text
stringlengths 2.5k
6.39M
| kind
stringclasses 3
values |
|---|---|
# Cat Dog Classification
## 1. 下载数据
我们将使用包含猫与狗图片的数据集。它是Kaggle.com在2013年底计算机视觉竞赛提供的数据集的一部分,当时卷积神经网络还不是主流。可以在以下位置下载原始数据集: `https://www.kaggle.com/c/dogs-vs-cats/data`。
图片是中等分辨率的彩色JPEG。看起来像这样:

不出所料,2013年的猫狗大战的Kaggle比赛是由使用卷积神经网络的参赛者赢得的。最佳成绩达到了高达95%的准确率。在本例中,我们将非常接近这个准确率,即使我们将使用不到10%的训练集数据来训练我们的模型。
原始数据集的训练集包含25,000张狗和猫的图像(每个类别12,500张),543MB大(压缩)。
在下载并解压缩之后,我们将创建一个包含三个子集的新数据集:
* 每个类有1000个样本的训练集,
* 每个类500个样本的验证集,
* 最后是每个类500个样本的测试集。
数据已经提前处理好。
### 1.1 加载数据集目录
```
import os, shutil
# The directory where we will
# store our smaller dataset
base_dir = './data/cats_and_dogs_small'
# Directories for our training,
# validation and test splits
train_dir = os.path.join(base_dir, 'train')
validation_dir = os.path.join(base_dir, 'validation')
test_dir = os.path.join(base_dir, 'test')
# Directory with our training cat pictures
train_cats_dir = os.path.join(train_dir, 'cats')
# Directory with our training dog pictures
train_dogs_dir = os.path.join(train_dir, 'dogs')
# Directory with our validation cat pictures
validation_cats_dir = os.path.join(validation_dir, 'cats')
# Directory with our validation dog pictures
validation_dogs_dir = os.path.join(validation_dir, 'dogs')
# Directory with our validation cat pictures
test_cats_dir = os.path.join(test_dir, 'cats')
# Directory with our validation dog pictures
test_dogs_dir = os.path.join(test_dir, 'dogs')
```
## 2. 模型一
### 2.1 数据处理
```
from keras.preprocessing.image import ImageDataGenerator
# All images will be rescaled by 1./255
train_datagen = ImageDataGenerator(rescale=1./255)
validation_datagen = ImageDataGenerator(rescale=1./255)
test_datagen = ImageDataGenerator(rescale=1./255)
# 150*150
train_generator = train_datagen.flow_from_directory(
# This is the target directory
train_dir,
# All images will be resized to 150x150
target_size=(150, 150),
batch_size=20,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
validation_generator = validation_datagen.flow_from_directory(
validation_dir,
target_size=(150, 150),
batch_size=20,
class_mode='binary')
test_generator = test_datagen.flow_from_directory(
test_dir,
target_size=(150, 150),
batch_size=20,
class_mode='binary')
print('train_dir: ',train_dir)
print('validation_dir: ',validation_dir)
print('test_dir: ',test_dir)
for data_batch, labels_batch in train_generator:
print('data batch shape:', data_batch.shape)
print('labels batch shape:', labels_batch.shape)
break
labels_batch
```
### 2.2 构建模型
```
from keras import layers
from keras import models
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu', input_shape=(150, 150, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Flatten())
model.add(layers.Dense(512, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
from keras import optimizers
model.compile(optimizer=optimizers.RMSprop(lr=1e-4),
loss='binary_crossentropy',
metrics=['acc'])
history = model.fit_generator(train_generator,
steps_per_epoch=100,
epochs=30,
validation_data=validation_generator,
validation_steps=50)
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper right')
plt.show()
plt.plot(history.history['acc'])
plt.plot(history.history['val_acc'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper right')
plt.show()
val_loss_min = history.history['val_loss'].index(min(history.history['val_loss']))
val_acc_max = history.history['val_acc'].index(max(history.history['val_acc']))
print('validation set min loss: ', val_loss_min)
print('validation set max accuracy: ', val_acc_max)
from keras import layers
from keras import models
# vgg的做法
model = models.Sequential()
model.add(layers.Conv2D(32, 3, activation='relu', padding="same", input_shape=(64, 64, 3)))
model.add(layers.Conv2D(32, 3, activation='relu', padding="same"))
model.add(layers.MaxPooling2D(pool_size=(2, 2)))
model.add(layers.Conv2D(64, 3, activation='relu', padding="same"))
model.add(layers.Conv2D(64, 3, activation='relu', padding="same"))
model.add(layers.MaxPooling2D(pool_size=(2, 2)))
model.add(layers.Conv2D(128, 3, activation='relu', padding="same"))
model.add(layers.Conv2D(128, 3, activation='relu', padding="same"))
model.add(layers.MaxPooling2D(pool_size=(2, 2)))
model.add(layers.Conv2D(256, 3, activation='relu', padding="same"))
model.add(layers.Conv2D(256, 3, activation='relu', padding="same"))
model.add(layers.MaxPooling2D(pool_size=2))
model.add(layers.Flatten())
model.add(layers.Dense(256, activation='relu'))
model.add(layers.Dropout(0.5))
model.add(layers.Dense(256, activation='relu'))
model.add(layers.Dropout(0.5))
model.add(layers.Dense(1, activation='sigmoid'))
model.summary()
from keras import optimizers
model.compile(optimizer=optimizers.RMSprop(lr=1e-4),
loss='binary_crossentropy',
metrics=['acc'])
# model.compile(loss='binary_crossentropy',
# optimizer='adam',
# metrics=['accuracy'])
```
### 2.3 训练模型
```
history = model.fit_generator(train_generator,
steps_per_epoch=100,
epochs=30,
validation_data=validation_generator,
validation_steps=50)
```
### 2.4 画出表现
```
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
val_loss_min = val_loss.index(min(val_loss))
val_acc_max = val_acc.index(max(val_acc))
print('validation set min loss: ', val_loss_min)
print('validation set max accuracy: ', val_acc_max)
```
### 2.5 测试集表现
```
scores = model.evaluate_generator(test_generator, verbose=0)
print("Large CNN Error: %.2f%%" % (100 - scores[1] * 100))
```
## 3. 模型二 使用数据增强来防止过拟合
### 3.1 数据增强示例
```
datagen = ImageDataGenerator(
rotation_range=40, # 角度值(在 0~180 范围内),表示图像随机旋转的角度范围
width_shift_range=0.2, # 图像在水平或垂直方向上平移的范围
height_shift_range=0.2, # (相对于总宽度或总高度的比例)
shear_range=0.2, # 随机错切变换的角度
zoom_range=0.2, # 图像随机缩放的范围
horizontal_flip=True, # 随机将一半图像水平翻转
fill_mode='nearest') # 用于填充新创建像素的方法,
# 这些新像素可能来自于旋转或宽度/高度平移
# This is module with image preprocessing utilities
from keras.preprocessing import image
fnames = [os.path.join(train_cats_dir, fname) for fname in os.listdir(train_cats_dir)]
# We pick one image to "augment"
img_path = fnames[3]
# Read the image and resize it
img = image.load_img(img_path, target_size=(150, 150))
imgplot_oringe = plt.imshow(img)
# Convert it to a Numpy array with shape (150, 150, 3)
x = image.img_to_array(img)
# Reshape it to (1, 150, 150, 3)
x = x.reshape((1,) + x.shape)
# The .flow() command below generates batches of randomly transformed images.
# It will loop indefinitely, so we need to `break` the loop at some point!
i = 0
for batch in datagen.flow(x, batch_size=1):
plt.figure(i)
imgplot = plt.imshow(image.array_to_img(batch[0]))
i += 1
if i % 4 == 0:
break
plt.show()
```
### 3.2 定义数据增强
```
train_datagen = ImageDataGenerator(
rescale=1./255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,)
# Note that the validation data should not be augmented!
test_datagen = ImageDataGenerator(rescale=1./255) # 注意,不能增强验证数据
train_generator = train_datagen.flow_from_directory(
# This is the target directory
train_dir,
# All images will be resized to 150x150
target_size=(150, 150),
batch_size=32,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(150, 150),
batch_size=32,
class_mode='binary')
```
### 3.3 训练网络
```
model = models.Sequential()
model.add(layers.Conv2D(32, 3, activation='relu', padding="same", input_shape=(150, 150, 3)))
model.add(layers.Conv2D(32, 3, activation='relu', padding="same"))
model.add(layers.MaxPooling2D(pool_size=(2, 2)))
model.add(layers.Conv2D(64, 3, activation='relu', padding="same"))
model.add(layers.Conv2D(64, 3, activation='relu', padding="same"))
model.add(layers.MaxPooling2D(pool_size=(2, 2)))
model.add(layers.Conv2D(128, 3, activation='relu', padding="same"))
model.add(layers.Conv2D(128, 3, activation='relu', padding="same"))
model.add(layers.MaxPooling2D(pool_size=(2, 2)))
model.add(layers.Conv2D(256, 3, activation='relu', padding="same"))
model.add(layers.Conv2D(256, 3, activation='relu', padding="same"))
model.add(layers.MaxPooling2D(pool_size=2))
model.add(layers.Flatten())
model.add(layers.Dense(256, activation='relu'))
model.add(layers.Dropout(0.5))
model.add(layers.Dense(256, activation='relu'))
model.add(layers.Dropout(0.5))
model.add(layers.Dense(1, activation='sigmoid'))
# model.compile(optimizer=optimizers.RMSprop(lr=1e-4),
# loss='binary_crossentropy',
# metrics=['acc'])
model.compile(loss='binary_crossentropy',
optimizer='adam',
metrics=['accuracy'])
history = model.fit_generator(train_generator,
steps_per_epoch=100, # 训练集分成100批送进去,相当于每批送20个
epochs=100, # 循环100遍
validation_data=validation_generator,
validation_steps=50, # 验证集分50批送进去,每批20个
verbose=0)
```
### 3.4 画出表现
```
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
val_loss_min = val_loss.index(min(val_loss))
val_acc_max = val_acc.index(max(val_acc))
print('validation set min loss: ', val_loss_min)
print('validation set max accuracy: ', val_acc_max)
# train_datagen = ImageDataGenerator(rotation_range=40,
# width_shift_range=0.2,
# height_shift_range=0.2,
# shear_range=0.2,
# zoom_range=0.2,
# horizontal_flip=True,
# fill_mode='nearest')
# train_datagen.fit(train_X)
# train_generator = train_datagen.flow(train_X, train_y,
# batch_size = 64)
# history = model_vgg16.fit_generator(train_generator,
# validation_data = (test_X, test_y),
# steps_per_epoch = train_X.shape[0] / 100,
# epochs = 10)
```
## 4. 使用预训练的VGG-16
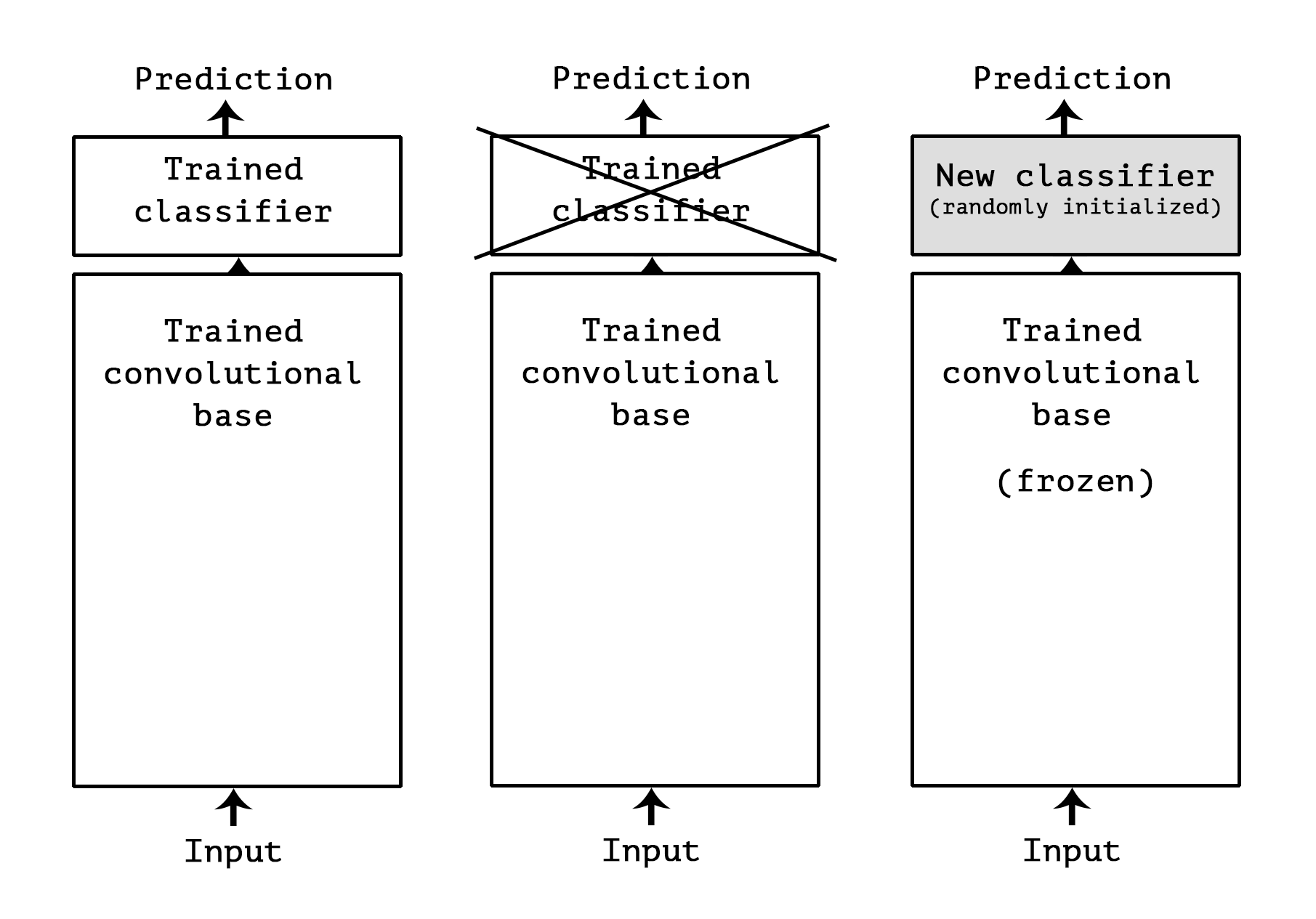
```
from keras.applications import VGG16
conv_base = VGG16(weights='imagenet',
include_top=False, # 不要分类层
input_shape=(150, 150, 3))
conv_base.summary()
from keras import models
from keras import layers
model = models.Sequential()
model.add(conv_base)
model.add(layers.Flatten())
model.add(layers.Dense(256, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
# model = models.Sequential()
# model.add(conv_base)
# model.add(layers.Dense(256, activation='relu'))
# model.add(layers.Dropout(0.5))
# model.add(layers.Dense(256, activation='relu'))
# model.add(layers.Dropout(0.5))
# model.add(layers.Dense(1, activation='sigmoid'))
print('This is the number of trainable weights '
'before freezing the conv base:', len(model.trainable_weights))
conv_base.trainable = False
print('This is the number of trainable weights '
'after freezing the conv base:', len(model.trainable_weights))
from keras.preprocessing.image import ImageDataGenerator
from keras import optimizers
train_datagen = ImageDataGenerator(
rescale=1./255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest')
# Note that the validation data should not be augmented!
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(
# This is the target directory
train_dir,
# All images will be resized to 150x150
target_size=(150, 150),
batch_size=20,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(150, 150),
batch_size=20,
class_mode='binary')
model.compile(loss='binary_crossentropy',
optimizer=optimizers.RMSprop(lr=2e-5),
metrics=['acc'])
history = model.fit_generator(
train_generator,
steps_per_epoch=100,
epochs=30,
validation_data=validation_generator,
validation_steps=50,
verbose=2)
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
val_loss_min = val_loss.index(min(val_loss))
val_acc_max = val_acc.index(max(val_acc))
print('validation set min loss: ', val_loss_min)
print('validation set max accuracy: ', val_acc_max)
```
## Fine-tuning
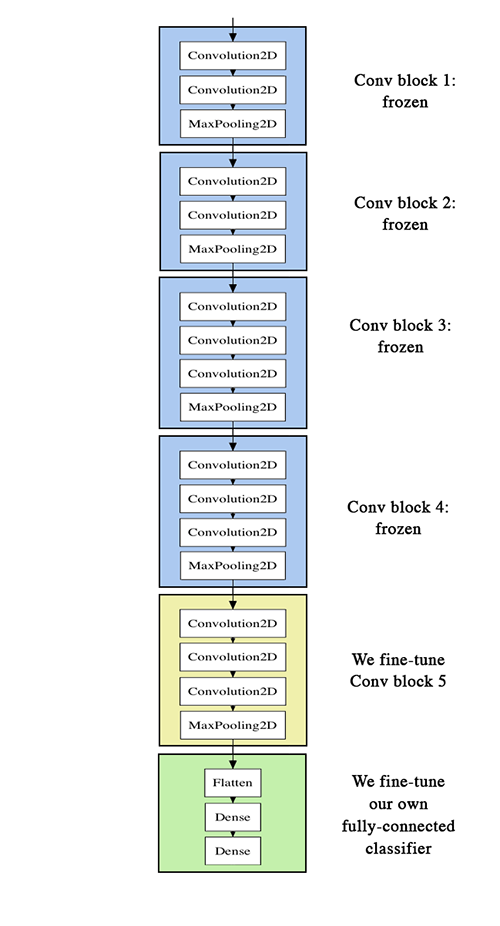
```
conv_base.summary()
conv_base.trainable = True
set_trainable = False
for layer in conv_base.layers:
if layer.name == 'block5_conv1':
set_trainable = True
if set_trainable:
layer.trainable = True
else:
layer.trainable = False
model.summary()
model.compile(optimizer=optimizers.RMSprop(lr=1e-5),
loss='binary_crossentropy',
metrics=['acc'])
history = model.fit_generator(train_generator,
steps_per_epoch=100,
epochs=100,
validation_data=validation_generator,
validation_steps=50,
verbose=0)
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
def smooth_curve(points, factor=0.8):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
plt.plot(epochs,
smooth_curve(acc), 'bo', label='Smoothed training acc')
plt.plot(epochs,
smooth_curve(val_acc), 'b', label='Smoothed validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs,
smooth_curve(loss), 'bo', label='Smoothed training loss')
plt.plot(epochs,
smooth_curve(val_loss), 'b', label='Smoothed validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
smooth_val_loss = smooth_curve(val_loss)
smooth_val_loss.index(min(smooth_val_loss))
test_generator = test_datagen.flow_from_directory(test_dir,
target_size=(150, 150),
batch_size=20,
class_mode='binary')
test_loss, test_acc = model.evaluate_generator(test_generator, steps=50)
print('test acc:', test_acc)
# plt.plot(history.history['loss'])
# plt.plot(history.history['val_loss'])
# plt.title('model loss')
# plt.ylabel('loss')
# plt.xlabel('epoch')
# plt.legend(['train', 'test'], loc='upper right')
# plt.show()
# plt.plot(history.history['acc'])
# plt.plot(history.history['val_acc'])
# plt.title('model accuracy')
# plt.ylabel('accuracy')
# plt.xlabel('epoch')
# plt.legend(['train', 'test'], loc='upper right')
# plt.show()
```
|
github_jupyter
|
# Widget Events
In this lecture we will discuss widget events, such as button clicks!
## Special events
The `Button` is not used to represent a data type. Instead the button widget is used to handle mouse clicks. The `on_click` method of the `Button` can be used to register a function to be called when the button is clicked. The docstring of the `on_click` can be seen below.
```
import ipywidgets as widgets
print(widgets.Button.on_click.__doc__)
```
### Example #1 - on_click
Since button clicks are stateless, they are transmitted from the front-end to the back-end using custom messages. By using the `on_click` method, a button that prints a message when it has been clicked is shown below.
```
from IPython.display import display
button = widgets.Button(description="Click Me!")
display(button)
def on_button_clicked(b):
print("Button clicked.")
button.on_click(on_button_clicked)
```
### Example #2 - on_submit
The `Text` widget also has a special `on_submit` event. The `on_submit` event fires when the user hits <kbd>enter</kbd>.
```
text = widgets.Text()
display(text)
def handle_submit(sender):
print(text.value)
text.on_submit(handle_submit)
```
## Traitlet events
Widget properties are IPython traitlets and traitlets are eventful. To handle changes, the `observe` method of the widget can be used to register a callback. The docstring for `observe` can be seen below.
```
print(widgets.Widget.observe.__doc__)
```
### Signatures
Mentioned in the docstring, the callback registered must have the signature `handler(change)` where `change` is a dictionary holding the information about the change.
Using this method, an example of how to output an `IntSlider`’s value as it is changed can be seen below.
```
int_range = widgets.IntSlider()
display(int_range)
def on_value_change(change):
print(change['new'])
int_range.observe(on_value_change, names='value')
```
# Linking Widgets
Often, you may want to simply link widget attributes together. Synchronization of attributes can be done in a simpler way than by using bare traitlets events.
## Linking traitlets attributes in the kernel¶
The first method is to use the `link` and `dlink` functions from the `traitlets` module. This only works if we are interacting with a live kernel.
```
import traitlets
# Create Caption
caption = widgets.Label(value = 'The values of slider1 and slider2 are synchronized')
# Create IntSliders
slider1 = widgets.IntSlider(description='Slider 1')
slider2 = widgets.IntSlider(description='Slider 2')
# Use trailets to link
l = traitlets.link((slider1, 'value'), (slider2, 'value'))
# Display!
display(caption, slider1, slider2)
# Create Caption
caption = widgets.Label(value='Changes in source values are reflected in target1')
# Create Sliders
source = widgets.IntSlider(description='Source')
target1 = widgets.IntSlider(description='Target 1')
# Use dlink
dl = traitlets.dlink((source, 'value'), (target1, 'value'))
display(caption, source, target1)
```
Function `traitlets.link` and `traitlets.dlink` return a `Link` or `DLink` object. The link can be broken by calling the `unlink` method.
```
# May get an error depending on order of cells being run!
l.unlink()
dl.unlink()
```
### Registering callbacks to trait changes in the kernel
Since attributes of widgets on the Python side are traitlets, you can register handlers to the change events whenever the model gets updates from the front-end.
The handler passed to observe will be called with one change argument. The change object holds at least a `type` key and a `name` key, corresponding respectively to the type of notification and the name of the attribute that triggered the notification.
Other keys may be passed depending on the value of `type`. In the case where type is `change`, we also have the following keys:
* `owner` : the HasTraits instance
* `old` : the old value of the modified trait attribute
* `new` : the new value of the modified trait attribute
* `name` : the name of the modified trait attribute.
```
caption = widgets.Label(value='The values of range1 and range2 are synchronized')
slider = widgets.IntSlider(min=-5, max=5, value=1, description='Slider')
def handle_slider_change(change):
caption.value = 'The slider value is ' + (
'negative' if change.new < 0 else 'nonnegative'
)
slider.observe(handle_slider_change, names='value')
display(caption, slider)
```
## Linking widgets attributes from the client side
When synchronizing traitlets attributes, you may experience a lag because of the latency due to the roundtrip to the server side. You can also directly link widget attributes in the browser using the link widgets, in either a unidirectional or a bidirectional fashion.
Javascript links persist when embedding widgets in html web pages without a kernel.
```
# NO LAG VERSION
caption = widgets.Label(value = 'The values of range1 and range2 are synchronized')
range1 = widgets.IntSlider(description='Range 1')
range2 = widgets.IntSlider(description='Range 2')
l = widgets.jslink((range1, 'value'), (range2, 'value'))
display(caption, range1, range2)
# NO LAG VERSION
caption = widgets.Label(value = 'Changes in source_range values are reflected in target_range')
source_range = widgets.IntSlider(description='Source range')
target_range = widgets.IntSlider(description='Target range')
dl = widgets.jsdlink((source_range, 'value'), (target_range, 'value'))
display(caption, source_range, target_range)
```
Function `widgets.jslink` returns a `Link` widget. The link can be broken by calling the `unlink` method.
```
l.unlink()
dl.unlink()
```
### The difference between linking in the kernel and linking in the client
Linking in the kernel means linking via python. If two sliders are linked in the kernel, when one slider is changed the browser sends a message to the kernel (python in this case) updating the changed slider, the link widget in the kernel then propagates the change to the other slider object in the kernel, and then the other slider’s kernel object sends a message to the browser to update the other slider’s views in the browser. If the kernel is not running (as in a static web page), then the controls will not be linked.
Linking using jslink (i.e., on the browser side) means contructing the link in Javascript. When one slider is changed, Javascript running in the browser changes the value of the other slider in the browser, without needing to communicate with the kernel at all. If the sliders are attached to kernel objects, each slider will update their kernel-side objects independently.
To see the difference between the two, go to the [ipywidgets documentation](http://ipywidgets.readthedocs.io/en/latest/examples/Widget%20Events.html) and try out the sliders near the bottom. The ones linked in the kernel with `link` and `dlink` are no longer linked, but the ones linked in the browser with `jslink` and `jsdlink` are still linked.
## Continuous updates
Some widgets offer a choice with their `continuous_update` attribute between continually updating values or only updating values when a user submits the value (for example, by pressing Enter or navigating away from the control). In the next example, we see the “Delayed” controls only transmit their value after the user finishes dragging the slider or submitting the textbox. The “Continuous” controls continually transmit their values as they are changed. Try typing a two-digit number into each of the text boxes, or dragging each of the sliders, to see the difference.
```
import traitlets
a = widgets.IntSlider(description="Delayed", continuous_update=False)
b = widgets.IntText(description="Delayed", continuous_update=False)
c = widgets.IntSlider(description="Continuous", continuous_update=True)
d = widgets.IntText(description="Continuous", continuous_update=True)
traitlets.link((a, 'value'), (b, 'value'))
traitlets.link((a, 'value'), (c, 'value'))
traitlets.link((a, 'value'), (d, 'value'))
widgets.VBox([a,b,c,d])
```
Sliders, `Text`, and `Textarea` controls default to `continuous_update=True`. `IntText` and other text boxes for entering integer or float numbers default to `continuous_update=False` (since often you’ll want to type an entire number before submitting the value by pressing enter or navigating out of the box).
# Conclusion
You should now feel comfortable linking Widget events!
|
github_jupyter
|
# Broadcast Variables
We already saw so called *broadcast joins* which is a specific impementation of a join suitable for small lookup tables. The term *broadcast* is also used in a different context in Spark, there are also *broadcast variables*.
### Origin of Broadcast Variables
Broadcast variables where introduced fairly early with Spark and were mainly targeted at the RDD API. Nontheless they still have their place with the high level DataFrames API in conjunction with user defined functions (UDFs).
### Weather Example
As usual, we'll use the weather data example. This time we'll manually implement a join using a UDF (actually this would be again a manual broadcast join).
# 1 Load Data
First we load the weather data, which consists of the measurement data and some station metadata.
```
storageLocation = "s3://dimajix-training/data/weather"
```
## 1.1 Load Measurements
Measurements are stored in multiple directories (one per year). But we will limit ourselves to a single year in the analysis to improve readability of execution plans.
```
from pyspark.sql.functions import *
from functools import reduce
# Read in all years, store them in an Python array
raw_weather_per_year = [spark.read.text(storageLocation + "/" + str(i)).withColumn("year", lit(i)) for i in range(2003,2015)]
# Union all years together
raw_weather = reduce(lambda l,r: l.union(r), raw_weather_per_year)
```
Use a single year to keep execution plans small
```
raw_weather = spark.read.text(storageLocation + "/2003").withColumn("year", lit(2003))
```
### Extract Measurements
Measurements were stored in a proprietary text based format, with some values at fixed positions. We need to extract these values with a simple SELECT statement.
```
weather = raw_weather.select(
col("year"),
substring(col("value"),5,6).alias("usaf"),
substring(col("value"),11,5).alias("wban"),
substring(col("value"),16,8).alias("date"),
substring(col("value"),24,4).alias("time"),
substring(col("value"),42,5).alias("report_type"),
substring(col("value"),61,3).alias("wind_direction"),
substring(col("value"),64,1).alias("wind_direction_qual"),
substring(col("value"),65,1).alias("wind_observation"),
(substring(col("value"),66,4).cast("float") / lit(10.0)).alias("wind_speed"),
substring(col("value"),70,1).alias("wind_speed_qual"),
(substring(col("value"),88,5).cast("float") / lit(10.0)).alias("air_temperature"),
substring(col("value"),93,1).alias("air_temperature_qual")
)
```
## 1.2 Load Station Metadata
We also need to load the weather station meta data containing information about the geo location, country etc of individual weather stations.
```
stations = spark.read \
.option("header", True) \
.csv(storageLocation + "/isd-history")
```
### Convert Station Metadata
We convert the stations DataFrame to a normal Python map, since we want to discuss broadcast variables. This means that the variable `py_stations` contains a normal Python object which only lives on the driver. It has no connection to Spark any more.
The resulting map converts a given station id (usaf and wban) to a country.
```
py_stations = stations.select(concat(stations["usaf"], stations["wban"]).alias("key"), stations["ctry"]).collect()
py_stations = {key:value for (key,value) in py_stations}
# Inspect result
list(py_stations.items())[0:10]
```
# 2 Using Broadcast Variables
In the following section, we want to use a Spark broadcast variable inside a UDF. Technically this is not required, as Spark also has other mechanisms of distributing data, so we'll start with a simple implementation *without* using a broadcast variable.
## 2.1 Create a UDF
For the initial implementation, we create a simple Python UDF which looks up the country for a given station id, which consists of the usaf and wban code. This way we will replace the `JOIN` of our original solution with a UDF implemented in Python.
```
def lookup_country(usaf, wban):
return py_stations.get(usaf + wban)
# Test lookup with an existing station
print(lookup_country("007026", "99999"))
# Test lookup with a non-existing station (better should not throw an exception)
print(lookup_country("123", "456"))
```
## 2.2 Not using a broadcast variable
Now that we have a simple Python function providing the required functionality, we convert it to a PySpark UDF using a Python decorator.
```
@udf('string')
def lookup_country(usaf, wban):
return py_stations.get(usaf + wban)
```
### Replace JOIN by UDF
Now we can perform the lookup by using the UDF instead of the original `JOIN`.
```
result = weather.withColumn('country', lookup_country(weather["usaf"], weather["wban"]))
result.limit(10).toPandas()
```
### Remarks
Since the code is actually executed not on the driver, but istributed on the executors, the executors also require access to the Python map. PySpark automatically serializes the map and sends it to the executors on the fly.
### Inspect Plan
We can also inspect the execution plan, which is different from the original implementation. Instead of the broadcast join, it now contains a `BatchEvalPython` step which looks up the stations country from the station id.
```
result.explain()
```
## 2.2 Using a Broadcast Variable
Now let us change the implementation to use a so called *broadcast variable*. While the original implementation implicitly sent the Python map to all executors, a broadcast variable makes the process of sending (*broadcasting*) a Python variable to all executors more explicit.
A Python variable can be broadcast using the `broadcast` method of the underlying Spark context (the Spark session does not export this functionality). Once the data is encapsulated in the broadcast variable, all executors can access the original data via the `value` member variable.
```
# First create a broadcast variable from the original Python map
bc_stations = spark.sparkContext.broadcast(py_stations)
@udf('string')
def lookup_country(usaf, wban):
# Access the broadcast variables value and perform lookup
return bc_stations.value.get(usaf + wban)
```
### Replace JOIN by UDF
Again we replace the original `JOIN` by the UDF we just defined above
```
result = weather.withColumn('country', lookup_country(weather["usaf"], weather["wban"]))
result.limit(10).toPandas()
```
### Remarks
Actually there is no big difference to the original implementation. But Spark handles a broadcast variable slightly more efficiently, especially if the variable is used in multiple UDFs. In this case the data will be broadcast only a single time, while not using a broadcast variable would imply sending the data around for every UDF.
### Execution Plan
The execution plan does not differ at all, since it does not provide information on broadcast variables.
```
result.explain()
```
## 2.3 Pandas UDFs
Since we already learnt that Pandas UDFs are executed more efficiently than normal UDFs, we want to provide a better implementation using Pandas. Of course Pandas UDFs can also access broadcast variables.
```
from pyspark.sql.functions import pandas_udf, PandasUDFType
@pandas_udf('string', PandasUDFType.SCALAR)
def lookup_country(usaf, wban):
# Create helper function
def lookup(key):
# Perform lookup by accessing the Python map
return bc_stations.value.get(key)
# Create key from both incoming Pandas series
usaf_wban = usaf + wban
# Perform lookup
return usaf_wban.apply(lookup)
```
### Replace JOIN by Pandas UDF
Again, we replace the original `JOIN` by the Pandas UDF.
```
result = weather.withColumn('country', lookup_country(weather["usaf"], weather["wban"]))
result.limit(10).toPandas()
```
### Execution Plan
Again, let's inspect the execution plan.
```
result.explain(True)
```
|
github_jupyter
|
# Sudoku
This tutorial includes everything you need to set up decision optimization engines, build constraint programming models.
When you finish this tutorial, you'll have a foundational knowledge of _Prescriptive Analytics_.
>This notebook is part of the **[Prescriptive Analytics for Python](https://rawgit.com/IBMDecisionOptimization/docplex-doc/master/docs/index.html)**
>It requires a **local installation of CPLEX Optimizers**.
Table of contents:
- [Describe the business problem](#Describe-the-business-problem)
* [How decision optimization (prescriptive analytics) can help](#How--decision-optimization-can-help)
* [Use decision optimization](#Use-decision-optimization)
* [Step 1: Download the library](#Step-1:-Download-the-library)
* [Step 2: Model the Data](#Step-2:-Model-the-data)
* [Step 3: Set up the prescriptive model](#Step-3:-Set-up-the-prescriptive-model)
* [Define the decision variables](#Define-the-decision-variables)
* [Express the business constraints](#Express-the-business-constraints)
* [Express the objective](#Express-the-objective)
* [Solve with Decision Optimization solve service](#Solve-with-Decision-Optimization-solve-service)
* [Step 4: Investigate the solution and run an example analysis](#Step-4:-Investigate-the-solution-and-then-run-an-example-analysis)
* [Summary](#Summary)
****
### Describe the business problem
* Sudoku is a logic-based, combinatorial number-placement puzzle.
* The objective is to fill a 9x9 grid with digits so that each column, each row,
and each of the nine 3x3 sub-grids that compose the grid contains all of the digits from 1 to 9.
* The puzzle setter provides a partially completed grid, which for a well-posed puzzle has a unique solution.
#### References
* See https://en.wikipedia.org/wiki/Sudoku for details
*****
## How decision optimization can help
* Prescriptive analytics technology recommends actions based on desired outcomes, taking into account specific scenarios, resources, and knowledge of past and current events. This insight can help your organization make better decisions and have greater control of business outcomes.
* Prescriptive analytics is the next step on the path to insight-based actions. It creates value through synergy with predictive analytics, which analyzes data to predict future outcomes.
* Prescriptive analytics takes that insight to the next level by suggesting the optimal way to handle that future situation. Organizations that can act fast in dynamic conditions and make superior decisions in uncertain environments gain a strong competitive advantage.
<br/>
+ For example:
+ Automate complex decisions and trade-offs to better manage limited resources.
+ Take advantage of a future opportunity or mitigate a future risk.
+ Proactively update recommendations based on changing events.
+ Meet operational goals, increase customer loyalty, prevent threats and fraud, and optimize business processes.
## Use decision optimization
### Step 1: Download the library
Run the following code to install Decision Optimization CPLEX Modeling library. The *DOcplex* library contains the two modeling packages, Mathematical Programming and Constraint Programming, referred to earlier.
```
import sys
try:
import docplex.cp
except:
if hasattr(sys, 'real_prefix'):
#we are in a virtual env.
!pip install docplex
else:
!pip install --user docplex
```
Note that the more global package <i>docplex</i> contains another subpackage <i>docplex.mp</i> that is dedicated to Mathematical Programming, another branch of optimization.
```
from docplex.cp.model import *
from sys import stdout
```
### Step 2: Model the data
#### Grid range
```
GRNG = range(9)
```
#### Different problems
_zero means cell to be filled with appropriate value_
```
SUDOKU_PROBLEM_1 = ( (0, 0, 0, 0, 9, 0, 1, 0, 0),
(2, 8, 0, 0, 0, 5, 0, 0, 0),
(7, 0, 0, 0, 0, 6, 4, 0, 0),
(8, 0, 5, 0, 0, 3, 0, 0, 6),
(0, 0, 1, 0, 0, 4, 0, 0, 0),
(0, 7, 0, 2, 0, 0, 0, 0, 0),
(3, 0, 0, 0, 0, 1, 0, 8, 0),
(0, 0, 0, 0, 0, 0, 0, 5, 0),
(0, 9, 0, 0, 0, 0, 0, 7, 0),
)
SUDOKU_PROBLEM_2 = ( (0, 7, 0, 0, 0, 0, 0, 4, 9),
(0, 0, 0, 4, 0, 0, 0, 0, 0),
(4, 0, 3, 5, 0, 7, 0, 0, 8),
(0, 0, 7, 2, 5, 0, 4, 0, 0),
(0, 0, 0, 0, 0, 0, 8, 0, 0),
(0, 0, 4, 0, 3, 0, 5, 9, 2),
(6, 1, 8, 0, 0, 0, 0, 0, 5),
(0, 9, 0, 1, 0, 0, 0, 3, 0),
(0, 0, 5, 0, 0, 0, 0, 0, 7),
)
SUDOKU_PROBLEM_3 = ( (0, 0, 0, 0, 0, 6, 0, 0, 0),
(0, 5, 9, 0, 0, 0, 0, 0, 8),
(2, 0, 0, 0, 0, 8, 0, 0, 0),
(0, 4, 5, 0, 0, 0, 0, 0, 0),
(0, 0, 3, 0, 0, 0, 0, 0, 0),
(0, 0, 6, 0, 0, 3, 0, 5, 4),
(0, 0, 0, 3, 2, 5, 0, 0, 6),
(0, 0, 0, 0, 0, 0, 0, 0, 0),
(0, 0, 0, 0, 0, 0, 0, 0, 0)
)
try:
import numpy as np
import matplotlib.pyplot as plt
VISU_ENABLED = True
except ImportError:
VISU_ENABLED = False
def print_grid(grid):
""" Print Sudoku grid """
for l in GRNG:
if (l > 0) and (l % 3 == 0):
stdout.write('\n')
for c in GRNG:
v = grid[l][c]
stdout.write(' ' if (c % 3 == 0) else ' ')
stdout.write(str(v) if v > 0 else '.')
stdout.write('\n')
def draw_grid(values):
%matplotlib inline
fig, ax = plt.subplots(figsize =(4,4))
min_val, max_val = 0, 9
R = range(0,9)
for l in R:
for c in R:
v = values[c][l]
s = " "
if v > 0:
s = str(v)
ax.text(l+0.5,8.5-c, s, va='center', ha='center')
ax.set_xlim(min_val, max_val)
ax.set_ylim(min_val, max_val)
ax.set_xticks(np.arange(max_val))
ax.set_yticks(np.arange(max_val))
ax.grid()
plt.show()
def display_grid(grid, name):
stdout.write(name)
stdout.write(":\n")
if VISU_ENABLED:
draw_grid(grid)
else:
print_grid(grid)
display_grid(SUDOKU_PROBLEM_1, "PROBLEM 1")
display_grid(SUDOKU_PROBLEM_2, "PROBLEM 2")
display_grid(SUDOKU_PROBLEM_3, "PROBLEM 3")
```
#### Choose your preferred problem (SUDOKU_PROBLEM_1 or SUDOKU_PROBLEM_2 or SUDOKU_PROBLEM_3)
If you change the problem, ensure to re-run all cells below this one.
```
problem = SUDOKU_PROBLEM_3
```
### Step 3: Set up the prescriptive model
```
mdl = CpoModel(name="Sudoku")
```
#### Define the decision variables
```
grid = [[integer_var(min=1, max=9, name="C" + str(l) + str(c)) for l in GRNG] for c in GRNG]
```
#### Express the business constraints
Add alldiff constraints for lines
```
for l in GRNG:
mdl.add(all_diff([grid[l][c] for c in GRNG]))
```
Add alldiff constraints for columns
```
for c in GRNG:
mdl.add(all_diff([grid[l][c] for l in GRNG]))
```
Add alldiff constraints for sub-squares
```
ssrng = range(0, 9, 3)
for sl in ssrng:
for sc in ssrng:
mdl.add(all_diff([grid[l][c] for l in range(sl, sl + 3) for c in range(sc, sc + 3)]))
```
Initialize known cells
```
for l in GRNG:
for c in GRNG:
v = problem[l][c]
if v > 0:
grid[l][c].set_domain((v, v))
```
#### Solve with Decision Optimization solve service
```
print("\nSolving model....")
msol = mdl.solve(TimeLimit=10)
```
### Step 4: Investigate the solution and then run an example analysis
```
display_grid(problem, "Initial problem")
if msol:
sol = [[msol[grid[l][c]] for c in GRNG] for l in GRNG]
stdout.write("Solve time: " + str(msol.get_solve_time()) + "\n")
display_grid(sol, "Solution")
else:
stdout.write("No solution found\n")
```
## Summary
You learned how to set up and use the IBM Decision Optimization CPLEX Modeling for Python to formulate and solve a Constraint Programming model.
#### References
* [CPLEX Modeling for Python documentation](https://rawgit.com/IBMDecisionOptimization/docplex-doc/master/docs/index.html)
* [Decision Optimization on Cloud](https://developer.ibm.com/docloud/)
* Need help with DOcplex or to report a bug? Please go [here](https://developer.ibm.com/answers/smartspace/docloud)
* Contact us at [email protected]
Copyright © 2017, 2018 IBM. IPLA licensed Sample Materials.
|
github_jupyter
|
# RadarCOVID-Report
## Data Extraction
```
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import pycountry
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
```
### Constants
```
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
```
### Parameters
```
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
environment_enable_multi_backend_download = \
os.environ.get("RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD")
if environment_enable_multi_backend_download:
report_backend_identifiers = None
else:
report_backend_identifiers = [report_backend_identifier]
report_backend_identifiers
environment_invalid_shared_diagnoses_dates = \
os.environ.get("RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES")
if environment_invalid_shared_diagnoses_dates:
invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(",")
else:
invalid_shared_diagnoses_dates = []
invalid_shared_diagnoses_dates
```
### COVID-19 Cases
```
report_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=report_backend_identifier)
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe():
return pd.read_csv("https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv")
confirmed_df_ = download_cases_dataframe()
confirmed_df_.iloc[0]
confirmed_df = confirmed_df_.copy()
confirmed_df = confirmed_df[["date", "new_cases", "iso_code"]]
confirmed_df.rename(
columns={
"date": "sample_date",
"iso_code": "country_code",
},
inplace=True)
def convert_iso_alpha_3_to_alpha_2(x):
try:
return pycountry.countries.get(alpha_3=x).alpha_2
except Exception as e:
logging.info(f"Error converting country ISO Alpha 3 code '{x}': {repr(e)}")
return None
confirmed_df["country_code"] = confirmed_df.country_code.apply(convert_iso_alpha_3_to_alpha_2)
confirmed_df.dropna(inplace=True)
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_days_df["sample_date_string"] = \
confirmed_days_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_days_df.tail()
def sort_source_regions_for_display(source_regions: list) -> list:
if report_backend_identifier in source_regions:
source_regions = [report_backend_identifier] + \
list(sorted(set(source_regions).difference([report_backend_identifier])))
else:
source_regions = list(sorted(source_regions))
return source_regions
report_source_regions = report_backend_client.source_regions_for_date(
date=extraction_datetime.date())
report_source_regions = sort_source_regions_for_display(
source_regions=report_source_regions)
report_source_regions
def get_cases_dataframe(source_regions_for_date_function, columns_suffix=None):
source_regions_at_date_df = confirmed_days_df.copy()
source_regions_at_date_df["source_regions_at_date"] = \
source_regions_at_date_df.sample_date.apply(
lambda x: source_regions_for_date_function(date=x))
source_regions_at_date_df.sort_values("sample_date", inplace=True)
source_regions_at_date_df["_source_regions_group"] = source_regions_at_date_df. \
source_regions_at_date.apply(lambda x: ",".join(sort_source_regions_for_display(x)))
source_regions_at_date_df.tail()
#%%
source_regions_for_summary_df_ = \
source_regions_at_date_df[["sample_date", "_source_regions_group"]].copy()
source_regions_for_summary_df_.rename(columns={"_source_regions_group": "source_regions"}, inplace=True)
source_regions_for_summary_df_.tail()
#%%
confirmed_output_columns = ["sample_date", "new_cases", "covid_cases"]
confirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)
for source_regions_group, source_regions_group_series in \
source_regions_at_date_df.groupby("_source_regions_group"):
source_regions_set = set(source_regions_group.split(","))
confirmed_source_regions_set_df = \
confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()
confirmed_source_regions_group_df = \
confirmed_source_regions_set_df.groupby("sample_date").new_cases.sum() \
.reset_index().sort_values("sample_date")
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df.merge(
confirmed_days_df[["sample_date_string"]].rename(
columns={"sample_date_string": "sample_date"}),
how="right")
confirmed_source_regions_group_df["new_cases"] = \
confirmed_source_regions_group_df["new_cases"].clip(lower=0)
confirmed_source_regions_group_df["covid_cases"] = \
confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[confirmed_output_columns]
confirmed_source_regions_group_df = confirmed_source_regions_group_df.replace(0, np.nan)
confirmed_source_regions_group_df.fillna(method="ffill", inplace=True)
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[
confirmed_source_regions_group_df.sample_date.isin(
source_regions_group_series.sample_date_string)]
confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)
result_df = confirmed_output_df.copy()
result_df.tail()
#%%
result_df.rename(columns={"sample_date": "sample_date_string"}, inplace=True)
result_df = confirmed_days_df[["sample_date_string"]].merge(result_df, how="left")
result_df.sort_values("sample_date_string", inplace=True)
result_df.fillna(method="ffill", inplace=True)
result_df.tail()
#%%
result_df[["new_cases", "covid_cases"]].plot()
if columns_suffix:
result_df.rename(
columns={
"new_cases": "new_cases_" + columns_suffix,
"covid_cases": "covid_cases_" + columns_suffix},
inplace=True)
return result_df, source_regions_for_summary_df_
confirmed_eu_df, source_regions_for_summary_df = get_cases_dataframe(
report_backend_client.source_regions_for_date)
confirmed_es_df, _ = get_cases_dataframe(
lambda date: [spain_region_country_code],
columns_suffix=spain_region_country_code.lower())
```
### Extract API TEKs
```
raw_zip_path_prefix = "Data/TEKs/Raw/"
base_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=base_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
```
### Dump API TEKs
```
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_base_df = tek_list_df[tek_list_df.region == report_backend_identifier]
tek_list_base_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_base_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_base_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_base_df.head()
```
### Load TEK Dumps
```
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
```
### Daily New TEKs
```
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \
.groupby(["upload_date"]).shared_teks.max().reset_index() \
.sort_values(["upload_date"], ascending=False) \
.rename(columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
invalid_shared_diagnoses_dates_mask = \
estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)
estimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0
estimated_shared_diagnoses_df.head()
```
### Hourly New TEKs
```
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
```
### Official Statistics
```
import requests
import pandas.io.json
official_stats_response = requests.get("https://radarcovid.covid19.gob.es/kpi/statistics/basics")
official_stats_response.raise_for_status()
official_stats_df_ = pandas.io.json.json_normalize(official_stats_response.json())
official_stats_df = official_stats_df_.copy()
official_stats_df["date"] = pd.to_datetime(official_stats_df["date"], dayfirst=True)
official_stats_df.head()
official_stats_column_map = {
"date": "sample_date",
"applicationsDownloads.totalAcummulated": "app_downloads_es_accumulated",
"communicatedContagions.totalAcummulated": "shared_diagnoses_es_accumulated",
}
accumulated_suffix = "_accumulated"
accumulated_values_columns = \
list(filter(lambda x: x.endswith(accumulated_suffix), official_stats_column_map.values()))
interpolated_values_columns = \
list(map(lambda x: x[:-len(accumulated_suffix)], accumulated_values_columns))
official_stats_df = \
official_stats_df[official_stats_column_map.keys()] \
.rename(columns=official_stats_column_map)
official_stats_df["extraction_date"] = extraction_date
official_stats_df.head()
official_stats_path = "Data/Statistics/Current/RadarCOVID-Statistics.json"
previous_official_stats_df = pd.read_json(official_stats_path, orient="records", lines=True)
previous_official_stats_df["sample_date"] = pd.to_datetime(previous_official_stats_df["sample_date"], dayfirst=True)
official_stats_df = official_stats_df.append(previous_official_stats_df)
official_stats_df.head()
official_stats_df = official_stats_df[~(official_stats_df.shared_diagnoses_es_accumulated == 0)]
official_stats_df.sort_values("extraction_date", ascending=False, inplace=True)
official_stats_df.drop_duplicates(subset=["sample_date"], keep="first", inplace=True)
official_stats_df.head()
official_stats_stored_df = official_stats_df.copy()
official_stats_stored_df["sample_date"] = official_stats_stored_df.sample_date.dt.strftime("%Y-%m-%d")
official_stats_stored_df.to_json(official_stats_path, orient="records", lines=True)
official_stats_df.drop(columns=["extraction_date"], inplace=True)
official_stats_df = confirmed_days_df.merge(official_stats_df, how="left")
official_stats_df.sort_values("sample_date", ascending=False, inplace=True)
official_stats_df.head()
official_stats_df[accumulated_values_columns] = \
official_stats_df[accumulated_values_columns] \
.astype(float).interpolate(limit_area="inside")
official_stats_df[interpolated_values_columns] = \
official_stats_df[accumulated_values_columns].diff(periods=-1)
official_stats_df.drop(columns="sample_date", inplace=True)
official_stats_df.head()
```
### Data Merge
```
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
official_stats_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_eu_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df = confirmed_es_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df = result_summary_df.merge(source_regions_for_summary_df, how="left")
result_summary_df.set_index(["sample_date", "source_regions"], inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(result_summary_df.shared_diagnoses_es / result_summary_df.covid_cases_es).fillna(0)
result_summary_df.head(daily_plot_days)
def compute_aggregated_results_summary(days) -> pd.DataFrame:
aggregated_result_summary_df = result_summary_df.copy()
aggregated_result_summary_df["covid_cases_for_ratio"] = \
aggregated_result_summary_df.covid_cases.mask(
aggregated_result_summary_df.shared_diagnoses == 0, 0)
aggregated_result_summary_df["covid_cases_for_ratio_es"] = \
aggregated_result_summary_df.covid_cases_es.mask(
aggregated_result_summary_df.shared_diagnoses_es == 0, 0)
aggregated_result_summary_df = aggregated_result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(days).agg({
"covid_cases": "sum",
"covid_cases_es": "sum",
"covid_cases_for_ratio": "sum",
"covid_cases_for_ratio_es": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum",
"shared_diagnoses_es": "sum",
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
aggregated_result_summary_df = aggregated_result_summary_df.fillna(0).astype(int)
aggregated_result_summary_df["teks_per_shared_diagnosis"] = \
(aggregated_result_summary_df.shared_teks_by_upload_date /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case"] = \
(aggregated_result_summary_df.shared_diagnoses /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(aggregated_result_summary_df.shared_diagnoses_es /
aggregated_result_summary_df.covid_cases_for_ratio_es).fillna(0)
return aggregated_result_summary_df
aggregated_result_with_7_days_window_summary_df = compute_aggregated_results_summary(days=7)
aggregated_result_with_7_days_window_summary_df.head()
last_7_days_summary = aggregated_result_with_7_days_window_summary_df.to_dict(orient="records")[1]
last_7_days_summary
aggregated_result_with_14_days_window_summary_df = compute_aggregated_results_summary(days=13)
last_14_days_summary = aggregated_result_with_14_days_window_summary_df.to_dict(orient="records")[1]
last_14_days_summary
```
## Report Results
```
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"source_regions": "Source Countries",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases (Source Countries)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date (Source Countries)",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date (Source Countries)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date (Source Countries)",
"shared_diagnoses": "Shared Diagnoses (Source Countries – Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis (Source Countries)",
"shared_diagnoses_per_covid_case": "Usage Ratio (Source Countries)",
"covid_cases_es": "COVID-19 Cases (Spain)",
"app_downloads_es": "App Downloads (Spain – Official)",
"shared_diagnoses_es": "Shared Diagnoses (Spain – Official)",
"shared_diagnoses_per_covid_case_es": "Usage Ratio (Spain)",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
"covid_cases_es",
"app_downloads_es",
"shared_diagnoses_es",
"shared_diagnoses_per_covid_case_es",
]
summary_percentage_columns= [
"shared_diagnoses_per_covid_case_es",
"shared_diagnoses_per_covid_case",
]
```
### Daily Summary Table
```
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
```
### Daily Summary Plots
```
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.droplevel(level=["source_regions"]) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 30), legend=False)
ax_ = summary_ax_list[0]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
for percentage_column in summary_percentage_columns:
percentage_column_index = summary_columns.index(percentage_column)
summary_ax_list[percentage_column_index].yaxis \
.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
```
### Daily Generation to Upload Period Table
```
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
```
### Hourly Summary Plots
```
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
```
### Publish Results
```
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case_es"]: lambda x: f"{x:.2%}" if x != 0 else "",
}
general_columns = \
list(filter(lambda x: x not in display_formatters, display_column_name_mapping.values()))
general_formatter = lambda x: f"{x}" if x != 0 else ""
display_formatters.update(dict(map(lambda x: (x, general_formatter), general_columns)))
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index.get_level_values("sample_date") == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.item()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.item()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.item()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.item()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.item()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
if len(report_source_regions) == 1:
display_brief_source_regions = report_source_regions[0]
else:
display_brief_source_regions = f"{len(report_source_regions)} 🇪🇺"
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
df = df.copy()
df_styler = df.style.format(display_formatters)
media_path = get_temporary_image_path()
dfi.export(df_styler, media_path)
return media_path
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
```
### Save Results
```
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
```
### Publish Results as JSON
```
def generate_summary_api_results(df: pd.DataFrame) -> list:
api_df = df.reset_index().copy()
api_df["sample_date_string"] = \
api_df["sample_date"].dt.strftime("%Y-%m-%d")
api_df["source_regions"] = \
api_df["source_regions"].apply(lambda x: x.split(","))
return api_df.to_dict(orient="records")
summary_api_results = \
generate_summary_api_results(df=result_summary_df)
today_summary_api_results = \
generate_summary_api_results(df=extraction_date_result_summary_df)[0]
summary_results = dict(
backend_identifier=report_backend_identifier,
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=today_summary_api_results,
last_7_days=last_7_days_summary,
last_14_days=last_14_days_summary,
daily_results=summary_api_results)
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
```
### Publish on README
```
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
```
### Publish on Twitter
```
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
def format_shared_diagnoses_per_covid_case(value) -> str:
if value == 0:
return "–"
return f"≤{value:.2%}"
display_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=shared_diagnoses_per_covid_case)
display_last_14_days_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case"])
display_last_14_days_shared_diagnoses_per_covid_case_es = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case_es"])
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: {display_shared_diagnoses_per_covid_case}
Last 14 Days:
- Usage Ratio (Estimation): {display_last_14_days_shared_diagnoses_per_covid_case}
- Usage Ratio (Official): {display_last_14_days_shared_diagnoses_per_covid_case_es}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
```
|
github_jupyter
|
## Dragon Real Estate - Price Predictor
```
import pandas as pd
housing = pd.read_csv("data.csv")
housing.head()
housing.info()
housing['CHAS'].value_counts()
housing.describe()
%matplotlib inline
# # For plotting histogram
# import matplotlib.pyplot as plt
# housing.hist(bins=50, figsize=(20, 15))
```
## Train-Test Splitting
```
# For learning purpose
import numpy as np
def split_train_test(data, test_ratio):
np.random.seed(42)
shuffled = np.random.permutation(len(data))
print(shuffled)
test_set_size = int(len(data) * test_ratio)
test_indices = shuffled[:test_set_size]
train_indices = shuffled[test_set_size:]
return data.iloc[train_indices], data.iloc[test_indices]
# train_set, test_set = split_train_test(housing, 0.2)
# print(f"Rows in train set: {len(train_set)}\nRows in test set: {len(test_set)}\n")
from sklearn.model_selection import train_test_split
train_set, test_set = train_test_split(housing, test_size=0.2, random_state=42)
print(f"Rows in train set: {len(train_set)}\nRows in test set: {len(test_set)}\n")
from sklearn.model_selection import StratifiedShuffleSplit
split = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=42)
for train_index, test_index in split.split(housing, housing['CHAS']):
strat_train_set = housing.loc[train_index]
strat_test_set = housing.loc[test_index]
strat_test_set['CHAS'].value_counts()
strat_train_set['CHAS'].value_counts()
# 95/7
# 376/28
housing = strat_train_set.copy()
```
## Looking for Correlations
```
corr_matrix = housing.corr()
corr_matrix['MEDV'].sort_values(ascending=False)
# from pandas.plotting import scatter_matrix
# attributes = ["MEDV", "RM", "ZN", "LSTAT"]
# scatter_matrix(housing[attributes], figsize = (12,8))
housing.plot(kind="scatter", x="RM", y="MEDV", alpha=0.8)
```
## Trying out Attribute combinations
```
housing["TAXRM"] = housing['TAX']/housing['RM']
housing.head()
corr_matrix = housing.corr()
corr_matrix['MEDV'].sort_values(ascending=False)
housing.plot(kind="scatter", x="TAXRM", y="MEDV", alpha=0.8)
housing = strat_train_set.drop("MEDV", axis=1)
housing_labels = strat_train_set["MEDV"].copy()
```
## Missing Attributes
```
# To take care of missing attributes, you have three options:
# 1. Get rid of the missing data points
# 2. Get rid of the whole attribute
# 3. Set the value to some value(0, mean or median)
a = housing.dropna(subset=["RM"]) #Option 1
a.shape
# Note that the original housing dataframe will remain unchanged
housing.drop("RM", axis=1).shape # Option 2
# Note that there is no RM column and also note that the original housing dataframe will remain unchanged
median = housing["RM"].median() # Compute median for Option 3
housing["RM"].fillna(median) # Option 3
# Note that the original housing dataframe will remain unchanged
housing.shape
housing.describe() # before we started filling missing attributes
from sklearn.impute import SimpleImputer
imputer = SimpleImputer(strategy="median")
imputer.fit(housing)
imputer.statistics_
X = imputer.transform(housing)
housing_tr = pd.DataFrame(X, columns=housing.columns)
housing_tr.describe()
```
## Scikit-learn Design
Primarily, three types of objects
1. Estimators - It estimates some parameter based on a dataset. Eg. imputer. It has a fit method and transform method. Fit method - Fits the dataset and calculates internal parameters
2. Transformers - transform method takes input and returns output based on the learnings from fit(). It also has a convenience function called fit_transform() which fits and then transforms.
3. Predictors - LinearRegression model is an example of predictor. fit() and predict() are two common functions. It also gives score() function which will evaluate the predictions.
## Feature Scaling
Primarily, two types of feature scaling methods:
1. Min-max scaling (Normalization)
(value - min)/(max - min)
Sklearn provides a class called MinMaxScaler for this
2. Standardization
(value - mean)/std
Sklearn provides a class called StandardScaler for this
## Creating a Pipeline
```
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
my_pipeline = Pipeline([
('imputer', SimpleImputer(strategy="median")),
# ..... add as many as you want in your pipeline
('std_scaler', StandardScaler()),
])
housing_num_tr = my_pipeline.fit_transform(housing)
housing_num_tr.shape
```
## Selecting a desired model for Dragon Real Estates
```
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
# model = LinearRegression()
# model = DecisionTreeRegressor()
model = RandomForestRegressor()
model.fit(housing_num_tr, housing_labels)
some_data = housing.iloc[:5]
some_labels = housing_labels.iloc[:5]
prepared_data = my_pipeline.transform(some_data)
model.predict(prepared_data)
list(some_labels)
```
## Evaluating the model
```
from sklearn.metrics import mean_squared_error
housing_predictions = model.predict(housing_num_tr)
mse = mean_squared_error(housing_labels, housing_predictions)
rmse = np.sqrt(mse)
rmse
```
## Using better evaluation technique - Cross Validation
```
# 1 2 3 4 5 6 7 8 9 10
from sklearn.model_selection import cross_val_score
scores = cross_val_score(model, housing_num_tr, housing_labels, scoring="neg_mean_squared_error", cv=10)
rmse_scores = np.sqrt(-scores)
rmse_scores
def print_scores(scores):
print("Scores:", scores)
print("Mean: ", scores.mean())
print("Standard deviation: ", scores.std())
print_scores(rmse_scores)
```
Quiz: Convert this notebook into a python file and run the pipeline using Visual Studio Code
## Saving the model
```
from joblib import dump, load
dump(model, 'Dragon.joblib')
```
## Testing the model on test data
```
X_test = strat_test_set.drop("MEDV", axis=1)
Y_test = strat_test_set["MEDV"].copy()
X_test_prepared = my_pipeline.transform(X_test)
final_predictions = model.predict(X_test_prepared)
final_mse = mean_squared_error(Y_test, final_predictions)
final_rmse = np.sqrt(final_mse)
# print(final_predictions, list(Y_test))
final_rmse
prepared_data[0]
```
## Using the model
```
from joblib import dump, load
import numpy as np
model = load('Dragon.joblib')
features = np.array([[-5.43942006, 4.12628155, -1.6165014, -0.67288841, -1.42262747,
-11.44443979304, -49.31238772, 7.61111401, -26.0016879 , -0.5778192 ,
-0.97491834, 0.41164221, -66.86091034]])
model.predict(features)
```
|
github_jupyter
|
# Replacing scalar values I
In this exercise, we will replace a list of values in our dataset by using the .replace() method with another list of desired values.
We will apply the functions in the poker_hands DataFrame. Remember that in the poker_hands DataFrame, each row of columns R1 to R5 represents the rank of each card from a player's poker hand spanning from 1 (Ace) to 13 (King). The Class feature classifies each hand as a category, and the Explanation feature briefly explains each hand.
The poker_hands DataFrame is already loaded for you, and you can explore the features Class and Explanation.
Remember you can always explore the dataset and see how it changes in the IPython Shell, and refer to the slides in the Slides tab.
```
import pandas as pd
poker_hands = pd.read_csv('../datasets/poker_hand.csv')
poker_hands
# Replace Class 1 to -2
poker_hands['Class'].replace(1, -2, inplace=True)
# Replace Class 2 to -3
poker_hands['Class'].replace(2, -3, inplace=True)
print(poker_hands[['Class']])
```
# Replace scalar values II
As discussed in the video, in a pandas DataFrame, it is possible to replace values in a very intuitive way: we locate the position (row and column) in the Dataframe and assign in the new value you want to replace with. In a more pandas-ian way, the .replace() function is available that performs the same task.
You will be using the names DataFrame which includes, among others, the most popular names in the US by year, gender and ethnicity.
Your task is to replace all the babies that are classified as FEMALE to GIRL using the following methods:
- intuitive scalar replacement
- using the .replace() function
```
names = pd.read_csv('../datasets/Popular_Baby_Names.csv')
names.head()
import time
start_time = time.time()
# Replace all the entries that has 'FEMALE' as a gender with 'GIRL'
names['Gender'].loc[names['Gender'] == 'FEMALE'] = 'GIRL'
print("Time using .loc[]: {} sec".format(time.time() - start_time))
start_time = time.time()
# Replace all the entries that has 'FEMALE' as a gender with 'GIRL'
names['Gender'].replace('FEMALE', 'GIRL', inplace=True)
print("Time using .replace(): {} sec".format(time.time() - start_time))
```
# Replace multiple values I
In this exercise, you will apply the .replace() function for the task of replacing multiple values with one or more values. You will again use the names dataset which contains, among others, the most popular names in the US by year, gender and Ethnicity.
Thus you want to replace all ethnicities classified as black or white non-hispanics to non-hispanic. Remember, the ethnicities are stated in the dataset as follows: ```['BLACK NON HISP', 'BLACK NON HISPANIC', 'WHITE NON HISP' , 'WHITE NON HISPANIC']``` and should be replaced to 'NON HISPANIC'
```
start_time = time.time()
# Replace all non-Hispanic ethnicities with 'NON HISPANIC'
names['Ethnicity'].loc[(names["Ethnicity"] == 'BLACK NON HISP') |
(names["Ethnicity"] == 'BLACK NON HISPANIC') |
(names["Ethnicity"] == 'WHITE NON HISP') |
(names["Ethnicity"] == 'WHITE NON HISPANIC')] = 'NON HISPANIC'
print("Time using .loc[]: {0} sec".format(time.time() - start_time))
start_time = time.time()
# Replace all non-Hispanic ethnicities with 'NON HISPANIC'
names['Ethnicity'].replace(['BLACK NON HISP', 'BLACK NON HISPANIC', 'WHITE NON HISP' , 'WHITE NON HISPANIC'], 'NON HISPANIC', inplace=True)
print("Time using .replace(): {} sec".format(time.time() - start_time))
```
# Replace multiple values II
As discussed in the video, instead of using the .replace() function multiple times to replace multiple values, you can use lists to map the elements you want to replace one to one with those you want to replace them with.
As you have seen in our popular names dataset, there are two names for the same ethnicity. We want to standardize the naming of each ethnicity by replacing
- 'ASIAN AND PACI' to 'ASIAN AND PACIFIC ISLANDER'
- 'BLACK NON HISP' to 'BLACK NON HISPANIC'
- 'WHITE NON HISP' to 'WHITE NON HISPANIC'
In the DataFrame names, you are going to replace all the values on the left by the values on the right.
```
start_time = time.time()
# Replace ethnicities as instructed
names['Ethnicity'].replace(['ASIAN AND PACI','BLACK NON HISP', 'WHITE NON HISP'], ['ASIAN AND PACIFIC ISLANDER','BLACK NON HISPANIC','WHITE NON HISPANIC'], inplace=True)
print("Time using .replace(): {} sec".format(time.time() - start_time))
```
# Replace single values I
In this exercise, we will apply the following replacing technique of replacing multiple values using dictionaries on a different dataset.
We will apply the functions in the data DataFrame. Each row represents the rank of 5 cards from a playing card deck, spanning from 1 (Ace) to 13 (King) (features R1, R2, R3, R4, R5). The feature 'Class' classifies each row to a category (from 0 to 9) and the feature 'Explanation' gives a brief explanation of what each class represents.
The purpose of this exercise is to categorize the two types of flush in the game ('Royal flush' and 'Straight flush') under the 'Flush' name.
```
# Replace Royal flush or Straight flush to Flush
poker_hands.replace({'Royal flush':'Flush', 'Straight flush':'Flush'}, inplace=True)
print(poker_hands['Explanation'].head())
```
# Replace single values II
For this exercise, we will be using the names DataFrame. In this dataset, the column 'Rank' shows the ranking of each name by year. For this exercise, you will use dictionaries to replace the first ranked name of every year as 'FIRST', the second name as 'SECOND' and the third name as 'THIRD'.
You will use dictionaries to replace one single value per key.
You can already see the first 5 names of the data, which correspond to the 5 most popular names for all the females belonging to the 'ASIAN AND PACIFIC ISLANDER' ethnicity in 2011.
```
# Replace the number rank by a string
names['Rank'].replace({1:'FIRST', 2:'SECOND', 3:'THIRD'}, inplace=True)
print(names.head())
```
# Replace multiple values III
As you saw in the video, you can use dictionaries to replace multiple values with just one value, even from multiple columns. To show the usefulness of replacing with dictionaries, you will use the names dataset one more time.
In this dataset, the column 'Rank' shows which rank each name reached every year. You will change the rank of the first three ranked names of every year to 'MEDAL' and those from 4th and 5th place to 'ALMOST MEDAL'.
You can already see the first 5 names of the data, which correspond to the 5 most popular names for all the females belonging to the 'ASIAN AND PACIFIC ISLANDER' ethnicity in 2011.
```
# Replace the rank of the first three ranked names to 'MEDAL'
names.replace({'Rank': {1:'MEDAL', 2:'MEDAL', 3:'MEDAL'}}, inplace=True)
# Replace the rank of the 4th and 5th ranked names to 'ALMOST MEDAL'
names.replace({'Rank': {4:'ALMOST MEDAL', 5:'ALMOST MEDAL'}}, inplace=True)
print(names.head())
```
# Most efficient method for scalar replacement
If you want to replace a scalar value with another scalar value, which technique is the most efficient??
Replace using dictionaries.
|
github_jupyter
|
```
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
def plot_series(time, series, format="-", start=0, end=None):
plt.plot(time[start:end], series[start:end], format)
plt.xlabel("Time")
plt.ylabel("Value")
plt.grid(True)
!wget --no-check-certificate \
https://raw.githubusercontent.com/jbrownlee/Datasets/master/daily-min-temperatures.csv \
-O /tmp/daily-min-temperatures.csv
import csv
time_step = []
temps = []
with open('/tmp/daily-min-temperatures.csv') as csvfile:
reader = csv.reader(csvfile, delimiter=',')
next(reader)
step=0
for row in reader:
temps.append(float(row[1]))
time_step.append(step)
step = step + 1
series = np.array(temps)
time = np.array(time_step)
plt.figure(figsize=(10, 6))
plot_series(time, series)
split_time = 2500
time_train = time[:split_time]
x_train = series[:split_time]
time_valid = time[split_time:]
x_valid = series[split_time:]
window_size = 30
batch_size = 32
shuffle_buffer_size = 1000
def windowed_dataset(series, window_size, batch_size, shuffle_buffer):
series = tf.expand_dims(series, axis=-1)
ds = tf.data.Dataset.from_tensor_slices(series)
ds = ds.window(window_size + 1, shift=1, drop_remainder=True)
ds = ds.flat_map(lambda w: w.batch(window_size + 1))
ds = ds.shuffle(shuffle_buffer)
ds = ds.map(lambda w: (w[:-1], w[1:]))
return ds.batch(batch_size).prefetch(1)
def model_forecast(model, series, window_size):
ds = tf.data.Dataset.from_tensor_slices(series)
ds = ds.window(window_size, shift=1, drop_remainder=True)
ds = ds.flat_map(lambda w: w.batch(window_size))
ds = ds.batch(32).prefetch(1)
forecast = model.predict(ds)
return forecast
tf.keras.backend.clear_session()
tf.random.set_seed(51)
np.random.seed(51)
window_size = 64
batch_size = 256
train_set = windowed_dataset(x_train, window_size, batch_size, shuffle_buffer_size)
print(train_set)
print(x_train.shape)
model = tf.keras.models.Sequential([
tf.keras.layers.Conv1D(filters=32, kernel_size=5,
strides=1, padding="causal",
activation="relu",
input_shape=[None, 1]),
tf.keras.layers.LSTM(64, return_sequences=True),
tf.keras.layers.LSTM(64, return_sequences=True),
tf.keras.layers.Dense(30, activation="relu"),
tf.keras.layers.Dense(10, activation="relu"),
tf.keras.layers.Dense(1),
tf.keras.layers.Lambda(lambda x: x * 400)
])
lr_schedule = tf.keras.callbacks.LearningRateScheduler(
lambda epoch: 1e-8 * 10**(epoch / 20))
optimizer = tf.keras.optimizers.SGD(lr=1e-8, momentum=0.9)
model.compile(loss=tf.keras.losses.Huber(),
optimizer=optimizer,
metrics=["mae"])
history = model.fit(train_set, epochs=100, callbacks=[lr_schedule])
plt.semilogx(history.history["lr"], history.history["loss"])
plt.axis([1e-8, 1e-4, 0, 60])
tf.keras.backend.clear_session()
tf.random.set_seed(51)
np.random.seed(51)
train_set = windowed_dataset(x_train, window_size=60, batch_size=100, shuffle_buffer=shuffle_buffer_size)
model = tf.keras.models.Sequential([
tf.keras.layers.Conv1D(filters=32, kernel_size=5,
strides=1, padding="causal",
activation="relu",
input_shape=[None, 1]),
tf.keras.layers.LSTM(64, return_sequences=True),
tf.keras.layers.LSTM(64, return_sequences=True),
tf.keras.layers.Dense(30, activation="relu"),
tf.keras.layers.Dense(10, activation="relu"),
tf.keras.layers.Dense(1),
tf.keras.layers.Lambda(lambda x: x * 400)
])
optimizer = tf.keras.optimizers.SGD(lr=5e-5, momentum=0.9)
model.compile(loss=tf.keras.losses.Huber(),
optimizer=optimizer,
metrics=["mae"])
history = model.fit(train_set,epochs=150)
# EXPECTED OUTPUT SHOULD SEE AN MAE OF <2 WITHIN ABOUT 30 EPOCHS
rnn_forecast = model_forecast(model, series[..., np.newaxis], window_size)
rnn_forecast = rnn_forecast[split_time - window_size:-1, -1, 0]
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid)
plot_series(time_valid, rnn_forecast)
# EXPECTED OUTPUT. PLOT SHOULD SHOW PROJECTIONS FOLLOWING ORIGINAL DATA CLOSELY
tf.keras.metrics.mean_absolute_error(x_valid, rnn_forecast).numpy()
# EXPECTED OUTPUT MAE < 2 -- I GOT 1.789626
print(rnn_forecast)
# EXPECTED OUTPUT -- ARRAY OF VALUES IN THE LOW TEENS
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/christianadriano/PCA_AquacultureSystem/blob/master/PCA_KMeans_All_Piscicultura.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import pandas as pd #tables for data wrangling
import numpy as np #basic statistical methods
import io #for uploading data
#Manual option
from google.colab import files
uploaded = files.upload() #choose file dados_relativizados_centralizados_piscicultura.csv
#Upload data from cvs file
df = pd.read_csv(io.StringIO(uploaded['dados_relativizados_centralizados_piscicultura.csv'].decode('utf-8')))
#print(df)
column_names = df.columns
#Select fatores Ambientais
feature_names = [name for name in column_names if name.startswith("E")]
#feature_names = list(df.columns["A2_DA":"A4_EUC"])
#print(feature_names)
list_names = ['fazenda'] + feature_names
df_cultivo = df[list_names]
df_cultivo.head()
#Look at correlations
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
corr = df_cultivo.corr()
# using a styled panda's dataframe from https://stackoverflow.com/a/42323184/1215012
cmap = 'coolwarm'
corr.style.background_gradient(cmap, axis=1)\
.set_properties(**{'max-width': '80px', 'font-size': '10pt'})\
.set_precision(2)\
#smaller chart
sns.heatmap(corr, xticklabels=corr.columns, yticklabels=corr.columns, cmap='coolwarm')
#check which ones are statiscally significant
from scipy.stats import pearsonr
import pandas as pd
def calculate_pvalues(df):
df = df.dropna()._get_numeric_data()
dfcols = pd.DataFrame(columns=df.columns)
pvalues = dfcols.transpose().join(dfcols, how='outer')
for r in df.columns:
for c in df.columns:
pvalues[r][c] = round(pearsonr(df[r], df[c])[1], 4)
return pvalues
p_values = calculate_pvalues(df_cultivo)
#Plot p-values
def highlight_significant(val):
'''
highlight in blue only the statistically significant cells
'''
color = 'blue' if val < 0.05 else 'grey'
return 'color: %s' % color
p_values.style.applymap(highlight_significant)
#Smaller plot of p-values
import matplotlib.pyplot as plt
from matplotlib import colors
import numpy as np
np.random.seed(101)
zvals = np.random.rand(100, 100) * 10
# make a color map of fixed colors
cmap_discrete = colors.ListedColormap(['lightblue', 'white'])
bounds=[0,0.05,1]
norm_binary = colors.BoundaryNorm(bounds, cmap_discrete.N)
# tell imshow about color map so that only set colors are used
img = plt.imshow(zvals, interpolation='nearest', origin='lower',
cmap=cmap_discrete, norm=norm_binary)
sns.heatmap(p_values, xticklabels=p_values.columns, yticklabels=p_values.columns, cmap=cmap_discrete, norm=norm_binary)
```
**PCA**
Now we do the PCA
```
#Normalize the data to have MEAN==0
from sklearn.preprocessing import StandardScaler
x = df_cultivo.iloc[:,1:].values
x = StandardScaler().fit_transform(x) # normalizing the features
#print(x)
#Run PCA
from sklearn.decomposition import PCA
pca = PCA(n_components=2)
principalComponents = pca.fit_transform(x)
principalDf = pd.DataFrame(data = principalComponents
, columns = ['principal component 1', 'principal component 2'])
finalDf = pd.concat([principalDf, df_cultivo[['fazenda']]], axis = 1)
#Visualize results of PCA in Two Dimensions
import matplotlib.pyplot as plt
fig = plt.figure(figsize = (12,12))
ax = fig.add_subplot(1,1,1)
ax.set_xlabel('Principal Component 1', fontsize = 15)
ax.set_ylabel('Principal Component 2', fontsize = 15)
ax.set_title('2 component PCA', fontsize = 20)
targets = df_cultivo['fazenda'].to_numpy()
print(targets)
for target in targets:
indicesToKeep = finalDf['fazenda'] == target
x = finalDf.loc[indicesToKeep, 'principal component 1']
y = finalDf.loc[indicesToKeep, 'principal component 2']
ax.scatter(x,y,s = 100)
ax.annotate(target, (x+0.1,y))
#for name in targets:
ax.legend(targets, loc='top right')
ax.grid()
variance_list =pca.explained_variance_ratio_
print("variance explained by each component:", variance_list)
print("total variance explained:", sum(variance_list))
#principal components for each indicador
#print(principalComponents)
#print(targets)
df_clustering = pd.DataFrame({'fazenda': targets, 'pc1':list(principalComponents[:,0]), 'pc2': list(principalComponents[:,1])}, columns=['fazenda', 'pc1','pc2'])
#df_clustering
#Find clusters
from sklearn.cluster import KMeans
#4 clusters
model = KMeans(4)
model.fit(df_clustering.iloc[:,1:3])
#print(model.cluster_centers_)
#Plot clusters
plt.scatter(df_clustering.iloc[:,1],df_clustering.iloc[:,2], c=model.labels_.astype(float));
plt.scatter(model.cluster_centers_[:,0], model.cluster_centers_[:,1], s=50, marker="x", color="grey"); # Show the
#To which cluster each point belongs?
df1= df_clustering.assign(cluster=pd.Series(model.labels_).values)
df1.sort_values(by='cluster')
#5 clusters
model = KMeans(5)
model.fit(df_clustering.iloc[:,1:3])
#print(model.cluster_centers_)
#Plot clusters
plt.scatter(df_clustering.iloc[:,1],df_clustering.iloc[:,2], c=model.labels_.astype(float));
plt.scatter(model.cluster_centers_[:,0], model.cluster_centers_[:,1], s=50, marker="x", color="grey"); # Show the
```
In my view, we have two large clusters and three outliers, as the graph above shows.
```
#To which cluster each point belongs?
df1= df_clustering.assign(cluster=pd.Series(model.labels_).values)
df1.sort_values(by='cluster')
#6 clusters
model = KMeans(6)
model.fit(df_clustering.iloc[:,1:3])
#print(model.cluster_centers_)
#Plot clusters
plt.scatter(df_clustering.iloc[:,1],df_clustering.iloc[:,2], c=model.labels_.astype(float));
plt.scatter(model.cluster_centers_[:,0], model.cluster_centers_[:,1], s=50, marker="x", color="grey"); # Show the
#To which cluster each point belongs?
df1= df_clustering.assign(cluster=pd.Series(model.labels_).values)
df1.sort_values(by='cluster')
```
Now we analyze 3 Principal Components
```
#Normalize the data to have MEAN==0
from sklearn.preprocessing import StandardScaler
x = df_cultivo.iloc[:,1:].values
x = StandardScaler().fit_transform(x) # normalizing the features
#print(x)
#Run PCA
from sklearn.decomposition import PCA
pca = PCA(n_components=3)
principalComponents = pca.fit_transform(x)
principalDf = pd.DataFrame(data = principalComponents
, columns = ['principal component 1', 'principal component 2','principal component 3'])
finalDf = pd.concat([principalDf, df_cultivo[['fazenda']]], axis = 1)
variance_list =pca.explained_variance_ratio_
print("variance explained by each component:", variance_list)
print("total variance explained:", sum(variance_list))
```
Now we search for clusters for 3 principal components
```
#Find clusters
from sklearn.cluster import KMeans
#4 clusters
model = KMeans(4)
model.fit(df_clustering.iloc[:,1:4])
#print(model.cluster_centers_)
#principal components for each indicador
#print(principalComponents)
#print(targets)
df_clustering = pd.DataFrame({'fazenda': targets, 'pc1':list(principalComponents[:,0]),
'pc2': list(principalComponents[:,1]),'pc3': list(principalComponents[:,2])},
columns=['fazenda', 'pc1','pc2','pc3'])
#df_clustering
#4 clusters
from sklearn.cluster import KMeans
model = KMeans(4)
model.fit(df_clustering.iloc[:,1:4])
#print(model.cluster_centers_)
#Plot clusters
fig = plt.figure(figsize = (12,12))
ax = fig.add_subplot(111, projection='3d')
ax.set_xlabel('Principal Component 1', fontsize = 15)
ax.set_ylabel('Principal Component 2', fontsize = 15)
ax.set_zlabel('Principal Component 3', fontsize = 15)
ax.set_title('3-components PCA', fontsize = 20)
targets = df_cultivo['fazenda'].to_numpy()
for target in targets:
indicesToKeep = finalDf['fazenda'] == target
x = finalDf.loc[indicesToKeep, 'principal component 1']
y = finalDf.loc[indicesToKeep, 'principal component 2']
z = finalDf.loc[indicesToKeep, 'principal component 3']
ax.scatter(x,y,z,s = 100)
ax.scatter(model.cluster_centers_[:,0], model.cluster_centers_[:,1],model.cluster_centers_[:,2], s=150, marker='x', color="black"); # Show the
ax.legend(targets)
ax.grid()
```
Now we search for clusters for the 3 principal components
```
#To which cluster each point belongs?
df1= df_clustering.assign(cluster=pd.Series(model.labels_).values)
df1.sort_values(by='cluster')
```
Comparing k-means of PC12 with PC123, we see that the cluster membership changes completely.
```
#5 clusters
from sklearn.cluster import KMeans
model = KMeans(5)
model.fit(df_clustering.iloc[:,1:4])
#print(model.cluster_centers_)
#Plot clusters
#plt.scatter(df_clustering.iloc[:,1],df_clustering.iloc[:,2], c=model.labels_.astype(float));
#plt.scatter(model.cluster_centers_[:,0], model.cluster_centers_[:,1], s=50, color="red"); # Show the
fig = plt.figure(figsize = (12,12))
ax = fig.add_subplot(111, projection='3d')
ax.set_xlabel('Principal Component 1', fontsize = 15)
ax.set_ylabel('Principal Component 2', fontsize = 15)
ax.set_zlabel('Principal Component 3', fontsize = 15)
ax.set_title('3-components PCA', fontsize = 20)
targets = df_cultivo['fazenda'].to_numpy()
for target in targets:
indicesToKeep = finalDf['fazenda'] == target
x = finalDf.loc[indicesToKeep, 'principal component 1']
y = finalDf.loc[indicesToKeep, 'principal component 2']
z = finalDf.loc[indicesToKeep, 'principal component 3']
ax.scatter(x,y,z,s = 100)
#ax.annotate(target, (x,y))
ax.scatter(model.cluster_centers_[:,0], model.cluster_centers_[:,1],model.cluster_centers_[:,2], s=150, marker='x', color="black"); # Show the
#for name in targets:
ax.legend(targets)
ax.grid()
#To which cluster each point belongs?
df1= df_clustering.assign(cluster=pd.Series(model.labels_).values)
df1.sort_values(by='cluster')
#6 clusters
from sklearn.cluster import KMeans
model = KMeans(6)
model.fit(df_clustering.iloc[:,1:4])
#print(model.cluster_centers_)
#Plot clusters
#plt.scatter(df_clustering.iloc[:,1],df_clustering.iloc[:,2], c=model.labels_.astype(float));
#plt.scatter(model.cluster_centers_[:,0], model.cluster_centers_[:,1], s=50, color="red"); # Show the
fig = plt.figure(figsize = (12,12))
ax = fig.add_subplot(111, projection='3d')
ax.set_xlabel('Principal Component 1', fontsize = 15)
ax.set_ylabel('Principal Component 2', fontsize = 15)
ax.set_zlabel('Principal Component 3', fontsize = 15)
ax.set_title('3-components PCA', fontsize = 20)
targets = df_cultivo['fazenda'].to_numpy()
for target in targets:
indicesToKeep = finalDf['fazenda'] == target
x = finalDf.loc[indicesToKeep, 'principal component 1']
y = finalDf.loc[indicesToKeep, 'principal component 2']
z = finalDf.loc[indicesToKeep, 'principal component 3']
ax.scatter(x,y,z,s = 100)
#ax.annotate(target, (x,y))
ax.scatter(model.cluster_centers_[:,0], model.cluster_centers_[:,1],model.cluster_centers_[:,2], s=150, marker='x', color="black"); # Show the
#for name in targets:
ax.legend(targets)
ax.grid()
#To which cluster each point belongs?
df1= df_clustering.assign(cluster=pd.Series(model.labels_).values)
df1.sort_values(by='cluster')
```
|
github_jupyter
|
```
import torch as t
import torchvision as tv
import numpy as np
import time
```
# 不是逻辑回归
```
# 超参数
EPOCH = 5
BATCH_SIZE = 100
DOWNLOAD_MNIST = True # 下过数据的话, 就可以设置成 False
N_TEST_IMG = 10 # 到时候显示 5张图片看效果, 如上图一
class DNN(t.nn.Module):
def __init__(self):
super(DNN, self).__init__()
train_data = tv.datasets.FashionMNIST(
root="./fashionmnist/",
train=True,
transform=tv.transforms.ToTensor(),
download=DOWNLOAD_MNIST
)
test_data = tv.datasets.FashionMNIST(
root="./fashionmnist/",
train=False,
transform=tv.transforms.ToTensor(),
download=DOWNLOAD_MNIST
)
print(test_data)
# Data Loader for easy mini-batch return in training, the image batch shape will be (50, 1, 28, 28)
self.train_loader = t.utils.data.DataLoader(
dataset=train_data,
batch_size=BATCH_SIZE,
shuffle=True)
self.test_loader = t.utils.data.DataLoader(
dataset=test_data,
batch_size=1000,
shuffle=True)
self.cnn = t.nn.Sequential(
t.nn.Conv2d(
in_channels=1, # input height
out_channels=32, # n_filters
kernel_size=5, # filter size
stride=1, # filter movement/step
padding=2, # 如果想要 con2d 出来的图片长宽没有变化, padding=(kernel_size-1)/2 当 stride=1
), # output shape (16, 28, 28)
t.nn.ELU(), # activation
t.nn.MaxPool2d(kernel_size=2),
t.nn.Conv2d(
in_channels=32, # input height
out_channels=64, # n_filters
kernel_size=3, # filter size
stride=1, # filter movement/step
padding=1, # 如果想要 con2d 出来的图片长宽没有变化, padding=(kernel_size-1)/2 当 stride=1
), # output shape (64, 14, 14)
t.nn.ELU(), # activation
t.nn.MaxPool2d(kernel_size=2) # output shape (64, 7, 7)
)
self.dnn = t.nn.Sequential(
t.nn.Linear(7*7*64,256),
t.nn.Dropout(0.5),
t.nn.ELU(),
t.nn.Linear(256,10),
)
self.lr = 0.001
self.loss = t.nn.CrossEntropyLoss()
self.opt = t.optim.Adam(self.parameters(), lr = self.lr)
def forward(self,x):
cnn1 = self.cnn(x)
#print(cnn1.shape)
cnn1 = cnn1.view(-1,7*7*64)
#print(cnn1.shape)
out = self.dnn(cnn1)
#print(out.shape)
return(out)
def train():
use_gpu = t.cuda.is_available()
model = DNN()
if(use_gpu):
model.cuda()
print(model)
loss = model.loss
opt = model.opt
dataloader = model.train_loader
testloader = model.test_loader
for e in range(EPOCH):
step = 0
ts = time.time()
for (x, y) in (dataloader):
model.train()# train model dropout used
step += 1
b_x = x.view(-1,1,28,28) # batch x, shape (batch, 28*28)
#print(b_x.shape)
b_y = y
if(use_gpu):
b_x = b_x.cuda()
b_y = b_y.cuda()
out = model(b_x)
losses = loss(out,b_y)
opt.zero_grad()
losses.backward()
opt.step()
if(step%100 == 0):
if(use_gpu):
print(e,step,losses.data.cpu().numpy())
else:
print(e,step,losses.data.numpy())
model.eval() # train model dropout not use
for (tx,ty) in testloader:
t_x = tx.view(-1,1, 28,28) # batch x, shape (batch, 28*28)
t_y = ty
if(use_gpu):
t_x = t_x.cuda()
t_y = t_y.cuda()
t_out = model(t_x)
if(use_gpu):
acc = (np.argmax(t_out.data.cpu().numpy(),axis=1) == t_y.data.cpu().numpy())
else:
acc = (np.argmax(t_out.data.numpy(),axis=1) == t_y.data.numpy())
print(time.time() - ts ,np.sum(acc)/1000)
ts = time.time()
break#只测试前1000个
t.save(model, './model.pkl') # 保存整个网络
t.save(model.state_dict(), './model_params.pkl') # 只保存网络中的参数 (速度快, 占内存少)
#加载参数的方式
"""net = DNN()
net.load_state_dict(t.load('./model_params.pkl'))
net.eval()"""
#加载整个模型的方式
net = t.load('./model.pkl')
net.cpu()
net.eval()
for (tx,ty) in testloader:
t_x = tx.view(-1, 1,28,28) # batch x, shape (batch, 28*28)
t_y = ty
t_out = net(t_x)
#acc = (np.argmax(t_out.data.CPU().numpy(),axis=1) == t_y.data.CPU().numpy())
acc = (np.argmax(t_out.data.numpy(),axis=1) == t_y.data.numpy())
print(np.sum(acc)/1000)
train()
```
|
github_jupyter
|
## Dependencies
```
import os
import sys
import cv2
import shutil
import random
import warnings
import numpy as np
import pandas as pd
import seaborn as sns
import multiprocessing as mp
import matplotlib.pyplot as plt
from tensorflow import set_random_seed
from sklearn.utils import class_weight
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix, cohen_kappa_score
from keras import backend as K
from keras.models import Model
from keras.utils import to_categorical
from keras import optimizers, applications
from keras.preprocessing.image import ImageDataGenerator
from keras.layers import Dense, Dropout, GlobalAveragePooling2D, Input
from keras.callbacks import EarlyStopping, ReduceLROnPlateau, Callback, LearningRateScheduler
def seed_everything(seed=0):
random.seed(seed)
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
set_random_seed(0)
seed = 0
seed_everything(seed)
%matplotlib inline
sns.set(style="whitegrid")
warnings.filterwarnings("ignore")
sys.path.append(os.path.abspath('../input/efficientnet/efficientnet-master/efficientnet-master/'))
from efficientnet import *
```
## Load data
```
hold_out_set = pd.read_csv('../input/aptos-data-split/hold-out.csv')
X_train = hold_out_set[hold_out_set['set'] == 'train']
X_val = hold_out_set[hold_out_set['set'] == 'validation']
test = pd.read_csv('../input/aptos2019-blindness-detection/test.csv')
print('Number of train samples: ', X_train.shape[0])
print('Number of validation samples: ', X_val.shape[0])
print('Number of test samples: ', test.shape[0])
# Preprocecss data
X_train["id_code"] = X_train["id_code"].apply(lambda x: x + ".png")
X_val["id_code"] = X_val["id_code"].apply(lambda x: x + ".png")
test["id_code"] = test["id_code"].apply(lambda x: x + ".png")
display(X_train.head())
```
# Model parameters
```
# Model parameters
FACTOR = 2
BATCH_SIZE = 8 * FACTOR
EPOCHS = 10
WARMUP_EPOCHS = 3
LEARNING_RATE = 1e-4 * FACTOR
WARMUP_LEARNING_RATE = 1e-3 * FACTOR
HEIGHT = 256
WIDTH = 256
CHANNELS = 3
TTA_STEPS = 5
ES_PATIENCE = 5
RLROP_PATIENCE = 3
DECAY_DROP = 0.5
LR_WARMUP_EPOCHS_1st = 2
LR_WARMUP_EPOCHS_2nd = 3
STEP_SIZE = len(X_train) // BATCH_SIZE
TOTAL_STEPS_1st = WARMUP_EPOCHS * STEP_SIZE
TOTAL_STEPS_2nd = EPOCHS * STEP_SIZE
WARMUP_STEPS_1st = LR_WARMUP_EPOCHS_1st * STEP_SIZE
WARMUP_STEPS_2nd = LR_WARMUP_EPOCHS_2nd * STEP_SIZE
```
# Pre-procecess images
```
train_base_path = '../input/aptos2019-blindness-detection/train_images/'
test_base_path = '../input/aptos2019-blindness-detection/test_images/'
train_dest_path = 'base_dir/train_images/'
validation_dest_path = 'base_dir/validation_images/'
test_dest_path = 'base_dir/test_images/'
# Making sure directories don't exist
if os.path.exists(train_dest_path):
shutil.rmtree(train_dest_path)
if os.path.exists(validation_dest_path):
shutil.rmtree(validation_dest_path)
if os.path.exists(test_dest_path):
shutil.rmtree(test_dest_path)
# Creating train, validation and test directories
os.makedirs(train_dest_path)
os.makedirs(validation_dest_path)
os.makedirs(test_dest_path)
def crop_image(img, tol=7):
if img.ndim ==2:
mask = img>tol
return img[np.ix_(mask.any(1),mask.any(0))]
elif img.ndim==3:
gray_img = cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)
mask = gray_img>tol
check_shape = img[:,:,0][np.ix_(mask.any(1),mask.any(0))].shape[0]
if (check_shape == 0): # image is too dark so that we crop out everything,
return img # return original image
else:
img1=img[:,:,0][np.ix_(mask.any(1),mask.any(0))]
img2=img[:,:,1][np.ix_(mask.any(1),mask.any(0))]
img3=img[:,:,2][np.ix_(mask.any(1),mask.any(0))]
img = np.stack([img1,img2,img3],axis=-1)
return img
def circle_crop(img):
img = crop_image(img)
height, width, depth = img.shape
largest_side = np.max((height, width))
img = cv2.resize(img, (largest_side, largest_side))
height, width, depth = img.shape
x = width//2
y = height//2
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def preprocess_image(base_path, save_path, image_id, HEIGHT, WIDTH, sigmaX=10):
image = cv2.imread(base_path + image_id)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = circle_crop(image)
image = cv2.resize(image, (HEIGHT, WIDTH))
image = cv2.addWeighted(image, 4, cv2.GaussianBlur(image, (0,0), sigmaX), -4 , 128)
cv2.imwrite(save_path + image_id, image)
# Pre-procecss train set
for i, image_id in enumerate(X_train['id_code']):
preprocess_image(train_base_path, train_dest_path, image_id, HEIGHT, WIDTH)
# Pre-procecss validation set
for i, image_id in enumerate(X_val['id_code']):
preprocess_image(train_base_path, validation_dest_path, image_id, HEIGHT, WIDTH)
# Pre-procecss test set
for i, image_id in enumerate(test['id_code']):
preprocess_image(test_base_path, test_dest_path, image_id, HEIGHT, WIDTH)
```
# Data generator
```
datagen=ImageDataGenerator(rescale=1./255,
rotation_range=360,
horizontal_flip=True,
vertical_flip=True)
train_generator=datagen.flow_from_dataframe(
dataframe=X_train,
directory=train_dest_path,
x_col="id_code",
y_col="diagnosis",
class_mode="raw",
batch_size=BATCH_SIZE,
target_size=(HEIGHT, WIDTH),
seed=seed)
valid_generator=datagen.flow_from_dataframe(
dataframe=X_val,
directory=validation_dest_path,
x_col="id_code",
y_col="diagnosis",
class_mode="raw",
batch_size=BATCH_SIZE,
target_size=(HEIGHT, WIDTH),
seed=seed)
test_generator=datagen.flow_from_dataframe(
dataframe=test,
directory=test_dest_path,
x_col="id_code",
batch_size=1,
class_mode=None,
shuffle=False,
target_size=(HEIGHT, WIDTH),
seed=seed)
def cosine_decay_with_warmup(global_step,
learning_rate_base,
total_steps,
warmup_learning_rate=0.0,
warmup_steps=0,
hold_base_rate_steps=0):
"""
Cosine decay schedule with warm up period.
In this schedule, the learning rate grows linearly from warmup_learning_rate
to learning_rate_base for warmup_steps, then transitions to a cosine decay
schedule.
:param global_step {int}: global step.
:param learning_rate_base {float}: base learning rate.
:param total_steps {int}: total number of training steps.
:param warmup_learning_rate {float}: initial learning rate for warm up. (default: {0.0}).
:param warmup_steps {int}: number of warmup steps. (default: {0}).
:param hold_base_rate_steps {int}: Optional number of steps to hold base learning rate before decaying. (default: {0}).
:param global_step {int}: global step.
:Returns : a float representing learning rate.
:Raises ValueError: if warmup_learning_rate is larger than learning_rate_base, or if warmup_steps is larger than total_steps.
"""
if total_steps < warmup_steps:
raise ValueError('total_steps must be larger or equal to warmup_steps.')
learning_rate = 0.5 * learning_rate_base * (1 + np.cos(
np.pi *
(global_step - warmup_steps - hold_base_rate_steps
) / float(total_steps - warmup_steps - hold_base_rate_steps)))
if hold_base_rate_steps > 0:
learning_rate = np.where(global_step > warmup_steps + hold_base_rate_steps,
learning_rate, learning_rate_base)
if warmup_steps > 0:
if learning_rate_base < warmup_learning_rate:
raise ValueError('learning_rate_base must be larger or equal to warmup_learning_rate.')
slope = (learning_rate_base - warmup_learning_rate) / warmup_steps
warmup_rate = slope * global_step + warmup_learning_rate
learning_rate = np.where(global_step < warmup_steps, warmup_rate,
learning_rate)
return np.where(global_step > total_steps, 0.0, learning_rate)
class WarmUpCosineDecayScheduler(Callback):
"""Cosine decay with warmup learning rate scheduler"""
def __init__(self,
learning_rate_base,
total_steps,
global_step_init=0,
warmup_learning_rate=0.0,
warmup_steps=0,
hold_base_rate_steps=0,
verbose=0):
"""
Constructor for cosine decay with warmup learning rate scheduler.
:param learning_rate_base {float}: base learning rate.
:param total_steps {int}: total number of training steps.
:param global_step_init {int}: initial global step, e.g. from previous checkpoint.
:param warmup_learning_rate {float}: initial learning rate for warm up. (default: {0.0}).
:param warmup_steps {int}: number of warmup steps. (default: {0}).
:param hold_base_rate_steps {int}: Optional number of steps to hold base learning rate before decaying. (default: {0}).
:param verbose {int}: quiet, 1: update messages. (default: {0}).
"""
super(WarmUpCosineDecayScheduler, self).__init__()
self.learning_rate_base = learning_rate_base
self.total_steps = total_steps
self.global_step = global_step_init
self.warmup_learning_rate = warmup_learning_rate
self.warmup_steps = warmup_steps
self.hold_base_rate_steps = hold_base_rate_steps
self.verbose = verbose
self.learning_rates = []
def on_batch_end(self, batch, logs=None):
self.global_step = self.global_step + 1
lr = K.get_value(self.model.optimizer.lr)
self.learning_rates.append(lr)
def on_batch_begin(self, batch, logs=None):
lr = cosine_decay_with_warmup(global_step=self.global_step,
learning_rate_base=self.learning_rate_base,
total_steps=self.total_steps,
warmup_learning_rate=self.warmup_learning_rate,
warmup_steps=self.warmup_steps,
hold_base_rate_steps=self.hold_base_rate_steps)
K.set_value(self.model.optimizer.lr, lr)
if self.verbose > 0:
print('\nBatch %02d: setting learning rate to %s.' % (self.global_step + 1, lr))
```
# Model
```
def create_model(input_shape):
input_tensor = Input(shape=input_shape)
base_model = EfficientNetB5(weights=None,
include_top=False,
input_tensor=input_tensor)
# base_model.load_weights('../input/efficientnet-keras-weights-b0b5/efficientnet-b5_imagenet_1000_notop.h5')
x = GlobalAveragePooling2D()(base_model.output)
final_output = Dense(1, activation='linear', name='final_output')(x)
model = Model(input_tensor, final_output)
model.load_weights('../input/aptos-pretrain-olddata-effnetb5/effNetB5_img224_oldData.h5')
return model
```
# Train top layers
```
model = create_model(input_shape=(HEIGHT, WIDTH, CHANNELS))
for layer in model.layers:
layer.trainable = False
for i in range(-2, 0):
model.layers[i].trainable = True
cosine_lr_1st = WarmUpCosineDecayScheduler(learning_rate_base=WARMUP_LEARNING_RATE,
total_steps=TOTAL_STEPS_1st,
warmup_learning_rate=0.0,
warmup_steps=WARMUP_STEPS_1st,
hold_base_rate_steps=(2 * STEP_SIZE))
metric_list = ["accuracy"]
callback_list = [cosine_lr_1st]
optimizer = optimizers.Adam(lr=WARMUP_LEARNING_RATE)
model.compile(optimizer=optimizer, loss='mean_squared_error', metrics=metric_list)
model.summary()
STEP_SIZE_TRAIN = train_generator.n//train_generator.batch_size
STEP_SIZE_VALID = valid_generator.n//valid_generator.batch_size
history_warmup = model.fit_generator(generator=train_generator,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_generator,
validation_steps=STEP_SIZE_VALID,
epochs=WARMUP_EPOCHS,
callbacks=callback_list,
verbose=2).history
```
# Fine-tune the complete model
```
for layer in model.layers:
layer.trainable = True
es = EarlyStopping(monitor='val_loss', mode='min', patience=ES_PATIENCE, restore_best_weights=True, verbose=1)
cosine_lr_2nd = WarmUpCosineDecayScheduler(learning_rate_base=LEARNING_RATE,
total_steps=TOTAL_STEPS_2nd,
warmup_learning_rate=0.0,
warmup_steps=WARMUP_STEPS_2nd,
hold_base_rate_steps=(2 * STEP_SIZE))
callback_list = [es, cosine_lr_2nd]
optimizer = optimizers.Adam(lr=LEARNING_RATE)
model.compile(optimizer=optimizer, loss='mean_squared_error', metrics=metric_list)
model.summary()
history = model.fit_generator(generator=train_generator,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_generator,
validation_steps=STEP_SIZE_VALID,
epochs=EPOCHS,
callbacks=callback_list,
verbose=2).history
fig, (ax1, ax2) = plt.subplots(2, 1, sharex='col', figsize=(20, 6))
ax1.plot(cosine_lr_1st.learning_rates)
ax1.set_title('Warm up learning rates')
ax2.plot(cosine_lr_2nd.learning_rates)
ax2.set_title('Fine-tune learning rates')
plt.xlabel('Steps')
plt.ylabel('Learning rate')
sns.despine()
plt.show()
```
# Model loss graph
```
fig, (ax1, ax2) = plt.subplots(2, 1, sharex='col', figsize=(20, 14))
ax1.plot(history['loss'], label='Train loss')
ax1.plot(history['val_loss'], label='Validation loss')
ax1.legend(loc='best')
ax1.set_title('Loss')
ax2.plot(history['acc'], label='Train accuracy')
ax2.plot(history['val_acc'], label='Validation accuracy')
ax2.legend(loc='best')
ax2.set_title('Accuracy')
plt.xlabel('Epochs')
sns.despine()
plt.show()
# Create empty arays to keep the predictions and labels
df_preds = pd.DataFrame(columns=['label', 'pred', 'set'])
train_generator.reset()
valid_generator.reset()
# Add train predictions and labels
for i in range(STEP_SIZE_TRAIN + 1):
im, lbl = next(train_generator)
preds = model.predict(im, batch_size=train_generator.batch_size)
for index in range(len(preds)):
df_preds.loc[len(df_preds)] = [lbl[index], preds[index][0], 'train']
# Add validation predictions and labels
for i in range(STEP_SIZE_VALID + 1):
im, lbl = next(valid_generator)
preds = model.predict(im, batch_size=valid_generator.batch_size)
for index in range(len(preds)):
df_preds.loc[len(df_preds)] = [lbl[index], preds[index][0], 'validation']
df_preds['label'] = df_preds['label'].astype('int')
def classify(x):
if x < 0.5:
return 0
elif x < 1.5:
return 1
elif x < 2.5:
return 2
elif x < 3.5:
return 3
return 4
# Classify predictions
df_preds['predictions'] = df_preds['pred'].apply(lambda x: classify(x))
train_preds = df_preds[df_preds['set'] == 'train']
validation_preds = df_preds[df_preds['set'] == 'validation']
```
# Model Evaluation
## Confusion Matrix
### Original thresholds
```
labels = ['0 - No DR', '1 - Mild', '2 - Moderate', '3 - Severe', '4 - Proliferative DR']
def plot_confusion_matrix(train, validation, labels=labels):
train_labels, train_preds = train
validation_labels, validation_preds = validation
fig, (ax1, ax2) = plt.subplots(1, 2, sharex='col', figsize=(24, 7))
train_cnf_matrix = confusion_matrix(train_labels, train_preds)
validation_cnf_matrix = confusion_matrix(validation_labels, validation_preds)
train_cnf_matrix_norm = train_cnf_matrix.astype('float') / train_cnf_matrix.sum(axis=1)[:, np.newaxis]
validation_cnf_matrix_norm = validation_cnf_matrix.astype('float') / validation_cnf_matrix.sum(axis=1)[:, np.newaxis]
train_df_cm = pd.DataFrame(train_cnf_matrix_norm, index=labels, columns=labels)
validation_df_cm = pd.DataFrame(validation_cnf_matrix_norm, index=labels, columns=labels)
sns.heatmap(train_df_cm, annot=True, fmt='.2f', cmap="Blues",ax=ax1).set_title('Train')
sns.heatmap(validation_df_cm, annot=True, fmt='.2f', cmap=sns.cubehelix_palette(8),ax=ax2).set_title('Validation')
plt.show()
plot_confusion_matrix((train_preds['label'], train_preds['predictions']), (validation_preds['label'], validation_preds['predictions']))
```
## Quadratic Weighted Kappa
```
def evaluate_model(train, validation):
train_labels, train_preds = train
validation_labels, validation_preds = validation
print("Train Cohen Kappa score: %.3f" % cohen_kappa_score(train_preds, train_labels, weights='quadratic'))
print("Validation Cohen Kappa score: %.3f" % cohen_kappa_score(validation_preds, validation_labels, weights='quadratic'))
print("Complete set Cohen Kappa score: %.3f" % cohen_kappa_score(np.append(train_preds, validation_preds), np.append(train_labels, validation_labels), weights='quadratic'))
evaluate_model((train_preds['label'], train_preds['predictions']), (validation_preds['label'], validation_preds['predictions']))
```
## Apply model to test set and output predictions
```
def apply_tta(model, generator, steps=10):
step_size = generator.n//generator.batch_size
preds_tta = []
for i in range(steps):
generator.reset()
preds = model.predict_generator(generator, steps=step_size)
preds_tta.append(preds)
return np.mean(preds_tta, axis=0)
preds = apply_tta(model, test_generator, TTA_STEPS)
predictions = [classify(x) for x in preds]
results = pd.DataFrame({'id_code':test['id_code'], 'diagnosis':predictions})
results['id_code'] = results['id_code'].map(lambda x: str(x)[:-4])
# Cleaning created directories
if os.path.exists(train_dest_path):
shutil.rmtree(train_dest_path)
if os.path.exists(validation_dest_path):
shutil.rmtree(validation_dest_path)
if os.path.exists(test_dest_path):
shutil.rmtree(test_dest_path)
```
# Predictions class distribution
```
fig = plt.subplots(sharex='col', figsize=(24, 8.7))
sns.countplot(x="diagnosis", data=results, palette="GnBu_d").set_title('Test')
sns.despine()
plt.show()
results.to_csv('submission.csv', index=False)
display(results.head())
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from tqdm import tqdm
%matplotlib inline
from torch.utils.data import Dataset, DataLoader
import torch
import torchvision
import torch.nn as nn
import torch.optim as optim
from torch.nn import functional as F
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
```
# Generate dataset
```
y = np.random.randint(0,3,500)
idx= []
for i in range(3):
print(i,sum(y==i))
idx.append(y==i)
x = np.zeros((500,))
np.random.seed(12)
x[idx[0]] = np.random.uniform(low =-1,high =0,size= sum(idx[0]))
x[idx[1]] = np.random.uniform(low =0,high =1,size= sum(idx[1]))
x[idx[2]] = np.random.uniform(low =2,high =3,size= sum(idx[2]))
x[idx[0]][0], x[idx[2]][5]
print(x.shape,y.shape)
idx= []
for i in range(3):
idx.append(y==i)
for i in range(3):
y= np.zeros(x[idx[i]].shape[0])
plt.scatter(x[idx[i]],y,label="class_"+str(i))
plt.legend()
bg_idx = [ np.where(idx[2] == True)[0]]
bg_idx = np.concatenate(bg_idx, axis = 0)
bg_idx.shape
np.unique(bg_idx).shape
x = x - np.mean(x[bg_idx], axis = 0, keepdims = True)
np.mean(x[bg_idx], axis = 0, keepdims = True), np.mean(x, axis = 0, keepdims = True)
x = x/np.std(x[bg_idx], axis = 0, keepdims = True)
np.std(x[bg_idx], axis = 0, keepdims = True), np.std(x, axis = 0, keepdims = True)
for i in range(3):
y= np.zeros(x[idx[i]].shape[0])
plt.scatter(x[idx[i]],y,label="class_"+str(i))
plt.legend()
foreground_classes = {'class_0','class_1' }
background_classes = {'class_2'}
fg_class = np.random.randint(0,2)
fg_idx = np.random.randint(0,9)
a = []
for i in range(9):
if i == fg_idx:
b = np.random.choice(np.where(idx[fg_class]==True)[0],size=1)
a.append(x[b])
print("foreground "+str(fg_class)+" present at " + str(fg_idx))
else:
bg_class = np.random.randint(2,3)
b = np.random.choice(np.where(idx[bg_class]==True)[0],size=1)
a.append(x[b])
print("background "+str(bg_class)+" present at " + str(i))
a = np.concatenate(a,axis=0)
print(a.shape)
print(fg_class , fg_idx)
a.shape
np.reshape(a,(9,1))
a=np.reshape(a,(3,3))
plt.imshow(a)
desired_num = 2000
mosaic_list_of_images =[]
mosaic_label = []
fore_idx=[]
for j in range(desired_num):
np.random.seed(j)
fg_class = np.random.randint(0,2)
fg_idx = 0
a = []
for i in range(9):
if i == fg_idx:
b = np.random.choice(np.where(idx[fg_class]==True)[0],size=1)
a.append(x[b])
# print("foreground "+str(fg_class)+" present at " + str(fg_idx))
else:
bg_class = np.random.randint(2,3)
b = np.random.choice(np.where(idx[bg_class]==True)[0],size=1)
a.append(x[b])
# print("background "+str(bg_class)+" present at " + str(i))
a = np.concatenate(a,axis=0)
mosaic_list_of_images.append(np.reshape(a,(9,1)))
mosaic_label.append(fg_class)
fore_idx.append(fg_idx)
mosaic_list_of_images = np.concatenate(mosaic_list_of_images,axis=1).T
mosaic_list_of_images.shape, mosaic_list_of_images[0]
for j in range(9):
print(mosaic_list_of_images[0][j])
class MosaicDataset(Dataset):
"""MosaicDataset dataset."""
def __init__(self, mosaic_list_of_images, mosaic_label, fore_idx):
"""
Args:
csv_file (string): Path to the csv file with annotations.
root_dir (string): Directory with all the images.
transform (callable, optional): Optional transform to be applied
on a sample.
"""
self.mosaic = mosaic_list_of_images
self.label = mosaic_label
self.fore_idx = fore_idx
def __len__(self):
return len(self.label)
def __getitem__(self, idx):
return self.mosaic[idx] , self.label[idx], self.fore_idx[idx]
batch = 250
msd1 = MosaicDataset(mosaic_list_of_images[0:1000], mosaic_label[0:1000] , fore_idx[0:1000])
train_loader = DataLoader( msd1 ,batch_size= batch ,shuffle=True)
batch = 250
msd2 = MosaicDataset(mosaic_list_of_images[1000:2000], mosaic_label[1000:2000] , fore_idx[1000:2000])
test_loader = DataLoader( msd2 ,batch_size= batch ,shuffle=True)
class Focus(nn.Module):
def __init__(self):
super(Focus, self).__init__()
self.fc1 = nn.Linear(1, 1)
# self.fc2 = nn.Linear(2, 1)
def forward(self,z): #y is avg image #z batch of list of 9 images
y = torch.zeros([batch], dtype=torch.float64)
x = torch.zeros([batch,9],dtype=torch.float64)
y = y.to("cuda")
x = x.to("cuda")
# print(x.shape, z.shape)
for i in range(9):
# print(z[:,i].shape)
# print(self.helper(z[:,i])[:,0].shape)
x[:,i] = self.helper(z[:,i])[:,0]
# print(x.shape, z.shape)
x = F.softmax(x,dim=1)
# print(x.shape, z.shape)
# x1 = x[:,0]
# print(torch.mul(x[:,0],z[:,0]).shape)
for i in range(9):
# x1 = x[:,i]
y = y + torch.mul(x[:,i],z[:,i])
# print(x.shape, y.shape)
return x, y
def helper(self, x):
x = x.view(-1, 1)
# x = F.relu(self.fc1(x))
x = (self.fc1(x))
return x
class Classification(nn.Module):
def __init__(self):
super(Classification, self).__init__()
self.fc1 = nn.Linear(1, 2)
def forward(self, x):
x = x.view(-1, 1)
x = self.fc1(x)
# print(x.shape)
return x
torch.manual_seed(12)
focus_net = Focus().double()
focus_net = focus_net.to("cuda")
torch.manual_seed(12)
classify = Classification().double()
classify = classify.to("cuda")
focus_net.fc1.weight, focus_net.fc1.bias
classify.fc1.weight, classify.fc1.bias
focus_net.fc1.weight = torch.nn.Parameter(torch.tensor(np.array([[0.0]])))
focus_net.fc1.bias = torch.nn.Parameter(torch.tensor(np.array([0.0])))
focus_net.fc1.weight, focus_net.fc1.bias
classify.fc1.weight = torch.nn.Parameter(torch.tensor(np.array([[0.0],[0.0]])))
classify.fc1.bias = torch.nn.Parameter(torch.tensor(np.array([0.0, 0.0])))
classify.fc1.weight, classify.fc1.bias
focus_net = focus_net.to("cuda")
classify = classify.to("cuda")
focus_net.fc1.weight, focus_net.fc1.bias
classify.fc1.weight, classify.fc1.bias
import torch.optim as optim
criterion = nn.CrossEntropyLoss()
optimizer_classify = optim.SGD(classify.parameters(), lr=0.01, momentum=0.9)
optimizer_focus = optim.SGD(focus_net.parameters(), lr=0.01, momentum=0.9)
# optimizer_classify = optim.Adam(classify.parameters(), lr=0.01)
# optimizer_focus = optim.Adam(focus_net.parameters(), lr=0.01)
col1=[]
col2=[]
col3=[]
col4=[]
col5=[]
col6=[]
col7=[]
col8=[]
col9=[]
col10=[]
col11=[]
col12=[]
col13=[]
correct = 0
total = 0
count = 0
flag = 1
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
with torch.no_grad():
for data in train_loader:
inputs, labels , fore_idx = data
inputs = inputs.double()
inputs, labels , fore_idx = inputs.to("cuda"),labels.to("cuda"), fore_idx.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
# print(outputs.shape)
_, predicted = torch.max(outputs.data, 1)
# print(predicted.shape)
for j in range(labels.size(0)):
count += 1
focus = torch.argmax(alphas[j])
if alphas[j][focus] >= 0.5 :
argmax_more_than_half += 1
else:
argmax_less_than_half += 1
# print(focus, fore_idx[j], predicted[j])
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true += 1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false += 1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false += 1
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 1000 train images: %d %%' % ( 100 * correct / total))
print("total correct", correct)
print("total train set images", total)
print("focus_true_pred_true %d =============> FTPT : %d %%" % (focus_true_pred_true , (100 * focus_true_pred_true / total) ) )
print("focus_false_pred_true %d =============> FFPT : %d %%" % (focus_false_pred_true, (100 * focus_false_pred_true / total) ) )
print("focus_true_pred_false %d =============> FTPF : %d %%" %( focus_true_pred_false , ( 100 * focus_true_pred_false / total) ) )
print("focus_false_pred_false %d =============> FFPF : %d %%" % (focus_false_pred_false, ( 100 * focus_false_pred_false / total) ) )
print("argmax_more_than_half ==================> ",argmax_more_than_half)
print("argmax_less_than_half ==================> ",argmax_less_than_half)
print(count)
print("="*100)
col1.append(0)
col2.append(argmax_more_than_half)
col3.append(argmax_less_than_half)
col4.append(focus_true_pred_true)
col5.append(focus_false_pred_true)
col6.append(focus_true_pred_false)
col7.append(focus_false_pred_false)
correct = 0
total = 0
count = 0
flag = 1
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
with torch.no_grad():
for data in test_loader:
inputs, labels , fore_idx = data
inputs = inputs.double()
inputs, labels , fore_idx = inputs.to("cuda"),labels.to("cuda"), fore_idx.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
for j in range(labels.size(0)):
focus = torch.argmax(alphas[j])
if alphas[j][focus] >= 0.5 :
argmax_more_than_half += 1
else:
argmax_less_than_half += 1
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true += 1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false += 1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false += 1
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 1000 test images: %d %%' % (
100 * correct / total))
print("total correct", correct)
print("total train set images", total)
print("focus_true_pred_true %d =============> FTPT : %d %%" % (focus_true_pred_true , (100 * focus_true_pred_true / total) ) )
print("focus_false_pred_true %d =============> FFPT : %d %%" % (focus_false_pred_true, (100 * focus_false_pred_true / total) ) )
print("focus_true_pred_false %d =============> FTPF : %d %%" %( focus_true_pred_false , ( 100 * focus_true_pred_false / total) ) )
print("focus_false_pred_false %d =============> FFPF : %d %%" % (focus_false_pred_false, ( 100 * focus_false_pred_false / total) ) )
print("argmax_more_than_half ==================> ",argmax_more_than_half)
print("argmax_less_than_half ==================> ",argmax_less_than_half)
col8.append(argmax_more_than_half)
col9.append(argmax_less_than_half)
col10.append(focus_true_pred_true)
col11.append(focus_false_pred_true)
col12.append(focus_true_pred_false)
col13.append(focus_false_pred_false)
nos_epochs = 1000
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
for epoch in range(nos_epochs): # loop over the dataset multiple times
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
running_loss = 0.0
epoch_loss = []
cnt=0
iteration = desired_num // batch
#training data set
for i, data in enumerate(train_loader):
inputs , labels , fore_idx = data
inputs, labels = inputs.to("cuda"), labels.to("cuda")
inputs = inputs.double()
# zero the parameter gradients
optimizer_focus.zero_grad()
optimizer_classify.zero_grad()
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
# print(outputs)
# print(outputs.shape,labels.shape , torch.argmax(outputs, dim=1))
loss = criterion(outputs, labels)
loss.backward()
optimizer_focus.step()
optimizer_classify.step()
running_loss += loss.item()
mini = 3
if cnt % mini == mini-1: # print every 40 mini-batches
print('[%d, %5d] loss: %.3f' %(epoch + 1, cnt + 1, running_loss / mini))
epoch_loss.append(running_loss/mini)
running_loss = 0.0
cnt=cnt+1
if epoch % 5 == 0:
for j in range (batch):
focus = torch.argmax(alphas[j])
if(alphas[j][focus] >= 0.5):
argmax_more_than_half +=1
else:
argmax_less_than_half +=1
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true +=1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false +=1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false +=1
if(np.mean(epoch_loss) <= 0.001):
break;
if epoch % 5 == 0:
col1.append(epoch + 1)
col2.append(argmax_more_than_half)
col3.append(argmax_less_than_half)
col4.append(focus_true_pred_true)
col5.append(focus_false_pred_true)
col6.append(focus_true_pred_false)
col7.append(focus_false_pred_false)
# print("="*20)
# print("Train FTPT : ", col4)
# print("Train FFPT : ", col5)
#************************************************************************
#testing data set
# focus_net.eval()
with torch.no_grad():
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
for data in test_loader:
inputs, labels , fore_idx = data
inputs = inputs.double()
inputs, labels = inputs.to("cuda"), labels.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
for j in range (batch):
focus = torch.argmax(alphas[j])
if(alphas[j][focus] >= 0.5):
argmax_more_than_half +=1
else:
argmax_less_than_half +=1
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true +=1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false +=1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false +=1
col8.append(argmax_more_than_half)
col9.append(argmax_less_than_half)
col10.append(focus_true_pred_true)
col11.append(focus_false_pred_true)
col12.append(focus_true_pred_false)
col13.append(focus_false_pred_false)
# print("Test FTPT : ", col10)
# print("Test FFPT : ", col11)
# print("="*20)
print('Finished Training')
df_train = pd.DataFrame()
df_test = pd.DataFrame()
columns = ["epochs", "argmax > 0.5" ,"argmax < 0.5", "focus_true_pred_true", "focus_false_pred_true", "focus_true_pred_false", "focus_false_pred_false" ]
df_train[columns[0]] = col1
df_train[columns[1]] = col2
df_train[columns[2]] = col3
df_train[columns[3]] = col4
df_train[columns[4]] = col5
df_train[columns[5]] = col6
df_train[columns[6]] = col7
df_test[columns[0]] = col1
df_test[columns[1]] = col8
df_test[columns[2]] = col9
df_test[columns[3]] = col10
df_test[columns[4]] = col11
df_test[columns[5]] = col12
df_test[columns[6]] = col13
df_train
# plt.figure(12,12)
plt.plot(col1,np.array(col2)/10, label='argmax > 0.5')
plt.plot(col1,np.array(col3)/10, label='argmax < 0.5')
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("epochs")
plt.ylabel("training data")
plt.title("On Training set")
plt.show()
plt.plot(col1,np.array(col4)/10, label ="focus_true_pred_true ")
plt.plot(col1,np.array(col5)/10, label ="focus_false_pred_true ")
plt.plot(col1,np.array(col6)/10, label ="focus_true_pred_false ")
plt.plot(col1,np.array(col7)/10, label ="focus_false_pred_false ")
plt.title("On Training set")
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("epochs")
plt.ylabel("training data")
plt.show()
df_test
# plt.figure(12,12)
plt.plot(col1,np.array(col8)/10, label='argmax > 0.5')
plt.plot(col1,np.array(col9)/10, label='argmax < 0.5')
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("epochs")
plt.ylabel("Testing data")
plt.title("On Testing set")
plt.show()
plt.plot(col1,np.array(col10)/10, label ="focus_true_pred_true ")
plt.plot(col1,np.array(col11)/10, label ="focus_false_pred_true ")
plt.plot(col1,np.array(col12)/10, label ="focus_true_pred_false ")
plt.plot(col1,np.array(col13)/10, label ="focus_false_pred_false ")
plt.title("On Testing set")
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("epochs")
plt.ylabel("Testing data")
plt.show()
correct = 0
total = 0
count = 0
flag = 1
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
with torch.no_grad():
for data in train_loader:
inputs, labels , fore_idx = data
inputs = inputs.double()
inputs, labels , fore_idx = inputs.to("cuda"),labels.to("cuda"), fore_idx.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
for j in range(labels.size(0)):
focus = torch.argmax(alphas[j])
if alphas[j][focus] >= 0.5 :
argmax_more_than_half += 1
else:
argmax_less_than_half += 1
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true += 1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false += 1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false += 1
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 1000 train images: %d %%' % (
100 * correct / total))
print("total correct", correct)
print("total train set images", total)
print("focus_true_pred_true %d =============> FTPT : %d %%" % (focus_true_pred_true , (100 * focus_true_pred_true / total) ) )
print("focus_false_pred_true %d =============> FFPT : %d %%" % (focus_false_pred_true, (100 * focus_false_pred_true / total) ) )
print("focus_true_pred_false %d =============> FTPF : %d %%" %( focus_true_pred_false , ( 100 * focus_true_pred_false / total) ) )
print("focus_false_pred_false %d =============> FFPF : %d %%" % (focus_false_pred_false, ( 100 * focus_false_pred_false / total) ) )
print("argmax_more_than_half ==================> ",argmax_more_than_half)
print("argmax_less_than_half ==================> ",argmax_less_than_half)
correct = 0
total = 0
count = 0
flag = 1
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
with torch.no_grad():
for data in test_loader:
inputs, labels , fore_idx = data
inputs = inputs.double()
inputs, labels , fore_idx = inputs.to("cuda"),labels.to("cuda"), fore_idx.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
for j in range(labels.size(0)):
focus = torch.argmax(alphas[j])
if alphas[j][focus] >= 0.5 :
argmax_more_than_half += 1
else:
argmax_less_than_half += 1
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true += 1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false += 1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false += 1
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 1000 test images: %d %%' % (
100 * correct / total))
print("total correct", correct)
print("total train set images", total)
print("focus_true_pred_true %d =============> FTPT : %d %%" % (focus_true_pred_true , (100 * focus_true_pred_true / total) ) )
print("focus_false_pred_true %d =============> FFPT : %d %%" % (focus_false_pred_true, (100 * focus_false_pred_true / total) ) )
print("focus_true_pred_false %d =============> FTPF : %d %%" %( focus_true_pred_false , ( 100 * focus_true_pred_false / total) ) )
print("focus_false_pred_false %d =============> FFPF : %d %%" % (focus_false_pred_false, ( 100 * focus_false_pred_false / total) ) )
print("argmax_more_than_half ==================> ",argmax_more_than_half)
print("argmax_less_than_half ==================> ",argmax_less_than_half)
correct = 0
total = 0
with torch.no_grad():
for data in train_loader:
inputs, labels , fore_idx = data
inputs = inputs.double()
inputs, labels = inputs.to("cuda"), labels.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 1000 train images: %d %%' % ( 100 * correct / total))
print("total correct", correct)
print("total train set images", total)
correct = 0
total = 0
with torch.no_grad():
for data in test_loader:
inputs, labels , fore_idx = data
inputs = inputs.double()
inputs, labels = inputs.to("cuda"), labels.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 1000 test images: %d %%' % ( 100 * correct / total))
print("total correct", correct)
print("total train set images", total)
focus_net.fc1.weight, focus_net.fc1.bias
classify.fc1.weight, classify.fc1.bias
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/AliaksandrSiarohin/first-order-model/blob/master/demo.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Demo for paper "First Order Motion Model for Image Animation"
To try the demo, press the 2 play buttons in order and scroll to the bottom. Note that it may take several minutes to load.
```
!pip install ffmpy &> /dev/null
!git init -q .
!git remote add origin https://github.com/AliaksandrSiarohin/first-order-model
!git pull -q origin master
!git clone -q https://github.com/graphemecluster/first-order-model-demo demo
import IPython.display
import PIL.Image
import cv2
import imageio
import io
import ipywidgets
import numpy
import os.path
import requests
import skimage.transform
import warnings
from base64 import b64encode
from demo import load_checkpoints, make_animation
from ffmpy import FFmpeg
from google.colab import files, output
from IPython.display import HTML, Javascript
from skimage import img_as_ubyte
warnings.filterwarnings("ignore")
os.makedirs("user", exist_ok=True)
display(HTML("""
<style>
.widget-box > * {
flex-shrink: 0;
}
.widget-tab {
min-width: 0;
flex: 1 1 auto;
}
.widget-tab .p-TabBar-tabLabel {
font-size: 15px;
}
.widget-upload {
background-color: tan;
}
.widget-button {
font-size: 18px;
width: 160px;
height: 34px;
line-height: 34px;
}
.widget-dropdown {
width: 250px;
}
.widget-checkbox {
width: 650px;
}
.widget-checkbox + .widget-checkbox {
margin-top: -6px;
}
.input-widget .output_html {
text-align: center;
width: 266px;
height: 266px;
line-height: 266px;
color: lightgray;
font-size: 72px;
}
div.stream {
display: none;
}
.title {
font-size: 20px;
font-weight: bold;
margin: 12px 0 6px 0;
}
.warning {
display: none;
color: red;
margin-left: 10px;
}
.warn {
display: initial;
}
.resource {
cursor: pointer;
border: 1px solid gray;
margin: 5px;
width: 160px;
height: 160px;
min-width: 160px;
min-height: 160px;
max-width: 160px;
max-height: 160px;
-webkit-box-sizing: initial;
box-sizing: initial;
}
.resource:hover {
border: 6px solid crimson;
margin: 0;
}
.selected {
border: 6px solid seagreen;
margin: 0;
}
.input-widget {
width: 266px;
height: 266px;
border: 1px solid gray;
}
.input-button {
width: 268px;
font-size: 15px;
margin: 2px 0 0;
}
.output-widget {
width: 256px;
height: 256px;
border: 1px solid gray;
}
.output-button {
width: 258px;
font-size: 15px;
margin: 2px 0 0;
}
.uploaded {
width: 256px;
height: 256px;
border: 6px solid seagreen;
margin: 0;
}
.label-or {
align-self: center;
font-size: 20px;
margin: 16px;
}
.loading {
align-items: center;
width: fit-content;
}
.loader {
margin: 32px 0 16px 0;
width: 48px;
height: 48px;
min-width: 48px;
min-height: 48px;
max-width: 48px;
max-height: 48px;
border: 4px solid whitesmoke;
border-top-color: gray;
border-radius: 50%;
animation: spin 1.8s linear infinite;
}
.loading-label {
color: gray;
}
.comparison-widget {
width: 256px;
height: 256px;
border: 1px solid gray;
margin-left: 2px;
}
.comparison-label {
color: gray;
font-size: 14px;
text-align: center;
position: relative;
bottom: 3px;
}
@keyframes spin {
from { transform: rotate(0deg); }
to { transform: rotate(360deg); }
}
</style>
"""))
def thumbnail(file):
return imageio.get_reader(file, mode='I', format='FFMPEG').get_next_data()
def create_image(i, j):
image_widget = ipywidgets.Image(
value=open('demo/images/%d%d.png' % (i, j), 'rb').read(),
format='png'
)
image_widget.add_class('resource')
image_widget.add_class('resource-image')
image_widget.add_class('resource-image%d%d' % (i, j))
return image_widget
def create_video(i):
video_widget = ipywidgets.Image(
value=cv2.imencode('.png', cv2.cvtColor(thumbnail('demo/videos/%d.mp4' % i), cv2.COLOR_RGB2BGR))[1].tostring(),
format='png'
)
video_widget.add_class('resource')
video_widget.add_class('resource-video')
video_widget.add_class('resource-video%d' % i)
return video_widget
def create_title(title):
title_widget = ipywidgets.Label(title)
title_widget.add_class('title')
return title_widget
def download_output(button):
complete.layout.display = 'none'
loading.layout.display = ''
files.download('output.mp4')
loading.layout.display = 'none'
complete.layout.display = ''
def convert_output(button):
complete.layout.display = 'none'
loading.layout.display = ''
FFmpeg(inputs={'output.mp4': None}, outputs={'scaled.mp4': '-vf "scale=1080x1080:flags=lanczos,pad=1920:1080:420:0" -y'}).run()
files.download('scaled.mp4')
loading.layout.display = 'none'
complete.layout.display = ''
def back_to_main(button):
complete.layout.display = 'none'
main.layout.display = ''
label_or = ipywidgets.Label('or')
label_or.add_class('label-or')
image_titles = ['Peoples', 'Cartoons', 'Dolls', 'Game of Thrones', 'Statues']
image_lengths = [8, 4, 8, 9, 4]
image_tab = ipywidgets.Tab()
image_tab.children = [ipywidgets.HBox([create_image(i, j) for j in range(length)]) for i, length in enumerate(image_lengths)]
for i, title in enumerate(image_titles):
image_tab.set_title(i, title)
input_image_widget = ipywidgets.Output()
input_image_widget.add_class('input-widget')
upload_input_image_button = ipywidgets.FileUpload(accept='image/*', button_style='primary')
upload_input_image_button.add_class('input-button')
image_part = ipywidgets.HBox([
ipywidgets.VBox([input_image_widget, upload_input_image_button]),
label_or,
image_tab
])
video_tab = ipywidgets.Tab()
video_tab.children = [ipywidgets.HBox([create_video(i) for i in range(5)])]
video_tab.set_title(0, 'All Videos')
input_video_widget = ipywidgets.Output()
input_video_widget.add_class('input-widget')
upload_input_video_button = ipywidgets.FileUpload(accept='video/*', button_style='primary')
upload_input_video_button.add_class('input-button')
video_part = ipywidgets.HBox([
ipywidgets.VBox([input_video_widget, upload_input_video_button]),
label_or,
video_tab
])
model = ipywidgets.Dropdown(
description="Model:",
options=[
'vox',
'vox-adv',
'taichi',
'taichi-adv',
'nemo',
'mgif',
'fashion',
'bair'
]
)
warning = ipywidgets.HTML('<b>Warning:</b> Upload your own images and videos (see README)')
warning.add_class('warning')
model_part = ipywidgets.HBox([model, warning])
relative = ipywidgets.Checkbox(description="Relative keypoint displacement (Inherit object proporions from the video)", value=True)
adapt_movement_scale = ipywidgets.Checkbox(description="Adapt movement scale (Don’t touch unless you know want you are doing)", value=True)
generate_button = ipywidgets.Button(description="Generate", button_style='primary')
main = ipywidgets.VBox([
create_title('Choose Image'),
image_part,
create_title('Choose Video'),
video_part,
create_title('Settings'),
model_part,
relative,
adapt_movement_scale,
generate_button
])
loader = ipywidgets.Label()
loader.add_class("loader")
loading_label = ipywidgets.Label("This may take several minutes to process…")
loading_label.add_class("loading-label")
loading = ipywidgets.VBox([loader, loading_label])
loading.add_class('loading')
output_widget = ipywidgets.Output()
output_widget.add_class('output-widget')
download = ipywidgets.Button(description='Download', button_style='primary')
download.add_class('output-button')
download.on_click(download_output)
convert = ipywidgets.Button(description='Convert to 1920×1080', button_style='primary')
convert.add_class('output-button')
convert.on_click(convert_output)
back = ipywidgets.Button(description='Back', button_style='primary')
back.add_class('output-button')
back.on_click(back_to_main)
comparison_widget = ipywidgets.Output()
comparison_widget.add_class('comparison-widget')
comparison_label = ipywidgets.Label('Comparison')
comparison_label.add_class('comparison-label')
complete = ipywidgets.HBox([
ipywidgets.VBox([output_widget, download, convert, back]),
ipywidgets.VBox([comparison_widget, comparison_label])
])
display(ipywidgets.VBox([main, loading, complete]))
display(Javascript("""
var images, videos;
function deselectImages() {
images.forEach(function(item) {
item.classList.remove("selected");
});
}
function deselectVideos() {
videos.forEach(function(item) {
item.classList.remove("selected");
});
}
function invokePython(func) {
google.colab.kernel.invokeFunction("notebook." + func, [].slice.call(arguments, 1), {});
}
setTimeout(function() {
(images = [].slice.call(document.getElementsByClassName("resource-image"))).forEach(function(item) {
item.addEventListener("click", function() {
deselectImages();
item.classList.add("selected");
invokePython("select_image", item.className.match(/resource-image(\d\d)/)[1]);
});
});
images[0].classList.add("selected");
(videos = [].slice.call(document.getElementsByClassName("resource-video"))).forEach(function(item) {
item.addEventListener("click", function() {
deselectVideos();
item.classList.add("selected");
invokePython("select_video", item.className.match(/resource-video(\d)/)[1]);
});
});
videos[0].classList.add("selected");
}, 1000);
"""))
selected_image = None
def select_image(filename):
global selected_image
selected_image = resize(PIL.Image.open('demo/images/%s.png' % filename).convert("RGB"))
input_image_widget.clear_output(wait=True)
with input_image_widget:
display(HTML('Image'))
input_image_widget.remove_class('uploaded')
output.register_callback("notebook.select_image", select_image)
selected_video = None
def select_video(filename):
global selected_video
selected_video = 'demo/videos/%s.mp4' % filename
input_video_widget.clear_output(wait=True)
with input_video_widget:
display(HTML('Video'))
input_video_widget.remove_class('uploaded')
output.register_callback("notebook.select_video", select_video)
def resize(image, size=(256, 256)):
w, h = image.size
d = min(w, h)
r = ((w - d) // 2, (h - d) // 2, (w + d) // 2, (h + d) // 2)
return image.resize(size, resample=PIL.Image.LANCZOS, box=r)
def upload_image(change):
global selected_image
for name, file_info in upload_input_image_button.value.items():
content = file_info['content']
if content is not None:
selected_image = resize(PIL.Image.open(io.BytesIO(content)).convert("RGB"))
input_image_widget.clear_output(wait=True)
with input_image_widget:
display(selected_image)
input_image_widget.add_class('uploaded')
display(Javascript('deselectImages()'))
upload_input_image_button.observe(upload_image, names='value')
def upload_video(change):
global selected_video
for name, file_info in upload_input_video_button.value.items():
content = file_info['content']
if content is not None:
selected_video = 'user/' + name
preview = resize(PIL.Image.fromarray(thumbnail(content)).convert("RGB"))
input_video_widget.clear_output(wait=True)
with input_video_widget:
display(preview)
input_video_widget.add_class('uploaded')
display(Javascript('deselectVideos()'))
with open(selected_video, 'wb') as video:
video.write(content)
upload_input_video_button.observe(upload_video, names='value')
def change_model(change):
if model.value.startswith('vox'):
warning.remove_class('warn')
else:
warning.add_class('warn')
model.observe(change_model, names='value')
def generate(button):
main.layout.display = 'none'
loading.layout.display = ''
filename = model.value + ('' if model.value == 'fashion' else '-cpk') + '.pth.tar'
if not os.path.isfile(filename):
download = requests.get(requests.get('https://cloud-api.yandex.net/v1/disk/public/resources/download?public_key=https://yadi.sk/d/lEw8uRm140L_eQ&path=/' + filename).json().get('href'))
with open(filename, 'wb') as checkpoint:
checkpoint.write(download.content)
reader = imageio.get_reader(selected_video, mode='I', format='FFMPEG')
fps = reader.get_meta_data()['fps']
driving_video = []
for frame in reader:
driving_video.append(frame)
generator, kp_detector = load_checkpoints(config_path='config/%s-256.yaml' % model.value, checkpoint_path=filename)
predictions = make_animation(
skimage.transform.resize(numpy.asarray(selected_image), (256, 256)),
[skimage.transform.resize(frame, (256, 256)) for frame in driving_video],
generator,
kp_detector,
relative=relative.value,
adapt_movement_scale=adapt_movement_scale.value
)
if selected_video.startswith('user/') or selected_video == 'demo/videos/0.mp4':
imageio.mimsave('temp.mp4', [img_as_ubyte(frame) for frame in predictions], fps=fps)
FFmpeg(inputs={'temp.mp4': None, selected_video: None}, outputs={'output.mp4': '-c copy -y'}).run()
else:
imageio.mimsave('output.mp4', [img_as_ubyte(frame) for frame in predictions], fps=fps)
loading.layout.display = 'none'
complete.layout.display = ''
with output_widget:
display(HTML('<video id="left" controls src="data:video/mp4;base64,%s" />' % b64encode(open('output.mp4', 'rb').read()).decode()))
with comparison_widget:
display(HTML('<video id="right" muted src="data:video/mp4;base64,%s" />' % b64encode(open(selected_video, 'rb').read()).decode()))
display(Javascript("""
(function(left, right) {
left.addEventListener("play", function() {
right.play();
});
left.addEventListener("pause", function() {
right.pause();
});
left.addEventListener("seeking", function() {
right.currentTime = left.currentTime;
});
})(document.getElementById("left"), document.getElementById("right"));
"""))
generate_button.on_click(generate)
loading.layout.display = 'none'
complete.layout.display = 'none'
select_image('00')
select_video('0')
```
|
github_jupyter
|
# Installing Tensorflow
We will creat an environment for tensorflow that will activate every time we use th package
### NOTE: it will take some time!
```
%pip install --upgrade pip
%pip install tensorflow==2.5.0
```
#### If you see the message below, restart the kernel please from the panel above (Kernels>restart)!
'Note: you may need to restart the kernel to use updated packages.'
