')
print align2 == align
print int(stride2) == int(stride), stride2, stride
```
Actual Generation
----
```
dist_pattern = random_distort(simple)
align = align_frame_to_frame(dist_pattern, stride=1.)
print align
dist_pattern.play()
num_samples = 10
stride = 0.1
for i in xrange(num_samples):
base_name = 'generated/sample-{}'.format(i)
align_name = '{}.txt'.format(base_name)
midi_name = '{}.mid'.format(base_name)
wav_name = '{}.wav'.format(base_name)
distorted = random_distort(simple)
align = align_frame_to_frame(distorted, stride)
write_align(align_name, align, stride)
midi.write_midifile(midi_name, distorted)
# Convert to wav using timidity
print wav_name
subprocess.check_call(['timidity', '-Ow', midi_name, '-o', wav_name])
print 'Done generating {}'.format(base_name)
```
|
github_jupyter
|
# SciPy를 이용한 최적화
- fun: 2.0
hess_inv: array([[ 0.5]])
jac: array([ 0.])
message: 'Optimization terminated successfully.'
nfev: 9 # SciPy는 Sympy가 아니라서, Symbolic을 활용하지 못하기에 수치 미분을 함 - 1위치에서 3번 계산됨 nit가 2 이라는거는 2번 뛰엇나느 것이며, 3곳에서 9번함수를돌림..
nit: 2
njev: 3
status: 0
success: True
x: array([ 1.99999999])
def f1p(x):
return 2 * (x - 2)
result = sp.optimize.minimize(f1, x0, jac=f1p) ## 여기 flp 값을 미리 설정해줘야 1자리에서 3번 계산안하게 됨, 더 빨리됨
print(result)
fun: 2.0
hess_inv: array([[ 0.5]])
jac: array([ 0.])
message: 'Optimization terminated successfully.'
nfev: 3
nit: 2
njev: 3
status: 0
success: True
x: array([ 2.])
```
# 연습문제 1
# 2차원 RosenBerg 함수에 대해
# 1) 최적해에 수렴할 수 있도록 초기점을 변경하여 본다.
# 2) 그레디언트 벡터 함수를 구현하여 jac 인수로 주는 방법으로 계산 속도를 향상시킨다.
# 1) 최적해에 수렴할 수 있도록 초기점을 변경하여 본다.
x0 = 1 # 초기값 설정
result = sp.optimize.minimize(f1, x0)
print(result)
%matplotlib inline
def f1(x):
return (x - 2) ** 2 + 2
xx = np.linspace(-1, 4, 100)
plt.plot(xx, f1(xx))
plt.plot(2, 2, 'ro', markersize=20)
plt.ylim(0, 10)
plt.show()
def f2(x, y):
return (1 - x)**2 + 100.0 * (y - x**2)**2
xx = np.linspace(-4, 4, 800)
yy = np.linspace(-3, 3, 600)
X, Y = np.meshgrid(xx, yy)
Z = f2(X, Y)
plt.contour(X, Y, Z, colors="gray", levels=[0.4, 3, 15, 50, 150, 500, 1500, 5000])
plt.plot(1, 1, 'ro', markersize=20)
plt.xlim(-4, 4)
plt.ylim(-3, 3)
plt.xticks(np.linspace(-4, 4, 9))
plt.yticks(np.linspace(-3, 3, 7))
plt.show()
def f1d(x):
"""derivative of f1(x)"""
return 2 * (x - 2.0)
xx = np.linspace(-1, 4, 100)
plt.plot(xx, f1(xx), 'k-')
# step size
mu = 0.4
# k = 0
x = 0
plt.plot(x, f1(x), 'go', markersize=10)
plt.plot(xx, f1d(x) * (xx - x) + f1(x), 'b--')
print("x = {}, g = {}".format(x, f1d(x)))
# k = 1
x = x - mu * f1d(x)
plt.plot(x, f1(x), 'go', markersize=10)
plt.plot(xx, f1d(x) * (xx - x) + f1(x), 'b--')
print("x = {}, g = {}".format(x, f1d(x)))
# k = 2
x = x - mu * f1d(x)
plt.plot(x, f1(x), 'go', markersize=10)
plt.plot(xx, f1d(x) * (xx - x) + f1(x), 'b--')
print("x = {}, g = {}".format(x, f1d(x)))
plt.ylim(0, 10)
plt.show()
# 1)
def f2g(x, y):
"""gradient of f2(x)"""
return np.array((2.0 * (x - 1) - 400.0 * x * (y - x**2), 200.0 * (y - x**2)))
xx = np.linspace(-4, 4, 800)
yy = np.linspace(-3, 3, 600)
X, Y = np.meshgrid(xx, yy)
Z = f2(X, Y)
levels=np.logspace(-1, 3, 10)
plt.contourf(X, Y, Z, alpha=0.2, levels=levels)
plt.contour(X, Y, Z, colors="green", levels=levels, zorder=0)
plt.plot(1, 1, 'ro', markersize=10)
mu = 8e-4 # step size
s = 0.95 # for arrowr head drawing
x, y = 0, 0 # x = 1 , y = 1 에서 시작
for i in range(5):
g = f2g(x, y)
plt.arrow(x, y, -s * mu * g[0], -s * mu * g[1], head_width=0.04, head_length=0.04, fc='k', ec='k', lw=2)
x = x - mu * g[0]
y = y - mu * g[1]
plt.xlim(-3, 3)
plt.ylim(-2, 2)
plt.xticks(np.linspace(-3, 3, 7))
plt.yticks(np.linspace(-2, 2, 5))
plt.show()
x0 = -0.5 # 초기값
result = sp.optimize.minimize(f1, x0)
print(result)
def f1p(x):
return 2 * (x - 2)
result = sp.optimize.minimize(f1, x0, jac=f1p)
print(result)
def f2(x):
return (1 - x[0])**2 + 400.0 * (x[1] - x[0]**2)**2
x0 = (0.7, 0.7)
result = sp.optimize.minimize(f2, x0)
print(result)
def f2p(x):
return np.array([2*x[0]-2-1600*x[0]*x[1]+1600*x[0]**3, 800*x[1]-800*x[0]**2])
result = sp.optimize.minimize(f2, x0, jac=f2p)
print(result)
```
|
github_jupyter
|
# RadarCOVID-Report
## Data Extraction
```
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import pycountry
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
```
### Constants
```
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
```
### Parameters
```
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
environment_enable_multi_backend_download = \
os.environ.get("RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD")
if environment_enable_multi_backend_download:
report_backend_identifiers = None
else:
report_backend_identifiers = [report_backend_identifier]
report_backend_identifiers
environment_invalid_shared_diagnoses_dates = \
os.environ.get("RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES")
if environment_invalid_shared_diagnoses_dates:
invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(",")
else:
invalid_shared_diagnoses_dates = []
invalid_shared_diagnoses_dates
```
### COVID-19 Cases
```
report_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=report_backend_identifier)
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe():
return pd.read_csv("https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv")
confirmed_df_ = download_cases_dataframe()
confirmed_df_.iloc[0]
confirmed_df = confirmed_df_.copy()
confirmed_df = confirmed_df[["date", "new_cases", "iso_code"]]
confirmed_df.rename(
columns={
"date": "sample_date",
"iso_code": "country_code",
},
inplace=True)
def convert_iso_alpha_3_to_alpha_2(x):
try:
return pycountry.countries.get(alpha_3=x).alpha_2
except Exception as e:
logging.info(f"Error converting country ISO Alpha 3 code '{x}': {repr(e)}")
return None
confirmed_df["country_code"] = confirmed_df.country_code.apply(convert_iso_alpha_3_to_alpha_2)
confirmed_df.dropna(inplace=True)
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_days_df["sample_date_string"] = \
confirmed_days_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_days_df.tail()
def sort_source_regions_for_display(source_regions: list) -> list:
if report_backend_identifier in source_regions:
source_regions = [report_backend_identifier] + \
list(sorted(set(source_regions).difference([report_backend_identifier])))
else:
source_regions = list(sorted(source_regions))
return source_regions
report_source_regions = report_backend_client.source_regions_for_date(
date=extraction_datetime.date())
report_source_regions = sort_source_regions_for_display(
source_regions=report_source_regions)
report_source_regions
def get_cases_dataframe(source_regions_for_date_function, columns_suffix=None):
source_regions_at_date_df = confirmed_days_df.copy()
source_regions_at_date_df["source_regions_at_date"] = \
source_regions_at_date_df.sample_date.apply(
lambda x: source_regions_for_date_function(date=x))
source_regions_at_date_df.sort_values("sample_date", inplace=True)
source_regions_at_date_df["_source_regions_group"] = source_regions_at_date_df. \
source_regions_at_date.apply(lambda x: ",".join(sort_source_regions_for_display(x)))
source_regions_at_date_df.tail()
#%%
source_regions_for_summary_df_ = \
source_regions_at_date_df[["sample_date", "_source_regions_group"]].copy()
source_regions_for_summary_df_.rename(columns={"_source_regions_group": "source_regions"}, inplace=True)
source_regions_for_summary_df_.tail()
#%%
confirmed_output_columns = ["sample_date", "new_cases", "covid_cases"]
confirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)
for source_regions_group, source_regions_group_series in \
source_regions_at_date_df.groupby("_source_regions_group"):
source_regions_set = set(source_regions_group.split(","))
confirmed_source_regions_set_df = \
confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()
confirmed_source_regions_group_df = \
confirmed_source_regions_set_df.groupby("sample_date").new_cases.sum() \
.reset_index().sort_values("sample_date")
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df.merge(
confirmed_days_df[["sample_date_string"]].rename(
columns={"sample_date_string": "sample_date"}),
how="right")
confirmed_source_regions_group_df["covid_cases"] = \
confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[confirmed_output_columns]
confirmed_source_regions_group_df = confirmed_source_regions_group_df.replace(0, np.nan)
confirmed_source_regions_group_df.fillna(method="ffill", inplace=True)
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[
confirmed_source_regions_group_df.sample_date.isin(
source_regions_group_series.sample_date_string)]
confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)
result_df = confirmed_output_df.copy()
result_df.tail()
#%%
result_df.rename(columns={"sample_date": "sample_date_string"}, inplace=True)
result_df = confirmed_days_df[["sample_date_string"]].merge(result_df, how="left")
result_df.sort_values("sample_date_string", inplace=True)
result_df.fillna(method="ffill", inplace=True)
result_df.tail()
#%%
result_df[["new_cases", "covid_cases"]].plot()
if columns_suffix:
result_df.rename(
columns={
"new_cases": "new_cases_" + columns_suffix,
"covid_cases": "covid_cases_" + columns_suffix},
inplace=True)
return result_df, source_regions_for_summary_df_
confirmed_eu_df, source_regions_for_summary_df = get_cases_dataframe(
report_backend_client.source_regions_for_date)
confirmed_es_df, _ = get_cases_dataframe(
lambda date: [spain_region_country_code],
columns_suffix=spain_region_country_code.lower())
```
### Extract API TEKs
```
raw_zip_path_prefix = "Data/TEKs/Raw/"
base_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=base_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
```
### Dump API TEKs
```
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_base_df = tek_list_df[tek_list_df.region == report_backend_identifier]
tek_list_base_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_base_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_base_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_base_df.head()
```
### Load TEK Dumps
```
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
```
### Daily New TEKs
```
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \
.groupby(["upload_date"]).shared_teks.max().reset_index() \
.sort_values(["upload_date"], ascending=False) \
.rename(columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
invalid_shared_diagnoses_dates_mask = \
estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)
estimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0
estimated_shared_diagnoses_df.head()
```
### Hourly New TEKs
```
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
```
### Official Statistics
```
import requests
import pandas.io.json
official_stats_response = requests.get("https://radarcovid.covid19.gob.es/kpi/statistics/basics")
official_stats_response.raise_for_status()
official_stats_df_ = pandas.io.json.json_normalize(official_stats_response.json())
official_stats_df = official_stats_df_.copy()
official_stats_df["date"] = pd.to_datetime(official_stats_df["date"], dayfirst=True)
official_stats_df.head()
official_stats_column_map = {
"date": "sample_date",
"applicationsDownloads.totalAcummulated": "app_downloads_es_accumulated",
"communicatedContagions.totalAcummulated": "shared_diagnoses_es_accumulated",
}
accumulated_suffix = "_accumulated"
accumulated_values_columns = \
list(filter(lambda x: x.endswith(accumulated_suffix), official_stats_column_map.values()))
interpolated_values_columns = \
list(map(lambda x: x[:-len(accumulated_suffix)], accumulated_values_columns))
official_stats_df = \
official_stats_df[official_stats_column_map.keys()] \
.rename(columns=official_stats_column_map)
official_stats_df["extraction_date"] = extraction_date
official_stats_df.head()
official_stats_path = "Data/Statistics/Current/RadarCOVID-Statistics.json"
previous_official_stats_df = pd.read_json(official_stats_path, orient="records", lines=True)
previous_official_stats_df["sample_date"] = pd.to_datetime(previous_official_stats_df["sample_date"], dayfirst=True)
official_stats_df = official_stats_df.append(previous_official_stats_df)
official_stats_df.head()
official_stats_df = official_stats_df[~(official_stats_df.shared_diagnoses_es_accumulated == 0)]
official_stats_df.sort_values("extraction_date", ascending=False, inplace=True)
official_stats_df.drop_duplicates(subset=["sample_date"], keep="first", inplace=True)
official_stats_df.head()
official_stats_stored_df = official_stats_df.copy()
official_stats_stored_df["sample_date"] = official_stats_stored_df.sample_date.dt.strftime("%Y-%m-%d")
official_stats_stored_df.to_json(official_stats_path, orient="records", lines=True)
official_stats_df.drop(columns=["extraction_date"], inplace=True)
official_stats_df = confirmed_days_df.merge(official_stats_df, how="left")
official_stats_df.sort_values("sample_date", ascending=False, inplace=True)
official_stats_df.head()
official_stats_df[accumulated_values_columns] = \
official_stats_df[accumulated_values_columns] \
.astype(float).interpolate(limit_area="inside")
official_stats_df[interpolated_values_columns] = \
official_stats_df[accumulated_values_columns].diff(periods=-1)
official_stats_df.drop(columns="sample_date", inplace=True)
official_stats_df.head()
```
### Data Merge
```
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
official_stats_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_eu_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df = confirmed_es_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df = result_summary_df.merge(source_regions_for_summary_df, how="left")
result_summary_df.set_index(["sample_date", "source_regions"], inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(result_summary_df.shared_diagnoses_es / result_summary_df.covid_cases_es).fillna(0)
result_summary_df.head(daily_plot_days)
def compute_aggregated_results_summary(days) -> pd.DataFrame:
aggregated_result_summary_df = result_summary_df.copy()
aggregated_result_summary_df["covid_cases_for_ratio"] = \
aggregated_result_summary_df.covid_cases.mask(
aggregated_result_summary_df.shared_diagnoses == 0, 0)
aggregated_result_summary_df["covid_cases_for_ratio_es"] = \
aggregated_result_summary_df.covid_cases_es.mask(
aggregated_result_summary_df.shared_diagnoses_es == 0, 0)
aggregated_result_summary_df = aggregated_result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(days).agg({
"covid_cases": "sum",
"covid_cases_es": "sum",
"covid_cases_for_ratio": "sum",
"covid_cases_for_ratio_es": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum",
"shared_diagnoses_es": "sum",
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
aggregated_result_summary_df = aggregated_result_summary_df.fillna(0).astype(int)
aggregated_result_summary_df["teks_per_shared_diagnosis"] = \
(aggregated_result_summary_df.shared_teks_by_upload_date /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case"] = \
(aggregated_result_summary_df.shared_diagnoses /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(aggregated_result_summary_df.shared_diagnoses_es /
aggregated_result_summary_df.covid_cases_for_ratio_es).fillna(0)
return aggregated_result_summary_df
aggregated_result_with_7_days_window_summary_df = compute_aggregated_results_summary(days=7)
aggregated_result_with_7_days_window_summary_df.head()
last_7_days_summary = aggregated_result_with_7_days_window_summary_df.to_dict(orient="records")[1]
last_7_days_summary
aggregated_result_with_14_days_window_summary_df = compute_aggregated_results_summary(days=14)
last_14_days_summary = aggregated_result_with_14_days_window_summary_df.to_dict(orient="records")[1]
last_14_days_summary
```
## Report Results
```
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"source_regions": "Source Countries",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases (Source Countries)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date (Source Countries)",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date (Source Countries)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date (Source Countries)",
"shared_diagnoses": "Shared Diagnoses (Source Countries – Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis (Source Countries)",
"shared_diagnoses_per_covid_case": "Usage Ratio (Source Countries)",
"covid_cases_es": "COVID-19 Cases (Spain)",
"app_downloads_es": "App Downloads (Spain – Official)",
"shared_diagnoses_es": "Shared Diagnoses (Spain – Official)",
"shared_diagnoses_per_covid_case_es": "Usage Ratio (Spain)",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
"covid_cases_es",
"app_downloads_es",
"shared_diagnoses_es",
"shared_diagnoses_per_covid_case_es",
]
summary_percentage_columns= [
"shared_diagnoses_per_covid_case_es",
"shared_diagnoses_per_covid_case",
]
```
### Daily Summary Table
```
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
```
### Daily Summary Plots
```
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.droplevel(level=["source_regions"]) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 30), legend=False)
ax_ = summary_ax_list[0]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
for percentage_column in summary_percentage_columns:
percentage_column_index = summary_columns.index(percentage_column)
summary_ax_list[percentage_column_index].yaxis \
.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
```
### Daily Generation to Upload Period Table
```
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
```
### Hourly Summary Plots
```
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
```
### Publish Results
```
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case_es"]: lambda x: f"{x:.2%}" if x != 0 else "",
}
general_columns = \
list(filter(lambda x: x not in display_formatters, display_column_name_mapping.values()))
general_formatter = lambda x: f"{x}" if x != 0 else ""
display_formatters.update(dict(map(lambda x: (x, general_formatter), general_columns)))
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index.get_level_values("sample_date") == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.item()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.item()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.item()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.item()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.item()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
if len(report_source_regions) == 1:
display_brief_source_regions = report_source_regions[0]
else:
display_brief_source_regions = f"{len(report_source_regions)} 🇪🇺"
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
df = df.copy()
df_styler = df.style.format(display_formatters)
media_path = get_temporary_image_path()
dfi.export(df_styler, media_path)
return media_path
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
```
### Save Results
```
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
```
### Publish Results as JSON
```
def generate_summary_api_results(df: pd.DataFrame) -> list:
api_df = df.reset_index().copy()
api_df["sample_date_string"] = \
api_df["sample_date"].dt.strftime("%Y-%m-%d")
api_df["source_regions"] = \
api_df["source_regions"].apply(lambda x: x.split(","))
return api_df.to_dict(orient="records")
summary_api_results = \
generate_summary_api_results(df=result_summary_df)
today_summary_api_results = \
generate_summary_api_results(df=extraction_date_result_summary_df)[0]
summary_results = dict(
backend_identifier=report_backend_identifier,
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=today_summary_api_results,
last_7_days=last_7_days_summary,
last_14_days=last_14_days_summary,
daily_results=summary_api_results)
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
```
### Publish on README
```
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
```
### Publish on Twitter
```
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
def format_shared_diagnoses_per_covid_case(value) -> str:
if value == 0:
return "–"
return f"≤{value:.2%}"
display_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=shared_diagnoses_per_covid_case)
display_last_14_days_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case"])
display_last_14_days_shared_diagnoses_per_covid_case_es = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case_es"])
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
Source Countries: {display_brief_source_regions}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: {display_shared_diagnoses_per_covid_case}
Last 14 Days:
- Usage Ratio: {display_last_14_days_shared_diagnoses_per_covid_case}
- Usage Ratio (Spain): {display_last_14_days_shared_diagnoses_per_covid_case_es}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import math as m
%matplotlib inline
import torch
import torchvision
import torchvision.transforms as transforms
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
import random
from torch.utils.data import Dataset, DataLoader
from mpl_toolkits.mplot3d import Axes3D
# point = np.array([1, 2, 3])
# normal = np.array([1, 1, 2])
# point2 = np.array([10, 50, 50])
# # a plane is a*x+b*y+c*z+d=0
# # [a,b,c] is the normal. Thus, we have to calculate
# # d and we're set
# d = -point.dot(normal)
# # create x,y
# xx, yy = np.meshgrid(range(10), range(10))
# # calculate corresponding z
# z = (-normal[0] * xx - normal[1] * yy - d) * 1. /normal[2]
# # plot the surface
# plt3d = plt.figure().gca(projection='3d')
# plt3d.plot_surface(xx, yy, z, alpha=1)
# ax = plt.gca()
# #and i would like to plot this point :
# ax.scatter(point2[0] , point2[1] , point2[2], color='green')
# plt.show()
# # plot the surface
# plt3d = plt.figure().gca(projection='3d')
# plt3d.plot_surface(xx, yy, z, alpha=0.2)
# # Ensure that the next plot doesn't overwrite the first plot
# ax = plt.gca()
# # ax.hold(True)
# ax.scatter(points2[0], point2[1], point2[2], color='green')
y = np.random.randint(0,10,1000)
idx= []
for i in range(10):
print(i,sum(y==i))
idx.append(y==i)
x = np.zeros((1000,2))
x0=x[idx[0],:] = np.random.multivariate_normal(mean = [2,2],cov=[[0.01,0],[0,0.01]],size=sum(idx[0]))
x[idx[1],:] = np.random.multivariate_normal(mean = [0,-2],cov=[[0.01,0],[0,0.01]],size=sum(idx[1]))
x[idx[2],:] = np.random.multivariate_normal(mean = [-2,2],cov=[[0.01,0],[0,0.01]],size=sum(idx[2]))
x[idx[3],:] = np.random.multivariate_normal(mean = [0,0],cov=[[0.01,0],[0,0.01]],size=sum(idx[3]))
x[idx[4],:] = np.random.multivariate_normal(mean =[-2,-4] ,cov=[[0.01,0],[0,0.01]],size=sum(idx[4]))
x[idx[5],:] = np.random.multivariate_normal(mean = [2,-4],cov=[[0.01,0],[0,0.01]],size=sum(idx[5]))
x[idx[6],:] = np.random.multivariate_normal(mean = [-4,0],cov=[[0.01,0],[0,0.01]],size=sum(idx[6]))
x[idx[7],:] = np.random.multivariate_normal(mean = [-2,4],cov=[[0.01,0],[0,0.01]],size=sum(idx[7]))
x[idx[8],:] = np.random.multivariate_normal(mean = [2,4],cov=[[0.01,0],[0,0.01]],size=sum(idx[8]))
x[idx[9],:] = np.random.multivariate_normal(mean = [4,0],cov=[[0.01,0],[0,0.01]],size=sum(idx[9]))
idx= []
for i in range(10):
#print(i,sum(y==i))
idx.append(y==i)
for i in range(10):
plt.scatter(x[idx[i],0],x[idx[i],1],label="class_"+str(i))
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
z = np.zeros((1000,1))
x = np.concatenate((x, z) , axis =1)
x
x.shape
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
for i in range(10):
ax.scatter(x[idx[i],0],x[idx[i],1],x[idx[i],2],label="class_"+str(i))
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
normal = np.array([0,0,1])
# a plane is a*x+b*y+c*z+d=0
# [a,b,c] is the normal. Thus, we have to calculate
# d and we're set
d = 0
# create x,y
xx, yy = np.meshgrid(range(5), range(5))
# calculate corresponding z
z = (-normal[0] * xx - normal[1] * yy - d) * 1. /normal[2]
# plot the surface
plt3d = plt.figure().gca(projection='3d')
plt3d.plot_surface(xx, yy, z, alpha=0.5)
# fig = plt.figure()
ax = plt.gca()
for i in range(10):
ax.scatter(x[idx[i],0],x[idx[i],1],x[idx[i],2],label="class_"+str(i))
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
```
## angle = pi/2, p1: z = 0, p2: 2x + 3y = 0
## angle = pi/6, p1: z = 0, p2: 2x + 3y + sqrt(39)z = 0
## angle = pi/3, p1: z = 0, p2: 2x + 3y + sqrt(13/3)z = 0
```
angle = np.pi/4
angle
a = 2
b = 3
if(angle == np.pi/2):
c=0
else:
c = np.sqrt(a*a + b*b )/m.tan(angle)
print(c)
x[idx[0],:]
x[idx[0],2]
if(angle == np.pi/2):
for i in range(3):
x[idx[i],2] = (i+2)*(x[idx[i],0] + x[idx[i],1])/(x[idx[i],0] + x[idx[i],1])
else:
for i in range(3):
x[idx[i],2] = (2*x[idx[i],0] + 3*x[idx[i],1])/c
x[idx[0],:]
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
for i in range(10):
ax.scatter(x[idx[i],0],x[idx[i],1],x[idx[i],2],label="class_"+str(i))
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
normal = np.array([a,b,c])
# a plane is a*x+b*y+c*z+d=0
# [a,b,c] is the normal. Thus, we have to calculate
# d and we're set
d = 0
# create x,y
xx, yy = np.meshgrid(range(5), range(5))
# calculate corresponding z
z = -(-normal[0] * xx - normal[1] * yy - d) * 1. /normal[2]
# plot the surface
plt3d = plt.figure().gca(projection='3d')
plt3d.plot_surface(xx, yy, z, alpha=0.5)
# fig = plt.figure()
ax = plt.gca()
for i in range(10):
ax.scatter(x[idx[i],0],x[idx[i],1],x[idx[i],2],label="class_"+str(i))
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
# true_data = 30
# true_class_size = int(true_data/2)
# corruption_size = 240
# corrupted_class_size = int(corruption_size/8)
# x0 = np.random.uniform(low=[2,1.5], high = [2.5,2],size =(true_class_size,2) )
# x1 = np.random.uniform(low=[2.5,2], high = [3,2.5],size =(true_class_size,2) )
# x2 = np.random.uniform(low = [0,1.5] , high = [1,2.5],size=(corrupted_class_size,2))
# x3 = np.random.uniform(low = [0,3] , high = [1,4],size=(corrupted_class_size,2))
# x4 = np.random.uniform(low = [2,0] , high = [3,1],size=(corrupted_class_size,2))
# x5 = np.random.uniform(low = [0,0] , high = [1,1],size=(corrupted_class_size,2))
# x6 = np.random.uniform(low = [2,3] , high = [3,4],size=(corrupted_class_size,2))
# x7 = np.random.uniform(low = [4,0] , high = [5,1],size=(corrupted_class_size,2))
# x8 = np.random.uniform(low = [4,1.5] , high = [5,2.5],size=(corrupted_class_size,2))
# x9 = np.random.uniform(low = [4,3] , high = [5,4],size=(corrupted_class_size,2))
# z = np.zeros((true_class_size,1))
# x0 = np.concatenate((x0, z) , axis =1)
# x1 = np.concatenate((x1, z) , axis =1)
# z= np.zeros((corrupted_class_size,1))
# x2 = np.concatenate((x2, z) , axis =1)
# x3 = np.concatenate((x3, z) , axis =1)
# x4 = np.concatenate((x4, z) , axis =1)
# x5 = np.concatenate((x5, z) , axis =1)
# x6 = np.concatenate((x6, z) , axis =1)
# x7 = np.concatenate((x7, z) , axis =1)
# x8 = np.concatenate((x8, z) , axis =1)
# x9 = np.concatenate((x9, z) , axis =1)
# x0.shape , x1.shape , x2.shape, x3.shape
# fig = plt.figure()
# ax = fig.add_subplot(111, projection='3d')
# # ax = plt.gca()
# ax.scatter(x0[:,0], x0[:, 1], x0[:,2])
# ax.scatter(x1[:,0],x1[:,1], x1[:,2])
# ax.scatter(x2[:,0],x2[:,1], x2[:,2])
# ax.scatter(x3[:,0],x3[:,1], x3[:,2])
# ax.scatter(x4[:,0],x4[:,1], x4[:,2])
# ax.scatter(x5[:,0],x5[:,1], x5[:,2])
# ax.scatter(x6[:,0],x6[:,1], x6[:,2])
# ax.scatter(x7[:,0],x7[:,1], x7[:,2])
# ax.scatter(x8[:,0],x8[:,1], x8[:,2])
# ax.scatter(x9[:,0],x9[:,1], x9[:,2])
# import plotly.express as px
# fig = px.scatter_3d(x0, x='sepal_length', y='sepal_width', z='petal_width',
# color='petal_length', symbol='species')
# fig.show()
x.shape,y.shape
classes = ('0', '1', '2','3', '4', '5', '6', '7','8', '9')
foreground_classes = {'0', '1', '2'}
background_classes = {'3', '4', '5', '6', '7','8', '9'}
class sub_clust_data(Dataset):
def __init__(self,x, y):
self.x = torch.Tensor(x)
self.y = torch.Tensor(y).type(torch.LongTensor)
#self.fore_idx = fore_idx
def __len__(self):
return len(self.y)
def __getitem__(self, idx):
return self.x[idx] , self.y[idx] #, self.fore_idx[idx]
trainset = sub_clust_data(x,y)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=10, shuffle=True)
dataiter = iter(trainloader)
background_data=[]
background_label=[]
foreground_data=[]
foreground_label=[]
batch_size=10
for i in range(100): #5000*batch_size = 50000 data points
images, labels = dataiter.next()
for j in range(batch_size):
if(classes[labels[j]] in background_classes):
img = images[j].tolist()
background_data.append(img)
background_label.append(labels[j])
else:
img = images[j].tolist()
foreground_data.append(img)
foreground_label.append(labels[j])
np.shape(foreground_data),np.shape(foreground_label)
np.shape(background_data),np.shape(background_label)
foreground_data = torch.tensor(foreground_data)
foreground_label = torch.tensor(foreground_label).type(torch.LongTensor)
background_data = torch.tensor(background_data)
background_label = torch.tensor(background_label).type(torch.LongTensor)
def create_mosaic_img(bg_idx,fg_idx,fg):
image_list=[]
j=0
for i in range(9):
if i != fg:
image_list.append(background_data[bg_idx[j]].type("torch.DoubleTensor"))
j+=1
else:
image_list.append(foreground_data[fg_idx].type("torch.DoubleTensor"))
label = foreground_label[fg_idx] #-7 # minus 7 because our fore ground classes are 7,8,9 but we have to store it as 0,1,2
#image_list = np.concatenate(image_list ,axis=0)
image_list = torch.stack(image_list)
return image_list,label
desired_num = 1000
mosaic_list_of_images =[] # list of mosaic images, each mosaic image is saved as list of 9 images
fore_idx =[] # list of indexes at which foreground image is present in a mosaic image i.e from 0 to 9
mosaic_label=[] # label of mosaic image = foreground class present in that mosaic
for i in range(desired_num):
bg_idx = np.random.randint(0,702,8)
fg_idx = np.random.randint(0,298)
fg = np.random.randint(0,9)
fore_idx.append(fg)
image_list,label = create_mosaic_img(bg_idx,fg_idx,fg)
mosaic_list_of_images.append(image_list)
mosaic_label.append(label)
def create_avg_image_from_mosaic_dataset(mosaic_dataset,labels,foreground_index,dataset_number):
avg_image_dataset = []
for i in range(len(mosaic_dataset)):
img = torch.zeros([3], dtype=torch.float64)
for j in range(9):
if j == foreground_index[i]:
img = img + mosaic_dataset[i][j]*dataset_number/9
else :
img = img + mosaic_dataset[i][j]*(9-dataset_number)/(8*9)
avg_image_dataset.append(img)
return avg_image_dataset , labels , foreground_index
avg_image_dataset_1 , labels_1, fg_index_1 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images[0:1000], mosaic_label[0:1000], fore_idx[0:1000] , 1)
avg_image_dataset_2 , labels_2, fg_index_2 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images[0:1000], mosaic_label[0:1000], fore_idx[0:1000] , 2)
avg_image_dataset_3 , labels_3, fg_index_3 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images[0:1000], mosaic_label[0:1000], fore_idx[0:1000] , 3)
avg_image_dataset_4 , labels_4, fg_index_4 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images[0:1000], mosaic_label[0:1000], fore_idx[0:1000] , 4)
avg_image_dataset_5 , labels_5, fg_index_5 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images[0:1000], mosaic_label[0:1000], fore_idx[0:1000] , 5)
avg_image_dataset_6 , labels_6, fg_index_6 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images[0:1000], mosaic_label[0:1000], fore_idx[0:1000] , 6)
avg_image_dataset_7 , labels_7, fg_index_7 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images[0:1000], mosaic_label[0:1000], fore_idx[0:1000] , 7)
avg_image_dataset_8 , labels_8, fg_index_8 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images[0:1000], mosaic_label[0:1000], fore_idx[0:1000] , 8)
avg_image_dataset_9 , labels_9, fg_index_9 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images[0:1000], mosaic_label[0:1000], fore_idx[0:1000] , 9)
#test_dataset_10 , labels_10 , fg_index_10 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images[10000:20000], mosaic_label[10000:20000], fore_idx[10000:20000] , 9)
class MosaicDataset(Dataset):
"""MosaicDataset dataset."""
def __init__(self, mosaic_list_of_images, mosaic_label):
"""
Args:
csv_file (string): Path to the csv file with annotations.
root_dir (string): Directory with all the images.
transform (callable, optional): Optional transform to be applied
on a sample.
"""
self.mosaic = mosaic_list_of_images
self.label = mosaic_label
#self.fore_idx = fore_idx
def __len__(self):
return len(self.label)
def __getitem__(self, idx):
return self.mosaic[idx] , self.label[idx] #, self.fore_idx[idx]
batch = 256
# training_data = avg_image_dataset_5 #just change this and training_label to desired dataset for training
# training_label = labels_5
traindata_1 = MosaicDataset(avg_image_dataset_1, labels_1 )
trainloader_1 = DataLoader( traindata_1 , batch_size= batch ,shuffle=True)
traindata_2 = MosaicDataset(avg_image_dataset_2, labels_2 )
trainloader_2 = DataLoader( traindata_2 , batch_size= batch ,shuffle=True)
traindata_3 = MosaicDataset(avg_image_dataset_3, labels_3 )
trainloader_3 = DataLoader( traindata_3 , batch_size= batch ,shuffle=True)
traindata_4 = MosaicDataset(avg_image_dataset_4, labels_4 )
trainloader_4 = DataLoader( traindata_4 , batch_size= batch ,shuffle=True)
traindata_5 = MosaicDataset(avg_image_dataset_5, labels_5 )
trainloader_5 = DataLoader( traindata_5 , batch_size= batch ,shuffle=True)
traindata_6 = MosaicDataset(avg_image_dataset_6, labels_6 )
trainloader_6 = DataLoader( traindata_6 , batch_size= batch ,shuffle=True)
traindata_7 = MosaicDataset(avg_image_dataset_7, labels_7 )
trainloader_7 = DataLoader( traindata_7 , batch_size= batch ,shuffle=True)
traindata_8 = MosaicDataset(avg_image_dataset_8, labels_8 )
trainloader_8 = DataLoader( traindata_8 , batch_size= batch ,shuffle=True)
traindata_9 = MosaicDataset(avg_image_dataset_9, labels_9 )
trainloader_9 = DataLoader( traindata_9 , batch_size= batch ,shuffle=True)
testdata_1 = MosaicDataset(avg_image_dataset_1, labels_1 )
testloader_1 = DataLoader( testdata_1 , batch_size= batch ,shuffle=False)
testdata_2 = MosaicDataset(avg_image_dataset_2, labels_2 )
testloader_2 = DataLoader( testdata_2 , batch_size= batch ,shuffle=False)
testdata_3 = MosaicDataset(avg_image_dataset_3, labels_3 )
testloader_3 = DataLoader( testdata_3 , batch_size= batch ,shuffle=False)
testdata_4 = MosaicDataset(avg_image_dataset_4, labels_4 )
testloader_4 = DataLoader( testdata_4 , batch_size= batch ,shuffle=False)
testdata_5 = MosaicDataset(avg_image_dataset_5, labels_5 )
testloader_5 = DataLoader( testdata_5 , batch_size= batch ,shuffle=False)
testdata_6 = MosaicDataset(avg_image_dataset_6, labels_6 )
testloader_6 = DataLoader( testdata_6 , batch_size= batch ,shuffle=False)
testdata_7 = MosaicDataset(avg_image_dataset_7, labels_7 )
testloader_7 = DataLoader( testdata_7 , batch_size= batch ,shuffle=False)
testdata_8 = MosaicDataset(avg_image_dataset_8, labels_8 )
testloader_8 = DataLoader( testdata_8 , batch_size= batch ,shuffle=False)
testdata_9 = MosaicDataset(avg_image_dataset_9, labels_9 )
testloader_9 = DataLoader( testdata_9 , batch_size= batch ,shuffle=False)
class Net(nn.Module):
def __init__(self):
super(Net,self).__init__()
self.linear1 = nn.Linear(3,64)
self.linear2 = nn.Linear(64,128)
self.linear3 = nn.Linear(128,3)
def forward(self,x):
x = F.relu(self.linear1(x))
x = F.relu(self.linear2(x))
x = self.linear3(x)
return x
def test_all(number, testloader,inc):
correct = 0
total = 0
out = []
pred = []
with torch.no_grad():
for data in testloader:
images, labels = data
images, labels = images.to("cuda"),labels.to("cuda")
out.append(labels.cpu().numpy())
outputs= inc(images)
_, predicted = torch.max(outputs.data, 1)
pred.append(predicted.cpu().numpy())
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 1000 test dataset %d: %d %%' % (number , 100 * correct / total))
def train_all(trainloader, ds_number, testloader_list):
print("--"*40)
print("training on data set ", ds_number)
inc = Net().double()
inc = inc.to("cuda")
criterion_inception = nn.CrossEntropyLoss()
optimizer_inception = optim.SGD(inc.parameters(), lr=0.01, momentum=0.9)
acti = []
loss_curi = []
epochs = 70
for epoch in range(epochs): # loop over the dataset multiple times
ep_lossi = []
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
# get the inputs
inputs, labels = data
inputs, labels = inputs.to("cuda"),labels.to("cuda")
# zero the parameter gradients
optimizer_inception.zero_grad()
# forward + backward + optimize
outputs = inc(inputs)
loss = criterion_inception(outputs, labels)
loss.backward()
optimizer_inception.step()
# print statistics
running_loss += loss.item()
mini=4
if i % mini == mini-1: # print every 10 mini-batches
print('[%d, %5d] loss: %.3f' %
(epoch + 1, i + 1, running_loss / mini))
ep_lossi.append(running_loss/mini) # loss per minibatch
running_loss = 0.0
if(np.mean(ep_lossi)<=0.01):
break
loss_curi.append(np.mean(ep_lossi)) #loss per epoch
# if (epoch%5 == 0):
# _,actis= inc(inputs)
# acti.append(actis)
print('Finished Training')
# torch.save(inc.state_dict(),"/content/drive/My Drive/Research/Experiments on CIFAR mosaic/Exp_2_Attention_models_on_9_datasets_made_from_10k_mosaic/weights/train_dataset_"+str(ds_number)+"_"+str(epochs)+".pt")
correct = 0
total = 0
with torch.no_grad():
for data in trainloader:
images, labels = data
images, labels = images.to("cuda"), labels.to("cuda")
outputs = inc(images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 train images: %d %%' % ( 100 * correct / total))
for i, j in enumerate(testloader_list):
test_all(i+1, j,inc)
print("--"*40)
return loss_curi
train_loss_all=[]
testloader_list= [ testloader_1, testloader_2, testloader_3, testloader_4, testloader_5, testloader_6,
testloader_7, testloader_8, testloader_9]
train_loss_all.append(train_all(trainloader_1, 1, testloader_list))
train_loss_all.append(train_all(trainloader_2, 2, testloader_list))
train_loss_all.append(train_all(trainloader_3, 3, testloader_list))
train_loss_all.append(train_all(trainloader_4, 4, testloader_list))
train_loss_all.append(train_all(trainloader_5, 5, testloader_list))
train_loss_all.append(train_all(trainloader_6, 6, testloader_list))
train_loss_all.append(train_all(trainloader_7, 7, testloader_list))
train_loss_all.append(train_all(trainloader_8, 8, testloader_list))
train_loss_all.append(train_all(trainloader_9, 9, testloader_list))
```
|
github_jupyter
|
Copyright (c) Microsoft Corporation. All rights reserved.
Licensed under the MIT License.

# Reinforcement Learning in Azure Machine Learning - Training a Minecraft agent using custom environments
This tutorial will show how to set up a more complex reinforcement
learning (RL) training scenario. It demonstrates how to train an agent to
navigate through a lava maze in the Minecraft game using Azure Machine
Learning.
**Please note:** This notebook trains an agent on a randomly generated
Minecraft level. As a result, on rare occasions, a training run may fail
to produce a model that can solve the maze. If this happens, you can
re-run the training step as indicated below.
**Please note:** This notebook uses 1 NC6 type node and 8 D2 type nodes
for up to 5 hours of training, which corresponds to approximately $9.06 (USD)
as of May 2020.
Minecraft is currently one of the most popular video
games and as such has been a study object for RL. [Project
Malmo](https://www.microsoft.com/en-us/research/project/project-malmo/) is
a platform for artificial intelligence experimentation and research built on
top of Minecraft. We will use Minecraft [gym](https://gym.openai.com) environments from Project
Malmo's 2019 MineRL competition, which are part of the
[MineRL](http://minerl.io/docs/index.html) Python package.
Minecraft environments require a display to run, so we will demonstrate
how to set up a virtual display within the docker container used for training.
Learning will be based on the agent's visual observations. To
generate the necessary amount of sample data, we will run several
instances of the Minecraft game in parallel. Below, you can see a video of
a trained agent navigating a lava maze. Starting from the green position,
it moves to the blue position by moving forward, turning left or turning right:
<table style="width:50%">
<tr>
<th style="text-align: center;">
<img src="./images/lava_maze_minecraft.gif" alt="Minecraft lava maze" align="middle" margin-left="auto" margin-right="auto"/>
</th>
</tr>
<tr style="text-align: center;">
<th>Fig 1. Video of a trained Minecraft agent navigating a lava maze.</th>
</tr>
</table>
The tutorial will cover the following steps:
- Initializing Azure Machine Learning resources for training
- Training the RL agent with Azure Machine Learning service
- Monitoring training progress
- Reviewing training results
## Prerequisites
The user should have completed the Azure Machine Learning introductory tutorial.
You will need to make sure that you have a valid subscription id, a resource group and a
workspace. For detailed instructions see [Tutorial: Get started creating
your first ML experiment.](https://docs.microsoft.com/en-us/azure/machine-learning/tutorial-1st-experiment-sdk-setup)
While this is a standalone notebook, we highly recommend going over the
introductory notebooks for RL first.
- Getting started:
- [RL using a compute instance with Azure Machine Learning service](../cartpole-on-compute-instance/cartpole_ci.ipynb)
- [Using Azure Machine Learning compute](../cartpole-on-single-compute/cartpole_sc.ipynb)
- [Scaling RL training runs with Azure Machine Learning service](../atari-on-distributed-compute/pong_rllib.ipynb)
## Initialize resources
All required Azure Machine Learning service resources for this tutorial can be set up from Jupyter.
This includes:
- Connecting to your existing Azure Machine Learning workspace.
- Creating an experiment to track runs.
- Setting up a virtual network
- Creating remote compute targets for [Ray](https://docs.ray.io/en/latest/index.html).
### Azure Machine Learning SDK
Display the Azure Machine Learning SDK version.
```
import azureml.core
print("Azure Machine Learning SDK Version: ", azureml.core.VERSION)
```
### Connect to workspace
Get a reference to an existing Azure Machine Learning workspace.
```
from azureml.core import Workspace
ws = Workspace.from_config()
print(ws.name, ws.location, ws.resource_group, sep=' | ')
```
### Create an experiment
Create an experiment to track the runs in your workspace. A
workspace can have multiple experiments and each experiment
can be used to track multiple runs (see [documentation](https://docs.microsoft.com/en-us/python/api/azureml-core/azureml.core.experiment.experiment?view=azure-ml-py)
for details).
```
from azureml.core import Experiment
exp = Experiment(workspace=ws, name='minecraft-maze')
```
### Create Virtual Network
If you are using separate compute targets for the Ray head and worker, a virtual network must be created in the resource group. If you have alraeady created a virtual network in the resource group, you can skip this step.
To do this, you first must install the Azure Networking API.
`pip install --upgrade azure-mgmt-network`
```
# If you need to install the Azure Networking SDK, uncomment the following line.
#!pip install --upgrade azure-mgmt-network
from azure.mgmt.network import NetworkManagementClient
# Virtual network name
vnet_name ="your_vnet"
# Default subnet
subnet_name ="default"
# The Azure subscription you are using
subscription_id=ws.subscription_id
# The resource group for the reinforcement learning cluster
resource_group=ws.resource_group
# Azure region of the resource group
location=ws.location
network_client = NetworkManagementClient(ws._auth_object, subscription_id)
async_vnet_creation = network_client.virtual_networks.create_or_update(
resource_group,
vnet_name,
{
'location': location,
'address_space': {
'address_prefixes': ['10.0.0.0/16']
}
}
)
async_vnet_creation.wait()
print("Virtual network created successfully: ", async_vnet_creation.result())
```
### Set up Network Security Group on Virtual Network
Depending on your Azure setup, you may need to open certain ports to make it possible for Azure to manage the compute targets that you create. The ports that need to be opened are described [here](https://docs.microsoft.com/en-us/azure/machine-learning/how-to-enable-virtual-network).
A common situation is that ports `29876-29877` are closed. The following code will add a security rule to open these ports. Or you can do this manually in the [Azure portal](https://portal.azure.com).
You may need to modify the code below to match your scenario.
```
import azure.mgmt.network.models
security_group_name = vnet_name + '-' + "nsg"
security_rule_name = "AllowAML"
# Create a network security group
nsg_params = azure.mgmt.network.models.NetworkSecurityGroup(
location=location,
security_rules=[
azure.mgmt.network.models.SecurityRule(
name=security_rule_name,
access=azure.mgmt.network.models.SecurityRuleAccess.allow,
description='Reinforcement Learning in Azure Machine Learning rule',
destination_address_prefix='*',
destination_port_range='29876-29877',
direction=azure.mgmt.network.models.SecurityRuleDirection.inbound,
priority=400,
protocol=azure.mgmt.network.models.SecurityRuleProtocol.tcp,
source_address_prefix='BatchNodeManagement',
source_port_range='*'
),
],
)
async_nsg_creation = network_client.network_security_groups.create_or_update(
resource_group,
security_group_name,
nsg_params,
)
async_nsg_creation.wait()
print("Network security group created successfully:", async_nsg_creation.result())
network_security_group = network_client.network_security_groups.get(
resource_group,
security_group_name,
)
# Define a subnet to be created with network security group
subnet = azure.mgmt.network.models.Subnet(
id='default',
address_prefix='10.0.0.0/24',
network_security_group=network_security_group
)
# Create subnet on virtual network
async_subnet_creation = network_client.subnets.create_or_update(
resource_group_name=resource_group,
virtual_network_name=vnet_name,
subnet_name=subnet_name,
subnet_parameters=subnet
)
async_subnet_creation.wait()
print("Subnet created successfully:", async_subnet_creation.result())
```
### Review the virtual network security rules
Ensure that the virtual network is configured correctly with required ports open. It is possible that you have configured rules with broader range of ports that allows ports 29876-29877 to be opened. Kindly review your network security group rules.
```
from files.networkutils import *
check_vnet_security_rules(ws._auth_object, ws.subscription_id, ws.resource_group, vnet_name, True)
```
### Create or attach an existing compute resource
A compute target is a designated compute resource where you
run your training script. For more information, see [What
are compute targets in Azure Machine Learning service?](https://docs.microsoft.com/en-us/azure/machine-learning/concept-compute-target).
#### GPU target for Ray head
In the experiment setup for this tutorial, the Ray head node
will run on a GPU-enabled node. A maximum cluster size
of 1 node is therefore sufficient. If you wish to run
multiple experiments in parallel using the same GPU
cluster, you may elect to increase this number. The cluster
will automatically scale down to 0 nodes when no training jobs
are scheduled (see `min_nodes`).
The code below creates a compute cluster of GPU-enabled NC6
nodes. If the cluster with the specified name is already in
your workspace the code will skip the creation process.
Note that we must specify a Virtual Network during compute
creation to allow communication between the cluster running
the Ray head node and the additional Ray compute nodes. For
details on how to setup the Virtual Network, please follow the
instructions in the "Prerequisites" section above.
**Note: Creation of a compute resource can take several minutes**
```
from azureml.core.compute import ComputeTarget, AmlCompute
from azureml.core.compute_target import ComputeTargetException
gpu_cluster_name = 'gpu-cl-nc6-vnet'
try:
gpu_cluster = ComputeTarget(workspace=ws, name=gpu_cluster_name)
print('Found existing compute target')
except ComputeTargetException:
print('Creating a new compute target...')
compute_config = AmlCompute.provisioning_configuration(
vm_size='Standard_NC6',
min_nodes=0,
max_nodes=1,
vnet_resourcegroup_name=ws.resource_group,
vnet_name=vnet_name,
subnet_name=subnet_name)
gpu_cluster = ComputeTarget.create(ws, gpu_cluster_name, compute_config)
gpu_cluster.wait_for_completion(show_output=True, min_node_count=None, timeout_in_minutes=20)
print('Cluster created.')
```
#### CPU target for additional Ray nodes
The code below creates a compute cluster of D2 nodes. If the cluster with the specified name is already in your workspace the code will skip the creation process.
This cluster will be used to start additional Ray nodes
increasing the clusters CPU resources.
**Note: Creation of a compute resource can take several minutes**
```
cpu_cluster_name = 'cpu-cl-d2-vnet'
try:
cpu_cluster = ComputeTarget(workspace=ws, name=cpu_cluster_name)
print('Found existing compute target')
except ComputeTargetException:
print('Creating a new compute target...')
compute_config = AmlCompute.provisioning_configuration(
vm_size='STANDARD_D2',
min_nodes=0,
max_nodes=10,
vnet_resourcegroup_name=ws.resource_group,
vnet_name=vnet_name,
subnet_name=subnet_name)
cpu_cluster = ComputeTarget.create(ws, cpu_cluster_name, compute_config)
cpu_cluster.wait_for_completion(show_output=True, min_node_count=None, timeout_in_minutes=20)
print('Cluster created.')
```
## Training the agent
### Training environments
This tutorial uses custom docker images (CPU and GPU respectively)
with the necessary software installed. The
[Environment](https://docs.microsoft.com/en-us/azure/machine-learning/how-to-use-environments)
class stores the configuration for the training environment. The docker
image is set via `env.docker.base_image` which can point to any
publicly available docker image. `user_managed_dependencies`
is set so that the preinstalled Python packages in the image are preserved.
Note that since Minecraft requires a display to start, we set the `interpreter_path`
such that the Python process is started via **xvfb-run**.
```
import os
from azureml.core import Environment
max_train_time = os.environ.get("AML_MAX_TRAIN_TIME_SECONDS", 5 * 60 * 60)
def create_env(env_type):
env = Environment(name='minecraft-{env_type}'.format(env_type=env_type))
env.docker.enabled = True
env.docker.base_image = 'akdmsft/minecraft-{env_type}'.format(env_type=env_type)
env.python.interpreter_path = "xvfb-run -s '-screen 0 640x480x16 -ac +extension GLX +render' python"
env.environment_variables["AML_MAX_TRAIN_TIME_SECONDS"] = str(max_train_time)
env.python.user_managed_dependencies = True
return env
cpu_minecraft_env = create_env('cpu')
gpu_minecraft_env = create_env('gpu')
```
### Training script
As described above, we use the MineRL Python package to launch
Minecraft game instances. MineRL provides several OpenAI gym
environments for different scenarios, such as chopping wood.
Besides predefined environments, MineRL lets its users create
custom Minecraft environments through
[minerl.env](http://minerl.io/docs/api/env.html). In the helper
file **minecraft_environment.py** provided with this tutorial, we use the
latter option to customize a Minecraft level with a lava maze
that the agent has to navigate. The agent receives a negative
reward of -1 for falling into the lava, a negative reward of
-0.02 for sending a command (i.e. navigating through the maze
with fewer actions yields a higher total reward) and a positive reward
of 1 for reaching the goal. To encourage the agent to explore
the maze, it also receives a positive reward of 0.1 for visiting
a tile for the first time.
The agent learns purely from visual observations and the image
is scaled to an 84x84 format, stacking four frames. For the
purposes of this example, we use a small action space of size
three: move forward, turn 90 degrees to the left, and turn 90
degrees to the right.
The training script itself registers the function to create training
environments with the `tune.register_env` function and connects to
the Ray cluster Azure Machine Learning service started on the GPU
and CPU nodes. Lastly, it starts a RL training run with `tune.run()`.
We recommend setting the `local_dir` parameter to `./logs` as this
directory will automatically become available as part of the training
run's files in the Azure Portal. The Tensorboard integration
(see "View the Tensorboard" section below) also depends on the files'
availability. For a list of common parameter options, please refer
to the [Ray documentation](https://docs.ray.io/en/latest/rllib-training.html#common-parameters).
```python
# Taken from minecraft_environment.py and minecraft_train.py
# Define a function to create a MineRL environment
def create_env(config):
mission = config['mission']
port = 1000 * config.worker_index + config.vector_index
print('*********************************************')
print(f'* Worker {config.worker_index} creating from mission: {mission}, port {port}')
print('*********************************************')
if config.worker_index == 0:
# The first environment is only used for checking the action and observation space.
# By using a dummy environment, there's no need to spin up a Minecraft instance behind it
# saving some CPU resources on the head node.
return DummyEnv()
env = EnvWrapper(mission, port)
env = TrackingEnv(env)
env = FrameStack(env, 2)
return env
def stop(trial_id, result):
return result["episode_reward_mean"] >= 1 \
or result["time_total_s"] > 5 * 60 * 60
if __name__ == '__main__':
tune.register_env("Minecraft", create_env)
ray.init(address='auto')
tune.run(
run_or_experiment="IMPALA",
config={
"env": "Minecraft",
"env_config": {
"mission": "minecraft_missions/lava_maze-v0.xml"
},
"num_workers": 10,
"num_cpus_per_worker": 2,
"rollout_fragment_length": 50,
"train_batch_size": 1024,
"replay_buffer_num_slots": 4000,
"replay_proportion": 10,
"learner_queue_timeout": 900,
"num_sgd_iter": 2,
"num_data_loader_buffers": 2,
"exploration_config": {
"type": "EpsilonGreedy",
"initial_epsilon": 1.0,
"final_epsilon": 0.02,
"epsilon_timesteps": 500000
},
"callbacks": {"on_train_result": callbacks.on_train_result},
},
stop=stop,
checkpoint_at_end=True,
local_dir='./logs'
)
```
### Submitting a training run
Below, you create the training run using a `ReinforcementLearningEstimator`
object, which contains all the configuration parameters for this experiment:
- `source_directory`: Contains the training script and helper files to be
copied onto the node running the Ray head.
- `entry_script`: The training script, described in more detail above..
- `compute_target`: The compute target for the Ray head and training
script execution.
- `environment`: The Azure machine learning environment definition for
the node running the Ray head.
- `worker_configuration`: The configuration object for the additional
Ray nodes to be attached to the Ray cluster:
- `compute_target`: The compute target for the additional Ray nodes.
- `node_count`: The number of nodes to attach to the Ray cluster.
- `environment`: The environment definition for the additional Ray nodes.
- `max_run_duration_seconds`: The time after which to abort the run if it
is still running.
- `shm_size`: The size of docker container's shared memory block.
For more details, please take a look at the [online documentation](https://docs.microsoft.com/en-us/python/api/azureml-contrib-reinforcementlearning/?view=azure-ml-py)
for Azure Machine Learning service's reinforcement learning offering.
We configure 8 extra D2 (worker) nodes for the Ray cluster, giving us a total of
22 CPUs and 1 GPU. The GPU and one CPU are used by the IMPALA learner,
and each MineRL environment receives 2 CPUs allowing us to spawn a total
of 10 rollout workers (see `num_workers` parameter in the training script).
Lastly, the `RunDetails` widget displays information about the submitted
RL experiment, including a link to the Azure portal with more details.
```
from azureml.contrib.train.rl import ReinforcementLearningEstimator, WorkerConfiguration
from azureml.widgets import RunDetails
worker_config = WorkerConfiguration(
compute_target=cpu_cluster,
node_count=8,
environment=cpu_minecraft_env)
rl_est = ReinforcementLearningEstimator(
source_directory='files',
entry_script='minecraft_train.py',
compute_target=gpu_cluster,
environment=gpu_minecraft_env,
worker_configuration=worker_config,
max_run_duration_seconds=6 * 60 * 60,
shm_size=1024 * 1024 * 1024 * 30)
train_run = exp.submit(rl_est)
RunDetails(train_run).show()
# If you wish to cancel the run before it completes, uncomment and execute:
#train_run.cancel()
```
## Monitoring training progress
### View the Tensorboard
The Tensorboard can be displayed via the Azure Machine Learning service's
[Tensorboard API](https://docs.microsoft.com/en-us/azure/machine-learning/how-to-monitor-tensorboard).
When running locally, please make sure to follow the instructions in the
link and install required packages. Running this cell will output a URL
for the Tensorboard.
Note that the training script sets the log directory when starting RLlib
via the `local_dir` parameter. `./logs` will automatically appear in
the downloadable files for a run. Since this script is executed on the
Ray head node run, we need to get a reference to it as shown below.
The Tensorboard API will continuously stream logs from the run.
**Note: It may take a couple of minutes after the run is in "Running" state
before Tensorboard files are available and the board will refresh automatically**
```
import time
from azureml.tensorboard import Tensorboard
head_run = None
timeout = 60
while timeout > 0 and head_run is None:
timeout -= 1
try:
head_run = next(r for r in train_run.get_children() if r.id.endswith('head'))
except StopIteration:
time.sleep(1)
tb = Tensorboard([head_run], port=6007)
tb.start()
```
## Review results
Please ensure that the training run has completed before continuing with this section.
```
train_run.wait_for_completion()
print('Training run completed.')
```
**Please note:** If the final "episode_reward_mean" metric from the training run is negative,
the produced model does not solve the problem of navigating the maze well. You can view
the metric on the Tensorboard or in "Metrics" section of the head run in the Azure Machine Learning
portal. We recommend training a new model by rerunning the notebook starting from "Submitting a training run".
### Export final model
The key result from the training run is the final checkpoint
containing the state of the IMPALA trainer (model) upon meeting the
stopping criteria specified in `minecraft_train.py`.
Azure Machine Learning service offers the [Model.register()](https://docs.microsoft.com/en-us/python/api/azureml-core/azureml.core.model.model?view=azure-ml-py)
API which allows you to persist the model files from the
training run. We identify the directory containing the
final model written during the training run and register
it with Azure Machine Learning service. We use a Dataset
object to filter out the correct files.
```
import re
import tempfile
from azureml.core import Dataset
path_prefix = os.path.join(tempfile.gettempdir(), 'tmp_training_artifacts')
run_artifacts_path = os.path.join('azureml', head_run.id)
datastore = ws.get_default_datastore()
run_artifacts_ds = Dataset.File.from_files(datastore.path(os.path.join(run_artifacts_path, '**')))
cp_pattern = re.compile('.*checkpoint-\\d+$')
checkpoint_files = [file for file in run_artifacts_ds.to_path() if cp_pattern.match(file)]
# There should only be one checkpoint with our training settings...
final_checkpoint = os.path.dirname(os.path.join(run_artifacts_path, os.path.normpath(checkpoint_files[-1][1:])))
datastore.download(target_path=path_prefix, prefix=final_checkpoint.replace('\\', '/'), show_progress=True)
print('Download complete.')
from azureml.core.model import Model
model_name = 'final_model_minecraft_maze'
model = Model.register(
workspace=ws,
model_path=os.path.join(path_prefix, final_checkpoint),
model_name=model_name,
description='Model of an agent trained to navigate a lava maze in Minecraft.')
```
Models can be used through a varity of APIs. Please see the
[documentation](https://docs.microsoft.com/en-us/azure/machine-learning/how-to-deploy-and-where)
for more details.
### Test agent performance in a rollout
To observe the trained agent's behavior, it is a common practice to
view its behavior in a rollout. The previous reinforcement learning
tutorials explain rollouts in more detail.
The provided `minecraft_rollout.py` script loads the final checkpoint
of the trained agent from the model registered with Azure Machine Learning
service. It then starts a rollout on 4 different lava maze layouts, that
are all larger and thus more difficult than the maze the agent was trained
on. The script further records videos by replaying the agent's decisions
in [Malmo](https://github.com/microsoft/malmo). Malmo supports multiple
agents in the same environment, thus allowing us to capture videos that
depict the agent from another agent's perspective. The provided
`malmo_video_recorder.py` file and the Malmo Github repository have more
details on the video recording setup.
You can view the rewards for each rollout episode in the logs for the 'head'
run submitted below. In some episodes, the agent may fail to reach the goal
due to the higher level of difficulty - in practice, we could continue
training the agent on harder tasks starting with the final checkpoint.
```
script_params = {
'--model_name': model_name
}
rollout_est = ReinforcementLearningEstimator(
source_directory='files',
entry_script='minecraft_rollout.py',
script_params=script_params,
compute_target=gpu_cluster,
environment=gpu_minecraft_env,
shm_size=1024 * 1024 * 1024 * 30)
rollout_run = exp.submit(rollout_est)
RunDetails(rollout_run).show()
```
### View videos captured during rollout
To inspect the agent's training progress you can view the videos captured
during the rollout episodes. First, ensure that the training run has
completed.
```
rollout_run.wait_for_completion()
head_run_rollout = next(r for r in rollout_run.get_children() if r.id.endswith('head'))
print('Rollout completed.')
```
Next, you need to download the video files from the training run. We use a
Dataset to filter out the video files which are in tgz archives.
```
rollout_run_artifacts_path = os.path.join('azureml', head_run_rollout.id)
datastore = ws.get_default_datastore()
rollout_run_artifacts_ds = Dataset.File.from_files(datastore.path(os.path.join(rollout_run_artifacts_path, '**')))
video_archives = [file for file in rollout_run_artifacts_ds.to_path() if file.endswith('.tgz')]
video_archives = [os.path.join(rollout_run_artifacts_path, os.path.normpath(file[1:])) for file in video_archives]
datastore.download(
target_path=path_prefix,
prefix=os.path.dirname(video_archives[0]).replace('\\', '/'),
show_progress=True)
print('Download complete.')
```
Next, unzip the video files and rename them by the Minecraft mission seed used
(see `minecraft_rollout.py` for more details on how the seed is used).
```
import tarfile
import shutil
training_artifacts_dir = './training_artifacts'
video_dir = os.path.join(training_artifacts_dir, 'videos')
video_files = []
for tar_file_path in video_archives:
seed = tar_file_path[tar_file_path.index('rollout_') + len('rollout_'): tar_file_path.index('.tgz')]
tar = tarfile.open(os.path.join(path_prefix, tar_file_path).replace('\\', '/'), 'r')
tar_info = next(t_info for t_info in tar.getmembers() if t_info.name.endswith('mp4'))
tar.extract(tar_info, video_dir)
tar.close()
unzipped_folder = os.path.join(video_dir, next(f_ for f_ in os.listdir(video_dir) if not f_.endswith('mp4')))
video_file = os.path.join(unzipped_folder,'video.mp4')
final_video_path = os.path.join(video_dir, '{seed}.mp4'.format(seed=seed))
shutil.move(video_file, final_video_path)
video_files.append(final_video_path)
shutil.rmtree(unzipped_folder)
# Clean up any downloaded 'tmp' files
shutil.rmtree(path_prefix)
print('Local video files:\n', video_files)
```
Finally, run the cell below to display the videos in-line. In some cases,
the agent may struggle to find the goal since the maze size was increased
compared to training.
```
from IPython.core.display import display, HTML
index = 0
while index < len(video_files) - 1:
display(
HTML('\
<video controls alt="cannot display video" autoplay loop width=49%> \
<source src="{f1}" type="video/mp4"> \
</video> \
<video controls alt="cannot display video" autoplay loop width=49%> \
<source src="{f2}" type="video/mp4"> \
</video>'.format(f1=video_files[index], f2=video_files[index + 1]))
)
index += 2
if index < len(video_files):
display(
HTML('\
<video controls alt="cannot display video" autoplay loop width=49%> \
<source src="{f1}" type="video/mp4"> \
</video>'.format(f1=video_files[index]))
)
```
## Cleaning up
Below, you can find code snippets for your convenience to clean up any resources created as part of this tutorial you don't wish to retain.
```
# to stop the Tensorboard, uncomment and run
#tb.stop()
# to delete the gpu compute target, uncomment and run
#gpu_cluster.delete()
# to delete the cpu compute target, uncomment and run
#cpu_cluster.delete()
# to delete the registered model, uncomment and run
#model.delete()
# to delete the local video files, uncomment and run
#shutil.rmtree(training_artifacts_dir)
```
## Next steps
This is currently the last introductory tutorial for Azure Machine Learning
service's Reinforcement
Learning offering. We would love to hear your feedback to build the features
you need!
|
github_jupyter
|
```
import csv
import tensorflow as tf
import numpy as np
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/bbc-text.csv \
-O /tmp/bbc-text.csv
vocab_size = 1000
embedding_dim = 16
max_length = 120
trunc_type='post'
padding_type='post'
oov_tok = "<OOV>"
training_portion = .8
sentences = []
labels = []
stopwords = [ "a", "about", "above", "after", "again", "against", "all", "am", "an", "and", "any", "are", "as", "at", "be", "because", "been", "before", "being", "below", "between", "both", "but", "by", "could", "did", "do", "does", "doing", "down", "during", "each", "few", "for", "from", "further", "had", "has", "have", "having", "he", "he'd", "he'll", "he's", "her", "here", "here's", "hers", "herself", "him", "himself", "his", "how", "how's", "i", "i'd", "i'll", "i'm", "i've", "if", "in", "into", "is", "it", "it's", "its", "itself", "let's", "me", "more", "most", "my", "myself", "nor", "of", "on", "once", "only", "or", "other", "ought", "our", "ours", "ourselves", "out", "over", "own", "same", "she", "she'd", "she'll", "she's", "should", "so", "some", "such", "than", "that", "that's", "the", "their", "theirs", "them", "themselves", "then", "there", "there's", "these", "they", "they'd", "they'll", "they're", "they've", "this", "those", "through", "to", "too", "under", "until", "up", "very", "was", "we", "we'd", "we'll", "we're", "we've", "were", "what", "what's", "when", "when's", "where", "where's", "which", "while", "who", "who's", "whom", "why", "why's", "with", "would", "you", "you'd", "you'll", "you're", "you've", "your", "yours", "yourself", "yourselves" ]
print(len(stopwords))
# Expected Output
# 153
with open("/tmp/bbc-text.csv", 'r') as csvfile:
reader = csv.reader(csvfile, delimiter=',')
next(reader)
for row in reader:
labels.append(row[0])
sentence = row[1]
for word in stopwords:
token = " " + word + " "
sentence = sentence.replace(token, " ")
sentences.append(sentence)
print(len(labels))
print(len(sentences))
print(sentences[0])
# Expected Output
# 2225
# 2225
# tv future hands viewers home theatre systems plasma high-definition tvs digital video recorders moving living room way people watch tv will radically different five years time. according expert panel gathered annual consumer electronics show las vegas discuss new technologies will impact one favourite pastimes. us leading trend programmes content will delivered viewers via home networks cable satellite telecoms companies broadband service providers front rooms portable devices. one talked-about technologies ces digital personal video recorders (dvr pvr). set-top boxes like us s tivo uk s sky+ system allow people record store play pause forward wind tv programmes want. essentially technology allows much personalised tv. also built-in high-definition tv sets big business japan us slower take off europe lack high-definition programming. not can people forward wind adverts can also forget abiding network channel schedules putting together a-la-carte entertainment. us networks cable satellite companies worried means terms advertising revenues well brand identity viewer loyalty channels. although us leads technology moment also concern raised europe particularly growing uptake services like sky+. happens today will see nine months years time uk adam hume bbc broadcast s futurologist told bbc news website. likes bbc no issues lost advertising revenue yet. pressing issue moment commercial uk broadcasters brand loyalty important everyone. will talking content brands rather network brands said tim hanlon brand communications firm starcom mediavest. reality broadband connections anybody can producer content. added: challenge now hard promote programme much choice. means said stacey jolna senior vice president tv guide tv group way people find content want watch simplified tv viewers. means networks us terms channels take leaf google s book search engine future instead scheduler help people find want watch. kind channel model might work younger ipod generation used taking control gadgets play them. might not suit everyone panel recognised. older generations comfortable familiar schedules channel brands know getting. perhaps not want much choice put hands mr hanlon suggested. end kids just diapers pushing buttons already - everything possible available said mr hanlon. ultimately consumer will tell market want. 50 000 new gadgets technologies showcased ces many enhancing tv-watching experience. high-definition tv sets everywhere many new models lcd (liquid crystal display) tvs launched dvr capability built instead external boxes. one example launched show humax s 26-inch lcd tv 80-hour tivo dvr dvd recorder. one us s biggest satellite tv companies directtv even launched branded dvr show 100-hours recording capability instant replay search function. set can pause rewind tv 90 hours. microsoft chief bill gates announced pre-show keynote speech partnership tivo called tivotogo means people can play recorded programmes windows pcs mobile devices. reflect increasing trend freeing multimedia people can watch want want.
train_size = int(len(sentences) * training_portion)
train_sentences = sentences[:train_size]
train_labels = labels[:train_size]
validation_sentences = sentences[train_size:]
validation_labels = labels[train_size:]
print(train_size)
print(len(train_sentences))
print(len(train_labels))
print(len(validation_sentences))
print(len(validation_labels))
# Expected output (if training_portion=.8)
# 1780
# 1780
# 1780
# 445
# 445
tokenizer = Tokenizer(num_words = vocab_size, oov_token=oov_tok)
tokenizer.fit_on_texts(train_sentences)
word_index = tokenizer.word_index
train_sequences = tokenizer.texts_to_sequences(train_sentences)
train_padded = pad_sequences(train_sequences, padding=padding_type, maxlen=max_length)
print(len(train_sequences[0]))
print(len(train_padded[0]))
print(len(train_sequences[1]))
print(len(train_padded[1]))
print(len(train_sequences[10]))
print(len(train_padded[10]))
# Expected Ouput
# 449
# 120
# 200
# 120
# 192
# 120
validation_sequences = tokenizer.texts_to_sequences(validation_sentences)
validation_padded = pad_sequences(validation_sequences, padding=padding_type, maxlen=max_length)
print(len(validation_sequences))
print(validation_padded.shape)
# Expected output
# 445
# (445, 120)
label_tokenizer = Tokenizer()
label_tokenizer.fit_on_texts(labels)
training_label_seq = np.array(label_tokenizer.texts_to_sequences(train_labels))
validation_label_seq = np.array(label_tokenizer.texts_to_sequences(validation_labels))
model = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.GlobalAveragePooling1D(),
tf.keras.layers.Dense(24, activation='relu'),
tf.keras.layers.Dense(6, activation = 'softmax')
])
model.compile(loss='sparse_categorical_crossentropy',optimizer='adam',metrics=['accuracy'])
model.summary()
num_epochs = 30
history = model.fit(train_padded, training_label_seq, epochs=num_epochs, validation_data=(validation_padded, validation_label_seq), verbose=2)
import matplotlib.pyplot as plt
def plot_graphs(history, string):
plt.plot(history.history[string])
plt.plot(history.history['val_'+string])
plt.xlabel("Epochs")
plt.ylabel(string)
plt.legend([string, 'val_'+string])
plt.show()
plot_graphs(history, "accuracy")
plot_graphs(history, "loss")
reverse_word_index = dict([(value, key) for (key, value) in word_index.items()])
def decode_sentence(text):
return ' '.join([reverse_word_index.get(i, '?') for i in text])
e = model.layers[0]
weights = e.get_weights()[0]
print(weights.shape) # shape: (vocab_size, embedding_dim)
# Expected output
# (1000, 16)
```
|
github_jupyter
|
# Learning Tree-augmented Naive Bayes (TAN) Structure from Data
In this notebook, we show an example for learning the structure of a Bayesian Network using the TAN algorithm. We will first build a model to generate some data and then attempt to learn the model's graph structure back from the generated data.
For comparison of Naive Bayes and TAN classifier, refer to the blog post [Classification with TAN and Pgmpy](https://loudly-soft.blogspot.com/2020/08/classification-with-tree-augmented.html).
## First, create a Naive Bayes graph
```
import networkx as nx
import matplotlib.pyplot as plt
from pgmpy.models import BayesianNetwork
# class variable is A and feature variables are B, C, D, E and R
model = BayesianNetwork([("A", "R"), ("A", "B"), ("A", "C"), ("A", "D"), ("A", "E")])
nx.draw_circular(
model, with_labels=True, arrowsize=30, node_size=800, alpha=0.3, font_weight="bold"
)
plt.show()
```
## Second, add interaction between the features
```
# feature R correlates with other features
model.add_edges_from([("R", "B"), ("R", "C"), ("R", "D"), ("R", "E")])
nx.draw_circular(
model, with_labels=True, arrowsize=30, node_size=800, alpha=0.3, font_weight="bold"
)
plt.show()
```
## Then, parameterize our graph to create a Bayesian network
```
from pgmpy.factors.discrete import TabularCPD
# add CPD to each edge
cpd_a = TabularCPD("A", 2, [[0.7], [0.3]])
cpd_r = TabularCPD(
"R", 3, [[0.6, 0.2], [0.3, 0.5], [0.1, 0.3]], evidence=["A"], evidence_card=[2]
)
cpd_b = TabularCPD(
"B",
3,
[
[0.1, 0.1, 0.2, 0.2, 0.7, 0.1],
[0.1, 0.3, 0.1, 0.2, 0.1, 0.2],
[0.8, 0.6, 0.7, 0.6, 0.2, 0.7],
],
evidence=["A", "R"],
evidence_card=[2, 3],
)
cpd_c = TabularCPD(
"C",
2,
[[0.7, 0.2, 0.2, 0.5, 0.1, 0.3], [0.3, 0.8, 0.8, 0.5, 0.9, 0.7]],
evidence=["A", "R"],
evidence_card=[2, 3],
)
cpd_d = TabularCPD(
"D",
3,
[
[0.3, 0.8, 0.2, 0.8, 0.4, 0.7],
[0.4, 0.1, 0.4, 0.1, 0.1, 0.1],
[0.3, 0.1, 0.4, 0.1, 0.5, 0.2],
],
evidence=["A", "R"],
evidence_card=[2, 3],
)
cpd_e = TabularCPD(
"E",
2,
[[0.5, 0.6, 0.6, 0.5, 0.5, 0.4], [0.5, 0.4, 0.4, 0.5, 0.5, 0.6]],
evidence=["A", "R"],
evidence_card=[2, 3],
)
model.add_cpds(cpd_a, cpd_r, cpd_b, cpd_c, cpd_d, cpd_e)
```
## Next, generate sample data from our Bayesian network
```
from pgmpy.sampling import BayesianModelSampling
# sample data from BN
inference = BayesianModelSampling(model)
df_data = inference.forward_sample(size=10000)
print(df_data)
```
## Now we are ready to learn the TAN structure from sample data
```
from pgmpy.estimators import TreeSearch
# learn graph structure
est = TreeSearch(df_data, root_node="R")
dag = est.estimate(estimator_type="tan", class_node="A")
nx.draw_circular(
dag, with_labels=True, arrowsize=30, node_size=800, alpha=0.3, font_weight="bold"
)
plt.show()
```
## To parameterize the learned graph from data, check out the other tutorials for more info
```
from pgmpy.estimators import BayesianEstimator
# there are many choices of parametrization, here is one example
model = BayesianNetwork(dag.edges())
model.fit(
df_data, estimator=BayesianEstimator, prior_type="dirichlet", pseudo_counts=0.1
)
model.get_cpds()
```
|
github_jupyter
|
**This notebook is an exercise in the [Intermediate Machine Learning](https://www.kaggle.com/learn/intermediate-machine-learning) course. You can reference the tutorial at [this link](https://www.kaggle.com/alexisbcook/introduction).**
---
As a warm-up, you'll review some machine learning fundamentals and submit your initial results to a Kaggle competition.
# Setup
The questions below will give you feedback on your work. Run the following cell to set up the feedback system.
```
# Set up code checking
import os
if not os.path.exists("../input/train.csv"):
os.symlink("../input/home-data-for-ml-course/train.csv", "../input/train.csv")
os.symlink("../input/home-data-for-ml-course/test.csv", "../input/test.csv")
from learntools.core import binder
binder.bind(globals())
from learntools.ml_intermediate.ex1 import *
print("Setup Complete")
```
You will work with data from the [Housing Prices Competition for Kaggle Learn Users](https://www.kaggle.com/c/home-data-for-ml-course) to predict home prices in Iowa using 79 explanatory variables describing (almost) every aspect of the homes.

Run the next code cell without changes to load the training and validation features in `X_train` and `X_valid`, along with the prediction targets in `y_train` and `y_valid`. The test features are loaded in `X_test`. (_If you need to review **features** and **prediction targets**, please check out [this short tutorial](https://www.kaggle.com/dansbecker/your-first-machine-learning-model). To read about model **validation**, look [here](https://www.kaggle.com/dansbecker/model-validation). Alternatively, if you'd prefer to look through a full course to review all of these topics, start [here](https://www.kaggle.com/learn/machine-learning).)_
```
import pandas as pd
from sklearn.model_selection import train_test_split
# Read the data
X_full = pd.read_csv('../input/train.csv', index_col='Id')
X_test_full = pd.read_csv('../input/test.csv', index_col='Id')
# Obtain target and predictors
y = X_full.SalePrice
features = ['LotArea', 'YearBuilt', '1stFlrSF', '2ndFlrSF', 'FullBath', 'BedroomAbvGr', 'TotRmsAbvGrd']
X = X_full[features].copy()
X_test = X_test_full[features].copy()
# Break off validation set from training data
X_train, X_valid, y_train, y_valid = train_test_split(X, y, train_size=0.8, test_size=0.2,
random_state=0)
```
Use the next cell to print the first several rows of the data. It's a nice way to get an overview of the data you will use in your price prediction model.
```
X_train.head()
```
The next code cell defines five different random forest models. Run this code cell without changes. (_To review **random forests**, look [here](https://www.kaggle.com/dansbecker/random-forests)._)
```
from sklearn.ensemble import RandomForestRegressor
# Define the models
model_1 = RandomForestRegressor(n_estimators=50, random_state=0)
model_2 = RandomForestRegressor(n_estimators=100, random_state=0)
model_3 = RandomForestRegressor(n_estimators=100, criterion='mae', random_state=0)
model_4 = RandomForestRegressor(n_estimators=200, min_samples_split=20, random_state=0)
model_5 = RandomForestRegressor(n_estimators=100, max_depth=7, random_state=0)
models = [model_1, model_2, model_3, model_4, model_5]
```
To select the best model out of the five, we define a function `score_model()` below. This function returns the mean absolute error (MAE) from the validation set. Recall that the best model will obtain the lowest MAE. (_To review **mean absolute error**, look [here](https://www.kaggle.com/dansbecker/model-validation).)_
Run the code cell without changes.
```
from sklearn.metrics import mean_absolute_error
# Function for comparing different models
def score_model(model, X_t=X_train, X_v=X_valid, y_t=y_train, y_v=y_valid):
model.fit(X_t, y_t)
preds = model.predict(X_v)
return mean_absolute_error(y_v, preds)
for i in range(0, len(models)):
mae = score_model(models[i])
print("Model %d MAE: %d" % (i+1, mae))
```
# Step 1: Evaluate several models
Use the above results to fill in the line below. Which model is the best model? Your answer should be one of `model_1`, `model_2`, `model_3`, `model_4`, or `model_5`.
```
# Fill in the best model
best_model = model_3
# Check your answer
step_1.check()
# Lines below will give you a hint or solution code
#step_1.hint()
#step_1.solution()
```
# Step 2: Generate test predictions
Great. You know how to evaluate what makes an accurate model. Now it's time to go through the modeling process and make predictions. In the line below, create a Random Forest model with the variable name `my_model`.
```
# Define a model
my_model = best_model
# Check your answer
step_2.check()
# Lines below will give you a hint or solution code
#step_2.hint()
#step_2.solution()
```
Run the next code cell without changes. The code fits the model to the training and validation data, and then generates test predictions that are saved to a CSV file. These test predictions can be submitted directly to the competition!
```
# Fit the model to the training data
my_model.fit(X, y)
# Generate test predictions
preds_test = my_model.predict(X_test)
# Save predictions in format used for competition scoring
output = pd.DataFrame({'Id': X_test.index,
'SalePrice': preds_test})
output.to_csv('submission.csv', index=False)
```
# Submit your results
Once you have successfully completed Step 2, you're ready to submit your results to the leaderboard! First, you'll need to join the competition if you haven't already. So open a new window by clicking on [this link](https://www.kaggle.com/c/home-data-for-ml-course). Then click on the **Join Competition** button.

Next, follow the instructions below:
1. Begin by clicking on the blue **Save Version** button in the top right corner of the window. This will generate a pop-up window.
2. Ensure that the **Save and Run All** option is selected, and then click on the blue **Save** button.
3. This generates a window in the bottom left corner of the notebook. After it has finished running, click on the number to the right of the **Save Version** button. This pulls up a list of versions on the right of the screen. Click on the ellipsis **(...)** to the right of the most recent version, and select **Open in Viewer**. This brings you into view mode of the same page. You will need to scroll down to get back to these instructions.
4. Click on the **Output** tab on the right of the screen. Then, click on the file you would like to submit, and click on the blue **Submit** button to submit your results to the leaderboard.
You have now successfully submitted to the competition!
If you want to keep working to improve your performance, select the blue **Edit** button in the top right of the screen. Then you can change your code and repeat the process. There's a lot of room to improve, and you will climb up the leaderboard as you work.
# Keep going
You've made your first model. But how can you quickly make it better?
Learn how to improve your competition results by incorporating columns with **[missing values](https://www.kaggle.com/alexisbcook/missing-values)**.
---
*Have questions or comments? Visit the [Learn Discussion forum](https://www.kaggle.com/learn-forum/161289) to chat with other Learners.*
|
github_jupyter
|
<a href="https://colab.research.google.com/github/magenta/ddsp/blob/master/ddsp/colab/demos/timbre_transfer.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
##### Copyright 2020 Google LLC.
Licensed under the Apache License, Version 2.0 (the "License");
```
# Copyright 2020 Google LLC. All Rights Reserved.
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
```
# DDSP Timbre Transfer Demo
This notebook is a demo of timbre transfer using DDSP (Differentiable Digital Signal Processing).
The model here is trained to generate audio conditioned on a time series of fundamental frequency and loudness.
* [DDSP ICLR paper](https://openreview.net/forum?id=B1x1ma4tDr)
* [Audio Examples](http://goo.gl/magenta/ddsp-examples)
This notebook extracts these features from input audio (either uploaded files, or recorded from the microphone) and resynthesizes with the model.
<img src="https://magenta.tensorflow.org/assets/ddsp/ddsp_cat_jamming.png" alt="DDSP Tone Transfer" width="700">
By default, the notebook will download pre-trained models. You can train a model on your own sounds by using the [Train Autoencoder Colab](https://github.com/magenta/ddsp/blob/master/ddsp/colab/demos/train_autoencoder.ipynb).
Have fun! And please feel free to hack this notebook to make your own creative interactions.
### Instructions for running:
* Make sure to use a GPU runtime, click: __Runtime >> Change Runtime Type >> GPU__
* Press ▶️ on the left of each of the cells
* View the code: Double-click any of the cells
* Hide the code: Double click the right side of the cell
```
#@title #Install and Import
#@markdown Install ddsp, define some helper functions, and download the model. This transfers a lot of data and _should take a minute or two_.
%tensorflow_version 2.x
print('Installing from pip package...')
!pip install -qU ddsp
# Ignore a bunch of deprecation warnings
import warnings
warnings.filterwarnings("ignore")
import copy
import os
import time
import crepe
import ddsp
import ddsp.training
from ddsp.colab import colab_utils
from ddsp.colab.colab_utils import (
auto_tune, detect_notes, fit_quantile_transform,
get_tuning_factor, download, play, record,
specplot, upload, DEFAULT_SAMPLE_RATE)
import gin
from google.colab import files
import librosa
import matplotlib.pyplot as plt
import numpy as np
import pickle
import tensorflow.compat.v2 as tf
import tensorflow_datasets as tfds
# Helper Functions
sample_rate = DEFAULT_SAMPLE_RATE # 16000
print('Done!')
#@title Record or Upload Audio
#@markdown * Either record audio from microphone or upload audio from file (.mp3 or .wav)
#@markdown * Audio should be monophonic (single instrument / voice)
#@markdown * Extracts fundmanetal frequency (f0) and loudness features.
record_or_upload = "Record" #@param ["Record", "Upload (.mp3 or .wav)"]
record_seconds = 5#@param {type:"number", min:1, max:10, step:1}
if record_or_upload == "Record":
audio = record(seconds=record_seconds)
else:
# Load audio sample here (.mp3 or .wav3 file)
# Just use the first file.
filenames, audios = upload()
audio = audios[0]
audio = audio[np.newaxis, :]
print('\nExtracting audio features...')
# Plot.
specplot(audio)
play(audio)
# Setup the session.
ddsp.spectral_ops.reset_crepe()
# Compute features.
start_time = time.time()
audio_features = ddsp.training.metrics.compute_audio_features(audio)
audio_features['loudness_db'] = audio_features['loudness_db'].astype(np.float32)
audio_features_mod = None
print('Audio features took %.1f seconds' % (time.time() - start_time))
TRIM = -15
# Plot Features.
fig, ax = plt.subplots(nrows=3,
ncols=1,
sharex=True,
figsize=(6, 8))
ax[0].plot(audio_features['loudness_db'][:TRIM])
ax[0].set_ylabel('loudness_db')
ax[1].plot(librosa.hz_to_midi(audio_features['f0_hz'][:TRIM]))
ax[1].set_ylabel('f0 [midi]')
ax[2].plot(audio_features['f0_confidence'][:TRIM])
ax[2].set_ylabel('f0 confidence')
_ = ax[2].set_xlabel('Time step [frame]')
#@title Load a model
#@markdown Run for ever new audio input
model = 'Violin' #@param ['Violin', 'Flute', 'Flute2', 'Trumpet', 'Tenor_Saxophone', 'Upload your own (checkpoint folder as .zip)']
MODEL = model
def find_model_dir(dir_name):
# Iterate through directories until model directory is found
for root, dirs, filenames in os.walk(dir_name):
for filename in filenames:
if filename.endswith(".gin") and not filename.startswith("."):
model_dir = root
break
return model_dir
if model in ('Violin', 'Flute', 'Flute2', 'Trumpet', 'Tenor_Saxophone'):
# Pretrained models.
PRETRAINED_DIR = '/content/pretrained'
# Copy over from gs:// for faster loading.
!rm -r $PRETRAINED_DIR &> /dev/null
!mkdir $PRETRAINED_DIR &> /dev/null
GCS_CKPT_DIR = 'gs://ddsp/models/tf2'
model_dir = os.path.join(GCS_CKPT_DIR, 'solo_%s_ckpt' % model.lower())
!gsutil cp $model_dir/* $PRETRAINED_DIR &> /dev/null
model_dir = PRETRAINED_DIR
gin_file = os.path.join(model_dir, 'operative_config-0.gin')
else:
# User models.
UPLOAD_DIR = '/content/uploaded'
!mkdir $UPLOAD_DIR
uploaded_files = files.upload()
for fnames in uploaded_files.keys():
print("Unzipping... {}".format(fnames))
!unzip -o "/content/$fnames" -d $UPLOAD_DIR &> /dev/null
model_dir = find_model_dir(UPLOAD_DIR)
gin_file = os.path.join(model_dir, 'operative_config-0.gin')
# Load the dataset statistics.
DATASET_STATS = None
dataset_stats_file = os.path.join(model_dir, 'dataset_statistics.pkl')
print(f'Loading dataset statistics from {dataset_stats_file}')
try:
if tf.io.gfile.exists(dataset_stats_file):
with tf.io.gfile.GFile(dataset_stats_file, 'rb') as f:
DATASET_STATS = pickle.load(f)
except Exception as err:
print('Loading dataset statistics from pickle failed: {}.'.format(err))
# Parse gin config,
with gin.unlock_config():
gin.parse_config_file(gin_file, skip_unknown=True)
# Assumes only one checkpoint in the folder, 'ckpt-[iter]`.
ckpt_files = [f for f in tf.io.gfile.listdir(model_dir) if 'ckpt' in f]
ckpt_name = ckpt_files[0].split('.')[0]
ckpt = os.path.join(model_dir, ckpt_name)
# Ensure dimensions and sampling rates are equal
time_steps_train = gin.query_parameter('DefaultPreprocessor.time_steps')
n_samples_train = gin.query_parameter('Additive.n_samples')
hop_size = int(n_samples_train / time_steps_train)
time_steps = int(audio.shape[1] / hop_size)
n_samples = time_steps * hop_size
# print("===Trained model===")
# print("Time Steps", time_steps_train)
# print("Samples", n_samples_train)
# print("Hop Size", hop_size)
# print("\n===Resynthesis===")
# print("Time Steps", time_steps)
# print("Samples", n_samples)
# print('')
gin_params = [
'RnnFcDecoder.input_keys = ("f0_scaled", "ld_scaled")',
'Additive.n_samples = {}'.format(n_samples),
'FilteredNoise.n_samples = {}'.format(n_samples),
'DefaultPreprocessor.time_steps = {}'.format(time_steps),
]
with gin.unlock_config():
gin.parse_config(gin_params)
# Trim all input vectors to correct lengths
for key in ['f0_hz', 'f0_confidence', 'loudness_db']:
audio_features[key] = audio_features[key][:time_steps]
audio_features['audio'] = audio_features['audio'][:, :n_samples]
# Set up the model just to predict audio given new conditioning
model = ddsp.training.models.Autoencoder()
model.restore(ckpt)
# Build model by running a batch through it.
start_time = time.time()
_ = model(audio_features, training=False)
print('Restoring model took %.1f seconds' % (time.time() - start_time))
#@title Modify conditioning
#@markdown These models were not explicitly trained to perform timbre transfer, so they may sound unnatural if the incoming loudness and frequencies are very different then the training data (which will always be somewhat true).
#@markdown ## Note Detection
#@markdown You can leave this at 1.0 for most cases
threshold = 1 #@param {type:"slider", min: 0.0, max:2.0, step:0.01}
#@markdown ## Automatic
ADJUST = True #@param{type:"boolean"}
#@markdown Quiet parts without notes detected (dB)
quiet = 20 #@param {type:"slider", min: 0, max:60, step:1}
#@markdown Force pitch to nearest note (amount)
autotune = 0 #@param {type:"slider", min: 0.0, max:1.0, step:0.1}
#@markdown ## Manual
#@markdown Shift the pitch (octaves)
pitch_shift = 0 #@param {type:"slider", min:-2, max:2, step:1}
#@markdown Adjsut the overall loudness (dB)
loudness_shift = 0 #@param {type:"slider", min:-20, max:20, step:1}
audio_features_mod = {k: v.copy() for k, v in audio_features.items()}
## Helper functions.
def shift_ld(audio_features, ld_shift=0.0):
"""Shift loudness by a number of ocatves."""
audio_features['loudness_db'] += ld_shift
return audio_features
def shift_f0(audio_features, pitch_shift=0.0):
"""Shift f0 by a number of ocatves."""
audio_features['f0_hz'] *= 2.0 ** (pitch_shift)
audio_features['f0_hz'] = np.clip(audio_features['f0_hz'],
0.0,
librosa.midi_to_hz(110.0))
return audio_features
mask_on = None
if ADJUST and DATASET_STATS is not None:
# Detect sections that are "on".
mask_on, note_on_value = detect_notes(audio_features['loudness_db'],
audio_features['f0_confidence'],
threshold)
if np.any(mask_on):
# Shift the pitch register.
target_mean_pitch = DATASET_STATS['mean_pitch']
pitch = ddsp.core.hz_to_midi(audio_features['f0_hz'])
mean_pitch = np.mean(pitch[mask_on])
p_diff = target_mean_pitch - mean_pitch
p_diff_octave = p_diff / 12.0
round_fn = np.floor if p_diff_octave > 1.5 else np.ceil
p_diff_octave = round_fn(p_diff_octave)
audio_features_mod = shift_f0(audio_features_mod, p_diff_octave)
# Quantile shift the note_on parts.
_, loudness_norm = colab_utils.fit_quantile_transform(
audio_features['loudness_db'],
mask_on,
inv_quantile=DATASET_STATS['quantile_transform'])
# Turn down the note_off parts.
mask_off = np.logical_not(mask_on)
loudness_norm[mask_off] -= quiet * (1.0 - note_on_value[mask_off][:, np.newaxis])
loudness_norm = np.reshape(loudness_norm, audio_features['loudness_db'].shape)
audio_features_mod['loudness_db'] = loudness_norm
# Auto-tune.
if autotune:
f0_midi = np.array(ddsp.core.hz_to_midi(audio_features_mod['f0_hz']))
tuning_factor = get_tuning_factor(f0_midi, audio_features_mod['f0_confidence'], mask_on)
f0_midi_at = auto_tune(f0_midi, tuning_factor, mask_on, amount=autotune)
audio_features_mod['f0_hz'] = ddsp.core.midi_to_hz(f0_midi_at)
else:
print('\nSkipping auto-adjust (no notes detected or ADJUST box empty).')
else:
print('\nSkipping auto-adujst (box not checked or no dataset statistics found).')