#### Let's check if you have everything!
```
import tensorflow as tf
print(tf.__version__)
reachout='Please repeat the steps above. If it still does not work, reach out to me ([email protected])'
try:
import tensorflow
print('tensorflow is all good!')
except:
print("An exception occurred in tensorflow installation."+reachout)
try:
import keras
print('keras is all good!')
except:
print("An exception occurred in keras installation."+reachout)
```
### Now let's explore tensorflow!
From its name tensorflow stores constants as tensor objects! Let's create our first constant!
```
import tensorflow as tf
import tensorflow.compat.v1 as tf
tf.disable_v2_behavior()
myfirstconst = tf.constant('Hello World')
myfirstconst
x = tf.constant(130.272)
x
```
### TF Sessions
Let's create a TensorFlow Session. It can be thought of as a class for running TensorFlow operations. The session encapsulates the environment in which operations take place.
Let's do a quick example:
```
a = tf.constant(1)
b = tf.constant(5)
with tf.Session() as Session:
print('TF simple Operations')
print('Multiply',Session.run(a*b))
print('Divide',Session.run(a/b))
print('Add',Session.run(a+b))
print('Subtract',Session.run(b-a))
```
#### Now let's multiply a matrix
```
import numpy as np
m = np.array([[1.0,2.0]])
n = np.array([[3.0],[4.0]])
multi = tf.matmul(m,n)
multi
with tf.Session() as Session:
res = Session.run(multi)
print(res)
```
### TF Variables
Sometimes you want to define a variable rsulting from operations. **tf.variable is ideal for this case!**
Let's see how to use it!
```
#We have to start a session!
sess = tf.InteractiveSession()
atensor = tf.random_uniform((2,2),0,1)
atensor
var = tf.Variable(initial_value=atensor)
var
try:
with tf.Session() as Session:
res = Session.run(var)
print(res)
except:
print("error!")
initialize = tf.global_variables_initializer()
initialize.run()
var.eval()
sess.run(var)
```
## Now let's custom build our first neural networks!
```
xd = np.linspace(0,10,100) + np.random.uniform(-3,.5,100)
yd = np.linspace(0,10,100) + np.random.uniform(-.5,2,100)
import matplotlib.pyplot as plt
plt.plot(xd,yd,'o')
```
### Let's define our variables here
$y=m*x+b$
```
#Let's intialize with a guess
m = tf.Variable(1.0)
b = tf.Variable(0.1)
#Let's build or objective function!
#initialize error
e=0
for x,y in zip(xd,yd):
#our model
y_pred = m*x + b
# our error
e += (y-y_pred)**2
## tensorflow optimizer
optimizer = tf.train.GradientDescentOptimizer(learning_rate=0.0001)
## we want to minimize error
training = optimizer.minimize(e)
## initilize our variables with tensorflow
initalize = tf.global_variables_initializer()
#start the session for 1000 epochs!
with tf.Session() as sess:
sess.run(initalize)
epochs = 100
for i in range(epochs):
sess.run(training)
# Get results
mf, bf = sess.run([m,b])
print("The slope is {} and the intercept is {}".format(mf, bf))
#Let's evalute our results
x_v = np.linspace(-3,11,300)
y_v = mf*x_v + bf
plt.plot(x_v,y_v,'r')
plt.plot(xd,yd,'o')
```
|
github_jupyter
|
# Time Series Analysis 1
In the first lecture, we are mainly concerned with how to manipulate and smooth time series data.
```
%matplotlib inline
import matplotlib.pyplot as plt
import os
import time
import numpy as np
import pandas as pd
! python3 -m pip install --quiet gmaps
import gmaps
import gmaps.datasets
```
## Dates and times
### Timestamps
```
now = pd.to_datetime('now')
now
now.year, now.month, now.week, now.day, now.hour, now.minute, now.second, now.microsecond
now.month_name(), now.day_name()
```
### Formatting timestamps
See format [codes](https://docs.python.org/3/library/datetime.html#strftime-and-strptime-behavior)
```
now.strftime('%I:%m%p %d-%b-%Y')
```
### Parsing time strings
#### `pandas` can handle standard formats
```
ts = pd.to_datetime('6-Dec-2018 4:45 PM')
ts
```
#### For unusual formats, use `strptime`
```
from datetime import datetime
ts = datetime.strptime('10:11PM 02-Nov-2018', '%I:%m%p %d-%b-%Y')
ts
```
### Intervals
```
then = pd.to_datetime('now')
time.sleep(5)
now = pd.to_datetime('now')
now - then
```
### Date ranges
A date range is just a collection of time stamps.
```
dates = pd.date_range(then, now, freq='s')
dates
(then - pd.to_timedelta('1.5s')) in dates
```
### Periods
Periods are intervals, not a collection of timestamps.
```
span = dates.to_period()
span
(then + pd.to_timedelta('1.5s')) in span
```
## Lag and lead with `shift`
We will use a periodic time series as an example. Periodicity is important because many biological phenomena are linked to natural periods (seasons, diurnal, menstrual cycle) or are intrinsically periodic (e.g. EEG, EKG measurements).
```
index = pd.date_range('1-1-2018', '31-1-2018', freq='12h')
```
You can shift by periods or by frequency. Shifting by frequency maintains boundary data.
```
wave = pd.Series(np.sin(np.arange(len(index))), index=index)
wave.shift(periods=1).head(3)
wave.shift(periods=1).tail(3)
wave.shift(freq=pd.Timedelta(1, freq='D')).head(3)
wave.shift(freq=pd.Timedelta(1, freq='D')).tail(3)
```
#### Visualizing shifts
```
wave.plot()
pass
wave.plot(c='blue')
wave.shift(-1).plot(c='red')
pass
wave.plot(c='blue')
wave.shift(1).plot(c='red')
pass
(wave - wave.shift(-6)).plot(c='blue')
(wave - wave.shift(-3)).plot(c='red')
pass
```
Embedding the time series with its lagged version reveals its periodic nature.
```
plt.scatter(wave, wave.shift(-1))
pass
```
### Find percent change from previous period
```
wave.pct_change().head()
```
`pct_change` is just a convenience wrapper around the use of `shift`
```
((wave - wave.shift(-1, freq='12h'))/wave).head()
```
## Resampling and window functions
The `resample` and window method have the same syntax as `groupby`, in that you can apply an aggregate function to the new intervals.
### Resampling
Sometimes there is a need to generate new time intervals, for example, to regularize irregularly timed observations.
#### Down-sampling
```
index = pd.date_range(pd.to_datetime('1-1-2018'), periods=365, freq='d')
series = pd.Series(np.arange(len(index)), index=index)
series.head()
sereis_weekly_average = series.resample('w').mean()
sereis_weekly_average.head()
sereis_monthly_sum = series.resample('m').sum()
sereis_monthly_sum.head()
sereis_10day_median = series.resample('10d').median()
sereis_10day_median.head()
```
#### Up-sampling
For up-sampling, we need to figure out what we want to do with the missing values. The usual choices are forward fill, backward fill, or interpolation using one of many built-in methods.
```
upsampled = series.resample('12h')
upsampled.asfreq()[:5]
upsampled.ffill().head()
upsampled.bfill().head()
upsampled.interpolate('linear').head()
```
### Window functions
Window functions are typically used to smooth time series data. There are 3 variants - rolling, expanding and exponentially weighted. We use the Nile flooding data for these examples.
```
df = pd.read_csv('data/nile.csv', index_col=0)
df.head()
df.plot()
pass
```
#### Rolling windows generate windows of a specified width
```
ts = pd.DataFrame(dict(ts=np.arange(5)))
ts['rolling'] = ts.rolling(window=3).sum()
ts
rolling10 = df.rolling(window=10)
rolling100 = df.rolling(window=100)
df.plot()
plt.plot(rolling10.mean(), c='orange')
plt.plot(rolling100.mean(), c='red')
pass
```
#### Expanding windows grow as the time series progresses
```
ts['expanding'] = ts.ts.expanding().sum()
ts
df.plot()
plt.plot(df.expanding(center=True).mean(), c='orange')
plt.plot(df.expanding().mean(), c='red')
pass
```
#### Exponentially weighted windows place more weight on center of mass
```
n = 10
xs = np.arange(n, dtype='float')[::-1]
xs
```
Exponentially weighted windows without adjustment.
```
pd.Series(xs).ewm(alpha=0.8, adjust=False).mean()
```
Re-implementation for insight.
```
α = 0.8
ys = np.zeros_like(xs)
ys[0] = xs[0]
for i in range(1, len(xs)):
ys[i] = (1-α)*ys[i-1] + α*xs[i]
ys
```
Exponentially weighted windows with adjustment (default)
```
pd.Series(xs).ewm(alpha=0.8, adjust=True).mean()
```
Re-implementation for insight.
```
α = 0.8
ys = np.zeros_like(xs)
ys[0] = xs[0]
for i in range(1, len(xs)):
ws = np.array([(1-α)**(i-t) for t in range(i+1)])
ys[i] = (ws * xs[:len(ws)]).sum()/ws.sum()
ys
df.plot()
plt.plot(df.ewm(alpha=0.8).mean(), c='orange')
plt.plot(df.ewm(alpha=0.2).mean(), c='red')
pass
```
Alternatives to $\alpha$
Using `span`
$$
\alpha = \frac{2}{\text{span} + 1}
$$
Using `halflife`
$$
\alpha = 1 - e^\frac{-\log{2}}{t_{1/2}}
$$
Using `com`
$$
\alpha = \frac{1}{1 + \text{com}}
$$
```
df.plot()
plt.plot(df.ewm(span=10).mean(), c='orange')
plt.plot(1+ df.ewm(alpha=2/11).mean(), c='red') # offfset for visibility
pass
```
## Correlation between time series
Suppose we had a reference time series. It is often of interest to know how any particular time series is correlated with the reference. Often the reference might be a population average, and we want to see where a particular time series deviates in behavior.
```
! python3 -m pip install --quiet pandas_datareader
import pandas_datareader.data as web
```
We will look at the correlation of some stocks.
```
QQQ tracks Nasdaq
MSFT is Microsoft
GOOG is Gogole
BP is British Petroleum
```
We expect that the technology stocks should be correlated with Nasdaq, but maybe not BP.
```
df = web.DataReader(['QQQ', 'MSFT','GOOG', 'BP'], 'stooq')
# api_key=os.environ['IEX_SECRET_KEY'])
df = df[['Close']].reset_index()
df
df = df.set_index(( 'Date', ''))
df.head()
df.columns
df.rolling(100).corr(df[('Close', 'QQQ')]).plot()
pass
```
## Visualizing space and time data
Being able to visualize events in space and time can be impressive. With Python, often you need a trivial amount of code to produce an impressive visualization.
For example, lets generate a heatmap of crimes in Sacramento in 2006, and highlight the crimes committed 10 seconds before midnight.
See the [gmaps](https://github.com/pbugnion/gmaps) package for more information.
```
sacramento_crime = pd.read_csv('data/SacramentocrimeJanuary2006.csv', index_col=0)
sacramento_crime.index = pd.to_datetime(sacramento_crime.index)
sacramento_crime.head()
gmaps.configure(api_key=os.environ["GOOGLE_API_KEY"])
locations = sacramento_crime[['latitude', 'longitude']]
late_locations = sacramento_crime.between_time('23:59', '23:59:59')[['latitude', 'longitude']]
fig = gmaps.figure()
fig.add_layer(gmaps.heatmap_layer(locations))
markers = gmaps.marker_layer(late_locations)
fig.add_layer(markers)
fig
```
|
github_jupyter
|
## BERT model for MITMovies Dataset
I was going to make this repository a package with setup.py and everything but because of my deadlines and responsibilities at my current workplace I haven't got the time to do that so I shared the structure of the project in README.md file.
```
# If any issues open the one that gives error
# !pip install transformers
# !pip install torch==1.5.1
# !pip install tqdm
# !pip install tensorboard
# !pip install seqeval
# ! pip install tqdm
# ! pip install seaborn
# !pip install gensim
import os
import sys
import json
import numpy as np
from tqdm import tqdm
sys.path.append("..")
import torch
from torch import nn
import torch.nn.functional as F
from torch.utils import tensorboard
from seqeval.metrics import classification_report
from transformers import Trainer, TrainingArguments
from torch.utils.data import (DataLoader, RandomSampler, SequentialSampler,
TensorDataset)
from transformers import (WEIGHTS_NAME, AdamW, BertConfig,
BertForTokenClassification, BertTokenizerFast,
get_linear_schedule_with_warmup)
from src.namedentityrecognizer.models.bertner import BertNerModel
from src.namedentityrecognizer.data.analyze_dataset import Analyzer
from src.namedentityrecognizer.data.build_dataset import BuildData
from src.namedentityrecognizer.data.make_dataset import MakeData
from src.namedentityrecognizer.utils.processors import NerPreProcessor, NerDataset
from sklearn.metrics import accuracy_score, f1_score, precision_score, recall_score
# Some initializers
train = True
num_train_epochs = 5
train_batch_size = 32
eval_batch_size = 8
# Weight decay for regularization
weight_decay = 0.01
# Now 1 but if batches wont fit RAM can be increased
gradient_accumulation_steps = 1
# %10 warm up
warmup_proportion = 0.1
# Adam variables
adam_epsilon = 1e-8
learning_rate = 5e-5
# 16 floating point instead of 32
fp16 = False
if fp16:
# Apex AMP optimization level selected in ['O0', 'O1', 'O2', and 'O3']
fp16_opt_level
# max seq length (for engtrain.bio since the lengths are pretty short 128 is alright)
max_seq_length = 128
# For gradient clipping
max_grad_norm = 1.0
# For having flexibility over hardware
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# Data path
data_dir = "/content/drive/MyDrive/MovieEntityRecognizer/data/modified"
# Tensorboard Name
tensorboard_writer = tensorboard.SummaryWriter("bert_base_uncased_default")
validate = True
test = True
# For downloading data, doesn't require ssl so if downloaded no need to run it again
# dataset_names = ["engtrain.bio", "engtest.bio", "trivia10k13train.n,bio", "trivia10k13test.bio"]
# (MakeData.download_data(os.path.join("http://groups.csail.mit.edu/sls/downloads/movie", dataset_name) for dataset_name in dataset_names)
# Count also word frequencies and lengths or sentences
train_labels = Analyzer.count_labels("/home/karaz/Desktop/MovieEntityRecognizer/data/raw/mitmovies/engtrain.bio", without_o=True)
Analyzer.plot_data(train_labels)
test_labels = Analyzer.count_labels("/home/karaz/Desktop/MovieEntityRecognizer/data/raw/mitmovies/engtest.bio", without_o=True)
Analyzer.plot_data(test_labels)
# Get distinct labels
label_list = sorted(list(train_labels.keys()))
label_list.append("O")
num_labels = len(label_list)
label_map = {label: id for id, label in enumerate(label_list)}
print(f"Size of labels of regular dataset: {len(label_list)}\n{label_map}")
# model configurations and tokenizer
config = BertConfig.from_pretrained("bert-large-uncased", num_labels=num_labels, finetuning_task="ner")
tokenizer = BertTokenizerFast.from_pretrained("bert-large-uncased")
# Change home karaz desktop path to your home directory (basically where the repository is)
dataset = BuildData.create_dataset("/home/karaz/Desktop/MovieEntityRecognizer/data/modified/mitmovies")
id2label = {id: label for (label,id) in label_map.items()}
id2label[-100] = 'X'
id2label
if train:
num_train_optimization_steps = int(
len(dataset['train_instances']) / train_batch_size / gradient_accumulation_steps) * num_train_epochs
print(f"Number of training steps {num_train_optimization_steps}")
print(f"Number of training instances {len(dataset['train_instances'])}")
if test:
test_steps = int(
len(dataset['test_instances']) / eval_batch_size)
print(f"Number of test steps {test_steps}")
print(f"Number of test instances {len(dataset['test_instances'])}")
# Tokenize the datasets
train_tokens = tokenizer(dataset["train_instances"], is_split_into_words=True, return_offsets_mapping=True,
padding=True, truncation=True)
test_tokens = tokenizer(dataset['test_instances'], is_split_into_words=True, return_offsets_mapping=True,
padding=True, truncation=True)
# Encode labels and give -100 to tokens which you dont want to backpropagate (basically mask them out)
train_labels = NerPreProcessor.convert_labels(dataset["train_labels"],
label_map,
train_tokens)
test_labels = NerPreProcessor.convert_labels(dataset['test_labels'],
label_map,
test_tokens)
# Get rid of unnecessary data and create final data
if train_tokens["offset_mapping"]:
train_tokens.pop("offset_mapping")
if test_tokens["offset_mapping"]:
test_tokens.pop("offset_mapping")
train_dataset = NerDataset(train_tokens, train_labels)
test_dataset = NerDataset(test_tokens, test_labels)
# Model initialization for high level api of huggingface
def model_init():
model = BertForTokenClassification.from_pretrained('bert-large-uncased', num_labels=len(label_map))
return model
# I left the compute metrics here in order to show how the evaluation
def compute_metrics(p):
predictions, labels = p
predictions = np.argmax(predictions, axis=2)
# Remove ignored index (special tokens)
true_predictions = [
[id2label[p] for (p, l) in zip(prediction, label) if l != -100]
for prediction, label in zip(predictions, labels)
]
true_labels = [
[id2label[l] for (p, l) in zip(prediction, label) if l != -100]
for prediction, label in zip(predictions, labels)
]
preds_stretched = [label for doc in true_predictions for label in doc]
trues_stretched = [label for doc in true_labels for label in doc]
return {
"accuracy_score": accuracy_score(trues_stretched, preds_stretched),
"precision": precision_score(trues_stretched, preds_stretched, labels=np.unique(preds_stretched), average='macro'),
"recall": recall_score(trues_stretched, preds_stretched, labels=np.unique(preds_stretched), average='macro'),
"f1_macro": f1_score(trues_stretched, preds_stretched, labels=np.unique(preds_stretched), average='macro'),
"f1_micro": f1_score(trues_stretched, preds_stretched, average='micro'),
}
model_name = "bert-large-uncased-micro-10epoch"
training_args = TrainingArguments(
output_dir = "/home/kemalaraz/Desktop/MovieEntityRecognizer/pretrained_models/" + model_name, # output directory
overwrite_output_dir = True,
evaluation_strategy='epoch',
num_train_epochs = 10, # total number of training epochs
per_device_train_batch_size=16, # batch size per device during training
per_device_eval_batch_size=64, # batch size for evaluation
warmup_steps=500, # number of warmup steps for learning rate scheduler
weight_decay=0.01, # strength of weight decay
logging_dir = "/home/kemalaraz/Desktop/MovieEntityRecognizer/pretrained_models/" + model_name + '/logs', # directory for storing logs
logging_steps=10,
load_best_model_at_end=True,
learning_rate = 5e-5,
seed = 42
)
# The high level api of the trainer
trainer = Trainer(
model_init = model_init,
args = training_args,
train_dataset = train_dataset,
eval_dataset = test_dataset,
compute_metrics = compute_metrics
)
training_results = trainer.train()
evaluate_results_with_best_epoch = trainer.evaluate()
# For basic inference
model = BertForTokenClassification.from_pretrained(path_to_the_model, num_labels=len(label_map))
tokens = tokenizer.tokenize(tokenizer.decode(tokenizer.encode("list the five star movies starring john lennon")))
inputs = tokenizer.encode("list the four star movies starring john lennon", return_tensors="pt")
outputs = model(inputs)[0]
predictions = torch.argmax(outputs, dim=2)
print([(token, label_list[prediction]) for token, prediction in zip(tokens, predictions[0].tolist())])
```
## Attachments





|
github_jupyter
|
```
# Import common packages and create database connection
import pandas as pd
import sqlite3 as db
conn = db.connect('Db-IMDB.db')
```
1.List all the directors who directed a 'Comedy' movie in a leap year. (You need to check that the genre is 'Comedy’ and year is a leap year) Your query should return director name, the movie name, and the year.
```
%%time
# List all the distinct directors who directed a 'Comedy' movie in a leap year.
# citation https://stackoverflow.com/questions/6534788/check-for-leap-year
# https://www.mathsisfun.com/leap-years.html
result = pd.read_sql_query(
'''
SELECT DISTINCT trim(P.NAME) as director, M.title as movie, M.year, G.Name
FROM Movie M
JOIN M_Director MD ON M.MID = MD.MID
JOIN Person P on trim(MD.PID) = trim(P.PID)
JOIN M_Genre MG on M.MID = MG.MID
JOIN Genre G on MG.GID = G.GID
WHERE G.Name LIKE '%Comedy%'
AND (((M.year % 4 = 0) AND (M.year % 100 != 0)) OR (M.year % 400 = 0))
GROUP BY director
ORDER BY director
'''
, conn);
result
%%time
# List all the directors who directed a 'Comedy' movie in a leap year. A director can direct multiple movies in leap year.
# citation https://stackoverflow.com/questions/6534788/check-for-leap-year
# https://www.mathsisfun.com/leap-years.html
result = pd.read_sql_query(
'''
SELECT DISTINCT trim(P.NAME) as director, M.title as movie, M.year, G.Name
FROM Movie M
JOIN M_Director MD ON M.MID = MD.MID
JOIN Person P on trim(MD.PID) = trim(P.PID)
JOIN M_Genre MG on M.MID = MG.MID
JOIN Genre G on MG.GID = G.GID
WHERE G.Name LIKE '%Comedy%'
AND (((M.year % 4 = 0) AND (M.year % 100 != 0)) OR (M.year % 400 = 0))
ORDER BY director
'''
, conn);
result
```
2.List the names of all the actors who played in the movie 'Anand' (1971)
```
%%time
result = pd.read_sql_query(
'''
SELECT p.Name FROM Movie m
JOIN M_Cast mc ON m.MID=mc.MID
JOIN Person p ON trim(mc.PID)=trim(p.PID)
WHERE m.title='Anand' AND m.year=1971
'''
, conn)
result
```
3. List all the actors who acted in a film before 1970 andin a film after 1990. (That is: < 1970 and > 1990.)
```
%%time
result = pd.read_sql_query(
'''
SELECT DISTINCT trim(p.PID) as pid, p.Name
FROM Movie m
JOIN M_Cast mc ON m.MID = mc.MID
JOIN Person p ON trim(mc.PID) = trim(p.PID)
WHERE m.year > 1990
AND trim(p.PID) IN (SELECT DISTINCT trim(p.PID) as pid
FROM Movie m
JOIN M_Cast mc ON m.MID = mc.MID
JOIN Person p ON trim(mc.PID) = trim(p.PID)
WHERE m.year < 1970)
GROUP BY trim(p.PID)
''', conn)
result
```
4. List all directors who directed 10 movies or more, in descending order of the number of movies they directed. Return the directors' names and the number of movies each of them directed.
```
%%time
result = pd.read_sql_query(
'''
SELECT p.Name, count(md.ID) movieCount FROM M_Director md JOIN Person p ON md.PID=p.PID
GROUP BY md.PID HAVING movieCount >= 10 ORDER BY movieCount DESC
''', conn)
result
```
5a. For each year, count the number of movies in that year that had only female actors.
```
%%time
result = pd.read_sql_query(
'''
SELECT count(m.year) movie_count, m.year as movie_year
FROM Movie m where m.MID not in
(SELECT mc.MID FROM Person p JOIN M_Cast mc ON trim(p.PID)=trim(mc.PID) WHERE p.Gender='Male')
GROUP BY movie_year
ORDER BY movie_count DESC
''', conn)
result
```
5b.Now include a small change: report for each year the percentage of movies in that year with only female actors, and the total number of movies made that year. For example, one answer will be:1990 31.81 13522 meaning that in 1990 there were 13,522 movies, and 31.81% had only female actors. You do not need to round your answer.
```
%%time
result = pd.read_sql_query(
'''
SELECT y.allMov as 'movie_count', x.year as movie_year, ((x.Movies_Cnt*100.0)/y.allMov) as Percent FROM
(SELECT count(*) Movies_Cnt , m.year
FROM Movie m where m.MID not in
(SELECT mc.MID FROM Person p JOIN M_Cast mc ON trim(p.PID) = trim(mc.PID) WHERE p.Gender='Male')
GROUP BY m.year) x INNER JOIN
(SELECT count(*) allMov, m.year
FROM Movie m
GROUP BY m.year) y on x.year=y.year
''', conn)
result
```
6. Find the film(s) with the largest cast. Return the movie title and the size of the cast. By "cast size" we mean the number of distinct actors that played in that movie: if an actor played multiple roles, or if it simply occurs multiple times in casts,we still count her/him only once.
```
%%time
result = pd.read_sql_query(
'''
SELECT count(DISTINCT mc.PId) as cast_count, m.title FROM Movie m
JOIN M_Cast mc ON m.MID=mc.MID
JOIN Person p ON trim(mc.PID)=trim(p.PID)
GROUP BY m.MID
ORDER BY cast_count DESC limit 1
''', conn)
result
```
7. A decade is a sequence of 10 consecutive years. For example,say in your database you have movie information starting from 1965. Then the first decade is 1965, 1966, ..., 1974; the second one is 1967, 1968, ..., 1976 and so on. Find the decade D with the largest number of films and the total number of films in D.
```
%%time
# citation https://stackoverflow.com/questions/25955049/sql-how-to-sum-up-count-for-many-decades?rq=1
# result = pd.read_sql_query(
# '''
# SELECT (ROUND(m.year / 10) * 10) AS Decade, COUNT(1) AS total_movies
# FROM Movie m
# GROUP BY ROUND(m.year/ 10)
# ORDER BY total_movies DESC LIMIT 1
# ''', conn)
# result
result = pd.read_sql_query('''
SELECT d_year.year AS start, d_year.year+9 AS end, count(1) AS total_movies FROM
(SELECT DISTINCT(year) FROM Movie) d_year
JOIN Movie m WHERE m.year>=start AND m.year<=end
GROUP BY end
ORDER BY total_movies DESC
LIMIT 1
''', conn)
result
```
8. Find the actors that were never unemployed for more than 3 years at a stretch. (Assume that the actors remain unemployed between two consecutive movies).
```
%%time
# citation https://stackoverflow.com/questions/57733454/to-find-actors-who-were-never-unemployed-for-more-than-3-years-in-a-stretch
# Here I am using window function (LEAD) that provides comparing current row with next row
result = pd.read_sql_query(
'''
SELECT *, (next_year - year) AS gap FROM (SELECT *
, LEAD(year, 1, 0) OVER (PARTITION BY Name ORDER BY year ASC) AS next_year
FROM (SELECT p.Name, m.title, m.year FROM Movie m
JOIN M_Cast mc ON m.MID=mc.MID
JOIN Person p ON trim(mc.PID)=trim(p.PID)))
WHERE gap <=3 and gap >=0
GROUP BY Name
ORDER BY Name ASC
''', conn)
result
```
9. Find all the actors that made more movies with Yash Chopra than any other director.
```
# %%time
# The following query is correct but didn't give the results,
# Running below query gives "database or disk is full" error
# result = pd.read_sql_query(
# '''
# SELECT P1.PID, P1.Name, count(Movie.MID) AS movies_with_yc from Person as P1
# JOIN M_Cast
# JOIN Movie
# JOIN M_Director ON (trim(Movie.MID) = trim(M_Director.MID))
# JOIN Person as P2 ON (trim(M_Director.PID) = trim(P2.PID)) where P2.Name = 'Yash Chopra'
# GROUP BY P1.PID HAVING count(Movie.MID) >
# (
# SELECT count(Movie.MID) FROM Person AS P3
# JOIN M_Cast
# JOIN Movie
# JOIN M_Director ON (trim(Movie.MID) = trim(M_Director.MID))
# JOIN Person AS P4 ON (trim(M_Director.PID) = trim(P4.PID))
# WHERE P1.PID = P3.PID AND P4.Name != 'Yash Chopra'
# GROUP BY P4.PID
# )
# ORDER BY movies_with_yc DESC;
# ''', conn)
# result
%%time
result = pd.read_sql_query(
'''
SELECT Director, Actor, Count(1) AS Movies_with_YashChopra
FROM
(
SELECT p.Name AS Director, m.title AS Movie
FROM Person p
JOIN M_Director md ON trim(md.PID)=trim(p.PID)
JOIN Movie m ON trim(md.MID)=m.MID and p.Name LIKE 'Yash%'
GROUP BY p.Name, m.title
) t1
JOIN
(
SELECT p.Name AS Actor, m.title AS Movie
FROM Person p
JOIN M_Cast mc ON trim(mc.PID)=trim(p.PID)
JOIN Movie m ON trim(mc.MID)=m.MID
GROUP BY p.Name, m.title
) t2
ON t1.Movie=t2.Movie
GROUP BY t1.Director, t2.Actor
ORDER By Movies_with_YashChopra DESC
''', conn)
result
```
10. The Shahrukh number of an actor is the length of the shortest path between the actor and Shahrukh Khan in the "co-acting" graph. That is, Shahrukh Khan has Shahrukh number 0; all actors who acted in the same film as Shahrukh have Shahrukh number 1; all actors who acted in the same film as some actor with Shahrukh number 1 have Shahrukh number 2, etc. Return all actors whoseShahrukh number is 2
```
%%time
result = pd.read_sql_query(
'''
SELECT Name FROM Person WHERE trim(Name) LIKE '%shah rukh khan%'
''', conn)
result
```
<h2>Using below steps we can get Shah Rukh Khan 2nd Degree Connection</h2>
- Logic to Build following Query
- Select movies in which Shah Rukh Khan worked
- Select Shah Rukh level 1 i.e. 1st Degree connection of Shah Rukh Khan
- Select movies in which Shah Rukh level 1 worked but exclude movies with Shah Rukh Khan
- Select Shah Rukh level 2 who worked in some movie with Shah Rukh level 1
```
%%time
result = pd.read_sql_query('''
SELECT DISTINCT P.Name
FROM Person p
JOIN M_Cast mc
ON trim(p.PID) = trim(mc.PID)
WHERE mc.MID IN (SELECT mc.MID
FROM M_Cast mc
WHERE trim(mc.PID) IN (
SELECT trim(p.PID) as pid
FROM Person p
JOIN M_Cast mc
ON trim(p.PID) = trim(mc.PID)
WHERE mc.MID IN (
SELECT mc.MID
FROM Person p
JOIN M_Cast mc
ON trim(p.PID) = trim(mc.PID)
WHERE trim(p.Name) LIKE '%shah rukh khan%'
)
AND trim(p.Name) NOT LIKE '%shah rukh khan%'
)
AND mc.MID NOT IN (SELECT mc.MID
FROM Person p
JOIN M_Cast mc
ON trim(p.PID) = trim(mc.PID)
WHERE trim(p.Name) LIKE '%shah rukh khan%'))
''', conn)
result
```
|
github_jupyter
|
```
!pip install torchvision==0.2.2
!pip install https://download.pytorch.org/whl/cu100/torch-1.1.0-cp36-cp36m-linux_x86_64.whl
!pip install typing
!pip install opencv-python
!pip install slackweb
!pip list | grep torchvision
!pip list | grep torch
# import cv2
import audioread
import logging
import os
import random
import time
import warnings
import glob
from tqdm import tqdm
import librosa
import numpy as np
import pandas as pd
import soundfile as sf
import torch
import torch.optim as optim
import torch.nn as nn
import torch.nn.functional as F
import torch.utils.data as data
from contextlib import contextmanager
from pathlib import Path
from typing import Optional
from fastprogress import progress_bar
from sklearn.metrics import f1_score
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from torchvision import models
import matplotlib.pyplot as plt
import slackweb
def set_seed(seed: int = 42):
random.seed(seed)
np.random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed) # type: ignore
torch.backends.cudnn.deterministic = True # type: ignore
torch.backends.cudnn.benchmark = True # type: ignore
def get_logger(out_file=None):
logger = logging.getLogger()
formatter = logging.Formatter("%(asctime)s - %(levelname)s - %(message)s")
logger.handlers = []
logger.setLevel(logging.INFO)
handler = logging.StreamHandler()
handler.setFormatter(formatter)
handler.setLevel(logging.INFO)
logger.addHandler(handler)
if out_file is not None:
fh = logging.FileHandler(out_file)
fh.setFormatter(formatter)
fh.setLevel(logging.INFO)
logger.addHandler(fh)
logger.info("logger set up")
return logger
@contextmanager
def timer(name: str, logger: Optional[logging.Logger] = None):
t0 = time.time()
msg = f"[{name}] start"
if logger is None:
print(msg)
else:
logger.info(msg)
yield
msg = f"[{name}] done in {time.time() - t0:.2f} s"
if logger is None:
print(msg)
else:
logger.info(msg)
logger = get_logger("main.log")
set_seed(1213)
DATA_PATH = '/home/knikaido/work/Cornell-Birdcall-Identification/data/birdsong_recognition/'
TRAIN_PATH = DATA_PATH + 'train_audio/'
MEL_PATH = '/home/knikaido/work/Cornell-Birdcall-Identification/data/feature/08_06_melspectrogram_small/'
class ResNet(nn.Module):
def __init__(self, base_model_name: str, pretrained=False,
num_classes=264):
super().__init__()
base_model = models.__getattribute__(base_model_name)(
pretrained=pretrained)
layers = list(base_model.children())[:-2]
layers.append(nn.AdaptiveMaxPool2d(1))
self.encoder = nn.Sequential(*layers)
in_features = base_model.fc.in_features
self.classifier = nn.Sequential(
nn.Linear(in_features, 1024), nn.ReLU(), nn.Dropout(p=0.2),
nn.Linear(1024, 1024), nn.ReLU(), nn.Dropout(p=0.2),
nn.Linear(1024, num_classes))
def forward(self, x):
batch_size = x.size(0)
x = self.encoder(x).view(batch_size, -1)
x = self.classifier(x)
multiclass_proba = F.softmax(x, dim=1)
multilabel_proba = torch.sigmoid(x)
return {
"logits": x,
"multiclass_proba": multiclass_proba,
"multilabel_proba": multilabel_proba
}
model_config = {
"base_model_name": "resnet50",
"pretrained": False,
"num_classes": 264
}
BIRD_CODE = {
'aldfly': 0, 'ameavo': 1, 'amebit': 2, 'amecro': 3, 'amegfi': 4,
'amekes': 5, 'amepip': 6, 'amered': 7, 'amerob': 8, 'amewig': 9,
'amewoo': 10, 'amtspa': 11, 'annhum': 12, 'astfly': 13, 'baisan': 14,
'baleag': 15, 'balori': 16, 'banswa': 17, 'barswa': 18, 'bawwar': 19,
'belkin1': 20, 'belspa2': 21, 'bewwre': 22, 'bkbcuc': 23, 'bkbmag1': 24,
'bkbwar': 25, 'bkcchi': 26, 'bkchum': 27, 'bkhgro': 28, 'bkpwar': 29,
'bktspa': 30, 'blkpho': 31, 'blugrb1': 32, 'blujay': 33, 'bnhcow': 34,
'boboli': 35, 'bongul': 36, 'brdowl': 37, 'brebla': 38, 'brespa': 39,
'brncre': 40, 'brnthr': 41, 'brthum': 42, 'brwhaw': 43, 'btbwar': 44,
'btnwar': 45, 'btywar': 46, 'buffle': 47, 'buggna': 48, 'buhvir': 49,
'bulori': 50, 'bushti': 51, 'buwtea': 52, 'buwwar': 53, 'cacwre': 54,
'calgul': 55, 'calqua': 56, 'camwar': 57, 'cangoo': 58, 'canwar': 59,
'canwre': 60, 'carwre': 61, 'casfin': 62, 'caster1': 63, 'casvir': 64,
'cedwax': 65, 'chispa': 66, 'chiswi': 67, 'chswar': 68, 'chukar': 69,
'clanut': 70, 'cliswa': 71, 'comgol': 72, 'comgra': 73, 'comloo': 74,
'commer': 75, 'comnig': 76, 'comrav': 77, 'comred': 78, 'comter': 79,
'comyel': 80, 'coohaw': 81, 'coshum': 82, 'cowscj1': 83, 'daejun': 84,
'doccor': 85, 'dowwoo': 86, 'dusfly': 87, 'eargre': 88, 'easblu': 89,
'easkin': 90, 'easmea': 91, 'easpho': 92, 'eastow': 93, 'eawpew': 94,
'eucdov': 95, 'eursta': 96, 'evegro': 97, 'fiespa': 98, 'fiscro': 99,
'foxspa': 100, 'gadwal': 101, 'gcrfin': 102, 'gnttow': 103, 'gnwtea': 104,
'gockin': 105, 'gocspa': 106, 'goleag': 107, 'grbher3': 108, 'grcfly': 109,
'greegr': 110, 'greroa': 111, 'greyel': 112, 'grhowl': 113, 'grnher': 114,
'grtgra': 115, 'grycat': 116, 'gryfly': 117, 'haiwoo': 118, 'hamfly': 119,
'hergul': 120, 'herthr': 121, 'hoomer': 122, 'hoowar': 123, 'horgre': 124,
'horlar': 125, 'houfin': 126, 'houspa': 127, 'houwre': 128, 'indbun': 129,
'juntit1': 130, 'killde': 131, 'labwoo': 132, 'larspa': 133, 'lazbun': 134,
'leabit': 135, 'leafly': 136, 'leasan': 137, 'lecthr': 138, 'lesgol': 139,
'lesnig': 140, 'lesyel': 141, 'lewwoo': 142, 'linspa': 143, 'lobcur': 144,
'lobdow': 145, 'logshr': 146, 'lotduc': 147, 'louwat': 148, 'macwar': 149,
'magwar': 150, 'mallar3': 151, 'marwre': 152, 'merlin': 153, 'moublu': 154,
'mouchi': 155, 'moudov': 156, 'norcar': 157, 'norfli': 158, 'norhar2': 159,
'normoc': 160, 'norpar': 161, 'norpin': 162, 'norsho': 163, 'norwat': 164,
'nrwswa': 165, 'nutwoo': 166, 'olsfly': 167, 'orcwar': 168, 'osprey': 169,
'ovenbi1': 170, 'palwar': 171, 'pasfly': 172, 'pecsan': 173, 'perfal': 174,
'phaino': 175, 'pibgre': 176, 'pilwoo': 177, 'pingro': 178, 'pinjay': 179,
'pinsis': 180, 'pinwar': 181, 'plsvir': 182, 'prawar': 183, 'purfin': 184,
'pygnut': 185, 'rebmer': 186, 'rebnut': 187, 'rebsap': 188, 'rebwoo': 189,
'redcro': 190, 'redhea': 191, 'reevir1': 192, 'renpha': 193, 'reshaw': 194,
'rethaw': 195, 'rewbla': 196, 'ribgul': 197, 'rinduc': 198, 'robgro': 199,
'rocpig': 200, 'rocwre': 201, 'rthhum': 202, 'ruckin': 203, 'rudduc': 204,
'rufgro': 205, 'rufhum': 206, 'rusbla': 207, 'sagspa1': 208, 'sagthr': 209,
'savspa': 210, 'saypho': 211, 'scatan': 212, 'scoori': 213, 'semplo': 214,
'semsan': 215, 'sheowl': 216, 'shshaw': 217, 'snobun': 218, 'snogoo': 219,
'solsan': 220, 'sonspa': 221, 'sora': 222, 'sposan': 223, 'spotow': 224,
'stejay': 225, 'swahaw': 226, 'swaspa': 227, 'swathr': 228, 'treswa': 229,
'truswa': 230, 'tuftit': 231, 'tunswa': 232, 'veery': 233, 'vesspa': 234,
'vigswa': 235, 'warvir': 236, 'wesblu': 237, 'wesgre': 238, 'weskin': 239,
'wesmea': 240, 'wessan': 241, 'westan': 242, 'wewpew': 243, 'whbnut': 244,
'whcspa': 245, 'whfibi': 246, 'whtspa': 247, 'whtswi': 248, 'wilfly': 249,
'wilsni1': 250, 'wiltur': 251, 'winwre3': 252, 'wlswar': 253, 'wooduc': 254,
'wooscj2': 255, 'woothr': 256, 'y00475': 257, 'yebfly': 258, 'yebsap': 259,
'yehbla': 260, 'yelwar': 261, 'yerwar': 262, 'yetvir': 263
}
INV_BIRD_CODE = {v: k for k, v in BIRD_CODE.items()}
train_path = DATA_PATH + 'train.csv'
train = pd.read_csv(train_path)
le = LabelEncoder()
encoded = le.fit_transform(train['channels'].values)
decoded = le.inverse_transform(encoded)
train['channels'] = encoded
for i in tqdm(range(len(train))):
train['ebird_code'][i] = BIRD_CODE[train['ebird_code'][i]]
train['filename'] = train['filename'].str.replace(".mp3", "")
train.head()
mel_list = sorted(glob.glob(MEL_PATH + '*.npy'))
mel_list = pd.Series(mel_list)
len(mel_list)
import joblib
target_list = joblib.load(MEL_PATH+'target_list.pkl')
for i in tqdm(range(len(target_list))):
target_list[i] = BIRD_CODE[target_list[i]]
len(target_list)
X_train_mel, X_valid_mel, target_train, taret_valid = train_test_split(mel_list, target_list, test_size=0.2, stratify=target_list)
class TrainDateset(torch.utils.data.Dataset):
def __init__(self, mel_list, train, transform=None):
self.transform = transform
self.mel_list = mel_list
self.data_num = len(mel_list)
def __len__(self):
return self.data_num
def __getitem__(self, idx):
if self.transform:
pass
# out_data = self.transform(self.data)[0][idx]
# out_label = self.label[idx]
else:
# print(idx)
out_data = np.array(np.load(mel_list[idx]))
out_mel_list = mel_list[idx]
out_label = target_list[idx]
# out_label = self.label[idx]
return out_data, out_label
train_dataset = TrainDateset(X_train_mel, target_train)
train_dataloader = torch.utils.data.DataLoader(train_dataset, batch_size=128, shuffle=True)
valid_dataset = TrainDateset(X_valid_mel, taret_valid)
valid_dataloader = torch.utils.data.DataLoader(valid_dataset, batch_size=128, shuffle=True)
WEIGHT_DECAY = 0.005
LEARNING_RATE = 0.0001
EPOCH = 100
device = 'cuda' if torch.cuda.is_available() else 'cpu'
print(torch.cuda.is_available())
net = ResNet('resnet50')
net = net.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = optim.Adam(net.parameters(), lr=LEARNING_RATE, weight_decay=WEIGHT_DECAY)
%%time
train_losses = []
valid_losses = []
for epoch in tqdm(range(EPOCH)): # loop over the dataset multiple times
train_loss = 0.0
valid_loss = 0.0
net.train()
for i, data in enumerate(train_dataloader):
# 第二引数は,スタート位置で,0なのでenumerate(trainloader)と同じ
# https://docs.python.org/3/library/functions.html#enumerate
# get the inputs
inputs, labels = data
inputs = inputs.to(device)
labels = labels.to(device)
# wrap them in Variable
# inputs, labels = Variable(inputs), Variable(labels)
# zero the parameter gradients
optimizer.zero_grad()
# forward + backward + optimize
outputs = net(inputs)
loss = criterion(outputs['logits'], labels)
loss.backward()
optimizer.step()
# print statistics
# running_loss += loss.data[0]
train_loss += loss.to('cpu').detach().numpy().copy()
print('[%d, %5d] train loss: %.3f' %
(epoch + 1, i + 1, train_loss / (i+1)))
train_losses.append(train_loss / (i+1))
net.eval()
for i, data in enumerate(valid_dataloader):
# 第二引数は,スタート位置で,0なのでenumerate(trainloader)と同じ
# https://docs.python.org/3/library/functions.html#enumerate
# get the inputs
inputs, labels = data
inputs = inputs.to(device)
labels = labels.to(device)
# wrap them in Variable
# inputs, labels = Variable(inputs), Variable(labels)
# zero the parameter gradients
optimizer.zero_grad()
# forward + backward + optimize
outputs = net(inputs)
loss = criterion(outputs['logits'], labels)
# print statistics
# running_loss += loss.data[0]
valid_loss += loss.to('cpu').detach().numpy().copy()
print('[%d, %5d] valid loss: %.3f' %
(epoch + 1, i + 1, valid_loss / (i+1)))
valid_losses.append(valid_loss / (i+1))
# break
```
https://qiita.com/derodero24/items/f4cc46f144f404054501
```
import cloudpickle
with open('model.pkl', 'wb') as f:
cloudpickle.dump(net, f)
slack = slackweb.Slack(url="https://hooks.slack.com/services/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")
slack.notify(text="おわた")
with open('model.pkl', 'rb') as f:
net = cloudpickle.load(f)
```
## plot loss
```
plt.figure(figsize=(16,5), dpi= 80)
plt.plot(train_losses, color='tab:red', label='valid')
plt.plot(valid_losses, color='tab:blue', label='train')
plt.legend()
```
|
github_jupyter
|
Python programmers will often suggest that there many ways the language can be used to solve a particular
problem. But that some are more appropriate than others. The best solutions are celebrated as Idiomatic
Python and there are lots of great examples of this on StackOverflow and other websites.
A sort of sub-language within Python, Pandas has its own set of idioms. We've alluded to some of these
already, such as using vectorization whenever possible, and not using iterative loops if you don't need to.
Several developers and users within the Panda's community have used the term __pandorable__ for these
idioms. I think it's a great term. So, I wanted to share with you a couple of key features of how you can
make your code pandorable.
```
# Let's start by bringing in our data processing libraries
import pandas as pd
import numpy as np
# And we'll bring in some timing functionality too, from the timeit module
import timeit
# And lets look at some census data from the US
df = pd.read_csv('datasets/census.csv')
df.head()
# The first of the pandas idioms I would like to talk about is called method chaining. The general idea behind
# method chaining is that every method on an object returns a reference to that object. The beauty of this is
# that you can condense many different operations on a DataFrame, for instance, into one line or at least one
# statement of code.
# Here's the pandorable way to write code with method chaining. In this code I'm going to pull out the state
# and city names as a multiple index, and I'm going to do so only for data which has a summary level of 50,
# which in this dataset is county-level data. I'll rename a column too, just to make it a bit more readable.
(df.where(df['SUMLEV']==50)
.dropna()
.set_index(['STNAME','CTYNAME'])
.rename(columns={'ESTIMATESBASE2010': 'Estimates Base 2010'}))
# Lets walk through this. First, we use the where() function on the dataframe and pass in a boolean mask which
# is only true for those rows where the SUMLEV is equal to 50. This indicates in our source data that the data
# is summarized at the county level. With the result of the where() function evaluated, we drop missing
# values. Remember that .where() doesn't drop missing values by default. Then we set an index on the result of
# that. In this case I've set it to the state name followed by the county name. Finally. I rename a column to
# make it more readable. Note that instead of writing this all on one line, as I could have done, I began the
# statement with a parenthesis, which tells python I'm going to span the statement over multiple lines for
# readability.
# Here's a more traditional, non-pandorable way, of writing this. There's nothing wrong with this code in the
# functional sense, you might even be able to understand it better as a new person to the language. It's just
# not as pandorable as the first example.
# First create a new dataframe from the original
df = df[df['SUMLEV']==50] # I'll use the overloaded indexing operator [] which drops nans
# Update the dataframe to have a new index, we use inplace=True to do this in place
df.set_index(['STNAME','CTYNAME'], inplace=True)
# Set the column names
df.rename(columns={'ESTIMATESBASE2010': 'Estimates Base 2010'})
# Now, the key with any good idiom is to understand when it isn't helping you. In this case, you can actually
# time both methods and see which one runs faster
# We can put the approach into a function and pass the function into the timeit function to count the time the
# parameter number allows us to choose how many times we want to run the function. Here we will just set it to
# 10
# Lets write a wrapper for our first function
def first_approach():
global df
# And we'll just paste our code right here
return (df.where(df['SUMLEV']==50)
.dropna()
.set_index(['STNAME','CTYNAME'])
.rename(columns={'ESTIMATESBASE2010': 'Estimates Base 2010'}))
# Read in our dataset anew
df = pd.read_csv('datasets/census.csv')
# And now lets run it
timeit.timeit(first_approach, number=10)
# Now let's test the second approach. As you may notice, we use our global variable df in the function.
# However, changing a global variable inside a function will modify the variable even in a global scope and we
# do not want that to happen in this case. Therefore, for selecting summary levels of 50 only, I create a new
# dataframe for those records
# Let's run this for once and see how fast it is
def second_approach():
global df
new_df = df[df['SUMLEV']==50]
new_df.set_index(['STNAME','CTYNAME'], inplace=True)
return new_df.rename(columns={'ESTIMATESBASE2010': 'Estimates Base 2010'})
# Read in our dataset anew
df = pd.read_csv('datasets/census.csv')
# And now lets run it
timeit.timeit(second_approach, number=10)
# As you can see, the second approach is much faster! So, this is a particular example of a classic time
# readability trade off.
# You'll see lots of examples on stack overflow and in documentation of people using method chaining in their
# pandas. And so, I think being able to read and understand the syntax is really worth your time. But keep in
# mind that following what appears to be stylistic idioms might have performance issues that you need to
# consider as well.
# Here's another pandas idiom. Python has a wonderful function called map, which is sort of a basis for
# functional programming in the language. When you want to use map in Python, you pass it some function you
# want called, and some iterable, like a list, that you want the function to be applied to. The results are
# that the function is called against each item in the list, and there's a resulting list of all of the
# evaluations of that function.
# Pandas has a similar function called applymap. In applymap, you provide some function which should operate
# on each cell of a DataFrame, and the return set is itself a DataFrame. Now I think applymap is fine, but I
# actually rarely use it. Instead, I find myself often wanting to map across all of the rows in a DataFrame.
# And pandas has a function that I use heavily there, called apply. Let's look at an example.
# Let's take a look at our census DataFrame. In this DataFrame, we have five columns for population estimates,
# with each column corresponding with one year of estimates. It's quite reasonable to want to create some new
# columns for minimum or maximum values, and the apply function is an easy way to do this.
# First, we need to write a function which takes in a particular row of data, finds a minimum and maximum
# values, and returns a new row of data nd returns a new row of data. We'll call this function min_max, this
# is pretty straight forward. We can create some small slice of a row by projecting the population columns.
# Then use the NumPy min and max functions, and create a new series with a label values represent the new
# values we want to apply.
def min_max(row):
data = row[['POPESTIMATE2010',
'POPESTIMATE2011',
'POPESTIMATE2012',
'POPESTIMATE2013',
'POPESTIMATE2014',
'POPESTIMATE2015']]
return pd.Series({'min': np.min(data), 'max': np.max(data)})
# Then we just need to call apply on the DataFrame.
# Apply takes the function and the axis on which to operate as parameters. Now, we have to be a bit careful,
# we've talked about axis zero being the rows of the DataFrame in the past. But this parameter is really the
# parameter of the index to use. So, to apply across all rows, which is applying on all columns, you pass axis
# equal to 'columns'.
df.apply(min_max, axis='columns').head()
# Of course there's no need to limit yourself to returning a new series object. If you're doing this as part
# of data cleaning your likely to find yourself wanting to add new data to the existing DataFrame. In that
# case you just take the row values and add in new columns indicating the max and minimum scores. This is a
# regular part of my workflow when bringing in data and building summary or descriptive statistics, and is
# often used heavily with the merging of DataFrames.
# Here's an example where we have a revised version of the function min_max Instead of returning a separate
# series to display the min and max we add two new columns in the original dataframe to store min and max
def min_max(row):
data = row[['POPESTIMATE2010',
'POPESTIMATE2011',
'POPESTIMATE2012',
'POPESTIMATE2013',
'POPESTIMATE2014',
'POPESTIMATE2015']]
# Create a new entry for max
row['max'] = np.max(data)
# Create a new entry for min
row['min'] = np.min(data)
return row
# Now just apply the function across the dataframe
df.apply(min_max, axis='columns')
# Apply is an extremely important tool in your toolkit. The reason I introduced apply here is because you
# rarely see it used with large function definitions, like we did. Instead, you typically see it used with
# lambdas. To get the most of the discussions you'll see online, you're going to need to know how to at least
# read lambdas.
# Here's You can imagine how you might chain several apply calls with lambdas together to create a readable
# yet succinct data manipulation script. One line example of how you might calculate the max of the columns
# using the apply function.
rows = ['POPESTIMATE2010', 'POPESTIMATE2011', 'POPESTIMATE2012', 'POPESTIMATE2013','POPESTIMATE2014',
'POPESTIMATE2015']
# Now we'll just apply this across the dataframe with a lambda
df.apply(lambda x: np.max(x[rows]), axis=1).head()
# If you don't remember lambdas just pause the video for a moment and look up the syntax. A lambda is just an
# unnamed function in python, in this case it takes a single parameter, x, and returns a single value, in this
# case the maximum over all columns associated with row x.
# The beauty of the apply function is that it allows flexibility in doing whatever manipulation that you
# desire, as the function you pass into apply can be any customized however you want. Let's say we want to
# divide the states into four categories: Northeast, Midwest, South, and West We can write a customized
# function that returns the region based on the state the state regions information is obtained from Wikipedia
def get_state_region(x):
northeast = ['Connecticut', 'Maine', 'Massachusetts', 'New Hampshire',
'Rhode Island','Vermont','New York','New Jersey','Pennsylvania']
midwest = ['Illinois','Indiana','Michigan','Ohio','Wisconsin','Iowa',
'Kansas','Minnesota','Missouri','Nebraska','North Dakota',
'South Dakota']
south = ['Delaware','Florida','Georgia','Maryland','North Carolina',
'South Carolina','Virginia','District of Columbia','West Virginia',
'Alabama','Kentucky','Mississippi','Tennessee','Arkansas',
'Louisiana','Oklahoma','Texas']
west = ['Arizona','Colorado','Idaho','Montana','Nevada','New Mexico','Utah',
'Wyoming','Alaska','California','Hawaii','Oregon','Washington']
if x in northeast:
return "Northeast"
elif x in midwest:
return "Midwest"
elif x in south:
return "South"
else:
return "West"
# Now we have the customized function, let's say we want to create a new column called Region, which shows the
# state's region, we can use the customized function and the apply function to do so. The customized function
# is supposed to work on the state name column STNAME. So we will set the apply function on the state name
# column and pass the customized function into the apply function
df['state_region'] = df['STNAME'].apply(lambda x: get_state_region(x))
# Now let's see the results
df[['STNAME','state_region']].head()
```
So there are a couple of Pandas idioms. But I think there's many more, and I haven't talked about them here.
So here's an unofficial assignment for you. Go look at some of the top ranked questions on pandas on Stack
Overflow, and look at how some of the more experienced authors, answer those questions. Do you see any
interesting patterns? Feel free to share them with myself and others in the class.
|
github_jupyter
|
# 讀取字典
```
import pandas as pd
import numpy as np
import os
filepath = '/Volumes/backup_128G/z_repository/Yumin_data/玉敏_俄羅斯課本的研究'
file_dic = '華語八千詞(內含注音字型檔)/Chinese_8000W_20190515_v1.xlsx'
book_file = '實用漢語教科書2010_生詞表.xlsx'
to_file = 'processed/chinese_8000Words_results.xlsx'
# write_level_doc = '{0}/{1}'.format(filepath, to_level_doc)
read_dic = '{0}/{1}'.format(filepath, file_dic)
read_book = '{0}/{1}'.format(filepath, book_file)
write_file = '{0}/{1}'.format(filepath, to_file)
dicDf = pd.DataFrame()
with pd.ExcelFile(read_dic) as reader:
# read sheet by sheet
for sheet in reader.sheet_names:
# print(sheet)
sheetDf = pd.read_excel(reader, sheet, header=None)
sheetDf = sheetDf.fillna(0)
dicDf = dicDf.append(sheetDf, ignore_index=True)
# change to lowercase
len(dicDf.index)
dicDf.head()
dicList = {}
for idx in range(0, len(dicDf)):
row = dicDf.loc[idx]
dicWord = row[0]
dicLevel = row[1]
if dicWord not in dicList:
dicList[dicWord] = [dicLevel]
else:
# print(dicWord, dicLevel)
dicList[dicWord].append(dicLevel)
# dicList
```
# 讀取待分析檔
```
bookDf = pd.read_excel(read_book)
bookDf.head()
wordDifferentLevel = []
def wordLevel(word):
foundLevel = 9
if word in dicList:
foundLevel = dicList[word][0]
return foundLevel
levelList = []
for idx in range(0, len(bookDf)):
row = bookDf.loc[idx]
chapter = row[0]
wtype = row[1]
word = row[3]
level = wordLevel(word)
levelList.append([word, level, wtype, chapter])
# print(chapter, wtype, word)
levelDf = pd.DataFrame(levelList)
levelDf = levelDf.sort_values(by=[1, 3, 2, 0])
levelDf.head()
# levelDf.loc[levelDf[2] == 'A']
# levelDf.loc[levelDf[2] == 'B']
levelDf[~levelDf[2].isin(['A', 'B'])]
def statsLevel(INdf):
levelCountList = []
for level in range(1, 10):
levelCount = INdf[1].loc[INdf[1] == level].count()
levelCountList.append(levelCount)
levelCountDf = pd.DataFrame(levelCountList)
return levelCountDf
def statsLessonLevel(INdf):
levels = list(range(1, 10))
statDf = pd.DataFrame(levels)
lessons = INdf[3].unique()
lessons = np.sort(lessons)
for lesson in lessons:
lessonDf = INdf.loc[INdf[3] == lesson]
statDf[lesson] = statsLevel(lessonDf)
return statDf
headers = ['Word', 'Level', 'A/B', 'Lesson']
with pd.ExcelWriter(write_file) as writer:
# 1.列出每一個詞的等級
levelDf.to_excel(writer, 'All', index=False, header=headers)
# 2.統計每一個等級共有多少字
levels = list(range(1, 10))
levelCountDf = pd.DataFrame(levels)
## A.主要詞彙的統計
major = levelDf.loc[levelDf[2] == 'A']
levelCountDf['A'] = statsLevel(major)
## B.補充詞彙的統計
minor = levelDf.loc[levelDf[2] == 'B']
levelCountDf['B'] = statsLevel(minor)
## C.主要詞彙+補充詞彙的統計
levelCountDf['A/B'] = statsLevel(levelDf)
levelCountDf.to_excel(writer, 'Stats', index=False, header=['Level', 'A', 'B', 'A/B'])
# 3.統計每一個等級共有多少字 by lesson
lessonDf = statsLessonLevel(levelDf)
lessonDf.T.to_excel(writer, 'lessons', header=False)
# 4.列出不在8000詞的生詞有哪些
wordsNotIn = levelDf.loc[levelDf[1] == 9]
wordsNotInDf = pd.DataFrame(wordsNotIn)
wordsNotInDf.to_excel(writer, 'WordsNotIn', index=False, header=headers)
writer.save()
```
|
github_jupyter
|
# The Great Pyramid
This is an estimate of the number of people needed to raise stones to the top of the [great pyramid](https://en.wikipedia.org/wiki/Great_Pyramid_of_Giza) using basic physics, such as force, energy, and power. It relies solely on reasonable estimates of known dimensions of the great pyramid and typical human labor capacity. The analysis will show that it is possible for crews of workers to raise 2.5 ton limestones to almost any level using ropes alone. Each crew would stand on an unfinished level and pull wooden sleds carrying stones up the 51.86 degree incline of the pyramid. This solution does not require ramps, pulleys, levers or any other mechanical advantage. It only requires coordination, rope, and well fed crews. If a crew tires after raising a set of stones, they could be quickly replaced by another well rested crew. The analysis will estimate the minimum crew size, number of crews required, the rate at which stones can be raised, and the maneuvering area available at each level.
The dimensions of the great pyramid are shown below:

| Parameter | Value |
| ----- | ----:|
| Total number of stones| 2.5 million |
| Average mass of each stone | 2.5 tons |
| Total build time | 20 years |
| Power available per worker | 200 Calories/day |
| Active build time | 3 months/year |
| Pyramid slope | 51.86 degrees |
| Pyramid height | 146.5 meters |
| Pyramid base | 230 m |
| Coefficient of friction | 0.3 |
| Number of layers | 210 |
| Course | Height | Amount of Material |
| ------ |:------:| ------------------:|
| 1 | 15m | 30% |
| 2 | 30m | 22% |
| 3 | 50m | 30% |
| 4 | 100m | 15% |
| 5 | 146m | 3% |
```
from math import *
import pandas as pd
import matplotlib.pyplot as plt
# All values are in SI (MKS) units
lbm_per_kg = 2.20462
newtons_per_lbf = 4.44822
joules_per_kcal = 4184
sec_per_day = 24 * 3600
watts_per_hp = 746
# Total number of stones
N_s = 2.5e6
# Mass of one stone in kg
m_s = 2.5 * 2000 / lbm_per_kg
# Total build time in seconds
T_b = 20 * 365.25 * sec_per_day
# Average available power per crew member in kilocalories (nutrition calorie)
P_w_kcal = 200
# Average available power on crew member in Watts
P_w = P_w_kcal * joules_per_kcal / sec_per_day
# Pyramid slope in radians
theta = 51.86*pi/180
# Pyramid base length in meters
l_b = 230
# Coefficient of friction between limestone and wood sleds
mu = 0.3
# Acceleration of gravity in m/2^s
g = 9.81
# Number of layers
N_l = 210
# Height of pyramid in meters
h_max = 146.5
```
# Pulling Force
It is possible for a crew of men to stand on top of one flat level and simply pull a single stone up the side of a pyramid covered with smooth casing stones. It is expected that smooth casing stones were added at the same time each layer of rough blocks were added, which is very likely. This simple approach does not require large ramps, elaborate machines, deep knowledge, or alien intervention. It just requires many crews of workers pulling on ropes attached to rough stones. Of course, a number of additional crews are needed to place stones and align stones properly, but the solutions to those problems are well documented.
This analysis focuses solely on the rigging problem of raising stones to the proper level just prior to final placement.
The [force required](https://en.wikipedia.org/wiki/Inclined_plane) to pull one stone up the side of the pyramid is
$$ F_p = m_s g (sin \theta + \mu cos \theta)$$
Where $m_s$ is the mass of one stone, $g$ is acceleration of gravity, $\theta$ is the pyramid slope, and $\mu$ is the coefficient of friction.
Given the parameters above, the pulling force is
```
F_p = m_s * g * (sin(theta) + mu*cos(theta))
print('%.4f N' % F_p)
print('%.4f lbf' % (F_p / newtons_per_lbf) )
```
$$ F_p \approx 21620 N $$
or
$$ F_p \approx 4860 lbf $$
This is slightly less than the 5000 lb weight of each stone, which is due to the slope of incline and static friction. Dynamic friction is perhaps lower, so the actual pulling force while in motion may be less.
# Energy to Raise Stones
Energy is force times distance moved. The distance along the slope up to a height $h$ is
$$ d = \frac{h}{sin \theta} $$
Given the force derived earlier, energy required to raise a single stone to a height $h$ is
$$ E_s = \frac{F_p h}{sin \theta} $$
For all stones the total energy is
$$ E_t = \frac{F_p}{sin \theta} \sum_{i=1}^{m} h N_{blocks} $$
An approximate estimate for comparison is:
$$ E_t = \frac{F_p N_s}{sin \theta} (15m \times 0.3 + 30m \times 0.22 + 50m \times 0.3 + 100m \times 0.15 + 146m \times 0.03) $$
The total energy is estimate in two steps:
* Compute the total volume to get average block volume
* Compute energy per layer given average block volume
The iterative computation will be compared with the approximate estimate.
The total energy is
```
dh = h_max / N_l
total_volume = 0
h = 0
tan_theta2 = tan(theta)**2
for i in range(N_l):
th = (h_max - h)**2
A_f = 4 * th / tan_theta2
total_volume += dh * A_f
h += dh
print('Total volume: %.3e m^3' % total_volume)
block_volume = total_volume/N_s
print('Block volume: %.3e m^3' % block_volume)
E_t = 0
h = 0
for i in range(N_l):
th = (h_max - h)**2
A_f = 4 * th / tan_theta2
num_blocks = dh * A_f / block_volume
E_t += F_p * num_blocks * h / sin(theta)
h += dh
print('Total energy: %.2e Joules' % E_t)
print('Total energy: %.2e kcal' % (E_t/joules_per_kcal))
E_t_approx = F_p * N_s * (15*0.3 + 30*0.22 + 50*0.3 + 100*0.15 + 146*0.03) / sin(theta)
print('Approximate: %.2e Joules' % E_t_approx)
print('Approximate: %.2e kcal' % (E_t_approx/joules_per_kcal))
```
The iterative estimate is somewhat less than the approximate energy, which is reasonable.
$$ E_t \approx 2.5 \times 10^{12} J $$
or
$$ E_t \approx 5.97 \times 10^8 {kcal} $$
# Average Power
The average power required to raise all stones is
$$ P_{avg} = \frac{E_t}{T_b} $$
```
P_avg = E_t/T_b
print('%.2f W' % (P_avg))
print('%.2f HP' % (P_avg/watts_per_hp))
```
In watts, the value is:
$$ P_{avg} \approx 3960 W $$
In horse power:
$$ P_{avg} \approx 5.31 {HP} $$
This surprisingly modest number is due to the 20 year build time for the pyramid. Even though the size of the pyramid is staggering, the build time is equally large. By inspection, we can imagine the number of workers needed to deliver this power, which is not as large as might be expected.
5.3 horse power would be easily available using a few draught animals, but that would require coaxing animals to climb to high levels and repeatedly pulling over a significant distance. This presents several logistical challenges, which might explain why there is little evidence of animal power used to raise stones. Humans can stand in one place and pull ropes hand over hand with upper body power or two crews could alternate pulling one set of ropes using lower body power. Perhaps different techniques were used depending on available maneuvering area.
# Workforce Size
Human are not efficient machines, perhaps 20% thermal efficiency. Given a modest diet where 1000 calories are burned, one worker might deliver 200 calories/day of mechanical work. This is an average power of 9.7 Watts. Assuming work is performed during only one season (one quarter of a year), the total number of workers required to raise all blocks is given by
$$ N_w = 4 \frac{P_{avg}}{P_w} $$
The approximate number of workers is
```
N_w = 4 * P_avg / P_w
print('%d workers' % N_w)
```
$$ N_w \approx 1635 $$
Other estimates of total workforce are about 10 times this value, which makes sense given resting time, and many other tasks, such as cutting and transporting stones, finish work, food preparation, management, accounting, and other support activities.
To lift a single stone, a crew of workers would be required to raise each stone. Assuming each worker can pull 75 lbs, the size of a single lifting crew is
$$ N_{lc} = \frac{F_p}{75 lbf} $$
The number of workers in a lifting crew is
```
F_1p = 75 * newtons_per_lbf
N_lc = F_p / F_1p
print('%.1f workers per lifting crew' % N_lc)
```
$$ N_{lc} \approx 65 $$
That's 65 workers per lifting crew. The total number of crews is
$$ N_c = \frac{N_w}{N_{lc}} $$
```
N_c = N_w / N_lc
print('%.1f crews' % N_c)
```
Roughly 25 concurrent crews of 65 people are required just to raise all stones over 20 years.
# Stone Raising Rate
Assuming all 25 crews are operating concurrently, it is possible to estimate the block raising rate. 200 calories per day of worker output is an average number. Humans are not machines and need rest, so in practice, crews may only raise blocks as little as 4 hours per day. Assuming all 200 calories per worker is delivered in a four hour shift, the available peak crew power would be six times the average daily power:
$$ P_{cp} = 6 N_{lc} P_w$$
```
P_cp = 6 * N_lc * P_w
print('%.2f W' % (P_cp))
print('%.2f HP' % (P_cp/watts_per_hp))
```
This value is about 3.8 kW or just a little over 5 horsepower for a crew of 65 workers. This suggests about 13 humans can do the same amount of work as one horse for four hours a day, which seems reasonable.
The average velocity of a single block raised by a crew is given by
$$ v_{bc} = \frac{P_{cp}}{F_p} $$
```
feet_per_meter = 3.28084
v_bc = P_cp / F_p
print('%.3f m/s' % (v_bc))
print('%.3f ft/s' % (v_bc * feet_per_meter))
```
The rate along the slope is about 0.17 $m/s$ or 0.57 $ft/s$.
To raise one stone to a height h, the time required is
$$ t = \frac{h}{v_{bc} sin \theta} $$
```
h = 30
t = h/(v_bc * sin(theta))
print('%.1f seconds' % (t))
print('%.1f minutes' % (t/60))
```
To raise one block to a height of 30m, which includes more than 50% of all stones, the time is about 219 seconds or 3.6 minutes. With all 25 crews operating concurrently, one stone could be raised every nine seconds or less.
# Logistics
Fitting 1635 workers on a level at one time requires room to maneuver. The area available is reduced higher up the pyramid. Assuming all 25 crews are operating concurrently and each worker requires at least $1 m^2$, the minimum area required is $A_c \approx 1635 m^2$.
The available area at a height $h$ is
$$ A_l = \left(\frac{2 (h_{max} - h)}{tan \theta}\right)^2 $$
Where $l_b$ is the length of the base of the pyramid.
The fraction of available maneuvering area is
$$ r_m = \frac{A_l-A_c}{A_l} $$
A plot of available maneuvering area and completed volume is shown below.
```
A_c = N_w
dh = h_max / N_l
h = 0
tan_theta2 = tan(theta)**2
heights = []
areas = []
volumes = []
volume = 0
for i in range(N_l):
th = (h_max - h)**2
A_l = 4 * th / tan_theta2
volume += dh * A_l
r_a = (A_l-A_c)/A_l
heights.append(h)
areas.append(100*r_a)
volumes.append(100*(volume/total_volume))
h += dh
limit = -40
plt.plot(heights[0:limit], areas[0:limit], label='Maneuvering area', color='blue')
plt.plot(heights[0:limit], volumes[0:limit], label='Completed volume', color='red')
plt.ylabel('Percentage (%)')
plt.xlabel('Height (m)')
plt.legend(loc='best')
plt.show()
limit = -66
print('At a height of %.1f m, %.1f %% of the pyramid is complete.' % (heights[limit], volumes[limit]))
```
Even at a height of 100m, where only 3% of the pyramid remains, more than two times the area required by all 25 lifting crews is still available. This should leave sufficient room for others to position stones after they have been lifted. At 117m, there is just enough room for all 25 crews, so stone placement will slow down. Fortunately, fewer stones are required at the highest levels.
# Ramps and Stone Size
This theory might explain why there is little evidence of external or internal ramps, simply because a smooth pyramid can act as the ramp itself. It might also explain how large granite blocks were hauled up to the kings chamber. Considering the required rate of block raising, a wide ramp is needed. Narrow ramps that can only support one or two blocks side by side seem like a bottleneck. Ramps with right angles require more time to rotate and orient blocks. Using the sides of the pyramid offers the largest ramp possible on all four sides, so the only limitation would be the number of workers that could be on top at any one time. Even if one set of crews becomes fatigued raising stones, they could be relieved by another crew later in the day. It is possible that two or more shifts of lifting crews were used to minimize fatigue or injury. If ropes were long enough, it is possible that workers could have walked down the opposite slope of the pyramid, using their own weight to counter the weight of stones they were attempting to lift.
A similar energy analysis can be done using conventional shallow ramps to raise stones. Interestingly, a ramp with a 7% grade requires almost 5 times more energy to raise all 2.5 million stones than using the side of pyramid. Although a shallow ramp reduces the amount of force required to move stones, the distance travelled is much farther, so more energy is lost in friction. Additionally, a conventional ramp requires workers to climb the height of the pyramid along with the stone they are pulling, so they must lift their own weight in addition to the weight of the stone. This requires more energy, which is not used to lift stones. Clearly, it's a highly inefficient strategy.
The Egyptians were free to decide how big to make the rough limestone blocks. They could have made them small enough for one person to carry, but they chose not to. After many pyramid construction attempts, they decided that 2.5 ton blocks were small enough to handle without too much difficulty, so raising these stones had to be straightforward. It seems that simply dragging blocks up the side of a smooth pyramid is a straightforward solution that they could have easily developed on their own. It probably seemed so obvious to them that it made no sense to document it.
# Summary
A crew of about 65 workers can raise 2.5 ton stones using simple ropes alone. Over a 20 year period, 25 concurrent crews totalling roughly 1625 workers are sufficient to raise all 2.5 million stones. There are a number of factors that could reduce the number of workers required. Friction could be reduced using available lubricants or particularly strong, well fed workers could have been selected for this critical role.
Building the pyramids seems staggering to us today, but that may be due more to our short attention span and availability of powerful machines to do the heavy lifting. We don't stop to consider that a large, organized workforce, all pulling together at the same time, can do a lot of work. It's not magic, just dedication and arithmetic.
In the modern day, we expect a return on our investment in a reasonable time, perhaps five or ten years for large public works projects. For the pharoahs, 20 years was a completely acceptable delivery schedule for their investment and exit strategy. To achieve higher rates of return, we build powerful machines that could be operated by a single person. We just don't accept slow progress over a long period of time because our expectations and labor costs are so high. The pharoahs on the other hand, were in the opposite position. They had a large workforce that was willing dedicate themselves to a single cause over a significant part of their lifetime. This dedication is perhaps the real achievement we should admire.
Copyright (c) Madhu Siddalingaiah 2020
|
github_jupyter
|
# Time series analysis and visualization
```
# Hide all warnings
import warnings
warnings.simplefilter('ignore')
import numpy as np
import pandas as pd
%matplotlib inline
import matplotlib.pyplot as plt
import statsmodels as sm
import statsmodels.api
from tqdm import tqdm
from pylab import rcParams # Run-Control (default) parameters
rcParams['figure.figsize'] = 16, 8
rcParams['lines.linewidth'] = 4
rcParams['font.size'] = 26
```
<br>
## Time series analysis is for
* compact **dynamics description** of observable processes
* interpretation of dynamics and **estimation of impulse response**
* **forecasting** and simulation
* solution **optimal control** problems
<br>
## The objective of time series analysis
Construct a model of time series for _current value_ of **endogeneous** variable $y_t$
* by the _history_ of itself $$y_{:t} = (y_{t-1}, y_{t-2}, \ldots)$$
* by _current value_ of **exogeneous** variables $x_t$ and possibly by its _history_ too
$$
y_t \approx \text{model}\bigl( t,\, y_{:t},\, x_t,\, x_{:t} \bigr)
\,. $$
Usually one forecasts a single time step ahead.
<br>
## Difference from other Machine Learning tasks
* Data are sequential
* order of **time** has to be respected strictly due to not break the causality
* Much attention to **extrapolation** — a forecast of future values related to observed sample
* It is important to be sure that data do not leak from future to current and to past observations of train subsample during feature engineering and training the model
Thus features of the model can depend only on
* **endogeneous** variables $y_{t-1}, y_{t-2}, \ldots$, i.e. they are available to the moment $t-1$ _inclusively_
* **exogeneous** variables $x_t, x_{t-1}, \ldots$, i.e. they are available to the moment $t$ _inclusively_
<br>
## $CO_2$ concentration in atmosphere [dataset](https://www.co2.earth/weekly-co2)
```
dataset = pd.read_csv('./mauna_loa_atmospheric_c02.csv',
index_col=None, usecols=['date', 'WMLCO2'])
dataset.head()
```
When you loads a time series within `Pandas` you have to set format of date and time explicitly
```
dataset['date'] = pd.to_datetime(dataset['date'], format='%Y-%m-%d')
```
Create the index for loaded data: it will be **weekly periodical index**. We will get data with regular frequency.
```
dataset = dataset.set_index('date').to_period('W')
dataset.head()
```
Plot dynamics of the time series
```
dataset.plot()
plt.grid(which='major', axis='both')
```
Aggregate weekly data to monthly
```
dataset = dataset.to_timestamp()
dataset = dataset.resample('M').mean()
dataset.head()
dataset.plot()
plt.grid(which='major', axis='both')
```
Create summary statistics
```
print('Series {1}, Observations {0}'.format(*dataset.shape))
dataset.describe().T.head()
dataset.loc['1960':'1967'].plot()
plt.grid(which='major', axis='both')
```
### Missed values
```
maginfy_slice = slice('1960', '1967')
```
Missed values can be filled by
1) last known observable
* **+** doesn't look through the future
* **-** can't fill the beginning of the series
* **-** doesn't account specificity of the series
```
dataset_ff = dataset.fillna(method='ffill')
dataset_ff.loc[maginfy_slice].plot()
plt.grid(which='major', axis='both')
```
2) iterpolation of the neighboring values
* **+** smooth peaks
* **-** doesn't fill the ends of the series
* **-** slightly look through the future
```
dataset_linterp = dataset.interpolate(method='linear')
dataset_pinterp = dataset.interpolate(method='polynomial', order=2)
ax = dataset_pinterp.loc[maginfy_slice].plot()
dataset_linterp.loc[maginfy_slice].plot(ax=ax, linewidth=4, alpha=0.7)
plt.grid(which='major', axis='both')
```
3) exlude at all
* **+** doesn't change the values
* **-** break the regularity and related periodicity
* **-** deplete the sampling
```
dataset_drop = dataset.dropna()
dataset_drop.loc[maginfy_slice].plot()
plt.grid(which='major', axis='both')
```
4) estimate by probabilty model
* **+** filling based on extracted patterns (learned dependencies)
* **-** it is needed to specify the model and to train it
5) smooth by splines or by local kernel model
* **+** explicitly accounts close in time observations
* **+** allows to increase the frequency of observations ("_resolution_")
* **+** allows to fill missed boundary values
* **-** look through the future far
* **-** it is needed to define th kernel and the model for extrapolation
Looking into the future can be ignorred if **missed values are minority**.
But if missed values are majority then it is needed to understand why it is happened in the sampling.
```
full_dataset = dataset_pinterp
```
Prepare train and test samplings in the ratio 3 to 1
```
holdout = full_dataset.loc['1991-01-01':]
dataset = full_dataset.loc[:'1990-12-31']
print(len(dataset), len(holdout))
```
Make sure the parts don't intersect
```
pd.concat([
dataset.tail(),
holdout.head()
], axis=1)
```
Store the bounds of the intervals explicitly
```
holdout_slice = slice(*holdout.index[[0, -1]])
print('Train sample from {} to {}'.format(*dataset.index[[0, -1]]))
print('Test sample from {} to {}'.format(holdout_slice.start, holdout_slice.stop))
```
Select the column of target variable
```
target_column = 'WMLCO2'
fig = plt.figure()
ax = fig.add_subplot(111, xlabel='Date', ylabel='value', title=target_column) # 111 means 1 row 1 column 1st axes on the "grid"
# plot dynamics of entire time series
full_dataset[target_column].plot(ax=ax)
# highlight delayed interval for testing
ax.axvspan(holdout_slice.start, holdout_slice.stop,
color='C1', alpha=0.25, zorder=-99)
ax.grid(which='major', axis='both');
```
<br>
# A property
**Stationarity** is a property of a process $\{y_t\}_{t\geq0}$ meaning
> probabilistic interconnections in the set $(y_{t_1},\,\ldots,\,y_{t_m})$ are invariant with respect to shift $s \neq 0$.
That means
* **there are no special moments** in the time when statistical properties of observables are changing
* patterns are stable in time and are determined by **indentation of observables** relative to each other:
* mean, dispersion, and autocorrelation doesn't depend on moment of time
## A ghost property
Stochastic processes in real problems are **almost always non-stationary**
* mean depends on time (there is a trend in the dynamics)
* calendar events (holidays or vacations)
* season periodicity
* daily rhythm of power grid load
* season temperature
* yearly peak of monthly inflation in the beginning of year
* unpredictable structural drift
* political decisions
* blackouts
* hysteresis
Thus majority of time series especially economic, climatic, and financial are non-stationary.
<br>
# Visualization and diagnosis of non-stationarity
Visualization in time series analysis allows to
* get preliminary picture of correlations
* select reasonable strategy of validation a model
* estimate if there is structural drift
* leaps and gaps
* clusters of intensive oscillations or periods of plateau
* diagnose non-stationarity: trend, seasonality, etc.
### A plot of moving statistics
Moving statistics of a series within window of length $N$ allow to discover changes in time
* **moving average** of time series level
$$
m_t = \frac1{N} \sum_{s=t-N+1}^t y_s
$$
* **moving standard deviation** (scatter)
$$
s_t = \sqrt{s^2_t}
\,, \quad
s^2_t = \frac1{N-1} \sum_{s=t-N+1}^t (y_s - m_t)^2
$$
```
rcParams['figure.figsize'] = 16, 10
def rolling_diagnostics(series, window=500):
rolling = series.rolling(window)
# Create top and bottom plots
fig = plt.figure()
ax_top = fig.add_subplot(211, title='Moving average', xlabel='Date', ylabel='value')
ax_bottom = fig.add_subplot(212, title='Moving standard deviation',
sharex=ax_top, xlabel='Date', ylabel='std.')
# Plot the graphs
# series itself and moving average
rolling.mean().plot(ax=ax_top)
series.plot(ax=ax_top, color='black', lw=2, alpha=.25, zorder=-10)
ax_top.grid(which='major', axis='both')
# moving std.
rolling.std().plot(ax=ax_bottom)
ax_bottom.grid(which='major', axis='both')
fig.tight_layout()
return fig
rolling_diagnostics(dataset[target_column], window=36);
```
The graphs show the trend in the dynamics of time series
<br>
### Rough estimate of seasonality
It is disarable to make season normalization relatively to trend.
Let's discover seasonality, for example monthly
```
def monthly_seasonality_diagnostics(series, fraction=0.66, period='month'):
# Use non-parametric local linear regression to preliminary estimate the trend
trend = sm.api.nonparametric.lowess(series, np.r_[:len(series)],
frac=fraction, it=5)
# Aggregate by months and calculate average and standard deviation
by = getattr(series.index, period, 'month')
season_groupby = (series - trend[:, 1]).groupby(by)
seas_mean, seas_std = season_groupby.mean(), season_groupby.std()
# Create subplots
fig = plt.figure()
ax_top = fig.add_subplot(211, title='Trend', xlabel='Date')
ax_bottom = fig.add_subplot(212, title='Seasonality', xlabel=period)
# Plot the graphs
# The series and the trend
pd.Series(trend[:, 1], index=series.index).plot(ax=ax_top)
series.plot(ax=ax_top, color="black", lw=2, alpha=.25, zorder=-10)
ax_top.grid(which="major", axis="both")
# Seasonality and 90% normal confidence interval
ax_bottom.plot(1 + np.r_[:len(seas_mean)], seas_mean, lw=2)
ax_bottom.fill_between(1 + np.r_[:len(seas_mean)],
seas_mean - 1.96 * seas_std,
seas_mean + 1.96 * seas_std,
zorder=-10, color="C1", alpha=0.15)
ax_bottom.grid(which="major", axis="both")
fig.tight_layout()
return fig
monthly_seasonality_diagnostics(dataset[target_column], fraction=0.33, period='month');
```
The graph shows the **monthly** seasonality in the dynamics
```
## TODO: check visually if there is weekly seasonality
```
<br>
### Total vs. partial autocorrelations
The functions estimate influence of observation of $h$ steps (_lags_) on the current observation, but they does it differently
* **total autocorrelation** $\rho_h$
* shows cumulative impact $y_{t-h}$ to $y_t$ **via** influence on all intermediate $y_{t-j}$, $j=1,\,...,\,h-1$
* **partial autocorrelation** $\phi_h$
* shows **net** (pure) impract $y_{t-h}$ to $y_t$ **excluding** influence on all intermediate $y_{t-j}$, $j=1,\,...,\,h-1$
```
from statsmodels.tsa.stattools import acf, pacf
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
def correlation_diagnostics(series, lags=60):
fig = plt.figure(figsize=(20, 6))
ax_left, ax_right = fig.subplots(
nrows=1, ncols=2, sharey=True, sharex=True,
subplot_kw={'xlabel': 'lag', 'ylim': (-1.1, 1.1)})
# Use intrinsic statsmodels functions
plot_acf(series, ax_left, lags=lags, zero=False, alpha=0.05,
title='Sample autocorrelation', marker=None)
plot_pacf(series, ax_right, lags=lags, zero=False, alpha=0.05,
title='Sample partial autocorrelation', marker=None)
fig.tight_layout()
return fig
```
Let's explore sample autocorrelations of the series
```
correlation_diagnostics(dataset[target_column], lags=250);
```
* On the **left plot** autocorrelation of small lags is near to $1.0$ and decreases pretty slowly
* On the **right plot** observations with lag $1$, $110$, $215$ has statistically non-null net effect
It is indication of very typical kind of non-stationarity: $y_t = y_{t-1} + \ldots$.
That means it is observed strong dependance of the past (the history of a process).
---
# Key steps of model construction for time series
* Stationarize a time series
* Estimate parameter of the model
* Visualize remains after stationarization
* check if respect the model requirements
* Validation of the model
|
github_jupyter
|
<center>
<img src="../../img/ods_stickers.jpg">
## Открытый курс по машинному обучению
<center>Автор материала: Ефремова Дина (@ldinka).
# <center>Исследование возможностей BigARTM</center>
## <center>Тематическое моделирование с помощью BigARTM</center>
#### Интро
BigARTM — библиотека, предназначенная для тематической категоризации текстов; делает разбиение на темы без «учителя».
Я собираюсь использовать эту библиотеку для собственных нужд в будущем, но так как она не предназначена для обучения с учителем, решила, что для начала ее стоит протестировать на какой-нибудь уже размеченной выборке. Для этих целей был использован датасет "20 news groups".
Идея экперимента такова:
- делим выборку на обучающую и тестовую;
- обучаем модель на обучающей выборке;
- «подгоняем» выделенные темы под действительные;
- смотрим, насколько хорошо прошло разбиение;
- тестируем модель на тестовой выборке.
#### Поехали!
**Внимание!** Данный проект был реализован с помощью Python 3.6 и BigARTM 0.9.0. Методы, рассмотренные здесь, могут отличаться от методов в других версиях библиотеки.
<img src="../../img/bigartm_logo.png"/>
### <font color="lightgrey">Не</font>множко теории
У нас есть словарь терминов $W = \{w \in W\}$, который представляет из себя мешок слов, биграмм или n-грамм;
Есть коллекция документов $D = \{d \in D\}$, где $d \subset W$;
Есть известное множество тем $T = \{t \in T\}$;
$n_{dw}$ — сколько раз термин $w$ встретился в документе $d$;
$n_{d}$ — длина документа $d$.
Мы считаем, что существует матрица $\Phi$ распределения терминов $w$ в темах $t$: (фи) $\Phi = (\phi_{wt})$
и матрица распределения тем $t$ в документах $d$: (тета) $\Theta = (\theta_{td})$,
переумножение которых дает нам тематическую модель, или, другими словами, представление наблюдаемого условного распределения $p(w|d)$ терминов $w$ в документах $d$ коллекции $D$:
<center>$\large p(w|d) = \Phi \Theta$</center>
<center>$$\large p(w|d) = \sum_{t \in T} \phi_{wt} \theta_{td}$$</center>
где $\phi_{wt} = p(w|t)$ — вероятности терминов $w$ в каждой теме $t$
и $\theta_{td} = p(t|d)$ — вероятности тем $t$ в каждом документе $d$.
<img src="../../img/phi_theta.png"/>
Нам известны наблюдаемые частоты терминов в документах, это:
<center>$ \large \hat{p}(w|d) = \frac {n_{dw}} {n_{d}} $</center>
Таким образом, наша задача тематического моделирования становится задачей стохастического матричного разложения матрицы $\hat{p}(w|d)$ на стохастические матрицы $\Phi$ и $\Theta$.
Напомню, что матрица является стохастической, если каждый ее столбец представляет дискретное распределение вероятностей, сумма значений каждого столбца равна 1.
Воспользовавшись принципом максимального правдоподобия, т. е. максимизируя логарифм правдоподобия, мы получим:
<center>$
\begin{cases}
\sum_{d \in D} \sum_{w \in d} n_{dw} \ln \sum_{t \in T} \phi_{wt} \theta_{td} \rightarrow \max\limits_{\Phi,\Theta};\\
\sum_{w \in W} \phi_{wt} = 1, \qquad \phi_{wt}\geq0;\\
\sum_{t \in T} \theta_{td} = 1, \quad\quad\;\; \theta_{td}\geq0.
\end{cases}
$</center>
Чтобы из множества решений выбрать наиболее подходящее, введем критерий регуляризации $R(\Phi, \Theta)$:
<center>$
\begin{cases}
\sum_{d \in D} \sum_{w \in d} n_{dw} \ln \sum_{t \in T} \phi_{wt} \theta_{td} + R(\Phi, \Theta) \rightarrow \max\limits_{\Phi,\Theta};\\
\sum_{w \in W} \phi_{wt} = 1, \qquad \phi_{wt}\geq0;\\
\sum_{t \in T} \theta_{td} = 1, \quad\quad\;\; \theta_{td}\geq0.
\end{cases}
$</center>
Два наиболее известных частных случая этой системы уравнений:
- **PLSA**, вероятностный латентный семантический анализ, когда $R(\Phi, \Theta) = 0$
- **LDA**, латентное размещение Дирихле:
$$R(\Phi, \Theta) = \sum_{t,w} (\beta_{w} - 1) \ln \phi_{wt} + \sum_{d,t} (\alpha_{t} - 1) \ln \theta_{td} $$
где $\beta_{w} > 0$, $\alpha_{t} > 0$ — параметры регуляризатора.
Однако оказывается запас неединственности решения настолько большой, что на модель можно накладывать сразу несколько ограничений, такой подход называется **ARTM**, или аддитивной регуляризацией тематических моделей:
<center>$
\begin{cases}
\sum_{d,w} n_{dw} \ln \sum_{t} \phi_{wt} \theta_{td} + \sum_{i=1}^k \tau_{i} R_{i}(\Phi, \Theta) \rightarrow \max\limits_{\Phi,\Theta};\\
\sum_{w \in W} \phi_{wt} = 1, \qquad \phi_{wt}\geq0;\\
\sum_{t \in T} \theta_{td} = 1, \quad\quad\;\; \theta_{td}\geq0.
\end{cases}
$</center>
где $\tau_{i}$ — коэффициенты регуляризации.
Теперь давайте познакомимся с библиотекой BigARTM и разберем еще некоторые аспекты тематического моделирования на ходу.
Если Вас очень сильно заинтересовала теоретическая часть категоризации текстов и тематического моделирования, рекомендую посмотреть видеолекции из курса Яндекса на Coursera «Поиск структуры в данных» четвертой недели: <a href="https://www.coursera.org/learn/unsupervised-learning/home/week/4">Тематическое моделирование</a>.
### BigARTM
#### Установка
Естественно, для начала работы с библиотекой ее надо установить. Вот несколько видео, которые рассказывают, как это сделать в зависимости от вашей операционной системы:
- <a href="https://www.coursera.org/learn/unsupervised-learning/lecture/qmsFm/ustanovka-bigartm-v-windows">Установка BigARTM в Windows</a>
- <a href="https://www.coursera.org/learn/unsupervised-learning/lecture/zPyO0/ustanovka-bigartm-v-linux-mint">Установка BigARTM в Linux</a>
- <a href="https://www.coursera.org/learn/unsupervised-learning/lecture/nuIhL/ustanovka-bigartm-v-mac-os-x">Установка BigARTM в Mac OS X</a>
Либо можно воспользоваться инструкцией с официального сайта, которая, скорее всего, будет гораздо актуальнее: <a href="https://bigartm.readthedocs.io/en/stable/installation/index.html">здесь</a>. Там же указано, как можно установить BigARTM в качестве <a href="https://bigartm.readthedocs.io/en/stable/installation/docker.html">Docker-контейнера</a>.
#### Использование BigARTM
```
import artm
import re
import numpy as np
import seaborn as sns; sns.set()
from sklearn.metrics import classification_report, confusion_matrix
from sklearn.preprocessing import normalize
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from matplotlib import pyplot as plt
%matplotlib inline
artm.version()
```
Скачаем датасет ***the 20 news groups*** с заранее известным количеством категорий новостей:
```
from sklearn.datasets import fetch_20newsgroups
newsgroups = fetch_20newsgroups('../../data/news_data')
newsgroups['target_names']
```
Приведем данные к формату *Vowpal Wabbit*. Так как BigARTM не рассчитан на обучение с учителем, то мы поступим следующим образом:
- обучим модель на всем корпусе текстов;
- выделим ключевые слова тем и по ним определим, к какой теме они скорее всего относятся;
- сравним наши полученные результаты разбиения с истинными значенями.
```
TEXT_FIELD = "text"
def to_vw_format(document, label=None):
return str(label or '0') + ' |' + TEXT_FIELD + ' ' + ' '.join(re.findall('\w{3,}', document.lower())) + '\n'
all_documents = newsgroups['data']
all_targets = newsgroups['target']
len(newsgroups['target'])
train_documents, test_documents, train_labels, test_labels = \
train_test_split(all_documents, all_targets, random_state=7)
with open('../../data/news_data/20news_train_mult.vw', 'w') as vw_train_data:
for text, target in zip(train_documents, train_labels):
vw_train_data.write(to_vw_format(text, target))
with open('../../data/news_data/20news_test_mult.vw', 'w') as vw_test_data:
for text in test_documents:
vw_test_data.write(to_vw_format(text))
```
Загрузим данные в необходимый для BigARTM формат:
```
batch_vectorizer = artm.BatchVectorizer(data_path="../../data/news_data/20news_train_mult.vw",
data_format="vowpal_wabbit",
target_folder="news_batches")
```
Данные в BigARTM загружаются порционно, укажем в
- *data_path* путь к обучающей выборке,
- *data_format* — формат наших данных, может быть:
* *bow_n_wd* — это вектор $n_{wd}$ в виду массива *numpy.ndarray*, также необходимо передать соответствующий словарь терминов, где ключ — это индекс вектора *numpy.ndarray* $n_{wd}$, а значение — соответствующий токен.
```python
batch_vectorizer = artm.BatchVectorizer(data_format='bow_n_wd',
n_wd=n_wd,
vocabulary=vocabulary)
```
* *vowpal_wabbit* — формат Vowpal Wabbit;
* *bow_uci* — UCI формат (например, с *vocab.my_collection.txt* и *docword.my_collection.txt* файлами):
```python
batch_vectorizer = artm.BatchVectorizer(data_path='',
data_format='bow_uci',
collection_name='my_collection',
target_folder='my_collection_batches')
```
* *batches* — данные, уже сконверченные в батчи с помощью BigARTM;
- *target_folder* — путь для сохранения батчей.
Пока это все параметры, что нам нужны для загрузки наших данных.
После того, как BigARTM создал батчи из данных, можно использовать их для загрузки:
```
batch_vectorizer = artm.BatchVectorizer(data_path="news_batches", data_format='batches')
```
Инициируем модель с известным нам количеством тем. Количество тем — это гиперпараметр, поэтому если он заранее нам неизвестен, то его необходимо настраивать, т. е. брать такое количество тем, при котором разбиение кажется наиболее удачным.
**Важно!** У нас 20 предметных тем, однако некоторые из них довольно узкоспециализированны и смежны, как например 'comp.os.ms-windows.misc' и 'comp.windows.x', или 'comp.sys.ibm.pc.hardware' и 'comp.sys.mac.hardware', тогда как другие размыты и всеобъемлющи: talk.politics.misc' и 'talk.religion.misc'.
Скорее всего, нам не удастся в чистом виде выделить все 20 тем — некоторые из них окажутся слитными, а другие наоборот раздробятся на более мелкие. Поэтому мы попробуем построить 40 «предметных» тем и одну фоновую. Чем больше вы будем строить категорий, тем лучше мы сможем подстроиться под данные, однако это довольно трудоемкое занятие сидеть потом и распределять в получившиеся темы по реальным категориям (<strike>я правда очень-очень задолбалась!</strike>).
Зачем нужны фоновые темы? Дело в том, что наличие общей лексики в темах приводит к плохой ее интерпретируемости. Выделив общую лексику в отдельную тему, мы сильно снизим ее количество в предметных темах, таким образом оставив там лексическое ядро, т. е. ключевые слова, которые данную тему характеризуют. Также этим преобразованием мы снизим коррелированность тем, они станут более независимыми и различимыми.
```
T = 41
model_artm = artm.ARTM(num_topics=T,
topic_names=[str(i) for i in range(T)],
class_ids={TEXT_FIELD:1},
num_document_passes=1,
reuse_theta=True,
cache_theta=True,
seed=4)
```
Передаем в модель следующие параметры:
- *num_topics* — количество тем;
- *topic_names* — названия тем;
- *class_ids* — название модальности и ее вес. Дело в том, что кроме самих текстов, в данных может содержаться такая информация, как автор, изображения, ссылки на другие документы и т. д., по которым также можно обучать модель;
- *num_document_passes* — количество проходов при обучении модели;
- *reuse_theta* — переиспользовать ли матрицу $\Theta$ с предыдущей итерации;
- *cache_theta* — сохранить ли матрицу $\Theta$ в модели, чтобы в дальнейшем ее использовать.
Далее необходимо создать словарь; передадим ему какое-нибудь название, которое будем использовать в будущем для работы с этим словарем.
```
DICTIONARY_NAME = 'dictionary'
dictionary = artm.Dictionary(DICTIONARY_NAME)
dictionary.gather(batch_vectorizer.data_path)
```
Инициализируем модель с тем именем словаря, что мы передали выше, можно зафиксировать *random seed* для вопроизводимости результатов:
```
np.random.seed(1)
model_artm.initialize(DICTIONARY_NAME)
```
Добавим к модели несколько метрик:
- перплексию (*PerplexityScore*), чтобы индентифицировать сходимость модели
* Перплексия — это известная в вычислительной лингвистике мера качества модели языка. Можно сказать, что это мера неопределенности или различности слов в тексте.
- специальный *score* ключевых слов (*TopTokensScore*), чтобы в дальнейшем мы могли идентифицировать по ним наши тематики;
- разреженность матрицы $\Phi$ (*SparsityPhiScore*);
- разреженность матрицы $\Theta$ (*SparsityThetaScore*).
```
model_artm.scores.add(artm.PerplexityScore(name='perplexity_score',
dictionary=DICTIONARY_NAME))
model_artm.scores.add(artm.SparsityPhiScore(name='sparsity_phi_score', class_id="text"))
model_artm.scores.add(artm.SparsityThetaScore(name='sparsity_theta_score'))
model_artm.scores.add(artm.TopTokensScore(name="top_words", num_tokens=15, class_id=TEXT_FIELD))
```
Следующая операция *fit_offline* займет некоторое время, мы будем обучать модель в режиме *offline* в 40 проходов. Количество проходов влияет на сходимость модели: чем их больше, тем лучше сходится модель.
```
%%time
model_artm.fit_offline(batch_vectorizer=batch_vectorizer, num_collection_passes=40)
```
Построим график сходимости модели и увидим, что модель сходится довольно быстро:
```
plt.plot(model_artm.score_tracker["perplexity_score"].value);
```
Выведем значения разреженности матриц:
```
print('Phi', model_artm.score_tracker["sparsity_phi_score"].last_value)
print('Theta', model_artm.score_tracker["sparsity_theta_score"].last_value)
```
После того, как модель сошлась, добавим к ней регуляризаторы. Для начала сглаживающий регуляризатор — это *SmoothSparsePhiRegularizer* с большим положительным коэффициентом $\tau$, который нужно применить только к фоновой теме, чтобы выделить в нее как можно больше общей лексики. Пусть тема с последним индексом будет фоновой, передадим в *topic_names* этот индекс:
```
model_artm.regularizers.add(artm.SmoothSparsePhiRegularizer(name='SparsePhi',
tau=1e5,
dictionary=dictionary,
class_ids=TEXT_FIELD,
topic_names=str(T-1)))
```
Дообучим модель, сделав 20 проходов по ней с новым регуляризатором:
```
%%time
model_artm.fit_offline(batch_vectorizer=batch_vectorizer, num_collection_passes=20)
```
Выведем значения разреженности матриц, заметим, что значение для $\Theta$ немного увеличилось:
```
print('Phi', model_artm.score_tracker["sparsity_phi_score"].last_value)
print('Theta', model_artm.score_tracker["sparsity_theta_score"].last_value)
```
Теперь добавим к модели разреживающий регуляризатор, это тот же *SmoothSparsePhiRegularizer* резуляризатор, только с отрицательным значением $\tau$ и примененный ко всем предметным темам:
```
model_artm.regularizers.add(artm.SmoothSparsePhiRegularizer(name='SparsePhi2',
tau=-5e5,
dictionary=dictionary,
class_ids=TEXT_FIELD,
topic_names=[str(i) for i in range(T-1)]),
overwrite=True)
%%time
model_artm.fit_offline(batch_vectorizer=batch_vectorizer, num_collection_passes=20)
```
Видим, что значения разреженности увеличились еще больше:
```
print(model_artm.score_tracker["sparsity_phi_score"].last_value)
print(model_artm.score_tracker["sparsity_theta_score"].last_value)
```
Посмотрим, сколько категорий-строк матрицы $\Theta$ после регуляризации осталось, т. е. не занулилось/выродилось. И это одна категория:
```
len(model_artm.score_tracker["top_words"].last_tokens.keys())
```
Теперь выведем ключевые слова тем, чтобы определить, каким образом прошло разбиение, и сделать соответствие с нашим начальным списком тем:
```
for topic_name in model_artm.score_tracker["top_words"].last_tokens.keys():
tokens = model_artm.score_tracker["top_words"].last_tokens
res_str = topic_name + ': ' + ', '.join(tokens[topic_name])
print(res_str)
```
Далее мы будем подгонять разбиение под действительные темы с помощью *confusion matrix*.
```
target_dict = {
'alt.atheism': 0,
'comp.graphics': 1,
'comp.os.ms-windows.misc': 2,
'comp.sys.ibm.pc.hardware': 3,
'comp.sys.mac.hardware': 4,
'comp.windows.x': 5,
'misc.forsale': 6,
'rec.autos': 7,
'rec.motorcycles': 8,
'rec.sport.baseball': 9,
'rec.sport.hockey': 10,
'sci.crypt': 11,
'sci.electronics': 12,
'sci.med': 13,
'sci.space': 14,
'soc.religion.christian': 15,
'talk.politics.guns': 16,
'talk.politics.mideast': 17,
'talk.politics.misc': 18,
'talk.religion.misc': 19
}
mixed = [
'comp.sys.ibm.pc.hardware',
'talk.politics.mideast',
'sci.electronics',
'rec.sport.hockey',
'sci.med',
'rec.motorcycles',
'comp.graphics',
'rec.sport.hockey',
'talk.politics.mideast',
'talk.religion.misc',
'rec.autos',
'comp.graphics',
'sci.space',
'soc.religion.christian',
'comp.os.ms-windows.misc',
'sci.crypt',
'comp.windows.x',
'misc.forsale',
'sci.space',
'sci.crypt',
'talk.religion.misc',
'alt.atheism',
'comp.os.ms-windows.misc',
'alt.atheism',
'sci.med',
'comp.os.ms-windows.misc',
'soc.religion.christian',
'talk.politics.guns',
'rec.autos',
'rec.autos',
'talk.politics.mideast',
'rec.sport.baseball',
'talk.religion.misc',
'talk.politics.misc',
'rec.sport.hockey',
'comp.sys.mac.hardware',
'misc.forsale',
'sci.space',
'talk.politics.guns',
'rec.autos',
'-'
]
```
Построим небольшой отчет о правильности нашего разбиения:
```
theta_train = model_artm.get_theta()
model_labels = []
keys = np.sort([int(i) for i in theta_train.keys()])
for i in keys:
max_val = 0
max_idx = 0
for j in theta_train[i].keys():
if j == str(T-1):
continue
if theta_train[i][j] > max_val:
max_val = theta_train[i][j]
max_idx = j
topic = mixed[int(max_idx)]
if topic == '-':
print(i, '-')
label = target_dict[topic]
model_labels.append(label)
print(classification_report(train_labels, model_labels))
print(classification_report(train_labels, model_labels))
mat = confusion_matrix(train_labels, model_labels)
sns.heatmap(mat.T, annot=True, fmt='d', cbar=False)
plt.xlabel('True label')
plt.ylabel('Predicted label');
accuracy_score(train_labels, model_labels)
```
Нам удалось добиться 80% *accuracy*. По матрице ответов мы видим, что для модели темы *comp.sys.ibm.pc.hardware* и *comp.sys.mac.hardware* практически не различимы (<strike>честно говоря, для меня тоже</strike>), в остальном все более или менее прилично.
Проверим модель на тестовой выборке:
```
batch_vectorizer_test = artm.BatchVectorizer(data_path="../../data/news_data/20news_test_mult.vw",
data_format="vowpal_wabbit",
target_folder="news_batches_test")
theta_test = model_artm.transform(batch_vectorizer_test)
test_score = []
for i in range(len(theta_test.keys())):
max_val = 0
max_idx = 0
for j in theta_test[i].keys():
if j == str(T-1):
continue
if theta_test[i][j] > max_val:
max_val = theta_test[i][j]
max_idx = j
topic = mixed[int(max_idx)]
label = target_dict[topic]
test_score.append(label)
print(classification_report(test_labels, test_score))
mat = confusion_matrix(test_labels, test_score)
sns.heatmap(mat.T, annot=True, fmt='d', cbar=False)
plt.xlabel('True label')
plt.ylabel('Predicted label');
accuracy_score(test_labels, test_score)
```
Итого почти 77%, незначительно хуже, чем на обучающей.
**Вывод:** безумно много времени пришлось потратить на подгонку категорий к реальным темам, но в итоге я осталась довольна результатом. Такие смежные темы, как *alt.atheism*/*soc.religion.christian*/*talk.religion.misc* или *talk.politics.guns*/*talk.politics.mideast*/*talk.politics.misc* разделились вполне неплохо. Думаю, что я все-таки попробую использовать BigARTM в будущем для своих <strike>корыстных</strike> целей.
|
github_jupyter
|
# SIR-X
This notebook exemplifies how Open-SIR can be used to fit the SIR-X model by [Maier and Dirk (2020)](https://science.sciencemag.org/content/early/2020/04/07/science.abb4557.full) to existing data and make predictions. The SIR-X model is a standard generalization of the Susceptible-Infectious-Removed (SIR) model, which includes the influence of exogenous factors such as policy changes, lockdown of the whole population and quarantine of the infectious individuals.
The Open-SIR implementation of the SIR-X model will be validated reproducing the parameter fitting published in the [supplementary material](https://science.sciencemag.org/cgi/content/full/science.abb4557/DC1) of the original article published by [Maier and Brockmann (2020)](https://science.sciencemag.org/content/early/2020/04/07/science.abb4557.full). For simplicity, the validation will be performed only for the city of Guangdong, China.
## Import modules
```
# Uncomment this cell to activate black code formatter in the notebook
# %load_ext nb_black
# Import packages
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
```
## Data sourcing
We will source data from the repository of the [John Hopkins University COVID-19 dashboard] (https://coronavirus.jhu.edu/map.html) published formally as a correspondence in [The Lancet](https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(20)30120-1/fulltext#seccestitle10). This time series data contains the number of reported cases $C(t)$ per day for a number of cities.
```
# Source data from John Hokpins university reposotiry
# jhu_link = "https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/who_covid_19_situation_reports/who_covid_19_sit_rep_time_series/who_covid_19_sit_rep_time_series.csv"
jhu_link = "https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv"
jhu_df = pd.read_csv(jhu_link)
# Explore the dataset
jhu_df.head(10)
```
It is observed that the column "Province/States" contains the name of the cities, and since the forth column a time series stamp (or index) is provided to record daily data of reported cases. Additionally, there are many days without recorded data for a number of chinese cities. This won't be an issue for parameter fitting as Open-SIR doesn't require uniform spacement of the observed data.
### Data preparation
In the following lines, the time series for Guangdong reported cases $C(t)$ is extracted from the original dataframe. Thereafter, the columns are converted to a pandas date time index in order to perform further data preparation steps.
```
China = jhu_df[jhu_df[jhu_df.columns[1]] == "China"]
city_name = "Guangdong"
city = China[China["Province/State"] == city_name]
city = city.drop(columns=["Province/State", "Country/Region", "Lat", "Long"])
time_index = pd.to_datetime(city.columns)
data = city.values
# Visualize the time
ts = pd.Series(data=city.values[0], index=time_index)
```
Using the function ts.plot() a quick visualization of the dataset is obtained:
```
ts.plot()
plt.title("Guangdong COVID-19 cases")
plt.ylabel("$C(t)$: Number of reported cases", size=12)
plt.show()
```
Data cleaning
```
ts_clean = ts.dropna()
# Extract data
ts_fit = ts_clean["2020-01-21":"2020-02-12"]
# Convert index to numeric
ts_num = pd.to_numeric(ts_fit.index)
t0 = ts_num[0]
# Convert datetime to days
t_days = (ts_num - t0) / (10 ** 9 * 86400)
t_days = t_days.astype(int).values
# t_days is an input for SIR
# Define the X number
nX = ts_fit.values # Number of infected
N = 104.3e6 # Population size of Guangdong
```
Exploration of the dataset
```
ts_fit.plot(style="ro")
plt.xlabel("Number of infected")
plt.show()
```
### Setting up SIR and SIR-X models
The population $N$ of the city is a necessary input for the model. In this notebook, this was hardocded, but it can be sourced directly from a web source.
Note that whilst the SIR model estimates directly the number of infected people, $N I(t)$, SIR-X estimates the number of infected people based on the number of tested cases that are in quarantine or in an hospital $N X(t)$
```
# These lines are required only if opensir wasn't installed using pip install, or if opensir is running in the pipenv virtual environment
import sys
path_opensir = "../../"
sys.path.append(path_opensir)
# Import SIR and SIRX models
from opensir.models import SIR, SIRX
nX = ts_fit.values # Number of observed infections of the time series
N = 104.3e6 # Population size of Guangdong
params = [0.95, 0.38]
w0 = (N - nX[0], nX[0], 0)
G_sir = SIR()
G_sir.set_params(p=params, initial_conds=w0)
G_sir.fit_input = 2
G_sir.fit(t_days, nX)
G_sir.solve(t_days[-1], t_days[-1] + 1)
t_SIR = G_sir.fetch()[:, 0]
I_SIR = G_sir.fetch()[:, 2]
```
### Try to fit a SIR model to Guangdong data
```
ax = plt.axes()
ax.tick_params(axis="both", which="major", labelsize=14)
plt.plot(t_SIR, I_SIR)
plt.plot(t_days, nX, "ro")
plt.show()
```
The SIR model is clearly not appropriate to fit this data, as it cannot resolve the effect of exogeneous containment efforts such as quarantines or lockdown. We will repeat the process with a SIR-X model.
### Fit SIR-X to Guangdong Data
```
g_sirx = SIRX()
params = [6.2 / 8, 1 / 8, 0.05, 0.05, 5]
# X_0 can be directly ontained from the statistics
n_x0 = nX[0] # Number of people tested positive
n_i0 = nX[0]
w0 = (N - n_x0 - n_i0, n_i0, 0, n_x0)
g_sirx.set_params(p=params, initial_conds=w0)
# Fit all parameters
fit_index = [False, False, True, True, True]
g_sirx.fit(t_days, nX, fit_index=fit_index)
g_sirx.solve(t_days[-1], t_days[-1] + 1)
t_sirx = g_sirx.fetch()[:, 0]
inf_sirx = g_sirx.fetch()[:, 4]
plt.figure(figsize=[6, 6])
ax = plt.axes()
plt.plot(t_sirx, inf_sirx, "b-", linewidth=2)
plt.plot(t_SIR, I_SIR, "g-", linewidth=2)
plt.plot(t_days, nX, "ro")
plt.legend(
["SIR-X model fit", "SIR model fit", "Number of reported cases"], fontsize=13
)
plt.title("SARS-CoV-2 evolution in Guangdong, China", size=15)
plt.xlabel("Days", fontsize=14)
plt.ylabel("COVID-19 confirmed cases", fontsize=14)
ax.tick_params(axis="both", which="major", labelsize=14)
plt.show()
```
After fitting the parameters, the effective infectious period $T_{I,eff}$ and the effective reproduction rate $R_{0,eff}$ can be obtained from the model properties
$$T_{I,eff} = (\beta + \kappa + \kappa_0)^{-1}$$
$$R_{0,eff} = \alpha T_{I,eff}$$
Aditionally, the Public containment leverage $P$ and the quarantine probability $Q$ can be calculated through:
$$P = \frac{\kappa_0}{\kappa_0 + \kappa}$$
$$Q = \frac{\kappa_0 + \kappa}{\beta + \kappa_0 + \kappa}$$
```
print("Effective infectious period T_I_eff = %.2f days " % g_sirx.t_inf_eff)
print(
"Effective reproduction rate R_0_eff = %.2f, Maier and Brockmann = %.2f"
% (g_sirx.r0_eff, 3.02)
)
print(
"Public containment leverage = %.2f, Maier and Brockmann = %.2f"
% (g_sirx.pcl, 0.75)
)
print(
"Quarantine probability = %.2f, Maier and Brockmann = %.2f" % (g_sirx.q_prob, 0.51)
)
```
### Make predictions using `model.predict`
```
# Make predictions and visualize
# Obtain the results 14 days after the train data ends
sirx_pred = g_sirx.predict(14)
print("T n_S \t n_I \tn_R \tn_X")
for i in sirx_pred:
print(*i.astype(int))
```
Prepare date time index to plot predictions
```
# Import datetime module from the standard library
import datetime
# Obtain the last day from the data used to train the model
last_time = ts_fit.index[-1]
# Create a date time range based on the number of rows of the prediction
numdays = sirx_pred.shape[0]
day_zero = datetime.datetime(last_time.year, last_time.month, last_time.day)
date_list = [day_zero + datetime.timedelta(days=x) for x in range(numdays)]
```
Plot predictions
```
# Extract figure and axes
fig, ax = plt.subplots(figsize=[5, 5])
# Create core plot attributes
plt.plot(date_list, sirx_pred[:, 4], color="blue", linewidth=2)
plt.title("Prediction of Guangdong Cases", size=14)
plt.ylabel("Number of infected", size=14)
# Remove trailing space
plt.xlim(date_list[0], date_list[-1])
# Limit the amount of data displayed
ax.xaxis.set_major_locator(plt.MaxNLocator(3))
# Increase the size of the ticks
ax.tick_params(labelsize=12)
plt.show()
```
### Calculation of predictive confidence intervals
The confidence intervals on the predictions of the SIR-X model can be calculated using a block cross validation. This technique is widely used in Time Series Analysis. In the open-sir API, the function `model.ci_block_cv` calculates the average mean squared error of the predictions, a list of the rolling mean squared errors and the list of parameters which shows how much each parameter changes taking different number of days for making predictions.
The three first parameters are the same as the fit function, while the last two parameters are the `lags` and the `min_sample`. The `lags` parameter indicates how many periods in the future will be forecasted in order to calculate the mean squared error of the model prediction. The `min_sample` parameter indicates the initial number of observations and days that will be taken to perform the block cross validation.
In the following example, `model.ci_block_cv` is used to estimate the average mean squared error of *1-day* predictions taking *6* observations as the starting point of the cross validation. For Guangdong, a `min_sample=6` higher than the default 3 is required to handle well the missing data. This way, both the data on the four first days, and two days after the data starts again, are considered for cross validation.
```
# Calculate confidence intervals
mse_avg, mse_list, p_list, pred_data = g_sirx.block_cv(lags=1, min_sample=6)
```
If it is assumed that the residuals distribute normally, then a good estimation of a 95% confidence interval on the one-day prediction of the number of confirmed cases is
$$\sigma \sim \mathrm{MSE} \rightarrow n_{X,{t+1}} \sim \hat{n}_{X,{t+1}} \pm 2 \sigma$$
Where $n_{X,{t+1}}$ is the real number of confirmed cases in the next day, and $\hat{n}_{X,{t+1}}$ is the estimation using the SIR-X model using cross validation. We can use the `PredictionResults` instance `pred_data` functionality to explore the mean-squared errors and the predictions confidence intervals:
```
pred_data.print_mse()
```
The predictive accuracy of the model is quite impressive, even for 9-day predictions. Let's take advantage of the relatively low mean squared error to forecast a 10 days horizon with confidence intervals using `pred_data.plot_predictions(n_days=9)`
```
pred_data.plot_pred_ci(n_days=9)
```
If it is assumed that the residuals distribute normally, then a good estimation of a 95% confidence interval on the one-day prediction of the number of confirmed cases is
$$\sigma \sim \mathrm{MSE} \rightarrow n_{X,{t+1}} \sim \hat{n}_{X,{t+1}} \pm 2 \sigma$$
Where $n_{X,{t+1}}$ is the real number of confirmed cases in the next day, and $\hat{n}_{X,{t+1}}$ is the estimation using the SIR-X model using cross validation. We use solve to make a 1-day prediction and append the 95% confidence interval.
```
# Predict
g_sirx.solve(t_days[-1] + 1, t_days[-1] + 2)
n_X_tplusone = g_sirx.fetch()[-1, 4]
print("Estimation of n_X_{t+1} = %.0f +- %.0f " % (n_X_tplusone, 2 * mse_avg[0]))
# Transform parameter list into a DataFrame
par_block_cv = pd.DataFrame(p_list)
# Rename dataframe columns based on SIR-X parameter names
par_block_cv.columns = g_sirx.PARAMS
# Add the day. Note that we take the days from min_sample until the end of the array, as days
# 0,1,2 are used for the first sampling in the block cross-validation
par_block_cv["Day"] = t_days[5:]
# Explore formatted dataframe for parametric analysis
par_block_cv.head(len(p_list))
plt.figure(figsize=[5, 5])
ax = plt.axes()
ax.tick_params(axis="both", which="major", labelsize=14)
plt.plot(mse_list[0], "ro")
plt.xlabel("Number of days used to predict the next day", size=14)
plt.ylabel("MSE", size=14)
plt.show()
```
There is an outlier on day 1, as this is when the missing date starts. A more reliable approach would be to take the last 8 values of the mean squared error to calculate a new average assuming that there will be no more missing data.
#### Variation of fitted parameters
Finally, it is possible to observe how the model parameters change as more days and number of confirmed cases are introduced in the block cross validation.
It is clear to observe that after day 15 all parameters except kappa begin to converge. Therefore, care must be taken when performing inference over the parameter kappa.
### Long term prediction
Now we can use the model to predict when the peak will occur and what will be the maximum number of infected
```
# Predict
plt.figure(figsize=[6, 6])
ax = plt.axes()
ax.tick_params(axis="both", which="major", labelsize=14)
g_sirx.solve(40, 41)
# Plot
plt.plot(g_sirx.fetch()[:, 4], "b-", linewidth=2) # X(t)
plt.plot(g_sirx.fetch()[:, 2], "b--", linewidth=2) # I(t)
plt.xlabel("Day", size=14)
plt.ylabel("Number of people", size=14)
plt.legend(["X(t): Confirmed", "I(t) = Infected"], fontsize=13)
plt.title(city_name)
plt.show()
```
The model was trained with a limited amount of data. It is clear to observe that since the measures took place in Guangdong, at least 6 weeks of quarantine were necessary to control the pandemics. Note that a limitation of this model is that it predicts an equilibrium where the number of infected, denoted by the yellow line in the figure above, is 0 after a short time. In reality, this amount will decrease to a small number.
After the peak of infections is reached, it is necessary to keep the quarantine and effective contact tracing for at least 30 days more.
### Validate long term plot using model.plot()
The function `model.plot()` offers a handy way to visualize model fitting and predictions. Custom visualizations can be validated against the `model.plot()` function.
```
g_sirx.plot()
```
|
github_jupyter
|
# Huggingface SageMaker-SDK - BERT Japanese QA example
1. [Introduction](#Introduction)
2. [Development Environment and Permissions](#Development-Environment-and-Permissions)
1. [Installation](#Installation)
2. [Permissions](#Permissions)
3. [Uploading data to sagemaker_session_bucket](#Uploading-data-to-sagemaker_session_bucket)
3. [(Optional) Deepen your understanding of SQuAD](#(Optional)-Deepen-your-understanding-of-SQuAD)
4. [Fine-tuning & starting Sagemaker Training Job](#Fine-tuning-\&-starting-Sagemaker-Training-Job)
1. [Creating an Estimator and start a training job](#Creating-an-Estimator-and-start-a-training-job)
2. [Estimator Parameters](#Estimator-Parameters)
3. [Download fine-tuned model from s3](#Download-fine-tuned-model-from-s3)
4. [Question Answering on Local](#Question-Answering-on-Local)
5. [_Coming soon_:Push model to the Hugging Face hub](#Push-model-to-the-Hugging-Face-hub)
# Introduction
このnotebookはHuggingFaceの[run_squad.py](https://github.com/huggingface/transformers/blob/master/examples/legacy/question-answering/run_squad.py)を日本語データで動作する様に変更を加えたものです。
データは[運転ドメインQAデータセット](https://nlp.ist.i.kyoto-u.ac.jp/index.php?Driving%20domain%20QA%20datasets)を使用します。
このデモでは、AmazonSageMakerのHuggingFace Estimatorを使用してSageMakerのトレーニングジョブを実行します。
_**NOTE: このデモは、SagemakerNotebookインスタンスで動作検証しています**_
_**データセットは各自許諾に同意の上ダウンロードしていただけますようお願いいたします(データサイズは約4MBです)**_
# Development Environment and Permissions
## Installation
このNotebookはSageMakerの`conda_pytorch_p36`カーネルを利用しています。
日本語処理のため、`transformers`ではなく`transformers[ja]`をインスールします。
**_Note: このnotebook上で推論テストを行う場合、(バージョンが古い場合は)pytorchのバージョンアップが必要になります。_**
```
# localで推論のテストを行う場合
!pip install torch==1.7.1
!pip install "sagemaker>=2.31.0" "transformers[ja]==4.6.1" "datasets[s3]==1.6.2" --upgrade
```
## Permissions
ローカル環境でSagemakerを使用する場合はSagemakerに必要な権限を持つIAMロールにアクセスする必要があります。[こちら](https://docs.aws.amazon.com/sagemaker/latest/dg/sagemaker-roles.html)を参照してください
```
import sagemaker
sess = sagemaker.Session()
# sagemaker session bucket -> used for uploading data, models and logs
# sagemaker will automatically create this bucket if it not exists
sagemaker_session_bucket=None
if sagemaker_session_bucket is None and sess is not None:
# set to default bucket if a bucket name is not given
sagemaker_session_bucket = sess.default_bucket()
role = sagemaker.get_execution_role()
sess = sagemaker.Session(default_bucket=sagemaker_session_bucket)
print(f"sagemaker role arn: {role}")
print(f"sagemaker bucket: {sess.default_bucket()}")
print(f"sagemaker session region: {sess.boto_region_name}")
```
# データの準備
事前にデータ(`DDQA-1.0.tar.gz`)をこのnotobookと同じ階層に配置してください
以下では、データをダウンロードして解凍 (unzip) します。
```
# Unzip
!tar -zxvf DDQA-1.0.tar.gz
```
## Uploading data to `sagemaker_session_bucket`
S3へデータをアップロードします。
```
s3_prefix = 'samples/datasets/driving-domain-qa'
input_train = sess.upload_data(
path='./DDQA-1.0/RC-QA/DDQA-1.0_RC-QA_train.json',
key_prefix=f'{s3_prefix}/train'
)
input_validation = sess.upload_data(
path='./DDQA-1.0/RC-QA/DDQA-1.0_RC-QA_dev.json',
key_prefix=f'{s3_prefix}/valid'
)
# データのUpload path
print(input_train)
print(input_validation)
```
# (Optional) Deepen your understanding of SQuAD
**このセクションはオプションであり、Fine-tuning & starting Sagemaker Training Jobまでスキップできます**
## 運転ドメインQAデータセットについて
運転ドメインQAデータセットはSQuAD2.0形式となっており、`run_squad.py`でそのまま実行できます。
トレーニングジョブの実行とは関連しませんが、ここでは少しデータについて理解を深めたいと思います。
QAデータセットの形式(README_ja.txt)
--------------------
本QAデータセットの形式はSQuAD2.0と同じです。SQuAD2.0の問題は、「文章」、「質問」、「答え」の三つ組になっており、「答え」は「文章」の中の一部になっています。一部の問題は、「文章」の中に「答え」が無いなど、答えられない問題になっています。詳細は以下の論文をご参照ください。
Pranav Rajpurkar, Robin Jia, and Percy Liang.
Know what you don’t know: Unanswerable questions for SQuAD,
In ACL2018, pages 784–789.
https://www.aclweb.org/anthology/P18-2124.pdf
以下に、jsonファイル中のQAデータセットを例示します。
注)jsonファイル中の"context"は「文章」
```json
{
"version": "v2.0",
"data": [
{
"title": "運転ドメイン",
"paragraphs": [
{
"context": "著者は以下の文章を書きました。本日お昼頃、梅田方面へ自転車で出かけました。ちょっと大きな交差点に差し掛かりました。自転車にまたがった若い女性が信号待ちしています。その後で私も止まって信号が青になるのを待っていました。",
"qas": [
{
"id": "55604556390008_00",
"question": "待っていました、の主語は何か?",
"answers": [
{
"text": "私",
"answer_start": 85
},
{
"text": "著者",
"answer_start": 0
}
],
"is_impossible": false
}
]
}
]
}
]
}
```
参考文献
--------
高橋 憲生、柴田 知秀、河原 大輔、黒橋 禎夫
ドメインを限定した機械読解モデルに基づく述語項構造解析
言語処理学会 第25回年次大会 発表論文集 (2019年3月)
https://www.anlp.jp/proceedings/annual_meeting/2019/pdf_dir/B1-4.pdf
※データセットの構築方法について記載
Norio Takahashi, Tomohide Shibata, Daisuke Kawahara and Sadao Kurohashi.
Machine Comprehension Improves Domain-Specific Japanese Predicate-Argument Structure Analysis,
In Proceedings of 2019 Conference on Empirical Methods in Natural Language Processing and 9th International Joint Conference on Natural Language Processing, Workshop MRQA: Machine Reading for Question Answering, 2019.
https://mrqa.github.io/assets/papers/42_Paper.pdf
※データセットの構築方法、文章中に答えが無い問題について記載
```
# データの読み込み
import json
with open("./DDQA-1.0/RC-QA/DDQA-1.0_RC-QA_train.json", "r") as f:
squad = json.load(f)
squad['data'][0]['paragraphs'][0]
```
SQuAD2.0形式は少し複雑なjson形式となっています。
次に`run_squad.py`内でどのような前処理が実行されているかについて少し触れます。
このparagraphsにはコンテキストが1つと質問が2つ、回答が6つ含まれていますが、後の処理ではここから
**2つの「コンテキスト」、「質問」、「答え」の三つ組**が作成されます。
回答は1番目のものが使用されます。
```
from transformers.data.processors.squad import SquadV2Processor
from transformers import squad_convert_examples_to_features
data_dir = './DDQA-1.0/RC-QA'
train_file = 'DDQA-1.0_RC-QA_train.json'
max_seq_length = 384 # トークン化後の最大入力シーケンス長。これより長いシーケンスは切り捨てられ、これより短いシーケンスはパディングされます
doc_stride = 128 # 長いドキュメントをチャンクに分割する場合、チャンク間でどのくらいのストライドを取るか
max_query_length = 64 # 質問のトークンの最大数。 これより長い質問はこの長さに切り捨てられます
threads = 1
from transformers import AutoTokenizer
# Tokenizer
tokenizer = AutoTokenizer.from_pretrained('cl-tohoku/bert-base-japanese-whole-word-masking')
# jsonファイルを読みこみ、複雑な構造を分解します
processor = SquadV2Processor()
examples = processor.get_train_examples(data_dir, filename=train_file)
# QuestionAnsweringモデルへ入力できるようにトークナイズします
# 以下の実行に数分時間がかかります
features, dataset = squad_convert_examples_to_features(
examples=examples,
tokenizer=tokenizer,
max_seq_length=max_seq_length,
doc_stride=doc_stride,
max_query_length=max_query_length,
is_training=True,
return_dataset="pt",
threads=threads,
)
```
`dataset`は後に`dataloader`に渡され、以下のように使用されます。
```python
for _ in train_iterator:
epoch_iterator = tqdm(train_dataloader, desc="Iteration", disable=args.local_rank not in [-1, 0])
for step, batch in enumerate(epoch_iterator):
# Skip past any already trained steps if resuming training
if steps_trained_in_current_epoch > 0:
steps_trained_in_current_epoch -= 1
continue
model.train()
batch = tuple(t.to(args.device) for t in batch)
inputs = {
"input_ids": batch[0],
"attention_mask": batch[1],
"token_type_ids": batch[2],
"start_positions": batch[3],
"end_positions": batch[4],
}
```
`input_ids`, `attention_mask`, `token_type_ids`はTransformerベースのモデルで一般的な入力形式です
QuestionAnsweringモデル特有のものとして`start_positions`, `end_positions`が挙げられます
```
# 参考に一つ目の中身を見てみます
i = 0
dataset[i]
# すでに テキスト→トークン化→ID化されているため、逆の操作で元に戻します。
# 質問と文章が含まれていることが確認できます
tokenizer.convert_tokens_to_string(tokenizer.convert_ids_to_tokens(dataset[i][0]))
# ID化→トークン化まで
tokenizer.convert_ids_to_tokens(dataset[i][0])
# 回答は、start_positionsのトークンで始まり、end_positionsでトークンで終わるように表現されます
# 試しに該当箇所のトークンを文字に戻してみます。
print(tokenizer.convert_tokens_to_string(tokenizer.convert_ids_to_tokens([dataset[i][0][dataset[i][3]]])))
print(tokenizer.convert_tokens_to_string(tokenizer.convert_ids_to_tokens([dataset[i][0][dataset[i][4]]])))
```
これから実行する`QuestionAnswering`は、**「コンテキスト」**内から**「質問」**に対する**「答え」**となる`start_positions`と`end_positions`を予測し、そのスパンを抽出するタスクとなります。
# Fine-tuning & starting Sagemaker Training Job
`HuggingFace`のトレーニングジョブを作成するためには`HuggingFace` Estimatorが必要になります。
Estimatorは、エンドツーエンドのAmazonSageMakerトレーニングおよびデプロイタスクを処理します。 Estimatorで、どのFine-tuningスクリプトを`entry_point`として使用するか、どの`instance_type`を使用するか、どの`hyperparameters`を渡すかなどを定義します。
```python
huggingface_estimator = HuggingFace(
entry_point='train.py',
source_dir='./scripts',
base_job_name='huggingface-sdk-extension',
instance_type='ml.p3.2xlarge',
instance_count=1,
transformers_version='4.4',
pytorch_version='1.6',
py_version='py36',
role=role,
hyperparameters={
'epochs': 1,
'train_batch_size': 32,
'model_name':'distilbert-base-uncased'
}
)
```
SageMakerトレーニングジョブを作成すると、SageMakerは`huggingface`コンテナを実行するために必要なec2インスタンスの起動と管理を行います。
Fine-tuningスクリプト`train.py`をアップロードし、`sagemaker_session_bucket`からコンテナ内の`/opt/ml/input/data`にデータをダウンロードして、トレーニングジョブを実行します。
```python
/opt/conda/bin/python train.py --epochs 1 --model_name distilbert-base-uncased --train_batch_size 32
```
`HuggingFace estimator`で定義した`hyperparameters`は、名前付き引数として渡されます。
またSagemakerは、次のようなさまざまな環境変数を通じて、トレーニング環境に関する有用なプロパティを提供しています。
* `SM_MODEL_DIR`:トレーニングジョブがモデルアーティファクトを書き込むパスを表す文字列。トレーニング後、このディレクトリのアーティファクトはモデルホスティングのためにS3にアップロードされます。
* `SM_NUM_GPUS`:ホストで使用可能なGPUの数を表す整数。
* `SM_CHANNEL_XXXX`:指定されたチャネルの入力データを含むディレクトリへのパスを表す文字列。たとえば、HuggingFace estimatorのfit呼び出しで`train`と`test`という名前の2つの入力チャネルを指定すると、環境変数`SM_CHANNEL_TRAIN`と`SM_CHANNEL_TEST`が設定されます。
このトレーニングジョブをローカル環境で実行するには、`instance_type='local'`、GPUの場合は`instance_type='local_gpu'`で定義できます。
**_Note:これはSageMaker Studio内では機能しません_**
```
# requirements.txtはトレーニングジョブの実行前に実行されます(コンテナにライブラリを追加する際に使用します)
# 残念なことにSageMakerのHuggingFaceコンテナは日本語処理(トークナイズ)に必要なライブラリが組み込まれていません
# したがってtransformers[ja]==4.6.1をジョブ実行前にインストールしています(fugashiとipadic)でも構いません
# tensorboardも組み込まれていないため、インストールします
!pygmentize ./scripts/requirements.txt
# トレーニングジョブで実行されるコード
!pygmentize ./scripts/run_squad.py
from sagemaker.huggingface import HuggingFace
# hyperparameters, which are passed into the training job
hyperparameters={
'model_type': 'bert',
'model_name_or_path': 'cl-tohoku/bert-base-japanese-whole-word-masking',
'output_dir': '/opt/ml/model',
'data_dir':'/opt/ml/input/data',
'train_file': 'train/DDQA-1.0_RC-QA_train.json',
'predict_file': 'validation/DDQA-1.0_RC-QA_dev.json',
'version_2_with_negative': 'True',
'do_train': 'True',
'do_eval': 'True',
'fp16': 'True',
'per_gpu_train_batch_size': 16,
'per_gpu_eval_batch_size': 16,
'max_seq_length': 384,
'doc_stride': 128,
'max_query_length': 64,
'learning_rate': 5e-5,
'num_train_epochs': 2,
#'max_steps': 100, # If > 0: set total number of training steps to perform. Override num_train_epochs.
'save_steps': 1000,
}
# metric definition to extract the results
metric_definitions=[
{"Name": "train_runtime", "Regex": "train_runtime.*=\D*(.*?)$"},
{'Name': 'train_samples_per_second', 'Regex': "train_samples_per_second.*=\D*(.*?)$"},
{'Name': 'epoch', 'Regex': "epoch.*=\D*(.*?)$"},
{'Name': 'f1', 'Regex': "f1.*=\D*(.*?)$"},
{'Name': 'exact_match', 'Regex': "exact_match.*=\D*(.*?)$"}]
```
## Creating an Estimator and start a training job
```
# estimator
huggingface_estimator = HuggingFace(
entry_point='run_squad.py',
source_dir='./scripts',
metric_definitions=metric_definitions,
instance_type='ml.p3.8xlarge',
instance_count=1,
volume_size=200,
role=role,
transformers_version='4.6',
pytorch_version='1.7',
py_version='py36',
hyperparameters=hyperparameters
)
# starting the train job with our uploaded datasets as input
huggingface_estimator.fit({'train': input_train, 'validation': input_validation})
# ml.p3.8xlarge, 2 epochでの実行時間の目安
# Training seconds: 758
# Billable seconds: 758
```
## Estimator Parameters
```
# container image used for training job
print(f"container image used for training job: \n{huggingface_estimator.image_uri}\n")
# s3 uri where the trained model is located
print(f"s3 uri where the trained model is located: \n{huggingface_estimator.model_data}\n")
# latest training job name for this estimator
print(f"latest training job name for this estimator: \n{huggingface_estimator.latest_training_job.name}\n")
# access the logs of the training job
huggingface_estimator.sagemaker_session.logs_for_job(huggingface_estimator.latest_training_job.name)
```
## Download-fine-tuned-model-from-s3
```
import os
OUTPUT_DIR = './output/'
if not os.path.exists(OUTPUT_DIR):
os.makedirs(OUTPUT_DIR)
from sagemaker.s3 import S3Downloader
# 学習したモデルのダウンロード
S3Downloader.download(
s3_uri=huggingface_estimator.model_data, # s3 uri where the trained model is located
local_path='.', # local path where *.targ.gz is saved
sagemaker_session=sess # sagemaker session used for training the model
)
# OUTPUT_DIRに解凍します
!tar -zxvf model.tar.gz -C output
```
## Question Answering on Local
```
from transformers import AutoTokenizer, AutoModelForQuestionAnswering
import torch
model = AutoModelForQuestionAnswering.from_pretrained('./output')
tokenizer = AutoTokenizer.from_pretrained('cl-tohoku/bert-base-japanese-whole-word-masking')
```
以下のセルは`./DDQA-1.0/RC-QA/DDQA-1.0_RC-QA_dev.json`からコピーしたものです
```
context = '実は先週、CBR600RRで事故りました。たまにはCBRにも乗らなきゃなーと思い久々にCBRで出勤したところ、家から10分ほど走ったところにある片側一車線の交差点で対向右折車と衝突してしまいました。自分が直進青信号で交差点へ進入したところで対向右折車線の車が突然右折を開始。とっさに急ブレーキはかけましたが、止まることはできずに右折車に衝突、自分は空中で一回転して左斜め数メートル先の路上へと飛ばされました。'
question='何に乗っていて事故りましたか?'
#context = 'まぁ,何回か改正してるわけで,自転車を走らせる領域を変更しないって言うのは,怠慢っていうか責任逃れっていうか,道交法に携わってるヤツはみんな馬鹿なのか.大体の人はここまで極端な意見ではないだろうけど,自転車は歩道を走るほうが自然だとは考えているだろう.というのも, みんな自転車乗ってる時歩道を走るでしょ?自転車で歩道走ってても歩行者にそこまで危険な目に合わせないと考えているし,車道に出たら明らかに危険な目に合うと考えている.'
#question='大体の人は自転車はどこを走るのが自然だと思っている?'
#context = '幸いけが人が出なくて良かったものの、タイヤの脱落事故が後を絶たない。先日も高速道路でトラックのタイヤがはずれ、中央分離帯を越え、反対車線を通行していた観光バスに直撃した。不幸にもバスを運転していた運転手さんがお亡くなりになった。もし、僕がこんな場面に遭遇していたら、この運転手さんのように、乗客の安全を考えて冷静に止まっただろうか?'
#question = '後を絶たないのは何ですか?'
#context = '右折待ちの一般ドライバーの方は、直進車線からの右折タクシーに驚いて右折のタイミングを失ってしまい、更なる混雑を招いているようでした」と述べていました。2004年8月6日付けには、ある女性が「道を譲っても挨拶をしない人が多い。特に女性の方。そのため意地悪ですが対向車のドライバーが女性だと譲りません。私はまだ人間が出来ていないので受け流すことが出来ません」ということを言っていましたが、その気持ち良く分かります。私は横断歩道の歩行者に対しては特別真面目で、歩行者がいるかどうかを常に注意して、いるときは必ず止まるよう心掛けています。それでも気付かずに止まることができなかったときは、「ああ、悪いことしちゃったな…」と、バックミラーを見ながら思います。'
#question = '歩行者がいるかどうかを常に注意しているのは誰ですか?'