# Manual Shifts.
audio_features_mod = shift_ld(audio_features_mod, loudness_shift)
audio_features_mod = shift_f0(audio_features_mod, pitch_shift)
# Plot Features.
has_mask = int(mask_on is not None)
n_plots = 3 if has_mask else 2
fig, axes = plt.subplots(nrows=n_plots,
ncols=1,
sharex=True,
figsize=(2*n_plots, 8))
if has_mask:
ax = axes[0]
ax.plot(np.ones_like(mask_on[:TRIM]) * threshold, 'k:')
ax.plot(note_on_value[:TRIM])
ax.plot(mask_on[:TRIM])
ax.set_ylabel('Note-on Mask')
ax.set_xlabel('Time step [frame]')
ax.legend(['Threshold', 'Likelihood','Mask'])
ax = axes[0 + has_mask]
ax.plot(audio_features['loudness_db'][:TRIM])
ax.plot(audio_features_mod['loudness_db'][:TRIM])
ax.set_ylabel('loudness_db')
ax.legend(['Original','Adjusted'])
ax = axes[1 + has_mask]
ax.plot(librosa.hz_to_midi(audio_features['f0_hz'][:TRIM]))
ax.plot(librosa.hz_to_midi(audio_features_mod['f0_hz'][:TRIM]))
ax.set_ylabel('f0 [midi]')
_ = ax.legend(['Original','Adjusted'])
#@title #Resynthesize Audio
af = audio_features if audio_features_mod is None else audio_features_mod
# Run a batch of predictions.
start_time = time.time()
audio_gen = model(af, training=False)
print('Prediction took %.1f seconds' % (time.time() - start_time))
# Plot
print('Original')
play(audio)
print('Resynthesis')
play(audio_gen)
specplot(audio)
plt.title("Original")
specplot(audio_gen)
_ = plt.title("Resynthesis")
```
|
github_jupyter
|
```
import numpy as np
import copy
from sklearn import preprocessing
import tensorflow as tf
from tensorflow import keras
import os
import pandas as pd
from matplotlib import pyplot as plt
from numpy.random import seed
np.random.seed(2095)
data = pd.read_excel('Dataset/CardiacPrediction.xlsx')
data.drop(['SEQN','Annual-Family-Income','Height','Ratio-Family-Income-Poverty','X60-sec-pulse',
'Health-Insurance','Glucose','Vigorous-work','Total-Cholesterol','CoronaryHeartDisease','Blood-Rel-Stroke','Red-Cell-Distribution-Width','Triglycerides','Mean-Platelet-Vol','Platelet-count','Lymphocyte','Monocyte','Eosinophils','Mean-cell-Hemoglobin','White-Blood-Cells','Red-Blood-Cells','Basophils','Mean-Cell-Vol','Mean-Cell-Hgb-Conc.','Hematocrit','Segmented-Neutrophils'], axis = 1, inplace=True)
#data['Diabetes'] = data['Diabetes'].replace('3','1')
#data = data.astype(float)
data['Diabetes'].loc[(data['Diabetes'] == 3 )] = 1
#data= data["Diabetes"].replace({"3": "1"},inplace=True)
data["Diabetes"].value_counts()
data["Diabetes"].describe()
#del data['Basophils']
#del data['Health-Insurance']
#del data['Platelet-count']
data.shape
data.columns
data = data[['Gender', 'Age', 'Systolic', 'Diastolic', 'Weight', 'Body-Mass-Index',
'Hemoglobin', 'Albumin', 'ALP', 'AST', 'ALT', 'Cholesterol',
'Creatinine', 'GGT', 'Iron', 'LDH', 'Phosphorus',
'Bilirubin', 'Protein', 'Uric.Acid', 'HDL',
'Glycohemoglobin', 'Moderate-work',
'Blood-Rel-Diabetes', 'Diabetes']]
data.columns
data.isnull().sum()
data.describe()
data.shape
data['Diabetes'].describe()
data.columns
data["Diabetes"].value_counts().sort_index().plot.barh()
#data["Gender"].value_counts().sort_index().plot.barh()
#balanced
#data.corr()
data.columns
data.shape
data.info()
data = data.astype(float)
data.info()
import seaborn as sns
plt.subplots(figsize=(12,8))
sns.heatmap(data.corr(),cmap='inferno', annot=True)
plt.subplots(figsize=(25,15))
data.boxplot(patch_artist=True, sym="k.")
plt.xticks(rotation=90)
minimum = 0
maximum = 0
def detect_outlier(feature):
first_q = np.percentile(feature, 25)
third_q = np.percentile(feature, 75)
IQR = third_q - first_q
IQR *= 1.5
minimum = first_q - IQR
maximum = third_q + IQR
flag = False
if(minimum > np.min(feature)):
flag = True
if(maximum < np.max(feature)):
flag = True
return flag
def remove_outlier(feature):
first_q = np.percentile(X[feature], 25)
third_q = np.percentile(X[feature], 75)
IQR = third_q - first_q
IQR *= 1.5
minimum = first_q - IQR # the acceptable minimum value
maximum = third_q + IQR # the acceptable maximum value
median = X[feature].median()
"""
# any value beyond the acceptance range are considered
as outliers.
# we replace the outliers with the median value of that
feature.
"""
X.loc[X[feature] < minimum, feature] = median
X.loc[X[feature] > maximum, feature] = median
# taking all the columns except the last one
# last column is the label
X = data.iloc[:, :-1]
for i in range(len(X.columns)):
remove_outlier(X.columns[i])
X = data.iloc[:, :-1]
for i in range(len(X.columns)):
if(detect_outlier(X[X.columns[i]])):
print(X.columns[i], "Contains Outlier")
for i in range (50):
for i in range(len(X.columns)):
remove_outlier(X.columns[i])
plt.subplots(figsize=(15,6))
X.boxplot(patch_artist=True, sym="k.")
plt.xticks(rotation=90)
for i in range(len(X.columns)):
if(detect_outlier(X[X.columns[i]])):
print(X.columns[i], "Contains Outlier")
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
sns.set()
from sklearn.preprocessing import MinMaxScaler, StandardScaler, LabelEncoder
from sklearn.svm import SVC
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import LogisticRegression
#from xgboost import XGBClassifier, plot_importance
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score,confusion_matrix
scaler = StandardScaler()
scaled_data = scaler.fit_transform(X)
scaled_df = pd.DataFrame(data = scaled_data, columns = X.columns)
scaled_df.head()
label = data["Diabetes"]
encoder = LabelEncoder()
label = encoder.fit_transform(label)
X = scaled_df
y = label
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=420)
print(X_train.shape, y_test.shape)
print(y_train.shape, y_test.shape)
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_classif
xnew2=SelectKBest(f_classif, k=20).fit_transform(X, y)
import sklearn.feature_selection as fs
import matplotlib.pyplot as plt
df2 = fs.SelectKBest(k='all')
df2.fit(X, y)
names = X.columns.values[df2.get_support()]
scores = df2.scores_[df2.get_support()]
names_scores = list(zip(names, scores))
ns_df = pd.DataFrame(data = names_scores, columns=
['Features','F_Scores'])
ns_df_sorted = ns_df.sort_values(['F_Scores','Features'], ascending =
[False, True])
print(ns_df_sorted)
#import statsmodels.api as sm
#import pandas
#from patsy import dmatrices
#logit_model = sm.OLS(y_train, X_train)
#result = logit_model.fit()
#print(result.summary2())
#np.exp(result.params)
#params = result.params
#conf = result.conf_int()
#conf['Odds Ratio'] = params.sort_index()
#conf.columns = ['5%', '95%', 'Odds Ratio']
#print(np.exp(conf))
#result.pvalues.sort_values()
#from sklearn.utils import class_weight
#class_weights = class_weight.compute_class_weight('balanced',
# np.unique(y_train),
# y_train)
#model.fit(X_train, y_train, class_weight=class_weights)
from sklearn.model_selection import GridSearchCV
weights = np.linspace(0.05, 0.95, 20)
gsc = GridSearchCV(
estimator=LogisticRegression(),
param_grid={
'class_weight': [{0: x, 1: 1.0-x} for x in weights]
},
scoring='accuracy',
cv=15
)
grid_result = gsc.fit(X, y)
print("Best parameters : %s" % grid_result.best_params_)
# Plot the weights vs f1 score
dataz = pd.DataFrame({ 'score': grid_result.cv_results_['mean_test_score'],
'weight': weights })
dataz.plot(x='weight')
class_weight = {0: 0.5236842105263158,
1: 0.47631578947368425}
#LR
'''
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import confusion_matrix, classification_report
from mlxtend.plotting import plot_decision_regions, plot_confusion_matrix
from matplotlib import pyplot as plt
lr = LogisticRegression(class_weight='balanced',random_state=420)
# Fit..
lr.fit(X_train, y_train)
# Predict..
y_pred = lr.predict(X_test)
# Evaluate the model
print(classification_report(y_test, y_pred))
plot_confusion_matrix(confusion_matrix(y_test, y_pred))
from sklearn.metrics import roc_curve, auc
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
roc_auc
'''
'''
from sklearn.svm import SVC
clf_svc_rbf = SVC(kernel="rbf",class_weight='balanced',random_state=4200)
clf_svc_rbf.fit(X_train,y_train)
y_pred_clf_svc_rbf = clf_svc_rbf.predict(X_test)
import matplotlib.pyplot as plt
cm = confusion_matrix(y_test,y_pred_clf_svc_rbf)
#plt.figure(figsize=(5,5))
#sns.heatmap(cm,annot=True)
#plt.show()
#print(classification_report(y_test,y_pred_clf_svc_rbf))
print(classification_report(y_test, y_pred_clf_svc_rbf))
plot_confusion_matrix(confusion_matrix(y_test, y_pred_clf_svc_rbf))
from sklearn.metrics import roc_curve, auc
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred_clf_svc_rbf)
roc_auc = auc(false_positive_rate, true_positive_rate)
roc_auc
'''
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report
rd = RandomForestClassifier(class_weight='balanced',random_state=4200)
rd.fit(X_train,y_train)
y_pred_rd = rd.predict(X_test)
import matplotlib.pyplot as plt
cm = confusion_matrix(y_test,y_pred_rd)
#plt.figure(figsize=(5,5))
#sns.heatmap(cm,annot=True,linewidths=.3)
#plt.show()
print(classification_report(y_test,y_pred_rd))
plot_confusion_matrix(confusion_matrix(y_test, y_pred_rd))
from sklearn.metrics import roc_curve, auc
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred_rd)
roc_auc = auc(false_positive_rate, true_positive_rate)
roc_auc
#CV appraoach
```
## SVM
```
# evaluate a logistic regression model using k-fold cross-validation
from numpy import mean
from numpy import std
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import ShuffleSplit
from sklearn.linear_model import LogisticRegression
# create dataset
#X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=1)
# prepare the cross-validation procedure
#cv = KFold(n_splits=5, test_size= 0.2, random_state=0)
cv = ShuffleSplit(n_splits=10, test_size=0.2, random_state=42)
# create model
model = SVC(kernel='rbf', C=1, class_weight=class_weight)
# evaluate model
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1)
# report performance
print('Accuracy: %.4f (%.4f)' % (mean(scores), std(scores)))
scores
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm, datasets
from sklearn.metrics import auc
from sklearn.metrics import plot_roc_curve
from sklearn.model_selection import StratifiedKFold
# #############################################################################
# Classification and ROC analysis
# Run classifier with cross-validation and plot ROC curves
cv = ShuffleSplit(n_splits=10, test_size=0.2, random_state=42)
classifier = svm.SVC(kernel='rbf', probability=True, class_weight=class_weight,
random_state=42)
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
fig, ax = plt.subplots()
for i, (train, test) in enumerate(cv.split(X, y)):
classifier.fit(X, y)
viz = plot_roc_curve(classifier, X, y,
name='ROC fold {}'.format(i),
alpha=0.3, lw=1, ax=ax)
interp_tpr = np.interp(mean_fpr, viz.fpr, viz.tpr)
interp_tpr[0] = 0.0
tprs.append(interp_tpr)
aucs.append(viz.roc_auc)
ax.plot([0, 1], [0, 1], linestyle='--', lw=2, color='r',
label='Chance', alpha=.8)
mean_tpr = np.mean(tprs, axis=0)
mean_tpr[-1] = 1.0
mean_auc = auc(mean_fpr, mean_tpr)
std_auc = np.std(aucs)
ax.plot(mean_fpr, mean_tpr, color='b',
label=r'Mean ROC (AUC = %0.4f $\pm$ %0.4f)' % (mean_auc, std_auc),
lw=2, alpha=.8)
std_tpr = np.std(tprs, axis=0)
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
ax.fill_between(mean_fpr, tprs_lower, tprs_upper, color='grey', alpha=.2,
label=r'$\pm$ 1 std. dev.')
ax.set(xlim=[-0.05, 1.05], ylim=[-0.05, 1.05],
title="Receiver operating characteristic")
ax.legend(loc="lower right")
plt.show()
```
# LR
```
from numpy import mean
from numpy import std
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import ShuffleSplit
from sklearn.linear_model import LogisticRegression
# create dataset
#X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=1)
# prepare the cross-validation procedure
#cv = KFold(n_splits=5, test_size= 0.2, random_state=0)
cv = ShuffleSplit(n_splits=10, test_size=0.2, random_state=42)
# create model
model = LogisticRegression(class_weight=class_weight)
# evaluate model
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1)
# report performance
print('Accuracy: %.4f (%.4f)' % (mean(scores), std(scores)))
scores
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm, datasets
from sklearn.metrics import auc
from sklearn.metrics import plot_roc_curve
from sklearn.model_selection import StratifiedKFold
# #############################################################################
# Data IO and generation
# Import some data to play with
#iris = datasets.load_iris()
#X = iris.data
#y = iris.target
#X, y = X[y != 2], y[y != 2]
#n_samples, n_features = X.shape
# Add noisy features
#random_state = np.random.RandomState(0)
#X = np.c_[X, random_state.randn(n_samples, 200 * n_features)]
# #############################################################################
# Classification and ROC analysis
# Run classifier with cross-validation and plot ROC curves
cv = ShuffleSplit(n_splits=10, test_size=0.2, random_state=42)
classifier = LogisticRegression(class_weight=class_weight,random_state=42)
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
fig, ax = plt.subplots()
for i, (train, test) in enumerate(cv.split(X, y)):
classifier.fit(X, y)
viz = plot_roc_curve(classifier, X, y,
name='ROC fold {}'.format(i),
alpha=0.3, lw=1, ax=ax)
interp_tpr = np.interp(mean_fpr, viz.fpr, viz.tpr)
interp_tpr[0] = 0.0
tprs.append(interp_tpr)
aucs.append(viz.roc_auc)
ax.plot([0, 1], [0, 1], linestyle='--', lw=2, color='r',
label='Chance', alpha=.8)
mean_tpr = np.mean(tprs, axis=0)
mean_tpr[-1] = 1.0
mean_auc = auc(mean_fpr, mean_tpr)
std_auc = np.std(aucs)
ax.plot(mean_fpr, mean_tpr, color='b',
label=r'Mean ROC (AUC = %0.4f $\pm$ %0.4f)' % (mean_auc, std_auc),
lw=2, alpha=.8)
std_tpr = np.std(tprs, axis=0)
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
ax.fill_between(mean_fpr, tprs_lower, tprs_upper, color='grey', alpha=.2,
label=r'$\pm$ 1 std. dev.')
ax.set(xlim=[-0.05, 1.05], ylim=[-0.05, 1.05],
title="Receiver operating characteristic example")
ax.legend(loc="lower right")
plt.show()
```
## RF
```
from numpy import mean
from numpy import std
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import ShuffleSplit
# create dataset
#X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=1)
# prepare the cross-validation procedure
#cv = KFold(n_splits=5, test_size= 0.2, random_state=0)
cv = ShuffleSplit(n_splits=10, test_size=0.2, random_state=42)
# create model
model = RandomForestClassifier(class_weight=class_weight)
# evaluate model
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1)
# report performance
print('Accuracy: %.4f (%.4f)' % (mean(scores), std(scores)))
scores
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm, datasets
from sklearn.metrics import auc
from sklearn.metrics import plot_roc_curve
from sklearn.model_selection import StratifiedKFold
# #############################################################################
# Data IO and generation
# Import some data to play with
#iris = datasets.load_iris()
#X = iris.data
#y = iris.target
#X, y = X[y != 2], y[y != 2]
#n_samples, n_features = X.shape
# Add noisy features
#random_state = np.random.RandomState(0)
#X = np.c_[X, random_state.randn(n_samples, 200 * n_features)]
# #############################################################################
# Classification and ROC analysis
# Run classifier with cross-validation and plot ROC curves
cv = ShuffleSplit(n_splits=10, test_size=0.2, random_state=42)
classifier = RandomForestClassifier(class_weight=class_weight,random_state=42)
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
fig, ax = plt.subplots()
for i, (train, test) in enumerate(cv.split(X, y)):
classifier.fit(X, y)
#viz = plot_roc_curve(classifier, X, y,
# name='ROC fold {}'.format(i),
# alpha=0.3, lw=1, ax=ax)
interp_tpr = np.interp(mean_fpr, viz.fpr, viz.tpr)
interp_tpr[0] = 0.0
tprs.append(interp_tpr)
aucs.append(viz.roc_auc)
ax.plot([0, 1], [0, 1], linestyle='--', lw=2, color='r',
label='Chance', alpha=.8)
mean_tpr = np.mean(tprs, axis=0)
mean_tpr[-1] = 1.0
mean_auc = auc(mean_fpr, mean_tpr)
std_auc = np.std(aucs)
ax.plot(mean_fpr, mean_tpr, color='b',
label=r'Mean ROC (AUC = %0.4f $\pm$ %0.4f)' % (mean_auc, std_auc),
lw=2, alpha=.8)
std_tpr = np.std(tprs, axis=0)
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
ax.fill_between(mean_fpr, tprs_lower, tprs_upper, color='grey', alpha=.2,
label=r'$\pm$ 1 std. dev.')
ax.set(xlim=[-0.05, 1.05], ylim=[-0.05, 1.05],
title="Receiver operating characteristic of Random Forest Classifier")
ax.legend(loc="lower right")
plt.show()
```
## DT
```
from numpy import mean
from numpy import std
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import ShuffleSplit
from sklearn.tree import DecisionTreeClassifier
# create dataset
#X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=1)
# prepare the cross-validation procedure
#cv = KFold(n_splits=5, test_size= 0.2, random_state=0)
cv = ShuffleSplit(n_splits=10, test_size=0.2, random_state=42)
# create model
model = DecisionTreeClassifier(class_weight=class_weight)
# evaluate model
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1)
# report performance
print('Accuracy: %.4f (%.4f)' % (mean(scores), std(scores)))
scores
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm, datasets
from sklearn.metrics import auc
from sklearn.metrics import plot_roc_curve
from sklearn.model_selection import StratifiedKFold
# #############################################################################
# Data IO and generation
# Import some data to play with
#iris = datasets.load_iris()
#X = iris.data
#y = iris.target
#X, y = X[y != 2], y[y != 2]
#n_samples, n_features = X.shape
# Add noisy features
#random_state = np.random.RandomState(0)
#X = np.c_[X, random_state.randn(n_samples, 200 * n_features)]
# #############################################################################
# Classification and ROC analysis
# Run classifier with cross-validation and plot ROC curves
cv = ShuffleSplit(n_splits=10, test_size=0.2, random_state=42)
classifier = DecisionTreeClassifier(class_weight=class_weight,random_state=42)
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
fig, ax = plt.subplots()
for i, (train, test) in enumerate(cv.split(X, y)):
classifier.fit(X, y)
viz = plot_roc_curve(classifier, X, y,
name='ROC fold {}'.format(i),
alpha=0.3, lw=1, ax=ax)
interp_tpr = np.interp(mean_fpr, viz.fpr, viz.tpr)
interp_tpr[0] = 0.0
tprs.append(interp_tpr)
aucs.append(viz.roc_auc)
ax.plot([0, 1], [0, 1], linestyle='--', lw=2, color='r',
label='Chance', alpha=.8)
mean_tpr = np.mean(tprs, axis=0)
mean_tpr[-1] = 1.0
mean_auc = auc(mean_fpr, mean_tpr)
std_auc = np.std(aucs)
ax.plot(mean_fpr, mean_tpr, color='b',
label=r'Mean ROC (AUC = %0.4f $\pm$ %0.4f)' % (mean_auc, std_auc),
lw=2, alpha=.8)
std_tpr = np.std(tprs, axis=0)
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
ax.fill_between(mean_fpr, tprs_lower, tprs_upper, color='grey', alpha=.2,
label=r'$\pm$ 1 std. dev.')
ax.set(xlim=[-0.05, 1.05], ylim=[-0.05, 1.05],
title="Receiver operating characteristic example")
ax.legend(loc="lower right")
plt.show()
#from sklearn.model_selection import cross_val_score
#from sklearn import svm
#clf = svm.SVC(kernel='rbf', C=1, class_weight=class_weight)
#scores = cross_val_score(clf, X, y, cv=5)
#print("Accuracy: %0.4f (+/- %0.4f)" % (scores.mean(), scores.std() * 2))
#clf.score(X_test, y_test)
```
## ANN
```
import keras
from keras.models import Sequential
from keras.layers import Dense,Dropout
classifier=Sequential()
classifier.add(Dense(units=256, kernel_initializer='uniform',activation='relu',input_dim=24))
classifier.add(Dense(units=128, kernel_initializer='uniform',activation='relu'))
classifier.add(Dropout(p=0.5))
classifier.add(Dense(units=64, kernel_initializer='uniform',activation='relu'))
classifier.add(Dropout(p=0.4))
classifier.add(Dense(units=32, kernel_initializer='uniform',activation='relu'))
classifier.add(Dense(units=1, kernel_initializer='uniform',activation='sigmoid'))
classifier.compile(optimizer='adam',loss="binary_crossentropy",metrics=['accuracy'])
classifier.fit(X_train,y_train,batch_size=10,epochs=100,class_weight=class_weight,validation_data=(X_test, y_test))
#clf_svc_rbf.fit(X_train,y_train)
from sklearn.metrics import confusion_matrix,classification_report,roc_auc_score,auc,f1_score
y_pred = classifier.predict(X_test)>0.9
import matplotlib.pyplot as plt
cm = confusion_matrix(y_test,y_pred)
#plt.figure(figsize=(5,5))
#sns.heatmap(cm,annot=True)
#plt.show()
#print(classification_report(y_test,y_pred_clf_svc_rbf))
print(classification_report(y_test, y_pred))
#plot_confusion_matrix(confusion_matrix(y_test, y_pred))
from sklearn.metrics import roc_curve, auc
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
roc_auc
from sklearn.metrics import roc_curve,roc_auc_score
from sklearn.metrics import auc
fpr , tpr , thresholds = roc_curve ( y_test , y_pred)
auc_keras = auc(fpr, tpr)
print("AUC Score:",auc_keras)
plt.figure()
lw = 2
plt.plot(fpr, tpr, color='darkorange',
lw=lw, label='ROC curve (area = %0.4f)' % auc_keras)
plt.plot([0, 1], [0, 1], color='navy', lw=lw, linestyle='--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic example')
plt.legend(loc="lower right")
plt.show()
#from sklearn.tree import DecisionTreeClassifier
#from sklearn.model_selection import cross_val_score
#dt = DecisionTreeClassifier(class_weight=class_weight)
#scores = cross_val_score(clf, X, y, cv=5)
#print("Accuracy: %0.4f (+/- %0.4f)" % (scores.mean(), scores.std() * 2))
'''
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import confusion_matrix,classification_report,roc_auc_score,auc,f1_score
lr = LogisticRegression()
lr.fit(X_train,y_train)
y_pred_logistic = lr.predict(X_test)
import matplotlib.pyplot as plt
cm = confusion_matrix(y_test,y_pred_logistic)
plt.figure(figsize=(5,5))
sns.heatmap(cm,annot=True,linewidths=.3)
plt.show()
print(classification_report(y_test,y_pred_logistic))
from sklearn.metrics import roc_curve, auc
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred_logistic)
roc_auc = auc(false_positive_rate, true_positive_rate)
roc_auc
print(f1_score(y_test, y_pred_logistic,average="macro"))
'''
from sklearn import datasets
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import VotingClassifier
clf1 = SVC(kernel='rbf', C=1, class_weight=class_weight,random_state=42)
clf2 = LogisticRegression(class_weight=class_weight,random_state=42)
clf3 = RandomForestClassifier(class_weight=class_weight,random_state=42)
clf4 = DecisionTreeClassifier(class_weight=class_weight,random_state=42)
#clf5 = Sequential()
eclf = VotingClassifier( estimators=[('svm', clf1), ('lr', clf2), ('rf', clf3), ('dt',clf4)],
voting='hard')
for clf, label in zip([clf1, clf2, clf3,clf4 ,eclf], ['SVM', 'LR', 'RF','DT', 'Ensemble']):
scores = cross_val_score(clf, X, y, scoring='accuracy', cv=10)
print("Accuracy: %0.4f (+/- %0.4f) [%s]" % (scores.mean(), scores.std(), label))
scores
```
|
github_jupyter
|
# Importing libraries
```
import nltk
import glob
import os
import numpy as np
import string
import pickle
from gensim.models import Doc2Vec
from gensim.models.doc2vec import LabeledSentence
from tqdm import tqdm
from sklearn import utils
from sklearn.svm import LinearSVC
from sklearn.neural_network import MLPClassifier
from sklearn.metrics import roc_curve
from sklearn.metrics import roc_auc_score
from matplotlib import pyplot
from nltk import sent_tokenize
from nltk.tokenize import word_tokenize
from nltk.corpus import stopwords
from nltk.stem.porter import PorterStemmer
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.naive_bayes import BernoulliNB
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
from collections import Counter
from collections import defaultdict
X_train_text = []
Y_train = []
X_test_text =[]
Y_test =[]
Vocab = {}
VocabFile = "aclImdb/imdb.vocab"
```
# Create Vocabulary Function
```
def CreateVocab():
with open(VocabFile, encoding='latin-1') as f:
words = f.read().splitlines()
stop_words = set(stopwords.words('english'))
i=0
for word in words:
if word not in stop_words:
Vocab[word] = i
i+=1
print(len(Vocab))
```
# Cleaning Data
```
def clean_review(text):
tokens = word_tokenize(text)
tokens = [w.lower() for w in tokens]
table = str.maketrans('', '', string.punctuation)
stripped = [w.translate(table) for w in tokens]
words = [word for word in stripped if word.isalpha()]
stop_words = set(stopwords.words('english'))
words = [w for w in words if not w in stop_words]
return words
```
# Generating Word Matrices
```
def BoWMatrix(docs):
vectorizer = CountVectorizer(binary=True,vocabulary = Vocab)
Doc_Term_matrix = vectorizer.fit_transform(docs)
return Doc_Term_matrix
def TfidfMatrix(docs):
vectorizer = TfidfVectorizer(vocabulary = Vocab,norm = 'l1')
Doc_Term_matrix = vectorizer.fit_transform(docs)
return Doc_Term_matrix
```
# ROC Curve Function
```
def ROC(Y_train, pred1, Y_test, pred2):
fpr1, tpr1, thresholds1 = roc_curve(Y_train, pred1)
pyplot.plot([0, 1], [0, 1], linestyle='--')
pyplot.plot(fpr1, tpr1, marker='.', color='blue', label="Train", linewidth=1.0)
fpr2, tpr2, thresholds2 = roc_curve(Y_test, pred2)
pyplot.plot(fpr2, tpr2, marker='.', color='red', label="Test", linewidth=1.0)
pyplot.legend()
pyplot.show()
def ROC2(X, pred, pred1, pred2):
fpr, tpr, thresholds = roc_curve(X, pred)
pyplot.plot([0, 1], [0, 1], linestyle='--')
pyplot.plot(fpr, tpr, marker='.')
fpr1, tpr1, thresholds1 = roc_curve(X, pred1)
pyplot.plot([0, 1], [0, 1], linestyle='--')
pyplot.plot(fpr1, tpr1, marker='.')
fpr2, tpr2, thresholds2 = roc_curve(X, pred2)
pyplot.plot([0, 1], [0, 1], linestyle='--')
pyplot.plot(fpr2, tpr2, marker='.')
pyplot.show()
```
# Naive Bayes Function
```
def NB(X,Y_train,Xtest,Y_test,mtype):
if mtype == "Bow":
model = BernoulliNB()
elif mtype == "Tfidf":
model = MultinomialNB()
else:
model = GaussianNB()
model.fit(X,Y_train)
pred1 = model.predict(X)
pred2 = model.predict(Xtest)
acc1 = accuracy_score(Y_train,pred1)
acc2 = accuracy_score(Y_test,pred2)
print("NaiveBayes + " + mtype + " Train Accuracy: " + str(acc1*100) + "%")
print("NaiveBayes + " + mtype + " Test Accuracy: " + str(acc2*100) + "%")
prob1 = model.predict_proba(X)
prob1 = prob1[:, 1]
prob2 = model.predict_proba(Xtest)
prob2 = prob2[:, 1]
#ROC(Y_train, pred1, Y_test, pred2)
ROC(Y_train, prob1, Y_test, prob2)
```
# Logistic Regression Function
```
def LR(X,Y_train,Xtest,Y_test,mtype):
model = LogisticRegression()
model.fit(X,Y_train)
pred1 = model.predict(X)
pred2 = model.predict(Xtest)
acc1 = accuracy_score(Y_train,pred1)
acc2 = accuracy_score(Y_test,pred2)
print("LogisticRegression + " + mtype + " Train Accuracy: " + str(acc1*100) + "%")
print("LogisticRegression + " + mtype + " Test Accuracy: " + str(acc2*100) + "%")
prob1 = model.predict_proba(X)
prob1 = prob1[:, 1]
prob2 = model.predict_proba(Xtest)
prob2 = prob2[:, 1]
#ROC(Y_train, pred1, Y_test, pred2)
ROC(Y_train, prob1, Y_test, prob2)
```
# Random Forest Function
```
def RF(X,Y_train,Xtest,Y_test,mtype):
if mtype == "Bow":
n = 400
md = 100
elif mtype == "Tfidf":
n = 400
md = 100
else:
n = 100
md = 10
model = RandomForestClassifier(n_estimators=n, bootstrap=True,
max_depth=md, max_features='auto',
min_samples_leaf=4, min_samples_split=10)
model.fit(X,Y_train)
pred1 = model.predict(X)
pred2 = model.predict(Xtest)
acc1 = accuracy_score(Y_train,pred1)
acc2 = accuracy_score(Y_test,pred2)
print("RandomForest + " + mtype + " Train Accuracy: " + str(acc1*100) + "%")
print("RandomForest + " + mtype + " Test Accuracy: " + str(acc2*100) + "%")
prob1 = model.predict_proba(X)
prob1 = prob1[:, 1]
prob2 = model.predict_proba(Xtest)
prob2 = prob2[:, 1]
#ROC(Y_train, pred1, Y_test, pred2)
ROC(Y_train, prob1, Y_test, prob2)
```
# Support Vector Machine Function
```
def SVM(X,Y_train,Xtest,Y_test,mtype):
model = LinearSVC()
model.fit(X,Y_train)
pred1 = model.predict(X)
pred2 = model.predict(Xtest)
acc1 = accuracy_score(Y_train,pred1)
acc2 = accuracy_score(Y_test,pred2)
print("SVM + " + mtype + " Train Accuracy: " + str(acc1*100) + "%")
print("SVM + " + mtype + " Test Accuracy: " + str(acc2*100) + "%")
ROC(Y_train, pred1, Y_test, pred2)
```
# Forward Feed Neural Network Function
```
def NN(X,Y_train,Xtest,Y_test,mtype):
model = MLPClassifier(hidden_layer_sizes=(10,10),activation='relu',max_iter=200)
model.fit(X,Y_train)
pred1 = model.predict(X)
pred2 = model.predict(Xtest)
acc1 = accuracy_score(Y_train,pred1)
acc2 = accuracy_score(Y_test,pred2)
print("FFN + " + mtype + " Train Accuracy: " + str(acc1*100) + "%")
print("FFN + " + mtype + " Test Accuracy: " + str(acc2*100) + "%")
prob1 = model.predict_proba(X)
prob1 = prob1[:, 1]
prob2 = model.predict_proba(Xtest)
prob2 = prob2[:, 1]
#ROC(Y_train, pred1, Y_test, pred2)
ROC(Y_train, prob1, Y_test, prob2)
```
# Loading Data
```
path1 = 'aclImdb/train/pos/*.txt'
path2 = 'aclImdb/train/neg/*.txt'
path3 = 'aclImdb/test/pos/*.txt'
path4 = 'aclImdb/test/neg/*.txt'
files1 = glob.glob(path1)
files2 = glob.glob(path2)
files3 = glob.glob(path3)
files4 = glob.glob(path4)
#Positive labels
for i,filename in enumerate(files1):
f = open(filename,"r+", encoding='latin-1')
text = f.read()
f.close()
X_train_text.append(text)
Y_train.append(1)
#Neg labels
for j,filename in enumerate(files2):
f = open(filename,"r+", encoding='latin-1')
text = f.read()
f.close()
X_train_text.append(text)
Y_train.append(0)
#Test labels +
for k,filename in enumerate(files3):
f = open(filename,"r+", encoding='latin-1')
text = f.read()
f.close()
X_test_text.append(text)
Y_test.append(1)
#Test labels +
for l,filename in enumerate(files4):
f = open(filename,"r+", encoding='latin-1')
text = f.read()
f.close()
X_test_text.append(text)
Y_test.append(0)
CreateVocab();
```
# Generating Word Matrix for Test & Train Data
```
def Getbowvec(X_train_text,Y_train,X_test_text,Y_test):
X = BoWMatrix(X_train_text)
Xtest = BoWMatrix(X_test_text)
return X,Xtest
def Gettfidfvec(X_train_text,Y_train,X_test_text,Y_test):
X = TfidfMatrix(X_train_text)
Xtest = TfidfMatrix(X_test_text)
return X,Xtest
```
# Doc2Vec Representation
```
'''
def LabelRev(reviews,label_string):
result = []
prefix = label_string
for i, t in enumerate(reviews):
# print(t)
result.append(LabeledSentence(t, [prefix + '_%s' % i]))
return result
LabelledXtrain = LabelRev(X_train_text,"review")
LabelledXtest = LabelRev(X_test_text,"test")
LabelledData = LabelledXtrain + LabelledXtest
modeld2v = Doc2Vec(dm=1, min_count=2, alpha=0.065, min_alpha=0.065)
modeld2v.build_vocab([x for x in tqdm(LabelledData)])
print("Training the Doc2Vec Model.....")
for epoch in range(50):
print("epoch : ",epoch)
modeld2v.train(utils.shuffle([x for x in tqdm(LabelledData)]), total_examples=len(LabelledData), epochs=1)
modeld2v.alpha -= 0.002
modeld2v.min_alpha = modeld2v.alpha
print("Saving Doc2Vec1 Model....")
modeld2v.save('doc2vec1.model')
#print("Saving Doc2Vec Model....")
#modeld2v.save('doc2vec.model')
'''
def Doc2vec(X_train_text,Y_train,X_test_text,Y_test):
model = Doc2Vec.load('doc2vec.model')
#model = Doc2Vec.load('doc2vec1.model')
X = []
Xtest =[]
for i,l in enumerate(X_train_text):
temp = "review" + "_" + str(i)
X.append(model.docvecs[temp])
for i,l in enumerate(X_test_text):
temp = "test" + "_" + str(i)
Xtest.append(model.docvecs[temp])
return X,Xtest
print("Bag of Words is being built...")
X,Xtest = Getbowvec(X_train_text,Y_train,X_test_text,Y_test)
print("Tf-idf is being built...")
X1,Xtest1 = Gettfidfvec(X_train_text,Y_train,X_test_text,Y_test)
print("Doc2Vec is being built...")
X2,Xtest2 = Doc2vec(X_train_text,Y_train,X_test_text,Y_test)
len(X[0])
```
# Applying Classification Algorithms
```
print("Naive Bayes:")
NB(X,Y_train,Xtest,Y_test,"Bow")
NB(X1,Y_train,Xtest1,Y_test,"Tfidf")
NB(X2,Y_train,Xtest2,Y_test,"Doc2Vec")
print("Logistic Regression:")
LR(X,Y_train,Xtest,Y_test,"Bow")
LR(X1,Y_train,Xtest1,Y_test,"Tfidf")
LR(X2,Y_train,Xtest2,Y_test,"Doc2Vec")
print("Random Forest:")
RF(X,Y_train,Xtest,Y_test,"Bow")
RF(X1,Y_train,Xtest1,Y_test,"Tfidf")
RF(X2,Y_train,Xtest2,Y_test,"Doc2Vec")
print("SVM:")
SVM(X,Y_train,Xtest,Y_test,"Bow")
SVM(X1,Y_train,Xtest1,Y_test,"Tfidf")
SVM(X2,Y_train,Xtest2,Y_test,"Doc2Vec")
print("Neural Networks:")
NN(X,Y_train,Xtest,Y_test,"Bow")
NN(X1,Y_train,Xtest1,Y_test,"Tfidf")
NN(X2,Y_train,Xtest2,Y_test,"Doc2Vec")
```
|
github_jupyter
|
# Lab 2: Object-Oriented Python
## Overview
After have covered rules, definitions, and semantics, we'll be playing around with actual classes, writing a fair chunk of code and building several classes to solve a variety of problems.
Recall our starting definitions:
- An *object* has identity
- A *name* is a reference to an object
- A *namespace* is an associative mapping from names to objects
- An *attribute* is any name following a dot ('.')
## Course class
### Basic Class
Let’s create a class to represent courses!
A course will have three attributes to start: a
1. department (like `"AI"` `"CHEM"`),
2. a course code (like `"42"` or `"92SI"`),
3. and a title (like `"IAP"`).
```Python
class Course:
def __init__(self, department, code, title):
self.department = department
self.code = code
self.title = title
```
You can assume that all arguments to this constructor will be strings.
Running the following code cell will create a class object `Course` and print some information about it.
*Note: If you change the content of this class definition, you will need to re-execute the code cell for it to have any effect. Any instance objects of the old class object will not be automatically updated, so you may need to rerun instantiations of this class object as well.*
```
class Course:
def __init__(self, department, code, title):
self.department = department
self.code = code
self.title = title
print(Course)
print(Course.mro())
print(Course.__init__)
```
We create an instance of the class by instantiating the class object, supplying some arguments.
```Python
iap = Course("AI", "91256", "IAP: Introduction to Algorithms and Programming")
```
Print out the three attributes of the `unbo_python` instance object.
```
iap = Course("AI", "91256", "IAP: Introduction to Algorithms and Programming")
print(iap.department) # Print out the department
print(iap.code) # Print out the code
print(iap.title) # Print out the title
```
### Inheritance
Let's explore inheritance by creating a `AICourse` class that takes an additional parameter `recorded` that defaults to `False`.
```
class AICourse(Course):
def __init__(self, department, code, title, recorded=False):
super().__init__(department, code, title)
self.is_recorded = recorded
```
The `super()` concretely lets us treat the `self` object as an instance object of the immediate superclass (as measured by MRO), so we can call the superclass's `__init__` method.
We can instantiate our new class:
```Python
a = Course("AI", "91254", "Image Processing and Computer Vision")
b = AICourse("AI", "91247", "Cognition and Neuroscience")
x = AICourse("AI", "91247X", "Cognition and Neuroscience", recorded=True)
print(a.code) # => "91254"
print(b.code) # => "91247"
```
Read through the following statements and try to predict their output.
```Python
type(a)
isinstance(a, Course)
isinstance(b, Course)
isinstance(x, Course)
isinstance(x, AICourse)
issubclass(x, AICourse)
issubclass(Course, AICourse)
type(a) == type(b)
type(b) == type(x)
a == b
b == x
```
```
a = Course("AI", "91254", "Image Processing and Computer Vision")
b = AICourse("AI", "91247", "Cognition and Neuroscience")
x = AICourse("AI", "91247X", "Cognition and Neuroscience", recorded=True)
print("1.", type(a))
print("2.", isinstance(a, Course))
print("3.", isinstance(b, Course))
print("4.", isinstance(x, Course))
print("5.", isinstance(x, AICourse))
print("6.", issubclass(Course, AICourse))
print("7.", issubclass(AICourse, Course))
print("8.", type(a) == type(b))
print("9.", type(b) == type(x))
print("10.", a == b)
print("11.", b == x)
```
### Additional Attributes
Let's add more functionality to the `Course` class!
* Add a attribute `students` to the instances of the `Course` class that tracks whether students are present. Initially, students should be an empty set.
* Create a method `mark_attendance(*students)` that takes a variadic number of `students` and marks them as present.
* Create a method `is_present(student)` that takes a student’s name as a parameter and returns `True` if the student is present and `False` otherwise.
```
class Course:
def __init__(self, department, code, title):
self.department = department
self.code = code
self.title = title
self.students = {}
def mark_attendance(self, student):
if student in self.students:
self.students[student] += 1
else:
self.students[student] = 1
def is_present(self, student):
return student in self.students
```
### Implementing Prerequisites
Now, we'll focus on `AICourse`. We want to implement functionality to determine if one computer science course is a prerequisite of another. In our implementation, we will assume that the ordering for courses is determined first by the numeric part of the course code: for example, `140` comes before `255`. If there is a tie, the ordering is determined by the default string ordering of the letters that follow. For example, `91254 > 91247`. After implementing, you should be able to see:
```Python
>>> ai91245 = Course("AI", "91254", "Image Processing and Computer Vision")
>>> ai91247 = AICourse("AI", "91247", "Cognition and Neuroscience")
>>> ai91247 > ai91245
True
```
To accomplish this, you will need to implement a magic method `__le__` that will add functionality to determine if a course is a prerequisite for another course. Read up on [total ordering](https://docs.python.org/3/library/functools.html#functools.total_ordering) to figure out what `__le__` should return based on the argument you pass in.
To give a few hints on how to add this piece of functionality might be implemented, consider how you might extract the actual `int` number from the course code attribute.
Additionally, you should implement a `__eq__` on `Course`s. Two classes are equivalent if they are in the same department and have the same course code: the course title doesn't matter here.
```
class Course:
def __init__(self, department, code, title):
self.department = department
self.code = code
self.title = title
self.students = {}
def mark_attendance(self, student):
if student in self.students:
self.students[student] += 1
else:
self.students[student] = 1
def is_present(self, student):
return student in self.students
def __le__(self, other):
mycode = int(self.code)
othercode = int(other.code)
return mycode < othercode
def __eq__(self, other):
mycode = int(self.code)
othercode = int(other.code)
mydepartment = self.department
otherdepartment = other.department
return (mycode == othercode) and (mydepartment == otherdepartment)
c1 = Course(...)
c1.mark_atte
student1 = "Mark"
```
#### Sorting
Now that we've written a `__le__` method and an `__eq__` method, we've implemented everything we need to speak about an "ordering" of `Course`s.
##### Let Python do all the rest (Optional)
Using the [`functools.total_ordering` decorator](https://docs.python.org/3/library/functools.html#functools.total_ordering), get back to the Course class definition and "decorate" it by adding `@total_ordering` before the very class definition, so that all of the comparison methods are implemented.
Then, you should be able to run:
```
# Let's make ai91245 an AI course
ai91245 = AICourse("AI", "91254", "Image Processing and Computer Vision")
ai91247 = AICourse("AI", "91247", "Cognition and Neuroscience")
ai91762 = AICourse("AI", "107", "Combinatorial Decision Making and Optimization")
ai91249 = AICourse("AI", "110", "Machine Learning and Deep Learning")
courses = [ai91247, ai91245, ai91762, ai91249]
courses.sort()
courses # => [ai91245, ai91247, ai91249, ai91762]
```
### Instructors (optional)
Allow the class to take a splat argument `instructors` that will take any number of strings and store them as a list of instructors.
Modify the way you track attendance in the `Course` class to map a Python date object (you can use the `datetime` module) to a data structure tracking what students are there on that day.
```
class CourseWithInstructors:
pass
```
### Catalog (optional)
Implement a class called `CourseCatalog` that is constructed from a list of `Course`s. Write a method for the `CourseCatalog` which returns a list of courses in a given department. Additionally, write a method for `CourseCatalog` that returns all courses that contain a given piece of search text in their title.
Feel free to implement any other interesting methods you'd like.
```
class CourseCatalog:
def __init__(self, courses):
pass
def courses_by_department(self, department_name):
pass
def courses_by_search_term(self, search_snippet):
pass
```
## Inheritance
Consider the following code:
```Python
"""Examples of Single Inheritance"""
class Transportation:
wheels = 0
def __init__(self):
self.wheels = -1
def travel_one(self):
print("Travelling on generic transportation")
def travel(self, distance):
for _ in range(distance):
self.travel_one()
def is_car(self):
return self.wheels == 4
class Bike(Transportation):
def travel_one(self):
print("Biking one km")
class Car(Transportation):
wheels = 4
def travel_one(self):
print("Driving one km")
def make_sound(self):
print("VROOM")
class Ferrari(Car):
pass
t = Transportation()
b = Bike()
c = Car()
f = Ferrari()
```
Predict the outcome of each of the following lines of code.
```Python
isinstance(t, Transportation)
isinstance(b, Bike)
isinstance(b, Transportation)
isinstance(b, Car)
isinstance(b, t)
isinstance(c, Car)
isinstance(c, Transportation)
isinstance(f, Ferrari)
isinstance(f, Car)
isinstance(f, Transportation)
issubclass(Bike, Transportation)
issubclass(Car, Transportation)
issubclass(Ferrari, Car)
issubclass(Ferrari, Transportation)
issubclass(Transportation, Transportation)
b.travel(5)
c.is_car()
f.is_car()
b.is_car()
b.make_sound()
c.travel(10)
f.travel(4)
```
```
class Transportation:
wheels = 0
def __init__(self):
self.wheels = -1
def travel_one(self):
print("Travelling on generic transportation")
def travel(self, distance):
for _ in range(distance):
self.travel_one()
def is_car(self):
return self.wheels == 4
class Bike(Transportation):
wheels = 2
def travel_one(self):
print("Biking one km")
class Car(Transportation):
wheels = 4
def travel_one(self):
print("Driving one km")
def make_sound(self):
print("VROOM")
class Ferrari(Car):
pass
t = Transportation()
b = Bike()
c = Car()
f = Ferrari()
print("1.", isinstance(t, Transportation))
print("2.", isinstance(b, Bike))
print("3.", isinstance(b, Transportation))
print("4.", isinstance(b, Car))
print("5.", isinstance(b, type(Car)))
print("6.", isinstance(c, Car))
print("7.", isinstance(c, Transportation))
print("8.", isinstance(f, Ferrari))
print("9.", isinstance(f, Car))
print("10.", isinstance(f, Transportation))
print("11.", issubclass(Bike, Transportation))
print("12.", issubclass(Car, Transportation))
print("13.", issubclass(Ferrari, Car))
print("14.", issubclass(Ferrari, Transportation))
print("15.", issubclass(Transportation, Transportation))
b.travel(5)
print("16.", c.is_car()) # => c.wheels ?
print("17.", f.is_car()) # => f.wheels ?
print("18.", b.is_car()) # => b.wheels ?
# b.make_sound()
c.travel(10)
f.travel(4)
```
## SimpleGraph
In this part, you'll build the implementation for a `SimpleGraph` class in Python.
In particular, you will need to define a `Vertex` class, an `Edge` class, and a `SimpleGraph` class. The specification is as follows:
A `Vertex` has attributes:
* `name`, a string representing the label of the vertex.
* `edges`, a set representing edges outbound from this vertex to its neighbors
A new Vertex should be initialized with an optional `name`, which defaults to `""`, and should be initialized with an empty edge set.
An `Edge` has attributes:
* `start`, a `Vertex` representing the start point of the edge.
* `end`, a `Vertex` representing the end point of the edge.
* `cost`, a `float` (used for graph algorithms) representing the weight of the edge.
* `visited`, a `bool` (used for graph algorithms) representing whether this edge has been visited before.
Note that for our purposes, an `Edge` is directed.
An `Edge` requires a `start` and `end` vertex in order to be instantiated. `cost` should default to 1, and `visited` should default to `False`, but both should be able to be set via an initializer.
A `SimpleGraph` has attributes
* `verts`, a collection of `Vertex`s (you need to decide the collection type)
* `edges`, a collection of `Edge`s (you need to decide the collection type)
as well as several methods:
* `graph.add_vertex(v)`
* `graph.add_edge(v_1, v_2)`
* `graph.contains_vertex(v)`
* `graph.contains_edge(v_1, v_2)`
* `graph.get_neighbors(v)`
* `graph.is_empty()`
* `graph.size()`
* `graph.remove_vertex(v)`
* `graph.remove_edge(v_1, v_2)`
* `graph.is_neighbor(v1, v2)`
* `graph.is_reachable(v1, v2) # Use any algorithm you like`
* `graph.clear_all()`
The actual implementation details are up to you.
*Note: debugging will significantly easier if you write `__str__` or `__repr__` methods on your custom classes.*
```
class Vertex:
pass
class Edge:
pass
class SimpleGraph:
pass
```
### Challenge: Graph Algorithms
If you're feeling up to the challenge, and you have sufficient time, implement other graph algorithms, including those covered in ai91247/X, using your SimpleGraph. The point isn't to check whether you still know your graph algorithms - rather, these algorithms will serve to test the correctness of your graph implementation. The particulars are up to you.
As some suggestions:
* Longest path
* D'ijkstras Algorithm
* A*
* Max Flow
* K-Clique
* Largest Connected Component
* is_bipartite
* hamiltonian_path_exists
```
graph = SimpleGraph()
# Your extension code here
```
### Challenge: Using Magic Methods
See if you can rewrite the `SimpleGraph` class using magic methods to emulate the behavior and operators of standard Python. In particular,
```
graph[v] # returns neighbors of v
graph[v] = v_2 # Insert an edge from v to v2
len(graph)
# etc
```
## Timed Key-Value Store (challenge)
Let's build an interesting data structure straight out of an interview programming challenge from [Stripe](https://stripe.com/). This is more of an algorithms challenge than a Python challenge, but we hope you're still interested in tackling it.
At a high-level, we'll be building a key-value store (think `dict` or Java's `HashMap`) that has a `get` method that takes an optional second parameter as a `time` object in Python to return the most recent value before that period in time. If no key-value pair was added to the map before that period in time, return `None`.
For consistency’s sake, let’s call this class `TimedKVStore` and put it into a file called `kv_store.py`
You’ll need some sort of `time` object to track when key-value pairs are getting added to this map. Consider using [the `time` module](https://docs.python.org/3/library/time.html).
To give you an idea of how this class works, this is what should happen after you implement `TimedKVStore`.
```Python
d = TimedKVStore()
t0 = time.time()
d.put("1", 1)
t1 = time.time()
d.put("1", 1.1)
d.get("1") # => 1.1
d.get("1", t1) # => 1
d.get("1", t0) # => None
```
```
class TimedKVStore:
pass
d = TimedKVStore()
t0 = time.time()
d.put("1", 1)
t1 = time.time()
d.put("1", 1.1)
print(d.get("1")) # => 1.1
print(d.get("1", t1)) # => 1
print(d.get("1", t0)) # => None
```
### Remove (challenge)
Implement a method on a `TimedKVStore` to `remove(key)` that takes a key and removes that entire key from the key-value store.
Write another `remove(key, time)` method that takes a key and removes all memory of values before that time method.
## Bloom Filter (challenge)
A bloom filter is a fascinating data structure that support insertion and probabilistic set membership. Read up on Wikipedia!
Write a class `BloomFilter` to implement a bloom filter data structure. Override the `__contains__` method so that membership can be tested with `x in bloom_filter`.
```
class BloomFilter:
pass
```
## Silencer Context Manager (challenge)
In some cases, you may want to suppress the output a given code block. Maybe it's untrusted code, or maybe it's littered with `print`s that you don't want to comment out. We can use the context manager syntax in Python to define a class that serves as a context manager. We want to use this as:
```Python
with Silencer():
noisy_code()
```
Our class will look something like
```Python
class Silencer:
def __init__(self):
pass
def __enter__(self):
pass
def __exit__(self, *exc):
pass
```
The `__enter__` method is called when the with block is entered, and `__exit__` is called when leaving the block, with any relevant information about an active exception passed in. Write the `__enter__` method to redirect standard output and standard error to `stringio.StringIO()` objects to capture the output, and make sure that `__exit__` restored the saved stdout and stderr. What would a `__str__` method on a `Silencer` object look like?
Recall that the with statement in Python is *almost* implemented as:
```Python
with open(filename) as f:
raw = f.read()
# is (almost) equivalent to
f = open(filename)
f.__enter__()
try:
raw = f.read()
finally:
f.__exit__() # Closes the file
```
```
class Silencer:
pass
```
## Magic Methods
### Reading
Python provides an enormous number of special methods that a class can override to interoperator with builtin Python operations. You can skim through an [approximate visual list](http://diveintopython3.problemsolving.io/special-method-names.html) from Dive into Python3, or a [more verbose explanation](https://rszalski.github.io/magicmethods/), or the [complete Python documentation](https://docs.python.org/3/reference/datamodel.html#specialnames) on special methods. Fair warning, there are a lot of them, so it's probably better to skim than to really take a deep dive, unless you're loving this stuff.
### Writing (Polynomial Class)
We will write a `Polynomial` class that acts like a number. As a a reminder, a [polynomial](https://en.wikipedia.org/wiki/Polynomial) is a mathematical object that looks like $1 + x + x^2$ or $4 - 10x + x^3$ or $-4 - 2x^{10}$. A mathematical polynomial can be evaluated at a given value of $x$. For example, if $f(x) = 1 + x + x^2$, then $f(5) = 1 + 5 + 5^2 = 1 + 5 + 25 = 31$.
Polynomials are also added componentwise: If $f(x) = 1 + 4x + 4x^3$ and $g(x) = 2 + 3x^2 + 5x^3$, then $(f + g)(x) = (1 + 2) + 4x + 3x^2 + (4 + 5)x^3 = 3 + 4 + 3x^2 + 9x^3$.
Construct a polynomial with a variadic list of coefficients: the zeroth argument is the coordinate of the $x^0$'s place, the first argument is the coordinate of the $x^1$'s place, and so on. For example, `f = Polynomial(1, 3, 5)` should construct a `Polynomial` representing $1 + 3x + 5x^2$.
You will need to override the addition special method (`__add__`) and the callable special method (`__call__`).
You should be able to emulate the following code:
```Python
f = Polynomial(1, 5, 10)
g = Polynomial(1, 3, 5)
print(f(5)) # => Invokes `f.__call__(5)`
print(g(2)) # => Invokes `g.__call__(2)`
h = f + g # => Invokes `f.__add__(g)`
print(h(3)) # => Invokes `h.__call__(3)`
```
Lastly, implement a method to convert a `Polynomial` to an informal string representation. For example, the polynomial `Polynomial(1, 3, 5)` should be represented by the string `"1 * x^0 + 3 * x^1 + 5 * x^2"`.
```
class Polynomial:
def __init__(self):
pass
def __call__(self, x):
"""Implement `self(x)`."""
pass
def __add__(self, other):
"""Implement `self + other`."""
pass
def __str__(self):
"""Implement `str(x)`."""
pass
```
#### Polynomial Extensions (optional)
If you are looking for more, implement additional operations on our `Polynomial` class. You may want to implement `__sub__`, `__mul__`, and `__div__`.
You can also implement more complicated mathematical operations, such as `f.derivative()`, which returns a new function that is the derivative of `f`, or `.zeros()`, which returns a collection of the function's zeros.
If you need even more, write a `classmethod` to construct a polynomial from a string representation of it. You should be able to write:
```
f = Polynomial.parse("1 * x^0 + 3 * x^1 + 5 * x^2")
```
#### Challenge (`MultivariatePolynomial`)
Write a class called `MultivariatePolynomial` that represents a polynomial in many variables. For example, $f(x, y, z) = 4xy + 10x^2z - 5x^3yz + y^4z^3$ is a polynomial in three variables.
How would you provide coefficients to the constructor? How would you define the arguments to the callable? How would you implement the mathematical operations efficiently?
|
github_jupyter
|
## KITTI Object Detection finetuning
### This notebook is used to lunch the finetuning of FPN on KITTI object detection benchmark, the code fetches COCO weights for weight initialization
```
data_path = "../datasets/KITTI/data_object_image_2/training"
import detectron2
from detectron2.utils.logger import setup_logger
setup_logger()
import numpy as np
import cv2
import random
from detectron2.engine import DefaultPredictor
from detectron2.config import get_cfg
from detectron2.utils.visualizer import Visualizer
from detectron2.data import MetadataCatalog
from detectron2.modeling import build_model
from detectron2.evaluation import COCOEvaluator,PascalVOCDetectionEvaluator
import matplotlib.pyplot as plt
import torch.tensor as tensor
from detectron2.data import build_detection_test_loader
from detectron2.evaluation import inference_on_dataset
import torch
from detectron2.structures.instances import Instances
from detectron2.modeling import build_model
%matplotlib inline
```
## Dataset Parsing
```
import os
import numpy as np
import json
from detectron2.structures import BoxMode
def get_kitti_dicts(img_dir):
dataset_dicts = []
with open('../datasets/KITTI/kitti_train.txt') as f:
for line in f:
record = {}
image_path = os.path.join(img_dir, 'image_2/%s.png'%line.replace('\n',''))
height, width = cv2.imread(image_path).shape[:2]
record["file_name"] = image_path
record["image_id"] = int(line)
record["height"] = height
record["width"] = width
objs = []
ann_path = os.path.join(img_dir,'label_2/%s.txt'%line.replace('\n',''))
with open(ann_path) as ann_file:
for ann_line in ann_file:
line_items = ann_line.split(' ')
if(line_items[0]=='Car'):
class_id=2
elif(line_items[0]=='Pedestrian'):
class_id=0
elif(line_items[0]=='Cyclist'):
class_id=1
else:
continue
obj = {'bbox':[np.round(float(line_items[4])),np.round(float(line_items[5])),
np.round(float(line_items[6])),np.round(float(line_items[7]))],"category_id": class_id,"iscrowd": 0,"bbox_mode": BoxMode.XYXY_ABS}
objs.append(obj)
record["annotations"] = objs
dataset_dicts.append(record)
items+=1
return dataset_dicts
def get_kitti_val(img_dir):
dataset_dicts = []
items = 0
with open('kitti_val.txt') as f:
for line in f:
record = {}
image_path = os.path.join(img_dir, 'image_2/%s.png'%line.replace('\n','').zfill(6))
height, width = cv2.imread(image_path).shape[:2]
record["file_name"] = image_path
record["image_id"] = int(line)
record["height"] = height
record["width"] = width
objs = []
ann_path = os.path.join(img_dir,'label_2/%s.txt'%line.replace('\n','').zfill(6))
with open(ann_path) as ann_file:
for ann_line in ann_file:
line_items = ann_line.split(' ')
if(line_items[0]=='Car'):
class_id=2
elif(line_items[0]=='Pedestrian'):
class_id=0
elif(line_items[0]=='Cyclist'):
class_id=1
else:
continue
obj = {'bbox':[np.round(float(line_items[4])),np.round(float(line_items[5])),
np.round(float(line_items[6])),np.round(float(line_items[7]))],"category_id": class_id,"iscrowd": 0,"bbox_mode": BoxMode.XYXY_ABS}
objs.append(obj)
record["annotations"] = objs
dataset_dicts.append(record)
items+=1
return dataset_dicts
from detectron2.data import DatasetCatalog, MetadataCatalog
for d in ["train", "val"]:
DatasetCatalog.register("kitti_" + d, lambda d=d: get_kitti_dicts(data_path))
```
## Training Parameters
```
from detectron2.engine import DefaultTrainer
from detectron2.config import get_cfg
import os
cfg = get_cfg())
cfg.merge_from_file("../configs/COCO-Detection/faster_rcnn_R_50_FPN_3x.yaml")
cfg.DATASETS.TRAIN = ("kitti_train",)
cfg.DATASETS.TEST = ()
cfg.DATALOADER.NUM_WORKERS = 2
#load coco weights
cfg.MODEL.WEIGHTS="https://dl.fbaipublicfiles.com/detectron2/COCO-Detection/faster_rcnn_R_50_FPN_3x/137849458/model_final_280758.pkl"
cfg.SOLVER.IMS_PER_BATCH = 2
cfg.SOLVER.BASE_LR = 0.0025 # pick a good LR
cfg.SOLVER.MAX_ITER = 20000
cfg.MODEL.ROI_HEADS.BATCH_SIZE_PER_IMAGE = 512 #(default: 512)
cfg.OUTPUT_DIR='../models/KITTI/KITTI_DET'
```
### Initialize the trainer and load the dataset
```
os.makedirs(cfg.OUTPUT_DIR, exist_ok=True)
trainer = DefaultTrainer(cfg,False)
```
### Begin Training
```
trainer.resume_or_load(resume=False)
trainer.train()
```
|
github_jupyter
|
# SP LIME
## Regression explainer with boston housing prices dataset
```
from sklearn.datasets import load_boston
import sklearn.ensemble
import sklearn.linear_model
import sklearn.model_selection
import numpy as np
from sklearn.metrics import r2_score
np.random.seed(1)
#load example dataset
boston = load_boston()
#print a description of the variables
print(boston.DESCR)
#train a regressor
rf = sklearn.ensemble.RandomForestRegressor(n_estimators=1000)
train, test, labels_train, labels_test = sklearn.model_selection.train_test_split(boston.data, boston.target, train_size=0.80, test_size=0.20)
rf.fit(train, labels_train);
#train a linear regressor
lr = sklearn.linear_model.LinearRegression()
lr.fit(train,labels_train)
#print the R^2 score of the random forest
print("Random Forest R^2 Score: " +str(round(r2_score(rf.predict(test),labels_test),3)))
print("Linear Regression R^2 Score: " +str(round(r2_score(lr.predict(test),labels_test),3)))
# import lime tools
import lime
import lime.lime_tabular
# generate an "explainer" object
categorical_features = np.argwhere(np.array([len(set(boston.data[:,x])) for x in range(boston.data.shape[1])]) <= 10).flatten()
explainer = lime.lime_tabular.LimeTabularExplainer(train, feature_names=boston.feature_names, class_names=['price'], categorical_features=categorical_features, verbose=False, mode='regression',discretize_continuous=False)
#generate an explanation
i = 13
exp = explainer.explain_instance(test[i], rf.predict, num_features=14)
%matplotlib inline
fig = exp.as_pyplot_figure();
print("Input feature names: ")
print(boston.feature_names)
print('\n')
print("Input feature values: ")
print(test[i])
print('\n')
print("Predicted: ")
print(rf.predict(test)[i])
```
# SP-LIME pick step
### Maximize the 'coverage' function:
$c(V,W,I) = \sum_{j=1}^{d^{\prime}}{\mathbb{1}_{[\exists i \in V : W_{ij}>0]}I_j}$
$W = \text{Explanation Matrix, } n\times d^{\prime}$
$V = \text{Set of chosen explanations}$
$I = \text{Global feature importance vector, } I_j = \sqrt{\sum_i{|W_{ij}|}}$
```
import lime
import warnings
from lime import submodular_pick
sp_obj = submodular_pick.SubmodularPick(explainer, train, rf.predict, sample_size=20, num_features=14, num_exps_desired=5)
[exp.as_pyplot_figure() for exp in sp_obj.sp_explanations];
import pandas as pd
W=pd.DataFrame([dict(this.as_list()) for this in sp_obj.explanations])
W.head()
im=W.hist('NOX',bins=20)
```
## Text explainer using the newsgroups
```
# run the text explainer example notebook, up to single explanation
import sklearn
import numpy as np
import sklearn
import sklearn.ensemble
import sklearn.metrics
# from __future__ import print_function
from sklearn.datasets import fetch_20newsgroups
categories = ['alt.atheism', 'soc.religion.christian']
newsgroups_train = fetch_20newsgroups(subset='train', categories=categories)
newsgroups_test = fetch_20newsgroups(subset='test', categories=categories)
class_names = ['atheism', 'christian']
vectorizer = sklearn.feature_extraction.text.TfidfVectorizer(lowercase=False)
train_vectors = vectorizer.fit_transform(newsgroups_train.data)
test_vectors = vectorizer.transform(newsgroups_test.data)
rf = sklearn.ensemble.RandomForestClassifier(n_estimators=500)
rf.fit(train_vectors, newsgroups_train.target)
pred = rf.predict(test_vectors)
sklearn.metrics.f1_score(newsgroups_test.target, pred, average='binary')
from lime import lime_text
from sklearn.pipeline import make_pipeline
c = make_pipeline(vectorizer, rf)
from lime.lime_text import LimeTextExplainer
explainer = LimeTextExplainer(class_names=class_names)
idx = 83
exp = explainer.explain_instance(newsgroups_test.data[idx], c.predict_proba, num_features=6)
print('Document id: %d' % idx)
print('Probability(christian) =', c.predict_proba([newsgroups_test.data[idx]])[0,1])
print('True class: %s' % class_names[newsgroups_test.target[idx]])
sp_obj = submodular_pick.SubmodularPick(explainer, newsgroups_test.data, c.predict_proba, sample_size=2, num_features=6,num_exps_desired=2)
[exp.as_pyplot_figure(label=exp.available_labels()[0]) for exp in sp_obj.sp_explanations];
from sklearn.datasets import load_iris
iris=load_iris()
from sklearn.model_selection import train_test_split as tts
Xtrain,Xtest,ytrain,ytest=tts(iris.data,iris.target,test_size=.2)
from sklearn.ensemble import RandomForestClassifier
rf=RandomForestClassifier()
rf.fit(Xtrain,ytrain)
rf.score(Xtest,ytest)
explainer = lime.lime_tabular.LimeTabularExplainer(Xtrain,
feature_names=iris.feature_names,
class_names=iris.target_names,
verbose=False,
mode='classification',
discretize_continuous=False)
exp=explainer.explain_instance(Xtrain[i],rf.predict_proba,top_labels=3)
exp.available_labels()
sp_obj = submodular_pick.SubmodularPick(data=Xtrain,explainer=explainer,num_exps_desired=5,predict_fn=rf.predict_proba, sample_size=20, num_features=4, top_labels=3)
import pandas as pd
df=pd.DataFrame({})
for this_label in range(3):
dfl=[]
for i,exp in enumerate(sp_obj.sp_explanations):
l=exp.as_list(label=this_label)
l.append(("exp number",i))
dfl.append(dict(l))
dftest=pd.DataFrame(dfl)
df=df.append(pd.DataFrame(dfl,index=[iris.target_names[this_label] for i in range(len(sp_obj.sp_explanations))]))
df
```
|
github_jupyter
|
# Practice: Basic Statistics I: Averages
For this practice, let's use the Boston dataset.
```
# Import the numpy package so that we can use the method mean to calculate averages
import numpy as np
# Import the load_boston method
from sklearn.datasets import load_boston
# Import pandas, so that we can work with the data frame version of the Boston data
import pandas as pd
# Load the Boston data
boston = load_boston()
# This will provide the characteristics for the Boston dataset
print(boston.DESCR)
# Here, I'm including the prices of Boston's houses, which is boston['target'], as a column with the other
# features in the Boston dataset.
boston_data = np.concatenate((boston['data'], pd.DataFrame(boston['target'])), axis = 1)
# Convert the Boston data to a data frame format, so that it's easier to view and process
boston_df = pd.DataFrame(boston_updated, columns = np.concatenate((boston['feature_names'], 'MEDV'), axis = None))
boston_df
# Determine the mean of each feature
averages_column = np.mean(boston_df, axis = 0)
print(averages_column)
# Determine the mean of each row
averages_row = np.mean(boston_df, axis = 1)
print(averages_row)
```
So we can determine the averages by row, but should we do this? Why or why not?
**Answer:** It's very hard to interpret a these values, because taking an average across different features does not make sense.
Let's put together what you have learned about averages and subsetting to do the next problems.
We will determine the average price for houses along the Charles River and that for houses NOT along the river.
```
# Use the query method to define a subset of boston_df that only include houses are along the river (CHAS = 1).
along_river = boston_df.query('CHAS == 1')
along_river
```
What do you notice about the CHAS column?
**Answer:** It's all 1.0! This means that we successfully subsetting all houses that are along the Charles River. Great work!
```
# Now determine the average price for these houses. 'MEDV' is the column name for the prices.
averages_price_along_river = np.mean(along_river['MEDV'])
averages_price_along_river
```
Now try determining the average for houses NOT along the River.
```
# Determine the average price for houses that are NOT along the Charles River (when CHAS = 0).
not_along_river = boston_df.query('CHAS == 0')
averages_price_not_along_river = np.mean(not_along_river['MEDV'])
averages_price_not_along_river
```
Good work! You're becoming an expert in subsetting and determining averages on subsetted data. This will be integral for your capstone projects and future careers as data scientists!
|
github_jupyter
|
# 第2章 スカラー移流方程式(数値計算法の基礎)
## 2.2 [3] 空間微分項に対する1次精度風上差分の利用
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
```
(1) $\Delta t = 0.05, \Delta x = 0.1$
初期化
```
c = 1
dt = 0.05
dx = 0.1
jmax = 21
nmax = 6
x = np.linspace(0, dx * (jmax - 1), jmax)
q = np.zeros(jmax)
for j in range(jmax):
if (j < jmax / 2):
q[j] = 1
else:
q[j] = 0
```
メインループ(計算+可視化)
```
plt.figure(figsize=(7,7), dpi=100) # グラフのサイズ
plt.rcParams["font.size"] = 22 # グラフの文字サイズ
# 初期分布の可視化
plt.plot(x, q, marker='o', lw=2, label='n=0')
for n in range(1, nmax + 1):
qold = q.copy()
for j in range(1, jmax-1):
q[j] = qold[j] - dt * c * (qold[j] - qold[j - 1]) / dx # 式(2.9)
# 各ステップの可視化
if n % 2 == 0:
plt.plot(x, q, marker='o', lw=2, label=f'n={n}')
# グラフの後処理
plt.grid(color='black', linestyle='dashed', linewidth=0.5)
plt.xlim([0, 2.0])
plt.ylim([0, 1.2])
plt.xlabel('x')
plt.ylabel('q')
plt.legend()
plt.show()
```
(2) $\Delta t = 0.1, \Delta x = 0.1$ (省略. (1)のdt, dxを変更してみよう)
(3) $\Delta t = 0.2, \Delta x = 0.1$ (省略. (1)のdt, dxを変更してみよう)
(4) $\Delta t = 0.025, \Delta x = 0.05$
```
c = 1
dt = 0.025
dx = 0.05
jmax = 20 * 2 + 1
nmax = 6 * 2
x = np.linspace(0, dx * (jmax - 1), jmax)
q = np.zeros(jmax)
for j in range(jmax):
if (j < jmax / 2):
q[j] = 1
else:
q[j] = 0
plt.figure(figsize=(7,7), dpi=100) # グラフのサイズ
plt.rcParams["font.size"] = 22 # グラフの文字サイズ
# 初期分布の可視化
plt.plot(x, q, marker='o', lw=2, label='n=0')
for n in range(1, nmax + 1):
qold = q.copy()
for j in range(1, jmax-1):
q[j] = qold[j] - dt * c * (qold[j] - qold[j - 1]) / dx # 式(2.9)
# 各ステップの可視化
if n % (2 * 2) == 0:
plt.plot(x, q, marker='o', lw=2, label=f'n={n}')
# グラフの後処理
plt.grid(color='black', linestyle='dashed', linewidth=0.5)
plt.xlim([0, 2.0])
plt.ylim([0, 1.2])
plt.xlabel('x')
plt.ylabel('q')
plt.legend()
plt.show()
```
(5) $\Delta t = 0.01, \Delta x = 0.02$
```
c = 1
dt = 0.01
dx = 0.02
jmax = 20 * 5 + 1
nmax = 6 * 5
x = np.linspace(0, dx * (jmax - 1), jmax)
q = np.zeros(jmax)
for j in range(jmax):
if (j < jmax / 2):
q[j] = 1
else:
q[j] = 0
plt.figure(figsize=(7,7), dpi=100) # グラフのサイズ
plt.rcParams["font.size"] = 22 # グラフの文字サイズ
# 初期分布の可視化
plt.plot(x, q, marker='o', lw=2, label='n=0')
for n in range(1, nmax + 1):
qold = q.copy()
for j in range(1, jmax-1):
q[j] = qold[j] - dt * c * (qold[j] - qold[j - 1]) / dx # 式(2.9)
# 各ステップの可視化
if n % (2 * 5) == 0:
plt.plot(x, q, marker='o', lw=2, label=f'n={n}')
# グラフの後処理
plt.grid(color='black', linestyle='dashed', linewidth=0.5)
plt.xlim([0, 2.0])
plt.ylim([0, 1.2])
plt.xlabel('x')
plt.ylabel('q')
plt.legend()
plt.show()
```
|
github_jupyter
|
```
from functools import wraps
import time
def show_args(function):
@wraps(function)
def wrapper(*args, **kwargs):
print('hi from decorator - args:')
print(args)
result = function(*args, **kwargs)
print('hi again from decorator - kwargs:')
print(kwargs)
return result
# return wrapper as a decorated function
return wrapper
@show_args
def get_profile(name, active=True, *sports, **awards):
print('\n\thi from the get_profile function\n')
get_profile('bob', True, 'basketball', 'soccer',
pythonista='special honor of the community', topcoder='2017 code camp')
```
### Using @wraps
```
def timeit(func):
'''Decorator to time a function'''
@wraps(func)
def wrapper(*args, **kwargs):
# before calling the decorated function
print('== starting timer')
start = time.time()
# call the decorated function
func(*args, **kwargs)
# after calling the decorated function
end = time.time()
print(f'== {func.__name__} took {int(end-start)} seconds to complete')
return wrapper
@timeit
def generate_report():
'''Function to generate revenue report'''
time.sleep(2)
print('(actual function) Done, report links ...')
generate_report()
```
### stacking decorators
```
def timeit(func):
'''Decorator to time a function'''
@wraps(func)
def wrapper(*args, **kwargs):
# before calling the decorated function
print('== starting timer')
start = time.time()
# call the decorated function
func(*args, **kwargs)
# after calling the decorated function
end = time.time()
print(f'== {func.__name__} took {int(end-start)} seconds to complete')
return wrapper
def print_args(func):
'''Decorator to print function arguments'''
@wraps(func)
def wrapper(*args, **kwargs):
# before
print()
print('*** args:')
for arg in args:
print(f'- {arg}')
print('**** kwargs:')
for k, v in kwargs.items():
print(f'- {k}: {v}')
print()
# call func
func(*args, **kwargs)
return wrapper
def generate_report(*months, **parameters):
time.sleep(2)
print('(actual function) Done, report links ...')
@timeit
@print_args
def generate_report(*months, **parameters):
time.sleep(2)
print('(actual function) Done, report links ...')
parameters = dict(split_geos=True, include_suborgs=False, tax_rate=33)
generate_report('October', 'November', 'December', **parameters)
```
### Passing arguments to a decorator
Another powerful capability of decs is the ability to pass arguments to them like normal functions, afterall they're functions too. Let's write a simple decorator to return a noun in a format:
```
def noun(i):
def tag(func):
def wrapper(name):
return "My {0} is {1}".format(i, func(name))
return wrapper
return tag
@noun("name")
def say_something(something):
return something
print(say_something('Ant'))
@noun("age")
def say_something(something):
return something
print(say_something(44))
def noun(i):
def tag(func):
def wrapper(name):
return "<{0}>{1}</{0}>".format(i, func(name),i)
return wrapper
return tag
@noun("p")
@noun("strong")
def say_something(something):
return something
# print(say_something('Coding with PyBites!'))
print(say_something('abc'))
def make_html(i):
#@wraps(element)
def tag(func):
def wrapper(*args):
return "<{0}>{1}</{0}>".format(i, func(*args), i)
return wrapper
return tag
@make_html("p")
@make_html("strong")
def get_text(text='I can code with PyBites'):
return text
print(get_text('Some random text here'))
# how do I get default text to print though?
print(get_text)
print(get_text('text'))
print(get_text())
@make_html('p')
@make_html('strong')
def get_text(text='I code with PyBites'):
return text
from functools import wraps
def make_html(element):
pass
from functools import wraps
def exponential_backoff(func):
@wraps(func)
def function_wrapper(*args, **kwargs):
pass
return function_wrapper
@exponential_backoff
def test():
pass
print(test) # <function exponential_backoff.<locals>.function_wrapper at 0x7fcc343a4268>
# uncomment `@wraps(func)` line:
print(test) # <function test at 0x7fcc343a4400>
```
```
@exponential_backoff()
def test():
pass```
equals to:
```
def test():
pass
test = exponential_backoff()(test)```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/yukinaga/object_detection/blob/main/section_3/03_exercise.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# 演習
RetinaNetで、物体の領域を出力する`regression_head`も訓練対象に加えてみましょう。
モデルを構築するコードに、追記を行なってください。
## 各設定
```
import torch
from torch.utils.data import DataLoader
import torchvision
import torchvision.transforms as transforms
from torchvision.utils import draw_bounding_boxes
import numpy as np
import matplotlib.pyplot as plt
import math
# インデックスを物体名に変換
index2name = [
"person",
"bird",
"cat",
"cow",
"dog",
"horse",
"sheep",
"aeroplane",
"bicycle",
"boat",
"bus",
"car",
"motorbike",
"train",
"bottle",
"chair",
"diningtable",
"pottedplant",
"sofa",
"tvmonitor",
]
print(index2name)
# 物体名をインデックスに変換
name2index = {}
for i in range(len(index2name)):
name2index[index2name[i]] = i
print(name2index)
```
## ターゲットを整える関数
```
def arrange_target(target):
objects = target["annotation"]["object"]
box_dics = [obj["bndbox"] for obj in objects]
box_keys = ["xmin", "ymin", "xmax", "ymax"]
# バウンディングボックス
boxes = []
for box_dic in box_dics:
box = [int(box_dic[key]) for key in box_keys]
boxes.append(box)
boxes = torch.tensor(boxes)
# 物体名
labels = [name2index[obj["name"]] for obj in objects] # 物体名はインデックスに変換
labels = torch.tensor(labels)
dic = {"boxes":boxes, "labels":labels}
return dic
```
## データセットの読み込み
```
dataset_train=torchvision.datasets.VOCDetection(root="./VOCDetection/2012",
year="2012",image_set="train",
download=True,
transform=transforms.ToTensor(),
target_transform=transforms.Lambda(arrange_target)
)
dataset_test=torchvision.datasets.VOCDetection(root="./VOCDetection/2012",
year="2012",image_set="val",
download=True,
transform=transforms.ToTensor(),
target_transform=transforms.Lambda(arrange_target)
)
```
## DataLoaderの設定
```
data_loader_train = DataLoader(dataset_train, batch_size=1, shuffle=True)
data_loader_test = DataLoader(dataset_test, batch_size=1, shuffle=True)
```
## ターゲットの表示
```
def show_boxes(image, boxes, names):
drawn_boxes = draw_bounding_boxes(image, boxes, labels=names)
plt.figure(figsize = (16,16))
plt.imshow(np.transpose(drawn_boxes, (1, 2, 0))) # チャンネルを一番後ろに
plt.tick_params(labelbottom=False, labelleft=False, bottom=False, left=False) # ラベルとメモリを非表示に
plt.show()
dataiter = iter(data_loader_train) # イテレータ
image, target = dataiter.next() # バッチを取り出す
print(target)
image = image[0]
image = (image*255).to(torch.uint8) # draw_bounding_boxes関数の入力は0-255
boxes = target["boxes"][0]
labels = target["labels"][0]
names = [index2name[label.item()] for label in labels]
show_boxes(image, boxes, names)
```
# モデルの構築
以下のセルのコードに追記を行い、物体領域の座標を出力する`regression_head`のパラメータも訓練可能にしましょう。
PyTorchの公式ドキュメントに記載されている、RetinaNetのコードを参考にしましょう。
https://pytorch.org/vision/stable/_modules/torchvision/models/detection/retinanet.html#retinanet_resnet50_fpn
```
model = torchvision.models.detection.retinanet_resnet50_fpn(pretrained=True)
num_classes=len(index2name)+1 # 分類数: 背景も含めて分類するため1を加える
num_anchors = model.head.classification_head.num_anchors # アンカーの数
# 分類数を設定
model.head.classification_head.num_classes = num_classes
# 分類結果を出力する層の入れ替え
cls_logits = torch.nn.Conv2d(256, num_anchors*num_classes, kernel_size=3, stride=1, padding=1)
torch.nn.init.normal_(cls_logits.weight, std=0.01) # RetinaNetClassificationHeadクラスより
torch.nn.init.constant_(cls_logits.bias, -math.log((1 - 0.01) / 0.01)) # RetinaNetClassificationHeadクラスより
model.head.classification_head.cls_logits = cls_logits # 層の入れ替え
# 全てのパラメータを更新不可に
for p in model.parameters():
p.requires_grad = False
# classification_headのパラメータを更新可能に
for p in model.head.classification_head.parameters():
p.requires_grad = True
# regression_headのパラメータを更新可能に
# ------- 以下にコードを書く -------
# ------- ここまで -------
model.cuda() # GPU対応
```
## 訓練
```
# 最適化アルゴリズム
params = [p for p in model.parameters() if p.requires_grad]
optimizer = torch.optim.SGD(params, lr=0.001, momentum=0.9)
model.train() # 訓練モード
epochs = 3
for epoch in range(epochs):
for i, (image, target) in enumerate(data_loader_train):
image = image.cuda() # GPU対応
boxes = target["boxes"][0].cuda()
labels = target["labels"][0].cuda()
target = [{"boxes":boxes, "labels":labels}] # ターゲットは辞書を要素に持つリスト
loss_dic = model(image, target)
loss = sum(loss for loss in loss_dic.values()) # 誤差の合計を計算
optimizer.zero_grad()
loss.backward()
optimizer.step()
if i%100 == 0: # 100回ごとに経過を表示
print("epoch:", epoch, "iteration:", i, "loss:", loss.item())
```
## 訓練したモデルの使用
```
dataiter = iter(data_loader_test) # イテレータ
image, target = dataiter.next() # バッチを取り出す
image = image.cuda() # GPU対応
model.eval()
predictions = model(image)
print(predictions)
image = (image[0]*255).to(torch.uint8).cpu() # draw_bounding_boxes関数の入力は0-255
boxes = predictions[0]["boxes"].cpu()
labels = predictions[0]["labels"].cpu().detach().numpy()
labels = np.where(labels>=len(index2name), 0, labels) # ラベルが範囲外の場合は0に
names = [index2name[label.item()] for label in labels]
print(names)
show_boxes(image, boxes, names)
```
## スコアによる選別
```
boxes = []
names = []
for i, box in enumerate(predictions[0]["boxes"]):
score = predictions[0]["scores"][i].cpu().detach().numpy()
if score > 0.5: # スコアが0.5より大きいものを抜き出す
boxes.append(box.cpu().tolist())
label = predictions[0]["labels"][i].item()
if label >= len(index2name): # ラベルが範囲外の場合は0に
label = 0
name = index2name[label]
names.append(name)
boxes = torch.tensor(boxes)
show_boxes(image, boxes, names)
```
# 解答例
以下は、どうしても手がかりがないときのみ参考にしましょう。
```
model = torchvision.models.detection.retinanet_resnet50_fpn(pretrained=True)
num_classes=len(index2name)+1 # 分類数: 背景も含めて分類するため1を加える
num_anchors = model.head.classification_head.num_anchors # アンカーの数
# 分類数を設定
model.head.classification_head.num_classes = num_classes
# 分類結果を出力する層の入れ替え
cls_logits = torch.nn.Conv2d(256, num_anchors*num_classes, kernel_size=3, stride=1, padding=1)
torch.nn.init.normal_(cls_logits.weight, std=0.01) # RetinaNetClassificationHeadクラスより
torch.nn.init.constant_(cls_logits.bias, -math.log((1 - 0.01) / 0.01)) # RetinaNetClassificationHeadクラスより
model.head.classification_head.cls_logits = cls_logits # 層の入れ替え
# 全てのパラメータを更新不可に
for p in model.parameters():
p.requires_grad = False
# classification_headのパラメータを更新可能に
for p in model.head.classification_head.parameters():
p.requires_grad = True
# regression_headのパラメータを更新可能に
# ------- 以下にコードを書く -------
for p in model.head.regression_head.parameters():
p.requires_grad = True
# ------- ここまで -------
model.cuda() # GPU対応
```
|
github_jupyter
|
### Лекция 7. Исключения
https://en.cppreference.com/w/cpp/language/exceptions
https://en.cppreference.com/w/cpp/error
https://apprize.info/c/professional/13.html
<br />
##### Зачем нужны исключения
Для обработки исключительных ситуаций.
Как вариант - обработка ошибок.
<br />
###### Как пользоваться исключениями
Нужно определиться с двумя точками в программе:
1. Момент детекции ошибки
2. Момент обработки ошибки
В момент возникновения ошибки бросаем (`throw`) любой объект, в который вкладываем описание исключительной ситуации:
```c++
double my_sqrt(double x)
{
if (x < 0)
throw std::invalid_argument("sqrt of negative doubles can not be represented in terms of real numbers");
...
}
```
В вызывающем коде оборачиваем бросающий блок в `try`-`catch`:
*(обратить внимание как брошенное исключение будет обрабатываться)*
```c++
void run_dialogue()
{
std::cout << "enter x: ";
double x;
std::cin >> x;
std::cout << "sqrt(x) = " << my_sqrt(x) << std::endl;
}
int main()
{
try
{
run_dialogue();
}
catch(const std::invalid_argument& e)
{
std::cout << "invalid argument: " << e.what() << std::endl;
return 1;
}
catch(const std::exception& e)
{
std::cout << "common exception: " << e.what() << std::endl;
return 1;
}
return 0;
}
```
**Замечание**: бросать можно объекты любого типа, но рекомендуется бросать именно специальные объекты-исключения для читабельности.
**Вопрос:** `std::invalid_argument` - наследник `std::exception`. Что будет, если поменять блоки-обработчики местами?
<br />
##### Стратегии обработки ошибок
Есть как минимум три стратегии обработки ошибок:
* через коды возврата
* через исключения
* сделать вид, что ошибок не существует
Типичная реализация функции, когда ошибки обрабатываются через коды возврата:
```c++
int get_youngest_student(std::string& student_name)
{
int err_code = 0;
// вытащить всех студентов из БД (пример: проброс ошибки)
std::vector<Student> students;
err_code = fetch_students(students);
if (err_code != ErrCode::OK)
return err_code;
// найти самого молодого (пример: ручное управление)
auto youngest_it = std::min_element(students.begin(),
students.end(),
[](const auto& lhs, const auto& rhs){
return lhs.age < rhs.age;
});
if (youngest_it == students.end())
return ErrCode::NoStudents;
// вытащить из базы его имя (пример: частичная обработка)
err_code = fetch_student_name(youngest_it->id, student_name);
if (err_code != ErrCode::OK)
if (err_code == ErrCode::NoObject)
return ErrCode::CorruptedDatabase;
else
return err_code;
return ErrCode::OK;
}
```
Типичная реализация в случае использования исключений:
```c++
std::string get_youngest_student()
{
// вытащить всех студентов из БД (пример: проброс ошибки)
std::vector<Student> students = fetch_students();
// найти самого молодого (пример: ручное управление)
auto youngest_it = std::min_element(students.begin(),
students.end(),
[](const auto& lhs, const auto& rhs){
return lhs.age < rhs.age;
});
if (youngest_it == students.end())
throw std::runtime_error("students set is empty");
// вытащить из базы его имя (пример: частичная обработка)
try
{
return fetch_student_name(youngest_it->id);
}
catch(const MyDBExceptions::NoObjectException& exception)
{
throw MyDBExceptions::CorruptedDatabase();
}
}
```
Типичная реализация в случае игнорирования ошибок
```c++
std::string get_youngest_student()
{
// вытащить всех студентов из БД (пример: проброс ошибки)
std::vector<Student> students = fetch_students(); // не кидает исключений,
// никак не узнать, что проблемы с доступом к базе
// найти самого молодого (пример: ручное управление)
auto youngest_it = std::min_element(students.begin(),
students.end(),
[](const auto& lhs, const auto& rhs){
return lhs.age < rhs.age;
});
if (youngest_it == students.end())
return "UNK"; // не отделить ситуацию, когда нет студентов в базе вообще
// от ситуации, когда в базе имя UNK у студента
// вытащить из базы его имя (пример: частичная обработка)
return fetch_student_name(youngest_it->id); // # не кидает исключений
// не отделить ситуацию, когда в таблице имён пропущен студент
// от ситуации, когда студент есть с именем UNK
}
```
**На практике: разумный баланс между детализацией ошибок и сложностью программы.**
<br />
##### как бросать и как ловить исключения, размотка стека
В момент исключительной ситуации:
```c++
double my_sqrt(double x)
{
if (x < 0)
throw std::invalid_argument("sqrt can not be calculated for negatives in terms of double");
...;
}
```
Далее начинается размотка стека и поиск соответствующего catch-блока:
(объяснить на примере, показать 3 ошибки в коде кроме "oooops")
<details>
<summary>Подсказка</summary>
<p>
1. утечка `logger`
2. `front` без проверок
3. порядок обработчиков
</p>
</details>
```c++
double get_radius_of_first_polyline_point()
{
auto* logger = new Logger();
std::vector<Point> polyline = make_polyline();
Point p = polyline.front();
logger->log("point is " + std::to_string(p.x) + " " + std::to_string(p.y));
double r = my_sqrt(p.x * p.x - p.y * p.y); // ooops
logger->log("front radius is " + std::to_string(r));
delete logger;
return r;
}
void func()
{
try
{
std::cout << get_radius_of_first_polyline_point() << std::endl;
}
catch (const std::exception& e)
{
std::cout << "unknown exception: " << e.what() << std::endl;
}
catch (const std::invalid_argument& e)
{
std::cout << "aren't we trying to calculate sqrt of negatve? " << e.what() << std::endl;
}
catch (...) // you should never do that
{
std::cout << "what?" << std::endl;
}
}
```
__Вопрос__:
* какие операции в коде могут кинуть исключение?
* Как в `catch (const std::invalid_argument& e)` отличить подсчёт корня из отрицательного числа от других `std::invalid_argument`?
* Что будет в таком варианте?
```c++
catch (const std::invalid_argument e)
{
std::cout << "aren't we trying to calculate sqrt of negatve? " << e.what() << std::endl;
}
```
* а в таком?
```c++
catch (std::invalid_argument& e)
{
std::cout << "aren't we trying to calculate sqrt of negatve? " << e.what() << std::endl;
}
```
<br />
##### noexcept
Если функция не бросает исключений, желательно пометить её `noexcept`.
Вызов такой функции будет чуть дешевле и объём бинарного файла чуть меньше (не нужно генерировать кода поддержки исключений).
Что будет если `noexcept` - функция попытается бросить исключение?
```c++
int get_sum(const std::vector<int>& v) noexcept
{
return std::reduce(v.begin(), v.end());
}
int get_min(const std::vector<int>& v) noexcept
{
if (v.empty())
throw std::invalid_argument("can not find minimum in empty sequence");
return *std::min_element(v.begin(), v.end());
}
```
<details>
<summary>Ответ</summary>
Вызов std::terminate. Дальше продолжать выполнение программы нельзя, т.к. внешнему коду пообещали, что функция исключений не бросает.
При этом, если компилятор не может доказать, что тело функции не бросает исключений, он генерирует try-catch блок на всю функцию с std::terminate в catch.
</details>
<br />
##### стандартные и собственные классы исключений
Для стандартных исключений в С++ выделен базовый класс `std::exception`:
```c++
class exception
{
public:
exception() noexcept;
virtual ~exception() noexcept;
exception(const exception&) noexcept;
exception& operator=(const exception&) noexcept;
virtual const char* what() const noexcept;
};
```
Остальные стандартные исключения наследуются от него:

Как бросать стандартные исключения:
```c++
// проверить на ноль перед делением
if (den == 0)
throw std::runtime_error("unexpected integer division by zero");
// проверить аргументы на корректность
bool binsearch(const int* a, const int n, const int value)
{
if (n < 0)
throw std::invalid_argument("unexpexted negative array size");
...;
}
```
<br />
Зачем свои классы исключений?
* бОльшая детализация ошибки
* возможность добавить информацию к классу исключения
Рекомендации:
* свои классы наследовать от `std::exception`
* если возможно - организовать свои исключения в иерархию (чтобы была возможность ловить общую ошибку библиотеки или более детальные)
* если возможно - предоставить информацию для анализа и восстановления
Рассмотрим пример - вы пишете свой читатель-писатель json-ов (зачем? их миллионы уже!)
```c++
namespace myjson
{
// общий наследник исключений вашей библиотеки парсинга,
// чтобы пользователи могли просто отловить события
// "в этой билиотеке что-то пошло не так"
class MyJsonException : public std::exception {};
// исключение для случая, когда при запросе к полю у объекта
// поле отсутствовало
//
// можно дать обработчику исключения больше информации
// для более разумной обработки ситуации или хотя бы
// более подробного логирования проблемы
class FieldNotFound : public MyJsonException
{
public:
FieldNotFound(std::string parent_name, std::string field_name);
const std::string& parent_name() const noexcept;
const std::string& field_name() const noexcept;
private:
std::string parent_name_;
std::string field_name_;
};
// исключение для ошибок при парсинге json-строки
//
// можно дать больше информации, на каком символе
// обломился парсинг строки
class ParsingException : public MyJsonException
{
public:
ParsingException(int symbol_ix);
int symbol_ix() const noexcept;
private:
int symbol_ix_;
};
// исключение для ошибок при парсинге int-а - сужение |ParsingException|
class IntegerOverflowOnParsing : public ParsingException {};
// и т.д.
} // namespace myjson
```
Опять же, на практике выбирают баланс между детализацией и сложностью.
Чем глубже детализация, тем сложнее программа, но с определённого уровня пользы от большей детализации мало.
<br />
##### исключения в конструкторах и деструкторах
Почему исключения полезны в конструкторах?
Потому что у конструктора нет другого способа сообщить об ошибке!
```c++
std::vector<int> v = {1, 2, 3, 4, 5};
// нет другого (нормального) способа сообщить вызываемому коду,
// что памяти не хватило и вектор не создан, только бросить
// исключение
```
**Пример**: на порядок вызова конструкторов и деструкторов
```c++
class M { ...; };
class B { ...; };
class D : public B {
public:
D() {
throw std::runtime_error("error");
}
private:
M m_;
};
```
Какие конструкторы и деструкторы будут вызваны?
```c++
try {
D d;
}
catch (const std::exception& e) {
}
```
А если так?
```c++
class M { ...; };
class B { ...; };
class D : public B {
public:
D() : D(0) {
throw std::runtime_error("error");
}
D(int x) {}
private:
M m_;
};
```
Что с исключениями из деструкторов?
```c++
class D
{
public:
D() {}
~D() {
throw std::runtime_error("can not free resource");
}
};
```
* Бросать исключения из деструкторов - "плохо"
* По умолчанию деструктор - `noexcept` (если нет специфических проблем с базовыми классами и членами)
* Если при размотке стека из деструктора объекта бросается исключение, программа завершается с `std::terminate` по стандарту: https://en.cppreference.com/w/cpp/language/destructor (раздел Exceptions) "you can not fail to fail"
__Упражение__: чтобы понять, почему деструктору нельзя кидать исключения, попробуйте на досуге представить, как корректно реализовать `resize` у `std::vector<T>`, если `~T` иногда кидает исключение
<br />
##### гарантии при работе с исключениями
* `nothrow` - функция не бросает исключений
```c++
int swap(int& x, int& y)
```
* `strong` - функция отрабатывает как транзакция: если из функции вылетает исключение, состояние программы откатывается на момент как до вызова функции.
```c++
std::vector::push_back
```
* `basic` - если из функции вылетает исключение, программа ещё корректна (инварианты сохранены). Может потребоваться очистка.
```c++
void write_to_csv(const char* filename)
{
std::ofsteam ofs(filename);
ofs << "id,name,age" << std::endl;
std::vector<std::string> names = ...; // bad_alloc
ofs << ...;
}
```
* `no exception guarantee` - если из функции вылетает исключение, молитесь
```
любой production код (история про обработку ошибок в файловых системах)
```
* `exception-neutral` - только для шаблонных компонент - пропускает сквозь себя все исключения, которые кидают шаблонные параметры
```c++
std::min_element(v.begin(), v.end())
```
<br />
##### стоимость исключений
Зависит от реализации.
"В интернете пишут, что" исключения в основных компиляторах реализованы по принципу (бесплатно, пока не вылетел exception, дорого, если вылетел). При этом при выбросе исключений формируются специальные exception frame-ы, осуществляется поиск handler-ов и cleanup-процедур по заранее сгенерированным компилятором таблицам.
Подробнее:
* https://stackoverflow.com/questions/13835817/are-exceptions-in-c-really-slow
* https://mortoray.com/2013/09/12/the-true-cost-of-zero-cost-exceptions/
При этом код обслуживания исключений тоже надо сгенерировать. Статья как в microsoft провели исследование сколько занимает код обслуживания механизма исключений (спойлер: для конкретной билиотеки в районе 26% - зависит от кол-ва исключений, кол-ва бросающих исключения функций, кол-ва вызовов throw, сложности объектов на стеке и т.д.) и как его сократили где-то в 2 раза:
https://devblogs.microsoft.com/cppblog/making-cpp-exception-handling-smaller-x64/
<br />
##### noexcept-move-операции
Пояснить на классическом примере `std::vector::push_back` каким образом объявление move-операций `noexcept` позволяет ускорить программу:

Аналогично допустимы оптимизации при работе с std::any для nothrow move constructible типов<br />
https://en.cppreference.com/w/cpp/utility/any
<br />
##### правила хорошего тона при реализации исключений
* Деструкторы не должны бросать исключений. Можете помечать их `noexcept` или знать, что компилятор во многих случаях автоматически добавляет `noexcept` к деструкторам.
* Реализовывать и помечать move-операции как `noexcept`
* Реализовывать и помечать default constructor как `noexcept` (cppcoreguildelines для скорости)
* `noexcept` everything you can!
* Цитата с cppcoreguidelines: `If you know that your application code cannot respond to an allocation failure, it may be appropriate to add noexcept even on functions that allocate.` Объяснить её смысл про восстановление после ошибки и почему "нелогичность" здесь полезна.
* Пользовательские классы исключений наследовать от `std::exception` или его подклассов
* Ловить исключений по const-ссылкам
* `throw;` вместо `throw e;` из catch-блока, когда нужен rethrow
* Исключения являются частью контракта (или спецификации) функции! Желательно их протестировать.
* Использовать исключения для исключительных ситуаций, а не для естественного потока выполнения.
* Плохой код:
* исключение чтобы прервать цикл
* исключение чтобы сделать особое возвращаемое значение из функции
* Приемлемо:
* исключение чтобы сообщить об ошибке
* исключение чтобы сообщить о нарушении контракта (объяснить, почему это не лучший вариант использования исключений)
* Исключения служат для того чтобы восстановиться после ошибки и продолжить работу:
* пропустить некритичные действия. Пример:
* отобразить телефон организации в информационном листе)
* fallback с восстановлением или откатом:
* memory-consuming алгоритмы
* message box-ы об ошибке (не удалось открыть документ в msword)
* красиво умереть на критических ошибках:
* memory allocation on game start
* некорректная команда в текстовом интерпретаторе
* Глобальный try-catch (плюсы: программа завершается без падений, деструкторы объектов на стеке будут позваны (если нет catch-блока, вызов процедура размотки стека может не выполняться - лазейка стандарта). минус: не будет создан crashdump для анализа проблемы):
```c++
int main() {
try {
...
} catch(const std::exception& e) {
std::cout << "Until your last breath!\n";
std::cout << "ERROR: " << e.what() << std::endl;
return 1;
}
return 0;
}
```
<br />
**Полезные материалы**:
* [C++ Russia: Роман Русяев — Исключения C++ через призму компиляторных оптимизаций.](https://www.youtube.com/watch?v=ItemByR4PRg)
* [CppCon 2018: James McNellis “Unwinding the Stack: Exploring How C++ Exceptions Work on Windows”](https://youtu.be/COEv2kq_Ht8)
|
github_jupyter
|
```
import numpy as np
from sklearn.datasets import load_iris
# Loading the dataset
iris = load_iris()
X_raw = iris['data']
y_raw = iris['target']
# Isolate our examples for our labeled dataset.
n_labeled_examples = X_raw.shape[0]
training_indices = np.random.randint(low=0, high=len(X_raw)+1, size=3)
# Defining the training data
X_training = X_raw[training_indices]
y_training = y_raw[training_indices]
# Isolate the non-training examples we'll be querying.
X_pool = np.delete(X_raw, training_indices, axis=0)
y_pool = np.delete(y_raw, training_indices, axis=0)
from sklearn.decomposition import PCA
# Define our PCA transformer and fit it onto our raw dataset.
pca = PCA(n_components=2)
pca.fit(X=X_raw)
from modAL.models import ActiveLearner
from modAL.batch import uncertainty_batch_sampling
from sklearn.neighbors import KNeighborsClassifier
# Specify our core estimator.
knn = KNeighborsClassifier(n_neighbors=3)
learner = ActiveLearner(
estimator=knn,
query_strategy=uncertainty_batch_sampling,
X_training=X_training, y_training=y_training
)
from modAL.batch import ranked_batch
from modAL.uncertainty import classifier_uncertainty
from sklearn.metrics.pairwise import pairwise_distances
uncertainty = classifier_uncertainty(learner, X_pool)
distance_scores = pairwise_distances(X_pool, X_training, metric='euclidean').min(axis=1)
similarity_scores = 1 / (1 + distance_scores)
alpha = len(X_training)/len(X_raw)
scores = alpha * (1 - similarity_scores) + (1 - alpha) * uncertainty
import matplotlib.pyplot as plt
%matplotlib inline
transformed_pool = pca.transform(X_pool)
transformed_training = pca.transform(X_training)
with plt.style.context('seaborn-white'):
plt.figure(figsize=(8, 8))
plt.scatter(transformed_pool[:, 0], transformed_pool[:, 1], c=scores, cmap='viridis')
plt.colorbar()
plt.scatter(transformed_training[:, 0], transformed_training[:, 1], c='r', s=200, label='labeled')
plt.title('Scores of the first instance')
plt.legend()
query_idx, query_instances = learner.query(X_pool, n_instances=5)
transformed_batch = pca.transform(query_instances)
with plt.style.context('seaborn-white'):
plt.figure(figsize=(8, 8))
plt.scatter(transformed_pool[:, 0], transformed_pool[:, 1], c='0.8', label='unlabeled')
plt.scatter(transformed_training[:, 0], transformed_training[:, 1], c='r', s=100, label='labeled')
plt.scatter(transformed_batch[:, 0], transformed_batch[:, 1], c='k', s=100, label='queried')
plt.title('The instances selected for labeling')
plt.legend()
```
|
github_jupyter
|
```
import pandas as pd
import numpy as np
import os
from sklearn.metrics import mean_squared_error, mean_absolute_error
import matplotlib.pyplot as plt
import pickle
import random
import train
from model import NNModelEx
pd.set_option('display.max_columns', 999)
# For this model, the data preprocessing part is already completed with the exception of scaling.
# so we just need to scale here.
def get_ref_X_y(df):
X_cols = [c for c in df.columns if c.startswith('tc2x_')]
y_cols = [c for c in df.columns if c.startswith('y')]
return (df[X_cols], df[y_cols])
raw_data = {} # loads raw data and stores as a dict cache
def dataset_key(dataset='', validation=False):
return dataset+('test' if validation else 'train')
def load_data(raw, dataset='', validation=False):
'''
Return dataframe matching data set and validation. Dictionary input will be updated.
Parameters
----------
raw : dict
dictionary which caches the dataframes and will be updated accordingly
dataset : str
which dataset to use? valid input includes: empty str for full set, sample_, and secret_
validation : bool
load validation set? if true then use _test, otherwise use _train. Note secret_ doesn't have _train
'''
key = dataset+('test' if validation else 'train')
if key not in raw:
print(f"Loading data to cache for: {key}")
raw[key] = pd.read_pickle(f'./data/{key}.pkl')
return raw[key]
configurations = {
'dataset' : 't3/', # '', 'sample_', 'secret_'
'model_identifier' : "tc2_4",
'model_path' : f"./models",
'model': NNModelEx,
'device' : 'cpu',
'random_seed' : 0,
'lr' : 3e-3,
'weight_decay' : 0.3, #Adam
'max_epochs' : 50000,
'do_validate' : True,
'model_definition' : [
('l', (600,)), ('r', (True,)),
('l', (600,)), ('r', (True,)),
('l', (600,)), ('r', (True,)),
('l', (600,)), ('r', (True,)),
('l', (600,)), ('r', (True,)),
('l', (600,)), ('r', (True,)),
('l', (600,)), ('r', (True,)),
('l', (1,)), ('r', (True,)),
],
'train_params' : {
'batch_size': 10000,
'shuffle': True,
'num_workers': 3,
'pin_memory': True,
},
'test_params' : {
'batch_size': 200000,
'num_workers': 1,
'pin_memory': True,
},
}
%%time
train_df = load_data(raw_data,dataset=configurations['dataset'],validation=False)
test_df = load_data(raw_data,dataset=configurations['dataset'],validation=True)
X_train, y_train = get_ref_X_y(train_df)
X_test, y_test = get_ref_X_y(test_df)
import torch
model, _, _, mean_losses, _ = train.load_model_with_config(configurations)
tl, vl = zip(*mean_losses)
fig,ax = plt.subplots()
ax.plot(tl, label="Training Loss")
ax.plot(vl, label="Validation Loss")
fig.legend()
plt.show()
mean_losses
trained_model = model
y_train_pred = train.predict(trained_model, X_train, y_train, device="cpu") # get predictions for each train
y_train_pred_df = pd.DataFrame(y_train_pred, columns=y_train.columns) # put results into a dataframe
print(f' Train set MAE (L1) loss: {mean_absolute_error(y_train, y_train_pred_df)}')
print(f' Train set MSE (L2) loss: {mean_squared_error(y_train, y_train_pred_df)}')
# random.seed(0)
# sample = random.sample(list(y_train_pred_df.index), 10)
print("Train")
train_res = pd.concat([y_train, y_train_pred_df], axis=1)
train_res.columns = ['Ground Truth', 'Pred']
train_res['binarize'] = (train_res['Pred'] > 0.5).astype(float)
train_res['correct'] = train_res['Ground Truth'] == train_res['binarize']
display(train_res)
train_res[train_res['Ground Truth']==1]['correct'].value_counts()
train_res[train_res['Ground Truth']==0]['correct'].value_counts()
from sklearn.metrics import confusion_matrix
confusion_matrix(y_train, (y_train_pred > 0.5))
```
|
github_jupyter
|
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
credit_df = pd.read_csv('German Credit Data.csv')
credit_df
credit_df.info()
X_features = list(credit_df.columns)
X_features.remove('status')
X_features
encoded_df = pd.get_dummies(credit_df[X_features],drop_first = True)
encoded_df
import statsmodels.api as sm
Y = credit_df.status
X = sm.add_constant(encoded_df)
from sklearn.model_selection import train_test_split
X_train,X_test,y_train,y_test = train_test_split(X,Y,test_size = 0.3,random_state = 42)
logit = sm.Logit(y_train,X_train)
logit_model =logit.fit()
logit_model.summary2()
```
## Models Diagnostics using CHI SQUARE TEST
The model summary suggests that as per the Wald's test, only 8 features are statistically significant at a significant value of alpha= 0.05 as p-values are less than 0.05.
p-value for liklelihood ratio test (almost 0.00) indicates that the overall model is statiscally significant.
```
def get_significant_vars(lm):
var_p_values_df = pd.DataFrame(lm.pvalues)
var_p_values_df['vars'] = var_p_values_df.index
var_p_values_df.columns = ['pvals','vars']
return list(var_p_values_df[var_p_values_df['pvals']<=0.05]['vars'])
significant_vars = get_significant_vars(logit_model)
significant_vars
final_logit = sm.Logit(y_train,sm.add_constant(X_train[significant_vars])).fit()
final_logit.summary2()
```
The negative sign in Coefficient value indicates that as the values of this variable increases, the probability of being a bad credit decreases.
A positive value indicates that the probability of a bad credit increases as the corresponding value of the variable increases.
```
y_pred_df = pd.DataFrame({'actual':y_test,'predicted_prob':final_logit.predict(sm.add_constant(X_test[significant_vars]))})
y_pred_df
```
To understand how many observations the model has classified correctly and how many not, a cut-off probability needs to be assumed.
let the assumption be 0.5 for now.
```
y_pred_df['predicted'] = y_pred_df['predicted_prob'].map(lambda x: 1 if x>0.5 else 0)
y_pred_df.sample(5)
```
## Creating a Confusion Matrix
```
from sklearn import metrics
def draw_cm(actual,predicted):
cm = metrics.confusion_matrix(actual,predicted,[1,0])
sns.heatmap(cm,annot=True,fmt= '.2f',
xticklabels=['Bad Credit','Good Credit'],
yticklabels=['Bad Credit','Good Credit'])
plt.ylabel('True Label')
plt.xlabel('Predicted Label')
plt.show()
draw_cm(y_pred_df['actual'],y_pred_df['predicted'])
```
# Building Decision Tree using Gini Criterion
```
Y = credit_df['status']
X = encoded_df
from sklearn.model_selection import train_test_split
X_train,X_test,y_train,y_test = train_test_split(X,Y,test_size= 0.3,random_state=42)
from sklearn.tree import DecisionTreeClassifier
clf_tree = DecisionTreeClassifier(criterion= 'gini',max_depth = 3)
clf_tree.fit(X_train,y_train)
tree_predict = clf_tree.predict(X_test)
metrics.roc_auc_score(y_test,tree_predict)
```
# Displaying the Tree
```
Y = credit_df.status
X =encoded_df
from sklearn.tree import DecisionTreeClassifier,plot_tree
from sklearn.model_selection import train_test_split
X_train,X_test,y_train,y_test = train_test_split(X,Y,test_size = 0.3,random_state = 42)
```
### Using Gini Criterion
```
clf_tree = DecisionTreeClassifier(criterion='gini',max_depth=3)
clf_tree.fit(X_train,y_train)
tree_predict = clf_tree.predict(X_test)
metrics.roc_auc_score(tree_predict,y_test)
## Displaying the Tree
plt.figure(figsize = (20,10))
plot_tree(clf_tree,feature_names=X.columns)
plt.show()
```
### Using Entropy criterion
```
clf_tree_ent = DecisionTreeClassifier(criterion='entropy',max_depth=3)
clf_tree_ent.fit(X_train,y_train)
tree_predict = clf_tree_ent.predict(X_test)
metrics.roc_auc_score(tree_predict,y_test)
## Displaying the Tree
plt.figure(figsize = (20,10))
plot_tree(clf_tree_ent,feature_names=X.columns)
plt.show()
from sklearn.model_selection import GridSearchCV
tuned_params = [{'criterion':['gini','entropy'],
'max_depth':range(2,10)}]
clf_ = DecisionTreeClassifier()
clf = GridSearchCV(clf_tree,
tuned_params,cv =10,
scoring='roc_auc')
clf.fit(X_train,y_train)
score = clf.best_score_*100
print("Best Score is:",score)
best_params = clf.best_params_
print("Best Params is:",best_params)
```
The tree with gini and max_depth = 4 is the best model. Finally, we can build a model with these params and measure the accuracy of the test.
|
github_jupyter
|
# Comparing Training and Test and Parking and Sensor Datasets
```
import sys
import pandas as pd
import numpy as np
import datetime as dt
import time
import matplotlib.pyplot as plt
sys.path.append('../')
from common import reorder_street_block, process_sensor_dataframe, get_train, \
feat_eng, add_tt_gps, get_parking, get_test, plot_dataset_overlay, \
parking_join_addr, tt_join_nh
%matplotlib inline
```
### Import City data
```
train = get_train()
train = feat_eng(train)
```
### Import Scraped City Data
```
city_stats = pd.read_csv('../ref_data/nh_city_stats.txt',delimiter='|')
city_stats.head()
```
### Import Parking data with Addresses
```
clean_park = parking_join_addr(True)
clean_park.min_join_dist.value_counts()
clean_park.head(25)
plot_dataset_overlay()
```
### Prototyping Below (joining and Mapping Code)
```
from multiprocessing import cpu_count, Pool
# simple example of parallelizing filling nulls
def parallelize(data, func):
cores = cpu_count()
data_split = np.array_split(data, cores)
pool = Pool(cores)
data = np.concatenate(pool.map(func, data_split), axis=0)
pool.close()
pool.join()
return data
def closest_point(park_dist):
output = np.zeros((park_dist.shape[0], 3), dtype=int)
for i, point in enumerate(park_dist):
x,y, id_ = point
dist = np.sqrt(np.power(gpspts.iloc[:,0]-x,2) + np.power(gpspts.iloc[:,1]-y,2))
output[i,:] = (id_,np.argmin(dist),np.min(dist))
return output
def parking_join_addr(force=False):
save_path = DATA_PATH + 'P_parking_clean.feather'
if os.path.isfile(save_path) and force==False:
print('loading cached copy')
join_parking_df = pd.read_feather(save_path)
return join_parking_df
else:
parking_df = get_parking()
park_dist = parking_df.groupby(['lat','lon'])[['datetime']].count().reset_index()[['lat','lon']]
park_dist['id'] =park_dist.index
gps2addr = pd.read_csv('../ref_data/clean_parking_gps2addr.txt', delimiter='|')
keep_cols = ['full_addr','jlat','jlon','nhood','road','zipcode']
gpspts = gps2addr[['lat','lon']]
lkup = parallelize(park_dist.values, closest_point)
lkup_df = pd.DataFrame(lkup)
lkup_df.columns = ['parking_idx','addr_idx','min_join_dist']
tmp = park_dist.merge(lkup_df, how='left', left_index=True, right_on='parking_idx')
tmp = tmp.merge(gps2addr[keep_cols], how='left', left_on='addr_idx', right_index=True)
join_parking_df = parking_df.merge(tmp, how='left', on=['lat','lon'])
join_parking_df.to_feather(save_path)
return join_parking_df
print("loading parking data 1.7M")
parking_df = get_parking()
park_dist = parking_df.groupby(['lat','lon'])[['datetime']].count().reset_index()[['lat','lon']]
park_dist['id'] =park_dist.index
print("loading address data 30K")
gps2addr = pd.read_csv('../ref_data/clean_parking_gps2addr.txt', delimiter='|')
keep_cols = ['full_addr','jlat','jlon','nhood','road','zipcode']
gpspts = gps2addr[['lat','lon']]
x,y,id_= park_dist.iloc[0,:]
dist = np.sqrt(np.power(gpspts.iloc[:,0]-x,2) + np.power(gpspts.iloc[:,1]-y,2))
np.log(dist)
dist = np.sqrt(np.power(gpspts.iloc[:,0]-x,2) + np.power(gpspts.iloc[:,1]-y,2))
join_parking_df
lkup_df = pd.DataFrame(lkup)
lkup_df.columns = ['parking_idx','addr_idx']
tmp = park_dist.merge(lkup_df, how='left', left_index=True, right_on='parking_idx')
keep_cols = ['full_addr','jlat','jlon','nhood','road','zipcode']
tmp = tmp.merge(gps2addr[keep_cols], how='left', left_on='addr_idx', right_index=True)
tmp = parking_df.merge(tmp, how='left', on=['lat','lon'])
tmp.isna().sum()
gpspts = gps2addr[['lat','lon']]
park_dist['id'] =park_dist.index
park_dist.head()
park_dist.shape, gpspts.shape
np.concatenate()
from multiprocessing import cpu_count, Pool
# simple example of parallelizing filling nulls
def parallelize(data, func):
cores = cpu_count()
data_split = np.array_split(data, cores)
pool = Pool(cores)
data = np.concatenate(pool.map(func, data_split), axis=0)
pool.close()
pool.join()
return data
def closest_point(park_dist):
output = np.zeros((park_dist.shape[0], 2), dtype=int)
for i, point in enumerate(park_dist):
x,y, id_ = point
dist = np.sqrt(np.power(gpspts.iloc[:,0]-x,2) + np.power(gpspts.iloc[:,1]-y,2))
output[i,:] = (id_,np.argmin(dist))
return output
lkup
122.465370178
gps2addr[(gps2addr['lon'] <= -122.46537) & (gps2addr['lon'] > -122.4654) ].sort_values('lon')
```
|
github_jupyter
|
This notebook is part of the $\omega radlib$ documentation: https://docs.wradlib.org.
Copyright (c) $\omega radlib$ developers.
Distributed under the MIT License. See LICENSE.txt for more info.
# Export a dataset in GIS-compatible format
In this notebook, we demonstrate how to export a gridded dataset in GeoTIFF and ESRI ASCII format. This will be exemplified using RADOLAN data from the German Weather Service.
```
import wradlib as wrl
import numpy as np
import warnings
warnings.filterwarnings('ignore')
```
### Step 1: Read the original data
```
# We will export this RADOLAN dataset to a GIS compatible format
wdir = wrl.util.get_wradlib_data_path() + '/radolan/grid/'
filename = 'radolan/misc/raa01-sf_10000-1408102050-dwd---bin.gz'
filename = wrl.util.get_wradlib_data_file(filename)
data_raw, meta = wrl.io.read_radolan_composite(filename)
```
### Step 2: Get the projected coordinates of the RADOLAN grid
```
# This is the RADOLAN projection
proj_osr = wrl.georef.create_osr("dwd-radolan")
# Get projected RADOLAN coordinates for corner definition
xy_raw = wrl.georef.get_radolan_grid(900, 900)
```
### Step 3: Check Origin and Row/Column Order
We know, that `wrl.read_radolan_composite` returns a 2D-array (rows, cols) with the origin in the lower left corner. Same applies to `wrl.georef.get_radolan_grid`. For the next step, we need to flip the data and the coords up-down. The coordinate corner points also need to be adjusted from lower left corner to upper right corner.
```
data, xy = wrl.georef.set_raster_origin(data_raw, xy_raw, 'upper')
```
### Step 4a: Export as GeoTIFF
For RADOLAN grids, this projection will probably not be recognized by
ESRI ArcGIS.
```
# create 3 bands
data = np.stack((data, data+100, data+1000))
ds = wrl.georef.create_raster_dataset(data, xy, projection=proj_osr)
wrl.io.write_raster_dataset(wdir + "geotiff.tif", ds, 'GTiff')
```
### Step 4b: Export as ESRI ASCII file (aka Arc/Info ASCII Grid)
```
# Export to Arc/Info ASCII Grid format (aka ESRI grid)
# It should be possible to import this to most conventional
# GIS software.
# only use first band
proj_esri = proj_osr.Clone()
proj_esri.MorphToESRI()
ds = wrl.georef.create_raster_dataset(data[0], xy, projection=proj_esri)
wrl.io.write_raster_dataset(wdir + "aaigrid.asc", ds, 'AAIGrid', options=['DECIMAL_PRECISION=2'])
```
### Step 5a: Read from GeoTIFF
```
ds1 = wrl.io.open_raster(wdir + "geotiff.tif")
data1, xy1, proj1 = wrl.georef.extract_raster_dataset(ds1, nodata=-9999.)
np.testing.assert_array_equal(data1, data)
np.testing.assert_array_equal(xy1, xy)
```
### Step 5b: Read from ESRI ASCII file (aka Arc/Info ASCII Grid)
```
ds2 = wrl.io.open_raster(wdir + "aaigrid.asc")
data2, xy2, proj2 = wrl.georef.extract_raster_dataset(ds2, nodata=-9999.)
np.testing.assert_array_almost_equal(data2, data[0], decimal=2)
np.testing.assert_array_almost_equal(xy2, xy)
```
|
github_jupyter
|
# FAO Economic and Employment Stats
Two widgets for the 'People' tab.
- No of people employed full time (```forempl``` x 1000)
- ...of which are female (```femempl``` x 1000)
- Net USD generate by forest ({```usdrev``` - ```usdexp```} x 1000)
- GDP in USD in 2012 (```gdpusd2012``` x 1000) **NOTE: GDP in year=9999**
- Total Population (```totpop1000``` x 1000)
```
#Import Global Metadata etc
%run '0.Importable_Globals.ipynb'
# First, get the FAO data from a carto table
sql = ("SELECT fao.country, fao.forempl, fao.femempl, fao.usdrev, fao.usdexp, "
"fao.gdpusd2012, fao.totpop1000, fao.year "
"FROM table_7_economics_livelihood as fao "
"WHERE fao.year = 2000 or fao.year = 2005 or fao.year = 2010 or fao.year = 9999"
)
account = 'wri-01'
urlCarto = "https://{0}.carto.com/api/v2/sql".format(account)
sql = {"q": sql}
r = requests.get(urlCarto, params=sql)
print(r.url,'\n')
pprint(r.json().get('rows')[0:3])
try:
fao_data = r.json().get('rows')
except:
fao_data = None
```
# Widget 1: "Forestry Sector Employment"
Display a pie chart showing the number of male and female employees employed in the Forestry Sector in a given year as well as a dynamic entence.
On hover the segments of the pie chart should show the number of male or female employees as well as the % of the total.
**If no data for female employees - DO NOT SHOW pie chart!**
User Variables:
- adm0 (see whitelist below, not available for all countries)
- year (2000, 2005, 2010)
### NOTE
Both widgets will use the same requests since it is easier to request all the relevent data in one go.
```
# First, get ALL data from the FAO data from a carto table
sql = ("SELECT fao.country, fao.forempl, fao.femempl, fao.usdrev, fao.usdexp, "
"fao.gdpusd2012, fao.year "
"FROM table_7_economics_livelihood as fao "
"WHERE fao.year = 2000 or fao.year = 2005 or fao.year = 2010 or fao.year = 9999"
)
account = 'wri-01'
urlCarto = "https://{0}.carto.com/api/v2/sql".format(account)
sql = {"q": sql}
r = requests.get(urlCarto, params=sql)
print(r.url,'\n')
try:
fao_data = r.json().get('rows')
except:
fao_data = None
fao_data[0:3]
# Build a whitelist for this widget (not all countries have data!)
empl_whitelist = []
for d in fao_data:
if d.get('iso') not in empl_whitelist:
empl_whitelist.append(d.get('country'))
empl_whitelist[0:3]
adm0 = 'GBR'
year = 2010 #2000, 2005, 2010
# Retrieve data for relevent country by filtering by iso
iso_filter = list(filter(lambda x: x.get('country') == adm0, fao_data))
iso_filter
# Sanitise data. May have empty fields, and scales numbers by 1000
empl_data = []
for d in iso_filter:
if d.get('year') != 9999:
try:
empl_data.append({
'male': (d.get('forempl') - d.get('femempl'))*1000,
'female': d.get('femempl')*1000,
'year': d.get('year')
})
except:
empl_data.append({
'male': d.get('forempl'),
'female': None,
'year': d.get('year')
})
empl_data
# Create a list for male and female data respectively for the user selected year
for i in empl_data:
if i.get('year') == year:
male_data = i.get('male')
female_data = i.get('female')
if female_data:
labels = ['Male', 'Female']
data = [male_data, female_data]
colors = ['lightblue', 'pink']
fig1, ax1 = plt.subplots()
ax1.pie(data, labels=labels, autopct='%1.1f%%',
shadow=False, startangle=90, colors=colors)
ax1.axis('equal')
centre_circle = plt.Circle((0,0),0.75,color='black', fc='white',linewidth=0.5)
fig1 = plt.gcf()
fig1.gca().add_artist(centre_circle)
plt.title(f'Forestry Employment by Gender in {adm0}')
plt.show()
else:
print(f'No data for {adm0} in {year}')
if female_data:
print(f"According to the FAO there were {male_data + female_data} people employed in {iso_to_countries[adm0]}'s ", end="")
print(f"Forestry sector in {year}, of which {female_data} were female.", end="")
else:
print(f"According to the FAO there were {male_data} people employed in {iso_to_countries[adm0]}'s ", end="")
print(f"Forestry sector in {year}.", end="")
```
# Widget 2: "Economic Impact of X's Forestry Sector"
Displays a bar chart and ranked list (side by side) as well as a dynamic sentence.
The bar chart will display revenue and expenditure bars side-by-side, and display 'contribution relative to GDP' on hover.
The ranked list will show countries with similar contributions (and sort by net or % as described below)
User Variables:
- adm0 (see whitelist)
- year (2000, 2005, 2010)
- net contribution in USD or as a % of the country's GDP
Maths:
```
[net contribution (USD) = (revenue - expenditure)\*1000]
[net contribution (%) = 100\*(revenue - expenditure)\*1000/GDP]
```
### NOTE
Both widgets will use the same requests since it is easier to request all the relevent data in one go.
```
# First, get ALL data from the FAO data from a carto table
sql = ("SELECT fao.country, fao.forempl, fao.femempl, fao.usdrev, fao.usdexp, "
"fao.gdpusd2012, fao.year "
"FROM table_7_economics_livelihood as fao "
"WHERE fao.year = 2000 or fao.year = 2005 or fao.year = 2010 or fao.year = 9999"
)
account = 'wri-01'
urlCarto = "https://{0}.carto.com/api/v2/sql".format(account)
sql = {"q": sql}
r = requests.get(urlCarto, params=sql)
print(r.url,'\n')
try:
fao_data = r.json().get('rows')
except:
fao_data = None
fao_data[0:3]
#Sanitise data. Note that some revenue, expenditure, and GDP
# values from the table may come back as None, 0 or empty strings...
#Hence we have to acount for all of these!
econ_data = []
gdp = []
#Get GDP of each country (found in element with 'year' = 9999)
for d in fao_data:
if d.get('gdpusd2012') and d.get('gdpusd2012') != '-9999' and d.get('year') == 9999:
gdp.append({
'gdp': float(d.get('gdpusd2012')),
'iso': d.get('country')
})
#Build data structure
for d in fao_data:
if d.get('year') != 9999:
for g in gdp:
if g.get('iso') == d.get('country'):
tmp_gdp = g.get('gdp')
break
if d.get('usdrev') and d.get('usdrev') != '' and d.get('usdexp') and d.get('usdexp') != '':
net = (d.get('usdrev') - int(d.get('usdexp')))*1000
econ_data.append({
'iso': d.get('country'),
'rev': d.get('usdrev')*1000,
'exp': int(d.get('usdexp'))*1000,
'net_usd': net,
'gdp': tmp_gdp,
'net_perc': 100*net/tmp_gdp,
'year': d.get('year')
})
econ_data[0:3]
```
### Get available Countries and Build Whitelist
```
# Build whitelist of countries with the data we want to analyse
econ_whitelist = []
for e in econ_data:
if e.get('iso') not in econ_whitelist:
econ_whitelist.append(e.get('iso'))
econ_whitelist[0:3]
```
# Do Ranking (*using functional python!*)
```
adm0 = 'BRA'
year = 2010 #2000, 2005, 2010
#Filter the data for year of interest
# NOTE: IF year equals 2010 ignore Lebanon (LBN) - mistake in data!
if year == 2010:
in_year = list(filter(lambda x: x.get('year') == year and x.get('iso') != 'LBN', econ_data))
else:
in_year = list(filter(lambda x: x.get('year') == year, econ_data))
in_year[0:3]
```
### Net Revenue in USD
```
# Order by net revenue ('net_usd')
rank_list_net = sorted(in_year, key=lambda k: k['net_usd'], reverse=True)
rank_list_net[0:3]
# Get country's rank and print adjacent values ('net_usd' and 'iso' in this case)
rank = 1
for i in rank_list_net:
if i.get('iso') == adm0:
print('RANK =', rank)
break
else:
rank += 1
if rank == 1:
bottom_bound = -1
upper_bound = 4
elif rank == 2:
bottom_bound = 2
upper_bound = 3
elif rank == len(rank_list_net):
bottom_bound = 5
upper_bound = -1
elif rank == len(rank_list_net)-1:
bottom_bound = 4
upper_bound = 0
else:
bottom_bound = 3
upper_bound = 2
rank_list_net[rank-bottom_bound:rank+upper_bound]
```
### Net Revenue as a percentage of Nations GDP
```
# Order by net revenue per GDP ('net_perc')
rank_list_perc = sorted(in_year, key=lambda k: k['net_perc'], reverse=True)
rank_list_perc[0:3]
# Get country's rank and print adjacent values ('net_perc' and 'iso' in this case)
rank = 1
for i in rank_list_perc:
if i.get('iso') == adm0:
print('RANK =',rank)
break
else:
rank += 1
if rank == 1:
bottom_bound = -1
upper_bound = 4
elif rank == 2:
bottom_bound = 2
upper_bound = 3
elif rank == len(rank_list_perc):
bottom_bound = 5
upper_bound = -1
elif rank == len(rank_list_perc)-1:
bottom_bound = 4
upper_bound = 0
else:
bottom_bound = 3
upper_bound = 2
rank_list_perc[rank-bottom_bound:rank+upper_bound]
```
# Graph and Dynamic Sentence
```
# Get data for iso and year of interest
iso_and_year = list(filter(lambda x: x.get('year') == year and x.get('iso') == adm0, econ_data))
iso_and_year[0:3]
# Graph
bars = ['Revenue', 'Expenditure']
colors = ['blue','red']
width = 0.35
fig, ax = plt.subplots()
rects1 = ax.bar(bars, [iso_and_year[0].get('rev'), iso_and_year[0].get('exp')], color=colors)
# add some text for labels, title and axes ticks
ax.set_ylabel('USD')
ax.set_title(f'Forestry Revenue vs Expenditure for {adm0} in {year}')
plt.show()
# Dynamic Sentence
print(f"According to the FAO the forestry sector contributed a net ", end="")
print(f"{iso_and_year[0].get('net_usd')/1e9} billion USD to the economy in {year}, ", end="")
print(f"which is approximately {iso_and_year[0].get('net_perc')}% of {iso_to_countries[adm0]}'s GDP.", end="")
```
|
github_jupyter
|
```
import numpy as np
import tensorflow as tf
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_auc_score
import json
import pickle
from sklearn.externals import joblib
import sys
sys.path.append('../src/')
from TFExpMachine import TFExpMachine, simple_batcher
```
# Load data (see movielens-prepare.ipynb)
```
X_tr, y_tr, s_features = joblib.load('tmp/train_categotical.jl')
X_te, y_te, s_features = joblib.load('tmp/test_categorical.jl')
```
# Prepare init from LogReg
```
from sklearn.preprocessing import OneHotEncoder
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_auc_score
target_rank = 10
oh = OneHotEncoder()
oh.fit(np.vstack((X_tr, X_te))-1)
X_tr_sp = oh.transform(X_tr-1)
X_te_sp = oh.transform(X_te-1)
logreg = LogisticRegression()
logreg.fit(X_tr_sp, y_tr)
y_pred = logreg.predict_proba(X_te_sp)[:, 1]
print(roc_auc_score(y_te, y_pred))
target_rank = 10
num_features = len(s_features)
w_cores = [None] * num_features
coef = logreg.coef_[0]
intercept = logreg.intercept_[0]
# see paper for details about initialization
begin_feature = [0] + list(np.cumsum(s_features))
for i in range(num_features):
n_factors = s_features[i]
if i == 0:
tmp = np.zeros((n_factors+1, 1, target_rank))
for local_j, global_j in enumerate([-1] + list(range(begin_feature[i], s_features[i]))):
if local_j==0:
tmp[local_j,:1,:2] = [1, 0]
else:
tmp[local_j,:1,:2] = [0, coef[global_j]]
w_cores[i] = tmp.astype(np.float32)
elif i == num_features-1:
tmp = np.zeros((n_factors+1, target_rank, 1))
for local_j, global_j in enumerate([-1] + list(range(begin_feature[i], s_features[i]))):
if local_j==0:
tmp[local_j,:2,:1] = np.array([[intercept], [1]])
else:
tmp[local_j,:2,:1] = [[coef[global_j]], [0]]
w_cores[i] = tmp.astype(np.float32)
else:
tmp = np.zeros((n_factors+1, target_rank, target_rank))
for local_j, global_j in enumerate([-1] + list(range(begin_feature[i], s_features[i]))):
if local_j==0:
tmp[local_j,:2,:2] = np.eye(2)
else:
tmp[local_j,:2,:2] = [[0, coef[global_j]], [0,0]]
w_cores[i] = tmp.astype(np.float32)
```
# Init model
```
model.destroy()
model = TFExpMachine(rank=target_rank, s_features=s_features, init_std=0.001, reg=0.012, exp_reg=1.8)
model.init_from_cores(w_cores)
model.build_graph()
model.initialize_session()
```
# Learning
```
epoch_hist = []
for epoch in range(50):
# train phase
loss_hist = []
penalty_hist = []
for x, y in simple_batcher(X_tr, y_tr, 256):
fd = {model.X: x, model.Y: 2*y-1}
run_ops = [model.trainer, model.outputs, model.loss, model.penalty]
_, outs, batch_loss, penalty = model.session.run(run_ops, fd)
loss_hist.append(batch_loss)
penalty_hist.append(penalty)
epoch_train_loss = np.mean(loss_hist)
epoch_train_pen = np.mean(penalty_hist)
epoch_stats = {
'epoch': epoch,
'train_logloss': float(epoch_train_loss)
}
# test phase
if epoch%2==0 and epoch>0:
fd = {model.X: X_te, model.Y: 2*y_te-1}
run_ops = [model.outputs, model.loss, model.penalty, model.penalized_loss]
outs, raw_loss, raw_penalty, loss = model.session.run(run_ops, fd)
epoch_test_loss = roc_auc_score(y_te, outs)
epoch_stats['test_auc'] = float(epoch_test_loss),
epoch_stats['penalty'] = float(raw_penalty)
print('{}: te_auc: {:.4f}'.format(epoch, epoch_test_loss))
epoch_hist.append(epoch_stats)
# dump to json
json.dump(epoch_hist, open('./tmp/ExM_rank10_ereg1.8.json', 'w'))
# Draw plot
%pylab inline
plot([x['epoch'] for x in epoch_hist if 'test_auc' in x], [x['test_auc'] for x in epoch_hist if 'test_auc' in x])
grid()
ylim(0.775, 0.785)
xlabel('epoch')
ylabel('test auc')
# release resources
model.destroy()
```
|
github_jupyter
|
# Python 101
```
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
```
### First code in Python
#### Running (executing) a cell
Jupyter Notebooks allow code to be separated into sections that can be executed independent of one another. These sections are called "cells".
Running a cell means that you will execute the cell’s contents. To execute a cell, you can just select the cell and click the Run button that is in the row of buttons along the top. It’s towards the middle. If you prefer using your keyboard, you can just press SHIFT + ENTER
To automatically run all cells in a notebook, navigate to the "Run" tab of the menu bar at the top of JupyterLab and select "Run All Cells" (or the option that best suits your needs). When a cell is run, the cell's content is executed. Any output produced from running the cell will appear directly below it.
```
print('Hello World')
```
#### Cell status
The [ ]: symbol to the left of each Code cell describes the state of the cell:
[ ]: means that the cell has not been run yet.
[*]: means that the cell is currently running.
[1]: means that the cell has finished running and was the first cell run.
For more information on jupyter notebooks have a look at the jupyter_introduction.ipynb notebook in the additional content section
### Mathematical Operations
Now we can try some basic mathematical operations
```
22 / 9
243 + 3
4454 - 32
222 / 2
```
### Variable Assignment
In python the '=' is used to assign a value to a variable. Besides a single equal sign you can also use combinations with other operators.
```
x = 5
```
### Functions
Functions are named pieces of code, to perform a particular job. Functions in Python are excuted by specifying their name, followed by parentheses.
```
abs(-7)
```
### Python libraries
One of the main advantages in python is the extensive standard library (already included in python) and the huge number of third party libraries.
In order to use these libraries you have to import them. Therefor you just need the 'import' command.
```
import math
math.ceil(3.445)
```
When using the import command, python only loads the name of this module (e.g. math) and not the names of the single functions. <br>
If you want to use individual classes or functions within the module you have to enter the name of the module and the name of the function separated by a dot:
```
import math
math.ceil(1.34) # math = module name , ceil = function name
```
You can also assign a function to variable to a variable name
```
import math
ceil = math.ceil
ceil(1.34)
```
If you want to load only one or more specific function you can use the term from ... import ...
```
from math import ceil
from math import ceil, fabs, trunc
from math import * # import all functions of the module
```
You can also assign a new name to the a module or function while importing
```
import math as m
print(m.ceil(1.34))
from math import ceil as c
print(c(1.34))
```
### Installing libraries
If you want to install a external library you can do this via pip or conda
```
!pip install geopandas
!conda install geopandas
```
### Help
If you want to know more about a function/library or what they are exactly doing you can use the 'help()' function.
```
import geopandas as gpd
help(gpd)
import geopandas as gpd
help(gpd.read_file)
```
### Data types
These classes are the basic building blocks of Python
|Type | Meaning | Mutability | Examples |
|-----|---------|------------|----------|
| int | Integer | immutable | 1, -10, 0 |
| float | Float | immutable | 3.2, 100.0, -9.9 |
| bool | Boolean | immutable | True, False |
| str | String | immutable | "Hello!", "a", "I like Python" |
| list | List | mutable | [1,2,3] |
| tuple | Tuple | immutable | (1, 2) |
| dict | Dictionary | mutable | {"a": 2} |
| set | Set | mutable | {"a", "b"} |
### Numbers
Python can handle several types of numbers, but the two most common are:
- int, which represents integer values like 100, and
- float, which represents numbers that have a fraction part, like 0.5
```
population = 127880
latitude = 49.79391
longitude = 9.95121
print(type(population))
print(type(latitude))
area = 87.63
density = population / area
print(density)
87**2
```
Below is a list of operations for these build-in numeric types:
| Operation | Result |
|---------------|--------------|
|x + y |sum of x and y|
|x - y |difference of x and y|
|x * y |product of x and y|
|x / y |quotient of x and y|
|x // y |(floored) quotient of x and y |
|x % y |remainder of x / y |
|-x |x negated |
|+x |x unchanged |
|abs(x) |absolute value or magnitude of x |
|int(x) |x converted to integer |
|long(x) |x converted to long integer |
|float(x) |x converted to floating point |
|complex(re,im) |a complex number with real part re, |
| |imaginary part im (im defaults to zero) |
|c.conjugate() |conjugate of the complex number c |
|divmod(x, y) |the pair (x // y, x % y) |
|pow(x, y) |x to the power y |
|x ** y |x to the power y |
### Booleans and comparison
Another type in python is the so called boolean type. This type has two values: True and false. Booleans can be assign to a variable name or created for example when comparing values using the equal operator '=='. Another way to compare values is the not equal operator '!=' and the operators for greater '>' or smaller '<'.
```
x = True
print(x)
y = False
print(y)
city_1 = 'Wuerzburg'
pop_1 = 127880
region_1 = 'Bavaria'
city_2 = 'Munich'
pop_2 = 1484226
region_2 = 'Bavaria'
print(pop_1 == pop_2)
print(region_1 == region_2)
print(pop_1 >= pop_2)
print(city_1 != city_2)
```
### Strings
If you want to use text in python, you have to use 'strings'. A string is created by writing your desired text between two single '(...)' or double quotation marks "(...)". For printing text (or numbers) use the 'print()' function.
```
type("Spatial Python")
```
Other data types can be converted to a string using the str function:
```
"Sentinel2_" + "B" + str(1) + ".tif"
pop = 127880
'The population of Würzburg is ' + str(pop)
```
Of course strings can be also converted to numbers
```
int("11")
float("42.2")
```
Strings can be concatenated with the + operator
```
"Sentinel2_" + "B" + str(1) + ".tif"
```
Besides the + operator Python also has some more advanced formatting methods
```
x=1
f"Sentinel2_B{x}.tif"
"Sentinel2_B{x}.tif".format(x=1)
```
Python also provides many built-in functions and methods for strings. Below are just a few examples
| Function/Methods Name | Description |
|---------------|------------|
| capitalize() | Converts the first character of the string to a capital (uppercase) letter |
| count()| Returns the number of occurrences of a substring in the string. |
| encode()| Encodes strings with the specified encoded scheme |
| endswith()| Returns “True” if a string ends with the given suffix |
| find()| Returns the lowest index of the substring if it is found |
| format()| Formats the string for printing it to console |
| index()| Returns the position of the first occurrence of a substring in a string |
| isalnum()| Checks whether all the characters in a given string is alphanumeric or not |
| isalpha()| Returns “True” if all characters in the string are alphabets |
| isdecimal()| Returns true if all characters in a string are decimal |
| isnumeric()| Returns “True” if all characters in the string are numeric characters |
| isprintable()| Returns “True” if all characters in the string are printable or the string is empty |
| supper()| Checks if all characters in the string are uppercase |
| join()| Returns a concatenated String |
| lower()| Converts all uppercase characters in a string into lowercase |
| replace()| Replaces all occurrences of a substring with another substring |
| startswith()| Returns “True” if a string starts with the given prefix |
| strip()| Returns the string with both leading and trailing characters |
| swapcase()| Converts all uppercase characters to lowercase and vice versa |
| title()| Convert string to title case |
| translate()| Modify string according to given translation mappings |
| upper()| Converts all lowercase characters in a string into uppercase |
| zfill()| Returns a copy of the string with ‘0’ characters padded to the left side of the string |
```
string = "Hello World"
string.upper()
string.replace('Hello', 'My')
string.find('l')
string.count('l')
```
Strings in python can be accesssed by index or sliced
```
#string[2] # get third character
string[1:5] # slice from 1 (included) to 5 (excluded) postion 2 - 5
string[-5] # count from behind
string[2:] # from 2 (included) to end
string[:2] # from 0 to 1
string[-5] # last character
```
<img src="images/indexing.png" width=600 />
### Lists
Another data type are so called lists. Lists can be created putting several comma-separated values between square brackets. You can use lists to generate sequences of values, which can be of the same or different datatype.
```
letter_list = ['a','b','c','d','e','f'] #list of stringd
letter_list
list_of_numbers = [1,2,3,4,5,6,7] #list of numbers
list_of_numbers
mixed_list = ['hello', 2.45, 3, 'a', -.6545+0J] #mixing different data types
mixed_list
```
Similar to strings values in a list can be done using indexing or slicing.
```
random = [1, 2, 3, 4, 'a', 'b','c','d']
random[2]
print(random[1:5]) # slice from 1 (included) to 5 (excluded)
print(random[-5]) # count from behind
print(random[2:]) # from 2 (included) to end
print(random[:2]) # from begin to 2 (!not included!)
```
You can also update lists with one or more elements by giving the slice on the left-hand side. It´s also possible to append new elements to the list or delete list elements with the function.
```
cities = ['Berlin', 'Paris','London','Madrid','Lisboa']
cities[3]
# Update list
cities[3] = 'Rome'
cities
# deleting elemants
del(cities[3])
cities
# append elemnts
cities.append('Vienna')
cities
```
There are many different ways to interact with lists. Exploring them is part of the fun of python.
| Function/Method Name | Description |
|---------------|-------------|
| list.append(x) | Add an item to the end of the list. Equivalent to a[len(a):] = [x]. |
| list.extend(L) | Extend the list by appending all the items in the given list. Equivalent to a[len(a):] = L. |
| list.insert(i, x) | Insert an item at a given position. The first argument is the index of the element before which to insert, so a.insert(0, x) inserts at the front of the list, and a.insert(len(a), x) is equivalent to a.append(x). |
| list.remove(x) | Remove the first item from the list whose value is x. It is an error if there is no such item. |
| list.pop([i]) | Remove the item at the given position in the list, and return it. If no index is specified, a.pop() removes and returns the last item in the list. |
| list.clear() | Remove all items from the list. Equivalent to del a[:]. |
| list.index(x) | Return the index in the list of the first item whose value is x. It is an error if there is no such item. |
| list.count(x) | Return the number of times x appears in the list. |
| list.sort() | Sort the items of the list in place. |
| list.reverse() | Reverse the elements of the list in place. |
| list.copy() | Return a shallow copy of the list. Equivalent to a[:]. |
| len(list) | Returns the number of items in the list. |
| max(list) | Returns the largest item or the largest of two or more arguments. |
| min(list) | Returns the smallest item or the smallest of two or more arguments. |
```
temp = [3.4,4.3,5.6,0.21,3.0]
len(temp)
min(temp)
temp.sort()
temp
```
### Tuples
Tuples are sequences, just like lists. The difference is that tuples are immutable. Which means that they can´t be changed like lists. Tuples can be created without brackets (optionally you can use parenthesis).
```
tup1 = "a", "b","c"
tup2 = (1, 2, 3, 4, 5 )
tup3 = ("a", "b", "c", "d",2)
tup1
tup2
tup3
```
You can access elements in the same way as lists. But due to the fact that tuples are **immutable**, you cannot update or change the values of tuple elements.
```
# Access elements
tup1 = 1, 2, 3, 4, 'a', 'b','c','d'
tup1[2]
tup1[1:5] # slice from 1 (included) to 5 (included)
tup1[-5] # count from behind
tup1[2:] # from 2 (included) to end
tup1[:2] # from begin to 2 (!not included!)
tup1 = (123,4554,5,454, 34.56)
tup2 = ('abc','def', 'ghi' ,'xyz')
tup1[0] = 10 #This action is not allowed for tuples
```
Tuples also have some built-in functions
```
tup1 = [1,2,1,4,3,4,5,6,7,8,8,9]
len(tup1)
min(tup1)
max(tup1)
tup1.count(8)
```
#### Why using tuples at all ?
- Tuples are faster than lists and need less memory
- It makes your code safer if you “write-protect” data that does not need to be changed.
- Tuples can be used as dictionary keys
### Dictionaries
Strings, lists and tuples are so called sequential datatypes. Dictionaries belong to python’s built-in mapping type. Sequential datatypes use integers as indices to access the values they contain within them. Dictionaries allows to use map keys. The values of a dictionary can be of any type, but the keys must be of an immutable data type (strings, numbers, tuples).
Dictionaries are constructed with curly brackets {}. Key and values are separated by colons ':'and square brackets are used to index it. It´s not allowed more entries per key, which also means that duplicate keys are also not allowed.
<img src="images/dict.png" width=600 />
```
city_temp = {'City': 'Dublin', 'MaxTemp': 15.5, 'MinTemp': 5.0}
```
Dictionary values are accessible through the keys
```
city_temp['City']
city_temp['MaxTemp']
city_temp['MinTemp']
```
Dictionaries can be updated and elements can be removed.
```
# Update dictionaries
city_temp = {'City': 'Dublin', 'MaxTemp': 15.5, 'MinTemp': 5.0}
city_temp['MaxTemp'] = 15.65; # update existing entry
city_temp['Population'] = 544107; # Add new entry
city_temp['MaxTemp']
city_temp['Population']
```
Of course we can also use more than one value per key
```
city_temp = {'City': ['Dublin','London'], 'MaxTemp': [15.5,12.5], 'MinTemp': [15.5,12.5]}
```
If we access a key with multiple values we get back a list
```
city_temp['MaxTemp']
city_temp['City'][1][0]
```
Few examples of built-in functions and methods.
| Function/Method | Description|
| ----------------| -----------|
| clear()| Removes all the elements from the dictionary|
| copy()| Returns a copy of the dictionary|
| fromkeys()| Returns a dictionary with the specified keys and value|
| get() | Returns the value of the specified key|
| items()| Returns a list containing a tuple for each key value pair|
| keys()| Returns a list containing the dictionary's keys|
| pop() | Removes the element with the specified key|
| popitem()| Removes the last inserted key-value pair|
| setdefault()| Returns the value of the specified key. If the key does not exist: insert the key, with the specified value|
| update()| Updates the dictionary with the specified key-value pairs|
| values()| Returns a list of all the values in the dictionary|
```
city_temp.keys()
city_temp.values()
len(city_temp)
len(city_temp['City'])
max(city_temp['MaxTemp'])
min(city_temp['MinTemp'])
```
### Indentation
A python program is structured through indentation. Indentations are used to separate different code block. This make it´s easier to read and understand your own code and the code of others. While in other programming languages indentation is a matter of style, in python it´s a language requirement.
```
def letterGrade(score):
if score >= 90:
letter = 'A'
else: # grade must be B, C, D or F
if score >= 80:
letter = 'B'
else: # grade must be C, D or F
if score >= 70:
letter = 'C'
else: # grade must D or F
if score >= 60:
letter = 'D'
else:
letter = 'F'
return letter
letterGrade(9)
```
## Control flow statements
### While, if, else
Decision making is required when we want to execute a code only if a certain condition holds. This means e.g. that some statements are only carried out if an expression is True. The 'while' statement repeatedly tests the given expression and executes the code block as long as the expression is True
```
password = "datacube"
attempt = input("Enter password: ")
if attempt == password:
print("Welcome")
```
In this case, the if statement is used to evaluates the input of the user and the following code block will only be executed if the expression is True. If the expression is False, the statement(s) is not executed.
But if we want that the the program does something else, even when the if-statement evaluates to false, we can add the 'else' statement.
```
password = "python2017"
attempt = input("Enter password: ")
if attempt == password:
print("Welcome")
else:
print("Incorrect password!")
```
You can also use multiple if...else statements nested into each other
```
passlist = ['1223','hamster','mydog','python','snow' ]
name = input("What is your username? ")
if name == 'Steve':
password = input("What’s the password? ")
#did they enter the correct password?
if password in passlist:
print("Welcome {0}".format(name))
else:
print("Incorrect password")
else:
print("No valid username")
```
In the next example will want a program that evaluates more than two possible outcomes. For this, we will use an else if statement. In python else if statment is written as 'elif'.
```
name = input("What is your username? ")
password = input("What’s the password? ")
if name == 'Steve':
if password == 'kingofthehill':
print("Welcome {0}".format(name))
else:
print("Incorrect password")
elif name == 'Insa':
if password == 'IOtte123':
print("Welcome {0}".format(name))
else:
print("Incorrect password")
elif name == 'Johannes':
if password == 'RadarLove':
print("Welcome {0}".format(name))
else:
print("Incorrect password")
else:
print("No valid username")
```
Sometimes you want that a specific code block is carried out repeatedly. This can be accomplished creating so called loops. A loop allows you to execute a statement or even a group of statements multiple times. For example, the 'while' statement, allows you to run the code within the loop as long as the expression is True.
```
count = 0
while count < 9:
print('The count is:', count)
count += 1
print("Count maximum is reached!")
```
You can also create infinite loops
```
var = 1
while var == 1 : # This constructs an infinite loop
num = int(input("Enter a number :"))
print("You entered: ", num)
print('Thanks')
```
Just like the 'if-statement', you can also combine 'while' with 'else'
```
count = 0
while count < 12:
print(count, " is less than 12")
count = count + 1
else:
print(count, " is not less than 12")
```
Another statement you can use is break(). It terminates the enclosing loop. A premature termination of the current loop can be useful when some external condition is triggered requiring an exit from a loop.
```
import random
number = random.randint(1, 15)
number_of_guesses = 0
while number_of_guesses < 5:
print('Guess a number between 1 and 15:')
guess = input()
guess = int(guess)
number_of_guesses = number_of_guesses + 1
if guess < number:
print('Your guess is too low')
if guess > number:
print('Your guess is too high')
if guess == number:
break
if guess == number:
print('You guessed the number in ' , number_of_guesses,' tries!')
else:
print('You did not guess the number. The number was ' , number)
```
Sometimes, you want to perform code on each item on a list. This can be accomplished with a while loop and counter variable.
```
words = ['one', 'two', 'three', 'four','five' ]
count = 0
max_count = len(words) - 1
while count <= max_count:
word = words[count]
print(word +'!')
count = count + 1
```
Using a while loop for iterating through list requires quite a lot of code. Python also provides the for-loop as shortcut that accomplishes the same task. Let´s do the same code as above with a for-loop
```
words = ['one', 'two', 'three', 'four','five' ]
for index, value in enumerate(words):
print(index)
print(value + '!')
```
If you want to repeat some code a certain numbers of time, you can combine the for-loop with an range object.
```
range(9)
for i in range(9):
print(str(i) + " !")
```
Now we can use our gained knowledge to program e.g. a simple calculator.
```
print("1.Add")
print("2.Subtract")
print("3.Multiply")
print("4.Divide")
# Take input from the user
choice = input("Enter choice(1/2/3/4):")
num1 = int(input("Enter first number: "))
num2 = int(input("Enter second number: "))
if choice == '1':
result = num1 + num2
print("{0} + {1} = {2}".format(num1,num2,result))
elif choice == '2':
result = num1 - num2
print("{0} - {1} = {2}".format(num1,num2,result))
elif choice == '3':
result = num1*num2
print("{0} * {1} = {2}".format(num1,num2,result))
elif choice == '4':
result = num1/num2
print("{0} / {1} = {2}".format(num1,num2,result))
else:
print("Invalid input")
```
#### Comprehensions
Comprehensions are constructs that allow sequences to be built from other sequences. Let's assume we have a list with temperature values in Celsius and we want to convert them to Fahrenheit
```
T_in_celsius = [3, 12, 18, 9, 10, 20]
```
We could write a for loop for this problem
```
fahrenheit = []
for temp in T_in_celsius:
temp_fahr = (temp * 9 / 5) + 32
fahrenheit.append(temp_fahr)
fahrenheit
```
Or, we could use a list comprehension:
```
fahrenheit = [(temp * 9 / 5) + 32 for temp in T_in_celsius]
fahrenheit
```
We can also go one step further and also include a if statement
```
# Pythagorean triple
# consists of three positive integers a, b, and c, such that a**2 + b**2 = c**2
[(a,b,c) for a in range(1,30) for b in range(1,30) for c in range(1,30) if a**2 + b**2 == c**2]
```
You can even create nested comprehensions
```
matrix = [[j * j+i for j in range(4)] for i in range(3)]
matrix
```
Of course you can use comprehensions also for dictionaries
```
fruits = ['apple', 'mango', 'banana','cherry']
{f:len(f) for f in fruits}
```
### Functions
In this chapter, we will learn how to write your own functions. A function can be used as kind of a structuring element in programming languages to group a set of statements so you can reuse them. Decreasing code size by using functions make it more readable and easier to maintain. And of course it saves a lot of typing. In python a function call is a statement consisting of a function name followed by information between in parentheses. You have already used functions in the previous chapters
A function consists of several parts:
- **Name**: What you call the function by
- **Parameters**: You can provide functions with variables.
- **Docstring**: A docstring allows you to write a little documentation were you explain how the function works
- **Body**: This is were the magic happens, as here is the place for the code itself
- **Return values**: You usually create functions to do something that create a result.
<img src="images/function.png" width=600 />
Let´s start with a very simple function
```
def my_function():
print("I love python!")
my_function()
```
You can also create functions which receive arguments
```
def function1(value, value2 = 5):
return value**2 + value2
function1(value = 4, value2 = 10000)
```
Or create functions inside functions
```
def area(width, height, func):
print("Area: {0}".format(func(width*height)))
x = area(width = 4, height = 6, func=function1)
```
If the function should return a result (not only print) you can use the 'return' statement. The return statement exits the function and can contain a expression which gets evaluated or a value is returned. If there is no expression or value the function returns the 'None' object
```
def fahrenheit(T_in_celsius):
""" returns the temperature in degrees Fahrenheit """
return (T_in_celsius * 9 / 5) + 32
x = fahrenheit(35)
x
```
Now we can rewrite our simple calculator. But this time we define our functions in front.
```
def add(x,y):
return x+y
def diff(x,y):
return x-y
def multiply(x,y):
return x*y
def divide(x,y):
return x/y
print("1.Add")
print("2.Subtract")
print("3.Multiply")
print("4.Divide")
# Take input from the user
choice = eval(input("Enter choice(1/2/3/4):"))
num1 = eval(input("Enter first number: "))
num2 = eval(input("Enter second number: "))
if choice == 1:
result = add(num1,num2)
print("{0} + {1} = {2}".format(num1,num2,result))
elif choice == 2:
result = diff(num1,num2)
print("{0} - {1} = {2}".format(num1,num2,result))
elif choice == 3:
result = multiply(num1,num2)
print("{0} * {1} = {2}".format(num1,num2,result))
elif choice == 4:
result = divide(num1,num2)
print("{0} / {1} = {2}".format(num1,num2,result))
else:
print("Invalid input")
```
# Literature
For this script I mainly used following sources:
<br>[1] https://docs.python.org/3/
<br>[2] https://www.tutorialspoint.com/python/python_lists.htm
<br>[3] https://www.datacamp.com
<br>[4] https://anh.cs.luc.edu/python/hands-on/3.1/handsonHtml/index.html
<br>[5] Python - kurz und gut (2014) Mark Lutz
|
github_jupyter
|
# Working with Data in OpenCV
Now that we have whetted our appetite for machine learning, it is time to delve a little
deeper into the different parts that make up a typical machine learning system.
Machine learning is all about building mathematical models in order
to understand data. The learning aspect enters this process when we give a machine
learning model the capability to adjust its **internal parameters**; we can tweak these
parameters so that the model explains the data better. In a sense, this can be understood as
the model learning from the data. Once the model has learned enough—whatever that
means—we can ask it to explain newly observed data.
Hence machine learning problems are always split into (at least) two
distinct phases:
- A **training phase**, during which we aim to train a machine learning model on a set of data that we call the **training dataset**.
- A **test phase**, during which we evaluate the learned (or finalized) machine learning model on a new set of never-before-seen data that we call the **test dataset**.
The importance of splitting our data into a training set and test set cannot be understated.
We always evaluate our models on an independent test set because we are interested in
knowing how well our models generalize to new data. In the end, isn't this what learning is
all about—be it machine learning or human learning?
Machine learning is also all about the **data**.
Data can be anything from images and movies to text
documents and audio files. Therefore, in its raw form, data might be made of pixels, letters,
words, or even worse: pure bits. It is easy to see that data in such a raw form might not be
very convenient to work with. Instead, we have to find ways to **preprocess** the data in order
to bring it into a form that is easy to parse.
In this chapter, we want to learn how data fits in with machine learning, and how to work with data using the tools of our choice: OpenCV and Python.
In specific, we want to address the following questions:
- What does a typical machine learning workflow look like?
- What are training data, validation data, and test data - and what are they good for?
- How do I load, store, and work with such data in OpenCV using Python?
## Outline
- [Dealing with Data Using Python's NumPy Package](02.01-Dealing-with-Data-Using-Python-NumPy.ipynb)
- [Loading External Datasets in Python](02.02-Loading-External-Datasets-in-Python.ipynb)
- [Visualizing Data Using Matplotlib](02.03-Visualizing-Data-Using-Matplotlib.ipynb)
- [Visualizing Data from an External Dataset](02.04-Visualizing-Data-from-an-External-Dataset.ipynb)
- [Dealing with Data Using OpenCV's TrainData container in C++](02.05-Dealing-with-Data-Using-the-OpenCV-TrainData-Container-in-C%2B%2B.ipynb)
## Starting a new IPython or Jupyter session
Before we can get started, we need to open an IPython shell or start a Jupyter Notebook:
1. Open a terminal like we did in the previous chapter, and navigate to the `Machine-Learning-for-OpenCV-Second-Edition` directory:
```
$ cd Desktop/Machine-Learning-for-OpenCV-Second-Edition
```
2. Activate the conda environment we created in the previous chapter:
```
$ source activate OpenCV-ML # Mac OS X / Linux
$ activate OpenCV-ML # Windows
```
3. Start a new IPython or Jupyter session:
```
$ ipython # for an IPython session
$ jupyter notebook # for a Jupyter session
```
If you chose to start an IPython session, the program should have greeted you with a
welcome message such as the following:
$ ipython
Python 3.6.0 | packaged by conda-forge | (default, Feb 9 2017, 14:36:55)
Type 'copyright', 'credits' or 'license' for more information
IPython 7.2.0 -- An enhanced Interactive Python. Type '?' for help.
In [1]:
The line starting with `In [1]` is where you type in your regular Python commands. In
addition, you can also use the Tab key while typing the names of variables and functions in
order to have IPython automatically complete them.
If you chose to start a Jupyter session, a new window should have opened in your web
browser that is pointing to http://localhost:8888. You want to create a new notebook by
clicking on New in the top-right corner and selecting Notebooks (Python3).
This will open a new window that contains an empty page with the same command line as in an IPython session:
In [ ]:
|
github_jupyter
|
<a href="https://colab.research.google.com/github/alijablack/data-science/blob/main/Wikipedia_NLP_Sentiment_Analysis.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Natural Language Processing
## Problem Statement
Use natural language processing on Wikipedia articles to identify the overall sentiment analysis for a page and number of authors.
## Data Collection
```
from google.colab import drive
drive.mount('/content/drive')
!python -m textblob.download_corpora
from textblob import TextBlob
import numpy as np
import pandas as pd
from sklearn.neighbors import NearestNeighbors
from sklearn.feature_extraction.text import CountVectorizer
people_path = '/content/drive/My Drive/Copy of people_db.csv'
people_df = pd.read_csv(people_path)
```
## Exploratory Data Analysis
## Part 1 of Project
This dataset from dbpedia includes over 42,000 entries.
```
people_df.info
```
Explore the first 100 to decide who to choose.
```
people_df.head(100).T
```
Select a person, Armen Ra, from the list to use as the input for sentiment analysis. Output Armen Ra's overview from the database.
```
my_person = [people_df.iloc[96]['text']]
my_person
```
### Data Processing
#### Vector Analysis
```
vect_people = CountVectorizer(stop_words='english')
word_weight = vect_people.fit_transform(people_df['text'])
word_weight
```
#### Nearest Neighbors
Fit the nearest neighbors model with content from people dataframe.
```
nn = NearestNeighbors(metric='euclidean')
nn.fit(word_weight)
ra_index = people_df[people_df['name'] == 'Armen Ra'].index[0]
ra_index
```
Use the nearest neighbor model to output people with overviews similar to Armen Ra's page.
```
distances, indices = nn.kneighbors(word_weight[ra_index], n_neighbors=11)
distances
```
Show the index of 10 similar overviews.
```
indices
```
Output the 10 people with overviews closest to Armen Ra.
```
people_df.iloc[indices[0],:]
top_ten = people_df.iloc[indices[0],1:11]
top_ten.head(11)
df2 = people_df[['text','name']]
# For each row, combine all the columns into one column
df3 = df2.apply(lambda x: ','.join(x.astype(str)), axis=1)
# Store them in a pandas dataframe
df_clean = pd.DataFrame({'clean': df3})
# Create the list of list format of the custom corpus for gensim modeling
sent = [row.split(',') for row in df_clean['clean']]
# show the example of list of list format of the custom corpus for gensim modeling
sent[:2]
```
Another way to output the 10 people with overviews closest to Armen Ra's page.
```
import gensim
from gensim.models import Word2Vec
model = Word2Vec(sent, min_count=1,size= 50,workers=3, window =3, sg = 1)
model['Armen Ra']
model.most_similar('Armen Ra'[:10])
```
This method outputs a different set of people than the nearest neighbors method. The nearest neighbors method output appears more closely aligned with the substance of Armen Ra's overview by similarly outputting people in creative industries. Whereas the similarity method outputs people with overviews that share a similar tone and format as Armen Ra's overview that is brief, informational, neutral.
#### Sentiment Analysis
Make Armen Ra's overview a string.
```
df2 = pd.DataFrame(my_person)
# For each row, combine all the columns into one column
df3 = df2.apply(lambda x: ','.join(x.astype(str)), axis=1)
# Store them in a pandas dataframe
df_clean = pd.DataFrame({'clean': df3})
# Create the list of list format of the custom corpus for gensim modeling
sent1 = [row.split(',') for row in df_clean['clean']]
# show the example of list of list format of the custom corpus for gensim modeling
sent1[:2]
```
Assign tags to each word in the overview.
```
!python -m textblob.download_corpora
from textblob import TextBlob
wiki = TextBlob(str(sent1))
wiki.tags
```
Identify the nouns in the overview.
```
wiki.noun_phrases
zen = TextBlob(str(sent1))
```
Identify the words in the overview.
```
zen.words
```
Identify the sentences in the overview.
```
zen.sentences
sentence = TextBlob(str(sent1))
sentence.words
sentence.words[-1].pluralize()
sentence.words[-1].singularize()
b = TextBlob(str(sentence))
print(b.correct())
```
Output the sentiment for Armen Ra's overview.
```
for sentence in zen.sentences:
print(sentence.sentiment[0])
```
## Part 2 of Project
### Data Collection
Install Wikipedia API. Wikipedia will be the main datasource for this step to access the full content of Armen Ra's page.
```
!pip install wikipedia
import wikipedia
```
### Data Processing
Produce the entire page of Armen Ra
```
#search wikipedia for Armen Ra
print(wikipedia.search('Armen Ra'))
#output the summary for Armen Ra
print(wikipedia.summary("Armen Ra"))
#output the page for Armen Ra
print(wikipedia.page("Armen Ra"))
#output the page content for Armen Ra
print(wikipedia.page('Armen Ra').content)
#output the url for Armen Ra's Wikipedia page
print(wikipedia.page('Armen Ra').url)
ra_df = pd.read_html('https://en.wikipedia.org/wiki/Armen_Ra')
type(ra_df)
page = wikipedia.page('Armen Ra')
page.summary
page.content
type(page.content)
wiki1 = TextBlob(page.content)
wiki1.tags
wiki1.noun_phrases
```
##### Sentiment Analysis
Produce the sentiment for Armen Ra's page.
```
testimonial = TextBlob(page.content)
testimonial.sentiment
```
Sentiment analysis shows a primarily neutral and objective tone throughout the page.
```
zen = TextBlob(page.content)
```
Process Armen Ra's page into words and sentences to determine how the sentiment changes throughout the page.
```
zen.words
zen.sentences
```
Determine any changes in sentiment throughout the page.
```
for sentence in zen.sentences:
print(sentence.sentiment[0])
```
Estimate 6 or 7 authors contributed to the Wikipedia article based on changes in the sentiment analysis.
Output a summary of the Armen Ra page
```
page.summary
sentence = TextBlob(page.content)
sentence.words
sentence.words[2].singularize()
sentence.words[2].pluralize()
b = TextBlob(page.content)
print(b.correct())
```
Consider algorithmic bias and errors in the natural language processing tools as Armen Ra's name is being shortened to 'Men A' or 'A'.
```
blob = TextBlob(page.content)
blob.ngrams(n=3)
#The sentiment of Armen Ra's page is in a informational, neutral tone
testimonial = TextBlob(page.content)
testimonial.sentiment
```
### Communication of Results
Ultimately, the sentiment analysis for Armen Ra's page shows the tone is primarily informational, objective, and neutral. When using Nearest Neighbors or Model Most Similar to identify Wikipedia pages similar to Armen Ra's, there were different results presented based on the method was used. Nearest Neighbors presented pages of individuals that had similarly neutral tones, while Most Similar showed individuals in similar industries as Armen Ra. The natural language processing tools at times output errors in Armen Ra's name and typos throughout the content. Consider further analysis into algorithmic bias present within the natural language processing tools and alternative data analysis and visualization methods available.
## Live Coding
In addition to presenting our slides to each other, at the end of the presentation each analyst will demonstrate their code using a famous person randomly selected from the database.