# 推論
inputs = tokenizer.encode_plus(question, context, add_special_tokens=True, return_tensors="pt")
input_ids = inputs["input_ids"].tolist()[0]
output = model(**inputs)
answer_start = torch.argmax(output.start_logits)
answer_end = torch.argmax(output.end_logits) + 1
answer = tokenizer.convert_tokens_to_string(tokenizer.convert_ids_to_tokens(input_ids[answer_start:answer_end]))
# 結果
print("質問: "+question)
print("回答: "+answer)
```
|
github_jupyter
|
```
from google.colab import drive
drive.mount('gdrive')
%cd /content/gdrive/My\ Drive/colab
from __future__ import print_function
import json
import keras
import pickle
import os.path
from keras.datasets import cifar10
from keras.preprocessing.image import ImageDataGenerator
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Flatten
from keras.layers import Conv2D, MaxPooling2D, BatchNormalization
from keras.callbacks import ModelCheckpoint
from keras.callbacks import LambdaCallback
from keras import optimizers
from keras import regularizers
from keras.utils import plot_model
import numpy as np
import matplotlib.pyplot as plt
def build_model(x_shape, weight_decay, num_classes):
# Build the network of vgg for 10 classes with massive dropout and weight decay as described in the paper.
model = Sequential()
weight_decay = weight_decay
model.add(Conv2D(64, (3, 3), padding='same',
input_shape=x_shape, kernel_regularizer=regularizers.l2(weight_decay)))
model.add(Activation('relu'))
model.add(BatchNormalization())
model.add(Dropout(0.3))
model.add(Conv2D(64, (3, 3), padding='same', kernel_regularizer=regularizers.l2(weight_decay)))
model.add(Activation('relu'))
model.add(BatchNormalization())
model.add(Flatten())
model.add(Dense(200, kernel_regularizer=regularizers.l2(weight_decay)))
model.add(Activation('relu'))
model.add(BatchNormalization())
model.add(Dropout(0.5))
model.add(Dense(100, kernel_regularizer=regularizers.l2(weight_decay)))
model.add(Activation('relu'))
model.add(BatchNormalization())
model.add(Dropout(0.5))
model.add(Dense(num_classes))
model.add(Activation('softmax'))
return model
def normalize(X_train, X_test):
# this function normalize inputs for zero mean and unit variance
# it is used when training a model.
# Input: training set and test set
# Output: normalized training set and test set according to the trianing set statistics.
mean = np.mean(X_train, axis=(0, 1, 2, 3))
std = np.std(X_train, axis=(0, 1, 2, 3))
X_train = (X_train - mean) / (std + 1e-7)
X_test = (X_test - mean) / (std + 1e-7)
return X_train, X_test
def normalize_production(x):
# this function is used to normalize instances in production according to saved training set statistics
# Input: X - a training set
# Output X - a normalized training set according to normalization constants.
# these values produced during first training and are general for the standard cifar10 training set normalization
mean = 120.707
std = 64.15
return (x - mean)/(std+1e-7)
def predict(x, normalize=True, batch_size=50):
if normalize:
x = normalize_production(x)
return model.predict(x, batch_size)
def updateEpoch(epoch, logs):
to_save = num_epoch + epoch + 1
report_data['acc'].append(logs['acc'])
report_data['loss'].append(logs['loss'])
report_data['val_acc'].append(logs['val_acc'])
report_data['val_loss'].append(logs['val_loss'])
with open(epoch_file, "w") as file:
file.write(str(to_save))
with open(data_file, "wb") as file:
pickle.dump(report_data, file)
with open(all_file, "a+") as file:
all_data = [to_save, report_data['acc'], report_data['val_acc'],
report_data['loss'], report_data['val_loss']]
file.write(json.dumps(all_data))
print(epoch, logs)
def train(model):
# training parameters
batch_size = 128
maxepoches = 100
learning_rate = 0.1
lr_decay = 1e-6
lr_drop = 20
# The data, shuffled and split between train and test sets:
(x_train, y_train), (x_test, y_test) = cifar10.load_data()
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
x_train, x_test = normalize(x_train, x_test)
y_train = keras.utils.to_categorical(y_train, num_classes)
y_test = keras.utils.to_categorical(y_test, num_classes)
def lr_scheduler(epoch):
return learning_rate * (0.5 ** (epoch // lr_drop))
# data augmentation
datagen = ImageDataGenerator(
featurewise_center=False, # set input mean to 0 over the dataset
samplewise_center=False, # set each sample mean to 0
featurewise_std_normalization=False, # divide inputs by std of the dataset
samplewise_std_normalization=False, # divide each input by its std
zca_whitening=False, # apply ZCA whitening
rotation_range=15, # randomly rotate images in the range (degrees, 0 to 180)
width_shift_range=0.1, # randomly shift images horizontally (fraction of total width)
height_shift_range=0.1, # randomly shift images vertically (fraction of total height)
horizontal_flip=True, # randomly flip images
vertical_flip=False) # randomly flip images
# (std, mean, and principal components if ZCA whitening is applied).
datagen.fit(x_train)
# optimization details
sgd = optimizers.SGD(lr=learning_rate, decay=lr_decay, momentum=0.9)
model.compile(loss='categorical_crossentropy', optimizer=sgd, metrics=['accuracy'])
plot_model(model, to_file='model.png')
reduce_lr = keras.callbacks.LearningRateScheduler(lr_scheduler)
checkpoint = ModelCheckpoint(filepath, monitor='val_acc', verbose=1, save_best_only=True, mode='max')
lambdaCall = LambdaCallback(on_epoch_end=updateEpoch)
callbacks_list = [reduce_lr,checkpoint,lambdaCall]
# training process in a for loop with learning rate drop every 20 epoches.
history = model.fit_generator(
datagen.flow(x_train, y_train, batch_size=batch_size),
steps_per_epoch=x_train.shape[0] // batch_size,
epochs=maxepoches,
validation_data=(x_test, y_test),
callbacks=callbacks_list,
verbose=1)
model.save_weights('cifar10vgg_3.h5')
# summarize history for accuracy
plt.plot(report_data['acc'])
plt.plot(report_data['val_acc'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
# summarize history for loss
plt.plot(report_data['loss'])
plt.plot(report_data['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
return history
num_classes = 10
weight_decay = 0.0005
x_shape = [32, 32, 3]
train_bool = True
epoch_file="hw1_3_epoch_num.txt"
data_file="hw1_3_data.txt"
filepath="hw1_3_weights.best.hdf5"
all_file="hw1_3_all.txt"
model = build_model(x_shape, weight_decay, num_classes)
num_epoch = 0
if not os.path.isfile(epoch_file):
with open(epoch_file, "w+") as file:
file.write(str(num_epoch))
else:
with open(epoch_file, "r") as file:
num_epoch = int(file.read())
if os.path.isfile(filepath):
model.load_weights(filepath)
if os.path.isfile(data_file):
with open(data_file, "rb") as file:
report_data = pickle.load(file)
# print the model summary
model.summary()
report_data = {
"acc":[],
"val_acc":[],
"loss":[],
"val_loss":[]
}
if train_bool:
history = train(model)
else:
model.load_weights('cifar10vgg_3.h5')
(x_train, y_train), (x_test, y_test) = cifar10.load_data()
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
y_train = keras.utils.to_categorical(y_train, 10)
y_test = keras.utils.to_categorical(y_test, 10)
predicted_x = model.predict(x_test)
residuals = np.argmax(predicted_x, 1) != np.argmax(y_test, 1)
loss = sum(residuals)/len(residuals)
print("the validation 0/1 loss is: ", loss)
```
|
github_jupyter
|
```
# -*- coding: utf-8 -*-
# This work is part of the Core Imaging Library (CIL) developed by CCPi
# (Collaborative Computational Project in Tomographic Imaging), with
# substantial contributions by UKRI-STFC and University of Manchester.
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# Copyright 2019 UKRI-STFC, The University of Manchester
# Authored by: Evangelos Papoutsellis (UKRI-STFC)
```
<h1><center>Primal Dual Hybrid Gradient Algorithm </center></h1>
In this demo, we learn how to use the **Primal Dual Hybrid Algorithm (PDHG)** introduced by [Chambolle & Pock](https://hal.archives-ouvertes.fr/hal-00490826/document) for Tomography Reconstruction. We will solve the following minimisation problem under three different regularisation terms, i.e.,
* $\|\cdot\|_{1}$ or
* Tikhonov regularisation or
* with $L=\nabla$ and Total variation:
<a id='all_reg'></a>
$$\begin{equation}
u^{*} =\underset{u}{\operatorname{argmin}} \frac{1}{2} \| \mathcal{A} u - g\|^{2} +
\underbrace{
\begin{cases}
\alpha\,\|u\|_{1}, & \\[10pt]
\alpha\,\|\nabla u\|_{2}^{2}, & \\[10pt]
\alpha\,\mathrm{TV}(u) + \mathbb{I}_{\{u\geq 0\}}(u).
\end{cases}}_{Regularisers}
\tag{1}
\end{equation}$$
where,
1. $g$ is the Acqusisition data obtained from the detector.
1. $\mathcal{A}$ is the projection operator ( _Radon transform_ ) that maps from an image-space to an acquisition space, i.e., $\mathcal{A} : \mathbb{X} \rightarrow \mathbb{Y}, $ where $\mathbb{X}$ is an __ImageGeometry__ and $\mathbb{Y}$ is an __AcquisitionGeometry__.
1. $\alpha$: regularising parameter that measures the trade-off between the fidelity and the regulariser terms.
1. The total variation (isotropic) is defined as $$\mathrm{TV}(u) = \|\nabla u \|_{2,1} = \sum \sqrt{ (\partial_{y}u)^{2} + (\partial_{x}u)^{2} }$$
1. $\mathbb{I}_{\{u\geq 0\}}(u) : =
\begin{cases}
0, & \mbox{ if } u\geq 0\\
\infty , & \mbox{ otherwise}
\,
\end{cases}
$, $\quad$ a non-negativity constraint for the minimiser $u$.
<h2><center><u> Learning objectives </u></center></h2>
- Load the data using the CIL reader: `TXRMDataReader`.
- Preprocess the data using the CIL processors: `Binner`, `TransmissionAbsorptionConverter`.
- Run FBP and SIRT reconstructions.
- Setup PDHG for 3 different regularisers: $L^{1}$, Tikhonov and Total variation.
<!---
1. Brief intro for non-smooth minimisation problems using PDHG algorithm.
1. Setup and run PDHG with (__non-smooth__) $L^{1}$ norm regulariser. __(No BlockFramework)__
1. Use __BlockFunction__ and __Block Framework__ to setup PDHG for Tikhonov and TV reconstructions.
1. Run Total variation reconstruction with different regularising parameters and compared with FBP and SIRT reconstructions.
At the end of this demo, we will be able to reproduce all the reconstructions presented in the figure below. One can observe that the __Tikhonov regularisation__ with $L = \nabla$ was able to remove the noise but could not preserve the edges. However, this can be achieved with the the total variation reconstruction.
<img src="CIL-Demos/Notebooks/images/recon_all_tomo.jpeg" width="1500"/>
--->
<!-- <h2><center><u> Prerequisites </u></center></h2>
- AcquisitionData, AcquisitionGeometry, AstraProjectorSimple.
- BlockOperator, Gradient.
- FBP, SIRT, CGLS, Tikhonov. -->
We first import all the necessary libraries for this notebook.
<!---
In order to use the PDHG algorithm for the problem above, we need to express our minimisation problem into the following form:
<a id='PDHG_form'></a>
$$\min_{u} \mathcal{F}(K u) + \mathcal{G}(u)$$
where we assume that:
1. $\mathcal{F}$, $\mathcal{G}$ are __convex__ functionals
- $\mathcal{F}: Y \rightarrow \mathbb{R}$
- $\mathcal{G}: X \rightarrow \mathbb{R}$
2. $K$ is a continuous linear operator acting from a space X to another space Y :
$$K : X \rightarrow Y \quad $$
with operator norm defined as $$\| K \| = \max\{ \|K x\|_{Y} : \|x\|_{X}\leq 1 \}.$$
**Note**: The Gradient operator has $\|\nabla\| = \sqrt{8} $ and for the projection operator we use the [Power Method](https://en.wikipedia.org/wiki/Power_iteration) to approximate the greatest eigenvalue of $K$.
--->
```
# Import libraries
from cil.framework import BlockDataContainer
from cil.optimisation.functions import L2NormSquared, L1Norm, BlockFunction, MixedL21Norm, IndicatorBox, TotalVariation
from cil.optimisation.operators import GradientOperator, BlockOperator
from cil.optimisation.algorithms import PDHG, SIRT
from cil.plugins.astra.operators import ProjectionOperator
from cil.plugins.astra.processors import FBP
from cil.plugins.ccpi_regularisation.functions import FGP_TV
from cil.utilities.display import show2D, show_geometry
from cil.utilities.jupyter import islicer
from cil.io import TXRMDataReader
from cil.processors import Binner, TransmissionAbsorptionConverter, Slicer
import matplotlib.pyplot as plt
import numpy as np
import os
```
# Data information
In this demo, we use the **Walnut** found in [Jørgensen_et_all](https://zenodo.org/record/4822516#.YLXyAJMzZp8). In total, there are 6 individual micro Computed Tomography datasets in the native Zeiss TXRM/TXM format. The six datasets were acquired at the 3D Imaging Center at Technical University of Denmark in 2014 (HDTomo3D in 2016) as part of the ERC-funded project High-Definition Tomography (HDTomo) headed by Prof. Per Christian Hansen.
# Load walnut data
```
reader = TXRMDataReader()
pathname = os.path.abspath("/mnt/materials/SIRF/Fully3D/CIL/Walnut/valnut_2014-03-21_643_28/tomo-A")
data_name = "valnut_tomo-A.txrm"
filename = os.path.join(pathname,data_name )
reader.set_up(file_name=filename, angle_unit='radian')
data3D = reader.read()
# reorder data to match default order for Astra/Tigre operator
data3D.reorder('astra')
# Get Image and Acquisition geometries
ag3D = data3D.geometry
ig3D = ag3D.get_ImageGeometry()
```
## Acquisition and Image geometry information
```
print(ag3D)
print(ig3D)
```
# Show Acquisition geometry and full 3D sinogram.
```
show_geometry(ag3D)
show2D(data3D, slice_list = [('vertical',512), ('angle',800), ('horizontal',512)], cmap="inferno", num_cols=3, size=(15,15))
```
# Slice through projections
```
islicer(data3D, direction=1, cmap="inferno")
```
## For demonstration purposes, we extract the central slice and select only 160 angles from the total 1601 angles.
1. We use the `Slicer` processor with step size of 10.
1. We use the `Binner` processor to crop and bin the acquisition data in order to reduce the field of view.
1. We use the `TransmissionAbsorptionConverter` to convert from transmission measurements to absorption based on the Beer-Lambert law.
**Note:** To avoid circular artifacts in the reconstruction space, we subtract the mean value of a background Region of interest (ROI), i.e., ROI that does not contain the walnut.
```
# Extract vertical slice
data2D = data3D.subset(vertical='centre')
# Select every 10 angles
sliced_data = Slicer(roi={'angle':(0,1601,10)})(data2D)
# Reduce background regions
binned_data = Binner(roi={'horizontal':(120,-120,2)})(sliced_data)
# Create absorption data
absorption_data = TransmissionAbsorptionConverter()(binned_data)
# Remove circular artifacts
absorption_data -= np.mean(absorption_data.as_array()[80:100,0:30])
# Get Image and Acquisition geometries for one slice
ag2D = absorption_data.geometry
ag2D.set_angles(ag2D.angles, initial_angle=0.2, angle_unit='radian')
ig2D = ag2D.get_ImageGeometry()
print(" Acquisition Geometry 2D: {} with labels {}".format(ag2D.shape, ag2D.dimension_labels))
print(" Image Geometry 2D: {} with labels {}".format(ig2D.shape, ig2D.dimension_labels))
```
## Define Projection Operator
We can define our projection operator using our __astra__ __plugin__ that wraps the Astra-Toolbox library.
```
A = ProjectionOperator(ig2D, ag2D, device = "gpu")
```
## FBP and SIRT reconstuctions
Now, let's perform simple reconstructions using the **Filtered Back Projection (FBP)** and **Simultaneous Iterative Reconstruction Technique [SIRT](../appendix.ipynb/#SIRT) .**
Recall, for FBP we type
```python
fbp_recon = FBP(ig, ag, device = 'gpu')(absorption_data)
```
For SIRT, we type
```python
x_init = ig.allocate()
sirt = SIRT(initial = x_init, operator = A, data=absorption_data,
max_iteration = 50, update_objective_interval=10)
sirt.run(verbose=1)
sirt_recon = sirt.solution
```
**Note**: In SIRT, a non-negative constraint can be used with
```python
constraint=IndicatorBox(lower=0)
```
## Exercise 1: Run FBP and SIRT reconstructions
Use the code blocks described above and run FBP (`fbp_recon`) and SIRT (`sirt_recon`) reconstructions.
**Note**: To display the results, use
```python
show2D([fbp_recon,sirt_recon], title = ['FBP reconstruction','SIRT reconstruction'], cmap = 'inferno')
```
```
# Setup and run the FBP algorithm
fbp_recon = FBP(..., ..., device = 'gpu')(absorption_data)
# Setup and run the SIRT algorithm, with non-negative constraint
x_init = ig2D.allocate()
sirt = SIRT(initial = x_init,
operator = ...,
data= ...,
constraint = ...,
max_iteration = 300,
update_objective_interval=100)
sirt.run(verbose=1)
sirt_recon = sirt.solution
# Show reconstructions
show2D([fbp_recon,sirt_recon],
title = ['FBP reconstruction','SIRT reconstruction'],
cmap = 'inferno', fix_range=(0,0.05))
```
## Exercise 1: Solution
```
# Setup and run the FBP algorithm
fbp_recon = FBP(ig2D, ag2D, device = 'gpu')(absorption_data)
# Setup and run the SIRT algorithm, with non-negative constraint
x_init = ig2D.allocate()
sirt = SIRT(initial = x_init,
operator = A ,
data = absorption_data,
constraint = IndicatorBox(lower=0),
max_iteration = 300,
update_objective_interval=100)
sirt.run(verbose=1)
sirt_recon = sirt.solution
# Show reconstructions
show2D([fbp_recon,sirt_recon],
title = ['FBP reconstruction','SIRT reconstruction'],
cmap = 'inferno', fix_range=(0,0.05))
```
<h2><center> Why PDHG? </center></h2>
In the previous notebook, we presented the __Tikhonov regularisation__ for tomography reconstruction, i.e.,
<a id='Tikhonov'></a>
$$\begin{equation}
u^{*} =\underset{u}{\operatorname{argmin}} \frac{1}{2} \| \mathcal{A} u - g\|^{2} + \alpha\|L u\|^{2}_{2}
\tag{Tikhonov}
\end{equation}$$
where we can use either the `GradientOperator` ($L = \nabla) $ or the `IdentityOperator` ($L = \mathbb{I}$). Due to the $\|\cdot\|^{2}_{2}$ terms, one can observe that the above objective function is differentiable. As shown in the previous notebook, we can use the standard `GradientDescent` algorithm namely
```python
f1 = LeastSquares(A, absorption_data)
D = GradientOperator(ig2D)
f2 = OperatorCompositionFunction(L2NormSquared(),D)
f = f1 + alpha_tikhonov*f2
gd = GD(x_init=ig2D.allocate(), objective_function=f, step_size=None,
max_iteration=1000, update_objective_interval = 10)
gd.run(100, verbose=1)
```
However, this is not always the case. Consider for example an $L^{1}$ norm for the fidelity, i.e., $\|\mathcal{A} u - g\|_{1}$ or an $L^{1}$ norm of the regulariser i.e., $\|u\|_{1}$ or a non-negativity constraint $\mathbb{I}_{\{u>0\}}(u)$. An alternative is to use **Proximal Gradient Methods**, discused in the previous notebook, e.g., the `FISTA` algorithm, where we require one of the functions to be differentiable and the other to have a __simple__ proximal method, i.e., "easy to solve". For more information, we refer to [Parikh_Boyd](https://web.stanford.edu/~boyd/papers/pdf/prox_algs.pdf#page=30).
Using the __PDHG algorithm__, we can solve minimisation problems where the objective is not differentiable, and the only required assumption is convexity with __simple__ proximal problems.
<h2><center> $L^{1}$ regularisation </center></h2>
Let $L=$`IdentityOperator` in [Tikhonov regularisation](#Tikhonov) and replace the
$$\alpha^{2}\|L u\|^{2}_{2}\quad\mbox{ with }\quad \alpha\|u\|_{1}, $$
which results to a non-differentiable objective function. Hence, we have
<a id='Lasso'></a>
$$\begin{equation}
u^{*} =\underset{u}{\operatorname{argmin}} \frac{1}{2} \| \mathcal{A} u - g\|^{2} + \alpha\|u\|_{1}
\tag{$L^{2}-L^{1}$}
\end{equation}$$
<h2><center> How to setup and run PDHG? </center></h2>
In order to use the PDHG algorithm for the problem above, we need to express our minimisation problem into the following form:
<a id='PDHG_form'></a>
$$\begin{equation}
\min_{u\in\mathbb{X}} \mathcal{F}(K u) + \mathcal{G}(u)
\label{PDHG_form}
\tag{2}
\end{equation}$$
where we assume that:
1. $\mathcal{F}$, $\mathcal{G}$ are __convex__ functionals:
- $\mathcal{F}: \mathbb{Y} \rightarrow \mathbb{R}$
- $\mathcal{G}: \mathbb{X} \rightarrow \mathbb{R}$
1. $K$ is a continuous linear operator acting from a space $\mathbb{X}$ to another space $\mathbb{Y}$ :
$$K : \mathbb{X} \rightarrow \mathbb{Y} \quad $$
with operator norm defined as $$\| K \| = \max\{ \|K x\|_{\mathbb{Y}} : \|x\|_{\mathbb{X}}\leq 1 \}.$$
We can write the problem [($L^{2}-L^{1})$](#Lasso) into [(2)](#PDHG_form), if we let
1. $K = \mathcal{A} \quad \Longleftrightarrow \quad $ `K = A`
1. $\mathcal{F}: Y \rightarrow \mathbb{R}, \mbox{ with } \mathcal{F}(z) := \frac{1}{2}\| z - g \|^{2}, \quad \Longleftrightarrow \quad$ ` F = 0.5 * L2NormSquared(absorption_data)`
1. $\mathcal{G}: X \rightarrow \mathbb{R}, \mbox{ with } \mathcal{G}(z) := \alpha\|z\|_{1}, \quad \Longleftrightarrow \quad$ ` G = alpha * L1Norm()`
Hence, we can verify that with the above setting we have that [($L^{2}-L^{1})$](#Lasso)$\Rightarrow$[(2)](#PDHG_form) for $x=u$, $$\underset{u}{\operatorname{argmin}} \frac{1}{2}\|\mathcal{A} u - g\|^{2}_{2} + \alpha\|u\|_{1} =
\underset{u}{\operatorname{argmin}} \mathcal{F}(\mathcal{A}u) + \mathcal{G}(u) = \underset{x}{\operatorname{argmin}} \mathcal{F}(Kx) + \mathcal{G}(x) $$
The algorithm is described in the [Appendix](../appendix.ipynb/#PDHG) and for every iteration, we solve two (proximal-type) subproblems, i.e., __primal & dual problems__ where
$\mbox{prox}_{\tau \mathcal{G}}(x)$ and $\mbox{prox}_{\sigma \mathcal{F^{*}}}(x)$ are the **proximal operators** of $\mathcal{G}$ and $\mathcal{F}^{*}$ (convex conjugate of $\mathcal{F}$), i.e.,
$$\begin{equation}
\mbox{prox}_{\lambda \mathcal{F}}(x) = \underset{z}{\operatorname{argmin}} \frac{1}{2}\|z - x \|^{2} + \lambda
\mathcal{F}(z) \end{equation}
$$
One application of the proximal operator is similar to a gradient step but is defined for convex and not necessarily differentiable functions.
To setup and run PDHG in CIL:
```python
pdhg = PDHG(f = F, g = G, operator = K,
max_iterations = 500, update_objective_interval = 100)
pdhg.run(verbose=1)
```
**Note:** To monitor convergence, we use `pdhg.run(verbose=1)` that prints the objective value of the primal problem, or `pdhg.run(verbose=2)` that prints the objective value of the primal and dual problems, as well as the primal dual gap. Nothing is printed with `verbose=0`.
<a id='sigma_tau'></a>
### Define operator $K$, functions $\mathcal{F}$ and $\mathcal{G}$
```
K = A
F = 0.5 * L2NormSquared(b=absorption_data)
alpha = 0.01
G = alpha * L1Norm()
```
### Setup and run PDHG
```
# Setup and run PDHG
pdhg_l1 = PDHG(f = F, g = G, operator = K,
max_iteration = 500,
update_objective_interval = 100)
pdhg_l1.run(verbose=1)
# Show reconstuction and ground truth
show2D([pdhg_l1.solution,fbp_recon], fix_range=(0,0.05), title = ['L1 regularisation', 'FBP'], cmap = 'inferno')
# Plot middle line profile
plt.figure(figsize=(30,8))
plt.rcParams.update({'font.size': 15})
plt.rcParams.update({'lines.linewidth': 5})
plt.plot(fbp_recon.subset(horizontal_y = int(ig2D.voxel_num_y/2)).as_array(), label = 'FBP')
plt.plot(pdhg_l1.solution.subset(horizontal_y = int(ig2D.voxel_num_y/2)).as_array(), label = 'L1 regularisation')
plt.legend()
plt.title('Middle Line Profiles')
plt.show()
```
<h2><center> PDHG for Total Variation Regularisation </center></h2>
Now, we continue with the setup of the PDHG algorithm using the Total variation regulariser appeared in [(1)](#all_reg).
Similarly, to the [($L^{2}-L^{1}$)](#Lasso) problem, we need to express [($L^{2}-TV$)](#all_reg) in the general form of [PDHG](#PDHG_form). This can be done using two different formulations:
1. Explicit formulation: All the subproblems in the PDHG algorithm have a closed form solution.
1. Implicit formulation: One of the subproblems in the PDHG algorithm is not solved explicitly but an inner solver is used.
---
<h2><center> ($L^{2}-TV$) with Explicit PDHG </center></h2>
For the setup of the **($L^{2}-TV$) Explicit PDHG**, we let
$$\begin{align}
& f_{1}: \mathbb{Y} \rightarrow \mathbb{R}, \quad f_{1}(z_{1}) = \alpha\,\|z_{1}\|_{2,1}, \mbox{ ( the TV term ) }\\
& f_{2}: \mathbb{X} \rightarrow \mathbb{R}, \quad f_{2}(z_{2}) = \frac{1}{2}\|z_{2} - g\|_{2}^{2}, \mbox{ ( the data-fitting term ). }
\end{align}$$
```python
f1 = alpha * MixedL21Norm()
f2 = 0.5 * L2NormSquared(b=absorption_data)
```
For $z = (z_{1}, z_{2})\in \mathbb{Y}\times \mathbb{X}$, we define a separable function, e.g., [BlockFunction,](../appendix.ipynb/#BlockFunction)
$$\mathcal{F}(z) : = \mathcal{F}(z_{1},z_{2}) = f_{1}(z_{1}) + f_{2}(z_{2})$$
```python
F = BlockFunction(f1, f2)
```
In order to obtain an element $z = (z_{1}, z_{2})\in \mathbb{Y}\times \mathbb{X}$, we need to define a `BlockOperator` $K$, using the two operators involved in [$L^{2}-TV$](#TomoTV), i.e., the `GradientOperator` $\nabla$ and the `ProjectionOperator` $\mathcal{A}$.
$$ \mathcal{K} =
\begin{bmatrix}
\nabla\\
\mathcal{A}
\end{bmatrix}
$$
```python
Grad = GradientOperator(ig)
K = BlockOperator(Grad, A)
```
Finally, we enforce a non-negativity constraint by letting $\mathcal{G} = \mathbb{I}_{\{u>0\}}(u)$ $\Longleftrightarrow$ `G = IndicatorBox(lower=0)`
Again, we can verify that with the above setting we can express our problem into [(2)](#PDHG_form), for $x=u$
$$
\begin{align}
\underset{u}{\operatorname{argmin}}\alpha\|\nabla u\|_{2,1} + \frac{1}{2}\|\mathcal{A} u - g\|^{2}_{2} + \mathbb{I}_{\{u>0\}}(u) = \underset{u}{\operatorname{argmin}} f_{1}(\nabla u) + f_{2}(\mathcal{A}u) + \mathbb{I}_{\{u>0\}}(u) \\ = \underset{u}{\operatorname{argmin}} F(
\begin{bmatrix}
\nabla \\
\mathcal{A}
\end{bmatrix}u) + \mathbb{I}_{\{u>0\}}(u) =
\underset{u}{\operatorname{argmin}} \mathcal{F}(Ku) + \mathcal{G}(u) = \underset{x}{\operatorname{argmin}} \mathcal{F}(Kx) + \mathcal{G}(x)
\end{align}
$$
```
# Define BlockFunction F
alpha_tv = 0.0003
f1 = alpha_tv * MixedL21Norm()
f2 = 0.5 * L2NormSquared(b=absorption_data)
F = BlockFunction(f1, f2)
# Define BlockOperator K
Grad = GradientOperator(ig2D)
K = BlockOperator(Grad, A)
# Define Function G
G = IndicatorBox(lower=0)
# Setup and run PDHG
pdhg_tv_explicit = PDHG(f = F, g = G, operator = K,
max_iteration = 1000,
update_objective_interval = 200)
pdhg_tv_explicit.run(verbose=1)
# Show reconstuction and ground truth
show2D([pdhg_tv_explicit.solution,fbp_recon], fix_range=(0,0.055), title = ['TV regularisation','FBP'], cmap = 'inferno')
# Plot middle line profile
plt.figure(figsize=(30,8))
plt.rcParams.update({'font.size': 15})
plt.rcParams.update({'lines.linewidth': 5})
plt.plot(fbp_recon.subset(horizontal_y = int(ig2D.voxel_num_y/2)).as_array(), label = 'FBP')
plt.plot(pdhg_tv_explicit.solution .subset(horizontal_y = int(ig2D.voxel_num_y/2)).as_array(), label = 'TV regularisation')
plt.legend()
plt.title('Middle Line Profiles')
plt.show()
```
## Speed of PDHG convergence
The PDHG algorithm converges when $\sigma\tau\|K\|^{2}<1$, where the variable $\sigma$, $\tau$ are called the _primal and dual stepsizes_. When we setup the PDHG algorithm, the default values of $\sigma$ and $\tau$ are used:
- $\sigma=1.0$
- $\tau = \frac{1.0}{\sigma\|K\|^{2}}$,
and are not passed as arguments in the setup of PDHG. However, **the speed of the algorithm depends heavily on the choice of these stepsizes.** For the following, we encourage you to use different values, such as:
- $\sigma=\frac{1}{\|K\|}$
- $\tau =\frac{1}{\|K\|}$
where $\|K\|$ is the operator norm of $K$.
```python
normK = K.norm()
sigma = 1./normK
tau = 1./normK
PDHG(f = F, g = G, operator = K, sigma=sigma, tau=tau,
max_iteration = 2000,
update_objective_interval = 500)
```
The operator norm is computed using the [Power Method](https://en.wikipedia.org/wiki/Power_iteration) to approximate the greatest eigenvalue of $K$.
## Exercise 2: Setup and run PDHG algorithm for Tikhonov regularisation
Use exactly the same code as above and replace:
$$f_{1}(z_{1}) = \alpha\,\|z_{1}\|_{2,1} \mbox{ with } f_{1}(z_{1}) = \alpha\,\|z_{1}\|_{2}^{2}.$$
```
# Define BlockFunction F
alpha_tikhonov = 0.05
f1 = ...
F = BlockFunction(f1, f2)
# Setup and run PDHG
pdhg_tikhonov_explicit = PDHG(f = F, g = G, operator = K,
max_iteration = 500,
update_objective_interval = 100)
pdhg_tikhonov_explicit.run(verbose=1)
```
## Exercise 2: Solution
```
# Define BlockFunction F
alpha_tikhonov = 0.05
f1 = alpha_tikhonov * L2NormSquared()
F = BlockFunction(f1, f2)
# Setup and run PDHG
pdhg_tikhonov_explicit = PDHG(f = F, g = G, operator = K,
max_iteration = 1000,
update_objective_interval = 200)
pdhg_tikhonov_explicit.run(verbose=1)
# Show reconstuction and ground truth
show2D([pdhg_tikhonov_explicit.solution,fbp_recon], fix_range=(0,0.055), title = ['Tikhonov regularisation','FBP'], cmap = 'inferno')
# Plot middle line profile
plt.figure(figsize=(30,8))
plt.rcParams.update({'font.size': 15})
plt.rcParams.update({'lines.linewidth': 5})
plt.plot(fbp_recon.subset(horizontal_y = int(ig2D.voxel_num_y/2)).as_array(), label = 'FBP')
plt.plot(pdhg_tikhonov_explicit.solution .subset(horizontal_y = int(ig2D.voxel_num_y/2)).as_array(), label = 'Tikhonov regularisation')
plt.legend()
plt.title('Middle Line Profiles')
plt.show()
```
---
<h2><center> ($L^{2}-TV$) with Implicit PDHG </center></h2>
In the implicit PDHG, one of the proximal subproblems, i.e., $\mathrm{prox}_{\tau\mathcal{F}^{*}}$ or $\mathrm{prox}_{\sigma\mathcal{G}}$ are not solved exactly and an iterative solver is used. For the setup of the **Implicit PDHG**, we let
$$\begin{align}
& \mathcal{F}: \mathbb{Y} \rightarrow \mathbb{R}, \quad \mathcal{F}(z_{1}) = \frac{1}{2}\|z_{1} - g\|_{2}^{2}\\
& \mathcal{G}: \mathbb{X} \rightarrow \mathbb{R}, \quad \mathcal{G}(z_{2}) = \alpha\, \mathrm{TV}(z_{2}) = \|\nabla z_{2}\|_{2,1}
\end{align}$$
For the function $\mathcal{G}$, we can use the `TotalVariation` `Function` class from `CIL`. Alternatively, we can use the `FGP_TV` `Function` class from our `cil.plugins.ccpi_regularisation` that wraps regularisation routines from the [CCPi-Regularisation Toolkit](https://github.com/vais-ral/CCPi-Regularisation-Toolkit). For these functions, the `proximal` method implements an iterative solver, namely the **Fast Gradient Projection (FGP)** algorithm that solves the **dual** problem of
$$\begin{equation}
\mathrm{prox}_{\tau G}(u) = \underset{z}{\operatorname{argmin}} \frac{1}{2} \| u - z\|^{2} + \tau\,\alpha\,\mathrm{TV}(z) + \mathbb{I}_{\{z>0\}}(z),
\end{equation}
$$
for every PDHG iteration. Hence, we need to specify the number of iterations for the FGP algorithm. In addition, we can enforce a non-negativity constraint using `lower=0.0`. For the `FGP_TV` class, we can either use `device=cpu` or `device=gpu` to speed up this inner solver.
```python
G = alpha * FGP_TV(max_iteration=100, nonnegativity = True, device = 'gpu')
G = alpha * TotalVariation(max_iteration=100, lower=0.)
```
## Exercise 3: Setup and run implicit PDHG algorithm with the Total variation regulariser
- Using the TotalVariation class, from CIL. This solves the TV denoising problem (using the FGP algorithm) in CPU.
- Using the FGP_TV class from the CCPi regularisation plugin.
**Note:** In the FGP_TV implementation no pixel size information is included when in the forward and backward of the finite difference operator. Hence, we need to divide our regularisation parameter by the pixel size, e.g., $$\frac{\alpha}{\mathrm{ig2D.voxel\_size\_y}}$$
## $(L^{2}-TV)$ Implicit PDHG: using FGP_TV
```
F = 0.5 * L2NormSquared(b=absorption_data)
G = (alpha_tv/ig2D.voxel_size_y) * ...
K = A
# Setup and run PDHG
pdhg_tv_implicit_regtk = PDHG(f = F, g = G, operator = K,
max_iteration = 1000,
update_objective_interval = 200)
pdhg_tv_implicit_regtk.run(verbose=1)
```
## Exercise 3: Solution
```
F = 0.5 * L2NormSquared(b=absorption_data)
G = (alpha_tv/ig2D.voxel_size_y) * FGP_TV(max_iteration=100, device='gpu')
K = A
# Setup and run PDHG
pdhg_tv_implicit_regtk = PDHG(f = F, g = G, operator = K,
max_iteration = 1000,
update_objective_interval = 200)
pdhg_tv_implicit_regtk.run(verbose=1)
# Show reconstuction and ground truth
show2D([pdhg_tv_implicit_regtk.solution,pdhg_tv_explicit.solution,
(pdhg_tv_explicit.solution-pdhg_tv_implicit_regtk.solution).abs()],
fix_range=[(0,0.055),(0,0.055),(0,1e-3)],
title = ['TV (Implicit CCPi-RegTk)','TV (Explicit)', 'Absolute Difference'],
cmap = 'inferno', num_cols=3)
# Plot middle line profile
plt.figure(figsize=(30,8))
plt.rcParams.update({'font.size': 15})
plt.rcParams.update({'lines.linewidth': 5})
plt.plot(pdhg_tv_explicit.solution.subset(horizontal_y = int(ig2D.voxel_num_y/2)).as_array(), label = 'TV (explicit)')
plt.plot(pdhg_tv_implicit_regtk.solution.subset(horizontal_y = int(ig2D.voxel_num_y/2)).as_array(), label = 'TV (implicit)')
plt.legend()
plt.title('Middle Line Profiles')
plt.show()
```
In the above comparison between explicit and implicit TV reconstructions, we observe some differences in the reconstructions and in the middle line profiles. This is due to a) the number of iterations and b) $\sigma, \tau$ values used in both the explicit and implicit setup of the PDHG algorithm. You can try more iterations with different values of $\sigma$ and $\tau$ for both cases in order to be sure that converge to the same solution.
For example, you can use:
* max_iteration = 2000
* $\sigma=\tau=\frac{1}{\|K\|}$
## $(L^{2}-TV)$ Implicit PDHG: using TotalVariation
```
G = alpha_tv * TotalVariation(max_iteration=100, lower=0.)
# Setup and run PDHG
pdhg_tv_implicit_cil = PDHG(f = F, g = G, operator = K,
max_iteration = 500,
update_objective_interval = 100)
pdhg_tv_implicit_cil.run(verbose=1)
# Show reconstuction and ground truth
show2D([pdhg_tv_implicit_regtk.solution,
pdhg_tv_implicit_cil.solution,
(pdhg_tv_implicit_cil.solution-pdhg_tv_implicit_regtk.solution).abs()],
fix_range=[(0,0.055),(0,0.055),(0,1e-3)], num_cols=3,
title = ['TV (CIL)','TV (CCPI-RegTk)', 'Absolute Difference'],
cmap = 'inferno')
# Plot middle line profile
plt.figure(figsize=(30,8))
plt.rcParams.update({'font.size': 15})
plt.rcParams.update({'lines.linewidth': 5})
plt.plot(pdhg_tv_implicit_regtk.solution.subset(horizontal_y = int(ig2D.voxel_num_y/2)).as_array(), label = 'TV (CCPi-RegTk)')
plt.plot(pdhg_tv_implicit_cil.solution.subset(horizontal_y = int(ig2D.voxel_num_y/2)).as_array(), label = 'TV (CIL)')
plt.legend()
plt.title('Middle Line Profiles')
plt.show()
```
# FBP reconstruction with all the projection angles.
```
binned_data3D = Binner(roi={'horizontal':(120,-120,2)})(data3D)
absorption_data3D = TransmissionAbsorptionConverter()(binned_data3D.subset(vertical=512))
absorption_data3D -= np.mean(absorption_data3D.as_array()[80:100,0:30])
ag3D = absorption_data3D.geometry
ag3D.set_angles(ag3D.angles, initial_angle=0.2, angle_unit='radian')
ig3D = ag3D.get_ImageGeometry()
fbp_recon3D = FBP(ig3D, ag3D)(absorption_data3D)
```
# Show all reconstructions
- FBP (1601 projections)
- FBP (160 projections)
- SIRT (160 projections)
- $L^{1}$ regularisation (160 projections)
- Tikhonov regularisation (160 projections)
- Total variation regularisation (160 projections)
```
show2D([fbp_recon3D,
fbp_recon,
sirt_recon,
pdhg_l1.solution,
pdhg_tikhonov_explicit.solution,
pdhg_tv_explicit.solution],
title=['FBP 1601 projections', 'FBP', 'SIRT','$L^{1}$','Tikhonov','TV'],
cmap="inferno",num_cols=3, size=(25,20), fix_range=(0,0.05))
```
## Zoom ROIs
```
show2D([fbp_recon3D.as_array()[175:225,150:250],
fbp_recon.as_array()[175:225,150:250],
sirt_recon.as_array()[175:225,150:250],
pdhg_l1.solution.as_array()[175:225,150:250],
pdhg_tikhonov_explicit.solution.as_array()[175:225,150:250],
pdhg_tv_implicit_regtk.solution.as_array()[175:225,150:250]],
title=['FBP 1601 projections', 'FBP', 'SIRT','$L^{1}$','Tikhonov','TV'],
cmap="inferno",num_cols=3, size=(25,20), fix_range=(0,0.05))
```
<h1><center>Conclusions</center></h1>
In the PDHG algorithm, the step-sizes $\sigma, \tau$ play a significant role in terms of the convergence speed. In the above problems, we used the default values:
* $\sigma = 1.0$, $\tau = \frac{1.0}{\sigma\|K\|^{2}}$
and we encourage you to try different values provided that $\sigma\tau\|K\|^{2}<1$ is satisfied. Certainly, these values are not the optimal ones and there are sevelar accelaration methods in the literature to tune these parameters appropriately, see for instance [Chambolle_Pock2010](https://hal.archives-ouvertes.fr/hal-00490826/document), [Chambolle_Pock2011](https://ieeexplore.ieee.org/document/6126441), [Goldstein et al](https://arxiv.org/pdf/1305.0546.pdf), [Malitsky_Pock](https://arxiv.org/pdf/1608.08883.pdf).
In the following notebook, we are going to present a stochastic version of PDHG, namely **SPDHG** introduced in [Chambolle et al](https://arxiv.org/pdf/1706.04957.pdf) which is extremely useful to reconstruct large datasets, e.g., 3D walnut data. The idea behind SPDHG is to split our initial dataset into smaller chunks and apply forward and backward operations to these randomly selected subsets of the data. SPDHG has been used for different imaging applications and produces significant computational improvements
over the PDHG algorithm, see [Ehrhardt et al](https://arxiv.org/abs/1808.07150) and [Papoutsellis et al](https://arxiv.org/pdf/2102.06126.pdf).
|
github_jupyter
|
# Isolated skyrmion in confined helimagnetic nanostructure
**Authors**: Marijan Beg, Marc-Antonio Bisotti, Weiwei Wang, Ryan Pepper, David Cortes-Ortuno
**Date**: 26 June 2016 (Updated 24 Jan 2019)
This notebook can be downloaded from the github repository, found [here](https://github.com/computationalmodelling/fidimag/blob/master/doc/ipynb/isolated_skyrmion.ipynb).
## Problem specification
A thin film disk sample with thickness $t=10 \,\text{nm}$ and diameter $d=100 \,\text{nm}$ is simulated. The material is FeGe with material parameters [1]:
- exchange energy constant $A = 8.78 \times 10^{-12} \,\text{J/m}$,
- magnetisation saturation $M_\text{s} = 3.84 \times 10^{5} \,\text{A/m}$, and
- Dzyaloshinskii-Moriya energy constant $D = 1.58 \times 10^{-3} \,\text{J/m}^{2}$.
It is expected that when the system is initialised in the uniform out-of-plane direction $\mathbf{m}_\text{init} = (0, 0, 1)$, it relaxes to the isolated Skyrmion (Sk) state (See Supplementary Information in Ref. 1). (Note that LLG dynamics is important, which means that artificially disable the precession term in LLG may lead to other states).
## Simulation using the LLG equation
```
from fidimag.micro import Sim
from fidimag.common import CuboidMesh
from fidimag.micro import UniformExchange, Demag, DMI
from fidimag.common import plot
import time
%matplotlib inline
```
The cuboidal thin film mesh which contains the disk is created:
```
d = 100 # diameter (nm)
t = 10 # thickness (nm)
# Mesh discretisation.
dx = dy = 2.5 # nm
dz = 2
mesh = CuboidMesh(nx=int(d/dx), ny=int(d/dy), nz=int(t/dz), dx=dx, dy=dy, dz=dz, unit_length=1e-9)
```
Since the disk geometry is simulated, it is required to set the saturation magnetisation to zero in the regions of the mesh outside the disk. In order to do that, the following function is created:
```
def Ms_function(Ms):
def wrapped_function(pos):
x, y, z = pos[0], pos[1], pos[2]
r = ((x-d/2.)**2 + (y-d/2.)**2)**0.5 # distance from the centre
if r <= d/2:
# Mesh point is inside the disk.
return Ms
else:
# Mesh point is outside the disk.
return 0
return wrapped_function
```
To reduce the relaxation time, we define a state using a python function.
```
def init_m(pos):
x,y,z = pos
x0, y0 = d/2., d/2.
r = ((x-x0)**2 + (y-y0)**2)**0.5
if r<10:
return (0,0, 1)
elif r<30:
return (0,0, -1)
elif r<60:
return (0, 0, 1)
else:
return (0, 0, -1)
```
Having the magnetisation saturation function, the simulation object can be created:
```
# FeGe material paremeters.
Ms = 3.84e5 # saturation magnetisation (A/m)
A = 8.78e-12 # exchange energy constant (J/m)
D = 1.58e-3 # Dzyaloshinkii-Moriya energy constant (J/m**2)
alpha = 1 # Gilbert damping
gamma = 2.211e5 # gyromagnetic ration (m/As)
# Create simulation object.
sim = Sim(mesh)
# sim = Sim(mesh, driver='steepest_descent')
sim.Ms = Ms_function(Ms)
sim.driver.alpha = alpha
sim.driver.gamma = gamma
# Add energies.
sim.add(UniformExchange(A=A))
sim.add(DMI(D=D))
sim.add(Demag())
# Since the magnetisation dynamics is not important in this stage,
# the precession term in LLG equation can be set to artificially zero.
# sim.driver.do_precession = False
# Initialise the system.
sim.set_m(init_m)
```
This is the initial configuration used before relaxation:
```
plot(sim, component='all', z=0.0, cmap='RdBu')
```
Now the system is relaxed to find a metastable state of the system:
```
# Relax the system to its equilibrium.
start = time.time()
sim.driver.relax(dt=1e-13, stopping_dmdt=0.1, max_steps=10000,
save_m_steps=None, save_vtk_steps=None, printing=False)
end = time.time()
#NBVAL_IGNORE_OUTPUT
print('Timing: ', end - start)
sim.save_vtk()
```
The magnetisation components of obtained equilibrium configuration can be plotted in the following way:
We plot the magnetisation at the bottom of the sample:
```
plot(sim, component='all', z=0.0, cmap='RdBu')
```
and at the top of the sample:
```
plot(sim, component='all', z=10.0, cmap='RdBu')
```
and we plot the xy spin angle through the middle of the sample:
```
plot(sim, component='angle', z=5.0, cmap='hsv')
```
## Simulation using Steepest Descent
An alternative method for the minimisation of the energy is using a SteepestDescent method:
```
# Create simulation object.
sim = Sim(mesh, driver='steepest_descent')
sim.Ms = Ms_function(Ms)
sim.driver.gamma = gamma
# Add energies.
sim.add(UniformExchange(A=A))
sim.add(DMI(D=D))
sim.add(Demag())
# The maximum timestep:
sim.driver.tmax = 1
# Initialise the system.
sim.set_m(init_m)
```
In this case the driver has a `minimise` method
```
start = time.time()
sim.driver.minimise(max_steps=10000, stopping_dm=0.5e-4, initial_t_step=1e-2)
end = time.time()
#NBVAL_IGNORE_OUTPUT
print('Timing: ', end - start)
```
And the final state is equivalent to the one found with the LLG technique
```
plot(sim, component='all', z=0.0, cmap='RdBu')
```
## References
[1] Beg, M. et al. Ground state search, hysteretic behaviour, and reversal mechanism of skyrmionic textures in confined helimagnetic nanostructures. *Sci. Rep.* **5**, 17137 (2015).
|
github_jupyter
|
```
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
```
## 1. 加载并可视化数据
```
path = 'LogiReg_data.txt'
pdData = pd.read_csv(path, header=None, names=['Exam 1', 'Exam 2', 'Admitted'])
pdData.head()
pdData.shape
positive = pdData[pdData['Admitted'] == 1]
negative = pdData[pdData['Admitted'] == 0]
fig, ax = plt.subplots(figsize=(10, 5))
ax.scatter(positive['Exam 1'], positive['Exam 2'], s=30, c='b', marker='o', label='Admitted')
ax.scatter(negative['Exam 1'], negative['Exam 2'], s=30, c='r', marker='x', label='Not Admitted')
ax.legend()
ax.set_xlabel('Exam 1 Score')
ax.set_ylabel('Exam 2 Score')
```
## 2. Sigmoid函数
$$
g(z) = \frac{1}{1+e^{-z}}
$$
```
def sigmoid(z):
return 1 / (1 + np.exp(-z))
nums = np.arange(-10, 10, step=1)
fig, ax = plt.subplots(figsize=(12, 4))
ax.plot(nums, sigmoid(nums), 'r')
```
## 3. 建立Model
$$
\begin{array}{ccc}
\begin{pmatrix}\theta_{0} & \theta_{1} & \theta_{2}\end{pmatrix} & \times & \begin{pmatrix}1\\
x_{1}\\
x_{2}
\end{pmatrix}\end{array}=\theta_{0}+\theta_{1}x_{1}+\theta_{2}x_{2}
$$
```
def model(X, theta):
return sigmoid(np.dot(X, theta.T))
# 在第0列插入1
pdData.insert(0, 'Ones', 1)
# 获取<training data, y>
orig_data = pdData.values
cols = orig_data.shape[1]
X = orig_data[:, 0:cols-1]
y = orig_data[:, cols-1:cols]
# 初始化参数
theta = np.zeros([1, 3])
X[:5]
y[:5]
theta
```
## 4. 建立Loss Function
将对数似然函数去负号
$$
D(h_\theta(x), y) = -y\log(h_\theta(x)) - (1-y)\log(1-h_\theta(x))
$$
求平均损失
$$
J(\theta)=\frac{1}{n}\sum_{i=1}^{n} D(h_\theta(x_i), y_i)
$$
```
def cost(X, y, theta):
left = np.multiply(-y, np.log(model(X, theta)))
right = np.multiply(1 - y, np.log(1 - model(X, theta)))
return np.sum(left - right) / (len(X))
cost(X, y, theta)
```
## 5. 计算梯度
$$
\frac{\partial J}{\partial \theta_j}=-\frac{1}{m}\sum_{i=1}^n (y_i - h_\theta (x_i))x_{ij}
$$
```
def gradient(X, y, theta):
grad = np.zeros(theta.shape)
error = (model(X, theta) - y).ravel()
# 对于每一个参数,取出相关列的数据进行更新
for j in range(len(theta.ravel())):
term = np.multiply(error, X[:, j])
grad[0, j] = np.sum(term) / len(X)
return grad
```
## 6. 梯度下降
```
import time
import numpy.random
STOP_ITER = 0
STOP_COST = 1
STOP_GRAD = 2
def stopCriterion(dtype, value, threshold):
if dtype == STOP_ITER:
return value > threshold
elif dtype == STOP_COST:
return abs(value[-1] - value[-2]) < threshold
elif dtype == STOP_GRAD:
return np.linalg.norm(value) < threshold
def shuffleData(data):
# 洗牌操作
np.random.shuffle(data)
cols = data.shape[1]
X = data[:, 0:cols-1]
y = data[:, cols-1:]
return X, y
def descent(data, theta, batchSize, stopType, thresh, alpha):
i = 0
k = 0
init_time = time.time()
X, y = shuffleData(data)
grad = np.zeros(theta.shape)
costs = [cost(X, y, theta)]
while True:
grad = gradient(X[k: k+batchSize], y[k: k+batchSize], theta)
k += batchSize
if k >= n:
k = 0
X, y = shuffleData(data)
theta = theta - alpha*grad
costs.append(cost(X, y, theta))
i += 1
if stopType == STOP_ITER:
value = i
elif stopType == STOP_COST:
value = costs
elif stopType == STOP_GRAD:
value = grad
if stopCriterion(stopType, value, thresh):
break
return theta, i-1, costs, grad, time.time()-init_time
def runExpe(data, theta, batchSize, stopType, thresh, alpha):
theta, iter, costs, grad, dur = descent(data, theta, batchSize, stopType, thresh, alpha)
name = "Original" if (data[:,1]>2).sum() > 1 else "Scaled"
name += " data - learning rate: {} - ".format(alpha)
if batchSize == n:
strDescType = "Gradient"
elif batchSize == 1:
strDescType = "Stochastic"
else:
strDescType = "Mini-batch ({})".format(batchSize)
name += strDescType + " descent - Stop: "
if stopType == STOP_ITER:
strStop = "{} iterations".format(thresh)
elif stopType == STOP_COST:
strStop = "costs change < {}".format(thresh)
else:
strStop = "gradient norm < {}".format(thresh)
name += strStop
print ("***{}\nTheta: {} - Iter: {} - Last cost: {:03.2f} - Duration: {:03.2f}s".format(
name, theta, iter, costs[-1], dur))
fig, ax = plt.subplots(figsize=(12,4))
ax.plot(np.arange(len(costs)), costs, 'r')
ax.set_xlabel('Iterations')
ax.set_ylabel('Cost')
ax.set_title(name.upper() + ' - Error vs. Iteration')
return theta
```
## 7. 不同的停止策略
### 设定迭代次数
```
#选择的梯度下降方法是基于所有样本的
n=100
runExpe(orig_data, theta, n, STOP_ITER, thresh=5000, alpha=0.000001)
```
### 根据损失值停止
```
runExpe(orig_data, theta, n, STOP_COST, thresh=0.000001, alpha=0.001)
```
### 根据梯度变化停止
```
runExpe(orig_data, theta, n, STOP_GRAD, thresh=0.05, alpha=0.001)
```
## 8. 不同的梯度下降方法
### Stochastic descent
```
runExpe(orig_data, theta, 1, STOP_ITER, thresh=5000, alpha=0.001)
# 降低学习率
runExpe(orig_data, theta, 1, STOP_ITER, thresh=15000, alpha=0.000002)
```
结论: 速度快,但稳定性差,需要很小的学习率
### Mini-batch descent
```
runExpe(orig_data, theta, 16, STOP_ITER, thresh=15000, alpha=0.001)
from sklearn import preprocessing as pp
# 对数据进行标准化 将数据按其属性(按列进行)减去其均值,然后除以其方差。最后得到的结果是,对每个属性/每列来说所有数据都聚集在0附近,方差值为1
scaled_data = orig_data.copy()
scaled_data[:, 1:3] = pp.scale(orig_data[:, 1:3])
runExpe(scaled_data, theta, n, STOP_ITER, thresh=5000, alpha=0.001)
```
结论: 原始数据为0.61,而预处理后0.38。数据做预处理非常重要
```
runExpe(scaled_data, theta, n, STOP_GRAD, thresh=0.02, alpha=0.001)
theta = runExpe(scaled_data, theta, 1, STOP_GRAD, thresh=0.002/5, alpha=0.001)
runExpe(scaled_data, theta, 16, STOP_GRAD, thresh=0.002*2, alpha=0.001)
```
## 9. 测试精度
```
def predict(X, theta):
return [1 if x >= 0.5 else 0 for x in model(X, theta)]
scaled_X = scaled_data[:, :3]
y = scaled_data[:, 3]
predictions = predict(scaled_X, theta)
correct = [1 if ((a == 1 and b == 1) or (a == 0 and b == 0)) else 0 for (a, b) in zip(predictions, y)]
accuracy = (sum(map(int, correct)) % len(correct))
print ('accuracy = {0}%'.format(accuracy))
```
|
github_jupyter
|
##### Copyright 2020 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# MinDiff Data Preparation
<div class="devsite-table-wrapper"><table class="tfo-notebook-buttons" align="left">
<td><a target="_blank" href="https://tensorflow.org/responsible_ai/model_remediation/min_diff/guide/min_diff_data_preparation.ipynb">
<img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/model-remediation/blob/master/docs/min_diff/guide/min_diff_data_preparation.ipynb">
<img src="https://www.tensorflow.org/images/colab_logo_32px.png">Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/model-remediation/blob/master/docs/min_diff/guide/min_diff_data_preparation.ipynb">
<img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png">View source on GitHub</a>
</td>
<td>
<a target="_blank" href="https://storage.googleapis.com/tensorflow_docs/model-remediation/docs/min_diff/guide/min_diff_data_preparation.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table></div>
##Introduction
When implementing MinDiff, you will need to make complex decisions as you choose and shape your input before passing it on to the model. These decisions will largely determine the behavior of MinDiff within your model.
This guide will cover the technical aspects of this process, but will not discuss how to evaluate a model for fairness, or how to identify particular slices and metrics for evaluation. Please see the [Fairness Indicators guidance](https://www.tensorflow.org/responsible_ai/fairness_indicators/guide/guidance) for details on this.
To demonstrate MinDiff, this guide uses the [UCI income dataset](https://archive.ics.uci.edu/ml/datasets/census+income). The model task is to predict whether an individual has an income exceeding $50k, based on various personal attributes. This guide assumes there is a problematic gap in the FNR (false negative rate) between `"Male"` and `"Female"` slices and the model owner (you) has decided to apply MinDiff to address the issue. For more information on the scenarios in which one might choose to apply MinDiff, see the [requirements page](https://www.tensorflow.org/responsible_ai/model_remediation/min_diff/guide/requirements).
Note: We recognize the limitations of the categories used in the original dataset, and acknowledge that these terms do not encompass the full range of vocabulary used in describing gender. Further, we acknowledge that this task doesn’t represent a real-world use case, and is used only to demonstrate the technical details of the MinDiff library.
MinDiff works by penalizing the difference in distribution scores between examples in two sets of data. This guide will demonstrate how to choose and construct these additional MinDiff sets as well as how to package everything together so that it can be passed to a model for training.
##Setup
```
!pip install --upgrade tensorflow-model-remediation
import tensorflow as tf
from tensorflow_model_remediation import min_diff
from tensorflow_model_remediation.tools.tutorials_utils import uci as tutorials_utils
```
## Original Data
For demonstration purposes and to reduce runtimes, this guide uses only a sample fraction of the UCI Income dataset. In a real production setting, the full dataset would be utilized.
```
# Sampled at 0.3 for reduced runtimes.
train = tutorials_utils.get_uci_data(split='train', sample=0.3)
print(len(train), 'train examples')
```
### Converting to `tf.data.Dataset`
`MinDiffModel` requires that the input be a `tf.data.Dataset`. If you were using a different format of input prior to integrating MinDiff, you will have to convert your input data.
Use `tf.data.Dataset.from_tensor_slices` to convert to `tf.data.Dataset`.
```
dataset = tf.data.Dataset.from_tensor_slices((x, y, weights))
dataset.shuffle(...) # Optional.
dataset.batch(batch_size)
```
See [`Model.fit`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#fit) documentation for details on equivalences between the two methods of input.
In this guide, the input is downloaded as a Pandas DataFrame and therefore, needs this conversion.
```
# Function to convert a DataFrame into a tf.data.Dataset.
def df_to_dataset(dataframe, shuffle=True):
dataframe = dataframe.copy()
labels = dataframe.pop('target')
ds = tf.data.Dataset.from_tensor_slices((dict(dataframe), labels))
if shuffle:
ds = ds.shuffle(buffer_size=5000) # Reasonable but arbitrary buffer_size.
return ds
# Convert the train DataFrame into a Dataset.
original_train_ds = df_to_dataset(train)
```
Note: The training dataset has not been batched yet but it will be later.
## Creating MinDiff data
During training, MinDiff will encourage the model to reduce differences in predictions between two additional datasets (which may include examples from the original dataset). The selection of these two datasets is the key decision which will determine the effect MinDiff has on the model.
The two datasets should be picked such that the disparity in performance that you are trying to remediate is evident and well-represented. Since the goal is to reduce a gap in FNR between `"Male"` and `"Female"` slices, this means creating one dataset with only _positively_ labeled `"Male"` examples and another with only _positively_ labeled `"Female"` examples; these will be the MinDiff datasets.
Note: The choice of using only _positively_ labeled examples is directly tied to the target metric. This guide is concerned with _false negatives_ which, by definition, are _positively_ labeled examples that were incorrectly classified.
First, examine the data present.
```
female_pos = train[(train['sex'] == ' Female') & (train['target'] == 1)]
male_pos = train[(train['sex'] == ' Male') & (train['target'] == 1)]
print(len(female_pos), 'positively labeled female examples')
print(len(male_pos), 'positively labeled male examples')
```
It is perfectly acceptable to create MinDiff datasets from subsets of the original dataset.
While there aren't 5,000 or more positive `"Male"` examples as recommended in the [requirements guidance](https://www.tensorflow.org/responsible_ai/model_remediation/min_diff/guide/requirements#how_much_data_do_i_need), there are over 2,000 and it is reasonable to try with that many before collecting more data.
```
min_diff_male_ds = df_to_dataset(male_pos)
```
Positive `"Female"` examples, however, are much scarcer at 385. This is probably too small for good performance and so will require pulling in additional examples.
Note: Since this guide began by reducing the dataset via sampling, this problem (and the corresponding solution) may seem contrived. However, it serves as a good example of how to approach concerns about the size of your MinDiff datasets.
```
full_uci_train = tutorials_utils.get_uci_data(split='train')
augmented_female_pos = full_uci_train[((full_uci_train['sex'] == ' Female') &
(full_uci_train['target'] == 1))]
print(len(augmented_female_pos), 'positively labeled female examples')
```
Using the full dataset has more than tripled the number of examples that can be used for MinDiff. It’s still low but it is enough to try as a first pass.
```
min_diff_female_ds = df_to_dataset(augmented_female_pos)
```
Both the MinDiff datasets are significantly smaller than the recommended 5,000 or more examples. While it is reasonable to attempt to apply MinDiff with the current data, you may need to consider collecting additional data if you observe poor performance or overfitting during training.
### Using `tf.data.Dataset.filter`
Alternatively, you can create the two MinDiff datasets directly from the converted original `Dataset`.
Note: When using `.filter` it is recommended to use `.cache()` if the dataset can easily fit in memory for runtime performance. If it is too large to do so, consider storing your filtered datasets in your file system and reading them in.
```
# Male
def male_predicate(x, y):
return tf.equal(x['sex'], b' Male') and tf.equal(y, 0)
alternate_min_diff_male_ds = original_train_ds.filter(male_predicate).cache()
# Female
def female_predicate(x, y):
return tf.equal(x['sex'], b' Female') and tf.equal(y, 0)
full_uci_train_ds = df_to_dataset(full_uci_train)
alternate_min_diff_female_ds = full_uci_train_ds.filter(female_predicate).cache()
```
The resulting `alternate_min_diff_male_ds` and `alternate_min_diff_female_ds` will be equivalent in output to `min_diff_male_ds` and `min_diff_female_ds` respectively.
## Constructing your Training Dataset
As a final step, the three datasets (the two newly created ones and the original) need to be merged into a single dataset that can be passed to the model.
### Batching the datasets
Before merging, the datasets need to batched.
* The original dataset can use the same batching that was used before integrating MinDiff.
* The MinDiff datasets do not need to have the same batch size as the original dataset. In all likelihood, a smaller one will perform just as well. While they don't even need to have the same batch size as each other, it is recommended to do so for best performance.
While not strictly necessary, it is recommended to use `drop_remainder=True` for the two MinDiff datasets as this will ensure that they have consistent batch sizes.
Warning: The 3 datasets must be batched **before** they are merged together. Failing to do so will likely result in unintended input shapes that will cause errors downstream.
```
original_train_ds = original_train_ds.batch(128) # Same as before MinDiff.
# The MinDiff datasets can have a different batch_size from original_train_ds
min_diff_female_ds = min_diff_female_ds.batch(32, drop_remainder=True)
# Ideally we use the same batch size for both MinDiff datasets.
min_diff_male_ds = min_diff_male_ds.batch(32, drop_remainder=True)
```
### Packing the Datasets with `pack_min_diff_data`
Once the datasets are prepared, pack them into a single dataset which will then be passed along to the model. A single batch from the resulting dataset will contain one batch from each of the three datasets you prepared previously.
You can do this by using the provided `utils` function in the `tensorflow_model_remediation` package:
```
train_with_min_diff_ds = min_diff.keras.utils.pack_min_diff_data(
original_dataset=original_train_ds,
sensitive_group_dataset=min_diff_female_ds,
nonsensitive_group_dataset=min_diff_male_ds)
```
And that's it! You will be able to use other `util` functions in the package to unpack individual batches if needed.
```
for inputs, original_labels in train_with_min_diff_ds.take(1):
# Unpacking min_diff_data
min_diff_data = min_diff.keras.utils.unpack_min_diff_data(inputs)
min_diff_examples, min_diff_membership = min_diff_data
# Unpacking original data
original_inputs = min_diff.keras.utils.unpack_original_inputs(inputs)
```
With your newly formed data, you are now ready to apply MinDiff in your model! To learn how this is done, please take a look at the other guides starting with [Integrating MinDiff with MinDiffModel](./integrating_min_diff_with_min_diff_model).
### Using a Custom Packing Format (optional)
You may decide to pack the three datasets together in whatever way you choose. The only requirement is that you will need to ensure the model knows how to interpret the data. The default implementation of `MinDiffModel` assumes that the data was packed using `min_diff.keras.utils.pack_min_diff_data`.
One easy way to format your input as you want is to transform the data as a final step after you have used `min_diff.keras.utils.pack_min_diff_data`.
```
# Reformat input to be a dict.
def _reformat_input(inputs, original_labels):
unpacked_min_diff_data = min_diff.keras.utils.unpack_min_diff_data(inputs)
unpacked_original_inputs = min_diff.keras.utils.unpack_original_inputs(inputs)
return {
'min_diff_data': unpacked_min_diff_data,
'original_data': (unpacked_original_inputs, original_labels)}
customized_train_with_min_diff_ds = train_with_min_diff_ds.map(_reformat_input)
```
Your model will need to know how to read this customized input as detailed in the [Customizing MinDiffModel guide](./customizing_min_diff_model#customizing_default_behaviors_of_mindiffmodel).
```
for batch in customized_train_with_min_diff_ds.take(1):
# Customized unpacking of min_diff_data
min_diff_data = batch['min_diff_data']
# Customized unpacking of original_data
original_data = batch['original_data']
```
## Additional Resources
* For an in depth discussion on fairness evaluation see the [Fairness Indicators guidance](https://www.tensorflow.org/responsible_ai/fairness_indicators/guide/guidance)
* For general information on Remediation and MinDiff, see the [remediation overview](https://www.tensorflow.org/responsible_ai/model_remediation).
* For details on requirements surrounding MinDiff see [this guide](https://www.tensorflow.org/responsible_ai/model_remediation/min_diff/guide/requirements).
* To see an end-to-end tutorial on using MinDiff in Keras, see [this tutorial](https://www.tensorflow.org/responsible_ai/model_remediation/min_diff/tutorials/min_diff_keras).
## Utility Functions for other Guides
This guide outlines the process and decision making that you can follow whenever applying MinDiff. The rest of the guides build off this framework. To make this easier, logic found in this guide has been factored out into helper functions:
* `get_uci_data`: This function is already used in this guide. It returns a `DataFrame` containing the UCI income data from the indicated split sampled at whatever rate is indicated (100% if unspecified).
* `df_to_dataset`: This function converts a `DataFrame` into a `tf.data.Dataset` as detailed in this guide with the added functionality of being able to pass the batch_size as a parameter.
* `get_uci_with_min_diff_dataset`: This function returns a `tf.data.Dataset` containing both the original data and the MinDiff data packed together using the Model Remediation Library util functions as described in this guide.
Warning: These utility functions are **not** part of the official `tensorflow-model-remediation` package API and are subject to change at any time.
The rest of the guides will build off of these to show how to use other parts of the library.
|
github_jupyter
|
# VIME: Self/Semi Supervised Learning for Tabular Data
# Setup
```
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
import tensorflow as tf
import umap
from sklearn.metrics import (average_precision_score, mean_squared_error,
roc_auc_score)
from sklearn.model_selection import train_test_split
from tqdm import tqdm
from vime import VIME, VIME_Self
from vime_data import (
labelled_loss_fn, mask_generator_tf,
pretext_generator_tf, semi_supervised_generator,
to_vime_dataset, unlabelled_loss_fn
)
%matplotlib inline
%load_ext autoreload
%autoreload 2
plt.rcParams["figure.figsize"] = (20,10)
```
# Data
The example data is taken from [Kaggle](https://www.kaggle.com/c/ieee-fraud-detection) but it's already pre-processed and ready to be used. You can checkout the pre-processing notebook in the same folder to get some understanding about what transformations were done to the features.
```
train = pd.read_csv("fraud_train_preprocessed.csv")
test = pd.read_csv("fraud_test_preprocessed.csv")
# Drop nan columns as they are not useful for reconstruction error
nan_columns = [f for f in train.columns if 'nan' in f]
train = train.drop(nan_columns, axis=1)
test = test.drop(nan_columns, axis=1)
# Also, using only numerical columns because NNs have issue with one-hot encoding
num_cols = train.columns[:-125]
# Validation size is 10%
val_size = int(train.shape[0] * 0.1)
X_train = train.iloc[:-val_size, :]
X_val = train.iloc[-val_size:, :]
# Labelled 1% of data, everything else is unlabelled
X_train_labelled = train.sample(frac=0.01)
y_train_labelled = X_train_labelled.pop('isFraud')
X_val_labelled = X_val.sample(frac=0.01)
y_val_labelled = X_val_labelled.pop('isFraud')
X_train_unlabelled = X_train.loc[~X_train.index.isin(X_train_labelled.index), :].drop('isFraud', axis=1)
X_val_unlabelled = X_val.loc[~X_val.index.isin(X_val_labelled.index), :].drop('isFraud', axis=1)
X_train_labelled = X_train_labelled[num_cols]
X_val_labelled = X_val_labelled[num_cols]
X_train_unlabelled = X_train_unlabelled[num_cols]
X_val_unlabelled = X_val_unlabelled[num_cols]
X_val_labelled.shape, X_train_labelled.shape
print("Labelled Fraudsters", y_train_labelled.sum())
print(
"Labelled Proportion:",
np.round(X_train_labelled.shape[0] / (X_train_unlabelled.shape[0] + X_train_labelled.shape[0]), 5)
)
```
The following model will be trained with these hyperparameters:
```
vime_params = {
'alpha': 4,
'beta': 10,
'k': 5,
'p_m': 0.36
}
```
## Self-Supervised Learning
### Data Prep
The model needs 1 input - corrupted X, and 2 outputs - mask and original X.
```
batch_size = 1024
# Datasets
train_ds, train_m = to_vime_dataset(X_train_unlabelled, vime_params['p_m'], batch_size=batch_size, shuffle=True)
val_ds, val_m = to_vime_dataset(X_val_unlabelled, vime_params['p_m'], batch_size=batch_size)
num_features = X_train_unlabelled.shape[1]
print('Proportion Corrupted:', np.round(train_m.numpy().mean(), 2))
# Training
vime_s = VIME_Self(num_features)
vime_s.compile(
optimizer=tf.keras.optimizers.Adam(learning_rate=0.001),
loss={
'mask': 'binary_crossentropy',
'feature': 'mean_squared_error'},
loss_weights={'mask':1, 'feature': vime_params['alpha']}
)
cbs = [tf.keras.callbacks.EarlyStopping(
monitor="val_loss", patience=10, restore_best_weights=True
)]
vime_s.fit(
train_ds,
validation_data=val_ds,
epochs=1000,
callbacks=cbs
)
vime_s.save('./vime_self')
vime_s = tf.keras.models.load_model('./vime_self')
```
### Evaluation
All the evaluation will be done on the validation set
```
val_self_preds = vime_s.predict(val_ds)
```
To evaluate the mask reconstruction ability we can simply check the ROC AUC score for mask predictions across all the features.
```
feature_aucs = []
for i in tqdm(range(X_val_unlabelled.shape[1])):
roc = roc_auc_score(val_m.numpy()[:, i], val_self_preds['mask'][:, i])
feature_aucs.append(roc)
self_metrics = pd.DataFrame({"metric": 'mask_auc',
"metric_values": feature_aucs})
```
Now, we can evaluate the feature reconstruction ability using RMSE and correlation coefficients
```
feature_corrs = []
for i in tqdm(range(X_val_unlabelled.shape[1])):
c = np.corrcoef(X_val_unlabelled.values[:, i], val_self_preds['feature'][:, i])[0, 1]
feature_corrs.append(c)
self_metrics = pd.concat([
self_metrics,
pd.DataFrame({"metric": 'feature_correlation',
"metric_values": feature_corrs})
])
```
From the plot and table above, we can see that the model has learned to reconstruct most of the features. Half of the features are reconstructed with relatively strong correlation with original data. Only a handful of features are not properly reconstructed. Let's check the RMSE across all the features
```
rmses = []
for i in tqdm(range(X_val_unlabelled.shape[1])):
mse = mean_squared_error(X_val_unlabelled.values[:, i], val_self_preds['feature'][:, i])
rmses.append(np.sqrt(mse))
self_metrics = pd.concat([
self_metrics,
pd.DataFrame({"metric": 'RMSE',
"metric_values": rmses})
])
sns.boxplot(x=self_metrics['metric'], y=self_metrics['metric_values'])
plt.title("Self-Supervised VIME Evaluation")
```
RMSE distribution further indicates that mjority of the features are well-reconstructed.
Another way to evaluate the self-supervised model is to look at the embeddings. Since the whole point of corrupting the dataset is to learn to generate robust embeddings, we can assume that if a sample was corrupted 5 times, all 5 embeddings should be relatively close to each other in the vector space. Let's check this hypothesis by corrupting 10 different samples 5 times and projecting their embeddings to 2-dimensional space using UMAP.
```
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Input, Dense, Dropout
def generate_k_corrupted(x, k, p_m):
x_u_list = []
for i in range(k):
mask = mask_generator_tf(p_m, x)
_, x_corr = pretext_generator_tf(mask, tf.constant(x, dtype=tf.float32))
x_u_list.append(x_corr)
# Prepare input with shape (n, k, d)
x_u_corrupted = np.zeros((x.shape[0], k, x.shape[1]))
for i in range(x.shape[0]):
for j in range(k):
x_u_corrupted[i, j, :] = x_u_list[j][i, :]
return x_u_corrupted
vime_s = tf.keras.models.load_model('./vime_self')
# Sequential model to produce embeddings
encoding_model = Sequential(
[
Input(num_features),
vime_s.encoder
]
)
dense_model = Sequential(
[
Input(num_features),
Dense(num_features, activation="relu"),
]
)
# Create corrupted sample
samples = X_val_unlabelled.sample(10)
sample_corrupted = generate_k_corrupted(
x=samples,
k=5,
p_m=0.4
)
val_encoding = encoding_model.predict(sample_corrupted, batch_size=batch_size)
random_encoding = dense_model.predict(sample_corrupted, batch_size=batch_size)
fig, axs = plt.subplots(1, 2)
# Project corrupted samples
u = umap.UMAP(n_neighbors=5, min_dist=0.8)
corrupted_umap = u.fit_transform(val_encoding.reshape(-1, val_encoding.shape[2]))
sample_ids = np.array([np.repeat(i, 5) for i in range(10)]).ravel()
sns.scatterplot(corrupted_umap[:, 0], corrupted_umap[:, 1], hue=sample_ids, palette="tab10", ax=axs[0])
axs[0].set_title('VIME Embeddings of Corrupted Samples')
plt.legend(title='Sample ID')
# Project corrupted samples
u = umap.UMAP(n_neighbors=5, min_dist=0.8)
corrupted_umap = u.fit_transform(random_encoding.reshape(-1, random_encoding.shape[2]))
sample_ids = np.array([np.repeat(i, 5) for i in range(10)]).ravel()
sns.scatterplot(corrupted_umap[:, 0], corrupted_umap[:, 1], hue=sample_ids, palette="tab10", ax=axs[1])
axs[1].set_title('Not-trained Embeddings of Corrupted Samples')
plt.legend(title='Sample ID')
plt.show()
```
As you can see, the embeddings indeed put the same samples closer to each other, even though some of their values were corrupted. According to the authors, this means that the model has learned useful information about the feature correlations which can be helpful in the downstream tasks. Now, we can use this encoder in the next semi-supervised part.
## Semi-Supervised Learning
```
semi_batch_size = 512
num_features = X_train_unlabelled.shape[1]
```
Since we have different number of labelled and unlabelled examples we need to use generators. They will shuffle and select appropriate number of rows for each training iteration.
```
def train_semi_generator():
return semi_supervised_generator(
X_train_labelled.values,
X_train_unlabelled.values,
y_train_labelled.values,
bs=semi_batch_size
)
def val_semi_generator():
return semi_supervised_generator(
X_val_labelled.values,
X_val_unlabelled.values,
y_val_labelled.values,
bs=semi_batch_size
)
semi_train_dataset = tf.data.Dataset.from_generator(
train_semi_generator,
output_signature=(
tf.TensorSpec(shape=(semi_batch_size, num_features), dtype=tf.float32),
tf.TensorSpec(shape=(semi_batch_size), dtype=tf.float32),
tf.TensorSpec(shape=(semi_batch_size, num_features), dtype=tf.float32)
)
)
semi_val_dataset = tf.data.Dataset.from_generator(
val_semi_generator,
output_signature=(
tf.TensorSpec(shape=(semi_batch_size, num_features), dtype=tf.float32),
tf.TensorSpec(shape=(semi_batch_size), dtype=tf.float32),
tf.TensorSpec(shape=(semi_batch_size, num_features), dtype=tf.float32)
)
)
```
## Self Supervised VIME
```
def train_vime_semi(encoder, train_dataset, val_dataset, train_params, vime_params):
# Model
vime = VIME(encoder)
# Training parameters
iterations = train_params['iterations']
optimizer = tf.keras.optimizers.Adam(train_params['learning_rate'])
early_stop = train_params['early_stop']
# Set metrics to track
best_loss = 1e10
no_improve = 0
# Begining training loop
for it in range(iterations):
# Grab a batch for iteration
it_train = iter(train_dataset)
X_l, y_l, X_u = next(it_train)
# Generate unlabelled batch with k corrupted examples per sample
X_u_corrupted = generate_k_corrupted(X_u, vime_params['k'], vime_params['p_m'])
with tf.GradientTape() as tape:
# Predict labelled & unlabelled
labelled_preds = vime(X_l)
unlabelled_preds = vime(X_u_corrupted)
# Calculate losses
labelled_loss = labelled_loss_fn(y_l, labelled_preds)
unlabelled_loss = unlabelled_loss_fn(unlabelled_preds)
# Total loss
semi_supervised_loss = unlabelled_loss + vime_params['beta'] * labelled_loss
if it % 10 == 0:
val_iter_losses = []
print(f"\nMetrics for Iteration {it}")
for i in range(5):
# Grab a batch
it_val = iter(val_dataset)
X_l_val, y_l_val, X_u_val = next(it_val)
# Generate unlabelled batch with k corrupted examples per sample
X_u_corrupted = generate_k_corrupted(X_u_val, vime_params['k'], vime_params['p_m'])
# Predict labelled & unlabelled
labelled_preds_val = vime(X_l_val)
unlabelled_preds_val = vime(X_u_corrupted)
# Calculate losses
labelled_loss_val = labelled_loss_fn(y_l_val, labelled_preds_val)
unlabelled_loss_val = unlabelled_loss_fn(unlabelled_preds_val)
semi_supervised_loss_val = unlabelled_loss_val + vime_params['beta'] * labelled_loss_val
val_iter_losses.append(semi_supervised_loss_val)
# Average loss over 5 validation iterations
semi_supervised_loss_val = np.mean(val_iter_losses)
print(f"Train Loss {np.round(semi_supervised_loss, 5)}, Val Loss {np.round(semi_supervised_loss_val, 5)}")
# Update metrics if val_loss is better
if semi_supervised_loss_val < best_loss:
best_loss = semi_supervised_loss_val
no_improve = 0
vime.save('./vime')
else:
no_improve += 1
print(f"Validation loss not improved {no_improve} times")
# Early stopping
if no_improve == early_stop:
break
# Update weights
grads = tape.gradient(semi_supervised_loss, vime.trainable_weights)
optimizer.apply_gradients(zip(grads, vime.trainable_weights))
vime = tf.keras.models.load_model('./vime')
return vime
train_params = {
'num_features': num_features,
'iterations': 1000,
'early_stop': 20,
'learning_rate': 0.001
}
vime_self = tf.keras.models.load_model('./vime_self')
vime_semi = train_vime_semi(
encoder = vime_self.encoder,
train_dataset = semi_train_dataset,
val_dataset = semi_val_dataset,
train_params = train_params,
vime_params = vime_params
)
test_ds = tf.data.Dataset.from_tensor_slices(test[num_cols]).batch(batch_size)
vime_tuned_preds = vime_semi.predict(test_ds)
pr = average_precision_score(test['isFraud'], vime_tuned_preds)
print(pr)
```
## Evaluation
Re-training the model 10 times to get distribution of PR AUC scores.
```
vime_prs = []
test_ds = tf.data.Dataset.from_tensor_slices(test[num_cols]).batch(batch_size)
for i in range(10):
train_params = {
'num_features': num_features,
'iterations': 1000,
'early_stop': 10,
'learning_rate': 0.001
}
vime_self = tf.keras.models.load_model('./vime_self')
vime_self.encoder.trainable = False
vime_semi = train_vime_semi(
encoder = vime_self.encoder,
train_dataset = semi_train_dataset,
val_dataset = semi_val_dataset,
train_params = train_params,
vime_params = vime_params
)
# fine-tune
vime_semi = tf.keras.models.load_model('./vime')
vime_semi.encoder.trainable
vime_tuned_preds = vime_semi.predict(test_ds)
pr = average_precision_score(test['isFraud'], vime_tuned_preds)
vime_prs.append(pr)
print('VIME Train', i, "PR AUC:", pr)
```
### Compare with MLP and RF
```
mlp_prs = []
for i in range(10):
base_mlp = Sequential([
Input(shape=num_features),
Dense(num_features),
Dense(128),
Dropout(0.2),
Dense(128),
Dropout(0.2),
Dense(1, activation='sigmoid')
])
base_mlp.compile(
optimizer=tf.keras.optimizers.Adam(learning_rate=0.0001),
loss='binary_crossentropy'
)
# Early stopping based on validation loss
cbs = [tf.keras.callbacks.EarlyStopping(
monitor="val_loss", patience=20, restore_best_weights=True
)]
base_mlp.fit(
x=X_train_labelled.values,
y=y_train_labelled,
validation_data=(X_val_labelled.values, y_val_labelled),
epochs=1000,
callbacks=cbs
)
base_mlp_preds = base_mlp.predict(test_ds)
mlp_prs.append(average_precision_score(test['isFraud'], base_mlp_preds))
from lightgbm import LGBMClassifier
train_tree_X = pd.concat([X_train_labelled, X_val_labelled])
train_tree_y = pd.concat([y_train_labelled, y_val_labelled])
rf_prs = []
for i in tqdm(range(10)):
rf = RandomForestClassifier(max_depth=4)
rf.fit(train_tree_X.values, train_tree_y)
rf_preds = rf.predict_proba(test[X_train_labelled.columns])
rf_prs.append(average_precision_score(test['isFraud'], rf_preds[:, 1]))
metrics_df = pd.DataFrame({"MLP": mlp_prs,
"VIME": vime_prs,
"RF": rf_prs})
metrics_df.boxplot()
plt.ylabel("PR AUC")
plt.show()
metrics_df.describe()
```
|
github_jupyter
|
```
JSON_PATH = 'by-article-train_attn-data.json'
from json import JSONDecoder
data = JSONDecoder().decode(open(JSON_PATH).read())
word = 'Sponsored'
hyper_count = dict()
main_count = dict()
for i, article in enumerate(data):
if word in article['normalizedText'][-1]:
energies = [e for w, e in article['activations'][-1][0] if w == word]
if article['hyperpartisan'] == 'true':
hyper_count[i] = {
'energies': energies,
'truth': article['hyperpartisan'],
'prediction': article['prediction'],
'pred_value': article['pred_value'],
'last_sent_e': article['activations'][-1][-1],
}
elif article['hyperpartisan'] == 'false':
main_count[i] = {
'energies': energies,
'truth': article['hyperpartisan'],
'prediction': article['prediction'],
'pred_value': article['pred_value'],
'last_sent_e': article['activations'][-1][-1],
}
else:
raise RuntimeError('json format invalid')
# Average word energy of 1st 'Sponsored' tag
avg_final_e = [el['energies'][0] * el['last_sent_e'] for el in hyper_count.values()]
print('AVG:', sum(avg_final_e) / len(avg_final_e))
avg_final_e
# Average final energy of 1st 'Sponsored' tag (word_e * sentence_e)
avg_final_e = [el['energies'][0] * el['last_sent_e'] for el in hyper_count.values()]
print('AVG:', sum(avg_final_e) / len(avg_final_e))
avg_final_e
### ### ###
hyper_articles = [el for el in data if el['hyperpartisan'] == 'true']
main_articles = [el for el in data if el['hyperpartisan'] == 'false']
assert len(hyper_articles) + len(main_articles) == len(data)
hyper_sent_att = [activ[-1] for a in hyper_articles for activ in a['activations']]
main_sent_att = [activ[-1] for a in main_articles for activ in a['activations']]
import seaborn as sns
import matplotlib.pyplot as plt
sns.distplot(hyper_sent_att, hist=False, rug=False, label="hyperpartisan")
sns.distplot(main_sent_att, hist=False, rug=False, label="mainstream")
plt.gcf().savefig('imgs/sentence_energy_distribution.png', dpi=400)
plt.show()
## Describe distribution
from scipy import stats
print('Hyperpartisan Sentence Energy distribution:')
print(stats.describe(hyper_sent_att), end='\n\n')
print('Mainstream Sentence Energy distribution:')
print(stats.describe(main_sent_att), end='\n\n')
## Average attention on most important sentence
hyper_most_imp_sent = [max(activ[-1] for activ in a['activations']) for a in hyper_articles]
main_most_imp_sent = [max(activ[-1] for activ in a['activations']) for a in main_articles]
print('Avg Hyperpartisan:', sum(hyper_most_imp_sent) / len(hyper_most_imp_sent))
print('Avg Mainstream:', sum(main_most_imp_sent) / len(main_most_imp_sent))
sns.distplot(hyper_most_imp_sent, hist=False, rug=False, label="hyperpartisan")
sns.distplot(main_most_imp_sent, hist=False, rug=False, label="mainstream")
plt.gcf().savefig('imgs/most_important_sentence_energy_distribution.png', dpi=400)
plt.show()
## Number of sentences with attention above a given threshold of importance
THRESHOLD = 0.3
hyper_important_sentences = [sum(1 for activ in a['activations'] if activ[-1] > THRESHOLD) for a in hyper_articles]
main_important_sentences = [sum(1 for activ in a['activations'] if activ[-1] > THRESHOLD) for a in main_articles]
print('Average number of sentences above {}:'.format(THRESHOLD))
print('\thyperpartisan: {}'.format(sum(hyper_important_sentences) / len(hyper_important_sentences)))
print('\tmainstream: {}'.format(sum(main_important_sentences) / len(main_important_sentences)))
### ### ###
## Calculating statistical significance that the two distributions are distinct
## Welch's t-test: https://en.wikipedia.org/wiki/Welch%27s_t-test
t_val, p_val = stats.ttest_ind(hyper_sent_att, main_sent_att, equal_var=False)
print('p-value for the hypothesis that the two distributions have equal mean:', p_val)
## Statistical significance of hypothesis:
## attention of most important sentence of a mainstream article is larger than that of a hyperpartisan article
from statsmodels.stats import weightstats as stests
_, p_val = stests.ztest(hyper_most_imp_sent, main_most_imp_sent, value=0)
print(p_val)
```
|
github_jupyter
|
```
import detectron2
from detectron2.utils.logger import setup_logger
setup_logger()
import numpy as np
import random
from detectron2.engine import DefaultPredictor
from detectron2.config import get_cfg
from detectron2.utils.visualizer import Visualizer
from detectron2.data import MetadataCatalog
from detectron2.modeling import build_model
from detectron2.evaluation import COCOEvaluator,PascalVOCDetectionEvaluator
import matplotlib.pyplot as plt
import torch.tensor as tensor
from detectron2.data import build_detection_test_loader
from detectron2.evaluation import inference_on_dataset
import torch
from detectron2.structures.instances import Instances
from detectron2.modeling import build_model
from detectron2.modeling.meta_arch.tracker import Tracker
from detectron2.modeling.meta_arch.soft_tracker import SoftTracker
%matplotlib inline
```
## Loading Weights
```
cfg = get_cfg()
cfg.merge_from_file("../configs/COCO-Detection/faster_rcnn_R_50_FPN_3x_Video.yaml")
cfg.MODEL.ROI_HEADS.SCORE_THRESH_TEST = 0.4 # set threshold for this model
cfg.MODEL.WEIGHTS = '/media/DATA/Users/Issa/models_pub/kitti_jde.pth'
#cfg.MODEL.WEIGHTS = "../models_pub/kitti_jde.pth"
print(cfg.MODEL)
```
## functions to validate annotated data using devkit_tracking from KITTI
```
from contextlib import contextmanager
import sys, os
@contextmanager
def suppress_stdout():
with open(os.devnull, "w") as devnull:
old_stdout = sys.stdout
sys.stdout = devnull
try:
yield
finally:
sys.stdout = old_stdout
def print_val_results(results_name):
with suppress_stdout():
print("Now you don't")
os.system('python2 /home/issa/devkit_tracking/python/validate_tracking.py val')
labels = {1:'MOTA',2:'MOTP',3:'MOTAL',4:'MODA',5:'MODP',7:'R',8:'P',12:'MT',13:'PT',14:'ML',18:'FP',19:'FN',22:'IDs'}
summary_heading = 'Metric\t'
for label in labels.keys():
summary_heading+=labels[label] + '\t'
summary_cars = 'Cars\t'
summary_peds = 'Peds\t'
with open('/home/issa/devkit_tracking/python/results/'+results_name+'/summary_car.txt') as f:
i=0
for line in f:
if(i==0):
i+=1
continue
if(i in labels.keys()):
summary_cars+= str(round(float(line[len(line)-9:len(line)-1].strip()),2))+'\t'
i+=1
print(summary_heading)
print(summary_cars)
def print_test_results(results_name):
#with suppress_stdout():
print("Now you don't")
os.system('python2 ../devkit_tracking/python/evaluate_tracking.py test')
labels = {1:'MOTA',2:'MOTP',3:'MOTAL',4:'MODA',5:'MODP',7:'R',8:'P',12:'MT',13:'PT',14:'ML',18:'FP',19:'FN',22:'IDs'}
summary_heading = 'Metric\t'
for label in labels.keys():
summary_heading+=labels[label] + '\t'
summary_cars = 'Cars\t'
summary_peds = 'Peds\t'
with open('../devkit_tracking/python/results/'+results_name+'/summary_car.txt') as f:
i=0
for line in f:
if(i==0):
i+=1
continue
if(i in labels.keys()):
summary_cars+= str(round(float(line[len(line)-9:len(line)-1].strip()),2))+'\t'
i+=1
print(summary_heading)
print(summary_cars)
```
## Inference : Joint Detection and Tracking
```
import json
import os
import cv2 as cv2
import time
from tqdm.notebook import tqdm
colors = [[0,0,128],[0,255,0],[0,0,255],[255,0,0],[0,128,128],[128,0,128],[128,128,0],[255,255,0],[0,255,255],[255,255,0],[128,0,0],[0,128,0]
,[0,128,255],[0,255,128],[255,0,128],[128,255,0],[255,128,0],[128,255,255],[128,0,255],[128,128,128],[128,255,128]]
#dirC = '/../datasets/KITTI/tracking/data_tracking_image_2/training/image_02/'
dirC = '/media/DATA/Datasets/KITTI/tracking/data_tracking_image_2/training/image_02/'
#dirDets = '../datasets/KITTI/tracking/data_tracking_det_2_lsvm/training/det_02/'
names = []
arr = {2:'Car'}
if(not os.path.exists("../results")):
os.mkdir('../results')
os.mkdir('../results/KITTI')
else:
if(not os.path.exists("../results/KITTI")):
os.mkdir('../results/KITTI')
output_path = '/home/issa/devkit_tracking/python/results'
settings = [
dict(props=20, #number of proposals to use by rpn
st=1.05, #acceptance distance percentage for soft tracker
sup_fp = True, # fp suppression based on Intersection over Union for new detections
alpha = 0.6, # the percentage of the new embedding in track embedding update (emb = alpha * emb(t) +(1-alpha) emb(t-1))
fp_thresh=0.95, # iou threshold above which the new detection is considered a fp
T=True, #use past tracks as proposals
D='cosine', # distance metric for embeddings
Re=True, #use the embedding head
A=True, # use appearance information
K=True, # use kalman for motion prediction
E=False, #use raw FPN features as appearance descriptors
measurement=0.001, #measruement noise for the kalman filter
process=1, #process noise for the kalman filter
dist_thresh=1.5, # the normalization factor for the appearance distance
track_life=7, #frames for which a track is kept in memory without an update
track_vis=2, #frames for which a track is displayed without an update
),
]
train_folders = ['0000','0002','0003','0004','0005','0009','0011','0017','0020']
val_folders = ['0001','0006','0008','0016','0018','0019']
test_folders = ['0014','0015','0016','0018','0019','0001','0006','0008','0010','0012','0013']
submission_folders = ['0000','0001','0002','0003','0004','0005','0006','0007',
'0008','0009','0010','0011','0012','0013','0014','0015','0016','0017',
'0018','0019','0020','0021','0022','0023','0024','0025','0026','0027','0028']
final_test_folders = ['0014']
for setting in settings:
test_name = 'val'
exp_name = output_path+ '/'+test_name
if(not os.path.exists(exp_name)):
os.mkdir(exp_name)
os.mkdir(exp_name+'/data')
avg=0
for folder_name in val_folders:
dets = {}
public_det=False
if public_det==True:
with open(dirDets+folder_name+'.txt') as det_file:
for line in det_file:
parts = line.split(' ')
if(parts[0] not in dets):
dets[parts[0]] = []
if(parts[2] =='Car' and float(parts[17])>-1):
dets[parts[0]].append([float(parts[6])
,float(parts[7]),float(parts[8])
,float(parts[9]),float(parts[6]),float(parts[17])])
predictor = DefaultPredictor(cfg,True)
predictor.model.tracker = Tracker()
predictor.model.tracking_proposals = setting['T']
predictor.model.tracker.track_life = setting['track_life']
predictor.model.tracker.track_visibility = setting['track_vis']
predictor.model.tracker.use_appearance = setting['A']
predictor.model.tracker.use_kalman = setting['K']
predictor.model.tracker.embed = setting['E']
predictor.model.tracker.reid = setting['Re']
predictor.model.tracker.dist = setting['D']
predictor.model.tracker.measurement_noise=setting['measurement']
predictor.model.tracker.process_noise = setting['process']
predictor.model.tracker.dist_thresh = setting['dist_thresh']
predictor.model.use_reid = setting['Re']
predictor.model.tracker.soft_thresh = setting['st']
predictor.model.tracker.suppress_fp = setting['sup_fp']
predictor.model.tracker.fp_thresh = setting['fp_thresh']
predictor.model.tracker.embed_alpha = setting['alpha']
max_distance = 0.2
output_file = open('%s/data/%s.txt'%(exp_name,folder_name),'w')
frames = {}
frame_counter = 0
prev_path = 0
elapsed = 0
predictor.model.prev_path = 0
for photo_name in sorted(os.listdir(dirC+folder_name+'/')):
frames[frame_counter] = {}
img_path = dirC+folder_name+'/'+photo_name
img = cv2.imread(img_path)
inp = {}
inp['width'] = img.shape[1]
inp['height'] = img.shape[0]
inp['file_name'] = photo_name
inp['image_id'] = photo_name
predictor.model.photo_name = img_path
start = time.time()
outputs = predictor(img,setting['props'])
end = time.time()
elapsed +=(end-start)
for i in outputs:
if(i.pred_class in arr):
output_file.write("%d %d %s 0 0 -0.20 %d %d %d %d 1.89 0.48 1.20 1.84 1.47 8.41 0.01 %f\n"%(frame_counter
,i.track_id,arr[i.pred_class],i.xmin,i.ymin,i.xmax,i.ymax,i.conf))
frame_counter +=1
predictor.model.prev_path = img_path
avg += (frame_counter/elapsed)
output_file.close()
print(setting)
print('avg_time :',avg/len(val_folders))
print_val_results(test_name)
```
|
github_jupyter
|
# Amazon SageMaker로 다중 노드들 간 분산 RL을 이용해 Roboschool 에이전트 훈련
---
이 노트북은 `rl_roboschool_ray.ipynb` 의 확장으로, Ray와 TensorFlow를 사용한 강화 학습의 수평(horizontal) 스케일링을 보여줍니다.
## 해결해야 할 Roboschool 문제 선택
Roboschool은 가상 로봇 시스템에 대한 RL 정책을 훈련시키는 데 주로 사용되는 [오픈 소스](https://github.com/openai/roboschool/tree/master/roboschool) 물리 시뮬레이터입니다. Roboschool은 다양한 로봇 문제에 해당하는 [다양한](https://github.com/openai/roboschool/blob/master/roboschool/__init__.py) gym 환경을 정의합니다. 아래는 다양한 난이도 중 몇 가지를 보여줍니다.
- **Reacher (쉬움)** - 2개의 조인트만 있는 매우 간단한 로봇이 목표물에 도달합니다.
- **호퍼 (중간)** - 한쪽 다리와 발이 달린 간단한 로봇이 트랙을 뛰어 내리는 법을 배웁니다.
- **휴머노이드 (어려움)** - 두 개의 팔, 두 개의 다리 등이 있는 복잡한 3D 로봇은 넘어지지 않고 균형을 잡은 다음 트랙에서 달리는 법을 배웁니다.
간단한 문제들은 적은 계산 리소스 상에서 더 빨리 훈련됩니다. 물론 더 복잡한 문제들은 훈련이 느리지만 더 재미있습니다.
```
# Uncomment the problem to work on
#roboschool_problem = 'reacher'
#roboschool_problem = 'hopper'
roboschool_problem = 'humanoid'
```
## 전제 조건(Pre-requisites)
### 라이브러리 임포트
시작하기 위해, 필요한 Python 라이브러리를 가져와서 권한 및 구성을 위한 몇 가지 전제 조건으로 환경을 설정합니다.
```
import sagemaker
import boto3
import sys
import os
import glob
import re
import subprocess
from IPython.display import HTML, Markdown
import time
from time import gmtime, strftime
sys.path.append("common")
from misc import get_execution_role, wait_for_s3_object
from docker_utils import build_and_push_docker_image
from sagemaker.rl import RLEstimator, RLToolkit, RLFramework
from markdown_helper import generate_help_for_s3_endpoint_permissions, create_s3_endpoint_manually
```
### S3 버킷 설정
체크포인트(checkpoint) 및 메타데이터에 사용하려는 S3 버킷에 대한 연결 및 인증을 설정합니다.
```
sage_session = sagemaker.session.Session()
s3_bucket = sage_session.default_bucket()
s3_output_path = 's3://{}/'.format(s3_bucket)
print("S3 bucket path: {}".format(s3_output_path))
```
### 변수 설정
훈련 작업의 작업 접두사(job prefix)와 *컨테이너의 이미지 경로(BYOC 인 경우에만)와 같은 변수*를 정의합니다.
```
# create a descriptive job name
job_name_prefix = 'rl-roboschool-distributed-' + roboschool_problem
aws_region = boto3.Session().region_name
```
### 훈련이 진행되는 위치 구성
SageMaker 노트북 인스턴스 또는 로컬 노트북 인스턴스를 사용하여 RL 훈련 작업을 훈련할 수 있습니다. 로컬 모드는 SageMaker Python SDK를 사용하여 SageMaker에 배포하기 전에 로컬 컨테이너에서 코드를 실행합니다. 이렇게 하면 , 익숙한 Python SDK 인터페이스를 사용하면서 반복 테스트 및 디버깅 속도를 높일 수 있습니다. 여러분은 `local_mode = True` 만 설정하면 됩니다.
```
# run in local_mode on this machine, or as a SageMaker TrainingJob?
local_mode = False
if local_mode:
instance_type = 'local'
else:
# If on SageMaker, pick the instance type
instance_type = "ml.c5.2xlarge"
train_instance_count = 3
```
### IAM 역할 생성
SageMaker 노트북 `role = sagemaker.get_execution_role()`을 실행할 때 실행 역할(execution role)을 얻거나 로컬 시스템에서 실행할 때 utils 메소드 `role = get_execution_role()`을 사용하여 실행 역할을 작성하세요.
```
try:
role = sagemaker.get_execution_role()
except:
role = get_execution_role()
print("Using IAM role arn: {}".format(role))
```
### `로컬` 모드용 도커 설치
로컬 모드에서 작업하려면 도커(docker)가 설치되어 있어야 합니다. 로컬 머신에서 실행할 때는 docker 또는 docker-compose(로컬 CPU 머신의 경우) 및 nvidia-docker(로컬 GPU 머신의 경우)가 설치되어 있는지 확인하세요. 또는, SageMaker 노트북 인스턴스에서 실행할 때 다음 스크립트를 실행하여 관련 패키지들을 설치할 수 있습니다.
참고로, 한 번에 하나의 로컬 노트북만 실행할 수 있습니다.
```
# only run from SageMaker notebook instance
if local_mode:
!/bin/bash ./common/setup.sh
```
## 도커 컨테이너 빌드
Roboschool이 설치된 사용자 정의 도커 컨테이너를 빌드해야 합니다. 컨테이너 빌드 작업은 아래 과정을 거쳐 처리됩니다.
1. 기본 컨테이너 이미지 가져오기
2. Roboschool 및 의존성 패키지 설치
3. 새 컨테이너 이미지를 ECR에 업로드
인터넷 연결이 느린 컴퓨터에서 실행 중인 경우, 이 단계에서 시간이 오래 걸릴 수 있습니다. 노트북 인스턴스가 SageMaker 또는 EC2 인 경우 인스턴스 유형에 따라 3-10 분이 걸립니다.
```
%%time
cpu_or_gpu = 'gpu' if instance_type.startswith('ml.p') else 'cpu'
repository_short_name = "sagemaker-roboschool-ray-%s" % cpu_or_gpu
docker_build_args = {
'CPU_OR_GPU': cpu_or_gpu,
'AWS_REGION': boto3.Session().region_name,
}
custom_image_name = build_and_push_docker_image(repository_short_name, build_args=docker_build_args)
print("Using ECR image %s" % custom_image_name)
```
## 훈련 코드 작성
훈련 코드는 `/src` 디렉토리에 업로드된 `“train-{roboschool_problem}.py”` 파일에 작성됩니다. 먼저 환경 파일과 사전 설정 파일을 가져온 다음, `main()` 함수를 정의하세요.
```
!pygmentize src/train-{roboschool_problem}.py
```
## Ray 동종 스케일링 - train_instance_count > 1 지정
동종(Homogeneous) 스케일링을 통해 동일한 유형의 여러 인스턴스를 사용할 수 있습니다.
```
metric_definitions = RLEstimator.default_metric_definitions(RLToolkit.RAY)
estimator = RLEstimator(entry_point="train-%s.py" % roboschool_problem,
source_dir='src',
dependencies=["common/sagemaker_rl"],
image_name=custom_image_name,
role=role,
train_instance_type=instance_type,
train_instance_count=train_instance_count,
output_path=s3_output_path,
base_job_name=job_name_prefix,
metric_definitions=metric_definitions,
hyperparameters={
# Attention scientists! You can override any Ray algorithm parameter here:
# 3 m4.2xl with 8 cores each. We have to leave 1 core for ray scheduler.
# Don't forget to change this on the basis of instance type.
"rl.training.config.num_workers": (8 * train_instance_count) - 1
#"rl.training.config.horizon": 5000,
#"rl.training.config.num_sgd_iter": 10,
}
)
estimator.fit(wait=local_mode)
job_name = estimator.latest_training_job.job_name
print("Training job: %s" % job_name)
```
## 시각화
RL 훈련에는 시간이 오래 걸릴 수 있습니다. 따라서 훈련 작업이 동작하는 동안 훈련 작업의 진행 상황을 추적할 수 있는 다양한 방법들이 있습니다. 훈련 도중 일부 중간 출력이 S3에 저장되므로, 이를 캡처하도록 설정합니다.
```
print("Job name: {}".format(job_name))
s3_url = "s3://{}/{}".format(s3_bucket,job_name)
if local_mode:
output_tar_key = "{}/output.tar.gz".format(job_name)
else:
output_tar_key = "{}/output/output.tar.gz".format(job_name)
intermediate_folder_key = "{}/output/intermediate/".format(job_name)
output_url = "s3://{}/{}".format(s3_bucket, output_tar_key)
intermediate_url = "s3://{}/{}".format(s3_bucket, intermediate_folder_key)
print("S3 job path: {}".format(s3_url))
print("Output.tar.gz location: {}".format(output_url))
print("Intermediate folder path: {}".format(intermediate_url))
tmp_dir = "/tmp/{}".format(job_name)
os.system("mkdir {}".format(tmp_dir))
print("Create local folder {}".format(tmp_dir))
```
### 훈련 롤아웃 비디오 가져오기
특정 롤아웃의 비디오는 훈련 중 S3에 기록됩니다. 여기에서는 S3에서 마지막 10개의 비디오 클립을 가져 와서 마지막 비디오를 렌더링합니다.
```
recent_videos = wait_for_s3_object(s3_bucket, intermediate_folder_key, tmp_dir,
fetch_only=(lambda obj: obj.key.endswith(".mp4") and obj.size>0), limit=10)
last_video = sorted(recent_videos)[-1] # Pick which video to watch
os.system("mkdir -p ./src/tmp_render_homogeneous/ && cp {} ./src/tmp_render_homogeneous/last_video.mp4".format(last_video))
HTML('<video src="./src/tmp_render_homogeneous/last_video.mp4" controls autoplay></video>')
```
### 훈련 작업에 대한 지표 plot
CloudWatch 지표에 기록된 알고리즘 지표를 사용하여 실행 중인 훈련의 보상 지표를 볼 수 있습니다. 시간이 지남에 따라, 모델의 성능을 볼 수 있도록 이를 plot할 수 있습니다.
```
%matplotlib inline
from sagemaker.analytics import TrainingJobAnalytics
df = TrainingJobAnalytics(job_name, ['episode_reward_mean']).dataframe()
num_metrics = len(df)
if num_metrics == 0:
print("No algorithm metrics found in CloudWatch")
else:
plt = df.plot(x='timestamp', y='value', figsize=(12,5), legend=True, style='b-')
plt.set_ylabel('Mean reward per episode')
plt.set_xlabel('Training time (s)')
```
### 훈련 진행 상황 모니터링
위의 시각화 셀을 반복해서 실행하여 최신 비디오를 얻거나, 훈련 작업이 진행됨에 따라 최신 지표를 볼 수 있습니다.
## Ray 이기종(heterogeneous) 스케일링
RL 훈련을 확장하기 위해 롤아웃 작업자 수를 늘릴 수 있습니다. 그러나, 롤아웃이 많을수록 훈련 중 종종 병목 현상이 발생할 수 있습니다. 이를 방지하기 위해 하나 이상의 GPU가 있는 인스턴스를 훈련용으로 사용하고 여러 개의 CPU 인스턴스들을 롤아웃에 사용할 수 있습니다.
SageMaker는 훈련 작업에서 단일 유형의 인스턴스를 지원하므로, 두 개의 SageMaker 작업을 서로 통신하도록 함으로써 위의 목표를 달성할 수 있습니다. 이름 지정을 위해 `기본 클러스터(Primary cluster)`를 사용하여 하나 이상의 GPU 인스턴스를 참조하고 `보조 클러스터(Secondary cluster)`를 사용하여 CPU 인스턴스 클러스터를 참조합니다.
> local_mode는 이 유형의 스케일링을 테스트하는 데 사용할 수 없습니다.
SageMaker 작업을 구성하기 전에 먼저 VPC 모드에서 SageMaker를 실행해야 합니다. VPC 모드에서는 두 SageMaker 작업이 네트워크를 통해 통신할 수 있습니다.
작업 시작 스크립트에 서브넷(subnet)과 보안 그룹(security group)을 제공하면 됩니다. 이 예에서는 기본 VPC 구성을 사용합니다.
```
ec2 = boto3.client('ec2')
default_vpc = [vpc['VpcId'] for vpc in ec2.describe_vpcs()['Vpcs'] if vpc["IsDefault"] == True][0]
default_security_groups = [group["GroupId"] for group in ec2.describe_security_groups()['SecurityGroups'] \
if group["GroupName"] == "default" and group["VpcId"] == default_vpc]
default_subnets = [subnet["SubnetId"] for subnet in ec2.describe_subnets()["Subnets"] \
if subnet["VpcId"] == default_vpc and subnet['DefaultForAz']==True]
print("Using default VPC:", default_vpc)
print("Using default security group:", default_security_groups)
print("Using default subnets:", default_subnets)
```
VPC 모드에서 실행 중인 SageMaker 작업은 S3 리소스에 액세스할 수 없습니다. 따라서, SageMaker 컨테이너에서 S3에 액세스할 수 있도록 VPC S3 엔드포인트를 생성해야 합니다. VPC 모드에 대한 자세한 내용을 보려면 [이 링크](https://docs.aws.amazon.com/sagemaker/latest/dg/train-vpc.html)를 방문하세요.
```
try:
route_tables = [route_table["RouteTableId"] for route_table in ec2.describe_route_tables()['RouteTables']\
if route_table['VpcId'] == default_vpc]
except Exception as e:
if "UnauthorizedOperation" in str(e):
display(Markdown(generate_help_for_s3_endpoint_permissions(role)))
else:
display(Markdown(create_s3_endpoint_manually(aws_region, default_vpc)))
raise e
print("Trying to attach S3 endpoints to the following route tables:", route_tables)
assert len(route_tables) >= 1, "No route tables were found. Please follow the VPC S3 endpoint creation "\
"guide by clicking the above link."
try:
ec2.create_vpc_endpoint(DryRun=False,
VpcEndpointType="Gateway",
VpcId=default_vpc,
ServiceName="com.amazonaws.{}.s3".format(aws_region),
RouteTableIds=route_tables)
print("S3 endpoint created successfully!")
except Exception as e:
if "RouteAlreadyExists" in str(e):
print("S3 endpoint already exists.")
elif "UnauthorizedOperation" in str(e):
display(Markdown(generate_help_for_s3_endpoint_permissions(role)))
raise e
else:
display(Markdown(create_s3_endpoint_manually(aws_region, default_vpc)))
raise e
```
### 인스턴스 유형 구성
1 Volta (V100) GPU와 40개의 CPU 코어로 클러스터를 구성해 보겠습니다. ml.p3.2xlarge에는 8개의 CPU 코어가 있고 ml.c5.4xlarge에는 16개의 CPU 코어가 있으므로 1개의 ml.p3.2xlarge 인스턴스와 2개의 ml.c5.4xlarge 인스턴스를 사용하여 이 작업을 수행할 수 있습니다.
```
%%time
# Build CPU image
cpu_repository_short_name = "sagemaker-roboschool-ray-%s" % "cpu"
docker_build_args = {
'CPU_OR_GPU': "cpu",
'AWS_REGION': boto3.Session().region_name,
}
cpu_image_name = build_and_push_docker_image(repository_short_name, build_args=docker_build_args)
print("Using CPU ECR image %s" % cpu_image_name)
# Build GPU image
gpu_repository_short_name = "sagemaker-roboschool-ray-%s" % "gpu"
docker_build_args = {
'CPU_OR_GPU': "gpu",
'AWS_REGION': boto3.Session().region_name,
}
gpu_image_name = build_and_push_docker_image(repository_short_name, build_args=docker_build_args)
print("Using GPU ECR image %s" % gpu_image_name)
primary_cluster_instance_type = "ml.p3.2xlarge"
primary_cluster_instance_count = 1
secondary_cluster_instance_type = "ml.c5.4xlarge"
secondary_cluster_instance_count = 2
total_cpus = 40 - 1 # Leave one for ray scheduler
total_gpus = 1
primary_cluster_instance_type = "ml.p3.16xlarge"
primary_cluster_instance_count = 1
secondary_cluster_instance_type = "ml.c5.4xlarge"
secondary_cluster_instance_count = 2
total_cpus = 40 - 1 # Leave one for ray scheduler
total_gpus = 8
```
다음으로, 훈련하려는 roboschool 에이전트를 선택합니다. 이기종(heterogeneous) 훈련의 경우 인스턴스 간 동기화를 지원하는 몇 가지 추가 파라메터들을 훈련 작업에 전달합니다.
- s3_bucket, s3_prefix: 마스터 IP 주소와 같은 메타데이터 저장에 사용
- rl_cluster_type: "기본" 또는 "보조"
- aws_region: VPC 모드에서 S3에 연결하는 데 필요
- rl_num_instances_secondary: 보조 클러스터의 노드 수
- subnets, security_group_ids: VPC 모드에 필요
```
roboschool_problem = 'reacher'
job_name_prefix = 'rl-roboschool-distributed-'+ roboschool_problem
s3_output_path = 's3://{}/'.format(s3_bucket) # SDK appends the job name and output folder
# We explicitly need to specify these params so that the two jobs can synchronize using the metadata stored here
s3_bucket = sage_session.default_bucket()
s3_prefix = "dist-ray-%s-1GPU-40CPUs" % (roboschool_problem)
# Make sure that the prefix is empty
!aws s3 rm --recursive s3://{s3_bucket}/{s3_prefix}
```
### 기본 클러스터 시작 (1 GPU 훈련 인스턴스)
```
primary_cluster_estimator = RLEstimator(entry_point="train-%s.py" % roboschool_problem,
source_dir='src',
dependencies=["common/sagemaker_rl"],
image_name=gpu_image_name,
role=role,
train_instance_type=primary_cluster_instance_type,
train_instance_count=primary_cluster_instance_count,
output_path=s3_output_path,
base_job_name=job_name_prefix,
metric_definitions=metric_definitions,
train_max_run=int(3600 * .5), # Maximum runtime in seconds
hyperparameters={
"s3_prefix": s3_prefix, # Important for syncing
"s3_bucket": s3_bucket, # Important for syncing
"aws_region": boto3.Session().region_name, # Important for S3 connection
"rl_cluster_type": "primary", # Important for syncing
"rl_num_instances_secondary": secondary_cluster_instance_count, # Important for syncing
"rl.training.config.num_workers": total_cpus,
"rl.training.config.train_batch_size": 20000,
"rl.training.config.num_gpus": total_gpus,
},
subnets=default_subnets, # Required for VPC mode
security_group_ids=default_security_groups # Required for VPC mode
)
primary_cluster_estimator.fit(wait=False)
primary_job_name = primary_cluster_estimator.latest_training_job.job_name
print("Primary Training job: %s" % primary_job_name)
```
### 보조 클러스터 시작 (2 CPU 인스턴스)
```
secondary_cluster_estimator = RLEstimator(entry_point="train-%s.py" % roboschool_problem,
source_dir='src',
dependencies=["common/sagemaker_rl"],
image_name=cpu_image_name,
role=role,
train_instance_type=secondary_cluster_instance_type,
train_instance_count=secondary_cluster_instance_count,
output_path=s3_output_path,
base_job_name=job_name_prefix,
metric_definitions=metric_definitions,
train_max_run=3600, # Maximum runtime in seconds
hyperparameters={
"s3_prefix": s3_prefix, # Important for syncing
"s3_bucket": s3_bucket, # Important for syncing
"aws_region": boto3.Session().region_name, # Important for S3 connection
"rl_cluster_type": "secondary", # Important for syncing
},
subnets=default_subnets, # Required for VPC mode
security_group_ids=default_security_groups # Required for VPC mode
)
secondary_cluster_estimator.fit(wait=False)
secondary_job_name = secondary_cluster_estimator.latest_training_job.job_name
print("Secondary Training job: %s" % secondary_job_name)
```
### 시각화
```
print("Job name: {}".format(primary_job_name))
s3_url = "s3://{}/{}".format(s3_bucket,primary_job_name)
if local_mode:
output_tar_key = "{}/output.tar.gz".format(primary_job_name)
else:
output_tar_key = "{}/output/output.tar.gz".format(primary_job_name)
intermediate_folder_key = "{}/output/intermediate/".format(primary_job_name)
output_url = "s3://{}/{}".format(s3_bucket, output_tar_key)
intermediate_url = "s3://{}/{}".format(s3_bucket, intermediate_folder_key)
print("S3 job path: {}".format(s3_url))
print("Output.tar.gz location: {}".format(output_url))
print("Intermediate folder path: {}".format(intermediate_url))
tmp_dir = "/tmp/{}".format(primary_job_name)
os.system("mkdir {}".format(tmp_dir))
print("Create local folder {}".format(tmp_dir))
```
### 훈련 롤아웃 비디오 가져오기
특정 롤아웃의 비디오는 훈련 중 S3에 기록됩니다. 여기에서는 S3에서 마지막 10개의 비디오 클립을 가져 와서 마지막 비디오를 렌더링합니다.
```
recent_videos = wait_for_s3_object(s3_bucket, intermediate_folder_key, tmp_dir,
fetch_only=(lambda obj: obj.key.endswith(".mp4") and obj.size>0), limit=10)
last_video = sorted(recent_videos)[-1] # Pick which video to watch
os.system("mkdir -p ./src/tmp_render_heterogeneous/ && cp {} ./src/tmp_render_heterogeneous/last_video.mp4".format(last_video))
HTML('<video src="./src/tmp_render_heterogeneous/last_video.mp4" controls autoplay></video>')
```
### 훈련 작업에 대한 지표 plot
CloudWatch 지표에 기록된 알고리즘 지표를 사용하여 실행 중인 훈련의 보상 지표를 볼 수 있습니다. 시간이 지남에 따라, 모델의 성능을 볼 수 있도록 이를 plot할 수 있습니다.
```
%matplotlib inline
from sagemaker.analytics import TrainingJobAnalytics
df = TrainingJobAnalytics(primary_job_name, ['episode_reward_mean']).dataframe()
num_metrics = len(df)
if num_metrics == 0:
print("No algorithm metrics found in CloudWatch")
else:
plt = df.plot(x='timestamp', y='value', figsize=(12,5), legend=True, style='b-')
plt.set_ylabel('Mean reward per episode')
plt.set_xlabel('Training time (s)')
```
위의 시각화 셀을 반복해서 실행하여 최신 비디오를 얻거나, 훈련 작업이 진행됨에 따라 최신 지표를 볼 수 있습니다.
|
github_jupyter
|
```
import pickle
import os
import numpy as np
from tqdm.notebook import tqdm
from quchem_ibm.exp_analysis import *
def dict_of_M_to_list(M_dict, PauliOP):
P_Qubit_list, _ = zip(*(list(*PauliOP.terms.keys())))
list_of_M_bitstrings=None
for bit_string, N_obtained in M_dict.items():
M_string = np.take(list(bit_string[::-1]), P_Qubit_list) # only take terms measured! Note bitstring reversed!
array_meas = np.repeat(''.join(M_string), N_obtained)
if list_of_M_bitstrings is None:
list_of_M_bitstrings=array_meas
else:
list_of_M_bitstrings=np.hstack((list_of_M_bitstrings,array_meas))
# randomly shuffle (seed means outcome will always be the SAME!)
# np.random.seed(42)
np.random.shuffle(list_of_M_bitstrings)
return list_of_M_bitstrings
# # input for exp
base_dir = os.getcwd()
input_file = os.path.join(base_dir, 'LiH_simulation_RESULTS_time=2020Oct07-163210198971.pickle')
with open(input_file, 'rb') as handle:
LiH_data = pickle.load(handle)
experimental_data_STANDARD = LiH_data['experiment_data'].copy()
del LiH_data
STANDARD_data = experimental_data_STANDARD[101852100]['standard'].copy()
del experimental_data_STANDARD
len(STANDARD_data)
STANDARD_Hist_data_sim={}
for exp_instance in STANDARD_data: #each exp repeated 10 times!
for exp_dict_key in exp_instance:
exp_dict= exp_instance[exp_dict_key]
P=exp_dict['qubitOp']
coeff = exp_dict['coeff']
measured_dict_sim = exp_dict['measurement_dict']
M_list_sim = dict_of_M_to_list(measured_dict_sim, P)
if exp_dict_key in STANDARD_Hist_data_sim.keys():
STANDARD_Hist_data_sim[exp_dict_key]={'P':list(P.terms.items())[0] ,'coeff': coeff.real, 'Measurements': np.hstack((STANDARD_Hist_data_sim[exp_dict_key]['Measurements'],M_list_sim))}
else:
STANDARD_Hist_data_sim[exp_dict_key]={'P':list(P.terms.items())[0] ,'coeff': coeff.real, 'Measurements': M_list_sim}
del exp_dict
del STANDARD_data
# for key in STANDARD_Hist_data_sim:
# STANDARD_Hist_data_sim[key]['Measurements']=STANDARD_Hist_data_sim[key]['Measurements'].tolist()
# STANDARD_Hist_data_sim[key]['P']=(STANDARD_Hist_data_sim[key]['P'][0], STANDARD_Hist_data_sim[key]['P'][1].real)
# import json
# with open("STANDARD_Hist_data_sim", "w") as write_file:
# json.dump(STANDARD_Hist_data_sim, write_file)
STANDARD_Hist_data_sim[0]['Measurements'].shape
# ### save output
# np.save('Standard_hist_data', STANDARD_Hist_data_sim)
import matplotlib.pyplot as plt
fci_energy= -7.971184315565538
```
# Histogram
```
def Get_Hist_data(Histogram_data, I_term):
E_list=[]
for m_index in tqdm(range(Histogram_data[0]['Measurements'].shape[0])):
E=I_term
for M_dict_key in Histogram_data:
coeff = Histogram_data[M_dict_key]['coeff']
parity = 1 if sum(map(int, Histogram_data[M_dict_key]['Measurements'][m_index])) % 2 == 0 else -1
E+=coeff*parity
E_list.append(E)
return E_list
I_term = -4.142299396835105
E_list_STANDARD_sim=Get_Hist_data(STANDARD_Hist_data_sim, I_term)
import json
with open("E_list_STANDARD_sim.json", "w") as write_file:
json.dump(E_list_STANDARD_sim, write_file)
E_list_STANDARD_sim=np.array(E_list_STANDARD_sim)
def gaussian(x, mean, amplitude, standard_deviation):
return amplitude * np.exp( - ((x - mean)**2 / (2*standard_deviation**2)))
from scipy.optimize import curve_fit
# from matplotlib import pyplot
# %matplotlib inline
# # bins_standard = len(set(E_list_STANDARD_sim))
# bins_standard = 1000
# bin_heights_STANDARD, bin_borders_STANDARD, _=pyplot.hist(E_list_STANDARD_sim,
# bins_standard, alpha=0.7,
# label='$E$ standard VQE - sim',
# color='g',
# density=False)
# bin_centers_STANDARD = bin_borders_STANDARD[:-1] + np.diff(bin_borders_STANDARD) / 2
# popt, _ = curve_fit(gaussian, bin_centers_STANDARD, bin_heights_STANDARD, p0=[fci_energy, 0., 1.], **{'maxfev':10000})
# mean_STANDARD, amplitude_STANDARD, standard_deviation_STANDARD= popt
# x_interval_for_fit = np.linspace(bin_borders_STANDARD[0], bin_borders_STANDARD[-1], 10000)
# pyplot.plot(x_interval_for_fit, gaussian(x_interval_for_fit, *popt), label='Gaussian fit', color='g')
# pyplot.axvline(mean_STANDARD, color='g', linestyle='dashed', linewidth=1,
# label='$E_{average}$ standard VQE - sim') # mean of GAUSSIAN FIT
# # pyplot.axvline(E_list_STANDARD_sim.mean(), color='g', linestyle='dashed', linewidth=1,
# # label='$E_{average}$ standard VQE - sim') # mean of DATA
# pyplot.errorbar(mean_STANDARD,65_000,
# xerr=standard_deviation_STANDARD, linestyle="None", color='g',
# uplims=True, lolims=True, label='$\sigma_{E_{av}}$standard VQE - sim')
# pyplot.axvline(fci_energy, color='k', linestyle='solid', linewidth=2,
# label='$E_{FCI}$', alpha=0.4)
# pyplot.legend(loc='upper right')
# # pyplot.legend(bbox_to_anchor=(0.865,1.9), loc="upper left")
# pyplot.ylabel('Frequency')
# pyplot.xlabel('Energy')
# pyplot.tight_layout()
# file_name = 'LiH_Histogram_STANDARD_sim_Gaussian.jpeg'
# pyplot.savefig(file_name, dpi=300,transparent=True,) # edgecolor='black', facecolor='white')
# pyplot.show()
def normal_dist(x, mean, standard_deviation):
return (1/(np.sqrt(2*np.pi)*standard_deviation)) * np.exp( - ((x - mean)**2 / (2*standard_deviation**2)))
plt.plot(x, normal_dist(x, av, sig))
# from scipy.stats import norm
# x=np.linspace(-10, 10, 1000)
# av=2
# sig=1
# plt.plot(x, norm.pdf(x, av, sig))
len(set(np.around(E_list_STANDARD_sim, 5)))
E_list_STANDARD_sim.shape
E_list_STANDARD_sim.shape[0]**(1/3)
# https://stats.stackexchange.com/questions/798/calculating-optimal-number-of-bins-in-a-histogram
from scipy.stats import iqr
bin_width = 2 * iqr(E_list_STANDARD_sim) / E_list_STANDARD_sim.shape[0]**(1/3)
np.ceil((max(E_list_STANDARD_sim)-min(E_list_STANDARD_sim))/bin_width)
from matplotlib import pyplot
%matplotlib inline
# bins = len(set(E_list_SEQ_ROT_sim))
# bins_standard = len(set(E_list_STANDARD_sim))
# bins_standard = 150_000
bins_standard = 2500
bin_heights_STANDARD, bin_borders_STANDARD, _=pyplot.hist(E_list_STANDARD_sim,
bins_standard, alpha=0.7,
label='$E$ standard VQE - sim',
color='g',
density=True)
#### ,hatch='-')
###### Gaussian fit
bin_centers_STANDARD = bin_borders_STANDARD[:-1] + np.diff(bin_borders_STANDARD) / 2
popt, _ = curve_fit(gaussian, bin_centers_STANDARD, bin_heights_STANDARD, p0=[fci_energy, 0., 1.])#, **{'maxfev':10000})
mean_STANDARD, amplitude_STANDARD, standard_deviation_STANDARD= popt
x_interval_for_fit = np.linspace(bin_borders_STANDARD[0], bin_borders_STANDARD[-1], 10000)
pyplot.plot(x_interval_for_fit, gaussian(x_interval_for_fit, *popt), label='Gaussian fit', color='olive',
linewidth=3)
### normal fit
# popt_norm, _ = curve_fit(normal_dist, bin_centers_STANDARD, bin_heights_STANDARD, p0=[fci_energy, standard_deviation_STANDARD])#, **{'maxfev':10000})
# mean_norm, standard_deviation_norm= popt_norm
# pyplot.plot(x_interval_for_fit, normal_dist(x_interval_for_fit, *popt_norm), label='Normal fit', color='b',
# linestyle='--')
# pyplot.plot(x_interval_for_fit, normal_dist(x_interval_for_fit, mean_STANDARD, standard_deviation_STANDARD),
# label='Normal fit', color='b', linestyle='--')
#### Average energy from data
pyplot.axvline(E_list_STANDARD_sim.mean(), color='g', linestyle='--', linewidth=2,
label='$E_{average}$ standard VQE - sim') # mean of DATA
##############
# chemical accuracy
pyplot.axvline(fci_energy, color='k', linestyle='solid', linewidth=3,
label='$E_{FCI}$', alpha=0.3)
# # chemical accuracy
# pyplot.fill_between([fci_energy-1.6e-3, fci_energy+1.6e-3],
# [0, np.ceil(max(bin_heights_STANDARD))] ,
# color='k',
# label='chemical accuracy',
# alpha=0.5)
pyplot.rcParams["font.family"] = "Times New Roman"
# pyplot.legend(loc='upper right')
# # pyplot.legend(bbox_to_anchor=(0.865,1.9), loc="upper left")
pyplot.ylabel('Probability Density', fontsize=20)
pyplot.xlabel('Energy / Hartree', fontsize=20)
pyplot.xticks(np.arange(-9.5,-5.5,0.5), fontsize=20)
pyplot.yticks(np.arange(0,2.5,0.5), fontsize=20)
# pyplot.xlim(np.floor(min(bin_borders_STANDARD)), np.ceil(max(bin_borders_STANDARD)))
pyplot.xlim(-9.5, -6.5)
pyplot.tight_layout()
file_name = 'LiH_Histogram_STANDARD_sim_Gaussian.jpeg'
pyplot.savefig(file_name, dpi=300,transparent=True,) # edgecolor='black', facecolor='white')
pyplot.show()
from matplotlib import pyplot
%matplotlib inline
# bins = len(set(E_list_SEQ_ROT_sim))
# bins_standard = len(set(E_list_STANDARD_sim))
# bins_standard = 5000
bins_standard = 150_000
bin_heights_STANDARD, bin_borders_STANDARD, _=pyplot.hist(E_list_STANDARD_sim,
bins_standard, alpha=0.7,
label='$E$ standard VQE - sim',
color='g',
density=True)
##############
pyplot.rcParams["font.family"] = "Times New Roman"
# pyplot.legend(loc='upper right')
# # pyplot.legend(bbox_to_anchor=(0.865,1.9), loc="upper left")
pyplot.ylabel('Probability Density', fontsize=20)
pyplot.xlabel('Energy / Hartree', fontsize=20)
pyplot.xticks(np.arange(-9.5,-5.5,0.5), fontsize=20)
pyplot.yticks(np.arange(0,3,0.5), fontsize=20)
# pyplot.xlim(np.floor(min(bin_borders_STANDARD)), np.ceil(max(bin_borders_STANDARD)))
pyplot.xlim(-9.5, -6.5)
pyplot.tight_layout()
# file_name = 'LiH_Histogram_STANDARD_sim_Gaussian.jpeg'
# pyplot.savefig(file_name, dpi=300,transparent=True,) # edgecolor='black', facecolor='white')
pyplot.show()
from scipy import stats
print(stats.shapiro(E_list_STANDARD_sim))
print(stats.kstest(E_list_STANDARD_sim, 'norm'))
```
# XY Z comparison
```
i_list_XY=[]
STANDARD_Hist_data_XY={}
i_list_Z=[]
STANDARD_Hist_data_Z={}
amplitude_min=0.00
XY_terms=[]
Z_amp_sum=0
for key in STANDARD_Hist_data_sim:
Pword, const = STANDARD_Hist_data_sim[key]['P']
coeff=STANDARD_Hist_data_sim[key]['coeff']
if np.abs(coeff)>amplitude_min:
qubitNos, qubitPstrs = zip(*(list(Pword)))
# XY terms only!
if ('X' in qubitPstrs) or ('Y' in qubitPstrs):
i_list_XY.append(key)
STANDARD_Hist_data_XY[key]=STANDARD_Hist_data_sim[key]
XY_terms.append(STANDARD_Hist_data_sim[key]['P'])
else:
i_list_Z.append(key)
STANDARD_Hist_data_Z[key]=STANDARD_Hist_data_sim[key]
Z_amp_sum+=coeff
Z_amp_sum
def Get_Hist_data(Histogram_data, I_term):
E_list=[]
for m_index in tqdm(range(Histogram_data[list(Histogram_data.keys())[0]]['Measurements'].shape[0])):
E=I_term
for M_dict_key in Histogram_data:
coeff = Histogram_data[M_dict_key]['coeff']
parity = 1 if sum(map(int, Histogram_data[M_dict_key]['Measurements'][m_index])) % 2 == 0 else -1
E+=coeff*parity
E_list.append(E)
return E_list
I_term = -4.142299396835105
E_list_STANDARD_XY=Get_Hist_data(STANDARD_Hist_data_XY, 0)
E_list_STANDARD_Z=Get_Hist_data(STANDARD_Hist_data_Z, 0)
print(len(set(np.around(E_list_STANDARD_XY, 5))))
print(len(set(np.around(E_list_STANDARD_Z, 5))))
from matplotlib import pyplot
%matplotlib inline
# bins_standard = len(set(E_list_STANDARD_sim))
# bins_standard = 1000
bins_standard=8_000
# bin_heights_XY, bin_borders_XY, _=pyplot.hist(E_list_STANDARD_XY,
# bins_standard, alpha=0.7,
# label='$XY$ terms',
# color='b',
# density=False)
bin_heights_Z, bin_borders_Z, _=pyplot.hist(E_list_STANDARD_Z,
bins_standard, alpha=0.7,
label='$Z$ terms',
color='g',
density=True)
pyplot.rcParams["font.family"] = "Times New Roman"
pyplot.ylabel('Probability Density', fontsize=20)
pyplot.xlabel('Energy / Hartree', fontsize=20)
pyplot.xticks(np.arange(-4.2,-3.0,0.2), fontsize=20)
pyplot.xlim((-4.2, -3.2))
pyplot.yticks(np.arange(0,1200,200), fontsize=20)
pyplot.ylim((0, 1000))
pyplot.tight_layout()
file_name = 'LiH_standard_Z.jpeg'
pyplot.savefig(file_name, dpi=300,transparent=True,) # edgecolor='black', facecolor='white')
pyplot.show()
np.where(bin_heights_Z==max(bin_heights_Z))[0]
print(bin_heights_Z[2334])
print('left sum:',sum(bin_heights_Z[:2334]))
print('right sum:', sum(bin_heights_Z[2335:]))
# therefore slighlt more likely to get more +ve energy!!!
bin_borders_Z[583]
print(len(np.where(np.array(E_list_STANDARD_Z)>-3.8)[0]))
print(len(np.where(np.array(E_list_STANDARD_Z)<-3.89)[0]))
len(E_list_STANDARD_Z)
from matplotlib import pyplot
%matplotlib inline
# bins_standard = len(set(E_list_STANDARD_sim))
# bins_standard = 1000
bins_standard = 5000
bin_heights_XY, bin_borders_XY, _=pyplot.hist(E_list_STANDARD_XY,
bins_standard, alpha=0.7,
label='$XY$ terms',
color='g',
density=True)
pyplot.rcParams["font.family"] = "Times New Roman"
pyplot.ylabel('Probability Density', fontsize=20)
pyplot.xlabel('Energy / Hartree', fontsize=20)
pyplot.xticks(np.arange(-0.8,0.9,0.2), fontsize=20)
pyplot.xlim((-0.8, 0.8))
pyplot.yticks(np.arange(0,3,0.5), fontsize=20)
pyplot.tight_layout()
file_name = 'LiH_standard_XY.jpeg'
pyplot.savefig(file_name, dpi=300,transparent=True,) # edgecolor='black', facecolor='white')
pyplot.show()
```
|
github_jupyter
|
##### Copyright 2018 The TensorFlow Authors.
Licensed under the Apache License, Version 2.0 (the "License");
```
#@title Licensed under the Apache License, Version 2.0 (the "License"); { display-mode: "form" }
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# AutoGraph: Easy control flow for graphs
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/versions/master/guide/autograph"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/models/blob/master/samples/core/guide/autograph.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/models/blob/master/samples/core/guide/autograph.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
</table>
[AutoGraph](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/contrib/autograph/) helps you write complicated graph code using normal Python. Behind the scenes, AutoGraph automatically transforms your code into the equivalent [TensorFlow graph code](https://www.tensorflow.org/guide/graphs). AutoGraph already supports much of the Python language, and that coverage continues to grow. For a list of supported Python language features, see the [Autograph capabilities and limitations](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/contrib/autograph/LIMITATIONS.md).
## Setup
To use AutoGraph, install the latest version of TensorFlow:
```
! pip install -U tf-nightly
```
Import TensorFlow, AutoGraph, and any supporting modules:
```
from __future__ import division, print_function, absolute_import
import tensorflow as tf
import tensorflow.keras.layers as layers
from tensorflow.contrib import autograph
import numpy as np
import matplotlib.pyplot as plt
```
We'll enable [eager execution](https://www.tensorflow.org/guide/eager) for demonstration purposes, but AutoGraph works in both eager and [graph execution](https://www.tensorflow.org/guide/graphs) environments:
```
tf.enable_eager_execution()
```
Note: AutoGraph converted code is designed to run during graph execution. When eager exectuon is enabled, use explicit graphs (as this example shows) or `tf.contrib.eager.defun`.
## Automatically convert Python control flow
AutoGraph will convert much of the Python language into the equivalent TensorFlow graph building code.
Note: In real applications batching is essential for performance. The best code to convert to AutoGraph is code where the control flow is decided at the _batch_ level. If making decisions at the individual _example_ level, you must index and batch the examples to maintain performance while applying the control flow logic.
AutoGraph converts a function like:
```
def square_if_positive(x):
if x > 0:
x = x * x
else:
x = 0.0
return x
```
To a function that uses graph building:
```
print(autograph.to_code(square_if_positive))
```
Code written for eager execution can run in a `tf.Graph` with the same results, but with the benfits of graph execution:
```
print('Eager results: %2.2f, %2.2f' % (square_if_positive(tf.constant(9.0)),
square_if_positive(tf.constant(-9.0))))
```
Generate a graph-version and call it:
```
tf_square_if_positive = autograph.to_graph(square_if_positive)
with tf.Graph().as_default():
# The result works like a regular op: takes tensors in, returns tensors.
# You can inspect the graph using tf.get_default_graph().as_graph_def()
g_out1 = tf_square_if_positive(tf.constant( 9.0))
g_out2 = tf_square_if_positive(tf.constant(-9.0))
with tf.Session() as sess:
print('Graph results: %2.2f, %2.2f\n' % (sess.run(g_out1), sess.run(g_out2)))
```
AutoGraph supports common Python statements like `while`, `for`, `if`, `break`, and `return`, with support for nesting. Compare this function with the complicated graph verson displayed in the following code blocks:
```
# Continue in a loop
def sum_even(items):
s = 0
for c in items:
if c % 2 > 0:
continue
s += c
return s
print('Eager result: %d' % sum_even(tf.constant([10,12,15,20])))
tf_sum_even = autograph.to_graph(sum_even)
with tf.Graph().as_default(), tf.Session() as sess:
print('Graph result: %d\n\n' % sess.run(tf_sum_even(tf.constant([10,12,15,20]))))
print(autograph.to_code(sum_even))
```
## Decorator
If you don't need easy access to the original Python function, use the `convert` decorator:
```
@autograph.convert()
def fizzbuzz(i, n):
while i < n:
msg = ''
if i % 3 == 0:
msg += 'Fizz'
if i % 5 == 0:
msg += 'Buzz'
if msg == '':
msg = tf.as_string(i)
print(msg)
i += 1
return i
with tf.Graph().as_default():
final_i = fizzbuzz(tf.constant(10), tf.constant(16))
# The result works like a regular op: takes tensors in, returns tensors.
# You can inspect the graph using tf.get_default_graph().as_graph_def()
with tf.Session() as sess:
sess.run(final_i)
```
## Examples
Let's demonstrate some useful Python language features.
### Assert
AutoGraph automatically converts the Python `assert` statement into the equivalent `tf.Assert` code:
```
@autograph.convert()
def inverse(x):
assert x != 0.0, 'Do not pass zero!'
return 1.0 / x
with tf.Graph().as_default(), tf.Session() as sess:
try:
print(sess.run(inverse(tf.constant(0.0))))
except tf.errors.InvalidArgumentError as e:
print('Got error message:\n %s' % e.message)
```
### Print
Use the Python `print` function in-graph:
```
@autograph.convert()
def count(n):
i=0
while i < n:
print(i)
i += 1
return n
with tf.Graph().as_default(), tf.Session() as sess:
sess.run(count(tf.constant(5)))
```
### Lists
Append to lists in loops (tensor list ops are automatically created):
```
@autograph.convert()
def arange(n):
z = []
# We ask you to tell us the element dtype of the list
autograph.set_element_type(z, tf.int32)
for i in range(n):
z.append(i)
# when you're done with the list, stack it
# (this is just like np.stack)
return autograph.stack(z)
with tf.Graph().as_default(), tf.Session() as sess:
sess.run(arange(tf.constant(10)))
```
### Nested control flow
```
@autograph.convert()
def nearest_odd_square(x):
if x > 0:
x = x * x
if x % 2 == 0:
x = x + 1
return x
with tf.Graph().as_default():
with tf.Session() as sess:
print(sess.run(nearest_odd_square(tf.constant(4))))
print(sess.run(nearest_odd_square(tf.constant(5))))
print(sess.run(nearest_odd_square(tf.constant(6))))
```
### While loop
```
@autograph.convert()
def square_until_stop(x, y):
while x < y:
x = x * x
return x
with tf.Graph().as_default():
with tf.Session() as sess:
print(sess.run(square_until_stop(tf.constant(4), tf.constant(100))))
```
### For loop
```
@autograph.convert()
def fizzbuzz_each(nums):
result = []
autograph.set_element_type(result, tf.string)
for num in nums:
result.append(fizzbuzz(num))
return autograph.stack(result)
with tf.Graph().as_default():
with tf.Session() as sess:
print(sess.run(fizzbuzz_each(tf.constant(np.arange(10)))))
```
### Break
```
@autograph.convert()
def argwhere_cumsum(x, threshold):
current_sum = 0.0
idx = 0
for i in range(len(x)):
idx = i
if current_sum >= threshold:
break
current_sum += x[i]
return idx
N = 10
with tf.Graph().as_default():
with tf.Session() as sess:
idx = argwhere_cumsum(tf.ones(N), tf.constant(float(N/2)))
print(sess.run(idx))
```
## Interoperation with `tf.Keras`
Now that you've seen the basics, let's build some model components with autograph.
It's relatively simple to integrate `autograph` with `tf.keras`.
### Stateless functions
For stateless functions, like `collatz` shown below, the easiest way to include them in a keras model is to wrap them up as a layer uisng `tf.keras.layers.Lambda`.
```
import numpy as np
@autograph.convert()
def collatz(x):
x=tf.reshape(x,())
assert x>0
n = tf.convert_to_tensor((0,))
while not tf.equal(x,1):
n+=1
if tf.equal(x%2, 0):
x = x//2
else:
x = 3*x+1
return n
with tf.Graph().as_default():
model = tf.keras.Sequential([
tf.keras.layers.Lambda(collatz, input_shape=(1,), output_shape=(), )
])
result = model.predict(np.array([6171])) #261
result
```
### Custom Layers and Models
<!--TODO(markdaoust) link to full examples or these referenced models.-->
The easiest way to use AutoGraph with Keras layers and models is to `@autograph.convert()` the `call` method. See the [TensorFlow Keras guide](https://tensorflow.org/guide/keras#build_advanced_models) for details on how to build on these classes.
Here is a simple example of the [stocastic network depth](https://arxiv.org/abs/1603.09382) technique :
```
# `K` is used to check if we're in train or test mode.
import tensorflow.keras.backend as K
class StocasticNetworkDepth(tf.keras.Sequential):
def __init__(self, pfirst=1.0, plast=0.5, *args,**kwargs):
self.pfirst = pfirst
self.plast = plast
super().__init__(*args,**kwargs)
def build(self,input_shape):
super().build(input_shape.as_list())
self.depth = len(self.layers)
self.plims = np.linspace(self.pfirst, self.plast, self.depth+1)[:-1]
@autograph.convert()
def call(self, inputs):
training = tf.cast(K.learning_phase(), dtype=bool)
if not training:
count = self.depth
return super(StocasticNetworkDepth, self).call(inputs), count
p = tf.random_uniform((self.depth,))
keeps = p<=self.plims
x = inputs
count = tf.reduce_sum(tf.cast(keeps, tf.int32))
for i in range(self.depth):
if keeps[i]:
x = self.layers[i](x)
# return both the final-layer output and the number of layers executed.
return x, count
```
Let's try it on mnist-shaped data:
```
train_batch = np.random.randn(64, 28,28,1).astype(np.float32)
```
Build a simple stack of `conv` layers, in the stocastic depth model:
```
with tf.Graph().as_default() as g:
model = StocasticNetworkDepth(
pfirst=1.0, plast=0.5)
for n in range(20):
model.add(
layers.Conv2D(filters=16, activation=tf.nn.relu,
kernel_size=(3,3), padding='same'))
model.build(tf.TensorShape((None, None, None,1)))
init = tf.global_variables_initializer()
```
Now test it to ensure it behaves as expected in train and test modes:
```
# Use an explicit session here so we can set the train/test switch, and
# inspect the layer count returned by `call`
with tf.Session(graph=g) as sess:
init.run()
for phase, name in enumerate(['test','train']):
K.set_learning_phase(phase)
result, count = model(tf.convert_to_tensor(train_batch, dtype=tf.float32))
result1, count1 = sess.run((result, count))
result2, count2 = sess.run((result, count))
delta = (result1 - result2)
print(name, "sum abs delta: ", abs(delta).mean())
print(" layers 1st call: ", count1)
print(" layers 2nd call: ", count2)
print()
```
## Advanced example: An in-graph training loop
The previous section showed that AutoGraph can be used inside Keras layers and models. Keras models can also be used in AutoGraph code.
Since writing control flow in AutoGraph is easy, running a training loop in a TensorFlow graph should also be easy.
This example shows how to train a simple Keras model on MNIST with the entire training process—loading batches, calculating gradients, updating parameters, calculating validation accuracy, and repeating until convergence—is performed in-graph.
### Download data
```
(train_images, train_labels), (test_images, test_labels) = tf.keras.datasets.mnist.load_data()
```
### Define the model
```
def mlp_model(input_shape):
model = tf.keras.Sequential((
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(100, activation='relu', input_shape=input_shape),
tf.keras.layers.Dense(100, activation='relu'),
tf.keras.layers.Dense(10, activation='softmax')))
model.build()
return model
def predict(m, x, y):
y_p = m(x)
losses = tf.keras.losses.categorical_crossentropy(y, y_p)
l = tf.reduce_mean(losses)
accuracies = tf.keras.metrics.categorical_accuracy(y, y_p)
accuracy = tf.reduce_mean(accuracies)
return l, accuracy
def fit(m, x, y, opt):
l, accuracy = predict(m, x, y)
# Autograph automatically adds the necessary `tf.control_dependencies` here.
# (Without them nothing depends on `opt.minimize`, so it doesn't run.)
# This makes it much more like eager-code.
opt.minimize(l)
return l, accuracy
def setup_mnist_data(is_training, batch_size):
if is_training:
ds = tf.data.Dataset.from_tensor_slices((train_images, train_labels))
ds = ds.shuffle(batch_size * 10)
else:
ds = tf.data.Dataset.from_tensor_slices((test_images, test_labels))
ds = ds.repeat()
ds = ds.batch(batch_size)
return ds
def get_next_batch(ds):
itr = ds.make_one_shot_iterator()
image, label = itr.get_next()
x = tf.to_float(image)/255.0
y = tf.one_hot(tf.squeeze(label), 10)
return x, y
```
### Define the training loop
```
# Use `recursive = True` to recursively convert functions called by this one.
@autograph.convert(recursive=True)
def train(train_ds, test_ds, hp):
m = mlp_model((28 * 28,))
opt = tf.train.AdamOptimizer(hp.learning_rate)
# We'd like to save our losses to a list. In order for AutoGraph
# to convert these lists into their graph equivalent,
# we need to specify the element type of the lists.
train_losses = []
autograph.set_element_type(train_losses, tf.float32)
test_losses = []
autograph.set_element_type(test_losses, tf.float32)
train_accuracies = []
autograph.set_element_type(train_accuracies, tf.float32)
test_accuracies = []
autograph.set_element_type(test_accuracies, tf.float32)
# This entire training loop will be run in-graph.
i = tf.constant(0)
while i < hp.max_steps:
train_x, train_y = get_next_batch(train_ds)
test_x, test_y = get_next_batch(test_ds)
step_train_loss, step_train_accuracy = fit(m, train_x, train_y, opt)
step_test_loss, step_test_accuracy = predict(m, test_x, test_y)
if i % (hp.max_steps // 10) == 0:
print('Step', i, 'train loss:', step_train_loss, 'test loss:',
step_test_loss, 'train accuracy:', step_train_accuracy,
'test accuracy:', step_test_accuracy)
train_losses.append(step_train_loss)
test_losses.append(step_test_loss)
train_accuracies.append(step_train_accuracy)
test_accuracies.append(step_test_accuracy)
i += 1
# We've recorded our loss values and accuracies
# to a list in a graph with AutoGraph's help.
# In order to return the values as a Tensor,
# we need to stack them before returning them.
return (autograph.stack(train_losses), autograph.stack(test_losses),
autograph.stack(train_accuracies), autograph.stack(test_accuracies))
```
Now build the graph and run the training loop:
```
with tf.Graph().as_default() as g:
hp = tf.contrib.training.HParams(
learning_rate=0.005,
max_steps=500,
)
train_ds = setup_mnist_data(True, 50)
test_ds = setup_mnist_data(False, 1000)
(train_losses, test_losses, train_accuracies,
test_accuracies) = train(train_ds, test_ds, hp)
init = tf.global_variables_initializer()
with tf.Session(graph=g) as sess:
sess.run(init)
(train_losses, test_losses, train_accuracies,
test_accuracies) = sess.run([train_losses, test_losses, train_accuracies,
test_accuracies])
plt.title('MNIST train/test losses')
plt.plot(train_losses, label='train loss')
plt.plot(test_losses, label='test loss')
plt.legend()
plt.xlabel('Training step')
plt.ylabel('Loss')
plt.show()
plt.title('MNIST train/test accuracies')
plt.plot(train_accuracies, label='train accuracy')
plt.plot(test_accuracies, label='test accuracy')
plt.legend(loc='lower right')
plt.xlabel('Training step')
plt.ylabel('Accuracy')
plt.show()
```
|
github_jupyter
|
<script async src="https://www.googletagmanager.com/gtag/js?id=UA-59152712-8"></script>
<script>
window.dataLayer = window.dataLayer || [];
function gtag(){dataLayer.push(arguments);}
gtag('js', new Date());
gtag('config', 'UA-59152712-8');
</script>
# Generating C code for the right-hand sides of Maxwell's equations, in ***curvilinear*** coordinates, using a reference metric formalism
## Author: Ian Ruchlin
### Formatting improvements courtesy Brandon Clark
[comment]: <> (Abstract: TODO)
### The following formulations of Maxwell's equations, called System I and System II, are described in [Illustrating Stability Properties of Numerical Relativity in Electrodynamics](https://arxiv.org/abs/gr-qc/0201051) by Knapp et al.
**Notebook Status:** <font color='red'><b> In progress </b></font>
**Validation Notes:** This module has not yet undergone validation testing. Do ***not*** use it until after appropriate validation testing has been performed.
## Introduction:
[Maxwell's equations](https://en.wikipedia.org/wiki/Maxwell%27s_equations) are subject to the Gauss' law constraint
$$\mathcal{C} \equiv \hat{D}_{i} E^{i} - 4 \pi \rho = 0 \; ,$$
where $E^{i}$ is the electric vector field, $\hat{D}_{i}$ is the [covariant derivative](https://en.wikipedia.org/wiki/Covariant_derivative) associated with the reference metric $\hat{\gamma}_{i j}$ (which is taken to represent flat space), and $\rho$ is the electric charge density. We use $\mathcal{C}$ as a measure of numerical error. Maxwell's equations are also required to satisfy $\hat{D}_{i} B^{i} = 0$, where $B^{i}$ is the magnetic vector field. The magnetic constraint implies that the magnetic field can be expressed as
$$B_{i} = \epsilon_{i j k} \hat{D}^{j} A^{k} \; ,$$
where $\epsilon_{i j k}$ is the totally antisymmetric [Levi-Civita tensor](https://en.wikipedia.org/wiki/Levi-Civita_symbol) and $A^{i}$ is the vector potential field. Together with the scalar potential $\psi$, the electric field can be expressed in terms of the potential fields as
$$E_{i} = -\hat{D}_{i} \psi - \partial_{t} A_{i} \; .$$
For now, we work in vacuum, where the electric charge density and the electric current density vector both vanish ($\rho = 0$ and $j_{i} = 0$).
In addition to the Gauss constraints, the electric and magnetic fields obey two independent [electromagnetic invariants](https://en.wikipedia.org/wiki/Classification_of_electromagnetic_fields#Invariants)
\begin{align}
\mathcal{P} &\equiv B_{i} B^{i} - E_{i} E^{i} \; , \\
\mathcal{Q} &\equiv E_{i} B^{i} \; .
\end{align}
In vacuum, these satisfy $\mathcal{P} = \mathcal{Q} = 0$.
<a id='toc'></a>
# Table of Contents:
$$\label{toc}$$
This notebook is organized as follows
1. [Step 1](#sys1): System I
1. [Step 2](#sys2): System II
1. [Step 3](#latex_pdf_output): Output this notebook to $\LaTeX$-formatted PDF file
<a id='sys1'></a>
# Step 1: System I \[Back to [top](#toc)\]
$$\label{sys1}$$
In terms of the above definitions, the evolution Maxwell's equations take the form
\begin{align}
\partial_{t} A_{i} &= -E_{i} - \hat{D}_{i} \psi \; , \\
\partial_{t} E_{i} &= -\hat{D}_{j} \hat{D}^{j} A_{i} + \hat{D}_{i} \hat{D}_{j} A^{j}\; , \\
\partial_{t} \psi &= -\hat{D}_{i} A^{i} \; .
\end{align}
Note that this coupled system contains mixed second derivatives in the second term on the right hand side of the $E^{i}$ evolution equation. We will revisit this fact when building System II.
It can be shown that the Gauss constraint satisfies the evolution equation
$$\partial_{t} \mathcal{C} = 0 \; .$$
This implies that any constraint violating numerical error remains fixed in place during the evolution. This becomes problematic when the violations grow large and spoil the physics of the simulation.
```
import NRPy_param_funcs as par # NRPy+: parameter interface
import indexedexp as ixp # NRPy+: Symbolic indexed expression (e.g., tensors, vectors, etc.) support
import grid as gri # NRPy+: Functions having to do with numerical grids
import finite_difference as fin # NRPy+: Finite difference C code generation module
import reference_metric as rfm # NRPy+: Reference metric support
from outputC import lhrh # NRPy+: Core C code output module
par.set_parval_from_str("reference_metric::CoordSystem", "Spherical")
par.set_parval_from_str("grid::DIM", 3)
rfm.reference_metric()
# The name of this module ("maxwell") is given by __name__:
thismodule = __name__
# Step 0: Read the spatial dimension parameter as DIM.
DIM = par.parval_from_str("grid::DIM")
# Step 1: Set the finite differencing order to 4.
par.set_parval_from_str("finite_difference::FD_CENTDERIVS_ORDER", 4)
# Step 2: Register gridfunctions that are needed as input.
psi = gri.register_gridfunctions("EVOL", ["psi"])
# Step 3a: Declare the rank-1 indexed expressions E_{i}, A_{i},
# and \partial_{i} \psi. Derivative variables like these
# must have an underscore in them, so the finite
# difference module can parse the variable name properly.
ED = ixp.register_gridfunctions_for_single_rank1("EVOL", "ED")
AD = ixp.register_gridfunctions_for_single_rank1("EVOL", "AD")
psi_dD = ixp.declarerank1("psi_dD")
# Step 3b: Declare the rank-2 indexed expression \partial_{j} A_{i},
# which is not symmetric in its indices.
# Derivative variables like these must have an underscore
# in them, so the finite difference module can parse the
# variable name properly.
AD_dD = ixp.declarerank2("AD_dD", "nosym")
# Step 3c: Declare the rank-3 indexed expression \partial_{jk} A_{i},
# which is symmetric in the two {jk} indices.
AD_dDD = ixp.declarerank3("AD_dDD", "sym12")
# Step 4: Calculate first and second covariant derivatives, and the
# necessary contractions.
# First covariant derivative
# D_{j} A_{i} = A_{i,j} - \Gamma^{k}_{ij} A_{k}
AD_dHatD = ixp.zerorank2()
for i in range(DIM):
for j in range(DIM):
AD_dHatD[i][j] = AD_dD[i][j]
for k in range(DIM):
AD_dHatD[i][j] -= rfm.GammahatUDD[k][i][j] * AD[k]
# Second covariant derivative
# D_{k} D_{j} A_{i} = \partial_{k} D_{j} A_{i} - \Gamma^{l}_{jk} D_{l} A_{i}
# - \Gamma^{l}_{ik} D_{j} A_{l}
# = A_{i,jk}
# - \Gamma^{l}_{ij,k} A_{l}
# - \Gamma^{l}_{ij} A_{l,k}
# - \Gamma^{l}_{jk} A_{i;\hat{l}}
# - \Gamma^{l}_{ik} A_{l;\hat{j}}
AD_dHatDD = ixp.zerorank3()
for i in range(DIM):
for j in range(DIM):
for k in range(DIM):
AD_dHatDD[i][j][k] = AD_dDD[i][j][k]
for l in range(DIM):
AD_dHatDD[i][j][k] += - rfm.GammahatUDDdD[l][i][j][k] * AD[l] \
- rfm.GammahatUDD[l][i][j] * AD_dD[l][k] \
- rfm.GammahatUDD[l][j][k] * AD_dHatD[i][l] \
- rfm.GammahatUDD[l][i][k] * AD_dHatD[l][j]
# Covariant divergence
# D_{i} A^{i} = ghat^{ij} D_{j} A_{i}
DivA = 0
# Gradient of covariant divergence
# DivA_dD_{i} = ghat^{jk} A_{k;\hat{j}\hat{i}}
DivA_dD = ixp.zerorank1()
# Covariant Laplacian
# LapAD_{i} = ghat^{jk} A_{i;\hat{j}\hat{k}}
LapAD = ixp.zerorank1()
for i in range(DIM):
for j in range(DIM):
DivA += rfm.ghatUU[i][j] * AD_dHatD[i][j]
for k in range(DIM):
DivA_dD[i] += rfm.ghatUU[j][k] * AD_dHatDD[k][j][i]
LapAD[i] += rfm.ghatUU[j][k] * AD_dHatDD[i][j][k]
# Step 5: Define right-hand sides for the evolution.
AD_rhs = ixp.zerorank1()
ED_rhs = ixp.zerorank1()
for i in range(DIM):
AD_rhs[i] = -ED[i] - psi_dD[i]
ED_rhs[i] = -LapAD[i] + DivA_dD[i]
psi_rhs = -DivA
# Step 6: Generate C code for System I Maxwell's evolution equations,
# print output to the screen (standard out, or stdout).
lhrh_list = []
for i in range(DIM):
lhrh_list.append(lhrh(lhs=gri.gfaccess("rhs_gfs", "AD" + str(i)), rhs=AD_rhs[i]))
lhrh_list.append(lhrh(lhs=gri.gfaccess("rhs_gfs", "ED" + str(i)), rhs=ED_rhs[i]))
lhrh_list.append(lhrh(lhs=gri.gfaccess("rhs_gfs", "psi"), rhs=psi_rhs))
fin.FD_outputC("stdout", lhrh_list)
```
<a id='sys2'></a>
# Step 2: System II \[Back to [top](#toc)\]
$$\label{sys2}$$
Define the auxiliary variable
$$\Gamma \equiv \hat{D}_{i} A^{i} \; .$$
Substituting this into Maxwell's equations yields the system
\begin{align}
\partial_{t} A_{i} &= -E_{i} - \hat{D}_{i} \psi \; , \\
\partial_{t} E_{i} &= -\hat{D}_{j} \hat{D}^{j} A_{i} + \hat{D}_{i} \Gamma \; , \\
\partial_{t} \psi &= -\Gamma \; , \\
\partial_{t} \Gamma &= -\hat{D}_{i} \hat{D}^{i} \psi \; .
\end{align}
It can be shown that the Gauss constraint now satisfies the wave equation
$$\partial_{t}^{2} \mathcal{C} = \hat{D}_{i} \hat{D}^{i} \mathcal{C} \; .$$
Thus, any constraint violation introduced by numerical error propagates away at the speed of light. This property increases the stability of of the simulation, compared to System I above. A similar trick is used in the [BSSN formulation](Tutorial-BSSNCurvilinear.ipynb) of Einstein's equations.
```
# We inherit here all of the definitions from System I, above
# Step 7a: Register the scalar auxiliary variable \Gamma
Gamma = gri.register_gridfunctions("EVOL", ["Gamma"])
# Step 7b: Declare the ordinary gradient \partial_{i} \Gamma
Gamma_dD = ixp.declarerank1("Gamma_dD")
# Step 8a: Construct the second covariant derivative of the scalar \psi
# \psi_{;\hat{i}\hat{j}} = \psi_{,i;\hat{j}}
# = \psi_{,ij} - \Gamma^{k}_{ij} \psi_{,k}
psi_dDD = ixp.declarerank2("psi_dDD", "sym01")
psi_dHatDD = ixp.zerorank2()
for i in range(DIM):
for j in range(DIM):
psi_dHatDD[i][j] = psi_dDD[i][j]
for k in range(DIM):
psi_dHatDD[i][j] += - rfm.GammahatUDD[k][i][j] * psi_dD[k]
# Step 8b: Construct the covariant Laplacian of \psi
# Lappsi = ghat^{ij} D_{j} D_{i} \psi
Lappsi = 0
for i in range(DIM):
for j in range(DIM):
Lappsi += rfm.ghatUU[i][j] * psi_dHatDD[i][j]
# Step 9: Define right-hand sides for the evolution.
AD_rhs = ixp.zerorank1()
ED_rhs = ixp.zerorank1()
for i in range(DIM):
AD_rhs[i] = -ED[i] - psi_dD[i]
ED_rhs[i] = -LapAD[i] + Gamma_dD[i]
psi_rhs = -Gamma
Gamma_rhs = -Lappsi
# Step 10: Generate C code for System II Maxwell's evolution equations,
# print output to the screen (standard out, or stdout).
lhrh_list = []
for i in range(DIM):
lhrh_list.append(lhrh(lhs=gri.gfaccess("rhs_gfs", "AD" + str(i)), rhs=AD_rhs[i]))
lhrh_list.append(lhrh(lhs=gri.gfaccess("rhs_gfs", "ED" + str(i)), rhs=ED_rhs[i]))
lhrh_list.append(lhrh(lhs=gri.gfaccess("rhs_gfs", "psi"), rhs=psi_rhs))
lhrh_list.append(lhrh(lhs=gri.gfaccess("rhs_gfs", "Gamma"), rhs=Gamma_rhs))
fin.FD_outputC("stdout", lhrh_list)
```
<a id='latex_pdf_output'></a>
# Step 3: Output this notebook to $\LaTeX$-formatted PDF file \[Back to [top](#toc)\]
$$\label{latex_pdf_output}$$
The following code cell converts this Jupyter notebook into a proper, clickable $\LaTeX$-formatted PDF file. After the cell is successfully run, the generated PDF may be found in the root NRPy+ tutorial directory, with filename
[Tutorial-MaxwellCurvilinear.pdf](Tutorial-MaxwellCurvilinear.pdf) (Note that clicking on this link may not work; you may need to open the PDF file through another means.)
```
import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface
cmd.output_Jupyter_notebook_to_LaTeXed_PDF("Tutorial-MaxwellCurvilinear")
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.autograd import Variable
import matplotlib.pyplot as plt
%matplotlib inline
data_raw = pd.read_csv('../input/sign_mnist_train.csv', sep=",")
test_data_raw = pd.read_csv('../input/sign_mnist_test.csv', sep=",")
labels = data_raw['label']
data_raw.drop('label', axis=1, inplace=True)
labels_test = test_data_raw['label']
test_data_raw.drop('label', axis=1, inplace=True)
data = data_raw.values
labels = labels.values
test_data = test_data_raw.values
labels_test = labels_test.values
pixels = data[10].reshape(28, 28)
plt.subplot(221)
sns.heatmap(data=pixels)
pixels = data[12].reshape(28, 28)
plt.subplot(222)
sns.heatmap(data=pixels)
pixels = data[20].reshape(28, 28)
plt.subplot(223)
sns.heatmap(data=pixels)
pixels = data[32].reshape(28, 28)
plt.subplot(224)
sns.heatmap(data=pixels)
reshaped = []
for i in data:
reshaped.append(i.reshape(1, 28, 28))
data = np.array(reshaped)
reshaped_test = []
for i in test_data:
reshaped_test.append(i.reshape(1,28,28))
test_data = np.array(reshaped_test)
x = torch.FloatTensor(data)
y = torch.LongTensor(labels.tolist())
test_x = torch.FloatTensor(test_data)
test_y = torch.LongTensor(labels_test.tolist())
class Network(nn.Module):
def __init__(self):
super(Network, self).__init__()
self.conv1 = nn.Conv2d(1, 10, 3)
self.pool1 = nn.MaxPool2d(2)
self.conv2 = nn.Conv2d(10, 20, 3)
self.pool2 = nn.MaxPool2d(2)
self.conv3 = nn.Conv2d(20, 30, 3)
self.dropout1 = nn.Dropout2d()
self.fc3 = nn.Linear(30 * 3 * 3, 270)
self.fc4 = nn.Linear(270, 26)
self.softmax = nn.LogSoftmax(dim=1)
def forward(self, x):
x = self.conv1(x)
x = F.relu(x)
x = self.pool1(x)
x = self.conv2(x)
x = F.relu(x)
x = self.pool2(x)
x = self.conv3(x)
x = F.relu(x)
x = self.dropout1(x)
x = x.view(-1, 30 * 3 * 3)
x = F.relu(self.fc3(x))
x = F.relu(self.fc4(x))
return self.softmax(x)
def test(self, predictions, labels):
self.eval()
correct = 0
for p, l in zip(predictions, labels):
if p == l:
correct += 1
acc = correct / len(predictions)
print("Correct predictions: %5d / %5d (%5f)" % (correct, len(predictions), acc))
def evaluate(self, predictions, labels):
correct = 0
for p, l in zip(predictions, labels):
if p == l:
correct += 1
acc = correct / len(predictions)
return(acc)
!pip install torchsummary
from torchsummary import summary
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model = Network().to(device)
summary(model, (1, 28, 28))
net = Network()
optimizer = optim.SGD(net.parameters(),0.001, momentum=0.7)
loss_func = nn.CrossEntropyLoss()
loss_log = []
acc_log = []
for e in range(50):
for i in range(0, x.shape[0], 100):
x_mini = x[i:i + 100]
y_mini = y[i:i + 100]
optimizer.zero_grad()
net_out = net(Variable(x_mini))
loss = loss_func(net_out, Variable(y_mini))
loss.backward()
optimizer.step()
if i % 1000 == 0:
#pred = net(Variable(test_data_formated))
loss_log.append(loss.item())
acc_log.append(net.evaluate(torch.max(net(Variable(test_x[:500])).data, 1)[1], test_y[:500]))
print('Epoch: {} - Loss: {:.6f}'.format(e + 1, loss.item()))
plt.figure(figsize=(10,8))
plt.plot(loss_log[2:])
plt.plot(acc_log)
plt.plot(np.ones(len(acc_log)), linestyle='dashed')
plt.show()
predictions = net(Variable(test_x))
net.test(torch.max(predictions.data, 1)[1], test_y)
```
|
github_jupyter
|
# Remuestreo Bootstrap
Entre los métodos inferenciales que permiten cuantificar el grado de confianza que se puede tener de un estadı́sitico, y saber cuán acertados son los resultados sobre los parámetros de la población, se encuentran las técnias de remuestreo.
Estas técnicas tienen la ventaja de que no necesitan datos distribuidos normalmente, muestras muy grandes y fórmulas complicadas. Además permiten obtener resultados muchas veces más exactos que otros métodos.
El bootstrap es un mecanismo que se centra en el remuestreo de datos dentro de una muestra aleatoria, diseñado para aproximar la precisión de un estimador.
El método se basa en: dada una muestra aleatoria con 'n' observaciones, se construyen con ella 'B' "muestras Bootstrap" del mismo tamaño con reposición (es decir los valores se pueden repeitir).
Para cada una de las B nuevas muestras, se realiza una estimación del parámetro de interés $\theta$.
Luego, se usan los B valores bootstrap estimados para aproximar la distribución del estimador del parámetro.
Esta distribución se utiliza para hacer más inferencias estadísticas, como la estimación del error estándar de $\theta$ o un intervalo de confianza para el mismo.
EL intervalo de confianza que se calcula a partir de los datos de la muestra, es un intervalo en donde se estima que estará cierto valor desconocido, como el parámtero poblacional, con un determinado nivel de confianza.Se denomina nivel de significancia a $\alpha$ y representa la probabilidad de que el intervalo contenga el parámetro poblacional.
En este ejercicio se quiere diseñar una función que por medio del método de boostrap resampling estime la varianza de una V.A. a partir de una muestra de datos. Se toma como 'muestra' a las magnitudes de estrellas pertenecientes a cúmulos globulares los cuales se encuentran en la columna número 6 (contando desde cero) del archivo 'cumulos_globulares.dat'.
Primero para estimar la varianza, se calcula la varianza muestral.
```
from math import *
import numpy as np
import matplotlib.pyplot as plt
import random
import seaborn as sns
sns.set()
muestra = np.genfromtxt('cumulos_globulares.dat', usecols=6) #se carga el archivo
muestra = muestra[~np.isnan(muestra)] #tiene NaNs, así que usa solo los numéricos.
n=len(muestra) #defino n como el tamaño de la muestra
xm= sum(muestra)/n #Calculo la media muestral
s2= sum((muestra-xm)**2)/(n-1) #Calculo varianza muestral
print('Varianza muestral:', s2)
```
A continuación, se realizan remuestreos para aplicar el método de bootstrap y calcular el intervalo de confianza.
Se define la función 'boot' que realiza realiza 'B' muestras nuevas aleatorias del mismo tamaño que la original utilizando la función 'np.random.choice'. Para cada muestra se calcula la varianza muestral y se guardan en una lista.
Abajo se grafica la distribución obtenida para la varianza para verla visualmente.
```
def boot(muestra, n, B=1000): #defino función con B=cantidad de muestras bootstraps
var_mues=[]
for i in range(B):
muestra_nueva=np.random.choice(muestra, size=n) #genera una muestra aleatoria a partir de un array de tamaño n
xm= sum(muestra_nueva)/n #calculo media muestral
s2= sum((muestra_nueva-xm)**2)/(n-1) #calculo varianza muestral
var_mues.append(s2)
return var_mues
#Grafico el histograma de las varianzas calculadas
var = boot(muestra, n) # varianzas muestrales de las distintas muestras
plt.hist(var, color='gold')
plt.title('Distribución muestral de la varianza')
plt.xlabel('$S^2$')
plt.ylabel('Frecuencia absoluta')
plt.show()
```
A continuación, se quiere calcular los intervalos de confidencia del estimador de la varianza con un nivel de significancia $\alpha$ dado. El intervalo de confianza va a estar definido entre los valores $(q_1, q_2)$, tal que el área bajo la curva de la distribución encerrada entre ellos es igual a $\alpha$.
Como en el histograma formado para la varianza se ve que la distribución que se forma es simétrica, se pide que el intervalo de confianza sea simétrico. Por lo tanto, las colas de la distribución (es decir $S^2<q_1$ y $S^2>q_2$), van a tener un área bajo la curva de valor $\frac{1-\alpha}{2}$ cada una.
Luego, se buscan los valores de $q_1$ y $q_2$ que cumplan con lo siguiente:
$$\frac{N(S^2<q_1)}{B}=\frac{1-\alpha}{2}$$
$$\frac{N(S^2>q_2)}{B}=\frac{1-\alpha}{2}$$
donde N() indica el número de valores de $S^2$ que cumplen esa codición.
Programa para calcular q1:
```
def IC_q1(var, a): #a es alpha
var.sort() #ordeno los valores de menor a mayor
suma=0
y=(1-a)/2 #condición que quiero que se cumpla
for i in range(len(var)):
x=var[i] #defino como x el elemento i de la varianza
suma=suma+x #los sumo
t=suma/(len(var)) #divido por la cantidad de muestras
if t<= y:
None
else:
q1=x
break
return q1
```
Programa para calcular q2:
```
def IC_q2(var, a):
var.sort(reverse=True) #ordeno los valores de mayor a menor
suma=0
y=(1-a)/2
for i in range(len(var)):
x=var[i]
suma=suma+x
t=suma/(len(var))
if t<= y:
None
else:
q2=x
break
return q2
```
Como ejemplo, se toma el valor de $\alpha$=0.95 y 0.9 para computar el valor final obtenido para la varianza con su intervalo de confianza.
```
q1=IC_q1(var, a=0.95)
print('Valor de q1=', q1)
q2=IC_q2(var, a=0.95)
print('Valor de q2=', q2)
print('El valor que se obtiene para la varianza es ', s2, 'con un intervalo de confianza de (', q1, ',', q2,').')
q1=IC_q1(var, a=0.9)
print('Valor de q1=', q1)
q2=IC_q2(var, a=0.9)
print('Valor de q2=', q2)
print('El valor que se obtiene para la varianza es ', s2, 'con un intervalo de confianza de (', q1, ',', q2,').')
```
## Conclusiones
Por medio del método de remuestreo bootstrap se puede conocer la varianza de una variable aleatoria y una estimación de su incerteza de la cual no se tiene conocimiento sobre su distribución. Además se puede calcular un intervalo de confianza para un determinado valor de $\alpha$ mediante el calculo de los límites inferiores y superiores del intervalo.
Se puede ver que la distribución de la varianza tiene forma de campana centrada en el valor estimado de la varianza muestral, por lo que el intervalo de confianza es simétrico.
También se ve, con los últimos ejemplos que si el valor de $\alpha$ decrece, el IC también.
|
github_jupyter
|
# Data Bootcamp: Demography
We love demography, specifically the dynamics of population growth and decline. You can drill down seemingly without end, as this [terrific graphic](http://www.bloomberg.com/graphics/dataview/how-americans-die/) about causes of death suggests.
We take a look here at the UN's [population data](http://esa.un.org/unpd/wpp/Download/Standard/Population/): the age distribution of the population, life expectancy, fertility (the word we use for births), and mortality (deaths). Explore the website, it's filled with interesting data. There are other sources that cover longer time periods, and for some countries you can get detailed data on specific things (causes of death, for example).
We use a number of countries as examples, but Japan and China are the most striking. The code is written so that the country is easily changed.
This IPython notebook was created by Dave Backus, Chase Coleman, and Spencer Lyon for the NYU Stern course [Data Bootcamp](http://databootcamp.nyuecon.com/).
## Preliminaries
Import statements and a date check for future reference.
```
# import packages
import pandas as pd # data management
import matplotlib.pyplot as plt # graphics
import matplotlib as mpl # graphics parameters
import numpy as np # numerical calculations
# IPython command, puts plots in notebook
%matplotlib inline
# check Python version
import datetime as dt
import sys
print('Today is', dt.date.today())
print('What version of Python are we running? \n', sys.version, sep='')
```
## Population by age
We have both "estimates" of the past (1950-2015) and "projections" of the future (out to 2100). Here we focus on the latter, specifically what the UN refers to as the medium variant: their middle of the road projection. It gives us a sense of how Japan's population might change over the next century.
It takes a few seconds to read the data.
What are the numbers? Thousands of people in various 5-year age categories.
```
url1 = 'http://esa.un.org/unpd/wpp/DVD/Files/'
url2 = '1_Indicators%20(Standard)/EXCEL_FILES/1_Population/'
url3 = 'WPP2015_POP_F07_1_POPULATION_BY_AGE_BOTH_SEXES.XLS'
url = url1 + url2 + url3
cols = [2, 5] + list(range(6,28))
#est = pd.read_excel(url, sheetname=0, skiprows=16, parse_cols=cols, na_values=['…'])
prj = pd.read_excel(url, sheetname=1, skiprows=16, parse_cols=cols, na_values=['…'])
prj.head(3)[list(range(6))]
# rename some variables
pop = prj
names = list(pop)
pop = pop.rename(columns={names[0]: 'Country',
names[1]: 'Year'})
# select country and years
country = ['Japan']
years = [2015, 2055, 2095]
pop = pop[pop['Country'].isin(country) & pop['Year'].isin(years)]
pop = pop.drop(['Country'], axis=1)
# set index = Year
# divide by 1000 to convert numbers from thousands to millions
pop = pop.set_index('Year')/1000
pop.head()[list(range(8))]
# transpose (T) so that index = age
pop = pop.T
pop.head(3)
ax = pop.plot(kind='bar',
color='blue',
alpha=0.5, subplots=True, sharey=True, figsize=(8,6))
for axnum in range(len(ax)):
ax[axnum].set_title('')
ax[axnum].set_ylabel('Millions')
ax[0].set_title('Population by age', fontsize=14, loc='left')
```
**Exercise.** What do you see here? What else would you like to know?
**Exercise.** Adapt the preceeding code to do the same thing for China. Or some other country that sparks your interest.
## Fertility: aka birth rates
We might wonder, why is the population falling in Japan? Other countries? Well, one reason is that birth rates are falling. Demographers call this fertility. Here we look at the fertility using the same [UN source](http://esa.un.org/unpd/wpp/Download/Standard/Fertility/) as the previous example. We look at two variables: total fertility and fertility by age of mother. In both cases we explore the numbers to date, but the same files contain projections of future fertility.
```
# fertility overall
uft = 'http://esa.un.org/unpd/wpp/DVD/Files/'
uft += '1_Indicators%20(Standard)/EXCEL_FILES/'
uft += '2_Fertility/WPP2015_FERT_F04_TOTAL_FERTILITY.XLS'
cols = [2] + list(range(5,18))
ftot = pd.read_excel(uft, sheetname=0, skiprows=16, parse_cols=cols, na_values=['…'])
ftot.head(3)[list(range(6))]
# rename some variables
names = list(ftot)
f = ftot.rename(columns={names[0]: 'Country'})
# select countries
countries = ['China', 'Japan', 'Germany', 'United States of America']
f = f[f['Country'].isin(countries)]
# shape
f = f.set_index('Country').T
f = f.rename(columns={'United States of America': 'United States'})
f.tail(3)
fig, ax = plt.subplots()
f.plot(ax=ax, kind='line', alpha=0.5, lw=3, figsize=(6.5, 4))
ax.set_title('Fertility (births per woman, lifetime)', fontsize=14, loc='left')
ax.legend(loc='best', fontsize=10, handlelength=2, labelspacing=0.15)
ax.set_ylim(ymin=0)
ax.hlines(2.1, -1, 13, linestyles='dashed')
ax.text(8.5, 2.4, 'Replacement = 2.1')
```
**Exercise.** What do you see here? What else would you like to know?
**Exercise.** Add Canada to the figure. How does it compare to the others? What other countries would you be interested in?
## Life expectancy
One of the bottom line summary numbers for mortality is life expectancy: if mortaility rates fall, people live longer, on average. Here we look at life expectancy at birth. There are also numbers for life expectancy given than you live to some specific age; for example, life expectancy given that you survive to age 60.
```
# life expectancy at birth, both sexes
ule = 'http://esa.un.org/unpd/wpp/DVD/Files/1_Indicators%20(Standard)/EXCEL_FILES/3_Mortality/'
ule += 'WPP2015_MORT_F07_1_LIFE_EXPECTANCY_0_BOTH_SEXES.XLS'
cols = [2] + list(range(5,34))
le = pd.read_excel(ule, sheetname=0, skiprows=16, parse_cols=cols, na_values=['…'])
le.head(3)[list(range(10))]
# rename some variables
oldname = list(le)[0]
l = le.rename(columns={oldname: 'Country'})
l.head(3)[list(range(8))]
# select countries
countries = ['China', 'Japan', 'Germany', 'United States of America']
l = l[l['Country'].isin(countries)]
# shape
l = l.set_index('Country').T
l = l.rename(columns={'United States of America': 'United States'})
l.tail()
fig, ax = plt.subplots()
l.plot(ax=ax, kind='line', alpha=0.5, lw=3, figsize=(6, 8), grid=True)
ax.set_title('Life expectancy at birth', fontsize=14, loc='left')
ax.set_ylabel('Life expectancy in years')
ax.legend(loc='best', fontsize=10, handlelength=2, labelspacing=0.15)
ax.set_ylim(ymin=0)
```
**Exercise.** What other countries would you like to see? Can you add them? The code below generates a list.
```
countries = le.rename(columns={oldname: 'Country'})['Country']
```
**Exercise.** Why do you think the US is falling behind? What would you look at to verify your conjecture?
## Mortality: aka death rates
Another thing that affects the age distribution of the population is the mortality rate: if mortality rates fall people live longer, on average. Here we look at how mortality rates have changed over the past 60+ years. Roughly speaking, people live an extra five years every generation. Which is a lot. Some of you will live to be a hundred. (Look at the 100+ agen category over time for Japan.)
The experts look at mortality rates by age. The UN has a [whole page](http://esa.un.org/unpd/wpp/Download/Standard/Mortality/) devoted to mortality numbers. We take 5-year mortality rates from the Abridged Life Table.
The numbers are percentages of people in a given age group who die over a 5-year period. 0.1 means that 90 percent of an age group is still alive in five years.
```
# mortality overall
url = 'http://esa.un.org/unpd/wpp/DVD/Files/'
url += '1_Indicators%20(Standard)/EXCEL_FILES/3_Mortality/'
url += 'WPP2015_MORT_F17_1_ABRIDGED_LIFE_TABLE_BOTH_SEXES.XLS'
cols = [2, 5, 6, 7, 9]
mort = pd.read_excel(url, sheetname=0, skiprows=16, parse_cols=cols, na_values=['…'])
mort.tail(3)
# change names
names = list(mort)
m = mort.rename(columns={names[0]: 'Country', names[2]: 'Age', names[3]: 'Interval', names[4]: 'Mortality'})
m.head(3)
```
**Comment.** At this point, we need to pivot the data. That's not something we've done before, so take it as simply something we can do easily if we have to. We're going to do this twice to produce different graphs:
* Compare countries for the same period.
* Compare different periods for the same country.
```
# compare countries for most recent period
countries = ['China', 'Japan', 'Germany', 'United States of America']
mt = m[m['Country'].isin(countries) & m['Interval'].isin([5]) & m['Period'].isin(['2010-2015'])]
print('Dimensions:', mt.shape)
mp = mt.pivot(index='Age', columns='Country', values='Mortality')
mp.head(3)
fig, ax = plt.subplots()
mp.plot(ax=ax, kind='line', alpha=0.5, linewidth=3,
# logy=True,
figsize=(6, 4))
ax.set_title('Mortality by age', fontsize=14, loc='left')
ax.set_ylabel('Mortality Rate (log scale)')
ax.legend(loc='best', fontsize=10, handlelength=2, labelspacing=0.15)
```
**Exercises.**
* What country's old people have the lowest mortality?
* What do you see here for the US? Why is our life expectancy shorter?
* What other countries would you like to see? Can you adapt the code to show them?
* Anything else cross your mind?
```
# compare periods for the one country -- countries[0] is China
mt = m[m['Country'].isin([countries[0]]) & m['Interval'].isin([5])]
print('Dimensions:', mt.shape)
mp = mt.pivot(index='Age', columns='Period', values='Mortality')
mp = mp[[0, 6, 12]]
mp.head(3)
fig, ax = plt.subplots()
mp.plot(ax=ax, kind='line', alpha=0.5, linewidth=3,
# logy=True,
figsize=(6, 4))
ax.set_title('Mortality over time', fontsize=14, loc='left')
ax.set_ylabel('Mortality Rate (log scale)')
ax.legend(loc='best', fontsize=10, handlelength=2, labelspacing=0.15)
```
**Exercise.** What do you see? What else would you like to know?
**Exercise.** Repeat this graph for the United States? How does it compare?
|
github_jupyter
|
# Model understanding and interpretability
In this colab, we will
- Will learn how to interpret model results and reason about the features
- Visualize the model results
```
import time
# We will use some np and pandas for dealing with input data.
import numpy as np
import pandas as pd
# And of course, we need tensorflow.
import tensorflow as tf
from matplotlib import pyplot as plt
from IPython.display import clear_output
tf.__version__
```
Below we demonstrate both *local* and *global* model interpretability for gradient boosted trees.
Local interpretability refers to an understanding of a model’s predictions at the individual example level, while global interpretability refers to an understanding of the model as a whole.
For local interpretability, we show how to create and visualize per-instance contributions using the technique outlined in [Palczewska et al](https://arxiv.org/pdf/1312.1121.pdf) and by Saabas in [Interpreting Random Forests](http://blog.datadive.net/interpreting-random-forests/) (this method is also available in scikit-learn for Random Forests in the [`treeinterpreter`](https://github.com/andosa/treeinterpreter) package). To distinguish this from feature importances, we refer to these values as directional feature contributions (DFCs).
For global interpretability we show how to retrieve and visualize gain-based feature importances, [permutation feature importances](https://www.stat.berkeley.edu/~breiman/randomforest2001.pdf) and also show aggregated DFCs.
# Setup
## Load dataset
We will be using the titanic dataset, where the goal is to predict passenger survival given characteristiscs such as gender, age, class, etc.
```
tf.logging.set_verbosity(tf.logging.ERROR)
tf.set_random_seed(123)
# Load dataset.
dftrain = pd.read_csv('https://storage.googleapis.com/tf-datasets/titanic/train.csv')
dfeval = pd.read_csv('https://storage.googleapis.com/tf-datasets/titanic/eval.csv')
y_train = dftrain.pop('survived')
y_eval = dfeval.pop('survived')
# Feature columns.
fcol = tf.feature_column
CATEGORICAL_COLUMNS = ['sex', 'n_siblings_spouses', 'parch', 'class', 'deck',
'embark_town', 'alone']
NUMERIC_COLUMNS = ['age', 'fare']
def one_hot_cat_column(feature_name, vocab):
return fcol.indicator_column(
fcol.categorical_column_with_vocabulary_list(feature_name,
vocab))
fc = []
for feature_name in CATEGORICAL_COLUMNS:
# Need to one-hot encode categorical features.
vocabulary = dftrain[feature_name].unique()
fc.append(one_hot_cat_column(feature_name, vocabulary))
for feature_name in NUMERIC_COLUMNS:
fc.append(fcol.numeric_column(feature_name,
dtype=tf.float32))
# Input functions.
def make_input_fn(X, y, n_epochs=None):
def input_fn():
dataset = tf.data.Dataset.from_tensor_slices((X.to_dict(orient='list'), y))
# For training, cycle thru dataset as many times as need (n_epochs=None).
dataset = (dataset
.repeat(n_epochs)
.batch(len(y))) # Use entire dataset since this is such a small dataset.
return dataset
return input_fn
# Training and evaluation input functions.
train_input_fn = make_input_fn(dftrain, y_train)
eval_input_fn = make_input_fn(dfeval, y_eval, n_epochs=1)
```
# Interpret model
## Local interpretability
Output directional feature contributions (DFCs) to explain individual predictions, using the approach outlined in [Palczewska et al](https://arxiv.org/pdf/1312.1121.pdf) and by Saabas in [Interpreting Random Forests](http://blog.datadive.net/interpreting-random-forests/). The DFCs are generated with:
`pred_dicts = list(est.experimental_predict_with_explanations(pred_input_fn))`
```
params = {
'n_trees': 50,
'max_depth': 3,
'n_batches_per_layer': 1,
# You must enable center_bias = True to get DFCs. This will force the model to
# make an initial prediction before using any features (e.g. use the mean of
# the training labels for regression or log odds for classification when
# using cross entropy loss).
'center_bias': True
}
est = tf.estimator.BoostedTreesClassifier(fc, **params)
# Train model.
est.train(train_input_fn)
# Evaluation.
results = est.evaluate(eval_input_fn)
clear_output()
pd.Series(results).to_frame()
```
## Local interpretability
Next you will output the directional feature contributions (DFCs) to explain individual predictions using the approach outlined in [Palczewska et al](https://arxiv.org/pdf/1312.1121.pdf) and by Saabas in [Interpreting Random Forests](http://blog.datadive.net/interpreting-random-forests/) (this method is also available in scikit-learn for Random Forests in the [`treeinterpreter`](https://github.com/andosa/treeinterpreter) package). The DFCs are generated with:
`pred_dicts = list(est.experimental_predict_with_explanations(pred_input_fn))`
(Note: The method is named experimental as we may modify the API before dropping the experimental prefix.)
```
import matplotlib.pyplot as plt
import seaborn as sns
sns_colors = sns.color_palette('colorblind')
pred_dicts = list(est.experimental_predict_with_explanations(eval_input_fn))
def clean_feature_names(df):
"""Boilerplate code to cleans up feature names -- this is unneed in TF 2.0"""
df.columns = [v.split(':')[0].split('_indi')[0] for v in df.columns.tolist()]
df = df.T.groupby(level=0).sum().T
return df
# Create DFC Pandas dataframe.
labels = y_eval.values
probs = pd.Series([pred['probabilities'][1] for pred in pred_dicts])
df_dfc = pd.DataFrame([pred['dfc'] for pred in pred_dicts])
df_dfc.columns = est._names_for_feature_id
df_dfc = clean_feature_names(df_dfc)
df_dfc.describe()
# Sum of DFCs + bias == probabality.
bias = pred_dicts[0]['bias']
dfc_prob = df_dfc.sum(axis=1) + bias
np.testing.assert_almost_equal(dfc_prob.values,
probs.values)
```
Plot results
```
import seaborn as sns # Make plotting nicer.
sns_colors = sns.color_palette('colorblind')
def plot_dfcs(example_id):
label, prob = labels[ID], probs[ID]
example = df_dfc.iloc[ID] # Choose ith example from evaluation set.
TOP_N = 8 # View top 8 features.
sorted_ix = example.abs().sort_values()[-TOP_N:].index
ax = example[sorted_ix].plot(kind='barh', color='g', figsize=(10,5))
ax.grid(False, axis='y')
plt.title('Feature contributions for example {}\n pred: {:1.2f}; label: {}'.format(ID, prob, label))
plt.xlabel('Contribution to predicted probability')
ID = 102
plot_dfcs(ID)
```
**??? ** How would you explain the above plot in plain english?
### Prettier plotting
Color codes based on directionality and adds feature values on figure. Please do not worry about the details of the plotting code :)
```
def plot_example_pretty(example):
"""Boilerplate code for better plotting :)"""
def _get_color(value):
"""To make positive DFCs plot green, negative DFCs plot red."""
green, red = sns.color_palette()[2:4]
if value >= 0: return green
return red
def _add_feature_values(feature_values, ax):
"""Display feature's values on left of plot."""
x_coord = ax.get_xlim()[0]
OFFSET = 0.15
for y_coord, (feat_name, feat_val) in enumerate(feature_values.items()):
t = plt.text(x_coord, y_coord - OFFSET, '{}'.format(feat_val), size=12)
t.set_bbox(dict(facecolor='white', alpha=0.5))
from matplotlib.font_manager import FontProperties
font = FontProperties()
font.set_weight('bold')
t = plt.text(x_coord, y_coord + 1 - OFFSET, 'feature\nvalue',
fontproperties=font, size=12)
TOP_N = 8 # View top 8 features.
sorted_ix = example.abs().sort_values()[-TOP_N:].index # Sort by magnitude.
example = example[sorted_ix]
colors = example.map(_get_color).tolist()
ax = example.to_frame().plot(kind='barh',
color=[colors],
legend=None,
alpha=0.75,
figsize=(10,6))
ax.grid(False, axis='y')
ax.set_yticklabels(ax.get_yticklabels(), size=14)
_add_feature_values(dfeval.iloc[ID].loc[sorted_ix], ax)
ax.set_title('Feature contributions for example {}\n pred: {:1.2f}; label: {}'.format(ID, probs[ID], labels[ID]))
ax.set_xlabel('Contribution to predicted probability', size=14)
plt.show()
return ax
# Plot results.
ID = 102
example = df_dfc.iloc[ID] # Choose ith example from evaluation set.
ax = plot_example_pretty(example)
```
## Global feature importances
1. Gain-based feature importances using `est.experimental_feature_importances`
2. Aggregate DFCs using `est.experimental_predict_with_explanations`
3. Permutation importances
Gain-based feature importances measure the loss change when splitting on a particular feature, while permutation feature importances are computed by evaluating model performance on the evaluation set by shuffling each feature one-by-one and attributing the change in model performance to the shuffled feature.
In general, permutation feature importance are preferred to gain-based feature importance, though both methods can be unreliable in situations where potential predictor variables vary in their scale of measurement or their number of categories and when features are correlated ([source](https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-9-307)). Check out [this article](http://explained.ai/rf-importance/index.html) for an in-depth overview and great discussion on different feature importance types.
## 1. Gain-based feature importances
```
features, importances = est.experimental_feature_importances(normalize=True)
df_imp = pd.DataFrame(importances, columns=['importances'], index=features)
# For plotting purposes. This is not needed in TF 2.0.
df_imp = clean_feature_names(df_imp.T).T.sort_values('importances', ascending=False)
# Visualize importances.
N = 8
ax = df_imp.iloc[0:N][::-1]\
.plot(kind='barh',
color=sns_colors[0],
title='Gain feature importances',
figsize=(10, 6))
ax.grid(False, axis='y')
plt.tight_layout()
```
**???** What does the x axis represent? -- A. It represents relative importance. Specifically, the average reduction in loss that occurs when a split occurs on that feature.
**???** Can we completely trust these results and the magnitudes? -- A. The results can be misleading because variables are correlated.
### 2. Average absolute DFCs
We can also average the absolute values of DFCs to understand impact at a global level.
```
# Plot.
dfc_mean = df_dfc.abs().mean()
sorted_ix = dfc_mean.abs().sort_values()[-8:].index # Average and sort by absolute.
ax = dfc_mean[sorted_ix].plot(kind='barh',
color=sns_colors[1],
title='Mean |directional feature contributions|',
figsize=(10, 6))
ax.grid(False, axis='y')
```
We can also see how DFCs vary as a feature value varies.
```
age = pd.Series(df_dfc.age.values, index=dfeval.age.values).sort_index()
sns.jointplot(age.index.values, age.values);
```
# Visualizing the model's prediction surface
Lets first simulate/create training data using the following formula:
$z=x* e^{-x^2 - y^2}$
Where $z$ is the dependent variable we are trying to predict and $x$ and $y$ are the features.
```
from numpy.random import uniform, seed
from matplotlib.mlab import griddata
# Create fake data
seed(0)
npts = 5000
x = uniform(-2, 2, npts)
y = uniform(-2, 2, npts)
z = x*np.exp(-x**2 - y**2)
# Prep data for training.
df = pd.DataFrame({'x': x, 'y': y, 'z': z})
xi = np.linspace(-2.0, 2.0, 200),
yi = np.linspace(-2.1, 2.1, 210),
xi,yi = np.meshgrid(xi, yi)
df_predict = pd.DataFrame({
'x' : xi.flatten(),
'y' : yi.flatten(),
})
predict_shape = xi.shape
def plot_contour(x, y, z, **kwargs):
# Grid the data.
plt.figure(figsize=(10, 8))
# Contour the gridded data, plotting dots at the nonuniform data points.
CS = plt.contour(x, y, z, 15, linewidths=0.5, colors='k')
CS = plt.contourf(x, y, z, 15,
vmax=abs(zi).max(), vmin=-abs(zi).max(), cmap='RdBu_r')
plt.colorbar() # Draw colorbar.
# Plot data points.
plt.xlim(-2, 2)
plt.ylim(-2, 2)
```
We can visualize our function:
```
zi = griddata(x, y, z, xi, yi, interp='linear')
plot_contour(xi, yi, zi)
plt.scatter(df.x, df.y, marker='.')
plt.title('Contour on training data')
plt.show()
def predict(est):
"""Predictions from a given estimator."""
predict_input_fn = lambda: tf.data.Dataset.from_tensors(dict(df_predict))
preds = np.array([p['predictions'][0] for p in est.predict(predict_input_fn)])
return preds.reshape(predict_shape)
```
First let's try to fit a linear model to the data.
```
fc = [tf.feature_column.numeric_column('x'),
tf.feature_column.numeric_column('y')]
train_input_fn = make_input_fn(df, df.z)
est = tf.estimator.LinearRegressor(fc)
est.train(train_input_fn, max_steps=500);
plot_contour(xi, yi, predict(est))
```
Not very good at all...
**???** Why is the linear model not performing well for this problem? Can you think of how to improve it just using a linear model?
Next let's try to fit a GBDT model to it and try to understand what the model does
```
for n_trees in [1,2,3,10,30,50,100,200]:
est = tf.estimator.BoostedTreesRegressor(fc,
n_batches_per_layer=1,
max_depth=4,
n_trees=n_trees)
est.train(train_input_fn)
plot_contour(xi, yi, predict(est))
plt.text(-1.8, 2.1, '# trees: {}'.format(n_trees), color='w', backgroundcolor='black', size=20)
```
Copyright 2019 Google Inc. Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License
|
github_jupyter
|
#**Exploratory Data Analysis**
### Setting Up Environment
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.style as style
import seaborn as sns
from scipy.stats import pointbiserialr
from scipy.stats import pearsonr
from scipy.stats import chi2_contingency
from sklearn.impute import SimpleImputer
plt.rcParams["figure.figsize"] = (15,8)
application_data_raw = pd.read_csv('application_data.csv', encoding = 'unicode_escape')
application_data_raw.info()
#application_data_raw.describe()
df = application_data_raw.copy()
```
### Data Cleaning
```
# drop the customer id column
df = df.drop(columns=['SK_ID_CURR'])
# remove invalid values in gender column
df['CODE_GENDER'] = df['CODE_GENDER'].replace("XNA", None)
# drop columns filled >25% with null values
num_missing_values = df.isnull().sum()
nulldf = round(num_missing_values/len(df)*100, 2)
cols_to_keep = nulldf[nulldf<=0.25].index.to_list()
df = df.loc[:, cols_to_keep] # 61 of 121 attributes were removed due to null values.
# impute remaining columns with null values
num_missing_values = df.isnull().sum()
missing_cols = num_missing_values[num_missing_values>0].index.tolist()
for col in missing_cols:
imp_mean = SimpleImputer(strategy='most_frequent')
imp_mean.fit(df[[col]])
df[col] = imp_mean.transform(df[[col]]).ravel()
df.info()
```
### Data Preprocessing
```
continuous_vars = ['CNT_CHILDREN', 'AMT_INCOME_TOTAL', 'AMT_CREDIT', 'AMT_ANNUITY', 'AMT_GOODS_PRICE', 'REGION_POPULATION_RELATIVE',
'DAYS_REGISTRATION', 'DAYS_ID_PUBLISH', 'CNT_FAM_MEMBERS', 'REGION_RATING_CLIENT', 'REGION_RATING_CLIENT_W_CITY',
'HOUR_APPR_PROCESS_START', 'EXT_SOURCE_2', 'DAYS_LAST_PHONE_CHANGE', 'YEARS_BIRTH', 'YEARS_EMPLOYED']
#categorical_variables = df.select_dtypes(include=["category"]).columns.tolist()
#len(categorical_variables)
categorical_vars = ['NAME_CONTRACT_TYPE', 'CODE_GENDER', 'FLAG_OWN_CAR', 'FLAG_OWN_REALTY', 'NAME_INCOME_TYPE','NAME_EDUCATION_TYPE',
'NAME_FAMILY_STATUS', 'NAME_HOUSING_TYPE', 'FLAG_MOBIL', 'FLAG_EMP_PHONE', 'FLAG_WORK_PHONE', 'FLAG_CONT_MOBILE', 'FLAG_PHONE',
'FLAG_EMAIL', 'WEEKDAY_APPR_PROCESS_START', 'REG_REGION_NOT_LIVE_REGION','REG_REGION_NOT_WORK_REGION',
'LIVE_REGION_NOT_WORK_REGION', 'REG_CITY_NOT_LIVE_CITY', 'REG_CITY_NOT_WORK_CITY', 'LIVE_CITY_NOT_WORK_CITY',
'ORGANIZATION_TYPE', 'FLAG_DOCUMENT_2', 'FLAG_DOCUMENT_3', 'FLAG_DOCUMENT_4', 'FLAG_DOCUMENT_5', 'FLAG_DOCUMENT_6',
'FLAG_DOCUMENT_7', 'FLAG_DOCUMENT_8', 'FLAG_DOCUMENT_9', 'FLAG_DOCUMENT_10', 'FLAG_DOCUMENT_11', 'FLAG_DOCUMENT_12',
'FLAG_DOCUMENT_13', 'FLAG_DOCUMENT_14', 'FLAG_DOCUMENT_15', 'FLAG_DOCUMENT_16', 'FLAG_DOCUMENT_17', 'FLAG_DOCUMENT_18',
'FLAG_DOCUMENT_19', 'FLAG_DOCUMENT_20', 'FLAG_DOCUMENT_21']
# plot to see distribution of categorical variables
n_cols = 4
fig, axes = plt.subplots(nrows=int(np.ceil(len(categorical_vars)/n_cols)),
ncols=n_cols,
figsize=(15,45))
for i in range(len(categorical_vars)):
var = categorical_vars[i]
dist = df[var].value_counts()
labels = dist.index
counts = dist.values
ax = axes.flatten()[i]
ax.bar(labels, counts)
ax.tick_params(axis='x', labelrotation = 90)
ax.title.set_text(var)
plt.tight_layout()
plt.show()
# This gives us an idea about which features may already be more useful
# Remove all FLAG_DOCUMENT features except for FLAG_DOCUMENT_3 as most did not submit, insignificant on model
vars_to_drop = []
vars_to_drop = ["FLAG_DOCUMENT_2"]
vars_to_drop += ["FLAG_DOCUMENT_{}".format(i) for i in range(4,22)]
# Unit conversions
df['AMT_INCOME_TOTAL'] = df['AMT_INCOME_TOTAL']/100000 # yearly income to be expressed in hundred thousands
df['YEARS_BIRTH'] = round((df['DAYS_BIRTH']*-1)/365).astype('int64') # days of birth changed to years of birth
df['YEARS_EMPLOYED'] = round((df['DAYS_EMPLOYED']*-1)/365).astype('int64') # days employed change to years employed
df.loc[df['YEARS_EMPLOYED']<0, 'YEARS_EMPLOYED'] = 0
df = df.drop(columns=['DAYS_BIRTH', 'DAYS_EMPLOYED'])
# Encoding categorical variables
def encode_cat(df, var_list):
for var in var_list:
df[var] = df[var].astype('category')
d = dict(zip(df[var], df[var].cat.codes))
df[var] = df[var].map(d)
print(var+" Category Codes")
print(d)
return df
already_coded = ['FLAG_MOBIL', 'FLAG_EMP_PHONE', 'FLAG_WORK_PHONE', 'FLAG_CONT_MOBILE', 'FLAG_PHONE', 'FLAG_EMAIL', 'REG_REGION_NOT_LIVE_REGION',
'REG_REGION_NOT_WORK_REGION', 'LIVE_REGION_NOT_WORK_REGION', 'REG_CITY_NOT_LIVE_CITY', 'REG_CITY_NOT_WORK_CITY',
'LIVE_CITY_NOT_WORK_CITY', 'FLAG_DOCUMENT_2', 'FLAG_DOCUMENT_3', 'FLAG_DOCUMENT_4', 'FLAG_DOCUMENT_5', 'FLAG_DOCUMENT_6',
'FLAG_DOCUMENT_7', 'FLAG_DOCUMENT_8', 'FLAG_DOCUMENT_9', 'FLAG_DOCUMENT_10', 'FLAG_DOCUMENT_11', 'FLAG_DOCUMENT_12',
'FLAG_DOCUMENT_13', 'FLAG_DOCUMENT_14', 'FLAG_DOCUMENT_15', 'FLAG_DOCUMENT_16', 'FLAG_DOCUMENT_17', 'FLAG_DOCUMENT_18',
'FLAG_DOCUMENT_19', 'FLAG_DOCUMENT_20', 'FLAG_DOCUMENT_21']
vars_to_encode = ['NAME_CONTRACT_TYPE', 'CODE_GENDER', 'FLAG_OWN_CAR', 'FLAG_OWN_REALTY', 'NAME_INCOME_TYPE','NAME_EDUCATION_TYPE',
'NAME_FAMILY_STATUS', 'NAME_HOUSING_TYPE', 'WEEKDAY_APPR_PROCESS_START', 'ORGANIZATION_TYPE']
for var in already_coded:
df[var] = df[var].astype('category')
df = encode_cat(df, vars_to_encode)
# removing rows with all 0
df = df[df.T.any()]
df.describe()
```
### Checking for correlations between variables
```
X = df.iloc[:, 1:]
# getting correlation matrix of continuous and categorical variables
cont = ['TARGET'] + continuous_vars
cat = ['TARGET'] + categorical_vars
cont_df = df.loc[:, cont]
cat_df = df.loc[:, cat]
cont_corr = cont_df.corr()
cat_corr = cat_df.corr()
plt.figure(figsize=(10,10));
sns.heatmap(cont_corr,
xticklabels = cont_corr.columns,
yticklabels = cont_corr.columns,
cmap="PiYG",
linewidth = 1);
# Find Point biserial correlation
for cat_var in categorical_vars:
for cont_var in continuous_vars:
data_cat = df[cat_var].to_numpy()
data_cont = df[cont_var].to_numpy()
corr, p_val = pointbiserialr(x=data_cat, y=data_cont)
if np.abs(corr) >= 0.8:
print(f'Categorical variable: {cat_var}, Continuous variable: {cont_var}, correlation: {corr}')
# Find Pearson correlation
total_len = len(continuous_vars)
for idx1 in range(total_len-1):
for idx2 in range(idx1+1, total_len):
cont_var1 = continuous_vars[idx1]
cont_var2 = continuous_vars[idx2]
data_cont1 = X[cont_var1].to_numpy()
data_cont2 = X[cont_var2].to_numpy()
corr, p_val = pearsonr(x=data_cont1, y=data_cont2)
if np.abs(corr) >= 0.8:
print(f' Continuous var 1: {cont_var1}, Continuous var 2: {cont_var2}, correlation: {corr}')
sns.scatterplot(data=X, x='CNT_CHILDREN',y='CNT_FAM_MEMBERS');
# Find Cramer's V correlation
total_len = len(categorical_vars)
for idx1 in range(total_len-1):
for idx2 in range(idx1+1, total_len):
cat_var1 = categorical_vars[idx1]
cat_var2 = categorical_vars[idx2]
c_matrix = pd.crosstab(X[cat_var1], X[cat_var2])
""" calculate Cramers V statistic for categorial-categorial association.
uses correction from Bergsma and Wicher,
Journal of the Korean Statistical Society 42 (2013): 323-328
"""
chi2 = chi2_contingency(c_matrix)[0]
n = c_matrix.sum().sum()
phi2 = chi2/n
r,k = c_matrix.shape
phi2corr = max(0, phi2-((k-1)*(r-1))/(n-1))
rcorr = r-((r-1)**2)/(n-1)
kcorr = k-((k-1)**2)/(n-1)
corr = np.sqrt(phi2corr/min((kcorr-1),(rcorr-1)))
if corr >= 0.8:
print(f'categorical variable 1 {cat_var1}, categorical variable 2: {cat_var2}, correlation: {corr}')
corr, p_val = pearsonr(x=df['REGION_RATING_CLIENT_W_CITY'], y=df['REGION_RATING_CLIENT'])
print(corr)
# High collinearity of 0.95 between variables suggests that one of it should be removed, we shall remove the REGION_RATING_CLIENT_W_CITY.
# Drop highly correlated variables
vars_to_drop += ['CNT_FAM_MEMBERS', 'REG_REGION_NOT_WORK_REGION', 'REG_CITY_NOT_WORK_CITY', 'AMT_GOODS_PRICE', 'REGION_RATING_CLIENT_W_CITY']
features_to_keep = [x for x in df.columns if x not in vars_to_drop]
features_to_keep
new_df = df.loc[:, features_to_keep]
new_df
# Checking correlation of X continuous columns vs TARGET column
plt.figure(figsize=(10,10))
df_corr = new_df.corr()
ax = sns.heatmap(df_corr,
xticklabels=df_corr.columns,
yticklabels=df_corr.columns,
annot = True,
cmap ="RdYlGn")
# No particular feature found to be significantly correlated with the target
# REGION_RATING_CLIENT and REGION_POPULATION_RELATIVE have multicollinearity
features_to_keep.remove('REGION_POPULATION_RELATIVE')
features_to_keep
# These are our final list of features
```
###Plots
```
ax1 = sns.boxplot(y='AMT_CREDIT', x= 'TARGET', data=new_df)
ax1.set_title("Target by amount credit of the loan", fontsize=20);
```
The amount credit of an individual does not seem to have a siginifcant effect on whether a person finds it difficult to pay. But they are crucial for our business reccomendations so we keep them.
```
ax2 = sns.barplot(x='CNT_CHILDREN', y= 'TARGET', data=new_df)
ax2.set_title("Target by number of children", fontsize=20);
```
From these plots, we can see that number of children has quite a significant effect on whether one defaults or not, with an increasing number of children proving more difficulty to return the loan.
```
ax3 = sns.barplot(x='NAME_FAMILY_STATUS', y= 'TARGET', data=new_df);
ax3.set_title("Target by family status", fontsize=20);
plt.xticks(np.arange(6), ['Civil marriage', 'Married', 'Separated', 'Single / not married',
'Unknown', 'Widow'], rotation=20);
```
Widows have the lowest likelihood of finding it difficult to pay, a possible target for our reccomendation strategy.
```
new_df['YEARS_BIRTH_CAT'] = pd.cut(df.YEARS_BIRTH, bins= [21, 25, 35, 45, 55, 69], labels= ["25 and below", "26-35", "36-45", "46-55", "Above 55"])
ax4 = sns.barplot(x='YEARS_BIRTH_CAT', y= 'TARGET', data=new_df);
ax4.set_title("Target by age", fontsize=20);
```
Analysis of age groups on ability to pay shows clear trend that the older you are, the better able you are to pay your loans. We will use this to craft our reccomendations.
```
ax5 = sns.barplot(y='TARGET', x= 'NAME_INCOME_TYPE', data=new_df);
ax5.set_title("Target by income type", fontsize=20);
plt.xticks(np.arange(0, 8),['Businessman', 'Commercial associate', 'Maternity leave', 'Pensioner',
'State servant', 'Student', 'Unemployed', 'Working'], rotation=20);
ax6 = sns.barplot(x='NAME_EDUCATION_TYPE', y= 'TARGET', data=new_df);
ax6.set_title("Target by education type", fontsize=20);
plt.xticks(np.arange(5), ['Academic Degree', 'Higher education', 'Incomplete higher', 'Lower secondary', 'Secondary / secondary special'], rotation=20);
ax7 = sns.barplot(x='ORGANIZATION_TYPE', y= 'TARGET', data=new_df);
ax7.set_title("Target by organization type", fontsize=20);
plt.xticks(np.arange(58), ['Unknown','Advertising','Agriculture', 'Bank', 'Business Entity Type 1', 'Business Entity Type 2',
'Business Entity Type 3', 'Cleaning', 'Construction', 'Culture', 'Electricity', 'Emergency', 'Government', 'Hotel', 'Housing', 'Industry: type 1', 'Industry: type 10', 'Industry: type 11', 'Industry: type 12', 'Industry: type 13', 'Industry: type 2', 'Industry: type 3', 'Industry: type 4', 'Industry: type 5', 'Industry: type 6', 'Industry: type 7', 'Industry: type 8', 'Industry: type 9', 'Insurance', 'Kindergarten', 'Legal Services', 'Medicine', 'Military', 'Mobile', 'Other', 'Police', 'Postal', 'Realtor', 'Religion', 'Restaurant', 'School', 'Security', 'Security Ministries', 'Self-employed', 'Services', 'Telecom', 'Trade: type 1', 'Trade: type 2', 'Trade: type 3', 'Trade: type 4', 'Trade: type 5', 'Trade: type 6', 'Trade: type 7', 'Transport: type 1', 'Transport: type 2', 'Transport: type 3', 'Transport: type 4','University'], rotation=90);
ax8 = sns.barplot(x='NAME_CONTRACT_TYPE', y= 'TARGET', data=new_df);
ax8.set_title("Target by contract type", fontsize=20);
plt.xticks(np.arange(2), ['Cash Loan', 'Revolving Loan']);
```
People who get revolving loans are more likely to pay back their loans than cash loans, perhaps due to the revolving loans being of a lower amount, and also its higher interest rate and recurring nature.
```
ax9 = sns.barplot(x='CODE_GENDER', y= 'TARGET', data=new_df);
ax9.set_title("Target by gender", fontsize=20);
plt.xticks(np.arange(2), ['Female', 'Male']);
```
Males find it harder to pay back their loans than females in general.
```
# Splitting Credit into bins of 10k
new_df['Credit_Category'] = pd.cut(new_df.AMT_CREDIT, bins= [0, 100000, 200000, 300000, 400000, 500000, 600000, 700000, 800000, 900000, 1000000, 4.050000e+06], labels= ["0-100k", "100-200k", "200-300k", "300-400k", "400-500k", "500-600k", "600-700k", "700-800k", "800-900k","900-1 million", "Above 1 million"])
setorder= new_df.groupby('Credit_Category')['TARGET'].mean().sort_values(ascending=False)
ax10 = sns.barplot(x='Credit_Category', y= 'TARGET', data=new_df, order = setorder.index);
ax10.set_title("Target by Credit Category", fontsize=20);
plt.show()
#No. of people who default
print(new_df.loc[new_df["TARGET"]==0, 'Credit_Category',].value_counts().sort_index())
#No. of people who repayed
print(new_df.loc[new_df["TARGET"]==1, 'Credit_Category',].value_counts().sort_index())
new_df['Credit_Category'].value_counts().sort_index()
# This will be useful for our first recommendation
#temp = new_df["Credit_Category"].value_counts()
#df1 = pd.DataFrame({"Credit_Category": temp.index,'Number of contracts': temp.values})
## Calculate the percentage of target=1 per category value
#cat_perc = new_df[["Credit_Category", 'TARGET']].groupby(["Credit_Category"],as_index=False).mean()
#cat_perc["TARGET"] = cat_perc["TARGET"]*100
#cat_perc.sort_values(by='TARGET', ascending=False, inplace=True)
#fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(12,6))
#s = sns.countplot(ax=ax1,
# x = "Credit_Category",
# data=new_df,
# hue ="TARGET",
# order=cat_perc["Credit_Category"],
# palette=['g','r'])
#ax1.set_title("Credit Category", fontdict={'fontsize' : 10, 'fontweight' : 3, 'color' : 'Blue'})
#ax1.legend(['Repayer','Defaulter'])
## If the plot is not readable, use the log scale.
##if ylog:
## ax1.set_yscale('log')
## ax1.set_ylabel("Count (log)",fontdict={'fontsize' : 10, 'fontweight' : 3, 'color' : 'Blue'})
#s.set_xticklabels(s.get_xticklabels(),rotation=90)
#s = sns.barplot(ax=ax2, x = "Credit_Category", y='TARGET', order=cat_perc["Credit_Category"], data=cat_perc, palette='Set2')
#s.set_xticklabels(s.get_xticklabels(),rotation=90)
#plt.ylabel('Percent of Defaulters [%]', fontsize=10)
#plt.tick_params(axis='both', which='major', labelsize=10)
#ax2.set_title("Credit Category" + " Defaulter %", fontdict={'fontsize' : 15, 'fontweight' : 5, 'color' : 'Blue'})
#plt.show();
new_df.info()
```
|
github_jupyter
|
```
import warnings
import sys
sys.path.insert(0, '../src')
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from felix_ml_tools import macgyver as mg
from preprocess import *
from sklearn.linear_model import LassoCV, Lasso
warnings.filterwarnings('ignore')
pd.set_option("max_columns", None)
pd.set_option("max_rows", None)
pd.set_option('display.float_format', lambda x: '%.3f' % x)
%matplotlib inline
# todas as UFs do Nordeste
UFs_NORDESTE = ['AL', 'BA', 'CE', 'MA', 'PB', 'PE', 'PI', 'RN', 'SE']
UFs_NORDESTE_NAMES = ['Alagoas', 'Bahia', 'Ceará', 'Maranhão', 'Paraíba', 'Pernambuco', 'Piauí', 'Rio Grande do Norte', 'Sergipe']
```
# Intro
Ingesting some base data
-----------
## Compreender os índices do CRAS
```
idcras = pd.read_excel(
'../../dataset/GAMMAChallenge21Dados/6. Assistencia Social/IDCRAS.xlsx',
sheet_name = 'dados'
)
inspect(idcras);
idcras = generates_normalized_column(idcras,
column_to_normalize='nome_municipio',
column_normalized='nome_municipio_norm')
idcras = keep_last_date(idcras, column_dates='ano')
columns_to_keep = ['uf', 'nome_municipio', 'nome_municipio_norm', 'cod_ibge', 'ind_estru_fisic', 'ind_servi', 'ind_rh', 'id_cras.1']
idcras = idcras[columns_to_keep]
# há cidades com mais de um nucleo CRAS
idcras = (idcras[idcras['uf'].isin(UFs_NORDESTE)]
.groupby(['uf', 'nome_municipio', 'nome_municipio_norm', 'cod_ibge'])[
['ind_estru_fisic', 'ind_servi', 'ind_rh', 'id_cras.1']
]
.mean())
```
Como são os índices médios das unidades públicas CRAS para estados e municipios?
```
idcras_cities = (
idcras.groupby(['nome_municipio',
'nome_municipio_norm',
'cod_ibge'])[['ind_estru_fisic',
'ind_servi', 'ind_rh',
'id_cras.1']]
.mean()
.reset_index()
)
inspect(idcras_cities);
idcras_states = (
idcras.groupby(['uf'])[['ind_estru_fisic',
'ind_servi',
'ind_rh',
'id_cras.1']]
.mean()
.sort_values('uf')
.reset_index()
)
inspect(idcras_states);
```
-----------
### Registros CRAS RMA
```
cras_rma = pd.read_excel(
'../../dataset/GAMMAChallenge21Dados/6. Assistencia Social/RMA.xlsx',
sheet_name = 'dados_CRAS'
)
cras_rma = generates_normalized_column(cras_rma,
column_to_normalize='nome_municipio',
column_normalized='nome_municipio_norm')
cras_rma = keep_last_date(cras_rma, column_dates='ano')
inspect(cras_rma);
cras_rma_states = cras_rma[cras_rma['uf'].isin(UFs_NORDESTE)]
cras_rma_states_grouped = (
cras_rma_states
.groupby(['uf', 'nome_municipio_norm'])
[['cod_ibge', 'a1', 'a2', 'b1', 'b2', 'b3', 'b5', 'b6', 'c1', 'c6', 'd1', 'd4']]
.mean()
.fillna(0)
.astype(int)
)
registry_cras_rma_states_grouped = (
cras_rma_states_grouped.groupby(['uf'])[['a1', 'a2', 'b1', 'b2', 'b3', 'b5', 'b6', 'c1', 'c6', 'd1', 'd4']]
.mean()
.sort_values('uf')
)
registry_cras_rma_states_grouped = (
registry_cras_rma_states_grouped
.rename(columns = {
'a1': 'a1 Total de famílias em acompanhamento pelo PAIF',
'a2': 'a2 Novas famílias inseridas no acompanhamento do PAIF no mês de referência',
'b1': 'b1 PAIF Famílias em situação de extrema pobreza',
'b2': 'b2 PAIF Famílias beneficiárias do Programa Bolsa Família',
'b3': 'b3 PAIF Famílias beneficiárias do Programa Bolsa Família, em descumprimento de condicionalidades',
'b5': 'b5 PAIF Famílias com crianças/adolescentes em situação de trabalho infantil',
'b6': 'b6 PAIF Famílias com crianças e adolescentes em Serviço de Acolhimento',
'c1': 'c1 Total de atendimentos individualizados realizados, no mês',
'c6': 'c6 Visitas domiciliares',
'd1': 'd1 Famílias participando regularmente de grupos no âmbito do PAIF',
'd4': 'd4 Adolescentes de 15 a 17 anos em Serviços de Convivência e Fortalecimentos de Vínculos'
},
inplace = False)
)
registry_cras_rma_states_grouped
```
## Target Population
```
sim_pf_homcidios = pd.read_parquet('../../dataset/handled/sim_pf_homcidios.parquet')
sim_pf_homcidios = sim_pf_homcidios[sim_pf_homcidios['uf'].isin(UFs_NORDESTE)]
ibge_municipio_pib = pd.read_parquet('../../dataset/handled/ibge_municipio_pib.parquet')
ibge_estados = pd.read_excel(
'../../dataset/GAMMAChallenge21Dados/3. Atlas de Desenvolvimento Humano/Atlas 2013_municipal, estadual e Brasil.xlsx',
sheet_name = 'UF 91-00-10'
)
ibge_estados = keep_last_date(ibge_estados, 'ANO')
ibge_estados = ibge_estados[ibge_estados.UFN.isin(UFs_NORDESTE_NAMES)]
inspect(ibge_estados);
mortes_estados_norm = dict()
for estado in ibge_estados.UFN.sort_values().to_list():
mortes_estados_norm[estado] = (
(sim_pf_homcidios[sim_pf_homcidios.nomeUf == estado]
.groupby('uf')['uf']
.count()
.values[0]) / (ibge_estados[ibge_estados.UFN == estado]
.POP
.values[0])*1000
)
mortes_estados_norm = pd.DataFrame.from_dict(mortes_estados_norm, orient='index', columns=['mortes_norm'])
mortes_estados_norm['mortes_norm']
homicidios_municipios = sim_pf_homcidios.groupby(['uf','municipioCodigo','nomeMunicipioNorm'])[['contador']].count().astype(int).reset_index()
inspect(homicidios_municipios);
ibge_municipio_populacao_estimada = pd.read_parquet('../../dataset/handled/ibge_municipio_populacao_estimada.parquet')
ibge_municipio_populacao_estimada = generates_normalized_column(ibge_municipio_populacao_estimada, 'nomeMunicipio', 'nome_municipio_norm')
inspect(ibge_municipio_populacao_estimada);
cras_rma_municipios = cras_rma.groupby(['uf','cod_ibge','nome_municipio_norm'])[[
'a1', 'a2',
'b1', 'b2', 'b3', 'b4', 'b5', 'b6',
'c1', 'c2', 'c3', 'c4', 'c5', 'c6',
'd1', 'd2', 'd3', 'd4', 'd5', 'd6', 'd7'
]].mean().fillna(0).astype(int).reset_index()
inspect(cras_rma_municipios);
list_pop = list()
list_mortes_norm = list()
for cod in cras_rma_municipios.cod_ibge:
populacao = ibge_municipio_populacao_estimada[ibge_municipio_populacao_estimada['municipioCodigo'].str.contains(str(cod))]['populacaoEstimada'].astype(int).values[0]
list_pop.append(populacao)
if homicidios_municipios[(homicidios_municipios['municipioCodigo'].str.contains(str(cod)))&(ibge_municipio_populacao_estimada['faixaPopulacaoEstimada'] == '0:10000')].any().any():
mortes = homicidios_municipios[(homicidios_municipios['municipioCodigo'].str.contains(str(cod)))]['contador'].astype(int).values[0]
mortes /= populacao
mortes *= 10000
else:
mortes = None
list_mortes_norm.append(mortes)
```
## Hypothesis
### Hypothesis 2
```
plot_grouped_df(sim_pf_homcidios.groupby('uf')['uf'],
xlabel='Quantidade de Homicidios',
ylabel='UF',
figsize=(17,3))
mortes_estados_norm.plot(kind='bar',
figsize=(17,3),
rot=0,
grid=True).set_ylabel("Quantidade de Homicidios Normalizado")
registry_cras_rma_states_grouped['mortes'] = mortes_estados_norm['mortes_norm'].values
registry_cras_rma_states_grouped
```
#### (2.1) Estados com CRAS suportam moradores com dificuldade
```
registry_cras_rma_states_grouped.plot(kind='bar',
title='Registro de Atividades - CRAS',
figsize=(17,5),
ylabel='Registros',
xlabel='UF',
rot=0,
grid=True)
```
```
X = registry_cras_rma_states_grouped.iloc[:,:-1]
y = registry_cras_rma_states_grouped.iloc[:,-1:]
reg = LassoCV()
reg.fit(X, y)
print("Melhor valor de alpha usando LassoCV: %f" % reg.alpha_)
print("Melhor score usando LassoCV: %f" % reg.score(X,y))
coef = pd.Series(reg.coef_, index = X.columns)
print("LASSO manteve " + str(sum(coef != 0)) + " variaveis, e elimina " + str(sum(coef == 0)) + " variaveis")
imp_coef = coef.sort_values()
import matplotlib
matplotlib.rcParams['figure.figsize'] = (5, 5)
imp_coef.plot(kind = "barh")
plt.title("Feature importance usando LASSO")
trabalhos_cras_estados_totfam = (
registry_cras_rma_states_grouped[['a1 Total de famílias em acompanhamento pelo PAIF',
'c1 Total de atendimentos individualizados realizados, no mês']]
)
trabalhos_cras_estados_extpobres = (
registry_cras_rma_states_grouped[['b1 PAIF Famílias em situação de extrema pobreza']]
)
trabalhos_cras_estados_acompanha = (
registry_cras_rma_states_grouped[['b5 PAIF Famílias com crianças/adolescentes em situação de trabalho infantil',
'b6 PAIF Famílias com crianças e adolescentes em Serviço de Acolhimento']]
)
trabalhos_cras_estados_atendimento = (
registry_cras_rma_states_grouped[['d1 Famílias participando regularmente de grupos no âmbito do PAIF',
'd4 Adolescentes de 15 a 17 anos em Serviços de Convivência e Fortalecimentos de Vínculos']]
)
def plot_cras_registries(df):
df.plot(kind='bar',
title='Registro de Atividades - CRAS',
figsize=(10,5),
ylabel='Registros',
xlabel='UF',
fontsize=12,
colormap='Pastel2',
rot=0,
grid=True)
plot_cras_registries(trabalhos_cras_estados_totfam)
plot_cras_registries(trabalhos_cras_estados_extpobres)
plot_cras_registries(trabalhos_cras_estados_acompanha)
plot_cras_registries(trabalhos_cras_estados_atendimento)
```
## Recomendação
O CRAS demonstra que, se rankear-mos os estados entra os mais e menos violentos, onde ocorre maior média do índice de registros apresentado por unidades públicas CRAS, o estado tende a controlar o números de homicidios
- índices na identificação de famílias em situação de extrema pobreza
- índices na identificação de trabalho infantil e acolhimento
- índices na participação das famílias em grupos no âmbito do PAIF
---------------
#### (2.2) Estados com CRAS suportam moradores com dificuldade
```
#cras_municipios['populacao'] = list_pop
cras_rma_municipios['Mortes/Populacao'] = list_mortes_norm
inspect(cras_rma_municipios);
cras_rma_municipios = cras_rma_municipios.dropna()
cras_rma_municipios_mortes = (
cras_rma_municipios[['uf',
'nome_municipio_norm',
'a1', 'a2',
'b1', 'b2', 'b3', 'b4', 'b5', 'b6',
'c1', 'c2', 'c3', 'c4', 'c5', 'c6', 'd1',
'd2', 'd3', 'd4', 'd5', 'd6', 'd7',
'Mortes/Populacao']]
.reset_index(drop=True)
)
inspect(cras_rma_municipios_mortes);
corr = cras_rma_municipios_mortes.corr()
corr.style.background_gradient(cmap='hot')
```
### Não é possível identificar, através de correlação, que alguma ação registrada pelo CRAS tem influência nos níveis de violência das cidades
```
cor_target = abs(corr["Mortes/Populacao"])
relevant_features = cor_target[cor_target>=0.07]
relevant_features
X = cras_rma_municipios_mortes.drop(['uf','nome_municipio_norm','Mortes/Populacao'],1)
y = cras_rma_municipios_mortes['Mortes/Populacao']
reg = LassoCV()
reg.fit(X, y)
print("Melhor valor de alpha usando LassoCV: %f" % reg.alpha_)
print("Melhor score usando LassoCV: %f" % reg.score(X,y))
coef = pd.Series(reg.coef_, index = X.columns)
print("LASSO manteve " + str(sum(coef != 0)) + " variaveis, e elimina " + str(sum(coef == 0)) + " variaveis")
imp_coef = coef.sort_values()
import matplotlib
matplotlib.rcParams['figure.figsize'] = (5, 5)
imp_coef.plot(kind = "barh")
plt.title("Feature importance usando LASSO")
```
```
cras_municipios_feat_select = cras_rma_municipios_mortes[['uf', 'nome_municipio_norm', 'Mortes/Populacao', 'c1', 'c3', 'd6']].reset_index(drop=True)
corr = cras_municipios_feat_select.sort_values(by='Mortes/Populacao', ascending=False).corr()
corr.style.background_gradient(cmap='hot')
```
## Recomendação
Não é possível recomendar uma ação do CRAS a nível municipal até este instante.
|
github_jupyter
|
```
import pandas as pd
from pandas import DataFrame
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from datetime import datetime, timedelta
from statsmodels.tsa.arima_model import ARIMA
from statsmodels.tsa.statespace.sarimax import SARIMAX
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
from statsmodels.tsa.stattools import adfuller
from statsmodels.tsa.seasonal import seasonal_decompose
from scipy import stats
import statsmodels.api as sm
from itertools import product
from math import sqrt
from sklearn.metrics import mean_squared_error
import warnings
warnings.filterwarnings('ignore')
%matplotlib inline
colors = ["windows blue", "amber", "faded green", "dusty purple"]
sns.set(rc={"figure.figsize": (20,10), "axes.titlesize" : 18, "axes.labelsize" : 12,
"xtick.labelsize" : 14, "ytick.labelsize" : 14 })
dateparse = lambda dates: pd.datetime.strptime(dates, '%m/%d/%Y')
df = pd.read_csv('BTCUSDTEST.csv', parse_dates=['Date'], index_col='Date', date_parser=dateparse)
df = df.loc[:, ~df.columns.str.contains('^Unnamed')]
df.sample(5)
# Extract the bitcoin data only
btc=df[df['Symbol']=='BTCUSD']
# Drop some columns
btc.drop(['Volume', 'Market Cap'],axis=1,inplace=True)
# Resampling to monthly frequency
btc_month = btc.resample('M').mean()
#seasonal_decompose(btc_month.close, freq=12).plot()
seasonal_decompose(btc_month.Close, model='additive').plot()
print("Dickey–Fuller test: p=%f" % adfuller(btc_month.Close)[1])
# Box-Cox Transformations
btc_month['close_box'], lmbda = stats.boxcox(btc_month.Close)
print("Dickey–Fuller test: p=%f" % adfuller(btc_month.close_box)[1])
# Seasonal differentiation (12 months)
btc_month['box_diff_seasonal_12'] = btc_month.close_box - btc_month.close_box.shift(12)
print("Dickey–Fuller test: p=%f" % adfuller(btc_month.box_diff_seasonal_12[12:])[1])
# Seasonal differentiation (3 months)
btc_month['box_diff_seasonal_3'] = btc_month.close_box - btc_month.close_box.shift(3)
print("Dickey–Fuller test: p=%f" % adfuller(btc_month.box_diff_seasonal_3[3:])[1])
# Regular differentiation
btc_month['box_diff2'] = btc_month.box_diff_seasonal_12 - btc_month.box_diff_seasonal_12.shift(1)
# STL-decomposition
seasonal_decompose(btc_month.box_diff2[13:]).plot()
print("Dickey–Fuller test: p=%f" % adfuller(btc_month.box_diff2[13:])[1])
#autocorrelation_plot(btc_month.close)
plot_acf(btc_month.Close[13:].values.squeeze(), lags=12)
plt.tight_layout()
# Initial approximation of parameters using Autocorrelation and Partial Autocorrelation Plots
ax = plt.subplot(211)
# Plot the autocorrelation function
#sm.graphics.tsa.plot_acf(btc_month.box_diff2[13:].values.squeeze(), lags=48, ax=ax)
plot_acf(btc_month.box_diff2[13:].values.squeeze(), lags=12, ax=ax)
ax = plt.subplot(212)
#sm.graphics.tsa.plot_pacf(btc_month.box_diff2[13:].values.squeeze(), lags=48, ax=ax)
plot_pacf(btc_month.box_diff2[13:].values.squeeze(), lags=12, ax=ax)
plt.tight_layout()
# Initial approximation of parameters
qs = range(0, 3)
ps = range(0, 3)
d=1
parameters = product(ps, qs)
parameters_list = list(parameters)
len(parameters_list)
# Model Selection
results = []
best_aic = float("inf")
warnings.filterwarnings('ignore')
for param in parameters_list:
try:
model = SARIMAX(btc_month.close_box, order=(param[0], d, param[1])).fit(disp=-1)
except ValueError:
print('bad parameter combination:', param)
continue
aic = model.aic
if aic < best_aic:
best_model = model
best_aic = aic
best_param = param
results.append([param, model.aic])
# Best Models
result_table = pd.DataFrame(results)
result_table.columns = ['parameters', 'aic']
print(result_table.sort_values(by = 'aic', ascending=True).head())
print(best_model.summary())
print("Dickey–Fuller test:: p=%f" % adfuller(best_model.resid[13:])[1])
best_model.plot_diagnostics(figsize=(15, 12))
plt.show()
# Inverse Box-Cox Transformation Function
def invboxcox(y,lmbda):
if lmbda == 0:
return(np.exp(y))
else:
return(np.exp(np.log(lmbda*y+1)/lmbda))
# Prediction
btc_month_pred = btc_month[['Close']]
date_list = [datetime(2019, 10, 31), datetime(2019, 11, 30), datetime(2020, 7, 31)]
future = pd.DataFrame(index=date_list, columns= btc_month.columns)
btc_month_pred = pd.concat([btc_month_pred, future])
#btc_month_pred['forecast'] = invboxcox(best_model.predict(start=0, end=75), lmbda)
btc_month_pred['forecast'] = invboxcox(best_model.predict(start=datetime(2015, 10, 31), end=datetime(2020, 7, 31)), lmbda)
btc_month_pred.Close.plot(linewidth=3)
btc_month_pred.forecast.plot(color='r', ls='--', label='Predicted Close', linewidth=3)
plt.legend()
plt.grid()
plt.title('Bitcoin monthly forecast')
plt.ylabel('USD')
#from google.colab import files
#uploaded = files.upload()
```
|
github_jupyter
|
```
# Install a pip package in the current Jupyter kernel
import sys
!{sys.executable} -m pip install pip install python-slugify
!{sys.executable} -m pip install pip install bs4
!{sys.executable} -m pip install pip install lxml
import requests, random, logging, urllib.request, json
from bs4 import BeautifulSoup
from tqdm import tqdm
from slugify import slugify
logging.basicConfig(filename='app.log', filemode='w', format='%(name)s - %(levelname)s - %(message)s')
url = 'https://www.investopedia.com/financial-term-dictionary-4769738'
master_links = []
page = urllib.request.urlopen(url).read().decode('utf8','ignore')
soup = BeautifulSoup(page,"lxml")
for link in soup.find_all('a',{'class': 'terms-bar__link mntl-text-link'}, href = True):
master_links.append(link.get('href'))
master_links = master_links[0:27]
with open('URL_INDEX_BY_ALPHA.txt', 'w') as f:
for item in master_links:
f.write("%s\n" % item)
list_alpha = []
for articleIdx in master_links:
page = urllib.request.urlopen(articleIdx).read().decode('utf8','ignore')
soup = BeautifulSoup(page,"lxml")
for link in soup.find_all('a',{'class': 'dictionary-top300-list__list mntl-text-link'}, href = True):
list_alpha.append(link.get('href'))
with open('FULL_URL_INDEX.txt', 'w') as f:
for item in list_alpha:
f.write("%s\n" % item)
logf = open("error.log", "w")
# for article in tqdm(random.sample(list_alpha, 10)):
data = {} #json file
for article in tqdm(list_alpha):
list_related = []
body = []
try:
page = urllib.request.urlopen(article, timeout = 3).read().decode('utf8','ignore')
soup = BeautifulSoup(page,"lxml")
myTags = soup.find_all('p', {'class': 'comp mntl-sc-block finance-sc-block-html mntl-sc-block-html'})
for link in soup.find_all('a',{'class': 'related-terms__title mntl-text-link'}, href = True):
list_related.append(link.get('href'))
title = slugify(soup.find('title').get_text(strip=True)) + '.json'
data['name'] = soup.find('title').get_text(strip=True)
data['@id'] = article
data['related'] = list_related
post = ''
for tag in myTags:
# body.append(str(tag.get_text(strip=True).encode('utf8', errors='replace'))) #get text content
body.append(tag.decode_contents()) # get html content
f = 'data/' + title
data['body'] = body
w = open(f, 'w')
w.write(json.dumps(data))
w.close()
except:
logf.write("Failed to extract: {0}\n".format(str(article)))
logging.error("Exception occurred", exc_info=True)
finally:
pass
```
# create RDF from JSON
```
# Install a pip package in the current Jupyter kernel
import sys
!{sys.executable} -m pip install pip install rdflib
import os, json, rdflib
from rdflib import URIRef, BNode, Literal, Namespace, Graph
from rdflib.namespace import CSVW, DC, DCAT, DCTERMS, DOAP, FOAF, ODRL2, ORG, OWL, \
PROF, PROV, RDF, RDFS, SDO, SH, SKOS, SOSA, SSN, TIME, \
VOID, XMLNS, XSD
path_to_json = 'data/'
json_files = [pos_json for pos_json in os.listdir(path_to_json) if pos_json.endswith('.json')]
for link in json_files:
with open('data/'+link) as f:
data = json.load(f)
#create RDF graph
g = Graph()
INVP = Namespace("https://www.investopedia.com/vocab/")
g.bind("rdfs", RDFS)
g.bind("schema", SDO)
g.bind("invp", INVP)
rdf_content = ''
termS = URIRef(data['@id'])
g.add((termS, RDF.type, INVP.Term))
g.add((termS, SDO.url, termS))
g.add((termS, RDFS.label, Literal(data['name'])))
for rel in data['related']:
g.add((termS, INVP.relates_to, URIRef(rel)))
separator = ''
content= separator.join(data['body'])
g.add((termS, INVP.description, Literal(content)))
output = '# '+data['name']+ '\n' + g.serialize(format="turtle").decode("utf-8")
w = open('rdf/'+link.replace('.json','')+'.ttl', 'wb')
w.write(output.encode("utf8"))
w.close()
```
|
github_jupyter
|
# Introduction to Python
> Defining Functions with Python
Kuo, Yao-Jen
## TL; DR
> In this lecture, we will talk about defining functions with Python.
## Encapsulations
## What is encapsulation?
> Encapsulation refers to one of two related but distinct notions, and sometimes to the combination thereof:
> 1. A language mechanism for restricting direct access to some of the object's components.
> 2. A language construct that facilitates the bundling of data with the methods (or other functions) operating on that data.
Source: <https://en.wikipedia.org/wiki/Encapsulation_(computer_programming)>
## Why encapsulation?
As our codes piled up, we need a mechanism making them:
- more structured
- more reusable
- more scalable
## Python provides several tools for programmers organizing their codes
- Functions
- Classes
- Modules
- Libraries
## How do we decide which tool to adopt?
Simply put, that depends on **scale** and project spec.
## These components are mixed and matched with great flexibility
- A couple lines of code assembles a function
- A couple of functions assembles a class
- A couple of classes assembles a module
- A couple of modules assembles a library
- A couple of libraries assembles a larger library
## Codes, assemble!

Source: <https://giphy.com/>
## Functions
## What is a function
> A function is a named sequence of statements that performs a computation, either mathematical, symbolic, or graphical. When we define a function, we specify the name and the sequence of statements. Later, we can call the function by name.
## Besides built-in functions or library-powered functions, we sometimes need to self-define our own functions
- `def` the name of our function
- `return` the output of our function
```python
def function_name(INPUTS, ARGUMENTS, ...):
"""
docstring: print documentation when help() is called
"""
# sequence of statements
return OUTPUTS
```
## The principle of designing of a function is about mapping the relationship of inputs and outputs
- The one-on-one relationship
- The many-on-one relationship
- The one-on-many relationship
- The many-on-many releationship
## The one-on-one relationship
Using scalar as input and output.
```
def absolute(x):
"""
Return the absolute value of the x.
"""
if x >= 0:
return x
else:
return -x
```
## Once the function is defined, call as if it is a built-in function
```
help(absolute)
print(absolute(-5566))
print(absolute(5566))
print(absolute(0))
```
## The many-on-one relationship relationship
- Using scalars or structures for fixed inputs
- Using `*args` or `**kwargs` for flexible inputs
## Using scalars for fixed inputs
```
def product(x, y):
"""
Return the product values of x and y.
"""
return x*y
print(product(5, 6))
```
## Using structures for fixed inputs
```
def product(x):
"""
x: an iterable.
Return the product values of x.
"""
prod = 1
for i in x:
prod *= i
return prod
print(product([5, 5, 6, 6]))
```
## Using `*args` for flexible inputs
- As in flexible arguments
- Getting flexible `*args` as a `tuple`
```
def plain_return(*args):
"""
Return args.
"""
return args
print(plain_return(5, 5, 6, 6))
```
## Using `**kwargs` for flexible inputs
- AS in keyword arguments
- Getting flexible `**kwargs` as a `dict`
```
def plain_return(**kwargs):
"""
Retrun kwargs.
"""
return kwargs
print(plain_return(TW='Taiwan', JP='Japan', CN='China', KR='South Korea'))
```
## The one-on-many relationship
- Using default `tuple` with comma
- Using preferred data structure
## Using default `tuple` with comma
```
def as_integer_ratio(x):
"""
Return x as integer ratio.
"""
x_str = str(x)
int_part = int(x_str.split(".")[0])
decimal_part = x_str.split(".")[1]
n_decimal = len(decimal_part)
denominator = 10**(n_decimal)
numerator = int(decimal_part)
while numerator % 2 == 0 and denominator % 2 == 0:
denominator /= 2
numerator /= 2
while numerator % 5 == 0 and denominator % 5 == 0:
denominator /= 5
numerator /= 5
final_numerator = int(int_part*denominator + numerator)
final_denominator = int(denominator)
return final_numerator, final_denominator
print(as_integer_ratio(3.14))
print(as_integer_ratio(0.56))
```
## Using preferred data structure
```
def as_integer_ratio(x):
"""
Return x as integer ratio.
"""
x_str = str(x)
int_part = int(x_str.split(".")[0])
decimal_part = x_str.split(".")[1]
n_decimal = len(decimal_part)
denominator = 10**(n_decimal)
numerator = int(decimal_part)
while numerator % 2 == 0 and denominator % 2 == 0:
denominator /= 2
numerator /= 2
while numerator % 5 == 0 and denominator % 5 == 0:
denominator /= 5
numerator /= 5
final_numerator = int(int_part*denominator + numerator)
final_denominator = int(denominator)
integer_ratio = {
'numerator': final_numerator,
'denominator': final_denominator
}
return integer_ratio
print(as_integer_ratio(3.14))
print(as_integer_ratio(0.56))
```
## The many-on-many relationship
A mix-and-match of one-on-many and many-on-one relationship.
## Handling errors
## Coding mistakes are common, they happen all the time

Source: Google Search
## How does a function designer handle errors?
Python mistakes come in three basic flavors:
- Syntax errors
- Runtime errors
- Semantic errors
## Syntax errors
Errors where the code is not valid Python (generally easy to fix).
```
# Python does not need curly braces to create a code block
for (i in range(10)) {print(i)}
```
## Runtime errors
Errors where syntactically valid code fails to execute, perhaps due to invalid user input (sometimes easy to fix)
- `NameError`
- `TypeError`
- `ZeroDivisionError`
- `IndexError`
- ...etc.
```
print('5566'[4])
```
## Semantic errors
Errors in logic: code executes without a problem, but the result is not what you expect (often very difficult to identify and fix)
```
def product(x):
"""
x: an iterable.
Return the product values of x.
"""
prod = 0 # set
for i in x:
prod *= i
return prod
print(product([5, 5, 6, 6])) # expecting 900
```
## Using `try` and `except` to catch exceptions
```python
try:
# sequence of statements if everything is fine
except TYPE_OF_ERROR:
# sequence of statements if something goes wrong
```
```
try:
exec("""for (i in range(10)) {print(i)}""")
except SyntaxError:
print("Encountering a SyntaxError.")
try:
print('5566'[4])
except IndexError:
print("Encountering a IndexError.")
try:
print(5566 / 0)
except ZeroDivisionError:
print("Encountering a ZeroDivisionError.")
# it is optional to specify the type of error
try:
print(5566 / 0)
except:
print("Encountering a whatever error.")
```
## Scope
## When it comes to defining functions, it is vital to understand the scope of a variable
## What is scope?
> In computer programming, the scope of a name binding, an association of a name to an entity, such as a variable, is the region of a computer program where the binding is valid.
Source: <https://en.wikipedia.org/wiki/Scope_(computer_science)>
## Simply put, now we have a self-defined function, so the programming environment is now split into 2:
- Global
- Local
## A variable declared within the indented block of a function is a local variable, it is only valid inside the `def` block
```
def check_odd_even(x):
mod = x % 2 # local variable, declared inside def block
if mod == 0:
return '{} is a even number.'.format(x)
else:
return '{} is a odd number.'.format(x)
print(check_odd_even(0))
print(x)
print(mod)
```
## A variable declared outside of the indented block of a function is a glocal variable, it is valid everywhere
```
x = 0
mod = x % 2
def check_odd_even():
if mod == 0:
return '{} is a even number.'.format(x)
else:
return '{} is a odd number.'.format(x)
print(check_odd_even())
print(x)
print(mod)
```
## Although global variable looks quite convenient, it is HIGHLY recommended NOT using global variable directly in a indented function block.
|
github_jupyter
|
<br><br><font color="gray">DOING COMPUTATIONAL SOCIAL SCIENCE<br>MODULE 10 <strong>PROBLEM SETS</strong></font>
# <font color="#49699E" size=40>MODULE 10 </font>
# What You Need to Know Before Getting Started
- **Every notebook assignment has an accompanying quiz**. Your work in each notebook assignment will serve as the basis for your quiz answers.
- **You can consult any resources you want when completing these exercises and problems**. Just as it is in the "real world:" if you can't figure out how to do something, look it up. My recommendation is that you check the relevant parts of the assigned reading or search for inspiration on [https://stackoverflow.com](https://stackoverflow.com).
- **Each problem is worth 1 point**. All problems are equally weighted.
- **The information you need for each problem set is provided in the blue and green cells.** General instructions / the problem set preamble are in the blue cells, and instructions for specific problems are in the green cells. **You have to execute all of the code in the problem set, but you are only responsible for entering code into the code cells that immediately follow a green cell**. You will also recognize those cells because they will be incomplete. You need to replace each blank `▰▰#▰▰` with the code that will make the cell execute properly (where # is a sequentially-increasing integer, one for each blank).
- Most modules will contain at least one question that requires you to load data from disk; **it is up to you to locate the data, place it in an appropriate directory on your local machine, and replace any instances of the `PATH_TO_DATA` variable with a path to the directory containing the relevant data**.
- **The comments in the problem cells contain clues indicating what the following line of code is supposed to do.** Use these comments as a guide when filling in the blanks.
- **You can ask for help**. If you run into problems, you can reach out to John ([email protected]) or Pierson ([email protected]) for help. You can ask a friend for help if you like, regardless of whether they are enrolled in the course.
Finally, remember that you do not need to "master" this content before moving on to other course materials, as what is introduced here is reinforced throughout the rest of the course. You will have plenty of time to practice and cement your new knowledge and skills.
<div class='alert alert-block alert-danger'>As you complete this assignment, you may encounter variables that can be assigned a wide variety of different names. Rather than forcing you to employ a particular convention, we leave the naming of these variables up to you. During the quiz, submit an answer of 'USER_DEFINED' (without the quotation marks) to fill in any blank that you assigned an arbitrary name to. In most circumstances, this will occur due to the presence of a local iterator in a for-loop.</b></div>
## Package Imports
```
import pandas as pd
import numpy as np
from numpy.random import seed as np_seed
import graphviz
from graphviz import Source
from pyprojroot import here
import matplotlib.pyplot as plt
import matplotlib as mpl
import seaborn as sns
from sklearn import preprocessing
from sklearn.model_selection import train_test_split, cross_val_score, ShuffleSplit
from sklearn.linear_model import LinearRegression, Ridge, Lasso, LogisticRegression
from sklearn.tree import DecisionTreeClassifier, export_graphviz
from sklearn.ensemble import BaggingClassifier, RandomForestClassifier, GradientBoostingClassifier
from sklearn.preprocessing import LabelEncoder, LabelBinarizer
from sklearn.neighbors import KNeighborsClassifier
from sklearn.feature_extraction.text import TfidfVectorizer
import tensorflow as tf
from tensorflow import keras
from tensorflow.random import set_seed
import spacy
from time import time
set_seed(42)
np_seed(42)
```
## Defaults
```
x_columns = [
# Religion and Morale
'v54', # Religious services? - 1=More than Once Per Week, 7=Never
'v149', # Do you justify: claiming state benefits? - 1=Never, 10=Always
'v150', # Do you justify: cheating on tax? - 1=Never, 10=Always
'v151', # Do you justify: taking soft drugs? - 1=Never, 10=Always
'v152', # Do you justify: taking a bribe? - 1=Never, 10=Always
'v153', # Do you justify: homosexuality? - 1=Never, 10=Always
'v154', # Do you justify: abortion? - 1=Never, 10=Always
'v155', # Do you justify: divorce? - 1=Never, 10=Always
'v156', # Do you justify: euthanasia? - 1=Never, 10=Always
'v157', # Do you justify: suicide? - 1=Never, 10=Always
'v158', # Do you justify: having casual sex? - 1=Never, 10=Always
'v159', # Do you justify: public transit fare evasion? - 1=Never, 10=Always
'v160', # Do you justify: prostitution? - 1=Never, 10=Always
'v161', # Do you justify: artificial insemination? - 1=Never, 10=Always
'v162', # Do you justify: political violence? - 1=Never, 10=Always
'v163', # Do you justify: death penalty? - 1=Never, 10=Always
# Politics and Society
'v97', # Interested in Politics? - 1=Interested, 4=Not Interested
'v121', # How much confidence in Parliament? - 1=High, 4=Low
'v126', # How much confidence in Health Care System? - 1=High, 4=Low
'v142', # Importance of Democracy - 1=Unimportant, 10=Important
'v143', # Democracy in own country - 1=Undemocratic, 10=Democratic
'v145', # Political System: Strong Leader - 1=Good, 4=Bad
# 'v208', # How often follow politics on TV? - 1=Daily, 5=Never
# 'v211', # How often follow politics on Social Media? - 1=Daily, 5=Never
# National Identity
'v170', # How proud are you of being a citizen? - 1=Proud, 4=Not Proud
'v184', # Immigrants: impact on development of country - 1=Bad, 5=Good
'v185', # Immigrants: take away jobs from Nation - 1=Take, 10=Do Not Take
'v198', # European Union Enlargement - 1=Should Go Further, 10=Too Far Already
]
y_columns = [
# Overview
'country',
# Socio-demographics
'v226', # Year of Birth by respondent
'v261_ppp', # Household Monthly Net Income, PPP-Corrected
]
```
## Problem 1:
<div class="alert alert-block alert-info">
In this assignment, we're going to continue our exploration of the European Values Survey dataset. By wielding the considerable power of Artificial Neural Networks, we'll aim to create a model capable of predicting an individual survey respondent's country of residence. As with all machine/deep learning projects, our first task will involve loading and preparing the data.
</div>
<div class="alert alert-block alert-success">
Load the EVS dataset and use it to create a feature matrix (using all columns from x_columns) and (with the assistance of Scikit Learn's LabelBinarizer) a target array (representing each respondent's country of residence).
</div>
```
# Load EVS Dataset
df = pd.read_csv(PATH_TO_DATA/"evs_module_08.csv")
# Create Feature Matrix (using all columns from x_columns)
X = df[x_columns]
# Initialize LabelBinarizer
country_encoder = ▰▰1▰▰()
# Fit the LabelBinarizer instance to the data's 'country' column and store transformed array as target
y = country_encoder.▰▰2▰▰(np.array(▰▰3▰▰))
```
## Problem 2:
<div class="alert alert-block alert-info">
As part of your work in the previous module, you were introduced to the concept of the train-validate-test split. Up until now, we had made extensive use of Scikit Learn's preprocessing and cross-validation suites in order to easily get the most out of our data. Since we're using TensorFlow for our Artificial Neural Networks, we're going to have to change course a little: we can still use the <code>train_test_split</code> function, but we must now use it twice: the first iteration will produce our test set and a 'temporary' dataset; the second iteration will split the 'temporary' data into training and validation sets. Throughout this process, we must take pains to ensure that each of the data splits are shuffled and stratified.
</div>
<div class="alert alert-block alert-success">
Create shuffled, stratified splits for testing (10% of original dataset), validation (10% of data remaining from test split), and training (90% of data remaining from test split) sets. Submit the number of observations in the <code>X_valid</code> set, as an integer.
</div>
```
# Split into temporary and test sets
X_t, X_test, y_t, y_test = ▰▰1▰▰(
▰▰2▰▰,
▰▰3▰▰,
test_size = ▰▰4▰▰,
shuffle = ▰▰5▰▰,
stratify = y,
random_state = 42
)
# Split into training and validation sets
X_train, X_valid, y_train, y_valid = train_test_split(
▰▰6▰▰,
▰▰7▰▰,
test_size = ▰▰8▰▰,
shuffle = ▰▰9▰▰,
stratify = ▰▰10▰▰,
random_state = 42,
)
len(X_valid)
```
## Problem 3:
<div class="alert alert-block alert-info">
As you work with Keras and Tensorflow, you'll rapidly discover that both packages are very picky about the 'shape' of the data you're using. What's more, you can't always rely on them to correctly infer your data's shape. As such, it's usually a good idea to store the two most important shapes -- number of variables in the feature matrix and number of unique categories in the target -- as explicit, named variables; doing so will save you the trouble of trying to retrieve them later (or as part of your model specification, which can get messy). We'll start with the number of variables in the feature matrix.
</div>
<div class="alert alert-block alert-success">
Store the number of variables in the feature matrix, as an integer, in the <code>num_vars</code> variable. Submit the resulting number as an integer.
</div>
```
# The code we've provided here is just a suggestion; feel free to use any approach you like
num_vars = np.▰▰1▰▰(▰▰2▰▰).▰▰3▰▰[1]
print(num_vars)
```
## Problem 4:
<div class="alert alert-block alert-info">
Now, for the number of categories (a.k.a. labels) in the target.
</div>
<div class="alert alert-block alert-success">
Store the number of categories in the target, as an integer, in the <code>num_vars</code> variable. Submit the resulting number as an integer.
</div>
```
# The code we've provided here is just a suggestion; feel free to use any approach you like
num_labels = ▰▰1▰▰.▰▰2▰▰[1]
print(num_labels)
```
## Problem 5:
<div class="alert alert-block alert-info">
Everything is now ready for us to begin building an Artifical Neural Network! Aside from specifying that the ANN must be built using Keras's <code>Sequential</code> API, we're going to give you the freedom to tackle the creation of your ANN in whichever manner you like. Feel free to use the 'add' method to build each layer one at a time, or pass all of the layers to your model at instantiation as a list, or any other approach you may be familiar with. Kindly ensure that your model matches the specifications below <b>exactly</b>!
</div>
<div class="alert alert-block alert-success">
Using Keras's <code>Sequential</code> API, create a new ANN. Your ANN should have the following layers, in this order:
<ol>
<li> Input layer with one argument: number of variables in the feature matrix
<li> Dense layer with 400 neurons and the "relu" activation function
<li> Dense layer with 10 neurons and the "relu" activation function
<li> Dense layer with neurons equal to the number of labels in the target and the "softmax" activation function
</ol>
Submit the number of hidden layers in your model.
</div>
```
# Create your ANN!
nn_model = keras.models.Sequential()
```
## Problem 6:
<div class="alert alert-block alert-info">
Even though we've specified all of the layers in our model, it isn't yet ready to go. We must first 'compile' the model, during which time we'll specify a number of high-level arguments. Just as in the textbook, we'll go with a fairly standard set of arguments: we'll use Stochastic Gradient Descent as our optimizer, and our only metric will be Accuracy (an imperfect but indispensably simple measure). It'll be up to you to figure out what loss function we should use: you might have to go digging in the textbook to find it!
</div>
<div class="alert alert-block alert-success">
Compile the model according to the specifications outlined in the blue text above. Submit the name of the loss function <b>exactly</b> as it appears in your code (you should only need to include a single underscore -- no other punctuation, numbers, or special characters).
</div>
```
nn_model.▰▰1▰▰(
loss=keras.losses.▰▰2▰▰,
optimizer=▰▰3▰▰,
metrics=[▰▰4▰▰]
)
```
## Problem 7:
<div class="alert alert-block alert-info">
Everything is prepared. All that remains is to train the model!
</div>
<div class="alert alert-block alert-success">
Train your neural network for 100 epochs. Be sure to include the validation data variables.
</div>
```
np_seed(42)
tf.random.set_seed(42)
history = nn_model.▰▰1▰▰(▰▰2▰▰, ▰▰3▰▰, epochs=▰▰4▰▰, validation_data = (▰▰5▰▰, ▰▰6▰▰))
```
## Problem 8:
<div class="alert alert-block alert-info">
For some Neural Networks, 100 epochs is more than ample time to reach a best solution. For others, 100 epochs isn't enough time for the learning process to even get underway. One good method for assessing the progress of your model at a glance involves visualizing how your loss scores and metric(s) -- for both your training and validation sets) -- changed during training.
</div>
<div class="alert alert-block alert-success">
After 100 epochs of training, is the model still appreciably improving? (If it is still improving, you shouldn't see much evidence of overfitting). Submit your answer as a boolean value (True = still improving, False = not still improving).
</div>
```
pd.DataFrame(history.history).plot(figsize = (8, 8))
plt.grid(True)
plt.show()
```
## Problem 9:
<div class="alert alert-block alert-info">
Regardless of whether this model is done or not, it's time to dig into what our model has done. Here, we'll continue re-tracing the steps taken in the textbook, producing a (considerably more involved) confusion matrix, visualizing it as a heatmap, and peering into our model's soul. The first step in this process involves creating the confusion matrix.
</div>
<div class="alert alert-block alert-success">
Using the held-back test data, create a confusion matrix.
</div>
```
y_pred = np.argmax(nn_model.predict(▰▰1▰▰), axis=1)
y_true = np.argmax(▰▰2▰▰, axis=1)
conf_mat = tf.math.confusion_matrix(▰▰3▰▰, ▰▰4▰▰)
```
## Problem 10:
<div class="alert alert-block alert-info">
Finally, we're ready to visualize the matrix we created above. Rather than asking you to recreate the baroque visualization code, we're going to skip straight to interpretation.
</div>
<div class="alert alert-block alert-success">
Plot the confusion matrix heatmap and examine it. Based on what you know about the dataset, should the sum of the values in a column (representing the number of observations from a country) be the same for each country? If so, submit the integer that each column adds up to. If not, submit 0.
</div>
```
sns.set(rc={'figure.figsize':(12,12)})
plt.figure()
sns.heatmap(
np.array(conf_mat).T,
xticklabels=country_encoder.classes_,
yticklabels=country_encoder.classes_,
square=True,
annot=True,
fmt='g',
)
plt.xlabel("Observed")
plt.ylabel("Predicted")
plt.show()
```
## Problem 11:
<div class="alert alert-block alert-success">
Based on what you know about the dataset, should the sum of the values in a row (representing the number of observations your model <b>predicted</b> as being from a country) be the same for each country? If so, submit the integer that each row adds up to. If not, submit 0.
</div>
```
```
## Problem 12:
<div class="alert alert-block alert-success">
If your model was built and run to the specifications outlined in the assignment, your results should include at least three countries whose observations the model struggled to identify (fewer than 7 accurate predictions each). Submit the name of one such country.<br><br>As a result of the randomness inherent to these models, it is possible that your interpretation will be correct, but will be graded as incorrect. If you feel that your interpretation was erroneously graded, please email a screenshot of your confusion matrix heatmap to Pierson along with an explanation of how you arrived at the answer you did.
</div>
```
```
|
github_jupyter
|
# A Câmara de Vereadores e o COVID-19
Você também fica curioso(a) para saber o que a Câmara de Vereadores
de Feira de Santana fez em relação ao COVID-19? O que discutiram?
O quão levaram a sério o vírus? Vamos responder essas perguntas e
também te mostrar como fazer essa análise. Vem com a gente!
Desde o início do ano o mundo tem falado a respeito do COVID-19.
Por isso, vamos basear nossa análise no período de 1 de Janeiro
a 27 de Março de 2020. Para entender o que se passou na Câmara
vamos utilizar duas fontes de dados:
* [Diário Oficial](diariooficial.feiradesantana.ba.gov.br/)
* [Atas das sessões](https://www.feiradesantana.ba.leg.br/atas)
Nas atas temos acesso ao que foi falado nos discursos e no Diário
Oficial temos acesso ao que realmente virou (ou não) lei. Você
pode fazer _download_ dos dados [aqui]()
e [aqui](https://www.kaggle.com/anapaulagomes/dirios-oficiais-de-feira-de-santana).
Procuramos mas não foi encontrada nenhuma menção ao vírus na
[Agenda da Câmara Municipal](https://www.feiradesantana.ba.leg.br/agenda).
Lembrando que as atas foram disponibilizadas no site da casa apenas
depois de uma reunião nossa com o presidente em exercício José Carneiro.
Uma grande vitória dos dados abertos e da transparência na cidade.
Os dados são coletados por nós e todo código está [aberto e disponível
no Github](https://github.com/DadosAbertosDeFeira/maria-quiteria/).
## Vamos começar por as atas
As atas trazem uma descrição do que foi falado durante as sessões.
Se você quer acompanhar o posicionamento do seu vereador(a), é uma
boa maneira. Vamos ver se encontramos alguém falando sobre *coronavírus*
ou *vírus*.
Se você não é uma pessoa técnica, não se preocupe. Vamos continuar o texto
junto com o código.
```
import pandas as pd
# esse arquivo pode ser baixado aqui: https://www.kaggle.com/anapaulagomes/atas-da-cmara-de-vereadores
atas = pd.read_csv('atas-28.03.2020.csv')
atas.describe()
```
Explicando um pouco sobre os dados (colunas):
* `crawled_at`: data da coleta
* `crawled_from`: fonte da coleta (site de onde a informação foi retirada)
* `date`: data da sessão
* `event_type`: tipo do evento: sessão ordinária, ordem do dia, solene, etc
* `file_content`: conteúdo do arquivo da ata
* `file_urls`: url(s) do arquivo da ata
* `title`: título da ata
### Filtrando por data
Bom, vamos então filtrar os dados e pegar apenas as atas entre 1 de Janeiro
e 28 de Março:
```
atas["date"] = pd.to_datetime(atas["date"])
atas["date"]
atas = atas[atas["date"].isin(pd.date_range("2020-01-01", "2020-03-28"))]
atas = atas.sort_values('date', ascending=True) # aqui ordenados por data em ordem crescente
atas.head()
```
Bom, quantas atas temos entre Janeiro e Março?
```
len(atas)
```
Apenas 21 atas, afinal os trabalhos na casa começaram no início de Fevereiro
e pausaram por uma semana por conta do coronavírus.
Fonte:
https://www.feiradesantana.ba.leg.br/agenda
https://www.feiradesantana.ba.leg.br/noticia/2029/c-mara-municipal-suspende-sess-es-ordin-rias-da-pr-xima-semana
### Filtrando conteúdo relacionado ao COVID-19
Agora que temos nossos dados filtrados por data, vamos ao conteúdo.
Na coluna `file_content` podemos ver o conteúdo das atas. Nelas vamos
buscar por as palavras:
- COVID, COVID-19
- coronavírus, corona vírus
```
termos_covid = "COVID-19|coronavírus"
atas = atas[atas['file_content'].str.contains(termos_covid, case=False)]
atas
len(atas)
```
Doze atas mencionando termos que representam o COVID-19 foram encontradas.
Vamos ver o que elas dizem?
Atenção: o conteúdo das atas é grande para ser mostrado aqui. Por isso vamos
destacar as partes que contém os termos encontrados.
```
import re
padrao = r'[A-Z][^\\.;]*(coronavírus|covid)[^\\.;]*'
def trecho_encontrado(conteudo_do_arquivo):
frases_encontradas = []
for encontrado in re.finditer(padrao, conteudo_do_arquivo, re.IGNORECASE):
frases_encontradas.append(encontrado.group().strip().replace('\n', ''))
return '\n'.join(frases_encontradas)
atas['trecho'] = atas['file_content'].apply(trecho_encontrado)
pd.set_option('display.max_colwidth', 100)
atas[['date', 'event_type', 'title', 'file_urls', 'trecho']]
```
Já que não temos tantos dados assim (apenas 12 atas) podemos fazer parte dessa análise manual,
para ter certeza que nenhum vereador foi deixado de fora. Vamos usar o próximo comando
para exportar os dados para um novo CSV. Nesse CSV vai ter os dados filtrados por data
e também o trecho, além do conteúdo da ata completo.
```
def converte_para_arquivo(df, nome_do_arquivo):
conteudo_do_csv = df.to_csv(index=False) # convertemos o conteudo para CSV
arquivo = open(nome_do_arquivo, 'w') # criamos um arquivo
arquivo.write(conteudo_do_csv)
arquivo.close()
converte_para_arquivo(atas, 'analise-covid19-atas-camara.csv')
```
### Quem levantou a bola do COVID-19 na Câmara?
Uma planilha com a análise do quem-disse-o-que pode ser encontrada [aqui](https://docs.google.com/spreadsheets/d/1h7ioFnHH8sGSxglThTpQX8W_rK9cgI_QRB3u5aAcNMI/edit?usp=sharing).
## E o que dizem os diários oficiais?
No diário oficial do município você encontra informações sobre o que virou
realidade: as decretas, medidas, nomeações, vetos.
Em Feira de Santana os diários dos poderes executivo e legislativo estão juntos.
Vamos filtrar os diários do legislativo, separar por data, como fizemos com as
atas, e ver o que foi feito.
```
# esse arquivo pode ser baixado aqui: https://www.kaggle.com/anapaulagomes/dirios-oficiais-de-feira-de-santana
diarios = pd.read_csv('gazettes-28.03.2020.csv')
diarios = diarios[diarios['power'] == 'legislativo']
diarios["date"] = pd.to_datetime(diarios["date"])
diarios = diarios[diarios["date"].isin(pd.date_range("2020-01-01", "2020-03-28"))]
diarios = diarios.sort_values('date', ascending=True) # aqui ordenados por data em ordem crescente
diarios.head()
```
O que é importante saber sobre as colunas dessa base de dados:
* `date`: quando o diário foi publicado
* `power`: poder executivo ou legislativo (sim, os diários são unificados)
* `year_and_edition`: ano e edição do diário
* `events`:
* `crawled_at`: quando foi feita essa coleta
* `crawled_from`: qual a fonte dos dados
* `file_urls`: url dos arquivos
* `file_content`: o conteúdo do arquivo do diário
Vamos filtrar o conteúdo dos arquivos que contém os termos relacionados ao COVID-19
(os mesmos que utilizamos com as atas).
```
diarios = diarios[diarios['file_content'].str.contains(termos_covid, case=False)]
diarios['trecho'] = diarios['file_content'].apply(trecho_encontrado)
diarios[['date', 'power', 'year_and_edition', 'file_urls', 'trecho']]
```
Apenas 4 diários com menção ao termo, entre 1 de Janeiro e 28 de Março de 2020
foram encontrados. O que será que eles dizem? Vamos exportar os resultados para
um novo CSV e continuar no Google Sheets.
```
converte_para_arquivo(diarios, 'analise-covid19-diarios-camara.csv')
```
### O que encontramos nos diários
Os 4 diários tratam de suspender licitações por conta da situação na cidade.
Apenas um dos diários, o diário do dia 17 de Março de 2020, [Ano VI - Edição Nº 733](http://www.diariooficial.feiradesantana.ba.gov.br/atos/legislativo/2CI2L71632020.pdf),
traz instruções a respeito do que será feito na casa. Aqui citamos o trecho que fala
a respeito das medidas:
> Art. 1º **Qualquer servidor, estagiário, terceirizado, vereador que apresentar febre ou sintomas respiratórios (tosse seca, dor de garganta, mialgia, cefaleia e prostração, dificuldade para respirar e batimento das asas nasais) passa a ser considerado um caso suspeito e deverá notificar imediatamente em até 24 horas à Vigilância Sanitária Epidemiológica/Secretaria Municipal de Saúde**.
§ 1ºQualquer servidor, estagiário, terceirizado, a partir dos 60 (sessenta)anos; portador de doença crônica e gestante estarão liberados das atividades laborais na Câmara Municipal de Feira de Santana sem prejuízo a sua remuneração.
§ 2º A(o) vereador(a) a partir dos 60 (sessenta)anos; portador de doença crônica e gestante será facultado as atividades laborais na Câmara Municipal de Feira de Santana sem prejuízo a sua remuneração
§ 3º Será considerado falta justificada ao serviço público ou à atividade laboral privada o período de ausência decorrente de afastamento por orientação médica.
> Art. 2º **Fica interditado o elevador no prédio anexo por tempo indeterminado**, até que sejam efetivamente contida a propagação do Coronavírus no Município e estabilizada a situação. _Parágrafo único_ O uso do elevador nesse período só será autorizado para transporte de portadores de necessidades especiais, como cadeirantes e pessoas com mobilidade reduzida.
> Art. 3º Será liberada a catraca para acesso ao prédio anexo.
> Art. 4º O portãolateral (acesso a rua do prédio anexo) será fechado com entrada apenas pela portaria principal.
> Art. 5º Será disponibilizado nas áreas comuns dispensadores para álcool em gel.
> Art.6º Será intensificada a limpeza nos banheiros, elevadores, corrimãos, maçanetase áreas comuns com grande circulação de pessoas.
> Art.7º Na cozinha e copa só será permitido simultaneamente à permanência de uma pessoa.
> Art. 8º **Ficam suspensas as Sessões Solenes, Especiais e Audiências Públicas por tempo indeterminado**, até que sejam efetivamente contida a propagação do Coronavírus no Município e estabilizada a situação.
> Art. 9º Nesse período de contenção é recomendado que o público/visitante assista a Sessão Ordinária on-line via TV Câmara. Parágrafo - Na Portaria do Prédio Sede **haverá um servidor orientando as pessoas a assistirem os trabalhos legislativos pela TV Câmara** disponível em https://www.feiradesantana.ba.leg.br/ distribuindo panfletos informativos sobre sintomas e métodos de prevenção.
> Art. 10º No âmbito dos gabinetes dos respectivos Vereadores, **fica a critério de cada qual adotar restrições ao atendimento presencial do público externo ou visitação à sua respectiva área**.
> Art.11º A Gerência Administrativa fica autorizada a adotar outras providências administrativas necessárias para evitar a propagação interna do vírus COVID-19, devendo as medidas serem submetidas ao conhecimento da Presidência.
> Art.12º A validade desta Portaria será enquanto valer oestado de emergência de saúde pública realizado pela Secretaria Municipal de Saúde e pelo Comitê de Ações de Enfrentamento ao Coronavírus no Município de Feira de Santana.
> Art.13º Esta Portaria entra em vigor na data de sua publicação, revogadas disposições em contrário.
## Conclusão
Caso os edis da casa tenham feito mais nós não teríamos como saber sem ser por as atas,
pelo diário oficial ou por notícias do site. O ideal seria ter uma página com projetos
e iniciativas de cada vereador. Dessa forma, seria mais fácil se manter atualizado a
respeito do trabalho de cada um na cidade.
As análises manuais e o que encontramos pode ser visto [aqui](https://docs.google.com/spreadsheets/d/1h7ioFnHH8sGSxglThTpQX8W_rK9cgI_QRB3u5aAcNMI/edit?usp=sharing).
Um texto sobre a análise e as descobertas podem ser encontradas no blog do [Dados Abertos de Feira](https://medium.com/@dadosabertosdefeira).
Gostou do que viu? Achou interessante? Compartilhe com outras pessoas e não esqueça de mencionar
o projeto.
|
github_jupyter
|
# 12 - Beginner Exercises
* Conditional Statements
## 🍼 🍼 🍼
1.Create a Python program that receive a number from the user and determines if a given integer is even or odd.
```
# Write your own code in this cell
n =
```
## 🍼🍼
2.Write a Python code that would read any integer day number and show the day's name in the word
```
# Write your own code in this cell
```
## 🍼🍼
3.Create a Python program that receive three angles from the user to determine whether the supplied angle values may be used to construct a triangle.
```
# Write your own code in this cell
a =
b =
c =
```
## 🍼
4.Write a Python program that receive three numbers from the user to calculate the root of a Quadratic Equation.
$$ax^2+bx+c$$
As you know, the roots of the quadratic equation can be obtained using the following formula:
$$x= \frac{-b \pm \sqrt{ \Delta } }{2a} $$
* If the delta is a number greater than zero ($\Delta > 0$), our quadratic equation has two roots as follows:
$$x_{1}= \frac{-b+ \sqrt{ \Delta } }{2a} $$
$$x_{2}= \frac{-b- \sqrt{ \Delta } }{2a} $$
* If the delta is zero, then there is exactly one real root:
$$x= \frac{-b}{2a} $$
* If the delta is negative, then there are no real roots
We also know that Delta equals:
$$ \Delta = b^{2} - 4ac$$
```
# Write your own code in this cell
a =
b =
c =
```
## 🍼
5.Create an application that takes the user's systolic and diastolic blood pressures and tells them category of their current condition .
|BLOOD PRESSURE CATEGORY | SYSTOLIC mm Hg |and/or| DIASTOLIC mm Hg |
|---|---|---|---|
|**NORMAL**| LESS THAN 120 | and | LESS THAN 80|
|**ELEVATED**| 120 – 129 | and | LESS THAN 80 |
|**HIGH BLOOD PRESSURE (HYPERTENSION) STAGE 1**| 130 – 139 | or | 80 – 89 |
|**HIGH BLOOD PRESSURE (HYPERTENSION) STAGE 2**| 140 OR HIGHER | or | 90 OR HIGHER |
|**HYPERTENSIVE CRISIS *(consult your doctor immediately)***| HIGHER THAN 180 | and/or | HIGHER THAN 120|
```
# Write your own code in this cell
```
## 🍼
6.Create an application that initially displays a list of geometric shapes to the user.
By entering a number, the user can pick one of those shapes.
Then, based on the geometric shape selected, obtain the shape parameters from the user.
Finally, as the output, display the shape's area.
```
# Write your own code in this cell
```
## 🌶️🌶️
7.Create a lambda function that accepts a number and returns a string based on this logic,
* If the given value is between 3 to 8 then return "Severe"
* else if it’s between 9 to 15 then return "Mild to Moderate"
* else returns the "The number is not valid"
```The above numbers are derived from the Glasgow Coma Scale.```
```
# Write your own code in this cell
```
## 🌶️
**```You can nest expressions to evaluate inside expressions in an f-string```**
8.You are given a set of prime numbers less than 1000.
Create a program that receives a number and then uses a single print() function with F-String to determine whether or not it was a prime number.
for example:
"997 is a prime number,"
"998 is not a prime number."
```
# Write your own code in this cell
P = {2, 3, 5, 7, 11, 13, 17, 19, 23, 29, 31, 37, 41,
43, 47, 53, 59, 61, 67, 71, 73, 79, 83, 89, 97,
101, 103, 107, 109, 113, 127, 131, 137, 139, 149,
151, 157, 163, 167, 173, 179, 181, 191, 193, 197,
199, 211, 223, 227, 229, 233, 239, 241, 251, 257,
263, 269, 271, 277, 281, 283, 293, 307, 311, 313,
317, 331, 337, 347, 349, 353, 359, 367, 373, 379,
383, 389, 397, 401, 409, 419, 421, 431, 433, 439,
443, 449, 457, 461, 463, 467, 479, 487, 491, 499,
503, 509, 521, 523, 541, 547, 557, 563, 569, 571,
577, 587, 593, 599, 601, 607, 613, 617, 619, 631,
641, 643, 647, 653, 659, 661, 673, 677, 683, 691,
701, 709, 719, 727, 733, 739, 743, 751, 757, 761,
769, 773, 787, 797, 809, 811, 821, 823, 827, 829,
839, 853, 857, 859, 863, 877, 881, 883, 887, 907,
911, 919, 929, 937, 941, 947, 953, 967, 971, 977,
983, 991, 997}
```
|
github_jupyter
|
```
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
%config InlineBackend.figure_format='retina'
dir_cat = './'
#vit_df = pd.read_csv(dir_cat+'gz2_vit_09172021_0000_predictions.csv')
#resnet_df = pd.read_csv(dir_cat+'gz2_resnet50_A_predictions.csv')
df = pd.read_csv(dir_cat+'gz2_predictions.csv')
df_vTrT = df[df.vitTresT == 1]
df_vTrF = df[df.vitTresF == 1]
df_vFrT = df[df.vitFresT == 1]
df_vFrF = df[df.vitFresF == 1]
print(f'Number of galaxies in test set : {len(df)}\n')
print(f'ViT True , resnet True galaxies: {len(df_vTrT)}')
print(f'ViT True , resnet False galaxies: {len(df_vTrF)}')
print(f'ViT False, resnet True galaxies: {len(df_vFrT)}')
print(f'ViT False, resnet False galaxies: {len(df_vFrF)}')
df.head()
df_stats = df.groupby(['class'])['class'].agg('count').to_frame('count').reset_index()
df_stats['test_set'] = df_stats['count']/df_stats['count'].sum()
df_stats['vitT_resT'] = df_vTrT.groupby('class').size() #/ df_stats['count'].sum()
df_stats['vitT_resF'] = df_vTrF.groupby('class').size() #/ df_stats['count'].sum()
df_stats['vitF_resT'] = df_vFrT.groupby('class').size() #/ df_stats['count'].sum()
df_stats['vitF_resF'] = df_vFrF.groupby('class').size() #/ df_stats['count'].sum()
print(df_stats)
###### plot ######
#ax = df_stats.plot.bar(x='class', y=['test_set', 'vitT_resT', 'vitT_resF', 'vitF_resT', 'vitF_resF'], rot=20, color=['gray', 'orange', 'red', 'blue', 'skyblue'])
#ax = df_stats.plot.bar(x='class', y=['test_set', 'vitT_resT'], rot=20, color=['gray', 'orange', 'red', 'blue', 'skyblue'])
ax = df_stats.plot.bar(x='class', y=['vitT_resF', 'vitF_resT'], rot=20, color=['red', 'blue', 'skyblue'])
#ax = df_stats.plot.bar(x='class', y=['vitT_resT', 'vitF_resF'], rot=20, color=['orange', 'skyblue'])
ax.set_xticklabels(['Round','In-between','Cigar-shaped','Edge-on','Barred','UnBarred','Irregular','Merger'])
ax.set_ylabel('class fraction')
ax.set_xlabel('galaxy morphology class')
df_vFrT.groupby('class').size()
df_stats = df.groupby(['class'])['class'].agg('count').to_frame('count').reset_index()
df_stats['test_set'] = df_stats['count']/df_stats['count'].sum()
df_stats['vitT_resT'] = df_vTrT.groupby('class').size() / df_vTrT.groupby('class').size().sum()
df_stats['vitT_resF'] = df_vTrF.groupby('class').size() / df_vTrF.groupby('class').size().sum()
df_stats['vitF_resT'] = df_vFrT.groupby('class').size() / df_vFrT.groupby('class').size().sum()
df_stats['vitF_resF'] = df_vFrF.groupby('class').size() / df_vFrF.groupby('class').size().sum()
print(df_stats)
###### plot ######
ax = df_stats.plot.bar(x='class', y=['test_set', 'vitT_resT', 'vitT_resF', 'vitF_resT', 'vitF_resF'], rot=20,
color=['gray', 'orange', 'red', 'blue', 'skyblue'])
ax.set_xticklabels(['Round','In-between','Cigar-shaped','Edge-on','Barred','UnBarred','Irregular','Merger'])
ax.set_ylabel('class fraction')
ax.set_xlabel('galaxy morphology class')
```
# color, size distributions
```
fig, ax = plt.subplots(figsize=(7.2, 5.5))
plt.rc('font', size=16)
#tag = 'model_g_r'
tag = 'dered_g_r'
bins = np.linspace(df[tag].min(), df[tag].max(),80)
ax.hist(df[tag] , bins=bins, color='lightgray' , label='full test set', density=True)
ax.hist(df_vTrT[tag], bins=bins, color='royalblue' , label=r'ViT $\bf{T}$ CNN $\bf{T}$', histtype='step' , lw=2, ls='-.', density=True)
ax.hist(df_vTrF[tag], bins=bins, color='firebrick' , label=r'ViT $\bf{T}$ CNN $\bf{F}$', histtype='step', lw=3, density=True)
ax.hist(df_vFrT[tag], bins=bins, color='orange' , label=r'ViT $\bf{F}$ CNN $\bf{T}$', histtype='step' , lw=3, ls='--', density=True)
ax.hist(df_vFrF[tag], bins=bins, color='forestgreen', label=r'ViT $\bf{F}$ CNN $\bf{F}$', histtype='step', lw=2, ls=':', density=True)
#r"$\bf{" + str(number) + "}$"
ax.set_xlabel('g-r')
ax.set_ylabel('pdf')
ax.set_xlim(-0.25, 2.2)
ax.legend(fontsize=14.5)
from scipy.stats import ks_2samp
ks_2samp(df_vTrF['dered_g_r'], df_vFrT['dered_g_r'])
fig, ax = plt.subplots(figsize=(7.2, 5.5))
plt.rc('font', size=16)
tag = 'petroR50_r'
bins = np.linspace(df[tag].min(), df[tag].max(),50)
ax.hist(df[tag] , bins=bins, color='lightgray' , label='full test set', density=True)
ax.hist(df_vTrT[tag], bins=bins, color='royalblue' , label=r'ViT $\bf{T}$ CNN $\bf{T}$', histtype='step' , lw=2, ls='-.', density=True)
ax.hist(df_vTrF[tag], bins=bins, color='firebrick' , label=r'ViT $\bf{T}$ CNN $\bf{F}$', histtype='step', lw=3, density=True)
ax.hist(df_vFrT[tag], bins=bins, color='orange' , label=r'ViT $\bf{F}$ CNN $\bf{T}$', histtype='step' , lw=3, ls='--', density=True)
ax.hist(df_vFrF[tag], bins=bins, color='forestgreen', label=r'ViT $\bf{F}$ CNN $\bf{F}$', histtype='step', lw=2, ls=':', density=True)
#r"$\bf{" + str(number) + "}$"
ax.set_xlabel('50% light radius')
ax.set_ylabel('pdf')
ax.set_xlim(0.5, 10)
ax.legend(fontsize=14.5)
fig, ax = plt.subplots(figsize=(7.2, 5.5))
plt.rc('font', size=16)
tag = 'petroR90_r'
bins = np.linspace(df[tag].min(), df[tag].max(),50)
ax.hist(df[tag] , bins=bins, color='lightgray' , label='full test set', density=True)
ax.hist(df_vTrT[tag], bins=bins, color='royalblue' , label=r'ViT $\bf{T}$ CNN $\bf{T}$', histtype='step' , lw=2, ls='-.', density=True)
ax.hist(df_vTrF[tag], bins=bins, color='firebrick' , label=r'ViT $\bf{T}$ CNN $\bf{F}$', histtype='step', lw=3, density=True)
ax.hist(df_vFrT[tag], bins=bins, color='orange' , label=r'ViT $\bf{F}$ CNN $\bf{T}$', histtype='step' , lw=3, ls='--', density=True)
ax.hist(df_vFrF[tag], bins=bins, color='forestgreen', label=r'ViT $\bf{F}$ CNN $\bf{F}$', histtype='step', lw=2, ls=':', density=True)
#r"$\bf{" + str(number) + "}$"
ax.set_xlabel('90% light radius')
ax.set_ylabel('pdf')
ax.set_xlim(2.5, 25)
ax.legend(fontsize=14.5)
ks_2samp(df_vTrF['petroR90_r'], df_vFrT['petroR90_r'])
fig, ax = plt.subplots(figsize=(7.2, 5.5))
plt.rc('font', size=16)
tag = 'dered_r'
bins = np.linspace(df[tag].min(), df[tag].max(),20)
ax.hist(df[tag] , bins=bins, color='lightgray' , label='full test set', density=True)
ax.hist(df_vTrT[tag], bins=bins, color='royalblue' , label=r'ViT $\bf{T}$ CNN $\bf{T}$', histtype='step' , lw=2, ls='-.', density=True)
ax.hist(df_vTrF[tag], bins=bins, color='firebrick' , label=r'ViT $\bf{T}$ CNN $\bf{F}$', histtype='step' , lw=3, density=True)
ax.hist(df_vFrT[tag], bins=bins, color='orange' , label=r'ViT $\bf{F}$ CNN $\bf{T}$', histtype='step' , lw=3, ls='--', density=True)
ax.hist(df_vFrF[tag], bins=bins, color='forestgreen', label=r'ViT $\bf{F}$ CNN $\bf{F}$', histtype='step' , lw=2, ls=':', density=True)
#r"$\bf{" + str(number) + "}$"
ax.set_xlabel('r-band (apparent) magnitude')
ax.set_ylabel('pdf')
#ax.set_xlim(2.5, 25)
ax.legend(fontsize=14.5)
```
### check galaxy image
```
dir_image = '/home/hhg/Research/galaxyClassify/catalog/galaxyZoo_kaggle/gz2_images/images'
galaxyID = 241961
current_IMG = plt.imread(dir_image+f'/{galaxyID}.jpg')
plt.imshow(current_IMG)
plt.axis('off')
```
|
github_jupyter
|
# Intro to Reinforcement Learning
Reinforcement learning requires us to model our problem using the following two constructs:
* An agent, the thing that makes decisions.