```
Roddy = people_df[people_df['name'].str.contains('Roddy Piper')]
Roddy
wikipedia.search('Roddy Piper')
wikipedia.summary('Roddy Piper')
wikipedia.page('Roddy Piper')
wikipedia.page('Roddy Piper').url
famous_page = wikipedia.page('Roddy Piper')
famous_page.summary
testimonial = TextBlob(famous_page.content)
testimonial.sentiment
```
Nearest Neighbors
```
people_df1 = [people_df.iloc[32819]['text']]
people_df1
nn = NearestNeighbors(metric='euclidean')
nn.fit(word_weight)
roddy_index = people_df[people_df['name'] == 'Roddy Piper'].index[0]
roddy_index
distances, indices = nn.kneighbors(word_weight[roddy_index], n_neighbors=11)
distances
indices
people_df.iloc[indices[0],:]
people_df.iloc[2037]['text']
people_df.iloc[18432]['text']
people_df.iloc[21038]['text']
people_df.iloc[35633]['text']
```
|
github_jupyter
|
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#Libraries-&-settings" data-toc-modified-id="Libraries-&-settings-1"><span class="toc-item-num">1 </span>Libraries & settings</a></span></li><li><span><a href="#Metrics" data-toc-modified-id="Metrics-2"><span class="toc-item-num">2 </span>Metrics</a></span><ul class="toc-item"><li><span><a href="#Crowd-related" data-toc-modified-id="Crowd-related-2.1"><span class="toc-item-num">2.1 </span>Crowd-related</a></span></li><li><span><a href="#Path-efficiency-related" data-toc-modified-id="Path-efficiency-related-2.2"><span class="toc-item-num">2.2 </span>Path efficiency-related</a></span></li><li><span><a href="#Control-related" data-toc-modified-id="Control-related-2.3"><span class="toc-item-num">2.3 </span>Control-related</a></span></li></ul></li><li><span><a href="#Pipeline" data-toc-modified-id="Pipeline-3"><span class="toc-item-num">3 </span>Pipeline</a></span><ul class="toc-item"><li><span><a href="#Result-loading" data-toc-modified-id="Result-loading-3.1"><span class="toc-item-num">3.1 </span>Result loading</a></span></li><li><span><a href="#Mean-Std-statistics" data-toc-modified-id="Mean-Std-statistics-3.2"><span class="toc-item-num">3.2 </span>Mean-Std statistics</a></span></li><li><span><a href="#ANOVA-test-for-controller-comparison" data-toc-modified-id="ANOVA-test-for-controller-comparison-3.3"><span class="toc-item-num">3.3 </span>ANOVA test for controller comparison</a></span></li><li><span><a href="#Visualize-with-grouping-by-date" data-toc-modified-id="Visualize-with-grouping-by-date-3.4"><span class="toc-item-num">3.4 </span>Visualize with grouping by date</a></span><ul class="toc-item"><li><span><a href="#Palette-settings" data-toc-modified-id="Palette-settings-3.4.1"><span class="toc-item-num">3.4.1 </span>Palette settings</a></span></li><li><span><a href="#Crowd-related-metrics" data-toc-modified-id="Crowd-related-metrics-3.4.2"><span class="toc-item-num">3.4.2 </span>Crowd-related metrics</a></span><ul class="toc-item"><li><span><a href="#4-in-1-plotting" data-toc-modified-id="4-in-1-plotting-3.4.2.1"><span class="toc-item-num">3.4.2.1 </span>4-in-1 plotting</a></span></li><li><span><a href="#Individual-figures" data-toc-modified-id="Individual-figures-3.4.2.2"><span class="toc-item-num">3.4.2.2 </span>Individual figures</a></span></li></ul></li><li><span><a href="#Path-efficiency-related-metrics" data-toc-modified-id="Path-efficiency-related-metrics-3.4.3"><span class="toc-item-num">3.4.3 </span>Path efficiency-related metrics</a></span><ul class="toc-item"><li><span><a href="#2-in-1-plotting" data-toc-modified-id="2-in-1-plotting-3.4.3.1"><span class="toc-item-num">3.4.3.1 </span>2-in-1 plotting</a></span></li><li><span><a href="#Individual-figures" data-toc-modified-id="Individual-figures-3.4.3.2"><span class="toc-item-num">3.4.3.2 </span>Individual figures</a></span></li></ul></li><li><span><a href="#Control-related-metrics" data-toc-modified-id="Control-related-metrics-3.4.4"><span class="toc-item-num">3.4.4 </span>Control-related metrics</a></span><ul class="toc-item"><li><span><a href="#4-in-1-plotting" data-toc-modified-id="4-in-1-plotting-3.4.4.1"><span class="toc-item-num">3.4.4.1 </span>4-in-1 plotting</a></span></li><li><span><a href="#Individual-figures" data-toc-modified-id="Individual-figures-3.4.4.2"><span class="toc-item-num">3.4.4.2 </span>Individual figures</a></span></li></ul></li></ul></li><li><span><a href="#Visualize-without-grouping-by-date" data-toc-modified-id="Visualize-without-grouping-by-date-3.5"><span class="toc-item-num">3.5 </span>Visualize without grouping by date</a></span><ul class="toc-item"><li><span><a href="#Palette-settings" data-toc-modified-id="Palette-settings-3.5.1"><span class="toc-item-num">3.5.1 </span>Palette settings</a></span></li><li><span><a href="#Crowd-related-metrics" data-toc-modified-id="Crowd-related-metrics-3.5.2"><span class="toc-item-num">3.5.2 </span>Crowd-related metrics</a></span><ul class="toc-item"><li><span><a href="#4-in-1-plotting" data-toc-modified-id="4-in-1-plotting-3.5.2.1"><span class="toc-item-num">3.5.2.1 </span>4-in-1 plotting</a></span></li><li><span><a href="#Individual-figures" data-toc-modified-id="Individual-figures-3.5.2.2"><span class="toc-item-num">3.5.2.2 </span>Individual figures</a></span></li></ul></li><li><span><a href="#Path-efficiency-related-metrics" data-toc-modified-id="Path-efficiency-related-metrics-3.5.3"><span class="toc-item-num">3.5.3 </span>Path efficiency-related metrics</a></span><ul class="toc-item"><li><span><a href="#2-in-1-plotting" data-toc-modified-id="2-in-1-plotting-3.5.3.1"><span class="toc-item-num">3.5.3.1 </span>2-in-1 plotting</a></span></li><li><span><a href="#Individual-figures" data-toc-modified-id="Individual-figures-3.5.3.2"><span class="toc-item-num">3.5.3.2 </span>Individual figures</a></span></li></ul></li><li><span><a href="#Control-related-metrics" data-toc-modified-id="Control-related-metrics-3.5.4"><span class="toc-item-num">3.5.4 </span>Control-related metrics</a></span><ul class="toc-item"><li><span><a href="#4-in-1-plotting" data-toc-modified-id="4-in-1-plotting-3.5.4.1"><span class="toc-item-num">3.5.4.1 </span>4-in-1 plotting</a></span></li><li><span><a href="#Individual-figures" data-toc-modified-id="Individual-figures-3.5.4.2"><span class="toc-item-num">3.5.4.2 </span>Individual figures</a></span></li></ul></li></ul></li></ul></li></ul></div>
# Controller comparison analysis
> Analysis of different control methods on 2021-04-10 and 2021-04-10 data
## Libraries & settings
```
import math
import datetime
import collections
import sys, os, fnmatch
from pathlib import Path
import numpy as np
import pandas as pd
import scipy.stats as stats
import matplotlib as mpl
import matplotlib.pyplot as plt
use_serif_font = True
if use_serif_font:
plt.style.use('./styles/serif.mplstyle')
else:
plt.style.use('./styles/sans_serif.mplstyle')
plt.ioff()
import seaborn as sns
sns.set_context("paper", font_scale=1.2, rc={"lines.linewidth": 1.3})
from qolo.utils.notebook_util import (
walk,
values2colors,
values2color_list,
violinplot,
categorical_plot,
barplot_annotate_brackets,
import_eval_res,
)
from qolo.core.crowdbot_data import CrowdBotDatabase, CrowdBotData
from qolo.metrics.metric_qolo_perf import compute_rel_jerk
```
## Metrics
### Crowd-related
1. Crowd Density (within an area of 2.5, 5m, 10m around the robot):
2. Minimum distance to pedestrians:
3. Number of violations to the virtual boundary set to the robot controller
```
crowd_metrics = (
'avg_crowd_density2_5',
'std_crowd_density2_5',
'max_crowd_density2_5',
'avg_crowd_density5',
'std_crowd_density5',
'max_crowd_density5',
'avg_min_dist',
'virtual_collision',
)
```
### Path efficiency-related
1. Relative time to goal (normalized by the goal distance)
2. Relative path length (normalized by the goal distance in straight line):
```
path_metrics = (
'rel_duration2goal',
'rel_path_length2goal',
'path_length2goal',
'duration2goal',
'min_dist2goal',
)
```
### Control-related
1. Agreement
2. Fluency
3. Contribution
4. Relative Jerk (smoothness of the path as added sum of linear and angular jerk)
```
control_metrics = (
'rel_jerk',
'avg_fluency',
'contribution',
'avg_agreement',
)
```
## Pipeline
```
qolo_dataset = CrowdBotData()
bagbase = qolo_dataset.bagbase_dir
outbase = qolo_dataset.outbase_dir
```
### Result loading
```
chosen_dates = ['0410', '0424']
chosen_type = ['mds', 'rds', 'shared_control']
eval_dirs = []
for root, dirs, files in walk(outbase, topdown=False, maxdepth=1):
for dir_ in dirs:
if any(s in dir_ for s in chosen_dates) and any(s in dir_ for s in chosen_type):
dir_ = dir_.replace("_processed", "")
eval_dirs.append(dir_)
print("{}/ is available!".format(dir_))
eval_res_df = import_eval_res(eval_dirs)
eval_res_df.head()
```
### Mean-Std statistics
```
for ctrl in chosen_type:
print(ctrl, ":", len(eval_res_df[eval_res_df.control_type == ctrl]))
frames_stat = []
for ctrl in chosen_type:
eval_res_df_ = eval_res_df[eval_res_df.control_type == ctrl]
stat_df = eval_res_df_.drop(['date'], axis=1).agg(['mean', 'std'])
if ctrl == 'shared_control':
stat_df.index = 'sc_'+stat_df.index.values
else:
stat_df.index = ctrl+'_'+stat_df.index.values
frames_stat.append(stat_df)
stat_df_all = pd.concat(frames_stat) # , ignore_index=True
stat_df_all.index.name = 'Metrics'
stat_df_all
export_metrics = (
'avg_crowd_density2_5',
'max_crowd_density2_5',
# 'avg_crowd_density5',
'avg_min_dist',
'rel_duration2goal',
'rel_path_length2goal',
'rel_jerk',
'contribution',
'avg_fluency',
'avg_agreement',
'virtual_collision',
)
export_control_df = stat_df_all[list(export_metrics)]
metrics_len = len(export_control_df.loc['mds_mean'])
methods = ['MDS', 'RDS', 'shared_control']
for idxx, method in enumerate(methods):
str_out = []
for idx in range(metrics_len):
avg = "${:0.2f}".format(round(export_control_df.iloc[2*idxx,idx],2))
std = "{:0.2f}$".format(round(export_control_df.iloc[2*idxx+1,idx],2))
str_out.append(avg+" \pm "+std)
export_control_df.loc[method] = str_out
export_contro_str_df = export_control_df.iloc[6:9]
export_contro_str_df
# print(export_contro_str_df.to_latex())
# print(export_contro_str_df.T.to_latex())
```
### ANOVA test for controller comparison
```
anova_metrics = (
'avg_crowd_density2_5',
'max_crowd_density2_5',
'avg_crowd_density5',
'avg_min_dist',
'virtual_collision',
'rel_duration2goal',
'rel_path_length2goal',
'rel_jerk',
'contribution',
'avg_fluency',
'avg_agreement',
)
mds_anova_ = eval_res_df[eval_res_df.control_type=='mds']
mds_metrics = mds_anova_[list(anova_metrics)].values
rds_anova_ = eval_res_df[eval_res_df.control_type=='rds']
rds_metrics = rds_anova_[list(anova_metrics)].values
shared_control_anova_ = eval_res_df[eval_res_df.control_type=='shared_control']
shared_control_metrics = shared_control_anova_[list(anova_metrics)].values
fvalue12, pvalue12 = stats.f_oneway(mds_metrics, rds_metrics)
fvalue23, pvalue23 = stats.f_oneway(mds_metrics, shared_control_metrics)
fvalue13, pvalue13 = stats.f_oneway(rds_metrics, shared_control_metrics)
# total
fvalue, pvalue = stats.f_oneway(mds_metrics, rds_metrics, shared_control_metrics)
statP_df = pd.DataFrame(
data=np.vstack((pvalue12, pvalue23, pvalue13, pvalue)),
index=['mds-rds', 'mds-shared', 'rds-shared', 'total'],
)
statP_df.columns = list(anova_metrics)
statP_df.index.name = 'Metrics'
statF_df = pd.DataFrame(
data=np.vstack((fvalue12, fvalue23, fvalue13, fvalue)),
index=['mds-rds', 'mds-shared', 'rds-shared', 'total'],
)
statF_df.columns = list(anova_metrics)
statF_df.index.name = 'Metrics'
statP_df
statF_df
# print(statF_df.T.to_latex())
# print(statP_df.T.to_latex())
# print(stat_df_all.T.to_latex())
```
### Visualize with grouping by date
#### Palette settings
```
dates=['0410', '0424']
value_unique, color_unique = values2color_list(
dates, cmap_name='hot', range=(0.55, 0.75)
)
value_unique, point_color_unique = values2color_list(
dates, cmap_name='hot', range=(0.3, 0.6)
)
# creating a dictionary with one specific color per group:
box_pal = {value_unique[i]: color_unique[i] for i in range(len(value_unique))}
# original: (0.3, 0.6)
scatter_pal = {value_unique[i]: point_color_unique[i] for i in range(len(value_unique))}
# black
# scatter_pal = {value_unique[i]: (0.0, 0.0, 0.0, 1.0) for i in range(len(value_unique))}
# gray
# scatter_pal = {value_unique[i]: (0.3, 0.3, 0.3, 0.8) for i in range(len(value_unique))}
box_pal, scatter_pal
```
#### Crowd-related metrics
```
crowd_metrics_df = eval_res_df[['seq', 'control_type'] + list(crowd_metrics) + ['date']]
for ctrl in chosen_type:
print("###", ctrl)
print("# mean")
print(crowd_metrics_df[crowd_metrics_df.control_type == ctrl].mean(numeric_only=True))
# print("# std")
# print(crowd_metrics_df[crowd_metrics_df.control_type == ctrl].std(numeric_only=True))
print()
print("# max value in each metrics")
print(crowd_metrics_df.max(numeric_only=True))
print("# min value in each metrics")
print(crowd_metrics_df.min(numeric_only=True))
```
##### 4-in-1 plotting
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
fig, axes = plt.subplots(2, 2, figsize=(16, 10))
categorical_plot(
axes=axes[0,0],
df=crowd_metrics_df,
metric='avg_crowd_density2_5',
category='control_type',
title='Mean crowd density within 2.5 m',
xlabel='',
ylabel='Density [1/$m^2$]',
ylim=[0.0, 0.25],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
axes[0,0].set_ylabel("Density [1/$m^2$]", fontsize=16)
axes[0,0].tick_params(axis='x', labelsize=16)
axes[0,0].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=axes[0,1],
df=crowd_metrics_df,
metric='max_crowd_density2_5',
category='control_type',
title='Max crowd density within 2.5 m',
xlabel='',
ylabel='Density [1/$m^2$]',
ylim=[0.3, 0.90],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
axes[0,1].set_ylabel("Density [1/$m^2$]", fontsize=16)
axes[0,1].tick_params(axis='x', labelsize=16)
axes[0,1].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=axes[1,0],
df=crowd_metrics_df,
metric='virtual_collision',
category='control_type',
title='Virtual collision with Qolo',
xlabel='',
ylabel='',
ylim=[-0.1, 20],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
axes[1,0].set_ylabel("Virtual collision", fontsize=16)
axes[1,0].tick_params(axis='x', labelsize=16)
axes[1,0].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=axes[1,1],
df=crowd_metrics_df,
metric='avg_min_dist',
category='control_type',
title='Min. distance of Pedestrain from qolo',
xlabel='',
ylabel='Distance [m]',
ylim=[0.6, 2.0],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
axes[1,1].set_ylabel("Distance [m]", fontsize=16)
axes[1,1].tick_params(axis='x', labelsize=16)
axes[1,1].tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/comp_crowd_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
```
##### Individual figures
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig1, control_axes1 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes1,
df=crowd_metrics_df,
metric='avg_crowd_density2_5',
category='control_type',
title='',
xlabel='',
ylabel='Density [1/$m^2$]',
ylim=[0.0, 0.25],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes1.set_ylabel("Density [1/$m^2$]", fontsize=16)
control_axes1.tick_params(axis='x', labelsize=16)
control_axes1.tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/pub/control_boxplot_mean_density_2_5_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig2, control_axes2 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes2,
df=crowd_metrics_df,
metric='max_crowd_density2_5',
category='control_type',
title='',
xlabel='',
ylabel='Density [1/$m^2$]',
ylim=[0.3, 0.90],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes2.set_ylabel("Density [1/$m^2$]", fontsize=16)
control_axes2.tick_params(axis='x', labelsize=16)
control_axes2.tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/pub/control_boxplot_max_density_2_5_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig3, control_axes3 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes3,
df=crowd_metrics_df,
metric='virtual_collision',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[-0.1, 20],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes3.set_ylabel("Virtual collision", fontsize=16)
control_axes3.tick_params(axis='x', labelsize=16)
control_axes3.tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/pub/control_boxplot_virtual_collision_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig4, control_axes4 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes4,
df=crowd_metrics_df,
metric='avg_min_dist',
category='control_type',
title='',
xlabel='',
ylabel='Distance [m]',
ylim=[0.6, 2.0],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes4.set_ylabel("Distance [m]", fontsize=16)
control_axes4.tick_params(axis='x', labelsize=16)
control_axes4.tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/pub/control_boxplot_mean_min_dist_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
```
#### Path efficiency-related metrics
```
path_metrics_df = eval_res_df[['seq', 'control_type'] + list(path_metrics) + ['date']]
print("# max value in each metrics")
print(path_metrics_df.max(numeric_only=True))
print("# min value in each metrics")
print(path_metrics_df.min(numeric_only=True))
```
##### 2-in-1 plotting
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
path_fig, path_axes = plt.subplots(1, 2, figsize=(16, 5))
categorical_plot(
axes=path_axes[0],
df=path_metrics_df,
metric='rel_duration2goal',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.0, 1.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
path_axes[0].set_ylabel("Relative time to the goal", fontsize=16)
path_axes[0].tick_params(axis='x', labelsize=16)
path_axes[0].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=path_axes[1],
df=path_metrics_df,
metric='rel_path_length2goal',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.0, 3.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
path_axes[1].set_ylabel("Relative path length to the goal", fontsize=16)
path_axes[1].tick_params(axis='x', labelsize=16)
path_axes[1].tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/comp_path_efficiency_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
```
##### Individual figures
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig5, control_axes5 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes5,
df=path_metrics_df,
metric='rel_duration2goal',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.0, 1.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes5.set_ylabel("Relative time to the goal", fontsize=16)
control_axes5.tick_params(axis='x', labelsize=16)
control_axes5.tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/pub/control_boxplot_rel_time2goal_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig6, control_axes6 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes6,
df=path_metrics_df,
metric='rel_path_length2goal',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[1.0, 2.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes6.set_ylabel("Relative path length to the goal", fontsize=16)
control_axes6.tick_params(axis='x', labelsize=16)
control_axes6.tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/pub/control_boxplot_rel_path_length2goal_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
```
#### Control-related metrics
```
control_metrics_df = eval_res_df[['seq', 'control_type'] + list(control_metrics) + ['date']]
print("# max value in each metrics")
print(control_metrics_df.max(numeric_only=True))
print("# min value in each metrics")
print(control_metrics_df.min(numeric_only=True))
```
##### 4-in-1 plotting
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig, control_axes = plt.subplots(2, 2, figsize=(16, 12))
categorical_plot(
axes=control_axes[0,0],
df=control_metrics_df,
metric='avg_fluency',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.90, 1.02],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes[0,0].set_ylabel("Average control fluency", fontsize=16)
control_axes[0,0].tick_params(axis='x', labelsize=16)
control_axes[0,0].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=control_axes[0,1],
df=control_metrics_df,
metric='rel_jerk',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0, 0.35],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes[0,1].set_ylabel("Relative jerk", fontsize=16)
control_axes[0,1].tick_params(axis='x', labelsize=16)
control_axes[0,1].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=control_axes[1,0],
df=control_metrics_df,
metric='contribution',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.0, 1.2],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes[1,0].set_ylabel("Contribution", fontsize=16)
control_axes[1,0].tick_params(axis='x', labelsize=16)
control_axes[1,0].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=control_axes[1,1],
df=control_metrics_df,
metric='avg_agreement',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.5, 1.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes[1,1].set_ylabel("Average agreement", fontsize=16)
control_axes[1,1].tick_params(axis='x', labelsize=16)
control_axes[1,1].tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/comp_control_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
```
##### Individual figures
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig7, control_axes7 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes7,
df=control_metrics_df,
metric='avg_fluency',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.90, 1.02],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes7.set_ylabel("Average control fluency", fontsize=16)
control_axes7.tick_params(axis='x', labelsize=16)
control_axes7.tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/pub/control_boxplot_avg_fluency_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig8, control_axes8 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes8,
df=control_metrics_df,
metric='rel_jerk',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0, 0.35],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes8.set_ylabel("Relative jerk", fontsize=16)
control_axes8.tick_params(axis='x', labelsize=16)
control_axes8.tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/pub/control_boxplot_rel_jerk_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig9, control_axes9 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes9,
df=control_metrics_df,
metric='contribution',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.0, 1.2],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes9.set_ylabel("Contribution", fontsize=16)
control_axes9.tick_params(axis='x', labelsize=16)
control_axes9.tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/pub/control_boxplot_contribution_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig10, control_axes10 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes10,
df=control_metrics_df,
metric='avg_agreement',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.5, 1.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes10.set_ylabel("Average agreement", fontsize=16)
control_axes10.tick_params(axis='x', labelsize=16)
control_axes10.tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/pub/control_boxplot_avg_agreement_group_by_date.pdf", dpi=300)
plt.show()
plt.close()
crowd_metrics_df0424 = crowd_metrics_df[crowd_metrics_df.date=='0424'].sort_values('control_type', ascending=False)
print("Sequence on 0424")
print(crowd_metrics_df0424['control_type'].value_counts())
crowd_metrics_df0410 = crowd_metrics_df[crowd_metrics_df.date=='0410'].sort_values(by=['control_type'], ascending=False, ignore_index=True).reindex()
print("Sequence on 0410")
print(crowd_metrics_df0410['control_type'].value_counts())
```
### Visualize without grouping by date
#### Palette settings
```
control_methods=['mds', 'rds', 'shared_control']
value_unique, color_unique = values2color_list(
eval_res_df['control_type'].values, cmap_name='hot', range=(0.55, 0.75)
)
value_unique, point_color_unique = values2color_list(
eval_res_df['control_type'].values, cmap_name='hot', range=(0.35, 0.5)
)
# creating a dictionary with one specific color per group:
box_pal = {value_unique[i]: color_unique[i] for i in range(len(value_unique))}
# original: (0.3, 0.6)
# scatter_pal = {value_unique[i]: point_color_unique[i] for i in range(len(value_unique))}
# black
# scatter_pal = {value_unique[i]: (0.0, 0.0, 0.0, 1.0) for i in range(len(value_unique))}
# gray
scatter_pal = {value_unique[i]: (0.3, 0.3, 0.3, 0.8) for i in range(len(value_unique))}
box_pal, scatter_pal
```
#### Crowd-related metrics
##### 4-in-1 plotting
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
fig, axes = plt.subplots(2, 2, figsize=(16, 10))
categorical_plot(
axes=axes[0,0],
df=crowd_metrics_df,
metric='avg_crowd_density2_5',
category='control_type',
title='Mean crowd density within 2.5 m',
xlabel='',
ylabel='Density [1/$m^2$]',
ylim=[0.05, 0.20],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
#group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
axes[0,0].set_ylabel("Density [1/$m^2$]", fontsize=16)
axes[0,0].tick_params(axis='x', labelsize=16)
axes[0,0].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=axes[0,1],
df=crowd_metrics_df,
metric='max_crowd_density2_5',
category='control_type',
title='Max crowd density within 2.5 m',
xlabel='',
ylabel='Density [1/$m^2$]',
ylim=[0.3, 0.90],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
#group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
axes[0,1].set_ylabel("Density [1/$m^2$]", fontsize=16)
axes[0,1].tick_params(axis='x', labelsize=16)
axes[0,1].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=axes[1,0],
df=crowd_metrics_df,
metric='virtual_collision',
category='control_type',
title='Virtual collision with Qolo',
xlabel='',
ylabel='',
ylim=[-0.1, 20],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
#group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
axes[1,0].set_ylabel("Virtual collision", fontsize=16)
axes[1,0].tick_params(axis='x', labelsize=16)
axes[1,0].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=axes[1,1],
df=crowd_metrics_df,
metric='avg_min_dist',
category='control_type',
title='Min. distance of Pedestrain from qolo',
xlabel='',
ylabel='Distance [m]',
ylim=[0.6, 1.6],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
#group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
axes[1,1].set_ylabel("Distance [m]", fontsize=16)
axes[1,1].tick_params(axis='x', labelsize=16)
axes[1,1].tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/comp_crowd.pdf", dpi=300)
plt.show()
plt.close()
```
##### Individual figures
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig1, control_axes1 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes1,
df=crowd_metrics_df,
metric='avg_crowd_density2_5',
category='control_type',
title='',
xlabel='',
ylabel='Density [1/$m^2$]',
ylim=[0.05, 0.20],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes1.set_ylabel("Density [1/$m^2$]", fontsize=16)
control_axes1.tick_params(axis='x', labelsize=16)
control_axes1.tick_params(axis='y', labelsize=14)
control_axes1.set_xticks([0,1,2])
control_axes1.set_xticklabels(['MDS','RDS','SC'], fontsize=16)
plt.savefig("./pdf/pub/control_boxplot_mean_density_2_5.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig2, control_axes2 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes2,
df=crowd_metrics_df,
metric='max_crowd_density2_5',
category='control_type',
title='',
xlabel='',
ylabel='Density [1/$m^2$]',
ylim=[0.2, 0.90],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes2.set_ylabel("Density [1/$m^2$]", fontsize=16)
control_axes2.tick_params(axis='x', labelsize=16)
control_axes2.tick_params(axis='y', labelsize=14)
control_axes2.set_xticks([0,1,2])
control_axes2.set_xticklabels(['MDS','RDS','SC'], fontsize=16)
plt.savefig("./pdf/pub/control_boxplot_max_density_2_5.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig3, control_axes3 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes3,
df=crowd_metrics_df,
metric='virtual_collision',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[-0.1, 15],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes3.set_ylabel("Virtual collision", fontsize=16)
control_axes3.tick_params(axis='x', labelsize=16)
control_axes3.tick_params(axis='y', labelsize=14)
control_axes3.set_xticks([0,1,2])
control_axes3.set_xticklabels(['MDS','RDS','SC'], fontsize=16)
plt.savefig("./pdf/pub/control_boxplot_virtual_collision.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig4, control_axes4 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes4,
df=crowd_metrics_df,
metric='avg_min_dist',
category='control_type',
title='',
xlabel='',
ylabel='Distance [m]',
ylim=[0.6, 1.6],
kind='box',
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes4.set_ylabel("Distance [m]", fontsize=16)
control_axes4.tick_params(axis='x', labelsize=16)
control_axes4.tick_params(axis='y', labelsize=14)
control_axes4.set_xticks([0,1,2])
control_axes4.set_xticklabels(['MDS','RDS','SC'], fontsize=16)
plt.savefig("./pdf/pub/control_boxplot_mean_min_dist.pdf", dpi=300)
plt.show()
plt.close()
```
#### Path efficiency-related metrics
##### 2-in-1 plotting
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
path_fig, path_axes = plt.subplots(1, 2, figsize=(16, 5))
categorical_plot(
axes=path_axes[0],
df=path_metrics_df,
metric='rel_duration2goal',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.0, 1.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
path_axes[0].set_ylabel("Relative time to the goal", fontsize=16)
path_axes[0].tick_params(axis='x', labelsize=16)
path_axes[0].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=path_axes[1],
df=path_metrics_df,
metric='rel_path_length2goal',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[1.0, 2.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
path_axes[1].set_ylabel("Relative path length to the goal", fontsize=16)
path_axes[1].tick_params(axis='x', labelsize=16)
path_axes[1].tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/comp_path_efficiency.pdf", dpi=300)
plt.show()
plt.close()
```
##### Individual figures
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig5, control_axes5 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes5,
df=path_metrics_df,
metric='rel_duration2goal',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.0, 1.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes5.set_ylabel("Relative time to the goal", fontsize=16)
control_axes5.tick_params(axis='x', labelsize=16)
control_axes5.tick_params(axis='y', labelsize=14)
control_axes5.set_xticks([0,1,2])
control_axes5.set_xticklabels(['MDS','RDS','SC'], fontsize=16)
plt.savefig("./pdf/pub/control_boxplot_rel_time2goal.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig6, control_axes6 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes6,
df=path_metrics_df,
metric='rel_path_length2goal',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[1.0, 2.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes6.set_ylabel("Relative path length to the goal", fontsize=16)
control_axes6.tick_params(axis='x', labelsize=16)
control_axes6.tick_params(axis='y', labelsize=14)
control_axes6.set_xticks([0,1,2])
control_axes6.set_xticklabels(['MDS','RDS','SC'], fontsize=16)
plt.savefig("./pdf/pub/control_boxplot_rel_path_length2goal.pdf", dpi=300)
plt.show()
plt.close()
```
#### Control-related metrics
##### 4-in-1 plotting
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig, control_axes = plt.subplots(2, 2, figsize=(16, 12))
categorical_plot(
axes=control_axes[0,0],
df=control_metrics_df,
metric='avg_fluency',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.90, 1.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes[0,0].set_ylabel("Average control fluency", fontsize=16)
control_axes[0,0].tick_params(axis='x', labelsize=16)
control_axes[0,0].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=control_axes[0,1],
df=control_metrics_df,
metric='rel_jerk',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0, 0.3],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes[0,1].set_ylabel("Relative jerk", fontsize=16)
control_axes[0,1].tick_params(axis='x', labelsize=16)
control_axes[0,1].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=control_axes[1,0],
df=control_metrics_df,
metric='contribution',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.0, 1.2],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes[1,0].set_ylabel("Contribution", fontsize=16)
control_axes[1,0].tick_params(axis='x', labelsize=16)
control_axes[1,0].tick_params(axis='y', labelsize=14)
categorical_plot(
axes=control_axes[1,1],
df=control_metrics_df,
metric='avg_agreement',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.5, 1.0],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes[1,1].set_ylabel("Average agreement", fontsize=16)
control_axes[1,1].tick_params(axis='x', labelsize=16)
control_axes[1,1].tick_params(axis='y', labelsize=14)
plt.savefig("./pdf/comp_control.pdf", dpi=300)
plt.show()
plt.close()
```
##### Individual figures
```
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig7, control_axes7 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes7,
df=control_metrics_df,
metric='avg_fluency',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.90, 1.06],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes7.set_ylabel("Average control fluency", fontsize=16)
control_axes7.tick_params(axis='x', labelsize=16)
control_axes7.tick_params(axis='y', labelsize=14)
control_axes7.set_xticks([0,1,2])
control_axes7.set_xticklabels(['MDS','RDS','SC'], fontsize=16)
# significance
bars = [0, 1, 2]
heights = [0.99, 1.0, 1.03]
barplot_annotate_brackets(0, 1, 3.539208e-04, bars, heights, line_y=1.00)
barplot_annotate_brackets(0, 2, 4.194127e-03, bars, heights, line_y=1.03)
barplot_annotate_brackets(1, 2, 7.744226e-10, bars, heights, line_y=1.015)
plt.savefig("./pdf/pub/control_boxplot_avg_fluency.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig8, control_axes8 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes8,
df=control_metrics_df,
metric='rel_jerk',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0, 0.30],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes8.set_ylabel("Relative jerk", fontsize=16)
control_axes8.tick_params(axis='x', labelsize=16)
control_axes8.tick_params(axis='y', labelsize=14)
control_axes8.set_xticks([0,1,2])
control_axes8.set_xticklabels(['MDS','RDS','SC'], fontsize=16)
# significance
bars = [0, 1, 2]
heights = [0.99, 1.0, 1.03]
barplot_annotate_brackets(0, 1, 1.022116e-02, bars, heights, line_y=0.265)
barplot_annotate_brackets(0, 2, 2.421626e-01, bars, heights, line_y=0.30)
barplot_annotate_brackets(1, 2, 2.126847e-07, bars, heights, line_y=0.19)
plt.savefig("./pdf/pub/control_boxplot_rel_jerk.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig9, control_axes9 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes9,
df=control_metrics_df,
metric='contribution',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.0, 1.4],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes9.set_ylabel("Contribution", fontsize=16)
control_axes9.tick_params(axis='x', labelsize=16)
control_axes9.tick_params(axis='y', labelsize=14)
control_axes9.set_xticks([0,1,2])
control_axes9.set_xticklabels(['MDS','RDS','SC'], fontsize=16)
# significance
bars = [0, 1, 2]
heights = [0.99, 1.0, 1.03]
barplot_annotate_brackets(0, 1, 1.701803e-10, bars, heights, line_y=1.15)
barplot_annotate_brackets(0, 2, 1.271729e-01, bars, heights, line_y=1.2)
barplot_annotate_brackets(1, 2, 3.495410e-09, bars, heights, line_y=1.25)
plt.savefig("./pdf/pub/control_boxplot_contribution.pdf", dpi=300)
plt.show()
plt.close()
mpl.rcParams['font.family'] = ['serif']
mpl.rcParams['font.serif'] = ['Times New Roman']
mpl.rcParams['mathtext.fontset'] = 'cm'
control_fig10, control_axes10 = plt.subplots(figsize=(6, 5))
categorical_plot(
axes=control_axes10,
df=control_metrics_df,
metric='avg_agreement',
category='control_type',
title='',
xlabel='',
ylabel='',
ylim=[0.5, 1.1],
lgd_labels=['April 10, 2021', 'April 24, 2021'],
lgd_font="Times New Roman",
kind='box',
# group='date',
loc='upper left',
box_palette=box_pal,
scatter_palette=scatter_pal,
)
control_axes10.set_ylabel("Average agreement", fontsize=16)
control_axes10.tick_params(axis='x', labelsize=16)
control_axes10.tick_params(axis='y', labelsize=14)
control_axes10.set_xticks([0,1,2])
control_axes10.set_xticklabels(['MDS','RDS','SC'], fontsize=16)
# significance
bars = [0, 1, 2]
heights = [0.99, 1.0, 1.03]
barplot_annotate_brackets(0, 1, 5.248126e-02, bars, heights, line_y=0.82)
barplot_annotate_brackets(0, 2, 4.394447e-12, bars, heights, line_y=1.0)
barplot_annotate_brackets(1, 2, 3.542947e-15, bars, heights, line_y=0.94)
plt.savefig("./pdf/pub/control_boxplot_avg_agreement.pdf", dpi=300)
plt.show()
plt.close()
```
|
github_jupyter
|
# Introduction to Modeling Libraries
```
import numpy as np
import pandas as pd
np.random.seed(12345)
import matplotlib.pyplot as plt
plt.rc('figure', figsize=(10, 6))
PREVIOUS_MAX_ROWS = pd.options.display.max_rows
pd.options.display.max_rows = 20
np.set_printoptions(precision=4, suppress=True)
```
## Interfacing Between pandas and Model Code
```
import pandas as pd
import numpy as np
data = pd.DataFrame({
'x0': [1, 2, 3, 4, 5],
'x1': [0.01, -0.01, 0.25, -4.1, 0.],
'y': [-1.5, 0., 3.6, 1.3, -2.]})
data
data.columns
data.values
df2 = pd.DataFrame(data.values, columns=['one', 'two', 'three'])
df2
model_cols = ['x0', 'x1']
data.loc[:, model_cols].values
data['category'] = pd.Categorical(['a', 'b', 'a', 'a', 'b'],
categories=['a', 'b'])
data
dummies = pd.get_dummies(data.category, prefix='category')
data_with_dummies = data.drop('category', axis=1).join(dummies)
data_with_dummies
```
## Creating Model Descriptions with Patsy
y ~ x0 + x1
```
data = pd.DataFrame({
'x0': [1, 2, 3, 4, 5],
'x1': [0.01, -0.01, 0.25, -4.1, 0.],
'y': [-1.5, 0., 3.6, 1.3, -2.]})
data
import patsy
y, X = patsy.dmatrices('y ~ x0 + x1', data)
y
X
np.asarray(y)
np.asarray(X)
patsy.dmatrices('y ~ x0 + x1 + 0', data)[1]
coef, resid, _, _ = np.linalg.lstsq(X, y)
coef
coef = pd.Series(coef.squeeze(), index=X.design_info.column_names)
coef
```
### Data Transformations in Patsy Formulas
```
y, X = patsy.dmatrices('y ~ x0 + np.log(np.abs(x1) + 1)', data)
X
y, X = patsy.dmatrices('y ~ standardize(x0) + center(x1)', data)
X
new_data = pd.DataFrame({
'x0': [6, 7, 8, 9],
'x1': [3.1, -0.5, 0, 2.3],
'y': [1, 2, 3, 4]})
new_X = patsy.build_design_matrices([X.design_info], new_data)
new_X
y, X = patsy.dmatrices('y ~ I(x0 + x1)', data)
X
```
### Categorical Data and Patsy
```
data = pd.DataFrame({
'key1': ['a', 'a', 'b', 'b', 'a', 'b', 'a', 'b'],
'key2': [0, 1, 0, 1, 0, 1, 0, 0],
'v1': [1, 2, 3, 4, 5, 6, 7, 8],
'v2': [-1, 0, 2.5, -0.5, 4.0, -1.2, 0.2, -1.7]
})
y, X = patsy.dmatrices('v2 ~ key1', data)
X
y, X = patsy.dmatrices('v2 ~ key1 + 0', data)
X
y, X = patsy.dmatrices('v2 ~ C(key2)', data)
X
data['key2'] = data['key2'].map({0: 'zero', 1: 'one'})
data
y, X = patsy.dmatrices('v2 ~ key1 + key2', data)
X
y, X = patsy.dmatrices('v2 ~ key1 + key2 + key1:key2', data)
X
```
## Introduction to statsmodels
### Estimating Linear Models
```
import statsmodels.api as sm
import statsmodels.formula.api as smf
def dnorm(mean, variance, size=1):
if isinstance(size, int):
size = size,
return mean + np.sqrt(variance) * np.random.randn(*size)
# For reproducibility
np.random.seed(12345)
N = 100
X = np.c_[dnorm(0, 0.4, size=N),
dnorm(0, 0.6, size=N),
dnorm(0, 0.2, size=N)]
eps = dnorm(0, 0.1, size=N)
beta = [0.1, 0.3, 0.5]
y = np.dot(X, beta) + eps
X[:5]
y[:5]
X_model = sm.add_constant(X)
X_model[:5]
model = sm.OLS(y, X)
results = model.fit()
results.params
print(results.summary())
data = pd.DataFrame(X, columns=['col0', 'col1', 'col2'])
data['y'] = y
data[:5]
results = smf.ols('y ~ col0 + col1 + col2', data=data).fit()
results.params
results.tvalues
results.predict(data[:5])
```
### Estimating Time Series Processes
```
init_x = 4
import random
values = [init_x, init_x]
N = 1000
b0 = 0.8
b1 = -0.4
noise = dnorm(0, 0.1, N)
for i in range(N):
new_x = values[-1] * b0 + values[-2] * b1 + noise[i]
values.append(new_x)
MAXLAGS = 5
model = sm.tsa.AR(values)
results = model.fit(MAXLAGS)
results.params
```
## Introduction to scikit-learn
```
train = pd.read_csv('datasets/titanic/train.csv')
test = pd.read_csv('datasets/titanic/test.csv')
train[:4]
train.isnull().sum()
test.isnull().sum()
impute_value = train['Age'].median()
train['Age'] = train['Age'].fillna(impute_value)
test['Age'] = test['Age'].fillna(impute_value)
train['IsFemale'] = (train['Sex'] == 'female').astype(int)
test['IsFemale'] = (test['Sex'] == 'female').astype(int)
predictors = ['Pclass', 'IsFemale', 'Age']
X_train = train[predictors].values
X_test = test[predictors].values
y_train = train['Survived'].values
X_train[:5]
y_train[:5]
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
model.fit(X_train, y_train)
y_predict = model.predict(X_test)
y_predict[:10]
```
(y_true == y_predict).mean()
```
from sklearn.linear_model import LogisticRegressionCV
model_cv = LogisticRegressionCV(10)
model_cv.fit(X_train, y_train)
from sklearn.model_selection import cross_val_score
model = LogisticRegression(C=10)
scores = cross_val_score(model, X_train, y_train, cv=4)
scores
```
## Continuing Your Education
```
pd.options.display.max_rows = PREVIOUS_MAX_ROWS
```
|
github_jupyter
|
```
import torch
# Check if pytorch is using GPU:
print('Used device name: {}'.format(torch.cuda.get_device_name(0)))
```
Import your google drive if necessary.
```
from google.colab import drive
drive.mount('/content/drive')
import sys
import os
ROOT_DIR = 'your_dir'
sys.path.insert(0, ROOT_DIR)
import pickle
import numpy as np
import pandas as pd
import torch
import matplotlib.pyplot as plt
import matplotlib.cm as cm
from mpl_toolkits.mplot3d import Axes3D
from sklearn.manifold import TSNE
% matplotlib inline
```
After trraining preprocessing the data and training the model, load all the needed files.
```
resources_dir = os.path.join(ROOT_DIR, 'resources', '')
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
vocabulary = pickle.load(open(os.path.join(os.path.join(resources_dir, 'vocabulary'), 'vocabulary.pickle'), 'rb'))
word2vec_path = 'your_path/idx2vec.pickle'
word2idx = pickle.load(open(os.path.join(os.path.join(resources_dir, 'word2idx'), 'word2idx.pickle'), 'rb'))
idx2word = pickle.load(open(os.path.join(os.path.join(resources_dir, 'idx2word'), 'idx2word.pickle'), 'rb'))
word_count = pickle.load(open(os.path.join(os.path.join(resources_dir, 'word_counts'), 'word_counts.pickle'), 'rb'))
embeddings_weigths = pickle.load(open(word2vec_path, 'rb'))
embeddings_weigths = torch.tensor(embeddings_weigths).to(device)
embeddings_weigths[1]
```
Define the cosine similarity between two vectors.
```
def cosine_sim(x_vector, y_vector):
dot_prod = torch.dot(x_vector.T, y_vector)
vector_norms = torch.sqrt(torch.sum(x_vector**2)) * torch.sqrt(torch.sum(y_vector**2))
similarity = dot_prod / vector_norms
return similarity
```
Plot results from t-SNE for a group of selected words.
```
test_words = ['frodo', 'gandalf', 'gimli', 'saruman', 'sauron', 'aragorn', 'ring', 'bilbo',
'shire', 'gondor', 'sam', 'pippin', 'baggins', 'legolas',
'gollum', 'elrond', 'isengard', 'king', 'merry', 'elf']
test_idx = [word2idx[word] for word in test_words]
test_embds = embeddings_weigths[test_idx]
tsne = TSNE(perplexity=5, n_components=2, init='pca', n_iter=10000, random_state=12,
verbose=1)
test_embds_2d = tsne.fit_transform(test_embds.cpu().numpy())
plt.figure(figsize = (9, 9), dpi=120)
for idx, word in enumerate(test_words):
plt.scatter(test_embds_2d[idx][0], test_embds_2d[idx][1])
plt.annotate(word, xy = (test_embds_2d[idx][0], test_embds_2d[idx][1]), \
ha='right',va='bottom')
plt.show()
```
Compute cosine similarities for a group of selected words.
```
words = ['frodo', 'gandalf', 'gimli', 'saruman', 'sauron', 'aragorn', 'ring', 'bilbo',
'shire', 'gondor', 'sam', 'pippin', 'baggins', 'legolas',
'gollum', 'elrond', 'isengard', 'king', 'merry', 'elf']
words_idx = [word2idx[word] for word in words]
embeddings_words = [embeddings_weigths[idx] for idx in words_idx]
top_num = 5
t = tqdm(embeddings_words)
t.set_description('Checking words for similarities')
similarities = {}
for idx_1, word_1 in enumerate(t):
key_word = words[idx_1]
similarities[key_word] = []
for idx_2, word_2 in enumerate(embeddings_weigths):
# the first two elements in vocab are padding word and unk word
if idx_2 > 1:
similarity = float(cosine_sim(word_1, word_2))
if word2idx[key_word] != idx_2:
similarities[key_word].append([idx2word[idx_2], similarity])
similarities[key_word].sort(key= lambda x: x[1])
similarities[key_word] = similarities[key_word][:-top_num-1:-1]
for key in similarities:
for item in similarities[key]:
item[1] = round(item[1], 4)
```
Format the results and convert them into a pandas dataframe.
```
formated_sim = {}
for key in similarities:
temp_list = []
for items in similarities[key]:
string = '"{}": {}'.format(items[0], items[1])
temp_list.append(string)
formated_sim[key] = temp_list
df = pd.DataFrame(data=formated_sim)
df
```
|
github_jupyter
|
# Notebook version of NSGA-II constrained, without scoop
```
%matplotlib inline
#!/usr/bin/env python
# This file is part of DEAP.
#
# DEAP is free software: you can redistribute it and/or modify
# it under the terms of the GNU Lesser General Public License as
# published by the Free Software Foundation, either version 3 of
# the License, or (at your option) any later version.
#
# DEAP is distributed in the hope that it will be useful,
# but WITHOUT ANY WARRANTY; without even the implied warranty of
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
# GNU Lesser General Public License for more details.
#
# You should have received a copy of the GNU Lesser General Public
# License along with DEAP. If not, see <http://www.gnu.org/licenses/>.
import array
import random
import json
import time
import numpy
from math import sqrt, cos, atan
#from scoop import futures
from deap import algorithms
#from deap import base
from deap import benchmarks
from deap.benchmarks.tools import diversity, convergence
from deap import creator
from deap import base, tools
from xopt import fitness_with_constraints # Chris' custom routines
from deap.benchmarks.tools import diversity, convergence, hypervolume
creator.create("FitnessMin", fitness_with_constraints.FitnessWithConstraints, weights=(-1.0, -1.0, 1.0, 1.0))
creator.create("Individual", array.array, typecode='d', fitness=creator.FitnessMin)
toolbox = base.Toolbox()
def uniform(low, up, size=None):
try:
return [random.uniform(a, b) for a, b in zip(low, up)]
except TypeError:
return [random.uniform(a, b) for a, b in zip([low] * size, [up] * size)]
NDIM = 2
N_CONSTRAINTS = 2
#BOUND_LOW, BOUND_UP = [0.1, 0.0] , [1.0, 1.0]
def CONSTR(individual):
#time.sleep(.01)
x1=individual[0]
x2=individual[1]
objectives = (x1, (1.0+x2)/x1)
constraints = (x2+9*x1-6.0, -x2+9*x1-1.0)
return (objectives, constraints)
BOUND_LOW, BOUND_UP = [0.0, 0.0], [3.14159, 3.14159]
def TNK(individual):
x1=individual[0]
x2=individual[1]
objectives = (x1, x2)
constraints = (x1**2+x2**2-1.0 - 0.1*cos(16*atan(x1/x2)), 0.5-(x1-0.5)**2-(x2-0.5)**2 )
return (objectives, constraints, (x1, x2))
#BOUND_LOW, BOUND_UP = [-20.0, -20.0], [20.0, 20.0]
def SRN(individual):
x1=individual[0]
x2=individual[1]
objectives = ( (x1-2.0)**2 + (x2-1.0)**2+2.0, 9*x1-(x2-1.0)**2 )
constraints = (225.0-x1**2-x2**2, -10.0 -x1 - 3*x2 )
return (objectives, constraints)
toolbox.register("attr_float", uniform, BOUND_LOW, BOUND_UP, NDIM)
toolbox.register("individual", tools.initIterate, creator.Individual, toolbox.attr_float)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
# scoop map function
#toolbox.register('map', futures.map)
toolbox.register('map', map)
#toolbox.register("evaluate", CONSTR)
toolbox.register("evaluate", TNK)
#toolbox.register("evaluate", SRN)
toolbox.register("mate", tools.cxSimulatedBinaryBounded, low=BOUND_LOW, up=BOUND_UP, eta=20.0)
toolbox.register("mutate", tools.mutPolynomialBounded, low=BOUND_LOW, up=BOUND_UP, eta=20.0, indpb=1.0/NDIM)
toolbox.register("select", tools.selNSGA2)
def main(seed=None):
random.seed(seed)
NGEN = 50
MU = 100
CXPB = 0.9
stats = tools.Statistics(lambda ind: ind.fitness.values)
stats.register("avg", numpy.mean, axis=0)
stats.register("std", numpy.std, axis=0)
stats.register("min", numpy.min, axis=0)
stats.register("max", numpy.max, axis=0)
logbook = tools.Logbook()
logbook.header = "gen", "evals", "std", "min", "avg", "max"
pop = toolbox.population(n=MU)
# Evaluate the individuals with an invalid fitness
invalid_ind = [ind for ind in pop if not ind.fitness.valid]
evaluate_result = toolbox.map(toolbox.evaluate, invalid_ind)
for ind, fit in zip(invalid_ind, evaluate_result):
ind.fitness.values = fit[0]
ind.fitness.cvalues = fit[1]
ind.fitness.n_constraints = len(fit[1])
# This is just to assign the crowding distance to the individuals
# no actual selection is done
pop = toolbox.select(pop, len(pop))
record = stats.compile(pop)
logbook.record(gen=0, evals=len(invalid_ind), **record)
print(logbook.stream)
# Begin the generational process
for gen in range(1, NGEN):
# Vary the population
offspring = tools.selTournamentDCD(pop, len(pop))
offspring = [toolbox.clone(ind) for ind in offspring]
for ind1, ind2 in zip(offspring[::2], offspring[1::2]):
if random.random() <= CXPB:
toolbox.mate(ind1, ind2)
toolbox.mutate(ind1)
toolbox.mutate(ind2)
del ind1.fitness.values, ind2.fitness.values
# Evaluate the individuals with an invalid fitness
invalid_ind = [ind for ind in offspring if not ind.fitness.valid]
fitnesses = toolbox.map(toolbox.evaluate, invalid_ind)
for ind, fit in zip(invalid_ind, fitnesses):
ind.fitness.values = fit[0]
ind.fitness.cvalues = fit[1]
ind.fitness.n_constraints = len(fit[1])
# Allow for additional info to be saved (for example, a dictionary of properties)
if len(fit) > 2:
ind.fitness.info = fit[2]
# Select the next generation population
pop = toolbox.select(pop + offspring, MU)
record = stats.compile(pop)
logbook.record(gen=gen, evals=len(invalid_ind), **record)
print(logbook.stream, hypervolume(pop, [1.0,1.0]))
return pop, logbook
#if __name__ == "__main__":
# #optimal_front = json.load(open("pareto_front/zdt4_front.json"))
# # Use 500 of the 1000 points in the json file
# #optimal_front = sorted(optimal_front[i] for i in range(0, len(optimal_front), 2))
pop, stats = main()
pop.sort(key=lambda x: x.fitness.values)
print(stats)
#print("Convergence: ", convergence(pop, optimal_front))
#print("Diversity: ", diversity(pop, optimal_front[0], optimal_front[-1]))
import matplotlib.pyplot as plt
import numpy
front = numpy.array([ind.fitness.values for ind in pop])
#optimal_front = numpy.array(optimal_front)
#plt.scatter(optimal_front[:,0], optimal_front[:,1], c="r")
plt.scatter(front[:,0], front[:,1], c="b")
plt.axis("tight")
plt.show()
pop[0]
[float(x) for x in pop[0]]
pop[0].fitness.info
```
# Hypervolume
```
from deap.benchmarks.tools import diversity, convergence, hypervolume
print("Final population hypervolume is %f" % hypervolume(pop, [1.0,1.0]))
```
|
github_jupyter
|

# 1. Introduction
This notebook demonstrates how to create two parallel video pipelines using the GStreamer multimedia framework:
* The first pipeline captures video from a V4L2 device and displays the output on a monitor using a DRM/KMS display device.
* The second pipeline decodes a VP9 encoded video file and displays the output on the same monitor using the same DRM/KMS display device.
The display device contains a video mixer which allows targeting different video planes for the individual pipelines with programmable x/y-offsets as well as width and height.
Refer to:
* nb1 for more details on the video file decode pipeline
* nb2 for more details on the V4L2 capture pipeline
* nb3 for more details on the video mixer configuration and display pipeline
In this notebook, you will:
1. Create two parallel GStreamer video pipelines using the ``parse_launch()`` API
2. Create a GStreamer pipeline graph and view it inside this notebook.
# 2. Imports and Initialization
Import all python modules required for this notebook.
```
from IPython.display import Image, display, clear_output
import pydot
import sys
import time
import gi
gi.require_version('Gst', '1.0')
gi.require_version("GstApp", "1.0")
from gi.repository import GObject, GLib, Gst, GstApp
```
This is the VMK180 TRD notebook 4 (nb4).
```
nb = "nb4"
```
Create a directory for saving the pipeline graph as dot file. Set the GStreamer debug dot directory environement variable to point to that directory.
```
dotdir = "/home/root/gst-dot/" + nb
!mkdir -p $dotdir
%env GST_DEBUG_DUMP_DOT_DIR = $dotdir
```
Initialize the GStreamer library. Optionally enable debug (default off) and set the debug level.
```
Gst.init(None)
Gst.debug_set_active(False)
Gst.debug_set_default_threshold(1)
```
# 3. Create String Representation of the First GStreamer Pipeline
The first pipeline consist of the following elements:
* ``xlnxvideosrc``
* ``caps``
* ``kmssink``
Describe the ``xlnxvideosrc`` element and its properties as string representation.
```
src_types = ["vivid", "usbcam", "mipi"]
src_type = src_types[1] # Change the source type to vivid, usbcam, or mipi via list index
io_mode = "mmap"
if src_type == "mipi":
io_mode = "dmabuf"
src_1 = "xlnxvideosrc src-type=" + src_type + " io-mode=" + io_mode
```
Describe the ``caps`` filter element as string representation.
```
width = 1280
height = 720
fmt = "YUY2"
caps = "video/x-raw, width=" + str(width) + ", height=" + str(height) + ", format=" + fmt
```
Describe the ``kmssink`` element and its properties as string representation.
```
driver_name = "xlnx"
plane_id_1 = 39
xoff_1 = 0
yoff_1 = 0
render_rectangle_1 = "<" + str(xoff_1) + "," + str(yoff_1) + "," + str(width) + "," + str(height) + ">"
sink_1 = "kmssink" + " driver-name=" + driver_name + " plane-id=" + str(plane_id_1) + " render-rectangle=" + render_rectangle_1
```
Create a string representation of the first pipeline by concatenating the individual element strings.
```
pipe_1 = src_1 + " ! " + caps + " ! " + sink_1
print(pipe_1)
```
# 4. Create String Representation of the Second GStreamer Pipeline
The second pipeline consist of the following elements:
* ``multifilesrc``
* ``decodebin``
* ``videoconvert``
* ``kmssink``
Describe the ``multifilesrc`` element and its properties as string representation.
```
file_name = "/usr/share/movies/Big_Buck_Bunny_4K.webm.360p.vp9.webm"
loop = True
src_2 = "multifilesrc location=" + file_name + " loop=" + str(loop)
```
Describe the ``decodebin`` and ``videoconvert`` elements as string representations.
```
dec = "decodebin"
cvt = "videoconvert"
```
Describe the ``kmssink`` element and its properties as string representation.
**Note:** The same ``kmssink`` element and ``driver-name`` property are used as in pipeline 1, only the ``plane-id`` and the ``render-rectangle`` properties are set differently. The output of this pipeline is shown on a different plane and the x/y-offsets are set such that the planes of pipeline 1 and 2 don't overlap.
```
driver_name = "xlnx"
plane_id_2 = 38
xoff_2 = 0
yoff_2 = 720
width_2 = 640
height_2 = 360
render_rectangle_2 = "<" + str(xoff_2) + "," + str(yoff_2) + "," + str(width_2) + "," + str(height_2) + ">"
sink_2 = "kmssink" + " driver-name=" + driver_name + " plane-id=" + str(plane_id_2) + " render-rectangle=" + render_rectangle_2
```
Create a string representation of the second pipeline by concatenating the individual element strings.
```
pipe_2 = src_2 + " ! " + dec + " ! " + cvt + " ! "+ sink_2
print(pipe_2)
```
# 5. Create and Run the GStreamer Pipelines
Parse the string representations of the first and second pipeline as a single pipeline graph.
```
pipeline = Gst.parse_launch(pipe_1 + " " + pipe_2)
```
The ``bus_call`` function listens on the bus for ``EOS`` and ``ERROR`` events. If any of these events occur, stop the pipeline (set to ``NULL`` state) and quit the main loop.
In case of an ``ERROR`` event, parse and print the error message.
```
def bus_call(bus, message, loop):
t = message.type
if t == Gst.MessageType.EOS:
sys.stdout.write("End-of-stream\n")
pipeline.set_state(Gst.State.NULL)
loop.quit()
elif t == Gst.MessageType.ERROR:
err, debug = message.parse_error()
sys.stderr.write("Error: %s: %s\n" % (err, debug))
pipeline.set_state(Gst.State.NULL)
loop.quit()
return True
```
Start the pipeline (set to ``PLAYING`` state), create the main loop and listen to messages on the bus. Register the ``bus_call`` callback function with the ``message`` signal of the bus. Start the main loop.
The video will be displayed on the monitor.
To stop the pipeline, click the square shaped icon labelled 'Interrupt the kernel' in the top menu bar. Create a dot graph of the pipeline topology before stopping the pipeline. Quit the main loop.
```
pipeline.set_state(Gst.State.PLAYING);
loop = GLib.MainLoop()
bus = pipeline.get_bus()
bus.add_signal_watch()
bus.connect("message", bus_call, loop)
try:
loop.run()
except:
sys.stdout.write("Interrupt caught\n")
Gst.debug_bin_to_dot_file(pipeline, Gst.DebugGraphDetails.ALL, nb)
pipeline.set_state(Gst.State.NULL)
loop.quit()
pass
```
# 6. View the Pipeline dot Graph
Register dot plugins for png export to work.
```
!dot -c
```
Convert the dot file to png and display the pipeline graph. The image will be displayed below the following code cell. Double click on the generate image file to zoom in.
**Note:** This step may take a few seconds. Also, compared to previous notebooks, two disjoint graphs are displayed in the same image as we have created two parallel pipelines in this example.
```
dotfile = dotdir + "/" + nb + ".dot"
graph = pydot.graph_from_dot_file(dotfile, 'utf-8')
display(Image(graph[0].create(None, 'png', 'utf-8')))
```
# 7. Summary
In this notebook you learned how to:
1. Create two parallel GStreamer pipelines from a string representation using the ``parse_launch()`` API
2. Export the pipeline topology as a dot file image and display it in the notebook
<center>Copyright© 2019 Xilinx</center>
|
github_jupyter
|
# Running a Federated Cycle with Synergos
In a federated learning system, there are many contributory participants, known as Worker nodes, which receive a global model to train on, with their own local dataset. The dataset does not leave the individual Worker nodes at any point, and remains private to the node.
The job to synchronize, orchestrate and initiate an federated learning cycle, falls on a Trusted Third Party (TTP). The TTP pushes out the global model architecture and parameters for the individual nodes to train on, calling upon the required data, based on tags, e.g "training", which points to relevant data on the individual nodes. At no point does the TTP receive, copy or access the Worker nodes' local datasets.

This tutorial aims to give you an understanding of how to use the synergos package to run a full federated learning cycle on a `Synergos Cluster` grid.
In a `Synergos Cluster` Grid, with the inclusion of a new director and queue component, you will be able to parallelize your jobs, where the number of concurrent jobs possible is equal to the number of sub-grids. This is done alongside all quality-of-life components supported in a `Synergos Plus` grid.
In this tutorial, you will go through the steps required by each participant (TTP and Worker), by simulating each of them locally with docker containers. Specifically, we will simulate a Director and 2 sub-grids, each of which has a TTP and 2 Workers, allowing us to perform 2 concurrent federated operations at any time.
At the end of this, we will have:
- Connected the participants
- Trained the model
- Evaluate the model
## About the Dataset and Task
The dataset used in this notebook is on a small subset of Imagenette images, comprising 3 classes, and all images are 28 x 28 pixels. The dataset is available in the same directory as this notebook. Within the dataset directory, `data1` is for Worker 1 and `data2` is for Worker 2. The task to be carried out will be a multi-classification.
The dataset we have provided is a processed subset of the [original Imagenette dataset](https://github.com/fastai/imagenette).
## Initiating the docker containers
Before we begin, we have to start the docker containers.
### A. Initialization via `Synergos Simulator`
In `Synergos Simulator`, a sandboxed environment has been created for you!
By running:
`docker-compose -f docker-compose-syncluster.yml up --build`
the following components will be started:
- Director
- Sub-Grid 1
- TTP_1 (Cluster)
- Worker_1_n1
- Worker_2_n1
- Sub-Grid 2
- TTP_2 (Cluster)
- Worker_1_n2
- Worker_2_n2
- Synergos UI
- Synergos Logger
- Synergos MLOps
- Synergos MQ
Refer to [this](https://github.com/aimakerspace/synergos_simulator) for all the pre-allocated host & port mappings.
### B. Manual Initialization
Firstly, pull the required docker images with the following commands:
1. Synergos Director:
`docker pull gcr.io/synergos-aisg/synergos_director:v0.1.0`
2. Synergos TTP (Cluster):
`docker pull gcr.io/synergos-aisg/synergos_ttp_cluster:v0.1.0`
3. Synergos Worker:
`docker pull gcr.io/synergos-aisg/synergos_worker:v0.1.0`
4. Synergos MLOps:
`docker pull gcr.io/synergos-aisg/synergos_mlops:v0.1.0`
5. Synergos MQ:
`docker pull gcr.io/synergos-aisg/synergos_mq:v0.1.0`
Next, in <u>separate</u> CLI terminals, run the following command(s):
**Note: For Windows users, it is advisable to use powershell or command prompt based interfaces**
#### Director
```
docker run --rm
-p 5000:5000
-v <directory imagenette/orchestrator_data>:/orchestrator/data
-v <directory imagenette/orchestrator_outputs>:/orchestrator/outputs
-v <directory imagenette/mlflow>:/mlflow
--name director
gcr.io/synergos-aisg/synergos_director:v0.1.0
--id ttp
--logging_variant graylog <IP Synergos Logger> <TTP port>
--queue rabbitmq <IP Synergos Logger> <AMQP port>
```
#### Sub-Grid 1
- **TTP_1**
```
docker run --rm
-p 6000:5000
-p 9020:8020
-v <directory imagenette/orchestrator_data>:/orchestrator/data
-v <directory imagenette/orchestrator_outputs>:/orchestrator/outputs
--name ttp_1
gcr.io/synergos-aisg/synergos_ttp_cluster:v0.1.0
--id ttp
--logging_variant graylog <IP Synergos Logger> <TTP port>
--queue rabbitmq <IP Synergos Logger> <AMQP port>
```
- **WORKER_1 Node 1**
```
docker run --rm
-p 5001:5000
-p 8021:8020
-v <directory imagenette/data1>:/worker/data
-v <directory imagenette/outputs_1>:/worker/outputs
--name worker_1_n1
gcr.io/synergos-aisg/synergos_worker:v0.1.0
--id worker_1_n1
--logging_variant graylog <IP Synergos Logger> <Worker port>
--queue rabbitmq <IP Synergos Logger> <AMQP port>
```
- **WORKER_2 Node 1**
```
docker run --rm
-p 5002:5000
-p 8022:8020
-v <directory imagenette/data2>:/worker/data
-v <directory imagenette/outputs_2>:/worker/outputs
--name worker_2_n1
gcr.io/synergos-aisg/synergos_worker:v0.1.0
--id worker_2_n1
--logging_variant graylog <IP Synergos Logger> <Worker port>
--queue rabbitmq <IP Synergos Logger> <AMQP port>
```
#### Sub-Grid 2
- **TTP_2**
```
docker run --rm
-p 7000:5000
-p 10020:8020
-v <directory imagenette/orchestrator_data>:/orchestrator/data
-v <directory imagenette/orchestrator_outputs>:/orchestrator/outputs
--name ttp_2
gcr.io/synergos-aisg/synergos_ttp_cluster:v0.1.0
--id ttp
--logging_variant graylog <IP Synergos Logger> <TTP port>
--queue rabbitmq <IP Synergos Logger> <AMQP port>
```
- **WORKER_1 Node 2**
```
docker run --rm
-p 5003:5000
-p 8023:8020
-v <directory imagenette/data1>:/worker/data
-v <directory imagenette/outputs_1>:/worker/outputs
--name worker_1_n2
gcr.io/synergos-aisg/synergos_worker:v0.1.0
--id worker_1_n2
--logging_variant graylog <IP Synergos Logger> <Worker port>
--queue rabbitmq <IP Synergos Logger> <AMQP port>
```
- **WORKER_2 Node 2**
```
docker run --rm
-p 5004:5000
-p 8024:8020
-v <directory imagenette/data2>:/worker/data
-v <directory imagenette/outputs_2>:/worker/outputs
--name worker_2_n2
gcr.io/synergos-aisg/synergos_worker:v0.1.0
--id worker_2_n2
--logging_variant graylog <IP Synergos Logger> <Worker port>
--queue rabbitmq <IP Synergos Logger> <AMQP port>
```
#### Synergos MLOps
```
docker run --rm
-p 5500:5500
-v /path/to/mlflow_test/:/mlflow # <-- IMPT! Same as orchestrator's
--name synmlops
gcr.io/synergos-aisg/synergos_mlops:v0.1.0
```
#### Synergos MQ
```
docker run --rm
-p 15672:15672 # UI port
-p 5672:5672 # AMQP port
--name synergos_mq
gcr.io/synergos-aisg/synergos_mq:v0.1.0
```
#### Synergos UI
- Refer to these [instructions](https://github.com/aimakerspace/synergos_ui) to deploy `Synergos UI`.
#### Synergos Logger
- Refer to these [instructions](https://github.com/aimakerspace/synergos_logger) to deploy `Synergos Logger`.
Once ready, for each terminal, you should see a REST server running on http://0.0.0.0:5000 of the container.
You are now ready for the next step.
## Configurations
### A. Configuring `Synergos Simulator`
All hosts & ports have already been pre-allocated!
Refer to [this](https://github.com/aimakerspace/synergos_simulator) for all the pre-allocated host & port mappings.
### B. Configuring your manual setup
In a new terminal, run `docker inspect bridge` and find the IPv4Address for each container. Ideally, the containers should have the following addresses:
- director address: `172.17.0.2`
- Sub-Grid 1
- ttp_1 address: `172.17.0.3`
- worker_1_n1 address: `172.17.0.4`
- worker_2_n1 address: `172.17.0.5`
- Sub-Grid 2
- ttp_2 address: `172.17.0.6`
- worker_1_n2 address: `172.17.0.7`
- worker_2_n2 address: `172.17.0.8`
- UI address: `172.17.0.9`
- Logger address: `172.17.0.14`
- MLOps address: `172.17.0.15`
- MQ address: `172.17.0.16`
If not, just note the relevant IP addresses for each docker container.
Run the following cells below.
**Note: For Windows users, `host` should be Docker Desktop VM's IP. Follow [this](https://stackoverflow.com/questions/58073936/how-to-get-ip-address-of-docker-desktop-vm) on instructions to find IP**
```
import time
from synergos import Driver
host = "172.20.0.2"
port = 5000
# Initiate Driver
driver = Driver(host=host, port=port)
```
## Phase 1: Registration
Submitting Orchestrator & Participant metadata
#### 1A. Orchestrator creates a collaboration
```
collab_task = driver.collaborations
collab_task.configure_logger(
host="172.20.0.14",
port=9000,
sysmetrics_port=9100,
director_port=9200,
ttp_port=9300,
worker_port=9400,
ui_port=9000,
secure=False
)
collab_task.configure_mlops(
host="172.20.0.15",
port=5500,
ui_port=5500,
secure=False
)
collab_task.configure_mq(
host="172.20.0.16",
port=5672,
ui_port=15672,
secure=False
)
collab_task.create('imagenette_syncluster_collaboration')
```
#### 1B. Orchestrator creates a project
```
driver.projects.create(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
action="classify",
incentives={
'tier_1': [],
'tier_2': [],
}
)
```
#### 1C. Orchestrator creates an experiment
```
driver.experiments.create(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment",
model=[
{
"activation": "relu",
"is_input": True,
"l_type": "Conv2d",
"structure": {
"in_channels": 1,
"out_channels": 4,
"kernel_size": 3,
"stride": 1,
"padding": 1
}
},
{
"activation": None,
"is_input": False,
"l_type": "Flatten",
"structure": {}
},
{
"activation": "softmax",
"is_input": False,
"l_type": "Linear",
"structure": {
"bias": True,
"in_features": 4 * 28 * 28,
"out_features": 3
}
}
]
)
```
#### 1D. Orchestrator creates a run
```
driver.runs.create(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment",
run_id="imagenette_syncluster_run",
rounds=2,
epochs=1,
base_lr=0.0005,
max_lr=0.005,
criterion="NLLLoss"
)
```
#### 1E. Participants registers their servers' configurations and roles
```
participant_resp_1 = driver.participants.create(
participant_id="worker_1",
)
display(participant_resp_1)
participant_resp_2 = driver.participants.create(
participant_id="worker_2",
)
display(participant_resp_2)
registration_task = driver.registrations
# Add and register worker_1 node
registration_task.add_node(
host='172.20.0.4',
port=8020,
f_port=5000,
log_msgs=True,
verbose=True
)
registration_task.add_node(
host='172.20.0.7',
port=8020,
f_port=5000,
log_msgs=True,
verbose=True
)
registration_task.create(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
participant_id="worker_1",
role="host"
)
registration_task = driver.registrations
# Add and register worker_2 node
registration_task.add_node(
host='172.20.0.5',
port=8020,
f_port=5000,
log_msgs=True,
verbose=True
)
registration_task.add_node(
host='172.20.0.8',
port=8020,
f_port=5000,
log_msgs=True,
verbose=True
)
registration_task.create(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
participant_id="worker_2",
role="guest"
)
```
#### 1F. Participants registers their tags for a specific project
```
# Worker 1 declares their data tags
driver.tags.create(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
participant_id="worker_1",
train=[["imagenette", "dataset", "data1", "train"]],
evaluate=[["imagenette", "dataset", "data1", "evaluate"]]
)
# Worker 2 declares their data tags
driver.tags.create(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
participant_id="worker_2",
train=[["imagenette", "dataset", "data2", "train"]],
evaluate=[["imagenette", "dataset", "data2", "evaluate"]]
)
stop!
```
## Phase 2:
Alignment, Training & Optimisation
#### 2A. Perform multiple feature alignment to dynamically configure datasets and models for cross-grid compatibility
```
driver.alignments.create(
collab_id='imagenette_syncluster_collaboration',
project_id="imagenette_syncluster_project",
verbose=False,
log_msg=False
)
# Important! MUST wait for alignment process to first complete before proceeding on
while True:
align_resp = driver.alignments.read(
collab_id='imagenette_syncluster_collaboration',
project_id="imagenette_syncluster_project"
)
align_data = align_resp.get('data')
if align_data:
display(align_resp)
break
time.sleep(5)
```
#### 2B. Trigger training across the federated grid
```
model_resp = driver.models.create(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment",
run_id="imagenette_syncluster_run",
log_msg=False,
verbose=False
)
display(model_resp)
# Important! MUST wait for training process to first complete before proceeding on
while True:
train_resp = driver.models.read(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment",
run_id="imagenette_syncluster_run"
)
train_data = train_resp.get('data')
if train_data:
display(train_data)
break
time.sleep(5)
```
#### 2C. Perform hyperparameter tuning once ideal model is found (experimental)
```
optim_parameters = {
'search_space': {
"rounds": {"_type": "choice", "_value": [1, 2]},
"epochs": {"_type": "choice", "_value": [1, 2]},
"batch_size": {"_type": "choice", "_value": [32, 64]},
"lr": {"_type": "choice", "_value": [0.0001, 0.1]},
"criterion": {"_type": "choice", "_value": ["NLLLoss"]},
"mu": {"_type": "uniform", "_value": [0.0, 1.0]},
"base_lr": {"_type": "choice", "_value": [0.00005]},
"max_lr": {"_type": "choice", "_value": [0.2]}
},
'backend': "tune",
'optimize_mode': "max",
'metric': "accuracy",
'trial_concurrency': 1,
'max_exec_duration': "1h",
'max_trial_num': 2,
'max_concurrent': 1,
'is_remote': True,
'use_annotation': True,
'auto_align': True,
'dockerised': True,
'verbose': True,
'log_msgs': True
}
driver.optimizations.create(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment",
**optim_parameters
)
driver.optimizations.read(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment"
)
```
## Phase 3: EVALUATE
Validation & Predictions
#### 3A. Perform validation(s) of combination(s)
```
# Orchestrator performs post-mortem validation
driver.validations.create(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment",
run_id="imagenette_syncluster_run",
log_msg=False,
verbose=False
)
# Run this cell again after validation has completed to retrieve your validation statistics
# NOTE: You do not need to wait for validation/prediction requests to complete to proceed
driver.validations.read(
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment",
run_id="imagenette_syncluster_run",
)
```
#### 3B. Perform prediction(s) of combination(s)
```
# Worker 1 requests for inferences
driver.predictions.create(
tags={
"imagenette_syncluster_project": [
["imagenette", "dataset", "data1", "predict"]
]
},
participant_id="worker_1",
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment",
run_id="imagenette_syncluster_run",
log_msg=False,
verbose=False
)
# Run this cell again after prediction has completed to retrieve your predictions for worker 1
# NOTE: You do not need to wait for validation/prediction requests to complete to proceed
driver.predictions.read(
participant_id="worker_1",
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment",
run_id="imagenette_syncluster_run",
)
# Worker 2 requests for inferences
driver.predictions.create(
tags={
"imagenette_syncluster_project": [
["imagenette", "dataset", "data2", "predict"]
]
},
participant_id="worker_2",
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment",
run_id="imagenette_syncluster_run",
log_msg=False,
verbose=False
)
# Run this cell again after prediction has completed to retrieve your predictions for worker 2
# NOTE: You do not need to wait for validation/prediction requests to complete to proceed
driver.predictions.read(
participant_id="worker_2",
collab_id="imagenette_syncluster_collaboration",
project_id="imagenette_syncluster_project",
expt_id="imagenette_syncluster_experiment",
run_id="imagenette_syncluster_run",
)
```
|
github_jupyter
|
```
import torch
import torch.nn as nn
import numpy as np
from copy import deepcopy
device = "cuda" if torch.cuda.is_available() else "cpu"
class RBF(nn.Module):
def __init__(self):
super(RBF, self).__init__()
torch.cuda.manual_seed(0)
self.rbf_clt = self.init_clt()
self.rbf_std = self.init_std()
def init_clt(self):
return nn.Parameter(torch.rand(1))
def init_std(self):
return nn.Parameter(torch.rand(1))
def rbf(self, x, cluster, std):
return torch.exp(-(x - cluster) * (x - cluster) / 2 * (std * std))
def forward(self, x):
x = self.rbf(x, self.rbf_clt, self.rbf_std)
return x
class RBFnetwork(nn.Module):
def __init__(self, timelag):
super(RBFnetwork, self).__init__()
torch.cuda.manual_seed(0)
device = "cuda" if torch.cuda.is_available() else "cpu"
self.timelag = timelag
self.init_weight = nn.Parameter(torch.rand(self.timelag))
self.rbf_list = [RBF().to(device) for i in range(self.timelag)]
def forward(self, x):
for j in range(self.timelag):
if j ==0:
y = sum([self.init_weight[i] * self.rbf_list[i](x[j]) for i in range(self.timelag)])
else:
y = torch.cat([y, sum([self.init_weight[i] * self.rbf_list[i](x[j]) for i in range(self.timelag)])])
return y
def restore_parameters(model, best_model):
'''Move parameter values from best_model to model.'''
for params, best_params in zip(model.parameters(), best_model.parameters()):
params.data = best_params
def train_RBFlayer(model, input_, target, lr, epochs, lookback = 5, device = device):
model.to(device)
loss_fn = nn.MSELoss(reduction='mean')
optimizer = torch.optim.Adam(model.parameters(), lr = lr)
train_loss_list = []
best_it = None
best_model = None
best_loss = np.inf
target_list = []
for j in range(len(target) - 2):
target_list.append((target[j+2] - target[j])/2)
loss_list = []
cause_list = []
for epoch in range(epochs):
cause = model(input_)
cause_list.append(cause)
grad = []
for i in range(len(cause) - 2):
grad.append((cause[i+2] - cause[i])/2)
loss1 = sum([loss_fn(grad[i], target_list[i]) for i in range(len(grad))])
loss2 = sum([loss_fn(cause[i], target[i]) for i in range(len(input_))])
loss = loss1 + loss2
loss.backward()
optimizer.step()
model.zero_grad()
loss_list.append(loss)
mean_loss = loss / len(grad)
train_loss_list.append(mean_loss)
if mean_loss < best_loss:
best_loss = mean_loss
best_it = epoch
best_model = deepcopy(model)
elif (epoch - best_it) == lookback:
if verbose:
print('Stopping early')
break
print("epoch {} cause loss {} :".format(epoch, loss / len(input_)))
print('gradient loss :', loss1/len(grad))
print('value loss :', loss2/len(input_))
best_cause = cause_list[best_it]
restore_parameters(model, best_model)
return best_model, loss_list, best_cause
```
# data generation
```
import random as rand
import numpy as np
def data_gen(timelag):
data = []
clt_list = []
std_list = []
for i in range(timelag):
clt = rand.random()
std = rand.random()
data_i = np.exp(-(i - clt) * (i - clt) / 2 * (std * std))
data.append(data_i)
clt_list.append(clt)
std_list.append(std)
return torch.tensor(data, device = device).float(), torch.tensor(clt_list, device = device).float(), torch.tensor(std_list, device = device).float()
data, clt_list, std_list = data_gen(10)
data
clt_list
std_list
```
# test1
```
import time
cause_list = []
start = time.time()
model = RBFnetwork(10)
best_model, loss_list, best_cause = train_RBFlayer(model, data, data, 0.001, 1000, device)
cause_list.append(best_cause.cpu().detach().numpy())
print("time :", time.time() - start)
print('-------------------------------------------------------------------------------------------')
import matplotlib.pyplot as plt
plt.plot(cause_list[0])
plt.plot(cause_list[0])
plt.plot(data.cpu().detach().numpy())
plt.show()
```
|
github_jupyter
|
```
import os
import numpy as np
import sys
import matplotlib.pyplot as plt
from matplotlib import rc
from matplotlib.pyplot import cm
from library.trajectory import Trajectory
# uzh trajectory toolbox
sys.path.append(os.path.abspath('library/rpg_trajectory_evaluation/src/rpg_trajectory_evaluation'))
import plot_utils as pu
%matplotlib inline
rc('font', **{'family': 'serif', 'serif': ['Cardo']})
rc('text', usetex=True)
```
### Parameters (to specify/set)
```
# directory where the data is saved
DATA_DIR = '/home/mayankm/my_projects/multiview_deeptam_3DV/multi-camera-deeptam/resources/data/cvg_cams'
# directory to save the output
RESULTS_DIR = os.path.abspath('eval')
# format in which to save the plots
FORMAT = '.png'
# set the camera indices to plot
CAM_IDXS = [0, 2, 4, 6, 8]
# set the reference camera (in case groundtruth is not available)
REF_CAM_ID = 0
# evaluation parameters
align_type = 'none' # choose from ['posyaw', 'sim3', 'se3', 'none']
align_num_frames = -1
```
### Variables to allow the plots to look nice
```
N = len(CAM_IDXS)
ALGORITHM_CONFIGS = []
for i in range(N):
ALGORITHM_CONFIGS.append('cam_%d' % CAM_IDXS[i])
# These are the labels that will be displayed for items in ALGORITHM_CONFIGS
PLOT_LABELS = { 'cam_0': 'Camera 0',
'cam_2': 'Camera 2',
'cam_4': 'Camera 4',
'cam_6': 'Camera 6',
'cam_8': 'Camera 8'}
PLOT_LABELS['cam_%d' % REF_CAM_ID] = PLOT_LABELS['cam_%d' % REF_CAM_ID] + ' (ref)'
# assgin colors to different configurations
COLORS = {}
color = iter(cm.plasma(np.linspace(0, 0.75, N)))
for i in range(N):
COLORS['cam_%d' % CAM_IDXS[i]] = next(color)
```
### Defining the txt files with the pose information
```
# file name for reference trajectory
ref_traj_file = os.path.join(DATA_DIR, 'cam_%d' % REF_CAM_ID, 'groundtruth.txt')
# file names for camera trajectories
estimated_traj_files = []
for i in range(N):
# path to camera trajectory
estimated_traj_file = os.path.join(DATA_DIR, 'cam_%d' % CAM_IDXS[i], 'groundtruth.txt')
assert os.path.exists(estimated_traj_file), "No corresponding file exists: %s!" % estimated_traj_file
estimated_traj_files.append(estimated_traj_file)
```
# Main
```
print("Going to analyze the results in {0}.".format(DATA_DIR))
print("The plots will saved in {0}.".format(RESULTS_DIR))
if not os.path.exists(plots_dir):
os.makedirs(plots_dir)
os.makedies(RESULTS_DIR, )
print("#####################################")
print(">>> Start loading and preprocessing all trajectories...")
print("#####################################")
config_trajectories_list = []
for i in range(N):
# create instance of trajectory object
cur_traj = Trajectory(RESULTS_DIR, run_name='cam_%d' % CAM_IDXS[i], gt_traj_file=ref_traj_file, estimated_traj_file=estimated_traj_files[i], \
align_type=align_type, align_num_frames=align_num_frames)
config_trajectories_list.append(cur_traj)
print("#####################################")
print(">>> Start plotting results....")
print("#####################################")
p_gt_0 = config_trajectories_list[0].p_gt
fig1 = plt.figure(figsize=(10, 10))
ax1 = fig1.add_subplot(111, aspect='equal',
xlabel='x [m]', ylabel='y [m]')
fig2 = plt.figure(figsize=(8, 8))
ax2 = fig2.add_subplot(111, aspect='equal',
xlabel='x [m]', ylabel='z [m]')
# pu.plot_trajectory_top(ax1, p_gt_0, 'k', 'Groundtruth')
# pu.plot_trajectory_side(ax2, p_gt_0,'k', 'Groundtruth')
for i in range(N):
traj = config_trajectories_list[i]
p_es_0 = traj.p_es_aligned
alg = ALGORITHM_CONFIGS[i]
print('Plotting for %s' % alg)
# plot trajectory
pu.plot_trajectory_top(ax1, p_es_0, COLORS[alg], PLOT_LABELS[alg])
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
fig1.tight_layout()
# plot trajectory side
pu.plot_trajectory_side(ax2, p_es_0, COLORS[alg], PLOT_LABELS[alg])
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
fig2.tight_layout()
fig1.savefig(RESULTS_DIR + '/plots/trajectory_top_' + align_type + FORMAT,bbox_inches="tight")
plt.close(fig1)
fig2.savefig(RESULTS_DIR + '/plots/trajectory_side_' + align_type + FORMAT, bbox_inches="tight")
plt.close(fig2)
```
|
github_jupyter
|
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#Goal" data-toc-modified-id="Goal-1"><span class="toc-item-num">1 </span>Goal</a></span></li><li><span><a href="#Var" data-toc-modified-id="Var-2"><span class="toc-item-num">2 </span>Var</a></span><ul class="toc-item"><li><span><a href="#Init" data-toc-modified-id="Init-2.1"><span class="toc-item-num">2.1 </span>Init</a></span></li></ul></li><li><span><a href="#DeepMAsED-SM" data-toc-modified-id="DeepMAsED-SM-3"><span class="toc-item-num">3 </span>DeepMAsED-SM</a></span><ul class="toc-item"><li><span><a href="#Config" data-toc-modified-id="Config-3.1"><span class="toc-item-num">3.1 </span>Config</a></span></li><li><span><a href="#Run" data-toc-modified-id="Run-3.2"><span class="toc-item-num">3.2 </span>Run</a></span></li></ul></li><li><span><a href="#Summary" data-toc-modified-id="Summary-4"><span class="toc-item-num">4 </span>Summary</a></span><ul class="toc-item"><li><span><a href="#Communities" data-toc-modified-id="Communities-4.1"><span class="toc-item-num">4.1 </span>Communities</a></span></li><li><span><a href="#Feature-tables" data-toc-modified-id="Feature-tables-4.2"><span class="toc-item-num">4.2 </span>Feature tables</a></span><ul class="toc-item"><li><span><a href="#No.-of-contigs" data-toc-modified-id="No.-of-contigs-4.2.1"><span class="toc-item-num">4.2.1 </span>No. of contigs</a></span></li><li><span><a href="#Misassembly-types" data-toc-modified-id="Misassembly-types-4.2.2"><span class="toc-item-num">4.2.2 </span>Misassembly types</a></span></li></ul></li></ul></li><li><span><a href="#sessionInfo" data-toc-modified-id="sessionInfo-5"><span class="toc-item-num">5 </span>sessionInfo</a></span></li></ul></div>
# Goal
* Replicate metagenome assemblies using intra-spec training genome dataset
* Richness = 0.1 (10% of all ref genomes used)
# Var
```
ref_dir = '/ebio/abt3_projects/databases_no-backup/DeepMAsED/GTDB_ref_genomes/intraSpec/'
ref_file = file.path(ref_dir, 'GTDBr86_genome-refs_train_clean.tsv')
work_dir = '/ebio/abt3_projects/databases_no-backup/DeepMAsED/train_runs/intra-species/diff_richness/n1000_r6_rich0p1/'
# params
pipeline_dir = '/ebio/abt3_projects/databases_no-backup/bin/deepmased/DeepMAsED-SM/'
```
## Init
```
library(dplyr)
library(tidyr)
library(ggplot2)
library(data.table)
source('/ebio/abt3_projects/software/dev/DeepMAsED/bin/misc_r_functions/init.R')
#' "cat {file}" in R
cat_file = function(file_name){
cmd = paste('cat', file_name, collapse=' ')
system(cmd, intern=TRUE) %>% paste(collapse='\n') %>% cat
}
```
# DeepMAsED-SM
## Config
```
config_file = file.path(work_dir, 'config.yaml')
cat_file(config_file)
```
## Run
```
(snakemake_dev) @ rick:/ebio/abt3_projects/databases_no-backup/bin/deepmased/DeepMAsED-SM
$ screen -L -S DM-intraS-rich0.1 ./snakemake_sge.sh /ebio/abt3_projects/databases_no-backup/DeepMAsED/train_runs/intra-species/diff_richness/n1000_r6_rich0p1/config.yaml cluster.json /ebio/abt3_projects/databases_no-backup/DeepMAsED/train_runs/intra-species/diff_richness/n1000_r6_rich0p1/SGE_log 20
```
# Summary
## Communities
```
comm_files = list.files(file.path(work_dir, 'MGSIM'), 'comm_wAbund.txt', full.names=TRUE, recursive=TRUE)
comm_files %>% length %>% print
comm_files %>% head
comms = list()
for(F in comm_files){
df = read.delim(F, sep='\t')
df$Rep = basename(dirname(F))
comms[[F]] = df
}
comms = do.call(rbind, comms)
rownames(comms) = 1:nrow(comms)
comms %>% dfhead
p = comms %>%
mutate(Perc_rel_abund = ifelse(Perc_rel_abund == 0, 1e-5, Perc_rel_abund)) %>%
group_by(Taxon) %>%
summarize(mean_perc_abund = mean(Perc_rel_abund),
sd_perc_abund = sd(Perc_rel_abund)) %>%
ungroup() %>%
mutate(neg_sd_perc_abund = mean_perc_abund - sd_perc_abund,
pos_sd_perc_abund = mean_perc_abund + sd_perc_abund,
neg_sd_perc_abund = ifelse(neg_sd_perc_abund <= 0, 1e-5, neg_sd_perc_abund)) %>%
mutate(Taxon = Taxon %>% reorder(-mean_perc_abund)) %>%
ggplot(aes(Taxon, mean_perc_abund)) +
geom_linerange(aes(ymin=neg_sd_perc_abund, ymax=pos_sd_perc_abund),
size=0.3, alpha=0.3) +
geom_point(size=0.5, alpha=0.4, color='red') +
labs(y='% abundance') +
theme_bw() +
theme(
axis.text.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.major.y = element_blank(),
panel.grid.minor.x = element_blank(),
panel.grid.minor.y = element_blank()
)
dims(10,2.5)
plot(p)
dims(10,2.5)
plot(p + scale_y_log10())
```
## Feature tables
```
feat_files = list.files(file.path(work_dir, 'map'), 'features.tsv.gz', full.names=TRUE, recursive=TRUE)
feat_files %>% length %>% print
feat_files %>% head
feats = list()
for(F in feat_files){
cmd = glue::glue('gunzip -c {F}', F=F)
df = fread(cmd, sep='\t') %>%
distinct(contig, assembler, Extensive_misassembly)
df$Rep = basename(dirname(dirname(F)))
feats[[F]] = df
}
feats = do.call(rbind, feats)
rownames(feats) = 1:nrow(feats)
feats %>% dfhead
```
### No. of contigs
```
feats_s = feats %>%
group_by(assembler, Rep) %>%
summarize(n_contigs = n_distinct(contig)) %>%
ungroup
feats_s$n_contigs %>% summary
```
### Misassembly types
```
p = feats %>%
mutate(Extensive_misassembly = ifelse(Extensive_misassembly == '', 'None',
Extensive_misassembly)) %>%
group_by(Extensive_misassembly, assembler, Rep) %>%
summarize(n = n()) %>%
ungroup() %>%
ggplot(aes(Extensive_misassembly, n, color=assembler)) +
geom_boxplot() +
scale_y_log10() +
labs(x='metaQUAST extensive mis-assembly', y='Count') +
coord_flip() +
theme_bw() +
theme(
axis.text.x = element_text(angle=45, hjust=1)
)
dims(8,4)
plot(p)
```
# sessionInfo
```
sessionInfo()
pipelineInfo(pipeline_dir)
```
|
github_jupyter
|
# Distributed Object Tracker RL training with Amazon SageMaker RL and RoboMaker
---
## Introduction
In this notebook, we show you how you can apply reinforcement learning to train a Robot (named Waffle) track and follow another Robot (named Burger) by using the [Clipped PPO](https://coach.nervanasys.com/algorithms/policy_optimization/cppo/index.html) algorithm implementation in [coach](https://ai.intel.com/r-l-coach/) toolkit, [Tensorflow](https://www.tensorflow.org/) as the deep learning framework, and [AWS RoboMaker](https://console.aws.amazon.com/robomaker/home#welcome) as the simulation environment.

---
## How it works?
The reinforcement learning agent (i.e. Waffle) learns to track and follow Burger by interacting with its environment, e.g., visual world around it, by taking an action in a given state to maximize the expected reward. The agent learns the optimal plan of actions in training by trial-and-error through multiple episodes.
This notebook shows an example of distributed RL training across SageMaker and two RoboMaker simulation envrionments that perform the **rollouts** - execute a fixed number of episodes using the current model or policy. The rollouts collect agent experiences (state-transition tuples) and share this data with SageMaker for training. SageMaker updates the model policy which is then used to execute the next sequence of rollouts. This training loop continues until the model converges, i.e. the car learns to drive and stops going off-track. More formally, we can define the problem in terms of the following:
1. **Objective**: Learn to drive toward and reach the Burger.
2. **Environment**: A simulator with Burger hosted on AWS RoboMaker.
3. **State**: The driving POV image captured by the Waffle's head camera.
4. **Action**: Six discrete steering wheel positions at different angles (configurable)
5. **Reward**: Reward is inversely proportional to distance from Burger. Waffle gets more reward as it get closer to the Burger. It gets a reward of 0 if the action takes it away from Burger.
---
## Prequisites
### Imports
To get started, we'll import the Python libraries we need, set up the environment with a few prerequisites for permissions and configurations.
You can run this notebook from your local host or from a SageMaker notebook instance. In both of these scenarios, you can run the following to launch a training job on `SageMaker` and a simulation job on `RoboMaker`.
```
import sagemaker
import boto3
import sys
import os
import glob
import re
import subprocess
from IPython.display import Markdown
import time
from time import gmtime, strftime
sys.path.append("common")
from misc import get_execution_role
from sagemaker.rl import RLEstimator, RLToolkit, RLFramework
from markdown_helper import *
```
### Setup S3 bucket
```
# S3 bucket
sage_session = sagemaker.session.Session()
s3_bucket = sage_session.default_bucket()
s3_output_path = 's3://{}/'.format(s3_bucket) # SDK appends the job name and output folder
print("S3 bucket path: {}".format(s3_output_path))
```
### Define Variables
We define variables such as the job prefix for the training jobs and s3_prefix for storing metadata required for synchronization between the training and simulation jobs
```
# create unique job name
job_name_prefix = 'rl-object-tracker'
# create unique job name
job_name = s3_prefix = job_name_prefix + "-sagemaker-" + strftime("%y%m%d-%H%M%S", gmtime())
# Duration of job in seconds (5 hours)
job_duration_in_seconds = 3600 * 5
aws_region = sage_session.boto_region_name
print("S3 bucket path: {}{}".format(s3_output_path, job_name))
if aws_region not in ["us-west-2", "us-east-1", "eu-west-1"]:
raise Exception("This notebook uses RoboMaker which is available only in US East (N. Virginia), US West (Oregon) and EU (Ireland). Please switch to one of these regions.")
print("Model checkpoints and other metadata will be stored at: {}{}".format(s3_output_path, job_name))
```
### Create an IAM role
Either get the execution role when running from a SageMaker notebook `role = sagemaker.get_execution_role()` or, when running from local machine, use utils method `role = get_execution_role('role_name')` to create an execution role.
```
try:
role = sagemaker.get_execution_role()
except:
role = get_execution_role('sagemaker')
print("Using IAM role arn: {}".format(role))
```
### Permission setup for invoking AWS RoboMaker from this notebook
In order to enable this notebook to be able to execute AWS RoboMaker jobs, we need to add one trust relationship to the default execution role of this notebook.
```
display(Markdown(generate_help_for_robomaker_trust_relationship(role)))
```
## Configure VPC
Since SageMaker and RoboMaker have to communicate with each other over the network, both of these services need to run in VPC mode. This can be done by supplying subnets and security groups to the job launching scripts.
We will use the default VPC configuration for this example.
```
ec2 = boto3.client('ec2')
default_vpc = [vpc['VpcId'] for vpc in ec2.describe_vpcs()['Vpcs'] if vpc["IsDefault"] == True][0]
default_security_groups = [group["GroupId"] for group in ec2.describe_security_groups()['SecurityGroups'] \
if group["GroupName"] == "default" and group["VpcId"] == default_vpc]
default_subnets = [subnet["SubnetId"] for subnet in ec2.describe_subnets()["Subnets"] \
if subnet["VpcId"] == default_vpc and subnet['DefaultForAz']==True]
print("Using default VPC:", default_vpc)
print("Using default security group:", default_security_groups)
print("Using default subnets:", default_subnets)
```
A SageMaker job running in VPC mode cannot access S3 resourcs. So, we need to create a VPC S3 endpoint to allow S3 access from SageMaker container. To learn more about the VPC mode, please visit [this link.](https://docs.aws.amazon.com/sagemaker/latest/dg/train-vpc.html)
> The cell below should be executed to create the VPC S3 endpoint only if your are running this example for the first time. If the execution fails due to insufficient premissions or some other reasons, please create a VPC S3 endpoint manually by following [create-s3-endpoint.md](create-s3-endpoint.md) (can be found in the same folder as this notebook).
```
try:
route_tables = [route_table["RouteTableId"] for route_table in ec2.describe_route_tables()['RouteTables']\
if route_table['VpcId'] == default_vpc]
except Exception as e:
if "UnauthorizedOperation" in str(e):
display(Markdown(generate_help_for_s3_endpoint_permissions(role)))
else:
display(Markdown(create_s3_endpoint_manually(aws_region, default_vpc)))
raise e
print("Trying to attach S3 endpoints to the following route tables:", route_tables)
assert len(route_tables) >= 1, "No route tables were found. Please follow the VPC S3 endpoint creation "\
"guide by clicking the above link."
try:
ec2.create_vpc_endpoint(DryRun=False,
VpcEndpointType="Gateway",
VpcId=default_vpc,
ServiceName="com.amazonaws.{}.s3".format(aws_region),
RouteTableIds=route_tables)
print("S3 endpoint created successfully!")
except Exception as e:
if "RouteAlreadyExists" in str(e):
print("S3 endpoint already exists.")
elif "UnauthorizedOperation" in str(e):
display(Markdown(generate_help_for_s3_endpoint_permissions(role)))
raise e
else:
display(Markdown(create_s3_endpoint_manually(aws_region, default_vpc)))
raise e
```
## Setup the environment
The environment is defined in a Python file called “object_tracker_env.py” and the file can be found at `src/robomaker/environments/`. This file implements the gym interface for our Gazebo based RoboMakersimulator. This is a common environment file used by both SageMaker and RoboMaker. The environment variable - `NODE_TYPE` defines which node the code is running on. So, the expressions that have `rospy` dependencies are executed on RoboMaker only.
We can experiment with different reward functions by modifying `reward_function` in this file. Action space and steering angles can be changed by modifying the step method in `TurtleBot3ObjectTrackerAndFollowerDiscreteEnv` class.
### Configure the preset for RL algorithm
The parameters that configure the RL training job are defined in `src/robomaker/presets/object_tracker.py`. Using the preset file, you can define agent parameters to select the specific agent algorithm. We suggest using Clipped PPO for this example.
You can edit this file to modify algorithm parameters like learning_rate, neural network structure, batch_size, discount factor etc.
```
!pygmentize src/robomaker/presets/object_tracker.py
```
### Training Entrypoint
The training code is written in the file “training_worker.py” which is uploaded in the /src directory. At a high level, it does the following:
- Uploads SageMaker node's IP address.
- Starts a Redis server which receives agent experiences sent by rollout worker[s] (RoboMaker simulator).
- Trains the model everytime after a certain number of episodes are received.
- Uploads the new model weights on S3. The rollout workers then update their model to execute the next set of episodes.
```
# Uncomment the line below to see the training code
#!pygmentize src/training_worker.py
```
## Train the model using Python SDK/ script mode
```
s3_location = "s3://%s/%s" % (s3_bucket, s3_prefix)
!aws s3 rm --recursive {s3_location}
# Make any changes to the envrironment and preset files below and upload these files if you want to use custom environment and preset
!aws s3 cp src/robomaker/environments/ {s3_location}/environments/ --recursive --exclude ".ipynb_checkpoints*"
!aws s3 cp src/robomaker/presets/ {s3_location}/presets/ --recursive --exclude ".ipynb_checkpoints*"
```
First, we define the following algorithm metrics that we want to capture from cloudwatch logs to monitor the training progress. These are algorithm specific parameters and might change for different algorithm. We use [Clipped PPO](https://coach.nervanasys.com/algorithms/policy_optimization/cppo/index.html) for this example.
```
metric_definitions = [
# Training> Name=main_level/agent, Worker=0, Episode=19, Total reward=-102.88, Steps=19019, Training iteration=1
{'Name': 'reward-training',
'Regex': '^Training>.*Total reward=(.*?),'},
# Policy training> Surrogate loss=-0.32664725184440613, KL divergence=7.255815035023261e-06, Entropy=2.83156156539917, training epoch=0, learning_rate=0.00025
{'Name': 'ppo-surrogate-loss',
'Regex': '^Policy training>.*Surrogate loss=(.*?),'},
{'Name': 'ppo-entropy',
'Regex': '^Policy training>.*Entropy=(.*?),'},
# Testing> Name=main_level/agent, Worker=0, Episode=19, Total reward=1359.12, Steps=20015, Training iteration=2
{'Name': 'reward-testing',
'Regex': '^Testing>.*Total reward=(.*?),'},
]
```
We use the RLEstimator for training RL jobs.
1. Specify the source directory where the environment, presets and training code is uploaded.
2. Specify the entry point as the training code
3. Specify the choice of RL toolkit and framework. This automatically resolves to the ECR path for the RL Container.
4. Define the training parameters such as the instance count, instance type, job name, s3_bucket and s3_prefix for storing model checkpoints and metadata. **Only 1 training instance is supported for now.**
4. Set the RLCOACH_PRESET as "object_tracker" for this example.
5. Define the metrics definitions that you are interested in capturing in your logs. These can also be visualized in CloudWatch and SageMaker Notebooks.
```
RLCOACH_PRESET = "object_tracker"
instance_type = "ml.c5.4xlarge"
estimator = RLEstimator(entry_point="training_worker.py",
source_dir='src',
dependencies=["common/sagemaker_rl"],
toolkit=RLToolkit.COACH,
toolkit_version='0.11.0',
framework=RLFramework.TENSORFLOW,
role=role,
train_instance_type=instance_type,
train_instance_count=1,
output_path=s3_output_path,
base_job_name=job_name_prefix,
train_max_run=job_duration_in_seconds,
hyperparameters={"s3_bucket": s3_bucket,
"s3_prefix": s3_prefix,
"aws_region": aws_region,
"RLCOACH_PRESET": RLCOACH_PRESET,
},
metric_definitions = metric_definitions,
subnets=default_subnets,
security_group_ids=default_security_groups,
)
estimator.fit(job_name=job_name, wait=False)
```
### Start the Robomaker job
```
from botocore.exceptions import UnknownServiceError
robomaker = boto3.client("robomaker")
```
### Create Simulation Application
We first create a RoboMaker simulation application using the `object-tracker public bundle`. Please refer to [RoboMaker Sample Application Github Repository](https://github.com/aws-robotics/aws-robomaker-sample-application-objecttracker) if you want to learn more about this bundle or modify it.
```
bundle_s3_key = 'object-tracker/simulation_ws.tar.gz'
bundle_source = {'s3Bucket': s3_bucket,
's3Key': bundle_s3_key,
'architecture': "X86_64"}
simulation_software_suite={'name': 'Gazebo',
'version': '7'}
robot_software_suite={'name': 'ROS',
'version': 'Kinetic'}
rendering_engine={'name': 'OGRE',
'version': '1.x'}
simulation_application_bundle_location = "https://s3-us-west-2.amazonaws.com/robomaker-applications-us-west-2-11d8d0439f6a/object-tracker/object-tracker-1.0.74.0.1.0.105.0/simulation_ws.tar.gz"
!wget {simulation_application_bundle_location}
!aws s3 cp simulation_ws.tar.gz s3://{s3_bucket}/{bundle_s3_key}
!rm simulation_ws.tar.gz
app_name = "object-tracker-sample-application" + strftime("%y%m%d-%H%M%S", gmtime())
try:
response = robomaker.create_simulation_application(name=app_name,
sources=[bundle_source],
simulationSoftwareSuite=simulation_software_suite,
robotSoftwareSuite=robot_software_suite,
renderingEngine=rendering_engine
)
simulation_app_arn = response["arn"]
print("Created a new simulation app with ARN:", simulation_app_arn)
except Exception as e:
if "AccessDeniedException" in str(e):
display(Markdown(generate_help_for_robomaker_all_permissions(role)))
raise e
else:
raise e
```
### Launch the Simulation job on RoboMaker
We create [AWS RoboMaker](https://console.aws.amazon.com/robomaker/home#welcome) Simulation Jobs that simulates the environment and shares this data with SageMaker for training.
```
num_simulation_workers = 1
envriron_vars = {
"MODEL_S3_BUCKET": s3_bucket,
"MODEL_S3_PREFIX": s3_prefix,
"ROS_AWS_REGION": aws_region,
"MARKOV_PRESET_FILE": "object_tracker.py",
"NUMBER_OF_ROLLOUT_WORKERS": str(num_simulation_workers)}
simulation_application = {"application":simulation_app_arn,
"launchConfig": {"packageName": "object_tracker_simulation",
"launchFile": "distributed_training.launch",
"environmentVariables": envriron_vars}
}
vpcConfig = {"subnets": default_subnets,
"securityGroups": default_security_groups,
"assignPublicIp": True}
responses = []
for job_no in range(num_simulation_workers):
response = robomaker.create_simulation_job(iamRole=role,
clientRequestToken=strftime("%Y-%m-%d-%H-%M-%S", gmtime()),
maxJobDurationInSeconds=job_duration_in_seconds,
failureBehavior="Continue",
simulationApplications=[simulation_application],
vpcConfig=vpcConfig
)
responses.append(response)
print("Created the following jobs:")
job_arns = [response["arn"] for response in responses]
for job_arn in job_arns:
print("Job ARN", job_arn)
```
### Visualizing the simulations in RoboMaker
You can visit the RoboMaker console to visualize the simulations or run the following cell to generate the hyperlinks.
```
display(Markdown(generate_robomaker_links(job_arns, aws_region)))
```
### Clean Up
Execute the cells below if you want to kill RoboMaker and SageMaker job. It also removes RoboMaker resources created during the run.
```
for job_arn in job_arns:
robomaker.cancel_simulation_job(job=job_arn)
sage_session.sagemaker_client.stop_training_job(TrainingJobName=estimator._current_job_name)
```
### Evaluation
```
envriron_vars = {"MODEL_S3_BUCKET": s3_bucket,
"MODEL_S3_PREFIX": s3_prefix,
"ROS_AWS_REGION": aws_region,
"NUMBER_OF_TRIALS": str(20),
"MARKOV_PRESET_FILE": "%s.py" % RLCOACH_PRESET
}
simulation_application = {"application":simulation_app_arn,
"launchConfig": {"packageName": "object_tracker_simulation",
"launchFile": "evaluation.launch",
"environmentVariables": envriron_vars}
}
vpcConfig = {"subnets": default_subnets,
"securityGroups": default_security_groups,
"assignPublicIp": True}
response = robomaker.create_simulation_job(iamRole=role,
clientRequestToken=strftime("%Y-%m-%d-%H-%M-%S", gmtime()),
maxJobDurationInSeconds=job_duration_in_seconds,
failureBehavior="Continue",
simulationApplications=[simulation_application],
vpcConfig=vpcConfig
)
print("Created the following job:")
print("Job ARN", response["arn"])
```
### Clean Up Simulation Application Resource
```
robomaker.delete_simulation_application(application=simulation_app_arn)
```
|
github_jupyter
|
# This jupyter notebook contains examples of
- some basic functions related to Global Distance Test (GDT) analyses
- local accuracy plot
```
%matplotlib notebook
import numpy as np
import matplotlib.pyplot as plt
import MDAnalysis as mda
import pyrexMD.misc as misc
import pyrexMD.core as core
import pyrexMD.topology as top
import pyrexMD.analysis.analyze as ana
import pyrexMD.analysis.gdt as gdt
```
We define MDAnalysis universes to handle data. In this case we define:
- ref: universe with reference structure
- mobile: universe with trajectory
```
pdb = "files/traj_rna/4tzx_ref.pdb"
tpr = "files/traj_rna/traj_rna.tpr"
traj = "files/traj_rna/traj_rna_cat.xtc"
ref = mda.Universe(pdb)
mobile = mda.Universe(tpr, traj)
tv = core.iPlayer(mobile)
tv()
```
# Global Distance Test (GDT) Analysis
first we norm and align the universes (shift res ids, atom ids) and run the Global Distance Test
```
# first norm and align universes
top.norm_and_align_universe(mobile, ref)
# run GDT using selection idnex string for correct mapping
GDT = gdt.GDT_rna(mobile, ref)
GDT_percent, GDT_resids, GDT_cutoff, RMSD, FRAME = GDT
```
Now we can calculate individual GDT scores
- TS: Total Score
- HA: High Accuracy
```
GDT_TS = gdt.get_GDT_TS(GDT_percent)
GDT_HA = gdt.get_GDT_HA(GDT_percent)
```
We can print the scores in a table to take a quick look on the content
```
frames = [i for i in range(len(GDT_TS))]
misc.cprint("GDT TS GDT HA frame", "blue")
_ = misc.print_table([GDT_TS, GDT_HA, frames], verbose_stop=10, spacing=10)
```
alternatively we can also first rank the scores and print the table sorted by rank
```
SCORES = gdt.GDT_rank_scores(GDT_percent, ranking_order="GDT_TS", verbose=False)
GDT_TS_ranked, GDT_HA_ranked, GDT_ndx_ranked = SCORES
misc.cprint("GDT TS GDT HA frame", "blue")
_ = misc.print_table([GDT_TS_ranked, GDT_HA_ranked, GDT_ndx_ranked], spacing=10, verbose_stop=10)
```
To plot the GDT_TS curve we can use a generalized PLOT function:
```
fig, ax = ana.PLOT(xdata=frames, ydata=GDT_TS, xlabel="Frame", ylabel="GDT TS")
```
Histrograms are often also important as they can be used to extract probabilities of protein conformations
```
hist = ana.plot_hist(GDT_TS, n_bins=20, xlabel="GDT TS", ylabel="Counts")
```
# Local Accuracy Plot
Figure showing local accuracy of models at specified frames to identify which parts of a structure are good or bad refined.
```
# edit text box positions of labels "Frame", "TS", "HA"
text_pos_kws = {"text_pos_Frame": [-33.6, -0.3],
"text_pos_TS": [-16.0, -0.3],
"text_pos_HA": [-7.4, -0.3],
"font_scale": 1.0,
"show_frames": True,
"vmax": 14}
# plot
A = gdt.plot_LA_rna(mobile, ref, GDT_TS_ranked, GDT_HA_ranked, GDT_ndx_ranked, **text_pos_kws)
```
|
github_jupyter
|
# Text Using Markdown
**If you double click on this cell**, you will see the text change so that all of the formatting is removed. This allows you to edit this block of text. This block of text is written using [Markdown](http://daringfireball.net/projects/markdown/syntax), which is a way to format text using headers, links, italics, and many other options. Hit _shift_ + _enter_ or _shift_ + _return_ on your keyboard to show the formatted text again. This is called "running" the cell, and you can also do it using the run button in the toolbar.
# Code cells
One great advantage of IPython notebooks is that you can show your Python code alongside the results, add comments to the code, or even add blocks of text using Markdown. These notebooks allow you to collaborate with others and share your work. The following cell is a code cell.
```
# Hit shift + enter or use the run button to run this cell and see the results
print('hello world')
# The last line of every code cell will be displayed by default,
# even if you don't print it. Run this cell to see how this works.
2 + 2 # The result of this line will not be displayed
3 + 3 # The result of this line will be displayed, because it is the last line of the cell
```
# Nicely formatted results
IPython notebooks allow you to display nicely formatted results, such as plots and tables, directly in
the notebook. You'll learn how to use the following libraries later on in this course, but for now here's a
preview of what IPython notebook can do.
```
# If you run this cell, you should see the values displayed as a table.
# Pandas is a software library for data manipulation and analysis. You'll learn to use it later in this course.
import pandas as pd
df = pd.DataFrame({'a': [2, 4, 6, 8], 'b': [1, 3, 5, 7]})
df
# If you run this cell, you should see a scatter plot of the function y = x^2
%pylab inline
import matplotlib.pyplot as plt
xs = range(-30, 31)
ys = [x ** 2 for x in xs]
plt.scatter(xs, ys)
```
# Creating cells
To create a new **code cell**, click "Insert > Insert Cell [Above or Below]". A code cell will automatically be created.
To create a new **markdown cell**, first follow the process above to create a code cell, then change the type from "Code" to "Markdown" using the dropdown next to the run, stop, and restart buttons.
# Re-running cells
If you find a bug in your code, you can always update the cell and re-run it. However, any cells that come afterward won't be automatically updated. Try it out below. First run each of the three cells. The first two don't have any output, but you will be able to tell they've run because a number will appear next to them, for example, "In [5]". The third cell should output the message "Intro to Data Analysis is awesome!"
```
class_name = "Intro to Data Analysis"
message = class_name + " is awesome!"
message
```
Once you've run all three cells, try modifying the first one to set `class_name` to your name, rather than "Intro to Data Analysis", so you can print that you are awesome. Then rerun the first and third cells without rerunning the second.
You should have seen that the third cell still printed "Intro to Data Analysis is awesome!" That's because you didn't rerun the second cell, so even though the `class_name` variable was updated, the `message` variable was not. Now try rerunning the second cell, and then the third.
You should have seen the output change to "*your name* is awesome!" Often, after changing a cell, you'll want to rerun all the cells below it. You can do that quickly by clicking "Cell > Run All Below".
One final thing to remember: if you shut down the kernel after saving your notebook, the cells' output will still show up as you left it at the end of your session when you start the notebook back up. However, the state of the kernel will be reset. If you are actively working on a notebook, remember to re-run your cells to set up your working environment to really pick up where you last left off.
|
github_jupyter
|
<h3> ABSTRACT </h3>
All CMEMS in situ data products can be found and downloaded after [registration](http://marine.copernicus.eu/services-portfolio/register-now/) via [CMEMS catalogue] (http://marine.copernicus.eu/services-portfolio/access-to-products/).
Such channel is advisable just for sporadic netCDF donwloading because when operational, interaction with the web user interface is not practical. In this context though, the use of scripts for ftp file transference is is a much more advisable approach.
As long as every line of such files contains information about the netCDFs contained within the different directories [see at tips why](https://github.com/CopernicusMarineInsitu/INSTACTraining/blob/master/tips/README.md), it is posible for users to loop over its lines to download only those that matches a number of specifications such as spatial coverage, time coverage, provider, data_mode, parameters or file_name related (region, data type, TS or PF, platform code, or/and platform category, timestamp).
<h3>PREREQUISITES</h3>
- [credentias](http://marine.copernicus.eu/services-portfolio/register-now/)
- aimed [in situ product name](http://cmems-resources.cls.fr/documents/PUM/CMEMS-INS-PUM-013.pdf)
- aimed [hosting distribution unit](https://github.com/CopernicusMarineInsitu/INSTACTraining/blob/master/tips/README.md)
- aimed [index file](https://github.com/CopernicusMarineInsitu/INSTACTraining/blob/master/tips/README.md)
i.e:
```
user = '' #type CMEMS user name within colons
password = '' #type CMEMS password within colons
product_name = 'INSITU_BAL_NRT_OBSERVATIONS_013_032' #type aimed CMEMS in situ product
distribution_unit = 'cmems.smhi.se' #type aimed hosting institution
index_file = 'index_history.txt' #type aimed index file name
#remember! platform category only for history and monthly directories
```
<h3>DOWNLOAD</h3>
1. Index file download
```
import ftplib
ftp=ftplib.FTP(distribution_unit,user,password)
ftp.cwd("Core")
ftp.cwd(product_name)
local_file = open(index_file, 'wb')
ftp.retrbinary('RETR ' + index_file, local_file.write)
local_file.close()
ftp.quit()
#ready when 221 Goodbye.!
```
<h3>QUICK VIEW</h3>
```
import numpy as np
import pandas as pd
from random import randint
index = np.genfromtxt(index_file, skip_header=6, unpack=False, delimiter=',', dtype=None,
names=['catalog_id', 'file_name', 'geospatial_lat_min', 'geospatial_lat_max',
'geospatial_lon_min', 'geospatial_lon_max',
'time_coverage_start', 'time_coverage_end',
'provider', 'date_update', 'data_mode', 'parameters'])
dataset = randint(0,len(index)) #ramdom line of the index file
values = [index[dataset]['catalog_id'], '<a href='+index[dataset]['file_name']+'>'+index[dataset]['file_name']+'</a>', index[dataset]['geospatial_lat_min'], index[dataset]['geospatial_lat_max'],
index[dataset]['geospatial_lon_min'], index[dataset]['geospatial_lon_max'], index[dataset]['time_coverage_start'],
index[dataset]['time_coverage_end'], index[dataset]['provider'], index[dataset]['date_update'], index[dataset]['data_mode'],
index[dataset]['parameters']]
headers = ['catalog_id', 'file_name', 'geospatial_lat_min', 'geospatial_lat_max',
'geospatial_lon_min', 'geospatial_lon_max',
'time_coverage_start', 'time_coverage_end',
'provider', 'date_update', 'data_mode', 'parameters']
df = pd.DataFrame(values, index=headers, columns=[dataset])
df.style
```
<h3>FILTERING CRITERIA</h3>
Regarding the above glimpse, it is posible to filter by 12 criteria. As example we will setup next a filter to only download those files that contains data within a defined boundingbox.
1. Aimed category
```
targeted_category = 'drifter'
```
2. netCDF filtering/selection
```
selected_netCDFs = [];
for netCDF in index:
file_name = netCDF['file_name']
folders = file_name.split('/')[3:len(file_name.split('/'))-1]
category = file_name.split('/')[3:len(file_name.split('/'))-1][len(file_name.split('/')[3:len(file_name.split('/'))-1])-1]
if (category == targeted_category):
selected_netCDFs.append(file_name)
print("total: " +str(len(selected_netCDFs)))
```
<h3> SELECTION DOWNLOAD </h3>
```
for nc in selected_netCDFs:
last_idx_slash = nc.rfind('/')
ncdf_file_name = nc[last_idx_slash+1:]
folders = nc.split('/')[3:len(nc.split('/'))-1]
host = nc.split('/')[2] #or distribution unit
ftp=ftplib.FTP(host,user,password)
for folder in folders:
ftp.cwd(folder)
local_file = open(ncdf_file_name, 'wb')
ftp.retrbinary('RETR '+ncdf_file_name, local_file.write)
local_file.close()
ftp.quit()
```
|
github_jupyter
|
# TensorFlow script mode training and serving
Script mode is a training script format for TensorFlow that lets you execute any TensorFlow training script in SageMaker with minimal modification. The [SageMaker Python SDK](https://github.com/aws/sagemaker-python-sdk) handles transferring your script to a SageMaker training instance. On the training instance, SageMaker's native TensorFlow support sets up training-related environment variables and executes your training script. In this tutorial, we use the SageMaker Python SDK to launch a training job and deploy the trained model.
Script mode supports training with a Python script, a Python module, or a shell script. In this example, we use a Python script to train a classification model on the [MNIST dataset](http://yann.lecun.com/exdb/mnist/). In this example, we will show how easily you can train a SageMaker using TensorFlow 1.x and TensorFlow 2.0 scripts with SageMaker Python SDK. In addition, this notebook demonstrates how to perform real time inference with the [SageMaker TensorFlow Serving container](https://github.com/aws/sagemaker-tensorflow-serving-container). The TensorFlow Serving container is the default inference method for script mode. For full documentation on the TensorFlow Serving container, please visit [here](https://github.com/aws/sagemaker-python-sdk/blob/master/src/sagemaker/tensorflow/deploying_tensorflow_serving.rst).
# Set up the environment
Let's start by setting up the environment:
```
import os
import sagemaker
from sagemaker import get_execution_role
sagemaker_session = sagemaker.Session()
role = get_execution_role()
region = sagemaker_session.boto_session.region_name
```
## Training Data
The MNIST dataset has been loaded to the public S3 buckets ``sagemaker-sample-data-<REGION>`` under the prefix ``tensorflow/mnist``. There are four ``.npy`` file under this prefix:
* ``train_data.npy``
* ``eval_data.npy``
* ``train_labels.npy``
* ``eval_labels.npy``
```
training_data_uri = 's3://sagemaker-sample-data-{}/tensorflow/mnist'.format(region)
```
# Construct a script for distributed training
This tutorial's training script was adapted from TensorFlow's official [CNN MNIST example](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/examples/tutorials/layers/cnn_mnist.py). We have modified it to handle the ``model_dir`` parameter passed in by SageMaker. This is an S3 path which can be used for data sharing during distributed training and checkpointing and/or model persistence. We have also added an argument-parsing function to handle processing training-related variables.
At the end of the training job we have added a step to export the trained model to the path stored in the environment variable ``SM_MODEL_DIR``, which always points to ``/opt/ml/model``. This is critical because SageMaker uploads all the model artifacts in this folder to S3 at end of training.
Here is the entire script:
```
!pygmentize 'mnist.py'
# TensorFlow 2.1 script
!pygmentize 'mnist-2.py'
```
# Create a training job using the `TensorFlow` estimator
The `sagemaker.tensorflow.TensorFlow` estimator handles locating the script mode container, uploading your script to a S3 location and creating a SageMaker training job. Let's call out a couple important parameters here:
* `py_version` is set to `'py3'` to indicate that we are using script mode since legacy mode supports only Python 2. Though Python 2 will be deprecated soon, you can use script mode with Python 2 by setting `py_version` to `'py2'` and `script_mode` to `True`.
* `distributions` is used to configure the distributed training setup. It's required only if you are doing distributed training either across a cluster of instances or across multiple GPUs. Here we are using parameter servers as the distributed training schema. SageMaker training jobs run on homogeneous clusters. To make parameter server more performant in the SageMaker setup, we run a parameter server on every instance in the cluster, so there is no need to specify the number of parameter servers to launch. Script mode also supports distributed training with [Horovod](https://github.com/horovod/horovod). You can find the full documentation on how to configure `distributions` [here](https://github.com/aws/sagemaker-python-sdk/tree/master/src/sagemaker/tensorflow#distributed-training).
```
from sagemaker.tensorflow import TensorFlow
mnist_estimator = TensorFlow(entry_point='mnist.py',
role=role,
instance_count=2,
instance_type='ml.p3.2xlarge',
framework_version='1.15.2',
py_version='py3',
distribution={'parameter_server': {'enabled': True}})
```
You can also initiate an estimator to train with TensorFlow 2.1 script. The only things that you will need to change are the script name and ``framewotk_version``
```
mnist_estimator2 = TensorFlow(entry_point='mnist-2.py',
role=role,
instance_count=2,
instance_type='ml.p3.2xlarge',
framework_version='2.1.0',
py_version='py3',
distribution={'parameter_server': {'enabled': True}})
```
## Calling ``fit``
To start a training job, we call `estimator.fit(training_data_uri)`.
An S3 location is used here as the input. `fit` creates a default channel named `'training'`, which points to this S3 location. In the training script we can then access the training data from the location stored in `SM_CHANNEL_TRAINING`. `fit` accepts a couple other types of input as well. See the API doc [here](https://sagemaker.readthedocs.io/en/stable/estimators.html#sagemaker.estimator.EstimatorBase.fit) for details.
When training starts, the TensorFlow container executes mnist.py, passing `hyperparameters` and `model_dir` from the estimator as script arguments. Because we didn't define either in this example, no hyperparameters are passed, and `model_dir` defaults to `s3://<DEFAULT_BUCKET>/<TRAINING_JOB_NAME>`, so the script execution is as follows:
```bash
python mnist.py --model_dir s3://<DEFAULT_BUCKET>/<TRAINING_JOB_NAME>
```
When training is complete, the training job will upload the saved model for TensorFlow serving.
```
mnist_estimator.fit(training_data_uri)
```
Calling fit to train a model with TensorFlow 2.1 script.
```
mnist_estimator2.fit(training_data_uri)
```
# Deploy the trained model to an endpoint
The `deploy()` method creates a SageMaker model, which is then deployed to an endpoint to serve prediction requests in real time. We will use the TensorFlow Serving container for the endpoint, because we trained with script mode. This serving container runs an implementation of a web server that is compatible with SageMaker hosting protocol. The [Using your own inference code]() document explains how SageMaker runs inference containers.
```
predictor = mnist_estimator.deploy(initial_instance_count=1, instance_type='ml.p2.xlarge')
```
Deployed the trained TensorFlow 2.1 model to an endpoint.
```
predictor2 = mnist_estimator2.deploy(initial_instance_count=1, instance_type='ml.p2.xlarge')
```
# Invoke the endpoint
Let's download the training data and use that as input for inference.
```
import numpy as np
!aws --region {region} s3 cp s3://sagemaker-sample-data-{region}/tensorflow/mnist/train_data.npy train_data.npy
!aws --region {region} s3 cp s3://sagemaker-sample-data-{region}/tensorflow/mnist/train_labels.npy train_labels.npy
train_data = np.load('train_data.npy')
train_labels = np.load('train_labels.npy')
```
The formats of the input and the output data correspond directly to the request and response formats of the `Predict` method in the [TensorFlow Serving REST API](https://www.tensorflow.org/serving/api_rest). SageMaker's TensforFlow Serving endpoints can also accept additional input formats that are not part of the TensorFlow REST API, including the simplified JSON format, line-delimited JSON objects ("jsons" or "jsonlines"), and CSV data.
In this example we are using a `numpy` array as input, which will be serialized into the simplified JSON format. In addtion, TensorFlow serving can also process multiple items at once as you can see in the following code. You can find the complete documentation on how to make predictions against a TensorFlow serving SageMaker endpoint [here](https://github.com/aws/sagemaker-python-sdk/blob/master/src/sagemaker/tensorflow/deploying_tensorflow_serving.rst#making-predictions-against-a-sagemaker-endpoint).
```
predictions = predictor.predict(train_data[:50])
for i in range(0, 50):
prediction = predictions['predictions'][i]['classes']
label = train_labels[i]
print('prediction is {}, label is {}, matched: {}'.format(prediction, label, prediction == label))
```
Examine the prediction result from the TensorFlow 2.1 model.
```
predictions2 = predictor2.predict(train_data[:50])
for i in range(0, 50):
prediction = np.argmax(predictions2['predictions'][i])
label = train_labels[i]
print('prediction is {}, label is {}, matched: {}'.format(prediction, label, prediction == label))
```
# Delete the endpoint
Let's delete the endpoint we just created to prevent incurring any extra costs.
```
predictor.delete_endpoint()
```
Delete the TensorFlow 2.1 endpoint as well.
```
predictor2.delete_endpoint()
```
|
github_jupyter
|
# Rekurrente Netze (RNNs)
## Sequentialle Daten
<img src="img/ag/Figure-22-001.png" style="width: 10%; margin-left: auto; margin-right: auto;"/>
## Floating Window
<img src="img/ag/Figure-22-002.png" style="width: 20%; margin-left: auto; margin-right: auto;"/>
## Verarbeitung mit MLP
<img src="img/ag/Figure-22-002.png" style="width: 20%; margin-left: 10%; margin-right: auto; float: left;"/>
<img src="img/ag/Figure-22-003.png" style="width: 35%; margin-left: 10%; margin-right: auto; float: right;"/>
## MLP berücksichtigt die Reihenfolge nicht!
<img src="img/ag/Figure-22-004.png" style="width: 25%; margin-left: auto; margin-right: auto;"/>
## RNNs: Netzwerke mit Speicher
<img src="img/ag/Figure-22-005.png" style="width: 15%; margin-left: auto; margin-right: auto;"/>
## Zustand: Reperatur-Roboter
<img src="img/ag/Figure-22-006.png" style="width: 35%; margin-left: auto; margin-right: auto;"/>
## Zustand: Reperatur-Roboter
<img src="img/ag/Figure-22-007.png" style="width: 35%; margin-left: auto; margin-right: auto;"/>
## Zustand: Reperatur-Roboter
<img src="img/ag/Figure-22-008.png" style="width: 35%; margin-left: auto; margin-right: auto;"/>
## Zustand: Reperatur-Roboter
<img src="img/ag/Figure-22-009.png" style="width: 85%; margin-left: auto; margin-right: auto;"/>
# Arbeitsweise RNN
<img src="img/ag/Figure-22-010.png" style="width: 85%; margin-left: auto; margin-right: auto;"/>
## State wird nach der Verarbeitung geschrieben
<img src="img/ag/Figure-22-011.png" style="width: 35%; margin-left: 10%; margin-right: auto; float: left;"/>
<img src="img/ag/Figure-22-012.png" style="width: 15%; margin-left: auto; margin-right: 10%; float: right;"/>
## Netzwerkstruktur (einzelner Wert)
Welche Operation ist sinnvoll?
<img src="img/ag/Figure-22-013.png" style="width: 35%; margin-left: auto; margin-right: auto;"/>
## Netzwerkstruktur (einzelner Wert)
<img src="img/ag/Figure-22-014.png" style="width: 35%; margin-left: auto; margin-right: auto;"/>
## Repräsentation in Diagrammen
<img src="img/ag/Figure-22-015.png" style="width: 10%; margin-left: auto; margin-right: auto;"/>
## Entfaltete Darstellung
<img src="img/ag/Figure-22-016.png" style="width: 45%; margin-left: auto; margin-right: auto;"/>
## Netzwerkstruktur für mehrere Werte
<img src="img/ag/Figure-22-018.png" style="width: 35%; margin-left: auto; margin-right: auto;"/>
## Darstellung der Daten
<img src="img/ag/Figure-22-019.png" style="width: 65%; margin-left: auto; margin-right: auto;"/>
# Darstellung der Daten
<img src="img/ag/Figure-22-020.png" style="width: 45%; margin-left: auto; margin-right: auto;"/>
# Darstellung der Daten
<img src="img/ag/Figure-22-021.png" style="width: 45%; margin-left: auto; margin-right: auto;"/>
## Arbeitsweise
<img src="img/ag/Figure-22-022.png" style="width: 55%; margin-left: auto; margin-right: auto;"/>
## Probleme
<div style="margin-top: 20pt; float:left;">
<ul>
<li>Verlust der Gradienten</li>
<li>Explosion der Gradienten</li>
<li>Vergessen</li>
</ul>
</div>
<img src="img/ag/Figure-22-023.png" style="width: 55%; margin-left: auto; margin-right: 5%; float: right;"/>
## LSTM
<img src="img/ag/Figure-22-029.png" style="width: 65%; margin-left: auto; margin-right: auto;"/>
## Gates
<img src="img/ag/Figure-22-024.png" style="width: 55%; margin-left: auto; margin-right: auto;"/>
## Gates
<img src="img/ag/Figure-22-025.png" style="width: 65%; margin-left: auto; margin-right: auto;"/>
## Forget-Gate
<img src="img/ag/Figure-22-026.png" style="width: 30%; margin-left: auto; margin-right: auto;"/>
## Remember Gate
<img src="img/ag/Figure-22-027.png" style="width: 65%; margin-left: auto; margin-right: auto;"/>
## Output Gate
<img src="img/ag/Figure-22-028.png" style="width: 55%; margin-left: auto; margin-right: auto;"/>
## LSTM
<img src="img/ag/Figure-22-029.png" style="width: 65%; margin-left: auto; margin-right: auto;"/>
## LSTM Funktionsweise
<img src="img/ag/Figure-22-030.png" style="width: 65%; margin-left: auto; margin-right: auto;"/>
## LSTM Funktionsweise
<img src="img/ag/Figure-22-031.png" style="width: 45%; margin-left: auto; margin-right: auto;"/>
## LSTM Funktionsweise
<img src="img/ag/Figure-22-032.png" style="width: 65%; margin-left: auto; margin-right: auto;"/>
## LSTM Funktionsweise
<img src="img/ag/Figure-22-033.png" style="width: 65%; margin-left: auto; margin-right: auto;"/>
## Verwendung von LSTMs
<img src="img/ag/Figure-22-034.png" style="width: 75%; margin-left: auto; margin-right: auto;"/>
## Darstellung von LSTM Layern
<img src="img/ag/Figure-22-035.png" style="width: 25%; margin-left: auto; margin-right: auto;"/>
## Conv/LSTM (Conv/RNN) Architektur
<img src="img/ag/Figure-22-036.png" style="width: 15%; margin-left: auto; margin-right: auto;"/>
## Tiefe RNN Netze
<img src="img/ag/Figure-22-037.png" style="width: 55%; margin-left: auto; margin-right: auto;"/>
## Bidirektionale RNNs
<img src="img/ag/Figure-22-038.png" style="width: 65%; margin-left: auto; margin-right: auto;"/>
## Tiefe Bidirektionale Netze
<img src="img/ag/Figure-22-039.png" style="width: 45%; margin-left: auto; margin-right: auto;"/>
# Anwendung: Generierung von Text
<img src="img/ag/Figure-22-040.png" style="width: 15%; margin-left: auto; margin-right: auto;"/>
## Trainieren mittels Sliding Window
<img src="img/ag/Figure-22-042.png" style="width: 25%; margin-left: auto; margin-right: auto;"/>
# Vortrainierte LSTM-Modelle
```
from fastai.text.all import *
path = untar_data(URLs.IMDB)
path.ls()
(path/'train').ls()
dls = TextDataLoaders.from_folder(untar_data(URLs.IMDB), valid='test')
dls.show_batch()
learn = text_classifier_learner(dls, AWD_LSTM, drop_mult=0.5, metrics=accuracy)
# learn.fine_tune(4, 1e-2)
learn.fine_tune(4, 1e-2)
learn.show_results()
learn.predict("I really liked that movie!")
```
# ULMFiT
Problem: Wir trainieren die oberen Layer des Classifiers auf unser Problem, aber das Language-Model bleibt auf Wikipedia spezialisiert!
Lösung: Fine-Tuning des Language-Models bevor wir den Classifier trainieren.
<img src="img/ulmfit.png" style="width: 75%; margin-left: auto; margin-right: auto;"/>
```
dls_lm = TextDataLoaders.from_folder(path, is_lm=True, valid_pct=0.1)
dls_lm.show_batch(max_n=5)
learn = language_model_learner(dls_lm, AWD_LSTM, metrics=[accuracy, Perplexity()], path=path, wd=0.1).to_fp16()
learn.fit_one_cycle(1, 1e-2)
learn.save('epoch-1')
learn = learn.load('epoch-1')
learn.unfreeze()
learn.fit_one_cycle(10, 1e-3)
learn.save_encoder('finetuned')
TEXT = "I liked this movie because"
N_WORDS = 40
N_SENTENCES = 2
preds = [learn.predict(TEXT, N_WORDS, temperature=0.75)
for _ in range(N_SENTENCES)]
print("\n".join(preds))
dls_clas = TextDataLoaders.from_folder(untar_data(URLs.IMDB), valid='test', text_vocab=dls_lm.vocab)
learn = text_classifier_learner(dls, AWD_LSTM, drop_mult=0.5, metrics=accuracy)
learn = learn.load_encoder('finetuned')
learn.fit_one_cycle(1, 2e-2)
learn.freeze_to(-2)
learn.fit_one_cycle(1, slice(1e-2/(2.6**4),1e-2))
learn.freeze_to(-3)
learn.fit_one_cycle(1, slice(5e-3/(2.6**4),5e-3))
learn.unfreeze()
learn.fit_one_cycle(2, slice(1e-3/(2.6**4),1e-3))
```
|
github_jupyter
|
# This notebook is copied from [here](https://github.com/warmspringwinds/tensorflow_notes/blob/master/tfrecords_guide.ipynb) with some small changes
---
### Introduction
In this post we will cover how to convert a dataset into _.tfrecord_ file.
Binary files are sometimes easier to use, because you don't have to specify
different directories for images and groundtruth annotations. While storing your data
in binary file, you have your data in one block of memory, compared to storing
each image and annotation separately. Openning a file is a considerably
time-consuming operation especially if you use _hdd_ and not _ssd_, because it
involves moving the disk reader head and that takes quite some time. Overall,
by using binary files you make it easier to distribute and make
the data better aligned for efficient reading.
The post consists of tree parts:
* in the first part, we demonstrate how you can get raw data bytes of any image using _numpy_ which is in some sense similar to what you do when converting your dataset to binary format.
* Second part shows how to convert a dataset to _tfrecord_ file without defining a computational graph and only by employing some built-in _tensorflow_ functions.
* Third part explains how to define a model for reading your data from created binary file and batch it in a random manner, which is necessary during training.
### Getting raw data bytes in numpy
Here we demonstrate how you can get raw data bytes of an image (any ndarray)
and how to restore the image back.
One important note is that **during this operation
the information about the dimensions of the image is lost and we have to
use it to recover the original image. This is one of the reasons why
we will have to store the raw image representation along with the dimensions
of the original image.**
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
cat_img = plt.imread('data/imgs/cat.jpg')
plt.imshow(cat_img)
# io.imshow(cat_img)
# Let's convert the picture into string representation
# using the ndarray.tostring() function
cat_string = cat_img.tostring()
# Now let's convert the string back to the image
# Important: the dtype should be specified
# otherwise the reconstruction will be errorness
# Reconstruction is 1d, so we need sizes of image
# to fully reconstruct it.
reconstructed_cat_1d = np.fromstring(cat_string, dtype=np.uint8)
# Here we reshape the 1d representation
# This is the why we need to store the sizes of image
# along with its serialized representation.
reconstructed_cat_img = reconstructed_cat_1d.reshape(cat_img.shape)
# Let's check if we got everything right and compare
# reconstructed array to the original one.
np.allclose(cat_img, reconstructed_cat_img)
```
### Creating a _.tfrecord_ file and reading it without defining a graph
Here we show how to write a small dataset (three images/annotations from _PASCAL VOC_) to
_.tfrrecord_ file and read it without defining a computational graph.
We also make sure that images that we read back from _.tfrecord_ file are equal to
the original images. Pay attention that we also write the sizes of the images along with
the image in the raw format. We showed an example on why we need to also store the size
in the previous section.
```
# Get some image/annotation pairs for example
filename_pairs = [
('data/VOC2012/JPEGImages/2007_000032.jpg',
'data/VOC2012/SegmentationClass/2007_000032.png'),
('data/VOC2012/JPEGImages/2007_000039.jpg',
'data/VOC2012/SegmentationClass/2007_000039.png'),
('data/VOC2012/JPEGImages/2007_000033.jpg',
'data/VOC2012/SegmentationClass/2007_000033.png')
]
%matplotlib inline
# Important: We are using PIL to read .png files later.
# This was done on purpose to read indexed png files
# in a special way -- only indexes and not map the indexes
# to actual rgb values. This is specific to PASCAL VOC
# dataset data. If you don't want thit type of behaviour
# consider using skimage.io.imread()
from PIL import Image
import numpy as np
import skimage.io as io
import tensorflow as tf
def _bytes_feature(value):
return tf.train.Feature(bytes_list=tf.train.BytesList(value=[value]))
def _int64_feature(value):
return tf.train.Feature(int64_list=tf.train.Int64List(value=[value]))
tfrecords_filename = 'pascal_voc_segmentation.tfrecords'
writer = tf.python_io.TFRecordWriter(tfrecords_filename)
# Let's collect the real images to later on compare
# to the reconstructed ones
original_images = []
for img_path, annotation_path in filename_pairs:
img = np.array(Image.open(img_path))
annotation = np.array(Image.open(annotation_path))
# The reason to store image sizes was demonstrated
# in the previous example -- we have to know sizes
# of images to later read raw serialized string,
# convert to 1d array and convert to respective
# shape that image used to have.
height = img.shape[0]
width = img.shape[1]
# Put in the original images into array
# Just for future check for correctness
original_images.append((img, annotation))
img_raw = img.tostring()
annotation_raw = annotation.tostring()
example = tf.train.Example(features=tf.train.Features(feature={
'height': _int64_feature(height),
'width': _int64_feature(width),
'image_raw': _bytes_feature(img_raw),
'mask_raw': _bytes_feature(annotation_raw)}))
writer.write(example.SerializeToString())
writer.close()
reconstructed_images = []
record_iterator = tf.python_io.tf_record_iterator(path=tfrecords_filename)
for string_record in record_iterator:
example = tf.train.Example()
example.ParseFromString(string_record)
height = int(example.features.feature['height']
.int64_list
.value[0])
width = int(example.features.feature['width']
.int64_list
.value[0])
img_string = (example.features.feature['image_raw']
.bytes_list
.value[0])
annotation_string = (example.features.feature['mask_raw']
.bytes_list
.value[0])
img_1d = np.fromstring(img_string, dtype=np.uint8)
reconstructed_img = img_1d.reshape((height, width, -1))
annotation_1d = np.fromstring(annotation_string, dtype=np.uint8)
# Annotations don't have depth (3rd dimension)
reconstructed_annotation = annotation_1d.reshape((height, width))
reconstructed_images.append((reconstructed_img, reconstructed_annotation))
# Let's check if the reconstructed images match
# the original images
for original_pair, reconstructed_pair in zip(original_images, reconstructed_images):
img_pair_to_compare, annotation_pair_to_compare = zip(original_pair,
reconstructed_pair)
print(np.allclose(*img_pair_to_compare))
print(np.allclose(*annotation_pair_to_compare))
```
### Defining the graph to read and batch images from _.tfrecords_
Here we define a graph to read and batch images from the file that we have created
previously. It is very important to randomly shuffle images during training and depending
on the application we have to use different batch size.
It is very important to point out that if we use batching -- we have to define
the sizes of images beforehand. This may sound like a limitation, but actually in the
Image Classification and Image Segmentation fields the training is performed on the images
of the same size.
The code provided here is partially based on [this official example](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/examples/how_tos/reading_data/fully_connected_reader.py) and code from [this stackoverflow question](http://stackoverflow.com/questions/35028173/how-to-read-images-with-different-size-in-a-tfrecord-file).
Also if you want to know how you can control the batching according to your need read [these docs](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/g3doc/api_docs/python/functions_and_classes/shard2/tf.train.shuffle_batch.md)
.
```
%matplotlib inline
import tensorflow as tf
import skimage.io as io
IMAGE_HEIGHT = 384
IMAGE_WIDTH = 384
tfrecords_filename = 'pascal_voc_segmentation.tfrecords'
def read_and_decode(filename_queue):
reader = tf.TFRecordReader()
_, serialized_example = reader.read(filename_queue)
features = tf.parse_single_example(
serialized_example,
# Defaults are not specified since both keys are required.
features={
'height': tf.FixedLenFeature([], tf.int64),
'width': tf.FixedLenFeature([], tf.int64),
'image_raw': tf.FixedLenFeature([], tf.string),
'mask_raw': tf.FixedLenFeature([], tf.string)
})
# Convert from a scalar string tensor (whose single string has
# length mnist.IMAGE_PIXELS) to a uint8 tensor with shape
# [mnist.IMAGE_PIXELS].
image = tf.decode_raw(features['image_raw'], tf.uint8)
annotation = tf.decode_raw(features['mask_raw'], tf.uint8)
height = tf.cast(features['height'], tf.int32)
width = tf.cast(features['width'], tf.int32)
image_shape = tf.stack([height, width, 3])
annotation_shape = tf.stack([height, width, 1])
image = tf.reshape(image, image_shape)
annotation = tf.reshape(annotation, annotation_shape)
image_size_const = tf.constant((IMAGE_HEIGHT, IMAGE_WIDTH, 3), dtype=tf.int32)
annotation_size_const = tf.constant((IMAGE_HEIGHT, IMAGE_WIDTH, 1), dtype=tf.int32)
# Random transformations can be put here: right before you crop images
# to predefined size. To get more information look at the stackoverflow
# question linked above.
resized_image = tf.image.resize_image_with_crop_or_pad(image=image,
target_height=IMAGE_HEIGHT,
target_width=IMAGE_WIDTH)
resized_annotation = tf.image.resize_image_with_crop_or_pad(image=annotation,
target_height=IMAGE_HEIGHT,
target_width=IMAGE_WIDTH)
images, annotations = tf.train.shuffle_batch( [resized_image, resized_annotation],
batch_size=2,
capacity=30,
num_threads=2,
min_after_dequeue=10)
return images, annotations
filename_queue = tf.train.string_input_producer(
[tfrecords_filename], num_epochs=10)
# Even when reading in multiple threads, share the filename
# queue.
image, annotation = read_and_decode(filename_queue)
# The op for initializing the variables.
init_op = tf.group(tf.global_variables_initializer(),
tf.local_variables_initializer())
with tf.Session() as sess:
sess.run(init_op)
coord = tf.train.Coordinator()
threads = tf.train.start_queue_runners(coord=coord)
# Let's read off 3 batches just for example
for i in range(3):
img, anno = sess.run([image, annotation])
print(img[0, :, :, :].shape)
print('current batch')
# We selected the batch size of two
# So we should get two image pairs in each batch
# Let's make sure it is random
io.imshow(img[0, :, :, :])
io.show()
io.imshow(anno[0, :, :, 0])
io.show()
io.imshow(img[1, :, :, :])
io.show()
io.imshow(anno[1, :, :, 0])
io.show()
coord.request_stop()
coord.join(threads)
```
### Conclusion and Discussion
In this post we covered how to convert a dataset into _.tfrecord_ format,
made sure that we have the same data and saw how to define a graph to
read and batch files from the created file.
|
github_jupyter
|
# Custom Models in pycalphad: Viscosity
## Viscosity Model Background
We are going to take a CALPHAD-based property model from the literature and use it to predict the viscosity of Al-Cu-Zr liquids.
For a binary alloy liquid under small undercooling, Gąsior suggested an entropy model of the form
$$\eta = (\sum_i x_i \eta_i ) (1 - 2\frac{S_{ex}}{R})$$
where $\eta_i$ is the viscosity of the element $i$, $x_i$ is the mole fraction, $S_{ex}$ is the excess entropy, and $R$ is the gas constant.
For more details on this model, see
1. M.E. Trybula, T. Gancarz, W. Gąsior, *Density, surface tension and viscosity of liquid binary Al-Zn and ternary Al-Li-Zn alloys*, Fluid Phase Equilibria 421 (2016) 39-48, [doi:10.1016/j.fluid.2016.03.013](http://dx.doi.org/10.1016/j.fluid.2016.03.013).
2. Władysław Gąsior, *Viscosity modeling of binary alloys: Comparative studies*, Calphad 44 (2014) 119-128, [doi:10.1016/j.calphad.2013.10.007](http://dx.doi.org/10.1016/j.calphad.2013.10.007).
3. Chenyang Zhou, Cuiping Guo, Changrong Li, Zhenmin Du, *Thermodynamic assessment of the phase equilibria and prediction of glass-forming ability of the Al–Cu–Zr system*, Journal of Non-Crystalline Solids 461 (2017) 47-60, [doi:10.1016/j.jnoncrysol.2016.09.031](https://doi.org/10.1016/j.jnoncrysol.2016.09.031).
```
from pycalphad import Database
```
## TDB Parameters
We can calculate the excess entropy of the liquid using the Al-Cu-Zr thermodynamic database from Zhou et al.
We add three new parameters to describe the viscosity (in Pa-s) of the pure elements Al, Cu, and Zr:
```
$ Viscosity test parameters
PARAMETER ETA(LIQUID,AL;0) 2.98150E+02 +0.000281*EXP(12300/(8.3145*T)); 6.00000E+03
N REF:0 !
PARAMETER ETA(LIQUID,CU;0) 2.98150E+02 +0.000657*EXP(21500/(8.3145*T)); 6.00000E+03
N REF:0 !
PARAMETER ETA(LIQUID,ZR;0) 2.98150E+02 +4.74E-3 - 4.97E-6*(T-2128) ; 6.00000E+03
N REF:0 !
```
Great! However, if we try to load the database now, we will get an error. This is because `ETA` parameters are not supported by default in pycalphad, so we need to tell pycalphad's TDB parser that "ETA" should be on the list of supported parameter types.
```
dbf = Database('alcuzr-viscosity.tdb')
```
### Adding the `ETA` parameter to the TDB parser
```
import pycalphad.io.tdb_keywords
pycalphad.io.tdb_keywords.TDB_PARAM_TYPES.append('ETA')
```
Now the database will load:
```
dbf = Database('alcuzr-viscosity.tdb')
```
## Writing the Custom Viscosity Model
Now that we have our `ETA` parameters in the database, we need to write a `Model` class to tell pycalphad how to compute viscosity. All custom models are subclasses of the pycalphad `Model` class.
When the `ViscosityModel` is constructed, the `build_phase` method is run and we need to construct the viscosity model after doing all the other initialization using a new method `build_viscosity`. The implementation of `build_viscosity` needs to do four things:
1. Query the Database for all the `ETA` parameters
2. Compute their weighted sum
3. Compute the excess entropy of the liquid
4. Plug all the values into the Gąsior equation and return the result
Since the `build_phase` method sets the attribute `viscosity` to the `ViscosityModel`, we can access the property using `viscosity` as the output in pycalphad caluclations.
```
from tinydb import where
import sympy
from pycalphad import Model, variables as v
class ViscosityModel(Model):
def build_phase(self, dbe):
super(ViscosityModel, self).build_phase(dbe)
self.viscosity = self.build_viscosity(dbe)
def build_viscosity(self, dbe):
if self.phase_name != 'LIQUID':
raise ValueError('Viscosity is only defined for LIQUID phase')
phase = dbe.phases[self.phase_name]
param_search = dbe.search
# STEP 1
eta_param_query = (
(where('phase_name') == phase.name) & \
(where('parameter_type') == 'ETA') & \
(where('constituent_array').test(self._array_validity))
)
# STEP 2
eta = self.redlich_kister_sum(phase, param_search, eta_param_query)
# STEP 3
excess_energy = self.GM - self.models['ref'] - self.models['idmix']
#liquid_mod = Model(dbe, self.components, self.phase_name)
## we only want the excess contributions to the entropy
#del liquid_mod.models['ref']
#del liquid_mod.models['idmix']
excess_entropy = -excess_energy.diff(v.T)
ks = 2
# STEP 4
result = eta * (1 - ks * excess_entropy / v.R)
self.eta = eta
return result
```
## Performing Calculations
Now we can create an instance of `ViscosityModel` for the liquid phase using the `Database` object we created earlier. We can verify this model has a `viscosity` attribute containing a symbolic expression for the viscosity.
```
mod = ViscosityModel(dbf, ['CU', 'ZR'], 'LIQUID')
print(mod.viscosity)
```
Finally we calculate and plot the viscosity.
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from pycalphad import calculate
mod = ViscosityModel(dbf, ['CU', 'ZR'], 'LIQUID')
temp = 2100
# NOTICE: we need to tell pycalphad about our model for this phase
models = {'LIQUID': mod}
res = calculate(dbf, ['CU', 'ZR'], 'LIQUID', P=101325, T=temp, model=models, output='viscosity')
fig = plt.figure(figsize=(6,6))
ax = fig.gca()
ax.scatter(res.X.sel(component='ZR'), 1000 * res.viscosity.values)
ax.set_xlabel('X(ZR)')
ax.set_ylabel('Viscosity (mPa-s)')
ax.set_xlim((0,1))
ax.set_title('Viscosity at {}K'.format(temp));
```
We repeat the calculation for Al-Cu.
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from pycalphad import calculate
temp = 1300
models = {'LIQUID': ViscosityModel} # we can also use Model class
res = calculate(dbf, ['CU', 'AL'], 'LIQUID', P=101325, T=temp, model=models, output='viscosity')
fig = plt.figure(figsize=(6,6))
ax = fig.gca()
ax.scatter(res.X.sel(component='CU'), 1000 * res.viscosity.values)
ax.set_xlabel('X(CU)')
ax.set_ylabel('Viscosity (mPa-s)')
ax.set_xlim((0,1))
ax.set_title('Viscosity at {}K'.format(temp));
```
|
github_jupyter
|
# Section 3.3
```
%run preamble.py
danish = pd.read_csv("../Data/danish.csv").x.values
```
# MLE of composite models
```
parms, BIC, AIC = mle_composite(danish, (1,1,1), "gam-par")
fit_gam_par = pd.DataFrame(np.append(parms, [AIC, BIC])).T
fit_gam_par.columns = ["shape", "tail", "thres", "AIC","BIC"]
print(fit_gam_par)
parms, BIC, AIC = mle_composite(danish, (1,1,1), "wei-par")
fit_wei_par = pd.DataFrame(np.append(parms, [AIC, BIC])).T
fit_wei_par.columns = ["shape", "tail", "thres", "AIC","BIC"]
print(fit_wei_par)
parms, BIC, AIC = mle_composite(danish, (0.5,1,1), "lnorm-par")
fit_lnorm_par = pd.DataFrame(np.append(parms, [AIC, BIC])).T
fit_lnorm_par.columns = ["shape", "tail", "thres", "AIC","BIC"]
print(fit_lnorm_par)
```
# Bayesian inference and model comparison using SMC
```
np.random.seed(333)
model_prior, a, b = "gamma", 0.1*np.array([1,1,1]), 0.1*np.array([1, 1, 1])
popSize, verbose, smc_method, paralell, nproc = 1000, True, "likelihood_anealing", True, 20
loss_models = ['lnorm-par', "wei-par", "gam-par"]
%time traces_like, res_df_like = fit_composite_models_smc(danish,loss_models, model_prior, a, b, popSize, verbose, smc_method, paralell, nproc)
np.random.seed(333)
model_prior, a, b = "gamma", np.array([0.1,0.1,0.1]), np.array([0.1, 0.1, 0.1])
popSize, verbose, smc_method, paralell, nproc = 1000, True, "data_by_batch", True, 20
loss_models = ['lnorm-par', "wei-par", "gam-par"]
%time traces_data, res_df_data = fit_composite_models_smc(danish,loss_models, model_prior, a, b, popSize, verbose, smc_method, paralell, nproc)
```
## Fitting the gamma-Pareto model
```
np.random.seed(333)
fig, axs = plt.subplots(1, 3, figsize=(5, 3.5))
loss_model = "gam-par"
parms_names = ['shape', 'tail', 'thres' ]
x_labs = ['Shape', 'Tail', 'Threshold']
for k in range(3):
# positions = np.linspace(min(trace_gibbs_gam_par[parms_names[k]]), max(trace_gibbs_gam_par[parms_names[k]]), 1000)
# kernel = st.gaussian_kde(trace_gibbs_gam_par[parms_names[k]])
# axs[k].plot(positions, kernel(positions), lw=3, label = "Gibbs", color = "blue")
positions = np.linspace(min(traces_like[loss_model][parms_names[k]].values),
max(traces_like[loss_model][parms_names[k]].values), 1000)
kernel = st.gaussian_kde(traces_like[loss_model][parms_names[k]].values)
axs[k].plot(positions, kernel(positions), lw=3, label = "SMC simulated annealing",
color = "blue", linestyle = "dotted")
positions = np.linspace(min(traces_data[loss_model][parms_names[k]].values),
max(traces_data[loss_model][parms_names[k]].values), 1000)
kernel = st.gaussian_kde(traces_data[loss_model][parms_names[k]].values)
axs[k].plot(positions, kernel(positions), lw=3, label = "SMC data by batches",
color = "blue", linestyle = "dashed")
axs[k].axvline(fit_gam_par[parms_names[k]].values, color = "black", linestyle = "dotted", label = "mle")
axs[k].set_yticks([])
axs[k].set_xlabel(x_labs[k])
axs[k].set_xticks(np.round(
traces_like[loss_model][parms_names[k]].quantile([0.05, 0.95]).values, 2))
handles, labels = axs[0].get_legend_handles_labels()
fig.legend(handles, labels, ncol = 2, borderaxespad=-0.2, loc='upper center',
frameon=False)
# fig.tight_layout()
sns.despine()
plt.savefig("../Figures/smc_posterior_danish_gamma_par_en.pdf")
```
## Fitting the Weibull-Pareto model
```
np.random.seed(333)
fig, axs = plt.subplots(1, 3, figsize=(5, 3.5))
loss_model = "wei-par"
for k in range(3):
# positions = np.linspace(min(trace_gibbs_wei_par[parms_names[k]]), max(trace_gibbs_wei_par[parms_names[k]]), 1000)
# kernel = st.gaussian_kde(trace_gibbs_wei_par[parms_names[k]])
# axs[k].plot(positions, kernel(positions), lw=3, label = "Gibbs", color = "green")
positions = np.linspace(min(traces_like[loss_model][parms_names[k]].values),
max(traces_like[loss_model][parms_names[k]].values), 1000)
kernel = st.gaussian_kde(traces_like[loss_model][parms_names[k]].values)
axs[k].plot(positions, kernel(positions), lw=3, label = "SMC simulated annealing",
color = "green", linestyle = "dotted")
positions = np.linspace(min(traces_data[loss_model][parms_names[k]].values),
max(traces_data[loss_model][parms_names[k]].values), 1000)
kernel = st.gaussian_kde(traces_data[loss_model][parms_names[k]].values)
axs[k].plot(positions, kernel(positions), lw=3, label = "SMC data by batches",
color = "green", linestyle = "dashed")
axs[k].axvline(fit_wei_par[parms_names[k]].values, color = "black", linestyle = "dotted", label = "mle")
axs[k].set_yticks([])
axs[k].set_xlabel(x_labs[k])
axs[k].set_xticks(np.round(
traces_like[loss_model][parms_names[k]].quantile([0.05, 0.95]).values, 2))
handles, labels = axs[0].get_legend_handles_labels()
fig.legend(handles, labels, ncol = 2, borderaxespad=-0.2, loc='upper center',
frameon=False)
sns.despine()
print(fit_gam_par[parms_names[0]].values)
plt.savefig("../Figures/smc_posterior_danish_weibull_par_en.pdf")
```
## Fitting the lognormal-Pareto model
```
np.random.seed(333)
fig, axs = plt.subplots(1, 3, figsize=(5, 3.5))
loss_model = "lnorm-par"
for k in range(3):
# positions = np.linspace(min(trace_gibbs_lnorm_par[parms_names[k]]), max(trace_gibbs_lnorm_par[parms_names[k]]), 1000)
# kernel = st.gaussian_kde(trace_gibbs_lnorm_par[parms_names[k]])
# axs[k].plot(positions, kernel(positions), lw=3, label = "Gibbs", color = "red")
positions = np.linspace(min(traces_like[loss_model][parms_names[k]].values),
max(traces_like[loss_model][parms_names[k]].values), 1000)
kernel = st.gaussian_kde(traces_like[loss_model][parms_names[k]].values)
axs[k].plot(positions, kernel(positions), lw=3, label = "SMC simulated annealing",
color = "red", linestyle = "dotted")
positions = np.linspace(min(traces_data[loss_model][parms_names[k]].values),
max(traces_data[loss_model][parms_names[k]].values), 1000)
kernel = st.gaussian_kde(traces_data[loss_model][parms_names[k]].values)
axs[k].plot(positions, kernel(positions), lw=3, label = "SMC data by batches",
color = "red", linestyle = "dashed")
axs[k].axvline(fit_lnorm_par[parms_names[k]].values, color = "black", linestyle = "dotted", label = "mle")
axs[k].set_yticks([])
axs[k].set_xlabel(x_labs[k])
axs[k].set_xticks(np.round(
traces_like[loss_model][parms_names[k]].quantile([0.05, 0.95]).values, 2))
handles, labels = axs[0].get_legend_handles_labels()
fig.legend(handles, labels, ncol = 2, borderaxespad=-0.2, loc='upper center',
frameon=False)
sns.despine()
print(fit_gam_par[parms_names[0]].values)
plt.savefig("../Figures/smc_posterior_danish_lnorm_par_en.pdf")
print(res_df_data.to_latex(index = False,float_format="%.2f", columns = ["loss_model","log_marg","model_evidence", "DIC", "WAIC"]))
res_df_data
print(res_df_like.to_latex(index = False, float_format="%.2f", columns = ["loss_model","log_marg","model_evidence", "DIC", "WAIC"]))
res_df_like
```
|
github_jupyter
|
# "[Prob] Basics of the Poisson Distribution"
> "Some useful facts about the Poisson distribution"
- toc:false
- branch: master
- badges: false
- comments: true
- author: Peiyi Hung
- categories: [category, learning, probability]
# Introduction
The Poisson distribution is an important discrete probability distribution prevalent in a variety of fields. In this post, I will present some useful facts about the Poisson distribution. Here's the concepts I will discuss in the post:
* PMF, expectation and variance of Poisson
* In what situation we can use it?
* Sum of indepentent Poisson is also a Poisson
* Relationship with the Binomial distribution
# PMF, Expectation and Variance
First, we define what's Poisson distribution.
Let X be a Poisson random variable with a parameter $\lambda$, where $\lambda >0$. The pmf of X would be:
$$P(X=x) = \frac{e^{-\lambda}\lambda^{x}}{x!}, \quad \text{for } k = 0, 1,2,3,\dots$$
where $x$ can only be non-negative integer.
This is a valid pmf since
$$\sum_{k=0}^{\infty} \frac{e^{-\lambda}\lambda^{k}}{k!} = e^{-\lambda}\sum_{k=0}^{\infty} \frac{\lambda^{k}}{k!}= e^{-\lambda}e^{\lambda}=1$$
where $\displaystyle\sum_{k=0}^{\infty} \frac{\lambda^{k}}{k!}$ is the Taylor expansion of $e^{\lambda}$.
The expectation and the variance of the Poisson distribution are both $\lambda$. The derivation of this result is just some pattern recognition of $\sum_{k=0}^{\infty} \frac{\lambda^{k}}{k!}=e^{\lambda}$, so I omit it here.
# In what situation can we use it?
The Poisson distribution is often applied to the situation where we are counting the number of successes or an event happening in a time interval or a particular region, and there are a large number of trials with a small probability of success. The parameter $\lambda$ is the rate parameter which indicates the average number of successes in a time interval or a region.
Here are some examples:
* The number of emails you receive in an hour.
* The number of chips in a chocolate chip cookie.
* The number of earthquakes in a year in some region of the world.
Also, let's consider an example probability problem.
**Example problem 1**
> Raindrops are falling at an average rate of 20 drops per square inch per minute. Find the probability that the region has no rain drops in a given 1-minute time interval.
The success in this problem is one raindrop. The average rate is 20, so $\lambda=20$. Let $X$ be the raindrops that region has in a minute. We would model $X$ with Pois$(20)$, so the probability we concerned would be
$$P(X=0) = \frac{e^{-20}20^0}{0!}=e^{-20} \approx 2.0611\times 10 ^{-9}$$
If we are concerned with raindrops in a 3-second time interval in 5 square inches, then $$\lambda = 20\times\frac{1}{20} \text{ minutes} \times5 \text{ square inches} = 5$$
Let $Y$ be raindrops in a 3-second time interval. $Y$ would be Pois$(5)$, so $P(Y=0) = e^{-5} \approx 0.0067$.
# Sum of Independent Poisson
The sum of independent Poisson would also be Poisson. Let $X$ be Pois$(\lambda_1)$ and $Y$ be Pois$(\lambda_2)$. If $T=X+Y$, then $T \sim \text{Pois}(\lambda_1 + \lambda_2)$.
To get pmf of $T$, we should first apply the law of total probability:
$$
P(X+Y=t) = \sum_{k=0}^{t}P(X+Y=t|X=k)P(X=k)
$$
Since they are independent, we got
$$
\sum_{k=0}^{t}P(X+Y=t|X=k)P(X=k) = \sum_{k=0}^{t}P(Y=t-k)P(X=k)
$$
Next, we plug in the pmf of Poisson:
$$
\sum_{k=0}^{t}P(Y=t-k)P(X=k) = \sum_{k=0}^{t}\frac{e^{-\lambda_2}\lambda_2^{t-k}}{(t-k)!}\frac{e^{-\lambda_2}\lambda_1^k}{k!} = \frac{e^{-(\lambda_1+\lambda_2)}}{t!}\sum_{k=0}^{t} {t \choose k}\lambda_1^{k}\lambda_2^{t-k}
$$
Finally, by Binomial theorem, we got
$$
P(X+Y=t) = \frac{e^{-(\lambda_1+\lambda_2)}(\lambda_1+\lambda_2)^t}{t!}
$$
which is the pmf of Pois$(\lambda_1 + \lambda_2)$.
# Relationship with the Binomial distribution
We can obtain Poisson from Binomial and can also obtain Binomial to Poisson. Let's first see how we get the Binomial distribution from the Poisson distribution
**From Poisson to Binomial**
If $X \sim$ Pois$(\lambda_1)$ and $Y \sim$ Pois$(\lambda_2)$, and they are independent, then the conditional distribution of $X$ given $X+Y=n$ is Bin$(n, \lambda_1/(\lambda_1 + \lambda_2))$. Let's derive the pmf of $X$ given $X+Y=n$.
By Bayes' rule and the indenpendence between $X$ and $Y$:
$$
P(X=x|X+Y=n) = \frac{P(X+Y=n|X=x)P(X=x)}{P(X+Y=n)} = \frac{P(Y=n-k)P(X=x)}{P(X+Y=n)}
$$
From the previous section, we know $X+Y \sim$ Poin$(\lambda_1 + \lambda_2)$. Use this fact, we get
$$
P(X=x|X+Y=n) = \frac{ \big(\frac{e^{-\lambda_2}\lambda_2^{n-k}}{(n-k)!}\big) \big( \frac{e^{\lambda_1\lambda_1^k}}{k!} \big)}{ \frac{e^{-(\lambda_1 + \lambda_2)}(\lambda_1 + \lambda_2)^n}{n!}} = {n\choose k}\bigg(\frac{\lambda_1}{\lambda_1+\lambda_2}\bigg)^k \bigg(\frac{\lambda_2}{\lambda_1+\lambda_2}\bigg)^{n-k}
$$
which is the Bin$(n, \lambda_1/(\lambda_1 + \lambda_2))$ pmf.
**From Binomial to Poisson**
We can approximate Binomial by Poisson when $n \rightarrow \infty$ and $p \rightarrow 0$, and $\lambda = np$.
The pmf of Binomial is
$$
P(X=k) = {n \choose k}p^{k}(1-p)^{n-k} = {n \choose k}\big(\frac{\lambda}{n}\big)^{k}\big(1-\frac{\lambda}{n}\big)^n\big(1-\frac{\lambda}{n}\big)^{-k}
$$
By some algebra manipulation, we got
$$
P(X=k) = \frac{\lambda^{k}}{k!}\frac{n(n-1)\dots(n-k+1)}{n^k}\big(1-\frac{\lambda}{n}\big)^n\big(1-\frac{\lambda}{n}\big)^{-k}
$$
When $n \rightarrow \infty$, we got:
$$
\frac{n(n-1)\dots(n-k+1)}{n^k} \rightarrow 1,\\
\big(1-\frac{\lambda}{n}\big)^n \rightarrow e^{-\lambda}, \text{and}\\
\big(1-\frac{\lambda}{n}\big)^{-k} \rightarrow 1
$$
Therefore, $P(X=k) = \frac{e^{-\lambda}\lambda^k}{k!}$ when $n \rightarrow \infty$.
Let's see an example on how to use Poisson to approximate Binomial.
**Example problem 2**
>Ten million people enter a certain lottery. For each person, the chance of winning is one in ten million, independently. Find a simple, good approximation for the PMF of the number of people who win the lottery.
Let $X$ be the number of people winning the lottery. $X$ would be Bin$(10000000, 1/10000000)$ and $E(X) = 1$. We can approximate the pmf of $X$ by Pois$(1)$:
$$
P(X=k) \approx \frac{1}{e\cdot k!}
$$
Let's see if this approximation is accurate by Python code.
```
#collapse-hide
from scipy.stats import binom
from math import factorial, exp
import numpy as np
import matplotlib.pyplot as plt
def pois(k):
return 1 / (exp(1) * factorial(k))
n = 10000000
p = 1/10000000
k = np.arange(10)
binomial = binom.pmf(k, n, p)
poisson = [pois(i) for i in k]
fig, ax = plt.subplots(ncols=2, nrows=1, figsize=(15, 4), dpi=120)
ax[0].plot(k, binomial)
ax[0].set_title("PMF of Binomial")
ax[0].set_xlabel(r"$X=k$")
ax[0].set_xticks(k)
ax[1].plot(k, poisson)
ax[1].set_title("Approximation by Poisson")
ax[1].set_xlabel(r"X=k")
ax[1].set_xticks(k)
plt.tight_layout();
```
The approximation is quite accurate since these two graphs are almost identical.
**Reference**
1. *Introduction to Probability* by Joe Blitzstein and Jessica Hwang.
|
github_jupyter
|
# Introduction
```
#r "BoSSSpad.dll"
using System;
using System.Collections.Generic;
using System.Linq;
using ilPSP;
using ilPSP.Utils;
using BoSSS.Platform;
using BoSSS.Platform.LinAlg;
using BoSSS.Foundation;
using BoSSS.Foundation.XDG;
using BoSSS.Foundation.Grid;
using BoSSS.Foundation.Grid.Classic;
using BoSSS.Foundation.Grid.RefElements;
using BoSSS.Foundation.IO;
using BoSSS.Solution;
using BoSSS.Solution.Control;
using BoSSS.Solution.GridImport;
using BoSSS.Solution.Statistic;
using BoSSS.Solution.Utils;
using BoSSS.Solution.AdvancedSolvers;
using BoSSS.Solution.Gnuplot;
using BoSSS.Application.BoSSSpad;
using BoSSS.Application.XNSE_Solver;
using static BoSSS.Application.BoSSSpad.BoSSSshell;
Init();
```
# Note:
- Setting Boundary values and initial values is similar;
- For most solvers, inital and boundary values are set the same way;
- We will use the incompressible solver as an example:
```
using BoSSS.Application.XNSE_Solver;
```
Create a control object:
```
var C = new XNSE_Control();
```
# 1 From Formulas
If the Formula is simple enough to be represented by C\# code,
it can be embedded in the control file.
However, the code bust be put into a string, since it is not
possible to serialize classes/objects from the notebook
into a control object:
```
string code =
"static class MyInitialValue {" // class must be static!
// Warning: static constants are allowed,
// but any changes outside of the current text box in BoSSSpad
// will not be recorded for the code that is passed to the solver.
+ " public static double alpha = 0.7;"
// a method, which should be used for an initial value,
// must be static!
+ " public static double VelocityX(double[] X, double t) {"
+ " double x = X[0];"
+ " double y = X[1];"
+ " return Math.Sin(x*y*alpha);"
+ " }"
+ "}";
var fo = new BoSSS.Solution.Control.Formula("MyInitialValue.VelocityX",
true, code);
```
Use the BoSSSpad-intrinsic **GetFormulaObject** to set tie inital value:
```
C.AddInitialValue("VelocityX", fo);
/// Deprecated:
/// Note: such a declaration is very restrictive;
/// \code{GetFormulaObject} works only for
/// \begin{itemize}
/// \item a static class
/// \item no dependence on any external parameters
/// \end{itemize}
/// E.g. the following code would only change the behavior in BoSSSpad,
/// but not the code that is passed to the solver:
//Deprecated:
//MyInitialValue.alpha = 0.5;
//MyInitialValue.VelocityX(new double[]{ 0.5, 0.5 }, 0.0);
C.InitialValues["VelocityX"].Evaluate(new double[]{ 0.5, 0.5 }, 0.0)
```
# 2 Advanced functions
Some more advanced mathematical functions, e.g.
Jacobian elliptic functions $\text{sn}(u|m)$, $\text{cn}(u|m)$ and $\text{dn}(u|m)$
are available throug the GNU Scientific Library, for which BoSSS provides
bindings, see e.g.
**BoSSS.Platform.GSL.gsl\_sf\_elljac\_e**
## 2.1 From MATLAB code
Asssume e.g. the following MATLAB code; obviously, this could
also be implemented in C\#, we yust use something smple for demonstration:
```
string[] MatlabCode = new string[] {
@"[n,d2] = size(X_values);",
@"u=zeros(2,n);",
@"for k=1:n",
@"X=[X_values(k,1),X_values(k,2)];",
@"",
@"u_x_main = -(-sqrt(X(1).^ 2 + X(2).^ 2) / 0.3e1 + 0.4e1 / 0.3e1 * (X(1).^ 2 + X(2).^ 2) ^ (-0.1e1 / 0.2e1)) * sin(atan2(X(2), X(1)));",
@"u_y_main = (-sqrt(X(1).^ 2 + X(2).^ 2) / 0.3e1 + 0.4e1 / 0.3e1 * (X(1).^ 2 + X(2).^ 2) ^ (-0.1e1 / 0.2e1)) * cos(atan2(X(2), X(1)));",
@"",
@"u(1,k)=u_x_main;",
@"u(2,k)=u_y_main;",
@"end" };
```
We can evaluate this code in **BoSSS** using the MATLAB connector;
We encapsulate it in a **ScalarFunction** which allows
**vectorized** evaluation
(multiple evaluatiuons in one function call) e
of some function.
This is much more efficient, since there will be significant overhead
for calling MATLAB (starting MATLAB, checking the license,
transfering data, etc.).
```
using ilPSP.Connectors.Matlab;
ScalarFunction VelocityXInitial =
delegate(MultidimensionalArray input, MultidimensionalArray output) {
int N = input.GetLength(0); // number of points which we evaluate
// at once.
var output_vec = MultidimensionalArray.Create(2, N); // the MATLAB code
// returns an entire vector.
using(var bmc = new BatchmodeConnector()) {
bmc.PutMatrix(input,"X_values");
foreach(var line in MatlabCode) {
bmc.Cmd(line);
}
bmc.GetMatrix(output_vec, "u");
bmc.Execute(); // Note: 'Execute' has to be *after* 'GetMatrix'
}
output.Set(output_vec.ExtractSubArrayShallow(0,-1)); // extract row 0 from
// 'output_vec' and store it in 'output'
};
```
We test our implementation:
```
var inputTest = MultidimensionalArray.Create(3,2); // set some test values for input
inputTest.SetColumn(0, GenericBlas.Linspace(1,2,3));
inputTest.SetColumn(1, GenericBlas.Linspace(2,3,3));
var outputTest = MultidimensionalArray.Create(3); // allocate memory for output
VelocityXInitial(inputTest, outputTest);
```
We recive the following velocity values for our input coordinates:
```
outputTest.To1DArray()
```
# Projecting the MATLAB function to a DG field
As for a standard calculation, we create a mesh, save it to some database
and set the mesh in the control object.
```
var nodes = GenericBlas.Linspace(1,2,11);
GridCommons grid = Grid2D.Cartesian2DGrid(nodes,nodes);
var db = CreateTempDatabase();
db.SaveGrid(ref grid);
C.SetGrid(grid);
```
We create a DG field for the $x$-velocity on our grid:
```
var gdata = new GridData(grid);
var b = new Basis(gdata, 3); // use DG degree 2
var VelX = new SinglePhaseField(b,"VelocityX"); // important: name the DG field
// equal to initial value name
```
Finally, we are able to project the MATLAB function onto the DG field:
```
//VelX.ProjectField(VelocityXInitial);
```
One might want to check the data visually, so it can be exported
in the usual fashion
```
//Tecplot("initial",0.0,2,VelX);
```
# Storing the initial value in the database and linking it in the control object
The DG field with the initial value can be stored in the database.
this will create a dummy session.
```
BoSSSshell.WorkflowMgm.Init("TestProject");
var InitalValueTS = db.SaveTimestep(VelX); // further fields an be
// appended
BoSSSshell.WorkflowMgm.Sessions
/// Now, we can use this timestep as a restart-value for the simulation:
C.SetRestart(InitalValueTS);
```
|
github_jupyter
|
# <img style="float: left; padding-right: 10px; width: 45px" src="https://raw.githubusercontent.com/Harvard-IACS/2018-CS109A/master/content/styles/iacs.png"> CS109B Data Science 2: Advanced Topics in Data Science
## Lab 2 - Smoothers and Generalized Additive Models
**Harvard University**<br>
**Spring 2019**<br>
**Instructors:** Mark Glickman and Pavlos Protopapas<br>
**Lab Instructors:** Will Claybaugh<br>
**Contributors:** Paul Tyklin and Will Claybaugh
---
```
## RUN THIS CELL TO PROPERLY HIGHLIGHT THE EXERCISES
import requests
from IPython.core.display import HTML
styles = requests.get("https://raw.githubusercontent.com/Harvard-IACS/2019-CS109B/master/content/styles/cs109.css").text
HTML(styles)
```
## Learning Goals
The main goal of this lab is to get familiar with calling R functions within Python. Along the way, we'll learn about the "formula" interface to statsmodels, which gives an intuitive way of specifying regression models, and we'll review the different approaches to fitting curves.
Key Skills:
- Importing (base) R functions
- Importing R library functions
- Populating vectors R understands
- Populating dataframes R understands
- Populating formulas R understands
- Running models in R
- Getting results back to Python
- Getting model predictions in R
- Plotting in R
- Reading R's documentation
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
```
## Linear/Polynomial Regression (Python, Review)
Hopefully, you remember working with Statsmodels during 109a
Reading data and (some) exploring in Pandas:
```
diab = pd.read_csv("data/diabetes.csv")
print("""
# Variables are:
# subject: subject ID number
# age: age diagnosed with diabetes
# acidity: a measure of acidity called base deficit
# y: natural log of serum C-peptide concentration
#
# Original source is Sockett et al. (1987)
# mentioned in Hastie and Tibshirani's book
# "Generalized Additive Models".
""")
display(diab.head())
display(diab.dtypes)
display(diab.describe())
```
Plotting with matplotlib:
```
ax0 = diab.plot.scatter(x='age',y='y',c='Red',title="Diabetes data") #plotting direclty from pandas!
ax0.set_xlabel("Age at Diagnosis")
ax0.set_ylabel("Log C-Peptide Concentration");
```
Linear regression with statsmodels.
- Previously, we worked from a vector of target values and a design matrix we built ourself (e.g. from PolynomialFeatures).
- Now, Statsmodels' *formula interface* can help build the target value and design matrix for you.
```
#Using statsmodels
import statsmodels.formula.api as sm
model1 = sm.ols('y ~ age',data=diab)
fit1_lm = model1.fit()
```
Build a data frame to predict values on (sometimes this is just the test or validation set)
- Very useful for making pretty plots of the model predcitions -- predict for TONS of values, not just whatever's in the training set
```
x_pred = np.linspace(0,16,100)
predict_df = pd.DataFrame(data={"age":x_pred})
predict_df.head()
```
Use `get_prediction(<data>).summary_frame()` to get the model's prediction (and error bars!)
```
prediction_output = fit1_lm.get_prediction(predict_df).summary_frame()
prediction_output.head()
```
Plot the model and error bars
```
ax1 = diab.plot.scatter(x='age',y='y',c='Red',title="Diabetes data with least-squares linear fit")
ax1.set_xlabel("Age at Diagnosis")
ax1.set_ylabel("Log C-Peptide Concentration")
ax1.plot(predict_df.age, prediction_output['mean'],color="green")
ax1.plot(predict_df.age, prediction_output['mean_ci_lower'], color="blue",linestyle="dashed")
ax1.plot(predict_df.age, prediction_output['mean_ci_upper'], color="blue",linestyle="dashed");
ax1.plot(predict_df.age, prediction_output['obs_ci_lower'], color="skyblue",linestyle="dashed")
ax1.plot(predict_df.age, prediction_output['obs_ci_upper'], color="skyblue",linestyle="dashed");
```
<div class="discussion"><b>Discussion</b></div>
- What are the dark error bars?
- What are the light error bars?
<div class="exercise"><b>Exercise 1</b></div>
1. Fit a 3rd degree polynomial model and plot the model+error bars
- Route1: Build a design df with a column for each of `age`, `age**2`, `age**3`
- Route2: Just edit the formula
**Answers**:
1.
```
# your code here
```
2.
```
# your code here
```
## Linear/Polynomial Regression, but make it R
This is the meat of the lab. After this section we'll know everything we need to in order to work with R models. The rest of the lab is just applying these concepts to run particular models. This section therefore is your 'cheat sheet' for working in R.
What we need to know:
- Importing (base) R functions
- Importing R Library functions
- Populating vectors R understands
- Populating DataFrames R understands
- Populating Formulas R understands
- Running models in R
- Getting results back to Python
- Getting model predictions in R
- Plotting in R
- Reading R's documentation
**Importing R functions**
```
# if you're on JupyterHub you may need to specify the path to R
#import os
#os.environ['R_HOME'] = "/usr/share/anaconda3/lib/R"
import rpy2.robjects as robjects
r_lm = robjects.r["lm"]
r_predict = robjects.r["predict"]
#r_plot = robjects.r["plot"] # more on plotting later
#lm() and predict() are two of the most common functions we'll use
```
**Importing R libraries**
```
from rpy2.robjects.packages import importr
#r_cluster = importr('cluster')
#r_cluster.pam;
```
**Populating vectors R understands**
```
r_y = robjects.FloatVector(diab['y'])
r_age = robjects.FloatVector(diab['age'])
# What happens if we pass the wrong type?
# How does r_age display?
# How does r_age print?
```
**Populating Data Frames R understands**
```
diab_r = robjects.DataFrame({"y":r_y, "age":r_age})
# How does diab_r display?
# How does diab_r print?
```
**Populating formulas R understands**
```
simple_formula = robjects.Formula("y~age")
simple_formula.environment["y"] = r_y #populate the formula's .environment, so it knows what 'y' and 'age' refer to
simple_formula.environment["age"] = r_age
```
**Running Models in R**
```
diab_lm = r_lm(formula=simple_formula) # the formula object is storing all the needed variables
simple_formula = robjects.Formula("y~age") # reset the formula
diab_lm = r_lm(formula=simple_formula, data=diab_r) #can also use a 'dumb' formula and pass a dataframe
```
**Getting results back to Python**
```
diab_lm #the result is already 'in' python, but it's a special object
print(diab_lm.names) # view all names
diab_lm[0] #grab the first element
diab_lm.rx2("coefficients") #use rx2 to get elements by name!
np.array(diab_lm.rx2("coefficients")) #r vectors can be converted to numpy (but rarely needed)
```
**Getting Predictions**
```
# make a df to predict on (might just be the validation or test dataframe)
predict_df = robjects.DataFrame({"age": robjects.FloatVector(np.linspace(0,16,100))})
# call R's predict() function, passing the model and the data
predictions = r_predict(diab_lm, predict_df)
x_vals = predict_df.rx2("age")
ax = diab.plot.scatter(x='age',y='y',c='Red',title="Diabetes data")
ax.set_xlabel("Age at Diagnosis")
ax.set_ylabel("Log C-Peptide Concentration");
ax.plot(x_vals,predictions); #plt still works with r vectors as input!
```
**Plotting in R**
```
%load_ext rpy2.ipython
```
- The above turns on the %R "magic"
- R's plot() command responds differently based on what you hand to it; Different models get different plots!
- For any specific model search for plot.modelname. E.g. for a GAM model, search plot.gam for any details of plotting a GAM model
- The %R "magic" runs R code in 'notebook' mode, so figures display nicely
- Ahead of the `plot(<model>)` code we pass in the variables R needs to know about (`-i` is for "input")
```
%R -i diab_lm plot(diab_lm);
```
**Reading R's documentation**
The documentation for the `lm()` funciton is [here](https://stat.ethz.ch/R-manual/R-devel/library/stats/html/lm.html), and a prettier version (same content) is [here](https://www.rdocumentation.org/packages/stats/versions/3.5.2/topics/lm). When googling, perfer rdocumentation.org when possible.
Sections:
- **Usage**: gives the function signature, including all optional arguments
- **Arguments**: What each function input controls
- **Details**: additional info on what the funciton *does* and how arguments interact. **Often the right place to start reading**
- **Value**: the structure of the object returned by the function
- **Refferences**: The relevant academic papers
- **See Also**: other functions of interest
<div class="exercise"><b>Exercise 2</b></div>
1. Add confidence intervals calculated in R to the linear regression plot above. Use the `interval=` argument to `r_predict()` (documentation [here](https://stat.ethz.ch/R-manual/R-devel/library/stats/html/predict.lm.html)). You will have to work with a matrix returned by R.
2. Fit a 5th degree polynomial to the diabetes data in R. Search the web for an easier method than writing out a formula with all 5 polynomial terms.
**Answers**
1.
```
# your code here
```
2.
```
# your code here
```
## Lowess Smoothing
Lowess Smoothing is implemented in both Python and R. We'll use it as another example as we transition languages.
<div class="discussion"><b>Discussion</b></div>
- What is lowess smoothing? Which 109a models is it related to?
- How explainable is lowess?
- What are the tunable parameters?
**In Python**
```
from statsmodels.nonparametric.smoothers_lowess import lowess as lowess
ss1 = lowess(diab['y'],diab['age'],frac=0.15)
ss2 = lowess(diab['y'],diab['age'],frac=0.25)
ss3 = lowess(diab['y'],diab['age'],frac=0.7)
ss4 = lowess(diab['y'],diab['age'],frac=1)
ss1[:10,:] # we get back simple a smoothed y value for each x value in the data
```
Notice the clean code to plot different models. We'll see even cleaner code in a minute
```
for cur_model, cur_frac in zip([ss1,ss2,ss3,ss4],[0.15,0.25,0.7,1]):
ax = diab.plot.scatter(x='age',y='y',c='Red',title="Lowess Fit, Fraction = {}".format(cur_frac))
ax.set_xlabel("Age at Diagnosis")
ax.set_ylabel("Log C-Peptide Concentration")
ax.plot(cur_model[:,0],cur_model[:,1],color="blue")
plt.show()
```
<div class="discussion"><b>Discussion</b></div>
1. Which model has high variance, which has high bias?
2. What makes a model high variance or high bias?
**In R**
We need to:
- Import the loess function
- Send data over to R
- Call the function and get results
```
r_loess = robjects.r['loess.smooth'] #extract R function
r_y = robjects.FloatVector(diab['y'])
r_age = robjects.FloatVector(diab['age'])
ss1_r = r_loess(r_age,r_y, span=0.15, degree=1)
ss1_r #again, a smoothed y value for each x value in the data
```
<div class="exercise"><b>Exercise 3</b></div>
Predict the output of
1. `ss1_r[0]`
2. `ss1_r.rx2("y")`
1.
*your answer here*
2.
*your answer here*
**Varying span**
Next, some extremely clean code to fit and plot models with various parameter settings. (Though the `zip()` method seen earlier is great when e.g. the label and the parameter differ)
```
for cur_frac in [0.15,0.25,0.7,1]:
cur_smooth = r_loess(r_age,r_y, span=cur_frac)
ax = diab.plot.scatter(x='age',y='y',c='Red',title="Lowess Fit, Fraction = {}".format(cur_frac))
ax.set_xlabel("Age at Diagnosis")
ax.set_ylabel("Log C-Peptide Concentration")
ax.plot(cur_smooth[0], cur_smooth[1], color="blue")
plt.show()
```
<div class="discussion"><b>Discussion</b></div>
- Mark wasn't kidding; the Python and R results differ for frac=.15. Thoughts?
- Why isn't the bottom plot a straight line? We're using 100% of the data in each window...
## Smoothing Splines
From this point forward, we're working with R functions; these models aren't (well) supported in Python.
For clarity: this is the fancy spline model that minimizes $MSE - \lambda\cdot\text{wiggle penalty}$ $=$ $\sum_{i=1}^N \left(y_i - f(x_i)\right)^2 - \lambda \int \left(f''(x)\right)^2$, across all possible functions $f$. The winner will always be a continuous, cubic polynomial with a knot at each data point
<div class="discussion"><b>Discussion</b></div>
- Any idea why the winner is cubic?
- How interpretable is this model?
- What are the tunable parameters?
```
r_smooth_spline = robjects.r['smooth.spline'] #extract R function
# run smoothing function
spline1 = r_smooth_spline(r_age, r_y, spar=0)
```
<div class="exercise"><b>Exercise 4</b></div>
1. We actually set the spar parameter, a scale-free value that translates to a $\lambda$ through a complex expression. Inspect the 'spline1' result and extract the implied value of $\lambda$
2. Working from the fitting/plotting loop examples above, produce a plot like the one below for spar = [0,.5,.9,2], including axes labels and title.
1.
```
# your answer here
```
2.
```
# your answer here
```
**CV**
R's `smooth_spline` funciton has built-in CV to find a good lambda. See package [docs](https://www.rdocumentation.org/packages/stats/versions/3.5.2/topics/smooth.spline).
```
spline_cv = r_smooth_spline(r_age, r_y, cv=True)
lambda_cv = spline_cv.rx2("lambda")[0]
ax19 = diab.plot.scatter(x='age',y='y',c='Red',title="smoothing spline with $\lambda=$"+str(np.round(lambda_cv,4))+", chosen by cross-validation")
ax19.set_xlabel("Age at Diagnosis")
ax19.set_ylabel("Log C-Peptide Concentration")
ax19.plot(spline_cv.rx2("x"),spline_cv.rx2("y"),color="darkgreen");
```
<div class="discussion"><b>Discussion</b></div>
- Does the selected model look reasonable?
- How would you describe the effect of age at diagnosis on C_peptide concentration?
- What are the costs/benefits of the (fancy) spline model, relative to the linear regression we fit above?
## Natural & Basis Splines
Here, we take a step backward on model complexity, but a step forward in coding complexity. We'll be working with R's formula interface again, so we will need to populate Formulas and DataFrames.
<div class="discussion"><b>Discussion</b></div>
- In what way are Natural and Basis splines less complex than the splines we were just working with?
- What makes a spline 'natural'?
- What makes a spline 'basis'?
- What are the tuning parameters?
```
#We will now work with a new dataset, called GAGurine.
#The dataset description (from the R package MASS) is below:
#Data were collected on the concentration of a chemical GAG
# in the urine of 314 children aged from zero to seventeen years.
# The aim of the study was to produce a chart to help a paediatrican
# to assess if a child's GAG concentration is ‘normal’.
#The variables are:
# Age: age of child in years.
# GAG: concentration of GAG (the units have been lost).
GAGurine = pd.read_csv("data/GAGurine.csv")
display(GAGurine.head())
ax31 = GAGurine.plot.scatter(x='Age',y='GAG',c='black',title="GAG in urine of children")
ax31.set_xlabel("Age");
ax31.set_ylabel("GAG");
```
Standard stuff: import function, convert variables to R format, call function
```
from rpy2.robjects.packages import importr
r_splines = importr('splines')
# populate R variables
r_gag = robjects.FloatVector(GAGurine['GAG'].values)
r_age = robjects.FloatVector(GAGurine['Age'].values)
r_quarts = robjects.FloatVector(np.quantile(r_age,[.25,.5,.75])) #woah, numpy functions run on R objects!
```
What happens when we call the ns or bs functions from r_splines?
```
ns_design = r_splines.ns(r_age, knots=r_quarts)
bs_design = r_splines.bs(r_age, knots=r_quarts)
print(ns_design)
```
`ns` and `bs` return design matrices, not model objects! That's because they're meant to work with `lm`'s formula interface. To get a model object we populate a formula including `ns(<var>,<knots>)` and fit to data
```
r_lm = robjects.r['lm']
r_predict = robjects.r['predict']
# populate the formula
ns_formula = robjects.Formula("Gag ~ ns(Age, knots=r_quarts)")
ns_formula.environment['Gag'] = r_gag
ns_formula.environment['Age'] = r_age
ns_formula.environment['r_quarts'] = r_quarts
# fit the model
ns_model = r_lm(ns_formula)
```
Predict like usual: build a dataframe to predict on and call `predict()`
```
# predict
predict_frame = robjects.DataFrame({"Age": robjects.FloatVector(np.linspace(0,20,100))})
ns_out = r_predict(ns_model, predict_frame)
ax32 = GAGurine.plot.scatter(x='Age',y='GAG',c='grey',title="GAG in urine of children")
ax32.set_xlabel("Age")
ax32.set_ylabel("GAG")
ax32.plot(predict_frame.rx2("Age"),ns_out, color='red')
ax32.legend(["Natural spline, knots at quartiles"]);
```
<div class="exercise"><b>Exercise 5</b></div>
1. Fit a basis spline model with the same knots, and add it to the plot above
2. Fit a basis spline with 8 knots placed at [2,4,6...14,16] and add it to the plot above
**Answers:**
1.
```
# your answer here
```
2.
```
# your answer here
#%R -i overfit_model plot(overfit_model)
# we'd get the same diagnostic plot we get from an lm model
```
## GAMs
We come, at last, to our most advanced model. The coding here isn't any more complex than we've done before, though the behind-the-scenes is awesome.
First, let's get our (multivariate!) data
```
kyphosis = pd.read_csv("data/kyphosis.csv")
print("""
# kyphosis - wherther a particular deformation was present post-operation
# age - patient's age in months
# number - the number of vertebrae involved in the operation
# start - the number of the topmost vertebrae operated on
""")
display(kyphosis.head())
display(kyphosis.describe(include='all'))
display(kyphosis.dtypes)
#If there are errors about missing R packages, run the code below:
#r_utils = importr('utils')
#r_utils.install_packages('codetools')
#r_utils.install_packages('gam')
```
To fit a GAM, we
- Import the `gam` library
- Populate a formula including `s(<var>)` on variables we want to fit smooths for
- Call `gam(formula, family=<string>)` where `family` is a string naming a probability distribution, chosen based on how the response variable is thought to occur.
- Rough `family` guidelines:
- Response is binary or "N occurances out of M tries", e.g. number of lab rats (out of 10) developing disease: chooose `"binomial"`
- Response is a count with no logical upper bound, e.g. number of ice creams sold: choose `"poisson"`
- Response is real, with normally-distributed noise, e.g. person's height: choose `"gaussian"` (the default)
```
#There is a Python library in development for using GAMs (https://github.com/dswah/pyGAM)
# but it is not yet as comprehensive as the R GAM library, which we will use here instead.
# R also has the mgcv library, which implements some more advanced/flexible fitting methods
r_gam_lib = importr('gam')
r_gam = r_gam_lib.gam
r_kyph = robjects.FactorVector(kyphosis[["Kyphosis"]].values)
r_Age = robjects.FloatVector(kyphosis[["Age"]].values)
r_Number = robjects.FloatVector(kyphosis[["Number"]].values)
r_Start = robjects.FloatVector(kyphosis[["Start"]].values)
kyph1_fmla = robjects.Formula("Kyphosis ~ s(Age) + s(Number) + s(Start)")
kyph1_fmla.environment['Kyphosis']=r_kyph
kyph1_fmla.environment['Age']=r_Age
kyph1_fmla.environment['Number']=r_Number
kyph1_fmla.environment['Start']=r_Start
kyph1_gam = r_gam(kyph1_fmla, family="binomial")
```
The fitted gam model has a lot of interesting data within it
```
print(kyph1_gam.names)
```
Remember plotting? Calling R's `plot()` on a gam model is the easiest way to view the fitted splines
```
%R -i kyph1_gam plot(kyph1_gam, residuals=TRUE,se=TRUE, scale=20);
```
Prediction works like normal (build a data frame to predict on, if you don't already have one, and call `predict()`). However, predict always reports the sum of the individual variable effects. If `family` is non-default this can be different from the actual prediction for that point.
For instance, we're doing a 'logistic regression' so the raw prediction is log odds, but we can get probability by using in `predict(..., type="response")`
```
kyph_new = robjects.DataFrame({'Age': robjects.IntVector((84,85,86)),
'Start': robjects.IntVector((5,3,1)),
'Number': robjects.IntVector((1,6,10))})
print("Raw response (so, Log odds):")
display(r_predict(kyph1_gam, kyph_new))
print("Scaled response (so, probabilty of kyphosis):")
display(r_predict(kyph1_gam, kyph_new, type="response"))
```
<div class="discussion"><b>Discussion</b></div>
<div class="exercise"><b>Exercise 6</b></div>
1. What lambda did we use?
2. What is the model telling us about the effects of age, starting vertebrae, and number of vertebae operated on
3. If we fit a logistic regression instead, which variables might want quadratic terms. What is the cost and benefit of a logistic regression model versus a GAM?
4. Critique the model:
- What is it assuming? Are the assumptions reasonable
- Are we using the right data?
- Does the model's story about the world make sense?
## Appendix
GAMs and smoothing splines support hypothesis tets to compare models. (We can always compare models via out-of-sample prediction quality (i.e. performance on a validation set), but statistical ideas like hypothesis tests yet information criteria allow us to use all data for training *and* still compare the quality of model A to model B)
```
r_anova = robjects.r["anova"]
kyph0_fmla = robjects.Formula("Kyphosis~1")
kyph0_fmla.environment['Kyphosis']=r_kyph
kyph0_gam = r_gam(kyph0_fmla, family="binomial")
print(r_anova(kyph0_gam, kyph1_gam, test="Chi"))
```
**Explicitly joining spline functions**
```
def h(x, xi, pow_arg): #pow is a reserved keyword in Python
if (x > xi):
return pow((x-xi),pow_arg)
else:
return 0
h = np.vectorize(h,otypes=[np.float]) #default behavior is to return ints, which gives incorrect answer
#also, vectorize does not play nicely with default arguments, so better to set directly (e.g., pow_arg=1)
xvals = np.arange(0,10.1,0.1)
ax20 = plt.plot(xvals,h(xvals,4,1),color="red")
_ = plt.title("Truncated linear basis function with knot at x=4")
_ = plt.xlabel("$x$")
_ = plt.ylabel("$(x-4)_+$") #note the use of TeX in the label
ax21 = plt.plot(xvals,h(xvals,4,3),color="red")
_ = plt.title("Truncated cubic basis function with knot at x=4")
_ = plt.xlabel("$x$")
_ = plt.ylabel("$(x-4)_+^3$")
ax22 = plt.plot(xvals,2+xvals+3*h(xvals,2,1)-4*h(xvals,5,1)+0.5*h(xvals,8,1),color="red")
_ = plt.title("Piecewise linear spline with knots at x=2, 5, and 8")
_ = plt.xlabel("$x$")
_ = plt.ylabel("$y$")
```
Comparing splines to the (noisy) model that generated them.
```
x = np.arange(0.1,10,9.9/100)
from scipy.stats import norm
#ppf (percent point function) is the rather unusual name for
#the quantile or inverse CDF function in SciPy
y = norm.ppf(x/10) + np.random.normal(0,0.4,100)
ax23 = plt.scatter(x,y,facecolors='none', edgecolors='black')
_ = plt.title("3 knots")
_ = plt.xlabel("$x$")
_ = plt.ylabel("$y$")
_ = plt.plot(x,sm.ols('y~x+h(x,2,1)+h(x,5,1)+h(x,8,1)',data={'x':x,'y':y}).fit().predict(),color="darkblue",linewidth=2)
_ = plt.plot(x,norm.ppf(x/10),color="red")
ax24 = plt.scatter(x,y,facecolors='none', edgecolors='black')
_ = plt.title("6 knots")
_ = plt.xlabel("$x$")
_ = plt.ylabel("$y$")
_ = plt.plot(x,sm.ols('y~x+h(x,1,1)+h(x,2,1)+h(x,3.5,1)+h(x,5,1)+h(x,6.5,1)+h(x,8,1)',data={'x':x,'y':y}).fit().predict(),color="darkblue",linewidth=2)
_ = plt.plot(x,norm.ppf(x/10),color="red")
ax25 = plt.scatter(x,y,facecolors='none', edgecolors='black')
_ = plt.title("9 knots")
_ = plt.xlabel("$x$")
_ = plt.ylabel("$y$")
_ = plt.plot(x,sm.ols('y~x+h(x,1,1)+h(x,2,1)+h(x,3,1)+h(x,4,1)+h(x,5,1)+h(x,6,1)+h(x,7,1)+h(x,8,1)+h(x,9,1)',data={'x':x,'y':y}).fit().predict(),color="darkblue",linewidth=2)
_ = plt.plot(x,norm.ppf(x/10),color="red")
regstr = 'y~x+'
for i in range(1,26):
regstr += 'h(x,'+str(i/26*10)+',1)+'
regstr = regstr[:-1] #drop last +
ax26 = plt.scatter(x,y,facecolors='none', edgecolors='black')
_ = plt.title("25 knots")
_ = plt.xlabel("$x$")
_ = plt.ylabel("$y$")
_ = plt.plot(x,sm.ols(regstr,data={'x':x,'y':y}).fit().predict(),color="darkblue",linewidth=2)
_ = plt.plot(x,norm.ppf(x/10),color="red")
```
### Exercise:
Try generating random data from different distributions and fitting polynomials of different degrees to it. What do you observe?
```
# try it here
#So, we see that increasing the number of knots results in a more polynomial-like fit
#Next, we look at cubic splines with increasing numbers of knots
ax27 = plt.scatter(x,y,facecolors='none', edgecolors='black')
_ = plt.title("3 knots")
_ = plt.xlabel("$x$")
_ = plt.ylabel("$y$")
_ = plt.plot(x,sm.ols('y~x+np.power(x,2)+np.power(x,3)+h(x,2,3)+h(x,5,3)+h(x,8,3)',data={'x':x,'y':y}).fit().predict(),color="darkblue",linewidth=2)
_ = plt.plot(x,norm.ppf(x/10),color="red")
ax28 = plt.scatter(x,y,facecolors='none', edgecolors='black')
_ = plt.title("6 knots")
_ = plt.xlabel("$x$")
_ = plt.ylabel("$y$")
_ = plt.plot(x,sm.ols('y~x+np.power(x,2)+np.power(x,3)+h(x,1,3)+h(x,2,3)+h(x,3.5,3)+h(x,5,3)+h(x,6.5,3)+h(x,8,3)',data={'x':x,'y':y}).fit().predict(),color="darkblue",linewidth=2)
_ = plt.plot(x,norm.ppf(x/10),color="red")
ax29 = plt.scatter(x,y,facecolors='none', edgecolors='black')
_ = plt.title("9 knots")
_ = plt.xlabel("$x$")
_ = plt.ylabel("$y$")
_ = plt.plot(x,sm.ols('y~x+np.power(x,2)+np.power(x,3)+h(x,1,3)+h(x,2,3)+h(x,3,3)+h(x,4,3)+h(x,5,3)+h(x,6,3)+h(x,7,3)+h(x,8,3)+h(x,9,3)',data={'x':x,'y':y}).fit().predict(),color="darkblue",linewidth=2)
_ = plt.plot(x,norm.ppf(x/10),color="red")
regstr2 = 'y~x+np.power(x,2)+np.power(x,3)+'
for i in range(1,26):
regstr2 += 'h(x,'+str(i/26*10)+',3)+'
regstr2 = regstr2[:-1] #drop last +
ax30 = plt.scatter(x,y,facecolors='none', edgecolors='black')
_ = plt.title("25 knots")
_ = plt.xlabel("$x$")
_ = plt.ylabel("$y$")
_ = plt.plot(x,sm.ols(regstr2,data={'x':x,'y':y}).fit().predict(),color="darkblue",linewidth=2)
_ = plt.plot(x,norm.ppf(x/10),color="red")
```
|
github_jupyter
|
### **Install ChEMBL client for getting the dataset**
#### **https://www.ebi.ac.uk/chembl/**
```
!pip install chembl_webresource_client
```
### **Import Libraries**
```
import pandas as pd
from chembl_webresource_client.new_client import new_client
```
### **Find Coronavirus Dataset**
#### **Search Target**
```
target = new_client.target
target_query = target.search ('acetylcholinesterase')
targets = pd.DataFrame.from_dict (target_query)
targets
```
#### **Fetch Bio-Activity data for the target**
```
selected_target = targets.target_chembl_id [0]
selected_target
activity = new_client.activity
res = activity.filter (target_chembl_id = selected_target).filter (standard_type = "IC50")
```
#### **A Higher Standard Value means we'll require more amount of the drug for same inhibition**
```
df = pd.DataFrame.from_dict (res)
df.head (3)
df.standard_type.unique ()
```
##### **Save the resulting Bio-Activity data to a CSV file**
```
import os
df.to_csv (os.path.join ('Datasets', 'Part-1_Bioactivity_Data.csv'), index = False)
```
### **Pre-Processing Data**
#### **Ignore values with Missing Standard Value data**
```
df2 = df [df.standard_value.notna ()]
df2 = df2 [df.canonical_smiles.notna ()]
df2
```
#### **Label Compounds as active or inactive**
##### Compounds with IC50 less than 1000nM are considered active, greater than 10000nM are considered to be inactive, in between 1000nM to 10000nM are considered intermediate
##### 1. IC50 value of the drug indicates the toxicity of the drug to other disease causing organisms.
##### 2. IC50 is a quantitative measure that shows how much a particular inhibitory drug/substance/extract/fraction is needed to inhibit a biological component by 50%.