* An environment, the world which encodes what decisions can be made, and the impact of those decisions.
The environment contains all the possible states, knows all the actions that can be taken from each state, and knows when rewards should be given and what the magnitude of those rewards should be. An agent gets this information from the environment by exploring and learns from experience which states provide the best rewards. Rewards slowly percolate outward to neighboring states iteratively, which helps the agent make decisions over longer time horizons.
## The Agent/Environment Interface

> Image Source: [Reinforcement Learning:An Introduction](http://incompleteideas.net/book/bookdraft2017nov5.pdf)
Reinforcement learning typically takes place over a number of episodes—which are rougly analogous to an epoch. In most situations for RL, there are some "terminal" states within the environment which indicate the end of an episode. In games for example, this happens when the game ends. For example when Mario gets killed or reaches the end of the level. Or in chess when someone is put into checkmate or conceeds. An episode ends when an agent reaches one of these terminal states.
An episode is compirsed of the agent making a series of decisions until it reaches one of these terminal states. Sometimes engineers may choose to terminate an episode after a maximum number of decisions, especially if there is a strong chance that the agent will never reach a terminal state.
## Markov Decsion Processes
Formally, the problems that reinforcement learning is best at solving are modeled by Markov Decision Processes (MDP). MDPs are a special kind of graph that are similar to State Machines. MDPs have two kinds of nodes, states and actions. From any given state an agent can select from only the available actions, those actions will take an agent to another state. These transitions from actions to states are frequently stochastic—meaning taking a particular action might lead you to one of several states based on some probabilistic function.
Transitions from state to state can be associated with a reward but they are not required to be. Many MDPs have terminal states, but those are not formally required either.

> Image Source: [Wikimedia commons, public domain](https://commons.wikimedia.org/wiki/File:Markov_Decision_Process_example.png)
This MDP has 3 states (larger green nodes S0, S1, S2), each state has exactly 2 actions available (smaller red nodes, a0, a1), and two transitions have rewards (from S2a1 -> S0 has -1 reward, from S1a0 -> S0 has +5 reward).
## Finding a Policy
The reinforcment learning algorithm we're going to focus on (Q-Learning) is a "policy based" agent. This means, it's goal is to discover which decision is the "best" decision to make at any given state. Sometimes, this goal is a little naive, for example if the state-spaces is evolving in real time, it may not be possible to determine a "best" policy. In the above MDP, though, there IS an optimal policy... What is it?
### Optimal Policy For the Above:
The only way to gain a positive reward is to take the transition S1a0 -> S0. That gives us +5 70% of the time.
Getting to S1 requires a risk though: the only way to get to S1 is by taking a1 from S2, which has a 30% chance of landing us back at S0 with a -1 reward.
We can easily oscilate infinitely between S0 and S2 with zero reward by taking only S0a1, S0a0, and S2a0 repeatedly. So the question is: is the risk worth the reward?
Say we're in S1, we can get +5 70% of the time by taking a0. That's an expected value of 3.5 if our policy is always take action a0. If we're in S2, then, we can get an expected value of 3.5 30% of the time, and a -1 30% of the time by always taking action a1:
`(.3 * 3.5) + (.3 * -1) = 1.05 - .3 = .75`
So intiutively we should go ahead and take the risky action to gain the net-positive reward. **But wait!** Both of our actions are *self-referential* and might lead us back to the original state... how do we account for that?
For the mathematical purists, we can use something called the Bellman Optimality Equation. Intuitively, the Bellman optimality equation expresses the fact that the value of a state under an optimal policy must equal the expected return for the best action from that state:
For the value of states:

For the state-action pairs:

> For a more complete treatment of these equations, see [Chapter 3.6 of this book](http://incompleteideas.net/book/bookdraft2017nov5.pdf)
Several bits of notation were just introduced:
* The Discount Factor (γ) — some value between 0 and 1, which is required for convergance.
* The expected return (Gt) — the value we want to optimize.
* The table of state-action pairs (Q) — these are the values of being in a state and taking a given action.
* The table of state values (V*) — these are based on the Q-values from the Q-table based on taking the best action.
* The policy is represented by π — our policy is what our agent thinks is the best action
* S and S' both represent states, the current state (S) and the next state (S') for any given state-action pair.
* r represents a reward for a given transition.
Solving this series of equations is computationally unrealistic for most problems of any real size. It is an iterative process that will only converge if the discount factor λ is between 0 and 1, and even then often it converges slowly. The most common algorithm for solving the Bellman equations directly is called Value Iteration, and it is much like what we did above, but we'd apply that logic repeatedly, for every state-action pair, and we'd have to apply a discount factor to the expected values we computed.
Value iteration is never used in practice. Instead we use Q-learning to experimentally explore states, essentially we attempt to partially solve the above Bellman equations. For a more complete treatment of value iteration, see the book linked above.
In my opinion, it is much easier, and much more helpful to see Q-Learning in action than it is to pour over the dense and confusing mathematical notation above. Q-Learning is actually wonderfully intuitive when you take a step back from the math.
|
github_jupyter
|
# Analysis of Microglia data
```
# Setup
import warnings
warnings.filterwarnings("ignore")
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from sccoda.util import comp_ana as mod
from sccoda.util import cell_composition_data as dat
from sccoda.model import other_models as om
```
Load and format data:
4 control samples, 2 samples for other conditions each; 8 cell types
```
cell_counts = pd.read_csv("../../sccoda_benchmark_data/cell_count_microglia_AD_WT_location.csv", index_col=0)
# Sort values such that wild type is considered the base category
print(cell_counts)
data_cer = dat.from_pandas(cell_counts.loc[cell_counts["location"] == "cerebellum"],
covariate_columns=["mouse_type", "location", "replicate"])
data_cor = dat.from_pandas(cell_counts.loc[cell_counts["location"] == "cortex"],
covariate_columns=["mouse_type", "location", "replicate"])
```
Plot data:
```
# Count data to ratios
counts = cell_counts.iloc[:, 3:]
rowsum = np.sum(counts, axis=1)
ratios = counts.div(rowsum, axis=0)
ratios["mouse_type"] = cell_counts["mouse_type"]
ratios["location"] = cell_counts["location"]
# Make boxplots
fig, ax = plt.subplots(figsize=(12,5), ncols=3)
df = pd.melt(ratios, id_vars=['location', "mouse_type"], value_vars=ratios.columns[:3])
df = df.sort_values(["location", "mouse_type"], ascending=[True, False])
print(df)
sns.set_context('notebook')
sns.set_style('ticks')
for i in range(3):
d = sns.boxplot(x='location', y = 'value', hue="mouse_type",
data=df.loc[df["variable"]==f"microglia {i+1}"], fliersize=1,
palette='Blues', ax=ax[i])
d.set_ylabel('Proportion')
loc, labels = plt.xticks()
d.set_xlabel('Location')
d.set_title(f"microglia {i+1}")
plt.legend(bbox_to_anchor=(1.33, 1), borderaxespad=0., title="Condition")
# plt.savefig(plot_path + "haber_boxes_blue.svg", format="svg", bbox_inches="tight")
# plt.savefig(plot_path + "haber_boxes_blue.png", format="png", bbox_inches="tight")
plt.show()
```
Analyze cerebellum data:
Apply scCODA for every cell type set as the reference.
We see no credible effects at the 0.2 FDR level.
```
# Use this formula to make a wild type -> treated comparison, not the other way
formula = "C(mouse_type, levels=['WT', 'AD'])"
# cerebellum
res_cer = []
effects_cer = pd.DataFrame(index=data_cer.var.index.copy(),
columns=data_cer.var.index.copy())
effects_cer.index.rename("cell type", inplace=True)
effects_cer.columns.rename("reference", inplace=True)
for ct in data_cer.var.index:
print(f"Reference: {ct}")
model = mod.CompositionalAnalysis(data=data_cer, formula=formula, reference_cell_type=ct)
results = model.sample_hmc()
_, effect_df = results.summary_prepare(est_fdr=0.2)
res_cer.append(results)
effects_cer[ct] = effect_df.loc[:, "Final Parameter"].array
# Column: Reference category
# Row: Effect
print(effects_cer)
for x in res_cer:
print(x.summary_extended())
```
Now with cortex data:
We see credible effects on all cell types, depending on the reference.
Only the effects on microglia 2 and 3 when using the other one as the reference are not considered credible at the 0.2 FDR level.
```
# cortex
res_cor = []
effects_cor = pd.DataFrame(index=data_cor.var.index.copy(),
columns=data_cor.var.index.copy())
effects_cor.index.rename("cell type", inplace=True)
effects_cor.columns.rename("reference", inplace=True)
for ct in data_cer.var.index:
print(f"Reference: {ct}")
model = mod.CompositionalAnalysis(data=data_cor, formula=formula, reference_cell_type=ct)
results = model.sample_hmc()
_, effect_df = results.summary_prepare(est_fdr=0.2)
res_cor.append(results)
effects_cor[ct] = effect_df.loc[:, "Final Parameter"].array
# Column: Reference category
# Row: Effect
effects_cor.index.name = "cell type"
effects_cor.columns.name = "reference"
print(effects_cor)
for x in res_cor:
print(x.summary_extended())
```
For validataion, apply ancom to the same dataset.
We see also no changes for the cerebellum data, and change in all microglia types for the cortex data.
```
cer_ancom = data_cer.copy()
cer_ancom.obs = cer_ancom.obs.rename(columns={"mouse_type": "x_0"})
ancom_cer = om.AncomModel(cer_ancom)
ancom_cer.fit_model(alpha=0.2)
print(ancom_cer.ancom_out)
cor_ancom = data_cor.copy()
cor_ancom.obs = cor_ancom.obs.rename(columns={"mouse_type": "x_0"})
cor_ancom.X = cor_ancom.X + 0.5
ancom_cor = om.AncomModel(cor_ancom)
ancom_cor.fit_model(alpha=0.2)
print(ancom_cor.ancom_out)
```
|
github_jupyter
|
# A project that shows the law of large numbers
```
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
```
# **Generating a population of random numbers**
```
# Creating 230,000 random numbers in a 1/f distribution
randint = np.logspace(np.log10(0.001),np.log10(100),230000)
fdist = np.zeros(230000)
for i in range(len(randint)):
fdist[i] = 1/randint[i]
if fdist[i]< 0:
print(fdist[i])
fdist[:40]
# Only taking every 1000th entry in fdist
fdist1000 = fdist[0::1000]
fdist1000[:40]
#Sorting the array in descending order
fdist1000[::-1].sort()
fdist1000[1]
#Creating the index
index = np.zeros(len(fdist1000))
for i in range(len(fdist1000)):
index[i] = i+1
index[:40]
# Graphing the random numbers
plt.plot(index,fdist1000, 'go--')
plt.xlabel('Sample')
plt.ylabel('Data Value')
plt.show()
# Shuffling fdist1000
shuffledist = fdist1000.copy()
np.random.shuffle(shuffledist)
plt.plot(index,shuffledist, 'go')
plt.xlabel('Sample')
plt.ylabel('Data Value')
plt.show()
```
# **Monte Carlo sampling**
```
### Randomly selecting 50 of the 230000 data points. Finding the mean. Repeat this 500 times. ###
mean = np.zeros(500)
for i in range(len(mean)):
fifty = np.random.choice(fdist, size=50, replace=False)
mean[i] = np.mean(fifty)
mean[:100]
# Calculating the real average
realmean = sum(fdist)/len(fdist)
print(realmean)
# Creating the index for mean to plot
meanindex = np.zeros(len(mean))
for i in range(len(mean)):
meanindex[i] = i+1
meanindex[:40]
# Plotting the Monte-Carlo sampling
plt.plot(meanindex,mean, 'ko',markerfacecolor='c' , label = 'Sample Means')
plt.axhline(y=realmean, color='r', linewidth='3', linestyle='-', label = 'True Mean')
plt.xlabel('Sample Number')
plt.ylabel('Mean Value')
plt.legend()
plt.show()
```
# **Cumulative averaging**
```
# Cumulative average of all sample means
cumemean = np.zeros(500)
cumesum = np.zeros(500)
length = len(mean)
for i in range(length):
cumesum[i] = np.sum(mean[:i+1])
#if i == 0:
#cumemean[i] = cumesum[i]/(i+1)
#else:
cumemean[i] = cumesum[i] / (i+1)
# Creating the index for cumulative mean to plot
cumemeanindex = np.zeros(len(cumemean))
for i in range(len(cumemean)):
cumemeanindex[i] = i+1
cumemeanindex[-1]
# Plotting of the Cumulative Average
plt.plot(cumemeanindex,cumemean, 'bo',markerfacecolor='w' , label = 'Cumulative Averages')
plt.axhline(y=realmean, color='r', linewidth='3', linestyle='-', label = 'True Mean')
plt.xlabel('Sample Number')
plt.ylabel('Mean Value')
plt.legend()
plt.show()
### Computing the square divergence for each point. Repeating 100 times ###
dfdivergence = pd.DataFrame(cumemean, columns = ['Original Cumulative Mean Run 1'])
# Finding the square divergence from mean (realmean) for the first run
divergence1 = (dfdivergence["Original Cumulative Mean Run 1"] - realmean)**2
divergence1[:20]
dfdivergence['Square Divergence 1'] = divergence1
dfdivergence
dfmeans = pd.DataFrame()
for i in range(99):
dfmeans[i] = np.zeros(500)
# (Sampling 50 random point 500 times) for 100 runs.
for i in range(99):
for j in range(500):
dffifty = np.random.choice(fdist, size=50, replace=False)
tempmean = np.mean(dffifty)
dfmeans.at[j, i] = tempmean
dfmeans[:5]
dfcumemean = pd.DataFrame()
dfcumesum = pd.DataFrame()
dfcumesum = dfmeans.cumsum(axis=0) # Finding the cumulative sum down each column
dfcumesum
for i in range(99):
for j in range(500):
dfcumemean.at[j,i] = dfcumesum.at[j,i]/(j+1) # Finding the cumulative mean down each column
dfcumemean
for i in range(99):
dfdivergence['Square Divergence' + ' ' + str(i+2)] = (dfcumemean[i] - realmean)**2 # finding the divergence down each column
dfdivergence[:4]
dfdivergence = dfdivergence.drop(['Original Cumulative Mean Run 1'], axis=1)
dfdivergence[:4]
dfdivergence.plot(figsize = (12,7), legend=False) # This plot shows the divergence from the real mean for 100 runs of cumulative averaging.
plt.ylim([-10,500])
plt.ylabel('Square Divergence from True Mean')
plt.xlabel('Sample Number')
plt.show()
```
|
github_jupyter
|
# GLM: Robust Regression with Outlier Detection
**A minimal reproducable example of Robust Regression with Outlier Detection using Hogg 2010 Signal vs Noise method.**
+ This is a complementary approach to the Student-T robust regression as illustrated in Thomas Wiecki's notebook in the [PyMC3 documentation](http://pymc-devs.github.io/pymc3/GLM-robust/), that approach is also compared here.
+ This model returns a robust estimate of linear coefficients and an indication of which datapoints (if any) are outliers.
+ The likelihood evaluation is essentially a copy of eqn 17 in "Data analysis recipes: Fitting a model to data" - [Hogg 2010](http://arxiv.org/abs/1008.4686).
+ The model is adapted specifically from Jake Vanderplas' [implementation](http://www.astroml.org/book_figures/chapter8/fig_outlier_rejection.html) (3rd model tested).
+ The dataset is tiny and hardcoded into this Notebook. It contains errors in both the x and y, but we will deal here with only errors in y.
**Note:**
+ Python 3.4 project using latest available [PyMC3](https://github.com/pymc-devs/pymc3)
+ Developed using [ContinuumIO Anaconda](https://www.continuum.io/downloads) distribution on a Macbook Pro 3GHz i7, 16GB RAM, OSX 10.10.5.
+ During development I've found that 3 data points are always indicated as outliers, but the remaining ordering of datapoints by decreasing outlier-hood is slightly unstable between runs: the posterior surface appears to have a small number of solutions with similar probability.
+ Finally, if runs become unstable or Theano throws weird errors, try clearing the cache `$> theano-cache clear` and rerunning the notebook.
**Package Requirements (shown as a conda-env YAML):**
```
$> less conda_env_pymc3_examples.yml
name: pymc3_examples
channels:
- defaults
dependencies:
- python=3.4
- ipython
- ipython-notebook
- ipython-qtconsole
- numpy
- scipy
- matplotlib
- pandas
- seaborn
- patsy
- pip
$> conda env create --file conda_env_pymc3_examples.yml
$> source activate pymc3_examples
$> pip install --process-dependency-links git+https://github.com/pymc-devs/pymc3
```
## Setup
```
%matplotlib inline
import warnings
warnings.filterwarnings('ignore')
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import optimize
import pymc3 as pm
import theano as thno
import theano.tensor as T
# configure some basic options
sns.set(style="darkgrid", palette="muted")
pd.set_option('display.notebook_repr_html', True)
plt.rcParams['figure.figsize'] = 12, 8
np.random.seed(0)
```
### Load and Prepare Data
We'll use the Hogg 2010 data available at https://github.com/astroML/astroML/blob/master/astroML/datasets/hogg2010test.py
It's a very small dataset so for convenience, it's hardcoded below
```
#### cut & pasted directly from the fetch_hogg2010test() function
## identical to the original dataset as hardcoded in the Hogg 2010 paper
dfhogg = pd.DataFrame(np.array([[1, 201, 592, 61, 9, -0.84],
[2, 244, 401, 25, 4, 0.31],
[3, 47, 583, 38, 11, 0.64],
[4, 287, 402, 15, 7, -0.27],
[5, 203, 495, 21, 5, -0.33],
[6, 58, 173, 15, 9, 0.67],
[7, 210, 479, 27, 4, -0.02],
[8, 202, 504, 14, 4, -0.05],
[9, 198, 510, 30, 11, -0.84],
[10, 158, 416, 16, 7, -0.69],
[11, 165, 393, 14, 5, 0.30],
[12, 201, 442, 25, 5, -0.46],
[13, 157, 317, 52, 5, -0.03],
[14, 131, 311, 16, 6, 0.50],
[15, 166, 400, 34, 6, 0.73],
[16, 160, 337, 31, 5, -0.52],
[17, 186, 423, 42, 9, 0.90],
[18, 125, 334, 26, 8, 0.40],
[19, 218, 533, 16, 6, -0.78],
[20, 146, 344, 22, 5, -0.56]]),
columns=['id','x','y','sigma_y','sigma_x','rho_xy'])
## for convenience zero-base the 'id' and use as index
dfhogg['id'] = dfhogg['id'] - 1
dfhogg.set_index('id', inplace=True)
## standardize (mean center and divide by 1 sd)
dfhoggs = (dfhogg[['x','y']] - dfhogg[['x','y']].mean(0)) / dfhogg[['x','y']].std(0)
dfhoggs['sigma_y'] = dfhogg['sigma_y'] / dfhogg['y'].std(0)
dfhoggs['sigma_x'] = dfhogg['sigma_x'] / dfhogg['x'].std(0)
## create xlims ylims for plotting
xlims = (dfhoggs['x'].min() - np.ptp(dfhoggs['x'])/5
,dfhoggs['x'].max() + np.ptp(dfhoggs['x'])/5)
ylims = (dfhoggs['y'].min() - np.ptp(dfhoggs['y'])/5
,dfhoggs['y'].max() + np.ptp(dfhoggs['y'])/5)
## scatterplot the standardized data
g = sns.FacetGrid(dfhoggs, size=8)
_ = g.map(plt.errorbar, 'x', 'y', 'sigma_y', 'sigma_x', marker="o", ls='')
_ = g.axes[0][0].set_ylim(ylims)
_ = g.axes[0][0].set_xlim(xlims)
plt.subplots_adjust(top=0.92)
_ = g.fig.suptitle('Scatterplot of Hogg 2010 dataset after standardization', fontsize=16)
```
**Observe**:
+ Even judging just by eye, you can see these datapoints mostly fall on / around a straight line with positive gradient
+ It looks like a few of the datapoints may be outliers from such a line
## Create Conventional OLS Model
The *linear model* is really simple and conventional:
$$\bf{y} = \beta^{T} \bf{X} + \bf{\sigma}$$
where:
$\beta$ = coefs = $\{1, \beta_{j \in X_{j}}\}$
$\sigma$ = the measured error in $y$ in the dataset `sigma_y`
### Define model
**NOTE:**
+ We're using a simple linear OLS model with Normally distributed priors so that it behaves like a ridge regression
```
with pm.Model() as mdl_ols:
## Define weakly informative Normal priors to give Ridge regression
b0 = pm.Normal('b0_intercept', mu=0, sd=100)
b1 = pm.Normal('b1_slope', mu=0, sd=100)
## Define linear model
yest = b0 + b1 * dfhoggs['x']
## Use y error from dataset, convert into theano variable
sigma_y = thno.shared(np.asarray(dfhoggs['sigma_y'],
dtype=thno.config.floatX), name='sigma_y')
## Define Normal likelihood
likelihood = pm.Normal('likelihood', mu=yest, sd=sigma_y, observed=dfhoggs['y'])
```
### Sample
```
with mdl_ols:
## take samples
traces_ols = pm.sample(2000, tune=1000)
```
### View Traces
**NOTE**: I'll 'burn' the traces to only retain the final 1000 samples
```
_ = pm.traceplot(traces_ols[-1000:], figsize=(12,len(traces_ols.varnames)*1.5),
lines={k: v['mean'] for k, v in pm.df_summary(traces_ols[-1000:]).iterrows()})
```
**NOTE:** We'll illustrate this OLS fit and compare to the datapoints in the final plot
---
---
## Create Robust Model: Student-T Method
I've added this brief section in order to directly compare the Student-T based method exampled in Thomas Wiecki's notebook in the [PyMC3 documentation](http://pymc-devs.github.io/pymc3/GLM-robust/)
Instead of using a Normal distribution for the likelihood, we use a Student-T, which has fatter tails. In theory this allows outliers to have a smaller mean square error in the likelihood, and thus have less influence on the regression estimation. This method does not produce inlier / outlier flags but is simpler and faster to run than the Signal Vs Noise model below, so a comparison seems worthwhile.
**Note:** we'll constrain the Student-T 'degrees of freedom' parameter `nu` to be an integer, but otherwise leave it as just another stochastic to be inferred: no need for prior knowledge.
### Define Model
```
with pm.Model() as mdl_studentt:
## Define weakly informative Normal priors to give Ridge regression
b0 = pm.Normal('b0_intercept', mu=0, sd=100)
b1 = pm.Normal('b1_slope', mu=0, sd=100)
## Define linear model
yest = b0 + b1 * dfhoggs['x']
## Use y error from dataset, convert into theano variable
sigma_y = thno.shared(np.asarray(dfhoggs['sigma_y'],
dtype=thno.config.floatX), name='sigma_y')
## define prior for Student T degrees of freedom
nu = pm.Uniform('nu', lower=1, upper=100)
## Define Student T likelihood
likelihood = pm.StudentT('likelihood', mu=yest, sd=sigma_y, nu=nu,
observed=dfhoggs['y'])
```
### Sample
```
with mdl_studentt:
## take samples
traces_studentt = pm.sample(2000, tune=1000)
```
#### View Traces
```
_ = pm.traceplot(traces_studentt[-1000:],
figsize=(12,len(traces_studentt.varnames)*1.5),
lines={k: v['mean'] for k, v in pm.df_summary(traces_studentt[-1000:]).iterrows()})
```
**Observe:**
+ Both parameters `b0` and `b1` show quite a skew to the right, possibly this is the action of a few samples regressing closer to the OLS estimate which is towards the left
+ The `nu` parameter seems very happy to stick at `nu = 1`, indicating that a fat-tailed Student-T likelihood has a better fit than a thin-tailed (Normal-like) Student-T likelihood.
+ The inference sampling also ran very quickly, almost as quickly as the conventional OLS
**NOTE:** We'll illustrate this Student-T fit and compare to the datapoints in the final plot
---
---
## Create Robust Model with Outliers: Hogg Method
Please read the paper (Hogg 2010) and Jake Vanderplas' code for more complete information about the modelling technique.
The general idea is to create a 'mixture' model whereby datapoints can be described by either the linear model (inliers) or a modified linear model with different mean and larger variance (outliers).
The likelihood is evaluated over a mixture of two likelihoods, one for 'inliers', one for 'outliers'. A Bernouilli distribution is used to randomly assign datapoints in N to either the inlier or outlier groups, and we sample the model as usual to infer robust model parameters and inlier / outlier flags:
$$
\mathcal{logL} = \sum_{i}^{i=N} log \left[ \frac{(1 - B_{i})}{\sqrt{2 \pi \sigma_{in}^{2}}} exp \left( - \frac{(x_{i} - \mu_{in})^{2}}{2\sigma_{in}^{2}} \right) \right] + \sum_{i}^{i=N} log \left[ \frac{B_{i}}{\sqrt{2 \pi (\sigma_{in}^{2} + \sigma_{out}^{2})}} exp \left( - \frac{(x_{i}- \mu_{out})^{2}}{2(\sigma_{in}^{2} + \sigma_{out}^{2})} \right) \right]
$$
where:
$\bf{B}$ is Bernoulli-distibuted $B_{i} \in [0_{(inlier)},1_{(outlier)}]$
### Define model
```
def logp_signoise(yobs, is_outlier, yest_in, sigma_y_in, yest_out, sigma_y_out):
'''
Define custom loglikelihood for inliers vs outliers.
NOTE: in this particular case we don't need to use theano's @as_op
decorator because (as stated by Twiecki in conversation) that's only
required if the likelihood cannot be expressed as a theano expression.
We also now get the gradient computation for free.
'''
# likelihood for inliers
pdfs_in = T.exp(-(yobs - yest_in + 1e-4)**2 / (2 * sigma_y_in**2))
pdfs_in /= T.sqrt(2 * np.pi * sigma_y_in**2)
logL_in = T.sum(T.log(pdfs_in) * (1 - is_outlier))
# likelihood for outliers
pdfs_out = T.exp(-(yobs - yest_out + 1e-4)**2 / (2 * (sigma_y_in**2 + sigma_y_out**2)))
pdfs_out /= T.sqrt(2 * np.pi * (sigma_y_in**2 + sigma_y_out**2))
logL_out = T.sum(T.log(pdfs_out) * is_outlier)
return logL_in + logL_out
with pm.Model() as mdl_signoise:
## Define weakly informative Normal priors to give Ridge regression
b0 = pm.Normal('b0_intercept', mu=0, sd=10, testval=pm.floatX(0.1))
b1 = pm.Normal('b1_slope', mu=0, sd=10, testval=pm.floatX(1.))
## Define linear model
yest_in = b0 + b1 * dfhoggs['x']
## Define weakly informative priors for the mean and variance of outliers
yest_out = pm.Normal('yest_out', mu=0, sd=100, testval=pm.floatX(1.))
sigma_y_out = pm.HalfNormal('sigma_y_out', sd=100, testval=pm.floatX(1.))
## Define Bernoulli inlier / outlier flags according to a hyperprior
## fraction of outliers, itself constrained to [0,.5] for symmetry
frac_outliers = pm.Uniform('frac_outliers', lower=0., upper=.5)
is_outlier = pm.Bernoulli('is_outlier', p=frac_outliers, shape=dfhoggs.shape[0],
testval=np.random.rand(dfhoggs.shape[0]) < 0.2)
## Extract observed y and sigma_y from dataset, encode as theano objects
yobs = thno.shared(np.asarray(dfhoggs['y'], dtype=thno.config.floatX), name='yobs')
sigma_y_in = thno.shared(np.asarray(dfhoggs['sigma_y'], dtype=thno.config.floatX),
name='sigma_y_in')
## Use custom likelihood using DensityDist
likelihood = pm.DensityDist('likelihood', logp_signoise,
observed={'yobs': yobs, 'is_outlier': is_outlier,
'yest_in': yest_in, 'sigma_y_in': sigma_y_in,
'yest_out': yest_out, 'sigma_y_out': sigma_y_out})
```
### Sample
```
with mdl_signoise:
## two-step sampling to create Bernoulli inlier/outlier flags
step1 = pm.Metropolis([frac_outliers, yest_out, sigma_y_out, b0, b1])
step2 = pm.step_methods.BinaryGibbsMetropolis([is_outlier])
## take samples
traces_signoise = pm.sample(20000, step=[step1, step2], tune=10000, progressbar=True)
```
### View Traces
```
traces_signoise[-10000:]['b0_intercept']
_ = pm.traceplot(traces_signoise[-10000:], figsize=(12,len(traces_signoise.varnames)*1.5),
lines={k: v['mean'] for k, v in pm.df_summary(traces_signoise[-1000:]).iterrows()})
```
**NOTE:**
+ During development I've found that 3 datapoints id=[1,2,3] are always indicated as outliers, but the remaining ordering of datapoints by decreasing outlier-hood is unstable between runs: the posterior surface appears to have a small number of solutions with very similar probability.
+ The NUTS sampler seems to work okay, and indeed it's a nice opportunity to demonstrate a custom likelihood which is possible to express as a theano function (thus allowing a gradient-based sampler like NUTS). However, with a more complicated dataset, I would spend time understanding this instability and potentially prefer using more samples under Metropolis-Hastings.
---
---
## Declare Outliers and Compare Plots
### View ranges for inliers / outlier predictions
At each step of the traces, each datapoint may be either an inlier or outlier. We hope that the datapoints spend an unequal time being one state or the other, so let's take a look at the simple count of states for each of the 20 datapoints.
```
outlier_melt = pd.melt(pd.DataFrame(traces_signoise['is_outlier', -1000:],
columns=['[{}]'.format(int(d)) for d in dfhoggs.index]),
var_name='datapoint_id', value_name='is_outlier')
ax0 = sns.pointplot(y='datapoint_id', x='is_outlier', data=outlier_melt,
kind='point', join=False, ci=None, size=4, aspect=2)
_ = ax0.vlines([0,1], 0, 19, ['b','r'], '--')
_ = ax0.set_xlim((-0.1,1.1))
_ = ax0.set_xticks(np.arange(0, 1.1, 0.1))
_ = ax0.set_xticklabels(['{:.0%}'.format(t) for t in np.arange(0,1.1,0.1)])
_ = ax0.yaxis.grid(True, linestyle='-', which='major', color='w', alpha=0.4)
_ = ax0.set_title('Prop. of the trace where datapoint is an outlier')
_ = ax0.set_xlabel('Prop. of the trace where is_outlier == 1')
```
**Observe**:
+ The plot above shows the number of samples in the traces in which each datapoint is marked as an outlier, expressed as a percentage.
+ In particular, 3 points [1, 2, 3] spend >=95% of their time as outliers
+ Contrastingly, points at the other end of the plot close to 0% are our strongest inliers.
+ For comparison, the mean posterior value of `frac_outliers` is ~0.35, corresponding to roughly 7 of the 20 datapoints. You can see these 7 datapoints in the plot above, all those with a value >50% or thereabouts.
+ However, only 3 of these points are outliers >=95% of the time.
+ See note above regarding instability between runs.
The 95% cutoff we choose is subjective and arbitrary, but I prefer it for now, so let's declare these 3 to be outliers and see how it looks compared to Jake Vanderplas' outliers, which were declared in a slightly different way as points with means above 0.68.
### Declare outliers
**Note:**
+ I will declare outliers to be datapoints that have value == 1 at the 5-percentile cutoff, i.e. in the percentiles from 5 up to 100, their values are 1.
+ Try for yourself altering cutoff to larger values, which leads to an objective ranking of outlier-hood.
```
cutoff = 5
dfhoggs['outlier'] = np.percentile(traces_signoise[-1000:]['is_outlier'],cutoff, axis=0)
dfhoggs['outlier'].value_counts()
```
### Posterior Prediction Plots for OLS vs StudentT vs SignalNoise
```
g = sns.FacetGrid(dfhoggs, size=8, hue='outlier', hue_order=[True,False],
palette='Set1', legend_out=False)
lm = lambda x, samp: samp['b0_intercept'] + samp['b1_slope'] * x
pm.plot_posterior_predictive_glm(traces_ols[-1000:],
eval=np.linspace(-3, 3, 10), lm=lm, samples=200, color='#22CC00', alpha=.2)
pm.plot_posterior_predictive_glm(traces_studentt[-1000:], lm=lm,
eval=np.linspace(-3, 3, 10), samples=200, color='#FFA500', alpha=.5)
pm.plot_posterior_predictive_glm(traces_signoise[-1000:], lm=lm,
eval=np.linspace(-3, 3, 10), samples=200, color='#357EC7', alpha=.3)
_ = g.map(plt.errorbar, 'x', 'y', 'sigma_y', 'sigma_x', marker="o", ls='').add_legend()
_ = g.axes[0][0].annotate('OLS Fit: Green\nStudent-T Fit: Orange\nSignal Vs Noise Fit: Blue',
size='x-large', xy=(1,0), xycoords='axes fraction',
xytext=(-160,10), textcoords='offset points')
_ = g.axes[0][0].set_ylim(ylims)
_ = g.axes[0][0].set_xlim(xlims)
```
**Observe**:
+ The posterior preditive fit for:
+ the **OLS model** is shown in **Green** and as expected, it doesn't appear to fit the majority of our datapoints very well, skewed by outliers
+ the **Robust Student-T model** is shown in **Orange** and does appear to fit the 'main axis' of datapoints quite well, ignoring outliers
+ the **Robust Signal vs Noise model** is shown in **Blue** and also appears to fit the 'main axis' of datapoints rather well, ignoring outliers.
+ We see that the **Robust Signal vs Noise model** also yields specific estimates of _which_ datapoints are outliers:
+ 17 'inlier' datapoints, in **Blue** and
+ 3 'outlier' datapoints shown in **Red**.
+ From a simple visual inspection, the classification seems fair, and agrees with Jake Vanderplas' findings.
+ Overall, it seems that:
+ the **Signal vs Noise model** behaves as promised, yielding a robust regression estimate and explicit labelling of inliers / outliers, but
+ the **Signal vs Noise model** is quite complex and whilst the regression seems robust and stable, the actual inlier / outlier labelling seems slightly unstable
+ if you simply want a robust regression without inlier / outlier labelling, the **Student-T model** may be a good compromise, offering a simple model, quick sampling, and a very similar estimate.
---
Example originally contributed by Jonathan Sedar 2015-12-21 [github.com/jonsedar](https://github.com/jonsedar)
|
github_jupyter
|
# Demystifying Neural Networks
---
# Exercises - ANN Weights
We will generate matrices that can be used as an ANN.
You can generate matrices with any function from `numpy.random`.
You can provide a tuple to the `size=` parameter to get an array
of that shape. For example, `np.random.normal(0, 1, (3, 6))`
generates a matrix of 3 rows and 6 columns.
```
import numpy as np
```
#### 1. Generate the following matrices with values from the normal distribution
A) $A_{2 \times 3}$
B) $B_{7 \times 5}$
```
A = np.random.normal(0, 1, (2, 3))
B = np.random.normal(0, 1, (7, 5))
print(A)
print(B)
```
#### 2. Generate matrices of the same size as the used in the `pytorch` network
$$
W_{25 \times 8}, W_{B\: 25 \times 1},
W'_{10 \times 25}, W'_{B\: 10 \times 1},
W'_{2 \times 10}, W'_{B\: 2 \times 1}
$$
```
W = np.random.normal(0, 1, (25, 8))
W_B = np.random.normal(0, 1, (25, 1))
Wx = np.random.normal(0, 1, (10, 25))
Wx_B = np.random.normal(0, 1, (10, 1))
Wxx = np.random.normal(0, 1, (2, 10))
Wxx_B = np.random.normal(0, 1, (2, 1))
weights = [W, W_B, Wx, Wx_B, Wxx, Wxx_B]
print([x.shape for x in weights])
```
---
Weight generation is a big topic in ANN research.
We will use one well accepted way of generating ways but there are plethora of others.
The way we will generate weight matrices is to:
If we need to generate a matrix of size $p \times n$,
we take all values for the matrix from the normal distribution
with mean and standard deviation as:
$$
\mu = 0 \\
\sigma = \frac{1}{n + p}
$$
In `numpy` the mean argument is `loc=` and standard deviation is called `scale=`
#### 3. Generate the same matrices as above but use the distribution describe above, then evaluate
$$
X = \left[
\begin{matrix}
102.50781 & 58.88243 & 0.46532 & -0.51509 & 1.67726 & 14.86015 & 10.57649 & 127.39358 \\
142.07812 & 45.28807 & -0.32033 & 0.28395 & 5.37625 & 29.00990 & 6.07627 & 37.83139 \\
138.17969 & 51.52448 & -0.03185 & 0.04680 & 6.33027 & 31.57635 & 5.15594 & 26.14331 \\
\end{matrix}
\right]
$$
(These are the first three rows in the pulsar dataset)
$$
\hat{Y}_{2 \times 3} = tanh(W''_{2 \times 10} \times
tanh(W'_{10 \times 25} \times
tanh(W_{25 \times 8} \times X^T + W_{B\: 25 \times 1})
+ W'_{B\: 10 \times 1})
+ W''_{B\: 2 \times 1})
$$
```
X = np.array([
[102.50781, 58.88243, 0.46532, -0.51509, 1.67726, 14.86015, 10.57649, 127.39358],
[142.07812, 45.28807, -0.32033, 0.28395, 5.37625, 29.00990, 6.07627, 37.83139],
[138.17969, 51.52448, -0.03185, 0.04680, 6.33027, 31.57635, 5.15594, 26.14331],
])
W = np.random.normal(0, 1/(8+25), (25, 8))
W_B = np.random.normal(0, 1/(25+1), (25, 1))
Wx = np.random.normal(0, 1/(10+25), (10, 25))
Wx_B = np.random.normal(0, 1/(10+1), (10, 1))
Wxx = np.random.normal(0, 1/(2+10), (2, 10))
Wxx_B = np.random.normal(0, 1/(2+1), (2, 1));
Y_hat = np.tanh(Wxx @ np.tanh(Wx @ np.tanh(W @ X.T + W_B) + Wx_B) + Wxx_B)
print(Y_hat.T)
```
|
github_jupyter
|
# Document Processing with AutoML and Vision API
## Problem Statement
Formally the brief for this Open Project could be stated as follows: Given a collection of varying pdf/png documents containing similar information, create a pipeline that will extract relevant entities from the documents and store the entities in a standardized, easily accessible format.
The data for this project is contained in the Cloud Storage bucket [gs://document-processing/patent_dataset.zip](https://storage.googleapis.com/document-processing/patent_dataset.zip). The file [gs://document-processing/ground_truth.csv](https://storage.googleapis.com/document-processing/ground_truth.csv) contains hand-labeled fields extracted from the patents.
The labels in the ground_truth.csv file are filename, category, publication_date, classification_1, classification_2, application_number, filing_date, priority, representative, applicant, inventor, titleFL, titleSL, abstractFL, and publication_number
Here is an example of two different patent formats:
<table><tr>
<td> <img src="eu_patent.png" alt="Drawing" style="width: 600px;"/> </td>
<td> <img src="us_patent.png" alt="Drawing" style="width: 600px;"/> </td>
</tr></table>
### Flexible Solution
There are many possible ways to develop a solution to this task which allows students to touch on various functionality and GCP tools that we discuss during the ASL, including the Vision API, AutoML Vision, BigQuery, Tensorflow, Cloud Composer, PubSub.
For students more interested in modeling with Tensorflow, they could build a classification model from scratch to regcognize the various type of document formats at hand. Knowing the document format (e.g. US or EU patents as in the example above), relevant entities can then be extracted using the Vision API and some basic regex extactors. It might also be possible to train a [conditional random field in Tensorflow](https://github.com/tensorflow/tensorflow/tree/master/tensorflow/contrib/crf) to learn how to tag and extract relevant entities from text and the given labels, instead of writing regex-based entity extractors for each document class.
Students more interested in productionization could work to use Cloud Functions to automate the extraction pipeline. Or incorporate PubSub so that when a new document is uploaded to a specific GCS bucket it is parsed and the entities uploaded to a BigQuery table.
Below is a solution outline that uses the Vision API and AutoML, uploading the extracted entities to a table in BigQuery.
## Install AutoML package
**Caution:** Run the following command and **restart the kernel** afterwards.
```
!pip freeze | grep google-cloud-automl==0.1.2 || pip install --upgrade google-cloud-automl==0.1.2
```
## Set the correct environment variables
The following variables should be updated according to your own enviroment:
```
PROJECT_ID = "asl-open-projects"
SERVICE_ACCOUNT = "entity-extractor"
ZONE = "us-central1"
AUTOML_MODEL_ID = "ICN6705037528556716784"
```
The following variables are computed from the one you set above, and should
not be modified:
```
import os
PWD = os.path.abspath(os.path.curdir)
SERVICE_KEY_PATH = os.path.join(PWD, "{0}.json".format(SERVICE_ACCOUNT))
SERVICE_ACCOUNT_EMAIL="{0}@{1}.iam.gserviceaccount.com".format(SERVICE_ACCOUNT, PROJECT_ID)
# Exporting the variables into the environment to make them available to all the subsequent cells
os.environ["PROJECT_ID"] = PROJECT_ID
os.environ["SERVICE_ACCOUNT"] = SERVICE_ACCOUNT
os.environ["SERVICE_KEY_PATH"] = SERVICE_KEY_PATH
os.environ["SERVICE_ACCOUNT_EMAIL"] = SERVICE_ACCOUNT_EMAIL
os.environ["ZONE"] = ZONE
```
## Switching the right project and zone
```
%%bash
gcloud config set project $PROJECT_ID
gcloud config set compute/region $ZONE
```
## Create a service account
```
%%bash
gcloud iam service-accounts list | grep $SERVICE_ACCOUNT ||
gcloud iam service-accounts create $SERVICE_ACCOUNT
```
## Grant service account project ownership
TODO: We should ideally restrict the permissions to AutoML and Vision roles only
```
%%bash
gcloud projects add-iam-policy-binding $PROJECT_ID \
--member "serviceAccount:$SERVICE_ACCOUNT_EMAIL" \
--role "roles/owner"
```
## Create service account keys if not existing
```
%%bash
test -f $SERVICE_KEY_PATH ||
gcloud iam service-accounts keys create $SERVICE_KEY_PATH \
--iam-account $SERVICE_ACCOUNT_EMAIL
echo "Service key: $(ls $SERVICE_KEY_PATH)"
```
## Make the key available to google clients for authentication
```
os.environ['GOOGLE_APPLICATION_CREDENTIALS'] = SERVICE_KEY_PATH
```
## Implement a document classifier with AutoML
Here is a simple wrapper around an already trained AutoML model trained directly
from the cloud console on the various document types:
```
from google.cloud import automl_v1beta1 as automl
class DocumentClassifier:
def __init__(self, project_id, model_id, zone):
self.client = automl.PredictionServiceClient()
self.model = self.client.model_path(project_id, zone, model_id)
def __call__(self, filename):
with open(filename, 'rb') as fp:
image = fp.read()
payload = {
'image': {
'image_bytes': image
}
}
response = self.client.predict(self.model, payload)
predicted_class = response.payload[0].display_name
return predicted_class
```
Let's see how to use that `DocumentClassifier`:
```
classifier = DocumentClassifier(PROJECT_ID, AUTOML_MODEL_ID, ZONE)
eu_image_label = classifier("./eu_patent.png")
us_image_label = classifier("./us_patent.png")
print("EU patent inferred label:", eu_image_label)
print("US patent inferred label:", us_image_label)
```
## Implement a document parser with Vision API
Documentation:
* https://cloud.google.com/vision/docs/base64
* https://stackoverflow.com/questions/49918950/response-400-from-google-vision-api-ocr-with-a-base64-string-of-specified-image
Here is a simple class wrapping calls to the OCR capabilities of Cloud Vision:
```
!pip freeze | grep google-api-python-client==1.7.7 || pip install --upgrade google-api-python-client==1.7.7
import base64
from googleapiclient.discovery import build
class DocumentParser:
def __init__(self):
self.client = build('vision', 'v1')
def __call__(self, filename):
with open(filename, 'rb') as fp:
image = fp.read()
encoded_image = base64.b64encode(image).decode('UTF-8')
payload = {
'requests': [{
'image': {
'content': encoded_image
},
'features': [{
'type': 'TEXT_DETECTION',
}]
}],
}
request = self.client.images().annotate(body=payload)
response = request.execute(num_retries=3)
return response['responses'][0]['textAnnotations'][0]['description']
```
Let's now see how to use our `DocumentParser`:
```
parser = DocumentParser()
eu_patent_text = parser("./eu_patent.png")
us_patent_text = parser("./us_patent.png")
print(eu_patent_text)
```
# Implement the rule-based extractors for each document categories
For each patent type, we now want to write a simple function that takes the
text extracted by the OCR system above and extract the name and date of the patent.
We will write two rule base extractors, one for each type of patent (us or eu), each of
which will yield a PatentInfo object collecting the extracted object into a `nametuple` instance.
```
from collections import namedtuple
PatentInfo = namedtuple('PatentInfo', ['filename', 'category', 'date', 'number'])
```
Here are two helper functions for text splitting and pattern matching:
```
!pip freeze | grep pandas==0.23.4 || pip install --upgrade pandas==0.23.4
import pandas as pd
import re
def split_text_into_lines(text, sep="\(..\)"):
lines = [line.strip() for line in re.split(sep, text)]
return lines
def extract_pattern_from_lines(lines, pattern):
"""Extracts the first line from `text` with a matching `pattern`.
"""
lines = pd.Series(lines)
mask = lines.str.contains(pattern)
return lines[mask].values[0] if mask.any() else None
```
### European patent extractor
```
def extract_info_from_eu_patent(filename, text):
lines = split_text_into_lines(text)
category = "eu"
number_paragraph = extract_pattern_from_lines(lines, "EP")
number_lines = number_paragraph.split('\n')
number = extract_pattern_from_lines(number_lines, 'EP')
date_paragraph = extract_pattern_from_lines(lines, 'Date of filing:')
date = date_paragraph.replace("Date of filing:", "").strip()
return PatentInfo(
filename=filename,
category=category,
date=date,
number=number
)
eu_patent_info = extract_info_from_eu_patent("./eu_patent.png", eu_patent_text)
eu_patent_info
```
### US patent extractor
```
def extract_info_from_us_patent(filename, text):
lines = split_text_into_lines(text)
category = "us"
number_paragraph = extract_pattern_from_lines(lines, "Patent No.:")
number = number_paragraph.replace("Patent No.:", "").strip()
date_paragraph = extract_pattern_from_lines(lines, "Date of Patent:")
date = date_paragraph.split('\n')[-1]
return PatentInfo(
filename=filename,
category=category,
date=date,
number=number
)
us_patent_info = extract_info_from_us_patent("./us_patent.png", us_patent_text)
us_patent_info
```
## Tie all together into a DocumentExtractor
```
class DocumentExtractor:
def __init__(self, classifier, parser):
self.classifier = classifier
self.parser = parser
def __call__(self, filename):
text = self.parser(filename)
label = self.classifier(filename)
if label == 'eu':
info = extract_info_from_eu_patent(filename, text)
elif label == 'us':
info = extract_info_from_us_patent(filename, text)
else:
raise ValueError
return info
extractor = DocumentExtractor(classifier, parser)
eu_patent_info = extractor("./eu_patent.png")
us_patent_info = extractor("./us_patent.png")
print(eu_patent_info)
print(us_patent_info)
```
## Upload found entites to BigQuery
Start by adding a dataset called patents to the current project
```
!pip freeze | grep google-cloud-bigquery==1.8.1 || pip install google-cloud-bigquery==1.8.1
```
Check to see if the dataset called "patents" exists in the current project. If not, create it.
```
from google.cloud import bigquery
client = bigquery.Client()
# Collect datasets and project information
datasets = list(client.list_datasets())
project = client.project
# Create a list of the datasets. If the 'patents' dataset
# does not exist, then create it.
if datasets:
all_datasets = []
for dataset in datasets:
all_datasets.append(dataset.dataset_id)
else:
print('{} project does not contain any datasets.'.format(project))
if datasets and 'patents' in all_datasets:
print('The dataset "patents" already exists in project {}.'.format(project))
else:
dataset_id = 'patents'
dataset_ref = client.dataset(dataset_id)
# Construct a Dataset object.
dataset = bigquery.Dataset(dataset_ref)
# Specify the geographic location where the dataset should reside.
dataset.location = "US"
# Send the dataset to the API for creation.
dataset = client.create_dataset(dataset) # API request
print('The dataset "patents" was created in project {}.'.format(project))
```
Upload the extracted entities to a table called "found_entities" in the "patents" dataset.
Start by creating an empty table in the patents dataset.
```
# Create an empty table in the patents dataset and define schema
dataset_ref = client.dataset('patents')
schema = [
bigquery.SchemaField('filename', 'STRING', mode='NULLABLE'),
bigquery.SchemaField('category', 'STRING', mode='NULLABLE'),
bigquery.SchemaField('date', 'STRING', mode='NULLABLE'),
bigquery.SchemaField('number', 'STRING', mode='NULLABLE'),
]
table_ref = dataset_ref.table('found_entities')
table = bigquery.Table(table_ref, schema=schema)
table = client.create_table(table) # API request
assert table.table_id == 'found_entities'
def upload_to_bq(patent_info, dataset_id, table_id):
"""Appends the information extracted in patent_info into the
dataset_id:table_id in BigQuery.
patent_info should be a namedtuple as created above and should
have components matching the schema set up for the table
"""
table_ref = client.dataset(dataset_id).table(table_id)
table = client.get_table(table_ref) # API request
rows_to_insert = [tuple(patent_info._asdict().values())]
errors = client.insert_rows(table, rows_to_insert) # API request
assert errors == []
upload_to_bq(eu_patent_info, 'patents', 'found_entities')
upload_to_bq(us_patent_info, 'patents', 'found_entities')
```
### Examine the resuts in BigQuery.
We can now query the BigQuery table to see what values have been uploaded.
```
%load_ext google.cloud.bigquery
%%bigquery
SELECT
*
FROM `asl-open-projects.patents.found_entities`
```
We can also look at the resulting entities in a dataframe.
```
dataset_id = 'patents'
table_id = 'found_entities'
sql = """
SELECT
*
FROM
`{}.{}.{}`
LIMIT 10
""".format(project, dataset_id, table_id)
df = client.query(sql).to_dataframe()
df.head()
```
## Pipeline Evaluation
TODO: We should include some section on how to evaluate the performance of the extractor. Here we can use the ground_truth table and explore different kinds of string metrics (e.g. Levenshtein distance) to measure accuracy of the entity extraction.
## Clean up
To remove the table "found_entities" from the "patents" dataset created above.
```
dataset_id = 'patents'
table_id = 'found_entities'
tables = list(client.list_tables(dataset_id)) # API request(s)
if tables:
num_tables = len(tables)
all_tables = []
for _ in range(num_tables):
all_tables.append(tables[_].table_id)
print('These tables were found in the {} dataset: {}'.format(dataset_id,all_tables))
if table_id in all_tables:
table_ref = client.dataset(dataset_id).table(table_id)
client.delete_table(table_ref) # API request
print('Table {} was deleted from dataset {}.'.format(table_id, dataset_id))
else:
print('{} dataset does not contain any tables.'.format(dataset_id))
```
### The next cells will remove the patents dataset and all of its tables. Not recommended as I recently uploaded a talbe of 'ground_truth' for entities in the files
To remove the "patents" dataset and all of its tables.
```
'''
client = bigquery.Client()
# Collect datasets and project information
datasets = list(client.list_datasets())
project = client.project
if datasets:
all_datasets = []
for dataset in datasets:
all_datasets.append(dataset.dataset_id)
if 'patents' in all_datasets:
# Delete the dataset "patents" and its contents
dataset_id = 'patents'
dataset_ref = client.dataset(dataset_id)
client.delete_dataset(dataset_ref, delete_contents=True)
print('Dataset {} deleted from project {}.'.format(dataset_id, project))
else: print('{} project does not contain the "patents" datasets.'.format(project))
else:
print('{} project does not contain any datasets.'.format(project))
'''
```
|
github_jupyter
|
First we need to download the dataset. In this case we use a datasets containing poems. By doing so we train the model to create its own poems.
```
from datasets import load_dataset
dataset = load_dataset("poem_sentiment")
print(dataset)
```
Before training we need to preprocess the dataset. We tokenize the entries in the dataset and remove all columns we don't need to train the adapter.
```
from transformers import BertTokenizer, GPT2LMHeadModel, TextGenerationPipeline
tokenizer = BertTokenizer.from_pretrained("uer/gpt2-chinese-cluecorpussmall")
from transformers import GPT2Tokenizer
def encode_batch(batch):
"""Encodes a batch of input data using the model tokenizer."""
encoding = tokenizer(batch["verse_text"])
# For language modeling the labels need to be the input_ids
#encoding["labels"] = encoding["input_ids"]
return encoding
#tokenizer = GPT2Tokenizer.from_pretrained("gpt2")
#tokenizer.pad_token = tokenizer.eos_token
# The GPT-2 tokenizer does not have a padding token. In order to process the data
# in batches we set one here
column_names = dataset["train"].column_names
dataset = dataset.map(encode_batch, remove_columns=column_names, batched=True)
```
Next we concatenate the documents in the dataset and create chunks with a length of `block_size`. This is beneficial for language modeling.
```
block_size = 50
# Main data processing function that will concatenate all texts from our dataset and generate chunks of block_size.
def group_texts(examples):
# Concatenate all texts.
concatenated_examples = {k: sum(examples[k], []) for k in examples.keys()}
total_length = len(concatenated_examples[list(examples.keys())[0]])
# We drop the small remainder, we could add padding if the model supported it instead of this drop, you can
# customize this part to your needs.
total_length = (total_length // block_size) * block_size
# Split by chunks of max_len.
result = {
k: [t[i : i + block_size] for i in range(0, total_length, block_size)]
for k, t in concatenated_examples.items()
}
result["labels"] = result["input_ids"].copy()
return result
dataset = dataset.map(group_texts,batched=True,)
dataset.set_format(type="torch", columns=["input_ids", "attention_mask", "labels"])
```
Next we create the model and add our new adapter.Let's just call it `poem` since it is trained to create new poems. Then we activate it and prepare it for training.
```
from transformers import AutoModelForCausalLM
model = AutoModelForCausalLM.from_pretrained("uer/gpt2-chinese-cluecorpussmall")
# add new adapter
model.add_adapter("poem")
# activate adapter for training
model.train_adapter("poem")
```
The last thing we need to do before we can start training is create the trainer. As trainingsargumnénts we choose a learningrate of 1e-4. Feel free to play around with the paraeters and see how they affect the result.
```
from transformers import Trainer, TrainingArguments
training_args = TrainingArguments(
output_dir="./examples",
do_train=True,
remove_unused_columns=False,
learning_rate=5e-4,
num_train_epochs=3,
)
trainer = Trainer(
model=model,
args=training_args,
tokenizer=tokenizer,
train_dataset=dataset["train"],
eval_dataset=dataset["validation"],
)
trainer.train()
```
Now that we have a trained udapter we save it for future usage.
```
PREFIX = "what a "
encoding = tokenizer(PREFIX, return_tensors="pt")
encoding = encoding.to(model.device)
output_sequence = model.generate(
input_ids=encoding["input_ids"][:,:-1],
attention_mask=encoding["attention_mask"][:,:-1],
do_sample=True,
num_return_sequences=5,
max_length = 50,
)
```
Lastly we want to see what the model actually created. Too de this we need to decode the tokens from ids back to words and remove the end of sentence tokens. You can easily use this code with an other dataset. Don't forget to share your adapters at [AdapterHub](https://adapterhub.ml/).
```
for generated_sequence_idx, generated_sequence in enumerate(output_sequence):
print("=== GENERATED SEQUENCE {} ===".format(generated_sequence_idx + 1))
generated_sequence = generated_sequence.tolist()
# Decode text
text = tokenizer.decode(generated_sequence, clean_up_tokenization_spaces=True)
# Remove EndOfSentence Tokens
text = text[: text.find(tokenizer.pad_token)]
print(text)
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
from Utils import load
from Utils import generator
from Utils import metrics
from train import *
from prune import *
from Layers import layers
from torch.nn import functional as F
import torch.nn as nn
def fc(input_shape, nonlinearity=nn.ReLU()):
size = np.prod(input_shape)
# Linear feature extractor
modules = [nn.Flatten()]
modules.append(layers.Linear(size, 5000))
modules.append(nonlinearity)
modules.append(layers.Linear(5000, 900))
modules.append(nonlinearity)
modules.append(layers.Linear(900, 400))
modules.append(nonlinearity)
modules.append(layers.Linear(400, 100))
modules.append(nonlinearity)
modules.append(layers.Linear(100, 30))
modules.append(nonlinearity)
modules.append(layers.Linear(30, 1))
model = nn.Sequential(*modules)
return model
from data import *
from models import *
from utils import *
from sklearn.model_selection import KFold
import os, shutil, pickle
ctx = mx.gpu(0) if mx.context.num_gpus() > 0 else mx.cpu(0)
loss_before_prune=[]
loss_after_prune=[]
loss_prune_posttrain=[]
NUM_PARA=[]
for datasetindex in range(10):#[0,1,4,5,6,7,8,9]:
dataset=str(datasetindex)+'.csv'
X, y= get_data(dataset)
np.random.seed(0)
kf = KFold(n_splits=5,random_state=0,shuffle=True)
kf.get_n_splits(X)
seed=0#[0,1,2,3,4]
chosenarmsList=[]
for train_index, test_index in kf.split(X):
X_tr, X_te = X[train_index], X[test_index]
y_tr, y_te = y[train_index], y[test_index]
X_test=nd.array(X_te).as_in_context(ctx) # Fix test data for all seeds
y_test=nd.array(y_te).as_in_context(ctx)
factor=np.max(y_te)-np.min(y_te) #normalize RMSE
print(factor)
#X_tr, X_te, y_tr, y_te = get_data(0.2,0)
#selected_interaction = detectNID(X_tr,y_tr,X_te,y_te,test_size,seed)
#index_Subsets=get_interaction_index(selected_interaction)
N=X_tr.shape[0]
p=X_tr.shape[1]
batch_size=500
n_epochs=300
if N<250:
batch_size=50
X_train=nd.array(X_tr).as_in_context(ctx)
y_train=nd.array(y_tr).as_in_context(ctx)
train_dataset = ArrayDataset(X_train, y_train)
# num_workers=4
train_data = DataLoader(train_dataset, batch_size=batch_size, shuffle=True)#,num_workers=num_workers)
#X_test=nd.array(X_te).as_in_context(ctx)
#y_test=nd.array(y_te).as_in_context(ctx)
print('start training FC')
FCnet=build_FC(train_data,ctx) # initialize the overparametrized network
FCnet.load_parameters('Selected_models/FCnet_'+str(datasetindex)+'_seed_'+str(seed),ctx=ctx)
import torch.nn as nn
model=fc(10)
loss = nn.MSELoss()
dataset=torch.utils.data.TensorDataset(torch.Tensor(X_tr),torch.Tensor(y_tr))
for i in range(6):
model[int(i*2+1)].weight.data=torch.Tensor(FCnet[i].weight.data().asnumpy())
model[int(i*2+1)].bias.data=torch.Tensor(FCnet[i].bias.data().asnumpy())
print("dataset:",datasetindex,"seed",seed)
print("before prune:",torch.sqrt(torch.mean((model(torch.Tensor(X_te))-torch.Tensor(y_te))**2))/factor)
loss_before_prune.append(torch.sqrt(loss(model(torch.Tensor(X_te)),torch.Tensor(y_te)))/factor)
print(torch.sqrt(loss(model(torch.Tensor(X_te)),torch.Tensor(y_te)))/factor)
# Prune ##
device = torch.device("cpu")
prune_loader = load.dataloader(dataset, 64, True, 4, 1)
prune_epochs=10
print('Pruning with {} for {} epochs.'.format('synflow', prune_epochs))
pruner = load.pruner('synflow')(generator.masked_parameters(model, False, False, False))
sparsity = 10**(-float(2.44715803134)) #280X #100X 10**(-float(2))
prune_loop(model, loss, pruner, prune_loader, device, sparsity,
'exponential', 'global', prune_epochs, False, False, False, False)
pruner.apply_mask()
print("after prune:",torch.sqrt(loss(model(torch.Tensor(X_te)),torch.Tensor(y_te)))/factor)
loss_after_prune.append(torch.sqrt(loss(model(torch.Tensor(X_te)),torch.Tensor(y_te)))/factor)
## post_train
train_loader = load.dataloader(dataset, 64, True, 4)
test_loader = load.dataloader(dataset, 200 , False, 4)
optimizer = torch.optim.Adam(generator.parameters(model), betas=(0.9, 0.99))
scheduler = torch.optim.lr_scheduler.MultiStepLR(optimizer,milestones=[30,80], gamma=0.1)
post_result = train_eval_loop(model, loss, optimizer, scheduler, train_loader,
test_loader, device, 100, True)
print("after post_train:",torch.sqrt(loss(model(torch.Tensor(X_te)),torch.Tensor(y_te)))/factor)
loss_prune_posttrain.append(torch.sqrt(loss(model(torch.Tensor(X_te)),torch.Tensor(y_te)))/factor)
num=0
for i in pruner.masked_parameters:
num=num+sum(sum(i[0]))
print(num)
NUM_PARA.append(num)
seed=seed+1
import mxnet.gluon.nn as nn
##synflow results 280X
a=0
for i in range(10):
print(sum(loss_prune_posttrain[5*i:5*i+5])/5)
a=a+sum(loss_prune_posttrain[5*i:5*i+5])/5
print("ave:",a/10)
##synflow results 100X
a=0
for i in range(10):
print(sum(loss_prune_posttrain[5*i:5*i+5])/5)
a=a+sum(loss_prune_posttrain[5*i:5*i+5])/5
print("ave:",a/10)
```
|
github_jupyter
|
```
import numpy as np
from matplotlib import pyplot
def flux(psi_l, psi_r, C):
return .5 * (C + abs(C)) * psi_l + \
.5 * (C - abs(C)) * psi_r
def upwind(psi, i, C):
return psi[i] - flux(psi[i ], psi[i+one], C[i]) + \
flux(psi[i-one], psi[i ], C[i-one])
def C_corr(C, nx, psi):
j = slice(0, nx-1)
return (abs(C[j]) - C[j]**2) * (psi[j+one] - psi[j]) / (psi[j+one] + psi[j] + 1e-10)
class shift:
def __radd__(self, i):
return slice(i.start+1, i.stop+1)
def __rsub__(self, i):
return slice(i.start-1, i.stop-1)
one = shift()
def psi_0(x):
a = 25
return np.where(np.abs(x)<np.pi/2*a, np.cos(x/a)**2, 0)
def plot(x, psi_0, psi_T_a, psi_T_n, n_corr_it):
pyplot.step(x, psi_0, label='initial', where='mid')
pyplot.step(x, psi_T_a, label='analytical', where='mid')
pyplot.step(x, psi_T_n, label='numerical', where='mid')
pyplot.grid()
pyplot.legend()
pyplot.title(f'MPDATA {n_corr_it} corrective iterations')
pyplot.show()
def solve(nx=75, nt=50, n_corr_it=2, make_plot=False):
T = 50
x_min, x_max = -50, 250
C = .1
dt = T / nt
x, dx = np.linspace(x_min, x_max, nx, endpoint=False, retstep=True)
v = C / dt * dx
assert C <= 1
i = slice(1, nx-2)
C_phys = np.full(nx-1, C)
psi = psi_0(x)
for _ in range(nt):
psi[i] = upwind(psi, i, C_phys)
C_iter = C_phys
for it in range(n_corr_it):
C_iter = C_corr(C_iter, nx, psi)
psi[i] = upwind(psi, i, C_iter)
psi_true = psi_0(x-v*nt*dt)
err = np.sqrt(sum(pow(psi-psi_true, 2)) / nt / nx)
if make_plot:
plot(x, psi_0(x), psi_T_a=psi_true, psi_T_n=psi, n_corr_it=n_corr_it)
return dx, dt, err
solve(nx=150, nt=500, n_corr_it=0, make_plot=True)
for n_it in [0,1,2]:
data = {'x':[], 'y':[]}
for nx in range(600,1000,50):
dx, dt, err = solve(nx=nx, nt=300, n_corr_it=n_it)
data['x'].append(np.log2(dx))
data['y'].append(np.log2(err))
pyplot.scatter(data['x'], data['y'], label=f"{n_it} corrective iters")
pyplot.plot(data['x'], data['y'])
def line(k, offset, label=False):
pyplot.plot(
data['x'],
k*np.array(data['x']) + offset,
label=f'$err \sim dx^{k}$' if label else '',
linestyle=':',
color='black'
)
line(k=2, offset=-10.1, label=True)
line(k=3, offset=-9.1, label=True)
line(k=2, offset=-14)
line(k=3, offset=-13)
pyplot.legend()
pyplot.gca().set_xlabel('$log_2(dx)$')
pyplot.gca().set_ylabel('$log_2(err)$')
pyplot.grid()
```
|
github_jupyter
|
```
from urllib.request import urlopen
from bs4 import BeautifulSoup
html = urlopen('http://en.wikipedia.org/wiki/Kevin_Bacon')
bs = BeautifulSoup(html, 'html.parser')
for link in bs.find_all('a'):
if 'href' in link.attrs:
print(link.attrs['href'])
```
## Retrieving Articles Only
```
from urllib.request import urlopen
from bs4 import BeautifulSoup
import re
html = urlopen('http://en.wikipedia.org/wiki/Kevin_Bacon')
bs = BeautifulSoup(html, 'html.parser')
for link in bs.find('div', {'id':'bodyContent'}).find_all(
'a', href=re.compile('^(/wiki/)((?!:).)*$')):
if 'href' in link.attrs:
print(link.attrs['href'])
```
## Random Walk
```
from urllib.request import urlopen
from bs4 import BeautifulSoup
import datetime
import random
import re
random.seed(datetime.datetime.now())
def getLinks(articleUrl):
html = urlopen('http://en.wikipedia.org{}'.format(articleUrl))
bs = BeautifulSoup(html, 'html.parser')
return bs.find('div', {'id':'bodyContent'}).find_all('a', href=re.compile('^(/wiki/)((?!:).)*$'))
links = getLinks('/wiki/Kevin_Bacon')
while len(links) > 0:
newArticle = links[random.randint(0, len(links)-1)].attrs['href']
print(newArticle)
links = getLinks(newArticle)
```
## Recursively crawling an entire site
```
from urllib.request import urlopen
from bs4 import BeautifulSoup
import re
pages = set()
def getLinks(pageUrl):
global pages
html = urlopen('http://en.wikipedia.org{}'.format(pageUrl))
bs = BeautifulSoup(html, 'html.parser')
for link in bs.find_all('a', href=re.compile('^(/wiki/)')):
if 'href' in link.attrs:
if link.attrs['href'] not in pages:
#We have encountered a new page
newPage = link.attrs['href']
print(newPage)
pages.add(newPage)
getLinks(newPage)
getLinks('')
```
## Collecting Data Across an Entire Site
```
from urllib.request import urlopen
from bs4 import BeautifulSoup
import re
pages = set()
def getLinks(pageUrl):
global pages
html = urlopen('http://en.wikipedia.org{}'.format(pageUrl))
bs = BeautifulSoup(html, 'html.parser')
try:
print(bs.h1.get_text())
print(bs.find(id ='mw-content-text').find_all('p')[0])
print(bs.find(id='ca-edit').find('span').find('a').attrs['href'])
except AttributeError:
print('This page is missing something! Continuing.')
for link in bs.find_all('a', href=re.compile('^(/wiki/)')):
if 'href' in link.attrs:
if link.attrs['href'] not in pages:
#We have encountered a new page
newPage = link.attrs['href']
print('-'*20)
print(newPage)
pages.add(newPage)
getLinks(newPage)
getLinks('')
```
## Crawling across the Internet
```
from urllib.request import urlopen
from urllib.parse import urlparse
from bs4 import BeautifulSoup
import re
import datetime
import random
pages = set()
random.seed(datetime.datetime.now())
#Retrieves a list of all Internal links found on a page
def getInternalLinks(bs, includeUrl):
includeUrl = '{}://{}'.format(urlparse(includeUrl).scheme, urlparse(includeUrl).netloc)
internalLinks = []
#Finds all links that begin with a "/"
for link in bs.find_all('a', href=re.compile('^(/|.*'+includeUrl+')')):
if link.attrs['href'] is not None:
if link.attrs['href'] not in internalLinks:
if(link.attrs['href'].startswith('/')):
internalLinks.append(includeUrl+link.attrs['href'])
else:
internalLinks.append(link.attrs['href'])
return internalLinks
#Retrieves a list of all external links found on a page
def getExternalLinks(bs, excludeUrl):
externalLinks = []
#Finds all links that start with "http" that do
#not contain the current URL
for link in bs.find_all('a', href=re.compile('^(http|www)((?!'+excludeUrl+').)*$')):
if link.attrs['href'] is not None:
if link.attrs['href'] not in externalLinks:
externalLinks.append(link.attrs['href'])
return externalLinks
def getRandomExternalLink(startingPage):
html = urlopen(startingPage)
bs = BeautifulSoup(html, 'html.parser')
externalLinks = getExternalLinks(bs, urlparse(startingPage).netloc)
if len(externalLinks) == 0:
print('No external links, looking around the site for one')
domain = '{}://{}'.format(urlparse(startingPage).scheme, urlparse(startingPage).netloc)
internalLinks = getInternalLinks(bs, domain)
return getRandomExternalLink(internalLinks[random.randint(0,
len(internalLinks)-1)])
else:
return externalLinks[random.randint(0, len(externalLinks)-1)]
def followExternalOnly(startingSite):
externalLink = getRandomExternalLink(startingSite)
print('Random external link is: {}'.format(externalLink))
followExternalOnly(externalLink)
followExternalOnly('http://oreilly.com')
```
## Collect all External Links from a Site
```
# Collects a list of all external URLs found on the site
allExtLinks = set()
allIntLinks = set()
def getAllExternalLinks(siteUrl):
html = urlopen(siteUrl)
domain = '{}://{}'.format(urlparse(siteUrl).scheme,
urlparse(siteUrl).netloc)
bs = BeautifulSoup(html, 'html.parser')
internalLinks = getInternalLinks(bs, domain)
externalLinks = getExternalLinks(bs, domain)
for link in externalLinks:
if link not in allExtLinks:
allExtLinks.add(link)
print(link)
for link in internalLinks:
if link not in allIntLinks:
allIntLinks.add(link)
getAllExternalLinks(link)
allIntLinks.add('http://oreilly.com')
getAllExternalLinks('http://oreilly.com')
```
|
github_jupyter
|
```
import hail as hl
```
# *set up dataset*
```
# read in the dataset Zan produced
# metadata from Alicia + sample QC metadata from Julia + densified mt from Konrad
# no samples or variants removed yet
mt = hl.read_matrix_table('gs://african-seq-data/hgdp_tgp/hgdp_tgp_dense_meta_preQC.mt') # 211358784 snps & 4151 samples
# read in variant QC metadata
var_meta = hl.read_table('gs://gcp-public-data--gnomad/release/3.1.1/ht/genomes/gnomad.genomes.v3.1.1.sites.ht')
# annotate variant QC metadata onto mt
mt = mt.annotate_rows(**var_meta[mt.locus, mt.alleles])
# read in the new dataset (including samples that were removed unknowngly)
mt_post = hl.read_matrix_table('gs://african-seq-data/hgdp_tgp/new_hgdp_tgp_postQC.mt') # (155648020, 4099)
```
# *gnomAD filter QC*
```
# editing the format of the filter names and putting them together in a set so that we won't have an issue later when filtering the matrixTable using difference()
# create a set of the gnomAD qc filters (column names under "sample filters") - looks like: {'sex_aneuploidy', 'insert_size', ...} but not in a certain order (randomly ordered)
all_sample_filters = set(mt['sample_filters'])
import re # for renaming purposes
# bad_sample_filters are filters that removed whole populations despite them passing all other gnomAD filters (mostly AFR and OCE popns)
# remove "fail_" from the filter names and pick those out (9 filters) - if the filter name starts with 'fail_' then replace it with ''
bad_sample_filters = {re.sub('fail_', '', x) for x in all_sample_filters if x.startswith('fail_')}
# this filters to only samples that passed all gnomad QC or only failed filters in bad_sample_filters
# 'qc_metrics_filters' is under 'sample_filters' and includes a set of all qc filters a particular sample failed
# if a sample passed all gnomAD qc filters then the column entry for that sample under 'qc_metrics_filters' is an empty set
# so this line goes through the 'qc_metrics_filters'column and sees if there are any samples that passed all the other qc filters except for the ones in the "bad_sample_filters" set (difference())
# if a sample has an empty set for the 'qc_metrics_filters' column or if it only failed the filters that are found in the bad_sample_filters set, then a value of zero is returned and we would keep that sample
# if a sample failed any filters that are not in the "bad_sample_filters" set, remove it
# same as gs://african-seq-data/hgdp_tgp/hgdp_tgp_dense_meta_filt.mt - 211358784 snps & 4120 samples
mt_filt = mt.filter_cols(mt['sample_filters']['qc_metrics_filters'].difference(bad_sample_filters).length() == 0)
# How many samples were removed by the initial QC?
print('Num of samples before initial QC = ' + str(mt.count()[1])) # 4151
print('Num of samples after initial QC = ' + str(mt_filt.count()[1])) # 4120
print('Samples removed = ' + str(mt.count()[1] - mt_filt.count()[1])) # 31
```
# *remove duplicate sample*
```
# duplicate sample - NA06985
mt_filt = mt_filt.distinct_by_col()
print('Num of samples after removal of duplicate sample = ' + str(mt_filt.count()[1])) # 4119
```
# *keep only PASS variants*
```
# subset to only PASS variants (those which passed variant QC) ~6min to run
mt_filt = mt_filt.filter_rows(hl.len(mt_filt.filters) !=0, keep=False)
print('Num of only PASS variants = ' + str(mt_filt.count()[0])) # 155648020
```
# *variant filter and ld pruning*
```
# run common variant statistics (quality control metrics) - more info https://hail.is/docs/0.2/methods/genetics.html#hail.methods.variant_qc
mt_var = hl.variant_qc(mt_filt)
# trying to get down to ~100-300k SNPs - might need to change values later accordingly
# AF: allele freq and call_rate: fraction of calls neither missing nor filtered
# mt.variant_qc.AF[0] is referring to the first element of the list under that column field
mt_var_filt = mt_var.filter_rows((mt_var.variant_qc.AF[0] > 0.05) & (mt_var.variant_qc.AF[0] < 0.95) & (mt_var.variant_qc.call_rate > 0.999))
# ~20min to run
mt_var_filt.count() # started with 155648020 snps and ended up with 6787034 snps
# LD pruning (~113 min to run)
pruned = hl.ld_prune(mt_var_filt.GT, r2=0.1, bp_window_size=500000)
# subset data even further
mt_var_pru_filt = mt_var_filt.filter_rows(hl.is_defined(pruned[mt_var_filt.row_key]))
# write out the output as a temp file - make sure to save the file on this step b/c the pruning step takes a while to run
# saving took ~23 min
mt_var_pru_filt.write('gs://african-seq-data/hgdp_tgp/filtered_n_pruned_output_updated.mt', overwrite=False)
# after saving the pruned file to the cloud, reading it back in for the next steps
mt_var_pru_filt = hl.read_matrix_table('gs://african-seq-data/hgdp_tgp/filtered_n_pruned_output_updated.mt')
# how many snps are left after filtering and prunning?
mt_var_pru_filt.count() # 248,634 snps
# between ~100-300k so we proceed without any value adjustments
```
# *run pc_relate*
```
# compute relatedness estimates between individuals using a variant of the PC-Relate method (https://hail.is/docs/0.2/methods/relatedness.html#hail.methods.pc_relate)
# only compute the kinship statistic using:
# a minimum minor allele frequency filter of 0.05,
# excluding sample-pairs with kinship less than 0.05, and
# 20 principal components to control for population structure
# a hail table is produced (~4min to run)
relatedness_ht = hl.pc_relate(mt_var_pru_filt.GT, min_individual_maf=0.05, min_kinship=0.05, statistics='kin', k=20).key_by()
# write out result - for Julia (~2hr 19min to run)
# includes i – first sample, j – second sample, and kin – kinship estimate
relatedness_ht.write('gs://african-seq-data/hgdp_tgp/relatedness.ht')
# read back in
relatedness_ht = hl.read_table('gs://african-seq-data/hgdp_tgp/relatedness.ht')
# identify related individuals in pairs to remove - returns a list of sample IDs (~2hr & 22 min to run) - previous one took ~13min
related_samples_to_remove = hl.maximal_independent_set(relatedness_ht.i, relatedness_ht.j, False)
# unkey table for exporting purposes - for Julia
unkeyed_tbl = related_samples_to_remove.expand_types()
# export sample IDs of related individuals
unkeyed_tbl.node.s.export('gs://african-seq-data/hgdp_tgp/related_sample_ids.txt', header=False)
# import back to see if format is correct
#tbl = hl.import_table('gs://african-seq-data/hgdp_tgp/related_sample_ids.txt', impute=True, no_header=True)
#tbl.show()
# using sample IDs (col_key of the matrixTable), pick out the samples that are not found in 'related_samples_to_remove' (had 'False' values for the comparison)
# subset the mt to those only
mt_unrel = mt_var_pru_filt.filter_cols(hl.is_defined(related_samples_to_remove[mt_var_pru_filt.col_key]), keep=False)
# do the same as above but this time for the samples with 'True' values (found in 'related_samples_to_remove')
mt_rel = mt_var_pru_filt.filter_cols(hl.is_defined(related_samples_to_remove[mt_var_pru_filt.col_key]), keep=True)
# write out mts of unrelated and related samples on to the cloud
# unrelated mt
mt_unrel.write('gs://african-seq-data/hgdp_tgp/unrel_updated.mt', overwrite=False)
# related mt
mt_rel.write('gs://african-seq-data/hgdp_tgp/rel_updated.mt', overwrite=False)
# read saved mts back in
# unrelated mt
mt_unrel = hl.read_matrix_table('gs://african-seq-data/hgdp_tgp/unrel_updated.mt')
# related mt
mt_rel = hl.read_matrix_table('gs://african-seq-data/hgdp_tgp/rel_updated.mt')
```
# PCA
# *run pca*
```
def run_pca(mt: hl.MatrixTable, reg_name:str, out_prefix: str, overwrite: bool = False):
"""
Runs PCA on a dataset
:param mt: dataset to run PCA on
:param reg_name: region name for saving output purposes
:param out_prefix: path for where to save the outputs
:return:
"""
pca_evals, pca_scores, pca_loadings = hl.hwe_normalized_pca(mt.GT, k=20, compute_loadings=True)
pca_mt = mt.annotate_rows(pca_af=hl.agg.mean(mt.GT.n_alt_alleles()) / 2)
pca_loadings = pca_loadings.annotate(pca_af=pca_mt.rows()[pca_loadings.key].pca_af)
pca_scores = pca_scores.transmute(**{f'PC{i}': pca_scores.scores[i - 1] for i in range(1, 21)})
pca_scores.export(out_prefix + reg_name + '_scores.txt.bgz') # save individual-level genetic region PCs
pca_loadings.write(out_prefix + reg_name + '_loadings.ht', overwrite) # save PCA loadings
```
# *project related individuals*
```
#if running on GCP, need to add "--packages gnomad" when starting a cluster in order for the import to work
from gnomad.sample_qc.ancestry import *
def project_individuals(pca_loadings, project_mt, reg_name:str, out_prefix: str, overwrite: bool = False):
"""
Project samples into predefined PCA space
:param pca_loadings: existing PCA space - unrelated samples
:param project_mt: matrixTable of data to project - related samples
:param reg_name: region name for saving output purposes
:param project_prefix: path for where to save PCA projection outputs
:return:
"""
ht_projections = pc_project(project_mt, pca_loadings)
ht_projections = ht_projections.transmute(**{f'PC{i}': ht_projections.scores[i - 1] for i in range(1, 21)})
ht_projections.export(out_prefix + reg_name + '_projected_scores.txt.bgz') # save output
#return ht_projections # return to user
```
# *global pca*
```
# run 'run_pca' function for global pca
run_pca(mt_unrel, 'global', 'gs://african-seq-data/hgdp_tgp/pca_preoutlier/', False)
# run 'project_relateds' function for global pca
loadings = hl.read_table('gs://african-seq-data/hgdp_tgp/pca_preoutlier/global_loadings.ht') # read in the PCA loadings that were obtained from 'run_pca' function
project_individuals(loadings, mt_rel, 'global', 'gs://african-seq-data/hgdp_tgp/pca_preoutlier/', False)
```
# *subcontinental pca*
```
# obtain a list of the genetic regions in the dataset - used the unrelated dataset since it had more samples
regions = mt_unrel['hgdp_tgp_meta']['Genetic']['region'].collect()
regions = list(dict.fromkeys(regions)) # 7 regions - ['EUR', 'AFR', 'AMR', 'EAS', 'CSA', 'OCE', 'MID']
# set argument values
subcont_pca_prefix = 'gs://african-seq-data/hgdp_tgp/pca_preoutlier/subcont_pca/subcont_pca_' # path for outputs
overwrite = False
# run 'run_pca' function for each region - nb freezes after printing the log for AMR
# don't restart it - just let it run and you can follow the progress through the SparkUI
# even after all the outputs are produced and the run is complete, the code chunk will seem as if it's still running (* in the left square bracket)
# can check if the run is complete by either checking the output files in the Google cloud bucket or using the SparkUI
# after checking the desired outputs are generated and the run is done, exit the current nb, open a new session, and proceed to the next step
# ~27min to run
for i in regions:
subcont_unrel = mt_unrel.filter_cols(mt_unrel['hgdp_tgp_meta']['Genetic']['region'] == i) # filter the unrelateds per region
run_pca(subcont_unrel, i, subcont_pca_prefix, overwrite)
# run 'project_relateds' function for each region (~2min to run)
for i in regions:
loadings = hl.read_table(subcont_pca_prefix + i + '_loadings.ht') # for each region, read in the PCA loadings that were obtained from 'run_pca' function
subcont_rel = mt_rel.filter_cols(mt_rel['hgdp_tgp_meta']['Genetic']['region'] == i) # filter the relateds per region
project_individuals(loadings, subcont_rel, i, subcont_pca_prefix, overwrite)
```
# *outlier removal*
#### After plotting the PCs, 22 outliers that need to be removed were identified (the table below will be completed for the final report)
| s | Genetic region | Population | Note |
| --- | --- | --- | -- |
| NA20314 | AFR | ASW | Clusters with AMR in global PCA |
| NA20299 | - | - | - |
| NA20274 | - | - | - |
| HG01880 | - | - | - |
| HG01881 | - | - | - |
| HG01628 | - | - | - |
| HG01629 | - | - | - |
| HG01630 | - | - | - |
| HG01694 | - | - | - |
| HG01696 | - | - | - |
| HGDP00013 | - | - | - |
| HGDP00150 | - | - | - |
| HGDP00029 | - | - | - |
| HGDP01298 | - | - | - |
| HGDP00130 | CSA | Makrani | Closer to AFR than most CSA |
| HGDP01303 | - | - | - |
| HGDP01300 | - | - | - |
| HGDP00621 | MID | Bedouin | Closer to AFR than most MID |
| HGDP01270 | MID | Mozabite | Closer to AFR than most MID |
| HGDP01271 | MID | Mozabite | Closer to AFR than most MID |
| HGDP00057 | - | - | - |
| LP6005443-DNA_B02 | - | - | - |
```
# read back in the unrelated and related mts to remove outliers and run pca
mt_unrel_unfiltered = hl.read_matrix_table('gs://african-seq-data/hgdp_tgp/unrel_updated.mt') # unrelated mt
mt_rel_unfiltered = hl.read_matrix_table('gs://african-seq-data/hgdp_tgp/rel_updated.mt') # related mt
# read the outliers file into a list
with hl.utils.hadoop_open('gs://african-seq-data/hgdp_tgp/pca_outliers_v2.txt') as file:
outliers = [line.rstrip('\n') for line in file]
# capture and broadcast the list as an expression
outliers_list = hl.literal(outliers)
# remove 22 outliers
mt_unrel = mt_unrel_unfiltered.filter_cols(~outliers_list.contains(mt_unrel_unfiltered['s']))
mt_rel = mt_rel_unfiltered.filter_cols(~outliers_list.contains(mt_rel_unfiltered['s']))
# sanity check
print('Unrelated: Before filtering ' + str(mt_unrel_unfiltered.count()[1]) + ' | After filtering ' + str(mt_unrel.count()[1]))
print('Related: Before filtering: ' + str(mt_rel_unfiltered.count()[1]) + ' | After filtering ' + str(mt_rel.count()[1]))
num_outliers = (mt_unrel_unfiltered.count()[1] - mt_unrel.count()[1]) + (mt_rel_unfiltered.count()[1] - mt_rel.count()[1])
print('Total samples removed = ' + str(num_outliers))
```
# rerun PCA
### - The following steps are similar to the ones prior to removing the outliers except now we are using the updated unrelated & related dataset and a new GCS bucket path to save the outputs
# *global pca*
```
# run 'run_pca' function for global pca - make sure the code block for the function (located above) is run prior to running this
run_pca(mt_unrel, 'global', 'gs://african-seq-data/hgdp_tgp/pca_postoutlier/', False)
# run 'project_relateds' function for global pca - make sure the code block for the function (located above) is run prior to running this
loadings = hl.read_table('gs://african-seq-data/hgdp_tgp/pca_postoutlier/global_loadings.ht') # read in the PCA loadings that were obtained from 'run_pca' function
project_individuals(loadings, mt_rel, 'global', 'gs://african-seq-data/hgdp_tgp/pca_postoutlier/', False)
```
# *subcontinental pca*
```
# obtain a list of the genetic regions in the dataset - used the unrelated dataset since it had more samples
regions = mt_unrel['hgdp_tgp_meta']['Genetic']['region'].collect()
regions = list(dict.fromkeys(regions)) # 7 regions - ['EUR', 'AFR', 'AMR', 'EAS', 'CSA', 'OCE', 'MID']
# set argument values
subcont_pca_prefix = 'gs://african-seq-data/hgdp_tgp/pca_postoutlier/subcont_pca/subcont_pca_' # path for outputs
overwrite = False
# run 'run_pca' function (located above) for each region
# notebook became slow and got stuck - don't restart it, just let it run and you can follow the progress through the SparkUI
# after checking the desired outputs are generated (GCS bucket) and the run is done (SparkUI), exit the current nb, open a new session, and proceed to the next step
# took roughly 25-27 min
for i in regions:
subcont_unrel = mt_unrel.filter_cols(mt_unrel['hgdp_tgp_meta']['Genetic']['region'] == i) # filter the unrelateds per region
run_pca(subcont_unrel, i, subcont_pca_prefix, overwrite)
# run 'project_relateds' function (located above) for each region - took ~3min
for i in regions:
loadings = hl.read_table(subcont_pca_prefix + i + '_loadings.ht') # for each region, read in the PCA loadings that were obtained from 'run_pca' function
subcont_rel = mt_rel.filter_cols(mt_rel['hgdp_tgp_meta']['Genetic']['region'] == i) # filter the relateds per region
project_individuals(loadings, subcont_rel, i, subcont_pca_prefix, overwrite)
```
# FST
### For FST, we are using the data we had prior to running pc_relate (*filtered_n_pruned_output_updated.mt*)
```
# read filtered and pruned mt (prior to pc_relate) back in for FST analysis
mt_var_pru_filt = hl.read_matrix_table('gs://african-seq-data/hgdp_tgp/filtered_n_pruned_output_updated.mt')
# num of samples before outlier removal
print('Before filtering: ' + str(mt_var_pru_filt.count()[1]))
# read the outliers file into a list
with hl.utils.hadoop_open('gs://african-seq-data/hgdp_tgp/pca_outliers_v2.txt') as file:
outliers = [line.rstrip('\n') for line in file]
# capture and broadcast the list as an expression
outliers_list = hl.literal(outliers)
# remove 22 outliers
mt_var_pru_filt = mt_var_pru_filt.filter_cols(~outliers_list.contains(mt_var_pru_filt['s']))
# sanity check
print('After filtering: ' + str(mt_var_pru_filt.count()[1]))
```
## *pair-wise comparison*
Formula to calculate number of pair-wise comparisons = (k * (k-1))/2
So in our case, since we have 78 populations, we would expect = (78 * (78-1))/2 = 6006/2 = 3003 pair-wise comparisons
```
pop = mt_var_pru_filt['hgdp_tgp_meta']['Population'].collect()
pop = list(dict.fromkeys(pop))
len(pop) # 78 populations in total
# example
ex = ['a','b','c']
# pair-wise comparison
ex_pair_com = [[x,y] for i, x in enumerate(ex) for j,y in enumerate(ex) if i<j]
ex_pair_com
# pair-wise comparison - creating list of lists
# enumerate gives index values for each population in the 'pop' list (ex. 0 CEU, 1 YRI, 2 LWK ...) and then by
# comparing those index values, we create a pair-wise comparison between the populations
# i < j so that it only does a single comparison among two different populations
# ex. for a comparison between populations CEU and YRI, it only keeps CEU-YRI and discards YRI-CEU, CEU-CEU and YRI-YRI
pair_com = [[x,y] for i, x in enumerate(pop) for j,y in enumerate(pop) if i<j]
# first 5 elements in the list
pair_com[0:5]
# sanity check
len(pair_com)
```
## *subset mt into popns according to the pair-wise comparisons and run common variant statistics*
```
pair_com[0]
## example - pair_com[0] = ['CEU', 'YRI'] and pair_com[0][0] = 'CEU'
CEU_mt = mt_var_pru_filt.filter_cols(mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pair_com[0][0])
YRI_mt = mt_var_pru_filt.filter_cols(mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pair_com[0][1])
CEU_YRI_mt = mt_var_pru_filt.filter_cols((mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pair_com[0][0]) | (mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pair_com[0][1]))
# sanity check
CEU_mt.count()[1] + YRI_mt.count()[1] == CEU_YRI_mt.count()[1] # 175 + 170 = 345
# run common variant statistics for each population and their combined mt
CEU_var = hl.variant_qc(CEU_mt) # individual
YRI_var = hl.variant_qc(YRI_mt) # individual
CEU_YRI_var = hl.variant_qc(CEU_YRI_mt) # total
```
### *Set up mt table for FST calculation - the next code is run for each population and their combos*
##### *population 1*
```
# drop certain fields first to make mt smaller
# drop all entry fields
# everything except for 's' (key) from the column fields
# everything from the row fields except for the keys -'locus' and 'alleles' and row field 'variant_qc'
CEU_interm = CEU_var.drop(*list(CEU_var.entry), *list(CEU_var.col)[1:], *list(CEU_var.row)[2:-1])
# only select the row field keys (locus and allele) and row fields 'AF' & 'AN' which are under 'variant_qc'
CEU_interm2 = CEU_interm.select_rows(CEU_interm['variant_qc']['AF'], CEU_interm['variant_qc']['AN'])
# quick look at the condensed mt
CEU_interm2.describe()
CEU_interm2.rows().show(5)
# only include the second entry of the array from the row field 'AF'
CEU_interm3 = CEU_interm2.transmute_rows(AF = CEU_interm2.AF[1])
# previous code
# key the rows only by 'locus' so that the 'allele' row field can be split into two row fields (one for each allele)
# also, only include the second entry of the array from 'AF' row field
#CEU_interm3 = CEU_interm2.key_rows_by('locus')
#CEU_interm3 = CEU_interm3.transmute_rows(AF = CEU_interm3.AF[1], A1 = CEU_interm3.alleles[0], A2 = CEU_interm3.alleles[1])
# add a row field with population name to keep track of which mt it came from
CEU_final = CEU_interm3.annotate_rows(pop = pair_com[0][0])
CEU_final.rows().show(5)
```
##### *population 2*
```
# drop fields
# drop all entry fields
# everything except for 's' (key) from the column fields
# everything from the row fields except for the keys -'locus' and 'alleles' and row field 'variant_qc'
CEU_YRI_interm = CEU_YRI_var.drop(*list(CEU_YRI_var.entry), *list(CEU_YRI_var.col)[1:], *list(CEU_YRI_var.row)[2:-1])
# only select the row field keys (locus and allele) and row fields 'AF' & 'AN' which are under 'variant_qc'
CEU_YRI_interm2 = CEU_YRI_interm.select_rows(CEU_YRI_interm['variant_qc']['AF'], CEU_YRI_interm['variant_qc']['AN'])
# quick look at the condensed mt
CEU_YRI_interm2.describe()
CEU_YRI_interm2.rows().show(5)
# only include the second entry of the array from the row field 'AF'
CEU_YRI_interm3 = CEU_YRI_interm2.transmute_rows(AF = CEU_YRI_interm2.AF[1])
# previous code
# key the rows only by 'locus' so that the 'allele' row field can be split into two row fields (one for each allele)
# also, only include the second entry of the array from 'AF' row field
#CEU_YRI_interm3 = CEU_YRI_interm2.key_rows_by('locus')
#CEU_YRI_interm3 = CEU_YRI_interm3.transmute_rows(AF = CEU_YRI_interm3.AF[1], A1 = CEU_YRI_interm3.alleles[0], A2 = CEU_YRI_interm3.alleles[1])
# add a row field with population name to keep track of which mt it came from
CEU_YRI_final = CEU_YRI_interm3.annotate_rows(pop = f'{pair_com[0][0]}-{pair_com[0][1]}')
CEU_YRI_final.rows().show(5)
```
### *FST formula pre-setup* - trial run
#### *Variables needed for FST calculation*
```
# converting lists into numpy arrarys cause it is easier to work with and more readable
# assign populations to formula variables
pop1 = CEU_final
pop2 = CEU_YRI_final
# number of alleles
n1 = np.array(pop1.AN.collect())
n2 = np.array(pop2.AN.collect())
# allele frequencies
FREQpop1 = np.array(pop1.AF.collect())
FREQpop2 = np.array(pop2.AF.collect())
```
#### *Weighted average allele frequency*
```
FREQ = ((n1*FREQpop1) + (n2*FREQpop2)) / (n1+n2)
# sanity checks
print(((n1[0]*FREQpop1[0]) + (n2[0]*FREQpop2[0])) / (n1[0]+n2[0]) == FREQ[0])
print(len(FREQ) == len(FREQpop1)) # length of output should be equal to the length of arrays we started with
```
#### *Filter to only freqs between 0 and 1*
```
INCLUDE=(FREQ>0) & (FREQ<1) # only include ave freq between 0 and 1 - started with FREQ = 248634
print(np.count_nonzero(INCLUDE)) # 246984 ave freq values were between 0 and 1 - returned True to the conditions above; 248634 - 246984 = 1650 were False
# subset allele frequencies
FREQpop1=FREQpop1[INCLUDE]
FREQpop2=FREQpop2[INCLUDE]
FREQ=FREQ[INCLUDE]
# sanity check
print(len(FREQpop1) == np.count_nonzero(INCLUDE)) # TRUE
# subset the number of alleles
n1 = n1[INCLUDE]
n2 = n2[INCLUDE]
# sanity check
print(len(n1) == np.count_nonzero(INCLUDE)) # TRUE
```
#### *FST Estimate - W&C ESTIMATOR*
```
## average sample size that incorporates variance
nc =((1/(s-1)) * (n1+n2)) - ((np.square(n1) + np.square(n2))/(n1+n2))
msa= (1/(s-1))*((n1*(np.square(FREQpop1-FREQ)))+(n2*(np.square(FREQpop2-FREQ))))
msw = (1/((n1-1)+(n2-1))) * ((n1*(FREQpop1*(1-FREQpop1))) + (n2*(FREQpop2*(1-FREQpop2))))
numer = msa-msw
denom = msa + ((nc-1)*msw)
FST_val = numer/denom
# sanity check using the first element
nc_0 =((1/(s-1)) * (n1[0]+n2[0])) - ((np.square(n1[0]) + np.square(n2[0]))/(n1[0]+n2[0]))
msa_0= (1/(s-1))*((n1[0]*(np.square(FREQpop1[0]-FREQ[0])))+(n2[0]*(np.square(FREQpop2[0]-FREQ[0]))))
msw_0 = (1/((n1[0]-1)+(n2[0]-1))) * ((n1[0]*(FREQpop1[0]*(1-FREQpop1[0]))) + (n2[0]*(FREQpop2[0]*(1-FREQpop2[0]))))
numer_0 = msa_0-msw_0
denom_0 = msa_0 + ((nc_0-1)*msw_0)
FST_0 = numer_0/denom_0
print(FST_0 == FST_val[0]) # TRUE
FST_val
```
## *Which FST value is for which locus-allele?* - actual run
```
# resetting variables for the actual FST run
# assign populations to formula variables
pop1 = CEU_final
pop2 = CEU_YRI_final
# number of alleles
n1 = np.array(pop1.AN.collect())
n2 = np.array(pop2.AN.collect())
# allele frequencies
FREQpop1 = np.array(pop1.AF.collect())
FREQpop2 = np.array(pop2.AF.collect())
# locus + alleles = keys - needed for reference purposes - these values are uniform across all populations
locus = np.array(hl.str(pop1.locus).collect())
alleles = np.array(hl.str(pop1.alleles).collect())
key = np.array([i + ' ' + j for i, j in zip(locus, alleles)])
s=2 # s is the number of populations - since we are calculating pair-wise FSTs, this is always 2
key_FST = {}
for i in range(len(key)):
FREQ = ((n1[i]*FREQpop1[i]) + (n2[i]*FREQpop2[i])) / (n1[i]+n2[i])
if (FREQ>0) & (FREQ<1): # only include ave freq between 0 and 1
## average sample size that incorporates variance
nc = ((1/(s-1)) * (n1[i]+n2[i])) - ((np.square(n1[i]) + np.square(n2[i]))/(n1[i]+n2[i]))
msa= (1/(s-1))*((n1[i]*(np.square(FREQpop1[i]-FREQ)))+(n2[i]*(np.square(FREQpop2[i]-FREQ))))
msw = (1/((n1[i]-1)+(n2[i]-1))) * ((n1[i]*(FREQpop1[i]*(1-FREQpop1[i]))) + (n2[i]*(FREQpop2[i]*(1-FREQpop2[i]))))
numer = msa-msw
denom = msa + ((nc-1)*msw)
FST = numer/denom
key_FST[key[i]] = FST
key_FST
# sanity checks
print(all(np.array(list(key_FST.values())) == FST_val)) # True
print(len(key_FST) == len(FST_val)) # True
```
## *other pair*
### population 3
```
# population - YRI
# same steps we did to CEU
YRI_interm = YRI_var.drop(*list(YRI_var.entry), *list(YRI_var.col)[1:], *list(YRI_var.row)[2:-1])
# only select the row field keys (locus and allele) and row fields 'AF' & 'AN' which are under 'variant_qc'
YRI_interm2 = YRI_interm.select_rows(YRI_interm['variant_qc']['AF'], YRI_interm['variant_qc']['AN'])
# only include the second entry of the array from the row field 'AF'
YRI_interm3 = YRI_interm2.transmute_rows(AF = YRI_interm2.AF[1])
# add a row field with population name to keep track of which mt it came from
YRI_final = YRI_interm3.annotate_rows(pop = pair_com[0][1])
YRI_final.rows().show(5)
```
### *FST*
```
# resetting variables for the actual FST run
# assign populations to formula variables
pop1 = YRI_final
pop2 = CEU_YRI_final
# number of alleles
n1 = np.array(pop1.AN.collect())
n2 = np.array(pop2.AN.collect())
# allele frequencies
FREQpop1 = np.array(pop1.AF.collect())
FREQpop2 = np.array(pop2.AF.collect())
# locus + alleles = keys - needed for reference purposes - these values are uniform across all populations
locus = np.array(hl.str(pop1.locus).collect())
alleles = np.array(hl.str(pop1.alleles).collect())
key = np.array([i + ' ' + j for i, j in zip(locus, alleles)])
s=2 # s is the number of populations - since we are calculating pair-wise FSTs, this is always 2
key_FST_YRI = {}
for i in range(len(key)):
FREQ = ((n1[i]*FREQpop1[i]) + (n2[i]*FREQpop2[i])) / (n1[i]+n2[i])
if (FREQ>0) & (FREQ<1): # only include ave freq between 0 and 1
## average sample size that incorporates variance
nc = ((1/(s-1)) * (n1[i]+n2[i])) - ((np.square(n1[i]) + np.square(n2[i]))/(n1[i]+n2[i]))
msa= (1/(s-1))*((n1[i]*(np.square(FREQpop1[i]-FREQ)))+(n2[i]*(np.square(FREQpop2[i]-FREQ))))
msw = (1/((n1[i]-1)+(n2[i]-1))) * ((n1[i]*(FREQpop1[i]*(1-FREQpop1[i]))) + (n2[i]*(FREQpop2[i]*(1-FREQpop2[i]))))
numer = msa-msw
denom = msa + ((nc-1)*msw)
FST = numer/denom
key_FST_YRI[key[i]] = FST
CEU
YRI
key_FST_YRI
```
## *three popn pairs*
```
## example using three sample pairs ['CEU', 'YRI'], ['CEU', 'LWK'], ['CEU', 'ESN'] and setting up the function
example_pairs = pair_com[0:3]
ex_dict = {} # empty dictionary to hold final outputs
for pairs in example_pairs:
l = [] # empty list to hold the subsetted datasets
l.append(mt_var_pru_filt.filter_cols(mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pairs[0])) # first population
l.append(mt_var_pru_filt.filter_cols(mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pairs[1])) # second population
l.append(mt_var_pru_filt.filter_cols((mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pairs[0]) | (mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pairs[1]))) # first + second = total population
# sanity check - the sample count of the first and second subset mts should be equal to the total subset mt
if l[0].count()[1] + l[1].count()[1] == l[2].count()[1]:
v = [] # empty list to hold output mts from running common variant statistics
# run common variant statistics for each population and their combined mt
v.append(hl.variant_qc(l[0])) # first population
v.append(hl.variant_qc(l[1])) # second population
v.append(hl.variant_qc(l[2])) # both/total population
# add to dictionary
ex_dict["-".join(pairs)] = v
# three mt subsets per comparison pair - set up as a dictionary
ex_dict
# population - YRI
# same steps we did to CEU
YRI_var == ex_dict['CEU-YRI'][0]
YRI_interm = ex_dict['CEU-YRI'][0].drop(*list(ex_dict['CEU-YRI'][0].entry)
YRI_interm = ex_dict['CEU-YRI'][0].drop(*list(ex_dict['CEU-YRI'][0].entry), *list(ex_dict['CEU-YRI'][0].col)[1:], *list(ex_dict['CEU-YRI'][0].row)[2:-1])
# only select the row field keys (locus and allele) and row fields 'AF' & 'AN' which are under 'variant_qc'
YRI_interm2 = YRI_interm.select_rows(YRI_interm['variant_qc']['AF'], YRI_interm['variant_qc']['AN'])
# only include the second entry of the array from the row field 'AF'
YRI_interm3 = YRI_interm2.transmute_rows(AF = YRI_interm2.AF[1])
# add a row field with population name to keep track of which mt it came from
YRI_final = YRI_interm3.annotate_rows(pop = pairs[0])
YRI_final.rows().show(5)
# same as CEU_var['variant_qc'].show(5)
ex_dict['CEU-YRI'][0]['variant_qc'].show(5)
len(ex_dict['CEU-YRI'])
a = ['CEU-YRI','CEU-LWK', 'CEU-ESN']
b = [0,1,2]
dc = {}
for i in a:
li = []
for j in b:
li.append(str(j) + i)
dc[i] = li
for i in range(len(v)-1):
print(i)
from collections import defaultdict
dd = defaultdict(list)
for d in (key_FST, key_FST_YRI):
print(d)
#for key, value in d.items():
#dd[key].append(value)
range(len(ex_dict[pair]))
final_dic = {}
for pair in ex_dict.keys(): # for each population pair
u = [] # list to hold updated mts
for i in range(len(ex_dict[pair])): # for each population (each mt)
# pop1
# drop certain fields and only keep the ones we need
interm = ex_dict[pair][i].drop(*list(ex_dict[pair][i].entry), *list(ex_dict[pair][i].col)[1:], *list(ex_dict[pair][i].row)[2:-1])
interm2 = interm.select_rows(interm['variant_qc']['AF'], interm['variant_qc']['AN'])
interm3 = interm2.transmute_rows(AF = interm2.AF[1])
#final = interm3.annotate_rows(pop = pair) # keep track of which mt it came from
u.append(interm3) # add updated mt to list
# variables for FST run
# assign populations to formula variables
pop1 = u[0]
pop2 = u[1]
total = u[2]
# number of alleles
n1 = np.array(pop1.AN.collect())
n2 = np.array(pop2.AN.collect())
total_n = np.array(total.AN.collect())
# allele frequencies
FREQpop1 = np.array(pop1.AF.collect())
FREQpop2 = np.array(pop2.AF.collect())
total_FREQ = np.array(total.AF.collect())
# locus + alleles = keys - needed for reference purposes during FST calculations - these values are uniform across all populations
locus = np.array(hl.str(pop1.locus).collect())
alleles = np.array(hl.str(pop1.alleles).collect())
key = np.array([i + ' ' + j for i, j in zip(locus, alleles)])
s=2 # s is the number of populations - since we are calculating pair-wise FSTs, this is always 2
# FST pop1 and total popn
key_pop1_total = {}
for i in range(len(key)):
FREQ = ((n1[i]*FREQpop1[i]) + (total_n[i]*total_FREQ[i])) / (n1[i]+total_n[i])
if (FREQ>0) & (FREQ<1): # only include ave freq between 0 and 1
## average sample size that incorporates variance
nc = ((1/(s-1)) * (n1[i]+total_n[i])) - ((np.square(n1[i]) + np.square(total_n[i]))/(n1[i]+total_n[i]))
msa= (1/(s-1))*((n1[i]*(np.square(FREQpop1[i]-FREQ)))+(total_n[i]*(np.square(total_FREQ[i]-FREQ))))
msw = (1/((n1[i]-1)+(total_n[i]-1))) * ((n1[i]*(FREQpop1[i]*(1-FREQpop1[i]))) + (total_n[i]*(total_FREQ[i]*(1-total_FREQ[i]))))
numer = msa-msw
denom = msa + ((nc-1)*msw)
FST = numer/denom
key_pop1_total[key[i]] = FST
# FST pop2 and total popn
key_pop2_total = {}
for i in range(len(key)):
FREQ = ((n2[i]*FREQpop2[i]) + (total_n[i]*total_FREQ[i])) / (n2[i]+total_n[i])
if (FREQ>0) & (FREQ<1): # only include ave freq between 0 and 1
## average sample size that incorporates variance
nc = ((1/(s-1)) * (n2[i]+total_n[i])) - ((np.square(n2[i]) + np.square(total_n[i]))/(n2[i]+total_n[i]))
msa= (1/(s-1))*((n2[i]*(np.square(FREQpop2[i]-FREQ)))+(total_n[i]*(np.square(total_FREQ[i]-FREQ))))
msw = (1/((n2[i]-1)+(total_n[i]-1))) * ((n2[i]*(FREQpop2[i]*(1-FREQpop2[i]))) + (total_n[i]*(total_FREQ[i]*(1-total_FREQ[i]))))
numer = msa-msw
denom = msa + ((nc-1)*msw)
FST = numer/denom
key_pop2_total[key[i]] = FST
# merge the two FST results together
from collections import defaultdict
dd = defaultdict(list)
for d in (key_pop1_total, key_pop2_total):
for key, value in d.items():
dd[key].append(value)
final_dic[pair] = dd
# convert to a table
import pandas as pd
df = pd.DataFrame(final_dic)
len(final_dic['CEU-YRI']) # 246984
## example - pair_com[0] = ['CEU', 'YRI'] and pair_com[0][0] = 'CEU'
CEU_mt = mt_var_pru_filt.filter_cols(mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pair_com[1][0])
LWK_mt = mt_var_pru_filt.filter_cols(mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pair_com[1][1])
CEU_LWK_mt = mt_var_pru_filt.filter_cols((mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pair_com[1][0]) | (mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pair_com[1][1]))
# run common variant statistics for each population and their combined mt
CEU_var = hl.variant_qc(CEU_mt) # individual
LWK_var = hl.variant_qc(LWK_mt) # individual
CEU_LWK_var = hl.variant_qc(CEU_YRI_mt) # total
LWK_var.count()
# population - YRI
# same steps we did to CEU
YRI_interm = YRI_var.drop(*list(YRI_var.entry), *list(YRI_var.col)[1:], *list(YRI_var.row)[2:-1])
# only select the row field keys (locus and allele) and row fields 'AF' & 'AN' which are under 'variant_qc'
YRI_interm2 = YRI_interm.select_rows(YRI_interm['variant_qc']['AF'], YRI_interm['variant_qc']['AN'])
# only include the second entry of the array from the row field 'AF'
YRI_interm3 = YRI_interm2.transmute_rows(AF = YRI_interm2.AF[1])
# add a row field with population name to keep track of which mt it came from
YRI_final = YRI_interm3.annotate_rows(pop = pair_com[0][1])
YRI_final.rows().show(5)
for pair in ex_dict.keys(): # for each population pair
for i in range(len(ex_dict[i])): # for each population
interm = ex_dict[pair][i].drop(*list(ex_dict[pair][i].entry), *list(ex_dict[pair][i].col)[1:], *list(ex_dict[pair][i].row)[2:-1])
# only select the row field keys (locus and allele) and row fields 'AF' & 'AN' which are under 'variant_qc'
interm2 = interm.select_rows(interm['variant_qc']['AF'], interm['variant_qc']['AN'])
# only include the second entry of the array from the row field 'AF'
interm3 = interm2.transmute_rows(AF = interm2.AF[1])
# add a row field with population name to keep track of which mt it came from
final = interm3.annotate_rows(pop = pair)
final.rows().show(5)
%%time
# actual function/run using all population pairs
dict = {} # empty dictionary to hold final outputs
for pairs in pair_com:
l = [] # empty list to hold the subsetted datasets
l.append(mt_var_pru_filt.filter_cols(mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pairs[0])) # first population
l.append(mt_var_pru_filt.filter_cols(mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pairs[1])) # second population
l.append(mt_var_pru_filt.filter_cols((mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pairs[0]) | (mt_var_pru_filt['hgdp_tgp_meta']['Population'] == pairs[1]))) # first + second = total population
# sanity check - the sample count of the first and second subset mts should be equal to the total subset mt
if l[0].count()[1] + l[1].count()[1] == l[2].count()[1]:
v = [] # empty list to hold output mts from running common variant statistics
# run common variant statistics for each population and their combined mt
v.append(hl.variant_qc(l[0])) # first population
v.append(hl.variant_qc(l[1])) # second population
v.append(hl.variant_qc(l[2])) # both/total population
# add to dictionary
dict["-".join(pairs)] = v
len(dict)
dict
dict['CEU-YRI'][0]['variant_qc'].show(5)
dict['CEU-YRI'][1]['variant_qc'].show(5)
dict['CEU-YRI'][2]['variant_qc'].show(5)
# accessing dictionary element with index
ex_dict[list(ex_dict)[0]][0]['variant_qc'].show(5)
for l in list(ex_dict):
print(ex_dict[l][1]['variant_qc']['AF'][1].show(5))
list(ex_dict)
CEU_af_freq = ex_dict[list(ex_dict)[0]][0]['variant_qc']['AF'][1]
play_mt = hl.utils.range_matrix_table(0, 6)
ex_dict[list(ex_dict)[0]][0].cols().show(5)
mt.select_rows(mt.r1, mt.r2,
r3=hl.coalesce(mt.r1, mt.r2))
mt.select_cols(mt.c2,
sum=mt.c2+mt.c1)
play_mt = ex_dict[list(ex_dict)[0]][0]
row_subsetted_mt.cols().show(5)
CEU_af_freq = CEU_af_freq.annotate_cols(AN=ex_dict[list(ex_dict)[0]][0]['variant_qc']['AN'])
mtA = mtA.annotate_rows(phenos = hl.dict(hl.agg.collect((mtA.pheno, mtA.value))))
mtB = mtB.annotate_cols(
phenos = mtA.rows()[mtB.col_key].phenos)
# additional stuff
CEU_var = hl.variant_qc(CEU_mt)
CEU__YRI_var = hl.variant_qc(CEU_YRI_mt)
a, b, c
a -> ac
B -> cb
a -> ab
CEU_var.row.show()
CEU_mt['variant_qc']['AF'][0]*CEU_mt['variant_qc']['AF'][1]
CEU_YRI_mt['variant_qc']['AF'][0]*CEU_YRI_mt['variant_qc']['AF'][1]
```
# junk code below
```
# this code is if the alleles were split into their separate columns and if we expect a mismatch across popns
# remove indels - only include single letter varients for each allele in both populations
# this is b/c the FST formula is set up for single letter alleles
#pop1 = CEU_final.filter_rows((CEU_final.A1.length() == 1) & (CEU_final.A2.length() == 1))
#pop2 = CEU_YRI_final.filter_rows((CEU_YRI_final.A1.length() == 1) & (CEU_YRI_final.A2.length() == 1))
# sanity check
#A1 = pop1.A1.collect()
#A1 = list(set(A1)) # OR can also do:
### from collections import OrderedDict
### A1 = list(OrderedDict.fromkeys(A1))
#print(A1)
#len(A1) == 4
# total # of snps at the beginning - 255666
# unique snps before removing indels - 2712
# total # of snps after removing indels - 221017 (34649 snps were indels for A1, A2 or both)
# unique snps after removing indels - 4 ['C', 'A', 'T', 'G'] - which is what we expect
## *use the same reference allele - A2 is minor allele here*
# get the minor alleles from both populations
#pop1_A2 = pop1.A2.collect()
#pop2_A2 = pop2.A2.collect()
# find values that are unequal
#import numpy as np
#switch1 = (np.array(pop1_A2) != np.array(pop2_A2))
#print(switch1.all()) # all comparisons returned 'FALSE' which means that all variants that were compared are the same
# sanity check
#print(len(pop1_A2) == len(pop2_A2) == len(switch1)) # True
### *if there is a variant mismatch among the minor alleles of the two populations*
# in case there was a comparison that didn't match correctly among the minor alleles of the two populations, we would adjust the allele frequency(AF) accordingly
#new_frq = pop2.AF.collect()
#new_frq = np.array(new_frq) # convert to numpy array for the next step
# explanation (with an example) for what this does is right below it
#new_frq[switch1] = 1-(new_frq[switch1])
# Example: for pop_1, A1 and A2 are 'T' and 'C' with AF of 0.25
# and for pop_2, A1 and A2 are 'C and 'T' with AF of 0.25
# then since the same reference allele is not used (alleles don't correctly align) in this case,
# we would subtract the AF of pop_2 from 1, to get the correct allele frequency
# the AF of pop_2 with A1 and A2 oriented the same way as pop_1: 'T' and 'C', would be 1-0.25 = 0.75 (w/c is the correct AF)
# if we wanted to convert array back to list
#pop2_frq = new_frq.tolist()
# junk code
#pop2.rows().show(5)
#p = pop2.filter_rows(str(pop2.locus) =='chr10:38960343')
p.row.show()
# for i in locus:
# if i =='chr1:94607079':
# print ("True")
sum(num == dup for num,dup in zip(locus, d))
# code to check if there are duplicates in a list and print them out
#import collections
#dup = [item for item, count in collections.Counter(key).items() if count > 1]
#print('Num of duplicate loci: ' + str(len(dup)))
#print(dup)
# which FST value is for which locus?
key_freq1 = {key[i]: FREQpop1[i] for i in range(len(key))}
key_freq2 = {key[i]: FREQpop2[i] for i in range(len(key))}
key_n1 = {key[i]: n1[i] for i in range(len(key))}
key_n2 = {key[i]: n2[i] for i in range(len(key))}
# for key,value in zip (locus, FREQpop1):
# print(dict(key, value))
#for v1,v2 in zip(list(locus_freq1.values())[0:5], list(locus_freq2.values())[0:5]):
#lq = ((n1*locus_freq1.values()) + (n2*locus_freq2.values())) / (n1+n2)
#print(key,value)
#locus #220945
#len(set(FREQpop1))
# check if there are duplicates in locus list and print them out - 72 duplicates
# import collections
# d = [item for item, count in collections.Counter(locus).items() if count > 1]
# list.sort(locus)
#locus
# from collections import Counter
# [k for k,v in Counter(locus).items() if v>1]
# where are each of the duplicated loci located?
from collections import defaultdict
D = defaultdict(list)
for i,item in enumerate(locus):
D[item].append(i)
D = {k:v for k,v in D.items() if len(v)>1}
locus[6202]
bad_locus = locus[INCLUDE=='FALSE']
# ave freq values that were not between 0 and 1 - returned FALSE to the conditions in the above chuck of code
print(np.count_nonzero(INCLUDE==0))
DONT_INCLUDE= (FREQ=='') & (FREQ>=1)
np.count_nonzero(DONT_INCLUDE)
# convert the output from the preimp_qc module (qced.mt) into a vcf file in Hail
import hail as hl
mt = hl.read_matrix_table('gs://nepal-geno/GWASpy/Preimp_QC/Nepal_PTSD_GSA_Updated_May2021_qced.mt')
hl.export_vcf(mt, 'gs://nepal-geno/Nepal_PTSD_GSA_Updated_May2021_qced.vcf.bgz')
```
|
github_jupyter
|
<a name="top"></a>
<div style="width:1000 px">
<div style="float:right; width:98 px; height:98px;">
<img src="https://raw.githubusercontent.com/Unidata/MetPy/master/metpy/plots/_static/unidata_150x150.png" alt="Unidata Logo" style="height: 98px;">
</div>
<h1>Advanced Pythonic Data Analysis</h1>
<h3>Unidata Python Workshop</h3>
<div style="clear:both"></div>
</div>
<hr style="height:2px;">
<div style="float:right; width:250 px"><img src="http://matplotlib.org/_images/date_demo.png" alt="METAR" style="height: 300px;"></div>
## Overview:
* **Teaching:** 45 minutes
* **Exercises:** 45 minutes
### Questions
1. How can we improve upon the versatility of the plotter developed in the basic time series notebook?
1. How can we iterate over all data file in a directory?
1. How can data processing functions be applied on a variable-by-variable basis?
### Objectives
1. <a href="#basicfunctionality">From Time Series Plotting Episode</a>
1. <a href="#parameterdict">Dictionaries of Parameters</a>
1. <a href="#multipledict">Multiple Dictionaries</a>
1. <a href="#functions">Function Application</a>
1. <a href="#glob">Glob and Multiple Files</a>
<a name="basicfunctionality"></a>
## From Time Series Plotting Episode
Here's the basic set of imports and data reading functionality that we established in the [Basic Time Series Plotting](../Time_Series/Basic%20Time%20Series%20Plotting.ipynb) notebook.
```
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.dates import DateFormatter, DayLocator
from siphon.simplewebservice.ndbc import NDBC
%matplotlib inline
def format_varname(varname):
"""Format the variable name nicely for titles and labels."""
parts = varname.split('_')
title = parts[0].title()
label = varname.replace('_', ' ').title()
return title, label
def read_buoy_data(buoy, days=7):
# Read in some data
df = NDBC.realtime_observations(buoy)
# Trim to the last 7 days
df = df[df['time'] > (pd.Timestamp.utcnow() - pd.Timedelta(days=days))]
return df
```
<a href="#top">Top</a>
<hr style="height:2px;">
<a name="parameterdict"></a>
## Dictionaries of Parameters
When we left off last time, we had created dictionaries that stored line colors and plot properties in a key value pair. To further simplify things, we can actually pass a dictionary of arguements to the plot call. Enter the dictionary of dictionaries. Each key has a value that is a dictionary itself with it's key value pairs being the arguements to each plot call. Notice that different variables can have different arguements!
```
df = read_buoy_data('42039')
# Dictionary of plotting parameters by variable name
styles = {'wind_speed': dict(color='tab:orange'),
'wind_gust': dict(color='tab:olive', linestyle='None', marker='o', markersize=2),
'pressure': dict(color='black')}
plot_variables = [['wind_speed', 'wind_gust'], ['pressure']]
fig, axes = plt.subplots(1, len(plot_variables), sharex=True, figsize=(14, 5))
for col, var_names in enumerate(plot_variables):
ax = axes[col]
for var_name in var_names:
title, label = format_varname(var_name)
ax.plot(df.time, df[var_name], **styles[var_name])
ax.set_ylabel(title)
ax.set_title('Buoy 42039 {}'.format(title))
ax.grid(True)
ax.set_xlabel('Time')
ax.xaxis.set_major_formatter(DateFormatter('%m/%d'))
ax.xaxis.set_major_locator(DayLocator())
```
<a href="#top">Top</a>
<hr style="height:2px;">
<a name="multipledict"></a>
## Multiple Dictionaries
We can even use multiple dictionaries to define styles for types of observations and then specific observation properties such as levels, sources, etc. One common use case of this would be plotting all temperature data as red, but with different linestyles for an isobaric level and the surface.
```
type_styles = {'Temperature': dict(color='red', marker='o'),
'Relative humidity': dict(color='green', marker='s')}
level_styles = {'isobaric': dict(linestyle='-', linewidth=2),
'surface': dict(linestyle=':', linewidth=3)}
my_style = type_styles['Temperature']
print(my_style)
my_style.update(level_styles['isobaric'])
print(my_style)
```
If we look back at the original entry in `type_styles` we see it was updated too! That may not be the expected or even the desired behavior.
```
type_styles['Temperature']
```
We can use the `copy` method to make a copy of the element and avoid update the original.
```
type_styles = {'Temperature': dict(color='red', marker='o'),
'Relative humidity': dict(color='green', marker='s')}
level_styles = {'isobaric': dict(linestyle='-', linewidth=2),
'surface': dict(linestyle=':', linewidth=3)}
my_style = type_styles['Temperature'].copy() # Avoids altering the original entry
my_style.update(level_styles['isobaric'])
print(my_style)
type_styles['Temperature']
```
Since we don't have data from different levels, we'll work with wind measurements and pressure data. Our <code>format_varname</code> function returns a title and full variable name label.
<div class="alert alert-success">
<b>EXERCISE</b>:
<ul>
<li>Create a type styles dictionary of dictionaries with the variable title as the key that has styles for `Wind` and `Pressure` data. The pressure should be a solid black line. Wind should be a solid line.</li>
<li>Create a variable style dictionary of dictionaries with the variable name as the key that specifies an orange line of width 2 for wind speed, olive line of width 0.5 for gusts, and no additional information for pressure.</li>
<li>Update the plotting code below to use the new type and variable styles dictionary.
</ul>
</div>
```
# Your code goes here (modify the skeleton below)
type_styles = {}
variable_styles = {}
fig, axes = plt.subplots(1, len(plot_variables), sharex=True, figsize=(14, 5))
for col, var_names in enumerate(plot_variables):
ax = axes[col]
for var_name in var_names:
title, label = format_varname(var_name)
ax.plot(df.time, df[var_name], **styles[var_name])
ax.set_ylabel(title)
ax.set_title('Buoy 42039 {}'.format(title))
ax.grid(True)
ax.set_xlabel('Time')
ax.xaxis.set_major_formatter(DateFormatter('%m/%d'))
ax.xaxis.set_major_locator(DayLocator())
```
#### Solution
```
# %load solutions/dict_args.py
```
<a href="#top">Top</a>
<hr style="height:2px;">
<a name="functions"></a>
## Function Application
There are times where we might want to apply a certain amount of pre-processing to the data before they are plotted. Maybe we want to do a unit conversion, scale the data, or filter it. We can create a dictionary in which functions are the values and variable names are the keys.
For example, let's define a function that uses the running median to filter the wind data (effectively a low-pass). We'll also make a do nothing function for data we don't want to alter.
```
from scipy.signal import medfilt
def filter_wind(a):
return medfilt(a, 7)
def donothing(a):
return a
converters = {'Wind': filter_wind, 'Pressure': donothing}
type_styles = {'Pressure': dict(color='black'),
'Wind': dict(linestyle='-')}
variable_styles = {'pressure': dict(),
'wind_speed': dict(color='tab:orange', linewidth=2),
'wind_gust': dict(color='tab:olive', linewidth=0.5)}
fig, axes = plt.subplots(1, len(plot_variables), sharex=True, figsize=(14, 5))
for col, var_names in enumerate(plot_variables):
ax = axes[col]
for var_name in var_names:
title, label = format_varname(var_name)
# Apply our pre-processing
var_data = converters[title](df[var_name])
style = type_styles[title].copy() # So the next line doesn't change the original
style.update(variable_styles[var_name])
ax.plot(df.time, var_data, **style)
ax.set_ylabel(title)
ax.set_title('Buoy 42039 {}'.format(title))
ax.grid(True)
ax.set_xlabel('Time')
ax.xaxis.set_major_formatter(DateFormatter('%m/%d'))
ax.xaxis.set_major_locator(DayLocator())
```
<div class="alert alert-success">
<b>EXERCISE</b>:
<ul>
<li>Write a function to convert the pressure data to bars. (**Hint**: 1 bar = 100000 Pa)</li>
<li>Apply your converter in the code below and replot the data.</li>
</ul>
</div>
```
# Your code goes here (modify the code below)
converters = {'Wind': filter_wind, 'Pressure': donothing}
type_styles = {'Pressure': dict(color='black'),
'Wind': dict(linestyle='-')}
variable_styles = {'pressure': dict(),
'wind_speed': dict(color='tab:orange', linewidth=2),
'wind_gust': dict(color='tab:olive', linewidth=0.5)}
fig, axes = plt.subplots(1, len(plot_variables), sharex=True, figsize=(14, 5))
for col, var_names in enumerate(plot_variables):
ax = axes[col]
for var_name in var_names:
title, label = format_varname(var_name)
# Apply our pre-processing
var_data = converters[title](df[var_name])
style = type_styles[title].copy() # So the next line doesn't change the original
style.update(variable_styles[var_name])
ax.plot(df.time, var_data, **style)
ax.set_ylabel(title)
ax.set_title('Buoy 42039 {}'.format(title))
ax.grid(True)
ax.set_xlabel('Time')
ax.xaxis.set_major_formatter(DateFormatter('%m/%d'))
ax.xaxis.set_major_locator(DayLocator())
```
#### Solution
<div class="alert alert-info">
<b>REMINDER</b>:
You should be using the unit library to convert between various physical units, this is simply for demonstration purposes!
</div>
```
# %load solutions/function_application.py
```
<a href="#top">Top</a>
<hr style="height:2px;">
<a name="glob"></a>
## Multiple Buoys
We can now use the techniques we've seen before to make a plot of multiple buoys in a single figure.
```
buoys = ['42039', '42022']
type_styles = {'Pressure': dict(color='black'),
'Wind': dict(linestyle='-')}
variable_styles = {'pressure': dict(),
'wind_speed': dict(color='tab:orange', linewidth=2),
'wind_gust': dict(color='tab:olive', linewidth=0.5)}
fig, axes = plt.subplots(len(buoys), len(plot_variables), sharex=True, figsize=(14, 10))
for row, buoy in enumerate(buoys):
df = read_buoy_data(buoy)
for col, var_names in enumerate(plot_variables):
ax = axes[row, col]
for var_name in var_names:
title, label = format_varname(var_name)
style = type_styles[title].copy() # So the next line doesn't change the original
style.update(variable_styles[var_name])
ax.plot(df.time, df[var_name], **style)
ax.set_ylabel(title)
ax.set_title('Buoy {} {}'.format(buoy, title))
ax.grid(True)
ax.set_xlabel('Time')
ax.xaxis.set_major_formatter(DateFormatter('%m/%d'))
ax.xaxis.set_major_locator(DayLocator())
```
<a href="#top">Top</a>
<hr style="height:2px;">
<div class="alert alert-success">
<b>EXERCISE</b>: As a final exercise, use a dictionary to allow all of the plots to share common y axis limits based on the variable title.
</div>
```
# Your code goes here
```
#### Solution
```
# %load solutions/final.py
```
<a href="#top">Top</a>
<hr style="height:2px;">
|
github_jupyter
|
```
# Copyright 2020 Google LLC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Vertex client library: AutoML text sentiment analysis model for batch prediction
<table align="left">
<td>
<a href="https://colab.research.google.com/github/GoogleCloudPlatform/vertex-ai-samples/blob/master/notebooks/community/gapic/automl/showcase_automl_text_sentiment_analysis_batch.ipynb">
<img src="https://cloud.google.com/ml-engine/images/colab-logo-32px.png" alt="Colab logo"> Run in Colab
</a>
</td>
<td>
<a href="https://github.com/GoogleCloudPlatform/vertex-ai-samples/blob/master/notebooks/community/gapic/automl/showcase_automl_text_sentiment_analysis_batch.ipynb">
<img src="https://cloud.google.com/ml-engine/images/github-logo-32px.png" alt="GitHub logo">
View on GitHub
</a>
</td>
</table>
<br/><br/><br/>
## Overview
This tutorial demonstrates how to use the Vertex client library for Python to create text sentiment analysis models and do batch prediction using Google Cloud's [AutoML](https://cloud.google.com/vertex-ai/docs/start/automl-users).
### Dataset
The dataset used for this tutorial is the [Crowdflower Claritin-Twitter dataset](https://data.world/crowdflower/claritin-twitter) from [data.world Datasets](https://data.world). The version of the dataset you will use in this tutorial is stored in a public Cloud Storage bucket.
### Objective
In this tutorial, you create an AutoML text sentiment analysis model from a Python script, and then do a batch prediction using the Vertex client library. You can alternatively create and deploy models using the `gcloud` command-line tool or online using the Google Cloud Console.
The steps performed include:
- Create a Vertex `Dataset` resource.
- Train the model.
- View the model evaluation.
- Make a batch prediction.
There is one key difference between using batch prediction and using online prediction:
* Prediction Service: Does an on-demand prediction for the entire set of instances (i.e., one or more data items) and returns the results in real-time.
* Batch Prediction Service: Does a queued (batch) prediction for the entire set of instances in the background and stores the results in a Cloud Storage bucket when ready.
### Costs
This tutorial uses billable components of Google Cloud (GCP):
* Vertex AI
* Cloud Storage
Learn about [Vertex AI
pricing](https://cloud.google.com/vertex-ai/pricing) and [Cloud Storage
pricing](https://cloud.google.com/storage/pricing), and use the [Pricing
Calculator](https://cloud.google.com/products/calculator/)
to generate a cost estimate based on your projected usage.
## Installation
Install the latest version of Vertex client library.
```
import os
import sys
# Google Cloud Notebook
if os.path.exists("/opt/deeplearning/metadata/env_version"):
USER_FLAG = "--user"
else:
USER_FLAG = ""
! pip3 install -U google-cloud-aiplatform $USER_FLAG
```
Install the latest GA version of *google-cloud-storage* library as well.
```
! pip3 install -U google-cloud-storage $USER_FLAG
```
### Restart the kernel
Once you've installed the Vertex client library and Google *cloud-storage*, you need to restart the notebook kernel so it can find the packages.
```
if not os.getenv("IS_TESTING"):
# Automatically restart kernel after installs
import IPython
app = IPython.Application.instance()
app.kernel.do_shutdown(True)
```
## Before you begin
### GPU runtime
*Make sure you're running this notebook in a GPU runtime if you have that option. In Colab, select* **Runtime > Change Runtime Type > GPU**
### Set up your Google Cloud project
**The following steps are required, regardless of your notebook environment.**
1. [Select or create a Google Cloud project](https://console.cloud.google.com/cloud-resource-manager). When you first create an account, you get a $300 free credit towards your compute/storage costs.
2. [Make sure that billing is enabled for your project.](https://cloud.google.com/billing/docs/how-to/modify-project)
3. [Enable the Vertex APIs and Compute Engine APIs.](https://console.cloud.google.com/flows/enableapi?apiid=ml.googleapis.com,compute_component)
4. [The Google Cloud SDK](https://cloud.google.com/sdk) is already installed in Google Cloud Notebook.
5. Enter your project ID in the cell below. Then run the cell to make sure the
Cloud SDK uses the right project for all the commands in this notebook.
**Note**: Jupyter runs lines prefixed with `!` as shell commands, and it interpolates Python variables prefixed with `$` into these commands.
```
PROJECT_ID = "[your-project-id]" # @param {type:"string"}
if PROJECT_ID == "" or PROJECT_ID is None or PROJECT_ID == "[your-project-id]":
# Get your GCP project id from gcloud
shell_output = !gcloud config list --format 'value(core.project)' 2>/dev/null
PROJECT_ID = shell_output[0]
print("Project ID:", PROJECT_ID)
! gcloud config set project $PROJECT_ID
```
#### Region
You can also change the `REGION` variable, which is used for operations
throughout the rest of this notebook. Below are regions supported for Vertex. We recommend that you choose the region closest to you.
- Americas: `us-central1`
- Europe: `europe-west4`
- Asia Pacific: `asia-east1`
You may not use a multi-regional bucket for training with Vertex. Not all regions provide support for all Vertex services. For the latest support per region, see the [Vertex locations documentation](https://cloud.google.com/vertex-ai/docs/general/locations)
```
REGION = "us-central1" # @param {type: "string"}
```
#### Timestamp
If you are in a live tutorial session, you might be using a shared test account or project. To avoid name collisions between users on resources created, you create a timestamp for each instance session, and append onto the name of resources which will be created in this tutorial.
```
from datetime import datetime
TIMESTAMP = datetime.now().strftime("%Y%m%d%H%M%S")
```
### Authenticate your Google Cloud account
**If you are using Google Cloud Notebook**, your environment is already authenticated. Skip this step.
**If you are using Colab**, run the cell below and follow the instructions when prompted to authenticate your account via oAuth.
**Otherwise**, follow these steps:
In the Cloud Console, go to the [Create service account key](https://console.cloud.google.com/apis/credentials/serviceaccountkey) page.
**Click Create service account**.
In the **Service account name** field, enter a name, and click **Create**.
In the **Grant this service account access to project** section, click the Role drop-down list. Type "Vertex" into the filter box, and select **Vertex Administrator**. Type "Storage Object Admin" into the filter box, and select **Storage Object Admin**.
Click Create. A JSON file that contains your key downloads to your local environment.
Enter the path to your service account key as the GOOGLE_APPLICATION_CREDENTIALS variable in the cell below and run the cell.
```
# If you are running this notebook in Colab, run this cell and follow the
# instructions to authenticate your GCP account. This provides access to your
# Cloud Storage bucket and lets you submit training jobs and prediction
# requests.
# If on Google Cloud Notebook, then don't execute this code
if not os.path.exists("/opt/deeplearning/metadata/env_version"):
if "google.colab" in sys.modules:
from google.colab import auth as google_auth
google_auth.authenticate_user()
# If you are running this notebook locally, replace the string below with the
# path to your service account key and run this cell to authenticate your GCP
# account.
elif not os.getenv("IS_TESTING"):
%env GOOGLE_APPLICATION_CREDENTIALS ''
```
### Create a Cloud Storage bucket
**The following steps are required, regardless of your notebook environment.**
This tutorial is designed to use training data that is in a public Cloud Storage bucket and a local Cloud Storage bucket for your batch predictions. You may alternatively use your own training data that you have stored in a local Cloud Storage bucket.
Set the name of your Cloud Storage bucket below. Bucket names must be globally unique across all Google Cloud projects, including those outside of your organization.
```
BUCKET_NAME = "gs://[your-bucket-name]" # @param {type:"string"}
if BUCKET_NAME == "" or BUCKET_NAME is None or BUCKET_NAME == "gs://[your-bucket-name]":
BUCKET_NAME = "gs://" + PROJECT_ID + "aip-" + TIMESTAMP
```
**Only if your bucket doesn't already exist**: Run the following cell to create your Cloud Storage bucket.
```
! gsutil mb -l $REGION $BUCKET_NAME
```
Finally, validate access to your Cloud Storage bucket by examining its contents:
```
! gsutil ls -al $BUCKET_NAME
```
### Set up variables
Next, set up some variables used throughout the tutorial.
### Import libraries and define constants
#### Import Vertex client library
Import the Vertex client library into our Python environment.
```
import time
from google.cloud.aiplatform import gapic as aip
from google.protobuf import json_format
from google.protobuf.json_format import MessageToJson, ParseDict
from google.protobuf.struct_pb2 import Struct, Value
```
#### Vertex constants
Setup up the following constants for Vertex:
- `API_ENDPOINT`: The Vertex API service endpoint for dataset, model, job, pipeline and endpoint services.
- `PARENT`: The Vertex location root path for dataset, model, job, pipeline and endpoint resources.
```
# API service endpoint
API_ENDPOINT = "{}-aiplatform.googleapis.com".format(REGION)
# Vertex location root path for your dataset, model and endpoint resources
PARENT = "projects/" + PROJECT_ID + "/locations/" + REGION
```
#### AutoML constants
Set constants unique to AutoML datasets and training:
- Dataset Schemas: Tells the `Dataset` resource service which type of dataset it is.
- Data Labeling (Annotations) Schemas: Tells the `Dataset` resource service how the data is labeled (annotated).
- Dataset Training Schemas: Tells the `Pipeline` resource service the task (e.g., classification) to train the model for.
```
# Text Dataset type
DATA_SCHEMA = "gs://google-cloud-aiplatform/schema/dataset/metadata/text_1.0.0.yaml"
# Text Labeling type
LABEL_SCHEMA = "gs://google-cloud-aiplatform/schema/dataset/ioformat/text_sentiment_io_format_1.0.0.yaml"
# Text Training task
TRAINING_SCHEMA = "gs://google-cloud-aiplatform/schema/trainingjob/definition/automl_text_sentiment_1.0.0.yaml"
```
#### Hardware Accelerators
Set the hardware accelerators (e.g., GPU), if any, for prediction.