###### Above Definition taken from https://www.researchgate.net/post/What-is-the-significance-of-IC50-value-when-the-drug-is-exogenously-administered-to-an-animal-tissue
```
bioactivity_class = []
for i in df2.standard_value :
if float (i) >= 10000 :
bioactivity_class.append ("inactive")
elif float (i) <= 1000 :
bioactivity_class.append ("active")
else :
bioactivity_class.append ("intermediate")
print (len (bioactivity_class))
```
#### **Append Chembl ID, Canonical Smiles and Standard Value to a list**
##### Canonical Smiles :-
##### 1. Simplified Molecular Input Line Entry Specification
##### 2. They can represent a Molecular Compound in a Single Line
```
selection = ['molecule_chembl_id', 'canonical_smiles', 'standard_value']
df3 = df2 [selection]
print (len (df3))
df3
import numpy as np
#print (df3.values.shape)
#print (np.array (bioactivity_class).shape)
df4 = df3.values
df4
bioactivity_class = np.matrix (bioactivity_class).T
#bioactivity_class
columns = list (df3.columns)
columns.append ('bioactivity_class')
print (columns)
print (bioactivity_class.shape)
print (df4.shape)
#df3 = pd.concat ([df3, pd.Series (np.array (bioactivity_class))], axis = 1)
#print (len (df3))
#df3
df4
#df3 = df3.rename (columns = {0 : 'bioactivity_class'})
df_final = np.concatenate ((df4, bioactivity_class), axis = 1)
#df_final = pd.DataFrame (df_final, columns)
df_final
#df3.head (3)
#print (len (df3))
df_final = pd.DataFrame (df_final, columns = columns)
df_final
```
#### **Save Pre-Processed data to a CSV file**
```
df_final.to_csv (os.path.join ('Datasets', 'Part-1_Bioactivity_Preprocessed_Data.csv'), index = False)
!dir
```
|
github_jupyter
|
# Facial Keypoint Detection
This project will be all about defining and training a convolutional neural network to perform facial keypoint detection, and using computer vision techniques to transform images of faces. The first step in any challenge like this will be to load and visualize the data you'll be working with.
Let's take a look at some examples of images and corresponding facial keypoints.
<img src='images/key_pts_example.png' width=50% height=50%/>
Facial keypoints (also called facial landmarks) are the small magenta dots shown on each of the faces in the image above. In each training and test image, there is a single face and **68 keypoints, with coordinates (x, y), for that face**. These keypoints mark important areas of the face: the eyes, corners of the mouth, the nose, etc. These keypoints are relevant for a variety of tasks, such as face filters, emotion recognition, pose recognition, and so on. Here they are, numbered, and you can see that specific ranges of points match different portions of the face.
<img src='images/landmarks_numbered.jpg' width=30% height=30%/>
---
## Load and Visualize Data
The first step in working with any dataset is to become familiar with your data; you'll need to load in the images of faces and their keypoints and visualize them! This set of image data has been extracted from the [YouTube Faces Dataset](https://www.cs.tau.ac.il/~wolf/ytfaces/), which includes videos of people in YouTube videos. These videos have been fed through some processing steps and turned into sets of image frames containing one face and the associated keypoints.
#### Training and Testing Data
This facial keypoints dataset consists of 5770 color images. All of these images are separated into either a training or a test set of data.
* 3462 of these images are training images, for you to use as you create a model to predict keypoints.
* 2308 are test images, which will be used to test the accuracy of your model.
The information about the images and keypoints in this dataset are summarized in CSV files, which we can read in using `pandas`. Let's read the training CSV and get the annotations in an (N, 2) array where N is the number of keypoints and 2 is the dimension of the keypoint coordinates (x, y).
---
```
# import the required libraries
import glob
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import cv2
key_pts_frame = pd.read_csv('data/training_frames_keypoints.csv')
n = 0
image_name = key_pts_frame.iloc[n, 0]
key_pts = key_pts_frame.iloc[n, 1:].as_matrix()
key_pts = key_pts.astype('float').reshape(-1, 2)
print('Image name: ', image_name)
print('Landmarks shape: ', key_pts.shape)
print('First 4 key pts: {}'.format(key_pts[:4]))
# print out some stats about the data
print('Number of images: ', key_pts_frame.shape[0])
```
## Look at some images
Below, is a function `show_keypoints` that takes in an image and keypoints and displays them. As you look at this data, **note that these images are not all of the same size**, and neither are the faces! To eventually train a neural network on these images, we'll need to standardize their shape.
```
def show_keypoints(image, key_pts):
"""Show image with keypoints"""
plt.imshow(image)
plt.scatter(key_pts[:, 0], key_pts[:, 1], s=20, marker='.', c='m')
# Display a few different types of images by changing the index n
# select an image by index in our data frame
n = 0
image_name = key_pts_frame.iloc[n, 0]
key_pts = key_pts_frame.iloc[n, 1:].as_matrix()
key_pts = key_pts.astype('float').reshape(-1, 2)
plt.figure(figsize=(5, 5))
show_keypoints(mpimg.imread(os.path.join('data/training/', image_name)), key_pts)
plt.show()
```
## Dataset class and Transformations
To prepare our data for training, we'll be using PyTorch's Dataset class. Much of this this code is a modified version of what can be found in the [PyTorch data loading tutorial](http://pytorch.org/tutorials/beginner/data_loading_tutorial.html).
#### Dataset class
``torch.utils.data.Dataset`` is an abstract class representing a
dataset. This class will allow us to load batches of image/keypoint data, and uniformly apply transformations to our data, such as rescaling and normalizing images for training a neural network.
Your custom dataset should inherit ``Dataset`` and override the following
methods:
- ``__len__`` so that ``len(dataset)`` returns the size of the dataset.
- ``__getitem__`` to support the indexing such that ``dataset[i]`` can
be used to get the i-th sample of image/keypoint data.
Let's create a dataset class for our face keypoints dataset. We will
read the CSV file in ``__init__`` but leave the reading of images to
``__getitem__``. This is memory efficient because all the images are not
stored in the memory at once but read as required.
A sample of our dataset will be a dictionary
``{'image': image, 'keypoints': key_pts}``. Our dataset will take an
optional argument ``transform`` so that any required processing can be
applied on the sample. We will see the usefulness of ``transform`` in the
next section.
```
from torch.utils.data import Dataset, DataLoader
class FacialKeypointsDataset(Dataset):
"""Face Landmarks dataset."""
def __init__(self, csv_file, root_dir, transform=None):
"""
Args:
csv_file (string): Path to the csv file with annotations.
root_dir (string): Directory with all the images.
transform (callable, optional): Optional transform to be applied
on a sample.
"""
self.key_pts_frame = pd.read_csv(csv_file)
self.root_dir = root_dir
self.transform = transform
def __len__(self):
return len(self.key_pts_frame)
def __getitem__(self, idx):
image_name = os.path.join(self.root_dir,
self.key_pts_frame.iloc[idx, 0])
image = mpimg.imread(image_name)
# if image has an alpha color channel, get rid of it
if(image.shape[2] == 4):
image = image[:,:,0:3]
key_pts = self.key_pts_frame.iloc[idx, 1:].as_matrix()
key_pts = key_pts.astype('float').reshape(-1, 2)
sample = {'image': image, 'keypoints': key_pts}
if self.transform:
sample = self.transform(sample)
return sample
```
Now that we've defined this class, let's instantiate the dataset and display some images.
```
# Construct the dataset
face_dataset = FacialKeypointsDataset(csv_file='data/training_frames_keypoints.csv',
root_dir='data/training/')
# print some stats about the dataset
print('Length of dataset: ', len(face_dataset))
# Display a few of the images from the dataset
num_to_display = 3
for i in range(num_to_display):
# define the size of images
fig = plt.figure(figsize=(20,10))
# randomly select a sample
rand_i = np.random.randint(0, len(face_dataset))
sample = face_dataset[rand_i]
# print the shape of the image and keypoints
print(i, sample['image'].shape, sample['keypoints'].shape)
ax = plt.subplot(1, num_to_display, i + 1)
ax.set_title('Sample #{}'.format(i))
# Using the same display function, defined earlier
show_keypoints(sample['image'], sample['keypoints'])
```
## Transforms
Now, the images above are not of the same size, and neural networks often expect images that are standardized; a fixed size, with a normalized range for color ranges and coordinates, and (for PyTorch) converted from numpy lists and arrays to Tensors.
Therefore, we will need to write some pre-processing code.
Let's create four transforms:
- ``Normalize``: to convert a color image to grayscale values with a range of [0,1] and normalize the keypoints to be in a range of about [-1, 1]
- ``Rescale``: to rescale an image to a desired size.
- ``RandomCrop``: to crop an image randomly.
- ``ToTensor``: to convert numpy images to torch images.
We will write them as callable classes instead of simple functions so
that parameters of the transform need not be passed everytime it's
called. For this, we just need to implement ``__call__`` method and
(if we require parameters to be passed in), the ``__init__`` method.
We can then use a transform like this:
tx = Transform(params)
transformed_sample = tx(sample)
Observe below how these transforms are generally applied to both the image and its keypoints.
```
import torch
from torchvision import transforms, utils
# tranforms
class Normalize(object):
"""Convert a color image to grayscale and normalize the color range to [0,1]."""
def __call__(self, sample):
image, key_pts = sample['image'], sample['keypoints']
image_copy = np.copy(image)
key_pts_copy = np.copy(key_pts)
# convert image to grayscale
image_copy = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
# scale color range from [0, 255] to [0, 1]
image_copy= image_copy/255.0
# scale keypoints to be centered around 0 with a range of [-1, 1]
# mean = 100, sqrt = 50, so, pts should be (pts - 100)/50
key_pts_copy = (key_pts_copy - 100)/50.0
return {'image': image_copy, 'keypoints': key_pts_copy}
class Rescale(object):
"""Rescale the image in a sample to a given size.
Args:
output_size (tuple or int): Desired output size. If tuple, output is
matched to output_size. If int, smaller of image edges is matched
to output_size keeping aspect ratio the same.
"""
def __init__(self, output_size):
assert isinstance(output_size, (int, tuple))
self.output_size = output_size
def __call__(self, sample):
image, key_pts = sample['image'], sample['keypoints']
h, w = image.shape[:2]
if isinstance(self.output_size, int):
if h > w:
new_h, new_w = self.output_size * h / w, self.output_size
else:
new_h, new_w = self.output_size, self.output_size * w / h
else:
new_h, new_w = self.output_size
new_h, new_w = int(new_h), int(new_w)
img = cv2.resize(image, (new_w, new_h))
# scale the pts, too
key_pts = key_pts * [new_w / w, new_h / h]
return {'image': img, 'keypoints': key_pts}
class RandomCrop(object):
"""Crop randomly the image in a sample.
Args:
output_size (tuple or int): Desired output size. If int, square crop
is made.
"""
def __init__(self, output_size):
assert isinstance(output_size, (int, tuple))
if isinstance(output_size, int):
self.output_size = (output_size, output_size)
else:
assert len(output_size) == 2
self.output_size = output_size
def __call__(self, sample):
image, key_pts = sample['image'], sample['keypoints']
h, w = image.shape[:2]
new_h, new_w = self.output_size
top = np.random.randint(0, h - new_h)
left = np.random.randint(0, w - new_w)
image = image[top: top + new_h,
left: left + new_w]
key_pts = key_pts - [left, top]
return {'image': image, 'keypoints': key_pts}
class ToTensor(object):
"""Convert ndarrays in sample to Tensors."""
def __call__(self, sample):
image, key_pts = sample['image'], sample['keypoints']
# if image has no grayscale color channel, add one
if(len(image.shape) == 2):
# add that third color dim
image = image.reshape(image.shape[0], image.shape[1], 1)
# swap color axis because
# numpy image: H x W x C
# torch image: C X H X W
image = image.transpose((2, 0, 1))
return {'image': torch.from_numpy(image),
'keypoints': torch.from_numpy(key_pts)}
```
## Test out the transforms
Let's test these transforms out to make sure they behave as expected. As you look at each transform, note that, in this case, **order does matter**. For example, you cannot crop a image using a value smaller than the original image (and the orginal images vary in size!), but, if you first rescale the original image, you can then crop it to any size smaller than the rescaled size.
```
# test out some of these transforms
rescale = Rescale(100)
crop = RandomCrop(50)
composed = transforms.Compose([Rescale(250),
RandomCrop(224)])
# apply the transforms to a sample image
test_num = 500
sample = face_dataset[test_num]
fig = plt.figure()
for i, tx in enumerate([rescale, crop, composed]):
transformed_sample = tx(sample)
ax = plt.subplot(1, 3, i + 1)
plt.tight_layout()
ax.set_title(type(tx).__name__)
show_keypoints(transformed_sample['image'], transformed_sample['keypoints'])
plt.show()
```
## Create the transformed dataset
Apply the transforms in order to get grayscale images of the same shape. Verify that your transform works by printing out the shape of the resulting data (printing out a few examples should show you a consistent tensor size).
```
# define the data tranform
# order matters! i.e. rescaling should come before a smaller crop
data_transform = transforms.Compose([Rescale(250),
RandomCrop(224),
Normalize(),
ToTensor()])
# create the transformed dataset
transformed_dataset = FacialKeypointsDataset(csv_file='data/training_frames_keypoints.csv',
root_dir='data/training/',
transform=data_transform)
# print some stats about the transformed data
print('Number of images: ', len(transformed_dataset))
# make sure the sample tensors are the expected size
for i in range(5):
sample = transformed_dataset[i]
print(i, sample['image'].size(), sample['keypoints'].size())
```
## Data Iteration and Batching
Right now, we are iterating over this data using a ``for`` loop, but we are missing out on a lot of PyTorch's dataset capabilities, specifically the abilities to:
- Batch the data
- Shuffle the data
- Load the data in parallel using ``multiprocessing`` workers.
``torch.utils.data.DataLoader`` is an iterator which provides all these
features, and we'll see this in use in the *next* notebook, Notebook 2, when we load data in batches to train a neural network!
---
|
github_jupyter
|
```
import warnings
import matplotlib as mpl
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
from astropy.io import fits
from astropy.table import Table
import pandas as pd
import numpy as np
np.seterr(divide='ignore')
warnings.filterwarnings("ignore", category=RuntimeWarning)
class HRCevt1:
'''
A more robust HRC EVT1 file. Includes explicit
columns for every status bit, as well as calculated
columns for the f_p, f_b plane for your boomerangs.
Check out that cool new filtering algorithm!
'''
def __init__(self, evt1file):
# Do a standard read in of the EVT1 fits table
self.filename = evt1file
self.hdulist = fits.open(evt1file)
self.data = Table(self.hdulist[1].data)
self.header = self.hdulist[1].header
self.gti = self.hdulist[2].data
self.hdulist.close() # Don't forget to close your fits file!
fp_u, fb_u, fp_v, fb_v = self.calculate_fp_fb()
self.gti.starts = self.gti['START']
self.gti.stops = self.gti['STOP']
self.gtimask = []
# for start, stop in zip(self.gti.starts, self.gti.stops):
# self.gtimask = (self.data["time"] > start) & (self.data["time"] < stop)
self.gtimask = (self.data["time"] > self.gti.starts[0]) & (
self.data["time"] < self.gti.stops[-1])
self.data["fp_u"] = fp_u
self.data["fb_u"] = fb_u
self.data["fp_v"] = fp_v
self.data["fb_v"] = fb_v
# Make individual status bit columns with legible names
self.data["AV3 corrected for ringing"] = self.data["status"][:, 0]
self.data["AU3 corrected for ringing"] = self.data["status"][:, 1]
self.data["Event impacted by prior event (piled up)"] = self.data["status"][:, 2]
# Bit 4 (Python 3) is spare
self.data["Shifted event time"] = self.data["status"][:, 4]
self.data["Event telemetered in NIL mode"] = self.data["status"][:, 5]
self.data["V axis not triggered"] = self.data["status"][:, 6]
self.data["U axis not triggered"] = self.data["status"][:, 7]
self.data["V axis center blank event"] = self.data["status"][:, 8]
self.data["U axis center blank event"] = self.data["status"][:, 9]
self.data["V axis width exceeded"] = self.data["status"][:, 10]
self.data["U axis width exceeded"] = self.data["status"][:, 11]
self.data["Shield PMT active"] = self.data["status"][:, 12]
# Bit 14 (Python 13) is hardware spare
self.data["Upper level discriminator not exceeded"] = self.data["status"][:, 14]
self.data["Lower level discriminator not exceeded"] = self.data["status"][:, 15]
self.data["Event in bad region"] = self.data["status"][:, 16]
self.data["Amp total on V or U = 0"] = self.data["status"][:, 17]
self.data["Incorrect V center"] = self.data["status"][:, 18]
self.data["Incorrect U center"] = self.data["status"][:, 19]
self.data["PHA ratio test failed"] = self.data["status"][:, 20]
self.data["Sum of 6 taps = 0"] = self.data["status"][:, 21]
self.data["Grid ratio test failed"] = self.data["status"][:, 22]
self.data["ADC sum on V or U = 0"] = self.data["status"][:, 23]
self.data["PI exceeding 255"] = self.data["status"][:, 24]
self.data["Event time tag is out of sequence"] = self.data["status"][:, 25]
self.data["V amp flatness test failed"] = self.data["status"][:, 26]
self.data["U amp flatness test failed"] = self.data["status"][:, 27]
self.data["V amp saturation test failed"] = self.data["status"][:, 28]
self.data["U amp saturation test failed"] = self.data["status"][:, 29]
self.data["V hyperbolic test failed"] = self.data["status"][:, 30]
self.data["U hyperbolic test failed"] = self.data["status"][:, 31]
self.data["Hyperbola test passed"] = np.logical_not(np.logical_or(
self.data['U hyperbolic test failed'], self.data['V hyperbolic test failed']))
self.data["Hyperbola test failed"] = np.logical_or(
self.data['U hyperbolic test failed'], self.data['V hyperbolic test failed'])
self.obsid = self.header["OBS_ID"]
self.obs_date = self.header["DATE"]
self.target = self.header["OBJECT"]
self.detector = self.header["DETNAM"]
self.grating = self.header["GRATING"]
self.exptime = self.header["EXPOSURE"]
self.numevents = len(self.data["time"])
self.goodtimeevents = len(self.data["time"][self.gtimask])
self.badtimeevents = self.numevents - self.goodtimeevents
self.hyperbola_passes = np.sum(np.logical_or(
self.data['U hyperbolic test failed'], self.data['V hyperbolic test failed']))
self.hyperbola_failures = np.sum(np.logical_not(np.logical_or(
self.data['U hyperbolic test failed'], self.data['V hyperbolic test failed'])))
if self.hyperbola_passes + self.hyperbola_failures != self.numevents:
print("Warning: Number of Hyperbola Test Failures and Passes ({}) does not equal total number of events ({}).".format(
self.hyperbola_passes + self.hyperbola_failures, self.numevents))
# Multidimensional columns don't grok with Pandas
self.data.remove_column('status')
self.data = self.data.to_pandas()
def __str__(self):
return "HRC EVT1 object with {} events. Data is packaged as a Pandas Dataframe".format(self.numevents)
def calculate_fp_fb(self):
'''
Calculate the Fine Position (fp) and normalized central tap
amplitude (fb) for the HRC U- and V- axes.
Parameters
----------
data : Astropy Table
Table object made from an HRC evt1 event list. Must include the
au1, au2, au3 and av1, av2, av3 columns.
Returns
-------
fp_u, fb_u, fp_v, fb_v: float
Calculated fine positions and normalized central tap amplitudes
for the HRC U- and V- axes
'''
a_u = self.data["au1"] # otherwise known as "a1"
b_u = self.data["au2"] # "a2"
c_u = self.data["au3"] # "a3"
a_v = self.data["av1"]
b_v = self.data["av2"]
c_v = self.data["av3"]
with np.errstate(invalid='ignore'):
# Do the U axis
fp_u = ((c_u - a_u) / (a_u + b_u + c_u))
fb_u = b_u / (a_u + b_u + c_u)
# Do the V axis
fp_v = ((c_v - a_v) / (a_v + b_v + c_v))
fb_v = b_v / (a_v + b_v + c_v)
return fp_u, fb_u, fp_v, fb_v
def threshold(self, img, bins):
nozero_img = img.copy()
nozero_img[img == 0] = np.nan
# This is a really stupid way to threshold
median = np.nanmedian(nozero_img)
thresh = median*5
thresh_img = nozero_img
thresh_img[thresh_img < thresh] = np.nan
thresh_img[:int(bins[1]/2), :] = np.nan
# thresh_img[:,int(bins[1]-5):] = np.nan
return thresh_img
def hyperscreen(self):
'''
Grant Tremblay's new algorithm. Screens events on a tap-by-tap basis.
'''
data = self.data
#taprange = range(data['crsu'].min(), data['crsu'].max() + 1)
taprange_u = range(data['crsu'].min() -1 , data['crsu'].max() + 1)
taprange_v = range(data['crsv'].min() - 1, data['crsv'].max() + 1)
bins = [200, 200] # number of bins
# Instantiate these empty dictionaries to hold our results
u_axis_survivals = {}
v_axis_survivals = {}
for tap in taprange_u:
# Do the U axis
tapmask_u = data[data['crsu'] == tap].index.values
if len(tapmask_u) < 2:
continue
keep_u = np.isfinite(data['fb_u'][tapmask_u])
hist_u, xbounds_u, ybounds_u = np.histogram2d(
data['fb_u'][tapmask_u][keep_u], data['fp_u'][tapmask_u][keep_u], bins=bins)
thresh_hist_u = self.threshold(hist_u, bins=bins)
posx_u = np.digitize(data['fb_u'][tapmask_u], xbounds_u)
posy_u = np.digitize(data['fp_u'][tapmask_u], ybounds_u)
hist_mask_u = (posx_u > 0) & (posx_u <= bins[0]) & (
posy_u > -1) & (posy_u <= bins[1])
# Values of the histogram where the points are
hhsub_u = thresh_hist_u[posx_u[hist_mask_u] -
1, posy_u[hist_mask_u] - 1]
pass_fb_u = data['fb_u'][tapmask_u][hist_mask_u][np.isfinite(
hhsub_u)]
u_axis_survivals["U Axis Tap {:02d}".format(
tap)] = pass_fb_u.index.values
for tap in taprange_v:
# Now do the V axis:
tapmask_v = data[data['crsv'] == tap].index.values
if len(tapmask_v) < 2:
continue
keep_v = np.isfinite(data['fb_v'][tapmask_v])
hist_v, xbounds_v, ybounds_v = np.histogram2d(
data['fb_v'][tapmask_v][keep_v], data['fp_v'][tapmask_v][keep_v], bins=bins)
thresh_hist_v = self.threshold(hist_v, bins=bins)
posx_v = np.digitize(data['fb_v'][tapmask_v], xbounds_v)
posy_v = np.digitize(data['fp_v'][tapmask_v], ybounds_v)
hist_mask_v = (posx_v > 0) & (posx_v <= bins[0]) & (
posy_v > -1) & (posy_v <= bins[1])
# Values of the histogram where the points are
hhsub_v = thresh_hist_v[posx_v[hist_mask_v] -
1, posy_v[hist_mask_v] - 1]
pass_fb_v = data['fb_v'][tapmask_v][hist_mask_v][np.isfinite(
hhsub_v)]
v_axis_survivals["V Axis Tap {:02d}".format(
tap)] = pass_fb_v.index.values
# Done looping over taps
u_all_survivals = np.concatenate(
[x for x in u_axis_survivals.values()])
v_all_survivals = np.concatenate(
[x for x in v_axis_survivals.values()])
# If the event passes both U- and V-axis tests, it survives
all_survivals = np.intersect1d(u_all_survivals, v_all_survivals)
survival_mask = np.isin(self.data.index.values, all_survivals)
failure_mask = np.logical_not(survival_mask)
num_survivals = sum(survival_mask)
num_failures = sum(failure_mask)
percent_tapscreen_rejected = round(
((num_failures / self.numevents) * 100), 2)
# Do a sanity check to look for lost events. Shouldn't be any.
if num_survivals + num_failures != self.numevents:
print("WARNING: Total Number of survivals and failures does \
not equal total events in the EVT1 file. Something is wrong!")
legacy_hyperbola_test_survivals = sum(
self.data['Hyperbola test passed'])
legacy_hyperbola_test_failures = sum(
self.data['Hyperbola test failed'])
percent_legacy_hyperbola_test_rejected = round(
((legacy_hyperbola_test_failures / self.goodtimeevents) * 100), 2)
percent_improvement_over_legacy_test = round(
(percent_tapscreen_rejected - percent_legacy_hyperbola_test_rejected), 2)
hyperscreen_results_dict = {"ObsID": self.obsid,
"Target": self.target,
"Exposure Time": self.exptime,
"Detector": self.detector,
"Number of Events": self.numevents,
"Number of Good Time Events": self.goodtimeevents,
"U Axis Survivals by Tap": u_axis_survivals,
"V Axis Survivals by Tap": v_axis_survivals,
"U Axis All Survivals": u_all_survivals,
"V Axis All Survivals": v_all_survivals,
"All Survivals (event indices)": all_survivals,
"All Survivals (boolean mask)": survival_mask,
"All Failures (boolean mask)": failure_mask,
"Percent rejected by Tapscreen": percent_tapscreen_rejected,
"Percent rejected by Hyperbola": percent_legacy_hyperbola_test_rejected,
"Percent improvement": percent_improvement_over_legacy_test
}
return hyperscreen_results_dict
def hyperbola(self, fb, a, b, h):
'''Given the normalized central tap amplitude, a, b, and h,
return an array of length len(fb) that gives a hyperbola.'''
hyperbola = b * np.sqrt(((fb - h)**2 / a**2) - 1)
return hyperbola
def legacy_hyperbola_test(self, tolerance=0.035):
'''
Apply the hyperbolic test.
'''
# Remind the user what tolerance they're using
# print("{0: <25}| Using tolerance = {1}".format(" ", tolerance))
# Set hyperbolic coefficients, depending on whether this is HRC-I or -S
if self.detector == "HRC-I":
a_u = 0.3110
b_u = 0.3030
h_u = 1.0580
a_v = 0.3050
b_v = 0.2730
h_v = 1.1
# print("{0: <25}| Using HRC-I hyperbolic coefficients: ".format(" "))
# print("{0: <25}| Au={1}, Bu={2}, Hu={3}".format(" ", a_u, b_u, h_u))
# print("{0: <25}| Av={1}, Bv={2}, Hv={3}".format(" ", a_v, b_v, h_v))
if self.detector == "HRC-S":
a_u = 0.2706
b_u = 0.2620
h_u = 1.0180
a_v = 0.2706
b_v = 0.2480
h_v = 1.0710
# print("{0: <25}| Using HRC-S hyperbolic coefficients: ".format(" "))
# print("{0: <25}| Au={1}, Bu={2}, Hu={3}".format(" ", a_u, b_u, h_u))
# print("{0: <25}| Av={1}, Bv={2}, Hv={3}".format(" ", a_v, b_v, h_v))
# Set the tolerance boundary ("width" of the hyperbolic region)
h_u_lowerbound = h_u * (1 + tolerance)
h_u_upperbound = h_u * (1 - tolerance)
h_v_lowerbound = h_v * (1 + tolerance)
h_v_upperbound = h_v * (1 - tolerance)
# Compute the Hyperbolae
with np.errstate(invalid='ignore'):
zone_u_fit = self.hyperbola(self.data["fb_u"], a_u, b_u, h_u)
zone_u_lowerbound = self.hyperbola(
self.data["fb_u"], a_u, b_u, h_u_lowerbound)
zone_u_upperbound = self.hyperbola(
self.data["fb_u"], a_u, b_u, h_u_upperbound)
zone_v_fit = self.hyperbola(self.data["fb_v"], a_v, b_v, h_v)
zone_v_lowerbound = self.hyperbola(
self.data["fb_v"], a_v, b_v, h_v_lowerbound)
zone_v_upperbound = self.hyperbola(
self.data["fb_v"], a_v, b_v, h_v_upperbound)
zone_u = [zone_u_lowerbound, zone_u_upperbound]
zone_v = [zone_v_lowerbound, zone_v_upperbound]
# Apply the masks
# print("{0: <25}| Hyperbolic masks for U and V axes computed".format(""))
with np.errstate(invalid='ignore'):
# print("{0: <25}| Creating U-axis mask".format(""), end=" |")
between_u = np.logical_not(np.logical_and(
self.data["fp_u"] < zone_u[1], self.data["fp_u"] > -1 * zone_u[1]))
not_beyond_u = np.logical_and(
self.data["fp_u"] < zone_u[0], self.data["fp_u"] > -1 * zone_u[0])
condition_u_final = np.logical_and(between_u, not_beyond_u)
# print(" Creating V-axis mask")
between_v = np.logical_not(np.logical_and(
self.data["fp_v"] < zone_v[1], self.data["fp_v"] > -1 * zone_v[1]))
not_beyond_v = np.logical_and(
self.data["fp_v"] < zone_v[0], self.data["fp_v"] > -1 * zone_v[0])
condition_v_final = np.logical_and(between_v, not_beyond_v)
mask_u = condition_u_final
mask_v = condition_v_final
hyperzones = {"zone_u_fit": zone_u_fit,
"zone_u_lowerbound": zone_u_lowerbound,
"zone_u_upperbound": zone_u_upperbound,
"zone_v_fit": zone_v_fit,
"zone_v_lowerbound": zone_v_lowerbound,
"zone_v_upperbound": zone_v_upperbound}
hypermasks = {"mask_u": mask_u, "mask_v": mask_v}
# print("{0: <25}| Hyperbolic masks created".format(""))
# print("{0: <25}| ".format(""))
return hyperzones, hypermasks
def boomerang(self, mask=None, show=True, plot_legacy_zone=True, title=None, cmap=None, savepath=None, create_subplot=False, ax=None, rasterized=True):
# You can plot the image on axes of a subplot by passing
# that axis to this function. Here are some switches to enable that.
if create_subplot is False:
self.fig, self.ax = plt.subplots(figsize=(12, 8))
elif create_subplot is True:
if ax is None:
self.ax = plt.gca()
else:
self.ax = ax
if cmap is None:
cmap = 'plasma'
if mask is not None:
self.ax.scatter(self.data['fb_u'], self.data['fp_u'],
c=self.data['sumamps'], cmap='bone', s=0.3, alpha=0.8, rasterized=rasterized)
frame = self.ax.scatter(self.data['fb_u'][mask], self.data['fp_u'][mask],
c=self.data['sumamps'][mask], cmap=cmap, s=0.5, rasterized=rasterized)
else:
frame = self.ax.scatter(self.data['fb_u'], self.data['fp_u'],
c=self.data['sumamps'], cmap=cmap, s=0.5, rasterized=rasterized)
if plot_legacy_zone is True:
hyperzones, hypermasks = self.legacy_hyperbola_test(
tolerance=0.035)
self.ax.plot(self.data["fb_u"], hyperzones["zone_u_lowerbound"],
'o', markersize=0.3, color='black', alpha=0.8, rasterized=rasterized)
self.ax.plot(self.data["fb_u"], -1 * hyperzones["zone_u_lowerbound"],
'o', markersize=0.3, color='black', alpha=0.8, rasterized=rasterized)
self.ax.plot(self.data["fb_u"], hyperzones["zone_u_upperbound"],
'o', markersize=0.3, color='black', alpha=0.8, rasterized=rasterized)
self.ax.plot(self.data["fb_u"], -1 * hyperzones["zone_u_upperbound"],
'o', markersize=0.3, color='black', alpha=0.8, rasterized=rasterized)
self.ax.grid(False)
if title is None:
self.ax.set_title('{} | {} | ObsID {} | {} ksec | {} counts'.format(
self.target, self.detector, self.obsid, round(self.exptime / 1000, 1), self.numevents))
else:
self.ax.set_title(title)
self.ax.set_ylim(-1.1, 1.1)
self.ax.set_xlim(-0.1, 1.1)
self.ax.set_ylabel(r'Fine Position $f_p$ $(C-A)/(A + B + C)$')
self.ax.set_xlabel(
r'Normalized Central Tap Amplitude $f_b$ $B / (A+B+C)$')
if create_subplot is False:
self.cbar = plt.colorbar(frame, pad=-0.005)
self.cbar.set_label("SUMAMPS")
if show is True:
plt.show()
if savepath is not None:
plt.savefig(savepath, dpi=150, bbox_inches='tight')
print('Saved boomerang figure to: {}'.format(savepath))
def image(self, masked_x=None, masked_y=None, xlim=None, ylim=None, detcoords=False, title=None, cmap=None, show=True, savepath=None, create_subplot=False, ax=None):
'''
Create a quicklook image, in detector or sky coordinates, of the
observation. The image will be binned to 400x400.
'''
# Create the 2D histogram
nbins = (400, 400)
if masked_x is not None and masked_y is not None:
x = masked_x
y = masked_y
img_data, yedges, xedges = np.histogram2d(y, x, nbins)
else:
if detcoords is False:
x = self.data['x'][self.gtimask]
y = self.data['y'][self.gtimask]
elif detcoords is True:
x = self.data['detx'][self.gtimask]
y = self.data['dety'][self.gtimask]
img_data, yedges, xedges = np.histogram2d(y, x, nbins)
extent = [xedges[0], xedges[-1], yedges[0], yedges[-1]]
# Create the Figure
styleplots()
# You can plot the image on axes of a subplot by passing
# that axis to this function. Here are some switches to enable that.
if create_subplot is False:
self.fig, self.ax = plt.subplots()
elif create_subplot is True:
if ax is None:
self.ax = plt.gca()
else:
self.ax = ax
self.ax.grid(False)
if cmap is None:
cmap = 'viridis'
self.ax.imshow(img_data, extent=extent, norm=LogNorm(),
interpolation=None, cmap=cmap, origin='lower')
if title is None:
self.ax.set_title("ObsID {} | {} | {} | {:,} events".format(
self.obsid, self.target, self.detector, self.goodtimeevents))
else:
self.ax.set_title("{}".format(title))
if detcoords is False:
self.ax.set_xlabel("Sky X")
self.ax.set_ylabel("Sky Y")
elif detcoords is True:
self.ax.set_xlabel("Detector X")
self.ax.set_ylabel("Detector Y")
if xlim is not None:
self.ax.set_xlim(xlim)
if ylim is not None:
self.ax.set_ylim(ylim)
if show is True:
plt.show(block=True)
if savepath is not None:
plt.savefig('{}'.format(savepath))
print("Saved image to {}".format(savepath))
def styleplots():
mpl.rcParams['agg.path.chunksize'] = 10000
# Make things pretty
plt.style.use('ggplot')
labelsizes = 10
plt.rcParams['font.size'] = labelsizes
plt.rcParams['axes.titlesize'] = 12
plt.rcParams['axes.labelsize'] = labelsizes
plt.rcParams['xtick.labelsize'] = labelsizes
plt.rcParams['ytick.labelsize'] = labelsizes
from astropy.io import fits
import os
os.listdir('../tests/data/')
fitsfile = '../tests/data/hrcS_evt1_testfile.fits.gz'
obs = HRCevt1(fitsfile)
obs.image(obs.data['detx'][obs.gtimask], obs.data['dety'][obs.gtimask], xlim=(26000, 41000), ylim=(31500, 34000))
results = obs.hyperscreen()
obs.image(obs.data['detx'][results['All Failures (boolean mask)']], obs.data['dety'][results['All Failures (boolean mask)']], xlim=(26000, 41000), ylim=(31500, 34000))
obs.data['crsv'].min()
obs.data['crsv'].max()
obs.data['crsv']
obs.numevents
from astropy.io import fits
header = fits.getheader(fitsfile, 1)
header
from hyperscreen import hypercore
```
|
github_jupyter
|
# Parameterizing with Continuous Variables
```
from IPython.display import Image
```
## Continuous Factors
1. Base Class for Continuous Factors
2. Joint Gaussian Distributions
3. Canonical Factors
4. Linear Gaussian CPD
In many situations, some variables are best modeled as taking values in some continuous space. Examples include variables such as position, velocity, temperature, and pressure. Clearly, we cannot use a table representation in this case.
Nothing in the formulation of a Bayesian network requires that we restrict attention to discrete variables. The only requirement is that the CPD, P(X | Y1, Y2, ... Yn) represent, for every assignment of values y1 ∈ Val(Y1), y2 ∈ Val(Y2), .....yn ∈ val(Yn), a distribution over X. In this case, X might be continuous, in which case the CPD would need to represent distributions over a continuum of values; we might also have X’s parents continuous, so that the CPD would also need to represent a continuum of different probability distributions. There exists implicit representations for CPDs of this type, allowing us to apply all the network machinery for the continuous case as well.
### Base Class for Continuous Factors
This class will behave as a base class for the continuous factor representations. All the present and future factor classes will be derived from this base class. We need to specify the variable names and a pdf function to initialize this class.
```
import numpy as np
from scipy.special import beta
# Two variable drichlet ditribution with alpha = (1,2)
def drichlet_pdf(x, y):
return (np.power(x, 1)*np.power(y, 2))/beta(x, y)
from pgmpy.factors.continuous import ContinuousFactor
drichlet_factor = ContinuousFactor(['x', 'y'], drichlet_pdf)
drichlet_factor.scope(), drichlet_factor.assignment(5,6)
```
This class supports methods like **marginalize, reduce, product and divide** just like what we have with discrete classes. One caveat is that when there are a number of variables involved, these methods prove to be inefficient and hence we resort to certain Gaussian or some other approximations which are discussed later.
```
def custom_pdf(x, y, z):
return z*(np.power(x, 1)*np.power(y, 2))/beta(x, y)
custom_factor = ContinuousFactor(['x', 'y', 'z'], custom_pdf)
custom_factor.scope(), custom_factor.assignment(1, 2, 3)
custom_factor.reduce([('y', 2)])
custom_factor.scope(), custom_factor.assignment(1, 3)
from scipy.stats import multivariate_normal
std_normal_pdf = lambda *x: multivariate_normal.pdf(x, [0, 0], [[1, 0], [0, 1]])
std_normal = ContinuousFactor(['x1', 'x2'], std_normal_pdf)
std_normal.scope(), std_normal.assignment([1, 1])
std_normal.marginalize(['x2'])
std_normal.scope(), std_normal.assignment(1)
sn_pdf1 = lambda x: multivariate_normal.pdf([x], [0], [[1]])
sn_pdf2 = lambda x1,x2: multivariate_normal.pdf([x1, x2], [0, 0], [[1, 0], [0, 1]])
sn1 = ContinuousFactor(['x2'], sn_pdf1)
sn2 = ContinuousFactor(['x1', 'x2'], sn_pdf2)
sn3 = sn1 * sn2
sn4 = sn2 / sn1
sn3.assignment(0, 0), sn4.assignment(0, 0)
```
The ContinuousFactor class also has a method **discretize** that takes a pgmpy Discretizer class as input. It will output a list of discrete probability masses or a Factor or TabularCPD object depending upon the discretization method used. Although, we do not have inbuilt discretization algorithms for multivariate distributions for now, the users can always define their own Discretizer class by subclassing the pgmpy.BaseDiscretizer class.
### Joint Gaussian Distributions
In its most common representation, a multivariate Gaussian distribution over X1………..Xn is characterized by an n-dimensional mean vector μ, and a symmetric n x n covariance matrix Σ. The density function is most defined as -
$$
p(x) = \dfrac{1}{(2\pi)^{n/2}|Σ|^{1/2}} exp[-0.5*(x-μ)^TΣ^{-1}(x-μ)]
$$
The class pgmpy.JointGaussianDistribution provides its representation. This is derived from the class pgmpy.ContinuousFactor. We need to specify the variable names, a mean vector and a covariance matrix for its inialization. It will automatically comute the pdf function given these parameters.
```
from pgmpy.factors.distributions import GaussianDistribution as JGD
dis = JGD(['x1', 'x2', 'x3'], np.array([[1], [-3], [4]]),
np.array([[4, 2, -2], [2, 5, -5], [-2, -5, 8]]))
dis.variables
dis.mean
dis.covariance
dis.pdf([0,0,0])
```
This class overrides the basic operation methods **(marginalize, reduce, normalize, product and divide)** as these operations here are more efficient than the ones in its parent class. Most of these operation involve a matrix inversion which is O(n^3) with repect to the number of variables.
```
dis1 = JGD(['x1', 'x2', 'x3'], np.array([[1], [-3], [4]]),
np.array([[4, 2, -2], [2, 5, -5], [-2, -5, 8]]))
dis2 = JGD(['x3', 'x4'], [1, 2], [[2, 3], [5, 6]])
dis3 = dis1 * dis2
dis3.variables
dis3.mean
dis3.covariance
```
The others methods can also be used in a similar fashion.
### Canonical Factors
While the Joint Gaussian representation is useful for certain sampling algorithms, a closer look reveals that it can also not be used directly in the sum-product algorithms. Why? Because operations like product and reduce, as mentioned above involve matrix inversions at each step.
So, in order to compactly describe the intermediate factors in a Gaussian network without the costly matrix inversions at each step, a simple parametric representation is used known as the Canonical Factor. This representation is closed under the basic operations used in inference: factor product, factor division, factor reduction, and marginalization. Thus, we can define a set of simple data structures that allow the inference process to be performed. Moreover, the integration operation required by marginalization is always well defined, and it is guaranteed to produce a finite integral under certain conditions; when it is well defined, it has a simple analytical solution.
A canonical form C (X; K,h, g) is defined as:
$$C(X; K,h,g) = exp(-0.5X^TKX + h^TX + g)$$
We can represent every Gaussian as a canonical form. Rewriting the joint Gaussian pdf we obtain,
N (μ; Σ) = C (K, h, g) where:
$$
K = Σ^{-1}
$$
$$
h = Σ^{-1}μ
$$
$$
g = -0.5μ^TΣ^{-1}μ - log((2π)^{n/2}|Σ|^{1/2}
$$
Similar to the JointGaussainDistribution class, the CanonicalFactor class is also derived from the ContinuousFactor class but with its own implementations of the methods required for the sum-product algorithms that are much more efficient than its parent class methods. Let us have a look at the API of a few methods in this class.
```
from pgmpy.factors.continuous import CanonicalDistribution
phi1 = CanonicalDistribution(['x1', 'x2', 'x3'],
np.array([[1, -1, 0], [-1, 4, -2], [0, -2, 4]]),
np.array([[1], [4], [-1]]), -2)
phi2 = CanonicalDistribution(['x1', 'x2'], np.array([[3, -2], [-2, 4]]),
np.array([[5], [-1]]), 1)
phi3 = phi1 * phi2
phi3.variables
phi3.h
phi3.K
phi3.g
```
This class also has a method, to_joint_gaussian to convert the canoncial representation back into the joint gaussian distribution.
```
phi = CanonicalDistribution(['x1', 'x2'], np.array([[3, -2], [-2, 4]]),
np.array([[5], [-1]]), 1)
jgd = phi.to_joint_gaussian()
jgd.variables
jgd.covariance
jgd.mean
```
### Linear Gaussian CPD
A linear gaussian conditional probability distribution is defined on a continuous variable. All the parents of this variable are also continuous. The mean of this variable, is linearly dependent on the mean of its parent variables and the variance is independent.
For example,
$$
P(Y ; x1, x2, x3) = N(β_1x_1 + β_2x_2 + β_3x_3 + β_0 ; σ^2)
$$
Let Y be a linear Gaussian of its parents X1,...,Xk:
$$
p(Y | x) = N(β_0 + β^T x ; σ^2)
$$
The distribution of Y is a normal distribution p(Y) where:
$$
μ_Y = β_0 + β^Tμ
$$
$$
{{σ^2}_Y = σ^2 + β^TΣβ}
$$
The joint distribution over {X, Y} is a normal distribution where:
$$Cov[X_i; Y] = {\sum_{j=1}^{k} β_jΣ_{i,j}}$$
Assume that X1,...,Xk are jointly Gaussian with distribution N (μ; Σ). Then:
For its representation pgmpy has a class named LinearGaussianCPD in the module pgmpy.factors.continuous. To instantiate an object of this class, one needs to provide a variable name, the value of the beta_0 term, the variance, a list of the parent variable names and a list of the coefficient values of the linear equation (beta_vector), where the list of parent variable names and beta_vector list is optional and defaults to None.
```
# For P(Y| X1, X2, X3) = N(-2x1 + 3x2 + 7x3 + 0.2; 9.6)
from pgmpy.factors.continuous import LinearGaussianCPD
cpd = LinearGaussianCPD('Y', [0.2, -2, 3, 7], 9.6, ['X1', 'X2', 'X3'])
print(cpd)
```
A Gaussian Bayesian is defined as a network all of whose variables are continuous, and where all of the CPDs are linear Gaussians. These networks are of particular interest as these are an alternate form of representaion of the Joint Gaussian distribution.
These networks are implemented as the LinearGaussianBayesianNetwork class in the module, pgmpy.models.continuous. This class is a subclass of the BayesianModel class in pgmpy.models and will inherit most of the methods from it. It will have a special method known as to_joint_gaussian that will return an equivalent JointGuassianDistribution object for the model.
```
from pgmpy.models import LinearGaussianBayesianNetwork
model = LinearGaussianBayesianNetwork([('x1', 'x2'), ('x2', 'x3')])
cpd1 = LinearGaussianCPD('x1', [1], 4)
cpd2 = LinearGaussianCPD('x2', [-5, 0.5], 4, ['x1'])
cpd3 = LinearGaussianCPD('x3', [4, -1], 3, ['x2'])
# This is a hack due to a bug in pgmpy (LinearGaussianCPD
# doesn't have `variables` attribute but `add_cpds` function
# wants to check that...)
cpd1.variables = [*cpd1.evidence, cpd1.variable]
cpd2.variables = [*cpd2.evidence, cpd2.variable]
cpd3.variables = [*cpd3.evidence, cpd3.variable]
model.add_cpds(cpd1, cpd2, cpd3)
jgd = model.to_joint_gaussian()
jgd.variables
jgd.mean
jgd.covariance
```
|
github_jupyter
|
##### Copyright 2018 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# tf.data を使ったテキストの読み込み
<table class="tfo-notebook-buttons" align="left">
<td><a target="_blank" href="https://www.tensorflow.org/tutorials/load_data/text"><img src="https://www.tensorflow.org/images/tf_logo_32px.png">TensorFlow.org で表示</a></td>
<td> <a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/ja/tutorials/load_data/text.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png">Google Colab で実行</a> </td>
<td> <a target="_blank" href="https://github.com/tensorflow/docs-l10n/blob/master/site/ja/tutorials/load_data/text.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png">GitHub でソースを表示</a> </td>
<td> <a href="https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/ja/tutorials/load_data/text.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png">ノートブックをダウンロード</a> </td>
</table>
このチュートリアルでは、テキストを読み込んで前処理する 2 つの方法を紹介します。
- まず、Keras ユーティリティとレイヤーを使用します。 TensorFlow を初めて使用する場合は、これらから始める必要があります。
- このチュートリアルでは、`tf.data.TextLineDataset` を使ってテキストファイルからサンプルを読み込む方法を例示します。`TextLineDataset` は、テキストファイルからデータセットを作成するために設計されています。この中では、元のテキストファイルの一行一行がサンプルです。これは、(たとえば、詩やエラーログのような) 基本的に行ベースのテキストデータを扱うのに便利でしょう。
```
# Be sure you're using the stable versions of both tf and tf-text, for binary compatibility.
!pip uninstall -y tensorflow tf-nightly keras
!pip install -q -U tf-nightly
!pip install -q -U tensorflow-text-nightly
import collections
import pathlib
import re
import string
import tensorflow as tf
from tensorflow.keras import layers
from tensorflow.keras import losses
from tensorflow.keras import preprocessing
from tensorflow.keras import utils
from tensorflow.keras.layers.experimental.preprocessing import TextVectorization
import tensorflow_datasets as tfds
import tensorflow_text as tf_text
```
## 例 1: StackOverflow の質問のタグを予測する
最初の例として、StackOverflow からプログラミングの質問のデータセットをダウンロードします。それぞれの質問 (「ディクショナリを値で並べ替えるにはどうすればよいですか?」) は、1 つのタグ (`Python`、`CSharp`、`JavaScript`、または`Java`) でラベルされています。このタスクでは、質問のタグを予測するモデルを開発します。これは、マルチクラス分類の例です。マルチクラス分類は、重要で広く適用できる機械学習の問題です。
### データセットをダウンロードして調査する
次に、データセットをダウンロードして、ディレクトリ構造を調べます。
```
data_url = 'https://storage.googleapis.com/download.tensorflow.org/data/stack_overflow_16k.tar.gz'
dataset_dir = utils.get_file(
origin=data_url,
untar=True,
cache_dir='stack_overflow',
cache_subdir='')
dataset_dir = pathlib.Path(dataset_dir).parent
list(dataset_dir.iterdir())
train_dir = dataset_dir/'train'
list(train_dir.iterdir())
```
`train/csharp`、`train/java`, `train/python` および `train/javascript` ディレクトリには、多くのテキストファイルが含まれています。それぞれが Stack Overflow の質問です。ファイルを出力してデータを調べます。
```
sample_file = train_dir/'python/1755.txt'
with open(sample_file) as f:
print(f.read())
```
### データセットを読み込む
次に、データをディスクから読み込み、トレーニングに適した形式に準備します。これを行うには、[text_dataset_from_directory](https://www.tensorflow.org/api_docs/python/tf/keras/preprocessing/text_dataset_from_directory) ユーティリティを使用して、ラベル付きの `tf.data.Dataset` を作成します。これは、入力パイプラインを構築するための強力なツールのコレクションです。
`preprocessing.text_dataset_from_directory` は、次のようなディレクトリ構造を想定しています。
```
train/
...csharp/
......1.txt
......2.txt
...java/
......1.txt
......2.txt
...javascript/
......1.txt
......2.txt
...python/
......1.txt
......2.txt
```
機械学習実験を実行するときは、データセットを[トレーニング](https://developers.google.com/machine-learning/glossary#training_set)、[検証](https://developers.google.com/machine-learning/glossary#validation_set)、および、[テスト](https://developers.google.com/machine-learning/glossary#test-set)の 3 つに分割することをお勧めします。Stack Overflow データセットはすでにトレーニングとテストに分割されていますが、検証セットがありません。以下の `validation_split` 引数を使用して、トレーニングデータの 80:20 分割を使用して検証セットを作成します。
```
batch_size = 32
seed = 42
raw_train_ds = preprocessing.text_dataset_from_directory(
train_dir,
batch_size=batch_size,
validation_split=0.2,
subset='training',
seed=seed)
```
上記のように、トレーニングフォルダには 8,000 の例があり、そのうち 80% (6,400 件) をトレーニングに使用します。この後で見ていきますが、`tf.data.Dataset` を直接 `model.fit` に渡すことでモデルをトレーニングできます。まず、データセットを繰り返し処理し、いくつかの例を出力します。
注意: 分類問題の難易度を上げるために、データセットの作成者は、プログラミングの質問で、*Python*、*CSharp*、*JavaScript*、*Java* という単語を *blank* に置き換えました。
```
for text_batch, label_batch in raw_train_ds.take(1):
for i in range(10):
print("Question: ", text_batch.numpy()[i])
print("Label:", label_batch.numpy()[i])
```
ラベルは、`0`、`1`、`2` または `3` です。これらのどれがどの文字列ラベルに対応するかを確認するには、データセットの `class_names` プロパティを確認します。
```
for i, label in enumerate(raw_train_ds.class_names):
print("Label", i, "corresponds to", label)
```
次に、検証およびテスト用データセットを作成します。トレーニング用セットの残りの 1,600 件のレビューを検証に使用します。
注意: `validation_split` および `subset` 引数を使用する場合は、必ずランダムシードを指定するか、`shuffle=False`を渡して、検証とトレーニング分割に重複がないようにします。
```
raw_val_ds = preprocessing.text_dataset_from_directory(
train_dir,
batch_size=batch_size,
validation_split=0.2,
subset='validation',
seed=seed)
test_dir = dataset_dir/'test'
raw_test_ds = preprocessing.text_dataset_from_directory(
test_dir, batch_size=batch_size)
```
### トレーニング用データセットを準備する
注意: このセクションで使用される前処理 API は、TensorFlow 2.3 では実験的なものであり、変更される可能性があります。
次に、`preprocessing.TextVectorization` レイヤーを使用して、データを標準化、トークン化、およびベクトル化します。
- 標準化とは、テキストを前処理することを指します。通常、句読点や HTML 要素を削除して、データセットを簡素化します。
- トークン化とは、文字列をトークンに分割することです(たとえば、空白で分割することにより、文を個々の単語に分割します)。
- ベクトル化とは、トークンを数値に変換して、ニューラルネットワークに入力できるようにすることです。
これらのタスクはすべて、このレイヤーで実行できます。これらの詳細については、[API doc](https://www.tensorflow.org/api_docs/python/tf/keras/layers/experimental/preprocessing/TextVectorization) をご覧ください。
- デフォルトの標準化では、テキストが小文字に変換され、句読点が削除されます。
- デフォルトのトークナイザーは空白で分割されます。
- デフォルトのベクトル化モードは `int` です。これは整数インデックスを出力します(トークンごとに1つ)。このモードは、語順を考慮したモデルを構築するために使用できます。`binary` などの他のモードを使用して、bag-of-word モデルを構築することもできます。
これらについてさらに学ぶために、2 つのモードを構築します。まず、`binary` モデルを使用して、bag-of-words モデルを構築します。次に、1D ConvNet で `int` モードを使用します。
```
VOCAB_SIZE = 10000
binary_vectorize_layer = TextVectorization(
max_tokens=VOCAB_SIZE,
output_mode='binary')
```
`int` の場合、最大語彙サイズに加えて、明示的な最大シーケンス長を設定する必要があります。これにより、レイヤーはシーケンスを正確に sequence_length 値にパディングまたは切り捨てます。
```
MAX_SEQUENCE_LENGTH = 250
int_vectorize_layer = TextVectorization(
max_tokens=VOCAB_SIZE,
output_mode='int',
output_sequence_length=MAX_SEQUENCE_LENGTH)
```
次に、`adapt` を呼び出して、前処理レイヤーの状態をデータセットに適合させます。これにより、モデルは文字列から整数へのインデックスを作成します。
注意: Adapt を呼び出すときは、トレーニング用データのみを使用することが重要です (テスト用セットを使用すると情報が漏洩します)。
```
# Make a text-only dataset (without labels), then call adapt
train_text = raw_train_ds.map(lambda text, labels: text)
binary_vectorize_layer.adapt(train_text)
int_vectorize_layer.adapt(train_text)
```
これらのレイヤーを使用してデータを前処理した結果を確認してください。
```
def binary_vectorize_text(text, label):
text = tf.expand_dims(text, -1)
return binary_vectorize_layer(text), label
def int_vectorize_text(text, label):
text = tf.expand_dims(text, -1)
return int_vectorize_layer(text), label
# Retrieve a batch (of 32 reviews and labels) from the dataset
text_batch, label_batch = next(iter(raw_train_ds))
first_question, first_label = text_batch[0], label_batch[0]
print("Question", first_question)
print("Label", first_label)
print("'binary' vectorized question:",
binary_vectorize_text(first_question, first_label)[0])
print("'int' vectorized question:",
int_vectorize_text(first_question, first_label)[0])
```
上記のように、`binary` モードは、入力に少なくとも 1 回存在するトークンを示す配列を返しますが、`int` モードは、各トークンを整数に置き換えて、順序を維持します。レイヤーで `.get_vocabulary()` を呼び出すことにより、各整数が対応するトークン (文字列) を検索できます
```
print("1289 ---> ", int_vectorize_layer.get_vocabulary()[1289])
print("313 ---> ", int_vectorize_layer.get_vocabulary()[313])
print("Vocabulary size: {}".format(len(int_vectorize_layer.get_vocabulary())))
```
モデルをトレーニングする準備がほぼ整いました。最後の前処理ステップとして、トレーニング、検証、およびデータセットのテストのために前に作成した `TextVectorization` レイヤーを適用します。
```
binary_train_ds = raw_train_ds.map(binary_vectorize_text)
binary_val_ds = raw_val_ds.map(binary_vectorize_text)
binary_test_ds = raw_test_ds.map(binary_vectorize_text)
int_train_ds = raw_train_ds.map(int_vectorize_text)
int_val_ds = raw_val_ds.map(int_vectorize_text)
int_test_ds = raw_test_ds.map(int_vectorize_text)
```
### パフォーマンスのためにデータセットを構成する
以下は、データを読み込むときに I/O がブロックされないようにするために使用する必要がある 2 つの重要な方法です。
`.cache()` はデータをディスクから読み込んだ後、データをメモリに保持します。これにより、モデルのトレーニング中にデータセットがボトルネックになることを回避できます。データセットが大きすぎてメモリに収まらない場合は、この方法を使用して、パフォーマンスの高いオンディスクキャッシュを作成することもできます。これは、多くの小さなファイルを読み込むより効率的です。
`.prefetch()` はトレーニング中にデータの前処理とモデルの実行をオーバーラップさせます。
以上の 2 つの方法とデータをディスクにキャッシュする方法についての詳細は、[データパフォーマンスガイド](https://www.tensorflow.org/guide/data_performance)を参照してください。
```
AUTOTUNE = tf.data.AUTOTUNE
def configure_dataset(dataset):
return dataset.cache().prefetch(buffer_size=AUTOTUNE)
binary_train_ds = configure_dataset(binary_train_ds)
binary_val_ds = configure_dataset(binary_val_ds)
binary_test_ds = configure_dataset(binary_test_ds)
int_train_ds = configure_dataset(int_train_ds)
int_val_ds = configure_dataset(int_val_ds)
int_test_ds = configure_dataset(int_test_ds)
```
### モデルをトレーニングする
ニューラルネットワークを作成します。`binary` のベクトル化されたデータの場合、単純な bag-of-words 線形モデルをトレーニングします。
```
binary_model = tf.keras.Sequential([layers.Dense(4)])
binary_model.compile(
loss=losses.SparseCategoricalCrossentropy(from_logits=True),
optimizer='adam',
metrics=['accuracy'])
history = binary_model.fit(
binary_train_ds, validation_data=binary_val_ds, epochs=10)
```
次に、`int` ベクトル化レイヤーを使用して、1D ConvNet を構築します。
```
def create_model(vocab_size, num_labels):
model = tf.keras.Sequential([
layers.Embedding(vocab_size, 64, mask_zero=True),
layers.Conv1D(64, 5, padding="valid", activation="relu", strides=2),
layers.GlobalMaxPooling1D(),
layers.Dense(num_labels)
])
return model
# vocab_size is VOCAB_SIZE + 1 since 0 is used additionally for padding.
int_model = create_model(vocab_size=VOCAB_SIZE + 1, num_labels=4)
int_model.compile(
loss=losses.SparseCategoricalCrossentropy(from_logits=True),
optimizer='adam',
metrics=['accuracy'])
history = int_model.fit(int_train_ds, validation_data=int_val_ds, epochs=5)
```
2 つのモデルを比較します。
```
print("Linear model on binary vectorized data:")
print(binary_model.summary())
print("ConvNet model on int vectorized data:")
print(int_model.summary())
```
テストデータで両方のモデルを評価します。
```
binary_loss, binary_accuracy = binary_model.evaluate(binary_test_ds)
int_loss, int_accuracy = int_model.evaluate(int_test_ds)
print("Binary model accuracy: {:2.2%}".format(binary_accuracy))
print("Int model accuracy: {:2.2%}".format(int_accuracy))
```
注意: このサンプルデータセットは、かなり単純な分類問題を表しています。より複雑なデータセットと問題は、前処理戦略とモデルアーキテクチャに微妙ながら重要な違いをもたらします。さまざまなアプローチを比較するために、さまざまなハイパーパラメータとエポックを試してみてください。
### モデルをエクスポートする
上記のコードでは、モデルにテキストをフィードする前に、`TextVectorization` レイヤーをデータセットに適用しました。モデルで生の文字列を処理できるようにする場合 (たとえば、展開を簡素化するため)、モデル内に `TextVectorization` レイヤーを含めることができます。これを行うには、トレーニングしたばかりの重みを使用して新しいモデルを作成します。
```
export_model = tf.keras.Sequential(
[binary_vectorize_layer, binary_model,
layers.Activation('sigmoid')])
export_model.compile(
loss=losses.SparseCategoricalCrossentropy(from_logits=False),
optimizer='adam',
metrics=['accuracy'])
# Test it with `raw_test_ds`, which yields raw strings
loss, accuracy = export_model.evaluate(raw_test_ds)
print("Accuracy: {:2.2%}".format(binary_accuracy))
```
これで、モデルは生の文字列を入力として受け取り、`model.predict` を使用して各ラベルのスコアを予測できます。最大スコアのラベルを見つける関数を定義します。
```
def get_string_labels(predicted_scores_batch):
predicted_int_labels = tf.argmax(predicted_scores_batch, axis=1)
predicted_labels = tf.gather(raw_train_ds.class_names, predicted_int_labels)
return predicted_labels
```
### 新しいデータで推論を実行する
```
inputs = [
"how do I extract keys from a dict into a list?", # python
"debug public static void main(string[] args) {...}", # java
]
predicted_scores = export_model.predict(inputs)
predicted_labels = get_string_labels(predicted_scores)
for input, label in zip(inputs, predicted_labels):
print("Question: ", input)
print("Predicted label: ", label.numpy())
```
モデル内にテキスト前処理ロジックを含めると、モデルを本番環境にエクスポートして展開を簡素化し、[トレーニング/テストスキュー](https://developers.google.com/machine-learning/guides/rules-of-ml#training-serving_skew)の可能性を減らすことができます。
`TextVectorization` レイヤーを適用する場所を選択する際に性能の違いに留意する必要があります。モデルの外部で使用すると、GPU でトレーニングするときに非同期 CPU 処理とデータのバッファリングを行うことができます。したがって、GPU でモデルをトレーニングしている場合は、モデルの開発中に最高のパフォーマンスを得るためにこのオプションを使用し、デプロイの準備ができたらモデル内に TextVectorization レイヤーを含めるように切り替えることをお勧めします。
モデルの保存の詳細については、この[チュートリアル](https://www.tensorflow.org/tutorials/keras/save_and_load)にアクセスしてください。
## テキストをデータセットに読み込む
以下に、`tf.data.TextLineDataset` を使用してテキストファイルから例を読み込み、`tf.text` を使用してデータを前処理する例を示します。この例では、ホーマーのイーリアスの 3 つの異なる英語翻訳を使用し、与えられた 1 行のテキストから翻訳者を識別するようにモデルをトレーニングします。
### データセットをダウンロードして調査する
3 つのテキストの翻訳者は次のとおりです。
- [ウィリアム・クーパー](https://en.wikipedia.org/wiki/William_Cowper) — [テキスト](https://storage.googleapis.com/download.tensorflow.org/data/illiad/cowper.txt)
- [エドワード、ダービー伯爵](https://en.wikipedia.org/wiki/Edward_Smith-Stanley,_14th_Earl_of_Derby) — [テキスト](https://storage.googleapis.com/download.tensorflow.org/data/illiad/derby.txt)
- [サミュエル・バトラー](https://en.wikipedia.org/wiki/Samuel_Butler_%28novelist%29) — [テキスト](https://storage.googleapis.com/download.tensorflow.org/data/illiad/butler.txt)
このチュートリアルで使われているテキストファイルは、ヘッダ、フッタ、行番号、章のタイトルの削除など、いくつかの典型的な前処理が行われています。前処理後のファイルをローカルにダウンロードします。
```
DIRECTORY_URL = 'https://storage.googleapis.com/download.tensorflow.org/data/illiad/'
FILE_NAMES = ['cowper.txt', 'derby.txt', 'butler.txt']
for name in FILE_NAMES:
text_dir = utils.get_file(name, origin=DIRECTORY_URL + name)
parent_dir = pathlib.Path(text_dir).parent
list(parent_dir.iterdir())
```
### データセットを読み込む
`TextLineDataset` を使用します。これは、テキストファイルから `tf.data.Dataset` を作成するように設計されています。テキストファイルでは各例は、元のファイルのテキスト行ですが、`text_dataset_from_directory` は、ファイルのすべての内容を 1 つの例として扱います。`TextLineDataset` は、主に行があるテキストデータ(詩やエラーログなど)に役立ちます。
これらのファイルを繰り返し処理し、各ファイルを独自のデータセットに読み込みます。各例には個別にラベルを付ける必要があるため、`tf.data.Dataset.map` を使用して、それぞれにラベラー関数を適用します。これにより、データセット内のすべての例が繰り返され、 (`example, label`) ペアが返されます。
```
def labeler(example, index):
return example, tf.cast(index, tf.int64)
labeled_data_sets = []
for i, file_name in enumerate(FILE_NAMES):
lines_dataset = tf.data.TextLineDataset(str(parent_dir/file_name))
labeled_dataset = lines_dataset.map(lambda ex: labeler(ex, i))
labeled_data_sets.append(labeled_dataset)
```
次に、これらのラベル付きデータセットを 1 つのデータセットに結合し、シャッフルします。
```
BUFFER_SIZE = 50000
BATCH_SIZE = 64
TAKE_SIZE = 5000
all_labeled_data = labeled_data_sets[0]
for labeled_dataset in labeled_data_sets[1:]:
all_labeled_data = all_labeled_data.concatenate(labeled_dataset)
all_labeled_data = all_labeled_data.shuffle(
BUFFER_SIZE, reshuffle_each_iteration=False)
```
前述の手順でいくつかの例を出力します。データセットはまだバッチ処理されていないため、`all_labeled_data` の各エントリは 1 つのデータポイントに対応します。
```
for text, label in all_labeled_data.take(10):
print("Sentence: ", text.numpy())
print("Label:", label.numpy())
```
### トレーニング用データセットを準備する
Keras `TextVectorization` レイヤーを使用してテキストデータセットを前処理する代わりに、[`tf.text` API](https://www.tensorflow.org/tutorials/tensorflow_text/intro) を使用してデータを標準化およびトークン化し、語彙を作成し、`StaticVocabularyTable` を使用してトークンを整数にマッピングし、モデルにフィードします。
tf.text はさまざまなトークナイザーを提供しますが、`UnicodeScriptTokenizer` を使用してデータセットをトークン化します。テキストを小文字に変換してトークン化する関数を定義します。`tf.data.Dataset.map` を使用して、トークン化をデータセットに適用します。
```
tokenizer = tf_text.UnicodeScriptTokenizer()
def tokenize(text, unused_label):
lower_case = tf_text.case_fold_utf8(text)
return tokenizer.tokenize(lower_case)
tokenized_ds = all_labeled_data.map(tokenize)
```
データセットを反復処理して、トークン化されたいくつかの例を出力できます。
```
for text_batch in tokenized_ds.take(5):
print("Tokens: ", text_batch.numpy())
```
次に、トークンを頻度で並べ替え、上位の `VOCAB_SIZE` トークンを保持することにより、語彙を構築します。
```
tokenized_ds = configure_dataset(tokenized_ds)
vocab_dict = collections.defaultdict(lambda: 0)
for toks in tokenized_ds.as_numpy_iterator():
for tok in toks:
vocab_dict[tok] += 1
vocab = sorted(vocab_dict.items(), key=lambda x: x[1], reverse=True)
vocab = [token for token, count in vocab]
vocab = vocab[:VOCAB_SIZE]
vocab_size = len(vocab)
print("Vocab size: ", vocab_size)
print("First five vocab entries:", vocab[:5])
```
トークンを整数に変換するには、`vocab` セットを使用して、`StaticVocabularyTable`を作成します。トークンを [`2`, `vocab_size + 2`] の範囲の整数にマップします。`TextVectorization` レイヤーと同様に、`0` はパディングを示すために予約されており、`1` は語彙外 (OOV) トークンを示すために予約されています。
```
keys = vocab
values = range(2, len(vocab) + 2) # reserve 0 for padding, 1 for OOV
init = tf.lookup.KeyValueTensorInitializer(
keys, values, key_dtype=tf.string, value_dtype=tf.int64)
num_oov_buckets = 1
vocab_table = tf.lookup.StaticVocabularyTable(init, num_oov_buckets)
```
最後に、トークナイザーとルックアップテーブルを使用して、データセットを標準化、トークン化、およびベクトル化する関数を定義します。
```
def preprocess_text(text, label):
standardized = tf_text.case_fold_utf8(text)
tokenized = tokenizer.tokenize(standardized)
vectorized = vocab_table.lookup(tokenized)
return vectorized, label
```
1 つの例でこれを試して、出力を確認します。
```
example_text, example_label = next(iter(all_labeled_data))
print("Sentence: ", example_text.numpy())
vectorized_text, example_label = preprocess_text(example_text, example_label)
print("Vectorized sentence: ", vectorized_text.numpy())
```
次に、`tf.data.Dataset.map` を使用して、データセットに対して前処理関数を実行します。
```
all_encoded_data = all_labeled_data.map(preprocess_text)
```
### データセットをトレーニングとテストに分割する
Keras `TextVectorization` レイヤーでも、ベクトル化されたデータをバッチ処理してパディングします。バッチ内のサンプルは同じサイズと形状である必要があるため、パディングが必要です。これらのデータセットのサンプルはすべて同じサイズではありません。テキストの各行には、異なる数の単語があります。`tf.data.Dataset` は、データセットの分割と埋め込みバッチ処理をサポートしています
```
train_data = all_encoded_data.skip(VALIDATION_SIZE).shuffle(BUFFER_SIZE)
validation_data = all_encoded_data.take(VALIDATION_SIZE)
train_data = train_data.padded_batch(BATCH_SIZE)
validation_data = validation_data.padded_batch(BATCH_SIZE)
```
`validation_data` および `train_data` は(`example, label`) ペアのコレクションではなく、バッチのコレクションです。各バッチは、配列として表される (*多くの例*、*多くのラベル*) のペアです。以下に示します。
```
sample_text, sample_labels = next(iter(validation_data))
print("Text batch shape: ", sample_text.shape)
print("Label batch shape: ", sample_labels.shape)
print("First text example: ", sample_text[0])
print("First label example: ", sample_labels[0])
```
パディングに 0 を使用し、語彙外 (OOV) トークンに 1 を使用するため、語彙のサイズが 2 つ増えました。
```
vocab_size += 2
```
以前と同じように、パフォーマンスを向上させるためにデータセットを構成します。
```
train_data = configure_dataset(train_data)
validation_data = configure_dataset(validation_data)
```
### モデルをトレーニングする
以前と同じように、このデータセットでモデルをトレーニングできます。
```
model = create_model(vocab_size=vocab_size, num_labels=3)
model.compile(
optimizer='adam',
loss=losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=['accuracy'])
history = model.fit(train_data, validation_data=validation_data, epochs=3)
loss, accuracy = model.evaluate(validation_data)
print("Loss: ", loss)
print("Accuracy: {:2.2%}".format(accuracy))
```
### モデルをエクスポートする
モデルが生の文字列を入力として受け取ることができるようにするには、カスタム前処理関数と同じ手順を実行する `TextVectorization` レイヤーを作成します。すでに語彙をトレーニングしているので、新しい語彙をトレーニングする `adapt` の代わりに、`set_vocaublary` を使用できます。
```
preprocess_layer = TextVectorization(
max_tokens=vocab_size,
standardize=tf_text.case_fold_utf8,
split=tokenizer.tokenize,
output_mode='int',
output_sequence_length=MAX_SEQUENCE_LENGTH)
preprocess_layer.set_vocabulary(vocab)
export_model = tf.keras.Sequential(
[preprocess_layer, model,
layers.Activation('sigmoid')])
export_model.compile(
loss=losses.SparseCategoricalCrossentropy(from_logits=False),
optimizer='adam',
metrics=['accuracy'])
# Create a test dataset of raw strings
test_ds = all_labeled_data.take(VALIDATION_SIZE).batch(BATCH_SIZE)
test_ds = configure_dataset(test_ds)
loss, accuracy = export_model.evaluate(test_ds)
print("Loss: ", loss)
print("Accuracy: {:2.2%}".format(accuracy))
```
エンコードされた検証セットのモデルと生の検証セットのエクスポートされたモデルの損失と正確度は、予想どおり同じです。
### 新しいデータで推論を実行する
```
inputs = [
"Join'd to th' Ionians with their flowing robes,", # Label: 1
"the allies, and his armour flashed about him so that he seemed to all", # Label: 2
"And with loud clangor of his arms he fell.", # Label: 0
]
predicted_scores = export_model.predict(inputs)
predicted_labels = tf.argmax(predicted_scores, axis=1)
for input, label in zip(inputs, predicted_labels):
print("Question: ", input)
print("Predicted label: ", label.numpy())
```
## TensorFlow Datasets (TFDS) を使用してより多くのデータセットをダウンロードする
[TensorFlow Datasets](https://www.tensorflow.org/datasets/catalog/overview) からさらに多くのデータセットをダウンロードできます。例として、[IMDB Large Movie Review データセット](https://www.tensorflow.org/datasets/catalog/imdb_reviews)をダウンロードし、それを使用して感情分類のモデルをトレーニングします。
```
train_ds = tfds.load(
'imdb_reviews',
split='train[:80%]',
batch_size=BATCH_SIZE,
shuffle_files=True,
as_supervised=True)
val_ds = tfds.load(
'imdb_reviews',
split='train[80%:]',
batch_size=BATCH_SIZE,
shuffle_files=True,
as_supervised=True)
```
いくつかの例を出力します。
```
for review_batch, label_batch in val_ds.take(1):
for i in range(5):
print("Review: ", review_batch[i].numpy())
print("Label: ", label_batch[i].numpy())
```
これで、以前と同じようにデータを前処理してモデルをトレーニングできます。
注意: これはバイナリ分類の問題であるため、モデルには `losses.SparseCategoricalCrossentropy` の代わりに `losses.BinaryCrossentropy` を使用します。
### トレーニング用データセットを準備する
```
vectorize_layer = TextVectorization(
max_tokens=VOCAB_SIZE,
output_mode='int',
output_sequence_length=MAX_SEQUENCE_LENGTH)
# Make a text-only dataset (without labels), then call adapt
train_text = train_ds.map(lambda text, labels: text)
vectorize_layer.adapt(train_text)
def vectorize_text(text, label):
text = tf.expand_dims(text, -1)
return vectorize_layer(text), label
train_ds = train_ds.map(vectorize_text)
val_ds = val_ds.map(vectorize_text)
# Configure datasets for performance as before
train_ds = configure_dataset(train_ds)
val_ds = configure_dataset(val_ds)
```
### モデルをトレーニングする
```
model = create_model(vocab_size=VOCAB_SIZE + 1, num_labels=1)
model.summary()
model.compile(
loss=losses.BinaryCrossentropy(from_logits=True),
optimizer='adam',
metrics=['accuracy'])
history = model.fit(train_ds, validation_data=val_ds, epochs=3)
loss, accuracy = model.evaluate(val_ds)
print("Loss: ", loss)
print("Accuracy: {:2.2%}".format(accuracy))
```
### モデルをエクスポートする
```
export_model = tf.keras.Sequential(
[vectorize_layer, model,
layers.Activation('sigmoid')])
export_model.compile(
loss=losses.SparseCategoricalCrossentropy(from_logits=False),
optimizer='adam',
metrics=['accuracy'])
# 0 --> negative review
# 1 --> positive review
inputs = [
"This is a fantastic movie.",
"This is a bad movie.",
"This movie was so bad that it was good.",
"I will never say yes to watching this movie.",
]
predicted_scores = export_model.predict(inputs)
predicted_labels = [int(round(x[0])) for x in predicted_scores]
for input, label in zip(inputs, predicted_labels):
print("Question: ", input)
print("Predicted label: ", label)
```
## まとめ
このチュートリアルでは、テキストを読み込んで前処理するいくつかの方法を示しました。次のステップとして、Web サイトで他のチュートリアルをご覧ください。また、[TensorFlow Datasets](https://www.tensorflow.org/datasets/catalog/overview) から新しいデータセットをダウンロードできます。
|
github_jupyter
|
```
# 使下面的代码支持python2和python3
from __future__ import division, print_function, unicode_literals
# 查看python的版本是否为3.5及以上
import sys
assert sys.version_info >= (3, 5)
# 查看sklearn的版本是否为0.20及以上
import sklearn
assert sklearn.__version__ >= "0.20"
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import os
# 在每一次的运行后获得的结果与这个notebook的结果相同
np.random.seed(42)
# 让matplotlib的图效果更好
%matplotlib inline
import matplotlib as mpl
mpl.rc('axes', labelsize=14)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
# 设置保存图片的途径
PROJECT_ROOT_DIR = "."
IMAGE_PATH = os.path.join(PROJECT_ROOT_DIR, "images")
os.makedirs(IMAGE_PATH, exist_ok=True)
def save_fig(fig_id, tight_layout=True):
'''
运行即可保存自动图片
:param fig_id: 图片名称
'''
path = os.path.join(PROJECT_ROOT_DIR, "images", fig_id + ".png")
print("Saving figure", fig_id)
if tight_layout:
plt.tight_layout()
plt.savefig(path, format='png', dpi=300)
# 忽略掉没用的警告 (Scipy issue #5998)
import warnings
warnings.filterwarnings(action="ignore", category=FutureWarning, module='sklearn', lineno=196)
# 读取数据集
df = pd.read_excel('Test_2.xlsx')
df.head()
# 查看数据集是否有空值,看需不需要插值
df.info()
'''
# 插值
df.fillna(0, inplace=True)
# 或者是参考之前在多项式回归里的插值方式
'''
# 将真实的分类标签与特征分开
data = df.drop('TRUE VALUE', axis=1)
labels = df['TRUE VALUE'].copy()
np.unique(labels)
labels
# 获取数据的数量和特征的数量
n_samples, n_features = data.shape
# 获取分类标签的数量
n_labels = len(np.unique(labels))
np.unique(labels)
labels.value_counts()
```
# KMeans算法聚类
```
from sklearn import metrics
def get_marks(estimator, data, name=None, kmeans=None, af=None):
"""
获取评分,有五种需要知道数据集的实际分类信息,有三种不需要,参考readme.txt
对于Kmeans来说,一般用轮廓系数和inertia即可
:param estimator: 模型
:param name: 初始方法
:param data: 特征数据集
"""
estimator.fit(data)
print(20 * '*', name, 20 * '*')
if kmeans:
print("Mean Inertia Score: ", estimator.inertia_)
elif af:
cluster_centers_indices = estimator.cluster_centers_indices_
print("The estimated number of clusters: ", len(cluster_centers_indices))
print("Homogeneity Score: ", metrics.homogeneity_score(labels, estimator.labels_))
print("Completeness Score: ", metrics.completeness_score(labels, estimator.labels_))
print("V Measure Score: ", metrics.v_measure_score(labels, estimator.labels_))
print("Adjusted Rand Score: ", metrics.adjusted_rand_score(labels, estimator.labels_))
print("Adjusted Mutual Info Score: ", metrics.adjusted_mutual_info_score(labels, estimator.labels_))
print("Calinski Harabasz Score: ", metrics.calinski_harabasz_score(data, estimator.labels_))
print("Silhouette Score: ", metrics.silhouette_score(data, estimator.labels_))
from sklearn.cluster import KMeans
# 使用k-means进行聚类,设置簇=2,设置不同的初始化方式('k-means++'和'random')
km1 = KMeans(init='k-means++', n_clusters=n_labels-1, n_init=10, random_state=42)
km2 = KMeans(init='random', n_clusters=n_labels-1, n_init=10, random_state=42)
print("n_labels: %d \t n_samples: %d \t n_features: %d" % (n_labels, n_samples, n_features))
get_marks(km1, data, name="k-means++", kmeans=True)
get_marks(km2, data, name="random", kmeans=True)
# 聚类后每个数据的类别
km1.labels_
# 类别的类型
np.unique(km1.labels_)
# 将聚类的结果写入原始表格中
df['km_clustering_label'] = km1.labels_
# 以csv形式导出原始表格
#df.to_csv('result.csv')
# 区别于data,df是原始数据集
df.head()
from sklearn.model_selection import GridSearchCV
# 使用GridSearchCV自动寻找最优参数,kmeans在这里是作为分类模型使用
params = {'init':('k-means++', 'random'), 'n_clusters':[2, 3, 4, 5, 6], 'n_init':[5, 10, 15]}
cluster = KMeans(random_state=42)
# 使用调整的兰德系数(adjusted_rand_score)作为评分,具体可参考readme.txt
km_best_model = GridSearchCV(cluster, params, cv=3, scoring='adjusted_rand_score',
verbose=1, n_jobs=-1)
# 由于选用的是外部评价指标,因此得有原数据集的真实分类信息
km_best_model.fit(data, labels)
# 最优模型的参数
km_best_model.best_params_
# 最优模型的评分
km_best_model.best_score_
# 获得的最优模型
km3 = km_best_model.best_estimator_
km3
# 获取最优模型的8种评分,具体含义参考readme.txt
get_marks(km3, data, name="k-means++", kmeans=True)
from sklearn.metrics import silhouette_score
from sklearn.metrics import calinski_harabasz_score
from matplotlib import pyplot as plt
def plot_scores(init, max_k, data, labels):
'''画出kmeans不同初始化方法的三种评分图
:param init: 初始化方法,有'k-means++'和'random'两种
:param max_k: 最大的簇中心数目
:param data: 特征的数据集
:param labels: 真实标签的数据集
'''
i = []
inertia_scores = []
y_silhouette_scores = []
y_calinski_harabaz_scores = []
for k in range(2, max_k):
kmeans_model = KMeans(n_clusters=k, random_state=1, init=init, n_init=10)
pred = kmeans_model.fit_predict(data)
i.append(k)
inertia_scores.append(kmeans_model.inertia_)
y_silhouette_scores.append(silhouette_score(data, pred))
y_calinski_harabaz_scores.append(calinski_harabasz_score(data, pred))
new = [inertia_scores, y_silhouette_scores, y_calinski_harabaz_scores]
for j in range(len(new)):
plt.figure(j+1)
plt.plot(i, new[j], 'bo-')
plt.xlabel('n_clusters')
if j == 0:
name = 'inertia'
elif j == 1:
name = 'silhouette'
else:
name = 'calinski_harabasz'
plt.ylabel('{}_scores'.format(name))
plt.title('{}_scores with {} init'.format(name, init))
save_fig('{} with {}'.format(name, init))
plot_scores('k-means++', 18, data, labels)
plot_scores('random', 10, data, labels)
from sklearn.metrics import silhouette_samples, silhouette_score
from matplotlib.ticker import FixedLocator, FixedFormatter
def plot_silhouette_diagram(clusterer, X, show_xlabels=True,
show_ylabels=True, show_title=True):
"""
画轮廓图表
:param clusterer: 训练好的聚类模型(这里是能提前设置簇数量的,可以稍微修改代码换成不能提前设置的)
:param X: 只含特征的数据集
:param show_xlabels: 为真,添加横坐标信息
:param show_ylabels: 为真,添加纵坐标信息
:param show_title: 为真,添加图表名
"""
y_pred = clusterer.labels_
silhouette_coefficients = silhouette_samples(X, y_pred)
silhouette_average = silhouette_score(X, y_pred)
padding = len(X) // 30
pos = padding
ticks = []
for i in range(clusterer.n_clusters):
coeffs = silhouette_coefficients[y_pred == i]
coeffs.sort()
color = mpl.cm.Spectral(i / clusterer.n_clusters)
plt.fill_betweenx(np.arange(pos, pos + len(coeffs)), 0, coeffs,
facecolor=color, edgecolor=color, alpha=0.7)
ticks.append(pos + len(coeffs) // 2)
pos += len(coeffs) + padding
plt.axvline(x=silhouette_average, color="red", linestyle="--")
plt.gca().yaxis.set_major_locator(FixedLocator(ticks))
plt.gca().yaxis.set_major_formatter(FixedFormatter(range(clusterer.n_clusters)))
if show_xlabels:
plt.gca().set_xticks([-0.1, 0, 0.2, 0.4, 0.6, 0.8, 1])
plt.xlabel("Silhouette Coefficient")
else:
plt.tick_params(labelbottom=False)
if show_ylabels:
plt.ylabel("Cluster")
if show_title:
plt.title("init:{} n_cluster:{}".format(clusterer.init, clusterer.n_clusters))
plt.figure(figsize=(15, 4))
plt.subplot(121)
plot_silhouette_diagram(km1, data)
plt.subplot(122)
plot_silhouette_diagram(km3, data, show_ylabels=False)
save_fig("silhouette_diagram")
```
# MiniBatch KMeans
```
from sklearn.cluster import MiniBatchKMeans
# 测试KMeans算法运行速度
%timeit KMeans(n_clusters=3).fit(data)
# 测试MiniBatchKMeans算法运行速度
%timeit MiniBatchKMeans(n_clusters=5).fit(data)
from timeit import timeit
times = np.empty((100, 2))
inertias = np.empty((100, 2))
for k in range(1, 101):
kmeans = KMeans(n_clusters=k, random_state=42)
minibatch_kmeans = MiniBatchKMeans(n_clusters=k, random_state=42)
print("\r Training: {}/{}".format(k, 100), end="")
times[k-1, 0] = timeit("kmeans.fit(data)", number=10, globals=globals())
times[k-1, 1] = timeit("minibatch_kmeans.fit(data)", number=10, globals=globals())
inertias[k-1, 0] = kmeans.inertia_
inertias[k-1, 1] = minibatch_kmeans.inertia_
plt.figure(figsize=(10, 4))
plt.subplot(121)
plt.plot(range(1, 101), inertias[:, 0], "r--", label="K-Means")
plt.plot(range(1, 101), inertias[:, 1], "b.-", label="Mini-batch K-Means")
plt.xlabel("$k$", fontsize=16)
plt.ylabel("Inertia", fontsize=14)
plt.legend(fontsize=14)
plt.subplot(122)
plt.plot(range(1, 101), times[:, 0], "r--", label="K-Means")
plt.plot(range(1, 101), times[:, 1], "b.-", label="Mini-batch K-Means")
plt.xlabel("$k$", fontsize=16)
plt.ylabel("Training time (seconds)", fontsize=14)
plt.axis([1, 100, 0, 6])
plt.legend(fontsize=14)
save_fig("minibatch_kmeans_vs_kmeans")
plt.show()
```
# 降维后聚类
```
from sklearn.decomposition import PCA
# 使用普通PCA进行降维,将特征从11维降至3维
pca1 = PCA(n_components=n_labels)
pca1.fit(data)
km4 = KMeans(init=pca1.components_, n_clusters=n_labels, n_init=10)
get_marks(km4, data, name="PCA-based KMeans", kmeans=True)
# 查看训练集的维度,已降至3个维度
len(pca1.components_)
# 使用普通PCA降维,将特征降至2维,作二维平面可视化
pca2 = PCA(n_components=2)
reduced_data = pca2.fit_transform(data)
# 使用k-means进行聚类,设置簇=3,初始化方法为'k-means++'
kmeans1 = KMeans(init="k-means++", n_clusters=3, n_init=3)
kmeans2 = KMeans(init="random", n_clusters=3, n_init=3)
kmeans1.fit(reduced_data)
kmeans2.fit(reduced_data)
# 训练集的特征维度降至2维
len(pca2.components_)
# 2维的特征值(降维后)
reduced_data
# 3个簇中心的坐标
kmeans1.cluster_centers_
from matplotlib.colors import ListedColormap
def plot_data(X, real_tag=None):
"""
画散点图
:param X: 只含特征值的数据集
:param real_tag: 有值,则给含有不同分类的散点上色
"""
try:
if not real_tag:
plt.plot(X[:, 0], X[:, 1], 'k.', markersize=2)
except ValueError:
types = list(np.unique(real_tag))
for i in range(len(types)):
plt.plot(X[:, 0][real_tag==types[i]], X[:, 1][real_tag==types[i]],
'.', label="{}".format(types[i]), markersize=3)
plt.legend()
def plot_centroids(centroids, circle_color='w', cross_color='k'):
"""
画出簇中心
:param centroids: 簇中心坐标
:param circle_color: 圆圈的颜色
:param cross_color: 叉的颜色
"""
plt.scatter(centroids[:, 0], centroids[:, 1],
marker='o', s=30, zorder=10, linewidths=8,
color=circle_color, alpha=0.9)
plt.scatter(centroids[:, 0], centroids[:, 1],
marker='x', s=50, zorder=11, linewidths=50,
color=cross_color, alpha=1)
def plot_centroids_labels(clusterer):
labels = np.unique(clusterer.labels_)
centroids = clusterer.cluster_centers_
for i in range(centroids.shape[0]):
t = str(labels[i])
plt.text(centroids[i, 0]-1, centroids[i, 1]-1, t, fontsize=25,
zorder=10, bbox=dict(boxstyle='round', fc='yellow', alpha=0.5))
def plot_decision_boundaried(clusterer, X, tag=None, resolution=1000,
show_centroids=True, show_xlabels=True,
show_ylabels=True, show_title=True,
show_centroids_labels=False):
"""
画出决策边界,并填色
:param clusterer: 训练好的聚类模型(能提前设置簇中心数量或不能提前设置都可以)
:param X: 只含特征值的数据集
:param tag: 只含真实分类信息的数据集,有值,则给散点上色
:param resolution: 类似图片分辨率,给最小的单位上色
:param show_centroids: 为真,画出簇中心
:param show_centroids_labels: 为真,标注出该簇中心的标签
"""
mins = X.min(axis=0) - 0.1
maxs = X.max(axis=0) + 0.1
xx, yy = np.meshgrid(np.linspace(mins[0], maxs[0], resolution),
np.linspace(mins[1], maxs[1], resolution))
Z = clusterer.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
# 可用color code或者color自定义填充颜色
# custom_cmap = ListedColormap(["#fafab0", "#9898ff", "#a0faa0"])
plt.contourf(xx, yy, Z, extent=(mins[0], maxs[0], mins[1], maxs[1]),
cmap="Pastel2")
plt.contour(xx, yy, Z, extent=(mins[0], maxs[0], mins[1], maxs[1]),
colors='k')
try:
if not tag:
plot_data(X)
except ValueError:
plot_data(X, real_tag=tag)
if show_centroids:
plot_centroids(clusterer.cluster_centers_)
if show_centroids_labels:
plot_centroids_labels(clusterer)
if show_xlabels:
plt.xlabel(r"$x_1$", fontsize=14)
else:
plt.tick_params(labelbottom=False)
if show_ylabels:
plt.ylabel(r"$x_2$", fontsize=14, rotation=0)
else:
plt.tick_params(labelleft=False)
if show_title:
plt.title("init:{} n_cluster:{}".format(clusterer.init, clusterer.n_clusters))
plt.figure(figsize=(15, 4))
plt.subplot(121)
plot_decision_boundaried(kmeans1, reduced_data, tag=labels)
plt.subplot(122)
plot_decision_boundaried(kmeans2, reduced_data, show_centroids_labels=True)
save_fig("real_tag_vs_non")
plt.show()
kmeans3 = KMeans(init="k-means++", n_clusters=3, n_init=3)
kmeans4 = KMeans(init="k-means++", n_clusters=4, n_init=3)
kmeans5 = KMeans(init="k-means++", n_clusters=5, n_init=3)
kmeans6 = KMeans(init="k-means++", n_clusters=6, n_init=3)
kmeans3.fit(reduced_data)
kmeans4.fit(reduced_data)
kmeans5.fit(reduced_data)
kmeans6.fit(reduced_data)
plt.figure(figsize=(15, 8))
plt.subplot(221)
plot_decision_boundaried(kmeans3, reduced_data, show_xlabels=False, show_centroids_labels=True)
plt.subplot(222)
plot_decision_boundaried(kmeans4, reduced_data, show_ylabels=False, show_xlabels=False)
plt.subplot(223)
plot_decision_boundaried(kmeans5, reduced_data, show_centroids_labels=True)
plt.subplot(224)
plot_decision_boundaried(kmeans6, reduced_data, show_ylabels=False)
save_fig("reduced_and_cluster")
plt.show()
```
# AP算法聚类
```
from sklearn.cluster import AffinityPropagation
# 使用AP聚类算法
af = AffinityPropagation(preference=-500, damping=0.8)
af.fit(data)
# 获取簇的坐标
cluster_centers_indices = af.cluster_centers_indices_
cluster_centers_indices
# 获取分类的类别数量
af_labels = af.labels_
np.unique(af_labels)
get_marks(af, data=data, af=True)
# 将AP聚类聚类的结果写入原始表格中
df['ap_clustering_label'] = af.labels_
# 以csv形式导出原始表格
df.to_csv('test2_result.csv')
# 最后两列为两种聚类算法的分类信息
df.head()
from sklearn.model_selection import GridSearchCV
# from sklearn.model_selection import RamdomizedSearchCV
# 使用GridSearchCV自动寻找最优参数,如果时间太久(约4.7min),可以使用随机搜索,这里是用AP做分类的工作
params = {'preference':[-50, -100, -150, -200], 'damping':[0.5, 0.6, 0.7, 0.8, 0.9]}
cluster = AffinityPropagation()
af_best_model = GridSearchCV(cluster, params, cv=5, scoring='adjusted_rand_score', verbose=1, n_jobs=-1)
af_best_model.fit(data, labels)
# 最优模型的参数设置
af_best_model.best_params_
# 最优模型的评分,使用调整的兰德系数(adjusted_rand_score)作为评分
af_best_model.best_score_
# 获取最优模型
af1 = af_best_model.best_estimator_
af1
# 最优模型的评分
get_marks(af1, data=data, af=True)
"""
from sklearn.externals import joblib
# 保存以pkl格式最优模型
joblib.dump(af1, "af1.pkl")
"""
"""
# 从pkl格式中导出最优模型
my_model_loaded = joblib.load("af1.pkl")
"""
"""
my_model_loaded
"""
from sklearn.decomposition import PCA
# 使用普通PCA进行降维,将特征从11维降至3维
pca3 = PCA(n_components=n_labels)
reduced_data = pca3.fit_transform(data)
af2 = AffinityPropagation(preference=-200, damping=0.8)
get_marks(af2, reduced_data, name="PCA-based AF", af=True)
```
# 基于聚类结果的分层抽样
```
# data2是去掉真实分类信息的数据集(含有聚类后的结果)
data2 = df.drop("TRUE VALUE", axis=1)
data2.head()
# 查看使用kmeans聚类后的分类标签值,两类
data2['km_clustering_label'].hist()
from sklearn.model_selection import StratifiedShuffleSplit
# 基于kmeans聚类结果的分层抽样
split = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=42)
for train_index, test_index in split.split(data2, data2["km_clustering_label"]):
strat_train_set = data2.loc[train_index]
strat_test_set = data2.loc[test_index]
def clustering_result_propotions(data):
"""
分层抽样后,训练集或测试集里不同分类标签的数量比
:param data: 训练集或测试集,纯随机取样或分层取样
"""
return data["km_clustering_label"].value_counts() / len(data)
# 经过分层抽样的测试集中,不同分类标签的数量比
clustering_result_propotions(strat_test_set)
# 经过分层抽样的训练集中,不同分类标签的数量比
clustering_result_propotions(strat_train_set)
# 完整的数据集中,不同分类标签的数量比
clustering_result_propotions(data2)
from sklearn.model_selection import train_test_split
# 纯随机取样
random_train_set, random_test_set = train_test_split(data2, test_size=0.2, random_state=42)
# 完整的数据集、分层抽样后的测试集、纯随机抽样后的测试集中,不同分类标签的数量比
compare_props = pd.DataFrame({
"Overall": clustering_result_propotions(data2),
"Stratified": clustering_result_propotions(strat_test_set),
"Random": clustering_result_propotions(random_test_set),
}).sort_index()
# 计算分层抽样和纯随机抽样后的测试集中不同分类标签的数量比,和完整的数据集中不同分类标签的数量比的误差
compare_props["Rand. %error"] = 100 * compare_props["Random"] / compare_props["Overall"] - 100
compare_props["Start. %error"] = 100 * compare_props["Stratified"] / compare_props["Overall"] - 100
compare_props
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import f1_score
def get_classification_marks(model, data, labels, train_index, test_index):
"""
获取分类模型(二元或多元分类器)的评分:F1值
:param data: 只含有特征值的数据集
:param labels: 只含有标签值的数据集
:param train_index: 分层抽样获取的训练集中数据的索引
:param test_index: 分层抽样获取的测试集中数据的索引
:return: F1评分值
"""
m = model(random_state=42)
m.fit(data.loc[train_index], labels.loc[train_index])
test_labels_predict = m.predict(data.loc[test_index])
score = f1_score(labels.loc[test_index], test_labels_predict, average="weighted")
return score
# 用分层抽样后的训练集训练分类模型后的评分值
start_marks = get_classification_marks(LogisticRegression, data, labels, strat_train_set.index, strat_test_set.index)
start_marks
# 用纯随机抽样后的训练集训练分类模型后的评分值
random_marks = get_classification_marks(LogisticRegression, data, labels, random_train_set.index, random_test_set.index)
random_marks
import numpy as np
from sklearn.metrics import f1_score
from sklearn.model_selection import StratifiedKFold
from sklearn.base import clone, BaseEstimator, TransformerMixin
class stratified_cross_val_score(BaseEstimator, TransformerMixin):
"""实现基于分层抽样的k折交叉验证"""
def __init__(self, model, data, labels, random_state=0, cv=5):
"""
:model: 训练的模型(回归或分类)
:data: 只含特征值的完整数据集
:labels: 只含标签值的完整数据集
:random_state: 模型的随机种子值
:cv: 交叉验证的次数
"""
self.model = model
self.data = data
self.labels = labels
self.random_state = random_state
self.cv = cv
self.score = [] # 储存每折测试集的模型评分
self.i = 0
def fit(self, X, y):
"""
:param X: 含有特征值和聚类结果的完整数据集
:param y: 含有聚类结果的完整数据集
:return: 每一折交叉验证的评分
"""
skfolds = StratifiedKFold(n_splits=self.cv, random_state=self.random_state)
for train_index, test_index in skfolds.split(X, y):
# 复制要训练的模型(分类或回归)
clone_model = clone(self.model)
strat_X_train_folds = self.data.loc[train_index]
strat_y_train_folds = self.labels.loc[train_index]
strat_X_test_fold = self.data.loc[test_index]
strat_y_test_fold = self.labels.loc[test_index]
# 训练模型
clone_model.fit(strat_X_train_folds, strat_y_train_folds)
# 预测值(这里是分类模型的分类结果)
test_labels_pred = clone_model.predict(strat_X_test_fold)
# 这里使用的是分类模型用的F1值,如果是回归模型可以换成相应的模型
score_fold = f1_score(labels.loc[test_index], test_labels_pred, average="weighted")
# 避免重复向列表里重复添加值
if self.i < self.cv:
self.score.append(score_fold)
else:
None
self.i += 1
return self.score
def transform(self, X, y=None):
return self
def mean(self):
"""返回交叉验证评分的平均值"""
return np.array(self.score).mean()
def std(self):
"""返回交叉验证评分的标准差"""
return np.array(self.score).std()
from sklearn.linear_model import SGDClassifier
# 分类模型
clf_model = SGDClassifier(max_iter=5, tol=-np.infty, random_state=42)
# 基于分层抽样的交叉验证,data是只含特征值的完整数据集,labels是只含标签值的完整数据集
clf_cross_val = stratified_cross_val_score(clf_model, data, labels, cv=5, random_state=42)
# data2是含有特征值和聚类结果的完整数据集
clf_cross_val_score = clf_cross_val.fit(data2, data2["km_clustering_label"])
# 每折交叉验证的评分
clf_cross_val.score
# 交叉验证评分的平均值
clf_cross_val.mean()
# 交叉验证评分的标准差
clf_cross_val.std()
```
|
github_jupyter
|
```
import pandas as pd
from datetime import datetime, timedelta
import time
import requests
import numpy as np
import json
import urllib
from pandas.io.json import json_normalize
import re
import os.path
import zipfile
from glob import glob
url ="https://api.usaspending.gov/api/v1/awards/?limit=100"
r = requests.get(url, verify=False)
r.raise_for_status()
type(r)
data = r.json()
meta = data['page_metadata']
data = data['results']
df_API_data = pd.io.json.json_normalize(data)
df_API_data.col
base_url = "https://api.usaspending.gov"
endpt_trans = "/api/v2/search/spending_by_award/?limit=10"
params = {
"filters": {
"time_period": [
{
"start_date": "2016-10-01",
"end_date": "2017-09-30"
}
]
}
}
url = base_url + endpt_trans
r = requests.post(url, json=params)
print(r.status_code, r.reason)
r.raise_for_status()
r.headers
r.request.headers
data = r.json()
meta = data['page_metadata']
data = data['results']
df_trans = pd.io.json.json_normalize(data)
currentFY = 2019
n_years_desired = 10
def download_latest_data(currentFY,n_years_desired):
#find latest datestamp on usaspending files
usaspending_base = 'https://files.usaspending.gov/award_data_archive/'
save_path = '../new_data/'
r = requests.get(usaspending_base, allow_redirects=True)
r.raise_for_status()
datestr = re.findall('_(\d{8}).zip',r.content)[0]
for FY in np.arange(currentFY-n_years_desired+1,currentFY+1):
doe_contracts_url = usaspending_base+str(FY)+'_089_Contracts_Full_' + datestr + '.zip'
doe_grants_url = usaspending_base+str(FY)+'_089_Assistance_Full_' + datestr + '.zip'
nsf_grants_url = usaspending_base+str(FY)+'_049_Assistance_Full_' + datestr + '.zip'
doe_sc_url = 'https://science.energy.gov/~/media/_/excel/universities/DOE-SC_Grants_FY'+str(FY)+'.xlsx'
for url in [doe_contracts_url,doe_grants_url,nsf_grants_url,doe_sc_url]:
filename = url.split('/')[-1]
if os.path.exists(save_path+filename): continue
if url == doe_sc_url:
verify='doe_cert.pem'
else:
verify=True
try:
r = requests.get(url, allow_redirects=True,verify=verify)
r.raise_for_status()
except:
print 'could not find', url
continue
# DOE website stupidly returns a 200 HTTP code when displaying 404 page :/
page_not_found_text = 'The page that you have requested was not found.'
if page_not_found_text in r.content:
print 'could not find', url
continue
open(save_path+filename, 'wb+').write(r.content)
zipper = zipfile.ZipFile(save_path+filename,'r')
zipper.extractall(path='../new_data')
print 'Data download complete'
def unzip_all():
for unzip_this in glob('../new_data/*.zip'):
zipper = zipfile.ZipFile(unzip_this,'r')
zipper.extractall(path='../new_data')
print 'Generating DOE Contract data...'
contract_file_list = glob('../new_data/*089_Contracts*.csv')
contract_df_list = []
for contract_file in contract_file_list:
contract_df_list.append(pd.read_csv(contract_file))
fulldata = pd.concat(contract_df_list,ignore_index=True)
print len(fulldata)
sc_awarding_offices = ['CHICAGO SERVICE CENTER (OFFICE OF SCIENCE)',
'OAK RIDGE OFFICE (OFFICE OF SCIENCE)',
'SC CHICAGO SERVICE CENTER',
'SC OAK RIDGE OFFICE']
sc_funding_offices = ['CHICAGO SERVICE CENTER (OFFICE OF SCIENCE)',
'OAK RIDGE OFFICE (OFFICE OF SCIENCE)',
'SCIENCE',
'SC OAK RIDGE OFFICE',
'SC CHICAGO SERVICE CENTER'
]
sc_contracts = fulldata[(fulldata['awarding_office_name'].isin(
sc_awarding_offices)) | (fulldata['funding_office_name'].isin(sc_funding_offices))]
print len(sc_contracts)
#sc_contracts.to_pickle('../cleaned_data/sc_contracts.pkl')
print 'Generating NSF Grant data...'
grant_file_list = glob('../new_data/*049_Assistance*.csv')
grant_df_list = []
for grant_file in grant_file_list:
grant_df_list.append(pd.read_csv(grant_file))
fulldata = pd.concat(grant_df_list,ignore_index=True)
len(fulldata)
mps_grants = fulldata[fulldata['cfda_title'] == 'MATHEMATICAL AND PHYSICAL SCIENCES']
len(mps_grants)
mps_grants['recipient_congressional_district'].unique()
mps_grants = mps_grants.dropna(subset=['principal_place_cd'])
strlist = []
for code in mps_grants['principal_place_cd'].values:
if code == 'ZZ':
code = '00'
if len(str(int(code))) < 2:
strlist.append('0' + str(int(code)))
else:
strlist.append(str(int(code)))
mps_grants['cong_dist'] = mps_grants['principal_place_state_code'] + strlist
pd.to_pickle(mps_grants, '../cleaned_data/nsf_mps_grants.pkl')
```
|
github_jupyter
|
# Understanding Classification and Logistic Regression with Python
## Introduction
This notebook contains a short introduction to the basic principles of classification and logistic regression. A simple Python simulation is used to illustrate these principles. Specifically, the following steps are performed:
- A data set is created. The label has binary `TRUE` and `FALSE` labels. Values for two features are generated from two bivariate Normal distribion, one for each label class.