Set the variable `DEPLOY_GPU/DEPLOY_NGPU` to use a container image supporting a GPU and the number of GPUs allocated to the virtual machine (VM) instance. For example, to use a GPU container image with 4 Nvidia Telsa K80 GPUs allocated to each VM, you would specify:
(aip.AcceleratorType.NVIDIA_TESLA_K80, 4)
For GPU, available accelerators include:
- aip.AcceleratorType.NVIDIA_TESLA_K80
- aip.AcceleratorType.NVIDIA_TESLA_P100
- aip.AcceleratorType.NVIDIA_TESLA_P4
- aip.AcceleratorType.NVIDIA_TESLA_T4
- aip.AcceleratorType.NVIDIA_TESLA_V100
Otherwise specify `(None, None)` to use a container image to run on a CPU.
```
if os.getenv("IS_TESTING_DEPOLY_GPU"):
DEPLOY_GPU, DEPLOY_NGPU = (
aip.AcceleratorType.NVIDIA_TESLA_K80,
int(os.getenv("IS_TESTING_DEPOLY_GPU")),
)
else:
DEPLOY_GPU, DEPLOY_NGPU = (aip.AcceleratorType.NVIDIA_TESLA_K80, 1)
```
#### Container (Docker) image
For AutoML batch prediction, the container image for the serving binary is pre-determined by the Vertex prediction service. More specifically, the service will pick the appropriate container for the model depending on the hardware accelerator you selected.
#### Machine Type
Next, set the machine type to use for prediction.
- Set the variable `DEPLOY_COMPUTE` to configure the compute resources for the VM you will use for prediction.
- `machine type`
- `n1-standard`: 3.75GB of memory per vCPU.
- `n1-highmem`: 6.5GB of memory per vCPU
- `n1-highcpu`: 0.9 GB of memory per vCPU
- `vCPUs`: number of \[2, 4, 8, 16, 32, 64, 96 \]
*Note: You may also use n2 and e2 machine types for training and deployment, but they do not support GPUs*
```
if os.getenv("IS_TESTING_DEPLOY_MACHINE"):
MACHINE_TYPE = os.getenv("IS_TESTING_DEPLOY_MACHINE")
else:
MACHINE_TYPE = "n1-standard"
VCPU = "4"
DEPLOY_COMPUTE = MACHINE_TYPE + "-" + VCPU
print("Deploy machine type", DEPLOY_COMPUTE)
```
# Tutorial
Now you are ready to start creating your own AutoML text sentiment analysis model.
## Set up clients
The Vertex client library works as a client/server model. On your side (the Python script) you will create a client that sends requests and receives responses from the Vertex server.
You will use different clients in this tutorial for different steps in the workflow. So set them all up upfront.
- Dataset Service for `Dataset` resources.
- Model Service for `Model` resources.
- Pipeline Service for training.
- Job Service for batch prediction and custom training.
```
# client options same for all services
client_options = {"api_endpoint": API_ENDPOINT}
def create_dataset_client():
client = aip.DatasetServiceClient(client_options=client_options)
return client
def create_model_client():
client = aip.ModelServiceClient(client_options=client_options)
return client
def create_pipeline_client():
client = aip.PipelineServiceClient(client_options=client_options)
return client
def create_job_client():
client = aip.JobServiceClient(client_options=client_options)
return client
clients = {}
clients["dataset"] = create_dataset_client()
clients["model"] = create_model_client()
clients["pipeline"] = create_pipeline_client()
clients["job"] = create_job_client()
for client in clients.items():
print(client)
```
## Dataset
Now that your clients are ready, your first step in training a model is to create a managed dataset instance, and then upload your labeled data to it.
### Create `Dataset` resource instance
Use the helper function `create_dataset` to create the instance of a `Dataset` resource. This function does the following:
1. Uses the dataset client service.
2. Creates an Vertex `Dataset` resource (`aip.Dataset`), with the following parameters:
- `display_name`: The human-readable name you choose to give it.
- `metadata_schema_uri`: The schema for the dataset type.
3. Calls the client dataset service method `create_dataset`, with the following parameters:
- `parent`: The Vertex location root path for your `Database`, `Model` and `Endpoint` resources.
- `dataset`: The Vertex dataset object instance you created.
4. The method returns an `operation` object.
An `operation` object is how Vertex handles asynchronous calls for long running operations. While this step usually goes fast, when you first use it in your project, there is a longer delay due to provisioning.
You can use the `operation` object to get status on the operation (e.g., create `Dataset` resource) or to cancel the operation, by invoking an operation method:
| Method | Description |
| ----------- | ----------- |
| result() | Waits for the operation to complete and returns a result object in JSON format. |
| running() | Returns True/False on whether the operation is still running. |
| done() | Returns True/False on whether the operation is completed. |
| canceled() | Returns True/False on whether the operation was canceled. |
| cancel() | Cancels the operation (this may take up to 30 seconds). |
```
TIMEOUT = 90
def create_dataset(name, schema, labels=None, timeout=TIMEOUT):
start_time = time.time()
try:
dataset = aip.Dataset(
display_name=name, metadata_schema_uri=schema, labels=labels
)
operation = clients["dataset"].create_dataset(parent=PARENT, dataset=dataset)
print("Long running operation:", operation.operation.name)
result = operation.result(timeout=TIMEOUT)
print("time:", time.time() - start_time)
print("response")
print(" name:", result.name)
print(" display_name:", result.display_name)
print(" metadata_schema_uri:", result.metadata_schema_uri)
print(" metadata:", dict(result.metadata))
print(" create_time:", result.create_time)
print(" update_time:", result.update_time)
print(" etag:", result.etag)
print(" labels:", dict(result.labels))
return result
except Exception as e:
print("exception:", e)
return None
result = create_dataset("claritin-" + TIMESTAMP, DATA_SCHEMA)
```
Now save the unique dataset identifier for the `Dataset` resource instance you created.
```
# The full unique ID for the dataset
dataset_id = result.name
# The short numeric ID for the dataset
dataset_short_id = dataset_id.split("/")[-1]
print(dataset_id)
```
### Data preparation
The Vertex `Dataset` resource for text has a couple of requirements for your text data.
- Text examples must be stored in a CSV or JSONL file.
#### CSV
For text sentiment analysis, the CSV file has a few requirements:
- No heading.
- First column is the text example or Cloud Storage path to text file.
- Second column the label (i.e., sentiment).
- Third column is the maximum sentiment value. For example, if the range is 0 to 3, then the maximum value is 3.
#### Location of Cloud Storage training data.
Now set the variable `IMPORT_FILE` to the location of the CSV index file in Cloud Storage.
```
IMPORT_FILE = "gs://cloud-samples-data/language/claritin.csv"
SENTIMENT_MAX = 4
```
#### Quick peek at your data
You will use a version of the Crowdflower Claritin-Twitter dataset that is stored in a public Cloud Storage bucket, using a CSV index file.
Start by doing a quick peek at the data. You count the number of examples by counting the number of rows in the CSV index file (`wc -l`) and then peek at the first few rows.
```
if "IMPORT_FILES" in globals():
FILE = IMPORT_FILES[0]
else:
FILE = IMPORT_FILE
count = ! gsutil cat $FILE | wc -l
print("Number of Examples", int(count[0]))
print("First 10 rows")
! gsutil cat $FILE | head
```
### Import data
Now, import the data into your Vertex Dataset resource. Use this helper function `import_data` to import the data. The function does the following:
- Uses the `Dataset` client.
- Calls the client method `import_data`, with the following parameters:
- `name`: The human readable name you give to the `Dataset` resource (e.g., claritin).
- `import_configs`: The import configuration.
- `import_configs`: A Python list containing a dictionary, with the key/value entries:
- `gcs_sources`: A list of URIs to the paths of the one or more index files.
- `import_schema_uri`: The schema identifying the labeling type.
The `import_data()` method returns a long running `operation` object. This will take a few minutes to complete. If you are in a live tutorial, this would be a good time to ask questions, or take a personal break.
```
def import_data(dataset, gcs_sources, schema):
config = [{"gcs_source": {"uris": gcs_sources}, "import_schema_uri": schema}]
print("dataset:", dataset_id)
start_time = time.time()
try:
operation = clients["dataset"].import_data(
name=dataset_id, import_configs=config
)
print("Long running operation:", operation.operation.name)
result = operation.result()
print("result:", result)
print("time:", int(time.time() - start_time), "secs")
print("error:", operation.exception())
print("meta :", operation.metadata)
print(
"after: running:",
operation.running(),
"done:",
operation.done(),
"cancelled:",
operation.cancelled(),
)
return operation
except Exception as e:
print("exception:", e)
return None
import_data(dataset_id, [IMPORT_FILE], LABEL_SCHEMA)
```
## Train the model
Now train an AutoML text sentiment analysis model using your Vertex `Dataset` resource. To train the model, do the following steps:
1. Create an Vertex training pipeline for the `Dataset` resource.
2. Execute the pipeline to start the training.
### Create a training pipeline
You may ask, what do we use a pipeline for? You typically use pipelines when the job (such as training) has multiple steps, generally in sequential order: do step A, do step B, etc. By putting the steps into a pipeline, we gain the benefits of:
1. Being reusable for subsequent training jobs.
2. Can be containerized and ran as a batch job.
3. Can be distributed.
4. All the steps are associated with the same pipeline job for tracking progress.
Use this helper function `create_pipeline`, which takes the following parameters:
- `pipeline_name`: A human readable name for the pipeline job.
- `model_name`: A human readable name for the model.
- `dataset`: The Vertex fully qualified dataset identifier.
- `schema`: The dataset labeling (annotation) training schema.
- `task`: A dictionary describing the requirements for the training job.
The helper function calls the `Pipeline` client service'smethod `create_pipeline`, which takes the following parameters:
- `parent`: The Vertex location root path for your `Dataset`, `Model` and `Endpoint` resources.
- `training_pipeline`: the full specification for the pipeline training job.
Let's look now deeper into the *minimal* requirements for constructing a `training_pipeline` specification:
- `display_name`: A human readable name for the pipeline job.
- `training_task_definition`: The dataset labeling (annotation) training schema.
- `training_task_inputs`: A dictionary describing the requirements for the training job.
- `model_to_upload`: A human readable name for the model.
- `input_data_config`: The dataset specification.
- `dataset_id`: The Vertex dataset identifier only (non-fully qualified) -- this is the last part of the fully-qualified identifier.
- `fraction_split`: If specified, the percentages of the dataset to use for training, test and validation. Otherwise, the percentages are automatically selected by AutoML.
```
def create_pipeline(pipeline_name, model_name, dataset, schema, task):
dataset_id = dataset.split("/")[-1]
input_config = {
"dataset_id": dataset_id,
"fraction_split": {
"training_fraction": 0.8,
"validation_fraction": 0.1,
"test_fraction": 0.1,
},
}
training_pipeline = {
"display_name": pipeline_name,
"training_task_definition": schema,
"training_task_inputs": task,
"input_data_config": input_config,
"model_to_upload": {"display_name": model_name},
}
try:
pipeline = clients["pipeline"].create_training_pipeline(
parent=PARENT, training_pipeline=training_pipeline
)
print(pipeline)
except Exception as e:
print("exception:", e)
return None
return pipeline
```
### Construct the task requirements
Next, construct the task requirements. Unlike other parameters which take a Python (JSON-like) dictionary, the `task` field takes a Google protobuf Struct, which is very similar to a Python dictionary. Use the `json_format.ParseDict` method for the conversion.
The minimal fields we need to specify are:
- `sentiment_max`: The maximum value for the sentiment (e.g., 4).
Finally, create the pipeline by calling the helper function `create_pipeline`, which returns an instance of a training pipeline object.
```
PIPE_NAME = "claritin_pipe-" + TIMESTAMP
MODEL_NAME = "claritin_model-" + TIMESTAMP
task = json_format.ParseDict(
{
"sentiment_max": SENTIMENT_MAX,
},
Value(),
)
response = create_pipeline(PIPE_NAME, MODEL_NAME, dataset_id, TRAINING_SCHEMA, task)
```
Now save the unique identifier of the training pipeline you created.
```
# The full unique ID for the pipeline
pipeline_id = response.name
# The short numeric ID for the pipeline
pipeline_short_id = pipeline_id.split("/")[-1]
print(pipeline_id)
```
### Get information on a training pipeline
Now get pipeline information for just this training pipeline instance. The helper function gets the job information for just this job by calling the the job client service's `get_training_pipeline` method, with the following parameter:
- `name`: The Vertex fully qualified pipeline identifier.
When the model is done training, the pipeline state will be `PIPELINE_STATE_SUCCEEDED`.
```
def get_training_pipeline(name, silent=False):
response = clients["pipeline"].get_training_pipeline(name=name)
if silent:
return response
print("pipeline")
print(" name:", response.name)
print(" display_name:", response.display_name)
print(" state:", response.state)
print(" training_task_definition:", response.training_task_definition)
print(" training_task_inputs:", dict(response.training_task_inputs))
print(" create_time:", response.create_time)
print(" start_time:", response.start_time)
print(" end_time:", response.end_time)
print(" update_time:", response.update_time)
print(" labels:", dict(response.labels))
return response
response = get_training_pipeline(pipeline_id)
```
# Deployment
Training the above model may take upwards of 180 minutes time.
Once your model is done training, you can calculate the actual time it took to train the model by subtracting `end_time` from `start_time`. For your model, you will need to know the fully qualified Vertex Model resource identifier, which the pipeline service assigned to it. You can get this from the returned pipeline instance as the field `model_to_deploy.name`.
```
while True:
response = get_training_pipeline(pipeline_id, True)
if response.state != aip.PipelineState.PIPELINE_STATE_SUCCEEDED:
print("Training job has not completed:", response.state)
model_to_deploy_id = None
if response.state == aip.PipelineState.PIPELINE_STATE_FAILED:
raise Exception("Training Job Failed")
else:
model_to_deploy = response.model_to_upload
model_to_deploy_id = model_to_deploy.name
print("Training Time:", response.end_time - response.start_time)
break
time.sleep(60)
print("model to deploy:", model_to_deploy_id)
```
## Model information
Now that your model is trained, you can get some information on your model.
## Evaluate the Model resource
Now find out how good the model service believes your model is. As part of training, some portion of the dataset was set aside as the test (holdout) data, which is used by the pipeline service to evaluate the model.
### List evaluations for all slices
Use this helper function `list_model_evaluations`, which takes the following parameter:
- `name`: The Vertex fully qualified model identifier for the `Model` resource.
This helper function uses the model client service's `list_model_evaluations` method, which takes the same parameter. The response object from the call is a list, where each element is an evaluation metric.
For each evaluation -- you probably only have one, we then print all the key names for each metric in the evaluation, and for a small set (`meanAbsoluteError` and `precision`) you will print the result.
```
def list_model_evaluations(name):
response = clients["model"].list_model_evaluations(parent=name)
for evaluation in response:
print("model_evaluation")
print(" name:", evaluation.name)
print(" metrics_schema_uri:", evaluation.metrics_schema_uri)
metrics = json_format.MessageToDict(evaluation._pb.metrics)
for metric in metrics.keys():
print(metric)
print("meanAbsoluteError", metrics["meanAbsoluteError"])
print("precision", metrics["precision"])
return evaluation.name
last_evaluation = list_model_evaluations(model_to_deploy_id)
```
## Model deployment for batch prediction
Now deploy the trained Vertex `Model` resource you created for batch prediction. This differs from deploying a `Model` resource for on-demand prediction.
For online prediction, you:
1. Create an `Endpoint` resource for deploying the `Model` resource to.
2. Deploy the `Model` resource to the `Endpoint` resource.
3. Make online prediction requests to the `Endpoint` resource.
For batch-prediction, you:
1. Create a batch prediction job.
2. The job service will provision resources for the batch prediction request.
3. The results of the batch prediction request are returned to the caller.
4. The job service will unprovision the resoures for the batch prediction request.
## Make a batch prediction request
Now do a batch prediction to your deployed model.
### Get test item(s)
Now do a batch prediction to your Vertex model. You will use arbitrary examples out of the dataset as a test items. Don't be concerned that the examples were likely used in training the model -- we just want to demonstrate how to make a prediction.
```
test_items = ! gsutil cat $IMPORT_FILE | head -n2
if len(test_items[0]) == 4:
_, test_item_1, test_label_1, _ = str(test_items[0]).split(",")
_, test_item_2, test_label_2, _ = str(test_items[1]).split(",")
else:
test_item_1, test_label_1, _ = str(test_items[0]).split(",")
test_item_2, test_label_2, _ = str(test_items[1]).split(",")
print(test_item_1, test_label_1)
print(test_item_2, test_label_2)
```
### Make the batch input file
Now make a batch input file, which you will store in your local Cloud Storage bucket. The batch input file can only be in JSONL format. For JSONL file, you make one dictionary entry per line for each data item (instance). The dictionary contains the key/value pairs:
- `content`: The Cloud Storage path to the file with the text item.
- `mime_type`: The content type. In our example, it is an `text` file.
For example:
{'content': '[your-bucket]/file1.txt', 'mime_type': 'text'}
```
import json
import tensorflow as tf
gcs_test_item_1 = BUCKET_NAME + "/test1.txt"
with tf.io.gfile.GFile(gcs_test_item_1, "w") as f:
f.write(test_item_1 + "\n")
gcs_test_item_2 = BUCKET_NAME + "/test2.txt"
with tf.io.gfile.GFile(gcs_test_item_2, "w") as f:
f.write(test_item_2 + "\n")
gcs_input_uri = BUCKET_NAME + "/test.jsonl"
with tf.io.gfile.GFile(gcs_input_uri, "w") as f:
data = {"content": gcs_test_item_1, "mime_type": "text/plain"}
f.write(json.dumps(data) + "\n")
data = {"content": gcs_test_item_2, "mime_type": "text/plain"}
f.write(json.dumps(data) + "\n")
print(gcs_input_uri)
! gsutil cat $gcs_input_uri
```
### Compute instance scaling
You have several choices on scaling the compute instances for handling your batch prediction requests:
- Single Instance: The batch prediction requests are processed on a single compute instance.
- Set the minimum (`MIN_NODES`) and maximum (`MAX_NODES`) number of compute instances to one.
- Manual Scaling: The batch prediction requests are split across a fixed number of compute instances that you manually specified.
- Set the minimum (`MIN_NODES`) and maximum (`MAX_NODES`) number of compute instances to the same number of nodes. When a model is first deployed to the instance, the fixed number of compute instances are provisioned and batch prediction requests are evenly distributed across them.
- Auto Scaling: The batch prediction requests are split across a scaleable number of compute instances.
- Set the minimum (`MIN_NODES`) number of compute instances to provision when a model is first deployed and to de-provision, and set the maximum (`MAX_NODES) number of compute instances to provision, depending on load conditions.
The minimum number of compute instances corresponds to the field `min_replica_count` and the maximum number of compute instances corresponds to the field `max_replica_count`, in your subsequent deployment request.
```
MIN_NODES = 1
MAX_NODES = 1
```
### Make batch prediction request
Now that your batch of two test items is ready, let's do the batch request. Use this helper function `create_batch_prediction_job`, with the following parameters:
- `display_name`: The human readable name for the prediction job.
- `model_name`: The Vertex fully qualified identifier for the `Model` resource.
- `gcs_source_uri`: The Cloud Storage path to the input file -- which you created above.
- `gcs_destination_output_uri_prefix`: The Cloud Storage path that the service will write the predictions to.
- `parameters`: Additional filtering parameters for serving prediction results.
The helper function calls the job client service's `create_batch_prediction_job` metho, with the following parameters:
- `parent`: The Vertex location root path for Dataset, Model and Pipeline resources.
- `batch_prediction_job`: The specification for the batch prediction job.
Let's now dive into the specification for the `batch_prediction_job`:
- `display_name`: The human readable name for the prediction batch job.
- `model`: The Vertex fully qualified identifier for the `Model` resource.
- `dedicated_resources`: The compute resources to provision for the batch prediction job.
- `machine_spec`: The compute instance to provision. Use the variable you set earlier `DEPLOY_GPU != None` to use a GPU; otherwise only a CPU is allocated.
- `starting_replica_count`: The number of compute instances to initially provision, which you set earlier as the variable `MIN_NODES`.
- `max_replica_count`: The maximum number of compute instances to scale to, which you set earlier as the variable `MAX_NODES`.
- `model_parameters`: Additional filtering parameters for serving prediction results. *Note*, text models do not support additional parameters.
- `input_config`: The input source and format type for the instances to predict.
- `instances_format`: The format of the batch prediction request file: `jsonl` only supported.
- `gcs_source`: A list of one or more Cloud Storage paths to your batch prediction requests.
- `output_config`: The output destination and format for the predictions.
- `prediction_format`: The format of the batch prediction response file: `jsonl` only supported.
- `gcs_destination`: The output destination for the predictions.
This call is an asychronous operation. You will print from the response object a few select fields, including:
- `name`: The Vertex fully qualified identifier assigned to the batch prediction job.
- `display_name`: The human readable name for the prediction batch job.
- `model`: The Vertex fully qualified identifier for the Model resource.
- `generate_explanations`: Whether True/False explanations were provided with the predictions (explainability).
- `state`: The state of the prediction job (pending, running, etc).
Since this call will take a few moments to execute, you will likely get `JobState.JOB_STATE_PENDING` for `state`.
```
BATCH_MODEL = "claritin_batch-" + TIMESTAMP
def create_batch_prediction_job(
display_name,
model_name,
gcs_source_uri,
gcs_destination_output_uri_prefix,
parameters=None,
):
if DEPLOY_GPU:
machine_spec = {
"machine_type": DEPLOY_COMPUTE,
"accelerator_type": DEPLOY_GPU,
"accelerator_count": DEPLOY_NGPU,
}
else:
machine_spec = {
"machine_type": DEPLOY_COMPUTE,
"accelerator_count": 0,
}
batch_prediction_job = {
"display_name": display_name,
# Format: 'projects/{project}/locations/{location}/models/{model_id}'
"model": model_name,
"model_parameters": json_format.ParseDict(parameters, Value()),
"input_config": {
"instances_format": IN_FORMAT,
"gcs_source": {"uris": [gcs_source_uri]},
},
"output_config": {
"predictions_format": OUT_FORMAT,
"gcs_destination": {"output_uri_prefix": gcs_destination_output_uri_prefix},
},
"dedicated_resources": {
"machine_spec": machine_spec,
"starting_replica_count": MIN_NODES,
"max_replica_count": MAX_NODES,
},
}
response = clients["job"].create_batch_prediction_job(
parent=PARENT, batch_prediction_job=batch_prediction_job
)
print("response")
print(" name:", response.name)
print(" display_name:", response.display_name)
print(" model:", response.model)
try:
print(" generate_explanation:", response.generate_explanation)
except:
pass
print(" state:", response.state)
print(" create_time:", response.create_time)
print(" start_time:", response.start_time)
print(" end_time:", response.end_time)
print(" update_time:", response.update_time)
print(" labels:", response.labels)
return response
IN_FORMAT = "jsonl"
OUT_FORMAT = "jsonl" # [jsonl]
response = create_batch_prediction_job(
BATCH_MODEL, model_to_deploy_id, gcs_input_uri, BUCKET_NAME, None
)
```
Now get the unique identifier for the batch prediction job you created.
```
# The full unique ID for the batch job
batch_job_id = response.name
# The short numeric ID for the batch job
batch_job_short_id = batch_job_id.split("/")[-1]
print(batch_job_id)
```
### Get information on a batch prediction job
Use this helper function `get_batch_prediction_job`, with the following paramter:
- `job_name`: The Vertex fully qualified identifier for the batch prediction job.
The helper function calls the job client service's `get_batch_prediction_job` method, with the following paramter:
- `name`: The Vertex fully qualified identifier for the batch prediction job. In this tutorial, you will pass it the Vertex fully qualified identifier for your batch prediction job -- `batch_job_id`
The helper function will return the Cloud Storage path to where the predictions are stored -- `gcs_destination`.
```
def get_batch_prediction_job(job_name, silent=False):
response = clients["job"].get_batch_prediction_job(name=job_name)
if silent:
return response.output_config.gcs_destination.output_uri_prefix, response.state
print("response")
print(" name:", response.name)
print(" display_name:", response.display_name)
print(" model:", response.model)
try: # not all data types support explanations
print(" generate_explanation:", response.generate_explanation)
except:
pass
print(" state:", response.state)
print(" error:", response.error)
gcs_destination = response.output_config.gcs_destination
print(" gcs_destination")
print(" output_uri_prefix:", gcs_destination.output_uri_prefix)
return gcs_destination.output_uri_prefix, response.state
predictions, state = get_batch_prediction_job(batch_job_id)
```
### Get the predictions
When the batch prediction is done processing, the job state will be `JOB_STATE_SUCCEEDED`.
Finally you view the predictions stored at the Cloud Storage path you set as output. The predictions will be in a JSONL format, which you indicated at the time you made the batch prediction job, under a subfolder starting with the name `prediction`, and under that folder will be a file called `predictions*.jsonl`.
Now display (cat) the contents. You will see multiple JSON objects, one for each prediction.
The first field `text_snippet` is the text file you did the prediction on, and the second field `annotations` is the prediction, which is further broken down into:
- `sentiment`: The predicted sentiment level.
```
def get_latest_predictions(gcs_out_dir):
""" Get the latest prediction subfolder using the timestamp in the subfolder name"""
folders = !gsutil ls $gcs_out_dir
latest = ""
for folder in folders:
subfolder = folder.split("/")[-2]
if subfolder.startswith("prediction-"):
if subfolder > latest:
latest = folder[:-1]
return latest
while True:
predictions, state = get_batch_prediction_job(batch_job_id, True)
if state != aip.JobState.JOB_STATE_SUCCEEDED:
print("The job has not completed:", state)
if state == aip.JobState.JOB_STATE_FAILED:
raise Exception("Batch Job Failed")
else:
folder = get_latest_predictions(predictions)
! gsutil ls $folder/prediction*.jsonl
! gsutil cat $folder/prediction*.jsonl
break
time.sleep(60)
```
# Cleaning up
To clean up all GCP resources used in this project, you can [delete the GCP
project](https://cloud.google.com/resource-manager/docs/creating-managing-projects#shutting_down_projects) you used for the tutorial.
Otherwise, you can delete the individual resources you created in this tutorial:
- Dataset
- Pipeline
- Model
- Endpoint
- Batch Job
- Custom Job
- Hyperparameter Tuning Job
- Cloud Storage Bucket
```
delete_dataset = True
delete_pipeline = True
delete_model = True
delete_endpoint = True
delete_batchjob = True
delete_customjob = True
delete_hptjob = True
delete_bucket = True
# Delete the dataset using the Vertex fully qualified identifier for the dataset
try:
if delete_dataset and "dataset_id" in globals():
clients["dataset"].delete_dataset(name=dataset_id)
except Exception as e:
print(e)
# Delete the training pipeline using the Vertex fully qualified identifier for the pipeline
try:
if delete_pipeline and "pipeline_id" in globals():
clients["pipeline"].delete_training_pipeline(name=pipeline_id)
except Exception as e:
print(e)
# Delete the model using the Vertex fully qualified identifier for the model
try:
if delete_model and "model_to_deploy_id" in globals():
clients["model"].delete_model(name=model_to_deploy_id)
except Exception as e:
print(e)
# Delete the endpoint using the Vertex fully qualified identifier for the endpoint
try:
if delete_endpoint and "endpoint_id" in globals():
clients["endpoint"].delete_endpoint(name=endpoint_id)
except Exception as e:
print(e)
# Delete the batch job using the Vertex fully qualified identifier for the batch job
try:
if delete_batchjob and "batch_job_id" in globals():
clients["job"].delete_batch_prediction_job(name=batch_job_id)
except Exception as e:
print(e)
# Delete the custom job using the Vertex fully qualified identifier for the custom job
try:
if delete_customjob and "job_id" in globals():
clients["job"].delete_custom_job(name=job_id)
except Exception as e:
print(e)
# Delete the hyperparameter tuning job using the Vertex fully qualified identifier for the hyperparameter tuning job
try:
if delete_hptjob and "hpt_job_id" in globals():
clients["job"].delete_hyperparameter_tuning_job(name=hpt_job_id)
except Exception as e:
print(e)
if delete_bucket and "BUCKET_NAME" in globals():
! gsutil rm -r $BUCKET_NAME
```
|
github_jupyter
|
# Prepare Dataset for Model Training and Evaluating
# Amazon Customer Reviews Dataset
https://s3.amazonaws.com/amazon-reviews-pds/readme.html
## Schema
- `marketplace`: 2-letter country code (in this case all "US").
- `customer_id`: Random identifier that can be used to aggregate reviews written by a single author.
- `review_id`: A unique ID for the review.
- `product_id`: The Amazon Standard Identification Number (ASIN). `http://www.amazon.com/dp/<ASIN>` links to the product's detail page.
- `product_parent`: The parent of that ASIN. Multiple ASINs (color or format variations of the same product) can roll up into a single parent.
- `product_title`: Title description of the product.
- `product_category`: Broad product category that can be used to group reviews (in this case digital videos).
- `star_rating`: The review's rating (1 to 5 stars).
- `helpful_votes`: Number of helpful votes for the review.
- `total_votes`: Number of total votes the review received.
- `vine`: Was the review written as part of the [Vine](https://www.amazon.com/gp/vine/help) program?
- `verified_purchase`: Was the review from a verified purchase?
- `review_headline`: The title of the review itself.
- `review_body`: The text of the review.
- `review_date`: The date the review was written.
```
import boto3
import sagemaker
import pandas as pd
sess = sagemaker.Session()
bucket = sess.default_bucket()
role = sagemaker.get_execution_role()
region = boto3.Session().region_name
```
## Download
Let's start by retrieving a subset of the Amazon Customer Reviews dataset.
```
!aws s3 cp 's3://amazon-reviews-pds/tsv/amazon_reviews_us_Digital_Software_v1_00.tsv.gz' ./data/
import csv
df = pd.read_csv(
"./data/amazon_reviews_us_Digital_Software_v1_00.tsv.gz",
delimiter="\t",
quoting=csv.QUOTE_NONE,
compression="gzip",
)
df.shape
df.head(5)
import matplotlib.pyplot as plt
%matplotlib inline
%config InlineBackend.figure_format='retina'
df[["star_rating", "review_id"]].groupby("star_rating").count().plot(kind="bar", title="Breakdown by Star Rating")
plt.xlabel("Star Rating")
plt.ylabel("Review Count")
```
# Balance the Dataset
```
from sklearn.utils import resample
five_star_df = df.query("star_rating == 5")
four_star_df = df.query("star_rating == 4")
three_star_df = df.query("star_rating == 3")
two_star_df = df.query("star_rating == 2")
one_star_df = df.query("star_rating == 1")
# Check which sentiment has the least number of samples
minority_count = min(
five_star_df.shape[0], four_star_df.shape[0], three_star_df.shape[0], two_star_df.shape[0], one_star_df.shape[0]
)
five_star_df = resample(five_star_df, replace=False, n_samples=minority_count, random_state=27)
four_star_df = resample(four_star_df, replace=False, n_samples=minority_count, random_state=27)
three_star_df = resample(three_star_df, replace=False, n_samples=minority_count, random_state=27)
two_star_df = resample(two_star_df, replace=False, n_samples=minority_count, random_state=27)
one_star_df = resample(one_star_df, replace=False, n_samples=minority_count, random_state=27)
df_balanced = pd.concat([five_star_df, four_star_df, three_star_df, two_star_df, one_star_df])
df_balanced = df_balanced.reset_index(drop=True)
df_balanced.shape
df_balanced[["star_rating", "review_id"]].groupby("star_rating").count().plot(
kind="bar", title="Breakdown by Star Rating"
)
plt.xlabel("Star Rating")
plt.ylabel("Review Count")
df_balanced.head(5)
```
# Split the Data into Train, Validation, and Test Sets
```
from sklearn.model_selection import train_test_split
# Split all data into 90% train and 10% holdout
df_train, df_holdout = train_test_split(df_balanced, test_size=0.10, stratify=df_balanced["star_rating"])
# Split holdout data into 50% validation and 50% test
df_validation, df_test = train_test_split(df_holdout, test_size=0.50, stratify=df_holdout["star_rating"])
# Pie chart, where the slices will be ordered and plotted counter-clockwise:
labels = ["Train", "Validation", "Test"]
sizes = [len(df_train.index), len(df_validation.index), len(df_test.index)]
explode = (0.1, 0, 0)
fig1, ax1 = plt.subplots()
ax1.pie(sizes, explode=explode, labels=labels, autopct="%1.1f%%", startangle=90)
# Equal aspect ratio ensures that pie is drawn as a circle.
ax1.axis("equal")
plt.show()
```
# Show 90% Train Data Split
```
df_train.shape
df_train[["star_rating", "review_id"]].groupby("star_rating").count().plot(
kind="bar", title="90% Train Breakdown by Star Rating"
)
```
# Show 5% Validation Data Split
```
df_validation.shape
df_validation[["star_rating", "review_id"]].groupby("star_rating").count().plot(
kind="bar", title="5% Validation Breakdown by Star Rating"
)
```
# Show 5% Test Data Split
```
df_test.shape
df_test[["star_rating", "review_id"]].groupby("star_rating").count().plot(
kind="bar", title="5% Test Breakdown by Star Rating"
)
```
# Select `star_rating` and `review_body` for Training
```
df_train = df_train[["star_rating", "review_body"]]
df_train.shape
df_train.head(5)
```
# Write a CSV With No Header for Comprehend
```
comprehend_train_path = "./amazon_reviews_us_Digital_Software_v1_00_comprehend.csv"
df_train.to_csv(comprehend_train_path, index=False, header=False)
```
# Upload Train Data to S3 for Comprehend
```
train_s3_prefix = "data"
comprehend_train_s3_uri = sess.upload_data(path=comprehend_train_path, key_prefix=train_s3_prefix)
comprehend_train_s3_uri
!aws s3 ls $comprehend_train_s3_uri
```
# Store the location of our train data in our notebook server to be used next
```
%store comprehend_train_s3_uri
%store
```
# Release Resources
```
%%html
<p><b>Shutting down your kernel for this notebook to release resources.</b></p>
<button class="sm-command-button" data-commandlinker-command="kernelmenu:shutdown" style="display:none;">Shutdown Kernel</button>
<script>
try {
els = document.getElementsByClassName("sm-command-button");
els[0].click();
}
catch(err) {
// NoOp
}
</script>
%%javascript
try {
Jupyter.notebook.save_checkpoint();
Jupyter.notebook.session.delete();
}
catch(err) {
// NoOp
}
```
|
github_jupyter
|
# Time Series Cross Validation
```
import pandas as pd
import numpy as np
#suppress ARIMA warnings
import warnings
warnings.filterwarnings('ignore')
```
Up till now we have used a single validation period to select our best model. The weakness of that approach is that it gives you a sample size of 1 (that's better than nothing, but generally poor statistics!). Time series cross validation is an approach to provide more data points when comparing models. In the classicial time series literature time series cross validation is called a **Rolling Forecast Origin**. There may also be benefit of taking a **sliding window** approach to cross validaiton. This second approach maintains a fixed sized training set. I.e. it drops older values from the time series during validation.
## Rolling Forecast Origin
The following code and output provide a simplified view of how rolling forecast horizons work in practice.
```
def rolling_forecast_origin(train, min_train_size, horizon):
'''
Rolling forecast origin generator.
'''
for i in range(len(train) - min_train_size - horizon + 1):
split_train = train[:min_train_size+i]
split_val = train[min_train_size+i:min_train_size+i+horizon]
yield split_train, split_val
full_series = [2502, 2414, 2800, 2143, 2708, 1900, 2333, 2222, 1234, 3456]
test = full_series[-2:]
train = full_series[:-2]
print('full training set: {0}'.format(train))
print('hidden test set: {0}'.format(test))
cv_rolling = rolling_forecast_origin(train, min_train_size=4, horizon=2)
cv_rolling
i = 0
for cv_train, cv_val in cv_rolling:
print(f'CV[{i+1}]')
print(f'Train:\t{cv_train}')
print(f'Val:\t{cv_val}')
print('-----')
i += 1
```
## Sliding Window Cross Validation
```
def sliding_window(train, window_size, horizon, step=1):
'''
sliding window generator.
Parameters:
--------
train: array-like
training data for time series method
window_size: int
lookback - how much data to include.
horizon: int
forecast horizon
step: int, optional (default=1)
step=1 means that a single additional data point is added to the time
series. increase step to run less splits.
Returns:
array-like, array-like
split_training, split_validation
'''
for i in range(0, len(train) - window_size - horizon + 1, step):
split_train = train[i:window_size+i]
split_val = train[i+window_size:window_size+i+horizon]
yield split_train, split_val
```
This code tests its with `step=1`
```
cv_sliding = sliding_window(train, window_size=4, horizon=1)
print('full training set: {0}\n'.format(train))
i = 0
for cv_train, cv_val in cv_sliding:
print(f'CV[{i+1}]')
print(f'Train:\t{cv_train}')
print(f'Val:\t{cv_val}')
print('-----')
i += 1
```
The following code tests it with `step=2`. Note that you get less splits. The code is less computationally expensive at the cost of less data. That is probably okay.
```
cv_sliding = sliding_window(train, window_size=4, horizon=1, step=2)
print('full training set: {0}\n'.format(train))
i = 0
for cv_train, cv_val in cv_sliding:
print(f'CV[{i+1}]')
print(f'Train:\t{cv_train}')
print(f'Val:\t{cv_val}')
print('-----')
i += 1
```
# Parallel Cross Validation Example using Naive1
```
from forecast_tools.baseline import SNaive, Naive1
from forecast_tools.datasets import load_emergency_dept
#optimised version of the functions above...
from forecast_tools.model_selection import (rolling_forecast_origin,
sliding_window,
cross_validation_score)
from sklearn.metrics import mean_absolute_error
train = load_emergency_dept()
model = Naive1()
#%%timeit runs the code multiple times to get an estimate of runtime.
#comment if out to run the code only once.
```
Run on a single core
```
%%time
cv = sliding_window(train, window_size=14, horizon=7, step=1)
results_1 = cross_validation_score(model, train, cv, mean_absolute_error,
n_jobs=1)
```
Run across multiple cores by setting `n_jobs=-1`
```
%%time
cv = sliding_window(train, window_size=14, horizon=7, step=1)
results_2 = cross_validation_score(model, train, cv, mean_absolute_error,
n_jobs=-1)
results_1.shape
results_2.shape
print(results_1.mean(), results_1.std())
```
just to illustrate that the results are the same - the difference is runtime.
```
print(results_2.mean(), results_2.std())
```
# Cross validation with multiple forecast horizons
```
horizons = [7, 14, 21]
cv = sliding_window(train, window_size=14, horizon=max(horizons), step=1)
#note that we now pass in the horizons list to cross_val_score
results_h = cross_validation_score(model, train, cv, mean_absolute_error,
horizons=horizons, n_jobs=-1)
#results are returned as numpy array - easy to cast to dataframe and display
pd.DataFrame(results_h, columns=['7days', '14days', '21days']).head()
```
## Cross validation example using ARIMA - does it speed up when CV run in Parallel?
```
#use ARIMA from pmdarima as that has a similar interface to baseline models.
from pmdarima import ARIMA, auto_arima
#ato_model = auto_arima(train, suppress_warnings=True, n_jobs=-1, m=7)
#auto_model
#create arima model - reasonably complex model
#order=(1, 1, 2), seasonal_order=(2, 0, 2, 7)
args = {'order':(1, 1, 2), 'seasonal_order':(2, 0, 2, 7)}
model = ARIMA(order=args['order'], seasonal_order=args['seasonal_order'],
enforce_stationarity=False, suppress_warnings=True)
%%time
cv = rolling_forecast_origin(train, min_train_size=320, horizon=7)
results_1 = cross_validation_score(model, train, cv, mean_absolute_error,
n_jobs=1)
```
comment out %%timeit to run the code only once!
you should see a big improvement in performance. mine went
from 12.3 seconds to 2.4 seconds.
```
%%time
cv = rolling_forecast_origin(train, min_train_size=320, horizon=7)
results_2 = cross_validation_score(model, train, cv, mean_absolute_error,
n_jobs=-1)
results_1.shape
results_2.shape
results_1.mean()
results_2.mean()
```
|
github_jupyter
|
##### Copyright 2018 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
#@title MIT License
#
# Copyright (c) 2017 François Chollet
#
# Permission is hereby granted, free of charge, to any person obtaining a
# copy of this software and associated documentation files (the "Software"),
# to deal in the Software without restriction, including without limitation
# the rights to use, copy, modify, merge, publish, distribute, sublicense,
# and/or sell copies of the Software, and to permit persons to whom the
# Software is furnished to do so, subject to the following conditions:
#
# The above copyright notice and this permission notice shall be included in
# all copies or substantial portions of the Software.
#
# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
# DEALINGS IN THE SOFTWARE.
```
# Classificação de texto com avaliações de filmes
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/pt/r1/tutorials/keras/basic_text_classification.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Execute em Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/pt/r1/tutorials/keras/basic_text_classification.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />Veja a fonte em GitHub</a>
</td>
</table>
Este *notebook* classifica avaliações de filmes como **positiva** ou **negativa** usando o texto da avaliação. Isto é um exemplo de classificação *binária* —ou duas-classes—, um importante e bastante aplicado tipo de problema de aprendizado de máquina.
Usaremos a base de dados [IMDB](https://www.tensorflow.org/api_docs/python/tf/keras/datasets/imdb) que contém avaliaçòes de mais de 50000 filmes do bando de dados [Internet Movie Database](https://www.imdb.com/). A base é dividida em 25000 avaliações para treinamento e 25000 para teste. Os conjuntos de treinamentos e testes são *balanceados*, ou seja, eles possuem a mesma quantidade de avaliações positivas e negativas.
O notebook utiliza [tf.keras](https://www.tensorflow.org/r1/guide/keras), uma API alto-nível para construir e treinar modelos com TensorFlow. Para mais tutoriais avançados de classificação de textos usando `tf.keras`, veja em [MLCC Text Classification Guide](https://developers.google.com/machine-learning/guides/text-classification/).
```
# keras.datasets.imdb está quebrado em 1.13 e 1.14, pelo np 1.16.3
!pip install tf_nightly
from __future__ import absolute_import, division, print_function, unicode_literals
import tensorflow as tf
from tensorflow import keras
import numpy as np
print(tf.__version__)
```
## Baixe a base de dados IMDB
A base de dados vem empacotada com TensorFlow. Ele já vem pré-processado de forma que as avaliações (sequências de palavras) foi convertida em sequências de inteiros, onde cada inteiro representa uma palavra específica no dicionário.
O código abaixo baixa a base de dados IMDB para a sua máquina (ou usa a cópia em *cache*, caso já tenha baixado):
```
imdb = keras.datasets.imdb
(train_data, train_labels), (test_data, test_labels) = imdb.load_data(num_words=10000)
```
O argumento `num_words=10000` mantém as 10000 palavras mais frequentes no conjunto de treinamento. As palavras mais raras são descartadas para preservar o tamanho dos dados de forma maleável.
## Explore os dados
Vamos parar um momento para entender o formato dos dados. O conjunto de dados vem pré-processado: cada exemplo é um *array* de inteiros representando as palavras da avaliação do filme. Cada *label* é um inteiro com valor ou de 0 ou 1, onde 0 é uma avaliação negativa e 1 é uma avaliação positiva.
```
print("Training entries: {}, labels: {}".format(len(train_data), len(train_labels)))
```
O texto das avaliações foi convertido para inteiros, onde cada inteiro representa uma palavra específica no dicionário. Isso é como se parece a primeira revisão:
```
print(train_data[0])
```
As avaliações dos filmes têm diferentes tamanhos. O código abaixo mostra o número de palavras da primeira e segunda avaliação. Sabendo que o número de entradas da rede neural tem que ser de mesmo também, temos que resolver isto mais tarde.
```
len(train_data[0]), len(train_data[1])
```
### Converta os inteiros de volta a palavras
É util saber como converter inteiros de volta a texto. Aqui, criaremos uma função de ajuda para consultar um objeto *dictionary* que contenha inteiros mapeados em strings:
```
# Um dicionário mapeando palavras em índices inteiros
word_index = imdb.get_word_index()
# Os primeiros índices são reservados
word_index = {k:(v+3) for k,v in word_index.items()}
word_index["<PAD>"] = 0
word_index["<START>"] = 1
word_index["<UNK>"] = 2 # unknown
word_index["<UNUSED>"] = 3
reverse_word_index = dict([(value, key) for (key, value) in word_index.items()])
def decode_review(text):
return ' '.join([reverse_word_index.get(i, '?') for i in text])
```
Agora, podemos usar a função `decode_review` para mostrar o texto da primeira avaliação:
```
decode_review(train_data[0])
```
## Prepare os dados
As avaliações—o *arrays* de inteiros— deve ser convertida em tensores (*tensors*) antes de alimentar a rede neural. Essa conversão pode ser feita de duas formas:
* Converter os arrays em vetores de 0s e 1s indicando a ocorrência da palavra, similar com *one-hot encoding*. Por exemplo, a sequência [3, 5] se tornaria um vetor de 10000 dimensões, onde todos seriam 0s, tirando 3 would become a 10,000-dimensional vector that is all zeros except for indices 3 and 5, which are ones. Then, make this the first layer in our network—a Dense layer—that can handle floating point vector data. This approach is memory intensive, though, requiring a `num_words * num_reviews` size matrix.
* Alternatively, we can pad the arrays so they all have the same length, then create an integer tensor of shape `max_length * num_reviews`. We can use an embedding layer capable of handling this shape as the first layer in our network.
In this tutorial, we will use the second approach.
Since the movie reviews must be the same length, we will use the [pad_sequences](https://keras.io/preprocessing/sequence/#pad_sequences) function to standardize the lengths:
```
train_data = keras.preprocessing.sequence.pad_sequences(train_data,
value=word_index["<PAD>"],
padding='post',
maxlen=256)
test_data = keras.preprocessing.sequence.pad_sequences(test_data,
value=word_index["<PAD>"],
padding='post',
maxlen=256)
```
Let's look at the length of the examples now:
```
len(train_data[0]), len(train_data[1])
```
And inspect the (now padded) first review:
```
print(train_data[0])
```
## Construindo o modelo
A rede neural é criada por camadas empilhadas —isso necessita duas decisões arquiteturais principais:
* Quantas camadas serão usadas no modelo?
* Quantas *hidden units* são usadas em cada camada?
Neste exemplo, os dados de entrada são um *array* de palavras-índices. As *labels* para predizer são ou 0 ou 1. Vamos construir um modelo para este problema:
```
# O formato de entrada é a contagem vocabulário usados pelas avaliações dos filmes (10000 palavras)
vocab_size = 10000
model = keras.Sequential()
model.add(keras.layers.Embedding(vocab_size, 16))
model.add(keras.layers.GlobalAveragePooling1D())
model.add(keras.layers.Dense(16, activation=tf.nn.relu))
model.add(keras.layers.Dense(1, activation=tf.nn.sigmoid))
model.summary()
```
As camadas são empilhadas sequencialmente para construir o classificador:
1. A primeira camada é uma camada `Embedding` layer (*`Embedding` layer*). Essa camada pega o vocabulário em inteiros e olha o vetor *embedding* em cada palavra-index. Esses vetores são aprendidos pelo modelo, ao longo do treinamento. Os vetores adicionam a dimensão ao *array* de saída. As dimensões resultantes são: `(batch, sequence, embedding)`.
2. Depois, uma camada `GlobalAveragePooling1D` retorna um vetor de saída com comprimento fixo para cada exemplo fazendo a média da sequência da dimensão. Isso permite o modelo de lidar com entradas de tamanhos diferentes da maneira mais simples possível.
3. Esse vetor de saída com tamanho fixo passa por uma camada *fully-connected* (`Dense`) layer com 16 *hidden units*.
4. A última camada é uma *densely connected* com um único nó de saída. Usando uma função de ativação `sigmoid`, esse valor é um float que varia entre 0 e 1, representando a probabilidade, ou nível de confiança.
### Hidden units
O modelo abaixo tem duas camadas intermediárias ou _"hidden"_ (hidden layers), entre a entrada e saída. O número de saídas (unidades— *units*—, nós ou neurônios) é a dimensão do espaço representacional para a camada. Em outras palavras, a quantidade de liberdade que a rede é permitida enquanto aprende uma representação interna.
Se o modelo tem mais *hidden units* (um espaço representacional de maior dimensão), e/ou mais camadas, então a rede pode aprender representações mais complexas. Entretanto, isso faz com que a rede seja computacionamente mais custosa e pode levar o aprendizado de padrões não desejados— padrões que melhoram a performance com os dados de treinamento, mas não com os de teste. Isso se chama *overfitting*, e exploraremos mais tarde.
### Função Loss e otimizadores (optimizer)
O modelo precisa de uma função *loss* e um otimizador (*optimizer*) para treinamento. Já que é um problema de classificação binário e o modelo tem com saída uma probabilidade (uma única camada com ativação sigmoide), usaremos a função loss `binary_crossentropy`.
Essa não é a única escolha de função loss, você poderia escolher, no lugar, a `mean_squared_error`. Mas, geralmente, `binary_crossentropy` é melhor para tratar probabilidades— ela mede a "distância" entre as distribuições de probabilidade, ou, no nosso caso, sobre a distribuição real e as previsões.
Mais tarde, quando explorarmos problemas de regressão (como, predizer preço de uma casa), veremos como usar outra função loss chamada *mean squared error*.
Agora, configure o modelo para usar o *optimizer* a função loss:
```
model.compile(optimizer='adam',
loss='binary_crossentropy',
metrics=['acc'])
```
### Crie um conjunto de validação
Quando treinando. queremos checar a acurácia do modelo com os dados que ele nunca viu. Crie uma conjunto de *validação* tirando 10000 exemplos do conjunto de treinamento original. (Por que não usar o de teste agora? Nosso objetivo é desenvolver e melhorar (tunar) nosso modelo usando somente os dados de treinamento, depois usar o de teste uma única vez para avaliar a acurácia).
```
x_val = train_data[:10000]
partial_x_train = train_data[10000:]
y_val = train_labels[:10000]
partial_y_train = train_labels[10000:]
```
## Treine o modelo
Treine o modelo em 40 *epochs* com *mini-batches* de 512 exemplos. Essas 40 iterações sobre todos os exemplos nos tensores `x_train` e `y_train`. Enquanto treina, monitore os valores do loss e da acurácia do modelo nos 10000 exemplos do conjunto de validação:
```
history = model.fit(partial_x_train,
partial_y_train,
epochs=40,
batch_size=512,
validation_data=(x_val, y_val),
verbose=1)
```
## Avalie o modelo
E vamos ver como o modelo se saiu. Dois valores serão retornados. Loss (um número que representa o nosso erro, valores mais baixos são melhores), e acurácia.
```
results = model.evaluate(test_data, test_labels)
print(results)
```
Está é uma aproximação ingênua que conseguiu uma acurácia de 87%. Com mais abordagens avançadas, o modelo deve chegar em 95%.
## Crie um gráfico de acurácia e loss por tempo
`model.fit()` retorna um objeto `History` que contém um dicionário de tudo o que aconteceu durante o treinamento:
```
history_dict = history.history
history_dict.keys()
```
Tem 4 entradas: uma para cada métrica monitorada durante a validação e treinamento. Podemos usá-las para plotar a comparação do loss de treinamento e validação, assim como a acurácia de treinamento e validação:
```
import matplotlib.pyplot as plt
acc = history_dict['acc']
val_acc = history_dict['val_acc']
loss = history_dict['loss']
val_loss = history_dict['val_loss']
epochs = range(1, len(acc) + 1)
# "bo" de "blue dot" ou "ponto azul"
plt.plot(epochs, loss, 'bo', label='Training loss')
# b de "solid blue line" "linha azul"
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
plt.clf() # limpa a figura
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend()
plt.show()
```
No gráfico, os pontos representam o loss e acurácia de treinamento, e as linhas são o loss e a acurácia de validação.
Note que o loss de treinamento *diminui* a cada *epoch* e a acurácia *aumenta*. Isso é esperado quando usado um gradient descent optimization—ele deve minimizar a quantidade desejada a cada iteração.
Esse não é o caso do loss e da acurácia de validação— eles parecem ter um pico depois de 20 epochs. Isso é um exemplo de *overfitting*: o modelo desempenha melhor nos dados de treinamento do que quando usado com dados nunca vistos. Depois desse ponto, o modelo otimiza além da conta e aprende uma representação *especifica* para os dados de treinamento e não *generaliza* para os dados de teste.
Para esse caso particular, podemos prevenir o *overfitting* simplesmente parando o treinamento após mais ou menos 20 epochs. Depois, você verá como fazer isso automaticamente com um *callback*.
|
github_jupyter
|
# Exploratory data analysis
Exploratory data analysis is an important part of any data science projects. According to [Forbs](https://www.forbes.com/sites/gilpress/2016/03/23/data-preparation-most-time-consuming-least-enjoyable-data-science-task-survey-says/?sh=67e543e86f63), it accounts for about 80% of the work of data scientists. Thus, we are going to pay out attention to that part.
In the notebook are given data description, cleaning, variables preparation, and CTR calculation and visualization.
---
```
import pandas as pd
import random
import seaborn as sns
import matplotlib.pyplot as plt
import gc
%matplotlib inline
```
Given that file occupies 5.9G and has 40 mln rows we are going to read only a few rows to glimpse at data.
```
filename = 'data/train.csv'
!echo 'Number of lines in "train.csv":'
!wc -l {filename}
!echo '"train.csv" file size:'
!du -h {filename}
dataset_5 = pd.read_csv('data/train.csv', nrows=5)
dataset_5.head()
print("Number of columns: {}\n".format(dataset_5.shape[1]))
```
---
## Data preparation
* Column `Hour` has a format `YYMMDDHH` and has to be converted.
* It is necessary to load only `click` and `hour` columns for `CTR` calculation.
* For data exploration purposes we also calculate `hour` and build distributions of `CTR` by `hour` and `weekday`
---
```
pd.to_datetime(dataset_5['hour'], format='%y%m%d%H')
# custom_date_parser = lambda x: pd.datetime.strptime(x, '%y%m%d%H')
# The commented part is for preliminary analysis and reads only 10% of data
# row_num = 40428967
# to read 10% of data
# skip = sorted(random.sample(range(1, row_num), round(0.9 * row_num)))
# data_set = pd.read_csv('data/train.csv',
# header=0,
# skiprows=skip,
# usecols=['click', 'hour'])
data_set = pd.read_csv('data/train.csv',
header=0,
usecols=['click', 'hour'])
data_set['hour'] = pd.to_datetime(data_set['hour'], format='%y%m%d%H')
data_set.isna().sum()
data_set.shape
round(100 * data_set.click.value_counts() / data_set.shape[0])
data_set.hour.dt.date.unique()
```
### Data preparation for CTR time series graph
```
df_CTR = data_set.groupby('hour').agg({
'click': ['count', 'sum']
}).reset_index()
df_CTR.columns = ['hour', 'impressions', 'clicks']
df_CTR['CTR'] = df_CTR['clicks'] / df_CTR['impressions']
del data_set; gc.collect();
from pandas.plotting import register_matplotlib_converters
register_matplotlib_converters()
plt.figure(figsize=[16, 8])
sns.lineplot(x='hour', y='CTR', data=df_CTR, linewidth=3)
plt.title('Hourly CTR for period 2014/10/21 and 2014/10/30', fontsize=20)
```
### Data preparation for CTR by hours graph
```
df_CTR['h'] = df_CTR.hour.dt.hour
df_CTR_h = df_CTR[['h', 'impressions',
'clicks']].groupby('h').sum().reset_index()
df_CTR_h['CTR'] = df_CTR_h['clicks'] / df_CTR_h['impressions']
df_CTR_h_melt = pd.melt(df_CTR_h,
id_vars='h',
value_vars=['impressions', 'clicks'],
value_name='count',
var_name='type')
plt.figure(figsize=[16, 8])
sns.set_style("white")
g1 = sns.barplot(x='h',
y='count',
hue='type',
data=df_CTR_h_melt,
palette="deep")
g1.legend(loc=1).set_title(None)
ax2 = plt.twinx()
sns.lineplot(x='h',
y='CTR',
data=df_CTR_h,
palette="deep",
marker='o',
ax=ax2,
label='CTR',
linewidth=5,
color='lightblue')
plt.title('CTR, Number of Imressions and Clicks by hours', fontsize=20)
ax2.legend(loc=5)
plt.tight_layout()
```
### Data preparation for CTR by weekday graph
```
df_CTR['weekday'] = df_CTR.hour.dt.day_name()
df_CTR['weekday_num'] = df_CTR.hour.dt.weekday
df_CTR_w = df_CTR[['weekday', 'impressions',
'clicks']].groupby('weekday').sum().reset_index()
df_CTR_w['CTR'] = df_CTR_w['clicks'] / df_CTR_w['impressions']
df_CTR_w_melt = pd.melt(df_CTR_w,
id_vars='weekday',
value_vars=['impressions', 'clicks'],
value_name='count',
var_name='type')
plt.figure(figsize=[16, 8])
sns.set_style("white")
g1 = sns.barplot(x='weekday',
y='count',
hue='type',
data=df_CTR_w_melt.sort_values('weekday'),
palette="deep",
order=[
'Monday', 'Tuesday', 'Wednesday', 'Thursday', 'Friday',
'Saturday', 'Sunday'
])
g1.legend(loc=1).set_title(None)
ax2 = plt.twinx()
sns.lineplot(x='weekday',
y='CTR',
data=df_CTR.sort_values(by='weekday_num'),
palette="deep",
marker='o',
ax=ax2,
label='CTR',
linewidth=5,
sort=False)
plt.title('CTR, Number of Imressions and Clicks by weekday', fontsize=20)
ax2.legend(loc=5)
plt.tight_layout()
```
### Normality test
```
from scipy.stats import normaltest, shapiro
def test_interpretation(stat, p, alpha=0.05):
"""
Outputs the result of statistical test comparing test-statistic and p-value
"""
print('Statistics=%.3f, p-value=%.3f, alpha=%.2f' % (stat, p, alpha))
if p > alpha:
print('Sample looks like from normal distribution (fail to reject H0)')
else:
print('Sample is not from Normal distribution (reject H0)')
stat, p = shapiro(df_CTR.CTR)
test_interpretation(stat, p)
stat, p = normaltest(df_CTR.CTR)
test_interpretation(stat, p)
```
---
## Summary
* Number of rows: 40428967
* Date duration: 10 days between 2014/10/21 and 2014/10/30. Each day has 24 hours
* No missing values in variables `click` and `hour`
* For simplicity, analysis is provided for 10% of the data. And as soon as the notebook is finalized, it will be re-run for all available data. And as soon as the hour aggregation takes place, the raw data source is deleted
* Three graphs are provided:
* CTR time serirs for all data duration
* CTR, impressions, and click counts by hour
* CTR, impressions, and click counts by weekday
* Average `CTR` value is **17%**
* Most of the `Impressions` and `Clicks` are appeared on Tuesday, Wednesday and Thursday. But highest `CTR` values is on Monday and Sunday
* The normality in `CTR` time-series is **rejected** by two tests
---
## Hypothesis:
There is a seasonality in `CTR` by an `hour` and `weekday`. For instance, `CTR` at hour 21 is lower than `CTR` at hour 14 which can be observed from graphs. Ideally, it is necessary to use 24-hour lag for anomaly detection. It can be implemented by comparing, for instance, hour 1 at day 10 with an average value of hour 1 at days 3, 4, 5, 6, 7, 8, 9 (one week), etc. One week is chosen because averaging of whole week smooth weekday seasonality: Monday and Sunday are different from Tuesday and Wednesday, but there is no big difference between whole weeks. Additional improvement can be done by the use of the median for central tendency instead of a simple averaging because averaging is biased towards abnormal values.
```
# save the final aggregated data frame to use for anomaly detection in the corresponding notebook
df_CTR.to_pickle('./data/CTR_aggregated.pkl')
```
|
github_jupyter
|
# TensorFlow Fold Quick Start
TensorFlow Fold is a library for turning complicated Python data structures into TensorFlow Tensors.
```
# boilerplate
import random
import tensorflow as tf
sess = tf.InteractiveSession()
import tensorflow_fold as td
```
The basic elements of Fold are *blocks*. We'll start with some blocks that work on simple data types.
```
scalar_block = td.Scalar()
vector3_block = td.Vector(3)
```
Blocks are functions with associated input and output types.
```
def block_info(block):
print("%s: %s -> %s" % (block, block.input_type, block.output_type))
block_info(scalar_block)
block_info(vector3_block)
```
We can use `eval()` to see what a block does with its input:
```
scalar_block.eval(42)
vector3_block.eval([1,2,3])
```
Not very exciting. We can compose simple blocks together with `Record`, like so:
```
record_block = td.Record({'foo': scalar_block, 'bar': vector3_block})
block_info(record_block)
```
We can see that Fold's type system is a bit richer than vanilla TF; we have tuple types! Running a record block does what you'd expect:
```
record_block.eval({'foo': 1, 'bar': [5, 7, 9]})
```
One useful thing you can do with blocks is wire them up to create pipelines using the `>>` operator, which performs function composition. For example, we can take our two tuple tensors and compose it with `Concat`, like so:
```
record2vec_block = record_block >> td.Concat()
record2vec_block.eval({'foo': 1, 'bar': [5, 7, 9]})
```
Note that because Python dicts are unordered, Fold always sorts the outputs of a record block by dictionary key. If you want to preserve order you can construct a Record block from an OrderedDict.
The whole point of Fold is to get your data into TensorFlow; the `Function` block lets you convert a TITO (Tensors In, Tensors Out) function to a block:
```
negative_block = record2vec_block >> td.Function(tf.negative)
negative_block.eval({'foo': 1, 'bar': [5, 7, 9]})
```
This is all very cute, but where's the beef? Things start to get interesting when our inputs contain sequences of indeterminate length. The `Map` block comes in handy here:
```
map_scalars_block = td.Map(td.Scalar())
```
There's no TF type for sequences of indeterminate length, but Fold has one:
```
block_info(map_scalars_block)
```
Right, but you've done the TF [RNN Tutorial](https://www.tensorflow.org/tutorials/recurrent/) and even poked at [seq-to-seq](https://www.tensorflow.org/tutorials/seq2seq/). You're a wizard with [dynamic rnns](https://www.tensorflow.org/api_docs/python/tf/nn/dynamic_rnn). What does Fold offer?
Well, how about jagged arrays?
```
jagged_block = td.Map(td.Map(td.Scalar()))
block_info(jagged_block)
```
The Fold type system is fully compositional; any block you can create can be composed with `Map` to create a sequence, or `Record` to create a tuple, or both to create sequences of tuples or tuples of sequences:
```
seq_of_tuples_block = td.Map(td.Record({'foo': td.Scalar(), 'bar': td.Scalar()}))
seq_of_tuples_block.eval([{'foo': 1, 'bar': 2}, {'foo': 3, 'bar': 4}])
tuple_of_seqs_block = td.Record({'foo': td.Map(td.Scalar()), 'bar': td.Map(td.Scalar())})
tuple_of_seqs_block.eval({'foo': range(3), 'bar': range(7)})
```
Most of the time, you'll eventually want to get one or more tensors out of your sequence, for wiring up to your particular learning task. Fold has a bunch of built-in reduction functions for this that do more or less what you'd expect:
```
((td.Map(td.Scalar()) >> td.Sum()).eval(range(10)),
(td.Map(td.Scalar()) >> td.Min()).eval(range(10)),
(td.Map(td.Scalar()) >> td.Max()).eval(range(10)))
```
The general form of such functions is `Reduce`:
```
(td.Map(td.Scalar()) >> td.Reduce(td.Function(tf.multiply))).eval(range(1,10))
```
If the order of operations is important, you should use `Fold` instead of `Reduce` (but if you can use `Reduce` you should, because it will be faster):
```
((td.Map(td.Scalar()) >> td.Fold(td.Function(tf.divide), tf.ones([]))).eval(range(1,5)),
(td.Map(td.Scalar()) >> td.Reduce(td.Function(tf.divide), tf.ones([]))).eval(range(1,5))) # bad, not associative!
```
Now, let's do some learning! This is the part where "magic" happens; if you want a deeper understanding of what's happening here you might want to jump right to our more formal [blocks tutorial](https://github.com/tensorflow/fold/blob/master/tensorflow_fold/g3doc/blocks.md) or learn more about [running blocks in TensorFlow](https://github.com/tensorflow/fold/blob/master/tensorflow_fold/g3doc/running.md)
```
def reduce_net_block():
net_block = td.Concat() >> td.FC(20) >> td.FC(1, activation=None) >> td.Function(lambda xs: tf.squeeze(xs, axis=1))
return td.Map(td.Scalar()) >> td.Reduce(net_block)
```
The `reduce_net_block` function creates a block (`net_block`) that contains a two-layer fully connected (FC) network that takes a pair of scalar tensors as input and produces a scalar tensor as output. This network gets applied in a binary tree to reduce a sequence of scalar tensors to a single scalar tensor.
One thing to notice here is that we are calling [`tf.squeeze`](https://www.tensorflow.org/versions/r1.0/api_docs/python/array_ops/shapes_and_shaping#squeeze) with `axis=1`, even though the Fold output type of `td.FC(1, activation=None)` (and hence the input type of the enclosing `Function` block) is a `TensorType` with shape `(1)`. This is because all Fold blocks actually run on TF tensors with an implicit leading batch dimension, which enables execution via [*dynamic batching*](https://arxiv.org/abs/1702.02181). It is important to bear this in mind when creating `Function` blocks that wrap functions that are not applied elementwise.
```
def random_example(fn):
length = random.randrange(1, 10)
data = [random.uniform(0,1) for _ in range(length)]
result = fn(data)
return data, result
```
The `random_example` function generates training data consisting of `(example, fn(example))` pairs, where `example` is a random list of numbers, e.g.:
```
random_example(sum)
random_example(min)
def train(fn, batch_size=100):
net_block = reduce_net_block()
compiler = td.Compiler.create((net_block, td.Scalar()))
y, y_ = compiler.output_tensors
loss = tf.nn.l2_loss(y - y_)
train = tf.train.AdamOptimizer().minimize(loss)
sess.run(tf.global_variables_initializer())
validation_fd = compiler.build_feed_dict(random_example(fn) for _ in range(1000))
for i in range(2000):
sess.run(train, compiler.build_feed_dict(random_example(fn) for _ in range(batch_size)))
if i % 100 == 0:
print(i, sess.run(loss, validation_fd))
return net_block
```
Now we're going to train a neural network to approximate a reduction function of our choosing. Calling `eval()` repeatedly is super-slow and cannot exploit batch-wise parallelism, so we create a [`Compiler`](https://github.com/tensorflow/fold/blob/master/tensorflow_fold/g3doc/py/td.md#compiler). See our page on [running blocks in TensorFlow](https://github.com/tensorflow/fold/blob/master/tensorflow_fold/g3doc/running.md) for more on Compilers and how to use them effectively.
```
sum_block = train(sum)
sum_block.eval([1, 1])
```
Breaking news: deep neural network learns to calculate 1 + 1!!!!
Of course we've done something a little sneaky here by constructing a model that can only represent associative functions and then training it to compute an associative function. The technical term for being sneaky in machine learning is [inductive bias](https://en.wikipedia.org/wiki/Inductive_bias).
```
min_block = train(min)
min_block.eval([2, -1, 4])
```
Oh noes! What went wrong? Note that we trained our network to compute `min` on positive numbers; negative numbers are outside of its input distribution.
```
min_block.eval([0.3, 0.2, 0.9])
```
Well, that's better. What happens if you train the network on negative numbers as well as on positives? What if you only train on short lists and then evaluate the net on long ones? What if you used a `Fold` block instead of a `Reduce`? ... Happy Folding!
|
github_jupyter
|
<table class="ee-notebook-buttons" align="left">
<td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/Image/connected_pixel_count.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
<td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Image/connected_pixel_count.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
<td><a target="_blank" href="https://mybinder.org/v2/gh/giswqs/earthengine-py-notebooks/master?filepath=Image/connected_pixel_count.ipynb"><img width=58px src="https://mybinder.org/static/images/logo_social.png" />Run in binder</a></td>
<td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Image/connected_pixel_count.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
</table>
## Install Earth Engine API and geemap
Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
**Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).
```
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as emap
except:
import geemap as emap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
```
## Create an interactive map
The default basemap is `Google Satellite`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py#L13) can be added using the `Map.add_basemap()` function.
```
Map = emap.Map(center=[40,-100], zoom=4)
Map.add_basemap('ROADMAP') # Add Google Map
Map
```
## Add Earth Engine Python script
```
# Add Earth Engine dataset
# Image.ConnectedPixelCount example.
# Split pixels of band 01 into "bright" (arbitrarily defined as
# reflectance > 0.3) and "dim". Highlight small (<30 pixels)
# standalone islands of "bright" or "dim" type.
img = ee.Image('MODIS/006/MOD09GA/2012_03_09') \
.select('sur_refl_b01') \
.multiply(0.0001)
# Create a threshold image.
bright = img.gt(0.3)
# Compute connected pixel counts stop searching for connected pixels
# once the size of the connected neightborhood reaches 30 pixels, and
# use 8-connected rules.
conn = bright.connectedPixelCount(**{
'maxSize': 30,
'eightConnected': True
})
# Make a binary image of small clusters.
smallClusters = conn.lt(30)
Map.setCenter(-107.24304, 35.78663, 8)
Map.addLayer(img, {'min': 0, 'max': 1}, 'original')
Map.addLayer(smallClusters.updateMask(smallClusters),
{'min': 0, 'max': 1, 'palette': 'FF0000'}, 'cc')
```
## Display Earth Engine data layers
```
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
```
|
github_jupyter
|
# Experimental Mathimatics: Chronicle of Matlab code - 2008 - 2015
##### Whereas the discovery of Chaos, Fractal Geometry and Non-Linear Dynamical Systems falls outside the domain of analytic function in mathematical terms the path to discovery is taken as experimental computer-programming.
##### Whereas existing discoveries have been most delightfully difference equations this first effor concentrates on guided random search for equations and parameters of that type.
### equation and parameters data were saved in tiff file headers, matlab/python extracted to:
```
import os
import pandas as pd
spreadsheets_directory = '../data/Matlab_Chronicle_2008-2012/'
images_dataframe_filename = os.path.join(spreadsheets_directory, 'Of_tiff_headers.df')
equations_dataframe_filename = os.path.join(spreadsheets_directory, 'Of_m_files.df')
Images_Chronicle_df = pd.read_csv(images_dataframe_filename, sep='\t', index_col=0)
Equations_Chronicle_df = pd.read_csv(equations_dataframe_filename, sep='\t', index_col=0)
def get_number_of_null_parameters(df, print_out=True):
""" Usage good_null_bad_dict = get_number_of_null_parameters(df, print_out=True)
function to show the number of Images with missing or bad parameters
because they are only reproducable with both the equation and parameter set
Args:
df = dataframe in format of historical images - (not shown in this notebook)
Returns:
pars_contidtion_dict:
pars_contidtion_dict['good_pars']: number of good parametrs
pars_contidtion_dict['null_pars']: number of null parameters
pars_contidtion_dict['bad_list']: list of row numbers with bad parameters - for numeric indexing
"""
null_pars = 0
good_pars = 0
bad_list = []
for n, row in df.iterrows():
if row['parameters'] == []:
null_pars += 1
bad_list.append(n)
else:
good_pars += 1
if print_out:
print('good_pars', good_pars, '\nnull_pars', null_pars)
return {'good_pars': good_pars, 'null_pars': null_pars, 'bad_list': bad_list}
def display_images_df_columns_definition():
""" display an explanation of the images """
cols = {}
cols['image_filename'] = 'the file name as found in the header'
cols['function_name'] = 'm-file and function name'
cols['parameters'] = 'function parameters used to produce the image'
cols['max_iter'] = 'escape time algorithm maximum number of iterations'
cols['max_dist'] = 'escape time algorithm quit distance'
cols['Colormap'] = 'Name of colormap if logged'
cols['Center'] = 'center of image location on complex plane'
cols['bounds_box'] = '(upper left corner) ULC, URC, LLC, LRC'
cols['Author'] = 'author name if provied'
cols['location'] = 'file - subdirectory location'
cols['date'] = 'date the image file was written'
for k, v in cols.items():
print('%18s: %s'%(k,v))
def display_equations_df_columns_definition():
""" display an explanation of the images """
cols = {}
cols['arg_in'] = 'Input signiture of the m-file'
cols['arg_out'] = 'Output signiture of the m-file'
cols['eq_string'] = 'The equation as written in MATLAB'
cols['while_test'] = 'The loop test code'
cols['param_iterator'] = 'If parameters are iterated in the while loop'
cols['internal_vars'] = 'Variables that were set inside the m-file'
cols['while_lines'] = 'The actual code of the while loop'
for k, v in cols.items():
print('%15s: %s'%(k,v))
print('\n\tdisplay_images_df_columns_definition\n')
display_images_df_columns_definition()
print('\n\tdisplay_equations_df_columns_definition\n')
display_equations_df_columns_definition()
print('shape:',
Images_Chronicle_df.shape,
'Number of unique files:',
Images_Chronicle_df['image_filename'].nunique())
print('Number of unique functions used:',
Images_Chronicle_df['function_name'].nunique())
par_stats_dict = get_number_of_null_parameters(Images_Chronicle_df) # show parameters data
print('\nFirst 5 lines')
Images_Chronicle_df.head() # show top 5 lines
print(Equations_Chronicle_df.shape)
Equations_Chronicle_df.head()
cols = list(Equations_Chronicle_df.columns)
for c in cols:
print(c)
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/DJCordhose/ux-by-tfjs/blob/master/notebooks/click-sequence-model.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Training on Sequences of Clicks on the Server
Make sure to run this from top to bottom to export model, otherwise names of layers cause tf.js to bail out
```
# Gives us a well defined version of tensorflow
try:
# %tensorflow_version only exists in Colab.
%tensorflow_version 2.x
except Exception:
pass
import tensorflow as tf
print(tf.__version__)
# a small sanity check, does tf seem to work ok?
hello = tf.constant('Hello TF!')
print("This works: {}".format(hello))
# this should return True even on Colab
tf.test.is_gpu_available()
tf.test.is_built_with_cuda()
tf.executing_eagerly()
```
## load data
```
import pandas as pd
print(pd.__version__)
import numpy as np
print(np.__version__)
# local
# URL = '../data/click-sequence.json'
# remote
URL = 'https://raw.githubusercontent.com/DJCordhose/ux-by-tfjs/master//data/click-sequence.json'
df = pd.read_json(URL, typ='series')
len(df)
df.head()
type(df[0])
df[0], df[1]
all_buttons = set()
for seq in df:
for button in seq:
all_buttons.add(button)
all_buttons.add('<START>')
all_buttons.add('<EMPTY>')
all_buttons = list(all_buttons)
all_buttons
from sklearn.preprocessing import LabelBinarizer, MultiLabelBinarizer, LabelEncoder
encoder = LabelEncoder()
encoder.fit(all_buttons)
encoder.classes_
transfomed_labels = [encoder.transform(seq) for seq in df if len(seq) != 0]
transfomed_labels
```
## pre-process data into chunks
```
empty = encoder.transform(['<EMPTY>'])
start = encoder.transform(['<START>'])
chunk_size = 5
# [ 1, 11, 7, 11, 6] => [[[0, 0, 0, 0, 1], 11], [[0, 0, 0, 1, 11], 7], [[0, 0, 1, 11, 7], 11], [[0, 1, 11, 7, 11], 6]]
def create_sequences(seq, chunk_size, empty, start):
# all sequences implicitly start
seq = np.append(start, seq)
# if sequence is too short, we pad it to minimum size at the beginning
seq = np.append(np.full(chunk_size - 1, empty), seq)
seqs = np.array([])
for index in range(chunk_size, len(seq)):
y = seq[index]
x = seq[index-chunk_size : index]
seqs = np.append(seqs, [x, y])
return seqs
# seq = transfomed_labels[0]
# seq = transfomed_labels[9]
seq = transfomed_labels[3]
seq
create_sequences(seq, chunk_size, empty, start)
seqs = np.array([])
for seq in transfomed_labels:
seqs = np.append(seqs, create_sequences(seq, chunk_size, empty, start))
seqs = seqs.reshape(-1, 2)
seqs.shape
X = seqs[:, 0]
# X = X.reshape(-1, chunk_size)
X = np.vstack(X).astype('int32')
X.dtype, X.shape
y = seqs[:, 1].astype('int32')
y.dtype, y.shape, y
```
## Training
```
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.layers import Dense, LSTM, GRU, SimpleRNN, Embedding, BatchNormalization, Dropout
from tensorflow.keras.models import Sequential, Model
embedding_dim = 2
n_buttons = len(encoder.classes_)
dropout = .6
recurrent_dropout = .6
model = Sequential()
model.add(Embedding(name='embedding',
input_dim=n_buttons,
output_dim=embedding_dim,
input_length=chunk_size))
model.add(SimpleRNN(units=50, activation='relu', name="RNN", recurrent_dropout=recurrent_dropout))
# model.add(GRU(units=25, activation='relu', name="RNN", recurrent_dropout=0.5))
# model.add(LSTM(units=25, activation='relu', name="RNN", recurrent_dropout=0.5))
model.add(BatchNormalization())
model.add(Dropout(dropout))
model.add(Dense(units=n_buttons, name='softmax', activation='softmax'))
model.compile(optimizer='adam', loss='sparse_categorical_crossentropy', metrics=['accuracy'])
model.summary()
%%time
EPOCHS = 1000
BATCH_SIZE = 100
history = model.fit(X, y,
batch_size=BATCH_SIZE,
epochs=EPOCHS, verbose=0, validation_split=0.2)
loss, accuracy = model.evaluate(X, y, batch_size=BATCH_SIZE)
accuracy
%matplotlib inline
import matplotlib.pyplot as plt
# plt.yscale('log')
plt.ylabel('loss')
plt.xlabel('epochs')
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.legend(['loss', 'val_loss'])
plt.ylabel('accuracy')
plt.xlabel('epochs')
# TF 2.0
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
# plt.plot(history.history['acc'])
# plt.plot(history.history['val_acc'])
plt.legend(['accuracy', 'val_accuracy'])
model.predict([[X[0]]])
model.predict([[X[0]]]).argmax()
y[0]
y_pred = model.predict([X]).argmax(axis=1)
y_pred
y
# TF 2.0
cm = tf.math.confusion_matrix(labels=tf.constant(y, dtype=tf.int64), predictions=tf.constant(y_pred, dtype=tf.int64))
cm
import seaborn as sns
classes = encoder.classes_
plt.figure(figsize=(8, 8))
sns.heatmap(cm, annot=True, fmt="d", xticklabels=classes, yticklabels=classes);
embedding_layer = model.get_layer('embedding')
embedding_model = Model(inputs=model.input, outputs=embedding_layer.output)
embeddings_2d = embedding_model.predict(X)
embeddings_2d.shape
encoder.classes_
encoded_classes = encoder.transform(encoder.classes_)
same_button_seqs = np.repeat(encoded_classes, 5).reshape(14, 5)
embeddings_2d = embedding_model.predict(same_button_seqs)
embeddings_2d.shape
only_first = embeddings_2d[:, 0, :]
only_first
# for printing only
plt.figure(figsize=(10,10))
# plt.figure(dpi=600)
# plt.figure(dpi=300)
plt.axis('off')
plt.scatter(only_first[:, 0], only_first[:, 1], s=200)
for name, x_pos, y_pos in zip(encoder.classes_, only_first[:, 0], only_first[:, 1]):
# print(name, (x_pos, y_pos))
plt.annotate(name, (x_pos, y_pos), rotation=-60, size=25)
from sklearn.decomposition import PCA
import numpy as np
embeddings_1d = PCA(n_components=1).fit_transform(only_first)
# for printing only
plt.figure(figsize=(25,5))
# plt.figure(dpi=300)
plt.axis('off')
plt.scatter(embeddings_1d, np.zeros(len(embeddings_1d)), s=80)
for name, x_pos in zip(encoder.classes_, embeddings_1d):
plt.annotate(name, (x_pos, 0), rotation=-45)
```
## Convert Model into tfjs format
* https://www.tensorflow.org/js/tutorials/conversion/import_keras
```
!pip install -q tensorflowjs
model.save('ux.h5', save_format='h5')
!ls -l
!tensorflowjs_converter --input_format keras ux.h5 tfjs
!ls -l tfjs
```
Download using _Files_ menu on the left
```
```
|
github_jupyter
|
### Introduction
This is an instruction set for running SQUID simulations using a handful of packages writen in python 3. Each package is created around a SQUID model and includes a solver and some utilities to use the solver indirectly to produce more complicated output.
This tutorial will walk through using the **noisy_squid.py** package. Includeded in each SQUID package are:
**basic model** - gives timeseries output of the state of the SQUID
**vj_timeseries()** - gives a plot and csv file of the timeseries state of the SQUID
**iv_curve()** - gives contour plot (discrete values of one parameter) of I-V curve and csv file
**vphi_curve()** - gives countour plot (discrete values of one parameter) of V-PHI curve and csv file
**transfer_fn()** - gives plots of average voltage surface and transfer function surface as well as csv files for each. Returns array of average voltage over the input parameter space in i and phia
Associated with each package:
**dev_package_name.ipynb** - a jupyter notebook used in develping model and utilities
**package_name.ipynb** - a refined jupyter notebook with helpful explanations of how the model and utilities work
**package_name.py** - a python 3 file containing only code, inteded for import into python session of some sort
### Packages
There are four packages, **noisy_single_junction.py**, **quiet_squid.py**, **noisy_squid.py**, and **noisy_squid_2nd_order.py**. The first is somewhat separate and documentation is provided in the jupyter notebook for that file.
Each package requires simulation inputs, namely the time step size tau, the number of steps to simulate **nStep**, an initial state vector **s**. Also required are a set of input parameters **i** and **phia**, and physical parameters **alpha**, **betaL**, **eta**, **rho** at a minimum. The two more detailed solvers require a noise parameter **Gamma**, and perhaps two more parameters **betaC** and **kappa**. These are all supplied as one array of values called par.
The other three are useful to simulate a two-junction SQUID in different circumstances.
**quiet_squid.py** simulates a two-junction SQUID with no thermal noise and no appreciable shot noise. This model is first order and so does not account for capacitive effects. This model assumes no noise, and no capacitance. Use this model if the dynamics of the system without noise are to be investigated.
**noisy_squid.py** is similar to the above, but includes the effects of thermal Johnson noise in the junctions. This model is also first order, assuming negligible effects from capacitance. It is sometimes necessary and or safe to assume negligible effects from capacitance, or zero capacitance. It will be necessary to use the first order model in this case, as setting capacitance to zero in the second order model will result in divide by zero errors.
**noisy_squid_2nd_order.py** is similar to the above, but includes second order effects due to capacitance. This model should be used if capacitance should be considered non-zero.
#### Prerequisites
The only prerequisite is to have python 3 installed on your machine. You may prefer to use some python environment like jupyter notebook or Spyder. The easiest way to get everything is by downloading and installing the latest distribution of **Anaconda**, which will install python 3, juputer notebook, and Spyder as well as providing convenient utilities for adding community provided libraries. Anaconda download link provided:
https://www.anaconda.com/distribution/
Working from the console is easy enough, but working in your prefered development environment will be easier.
### Prepare a file for output
This tutorial will work out of the console, but the same commands will work for a development environment.
Create a file folder in a convenient location on your computer. Place the relevant python file described above in the file. Output files including csv data files and png plots will be stored in this file folder. All file outputs are named like **somethingDatetime.csv** or .png.
### Open a python environment
These packages can be used directly from the console or within your favorite python development environment.
This tutorial will assume the user is working from the console. Open a command prompt. You can do this on Windows by typing "cmd" in the start search bar and launching **Command Prompt**. Change directory to the file folder created in the step above.
***cd "file\tree\folder"***
With the command prompt indicating you are in the folder, type "python" and hit enter. If there are multiple instances (different iterations) of python on your machine, this may need to be accounted for in launching the correct version. See the internet for help on this.
If you have a favorite python environment, be sure to launch in the folder or change the working directory of the development environment to the folder you created. If you do not wish to change the working directory, place the package .py file in the working directory.
### Load the relevant package
With python running, at the command prompt in the console, import the python file containing the model needed.
In this tutorial we will use the first order model including noise, **noisy_squid.py**. Type "import noisy_squid". Execute the command by hitting enter on the console. It may be easier to give the package a nickname, as we will have to type it every time we call a function within it. Do this by instead typing "import noisy_squid as nickname", as below.
```
import noisy_squid as ns
```
We need a standard package called **numpy** as well. This library includes some tools we need to create input, namely numpy arrays. Type "import numpy as np" and hit enter.
The code inside the packages aslo relies on other standard packages. Those are loaded from within the package.
```
import numpy as np
```
#### Getting Help
You can access the short help description of each model and utility by typing:
***?package_name.utiltiy_name()***
Example:
```
?ns.transfer_fn()
```
### noisy_squid.noisySQUID()
The model itself, **noisySQUID()** can be run and gives a timeseries output array for the state of the system.
Included in the output array are **delta_1**, **delta_2**, **j**, **v_1**, **v_2**, and **v** at each simulated moment in time.
To run, first we need to set up some parameters. To see detailed explanations of these, see the developement log in the jupyter notebook associated with the package.
Parameter definitions can be handled in the function call or defined before the function call.
An example of the former: Define values using parameter names, build a parameter array, and finally call the function. Remember, s and par are numpy arrays.
```
# define values
nStep = 8000
tau = .1
s = np.array([0.,0.])
alpha = 0.
betaL = 1.
eta = 0.
rho = 0.
i = 1.5
phia = .5
Gamma = .05
par = np.array([alpha,betaL,eta,rho,i,phia,Gamma])
```
Now we can call the simulation. Type
***noisy_squid.noisySQUID(nStep,tau,s,par)***
to call the function. We may wish to define a new variable to hold the simple array output. We can then show the output by typing the variable again. We do this below by letting **S** be the output. **S** will take on the form of the output, here an array.
```
S = noisy_squid.noisySQUID(nStep,tau,s,par)
```
The shortcut method is to call the function only, replacing varibles with values. This example includes a linebreak "\". You can just type it out all in one line if it will fit.
```
S = noisy_squid.noisySQUID(8000,.1,np.array([0.,0.]),\
np.array([0.,1.,0.,0.,1.5,.5,.05]))
```
We can check the output. Have a look at **S** by typing it.
```
S
```
This doesn't mean much. Since the voltage accross the circuit, **v**, is stored in **S** as the 7th row (index 6), we can plot the voltage timeseries. The time **theta** is stored as the first row (index 0). We will need the ploting package **matplotlib.pyplot** to do this.
```
import matplotlib.pyplot as plt
plt.plot(S[0],S[6])
plt.xlabel(r'time, $\theta$')
plt.ylabel(r'ave voltage, $v$')
```
This model will be useful if you wish to extend the existing utilites here or develop new utilities. To explore the nature of SQUID parameter configurations, it may be easier to start with the other utilities provided.
### noisy_squid.vj_timeseries()
use "**'package_name'.vj_timeseries()**" or "**'package_nickname'.vj_timeseries()**".
This utility does the same as the funciton described above, but gives a plot of the output and creates a csv of the timeseries output array. These are named **timeseries'datetime'.csv** and .png. The plot includes the voltage timeseries and the circulating current time series. The csv file contains metadata describing the parameters.
To run this, define parameters as described above, and call the function. This time,we will change the value of **nStep** to be shorter so we can see some detail in the trace.
This utility does not return anything to store. The only output is the plot and the csv file, so don't bother storing the function call output.
```
# define values
nStep = 800
tau = .1
s = np.array([0.,0.])
alpha = 0.
betaL = 1.
eta = 0.
rho = 0.
i = 1.5
phia = .5
Gamma = .05
par = np.array([alpha,betaL,eta,rho,i,phia,Gamma])
ns.vj_timeseries(nStep,tau,s,par)
```
### noisy_squid.iv_curve()
This utility is used to create plots and data of the average voltage output of the SQUID vs the applied bias current. We can sweep any of the other parameters and create contours. The utility outputs a data file and plot, **IV'datetime'.csv** and .png.
Define the parameter array as normal. The parameter to sweep will be passed separately as a list of values at which to draw contours. If a value for the parameter to sweep is passed in **par**, it will simply be ignored in favor of the values in the list.
A list is defined by square brackets and values are separated by a comma. This parameter list must be a list and not an array.
The name of the list should be different than parameter name it represents. In this case, I wish to look at three contours corresponding to values of the applied flux **phia**. I name a list **Phi** and give it values.
Place the parameter list in the function call by typing the **parameter_name=List_name**. In this case, **phia=Phi**.
This utility has no console output, so don't bother storing it in a variable.
```
Phi = [.2,.8,1.6]
ns.iv_curve(nStep,tau,s,par,phia=Phi)
```
This curve is very noisy. To get finer detail, increase **nStep** and decrease **tau**.
The underlying numerical method is accurate but slow. Computation time considerations must be considered from here on. I recommend testing a set of parameters with a small number of large steps first, then adjusting for more detail as needed.
From here on, the utilities are looking at average voltage. To get an accurate average voltage you need lots of voltage values, and thus large **nStep**. The error in the underlying Runge-Kutta 4th order method is determined by the size of the time step, smaller = less error. Thus, a more accurate timeseries is provided by a smaller time step **tau**. A more accurate timeseries will result in better convergence of the model to the expected physical output, thus finer detail.
Computation time will grow directly with the size of **nStep** but will be uneffected by the size of **tau**. If **tau** is larger than one, there will be instability in the method, it will likely not work. There is a minimum size for **tau** as well, to insure stability. Something on the order of 0.1 to 0.01 will usually suffice.
These parameters are your tradeoff control in detail vs computation time.
At any rate, the erratic effect of noise is best dampened by using a larger **nStep**.
Lets try it with 10 times as many time steps.
```
nStep = 8000
tau = .1
ns.iv_curve(nStep,tau,s,par,phia=Phi)
```
This looks better. To get a usable plot, it will probably be necessary to set **nStep** on the order of 10^4 to 10^6. Start lower if possible.
Lets try **nStep**=8\*10^5.
```
nStep = 80000
ns.iv_curve(nStep,tau,s,par,phia=Phi)
```
We can look at a sweep of a different parameter by reseting **phia** to say .2, and creating a list to represent a different parameter. Lets sweep **betaL**.
```
phia = .2
Beta = [.5,1.,2.]
ns.iv_curve(nStep,tau,s,par,betaL=Beta)
```
### noisy_squid.vphi_curve()
This utility is used to create plots and data of the average voltage output of the SQUID vs the applied magnatic flux. We can sweep any of the other parameters and create contours. The utility outputs a data file and plot, **VPhi'datetime'.csv** and .png.
Define the parameter array as normal. The parameter to sweep will be passed separately as a list of values at which to draw contours. If a value for the parameter to sweep is passed in **par**, it will simply be ignored in favor of the values in the list.
A list is defined by square brackets and values are separated by a comma. This parameter list must be a list and not an array.
The name of the list should be different than parameter name it represents. In this case, I wish to look at three contours corresponding to values of the inductance constant **betaL**. I named a list **Beta** above and gave it values.
Place the parameter list in the function call by typing the **parameter_name=List_name**. In this case, **betaL=Beta**.
This utility has no console output, so don't bother storing it in a variable.
This utility can be computationally expensive. See the notes on this in the **noisy_squid.iv_curve()** section.
```
ns.vphi_curve(nStep,tau,s,par,betaL=Beta)
```
### noisy_squid.transfer_fn()
This utility creates the average voltage surface in bias current / applied flux space. It also calculates the partial derivative of the average voltage surface with respec to applied flux and returns this as the transfer funcion. These are named **AveVsurface'datetime'.png** and .csv, and **TransferFn'datetime'.png** and .csv. This utility also returns an array of average voltage values over the surface which can be stored for further manipulation.
This utility requires us to define an axis for both **i** and **phia**. We do this by making an array for each. We can define the individual elements of the array, but there is an easier way. We can make an array of values evenly spaced across an interval using **np.arange(start, stop+step, step)** as below.
Pass the other parameters as in the instructions under **noisy_squid()**. You may want to start with a smaller value for **nStep**.
```
nStep = 800
i = np.arange(-3.,3.1,.1)
phia = np.arange(-1.,1.1,.1)
vsurf = ns.transfer_fn(nStep,tau,s,par,i,phia)
```
The average voltage surface looks ok, but not great. Noisy spots in the surface will negatively effect the transfer function determination. The transfer function surface has large derivatives in the corner which over saturate the plot hiding detail in most of the surface. To fix this, we need a truer average voltage surface. We need more time steps. If you have some time, try **nStep**=8000.
Note that it may be possible to clean the data from the csv file and recover some detail in plotting. Be careful...
```
nStep = 8000
vsurf = ns.transfer_fn(nStep,tau,s,par,i,phia)
```
|
github_jupyter
|
# Week 2: Tackle Overfitting with Data Augmentation
Welcome to this assignment! As in the previous week, you will be using the famous `cats vs dogs` dataset to train a model that can classify images of dogs from images of cats. For this, you will create your own Convolutional Neural Network in Tensorflow and leverage Keras' image preprocessing utilities, more so this time around since Keras provides excellent support for augmenting image data.
You will also need to create the helper functions to move the images around the filesystem as you did last week, so if you need to refresh your memory with the `os` module be sure to take a look a the [docs](https://docs.python.org/3/library/os.html).
Let's get started!
```
import warnings
warnings.filterwarnings('ignore')
import os
import zipfile
import random
import shutil
import tensorflow as tf
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from shutil import copyfile
import matplotlib.pyplot as plt
```
Download the dataset from its original source by running the cell below.
Note that the `zip` file that contains the images is unzipped under the `/tmp` directory.
```
# If the URL doesn't work, visit https://www.microsoft.com/en-us/download/confirmation.aspx?id=54765
# And right click on the 'Download Manually' link to get a new URL to the dataset
# Note: This is a very large dataset and will take some time to download
!wget --no-check-certificate \
"https://download.microsoft.com/download/3/E/1/3E1C3F21-ECDB-4869-8368-6DEBA77B919F/kagglecatsanddogs_3367a.zip" \
-O "/tmp/cats-and-dogs.zip"
local_zip = '/tmp/cats-and-dogs.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp')
zip_ref.close()
```
Now the images are stored within the `/tmp/PetImages` directory. There is a subdirectory for each class, so one for dogs and one for cats.
```
source_path = '/tmp/PetImages'
source_path_dogs = os.path.join(source_path, 'Dog')
source_path_cats = os.path.join(source_path, 'Cat')
# os.listdir returns a list containing all files under the given path
print(f"There are {len(os.listdir(source_path_dogs))} images of dogs.")
print(f"There are {len(os.listdir(source_path_cats))} images of cats.")
```
**Expected Output:**
```
There are 12501 images of dogs.
There are 12501 images of cats.
```
You will need a directory for cats-v-dogs, and subdirectories for training
and testing. These in turn will need subdirectories for 'cats' and 'dogs'. To accomplish this, complete the `create_train_test_dirs` below:
```
# Define root directory
root_dir = '/tmp/cats-v-dogs'
# Empty directory to prevent FileExistsError is the function is run several times
if os.path.exists(root_dir):
shutil.rmtree(root_dir)
# GRADED FUNCTION: create_train_test_dirs
def create_train_test_dirs(root_path):
### START CODE HERE
# HINT:
# Use os.makedirs to create your directories with intermediate subdirectories
# Don't hardcode the paths. Use os.path.join to append the new directories to the root_path parameter
try:
os.makedirs(os.path.join(root_dir))
os.makedirs(os.path.join(root_dir, "training"))
os.makedirs(os.path.join(root_dir, "training", "cats"))
os.makedirs(os.path.join(root_dir, "training", "dogs"))
os.makedirs(os.path.join(root_dir, "testing"))
os.makedirs(os.path.join(root_dir, "testing", "cats"))
os.makedirs(os.path.join(root_dir, "testing", "dogs"))
except:
pass
### END CODE HERE
try:
create_train_test_dirs(root_path=root_dir)
except FileExistsError:
print("You should not be seeing this since the upper directory is removed beforehand")
# Test your create_train_test_dirs function
for rootdir, dirs, files in os.walk(root_dir):
for subdir in dirs:
print(os.path.join(rootdir, subdir))
```
**Expected Output (directory order might vary):**
``` txt
/tmp/cats-v-dogs/training
/tmp/cats-v-dogs/testing
/tmp/cats-v-dogs/training/cats
/tmp/cats-v-dogs/training/dogs
/tmp/cats-v-dogs/testing/cats
/tmp/cats-v-dogs/testing/dogs
```
Code the `split_data` function which takes in the following arguments:
- SOURCE: directory containing the files
- TRAINING: directory that a portion of the files will be copied to (will be used for training)
- TESTING: directory that a portion of the files will be copied to (will be used for testing)
- SPLIT SIZE: to determine the portion
The files should be randomized, so that the training set is a random sample of the files, and the test set is made up of the remaining files.
For example, if `SOURCE` is `PetImages/Cat`, and `SPLIT` SIZE is .9 then 90% of the images in `PetImages/Cat` will be copied to the `TRAINING` dir
and 10% of the images will be copied to the `TESTING` dir.
All images should be checked before the copy, so if they have a zero file length, they will be omitted from the copying process. If this is the case then your function should print out a message such as `"filename is zero length, so ignoring."`. **You should perform this check before the split so that only non-zero images are considered when doing the actual split.**
Hints:
- `os.listdir(DIRECTORY)` returns a list with the contents of that directory.
- `os.path.getsize(PATH)` returns the size of the file
- `copyfile(source, destination)` copies a file from source to destination
- `random.sample(list, len(list))` shuffles a list
```
# GRADED FUNCTION: split_data
def split_data(SOURCE, TRAINING, TESTING, SPLIT_SIZE):
### START CODE HERE
all_files = []
for file_name in os.listdir(SOURCE):
file_path = SOURCE + file_name
if os.path.getsize(file_path):
all_files.append(file_name)
else:
print('{} is zero length, so ignoring'.format(file_name))
n_files = len(all_files)
split_point = int(n_files * SPLIT_SIZE)
shuffled = random.sample(all_files, n_files)
train_set = shuffled[:split_point]
test_set = shuffled[split_point:]
for file_name in train_set:
copyfile(SOURCE + file_name, TRAINING + file_name)
for file_name in test_set:
copyfile(SOURCE + file_name, TESTING + file_name)
### END CODE HERE
# Test your split_data function
# Define paths
CAT_SOURCE_DIR = "/tmp/PetImages/Cat/"
DOG_SOURCE_DIR = "/tmp/PetImages/Dog/"
TRAINING_DIR = "/tmp/cats-v-dogs/training/"
TESTING_DIR = "/tmp/cats-v-dogs/testing/"
TRAINING_CATS_DIR = os.path.join(TRAINING_DIR, "cats/")
TESTING_CATS_DIR = os.path.join(TESTING_DIR, "cats/")
TRAINING_DOGS_DIR = os.path.join(TRAINING_DIR, "dogs/")
TESTING_DOGS_DIR = os.path.join(TESTING_DIR, "dogs/")
# Empty directories in case you run this cell multiple times
if len(os.listdir(TRAINING_CATS_DIR)) > 0:
for file in os.scandir(TRAINING_CATS_DIR):
os.remove(file.path)
if len(os.listdir(TRAINING_DOGS_DIR)) > 0:
for file in os.scandir(TRAINING_DOGS_DIR):
os.remove(file.path)
if len(os.listdir(TESTING_CATS_DIR)) > 0:
for file in os.scandir(TESTING_CATS_DIR):
os.remove(file.path)
if len(os.listdir(TESTING_DOGS_DIR)) > 0:
for file in os.scandir(TESTING_DOGS_DIR):
os.remove(file.path)
# Define proportion of images used for training
split_size = .9
# Run the function
# NOTE: Messages about zero length images should be printed out
split_data(CAT_SOURCE_DIR, TRAINING_CATS_DIR, TESTING_CATS_DIR, split_size)
split_data(DOG_SOURCE_DIR, TRAINING_DOGS_DIR, TESTING_DOGS_DIR, split_size)
# Check that the number of images matches the expected output
print(f"\n\nThere are {len(os.listdir(TRAINING_CATS_DIR))} images of cats for training")
print(f"There are {len(os.listdir(TRAINING_DOGS_DIR))} images of dogs for training")
print(f"There are {len(os.listdir(TESTING_CATS_DIR))} images of cats for testing")
print(f"There are {len(os.listdir(TESTING_DOGS_DIR))} images of dogs for testing")
```
**Expected Output:**
```
666.jpg is zero length, so ignoring.
11702.jpg is zero length, so ignoring.
```
```
There are 11250 images of cats for training
There are 11250 images of dogs for training
There are 1250 images of cats for testing
There are 1250 images of dogs for testing
```
Now that you have successfully organized the data in a way that can be easily fed to Keras' `ImageDataGenerator`, it is time for you to code the generators that will yield batches of images, both for training and validation. For this, complete the `train_val_generators` function below.
Something important to note is that the images in this dataset come in a variety of resolutions. Luckily, the `flow_from_directory` method allows you to standarize this by defining a tuple called `target_size` that will be used to convert each image to this target resolution. **For this exercise use a `target_size` of (150, 150)**.
**Note:** So far, you have seen the term `testing` being used a lot for referring to a subset of images within the dataset. In this exercise, all of the `testing` data is actually being used as `validation` data. This is not very important within the context of the task at hand but it is worth mentioning to avoid confusion.
```
TRAINING_DIR = '/tmp/cats-v-dogs/training'
VALIDATION_DIR = '/tmp/cats-v-dogs/testing'
# GRADED FUNCTION: train_val_generators
def train_val_generators(TRAINING_DIR, VALIDATION_DIR):
### START CODE HERE
# Instantiate the ImageDataGenerator class (don't forget to set the arguments to augment the images)
train_datagen = ImageDataGenerator(rescale=1.0/255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest')
# Pass in the appropriate arguments to the flow_from_directory method
train_generator = train_datagen.flow_from_directory(directory=TRAINING_DIR,
batch_size=64,
class_mode='binary',
target_size=(150, 150))
# Instantiate the ImageDataGenerator class (don't forget to set the rescale argument)
validation_datagen = ImageDataGenerator(rescale=1.0/255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest')
# Pass in the appropriate arguments to the flow_from_directory method
validation_generator = validation_datagen.flow_from_directory(directory=VALIDATION_DIR,
batch_size=64,
class_mode='binary',
target_size=(150, 150))
### END CODE HERE
return train_generator, validation_generator
# Test your generators
train_generator, validation_generator = train_val_generators(TRAINING_DIR, TESTING_DIR)
```
**Expected Output:**
```
Found 22498 images belonging to 2 classes.
Found 2500 images belonging to 2 classes.
```
One last step before training is to define the architecture of the model that will be trained.
Complete the `create_model` function below which should return a Keras' `Sequential` model.
Aside from defining the architecture of the model, you should also compile it so make sure to use a `loss` function that is compatible with the `class_mode` you defined in the previous exercise, which should also be compatible with the output of your network. You can tell if they aren't compatible if you get an error during training.
**Note that you should use at least 3 convolution layers to achieve the desired performance.**
```
from tensorflow.keras.optimizers import RMSprop
# GRADED FUNCTION: create_model
def create_model():
# DEFINE A KERAS MODEL TO CLASSIFY CATS V DOGS
# USE AT LEAST 3 CONVOLUTION LAYERS
### START CODE HERE
model = tf.keras.models.Sequential([
tf.keras.layers.Conv2D(32, (3,3), input_shape = (150, 150, 3), activation = tf.nn.relu),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64, (3,3), activation = tf.nn.relu),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(128, (3, 3), activation = tf.nn.relu),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(512, activation = tf.nn.relu),
tf.keras.layers.Dense(128, activation = tf.nn.relu),
tf.keras.layers.Dense(1, activation = tf.nn.sigmoid)
])
model.compile( optimizer = RMSprop(lr=0.001),
loss = 'binary_crossentropy',
metrics = ['accuracy'])
### END CODE HERE
return model
```
Now it is time to train your model!
Note: You can ignore the `UserWarning: Possibly corrupt EXIF data.` warnings.
```
# Get the untrained model
model = create_model()
# Train the model
# Note that this may take some time.
history = model.fit(train_generator,
epochs=15,
verbose=1,
validation_data=validation_generator)
```
Once training has finished, you can run the following cell to check the training and validation accuracy achieved at the end of each epoch.
**To pass this assignment, your model should achieve a training and validation accuracy of at least 80% and the final testing accuracy should be either higher than the training one or have a 5% difference at maximum**. If your model didn't achieve these thresholds, try training again with a different model architecture, remember to use at least 3 convolutional layers or try tweaking the image augmentation process.
You might wonder why the training threshold to pass this assignment is significantly lower compared to last week's assignment. Image augmentation does help with overfitting but usually this comes at the expense of requiring more training time. To keep the training time reasonable, the same number of epochs as in the previous assignment are kept.
However, as an optional exercise you are encouraged to try training for more epochs and to achieve really good training and validation accuracies.
```
#-----------------------------------------------------------
# Retrieve a list of list results on training and test data
# sets for each training epoch
#-----------------------------------------------------------
acc=history.history['accuracy']
val_acc=history.history['val_accuracy']
loss=history.history['loss']
val_loss=history.history['val_loss']
epochs=range(len(acc)) # Get number of epochs
#------------------------------------------------
# Plot training and validation accuracy per epoch
#------------------------------------------------
plt.plot(epochs, acc, 'r', "Training Accuracy")
plt.plot(epochs, val_acc, 'b', "Validation Accuracy")
plt.title('Training and validation accuracy')
plt.show()
print("")
#------------------------------------------------
# Plot training and validation loss per epoch
#------------------------------------------------
plt.plot(epochs, loss, 'r', "Training Loss")
plt.plot(epochs, val_loss, 'b', "Validation Loss")
plt.show()
```
You will probably encounter that the model is overfitting, which means that it is doing a great job at classifying the images in the training set but struggles with new data. This is perfectly fine and you will learn how to mitigate this issue in the upcomming week.
Before closing the assignment, be sure to also download the `history.pkl` file which contains the information of the training history of your model. You can download this file by running the cell below:
```
def download_history():
import pickle
from google.colab import files
with open('history_augmented.pkl', 'wb') as f:
pickle.dump(history.history, f)
files.download('history_augmented.pkl')
download_history()
```
You will also need to submit this notebook for grading. To download it, click on the `File` tab in the upper left corner of the screen then click on `Download` -> `Download .ipynb`. You can name it anything you want as long as it is a valid `.ipynb` (jupyter notebook) file.
**Congratulations on finishing this week's assignment!**
You have successfully implemented a convolutional neural network that classifies images of cats and dogs, along with the helper functions needed to pre-process the images!
**Keep it up!**
|
github_jupyter
|
**Author**: _Pradip Kumar Das_
**License:** https://github.com/PradipKumarDas/Competitions/blob/main/LICENSE
**Profile & Contact:** [LinkedIn](https://www.linkedin.com/in/daspradipkumar/) | [GitHub](https://github.com/PradipKumarDas) | [Kaggle](https://www.kaggle.com/pradipkumardas) | [email protected] (Email)
# Ugam Sentiment Analysis | MachineHack
**Dec. 22, 2021 - Jan. 10, 2022**
https://machinehack.com/hackathon/uhack_sentiments_20_decode_code_words/overview
**Sections:**
- Dependencies
- Exploratory Data Analysis (EDA) & Preprocessing
- Modeling & Evaluation
- Submission
NOTE: Running this notebook over CPU will be intractable as it uses Transformers, and hence it is recommended to use GPU.
# Dependencies
```
# The following packages may need to be first installed on cloud hosted Data Science platforms such as Google Colab.
!pip install transformers
# Imports required packages
import pandas as pd
import numpy as np
from sklearn.model_selection import StratifiedKFold
import tensorflow as tf
# from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint, TensorBoard
import transformers
from transformers import TFAutoModelForSequenceClassification, AutoTokenizer
import matplotlib.pyplot as plt
import seaborn as sns
import datetime, gc
```
# Initialization
```
# Connects drive in Google Colab
from google.colab import drive
drive.mount('/content/drive/')
# Changes working directory to the project directory
cd "/content/drive/MyDrive/Colab/Ugam_Sentiment_Analysis/"
# Configures styles for plotting runtime
plt.style.use("seaborn-whitegrid")
plt.rc(
"figure",
autolayout=True,
figsize=(11, 4),
titlesize=18,
titleweight='bold',
)
plt.rc(
"axes",
labelweight="bold",
labelsize="large",
titleweight="bold",
titlesize=16,
titlepad=10,
)
%config InlineBackend.figure_format = 'retina'
# Sets Tranformer's level of verbosity to INFO level
transformers.logging.set_verbosity_error()
```
# Exploratory Data Analysis (EDA) & Preprocessing
```
# Loads train data set
train = pd.read_csv("./data/train.csv")
# Checks few rows from train data set
display(train)
# Sets dataframe's `Id` columns as its index
train.set_index("Id", drop=True, append=False, inplace=True)
# Loads test data set
test = pd.read_csv("./data/test.csv")
# Checks top few rows from test data set
display(test.head(5))
# Sets dataframe's `Id` columns as its index
test.set_index("Id", drop=True, append=False, inplace=True)
# Checks the distribution of review length (number of characters in review)
fig, ax = plt.subplots(1, 2, sharey=True)
fig.suptitle("Review Length")
train.Review.str.len().plot(kind='hist', bins=50, ax=ax[0])
ax[0].set_xlabel("Train Data")
ax[0].set_ylabel("No. of Reviews")
test.Review.str.len().plot(kind='hist', bins=50, ax=ax[1])
ax[1].set_xlabel("Test Data")
```
The above plot shows that lengthy reviews containing 1000+ characters are less compares to that of reviews having less than 1000 characters. Hence, first 512 characters from the reviews will be considered for analysis.
```
# Finds the distribution of each label
display(train.select_dtypes(["int"]).apply(pd.Series.value_counts))
# Let's find stratified cross validation on 'Polarity' label will have same distribution
sk_fold = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
cv_generator = sk_fold.split(train, train.Polarity)
for fold, (idx_train, idx_val) in enumerate(cv_generator):
display(train.iloc[idx_train].select_dtypes(["int"]).apply(pd.Series.value_counts))
```
It shows the same distribution is available in cross validation.
# Modeling & Evaluation
The approach is to use pretrained **Transfomer** model and to fine-tune, if required. As fine-tuning over cross-validation is time consuming even on GPUs, let's avoid it, and hence prepare one stratified validation set first to check pretrained model's or fine-tuned model's performance.
```
# Creates data set splitter and gets indexes for train and validation rows
cv_generator = sk_fold.split(train, train.Polarity)
idx_train, idx_val = next(cv_generator)
# Sets parameters for Transformer model fine-tuning
model_config = {
"model_name": "distilbert-base-uncased-finetuned-sst-2-english", # selected pretrained model
"max_length": 512, # maximum number of review characters allowed to input to model
}
# Creates tokenizer from pre-trained transformer model
tokenizer = AutoTokenizer.from_pretrained(model_config["model_name"])
# Tokenize reviews for train, validation and test data set
train_encodings = tokenizer(
train.iloc[idx_train].Review.to_list(),
max_length=model_config["max_length"],
truncation=True,
padding=True,
return_tensors="tf"
)
val_encodings = tokenizer(
train.iloc[idx_val].Review.to_list(),
max_length=model_config["max_length"],
truncation=True,
padding=True,
return_tensors="tf"
)
test_encodings = tokenizer(
test.Review.to_list(),
max_length=model_config["max_length"],
truncation=True,
padding=True,
return_tensors="tf"
)
# Performs target specific model fine-tuning
"""
NOTE:
1) It was observed that increasing number of epochs more than one during model fine-tuning
does not improve model performance, and hence epochs is set to 1.
2) As pretrained model being used is already used for predicting sentiment polarity, that model
will not be fine-tuned any further, and will be used directly to predict sentimen polarity
against the test data. Fine-tuning was already experimented and found to be not useful as it
decreases performance with higher log loss and lower accuracy on validation data.
"""
columns = train.select_dtypes(["int"]).columns.tolist()
columns.remove("Polarity")
# Fine-tunes models except that of Polarity
for column in columns:
print(f"Fine tuning model for {column.upper()}...")
print("======================================================\n")
model = TFAutoModelForSequenceClassification.from_pretrained(model_config["model_name"])
# Prepares tensorflow dataset for both train, validation and test data
train_encodings_dataset = tf.data.Dataset.from_tensor_slices((
{"input_ids": train_encodings["input_ids"], "attention_mask": train_encodings["attention_mask"]},
train.iloc[idx_train][[column]]
)).batch(16).prefetch(tf.data.AUTOTUNE)
val_encodings_dataset = tf.data.Dataset.from_tensor_slices((
{"input_ids": val_encodings["input_ids"], "attention_mask": val_encodings["attention_mask"]},
train.iloc[idx_val][[column]]
)).batch(16).prefetch(tf.data.AUTOTUNE)
test_encodings_dataset = tf.data.Dataset.from_tensor_slices(
{"input_ids": test_encodings["input_ids"], "attention_mask": test_encodings["attention_mask"]}
).batch(16).prefetch(tf.data.AUTOTUNE)
predictions = tf.nn.softmax(model.predict(val_encodings_dataset).logits)
print("Pretrained model's perfomance on validation data before fine-tuning:",
tf.keras.metrics.binary_crossentropy(train.iloc[idx_val][column], predictions[:,1], from_logits=False).numpy(), "(log loss)",
tf.keras.metrics.binary_accuracy(train.iloc[idx_val][column], predictions[:,1]).numpy(), "(accuracy)\n"
)
del predictions
print("Starting fine tuning...")
# Freezes model configuration before starting fine-tuning
model.compile(
optimizer=tf.keras.optimizers.Adam(learning_rate=5e-5),
loss=tf.keras.losses.binary_crossentropy,
metrics=[tf.keras.metrics.binary_crossentropy, tf.keras.metrics.binary_accuracy]
)
# Sets model file name to organize storing logs and fine-tuned models against
# model_filename = f"{column}" + "_" + datetime.datetime.now().strftime("%Y.%m.%d-%H:%M:%S")
# Fine tunes model
model.fit(
x=train_encodings_dataset,
validation_data=val_encodings_dataset,
batch_size=16,
epochs=1,
# callbacks=[
# EarlyStopping(monitor="val_loss", mode="min", patience=2, restore_best_weights=True, verbose=1),
# ModelCheckpoint(filepath=f"./models/{model_filename}", monitor="val_loss", mode="min", save_best_only=True, save_weights_only=True),
# TensorBoard(log_dir=f"./logs/{model_filename}", histogram_freq=1, update_freq='epoch')
# ],
use_multiprocessing=True)
print("\nFine tuning was completed.\n")
del train_encodings_dataset, val_encodings_dataset
print("Performing prediction on test data...", end="")
# Performs predictions on test data
predictions = tf.nn.softmax(model.predict(test_encodings_dataset).logits)
test[column] = predictions[:, 1]
del test_encodings_dataset
print("done\n")
del predictions, model
print("Skipping fine-tuning model for POLARITY (as it uses pretrained model) and continuing direct prediction on test data...")
print("======================================================================================================================\n")
print("Performing prediction on test data...", end="")
model = TFAutoModelForSequenceClassification.from_pretrained(model_config["model_name"])
# Prepares tensorflow dataset for test data
test_encodings_dataset = tf.data.Dataset.from_tensor_slices(
{"input_ids": test_encodings["input_ids"], "attention_mask": test_encodings["attention_mask"]}
).batch(16).prefetch(tf.data.AUTOTUNE)
# Performs predictions on test data
predictions = tf.nn.softmax(model.predict(test_encodings_dataset).logits)
test["Polarity"] = predictions[:, 1]
del test_encodings_dataset
del predictions, model
print("done\n")
print("Fine-tuning and test predictions were completed.")
```
# Submission
```
# Saves test predictions
test.select_dtypes(["float"]).to_csv("./submission.csv", index=False)
```
***Leaderboard score for this submission was 8.8942 as against highest of that was 2.74 on Jan 06, 2022 at 11:50 PM.***
|
github_jupyter
|
# SESSIONS ARE ALL YOU NEED
### Workshop on e-commerce personalization
This notebook showcases with working code the main ideas of our ML-in-retail workshop from June lst, 2021 at MICES (https://mices.co/). Please refer to the README in the repo for a bit of context!
While the code below is (well, should be!) fully functioning, please note we aim for functions which are pedagogically useful, more than terse code per se: it should be fairly easy to take these ideas and refactor the code to achieve more speed, better re-usability etc.
_If you want to use Google Colab, you can uncomment this cell:_
```
# if you need requirements....
# !pip install -r requirements.txt
# #from google.colab import drive
#drive.mount('/content/drive',force_remount=True)
#%cd drive/MyDrive/path_to_directory_containing_train_folder
#LOCAL_FOLDER = 'train'
```
## Basic import and some global vars to know where data is!
Here we import the libraries we need and set the working folders - make sure your current python interpreter has all the dependencies installed. If you want to use the same real-world data as I'm using, please download the open dataset you find at: https://github.com/coveooss/SIGIR-ecom-data-challenge.
```
import os
from random import choice
import time
import ast
import json
import numpy as np
import csv
from collections import Counter,defaultdict
# viz stuff
from sklearn.manifold import TSNE
from matplotlib import pyplot as plt
from IPython.display import Image
# gensim stuff for prod2vec
import gensim # gensim > 4
from gensim.similarities.annoy import AnnoyIndexer
# keras stuff for auto-encoder
from keras.layers.core import Dropout
from keras.layers.core import Dense
from keras.layers import Concatenate
from keras.models import Sequential
from keras.layers import Input
from keras.optimizers import SGD, Adam
from keras.models import Model
from keras.callbacks import EarlyStopping
from keras.utils import plot_model
from sklearn.model_selection import train_test_split
from keras import utils
import hashlib
from copy import deepcopy
%matplotlib inline
LOCAL_FOLDER = '/Users/jacopotagliabue/Documents/data_dump/train' # where is the dataset stored?
N_ROWS = 5000000 # how many rows we want to take (to avoid waiting too much for tutorial purposes)?
```
## Step 1: build a prod2vec space
For more information on prod2vec and its use, you can also check our blog post: https://blog.coveo.com/clothes-in-space-real-time-personalization-in-less-than-100-lines-of-code/ or latest NLP paper: https://arxiv.org/abs/2104.02061
```
def read_sessions_from_training_file(training_file: str, K: int = None):
"""
Read the training file containing product interactions, up to K rows.
:return: a list of lists, each list being a session (sequence of product IDs)
"""
user_sessions = []
current_session_id = None
current_session = []
with open(training_file) as csvfile:
reader = csv.DictReader(csvfile)
for idx, row in enumerate(reader):
# if a max number of items is specified, just return at the K with what you have
if K and idx >= K:
break
# just append "detail" events in the order we see them
# row will contain: session_id_hash, product_action, product_sku_hash
_session_id_hash = row['session_id_hash']
# when a new session begins, store the old one and start again
if current_session_id and current_session and _session_id_hash != current_session_id:
user_sessions.append(current_session)
# reset session
current_session = []
# check for the right type and append
if row['product_action'] == 'detail':
current_session.append(row['product_sku_hash'])
# update the current session id
current_session_id = _session_id_hash
# print how many sessions we have...
print("# total sessions: {}".format(len(user_sessions)))
# print first one to check
print("First session is: {}".format(user_sessions[0]))
assert user_sessions[0][0] == 'd5157f8bc52965390fa21ad5842a8502bc3eb8b0930f3f8eafbc503f4012f69c'
assert user_sessions[0][-1] == '63b567f4cef976d1411aecc4240984e46ebe8e08e327f2be786beb7ee83216d0'
return user_sessions
def train_product_2_vec_model(sessions: list,
min_c: int = 3,
size: int = 48,
window: int = 5,
iterations: int = 15,
ns_exponent: float = 0.75):
"""
Train CBOW to get product embeddings. We start with sensible defaults from the literature - please
check https://arxiv.org/abs/2007.14906 for practical tips on how to optimize prod2vec.
:param sessions: list of lists, as user sessions are list of interactions
:param min_c: minimum frequency of an event for it to be calculated for product embeddings
:param size: output dimension
:param window: window parameter for gensim word2vec
:param iterations: number of training iterations
:param ns_exponent: ns_exponent parameter for gensim word2vec
:return: trained product embedding model
"""
model = gensim.models.Word2Vec(sentences=sessions,
min_count=min_c,
vector_size=size,
window=window,
epochs=iterations,
ns_exponent=ns_exponent)
print("# products in the space: {}".format(len(model.wv.index_to_key)))
return model.wv
```
Get sessions from the training file, and train a prod2vec model with standard hyperparameters
```
# get sessions
sessions = read_sessions_from_training_file(
training_file=os.path.join(LOCAL_FOLDER, 'browsing_train.csv'),
K=N_ROWS)
# get a counter on all items for later use
sku_cnt = Counter([item for s in sessions for item in s])
# print out most common SKUs
sku_cnt.most_common(3)
# leave some sessions aside
idx = int(len(sessions) * 0.8)
train_sessions = sessions[0: idx]
test_sessions = sessions[idx:]
print("Train sessions # {}, test sessions # {}".format(len(train_sessions), len(test_sessions)))
# finally, train the p2vec, leaving all the default hyperparameters
prod2vec_model = train_product_2_vec_model(train_sessions)
```
Show how to get a prediction with knn
```
prod2vec_model.similar_by_word(sku_cnt.most_common(1)[0][0], topn=3)
```
Visualize the prod2vec space, color-coding for categories in the catalog
```
def plot_scatter_by_category_with_lookup(title,
skus,
sku_to_target_cat,
results,
custom_markers=None):
groups = {}
for sku, target_cat in sku_to_target_cat.items():
if sku not in skus:
continue
sku_idx = skus.index(sku)
x = results[sku_idx][0]
y = results[sku_idx][1]
if target_cat in groups:
groups[target_cat]['x'].append(x)
groups[target_cat]['y'].append(y)
else:
groups[target_cat] = {
'x': [x], 'y': [y]
}
# DEBUG print
print("Total of # groups: {}".format(len(groups)))
fig, ax = plt.subplots(figsize=(10, 10))
for group, data in groups.items():
ax.scatter(data['x'], data['y'],
alpha=0.3,
edgecolors='none',
s=25,
marker='o' if not custom_markers else custom_markers,
label=group)
plt.title(title)
plt.show()
return
def tsne_analysis(embeddings, perplexity=25, n_iter=1000):
tsne = TSNE(n_components=2, verbose=1, perplexity=perplexity, n_iter=n_iter)
return tsne.fit_transform(embeddings)
def get_sku_to_category_map(catalog_file, depth_index=1):
"""
For each SKU, get category from catalog file (if specified)
:return: dictionary, mapping SKU to a category
"""
sku_to_cats = dict()
with open(catalog_file) as csvfile:
reader = csv.DictReader(csvfile)
for row in reader:
_sku = row['product_sku_hash']
category_hash = row['category_hash']
if not category_hash:
continue
# pick only category at a certain depth in the tree
# e.g. x/xx/xxx, with depth=1, -> xx
branches = category_hash.split('/')
target_branch = branches[depth_index] if depth_index < len(branches) else None
if not target_branch:
continue
# if all good, store the mapping
sku_to_cats[_sku] = target_branch
return sku_to_cats
sku_to_category = get_sku_to_category_map(os.path.join(LOCAL_FOLDER, 'sku_to_content.csv'))
print("Total of # {} categories".format(len(set(sku_to_category.values()))))
print("Total of # {} SKU with a category".format(len(sku_to_category)))
# debug with a sample SKU
print(sku_to_category[sku_cnt.most_common(1)[0][0]])
skus = prod2vec_model.index_to_key
print("Total of # {} skus in the model".format(len(skus)))
embeddings = [prod2vec_model[s] for s in skus]
# print out tsne plot with standard params
tsne_results = tsne_analysis(embeddings)
assert len(tsne_results) == len(skus)
plot_scatter_by_category_with_lookup('Prod2vec', skus, sku_to_category, tsne_results)
# do a version with only top K categories
TOP_K = 5
cnt_categories = Counter(list(sku_to_category.values()))
top_categories = [c[0] for c in cnt_categories.most_common(TOP_K)]
# filter out SKUs outside of top categories
top_skus = []
top_tsne_results = []
for _s, _t in zip(skus, tsne_results):
if sku_to_category.get(_s, None) not in top_categories:
continue
top_skus.append(_s)
top_tsne_results.append(_t)
# re-plot tsne with filtered SKUs
print("Top SKUs # {}".format(len(top_skus)))
plot_scatter_by_category_with_lookup('Prod2vec (top {})'.format(TOP_K),
top_skus, sku_to_category, top_tsne_results)
```
### Bonus: faster inference
Gensim is awesome and support approximate, faster inference! You need to have installed ANNOY first, e.g. "pip install annoy". We re-run here on our prod space the original benchmark for word2vec from gensim!
See: https://radimrehurek.com/gensim/auto_examples/tutorials/run_annoy.html
```
# Set up the model and vector that we are using in the comparison
annoy_index = AnnoyIndexer(prod2vec_model, 100)
test_sku = sku_cnt.most_common(1)[0][0]
# test all is good
print(prod2vec_model.most_similar([test_sku], topn=2, indexer=annoy_index))
print(prod2vec_model.most_similar([test_sku], topn=2))
def avg_query_time(model, annoy_index=None, queries=5000):
"""Average query time of a most_similar method over random queries."""
total_time = 0
for _ in range(queries):
_v = model[choice(model.index_to_key)]
start_time = time.process_time()
model.most_similar([_v], topn=5, indexer=annoy_index)
total_time += time.process_time() - start_time
return total_time / queries
gensim_time = avg_query_time(prod2vec_model)
annoy_time = avg_query_time(prod2vec_model, annoy_index=annoy_index)
print("Gensim (s/query):\t{0:.5f}".format(gensim_time))
print("Annoy (s/query):\t{0:.5f}".format(annoy_time))
speed_improvement = gensim_time / annoy_time
print ("\nAnnoy is {0:.2f} times faster on average on this particular run".format(speed_improvement))
```
### Bonus: hyper tuning
For more info on hyper tuning in the context of product embeddings, please see our paper: https://arxiv.org/abs/2007.14906 and our data release: https://github.com/coveooss/fantastic-embeddings-sigir-2020.
We use the sessions we left out to simulate a small optimization loop...
```
def calculate_HR_on_NEP(model, sessions, k=10, min_length=3):
_count = 0
_hits = 0
for session in sessions:
# consider only decently-long sessions
if len(session) < min_length:
continue
# update the counter
_count += 1
# get the item to predict
target_item = session[-1]
# get model prediction using before-last item
query_item = session[-2]
# if model cannot make the prediction, it's a failure
if query_item not in model:
continue
predictions = model.similar_by_word(query_item, topn=k)
# debug
# print(target_item, query_item, predictions)
if target_item in [p[0] for p in predictions]:
_hits += 1
# debug
print("Total test cases: {}".format(_count))
return _hits / _count
# we simulate a test with 3 values for epochs in prod2ve
iterations_values = [1, 10]
# for each value we train a model, and use Next Event Prediction (NEP) to get a quality assessment
for i in iterations_values:
print("\n ======> Hyper value: {}".format(i))
cnt_model = train_product_2_vec_model(train_sessions, iterations=i)
# use hold-out to have NEP performance
_hr = calculate_HR_on_NEP(cnt_model, test_sessions)
print("HR: {}\n".format(_hr))
```
## Step 2: improving low-count vectors
For more information about prod2vec in the cold start scenario, please see our paper: https://dl.acm.org/doi/10.1145/3383313.3411477 and video: https://vimeo.com/455641121
```
def build_mapper(pro2vec_dims=48):
"""
Build a Keras model for content-based "fake" embeddings.
:return: a Keras model, mapping BERT-like catalog representations to the prod2vec space
"""
# input
description_input = Input(shape=(50,))
image_input = Input(shape=(50,))
# model
x = Dense(25, activation="relu")(description_input)
y = Dense(25, activation="relu")(image_input)
combined = Concatenate()([x, y])
combined = Dropout(0.3)(combined)
combined = Dense(25)(combined)
output = Dense(pro2vec_dims)(combined)
return Model(inputs=[description_input, image_input], outputs=output)
# get vectors representing text and images in the catalog
def get_sku_to_embeddings_map(catalog_file):
"""
For each SKU, get the text and image embeddings, as provided pre-computed by the dataset
:return: dictionary, mapping SKU to a tuple of embeddings
"""
sku_to_embeddings = dict()
with open(catalog_file) as csvfile:
reader = csv.DictReader(csvfile)
for row in reader:
_sku = row['product_sku_hash']
_description = row['description_vector']
_image = row['image_vector']
# skip when both vectors are not there
if not _description or not _image:
continue
# if all good, store the mapping
sku_to_embeddings[_sku] = (json.loads(_description), json.loads(_image))
return sku_to_embeddings
sku_to_embeddings = get_sku_to_embeddings_map(os.path.join(LOCAL_FOLDER, 'sku_to_content.csv'))
print("Total of # {} SKUs with embeddings".format(len(sku_to_embeddings)))
# print out an example
_d, _i = sku_to_embeddings['438630a8ba0320de5235ee1bedf3103391d4069646d640602df447e1042a61a3']
print(len(_d), len(_i), _d[:5], _i[:5])
# just make sure we have the SKUs in the model and a counter
skus = prod2vec_model.index_to_key
print("Total of # {} skus in the model".format(len(skus)))
print(sku_cnt.most_common(5))
# above which percentile of frequency we consider SKU popular enough to be our training set?
FREQUENT_PRODUCTS_PTILE = 80
_counts = [c[1] for c in sku_cnt.most_common()]
_counts[:3]
# make sure we have just SKUS in the prod2vec space for which we have embeddings
popular_threshold = np.percentile(_counts, FREQUENT_PRODUCTS_PTILE)
popular_skus = [s for s in skus if s in sku_to_embeddings and sku_cnt.get(s, 0) > popular_threshold]
product_embeddings = [prod2vec_model[s] for s in popular_skus]
description_embeddings = [sku_to_embeddings[s][0] for s in popular_skus]
image_embeddings = [sku_to_embeddings[s][1] for s in popular_skus]
# debug
print(popular_threshold, len(skus), len(popular_skus))
# print(description_embeddings[:1][:3])
# print(image_embeddings[:1][:3])
# train the mapper now
training_data_X = [np.array(description_embeddings), np.array(image_embeddings)]
training_data_y = np.array(product_embeddings)
es = EarlyStopping(monitor='val_loss', mode='min', verbose=1, patience=20, restore_best_weights=True)
# build and display model
rare_net = build_mapper()
plot_model(rare_net, show_shapes=True, show_layer_names=True, to_file='rare_net.png')
Image('rare_net.png')
# train!
rare_net.compile(loss='mse', optimizer='rmsprop')
rare_net.fit(training_data_X,
training_data_y,
batch_size=200,
epochs=20000,
validation_split=0.2,
callbacks=[es])
# rarest_skus = [_[0] for _ in sku_cnt.most_common()[-500:]]
# test_skus = [s for s in rarest_skus if s in sku_to_embeddings]
# get to rare vectors
test_skus = [s for s in skus if s in sku_to_embeddings and sku_cnt.get(s, 0) < popular_threshold/2]
print(len(skus), len(test_skus))
# prepare embeddings for prediction
rare_description_embeddings = [sku_to_embeddings[s][0] for s in test_skus]
rare_image_embeddings = [sku_to_embeddings[s][1] for s in test_skus]
# prepare embeddings for prediction
test_data_X = [np.array(rare_description_embeddings), np.array(rare_image_embeddings)]
predicted_embeddings = rare_net.predict(test_data_X)
# debug
# print(len(predicted_embeddings))
# print(predicted_embeddings[0][:10])
def calculate_HR_on_NEP_rare(model, sessions, rare_skus, k=10, min_length=3):
_count = 0
_hits = 0
_rare_hits = 0
_rare_count = 0
for session in sessions:
# consider only decently-long sessions
if len(session) < min_length:
continue
# update the counter
_count += 1
# get the item to predict
target_item = session[-1]
# get model prediction using before-last item
query_item = session[-2]
# if model cannot make the prediction, it's a failure
if query_item not in model:
continue
# increment counter if rare sku
if query_item in rare_skus:
_rare_count+=1
predictions = model.similar_by_word(query_item, topn=k)
# debug
# print(target_item, query_item, predictions)
if target_item in [p[0] for p in predictions]:
_hits += 1
# track hits if query is rare sku
if query_item in rare_skus:
_rare_hits+=1
# debug
print("Total test cases: {}".format(_count))
print("Total rare test cases: {}".format(_rare_count))
return _hits / _count, _rare_hits/_rare_count
# make copy of original prod2vec model
prod2vec_rare_model = deepcopy(prod2vec_model)
# update model with new vectors
prod2vec_rare_model.add_vectors(test_skus, predicted_embeddings, replace=True)
prod2vec_rare_model.fill_norms(force=True)
# check
assert np.array_equal(predicted_embeddings[0], prod2vec_rare_model[test_skus[0]])
# test new model
calculate_HR_on_NEP_rare(prod2vec_rare_model, test_sessions, test_skus)
# test original model
calculate_HR_on_NEP_rare(prod2vec_model, test_sessions, test_skus)
```
## Step 3: query scoping
For more information about query scoping, please see our paper: https://www.aclweb.org/anthology/2020.ecnlp-1.2/ and repository: https://github.com/jacopotagliabue/session-path
```
# get vectors representing text and images in the catalog
def get_query_to_category_dataset(search_file, cat_2_id, sku_to_category):
"""
For each query, get a label representing the category in items clicked after the query.
It uses as input a mapping "sku_to_category" to join the search file with catalog meta-data!
:return: two lists, matching query vectors to a label
"""
query_X = list()
query_Y = list()
with open(search_file) as csvfile:
reader = csv.DictReader(csvfile)
for row in reader:
_click_products = row['clicked_skus_hash']
if not _click_products: # or _click_product not in sku_to_category:
continue
# clean the string and extract SKUs from array
cleaned_skus = ast.literal_eval(_click_products)
for s in cleaned_skus:
if s in sku_to_category:
query_X.append(json.loads(row['query_vector']))
target_category_as_int = cat_2_id[sku_to_category[s]]
query_Y.append(utils.to_categorical(target_category_as_int, num_classes=len(cat_2_id)))
return query_X, query_Y
sku_to_category = get_sku_to_category_map(os.path.join(LOCAL_FOLDER, 'sku_to_content.csv'))
print("Total of # {} categories".format(len(set(sku_to_category.values()))))
cats = list(set(sku_to_category.values()))
cat_2_id = {c: idx for idx, c in enumerate(cats)}
print(cat_2_id[cats[0]])
query_X, query_Y = get_query_to_category_dataset(os.path.join(LOCAL_FOLDER, 'search_train.csv'),
cat_2_id,
sku_to_category)
print(len(query_X))
print(query_Y[0])
x_train, x_test, y_train, y_test = train_test_split(np.array(query_X), np.array(query_Y), test_size=0.2)
def build_query_scoping_model(input_d, target_classes):
print('Shape tensor {}, target classes {}'.format(input_d, target_classes))
# define model
model = Sequential()
model.add(Dense(64, activation='relu', input_dim=input_d))
model.add(Dropout(0.5))
model.add(Dense(64, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(target_classes, activation='softmax'))
return model
query_model = build_query_scoping_model(x_train[0].shape[0], y_train[0].shape[0])
# compile model
sgd = SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)
query_model.compile(loss='categorical_crossentropy', optimizer=sgd, metrics=['accuracy'])
# train first
query_model.fit(x_train, y_train, epochs=10, batch_size=32)
# compute and print eval score
score = query_model.evaluate(x_test, y_test, batch_size=32)
score
# get vectors representing text and images in the catalog
def get_query_info(search_file):
"""
For each query, extract relevant metadata of query and to match with session data
:return: list of queries with metadata
"""
queries = list()
with open(search_file) as csvfile:
reader = csv.DictReader(csvfile)
for row in reader:
_click_products = row['clicked_skus_hash']
if not _click_products: # or _click_product not in sku_to_category:
continue
# clean the string and extract SKUs from array
cleaned_skus = ast.literal_eval(_click_products)
queries.append({'session_id_hash' : row['session_id_hash'],
'server_timestamp_epoch_ms' : int(row['server_timestamp_epoch_ms']),
'clicked_skus' : cleaned_skus,
'query_vector' : json.loads(row['query_vector'])})
print("# total queries: {}".format(len(queries)))
return queries
def get_session_info_for_queries(training_file: str, query_info: list, K: int = None):
"""
Read the training file containing product interactions for sessions with query, up to K rows.
:return: dict of lists with session_id as key, each list being a session (sequence of product events with metadata)
"""
user_sessions = dict()
current_session_id = None
current_session = []
query_session_ids = set([ _['session_id_hash'] for _ in query_info])
with open(training_file) as csvfile:
reader = csv.DictReader(csvfile)
for idx, row in enumerate(reader):
# if a max number of items is specified, just return at the K with what you have
if K and idx >= K:
break
# just append "detail" events in the order we see them
# row will contain: session_id_hash, product_action, product_sku_hash
_session_id_hash = row['session_id_hash']
# when a new session begins, store the old one and start again
if current_session_id and current_session and _session_id_hash != current_session_id:
user_sessions[current_session_id] = current_session
# reset session
current_session = []
# check for the right type and append event info
if row['product_action'] == 'detail' and _session_id_hash in query_session_ids :
current_session.append({'product_sku_hash': row['product_sku_hash'],
'server_timestamp_epoch_ms' : int(row['server_timestamp_epoch_ms'])})
# update the current session id
current_session_id = _session_id_hash
# print how many sessions we have...
print("# total sessions: {}".format(len(user_sessions)))
return dict(user_sessions)
query_info = get_query_info(os.path.join(LOCAL_FOLDER, 'search_train.csv'))
session_info = get_session_info_for_queries(os.path.join(LOCAL_FOLDER, 'browsing_train.csv'), query_info)
def get_contextual_query_to_category_dataset(query_info, session_info, prod2vec_model, cat_2_id, sku_to_category):
"""
For each query, get a label representing the category in items clicked after the query.
It uses as input a mapping "sku_to_category" to join the search file with catalog meta-data!
It also creates a joint embedding for input by concatenating query vector and average session vector up till
when query was made
:return: two lists, matching query vectors to a label
"""
query_X = list()
query_Y = list()
for row in query_info:
query_timestamp = row['server_timestamp_epoch_ms']
cleaned_skus = row['clicked_skus']
session_id_hash = row['session_id_hash']
if session_id_hash not in session_info or not cleaned_skus: # or _click_product not in sku_to_category:
continue
session_skus = session_info[session_id_hash]
context_skus = [ e['product_sku_hash'] for e in session_skus if query_timestamp > e['server_timestamp_epoch_ms']
and e['product_sku_hash'] in prod2vec_model]
if not context_skus:
continue
context_vector = np.mean([prod2vec_model[sku] for sku in context_skus], axis=0).tolist()
for s in cleaned_skus:
if s in sku_to_category:
query_X.append(row['query_vector'] + context_vector)
target_category_as_int = cat_2_id[sku_to_category[s]]
query_Y.append(utils.to_categorical(target_category_as_int, num_classes=len(cat_2_id)))
return query_X, query_Y
context_query_X, context_query_Y = get_contextual_query_to_category_dataset(query_info,
session_info,
prod2vec_model,
cat_2_id,
sku_to_category)
print(len(context_query_X))
print(context_query_Y[0])
x_train, x_test, y_train, y_test = train_test_split(np.array(context_query_X), np.array(context_query_Y), test_size=0.2)
contextual_query_model = build_query_scoping_model(x_train[0].shape[0], y_train[0].shape[0])
# compile model
sgd = SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)
contextual_query_model.compile(loss='categorical_crossentropy', optimizer=sgd, metrics=['accuracy'])
# train first
contextual_query_model.fit(x_train, y_train, epochs=10, batch_size=32)
# compute and print eval score
score = contextual_query_model.evaluate(x_test, y_test, batch_size=32)
score
```
|
github_jupyter
|
We sometimes want to know where a value is in an array.
```
import numpy as np
```
By "where" we mean, which element contains a particular value.
Here is an array.
```
arr = np.array([2, 99, -1, 4, 99])
arr
```
As you know, we can get element using their *index* in the array. In
Python, array indices start at zero.
Here's the value at index (position) 0:
```
arr[0]
```
We might also be interested to find which positions hold particular values.
In our array above, by reading, and counting positions, we can see
that the values of 99 are in positions 1 and 4. We can ask for these
elements by passing a list or an array between the square brackets, to
index the array:
```
positions_with_99 = np.array([1, 4])
arr[positions_with_99]
```
Of course, we are already used to finding and then selecting elements according to various conditions, using *Boolean vectors*.
Here we identify the elements that contain 99. There is a `True` at the position where the array contains 99, and `False` otherwise.
```
contains_99 = arr == 99
contains_99
```
We can then get the 99 values with:
```
arr[contains_99]
```
## Enter "where"
Sometimes we really do need to know the index of the values that meet a certain condition.
In that case, you can use the Numpy [where
function](https://docs.scipy.org/doc/numpy/reference/generated/numpy.where.html).
`where` finds the index positions of the `True` values in Boolean
vectors.
```
indices = np.where(arr == 99)
indices
```
We can use the returned `indices` to index into the array, using square brackets.
```
arr[indices]
```
This also works in two or more dimensions. Here is a two-dimensional array, with some values of 99.
```
arr2d = np.array([[4, 99, 3], [8, 8, 99]])
arr2d
```
`where` now returns two index arrays, one for the rows, and one for the columns.
```
indices2d = np.where(arr2d == 99)
indices2d
```
Just as for the one-dimensional case, we can use the returned indices to index into the array, and get the elements.
```
arr2d[indices2d]
```
## Where summary
Numpy `where` returns the indices of `True` values in a Boolean array/
You can use these indices to index into an array, and get the matching elements.
## Argmin
Numpy has various *argmin* functions that are a shortcut for using `where`, for particular cases.
A typical case is where you want to know the index (position) of the minimum value in an array.
Here is our array:
```
arr
```
We can get the minimum value with Numpy `min`:
```
np.min(arr)
```
Sometimes we want to know the *index position* of the minimum value. Numpy `argmin` returns the index of the minimum value:
```
min_pos = np.argmin(arr)
min_pos
```
Therefore, we can get the minimum value again with:
```
arr[min_pos]
```
There is a matching `argmax` function that returns the position of the maximum value:
```
np.max(arr)
max_pos = np.argmax(arr)
max_pos
arr[max_pos]
```
We could also have found the position of the minimum value above, using `np.min` and `where`:
```
min_value = np.min(arr)
min_indices = np.where(arr == min_value)
arr[min_indices]
```
The `argmin` and `argmax` functions are not quite the same, in that they only return the *first* position of the minimum or maximum, if there are multiple values with the same value.
Compare:
```
np.argmax(arr)
```
to
```
max_value = np.max(arr)
np.where(arr == max_value)
```
|
github_jupyter
|
```
# from utils import *
import tensorflow as tf
import os
import sklearn.datasets
import numpy as np
import re
import collections
import random
from sklearn import metrics
import jieba
# 写入停用词
with open(r'stopwords.txt','r',encoding='utf-8') as f:
english_stopwords = f.read().split('\n')
def separate_dataset(trainset, ratio = 0.5):
datastring = []
datatarget = []
for i in range(len(trainset.data)):
# 提取每一条文本数据,并过滤None值行文本;
data_ = trainset.data[i].split('\n')
data_ = list(filter(None, data_))
# 抽取len(data_) * ratio个样本,并打乱某类样本顺序;
data_ = random.sample(data_, int(len(data_) * ratio))
# 去除停用词
for n in range(len(data_)):
data_[n] = clearstring(data_[n])
# 提取所有的词
datastring += data_
# 为每一个样本补上标签
for n in range(len(data_)):
datatarget.append(trainset.target[i])
return datastring, datatarget
def clearstring(string):
# 清洗样本,并去停用词
# 去除非中文字符
string = re.sub(r'^[\u4e00-\u9fa5a-zA-Z0-9]', '', string)
string = list(jieba.cut(string, cut_all=False))
string = filter(None, string)
string = [y.strip() for y in string if y.strip() not in english_stopwords]
string = ' '.join(string)
return string.lower()
def str_idx(corpus, dic, maxlen, UNK = 3):
# 词典索引
X = np.zeros((len(corpus), maxlen))
for i in range(len(corpus)):
for no, k in enumerate(corpus[i].split()[:maxlen][::-1]):
X[i, -1 - no] = dic.get(k, UNK)
return X
trainset = sklearn.datasets.load_files(container_path = 'dataset', encoding = 'UTF-8')
trainset.data, trainset.target = separate_dataset(trainset,1.0)
print(trainset.target_names)
print(len(trainset.data))
print(len(trainset.target))
import collections
def build_dataset(words, n_words, atleast=1):
# 四种填充词
count = [['PAD', 0], ['GO', 1], ['EOS', 2], ['UNK', 3]]
counter = collections.Counter(words).most_common(n_words)
# 过滤那些只有一个字的字符
counter = [i for i in counter if i[1] >= atleast]
count.extend(counter)
dictionary = dict()
# 构建词的索引
for word, _ in count:
dictionary[word] = len(dictionary)
data = list()
for word in words:
# 如果字典中没有出现的词,用unk表示
index = dictionary.get(word, 3)
data.append(index)
# 翻转字典
reversed_dictionary = dict(zip(dictionary.values(), dictionary.keys()))
return data, dictionary, reversed_dictionary
split = (' '.join(trainset.data)).split()
# 去重后的所有单词集合,组成词典
vocabulary_size = len(list(set(split)))
# data为所有词的词索引,词典,反向词典
data, dictionary, rev_dictionary = build_dataset(split, vocabulary_size)
len(dictionary)
def build_char_dataset(words):
# 四种填充词
count = []
dictionary = dict()
# 构建词的索引
for word in words:
dictionary[word] = len(dictionary)
data = list()
for word in words:
# 如果字典中没有出现的词,用unk表示
index = dictionary.get(word, 3)
data.append(index)
# 翻转字典
reversed_dictionary = dict(zip(dictionary.values(), dictionary.keys()))
return data, dictionary, reversed_dictionary
# 构建所有中文汉字的字典,3912个汉字
char_split = list(set(list(''.join(trainset.data))))
length = len(char_split)
char_data, char_dictionary, char_rev_dictionary = build_char_dataset(char_split)
# 给中文字符编码
class Vocabulary:
def __init__(self, dictionary, rev_dictionary):
self._dictionary = dictionary
self._rev_dictionary = rev_dictionary
# 起始符
@property
def start_string(self):
return self._dictionary['GO']
# 结束符
@property
def end_string(self):
return self._dictionary['EOS']
# 未知单词
@property
def unk(self):
return self._dictionary['UNK']
@property
def size(self):
return len(self._dictionary)
# 查询词语的数值索引
def word_to_id(self, word):
return self._dictionary.get(word, self.unk)
# 通过索引反查词语
def id_to_word(self, cur_id):
return self._rev_dictionary.get(cur_id, self._rev_dictionary[3])
# 将数字索引解码成字符串并拼接起来
def decode(self, cur_ids):
return ' '.join([self.id_to_word(cur_id) for cur_id in cur_ids])
# 将字符串编码成数字索引
def encode(self, sentence, reverse = False, split = True):
if split:
sentence = sentence.split()
# 将文本转化为数字索引
word_ids = [self.word_to_id(cur_word) for cur_word in sentence]
# 为所有的文本加上起始符和结束符,双向编码都支持
if reverse:
return np.array(
[self.end_string] + word_ids + [self.start_string],
dtype = np.int32,
)
else:
return np.array(
[self.start_string] + word_ids + [self.end_string],
dtype = np.int32,
)
# 英文,数字,字符用自制转码,中文字符,从0开始进行编码,考虑到中文短语的长度一般不超过8,故可取max_length=10
class UnicodeCharsVocabulary(Vocabulary):
def __init__(self, dictionary, rev_dictionary,char_dictionary, char_rev_dictionary, max_word_length, **kwargs):
super(UnicodeCharsVocabulary, self).__init__(
dictionary, rev_dictionary, **kwargs
)
# 最大单词长度
self._max_word_length = max_word_length
self._char_dictionary = char_dictionary
self._char_rev_dictionary = char_rev_dictionary
self.bos_char = 3912
self.eos_char = 3913
self.bow_char = 3914
self.eow_char = 3915
self.pad_char = 3916
self.unk_char = 3917
# 单词的数量
num_words = self.size
# 构建字符级别的词典表,[num_words,max_word_length]
self._word_char_ids = np.zeros(
[num_words, max_word_length], dtype = np.int32
)
# 构建bos和eos的mask,初始化一个_max_word_length的张量,全部用3916填充,第一个字符位用3914,第三个字符位用3915,
# 第二个字符作为输入进行传入
def _make_bos_eos(c):
r = np.zeros([self._max_word_length], dtype = np.int32)
r[:] = self.pad_char
r[0] = self.bow_char
r[1] = c
r[2] = self.eow_char
return r
# 张量化
self.bos_chars = _make_bos_eos(self.bos_char)
self.eos_chars = _make_bos_eos(self.eos_char)
# 遍历字典中的每个单词,并将每个单词都进行字符级别的编码
for i, word in enumerate(self._dictionary.keys()):
self._word_char_ids[i] = self._convert_word_to_char_ids(word)
# 对于起始符GO和结束符EOS进行编码
self._word_char_ids[self.start_string] = self.bos_chars
self._word_char_ids[self.end_string] = self.eos_chars
@property
def word_char_ids(self):
return self._word_char_ids
@property
def max_word_length(self):
return self._max_word_length
# 将单词转化为字符级别的索引
def _convert_word_to_char_ids(self, word):
# 对输入的单词进行张量化
code = np.zeros([self.max_word_length], dtype = np.int32)
code[:] = self.pad_char
# 截取maxlen-2个字符,并将所有字符转化为unicode字符集
word_encoded = [self._char_dictionary.get(item,self.unk_char) for item in list(word)][:(self.max_word_length - 2)]
# 第一个字符位为3914
code[0] = self.bow_char
# 遍历单词的每一个字符,k从1开始
for k, chr_id in enumerate(word_encoded, start = 1):
code[k] = chr_id
# 在单词的末尾补充一个单词末尾结束符3915
code[len(word_encoded) + 1] = self.eow_char
return code
# 将单词转化为自定义字符编码
def word_to_char_ids(self, word):
if word in self._dictionary:
return self._word_char_ids[self._dictionary[word]]
else:
return self._convert_word_to_char_ids(word)
# 将句子转化为自定义字符编码矩阵
def encode_chars(self, sentence, reverse = False, split = True):
if split:
sentence = sentence.split()
chars_ids = [self.word_to_char_ids(cur_word) for cur_word in sentence]
if reverse:
return np.vstack([self.eos_chars] + chars_ids + [self.bos_chars])
else:
return np.vstack([self.bos_chars] + chars_ids + [self.eos_chars])
def _get_batch(generator, batch_size, num_steps, max_word_length):
# generator: 生成器
# batch_size: 每个批次的字符串的数量
# num_steps: 窗口大小
# max_word_length: 最大单词长度,一般设置为50
# 初始化batch_size个字符串
cur_stream = [None] * batch_size
no_more_data = False
while True:
# 初始化单词矩阵输入0值化[batch_size,num_steps]
inputs = np.zeros([batch_size, num_steps], np.int32)
# 初始化字符级矩阵,输入0值化
if max_word_length is not None:
char_inputs = np.zeros(
[batch_size, num_steps, max_word_length], np.int32
)
else:
char_inputs = None
# 初始化单词矩阵输出0值化[batch_size,num_steps]
targets = np.zeros([batch_size, num_steps], np.int32)
for i in range(batch_size):
cur_pos = 0
while cur_pos < num_steps:
if cur_stream[i] is None or len(cur_stream[i][0]) <= 1:
try:
# 每一步都获取词索引,字符集编码器
cur_stream[i] = list(next(generator))
except StopIteration:
no_more_data = True
break
# how_many 取当前总num_steps与文本词向量数量的较小值,累加
how_many = min(len(cur_stream[i][0]) - 1, num_steps - cur_pos)
next_pos = cur_pos + how_many
# 赋值输入对应的词索引范围和字符级别索引范围
inputs[i, cur_pos:next_pos] = cur_stream[i][0][:how_many]
if max_word_length is not None:
char_inputs[i, cur_pos:next_pos] = cur_stream[i][1][
:how_many
]
# targets 我们的目标是预测下一个词来优化emlo,所以我们以向右滑动的1个词作为target,作为预测对象
targets[i, cur_pos:next_pos] = cur_stream[i][0][
1 : how_many + 1
]
cur_pos = next_pos
# 处理完之前那段,重新处理下一段,每段的长度取决于howmany,这里既是window的宽度。
cur_stream[i][0] = cur_stream[i][0][how_many:]
if max_word_length is not None:
cur_stream[i][1] = cur_stream[i][1][how_many:]
if no_more_data:
break
X = {
'token_ids': inputs,
'tokens_characters': char_inputs,
'next_token_id': targets,
}
yield X
class LMDataset:
def __init__(self, string, vocab, reverse = False):
self._vocab = vocab
self._string = string
self._reverse = reverse
self._use_char_inputs = hasattr(vocab, 'encode_chars')
self._i = 0
# 总文本的数量
self._nids = len(self._string)
def _load_string(self, string):
if self._reverse:
string = string.split()
string.reverse()
string = ' '.join(string)
# 将一段文本解析成词索引,会在起始和末尾增加一个标志位
ids = self._vocab.encode(string, self._reverse)
# 将一段文本解析成字符级编码
if self._use_char_inputs:
chars_ids = self._vocab.encode_chars(string, self._reverse)
else:
chars_ids = None
# 返回由词索引和字符集编码的元组
return list(zip([ids], [chars_ids]))[0]
# 生成器,循环生成每个样本的词索引和字符编码
def get_sentence(self):
while True:
if self._i == self._nids:
self._i = 0
ret = self._load_string(self._string[self._i])
self._i += 1
yield ret
@property
def max_word_length(self):
if self._use_char_inputs:
return self._vocab.max_word_length
else:
return None
# batch生成器,每次只拿batch_size个数据,要多少数据就即时处理多少数据
def iter_batches(self, batch_size, num_steps):
for X in _get_batch(
self.get_sentence(), batch_size, num_steps, self.max_word_length
):
yield X
@property
def vocab(self):
return self._vocab
# 双向编码
class BidirectionalLMDataset:
def __init__(self, string, vocab):
# 正向编码和反向编码
self._data_forward = LMDataset(string, vocab, reverse = False)
self._data_reverse = LMDataset(string, vocab, reverse = True)
def iter_batches(self, batch_size, num_steps):
max_word_length = self._data_forward.max_word_length
for X, Xr in zip(
_get_batch(
self._data_forward.get_sentence(),
batch_size,
num_steps,
max_word_length,
),
_get_batch(
self._data_reverse.get_sentence(),
batch_size,
num_steps,
max_word_length,
),
):
# 拼接成一个6个item的字典,前三个为正向,后三个为反向
for k, v in Xr.items():
X[k + '_reverse'] = v
yield X
# maxlens=10,很明显没有超过8的词语,其中两个用来做填充符
uni = UnicodeCharsVocabulary(dictionary, rev_dictionary,char_dictionary,char_rev_dictionary, 10)
bi = BidirectionalLMDataset(trainset.data, uni)
# 每次只输入16个样本数据
batch_size = 16
# 训练用的词典大小
n_train_tokens = len(dictionary)
# 语言模型参数配置项
options = {
# 开启双向编码机制
'bidirectional': True,
# 字符级别的CNN,字符级别词嵌入128维,一共配置7种类型的滤波器,每个词最大长度为50,编码的有效数量为3918个,设置两条高速通道
'char_cnn': {
'activation': 'relu',
'embedding': {'dim': 128},
'filters': [
[1, 32],
[2, 32],
[3, 64],
[4, 128],
[5, 256],
[6, 512],
[7, 1024],
],
'max_characters_per_token': 10,
'n_characters': 3918,
'n_highway': 2,
},
# 随机失活率设置为0.1
'dropout': 0.1,
# lstm单元,设置三层,嵌入维度为512维
'lstm': {
# 截断值
'cell_clip': 3,
'dim': 512,
'n_layers': 2,
'projection_dim': 256,
# 裁剪到[-3,3]之间
'proj_clip': 3,
'use_skip_connections': True,
},
# 一共迭代100轮
'n_epochs': 100,
# 训练词典的大小
'n_train_tokens': n_train_tokens,
# 每个batch的大小
'batch_size': batch_size,
# 所有词的数量
'n_tokens_vocab': uni.size,
# 推断区间为20
'unroll_steps': 20,
'n_negative_samples_batch': 0.001,
'sample_softmax': True,
'share_embedding_softmax': False,
}
# 构建ELMO语言模型
class LanguageModel:
def __init__(self, options, is_training):
self.options = options
self.is_training = is_training
self.bidirectional = options.get('bidirectional', False)
self.char_inputs = 'char_cnn' in self.options
self.share_embedding_softmax = options.get(
'share_embedding_softmax', False
)
if self.char_inputs and self.share_embedding_softmax:
raise ValueError(
'Sharing softmax and embedding weights requires ' 'word input'
)
self.sample_softmax = options.get('sample_softmax', False)
# 建立模型
self._build()
# 配置学习率
lr = options.get('learning_rate', 0.2)
# 配置优化器
self.optimizer = tf.train.AdagradOptimizer(
learning_rate = lr, initial_accumulator_value = 1.0
).minimize(self.total_loss)
def _build_word_embeddings(self):
# 建立词嵌入
# 加载所有的词
n_tokens_vocab = self.options['n_tokens_vocab']
batch_size = self.options['batch_size']
# 上下文推断的窗口大小,这里关联20个单词
unroll_steps = self.options['unroll_steps']
# 词嵌入维度128
projection_dim = self.options['lstm']['projection_dim']
# 词索引
self.token_ids = tf.placeholder(
tf.int32, shape = (None, unroll_steps), name = 'token_ids'
)
self.batch_size = tf.shape(self.token_ids)[0]
with tf.device('/cpu:0'):
# 对单词进行256维的单词编码,初始化数据服从(-1,1)的正态分布
self.embedding_weights = tf.get_variable(
'embedding',
[n_tokens_vocab, projection_dim],
dtype = tf.float32,
initializer = tf.random_uniform_initializer(-1.0, 1.0),
)
# 20个词对应的词嵌入
self.embedding = tf.nn.embedding_lookup(
self.embedding_weights, self.token_ids
)
# 启用双向编码机制
if self.bidirectional:
self.token_ids_reverse = tf.placeholder(
tf.int32,
shape = (None, unroll_steps),
name = 'token_ids_reverse',
)
with tf.device('/cpu:0'):
self.embedding_reverse = tf.nn.embedding_lookup(
self.embedding_weights, self.token_ids_reverse
)
def _build_word_char_embeddings(self):
batch_size = self.options['batch_size']
unroll_steps = self.options['unroll_steps']
projection_dim = self.options['lstm']['projection_dim']
cnn_options = self.options['char_cnn']
filters = cnn_options['filters']
# 求和所有的滤波器数量
n_filters = sum(f[1] for f in filters)
# 最大单词字符长度
max_chars = cnn_options['max_characters_per_token']
# 字符级别嵌入维度,128
char_embed_dim = cnn_options['embedding']['dim']
# 所有字符的类型,一共261种
n_chars = cnn_options['n_characters']
# 配置激活函数
if cnn_options['activation'] == 'tanh':
activation = tf.nn.tanh
elif cnn_options['activation'] == 'relu':
activation = tf.nn.relu
# [batch_size,unroll_steps,max_chars]
self.tokens_characters = tf.placeholder(
tf.int32,
shape = (None, unroll_steps, max_chars),
name = 'tokens_characters',
)
self.batch_size = tf.shape(self.tokens_characters)[0]
with tf.device('/cpu:0'):
# 字符级别词嵌入,嵌入维度128维
self.embedding_weights = tf.get_variable(
'char_embed',
[n_chars, char_embed_dim],
dtype = tf.float32,
initializer = tf.random_uniform_initializer(-1.0, 1.0),
)
self.char_embedding = tf.nn.embedding_lookup(
self.embedding_weights, self.tokens_characters
)
if self.bidirectional:
self.tokens_characters_reverse = tf.placeholder(
tf.int32,
shape = (None, unroll_steps, max_chars),
name = 'tokens_characters_reverse',
)
self.char_embedding_reverse = tf.nn.embedding_lookup(
self.embedding_weights, self.tokens_characters_reverse
)
# 构建卷积层网络,用于字符级别的CNN卷积
def make_convolutions(inp, reuse):
with tf.variable_scope('CNN', reuse = reuse) as scope:
convolutions = []
# 这里构建7层卷积网络
for i, (width, num) in enumerate(filters):
if cnn_options['activation'] == 'relu':
w_init = tf.random_uniform_initializer(
minval = -0.05, maxval = 0.05
)
elif cnn_options['activation'] == 'tanh':
w_init = tf.random_normal_initializer(
mean = 0.0,
stddev = np.sqrt(1.0 / (width * char_embed_dim)),
)
w = tf.get_variable(
'W_cnn_%s' % i,
[1, width, char_embed_dim, num],
initializer = w_init,
dtype = tf.float32,
)
b = tf.get_variable(
'b_cnn_%s' % i,
[num],
dtype = tf.float32,
initializer = tf.constant_initializer(0.0),
)
# 卷积,uroll_nums,characters_nums采用1*1,1*2,...,1*7的卷积核,采用valid卷积策略;
# width上,(uroll_nums-1/1)+1=uroll_nums
# height上,(characters_nums-7/1)+1,捕捉词与词之间的相关性
conv = (
tf.nn.conv2d(
inp, w, strides = [1, 1, 1, 1], padding = 'VALID'
)
+ b
)
# 最大池化,每个词的字符编码
conv = tf.nn.max_pool(
conv,
[1, 1, max_chars - width + 1, 1],
[1, 1, 1, 1],
'VALID',
)
conv = activation(conv)
# 删除第三维,输入为[batch_size,uroll_nums,1,nums]
# 输出为[batch_size,uroll_nums,nums]
conv = tf.squeeze(conv, squeeze_dims = [2])
# 收集每个卷积层,并进行拼接
convolutions.append(conv)
return tf.concat(convolutions, 2)
reuse = tf.get_variable_scope().reuse
# inp [batch_size,uroll_nums,characters_nums,embedding_size]
embedding = make_convolutions(self.char_embedding, reuse)
# [batch_size,20,2048] #经过验证无误
# 增加一维[1,batch_size,uroll_nums,nums++]
self.token_embedding_layers = [embedding]
if self.bidirectional:
embedding_reverse = make_convolutions(
self.char_embedding_reverse, True
)
# 高速网络的数量
n_highway = cnn_options.get('n_highway')
use_highway = n_highway is not None and n_highway > 0
# use_proj 为True
use_proj = n_filters != projection_dim
# 本来已经第三维是2048维了,这么做的原因是?
if use_highway or use_proj:
embedding = tf.reshape(embedding, [-1, n_filters])
if self.bidirectional:
embedding_reverse = tf.reshape(
embedding_reverse, [-1, n_filters]
)
if use_proj:
# 使用投影,将滤波器再投影到一个projection_dim维的向量空间内
assert n_filters > projection_dim
with tf.variable_scope('CNN_proj') as scope:
W_proj_cnn = tf.get_variable(
'W_proj',
[n_filters, projection_dim],
initializer = tf.random_normal_initializer(
mean = 0.0, stddev = np.sqrt(1.0 / n_filters)
),
dtype = tf.float32,
)
b_proj_cnn = tf.get_variable(
'b_proj',
[projection_dim],
initializer = tf.constant_initializer(0.0),
dtype = tf.float32,
)
def high(x, ww_carry, bb_carry, ww_tr, bb_tr):
carry_gate = tf.nn.sigmoid(tf.matmul(x, ww_carry) + bb_carry)
transform_gate = tf.nn.relu(tf.matmul(x, ww_tr) + bb_tr)
return carry_gate * transform_gate + (1.0 - carry_gate) * x
if use_highway:
# 高速网络的维度为2048维
highway_dim = n_filters
for i in range(n_highway):
with tf.variable_scope('CNN_high_%s' % i) as scope:
W_carry = tf.get_variable(
'W_carry',
[highway_dim, highway_dim],
initializer = tf.random_normal_initializer(
mean = 0.0, stddev = np.sqrt(1.0 / highway_dim)
),
dtype = tf.float32,
)
b_carry = tf.get_variable(
'b_carry',
[highway_dim],
initializer = tf.constant_initializer(-2.0),
dtype = tf.float32,
)
W_transform = tf.get_variable(
'W_transform',
[highway_dim, highway_dim],
initializer = tf.random_normal_initializer(
mean = 0.0, stddev = np.sqrt(1.0 / highway_dim)
),
dtype = tf.float32,
)
b_transform = tf.get_variable(
'b_transform',
[highway_dim],
initializer = tf.constant_initializer(0.0),
dtype = tf.float32,
)
embedding = high(
embedding, W_carry, b_carry, W_transform, b_transform
)
if self.bidirectional:
embedding_reverse = high(
embedding_reverse,
W_carry,
b_carry,
W_transform,
b_transform,
)
# 扩展一层和两层经过高速网络的参数
self.token_embedding_layers.append(
tf.reshape(
embedding, [self.batch_size, unroll_steps, highway_dim]
)
)
# 经过一层线性变换[bacth_size,unroll_nums,projection_dim]
if use_proj:
embedding = tf.matmul(embedding, W_proj_cnn) + b_proj_cnn
if self.bidirectional:
embedding_reverse = (
tf.matmul(embedding_reverse, W_proj_cnn) + b_proj_cnn
)
# 只经过线性变换的网络参数
self.token_embedding_layers.append(
tf.reshape(
embedding, [self.batch_size, unroll_steps, projection_dim]
)
)
# 确保矩阵尺寸相同
if use_highway or use_proj:
shp = [self.batch_size, unroll_steps, projection_dim]
embedding = tf.reshape(embedding, shp)
if self.bidirectional:
embedding_reverse = tf.reshape(embedding_reverse, shp)
# 经过线性变化的embdedding [bacth_size,unroll_nums,projection_dim]
# self.token_embedding_layers 由四个嵌入层参数组成
# [bacth_size,unroll_nums,nums++] 原始词嵌入
# [bacth_size,unroll_nums,highway_dim] 经过第一层高速网络的词嵌入
# [bacth_size,unroll_nums,highway_dim] 经过第二层高速网络的词嵌入
# [bacth_size,unroll_nums,projection_dim] 经过低微线性投影的词嵌入
# print(embedding)
# print(self.token_embedding_layers)
self.embedding = embedding
if self.bidirectional:
self.embedding_reverse = embedding_reverse
# 构建模型
def _build(self):
# 所有词的数量
n_tokens_vocab = self.options['n_tokens_vocab']
batch_size = self.options['batch_size']
# window长度
unroll_steps = self.options['unroll_steps']
# lstm编码长度
lstm_dim = self.options['lstm']['dim']
projection_dim = self.options['lstm']['projection_dim']
# lstm的层数
n_lstm_layers = self.options['lstm'].get('n_layers', 1)
dropout = self.options['dropout']
# 保有率
keep_prob = 1.0 - dropout
# 如果是字符级别的输入,则建立词,字符嵌入,否则建立词嵌入,实际上使用前者
if self.char_inputs:
self._build_word_char_embeddings()
else:
self._build_word_embeddings()
# 存储lstm的状态
self.init_lstm_state = []
self.final_lstm_state = []
# 双向
# lstm_inputs单元为[batch_size,uroll_nums,projection_dim]双向单元
if self.bidirectional:
lstm_inputs = [self.embedding, self.embedding_reverse]
else:
lstm_inputs = [self.embedding]
cell_clip = self.options['lstm'].get('cell_clip')
proj_clip = self.options['lstm'].get('proj_clip')
use_skip_connections = self.options['lstm'].get('use_skip_connections')
print(lstm_inputs)
lstm_outputs = []
for lstm_num, lstm_input in enumerate(lstm_inputs):
lstm_cells = []
for i in range(n_lstm_layers):
# 在进行LSTM编码后再接入一个num_proj的全连接层,[batch_size,projection_dim]
# [batch_size,num_proj]
lstm_cell = tf.nn.rnn_cell.LSTMCell(
# 隐含层的单元数
lstm_dim,
num_proj = lstm_dim // 2,
cell_clip = cell_clip,
proj_clip = proj_clip,
)
if use_skip_connections:
if i == 0:
pass
else:
# 将上一个单元的输出,和当前输入映射到下一个单元
lstm_cell = tf.nn.rnn_cell.ResidualWrapper(lstm_cell)
# 添加随机失活层
if self.is_training:
lstm_cell = tf.nn.rnn_cell.DropoutWrapper(
lstm_cell, input_keep_prob = keep_prob
)
lstm_cells.append(lstm_cell)
# 构建多层LSTM
if n_lstm_layers > 1:
lstm_cell = tf.nn.rnn_cell.MultiRNNCell(lstm_cells)
else:
lstm_cell = lstm_cells[0]
with tf.control_dependencies([lstm_input]):
# 初始化状态
self.init_lstm_state.append(
lstm_cell.zero_state(self.batch_size, tf.float32)
)
if self.bidirectional:
with tf.variable_scope('RNN_%s' % lstm_num):
# 从最后一步开始,获取最后一步的输出,和最终的隐含状态,确保正反向LSTM单元可以拼接起来
_lstm_output_unpacked, final_state = tf.nn.static_rnn(
lstm_cell,
# 将每个词对应的张量进行分离并作为LSTM的输入
tf.unstack(lstm_input, axis = 1),
initial_state = self.init_lstm_state[-1],
)
else:
_lstm_output_unpacked, final_state = tf.nn.static_rnn(
lstm_cell,
tf.unstack(lstm_input, axis = 1),
initial_state = self.init_lstm_state[-1],
)
self.final_lstm_state.append(final_state)
# [batch_size,num_proj]
# print(final_state)
# 将一个隐含层的输出拼接起来 [batch_size,20,256]
lstm_output_flat = tf.reshape(
tf.stack(_lstm_output_unpacked, axis = 1), [-1, projection_dim]
)
print(lstm_output_flat)
tf.add_to_collection(
'lstm_output_embeddings', _lstm_output_unpacked
)
lstm_outputs.append(lstm_output_flat)
self._build_loss(lstm_outputs)
# 构建损失函数
def _build_loss(self, lstm_outputs):
batch_size = self.options['batch_size']
unroll_steps = self.options['unroll_steps']
# 所有词的数量
n_tokens_vocab = self.options['n_tokens_vocab']
def _get_next_token_placeholders(suffix):
name = 'next_token_id' + suffix
id_placeholder = tf.placeholder(
tf.int32, shape = (None, unroll_steps), name = name
)
return id_placeholder
self.next_token_id = _get_next_token_placeholders('')
# 每次抽取[batch_size,unroll_nums]个词
print(self.next_token_id)
if self.bidirectional:
self.next_token_id_reverse = _get_next_token_placeholders(
'_reverse'
)
# softmax的维度为projection_dim(256)
softmax_dim = self.options['lstm']['projection_dim']
# 与词嵌入的权重共享
if self.share_embedding_softmax:
self.softmax_W = self.embedding_weights
# 初始化softmax的参数
with tf.variable_scope('softmax'), tf.device('/cpu:0'):
softmax_init = tf.random_normal_initializer(
0.0, 1.0 / np.sqrt(softmax_dim)
)
# softmax分布到每一个词中
if not self.share_embedding_softmax:
self.softmax_W = tf.get_variable(
'W',
[n_tokens_vocab, softmax_dim],
dtype = tf.float32,
initializer = softmax_init,
)
self.softmax_b = tf.get_variable(
'b',
[n_tokens_vocab],
dtype = tf.float32,
initializer = tf.constant_initializer(0.0),
)
self.individual_losses = []
if self.bidirectional:
next_ids = [self.next_token_id, self.next_token_id_reverse]
else:
next_ids = [self.next_token_id]
print(lstm_outputs)
self.output_scores = tf.identity(lstm_outputs, name = 'softmax_score')
print(self.output_scores)
for id_placeholder, lstm_output_flat in zip(next_ids, lstm_outputs):
next_token_id_flat = tf.reshape(id_placeholder, [-1, 1])
with tf.control_dependencies([lstm_output_flat]):
if self.is_training and self.sample_softmax:
losses = tf.nn.sampled_softmax_loss(
self.softmax_W,
self.softmax_b,
next_token_id_flat,
lstm_output_flat,
int(
self.options['n_negative_samples_batch']
* self.options['n_tokens_vocab']
),
self.options['n_tokens_vocab'],
num_true = 1,
)
else:
output_scores = (
tf.matmul(
lstm_output_flat, tf.transpose(self.softmax_W)
)
+ self.softmax_b
)
losses = tf.nn.sparse_softmax_cross_entropy_with_logits(
logits = self.output_scores,
labels = tf.squeeze(
next_token_id_flat, squeeze_dims = [1]
),
)
self.individual_losses.append(tf.reduce_mean(losses))
if self.bidirectional:
self.total_loss = 0.5 * (
self.individual_losses[0] + self.individual_losses[1]
)
else:
self.total_loss = self.individual_losses[0]
tf.reset_default_graph()
sess = tf.InteractiveSession()
model = LanguageModel(options, True)
sess.run(tf.global_variables_initializer())
from tqdm import tqdm
def _get_feed_dict_from_X(X, model, char_inputs, bidirectional):
feed_dict = {}
if not char_inputs:
token_ids = X['token_ids']
feed_dict[model.token_ids] = token_ids
else:
char_ids = X['tokens_characters']
feed_dict[model.tokens_characters] = char_ids
if bidirectional:
if not char_inputs:
feed_dict[model.token_ids_reverse] = X['token_ids_reverse']
else:
feed_dict[model.tokens_characters_reverse] = X['tokens_characters_reverse']
next_id_placeholders = [[model.next_token_id, '']]
if bidirectional:
next_id_placeholders.append([model.next_token_id_reverse, '_reverse'])
for id_placeholder, suffix in next_id_placeholders:
name = 'next_token_id' + suffix
feed_dict[id_placeholder] = X[name]
return feed_dict
bidirectional = options.get('bidirectional', False)
batch_size = options['batch_size']
unroll_steps = options['unroll_steps']
n_train_tokens = options.get('n_train_tokens')
n_tokens_per_batch = batch_size * unroll_steps
n_batches_per_epoch = int(n_train_tokens / n_tokens_per_batch)
n_batches_total = options['n_epochs'] * n_batches_per_epoch
init_state_tensors = model.init_lstm_state
final_state_tensors = model.final_lstm_state
char_inputs = 'char_cnn' in options
if char_inputs:
max_chars = options['char_cnn']['max_characters_per_token']
feed_dict = {
model.tokens_characters: np.zeros(
[batch_size, unroll_steps, max_chars], dtype = np.int32
)
}
else:
feed_dict = {model.token_ids: np.zeros([batch_size, unroll_steps])}
if bidirectional:
if char_inputs:
feed_dict.update(
{
model.tokens_characters_reverse: np.zeros(
[batch_size, unroll_steps, max_chars], dtype = np.int32
)
}
)
else:
feed_dict.update(
{
model.token_ids_reverse: np.zeros(
[batch_size, unroll_steps], dtype = np.int32
)
}
)
init_state_values = sess.run(init_state_tensors, feed_dict = feed_dict)
data_gen = bi.iter_batches(batch_size, unroll_steps)
pbar = tqdm(range(n_batches_total), desc = 'train minibatch loop')
for p in pbar:
batch = next(data_gen)
feed_dict = {t: v for t, v in zip(init_state_tensors, init_state_values)}
feed_dict.update(_get_feed_dict_from_X(batch, model, char_inputs, bidirectional))
score, loss, _, init_state_values = sess.run([model.output_scores,
model.total_loss, model.optimizer, final_state_tensors],
feed_dict = feed_dict)
pbar.set_postfix(cost = loss)
word_embed = model.softmax_W.eval()
from scipy.spatial.distance import cdist
from sklearn.neighbors import NearestNeighbors
word = '金轮'
nn = NearestNeighbors(10, metric = 'cosine').fit(word_embed)
distances, idx = nn.kneighbors(word_embed[dictionary[word]].reshape((1, -1)))
word_list = []
for i in range(1, idx.shape[1]):
word_list.append([rev_dictionary[idx[0, i]], 1 - distances[0, i]])
word_list
```
|
github_jupyter
|
# Base dos dados
Base dos Dados is a Brazilian project of consolidation of datasets in a common repository with easy to follow codes.
Download *clean, integrated and updated* datasets in an easy way through SQL, Python, R or CLI (Stata in development). With Base dos Dados you have freedom to:
- download whole tables
- download tables partially
- cross datasets and download final product
This Jupyter Notebook aims to help Python coders in main funcionalities of the platform. If you just heard about Base Dos Dados this is the place to come and start your journey.
This manual is divided into three sections:
- Installation and starting commands
- Using Base dos Dados with Pandas package
- Examples
It is recommended to have know about the package Pandas before using *basedosdados* since data will be stored as dataframes.
If you have any feedback about this material please send me an email and I will gladly help: [email protected]
### 1. Installation and starting commands
Base dos Dados are located in pip and to install it you have to run the following code:
```
#!pip install basedosdados==1.3.0a3
```
Using an exclamation mark before the command will pass the command to the shell (not to the Python interpreter). If you prefer you can type *pip install basedosdados* directly in your Command Prompt.<br>
Before using *basedosdados* you have to import it:
```
import basedosdados as bd
```
Now that you have installed and imported the package it is time to see what data is available. The following command will list all current datasets.<br>
<br>
Note: the first time you execute a command in *basedosdados* it will prompt you to login with your Google Cloud. This is because results are delivered through BigQuery (which belongs to Google). BigQuery provides a reliable and quick service for managing data. It is free unless you require more than 1TB per month. If this happens to you just follow the on-screen instructions.
```
bd.list_datasets()
```
Note: the first time you execute a command in *basedosdados* it will prompt you to login with your Google Cloud. This is because results are delivered through BigQuery (which belongs to Google). BigQuery provides a reliable and quick service for managing data. It is free unless you require more than 1TB per month. If the login screen appears to you just follow the on-screen instructions.
You can check the data you want with the *get_dataset_description*. We will use *br_ibge_pib* as example:
```
bd.get_dataset_description('br_ibge_pib')
```
Each of the *datasets* has tables inside and to see what are the available tables you can use the following command:
```
bd.list_dataset_tables('br_ibge_pib')
```
To analyze which information is in that table you can run the following command.
```
bd.get_table_description('br_ibge_pib', 'municipios')
```
With these three commands you are set and know what data is available. You are now able to create your dataframe and start your analysis! In this example we will generate a dataframe called *df* with information regarding Brazilian municipalities' GDP. The data is stored as a dataframe object and it will interact with the same commands used in Pandas.
```
df = bd.read_table('br_ibge_pib', 'municipios')
# Note: depending on how you configured your Google Cloud setup, it might be necessary to add project ID in the request.
# Like `bd.read_table('br_ibge_pib', 'municipios', billing_project_id=<YOUR_PROJECT_ID>)
```
### Example
In this section we will do quick exploratory example on the dataset we have been using. <br>
First by let's define again the table and take a look at it:
```
df.head()
```
#### Example 1: São Paulo
The cities are being indexed by their ID You can see the correspondence in the following website: https://www.ibge.gov.br/explica/codigos-dos-municipios.php <br>
<br>
We will take a look at São Paulo city with the following ID: 3550308<br>
Creating a dataframe with São Paulo information:
```
sao_paulo = df[df.id_municipio == 3550308]
```
Looking at the table created we can check that everything is working as expected.
```
sao_paulo
```
Pandas has some graph functions built in and we can use them to further visualize data:
```
sao_paulo.plot(x='ano', y='PIB')
```
Since the tables are in nominal prices it might be interesting to see results in logarithmic form. For that we can use the following code:
```
import numpy as np
#Adding new columns:
df['log_pib'] = df.PIB.apply(np.log10)
#Separating Sao Paulo again, now it will come with the new columns:
sao_paulo = df[df.id_municipio == 3550308]
```
Analyzing data the way we did before:
```
sao_paulo
sao_paulo.plot(x='ano',y='log_pib')
```
#### Example 2: 2015
If you want to check all value added to GDP by the services sector in the year 2015 you could use the following code:
```
# First separating 2015 data only:
municipios2015 = df[df.ano == 2015]
#Adding logarithmic column:
municipios2015['log_VA_servicos'] = municipios2015.VA_servicos.apply(np.log10)
#Visualizing dataframe as histogram:
municipios2015[municipios2015['log_VA_servicos'] > 0].hist('log_VA_servicos')
```
For the last piece of code we are using conditional statement '> 0' because it might be the case of a city with value of zero in the *VA_servicos* and when we calculate log it is equal to -inf.
|
github_jupyter
|
##### Copyright 2018 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Text classification with an RNN
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/beta/tutorials/text/text_classification_rnn"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/r2/tutorials/text/text_classification_rnn.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/r2/tutorials/text/text_classification_rnn.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/docs/site/en/r2/tutorials/text/text_classification_rnn.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table>
This text classification tutorial trains a [recurrent neural network](https://developers.google.com/machine-learning/glossary/#recurrent_neural_network) on the [IMDB large movie review dataset](http://ai.stanford.edu/~amaas/data/sentiment/) for sentiment analysis.
```
from __future__ import absolute_import, division, print_function, unicode_literals
try:
# %tensorflow_version only exists in Colab.
%tensorflow_version 2.x
except Exception:
pass
import tensorflow_datasets as tfds
import tensorflow as tf
```
Import `matplotlib` and create a helper function to plot graphs:
```
import matplotlib.pyplot as plt
def plot_graphs(history, string):
plt.plot(history.history[string])
plt.plot(history.history['val_'+string])
plt.xlabel("Epochs")
plt.ylabel(string)
plt.legend([string, 'val_'+string])
plt.show()
```
## Setup input pipeline
The IMDB large movie review dataset is a *binary classification* dataset—all the reviews have either a *positive* or *negative* sentiment.
Download the dataset using [TFDS](https://www.tensorflow.org/datasets). The dataset comes with an inbuilt subword tokenizer.
```
dataset, info = tfds.load('imdb_reviews/subwords8k', with_info=True,
as_supervised=True)
train_dataset, test_dataset = dataset['train'], dataset['test']
```
As this is a subwords tokenizer, it can be passed any string and the tokenizer will tokenize it.
```
tokenizer = info.features['text'].encoder
print ('Vocabulary size: {}'.format(tokenizer.vocab_size))
sample_string = 'TensorFlow is cool.'
tokenized_string = tokenizer.encode(sample_string)
print ('Tokenized string is {}'.format(tokenized_string))
original_string = tokenizer.decode(tokenized_string)
print ('The original string: {}'.format(original_string))
assert original_string == sample_string
```
The tokenizer encodes the string by breaking it into subwords if the word is not in its dictionary.
```
for ts in tokenized_string:
print ('{} ----> {}'.format(ts, tokenizer.decode([ts])))
BUFFER_SIZE = 10000
BATCH_SIZE = 64
train_dataset = train_dataset.shuffle(BUFFER_SIZE)
train_dataset = train_dataset.padded_batch(BATCH_SIZE, train_dataset.output_shapes)
test_dataset = test_dataset.padded_batch(BATCH_SIZE, test_dataset.output_shapes)
```
## Create the model
Build a `tf.keras.Sequential` model and start with an embedding layer. An embedding layer stores one vector per word. When called, it converts the sequences of word indices to sequences of vectors. These vectors are trainable. After training (on enough data), words with similar meanings often have similar vectors.
This index-lookup is much more efficient than the equivalent operation of passing a one-hot encoded vector through a `tf.keras.layers.Dense` layer.
A recurrent neural network (RNN) processes sequence input by iterating through the elements. RNNs pass the outputs from one timestep to their input—and then to the next.
The `tf.keras.layers.Bidirectional` wrapper can also be used with an RNN layer. This propagates the input forward and backwards through the RNN layer and then concatenates the output. This helps the RNN to learn long range dependencies.
```
model = tf.keras.Sequential([
tf.keras.layers.Embedding(tokenizer.vocab_size, 64),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(64)),
tf.keras.layers.Dense(64, activation='relu'),
tf.keras.layers.Dense(1, activation='sigmoid')
])
```
Compile the Keras model to configure the training process:
```
model.compile(loss='binary_crossentropy',
optimizer='adam',
metrics=['accuracy'])
```
## Train the model
```
history = model.fit(train_dataset, epochs=10,
validation_data=test_dataset)
test_loss, test_acc = model.evaluate(test_dataset)
print('Test Loss: {}'.format(test_loss))
print('Test Accuracy: {}'.format(test_acc))
```
The above model does not mask the padding applied to the sequences. This can lead to skewness if we train on padded sequences and test on un-padded sequences. Ideally the model would learn to ignore the padding, but as you can see below it does have a small effect on the output.
If the prediction is >= 0.5, it is positive else it is negative.
```
def pad_to_size(vec, size):
zeros = [0] * (size - len(vec))
vec.extend(zeros)
return vec
def sample_predict(sentence, pad):
tokenized_sample_pred_text = tokenizer.encode(sample_pred_text)
if pad:
tokenized_sample_pred_text = pad_to_size(tokenized_sample_pred_text, 64)
predictions = model.predict(tf.expand_dims(tokenized_sample_pred_text, 0))
return (predictions)
# predict on a sample text without padding.
sample_pred_text = ('The movie was cool. The animation and the graphics '
'were out of this world. I would recommend this movie.')
predictions = sample_predict(sample_pred_text, pad=False)
print (predictions)
# predict on a sample text with padding
sample_pred_text = ('The movie was cool. The animation and the graphics '
'were out of this world. I would recommend this movie.')
predictions = sample_predict(sample_pred_text, pad=True)
print (predictions)
plot_graphs(history, 'accuracy')
plot_graphs(history, 'loss')
```
## Stack two or more LSTM layers
Keras recurrent layers have two available modes that are controlled by the `return_sequences` constructor argument:
* Return either the full sequences of successive outputs for each timestep (a 3D tensor of shape `(batch_size, timesteps, output_features)`).
* Return only the last output for each input sequence (a 2D tensor of shape (batch_size, output_features)).
```
model = tf.keras.Sequential([
tf.keras.layers.Embedding(tokenizer.vocab_size, 64),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(
64, return_sequences=True)),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(32)),
tf.keras.layers.Dense(64, activation='relu'),
tf.keras.layers.Dense(1, activation='sigmoid')
])
model.compile(loss='binary_crossentropy',
optimizer='adam',
metrics=['accuracy'])
history = model.fit(train_dataset, epochs=10,
validation_data=test_dataset)
test_loss, test_acc = model.evaluate(test_dataset)
print('Test Loss: {}'.format(test_loss))
print('Test Accuracy: {}'.format(test_acc))
# predict on a sample text without padding.
sample_pred_text = ('The movie was not good. The animation and the graphics '
'were terrible. I would not recommend this movie.')
predictions = sample_predict(sample_pred_text, pad=False)
print (predictions)
# predict on a sample text with padding
sample_pred_text = ('The movie was not good. The animation and the graphics '
'were terrible. I would not recommend this movie.')
predictions = sample_predict(sample_pred_text, pad=True)
print (predictions)
plot_graphs(history, 'accuracy')
plot_graphs(history, 'loss')
```
Check out other existing recurrent layers such as [GRU layers](https://www.tensorflow.org/api_docs/python/tf/keras/layers/GRU).
|
github_jupyter
|
## MIDI Generator
```
## Uncomment command below to kill current job:
#!neuro kill $(hostname)
import random
import sys
import subprocess
import torch
sys.path.append('../midi-generator')
%load_ext autoreload
%autoreload 2
import IPython.display as ipd
from model.dataset import MidiDataset
from utils.load_model import load_model
from utils.generate_midi import generate_midi
from utils.seed import set_seed
from utils.write_notes import write_notes
```
Each `*.mid` file can be thought of as a sequence where notes and chords follow each other with specified time offsets between them. So, following this model a next note can be predicted with a `seq2seq` model. In this work, a simple `GRU`-based model is used.
Note that the number of available notes and chord in vocabulary is not specified and depends on a dataset which a model was trained on.
To listen to MIDI files from Jupyter notebook, let's define help function which transforms `*.mid` file to `*.wav` file.
```
def mid2wav(mid_path, wav_path):
subprocess.check_output(['timidity', mid_path, '-OwS', '-o', wav_path])
```
The next step is loading the model from the checkpoint. To make experiments reproducible let's also specify random seed.
You can also try to use the model, which was trained with label smoothing (see `../results/smoothing.ch`).
```
seed = 1234
set_seed(seed)
device = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
print(device)
model, vocab = load_model(checkpoint_path='../results/test.ch', device=device)
```
Let's also specify additional help function to avoid code duplication.
```
def dump_result(file_preffix, vocab, note_seq, offset_seq=None):
note_seq = vocab.decode(note_seq)
notes = MidiDataset.decode_notes(note_seq, offset_seq=offset_seq)
mid_path = file_preffix + '.mid'
wav_path = file_preffix + '.wav'
write_notes(mid_path, notes)
mid2wav(mid_path, wav_path)
return wav_path
```
# MIDI file generation
Let's generate a new file. Note that the parameter `seq_len` specifies the length of the output sequence of notes.
Function `generate_midi` return sequence of generated notes and offsets between them.
## Nucleus (`top-p`) Sampling
Sample from the most probable tokens, which sum of probabilities gives `top-p`. If `top-p == 0` the most probable token is sampled.
## Temperature
As `temperature` → 0 this approaches greedy decoding, while `temperature` → ∞ asymptotically approaches uniform sampling from the vocabulary.
```
note_seq, offset_seq = generate_midi(model, vocab, seq_len=128, top_p=0, temperature=1, device=device)
```
Let's listen to result midi.
```
# midi with constant offsets
ipd.Audio(dump_result('../results/output_without_offsets', vocab, note_seq, offset_seq=None))
# midi with generated offsets
ipd.Audio(dump_result('../results/output_with_offsets.mid', vocab, note_seq, offset_seq))
```
The result with constant offsets sounds better, doesn't it? :)
Be free to try different generation parameters (`top-p` and `temperature`) to understand their impact on the resulting sound.
You can also train your own model with different specs (e.g. different hidden size) or use label smoothing during training.
# Continue existing file
## Continue sampled notes
For beginning, let's continue sound that consists of sampled from `vocab` notes.
```
seed = 4321
set_seed(seed)
history_notes = random.choices(range(len(vocab)), k=20)
history_offsets = len(history_notes) * [0.5]
ipd.Audio(dump_result('../results/random_history', vocab, history_notes, history_offsets))
```
It sounds a little bit chaotic. Let's try to continue this with our model.
```
history = [*zip(history_notes, history_offsets)]
note_seq, offset_seq = generate_midi(model, vocab, seq_len=128, top_p=0, temperature=1, device=device,
history=history)
# midi with constant offsets
ipd.Audio(dump_result('../results/random_without_offsets', vocab, note_seq, offset_seq=None))
```
After the sampled part ends, the generated melody starts to sound better.
## Continue existed melody
```
raw_notest = MidiDataset.load_raw_notes('../data/mining.mid')
org_note_seq, org_offset_seq = MidiDataset.encode_notes(raw_notest)
org_note_seq = vocab.encode(org_note_seq)
```
Let's listen to it
```
ipd.Audio(dump_result('../results/original_sound', vocab, org_note_seq, org_offset_seq))
```
and take 20 first elements from the sequence as out history sequence.
```
history_notes = org_note_seq[:20]
history_offsets = org_offset_seq[:20]
history = [*zip(history_notes, history_offsets)]
note_seq, offset_seq = generate_midi(model, vocab, seq_len=128, top_p=0, temperature=1, device=device,
history=history)
# result melody without generated offsets
ipd.Audio(dump_result('../results/continue_rand_without_offsets', vocab, note_seq, offset_seq=None))
# result melody with generated offsets
ipd.Audio(dump_result('../results/continue_rand_with_offsets', vocab, note_seq, offset_seq))
```
You can try to overfit your model on one melody to get better results. Otherwise, you can use already pretrained model (`../results/onemelody.ch`)
# Model overfitted on one melody
Let's try the same thing which we did before. Let's continue melody, but this time do it with the model,
which was overfitted with this melody.
```
seed = 1234
set_seed(seed)
device = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
model, vocab = load_model(checkpoint_path='../results/onemelody.ch', device=device)
raw_notest = MidiDataset.load_raw_notes('../data/Final_Fantasy_Matouyas_Cave_Piano.mid')
org_note_seq, org_offset_seq = MidiDataset.encode_notes(raw_notest)
org_note_seq = vocab.encode(org_note_seq)
```
Let's listen to it.
```
ipd.Audio(dump_result('../results/onemelody_original_sound', vocab, org_note_seq, org_offset_seq))
end = 60
history_notes = org_note_seq[:end]
history_offsets = org_offset_seq[:end]
```
Listen to history part of loaded melody.
```
ipd.Audio(dump_result('../results/onemelody_history', vocab, history_notes, history_offsets))
```
Now we can try to continue the original melody with our model. But firstly, you can listen to the original tail part of the melody do refresh it in the memory and have reference to compare with.
```
tail_notes = org_note_seq[end:]
tail_offsets = org_offset_seq[end:]
ipd.Audio(dump_result('../results/onemelody_tail', vocab, tail_notes, tail_offsets))
history = [*zip(history_notes, history_offsets)]
note_seq, offset_seq = generate_midi(model, vocab, seq_len=128, top_p=0, temperature=1, device=device,
history=history)
# delete history part
note_seq = note_seq[end:]
offset_seq = offset_seq[end:]
# result melody without generated offsets
ipd.Audio(dump_result('../results/continue_onemelody_without_offsets', vocab, note_seq, offset_seq=None))
# result melody with generated offsets
ipd.Audio(dump_result('../results/continue_onemelody_with_offsets', vocab, note_seq, offset_seq))
```
As you can hear, this time, the model generated better offsets and the result melody does not sound so chaostic.
|
github_jupyter
|
# Sandbox - Tutorial
## Building a fiber bundle
A [fiber bundle](https://github.com/3d-pli/fastpli/wiki/FiberModel) consit out of multiple individual nerve fibers.
A fiber bundle is a list of fibers, where fibers are represented as `(n,4)-np.array`.
This makes desining individually fiber of any shape possible.
However since nerve fibers are often in nerve fiber bundles, this toolbox allows to fill fiber_bundles from a pattern of fibers.
Additionally this toolbox also allows to build parallell cubic shapes as well as different kinds of cylindric shapes to allow a faster building experience.
## General imports
First, we prepair all necesarry modules and defining a function to euqalice all three axis of an 3d plot.
You can change the `magic ipython` line from `inline` to `qt`.
This generate seperate windows allowing us also to rotate the resulting plots and therfore to investigate the 3d models from different views.
Make sure you have `PyQt5` installed if you use it.
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# %matplotlib qt
import fastpli.model.sandbox as sandbox
def set_3d_axes_equal(ax):
x_limits = ax.get_xlim3d()
y_limits = ax.get_ylim3d()
z_limits = ax.get_zlim3d()
x_range = abs(x_limits[1] - x_limits[0])
x_middle = np.mean(x_limits)
y_range = abs(y_limits[1] - y_limits[0])
y_middle = np.mean(y_limits)
z_range = abs(z_limits[1] - z_limits[0])
z_middle = np.mean(z_limits)
plot_radius = 0.5 * max([x_range, y_range, z_range])
ax.set_xlim3d([x_middle - plot_radius, x_middle + plot_radius])
ax.set_ylim3d([y_middle - plot_radius, y_middle + plot_radius])
ax.set_zlim3d([z_middle - plot_radius, z_middle + plot_radius])
```
## Designing a fiber bundle
The idea is to build design first a macroscopic struces, i. e. nerve fiber bundles, which can then at a later step be filled with individual nerve fibers.
We start by defining a fiber bundle as a trajectory of points (similar to fibers).
As an example we start with use a helical form.
```
t = np.linspace(0, 4 * np.pi, 50, True)
traj = np.array((42 * np.cos(t), 42 * np.sin(t), 10 * t)).T
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1, projection='3d')
ax.plot(
traj[:, 0],
traj[:, 1],
traj[:, 2],
)
plt.title("fb trajectory")
set_3d_axes_equal(ax)
plt.show()
```
### seed points
seed points are used to initialize the populating process of individual fibers inside the fiber bundle.
Seed points are a list of 3d points.
This toolbox provides two methods to build seed points pattern.
The first one is a 2d triangular grid.
It is defined by a `width`, `height` and an inside `spacing` between the seed point.
Additionally one can actiavte the `center` option so that the seed points are centered around a seed point at `(0,0,0)`.
The second method provides a circular shape instead of a rectangular.
However it can also be achievd by using an additional function `crop_circle` which returns only seed points along the first two dimensions with the defined `radius` around the center.
```
seeds = sandbox.seeds.triangular_grid(width=42,
height=42,
spacing=6,
center=True)
radius = 21
circ_seeds = sandbox.seeds.crop_circle(radius=radius, seeds=seeds)
fig, ax = plt.subplots(1, 1)
plt.title("seed points")
plt.scatter(seeds[:, 0], seeds[:, 1])
plt.scatter(circ_seeds[:, 0], circ_seeds[:, 1])
ax.set_aspect('equal', 'box')
# plot circle margin
t = np.linspace(0, 2 * np.pi, 42)
x = radius * np.cos(t)
y = radius * np.sin(t)
plt.plot(x, y)
plt.show()
```
### Generating a fiber bundle from seed points
The next step is to build a fiber bundle from the desined trajectory and seed points.
However one additional step is necesarry.
Since nerve fibers are not a line, but a 3d object, they need also a volume for the later `solving` and `simulation` steps of this toolbox.
This toolbox describes nerve fibers as tubes, which are defined by a list of points and radii, i. e. (n,4)-np.array).
The radii `[:,3]` can change along the fiber trajectories `[:,0:3]` allowiing for a change of thickness.
Now we have everything we need to build a fiber bundle from the desined trajectory and seed points.
The function `bundle` provides this funcionallity.
Additionally to the `traj` and `seeds` parameter the `radii` can be a single number if all fibers should have the same radii, or a list of numbers, if each fiber shell have a different radii.
An additional `scale` parameter allows to scale the seed points along the trajectory e. g. allowing for a fanning.
```
# populating fiber bundle
fiber_bundle = sandbox.build.bundle(
traj=traj,
seeds=circ_seeds,
radii=np.random.uniform(0.5, 0.8, circ_seeds.shape[0]),
scale=0.25 + 0.5 * np.linspace(0, 1, traj.shape[0]))
# plotting
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1, projection='3d')
for fiber in fiber_bundle:
ax.plot(fiber[:, 0], fiber[:, 1], fiber[:, 2])
plt.title("helical thinning out fiber bundle")
set_3d_axes_equal(ax)
plt.show()
```
## Additional macroscopic structures
In the development and using of this toolbox, it was found that it is usefull to have other patterns than filled fiber bundles to build macroscopic structures.
Depending on a brain sections, where the nerve fiber orientation is measured with the 3D-PLI technique, nerve fibers can be visibale as type of patterns.
### Cylindrical shapes
Radial shaped patterns can be quickly build with the following `cylinder` method.
A hollow cylinder is defined by a inner and outer radii `r_in` and `r_out`, along two points `p` and `q`.
Additionally the cylinder can be also only partial along its radius by defining two angles `alpha` and `beta`.
Again as for the `bundle` method, one needs seed points to defining a pattern.
Filling this cylindrig shape can be performed by three differet `mode`s:
- radial
- circular
- parallel
```
# plotting
seeds = sandbox.seeds.triangular_grid(width=200,
height=200,
spacing=5,
center=True)
fig, axs = plt.subplots(1, 3, figsize=(15,5), subplot_kw={'projection':'3d'}, constrained_layout=True)
for i, mode in enumerate(['radial', 'circular', 'parallel']):
# ax = fig.add_subplot(1, 1, 1, projection='3d')
fiber_bundle = sandbox.build.cylinder(p=(0, 80, 50),
q=(40, 80, 100),
r_in=20,
r_out=40,
seeds=seeds,
radii=1,
alpha=np.deg2rad(20),
beta=np.deg2rad(160),
mode=mode)
for fiber in fiber_bundle:
axs[i].plot(fiber[:, 0], fiber[:, 1], fiber[:, 2])
set_3d_axes_equal(axs[i])
axs[i].set_title(f'{mode}')
plt.show()
```
### Cubic shapes
The next method allows placing fibers inside a cube with a use definde direction.
The cube is definded by two 3d points `p` and `q`.
The direction of the fibers inside the cube is defined by spherical angels `phi` and `theta`.
Seed points again describe the pattern of fibers inside the cube.
The seed points (rotated its xy-plane according to `phi` and `theta`) are places at point `q` and `q`.
From the corresponding seed points are the starting and end point for each fiber.
```
# define cub corner points
p = np.array([0, 80, 50])
q = np.array([40, 180, 100])
# create seed points which will fill the cube
d = np.max(np.abs(p - q)) * np.sqrt(3)
seeds = sandbox.seeds.triangular_grid(width=d,
height=d,
spacing=10,
center=True)
# fill a cube with (theta, phi) directed fibers
fiber_bundle = sandbox.build.cuboid(p=p,
q=q,
phi=np.deg2rad(45),
theta=np.deg2rad(90),
seeds=seeds,
radii=1)
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1, projection='3d')
for fiber in fiber_bundle:
ax.plot(fiber[:, 0], fiber[:, 1], fiber[:, 2])
plt.title('cubic shape')
set_3d_axes_equal(ax)
plt.show()
```
## next
from here further anatomical more interesting examples are presented in the solver tutorial and `examples/crossing.py` example.
|
github_jupyter
|
# Effect of House Characteristics on Their Prices
## by Lubomir Straka
## Investigation Overview
In this investigation, I wanted to look at the key characteristics of houses that could be used to predict their prices. The main focus was on three aspects: above grade living area representing space characteristics, overall quality of house's material and finish representing physical characteristics, and neighborhood cluster representing location characteristics.
## Dataset Overview
The data consists of information regarding 1460 houses in Ames, Iowa, including their sale price, physical characteristics, space properties and location within the city as provided by [Kaggle](https://www.kaggle.com/c/house-prices-advanced-regression-techniques/data). The data set contains 1460 observations and a large number of explanatory variables (23 nominal, 23 ordinal, 14 discrete, and 20 continuous) involved in assessing home values. These 79 explanatory variables plus one response variable (sale price) describe almost every aspect of residential homes in Ames, Iowa.
In addition to some basic data type encoding, missing value imputing and cleaning, four outliers were removed from the analysis due to unusual sale conditions.
```
# Import all packages and set plots to be embedded inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
# Suppress warnings from final output
import warnings
warnings.simplefilter("ignore")
# Load the Ames Housing dataset
path = 'https://raw.githubusercontent.com/lustraka/Data_Analysis_Workouts/main/Communicate_Data_Findings/ames_train_data.csv'
ames = pd.read_csv(path, index_col='Id')
################
# Wrangle data #
################
# The numeric features are already encoded correctly (`float` for
# continuous, `int` for discrete), but the categoricals we'll need to
# do ourselves. Note in particular, that the `MSSubClass` feature is
# read as an `int` type, but is actually a (nominative) categorical.
# The categorical features nominative (unordered)
catn = ["MSSubClass", "MSZoning", "Street", "Alley", "LandContour", "LotConfig",
"Neighborhood", "Condition1", "Condition2", "BldgType", "HouseStyle",
"RoofStyle", "RoofMatl", "Exterior1st", "Exterior2nd", "MasVnrType",
"Foundation", "Heating", "CentralAir", "GarageType", "MiscFeature",
"SaleType", "SaleCondition"]
# The categorical features ordinal (ordered)
# Pandas calls the categories "levels"
five_levels = ["Po", "Fa", "TA", "Gd", "Ex"]
ten_levels = list(range(10))
cato = {
"OverallQual": ten_levels,
"OverallCond": ten_levels,
"ExterQual": five_levels,
"ExterCond": five_levels,
"BsmtQual": five_levels,
"BsmtCond": five_levels,
"HeatingQC": five_levels,
"KitchenQual": five_levels,
"FireplaceQu": five_levels,
"GarageQual": five_levels,
"GarageCond": five_levels,
"PoolQC": five_levels,
"LotShape": ["Reg", "IR1", "IR2", "IR3"],
"LandSlope": ["Sev", "Mod", "Gtl"],
"BsmtExposure": ["No", "Mn", "Av", "Gd"],
"BsmtFinType1": ["Unf", "LwQ", "Rec", "BLQ", "ALQ", "GLQ"],
"BsmtFinType2": ["Unf", "LwQ", "Rec", "BLQ", "ALQ", "GLQ"],
"Functional": ["Sal", "Sev", "Maj1", "Maj2", "Mod", "Min2", "Min1", "Typ"],
"GarageFinish": ["Unf", "RFn", "Fin"],
"PavedDrive": ["N", "P", "Y"],
"Utilities": ["NoSeWa", "NoSewr", "AllPub"],
"CentralAir": ["N", "Y"],
"Electrical": ["Mix", "FuseP", "FuseF", "FuseA", "SBrkr"],
"Fence": ["MnWw", "GdWo", "MnPrv", "GdPrv"],
}
# Add a None level for missing values
cato = {key: ["None"] + value for key, value in
cato.items()}
def encode_dtypes(df):
"""Encode nominal and ordinal categorical variables."""
global catn, cato
# Nominal categories
for name in catn:
df[name] = df[name].astype("category")
# Add a None category for missing values
if "None" not in df[name].cat.categories:
df[name].cat.add_categories("None", inplace=True)
# Ordinal categories
for name, levels in cato.items():
df[name] = df[name].astype(pd.CategoricalDtype(levels,
ordered=True))
return df
def impute_missing(df):
"""Impute zeros to numerical and None to categorical variables."""
for name in df.select_dtypes("number"):
df[name] = df[name].fillna(0)
for name in df.select_dtypes("category"):
df[name] = df[name].fillna("None")
return df
def clean_data(df):
"""Remedy typos and mistakes based on EDA."""
global cato
# YearRemodAdd: Remodel date (same as construction date if no remodeling or additions)
df.YearRemodAdd = np.where(df.YearRemodAdd < df.YearBuilt, df.YearBuilt, df.YearRemodAdd)
assert len(df.loc[df.YearRemodAdd < df.YearBuilt]) == 0, 'Check YearRemodAdd - should be greater or equal then YearBuilt'
# Check range of years
yr_max = 2022
# Some values of GarageYrBlt are corrupt. Fix them by replacing them with the YearBuilt
df.GarageYrBlt = np.where(df.GarageYrBlt > yr_max, df.YearBuilt, df.GarageYrBlt)
assert df.YearBuilt.max() < yr_max and df.YearBuilt.min() > 1800, 'Check YearBuilt min() and max()'
assert df.YearRemodAdd.max() < yr_max and df.YearRemodAdd.min() > 1900, 'Check YearRemodAdd min() and max()'
assert df.YrSold.max() < yr_max and df.YrSold.min() > 2000, 'Check YrSold min() and max()'
assert df.GarageYrBlt.max() < yr_max and df.GarageYrBlt.min() >= 0, 'Check GarageYrBlt min() and max()'
# Check values of ordinal catagorical variables
for k in cato.keys():
assert set(df[k].unique()).difference(df[k].cat.categories) == set(), f'Check values of {k}'
# Check typos in nominal categorical variables
df['Exterior2nd'] = df['Exterior2nd'].replace({'Brk Cmn':'BrkComm', 'CmentBd':'CemntBd', 'Wd Shng':'WdShing'})
# Renew a data type after replacement
df['Exterior2nd'] = df['Exterior2nd'].astype("category")
if "None" not in df['Exterior2nd'].cat.categories:
df['Exterior2nd'].cat.add_categories("None", inplace=True)
return df
def label_encode(df):
"""Encode categorical variables using their dtype setting."""
X = df.copy()
for colname in X.select_dtypes(["category"]):
X[colname] = X[colname].cat.codes
return X
# Pre-process data
ames = encode_dtypes(ames)
ames = impute_missing(ames)
ames = clean_data(ames)
# Add log transformed SalePrice to the dataset
ames['logSalePrice'] = ames.SalePrice.apply(np.log10)
```
## Distribution of House Prices
**Graph on left**
The values of the response variable *SalePrice* are distributed between \$34.900 and \$755.000 with one mode at \$140.000, which is lower than the median at \$163.000, which is lower than the average price of \$180.921. The distribution of *SalePrice* is asymmetric with relatively few large values and tails off to the right. It is also relatively peaked.
**Graph on right**
For analysis of the relationships with other variables would be more suitable a log transformation of *SalePrice*. The distribution of *logSalePrice* is almost symmetric with skewness close to zero although the distribution is still a bit peaked.
```
def log_trans(x, inverse=False):
"""Get log or tenth power of the argument."""
if not inverse:
return np.log10(x)
else:
return 10**x
# Plot SalePrice with a standard and log scale
fig, axs = plt.subplots(1, 2, figsize=[16,5])
# LEFT plot
sns.histplot(data=ames, x='SalePrice', ax=axs[0])
axs[0].set_title('Distribution of Sale Price')
xticks = np.arange(100000, 800000, 100000)
axs[0].set_xticks(xticks)
axs[0].set_xticklabels([f'{int(xtick/1000)}K' for xtick in xticks])
axs[0].set_xlabel('Price ($)')
# RIGHT plot
sns.histplot(data=ames, x='logSalePrice', ax=axs[1])
axs[1].set_title('Distribution of Sale Price After Log Transformation')
lticks = [50000, 100000, 200000, 500000]
axs[1].set_xticks(log_trans(lticks))
axs[1].set_xticklabels([f'{int(xtick/1000)}K' for xtick in lticks])
axs[1].set_xlabel('Price ($)')
plt.show()
```
## Distribution of Living Area
The distribution of above grade living area (*GrLivArea*) is asymmetrical with skewness to the right and some peakness. There was two partial sales (houses not completed), one abnormal sale (trade, foreclosure, or short sale), and one normal, just simply unusual sale (very large house priced relatively appropriately). These outliers (any houses with *GrLivArea* greater then 4000 square feet) had been removed. This is a distribution of the cleaned dataset.
```
# Remove outliers
ames = ames.query('GrLivArea < 4000').copy()
# Plot a distribution of GrLivArea
sns.histplot(data=ames, x='GrLivArea')
plt.title('Distribution of Above Grade Living Area')
plt.xlabel('GrLivArea (sq ft)')
plt.show()
```
## Sale Price vs Overall Quality
Overall Quality (*OverallQual*) represents physical aspects of the building and rates the overall material and finish of the house. The moset frequent value of this categorical variable is average rate (5). The violin plot of sale price versus overall quality illuminates positive correlation between these variables. The higher the quality, the higher the price.
Interestingly, it looks like the missing values occur exclusively in observations with the best overall quality.
```
# Set the base color
base_color = sns.color_palette()[0]
# Show violin plot
plt.figure(figsize=(10,4))
sns.violinplot(data=ames, x='OverallQual', y='logSalePrice', color=base_color, inner='quartile')
plt.ylabel('Price ($)')
plt.yticks(log_trans(lticks), [f'{int(xtick/1000)}K' for xtick in lticks])
plt.xlabel('Overall Quality')
plt.title('Sale Price vs Overall Quality')
plt.show()
```
## Sale Price vs Neighborhood
Neighborhood represent the place where the house is located. The most frequent in the dataset are houses from North Ames (15 %) followed by houses from College Creek (10 %) and Old Town (8 %). To get insight into the effect of the nominal variable *Neighborhood* I had to cluster neighborhoods according to unit price per square feet. The violin plot of sale price versus three neighborhood clusters shows a positive correlation between these variables.
```
# Add a variable with total surface area
ames['TotalSF'] = ames['TotalBsmtSF'] + ames['1stFlrSF'] + ames['2ndFlrSF'] + ames['GarageArea']
# Calculate price per square feet
ames['SalePricePerSF'] = ames.SalePrice / ames.TotalSF
# Cluster neighborhoods into three clusters
ngb_mean_df = ames.groupby('Neighborhood')['SalePricePerSF'].mean().dropna()
bins = np.linspace(ngb_mean_df.min(), ngb_mean_df.max(), 4)
clusters = ['LowUnitPriceCluster', 'AvgUnitPriceCluster', 'HighUnitPriceCluster']
# Create a dict 'Neighborhood' : 'Cluster'
ngb_clusters = pd.cut(ngb_mean_df, bins=bins, labels=clusters, include_lowest=True).to_dict()
# Add new feature to the dataset
ames['NgbCluster'] = ames.Neighborhood.apply(lambda c: ngb_clusters.get(c, "")).astype(pd.CategoricalDtype(clusters, ordered=True))
# Plot the new feature
sns.violinplot(data=ames, x='NgbCluster', y='logSalePrice', color=base_color, inner='quartile')
plt.xlabel('Neighborhood Cluster')
plt.ylabel('Price ($)')
plt.yticks(log_trans(lticks), [f'{int(xtick/1000)}K' for xtick in lticks])
plt.title('Sale Price vs Neighborhood Cluster')
plt.show()
```
## Sale Price vs Living Area and Location
Two previous figures showed a positive correlation between building and location variables and sale price. Now, let's look at the relation between a space variable *GrLivArea* and price. The scatter plot of sale price versus living area with colored neighborhood clusters justifies the approximately linear relationship between price on one side and living area and location on the other side.
```
# Combine space and location variables
ax = sns.scatterplot(data=ames, x='GrLivArea', y='logSalePrice', hue='NgbCluster')
plt.title('Sale Price vs Living Area with Neighborhood Clusters')
plt.xlabel('Living Area (sq ft)')
plt.ylabel('Price ($)')
plt.yticks(log_trans(lticks), [f'{int(xtick/1000)}K' for xtick in lticks])
handles, _ = ax.get_legend_handles_labels()
ax.legend(handles[::-1], ['High Unit Price', 'Average Unit Price', 'Low Unit Price'], title='Neighborhood Clusters')
plt.show()
```
## Sale Price vs Overall Quality by Location
Finally, let's look at combination of overall quality and neighborhood clusters. These violin plots of sale price versus overall quality show differences between neighborhood clusters. The most diverse cluster in regards to overall quality of houses is the one with lower unit price per square feet. As expected, houses in the cluster with high unit prices per square feet have better overall quality as wall as higer prices.
```
# Combine building and location variables
g = sns.FacetGrid(data=ames, row='NgbCluster', aspect=3)
g.map(sns.violinplot, 'OverallQual', 'logSalePrice', inner='quartile')
g.set_titles('{row_name}')
g.set(yticks=log_trans(lticks), yticklabels=[f'{int(xtick/1000)}K' for xtick in lticks])
g.set_ylabels('Price ($)')
g.fig.suptitle('Sale Price vs OverallQual For Neighborhood Clusters', y=1.01)
plt.show()
```
## Conclusion
The investigation justified that the three selected explanatory variables of house can be used to predict the house sale price (a response variable). These variable are namely:
- *above grade living area* representing space characteristics,
- *overall quality* of house's material and finish representing physical characteristics, and
- *neighborhood cluster* representing location characteristics; this variable was engineered by clustering neighborhoods according to unit price per square feet.
All these variables have a positive relationship with sale price of the house.
## Thanks For Your Attention
Finished 2021-11-18
```
# !jupyter nbconvert Part_II_slide_deck_ames.ipynb --to slides --post serve --no-input --no-prompt
```
|
github_jupyter
|
```
import pickle
with open('ldaseq1234.pickle', 'rb') as f:
ldaseq = pickle.load(f)
print(ldaseq.print_topic_times(topic=0))
topicdis = [[0.04461942257217848,
0.08583100499534332,
0.0327237321141309,
0.0378249089831513,
0.08521717043434086,
0.03543307086614173,
0.054356108712217424,
0.04057658115316231,
0.0499745999491999,
0.04468292269917873,
0.028257556515113032,
0.026013885361104057,
0.0668021336042672,
0.07567098467530269,
0.08409533485733638,
0.026966387266107866,
0.04533909067818136,
0.028172889679112693,
0.04121158242316485,
0.06623063246126493],
[0.04375013958058825,
0.07278290193626193,
0.025437166402394087,
0.03566563190923912,
0.0600978180762445,
0.03541997007392188,
0.07258190588918417,
0.0305960649440561,
0.06355941666480559,
0.0459164303102039,
0.0295464189204279,
0.02733546240257275,
0.03622395426223284,
0.12723049780020992,
0.07838845836031891,
0.03517430823860464,
0.0370726042387833,
0.030216405744020368,
0.04841771445161579,
0.06458672979431404],
[0.047832813411448426,
0.07557888863526846,
0.01995992797179741,
0.03816987496512719,
0.13469781125567476,
0.03291993202972431,
0.07570569885109944,
0.030586624058434146,
0.049760328692079435,
0.04080752745441173,
0.02835476425980877,
0.02495625047553831,
0.044434299627177966,
0.08879251312485734,
0.07190139237616983,
0.028405488346141164,
0.034416292576529964,
0.028608384691470746,
0.04149230261989906,
0.06261888457734155],
[0.042617561579875174,
0.07770737052741912,
0.03601886702558483,
0.04750107199009005,
0.06608223355090762,
0.07060841393110677,
0.0826861689456382,
0.0338272428414884,
0.042951069607889844,
0.04888274810615084,
0.04173614750583639,
0.03728143313164038,
0.04371337367192339,
0.04190290151984373,
0.06603458954690553,
0.03363666682548001,
0.045190337795988376,
0.035590070989565965,
0.03327933679546429,
0.0727523941112011],
[0.05211050194283281,
0.022701868313195296,
0.04215681056049396,
0.03776547612710917,
0.06246340554638846,
0.05240325757172513,
0.03425240858040134,
0.060094746367168786,
0.04189066907968276,
0.03837760153297493,
0.031431308883802626,
0.08609676904242296,
0.04383350188960451,
0.11209879171767712,
0.06754670782988237,
0.03071272688561239,
0.0415446851546282,
0.02789162718901368,
0.0347314632458615,
0.07989567253952201],
[0.052505147563486614,
0.03777739647677877,
0.03743422557767102,
0.03311599176389842,
0.11201670098375657,
0.06963509494394875,
0.02916952642415923,
0.043239533287577216,
0.03854953099977122,
0.03260123541523679,
0.03546099290780142,
0.07958705101807367,
0.03165751544269046,
0.1153054221002059,
0.06637497140242507,
0.02304964539007092,
0.03955044612216884,
0.030942576069549303,
0.031457332418210936,
0.06056966369251887],
[0.03823109185601696,
0.0364105636723971,
0.03279255196570954,
0.033691293727243395,
0.15926164907590912,
0.061321841729271326,
0.036203161727427755,
0.03440567820436005,
0.03157118495644559,
0.0335069364428262,
0.03426741024104715,
0.07637000506982532,
0.03892243167258146,
0.11098308521915472,
0.05643637369221551,
0.026086555745033876,
0.036525786975157855,
0.04528275798497488,
0.033046043231783194,
0.04468359681061898],
[0.02800869900733102,
0.048055000175383215,
0.02597425374443158,
0.0358483285979866,
0.1538987688098495,
0.06082289803220036,
0.04098705671893087,
0.035585253779508226,
0.03213020449682556,
0.03448033954189905,
0.03277912238240556,
0.06162966080886738,
0.07131081412887158,
0.11022834894243923,
0.029727454488056405,
0.02874530849907047,
0.04032060051211898,
0.06248903854923007,
0.029253919814795328,
0.03772492896979901],
[0.04004854368932039,
0.019997889404812157,
0.015882228788518363,
0.04353102574926129,
0.10579358379062896,
0.01978682988602786,
0.030656395103419165,
0.02532714225411566,
0.055878007598142675,
0.033241874208526805,
0.02643520472773322,
0.05730265934993668,
0.05566694807935838,
0.20409455466441537,
0.05925495989869143,
0.02543267201350781,
0.0408400168847615,
0.03197551709582102,
0.033030814689742505,
0.07582313212325877],
[0.035613691698095196,
0.026543180407695613,
0.03375184990690791,
0.020337041103738004,
0.10770038669021817,
0.02291497589153578,
0.02597030601040722,
0.11419296319281998,
0.04516159831956843,
0.02897789659617129,
0.023344631689502078,
0.05060390509380818,
0.04430228672363584,
0.21196352699670598,
0.03948059387979186,
0.028643719864419725,
0.0347543801021626,
0.025779347877977754,
0.031078436052895404,
0.048885281901943]]
```
## Fig.1.a.topic proportion over time (bar chart)
```
import matplotlib.pyplot as plt
from pandas.core.frame import DataFrame
import numpy as np
fig, ax = plt.subplots(1, figsize=(32,16))
year = ['19Q1','19Q2','19Q3','19Q4','20Q1','20Q2','20Q3','20Q4','21Q1','21Q2']
col = ['#63b2ee','#76da91','#f8cb7f','#f89588','#7cd6cf','#9192ab','#7898e1','#efa666','#eddd86','#9987ce',
'#95a2ff','#fa8080','#ffc076','#fae768','#87e885','#3cb9fc','#73abf5','#cb9bff','#90ed7d','#f7a35c']
topicdis1 = DataFrame(topicdis)
topic2 = topicdis1.T
topic2 = np.array(topic2)
for i in range(20):
data = topic2[i]
if i == 0:
plt.bar(year,data)
else:
bot = sum(topic2[k] for k in range(i))
plt.bar(year,data,color=col[i],bottom = bot)
# size for xticks and yticks
plt.xticks(fontsize=20)
plt.yticks(fontsize=20)
# x and y limits
# plt.xlim(-0.6, 2.5)
# plt.ylim(-0.0, 1)
# remove spines
ax.spines['right'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.spines['top'].set_visible(False)
# ax.spines['bottom'].set_visible(False)
#grid
ax.set_axisbelow(True)
ax.yaxis.grid(color='gray', linestyle='dashed', alpha=0.7)
# title and legend
legend_label = ['Topic1', 'Topic2', 'Topic3', 'Topic4','Topic5','Topic6','Topic7','Topic8','Topic9','Topic10','Topic11'
,'Topic12','Topic13','Topic14','Topic15','Topic16','Topic17','Topic18','Topic19','Topic20']
plt.legend(legend_label, ncol = 1, bbox_to_anchor=([1.1, 0.65, 0, 0]), loc = 'right', borderaxespad = 0., frameon = True, fontsize = 18)
plt.rcParams['axes.titley'] = 1.05 # y is in axes-relative coordinates.
plt.title('Topic proportions over 2019 - 2021\n', loc='center', fontsize = 40)
plt.show()
```
## Fig.1.b.topic proportion over time (line graph) - for the best topics
```
# best topics in seed1234
ntop = [0,7,8,14,19]
year = ['19Q1','19Q2','19Q3','19Q4','20Q1','20Q2','20Q3','20Q4','21Q1','21Q2']
col = ['#63b2ee','#76da91','#f8cb7f','#f89588','#7cd6cf','#9192ab','#7898e1','#efa666','#eddd86','#9987ce',
'#95a2ff','#fa8080','#ffc076','#fae768','#87e885','#3cb9fc','#73abf5','#cb9bff','#90ed7d','#fa8080']
fig, ax = plt.subplots(1, figsize=(32,16))
# plot proportions of each topic over years
for i in ntop:
ys = [item[i] for item in topicdis]
ax.plot(year, ys, label='Topic ' + str(i+1),color = col[i],linewidth=3)
# size for xticks and yticks
plt.xticks(fontsize=25)
plt.yticks(fontsize=25)
# remove spines
ax.spines['right'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.spines['top'].set_visible(False)
# x and y limits
# plt.xlim(-0.2, 2.2)
plt.ylim(-0.0, 0.125)
# remove spines
ax.spines['right'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.spines['top'].set_visible(False)
#grid
ax.set_axisbelow(True)
ax.yaxis.grid(color='gray', linestyle='dashed', alpha=0.7)
# title and legend
legend_label = ['Topic1: Lipstick', 'Topic8: Tom Ford', 'Topic9: Beauty & Face products', 'Topic15: Eye Products','Topic20: Makeup','Topic6','Topic7','Topic8','Topic9','Topic10','Topic11'
,'Topic12','Topic13','Topic14','Topic15','Topic16','Topic17','Topic18','Topic19','Topic20']
plt.rcParams['axes.titley'] = 1.1 # y is in axes-relative coordinates.
plt.legend(legend_label, ncol = 1, bbox_to_anchor=([0.28, 0.975, 0, 0]), frameon = True, fontsize = 25)
plt.title('Topic proportions over 2019 - 2021 (5 topics) \n', loc='center', fontsize = 40)
plt.show()
```
* Drop in 2020 Q4 because of delay reveal of purchase patterns. Purchased in Q4 but discuss online in Q121.
## Fig.2. topic key words over time
```
import matplotlib.pyplot as plt
# best topics are [0,7,8,14,19]
t = 14
topicEvolution0 = ldaseq.print_topic_times(topic=t)
fig, axes = plt.subplots(2,5, figsize=(30, 15), sharex=True)
axes = axes.flatten()
year = ['19Q1','19Q2','19Q3','19Q4','20Q1','20Q2','20Q3','20Q4','21Q1','21Q2']
title = ['Topic1:Lipstick', 'Topic2', 'Topic3', 'Topic4','Topic5','Topic6','Topic7','Topic8: Tom Ford','Topic9: Beauty & Face Products','Topic10','Topic11'
,'Topic12','Topic13','Topic14','Topic15: Eye Products','Topic16','Topic17','Topic18','Topic19','Topic20: Makeup']
for i in range(len(topicEvolution0)):
value = [item[1] for item in topicEvolution0[i]]
index = [item[0] for item in topicEvolution0[i]]
ax = axes[i]
ax.barh(index,value,height = 0.7)
plt.rcParams['axes.titley'] = 1.25 # y is in axes-relative coordinates.
ax.set_title(year[i],
fontdict={'fontsize': 30})
ax.invert_yaxis()
ax.tick_params(axis='both', which='major', labelsize=20)
for k in 'top right left'.split():
ax.spines[k].set_visible(False)
fig.suptitle(title[t], fontsize=40)
plt.subplots_adjust(top=0.90, bottom=0.05, wspace=0.90, hspace=0.3)
plt.show()
```
## Fig.3. Trend of key words under 1 topic over time
```
from ekphrasis.classes.segmenter import Segmenter
seg = Segmenter(corpus="twitter")
ss_keywords = []
for i in range(len(keywords)):
s_keywords = [] # legends are keywords with first letter capitalised
for j in range(len(keywords[i])):
s_keyword = seg.segment(keywords[i][j])
s_keyword = s_keyword.capitalize()
s_keywords.append(s_keyword)
ss_keywords.append(s_keywords)
ss_keywords
# CHANGE k!
keyTopics = [0,7,8,14,19]
k = 3 # keyTopics index
topicEvolution_k = ldaseq.print_topic_times(topic=key_topics[k])
# transfrom topicEvolutiont to dictionary
for i in range(len(topicEvolution_k)):
topicEvolution_k[i] = dict(topicEvolution_k[i])
# our most interested keywords under each topic, pick manually
keywords = [['nars', 'revlon', 'wetnwild', 'dior'], # keywords for topic 0
['tomford', 'tomfordbeauty', 'body', 'japan', 'yuta'], # keywords for topic 7
['ysl', 'yvessaintlaurent', 'nyx', 'charlottetilbury','ctilburymakeup','diormakeup'], # keywords for topic 8
['maybelline', 'makeupforever', 'mua', 'anastasiabeverlyhills',
'morphe', 'morphebrushes', 'hudabeauty', 'fentybeauty', 'jeffreestar'], # keywords for topic 14
['colourpop', 'colorpopcosmetics', 'wetnwildbeauty', 'nyx',
'nyxcosmetics', 'bhcosmetics', 'tarte','tartecosmetics',
'elfcosmetics','benefit', 'makeuprevolution', 'loreal',
'katvond', 'jeffreestarcosmetics', 'urbandecaycosmetics']] # keywords for topic 19
year = ['19Q1','19Q2','19Q3','19Q4','20Q1','20Q2','20Q3','20Q4','21Q1','21Q2']
col = ['#63b2ee','#76da91','#f8cb7f','#f89588','#7cd6cf','#9192ab','#7898e1','#efa666','#eddd86','#9987ce',
'#95a2ff','#fa8080','#ffc076','#fae768','#87e885','#3cb9fc','#73abf5','#cb9bff','#90ed7d','#fa8080']
fig, ax = plt.subplots(1, figsize=(32,16))
# plot the value of keywords
yss = []
for word in keywords[k]: # for each word in keywords
ys = ["0"] * 10
for j in range(len(topicEvolution_k)): # for each top-20-keywords dict (one dict for one time period)
if word in topicEvolution_k[j]: # if keyword is in top 20 keywords dict
ys[j] = topicEvolution_k[j].get(word) # assign the keyword value to jth position
else:
ys[j] = 0 # else assign 0 to jth position
if k == 0 or k == 1 :
ax.plot(year, ys, linewidth=3) # plot keyword values against year
else:
yss.append(ys)
# k = 2
# ['ysl', 'yvessaintlaurent', 'nyx', 'charlottetilbury','ctilburymakeup','diormakeup']
if k == 2:
yss = [[a + b for a, b in zip(yss[0], yss[1])],
yss[2],
[a + b for a, b in zip(yss[3], yss[4])],
yss[5]]
for i in range(len(yss)):
ax.plot(year, yss[i], linewidth = 3)
# k = 3
# ['maybelline', 'makeupforever', 'mua', 'anatasiabeverlyhills',
# 'morphe', 'morphe brushes', 'hudabeauty', 'fentybeauty', 'jeffreestar']
if k == 3:
for i in range(len(keywords[3])):
if i == 4:
ax.plot(year, [a + b for a, b in zip(yss[4], yss[5])])
elif i == 5:
continue
else:
ax.plot(year, yss[i], linewidth = 3)
# k = 4:
# ['colourpop', 'colorpopcosmetics', 'wetnwildbeauty', 'nyx',
# 'nyxcosmetics', 'bhcosmetics', 'tarte','tartecosmetics',
# 'elfcosmetics','benefit', 'makeuprevolution', 'loreal',
# 'katvond', 'jeffreestarcosmetics', 'urbandecaycosmetics']
if k == 4:
for i in range(len(keywords[4])):
if i == 0:
ax.plot(year, [a + b for a, b in zip(yss[0], yss[1])])
elif i == 1:
continue
elif i == 3:
ax.plot(year, [a + b for a, b in zip(yss[3], yss[4])])
elif i == 4:
continue
else:
ax.plot(year, yss[i], linewidth = 3)
# size for xticks and yticks
plt.xticks(fontsize=25)
plt.yticks(fontsize=25)
# remove spines
ax.spines['right'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.spines['top'].set_visible(False)
# grid
ax.set_axisbelow(True)
ax.yaxis.grid(color='gray', linestyle='dashed', alpha=0.7)
# legend
legends = [['Nars', 'Revlon', 'wet n wild', 'Dior'],
['Tom Ford', 'Tom Ford Beauty', 'Body', 'Japan', 'Yuta'],
['YSL','NYX', 'Charlotte Tilbury','Dior'],
['Maybelline', 'Make Up For Ever','Mua','Anastasia Beverlyhills',
'Morphe','Huda Beauty','Fenty Beauty','Jeffree Star'],
['Colourpop','wet n wild', 'NYX','BH Cosmetics','Tarte',
'e.l.f','Benefit','Makeup revolution','Loreal',
'Kat Von D','Jeffree Star','Urban Decay']]
plt.legend(legends[k], ncol = 1, bbox_to_anchor=([1, 1.05, 0, 0]), frameon = True, fontsize = 25)
# title
titles = ['Brand occupation over topic "Lipstick" from 2019 - 2021',
'Effect of celebrity collaboration on brand',
'Brand occupation over topic "Beauty & Face Products" from 2019 - 2021',
'Brand occupation over topic "Eye Products" from 2019 - 2021',
'Brand occupation over topic "Makeup" from 2019 - 2021']
plt.title(titles[k], loc='left', fontsize = 40)
plt.show()
year = ['19Q1','19Q2','19Q3','19Q4','20Q1','20Q2','20Q3','20Q4','21Q1','21Q2']
col = ['#63b2ee','#76da91','#f8cb7f','#f89588','#7cd6cf','#9192ab','#7898e1','#efa666','#eddd86','#9987ce',
'#95a2ff','#fa8080','#ffc076','#fae768','#87e885','#3cb9fc','#73abf5','#cb9bff','#90ed7d','#fa8080']
fig, ax = plt.subplots(1, figsize=(32,16))
# plot the value of keywords
for word in keywords[k]: # for each word in keywords
ys = ["0"] * 10
for j in range(len(topicEvolution_k)): # for each top-20-keywords dict (one dict for one time period)
if word in topicEvolution_k[j]: # if keyword is in top 20 keywords dict
ys[j] = topicEvolution_k[j].get(word) # assign the keyword value to jth position
else:
ys[j] = 0 # else assign 0 to jth position:
ax.plot(year, ys, linewidth=3) # plot keyword values against year
# size for xticks and yticks
plt.xticks(fontsize=25)
plt.yticks(fontsize=25)
# remove spines
ax.spines['right'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.spines['top'].set_visible(False)
# grid
ax.set_axisbelow(True)
ax.yaxis.grid(color='gray', linestyle='dashed', alpha=0.7)
# legend
plt.legend(keywords[k], ncol = 1, bbox_to_anchor=([1, 1.05, 0, 0]), frameon = True, fontsize = 25)
# title
titles = ['Brand occupation over topic "Lipstick" from 2019 - 2021',
'Effect of celebrity collaboration on brand',
'Brand occupation over topic "Beauty & Face Products" from 2019 - 2021',
'Brand occupation over topic "Eye Products" from 2019 - 2021',
'Brand occupation over topic "Makeup" from 2019 - 2021']
plt.title(titles[k], loc='left', fontsize = 40)
plt.show()
# ver 1
# for z in keywords[k]:
# print(z)
# ys = [item[z] for item in topicEvolution_k]
# print(ys)
# ax.plot(year, ys, linewidth=3)
```
|
github_jupyter
|
```
%load_ext notexbook
%texify
```
# PyTorch `nn` package
### `torch.nn`
Computational graphs and autograd are a very powerful paradigm for defining complex operators and automatically taking derivatives; however for large neural networks raw autograd can be a bit too low-level.
When building neural networks we frequently think of arranging the computation into layers, some of which
have learnable parameters which will be optimized during learning.
In TensorFlow, packages like **Keras**, (old **TensorFlow-Slim**, and **TFLearn**) provide higher-level abstractions over raw computational graphs that are useful for building neural networks.
In PyTorch, the `nn` package serves this same purpose.
The `nn` package defines a set of `Module`s, which are roughly equivalent to neural network layers.
A `Module` receives input `Tensor`s and computes output `Tensor`s, but may also hold internal state such as `Tensor`s containing learnable parameters.
The `nn` package also defines a set of useful `loss` functions that are commonly used when
training neural networks.
In this example we use the `nn` package to implement our two-layer network:
```
import torch
# N is batch size; D_in is input dimension;
# H is hidden dimension; D_out is output dimension.
N, D_in, H, D_out = 64, 1000, 100, 10
# Create random Tensors to hold inputs and outputs
x = torch.randn(N, D_in)
y = torch.randn(N, D_out)
# Use the nn package to define our model as a sequence of layers. nn.Sequential
# is a Module which contains other Modules, and applies them in sequence to
# produce its output. Each Linear Module computes output from input using a
# linear function, and holds internal Tensors for its weight and bias.
model = torch.nn.Sequential(
torch.nn.Linear(D_in, H), # xW+b
torch.nn.ReLU(),
torch.nn.Linear(H, D_out),
)
[p.shape for p in model.parameters()]
# The nn package also contains definitions of popular loss functions; in this
# case we will use Mean Squared Error (MSE) as our loss function.
mseloss = torch.nn.MSELoss(reduction='sum')
learning_rate = 1e-4
for t in range(500):
# Forward pass: compute predicted y by passing x to the model. Module objects
# override the __call__ operator so you can call them like functions. When
# doing so you pass a Tensor of input data to the Module and it produces
# a Tensor of output data.
y_pred = model(x)
# Compute and print loss. We pass Tensors containing the predicted and true
# values of y, and the loss function returns a Tensor containing the
# loss.
loss = mseloss(y_pred, y)
if t % 50 == 0:
print(t, loss.item())
# Zero the gradients before running the backward pass.
model.zero_grad()
# Backward pass: compute gradient of the loss with respect to all the learnable
# parameters of the model. Internally, the parameters of each Module are stored
# in Tensors with requires_grad=True, so this call will compute gradients for
# all learnable parameters in the model.
loss.backward()
# Update the weights using gradient descent. Each parameter is a Tensor, so
# we can access its gradients like we did before.
with torch.no_grad():
for param in model.parameters():
param -= learning_rate * param.grad
```
---
### `torch.optim`
Up to this point we have updated the weights of our models by manually mutating the Tensors holding learnable parameters (**using `torch.no_grad()` or `.data` to avoid tracking history in autograd**).
This is not a huge burden for simple optimization algorithms like stochastic gradient descent, but in practice we often train neural networks using more sophisticated optimizers like `AdaGrad`, `RMSProp`,
`Adam`.
The optim package in PyTorch abstracts the idea of an optimization algorithm and provides implementations of commonly used optimization algorithms.
Let's finally modify the previous example in order to use `torch.optim` and the `Adam` algorithm:
```
# N is batch size; D_in is input dimension;
# H is hidden dimension; D_out is output dimension.
N, D_in, H, D_out = 64, 1000, 100, 10
# Create random Tensors to hold inputs and outputs
x = torch.randn(N, D_in)
y = torch.randn(N, D_out)
# Use the nn package to define our model and loss function.
model = torch.nn.Sequential(
torch.nn.Linear(D_in, H),
torch.nn.ReLU(),
torch.nn.Linear(H, D_out),
)
loss_fn = torch.nn.MSELoss(reduction='sum')
```
##### Model and Optimiser (w/ Parameters) at a glance

<span class="fn"><i>Source:</i> [1] - _Deep Learning with PyTorch_
```
# Use the optim package to define an Optimizer that will update the weights of
# the model for us. Here we will use Adam; the optim package contains many other
# optimization algoriths. The first argument to the Adam constructor tells the
# optimizer which Tensors it should update.
learning_rate = 1e-4
optimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
device
for t in range(500):
# Forward pass: compute predicted y by passing x to the model.
y_pred = model(x)
loss = loss_fn(y_pred, y)
if t % 50 == 0:
print(t, loss.item())
optimizer.zero_grad()
loss.backward()
optimizer.step()
```
### Can we do better ?
---
##### The Learning Process

<span class="fn"><i>Source:</i> [1] - _Deep Learning with PyTorch_ </span>
Possible scenarios:
- Specify models that are more complex than a sequence of existing (pre-defined) modules;
- Customise the learning procedure (e.g. _weight sharing_ ?)
- ?
For these cases, **PyTorch** allows to define our own custom modules by subclassing `nn.Module` and defining a `forward` method which receives the input data (i.e. `Tensor`) and returns the output (i.e. `Tensor`).
It is in the `forward` method that **all** the _magic_ of Dynamic Graph and `autograd` operations happen!
### PyTorch: Custom Modules
Let's implement our **two-layers** model as a custom `nn.Module` subclass
```
class TwoLayerNet(torch.nn.Module):
def __init__(self, D_in, H, D_out):
"""
In the constructor we instantiate two nn.Linear modules and assign them as
member variables.
"""
super(TwoLayerNet, self).__init__()
self.linear1 = torch.nn.Linear(D_in, H)
self.hidden_activation = torch.nn.ReLU()
self.linear2 = torch.nn.Linear(H, D_out)
def forward(self, x):
"""
In the forward function we accept a Tensor of input data and we must return
a Tensor of output data. We can use Modules defined in the constructor as
well as arbitrary operators on Tensors.
"""
l1 = self.linear1(x)
h_relu = self.hidden_activation(l1)
y_pred = self.linear2(h_relu)
return y_pred
# N is batch size; D_in is input dimension;
# H is hidden dimension; D_out is output dimension.
N, D_in, H, D_out = 64, 1000, 100, 10
# Create random Tensors to hold inputs and outputs
x = torch.randn(N, D_in)
y = torch.randn(N, D_out)
# Construct our model by instantiating the class defined above
model = TwoLayerNet(D_in, H, D_out)
# Construct our loss function and an Optimizer. The call to model.parameters()
# in the SGD constructor will contain the learnable parameters of the two
# nn.Linear modules which are members of the model.
criterion = torch.nn.MSELoss(reduction='sum')
optimizer = torch.optim.SGD(model.parameters(), lr=1e-4)
for t in range(500):
# Forward pass: Compute predicted y by passing x to the model
y_pred = model(x)
# Compute and print loss
loss = criterion(y_pred, y)
if t % 50 == 0:
print(t, loss.item())
# Zero gradients, perform a backward pass, and update the weights.
optimizer.zero_grad()
loss.backward()
optimizer.step()
```
#### What happened really? Let's have a closer look
```python
>>> model = TwoLayerNet(D_in, H, D_out)
```
This calls `TwoLayerNet.__init__` **constructor** method (_implementation reported below_ ):
```python
def __init__(self, D_in, H, D_out):
"""
In the constructor we instantiate two nn.Linear modules and assign them as
member variables.
"""
super(TwoLayerNet, self).__init__()
self.linear1 = torch.nn.Linear(D_in, H)
self.hidden_activation = torch.nn.ReLU()
self.linear2 = torch.nn.Linear(H, D_out)
```
1. First thing, we call the `nn.Module` constructor which sets up the housekeeping
- If you forget to do that, you will get and error message reminding that you should call it before using any `nn.Module` capabilities
2. We create a class attribute for each layer (`OP/Tensor/`) that we intend to include in our model
- These can be also `Sequential` as in _Submodules_ or *Block of Layers*
- **Note**: We are **not** defining the Graph yet, just the layer!
```python
>>> y_pred = model(x)
```
1. First thing to notice: the `model` object is **callable**
- It means `nn.Module` is implementing a `__call__` method
- We **don't** need to re-implement that!
2. (in fact) The `nn.Module` class will call `self.forward` - in a [Template Method Pattern](https://en.wikipedia.org/wiki/Template_method_pattern) fashion
- for this reason, we have to define the `forward` method
- (needless to say) the `forward` method implements the **forward** pass of our model
`from torch.nn.modules.module.py`
```python
class Module(object):
# [...] omissis
def __call__(self, *input, **kwargs):
for hook in self._forward_pre_hooks.values():
result = hook(self, input)
if result is not None:
if not isinstance(result, tuple):
result = (result,)
input = result
if torch._C._get_tracing_state():
result = self._slow_forward(*input, **kwargs)
else:
result = self.forward(*input, **kwargs)
for hook in self._forward_hooks.values():
hook_result = hook(self, input, result)
if hook_result is not None:
result = hook_result
if len(self._backward_hooks) > 0:
var = result
while not isinstance(var, torch.Tensor):
if isinstance(var, dict):
var = next((v for v in var.values() if isinstance(v, torch.Tensor)))
else:
var = var[0]
grad_fn = var.grad_fn
if grad_fn is not None:
for hook in self._backward_hooks.values():
wrapper = functools.partial(hook, self)
functools.update_wrapper(wrapper, hook)
grad_fn.register_hook(wrapper)
return result
# [...] omissis
def forward(self, *input):
r"""Defines the computation performed at every call.
Should be overridden by all subclasses.
.. note::
Although the recipe for forward pass needs to be defined within
this function, one should call the :class:`Module` instance afterwards
instead of this since the former takes care of running the
registered hooks while the latter silently ignores them.
"""
raise NotImplementedError
```
**Take away messages** :
1. We don't need to implement the `__call__` method at all in our custom model subclass
2. We don't need to call the `forward` method directly.
- We could, but we would miss the flexibility of _forward_ and _backwar_ hooks
##### Last but not least
```python
>>> optimizer = torch.optim.SGD(model.parameters(), lr=1e-4)
```
Being `model` a subclass of `nn.Module`, `model.parameters()` will automatically capture all the `Layers/OP/Tensors/Parameters` that require gradient computation, so to feed to the `autograd` engine during the *backward* (optimisation) step.
###### `model.named_parameters`
```
for name_str, param in model.named_parameters():
print("{:21} {:19} {}".format(name_str, str(param.shape), param.numel()))
```
**WAIT**: What happened to `hidden_activation` ?
```python
self.hidden_activation = torch.nn.ReLU()
```
So, it looks that we are registering in the constructor a submodule (`torch.nn.ReLU`) that has no parameters.
Generalising, if we would've had **more** (hidden) layers, it would have required the definition of one of these submodules for each pair of layers (at least).
Looking back at the implementation of the `TwoLayerNet` class as a whole, it looks like a bit of a waste.
**Can we do any better here?** 🤔
---
Well, in this particular case, we could implement the `ReLU` activation _manually_, it is not that difficult, isn't it?
$\rightarrow$ As we already did before, we could use the [`torch.clamp`](https://pytorch.org/docs/stable/torch.html?highlight=clamp#torch.clamp) function
> `torch.clamp`: Clamp all elements in input into the range [ min, max ] and return a resulting tensor
`t.clamp(min=0)` is **exactly** the ReLU that we want.
```
class TwoLayerNet(torch.nn.Module):
def __init__(self, D_in, H, D_out):
"""
In the constructor we instantiate two nn.Linear modules and assign them as
member variables.
"""
super(TwoLayerNet, self).__init__()
self.linear1 = torch.nn.Linear(D_in, H)
self.linear2 = torch.nn.Linear(H, D_out)
def forward(self, x):
"""
In the forward function we accept a Tensor of input data and we must return
a Tensor of output data. We can use Modules defined in the constructor as
well as arbitrary operators on Tensors.
"""
h_relu = self.linear1(x).clamp(min=0)
y_pred = self.linear2(h_relu)
return y_pred
```
###### Sorted!
That was easy, wasn't it? **However**, what if we wanted *other* activation functions (e.g. `tanh`,
`sigmoid`, `LeakyReLU`)?
### Introducing the Functional API
PyTorch has functional counterparts of every `nn` module.
By _functional_ here we mean "having no internal state", or, in other words, "whose output value is solely and fully determined by the value input arguments".
Indeed, `torch.nn.functional` provides the many of the same modules we find in `nn`, but with all eventual parameters moved as an argument to the function call.
For instance, the functional counterpart of `nn.Linear` is `nn.functional.linear`, which is a function that has signature `linear(input, weight, bias=None)`.
The `weight` and `bias` parameters are **arguments** to the function.
Back to our `TwoLayerNet` model, it makes sense to keep using nn modules for `nn.Linear`, so that our model will be able to manage all of its `Parameter` instances during training.
However, we can safely switch to the functional counterparts of `nn.ReLU`, since it has no parameters.
```
class TwoLayerNet(torch.nn.Module):
def __init__(self, D_in, H, D_out):
"""
In the constructor we instantiate two nn.Linear modules and assign them as
member variables.
"""
super(TwoLayerNet, self).__init__()
self.linear1 = torch.nn.Linear(D_in, H)
self.linear2 = torch.nn.Linear(H, D_out)
def forward(self, x):
"""
In the forward function we accept a Tensor of input data and we must return
a Tensor of output data. We can use Modules defined in the constructor as
well as arbitrary operators on Tensors.
"""
h_relu = torch.nn.functional.relu(self.linear1(x)) # torch.relu would do as well
y_pred = self.linear2(h_relu)
return y_pred
model = TwoLayerNet(D_in, H, D_out)
criterion = torch.nn.MSELoss(reduction='sum')
optimizer = torch.optim.SGD(model.parameters(), lr=1e-4)
for t in range(500):
y_pred = model(x)
loss = criterion(y_pred, y)
if t % 50 == 0:
print(t, loss.item())
optimizer.zero_grad()
loss.backward()
optimizer.step()
```
$\rightarrow$ For the curious minds: [The difference and connection between torch.nn and torch.nn.function from relu's various implementations](https://programmer.group/5d5a404b257d7.html)
#### Clever advice and Rule of thumb
> With **quantization**, stateless bits like activations suddenly become stateful because information on the quantization needs to be captured. This means that if we aim to quantize our model, it might be worthwile to stick with the modular API if we go for non-JITed quantization. There is one style matter that will help you avoid surprises with (originally unforseen) uses: if you need several applications of stateless modules (like `nn.HardTanh` or `nn.ReLU`), it is likely a good idea to have a separate instance for each. Re-using the same module appears to be clever and will give correct results with our standard Python usage here, but tools analysing your model might trip over it.
<span class="fn"><i>Source:</i> [1] - _Deep Learning with PyTorch_ </span>
### Custom Graph flow: Example of Weight Sharing
As we already discussed, the definition of custom `nn.Module` in PyTorch requires the definition of layers (i.e. Parameters) in the constructor (`__init__`), and the implementation of the `forward` method in which the dynamic graph will be traversed defined by the call to each of those layers/parameters.
As an example of **dynamic graphs** we are going to implement a scenario in which we require parameters (i.e. _weights_) sharing between layers.
In order to do so, we will implement a very odd model: a fully-connected ReLU network that on each `forward` call chooses a `random` number (between 1 and 4) and uses that many hidden layers, reusing the same weights multiple times to compute the innermost hidden layers.
In order to do so, we will implement _weight sharing_ among the innermost layers by simply reusing the same `Module` multiple times when defining the forward pass.
```
import torch
import random
class DynamicNet(torch.nn.Module):
def __init__(self, D_in, H, D_out):
"""
In the constructor we construct three nn.Linear instances that we will use
in the forward pass.
"""
super(DynamicNet, self).__init__()
self.input_linear = torch.nn.Linear(D_in, H)
self.middle_linear = torch.nn.Linear(H, H)
self.output_linear = torch.nn.Linear(H, D_out)
def forward(self, x):
"""
For the forward pass of the model, we randomly choose either 0, 1, 2, or 3
and reuse the middle_linear Module that many times to compute hidden layer
representations.
Since each forward pass builds a dynamic computation graph, we can use normal
Python control-flow operators like loops or conditional statements when
defining the forward pass of the model.
Here we also see that it is perfectly safe to reuse the same Module many
times when defining a computational graph. This is a big improvement from Lua
Torch, where each Module could be used only once.
"""
h_relu = torch.relu(self.input_linear(x))
hidden_layers = random.randint(0, 3)
for _ in range(hidden_layers):
h_relu = torch.relu(self.middle_linear(h_relu))
y_pred = self.output_linear(h_relu)
return y_pred
# N is batch size; D_in is input dimension;
# H is hidden dimension; D_out is output dimension.
N, D_in, H, D_out = 64, 1000, 100, 10
# Create random Tensors to hold inputs and outputs
x = torch.randn(N, D_in)
y = torch.randn(N, D_out)
# Construct our model by instantiating the class defined above
model = DynamicNet(D_in, H, D_out)
# Construct our loss function and an Optimizer. Training this strange model with
# vanilla stochastic gradient descent is tough, so we use momentum
criterion = torch.nn.MSELoss(reduction='sum')
optimizer = torch.optim.SGD(model.parameters(), lr=1e-4, momentum=0.9)
for t in range(500):
for i in range(2):
start, end = int((N/2)*i), int((N/2)*(i+1))
x = x[start:end, ...]
y = y[start:end, ...]
# Forward pass: Compute predicted y by passing x to the model
y_pred = model(x)
# Compute and print loss
loss = criterion(y_pred, y)
if t % 50 == 0:
print(t, loss.item())
# Zero gradients, perform a backward pass, and update the weights.
optimizer.zero_grad()
loss.backward()
optimizer.step()
```
### Latest from the `torch` ecosystem
* $\rightarrow$: [Migration from Chainer to PyTorch](https://medium.com/pytorch/migration-from-chainer-to-pytorch-8ed92c12c8)
* $\rightarrow$: [PyTorch Lightning](https://pytorch-lightning.readthedocs.io/en/latest/introduction_guide.html)
- [fast.ai](https://docs.fast.ai/)
---
### References and Futher Reading:
1. [Deep Learning with PyTorch, Luca Antiga et. al.](https://www.manning.com/books/deep-learning-with-pytorch)
2. [(**Terrific**) PyTorch Examples Repo](https://github.com/jcjohnson/pytorch-examples) (*where most of the examples in this notebook have been adapted from*)
|
github_jupyter
|
## 13.2 유가증권시장 12개월 모멘텀
최근 투자 기간 기준으로 12개월 모멘텀 계산 날짜 구하기
```
from pykrx import stock
import FinanceDataReader as fdr
df = fdr.DataReader(symbol='KS11', start="2019-11")
start = df.loc["2019-11"]
end = df.loc["2020-09"]
df.loc["2020-11"].head()
start
start_date = start.index[0]
end_date = end.index[-1]
print(start_date, end_date)
```
가격 모멘텀 계산 시작일 기준으로 등락률을 계산합니다.
```
df1 = stock.get_market_ohlcv_by_ticker("20191101")
df2 = stock.get_market_ohlcv_by_ticker("20200929")
kospi = df1.join(df2, lsuffix="_l", rsuffix="_r")
kospi
```
12개월 등락률(가격 모멘텀, 최근 1개월 제외)을 기준으로 상위 20종목의 종목 코드를 가져와봅시다.
```
kospi['모멘텀'] = 100 * (kospi['종가_r'] - kospi['종가_l']) / kospi['종가_l']
kospi = kospi[['종가_l', '종가_r', '모멘텀']]
kospi.sort_values(by='모멘텀', ascending=False)[:20]
kospi_momentum20 = kospi.sort_values(by='모멘텀', ascending=False)[:20]
kospi_momentum20.rename(columns={"종가_l": "매수가", "종가_r": "매도가"}, inplace=True)
kospi_momentum20
df3 = stock.get_market_ohlcv_by_ticker("20201102")
df4 = stock.get_market_ohlcv_by_ticker("20210430")
pct_df = df3.join(df4, lsuffix="_l", rsuffix="_r")
pct_df
pct_df = pct_df[['종가_l', '종가_r']]
kospi_momentum20_result = kospi_momentum20.join(pct_df)
kospi_momentum20_result
kospi_momentum20_result['수익률'] = (kospi_momentum20_result['종가_r'] /
kospi_momentum20_result['종가_l'])
kospi_momentum20_result
수익률평균 = kospi_momentum20_result['수익률'].fillna(0).mean()
수익률평균
mom20_cagr = 수익률평균 ** (1/0.5) - 1 # 6개월
mom20_cagr * 100
df_ref = fdr.DataReader(
symbol='KS11',
start="2020-11-02", # 첫번째 거래일
end="2021-04-30"
)
df_ref
CAGR = ((df_ref['Close'].iloc[-1] / df_ref['Close'].iloc[0]) ** (1/0.5)) -1
CAGR * 100
```
## 13.3 대형주 12개월 모멘텀
대형주(시가총액 200위) 기준으로 상대 모멘텀이 큰 종목을 20개 선정하는 전략
```
df1 = stock.get_market_ohlcv_by_ticker("20191101", market="ALL")
df2 = stock.get_market_ohlcv_by_ticker("20200929", market="ALL")
all = df1.join(df2, lsuffix="_l", rsuffix="_r")
all
# 기본 필터링
# 우선주 제외
all2 = all.filter(regex="0$", axis=0).copy()
all2
all2['모멘텀'] = 100 * (all2['종가_r'] - all2['종가_l']) / all2['종가_l']
all2 = all2[['모멘텀']]
all2
cap = stock.get_market_cap_by_ticker(date="20200929", market="ALL")
cap = cap[['시가총액']]
cap
all3 = all2.join(other=cap)
all3
# 대형주 필터링
big = all3.sort_values(by='시가총액', ascending=False)[:200]
big
big.sort_values(by='모멘텀', ascending=False)
big_pct20 = big.sort_values(by='모멘텀', ascending=False)[:20]
big_pct20
df3 = stock.get_market_ohlcv_by_ticker("20201102", market="ALL")
df4 = stock.get_market_ohlcv_by_ticker("20211015", market="ALL")
pct_df = df3.join(df4, lsuffix="_l", rsuffix="_r")
pct_df['수익률'] = pct_df['종가_r'] / pct_df['종가_l']
pct_df = pct_df[['종가_l', '종가_r', '수익률']]
pct_df
big_mom_result = big_pct20.join(pct_df)
big_mom_result
평균수익률 = big_mom_result['수익률'].mean()
big_mom_cagr = (평균수익률 ** 1/1) -1
big_mom_cagr * 100
```
## 13.4 장기 백테스팅
```
import pandas as pd
import datetime
from dateutil.relativedelta import relativedelta
year = 2010
month = 11
period = 6
inv_start = f"{year}-{month}-01"
inv_start = datetime.datetime.strptime(inv_start, "%Y-%m-%d")
inv_end = inv_start + relativedelta(months=period-1)
mom_start = inv_start - relativedelta(months=12)
mom_end = inv_start - relativedelta(months=2)
print(mom_start.strftime("%Y-%m"), mom_end.strftime("%Y-%m"), "=>",
inv_start.strftime("%Y-%m"), inv_end.strftime("%Y-%m"))
df = fdr.DataReader(symbol='KS11')
df
def get_business_day(df, year, month, index=0):
str_month = f"{year}-{month}"
return df.loc[str_month].index[index]
df = fdr.DataReader(symbol='KS11')
get_business_day(df, 2010, 1, 0)
def momentum(df, year=2010, month=11, period=12):
# 투자 시작일, 종료일
str_day = f"{year}-{month}-01"
start = datetime.datetime.strptime(str_day, "%Y-%m-%d")
end = start + relativedelta(months=period-1)
inv_start = get_business_day(df, start.year, start.month, 0) # 첫 번째 거래일의 종가
inv_end = get_business_day(df, end.year, end.month, -1)
inv_start = inv_start.strftime("%Y%m%d")
inv_end = inv_end.strftime("%Y%m%d")
#print(inv_start, inv_end)
# 모멘텀 계산 시작일, 종료일
end = start - relativedelta(months=2) # 역추세 1개월 제외
start = start - relativedelta(months=period)
mom_start = get_business_day(df, start.year, start.month, 0) # 첫 번째 거래일의 종가
mom_end = get_business_day(df, end.year, end.month, -1)
mom_start = mom_start.strftime("%Y%m%d")
mom_end = mom_end.strftime("%Y%m%d")
print(mom_start, mom_end, " | ", inv_start, inv_end)
# momentum 계산
df1 = stock.get_market_ohlcv_by_ticker(mom_start)
df2 = stock.get_market_ohlcv_by_ticker(mom_end)
mon_df = df1.join(df2, lsuffix="l", rsuffix="r")
mon_df['등락률'] = (mon_df['종가r'] - mon_df['종가l'])/mon_df['종가l']*100
# 우선주 제외
mon_df = mon_df.filter(regex="0$", axis=0)
mon20 = mon_df.sort_values(by="등락률", ascending=False)[:20]
mon20 = mon20[['등락률']]
#print(mon20)
# 투자 기간 수익률
df3 = stock.get_market_ohlcv_by_ticker(inv_start)
df4 = stock.get_market_ohlcv_by_ticker(inv_end)
inv_df = df3.join(df4, lsuffix="l", rsuffix="r")
inv_df['수익률'] = inv_df['종가r'] / inv_df['종가l'] # 수익률 = 매도가 / 매수가
inv_df = inv_df[['수익률']]
# join
result_df = mon20.join(inv_df)
result = result_df['수익률'].fillna(0).mean()
return year, result
import time
data = []
for year in range(2010, 2021):
ret = momentum(df, year, month=11, period=6)
data.append(ret)
time.sleep(1)
import pandas as pd
ret_df = pd.DataFrame(data=data, columns=['year', 'yield'])
ret_df.set_index('year', inplace=True)
ret_df
cum_yield = ret_df['yield'].cumprod()
cum_yield
CAGR = cum_yield.iloc[-1] ** (1/11) - 1
CAGR * 100
buy_price = df.loc["2010-11"].iloc[0, 0]
sell_price = df.loc["2021-04"].iloc[-1, 0]
kospi_yield = sell_price / buy_price
kospi_cagr = kospi_yield ** (1/11)-1
kospi_cagr * 100
```
|
github_jupyter
|
```
import numpy
import pandas as pd
import sqlite3
import os
from pandas.io import sql
from tables import *
import re
import pysam
import matplotlib
import matplotlib.image as mpimg
import seaborn
import matplotlib.pyplot
%matplotlib inline
def vectorizeSequence(seq):
# the order of the letters is not arbitrary.
# Flip the matrix up-down and left-right for reverse compliment
ltrdict = {'a':[1,0,0,0],'c':[0,1,0,0],'g':[0,0,1,0],'t':[0,0,0,1], 'n':[0,0,0,0]}
return numpy.array([ltrdict[x] for x in seq])
def Generate_training_and_test_datasets(Gem_events_file_path,ARF_label):
#Make Maize genome
from Bio import SeqIO
for record in SeqIO.parse(open('/mnt/Data_DapSeq_Maize/MaizeGenome.fa'),'fasta'):
if record.id =='1':
chr1 = record.seq.tostring()
if record.id =='2':
chr2 = record.seq.tostring()
if record.id =='3':
chr3 = record.seq.tostring()
if record.id =='4':
chr4 = record.seq.tostring()
if record.id =='5':
chr5 = record.seq.tostring()
if record.id =='6':
chr6 = record.seq.tostring()
if record.id =='7':
chr7 = record.seq.tostring()
if record.id =='8':
chr8 = record.seq.tostring()
if record.id =='9':
chr9 = record.seq.tostring()
if record.id =='10':
chr10 = record.seq.tostring()
wholegenome = {'chr1':chr1,'chr2':chr2,'chr3':chr3,'chr4':chr4,'chr5':chr5,'chr6':chr6,'chr7':chr7,'chr8':chr8,'chr9':chr9,'chr10':chr10}
rawdata = open(Gem_events_file_path)
GEM_events=rawdata.read()
GEM_events=re.split(',|\t|\n',GEM_events)
GEM_events=GEM_events[0:(len(GEM_events)-1)] # this is to make sure the reshape step works
GEM_events= numpy.reshape(GEM_events,(-1,10))
#Build Negative dataset
import random
Bound_Sequences = []
for i in range(0,len(GEM_events)):
Bound_Sequences.append(wholegenome[GEM_events[i][0]][int(GEM_events[i][1]):int(GEM_events[i][2])])
Un_Bound_Sequences = []
count=0
while count<len(Bound_Sequences):
chro = numpy.random.choice(['chr1','chr2','chr3','chr4','chr5','chr6','chr7','chr8','chr9','chr10'])
index = random.randint(1,len(wholegenome[chro]))
absent=True
for i in range(len(GEM_events)):
if chro == GEM_events[i][0]:
if index>int(GEM_events[i][1]) and index<int(GEM_events[i][2]):
absent = False
if absent:
if wholegenome[chro][index:(index+201)].upper().count('R') == 0 and wholegenome[chro][index:(index+201)].upper().count('W') == 0 and wholegenome[chro][index:(index+201)].upper().count('M') == 0 and wholegenome[chro][index:(index+201)].upper().count('S') == 0 and wholegenome[chro][index:(index+201)].upper().count('K') == 0 and wholegenome[chro][index:(index+201)].upper().count('Y') == 0and wholegenome[chro][index:(index+201)].upper().count('B') == 0 and wholegenome[chro][index:(index+201)].upper().count('D') == 0and wholegenome[chro][index:(index+201)].upper().count('H') == 0 and wholegenome[chro][index:(index+201)].upper().count('V') == 0 and wholegenome[chro][index:(index+201)].upper().count('Z') == 0 and wholegenome[chro][index:(index+201)].upper().count('N') == 0 :
Un_Bound_Sequences.append(wholegenome[chro][index:(index+201)])
count=count+1
response = [0]*(len(Un_Bound_Sequences))
temp3 = numpy.array(Un_Bound_Sequences)
temp2 = numpy.array(response)
neg = pd.DataFrame({'sequence':temp3,'response':temp2})
#Build Positive dataset labeled with signal value
Bound_Sequences = []
Responses=[]
for i in range(0,len(GEM_events)):
Bound_Sequences.append(wholegenome[GEM_events[i][0]][int(GEM_events[i][1]):int(GEM_events[i][2])])
Responses.append(float(GEM_events[i][6]))
d = {'sequence' : pd.Series(Bound_Sequences, index=range(len(Bound_Sequences))),
'response' : pd.Series(Responses, index=range(len(Bound_Sequences)))}
pos = pd.DataFrame(d)
#Put positive and negative datasets together
LearningData = neg.append(pos)
LearningData = LearningData.reindex()
#one hot encode sequence data
counter2=0
LearningData_seq_OneHotEncoded =numpy.empty([len(LearningData),201,4])
for counter1 in LearningData['sequence']:
LearningData_seq_OneHotEncoded[counter2]=vectorizeSequence(counter1.lower())
counter2=counter2+1
#Create training and test datasets
from sklearn.cross_validation import train_test_split
sequence_train, sequence_test, response_train, response_test = train_test_split(LearningData_seq_OneHotEncoded, LearningData['response'], test_size=0.2, random_state=42)
#Saving datasets
numpy.save('/mnt/Data_DapSeq_Maize/'+ARF_label+'_seq_train.npy',sequence_train)
numpy.save('/mnt/Data_DapSeq_Maize/'+ARF_label+'_res_train.npy',response_train)
numpy.save('/mnt/Data_DapSeq_Maize/'+ARF_label+'_seq_test.npy',sequence_test)
numpy.save('/mnt/Data_DapSeq_Maize/'+ARF_label+'_res_test.npy',response_test)
def Generate_training_and_test_datasets_no_negative(Gem_events_file_path,ARF_label):
#Make Maize genome
from Bio import SeqIO
for record in SeqIO.parse(open('/mnt/Data_DapSeq_Maize/MaizeGenome.fa'),'fasta'):
if record.id =='1':
chr1 = record.seq.tostring()
if record.id =='2':
chr2 = record.seq.tostring()
if record.id =='3':
chr3 = record.seq.tostring()
if record.id =='4':
chr4 = record.seq.tostring()
if record.id =='5':
chr5 = record.seq.tostring()
if record.id =='6':
chr6 = record.seq.tostring()
if record.id =='7':
chr7 = record.seq.tostring()
if record.id =='8':
chr8 = record.seq.tostring()
if record.id =='9':
chr9 = record.seq.tostring()
if record.id =='10':
chr10 = record.seq.tostring()
wholegenome = {'chr1':chr1,'chr2':chr2,'chr3':chr3,'chr4':chr4,'chr5':chr5,'chr6':chr6,'chr7':chr7,'chr8':chr8,'chr9':chr9,'chr10':chr10}
rawdata = open(Gem_events_file_path)
GEM_events=rawdata.read()
GEM_events=re.split(',|\t|\n',GEM_events)
GEM_events=GEM_events[0:(len(GEM_events)-1)] # this is to make sure the reshape step works
GEM_events= numpy.reshape(GEM_events,(-1,10))
#Build Positive dataset labeled with signal value
Bound_Sequences = []
Responses=[]
for i in range(0,len(GEM_events)):
Bound_Sequences.append(wholegenome[GEM_events[i][0]][int(GEM_events[i][1]):int(GEM_events[i][2])])
Responses.append(float(GEM_events[i][6]))
d = {'sequence' : pd.Series(Bound_Sequences, index=range(len(Bound_Sequences))),
'response' : pd.Series(Responses, index=range(len(Bound_Sequences)))}
pos = pd.DataFrame(d)
LearningData = pos
#one hot encode sequence data
counter2=0
LearningData_seq_OneHotEncoded =numpy.empty([len(LearningData),201,4])
for counter1 in LearningData['sequence']:
LearningData_seq_OneHotEncoded[counter2]=vectorizeSequence(counter1.lower())
counter2=counter2+1
#Create training and test datasets
from sklearn.cross_validation import train_test_split
sequence_train, sequence_test, response_train, response_test = train_test_split(LearningData_seq_OneHotEncoded, LearningData['response'], test_size=0.2, random_state=42)
#Saving datasets
numpy.save('/mnt/Data_DapSeq_Maize/'+ARF_label+'no_negative_seq_train.npy',sequence_train)
numpy.save('/mnt/Data_DapSeq_Maize/'+ARF_label+'no_negative_res_train.npy',response_train)
numpy.save('/mnt/Data_DapSeq_Maize/'+ARF_label+'no_negative_seq_test.npy',sequence_test)
numpy.save('/mnt/Data_DapSeq_Maize/'+ARF_label+'no_negative_res_test.npy',response_test)
def Train_and_save_DanQ_model(ARF_label,number_backpropagation_cycles):
#Loading the data
sequence_train=numpy.load('/mnt/Data_DapSeq_Maize/'+ARF_label+'_seq_train.npy')
response_train=numpy.load('/mnt/Data_DapSeq_Maize/'+ARF_label+'_res_train.npy')
sequence_test=numpy.load('/mnt/Data_DapSeq_Maize/'+ARF_label+'_seq_test.npy')
response_test=numpy.load('/mnt/Data_DapSeq_Maize/'+ARF_label+'_res_test.npy')
#Setting up the model
import keras
import numpy as np
from keras import backend
backend._BACKEND="theano"
#DanQ model
from keras.preprocessing import sequence
from keras.optimizers import RMSprop
from keras.models import Sequential
from keras.layers.core import Dense
from keras.layers.core import Merge
from keras.layers.core import Dropout
from keras.layers.core import Activation
from keras.layers.core import Flatten
from keras.layers.convolutional import Convolution1D, MaxPooling1D
from keras.regularizers import l2, activity_l1
from keras.constraints import maxnorm
from keras.layers.recurrent import LSTM, GRU
from keras.callbacks import ModelCheckpoint, EarlyStopping
from keras.layers import Bidirectional
model = Sequential()
model.add(Convolution1D(nb_filter=20,filter_length=26,input_dim=4,input_length=201,border_mode="valid"))
model.add(Activation('relu'))
model.add(MaxPooling1D(pool_length=6, stride=6))
model.add(Dropout(0.2))
model.add(Bidirectional(LSTM(5)))
model.add(Dropout(0.5))
model.add(Dense(10))
model.add(Activation('relu'))
model.add(Dense(1))
#compile the model
model.compile(loss='mean_squared_error', optimizer='rmsprop')
model.fit(sequence_train, response_train, validation_split=0.2,batch_size=100, nb_epoch=number_backpropagation_cycles, verbose=1)
#evaulting correlation between model and test data
import scipy
correlation = scipy.stats.pearsonr(response_test,model.predict(sequence_test).flatten())
correlation_2 = (correlation[0]**2)*100
print('Percent of variability explained by model: '+str(correlation_2))
# saving the model
model.save('/mnt/Data_DapSeq_Maize/TrainedModel_DanQ_' +ARF_label+'.h5')
def Train_and_save_DanQ_model_no_negative(ARF_label,number_backpropagation_cycles,train_size):
#Loading the data
sequence_train=numpy.load('/mnt/Data_DapSeq_Maize/'+ARF_label+'no_negative_seq_train.npy')
response_train=numpy.load('/mnt/Data_DapSeq_Maize/'+ARF_label+'no_negative_res_train.npy')
sequence_train=sequence_train[0:train_size]
response_train=response_train[0:train_size]
sequence_test=numpy.load('/mnt/Data_DapSeq_Maize/'+ARF_label+'no_negative_seq_test.npy')
response_test=numpy.load('/mnt/Data_DapSeq_Maize/'+ARF_label+'no_negative_res_test.npy')
#Setting up the model
import keras
import numpy as np
from keras import backend
backend._BACKEND="theano"
#DanQ model
from keras.preprocessing import sequence
from keras.optimizers import RMSprop
from keras.models import Sequential
from keras.layers.core import Dense
from keras.layers.core import Merge
from keras.layers.core import Dropout
from keras.layers.core import Activation
from keras.layers.core import Flatten
from keras.layers.convolutional import Convolution1D, MaxPooling1D
from keras.regularizers import l2, activity_l1
from keras.constraints import maxnorm
from keras.layers.recurrent import LSTM, GRU
from keras.callbacks import ModelCheckpoint, EarlyStopping
from keras.layers import Bidirectional
model = Sequential()
model.add(Convolution1D(nb_filter=20,filter_length=26,input_dim=4,input_length=201,border_mode="valid"))
model.add(Activation('relu'))
model.add(MaxPooling1D(pool_length=6, stride=6))
model.add(Dropout(0.2))
model.add(Bidirectional(LSTM(5)))
model.add(Dropout(0.5))
model.add(Dense(10))
model.add(Activation('relu'))
model.add(Dense(1))
#compile the model
model.compile(loss='mean_squared_error', optimizer='rmsprop')
model.fit(sequence_train, response_train, validation_split=0.2,batch_size=100, nb_epoch=number_backpropagation_cycles, verbose=1)
#evaulting correlation between model and test data
import scipy
correlation = scipy.stats.pearsonr(response_test,model.predict(sequence_test).flatten())
correlation_2 = (correlation[0]**2)*100
print('Percent of variability explained by model: '+str(correlation_2))
# saving the model
model.save('/mnt/Data_DapSeq_Maize/TrainedModel_DanQ_no_negative_' +ARF_label+'.h5')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF27_smaller_GEM_events.txt','ARF27_smaller')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF34_smaller_GEM_events.txt','ARF34_smaller')
Train_and_save_DanQ_model('ARF27_smaller',35)
Train_and_save_DanQ_model('ARF34_smaller',35)
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF16_GEM_events.txt','ARF16')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF4_GEM_events.txt','ARF4')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF4_rep2_GEM_events.txt','ARF4_rep2')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF4_rep3_GEM_events.txt','ARF4_rep3')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF10_GEM_events.txt','ARF10')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF13_GEM_events.txt','ARF13')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF18_GEM_events.txt','ARF18')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF27_GEM_events.txt','ARF27')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF29_GEM_events.txt','ARF29')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF34_GEM_events.txt','ARF34')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF35_GEM_events.txt','ARF35')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF39_GEM_events.txt','ARF39')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF10_rep1_ear_GEM_events.txt','ARF10_rep1_ear')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF10_rep2_ear_GEM_events.txt','ARF10_rep2_ear')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF10_rep1_tassel_GEM_events.txt','ARF10_rep1_tassel')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF10_rep2_tassel_GEM_events.txt','ARF10_rep2_tassel')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF7_GEM_events.txt','ARF7')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF14_GEM_events.txt','ARF14')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF24_GEM_events.txt','ARF24')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF25_GEM_events.txt','ARF25')
Generate_training_and_test_datasets('/mnt/Data_DapSeq_Maize/ARF36_GEM_events.txt','ARF36')
Train_and_save_DanQ_model('ARF7',35)
Train_and_save_DanQ_model('ARF14',35)
Train_and_save_DanQ_model('ARF24',35)
Train_and_save_DanQ_model('ARF25',35)
Train_and_save_DanQ_model('ARF36',35)
Train_and_save_DanQ_model('ARF10_rep1_ear',35)
Train_and_save_DanQ_model('ARF10_rep2_ear',35)
Train_and_save_DanQ_model('ARF10_rep1_tassel',35)
Train_and_save_DanQ_model('ARF10_rep2_tassel',35)
Train_and_save_DanQ_model('ARF4',35)
Train_and_save_DanQ_model('ARF4_rep2',35)
Train_and_save_DanQ_model('ARF4_rep3',35)
Train_and_save_DanQ_model('ARF10',35)
Train_and_save_DanQ_model('ARF13',35)
Train_and_save_DanQ_model('ARF16',35)
Train_and_save_DanQ_model('ARF18',35)
Train_and_save_DanQ_model('ARF27',35)
Train_and_save_DanQ_model('ARF29',35)
Train_and_save_DanQ_model('ARF34',35)
Train_and_save_DanQ_model('ARF35',35)
Train_and_save_DanQ_model('ARF39',35)
```
# Creating dataset without a negative set
```
Generate_training_and_test_datasets_no_negative('/mnt/Data_DapSeq_Maize/ARF4_GEM_events.txt','ARF4')
Generate_training_and_test_datasets_no_negative('/mnt/Data_DapSeq_Maize/ARF39_GEM_events.txt','ARF39')
Generate_training_and_test_datasets_no_negative('/mnt/Data_DapSeq_Maize/ARF35_GEM_events.txt','ARF35')
Generate_training_and_test_datasets_no_negative('/mnt/Data_DapSeq_Maize/ARF34_GEM_events.txt','ARF34')
Generate_training_and_test_datasets_no_negative('/mnt/Data_DapSeq_Maize/ARF10_GEM_events.txt','ARF10')
Generate_training_and_test_datasets_no_negative('/mnt/Data_DapSeq_Maize/ARF13_GEM_events.txt','ARF13')
Generate_training_and_test_datasets_no_negative('/mnt/Data_DapSeq_Maize/ARF16_GEM_events.txt','ARF16')
Generate_training_and_test_datasets_no_negative('/mnt/Data_DapSeq_Maize/ARF18_GEM_events.txt','ARF18')
Generate_training_and_test_datasets_no_negative('/mnt/Data_DapSeq_Maize/ARF27_GEM_events.txt','ARF27')
Generate_training_and_test_datasets_no_negative('/mnt/Data_DapSeq_Maize/ARF29_GEM_events.txt','ARF29')
#finding the min length of the test set
List_of_ARFs =['ARF4','ARF10','ARF13','ARF16','ARF18','ARF27','ARF29','ARF34','ARF35','ARF39']
seq_test_sets = [None]*len(List_of_ARFs)
counter1=0
for ARF_label in List_of_ARFs:
seq_test_sets[counter1]=numpy.load('/mnt/Data_DapSeq_Maize/'+ARF_label+'no_negative_seq_test.npy')
print(len(seq_test_sets[counter1]))
counter1=counter1+1
#based on this the test set will only be: 5960 in size
Train_and_save_DanQ_model_no_negative('ARF4',35,5960)
Train_and_save_DanQ_model_no_negative('ARF39',35,5960)
Train_and_save_DanQ_model_no_negative('ARF35',35,5960)
Train_and_save_DanQ_model_no_negative('ARF34',35,5960)
Train_and_save_DanQ_model_no_negative('ARF10',35,5960)
Train_and_save_DanQ_model_no_negative('ARF13',35,5960)
Train_and_save_DanQ_model_no_negative('ARF16',35,5960)
Train_and_save_DanQ_model_no_negative('ARF18',35,5960)
Train_and_save_DanQ_model_no_negative('ARF27',35,5960)
Train_and_save_DanQ_model_no_negative('ARF29',35,5960)
```
|
github_jupyter
|
The following latitude and longitude formats are supported by the `output_format` parameter:
* Decimal degrees (dd): 41.5
* Decimal degrees hemisphere (ddh): "41.5° N"
* Degrees minutes (dm): "41° 30′ N"
* Degrees minutes seconds (dms): "41° 30′ 0″ N"
You can split a column of geographic coordinates into one column for latitude and another for longitude by setting the parameter ``split`` to True.
Invalid parsing is handled with the `errors` parameter:
* "coerce" (default): invalid parsing will be set to NaN
* "ignore": invalid parsing will return the input
* "raise": invalid parsing will raise an exception
After cleaning, a **report** is printed that provides the following information:
* How many values were cleaned (the value must have been transformed).
* How many values could not be parsed.
* A summary of the cleaned data: how many values are in the correct format, and how many values are NaN.
The following sections demonstrate the functionality of `clean_lat_long()` and `validate_lat_long()`.
### An example dataset with geographic coordinates
```
import pandas as pd
import numpy as np
df = pd.DataFrame({
"lat_long":
[(41.5, -81.0), "41.5;-81.0", "41.5,-81.0", "41.5 -81.0",
"41.5° N, 81.0° W", "41.5 S;81.0 E", "-41.5 S;81.0 E",
"23 26m 22s N 23 27m 30s E", "23 26' 22\" N 23 27' 30\" E",
"UT: N 39°20' 0'' / W 74°35' 0''", "hello", np.nan, "NULL"]
})
df
```
## 1. Default `clean_lat_long()`
By default, the `output_format` parameter is set to "dd" (decimal degrees) and the `errors` parameter is set to "coerce" (set to NaN when parsing is invalid).
```
from dataprep.clean import clean_lat_long
clean_lat_long(df, "lat_long")
```
Note (41.5, -81.0) is considered not cleaned in the report since it's resulting format is the same as the input. Also, "-41.5 S;81.0 E" is invalid because if a coordinate has a hemisphere it cannot contain a negative decimal value.
## 2. Output formats
This section demonstrates the supported latitudinal and longitudinal formats.
### decimal degrees hemisphere (ddh)
```
clean_lat_long(df, "lat_long", output_format="ddh")
```
### degrees minutes (dm)
```
clean_lat_long(df, "lat_long", output_format="dm")
```
### degrees minutes seconds (dms)
```
clean_lat_long(df, "lat_long", output_format="dms")
```
## 3. `split` parameter
The split parameter adds individual columns containing the cleaned latitude and longitude values to the given DataFrame.
```
clean_lat_long(df, "lat_long", split=True)
```
Split can be used along with different output formats.