- A plot is made of the data set, using color and shape to show the two label classes.
- A plot of a logistic function is computed.
- For each of three data sets a logistic regression model is computed, scored and a plot created using color to show class and shape to show correct and incorrect scoring.
## Create the data set
The code in the cell below computes the two class data set. The feature values for each label level are computed from a bivariate Normal distribution. Run this code and examine the first few rows of the data frame.
```
def sim_log_data(x1, y1, n1, sd1, x2, y2, n2, sd2):
import pandas as pd
import numpy.random as nr
wx1 = nr.normal(loc = x1, scale = sd1, size = n1)
wy1 = nr.normal(loc = y1, scale = sd1, size = n1)
z1 = [1]*n1
wx2 = nr.normal(loc = x2, scale = sd2, size = n2)
wy2 = nr.normal(loc = y2, scale = sd2, size = n2)
z2 = [0]*n2
df1 = pd.DataFrame({'x': wx1, 'y': wy1, 'z': z1})
df2 = pd.DataFrame({'x': wx2, 'y': wy2, 'z': z2})
return pd.concat([df1, df2], axis = 0, ignore_index = True)
sim_data = sim_log_data(1, 1, 50, 1, -1, -1, 50, 1)
sim_data.head()
```
## Plot the data set
The code in the cell below plots the data set using color to show the two classes of the labels. Execute this code and examine the results. Notice that the posion of the points from each class overlap with each other.
```
%matplotlib inline
def plot_class(df):
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(5, 5))
fig.clf()
ax = fig.gca()
df[df.z == 1].plot(kind = 'scatter', x = 'x', y = 'y', ax = ax,
alpha = 1.0, color = 'Red', marker = 'x', s = 40)
df[df.z == 0].plot(kind = 'scatter', x = 'x', y = 'y', ax = ax,
alpha = 1.0, color = 'DarkBlue', marker = 'o', s = 40)
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_title('Classes vs X and Y')
return 'Done'
plot_class(sim_data)
```
## Plot the logistic function
Logistic regression computes a binary {0,1} score using a logistic function. A value of the logistic function above the cutoff (typically 0.5) are scored as a 1 or true, and values less than the cutoff are scored as a 0 or false. Execute the code and examine the resulting logistic function.
```
def plot_logistic(upper = 6, lower = -6, steps = 100):
import matplotlib.pyplot as plt
import pandas as pd
import math as m
step = float(upper - lower) / float(steps)
x = [lower + x * step for x in range(101)]
y = [m.exp(z)/(1 + m.exp(z)) for z in x]
fig = plt.figure(figsize=(5, 4))
fig.clf()
ax = fig.gca()
ax.plot(x, y, color = 'r')
ax.axvline(0, 0.0, 1.0)
ax.axhline(0.5, lower, upper)
ax.set_xlabel('X')
ax.set_ylabel('Probabiltiy of positive response')
ax.set_title('Logistic function for two-class classification')
return 'done'
plot_logistic()
```
## Compute and score a logistic regression model
There is a considerable anount of code in the cell below.
The fist function uses scikit-learn to compute and scores a logsitic regression model. Notie that the features and the label must be converted to a numpy array which is required for scikit-learn.
The second function computes the evaluation of the logistic regression model in the following steps:
- Compute the elements of theh confusion matrix.
- Plot the correctly and incorrectly scored cases, using shape and color to identify class and classification correctness.
- Commonly used performance statistics are computed.
Execute this code and examine the results. Notice that most of the cases have been correctly classified. Classification errors appear along a boundary between those two classes.
```
def logistic_mod(df, logProb = 1.0):
from sklearn import linear_model
## Prepare data for model
nrow = df.shape[0]
X = df[['x', 'y']].as_matrix().reshape(nrow,2)
Y = df.z.as_matrix().ravel() #reshape(nrow,1)
## Compute the logistic regression model
lg = linear_model.LogisticRegression()
logr = lg.fit(X, Y)
## Compute the y values
temp = logr.predict_log_proba(X)
df['predicted'] = [1 if (logProb > p[1]/p[0]) else 0 for p in temp]
return df
def eval_logistic(df):
import matplotlib.pyplot as plt
import pandas as pd
truePos = df[((df['predicted'] == 1) & (df['z'] == df['predicted']))]
falsePos = df[((df['predicted'] == 1) & (df['z'] != df['predicted']))]
trueNeg = df[((df['predicted'] == 0) & (df['z'] == df['predicted']))]
falseNeg = df[((df['predicted'] == 0) & (df['z'] != df['predicted']))]
fig = plt.figure(figsize=(5, 5))
fig.clf()
ax = fig.gca()
truePos.plot(kind = 'scatter', x = 'x', y = 'y', ax = ax,
alpha = 1.0, color = 'DarkBlue', marker = '+', s = 80)
falsePos.plot(kind = 'scatter', x = 'x', y = 'y', ax = ax,
alpha = 1.0, color = 'Red', marker = 'o', s = 40)
trueNeg.plot(kind = 'scatter', x = 'x', y = 'y', ax = ax,
alpha = 1.0, color = 'DarkBlue', marker = 'o', s = 40)
falseNeg.plot(kind = 'scatter', x = 'x', y = 'y', ax = ax,
alpha = 1.0, color = 'Red', marker = '+', s = 80)
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_title('Classes vs X and Y')
TP = truePos.shape[0]
FP = falsePos.shape[0]
TN = trueNeg.shape[0]
FN = falseNeg.shape[0]
confusion = pd.DataFrame({'Positive': [FP, TP],
'Negative': [TN, FN]},
index = ['TrueNeg', 'TruePos'])
accuracy = float(TP + TN)/float(TP + TN + FP + FN)
precision = float(TP)/float(TP + FP)
recall = float(TP)/float(TP + FN)
print(confusion)
print('accracy = ' + str(accuracy))
print('precision = ' + str(precision))
print('recall = ' + str(recall))
return 'Done'
mod = logistic_mod(sim_data)
eval_logistic(mod)
```
## Moving the decision boundary
The example above uses a cutoff at the midpoint of the logistic function. However, you can change the trade-off between correctly classifying the positive cases and correctly classifing the negative cases. The code in the cell below computes and scores a logistic regressiion model for three different cutoff points.
Run the code in the cell and carefully compare the results for the three cases. Notice, that as the logistic cutoff changes the decision boundary moves on the plot, with progressively more positive cases are correctly classified. In addition, accuracy and precision decrease and recall increases.
```
def logistic_demo_prob():
logt = sim_log_data(0.5, 0.5, 50, 1, -0.5, -0.5, 50, 1)
probs = [1, 2, 4]
for p in probs:
logMod = logistic_mod(logt, p)
eval_logistic(logMod)
return 'Done'
logistic_demo_prob()
```
|
github_jupyter
|
# Implementation of Softmax Regression from Scratch
:label:`chapter_softmax_scratch`
Just as we implemented linear regression from scratch,
we believe that multiclass logistic (softmax) regression
is similarly fundamental and you ought to know
the gory details of how to implement it from scratch.
As with linear regression, after doing things by hand
we will breeze through an implementation in Gluon for comparison.
To begin, let's import our packages.
```
import sys
sys.path.insert(0, '..')
%matplotlib inline
import d2l
import torch
from torch.distributions import normal
```
We will work with the Fashion-MNIST dataset just introduced,
cuing up an iterator with batch size 256.
```
batch_size = 256
train_iter, test_iter = d2l.load_data_fashion_mnist(batch_size)
```
## Initialize Model Parameters
Just as in linear regression, we represent each example as a vector.
Since each example is a $28 \times 28$ image,
we can flatten each example, treating them as $784$ dimensional vectors.
In the future, we'll talk about more sophisticated strategies
for exploiting the spatial structure in images,
but for now we treat each pixel location as just another feature.
Recall that in softmax regression,
we have as many outputs as there are categories.
Because our dataset has $10$ categories,
our network will have an output dimension of $10$.
Consequently, our weights will constitute a $784 \times 10$ matrix
and the biases will constitute a $1 \times 10$ vector.
As with linear regression, we will initialize our weights $W$
with Gaussian noise and our biases to take the initial value $0$.
```
num_inputs = 784
num_outputs = 10
W = normal.Normal(loc = 0, scale = 0.01).sample((num_inputs, num_outputs))
b = torch.zeros(num_outputs)
```
Recall that we need to *attach gradients* to the model parameters.
More literally, we are allocating memory for future gradients to be stored
and notifiying PyTorch that we want gradients to be calculated with respect to these parameters in the first place.
```
W.requires_grad_(True)
b.requires_grad_(True)
```
## The Softmax
Before implementing the softmax regression model,
let's briefly review how `torch.sum` work
along specific dimensions in a PyTorch tensor.
Given a matrix `X` we can sum over all elements (default) or only
over elements in the same column (`dim=0`) or the same row (`dim=1`).
Note that if `X` is an array with shape `(2, 3)`
and we sum over the columns (`torch.sum(X, dim=0`),
the result will be a (1D) vector with shape `(3,)`.
If we want to keep the number of axes in the original array
(resulting in a 2D array with shape `(1,3)`),
rather than collapsing out the dimension that we summed over
we can specify `keepdim=True` when invoking `torch.sum`.
```
X = torch.tensor([[1, 2, 3], [4, 5, 6]])
torch.sum(X, dim=0, keepdim=True), torch.sum(X, dim=1, keepdim=True)
```
We are now ready to implement the softmax function.
Recall that softmax consists of two steps:
First, we exponentiate each term (using `torch.exp`).
Then, we sum over each row (we have one row per example in the batch)
to get the normalization constants for each example.
Finally, we divide each row by its normalization constant,
ensuring that the result sums to $1$.
Before looking at the code, let's recall
what this looks expressed as an equation:
$$
\mathrm{softmax}(\mathbf{X})_{ij} = \frac{\exp(X_{ij})}{\sum_k \exp(X_{ik})}
$$
The denominator, or normalization constant,
is also sometimes called the partition function
(and its logarithm the log-partition function).
The origins of that name are in [statistical physics](https://en.wikipedia.org/wiki/Partition_function_(statistical_mechanics))
where a related equation models the distribution
over an ensemble of particles).
```
def softmax(X):
X_exp = torch.exp(X)
partition = torch.sum(X_exp, dim=1, keepdim=True)
return X_exp / partition # The broadcast mechanism is applied here
```
As you can see, for any random input, we turn each element into a non-negative number. Moreover, each row sums up to 1, as is required for a probability.
Note that while this looks correct mathematically,
we were a bit sloppy in our implementation
because failed to take precautions against numerical overflow or underflow
due to large (or very small) elements of the matrix,
as we did in
:numref:`chapter_naive_bayes`.
```
# X = nd.random.normal(shape=(2, 5))
X = normal.Normal(loc = 0, scale = 1).sample((2, 5))
X_prob = softmax(X)
X_prob, torch.sum(X_prob, dim=1)
```
## The Model
Now that we have defined the softmax operation,
we can implement the softmax regression model.
The below code defines the forward pass through the network.
Note that we flatten each original image in the batch
into a vector with length `num_inputs` with the `view` function
before passing the data through our model.
```
def net(X):
return softmax(torch.matmul(X.reshape((-1, num_inputs)), W) + b)
```
## The Loss Function
Next, we need to implement the cross entropy loss function,
introduced in :numref:`chapter_softmax`.
This may be the most common loss function
in all of deep learning because, at the moment,
classification problems far outnumber regression problems.
Recall that cross entropy takes the negative log likelihood
of the predicted probability assigned to the true label $-\log p(y|x)$.
Rather than iterating over the predictions with a Python `for` loop
(which tends to be inefficient), we can use the `gather` function
which allows us to select the appropriate terms
from the matrix of softmax entries easily.
Below, we illustrate the `gather` function on a toy example,
with 3 categories and 2 examples.
```
y_hat = torch.tensor([[0.1, 0.3, 0.6], [0.3, 0.2, 0.5]])
y = torch.tensor([0, 2])
torch.gather(y_hat, 1, y.unsqueeze(dim=1)) # y has to be unsqueezed so that shape(y_hat) = shape(y)
```
Now we can implement the cross-entropy loss function efficiently
with just one line of code.
```
def cross_entropy(y_hat, y):
return -torch.gather(y_hat, 1, y.unsqueeze(dim=1)).log()
```
## Classification Accuracy
Given the predicted probability distribution `y_hat`,
we typically choose the class with highest predicted probability
whenever we must output a *hard* prediction. Indeed, many applications require that we make a choice. Gmail must catetegorize an email into Primary, Social, Updates, or Forums. It might estimate probabilities internally, but at the end of the day it has to choose one among the categories.
When predictions are consistent with the actual category `y`, they are correct. The classification accuracy is the fraction of all predictions that are correct. Although we cannot optimize accuracy directly (it is not differentiable), it's often the performance metric that we care most about, and we will nearly always report it when training classifiers.
To compute accuracy we do the following:
First, we execute `y_hat.argmax(dim=1)`
to gather the predicted classes
(given by the indices for the largest entires each row).
The result has the same shape as the variable `y`.
Now we just need to check how frequently the two match. The result is PyTorch tensor containing entries of 0 (false) and 1 (true). Since the attribute `mean` can only calculate the mean of floating types,
we also need to convert the result to `float`. Taking the mean yields the desired result.
```
def accuracy(y_hat, y):
return (y_hat.argmax(dim=1) == y).float().mean().item()
```
We will continue to use the variables `y_hat` and `y`
defined in the `gather` function,
as the predicted probability distribution and label, respectively.
We can see that the first example's prediction category is 2
(the largest element of the row is 0.6 with an index of 2),
which is inconsistent with the actual label, 0.
The second example's prediction category is 2
(the largest element of the row is 0.5 with an index of 2),
which is consistent with the actual label, 2.
Therefore, the classification accuracy rate for these two examples is 0.5.
```
accuracy(y_hat, y)
```
Similarly, we can evaluate the accuracy for model `net` on the data set
(accessed via `data_iter`).
```
# The function will be gradually improved: the complete implementation will be
# discussed in the "Image Augmentation" section
def evaluate_accuracy(data_iter, net):
acc_sum, n = 0.0, 0
for X, y in data_iter:
acc_sum += (net(X).argmax(dim=1) == y).sum().item()
n += y.size()[0] # y.size()[0] = batch_size
return acc_sum / n
```
Because we initialized the `net` model with random weights,
the accuracy of this model should be close to random guessing,
i.e. 0.1 for 10 classes.
```
evaluate_accuracy(test_iter, net)
```
## Model Training
The training loop for softmax regression should look strikingly familiar
if you read through our implementation
of linear regression earlier in this chapter.
Again, we use the mini-batch stochastic gradient descent
to optimize the loss function of the model.
Note that the number of epochs (`num_epochs`),
and learning rate (`lr`) are both adjustable hyper-parameters.
By changing their values, we may be able to increase the classification accuracy of the model. In practice we'll want to split our data three ways
into training, validation, and test data, using the validation data to choose the best values of our hyperparameters.
```
num_epochs, lr = 5, 0.1
# This function has been saved in the d2l package for future use
def train_ch3(net, train_iter, test_iter, loss, num_epochs, batch_size, params=None, lr=None, trainer=None):
for epoch in range(num_epochs):
train_l_sum, train_acc_sum, n = 0.0, 0.0, 0
for X, y in train_iter:
y_hat = net(X)
l = loss(y_hat, y).sum()
l.backward()
if trainer is None:
d2l.sgd(params, lr, batch_size)
else:
# This will be illustrated in the next section
trainer.step(batch_size)
train_l_sum += l.item()
train_acc_sum += (y_hat.argmax(dim=1) == y).sum().item()
n += y.size()[0]
test_acc = evaluate_accuracy(test_iter, net)
print('epoch %d, loss %.4f, train acc %.3f, test acc %.3f'
% (epoch + 1, train_l_sum / n, train_acc_sum / n, test_acc))
train_ch3(net, train_iter, test_iter, cross_entropy, num_epochs, batch_size, [W, b], lr)
```
## Prediction
Now that training is complete, our model is ready to classify some images.
Given a series of images, we will compare their actual labels
(first line of text output) and the model predictions
(second line of text output).
```
for X, y in test_iter:
break
true_labels = d2l.get_fashion_mnist_labels(y.numpy())
pred_labels = d2l.get_fashion_mnist_labels(net(X).argmax(dim=1).numpy())
titles = [truelabel + '\n' + predlabel for truelabel, predlabel in zip(true_labels, pred_labels)]
d2l.show_fashion_mnist(X[10:20], titles[10:20])
```
## Summary
With softmax regression, we can train models for multi-category classification. The training loop is very similar to that in linear regression: retrieve and read data, define models and loss functions,
then train models using optimization algorithms. As you'll soon find out, most common deep learning models have similar training procedures.
## Exercises
1. In this section, we directly implemented the softmax function based on the mathematical definition of the softmax operation. What problems might this cause (hint - try to calculate the size of $\exp(50)$)?
1. The function `cross_entropy` in this section is implemented according to the definition of the cross-entropy loss function. What could be the problem with this implementation (hint - consider the domain of the logarithm)?
1. What solutions you can think of to fix the two problems above?
1. Is it always a good idea to return the most likely label. E.g. would you do this for medical diagnosis?
1. Assume that we want to use softmax regression to predict the next word based on some features. What are some problems that might arise from a large vocabulary?
|
github_jupyter
|
<table align="left">
<td>
<a target="_blank" href="https://colab.research.google.com/github/ShopRunner/collie/blob/main/tutorials/05_hybrid_model.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/ShopRunner/collie/blob/main/tutorials/05_hybrid_model.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a>
</td>
<td>
<a target="_blank" href="https://raw.githubusercontent.com/ShopRunner/collie/main/tutorials/05_hybrid_model.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" /> Download notebook</a>
</td>
</table>
```
# for Collab notebooks, we will start by installing the ``collie`` library
!pip install collie --quiet
%reload_ext autoreload
%autoreload 2
%matplotlib inline
%env DATA_PATH data/
import os
import numpy as np
import pandas as pd
from pytorch_lightning.utilities.seed import seed_everything
from IPython.display import HTML
import joblib
import torch
from collie.metrics import mapk, mrr, auc, evaluate_in_batches
from collie.model import CollieTrainer, HybridPretrainedModel, MatrixFactorizationModel
from collie.movielens import get_movielens_metadata, get_recommendation_visualizations
```
## Load Data From ``01_prepare_data`` Notebook
If you're running this locally on Jupyter, you should be able to run the next cell quickly without a problem! If you are running this on Colab, you'll need to regenerate the data by running the cell below that, which should only take a few extra seconds to complete.
```
try:
# let's grab the ``Interactions`` objects we saved in the last notebook
train_interactions = joblib.load(os.path.join(os.environ.get('DATA_PATH', 'data/'),
'train_interactions.pkl'))
val_interactions = joblib.load(os.path.join(os.environ.get('DATA_PATH', 'data/'),
'val_interactions.pkl'))
except FileNotFoundError:
# we're running this notebook on Colab where results from the first notebook are not saved
# regenerate this data below
from collie.cross_validation import stratified_split
from collie.interactions import Interactions
from collie.movielens import read_movielens_df
from collie.utils import convert_to_implicit, remove_users_with_fewer_than_n_interactions
df = read_movielens_df(decrement_ids=True)
implicit_df = convert_to_implicit(df, min_rating_to_keep=4)
implicit_df = remove_users_with_fewer_than_n_interactions(implicit_df, min_num_of_interactions=3)
interactions = Interactions(
users=implicit_df['user_id'],
items=implicit_df['item_id'],
ratings=implicit_df['rating'],
allow_missing_ids=True,
)
train_interactions, val_interactions = stratified_split(interactions, test_p=0.1, seed=42)
print('Train:', train_interactions)
print('Val: ', val_interactions)
```
# Hybrid Collie Model Using a Pre-Trained ``MatrixFactorizationModel``
In this notebook, we will use this same metadata and incorporate it directly into the model architecture with a hybrid Collie model.
## Read in Data
```
# read in the same metadata used in notebooks ``03`` and ``04``
metadata_df = get_movielens_metadata()
metadata_df.head()
# and, as always, set our random seed
seed_everything(22)
```
## Train a ``MatrixFactorizationModel``
The first step towards training a Collie Hybrid model is to train a regular ``MatrixFactorizationModel`` to generate rich user and item embeddings. We'll use these embeddings in a ``HybridPretrainedModel`` a bit later.
```
model = MatrixFactorizationModel(
train=train_interactions,
val=val_interactions,
embedding_dim=30,
lr=1e-2,
)
trainer = CollieTrainer(model=model, max_epochs=10, deterministic=True)
trainer.fit(model)
mapk_score, mrr_score, auc_score = evaluate_in_batches([mapk, mrr, auc], val_interactions, model)
print(f'Standard MAP@10 Score: {mapk_score}')
print(f'Standard MRR Score: {mrr_score}')
print(f'Standard AUC Score: {auc_score}')
```
## Train a ``HybridPretrainedModel``
With our trained ``model`` above, we can now use these embeddings and additional side data directly in a hybrid model. The architecture essentially takes our user embedding, item embedding, and item metadata for each user-item interaction, concatenates them, and sends it through a simple feedforward network to output a recommendation score.
We can initially freeze the user and item embeddings from our previously-trained ``model``, train for a few epochs only optimizing our newly-added linear layers, and then train a model with everything unfrozen at a lower learning rate. We will show this process below.
```
# we will apply a linear layer to the metadata with ``metadata_layers_dims`` and
# a linear layer to the combined embeddings and metadata data with ``combined_layers_dims``
hybrid_model = HybridPretrainedModel(
train=train_interactions,
val=val_interactions,
item_metadata=metadata_df,
trained_model=model,
metadata_layers_dims=[8],
combined_layers_dims=[16],
lr=1e-2,
freeze_embeddings=True,
)
hybrid_trainer = CollieTrainer(model=hybrid_model, max_epochs=10, deterministic=True)
hybrid_trainer.fit(hybrid_model)
mapk_score, mrr_score, auc_score = evaluate_in_batches([mapk, mrr, auc], val_interactions, hybrid_model)
print(f'Hybrid MAP@10 Score: {mapk_score}')
print(f'Hybrid MRR Score: {mrr_score}')
print(f'Hybrid AUC Score: {auc_score}')
hybrid_model_unfrozen = HybridPretrainedModel(
train=train_interactions,
val=val_interactions,
item_metadata=metadata_df,
trained_model=model,
metadata_layers_dims=[8],
combined_layers_dims=[16],
lr=1e-4,
freeze_embeddings=False,
)
hybrid_model.unfreeze_embeddings()
hybrid_model_unfrozen.load_from_hybrid_model(hybrid_model)
hybrid_trainer_unfrozen = CollieTrainer(model=hybrid_model_unfrozen, max_epochs=10, deterministic=True)
hybrid_trainer_unfrozen.fit(hybrid_model_unfrozen)
mapk_score, mrr_score, auc_score = evaluate_in_batches([mapk, mrr, auc],
val_interactions,
hybrid_model_unfrozen)
print(f'Hybrid Unfrozen MAP@10 Score: {mapk_score}')
print(f'Hybrid Unfrozen MRR Score: {mrr_score}')
print(f'Hybrid Unfrozen AUC Score: {auc_score}')
```
Note here that while our ``MAP@10`` and ``MRR`` scores went down slightly from the frozen version of the model above, our ``AUC`` score increased. For implicit recommendation models, each evaluation metric is nuanced in what it represents for real world recommendations.
You can read more about each evaluation metric by checking out the [Mean Average Precision at K (MAP@K)](https://en.wikipedia.org/wiki/Evaluation_measures_(information_retrieval)#Mean_average_precision), [Mean Reciprocal Rank](https://en.wikipedia.org/wiki/Mean_reciprocal_rank), and [Area Under the Curve (AUC)](https://en.wikipedia.org/wiki/Receiver_operating_characteristic#Area_under_the_curve) Wikipedia pages.
```
user_id = np.random.randint(0, train_interactions.num_users)
display(
HTML(
get_recommendation_visualizations(
model=hybrid_model_unfrozen,
user_id=user_id,
filter_films=True,
shuffle=True,
detailed=True,
)
)
)
```
The metrics and results look great, and we should only see a larger difference compared to a standard model as our data becomes more nuanced and complex (such as with MovieLens 10M data).
If we're happy with this model, we can go ahead and save it for later!
## Save and Load a Hybrid Model
```
# we can save the model with...
os.makedirs('models', exist_ok=True)
hybrid_model_unfrozen.save_model('models/hybrid_model_unfrozen')
# ... and if we wanted to load that model back in, we can do that easily...
hybrid_model_loaded_in = HybridPretrainedModel(load_model_path='models/hybrid_model_unfrozen')
hybrid_model_loaded_in
```
While our model works and the results look great, it's not always possible to be able to fully train two separate models like we've done in this tutorial. Sometimes, it's easier (and even better) to train a single hybird model up from scratch, no pretrained ``MatrixFactorizationModel`` needed.
In the next tutorial, we'll cover multi-stage models in Collie, tackling this exact problem and more! See you there!
-----
|
github_jupyter
|
# Implicit Georeferencing
This workbook sets explicit georeferences from implicit georeferencing through names of extents given in dataset titles or keywords.
A file `sources.py` needs to contain the CKAN and SOURCE config as follows:
```
CKAN = {
"dpaw-internal":{
"url": "http://internal-data.dpaw.wa.gov.au/",
"key": "API-KEY"
}
}
```
## Configure CKAN and source
```
import ckanapi
from harvest_helpers import *
from secret import CKAN
ckan = ckanapi.RemoteCKAN(CKAN["dpaw-internal"]["url"], apikey=CKAN["dpaw-internal"]["key"])
print("Using CKAN {0}".format(ckan.address))
```
## Spatial extent name-geometry lookup
The fully qualified names and GeoJSON geometries of relevant spatial areas are contained in our custom dataschema.
```
# Getting the extent dictionary e
url = "https://raw.githubusercontent.com/datawagovau/ckanext-datawagovautheme/dpaw-internal/ckanext/datawagovautheme/datawagovau_dataset.json"
ds = json.loads(requests.get(url).content)
choice_dict = [x for x in ds["dataset_fields"] if x["field_name"] == "spatial"][0]["choices"]
e = dict([(x["label"], json.dumps(x["value"])) for x in choice_dict])
print("Extents: {0}".format(e.keys()))
```
## Name lookups
Relevant areas are listed under different synonyms. We'll create a dictionary of synonymous search terms ("s") and extent names (index "i").
```
# Creating a search term - extent index lookup
# m is a list of keys "s" (search term) and "i" (extent index)
m = [
{"s":"Eighty", "i":"MPA Eighty Mile Beach"},
{"s":"EMBMP", "i":"MPA Eighty Mile Beach"},
{"s":"Camden", "i":"MPA Lalang-garram / Camden Sound"},
{"s":"LCSMP", "i":"MPA Lalang-garram / Camden Sound"},
{"s":"Rowley", "i":"MPA Rowley Shoals"},
{"s":"RSMP", "i":"MPA Rowley Shoals"},
{"s":"Montebello", "i":"MPA Montebello Barrow"},
{"s":"MBIMPA", "i":"MPA Montebello Barrow"},
{"s":"Ningaloo", "i":"MPA Ningaloo"},
{"s":"NMP", "i":"MPA Ningaloo"},
{"s":"Shark bay", "i":"MPA Shark Bay Hamelin Pool"},
{"s":"SBMP", "i":"MPA Shark Bay Hamelin Pool"},
{"s":"Jurien", "i":"MPA Jurien Bay"},
{"s":"JBMP", "i":"MPA Jurien Bay"},
{"s":"Marmion", "i":"MPA Marmion"},
{"s":"Swan Estuary", "i":"MPA Swan Estuary"},
{"s":"SEMP", "i":"MPA Swan Estuary"},
{"s":"Shoalwater", "i":"MPA Shoalwater Islands"},
{"s":"SIMP", "i":"MPA Shoalwater Islands"},
{"s":"Ngari", "i":"MPA Ngari Capes"},
{"s":"NCMP", "i":"MPA Ngari Capes"},
{"s":"Walpole", "i":"MPA Walpole Nornalup"},
{"s":"WNIMP", "i":"MPA Walpole Nornalup"}
]
def add_spatial(dsdict, extent_string, force=False, debug=False):
"""Adds a given spatial extent to a CKAN dataset dict if
"spatial" is None, "" or force==True.
Arguments:
dsdict (ckanapi.action.package_show()) CKAN dataset dict
extent_string (String) GeoJSON geometry as json.dumps String
force (Boolean) Whether to force overwriting "spatial"
debug (Boolean) Debug noise
Returns:
(dict) The dataset with spatial extent replaced per above rules.
"""
if not dsdict.has_key("spatial"):
overwrite = True
if debug:
msg = "Spatial extent not given"
elif dsdict["spatial"] == "":
overwrite = True
if debug:
msg = "Spatial extent is empty"
elif force:
overwrite = True
msg = "Spatial extent was overwritten"
else:
overwrite = False
msg = "Spatial extent unchanged"
if overwrite:
dsdict["spatial"] = extent_string
print(msg)
return dsdict
def restore_extents(search_mapping, extents, ckan, debug=False):
"""Restore spatial extents for datasets
Arguments:
search_mapping (list) A list of dicts with keys "s" for ckanapi
package_search query parameter "q", and key "i" for the name
of the extent
e.g.:
m = [
{"s":"tags:marinepark_80_mile_beach", "i":"MPA Eighty Mile Beach"},
...
]
extents (dict) A dict with key "i" (extent name) and
GeoJSON Multipolygon geometry strings as value, e.g.:
{u'MPA Eighty Mile Beach': '{"type": "MultiPolygon", "coordinates": [ .... ]', ...}
ckan (ckanapi) A ckanapi instance
debug (boolean) Debug noise
Returns:
A list of dictionaries returned by ckanapi's package_update
"""
for x in search_mapping:
if debug:
print("\nSearching CKAN with '{0}'".format(x["s"]))
found = ckan.action.package_search(q=x["s"])["results"]
if debug:
print("Found datasets: {0}\n".format([d["title"] for d in found]))
fixed = [add_spatial(d, extents[x["i"]], force=True, debug=True) for d in found]
if debug:
print(fixed, "\n")
datasets_updated = upsert_datasets(fixed, ckan, debug=False)
restore_extents(m, e, ckan)
d = [ckan.action.package_show(id = x) for x in ckan.action.package_list()]
fix = [x["title"] for x in d if not x.has_key("spatial")]
len(fix)
d[0]
fix
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/xavoliva6/dpfl_pytorch/blob/main/experiments/exp_FedMNIST.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
#Experiments on FedMNIST
**Colab Support**<br/>
Only run the following lines if you want to run the code on Google Colab
```
# Enable access to files stored in Google Drive
from google.colab import drive
drive.mount('/content/gdrive/')
% cd /content/gdrive/My Drive/OPT4ML/src
```
# Main
```
# Install necessary requirements
!pip install -r ../requirements.txt
# Make sure cuda support is available
import torch
if torch.cuda.is_available():
device_name = "cuda:0"
else:
device_name = "cpu"
print("device_name: {}".format(device_name))
device = torch.device(device_name)
%load_ext autoreload
%autoreload 2
import sys
import warnings
warnings.filterwarnings("ignore")
from server import Server
from utils import plot_exp
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = [6, 6]
plt.rcParams['figure.dpi'] = 100
```
### First experiment : impact of federated learning
```
LR = 0.01
EPOCHS = 1
NR_TRAINING_ROUNDS = 30
BATCH_SIZE = 128
RANGE_NR_CLIENTS = [1,5,10]
experiment_losses, experiment_accs = [], []
for nr_clients in RANGE_NR_CLIENTS:
print(f"### Number of clients : {nr_clients} ###\n\n")
server = Server(
nr_clients=nr_clients,
nr_training_rounds=NR_TRAINING_ROUNDS,
data='MNIST',
epochs=EPOCHS,
lr=LR,
batch_size=BATCH_SIZE,
is_private=False,
epsilon=None,
max_grad_norm=None,
noise_multiplier=None,
is_parallel=True,
device=device,
verbose='server')
test_losses, test_accs = server.train()
experiment_losses.append(test_losses)
experiment_accs.append(test_accs)
names = [f'{i} clients' for i in RANGE_NR_CLIENTS]
title = 'First experiment : MNIST database'
fig = plot_exp(experiment_losses, experiment_accs, names, title)
fig.savefig("MNIST_exp1.pdf")
```
### Second experiment : impact of differential privacy
```
NR_CLIENTS = 10
NR_TRAINING_ROUNDS = 30
EPOCHS = 1
LR = 0.01
BATCH_SIZE = 128
MAX_GRAD_NORM = 1.2
NOISE_MULTIPLIER = None
RANGE_EPSILON = [10,50,100]
experiment_losses, experiment_accs = [], []
for epsilon in RANGE_EPSILON:
print(f"### ε : {epsilon} ###\n\n")
server = Server(
nr_clients=NR_CLIENTS,
nr_training_rounds=NR_TRAINING_ROUNDS,
data='MNIST',
epochs=EPOCHS,
lr=LR,
batch_size=BATCH_SIZE,
is_private=True,
epsilon=epsilon,
max_grad_norm=MAX_GRAD_NORM,
noise_multiplier=NOISE_MULTIPLIER,
is_parallel=True,
device=device,
verbose='server')
test_losses, test_accs = server.train()
experiment_losses.append(test_losses)
experiment_accs.append(test_accs)
names = [f'ε = {i}' for i in RANGE_EPSILON]
title = 'Second experiment : MNIST database'
fig = plot_exp(experiment_losses, experiment_accs, names, title)
plt.savefig('MNIST_exp2.pdf')
```
|
github_jupyter
|
```
# for reading and validating data
import emeval.input.spec_details as eisd
import emeval.input.phone_view as eipv
import emeval.input.eval_view as eiev
# Visualization helpers
import emeval.viz.phone_view as ezpv
import emeval.viz.eval_view as ezev
import emeval.viz.geojson as ezgj
import pandas as pd
# Metrics helpers
import emeval.metrics.dist_calculations as emd
# For computation
import numpy as np
import math
import scipy.stats as stats
import matplotlib.pyplot as plt
import geopandas as gpd
import shapely as shp
import folium
DATASTORE_URL = "http://cardshark.cs.berkeley.edu"
AUTHOR_EMAIL = "[email protected]"
sd_la = eisd.SpecDetails(DATASTORE_URL, AUTHOR_EMAIL, "unimodal_trip_car_bike_mtv_la")
sd_sj = eisd.SpecDetails(DATASTORE_URL, AUTHOR_EMAIL, "car_scooter_brex_san_jose")
sd_ucb = eisd.SpecDetails(DATASTORE_URL, AUTHOR_EMAIL, "train_bus_ebike_mtv_ucb")
import importlib
importlib.reload(eisd)
pv_la = eipv.PhoneView(sd_la)
pv_sj = eipv.PhoneView(sd_sj)
pv_ucb = eipv.PhoneView(sd_ucb)
```
### Validate distance calculations
Our x,y coordinates are in degrees (lon, lat). So when we calculate the distance between two points, it is also in degrees. In order for this to be meaningful, we need to convert it to a regular distance metric such as meters.
This is a complicated problem in general because our distance calculation applies 2-D spatial operations to a 3-D curved space. However, as documented in the shapely documentation, since our areas of interest are small, we can use a 2-D approximation and get reasonable results.
In order to get distances from degree-based calculations, we can use the following options:
- perform the calculations in degrees and then convert them to meters. As an approximation, we can use the fact that 360 degrees represents the circumference of the earth. Therefore `dist = degree_dist * (C/360)`
- convert degrees to x,y coordinates using utm (https://en.wikipedia.org/wiki/Universal_Transverse_Mercator_coordinate_system) and then calculate the distance
- since we calculate the distance from the ground truth linestring, calculate the closest ground truth point in (lon,lat) and then use the haversine formula (https://en.wikipedia.org/wiki/Haversine_formula) to calculate the distance between the two points
Let us quickly all three calculations for three selected test cases and:
- check whether they are largely consistent
- compare with other distance calculators to see which are closer
```
test_cases = {
"commuter_rail_aboveground": {
"section": pv_ucb.map()["android"]["ucb-sdb-android-3"]["evaluation_ranges"][0]["evaluation_trip_ranges"][0]["evaluation_section_ranges"][2],
"ground_truth": sd_ucb.get_ground_truth_for_leg("mtv_to_berkeley_sf_bart", "commuter_rail_aboveground")
},
"light_rail_below_above_ground": {
"section": pv_ucb.map()["android"]["ucb-sdb-android-3"]["evaluation_ranges"][0]["evaluation_trip_ranges"][2]["evaluation_section_ranges"][7],
"ground_truth": sd_ucb.get_ground_truth_for_leg("berkeley_to_mtv_SF_express_bus", "light_rail_below_above_ground")
},
"express_bus": {
"section": pv_ucb.map()["ios"]["ucb-sdb-ios-3"]["evaluation_ranges"][1]["evaluation_trip_ranges"][2]["evaluation_section_ranges"][4],
"ground_truth": sd_ucb.get_ground_truth_for_leg("berkeley_to_mtv_SF_express_bus", "express_bus")
},
}
for t in test_cases.values():
t["gt_shapes"] = gpd.GeoSeries(eisd.SpecDetails.get_shapes_for_leg(t["ground_truth"]))
importlib.reload(emd)
dist_checks = []
pct_checks = []
for (k, t) in test_cases.items():
location_gpdf = emd.filter_geo_df(emd.to_geo_df(t["section"]["location_df"]), t["gt_shapes"].filter(["start_loc","end_loc"]))
gt_linestring = emd.filter_ground_truth_linestring(t["gt_shapes"])
dc = emd.dist_using_circumference(location_gpdf, gt_linestring)
dcrs = emd.dist_using_crs_change(location_gpdf, gt_linestring)
dmuc = emd.dist_using_manual_utm_change(location_gpdf, gt_linestring)
dmmc = emd.dist_using_manual_mercator_change(location_gpdf, gt_linestring)
dup = emd.dist_using_projection(location_gpdf, gt_linestring)
dist_compare = pd.DataFrame({"dist_circumference": dc, "dist_crs_change": dcrs,
"dist_manual_utm": dmuc, "dist_manual_mercator": dmmc,
"dist_project": dup})
dist_compare["diff_c_mu"] = (dist_compare.dist_circumference - dist_compare.dist_manual_utm).abs()
dist_compare["diff_mu_pr"] = (dist_compare.dist_manual_utm - dist_compare.dist_project).abs()
dist_compare["diff_mm_pr"] = (dist_compare.dist_manual_mercator - dist_compare.dist_project).abs()
dist_compare["diff_c_pr"] = (dist_compare.dist_circumference - dist_compare.dist_project).abs()
dist_compare["diff_c_mu_pct"] = dist_compare.diff_c_mu / dist_compare.dist_circumference
dist_compare["diff_mu_pr_pct"] = dist_compare.diff_mu_pr / dist_compare.dist_circumference
dist_compare["diff_mm_pr_pct"] = dist_compare.diff_mm_pr / dist_compare.dist_circumference
dist_compare["diff_c_pr_pct"] = dist_compare.diff_c_pr / dist_compare.dist_circumference
match_dist = lambda t: {"key": k,
"threshold": t,
"diff_c_mu": len(dist_compare.query('diff_c_mu > @t')),
"diff_mu_pr": len(dist_compare.query('diff_mu_pr > @t')),
"diff_mm_pr": len(dist_compare.query('diff_mm_pr > @t')),
"diff_c_pr": len(dist_compare.query('diff_c_pr > @t')),
"total_entries": len(dist_compare)}
dist_checks.append(match_dist(1))
dist_checks.append(match_dist(5))
dist_checks.append(match_dist(10))
dist_checks.append(match_dist(50))
match_pct = lambda t: {"key": k,
"threshold": t,
"diff_c_mu_pct": len(dist_compare.query('diff_c_mu_pct > @t')),
"diff_mu_pr_pct": len(dist_compare.query('diff_mu_pr_pct > @t')),
"diff_mm_pr_pct": len(dist_compare.query('diff_mm_pr_pct > @t')),
"diff_c_pr_pct": len(dist_compare.query('diff_c_pr_pct > @t')),
"total_entries": len(dist_compare)}
pct_checks.append(match_pct(0.01))
pct_checks.append(match_pct(0.05))
pct_checks.append(match_pct(0.10))
pct_checks.append(match_pct(0.15))
pct_checks.append(match_pct(0.20))
pct_checks.append(match_pct(0.25))
# t = "commuter_rail_aboveground"
# gt_gj = eisd.SpecDetails.get_geojson_for_leg(test_cases[t]["ground_truth"])
# print(gt_gj.features[2])
# gt_gj.features[2] = ezgj.get_geojson_for_linestring(emd.filter_ground_truth_linestring(test_cases[t]["gt_shapes"]))
# curr_map = ezgj.get_map_for_geojson(gt_gj)
# curr_map.add_child(ezgj.get_fg_for_loc_df(emd.linestring_to_geo_df(test_cases[t]["gt_shapes"].loc["route"]),
# name="gt_points", color="green"))
# curr_map
pd.DataFrame(dist_checks)
pd.DataFrame(pct_checks)
manual_check_points = pd.concat([location_gpdf, dist_compare], axis=1)[["latitude", "fmt_time", "longitude", "dist_circumference", "dist_manual_utm", "dist_manual_mercator", "dist_project"]].sample(n=3, random_state=10); manual_check_points
# curr_map = ezpv.display_map_detail_from_df(manual_check_points)
# curr_map.add_child(folium.GeoJson(eisd.SpecDetails.get_geojson_for_leg(t["ground_truth"])))
```
### Externally calculated distance for these points is:
Distance calculated manually using
1. https://www.freemaptools.com/measure-distance.htm
1. Google Maps
Note that the error of my eyes + hand is ~ 2-3 m
- 1213: within margin of error
- 1053: 3987 (freemaptools), 4km (google)
- 1107: 15799.35 (freemaptools), 15.80km (google)
```
manual_check_points
```
### Results and method choice
We find that the `manual_utm` and `project` methods are pretty consistent, and are significantly different from the `circumference` method. The `circumference` method appears to be consistently greater than the other two and the difference appears to be around 25%. The manual checks also appear to be closer to the `manual_utm` and `project` values. The `manual_utm` and `project` values are consistently within ~ 5% of each other, so we could really use either one.
**We will use the utm approach** since it is correct, is consistent with the shapely documentation (https://shapely.readthedocs.io/en/stable/manual.html#coordinate-systems) and applicable to operations beyond distance calculation
> Even though the Earth is not flat – and for that matter not exactly spherical – there are many analytic problems that can be approached by transforming Earth features to a Cartesian plane, applying tried and true algorithms, and then transforming the results back to geographic coordinates. This practice is as old as the tradition of accurate paper maps.
## Spatial error calculation
```
def get_spatial_errors(pv):
spatial_error_df = pd.DataFrame()
for phone_os, phone_map in pv.map().items():
for phone_label, phone_detail_map in phone_map.items():
for (r_idx, r) in enumerate(phone_detail_map["evaluation_ranges"]):
run_errors = []
for (tr_idx, tr) in enumerate(r["evaluation_trip_ranges"]):
trip_errors = []
for (sr_idx, sr) in enumerate(tr["evaluation_section_ranges"]):
# This is a Shapely LineString
section_gt_leg = pv.spec_details.get_ground_truth_for_leg(tr["trip_id_base"], sr["trip_id_base"])
section_gt_shapes = gpd.GeoSeries(eisd.SpecDetails.get_shapes_for_leg(section_gt_leg))
if len(section_gt_shapes) == 1:
print("No ground truth route for %s %s, must be polygon, skipping..." % (tr["trip_id_base"], sr["trip_id_base"]))
assert section_gt_leg["type"] != "TRAVEL", "For %s, %s, %s, %s, %s found type %s" % (phone_os, phone_label, r_idx, tr_idx, sr_idx, section_gt_leg["type"])
continue
if len(sr['location_df']) == 0:
print("No sensed locations found, role = %s skipping..." % (r["eval_role_base"]))
# assert r["eval_role_base"] == "power_control", "Found no locations for %s, %s, %s, %s, %s" % (phone_os, phone_label, r_idx, tr_idx, sr_idx)
continue
print("Processing travel leg %s, %s, %s, %s, %s" %
(phone_os, phone_label, r["eval_role_base"], tr["trip_id_base"], sr["trip_id_base"]))
# This is a GeoDataFrame
section_geo_df = emd.to_geo_df(sr["location_df"])
# After this point, everything is in UTM so that 2-D inside/filtering operations work
utm_section_geo_df = emd.to_utm_df(section_geo_df)
utm_section_gt_shapes = section_gt_shapes.apply(lambda s: shp.ops.transform(emd.to_utm_coords, s))
filtered_us_gpdf = emd.filter_geo_df(utm_section_geo_df, utm_section_gt_shapes.loc["start_loc":"end_loc"])
filtered_gt_linestring = emd.filter_ground_truth_linestring(utm_section_gt_shapes)
meter_dist = filtered_us_gpdf.geometry.distance(filtered_gt_linestring)
ne = len(meter_dist)
curr_spatial_error_df = gpd.GeoDataFrame({"error": meter_dist,
"ts": section_geo_df.ts,
"geometry": section_geo_df.geometry,
"phone_os": np.repeat(phone_os, ne),
"phone_label": np.repeat(phone_label, ne),
"role": np.repeat(r["eval_role_base"], ne),
"timeline": np.repeat(pv.spec_details.CURR_SPEC_ID, ne),
"run": np.repeat(r_idx, ne),
"trip_id": np.repeat(tr["trip_id_base"], ne),
"section_id": np.repeat(sr["trip_id_base"], ne)})
spatial_error_df = pd.concat([spatial_error_df, curr_spatial_error_df], axis="index")
return spatial_error_df
spatial_errors_df = pd.DataFrame()
spatial_errors_df = pd.concat([spatial_errors_df, get_spatial_errors(pv_la)], axis="index")
spatial_errors_df = pd.concat([spatial_errors_df, get_spatial_errors(pv_sj)], axis="index")
spatial_errors_df = pd.concat([spatial_errors_df, get_spatial_errors(pv_ucb)], axis="index")
spatial_errors_df.head()
r2q_map = {"power_control": 0, "HAMFDC": 1, "MAHFDC": 2, "HAHFDC": 3, "accuracy_control": 4}
q2r_map = {0: "power", 1: "HAMFDC", 2: "MAHFDC", 3: "HAHFDC", 4: "accuracy"}
spatial_errors_df["quality"] = spatial_errors_df.role.apply(lambda r: r2q_map[r])
spatial_errors_df["label"] = spatial_errors_df.role.apply(lambda r: r.replace('_control', ''))
timeline_list = ["train_bus_ebike_mtv_ucb", "car_scooter_brex_san_jose", "unimodal_trip_car_bike_mtv_la"]
spatial_errors_df.head()
```
## Overall stats
```
ifig, ax_array = plt.subplots(nrows=1,ncols=2,figsize=(8,2), sharey=True)
spatial_errors_df.query("phone_os == 'android' & quality > 0").boxplot(ax = ax_array[0], column=["error"], by=["quality"], showfliers=False)
ax_array[0].set_title('android')
spatial_errors_df.query("phone_os == 'ios' & quality > 0").boxplot(ax = ax_array[1], column=["error"], by=["quality"], showfliers=False)
ax_array[1].set_title("ios")
for i, ax in enumerate(ax_array):
# print([t.get_text() for t in ax.get_xticklabels()])
ax.set_xticklabels([q2r_map[int(t.get_text())] for t in ax.get_xticklabels()])
ax.set_xlabel("")
ax_array[0].set_ylabel("Spatial error (meters)")
# ax_array[1][0].set_ylabel("Spatial error (meters)")
ifig.suptitle("Spatial trajectory error v/s quality (excluding outliers)", y = 1.1)
# ifig.tight_layout()
ifig, ax_array = plt.subplots(nrows=1,ncols=2,figsize=(8,2), sharey=True)
spatial_errors_df.query("phone_os == 'android' & quality > 0").boxplot(ax = ax_array[0], column=["error"], by=["quality"])
ax_array[0].set_title('android')
spatial_errors_df.query("phone_os == 'ios' & quality > 0").boxplot(ax = ax_array[1], column=["error"], by=["quality"])
ax_array[1].set_title("ios")
for i, ax in enumerate(ax_array):
# print([t.get_text() for t in ax.get_xticklabels()])
ax.set_xticklabels([q2r_map[int(t.get_text())] for t in ax.get_xticklabels()])
ax.set_xlabel("")
ax_array[0].set_ylabel("Spatial error (meters)")
# ax_array[1][0].set_ylabel("Spatial error (meters)")
ifig.suptitle("Spatial trajectory error v/s quality", y = 1.1)
# ifig.tight_layout()
```
### Split out results by timeline
```
ifig, ax_array = plt.subplots(nrows=2,ncols=3,figsize=(12,6), sharex=False, sharey=False)
timeline_list = ["train_bus_ebike_mtv_ucb", "car_scooter_brex_san_jose", "unimodal_trip_car_bike_mtv_la"]
for i, tl in enumerate(timeline_list):
spatial_errors_df.query("timeline == @tl & phone_os == 'android' & quality > 0").boxplot(ax = ax_array[0][i], column=["error"], by=["quality"])
ax_array[0][i].set_title(tl)
spatial_errors_df.query("timeline == @tl & phone_os == 'ios' & quality > 0").boxplot(ax = ax_array[1][i], column=["error"], by=["quality"])
ax_array[1][i].set_title("")
for i, ax in enumerate(ax_array[0]):
ax.set_xticklabels([q2r_map[int(t.get_text())] for t in ax.get_xticklabels()])
ax.set_xlabel("")
for i, ax in enumerate(ax_array[1]):
ax.set_xticklabels([q2r_map[int(t.get_text())] for t in ax.get_xticklabels()])
ax.set_xlabel("")
ax_array[0][0].set_ylabel("Spatial error (android)")
ax_array[1][0].set_ylabel("Spatial error (iOS)")
ifig.suptitle("Spatial trajectory error v/s quality over multiple timelines")
# ifig.tight_layout()
```
### Split out results by section for the most complex timeline (train_bus_ebike_mtv_ucb)
```
ifig, ax_array = plt.subplots(nrows=2,ncols=4,figsize=(25,10), sharex=True, sharey=True)
timeline_list = ["train_bus_ebike_mtv_ucb"]
for i, tl in enumerate(timeline_list):
for q in range(1,5):
sel_df = spatial_errors_df.query("timeline == @tl & phone_os == 'android' & quality == @q")
if len(sel_df) > 0:
sel_df.boxplot(ax = ax_array[2*i][q-1], column=["error"], by=["section_id"])
ax_array[2*i][q-1].tick_params(axis="x", labelrotation=45)
sel_df = spatial_errors_df.query("timeline == @tl & phone_os == 'ios' & quality == @q")
if len(sel_df) > 0:
sel_df.boxplot(ax = ax_array[2*i+1][q-1], column=["error"], by=["section_id"])
# ax_array[i][].set_title("")
def make_acronym(s):
ssl = s.split("_")
# print("After splitting %s, we get %s" % (s, ssl))
if len(ssl) == 0 or len(ssl[0]) == 0:
return ""
else:
return "".join([ss[0] for ss in ssl])
for q in range(1,5):
ax_array[0][q-1].set_title(q2r_map[q])
curr_ticks = [t.get_text() for t in ax_array[1][q-1].get_xticklabels()]
new_ticks = [make_acronym(t) for t in curr_ticks]
ax_array[1][q-1].set_xticklabels(new_ticks)
print(list(zip(curr_ticks, new_ticks)))
# fig.text(0,0,"%s"% list(zip(curr_ticks, new_ticks)))
timeline_list = ["train_bus_ebike_mtv_ucb"]
for i, tl in enumerate(timeline_list):
unique_sections = spatial_errors_df.query("timeline == @tl").section_id.unique()
ifig, ax_array = plt.subplots(nrows=2,ncols=len(unique_sections),figsize=(40,10), sharex=True, sharey=False)
for sid, s_name in enumerate(unique_sections):
sel_df = spatial_errors_df.query("timeline == @tl & phone_os == 'android' & section_id == @s_name & quality > 0")
if len(sel_df) > 0:
sel_df.boxplot(ax = ax_array[2*i][sid], column=["error"], by=["quality"])
ax_array[2*i][sid].set_title(s_name)
sel_df = spatial_errors_df.query("timeline == @tl & phone_os == 'ios' & section_id == @s_name & quality > 0")
if len(sel_df) > 0:
sel_df.boxplot(ax = ax_array[2*i+1][sid], column=["error"], by=["quality"])
ax_array[2*i+1][sid].set_title("")
# ax_array[i][].set_title("")
```
### Focus only on sections where the max error is > 1000 meters
```
timeline_list = ["train_bus_ebike_mtv_ucb"]
for i, tl in enumerate(timeline_list):
unique_sections = pd.Series(spatial_errors_df.query("timeline == @tl").section_id.unique())
sections_with_outliers_mask = unique_sections.apply(lambda s_name: spatial_errors_df.query("timeline == 'train_bus_ebike_mtv_ucb' & section_id == @s_name").error.max() > 1000)
sections_with_outliers = unique_sections[sections_with_outliers_mask]
ifig, ax_array = plt.subplots(nrows=2,ncols=len(sections_with_outliers),figsize=(17,4), sharex=True, sharey=False)
for sid, s_name in enumerate(sections_with_outliers):
sel_df = spatial_errors_df.query("timeline == @tl & phone_os == 'android' & section_id == @s_name & quality > 0")
if len(sel_df) > 0:
sel_df.boxplot(ax = ax_array[2*i][sid], column=["error"], by=["quality"])
ax_array[2*i][sid].set_title(s_name)
ax_array[2*i][sid].set_xlabel("")
sel_df = spatial_errors_df.query("timeline == @tl & phone_os == 'ios' & section_id == @s_name & quality > 0")
if len(sel_df) > 0:
sel_df.boxplot(ax = ax_array[2*i+1][sid], column=["error"], by=["quality"])
ax_array[2*i+1][sid].set_title("")
print([t.get_text() for t in ax_array[2*i+1][sid].get_xticklabels()])
ax_array[2*i+1][sid].set_xticklabels([q2r_map[int(t.get_text())] for t in ax_array[2*i+1][sid].get_xticklabels() if len(t.get_text()) > 0])
ax_array[2*i+1][sid].set_xlabel("")
ifig.suptitle("")
```
### Validation of outliers
#### (express bus iOS, MAHFDC)
ok, so it looks like the error is non-trivial across all runs, but run #1 is the worst and is responsible for the majority of the outliers. And this is borne out by the map, where on run #1, we end up with points in San Leandro!!
```
spatial_errors_df.query("phone_os == 'ios' & quality == 2 & section_id == 'express_bus' & error > 500").run.unique()
spatial_errors_df.query("phone_os == 'ios' & quality == 2 & section_id == 'express_bus'").boxplot(column="error", by="run")
gt_leg = sd_ucb.get_ground_truth_for_leg("berkeley_to_mtv_SF_express_bus", "express_bus"); print(gt_leg["id"])
curr_map = ezgj.get_map_for_geojson(sd_ucb.get_geojson_for_leg(gt_leg), name="ground_truth")
ezgj.get_fg_for_loc_df(emd.linestring_to_geo_df(eisd.SpecDetails.get_shapes_for_leg(gt_leg)["route"]),
name="gt_points", color="green").add_to(curr_map)
name_err_time = lambda lr: "%d: %d, %s, %s" % (lr["index"], lr["df_idx"], lr["error"], sd_ucb.fmt(lr["ts"], "MM-DD HH:mm:ss"))
error_df = emd.to_loc_df(spatial_errors_df.query("phone_os == 'ios' & quality == 2 & section_id == 'express_bus' & run == 1"))
gt_16k = lambda lr: lr["error"] == error_df.error.max()
folium.GeoJson(ezgj.get_geojson_for_loc_df(error_df, color="red"), name="sensed_values").add_to(curr_map)
ezgj.get_fg_for_loc_df(error_df, name="sensed_points", color="red", popupfn=name_err_time, stickyfn=gt_16k).add_to(curr_map)
folium.LayerControl().add_to(curr_map)
curr_map
importlib.reload(ezgj)
gt_leg = sd_ucb.get_ground_truth_for_leg("berkeley_to_mtv_SF_express_bus", "express_bus"); print(gt_leg["id"])
curr_map = ezgj.get_map_for_geojson(sd_ucb.get_geojson_for_leg(gt_leg), name="ground_truth")
ezgj.get_fg_for_loc_df(emd.linestring_to_geo_df(eisd.SpecDetails.get_shapes_for_leg(gt_leg)["route"]),
name="gt_points", color="green").add_to(curr_map)
name_err_time = lambda lr: "%d: %d, %s, %s" % (lr["index"], lr["df_idx"], lr["error"], sd_ucb.fmt(lr["ts"], "MM-DD HH:mm:ss"))
colors = ["red", "yellow", "blue"]
for run in range(3):
error_df = emd.to_loc_df(spatial_errors_df.query("phone_os == 'ios' & quality == 2 & section_id == 'express_bus' & run == @run"))
gt_16k = lambda lr: lr["error"] == error_df.error.max()
print("max error for run %d is %s" % (run, error_df.error.max()))
folium.GeoJson(ezgj.get_geojson_for_loc_df(error_df, color=colors[run]), name="sensed_values").add_to(curr_map)
ezgj.get_fg_for_loc_df(error_df, name="sensed_points", color=colors[run], popupfn=name_err_time, stickyfn=gt_16k).add_to(curr_map)
folium.LayerControl().add_to(curr_map)
curr_map
```
#### (commuter rail aboveground android, HAMFDC)
Run 0: Multiple outliers at the start in San Jose. After that, everything is fine.
```
spatial_errors_df.query("phone_os == 'android' & quality == 1 & section_id == 'commuter_rail_aboveground' & error > 500").run.unique()
spatial_errors_df.query("phone_os == 'android' & quality == 1 & section_id == 'commuter_rail_aboveground' & error > 500").boxplot(column="error", by="run")
gt_leg = sd_ucb.get_ground_truth_for_leg("mtv_to_berkeley_sf_bart", "commuter_rail_aboveground"); print(gt_leg["id"])
curr_map = ezgj.get_map_for_geojson(sd_ucb.get_geojson_for_leg(gt_leg), name="ground_truth")
ezgj.get_fg_for_loc_df(emd.linestring_to_geo_df(eisd.SpecDetails.get_shapes_for_leg(gt_leg)["route"]),
name="gt_points", color="green").add_to(curr_map)
name_err_time = lambda lr: "%d: %d, %s, %s" % (lr["index"], lr["df_idx"], lr["error"], sd_ucb.fmt(lr["ts"], "MM-DD HH:mm:ss"))
error_df = emd.to_loc_df(spatial_errors_df.query("phone_os == 'android' & quality == 1 & section_id == 'commuter_rail_aboveground' & run == 0"))
maxes = [error_df.error.max(), error_df[error_df.error < 10000].error.max(), error_df[error_df.error < 1000].error.max()]
gt_16k = lambda lr: lr["error"] in maxes
folium.GeoJson(ezgj.get_geojson_for_loc_df(error_df, color="red"), name="sensed_values").add_to(curr_map)
ezgj.get_fg_for_loc_df(error_df, name="sensed_points", color="red", popupfn=name_err_time, stickyfn=gt_16k).add_to(curr_map)
folium.LayerControl().add_to(curr_map)
curr_map
spatial_errors_df.query("phone_os == 'android' & quality == 1 & section_id == 'commuter_rail_aboveground' & error > 10000")
```
#### (walk_to_bus android, HAMFDC, HAHFDC)
Huge zig zag when we get out of the BART station
```
spatial_errors_df.query("phone_os == 'android' & (quality == 1 | quality == 3) & section_id == 'walk_to_bus' & error > 500").run.unique()
spatial_errors_df.query("phone_os == 'android' & (quality == 1 | quality == 3) & section_id == 'walk_to_bus' & error > 500")
spatial_errors_df.query("phone_os == 'android' & (quality == 1 | quality == 3) & section_id == 'walk_to_bus'").boxplot(column="error", by="run")
spatial_errors_df.query("phone_os == 'android' & (quality == 1 | quality == 3) & section_id == 'walk_to_bus'").error.max()
error_df
ucb_and_back = pv_ucb.map()["android"]["ucb-sdb-android-2"]["evaluation_ranges"][0]; ucb_and_back["trip_id"]
to_trip = ucb_and_back["evaluation_trip_ranges"][0]; print(to_trip["trip_id"])
wb_leg = to_trip["evaluation_section_ranges"][6]; print(wb_leg["trip_id"])
gt_leg = sd_ucb.get_ground_truth_for_leg(to_trip["trip_id_base"], wb_leg["trip_id_base"]); gt_leg["id"]
importlib.reload(ezgj)
gt_leg = sd_ucb.get_ground_truth_for_leg("mtv_to_berkeley_sf_bart", "walk_to_bus"); print(gt_leg["id"])
curr_map = ezgj.get_map_for_geojson(sd_ucb.get_geojson_for_leg(gt_leg), name="ground_truth")
ezgj.get_fg_for_loc_df(emd.linestring_to_geo_df(eisd.SpecDetails.get_shapes_for_leg(gt_leg)["route"]),
name="gt_points", color="green").add_to(curr_map)
name_err_time = lambda lr: "%d: %d, %s, %s" % (lr["index"], lr["df_idx"], lr["error"], sd_ucb.fmt(lr["ts"], "MM-DD HH:mm:ss"))
error_df = emd.to_loc_df(spatial_errors_df.query("phone_os == 'android' & quality == 3 & section_id == 'walk_to_bus'").sort_index(axis="index"))
maxes = [error_df.error.max(), error_df[error_df.error < 16000].error.max(), error_df[error_df.error < 5000].error.max()]
gt_16k = lambda lr: lr["error"] in maxes
print("Checking errors %s" % maxes)
folium.GeoJson(ezgj.get_geojson_for_loc_df(error_df, color="red"), name="sensed_values").add_to(curr_map)
ezgj.get_fg_for_loc_df(error_df, name="sensed_points", color="red", popupfn=name_err_time, stickyfn=gt_16k).add_to(curr_map)
folium.LayerControl().add_to(curr_map)
curr_map
```
#### (light_rail_below_above_ground, android, accuracy_control)
ok, so it looks like the error is non-trivial across all runs, but run #1 is the worst and is responsible for the majority of the outliers. And this is borne out by the map, where on run #1, we end up with points in San Leandro!!
```
spatial_errors_df.query("phone_os == 'android' & quality == 4 & section_id == 'light_rail_below_above_ground' & error > 100").run.unique()
spatial_errors_df.query("phone_os == 'android' & (quality == 4) & section_id == 'light_rail_below_above_ground'").boxplot(column="error", by="run")
ucb_and_back = pv_ucb.map()["android"]["ucb-sdb-android-2"]["evaluation_ranges"][0]; ucb_and_back["trip_id"]
back_trip = ucb_and_back["evaluation_trip_ranges"][2]; print(back_trip["trip_id"])
lt_leg = back_trip["evaluation_section_ranges"][7]; print(lt_leg["trip_id"])
gt_leg = sd_ucb.get_ground_truth_for_leg(back_trip["trip_id_base"], lt_leg["trip_id_base"]); gt_leg["id"]
import folium
gt_leg = sd_ucb.get_ground_truth_for_leg("berkeley_to_mtv_SF_express_bus", "light_rail_below_above_ground"); print(gt_leg["id"])
curr_map = ezgj.get_map_for_geojson(sd_ucb.get_geojson_for_leg(gt_leg), name="ground_truth")
ezgj.get_fg_for_loc_df(emd.linestring_to_geo_df(eisd.SpecDetails.get_shapes_for_leg(gt_leg)["route"]),
name="gt_points", color="green").add_to(curr_map)
name_err_time = lambda lr: "%d: %d, %s, %s" % (lr["index"], lr["df_idx"], lr["error"], sd_ucb.fmt(lr["ts"], "MM-DD HH:mm:ss"))
colors = ["red", "yellow", "blue"]
for run in range(3):
error_df = emd.to_loc_df(spatial_errors_df.query("phone_os == 'android' & quality == 4 & section_id == 'light_rail_below_above_ground' & run == @run"))
gt_16k = lambda lr: lr["error"] == error_df.error.max()
print("max error for run %d is %s" % (run, error_df.error.max()))
folium.GeoJson(ezgj.get_geojson_for_loc_df(error_df, color=colors[run]), name="sensed_values").add_to(curr_map)
ezgj.get_fg_for_loc_df(error_df, name="sensed_points", color=colors[run], popupfn=name_err_time, stickyfn=gt_16k).add_to(curr_map)
folium.LayerControl().add_to(curr_map)
curr_map
```
#### (subway, android, HAMFDC)
This is the poster child for temporal accuracy tracking
```
bart_leg = pv_ucb.map()["android"]["ucb-sdb-android-3"]["evaluation_ranges"][0]["evaluation_trip_ranges"][0]["evaluation_section_ranges"][5]
gt_leg = sd_ucb.get_ground_truth_for_leg("mtv_to_berkeley_sf_bart", "subway_underground"); gt_leg["id"]
gt_leg = sd_ucb.get_ground_truth_for_leg("mtv_to_berkeley_sf_bart", "subway_underground"); print(gt_leg["id"])
curr_map = ezgj.get_map_for_geojson(sd_ucb.get_geojson_for_leg(gt_leg), name="ground_truth")
ezgj.get_fg_for_loc_df(emd.linestring_to_geo_df(eisd.SpecDetails.get_shapes_for_leg(gt_leg)["route"]),
name="gt_points", color="green").add_to(curr_map)
name_err_time = lambda lr: "%d: %d, %s, %s" % (lr["index"], lr["df_idx"], lr["error"], sd_ucb.fmt(lr["ts"], "MM-DD HH:mm:ss"))
error_df = emd.to_loc_df(spatial_errors_df.query("phone_os == 'android' & quality == 1 & section_id == 'subway_underground' & run == 0").sort_index(axis="index"))
maxes = [error_df.error.max(), error_df[error_df.error < 16000].error.max(), error_df[error_df.error < 5000].error.max()]
gt_16k = lambda lr: lr["error"] in maxes
print("Checking errors %s" % maxes)
folium.GeoJson(ezgj.get_geojson_for_loc_df(error_df, color="red"), name="sensed_values").add_to(curr_map)
ezgj.get_fg_for_loc_df(error_df, name="sensed_points", color="red", popupfn=name_err_time, stickyfn=gt_16k).add_to(curr_map)
folium.LayerControl().add_to(curr_map)
curr_map
gt_leg = sd_ucb.get_ground_truth_for_leg("mtv_to_berkeley_sf_bart", "subway_underground"); gt_leg["id"]
eisd.SpecDetails.get_shapes_for_leg(gt_leg)["route"].is_simple
pd.concat([
error_df.iloc[40:50],
error_df.iloc[55:60],
error_df.iloc[65:75],
error_df.iloc[70:75]])
import pyproj
latlonProj = pyproj.Proj(init="epsg:4326")
xyProj = pyproj.Proj(init="epsg:3395")
xy = pyproj.transform(latlonProj, xyProj, -122.08355963230133, 37.39091642895306); xy
pyproj.transform(xyProj, latlonProj, xy[0], xy[1])
import pandas as pd
df = pd.DataFrame({"a": [1,2,3], "b": [4,5,6]}); df
pd.concat([pd.DataFrame([{"a": 10, "b": 14}]), df, pd.DataFrame([{"a": 20, "b": 24}])], axis='index').reset_index(drop=True)
```
|
github_jupyter
|
<img width="100" src="https://carbonplan-assets.s3.amazonaws.com/monogram/dark-small.png" style="margin-left:0px;margin-top:20px"/>
# Forest Emissions Tracking - Validation
_CarbonPlan ClimateTrace Team_
This notebook compares our estimates of country-level forest emissions to prior estimates from other
groups. The notebook currently compares againsts:
- Global Forest Watch (Zarin et al. 2016)
- Global Carbon Project (Friedlingstein et al. 2020)
```
import geopandas
import pandas as pd
from io import StringIO
import matplotlib.pyplot as plt
from carbonplan_styles.mpl import set_theme
set_theme()
# Input data
# ----------
# country shapes from GADM36
countries = geopandas.read_file("s3://carbonplan-climatetrace/inputs/shapes/countries.shp")
# CarbonPlan's emissions
emissions = pd.read_csv("s3://carbonplan-climatetrace/v0.1/country_rollups.csv")
# GFW emissions
gfw_emissions = pd.read_excel(
"s3://carbonplan-climatetrace/validation/gfw_global_emissions.xlsx",
sheet_name="Country co2 emissions",
).dropna(axis=0)
gfw_emissions = gfw_emissions[gfw_emissions["threshold"] == 10] # select threshold
# Global Carbon Project
gcp_emissions = (
pd.read_excel(
"s3://carbonplan-climatetrace/validation/Global_Carbon_Budget_2020v1.0.xlsx",
sheet_name="Land-Use Change Emissions",
skiprows=28,
)
.dropna(axis=1)
.set_index("Year")
)
gcp_emissions *= 3.664 # C->CO2
gcp_emissions.index = [pd.to_datetime(f"{y}-01-01") for y in gcp_emissions.index]
gcp_emissions = gcp_emissions[["GCB", "H&N", "BLUE", "OSCAR"]]
# Merge emissions dataframes with countries GeoDataFrame
gfw_counties = countries.merge(gfw_emissions.rename(columns={"country": "name"}), on="name")
trace_counties = countries.merge(emissions.rename(columns={"iso3_country": "alpha3"}), on="alpha3")
# reformat to "wide" format (time x country)
trace_wide = (
emissions.drop(columns=["end_date"])
.pivot(index="begin_date", columns="iso3_country")
.droplevel(0, axis=1)
)
trace_wide.index = pd.to_datetime(trace_wide.index)
gfw_wide = gfw_emissions.set_index("country").filter(regex="whrc_aboveground_co2_emissions_Mg_.*").T
gfw_wide.index = [pd.to_datetime(f"{l[-4:]}-01-01") for l in gfw_wide.index]
gfw_wide.head()
```
## Part 1 - Compare time-averaged country emissions (tropics only)
```
# Create a new dataframe with average emissions
avg_emissions = countries.set_index("alpha3")
avg_emissions["trace"] = trace_wide.mean().transpose()
avg_emissions = avg_emissions.set_index("name")
avg_emissions["gfw"] = gfw_wide.mean().transpose() / 1e9
# Scatter Plot
avg_emissions.plot.scatter("gfw", "trace")
plt.ylabel("Trace [Tg CO2e]")
plt.xlabel("GFW [Tg CO2e]")
```
## Part 2 - Maps of Tropical Emissions
```
avg_emissions_nonan = avg_emissions.dropna()
kwargs = dict(
legend=True,
legend_kwds={"orientation": "horizontal", "label": "Emissions [Tg CO2e]"},
lw=0.25,
cmap="Reds",
vmin=0,
vmax=1,
)
avg_emissions_nonan.plot("trace", **kwargs)
plt.title("Trace v0")
avg_emissions_nonan.plot("gfw", **kwargs)
plt.title("GFW Tropics")
kwargs = dict(
legend=True,
legend_kwds={
"orientation": "horizontal",
"label": "Emissions Difference [%]",
},
lw=0.25,
cmap="RdBu_r",
vmin=-40,
vmax=40,
)
avg_emissions_nonan["pdiff"] = (
(avg_emissions_nonan["trace"] - avg_emissions_nonan["gfw"]) / avg_emissions_nonan["gfw"]
) * 100
avg_emissions_nonan.plot("pdiff", **kwargs)
plt.title("% difference")
```
## Part 3 - Compare global emissions timeseries to Global Carbon Project
```
ax = gcp_emissions[["H&N", "BLUE", "OSCAR"]].loc["2000":].plot(ls="--")
gcp_emissions["GCB"].loc["2000":].plot(ax=ax, label="GCB", lw=3)
trace_wide.sum(axis=1).plot(ax=ax, label="Trace v0", c="k", lw=3)
plt.ylabel("Emissions [Tg CO2e]")
plt.legend()
```
# Part 4 - Compare global emissions with those of other inventories
#### load in the inventory file from climate trace which aggregated multiple inventories (e.g. GCP, EDGAR, CAIT) into one place
```
inventories_df = pd.read_csv(
"s3://carbonplan-climatetrace/validation/210623_all_inventory_data.csv"
)
```
The following inventories are included:
{'CAIT', 'ClimateTRACE', 'EDGAR', 'GCP', 'PIK-CR', 'PIK-TP', 'carbon monitor', 'unfccc',
'unfccc_nai'}
```
set(inventories_df["Data source"].values)
def select_inventory_timeseries(df, inventory=None, country=None, sector=None):
if inventory is not None:
df = df[df["Data source"] == inventory]
if country is not None:
df = df[df["Country"] == country]
if sector is not None:
df = df[df["Sector"] == sector]
return df
```
### access the different inventories and compare with our estimates. country-level comparisons are to-do.
```
select_inventory_timeseries(inventories_df, country="Brazil", inventory="CAIT")
select_inventory_timeseries(
inventories_df,
country="United States of America",
inventory="unfccc",
sector="4.A Forest Land",
)
```
### todo: compare our estimates with these and the same from xu2021
|
github_jupyter
|
*Call expressions* invoke [functions](functions), which are named operations.
The name of the function appears first, followed by expressions in
parentheses.
For example, `abs` is a function that returns the absolute value of the input
argument:
```
abs(-12)
```
`round` is a function that returns the input argument rounded to the nearest integer (counting number).
```
round(5 - 1.3)
max(2, 5, 4)
```
In this last example, the `max` function is *called* on three *arguments*: 2,
5, and 4. The value of each expression within parentheses is passed to the
function, and the function *returns* the final value of the full call
expression. You separate the expressions with commas: `,`. The `max` function
can take any number of arguments and returns the maximum.
Many functions, like `max` can accept a variable number of arguments.
`round` is an example. If you call `round` with one argument, it returns the number rounded to the nearest integer, as you have already seen:
```
round(3.3333)
```
You can also call round with two arguments, where the first argument is the number you want to round, and the second argument is the number of decimal places you want to round to. If you don't pass this second argument, `round` assumes you mean 0, corresponding to no decimal places, and rounding to the nearest integer:
```
# The same as above, rounding to 0 decimal places.
round(3.3333, 0)
```
You can also round to - say - 2 decimal places, like this:
```
# Rounding to 2 decimal places.
round(3.3333, 2)
```
A few functions are available by default, such as `abs` and `round`, but most
functions that are built into the Python language are stored in a collection
of functions called a *module*. An *import statement* is used to provide
access to a module, such as `math`.
```
import math
math.sqrt(5)
```
Operators and call expressions can be used together in an expression. The
*percent difference* between two values is used to compare values for which
neither one is obviously `initial` or `changed`. For example, in 2014 Florida
farms produced 2.72 billion eggs while Iowa farms produced 16.25 billion eggs
[^eggs]. The percent difference is 100 times the absolute value of the
difference between the values, divided by their average. In this case, the
difference is larger than the average, and so the percent difference is
greater than 100.
[^eggs]: <http://quickstats.nass.usda.gov>
```
florida = 2.72
iowa = 16.25
100*abs(florida-iowa)/((florida+iowa)/2)
```
Learning how different functions behave is an important part of learning a
programming language. A Jupyter notebook can assist in remembering the names
and effects of different functions. When editing a code cell, press the *tab*
key after typing the beginning of a name to bring up a list of ways to
complete that name. For example, press *tab* after `math.` to see all of the
functions available in the `math` module. Typing will narrow down the list of
options. To learn more about a function, place a `?` after its name. For
example, typing `math.sin?` will bring up a description of the `sin`
function in the `math` module. Try it now. You should get something like
this:
```
sqrt(x)
Return the square root of x.
```
The list of [Python's built-in
functions](https://docs.python.org/3/library/functions.html) is quite long and
includes many functions that are never needed in data science applications.
The list of [mathematical functions in the `math`
module](https://docs.python.org/3/library/math.html) is similarly long. This
text will introduce the most important functions in context, rather than
expecting the reader to memorize or understand these lists.
### Example ###
In 1869, a French civil engineer named Charles Joseph Minard created what is
still considered one of the greatest graphs of all time. It shows the
decimation of Napoleon's army during its retreat from Moscow. In 1812,
Napoleon had set out to conquer Russia, with over 350,000 men in his army.
They did reach Moscow but were plagued by losses along the way. The Russian
army kept retreating farther and farther into Russia, deliberately burning
fields and destroying villages as it retreated. This left the French army
without food or shelter as the brutal Russian winter began to set in. The
French army turned back without a decisive victory in Moscow. The weather got
colder and more men died. Fewer than 10,000 returned.

The graph is drawn over a map of eastern Europe. It starts at the
Polish-Russian border at the left end. The light brown band represents
Napoleon's army marching towards Moscow, and the black band represents the
army returning. At each point of the graph, the width of the band is
proportional to the number of soldiers in the army. At the bottom of the
graph, Minard includes the temperatures on the return journey.
Notice how narrow the black band becomes as the army heads back. The crossing
of the Berezina river was particularly devastating; can you spot it on the
graph?
The graph is remarkable for its simplicity and power. In a single graph,
Minard shows six variables:
- the number of soldiers
- the direction of the march
- the latitude and longitude of location
- the temperature on the return journey
- the location on specific dates in November and December
Tufte says that Minard's graph is "probably the best statistical graphic ever
drawn."
Here is a subset of Minard's data, adapted from *The Grammar of Graphics* by
Leland Wilkinson.

Each row of the column represents the state of the army in a particular
location. The columns show the longitude and latitude in degrees, the name of
the location, whether the army was advancing or in retreat, and an estimate of
the number of men.
In this table the biggest change in the number of men between two consecutive
locations is when the retreat begins at Moscow, as is the biggest percentage
change.
```
moscou = 100000
wixma = 55000
wixma - moscou
(wixma - moscou)/moscou
```
That's a 45% drop in the number of men in the fighting at Moscow. In other
words, almost half of Napoleon's men who made it into Moscow didn't get very
much farther.
As you can see in the graph, Moiodexno is pretty close to Kowno where the army
started out. Fewer than 10% of the men who marched into Smolensk during the
advance made it as far as Moiodexno on the way back.
```
smolensk_A = 145000
moiodexno = 12000
(moiodexno - smolensk_A)/smolensk_A
```
Yes, you could do these calculations by just using the numbers without names.
But the names make it much easier to read the code and interpret the results.
It is worth noting that bigger absolute changes don't always correspond to
bigger percentage changes.
The absolute loss from Smolensk to Dorogobouge during the advance was 5,000
men, whereas the corresponding loss from Smolensk to Orscha during the retreat
was smaller, at 4,000 men.
However, the percent change was much larger between Smolensk and Orscha
because the total number of men in Smolensk was much smaller during the
retreat.
```
dorogobouge = 140000
smolensk_R = 24000
orscha = 20000
abs(dorogobouge - smolensk_A)
abs(dorogobouge - smolensk_A)/smolensk_A
abs(orscha - smolensk_R)
abs(orscha - smolensk_R)/smolensk_R
```
{% data8page Calls %}
|
github_jupyter
|
## Don't worry if you don't understand everything at first! You're not supposed to. We will start using some "black boxes" and then we'll dig into the lower level details later.
## To start, focus on what things DO, not what they ARE.
# What is NLP?
Natural Language Processing is technique where computers try an understand human language and make meaning out of it.