```
clean_lat_long(df, "lat_long", split=True, output_format="dm")
```
## 4. `inplace` parameter
This just deletes the given column from the returned dataframe.
A new column containing cleaned coordinates is added with a title in the format `"{original title}_clean"`.
```
clean_lat_long(df, "lat_long", inplace=True)
```
### `inplace` and `split`
```
clean_lat_long(df, "lat_long", split=True, inplace=True)
```
## 5. Latitude and longitude coordinates in separate columns
### Clean latitude or longitude coordinates individually
```
df = pd.DataFrame({"lat": [" 30′ 0″ E", "41° 30′ N", "41 S", "80", "hello", "NA"]})
clean_lat_long(df, lat_col="lat")
```
### Combine and clean separate columns
Latitude and longitude values are counted separately in the report.
```
df = pd.DataFrame({"lat": ["30° E", "41° 30′ N", "41 S", "80", "hello", "NA"],
"long": ["30° E", "41° 30′ N", "41 W", "80", "hello", "NA"]})
clean_lat_long(df, lat_col="lat", long_col="long")
```
### Clean separate columns and split the output
```
clean_lat_long(df, lat_col="lat", long_col="long", split=True)
```
## 6. `validate_lat_long()`
`validate_lat_long()` returns True when the input is a valid latitude or longitude value otherwise it returns False.
Valid types are the same as `clean_lat_long()`.
```
from dataprep.clean import validate_lat_long
print(validate_lat_long("41° 30′ 0″ N"))
print(validate_lat_long("41.5 S;81.0 E"))
print(validate_lat_long("-41.5 S;81.0 E"))
print(validate_lat_long((41.5, 81)))
print(validate_lat_long(41.5, lat_long=False, lat=True))
df = pd.DataFrame({"lat_long":
[(41.5, -81.0), "41.5;-81.0", "41.5,-81.0", "41.5 -81.0",
"41.5° N, 81.0° W", "-41.5 S;81.0 E",
"23 26m 22s N 23 27m 30s E", "23 26' 22\" N 23 27' 30\" E",
"UT: N 39°20' 0'' / W 74°35' 0''", "hello", np.nan, "NULL"]
})
validate_lat_long(df["lat_long"])
```
### Validate only one coordinate
```
df = pd.DataFrame({"lat":
[41.5, "41.5", "41.5 ",
"41.5° N", "-41.5 S",
"23 26m 22s N", "23 26' 22\" N",
"UT: N 39°20' 0''", "hello", np.nan, "NULL"]
})
validate_lat_long(df["lat"], lat_long=False, lat=True)
```
|
github_jupyter
|
<h2> 25ppm - somehow more features detected than at 4ppm... I guess because more likely to pass over the #scans needed to define a feature </h2>
Enough retcor groups, loads of peak insertion problem (1000's). Does that mean data isn't centroided...?
```
import time
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
from sklearn import preprocessing
from sklearn.ensemble import RandomForestClassifier
from sklearn.cross_validation import StratifiedShuffleSplit
from sklearn.cross_validation import cross_val_score
#from sklearn.model_selection import StratifiedShuffleSplit
#from sklearn.model_selection import cross_val_score
from sklearn.ensemble import AdaBoostClassifier
from sklearn.metrics import roc_curve, auc
from sklearn.utils import shuffle
from scipy import interp
%matplotlib inline
def remove_zero_columns(X, threshold=1e-20):
# convert zeros to nan, drop all nan columns, the replace leftover nan with zeros
X_non_zero_colum = X.replace(0, np.nan).dropna(how='all', axis=1).replace(np.nan, 0)
#.dropna(how='all', axis=0).replace(np.nan,0)
return X_non_zero_colum
def zero_fill_half_min(X, threshold=1e-20):
# Fill zeros with 1/2 the minimum value of that column
# input dataframe. Add only to zero values
# Get a vector of 1/2 minimum values
half_min = X[X > threshold].min(axis=0)*0.5
# Add the half_min values to a dataframe where everything that isn't zero is NaN.
# then convert NaN's to 0
fill_vals = (X[X < threshold] + half_min).fillna(value=0)
# Add the original dataframe to the dataframe of zeros and fill-values
X_zeros_filled = X + fill_vals
return X_zeros_filled
toy = pd.DataFrame([[1,2,3,0],
[0,0,0,0],
[0.5,1,0,0]], dtype=float)
toy_no_zeros = remove_zero_columns(toy)
toy_filled_zeros = zero_fill_half_min(toy_no_zeros)
print toy
print toy_no_zeros
print toy_filled_zeros
```
<h2> Import the dataframe and remove any features that are all zero </h2>
```
### Subdivide the data into a feature table
data_path = '/home/irockafe/Dropbox (MIT)/Alm_Lab/projects/revo_healthcare/data/processed/MTBLS315/'\
'uhplc_pos/xcms_result_25.csv'
## Import the data and remove extraneous columns
df = pd.read_csv(data_path, index_col=0)
df.shape
df.head()
# Make a new index of mz:rt
mz = df.loc[:,"mz"].astype('str')
rt = df.loc[:,"rt"].astype('str')
idx = mz+':'+rt
df.index = idx
df
# separate samples from xcms/camera things to make feature table
not_samples = ['mz', 'mzmin', 'mzmax', 'rt', 'rtmin', 'rtmax',
'npeaks', 'uhplc_pos',
]
samples_list = df.columns.difference(not_samples)
mz_rt_df = df[not_samples]
# convert to samples x features
X_df_raw = df[samples_list].T
# Remove zero-full columns and fill zeroes with 1/2 minimum values
X_df = remove_zero_columns(X_df_raw)
X_df_zero_filled = zero_fill_half_min(X_df)
print "original shape: %s \n# zeros: %f\n" % (X_df_raw.shape, (X_df_raw < 1e-20).sum().sum())
print "zero-columns repalced? shape: %s \n# zeros: %f\n" % (X_df.shape,
(X_df < 1e-20).sum().sum())
print "zeros filled shape: %s \n#zeros: %f\n" % (X_df_zero_filled.shape,
(X_df_zero_filled < 1e-20).sum().sum())
# Convert to numpy matrix to play nicely with sklearn
X = X_df.as_matrix()
print X.shape
```
<h2> Get mappings between sample names, file names, and sample classes </h2>
```
# Get mapping between sample name and assay names
path_sample_name_map = '/home/irockafe/Dropbox (MIT)/Alm_Lab/projects/revo_healthcare/data/raw/'\
'MTBLS315/metadata/a_UPLC_POS_nmfi_and_bsi_diagnosis.txt'
# Index is the sample name
sample_df = pd.read_csv(path_sample_name_map,
sep='\t', index_col=0)
sample_df = sample_df['MS Assay Name']
sample_df.shape
print sample_df.head(10)
# get mapping between sample name and sample class
path_sample_class_map = '/home/irockafe/Dropbox (MIT)/Alm_Lab/projects/revo_healthcare/data/raw/'\
'MTBLS315/metadata/s_NMFI and BSI diagnosis.txt'
class_df = pd.read_csv(path_sample_class_map,
sep='\t')
# Set index as sample name
class_df.set_index('Sample Name', inplace=True)
class_df = class_df['Factor Value[patient group]']
print class_df.head(10)
# convert all non-malarial classes into a single classes
# (collapse non-malarial febril illness and bacteremia together)
class_map_df = pd.concat([sample_df, class_df], axis=1)
class_map_df.rename(columns={'Factor Value[patient group]': 'class'}, inplace=True)
class_map_df
binary_class_map = class_map_df.replace(to_replace=['non-malarial febrile illness', 'bacterial bloodstream infection' ],
value='non-malarial fever')
binary_class_map
# convert classes to numbers
le = preprocessing.LabelEncoder()
le.fit(binary_class_map['class'])
y = le.transform(binary_class_map['class'])
```
<h2> Plot the distribution of classification accuracy across multiple cross-validation splits - Kinda Dumb</h2>
Turns out doing this is kind of dumb, because you're not taking into account the prediction score your classifier assigned. Use AUC's instead. You want to give your classifier a lower score if it is really confident and wrong, than vice-versa
```
def rf_violinplot(X, y, n_iter=25, test_size=0.3, random_state=1,
n_estimators=1000):
cross_val_skf = StratifiedShuffleSplit(y, n_iter=n_iter, test_size=test_size,
random_state=random_state)
clf = RandomForestClassifier(n_estimators=n_estimators, random_state=random_state)
scores = cross_val_score(clf, X, y, cv=cross_val_skf)
sns.violinplot(scores,inner='stick')
rf_violinplot(X,y)
# TODO - Switch to using caret for this bs..?
# Do multi-fold cross validation for adaboost classifier
def adaboost_violinplot(X, y, n_iter=25, test_size=0.3, random_state=1,
n_estimators=200):
cross_val_skf = StratifiedShuffleSplit(y, n_iter=n_iter, test_size=test_size, random_state=random_state)
clf = AdaBoostClassifier(n_estimators=n_estimators, random_state=random_state)
scores = cross_val_score(clf, X, y, cv=cross_val_skf)
sns.violinplot(scores,inner='stick')
adaboost_violinplot(X,y)
# TODO PQN normalization, and log-transformation,
# and some feature selection (above certain threshold of intensity, use principal components), et
def pqn_normalize(X, integral_first=False, plot=False):
'''
Take a feature table and run PQN normalization on it
'''
# normalize by sum of intensities in each sample first. Not necessary
if integral_first:
sample_sums = np.sum(X, axis=1)
X = (X / sample_sums[:,np.newaxis])
# Get the median value of each feature across all samples
mean_intensities = np.median(X, axis=0)
# Divde each feature by the median value of each feature -
# these are the quotients for each feature
X_quotients = (X / mean_intensities[np.newaxis,:])
if plot: # plot the distribution of quotients from one sample
for i in range(1,len(X_quotients[:,1])):
print 'allquotients reshaped!\n\n',
#all_quotients = X_quotients.reshape(np.prod(X_quotients.shape))
all_quotients = X_quotients[i,:]
print all_quotients.shape
x = np.random.normal(loc=0, scale=1, size=len(all_quotients))
sns.violinplot(all_quotients)
plt.title("median val: %f\nMax val=%f" % (np.median(all_quotients), np.max(all_quotients)))
plt.plot( title="median val: ")#%f" % np.median(all_quotients))
plt.xlim([-0.5, 5])
plt.show()
# Define a quotient for each sample as the median of the feature-specific quotients
# in that sample
sample_quotients = np.median(X_quotients, axis=1)
# Quotient normalize each samples
X_pqn = X / sample_quotients[:,np.newaxis]
return X_pqn
# Make a fake sample, with 2 samples at 1x and 2x dilutions
X_toy = np.array([[1,1,1,],
[2,2,2],
[3,6,9],
[6,12,18]], dtype=float)
print X_toy
print X_toy.reshape(1, np.prod(X_toy.shape))
X_toy_pqn_int = pqn_normalize(X_toy, integral_first=True, plot=True)
print X_toy_pqn_int
print '\n\n\n'
X_toy_pqn = pqn_normalize(X_toy)
print X_toy_pqn
```
<h2> pqn normalize your features </h2>
```
X_pqn = pqn_normalize(X)
print X_pqn
```
<h2>Random Forest & adaBoost with PQN-normalized data</h2>
```
rf_violinplot(X_pqn, y)
# Do multi-fold cross validation for adaboost classifier
adaboost_violinplot(X_pqn, y)
```
<h2> RF & adaBoost with PQN-normalized, log-transformed data </h2>
Turns out a monotonic transformation doesn't really affect any of these things.
I guess they're already close to unit varinace...?
```
X_pqn_nlog = np.log(X_pqn)
rf_violinplot(X_pqn_nlog, y)
adaboost_violinplot(X_pqn_nlog, y)
def roc_curve_cv(X, y, clf, cross_val,
path='/home/irockafe/Desktop/roc.pdf',
save=False, plot=True):
t1 = time.time()
# collect vals for the ROC curves
tpr_list = []
mean_fpr = np.linspace(0,1,100)
auc_list = []
# Get the false-positive and true-positive rate
for i, (train, test) in enumerate(cross_val):
clf.fit(X[train], y[train])
y_pred = clf.predict_proba(X[test])[:,1]
# get fpr, tpr
fpr, tpr, thresholds = roc_curve(y[test], y_pred)
roc_auc = auc(fpr, tpr)
#print 'AUC', roc_auc
#sns.plt.plot(fpr, tpr, lw=10, alpha=0.6, label='ROC - AUC = %0.2f' % roc_auc,)
#sns.plt.show()
tpr_list.append(interp(mean_fpr, fpr, tpr))
tpr_list[-1][0] = 0.0
auc_list.append(roc_auc)
if (i % 10 == 0):
print '{perc}% done! {time}s elapsed'.format(perc=100*float(i)/cross_val.n_iter, time=(time.time() - t1))
# get mean tpr and fpr
mean_tpr = np.mean(tpr_list, axis=0)
# make sure it ends up at 1.0
mean_tpr[-1] = 1.0
mean_auc = auc(mean_fpr, mean_tpr)
std_auc = np.std(auc_list)
if plot:
# plot mean auc
plt.plot(mean_fpr, mean_tpr, label='Mean ROC - AUC = %0.2f $\pm$ %0.2f' % (mean_auc,
std_auc),
lw=5, color='b')
# plot luck-line
plt.plot([0,1], [0,1], linestyle = '--', lw=2, color='r',
label='Luck', alpha=0.5)
# plot 1-std
std_tpr = np.std(tpr_list, axis=0)
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
plt.fill_between(mean_fpr, tprs_lower, tprs_upper, color='grey', alpha=0.2,
label=r'$\pm$ 1 stdev')
plt.xlim([-0.05, 1.05])
plt.ylim([-0.05, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('ROC curve, {iters} iterations of {cv} cross validation'.format(
iters=cross_val.n_iter, cv='{train}:{test}'.format(test=cross_val.test_size, train=(1-cross_val.test_size)))
)
plt.legend(loc="lower right")
if save:
plt.savefig(path, format='pdf')
plt.show()
return tpr_list, auc_list, mean_fpr
rf_estimators = 1000
n_iter = 3
test_size = 0.3
random_state = 1
cross_val_rf = StratifiedShuffleSplit(y, n_iter=n_iter, test_size=test_size, random_state=random_state)
clf_rf = RandomForestClassifier(n_estimators=rf_estimators, random_state=random_state)
rf_graph_path = '''/home/irockafe/Dropbox (MIT)/Alm_Lab/projects/revolutionizing_healthcare/data/MTBLS315/\
isaac_feature_tables/uhplc_pos/rf_roc_{trees}trees_{cv}cviter.pdf'''.format(trees=rf_estimators, cv=n_iter)
print cross_val_rf.n_iter
print cross_val_rf.test_size
tpr_vals, auc_vals, mean_fpr = roc_curve_cv(X_pqn, y, clf_rf, cross_val_rf,
path=rf_graph_path, save=False)
# For adaboosted
n_iter = 3
test_size = 0.3
random_state = 1
adaboost_estimators = 200
adaboost_path = '''/home/irockafe/Dropbox (MIT)/Alm_Lab/projects/revolutionizing_healthcare/data/MTBLS315/\
isaac_feature_tables/uhplc_pos/adaboost_roc_{trees}trees_{cv}cviter.pdf'''.format(trees=adaboost_estimators,
cv=n_iter)
cross_val_adaboost = StratifiedShuffleSplit(y, n_iter=n_iter, test_size=test_size, random_state=random_state)
clf = AdaBoostClassifier(n_estimators=adaboost_estimators, random_state=random_state)
adaboost_tpr, adaboost_auc, adaboost_fpr = roc_curve_cv(X_pqn, y, clf, cross_val_adaboost,
path=adaboost_path)
```
<h2> Great, you can classify things. But make null models and do a sanity check to make
sure you arent just classifying garbage </h2>
```
# Make a null model AUC curve
def make_null_model(X, y, clf, cross_val, random_state=1, num_shuffles=5, plot=True):
'''
Runs the true model, then sanity-checks by:
Shuffles class labels and then builds cross-validated ROC curves from them.
Compares true AUC vs. shuffled auc by t-test (assumes normality of AUC curve)
'''
null_aucs = []
print y.shape
print X.shape
tpr_true, auc_true, fpr_true = roc_curve_cv(X, y, clf, cross_val)
# shuffle y lots of times
for i in range(0, num_shuffles):
#Iterate through the shuffled y vals and repeat with appropriate params
# Retain the auc vals for final plotting of distribution
y_shuffle = shuffle(y)
cross_val.y = y_shuffle
cross_val.y_indices = y_shuffle
print 'Number of differences b/t original and shuffle: %s' % (y == cross_val.y).sum()
# Get auc values for number of iterations
tpr, auc, fpr = roc_curve_cv(X, y_shuffle, clf, cross_val, plot=False)
null_aucs.append(auc)
#plot the outcome
if plot:
flattened_aucs = [j for i in null_aucs for j in i]
my_dict = {'true_auc': auc_true, 'null_auc': flattened_aucs}
df_poop = pd.DataFrame.from_dict(my_dict, orient='index').T
df_tidy = pd.melt(df_poop, value_vars=['true_auc', 'null_auc'],
value_name='auc', var_name='AUC_type')
#print flattened_aucs
sns.violinplot(x='AUC_type', y='auc',
inner='points', data=df_tidy)
# Plot distribution of AUC vals
plt.title("Distribution of aucs")
#sns.plt.ylabel('count')
plt.xlabel('AUC')
#sns.plt.plot(auc_true, 0, color='red', markersize=10)
plt.show()
# Do a quick t-test to see if odds of randomly getting an AUC that good
return auc_true, null_aucs
# Make a null model AUC curve & compare it to null-model
# Random forest magic!
rf_estimators = 1000
n_iter = 50
test_size = 0.3
random_state = 1
cross_val_rf = StratifiedShuffleSplit(y, n_iter=n_iter, test_size=test_size, random_state=random_state)
clf_rf = RandomForestClassifier(n_estimators=rf_estimators, random_state=random_state)
true_auc, all_aucs = make_null_model(X_pqn, y, clf_rf, cross_val_rf, num_shuffles=5)
# make dataframe from true and false aucs
flattened_aucs = [j for i in all_aucs for j in i]
my_dict = {'true_auc': true_auc, 'null_auc': flattened_aucs}
df_poop = pd.DataFrame.from_dict(my_dict, orient='index').T
df_tidy = pd.melt(df_poop, value_vars=['true_auc', 'null_auc'],
value_name='auc', var_name='AUC_type')
print df_tidy.head()
#print flattened_aucs
sns.violinplot(x='AUC_type', y='auc',
inner='points', data=df_tidy, bw=0.7)
plt.show()
```
<h2> Let's check out some PCA plots </h2>
```
from sklearn.decomposition import PCA
# Check PCA of things
def PCA_plot(X, y, n_components, plot_color, class_nums, class_names, title='PCA'):
pca = PCA(n_components=n_components)
X_pca = pca.fit(X).transform(X)
print zip(plot_color, class_nums, class_names)
for color, i, target_name in zip(plot_color, class_nums, class_names):
# plot one class at a time, first plot all classes y == 0
#print color
#print y == i
xvals = X_pca[y == i, 0]
print xvals.shape
yvals = X_pca[y == i, 1]
plt.scatter(xvals, yvals, color=color, alpha=0.8, label=target_name)
plt.legend(bbox_to_anchor=(1.01,1), loc='upper left', shadow=False)#, scatterpoints=1)
plt.title('PCA of Malaria data')
plt.show()
PCA_plot(X_pqn, y, 2, ['red', 'blue'], [0,1], ['malaria', 'non-malaria fever'])
PCA_plot(X, y, 2, ['red', 'blue'], [0,1], ['malaria', 'non-malaria fever'])
```
<h2> What about with all thre classes? </h2>
```
# convert classes to numbers
le = preprocessing.LabelEncoder()
le.fit(class_map_df['class'])
y_three_class = le.transform(class_map_df['class'])
print class_map_df.head(10)
print y_three_class
print X.shape
print y_three_class.shape
y_labels = np.sort(class_map_df['class'].unique())
print y_labels
colors = ['green', 'red', 'blue']
print np.unique(y_three_class)
PCA_plot(X_pqn, y_three_class, 2, colors, np.unique(y_three_class), y_labels)
PCA_plot(X, y_three_class, 2, colors, np.unique(y_three_class), y_labels)
```
|
github_jupyter
|
<h1>REGIONE CAMPANIA</h1>
Confronto dei dati relativi ai decessi registrati dall'ISTAT e i decessi causa COVID-19 registrati dalla Protezione Civile Italiana con i decessi previsti dal modello predittivo SARIMA.
<h2>DECESSI MENSILI REGIONE CAMPANIA ISTAT</h2>
Il DataFrame contiene i dati relativi ai decessi mensili della regione <b>Campania</b> dal <b>2015</b> al <b>30 settembre 2020</b>.
```
import matplotlib.pyplot as plt
import pandas as pd
decessi_istat = pd.read_csv('../../csv/regioni/campania.csv')
decessi_istat.head()
decessi_istat['DATA'] = pd.to_datetime(decessi_istat['DATA'])
decessi_istat.TOTALE = pd.to_numeric(decessi_istat.TOTALE)
```
<h3>Recupero dei dati inerenti al periodo COVID-19</h3>
```
decessi_istat = decessi_istat[decessi_istat['DATA'] > '2020-02-29']
decessi_istat.head()
```
<h3>Creazione serie storica dei decessi ISTAT</h3>
```
decessi_istat = decessi_istat.set_index('DATA')
decessi_istat = decessi_istat.TOTALE
decessi_istat
```
<h2>DECESSI MENSILI REGIONE ABRUZZO CAMPANIA DAL COVID</h2>
Il DataFrame contine i dati forniti dalla Protezione Civile relativi ai decessi mensili della regione <b>Campania</b> da <b> marzo 2020</b> al <b>30 settembre 2020</b>.
```
covid = pd.read_csv('../../csv/regioni_covid/campania.csv')
covid.head()
covid['data'] = pd.to_datetime(covid['data'])
covid.deceduti = pd.to_numeric(covid.deceduti)
covid = covid.set_index('data')
covid.head()
```
<h3>Creazione serie storica dei decessi COVID-19</h3>
```
covid = covid.deceduti
```
<h2>PREDIZIONE DECESSI MENSILI REGIONE SECONDO MODELLO SARIMA</h2>
Il DataFrame contiene i dati riguardanti i decessi mensili della regione <b>Campania</b> secondo la predizione del modello SARIMA applicato.
```
predictions = pd.read_csv('../../csv/pred/predictions_SARIMA_campania.csv')
predictions.head()
predictions.rename(columns={'Unnamed: 0': 'Data', 'predicted_mean':'Totale'}, inplace=True)
predictions.head()
predictions['Data'] = pd.to_datetime(predictions['Data'])
predictions.Totale = pd.to_numeric(predictions.Totale)
```
<h3>Recupero dei dati inerenti al periodo COVID-19</h3>
```
predictions = predictions[predictions['Data'] > '2020-02-29']
predictions.head()
predictions = predictions.set_index('Data')
predictions.head()
```
<h3>Creazione serie storica dei decessi secondo la predizione del modello</h3>
```
predictions = predictions.Totale
```
<h1>INTERVALLI DI CONFIDENZA
<h3>Limite massimo
```
upper = pd.read_csv('../../csv/upper/predictions_SARIMA_campania_upper.csv')
upper.head()
upper.rename(columns={'Unnamed: 0': 'Data', 'upper TOTALE':'Totale'}, inplace=True)
upper['Data'] = pd.to_datetime(upper['Data'])
upper.Totale = pd.to_numeric(upper.Totale)
upper.head()
upper = upper[upper['Data'] > '2020-02-29']
upper = upper.set_index('Data')
upper.head()
upper = upper.Totale
```
<h3>Limite minimo
```
lower = pd.read_csv('../../csv/lower/predictions_SARIMA_campania_lower.csv')
lower.head()
lower.rename(columns={'Unnamed: 0': 'Data', 'lower TOTALE':'Totale'}, inplace=True)
lower['Data'] = pd.to_datetime(lower['Data'])
lower.Totale = pd.to_numeric(lower.Totale)
lower.head()
lower = lower[lower['Data'] > '2020-02-29']
lower = lower.set_index('Data')
lower.head()
lower = lower.Totale
```
<h1> CONFRONTO DELLE SERIE STORICHE </h1>
Di seguito il confronto grafico tra le serie storiche dei <b>decessi totali mensili</b>, dei <b>decessi causa COVID-19</b> e dei <b>decessi previsti dal modello SARIMA</b> della regione <b>Campania</b>.
<br />
I mesi di riferimento sono: <b>marzo</b>, <b>aprile</b>, <b>maggio</b>, <b>giugno</b>, <b>luglio</b>, <b>agosto</b> e <b>settembre</b>.
```
plt.figure(figsize=(15,4))
plt.title('CAMPANIA - Confronto decessi totali, decessi causa covid e decessi del modello predittivo', size=18)
plt.plot(covid, label='decessi causa covid')
plt.plot(decessi_istat, label='decessi totali')
plt.plot(predictions, label='predizione modello')
plt.legend(prop={'size': 12})
plt.show()
plt.figure(figsize=(15,4))
plt.title("CAMPANIA - Confronto decessi totali ISTAT con decessi previsti dal modello", size=18)
plt.plot(predictions, label='predizione modello')
plt.plot(upper, label='limite massimo')
plt.plot(lower, label='limite minimo')
plt.plot(decessi_istat, label='decessi totali')
plt.legend(prop={'size': 12})
plt.show()
```
<h3>Calcolo dei decessi COVID-19 secondo il modello predittivo</h3>
Differenza tra i decessi totali rilasciati dall'ISTAT e i decessi secondo la previsione del modello SARIMA.
```
n = decessi_istat - predictions
n_upper = decessi_istat - lower
n_lower = decessi_istat - upper
plt.figure(figsize=(15,4))
plt.title("CAMPANIA - Confronto decessi accertati covid con decessi covid previsti dal modello", size=18)
plt.plot(covid, label='decessi covid accertati - Protezione Civile')
plt.plot(n, label='devessi covid previsti - modello SARIMA')
plt.plot(n_upper, label='limite massimo - modello SARIMA')
plt.plot(n_lower, label='limite minimo - modello SARIMA')
plt.legend(prop={'size': 12})
plt.show()
d = decessi_istat.sum()
print("Decessi 2020:", d)
d_m = predictions.sum()
print("Decessi attesi dal modello 2020:", d_m)
d_lower = lower.sum()
print("Decessi attesi dal modello 2020 - livello mimino:", d_lower)
```
<h3>Numero totale dei decessi accertati COVID-19 regione Campania </h3>
```
m = covid.sum()
print(int(m))
```
<h3>Numero totale dei decessi COVID-19 previsti dal modello per la regione Campania </h3>
<h4>Valore medio
```
total = n.sum()
print(int(total))
```
<h4>Valore massimo
```
total_upper = n_upper.sum()
print(int(total_upper))
```
<h4>Valore minimo
```
total_lower = n_lower.sum()
print(int(total_lower))
```
<h3>Calcolo del numero dei decessi COVID-19 non registrati secondo il modello predittivo SARIMA della regione Campania</h3>
<h4>Valore medio
```
x = decessi_istat - predictions - covid
x = x.sum()
print(int(x))
```
<h4>Valore massimo
```
x_upper = decessi_istat - lower - covid
x_upper = x_upper.sum()
print(int(x_upper))
```
<h4>Valore minimo
```
x_lower = decessi_istat - upper - covid
x_lower = x_lower.sum()
print(int(x_lower))
```
|
github_jupyter
|
## 1. Inspecting transfusion.data file
<p><img src="https://assets.datacamp.com/production/project_646/img/blood_donation.png" style="float: right;" alt="A pictogram of a blood bag with blood donation written in it" width="200"></p>
<p>Blood transfusion saves lives - from replacing lost blood during major surgery or a serious injury to treating various illnesses and blood disorders. Ensuring that there's enough blood in supply whenever needed is a serious challenge for the health professionals. According to <a href="https://www.webmd.com/a-to-z-guides/blood-transfusion-what-to-know#1">WebMD</a>, "about 5 million Americans need a blood transfusion every year".</p>
<p>Our dataset is from a mobile blood donation vehicle in Taiwan. The Blood Transfusion Service Center drives to different universities and collects blood as part of a blood drive. We want to predict whether or not a donor will give blood the next time the vehicle comes to campus.</p>
<p>The data is stored in <code>datasets/transfusion.data</code> and it is structured according to RFMTC marketing model (a variation of RFM). We'll explore what that means later in this notebook. First, let's inspect the data.</p>
```
# Print out the first 5 lines from the transfusion.data file
!head -n 5 datasets/transfusion.data
```
## 2. Loading the blood donations data
<p>We now know that we are working with a typical CSV file (i.e., the delimiter is <code>,</code>, etc.). We proceed to loading the data into memory.</p>
```
# Import pandas
import pandas as pd
# Read in dataset
transfusion = pd.read_csv('datasets/transfusion.data')
# Print out the first rows of our dataset
transfusion.head()
```
## 3. Inspecting transfusion DataFrame
<p>Let's briefly return to our discussion of RFM model. RFM stands for Recency, Frequency and Monetary Value and it is commonly used in marketing for identifying your best customers. In our case, our customers are blood donors.</p>
<p>RFMTC is a variation of the RFM model. Below is a description of what each column means in our dataset:</p>
<ul>
<li>R (Recency - months since the last donation)</li>
<li>F (Frequency - total number of donation)</li>
<li>M (Monetary - total blood donated in c.c.)</li>
<li>T (Time - months since the first donation)</li>
<li>a binary variable representing whether he/she donated blood in March 2007 (1 stands for donating blood; 0 stands for not donating blood)</li>
</ul>
<p>It looks like every column in our DataFrame has the numeric type, which is exactly what we want when building a machine learning model. Let's verify our hypothesis.</p>
```
# Print a concise summary of transfusion DataFrame
transfusion.info()
```
## 4. Creating target column
<p>We are aiming to predict the value in <code>whether he/she donated blood in March 2007</code> column. Let's rename this it to <code>target</code> so that it's more convenient to work with.</p>
```
# Rename target column as 'target' for brevity
transfusion.rename(
columns={'whether he/she donated blood in March 2007':'target'},
inplace=True
)
# Print out the first 2 rows
transfusion.head(2)
```
## 5. Checking target incidence
<p>We want to predict whether or not the same donor will give blood the next time the vehicle comes to campus. The model for this is a binary classifier, meaning that there are only 2 possible outcomes:</p>
<ul>
<li><code>0</code> - the donor will not give blood</li>
<li><code>1</code> - the donor will give blood</li>
</ul>
<p>Target incidence is defined as the number of cases of each individual target value in a dataset. That is, how many 0s in the target column compared to how many 1s? Target incidence gives us an idea of how balanced (or imbalanced) is our dataset.</p>
```
# Print target incidence proportions, rounding output to 3 decimal places
transfusion.target.value_counts(normalize=True).round(3)
```
## 6. Splitting transfusion into train and test datasets
<p>We'll now use <code>train_test_split()</code> method to split <code>transfusion</code> DataFrame.</p>
<p>Target incidence informed us that in our dataset <code>0</code>s appear 76% of the time. We want to keep the same structure in train and test datasets, i.e., both datasets must have 0 target incidence of 76%. This is very easy to do using the <code>train_test_split()</code> method from the <code>scikit learn</code> library - all we need to do is specify the <code>stratify</code> parameter. In our case, we'll stratify on the <code>target</code> column.</p>
```
# Import train_test_split method
from sklearn.model_selection import train_test_split
# Split transfusion DataFrame into
# X_train, X_test, y_train and y_test datasets,
# stratifying on the `target` column
X_train, X_test, y_train, y_test = train_test_split(
transfusion.drop(columns='target'),
transfusion.target,
test_size=0.25,
random_state=42,
stratify=transfusion.target
)
# Print out the first 2 rows of X_train
X_train.head(2)
```
## 7. Selecting model using TPOT
<p><a href="https://github.com/EpistasisLab/tpot">TPOT</a> is a Python Automated Machine Learning tool that optimizes machine learning pipelines using genetic programming.</p>
<p><img src="https://assets.datacamp.com/production/project_646/img/tpot-ml-pipeline.png" alt="TPOT Machine Learning Pipeline"></p>
<p>TPOT will automatically explore hundreds of possible pipelines to find the best one for our dataset. Note, the outcome of this search will be a <a href="https://scikit-learn.org/stable/modules/generated/sklearn.pipeline.Pipeline.html">scikit-learn pipeline</a>, meaning it will include any pre-processing steps as well as the model.</p>
<p>We are using TPOT to help us zero in on one model that we can then explore and optimize further.</p>
```
# Import TPOTClassifier and roc_auc_score
from tpot import TPOTClassifier
from sklearn.metrics import roc_auc_score
# Instantiate TPOTClassifier
tpot = TPOTClassifier(
generations=5,
population_size=20,
verbosity=2,
scoring='roc_auc',
random_state=42,
disable_update_check=True,
config_dict='TPOT light'
)
tpot.fit(X_train, y_train)
# AUC score for tpot model
tpot_auc_score = roc_auc_score(y_test, tpot.predict_proba(X_test)[:, 1])
print(f'\nAUC score: {tpot_auc_score:.4f}')
# Print best pipeline steps
print('\nBest pipeline steps:', end='\n')
for idx, (name, transform) in enumerate(tpot.fitted_pipeline_.steps, start=1):
# Print idx and transform
print(f'{idx}. {transform}')
```
## 8. Checking the variance
<p>TPOT picked <code>LogisticRegression</code> as the best model for our dataset with no pre-processing steps, giving us the AUC score of 0.7850. This is a great starting point. Let's see if we can make it better.</p>
<p>One of the assumptions for linear regression models is that the data and the features we are giving it are related in a linear fashion, or can be measured with a linear distance metric. If a feature in our dataset has a high variance that's an order of magnitude or more greater than the other features, this could impact the model's ability to learn from other features in the dataset.</p>
<p>Correcting for high variance is called normalization. It is one of the possible transformations you do before training a model. Let's check the variance to see if such transformation is needed.</p>
```
# X_train's variance, rounding the output to 3 decimal places
X_train.var().round(3)
```
## 9. Log normalization
<p><code>Monetary (c.c. blood)</code>'s variance is very high in comparison to any other column in the dataset. This means that, unless accounted for, this feature may get more weight by the model (i.e., be seen as more important) than any other feature.</p>
<p>One way to correct for high variance is to use log normalization.</p>
```
# Import numpy
import numpy as np
# Copy X_train and X_test into X_train_normed and X_test_normed
X_train_normed, X_test_normed = X_train.copy(), X_test.copy()
# Specify which column to normalize
col_to_normalize = 'Monetary (c.c. blood)'
# Log normalization
for df_ in [X_train_normed, X_test_normed]:
# Add log normalized column
df_['monetary_log'] = np.log(df_[col_to_normalize])
# Drop the original column
df_.drop(columns=col_to_normalize, inplace=True)
# Check the variance for X_train_normed
X_train_normed.var().round(3)
```
## 10. Training the linear regression model
<p>The variance looks much better now. Notice that now <code>Time (months)</code> has the largest variance, but it's not the <a href="https://en.wikipedia.org/wiki/Order_of_magnitude">orders of magnitude</a> higher than the rest of the variables, so we'll leave it as is.</p>
<p>We are now ready to train the linear regression model.</p>
```
# Importing modules
from sklearn import linear_model.LogisticRegression
# Instantiate LogisticRegression
logreg = LogisticRegression(
solver='liblinear',
random_state=42
)
# Train the model
fit(X_train_normed, y_train)
# AUC score for tpot model
logreg_auc_score = roc_auc_score(y_test, logreg.predict_proba(X_test_normed)[:, 1])
print(f'\nAUC score: {logreg_auc_score:.4f}')
```
## 11. Conclusion
<p>The demand for blood fluctuates throughout the year. As one <a href="https://www.kjrh.com/news/local-news/red-cross-in-blood-donation-crisis">prominent</a> example, blood donations slow down during busy holiday seasons. An accurate forecast for the future supply of blood allows for an appropriate action to be taken ahead of time and therefore saving more lives.</p>
<p>In this notebook, we explored automatic model selection using TPOT and AUC score we got was 0.7850. This is better than simply choosing <code>0</code> all the time (the target incidence suggests that such a model would have 76% success rate). We then log normalized our training data and improved the AUC score by 0.5%. In the field of machine learning, even small improvements in accuracy can be important, depending on the purpose.</p>
<p>Another benefit of using logistic regression model is that it is interpretable. We can analyze how much of the variance in the response variable (<code>target</code>) can be explained by other variables in our dataset.</p>
```
# Importing itemgetter
from operator import itemgetter
# Sort models based on their AUC score from highest to lowest
sorted(
[('tpot', tpot_auc_score), ('logreg', logreg_auc_score)],
key=itemgetter(1),
reverse=True
)
```
|
github_jupyter
|
# A Table based Q-Learning Reinforcement Agent in A Grid World
This is a simple example of a Q-Learning agent. The Q function is a table, and each decision is made by sampling the Q-values for a particular state thermally.
```
import numpy as np
import random
import gym
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
from IPython.display import clear_output
from tqdm import tqdm
env = gym.make('FrozenLake-v0')
Q = np.zeros([env.observation_space.n, env.action_space.n])
# Set learning parameters
decision_temperature = 0.01
l_rate = 0.5
y = .99
e = 0.1
num_episodes = 900
# create lists to contain total rewawrds and steps per episode
epi_length = []
rs = []
for i in tqdm(range(num_episodes)):
s = env.reset()
r_total = 0
done = False
number_jumps = 0
# limit numerb of jumps
while number_jumps < 99:
number_jumps += 1
softmax = np.exp(Q[s]/decision_temperature)
rand_n = np.random.rand() * np.sum(softmax)
# pick the next action randomly
acc = 0
for ind in range(env.action_space.n):
acc += softmax[ind]
if acc >= rand_n:
a = ind
break
#print(a, softmax, rand_n)
# a = np.argmax(Q[s, :] + np.random.randn(1, env.action_space.n) * (1./(i+1)))
s_next, r, done, _ = env.step(a)
Q_next_value = Q[s_next]
max_Q_next = np.max(Q[s_next,:])
# now update Q
Q[s, a] += l_rate * (r + y * max_Q_next \
- Q[s, a])
r_total += r
s = s_next
if done:
# be more conservative as we learn more
e = 1./((i/50) + 10)
break
if i%900 == 899:
clear_output(wait=True)
print("success rate: " + str(sum(rs[-200:])/2) + "%")
plt.figure(figsize=(8, 8))
plt.subplot(211)
plt.title("Jumps Per Episode", fontsize=18)
plt.plot(epi_length[-200:], "#23aaff")
plt.subplot(212)
plt.title('Reward For Each Episode (0/1)', fontsize=18)
plt.plot(rs[-200:], "o", color='#23aaff', alpha=0.1)
plt.figure(figsize=(6, 6))
plt.title('Decision Table', fontsize=18)
plt.xlabel("States", fontsize=15)
plt.ylabel('Actions', fontsize=15)
plt.imshow(Q.T)
plt.show()
epi_length.append(number_jumps)
rs.append(r_total)
def mv_avg(xs, n):
return [sum(xs[i:i+n])/n for i in range(len(xs)-n)]
# plt.plot(mv_avg(rs, 200))
plt.figure(figsize=(8, 8))
plt.subplot(211)
plt.title("Jumps Per Episode", fontsize=18)
plt.plot(epi_length, "#23aaff", linewidth=0.1, alpha=0.7,
label="raw data")
plt.plot(mv_avg(epi_length, 200), color="blue", alpha=0.3, linewidth=4,
label="Moving Average")
plt.legend(loc=(1.05, 0), frameon=False, fontsize=15)
plt.subplot(212)
plt.title('Reward For Each Episode (0/1)', fontsize=18)
#plt.plot(rs, "o", color='#23aaff', alpha=0.2, markersize=0.4, label="Reward")
plt.plot(mv_avg(rs, 200), color="red", alpha=0.5, linewidth=4, label="Moving Average")
plt.ylim(-0.1, 1.1)
plt.legend(loc=(1.05, 0), frameon=False, fontsize=15)
plt.savefig('./figures/Frozen-Lake-v0-thermal-table.png', dpi=300, bbox_inches='tight')
```
|
github_jupyter
|
# Binary classification with Support Vector Machines (SVM)
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import ipywidgets as widgets
from sklearn.linear_model import LogisticRegression
from sklearn.svm import LinearSVC, SVC
from ipywidgets import interact, interactive, fixed
from numpy.random import default_rng
plt.rcParams['figure.figsize'] = [9.5, 6]
rng = default_rng(seed=42)
```
## Two Gaussian distributions
Let's generate some data, two sets of Normally distributed points...
```
def plot_data():
plt.xlim(0.0, 1.0)
plt.ylim(0.0, 1.0)
plt.plot(x1, y1, 'bs', markersize=6)
plt.plot(x2, y2, 'rx', markersize=6)
s1=0.01
s2=0.01
n1=30
n2=30
x1, y1 = rng.multivariate_normal([0.5, 0.3], [[s1, 0], [0, s1]], n1).T
x2, y2 = rng.multivariate_normal([0.7, 0.7], [[s2, 0], [0, s2]], n2).T
plot_data()
plt.suptitle('generated data points')
plt.show()
```
## Separating hyperplane
Linear classifiers: separate the two distributions with a line (hyperplane)
```
def plot_line(slope, intercept, show_params=False):
x_vals = np.linspace(0.0, 1.0)
y_vals = slope*x_vals +intercept
plt.plot(x_vals, y_vals, '--')
if show_params:
plt.title('slope={:.4f}, intercept={:.4f}'.format(slope, intercept))
```
You can try out different parameters (slope, intercept) for the line. Note that there are many (in fact an infinite number) of lines that separate the two classes.
```
#plot_data()
#plot_line(-1.1, 1.1)
#plot_line(-0.23, 0.62)
#plot_line(-0.41, 0.71)
#plt.savefig('just_points2.png')
def do_plot_interactive(slope=-1.0, intercept=1.0):
plot_data()
plot_line(slope, intercept, True)
plt.suptitle('separating hyperplane (line)')
interactive_plot = interactive(do_plot_interactive, slope=(-2.0, 2.0), intercept=(0.5, 1.5))
output = interactive_plot.children[-1]
output.layout.height = '450px'
interactive_plot
```
## Logistic regression
Let's create a training set $\mathbf{X}$ with labels in $\mathbf{y}$ with our points (in shuffled order).
```
X = np.block([[x1, x2], [y1, y2]]).T
y = np.hstack((np.repeat(0, len(x1)), np.repeat(1, len(x2))))
rand_idx = rng.permutation(len(x1) + len(x2))
X = X[rand_idx]
y = y[rand_idx]
print(X.shape, y.shape)
print(X[:10,:])
print(y[:10].reshape(-1,1))
```
The task is now to learn a classification model $\mathbf{y} = f(\mathbf{X})$.
First, let's try [logistic regression](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html).
```
clf_lr = LogisticRegression(penalty='none')
clf_lr.fit(X, y)
w1 = clf_lr.coef_[0][0]
w2 = clf_lr.coef_[0][1]
b = clf_lr.intercept_[0]
plt.suptitle('Logistic regression')
plot_data()
plot_line(slope=-w1/w2, intercept=-b/w2, show_params=True)
```
## Linear SVM
```
clf_lsvm = SVC(C=1000, kernel='linear')
clf_lsvm.fit(X, y)
w1 = clf_lsvm.coef_[0][0]
w2 = clf_lsvm.coef_[0][1]
b = clf_lsvm.intercept_[0]
plt.suptitle('Linear SVM')
plot_data()
plot_line(slope=-w1/w2, intercept=-b/w2, show_params=True)
def plot_clf(clf):
# plot the decision function
ax = plt.gca()
xlim = ax.get_xlim()
ylim = ax.get_ylim()
# create grid to evaluate model
xx = np.linspace(xlim[0], xlim[1], 30)
yy = np.linspace(ylim[0], ylim[1], 30)
YY, XX = np.meshgrid(yy, xx)
xy = np.vstack([XX.ravel(), YY.ravel()]).T
Z = clf.decision_function(xy).reshape(XX.shape)
# plot decision boundary and margins
ax.contour(XX, YY, Z, colors='k', levels=[-1, 0, 1], alpha=0.5,
linestyles=['--', '-', '--'])
# plot support vectors
ax.scatter(clf.support_vectors_[:, 0], clf.support_vectors_[:, 1], s=100,
linewidth=1, facecolors='none', edgecolors='k')
```
Let's try different $C$ values. We'll also visualize the margins and support vectors.
```
def do_plot_svm(C=1000.0):
clf = SVC(C=C, kernel='linear')
clf.fit(X, y)
plot_data()
plot_clf(clf)
interactive_plot = interactive(do_plot_svm, C=widgets.FloatLogSlider(value=1000, base=10, min=-0.5, max=4, step=0.2))
output = interactive_plot.children[-1]
output.layout.height = '400px'
interactive_plot
#do_plot_svm()
#plt.savefig('linear-svm.png')
```
## Kernel SVM
```
clf_ksvm = SVC(C=10, kernel='rbf')
clf_ksvm.fit(X, y)
plot_data()
plot_clf(clf_ksvm)
plt.savefig('kernel-svm.png')
def do_plot_svm(C=1000.0):
clf = SVC(C=C, kernel='rbf')
clf.fit(X, y)
plot_data()
plot_clf(clf)
interactive_plot = interactive(do_plot_svm, C=widgets.FloatLogSlider(value=100, base=10, min=-1, max=3, step=0.2))
output = interactive_plot.children[-1]
output.layout.height = '400px'
interactive_plot
```
|
github_jupyter
|
We saw in this [journal entry](http://wiki.noahbrenowitz.com/doku.php?id=journal:2018-10:day-2018-10-24#run_110) that multiple-step trained neural network gives a very imbalanced estimate, but the two-step trained neural network gives a good answer. Where do these two patterns disagree?
```
%matplotlib inline
import matplotlib.pyplot as plt
import xarray as xr
import click
import torch
from uwnet.model import call_with_xr
import holoviews as hv
from holoviews.operation import decimate
hv.extension('bokeh')
def column_integrate(data_array, mass):
return (data_array * mass).sum('z')
def compute_apparent_sources(model_path, ds):
model = torch.load(model_path)
return call_with_xr(model, ds, drop_times=0)
def get_single_location(ds, location=(32,0)):
y, x = location
return ds.isel(y=slice(y,y+1), x=slice(x,x+1))
def dict_to_dataset(datasets, dim='key'):
"""Concatenate a dict of datasets along a new axis"""
keys, values = zip(*datasets.items())
idx = pd.Index(keys, name=dim)
return xr.concat(values, dim=idx)
def dataarray_to_table(dataarray):
return dataarray.to_dataset('key').to_dataframe().reset_index()
def get_apparent_sources(model_paths, data_path):
ds = xr.open_dataset(data_path)
location = get_single_location(ds, location=(32,0))
sources = {training_strategy: compute_apparent_sources(model_path, location)
for training_strategy, model_path in model_paths.items()}
return dict_to_dataset(sources)
model_paths = {
'multi': '../models/113/3.pkl',
'single': '../models/110/3.pkl'
}
data_path = "../data/processed/training.nc"
sources = get_apparent_sources(model_paths, data_path)
```
# Apparent moistening and heating
Here we scatter plot the apparent heating and moistening:
```
%%opts Scatter[width=500, height=500, color_index='z'](cmap='viridis', alpha=.2)
%%opts Curve(color='black')
lims = (-30, 40)
df = dataarray_to_table(sources.QT)
moisture_source = hv.Scatter(df, kdims=["multi", "single"]).groupby('z').redim.range(multi=lims, single=lims) \
*hv.Curve((lims, lims))
lims = (-30, 40)
df = dataarray_to_table(sources.SLI)
heating = hv.Scatter(df, kdims=["multi", "single"]).groupby('z').redim.range(multi=lims, single=lims) \
*hv.Curve((lims, lims))
moisture_source.relabel("Moistening (g/kg/day)") + heating.relabel("Heating (K/day)")
```
The multistep moistening is far too negative in the upper parts of the atmosphere, and the corresponding heating is too positive. Does this **happen because the moisture is negative in those regions**.
|
github_jupyter
|
# ACS Download
## ACS TOOL STEP 1 -> SETUP :
#### Uses: csa2tractcrosswalk.csv, VitalSignsCensus_ACS_Tables.xlsx
#### Creates: ./AcsDataRaw/ ./AcsDataClean/
### Import Modules & Construct Path Handlers
```
import os
import sys
import pandas as pd
pd.set_option('display.expand_frame_repr', False)
pd.set_option('display.precision', 2)
```
### Get Vital Signs Reference Table
```
acs_tables = pd.read_csv('https://raw.githubusercontent.com/bniajfi/bniajfi/main/vs_acs_table_ids.csv')
acs_tables.head()
```
### Get Tract/ CSA CrossWalk
```
file = 'https://raw.githubusercontent.com/bniajfi/bniajfi/main/CSA-to-Tract-2010.csv'
crosswalk = pd.read_csv( file )
crosswalk = dict(zip(crosswalk['TRACTCE10'], crosswalk['CSA2010'] ) )
```
### Get retrieve_acs_data function
```
!pip install dataplay geopandas VitalSigns
import VitalSigns as vs
from VitalSigns import acsDownload
help(acsDownload)
help(vs.acsDownload.retrieve_acs_data)
```
### Column Operations
```
import csv # 'quote all'
def fixColNamesForCSV(x): return str(x)[:] if str(x) in ["NAME","state","county","tract", "CSA"] else str(x)[12:]
```
## ACS TOOL STEP 2 -> Execute :
```
acs_tables.head()
```
### Save the ACS Data
```
# Set Index df.set_index("NAME", inplace = True)
# Save raw to '../../data/3_outputs/acs/raw/'+year+'/'+tableId+'_'+description+'_5y'+year+'_est.csv'
# Tract to CSA df['CSA'] = df.apply(lambda row: crosswalk.get(int(row['tract']), "empty"), axis=1)
# Save 4 use '../../data/2_cleaned/acs/'+tableId+'_'+description+'_5y'+year+'_est.csv'
year = '19'
count = 0
startFrom = 0
state = '24'
county = '510'
tract = '*'
tableId = 'B19001'
saveAcs = True
# For each ACS Table
for x, row in acs_tables.iterrows():
count += 1
# Grab its Meta Data
description = str(acs_tables.loc[x, 'shortname'])
tableId = str(acs_tables.loc[x, 'id'])
yearExists = int(acs_tables.loc[x, year+'_exists'])
# If the Indicator is valid for the year
# use startFrom to being at a specific count
if yearExists and count >= startFrom:
print(str(count)+') '+tableId + ' ' + description)
# retrieve the Python ACS indicator
print('sending retrieve_acs_data', year, tableId)
df = vs.acsDownload.retrieve_acs_data(state, county, tract, tableId, year, saveAcs)
df.set_index("NAME", inplace = True)
# Save the Data as Raw
# We do not want the id in the column names
saveThis = df.rename( columns = lambda x : ( fixColNamesForCSV(x) ) )
saveThis.to_csv('./AcsDataRaw/'+tableId+'_'+description+'_5y'+year+'_est.csv', quoting=csv.QUOTE_ALL)
# Match Tract to CSA
df['CSA'] = df.apply(lambda row: crosswalk.get(int(row['tract']), "empty"), axis=1)
# Save the data (again) as Cleaned for me to use in the next scripts
df.to_csv('./AcsDataClean/'+tableId+'_5y'+year+'_est.csv', quoting=csv.QUOTE_ALL)
```
# ACS Create Indicators
ACS TOOL STEP 1 -> SETUP :
Uses: ./AcsDataClean/ VitalSignsCensus_ACS_Tables.xlsx VitalSignsCensus_ACS_compare_data.xlsm
Creates: ./VSData/
### Get Vital Signs Reference Table
```
ls
file = 'VitalSignsCensus_ACS_Tables.xlsx'
xls = pd.ExcelFile(findFile('./', file))
indicators = pd.read_excel(xls, sheet_name='indicators', index_col=0 )
indicators.head(30)
```
## ACS TOOL STEP 2 -> Execute :
### Create ACS Indicators
#### Settings/ Get Data
```
flag = True;
year = '19'
vsTbl = pd.read_excel(xls, sheet_name=str('vs'+year), index_col=0 )
# Prepare the Compare Historic Data
file = 'VitalSignsCensus_ACS_compare_data.xlsm'
compare_table = pd.read_excel(findFile('./', file), None);
comparable = False
if( str('VS'+year) in compare_table.keys() ):
compare_table = compare_table[str('VS'+year)]
comparable = True
columnsNames = compare_table.iloc[0]
compare_table = compare_table.drop(compare_table.index[0])
compare_table.set_index(['CSA2010'], drop = True, inplace = True)
```
#### Create Indicators
```
# For Each Indicator
for x, row in indicators.iterrows():
# Grab its Meta Data
shortSource = str(indicators.loc[x, 'Short Source'])
shortName = str(indicators.loc[x, 'ShortName'])[:-2]
yearExists = int(float(indicators.loc[x, year+'_exists']))
indicator = str(indicators.loc[x, 'Indicator'])
indicator_number = str(indicators.index.tolist().index(x)+1 )
fileLocation = str(findFile( './', shortName+'.py') )
# If the Indicator is valid for the year, and uses ACS Data, and method exists
flag = True if fileLocation != str('None') else False
flag = True if flag and yearExists else False
flag = True if flag and shortSource in ['ACS', 'Census'] else False
if flag:
print(shortSource, shortName, yearExists, indicator, fileLocation, indicator_number )
# retrieve the Python ACS indicator
module = __import__( shortName )
result = getattr( module, shortName )( year )
# Put Baltimore City at the bottom of the list
idx = result.index.tolist()
idx.pop(idx.index('Baltimore City'))
result = result.reindex(idx+['Baltimore City'])
# Write the results back into the XL dataframe
vsTbl[ str(indicator_number + '_' +shortName ) ] = result
# Save the Data
result.to_csv('./VSData/vs'+ str(year)+'_'+shortName+'.csv')
# drop columns with any empty values
vsTbl = vsTbl.dropna(axis=1, how='any')
# Save the Data
file = 'VS'+str(year)+'_indicators.xlsx'
file = findFile( 'VSData', file)
# writer = pd.ExcelWriter(file)
#vsTbl.to_excel(writer, str(year+'New_VS_Values') )
# Save the Data
vsTbl.to_csv('./VSData/vs'+str(year+'_New_VS_Values')+'.csv')
# Include Historic Data if exist
if( comparable ):
# add historic indicator to excel doc
# compare_table.to_excel(writer,sheet_name = str(year+'Original_VS_Values') )
# compare sets
info = pd.DataFrame()
diff = pd.DataFrame()
simi = pd.DataFrame()
for column in vsTbl:
number = ''
plchld = ''
if str(column[0:3]).isdigit(): plchld = 3
elif str(column[0:2]).isdigit(): plchld = 2
else: number = plchld = 1
number = int(column[0:plchld])
if number == 98: twoNotThree = False;
new = pd.to_numeric(vsTbl[column], downcast='float')
old = pd.to_numeric(compare_table[number], downcast='float', errors='coerce')
info[str(number)+'_Error#_'] = old - new
diff[str(number)+'_Error#_'] = old - new
info[str(number)+'_Error%_'] = old / new
simi[str(number)+'_Error%_'] = old / new
info[str(number)+'_new_'+column[plchld:]] = vsTbl[column]
info[str(number)+'_old_'+columnsNames[number]] = compare_table[number]
#info.to_csv('./VSData/vs_comparisons_'+ str(year)+'.csv')
#diff.to_csv('./VSData/vs_differences_'+ str(year)+'.csv')
# Save the info dataframe
#info.to_excel(writer, str(year+'_ExpandedView') )
# Save the diff dataframe
#diff.to_excel(writer,sheet_name = str(year+'_Error') )
# Save the diff dataframe
#simi.to_excel(writer,sheet_name = str(year+'_Similarity_Ratio') )
info.to_csv('./VSData/vs'+str(year+'_ExpandedView')+'.csv')
diff.to_csv('./VSData/vs'+str(year+'_Error')+'.csv')
simi.to_csv('./VSData/vs'+str(year+'_Similarity_Ratio')+'.csv')
# writer.save()
```
#### Compare Historic Indicators
```
ls
# Quick test
shortName = str('hh25inc')
year = 19
# retrieve the Python ACS indicator
module = __import__( shortName )
result = getattr( module, shortName )( year )
result
# Delete Unassigned--Jail
df = df[df.index != 'Unassigned--Jail']
# Move Baltimore to Bottom
bc = df.loc[ 'Baltimore City' ]
df = df.drop( df.index[1] )
df.loc[ 'Baltimore City' ] = bc
vsTbl['18_fam']
```
|
github_jupyter
|
# Logarithmic Regularization: Dataset 1
```
# Import libraries and modules
import numpy as np
import pandas as pd
import xgboost as xgb
from xgboost import plot_tree
from sklearn.metrics import r2_score, classification_report, confusion_matrix, \
roc_curve, roc_auc_score, plot_confusion_matrix, f1_score, \
balanced_accuracy_score, accuracy_score, mean_squared_error, \
log_loss
from sklearn.datasets import make_friedman1
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.linear_model import LogisticRegression, LinearRegression, SGDClassifier, \
Lasso, lasso_path
from sklearn.preprocessing import StandardScaler, LabelBinarizer
from sklearn.impute import SimpleImputer
from sklearn.pipeline import Pipeline
from sklearn_pandas import DataFrameMapper
import scipy
from scipy import stats
import os
import shutil
from pathlib import Path
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import albumentations as A
from albumentations.pytorch import ToTensorV2
import cv2
import itertools
import time
import tqdm
import copy
import warnings
import torch
import torchvision
import torchvision.transforms as transforms
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import torchvision.models as models
from torch.utils.data import Dataset
import PIL
import joblib
import json
# import mysgd
# Import user-defined modules
import sys
import imp
sys.path.append('/Users/arbelogonzalezw/Documents/ML_WORK/LIBS/Lockdown')
import tools_general as tg
import tools_pytorch as tp
import lockdown as ld
imp.reload(tg)
imp.reload(tp)
imp.reload(ld)
```
## Read, clean, and save data
```
# Read X and y
X = pd.read_csv('/Users/arbelogonzalezw/Documents/ML_WORK/Project_Jerry_Lockdown/dataset_10LungCarcinoma/GDS3837_gene_profile.csv', index_col=0)
dfy = pd.read_csv('/Users/arbelogonzalezw/Documents/ML_WORK/Project_Jerry_Lockdown/dataset_10LungCarcinoma/GDS3837_output.csv', index_col=0)
# Change column names
cols = X.columns.tolist()
for i in range(len(cols)):
cols[i] = cols[i].lower()
cols[i] = cols[i].replace('-', '_')
cols[i] = cols[i].replace('.', '_')
cols[i] = cols[i].strip()
X.columns = cols
cols = dfy.columns.tolist()
for i in range(len(cols)):
cols[i] = cols[i].lower()
cols[i] = cols[i].replace('-', '_')
cols[i] = cols[i].replace('.', '_')
cols[i] = cols[i].strip()
dfy.columns = cols
# Set target
dfy['disease_state'] = dfy['disease_state'].str.replace(' ', '_')
dfy.replace({'disease_state': {"lung_cancer": 1, "control": 0}}, inplace=True)
Y = pd.DataFrame(dfy['disease_state'])
# Split and save data set
xtrain, xvalid, xtest, ytrain, yvalid, ytest = tg.split_data(X, Y)
tg.save_data(X, xtrain, xvalid, xtest, Y, ytrain, yvalid, ytest, 'dataset/')
tg.save_list(X.columns.to_list(), 'dataset/X.columns')
tg.save_list(Y.columns.to_list(), 'dataset/Y.columns')
#
print("- X size: {}\n".format(X.shape))
print("- xtrain size: {}".format(xtrain.shape))
print("- xvalid size: {}".format(xvalid.shape))
print("- xtest size: {}".format(xtest.shape))
```
## Load Data
```
# Select type of processor to be used
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
if device == torch.device('cuda'):
print("-Type of precessor to be used: 'gpu'")
!nvidia-smi
else:
print("-Type of precessor to be used: 'cpu'")
# Choose device
# torch.cuda.set_device(6)
# Read data
X, x_train, x_valid, x_test, Y, ytrain, yvalid, ytest = tp.load_data_clf('dataset/')
cols_X = tg.read_list('dataset/X.columns')
cols_Y = tg.read_list('dataset/Y.columns')
# Normalize data
xtrain, xvalid, xtest = tp.normalize_x(x_train, x_valid, x_test)
# Create dataloaders
dl_train, dl_valid, dl_test = tp.make_DataLoaders(xtrain, xvalid, xtest, ytrain, yvalid, ytest,
tp.dataset_tabular, batch_size=10000)
# NN architecture with its corresponding forward method
class MyNet(nn.Module):
# .Network architecture
def __init__(self, features, layer_sizes):
super(MyNet, self).__init__()
self.classifier = nn.Sequential(
nn.Linear(features, layer_sizes[0], bias=True),
nn.ReLU(inplace=True),
nn.Linear(layer_sizes[0], layer_sizes[1], bias=True)
)
# .Forward function
def forward(self, x):
x = self.classifier(x)
return x
```
## Lockout (Log, beta=0.7)
```
# TRAIN WITH LOCKDOWN
model = MyNet(n_features, n_layers)
model.load_state_dict(torch.load('./model_forward_valid_min.pth'))
model.eval()
regul_type = [('classifier.0.weight', 2), ('classifier.2.weight', 2)]
regul_path = [('classifier.0.weight', True), ('classifier.2.weight', False)]
lockout_s = ld.lockdown(model, lr=1e-2,
regul_type=regul_type,
regul_path=regul_path,
loss_type=2, tol_grads=1e-2)
lockout_s.train(dl_train, dl_valid, dl_test, epochs=5000, early_stop=15, tol_loss=1e-5, epochs2=100000,
train_how="decrease_t0")
# Save model, data
tp.save_model(lockout_s.model_best_valid, 'model_lockout_valid_min_log7_path.pth')
tp.save_model(lockout_s.model_last, 'model_lockout_last_log7_path.pth')
lockout_s.path_data.to_csv('data_lockout_log7_path.csv')
# Relevant plots
df = pd.read_csv('data_lockout_log7_path.csv')
df.plot('iteration', y=['t0_calc__classifier.0.weight', 't0_used__classifier.0.weight'],
figsize=(8,6))
plt.show()
# L1
nn = int(1e2)
data_tmp = pd.read_csv('data_lockout_l1.csv', index_col=0)
data_lockout_l1 = pd.DataFrame(columns=['sparcity', 'train_accu', 'valid_accu', 'test_accu', 't0_used'])
xgrid, step = np.linspace(0., 1., num=nn,endpoint=True, retstep=True)
for x in xgrid:
msk = (data_tmp['sparcity__classifier.0.weight'] >= x) & \
(data_tmp['sparcity__classifier.0.weight'] < x+step)
train_accu = data_tmp.loc[msk, 'train_accu'].mean()
valid_accu = data_tmp.loc[msk, 'valid_accu'].mean()
test_accu = data_tmp.loc[msk, 'test_accu'].mean()
t0_used = data_tmp.loc[msk, 't0_used__classifier.0.weight'].mean()
data_lockout_l1 = data_lockout_l1.append({'sparcity': x,
'train_accu': train_accu,
'valid_accu': valid_accu,
'test_accu': test_accu,
't0_used': t0_used}, ignore_index=True)
data_lockout_l1.dropna(axis='index', how='any', inplace=True)
# Log, beta=0.7
nn = int(1e2)
data_tmp = pd.read_csv('data_lockout_log7_path.csv', index_col=0)
data_lockout_log7 = pd.DataFrame(columns=['sparcity', 'train_accu', 'valid_accu', 'test_accu', 't0_used'])
xgrid, step = np.linspace(0., 1., num=nn,endpoint=True, retstep=True)
for x in xgrid:
msk = (data_tmp['sparcity__classifier.0.weight'] >= x) & \
(data_tmp['sparcity__classifier.0.weight'] < x+step)
train_accu = data_tmp.loc[msk, 'train_accu'].mean()
valid_accu = data_tmp.loc[msk, 'valid_accu'].mean()
test_accu = data_tmp.loc[msk, 'test_accu'].mean()
t0_used = data_tmp.loc[msk, 't0_used__classifier.0.weight'].mean()
data_lockout_log7 = data_lockout_log7.append({'sparcity': x,
'train_accu': train_accu,
'valid_accu': valid_accu,
'test_accu': test_accu,
't0_used': t0_used}, ignore_index=True)
data_lockout_log7.dropna(axis='index', how='any', inplace=True)
# Plot
fig, axes = plt.subplots(figsize=(9,6))
axes.plot(n_features*data_lockout_l1.loc[2:, 'sparcity'],
1.0 - data_lockout_l1.loc[2:, 'valid_accu'],
"-", linewidth=4, markersize=10, label="Lockout(L1)",
color="tab:orange")
axes.plot(n_features*data_lockout_log7.loc[3:,'sparcity'],
1.0 - data_lockout_log7.loc[3:, 'valid_accu'],
"-", linewidth=4, markersize=10, label=r"Lockout(Log, $\beta$=0.7)",
color="tab:green")
axes.grid(True, zorder=2)
axes.set_xlabel("number of selected features", fontsize=16)
axes.set_ylabel("Validation Error", fontsize=16)
axes.tick_params(axis='both', which='major', labelsize=14)
axes.set_yticks(np.linspace(5e-3, 4.5e-2, 5, endpoint=True))
# axes.ticklabel_format(axis='y', style='sci', scilimits=(0,0))
axes.set_xlim(0, 54800)
axes.legend(fontsize=16)
plt.tight_layout()
plt.savefig('error_vs_features_log_dataset10.pdf', bbox_inches='tight')
plt.show()
```
|
github_jupyter
|
<img src="logos/Icos_cp_Logo_RGB.svg" align="right" width="400"> <br clear="all" />
# Visualization of average footprints
For questions and feedback contact [email protected]
To use the tool, <span style="background-color: #FFFF00">run all the Notebook cells</span> (see image below).
<img src="network_characterization/screenshots_for_into_texts/how_to_run.PNG" align="left"> <br clear="all" />
#### STILT footprints
STILT is implemented as an <a href="https://www.icos-cp.eu/data-services/tools/stilt-footprint" target="blank">online tool</a> at the ICOS Carbon Portal. Output footprints are presented on a grid with 1/12×1/8 degrees cells (approximately 10km x 10km) where the cell values represent the cell area’s estimated surface influence (“sensitivity”) in ppm / (μmol/ (m²s)) on the atmospheric tracer concentration at the station. Individual footprints are generated every three hours (between 0:00 and 21:00 UTC) and are based on a 10-days backward simulation.
On the ICOS Carbon Portal JupyterHub there are Notebook tools that use STILT footprints such as <a href="https://exploredata.icos-cp.eu/user/jupyter/notebooks/icos_jupyter_notebooks/station_characterization.ipynb">station characterization</a> and <a href="https://exploredata.icos-cp.eu/user/jupyter/notebooks/icos_jupyter_notebooks/network_characterization.ipynb">network characterization</a>. In the station characterization tool there is a visualization method that aggregates the surface influence into bins in different directions and at different distance intervals of the station. Without aggregation of the surface influence it can be difficult to understand to what degree different regions fluxes can be expected to influence the concentration at the station. In this tool an additional method to aggregate and visualize the surface influence is presented.
Example of average footprint for Hylemossa (tall tower - 150 meters - atmospheric station located in Sweden) displayed using logarithmic scale:
<img src="network_characterization/screenshots_for_into_texts/hyltemossa_2018_log.PNG" align="left" width="400"> <br clear="all" />
#### Percent of the footprint sensitivity
This tool generates a map with decreasing intensity in color from 10 to 90% of the footprint sensitivity. This is achieved by including the sensitivity values of the footprint cells in descending order until the 10%, 20%, 30% etc. is reached. In terms of Carbon Portal tools, it is important for an understanding of results in the network characterization tool where the user decides what percent of the footprint should be used for the analysis: The 10 days’ backward simulation means that a footprint can have a very large extent. When averaging many footprints almost the entire STILT model domain (Europe) will have influenced the concentration at the station at some point in time. Deciding on a percent threshold limits the footprint coverage to areas with more significant influence. This tool shows how the user choice will influence the result.
Example using the 2018 average footprint for Hyltemossa:
<img src="network_characterization/screenshots_for_into_texts/hyltemossa_2018.PNG" align="left" width="400"> <br clear="all" />
```
%%javascript
IPython.OutputArea.prototype._should_scroll = function(lines) {
return false;
}
import sys
sys.path.append('./network_characterization')
import gui_percent_aggregate_footprints
```
|
github_jupyter
|
```
Copyright 2021 IBM Corporation
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
```
# Random Forest on Allstate Dataset
## Background
The goal of this competition is to predict Bodily Injury Liability Insurance claim payments based on the characteristics of the insured’s vehicle.
## Source
The raw dataset can be obtained directly from the [Allstate Claim Prediction Challenge](https://www.kaggle.com/c/ClaimPredictionChallenge).
In this example, we download the dataset directly from Kaggle using their API. In order for to work work, you must:
1. Login into Kaggle and accept the [competition rules](https://www.kaggle.com/c/ClaimPredictionChallenge/rules).
2. Folow [these instructions](https://www.kaggle.com/docs/api) to install your API token on your machine.
## Goal
The goal of this notebook is to illustrate how Snap ML can accelerate training of a random forest model on this dataset.
## Code
```
cd ../../
CACHE_DIR='cache-dir'
import numpy as np
import time
from datasets import Allstate
from sklearn.ensemble import RandomForestClassifier
from snapml import RandomForestClassifier as SnapRandomForestClassifier
from sklearn.metrics import roc_auc_score as score
dataset = Allstate(cache_dir=CACHE_DIR)
X_train, X_test, y_train, y_test = dataset.get_train_test_split()
print("Number of examples: %d" % (X_train.shape[0]))
print("Number of features: %d" % (X_train.shape[1]))
print("Number of classes: %d" % (len(np.unique(y_train))))
# the dataset is highly imbalanced
labels, sizes = np.unique(y_train, return_counts=True)
print("%6.2f %% of the training transactions belong to class 0" % (sizes[0]*100.0/(sizes[0]+sizes[1])))
print("%6.2f %% of the training transactions belong to class 1" % (sizes[1]*100.0/(sizes[0]+sizes[1])))
from sklearn.utils.class_weight import compute_sample_weight
w_train = compute_sample_weight('balanced', y_train)
w_test = compute_sample_weight('balanced', y_test)
model = RandomForestClassifier(max_depth=6, n_estimators=100, n_jobs=4, random_state=42)
t0 = time.time()
model.fit(X_train, y_train, sample_weight=w_train)
t_fit_sklearn = time.time()-t0
score_sklearn = score(y_test, model.predict_proba(X_test)[:,1], sample_weight=w_test)
print("Training time (sklearn): %6.2f seconds" % (t_fit_sklearn))
print("ROC AUC score (sklearn): %.4f" % (score_sklearn))
model = SnapRandomForestClassifier(max_depth=6, n_estimators=100, n_jobs=4, random_state=42, use_histograms=True)
t0 = time.time()
model.fit(X_train, y_train, sample_weight=w_train)
t_fit_snapml = time.time()-t0
score_snapml = score(y_test, model.predict_proba(X_test)[:,1], sample_weight=w_test)
print("Training time (snapml): %6.2f seconds" % (t_fit_snapml))
print("ROC AUC score (snapml): %.4f" % (score_snapml))
speed_up = t_fit_sklearn/t_fit_snapml
score_diff = (score_snapml-score_sklearn)/score_sklearn
print("Speed-up: %.1f x" % (speed_up))
print("Relative diff. in score: %.4f" % (score_diff))
```
## Disclaimer
Performance results always depend on the hardware and software environment.
Information regarding the environment that was used to run this notebook are provided below:
```
import utils
environment = utils.get_environment()
for k,v in environment.items():
print("%15s: %s" % (k, v))
```
## Record Statistics
Finally, we record the enviroment and performance statistics for analysis outside of this standalone notebook.
```
import scrapbook as sb
sb.glue("result", {
'dataset': dataset.name,
'n_examples_train': X_train.shape[0],
'n_examples_test': X_test.shape[0],
'n_features': X_train.shape[1],
'n_classes': len(np.unique(y_train)),
'model': type(model).__name__,
'score': score.__name__,
't_fit_sklearn': t_fit_sklearn,
'score_sklearn': score_sklearn,
't_fit_snapml': t_fit_snapml,
'score_snapml': score_snapml,
'score_diff': score_diff,
'speed_up': speed_up,
**environment,
})
```
|
github_jupyter
|
```
import pandas as pd
import numpy as np
import scanpy as sc
import os
from sklearn.cluster import KMeans
from sklearn.cluster import AgglomerativeClustering
from sklearn.metrics.cluster import adjusted_rand_score
from sklearn.metrics.cluster import adjusted_mutual_info_score
from sklearn.metrics.cluster import homogeneity_score
import rpy2.robjects as robjects
from rpy2.robjects import pandas2ri
df_metrics = pd.DataFrame(columns=['ARI_Louvain','ARI_kmeans','ARI_HC',
'AMI_Louvain','AMI_kmeans','AMI_HC',
'Homogeneity_Louvain','Homogeneity_kmeans','Homogeneity_HC'])
workdir = './output/'
path_fm = os.path.join(workdir,'feature_matrices/')
path_clusters = os.path.join(workdir,'clusters/')
path_metrics = os.path.join(workdir,'metrics/')
os.system('mkdir -p '+path_clusters)
os.system('mkdir -p '+path_metrics)
metadata = pd.read_csv('./input/metadata.tsv',sep='\t',index_col=0)
num_clusters = len(np.unique(metadata['label']))
print(num_clusters)
files = [x for x in os.listdir(path_fm) if x.startswith('FM')]
len(files)
files
def getNClusters(adata,n_cluster,range_min=0,range_max=3,max_steps=20):
this_step = 0
this_min = float(range_min)
this_max = float(range_max)
while this_step < max_steps:
print('step ' + str(this_step))
this_resolution = this_min + ((this_max-this_min)/2)
sc.tl.louvain(adata,resolution=this_resolution)
this_clusters = adata.obs['louvain'].nunique()
print('got ' + str(this_clusters) + ' at resolution ' + str(this_resolution))
if this_clusters > n_cluster:
this_max = this_resolution
elif this_clusters < n_cluster:
this_min = this_resolution
else:
return(this_resolution, adata)
this_step += 1
print('Cannot find the number of clusters')
print('Clustering solution from last iteration is used:' + str(this_clusters) + ' at resolution ' + str(this_resolution))
for file in files:
file_split = file.split('_')
method = file_split[1]
dataset = file_split[2].split('.')[0]
if(len(file_split)>3):
method = method + '_' + '_'.join(file_split[3:]).split('.')[0]
print(method)
pandas2ri.activate()
readRDS = robjects.r['readRDS']
df_rds = readRDS(os.path.join(path_fm,file))
fm_mat = pandas2ri.ri2py(robjects.r['data.frame'](robjects.r['as.matrix'](df_rds)))
fm_mat.fillna(0,inplace=True)
fm_mat.columns = metadata.index
adata = sc.AnnData(fm_mat.T)
adata.var_names_make_unique()
adata.obs = metadata.loc[adata.obs.index,]
df_metrics.loc[method,] = ""
#Louvain
sc.pp.neighbors(adata, n_neighbors=15,use_rep='X')
# sc.tl.louvain(adata)
getNClusters(adata,n_cluster=num_clusters)
#kmeans
kmeans = KMeans(n_clusters=num_clusters, random_state=2019).fit(adata.X)
adata.obs['kmeans'] = pd.Series(kmeans.labels_,index=adata.obs.index).astype('category')
#hierachical clustering
hc = AgglomerativeClustering(n_clusters=num_clusters).fit(adata.X)
adata.obs['hc'] = pd.Series(hc.labels_,index=adata.obs.index).astype('category')
#clustering metrics
#adjusted rank index
ari_louvain = adjusted_rand_score(adata.obs['label'], adata.obs['louvain'])
ari_kmeans = adjusted_rand_score(adata.obs['label'], adata.obs['kmeans'])
ari_hc = adjusted_rand_score(adata.obs['label'], adata.obs['hc'])
#adjusted mutual information
ami_louvain = adjusted_mutual_info_score(adata.obs['label'], adata.obs['louvain'],average_method='arithmetic')
ami_kmeans = adjusted_mutual_info_score(adata.obs['label'], adata.obs['kmeans'],average_method='arithmetic')
ami_hc = adjusted_mutual_info_score(adata.obs['label'], adata.obs['hc'],average_method='arithmetic')
#homogeneity
homo_louvain = homogeneity_score(adata.obs['label'], adata.obs['louvain'])
homo_kmeans = homogeneity_score(adata.obs['label'], adata.obs['kmeans'])
homo_hc = homogeneity_score(adata.obs['label'], adata.obs['hc'])
df_metrics.loc[method,['ARI_Louvain','ARI_kmeans','ARI_HC']] = [ari_louvain,ari_kmeans,ari_hc]
df_metrics.loc[method,['AMI_Louvain','AMI_kmeans','AMI_HC']] = [ami_louvain,ami_kmeans,ami_hc]
df_metrics.loc[method,['Homogeneity_Louvain','Homogeneity_kmeans','Homogeneity_HC']] = [homo_louvain,homo_kmeans,homo_hc]
adata.obs[['louvain','kmeans','hc']].to_csv(os.path.join(path_clusters ,method + '_clusters.tsv'),sep='\t')
df_metrics.to_csv(path_metrics+'clustering_scores.csv')
df_metrics
```
|
github_jupyter
|
# Analytic center computation using a infeasible start Newton method
# The set-up
```
import numpy as np
import pandas as pd
import accpm
import accpm
from IPython.display import display
%load_ext autoreload
%autoreload 1
%aimport accpm
```
$\DeclareMathOperator{\domain}{dom}
\newcommand{\transpose}{\text{T}}
\newcommand{\vec}[1]{\begin{pmatrix}#1\end{pmatrix}}$
# Theory
To test the $\texttt{analytic_center}$ function we consider the following example. Suppose we want to find the analytic center $x_{ac} \in \mathbb{R}^2$ of the inequalities $x_1 \leq c_1, x_1 \geq 0, x_2 \leq c_2, x_2 \geq 0$. This is a rectange with dimensions $c_1 \times c_2$ centered at at $(\frac{c_1}{2}, \frac{c_2}{2})$ so we should have $x_{ac} = (\frac{c_1}{2}, \frac{c_2}{2})$. Now, $x_{ac}$ is the solution of the minimization problem
\begin{equation*}
\min_{\domain \phi} \phi(x) = - \sum_{i=1}^{4}{\log{(b_i - a_i^\transpose x)}}
\end{equation*}
where
\begin{equation*}
\domain \phi = \{x \;|\; a_i^\transpose x < b_i, i = 1, 2, 3, 4\}
\end{equation*}
with
\begin{align*}
&a_1 = \begin{bmatrix}1\\0\end{bmatrix}, &&b_1 = c_1, \\
&a_2 = \begin{bmatrix}-1\\0\end{bmatrix}, &&b_2 = 0, \\
&a_3 = \begin{bmatrix}0\\1\end{bmatrix}, &&b_3 = c_2, \\
&a_4 = \begin{bmatrix}0\\-1\end{bmatrix}, &&b_4 = 0.
\end{align*}
So we solve
\begin{align*}
&\phantom{iff}\nabla \phi(x) = \sum_{i=1}^{4
} \frac{1}{b_i - a_i^\transpose x}a_i = 0 \\
&\iff \frac{1}{c_1-x_1}\begin{bmatrix}1\\0\end{bmatrix} + \frac{1}{x_1}\begin{bmatrix}-1\\0\end{bmatrix} + \frac{1}{c_2-x_2}\begin{bmatrix}0\\1\end{bmatrix} + \frac{1}{x_2}\begin{bmatrix}0\\-1\end{bmatrix} = 0 \\
&\iff \frac{1}{c_1-x_1} - \frac{1}{x_1} = 0, \frac{1}{c_2-x_2} - \frac{1}{x_2} = 0 \\
&\iff x_1 = \frac{c_1}{2}, x_2 = \frac{c_2}{2},
\end{align*}
as expected.
# Testing
We test $\texttt{analytic_center}$ for varying values of $c_1, c_2$ and algorithm parameters $\texttt{alpha, beta}$:
```
def get_results(A, test_input, alpha, beta, tol=10e-8):
expected = []
actual = []
result = []
for (c1, c2) in test_input:
b = np.array([c1, 0, c2, 0])
ac_expected = np.asarray((c1/2, c2/2))
ac_actual = accpm.analytic_center(A, b, alpha = alpha, beta = beta)
expected.append(ac_expected)
actual.append(ac_actual)
# if np.array_equal(ac_expected, ac_actual):
if np.linalg.norm(ac_expected - ac_actual) <= tol:
result.append(True)
else:
result.append(False)
results = pd.DataFrame([test_input, expected, actual, result])
results = results.transpose()
results.columns = ['test_input', 'expected', 'actual', 'result']
print('alpha =', alpha, 'beta =', beta)
display(results)
```
Here we have results for squares of varying sizes and for varying values of $\texttt{alpha}$ and $\texttt{beta}$. In general, the algorithm performs worse on large starting polyhedrons than small starting polyhedrons. This seems acceptable given that we are most concerned with smaller polyhedrons.
```
A = np.array([[1, 0],[-1,0],[0,1],[0,-1]])
test_input = [(1, 1), (5, 5), (20, 20), (10e2, 10e2), (10e4, 10e4),
(10e6, 10e6), (10e8, 10e8), (10e10, 10e10),
(0.5, 0.5), (0.1, 0.1), (0.01, 0.01),
(0.005, 0.005), (0.001, 0.001),(0.0005, 0.0005), (0.0001, 0.0001),
(0.00005, 0.00005), (0.00001, 0.00001), (0.00001, 0.00001)]
get_results(A, test_input, alpha=0.01, beta=0.7)
get_results(A, test_input, alpha=0.01, beta=0.99)
get_results(A, test_input, alpha=0.49, beta=0.7)
get_results(A, test_input, alpha=0.25, beta=0.7)
```
|
github_jupyter
|
```
import fluentpy as _
```
These are solutions for the Advent of Code puzzles of 2018 in the hopes that this might inspire the reader how to use the fluentpy api to solve problems.
See https://adventofcode.com/2018/ for the problems.
The goal of this is not to produce minimal code or neccessarily to be as clear as possible, but to showcase as many of the features of fluentpy. Pull requests to use more of fluentpy are welcome!
I do hope however that you find the solutions relatively succinct as your understanding of how fluentpy works grows.
# Day 1
https://adventofcode.com/2018/day/1
```
_(open('input/day1.txt')).read().replace('\n','').call(eval)._
day1_input = (
_(open('input/day1.txt'))
.readlines()
.imap(eval)
._
)
seen = set()
def havent_seen(number):
if number in seen:
return False
seen.add(number)
return True
(
_(day1_input)
.icycle()
.iaccumulate()
.idropwhile(havent_seen)
.get(0)
._
)
```
# Day 2
https://adventofcode.com/2018/day/2
```
day2 = open('input/day2.txt').readlines()
def has_two_or_three(code):
counts = _.lib.collections.Counter(code).values()
return 2 in counts, 3 in counts
twos, threes = _(day2).map(has_two_or_three).star_call(zip).to(tuple)
sum(twos) * sum(threes)
def is_different_by_only_one_char(codes):
# REFACT consider how to more effectively vectorize this function
# i.e. map ord, elementwise minus, count non zeroes == 1
code1, code2 = codes
diff_count = 0
for index, char in enumerate(code1):
if char != code2[index]:
diff_count += 1
return 1 == diff_count
(
_(day2)
.icombinations(r=2)
.ifilter(is_different_by_only_one_char)
.get(0)
.star_call(zip)
.filter(lambda pair: pair[0] == pair[1])
.star_call(zip)
.get(0)
.join('')
._
)
```
# Day 3
https://adventofcode.com/2018/day/3
```
line_regex = r'#(\d+) @ (\d+),(\d+): (\d+)x(\d+)'
class Entry(_.lib.collections.namedtuple('Entry', ['id', 'left', 'top', 'width', 'height'])._):
def coordinates(self):
return _.lib.itertools.product._(
range(claim.left, claim.left + claim.width),
range(claim.top, claim.top + claim.height)
)
def parse_day3(line):
return _(line).match(line_regex).groups().map(int).star_call(Entry)._
day3 = _(open('input/day3.txt')).read().splitlines().map(parse_day3)._
plane = dict()
for claim in day3:
for coordinate in claim.coordinates():
plane[coordinate] = plane.get(coordinate, 0) + 1
_(plane).values().filter(_.each != 1).len()._
for claim in day3:
if _(claim.coordinates()).imap(lambda each: plane[each] == 1).all()._:
print(claim.id)
```
# Day 4
https://adventofcode.com/2018/day/4
```
day4_lines = _(open('input/day4.txt')).read().splitlines().sort().self._
class Sleep(_.lib.collections.namedtuple('Sleep', ['duty_start', 'sleep_start', 'sleep_end'])._):
def minutes(self):
return (self.sleep_end - self.sleep_start).seconds // 60
class Guard:
def __init__(self, guard_id, sleeps=None):
self.id = guard_id
self.sleeps = sleeps or list()
def minutes_asleep(self):
return _(self.sleeps).map(_.each.minutes()._).sum()._
def minutes_and_sleep_counts(self):
distribution = dict()
for sleep in self.sleeps:
# problematic if the hour wraps, but it never does, see check below
for minute in range(sleep.sleep_start.minute, sleep.sleep_end.minute):
distribution[minute] = distribution.get(minute, 0) + 1
return _(distribution).items().sorted(key=_.each[1]._, reverse=True)._
def minute_most_asleep(self):
return _(self.minutes_and_sleep_counts()).get(0, tuple()).get(0, 0)._
def number_of_most_sleeps(self):
return _(self.minutes_and_sleep_counts()).get(0, tuple()).get(1, 0)._
guards = dict()
current_guard = current_duty_start = current_sleep_start = None
for line in day4_lines:
time = _.lib.datetime.datetime.fromisoformat(line[1:17])._
if 'Guard' in line:
guard_id = _(line[18:]).match(r'.*?(\d+).*?').group(1).call(int)._
current_guard = guards.setdefault(guard_id, Guard(guard_id))
current_duty_start = time
if 'falls asleep' in line:
current_sleep_start = time
if 'wakes up' in line:
current_guard.sleeps.append(Sleep(current_duty_start, current_sleep_start, time))
# confirm that we don't really have to do real date calculations but can just work with simplified values
for guard in guards.values():
for sleep in guard.sleeps:
assert sleep.sleep_start.minute < sleep.sleep_end.minute
assert sleep.sleep_start.hour == 0
assert sleep.sleep_end.hour == 0
guard = (
_(guards)
.values()
.sorted(key=Guard.minutes_asleep, reverse=True)
.get(0)
._
)
guard.id * guard.minute_most_asleep()
guard = (
_(guards)
.values()
.sorted(key=Guard.number_of_most_sleeps, reverse=True)
.get(0)
._
)
guard.id * guard.minute_most_asleep()
```
# Day 5
https://adventofcode.com/2018/day/5
```
day5 = _(open('input/day5.txt')).read().strip()._
def is_reacting(a_polymer, an_index):
if an_index+2 > len(a_polymer):
return False
first, second = a_polymer[an_index:an_index+2]
return first.swapcase() == second
def reduce(a_polymer):
for index in range(len(a_polymer) - 2, -1, -1):
if is_reacting(a_polymer, index):
a_polymer = a_polymer[:index] + a_polymer[index+2:]
return a_polymer
def fully_reduce(a_polymer):
last_polymer = current_polymer = a_polymer
while True:
last_polymer, current_polymer = current_polymer, reduce(current_polymer)
if last_polymer == current_polymer:
break
return current_polymer
len(fully_reduce(day5))
alphabet = _(range(26)).map(_.each + ord('a')).map(chr)._
shortest_length = float('inf')
for char in alphabet:
polymer = day5.replace(char, '').replace(char.swapcase(), '')
length = len(fully_reduce(polymer))
if length < shortest_length:
shortest_length = length
shortest_length
```
# Day 6
https://adventofcode.com/2018/day/6
```
Point = _.lib.collections.namedtuple('Point', ['x', 'y'])._
day6_coordinates = (
_(open('input/day6.txt'))
.read()
.splitlines()
.map(lambda each: _(each).split(', ').map(int).star_call(Point)._)
._
)
def manhatten_distance(first, second):
return abs(first.x - second.x) + abs(first.y - second.y)
def nearest_two_points_and_distances(a_point):
return (
_(day6_coordinates)
.imap(lambda each: (each, manhatten_distance(each, a_point)))
.sorted(key=_.each[1]._)
.slice(2)
._
)
def has_nearest_point(a_point):
(nearest_point, nearest_distance), (second_point, second_distance) \
= nearest_two_points_and_distances(a_point)
return nearest_distance < second_distance
def nearest_point(a_point):
return nearest_two_points_and_distances(a_point)[0][0]
def plane_extent():
all_x, all_y = _(day6_coordinates).imap(lambda each: (each.x, each.y)).star_call(zip).to(tuple)
min_x, min_y = min(all_x) - 1, min(all_y) - 1
max_x, max_y = max(all_x) + 2, max(all_y) + 2
return (
(min_x, min_y),
(max_x, max_y)
)
def compute_bounding_box():
(min_x, min_y), (max_x, max_y) = plane_extent()
return _.lib.itertools.chain(
(Point(x, min_y) for x in range(min_x, max_x)),
(Point(x, max_y) for x in range(min_x, max_x)),
(Point(min_x, y) for y in range(min_y, max_y)),
(Point(max_x, y) for y in range(min_y, max_y)),
).to(tuple)
bounding_box = compute_bounding_box()
def internal_points():
# no point on bounding box is nearest to it
external_points = _(bounding_box).map(nearest_point).to(set)
return set(day6_coordinates) - external_points
def points_by_number_of_nearest_points():
plane = dict()
(min_x, min_y), (max_x, max_y) = plane_extent()
for x in range(min_x, max_x):
for y in range(min_y, max_y):
point = Point(x,y)
if has_nearest_point(point):
plane[point] = nearest_point(point)
plane_points = _(plane).values().to(tuple)
counts = dict()
for point in internal_points():
counts[point] = plane_points.count(point)
return counts
points = points_by_number_of_nearest_points()
_(points).items().sorted(key=_.each[1]._, reverse=True).get(0)._
def total_distance(a_point):
return (
_(day6_coordinates)
.imap(lambda each: manhatten_distance(a_point, each))
.sum()
._
)
def number_of_points_with_total_distance_less(a_limit):
plane = dict()
(min_x, min_y), (max_x, max_y) = plane_extent()
for x in range(min_x, max_x):
for y in range(min_y, max_y):
point = Point(x,y)
plane[point] = total_distance(point)
return (
_(plane)
.values()
.ifilter(_.each < a_limit)
.len()
._
)
number_of_points_with_total_distance_less(10000)
```
# Day 7
https://adventofcode.com/2018/day/7
```
import fluentpy as _
day7_input = (
_(open('input/day7.txt'))
.read()
.findall(r'Step (\w) must be finished before step (\w) can begin.', flags=_.lib.re.M._)
._
)
def execute_in_order(dependencies):
prerequisites = dict()
_(dependencies).each(lambda each: prerequisites.setdefault(each[1], []).append(each[0]))
all_jobs = _(dependencies).flatten().call(set)._
ready_jobs = all_jobs - prerequisites.keys()
done_jobs = []
while 0 != len(ready_jobs):
current_knot = _(ready_jobs).sorted()[0]._
ready_jobs.discard(current_knot)
done_jobs.append(current_knot)
for knot in all_jobs.difference(done_jobs):
if set(done_jobs).issuperset(prerequisites.get(knot, [])):
ready_jobs.add(knot)
return _(done_jobs).join('')._
execute_in_order(day7_input)
def cached_property(cache_instance_variable_name):
def outer_wrapper(a_method):
@property
@_.lib.functools.wraps._(a_method)
def wrapper(self):
if not hasattr(self, cache_instance_variable_name):
setattr(self, cache_instance_variable_name, a_method(self))
return getattr(self, cache_instance_variable_name)
return wrapper
return outer_wrapper
class Jobs:
def __init__(self, dependencies, delays):
self.dependencies = dependencies
self.delays = delays
self._ready = self.all.difference(self.prerequisites.keys())
self._done = []
self._in_progress = set()
@cached_property('_prerequisites')
def prerequisites(self):
prerequisites = dict()
for prerequisite, job in self.dependencies:
prerequisites.setdefault(job, []).append(prerequisite)
return prerequisites
@cached_property('_all')
def all(self):
return _(self.dependencies).flatten().call(set)._
def can_start(self, a_job):
return set(self._done).issuperset(self.prerequisites.get(a_job, []))
def has_ready_jobs(self):
return 0 != len(self._ready)
def get_ready_job(self):
assert self.has_ready_jobs()
current_job = _(self._ready).sorted()[0]._
self._ready.remove(current_job)
self._in_progress.add(current_job)
return current_job, self.delays[current_job]
def set_job_done(self, a_job):
assert a_job in self._in_progress
self._done.append(a_job)
self._in_progress.remove(a_job)
for job in self.unstarted():
if self.can_start(job):
self._ready.add(job)
def unstarted(self):
return self.all.difference(self._in_progress.union(self._done))
def is_done(self):
return set(self._done) == self.all
def __repr__(self):
return f'<Jobs(in_progress={self._in_progress}, done={self._done})>'
@_.lib.dataclasses.dataclass._
class Worker:
id: int
delay: int
current_job: str
jobs: Jobs
def work_a_second(self):
self.delay -= 1
if self.delay <= 0:
self.finish_job_if_working()
self.accept_job_if_available()
def finish_job_if_working(self):
if self.current_job is None:
return
self.jobs.set_job_done(self.current_job)
self.current_job = None
def accept_job_if_available(self):
if not self.jobs.has_ready_jobs():
return
self.current_job, self.delay = self.jobs.get_ready_job()
def execute_in_parallel(dependencies, delays, number_of_workers):
jobs = Jobs(dependencies, delays)
workers = _(range(number_of_workers)).map(_(Worker).curry(
id=_,
delay=0, current_job=None, jobs=jobs,
)._)._
seconds = -1
while not jobs.is_done():
seconds += 1
_(workers).each(_.each.work_a_second()._)
return seconds
test_input = (('C', 'A'), ('C', 'F'), ('A', 'B'), ('A', 'D'), ('B', 'E'), ('D', 'E'), ('F', 'E'))
test_delays = _(range(1,27)).map(lambda each: (chr(ord('A') + each - 1), each)).call(dict)._
execute_in_parallel(test_input, test_delays, 2)
day7_delays = _(range(1,27)).map(lambda each: (chr(ord('A') + each - 1), 60 + each)).call(dict)._
assert 1107 == execute_in_parallel(day7_input, day7_delays, 5)
execute_in_parallel(day7_input, day7_delays, 5)
```
# Day 8
https://adventofcode.com/2018/day/8
```
import fluentpy as _
@_.lib.dataclasses.dataclass._
class Node:
children: tuple
metadata: tuple
@classmethod
def parse(cls, number_iterator):
child_count = next(number_iterator)
metadata_count = next(number_iterator)
return cls(
children=_(range(child_count)).map(lambda ignored: Node.parse(number_iterator))._,
metadata=_(range(metadata_count)).map(lambda ignored: next(number_iterator))._,
)
def sum_all_metadata(self):
return sum(self.metadata) + _(self.children).imap(_.each.sum_all_metadata()._).sum()._
def value(self):
if 0 == len(self.children):
return sum(self.metadata)
return (
_(self.metadata)
.imap(_.each - 1) # convert to indexes
.ifilter(_.each >= 0)
.ifilter(_.each < len(self.children))
.imap(self.children.__getitem__)
.imap(Node.value)
.sum()
._
)
test_input = (2,3,0,3,10,11,12,1,1,0,1,99,2,1,1,2)
test_node = Node.parse(iter(test_input))
assert 138 == test_node.sum_all_metadata()
assert 66 == test_node.value()
day8_input = _(open('input/day8.txt')).read().split(' ').map(int)._
node = Node.parse(iter(day8_input))
node.sum_all_metadata()
```
# Day 9
https://adventofcode.com/2018/day/9
```
class Marble:
def __init__(self, value):
self.value = value
self.prev = self.next = self
def insert_after(self, a_marble):
a_marble.next = self.next
a_marble.prev = self
a_marble.next.prev = a_marble.prev.next = a_marble
def remove(self):
self.prev.next = self.next
self.next.prev = self.prev
return self
class Circle:
def __init__(self):
self.current = None
def play_marble(self, marble):
if self.current is None:
self.current = marble
return 0 # normmal insert, no points, only happens once at the beginning
elif marble.value % 23 == 0:
removed = self.current.prev.prev.prev.prev.prev.prev.prev.remove()
self.current = removed.next
return marble.value + removed.value
else:
self.current.next.insert_after(marble)
self.current = marble
return 0 # normal insert, no points
def marble_game(player_count, marbles):
player_scores = [0] * player_count
circle = Circle()
for marble_value in range(marbles + 1):
player_scores[marble_value % player_count] += circle.play_marble(Marble(marble_value))
return max(player_scores)
assert 8317 == marble_game(player_count=10, marbles=1618)
assert 146373 == marble_game(player_count=13, marbles=7999)
assert 2764 == marble_game(player_count=17, marbles=1104)
assert 54718 == marble_game(player_count=21, marbles=6111)
assert 37305 == marble_game(player_count=30, marbles=5807)
marble_game(player_count=455, marbles=71223)
marble_game(player_count=455, marbles=71223*100)
```
# Day 10
https://adventofcode.com/2018/day/10
```
@_.lib.dataclasses.dataclass._
class Particle:
x: int
y: int
delta_x: int
delta_y: int
day10_input = (
_(open('input/day10.txt'))
.read()
.findall(r'position=<\s?(-?\d+),\s+(-?\d+)> velocity=<\s*(-?\d+),\s+(-?\d+)>')
.map(lambda each: _(each).map(int)._)
.call(list)
._
)
%matplotlib inline
def evolve(particles):
particles.x += particles.delta_x
particles.y += particles.delta_y
def devolve(particles):
particles.x -= particles.delta_x
particles.y -= particles.delta_y
def show(particles):
particles.y *= -1
particles.plot(x='x', y='y', kind='scatter', s=1)
particles.y *= -1
last_width = last_height = float('inf')
def particles_are_shrinking(particles):
global last_width, last_height
current_width = particles.x.max() - particles.x.min()
current_height = particles.y.max() - particles.y.min()
is_shrinking = current_width < last_width and current_height < last_height
last_width, last_height = current_width, current_height
return is_shrinking
particles = _.lib.pandas.DataFrame.from_records(
data=day10_input,
columns=['x', 'y', 'delta_x', 'delta_y']
)._
last_width = last_height = float('inf')
seconds = 0
while particles_are_shrinking(particles):
evolve(particles)
seconds += 1
devolve(particles)
show(particles)
seconds - 1
```
# Day 11
https://adventofcode.com/2018/day/11
```
import fluentpy as _
from pyexpect import expect
def power_level(x, y, grid_serial):
rack_id = x + 10
power_level = rack_id * y
power_level += grid_serial
power_level *= rack_id
power_level //= 100
power_level %= 10
return power_level - 5
assert 4 == power_level(x=3, y=5, grid_serial=8)
assert -5 == power_level( 122,79, grid_serial=57)
assert 0 == power_level(217,196, grid_serial=39)
assert 4 == power_level(101,153, grid_serial=71)
def power_levels(grid_serial):
return (
_(range(1, 301))
.product(repeat=2)
.star_map(_(power_level).curry(x=_, y=_, grid_serial=grid_serial)._)
.to(_.lib.numpy.array)
._
.reshape(300, -1)
.T
)
def compute_max_power(matrix, subset_size):
expect(matrix.shape[0]) == matrix.shape[1]
expect(subset_size) <= matrix.shape[0]
expect(subset_size) > 0
# +1 because 300 matrix by 300 subset should produce one value
width = matrix.shape[0] - subset_size + 1
height = matrix.shape[1] - subset_size + 1
output = _.lib.numpy.zeros((width, height))._
for x in range(width):
for y in range(height):
output[x,y] = matrix[y:y+subset_size, x:x+subset_size].sum()
return output
def coordinates_with_max_power(matrix, subset_size=3):
output = compute_max_power(matrix, subset_size=subset_size)
np = _.lib.numpy._
index = np.unravel_index(np.argmax(output), output.shape)
return (
_(index).map(_.each + 1)._, # turn back into coordinates
np.amax(output)
)
result = coordinates_with_max_power(power_levels(18))
assert ((33, 45), 29) == result, result
result = coordinates_with_max_power(power_levels(42))
assert ((21, 61), 30) == result, result
coordinates_with_max_power(power_levels(5034))
def find_best_subset(matrix):
best_max_power = best_subset_size = float('-inf')
best_coordinates = None
for subset_size in range(1, matrix.shape[0] + 1):
coordinates, max_power = coordinates_with_max_power(matrix, subset_size=subset_size)
if max_power > best_max_power:
best_max_power = max_power
best_subset_size = subset_size
best_coordinates = coordinates
return (
best_coordinates,
best_subset_size,
best_max_power,
)
result = coordinates_with_max_power(power_levels(18), subset_size=16)
expect(result) == ((90, 269), 113)
result = coordinates_with_max_power(power_levels(42), subset_size=12)
expect(result) == ((232, 251), 119)
result = find_best_subset(power_levels(18))
expect(result) == ((90, 269), 16, 113)
find_best_subset(power_levels(5034))
```
# Day 12
https://adventofcode.com/2018/day/12
```
import fluentpy as _
def parse(a_string):
is_flower = _.each == '#'
initial_state = (
_(a_string)
.match(r'initial state:\s*([#.]+)')
.group(1)
.map(is_flower)
.enumerate()
.call(dict)
._
)
patterns = dict(
_(a_string)
.findall('([#.]{5})\s=>\s([#.])')
.map(lambda each: (_(each[0]).map(is_flower)._, is_flower(each[1])))
._
)
return initial_state, patterns
def print_state(generation, state):
lowest_offset = min(state)
print(f'{generation:>5} {sum_state(state):>5} {lowest_offset:>5}: ', end='')
print(string_from_state(state))
def string_from_state(state):
lowest_offset, highest_offset = min(state), max(state)
return (
_(range(lowest_offset - 2, highest_offset + 3))
.map(lambda each: state.get(each, False))
.map(lambda each: each and '#' or '.')
.join()
._
)
def sum_state(state):
return (
_(state)
.items()
.map(lambda each: each[1] and each[0] or 0)
.sum()
._
)
def evolve(initial_state, patterns, number_of_generations,
show_sums=False, show_progress=False, show_state=False, stop_on_repetition=False):
current_state = dict(initial_state)
next_state = dict()
def surrounding_of(state, index):
return tuple(state.get(each, False) for each in range(index-2, index+3))
def compute_next_generation():
nonlocal current_state, next_state
first_key, last_key = min(current_state), max(current_state)
for index in range(first_key - 2, last_key + 2):
is_flower = patterns.get(surrounding_of(current_state, index), False)
if is_flower:
next_state[index] = is_flower
current_state, next_state = next_state, dict()
return current_state
seen = set()
for generation in range(number_of_generations):
if show_sums:
print(generation, sum_state(current_state))
if show_progress and generation % 1000 == 0: print('.', end='')
if show_state: print_state(generation, current_state)
if stop_on_repetition:
stringified = string_from_state(current_state)
if stringified in seen:
print(f'repetition on generation {generation}')
print(stringified)
return current_state
seen.add(stringified)
compute_next_generation()
return current_state
end_state = evolve(*parse("""initial state: #..#.#..##......###...###
...## => #
..#.. => #
.#... => #
.#.#. => #
.#.## => #
.##.. => #
.#### => #
#.#.# => #
#.### => #
##.#. => #
##.## => #
###.. => #
###.# => #
####. => #
"""), 20, show_state=True)
assert 325 == sum_state(end_state)
day12_input = open('input/day12.txt').read()
sum_state(evolve(*parse(day12_input), 20))
# still very much tooo slow
number_of_iterations = 50000000000
#number_of_iterations = 200
sum_state(evolve(*parse(day12_input), number_of_iterations, stop_on_repetition=True))
last_score = 11959
increment_per_generation = 11959 - 11873
last_generation = 135
generations_to_go = number_of_iterations - last_generation
end_score = last_score + generations_to_go * increment_per_generation
end_score
import fluentpy as _
# Numpy implementation for comparison
np = _.lib.numpy._
class State:
@classmethod
def parse_string(cls, a_string):
is_flower = lambda each: int(each == '#')
initial_state = (
_(a_string)
.match(r'initial state:\s*([#\.]+)')
.group(1)
.map(is_flower)
._
)
patterns = (
_(a_string)
.findall('([#.]{5})\s=>\s([#\.])')
.map(lambda each: (_(each[0]).map(is_flower)._, is_flower(each[1])))
._
)
return initial_state, patterns
@classmethod
def from_string(cls, a_string):
return cls(*cls.parse_string(a_string))
def __init__(self, initial_state, patterns):
self.type = np.uint8
self.patterns = self.trie_from_patterns(patterns)
self.state = np.zeros(len(initial_state) * 3, dtype=self.type)
self.zero = self.state.shape[0] // 2
self.state[self.zero:self.zero+len(initial_state)] = initial_state
def trie_from_patterns(self, patterns):
trie = np.zeros((2,) * 5, dtype=self.type)
for pattern, production in patterns:
trie[pattern] = production
return trie
@property
def size(self):
return self.state.shape[0]
def recenter_or_grow_if_neccessary(self):
# check how much empty space there is, and if re-centering the pattern might be good enough
if self.needs_resize() and self.is_region_empty(0, self.zero - self.size // 4):
self.move(- self.size // 4)
if self.needs_resize() and self.is_region_empty(self.zero + self.size // 4, -1):
self.move(self.size // 4)
if self.needs_resize():
self.grow()
def needs_resize(self):
return any(self.state[:4]) or any(self.state[-4:])
def is_region_empty(self, lower_limit, upper_limit):
return not any(self.state[lower_limit:upper_limit])
def move(self, move_by):
assert move_by != 0
new_state = np.zeros_like(self.state)
if move_by < 0:
new_state[:move_by] = self.state[-move_by:]
else:
new_state[move_by:] = self.state[:-move_by]
self.state = new_state
self.zero += move_by
def grow(self):
new_state = np.zeros(self.size * 2, dtype=self.type)
move_by = self.zero - (self.size // 2)
new_state[self.zero : self.zero + self.size] = self.state
self.state = new_state
self.zero -= move_by
def evolve_once(self):
self.state[2:-2] = self.patterns[
self.state[:-4],
self.state[1:-3],
self.state[2:-2],
self.state[3:-1],
self.state[4:]
]
self.recenter_or_grow_if_neccessary()
return self
def evolve(self, number_of_iterations, show_progress=False, show_state=False):
while number_of_iterations:
self.evolve_once()
number_of_iterations -= 1
if show_progress and number_of_iterations % 1000 == 0:
print('.', end='')
if show_state:
self.print()
return self
def __repr__(self):
return (
_(self.state)
.map(lambda each: each and '#' or '.')
.join()
._
)
def print(self):
print(f"{self.zero:>5} {self.sum():>5}", repr(self))
def sum(self):
return (
_(self.state)
.ienumerate()
.imap(lambda each: each[1] and (each[0] - self.zero) or 0)
.sum()
._
)
test = State.from_string("""initial state: #..#.#..##......###...###
...## => #
..#.. => #
.#... => #
.#.#. => #
.#.## => #
.##.. => #
.#### => #
#.#.# => #
#.### => #
##.#. => #
##.## => #
###.. => #
###.# => #
####. => #
""")
assert 325 == test.evolve(20, show_state=True).sum(), test.sum()
# Much faster initially, but then gets linearly slower as the size of the memory increases. And since it increases linearly with execution time
# Its still way too slow
day12_input = open('input/day12.txt').read()
state = State.from_string(day12_input)
#state.evolve(50000000000, show_progress=True)
#state.evolve(10000, show_progress=True).print()
```
# Day 13
https://adventofcode.com/2018/day/13
```
import fluentpy as _
from pyexpect import expect
Location = _.lib.collections.namedtuple('Location', ('x', 'y'))._
UP, RIGHT, DOWN, LEFT, STRAIGHT = '^>v<|'
UPDOWN, LEFTRIGHT, UPRIGHT, RIGHTDOWN, DOWNLEFT, LEFTUP = '|-\/\/'
MOVEMENT = {
'^' : Location(0, -1),
'>' : Location(1, 0),
'v' : Location(0, 1),
'<' : Location(-1, 0)
}
CURVE = {
'\\': { '^':'<', '<':'^', 'v':'>', '>':'v'},
'/': { '^':'>', '<':'v', 'v':'<', '>':'^'},
}
INTERSECTION = {
'^': { LEFT:'<', STRAIGHT:'^', RIGHT:'>' },
'>': { LEFT:'^', STRAIGHT:'>', RIGHT:'v' },
'v': { LEFT:'>', STRAIGHT:'v', RIGHT:'<' },
'<': { LEFT:'v', STRAIGHT:'<', RIGHT:'^' },
}
@_.lib.dataclasses.dataclass._
class Cart:
location: Location
orientation: str
world: str
program: iter = _.lib.dataclasses.field._(default_factory=lambda: _((LEFT, STRAIGHT, RIGHT)).icycle()._)
def tick(self):
move = MOVEMENT[self.orientation]
self.location = Location(self.location.x + move.x, self.location.y + move.y)
if self.world_at_current_location() in CURVE:
self.orientation = CURVE[self.world_at_current_location()][self.orientation]
if self.world_at_current_location() == '+':
self.orientation = INTERSECTION[self.orientation][next(self.program)]
return self
def world_at_current_location(self):
expect(self.location.y) < len(self.world)
expect(self.location.x) < len(self.world[self.location.y])
return self.world[self.location.y][self.location.x]
def __repr__(self):
return f'<Cart(location={self.location}, orientation={self.orientation})'
def parse_carts(world):
world = world.splitlines()
for line_number, line in enumerate(world):
for line_offset, character in _(line).enumerate():
if character in '<>^v':
yield Cart(location=Location(line_offset, line_number), orientation=character, world=world)
def crashed_carts(cart, carts):
carts = carts[:]
if cart not in carts:
return tuple() # crashed carts already removed
carts.remove(cart)
for first, second in _([cart]).icycle().zip(carts):
if first.location == second.location:
return first, second
def did_crash(cart, carts):
carts = carts[:]
if cart not in carts: # already removed because of crash
return True
carts.remove(cart)
for first, second in _([cart]).icycle().zip(carts):
if first.location == second.location:
return True
return False
def location_of_first_crash(input_string):
carts = list(parse_carts(input_string))
while True:
for cart in _(carts).sorted(key=_.each.location._)._:
cart.tick()
if did_crash(cart, carts):
return cart.location
def location_of_last_cart_after_crashes(input_string):
carts = list(parse_carts(input_string))
while True:
for cart in _(carts).sorted(key=_.each.location._)._:
cart.tick()
if did_crash(cart, carts):
_(crashed_carts(cart, carts)).each(carts.remove)
if 1 == len(carts):
return carts[0].location
expect(Cart(location=Location(0,0), orientation='>', world=['>-']).tick().location) == (1,0)
expect(Cart(location=Location(0,0), orientation='>', world=['>\\']).tick().location) == (1,0)
expect(Cart(location=Location(0,0), orientation='>', world=['>\\']).tick().orientation) == 'v'
expect(Cart(location=Location(0,0), orientation='>', world=['>+']).tick().orientation) == '^'
cart1, cart2 = parse_carts('>--<')
expect(cart1).has_attributes(location=(0,0), orientation='>')
expect(cart2).has_attributes(location=(3,0), orientation='<')
expect(location_of_first_crash('>--<')) == (2,0)
test_input = r"""/->-\
| | /----\
| /-+--+-\ |
| | | | v |
\-+-/ \-+--/
\------/
"""
expect(location_of_first_crash(test_input)) == (7,3)
day13_input = open('input/day13.txt').read()
location_of_first_crash(day13_input)
test_input = r"""/>-<\
| |
| /<+-\
| | | v
\>+</ |
| ^
\<->/
"""
expect(location_of_last_cart_after_crashes(test_input)) == (6,4)
location_of_last_cart_after_crashes(day13_input)
```
# Day 14
https://adventofcode.com/2018/day/14
```
import fluentpy as _
from pyexpect import expect
scores = bytearray([3,7])
elf1 = 0
elf2 = 1
def reset():
global scores, elf1, elf2
scores = bytearray([3,7])
elf1 = 0
elf2 = 1
def generation():
global scores, elf1, elf2
new_recipe = scores[elf1] + scores[elf2]
first_digit, second_digit = divmod(new_recipe, 10)
if first_digit: scores.append(first_digit)
scores.append(second_digit)
elf1 = (elf1 + 1 + scores[elf1]) % len(scores)
elf2 = (elf2 + 1 + scores[elf2]) % len(scores)
def next_10_after(how_many_generations):
reset()
while len(scores) < how_many_generations + 10:
generation()
return _(scores)[how_many_generations:how_many_generations+10].join()._
expect(next_10_after(9)) == '5158916779'
expect(next_10_after(5)) == '0124515891'
expect(next_10_after(18)) == '9251071085'
expect(next_10_after(2018)) == '5941429882'
day14_input = 894501
print(next_10_after(day14_input))
def generations_till_we_generate(a_number):
needle = _(a_number).str().map(int).call(bytearray)._
reset()
while needle not in scores[-len(needle) - 2:]: # at most two numbers get appended
generation()
return scores.rindex(needle)
expect(generations_till_we_generate('51589')) == 9
expect(generations_till_we_generate('01245')) == 5
expect(generations_till_we_generate('92510')) == 18
expect(generations_till_we_generate('59414')) == 2018
print(generations_till_we_generate(day14_input))
```
# Day 15
https://adventofcode.com/2018/day/15
```
expect = _.lib.pyexpect.expect._
np = _.lib.numpy._
inf = _.lib.math.inf._
dataclasses = _.lib.dataclasses._
def tuplify(a_function):
@_.lib.functools.wraps._(a_function)
def wrapper(*args, **kwargs):
return tuple(a_function(*args, **kwargs))
return wrapper
Location = _.lib.collections.namedtuple('Location', ['x', 'y'])._
NO_LOCATION = Location(-1,-1)
@dataclasses.dataclass
class Player:
type: chr
location: Location
level: 'Level' = dataclasses.field(repr=False)
hitpoints: int = 200
attack_power: int = dataclasses.field(default=3, repr=False)
distances: np.ndarray = dataclasses.field(default=None, repr=False)
predecessors: np.ndarray = dataclasses.field(default=None, repr=False)
def turn(self):
self.distances = self.predecessors = None
if self.hitpoints <= 0:
return # we're dead
if not self.is_in_range_of_enemies():
self.move_towards_enemy()
if self.is_in_range_of_enemies():
self.attack_weakest_enemy_in_range()
return self
def is_in_range_of_enemies(self):
adjacent_values = (
_(self).level.adjacent_locations(self.location)
.map(self.level.level.__getitem__)
._
)
return self.enemy_class() in adjacent_values
def enemy_class(self):
if self.type == 'E':
return 'G'
return 'E'
def move_towards_enemy(self):
targets = (
_(self).level.enemies_of(self)
.map(_.each.attack_positions()._).flatten(level=1)
.filter(self.is_location_reachable)
.sorted(key=self.distance_to_location)
.groupby(key=self.distance_to_location)
.get(0, []).get(1, [])
._
)
target = _(targets).sorted().get(0, None)._
if target is None:
return # no targets in range
self.move_to(self.one_step_towards(target))
def move_to(self, new_location):
self.level.level[self.location] = '.'
self.level.level[new_location] = self.type
self.location = new_location
self.distances = self.predecessors = None
def attack_positions(self):
return self.level.reachable_adjacent_locations(self.location)
def is_location_reachable(self, location):
self.ensure_distances()
return inf != self.distances[location]
def distance_to_location(self, location):
self.ensure_distances()
return self.distances[location]
def one_step_towards(self, location):
self.ensure_distances()
if 2 != len(self.predecessors[location]):
breakpoint()
while Location(*self.predecessors[location]) != self.location:
location = Location(*self.predecessors[location])
return location
def ensure_distances(self):
if self.distances is not None:
return
self.distances, self.predecessors = self.level.shortest_distances_from(self.location)
def attack_weakest_enemy_in_range(self):
adjacent_locations = _(self).level.adjacent_locations(self.location)._
target = (
_(self).level.enemies_of(self)
.filter(_.each.location.in_(adjacent_locations)._)
.sorted(key=_.each.hitpoints._)
.groupby(key=_.each.hitpoints._)
.get(0).get(1)
.sorted(key=_.each.location._)
.get(0)
._
)
target.damage_by(self.attack_power)
def damage_by(self, ammount):
self.hitpoints -= ammount
# REFACT this should happen on the level object
if self.hitpoints <= 0:
self.level.players = _(self).level.players.filter(_.each != self)._
self.level.level[self.location] = '.'
class Level:
def __init__(self, level_description):
self.level = np.array(_(level_description).strip().split('\n').map(tuple)._)
self.players = self.parse_players()
self.number_of_full_rounds = 0
@tuplify
def parse_players(self):
for row_number, row in enumerate(self.level):
for col_number, char in enumerate(row):
if char in 'GE':
yield Player(char, location=Location(row_number,col_number), level=self)
def enemies_of(self, player):
return _(self).players.filter(_.each.type != player.type)._
def adjacent_locations(self, location):
return (
_([
(location.x-1, location.y),
(location.x, location.y-1),
(location.x, location.y+1),
(location.x+1, location.y),
])
.star_map(Location)
._
)
def reachable_adjacent_locations(self, location):
return (
_(self).adjacent_locations(location)
.filter(self.is_location_in_level)
.filter(self.is_traversible)
._
)
def is_location_in_level(self, location):
x_size, y_size = self.level.shape
return 0 <= location.x < x_size \
and 0 <= location.y < y_size
def is_traversible(self, location):
return '.' == self.level[location]
def shortest_distances_from(self, location):
distances = np.full(fill_value=_.lib.math.inf._, shape=self.level.shape, dtype=float)
distances[location] = 0
predecessors = np.full(fill_value=NO_LOCATION, shape=self.level.shape, dtype=(int, 2))
next_locations = _.lib.collections.deque._([location])
while len(next_locations) > 0:
current_location = next_locations.popleft()
for location in self.reachable_adjacent_locations(current_location):
if distances[location] <= (distances[current_location] + 1):
continue
distances[location] = distances[current_location] + 1
predecessors[location] = current_location
next_locations.append(location)
return distances, predecessors
def __repr__(self):
return '\n'.join(''.join(line) for line in self.level)
def print(self):
print(
repr(self)
+ f'\nrounds: {self.number_of_full_rounds}'
+ '\n' + _(self).players_in_reading_order().join('\n')._
)
def round(self):
for player in self.players_in_reading_order():
if self.did_battle_end():
return self
player.turn()
self.number_of_full_rounds += 1
return self
def players_in_reading_order(self):
return _(self).players.sorted(key=_.each.location._)._
def run_battle(self):
while not self.did_battle_end():
self.round()
return self
def run_rounds(self, number_of_full_rounds):
for ignored in range(number_of_full_rounds):
self.round()
if self.did_battle_end():
break
return self
def did_battle_end(self):
return _(self).players.map(_.each.type._).call(set).len()._ == 1
def battle_summary(self):
number_of_remaining_hitpoints = _(self).players.map(_.each.hitpoints._).sum()._
return self.number_of_full_rounds * number_of_remaining_hitpoints
level = Level("""\
#######
#.G.E.#
#E....#
#######
""")
expect(level.players) == (Player('G',Location(1,2), level), Player('E', Location(1, 4), level), Player('E', Location(2,1), level))
expect(level.enemies_of(level.players[0])) == (Player('E', Location(x=1, y=4), level), Player('E', Location(x=2, y=1), level))
level.players[0].damage_by(200)
expect(level.players) == (Player('E', Location(1, 4), level), Player('E', Location(2,1), level))
expect(repr(level)) == '''\
#######
#...E.#
#E....#
#######'''
inf = _.lib.math.inf._
NO = [-1,-1]
distances, parents = Level('''
###
#G#
#.#
#E#
###
''').shortest_distances_from(Location(1,1))
expect(distances.tolist()) == [
[inf, inf, inf],
[inf, 0, inf],
[inf, 1, inf],
[inf, inf, inf],
[inf, inf, inf],
]
expect(parents.tolist()) == [
[NO, NO, NO],
[NO, NO, NO],
[NO, [1,1], NO],
[NO, NO, NO],
[NO, NO, NO],
]
distances, parents = Level('''
#######
#E..G.#
#...#.#
#.G.#G#
#######
''').shortest_distances_from(Location(1,1))
expect(distances.tolist()) == [
[inf, inf, inf, inf, inf, inf, inf],
[inf, 0, 1, 2, inf, inf, inf],
[inf, 1, 2, 3, inf, inf, inf],
[inf, 2, inf, 4, inf, inf, inf],
[inf, inf, inf, inf, inf, inf, inf]
]
expect(parents.tolist()) == [
[NO, NO, NO, NO, NO, NO, NO],
[NO, NO, [1, 1], [1, 2], NO, NO, NO],
[NO, [1, 1], [1, 2], [1, 3], NO, NO, NO],
[NO, [2, 1], NO, [2, 3], NO, NO, NO],
[NO, NO, NO, NO, NO, NO, NO]
]
distances, parents = Level('''
#######
#E..G.#
#.#...#
#.G.#G#
#######
''').shortest_distances_from(Location(1,1))
expect(distances[1:-1, 1:-1].tolist()) == [
[0,1,2,inf,6],
[1,inf,3,4,5],
[2,inf,4,inf,inf]
]
level = Level("""\
#######
#.G.E.#
#E....#
#######
""")
expect(level.players[0].location) == Location(1,2)
expect(level.players[0].turn().location) == Location(1,1)
expect(level.players[0].is_in_range_of_enemies()).is_true()
level = Level('''\
#######
#..G..#
#...EG#
#.#G#G#
#...#E#
#.....#
#######''')
expect(level.players[0].is_in_range_of_enemies()).is_false()
level = Level('''\
#########
#G..G..G#
#.......#
#.......#
#G..E..G#
#.......#
#.......#
#G..G..G#
#########''')
expect(level.round().__repr__()) == '''\
#########
#.G...G.#
#...G...#
#...E..G#
#.G.....#
#.......#
#G..G..G#
#.......#
#########'''
expect(level.round().__repr__()) == '''\
#########
#..G.G..#
#...G...#
#.G.E.G.#
#.......#
#G..G..G#
#.......#
#.......#
#########'''
expect(level.round().__repr__()) == '''\
#########
#.......#
#..GGG..#
#..GEG..#
#G..G...#
#......G#
#.......#
#.......#
#########'''
level = Level('''\
#######
#.G...#
#...EG#
#.#.#G#
#..G#E#
#.....#
#######''')
expect(level.round().__repr__()) == '''\
#######
#..G..#
#...EG#
#.#G#G#
#...#E#
#.....#
#######'''
expect(level.players[0]).has_attributes(
hitpoints=200, location=Location(1,3)
)
expect(level.round().__repr__()) == '''\
#######
#...G.#
#..GEG#
#.#.#G#
#...#E#
#.....#
#######'''
level.run_rounds(21)
expect(level.number_of_full_rounds) == 23
expect(repr(level)) == '''\
#######
#...G.#
#..G.G#
#.#.#G#
#...#E#
#.....#
#######'''
expect(level.players).has_len(5)
level.run_rounds(47-23)
expect(level.number_of_full_rounds) == 47
expect(repr(level)) == '''\
#######
#G....#
#.G...#
#.#.#G#
#...#.#
#....G#
#######'''
expect(_(level).players.map(_.each.type._).join()._) == 'GGGG'
expect(level.battle_summary()) == 27730
expect(level.did_battle_end()).is_true()
level = Level('''\
#######
#.G...#
#...EG#
#.#.#G#
#..G#E#
#.....#
#######''')
level.run_battle()
expect(level.battle_summary()) == 27730
level = Level('''\
#######
#G..#E#
#E#E.E#
#G.##.#
#...#E#
#...E.#
#######''').run_battle()
expect(repr(level)) == '''\
#######
#...#E#
#E#...#
#.E##.#
#E..#E#
#.....#
#######'''
expect(level.number_of_full_rounds) == 37
expect(level.battle_summary()) == 36334
level = Level('''\
#######
#E..EG#
#.#G.E#
#E.##E#
#G..#.#
#..E#.#
#######''').run_battle()
expect(level.battle_summary()) == 39514
_('input/day15.txt').call(open).read().call(Level).run_battle().battle_summary()._
def number_of_losses_with_attack_power(level_ascii, attack_power):
level = Level(level_ascii)
elves = lambda: _(level).players.filter(_.each.type == 'E')._
staring_number_of_elves = len(elves())
for elf in elves():
elf.attack_power = attack_power
level.run_battle()
return staring_number_of_elves - len(elves()), level.battle_summary()
def minimum_attack_power_for_no_losses(level_ascii):
for attack_power in range(4, 100):
number_of_losses, summary = number_of_losses_with_attack_power(level_ascii, attack_power)
if 0 == number_of_losses:
return attack_power, summary
expect(minimum_attack_power_for_no_losses('''\
#######
#.G...#
#...EG#
#.#.#G#
#..G#E#
#.....#
#######''')) == (15, 4988)
expect(minimum_attack_power_for_no_losses('''\
#######
#E..EG#
#.#G.E#
#E.##E#
#G..#.#
#..E#.#
#######''')) == (4, 31284)
expect(minimum_attack_power_for_no_losses('''\
#######
#E.G#.#
#.#G..#
#G.#.G#
#G..#.#
#...E.#
#######''')) == (15, 3478)
expect(minimum_attack_power_for_no_losses('''\
#######
#.E...#
#.#..G#
#.###.#
#E#G#G#
#...#G#
#######''')) == (12, 6474)
expect(minimum_attack_power_for_no_losses('''\
#########
#G......#
#.E.#...#
#..##..G#
#...##..#
#...#...#
#.G...G.#
#.....G.#
#########''')) == (34, 1140)
_('input/day15.txt').call(open).read().call(minimum_attack_power_for_no_losses)._
```
# Day 16
https://adventofcode.com/2018/day/16
## Registers
- four registers 0,1,2,3
- initialized to 0
## instructions
- 16 opcodes
- 1 opcode, 2 source registers (A, B), 1 output register (C)
- inputs can be register addresses, immediate values,
- output is always register
only have the opcode numbers, need to check validity
```
import fluentpy as _
expect = _.lib.pyexpect.expect._
operator = _.lib.operator._
def identity(*args):
return args[0]
def register(self, address):
return self.registers[address]
def immediate(self, value):
return value
def ignored(self, value):
return None
def make_operation(namespace, name, operation, a_resolver, b_resolver):
def instruction(self, a, b, c):
self.registers[c] = operation(a_resolver(self, a), b_resolver(self, b))
return self
instruction.__name__ = instruction.__qualname__ = name
namespace[name] = instruction
return instruction
class CPU:
def __init__(self, initial_registers=(0,0,0,0)):
self.registers = list(initial_registers)
operations = (
_([
('addr', operator.add, register, register),
('addi', operator.add, register, immediate),
('mulr', operator.mul, register, register),
('muli', operator.mul, register, immediate),
('banr', operator.and_, register, register),
('bani', operator.and_, register, immediate),
('borr', operator.or_, register, register),
('bori', operator.or_, register, immediate),
('setr', identity, register, ignored),
('seti', identity, immediate, ignored),
('gtir', operator.gt, immediate, register),
('gtri', operator.gt, register, immediate),
('gtrr', operator.gt, register, register),
('eqir', operator.eq, immediate, register),
('eqri', operator.eq, register, immediate),
('eqrr', operator.eq, register, register),
])
.star_map(_(make_operation).curry(locals())._)
._
)
def evaluate_program(self, instructions, opcode_map):
for instruction in instructions:
opcode, a,b,c = instruction
operation = opcode_map[opcode]
operation(self, a,b,c)
return self
@classmethod
def number_of_qualifying_instructions(cls, input_registers, instruction, expected_output_registers):
return len(cls.qualifying_instructions(input_registers, instruction, expected_output_registers))
@classmethod
def qualifying_instructions(cls, input_registers, instruction, expected_output_registers):
opcode, a, b, c = instruction
return (
_(cls)
.operations
.filter(lambda operation: operation(CPU(input_registers), a,b,c).registers == expected_output_registers)
._
)
expect(CPU([3, 2, 1, 1]).mulr(2, 1, 2).registers) == [3, 2, 2, 1]
expect(CPU.number_of_qualifying_instructions([3, 2, 1, 1], (9, 2, 1, 2), [3, 2, 2, 1])) == 3
day16_input = _(open('input/day16.txt')).read()._
test_input, test_program_input = day16_input.split('\n\n\n')
def parse_inputs(before, instruction, after):
return (
_(before).split(', ').map(int).to(list),
_(instruction).split(' ').map(int)._,
_(after).split(', ').map(int).to(list),
)
test_inputs = (
_(test_input)
.findall(r'Before: \[(.*)]\n(.*)\nAfter: \[(.*)\]')
.star_map(parse_inputs)
._
)
(
_(test_inputs)
.star_map(CPU.number_of_qualifying_instructions)
.filter(_.each >= 3)
.len()
._
)
def add_operations(mapping, opcode_and_operations):
opcode, operations = opcode_and_operations
mapping[opcode].append(operations)
return mapping
opcode_mapping = (
_(test_inputs)
.map(_.each[1][0]._) # opcodes
.zip(
_(test_inputs).star_map(CPU.qualifying_instructions)._
)
# list[tuple[opcode, list[list[functions]]]]
.reduce(add_operations, _.lib.collections.defaultdict(list)._)
# dict[opcode, list[list[function]]]
.items()
.star_map(lambda opcode, operations: (
opcode,
_(operations).map(set).reduce(set.intersection)._
))
.to(dict)
# dict[opcode, set[functions]]
)
def resolved_operations():
return (
_(opcode_mapping)
.values()
.filter(lambda each: len(each) == 1)
.reduce(set.union)
._
)
def has_unresolved_operations():
return 0 != (
_(opcode_mapping)
.values()
.map(len)
.filter(_.each > 1)
.len()
._
)
while has_unresolved_operations():
for opcode, matching_operations in opcode_mapping.items():
if len(matching_operations) == 1:
continue # already resolved
opcode_mapping[opcode] = matching_operations.difference(resolved_operations())
opcode_mapping = _(opcode_mapping).items().star_map(lambda opcode, operations: (opcode, list(operations)[0])).to(dict)
# dict[opcode, function]
opcode_mapping
test_program = _(test_program_input).strip().split('\n').map(lambda each: _(each).split(' ').map(int)._)._
CPU().evaluate_program(test_program, opcode_mapping).registers[0]
```
# Day 17
https://adventofcode.com/2018/day/17
```
import fluentpy as _
@_.lib.dataclasses.dataclass._
class ClayLine:
x_from: int
x_to: int
y_from: int
y_to: int
@classmethod
def from_string(cls, a_string):
first_var, first_value, second_var, second_value_start, second_value_end = \
_(a_string).fullmatch(r'(\w)=(\d+), (\w)=(\d+)..(\d+)').groups()._
first_value, second_value_start, second_value_end = _((first_value, second_value_start, second_value_end)).map(int)._
if 'x' == first_var:
return cls(first_value, first_value, second_value_start, second_value_end)
else:
return cls(second_value_start, second_value_end, first_value, first_value)
@property
def x_range(self):
return range(self.x_from, self.x_to + 1) # last coordinate is included
@property
def y_range(self):
return range(self.y_from, self.y_to + 1) # last coordinate is included
class Underground:
def __init__(self):
self.earth = dict()
self.earth[(500, 0)] = '+' # spring
self.min_x = self.max_x = 500
self.max_y = - _.lib.math.inf._
self.min_y = _.lib.math.inf._
def add_clay_line(self, clay_line):
for x in clay_line.x_range:
for y in clay_line.y_range:
self.set_earth(x,y, '#', should_adapt_depth=True)
return self
def set_earth(self, x,y, to_what, should_adapt_depth=False):
self.earth[(x,y)] = to_what
# whatever is set will expand the looked at area
if x > self.max_x:
self.max_x = x
if x < self.min_x:
self.min_x = x
if should_adapt_depth:
# only clay setting will expand y (depth)
if y > self.max_y:
self.max_y = y
if y < self.min_y:
self.min_y = y
def flood_fill_down_from_spring(self):
return self.flood_fill_down(500,1)
def flood_fill_down(self, x,y):
while self.can_flow_down(x,y):
if y > self.max_y:
return self
if '|' == self.earth.get((x,y), '.'):
# we've already been here
return self
self.set_earth(x,y, '|')
y += 1
while self.is_contained(x,y):
self.fill_container_level(x,y)
y -=1
self.mark_flowing_water_around(x,y)
for overflow_x in self.find_overflows(x, y):
self.flood_fill_down(overflow_x,y+1)
return self
def fill_container_level(self, x,y):
leftmost_free, rightmost_free = self.find_furthest_away_free_spots(x,y)
for mark_x in range(leftmost_free, rightmost_free+1):
self.set_earth(mark_x,y, '~')
def find_overflows(self, x,y):
leftmost_flow_border, rightmost_flow_border = self.find_flow_borders(x,y)
if self.can_flow_down(leftmost_flow_border, y):
yield leftmost_flow_border
if self.can_flow_down(rightmost_flow_border, y):
yield rightmost_flow_border
def is_blocked(self, x,y):
return self.earth.get((x,y), '.') in '#~'
def can_flow_down(self, x,y):
return not self.is_blocked(x, y+1)
def can_flow_left(self, x,y):
return not self.is_blocked(x-1, y)
def can_flow_right(self, x,y):
return not self.is_blocked(x+1, y)
def x_coordinates_towards(self, x, target_x):
if target_x < x:
return range(x, target_x-2, -1)
else:
return range(x, target_x+2)
def coordinates_towards(self, x,y, target_x):
return _(self.x_coordinates_towards(x, target_x)).map(lambda x: (x, y))._
def first_coordinate_that_satisfies(self, coordinates, a_test):
for x, y in coordinates:
if a_test(x,y):
return x
return None
def is_contained(self, x,y):
leftmost_flow_border, rightmost_flow_border = self.find_flow_borders(x,y)
if leftmost_flow_border is None or rightmost_flow_border is None:
return False
return not self.can_flow_down(leftmost_flow_border,y) and not self.can_flow_down(rightmost_flow_border,y)
def find_furthest_away_free_spots(self, x,y):
blocked_right = self.first_coordinate_that_satisfies(
self.coordinates_towards(x, y, self.max_x),
lambda x,y: not self.can_flow_right(x,y)
)
blocked_left = self.first_coordinate_that_satisfies(
self.coordinates_towards(x, y, self.min_x),
lambda x,y: not self.can_flow_left(x,y)
)
return (blocked_left, blocked_right)
def mark_flowing_water_around(self, x,y):
leftmost_free_spot, rightmost_free_spot = self.find_flow_borders(x,y)
for mark_x in range(leftmost_free_spot, rightmost_free_spot+1):
self.set_earth(mark_x, y, '|')
def find_flow_borders(self, x, y):
# REFACT there should be a fluent utility for this? no?
flow_border_right = self.first_coordinate_that_satisfies(
self.coordinates_towards(x,y, self.max_x),
lambda x,y: self.can_flow_down(x,y) or not self.can_flow_right(x,y)
)
flow_border_left = self.first_coordinate_that_satisfies(
self.coordinates_towards(x, y, self.min_x),
lambda x,y: self.can_flow_down(x,y) or not self.can_flow_left(x,y)
)
return (flow_border_left, flow_border_right)
def __str__(self):
return (
_(range(0, self.max_y+1))
.map(lambda y: (
_(range(self.min_x, self.max_x+1))
.map(lambda x: self.earth.get((x,y), '.'))
.join()
._
))
.join('\n')
._
)
def visualize(self):
print('min_x', self.min_x, 'max_x', self.max_x, 'min_y', self.min_y, 'max_y', self.max_y)
print(str(self))
return self
def number_of_water_reachable_tiles(self):
return (
_(self).earth.keys()
.filter(lambda coordinates: self.min_y <= coordinates[1] <= self.max_y)
.map(self.earth.get)
.filter(_.each.in_('~|')._)
.len()
._
)
def number_of_tiles_with_standing_water(self):
return (
_(self).earth.keys()
.filter(lambda coordinates: self.min_y <= coordinates[1] <= self.max_y)
.map(self.earth.get)
.filter(_.each.in_('~')._)
.len()
._
)
test_input = '''\
x=495, y=2..7
y=7, x=495..501
x=501, y=3..7
x=498, y=2..4
x=506, y=1..2
x=498, y=10..13
x=504, y=10..13
y=13, x=498..504'''
underground = _(test_input).splitlines().map(ClayLine.from_string).reduce(Underground.add_clay_line, Underground()).visualize()._
underground.flood_fill_down_from_spring().visualize()
underground.number_of_water_reachable_tiles()
underground = _(open('input/day17.txt')).read().splitlines().map(ClayLine.from_string).reduce(Underground.add_clay_line, Underground())._
underground.flood_fill_down_from_spring()
from IPython.display import display, HTML
display(HTML(f'<pre style="font-size:6px">{underground}</pre>'))
underground.number_of_water_reachable_tiles()
underground.number_of_tiles_with_standing_water()
```
# Day 18
https://adventofcode.com/2018/day/18
```
import fluentpy as _
from pyexpect import expect
class Area:
OPEN = '.'
TREES = '|'
LUMBERYARD = '#'
def __init__(self, area_description):
self.area = _(area_description).strip().splitlines().to(tuple)
self.generation = 0
self.cache = dict()
def evolve_to_generation(self, target_generation):
remaining_generations = target_generation - self.generation # so we can restart
while remaining_generations > 0:
if self.area in self.cache:
# looping pattern detected
last_identical_generation = self.cache[self.area]
generation_difference = self.generation - last_identical_generation
number_of_possible_jumps = remaining_generations // generation_difference
if number_of_possible_jumps > 0:
remaining_generations -= generation_difference * number_of_possible_jumps
continue # jump forward
self.cache[self.area] = self.generation
self.evolve()
self.generation += 1
remaining_generations -= 1
return self
def evolve(self):
new_area = []
for x, line in enumerate(self.area):
new_line = ''
for y, tile in enumerate(line):
new_line += self.next_tile(tile, self.counts_around(x,y))
new_area.append(new_line)
self.area = tuple(new_area)
return self
def next_tile(self, current_tile, counts):
if current_tile == self.OPEN and counts[self.TREES] >= 3:
return self.TREES
elif current_tile == self.TREES and counts[self.LUMBERYARD] >= 3:
return self.LUMBERYARD
elif current_tile == self.LUMBERYARD:
if counts[self.LUMBERYARD] >= 1 and counts[self.TREES] >= 1:
return self.LUMBERYARD
else:
return self.OPEN
else:
return current_tile
def counts_around(self, x,y):
return _.lib.collections.Counter(self.tiles_around(x,y))._
def tiles_around(self, x,y):
if x > 0:
line = self.area[x-1]
yield from line[max(0, y-1):y+2]
line = self.area[x]
if y > 0: yield line[y-1]
if y+1 < len(line): yield line[y+1]
if x+1 < len(self.area):
line = self.area[x+1]
yield from line[max(0, y-1):y+2]
def resource_value(self):
counts = _(self).area.join().call(_.lib.collections.Counter)._
return counts[self.TREES] * counts[self.LUMBERYARD]
test_input = '''\
.#.#...|#.
.....#|##|
.|..|...#.
..|#.....#
#.#|||#|#|
...#.||...
.|....|...
||...#|.#|
|.||||..|.
...#.|..|.
'''
test_area = _(test_input).call(Area).evolve_to_generation(10)._
expect(test_area.area) == _('''\
.||##.....
||###.....
||##......
|##.....##
|##.....##
|##....##|
||##.####|
||#####|||
||||#|||||
||||||||||
''').strip().splitlines().to(tuple)
expect(test_area.resource_value()) == 1147
area = _(open('input/day18.txt')).read().call(Area)._
area.evolve_to_generation(10).resource_value()
area.evolve_to_generation(1000000000).resource_value()
```
# Day 19
https://adventofcode.com/2018/day/19
|
github_jupyter
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.