NLP is a broad field, encompassing a variety of tasks, including:
1. Part-of-speech tagging: identify if each word is a noun, verb, adjective, etc.)
2. Named entity recognition NER): identify person names, organizations, locations, medical codes, time expressions, quantities, monetary values, etc)
3. Question answering
4. Speech recognition
5. Text-to-speech and Speech-to-text
6. Topic modeling
7. Sentiment classification
9. Language modeling
10. Translation
# What is NLU?
Natural Language Understanding is all about understanding the natural language.
Goals of NLU
1. Gain insights into cognition
2. Develop Artifical Intelligent agents as an assistant.
# What is NLG?
Natural language generation is the natural language processing task of generating natural language from a machine representation system such as a knowledge base or a logical form.
Example applications of NLG
1. Recommendation and Comparison
2. Report Generation –Summarization
3. Paraphrase
4. Prompt and response generation in dialogue systems
# Packages
1. [Flair](https://github.com/zalandoresearch/flair)
2. [Allen NLP](https://github.com/allenai/allennlp)
3. [Deep Pavlov](https://github.com/deepmipt/deeppavlov)
4. [Pytext](https://github.com/facebookresearch/PyText)
5. [NLTK](https://www.nltk.org/)
6. [Hugging Face Pytorch Transformer](https://github.com/huggingface/pytorch-transformers)
7. [Spacy](https://spacy.io/)
8. [torchtext](https://torchtext.readthedocs.io/en/latest/)
9. [Ekphrasis](https://github.com/cbaziotis/ekphrasis)
10. [Genism](https://radimrehurek.com/gensim/)
# NLP Pipeline
## Data Collection
### Sources
For Generative Training :- Where the model has to learn about the data and its distribution
1. News Article:- Archives
2. Wikipedia Article
3. Book Corpus
4. Crawling the Internet for webpages.
5. Reddit
Generative training on an abundant set of unsupervised data helps in performing Transfer learning for a downstream task where few parameters need to be learnt from sratch and less data is also required.
For Determinstic Training :- Where the model learns about Decision boundary within the data.
Generic
1. Kaggle Dataset
Sentiment
1. Product Reviews :- Amazon, Flipkart
Emotion:-
1. ISEAR
2. Twitter dataset
Question Answering:-
1. SQUAD
etc.
### For Vernacular text
In vernacular context we have crisis in data especially when it comes to state specific language in India. (Ex. Bengali, Gujurati etc.)
Few Sources are:-
1. News (Jagran.com, Danik bhaskar)
2. Moview reviews (Web Duniya)
3. Hindi Wikipedia
4. Book Corpus
6. IIT Bombay (English-Hindi Parallel Corpus)
### Tools
1. Scrapy :- Simple, Extensible framework for scraping and crawling websites. Has numerous feature into it.
2. Beautiful-Soup :- For Parsing Html and xml documents.
3. Excel
4. wikiextractor:- A tool for extracting plain text from Wikipedia dumps
### Data Annotation Tool
1. TagTog
2. Prodigy (Explosion AI)
3. Mechanical Turk
4. PyBossa
5. Chakki-works Doccano
6. WebAnno
7. Brat
## Data Preprocessing
1. Cleaning
2. Regex
1. Url Cleanup
2. HTML Tag
3. Date
4. Numbers
5. Lingos
6. Emoticons
3. Lemmatization
4. Stemming
5. Chunking
6. POS Tags
7. NER Tags
8. Stopwords
9. Tokenizers
10. Spell Correction
11. Word Segmentation
12. Word Processing
1. Elongated
2. Repeated
3. All Caps
### Feature Selection
1. Bag of Words
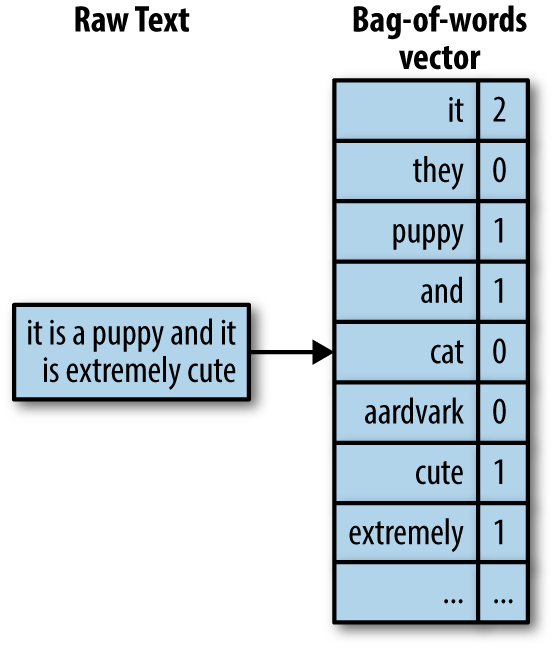
2. TF-IDF

3. Word Embeddings
1. Word2Vec
Word2Vec is a predictive model.

2. Glove
Glove is a Count-based models learn their vectors by essentially doing dimensionality reduction on the co-occurrence counts matrix.
3. FastText
Fastext is trained in a similar fashion how word2vec model is trained, the only difference is the fastext enchriches the word vectors with subword units.
[FastText works](https://www.quora.com/What-is-the-main-difference-between-word2vec-and-fastText)
4. ELMO
ELMo is a deep contextualized word representation that models both (1) complex characteristics of word use (e.g., syntax and semantics), and (2) how these uses vary across linguistic contexts (i.e., to model polysemy). These word vectors are learned functions of the internal states of a deep bidirectional language model (biLM), which is pre-trained on a large text corpus. They can be easily added to existing models and significantly improve the state of the art across a broad range of challenging NLP problems, including question answering, textual entailment and sentiment analysis.
ELMo representations are:
* Contextual: The representation for each word depends on the entire context in which it is used.
* Deep: The word representations combine all layers of a deep pre-trained neural network.
* Character based: ELMo representations are purely character based, allowing the network to use morphological clues to form robust representations for out-of-vocabulary tokens unseen in training.
### Modelling
1. RNN

RNN suffers from gradient vanishing problem and they do not persist long term dependencies.
2. LSTM
Long Short Term Memory networks – usually just called “LSTMs” – are a special kind of RNN, capable of learning long-term dependencies.
LSTMs are explicitly designed to avoid the long-term dependency problem. Remembering information for long periods of time is practically their default behavior, not something they struggle to learn!

3. BI-LSTM

4. GRU
5. CNNs
6. Seq-Seq

7. Seq-Seq Attention

8. Pointer Generator Network

8. Transformer

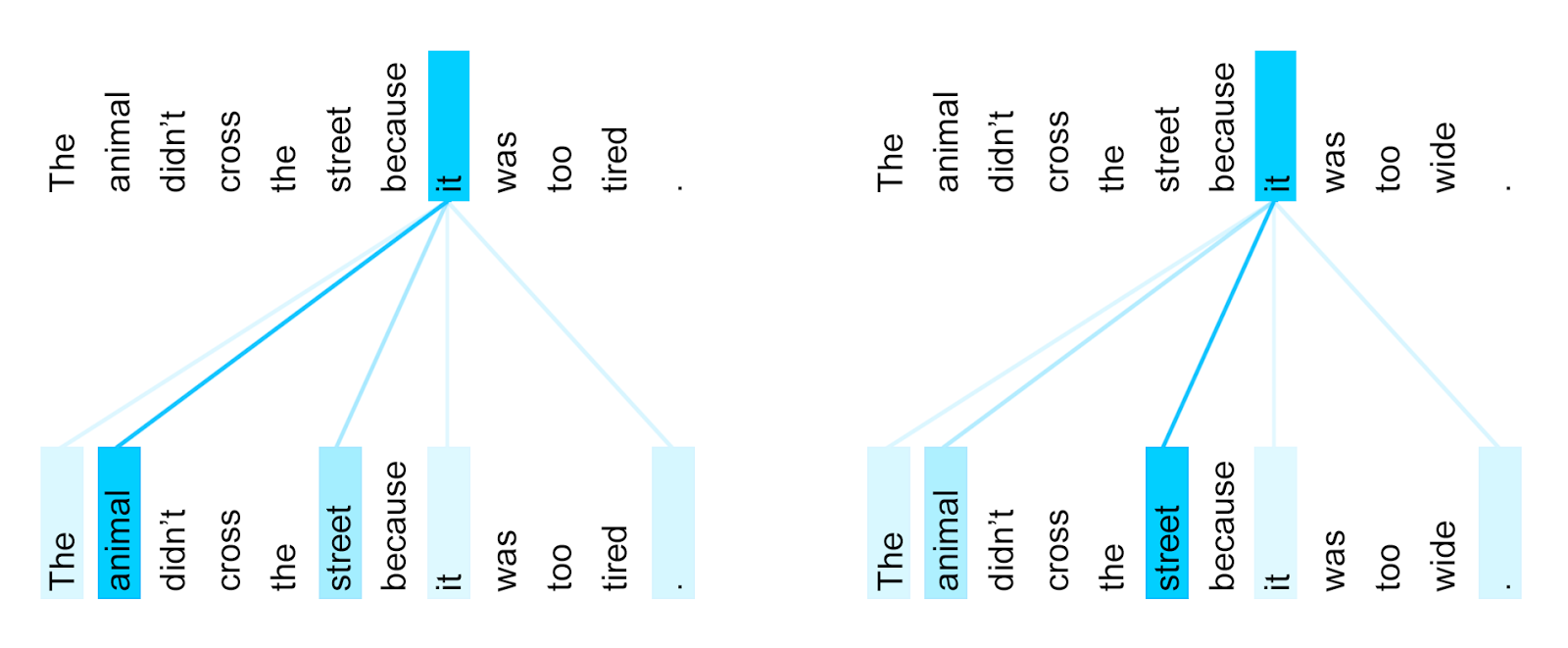
9. GPT

10. Transformer-XL

11. BERT
BERT’s key technical innovation is applying the bidirectional training of Transformer, a popular attention model, to language modelling.
BERT is given billions of sentences at training time. It’s then asked to predict a random selection of missing words from these sentences. After practicing with this corpus of text several times over, BERT adopts a pretty good understanding of how a sentence fits together grammatically. It’s also better at predicting ideas that are likely to show up together.
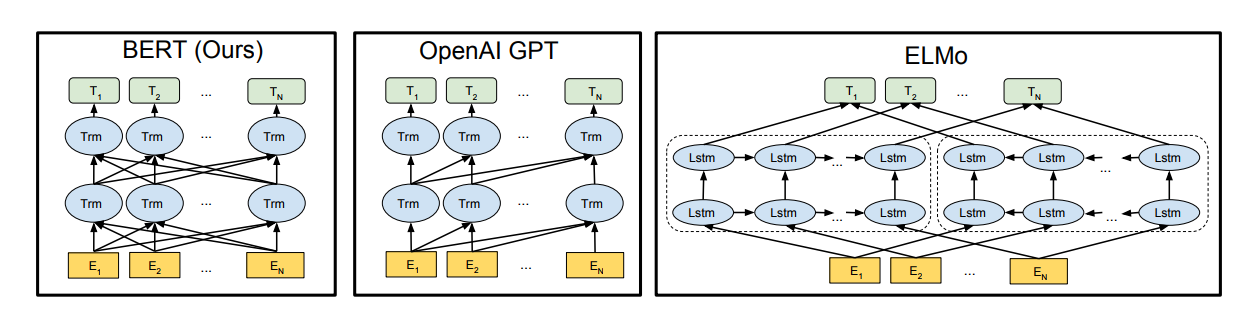
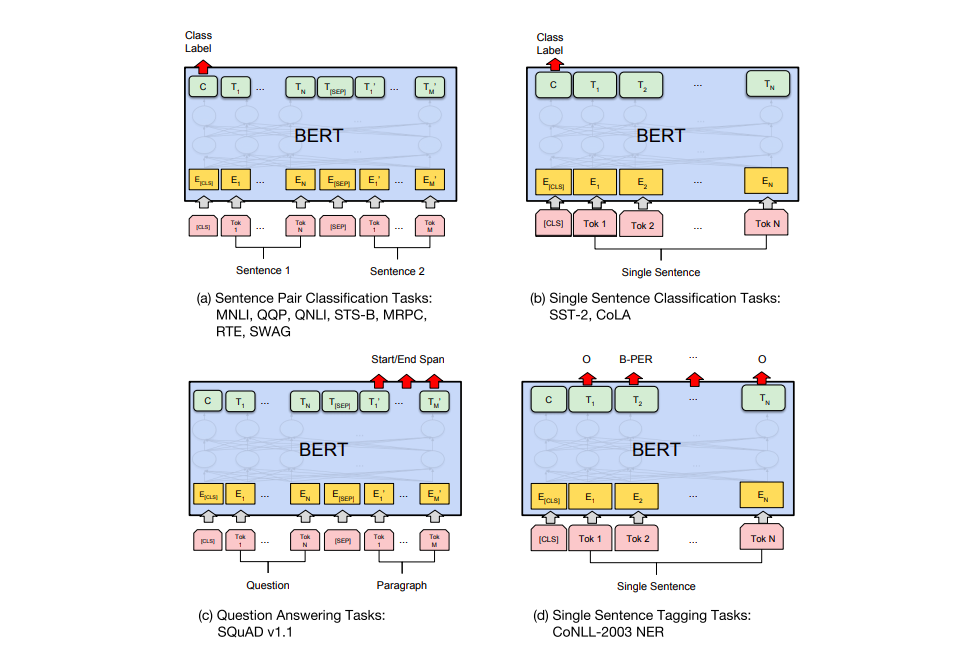
12. GPT-2

## Buisness Problem
1. Text Classification
1. Sentiment Classification
2. Emotion Classification
3. Reviews Rating
2. Topic Modeling
3. Named Entity Recognition
4. Part Of Speech Tagging
5. Language Model
6. Machine Translation
7. Question Answering
8. Text Summarization
9. Text Generation
10. Image Captioning
11. Optical Character Recognition
12. Chatbots
13. [Dependency Parsing](https://nlpprogress.com/english/dependency_parsing.html)
14. [Coreference Resolution](https://en.wikipedia.org/wiki/Coreference)
15. [Semantic Textual Similarity](https://nlpprogress.com/english/semantic_textual_similarity.html)
|
github_jupyter
|
To aid autoassociative recall (sparse recall using partial pattern), we need to two components -
1. each pattern remembers a soft mask of the contribution of each
element in activating it. For example, if an element varies a lot at high activation levels, that element should be masked out when determining activation. On the other hand, if an element has a very specific value every time the element has high activation, then that element is important and should be considered (masked-in).
2. Among the masked-in elements for a pattern, even a small subset (say 20%) perfect match should be able to activate the pattern. This can be achieved by considering number of elements that have similarity above a threshold, say 0.9. Sum up similarity of this subset and apply an activation curve that is a sharp sigmoid centered at a value that represents (# of masked-in element) * 0.2 * 0.9.
```
import math
import torch
import matplotlib.pyplot as plt
import pdb
import pandas as pd
import seaborn as sns
import numpy as np
# import plotly.graph_objects as go
from matplotlib.patches import Ellipse
%matplotlib inline
device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
print(device)
from sklearn.datasets import load_boston
def normalize(df):
df1 = (df - df.mean())/df.std()
return df1
def scale(df):
min = df.min()
max = df.max()
df1 = (df - min) / (max - min)
return df1
dataset = load_boston()
dataset = pd.DataFrame(dataset.data, columns=dataset.feature_names)
dataset = pd.DataFrame(np.c_[scale(normalize(dataset['LSTAT'])), scale(normalize(dataset['RM']))], columns = ['LSTAT','RM'])
dataset = torch.tensor(dataset.to_numpy()).float().to(device)
dataset1 = dataset[dataset[:,0] < 0.33]
dataset2 = dataset[(dataset[:,0] >= 0.33) & (dataset[:,0] < 0.66)]
dataset3 = dataset[dataset[:,0] >= 0.66]
# dataset = [[0.25, 0.4], [0.75, 0.75], [0.85, 0.65]]
original_dataset = dataset
print("dataset", dataset.shape)
# from https://kornia.readthedocs.io/en/latest/_modules/kornia/utils/grid.html
from typing import Optional
def create_meshgrid(
height: int,
width: int,
normalized_coordinates: Optional[bool] = True,
device: Optional[torch.device] = torch.device('cpu')) -> torch.Tensor:
"""Generates a coordinate grid for an image.
When the flag `normalized_coordinates` is set to True, the grid is
normalized to be in the range [-1,1] to be consistent with the pytorch
function grid_sample.
http://pytorch.org/docs/master/nn.html#torch.nn.functional.grid_sample
Args:
height (int): the image height (rows).
width (int): the image width (cols).
normalized_coordinates (Optional[bool]): whether to normalize
coordinates in the range [-1, 1] in order to be consistent with the
PyTorch function grid_sample.
Return:
torch.Tensor: returns a grid tensor with shape :math:`(1, H, W, 2)`.
"""
# generate coordinates
xs: Optional[torch.Tensor] = None
ys: Optional[torch.Tensor] = None
if normalized_coordinates:
xs = torch.linspace(-1, 1, width, device=device, dtype=torch.float)
ys = torch.linspace(-1, 1, height, device=device, dtype=torch.float)
else:
xs = torch.linspace(0, width - 1, width, device=device, dtype=torch.float)
ys = torch.linspace(0, height - 1, height, device=device, dtype=torch.float)
# generate grid by stacking coordinates
base_grid: torch.Tensor = torch.stack(
torch.meshgrid([xs, ys])).transpose(1, 2) # 2xHxW
return torch.unsqueeze(base_grid, dim=0).permute(0, 2, 3, 1) # 1xHxWx2
def add_gaussian_noise(tensor, mean=0., std=1.):
t = tensor + torch.randn(tensor.size()).to(device) * std + mean
t.to(device)
return t
def plot_patterns(patterns, pattern_lr, pattern_var, dataset):
patterns = patterns.cpu()
dataset = dataset.cpu()
assert len(patterns.shape) == 2 # (pattern count, 2)
assert patterns.shape[1] == 2 # 2D
rgba_colors = torch.zeros((patterns.shape[0], 4))
# for blue the last column needs to be one
rgba_colors[:,2] = 1.0
# the fourth column needs to be your alphas
alpha = (1.2 - pattern_lr.cpu()).clamp(0, 1) * 0.2
rgba_colors[:, 3] = alpha
# make ellipses
marker_list = []
min_size = 0.02
max_size = 2.0
for i in range(patterns.shape[0]):
pattern = patterns[i]
var = pattern_var[i].clamp(0.01, 2.0)
marker_list.append(Ellipse((pattern[0], pattern[1]), var[0], var[1], edgecolor='none', facecolor=rgba_colors[i], fill=True))
plt.figure(figsize=(7,7), dpi=100)
ax = plt.gca()
ax.cla() # clear things for fresh plot
ax.scatter(patterns[:, 0], patterns[:, 1], marker='.', c='b')
ax.scatter(dataset[:, 0], dataset[:, 1], marker='.', c='r', s=10)
ax.set_xlim(0, 1)
ax.set_ylim(0, 1)
for marker in marker_list:
ax.add_artist(marker)
plt.show()
grid_size = 4
patterns = create_meshgrid(grid_size, grid_size, normalized_coordinates=False).reshape(-1, 2) / (grid_size-1)
patterns = patterns.to(device)
pattern_lr = torch.ones((patterns.shape[0],)).to(device)
pattern_var = torch.ones_like(patterns).to(device) * 0 # start with high var indicating no specificity to any value
# patterns = torch.rand((50, 2))
# patterns = torch.tensor([[0.25, 0.30]])
# patterns
plot_patterns(patterns, pattern_lr, pattern_var, dataset)
original_patterns = patterns.clone().to(device)
def similarity(x, patterns, subset_threshold=0.2):
# Formula derivation https://www.desmos.com/calculator/iokn9kyuaq
# print("x", x)
dist_i = ((x - patterns) ** 2)
dist = dist_i.sum(dim=-1)
# print("patterns", patterns)
# print("dist", dist)
#dist = dist.sum(dim=-1) # TODO: use subset activation # TODO: apply mask (inverse variance)
winner_index = dist.min(dim=0)[1]
# print("winner_index", winner_index)
winning_pattern = patterns[winner_index]
a_scale = 0.2
a = a_scale * ((x - winning_pattern) ** -2)
a[a > 15000.0] = 15000.0
# print("a", a)
s = 0.8
sim = (-a * ((x - patterns) ** 2)).mean(dim=-1)
# print("sim1", sim)
# scale = 0.685
scale = 1.0
sim = (torch.exp(sim) - s * torch.exp(sim * 0.9)) / ((1 - s) * scale)
sim[sim>1.0] = 1.0
# print("sim", sim)
return sim, winner_index, dist, dist_i
sim, winner_index, dist, dist_i = similarity(dataset[0], patterns)
patterns = original_patterns
pattern_lr = torch.ones((patterns.shape[0],)).to(device)
pattern_var = torch.ones_like(patterns).to(device) * 10 # start with high var indicating no specificity to any value
def run_dataset(dataset, patterns, pattern_lr):
for x in dataset:
# print("-------")
sim, winner_index, dist, dist_i = similarity(x, patterns)
sim = sim.unsqueeze(-1)
# print("dist[winner_index]", dist[winner_index] * 100)
pattern_lr[winner_index] = 0.9 * pattern_lr[winner_index] + 0.1 * (1.0 - torch.exp(-dist[winner_index]))
pattern_var[winner_index] = 0.9 * pattern_var[winner_index] + 0.1 * (1.0 - torch.exp(-dist_i[winner_index])) * 100
# print("x", x)
# print("(x - patterns)", (x - patterns))
# print("sim", sim)
delta = (x - patterns) * sim * lr * pattern_lr.unsqueeze(-1)
# print("delta", delta)
patterns = patterns + delta
patterns.clamp_(0, 1)
pattern_lr.clamp(0, 1)
# print("patterns", patterns)
# print("pattern_lr", pattern_lr)
# print("pattern_var", pattern_var)
return patterns, pattern_lr
lr = 1
epochs = 10
noise = 0.0
for _ in range(2):
for i in range(epochs):
dataset = add_gaussian_noise(dataset1, std=noise)
if i % int(epochs / 2) == 0:
print("Iteration ", i)
plot_patterns(patterns, pattern_lr, pattern_var, dataset)
patterns, pattern_lr = run_dataset(dataset, patterns, pattern_lr)
for i in range(epochs):
dataset = add_gaussian_noise(dataset2, std=noise)
if i % int(epochs / 2) == 0:
print("Iteration ", i)
plot_patterns(patterns, pattern_lr, pattern_var, dataset)
patterns, pattern_lr = run_dataset(dataset, patterns, pattern_lr)
for i in range(epochs):
dataset = add_gaussian_noise(dataset3, std=noise)
if i % int(epochs / 2) == 0:
print("Iteration ", i)
plot_patterns(patterns, pattern_lr, pattern_var, dataset)
patterns, pattern_lr = run_dataset(dataset, patterns, pattern_lr)
plot_patterns(patterns, pattern_lr, pattern_var, original_dataset)
```
Notes -
- Patterns that see data (are winners) become "sticky", while rest of the pattern-pool remains more fluid to move towards subspaces that were previously unused. For example, learning an unrelated task. This could implications on meta-learning.
- Available pattern pool gets used to locally optimally represent data. This can be seen by using a small number of patterns (say 3x3) or a large number of patterns (say 100x100). The fact that a dense grid is not required should come in handy to fight the curse of dimentionality.
|
github_jupyter
|
Original samples in https://fslab.org/FSharp.Charting/FurtherSamples.html
```
#load "FSharp.Charting.Paket.fsx"
#load "FSharp.Charting.fsx"
```
## Sample data
```
open FSharp.Charting
open System
open System.Drawing
let data = [ for x in 0 .. 99 -> (x,x*x) ]
let data2 = [ for x in 0 .. 99 -> (x,sin(float x / 10.0)) ]
let data3 = [ for x in 0 .. 99 -> (x,cos(float x / 10.0)) ]
let timeSeriesData =
[ for x in 0 .. 99 -> (DateTime.Now.AddDays (float x),sin(float x / 10.0)) ]
let rnd = new System.Random()
let rand() = rnd.NextDouble()
let pointsWithSizes =
[ for i in 0 .. 30 -> (rand() * 10.0, rand() * 10.0, rand() / 100.0) ]
let pointsWithSizes2 =
[ for i in 0 .. 10 -> (rand() * 10.0, rand() * 10.0, rand() / 100.0) ]
let timeHighLowOpenClose =
[ for i in 0 .. 10 ->
let mid = rand() * 10.0
(DateTime.Now.AddDays (float i), mid + 0.5, mid - 0.5, mid + 0.25, mid - 0.25) ]
let timedPointsWithSizes =
[ for i in 0 .. 30 -> (DateTime.Now.AddDays(rand() * 10.0), rand() * 10.0, rand() / 100.0) ]
```
## Examples
```
Chart.Line(data).WithXAxis(MajorGrid=ChartTypes.Grid(Enabled=false))
Chart.Line [ DateTime.Now, 1; DateTime.Now.AddDays(1.0), 10 ]
Chart.Line [ for h in 1 .. 50 -> DateTime.Now.AddHours(float h), sqrt (float h) ]
Chart.Line [ for h in 1 .. 50 -> DateTime.Now.AddMinutes(float h), sqrt (float h) ]
Chart.Line(data,Title="Test Title")
Chart.Line(data,Title="Test Title").WithTitle(InsideArea=false)
Chart.Line(data,Title="Test Title").WithTitle(InsideArea=true)
Chart.Line(data,Title="Test Title")
|> Chart.WithTitle(InsideArea=true)
Chart.Line(data,Name="Test Data")
|> Chart.WithXAxis(Enabled=true,Title="X Axis")
Chart.Line(data,Name="Test Data")
|> Chart.WithXAxis(Enabled=false,Title="X Axis")
Chart.Line(data,Name="Test Data")
.WithXAxis(Enabled=false,Title="X Axis")
Chart.Line(data,Name="Test Data")
.WithXAxis(Enabled=true,Title="X Axis",Max=10.0, Min=0.0)
.WithYAxis(Max=100.0,Min=0.0)
Chart.Line(data,Name="Test Data").WithLegend(Title="Hello")
Chart.Line(data,Name="Test Data").WithLegend(Title="Hello",Enabled=false)
Chart.Line(data,Name="Test Data").With3D()
// TODO: x/y axis labels are a bit small by default
Chart.Line(data,Name="Test Data",XTitle="hello", YTitle="goodbye")
Chart.Line(data,Name="Test Data").WithXAxis(Title="XXX")
Chart.Line(data,Name="Test Data").WithXAxis(Title="XXX",Max=10.0,Min=4.0)
.WithYAxis(Title="YYY",Max=100.0,Min=4.0,Log=true)
Chart.Combine [ Chart.Line(data,Name="Test Data 1 With Long Name")
Chart.Line(data2,Name="Test Data 2") ]
|> Chart.WithLegend(Enabled=true,Title="Hello",Docking=ChartTypes.Docking.Left)
Chart.Combine [ Chart.Line(data,Name="Test Data 1")
Chart.Line(data2,Name="Test Data 2") ]
|> Chart.WithLegend(Docking=ChartTypes.Docking.Left, InsideArea=true)
Chart.Combine [ Chart.Line(data,Name="Test Data 1")
Chart.Line(data2,Name="Test Data 2") ]
|> Chart.WithLegend(InsideArea=true)
Chart.Rows
[ Chart.Line(data,Title="Chart 1", Name="Test Data 1")
Chart.Line(data2,Title="Chart 2", Name="Test Data 2") ]
|> Chart.WithLegend(Title="Hello",Docking=ChartTypes.Docking.Left)
// TODO: this title and docking left doesn't work
Chart.Columns
[ Chart.Line(data,Name="Test Data 1")
Chart.Line(data2,Name="Test Data 2")]
|> Chart.WithLegend(Title="Hello",Docking=ChartTypes.Docking.Left)
Chart.Combine [ Chart.Line(data,Name="Test Data 1")
Chart.Line(data2,Name="Test Data 2") ]
|> Chart.WithLegend(Title="Hello",Docking=ChartTypes.Docking.Bottom)
Chart.Line(data,Name="Test Data")
Chart.Line(data,Name="Test Data").WithLegend(Enabled=false)
Chart.Line(data,Name="Test Data").WithLegend(InsideArea=true)
Chart.Line(data,Name="Test Data").WithLegend(InsideArea=false)
Chart.Line(data).WithLegend().CopyAsBitmap()
Chart.Line(data)
Chart.Line(data,Name="Test Data").WithLegend(InsideArea=false)
Chart.Area(data)
Chart.Area(timeSeriesData)
Chart.Line(data)
Chart.Bar(data)
Chart.Bar(timeSeriesData)
Chart.Spline(data)
Chart.Spline(timeSeriesData)
Chart.Bubble(pointsWithSizes)
Chart.Bubble(pointsWithSizes)
.WithMarkers(Style=ChartTypes.MarkerStyle.Star10)
Chart.Bubble(pointsWithSizes)
.WithMarkers(Style=ChartTypes.MarkerStyle.Diamond)
Chart.Bubble(pointsWithSizes)
.WithMarkers(Style=ChartTypes.MarkerStyle.Cross,Color=Color.Red)
Chart.Bubble(pointsWithSizes)
.WithMarkers(Style=ChartTypes.MarkerStyle.Cross,Color=Color.Red,MaxPixelPointWidth=3)
Chart.Bubble(pointsWithSizes)
.WithMarkers(Style=ChartTypes.MarkerStyle.Cross,Size=3)
Chart.Bubble(pointsWithSizes)
.WithMarkers(Style=ChartTypes.MarkerStyle.Cross,PointWidth=0.1)
Chart.Bubble(pointsWithSizes)
.WithMarkers(Style=ChartTypes.MarkerStyle.Cross,PixelPointWidth=3)
Chart.Bubble(pointsWithSizes).WithMarkers(Style=ChartTypes.MarkerStyle.Circle)
Chart.Bubble(pointsWithSizes).WithMarkers(Style=ChartTypes.MarkerStyle.Square)
Chart.Bubble(pointsWithSizes).WithMarkers(Style=ChartTypes.MarkerStyle.Star6)
Chart.Combine [ Chart.Bubble(pointsWithSizes,UseSizeForLabel=true) .WithMarkers(Style=ChartTypes.MarkerStyle.Circle)
Chart.Bubble(pointsWithSizes2).WithMarkers(Style=ChartTypes.MarkerStyle.Star10) ]
Chart.Bubble(timedPointsWithSizes)
Chart.Candlestick(timeHighLowOpenClose)
Chart.Column(data)
Chart.Column(timeSeriesData)
Chart.Pie(Name="Pie", data=[ for i in 0 .. 10 -> i, i*i ])
Chart.Pie(Name="Pie", data=timeSeriesData)
Chart.Doughnut(data=[ for i in 0 .. 10 -> i, i*i ])
Chart.Doughnut(timeSeriesData)
Chart.FastPoint [ for x in 1 .. 10000 -> (rand(), rand()) ]
Chart.FastPoint timeSeriesData
Chart.Polar ([ for x in 1 .. 100 -> (360.0*rand(), rand()) ] |> Seq.sortBy fst)
Chart.Pyramid ([ for x in 1 .. 100 -> (360.0*rand(), rand()) ] |> Seq.sortBy fst)
Chart.Radar ([ for x in 1 .. 100 -> (360.0*rand(), rand()) ] |> Seq.sortBy fst)
Chart.Range ([ for x in 1.0 .. 10.0 -> (x, x + rand(), x-rand()) ])
Chart.RangeBar ([ for x in 1.0 .. 10.0 -> (x, x + rand(), x-rand()) ])
Chart.RangeColumn ([ for x in 1.0 .. 10.0 -> (x, x + rand(), x-rand()) ])
Chart.SplineArea ([ for x in 1.0 .. 10.0 -> (x, x + rand()) ])
Chart.SplineRange ([ for x in 1.0 .. 10.0 -> (x, x + rand(), x - rand()) ])
Chart.StackedBar ([ [ for x in 1.0 .. 10.0 -> (x, x + rand()) ];
[ for x in 1.0 .. 10.0 -> (x, x + rand()) ] ])
Chart.StackedColumn ([ [ for x in 1.0 .. 10.0 -> (x, x + rand()) ];
[ for x in 1.0 .. 10.0 -> (x, x + rand()) ] ])
Chart.StackedArea ([ [ for x in 1.0 .. 10.0 -> (x, x + rand()) ];
[ for x in 1.0 .. 10.0 -> (x, x + rand()) ] ])
Chart.StackedArea ([ [ for x in 1.0 .. 10.0 -> (DateTime.Now.AddDays x, x + rand()) ];
[ for x in 1.0 .. 10.0 -> (DateTime.Now.AddDays x, x + rand()) ] ])
Chart.StepLine(data,Name="Test Data").WithLegend(InsideArea=false)
Chart.StepLine(timeSeriesData,Name="Test Data").WithLegend(InsideArea=false)
Chart.Line(data,Name="SomeData").WithDataPointLabels(PointToolTip="Hello, I am #SERIESNAME")
Chart.Stock(timeHighLowOpenClose)
Chart.ThreeLineBreak(data,Name="SomeData").WithDataPointLabels(PointToolTip="Hello, I am #SERIESNAME")
Chart.Histogram([for x in 1 .. 100 -> rand()*10.],LowerBound=0.,UpperBound=10.,Intervals=10.)
// Example of .ApplyToChart() used to alter the settings on the window chart and to access the chart child objects.
// This can normally be done manually, in the chart property grid (right click the chart, then "Show Property Grid").
// This is useful when you want to try out carious settings first. But once you know what you want, .ApplyToChart()
// allows programmatic access to the window properties. The two examples below are: IsUserSelectionEnabled essentially
// allows zooming in and out along the given axes, and the longer fiddly example below does the same work as .WithDataPointLabels()
// but across all series objects.
[ Chart.Column(data);
Chart.Column(data2) |> Chart.WithSeries.AxisType( YAxisType = Windows.Forms.DataVisualization.Charting.AxisType.Secondary ) ]
|> Chart.Combine
|> fun c -> c.WithLegend()
.ApplyToChart( fun c -> c.ChartAreas.[0].CursorX.IsUserSelectionEnabled <- true )
.ApplyToChart( fun c -> let _ = [0 .. c.Series.Count-1] |> List.map ( fun s -> c.Series.[ s ].ToolTip <- "#SERIESNAME (#VALX, #VAL{0:00000})" ) in () )
```
|
github_jupyter
|
# Examples for Bounded Innovation Propagation (BIP) MM ARMA parameter estimation
```
import numpy as np
import scipy.signal as sps
import robustsp as rsp
import matplotlib.pyplot as plt
import matplotlib
# Fix random number generator for reproducibility
np.random.seed(1)
```
## Example 1: AR(1) with 30 percent isolated outliers
```
# Generate AR(1) observations
N = 300
a = np.random.randn(N)
x = sps.lfilter([1],[1,-.8],a)
p = 1
q = 0
```
### Generate isolated Outliers
```
cont_prob = 0.3 # outlier contamination probability
outlier_ind = np.where(np.sign(np.random.rand(N)-cont_prob)<0)# outlier index
outlier = 100*np.random.randn(N) # contaminating process
v = np.zeros(N) # additive outlier signal
v[outlier_ind] = outlier[outlier_ind]
v[0] = 0 # first sample should not be an outlier
x_ao = x+v # 30% of isolated additive outliers
```
### BIP MM Estimation
```
result = rsp.arma_est_bip_mm(x_ao,p,q)
print('Example: AR(1) with ar_coeff = -0.8')
print('30% of isolated additive outliers')
print('estimaed coefficients: %.3f' % result['ar_coeffs'])
%matplotlib inline
matplotlib.rcParams['figure.figsize'] = [10, 10]
plt.subplot(2,1,1)
plt.plot(x_ao,'-',lw=2,label='outlier contaminated AR(1)')
plt.plot(result['cleaned_signal'],'-.',c='y',label='cleaned')
plt.xlabel('samples')
plt.ylabel('Amplitude')
plt.title('BIP-AR(1) cleaned signal')
plt.legend()
plt.subplot(2,1,2)
plt.plot(x,lw=2,label='original AR(1)')
plt.plot(result['cleaned_signal'],'-.',label='cleaned')
plt.xlabel('samples')
plt.ylabel('Amplitude')
plt.title('BIP-AR(1) cleaned signal')
plt.legend()
plt.show()
```
# Example 2: ARMA(1,1) with 10% patchy outliers
## Generate ARMA(1,1) observations
```
N = 1000
a = np.random.randn(N)
x = sps.lfilter([1, 0.2],[1, -.8],a)
p = 1
q = 1
```
## Generate a patch of outliers of length 101 samples
```
v = 1000*np.random.randn(101)
```
## 10% of patch additive outliers
```
x_ao = np.array(x)
x_ao[99:200] += v
```
### BIP-MM estimation
```
result = rsp.arma_est_bip_mm(x_ao,p,q)
print('''Example 2: ARMA(1,1) with ar_coeff = -0.8, ma_coeff 0.2' \n
10 percent patchy additive outliers \n
estimated coefficients: \n
ar_coeff_est = %.3f \n
ma_coeff_est = %.3f''' %(result['ar_coeffs'],result['ma_coeffs']))
plt.subplot(2,1,1)
plt.plot(x_ao,'-',lw=2,label='outlier contaminated AR(1)')
plt.plot(result['cleaned_signal'],label='cleaned')
plt.xlabel('samples')
plt.ylabel('Amplitude')
plt.title('BIP-ARMA(1,1) cleaned signal')
plt.legend()
plt.subplot(2,1,2)
plt.plot(x,lw=2,label='original ARMA(1,1)')
plt.plot(result['cleaned_signal'],label='cleaned')
plt.xlabel('samples')
plt.ylabel('Amplitude')
plt.title('BIP-ARMA(1,1) cleaned signal')
plt.legend()
plt.show()
```
# Example 3: MA(2) with 20 % isolated Outliers
## Generate MA(2) observations
```
N = 500
a = np.random.randn(N)
x = sps.lfilter([1,-.7,.5],[1],a)
p=0
q=2
```
## Generate isolated Outliers
```
cont_prob = 0.2
outlier_ind = np.where(np.sign(np.random.rand(N)-(cont_prob))<0)
outlier = 100*np.random.randn(N)
v = np.zeros(N)
v[outlier_ind] = outlier[outlier_ind]
v[:2] = 0
```
## 20 % of isolated additive Outliers
```
x_ao = x+v
```
## BIP MM estimation
```
result = rsp.arma_est_bip_mm(x_ao,p,q)
print('''Example 3: MA(2) ma_coeff [-0.7 0.5]' \n
20 % of isolated additive Outliers \n
estimated coefficients: \n
ma_coeff_est = ''',result['ma_coeffs'])
plt.subplot(2,1,1)
plt.plot(x_ao,'-',lw=2,label='outlier contaminated AR(1)')
plt.plot(result['cleaned_signal'],label='cleaned')
plt.xlabel('samples')
plt.ylabel('Amplitude')
plt.title('BIP-MA(2) cleaned signal')
plt.legend()
plt.subplot(2,1,2)
plt.plot(x,lw=2,label='original MA(2)')
plt.plot(result['cleaned_signal'],label='cleaned')
plt.xlabel('samples')
plt.ylabel('Amplitude')
plt.title('BIP-MA(2) cleaned signal')
plt.legend()
plt.show()
```
|
github_jupyter
|
# Perceptron
### TODO
- **[ok]** Ajouter dans le code la fonction d'évaluation du réseau
- **[ok]** Plot de $\sum |E|$ par itération (i.e. num updates par itération)
- Critere d'arrêt + générale
- Lire l'article de rérérence
- Ajouter la preuve de convergence
- Ajouter notations et explications
- Tester l'autre version de la règle de mise à jours de $w$: if err then ...
- **[ok]** Décrire et illustrer les deux fonctions de transfert: signe et heaviside
- Plot de l'evolution de la courbe de niveau ($x_1 w_1 + x_2 w_2 + ... = 0$) dans l'espace des entrées: illustration avec 2 entrées seulement ou faire un graph de projection de type *scatter plot matrix*
- Plot de l'evolution de $w$ dans l'espace des $w$ illustration avec 2 entrées seulement ou faire un graph de projection de type *scatter plot matrix*
- Ajouter "Les limites du Perceptron"
Définition des macros LaTeX...
$$
\newcommand{\activthres}{\theta}
\newcommand{\activfunc}{f}
\newcommand{\pot}{p}
\newcommand{\learnrate}{\eta}
\newcommand{\it}{t}
\newcommand{\sigin}{s_i}
\newcommand{\sigout}{s_j}
\newcommand{\sigoutdes}{d_j}
\newcommand{\wij}{w_{ij}}
$$
Auteur: F. Rosenblatt
Reference: F. Rosenblatt 1958 *The Perceptron: a Probabilistic Model for Information Storage and Organization in the Brain* Psychological Review, 65, 386-408
Le modéle est constitué des éléments suivants:
- des *unités sensitives (S-units)*: réagissent à un stimuli extérieur (lumière, son, touché, ...)
- retournent `0` ou `1`:
- `1` si le signal d'entrée dépasse un seuil $\activthres$
- `0` sinon
- des *unités d'associations (A-units)*
- retournent `0` ou `1`:
- `1` si la somme des signaux d'entrée dépasse un seuil $\activthres$
- `0` sinon
- des *unités de réponse (R-units)*: sortie du réseau
- retournent `1`, `-1` ou une valeur indéterminée:
- `1` si la somme des signaux d'entrée est positive
- `-1` si elle est négative
- une valeur indéterminée si elle est égale à 0
- une *matrice d'intéractions*
Evaluation de la fonction:
$$
\pot = \sum \sigin \wij
$$
$$
\sigout = \activfunc(\pot - \activthres)
$$
Fonction de transfert: signe et heaviside
```
%matplotlib inline
#x = np.linspace(-5, 5, 300)
#y = np.array([-1 if xi < 0 else 1 for xi in x])
#plt.plot(x, y)
plt.hlines(y=-1, xmin=-5, xmax=0, color='red')
plt.hlines(y=1, xmin=0, xmax=5, color='red')
plt.hlines(y=0, xmin=-5, xmax=5, color='gray', linestyles='dotted')
plt.vlines(x=0, ymin=-2, ymax=2, color='gray', linestyles='dotted')
plt.title("Fonction signe")
plt.axis([-5, 5, -2, 2])
#x = np.linspace(-5, 5, 300)
#y = (x > 0).astype('float')
#plt.plot(x, y)
plt.hlines(y=0, xmin=-5, xmax=0, color='red')
plt.hlines(y=1, xmin=0, xmax=5, color='red')
plt.hlines(y=0, xmin=-5, xmax=5, color='gray', linestyles='dotted')
plt.vlines(x=0, ymin=-2, ymax=2, color='gray', linestyles='dotted')
plt.title("Fonction heaviside")
plt.axis([-5, 5, -2, 2])
```
Règle du Perceptron (mise à jour des poid $\wij$):
$$
\wij(\it + 1) = \wij(\it) + \learnrate (\sigoutdes - \sigout) \sigin
$$
* $\learnrate$: pas d'apprentissage, $\learnrate \in [0, 1]$. Géneralement, on lui donne une valeur proche de 1 au début de l'apprentissage et on diminue sa valeur à chaque itération.
Poids de depart des synapses du réseau
Nombre de neurones associatifs (A-units)
Nombre d'unités sensitives
Motif à apprendre
```
%matplotlib inline
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.patches as patches
import matplotlib.lines as mlines
import matplotlib.patches as mpatches
import itertools
# https://github.com/jeremiedecock/neural-network-figures.git
import nnfigs.core as fig
fig.draw_neural_network();
# Poids de depart des synapses du réseau
initial_weights = np.array([0., 0., 0., 0., 2.])
# Pas d'apprentissage eta=1
learning_rate = 1.
class Log:
def __init__(self):
self.input_signal = []
self.output_signal = []
self.desired_output_signal = []
self.error = []
self.weights = []
self.iteration = []
self.current_iteration = 0
def log(self, input_signal, output_signal, desired_output_signal, error, weights):
self.input_signal.append(input_signal)
self.output_signal.append(output_signal)
self.desired_output_signal.append(desired_output_signal)
self.error.append(error)
self.weights.append(weights)
self.iteration.append(self.current_iteration)
log = Log()
def sign_function(x):
y = 1. if x >= 0. else -1.
return y
def heaviside_function(x):
y = 1. if x >= 0. else 0.
return y
def activation_function(p):
return heaviside_function(p)
def evaluate_network(weights, input_signal): # TODO: find a better name
p = np.sum(input_signal * weights)
output_signal = activation_function(p)
return output_signal
def update_weights(weights, input_signal, desired_output_signal):
output_signal = evaluate_network(weights, input_signal)
error = desired_output_signal - output_signal
weights = weights + learning_rate * error * input_signal
log.log(input_signal, output_signal, desired_output_signal, error, weights)
return weights
def learn_examples(example_list, label_list, weights, num_iterations):
for it in range(num_iterations):
log.current_iteration = it
for input_signal, desired_output_signal in zip(example_list, label_list):
weights = update_weights(weights, np.array(input_signal + (-1,)), desired_output_signal)
return weights
```
Rappel: $\sigin \in \{0, 1\}$
```
example_list = tuple(reversed(tuple(itertools.product((0., 1.), repeat=4))))
# Motif à apprendre: (1 0 0 1)
label_list = [1. if x == (1., 0., 0., 1.) else 0. for x in example_list]
print(example_list)
print(label_list)
weights = learn_examples(example_list, label_list, initial_weights, 5)
weights
for input_signal, output_signal, desired_output_signal, error, weights, iteration in zip(log.input_signal, log.output_signal, log.desired_output_signal, log.error, log.weights, log.iteration):
print(iteration, input_signal, output_signal, desired_output_signal, error, weights)
plt.plot(log.error)
import pandas as pd
df = pd.DataFrame(np.array([log.iteration, log.error]).T, columns=["Iteration", "Error"])
abs_err_per_it = abs(df).groupby(["Iteration"]).sum()
abs_err_per_it.plot(title="Sum of absolute errors per iteration")
```
|
github_jupyter
|
## Load Weight
```
import torch
import numpy as np
path = './output/0210/Zero/checkpoint_400.pth'
import os
assert(os.path.isfile(path))
weight = torch.load(path)
input_dim = weight['input_dim']
branchNum = weight['branchNum']
IOScale = weight['IOScale']
state_dict = weight['state_dict']
# n_layers = weight['n_layers']
n_layers = 6
```
## Load Model
```
from model import Model
model = Model(branchNum, input_dim, n_layers)
model.load_state_dict(weight['state_dict'])
model = model.q_layer.layers
model.eval()
```
## Save to mat file
```
from inspect import isfunction
from scipy.io import savemat
name = 'SMINet'
v_names,d = [],{}
hdims = []
dim = 0
firstflag = False
for idx,layer in enumerate(model):
# handle Linear layer
if isinstance(layer,torch.nn.Linear):
layername = 'F_hid_lin_{dim}_kernel'.format(dim=dim)
d[layername] = layer.weight.detach().numpy().T
hdims.append(layer.weight.detach().numpy().T.shape[1])
layername = 'F_hid_lin_{dim}_bias'.format(dim=dim)
d[layername] = layer.bias.detach().numpy().T
lastlayer = idx
dim = dim+1
# find fist layer
if firstflag == False:
firstlayer = idx
firstflag = True
# handle normalization layer
if isinstance(layer,torch.nn.BatchNorm1d):
layername = 'F_bn_{dim}_mean'.format(dim=dim-1)
d[layername] = layer.running_mean.detach().numpy()
layername = 'F_bn_{dim}_sigma'.format(dim=dim-1)
sigma = torch.sqrt(layer.running_var+1e-5)
d[layername] = sigma.detach().numpy()
layername = 'F_bn_{dim}_kernel'.format(dim=dim-1)
d[layername] = layer.weight.detach().numpy()
layername = 'F_bn_{dim}_bias'.format(dim=dim-1)
d[layername] = layer.bias.detach().numpy()
# change name in last layer
lastlayername = 'F_hid_lin_{dim}_kernel'.format(dim=dim-1)
newlayername = 'F_y_pred_kernel'
d[newlayername] = d[lastlayername]
del d[lastlayername]
lastlayername = 'F_hid_lin_{dim}_bias'.format(dim=dim-1)
newlayername = 'F_y_pred_bias'
d[newlayername] = d[lastlayername]
del d[lastlayername]
xdim = model[firstlayer].weight.detach().numpy().shape[1]
ydim = model[lastlayer].weight.detach().numpy().shape[0]
d['xdim'] = xdim
d['ydim'] = ydim
d['name'] = name
d['hdims'] = np.array(hdims[:-1])
d['actv'] = 'leaky_relu'
d
# fix random seeds for reproducibility
SEED = 1
torch.manual_seed(SEED)
torch.cuda.manual_seed(SEED)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
np.random.seed(SEED)
from dataloader import *
data_path = './data/SorosimGrid'
train_data_loader = iter(ToyDataloader(os.path.join(data_path,'train'), IOScale, n_workers=1, batch=1))
x_vald = np.zeros((10,xdim))
y_vald = np.zeros((10,ydim))
for i in range(10):
(input,label) = next(train_data_loader)
output = model(input)
x_vald[i,:] = input.detach().numpy()
y_vald[i,:] = output.detach().numpy()
d['x_vald'] = x_vald
d['y_vald'] = y_vald
y_vald[-1,:],label
dir_path = 'nets/%s'%(name)
mat_path = os.path.join(dir_path,'weights.mat')
if not os.path.exists(dir_path):
os.makedirs(dir_path)
print ("[%s] created."%(dir_path))
savemat(mat_path,d) # save to a mat file
print ("[%s] saved. Size is[%.3f]MB."%(mat_path,os.path.getsize(mat_path) / 1000000))
```
|
github_jupyter
|
# OneHotEncoder
Performs One Hot Encoding.
The encoder can select how many different labels per variable to encode into binaries. When top_categories is set to None, all the categories will be transformed in binary variables.
However, when top_categories is set to an integer, for example 10, then only the 10 most popular categories will be transformed into binary, and the rest will be discarded.
The encoder has also the possibility to create binary variables from all categories (drop_last = False), or remove the binary for the last category (drop_last = True), for use in linear models.
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from feature_engine.encoding import OneHotEncoder
# Load titanic dataset from OpenML
def load_titanic():
data = pd.read_csv('https://www.openml.org/data/get_csv/16826755/phpMYEkMl')
data = data.replace('?', np.nan)
data['cabin'] = data['cabin'].astype(str).str[0]
data['pclass'] = data['pclass'].astype('O')
data['age'] = data['age'].astype('float')
data['fare'] = data['fare'].astype('float')
data['embarked'].fillna('C', inplace=True)
data.drop(labels=['boat', 'body', 'home.dest'], axis=1, inplace=True)
return data
data = load_titanic()
data.head()
X = data.drop(['survived', 'name', 'ticket'], axis=1)
y = data.survived
# we will encode the below variables, they have no missing values
X[['cabin', 'pclass', 'embarked']].isnull().sum()
''' Make sure that the variables are type (object).
if not, cast it as object , otherwise the transformer will either send an error (if we pass it as argument)
or not pick it up (if we leave variables=None). '''
X[['cabin', 'pclass', 'embarked']].dtypes
# let's separate into training and testing set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
X_train.shape, X_test.shape
```
One hot encoding consists in replacing the categorical variable by a
combination of binary variables which take value 0 or 1, to indicate if
a certain category is present in an observation.
Each one of the binary variables are also known as dummy variables. For
example, from the categorical variable "Gender" with categories 'female'
and 'male', we can generate the boolean variable "female", which takes 1
if the person is female or 0 otherwise. We can also generate the variable
male, which takes 1 if the person is "male" and 0 otherwise.
The encoder has the option to generate one dummy variable per category, or
to create dummy variables only for the top n most popular categories, that is,
the categories that are shown by the majority of the observations.
If dummy variables are created for all the categories of a variable, you have
the option to drop one category not to create information redundancy. That is,
encoding into k-1 variables, where k is the number if unique categories.
The encoder will encode only categorical variables (type 'object'). A list
of variables can be passed as an argument. If no variables are passed as
argument, the encoder will find and encode categorical variables (object type).
#### Note:
New categories in the data to transform, that is, those that did not appear
in the training set, will be ignored (no binary variable will be created for them).
### All binary, no top_categories
```
'''
Parameters
----------
top_categories: int, default=None
If None, a dummy variable will be created for each category of the variable.
Alternatively, top_categories indicates the number of most frequent categories
to encode. Dummy variables will be created only for those popular categories
and the rest will be ignored. Note that this is equivalent to grouping all the
remaining categories in one group.
variables : list
The list of categorical variables that will be encoded. If None, the
encoder will find and select all object type variables.
drop_last: boolean, default=False
Only used if top_categories = None. It indicates whether to create dummy
variables for all the categories (k dummies), or if set to True, it will
ignore the last variable of the list (k-1 dummies).
'''
ohe_enc = OneHotEncoder(top_categories=None,
variables=['pclass', 'cabin', 'embarked'],
drop_last=False)
ohe_enc.fit(X_train)
ohe_enc.encoder_dict_
train_t = ohe_enc.transform(X_train)
test_t = ohe_enc.transform(X_train)
test_t.head()
```
### Selecting top_categories to encode
```
ohe_enc = OneHotEncoder(top_categories=2,
variables=['pclass', 'cabin', 'embarked'],
drop_last=False)
ohe_enc.fit(X_train)
ohe_enc.encoder_dict_
train_t = ohe_enc.transform(X_train)
test_t = ohe_enc.transform(X_train)
test_t.head()
```
### Dropping the last category for linear models
```
ohe_enc = OneHotEncoder(top_categories=None,
variables=['pclass', 'cabin', 'embarked'],
drop_last=True)
ohe_enc.fit(X_train)
ohe_enc.encoder_dict_
train_t = ohe_enc.transform(X_train)
test_t = ohe_enc.transform(X_train)
test_t.head()
```
### Automatically select categorical variables
This encoder selects all the categorical variables, if None is passed to the variable argument when calling the encoder.
```
ohe_enc = OneHotEncoder(top_categories=None,
drop_last=True)
ohe_enc.fit(X_train)
train_t = ohe_enc.transform(X_train)
test_t = ohe_enc.transform(X_train)
test_t.head()
```
|
github_jupyter
|
```
from eva_cttv_pipeline.clinvar_xml_utils import *
from consequence_prediction.repeat_expansion_variants.clinvar_identifier_parsing import parse_variant_identifier
import os
import sys
import urllib
import requests
import xml.etree.ElementTree as ElementTree
from collections import Counter
import hgvs.parser
from hgvs.exceptions import HGVSParseError
import numpy as np
import pandas as pd
sys.path.append('../')
from gather_stats import counts
%matplotlib inline
import matplotlib.pyplot as plt
parser = hgvs.parser.Parser()
PROJECT_ROOT = '/home/april/projects/opentargets'
# dump of all records with no functional consequences: June consequence pred + ClinVar 6/26/2021
no_consequences_path = os.path.join(PROJECT_ROOT, 'no-consequences.xml.gz')
dataset = ClinVarDataset(no_consequences_path)
```
## Gather counts
Among records with no functional consequences
* how many of each variant type
* how many have hgvs, sequence location w/ start/stop position at least, cytogenic location
* of those with hgvs, how many can the library parse?
* how many can our code parse?
```
total_count, variant_type_hist, other_counts, exclusive_counts = counts(no_consequences_path, PROJECT_ROOT)
print(total_count)
plt.figure(figsize=(15,7))
plt.xticks(rotation='vertical')
plt.title('Variant Types (no functional consequences and incomplete coordinates)')
plt.bar(variant_type_hist.keys(), variant_type_hist.values())
variant_type_hist
plt.figure(figsize=(15,7))
plt.xticks(rotation='vertical')
plt.title('Variant Descriptors (no functional consequences and incomplete coordinates)')
plt.bar(other_counts.keys(), other_counts.values())
other_counts
def print_link_for_type(variant_type, min_score=-1):
for record in dataset:
if record.measure:
m = record.measure
if m.has_complete_coordinates:
continue
if m.variant_type == variant_type and record.score >= min_score:
print(f'https://www.ncbi.nlm.nih.gov/clinvar/{record.accession}/')
print_link_for_type('Microsatellite', min_score=1)
```
### Examples
Some hand-picked examples of complex variants from ClinVar. For each type I tried to choose at least one that seemed "typical" and one that was relatively high quality to get an idea of the variability, but no guarantees for how representative these are.
* Duplication
* https://www.ncbi.nlm.nih.gov/clinvar/variation/1062574/
* https://www.ncbi.nlm.nih.gov/clinvar/variation/89496/
* Deletion
* https://www.ncbi.nlm.nih.gov/clinvar/variation/1011851/
* Inversion
* https://www.ncbi.nlm.nih.gov/clinvar/variation/268016/
* https://www.ncbi.nlm.nih.gov/clinvar/variation/90611/
* Translocation
* https://www.ncbi.nlm.nih.gov/clinvar/variation/267959/
* https://www.ncbi.nlm.nih.gov/clinvar/variation/267873/
* https://www.ncbi.nlm.nih.gov/clinvar/variation/1012364/
* copy number gain
* https://www.ncbi.nlm.nih.gov/clinvar/variation/523250/
* https://www.ncbi.nlm.nih.gov/clinvar/variation/870516/
* copy number loss
* https://www.ncbi.nlm.nih.gov/clinvar/variation/1047901/
* https://www.ncbi.nlm.nih.gov/clinvar/variation/625801/
* Complex
* https://www.ncbi.nlm.nih.gov/clinvar/variation/267835/
* https://www.ncbi.nlm.nih.gov/clinvar/variation/585332/
### Appendix A: Marcos' questions
* What do the HGVS parser numbers mean?
* This is the number of records which had at least one HGVS descriptor for which the specified parser was able to extract _some_ information. For the official parser this means not throwing an exception; for our parser this means returning some non-`None` properties (though note our parser was originally written for the repeat expansion pipeline).
* What's the total number of HGVS we can parse with either parser?
* added to the above chart.
* From the variants with cytogenetic location, how many did not have any of the other descriptors, if any?
* see below
```
plt.figure(figsize=(10,7))
plt.title('Variant Descriptors (no functional consequences and incomplete coordinates)')
plt.bar(exclusive_counts.keys(), exclusive_counts.values())
exclusive_counts
```
### Appendix B: More HGVS parsing exploration
HGVS python library [doesn't support ranges](https://github.com/biocommons/hgvs/issues/225).
[VEP API](https://rest.ensembl.org/#VEP) has some limited support for HGVS.
```
def try_to_parse(hgvs):
try:
parser.parse_hgvs_variant(hgvs)
print(hgvs, 'SUCCESS')
except:
print(hgvs, 'FAILED')
try_to_parse('NC_000011.10:g.(?_17605796)_(17612832_?)del')
try_to_parse('NC_000011.10:g.(17605790_17605796)_(17612832_1761283)del')
try_to_parse('NC_000011.10:g.17605796_17612832del')
try_to_parse('NC_000011.10:g.?_17612832del')
def try_to_vep(hgvs):
safe_hgvs = urllib.parse.quote(hgvs)
vep_url = f'https://rest.ensembl.org/vep/human/hgvs/{safe_hgvs}?content-type=application/json'
resp = requests.get(vep_url)
print(resp.json())
try_to_vep('NC_000011.10:g.(?_17605796)_(17612832_?)del')
try_to_vep('NC_000011.10:g.(17605790_17605796)_(17612832_1761283)del')
try_to_vep('NC_000011.10:g.17605796_17612832del')
try_to_vep('NC_000011.10:g.?_17612832del')
```
|
github_jupyter
|
# GPU
```
gpu_info = !nvidia-smi
gpu_info = '\n'.join(gpu_info)
print(gpu_info)
```
# CFG
```
CONFIG_NAME = 'config41.yml'
debug = False
from google.colab import drive, auth
# ドライブのマウント
drive.mount('/content/drive')
# Google Cloudの権限設定
auth.authenticate_user()
def get_github_secret():
import json
with open('/content/drive/MyDrive/config/github.json') as f:
github_config = json.load(f)
return github_config
github_config = get_github_secret()
! rm -r kaggle-cassava
user_name = github_config["user_name"]
password = github_config["password"]
! git clone https://{user_name}:{password}@github.com/raijin0704/kaggle-cassava.git
import sys
sys.path.append('./kaggle-cassava')
from src.utils.envs.main import create_env
env_dict = create_env()
env_dict
# ====================================================
# CFG
# ====================================================
import yaml
CONFIG_PATH = f'./kaggle-cassava/config/{CONFIG_NAME}'
with open(CONFIG_PATH) as f:
config = yaml.load(f)
INFO = config['info']
TAG = config['tag']
CFG = config['cfg']
DATA_PATH = env_dict["data_path"]
env = env_dict["env"]
NOTEBOOK_PATH = env_dict["notebook_dir"]
OUTPUT_DIR = env_dict["output_dir"]
TITLE = env_dict["title"]
CFG['train'] = True
CFG['inference'] = False
CFG['debug'] = debug
if CFG['debug']:
CFG['epochs'] = 1
# 環境変数
import os
os.environ["GCLOUD_PROJECT"] = INFO['PROJECT_ID']
# 間違ったバージョンを実行しないかチェック
# assert INFO['TITLE'] == TITLE, f'{TITLE}, {INFO["TITLE"]}'
TITLE = INFO["TITLE"]
import os
if env=='colab':
!rm -r /content/input
! cp /content/drive/Shareddrives/便利用/kaggle/cassava/input.zip /content/input.zip
! unzip input.zip > /dev/null
! rm input.zip
train_num = len(os.listdir(DATA_PATH+"/train_images"))
assert train_num == 21397
```
# install apex
```
if CFG['apex']:
try:
import apex
except Exception:
! git clone https://github.com/NVIDIA/apex.git
% cd apex
!pip install --no-cache-dir --global-option="--cpp_ext" --global-option="--cuda_ext" .
%cd ..
```
# Library
```
# ====================================================
# Library
# ====================================================
import os
import datetime
import math
import time
import random
import glob
import shutil
from pathlib import Path
from contextlib import contextmanager
from collections import defaultdict, Counter
import scipy as sp
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import seaborn as sns
from sklearn import preprocessing
from sklearn.metrics import accuracy_score
from sklearn.model_selection import StratifiedKFold
from tqdm.auto import tqdm
from functools import partial
import cv2
from PIL import Image
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.optim import Adam, SGD
import torchvision.models as models
from torch.nn.parameter import Parameter
from torch.utils.data import DataLoader, Dataset
from torch.optim.lr_scheduler import CosineAnnealingWarmRestarts, CosineAnnealingLR, ReduceLROnPlateau
from albumentations import (
Compose, OneOf, Normalize, Resize, RandomResizedCrop, RandomCrop, HorizontalFlip, VerticalFlip,
RandomBrightness, RandomContrast, RandomBrightnessContrast, Rotate, ShiftScaleRotate, Cutout,
IAAAdditiveGaussianNoise, Transpose, CenterCrop
)
from albumentations.pytorch import ToTensorV2
from albumentations import ImageOnlyTransform
import timm
import mlflow
import warnings
warnings.filterwarnings('ignore')
if CFG['apex']:
from apex import amp
if CFG['debug']:
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
else:
device = torch.device('cuda')
from src.utils.logger import init_logger
from src.utils.utils import seed_torch, EarlyStopping
from src.utils.loss.bi_tempered_logistic_loss import bi_tempered_logistic_loss
from src.utils.augments.randaugment import RandAugment
from src.utils.augments.augmix import RandomAugMix
start_time = datetime.datetime.now()
start_time_str = start_time.strftime('%m%d%H%M')
```
# Directory settings
```
# ====================================================
# Directory settings
# ====================================================
if os.path.exists(OUTPUT_DIR):
shutil.rmtree(OUTPUT_DIR)
if not os.path.exists(OUTPUT_DIR):
os.makedirs(OUTPUT_DIR)
```
# save basic files
```
# with open(f'{OUTPUT_DIR}/{start_time_str}_TAG.json', 'w') as f:
# json.dump(TAG, f, indent=4)
# with open(f'{OUTPUT_DIR}/{start_time_str}_CFG.json', 'w') as f:
# json.dump(CFG, f, indent=4)
import shutil
notebook_path = f'{OUTPUT_DIR}/{start_time_str}_{TITLE}.ipynb'
shutil.copy2(NOTEBOOK_PATH, notebook_path)
```
# Data Loading
```
train = pd.read_csv(f'{DATA_PATH}/train.csv')
test = pd.read_csv(f'{DATA_PATH}/sample_submission.csv')
label_map = pd.read_json(f'{DATA_PATH}/label_num_to_disease_map.json',
orient='index')
if CFG['debug']:
train = train.sample(n=1000, random_state=CFG['seed']).reset_index(drop=True)
```
# Utils
```
# ====================================================
# Utils
# ====================================================
def get_score(y_true, y_pred):
return accuracy_score(y_true, y_pred)
logger_path = OUTPUT_DIR+f'{start_time_str}_train.log'
LOGGER = init_logger(logger_path)
seed_torch(seed=CFG['seed'])
def remove_glob(pathname, recursive=True):
for p in glob.glob(pathname, recursive=recursive):
if os.path.isfile(p):
os.remove(p)
def rand_bbox(size, lam):
W = size[2]
H = size[3]
cut_rat = np.sqrt(1. - lam)
cut_w = np.int(W * cut_rat)
cut_h = np.int(H * cut_rat)
# uniform
cx = np.random.randint(W)
cy = np.random.randint(H)
bbx1 = np.clip(cx - cut_w // 2, 0, W)
bby1 = np.clip(cy - cut_h // 2, 0, H)
bbx2 = np.clip(cx + cut_w // 2, 0, W)
bby2 = np.clip(cy + cut_h // 2, 0, H)
return bbx1, bby1, bbx2, bby2
```
# CV split
```
folds = train.copy()
Fold = StratifiedKFold(n_splits=CFG['n_fold'], shuffle=True, random_state=CFG['seed'])
for n, (train_index, val_index) in enumerate(Fold.split(folds, folds[CFG['target_col']])):
folds.loc[val_index, 'fold'] = int(n)
folds['fold'] = folds['fold'].astype(int)
print(folds.groupby(['fold', CFG['target_col']]).size())
```
# Dataset
```
# ====================================================
# Dataset
# ====================================================
class TrainDataset(Dataset):
def __init__(self, df, transform=None):
self.df = df
self.file_names = df['image_id'].values
self.labels = df['label'].values
self.transform = transform
def __len__(self):
return len(self.df)
def __getitem__(self, idx):
file_name = self.file_names[idx]
file_path = f'{DATA_PATH}/train_images/{file_name}'
image = cv2.imread(file_path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
if self.transform:
augmented = self.transform(image=image)
image = augmented['image']
label = torch.tensor(self.labels[idx]).long()
return image, label
class TestDataset(Dataset):
def __init__(self, df, transform=None):
self.df = df
self.file_names = df['image_id'].values
self.transform = transform
def __len__(self):
return len(self.df)
def __getitem__(self, idx):
file_name = self.file_names[idx]
file_path = f'{DATA_PATH}/test_images/{file_name}'
image = cv2.imread(file_path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
if self.transform:
augmented = self.transform(image=image)
image = augmented['image']
return image
# train_dataset = TrainDataset(train, transform=None)
# for i in range(1):
# image, label = train_dataset[i]
# plt.imshow(image)
# plt.title(f'label: {label}')
# plt.show()
```
# Transforms
```
def _get_train_augmentations(aug_list):
process = []
for aug in aug_list:
if aug == 'Resize':
process.append(Resize(CFG['size'], CFG['size']))
elif aug == 'RandomResizedCrop':
process.append(RandomResizedCrop(CFG['size'], CFG['size']))
elif aug =='CenterCrop':
process.append(CenterCrop(CFG['size'], CFG['size']))
elif aug == 'Transpose':
process.append(Transpose(p=0.5))
elif aug == 'HorizontalFlip':
process.append(HorizontalFlip(p=0.5))
elif aug == 'VerticalFlip':
process.append(VerticalFlip(p=0.5))
elif aug == 'ShiftScaleRotate':
process.append(ShiftScaleRotate(p=0.5))
elif aug == 'RandomBrightness':
process.append(RandomBrightness(limit=(-0.2,0.2), p=1))
elif aug == 'Cutout':
process.append(Cutout(max_h_size=CFG['CutoutSize'], max_w_size=CFG['CutoutSize'], p=0.5))
elif aug == 'RandAugment':
process.append(RandAugment(CFG['RandAugmentN'], CFG['RandAugmentM'], p=0.5))
elif aug == 'RandomAugMix':
process.append(RandomAugMix(severity=CFG['AugMixSeverity'],
width=CFG['AugMixWidth'],
alpha=CFG['AugMixAlpha'], p=0.5))
elif aug == 'Normalize':
process.append(Normalize(
mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225],
))
elif aug in ['mixup', 'cutmix', 'fmix']:
pass
else:
raise ValueError(f'{aug} is not suitable')
process.append(ToTensorV2())
return process
def _get_valid_augmentations(aug_list):
process = []
for aug in aug_list:
if aug == 'Resize':
process.append(Resize(CFG['size'], CFG['size']))
elif aug == 'RandomResizedCrop':
process.append(OneOf(
[RandomResizedCrop(CFG['size'], CFG['size'], p=0.5),
Resize(CFG['size'], CFG['size'], p=0.5)], p=1))
elif aug =='CenterCrop':
process.append(OneOf(
[CenterCrop(CFG['size'], CFG['size'], p=0.5),
Resize(CFG['size'], CFG['size'], p=0.5)], p=1))
# process.append(
# CenterCrop(CFG['size'], CFG['size'], p=1.))
elif aug == 'Transpose':
process.append(Transpose(p=0.5))
elif aug == 'HorizontalFlip':
process.append(HorizontalFlip(p=0.5))
elif aug == 'VerticalFlip':
process.append(VerticalFlip(p=0.5))
elif aug == 'ShiftScaleRotate':
process.append(ShiftScaleRotate(p=0.5))
elif aug == 'RandomBrightness':
process.append(RandomBrightness(limit=(-0.2,0.2), p=1))
elif aug == 'Cutout':
process.append(Cutout(max_h_size=CFG['CutoutSize'], max_w_size=CFG['CutoutSize'], p=0.5))
elif aug == 'RandAugment':
process.append(RandAugment(CFG['RandAugmentN'], CFG['RandAugmentM'], p=0.5))
elif aug == 'RandomAugMix':
process.append(RandomAugMix(severity=CFG['AugMixSeverity'],
width=CFG['AugMixWidth'],
alpha=CFG['AugMixAlpha'], p=0.5))
elif aug == 'Normalize':
process.append(Normalize(
mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225],
))
elif aug in ['mixup', 'cutmix', 'fmix']:
pass
else:
raise ValueError(f'{aug} is not suitable')
process.append(ToTensorV2())
return process
# ====================================================
# Transforms
# ====================================================
def get_transforms(*, data):
if data == 'train':
return Compose(
_get_train_augmentations(TAG['augmentation'])
)
elif data == 'valid':
try:
augmentations = TAG['valid_augmentation']
except KeyError:
augmentations = ['Resize', 'Normalize']
return Compose(
_get_valid_augmentations(augmentations)
)
num_fig = 5
train_dataset = TrainDataset(train, transform=get_transforms(data='train'))
valid_dataset = TrainDataset(train, transform=get_transforms(data='valid'))
origin_dataset = TrainDataset(train, transform=None)
fig, ax = plt.subplots(num_fig, 3, figsize=(10, num_fig*3))
for j, dataset in enumerate([train_dataset, valid_dataset, origin_dataset]):
for i in range(num_fig):
image, label = dataset[i]
if j < 2:
ax[i,j].imshow(image.transpose(0,2).transpose(0,1))
else:
ax[i,j].imshow(image)
ax[i,j].set_title(f'label: {label}')
```
# MODEL
```
# ====================================================
# MODEL
# ====================================================
class CustomModel(nn.Module):
def __init__(self, model_name, pretrained=False):
super().__init__()
self.model = timm.create_model(model_name, pretrained=pretrained)
if hasattr(self.model, 'classifier'):
n_features = self.model.classifier.in_features
self.model.classifier = nn.Linear(n_features, CFG['target_size'])
elif hasattr(self.model, 'fc'):
n_features = self.model.fc.in_features
self.model.fc = nn.Linear(n_features, CFG['target_size'])
elif hasattr(self.model, 'head'):
n_features = self.model.head.in_features
self.model.head = nn.Linear(n_features, CFG['target_size'])
def forward(self, x):
x = self.model(x)
return x
model = CustomModel(model_name=TAG['model_name'], pretrained=False)
train_dataset = TrainDataset(train, transform=get_transforms(data='train'))
train_loader = DataLoader(train_dataset, batch_size=4, shuffle=True,
num_workers=0, pin_memory=True, drop_last=True)
for image, label in train_loader:
output = model(image)
print(output)
break
```
# Helper functions
```
# ====================================================
# Helper functions
# ====================================================
class AverageMeter(object):
"""Computes and stores the average and current value"""
def __init__(self):
self.reset()
def reset(self):
self.val = 0
self.avg = 0
self.sum = 0
self.count = 0
def update(self, val, n=1):
self.val = val
self.sum += val * n
self.count += n
self.avg = self.sum / self.count
def asMinutes(s):
m = math.floor(s / 60)
s -= m * 60
return '%dm %ds' % (m, s)
def timeSince(since, percent):
now = time.time()
s = now - since
es = s / (percent)
rs = es - s
return '%s (remain %s)' % (asMinutes(s), asMinutes(rs))
# ====================================================
# loss
# ====================================================
def get_loss(criterion, y_preds, labels):
if TAG['criterion']=='CrossEntropyLoss':
loss = criterion(y_preds, labels)
elif TAG['criterion'] == 'bi_tempered_logistic_loss':
loss = criterion(y_preds, labels, t1=CFG['bi_tempered_loss_t1'], t2=CFG['bi_tempered_loss_t2'])
return loss
# ====================================================
# Helper functions
# ====================================================
def train_fn(train_loader, model, criterion, optimizer, epoch, scheduler, device):
batch_time = AverageMeter()
data_time = AverageMeter()
losses = AverageMeter()
scores = AverageMeter()
# switch to train mode
model.train()
start = end = time.time()
global_step = 0
for step, (images, labels) in enumerate(train_loader):
# measure data loading time
data_time.update(time.time() - end)
images = images.to(device)
labels = labels.to(device)
batch_size = labels.size(0)
r = np.random.rand(1)
is_aug = r < 0.5 # probability of augmentation
if is_aug & ('cutmix' in TAG['augmentation']) & (epoch+1>=CFG['heavy_aug_start_epoch']):
# generate mixed sample
# inference from https://github.com/clovaai/CutMix-PyTorch/blob/master/train.py
lam = np.random.beta(CFG['CutmixAlpha'], CFG['CutmixAlpha'])
rand_index = torch.randperm(images.size()[0]).to(device)
labels_a = labels
labels_b = labels[rand_index]
bbx1, bby1, bbx2, bby2 = rand_bbox(images.size(), lam)
images[:, :, bbx1:bbx2, bby1:bby2] = images[rand_index, :, bbx1:bbx2, bby1:bby2]
# adjust lambda to exactly match pixel ratio
lam = 1 - ((bbx2 - bbx1) * (bby2 - bby1) / (images.size()[-1] * images.size()[-2]))
# compute output
y_preds = model(images)
loss = get_loss(criterion, y_preds, labels_a) * lam + \
get_loss(criterion, y_preds, labels_b) * (1. - lam)
else:
y_preds = model(images)
loss = get_loss(criterion, y_preds, labels)
# record loss
losses.update(loss.item(), batch_size)
if CFG['gradient_accumulation_steps'] > 1:
loss = loss / CFG['gradient_accumulation_steps']
if CFG['apex']:
with amp.scale_loss(loss, optimizer) as scaled_loss:
scaled_loss.backward()
else:
loss.backward()
# clear memory
del loss, y_preds
torch.cuda.empty_cache()
grad_norm = torch.nn.utils.clip_grad_norm_(model.parameters(), CFG['max_grad_norm'])
if (step + 1) % CFG['gradient_accumulation_steps'] == 0:
optimizer.step()
optimizer.zero_grad()
global_step += 1
# measure elapsed time
batch_time.update(time.time() - end)
end = time.time()
if step % CFG['print_freq'] == 0 or step == (len(train_loader)-1):
print('Epoch: [{0}][{1}/{2}] '
'Data {data_time.val:.3f} ({data_time.avg:.3f}) '
'Elapsed {remain:s} '
'Loss: {loss.val:.4f}({loss.avg:.4f}) '
'Grad: {grad_norm:.4f} '
#'LR: {lr:.6f} '
.format(
epoch+1, step, len(train_loader), batch_time=batch_time,
data_time=data_time, loss=losses,
remain=timeSince(start, float(step+1)/len(train_loader)),
grad_norm=grad_norm,
#lr=scheduler.get_lr()[0],
))
return losses.avg
def valid_fn(valid_loader, model, criterion, device):
batch_time = AverageMeter()
data_time = AverageMeter()
losses = AverageMeter()
scores = AverageMeter()
# switch to evaluation mode
model.eval()
preds = []
start = end = time.time()
for step, (images, labels) in enumerate(valid_loader):
# measure data loading time
data_time.update(time.time() - end)
images = images.to(device)
labels = labels.to(device)
batch_size = labels.size(0)
# compute loss
with torch.no_grad():
y_preds = model(images)
loss = get_loss(criterion, y_preds, labels)
losses.update(loss.item(), batch_size)
# record accuracy
preds.append(y_preds.softmax(1).to('cpu').numpy())
if CFG['gradient_accumulation_steps'] > 1:
loss = loss / CFG['gradient_accumulation_steps']
# measure elapsed time
batch_time.update(time.time() - end)
end = time.time()
if step % CFG['print_freq'] == 0 or step == (len(valid_loader)-1):
print('EVAL: [{0}/{1}] '
'Data {data_time.val:.3f} ({data_time.avg:.3f}) '
'Elapsed {remain:s} '
'Loss: {loss.val:.4f}({loss.avg:.4f}) '
.format(
step, len(valid_loader), batch_time=batch_time,
data_time=data_time, loss=losses,
remain=timeSince(start, float(step+1)/len(valid_loader)),
))
predictions = np.concatenate(preds)
return losses.avg, predictions
def inference(model, states, test_loader, device):
model.to(device)
tk0 = tqdm(enumerate(test_loader), total=len(test_loader))
probs = []
for i, (images) in tk0:
images = images.to(device)
avg_preds = []
for state in states:
# model.load_state_dict(state['model'])
model.load_state_dict(state)
model.eval()
with torch.no_grad():
y_preds = model(images)
avg_preds.append(y_preds.softmax(1).to('cpu').numpy())
avg_preds = np.mean(avg_preds, axis=0)
probs.append(avg_preds)
probs = np.concatenate(probs)
return probs
```
# Train loop
```
# ====================================================
# scheduler
# ====================================================
def get_scheduler(optimizer):
if TAG['scheduler']=='ReduceLROnPlateau':
scheduler = ReduceLROnPlateau(optimizer, mode='min', factor=CFG['factor'], patience=CFG['patience'], verbose=True, eps=CFG['eps'])
elif TAG['scheduler']=='CosineAnnealingLR':
scheduler = CosineAnnealingLR(optimizer, T_max=CFG['T_max'], eta_min=CFG['min_lr'], last_epoch=-1)
elif TAG['scheduler']=='CosineAnnealingWarmRestarts':
scheduler = CosineAnnealingWarmRestarts(optimizer, T_0=CFG['T_0'], T_mult=1, eta_min=CFG['min_lr'], last_epoch=-1)
return scheduler
# ====================================================
# criterion
# ====================================================
def get_criterion():
if TAG['criterion']=='CrossEntropyLoss':
criterion = nn.CrossEntropyLoss()
elif TAG['criterion'] == 'bi_tempered_logistic_loss':
criterion = bi_tempered_logistic_loss
return criterion
# ====================================================
# Train loop
# ====================================================
def train_loop(folds, fold):
LOGGER.info(f"========== fold: {fold} training ==========")
if not CFG['debug']:
mlflow.set_tag('running.fold', str(fold))
# ====================================================
# loader
# ====================================================
trn_idx = folds[folds['fold'] != fold].index
val_idx = folds[folds['fold'] == fold].index
train_folds = folds.loc[trn_idx].reset_index(drop=True)
valid_folds = folds.loc[val_idx].reset_index(drop=True)
train_dataset = TrainDataset(train_folds,
transform=get_transforms(data='train'))
valid_dataset = TrainDataset(valid_folds,
transform=get_transforms(data='valid'))
train_loader = DataLoader(train_dataset,
batch_size=CFG['batch_size'],
shuffle=True,
num_workers=CFG['num_workers'], pin_memory=True, drop_last=True)
valid_loader = DataLoader(valid_dataset,
batch_size=CFG['batch_size'],
shuffle=False,
num_workers=CFG['num_workers'], pin_memory=True, drop_last=False)
# ====================================================
# model & optimizer & criterion
# ====================================================
best_model_path = OUTPUT_DIR+f'{TAG["model_name"]}_fold{fold}_best.pth'
latest_model_path = OUTPUT_DIR+f'{TAG["model_name"]}_fold{fold}_latest.pth'
model = CustomModel(TAG['model_name'], pretrained=True)
model.to(device)
# # 学習途中の重みがあれば読み込み
# if os.path.isfile(latest_model_path):
# state_latest = torch.load(latest_model_path)
# state_best = torch.load(best_model_path)
# model.load_state_dict(state_latest['model'])
# epoch_start = state_latest['epoch']+1
# # er_best_score = state_latest['score']
# er_counter = state_latest['counter']
# er_best_score = state_best['best_score']
# if 'val_loss_history' in state_latest.keys():
# val_loss_history = state_latest['val_loss_history']
# else:
# val_loss_history = []
# LOGGER.info(f'Load training model in epoch:{epoch_start}, best_score:{er_best_score:.3f}, counter:{er_counter}')
# # 学習済みモデルを再学習する場合
# elif os.path.isfile(best_model_path):
if os.path.isfile(best_model_path):
state_best = torch.load(best_model_path)
model.load_state_dict(state_best['model'])
epoch_start = 0 # epochは0からカウントしなおす
er_counter = 0
er_best_score = state_best['best_score']
val_loss_history = [] # 過去のval_lossも使用しない
LOGGER.info(f'Retrain model, best_score:{er_best_score:.3f}')
else:
epoch_start = 0
er_best_score = None
er_counter = 0
val_loss_history = []
optimizer = Adam(model.parameters(), lr=CFG['lr'], weight_decay=CFG['weight_decay'], amsgrad=False)
scheduler = get_scheduler(optimizer)
criterion = get_criterion()
# 再開時のepochまでschedulerを進める
# assert len(range(epoch_start)) == len(val_loss_history)
for _, val_loss in zip(range(epoch_start), val_loss_history):
if isinstance(scheduler, ReduceLROnPlateau):
scheduler.step(val_loss)
elif isinstance(scheduler, CosineAnnealingLR):
scheduler.step()
elif isinstance(scheduler, CosineAnnealingWarmRestarts):
scheduler.step()
# ====================================================
# apex
# ====================================================
if CFG['apex']:
model, optimizer = amp.initialize(model, optimizer, opt_level='O1', verbosity=0)
# ====================================================
# loop
# ====================================================
# best_score = 0.
# best_loss = np.inf
early_stopping = EarlyStopping(
patience=CFG['early_stopping_round'],
eps=CFG['early_stopping_eps'],
verbose=True,
save_path=best_model_path,
counter=er_counter, best_score=er_best_score,
val_loss_history = val_loss_history,
save_latest_path=latest_model_path)
for epoch in range(epoch_start, CFG['epochs']):
start_time = time.time()
# train
avg_loss = train_fn(train_loader, model, criterion, optimizer, epoch, scheduler, device)
# eval
avg_val_loss, preds = valid_fn(valid_loader, model, criterion, device)
valid_labels = valid_folds[CFG['target_col']].values
# scoring
score = get_score(valid_labels, preds.argmax(1))
# get learning rate
if hasattr(scheduler, 'get_last_lr'):
last_lr = scheduler.get_last_lr()[0]
else:
# ReduceLROnPlateauには関数get_last_lrがない
last_lr = optimizer.param_groups[0]['lr']
# log mlflow
if not CFG['debug']:
mlflow.log_metric(f"fold{fold} avg_train_loss", avg_loss, step=epoch)
mlflow.log_metric(f"fold{fold} avg_valid_loss", avg_val_loss, step=epoch)
mlflow.log_metric(f"fold{fold} score", score, step=epoch)
mlflow.log_metric(f"fold{fold} lr", last_lr, step=epoch)
# early stopping
early_stopping(avg_val_loss, model, preds, epoch)
if early_stopping.early_stop:
print(f'Epoch {epoch+1} - early stopping')
break
if isinstance(scheduler, ReduceLROnPlateau):
scheduler.step(avg_val_loss)
elif isinstance(scheduler, CosineAnnealingLR):
scheduler.step()
elif isinstance(scheduler, CosineAnnealingWarmRestarts):
scheduler.step()
elapsed = time.time() - start_time
LOGGER.info(f'Epoch {epoch+1} - avg_train_loss: {avg_loss:.4f} avg_val_loss: {avg_val_loss:.4f} time: {elapsed:.0f}s')
LOGGER.info(f'Epoch {epoch+1} - Accuracy: {score}')
# log mlflow
if not CFG['debug']:
mlflow.log_artifact(best_model_path)
if os.path.isfile(latest_model_path):
mlflow.log_artifact(latest_model_path)
check_point = torch.load(best_model_path)
valid_folds[[str(c) for c in range(5)]] = check_point['preds']
valid_folds['preds'] = check_point['preds'].argmax(1)
return valid_folds
def get_trained_fold_preds(folds, fold, best_model_path):
val_idx = folds[folds['fold'] == fold].index
valid_folds = folds.loc[val_idx].reset_index(drop=True)
check_point = torch.load(best_model_path)
valid_folds[[str(c) for c in range(5)]] = check_point['preds']
valid_folds['preds'] = check_point['preds'].argmax(1)
return valid_folds
def save_confusion_matrix(oof):
from sklearn.metrics import confusion_matrix
cm_ = confusion_matrix(oof['label'], oof['preds'], labels=[0,1,2,3,4])
label_name = ['0 (CBB)', '1 (CBSD)', '2 (CGM)', '3 (CMD)', '4 (Healthy)']
cm = pd.DataFrame(cm_, index=label_name, columns=label_name)
cm.to_csv(OUTPUT_DIR+'oof_confusion_matrix.csv', index=True)
# ====================================================
# main
# ====================================================
def get_result(result_df):
preds = result_df['preds'].values
labels = result_df[CFG['target_col']].values
score = get_score(labels, preds)
LOGGER.info(f'Score: {score:<.5f}')
return score
def main():
"""
Prepare: 1.train 2.test 3.submission 4.folds
"""
if CFG['train']:
# train
oof_df = pd.DataFrame()
for fold in range(CFG['n_fold']):
best_model_path = OUTPUT_DIR+f'{TAG["model_name"]}_fold{fold}_best.pth'
if fold in CFG['trn_fold']:
_oof_df = train_loop(folds, fold)
elif os.path.exists(best_model_path):
_oof_df = get_trained_fold_preds(folds, fold, best_model_path)
else:
_oof_df = None
if _oof_df is not None:
oof_df = pd.concat([oof_df, _oof_df])
LOGGER.info(f"========== fold: {fold} result ==========")
_ = get_result(_oof_df)
# CV result
LOGGER.info(f"========== CV ==========")
score = get_result(oof_df)
# save result
oof_df.to_csv(OUTPUT_DIR+'oof_df.csv', index=False)
save_confusion_matrix(oof_df)
# log mlflow
if not CFG['debug']:
mlflow.log_metric('oof score', score)
mlflow.delete_tag('running.fold')
mlflow.log_artifact(OUTPUT_DIR+'oof_df.csv')
if CFG['inference']:
# inference
model = CustomModel(TAG['model_name'], pretrained=False)
states = [torch.load(OUTPUT_DIR+f'{TAG["model_name"]}_fold{fold}_best.pth') for fold in CFG['trn_fold']]
test_dataset = TestDataset(test, transform=get_transforms(data='valid'))
test_loader = DataLoader(test_dataset, batch_size=CFG['batch_size'], shuffle=False,
num_workers=CFG['num_workers'], pin_memory=True)
predictions = inference(model, states, test_loader, device)
# submission
test['label'] = predictions.argmax(1)
test[['image_id', 'label']].to_csv(OUTPUT_DIR+'submission.csv', index=False)
```
# rerun
```
def _load_save_point(run_id):
# どこで中断したか取得
stop_fold = int(mlflow.get_run(run_id=run_id).to_dictionary()['data']['tags']['running.fold'])
# 学習対象のfoldを変更
CFG['trn_fold'] = [fold for fold in CFG['trn_fold'] if fold>=stop_fold]
# 学習済みモデルがあれば.pthファイルを取得(学習中も含む)
client = mlflow.tracking.MlflowClient()
artifacts = [artifact for artifact in client.list_artifacts(run_id) if ".pth" in artifact.path]
for artifact in artifacts:
client.download_artifacts(run_id, artifact.path, OUTPUT_DIR)
def check_have_run():
results = mlflow.search_runs(INFO['EXPERIMENT_ID'])
run_id_list = results[results['tags.mlflow.runName']==TITLE]['run_id'].tolist()
# 初めて実行する場合
if len(run_id_list) == 0:
run_id = None
# 既に実行されている場合
else:
assert len(run_id_list)==1
run_id = run_id_list[0]
_load_save_point(run_id)
return run_id
def push_github():
! cp {NOTEBOOK_PATH} kaggle-cassava/notebook/{TITLE}.ipynb
!git config --global user.email "[email protected]"
! git config --global user.name "Raijin Shibata"
!cd kaggle-cassava ;git add .; git commit -m {TITLE}; git pull; git remote set-url origin https://{user_name}:{password}@github.com/raijin0704/kaggle-cassava.git; git push origin master
def _load_save_point_copy(run_id):
# # どこで中断したか取得
# stop_fold = int(mlflow.get_run(run_id=run_id).to_dictionary()['data']['tags']['running.fold'])
# # 学習対象のfoldを変更
# CFG['trn_fold'] = [fold for fold in CFG['trn_fold'] if fold>=stop_fold]
# 学習済みモデルがあれば.pthファイルを取得(学習中も含む)
client = mlflow.tracking.MlflowClient()
artifacts = [artifact for artifact in client.list_artifacts(run_id) if ".pth" in artifact.path]
for artifact in artifacts:
client.download_artifacts(run_id, artifact.path, OUTPUT_DIR)
def check_have_run_copy(copy_from):
results = mlflow.search_runs(INFO['EXPERIMENT_ID'])
run_id_list = results[results['tags.mlflow.runName']==copy_from]['run_id'].tolist()
# 初めて実行する場合
if len(run_id_list) == 0:
run_id = None
# 既に実行されている場合
else:
assert len(run_id_list)==1
run_id = run_id_list[0]
_load_save_point_copy(run_id)
return run_id
if __name__ == '__main__':
if CFG['debug']:
mlflow.set_tracking_uri(INFO['TRACKING_URI'])
# 指定したrun_nameの学習済みモデルを取得
_ = check_have_run_copy(TAG['trained'])
main()
else:
mlflow.set_tracking_uri(INFO['TRACKING_URI'])
mlflow.set_experiment('single model')
# 指定したrun_nameの学習済みモデルを取得
_ = check_have_run_copy(TAG['trained'])
# 既に実行済みの場合は続きから実行する
run_id = check_have_run()
with mlflow.start_run(run_id=run_id, run_name=TITLE):
if run_id is None:
mlflow.log_artifact(CONFIG_PATH)
mlflow.log_param('device', device)
mlflow.set_tag('env', env)
mlflow.set_tags(TAG)
mlflow.log_params(CFG)
mlflow.log_artifact(notebook_path)
main()
mlflow.log_artifacts(OUTPUT_DIR)
# remove_glob(f'{OUTPUT_DIR}/*latest.pth')
push_github()
if env=="kaggle":
shutil.copy2(CONFIG_PATH, f'{OUTPUT_DIR}/{CONFIG_NAME}')
! rm -r kaggle-cassava
elif env=="colab":
shutil.copytree(OUTPUT_DIR, f'{INFO["SHARE_DRIVE_PATH"]}/{TITLE}')
shutil.copy2(CONFIG_PATH, f'{INFO["SHARE_DRIVE_PATH"]}/{TITLE}/{CONFIG_NAME}')
```
|
github_jupyter
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.